hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
4a8b4398fae9678d4f024fb8905fa8ea2c0aaf94
| 13,914 |
ipynb
|
Jupyter Notebook
|
notebook/models.ipynb
|
baasare/LoanChecker
|
09d24ef90266d368abf9ccb629793d13b3361e1a
|
[
"MIT"
] | null | null | null |
notebook/models.ipynb
|
baasare/LoanChecker
|
09d24ef90266d368abf9ccb629793d13b3361e1a
|
[
"MIT"
] | 9 |
2020-03-24T17:50:53.000Z
|
2022-02-10T09:44:04.000Z
|
notebook/models.ipynb
|
baasare/LoanChecker
|
09d24ef90266d368abf9ccb629793d13b3361e1a
|
[
"MIT"
] | null | null | null | 31.55102 | 157 | 0.581644 |
[
[
[
"#Load libraries\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.metrics import accuracy_score, precision_score, recall_score, confusion_matrix \nimport warnings # To ignore any warnings \n\nwarnings.filterwarnings(\"ignore\")\ndataset = pd.read_csv('loan_data_set.csv')\n\n\nprint(dataset['Loan_Status'].value_counts())\ndataset.describe()",
"_____no_output_____"
],
[
"dataset",
"_____no_output_____"
],
[
"sns.countplot(x = 'Loan_Status', data=dataset, palette='hls')\nsns.set(rc={'axes.facecolor':'#f8f9fa', 'figure.facecolor':'#f8f9fa'})\nplt.show()",
"_____no_output_____"
],
[
"import pandas as pd\ndataset = pd.read_csv('loan_data_set.csv')\ndataset.dtypes",
"_____no_output_____"
],
[
"print(dataset.columns[dataset.isnull().any()].tolist())\nmissing_values = dataset.isnull()\nmissing_values\n\nsns.heatmap(data = missing_values, yticklabels=False, cbar=False, cmap='viridis') #Heatmap of missing data values",
"_____no_output_____"
],
[
"\nsns.countplot(x='Loan_Status', data=dataset, hue='Education') #comparing those who had the loan and those who didint based their educational background",
"_____no_output_____"
],
[
"from sklearn.pipeline import Pipeline\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn import model_selection\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.svm import SVC, LinearSVC, NuSVC\nfrom sklearn.tree import DecisionTreeClassifier\n\n\n# encoding the categorical features\nvar_mod = ['Gender','Married','Dependents','Education','Self_Employed','Property_Area','Loan_Status']\nle = LabelEncoder()\nfor i in var_mod:\n dataset[i] = le.fit_transform(dataset[i].astype(str))\n\n\n# spliting the dataset into features and labels\nX = pd.DataFrame(dataset.iloc[:, 1:-1]) #excluding Loan_ID\ny = pd.DataFrame(dataset.iloc[:,-1]).values.ravel() #just labels\n\n\n# imputing missing values for the features\nimputer = SimpleImputer(strategy=\"mean\")\nimputer = imputer.fit(X)\nX = imputer.transform(X)\n\n# splitting dataset into train and test dataset\nx_train, x_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.2, random_state=7)",
"_____no_output_____"
],
[
"# Logistic Regression \nlogistic_reg_model = LogisticRegression(solver='liblinear')\nlogistic_reg_model.fit(x_train, y_train)\ny_pred = logistic_reg_model.predict(x_test)\n\nprint(\"Accuracy:\",accuracy_score(y_test, y_pred))\nprint(\"Precision:\",precision_score(y_test, y_pred))\nprint(\"Recall:\",recall_score(y_test, y_pred))\n\n\ny_single = logistic_reg_model.predict(x_test[0].reshape(1, -1))\n\n\n# confusion matrix\ncnf_matrix = confusion_matrix(y_test, y_pred)\n\n\nclass_names=[0,1] # name of classes\nfig, ax = plt.subplots()\ntick_marks = np.arange(len(class_names))\nplt.xticks(tick_marks, class_names)\nplt.yticks(tick_marks, class_names)\n\n\n# create heatmap\nsns.heatmap(pd.DataFrame(cnf_matrix), annot=True, cmap=\"YlGnBu\" ,fmt='g')\nax.xaxis.set_label_position(\"top\")\nplt.tight_layout()\nplt.title('Confusion matrix', y=1.1)\nplt.ylabel('Actual label')\nplt.xlabel('Predicted label')",
"_____no_output_____"
],
[
"model_decision_tree = DecisionTreeClassifier()\nmodel_decision_tree.fit(x_train,y_train)\npredictions = model_decision_tree.predict(x_test)\nprint(accuracy_score(y_test, predictions))\n\n\n\n\nmodel = RandomForestClassifier(n_estimators=100)\nmodel.fit(x_train,y_train)\npredictions = model.predict(x_test)\nprint(accuracy_score(y_test, predictions))\n\n\n\n\nmodel = KNeighborsClassifier(n_neighbors=9)\nmodel.fit(x_train,y_train)\npredictions = model.predict(x_test)\nprint(accuracy_score(y_test, predictions))\n\n\n\n\nmodel = SVC(gamma='scale', kernel='rbf')\nmodel.fit(x_train,y_train)\npredictions = model.predict(x_test)\nprint(accuracy_score(y_test, predictions))",
"_____no_output_____"
],
[
"dataset_test = dataset\n\nfeatures = ['LP001486', 'Male','Yes','1','Not Graduate','No',4583,1508,128,360,1,'Rural','N']\n\nnew_customer = pd.DataFrame({\n 'Loan_ID': [features[0]],\n 'Gender': [features[1]],\n 'Married': [features[2]],\n 'Dependents': [features[3]],\n 'Education': [features[4]],\n 'Self_Employed': [features[5]]\n 'ApplicantIncome': [features[6]],\n 'CoapplicantIncome': [features[7]],\n 'LoanAmount': [features[8]],\n 'Loan_Amount_Term': [features[9]],\n 'Credit_History': [features[10]],\n 'Property_Area':[features[11]],\n 'Loan_Status': [features[12]],\n})\n\n\n# append new single input to end of dataset\ndataset_test = dataset_test.append(new_customer)\n\n\n# encoding the categorical features\nvar_mod = ['Gender','Married','Dependents','Education','Self_Employed','Property_Area','Loan_Status']\nle = LabelEncoder()\nfor i in var_mod:\n dataset_test[i] = le.fit_transform(dataset_test[i].astype(str))\n\n \n# extrating encoded user input from encoded dataset \nuser = dataset_test[-1:] # last row contains user data\nuser = pd.DataFrame(user.iloc[:, 1:-1]) # exclude ID\nuser.values\n\ny_single = model_decision_tree.predict(user.values) # Test encoded input on decision tress model\nprint(y_single[0])",
"_____no_output_____"
],
[
"import pandas as pd\nfrom sklearn import model_selection\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.preprocessing import MinMaxScaler, OneHotEncoder, LabelEncoder\nfrom sklearn.compose import ColumnTransformer\nfrom sklearn.metrics import accuracy_score, precision_score, recall_score\n\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier, GradientBoostingClassifier\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.svm import SVC, LinearSVC, NuSVC\nfrom sklearn.tree import DecisionTreeClassifier\n\n\ndataset_1 = pd.read_csv('loan_data_set.csv')\ndataset_1 = dataset_1.drop('Loan_ID', axis=1)\n\n\nX = dataset_1.drop('Loan_Status', axis=1)\ny = dataset_1['Loan_Status']\n\n\nx_train, x_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.2, random_state=7)\n\n\n# numeric_features = ['ApplicantIncome','CoapplicantIncome','LoanAmount','Loan_Amount_Term']\nnumeric_features = dataset_1.select_dtypes(include=['int64', 'float64']).columns\nnumeric_features_steps = [('imputer', SimpleImputer(strategy='median')),('scaler', MinMaxScaler())]\nnumeric_transformer = Pipeline(steps=numeric_features_steps)\n\n\n\n# categorical_features = ['Gender','Married','Dependents','Education','Self_Employed','Property_Area']\ncategorical_features = dataset_1.select_dtypes(include=['object']).drop(['Loan_Status'], axis=1).columns\ncategorical_features_steps = [('imputer', SimpleImputer(strategy='constant', fill_value='missing')),('onehot', OneHotEncoder())]\ncategorical_transformer = Pipeline(steps=categorical_features_steps)\n\n\n\npreprocessor = ColumnTransformer(\n remainder = 'passthrough',\n transformers=[\n ('numeric', numeric_transformer, numeric_features),\n ('categorical', categorical_transformer, categorical_features)\n])\n\nclassifiers = {\n 'K-Nearnest Neighbour': KNeighborsClassifier(9),\n 'Logistic Regression(solver=liblinear)': LogisticRegression(solver='liblinear'),\n 'Support Vector Machine(gamma=auto, kernel=rbf)': SVC(gamma='auto', kernel='rbf'),\n 'Support Vector Machine(kernel=\"rbf\", C=0.025, probability=True)': SVC(gamma='auto', kernel=\"rbf\", C=0.025, \n probability=True),\n 'Nu Support Vector Machine(probability=True)': NuSVC(gamma='auto', probability=True),\n 'DecisionTreeClassifier': DecisionTreeClassifier(),\n 'Random Forest Classifier': RandomForestClassifier(n_estimators=100),\n 'AdaBoost Classifier': AdaBoostClassifier(),\n 'Gradient Boosting Classifier': GradientBoostingClassifier()\n}\n\n\npred_models = []\n\nfor name, classifier in classifiers.items():\n pipe = Pipeline(steps=[('preprocessor', preprocessor), ('classifier', classifier)])\n \n pipe.fit(x_train, y_train)\n pred_models.append(pipe)\n \ny_pred = pred_models[1].predict(x_test)\n# print(\"Accuracy: %.4f\" % pred_models[1].score(x_test, y_test))\n\nx_test\n\n\nfor name, classifier in classifiers.items():\n pipe = Pipeline(steps=[('preprocessor', preprocessor),('classifier', classifier)])\n pipe.fit(x_train, y_train)\n y_pred = pipe.predict(x_test)\n print(\"Classifier: \", name)\n print(\"Accuracy: %.4f\" % pipe.score(x_test, y_test))",
"_____no_output_____"
],
[
"features = ['LP001486', 'Male', 'Yes', '1', 'Graduate', 'No', 5483, 1508, 128, 360, 0, 'Urban', 'N']\n\nnew_customer = pd.DataFrame({\n 'Gender': [features[1]],\n 'Married': [features[2]],\n 'Dependents': [features[3]],\n 'Education': [features[4]],\n 'Self_Employed': [features[5]],\n 'ApplicantIncome': [features[6]],\n 'CoapplicantIncome': [features[7]],\n 'LoanAmount': [features[8]],\n 'Loan_Amount_Term': [features[9]],\n 'Credit_History': [features[10]],\n 'Property_Area':[features[11]],\n})\n\n\ny_pred_single = pred_models[1].predict(new_customer)\n# print(y_pred_single[0])\n\nif y_pred_single[0] == 'Y':\n print('Yes, you\\'re eligible')\nelse:\n print('Sorry, you\\'re not eligible')\n ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8b4d50c4711568d70b982781742c092a2e8294
| 22,319 |
ipynb
|
Jupyter Notebook
|
Misc/Python Exercise (doing what R does in python).ipynb
|
TensorMan/training-and-reference
|
68d2dea416e10bfe5b2a9b47b1794ce5c2b65371
|
[
"Apache-2.0"
] | null | null | null |
Misc/Python Exercise (doing what R does in python).ipynb
|
TensorMan/training-and-reference
|
68d2dea416e10bfe5b2a9b47b1794ce5c2b65371
|
[
"Apache-2.0"
] | null | null | null |
Misc/Python Exercise (doing what R does in python).ipynb
|
TensorMan/training-and-reference
|
68d2dea416e10bfe5b2a9b47b1794ce5c2b65371
|
[
"Apache-2.0"
] | null | null | null | 45.271805 | 414 | 0.468659 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a8b4d72109d4567944bdc11569cabc6d7377f43
| 104,334 |
ipynb
|
Jupyter Notebook
|
CNN/main_svm_epoch3.ipynb
|
ranery/Courses-only
|
8bc1254076c75b55b536498037ff0594a951d18f
|
[
"MIT"
] | 2 |
2021-03-31T21:46:58.000Z
|
2021-04-04T08:59:50.000Z
|
CNN/main_svm_epoch3.ipynb
|
ranery/Courses-only
|
8bc1254076c75b55b536498037ff0594a951d18f
|
[
"MIT"
] | null | null | null |
CNN/main_svm_epoch3.ipynb
|
ranery/Courses-only
|
8bc1254076c75b55b536498037ff0594a951d18f
|
[
"MIT"
] | null | null | null | 384.99631 | 94,526 | 0.916681 |
[
[
[
"import sys\nsys.path.append('C:\\\\Users\\dell-pc\\Desktop\\大四上\\Computer_Vision\\CNN')\nfrom data import *\nfrom network import three_layer_cnn",
"_____no_output_____"
],
[
"# data\ntrain_data, test_data = loaddata()",
"_____no_output_____"
],
[
"import numpy as np\nprint(train_data.keys())\nprint(\"Number of train items: %d\" % len(train_data['images']))\nprint(\"Number of test items: %d\" % len(test_data['labels']))\nprint(\"Edge length of picture : %f\" % np.sqrt(len(train_data['images'][0])))\nClass = set(train_data['labels'])\nprint(\"Total classes: \", Class)",
"dict_keys(['images', 'labels'])\nNumber of train items: 60000\nNumber of test items: 10000\nEdge length of picture : 28.000000\nTotal classes: {0, 1, 2, 3, 4, 5, 6, 7, 8, 9}\n"
],
[
"# reshape\ndef imageC(data_list):\n data = np.array(data_list).reshape(len(data_list), 1, 28, 28)\n return data\ndata = imageC(train_data['images'][0:3])\nprint(np.shape(data))",
"(3, 1, 28, 28)\n"
],
[
"# test\ndef test(cnn, test_batchSize):\n test_pred = []\n for i in range(int(len(test_data['images']) / test_batchSize)):\n out = cnn.inference(imageC(test_data['images'][i*test_batchSize:(i+1)*test_batchSize]))\n y = np.array(test_data['labels'][i*test_batchSize:(i+1)*test_batchSize])\n loss, pred = cnn.svm_loss(out, y, mode='test')\n test_pred.extend(pred)\n # accuracy\n count = 0\n for i in range(len(test_pred)):\n if test_pred[i] == test_data['labels'][i]:\n count += 1\n acc = count / len(test_pred)\n return acc, loss",
"_____no_output_____"
],
[
"# train\nprint('Begin training ...')\ncnn = three_layer_cnn()\ncnn.initial()\nepoch = 3\nbatchSize = 30\ntrain_loss = []\ntrain_acc = []\ntest_loss = []\ntest_acc = []\nfor i in range(epoch):\n for j in range(int(len(train_data['images']) / batchSize)):\n # for j in range(30):\n data = imageC(train_data['images'][j*batchSize:(j+1)*batchSize])\n label = np.array(train_data['labels'][j*batchSize:(j+1)*batchSize])\n output = cnn.forward(data)\n loss1, pred = cnn.svm_loss(output, label)\n train_loss.append(loss1)\n if j % 200 == 0:\n # train\n count = 0\n for k in range(batchSize):\n if pred[k] == label[k]:\n count += 1\n acc1 = count / batchSize\n train_acc.append(acc1)\n cnn.backward()\n if j % 200 == 0:\n # test\n acc2, loss2 = test(cnn, 10)\n test_loss.append(loss2)\n test_acc.append(acc2)\n print('Epoch: %d; Item: %d; Train loss: %f; Test loss: %f; Train acc: %f; Test acc: %f ' % (i, (j + 1) * batchSize, loss1, loss2, acc1, acc2))\nprint('End training!')\n",
"Begin training ...\nEpoch: 0; Item: 30; Train loss: 345.488145; Test loss: 89.517339; Train acc: 0.000000; Test acc: 0.105900 \nEpoch: 0; Item: 6030; Train loss: 26.794918; Test loss: 6.754075; Train acc: 0.800000; Test acc: 0.683400 \nEpoch: 0; Item: 12030; Train loss: 28.907206; Test loss: 3.671294; Train acc: 0.766667; Test acc: 0.726300 \nEpoch: 0; Item: 18030; Train loss: 26.445410; Test loss: 2.873742; Train acc: 0.833333; Test acc: 0.733500 \nEpoch: 0; Item: 24030; Train loss: 13.259348; Test loss: 6.427531; Train acc: 0.766667; Test acc: 0.694800 \nEpoch: 0; Item: 30030; Train loss: 22.563871; Test loss: 5.324414; Train acc: 0.733333; Test acc: 0.745500 \nEpoch: 0; Item: 36030; Train loss: 32.041588; Test loss: 2.870916; Train acc: 0.733333; Test acc: 0.749500 \nEpoch: 0; Item: 42030; Train loss: 25.343203; Test loss: 21.746450; Train acc: 0.733333; Test acc: 0.673200 \nEpoch: 0; Item: 48030; Train loss: 28.289382; Test loss: 38.737431; Train acc: 0.700000; Test acc: 0.723900 \nEpoch: 0; Item: 54030; Train loss: 60.662740; Test loss: 8.502795; Train acc: 0.600000; Test acc: 0.788200 \nEpoch: 1; Item: 30; Train loss: 19.494077; Test loss: 35.388683; Train acc: 0.866667; Test acc: 0.783600 \nEpoch: 1; Item: 6030; Train loss: 30.596390; Test loss: 21.238536; Train acc: 0.800000; Test acc: 0.736500 \nEpoch: 1; Item: 12030; Train loss: 32.444464; Test loss: 23.965808; Train acc: 0.766667; Test acc: 0.749300 \nEpoch: 1; Item: 18030; Train loss: 25.849059; Test loss: 22.973736; Train acc: 0.833333; Test acc: 0.727200 \nEpoch: 1; Item: 24030; Train loss: 7.510156; Test loss: 22.910779; Train acc: 0.933333; Test acc: 0.664300 \nEpoch: 1; Item: 30030; Train loss: 22.128428; Test loss: 7.789019; Train acc: 0.800000; Test acc: 0.786000 \nEpoch: 1; Item: 36030; Train loss: 74.703122; Test loss: 17.263572; Train acc: 0.700000; Test acc: 0.768300 \nEpoch: 1; Item: 42030; Train loss: 43.034299; Test loss: 2.612252; Train acc: 0.800000; Test acc: 0.699700 \nEpoch: 1; Item: 48030; Train loss: 45.003838; Test loss: 5.365515; Train acc: 0.766667; Test acc: 0.695100 \nEpoch: 1; Item: 54030; Train loss: 52.057511; Test loss: 5.770107; Train acc: 0.766667; Test acc: 0.805400 \nEpoch: 2; Item: 30; Train loss: 18.958401; Test loss: 8.012911; Train acc: 0.900000; Test acc: 0.808400 \nEpoch: 2; Item: 6030; Train loss: 6.489229; Test loss: 3.319428; Train acc: 0.866667; Test acc: 0.770000 \nEpoch: 2; Item: 12030; Train loss: 44.208407; Test loss: 5.674172; Train acc: 0.766667; Test acc: 0.794400 \nEpoch: 2; Item: 18030; Train loss: 32.268584; Test loss: 3.970969; Train acc: 0.833333; Test acc: 0.807600 \nEpoch: 2; Item: 24030; Train loss: 5.763226; Test loss: 2.588284; Train acc: 0.900000; Test acc: 0.759000 \nEpoch: 2; Item: 30030; Train loss: 24.317397; Test loss: 2.914502; Train acc: 0.800000; Test acc: 0.818900 \nEpoch: 2; Item: 36030; Train loss: 56.826149; Test loss: 2.053316; Train acc: 0.733333; Test acc: 0.797700 \nEpoch: 2; Item: 42030; Train loss: 75.065595; Test loss: 5.921901; Train acc: 0.666667; Test acc: 0.746800 \nEpoch: 2; Item: 48030; Train loss: 56.306741; Test loss: 0.930797; Train acc: 0.766667; Test acc: 0.762000 \nEpoch: 2; Item: 54030; Train loss: 33.781440; Test loss: 5.054786; Train acc: 0.833333; Test acc: 0.810800 \nEnd training!\n"
],
[
"# test\nacc, loss = test(cnn, 10)\nprint('Accuracy for 3-layers convolutional neural networks: %f' % acc)",
"Accuracy for 3-layers convolutional neural networks: 0.688000\n"
],
[
"# plot\nimport matplotlib.pyplot as plt\nfrom matplotlib.ticker import MultipleLocator, FormatStrFormatter\n\nax = plt.subplot(2, 1, 1)\nplt.title('Training loss (Batch Size: 30)')\nplt.xlabel('Iteration')\nplt.plot(train_loss, 'o')\n\nplt.subplot(2, 1, 2)\nplt.title('Accuracy')\nplt.xlabel('Iteration(x100)')\nplt.plot(train_acc, '-o', label='train')\nplt.plot(test_acc, '-o', label='test')\nplt.legend(loc='upper right', ncol=1)\nplt.gcf().set_size_inches(15, 15)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8b4e65e87fc87564e5b7743fc45b1583f528a1
| 467,377 |
ipynb
|
Jupyter Notebook
|
tools_pandas.ipynb
|
leader2345/handson-ml-master
|
a9a360e27084cb9db5ec91ffd1557a7d74daec90
|
[
"Apache-2.0"
] | null | null | null |
tools_pandas.ipynb
|
leader2345/handson-ml-master
|
a9a360e27084cb9db5ec91ffd1557a7d74daec90
|
[
"Apache-2.0"
] | null | null | null |
tools_pandas.ipynb
|
leader2345/handson-ml-master
|
a9a360e27084cb9db5ec91ffd1557a7d74daec90
|
[
"Apache-2.0"
] | null | null | null | 36.035235 | 20,024 | 0.504464 |
[
[
[
"**Tools - pandas**\n\n*The `pandas` library provides high-performance, easy-to-use data structures and data analysis tools. The main data structure is the `DataFrame`, which you can think of as an in-memory 2D table (like a spreadsheet, with column names and row labels). Many features available in Excel are available programmatically, such as creating pivot tables, computing columns based on other columns, plotting graphs, etc. You can also group rows by column value, or join tables much like in SQL. Pandas is also great at handling time series.*\n\nPrerequisites:\n* NumPy – if you are not familiar with NumPy, we recommend that you go through the [NumPy tutorial](tools_numpy.ipynb) now.",
"_____no_output_____"
],
[
"# Setup\nFirst, let's make sure this notebook works well in both python 2 and 3:",
"_____no_output_____"
]
],
[
[
"from __future__ import division, print_function, unicode_literals",
"_____no_output_____"
]
],
[
[
"Now let's import `pandas`. People usually import it as `pd`:",
"_____no_output_____"
]
],
[
[
"# import pandas as pd\n\n# Practice\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"# `Series` objects\nThe `pandas` library contains these useful data structures:\n* `Series` objects, that we will discuss now. A `Series` object is 1D array, similar to a column in a spreadsheet (with a column name and row labels).\n* `DataFrame` objects. This is a 2D table, similar to a spreadsheet (with column names and row labels).\n* `Panel` objects. You can see a `Panel` as a dictionary of `DataFrame`s. These are less used, so we will not discuss them here.",
"_____no_output_____"
],
[
"## Creating a `Series`\nLet's start by creating our first `Series` object!",
"_____no_output_____"
]
],
[
[
"# s = pd.Series([2,-1,3,5])\n# s\n\n# Practice\ns = pd.Series([2, -1, 3, 5])\ns",
"_____no_output_____"
]
],
[
[
"## Similar to a 1D `ndarray`\n`Series` objects behave much like one-dimensional NumPy `ndarray`s, and you can often pass them as parameters to NumPy functions:",
"_____no_output_____"
]
],
[
[
"# import numpy as np\n# np.exp(s)\n\n# Practice\nimport numpy as np\nnp.exp(s)",
"_____no_output_____"
]
],
[
[
"Arithmetic operations on `Series` are also possible, and they apply *elementwise*, just like for `ndarray`s:",
"_____no_output_____"
]
],
[
[
"# s + [1000,2000,3000,4000]\n\ns + [1000, 2000, 3000, 4000]",
"_____no_output_____"
]
],
[
[
"Similar to NumPy, if you add a single number to a `Series`, that number is added to all items in the `Series`. This is called * broadcasting*:",
"_____no_output_____"
]
],
[
[
"# s + 1000\n\n# Practice\n\ns + 1000",
"_____no_output_____"
]
],
[
[
"The same is true for all binary operations such as `*` or `/`, and even conditional operations:",
"_____no_output_____"
]
],
[
[
"# s < 0\n\n# Practice\nprint(s * 2 )\n\nprint(s < 0)",
"0 4\n1 -2\n2 6\n3 10\ndtype: int64\n0 False\n1 True\n2 False\n3 False\ndtype: bool\n"
]
],
[
[
"## Index labels\nEach item in a `Series` object has a unique identifier called the *index label*. By default, it is simply the rank of the item in the `Series` (starting at `0`) but you can also set the index labels manually:",
"_____no_output_____"
]
],
[
[
"# s2 = pd.Series([68, 83, 112, 68], index=[\"alice\", \"bob\", \"charles\", \"darwin\"])\n# s2\n\n# Practice\ns2 = pd.Series([68, 83, 112, 68], index=[\"alice\", \"bob\", \"charles\", \"darwin\"])\ns2",
"_____no_output_____"
]
],
[
[
"You can then use the `Series` just like a `dict`:",
"_____no_output_____"
]
],
[
[
"# s2[\"bob\"]\n\n# Practice\ns2[\"charles\"]",
"_____no_output_____"
]
],
[
[
"You can still access the items by integer location, like in a regular array:",
"_____no_output_____"
]
],
[
[
"# s2[1]\n\n# Practice\ns2[2]",
"_____no_output_____"
]
],
[
[
"To make it clear when you are accessing by label or by integer location, it is recommended to always use the `loc` attribute when accessing by label, and the `iloc` attribute when accessing by integer location:",
"_____no_output_____"
],
[
"My Notes: Gets the location of the index label",
"_____no_output_____"
]
],
[
[
"# s2.loc[\"bob\"]\n\n# Practice\ns2.loc[\"alice\"]",
"_____no_output_____"
],
[
"# s2.iloc[1]\n\n# Practice\ns2.iloc[3]",
"_____no_output_____"
]
],
[
[
"Slicing a `Series` also slices the index labels:",
"_____no_output_____"
]
],
[
[
"# s2.iloc[1:3]\n\n# Practice\ns2.iloc[2:3]",
"_____no_output_____"
]
],
[
[
"This can lead to unexpected results when using the default numeric labels, so be careful:",
"_____no_output_____"
]
],
[
[
"# surprise = pd.Series([1000, 1001, 1002, 1003])\n# surprise\n\n# Practice\n\nsurprise = pd.Series([1000, 1001, 1002, 1003])\nsurprise",
"_____no_output_____"
],
[
"# surprise_slice = surprise[2:]\n# surprise_slice\n\n# Practice\nsurprise_slice = surprise[2:]\nsurprise_slice",
"_____no_output_____"
]
],
[
[
"Oh look! The first element has index label `2`. The element with index label `0` is absent from the slice:",
"_____no_output_____"
]
],
[
[
"# try:\n# surprise_slice[0]\n# except KeyError as e:\n# print(\"Key error:\", e)\n\n# Practice\n\ntry:\n surprise_slice[0]\nexcept KeyError as e:\n print(\"Key error:\", e)",
"Key error: 0\n"
]
],
[
[
"But remember that you can access elements by integer location using the `iloc` attribute. This illustrates another reason why it's always better to use `loc` and `iloc` to access `Series` objects:",
"_____no_output_____"
]
],
[
[
"# surprise_slice.iloc[0]\n\n# Practice\nsurprise_slice.iloc[1]",
"_____no_output_____"
]
],
[
[
"## Init from `dict`\nYou can create a `Series` object from a `dict`. The keys will be used as index labels:",
"_____no_output_____"
]
],
[
[
"# weights = {\"alice\": 68, \"bob\": 83, \"colin\": 86, \"darwin\": 68}\n# s3 = pd.Series(weights)\n# s3\n\n# Practice\nweights = {\"alice\":68, \"bob\":83, \"colin\":86, \"darwin\":68}\ns3 = pd.Series(weights)\ns3",
"_____no_output_____"
]
],
[
[
"You can control which elements you want to include in the `Series` and in what order by explicitly specifying the desired `index`:",
"_____no_output_____"
]
],
[
[
"# s4 = pd.Series(weights, index = [\"colin\", \"alice\"])\n# s4\n\n# Practice\ns4 = pd.Series(weights, index=[\"colin\", \"alice\"])\ns4",
"_____no_output_____"
]
],
[
[
"## Automatic alignment\nWhen an operation involves multiple `Series` objects, `pandas` automatically aligns items by matching index labels.",
"_____no_output_____"
],
[
"My Notes:",
"_____no_output_____"
]
],
[
[
"s2",
"_____no_output_____"
],
[
"s3",
"_____no_output_____"
],
[
"# print(s2.keys())\n# print(s3.keys())\n\n# s2 + s3\n\n# Practice\nprint(s2.keys())\nprint(s3.keys())\n\ns2 + s3",
"Index(['alice', 'bob', 'charles', 'darwin'], dtype='object')\nIndex(['alice', 'bob', 'colin', 'darwin'], dtype='object')\n"
]
],
[
[
"The resulting `Series` contains the union of index labels from `s2` and `s3`. Since `\"colin\"` is missing from `s2` and `\"charles\"` is missing from `s3`, these items have a `NaN` result value. (ie. Not-a-Number means *missing*).\n\nAutomatic alignment is very handy when working with data that may come from various sources with varying structure and missing items. But if you forget to set the right index labels, you can have surprising results:",
"_____no_output_____"
]
],
[
[
"# s5 = pd.Series([1000,1000,1000,1000])\n# print(\"s2 =\", s2.values)\n# print(\"s5 =\", s5.values)\n\n# s2 + s5\n\n# Practice\ns5 = pd.Series([1000, 1000, 1000, 1000])\nprint(\"s2 =\", s2.values)\nprint(\"s5=\", s5.values)\n\ns2 + s5 # My Notes: Cannot add as they are not of the same index labels",
"s2 = [ 68 83 112 68]\ns5= [1000 1000 1000 1000]\n"
]
],
[
[
"Pandas could not align the `Series`, since their labels do not match at all, hence the full `NaN` result.",
"_____no_output_____"
],
[
"## Init with a scalar\nYou can also initialize a `Series` object using a scalar and a list of index labels: all items will be set to the scalar.",
"_____no_output_____"
]
],
[
[
"# meaning = pd.Series(42, [\"life\", \"universe\", \"everything\"])\n# meaning\n\n# Practice\nmeaning = pd.Series(42, [\"life\", \"universe\", \"everything\"])\nmeaning",
"_____no_output_____"
]
],
[
[
"## `Series` name\nA `Series` can have a `name`:",
"_____no_output_____"
]
],
[
[
"# s6 = pd.Series([83, 68], index=[\"bob\", \"alice\"], name=\"weights\")\n# s6\n\n# Practice\ns6 = pd.Series([83, 68], index=[\"bob\", \"alice\"], name=\"weights\")\ns6",
"_____no_output_____"
]
],
[
[
"## Plotting a `Series`\nPandas makes it easy to plot `Series` data using matplotlib (for more details on matplotlib, check out the [matplotlib tutorial](tools_matplotlib.ipynb)). Just import matplotlib and call the `plot()` method:",
"_____no_output_____"
]
],
[
[
"# %matplotlib inline\n# import matplotlib.pyplot as plt\n# temperatures = [4.4,5.1,6.1,6.2,6.1,6.1,5.7,5.2,4.7,4.1,3.9,3.5]\n# s7 = pd.Series(temperatures, name=\"Temperature\")\n# s7.plot()\n# plt.show()\n\n# Practice\n%matplotlib inline\nimport matplotlib.pyplot as plt\ntemperatures = [4.4, 5.1, 6.1, 6.2, 6.1, 6.1, 5.7, 5.2, 4.7, 4.1, 3.9, 3.5]\ns7 = pd.Series(temperatures, name=\"Temperature\")\ns7.plot()\nplt.show()",
"_____no_output_____"
]
],
[
[
"There are *many* options for plotting your data. It is not necessary to list them all here: if you need a particular type of plot (histograms, pie charts, etc.), just look for it in the excellent [Visualization](http://pandas.pydata.org/pandas-docs/stable/visualization.html) section of pandas' documentation, and look at the example code.",
"_____no_output_____"
],
[
"# Handling time\nMany datasets have timestamps, and pandas is awesome at manipulating such data:\n* it can represent periods (such as 2016Q3) and frequencies (such as \"monthly\"),\n* it can convert periods to actual timestamps, and *vice versa*,\n* it can resample data and aggregate values any way you like,\n* it can handle timezones.\n\n## Time range\nLet's start by creating a time series using `pd.date_range()`. This returns a `DatetimeIndex` containing one datetime per hour for 12 hours starting on October 29th 2016 at 5:30pm.",
"_____no_output_____"
]
],
[
[
"# dates = pd.date_range('2016/10/29 5:30pm', periods=12, freq='H')\n# dates\n\n# Practice\n\ndates = pd.date_range('2016/10/29 5:30pm', periods=12, freq='H')\ndates",
"_____no_output_____"
]
],
[
[
"This `DatetimeIndex` may be used as an index in a `Series`:",
"_____no_output_____"
]
],
[
[
"# temp_series = pd.Series(temperatures, dates)\n# temp_series\n\n# Practice\ntemp_series = pd.Series(temperatures, index=dates)\ntemp_series",
"_____no_output_____"
]
],
[
[
"Let's plot this series:",
"_____no_output_____"
]
],
[
[
"# temp_series.plot(kind=\"bar\")\n\n# plt.grid(True)\n# plt.show()\n\n# Practice\n\ntemp_series.plot(kind=\"bar\")\n\nplt.grid(True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Resampling\nPandas lets us resample a time series very simply. Just call the `resample()` method and specify a new frequency:",
"_____no_output_____"
]
],
[
[
"# temp_series_freq_2H = temp_series.resample(\"2H\")\n# temp_series_freq_2H\n\n# Practice\ntemp_series_freq_2H = temp_series.resample(\"2H\")\ntemp_series_freq_2H",
"_____no_output_____"
]
],
[
[
"The resampling operation is actually a deferred operation, which is why we did not get a `Series` object, but a `DatetimeIndexResampler` object instead. To actually perform the resampling operation, we can simply call the `mean()` method: Pandas will compute the mean of every pair of consecutive hours:",
"_____no_output_____"
]
],
[
[
"# temp_series_freq_2H = temp_series_freq_2H.mean()\n\n# Practice\ntemp_series_freq_2H = temp_series_freq_2H.mean()",
"_____no_output_____"
]
],
[
[
"Let's plot the result:",
"_____no_output_____"
]
],
[
[
"# temp_series_freq_2H.plot(kind=\"bar\")\n# plt.show()\n\n# Practice\ntemp_series_freq_2H.plot(kind=\"bar\")\nplt.grid(True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Note how the values have automatically been aggregated into 2-hour periods. If we look at the 6-8pm period, for example, we had a value of `5.1` at 6:30pm, and `6.1` at 7:30pm. After resampling, we just have one value of `5.6`, which is the mean of `5.1` and `6.1`. Rather than computing the mean, we could have used any other aggregation function, for example we can decide to keep the minimum value of each period:",
"_____no_output_____"
]
],
[
[
"# temp_series_freq_2H = temp_series.resample(\"2H\").min()\n# temp_series_freq_2H\n\n# Practice\ntemp_series_freq_2H = temp_series.resample(\"2H\").min()\ntemp_series_freq_2H",
"_____no_output_____"
]
],
[
[
"Or, equivalently, we could use the `apply()` method instead:",
"_____no_output_____"
]
],
[
[
"# temp_series_freq_2H = temp_series.resample(\"2H\").apply(np.min)\n# temp_series_freq_2H\n\n# Practice\ntemp_series_freq_2H = temp_series.resample(\"2H\").apply(np.min)\ntemp_series_freq_2H",
"_____no_output_____"
]
],
[
[
"## Upsampling and interpolation\nThis was an example of downsampling. We can also upsample (ie. increase the frequency), but this creates holes in our data:",
"_____no_output_____"
]
],
[
[
"temp_series_freq_15min = temp_series.resample(\"15Min\").mean()\ntemp_series_freq_15min.head(n=10) # `head` displays the top n values",
"_____no_output_____"
]
],
[
[
"One solution is to fill the gaps by interpolating. We just call the `interpolate()` method. The default is to use linear interpolation, but we can also select another method, such as cubic interpolation:",
"_____no_output_____"
]
],
[
[
"temp_series_freq_15min = temp_series.resample(\"15Min\").interpolate(method=\"cubic\")\ntemp_series_freq_15min.head(n=10)",
"_____no_output_____"
],
[
"temp_series.plot(label=\"Period: 1 hour\")\ntemp_series_freq_15min.plot(label=\"Period: 15 minutes\")\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Timezones\nBy default datetimes are *naive*: they are not aware of timezones, so 2016-10-30 02:30 might mean October 30th 2016 at 2:30am in Paris or in New York. We can make datetimes timezone *aware* by calling the `tz_localize()` method:",
"_____no_output_____"
]
],
[
[
"temp_series_ny = temp_series.tz_localize(\"America/New_York\")\ntemp_series_ny",
"_____no_output_____"
]
],
[
[
"Note that `-04:00` is now appended to all the datetimes. This means that these datetimes refer to [UTC](https://en.wikipedia.org/wiki/Coordinated_Universal_Time) - 4 hours.\n\nWe can convert these datetimes to Paris time like this:",
"_____no_output_____"
]
],
[
[
"temp_series_paris = temp_series_ny.tz_convert(\"Europe/Paris\")\ntemp_series_paris",
"_____no_output_____"
]
],
[
[
"You may have noticed that the UTC offset changes from `+02:00` to `+01:00`: this is because France switches to winter time at 3am that particular night (time goes back to 2am). Notice that 2:30am occurs twice! Let's go back to a naive representation (if you log some data hourly using local time, without storing the timezone, you might get something like this):",
"_____no_output_____"
]
],
[
[
"temp_series_paris_naive = temp_series_paris.tz_localize(None)\ntemp_series_paris_naive",
"_____no_output_____"
]
],
[
[
"Now `02:30` is really ambiguous. If we try to localize these naive datetimes to the Paris timezone, we get an error:",
"_____no_output_____"
]
],
[
[
"try:\n temp_series_paris_naive.tz_localize(\"Europe/Paris\")\nexcept Exception as e:\n print(type(e))\n print(e)",
"<class 'pytz.exceptions.AmbiguousTimeError'>\nCannot infer dst time from 2016-10-30 02:30:00, try using the 'ambiguous' argument\n"
]
],
[
[
"Fortunately using the `ambiguous` argument we can tell pandas to infer the right DST (Daylight Saving Time) based on the order of the ambiguous timestamps:",
"_____no_output_____"
]
],
[
[
"temp_series_paris_naive.tz_localize(\"Europe/Paris\", ambiguous=\"infer\")",
"_____no_output_____"
]
],
[
[
"## Periods\nThe `pd.period_range()` function returns a `PeriodIndex` instead of a `DatetimeIndex`. For example, let's get all quarters in 2016 and 2017:",
"_____no_output_____"
]
],
[
[
"# quarters = pd.period_range('2016Q1', periods=8, freq='Q')\n# quarters\n\n# Practice\nquarters = pd.period_range('2016Q1', periods=8, freq='Q')\nquarters",
"_____no_output_____"
]
],
[
[
"Adding a number `N` to a `PeriodIndex` shifts the periods by `N` times the `PeriodIndex`'s frequency:",
"_____no_output_____"
]
],
[
[
"# quarters + 3\n\n# Practice\nquarters + 3",
"_____no_output_____"
]
],
[
[
"The `asfreq()` method lets us change the frequency of the `PeriodIndex`. All periods are lengthened or shortened accordingly. For example, let's convert all the quarterly periods to monthly periods (zooming in):",
"_____no_output_____"
]
],
[
[
"# quarters.asfreq(\"M\")\n\n# Practice\nquarters.asfreq(\"M\")",
"_____no_output_____"
]
],
[
[
"By default, the `asfreq` zooms on the end of each period. We can tell it to zoom on the start of each period instead:",
"_____no_output_____"
]
],
[
[
"# quarters.asfreq(\"M\", how=\"start\")\n\n# Practice\nquarters.asfreq(\"M\", how=\"start\")",
"_____no_output_____"
]
],
[
[
"And we can zoom out:",
"_____no_output_____"
]
],
[
[
"# quarters.asfreq(\"A\")\n\n# Practice\nquarters.asfreq(\"A\")",
"_____no_output_____"
]
],
[
[
"Of course we can create a `Series` with a `PeriodIndex`:",
"_____no_output_____"
]
],
[
[
"# quarterly_revenue = pd.Series([300, 320, 290, 390, 320, 360, 310, 410], index = quarters)\n# quarterly_revenue\n\n# Practice\nquarterly_revenue = pd.Series([300, 320, 290, 390, 320, 360, 310, 410], index = quarters)\nquarterly_revenue",
"_____no_output_____"
],
[
"# quarterly_revenue.plot(kind=\"line\")\n# plt.show()\n\n# Practice\nquarterly_revenue.plot(kind=\"line\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can convert periods to timestamps by calling `to_timestamp`. By default this will give us the first day of each period, but by setting `how` and `freq`, we can get the last hour of each period:",
"_____no_output_____"
]
],
[
[
"# last_hours = quarterly_revenue.to_timestamp(how=\"end\", freq=\"H\")\n# last_hours\n\n# Practice\nlast_hours = quarterly_revenue.to_timestamp(how=\"end\", freq=\"H\")\nlast_hours",
"_____no_output_____"
]
],
[
[
"And back to periods by calling `to_period`:",
"_____no_output_____"
]
],
[
[
"# last_hours.to_period()\n\n# Practice\nlast_hours.to_period()",
"_____no_output_____"
]
],
[
[
"Pandas also provides many other time-related functions that we recommend you check out in the [documentation](http://pandas.pydata.org/pandas-docs/stable/timeseries.html). To whet your appetite, here is one way to get the last business day of each month in 2016, at 9am:",
"_____no_output_____"
]
],
[
[
"# months_2016 = pd.period_range(\"2016\", periods=12, freq=\"M\")\n# one_day_after_last_days = months_2016.asfreq(\"D\") + 1\n# last_bdays = one_day_after_last_days.to_timestamp() - pd.tseries.offsets.BDay()\n# last_bdays.to_period(\"H\") + 9",
"_____no_output_____"
]
],
[
[
"Practice",
"_____no_output_____"
]
],
[
[
"months_2016 = pd.period_range(\"2016\", periods=12, freq=\"M\")\nmonths_2016",
"_____no_output_____"
],
[
"one_day_after_last_days = months_2016.asfreq(\"D\") + 1\none_day_after_last_days",
"_____no_output_____"
],
[
"last_bdays = one_day_after_last_days.to_timestamp()\nlast_bdays",
"_____no_output_____"
],
[
"pd.tseries.offsets.BDay()",
"_____no_output_____"
],
[
"last_bdays = one_day_after_last_days.to_timestamp() - pd.tseries.offsets.BDay() # My Notes: Last Business days\nlast_bdays",
"_____no_output_____"
]
],
[
[
"My Notes: Adding 9:00am to the period",
"_____no_output_____"
]
],
[
[
"last_bdays.to_period(\"H\") + 9",
"_____no_output_____"
]
],
[
[
