markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
Analysis
|
df.groupby(by='raceethnicity').describe()
|
_____no_output_____
|
MIT
|
thecounted_visual.ipynb
|
KikiCS/thecounted
|
Check if there are any missing values:
|
df.isna().sum()
df = df[(df.raceethnicity != 'Arab-American') & (df.raceethnicity != 'Unknown')]
# no data available about the percentage of this ethnicity over population, so it is discarded
df.replace(to_replace=['Asian/Pacific Islander','Native American','Other'],value='Others',inplace=True)
# those categories all fall under Others in the population percentages found online
df.replace(to_replace=['Hispanic/Latino'],value='Latino',inplace=True)
# this value is renamed for consistency with population ethnicity data
df.groupby(by='raceethnicity').describe()
def givepercent (dtf,ethnicity):
# Function to compute percentages by ethnicity
return round(((dtf.raceethnicity == ethnicity).sum()/(dtf.shape[0])*100),2)
killed={"White":(df.raceethnicity == 'White').sum(),
"Latino": (df.raceethnicity == 'Latino').sum(),
"Black": (df.raceethnicity == 'Black').sum(),
"Others": (df.raceethnicity == 'Others').sum()
}
print(killed)
killedperc={"White": givepercent(df,'White'),
"Latino": givepercent(df,'Latino'),
"Black": givepercent(df,'Black'),
"Others": givepercent(df,'Others')
}
print(killedperc)
df.groupby(by='armed').describe()
|
_____no_output_____
|
MIT
|
thecounted_visual.ipynb
|
KikiCS/thecounted
|
The analysis is limited to the value *No*, but could consider *Disputed* and *Non-lethal firearm*, which constitute other 108 data points.
|
dfunarmed = df[(df.armed == 'No')]
dfunarmed.groupby(by='raceethnicity').describe()
unarmed={"White":(dfunarmed.raceethnicity == 'White').sum(),
"Latino": (dfunarmed.raceethnicity == 'Latino').sum(),
"Black": (dfunarmed.raceethnicity == 'Black').sum(),
"Others": (dfunarmed.raceethnicity == 'Others').sum()
}
print(unarmed)
unarmedperc={"White":givepercent(dfunarmed,'White'),
"Latino": givepercent(dfunarmed,'Latino'),
"Black": givepercent(dfunarmed,'Black'),
"Others": givepercent(dfunarmed,'Others')
}
print(unarmedperc)
def percent1ethn (portion,population,decimals):
# Function to compute the percentage of the portion killed over a given population
return round((portion/population*100),decimals)
killed1ethn={"White": percent1ethn(killed['White'],ethnestim['White'],6),
"Latino": percent1ethn(killed['Latino'],ethnestim['Latino'],6),
"Black": percent1ethn(killed['Black'],ethnestim['Black'],6),
"Others": percent1ethn(killed['Others'],ethnestim['Others'],6)
}
print(killed1ethn)
unarmedoverkilled={"White": percent1ethn(unarmed['White'],killed['White'],2),
"Latino": percent1ethn(unarmed['Latino'],killed['Latino'],2),
"Black": percent1ethn(unarmed['Black'],killed['Black'],2),
"Others": percent1ethn(unarmed['Others'],killed['Others'],2)
}
print(unarmedoverkilled)
|
{'White': 0.000582, 'Latino': 0.000664, 'Black': 0.001436, 'Others': 0.000313}
{'White': 17.36, 'Latino': 17.72, 'Black': 21.12, 'Others': 13.25}
|
MIT
|
thecounted_visual.ipynb
|
KikiCS/thecounted
|
**Hypothesis testing**
|
whitesample=ethnestim['White']
blacksample=ethnestim['Black']
wkilled=killed['White']
bkilled=killed['Black']
pw=wkilled/whitesample
pb=bkilled/blacksample
#happened by chance?
#Hnull pw-pb = 0 (no difference between white and black)
#Halt pw-pb != 0 (the two proportions are different)
#Significance level = 5%
# Test statistic: Z-statistic
difference=pb-pw
print(difference)
standarderror=np.sqrt(((pw*(1-pw))/whitesample)+((pb*(1-pb))/blacksample))
zstat=(difference)/standarderror
print(zstat)
# Z-score for significance level
zscore=1.96
if zstat > zscore:
print("The null hypothesis is rejected.")
else:
print("The null hypothesis is not rejected.")
whitesample=killed['White']
blacksample=killed['Black']
wunarmed=unarmed['White']
bunarmed=unarmed['Black']
pw=wunarmed/whitesample
pb=bunarmed/blacksample
#happened by chance?
#Hnull pw-pb = 0 (no difference between white and black)
#Halt pw-pb != 0 (the two proportions are different)
#Significance level = 5%
# Test statistic: Z-statistic
difference=pb-pw
print(difference)
standarderror=np.sqrt(((pw*(1-pw))/whitesample)+((pb*(1-pb))/blacksample))
zstat=(difference)/standarderror
print(zstat)
# Z-score for significance level
zscore=1.96
if zstat > zscore:
print("The null hypothesis is rejected.")
else:
print("The null hypothesis is not rejected.")
ethnicities = list(ethndic.keys())
populethn = list(ethndic.values())
killed = list(killedperc.values())
unarmed = list(unarmedperc.values())
data1 = {'ethnicities' : ethnicities,
'populethn' : populethn,
'killed' : killed,
'unarmed' : unarmed}
source = ColumnDataSource(data=data1)
|
_____no_output_____
|
MIT
|
thecounted_visual.ipynb
|
KikiCS/thecounted
|
Results
|
TOOLS = "pan,wheel_zoom,box_zoom,reset,save,box_select"
palette=Spectral4
titlefontsize='16pt'
cplot = figure(title="The Counted (data from 2015 and 2016)", tools=TOOLS,
x_range=ethnicities, y_range=(0, 75))#, sizing_mode='scale_both')
cplot.vbar(x=dodge('ethnicities', 0.25, range=cplot.x_range),top='populethn', source=source,
width=0.4,line_width=0 ,line_color=None, legend='Ethnicity % over population',
color=str(Spectral4[0]), name='populethn')
cplot.vbar(x=dodge('ethnicities', -0.25, range=cplot.x_range), top='killed', source=source,
width=0.4, line_width=0 ,line_color=None, legend="Killed % over total killed",
color=str(Spectral4[2]), name="killed")
cplot.vbar(x=dodge('ethnicities', 0.0, range=cplot.x_range), top='unarmed', source=source,
width=0.4, line_width=0 ,line_color=None, legend="Unarmed % over total unarmed",
color=str(Spectral4[1]), name="unarmed")
cplot.add_tools(HoverTool(names=["unarmed"],
tooltips=[
( 'Population', '@populethn{(00.00)}%' ),
( 'Killed', '@killed{(00.00)}%' ),
( 'Unarmed', '@unarmed{(00.00)}%' )], # Fields beginning with @ display values from ColumnDataSource.
mode='vline'))
#cplot.x_range.range_padding = 0.1
cplot.xgrid.grid_line_color = None
cplot.legend.location = "top_right"
cplot.xaxis.axis_label = "Ethnicity"
cplot.xaxis.axis_label_text_font_size='18pt'
cplot.xaxis.minor_tick_line_color = None
cplot.title.text_font_size=titlefontsize
cplot.legend.label_text_font_size='16pt'
cplot.xaxis.major_label_text_font_size='16pt'
cplot.yaxis.major_label_text_font_size='16pt'
perckillethn = list(killed1ethn.values())
data2 = {'ethnicities' : ethnicities,
'perckillethn' : perckillethn}
source = ColumnDataSource(data=dict(data2, color=PuBu4))
plot2 = figure(title="Killed % over population with same ethnicity",
tools=TOOLS, x_range=ethnicities, y_range=(0, max(perckillethn)*1.2))#, sizing_mode='scale_both')
plot2.vbar(x=dodge('ethnicities', 0.0, range=cplot.x_range), top='perckillethn', source=source,
width=0.4, line_width=0 ,line_color=None, legend="",
color='color', name="perckillethn")
plot2.add_tools(HoverTool(names=["perckillethn"],
tooltips=[
( 'Killed', '@perckillethn{(0.00000)}%' )],
#( 'Unarmed', '@unarmed{(00.00)}%' )], # Fields beginning with @ display values from ColumnDataSource.