"# `DataFrame` objects\nA DataFrame object represents a spreadsheet, with cell values, column names and row index labels. You can define expressions to compute columns based on other columns, create pivot-tables, group rows, draw graphs, etc. You can see `DataFrame`s as dictionaries of `Series`.\n\n## Creating a `DataFrame`\nYou can create a DataFrame by passing a dictionary of `Series` objects:",
"_____no_output_____"
]
],
[
[
"# people_dict = {\n# \"weight\": pd.Series([68, 83, 112], index=[\"alice\", \"bob\", \"charles\"]),\n# \"birthyear\": pd.Series([1984, 1985, 1992], index=[\"bob\", \"alice\", \"charles\"], name=\"year\"),\n# \"children\": pd.Series([0, 3], index=[\"charles\", \"bob\"]),\n# \"hobby\": pd.Series([\"Biking\", \"Dancing\"], index=[\"alice\", \"bob\"]),\n# }\n# people = pd.DataFrame(people_dict)\n# people\n\n# Practice\npeople_dict = {\n \"weight\": pd.Series([68, 83, 112], index=[\"alice\", \"bob\", \"charles\"]),\n \"birthyear\": pd.Series([1984, 1985, 1992], index=[\"bob\", \"alice\", \"charles\"], name=\"year\"),\n \"children\": pd.Series([0, 3], index=[\"charles\", \"bob\"]),\n \"hobby\": pd.Series([\"Biking\", \"Dancing\"], index=[\"alice\", \"bob\"]),\n}\npeople = pd.DataFrame(people_dict)\npeople",
"_____no_output_____"
]
],
[
[
"A few things to note:\n* the `Series` were automatically aligned based on their index,\n* missing values are represented as `NaN`,\n* `Series` names are ignored (the name `\"year\"` was dropped),\n* `DataFrame`s are displayed nicely in Jupyter notebooks, woohoo!",
"_____no_output_____"
],
[
"You can access columns pretty much as you would expect. They are returned as `Series` objects:",
"_____no_output_____"
]
],
[
[
"# people[\"birthyear\"]\n\n# Practice\npeople[\"birthyear\"]",
"_____no_output_____"
],
[
"# Practice\npeople.loc[\"bob\"]",
"_____no_output_____"
]
],
[
[
"You can also get multiple columns at once:",
"_____no_output_____"
]
],
[
[
"# people[[\"birthyear\", \"hobby\"]]\n\n# Practice\npeople[[\"birthyear\", \"hobby\", \"children\"]]",
"_____no_output_____"
]
],
[
[
"If you pass a list of columns and/or index row labels to the `DataFrame` constructor, it will guarantee that these columns and/or rows will exist, in that order, and no other column/row will exist. For example:",
"_____no_output_____"
]
],
[
[
"# d2 = pd.DataFrame(\n# people_dict,\n# columns=[\"birthyear\", \"weight\", \"height\"],\n# index=[\"bob\", \"alice\", \"eugene\"]\n# )\n# d2\n\n# Practice\nd2 = pd.DataFrame(\n people_dict,\n columns=[\"birthyear\", \"weight\", \"height\"],\n index = [\"bob\", \"alice\", \"eugene\", \"martin\"]\n)\nd2",
"_____no_output_____"
]
],
[
[
"Another convenient way to create a `DataFrame` is to pass all the values to the constructor as an `ndarray`, or a list of lists, and specify the column names and row index labels separately:",
"_____no_output_____"
]
],
[
[
"# values = [\n# [1985, np.nan, \"Biking\", 68],\n# [1984, 3, \"Dancing\", 83],\n# [1992, 0, np.nan, 112]\n# ]\n# d3 = pd.DataFrame(\n# values,\n# columns=[\"birthyear\", \"children\", \"hobby\", \"weight\"],\n# index=[\"alice\", \"bob\", \"charles\"]\n# )\n# d3\n\n# Practice\nvalues = [\n [1985, np.nan, \"Biking\", 68],\n [1984, 3, \"Dancing\", 83],\n [1992, 0, np.nan, 112]\n]\n\nd3 = pd.DataFrame(\n values,\n columns = [\"birthyear\", \"children\", \"hobby\", \"weight\"],\n index = [\"alice\", \"bob\", \"charles\"]\n)\nd3",
"_____no_output_____"
]
],
[
[
"To specify missing values, you can either use `np.nan` or NumPy's masked arrays:",
"_____no_output_____"
]
],
[
[
"# masked_array = np.ma.asarray(values, dtype=np.object)\n# masked_array[(0, 2), (1, 2)] = np.ma.masked\n# d3 = pd.DataFrame(\n# masked_array,\n# columns=[\"birthyear\", \"children\", \"hobby\", \"weight\"],\n# index=[\"alice\", \"bob\", \"charles\"]\n# )\n# d3\n\n# Practice\nmasked_array = np.ma.asarray(values, dtype=np.object)\nmasked_array[(0, 2), (1, 2)] = np.ma.masked\n\nd3 = pd.DataFrame(\n masked_array,\n columns = [\"birthyear\", \"children\", \"hobby\", \"weight\"],\n index=[\"alice\", \"bob\", \"charles\"]\n )\n\nd3",
"_____no_output_____"
],
[
"# Practice\nmasked_array[(0, 2), (1, 2)] = np.ma.masked",
"_____no_output_____"
]
],
[
[
"Instead of an `ndarray`, you can also pass a `DataFrame` object:",
"_____no_output_____"
]
],
[
[
"# d4 = pd.DataFrame(\n# d3,\n# columns=[\"hobby\", \"children\"],\n# index=[\"alice\", \"bob\"]\n# )\n# d4\n\n# Practice\nd4 = pd.DataFrame(\n d3,\n columns = [\"hobby\", \"children\"],\n index = [\"alice\", \"bob\"]\n)\n\nd4",
"_____no_output_____"
]
],
[
[
"It is also possible to create a `DataFrame` with a dictionary (or list) of dictionaries (or list):",
"_____no_output_____"
]
],
[
[
"# people = pd.DataFrame({\n# \"birthyear\": {\"alice\":1985, \"bob\": 1984, \"charles\": 1992},\n# \"hobby\": {\"alice\":\"Biking\", \"bob\": \"Dancing\"},\n# \"weight\": {\"alice\":68, \"bob\": 83, \"charles\": 112},\n# \"children\": {\"bob\": 3, \"charles\": 0}\n# })\n# people\n\n# Practice\npeople = pd.DataFrame({\n \"birthyear\": {\"alice\": 1985, \"bob\": 1984, \"charles\": 1992},\n \"hobby\": {\"alice\": \"Biking\", \"bob\": \"Dancing\"},\n \"weight\": {\"alice\": 68, \"bob\": 83, \"charles\": 112},\n})\npeople",
"_____no_output_____"
]
],
[
[
"## Multi-indexing\nIf all columns are tuples of the same size, then they are understood as a multi-index. The same goes for row index labels. For example:",
"_____no_output_____"
]
],
[
[
"# d5 = pd.DataFrame(\n# {\n# (\"public\", \"birthyear\"):\n# {(\"Paris\",\"alice\"):1985, (\"Paris\",\"bob\"): 1984, (\"London\",\"charles\"): 1992},\n# (\"public\", \"hobby\"):\n# {(\"Paris\",\"alice\"):\"Biking\", (\"Paris\",\"bob\"): \"Dancing\"},\n# (\"private\", \"weight\"):\n# {(\"Paris\",\"alice\"):68, (\"Paris\",\"bob\"): 83, (\"London\",\"charles\"): 112},\n# (\"private\", \"children\"):\n# {(\"Paris\", \"alice\"):np.nan, (\"Paris\",\"bob\"): 3, (\"London\",\"charles\"): 0}\n# }\n# )\n# d5\n\nd5 = pd.DataFrame(\n {\n (\"public\", \"birthyear\"):\n {(\"Paris\", \"alice\"): 1985, (\"Paris\", \"bob\"): 1984, (\"London\", \"charles\"): 1992},\n (\"public\", \"hobby\"):\n {(\"Paris\", \"alice\"): \"Biking\", (\"Paris\", \"bob\"): \"Dancing\"},\n (\"private\", \"weight\"):\n {(\"Paris\", \"alice\"): 68, (\"Paris\", \"bob\"): 83, (\"London\", \"charles\"): 112},\n (\"private\", \"children\"):\n {(\"Paris\", \"alice\"):np.nan, (\"Paris\", \"bob\"):3, (\"London\", \"charles\"): 0},\n }\n)\n\nd5\n\n# My Notes: Start from the top most columns",
"_____no_output_____"
]
],
[
[
"You can now get a `DataFrame` containing all the `\"public\"` columns very simply:",
"_____no_output_____"
]
],
[
[
"# d5[\"public\"]\n\n# Practice\nd5[\"private\"]",
"_____no_output_____"
],
[
"d5[\"public\", \"hobby\"] # Same result as d5[\"public\"][\"hobby\"]\n\n# Practice\n# d5[\"public\", \"hobby\"]\n# d5[\"public\"][\"hobby\"]\nd5[\"private\", \"children\"]",
"_____no_output_____"
]
],
[
[
"## Dropping a level\nLet's look at `d5` again:",
"_____no_output_____"
]
],
[
[
"# d5\n\n# Practice\nd5",
"_____no_output_____"
]
],
[
[
"There are two levels of columns, and two levels of indices. We can drop a column level by calling `droplevel()` (the same goes for indices):",
"_____no_output_____"
]
],
[
[
"# d5.columns = d5.columns.droplevel(level = 0)\n# d5\n\n# Practice\nd5.columns = d5.columns.droplevel(level=0)\nd5",
"_____no_output_____"
],
[
"# Practice\n# My Notes: Drop the top most index\nd5.index = d5.index.droplevel(level=0)\nd5",
"_____no_output_____"
]
],
[
[
"## Transposing\nYou can swap columns and indices using the `T` attribute:",
"_____no_output_____"
]
],
[
[
"# d6 = d5.T\n# d6\n\nd6 = d5.T\nd6",
"_____no_output_____"
]
],
[
[
"## Stacking and unstacking levels\nCalling the `stack()` method will push the lowest column level after the lowest index:",
"_____no_output_____"
]
],
[
[
"# d7 = d6.stack()\n# d7\n\nd7 =d6.stack()\nd7",
"_____no_output_____"
]
],
[
[
"Note that many `NaN` values appeared. This makes sense because many new combinations did not exist before (eg. there was no `bob` in `London`).\n\nCalling `unstack()` will do the reverse, once again creating many `NaN` values.",
"_____no_output_____"
]
],
[
[
"# d8 = d7.unstack()\n# d8\n\n# Practice\nd8 = d7.unstack()\nd8",
"_____no_output_____"
]
],
[
[
"If we call `unstack` again, we end up with a `Series` object:",
"_____no_output_____"
]
],
[
[
"# d9 = d8.unstack()\n# d9\n\n# Practice\nd9 = d8.unstack()\nd9",
"_____no_output_____"
]
],
[
[
"The `stack()` and `unstack()` methods let you select the `level` to stack/unstack. You can even stack/unstack multiple levels at once:",
"_____no_output_____"
]
],
[
[
"# d10 = d9.unstack(level = (0,1)) # My Notes: Alice, bob, charles (the names of the person goes to column). (0, 1) means get the (0) London, paris column \n# and the (1) names column transposes to the top.\n# d10\n\n# Practice\nd10 = d5.unstack(level = (0, 1))\nd10",
"_____no_output_____"
],
[
"d11 = d9.unstack(level = (0, 2)) # My Notes: (0, 2) means the (0) column London and Paris and \n# the (2) column birthyear, children and hobby column tranposes to the top\nd11",
"_____no_output_____"
],
[
"d12 = d9.unstack(level = (1, 0)) # My Notes: Moves the (1) names column and (0) city columns is transposed to the top.\nd12",
"_____no_output_____"
],
[
"d13 = d9.unstack(level = (1, 1))\nd13",
"_____no_output_____"
],
[
"d14 = d9.unstack(level = (1, 2))\nd14",
"_____no_output_____"
]
],
[
[
"## Most methods return modified copies\nAs you may have noticed, the `stack()` and `unstack()` methods do not modify the object they apply to. Instead, they work on a copy and return that copy. This is true of most methods in pandas.",
"_____no_output_____"
],
[
"## Accessing rows\nLet's go back to the `people` `DataFrame`:",
"_____no_output_____"
]
],
[
[
"# people\n\n# Practice\npeople",
"_____no_output_____"
]
],
[
[
"The `loc` attribute lets you access rows instead of columns. The result is a `Series` object in which the `DataFrame`'s column names are mapped to row index labels:",
"_____no_output_____"
]
],
[
[
"# people.loc[\"charles\"]\n\n# Practice\npeople.loc[\"charles\"]",
"_____no_output_____"
]
],
[
[
"You can also access rows by integer location using the `iloc` attribute:",
"_____no_output_____"
]
],
[
[
"# people.iloc[2]\n\n# Practice\npeople.iloc[2]",
"_____no_output_____"
]
],
[
[
"You can also get a slice of rows, and this returns a `DataFrame` object:",
"_____no_output_____"
]
],
[
[
"# people.iloc[1:3]\n\n# Practice\npeople.iloc[0:2]",
"_____no_output_____"
]
],
[
[
"Finally, you can pass a boolean array to get the matching rows:",
"_____no_output_____"
]
],
[
[
"people[np.array([True, False, True])]\n\n# My Notes: Only get the first and third indexes\n# Practice\n\npeople[np.array([True, False, True])]",
"_____no_output_____"
]
],
[
[
"This is most useful when combined with boolean expressions:",
"_____no_output_____"
]
],
[
[
"# people[people[\"birthyear\"] < 1990]\n\n# Practice\n# people[people[\"birthyear\"] < 1990]\n# people[people[\"weight\"] > 68]",
"_____no_output_____"
]
],
[
[
"## Adding and removing columns\nYou can generally treat `DataFrame` objects like dictionaries of `Series`, so the following work fine:",
"_____no_output_____"
]
],
[
[
"# people = pd.DataFrame({\n# \"birthyear\": {\"alice\":1985, \"bob\": 1984, \"charles\": 1992},\n# \"hobby\": {\"alice\":\"Biking\", \"bob\": \"Dancing\"},\n# \"weight\": {\"alice\":68, \"bob\": 83, \"charles\": 112},\n# \"children\": {\"bob\": 3, \"charles\": 0}\n# })\n# people\n\n# Practice\npeople = pd.DataFrame({\n \"birthyear\": {\"alice\": 1985, \"bob\": 1984, \"charles\": 1992},\n \"hobby\": {\"alice\": \"Biking\", \"bob\": \"Dancing\"},\n \"weight\": {\"alice\": 68, \"bob\": 83, \"charles\": 112},\n \"children\": {\"alice\": 0, \"bob\": 2, \"charles\": 4}\n})\npeople",
"_____no_output_____"
],
[
"# people\n\npeople",
"_____no_output_____"
],
[
"# people[\"age\"] = 2018 - people[\"birthyear\"] # adds a new column \"age\"\n# people[\"over 30\"] = people[\"age\"] > 30 # adds another column \"over 30\"\n# birthyears = people.pop(\"birthyear\")\n# del people[\"children\"]\n\n# people\n\n# Practice\npeople[\"age\"] = 2020 - people[\"birthyear\"] # adds a new column \"age\"\npeople[\"over 30\"] = people[\"age\"] > 30 # adds another column \"over 30\"\nbirthyears = people.pop(\"birthyear\") # My Notes: Removes the \"birthyear\" column\ndel people[\"children\"]\n\npeople",
"_____no_output_____"
],
[
"# birthyears\nbirthyears",
"_____no_output_____"
]
],
[
[
"When you add a new colum, it must have the same number of rows. Missing rows are filled with NaN, and extra rows are ignored:",
"_____no_output_____"
]
],
[
[
"# people[\"pets\"] = pd.Series({\"bob\": 0, \"charles\": 5, \"eugene\":1}) # alice is missing, eugene is ignored\n# people\n\n# Practice\npeople[\"pets\"] = pd.Series({\"bob\":0, \"charles\": 5, \"eugene\": 1}) # alice is missing, eugene is ignored\npeople",
"_____no_output_____"
]
],
[
[
"When adding a new column, it is added at the end (on the right) by default. You can also insert a column anywhere else using the `insert()` method:",
"_____no_output_____"
]
],
[
[
"people.insert(1, \"height\", [172, 181, 185])\npeople",
"_____no_output_____"
],
[
"# My Notes\npeople.insert(0, \"drinks\", pd.Series({\"alice\": \"pepsi\", \"bob\": \"coke\", \"charles\": \"water\"}))\npeople",
"_____no_output_____"
],
[
"# My Notes\ndel people[\"drinks\"]\npeople",
"_____no_output_____"
]
],
[
[
"## Assigning new columns\nYou can also create new columns by calling the `assign()` method. Note that this returns a new `DataFrame` object, the original is not modified:",
"_____no_output_____"
]
],
[
[
"# people.assign(\n# body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2,\n# has_pets = people[\"pets\"] > 0\n# )\n\n# Practice\npeople.assign(\n body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2,\n has_pets = people[\"pets\"] > 0\n)",
"_____no_output_____"
]
],
[
[
"Note that you cannot access columns created within the same assignment:",
"_____no_output_____"
]
],
[
[
"# try:\n# people.assign(\n# body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2,\n# overweight = people[\"body_mass_index\"] > 25 # My Notes: Just created in the assignment, but cannot be used\n# )\n# except KeyError as e:\n# print(\"Key error:\", e) \n\n# Practice\ntry:\n people.assign(\n body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2,\n overweight = people[\"body_mass_index\"] > 25\n )\n\nexcept KeyError as e:\n print(\"Key error:\", e)",
"Key error: 'body_mass_index'\n"
]
],
[
[
"The solution is to split this assignment in two consecutive assignments:",
"_____no_output_____"
]
],
[
[
"# d6 = people.assign(body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2)\n# d6.assign(overweight = d6[\"body_mass_index\"] > 25)\n\nd6 = people.assign(body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2)\nd6.assign(overweight = d6[\"body_mass_index\"] > 25)",
"_____no_output_____"
]
],
[
[
"Having to create a temporary variable `d6` is not very convenient. You may want to just chain the assigment calls, but it does not work because the `people` object is not actually modified by the first assignment:",
"_____no_output_____"
]
],
[
[
"# try:\n# (people\n# .assign(body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2)\n# .assign(overweight = people[\"body_mass_index\"] > 25)\n# )\n# except KeyError as e:\n# print(\"Key error:\", e)\n\n# Practice\ntry:\n (people\n .assign(body_mass_index = people[\"weight\"] / (people[\"height\"] / 100) ** 2)\n .assign(overweight = people[\"body_mass_index\"] > 25)\n )\n\nexcept KeyError as e:\n print(\"Key error:\", e)",
"Key error: 'body_mass_index'\n"
]
],
[
[
"But fear not, there is a simple solution. You can pass a function to the `assign()` method (typically a `lambda` function), and this function will be called with the `DataFrame` as a parameter:",
"_____no_output_____"
]
],
[
[
"# (people\n# .assign(body_mass_index = lambda df: df[\"weight\"] / (df[\"height\"] / 100) ** 2)\n# .assign(overweight = lambda df: df[\"body_mass_index\"] > 25)\n# )\n\n# Practice\n\n(people\n .assign(body_mass_index = lambda df: df[\"weight\"] / (df[\"height\"] / 100) ** 2)\n .assign(overweight = lambda df: df[\"body_mass_index\"] > 25)\n)",
"_____no_output_____"
]
],
[
[
"Problem solved!",
"_____no_output_____"
],
[
"## Evaluating an expression\nA great feature supported by pandas is expression evaluation. This relies on the `numexpr` library which must be installed.",
"_____no_output_____"
]
],
[
[
"# people.eval(\"weight / (height/100) ** 2 > 25\")\n\n# Practice\npeople.eval(\"weight / (height / 100) ** 2 > 25\")",
"_____no_output_____"
]
],
[
[
"Assignment expressions are also supported. Let's set `inplace=True` to directly modify the `DataFrame` rather than getting a modified copy:",
"_____no_output_____"
]
],
[
[
"# people.eval(\"body_mass_index = weight / (height/100) ** 2\", inplace=True)\n# people\n\n# Practice\npeople.eval(\"body_mass_index = weight / (height / 100) ** 2\", inplace=True)\npeople",
"_____no_output_____"
]
],
[
[
"You can use a local or global variable in an expression by prefixing it with `'@'`:",
"_____no_output_____"
]
],
[
[
"# overweight_threshold = 30\n# people.eval(\"overweight = body_mass_index > @overweight_threshold\", inplace=True)\n# people\n\n# Practice\noverweight_threshold = 30\npeople.eval(\"overweight = body_mass_index > @overweight_threshold\", inplace=True)\npeople",
"_____no_output_____"
]
],
[
[
"## Querying a `DataFrame`\nThe `query()` method lets you filter a `DataFrame` based on a query expression:",
"_____no_output_____"
]
],
[
[
"people",
"_____no_output_____"
],
[
"# people.query(\"age > 30 and pets == 0\")\n\n# Practice\npeople.query(\"age > 30 and pets == 0\")",
"_____no_output_____"
],
[
"# My Notes\nhobby_biking = \"Biking\"\npeople.query(\"hobby == @hobby_biking\")",
"_____no_output_____"
]
],
[
[
"## Sorting a `DataFrame`\nYou can sort a `DataFrame` by calling its `sort_index` method. By default it sorts the rows by their index label, in ascending order, but let's reverse the order:",
"_____no_output_____"
]
],
[
[
"# people.sort_index(ascending=False)\n\n# Practice\npeople.sort_index(ascending=False)",
"_____no_output_____"
]
],
[
[
"Note that `sort_index` returned a sorted *copy* of the `DataFrame`. To modify `people` directly, we can set the `inplace` argument to `True`. Also, we can sort the columns instead of the rows by setting `axis=1`:",
"_____no_output_____"
]
],
[
[
"# people.sort_index(axis=1, inplace=True)\n# people\n\n# Practice\npeople.sort_index(axis=1, inplace=True) # My Notes: Sorts the columns by columns by setting axis = 1, in alphabetical order\npeople",
"_____no_output_____"
]
],
[
[
"To sort the `DataFrame` by the values instead of the labels, we can use `sort_values` and specify the column to sort by:",
"_____no_output_____"
]
],
[
[
"# people.sort_values(by=\"age\", inplace=True)\n# people\n\n# Practice\npeople.sort_values(by=\"age\", inplace=True)\npeople",
"_____no_output_____"
]
],
[
[
"## Plotting a `DataFrame`\nJust like for `Series`, pandas makes it easy to draw nice graphs based on a `DataFrame`.\n\nFor example, it is trivial to create a line plot from a `DataFrame`'s data by calling its `plot` method:",
"_____no_output_____"
]
],
[
[
"# people.plot(kind = \"line\", x = \"body_mass_index\", y = [\"height\", \"weight\"])\n# plt.show()\n\n# Practice\npeople.plot(kind = \"line\", x = \"body_mass_index\", y = [\"height\", \"weight\"])\nplt.show()",
"_____no_output_____"
]
],
[
[
"You can pass extra arguments supported by matplotlib's functions. For example, we can create scatterplot and pass it a list of sizes using the `s` argument of matplotlib's `scatter()` function:",
"_____no_output_____"
]
],
[
[
"# people.plot(kind = \"scatter\", x = \"height\", y = \"weight\", s=[40, 120, 200])\n# plt.show()\n\n# Practice\npeople.plot(kind = \"scatter\", x = \"height\", y = \"weight\", s=[40, 120, 200])\nplt.show()",
"_____no_output_____"
]
],
[
[
"Again, there are way too many options to list here: the best option is to scroll through the [Visualization](http://pandas.pydata.org/pandas-docs/stable/visualization.html) page in pandas' documentation, find the plot you are interested in and look at the example code.",
"_____no_output_____"
],
[
"## Operations on `DataFrame`s\nAlthough `DataFrame`s do not try to mimick NumPy arrays, there are a few similarities. Let's create a `DataFrame` to demonstrate this:",
"_____no_output_____"
]
],
[
[
"# grades_array = np.array([[8,8,9],[10,9,9],[4, 8, 2], [9, 10, 10]])\n# grades = pd.DataFrame(grades_array, columns=[\"sep\", \"oct\", \"nov\"], index=[\"alice\",\"bob\",\"charles\",\"darwin\"])\n# grades\n\n# Practice\ngrades_array = np.array([[8,8,9], [10,9,9], [4,8,2], [9,10,10]])\ngrades = pd.DataFrame(grades_array, columns=[\"sep\", \"oct\", \"nov\"], index=[\"alice\", \"bob\", \"charles\", \"darwin\"])\ngrades",
"_____no_output_____"
]
],
[
[
"You can apply NumPy mathematical functions on a `DataFrame`: the function is applied to all values:",
"_____no_output_____"
]
],
[
[
"# np.sqrt(grades)\n\n# Practice\nnp.sqrt(grades)",
"_____no_output_____"
]
],
[
[
"Similarly, adding a single value to a `DataFrame` will add that value to all elements in the `DataFrame`. This is called *broadcasting*:",
"_____no_output_____"
]
],
[
[
"# grades + 1\n\n# Practice\ngrades + 1",
"_____no_output_____"
]
],
[
[
"Of course, the same is true for all other binary operations, including arithmetic (`*`,`/`,`**`...) and conditional (`>`, `==`...) operations:",
"_____no_output_____"
]
],
[
[
"# grades >= 5\n\n# Practice\ngrades >= 5",
"_____no_output_____"
]
],
[
[
"Aggregation operations, such as computing the `max`, the `sum` or the `mean` of a `DataFrame`, apply to each column, and you get back a `Series` object:",
"_____no_output_____"
]
],
[
[
"# grades.mean()\n\n# Practice\ngrades.mean()",
"_____no_output_____"
]
],
[
[
"The `all` method is also an aggregation operation: it checks whether all values are `True` or not. Let's see during which months all students got a grade greater than `5`:",
"_____no_output_____"
]
],
[
[
"# (grades > 5).all()\n\n# Practice\n(grades > 5).all()",
"_____no_output_____"
]
],
[
[
"Most of these functions take an optional `axis` parameter which lets you specify along which axis of the `DataFrame` you want the operation executed. The default is `axis=0`, meaning that the operation is executed vertically (on each column). You can set `axis=1` to execute the operation horizontally (on each row). For example, let's find out which students had all grades greater than `5`:",
"_____no_output_____"
]
],
[
[
"# (grades > 5).all(axis = 1)\n\n# Practice\n(grades > 5).all(axis = 1)",
"_____no_output_____"
]
],
[
[
"The `any` method returns `True` if any value is True. Let's see who got at least one grade 10:",
"_____no_output_____"
]
],
[
[
"# (grades == 10).any(axis = 1)\n\n# Practice\n(grades == 10).any(axis = 1)",
"_____no_output_____"
]
],
[
[
"If you add a `Series` object to a `DataFrame` (or execute any other binary operation), pandas attempts to broadcast the operation to all *rows* in the `DataFrame`. This only works if the `Series` has the same size as the `DataFrame`s rows. For example, let's substract the `mean` of the `DataFrame` (a `Series` object) from the `DataFrame`:",
"_____no_output_____"
]
],
[
[
"# My Notes\ngrades",
"_____no_output_____"
],
[
"pd.DataFrame(grades.mean(), columns=[\"mean_grades\"]) # My Notes: Convert series object to DataFrame for easier visualization",
"_____no_output_____"
],
[
"# grades - grades.mean() # equivalent to: grades - [7.75, 8.75, 7.50]\n\n# Practice\ngrades - grades.mean() # equivalent to: grades - [7.75, 8.75, 7.50]",
"_____no_output_____"
]
],
[
[
"We substracted `7.75` from all September grades, `8.75` from October grades and `7.50` from November grades. It is equivalent to substracting this `DataFrame`:",
"_____no_output_____"
]
],
[
[
"# pd.DataFrame([[7.75, 8.75, 7.50]]*4, index=grades.index, columns=grades.columns)\n\npd.DataFrame([[7.75, 8.75, 7.50]] * 4, index=grades.index, columns=grades.columns)",
"_____no_output_____"
]
],
[
[
"If you want to substract the global mean from every grade, here is one way to do it:",
"_____no_output_____"
]
],
[
[
"grades.values.mean()",
"_____no_output_____"
],
[
"# grades - grades.values.mean() # substracts the global mean (8.00) from all grades\n\n# Practice\ngrades - grades.values.mean() # substracts the global mean (800) from all grades",
"_____no_output_____"
]
],
[
[
"## Automatic alignment\nSimilar to `Series`, when operating on multiple `DataFrame`s, pandas automatically aligns them by row index label, but also by column names. Let's create a `DataFrame` with bonus points for each person from October to December:",
"_____no_output_____"
]
],
[
[
"# bonus_array = np.array([[0,np.nan,2],[np.nan,1,0],[0, 1, 0], [3, 3, 0]])\n# bonus_points = pd.DataFrame(bonus_array, columns=[\"oct\", \"nov\", \"dec\"], index=[\"bob\",\"colin\", \"darwin\", \"charles\"])\n# bonus_points\n\n# Practice\nbonus_array = np.array([[0, np.nan, 2], [np.nan, 1, 0], [0, 1, 0], [3, 3, 0]])\nbonus_points = pd.DataFrame(bonus_array, columns=[\"oct\", \"nov\", \"dec\"], index=[\"bob\", \"colin\", \"darwin\", \"charles\"])\nbonus_points.sort_index(axis=0, ascending=True)",
"_____no_output_____"
],
[
"# My Notes\ngrades.iloc[1:]",
"_____no_output_____"
],
[
"# grades + bonus_points\n\n# Practice\n# example = (grades + bonus_points)\n# example.sort_index(axis=1, ascending=False)\n(grades + bonus_points).sort_index(axis=1, ascending=False)",
"_____no_output_____"
]
],
[
[
"Looks like the addition worked in some cases but way too many elements are now empty. That's because when aligning the `DataFrame`s, some columns and rows were only present on one side, and thus they were considered missing on the other side (`NaN`). Then adding `NaN` to a number results in `NaN`, hence the result.\n\n## Handling missing data\nDealing with missing data is a frequent task when working with real life data. Pandas offers a few tools to handle missing data.\n \nLet's try to fix the problem above. For example, we can decide that missing data should result in a zero, instead of `NaN`. We can replace all `NaN` values by a any value using the `fillna()` method:",
"_____no_output_____"
]
],
[
[
"# (grades + bonus_points).fillna(0)\n\n# Practice\n(grades + bonus_points).fillna(0)",
"_____no_output_____"
]
],
[
[
"It's a bit unfair that we're setting grades to zero in September, though. Perhaps we should decide that missing grades are missing grades, but missing bonus points should be replaced by zeros:",
"_____no_output_____"
]
],
[
[
"# fixed_bonus_points = bonus_points.fillna(0)\n# fixed_bonus_points.insert(0, \"sep\", 0)\n# fixed_bonus_points.loc[\"alice\"] = 0\n# grades + fixed_bonus_points\n\n# Practice\nfixed_bonus_points = bonus_points.fillna(0) # My Notes: Fills the NaN values with 0's\nfixed_bonus_points.insert(0, \"sep\", 0) # My Notes: insert 0 values in \"sep\" column in position 0\nfixed_bonus_points.loc[\"alice\"] = 0 # My Notes: Set Alice's score to be 0 for all the months\ngrades + fixed_bonus_points",
"_____no_output_____"
]
],
[
[
"That's much better: although we made up some data, we have not been too unfair.\n\nAnother way to handle missing data is to interpolate. Let's look at the `bonus_points` `DataFrame` again:",
"_____no_output_____"
]
],
[
[
"# bonus_points\n\n# Practice\nbonus_points",
"_____no_output_____"
]
],
[
[
"Now let's call the `interpolate` method. By default, it interpolates vertically (`axis=0`), so let's tell it to interpolate horizontally (`axis=1`).",
"_____no_output_____"
]
],
[
[
"# bonus_points.interpolate(axis=1)\n\n# Practice\nbonus_points.interpolate(axis=1)",
"_____no_output_____"
]
],
[
[
"Bob had 0 bonus points in October, and 2 in December. When we interpolate for November, we get the mean: 1 bonus point. Colin had 1 bonus point in November, but we do not know how many bonus points he had in September, so we cannot interpolate, this is why there is still a missing value in October after interpolation. To fix this, we can set the September bonus points to 0 before interpolation.",
"_____no_output_____"
]
],
[
[
"# better_bonus_points = bonus_points.copy()\n# better_bonus_points.insert(0, \"sep\", 0)\n# better_bonus_points.loc[\"alice\"] = 0\n# better_bonus_points = better_bonus_points.interpolate(axis=1)\n# better_bonus_points\n\n# Practice\nbetter_bonus_points = bonus_points.copy()\nbetter_bonus_points.insert(0, \"sep\", 0)\nbetter_bonus_points.loc[\"alice\"] = 0\nbetter_bonus_points = better_bonus_points.interpolate(axis=1)\nbetter_bonus_points",
"_____no_output_____"
]
],
[
[
"Great, now we have reasonable bonus points everywhere. Let's find out the final grades:",
"_____no_output_____"
]
],
[
[
"# grades + better_bonus_points\n\n# Practice\ngrades + better_bonus_points",
"_____no_output_____"
]
],
[
[
"It is slightly annoying that the September column ends up on the right. This is because the `DataFrame`s we are adding do not have the exact same columns (the `grades` `DataFrame` is missing the `\"dec\"` column), so to make things predictable, pandas orders the final columns alphabetically. To fix this, we can simply add the missing column before adding:",
"_____no_output_____"
]
],
[
[
"# grades[\"dec\"] = np.nan\n# final_grades = grades + better_bonus_points\n# final_grades\n\n# Practice\ngrades[\"dec\"] = np.nan\nfinal_grades = grades + better_bonus_points\nfinal_grades",
"_____no_output_____"
]
],
[
[
"There's not much we can do about December and Colin: it's bad enough that we are making up bonus points, but we can't reasonably make up grades (well I guess some teachers probably do). So let's call the `dropna()` method to get rid of rows that are full of `NaN`s:",
"_____no_output_____"
]
],
[
[
"# final_grades_clean = final_grades.dropna(how=\"all\")\n# final_grades_clean\n\n# Practice\nfinal_grades_clean = final_grades.dropna(how=\"all\")\nfinal_grades_clean",
"_____no_output_____"
]
],
[
[
"Now let's remove columns that are full of `NaN`s by setting the `axis` argument to `1`:",
"_____no_output_____"
]
],
[
[
"# final_grades_clean = final_grades_clean.dropna(axis=1, how=\"all\")\n# final_grades_clean\n\n# Practice\nfinal_grades_clean = final_grades_clean.dropna(axis=1, how=\"all\")\nfinal_grades_clean",
"_____no_output_____"
]
],
[
[
"## Aggregating with `groupby`\nSimilar to the SQL language, pandas allows grouping your data into groups to run calculations over each group.\n\nFirst, let's add some extra data about each person so we can group them, and let's go back to the `final_grades` `DataFrame` so we can see how `NaN` values are handled:",
"_____no_output_____"
]
],
[
[
"# final_grades[\"hobby\"] = [\"Biking\", \"Dancing\", np.nan, \"Dancing\", \"Biking\"]\n# final_grades\n\n# Practice\nfinal_grades[\"hobby\"] = [\"Biking\", \"Dancing\", np.nan, \"Dancing\", \"Biking\"]\nfinal_grades",
"_____no_output_____"
]
],
[
[
"Now let's group data in this `DataFrame` by hobby:",
"_____no_output_____"
]
],
[
[
"# grouped_grades = final_grades.groupby(\"hobby\")\n# grouped_grades\n\n# Practice\ngrouped_grades = final_grades.groupby(\"hobby\")\ngrouped_grades",
"_____no_output_____"
]
],
[
[
"We are ready to compute the average grade per hobby:",
"_____no_output_____"
]
],
[
[
"# grouped_grades.mean()\n\n# Practice\ngrouped_grades.mean()",
"_____no_output_____"
]
],
[
[
"That was easy! Note that the `NaN` values have simply been skipped when computing the means.",
"_____no_output_____"
],
[
"## Pivot tables\nPandas supports spreadsheet-like [pivot tables](https://en.wikipedia.org/wiki/Pivot_table) that allow quick data summarization. To illustrate this, let's create a simple `DataFrame`:",
"_____no_output_____"
]
],
[
[
"# bonus_points\n\n# Practice\nbonus_points",
"_____no_output_____"
],
[
"# more_grades = final_grades_clean.stack().reset_index()\n# more_grades.columns = [\"name\", \"month\", \"grade\"]\n# more_grades[\"bonus\"] = [np.nan, np.nan, np.nan, 0, np.nan, 2, 3, 3, 0, 0, 1, 0]\n# more_grades\n\n# Practice\nmore_grades = final_grades_clean.stack().reset_index()\nmore_grades.columns = [\"name\", \"month\", \"grade\"]\nmore_grades[\"bonus\"] = [np.nan, np.nan, np.nan, 0, np.nan, 2, 3, 3, 0, 0, 1, 0]\nmore_grades",
"_____no_output_____"
]
],
[
[
"Now we can call the `pd.pivot_table()` function for this `DataFrame`, asking to group by the `name` column. By default, `pivot_table()` computes the mean of each numeric column:",
"_____no_output_____"
]
],
[
[
"# pd.pivot_table(more_grades, index=\"name\")\n\n# Practice\npd.pivot_table(more_grades, index=\"name\")",
"_____no_output_____"
]
],
[
[
"We can change the aggregation function by setting the `aggfunc` argument, and we can also specify the list of columns whose values will be aggregated:",
"_____no_output_____"
]
],
[
[
"# pd.pivot_table(more_grades, index=\"name\", values=[\"grade\",\"bonus\"], aggfunc=np.max)\n\n# Practice\npd.pivot_table(more_grades, index=\"name\", values=[\"grade\", \"bonus\"], aggfunc = np.max)",
"_____no_output_____"
]
],
[
[
"We can also specify the `columns` to aggregate over horizontally, and request the grand totals for each row and column by setting `margins=True`:",
"_____no_output_____"
]
],
[
[
"# pd.pivot_table(more_grades, index=\"name\", values=\"grade\", columns=\"month\", margins=True)\n\n# Practice\npd.pivot_table(more_grades, index=\"name\", values=\"grade\", columns=\"month\", margins=True)",
"_____no_output_____"
]
],
[
[
"Finally, we can specify multiple index or column names, and pandas will create multi-level indices:",
"_____no_output_____"
]
],
[
[
"# pd.pivot_table(more_grades, index=(\"name\", \"month\"), margins=True)\n\n# Practice\npd.pivot_table(more_grades, index=(\"name\", \"month\"), margins=True)",
"_____no_output_____"
],
[
"# My Notes\n# Note that NaN values are not included\npd.pivot_table(more_grades, index=(\"name\", \"month\", \"bonus\"), margins=True)",
"_____no_output_____"
]
],
[
[
"## Overview functions\nWhen dealing with large `DataFrames`, it is useful to get a quick overview of its content. Pandas offers a few functions for this. First, let's create a large `DataFrame` with a mix of numeric values, missing values and text values. Notice how Jupyter displays only the corners of the `DataFrame`:",
"_____no_output_____"
]
],
[
[
"# much_data = np.fromfunction(lambda x,y: (x+y*y)%17*11, (10000, 26))\n# large_df = pd.DataFrame(much_data, columns=list(\"ABCDEFGHIJKLMNOPQRSTUVWXYZ\"))\n# large_df[large_df % 16 == 0] = np.nan\n# large_df.insert(3,\"some_text\", \"Blabla\")\n# large_df\n\n# Practice\nmuch_data = np.fromfunction(lambda x,y: (x + y * y) % 17 * 11, (10000, 26))\nlarge_df = pd.DataFrame(much_data, columns=list(\"ABCDEFGHIJKLMNOPQRSTUVWXYZ\"))\nlarge_df[large_df % 16 == 0] = np.nan\nlarge_df.insert(3, \"some_text\", \"Blabla\") # My Notes: Insert column \"some_text\" in column 3 with text \"Blabla\"\nlarge_df",
"_____no_output_____"
],
[
"# My Notes\na = np.arange(0, 101)\na",
"_____no_output_____"
],
[
"# My Notes\na % 16 == 0",
"_____no_output_____"
]
],
[
[
"The `head()` method returns the top 5 rows:",
"_____no_output_____"
]
],
[
[
"# large_df.head()\n\n# Practice\nlarge_df.head()",
"_____no_output_____"
]
],
[
[
"Of course there's also a `tail()` function to view the bottom 5 rows. You can pass the number of rows you want:",
"_____no_output_____"
]
],
[
[
"# large_df.tail(n=2)\n\n# Practice\nlarge_df.tail(n=2)",
"_____no_output_____"
]
],
[
[
"The `info()` method prints out a summary of each columns contents:",
"_____no_output_____"
]
],
[
[
"# large_df.info()\n\n# Practice\nlarge_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 10000 entries, 0 to 9999\nData columns (total 27 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 A 8823 non-null float64\n 1 B 8824 non-null float64\n 2 C 8824 non-null float64\n 3 some_text 10000 non-null object \n 4 D 8824 non-null float64\n 5 E 8822 non-null float64\n 6 F 8824 non-null float64\n 7 G 8824 non-null float64\n 8 H 8822 non-null float64\n 9 I 8823 non-null float64\n 10 J 8823 non-null float64\n 11 K 8822 non-null float64\n 12 L 8824 non-null float64\n 13 M 8824 non-null float64\n 14 N 8822 non-null float64\n 15 O 8824 non-null float64\n 16 P 8824 non-null float64\n 17 Q 8824 non-null float64\n 18 R 8823 non-null float64\n 19 S 8824 non-null float64\n 20 T 8824 non-null float64\n 21 U 8824 non-null float64\n 22 V 8822 non-null float64\n 23 W 8824 non-null float64\n 24 X 8824 non-null float64\n 25 Y 8822 non-null float64\n 26 Z 8823 non-null float64\ndtypes: float64(26), object(1)\nmemory usage: 2.1+ MB\n"
]
],
[
[
"Finally, the `describe()` method gives a nice overview of the main aggregated values over each column:\n* `count`: number of non-null (not NaN) values\n* `mean`: mean of non-null values\n* `std`: [standard deviation](https://en.wikipedia.org/wiki/Standard_deviation) of non-null values\n* `min`: minimum of non-null values\n* `25%`, `50%`, `75%`: 25th, 50th and 75th [percentile](https://en.wikipedia.org/wiki/Percentile) of non-null values\n* `max`: maximum of non-null values",
"_____no_output_____"
]
],
[
[
"# large_df.describe()\n\nlarge_df.describe()",
"_____no_output_____"
]
],
[
[