mode='vline'))
#plot2.x_range.range_padding = 0.1
plot2.xgrid.grid_line_color = None
plot2.xaxis.axis_label = "Ethnicity"
plot2.xaxis.axis_label_text_font_size='18pt'
plot2.xaxis.minor_tick_line_color = None
plot2.title.text_font_size=titlefontsize
plot2.xaxis.major_label_text_font_size='16pt'
plot2.yaxis.major_label_text_font_size='16pt'
plot2.yaxis[0].formatter = NumeralTickFormatter(format="0.0000")
percunarmethn = list(unarmedoverkilled.values())
data3 = {'ethnicities' : ethnicities,
'percunarmethn' : percunarmethn}
source = ColumnDataSource(data=dict(data3, color=PuBu4))
plot3 = figure(title="Unarmed % over killed with same ethnicity",
tools=TOOLS, x_range=ethnicities, y_range=(0, max(percunarmethn)*1.2))#, sizing_mode='scale_both')
plot3.vbar(x=dodge('ethnicities', 0.0, range=cplot.x_range), top='percunarmethn', source=source,
width=0.4, line_width=0 ,line_color=None, legend="",
color='color', name="percunarmethn")
plot3.add_tools(HoverTool(names=["percunarmethn"],
tooltips=[
( 'Unarmed', '@percunarmethn{(00.00)}%' )],
#( 'Unarmed', '@unarmed{(00.00)}%' )], # Fields beginning with @ display values from ColumnDataSource.
mode='vline'))
#plot3.x_range.range_padding = 0.1
plot3.xgrid.grid_line_color = None
plot3.xaxis.axis_label = "Ethnicity"
plot3.xaxis.axis_label_text_font_size='18pt'
plot3.xaxis.minor_tick_line_color = None
plot3.title.text_font_size=titlefontsize
plot3.xaxis.major_label_text_font_size='16pt'
plot3.yaxis.major_label_text_font_size='16pt'
output_file("thecounted.html", title="The Counted Visualization")
output_notebook()
gplot=gridplot([cplot, plot2, plot3], sizing_mode='stretch_both', ncols=3)#, plot_width=800, plot_height=600)
show(gplot) # open a browser
export_png(gplot, filename="bokeh_thecounted.png")
|
_____no_output_____
|
MIT
|
thecounted_visual.ipynb
|
KikiCS/thecounted
|
ML 101 RDP and line simplificationRDP is a line simplifaction algorithm that can be used to reduce the number of points.
|
!pip install git+git://github.com/mariolpantunes/knee@main#egg=knee
%matplotlib inline
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [16, 9]
import numpy as np
import knee.linear_fit as lf
import knee.rdp as rdp
# generate some data
x = np.arange(0, 100, 0.1)
y = np.sin(x)
print(len(x))
plt.plot(x,y)
plt.show()
# generate the points
points = np.array([x,y]).T
reduced, removed = rdp.rdp(points, 0.01, cost=lf.Linear_Metrics.rpd)
reduced_points = points[reduced]
print(f'Lenght = {len(reduced_points)}')
plt.plot(x,y)
x = reduced_points[:,0]
y = reduced_points[:,1]
plt.plot(x,y, 'ro')
plt.show()
|
_____no_output_____
|
MIT
|
classes/01 preprocessing/01_preprocessing_03.ipynb
|
mariolpantunes/ml101
|
Knee detectionElbow/knee detection is a method to select te ideal cut-off point in a performance curve
|
# generate some data
x = np.arange(0.1, 10, 0.1)
y = 1/(x**2)
plt.plot(x,y)
plt.show()
import knee.lmethod as lmethod
# generate the points
points = np.array([x,y]).T
idx = lmethod.knee(points)
plt.plot(x,y)
plt.plot(x[idx], y[idx], 'ro')
plt.show()
import knee.dfdt as dfdt
# generate the points
points = np.array([x,y]).T
idx = dfdt.knee(points)
print(idx)
plt.plot(x,y)
plt.plot(x[idx], y[idx], 'ro')
plt.show()
|
_____no_output_____
|
MIT
|
classes/01 preprocessing/01_preprocessing_03.ipynb
|
mariolpantunes/ml101
|
Download MNIST
|
# For Google Colaboratory
import sys, os
if 'google.colab' in sys.modules:
from google.colab import drive
drive.mount('/content/gdrive')
file_name = 'generate_mnist.ipynb'
import subprocess
path_to_file = subprocess.check_output('find . -type f -name ' + str(file_name), shell=True).decode("utf-8")
print(path_to_file)
path_to_file = path_to_file.replace(file_name,"").replace('\n',"")
os.chdir(path_to_file)
!pwd
import torch
import torchvision
import torchvision.transforms as transforms
import numpy as np
import matplotlib.pyplot as plt
trainset = torchvision.datasets.MNIST(root='./temp', train=True,
download=True, transform=transforms.ToTensor())
testset = torchvision.datasets.MNIST(root='./temp', train=False,
download=True, transform=transforms.ToTensor())
idx=4
pic, label =trainset[idx]
pic=pic.squeeze()
plt.imshow(pic.numpy(), cmap='gray')
plt.show()
print(label)
train_data=torch.Tensor(60000,28,28)
train_label=torch.LongTensor(60000)
for idx , example in enumerate(trainset):
train_data[idx]=example[0].squeeze()
train_label[idx]=example[1]
torch.save(train_data,'train_data.pt')
torch.save(train_label,'train_label.pt')
test_data=torch.Tensor(10000,28,28)
test_label=torch.LongTensor(10000)
for idx , example in enumerate(testset):
test_data[idx]=example[0].squeeze()
test_label[idx]=example[1]
torch.save(test_data,'test_data.pt')
torch.save(test_label,'test_label.pt')
|
_____no_output_____
|
MIT
|
codes/data/mnist/generate_mnist.ipynb
|
NIRVANALAN/CE7454_2020
|
1. Write a Python Program to Find LCM?
|
# Given some numbers, LCM of these numbers is the smallest positive integer that is divisible by all the numbers numbers
def LCM(ls):
lar = max(ls)
flag=1
while(flag==1):
for i in ls:
if(lar%i!=0):
flag=1
break
else:
flag=0
continue
lcm=lar
lar += 1
return lcm
n=int(input("Enter the range of your input "))
ls=[]
for i in range(n):
a=int(input("Enter the number "))
ls.append(a)
print("The LCM of entered numbers is", LCM(ls))
|
Enter the range of your input 3
Enter the number 4
Enter the number 6
Enter the number 8
The LCM of entered numbers is 24
|
Apache-2.0
|
Python Basic Assignment -5.ipynb
|
MuhammedAbin/iNeuron-Assignment
|
2. Write a Python Program to Find HCF?
|
def HCF(ls):
lar=0
for i in range(2,min(ls)+1) :
count=0
for j in ls:
if j%i==0:
count=count+1
if count==len(ls):
lar=i
return lar
if count!=len(ls):
return 1
n=int(input("Enter the range of your input "))
ls=[]
for i in range(n):
a=int(input("Enter the number "))
ls.append(a)
print("The LCM of entered numbers is", HCF(ls))
|
Enter the range of your input 3
Enter the number 4
Enter the number 6
Enter the number 8
The LCM of entered numbers is 2
|
Apache-2.0
|
Python Basic Assignment -5.ipynb
|
MuhammedAbin/iNeuron-Assignment
|
3. Write a Python Program to Convert Decimal to Binary, Octal and Hexadecimal?
|
a=int(input("Enter the Decimal value: "))
def ToBinary(x):
binary=" "
while(x!=0 and x>0):
binary=binary+str(x%2)
x=int(x/2)
return(binary[::-1])
def ToOctal(x):
if(x<8):
print(x)
else:
octal=" "
while(x!=0 and x>0):
octal=octal+str(x%8)
x=int(x/8)
return(octal[::-1])
def ToHexa(x):
if(x<10):
print(x)
else:
ls={10:"A", 11:"B", 12:"C", 13:"D", 14:"E", 15:"F"}
Hexa=" "
while(x!=0 and x>0):
if(x%16) in ls.keys():
Hexa=Hexa+ls[x%16]
else:
Hexa=Hexa+str(x%16)
x=int(x/16)
return(Hexa[::-1])
print("Binary of",a,"is", ToBinary(a))
print("Octal of",a,"is", ToOctal(a))
print("Hexadecimal of",a,"is", ToHexa(a))
|
Enter the Decimal value: 15
Binary of 15 is 1111
Octal of 15 is 17
Hexadecimal of 15 is F
|
Apache-2.0
|
Python Basic Assignment -5.ipynb
|
MuhammedAbin/iNeuron-Assignment
|
4. Write a Python Program To Find ASCII value of a character?
|
a=input("Enter the character to find ASCII: ")
print("ASCII of ",a ,"is", ord(a))
|
Enter the character to find ASCII: g
ASCII of g is 103
|
Apache-2.0
|
Python Basic Assignment -5.ipynb
|
MuhammedAbin/iNeuron-Assignment
|
5.Write a Python Program to Make a Simple Calculator with 4 basic mathematical operations?