"# Saving & loading\nPandas can save `DataFrame`s to various backends, including file formats such as CSV, Excel, JSON, HTML and HDF5, or to a SQL database. Let's create a `DataFrame` to demonstrate this:",
"_____no_output_____"
]
],
[
[
"# my_df = pd.DataFrame(\n# [[\"Biking\", 68.5, 1985, np.nan], [\"Dancing\", 83.1, 1984, 3]], \n# columns=[\"hobby\",\"weight\",\"birthyear\",\"children\"],\n# index=[\"alice\", \"bob\"]\n# )\n# my_df\n\n# Practice\nmy_df = pd.DataFrame(\n [[\"Biking\", 68.5, 1985, np.nan], [\"Dancing\", 83.1, 1984, 3]],\n columns = [\"hobby\", \"weight\", \"birthyear\", \"children\"],\n index = [\"alice\", \"bob\"]\n)\nmy_df",
"_____no_output_____"
]
],
[
[
"## Saving\nLet's save it to CSV, HTML and JSON:",
"_____no_output_____"
]
],
[
[
"# my_df.to_csv(\"my_df.csv\")\n# my_df.to_html(\"my_df.html\")\n# my_df.to_json(\"my_df.json\")\n\n# Practice\nmy_df.to_csv(\"my_df.csv\")\nmy_df.to_html(\"my_df.html\")\nmy_df.to_json(\"my_df.json\")",
"_____no_output_____"
]
],
[
[
"Done! Let's take a peek at what was saved:",
"_____no_output_____"
]
],
[
[
"# for filename in (\"my_df.csv\", \"my_df.html\", \"my_df.json\"):\n# print(\"#\", filename)\n# with open(filename, \"rt\") as f:\n# print(f.read())\n# print()\n\n# Practice\nfor filename in (\"my_df.csv\", \"my_df.html\", \"my_df.json\"):\n print(\"#\", filename)\n with open(filename, \"rt\") as f:\n print(f.read())\n print()\n",
"# my_df.csv\n,hobby,weight,birthyear,children\nalice,Biking,68.5,1985,\nbob,Dancing,83.1,1984,3.0\n\n\n# my_df.html\n<table border=\"1\" class=\"dataframe\">\n <thead>\n <tr style=\"text-align: right;\">\n <th></th>\n <th>hobby</th>\n <th>weight</th>\n <th>birthyear</th>\n <th>children</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <th>alice</th>\n <td>Biking</td>\n <td>68.5</td>\n <td>1985</td>\n <td>NaN</td>\n </tr>\n <tr>\n <th>bob</th>\n <td>Dancing</td>\n <td>83.1</td>\n <td>1984</td>\n <td>3.0</td>\n </tr>\n </tbody>\n</table>\n\n# my_df.json\n{\"hobby\":{\"alice\":\"Biking\",\"bob\":\"Dancing\"},\"weight\":{\"alice\":68.5,\"bob\":83.1},\"birthyear\":{\"alice\":1985,\"bob\":1984},\"children\":{\"alice\":null,\"bob\":3.0}}\n\n"
]
],
[
[
"Note that the index is saved as the first column (with no name) in a CSV file, as `<th>` tags in HTML and as keys in JSON.\n\nSaving to other formats works very similarly, but some formats require extra libraries to be installed. For example, saving to Excel requires the openpyxl library:",
"_____no_output_____"
]
],
[
[
"# try:\n# my_df.to_excel(\"my_df.xlsx\", sheet_name='People')\n# except ImportError as e:\n# print(e)\n\n# Practice\ntry:\n my_df.to_excel(\"my_df.xlsx\", sheet_name='People')\nexcept ImportError as e:\n print(e)",
"No module named 'openpyxl'\n"
]
],
[
[
"## Loading\nNow let's load our CSV file back into a `DataFrame`:",
"_____no_output_____"
]
],
[
[
"# my_df_loaded = pd.read_csv(\"my_df.csv\", index_col=0)\n# my_df_loaded\n\n# Practice\nmy_df_loaded = pd.read_csv(\"my_df.csv\", index_col=0)\nmy_df_loaded",
"_____no_output_____"
]
],
[
[
"As you might guess, there are similar `read_json`, `read_html`, `read_excel` functions as well. We can also read data straight from the Internet. For example, let's load all U.S. cities from [simplemaps.com](http://simplemaps.com/):",
"_____no_output_____"
]
],
[
[
"# My Notes\nimport os\npath = os.path.join(\".\" + \"\\datasets\\simplemaps\\worldcities.csv\")\npath",
"_____no_output_____"
],
[
"# us_cities = None\n# try:\n# csv_url = \"http://simplemaps.com/files/cities.csv\"\n# us_cities = pd.read_csv(csv_url, index_col=0)\n# us_cities = us_cities.head()\n# except IOError as e:\n# print(e)\n# us_cities\n\n# Practice\n\nus_cities = None\ntry:\n csv_url = path\n us_cities = pd.read_csv(csv_url, index_col=0)\n us_cities = us_cities.head()\nexcept IOError as e:\n print(e)\nus_cities",
"_____no_output_____"
]
],
[
[
"There are more options available, in particular regarding datetime format. Check out the [documentation](http://pandas.pydata.org/pandas-docs/stable/io.html) for more details.",
"_____no_output_____"
],
[
"# Combining `DataFrame`s\n\n## SQL-like joins\nOne powerful feature of pandas is it's ability to perform SQL-like joins on `DataFrame`s. Various types of joins are supported: inner joins, left/right outer joins and full joins. To illustrate this, let's start by creating a couple simple `DataFrame`s:",
"_____no_output_____"
]
],
[
[
"# city_loc = pd.DataFrame(\n# [\n# [\"CA\", \"San Francisco\", 37.781334, -122.416728],\n# [\"NY\", \"New York\", 40.705649, -74.008344],\n# [\"FL\", \"Miami\", 25.791100, -80.320733],\n# [\"OH\", \"Cleveland\", 41.473508, -81.739791],\n# [\"UT\", \"Salt Lake City\", 40.755851, -111.896657]\n# ], columns=[\"state\", \"city\", \"lat\", \"lng\"])\n# city_loc\n\n# Practice\ncity_loc = pd.DataFrame(\n [\n [\"CA\", \"San Francisco\", 37.781334, -122.416728],\n [\"NY\", \"New York\", 40.705649, -74.008344],\n [\"FL\", \"Miami\", 25.791100, -80.320733],\n [\"OH\", \"Cleveland\", 41.473508, -81.739791],\n [\"UT\", \"Salt Lake City\", 40.755851, -111.896657]\n ], columns = [\"state\", \"city\", \"lat\", \"lng\"]\n)\n\ncity_loc",
"_____no_output_____"
],
[
"# city_pop = pd.DataFrame(\n# [\n# [808976, \"San Francisco\", \"California\"],\n# [8363710, \"New York\", \"New-York\"],\n# [413201, \"Miami\", \"Florida\"],\n# [2242193, \"Houston\", \"Texas\"]\n# ], index=[3,4,5,6], columns=[\"population\", \"city\", \"state\"])\n# city_pop\n\n# Practice\ncity_pop = pd.DataFrame(\n [\n [808976, \"San Francisco\", \"California\"],\n [8363710, \"New York\", \"New-York\"],\n [413201, \"Miami\", \"Florida\"],\n [2242193, \"Houston\", \"Texas\"]\n ], index = [3,4,5,6], columns=[\"population\", \"city\", \"state\"])\n\ncity_pop",
"_____no_output_____"
]
],
[
[
"Now let's join these `DataFrame`s using the `merge()` function:",
"_____no_output_____"
]
],
[
[
"# pd.merge(left=city_loc, right=city_pop, on=\"city\")\n\n# Practice\npd.merge(left=city_loc, right=city_pop, on=\"city\")",
"_____no_output_____"
]
],
[
[
"Note that both `DataFrame`s have a column named `state`, so in the result they got renamed to `state_x` and `state_y`.\n\nAlso, note that Cleveland, Salt Lake City and Houston were dropped because they don't exist in *both* `DataFrame`s. This is the equivalent of a SQL `INNER JOIN`. If you want a `FULL OUTER JOIN`, where no city gets dropped and `NaN` values are added, you must specify `how=\"outer\"`:",
"_____no_output_____"
]
],
[
[
"# all_cities = pd.merge(left=city_loc, right=city_pop, on=\"city\", how=\"outer\")\n# all_cities\n\n# Practice\nall_cities = pd.merge(left=city_loc, right=city_pop, on=\"city\", how=\"outer\")\nall_cities",
"_____no_output_____"
]
],
[
[
"Of course `LEFT OUTER JOIN` is also available by setting `how=\"left\"`: only the cities present in the left `DataFrame` end up in the result. Similarly, with `how=\"right\"` only cities in the right `DataFrame` appear in the result. For example:",
"_____no_output_____"
]
],
[
[
"# My Notes\npd.merge(left=city_loc, right=city_pop, on=\"city\", how=\"left\")",
"_____no_output_____"
],
[
"# pd.merge(left=city_loc, right=city_pop, on=\"city\", how=\"right\")\n\n# Practice\npd.merge(left=city_loc, right=city_pop, on=\"city\", how=\"right\")",
"_____no_output_____"
]
],
[
[
"If the key to join on is actually in one (or both) `DataFrame`'s index, you must use `left_index=True` and/or `right_index=True`. If the key column names differ, you must use `left_on` and `right_on`. For example:",
"_____no_output_____"
]
],
[
[
"# city_pop2 = city_pop.copy()\n# city_pop2.columns = [\"population\", \"name\", \"state\"]\n# pd.merge(left=city_loc, right=city_pop2, left_on=\"city\", right_on=\"name\")\n\n# Practice\ncity_pop2 = city_pop.copy()\ncity_pop2.columns = [\"population\", \"name\", \"state\"]\n# city_pop2\npd.merge(left=city_loc, right=city_pop2, left_on=\"city\", right_on=\"name\")",
"_____no_output_____"
]
],
[
[
"# Stopped here 21/5/2020 4:37PM",
"_____no_output_____"
],
[
"## Concatenation\nRather than joining `DataFrame`s, we may just want to concatenate them. That's what `concat()` is for:",
"_____no_output_____"
]
],
[
[
"city_loc",
"_____no_output_____"
],
[
"city_pop",
"_____no_output_____"
],
[
"# result_concat = pd.concat([city_loc, city_pop])\n# result_concat\n\n# Practice\nresult_concat = pd.concat([city_loc, city_pop])\nresult_concat\n\n# My Notes\n# Same info on the rows stack on top of one another",
"_____no_output_____"
]
],
[
[
"Note that this operation aligned the data horizontally (by columns) but not vertically (by rows). In this example, we end up with multiple rows having the same index (eg. 3). Pandas handles this rather gracefully:",
"_____no_output_____"
]
],
[
[
"# result_concat.loc[3]\n\n# My Notes: Notice in the index there are 2 3's and 4's.\n\n# Practice\nresult_concat.loc[3]",
"_____no_output_____"
]
],
[
[
"Or you can tell pandas to just ignore the index:",
"_____no_output_____"
]
],
[
[
"# pd.concat([city_loc, city_pop], ignore_index=True)\n\n# My Notes\n# \n\n# Practice\npd.concat([city_loc, city_pop], ignore_index=True)",
"_____no_output_____"
]
],
[
[
"Notice that when a column does not exist in a `DataFrame`, it acts as if it was filled with `NaN` values. If we set `join=\"inner\"`, then only columns that exist in *both* `DataFrame`s are returned:",
"_____no_output_____"
]
],
[
[
"# pd.concat([city_loc, city_pop], join=\"inner\")\n\n# My Notes\n# Similar columns of both data frames are only joined together.\n\n# Practice\npd.concat([city_loc, city_pop], join=\"inner\")",
"_____no_output_____"
]
],
[
[
"You can concatenate `DataFrame`s horizontally instead of vertically by setting `axis=1`:",
"_____no_output_____"
]
],
[
[
"# pd.concat([city_loc, city_pop], axis=1)\n\n# My Notes: Adding columns horizontally, notice the city and state columns\n\n# Practice\npd.concat([city_loc, city_pop], axis=1)",
"_____no_output_____"
],
[
"pd.concat([city_loc, city_pop], axis=0)\n\n# My Notes: Adding the dataframe vertically, aka appending the rows at the bottom\n",
"_____no_output_____"
]
],
[
[
"In this case it really does not make much sense because the indices do not align well (eg. Cleveland and San Francisco end up on the same row, because they shared the index label `3`). So let's reindex the `DataFrame`s by city name before concatenating:",
"_____no_output_____"
],
[
"My Notes: Your index is your first column without a name",
"_____no_output_____"
]
],
[
[
"pd.concat([city_loc.set_index(\"city\"), city_pop.set_index(\"city\")], axis=1)\n\n# Practice\n\npd.concat([city_loc.set_index(\"city\"), city_pop.set_index(\"city\")], axis=1)",
"_____no_output_____"
]
],
[
[
"This looks a lot like a `FULL OUTER JOIN`, except that the `state` columns were not renamed to `state_x` and `state_y`, and the `city` column is now the index.",
"_____no_output_____"
],
[
"The `append()` method is a useful shorthand for concatenating `DataFrame`s vertically:",
"_____no_output_____"
]
],
[
[
"# city_loc.append(city_pop)\ncity_loc.append(city_pop)",
"_____no_output_____"
]
],
[
[
"As always in pandas, the `append()` method does *not* actually modify `city_loc`: it works on a copy and returns the modified copy.",
"_____no_output_____"
],
[
"# Categories\nIt is quite frequent to have values that represent categories, for example `1` for female and `2` for male, or `\"A\"` for Good, `\"B\"` for Average, `\"C\"` for Bad. These categorical values can be hard to read and cumbersome to handle, but fortunately pandas makes it easy. To illustrate this, let's take the `city_pop` `DataFrame` we created earlier, and add a column that represents a category:",
"_____no_output_____"
]
],
[
[
"# city_eco = city_pop.copy()\n# city_eco[\"eco_code\"] = [17, 17, 34, 20]\n# city_eco\n\n# Practice\ncity_eco = city_pop.copy()\ncity_eco[\"eco_code\"] = [17, 17, 34, 20]\ncity_eco",
"_____no_output_____"
]
],
[
[
"Right now the `eco_code` column is full of apparently meaningless codes. Let's fix that. First, we will create a new categorical column based on the `eco_code`s:",
"_____no_output_____"
]
],
[
[
"# city_eco[\"economy\"] = city_eco[\"eco_code\"].astype('category')\n# city_eco[\"economy\"].cat.categories\n\n# Practice\ncity_eco[\"economy\"] = city_eco[\"eco_code\"].astype('category')\ncity_eco[\"economy\"].cat.categories",
"_____no_output_____"
]
],
[
[
"Now we can give each category a meaningful name:",
"_____no_output_____"
]
],
[
[
"# city_eco[\"economy\"].cat.categories = [\"Finance\", \"Energy\", \"Tourism\"]\n# city_eco\n\n# Practice\ncity_eco[\"economy\"].cat.categories = [\"Finance\", \"Energy\", \"Tourism\"]\ncity_eco",
"_____no_output_____"
]
],
[
[
"Note that categorical values are sorted according to their categorical order, *not* their alphabetical order:",
"_____no_output_____"
]
],
[
[
"# city_eco.sort_values(by=\"economy\", ascending=False)\n\n# Practice\ncity_eco.sort_values(by=\"economy\", ascending=False)",
"_____no_output_____"
]
],
[
[
"# What next?\nAs you probably noticed by now, pandas is quite a large library with *many* features. Although we went through the most important features, there is still a lot to discover. Probably the best way to learn more is to get your hands dirty with some real-life data. It is also a good idea to go through pandas' excellent [documentation](http://pandas.pydata.org/pandas-docs/stable/index.html), in particular the [Cookbook](http://pandas.pydata.org/pandas-docs/stable/cookbook.html).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a8b551d5d435e9001124da5cdffb0126f96ed0d
| 57,346 |
ipynb
|
Jupyter Notebook
|
C1_Neural Networks and Deep Learning/W4/Building your Deep Neural Network - Step by Step/Building_your_Deep_Neural_Network_Step_by_Step_v8a.ipynb
|
aurimas13/Deep-Learning-Specialization-solutions---notes
|
f2ca6b19b7a568de1e0e12c6341c0605e007e186
|
[
"MIT"
] | null | null | null |
C1_Neural Networks and Deep Learning/W4/Building your Deep Neural Network - Step by Step/Building_your_Deep_Neural_Network_Step_by_Step_v8a.ipynb
|
aurimas13/Deep-Learning-Specialization-solutions---notes
|
f2ca6b19b7a568de1e0e12c6341c0605e007e186
|
[
"MIT"
] | null | null | null |
C1_Neural Networks and Deep Learning/W4/Building your Deep Neural Network - Step by Step/Building_your_Deep_Neural_Network_Step_by_Step_v8a.ipynb
|
aurimas13/Deep-Learning-Specialization-solutions---notes
|
f2ca6b19b7a568de1e0e12c6341c0605e007e186
|
[
"MIT"
] | null | null | null | 37.752469 | 562 | 0.515799 |
[
[
[
"# Building your Deep Neural Network: Step by Step\n\nWelcome to your week 4 assignment (part 1 of 2)! You have previously trained a 2-layer Neural Network (with a single hidden layer). This week, you will build a deep neural network, with as many layers as you want!\n\n- In this notebook, you will implement all the functions required to build a deep neural network.\n- In the next assignment, you will use these functions to build a deep neural network for image classification.\n\n**After this assignment you will be able to:**\n- Use non-linear units like ReLU to improve your model\n- Build a deeper neural network (with more than 1 hidden layer)\n- Implement an easy-to-use neural network class\n\n**Notation**:\n- Superscript $[l]$ denotes a quantity associated with the $l^{th}$ layer. \n - Example: $a^{[L]}$ is the $L^{th}$ layer activation. $W^{[L]}$ and $b^{[L]}$ are the $L^{th}$ layer parameters.\n- Superscript $(i)$ denotes a quantity associated with the $i^{th}$ example. \n - Example: $x^{(i)}$ is the $i^{th}$ training example.\n- Lowerscript $i$ denotes the $i^{th}$ entry of a vector.\n - Example: $a^{[l]}_i$ denotes the $i^{th}$ entry of the $l^{th}$ layer's activations).\n\nLet's get started!",
"_____no_output_____"
],
[
"### <font color='darkblue'> Updates to Assignment <font>\n\n#### If you were working on a previous version\n* The current notebook filename is version \"4a\". \n* You can find your work in the file directory as version \"4\".\n* To see the file directory, click on the Coursera logo at the top left of the notebook.\n\n#### List of Updates\n* compute_cost unit test now includes tests for Y = 0 as well as Y = 1. This catches a possible bug before students get graded.\n* linear_backward unit test now has a more complete unit test that catches a possible bug before students get graded.\n",
"_____no_output_____"
],
[
"## 1 - Packages\n\nLet's first import all the packages that you will need during this assignment. \n- [numpy](www.numpy.org) is the main package for scientific computing with Python.\n- [matplotlib](http://matplotlib.org) is a library to plot graphs in Python.\n- dnn_utils provides some necessary functions for this notebook.\n- testCases provides some test cases to assess the correctness of your functions\n- np.random.seed(1) is used to keep all the random function calls consistent. It will help us grade your work. Please don't change the seed. ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport h5py\nimport matplotlib.pyplot as plt\nfrom testCases_v4a import *\nfrom dnn_utils_v2 import sigmoid, sigmoid_backward, relu, relu_backward\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (5.0, 4.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n%load_ext autoreload\n%autoreload 2\n\nnp.random.seed(1)",
"_____no_output_____"
]
],
[
[
"## 2 - Outline of the Assignment\n\nTo build your neural network, you will be implementing several \"helper functions\". These helper functions will be used in the next assignment to build a two-layer neural network and an L-layer neural network. Each small helper function you will implement will have detailed instructions that will walk you through the necessary steps. Here is an outline of this assignment, you will:\n\n- Initialize the parameters for a two-layer network and for an $L$-layer neural network.\n- Implement the forward propagation module (shown in purple in the figure below).\n - Complete the LINEAR part of a layer's forward propagation step (resulting in $Z^{[l]}$).\n - We give you the ACTIVATION function (relu/sigmoid).\n - Combine the previous two steps into a new [LINEAR->ACTIVATION] forward function.\n - Stack the [LINEAR->RELU] forward function L-1 time (for layers 1 through L-1) and add a [LINEAR->SIGMOID] at the end (for the final layer $L$). This gives you a new L_model_forward function.\n- Compute the loss.\n- Implement the backward propagation module (denoted in red in the figure below).\n - Complete the LINEAR part of a layer's backward propagation step.\n - We give you the gradient of the ACTIVATE function (relu_backward/sigmoid_backward) \n - Combine the previous two steps into a new [LINEAR->ACTIVATION] backward function.\n - Stack [LINEAR->RELU] backward L-1 times and add [LINEAR->SIGMOID] backward in a new L_model_backward function\n- Finally update the parameters.\n\n<img src=\"images/final outline.png\" style=\"width:800px;height:500px;\">\n<caption><center> **Figure 1**</center></caption><br>\n\n\n**Note** that for every forward function, there is a corresponding backward function. That is why at every step of your forward module you will be storing some values in a cache. The cached values are useful for computing gradients. In the backpropagation module you will then use the cache to calculate the gradients. This assignment will show you exactly how to carry out each of these steps. ",
"_____no_output_____"
],
[
"## 3 - Initialization\n\nYou will write two helper functions that will initialize the parameters for your model. The first function will be used to initialize parameters for a two layer model. The second one will generalize this initialization process to $L$ layers.\n\n### 3.1 - 2-layer Neural Network\n\n**Exercise**: Create and initialize the parameters of the 2-layer neural network.\n\n**Instructions**:\n- The model's structure is: *LINEAR -> RELU -> LINEAR -> SIGMOID*. \n- Use random initialization for the weight matrices. Use `np.random.randn(shape)*0.01` with the correct shape.\n- Use zero initialization for the biases. Use `np.zeros(shape)`.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_parameters\n\ndef initialize_parameters(n_x, n_h, n_y):\n \"\"\"\n Argument:\n n_x -- size of the input layer\n n_h -- size of the hidden layer\n n_y -- size of the output layer\n \n Returns:\n parameters -- python dictionary containing your parameters:\n W1 -- weight matrix of shape (n_h, n_x)\n b1 -- bias vector of shape (n_h, 1)\n W2 -- weight matrix of shape (n_y, n_h)\n b2 -- bias vector of shape (n_y, 1)\n \"\"\"\n \n np.random.seed(1)\n \n ### START CODE HERE ### (≈ 4 lines of code)\n W1 = np.random.randn(n_h, n_x) * 0.01 # None\n b1 = np.zeros((n_h, 1)) # None\n W2 = np.random.randn(n_y, n_h) * 0.01 # None\n b2 = np.zeros((n_y, 1)) # None\n ### END CODE HERE ###\n \n assert(W1.shape == (n_h, n_x))\n assert(b1.shape == (n_h, 1))\n assert(W2.shape == (n_y, n_h))\n assert(b2.shape == (n_y, 1))\n \n parameters = {\"W1\": W1,\n \"b1\": b1,\n \"W2\": W2,\n \"b2\": b2}\n \n return parameters ",
"_____no_output_____"
],
[
"parameters = initialize_parameters(3,2,1)\nprint(\"W1 = \" + str(parameters[\"W1\"]))\nprint(\"b1 = \" + str(parameters[\"b1\"]))\nprint(\"W2 = \" + str(parameters[\"W2\"]))\nprint(\"b2 = \" + str(parameters[\"b2\"]))",
"W1 = [[ 0.01624345 -0.00611756 -0.00528172]\n [-0.01072969 0.00865408 -0.02301539]]\nb1 = [[ 0.]\n [ 0.]]\nW2 = [[ 0.01744812 -0.00761207]]\nb2 = [[ 0.]]\n"
]
],
[
[
"**Expected output**:\n \n<table style=\"width:80%\">\n <tr>\n <td> **W1** </td>\n <td> [[ 0.01624345 -0.00611756 -0.00528172]\n [-0.01072969 0.00865408 -0.02301539]] </td> \n </tr>\n\n <tr>\n <td> **b1**</td>\n <td>[[ 0.]\n [ 0.]]</td> \n </tr>\n \n <tr>\n <td>**W2**</td>\n <td> [[ 0.01744812 -0.00761207]]</td>\n </tr>\n \n <tr>\n <td> **b2** </td>\n <td> [[ 0.]] </td> \n </tr>\n \n</table>",
"_____no_output_____"
],
[
"### 3.2 - L-layer Neural Network\n\nThe initialization for a deeper L-layer neural network is more complicated because there are many more weight matrices and bias vectors. When completing the `initialize_parameters_deep`, you should make sure that your dimensions match between each layer. Recall that $n^{[l]}$ is the number of units in layer $l$. Thus for example if the size of our input $X$ is $(12288, 209)$ (with $m=209$ examples) then:\n\n<table style=\"width:100%\">\n\n\n <tr>\n <td> </td> \n <td> **Shape of W** </td> \n <td> **Shape of b** </td> \n <td> **Activation** </td>\n <td> **Shape of Activation** </td> \n <tr>\n \n <tr>\n <td> **Layer 1** </td> \n <td> $(n^{[1]},12288)$ </td> \n <td> $(n^{[1]},1)$ </td> \n <td> $Z^{[1]} = W^{[1]} X + b^{[1]} $ </td> \n \n <td> $(n^{[1]},209)$ </td> \n <tr>\n \n <tr>\n <td> **Layer 2** </td> \n <td> $(n^{[2]}, n^{[1]})$ </td> \n <td> $(n^{[2]},1)$ </td> \n <td>$Z^{[2]} = W^{[2]} A^{[1]} + b^{[2]}$ </td> \n <td> $(n^{[2]}, 209)$ </td> \n <tr>\n \n <tr>\n <td> $\\vdots$ </td> \n <td> $\\vdots$ </td> \n <td> $\\vdots$ </td> \n <td> $\\vdots$</td> \n <td> $\\vdots$ </td> \n <tr>\n \n <tr>\n <td> **Layer L-1** </td> \n <td> $(n^{[L-1]}, n^{[L-2]})$ </td> \n <td> $(n^{[L-1]}, 1)$ </td> \n <td>$Z^{[L-1]} = W^{[L-1]} A^{[L-2]} + b^{[L-1]}$ </td> \n <td> $(n^{[L-1]}, 209)$ </td> \n <tr>\n \n \n <tr>\n <td> **Layer L** </td> \n <td> $(n^{[L]}, n^{[L-1]})$ </td> \n <td> $(n^{[L]}, 1)$ </td>\n <td> $Z^{[L]} = W^{[L]} A^{[L-1]} + b^{[L]}$</td>\n <td> $(n^{[L]}, 209)$ </td> \n <tr>\n\n</table>\n\nRemember that when we compute $W X + b$ in python, it carries out broadcasting. For example, if: \n\n$$ W = \\begin{bmatrix}\n j & k & l\\\\\n m & n & o \\\\\n p & q & r \n\\end{bmatrix}\\;\\;\\; X = \\begin{bmatrix}\n a & b & c\\\\\n d & e & f \\\\\n g & h & i \n\\end{bmatrix} \\;\\;\\; b =\\begin{bmatrix}\n s \\\\\n t \\\\\n u\n\\end{bmatrix}\\tag{2}$$\n\nThen $WX + b$ will be:\n\n$$ WX + b = \\begin{bmatrix}\n (ja + kd + lg) + s & (jb + ke + lh) + s & (jc + kf + li)+ s\\\\\n (ma + nd + og) + t & (mb + ne + oh) + t & (mc + nf + oi) + t\\\\\n (pa + qd + rg) + u & (pb + qe + rh) + u & (pc + qf + ri)+ u\n\\end{bmatrix}\\tag{3} $$",
"_____no_output_____"
],
[
"**Exercise**: Implement initialization for an L-layer Neural Network. \n\n**Instructions**:\n- The model's structure is *[LINEAR -> RELU] $ \\times$ (L-1) -> LINEAR -> SIGMOID*. I.e., it has $L-1$ layers using a ReLU activation function followed by an output layer with a sigmoid activation function.\n- Use random initialization for the weight matrices. Use `np.random.randn(shape) * 0.01`.\n- Use zeros initialization for the biases. Use `np.zeros(shape)`.\n- We will store $n^{[l]}$, the number of units in different layers, in a variable `layer_dims`. For example, the `layer_dims` for the \"Planar Data classification model\" from last week would have been [2,4,1]: There were two inputs, one hidden layer with 4 hidden units, and an output layer with 1 output unit. This means `W1`'s shape was (4,2), `b1` was (4,1), `W2` was (1,4) and `b2` was (1,1). Now you will generalize this to $L$ layers! \n- Here is the implementation for $L=1$ (one layer neural network). It should inspire you to implement the general case (L-layer neural network).\n```python\n if L == 1:\n parameters[\"W\" + str(L)] = np.random.randn(layer_dims[1], layer_dims[0]) * 0.01\n parameters[\"b\" + str(L)] = np.zeros((layer_dims[1], 1))\n```",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_parameters_deep\n\ndef initialize_parameters_deep(layer_dims):\n \"\"\"\n Arguments:\n layer_dims -- python array (list) containing the dimensions of each layer in our network\n \n Returns:\n parameters -- python dictionary containing your parameters \"W1\", \"b1\", ..., \"WL\", \"bL\":\n Wl -- weight matrix of shape (layer_dims[l], layer_dims[l-1])\n bl -- bias vector of shape (layer_dims[l], 1)\n \"\"\"\n \n np.random.seed(3)\n parameters = {}\n L = len(layer_dims) # number of layers in the network\n\n for l in range(1, L):\n ### START CODE HERE ### (≈ 2 lines of code)\n parameters['W' + str(l)] = np.random.randn(layer_dims[l], layer_dims[l-1]) * 0.01 # None\n parameters['b' + str(l)] = np.zeros((layer_dims[l], 1)) # None\n ### END CODE HERE ###\n \n assert(parameters['W' + str(l)].shape == (layer_dims[l], layer_dims[l-1]))\n assert(parameters['b' + str(l)].shape == (layer_dims[l], 1))\n\n \n return parameters",
"_____no_output_____"
],
[
"parameters = initialize_parameters_deep([5,4,3])\nprint(\"W1 = \" + str(parameters[\"W1\"]))\nprint(\"b1 = \" + str(parameters[\"b1\"]))\nprint(\"W2 = \" + str(parameters[\"W2\"]))\nprint(\"b2 = \" + str(parameters[\"b2\"]))",
"W1 = [[ 0.01788628 0.0043651 0.00096497 -0.01863493 -0.00277388]\n [-0.00354759 -0.00082741 -0.00627001 -0.00043818 -0.00477218]\n [-0.01313865 0.00884622 0.00881318 0.01709573 0.00050034]\n [-0.00404677 -0.0054536 -0.01546477 0.00982367 -0.01101068]]\nb1 = [[ 0.]\n [ 0.]\n [ 0.]\n [ 0.]]\nW2 = [[-0.01185047 -0.0020565 0.01486148 0.00236716]\n [-0.01023785 -0.00712993 0.00625245 -0.00160513]\n [-0.00768836 -0.00230031 0.00745056 0.01976111]]\nb2 = [[ 0.]\n [ 0.]\n [ 0.]]\n"
]
],
[
[
"**Expected output**:\n \n<table style=\"width:80%\">\n <tr>\n <td> **W1** </td>\n <td>[[ 0.01788628 0.0043651 0.00096497 -0.01863493 -0.00277388]\n [-0.00354759 -0.00082741 -0.00627001 -0.00043818 -0.00477218]\n [-0.01313865 0.00884622 0.00881318 0.01709573 0.00050034]\n [-0.00404677 -0.0054536 -0.01546477 0.00982367 -0.01101068]]</td> \n </tr>\n \n <tr>\n <td>**b1** </td>\n <td>[[ 0.]\n [ 0.]\n [ 0.]\n [ 0.]]</td> \n </tr>\n \n <tr>\n <td>**W2** </td>\n <td>[[-0.01185047 -0.0020565 0.01486148 0.00236716]\n [-0.01023785 -0.00712993 0.00625245 -0.00160513]\n [-0.00768836 -0.00230031 0.00745056 0.01976111]]</td> \n </tr>\n \n <tr>\n <td>**b2** </td>\n <td>[[ 0.]\n [ 0.]\n [ 0.]]</td> \n </tr>\n \n</table>",
"_____no_output_____"
],
[
"## 4 - Forward propagation module\n\n### 4.1 - Linear Forward \nNow that you have initialized your parameters, you will do the forward propagation module. You will start by implementing some basic functions that you will use later when implementing the model. You will complete three functions in this order:\n\n- LINEAR\n- LINEAR -> ACTIVATION where ACTIVATION will be either ReLU or Sigmoid. \n- [LINEAR -> RELU] $\\times$ (L-1) -> LINEAR -> SIGMOID (whole model)\n\nThe linear forward module (vectorized over all the examples) computes the following equations:\n\n$$Z^{[l]} = W^{[l]}A^{[l-1]} +b^{[l]}\\tag{4}$$\n\nwhere $A^{[0]} = X$. \n\n**Exercise**: Build the linear part of forward propagation.\n\n**Reminder**:\nThe mathematical representation of this unit is $Z^{[l]} = W^{[l]}A^{[l-1]} +b^{[l]}$. You may also find `np.dot()` useful. If your dimensions don't match, printing `W.shape` may help.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: linear_forward\n\ndef linear_forward(A, W, b):\n \"\"\"\n Implement the linear part of a layer's forward propagation.\n\n Arguments:\n A -- activations from previous layer (or input data): (size of previous layer, number of examples)\n W -- weights matrix: numpy array of shape (size of current layer, size of previous layer)\n b -- bias vector, numpy array of shape (size of the current layer, 1)\n\n Returns:\n Z -- the input of the activation function, also called pre-activation parameter \n cache -- a python tuple containing \"A\", \"W\" and \"b\" ; stored for computing the backward pass efficiently\n \"\"\"\n \n ### START CODE HERE ### (≈ 1 line of code)\n Z = np.dot(W, A) + b # None\n ### END CODE HERE ###\n \n assert(Z.shape == (W.shape[0], A.shape[1]))\n cache = (A, W, b)\n \n return Z, cache",
"_____no_output_____"
],
[
"A, W, b = linear_forward_test_case()\n\nZ, linear_cache = linear_forward(A, W, b)\nprint(\"Z = \" + str(Z))",
"Z = [[ 3.26295337 -1.23429987]]\n"
]
],
[
[
"**Expected output**:\n\n<table style=\"width:35%\">\n \n <tr>\n <td> **Z** </td>\n <td> [[ 3.26295337 -1.23429987]] </td> \n </tr>\n \n</table>",
"_____no_output_____"
],
[
"### 4.2 - Linear-Activation Forward\n\nIn this notebook, you will use two activation functions:\n\n- **Sigmoid**: $\\sigma(Z) = \\sigma(W A + b) = \\frac{1}{ 1 + e^{-(W A + b)}}$. We have provided you with the `sigmoid` function. This function returns **two** items: the activation value \"`a`\" and a \"`cache`\" that contains \"`Z`\" (it's what we will feed in to the corresponding backward function). To use it you could just call: \n``` python\nA, activation_cache = sigmoid(Z)\n```\n\n- **ReLU**: The mathematical formula for ReLu is $A = RELU(Z) = max(0, Z)$. We have provided you with the `relu` function. This function returns **two** items: the activation value \"`A`\" and a \"`cache`\" that contains \"`Z`\" (it's what we will feed in to the corresponding backward function). To use it you could just call:\n``` python\nA, activation_cache = relu(Z)\n```",
"_____no_output_____"
],
[
"For more convenience, you are going to group two functions (Linear and Activation) into one function (LINEAR->ACTIVATION). Hence, you will implement a function that does the LINEAR forward step followed by an ACTIVATION forward step.\n\n**Exercise**: Implement the forward propagation of the *LINEAR->ACTIVATION* layer. Mathematical relation is: $A^{[l]} = g(Z^{[l]}) = g(W^{[l]}A^{[l-1]} +b^{[l]})$ where the activation \"g\" can be sigmoid() or relu(). Use linear_forward() and the correct activation function.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: linear_activation_forward\n\ndef linear_activation_forward(A_prev, W, b, activation):\n \"\"\"\n Implement the forward propagation for the LINEAR->ACTIVATION layer\n\n Arguments:\n A_prev -- activations from previous layer (or input data): (size of previous layer, number of examples)\n W -- weights matrix: numpy array of shape (size of current layer, size of previous layer)\n b -- bias vector, numpy array of shape (size of the current layer, 1)\n activation -- the activation to be used in this layer, stored as a text string: \"sigmoid\" or \"relu\"\n\n Returns:\n A -- the output of the activation function, also called the post-activation value \n cache -- a python tuple containing \"linear_cache\" and \"activation_cache\";\n stored for computing the backward pass efficiently\n \"\"\"\n \n if activation == \"sigmoid\":\n # Inputs: \"A_prev, W, b\". Outputs: \"A, activation_cache\".\n ### START CODE HERE ### (≈ 2 lines of code)\n Z, linear_cache = linear_forward(A_prev, W, b) # None\n A, activation_cache = sigmoid(Z) # None\n ### END CODE HERE ###\n \n elif activation == \"relu\":\n # Inputs: \"A_prev, W, b\". Outputs: \"A, activation_cache\".\n ### START CODE HERE ### (≈ 2 lines of code)\n Z, linear_cache =linear_forward(A_prev, W, b) # None\n A, activation_cache = relu(Z) # None\n ### END CODE HERE ###\n \n assert (A.shape == (W.shape[0], A_prev.shape[1]))\n cache = (linear_cache, activation_cache)\n\n return A, cache",
"_____no_output_____"
],
[
"A_prev, W, b = linear_activation_forward_test_case()\n\nA, linear_activation_cache = linear_activation_forward(A_prev, W, b, activation = \"sigmoid\")\nprint(\"With sigmoid: A = \" + str(A))\n\nA, linear_activation_cache = linear_activation_forward(A_prev, W, b, activation = \"relu\")\nprint(\"With ReLU: A = \" + str(A))",
"With sigmoid: A = [[ 0.96890023 0.11013289]]\nWith ReLU: A = [[ 3.43896131 0. ]]\n"
]
],
[
[
"**Expected output**:\n \n<table style=\"width:35%\">\n <tr>\n <td> **With sigmoid: A ** </td>\n <td > [[ 0.96890023 0.11013289]]</td> \n </tr>\n <tr>\n <td> **With ReLU: A ** </td>\n <td > [[ 3.43896131 0. ]]</td> \n </tr>\n</table>\n",
"_____no_output_____"
],
[
"**Note**: In deep learning, the \"[LINEAR->ACTIVATION]\" computation is counted as a single layer in the neural network, not two layers. ",
"_____no_output_____"
],
[
"### d) L-Layer Model \n\nFor even more convenience when implementing the $L$-layer Neural Net, you will need a function that replicates the previous one (`linear_activation_forward` with RELU) $L-1$ times, then follows that with one `linear_activation_forward` with SIGMOID.\n\n<img src=\"images/model_architecture_kiank.png\" style=\"width:600px;height:300px;\">\n<caption><center> **Figure 2** : *[LINEAR -> RELU] $\\times$ (L-1) -> LINEAR -> SIGMOID* model</center></caption><br>\n\n**Exercise**: Implement the forward propagation of the above model.\n\n**Instruction**: In the code below, the variable `AL` will denote $A^{[L]} = \\sigma(Z^{[L]}) = \\sigma(W^{[L]} A^{[L-1]} + b^{[L]})$. (This is sometimes also called `Yhat`, i.e., this is $\\hat{Y}$.) \n\n**Tips**:\n- Use the functions you had previously written \n- Use a for loop to replicate [LINEAR->RELU] (L-1) times\n- Don't forget to keep track of the caches in the \"caches\" list. To add a new value `c` to a `list`, you can use `list.append(c)`.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: L_model_forward\n\ndef L_model_forward(X, parameters):\n \"\"\"\n Implement forward propagation for the [LINEAR->RELU]*(L-1)->LINEAR->SIGMOID computation\n \n Arguments:\n X -- data, numpy array of shape (input size, number of examples)\n parameters -- output of initialize_parameters_deep()\n \n Returns:\n AL -- last post-activation value\n caches -- list of caches containing:\n every cache of linear_activation_forward() (there are L-1 of them, indexed from 0 to L-1)\n \"\"\"\n\n caches = []\n A = X\n L = len(parameters) // 2 # number of layers in the neural network\n \n # Implement [LINEAR -> RELU]*(L-1). Add \"cache\" to the \"caches\" list.\n for l in range(1, L):\n A_prev = A \n ### START CODE HERE ### (≈ 2 lines of code)\n A, cache = linear_activation_forward(A_prev, parameters['W'+str(l)], \n parameters['b'+str(l)], \"relu\") # None\n caches.append(cache) # None\n ### END CODE HERE ###\n \n # Implement LINEAR -> SIGMOID. Add \"cache\" to the \"caches\" list.\n ### START CODE HERE ### (≈ 2 lines of code)\n AL, cache = linear_activation_forward(A, parameters['W'+str(L)], \n parameters['b'+str(L)], \"sigmoid\") # None\n caches.append(cache) # None\n ### END CODE HERE ###\n \n assert(AL.shape == (1,X.shape[1]))\n \n return AL, caches",
"_____no_output_____"
],
[
"X, parameters = L_model_forward_test_case_2hidden()\nAL, caches = L_model_forward(X, parameters)\nprint(\"AL = \" + str(AL))\nprint(\"Length of caches list = \" + str(len(caches)))",
"AL = [[ 0.03921668 0.70498921 0.19734387 0.04728177]]\nLength of caches list = 3\n"
]
],
[
[
"<table style=\"width:50%\">\n <tr>\n <td> **AL** </td>\n <td > [[ 0.03921668 0.70498921 0.19734387 0.04728177]]</td> \n </tr>\n <tr>\n <td> **Length of caches list ** </td>\n <td > 3 </td> \n </tr>\n</table>",
"_____no_output_____"
],
[
"Great! Now you have a full forward propagation that takes the input X and outputs a row vector $A^{[L]}$ containing your predictions. It also records all intermediate values in \"caches\". Using $A^{[L]}$, you can compute the cost of your predictions.",
"_____no_output_____"
],
[