|
class Calculator:
def Add(self,x,y):
return x+y
def Subtract(self,x,y):
return x-y
def Multiply(self,x,y):
return x*y
def Divide(self,x,y):
try:
return x/y
except Exception as es:
print("An error has occured",es)
Calc = Calculator()
print("Choose your Calculator Operation: ")
print("1 : Addition")
print("2 : Subtraction")
print("3 : Multipilication")
print("4 : Division")
a=int(input("Enter Your Selection: "))
x=int(input("Enter Your 1st number: "))
y=int(input("Enter Your 2nd number: "))
if(a==1):
print("You chose Addition: ")
print("The result of your operation is", Calc.Add(x,y))
elif(a==2):
print("You chose Subtraction: ")
print("The result of your operation is", Calc.Subtract(x,y))
elif(a==3):
print("You chose Multipilication: ")
print("The result of your operation is", Calc.Multiply(x,y))
elif(a==4):
print("You chose Division: ")
print("The result of your operation is", Calc.Divide(x,y))
else:
print("You chose a wrong operation")
|
Choose your Calculator Operation:
1 : Addition
2 : Subtraction
3 : Multipilication
4 : Division
Enter Your Selection: 1
Enter Your 1st number: 5
Enter Your 2nd number: 5
You chose addition:
The result of your opperation is 10
|
Apache-2.0
|
Python Basic Assignment -5.ipynb
|
MuhammedAbin/iNeuron-Assignment
|
Download the ULB Fraud DatasetAndrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. Calibrating Probability with Undersampling for Unbalanced Classification. In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015TERMS OF USE:This data is made available under the Open Database License: http://opendatacommons.org/licenses/odbl/1.0/. Any rights in individual contents of the database are licensed under the Database Contents License: http://opendatacommons.org/licenses/dbcl/1.0/. It is provided through the Google Cloud Public Datasets Program and is provided "AS IS" without any warranty, express or implied, from Google. Google disclaims all liability for any damages, direct or indirect, resulting from the use of the dataset.
|
import pandas as pd
from google.cloud import bigquery
client = bigquery.Client()
sql = """
SELECT * FROM `bigquery-public-data.ml_datasets.ulb_fraud_detection`
"""
df = client.query(sql).to_dataframe()
df.head()
df.to_csv('../dataset/creditcard.csv', index=False)
|
_____no_output_____
|
Apache-2.0
|
classification-fraud-detection/solution-0-setup/1.0 download [data].ipynb
|
nfaggian/ml-examples
|
Data Wrangling, Analysis and Visualization of @WeLoveDogs twitter data.
|
import pandas as pd
import numpy as np
import tweepy as ty
import requests
import json
import io
import time
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Gathering
|
df = pd.read_csv('twitter-archive-enhanced.csv')
df.head()
image_response = requests.get(r'https://d17h27t6h515a5.cloudfront.net/topher/2017/August/599fd2ad_image-predictions/image-predictions.tsv')
image_df = pd.read_csv(io.StringIO(image_response.content.decode('utf-8')), sep='\t')
image_df.head()
image_df.info()
CONSUMER_KEY = '<My Consumer Key>'
CONSUMER_SECRET = '<My Consumer Secret>'
ACCESS_TOKEN = '<My Access Token>'
ACCESS_TOKEN_SECRET = '<My Access Token Secret>'
auth = ty.OAuthHandler(CONSUMER_KEY, CONSUMER_SECRET)
auth.set_access_token(ACCESS_TOKEN, ACCESS_TOKEN_SECRET)
api = ty.API(auth, wait_on_rate_limit=True, wait_on_rate_limit_notify=True)
tweet_count = 0
for id in df['tweet_id']:
with open('tweet_json.txt', 'a') as file:
try:
start = time.time()
tweet = api.get_status(id, tweet_mode='extended')
st = json.dumps(tweet._json)
file.writelines(st + '\n')
tweet_count += 1
if tweet_count % 20 == 0 or tweet_count == len(df):
end = time.time()
print('Tweet id: {0}\tDownload Time: {1} sec\tTweets Downloaded: {2}.'.format(id, (end-start), tweet_count))
except Exception as e:
print('Exception occured for tweet {0} : {1}'.format(id, str(e)))
print('There are {} records for which tweets does not exist in Twitter database.'.format(len(df) - tweet_count))
tweet_ids = []
favorite_count = []
retweet_count = []
with open('tweet_json.txt', 'r') as file:
for line in file.readlines():
data = json.loads(line)
tweet_ids.append(data['id'])
favorite_count.append(data['favorite_count'])
retweet_count.append(data['retweet_count'])
favorite_retweet_df = pd.DataFrame(data={'tweet_id': tweet_ids, 'favorite_count': favorite_count,
'retweet_count': retweet_count})
favorite_retweet_df.head()
favorite_retweet_df.info()
|
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 2341 entries, 0 to 2340
Data columns (total 3 columns):
tweet_id 2341 non-null int64
favorite_count 2341 non-null int64
retweet_count 2341 non-null int64
dtypes: int64(3)
memory usage: 54.9 KB
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Assessing Quality Issues - The WeRateDogs Twitter archive contains some retweets also. We only need to consider original tweets for this project. - `tweet_id` column having integer datatype in all the dataframes. Conversion to string required. Since, We are not going to do any mathematical operations with it. - `rating_denominator` value should be 10 since we are giving rating out of 10. - For some tweets, The values of `rating_numerator` column are very high, possibly outliers. - `name` column has **None** string and *a*, *an*, *the* as values. - Extract dog **stages** from the tweet text (if present) for null values. - `timestamp` column is given as **string**. Convert it to date. - Since we are not using retweets, `in_reply_to_status_id`, `in_reply_to_user_id`, `source`, `retweeted_status_id`, `retweeted_status_user_id`, `retweeted_status_timestamp` columns are not required. - `image_df` contains tweets that do not belong to a dog. - `source` column in main dataframe is of no use in the analysis as it only tell us about the source of tweet. Tidiness Issues - A dog can have one stage at a time, still we are having 4 columns to store one piece of information. - We are having 3 predicted breeds of dogs in the image prediction file. But only required the one with higher probability, given that it is a breed of dog. - All 3 dataframes contains same tweet_id column, which we can use to merge them and use as one dataframe for our analysis.
|
sum(df.duplicated())
# name and stages having wrong values for no. of records
df.tail()
# tweet id is int64, retweet related columns are not required
# name and dog stages showing full data but most of them are None.
# timestamp having string datatype
df.info()
# rating_numerator is having min value of 0 and max of 1776 (outliers)
df.describe()
# denominator value should be 10
sum(df['rating_denominator'] != 10)
df.rating_numerator.value_counts()
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Cleaning
|
# Making a copy of the data so that original data will be unchanged
df_clean = df.copy()
image_df_clean = image_df.copy()
favorite_retweet_df_clean = favorite_retweet_df.copy()
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Quality Issues Define - Replacing all the **None** strings and *a*, *an* and *the* to **np.nan** in `name` column using pandas **_replace()_** function. Code
|
df_clean['name'] = df_clean['name'].replace({'None': np.nan, 'a': np.nan, 'an': np.nan, 'the': np.nan})
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
df_clean.info()
|
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 2356 entries, 0 to 2355
Data columns (total 17 columns):
tweet_id 2356 non-null int64
in_reply_to_status_id 78 non-null float64
in_reply_to_user_id 78 non-null float64
timestamp 2356 non-null object
source 2356 non-null object
text 2356 non-null object
retweeted_status_id 181 non-null float64
retweeted_status_user_id 181 non-null float64
retweeted_status_timestamp 181 non-null object
expanded_urls 2297 non-null object
rating_numerator 2356 non-null int64
rating_denominator 2356 non-null int64
name 1541 non-null object
doggo 2356 non-null object
floofer 2356 non-null object
pupper 2356 non-null object
puppo 2356 non-null object
dtypes: float64(4), int64(3), object(10)
memory usage: 313.0+ KB
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- `tweet_id` column having integer datatype in all the dataframes. Converting it to string using pandas **_astype()_** function.- `timestamp` column is given as string. Converting it to date using pandas **to_datetime()** function. Code
|
df_clean['tweet_id'] = df_clean['tweet_id'].astype(str)
image_df_clean['tweet_id'] = image_df_clean['tweet_id'].astype(str)
favorite_retweet_df_clean['tweet_id'] = favorite_retweet_df_clean['tweet_id'].astype(str)
df_clean.timestamp = pd.to_datetime(df_clean.timestamp)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
df_clean.dtypes
image_df_clean.dtypes
favorite_retweet_df_clean.dtypes
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- Converting `rating_denominator` column value to 10 value when it is not, using pandas boolean indexing. Since we are giving rating out of 10. Code
|
df_clean.loc[df_clean.rating_denominator != 10, 'rating_denominator'] = 10
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
sum(df_clean.rating_denominator != 10)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- Removing retweets since we are only concerned about original tweets. Since, there is null present in `in_reply_to_status_id`, `retweeted_status_id` columns for retweets. Droping these records using the pandas drop() function. Code
|
df_clean.drop(index=df_clean[df_clean.retweeted_status_id.notnull()].index, inplace=True)
df_clean.drop(index=df_clean[df_clean.in_reply_to_status_id.notnull()].index, inplace=True)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
sum(df_clean.retweeted_status_id.notnull())
sum(df_clean.in_reply_to_status_id.notnull())
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- Removing `in_reply_to_status_id`, `in_reply_to_user_id`, `retweeted_status_id`, `retweeted_status_user_id`, `retweeted_status_timestamp` columns since they are related to retweet info. Hence, not required. Code
|
df_clean.drop(columns=['in_reply_to_status_id', 'in_reply_to_user_id', 'retweeted_status_id',
'retweeted_status_user_id', 'retweeted_status_timestamp'], inplace=True)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
df_clean.info()
|
<class 'pandas.core.frame.DataFrame'>
Int64Index: 2097 entries, 0 to 2355
Data columns (total 12 columns):
tweet_id 2097 non-null object
timestamp 2097 non-null datetime64[ns]
source 2097 non-null object
text 2097 non-null object
expanded_urls 2094 non-null object
rating_numerator 2097 non-null int64
rating_denominator 2097 non-null int64
name 1425 non-null object
doggo 2097 non-null object
floofer 2097 non-null object
pupper 2097 non-null object
puppo 2097 non-null object
dtypes: datetime64[ns](1), int64(2), object(9)
memory usage: 213.0+ KB
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- Removing `source` column from df_clean using pandas **_drop_** function. Code
|
df_clean.drop(columns=['source'], inplace=True)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
df_clean.info()
|
<class 'pandas.core.frame.DataFrame'>
Int64Index: 2097 entries, 0 to 2355
Data columns (total 11 columns):
tweet_id 2097 non-null object
timestamp 2097 non-null datetime64[ns]
text 2097 non-null object
expanded_urls 2094 non-null object
rating_numerator 2097 non-null int64
rating_denominator 2097 non-null int64
name 1425 non-null object
doggo 2097 non-null object
floofer 2097 non-null object
pupper 2097 non-null object
puppo 2097 non-null object
dtypes: datetime64[ns](1), int64(2), object(8)
memory usage: 196.6+ KB
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Tidiness Issues Define- Replacing 3 predictions to one dog breed with higher probability, given that it is a breed of dog, with the use of pandas **_apply()_** function. Code
|
non_dog_ind = image_df_clean.query('not p1_dog and not p2_dog and not p3_dog').index
image_df_clean.drop(index=non_dog_ind, inplace=True)
def get_priority_dog(dog):
return dog['p1'] if dog['p1_dog'] else dog['p2'] if dog['p2_dog'] else dog['p3']
image_df_clean['dog_breed'] = image_df_clean.apply(get_priority_dog, axis=1)
image_df_clean.drop(columns=['p1', 'p1_conf', 'p1_dog', 'p2', 'p2_conf', 'p2_dog', 'p3', 'p3_conf', 'p3_dog', 'img_num'],
inplace=True)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
image_df_clean.head()
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- There are 4 column present for specifing dog breed, which can be done using one column only. Creating a column name `dog_stage` and adding present dog breed in it. Code
|
def get_dog_stage(dog):
if dog['doggo'] != 'None':
return dog['doggo']
elif dog['floofer'] != 'None':
return dog['floofer']
elif dog['pupper'] != 'None':
return dog['pupper']
else:
return dog['puppo'] # if last entry is also nan, we have to return nan anyway
df_clean['dog_stage'] = df_clean.apply(get_dog_stage, axis=1)
df_clean.drop(columns=['doggo', 'floofer', 'pupper', 'puppo'], inplace=True)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
df_clean.info()
|
<class 'pandas.core.frame.DataFrame'>
Int64Index: 2097 entries, 0 to 2355
Data columns (total 8 columns):
tweet_id 2097 non-null object
timestamp 2097 non-null datetime64[ns]
text 2097 non-null object
expanded_urls 2094 non-null object
rating_numerator 2097 non-null int64
rating_denominator 2097 non-null int64
name 1425 non-null object
dog_stage 2097 non-null object
dtypes: datetime64[ns](1), int64(2), object(5)
memory usage: 147.4+ KB
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- All 3 dataframes contains same `tweet_id` column, which we can use ot merge them and use as one dataframe for our analysis. Code
|
df_clean = pd.merge(df_clean, image_df_clean, on='tweet_id')
df_clean = pd.merge(df_clean, favorite_retweet_df_clean, on='tweet_id')
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
df_clean.head()
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Quality Issues Define- Try to extract dog dstage from tweet text using regular expressions and series **_str.extract()_** function. Code
|
stages = df_clean[df_clean.dog_stage == 'None'].text.str.extract(r'(doggo|pupper|floof|puppo|pup)', expand=True)
len(df_clean[df_clean.dog_stage == 'None'])
df_clean.loc[stages.index, 'dog_stage'] = stages[0]
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
len(df_clean[df_clean.dog_stage.isnull()])
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Define- Removing outliers from `rating_numerator` Code
|
df_clean.boxplot(column=['rating_numerator'], figsize=(20,8), vert=False)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
- As clear from the boxplot, `rating_numerator` has a number of outliers which can affect our analysis. So, removing all the rating points above 15 abd below 7 to reduce the effect of outliers.
|
df_clean.drop(index=df_clean.query('rating_numerator > 15 or rating_numerator < 7').index, inplace=True)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Test
|
df_clean.boxplot(column=['rating_numerator'], figsize=(20,8), vert=False)
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
- As we can see now, there are no outliers present in the datat anymore. Storing Data in a CSV file
|
df_clean.to_csv('twitter_archive_master.csv', index=False)
print('Save Done !')
|
Save Done !
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Data Analysis and Visualization
|
ax = df_clean.plot.scatter('rating_numerator', 'favorite_count', figsize=(10, 10), title='Rating VS. Favorites')
ax.set_xlabel('Ratings')
ax.set_ylabel('Favorites')
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Insight 1:- Number of favorite count is increasing with the rating. i.e. Dogs getting more rating in the tweets are likely to receive more likes (favorites).
|
df_clean.dog_breed.value_counts()
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Insight 2- Most of the pictures present in @WeLoveDogs twitter account are of Golden Retriever, followed by Labrador Retriever, Pembroke and Chihuahua .
|
df_clean.loc[df_clean.favorite_count.idxmax()][['dog_breed', 'favorite_count']]
df_clean.loc[df_clean.favorite_count.idxmin()][['dog_breed', 'favorite_count']]
df_clean.loc[df_clean.retweet_count.idxmax()][['dog_breed', 'retweet_count']]
df_clean.loc[df_clean.retweet_count.idxmin()][['dog_breed', 'retweet_count']]
|
_____no_output_____
|
MIT
|
Wrangling of @WeLoveDogs Twitter dataset using Python/wrangle_act.ipynb
|
navpreetmattu/dprojects
|
Moving average Run in Google Colab View source on GitHub Setup
|
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
keras = tf.keras
def plot_series(time, series, format="-", start=0, end=None, label=None):
plt.plot(time[start:end], series[start:end], format, label=label)
plt.xlabel("Time")
plt.ylabel("Value")
if label:
plt.legend(fontsize=14)
plt.grid(True)
def trend(time, slope=0):
return slope * time
def seasonal_pattern(season_time):
"""Just an arbitrary pattern, you can change it if you wish"""
return np.where(season_time < 0.4,
np.cos(season_time * 2 * np.pi),
1 / np.exp(3 * season_time))
def seasonality(time, period, amplitude=1, phase=0):
"""Repeats the same pattern at each period"""
season_time = ((time + phase) % period) / period
return amplitude * seasonal_pattern(season_time)
def white_noise(time, noise_level=1, seed=None):
rnd = np.random.RandomState(seed)
return rnd.randn(len(time)) * noise_level
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Trend and Seasonality
|
time = np.arange(4 * 365 + 1)
slope = 0.05
baseline = 10
amplitude = 40
series = baseline + trend(time, slope) + seasonality(time, period=365, amplitude=amplitude)
noise_level = 5
noise = white_noise(time, noise_level, seed=42)
series += noise
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Naive Forecast
|
split_time = 1000
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
naive_forecast = series[split_time - 1:-1]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, start=0, end=150, label="Series")
plot_series(time_valid, naive_forecast, start=1, end=151, label="Forecast")
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Now let's compute the mean absolute error between the forecasts and the predictions in the validation period:
|
keras.metrics.mean_absolute_error(x_valid, naive_forecast).numpy()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
That's our baseline, now let's try a moving average. Moving Average
|
def moving_average_forecast(series, window_size):
"""Forecasts the mean of the last few values.