"## 5 - Cost function\n\nNow you will implement forward and backward propagation. You need to compute the cost, because you want to check if your model is actually learning.\n\n**Exercise**: Compute the cross-entropy cost $J$, using the following formula: $$-\\frac{1}{m} \\sum\\limits_{i = 1}^{m} (y^{(i)}\\log\\left(a^{[L] (i)}\\right) + (1-y^{(i)})\\log\\left(1- a^{[L](i)}\\right)) \\tag{7}$$\n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: compute_cost\n\ndef compute_cost(AL, Y):\n \"\"\"\n Implement the cost function defined by equation (7).\n\n Arguments:\n AL -- probability vector corresponding to your label predictions, shape (1, number of examples)\n Y -- true \"label\" vector (for example: containing 0 if non-cat, 1 if cat), shape (1, number of examples)\n\n Returns:\n cost -- cross-entropy cost\n \"\"\"\n \n m = Y.shape[1]\n\n # Compute loss from aL and y.\n ### START CODE HERE ### (≈ 1 lines of code)\n cost = - 1 / m * np.sum(np.dot(Y, np.log(AL).T) + np.dot((1 - Y),np.log(1 - AL).T)) # None\n ### END CODE HERE ###\n \n cost = np.squeeze(cost) # To make sure your cost's shape is what we expect (e.g. this turns [[17]] into 17).\n assert(cost.shape == ())\n \n return cost",
"_____no_output_____"
],
[
"Y, AL = compute_cost_test_case()\n\nprint(\"cost = \" + str(compute_cost(AL, Y)))",
"cost = 0.279776563579\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n\n <tr>\n <td>**cost** </td>\n <td> 0.2797765635793422</td> \n </tr>\n</table>",
"_____no_output_____"
],
[
"## 6 - Backward propagation module\n\nJust like with forward propagation, you will implement helper functions for backpropagation. Remember that back propagation is used to calculate the gradient of the loss function with respect to the parameters. \n\n**Reminder**: \n<img src=\"images/backprop_kiank.png\" style=\"width:650px;height:250px;\">\n<caption><center> **Figure 3** : Forward and Backward propagation for *LINEAR->RELU->LINEAR->SIGMOID* <br> *The purple blocks represent the forward propagation, and the red blocks represent the backward propagation.* </center></caption>\n\n<!-- \nFor those of you who are expert in calculus (you don't need to be to do this assignment), the chain rule of calculus can be used to derive the derivative of the loss $\\mathcal{L}$ with respect to $z^{[1]}$ in a 2-layer network as follows:\n\n$$\\frac{d \\mathcal{L}(a^{[2]},y)}{{dz^{[1]}}} = \\frac{d\\mathcal{L}(a^{[2]},y)}{{da^{[2]}}}\\frac{{da^{[2]}}}{{dz^{[2]}}}\\frac{{dz^{[2]}}}{{da^{[1]}}}\\frac{{da^{[1]}}}{{dz^{[1]}}} \\tag{8} $$\n\nIn order to calculate the gradient $dW^{[1]} = \\frac{\\partial L}{\\partial W^{[1]}}$, you use the previous chain rule and you do $dW^{[1]} = dz^{[1]} \\times \\frac{\\partial z^{[1]} }{\\partial W^{[1]}}$. During the backpropagation, at each step you multiply your current gradient by the gradient corresponding to the specific layer to get the gradient you wanted.\n\nEquivalently, in order to calculate the gradient $db^{[1]} = \\frac{\\partial L}{\\partial b^{[1]}}$, you use the previous chain rule and you do $db^{[1]} = dz^{[1]} \\times \\frac{\\partial z^{[1]} }{\\partial b^{[1]}}$.\n\nThis is why we talk about **backpropagation**.\n!-->\n\nNow, similar to forward propagation, you are going to build the backward propagation in three steps:\n- LINEAR backward\n- LINEAR -> ACTIVATION backward where ACTIVATION computes the derivative of either the ReLU or sigmoid activation\n- [LINEAR -> RELU] $\\times$ (L-1) -> LINEAR -> SIGMOID backward (whole model)",
"_____no_output_____"
],
[
"### 6.1 - Linear backward\n\nFor layer $l$, the linear part is: $Z^{[l]} = W^{[l]} A^{[l-1]} + b^{[l]}$ (followed by an activation).\n\nSuppose you have already calculated the derivative $dZ^{[l]} = \\frac{\\partial \\mathcal{L} }{\\partial Z^{[l]}}$. You want to get $(dW^{[l]}, db^{[l]}, dA^{[l-1]})$.\n\n<img src=\"images/linearback_kiank.png\" style=\"width:250px;height:300px;\">\n<caption><center> **Figure 4** </center></caption>\n\nThe three outputs $(dW^{[l]}, db^{[l]}, dA^{[l-1]})$ are computed using the input $dZ^{[l]}$.Here are the formulas you need:\n$$ dW^{[l]} = \\frac{\\partial \\mathcal{J} }{\\partial W^{[l]}} = \\frac{1}{m} dZ^{[l]} A^{[l-1] T} \\tag{8}$$\n$$ db^{[l]} = \\frac{\\partial \\mathcal{J} }{\\partial b^{[l]}} = \\frac{1}{m} \\sum_{i = 1}^{m} dZ^{[l](i)}\\tag{9}$$\n$$ dA^{[l-1]} = \\frac{\\partial \\mathcal{L} }{\\partial A^{[l-1]}} = W^{[l] T} dZ^{[l]} \\tag{10}$$\n",
"_____no_output_____"
],
[
"**Exercise**: Use the 3 formulas above to implement linear_backward().",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: linear_backward\n\ndef linear_backward(dZ, cache):\n \"\"\"\n Implement the linear portion of backward propagation for a single layer (layer l)\n\n Arguments:\n dZ -- Gradient of the cost with respect to the linear output (of current layer l)\n cache -- tuple of values (A_prev, W, b) coming from the forward propagation in the current layer\n\n Returns:\n dA_prev -- Gradient of the cost with respect to the activation (of the previous layer l-1), same shape as A_prev\n dW -- Gradient of the cost with respect to W (current layer l), same shape as W\n db -- Gradient of the cost with respect to b (current layer l), same shape as b\n \"\"\"\n A_prev, W, b = cache\n m = A_prev.shape[1]\n\n ### START CODE HERE ### (≈ 3 lines of code)\n dW = 1 / m * np.dot(dZ, A_prev.T) # None\n db = 1 / m * np.sum(dZ, axis=1, keepdims=True) # None\n dA_prev = np.dot(W.T, dZ) # None\n ### END CODE HERE ###\n \n assert (dA_prev.shape == A_prev.shape)\n assert (dW.shape == W.shape)\n assert (db.shape == b.shape)\n \n return dA_prev, dW, db",
"_____no_output_____"
],
[
"# Set up some test inputs\ndZ, linear_cache = linear_backward_test_case()\n\ndA_prev, dW, db = linear_backward(dZ, linear_cache)\nprint (\"dA_prev = \"+ str(dA_prev))\nprint (\"dW = \" + str(dW))\nprint (\"db = \" + str(db))",
"dA_prev = [[-1.15171336 0.06718465 -0.3204696 2.09812712]\n [ 0.60345879 -3.72508701 5.81700741 -3.84326836]\n [-0.4319552 -1.30987417 1.72354705 0.05070578]\n [-0.38981415 0.60811244 -1.25938424 1.47191593]\n [-2.52214926 2.67882552 -0.67947465 1.48119548]]\ndW = [[ 0.07313866 -0.0976715 -0.87585828 0.73763362 0.00785716]\n [ 0.85508818 0.37530413 -0.59912655 0.71278189 -0.58931808]\n [ 0.97913304 -0.24376494 -0.08839671 0.55151192 -0.10290907]]\ndb = [[-0.14713786]\n [-0.11313155]\n [-0.13209101]]\n"
]
],
[
[
"** Expected Output**:\n \n```\ndA_prev = \n [[-1.15171336 0.06718465 -0.3204696 2.09812712]\n [ 0.60345879 -3.72508701 5.81700741 -3.84326836]\n [-0.4319552 -1.30987417 1.72354705 0.05070578]\n [-0.38981415 0.60811244 -1.25938424 1.47191593]\n [-2.52214926 2.67882552 -0.67947465 1.48119548]]\ndW = \n [[ 0.07313866 -0.0976715 -0.87585828 0.73763362 0.00785716]\n [ 0.85508818 0.37530413 -0.59912655 0.71278189 -0.58931808]\n [ 0.97913304 -0.24376494 -0.08839671 0.55151192 -0.10290907]]\ndb = \n [[-0.14713786]\n [-0.11313155]\n [-0.13209101]]\n```",
"_____no_output_____"
],
[
"### 6.2 - Linear-Activation backward\n\nNext, you will create a function that merges the two helper functions: **`linear_backward`** and the backward step for the activation **`linear_activation_backward`**. \n\nTo help you implement `linear_activation_backward`, we provided two backward functions:\n- **`sigmoid_backward`**: Implements the backward propagation for SIGMOID unit. You can call it as follows:\n\n```python\ndZ = sigmoid_backward(dA, activation_cache)\n```\n\n- **`relu_backward`**: Implements the backward propagation for RELU unit. You can call it as follows:\n\n```python\ndZ = relu_backward(dA, activation_cache)\n```\n\nIf $g(.)$ is the activation function, \n`sigmoid_backward` and `relu_backward` compute $$dZ^{[l]} = dA^{[l]} * g'(Z^{[l]}) \\tag{11}$$. \n\n**Exercise**: Implement the backpropagation for the *LINEAR->ACTIVATION* layer.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: linear_activation_backward\n\ndef linear_activation_backward(dA, cache, activation):\n \"\"\"\n Implement the backward propagation for the LINEAR->ACTIVATION layer.\n \n Arguments:\n dA -- post-activation gradient for current layer l \n cache -- tuple of values (linear_cache, activation_cache) we store for computing backward propagation efficiently\n activation -- the activation to be used in this layer, stored as a text string: \"sigmoid\" or \"relu\"\n \n Returns:\n dA_prev -- Gradient of the cost with respect to the activation (of the previous layer l-1), same shape as A_prev\n dW -- Gradient of the cost with respect to W (current layer l), same shape as W\n db -- Gradient of the cost with respect to b (current layer l), same shape as b\n \"\"\"\n linear_cache, activation_cache = cache\n \n if activation == \"relu\":\n ### START CODE HERE ### (≈ 2 lines of code)\n dZ = relu_backward(dA, activation_cache) # None\n dA_prev, dW, db = linear_backward(dZ, linear_cache) # None\n ### END CODE HERE ###\n \n elif activation == \"sigmoid\":\n ### START CODE HERE ### (≈ 2 lines of code)\n dZ = sigmoid_backward(dA, activation_cache) # None\n dA_prev, dW, db = linear_backward(dZ, linear_cache) # None\n ### END CODE HERE ###\n \n return dA_prev, dW, db",
"_____no_output_____"
],
[
"dAL, linear_activation_cache = linear_activation_backward_test_case()\n\ndA_prev, dW, db = linear_activation_backward(dAL, linear_activation_cache, activation = \"sigmoid\")\nprint (\"sigmoid:\")\nprint (\"dA_prev = \"+ str(dA_prev))\nprint (\"dW = \" + str(dW))\nprint (\"db = \" + str(db) + \"\\n\")\n\ndA_prev, dW, db = linear_activation_backward(dAL, linear_activation_cache, activation = \"relu\")\nprint (\"relu:\")\nprint (\"dA_prev = \"+ str(dA_prev))\nprint (\"dW = \" + str(dW))\nprint (\"db = \" + str(db))",
"sigmoid:\ndA_prev = [[ 0.11017994 0.01105339]\n [ 0.09466817 0.00949723]\n [-0.05743092 -0.00576154]]\ndW = [[ 0.10266786 0.09778551 -0.01968084]]\ndb = [[-0.05729622]]\n\nrelu:\ndA_prev = [[ 0.44090989 -0. ]\n [ 0.37883606 -0. ]\n [-0.2298228 0. ]]\ndW = [[ 0.44513824 0.37371418 -0.10478989]]\ndb = [[-0.20837892]]\n"
]
],
[
[
"**Expected output with sigmoid:**\n\n<table style=\"width:100%\">\n <tr>\n <td > dA_prev </td> \n <td >[[ 0.11017994 0.01105339]\n [ 0.09466817 0.00949723]\n [-0.05743092 -0.00576154]] </td> \n\n </tr> \n \n <tr>\n <td > dW </td> \n <td > [[ 0.10266786 0.09778551 -0.01968084]] </td> \n </tr> \n \n <tr>\n <td > db </td> \n <td > [[-0.05729622]] </td> \n </tr> \n</table>\n\n",
"_____no_output_____"
],
[
"**Expected output with relu:**\n\n<table style=\"width:100%\">\n <tr>\n <td > dA_prev </td> \n <td > [[ 0.44090989 0. ]\n [ 0.37883606 0. ]\n [-0.2298228 0. ]] </td> \n\n </tr> \n \n <tr>\n <td > dW </td> \n <td > [[ 0.44513824 0.37371418 -0.10478989]] </td> \n </tr> \n \n <tr>\n <td > db </td> \n <td > [[-0.20837892]] </td> \n </tr> \n</table>\n\n",
"_____no_output_____"
],
[
"### 6.3 - L-Model Backward \n\nNow you will implement the backward function for the whole network. Recall that when you implemented the `L_model_forward` function, at each iteration, you stored a cache which contains (X,W,b, and z). In the back propagation module, you will use those variables to compute the gradients. Therefore, in the `L_model_backward` function, you will iterate through all the hidden layers backward, starting from layer $L$. On each step, you will use the cached values for layer $l$ to backpropagate through layer $l$. Figure 5 below shows the backward pass. \n\n\n<img src=\"images/mn_backward.png\" style=\"width:450px;height:300px;\">\n<caption><center> **Figure 5** : Backward pass </center></caption>\n\n** Initializing backpropagation**:\nTo backpropagate through this network, we know that the output is, \n$A^{[L]} = \\sigma(Z^{[L]})$. Your code thus needs to compute `dAL` $= \\frac{\\partial \\mathcal{L}}{\\partial A^{[L]}}$.\nTo do so, use this formula (derived using calculus which you don't need in-depth knowledge of):\n```python\ndAL = - (np.divide(Y, AL) - np.divide(1 - Y, 1 - AL)) # derivative of cost with respect to AL\n```\n\nYou can then use this post-activation gradient `dAL` to keep going backward. As seen in Figure 5, you can now feed in `dAL` into the LINEAR->SIGMOID backward function you implemented (which will use the cached values stored by the L_model_forward function). After that, you will have to use a `for` loop to iterate through all the other layers using the LINEAR->RELU backward function. You should store each dA, dW, and db in the grads dictionary. To do so, use this formula : \n\n$$grads[\"dW\" + str(l)] = dW^{[l]}\\tag{15} $$\n\nFor example, for $l=3$ this would store $dW^{[l]}$ in `grads[\"dW3\"]`.\n\n**Exercise**: Implement backpropagation for the *[LINEAR->RELU] $\\times$ (L-1) -> LINEAR -> SIGMOID* model.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: L_model_backward\n\ndef L_model_backward(AL, Y, caches):\n \"\"\"\n Implement the backward propagation for the [LINEAR->RELU] * (L-1) -> LINEAR -> SIGMOID group\n \n Arguments:\n AL -- probability vector, output of the forward propagation (L_model_forward())\n Y -- true \"label\" vector (containing 0 if non-cat, 1 if cat)\n caches -- list of caches containing:\n every cache of linear_activation_forward() with \"relu\" (it's caches[l], for l in range(L-1) i.e l = 0...L-2)\n the cache of linear_activation_forward() with \"sigmoid\" (it's caches[L-1])\n \n Returns:\n grads -- A dictionary with the gradients\n grads[\"dA\" + str(l)] = ... \n grads[\"dW\" + str(l)] = ...\n grads[\"db\" + str(l)] = ... \n \"\"\"\n grads = {}\n L = len(caches) # the number of layers\n m = AL.shape[1]\n Y = Y.reshape(AL.shape) # after this line, Y is the same shape as AL\n \n # Initializing the backpropagation\n ### START CODE HERE ### (1 line of code)\n dAL = - (np.divide(Y, AL) - np.divide(1 - Y, 1 - AL)) # derivative of cost with respect to AL # None\n ### END CODE HERE ###\n \n # Lth layer (SIGMOID -> LINEAR) gradients. Inputs: \"dAL, current_cache\". Outputs: \"grads[\"dAL-1\"], grads[\"dWL\"], grads[\"dbL\"]\n ### START CODE HERE ### (approx. 2 lines)\n current_cache = caches[L-1] # None\n grads[\"dA\" + str(L-1)], grads[\"dW\" + str(L)], grads[\"db\" + str(L)] = linear_activation_backward(dAL, current_cache, \"sigmoid\") # None\n ### END CODE HERE ###\n \n # Loop from l=L-2 to l=0\n for l in reversed(range(L-1)):\n # lth layer: (RELU -> LINEAR) gradients.\n # Inputs: \"grads[\"dA\" + str(l + 1)], current_cache\". Outputs: \"grads[\"dA\" + str(l)] , grads[\"dW\" + str(l + 1)] , grads[\"db\" + str(l + 1)] \n ### START CODE HERE ### (approx. 5 lines)\n current_cache = caches[l] # None\n dA_prev_temp, dW_temp, db_temp = linear_activation_backward(grads[\"dA\" + str(l+1)], current_cache, \"relu\") # None\n grads[\"dA\" + str(l)] = dA_prev_temp # None\n grads[\"dW\" + str(l + 1)] = dW_temp # None\n grads[\"db\" + str(l + 1)] = db_temp # None\n ### END CODE HERE ###\n\n return grads",
"_____no_output_____"
],
[
"AL, Y_assess, caches = L_model_backward_test_case()\ngrads = L_model_backward(AL, Y_assess, caches)\nprint_grads(grads)",
"dW1 = [[ 0.41010002 0.07807203 0.13798444 0.10502167]\n [ 0. 0. 0. 0. ]\n [ 0.05283652 0.01005865 0.01777766 0.0135308 ]]\ndb1 = [[-0.22007063]\n [ 0. ]\n [-0.02835349]]\ndA1 = [[ 0.12913162 -0.44014127]\n [-0.14175655 0.48317296]\n [ 0.01663708 -0.05670698]]\n"
]
],
[
[
"**Expected Output**\n\n<table style=\"width:60%\">\n \n <tr>\n <td > dW1 </td> \n <td > [[ 0.41010002 0.07807203 0.13798444 0.10502167]\n [ 0. 0. 0. 0. ]\n [ 0.05283652 0.01005865 0.01777766 0.0135308 ]] </td> \n </tr> \n \n <tr>\n <td > db1 </td> \n <td > [[-0.22007063]\n [ 0. ]\n [-0.02835349]] </td> \n </tr> \n \n <tr>\n <td > dA1 </td> \n <td > [[ 0.12913162 -0.44014127]\n [-0.14175655 0.48317296]\n [ 0.01663708 -0.05670698]] </td> \n\n </tr> \n</table>\n\n",
"_____no_output_____"
],
[
"### 6.4 - Update Parameters\n\nIn this section you will update the parameters of the model, using gradient descent: \n\n$$ W^{[l]} = W^{[l]} - \\alpha \\text{ } dW^{[l]} \\tag{16}$$\n$$ b^{[l]} = b^{[l]} - \\alpha \\text{ } db^{[l]} \\tag{17}$$\n\nwhere $\\alpha$ is the learning rate. After computing the updated parameters, store them in the parameters dictionary. ",
"_____no_output_____"
],
[
"**Exercise**: Implement `update_parameters()` to update your parameters using gradient descent.\n\n**Instructions**:\nUpdate parameters using gradient descent on every $W^{[l]}$ and $b^{[l]}$ for $l = 1, 2, ..., L$. \n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: update_parameters\n\ndef update_parameters(parameters, grads, learning_rate):\n \"\"\"\n Update parameters using gradient descent\n \n Arguments:\n parameters -- python dictionary containing your parameters \n grads -- python dictionary containing your gradients, output of L_model_backward\n \n Returns:\n parameters -- python dictionary containing your updated parameters \n parameters[\"W\" + str(l)] = ... \n parameters[\"b\" + str(l)] = ...\n \"\"\"\n \n L = len(parameters) // 2 # number of layers in the neural network\n\n # Update rule for each parameter. Use a for loop.\n ### START CODE HERE ### (≈ 3 lines of code)\n for l in range(L):\n parameters[\"W\" + str(l+1)] += - learning_rate * grads[\"dW\" + str(l + 1)] # None\n parameters[\"b\" + str(l+1)] += - learning_rate * grads[\"db\" + str(l + 1)] # None\n ### END CODE HERE ###\n return parameters",
"_____no_output_____"
],
[
"parameters, grads = update_parameters_test_case()\nparameters = update_parameters(parameters, grads, 0.1)\n\nprint (\"W1 = \"+ str(parameters[\"W1\"]))\nprint (\"b1 = \"+ str(parameters[\"b1\"]))\nprint (\"W2 = \"+ str(parameters[\"W2\"]))\nprint (\"b2 = \"+ str(parameters[\"b2\"]))",
"W1 = [[-0.59562069 -0.09991781 -2.14584584 1.82662008]\n [-1.76569676 -0.80627147 0.51115557 -1.18258802]\n [-1.0535704 -0.86128581 0.68284052 2.20374577]]\nb1 = [[-0.04659241]\n [-1.28888275]\n [ 0.53405496]]\nW2 = [[-0.55569196 0.0354055 1.32964895]]\nb2 = [[-0.84610769]]\n"
]
],
[
[
"**Expected Output**:\n\n<table style=\"width:100%\"> \n <tr>\n <td > W1 </td> \n <td > [[-0.59562069 -0.09991781 -2.14584584 1.82662008]\n [-1.76569676 -0.80627147 0.51115557 -1.18258802]\n [-1.0535704 -0.86128581 0.68284052 2.20374577]] </td> \n </tr> \n \n <tr>\n <td > b1 </td> \n <td > [[-0.04659241]\n [-1.28888275]\n [ 0.53405496]] </td> \n </tr> \n <tr>\n <td > W2 </td> \n <td > [[-0.55569196 0.0354055 1.32964895]]</td> \n </tr> \n \n <tr>\n <td > b2 </td> \n <td > [[-0.84610769]] </td> \n </tr> \n</table>\n",
"_____no_output_____"
],
[
"\n## 7 - Conclusion\n\nCongrats on implementing all the functions required for building a deep neural network! \n\nWe know it was a long assignment but going forward it will only get better. The next part of the assignment is easier. \n\nIn the next assignment you will put all these together to build two models:\n- A two-layer neural network\n- An L-layer neural network\n\nYou will in fact use these models to classify cat vs non-cat images!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4a8b557dc551b188294eb41ee1485cb86949428d
| 18,072 |
ipynb
|
Jupyter Notebook
|
Resources/random_forest.ipynb
|
shumeiberk/supervised_ML
|
98763f4201fb96bd8423e300b5526ee405236a52
|
[
"MIT"
] | null | null | null |
Resources/random_forest.ipynb
|
shumeiberk/supervised_ML
|
98763f4201fb96bd8423e300b5526ee405236a52
|
[
"MIT"
] | null | null | null |
Resources/random_forest.ipynb
|
shumeiberk/supervised_ML
|
98763f4201fb96bd8423e300b5526ee405236a52
|
[
"MIT"
] | null | null | null | 28.504732 | 96 | 0.361664 |
[
[
[
"# Initial imports.\nimport pandas as pd\nfrom path import Path\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import confusion_matrix, accuracy_score, classification_report",
"_____no_output_____"
],
[
"df_loans = pd.read_csv(\"./Resources/loans_data_encoded.csv\")\ndf_loans.head()",
"_____no_output_____"
],
[
"# Define the features set.\nX = df_loans.copy()\nX = X.drop(\"bad\", axis=1)\nX.head()",
"_____no_output_____"
],
[
"# Define the target set.\ny = df_loans[\"bad\"].ravel()\ny[:5]",
"_____no_output_____"
],
[
"# Splitting into Train and Test sets.\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=78)",
"_____no_output_____"
],
[
"# Creating a StandardScaler instance.\nscaler = StandardScaler()\n# Fitting the Standard Scaler with the training data.\nX_scaler = scaler.fit(X_train)\n\n# Scaling the data.\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)",
"_____no_output_____"
],
[
"# Create a random forest classifier.\nrf_model = RandomForestClassifier(n_estimators=128, random_state=78) ",
"_____no_output_____"
],
[
"# Fitting the model\nrf_model = rf_model.fit(X_train_scaled, y_train)",
"_____no_output_____"
],
[
"# Making predictions using the testing data.\npredictions = rf_model.predict(X_test_scaled)",
"_____no_output_____"
],
[
"# Calculating the confusion matrix.\ncm = confusion_matrix(y_test, predictions)\n\n# Create a DataFrame from the confusion matrix.\ncm_df = pd.DataFrame(\n cm, index=[\"Actual 0\", \"Actual 1\"], columns=[\"Predicted 0\", \"Predicted 1\"])\n\ncm_df",
"_____no_output_____"
],
[
"# Calculating the accuracy score.\nacc_score = accuracy_score(y_test, predictions)",
"_____no_output_____"
],
[
"# Displaying results\nprint(\"Confusion Matrix\")\ndisplay(cm_df)\nprint(f\"Accuracy Score : {acc_score}\")\nprint(\"Classification Report\")\nprint(classification_report(y_test, predictions))",
"Confusion Matrix\n"
],
[
"# Calculate feature importance in the Random Forest model.\nimportances = rf_model.feature_importances_\nimportances",
"_____no_output_____"
],
[
"# We can sort the features by their importance.\nsorted(zip(rf_model.feature_importances_, X.columns), reverse=True)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8b55b7b2e76e9c7ab55d176aa1801885ffd833
| 14,147 |
ipynb
|
Jupyter Notebook
|
Chapter 09/Chapter 9.ipynb
|
PacktPublishing/Algorithmic-Short-Selling-with-Python
|
033c5259f2d6410a221c1949a11b44ee90ae3e57
|
[
"MIT"
] | 20 |
2021-10-01T22:06:16.000Z
|
2022-03-06T16:31:07.000Z
|
Chapter 09/Chapter 9.ipynb
|
aljamima/Algorithmic-Short-Selling-with-Python
|
a7b8deba5907964ec03a801bb5f698c2315aca90
|
[
"MIT"
] | null | null | null |
Chapter 09/Chapter 9.ipynb
|
aljamima/Algorithmic-Short-Selling-with-Python
|
a7b8deba5907964ec03a801bb5f698c2315aca90
|
[
"MIT"
] | 24 |
2021-10-02T10:52:52.000Z
|
2022-03-31T16:19:57.000Z
| 14,147 | 14,147 | 0.685092 |
[
[
[
"# Preliminary instruction\n\nTo follow the code in this chapter, the `yfinance` package must be installed in your environment. If you do not have this installed yet, review Chapter 4 for instructions on how to do so.",
"_____no_output_____"
],
[
"# Chapter 9: Risk is a Number",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\nimport pandas as pd\nimport numpy as np\nimport yfinance as yf\n%matplotlib inline\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"#### Mock Strategy: Turtle for dummies",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\ndef regime_breakout(df,_h,_l,window):\n hl = np.where(df[_h] == df[_h].rolling(window).max(),1,\n np.where(df[_l] == df[_l].rolling(window).min(), -1,np.nan))\n roll_hl = pd.Series(index= df.index, data= hl).fillna(method= 'ffill')\n return roll_hl\n\ndef turtle_trader(df, _h, _l, slow, fast):\n '''\n _slow: Long/Short direction\n _fast: trailing stop loss\n '''\n _slow = regime_breakout(df,_h,_l,window = slow)\n _fast = regime_breakout(df,_h,_l,window = fast)\n turtle = pd. Series(index= df.index, \n data = np.where(_slow == 1,np.where(_fast == 1,1,0), \n np.where(_slow == -1, np.where(_fast ==-1,-1,0),0)))\n return turtle",
"_____no_output_____"
]
],
[
[
"#### Run the strategy with Softbank in absolute\nPlot: Softbank turtle for dummies, positions, and returns\nPlot: Softbank cumulative returns and Sharpe ratios: rolling and cumulative",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\nticker = '9984.T' # Softbank\nstart = '2017-12-31'\nend = None\ndf = round(yf.download(tickers= ticker,start= start, end = end, \n interval = \"1d\",group_by = 'column',auto_adjust = True, \n prepost = True, treads = True, proxy = None),0)\nslow = 50\nfast = 20 \ndf['tt'] = turtle_trader(df, _h= 'High', _l= 'Low', slow= slow,fast= fast)\ndf['stop_loss'] = np.where(df['tt'] == 1, df['Low'].rolling(fast).min(),\n np.where(df['tt'] == -1, df['High'].rolling(fast).max(),np.nan))\n\ndf['tt_chg1D'] = df['Close'].diff() * df['tt'].shift()\ndf['tt_PL_cum'] = df['tt_chg1D'].cumsum()\n\ndf['tt_returns'] = df['Close'].pct_change() * df['tt'].shift()\ntt_log_returns = np.log(df['Close']/df['Close'].shift()) * df['tt'].shift()\ndf['tt_cumul'] = tt_log_returns.cumsum().apply(np.exp) - 1 \n\n\ndf[['Close','stop_loss','tt','tt_cumul']].plot(secondary_y=['tt','tt_cumul'],\n figsize=(20,8),style= ['k','r--','b:','b'],\n title= str(ticker)+' Close Price, Turtle L/S entries, cumulative returns')\n\ndf[['tt_PL_cum','tt_chg1D']].plot(secondary_y=['tt_chg1D'],\n figsize=(20,8),style= ['b','c:'],\n title= str(ticker) +' Daily P&L & Cumulative P&L')",
"_____no_output_____"
]
],
[
[
"#### Sharpe ratio: the right mathematical answer to the wrong question\nPlot: Softbank cumulative returns and Sharpe ratios: rolling and cumulative",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\nr_f = 0.00001 # risk free returns\n\ndef rolling_sharpe(returns, r_f, window):\n avg_returns = returns.rolling(window).mean()\n std_returns = returns.rolling(window).std(ddof=0)\n return (avg_returns - r_f) / std_returns\n\ndef expanding_sharpe(returns, r_f):\n avg_returns = returns.expanding().mean()\n std_returns = returns.expanding().std(ddof=0)\n return (avg_returns - r_f) / std_returns\n\nwindow= 252\ndf['sharpe_roll'] = rolling_sharpe(returns= tt_log_returns, r_f= r_f, window= window) * 252**0.5\n\ndf['sharpe']= expanding_sharpe(returns=tt_log_returns,r_f= r_f) * 252**0.5\n\ndf[window:][['tt_cumul','sharpe_roll','sharpe'] ].plot(figsize = (20,8),style = ['b','c-.','c'],grid=True,\n title = str(ticker)+' cumulative returns, Sharpe ratios: rolling & cumulative') \n\n",
"_____no_output_____"
]
],
[
[
"### Grit Index\n\nThis formula was originally invented by Peter G. Martin in 1987 and published as the Ulcer Index in his book The Investor's Guide to Fidelity Funds. Legendary trader Ed Seykota recycled it into the Seykota Lake ratio.\n\nInvestors react to drawdowns in three ways:\n1. Magnitude: never test the stomach of your investors\n2. Frequency: never test the nerves of your investors\n3. Duration: never test the patience of your investors\n\nThe Grit calculation sequence is as follows:\n1. Calculate the peak cumulative returns using rolling().max() or expanding().max()\n2. Calculate the squared drawdown from the peak and square them\n3. Calculate the least square sum by taking the square root of the squared drawdowns \n4. Divide the cumulative returns by the surface of losses\n\nPlot: Softbank cumulative returns and Grit ratios: rolling and cumulative",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\ndef rolling_grit(cumul_returns, window):\n tt_rolling_peak = cumul_returns.rolling(window).max()\n drawdown_squared = (cumul_returns - tt_rolling_peak) ** 2\n ulcer = drawdown_squared.rolling(window).sum() ** 0.5\n return cumul_returns / ulcer\n\ndef expanding_grit(cumul_returns):\n tt_peak = cumul_returns.expanding().max()\n drawdown_squared = (cumul_returns - tt_peak) ** 2\n ulcer = drawdown_squared.expanding().sum() ** 0.5\n return cumul_returns / ulcer\n\nwindow = 252\ndf['grit_roll'] = rolling_grit(cumul_returns= df['tt_cumul'] , window = window)\ndf['grit'] = expanding_grit(cumul_returns= df['tt_cumul'])\ndf[window:][['tt_cumul','grit_roll', 'grit'] ].plot(figsize = (20,8), \n secondary_y = 'tt_cumul',style = ['b','g-.','g'],grid=True,\n title = str(ticker) + ' cumulative returns & Grit Ratios: rolling & cumulative '+ str(window) + ' days') \n\n",
"_____no_output_____"
]
],
[
[
"### Common Sense Ratio\n\n1. Risk metric for trend following strategies: profit ratio, gain-to-pain ratio\n2. Risk metric for trend following strategies: tail ratio\n3. Combined risk metric: profit ratio * tail ratio\n\nPlot: Cumulative returns and common sense ratios: cumulative and rolling",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\ndef rolling_profits(returns,window):\n profit_roll = returns.copy()\n profit_roll[profit_roll < 0] = 0\n profit_roll_sum = profit_roll.rolling(window).sum().fillna(method='ffill')\n return profit_roll_sum\n\ndef rolling_losses(returns,window):\n loss_roll = returns.copy()\n loss_roll[loss_roll > 0] = 0\n loss_roll_sum = loss_roll.rolling(window).sum().fillna(method='ffill')\n return loss_roll_sum\n\ndef expanding_profits(returns): \n profit_roll = returns.copy() \n profit_roll[profit_roll < 0] = 0 \n profit_roll_sum = profit_roll.expanding().sum().fillna(method='ffill') \n return profit_roll_sum \n \ndef expanding_losses(returns): \n loss_roll = returns.copy() \n loss_roll[loss_roll > 0] = 0 \n loss_roll_sum = loss_roll.expanding().sum().fillna(method='ffill') \n return loss_roll_sum \n\ndef profit_ratio(profits, losses): \n pr = profits.fillna(method='ffill') / abs(losses.fillna(method='ffill'))\n return pr\n\n\ndef rolling_tail_ratio(cumul_returns, window, percentile,limit):\n left_tail = np.abs(cumul_returns.rolling(window).quantile(percentile))\n right_tail = cumul_returns.rolling(window).quantile(1-percentile)\n np.seterr(all='ignore')\n tail = np.maximum(np.minimum(right_tail / left_tail,limit),-limit)\n return tail\n\ndef expanding_tail_ratio(cumul_returns, percentile,limit):\n left_tail = np.abs(cumul_returns.expanding().quantile(percentile))\n right_tail = cumul_returns.expanding().quantile(1 - percentile)\n np.seterr(all='ignore')\n tail = np.maximum(np.minimum(right_tail / left_tail,limit),-limit)\n return tail\n\ndef common_sense_ratio(pr,tr):\n return pr * tr \n\n\n",
"_____no_output_____"
]
],
[
[
"#### Plot: Cumulative returns and profit ratios: cumulative and rolling",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\nwindow = 252\ndf['pr_roll'] = profit_ratio(profits= rolling_profits(returns = tt_log_returns,window = window), \n losses= rolling_losses(returns = tt_log_returns,window = window))\ndf['pr'] = profit_ratio(profits= expanding_profits(returns= tt_log_returns), \n losses= expanding_losses(returns = tt_log_returns))\n\ndf[window:] [['tt_cumul','pr_roll','pr'] ].plot(figsize = (20,8),secondary_y= ['tt_cumul'], \n style = ['r','y','y:'],grid=True) ",
"_____no_output_____"
]
],
[
[
"#### Plot: Cumulative returns and common sense ratios: cumulative and rolling",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\nwindow = 252\n\ndf['tr_roll'] = rolling_tail_ratio(cumul_returns= df['tt_cumul'], \n window= window, percentile= 0.05,limit=5)\ndf['tr'] = expanding_tail_ratio(cumul_returns= df['tt_cumul'], percentile= 0.05,limit=5)\n\ndf['csr_roll'] = common_sense_ratio(pr= df['pr_roll'],tr= df['tr_roll'])\ndf['csr'] = common_sense_ratio(pr= df['pr'],tr= df['tr'])\n\ndf[window:] [['tt_cumul','csr_roll','csr'] ].plot(secondary_y= ['tt_cumul'],style = ['b','r-.','r'], figsize = (20,8),\n title= str(ticker)+' cumulative returns, Common Sense Ratios: cumulative & rolling '+str(window)+ ' days')\n\n\n",
"_____no_output_____"
]
],
[
[
"### T-stat of gain expectancy, Van Tharp's System Quality Number (SQN)\n\nPlot: Softbank cumulative returns and t-stat (Van Tharp's SQN): cumulative and rolling",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\ndef expectancy(win_rate,avg_win,avg_loss): \n # win% * avg_win% - loss% * abs(avg_loss%) \n return win_rate * avg_win + (1-win_rate) * avg_loss \n\ndef t_stat(signal_count, trading_edge): \n sqn = (signal_count ** 0.5) * trading_edge / trading_edge.std(ddof=0) \n return sqn \n\n# Trade Count\ndf['trades'] = df.loc[(df['tt'].diff() !=0) & (pd.notnull(df['tt'])),'tt'].abs().cumsum()\nsignal_count = df['trades'].fillna(method='ffill')\nsignal_roll = signal_count.diff(window)\n\n# Rolling t_stat\nwindow = 252\nwin_roll = tt_log_returns.copy()\nwin_roll[win_roll < 0] = np.nan\nwin_rate_roll = win_roll.rolling(window,min_periods=0).count() / window\navg_win_roll = rolling_profits(returns = tt_log_returns,window = window) / window\navg_loss_roll = rolling_losses(returns = tt_log_returns,window = window) / window\n\nedge_roll= expectancy(win_rate= win_rate_roll,avg_win=avg_win_roll,avg_loss=avg_loss_roll)\ndf['sqn_roll'] = t_stat(signal_count= signal_roll, trading_edge=edge_roll)\n\n# Cumulative t-stat\ntt_win_count = tt_log_returns[tt_log_returns>0].expanding().count().fillna(method='ffill')\ntt_count = tt_log_returns[tt_log_returns!=0].expanding().count().fillna(method='ffill')\n\nwin_rate = (tt_win_count / tt_count).fillna(method='ffill')\navg_win = expanding_profits(returns= tt_log_returns) / tt_count\navg_loss = expanding_losses(returns= tt_log_returns) / tt_count\ntrading_edge = expectancy(win_rate,avg_win,avg_loss).fillna(method='ffill')\ndf['sqn'] = t_stat(signal_count, trading_edge)\n\ndf[window:][['tt_cumul','sqn','sqn_roll'] ].plot(figsize = (20,8),\n secondary_y= ['tt_cumul'], grid= True,style = ['b','y','y-.'], \n title= str(ticker)+' Cumulative Returns and SQN: cumulative & rolling'+ str(window)+' days')",
"_____no_output_____"
]
],
[
[
"### Robustness score\n\nCombined risk metric:\n1. The Grit Index integrates losses throughout the period\n2. The CSR combines risks endemic to the two types of strategies in a single measure\n3. The t-stat SQN incorporates trading frequency into the trading edge formula to show the most efficient use of capital.",
"_____no_output_____"
]
],
[
[
"# Chapter 9: Risk is a Number\n\ndef robustness_score(grit,csr,sqn): \n start_date = max(grit[pd.notnull(grit)].index[0],\n csr[pd.notnull(csr)].index[0],\n sqn[pd.notnull(sqn)].index[0])\n score = grit * csr * sqn / (grit[start_date] * csr[start_date] * sqn[start_date])\n return score\n\ndf['score_roll'] = robustness_score(grit = df['grit_roll'], csr = df['csr_roll'],sqn= df['sqn_roll'])\ndf['score'] = robustness_score(grit = df['grit'],csr = df['csr'],sqn = df['sqn'])\ndf[window:][['tt_cumul','score','score_roll']].plot(\n secondary_y= ['score'],figsize=(20,6),style = ['b','k','k-.'], \n title= str(ticker)+' Cumulative Returns and Robustness Score: cumulative & rolling '+ str(window)+' days')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8b659f5eebe92499581a3c414d5a2468dd6972
| 11,129 |
ipynb
|
Jupyter Notebook
|
4_boucles_tests.ipynb
|
pierrepo/ibi-python
|
63bc65b86800d975afafe1d0663670e282aee7c3
|
[
"BSD-3-Clause"
] | 1 |
2021-03-15T08:07:39.000Z
|
2021-03-15T08:07:39.000Z
|
4_boucles_tests.ipynb
|
pierrepo/ibi-python
|
63bc65b86800d975afafe1d0663670e282aee7c3
|
[
"BSD-3-Clause"
] | null | null | null |
4_boucles_tests.ipynb
|
pierrepo/ibi-python
|
63bc65b86800d975afafe1d0663670e282aee7c3
|
[
"BSD-3-Clause"
] | null | null | null | 21.157795 | 123 | 0.494025 |
[
[
[
"# Boucles\n\nhttps://python.sdv.univ-paris-diderot.fr/05_boucles_comparaisons/\n\nRépéter des actions\n\n## Itération sur les éléments d'une liste",
"_____no_output_____"
]
],
[
[
"placard = [\"farine\", \"oeufs\", \"lait\", \"sucre\"]\nfor ingredient in placard:\n print(ingredient)",
"_____no_output_____"
]
],
[
[
"Remarques :\n\n- La variable *ingredient* est appelée *variable d'itération* et change de valeur à chaque itération de la boucle.\n- La ligne débutant par `for` se termine toujours par `:`\n- Le bloc d'instructions `print(ingredient)` est indenté : décalage vers la droite du contenu du bloc d'instructions.",
"_____no_output_____"
]
],
[
[
"placard = [\"farine\", \"oeufs\", \"lait\", \"sucre\"]\n\nfor ingredient in placard:\n print(\"J'ajoute un ingrédient :\")\n print(ingredient)\nprint(\"Les crèpes sont prêtes !\")",
"_____no_output_____"
]
],
[
[
"Ici, le bloc d'instructions de la boucle `for` est composé des 2 instructions :\n```\nprint(\"J'ajoute un ingrédient :\")\nprint(ingredient)\n```\n\nL'instruction `print(\"Les crèpes sont prêtes !\")` est en dehors du bloc d'instructions.",
"_____no_output_____"
],
[
"## Itération sur les caractères d'une chaîne de caractères",
"_____no_output_____"
]
],
[
[
"sequence = \"ATCG\"\n\nfor base in sequence:\n print(base)",
"_____no_output_____"
],
[
"sequence = \"ATCG\"\n\nfor base in sequence:\n print(\"La base est : {}\".format(base))",
"_____no_output_____"
]
],
[
[
"# Tests\n\nhttps://python.sdv.univ-paris-diderot.fr/06_tests/\n\nPrendre des décisions",
"_____no_output_____"
]
],
[
[
"nombre = 2\n\nif nombre == 2:\n print(\"Gagné !\")",
"_____no_output_____"
]
],
[
[
"Remarques :\n\n- `:` après `if`\n- Un bloc d'instructions après `if`",
"_____no_output_____"
],
[
"## Tests à deux cas",
"_____no_output_____"
]
],
[
[
"nombre = 2\n\nif nombre == 2:\n print(\"Gagné !\")\nelse:\n print(\"Perdu !\")",
"_____no_output_____"
]
],
[
[
"Remarques :\n\n- `:` après `if` et `else`\n- Un bloc d'instructions après `if`\n- Un bloc d'instructions après `else`",
"_____no_output_____"
],
[
"## Tests à plusieurs cas",
"_____no_output_____"
]
],
[
[
"base = \"T\"",
"_____no_output_____"
],
[
"if base == \"A\":\n print(\"Choix d'une adénine\")\nelif base == \"T\":\n print(\"Choix d'une thymine\")\nelif base == \"C\":\n print(\"Choix d'une cytosine\")\nelif base == \"G\":\n print(\"Choix d'une guanine\")",
"_____no_output_____"
]
],
[
[
"On peut également définir un cas « par défaut » avec `else` :",
"_____no_output_____"
]
],
[
[
"base = \"P\"\n\nif base == \"A\":\n print(\"Choix d'une adénine\")\nelif base == \"T\":\n print(\"Choix d'une thymine\")\nelif base == \"C\":\n print(\"Choix d'une cytosine\")\nelif base == \"G\":\n print(\"Choix d'une guanine\")\nelse:\n print(\"Révise ta biologie !\")",
"_____no_output_____"
]
],
[
[
"## Tirage aléatoire",
"_____no_output_____"
]
],
[
[
"import random",
"_____no_output_____"
],
[
"random.choice([\"Sandra\", \"Julie\", \"Magali\", \"Benoist\", \"Hubert\"])",
"_____no_output_____"
],
[
"base = random.choice([\"A\", \"T\", \"C\", \"G\"])\n\nif base == \"A\":\n print(\"Choix d'une adénine\")\nelif base == \"T\":\n print(\"Choix d'une thymine\")\nelif base == \"C\":\n print(\"Choix d'une cytosine\")\nelif base == \"G\":\n print(\"Choix d'une guanine\")",
"_____no_output_____"
]
],
[
[
"Remarques :\n\n- `:` après `if` et `elif`\n- Un bloc d'instructions après `if`\n- Un bloc d'instructions après `elif`",
"_____no_output_____"
],
[
"## Attention à l'indentation !",
"_____no_output_____"
]
],
[
[
"nombres = [4, 5, 6]\n\nfor nb in nombres:\n if nb == 5:\n print(\"Le test est vrai\")\n print(\"car la variable nb vaut {}\".format(nb))",
"_____no_output_____"
],
[
"nombres = [4, 5, 6]\n\nfor nb in nombres:\n if nb == 5:\n print(\"Le test est vrai\")\n print(\"car la variable nb vaut {}\".format(nb))",
"_____no_output_____"
]
],
[
[
"# Exercices",
"_____no_output_____"
],
[
"## Notes d'un étudiant\n\nVoici les notes d'un étudiant :\n",
"_____no_output_____"
]
],
[
[
"notes = [14, 9, 6, 8, 12]",
"_____no_output_____"
]
],
[
[
"Calculez la moyenne de ces notes.",
"_____no_output_____"
],
[
"Utilisez l'écriture formatée pour afficher la valeur de la moyenne avec deux décimales.",
"_____no_output_____"
],
[
"## Séquence complémentaire\n\nLa liste ci-dessous représente la séquence d'un brin d'ADN :",
"_____no_output_____"
]
],
[
[
"sequence = [\"A\",\"C\",\"G\",\"T\",\"T\",\"A\",\"G\",\"C\",\"T\",\"A\",\"A\",\"C\",\"G\"]",
"_____no_output_____"
]
],
[
[
"Créez un code qui transforme cette séquence en sa séquence complémentaire.\n\nRappel : la séquence complémentaire s'obtient en remplaçant A par T, T par A, C par G et G par C.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a8b69766db6510005051a915ea09061892c48d6
| 47,490 |
ipynb
|
Jupyter Notebook
|
hierarchicalclustering.ipynb
|
abegpatel/Hierarchical-Clustering
|
a49dc4aabd23e5937fe29f6ae44a3b62906a88d4
|
[
"MIT"
] | 1 |
2021-06-12T06:56:13.000Z
|
2021-06-12T06:56:13.000Z
|
hierarchicalclustering.ipynb
|
abegpatel/Hierarchical-Clustering
|
a49dc4aabd23e5937fe29f6ae44a3b62906a88d4
|
[
"MIT"
] | null | null | null |
hierarchicalclustering.ipynb
|
abegpatel/Hierarchical-Clustering
|
a49dc4aabd23e5937fe29f6ae44a3b62906a88d4
|
[
"MIT"
] | null | null | null | 416.578947 | 26,120 | 0.938008 |
[
[
[
"# Hierarchical Clustering\n\n# Importing the libraries\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\n\n# Importing the dataset\ndataset = pd.read_csv('Hierarchical_Clustering/Mall_Customers.csv')\nX = dataset.iloc[:, [3, 4]].values\n# y = dataset.iloc[:, 3].values\n\n# Splitting the dataset into the Training set and Test set\n\"\"\"from sklearn.cross_validation import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 0)\"\"\"\n\n# Feature Scaling\n\"\"\"from sklearn.preprocessing import StandardScaler\nsc_X = StandardScaler()\nX_train = sc_X.fit_transform(X_train)\nX_test = sc_X.transform(X_test)\nsc_y = StandardScaler()\ny_train = sc_y.fit_transform(y_train)\"\"\"\n\n# Using the dendrogram to find the optimal number of clusters\nimport scipy.cluster.hierarchy as sch\ndendrogram = sch.dendrogram(sch.linkage(X, method = 'ward'))\nplt.title('Dendrogram')\nplt.xlabel('Customers')\nplt.ylabel('Euclidean distances')\nplt.show()\n\n# Fitting Hierarchical Clustering to the dataset\nfrom sklearn.cluster import AgglomerativeClustering\nhc = AgglomerativeClustering(n_clusters = 5, affinity = 'euclidean', linkage = 'ward')\ny_hc = hc.fit_predict(X)\n\n# Visualising the clusters\nplt.scatter(X[y_hc == 0, 0], X[y_hc == 0, 1], s = 100, c = 'red', label = 'Cluster 1')\nplt.scatter(X[y_hc == 1, 0], X[y_hc == 1, 1], s = 100, c = 'blue', label = 'Cluster 2')\nplt.scatter(X[y_hc == 2, 0], X[y_hc == 2, 1], s = 100, c = 'green', label = 'Cluster 3')\nplt.scatter(X[y_hc == 3, 0], X[y_hc == 3, 1], s = 100, c = 'cyan', label = 'Cluster 4')\nplt.scatter(X[y_hc == 4, 0], X[y_hc == 4, 1], s = 100, c = 'magenta', label = 'Cluster 5')\nplt.title('Clusters of customers')\nplt.xlabel('Annual Income (k$)')\nplt.ylabel('Spending Score (1-100)')\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
4a8b72099398d0c45af2be43fdecc9c7cdd0580d
| 44,587 |
ipynb
|
Jupyter Notebook
|
Notebooks/cifar-10-senet18-mish.ipynb
|
DEVESHTARASIA/Mish
|
21ee1ce37db046ab31de8fd72ecea0156b1c8f8c
|
[
"MIT"
] | 3 |
2020-02-07T08:26:23.000Z
|
2020-05-18T06:30:00.000Z
|
Notebooks/cifar-10-senet18-mish.ipynb
|
DEVESHTARASIA/Mish
|
21ee1ce37db046ab31de8fd72ecea0156b1c8f8c
|
[
"MIT"
] | null | null | null |
Notebooks/cifar-10-senet18-mish.ipynb
|
DEVESHTARASIA/Mish
|
21ee1ce37db046ab31de8fd72ecea0156b1c8f8c
|
[
"MIT"
] | null | null | null | 63.60485 | 469 | 0.595981 |
[
[
[
"import numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\n\nimport os\nprint(os.listdir(\"../input\"))\n\nimport time\n\n# import pytorch\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.optim import SGD,Adam,lr_scheduler\nfrom torch.utils.data import random_split\nimport torchvision\nfrom torchvision import transforms, datasets\nfrom torch.utils.data import DataLoader",
"['cifar-10-python.tar.gz']\n"
],
[
"# define transformations for train\ntrain_transform = transforms.Compose([\n transforms.RandomHorizontalFlip(p=.40),\n transforms.RandomRotation(30),\n transforms.ToTensor(),\n transforms.Normalize([0.5, 0.5, 0.5], [0.5, 0.5, 0.5])])\n\n# define transformations for test\ntest_transform = transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize([0.5, 0.5, 0.5], [0.5, 0.5, 0.5])])\n\n# define training dataloader\ndef get_training_dataloader(train_transform, batch_size=128, num_workers=0, shuffle=True):\n \"\"\" return training dataloader\n Args:\n train_transform: transfroms for train dataset\n path: path to cifar100 training python dataset\n batch_size: dataloader batchsize\n num_workers: dataloader num_works\n shuffle: whether to shuffle \n Returns: train_data_loader:torch dataloader object\n \"\"\"\n\n transform_train = train_transform\n cifar10_training = torchvision.datasets.CIFAR10(root='.', train=True, download=True, transform=transform_train)\n cifar10_training_loader = DataLoader(\n cifar10_training, shuffle=shuffle, num_workers=num_workers, batch_size=batch_size)\n\n return cifar10_training_loader\n\n# define test dataloader\ndef get_testing_dataloader(test_transform, batch_size=128, num_workers=0, shuffle=True):\n \"\"\" return training dataloader\n Args:\n test_transform: transforms for test dataset\n path: path to cifar100 test python dataset\n batch_size: dataloader batchsize\n num_workers: dataloader num_works\n shuffle: whether to shuffle \n Returns: cifar100_test_loader:torch dataloader object\n \"\"\"\n\n transform_test = test_transform\n cifar10_test = torchvision.datasets.CIFAR10(root='.', train=False, download=True, transform=transform_test)\n cifar10_test_loader = DataLoader(\n cifar10_test, shuffle=shuffle, num_workers=num_workers, batch_size=batch_size)\n\n return cifar10_test_loader",
"_____no_output_____"
],
[
"# implement mish activation function\ndef f_mish(input):\n '''\n Applies the mish function element-wise:\n mish(x) = x * tanh(softplus(x)) = x * tanh(ln(1 + exp(x)))\n '''\n return input * torch.tanh(F.softplus(input))\n\n# implement class wrapper for mish activation function\nclass mish(nn.Module):\n '''\n Applies the mish function element-wise:\n mish(x) = x * tanh(softplus(x)) = x * tanh(ln(1 + exp(x)))\n\n Shape:\n - Input: (N, *) where * means, any number of additional\n dimensions\n - Output: (N, *), same shape as the input\n\n Examples:\n >>> m = mish()\n >>> input = torch.randn(2)\n >>> output = m(input)\n\n '''\n def __init__(self):\n '''\n Init method.\n '''\n super().__init__()\n\n def forward(self, input):\n '''\n Forward pass of the function.\n '''\n return f_mish(input)",
"_____no_output_____"
],
[
"# implement swish activation function\ndef f_swish(input):\n '''\n Applies the swish function element-wise:\n swish(x) = x * sigmoid(x)\n '''\n return input * torch.sigmoid(input)\n\n# implement class wrapper for swish activation function\nclass swish(nn.Module):\n '''\n Applies the swish function element-wise:\n swish(x) = x * sigmoid(x)\n\n Shape:\n - Input: (N, *) where * means, any number of additional\n dimensions\n - Output: (N, *), same shape as the input\n\n Examples:\n >>> m = swish()\n >>> input = torch.randn(2)\n >>> output = m(input)\n\n '''\n def __init__(self):\n '''\n Init method.\n '''\n super().__init__()\n\n def forward(self, input):\n '''\n Forward pass of the function.\n '''\n return f_swish(input)",
"_____no_output_____"
],
[
"class BasicResidualSEBlock(nn.Module):\n\n expansion = 1\n\n def __init__(self, in_channels, out_channels, stride, r=16, activation = 'relu'):\n super().__init__()\n \n if activation == 'relu':\n f_activation = nn.ReLU(inplace=True)\n self.activation = F.relu\n \n if activation == 'swish':\n f_activation = swish()\n self.activation = f_swish\n \n if activation == 'mish':\n f_activation = mish()\n self.activation = f_mish\n\n self.residual = nn.Sequential(\n nn.Conv2d(in_channels, out_channels, 3, stride=stride, padding=1),\n nn.BatchNorm2d(out_channels),\n f_activation,\n \n nn.Conv2d(out_channels, out_channels * self.expansion, 3, padding=1),\n nn.BatchNorm2d(out_channels * self.expansion),\n f_activation\n )\n\n self.shortcut = nn.Sequential()\n if stride != 1 or in_channels != out_channels * self.expansion:\n self.shortcut = nn.Sequential(\n nn.Conv2d(in_channels, out_channels * self.expansion, 1, stride=stride),\n nn.BatchNorm2d(out_channels * self.expansion)\n )\n \n self.squeeze = nn.AdaptiveAvgPool2d(1)\n self.excitation = nn.Sequential(\n nn.Linear(out_channels * self.expansion, out_channels * self.expansion // r),\n f_activation,\n nn.Linear(out_channels * self.expansion // r, out_channels * self.expansion),\n nn.Sigmoid()\n )\n\n def forward(self, x):\n shortcut = self.shortcut(x)\n residual = self.residual(x)\n\n squeeze = self.squeeze(residual)\n squeeze = squeeze.view(squeeze.size(0), -1)\n excitation = self.excitation(squeeze)\n excitation = excitation.view(residual.size(0), residual.size(1), 1, 1)\n\n x = residual * excitation.expand_as(residual) + shortcut\n\n return self.activation(x)\n\nclass BottleneckResidualSEBlock(nn.Module):\n\n expansion = 4\n\n def __init__(self, in_channels, out_channels, stride, r=16, activation = 'relu'):\n super().__init__()\n \n if activation == 'relu':\n f_activation = nn.ReLU(inplace=True)\n self.activation = F.relu\n \n if activation == 'swish':\n f_activation = swish()\n self.activation = f_swish\n \n if activation == 'mish':\n f_activation = mish()\n self.activation = f_mish\n\n self.residual = nn.Sequential(\n nn.Conv2d(in_channels, out_channels, 1),\n nn.BatchNorm2d(out_channels),\n f_activation,\n\n nn.Conv2d(out_channels, out_channels, 3, stride=stride, padding=1),\n nn.BatchNorm2d(out_channels),\n f_activation,\n\n nn.Conv2d(out_channels, out_channels * self.expansion, 1),\n nn.BatchNorm2d(out_channels * self.expansion),\n f_activation\n )\n\n self.squeeze = nn.AdaptiveAvgPool2d(1)\n self.excitation = nn.Sequential(\n nn.Linear(out_channels * self.expansion, out_channels * self.expansion // r),\n f_activation,\n nn.Linear(out_channels * self.expansion // r, out_channels * self.expansion),\n nn.Sigmoid()\n )\n\n self.shortcut = nn.Sequential()\n if stride != 1 or in_channels != out_channels * self.expansion:\n self.shortcut = nn.Sequential(\n nn.Conv2d(in_channels, out_channels * self.expansion, 1, stride=stride),\n nn.BatchNorm2d(out_channels * self.expansion)\n )\n\n def forward(self, x):\n\n shortcut = self.shortcut(x)\n\n residual = self.residual(x)\n squeeze = self.squeeze(residual)\n squeeze = squeeze.view(squeeze.size(0), -1)\n excitation = self.excitation(squeeze)\n excitation = excitation.view(residual.size(0), residual.size(1), 1, 1)\n\n x = residual * excitation.expand_as(residual) + shortcut\n\n return self.activation(x)\n\nclass SEResNet(nn.Module):\n\n def __init__(self, block, block_num, class_num=10, activation = 'relu'):\n super().__init__()\n\n self.in_channels = 64\n \n if activation == 'relu':\n f_activation = nn.ReLU(inplace=True)\n self.activation = F.relu\n \n if activation == 'swish':\n f_activation = swish()\n self.activation = f_swish\n \n if activation == 'mish':\n f_activation = mish()\n self.activation = f_mish\n\n self.pre = nn.Sequential(\n nn.Conv2d(3, 64, 3, padding=1),\n nn.BatchNorm2d(64),\n f_activation\n )\n\n self.stage1 = self._make_stage(block, block_num[0], 64, 1, activation = activation)\n self.stage2 = self._make_stage(block, block_num[1], 128, 2, activation = activation)\n self.stage3 = self._make_stage(block, block_num[2], 256, 2, activation = activation)\n self.stage4 = self._make_stage(block, block_num[3], 516, 2, activation = activation)\n\n self.linear = nn.Linear(self.in_channels, class_num)\n \n def forward(self, x):\n x = self.pre(x)\n\n x = self.stage1(x)\n x = self.stage2(x)\n x = self.stage3(x)\n x = self.stage4(x)\n\n x = F.adaptive_avg_pool2d(x, 1)\n x = x.view(x.size(0), -1)\n\n x = self.linear(x)\n\n return x\n\n \n def _make_stage(self, block, num, out_channels, stride, activation = 'relu'):\n\n layers = []\n layers.append(block(self.in_channels, out_channels, stride, activation = activation))\n self.in_channels = out_channels * block.expansion\n\n while num - 1:\n layers.append(block(self.in_channels, out_channels, 1, activation = activation))\n num -= 1\n \n return nn.Sequential(*layers)\n \ndef seresnet18(activation = 'relu'):\n return SEResNet(BasicResidualSEBlock, [2, 2, 2, 2], activation = activation)\n\ndef seresnet34(activation = 'relu'):\n return SEResNet(BasicResidualSEBlock, [3, 4, 6, 3], activation = activation)\n\ndef seresnet50(activation = 'relu'):\n return SEResNet(BottleneckResidualSEBlock, [3, 4, 6, 3], activation = activation)\n\ndef seresnet101(activation = 'relu'):\n return SEResNet(BottleneckResidualSEBlock, [3, 4, 23, 3], activation = activation)\n\ndef seresnet152(activation = 'relu'):\n return SEResNet(BottleneckResidualSEBlock, [3, 8, 36, 3], activation = activation)",
"_____no_output_____"
],
[
"trainloader = get_training_dataloader(train_transform)\ntestloader = get_testing_dataloader(test_transform)",
"0it [00:00, ?it/s]"
],
[
"epochs = 100\nbatch_size = 128\nlearning_rate = 0.001\ndevice = torch.device('cuda:0' if torch.cuda.is_available() else \"cpu\")\ndevice",
"_____no_output_____"
],
[
"model = seresnet18(activation = 'mish')",
"_____no_output_____"
],
[
"# set loss function\ncriterion = nn.CrossEntropyLoss()\n\n# set optimizer, only train the classifier parameters, feature parameters are frozen\noptimizer = Adam(model.parameters(), lr=learning_rate)",
"_____no_output_____"
],
[
"train_stats = pd.DataFrame(columns = ['Epoch', 'Time per epoch', 'Avg time per step', 'Train loss', 'Train accuracy', 'Train top-3 accuracy','Test loss', 'Test accuracy', 'Test top-3 accuracy']) ",
"_____no_output_____"
],
[
"#train the model\nmodel.to(device)\n\nsteps = 0\nrunning_loss = 0\nfor epoch in range(epochs):\n \n since = time.time()\n \n train_accuracy = 0\n top3_train_accuracy = 0 \n for inputs, labels in trainloader:\n steps += 1\n # Move input and label tensors to the default device\n inputs, labels = inputs.to(device), labels.to(device)\n \n optimizer.zero_grad()\n \n logps = model.forward(inputs)\n loss = criterion(logps, labels)\n loss.backward()\n optimizer.step()\n\n running_loss += loss.item()\n \n # calculate train top-1 accuracy\n ps = torch.exp(logps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n train_accuracy += torch.mean(equals.type(torch.FloatTensor)).item()\n \n # Calculate train top-3 accuracy\n np_top3_class = ps.topk(3, dim=1)[1].cpu().numpy()\n target_numpy = labels.cpu().numpy()\n top3_train_accuracy += np.mean([1 if target_numpy[i] in np_top3_class[i] else 0 for i in range(0, len(target_numpy))])\n \n time_elapsed = time.time() - since\n \n test_loss = 0\n test_accuracy = 0\n top3_test_accuracy = 0\n model.eval()\n with torch.no_grad():\n for inputs, labels in testloader:\n inputs, labels = inputs.to(device), labels.to(device)\n logps = model.forward(inputs)\n batch_loss = criterion(logps, labels)\n\n test_loss += batch_loss.item()\n\n # Calculate test top-1 accuracy\n ps = torch.exp(logps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n test_accuracy += torch.mean(equals.type(torch.FloatTensor)).item()\n \n # Calculate test top-3 accuracy\n np_top3_class = ps.topk(3, dim=1)[1].cpu().numpy()\n target_numpy = labels.cpu().numpy()\n top3_test_accuracy += np.mean([1 if target_numpy[i] in np_top3_class[i] else 0 for i in range(0, len(target_numpy))])\n\n print(f\"Epoch {epoch+1}/{epochs}.. \"\n f\"Time per epoch: {time_elapsed:.4f}.. \"\n f\"Average time per step: {time_elapsed/len(trainloader):.4f}.. \"\n f\"Train loss: {running_loss/len(trainloader):.4f}.. \"\n f\"Train accuracy: {train_accuracy/len(trainloader):.4f}.. \"\n f\"Top-3 train accuracy: {top3_train_accuracy/len(trainloader):.4f}.. \"\n f\"Test loss: {test_loss/len(testloader):.4f}.. \"\n f\"Test accuracy: {test_accuracy/len(testloader):.4f}.. \"\n f\"Top-3 test accuracy: {top3_test_accuracy/len(testloader):.4f}\")\n\n train_stats = train_stats.append({'Epoch': epoch, 'Time per epoch':time_elapsed, 'Avg time per step': time_elapsed/len(trainloader), 'Train loss' : running_loss/len(trainloader), 'Train accuracy': train_accuracy/len(trainloader), 'Train top-3 accuracy':top3_train_accuracy/len(trainloader),'Test loss' : test_loss/len(testloader), 'Test accuracy': test_accuracy/len(testloader), 'Test top-3 accuracy':top3_test_accuracy/len(testloader)}, ignore_index=True)\n\n running_loss = 0\n model.train()",
"Epoch 1/100.. Time per epoch: 59.5058.. Average time per step: 0.1522.. Train loss: 1.4410.. Train accuracy: 0.4719.. Top-3 train accuracy: 0.7975.. Test loss: 1.1111.. Test accuracy: 0.5979.. Top-3 test accuracy: 0.8827\nEpoch 2/100.. Time per epoch: 58.5211.. Average time per step: 0.1497.. Train loss: 1.0474.. Train accuracy: 0.6266.. Top-3 train accuracy: 0.8901.. Test loss: 0.8924.. Test accuracy: 0.6863.. Top-3 test accuracy: 0.9165\nEpoch 3/100.. Time per epoch: 58.5008.. Average time per step: 0.1496.. Train loss: 0.8766.. Train accuracy: 0.6898.. Top-3 train accuracy: 0.9190.. Test loss: 0.7806.. Test accuracy: 0.7277.. Top-3 test accuracy: 0.9346\nEpoch 4/100.. Time per epoch: 58.4295.. Average time per step: 0.1494.. Train loss: 0.7576.. Train accuracy: 0.7317.. Top-3 train accuracy: 0.9367.. Test loss: 0.6503.. Test accuracy: 0.7715.. Top-3 test accuracy: 0.9529\nEpoch 5/100.. Time per epoch: 58.4691.. Average time per step: 0.1495.. Train loss: 0.6693.. Train accuracy: 0.7663.. Top-3 train accuracy: 0.9485.. Test loss: 0.5966.. Test accuracy: 0.7917.. Top-3 test accuracy: 0.9598\nEpoch 6/100.. Time per epoch: 58.0231.. Average time per step: 0.1484.. Train loss: 0.5977.. Train accuracy: 0.7916.. Top-3 train accuracy: 0.9565.. Test loss: 0.5645.. Test accuracy: 0.8021.. Top-3 test accuracy: 0.9642\nEpoch 7/100.. Time per epoch: 58.0686.. Average time per step: 0.1485.. Train loss: 0.5437.. Train accuracy: 0.8090.. Top-3 train accuracy: 0.9631.. Test loss: 0.4856.. Test accuracy: 0.8345.. Top-3 test accuracy: 0.9702\nEpoch 8/100.. Time per epoch: 58.1408.. Average time per step: 0.1487.. Train loss: 0.5034.. Train accuracy: 0.8250.. Top-3 train accuracy: 0.9669.. Test loss: 0.4975.. Test accuracy: 0.8338.. Top-3 test accuracy: 0.9693\nEpoch 9/100.. Time per epoch: 58.0765.. Average time per step: 0.1485.. Train loss: 0.4637.. Train accuracy: 0.8369.. Top-3 train accuracy: 0.9714.. Test loss: 0.4508.. Test accuracy: 0.8477.. Top-3 test accuracy: 0.9737\nEpoch 10/100.. Time per epoch: 58.1137.. Average time per step: 0.1486.. Train loss: 0.4342.. Train accuracy: 0.8477.. Top-3 train accuracy: 0.9747.. Test loss: 0.4432.. Test accuracy: 0.8466.. Top-3 test accuracy: 0.9739\nEpoch 11/100.. Time per epoch: 58.1785.. Average time per step: 0.1488.. Train loss: 0.4027.. Train accuracy: 0.8582.. Top-3 train accuracy: 0.9769.. Test loss: 0.4274.. Test accuracy: 0.8584.. Top-3 test accuracy: 0.9770\nEpoch 12/100.. Time per epoch: 58.2584.. Average time per step: 0.1490.. Train loss: 0.3746.. Train accuracy: 0.8684.. Top-3 train accuracy: 0.9801.. Test loss: 0.4125.. Test accuracy: 0.8625.. Top-3 test accuracy: 0.9777\nEpoch 13/100.. Time per epoch: 58.2688.. Average time per step: 0.1490.. Train loss: 0.3531.. Train accuracy: 0.8751.. Top-3 train accuracy: 0.9819.. Test loss: 0.4043.. Test accuracy: 0.8666.. Top-3 test accuracy: 0.9798\nEpoch 14/100.. Time per epoch: 58.3006.. Average time per step: 0.1491.. Train loss: 0.3286.. Train accuracy: 0.8850.. Top-3 train accuracy: 0.9837.. Test loss: 0.3827.. Test accuracy: 0.8715.. Top-3 test accuracy: 0.9799\nEpoch 15/100.. Time per epoch: 58.2233.. Average time per step: 0.1489.. Train loss: 0.3078.. Train accuracy: 0.8920.. Top-3 train accuracy: 0.9854.. Test loss: 0.4396.. Test accuracy: 0.8582.. Top-3 test accuracy: 0.9757\nEpoch 16/100.. Time per epoch: 58.0980.. Average time per step: 0.1486.. Train loss: 0.2919.. Train accuracy: 0.8972.. Top-3 train accuracy: 0.9872.. Test loss: 0.3837.. Test accuracy: 0.8750.. Top-3 test accuracy: 0.9812\nEpoch 17/100.. Time per epoch: 58.2424.. Average time per step: 0.1490.. Train loss: 0.2716.. Train accuracy: 0.9031.. Top-3 train accuracy: 0.9888.. Test loss: 0.3785.. Test accuracy: 0.8745.. Top-3 test accuracy: 0.9818\nEpoch 18/100.. Time per epoch: 58.3789.. Average time per step: 0.1493.. Train loss: 0.2576.. Train accuracy: 0.9101.. Top-3 train accuracy: 0.9896.. Test loss: 0.3831.. Test accuracy: 0.8791.. Top-3 test accuracy: 0.9806\nEpoch 19/100.. Time per epoch: 58.2301.. Average time per step: 0.1489.. Train loss: 0.2410.. Train accuracy: 0.9148.. Top-3 train accuracy: 0.9913.. Test loss: 0.3662.. Test accuracy: 0.8826.. Top-3 test accuracy: 0.9820\nEpoch 20/100.. Time per epoch: 58.4980.. Average time per step: 0.1496.. Train loss: 0.2275.. Train accuracy: 0.9191.. Top-3 train accuracy: 0.9917.. Test loss: 0.3608.. Test accuracy: 0.8839.. Top-3 test accuracy: 0.9832\nEpoch 21/100.. Time per epoch: 58.4391.. Average time per step: 0.1495.. Train loss: 0.2108.. Train accuracy: 0.9258.. Top-3 train accuracy: 0.9935.. Test loss: 0.3808.. Test accuracy: 0.8841.. Top-3 test accuracy: 0.9826\nEpoch 22/100.. Time per epoch: 58.6067.. Average time per step: 0.1499.. Train loss: 0.2002.. Train accuracy: 0.9291.. Top-3 train accuracy: 0.9934.. Test loss: 0.3754.. Test accuracy: 0.8851.. Top-3 test accuracy: 0.9840\nEpoch 23/100.. Time per epoch: 58.3566.. Average time per step: 0.1492.. Train loss: 0.1872.. Train accuracy: 0.9344.. Top-3 train accuracy: 0.9942.. Test loss: 0.3650.. Test accuracy: 0.8903.. Top-3 test accuracy: 0.9841\nEpoch 24/100.. Time per epoch: 58.4070.. Average time per step: 0.1494.. Train loss: 0.1795.. Train accuracy: 0.9364.. Top-3 train accuracy: 0.9946.. Test loss: 0.3997.. Test accuracy: 0.8849.. Top-3 test accuracy: 0.9781\nEpoch 25/100.. Time per epoch: 57.8835.. Average time per step: 0.1480.. Train loss: 0.1687.. Train accuracy: 0.9405.. Top-3 train accuracy: 0.9954.. Test loss: 0.3901.. Test accuracy: 0.8852.. Top-3 test accuracy: 0.9830\nEpoch 26/100.. Time per epoch: 58.0625.. Average time per step: 0.1485.. Train loss: 0.1586.. Train accuracy: 0.9443.. Top-3 train accuracy: 0.9957.. Test loss: 0.4094.. Test accuracy: 0.8796.. Top-3 test accuracy: 0.9830\nEpoch 27/100.. Time per epoch: 58.2370.. Average time per step: 0.1489.. Train loss: 0.1524.. Train accuracy: 0.9470.. Top-3 train accuracy: 0.9957.. Test loss: 0.3868.. Test accuracy: 0.8846.. Top-3 test accuracy: 0.9838\nEpoch 28/100.. Time per epoch: 57.9287.. Average time per step: 0.1482.. Train loss: 0.1429.. Train accuracy: 0.9490.. Top-3 train accuracy: 0.9964.. Test loss: 0.3967.. Test accuracy: 0.8890.. Top-3 test accuracy: 0.9826\nEpoch 29/100.. Time per epoch: 57.8662.. Average time per step: 0.1480.. Train loss: 0.1375.. Train accuracy: 0.9510.. Top-3 train accuracy: 0.9971.. Test loss: 0.3824.. Test accuracy: 0.8901.. Top-3 test accuracy: 0.9856\nEpoch 30/100.. Time per epoch: 57.8405.. Average time per step: 0.1479.. Train loss: 0.1247.. Train accuracy: 0.9558.. Top-3 train accuracy: 0.9971.. Test loss: 0.3922.. Test accuracy: 0.8924.. Top-3 test accuracy: 0.9844\nEpoch 31/100.. Time per epoch: 58.0472.. Average time per step: 0.1485.. Train loss: 0.1211.. Train accuracy: 0.9567.. Top-3 train accuracy: 0.9976.. Test loss: 0.4066.. Test accuracy: 0.8900.. Top-3 test accuracy: 0.9833\nEpoch 32/100.. Time per epoch: 58.0543.. Average time per step: 0.1485.. Train loss: 0.1149.. Train accuracy: 0.9600.. Top-3 train accuracy: 0.9974.. Test loss: 0.3933.. Test accuracy: 0.8939.. Top-3 test accuracy: 0.9845\nEpoch 33/100.. Time per epoch: 57.9562.. Average time per step: 0.1482.. Train loss: 0.1116.. Train accuracy: 0.9607.. Top-3 train accuracy: 0.9980.. Test loss: 0.3912.. Test accuracy: 0.8937.. Top-3 test accuracy: 0.9850\nEpoch 34/100.. Time per epoch: 57.9038.. Average time per step: 0.1481.. Train loss: 0.1007.. Train accuracy: 0.9650.. Top-3 train accuracy: 0.9981.. Test loss: 0.4046.. Test accuracy: 0.8919.. Top-3 test accuracy: 0.9843\nEpoch 35/100.. Time per epoch: 58.0285.. Average time per step: 0.1484.. Train loss: 0.1025.. Train accuracy: 0.9644.. Top-3 train accuracy: 0.9982.. Test loss: 0.4032.. Test accuracy: 0.8952.. Top-3 test accuracy: 0.9846\nEpoch 36/100.. Time per epoch: 58.0629.. Average time per step: 0.1485.. Train loss: 0.0977.. Train accuracy: 0.9651.. Top-3 train accuracy: 0.9986.. Test loss: 0.4092.. Test accuracy: 0.8944.. Top-3 test accuracy: 0.9836\nEpoch 37/100.. Time per epoch: 58.1191.. Average time per step: 0.1486.. Train loss: 0.0890.. Train accuracy: 0.9687.. Top-3 train accuracy: 0.9987.. Test loss: 0.4160.. Test accuracy: 0.8934.. Top-3 test accuracy: 0.9836\nEpoch 38/100.. Time per epoch: 57.9099.. Average time per step: 0.1481.. Train loss: 0.0885.. Train accuracy: 0.9680.. Top-3 train accuracy: 0.9987.. Test loss: 0.4340.. Test accuracy: 0.8904.. Top-3 test accuracy: 0.9847\nEpoch 39/100.. Time per epoch: 57.9002.. Average time per step: 0.1481.. Train loss: 0.0854.. Train accuracy: 0.9699.. Top-3 train accuracy: 0.9988.. Test loss: 0.4269.. Test accuracy: 0.8930.. Top-3 test accuracy: 0.9844\nEpoch 40/100.. Time per epoch: 57.7297.. Average time per step: 0.1476.. Train loss: 0.0851.. Train accuracy: 0.9698.. Top-3 train accuracy: 0.9990.. Test loss: 0.4152.. Test accuracy: 0.8968.. Top-3 test accuracy: 0.9842\nEpoch 41/100.. Time per epoch: 57.8792.. Average time per step: 0.1480.. Train loss: 0.0754.. Train accuracy: 0.9739.. Top-3 train accuracy: 0.9989.. Test loss: 0.4398.. Test accuracy: 0.8941.. Top-3 test accuracy: 0.9831\nEpoch 42/100.. Time per epoch: 57.6727.. Average time per step: 0.1475.. Train loss: 0.0767.. Train accuracy: 0.9727.. Top-3 train accuracy: 0.9989.. Test loss: 0.4618.. Test accuracy: 0.8874.. Top-3 test accuracy: 0.9828\nEpoch 43/100.. Time per epoch: 57.7151.. Average time per step: 0.1476.. Train loss: 0.0712.. Train accuracy: 0.9753.. Top-3 train accuracy: 0.9991.. Test loss: 0.4396.. Test accuracy: 0.8888.. Top-3 test accuracy: 0.9845\nEpoch 44/100.. Time per epoch: 57.8193.. Average time per step: 0.1479.. Train loss: 0.0692.. Train accuracy: 0.9754.. Top-3 train accuracy: 0.9992.. Test loss: 0.4268.. Test accuracy: 0.8986.. Top-3 test accuracy: 0.9850\nEpoch 45/100.. Time per epoch: 58.1237.. Average time per step: 0.1487.. Train loss: 0.0676.. Train accuracy: 0.9762.. Top-3 train accuracy: 0.9992.. Test loss: 0.4514.. Test accuracy: 0.8924.. Top-3 test accuracy: 0.9833\nEpoch 46/100.. Time per epoch: 58.0621.. Average time per step: 0.1485.. Train loss: 0.0666.. Train accuracy: 0.9770.. Top-3 train accuracy: 0.9991.. Test loss: 0.4390.. Test accuracy: 0.8951.. Top-3 test accuracy: 0.9843\nEpoch 47/100.. Time per epoch: 57.9843.. Average time per step: 0.1483.. Train loss: 0.0680.. Train accuracy: 0.9763.. Top-3 train accuracy: 0.9992.. Test loss: 0.4342.. Test accuracy: 0.8983.. Top-3 test accuracy: 0.9829\nEpoch 48/100.. Time per epoch: 57.8841.. Average time per step: 0.1480.. Train loss: 0.0618.. Train accuracy: 0.9782.. Top-3 train accuracy: 0.9994.. Test loss: 0.4199.. Test accuracy: 0.8967.. Top-3 test accuracy: 0.9866\nEpoch 49/100.. Time per epoch: 58.0302.. Average time per step: 0.1484.. Train loss: 0.0571.. Train accuracy: 0.9801.. Top-3 train accuracy: 0.9996.. Test loss: 0.4471.. Test accuracy: 0.8951.. Top-3 test accuracy: 0.9845\nEpoch 50/100.. Time per epoch: 57.8835.. Average time per step: 0.1480.. Train loss: 0.0586.. Train accuracy: 0.9793.. Top-3 train accuracy: 0.9992.. Test loss: 0.4552.. Test accuracy: 0.8962.. Top-3 test accuracy: 0.9838\nEpoch 51/100.. Time per epoch: 58.1047.. Average time per step: 0.1486.. Train loss: 0.0560.. Train accuracy: 0.9799.. Top-3 train accuracy: 0.9993.. Test loss: 0.4810.. Test accuracy: 0.8934.. Top-3 test accuracy: 0.9831\nEpoch 52/100.. Time per epoch: 57.9741.. Average time per step: 0.1483.. Train loss: 0.0541.. Train accuracy: 0.9812.. Top-3 train accuracy: 0.9994.. Test loss: 0.5113.. Test accuracy: 0.8890.. Top-3 test accuracy: 0.9824\nEpoch 53/100.. Time per epoch: 57.9915.. Average time per step: 0.1483.. Train loss: 0.0553.. Train accuracy: 0.9809.. Top-3 train accuracy: 0.9993.. Test loss: 0.4760.. Test accuracy: 0.8953.. Top-3 test accuracy: 0.9833\nEpoch 54/100.. Time per epoch: 58.2543.. Average time per step: 0.1490.. Train loss: 0.0533.. Train accuracy: 0.9806.. Top-3 train accuracy: 0.9995.. Test loss: 0.4760.. Test accuracy: 0.8912.. Top-3 test accuracy: 0.9816\nEpoch 55/100.. Time per epoch: 58.2874.. Average time per step: 0.1491.. Train loss: 0.0488.. Train accuracy: 0.9829.. Top-3 train accuracy: 0.9996.. Test loss: 0.4730.. Test accuracy: 0.8951.. Top-3 test accuracy: 0.9839\nEpoch 56/100.. Time per epoch: 58.4716.. Average time per step: 0.1495.. Train loss: 0.0482.. Train accuracy: 0.9832.. Top-3 train accuracy: 0.9994.. Test loss: 0.4794.. Test accuracy: 0.8935.. Top-3 test accuracy: 0.9835\nEpoch 57/100.. Time per epoch: 58.3004.. Average time per step: 0.1491.. Train loss: 0.0497.. Train accuracy: 0.9823.. Top-3 train accuracy: 0.9995.. Test loss: 0.4655.. Test accuracy: 0.8949.. Top-3 test accuracy: 0.9835\nEpoch 58/100.. Time per epoch: 58.3081.. Average time per step: 0.1491.. Train loss: 0.0492.. Train accuracy: 0.9828.. Top-3 train accuracy: 0.9996.. Test loss: 0.4524.. Test accuracy: 0.8982.. Top-3 test accuracy: 0.9835\nEpoch 59/100.. Time per epoch: 58.1601.. Average time per step: 0.1487.. Train loss: 0.0422.. Train accuracy: 0.9849.. Top-3 train accuracy: 0.9996.. Test loss: 0.4793.. Test accuracy: 0.8948.. Top-3 test accuracy: 0.9840\nEpoch 60/100.. Time per epoch: 58.6814.. Average time per step: 0.1501.. Train loss: 0.0453.. Train accuracy: 0.9842.. Top-3 train accuracy: 0.9996.. Test loss: 0.4736.. Test accuracy: 0.8952.. Top-3 test accuracy: 0.9848\nEpoch 61/100.. Time per epoch: 58.3997.. Average time per step: 0.1494.. Train loss: 0.0438.. Train accuracy: 0.9847.. Top-3 train accuracy: 0.9995.. Test loss: 0.4581.. Test accuracy: 0.8949.. Top-3 test accuracy: 0.9855\nEpoch 62/100.. Time per epoch: 58.2919.. Average time per step: 0.1491.. Train loss: 0.0433.. Train accuracy: 0.9849.. Top-3 train accuracy: 0.9996.. Test loss: 0.4634.. Test accuracy: 0.8973.. Top-3 test accuracy: 0.9843\nEpoch 63/100.. Time per epoch: 58.3452.. Average time per step: 0.1492.. Train loss: 0.0396.. Train accuracy: 0.9865.. Top-3 train accuracy: 0.9998.. Test loss: 0.4982.. Test accuracy: 0.8921.. Top-3 test accuracy: 0.9834\nEpoch 64/100.. Time per epoch: 59.3747.. Average time per step: 0.1519.. Train loss: 0.0429.. Train accuracy: 0.9846.. Top-3 train accuracy: 0.9998.. Test loss: 0.4714.. Test accuracy: 0.8915.. Top-3 test accuracy: 0.9828\nEpoch 65/100.. Time per epoch: 59.2911.. Average time per step: 0.1516.. Train loss: 0.0418.. Train accuracy: 0.9853.. Top-3 train accuracy: 0.9996.. Test loss: 0.4850.. Test accuracy: 0.8938.. Top-3 test accuracy: 0.9844\nEpoch 66/100.. Time per epoch: 59.4622.. Average time per step: 0.1521.. Train loss: 0.0385.. Train accuracy: 0.9869.. Top-3 train accuracy: 0.9996.. Test loss: 0.4844.. Test accuracy: 0.8976.. Top-3 test accuracy: 0.9845\nEpoch 67/100.. Time per epoch: 59.2766.. Average time per step: 0.1516.. Train loss: 0.0393.. Train accuracy: 0.9865.. Top-3 train accuracy: 0.9997.. Test loss: 0.4763.. Test accuracy: 0.9004.. Top-3 test accuracy: 0.9852\nEpoch 68/100.. Time per epoch: 59.3961.. Average time per step: 0.1519.. Train loss: 0.0372.. Train accuracy: 0.9875.. Top-3 train accuracy: 0.9998.. Test loss: 0.5198.. Test accuracy: 0.8917.. Top-3 test accuracy: 0.9815\nEpoch 69/100.. Time per epoch: 59.0566.. Average time per step: 0.1510.. Train loss: 0.0383.. Train accuracy: 0.9864.. Top-3 train accuracy: 0.9996.. Test loss: 0.4918.. Test accuracy: 0.8919.. Top-3 test accuracy: 0.9842\nEpoch 70/100.. Time per epoch: 58.8006.. Average time per step: 0.1504.. Train loss: 0.0380.. Train accuracy: 0.9868.. Top-3 train accuracy: 0.9998.. Test loss: 0.5179.. Test accuracy: 0.8926.. Top-3 test accuracy: 0.9837\nEpoch 71/100.. Time per epoch: 58.9499.. Average time per step: 0.1508.. Train loss: 0.0367.. Train accuracy: 0.9876.. Top-3 train accuracy: 0.9996.. Test loss: 0.4880.. Test accuracy: 0.8926.. Top-3 test accuracy: 0.9831\nEpoch 72/100.. Time per epoch: 58.7406.. Average time per step: 0.1502.. Train loss: 0.0354.. Train accuracy: 0.9882.. Top-3 train accuracy: 0.9995.. Test loss: 0.4815.. Test accuracy: 0.8993.. Top-3 test accuracy: 0.9832\nEpoch 73/100.. Time per epoch: 58.9139.. Average time per step: 0.1507.. Train loss: 0.0333.. Train accuracy: 0.9882.. Top-3 train accuracy: 0.9998.. Test loss: 0.4841.. Test accuracy: 0.8973.. Top-3 test accuracy: 0.9825\nEpoch 74/100.. Time per epoch: 59.0054.. Average time per step: 0.1509.. Train loss: 0.0346.. Train accuracy: 0.9881.. Top-3 train accuracy: 0.9998.. Test loss: 0.5128.. Test accuracy: 0.8955.. Top-3 test accuracy: 0.9825\nEpoch 75/100.. Time per epoch: 59.1839.. Average time per step: 0.1514.. Train loss: 0.0329.. Train accuracy: 0.9884.. Top-3 train accuracy: 0.9998.. Test loss: 0.5019.. Test accuracy: 0.9021.. Top-3 test accuracy: 0.9849\nEpoch 76/100.. Time per epoch: 57.9126.. Average time per step: 0.1481.. Train loss: 0.0318.. Train accuracy: 0.9892.. Top-3 train accuracy: 0.9999.. Test loss: 0.5108.. Test accuracy: 0.8955.. Top-3 test accuracy: 0.9832\nEpoch 77/100.. Time per epoch: 58.4083.. Average time per step: 0.1494.. Train loss: 0.0311.. Train accuracy: 0.9895.. Top-3 train accuracy: 0.9998.. Test loss: 0.4937.. Test accuracy: 0.8987.. Top-3 test accuracy: 0.9843\nEpoch 78/100.. Time per epoch: 58.2263.. Average time per step: 0.1489.. Train loss: 0.0337.. Train accuracy: 0.9883.. Top-3 train accuracy: 0.9997.. Test loss: 0.4887.. Test accuracy: 0.8967.. Top-3 test accuracy: 0.9823\nEpoch 79/100.. Time per epoch: 57.9680.. Average time per step: 0.1483.. Train loss: 0.0348.. Train accuracy: 0.9882.. Top-3 train accuracy: 0.9998.. Test loss: 0.4860.. Test accuracy: 0.9002.. Top-3 test accuracy: 0.9838\nEpoch 80/100.. Time per epoch: 58.1310.. Average time per step: 0.1487.. Train loss: 0.0305.. Train accuracy: 0.9897.. Top-3 train accuracy: 0.9998.. Test loss: 0.4966.. Test accuracy: 0.8963.. Top-3 test accuracy: 0.9841\nEpoch 81/100.. Time per epoch: 58.1362.. Average time per step: 0.1487.. Train loss: 0.0308.. Train accuracy: 0.9897.. Top-3 train accuracy: 0.9998.. Test loss: 0.4912.. Test accuracy: 0.8995.. Top-3 test accuracy: 0.9860\nEpoch 82/100.. Time per epoch: 57.8786.. Average time per step: 0.1480.. Train loss: 0.0280.. Train accuracy: 0.9898.. Top-3 train accuracy: 0.9999.. Test loss: 0.4977.. Test accuracy: 0.8957.. Top-3 test accuracy: 0.9846\nEpoch 83/100.. Time per epoch: 57.4329.. Average time per step: 0.1469.. Train loss: 0.0280.. Train accuracy: 0.9903.. Top-3 train accuracy: 0.9998.. Test loss: 0.5081.. Test accuracy: 0.8971.. Top-3 test accuracy: 0.9838\nEpoch 84/100.. Time per epoch: 57.7010.. Average time per step: 0.1476.. Train loss: 0.0336.. Train accuracy: 0.9882.. Top-3 train accuracy: 0.9999.. Test loss: 0.5063.. Test accuracy: 0.8948.. Top-3 test accuracy: 0.9842\nEpoch 85/100.. Time per epoch: 57.5576.. Average time per step: 0.1472.. Train loss: 0.0261.. Train accuracy: 0.9910.. Top-3 train accuracy: 0.9999.. Test loss: 0.4800.. Test accuracy: 0.8961.. Top-3 test accuracy: 0.9851\nEpoch 86/100.. Time per epoch: 57.5026.. Average time per step: 0.1471.. Train loss: 0.0275.. Train accuracy: 0.9907.. Top-3 train accuracy: 0.9998.. Test loss: 0.4948.. Test accuracy: 0.8989.. Top-3 test accuracy: 0.9844\nEpoch 87/100.. Time per epoch: 57.3774.. Average time per step: 0.1467.. Train loss: 0.0282.. Train accuracy: 0.9901.. Top-3 train accuracy: 0.9998.. Test loss: 0.4867.. Test accuracy: 0.8998.. Top-3 test accuracy: 0.9848\nEpoch 88/100.. Time per epoch: 57.2970.. Average time per step: 0.1465.. Train loss: 0.0262.. Train accuracy: 0.9907.. Top-3 train accuracy: 0.9998.. Test loss: 0.4963.. Test accuracy: 0.9001.. Top-3 test accuracy: 0.9854\nEpoch 89/100.. Time per epoch: 57.3851.. Average time per step: 0.1468.. Train loss: 0.0258.. Train accuracy: 0.9908.. Top-3 train accuracy: 1.0000.. Test loss: 0.5331.. Test accuracy: 0.8952.. Top-3 test accuracy: 0.9836\nEpoch 90/100.. Time per epoch: 57.2572.. Average time per step: 0.1464.. Train loss: 0.0291.. Train accuracy: 0.9900.. Top-3 train accuracy: 0.9998.. Test loss: 0.5104.. Test accuracy: 0.8941.. Top-3 test accuracy: 0.9841\nEpoch 91/100.. Time per epoch: 57.3480.. Average time per step: 0.1467.. Train loss: 0.0242.. Train accuracy: 0.9917.. Top-3 train accuracy: 0.9999.. Test loss: 0.4944.. Test accuracy: 0.9011.. Top-3 test accuracy: 0.9860\nEpoch 92/100.. Time per epoch: 57.3575.. Average time per step: 0.1467.. Train loss: 0.0241.. Train accuracy: 0.9916.. Top-3 train accuracy: 0.9998.. Test loss: 0.5331.. Test accuracy: 0.8958.. Top-3 test accuracy: 0.9850\nEpoch 93/100.. Time per epoch: 57.3366.. Average time per step: 0.1466.. Train loss: 0.0255.. Train accuracy: 0.9909.. Top-3 train accuracy: 0.9998.. Test loss: 0.4910.. Test accuracy: 0.9008.. Top-3 test accuracy: 0.9836\nEpoch 94/100.. Time per epoch: 57.6525.. Average time per step: 0.1474.. Train loss: 0.0250.. Train accuracy: 0.9913.. Top-3 train accuracy: 0.9999.. Test loss: 0.5234.. Test accuracy: 0.8969.. Top-3 test accuracy: 0.9846\nEpoch 95/100.. Time per epoch: 57.6418.. Average time per step: 0.1474.. Train loss: 0.0225.. Train accuracy: 0.9923.. Top-3 train accuracy: 0.9999.. Test loss: 0.5251.. Test accuracy: 0.8980.. Top-3 test accuracy: 0.9846\nEpoch 96/100.. Time per epoch: 57.6497.. Average time per step: 0.1474.. Train loss: 0.0272.. Train accuracy: 0.9908.. Top-3 train accuracy: 0.9999.. Test loss: 0.5212.. Test accuracy: 0.8975.. Top-3 test accuracy: 0.9840\nEpoch 97/100.. Time per epoch: 57.6144.. Average time per step: 0.1474.. Train loss: 0.0236.. Train accuracy: 0.9918.. Top-3 train accuracy: 0.9999.. Test loss: 0.4954.. Test accuracy: 0.9033.. Top-3 test accuracy: 0.9835\nEpoch 98/100.. Time per epoch: 57.7673.. Average time per step: 0.1477.. Train loss: 0.0249.. Train accuracy: 0.9916.. Top-3 train accuracy: 0.9998.. Test loss: 0.4987.. Test accuracy: 0.9027.. Top-3 test accuracy: 0.9850\nEpoch 99/100.. Time per epoch: 57.6581.. Average time per step: 0.1475.. Train loss: 0.0217.. Train accuracy: 0.9923.. Top-3 train accuracy: 0.9999.. Test loss: 0.5139.. Test accuracy: 0.8985.. Top-3 test accuracy: 0.9845\nEpoch 100/100.. Time per epoch: 57.5586.. Average time per step: 0.1472.. Train loss: 0.0233.. Train accuracy: 0.9921.. Top-3 train accuracy: 0.9998.. Test loss: 0.5443.. Test accuracy: 0.8991.. Top-3 test accuracy: 0.9841\n"