If window_size=1, then this is equivalent to naive forecast"""
forecast = []
for time in range(len(series) - window_size):
forecast.append(series[time:time + window_size].mean())
return np.array(forecast)
def moving_average_forecast(series, window_size):
"""Forecasts the mean of the last few values.
If window_size=1, then this is equivalent to naive forecast
This implementation is *much* faster than the previous one"""
mov = np.cumsum(series)
mov[window_size:] = mov[window_size:] - mov[:-window_size]
return mov[window_size - 1:-1] / window_size
moving_avg = moving_average_forecast(series, 30)[split_time - 30:]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, label="Series")
plot_series(time_valid, moving_avg, label="Moving average (30 days)")
keras.metrics.mean_absolute_error(x_valid, moving_avg).numpy()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
That's worse than naive forecast! The moving average does not anticipate trend or seasonality, so let's try to remove them by using differencing. Since the seasonality period is 365 days, we will subtract the value at time *t* – 365 from the value at time *t*.
|
time_a = np.array(range(1, 21))
a = np.array([1.1, 1.5, 1.6, 1.4, 1.5, 1.6,1.7, 1.8, 1.9, 2,
2.1, 2.5, 2.6, 2.4, 2.5, 2.6, 2.7, 2.8, 2.9, 3])
print("a: ", a, len(a))
print("time_a: ", time_a, len (time_a))
ma_a = moving_average_forecast(a, 10)
print("ma_a: ", ma_a)
diff_a = (a[10:] - a[:-10]) ### This removes TREND + SEASONALITY, so, Only Noise is left
diff_time = time_a[10:]
print("diff_time: ", diff_time)
print("diff_a: ", diff_a)
a[10:]
a[:-10]
diff_series = (series[365:] - series[:-365])
diff_time = time[365:]
plt.figure(figsize=(10, 6))
plot_series(diff_time, diff_series, label="Series(t) – Series(t–365)")
plt.show()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Focusing on the validation period:
|
plt.figure(figsize=(10, 6))
plot_series(time_valid, diff_series[split_time - 365:], label="Series(t) – Series(t–365)")
plt.show()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Great, the trend and seasonality seem to be gone, so now we can use the moving average:
|
diff_moving_avg = moving_average_forecast(diff_series, 50)[split_time - 365 - 50:]
plt.figure(figsize=(10, 6))
plot_series(time_valid, diff_series[split_time - 365:], label="Series(t) – Series(t–365)")
plot_series(time_valid, diff_moving_avg, label="Moving Average of Diff")
plt.show()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Now let's bring back the trend and seasonality by adding the past values from t – 365:
|
diff_moving_avg_plus_past = series[split_time - 365:-365] + diff_moving_avg
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, label="Series")
plot_series(time_valid, diff_moving_avg_plus_past, label="Forecasts")
plt.show()
keras.metrics.mean_absolute_error(x_valid, diff_moving_avg_plus_past).numpy()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Better than naive forecast, good. However the forecasts look a bit too random, because we're just adding past values, which were noisy. Let's use a moving averaging on past values to remove some of the noise:
|
diff_moving_avg_plus_smooth_past = moving_average_forecast(series[split_time - 370:-359], 11) + diff_moving_avg
# moving_average_forecast(series[split_time - 370:-359], 11) = Past series have been smoothed to reduce the effects of the past noise
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, label="Series")
plot_series(time_valid, diff_moving_avg_plus_smooth_past, label="Forecasts")
plt.show()
keras.metrics.mean_absolute_error(x_valid, diff_moving_avg_plus_smooth_past).numpy()
|
_____no_output_____
|
MIT
|
08-Time-Series-Forecasting/03_moving_average.ipynb
|
iVibudh/TensorFlow-for-DeepLearning
|
Approximate q-learningIn this notebook you will teach a __tensorflow__ neural network to do Q-learning. __Frameworks__ - we'll accept this homework in any deep learning framework. This particular notebook was designed for tensorflow, but you will find it easy to adapt it to almost any python-based deep learning framework.
|
#XVFB will be launched if you run on a server
import os
if os.environ.get("DISPLAY") is not str or len(os.environ.get("DISPLAY"))==0:
!bash ../xvfb start
%env DISPLAY=:1
import gym
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
env = gym.make("CartPole-v0").env
env.reset()
n_actions = env.action_space.n
state_dim = env.observation_space.shape
plt.imshow(env.render("rgb_array"))
state_dim
|
_____no_output_____
|
MIT
|
week4_approx/practice_approx_qlearning.ipynb
|
Innuendo1975/Practical_RL
|
Approximate (deep) Q-learning: building the networkTo train a neural network policy one must have a neural network policy. Let's build it.Since we're working with a pre-extracted features (cart positions, angles and velocities), we don't need a complicated network yet. In fact, let's build something like this for starters: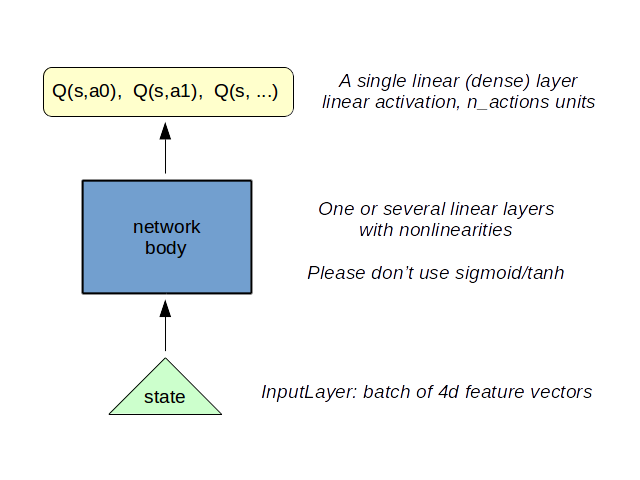For your first run, please only use linear layers (L.Dense) and activations. Stuff like batch normalization or dropout may ruin everything if used haphazardly. Also please avoid using nonlinearities like sigmoid & tanh: agent's observations are not normalized so sigmoids may become saturated from init.Ideally you should start small with maybe 1-2 hidden layers with < 200 neurons and then increase network size if agent doesn't beat the target score.
|
import tensorflow as tf
import keras
import keras.layers as L
tf.reset_default_graph()
sess = tf.InteractiveSession()
keras.backend.set_session(sess)
network = keras.models.Sequential()
network.add(L.InputLayer(state_dim))
# let's create a network for approximate q-learning following guidelines above
# <YOUR CODE: stack more layers!!!1 >
network.add(L.Dense(128, activation='relu'))
network.add(L.Dense(196, activation='relu'))
network.add(L.Dense(n_actions, activation='linear'))
import numpy as np
import random
def get_action(state, epsilon=0):
"""
sample actions with epsilon-greedy policy
recap: with p = epsilon pick random action, else pick action with highest Q(s,a)
"""
q_values = network.predict(state[None])[0]
n = len(q_values)
if random.random() < epsilon:
return random.randint(0, n - 1)
else:
return np.argmax(q_values)
# return <epsilon-greedily selected action>
assert network.output_shape == (None, n_actions), "please make sure your model maps state s -> [Q(s,a0), ..., Q(s, a_last)]"
assert network.layers[-1].activation == keras.activations.linear, "please make sure you predict q-values without nonlinearity"
# test epsilon-greedy exploration
s = env.reset()
assert np.shape(get_action(s)) == (), "please return just one action (integer)"
for eps in [0., 0.1, 0.5, 1.0]:
state_frequencies = np.bincount([get_action(s, epsilon=eps) for i in range(10000)], minlength=n_actions)
best_action = state_frequencies.argmax()
assert abs(state_frequencies[best_action] - 10000 * (1 - eps + eps / n_actions)) < 200
for other_action in range(n_actions):
if other_action != best_action:
assert abs(state_frequencies[other_action] - 10000 * (eps / n_actions)) < 200
print('e=%.1f tests passed'%eps)
|
e=0.0 tests passed
e=0.1 tests passed
e=0.5 tests passed
e=1.0 tests passed
|
MIT
|
week4_approx/practice_approx_qlearning.ipynb
|
Innuendo1975/Practical_RL
|
Q-learning via gradient descentWe shall now train our agent's Q-function by minimizing the TD loss:$$ L = { 1 \over N} \sum_i (Q_{\theta}(s,a) - [r(s,a) + \gamma \cdot max_{a'} Q_{-}(s', a')]) ^2 $$Where* $s, a, r, s'$ are current state, action, reward and next state respectively* $\gamma$ is a discount factor defined two cells above.The tricky part is with $Q_{-}(s',a')$. From an engineering standpoint, it's the same as $Q_{\theta}$ - the output of your neural network policy. However, when doing gradient descent, __we won't propagate gradients through it__ to make training more stable (see lectures).To do so, we shall use `tf.stop_gradient` function which basically says "consider this thing constant when doingbackprop".