],
[
"train_stats.to_csv('train_log_SENet18_Mish.csv')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8b77a6b4c164d7bf5a7b27cfce2c9302e6ab73
| 4,950 |
ipynb
|
Jupyter Notebook
|
source/pytorch/pytorch_with_examples/polynomial_custom_function.ipynb
|
alphajayGithub/ai.online
|
3e440d88111627827456aa8672516eb389a68e98
|
[
"MIT"
] | null | null | null |
source/pytorch/pytorch_with_examples/polynomial_custom_function.ipynb
|
alphajayGithub/ai.online
|
3e440d88111627827456aa8672516eb389a68e98
|
[
"MIT"
] | null | null | null |
source/pytorch/pytorch_with_examples/polynomial_custom_function.ipynb
|
alphajayGithub/ai.online
|
3e440d88111627827456aa8672516eb389a68e98
|
[
"MIT"
] | null | null | null | 91.666667 | 3,212 | 0.627677 |
[
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nPyTorch: Defining New autograd Functions\n----------------------------------------\n\nA third order polynomial, trained to predict $y=\\sin(x)$ from $-\\pi$\nto $\\pi$ by minimizing squared Euclidean distance. Instead of writing the\npolynomial as $y=a+bx+cx^2+dx^3$, we write the polynomial as\n$y=a+b P_3(c+dx)$ where $P_3(x)=\frac{1}{2}\\left(5x^3-3x\right)$ is\nthe `Legendre polynomial`_ of degree three.\n\n https://en.wikipedia.org/wiki/Legendre_polynomials\n\nThis implementation computes the forward pass using operations on PyTorch\nTensors, and uses PyTorch autograd to compute gradients.\n\nIn this implementation we implement our own custom autograd function to perform\n$P_3'(x)$. By mathematics, $P_3'(x)=\frac{3}{2}\\left(5x^2-1\right)$\n\n",
"_____no_output_____"
]
],
[
[
"import torch\nimport math\n\n\nclass LegendrePolynomial3(torch.autograd.Function):\n \"\"\"\n We can implement our own custom autograd Functions by subclassing\n torch.autograd.Function and implementing the forward and backward passes\n which operate on Tensors.\n \"\"\"\n\n @staticmethod\n def forward(ctx, input):\n \"\"\"\n In the forward pass we receive a Tensor containing the input and return\n a Tensor containing the output. ctx is a context object that can be used\n to stash information for backward computation. You can cache arbitrary\n objects for use in the backward pass using the ctx.save_for_backward method.\n \"\"\"\n ctx.save_for_backward(input)\n return 0.5 * (5 * input ** 3 - 3 * input)\n\n @staticmethod\n def backward(ctx, grad_output):\n \"\"\"\n In the backward pass we receive a Tensor containing the gradient of the loss\n with respect to the output, and we need to compute the gradient of the loss\n with respect to the input.\n \"\"\"\n input, = ctx.saved_tensors\n return grad_output * 1.5 * (5 * input ** 2 - 1)\n\n\ndtype = torch.float\ndevice = torch.device(\"cpu\")\n# device = torch.device(\"cuda:0\") # Uncomment this to run on GPU\n\n# Create Tensors to hold input and outputs.\n# By default, requires_grad=False, which indicates that we do not need to\n# compute gradients with respect to these Tensors during the backward pass.\nx = torch.linspace(-math.pi, math.pi, 2000, device=device, dtype=dtype)\ny = torch.sin(x)\n\n# Create random Tensors for weights. For this example, we need\n# 4 weights: y = a + b * P3(c + d * x), these weights need to be initialized\n# not too far from the correct result to ensure convergence.\n# Setting requires_grad=True indicates that we want to compute gradients with\n# respect to these Tensors during the backward pass.\na = torch.full((), 0.0, device=device, dtype=dtype, requires_grad=True)\nb = torch.full((), -1.0, device=device, dtype=dtype, requires_grad=True)\nc = torch.full((), 0.0, device=device, dtype=dtype, requires_grad=True)\nd = torch.full((), 0.3, device=device, dtype=dtype, requires_grad=True)\n\nlearning_rate = 5e-6\nfor t in range(2000):\n # To apply our Function, we use Function.apply method. We alias this as 'P3'.\n P3 = LegendrePolynomial3.apply\n\n # Forward pass: compute predicted y using operations; we compute\n # P3 using our custom autograd operation.\n y_pred = a + b * P3(c + d * x)\n\n # Compute and print loss\n loss = (y_pred - y).pow(2).sum()\n if t % 100 == 99:\n print(t, loss.item())\n\n # Use autograd to compute the backward pass.\n loss.backward()\n\n # Update weights using gradient descent\n with torch.no_grad():\n a -= learning_rate * a.grad\n b -= learning_rate * b.grad\n c -= learning_rate * c.grad\n d -= learning_rate * d.grad\n\n # Manually zero the gradients after updating weights\n a.grad = None\n b.grad = None\n c.grad = None\n d.grad = None\n\nprint(f'Result: y = {a.item()} + {b.item()} * P3({c.item()} + {d.item()} x)')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8b9c0c25bac8cf0154c5438db3cfa0ce47be13
| 11,734 |
ipynb
|
Jupyter Notebook
|
python_function.ipynb
|
SolbiChoi/learn_python
|
1949f2a9de2ae9ffe0b24201a92bf290ee0a064a
|
[
"Apache-2.0"
] | null | null | null |
python_function.ipynb
|
SolbiChoi/learn_python
|
1949f2a9de2ae9ffe0b24201a92bf290ee0a064a
|
[
"Apache-2.0"
] | null | null | null |
python_function.ipynb
|
SolbiChoi/learn_python
|
1949f2a9de2ae9ffe0b24201a92bf290ee0a064a
|
[
"Apache-2.0"
] | null | null | null | 23.51503 | 236 | 0.398756 |
[
[
[
"<a href=\"https://colab.research.google.com/github/SolbiChoi/learn_python/blob/master/python_function.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"np.__version__",
"_____no_output_____"
],
[
"def onehotcylinder(cylinders):\n if cylinders == 3:\n cylinder = np.array([1,0,0,0,0])\n elif cylinders == 4:\n cylinder = np.array([0,1,0,0,0])\n elif cylinders == 5:\n cylinder = np.array([0,1,0,0,0])\n elif cylinders == 6:\n cylinder = np.array([0,0,1,0,0])\n else:\n cylinder = np.array([0,0,0,1,0])\n return cylinder",
"_____no_output_____"
],
[
"displacement = 307.0\nhorsepower = 130.0\nweight = 3504.0\naccle = 12.0\ncylinders = 8\norigin = 1\n\n# if cylinder = 8, origon = 1 -> onehot encoding\n\nx_continuous = np.array([displacement,horsepower,weight,accle])\n# cylinder = np.array([0,0,0,0,1])\ncylinder = onehotcylinder(cylinders)\norg = np.array([1,0,0])",
"_____no_output_____"
],
[
"# np.concatenate((a, b), axis=None)\nresult = np.concatenate((x_continuous, cylinder, org), axis=None)\nresult.shape, result",
"_____no_output_____"
],
[
"# result.reshape(1,12) \nresult = result.reshape(-1, result.size)\nresult, result.shape",
"_____no_output_____"
],
[
"result.size\nresult",
"_____no_output_____"
]
],
[
[
"## function 이해",
"_____no_output_____"
]
],
[
[
"21 + 14",
"_____no_output_____"
],
[
"32 + 45",
"_____no_output_____"
],
[
"def sum():\n first = 21\n second = 14\n result = first + second\n return result",
"_____no_output_____"
],
[
"sum()",
"_____no_output_____"
],
[
"def sum(f01, s02):\n first = s02\n second = f01\n result = first + second\n return result, second, first",
"_____no_output_____"
],
[
"r01, r02, r03 = sum(4,5)\nprint(r01, r02)",
"9 4\n"
],
[
"def mul(a1,a2,a3):\n first = a1\n second = a2\n third = a3\n result = first*second*third\n return result",
"_____no_output_____"
],
[
"mul(21,32,2), mul(31,63,41)",
"_____no_output_____"
],
[
"def diffparam(*var_list,**var_dict):\n print(var_dict)\n result01 = len(var_list)\n return result01",
"_____no_output_____"
],
[
"diffparam(1,2,3,a=2,b=3)",
"{'a': 2, 'b': 3}\n"
],
[
"diffparam(1,2,3,4,5,a=2,b=3,c=4)",
"{'a': 2, 'b': 3, 'c': 4}\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8ba6d1339a0f5ac38e44dcc11cf04dfa2a09a2
| 1,025,439 |
ipynb
|
Jupyter Notebook
|
notebooks/GD1_Hectochelle_2018B.ipynb
|
AllenDowney/GD1-DR2
|
fc46112d40070be6dd460a496e419ab37eb5d7ad
|
[
"MIT"
] | 1 |
2019-02-13T21:19:57.000Z
|
2019-02-13T21:19:57.000Z
|
notebooks/GD1_Hectochelle_2018B.ipynb
|
AllenDowney/GD1-DR2
|
fc46112d40070be6dd460a496e419ab37eb5d7ad
|
[
"MIT"
] | 2 |
2018-05-08T14:48:23.000Z
|
2018-06-21T00:46:54.000Z
|
notebooks/GD1_Hectochelle_2018B.ipynb
|
AllenDowney/GD1-DR2
|
fc46112d40070be6dd460a496e419ab37eb5d7ad
|
[
"MIT"
] | 1 |
2020-06-12T17:50:58.000Z
|
2020-06-12T17:50:58.000Z
| 913.124666 | 506,136 | 0.950738 |
[
[
[
"from os import path\n\n# Third-party\nimport astropy\nimport astropy.coordinates as coord\nfrom astropy.table import Table, vstack\nfrom astropy.io import fits\nimport astropy.units as u\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport numpy as np\n%matplotlib inline\n\nfrom pyvo.dal import TAPService\nfrom pyia import GaiaData\n\nimport gala.coordinates as gc\nimport scipy.stats",
"_____no_output_____"
],
[
"plt.style.use('notebook')",
"_____no_output_____"
],
[
"t = Table.read('../data/gd1-all-ps1-red.fits')",
"_____no_output_____"
],
[
"# deredden\nbands = ['g', 'r', 'i', 'z', 'y']\nfor band in bands:\n t[band] = t[band] - t['A_{}'.format(band)]",
"_____no_output_____"
],
[
"g = GaiaData(t)\nc = coord.SkyCoord(ra=g.ra, dec=g.dec, pm_ra_cosdec=g.pmra, pm_dec=g.pmdec)",
"_____no_output_____"
],
[
"def gd1_dist(phi1):\n # 0, 10\n # -60, 7\n m = (10-7) / (60)\n return (m*phi1.wrap_at(180*u.deg).value + 10) * u.kpc",
"_____no_output_____"
],
[
"gd1_c = c.transform_to(gc.GD1)\n\ngd1_c_dist = gc.GD1(phi1=gd1_c.phi1, phi2=gd1_c.phi2,\n distance=gd1_dist(gd1_c.phi1),\n pm_phi1_cosphi2=gd1_c.pm_phi1_cosphi2,\n pm_phi2=gd1_c.pm_phi2,\n radial_velocity=[0]*len(gd1_c)*u.km/u.s)\n\n# Correct for reflex motion\nv_sun = coord.Galactocentric.galcen_v_sun\nobserved = gd1_c_dist.transform_to(coord.Galactic)\nrep = observed.cartesian.without_differentials()\nrep = rep.with_differentials(observed.cartesian.differentials['s'] + v_sun)\ngd1_c = coord.Galactic(rep).transform_to(gc.GD1)",
"_____no_output_____"
],
[
"wangle = 180*u.deg\n\npm_mask = ((gd1_c.pm_phi1_cosphi2 < -5*u.mas/u.yr) & (gd1_c.pm_phi1_cosphi2 > -10*u.mas/u.yr) & \n (gd1_c.pm_phi2 < 1*u.mas/u.yr) & (gd1_c.pm_phi2 > -2*u.mas/u.yr) & \n (g.bp_rp < 1.5*u.mag) & (g.bp_rp > 0*u.mag))",
"/home/ana/install/anaconda3/lib/python3.6/site-packages/astropy/units/quantity.py:639: RuntimeWarning: invalid value encountered in less\n result = super().__array_ufunc__(function, method, *arrays, **kwargs)\n/home/ana/install/anaconda3/lib/python3.6/site-packages/astropy/units/quantity.py:639: RuntimeWarning: invalid value encountered in greater\n result = super().__array_ufunc__(function, method, *arrays, **kwargs)\n"
],
[
"phi_mask_stream = ((np.abs(gd1_c.phi2)<1*u.deg) & (gd1_c.phi1.wrap_at(wangle)>-50*u.deg) & \n (gd1_c.phi1.wrap_at(wangle)<-10*u.deg))\nphi_mask_off = ((gd1_c.phi2<-2*u.deg) & (gd1_c.phi2>-3*u.deg)) | ((gd1_c.phi2<3*u.deg) & (gd1_c.phi2>2*u.deg))",
"_____no_output_____"
],
[
"iso = Table.read('../data/mist_12.0_-1.35.cmd', format='ascii.commented_header', header_start=12)\nphasecut = (iso['phase']>=0) & (iso['phase']<3)\niso = iso[phasecut]",
"_____no_output_____"
],
[
"# distance modulus\ndistance_app = 7.8*u.kpc\ndm = 5*np.log10((distance_app.to(u.pc)).value)-5\n\n# main sequence + rgb\ni_gi = iso['PS_g']-iso['PS_i']\ni_g = iso['PS_g']+dm\n\ni_left = i_gi - 0.4*(i_g/28)**5\ni_right = i_gi + 0.5*(i_g/28)**5\n\npoly = np.hstack([np.array([i_left, i_g]), np.array([i_right[::-1], i_g[::-1]])]).T\nind = (poly[:,1]<21.3) & (poly[:,1]>17.8)\npoly_main = poly[ind]",
"_____no_output_____"
],
[
"points = np.array([g.g - g.i, g.g]).T\npath_main = mpl.path.Path(poly_main)\ncmd_mask = path_main.contains_points(points)\n\npm1_min = -9*u.mas/u.yr\npm1_max = -4.5*u.mas/u.yr\npm2_min = -1.7*u.mas/u.yr\npm2_max = 1.*u.mas/u.yr\npm_mask = ((gd1_c.pm_phi1_cosphi2 < pm1_max) & (gd1_c.pm_phi1_cosphi2 > pm1_min) & \n (gd1_c.pm_phi2 < pm2_max) & (gd1_c.pm_phi2 > pm2_min))",
"_____no_output_____"
]
],
[
[
"## Define target fields",
"_____no_output_____"
]
],
[
[
"targets = {}\ntargets['phi1'] = np.array([-36.35, -39.5, -32.4, -29.8, -29.8])*u.deg\ntargets['phi2'] = np.array([0.2, 0.2, 1.1, 0, 1])*u.deg\nNf = len(targets['phi1'])",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,8))\nplt.plot(gd1_c.phi1[pm_mask & cmd_mask].wrap_at(wangle), gd1_c.phi2[pm_mask & cmd_mask],\n 'ko', ms=4)\n\nfor i in range(Nf):\n c = mpl.patches.Circle((targets['phi1'][i].value, targets['phi2'][i].value), \n radius=0.5, fc='none', ec='r', lw=2, zorder=2)\n plt.gca().add_patch(c)\n\nplt.gca().set_aspect('equal')\nplt.xlim(-45,-25)\nplt.ylim(-5,5)\n\nplt.xlabel('$\\phi_1$ [deg]')\nplt.ylabel('$\\phi_2$ [deg]')\n\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"### Show overall stream",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(13,10))\nplt.plot(gd1_c.phi1[pm_mask & cmd_mask].wrap_at(wangle), gd1_c.phi2[pm_mask & cmd_mask],\n 'ko', ms=0.7, alpha=0.7, rasterized=True)\n\nfor i in range(Nf):\n c = mpl.patches.Circle((targets['phi1'][i].value, targets['phi2'][i].value), \n radius=0.5, fc='none', ec='r', lw=1, zorder=2)\n plt.gca().add_patch(c)\n\nplt.gca().set_aspect('equal')\nplt.xlabel('$\\phi_1$ [deg]')\nplt.ylabel('$\\phi_2$ [deg]')\n\nplt.xlim(-90,10)\nplt.ylim(-12,12)\nplt.tight_layout()",
"_____no_output_____"
],
[
"targets_c = coord.SkyCoord(phi1=targets['phi1'], phi2=targets['phi2'], frame=gc.GD1)\nra_field = targets_c.icrs.ra.to_string(unit=u.hour, sep=':')\ndec_field = targets_c.icrs.dec.to_string(unit=u.degree, sep=':')\n\ntfield = Table(np.array([ra_field, dec_field]).T, names=('ra', 'dec'))\ntfield.write('../data/GD1_fields_2018B.txt', format='ascii.commented_header', overwrite=True)\ntfield",
"_____no_output_____"
]
],
[
[
"## Target priorities",
"_____no_output_____"
]
],
[
[
"iso = Table.read('/home/ana/data/isochrones/panstarrs/mist_12.6_-1.50.cmd', \n format='ascii.commented_header', header_start=12)\nphasecut = (iso['phase']>=0) & (iso['phase']<3)\niso = iso[phasecut]\n\n# distance modulus\ndistance_app = 7.8*u.kpc\ndm = 5*np.log10((distance_app.to(u.pc)).value)-5\n\n# main sequence + rgb\ni_gi = iso['PS_g']-iso['PS_i']\ni_g = iso['PS_g']+dm\n\ni_left_narrow = i_gi - 0.4*(i_g/28)**5\ni_right_narrow = i_gi + 0.5*(i_g/28)**5\npoly_narrow = np.hstack([np.array([i_left_narrow, i_g]), np.array([i_right_narrow[::-1], i_g[::-1]])]).T\n\ni_left_wide = i_gi - 0.6*(i_g/28)**3\ni_right_wide = i_gi + 0.7*(i_g/28)**3\npoly_wide = np.hstack([np.array([i_left_wide, i_g]), np.array([i_right_wide[::-1], i_g[::-1]])]).T\n\nind = (poly_wide[:,1]<18.3) & (poly_wide[:,1]>14)\npoly_low = poly_wide[ind]\n\nind = (poly_narrow[:,1]<20.5) & (poly_narrow[:,1]>14)\npoly_med = poly_narrow[ind]\n\nind = (poly_narrow[:,1]<20.5) & (poly_narrow[:,1]>17.5)\npoly_high = poly_narrow[ind]",
"_____no_output_____"
],
[
"plt.figure(figsize=(5,10))\n\nplt.plot(g.g[phi_mask_stream & pm_mask] - g.i[phi_mask_stream & pm_mask], g.g[phi_mask_stream & pm_mask], \n 'ko', ms=2, alpha=1, rasterized=True, label='')\n\nplt.plot(i_gi, i_g, 'r-')\n\npml = mpl.patches.Polygon(poly_low, color='moccasin', alpha=0.4, zorder=2)\nplt.gca().add_artist(pml)\npmm = mpl.patches.Polygon(poly_med, color='orange', alpha=0.3, zorder=2)\nplt.gca().add_artist(pmm)\npmh = mpl.patches.Polygon(poly_high, color='green', alpha=0.3, zorder=2)\nplt.gca().add_artist(pmh)\n\nplt.xlim(-0.2, 1.8)\nplt.ylim(21, 13)\nplt.xlabel('g - i')\nplt.ylabel('g')\n\nplt.tight_layout()",
"_____no_output_____"
],
[
"pm1_bmin = -12*u.mas/u.yr\npm1_bmax = 2*u.mas/u.yr\npm2_bmin = -5*u.mas/u.yr\npm2_bmax = 5*u.mas/u.yr\npm_broad_mask = ((gd1_c.pm_phi1_cosphi2 < pm1_bmax) & (gd1_c.pm_phi1_cosphi2 > pm1_bmin) & \n (gd1_c.pm_phi2 < pm2_bmax) & (gd1_c.pm_phi2 > pm2_bmin))",
"_____no_output_____"
],
[
"plt.plot(gd1_c.pm_phi1_cosphi2[phi_mask_stream].to(u.mas/u.yr), \n gd1_c.pm_phi2[phi_mask_stream].to(u.mas/u.yr), \n 'ko', ms=0.5, alpha=0.5, rasterized=True)\n\nrect_xy = [pm1_bmin.to(u.mas/u.yr).value, pm2_bmin.to(u.mas/u.yr).value]\nrect_w = pm1_bmax.to(u.mas/u.yr).value - pm1_bmin.to(u.mas/u.yr).value\nrect_h = pm2_bmax.to(u.mas/u.yr).value - pm2_bmin.to(u.mas/u.yr).value\npr = mpl.patches.Rectangle(rect_xy, rect_w, rect_h, color='orange', alpha=0.3)\nplt.gca().add_artist(pr)\n\nrect_xy = [pm1_min.to(u.mas/u.yr).value, pm2_min.to(u.mas/u.yr).value]\nrect_w = pm1_max.to(u.mas/u.yr).value - pm1_min.to(u.mas/u.yr).value\nrect_h = pm2_max.to(u.mas/u.yr).value - pm2_min.to(u.mas/u.yr).value\npr = mpl.patches.Rectangle(rect_xy, rect_w, rect_h, color='green', alpha=0.3)\nplt.gca().add_artist(pr)\n\nplt.xlim(-12,12)\nplt.ylim(-12,12)\nplt.xlabel('$\\mu_{\\phi_1}$ [mas yr$^{-1}$]')\nplt.ylabel('$\\mu_{\\phi_2}$ [mas yr$^{-1}$]')\n\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"## 2018C proposal",
"_____no_output_____"
]
],
[
[
"path_high = mpl.path.Path(poly_high)\nms_mask = path_high.contains_points(points)",
"_____no_output_____"
],
[
"plt.figure(figsize=(13,10))\nplt.plot(gd1_c.phi1[pm_mask & cmd_mask].wrap_at(wangle), gd1_c.phi2[pm_mask & cmd_mask],\n 'ko', ms=0.7, alpha=0.7, rasterized=True)\n\n# plt.annotate('Progenitor?', xy=(-13, 0.5), xytext=(-10, 7),\n# arrowprops=dict(color='0.3', shrink=0.05, width=1.5, headwidth=6, headlength=8, alpha=0.4),\n# fontsize='small')\n\n# plt.annotate('Blob', xy=(-14, -2), xytext=(-14, -10),\n# arrowprops=dict(color='0.3', shrink=0.08, width=1.5, headwidth=6, headlength=8, alpha=0.4),\n# fontsize='small')\n\nplt.annotate('Spur', xy=(-33, 2), xytext=(-42, 7),\n arrowprops=dict(color='0.3', shrink=0.08, width=1.5, headwidth=6, headlength=8, alpha=0.4),\n fontsize='small')\n\nplt.annotate('Gaps', xy=(-40, -2), xytext=(-35, -10),\n arrowprops=dict(color='0.3', shrink=0.08, width=1.5, headwidth=6, headlength=8, alpha=0.4),\n fontsize='small')\n\nplt.annotate('Gaps', xy=(-21, -1), xytext=(-35, -10),\n arrowprops=dict(color='0.3', shrink=0.08, width=1.5, headwidth=6, headlength=8, alpha=0.4),\n fontsize='small')\n\n# plt.axvline(-55, ls='--', color='0.3', alpha=0.4, dashes=(6,4), lw=2)\n# plt.text(-60, 9.5, 'Previously\\nundetected', fontsize='small', ha='right', va='top')\n\npr = mpl.patches.Rectangle([-50, -5], 25, 10, color='none', ec='darkorange', lw=2)\nplt.gca().add_artist(pr)\n\nplt.gca().set_aspect('equal')\nplt.xlabel('$\\phi_1$ [deg]')\nplt.ylabel('$\\phi_2$ [deg]')\n\nplt.xlim(-90,10)\nplt.ylim(-12,12)\nplt.tight_layout()\n\nax_inset = plt.axes([0.2,0.62,0.6,0.2])\nplt.sca(ax_inset)\n\nplt.plot(gd1_c.phi1[pm_mask & cmd_mask].wrap_at(wangle), gd1_c.phi2[pm_mask & cmd_mask],\n 'ko', ms=4, alpha=0.2, rasterized=True, label='All likely GD-1 members')\n\nplt.plot(gd1_c.phi1[pm_mask & cmd_mask & ms_mask].wrap_at(wangle), gd1_c.phi2[pm_mask & cmd_mask & ms_mask],\n 'ko', ms=4, alpha=1, rasterized=True, label='High priority targets')\n\nplt.text(-0.07, 0.5, 'GD-1 region for\\nHectochelle follow-up', transform=plt.gca().transAxes, ha='right')\nplt.legend(bbox_to_anchor=(1, 0.85), frameon=False, loc='upper left', handlelength=0.3, markerscale=1.5)\n\nfor pos in ['top', 'bottom', 'right', 'left']:\n plt.gca().spines[pos].set_edgecolor('orange')\n\nplt.gca().set_aspect('equal')\nplt.xlim(-50,-25)\nplt.ylim(-5,5)\nplt.setp(plt.gca().get_xticklabels(), visible=False)\nplt.setp(plt.gca().get_yticklabels(), visible=False)\nplt.gca().tick_params(bottom='off', left='off', right='off', top='off');\n\nplt.savefig('../plots/prop_fig1.pdf')",
"_____no_output_____"
],
[
"ts = Table.read('../data/gd1_4_vels.tab', format='ascii.commented_header', delimiter='\\t')\n# ts = Table.read('../data/gd1_both.tab', format='ascii.commented_header', delimiter='\\t')",
"_____no_output_____"
],
[
"vbins = np.arange(-200,200,10)\nfig, ax = plt.subplots(1,3,figsize=(15,5))\n\nplt.sca(ax[0])\nplt.plot(gd1_c.pm_phi1_cosphi2[phi_mask_stream].to(u.mas/u.yr), \n gd1_c.pm_phi2[phi_mask_stream].to(u.mas/u.yr), \n 'ko', ms=0.5, alpha=0.1, rasterized=True)\n\nrect_xy = [pm1_bmin.to(u.mas/u.yr).value, pm2_bmin.to(u.mas/u.yr).value]\nrect_w = pm1_bmax.to(u.mas/u.yr).value - pm1_bmin.to(u.mas/u.yr).value\nrect_h = pm2_bmax.to(u.mas/u.yr).value - pm2_bmin.to(u.mas/u.yr).value\npr = mpl.patches.Rectangle(rect_xy, rect_w, rect_h, color='k', alpha=0.1)\nplt.gca().add_artist(pr)\n\nrect_xy = [pm1_min.to(u.mas/u.yr).value, pm2_min.to(u.mas/u.yr).value]\nrect_w = pm1_max.to(u.mas/u.yr).value - pm1_min.to(u.mas/u.yr).value\nrect_h = pm2_max.to(u.mas/u.yr).value - pm2_min.to(u.mas/u.yr).value\npr = mpl.patches.Rectangle(rect_xy, rect_w, rect_h, color='w', alpha=1)\nplt.gca().add_artist(pr)\npr = mpl.patches.Rectangle(rect_xy, rect_w, rect_h, color='tab:blue', alpha=0.5)\nplt.gca().add_artist(pr)\n\nplt.xlim(-12,12)\nplt.ylim(-12,12)\nplt.xlabel('$\\mu_{\\phi_1}$ [mas yr$^{-1}$]')\nplt.ylabel('$\\mu_{\\phi_2}$ [mas yr$^{-1}$]')\n\nplt.sca(ax[1])\nplt.plot(g.g[phi_mask_stream & pm_mask] - g.i[phi_mask_stream & pm_mask], g.g[phi_mask_stream & pm_mask], \n 'ko', ms=2, alpha=0.5, rasterized=True, label='')\n\n# plt.plot(i_gi, i_g, 'r-')\n\n# pml = mpl.patches.Polygon(poly_low, color='moccasin', alpha=0.4, zorder=2)\n# plt.gca().add_artist(pml)\n# pmm = mpl.patches.Polygon(poly_med, color='orange', alpha=0.3, zorder=2)\n# plt.gca().add_artist(pmm)\npmh = mpl.patches.Polygon(poly_high, color='tab:blue', alpha=0.5, zorder=2)\nplt.gca().add_artist(pmh)\nplt.gca().set_facecolor('0.95')\n\nplt.xlim(-0.2, 1.8)\nplt.ylim(21, 13)\nplt.xlabel('g - i [mag]')\nplt.ylabel('g [mag]')\n\nplt.sca(ax[2])\nplt.hist(ts['VELOCITY'][ts['rank']==1], bins=vbins, alpha=0.5, color='tab:blue', label='Priority 1')\nplt.hist(ts['VELOCITY'][ts['rank']==5], bins=vbins, alpha=0.1, histtype='stepfilled', color='k', label='Priority 5')\n\nplt.legend(fontsize='small')\nplt.xlabel('Radial velocity [km s$^{-1}$]')\nplt.ylabel('Number')\nplt.tight_layout()\nplt.savefig('../plots/prop_fig3.pdf')",
"_____no_output_____"
]
],
[
[
"## Target list",
"_____no_output_____"
]
],
[
[
"# check total number of stars per field\nr_fov = 0.5*u.deg\nmag_mask = g.g<20.5*u.mag\nguide = (g.g>13*u.mag) & (g.g<15*u.mag)\n\nfor i in range(Nf):\n infield = (gd1_c.phi1.wrap_at(wangle) - targets['phi1'][i])**2 + (gd1_c.phi2 - targets['phi2'][i])**2 < r_fov**2\n print(i, np.sum(infield & pm_broad_mask & mag_mask), \n np.sum(infield & pm_mask & mag_mask), np.sum(infield & guide))",
"0 304 25 28\n1 297 30 32\n2 288 25 24\n3 296 42 29\n4 286 19 14\n"
],
[
"# plt.plot(g.g[infield]-g.i[infield],g.g[infield], 'k.')\nplt.plot(g.pmra[infield],g.pmdec[infield], 'k.')\n# plt.xlim(-1,3)\n# plt.ylim(22,12)",
"_____no_output_____"
],
[
"# find ra, dec corners for querying for guide stars\ncornersgd1 = astropy.coordinates.SkyCoord(phi1=np.array([-45,-45,-25,-25])*u.deg, \n phi2=np.array([-3,3,3,-3])*u.deg, frame=gc.GD1)\ncorners = cornersgd1.icrs",
"_____no_output_____"
],
[
"query ='''SELECT * FROM gaiadr2.gaia_source\nWHERE phot_g_mean_mag < 16 AND phot_g_mean_mag > 13 AND\n CONTAINS(POINT('ICRS', ra, dec), \n POLYGON('ICRS', \n {0.ra.degree}, {0.dec.degree}, \n {1.ra.degree}, {1.dec.degree}, \n {2.ra.degree}, {2.dec.degree}, \n {3.ra.degree}, {3.dec.degree})) = 1\n'''.format(corners[0], corners[1], corners[2], corners[3])",
"_____no_output_____"
],
[
"print(query)",
"SELECT * FROM gaiadr2.gaia_source\nWHERE phot_g_mean_mag < 16 AND phot_g_mean_mag > 13 AND\n CONTAINS(POINT('ICRS', ra, dec), \n POLYGON('ICRS', \n 148.3460521663554, 30.238971765560628, \n 142.6037429840887, 33.746595122410596, \n 159.50134916496032, 49.32687740059019, \n 165.54766971564723, 44.95304274266651)) = 1\n\n"
],
[
"spatial_mask = ((gd1_c.phi1.wrap_at(wangle)<-25*u.deg) & (gd1_c.phi1.wrap_at(wangle)>-45*u.deg) & \n (gd1_c.phi2<3*u.deg) & (gd1_c.phi2>-2*u.deg))\nshape_mask = spatial_mask & mag_mask & pm_broad_mask\nNout = np.sum(shape_mask)",
"_____no_output_____"
],
[
"points = np.array([g.g[shape_mask] - g.i[shape_mask], g.g[shape_mask]]).T\n\npm_mask = ((gd1_c.pm_phi1_cosphi2[shape_mask] < pm1_max) & (gd1_c.pm_phi1_cosphi2[shape_mask] > pm1_min) & \n (gd1_c.pm_phi2[shape_mask] < pm2_max) & (gd1_c.pm_phi2[shape_mask] > pm2_min))",
"_____no_output_____"
],
[
"path_med = mpl.path.Path(poly_med)\npath_low = mpl.path.Path(poly_low)\npath_high = mpl.path.Path(poly_high)\n\n# guide = (g.g[shape_mask]>13*u.mag) & (g.g[shape_mask]<15*u.mag)\n\npriority4 = pm_mask\npriority3 = path_low.contains_points(points) & pm_mask\npriority2 = path_main.contains_points(points) & pm_mask\npriority1 = path_high.contains_points(points) & pm_mask\n\n# set up output priorities\npriority = np.zeros(Nout, dtype=np.int64) + 5\n# priority[guide] = -1\npriority[priority4] = 4\npriority[priority3] = 3\npriority[priority2] = 2\npriority[priority1] = 1",
"_____no_output_____"
],
[
"ttype = np.empty(Nout, dtype='S10')\nnontarget = priority>-1\nttype[~nontarget] = 'guide'\nttype[nontarget] = 'target'\n\nname = np.arange(Nout)\n\nara = coord.Angle(t['ra'][shape_mask]*u.deg)\nadec = coord.Angle(t['dec'][shape_mask]*u.deg)\nra = ara.to_string(unit=u.hour, sep=':', precision=2)\ndec = adec.to_string(unit=u.degree, sep=':', precision=2)\n\ntcatalog = Table(np.array([ra, dec, name, priority, ttype, g.g[shape_mask]]).T, \n names=('ra', 'dec', 'object', 'rank', 'type', 'mag'), masked=True)\ntcatalog['rank'].mask = ~nontarget",
"_____no_output_____"
],
[
"tguide = Table.read('../data/guides.fits.gz')",
"_____no_output_____"
],
[
"plt.plot(tguide['ra'], tguide['dec'],'k.')",
"_____no_output_____"
],
[
"# add guides\nNguide = len(tguide)\nname_guides = np.arange(Nout, Nout+Nguide)\n\npriority_guides = np.zeros(Nguide, dtype='int') - 1\nnontarget_guides = priority_guides==-1\n\nttype_guides = np.empty(Nguide, dtype='S10')\nttype_guides[nontarget_guides] = 'guide'\n\nara_guides = coord.Angle(tguide['ra'])\nadec_guides = coord.Angle(tguide['dec'])\nra_guides = ara_guides.to_string(unit=u.hour, sep=':', precision=2)\ndec_guides = adec_guides.to_string(unit=u.degree, sep=':', precision=2)\n\ntguides_out = Table(np.array([ra_guides, dec_guides, name_guides, priority_guides, \n ttype_guides, tguide['phot_g_mean_mag']]).T,\n names=('ra', 'dec', 'object', 'rank', 'type', 'mag'), masked=True)\ntguides_out['rank'].mask = ~nontarget_guides",
"_____no_output_____"
],
[
"tguides_out",
"_____no_output_____"
],
[
"tcatalog = astropy.table.vstack([tcatalog, tguides_out])",
"_____no_output_____"
],
[
"tcatalog",
"_____no_output_____"
],
[
"tcatalog.write('../data/gd1_catalog.cat', format='ascii.fixed_width_two_line', \n fill_values=[(astropy.io.ascii.masked, '')], delimiter='\\t', overwrite=True)",
"_____no_output_____"
],
[
"# output cutout of the whole input catalog\nshape_mask_arr = np.array(shape_mask)\ntcat_input = t[shape_mask_arr]\ntcat_input['name'] = name\ntcat_input['priority'] = priority\ntcat_input['type'] = ttype\n\ntcat_input.write('../data/gd1_input_catalog.fits', overwrite=True)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8ba6f5cf83580676c23291cd718934728211de
| 70,176 |
ipynb
|
Jupyter Notebook
|
scripts/auth-token-result-script/result_analytics.ipynb
|
rdsea/ZETA
|
da4410b443ba918870773c43d117267bf4329471
|
[
"Apache-2.0"
] | 1 |
2021-11-09T05:32:54.000Z
|
2021-11-09T05:32:54.000Z
|
scripts/auth-token-result-script/result_analytics.ipynb
|
rdsea/ZETA
|
da4410b443ba918870773c43d117267bf4329471
|
[
"Apache-2.0"
] | 2 |
2021-08-31T21:20:15.000Z
|
2021-09-02T16:30:31.000Z
|
scripts/auth-token-result-script/result_analytics.ipynb
|
rdsea/ZETA
|
da4410b443ba918870773c43d117267bf4329471
|
[
"Apache-2.0"
] | null | null | null | 345.694581 | 34,648 | 0.929192 |
[
[
[
"import csv\nimport matplotlib\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"auth_csv_path = \"./auth_endpoint_values.csv\"\nservice_csv_path = \"./service_endpoint_values.csv\"",
"_____no_output_____"
],
[
"def convert_cpu_to_dict(file_path):\n data = []\n with open(file_path) as csv_file:\n csv_reader = csv.reader(csv_file, delimiter=',')\n csv_reader = list(csv_reader)\n for idx, row in enumerate(csv_reader):\n if idx == 0:\n pass #skip the first and last row\n else:\n data.append({'workers':row[0], 'cpu_utils': row[1]})\n return data\n\ndef convert_resp_to_dict(file_path):\n data = []\n with open(file_path) as csv_file:\n csv_reader = csv.reader(csv_file, delimiter=',')\n csv_reader = list(csv_reader)\n for idx, row in enumerate(csv_reader):\n if idx == 0:\n pass #skip the first and last row\n else:\n data.append({'workers':row[0], 'response_time': row[3]})\n return data\n\ndef convert_resp_95_to_dict(file_path):\n data = []\n with open(file_path) as csv_file:\n csv_reader = csv.reader(csv_file, delimiter=',')\n csv_reader = list(csv_reader)\n for idx, row in enumerate(csv_reader):\n if idx == 0:\n pass #skip the first and last row\n else:\n data.append({'workers':row[0], 'p95_response_time': row[4]})\n return data\n\n",
"_____no_output_____"
],
[
"auth_service_values = convert_cpu_to_dict(auth_csv_path)\nservice_endpoint_values = convert_cpu_to_dict(service_csv_path)",
"_____no_output_____"
],
[
"workers = [int(x['workers']) for x in auth_service_values]\nauth_cpu_utlis = [(float(x['cpu_utils']))/4 for x in auth_service_values]\nservice_cpu_utlis = [(float(x['cpu_utils']))/4 for x in service_endpoint_values]\ntotal_cpu = [x + y for x, y in zip(auth_cpu_utlis, service_cpu_utlis)]",
"_____no_output_____"
],
[
"plt.rc('font', size=14) \nfig, axs = plt.subplots()\naxs.set_ylim([0, 10])\naxs.set_facecolor('#fcfcfc')\n\naxs.set_xlabel('Parallel virtual users')\n\naxs.set_ylabel('CPU utilization as % $\\it{(4~cores)}$')\naxs.plot(workers, total_cpu, 'r', label='total CPU usage', marker='d', markersize=7)\naxs.plot(workers, service_cpu_utlis, linestyle='dotted',label='service-endpoint', marker='o', markersize=5)\naxs.plot(workers, auth_cpu_utlis, 'g--' ,label='auth-service', marker='x', mec='k', markersize=5)\n\naxs.legend()\naxs.grid(axis='both', color='#7D7D7D', linestyle='-', linewidth=0.5)\nplt.savefig(\"auth_token_cpu_util.pdf\")\nplt.show()\n",
"_____no_output_____"
],
[
"service_endpoint_resp_values = convert_resp_to_dict(service_csv_path)\nservice_endpoint_resp_values = [float(x['response_time']) for x in service_endpoint_resp_values]\n\n\n\nservice_endpoint_95_resp_values = convert_resp_95_to_dict(service_csv_path)\nservice_endpoint_95_resp_values = [float(x['p95_response_time']) for x in service_endpoint_95_resp_values]\n",
"_____no_output_____"
],
[
"#plt.rc('font', size=20) # controls default text sizes\nfig, axs = plt.subplots()\naxs.set_ylim([60, 180])\naxs.set_facecolor('#fcfcfc')\n\naxs.grid(axis='both', color='#7D7D7D', linestyle='-', linewidth=0.5, zorder=0)\n\naxs.set_xlabel('Parallel virtual users')\n\naxs.set_ylabel('Response time (ms)')\n\n#p1 = axs.bar(workers, service_endpoint_resp_values, 3, zorder=3, alpha=0.9)\naxs.plot(workers, service_endpoint_resp_values, label='avg response time', marker='x', markersize=7)\n\naxs.plot(workers, service_endpoint_95_resp_values, 'r', linestyle='dotted',label='p(95) response time', marker='o', markersize=5)\naxs.legend(loc='upper left')\n\nplt.savefig(\"auth_token_response_time.pdf\")\nplt.show()\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8bb07440855cd5f4ffe2984113eb7f6e8806c4
| 887,250 |
ipynb
|
Jupyter Notebook
|
huggingface_course/chap5.ipynb
|
TokisakiKurumi2001/pytorch_nlp_learn
|
e43f2ac6449414aab4de29fbaafbb29985ac1baa
|
[
"MIT"
] | null | null | null |
huggingface_course/chap5.ipynb
|
TokisakiKurumi2001/pytorch_nlp_learn
|
e43f2ac6449414aab4de29fbaafbb29985ac1baa
|
[
"MIT"
] | null | null | null |
huggingface_course/chap5.ipynb
|
TokisakiKurumi2001/pytorch_nlp_learn
|
e43f2ac6449414aab4de29fbaafbb29985ac1baa
|
[
"MIT"
] | null | null | null | 40.498905 | 1,339 | 0.509635 |
[
[
[
"!pip install transformers datasets",
"Collecting transformers\n Downloading transformers-4.17.0-py3-none-any.whl (3.8 MB)\n\u001b[K |████████████████████████████████| 3.8 MB 4.9 MB/s \n\u001b[?25hCollecting datasets\n Downloading datasets-2.0.0-py3-none-any.whl (325 kB)\n\u001b[K |████████████████████████████████| 325 kB 42.8 MB/s \n\u001b[?25hRequirement already satisfied: regex!=2019.12.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (2019.12.20)\nRequirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.7/dist-packages (from transformers) (21.3)\nRequirement already satisfied: numpy>=1.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (1.21.5)\nCollecting pyyaml>=5.1\n Downloading PyYAML-6.0-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (596 kB)\n\u001b[K |████████████████████████████████| 596 kB 44.5 MB/s \n\u001b[?25hCollecting huggingface-hub<1.0,>=0.1.0\n Downloading huggingface_hub-0.4.0-py3-none-any.whl (67 kB)\n\u001b[K |████████████████████████████████| 67 kB 5.6 MB/s \n\u001b[?25hCollecting tokenizers!=0.11.3,>=0.11.1\n Downloading tokenizers-0.11.6-cp37-cp37m-manylinux_2_12_x86_64.manylinux2010_x86_64.whl (6.5 MB)\n\u001b[K |████████████████████████████████| 6.5 MB 33.0 MB/s \n\u001b[?25hCollecting sacremoses\n Downloading sacremoses-0.0.49-py3-none-any.whl (895 kB)\n\u001b[K |████████████████████████████████| 895 kB 47.8 MB/s \n\u001b[?25hRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from transformers) (4.11.2)\nRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.7/dist-packages (from transformers) (4.63.0)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from transformers) (2.23.0)\nRequirement already satisfied: filelock in /usr/local/lib/python3.7/dist-packages (from transformers) (3.6.0)\nRequirement already satisfied: typing-extensions>=3.7.4.3 in /usr/local/lib/python3.7/dist-packages (from huggingface-hub<1.0,>=0.1.0->transformers) (3.10.0.2)\nRequirement already satisfied: pyparsing!=3.0.5,>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from packaging>=20.0->transformers) (3.0.7)\nCollecting fsspec[http]>=2021.05.0\n Downloading fsspec-2022.2.0-py3-none-any.whl (134 kB)\n\u001b[K |████████████████████████████████| 134 kB 46.7 MB/s \n\u001b[?25hRequirement already satisfied: pyarrow>=5.0.0 in /usr/local/lib/python3.7/dist-packages (from datasets) (6.0.1)\nCollecting xxhash\n Downloading xxhash-3.0.0-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (212 kB)\n\u001b[K |████████████████████████████████| 212 kB 39.6 MB/s \n\u001b[?25hCollecting aiohttp\n Downloading aiohttp-3.8.1-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (1.1 MB)\n\u001b[K |████████████████████████████████| 1.1 MB 48.8 MB/s \n\u001b[?25hCollecting responses<0.19\n Downloading responses-0.18.0-py3-none-any.whl (38 kB)\nRequirement already satisfied: pandas in /usr/local/lib/python3.7/dist-packages (from datasets) (1.3.5)\nRequirement already satisfied: dill in /usr/local/lib/python3.7/dist-packages (from datasets) (0.3.4)\nRequirement already satisfied: multiprocess in /usr/local/lib/python3.7/dist-packages (from datasets) (0.70.12.2)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2021.10.8)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2.10)\nCollecting urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1\n Downloading urllib3-1.25.11-py2.py3-none-any.whl (127 kB)\n\u001b[K |████████████████████████████████| 127 kB 50.4 MB/s \n\u001b[?25hCollecting yarl<2.0,>=1.0\n Downloading yarl-1.7.2-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (271 kB)\n\u001b[K |████████████████████████████████| 271 kB 44.5 MB/s \n\u001b[?25hCollecting aiosignal>=1.1.2\n Downloading aiosignal-1.2.0-py3-none-any.whl (8.2 kB)\nCollecting asynctest==0.13.0\n Downloading asynctest-0.13.0-py3-none-any.whl (26 kB)\nRequirement already satisfied: attrs>=17.3.0 in /usr/local/lib/python3.7/dist-packages (from aiohttp->datasets) (21.4.0)\nCollecting multidict<7.0,>=4.5\n Downloading multidict-6.0.2-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (94 kB)\n\u001b[K |████████████████████████████████| 94 kB 3.8 MB/s \n\u001b[?25hRequirement already satisfied: charset-normalizer<3.0,>=2.0 in /usr/local/lib/python3.7/dist-packages (from aiohttp->datasets) (2.0.12)\nCollecting frozenlist>=1.1.1\n Downloading frozenlist-1.3.0-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl (144 kB)\n\u001b[K |████████████████████████████████| 144 kB 48.1 MB/s \n\u001b[?25hCollecting async-timeout<5.0,>=4.0.0a3\n Downloading async_timeout-4.0.2-py3-none-any.whl (5.8 kB)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata->transformers) (3.7.0)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/dist-packages (from pandas->datasets) (2.8.2)\nRequirement already satisfied: pytz>=2017.3 in /usr/local/lib/python3.7/dist-packages (from pandas->datasets) (2018.9)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.7.3->pandas->datasets) (1.15.0)\nRequirement already satisfied: joblib in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (1.1.0)\nRequirement already satisfied: click in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (7.1.2)\nInstalling collected packages: multidict, frozenlist, yarl, urllib3, asynctest, async-timeout, aiosignal, pyyaml, fsspec, aiohttp, xxhash, tokenizers, sacremoses, responses, huggingface-hub, transformers, datasets\n Attempting uninstall: urllib3\n Found existing installation: urllib3 1.24.3\n Uninstalling urllib3-1.24.3:\n Successfully uninstalled urllib3-1.24.3\n Attempting uninstall: pyyaml\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ndatascience 0.10.6 requires folium==0.2.1, but you have folium 0.8.3 which is incompatible.\u001b[0m\nSuccessfully installed aiohttp-3.8.1 aiosignal-1.2.0 async-timeout-4.0.2 asynctest-0.13.0 datasets-2.0.0 frozenlist-1.3.0 fsspec-2022.2.0 huggingface-hub-0.4.0 multidict-6.0.2 pyyaml-6.0 responses-0.18.0 sacremoses-0.0.49 tokenizers-0.11.6 transformers-4.17.0 urllib3-1.25.11 xxhash-3.0.0 yarl-1.7.2\n"
],
[
"!wget https://github.com/crux82/squad-it/raw/master/SQuAD_it-train.json.gz\n!wget https://github.com/crux82/squad-it/raw/master/SQuAD_it-test.json.gz",
"--2022-03-21 13:42:25-- https://github.com/crux82/squad-it/raw/master/SQuAD_it-train.json.gz\nResolving github.com (github.com)... 140.82.113.4\nConnecting to github.com (github.com)|140.82.113.4|:443... connected.\nHTTP request sent, awaiting response... 302 Found\nLocation: https://raw.githubusercontent.com/crux82/squad-it/master/SQuAD_it-train.json.gz [following]\n--2022-03-21 13:42:25-- https://raw.githubusercontent.com/crux82/squad-it/master/SQuAD_it-train.json.gz\nResolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.110.133, ...\nConnecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 7725286 (7.4M) [application/octet-stream]\nSaving to: ‘SQuAD_it-train.json.gz’\n\nSQuAD_it-train.json 100%[===================>] 7.37M --.-KB/s in 0.08s \n\n2022-03-21 13:42:26 (88.6 MB/s) - ‘SQuAD_it-train.json.gz’ saved [7725286/7725286]\n\n--2022-03-21 13:42:26-- https://github.com/crux82/squad-it/raw/master/SQuAD_it-test.json.gz\nResolving github.com (github.com)... 140.82.112.4\nConnecting to github.com (github.com)|140.82.112.4|:443... connected.\nHTTP request sent, awaiting response... 302 Found\nLocation: https://raw.githubusercontent.com/crux82/squad-it/master/SQuAD_it-test.json.gz [following]\n--2022-03-21 13:42:26-- https://raw.githubusercontent.com/crux82/squad-it/master/SQuAD_it-test.json.gz\nResolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.111.133, 185.199.108.133, 185.199.109.133, ...\nConnecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.111.133|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 1051245 (1.0M) [application/octet-stream]\nSaving to: ‘SQuAD_it-test.json.gz’\n\nSQuAD_it-test.json. 100%[===================>] 1.00M --.-KB/s in 0.05s \n\n2022-03-21 13:42:26 (20.3 MB/s) - ‘SQuAD_it-test.json.gz’ saved [1051245/1051245]\n\n"
],
[
"!gzip -dkv SQuAD_it-*.json.gz",
"SQuAD_it-test.json.gz:\t 87.5% -- replaced with SQuAD_it-test.json\nSQuAD_it-train.json.gz:\t 82.3% -- replaced with SQuAD_it-train.json\n"
],
[
"from datasets import load_dataset\nsquad_it_dataset = load_dataset(\"json\", data_files=\"SQuAD_it-train.json\", field=\"data\")",
"Using custom data configuration default-03f2eaa6e4e43233\n"
],
[
"squad_it_dataset",
"_____no_output_____"
],
[
"squad_it_dataset[\"train\"][0]",
"_____no_output_____"
],
[
"data_files = {\"train\": \"SQuAD_it-train.json\", \"test\": \"SQuAD_it-test.json\"}\nsquad_it_dataset = load_dataset(\"json\", data_files=data_files, field=\"data\")\nsquad_it_dataset",
"Using custom data configuration default-cd8746634437af48\n"
],
[
"!wget \"https://archive.ics.uci.edu/ml/machine-learning-databases/00462/drugsCom_raw.zip\"\n!unzip drugsCom_raw.zip",
"--2022-03-21 13:42:39-- https://archive.ics.uci.edu/ml/machine-learning-databases/00462/drugsCom_raw.zip\nResolving archive.ics.uci.edu (archive.ics.uci.edu)... 128.195.10.252\nConnecting to archive.ics.uci.edu (archive.ics.uci.edu)|128.195.10.252|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 42989872 (41M) [application/x-httpd-php]\nSaving to: ‘drugsCom_raw.zip’\n\ndrugsCom_raw.zip 100%[===================>] 41.00M 38.6MB/s in 1.1s \n\n2022-03-21 13:42:41 (38.6 MB/s) - ‘drugsCom_raw.zip’ saved [42989872/42989872]\n\nArchive: drugsCom_raw.zip\n inflating: drugsComTest_raw.tsv \n inflating: drugsComTrain_raw.tsv \n"
],
[
"from datasets import load_dataset\n\ndata_files = {\"train\": \"drugsComTrain_raw.tsv\", \"test\": \"drugsComTest_raw.tsv\"}\n# \\t is the tab character in Python\ndrug_dataset = load_dataset(\"csv\", data_files=data_files, delimiter=\"\\t\")",
"Using custom data configuration default-3761173c276c0a9a\n"
],
[
"drug_sample = drug_dataset[\"train\"].shuffle(seed=42).select(range(1000))\ndrug_sample[:3]",
"_____no_output_____"
],
[
"for split in drug_dataset.keys():\n assert len(drug_dataset[split]) == len(drug_dataset[split].unique(\"Unnamed: 0\"))",
"_____no_output_____"
],
[
"drug_dataset = drug_dataset.rename_column(\n original_column_name=\"Unnamed: 0\", new_column_name=\"patient_id\"\n)\ndrug_dataset",
"_____no_output_____"
],
[
"def filter_nones(x):\n return x[\"condition\"] is not None",
"_____no_output_____"
],
[
"drug_dataset = drug_dataset.filter(lambda x: x[\"condition\"] is not None)",
"_____no_output_____"
],
[
"def lowercase_condition(example):\n return {\"condition\": example[\"condition\"].lower()}\n\n\ndrug_dataset = drug_dataset.map(lowercase_condition)\ndrug_dataset[\"train\"][\"condition\"][:3]",
"_____no_output_____"
],
[
"def compute_review_length(example):\n return {\"review_length\": len(example[\"review\"].split())}",
"_____no_output_____"
],
[
"drug_dataset = drug_dataset.map(compute_review_length)\n# Inspect the first training example\ndrug_dataset[\"train\"][0]",
"_____no_output_____"
],
[
"drug_dataset[\"train\"].sort(\"review_length\")[:3]",
"_____no_output_____"
],
[
"drug_dataset = drug_dataset.filter(lambda x: x[\"review_length\"] > 30)\nprint(drug_dataset.num_rows)",
"_____no_output_____"
],
[
"import html\n\ntext = \"I'm a transformer called BERT\"\nhtml.unescape(text)",
"_____no_output_____"
],
[
"drug_dataset = drug_dataset.map(lambda x: {\"review\": html.unescape(x[\"review\"])})",
"_____no_output_____"
],
[
"new_drug_dataset = drug_dataset.map(\n lambda x: {\"review\": [html.unescape(o) for o in x[\"review\"]]}, batched=True\n)",
"_____no_output_____"
],
[
"from transformers import AutoTokenizer\n\ntokenizer = AutoTokenizer.from_pretrained(\"bert-base-cased\")\n\n\ndef tokenize_function(examples):\n return tokenizer(examples[\"review\"], truncation=True)",
"_____no_output_____"
],
[
"%time tokenized_dataset = drug_dataset.map(tokenize_function, batched=True)",
"_____no_output_____"
],
[
"slow_tokenizer = AutoTokenizer.from_pretrained(\"bert-base-cased\", use_fast=False)\n\n\ndef slow_tokenize_function(examples):\n return slow_tokenizer(examples[\"review\"], truncation=True)\n\n\ntokenized_dataset = drug_dataset.map(slow_tokenize_function, batched=True, num_proc=8)",
" "
],
[
"def tokenize_and_split(examples):\n return tokenizer(\n examples[\"review\"],\n truncation=True,\n max_length=128,\n return_overflowing_tokens=True\n )\nresult = tokenize_and_split(drug_dataset[\"train\"][0])\n[len(inp) for inp in result[\"input_ids\"]]",
"_____no_output_____"
],
[
"tokenized_dataset = drug_dataset.map(\n tokenize_and_split, batched=True, remove_columns=drug_dataset[\"train\"].column_names\n)",
"_____no_output_____"
],
[
"drug_dataset.set_format(\"pandas\")",
"_____no_output_____"
],
[
"train_df = drug_dataset[\"train\"][:]\nfrequencies = (\n train_df[\"condition\"]\n .value_counts()\n .to_frame()\n .reset_index()\n .rename(columns={\"index\": \"condition\", \"condition\": \"frequency\"})\n)\nfrequencies.head()",
"_____no_output_____"
],
[
"from datasets import Dataset\nfreq_dataset = Dataset.from_pandas(frequencies)\nfreq_dataset",
"_____no_output_____"
],
[
"drug_dataset.reset_format()",
"_____no_output_____"
],
[
"drug_dataset_clean = drug_dataset[\"train\"].train_test_split(train_size=0.8, seed=42)\n# Rename the default \"test\" split to \"validation\"\ndrug_dataset_clean[\"validation\"] = drug_dataset_clean.pop(\"test\")\ndrug_dataset_clean[\"test\"] = drug_dataset[\"test\"]\ndrug_dataset_clean",
"_____no_output_____"
],
[
"drug_dataset_clean.save_to_disk(\"drug-reviews\")",
"_____no_output_____"
],
[
"from datasets import load_from_disk\ndrug_dataset_reloaded = load_from_disk(\"drug-reviews\")\ndrug_dataset_reloaded",
"_____no_output_____"
],
[
"for split, dataset in drug_dataset_clean.items():\n dataset.to_json(f\"drug-reviews-{split}.jsonl\")",
"_____no_output_____"
],
[
"data_files = {\n \"train\": \"drug-reviews-train.jsonl\",\n \"validation\": \"drug-reviews-validation.jsonl\",\n \"test\": \"drug-reviews-test.jsonl\",\n}\ndrug_dataset_reloaded = load_dataset(\"json\", data_files=data_files)",
"Using custom data configuration default-551cf3d148932df2\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8bb16972c752faaa430782e8055da9a237c52e
| 2,486 |
ipynb
|
Jupyter Notebook
|
handrecognition.ipynb
|
jagadeesh-vinnakota/tensorflow_basics
|
656f5c397bfaebde02b319d7f596c67bf68407fb
|
[
"MIT"
] | null | null | null |
handrecognition.ipynb
|
jagadeesh-vinnakota/tensorflow_basics
|
656f5c397bfaebde02b319d7f596c67bf68407fb
|
[
"MIT"
] | null | null | null |
handrecognition.ipynb
|
jagadeesh-vinnakota/tensorflow_basics
|
656f5c397bfaebde02b319d7f596c67bf68407fb
|
[
"MIT"
] | null | null | null | 26.731183 | 100 | 0.591714 |
[
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"import numpy as np\nimport tensorflow as tf\ndef get_input_fn(dataset_split, batch_size, capacity=10000, min_after_dequeue=3000):\n def _input_fn():\n images_batch, labels_batch = tf.train.shuffle_batch(\n tensors=[dataset_split.images, dataset_split.labels.astype(np.int32)],\n batch_size=batch_size,\n capacity=capacity,\n min_after_dequeue=min_after_dequeue,\n enqueue_many=True,\n num_threads=4)\n features_map = {'images': images_batch}\n return features_map, labels_batch\n return _input_fn\ndata = tf.contrib.learn.datasets.mnist.load_mnist()\ntrain_input_fn = get_input_fn(data.train, batch_size=256)\neval_input_fn = get_input_fn(data.validation, batch_size=5000)",
"_____no_output_____"
],
[
"image_column = tf.contrib.layers.real_valued_column('images', dimension=784)",
"_____no_output_____"
],
[
"import time\nimage_column = tf.contrib.layers.real_valued_column('images', dimension=784)\nestimator = tf.contrib.learn.LinearClassifier(feature_columns=[image_column], n_classes=10)\nstart = time.time()\nestimator.fit(input_fn=train_input_fn, steps=2000)\nend = time.time()\nprint('Elapsed time: {} seconds'.format(end - start))\neval_metrics = estimator.evaluate(input_fn=eval_input_fn, steps=1)\nprint(eval_metrics)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a8bb3097c19a1ab2e8c12b412dfc91f82cb758b
| 579,354 |
ipynb
|
Jupyter Notebook
|
notebooks/exploratory/.ipynb_checkpoints/exploratory_spectrogram-checkpoint.ipynb
|
MariusDgr/AudioMining
|
ef74567fcc1d9034777bde45bc4a4ead20e8aa75
|
[
"Apache-2.0"
] | null | null | null |
notebooks/exploratory/.ipynb_checkpoints/exploratory_spectrogram-checkpoint.ipynb
|
MariusDgr/AudioMining
|
ef74567fcc1d9034777bde45bc4a4ead20e8aa75
|
[
"Apache-2.0"
] | null | null | null |
notebooks/exploratory/.ipynb_checkpoints/exploratory_spectrogram-checkpoint.ipynb
|
MariusDgr/AudioMining
|
ef74567fcc1d9034777bde45bc4a4ead20e8aa75
|
[
"Apache-2.0"
] | null | null | null | 985.295918 | 129,508 | 0.954473 |
[
[
[
"import os \n\nimport numpy as np\n\n# matplotlib for displaying the output\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport seaborn as sns\n# sns.set()\nsns.set(style=\"ticks\", context=\"talk\")\n\nfrom scipy import signal\nfrom scipy.io import wavfile\n\n# and IPython.display for audio output\nimport IPython.display\n\n# Librosa for audio\nimport librosa\n# And the display module for visualization\nimport librosa.display",
"_____no_output_____"
],
[
"# Get data files\n\ntwo_up = os.path.abspath(os.path.join('.' ,\"../..\"))\nprint(\"Project root path is: \", two_up)\n\n\ndataDirName = \"data\"\nrawDataDirName = \"converted_wav\"\nclassName = \"violin\"\n# className = \"guitar\"\ndata_path = os.path.join(two_up, dataDirName, rawDataDirName, className)\n\nprint(data_path)\nroot_paths = []\n\n# Get all files from data_path \n# r=root, d=directories, f = files\n(_, d, allFiles) = next(os.walk(data_path))\nwavFiles = [f for f in allFiles if f.endswith(\".wav\")]\n",
"Project root path is: D:\\Programming\\AudioMining\nD:\\Programming\\AudioMining\\data\\converted_wav\\violin\n"
],
[
"print(wavFiles[0])",
"violin_A3_025_forte_arco-normal.wav\n"
]
],
[
[
"### Spectrogram ",
"_____no_output_____"
]
],
[
[
"\n\nfile = wavFiles[3]\nsample_rate, samples = wavfile.read(os.path.join(data_path, file))\nfrequencies, times, spectrogram = signal.spectrogram(samples, sample_rate)\n\n# all spectrogram\nplt.pcolormesh(times, frequencies, spectrogram)\nplt.ylabel('Frequency')\nplt.xlabel('Time')\nplt.show()\n\nprint(times[0], times[-1])\nprint(frequencies[0], frequencies[-1])\n\n# plot(times, frequencies)\n",
"_____no_output_____"
],
[
"plt.specgram(samples,Fs=sample_rate)\nplt.xlabel('Time')\nplt.ylabel('Frequency')\nplt.colorbar()\nplt.show()\n",
"C:\\Users\\Marius\\AppData\\Roaming\\Python\\Python36\\site-packages\\matplotlib\\axes\\_axes.py:7221: RuntimeWarning: divide by zero encountered in log10\n Z = 10. * np.log10(spec)\n"
]
],
[
[
"### Time Domain",
"_____no_output_____"
]
],
[
[
"zoom_left = 10000\nzoom_right = 30000\n\nplt.plot(samples)\nplt.axvline(x=zoom_left)\nplt.axvline(x=zoom_right)\nplt.show()\n\nplt.plot(samples[zoom_left:zoom_right])\nplt.show()",
"_____no_output_____"
]
],
[
[
"Librosa example",
"_____no_output_____"
]
],
[
[
"y, sr = librosa.load(os.path.join(data_path, file), sr=None)",
"_____no_output_____"
],
[
"S = librosa.feature.melspectrogram(y, sr=sr, n_mels=128)\n\n# Convert to log scale (dB). We'll use the peak power (max) as reference.\nlog_S = librosa.power_to_db(S, ref=np.max)\n\n# Make a new figure\nplt.figure(figsize=(12,4))\n\n# Display the spectrogram on a mel scale\n# sample rate and hop length parameters are used to render the time axis\nlibrosa.display.specshow(log_S, sr=sr, x_axis='time', y_axis='mel')\n\n# Put a descriptive title on the plot\nplt.title('mel power spectrogram')\n\n# draw a color bar\nplt.colorbar(format='%+02.0f dB')\n\n# Make the figure layout compact\nplt.tight_layout()",
"_____no_output_____"
],
[
"y_harmonic, y_percussive = librosa.effects.hpss(y)\n# What do the spectrograms look like?\n# Let's make and display a mel-scaled power (energy-squared) spectrogram\nS_harmonic = librosa.feature.melspectrogram(y_harmonic, sr=sr)\nS_percussive = librosa.feature.melspectrogram(y_percussive, sr=sr)\n\n# Convert to log scale (dB). We'll use the peak power as reference.\nlog_Sh = librosa.power_to_db(S_harmonic, ref=np.max)\nlog_Sp = librosa.power_to_db(S_percussive, ref=np.max)\n\n# Make a new figure\nplt.figure(figsize=(12,6))\n\nplt.subplot(2,1,1)\n# Display the spectrogram on a mel scale\nlibrosa.display.specshow(log_Sh, sr=sr, y_axis='mel')\n\n# Put a descriptive title on the plot\nplt.title('mel power spectrogram (Harmonic)')\n\n# draw a color bar\nplt.colorbar(format='%+02.0f dB')\n\nplt.subplot(2,1,2)\nlibrosa.display.specshow(log_Sp, sr=sr, x_axis='time', y_axis='mel')\n\n# Put a descriptive title on the plot\nplt.title('mel power spectrogram (Percussive)')\n\n# draw a color bar\nplt.colorbar(format='%+02.0f dB')\n\n# Make the figure layout compact\nplt.tight_layout()\n",
"_____no_output_____"
],
[
"# We'll use a CQT-based chromagram with 36 bins-per-octave in the CQT analysis. An STFT-based implementation also exists in chroma_stft()\n# We'll use the harmonic component to avoid pollution from transients\nC = librosa.feature.chroma_cqt(y=y_harmonic, sr=sr, bins_per_octave=36)\n\n# Make a new figure\nplt.figure(figsize=(12,4))\n\n# Display the chromagram: the energy in each chromatic pitch class as a function of time\n# To make sure that the colors span the full range of chroma values, set vmin and vmax\nlibrosa.display.specshow(C, sr=sr, x_axis='time', y_axis='chroma', vmin=0, vmax=1)\n\nplt.title('Chromagram')\nplt.colorbar()\n\nplt.tight_layout()",
"_____no_output_____"
],
[
"# Next, we'll extract the top 13 Mel-frequency cepstral coefficients (MFCCs)\nmfcc = librosa.feature.mfcc(S=log_S, n_mfcc=13)\n\n# Let's pad on the first and second deltas while we're at it\ndelta_mfcc = librosa.feature.delta(mfcc)\ndelta2_mfcc = librosa.feature.delta(mfcc, order=2)\n\n# How do they look? We'll show each in its own subplot\nplt.figure(figsize=(12, 6))\n\nplt.subplot(3,1,1)\nlibrosa.display.specshow(mfcc)\nplt.ylabel('MFCC')\nplt.colorbar()\n\nplt.subplot(3,1,2)\nlibrosa.display.specshow(delta_mfcc)\nplt.ylabel('MFCC-$\\Delta$')\nplt.colorbar()\n\nplt.subplot(3,1,3)\nlibrosa.display.specshow(delta2_mfcc, sr=sr, x_axis='time')\nplt.ylabel('MFCC-$\\Delta^2$')\nplt.colorbar()\n\nplt.tight_layout()\n\n# For future use, we'll stack these together into one matrix\nM = np.vstack([mfcc, delta_mfcc, delta2_mfcc])",
"_____no_output_____"
],
[
"# Now, let's run the beat tracker.\n# We'll use the percussive component for this part\nplt.figure(figsize=(12, 6))\ntempo, beats = librosa.beat.beat_track(y=y_percussive, sr=sr)\n\n# Let's re-draw the spectrogram, but this time, overlay the detected beats\nplt.figure(figsize=(12,4))\nlibrosa.display.specshow(log_S, sr=sr, x_axis='time', y_axis='mel')\n\n# Let's draw transparent lines over the beat frames\nplt.vlines(librosa.frames_to_time(beats),\n 1, 0.5 * sr,\n colors='w', linestyles='-', linewidth=2, alpha=0.5)\n\nplt.axis('tight')\n\nplt.colorbar(format='%+02.0f dB')\n\nplt.tight_layout();",
"_____no_output_____"
],
[
"print('Estimated tempo: %.2f BPM' % tempo)\nprint('First 5 beat frames: ', beats[:5])\n\n# Frame numbers are great and all, but when do those beats occur?\nprint('First 5 beat times: ', librosa.frames_to_time(beats[:5], sr=sr))\n\n# We could also get frame numbers from times by librosa.time_to_frames()",
"Estimated tempo: 132.51 BPM\nFirst 5 beat frames: [3]\nFirst 5 beat times: [0.03482993]\n"
],
[
"# feature.sync will summarize each beat event by the mean feature vector within that beat\n\nM_sync = librosa.util.sync(M, beats)\n\nplt.figure(figsize=(12,6))\n\n# Let's plot the original and beat-synchronous features against each other\nplt.subplot(2,1,1)\nlibrosa.display.specshow(M)\nplt.title('MFCC-$\\Delta$-$\\Delta^2$')\n\n# We can also use pyplot *ticks directly\n# Let's mark off the raw MFCC and the delta features\nplt.yticks(np.arange(0, M.shape[0], 13), ['MFCC', '$\\Delta$', '$\\Delta^2$'])\n\nplt.colorbar()\n\nplt.subplot(2,1,2)\n# librosa can generate axis ticks from arbitrary timestamps and beat events also\nlibrosa.display.specshow(M_sync, x_axis='time',\n x_coords=librosa.frames_to_time(librosa.util.fix_frames(beats)))\n\nplt.yticks(np.arange(0, M_sync.shape[0], 13), ['MFCC', '$\\Delta$', '$\\Delta^2$']) \nplt.title('Beat-synchronous MFCC-$\\Delta$-$\\Delta^2$')\nplt.colorbar()\n\nplt.tight_layout()",
"_____no_output_____"
],
[
"# Beat synchronization is flexible.\n# Instead of computing the mean delta-MFCC within each beat, let's do beat-synchronous chroma\n# We can replace the mean with any statistical aggregation function, such as min, max, or median.\n\nC_sync = librosa.util.sync(C, beats, aggregate=np.median)\n\nplt.figure(figsize=(12,6))\n\nplt.subplot(2, 1, 1)\nlibrosa.display.specshow(C, sr=sr, y_axis='chroma', vmin=0.0, vmax=1.0, x_axis='time')\n\nplt.title('Chroma')\nplt.colorbar()\n\nplt.subplot(2, 1, 2)\nlibrosa.display.specshow(C_sync, y_axis='chroma', vmin=0.0, vmax=1.0, x_axis='time', \n x_coords=librosa.frames_to_time(librosa.util.fix_frames(beats)))\n\n\nplt.title('Beat-synchronous Chroma (median aggregation)')\n\nplt.colorbar()\nplt.tight_layout()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8beb51ac572dda5e58db459233c580712e56d1
| 24,810 |
ipynb
|
Jupyter Notebook
|
notebooks/minst/sigmoid.ipynb
|
nlauchande/panha
|
15362f7ebaafc4b39989a57ce090bc15dd7e8152
|
[
"MIT"
] | null | null | null |
notebooks/minst/sigmoid.ipynb
|
nlauchande/panha
|
15362f7ebaafc4b39989a57ce090bc15dd7e8152
|
[
"MIT"
] | 6 |
2017-10-22T17:34:15.000Z
|
2017-10-22T18:11:50.000Z
|
notebooks/minst/sigmoid.ipynb
|
nlauchande/panha
|
15362f7ebaafc4b39989a57ce090bc15dd7e8152
|
[
"MIT"
] | null | null | null | 102.520661 | 14,280 | 0.794559 |
[
[
[
"# DLW Practical 1: MNIST\n# From linear to non-linear models with MNIST\n\n**Introduction**\n\nIn this practical we will experiment further with linear and non-linear models using the MNIST dataset. MNIST consists of images of handwritten digits that we want to classify correctly.\n\n**Learning objectives**:\n* Implement a linear classifier on the MNIST image data set in Tensorflow. \n* Modify the code to to make the classifier non-linear by introducing a hidden non-linear layer. \n\n**What is expected of you:**\n* Step through the code and make sure you understand each step. What test set accuracy do you get?\n* Modify the code to make the classifier non-linear by adding a non-linear activation function layer in Tensorflow. What accuracy do you get now?\n\n*Some parts of the code were adapted from the DL Indaba practicals.*",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\nfrom tensorflow.examples.tutorials.mnist import input_data\n\n\ndef display_mnist_images(gens, num_images):\n plt.rcParams['image.interpolation'] = 'nearest'\n plt.rcParams['image.cmap'] = 'gray'\n fig, axs = plt.subplots(1, num_images, figsize=(25, 3))\n for i in range(num_images):\n reshaped_img = (gens[i].reshape(28, 28) * 255).astype(np.uint8)\n axs.flat[i].imshow(reshaped_img)\n plt.show()\n\n\n# download MNIST dataset #\nmnist = input_data.read_data_sets(\"MNIST_data/\", one_hot=True)\n\n# visualize random MNIST images #\nbatch_xs, batch_ys = mnist.train.next_batch(10)\nlist_of_images = np.split(batch_xs, 10)\ndisplay_mnist_images(list_of_images, 10)\n\nx_dim, train_examples, n_classes = mnist.train.images.shape[1], mnist.train.num_examples, mnist.train.labels.shape[1]\n\n######################################\n# define the model (build the graph) #\n######################################\n\nx = tf.placeholder(tf.float32, [None, x_dim])\nW = tf.Variable(tf.random_normal([x_dim, n_classes]))\nb = tf.Variable(tf.ones([n_classes]))\ny = tf.placeholder(tf.float32, [None, n_classes])\ny_ = tf.add(tf.matmul(x, W), b)\nprob = tf.nn.softmax(y_)\n\n########################\n# define loss function #\n########################\n\ncross_entropy_loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=y_, labels=y))\n\nlearning_rate = 0.01\n\ntrain_step = tf.train.GradientDescentOptimizer(learning_rate).minimize(cross_entropy_loss)\n\n###########################\n# define model evaluation #\n###########################\n\nactual_class, predicted_class = tf.argmax(y, 1), tf.argmax(prob, 1)\ncorrect_prediction = tf.cast(tf.equal(predicted_class, actual_class), tf.float32)\nclassification_accuracy = tf.reduce_mean(correct_prediction)\n\n#########################\n# define training cycle #\n#########################\n\nnum_epochs = 50\nbatch_size = 20\n\n# initializing the variables before starting the session #\ninit = tf.global_variables_initializer()\n\n# launch the graph in a session (use the session as a context manager) #\nwith tf.Session() as sess:\n # run session #\n sess.run(init)\n # start main training cycle #\n for epoch in range(num_epochs):\n avg_cost = 0.\n avg_acc = 0.\n total_batch = int(mnist.train.num_examples / batch_size)\n # loop over all batches #\n for i in range(total_batch):\n batch_x, batch_y = mnist.train.next_batch(batch_size)\n # run optimization op (backprop), cost op and accuracy op (to get training losses) #\n _, c, a = sess.run([train_step, cross_entropy_loss, classification_accuracy], feed_dict={x: batch_x, y: batch_y})\n # compute avg training loss and avg training accuracy #\n avg_cost += c / total_batch\n avg_acc += a / total_batch\n # display logs per epoch step #\n if epoch % 1 == 0:\n print(\"Epoch {}: cross-entropy-loss = {:.4f}, training-accuracy = {:.3f}%\".format(epoch + 1, avg_cost, avg_acc * 100))\n print(\"Optimization Finished!\")\n # calculate test set accuracy #\n test_accuracy = classification_accuracy.eval({x: mnist.test.images, y: mnist.test.labels})\n print(\"Accuracy on test set = {:.3f}%\".format(test_accuracy * 100))",
"Extracting MNIST_data/train-images-idx3-ubyte.gz\nExtracting MNIST_data/train-labels-idx1-ubyte.gz\nExtracting MNIST_data/t10k-images-idx3-ubyte.gz\nExtracting MNIST_data/t10k-labels-idx1-ubyte.gz\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
4a8c07c00788e9a12f25d6e4b1248fde97a11b09
| 3,104 |
ipynb
|
Jupyter Notebook
|
Numpy/Ipynb/.ipynb_checkpoints/Numpy_Performace_Test-checkpoint.ipynb
|
BhagyeshVikani/Tutorials
|
21480ba9137c2dbe6559c7f8d01a14c336a9ae0a
|
[
"MIT"
] | 1 |
2017-03-23T19:28:37.000Z
|
2017-03-23T19:28:37.000Z
|
Numpy/Ipynb/Numpy_Performace_Test.ipynb
|
BhagyeshVikani/Tutorials
|
21480ba9137c2dbe6559c7f8d01a14c336a9ae0a
|
[
"MIT"
] | null | null | null |
Numpy/Ipynb/Numpy_Performace_Test.ipynb
|
BhagyeshVikani/Tutorials
|
21480ba9137c2dbe6559c7f8d01a14c336a9ae0a
|
[
"MIT"
] | null | null | null | 25.866667 | 362 | 0.569265 |
[
[
[
"# Numpy_Performace_Test\n\nNumpy is an optimized python library for linear Algebra. Numpy can give very high performace boost for vectorized operations. NumPy arrays facilitate advanced mathematical and other types of operations on large numbers of data. Typi-cally, such operations are executed more efficiently and with less code than is possible using Python’s built-in sequences.",
"_____no_output_____"
]
],
[
[
"import time # Library for tracking execution time\nimport numpy as np # Numpy Library for linear algebra",
"_____no_output_____"
],
[
"n = 300 # No. of elments in a dimension\n\nx = np.random.randn(n , n) # Randomly initializing array(nxn) with N(0,1) distribution",
"_____no_output_____"
],
[
"start = time.time()\n\ny = np.zeros((n , n)) # Allocate memory for output memory\nfor i in range(n):\n for j in range(n):\n for k in range(n):\n y[i, j] += x[i, k] * x[k, j]\n \nstop = time.time()\nprint(\"Python (None-Vectorized) implementation, Execution time = %f s\" % (stop - start))",
"Python (None-Vectorized) implementation, Execution time = 20.671624 s\n"
],
[
"start = time.time()\n\ny = np.matmul(x, x) # Numpy function for matrix multiplication\n\nstop = time.time()\nprint(\"Numpy (Vectorized) implementation, Execution time = %f s\" % (stop - start))",
"Numpy (Vectorized) implementation, Execution time = 0.006236 s\n"
]
],
[
[
"Non-vectorized python implementation takes ~20s for matrix multiplication of two array of just (300x300) dimensions whereas numpy implementation takes only ~0.006s for the same computation. So numpy giving speedup = ~2500.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a8c124cab5167c4c79927142f0ff918d9748b3c
| 309,733 |
ipynb
|
Jupyter Notebook
|
dali/data_loading.ipynb
|
evdcush/DALI
|
802723afd2fc815a73e97982b1a95383db861d60
|
[
"BSD-3-Clause"
] | null | null | null |
dali/data_loading.ipynb
|
evdcush/DALI
|
802723afd2fc815a73e97982b1a95383db861d60
|
[
"BSD-3-Clause"
] | null | null | null |
dali/data_loading.ipynb
|
evdcush/DALI
|
802723afd2fc815a73e97982b1a95383db861d60
|
[
"BSD-3-Clause"
] | null | null | null | 696.029213 | 167,720 | 0.949214 |
[
[
[
"# Data loading with ExternalSource operator\nIn this notebook, we will see how to use the `ExternalSource` operator, which allows us to use an external data source as input to the Pipeline.\n\nThis notebook derived from: https://docs.nvidia.com/deeplearning/dali/user-guide/docs/examples/general/data_loading/external_input.html",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport collections\nfrom random import shuffle\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom nvidia.dali.pipeline import Pipeline\nfrom nvidia.dali import ops, types\n\nbatch_size = 16",
"_____no_output_____"
]
],
[
[
"## Defining the data source\nWe use an infinite iterator as a data source, on the sample dogs & cats images.",
"_____no_output_____"
]
],
[
[
"class ExternalInputIterator:\n def __init__(self, batch_size):\n self.images_dir = 'data/images/'\n self.batch_size = batch_size\n with open(self.images_dir + 'file_list.txt') as file:\n self.files = [line.rstrip() for line in file if line]\n shuffle(self.files)\n \n def __iter__(self):\n \"\"\" (this is not typical __iter__ ?) \"\"\"\n self.i = 0\n self.n = len(self.files)\n return self\n \n def __next__(self):\n batch = []\n labels = []\n for _ in range(self.batch_size):\n jpg_fname, label = self.files[self.i].split(' ')\n file = open(self.images_dir + jpg_fname, 'rb')\n batch.append(np.frombuffer(file.read(), dtype=np.uint8))\n labels.append(np.array([label], dtype=np.uint8))\n self.i = (self.i + 1) % self.n\n return batch, labels",
"_____no_output_____"
]
],
[
[
"# Defining the pipeline\nThe next step is to define the Pipeline.\n\nThe `ExternalSource` op accepts an iterable or a callable. If the source provides multiple outputs (eg images and labels), that number must also be specified as `num_outputs` argument.\n\nInternally, the pipeline will call `source` (if callable) or run `next(source)`(if iterable) whenever more data is needed to keep the pipeline running.",
"_____no_output_____"
]
],
[
[
"ext_input_iter = ExternalInputIterator(batch_size)",
"_____no_output_____"
],
[
"class ExternalSourcePipeline(Pipeline):\n def __init__(self, batch_size, ext_inp_iter, \n num_threads, device_id):\n super().__init__(batch_size, num_threads, device_id, seed=12)\n self.source = ops.ExternalSource(source=ext_inp_iter, num_outputs=2)\n self.decode = ops.ImageDecoder(device='mixed', output_type=types.RGB)\n self.enhance = ops.BrightnessContrast(device='gpu', contrast=2)\n \n def define_graph(self):\n jpgs, labels = self.source()\n images = self.decode(jpgs)\n output = self.enhance(images)\n return output, labels",
"_____no_output_____"
]
],
[
[
"# Using the Pipeline",
"_____no_output_____"
]
],
[
[
"ext_pipe = ExternalSourcePipeline\npipe = ext_pipe(batch_size, ext_input_iter, num_threads=2, device_id=0)\npipe.build()\npipe_out = pipe.run()",
"_____no_output_____"
]
],
[
[
"Notice that labels are still on CPU and no `as_cpu()` call is needed to show them.",
"_____no_output_____"
]
],
[
[
"batch_cpu = pipe_out[0].as_cpu()\nlabels_cpu = pipe_out[1] # already on cpu!",
"_____no_output_____"
],
[
"print(type(batch_cpu))\ndir(batch_cpu)",
"<class 'nvidia.dali.backend_impl.TensorListCPU'>\n"
],
[
"img = batch_cpu.at(2)\nprint(img.shape)\nprint(labels_cpu.at(2))\nplt.axis('off')\nplt.imshow(img)",
"(427, 640, 3)\n[0]\n"
]
],
[
[
"## Interacting with the GPU input\nThe external source operator can also accept GPU data from CuPy, or any other data source that supports the [CUDA array interface](https://numba.pydata.org/numba-doc/latest/cuda/cuda_array_interface.html) (including PyTorch).\n\nFor the sake of this example, we will create an `ExternalInputGpuIterator` in such a way that it returns data on the GPU already.\n\n\nAs `ImageDecoder` does not accept data on the GPU, *we need to decode it outside of DALI on the CPU and then move it to the GPU.* In normal cases, the image, or other data, would already be on the GPU as a result of the operation of another library.",
"_____no_output_____"
]
],
[
[
"import imageio\nimport cupy as cp\n\nclass ExternalInputGpuIterator:\n def __init__(self, batch_size):\n self.images_dir = 'data/images/'\n self.batch_size = batch_size\n with open(self.images_dir + 'file_list.txt') as file:\n self.files = [line.rstrip() for line in file if line]\n shuffle(self.files)\n \n def __iter__(self):\n self.i = 0\n self.n = len(self.files)\n return self\n \n def __next__(self):\n batch = []\n labels = []\n for _ in range(self.batch_size):\n jpg_fname, label = self.files[self.i].split(' ')\n img = imageio.imread(self.images_dir + jpg_fname)\n img = cp.asarray(img)\n img = img * 0.6\n batch.append(img.astype(cp.uint8))\n labels.append(cp.asarray([label], dtype=np.uint8))\n self.i = (self.i + 1) % self.n\n return batch, labels",
"_____no_output_____"
]
],
[
[
"Now that we assume the image decoding was done outside of DALI for the GPU case (ie, the raw image is already on the GPU), let's modify our previous pipeline to use the GPU version of our external source iterator.",
"_____no_output_____"
]
],
[
[
"ext_iter_gpu = ExternalInputGpuIterator(batch_size)\nprint(type(next(iter(ext_iter_gpu))[0][0]))",
"<class 'cupy.core.core.ndarray'>\n"
],
[
"class ExternalSourceGpuPipeline(Pipeline):\n def __init__(self, batch_size, ext_inp_iter, num_threads, \n device_id):\n super().__init__(batch_size, num_threads, device_id, seed=12)\n self.source = ops.ExternalSource(device='gpu', \n source=ext_inp_iter,\n num_outputs=2)\n self.enhance = ops.BrightnessContrast(device='gpu', \n contrast=2)\n \n def define_graph(self):\n images, labels = self.source()\n output = self.enhance(images)\n return output, labels",
"_____no_output_____"
],
[
"pipe_gpu = ExternalSourceGpuPipeline(batch_size,ext_iter_gpu, \n num_threads=2, device_id=0)\npipe_gpu.build()",
"_____no_output_____"
],
[
"pipe_out_gpu = pipe_gpu.run()\nbatch_gpu = pipe_out_gpu[0].as_cpu() # dali.backend_impl.TensorListCPU\nlabels_gpu = pipe_out_gpu[1].as_cpu() # dali.backend_impl.TensorListCPU\n\n# show img\nimg = batch_gpu.at(2)\nprint(img.shape)\nprint(labels_gpu.at(2))\nplt.axis('off')\nplt.imshow(img)",
"(425, 640, 3)\n[0]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4a8c1421d2c9e2a82389a8f92cf63213bff07c45
| 10,319 |
ipynb
|
Jupyter Notebook
|
lessons/bmi/bmi-run-model-from-bmi.ipynb
|
csdms/espin
|
470dca76175e0a76d902d1e14e0003a49d31f4d0
|
[
"CC-BY-4.0"
] | 27 |
2020-08-07T23:16:44.000Z
|
2022-03-30T15:59:16.000Z
|
lessons/bmi/bmi-run-model-from-bmi.ipynb
|
csdms/espin
|
470dca76175e0a76d902d1e14e0003a49d31f4d0
|
[
"CC-BY-4.0"
] | 28 |
2020-07-09T21:28:49.000Z
|
2022-03-11T16:49:24.000Z
|
lessons/bmi/bmi-run-model-from-bmi.ipynb
|
csdms/espin
|
470dca76175e0a76d902d1e14e0003a49d31f4d0
|
[
"CC-BY-4.0"
] | 48 |
2020-08-09T23:03:15.000Z
|
2021-06-18T20:50:11.000Z
| 22.002132 | 441 | 0.495881 |
[
[
[
"<a href=\"https://bmi.readthedocs.io\"><img src=\"https://raw.githubusercontent.com/csdms/espin/main/media/bmi-logo-header-text.png\"></a>",
"_____no_output_____"
],
[
"# Run the `Heat` model through its BMI",
"_____no_output_____"
],
[
"`Heat` models the diffusion of temperature on a uniform rectangular plate with Dirichlet boundary conditions. This is the canonical example used in the [bmi-example-python](https://github.com/csdms/bmi-example-python) repository. View the source code for the [model](https://github.com/csdms/bmi-example-python/blob/master/heat/heat.py) and its [BMI](https://github.com/csdms/bmi-example-python/blob/master/heat/bmi_heat.py) on GitHub.",
"_____no_output_____"
],
[
"Start by importing `os`, `numpy` and the `Heat` BMI:",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\n\nfrom heat import BmiHeat",
"_____no_output_____"
]
],
[
[
"Create an instance of the model's BMI.",
"_____no_output_____"
]
],
[
[
"x = BmiHeat()",
"_____no_output_____"
]
],
[
[
"What's the name of this model?",
"_____no_output_____"
]
],
[
[
"print(x.get_component_name())",
"The 2D Heat Equation\n"
]
],
[
[
"Start the `Heat` model through its BMI using a configuration file:",
"_____no_output_____"
]
],
[
[
"cat heat.yaml",
"# Heat model configuration\r\nshape:\r\n - 6\r\n - 8\r\nspacing:\r\n - 1.0\r\n - 1.0\r\norigin:\r\n - 0.0\r\n - 0.0\r\nalpha: 1.0\r\n"
],
[
"x.initialize('heat.yaml')",
"_____no_output_____"
]
],
[
[
"Check the time information for the model.",