|
# Create placeholders for the <s, a, r, s'> tuple and a special indicator for game end (is_done = True)
states_ph = keras.backend.placeholder(dtype='float32', shape=(None,) + state_dim)
actions_ph = keras.backend.placeholder(dtype='int32', shape=[None])
rewards_ph = keras.backend.placeholder(dtype='float32', shape=[None])
next_states_ph = keras.backend.placeholder(dtype='float32', shape=(None,) + state_dim)
is_done_ph = keras.backend.placeholder(dtype='bool', shape=[None])
#get q-values for all actions in current states
predicted_qvalues = network(states_ph)
#select q-values for chosen actions
predicted_qvalues_for_actions = tf.reduce_sum(predicted_qvalues * tf.one_hot(actions_ph, n_actions), axis=1)
gamma = 0.99
# compute q-values for all actions in next states
# predicted_next_qvalues = <YOUR CODE - apply network to get q-values for next_states_ph>
predicted_next_qvalues = network(next_states_ph)
# compute V*(next_states) using predicted next q-values
next_state_values = tf.reduce_max(predicted_next_qvalues, axis=1)
# compute "target q-values" for loss - it's what's inside square parentheses in the above formula.
target_qvalues_for_actions = rewards_ph + gamma * next_state_values
# at the last state we shall use simplified formula: Q(s,a) = r(s,a) since s' doesn't exist
target_qvalues_for_actions = tf.where(is_done_ph, rewards_ph, target_qvalues_for_actions)
#mean squared error loss to minimize
loss = (predicted_qvalues_for_actions - tf.stop_gradient(target_qvalues_for_actions)) ** 2
loss = tf.reduce_mean(loss)
# training function that resembles agent.update(state, action, reward, next_state) from tabular agent
train_step = tf.train.AdamOptimizer(1e-4).minimize(loss)
assert tf.gradients(loss, [predicted_qvalues_for_actions])[0] is not None, "make sure you update q-values for chosen actions and not just all actions"
assert tf.gradients(loss, [predicted_next_qvalues])[0] is None, "make sure you don't propagate gradient w.r.t. Q_(s',a')"
assert predicted_next_qvalues.shape.ndims == 2, "make sure you predicted q-values for all actions in next state"
assert next_state_values.shape.ndims == 1, "make sure you computed V(s') as maximum over just the actions axis and not all axes"
assert target_qvalues_for_actions.shape.ndims == 1, "there's something wrong with target q-values, they must be a vector"
|
_____no_output_____
|
MIT
|
week4_approx/practice_approx_qlearning.ipynb
|
Innuendo1975/Practical_RL
|
Playing the game
|
def generate_session(t_max=1000, epsilon=0, train=False):
"""play env with approximate q-learning agent and train it at the same time"""
total_reward = 0
s = env.reset()
for t in range(t_max):
a = get_action(s, epsilon=epsilon)
next_s, r, done, _ = env.step(a)
if train:
sess.run(train_step,{
states_ph: [s], actions_ph: [a], rewards_ph: [r],
next_states_ph: [next_s], is_done_ph: [done]
})
total_reward += r
s = next_s
if done: break
return total_reward
epsilon = 0.5
for i in range(1000):
session_rewards = [generate_session(epsilon=epsilon, train=True) for _ in range(100)]
print("epoch #{}\tmean reward = {:.3f}\tepsilon = {:.3f}".format(i, np.mean(session_rewards), epsilon))
epsilon *= 0.99
assert epsilon >= 1e-4, "Make sure epsilon is always nonzero during training"
if np.mean(session_rewards) > 300:
print ("You Win!")
break
|
epoch #0 mean reward = 16.740 epsilon = 0.500
epoch #1 mean reward = 16.610 epsilon = 0.495
epoch #2 mean reward = 14.050 epsilon = 0.490
epoch #3 mean reward = 15.400 epsilon = 0.485
epoch #4 mean reward = 14.950 epsilon = 0.480
epoch #5 mean reward = 19.480 epsilon = 0.475
epoch #6 mean reward = 16.150 epsilon = 0.471
epoch #7 mean reward = 17.380 epsilon = 0.466
epoch #8 mean reward = 15.730 epsilon = 0.461
epoch #9 mean reward = 25.080 epsilon = 0.457
epoch #10 mean reward = 29.930 epsilon = 0.452
epoch #11 mean reward = 37.970 epsilon = 0.448
epoch #12 mean reward = 45.640 epsilon = 0.443
epoch #13 mean reward = 46.730 epsilon = 0.439
epoch #14 mean reward = 43.390 epsilon = 0.434
epoch #15 mean reward = 79.620 epsilon = 0.430
epoch #16 mean reward = 92.650 epsilon = 0.426
epoch #17 mean reward = 125.850 epsilon = 0.421
epoch #18 mean reward = 100.480 epsilon = 0.417
epoch #19 mean reward = 150.340 epsilon = 0.413
epoch #20 mean reward = 161.240 epsilon = 0.409
epoch #21 mean reward = 131.490 epsilon = 0.405
epoch #22 mean reward = 184.870 epsilon = 0.401
epoch #23 mean reward = 191.210 epsilon = 0.397
epoch #24 mean reward = 247.040 epsilon = 0.393
epoch #25 mean reward = 207.300 epsilon = 0.389
epoch #26 mean reward = 249.010 epsilon = 0.385
epoch #27 mean reward = 252.960 epsilon = 0.381
epoch #28 mean reward = 276.790 epsilon = 0.377
epoch #29 mean reward = 376.700 epsilon = 0.374
You Win!
|
MIT
|
week4_approx/practice_approx_qlearning.ipynb
|
Innuendo1975/Practical_RL
|
How to interpret resultsWelcome to the f.. world of deep f...n reinforcement learning. Don't expect agent's reward to smoothly go up. Hope for it to go increase eventually. If it deems you worthy.Seriously though,* __ mean reward__ is the average reward per game. For a correct implementation it may stay low for some 10 epochs, then start growing while oscilating insanely and converges by ~50-100 steps depending on the network architecture. * If it never reaches target score by the end of for loop, try increasing the number of hidden neurons or look at the epsilon.* __ epsilon__ - agent's willingness to explore. If you see that agent's already at < 0.01 epsilon before it's is at least 200, just reset it back to 0.1 - 0.5. Record videosAs usual, we now use `gym.wrappers.Monitor` to record a video of our agent playing the game. Unlike our previous attempts with state binarization, this time we expect our agent to act ~~(or fail)~~ more smoothly since there's no more binarization error at play.As you already did with tabular q-learning, we set epsilon=0 for final evaluation to prevent agent from exploring himself to death.
|
#record sessions
import gym.wrappers
env = gym.wrappers.Monitor(gym.make("CartPole-v0"),directory="videos",force=True)
sessions = [generate_session(epsilon=0, train=False) for _ in range(100)]
env.close()
#show video
from IPython.display import HTML
import os
video_names = list(filter(lambda s:s.endswith(".mp4"),os.listdir("./videos/")))
HTML("""
<video width="640" height="480" controls>
<source src="{}" type="video/mp4">
</video>
""".format("./videos/"+video_names[-1])) #this may or may not be _last_ video. Try other indices
|
_____no_output_____
|
MIT
|
week4_approx/practice_approx_qlearning.ipynb
|
Innuendo1975/Practical_RL
|
--- Submit to coursera
|
%load_ext autoreload
%autoreload 2
from submit2 import submit_cartpole
submit_cartpole(generate_session, '[email protected]', 'gMmaBajRboD6YXKK')
|
Submitted to Coursera platform. See results on assignment page!
|
MIT
|
week4_approx/practice_approx_qlearning.ipynb
|
Innuendo1975/Practical_RL
|
NOAA Wave Watch 3 and NDBC Buoy Data Comparison *Note: this notebook requires python3.*This notebook demostrates how to compare [WaveWatch III Global Ocean Wave Model](http://data.planetos.com/datasets/noaa_ww3_global_1.25x1d:noaa-wave-watch-iii-nww3-ocean-wave-model?utm_source=github&utm_medium=notebook&utm_campaign=ndbc-wavewatch-iii-notebook) and [NOAA NDBC buoy data](http://data.planetos.com/datasets/noaa_ndbc_stdmet_stations?utm_source=github&utm_medium=notebook&utm_campaign=ndbc-wavewatch-iii-notebook) using the Planet OS API.API documentation is available at http://docs.planetos.com. If you have questions or comments, join the [Planet OS Slack community](http://slack.planetos.com/) to chat with our development team.For general information on usage of IPython/Jupyter and Matplotlib, please refer to their corresponding documentation. https://ipython.org/ and http://matplotlib.org/. This notebook also makes use of the [matplotlib basemap toolkit.](http://matplotlib.org/basemap/index.html)
|
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import dateutil.parser
import datetime
from urllib.request import urlopen, Request
import simplejson as json
from datetime import date, timedelta, datetime
import matplotlib.dates as mdates
from mpl_toolkits.basemap import Basemap
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
**Important!** You'll need to replace apikey below with your actual Planet OS API key, which you'll find [on the Planet OS account settings page.](http://data.planetos.com/account/settings/?utm_source=github&utm_medium=notebook&utm_campaign=ww3-api-notebook) and NDBC buoy station name in which you are intrested.
|
dataset_id = 'noaa_ndbc_stdmet_stations'
## stations with wave height available: '46006', '46013', '46029'
## stations without wave height: icac1', '41047', 'bepb6', '32st0', '51004'
## stations too close to coastline (no point to compare to ww3)'sacv4', 'gelo1', 'hcef1'
station = '46029'
apikey = open('APIKEY').readlines()[0].strip() #'<YOUR API KEY HERE>'
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Let's first query the API to see what stations are available for the [NDBC Standard Meteorological Data dataset.](http://data.planetos.com/datasets/noaa_ndbc_stdmet_stations?utm_source=github&utm_medium=notebook&utm_campaign=ndbc-wavewatch-iii-notebook)
|
API_url = 'http://api.planetos.com/v1/datasets/%s/stations?apikey=%s' % (dataset_id, apikey)
request = Request(API_url)
response = urlopen(request)
API_data_locations = json.loads(response.read())
# print(API_data_locations)
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Now we'll use matplotlib to visualize the stations on a simple basemap.
|
m = Basemap(projection='merc',llcrnrlat=-80,urcrnrlat=80,\
llcrnrlon=-180,urcrnrlon=180,lat_ts=20,resolution='c')
fig=plt.figure(figsize=(15,10))
m.drawcoastlines()
##m.fillcontinents()
for i in API_data_locations['station']:
x,y=m(API_data_locations['station'][i]['SpatialExtent']['coordinates'][0],
API_data_locations['station'][i]['SpatialExtent']['coordinates'][1])
plt.scatter(x,y,color='r')
x,y=m(API_data_locations['station'][station]['SpatialExtent']['coordinates'][0],
API_data_locations['station'][station]['SpatialExtent']['coordinates'][1])
plt.scatter(x,y,s=100,color='b')
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Let's examine the last five days of data. For the WaveWatch III forecast, we'll use the reference time parameter to pull forecast data from the 18:00 model run from five days ago.
|
## Find suitable reference time values
atthemoment = datetime.utcnow()
atthemoment = atthemoment.strftime('%Y-%m-%dT%H:%M:%S')
before5days = datetime.utcnow() - timedelta(days=5)
before5days_long = before5days.strftime('%Y-%m-%dT%H:%M:%S')
before5days_short = before5days.strftime('%Y-%m-%d')
start = before5days_long
end = atthemoment
reftime_start = str(before5days_short) + 'T18:00:00'
reftime_end = reftime_start
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
API request for NOAA NDBC buoy station data
|
API_url = "http://api.planetos.com/v1/datasets/{0}/point?station={1}&apikey={2}&start={3}&end={4}&count=1000".format(dataset_id,station,apikey,start,end)
print(API_url)
request = Request(API_url)
response = urlopen(request)
API_data_buoy = json.loads(response.read())
buoy_variables = []
for k,v in set([(j,i['context']) for i in API_data_buoy['entries'] for j in i['data'].keys()]):
buoy_variables.append(k)
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Find buoy station coordinates to use them later for finding NOAA Wave Watch III data
|
for i in API_data_buoy['entries']:
#print(i['axes']['time'])
if i['context'] == 'time_latitude_longitude':
longitude = (i['axes']['longitude'])
latitude = (i['axes']['latitude'])
print ('Latitude: '+ str(latitude))
print ('Longitude: '+ str(longitude))
|
Latitude: 46.159000396728516
Longitude: -124.51399993896484
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
API request for NOAA WaveWatch III (NWW3) Ocean Wave Model near the point of selected station. Note that data may not be available at the requested reference time. If the response is empty, try removing the reference time parameters `reftime_start` and `reftime_end` from the query.
|
API_url = 'http://api.planetos.com/v1/datasets/noaa_ww3_global_1.25x1d/point?lat={0}&lon={1}&verbose=true&apikey={2}&count=100&end={3}&reftime_start={4}&reftime_end={5}'.format(latitude,longitude,apikey,end,reftime_start,reftime_end)
request = Request(API_url)
response = urlopen(request)
API_data_ww3 = json.loads(response.read())
print(API_url)
ww3_variables = []
for k,v in set([(j,i['context']) for i in API_data_ww3['entries'] for j in i['data'].keys()]):
ww3_variables.append(k)
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Manually review the list of WaveWatch and NDBC data variables to determine which parameters are equivalent for comparison.
|
print(ww3_variables)
print(buoy_variables)
|
['Wind_speed_surface', 'v-component_of_wind_surface', 'u-component_of_wind_surface', 'Primary_wave_direction_surface', 'Significant_height_of_combined_wind_waves_and_swell_surface', 'Significant_height_of_swell_waves_ordered_sequence_of_data', 'Mean_period_of_swell_waves_ordered_sequence_of_data', 'Mean_period_of_wind_waves_surface', 'Direction_of_swell_waves_ordered_sequence_of_data', 'Primary_wave_mean_period_surface', 'Secondary_wave_direction_surface', 'Secondary_wave_mean_period_surface', 'Wind_direction_from_which_blowing_surface', 'Direction_of_wind_waves_surface', 'Significant_height_of_wind_waves_surface']
['wind_spd', 'wave_height', 'air_temperature', 'dewpt_temperature', 'mean_wave_dir', 'average_wpd', 'dominant_wpd', 'sea_surface_temperature', 'air_pressure', 'wind_dir', 'water_level', 'visibility', 'gust']
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Next we'll build a dictionary of corresponding variables that we want to compare.
|
buoy_model = {'wave_height':'Significant_height_of_combined_wind_waves_and_swell_surface',
'mean_wave_dir':'Primary_wave_direction_surface',
'average_wpd':'Primary_wave_mean_period_surface',
'wind_spd':'Wind_speed_surface'}
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Read data from the JSON responses and convert the values to floats for plotting. Note that depending on the dataset, some variables have different timesteps than others, so a separate time array for each variable is recommended.
|
def append_data(in_string):
if in_string == None:
return np.nan
elif in_string == 'None':
return np.nan
else:
return float(in_string)
ww3_data = {}
ww3_times = {}
buoy_data = {}
buoy_times = {}
for k,v in buoy_model.items():
ww3_data[v] = []
ww3_times[v] = []
buoy_data[k] = []
buoy_times[k] = []
for i in API_data_ww3['entries']:
for j in i['data']:
if j in buoy_model.values():
ww3_data[j].append(append_data(i['data'][j]))
ww3_times[j].append(dateutil.parser.parse(i['axes']['time']))
for i in API_data_buoy['entries']:
for j in i['data']:
if j in buoy_model.keys():
buoy_data[j].append(append_data(i['data'][j]))
buoy_times[j].append(dateutil.parser.parse(i['axes']['time']))
for i in ww3_data:
ww3_data[i] = np.array(ww3_data[i])
ww3_times[i] = np.array(ww3_times[i])
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
Finally, let's plot the data using matplotlib.
|
buoy_label = "NDBC Station %s" % station
ww3_label = "WW3 at %s" % reftime_start
for k,v in buoy_model.items():
if np.abs(np.nansum(buoy_data[k]))>0:
fig=plt.figure(figsize=(10,5))
plt.title(k+' '+v)
plt.plot(ww3_times[v],ww3_data[v], label=ww3_label)
plt.plot(buoy_times[k],buoy_data[k],'*',label=buoy_label)
plt.legend(bbox_to_anchor=(1.5, 0.22), loc=1, borderaxespad=0.)
plt.xlabel('Time')
plt.ylabel(k)
fig.autofmt_xdate()
plt.grid()
|
_____no_output_____
|
MIT
|
api-examples/ndbc-wavewatch-iii.ipynb
|
evan1997123/PlanetOSDatathon
|
SCEE and Interconnect The SCEE module in SiPANN also has built in functionality to export any of it's models directly into a format readable by Lumerical Interconnect via the `export_interconnect()` function. This gives the user multiple options (Interconnect or Simphony) to cascade devices into complex structures. To export to a Interconnect file is as simple as a function call. First we declare all of our imports:
|
import numpy as np
from SiPANN import scee
|
_____no_output_____
|
MIT
|
examples/Tutorials/ExportToInterConnect.ipynb
|
joamatab/SiPANN
|
Then make our device and calculate it's scattering parameters (we arbitrarily choose a half ring resonator here)
|
r = 10000
w = 500
t = 220
wavelength = np.linspace(1500, 1600)
gap = 100
hr = scee.HalfRing(w, t, r, gap)
sparams = hr.sparams(wavelength)
|
_____no_output_____
|
MIT
|
examples/Tutorials/ExportToInterConnect.ipynb
|
joamatab/SiPANN
|
And then export. Note `export_interconnect` takes in wavelengths in nms, but the Lumerical file will have frequency in meters, as is standard in Interconnect. To export:
|
filename = "halfring_10microns_sparams.txt"
scee.export_interconnect(sparams, wavelength, filename)
|
_____no_output_____
|
MIT
|
examples/Tutorials/ExportToInterConnect.ipynb
|
joamatab/SiPANN
|
Aula 1
|
import pandas as pd
url_dados = 'https://github.com/alura-cursos/imersaodados3/blob/main/dados/dados_experimentos.zip?raw=true'
dados = pd.read_csv(url_dados, compression = 'zip')
dados
dados.head()
dados.shape
dados['tratamento']
dados['tratamento'].unique()
dados['tempo'].unique()
dados['dose'].unique()
dados['droga'].unique()
dados['g-0'].unique()
dados['tratamento'].value_counts()
dados['dose'].value_counts()
dados['tratamento'].value_counts(normalize = True)
dados['dose'].value_counts(normalize = True)
dados['tratamento'].value_counts().plot.pie()
dados['tempo'].value_counts().plot.pie()
dados['tempo'].value_counts().plot.bar()
dados_filtrados = dados[dados['g-0'] > 0]
dados_filtrados.head()
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Desafios Aula 1 Desafio 01: Investigar por que a classe tratamento é tão desbalanceada? Dependendo o tipo de pesquisa é possível usar o mesmo controle para mais de um caso. Repare que o grupo de controle é um grupo onde não estamos aplicando o efeito de uma determinada droga. Então, esse mesmo grupo pode ser utilizado como controle para cada uma das drogas estudadas. Um ponto relevante da base de dados que estamos trabalhando é que todos os dados de controle estão relacionados ao estudo de apenas uma droga.
|
print(f"Total de dados {len(dados['id'])}\n")
print(f"Quantidade de drogas {len(dados.groupby(['droga', 'tratamento']).count()['id'])}\n")
display(dados.query('tratamento == "com_controle"').value_counts('droga'))
print()
display(dados.query('droga == "cacb2b860"').value_counts('tratamento'))
print()
|
Total de dados 23814
Quantidade de drogas 3289
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Desafio 02: Plotar as 5 últimas linhas da tabela
|
dados.tail()
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Outra opção seria usar o seguinte comando:
|
dados[-5:]
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Desafio 03: Proporção das classes tratamento.
|
dados['tratamento'].value_counts(normalize = True)
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Desafio 04: Quantas tipos de drogas foram investigadas.
|
dados['droga'].unique().shape[0]
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Outra opção de solução:
|
len(dados['droga'].unique())
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Desafio 05: Procurar na documentação o método query(pandas). https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.query.html Desafio 06: Renomear as colunas tirando o hífen.
|
dados.columns
nome_das_colunas = dados.columns
novo_nome_coluna = []
for coluna in nome_das_colunas:
coluna = coluna.replace('-', '_')
novo_nome_coluna.append(coluna)
dados.columns = novo_nome_coluna
dados.head()
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Agora podemos comparar o resultado usando Query com o resultado usando máscara + slice
|
dados_filtrados = dados[dados['g_0'] > 0]
dados_filtrados.head()
dados_filtrados = dados.query('g_0 > 0')
dados_filtrados.head()
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Desafio 07: Deixar os gráficos bonitões. (Matplotlib.pyplot)
|
import matplotlib.pyplot as plt
valore_tempo = dados['tempo'].value_counts(ascending=True)
valore_tempo.sort_index()
plt.figure(figsize=(15, 10))
valore_tempo = dados['tempo'].value_counts(ascending=True)
ax = valore_tempo.sort_index().plot.bar()
ax.set_title('Janelas de tempo', fontsize=20)
ax.set_xlabel('Tempo', fontsize=18)
ax.set_ylabel('Quantidade', fontsize=18)
plt.xticks(rotation = 0, fontsize=16)
plt.yticks(fontsize=16)
plt.show()
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
Desafio 08: Resumo do que você aprendeu com os dados Nesta aula utilizei a biblioteca Pandas, diversas funcionalidades da mesma para explorar dados. Durante a análise de dados, descobri fatores importantes para a obtenção de insights e também aprendi como plotar os gráficos de pizza e de colunas discutindo pontos positivos e negativos. Para mais informações a base dados estudada na imersão é uma versão simplificada [deste desafio](https://www.kaggle.com/c/lish-moa/overview/description) do Kaggle (em inglês).Também recomendo acessar o[Connectopedia](https://clue.io/connectopedia/). O Connectopedia é um dicionário gratuito de termos e conceitos que incluem definições de viabilidade de células e expressão de genes. O desafio do Kaggle também está relacionado a estes artigos científicos:Corsello et al. “Discovering the anticancer potential of non-oncology drugs by systematic viability profiling,” Nature Cancer, 2020, https://doi.org/10.1038/s43018-019-0018-6Subramanian et al. “A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles,” Cell, 2017, https://doi.org/10.1016/j.cell.2017.10.049
|
_____no_output_____
|
MIT
|
Desafios_aula01respostas.ipynb
|
Adrianacms/ImersaoDados_Alura
|
|
Part 2 - ListsWe learned about **variables** in Part 1. Sometimes it makes sense to group lots of items of information together in a **list**. This is a good idea when the items are all connected in some way.For example, we might want to store the names of our friends. We could create several variables and assign the name of one friend to each of them (remember to do a Shift-Return on your keyboard, or click the Run button, to run the code):
|
friend_1 = "Fred"
friend_2 = "Jane"
friend_3 = "Rob"
friend_4 = "Sophie"
friend_5 = "Rachel"
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
This works OK. All the names are stored. But the problem is that they are all stored separately - Python does not know that they are all part of the same collection of friends.But there's a really nice way to solve this problem: **lists**. We can create a list like this:
|
friends = [ "Fred", "Jane", "Rob", "Sophie", "Rachel" ]
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
We create a list using square brackets `[` `]`, with each item in the list separated by a comma `,`. List methodsThere are some special **list** tools (called **methods**) that we can use on our `friends` list. We can add a name to the list using `append`:
|
friends.append("David")
print(friends)
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
We can put the names in alphabetical order using `sort`:
|
friends.sort()
print(friends)
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
We can reverse the order using `reverse`:
|
friends.reverse()
print(friends)
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
These **list** **methods** allow us to do lots of things like this without having to write much code ourselves. What is a list index?Let's make a new list. This time, we will start with an empty list, and then add to it:
|
shapes = []
print(shapes)
shapes.append("triangle")
shapes.append("square")
shapes.append("pentagon")
shapes.append("hexagon")
print(shapes)
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
These lists are great, but we might not always want to operate on the whole list at once. Instead, we might want to pick out one particular item in the list. In a **list**, each item is numbered, *starting from zero*. This number is called an **index**. So, remembering that the numbering *starts from zero*, we can access the first item in the list like this:
|
print(shapes[0])
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
Notice that the **index** is surrounded by square brackets `[` `]`. We can get the second item in `shapes` like this:
|
print(shapes[1])
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
and the last one like this:
|
print(shapes[3])
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
Python also has a special way of indexing the last item in a list, and this is especially useful if you're not sure how many items there are in your list. You can use a negative index:
|
print(shapes[-1])
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
We can ask Python to tell us the index of an item in the list:
|
idx = shapes.index("square")
print(idx)
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
Actually, the `index` method tells us the index of the first time that an item appears in a list. Displaying our data in a bar chart Let's have another look at our `shapes` list:
|
print(shapes)
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
and let's make another list that has the number of sides that each shape has:
|
sides=[3,4,5,6]
print(sides)
|
_____no_output_____
|
MIT
|
2 - Lists.ipynb
|
grahampullan/pythonical
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.