"_____no_output_____"
]
],
[
[
"print('Start time:', x.get_start_time())\nprint('End time:', x.get_end_time())\nprint('Current time:', x.get_current_time())\nprint('Time step:', x.get_time_step())\nprint('Time units:', x.get_time_units())",
"Start time: 0.0\nEnd time: 1.7976931348623157e+308\nCurrent time: 0.0\nTime step: 0.25\nTime units: s\n"
]
],
[
[
"Show the input and output variables for the component (aside on [Standard Names](https://csdms.colorado.edu/wiki/CSDMS_Standard_Names)):",
"_____no_output_____"
]
],
[
[
"print(x.get_input_var_names())\nprint(x.get_output_var_names())",
"('plate_surface__temperature',)\n('plate_surface__temperature',)\n"
]
],
[
[
"Next, get the identifier for the grid on which the temperature variable is defined:",
"_____no_output_____"
]
],
[
[
"grid_id = x.get_var_grid('plate_surface__temperature')\nprint('Grid id:', grid_id)",
"Grid id: 0\n"
]
],
[
[
"Then get the grid attributes:",
"_____no_output_____"
]
],
[
[
"print('Grid type:', x.get_grid_type(grid_id))\n\nrank = x.get_grid_rank(grid_id)\nprint('Grid rank:', rank)\n\nshape = np.ndarray(rank, dtype=int)\nx.get_grid_shape(grid_id, shape)\nprint('Grid shape:', shape)\n\nspacing = np.ndarray(rank, dtype=float)\nx.get_grid_spacing(grid_id, spacing)\nprint('Grid spacing:', spacing)",
"Grid type: uniform_rectilinear\nGrid rank: 2\nGrid shape: [6 8]\nGrid spacing: [1. 1.]\n"
]
],
[
[
"These commands are made somewhat un-Pythonic by the generic design of the BMI.",
"_____no_output_____"
],
[
"Through the model's BMI, zero out the initial temperature field, except for an impulse near the middle.\nNote that *set_value* expects a one-dimensional array for input.",
"_____no_output_____"
]
],
[
[
"temperature = np.zeros(shape)\ntemperature[3, 4] = 100.0\nx.set_value('plate_surface__temperature', temperature)",
"_____no_output_____"
]
],
[
[
"Check that the temperature field has been updated. Note that *get_value* expects a one-dimensional array to receive output.",
"_____no_output_____"
]
],
[
[
"temperature_flat = np.empty_like(temperature).flatten()\nx.get_value('plate_surface__temperature', temperature_flat)\nprint(temperature_flat.reshape(shape))",
"[[ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 100. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]\n [ 0. 0. 0. 0. 0. 0. 0. 0.]]\n"
]
],
[
[
"Now advance the model by a single time step:",
"_____no_output_____"
]
],
[
[
"x.update()",
"_____no_output_____"
]
],
[
[
"View the new state of the temperature field:",
"_____no_output_____"
]
],
[
[
"x.get_value('plate_surface__temperature', temperature_flat)\nprint(temperature_flat.reshape(shape))",
"[[ 0. 0. 0. 0. 0. 0. 0. 0. ]\n [ 0. 0. 0. 0. 0. 0. 0. 0. ]\n [ 0. 0. 0. 0. 12.5 0. 0. 0. ]\n [ 0. 0. 0. 12.5 50. 12.5 0. 0. ]\n [ 0. 0. 0. 0. 12.5 0. 0. 0. ]\n [ 0. 0. 0. 0. 0. 0. 0. 0. ]]\n"
]
],
[
[
"There's diffusion!",
"_____no_output_____"
],
[
"Advance the model to some distant time:",
"_____no_output_____"
]
],
[
[
"distant_time = 2.0\nwhile x.get_current_time() < distant_time:\n x.update()",
"_____no_output_____"
]
],
[
[
"View the final state of the temperature field:",
"_____no_output_____"
]
],
[
[
"np.set_printoptions(formatter={'float': '{: 5.1f}'.format})\nx.get_value('plate_surface__temperature', temperature_flat)\nprint(temperature_flat.reshape(shape))",
"[[ 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0]\n [ 0.0 0.2 0.9 2.1 2.8 2.1 0.9 0.0]\n [ 0.0 0.7 2.2 4.7 6.2 4.7 2.1 0.0]\n [ 0.0 0.9 3.0 6.1 7.9 6.1 2.8 0.0]\n [ 0.0 0.6 2.0 4.1 5.3 4.1 1.8 0.0]\n [ 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0]]\n"
]
],
[
[
"Note that temperature isn't conserved on the plate:",
"_____no_output_____"
]
],
[
[
"print(temperature_flat.sum())",
"74.10263419151306\n"
]
],
[
[
"End the model:",
"_____no_output_____"
]
],
[
[
"x.finalize()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8c18f3e2cd7d90f6e8504d8f02e405662b92b7
| 18,927 |
ipynb
|
Jupyter Notebook
|
cv_rnr_8s_proposed_gap.ipynb
|
iamsoroush/DeepEEGAbstractor
|
3b83570b8edca63a0132b98729cd7e5f4a9d355c
|
[
"MIT"
] | 1 |
2022-03-11T05:52:18.000Z
|
2022-03-11T05:52:18.000Z
|
cv_rnr_8s_proposed_gap.ipynb
|
iamsoroush/DeepEEGAbstractor
|
3b83570b8edca63a0132b98729cd7e5f4a9d355c
|
[
"MIT"
] | null | null | null |
cv_rnr_8s_proposed_gap.ipynb
|
iamsoroush/DeepEEGAbstractor
|
3b83570b8edca63a0132b98729cd7e5f4a9d355c
|
[
"MIT"
] | null | null | null | 38.547862 | 249 | 0.400697 |
[
[
[
"<a href=\"https://colab.research.google.com/github/iamsoroush/DeepEEGAbstractor/blob/master/cv_rnr_8s_proposed_gap.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#@title # Clone the repository and upgrade Keras {display-mode: \"form\"}\n\n!git clone https://github.com/iamsoroush/DeepEEGAbstractor.git\n!pip install --upgrade keras",
"_____no_output_____"
],
[
"#@title # Imports {display-mode: \"form\"}\n\nimport os\nimport pickle\nimport sys\nsys.path.append('DeepEEGAbstractor')\n\nimport numpy as np\n\nfrom src.helpers import CrossValidator\nfrom src.models import SpatioTemporalWFB, TemporalWFB, TemporalDFB, SpatioTemporalDFB\nfrom src.dataset import DataLoader, Splitter, FixedLenGenerator\n\nfrom google.colab import drive\ndrive.mount('/content/gdrive')",
"_____no_output_____"
],
[
"#@title # Set data path {display-mode: \"form\"}\n\n#@markdown ---\n#@markdown Type in the folder in your google drive that contains numpy _data_ folder:\n\nparent_dir = 'soroush'#@param {type:\"string\"}\ngdrive_path = os.path.abspath(os.path.join('gdrive/My Drive', parent_dir))\ndata_dir = os.path.join(gdrive_path, 'data')\ncv_results_dir = os.path.join(gdrive_path, 'cross_validation')\nif not os.path.exists(cv_results_dir):\n os.mkdir(cv_results_dir)\n\nprint('Data directory: ', data_dir)\nprint('Cross validation results dir: ', cv_results_dir)",
"_____no_output_____"
],
[
"#@title ## Set Parameters\n\nbatch_size = 80\nepochs = 50\nk = 10\nt = 10\ninstance_duration = 8 #@param {type:\"slider\", min:3, max:10, step:0.5}\ninstance_overlap = 2 #@param {type:\"slider\", min:0, max:3, step:0.5}\nsampling_rate = 256 #@param {type:\"number\"}\nn_channels = 20 #@param {type:\"number\"}\ntask = 'rnr'\ndata_mode = 'cross_subject'",
"_____no_output_____"
],
[
"#@title ## Spatio-Temporal WFB\n\nmodel_name = 'ST-WFB-GAP'\n\ntrain_generator = FixedLenGenerator(batch_size=batch_size,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=True)\n\ntest_generator = FixedLenGenerator(batch_size=8,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=False)\n\nparams = {'task': task,\n 'data_mode': data_mode,\n 'main_res_dir': cv_results_dir,\n 'model_name': model_name,\n 'epochs': epochs,\n 'train_generator': train_generator,\n 'test_generator': test_generator,\n 't': t,\n 'k': k,\n 'channel_drop': True}\n\nvalidator = CrossValidator(**params)\n\ndataloader = DataLoader(data_dir,\n task,\n data_mode,\n sampling_rate,\n instance_duration,\n instance_overlap)\ndata, labels = dataloader.load_data()\n\ninput_shape = (sampling_rate * instance_duration,\n n_channels)\n\nmodel_obj = SpatioTemporalWFB(input_shape,\n model_name=model_name,\n spatial_dropout_rate=0.2,\n dropout_rate=0.4)\n\nscores = validator.do_cv(model_obj,\n data,\n labels)",
"_____no_output_____"
],
[
"#@title ## Temporal WFB\n\nmodel_name = 'T-WFB-GAP'\n\ntrain_generator = FixedLenGenerator(batch_size=batch_size,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=True)\n\ntest_generator = FixedLenGenerator(batch_size=8,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=False)\n\nparams = {'task': task,\n 'data_mode': data_mode,\n 'main_res_dir': cv_results_dir,\n 'model_name': model_name,\n 'epochs': epochs,\n 'train_generator': train_generator,\n 'test_generator': test_generator,\n 't': t,\n 'k': k,\n 'channel_drop': True}\n\nvalidator = CrossValidator(**params)\n\ndataloader = DataLoader(data_dir,\n task,\n data_mode,\n sampling_rate,\n instance_duration,\n instance_overlap)\ndata, labels = dataloader.load_data()\n\ninput_shape = (sampling_rate * instance_duration,\n n_channels)\n\nmodel_obj = TemporalWFB(input_shape,\n model_name=model_name,\n spatial_dropout_rate=0.2,\n dropout_rate=0.4)\n\nscores = validator.do_cv(model_obj,\n data,\n labels)",
"_____no_output_____"
],
[
"#@title ## Spatio-Temporal DFB\n\nmodel_name = 'ST-DFB-GAP'\n\ntrain_generator = FixedLenGenerator(batch_size=batch_size,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=True)\n\ntest_generator = FixedLenGenerator(batch_size=8,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=False)\n\nparams = {'task': task,\n 'data_mode': data_mode,\n 'main_res_dir': cv_results_dir,\n 'model_name': model_name,\n 'epochs': epochs,\n 'train_generator': train_generator,\n 'test_generator': test_generator,\n 't': t,\n 'k': k,\n 'channel_drop': True}\n\nvalidator = CrossValidator(**params)\n\ndataloader = DataLoader(data_dir,\n task,\n data_mode,\n sampling_rate,\n instance_duration,\n instance_overlap)\ndata, labels = dataloader.load_data()\n\ninput_shape = (sampling_rate * instance_duration,\n n_channels)\n\nmodel_obj = SpatioTemporalDFB(input_shape,\n model_name=model_name,\n spatial_dropout_rate=0.2,\n dropout_rate=0.4)\n\nscores = validator.do_cv(model_obj,\n data,\n labels)",
"_____no_output_____"
],
[
"#@title ## Spatio-Temporal DFB (Normalized Kernels)\n\nmodel_name = 'ST-DFB-NK-GAP'\n\ntrain_generator = FixedLenGenerator(batch_size=batch_size,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=True)\n\ntest_generator = FixedLenGenerator(batch_size=8,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=False)\n\nparams = {'task': task,\n 'data_mode': data_mode,\n 'main_res_dir': cv_results_dir,\n 'model_name': model_name,\n 'epochs': epochs,\n 'train_generator': train_generator,\n 'test_generator': test_generator,\n 't': t,\n 'k': k,\n 'channel_drop': True}\n\nvalidator = CrossValidator(**params)\n\ndataloader = DataLoader(data_dir,\n task,\n data_mode,\n sampling_rate,\n instance_duration,\n instance_overlap)\ndata, labels = dataloader.load_data()\n\ninput_shape = (sampling_rate * instance_duration,\n n_channels)\n\nmodel_obj = SpatioTemporalDFB(input_shape,\n model_name=model_name,\n spatial_dropout_rate=0.2,\n dropout_rate=0.4,\n normalize_kernels=True)\n\nscores = validator.do_cv(model_obj,\n data,\n labels)",
"_____no_output_____"
],
[
"#@title ## Temporal DFB\n\nmodel_name = 'T-DFB-GAP'\n\ntrain_generator = FixedLenGenerator(batch_size=batch_size,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=True)\n\ntest_generator = FixedLenGenerator(batch_size=8,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=False)\n\nparams = {'task': task,\n 'data_mode': data_mode,\n 'main_res_dir': cv_results_dir,\n 'model_name': model_name,\n 'epochs': epochs,\n 'train_generator': train_generator,\n 'test_generator': test_generator,\n 't': t,\n 'k': k,\n 'channel_drop': True}\n\nvalidator = CrossValidator(**params)\n\ndataloader = DataLoader(data_dir,\n task,\n data_mode,\n sampling_rate,\n instance_duration,\n instance_overlap)\ndata, labels = dataloader.load_data()\n\ninput_shape = (sampling_rate * instance_duration,\n n_channels)\n\nmodel_obj = TemporalDFB(input_shape,\n model_name=model_name,\n spatial_dropout_rate=0.2,\n dropout_rate=0.4)\n\nscores = validator.do_cv(model_obj,\n data,\n labels)",
"_____no_output_____"
],
[
"#@title ## Temporal DFB (Normalized Kernels)\n\nmodel_name = 'T-DFB-NK-GAP'\n\ntrain_generator = FixedLenGenerator(batch_size=batch_size,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=True)\n\ntest_generator = FixedLenGenerator(batch_size=8,\n duration=instance_duration,\n overlap=instance_overlap,\n sampling_rate=sampling_rate,\n is_train=False)\n\nparams = {'task': task,\n 'data_mode': data_mode,\n 'main_res_dir': cv_results_dir,\n 'model_name': model_name,\n 'epochs': epochs,\n 'train_generator': train_generator,\n 'test_generator': test_generator,\n 't': t,\n 'k': k,\n 'channel_drop': True}\n\nvalidator = CrossValidator(**params)\n\ndataloader = DataLoader(data_dir,\n task,\n data_mode,\n sampling_rate,\n instance_duration,\n instance_overlap)\ndata, labels = dataloader.load_data()\n\ninput_shape = (sampling_rate * instance_duration,\n n_channels)\n\nmodel_obj = TemporalDFB(input_shape,\n model_name=model_name,\n spatial_dropout_rate=0.2,\n dropout_rate=0.4,\n normalize_kernels=True)\n\nscores = validator.do_cv(model_obj,\n data,\n labels)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8c22d93f31b00c3862e8c4a0fde49748cd459d
| 182,445 |
ipynb
|
Jupyter Notebook
|
Cat vs Dog-1.ipynb
|
Yoshibansal/ML-practical
|
f381b0a19781a81cb6f533be28c3710f866b9f7b
|
[
"MIT"
] | 1 |
2020-12-02T07:58:09.000Z
|
2020-12-02T07:58:09.000Z
|
Cat vs Dog-1.ipynb
|
Yoshibansal/ML-practical
|
f381b0a19781a81cb6f533be28c3710f866b9f7b
|
[
"MIT"
] | null | null | null |
Cat vs Dog-1.ipynb
|
Yoshibansal/ML-practical
|
f381b0a19781a81cb6f533be28c3710f866b9f7b
|
[
"MIT"
] | null | null | null | 338.487941 | 125,386 | 0.903818 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Yoshibansal/ML-practical/blob/main/Cat_vs_Dog_Part-1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##Cat vs Dog (Binary class classification)\nImageDataGenerator\n\n(Understanding overfitting)",
"_____no_output_____"
],
[
"Download dataset",
"_____no_output_____"
]
],
[
[
"!wget --no-check-certificate \\\n https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip \\\n -O /tmp/cats_and_dogs_filtered.zip",
"--2020-12-05 14:54:05-- https://storage.googleapis.com/mledu-datasets/cats_and_dogs_filtered.zip\nResolving storage.googleapis.com (storage.googleapis.com)... 172.217.7.208, 142.250.73.208, 142.250.73.240, ...\nConnecting to storage.googleapis.com (storage.googleapis.com)|172.217.7.208|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 68606236 (65M) [application/zip]\nSaving to: ‘/tmp/cats_and_dogs_filtered.zip’\n\n/tmp/cats_and_dogs_ 100%[===================>] 65.43M 235MB/s in 0.3s \n\n2020-12-05 14:54:05 (235 MB/s) - ‘/tmp/cats_and_dogs_filtered.zip’ saved [68606236/68606236]\n\n"
],
[
"#importing libraries\nimport os\nimport zipfile\nimport tensorflow as tf\nfrom tensorflow.keras.optimizers import RMSprop\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator",
"_____no_output_____"
],
[
"#unzip\nlocal_zip = '/tmp/cats_and_dogs_filtered.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp')\nzip_ref.close()",
"_____no_output_____"
],
[
"base_dir = '/tmp/cats_and_dogs_filtered'\ntrain_dir = os.path.join(base_dir, 'train')\nvalidation_dir = os.path.join(base_dir, 'validation')\n\n# Directory with our training cat pictures\ntrain_cats_dir = os.path.join(train_dir, 'cats')\n\n# Directory with our training dog pictures\ntrain_dogs_dir = os.path.join(train_dir, 'dogs')\n\n# Directory with our validation cat pictures\nvalidation_cats_dir = os.path.join(validation_dir, 'cats')\n\n# Directory with our validation dog pictures\nvalidation_dogs_dir = os.path.join(validation_dir, 'dogs')",
"_____no_output_____"
],
[
"INPUT_SHAPE = (150, 150)\nMODEL_INPUT_SHAPE = INPUT_SHAPE + (3,)\n\n#HYPERPARAMETERS\nLEARNING_RATE = 1e-4\nBATCH_SIZE = 20\nEPOCHS = 50",
"_____no_output_____"
],
[
"#model architecture\nmodel = tf.keras.models.Sequential([\n tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape = MODEL_INPUT_SHAPE),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Conv2D(64, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(128, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Conv2D(128, (3,3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2,2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(512, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid')\n])\n\nmodel.compile(loss='binary_crossentropy',\n optimizer=RMSprop(lr=LEARNING_RATE),\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"#summary of model (including type of layer, Ouput shape and number of parameters)\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d (Conv2D) (None, 148, 148, 32) 896 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 74, 74, 32) 0 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 72, 72, 64) 18496 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 36, 36, 64) 0 \n_________________________________________________________________\nconv2d_2 (Conv2D) (None, 34, 34, 128) 73856 \n_________________________________________________________________\nmax_pooling2d_2 (MaxPooling2 (None, 17, 17, 128) 0 \n_________________________________________________________________\nconv2d_3 (Conv2D) (None, 15, 15, 128) 147584 \n_________________________________________________________________\nmax_pooling2d_3 (MaxPooling2 (None, 7, 7, 128) 0 \n_________________________________________________________________\nflatten (Flatten) (None, 6272) 0 \n_________________________________________________________________\ndense (Dense) (None, 512) 3211776 \n_________________________________________________________________\ndense_1 (Dense) (None, 1) 513 \n=================================================================\nTotal params: 3,453,121\nTrainable params: 3,453,121\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"#plotting model and saving it architecture picture\ndot_img_file = '/tmp/model_1.png'\ntf.keras.utils.plot_model(model, to_file=dot_img_file, show_shapes=True)",
"_____no_output_____"
],
[
"# All images will be rescaled by 1./255\ntrain_datagen = ImageDataGenerator(rescale=1./255)\ntest_datagen = ImageDataGenerator(rescale=1./255)",
"_____no_output_____"
],
[
"# Flow training images in batches of 20 using train_datagen generator\ntrain_generator = train_datagen.flow_from_directory(\n train_dir, # This is the source directory for training images\n target_size=INPUT_SHAPE, # All images will be resized to 150x150\n batch_size=BATCH_SIZE,\n # Since we use binary_crossentropy loss, we need binary labels\n class_mode='binary')\n\n# Flow validation images in batches of 20 using test_datagen generator\nvalidation_generator = test_datagen.flow_from_directory(\n validation_dir,\n target_size=INPUT_SHAPE,\n batch_size=BATCH_SIZE,\n class_mode='binary')",
"Found 2000 images belonging to 2 classes.\nFound 1000 images belonging to 2 classes.\n"
],
[
"#Fitting data into model -> training model\nhistory = model.fit(\n train_generator,\n steps_per_epoch=100, # steps = 2000 images / batch_size\n epochs=EPOCHS,\n validation_data=validation_generator,\n validation_steps=50, # steps = 1000 images / batch_size\n verbose=1)",
"Epoch 1/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.6879 - accuracy: 0.5430 - val_loss: 0.6701 - val_accuracy: 0.5190\nEpoch 2/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.6447 - accuracy: 0.6365 - val_loss: 0.6158 - val_accuracy: 0.6660\nEpoch 3/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.5964 - accuracy: 0.6850 - val_loss: 0.5820 - val_accuracy: 0.7000\nEpoch 4/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.5585 - accuracy: 0.7110 - val_loss: 0.5762 - val_accuracy: 0.6860\nEpoch 5/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.5240 - accuracy: 0.7350 - val_loss: 0.5707 - val_accuracy: 0.7080\nEpoch 6/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.5023 - accuracy: 0.7520 - val_loss: 0.5986 - val_accuracy: 0.6730\nEpoch 7/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.4773 - accuracy: 0.7700 - val_loss: 0.5447 - val_accuracy: 0.7260\nEpoch 8/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.4528 - accuracy: 0.7940 - val_loss: 0.6390 - val_accuracy: 0.6600\nEpoch 9/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.4295 - accuracy: 0.8020 - val_loss: 0.5196 - val_accuracy: 0.7380\nEpoch 10/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.4122 - accuracy: 0.8125 - val_loss: 0.5803 - val_accuracy: 0.7150\nEpoch 11/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.3795 - accuracy: 0.8255 - val_loss: 0.5562 - val_accuracy: 0.7330\nEpoch 12/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.3613 - accuracy: 0.8310 - val_loss: 0.5210 - val_accuracy: 0.7430\nEpoch 13/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.3379 - accuracy: 0.8525 - val_loss: 0.5173 - val_accuracy: 0.7440\nEpoch 14/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.3202 - accuracy: 0.8635 - val_loss: 0.5308 - val_accuracy: 0.7460\nEpoch 15/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.2995 - accuracy: 0.8700 - val_loss: 0.5562 - val_accuracy: 0.7440\nEpoch 16/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.2835 - accuracy: 0.8850 - val_loss: 0.5315 - val_accuracy: 0.7450\nEpoch 17/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.2555 - accuracy: 0.9025 - val_loss: 0.5636 - val_accuracy: 0.7480\nEpoch 18/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.2349 - accuracy: 0.9120 - val_loss: 0.6298 - val_accuracy: 0.7330\nEpoch 19/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.2185 - accuracy: 0.9180 - val_loss: 0.5489 - val_accuracy: 0.7650\nEpoch 20/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.2081 - accuracy: 0.9215 - val_loss: 0.5464 - val_accuracy: 0.7610\nEpoch 21/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.1836 - accuracy: 0.9305 - val_loss: 0.5613 - val_accuracy: 0.7530\nEpoch 22/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.1654 - accuracy: 0.9415 - val_loss: 0.5568 - val_accuracy: 0.7600\nEpoch 23/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.1527 - accuracy: 0.9525 - val_loss: 0.5870 - val_accuracy: 0.7600\nEpoch 24/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.1387 - accuracy: 0.9530 - val_loss: 0.6485 - val_accuracy: 0.7500\nEpoch 25/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.1255 - accuracy: 0.9625 - val_loss: 0.6270 - val_accuracy: 0.7620\nEpoch 26/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.1046 - accuracy: 0.9680 - val_loss: 0.6643 - val_accuracy: 0.7620\nEpoch 27/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0938 - accuracy: 0.9715 - val_loss: 0.7531 - val_accuracy: 0.7430\nEpoch 28/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0832 - accuracy: 0.9750 - val_loss: 0.6733 - val_accuracy: 0.7720\nEpoch 29/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0738 - accuracy: 0.9785 - val_loss: 0.6778 - val_accuracy: 0.7640\nEpoch 30/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.0610 - accuracy: 0.9825 - val_loss: 0.8881 - val_accuracy: 0.7290\nEpoch 31/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0506 - accuracy: 0.9870 - val_loss: 0.7932 - val_accuracy: 0.7520\nEpoch 32/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0475 - accuracy: 0.9880 - val_loss: 0.7944 - val_accuracy: 0.7560\nEpoch 33/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0408 - accuracy: 0.9865 - val_loss: 0.8426 - val_accuracy: 0.7510\nEpoch 34/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0325 - accuracy: 0.9930 - val_loss: 0.9244 - val_accuracy: 0.7480\nEpoch 35/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0280 - accuracy: 0.9945 - val_loss: 1.6320 - val_accuracy: 0.6920\nEpoch 36/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0281 - accuracy: 0.9925 - val_loss: 0.9100 - val_accuracy: 0.7530\nEpoch 37/50\n100/100 [==============================] - 9s 92ms/step - loss: 0.0225 - accuracy: 0.9955 - val_loss: 1.0911 - val_accuracy: 0.7520\nEpoch 38/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0199 - accuracy: 0.9945 - val_loss: 1.0187 - val_accuracy: 0.7560\nEpoch 39/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0187 - accuracy: 0.9950 - val_loss: 0.9878 - val_accuracy: 0.7590\nEpoch 40/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0158 - accuracy: 0.9960 - val_loss: 1.0300 - val_accuracy: 0.7580\nEpoch 41/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0132 - accuracy: 0.9970 - val_loss: 1.0388 - val_accuracy: 0.7670\nEpoch 42/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0170 - accuracy: 0.9950 - val_loss: 1.0747 - val_accuracy: 0.7640\nEpoch 43/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0094 - accuracy: 0.9970 - val_loss: 1.1586 - val_accuracy: 0.7620\nEpoch 44/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0157 - accuracy: 0.9955 - val_loss: 1.1488 - val_accuracy: 0.7590\nEpoch 45/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0092 - accuracy: 0.9980 - val_loss: 1.1961 - val_accuracy: 0.7590\nEpoch 46/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0077 - accuracy: 0.9990 - val_loss: 1.2261 - val_accuracy: 0.7600\nEpoch 47/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0107 - accuracy: 0.9965 - val_loss: 1.2308 - val_accuracy: 0.7530\nEpoch 48/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0096 - accuracy: 0.9970 - val_loss: 1.3061 - val_accuracy: 0.7510\nEpoch 49/50\n100/100 [==============================] - 9s 90ms/step - loss: 0.0077 - accuracy: 0.9980 - val_loss: 1.2961 - val_accuracy: 0.7600\nEpoch 50/50\n100/100 [==============================] - 9s 91ms/step - loss: 0.0095 - accuracy: 0.9980 - val_loss: 1.4330 - val_accuracy: 0.7450\n"
],
[
"#PLOTTING model performance\n\nimport matplotlib.pyplot as plt\nacc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(len(acc))\n\nplt.plot(epochs, acc, 'bo', label='Training accuracy')\nplt.plot(epochs, val_acc, 'b', label='Validation accuracy')\nplt.title('Training and validation accuracy')\n\nplt.figure()\n\nplt.plot(epochs, loss, 'ro', label='Training Loss')\nplt.plot(epochs, val_loss, 'r', label='Validation Loss')\nplt.title('Training and validation loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"The Training Accuracy is close to 100%, and the validation accuracy is in the 70%-80% range. This is a great example of overfitting -- which in short means that it can do very well with images it has seen before, but not so well with images it hasn't.\n\nnext we see how we can do better to avoid overfitting -- and one simple method is to **augment** the images a bit.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8c31f4fa5d41fde90e596aa5bd574ab6792128
| 18,720 |
ipynb
|
Jupyter Notebook
|
Project_2_Project_Notebook.ipynb
|
jukkakansanaho/udacity-dend-project-2
|
fa868176e4e567390457788d96d2627405a16d41
|
[
"MIT"
] | 1 |
2021-06-22T11:22:59.000Z
|
2021-06-22T11:22:59.000Z
|
Project_2_Project_Notebook.ipynb
|
jukkakansanaho/udacity-dend-project-2
|
fa868176e4e567390457788d96d2627405a16d41
|
[
"MIT"
] | null | null | null |
Project_2_Project_Notebook.ipynb
|
jukkakansanaho/udacity-dend-project-2
|
fa868176e4e567390457788d96d2627405a16d41
|
[
"MIT"
] | 3 |
2019-09-19T21:38:18.000Z
|
2022-01-14T14:30:23.000Z
| 30.538336 | 210 | 0.523718 |
[
[
[
"# Part I. ETL Pipeline for Pre-Processing the Files",
"_____no_output_____"
],
[
"## PLEASE RUN THE FOLLOWING CODE FOR PRE-PROCESSING THE FILES",
"_____no_output_____"
],
[
"#### Import Python packages ",
"_____no_output_____"
]
],
[
[
"# Import Python packages \nimport pandas as pd\nimport cassandra\nimport re\nimport os\nimport glob\nimport numpy as np\nimport json\nimport csv",
"_____no_output_____"
]
],
[
[
"#### Creating list of filepaths to process original event csv data files",
"_____no_output_____"
]
],
[
[
"# checking your current working directory\nprint(os.getcwd())\n\n# Get current folder and subfolder event data\nfilepath = os.getcwd() + '/event_data'\n\n# Create a list of files and collect each filepath\nfile_path_list = []\nfor root, dirs, files in os.walk(filepath):\n for f in files :\n file_path_list.append(os.path.abspath(f))\n\n # get total number of files found\n num_files = len(file_path_list)\n print('{} files found in {}\\n'.format(num_files, filepath))\n\n # join the file path and roots with the subdirectories using glob\n file_path_list = glob.glob(os.path.join(root,'*'))\n print(file_path_list)",
"_____no_output_____"
]
],
[
[
"#### Processing the files to create the data file csv that will be used for Apache Casssandra tables",
"_____no_output_____"
]
],
[
[
"# initiating an empty list of rows that will be generated from each file\nfull_data_rows_list = [] \n \n# for every filepath in the file path list \nfor f in file_path_list:\n\n# reading csv file \n with open(f, 'r', encoding = 'utf8', newline='') as csvfile: \n # creating a csv reader object \n csvreader = csv.reader(csvfile) \n next(csvreader)\n \n # extracting each data row one by one and append it \n for line in csvreader:\n #print(line)\n full_data_rows_list.append(line) \n \n# uncomment the code below if you would like to get total number of rows \n#print(len(full_data_rows_list))\n\n# uncomment the code below if you would like to check to see what the list of event data rows will look like\n#print(full_data_rows_list)\n\n# creating a smaller event data csv file called event_datafile_full csv that will be used to insert data into the \\\n# Apache Cassandra tables\ncsv.register_dialect('myDialect', quoting=csv.QUOTE_ALL, skipinitialspace=True)\n\nwith open('event_datafile_new.csv', 'w', encoding = 'utf8', newline='') as f:\n writer = csv.writer(f, dialect='myDialect')\n writer.writerow(['artist','firstName','gender','itemInSession','lastName','length',\\\n 'level','location','sessionId','song','userId'])\n for row in full_data_rows_list:\n if (row[0] == ''):\n continue\n writer.writerow((row[0], row[2], row[3], row[4], row[5], row[6], row[7], row[8], row[12], row[13], row[16]))\n",
"_____no_output_____"
],
[
"# check the number of rows in your csv file\nwith open('event_datafile_new.csv', 'r', encoding = 'utf8') as f:\n print(sum(1 for line in f))",
"_____no_output_____"
]
],
[
[
"# Part II. Complete the Apache Cassandra coding portion of your project. \n\n## Now you are ready to work with the CSV file titled <font color=red>event_datafile_new.csv</font>, located within the Workspace directory. The event_datafile_new.csv contains the following columns: \n- artist \n- firstName of user\n- gender of user\n- item number in session\n- last name of user\n- length of the song\n- level (paid or free song)\n- location of the user\n- sessionId\n- song title\n- userId\n\nThe image below is a screenshot of what the denormalized data should appear like in the <font color=red>**event_datafile_new.csv**</font> after the code above is run:<br>\n\n<img src=\"images/image_event_datafile_new.jpg\">",
"_____no_output_____"
],
[
"## Begin writing your Apache Cassandra code in the cells below",
"_____no_output_____"
],
[
"#### Creating a Cluster",
"_____no_output_____"
]
],
[
[
"# This should make a connection to a Cassandra instance your local machine \n# (127.0.0.1)\n\nfrom cassandra.cluster import Cluster\ntry: \n # Connect to local Apache Cassandra instance\n cluster = Cluster(['127.0.0.1']) \n\n # Set session to connect andexecute queries.\n session = cluster.connect()\n\nexcept Exception as e:\n print(e)",
"_____no_output_____"
]
],
[
[
"#### Create Keyspace",
"_____no_output_____"
]
],
[
[
"# Create a Keyspace \ntry:\n session.execute(\"\"\"\n CREATE KEYSPACE IF NOT EXISTS sparkifydb \n WITH REPLICATION = \n { 'class' : 'SimpleStrategy', 'replication_factor' : 1 }\"\"\"\n)\n\nexcept Exception as e:\n print(e)",
"_____no_output_____"
]
],
[
[
"#### Set Keyspace",
"_____no_output_____"
]
],
[
[
"# Set KEYSPACE\ntry:\n session.set_keyspace('sparkifydb')\nexcept Exception as e:\n print(e)",
"_____no_output_____"
]
],
[
[
"### Now we need to create tables to run the following queries. Remember, with Apache Cassandra you model the database tables on the queries you want to run.",
"_____no_output_____"
],
[
"## Create queries to ask the following three questions of the data\n\n### 1. Give me the artist, song title and song's length in the music app history that was heard during sessionId = 338, and itemInSession = 4\n\n\n### 2. Give me only the following: name of artist, song (sorted by itemInSession) and user (first and last name) for userid = 10, sessionid = 182\n \n\n### 3. Give me every user name (first and last) in my music app history who listened to the song 'All Hands Against His Own'\n\n\n",
"_____no_output_____"
],
[
"### Query-1",
"_____no_output_____"
]
],
[
[
"## Query 1: Give me the artist, song title and song's length in the music app history that was heard during \\\n## sessionId = 338, and itemInSession = 4\n\n# CREATE TABLE:\n# This CQL query creates song_in_session table which contains the following columns (with data type):\n# * session_id INT, \n# * item_in_session INT, \n# * artist TEXT, \n# * song TEXT, \n# * length FLOAT\n#\n# To uniquely identify each row and allow efficient distribution in Cassandra cluster, \n# * session_id and item_in_session columns: are used as table's Primary Key (composite Partition Key). \n\nquery = \"CREATE TABLE IF NOT EXISTS song_in_session \"\nquery = query + \"(session_id int, item_in_session int, artist text, song text, length float, \\\n PRIMARY KEY(session_id, item_in_session))\"\ntry:\n session.execute(query)\nexcept Exception as e:\n print(e)",
"_____no_output_____"
],
[
"# INSERT data\n# Set new file name.\nfile = 'event_datafile_new.csv'\n\nwith open(file, encoding = 'utf8') as f:\n csvreader = csv.reader(f)\n next(csvreader) # skip header\n for line in csvreader:\n # Assign the INSERT statements into the `query` variable\n query = \"INSERT INTO song_in_session (session_id, item_in_session, artist, song, length)\"\n query = query + \" VALUES (%s, %s, %s, %s, %s)\"\n ## Assign column elements in the INSERT statement.\n session.execute(query, (int(line[8]), int(line[3]), line[0], line[9], float(line[5])))",
"_____no_output_____"
]
],
[
[
"#### Do a SELECT to verify that the data have been inserted into each table",
"_____no_output_____"
]
],
[
[
"# SELECT statement:\n# To answer Query-1, this CQL query \n# * matches session_id (=338) and item_in_session (=4) to \n# * return artist, song and length from song_in_session table.\n\nquery = \"SELECT artist, song, length \\\n FROM song_in_session \\\n WHERE session_id = 338 AND \\\n item_in_session = 4\"\n\ntry:\n songs = session.execute(query)\nexcept Exception as e:\n print(e)\n\nfor row in songs:\n print (row.artist, row.song, row.length)\n",
"_____no_output_____"
]
],
[
[
"### COPY AND REPEAT THE ABOVE THREE CELLS FOR EACH OF THE THREE QUESTIONS",
"_____no_output_____"
],
[
"### Query-2",
"_____no_output_____"
]
],
[
[
"## Query 2: Give me only the following: name of artist, song (sorted by itemInSession) and \n# user (first and last name) for userid = 10, sessionid = 182\n\n# CREATE TABLE\n# This CQL query creates artist_in_session table which contains the following columns (with data type):\n# * user_id INT,\n# * session_id INT, \n# * artist TEXT, \n# * song TEXT, \n# * item_in_session INT, \n# * first_name TEXT,\n# * last_name TEXT,\n#\n# To uniquely identify each row and allow efficient distribution in Cassandra cluster, \n# * user_id and session_id columns: are used as Composite Partition Key in table's Primary Key.\n# * item_in_session column: is used as Clustering Key in table's Primary Key and allows sorting order of the data.\n\nquery = \"CREATE TABLE IF NOT EXISTS artist_in_session \"\nquery = query + \"( user_id int, \\\n session_id int, \\\n artist text, \\\n song text, \\\n item_in_session int, \\\n first_name text, \\\n last_name text, \\\n PRIMARY KEY((user_id, session_id), item_in_session))\"\ntry:\n session.execute(query)\nexcept Exception as e:\n print(e) ",
"_____no_output_____"
],
[
"# INSERT data\nfile = 'event_datafile_new.csv'\n\nwith open(file, encoding = 'utf8') as f:\n csvreader = csv.reader(f)\n next(csvreader)\n for line in csvreader:\n query = \"INSERT INTO artist_in_session (user_id, \\\n session_id, \\\n artist, \\\n song, \\\n item_in_session, \\\n first_name, \\\n last_name)\"\n query = query + \" VALUES (%s, %s, %s, %s, %s, %s, %s)\"\n session.execute(query, (int(line[10]), int(line[8]), line[0], line[9], int(line[3]), line[1], line[4]))",
"_____no_output_____"
],
[
"# SELECT statement:\n# To answer Query-2, this CQL query \n# * matches user_id (=10) and session_id (=182) to \n# * return artist, song, first_name, and last_name (of user) from artist_in_session table.\n\nquery = \"SELECT artist, song, first_name, last_name \\\n FROM artist_in_session \\\n WHERE user_id = 10 AND \\\n session_id = 182\"\ntry:\n artists = session.execute(query)\nexcept Exception as e:\n print(e)\n\nfor row in artists:\n print (row.artist, row.song, row.first_name, row.last_name)",
"_____no_output_____"
]
],
[
[
"### Query-3",
"_____no_output_____"
]
],
[
[
"## Query 3: Give me every user name (first and last) in my music app history who listened \n# to the song 'All Hands Against His Own'\n\n# CREATE TABLE\n# CREATE TABLE\n# This CQL query creates artist_in_session table which contains the following columns (with data type):\n# * song TEXT,\n# * user_id INT,\n# * first_name TEXT,\n# * last_name TEXT,\n#\n# To uniquely identify each row and allow efficient distribution in Cassandra cluster, \n# * song, user_id columns: are used as Composite Partition Key in table's Primary Key.\n\nquery = \"CREATE TABLE IF NOT EXISTS user_and_song \"\nquery = query + \"( song text, \\\n user_id int, \\\n first_name text, \\\n last_name text, \\\n PRIMARY KEY(song, user_id))\"\ntry:\n session.execute(query)\nexcept Exception as e:\n print(e)",
"_____no_output_____"
],
[
"# INSERT data\nfile = 'event_datafile_new.csv'\n\nwith open(file, encoding = 'utf8') as f:\n csvreader = csv.reader(f)\n next(csvreader)\n for line in csvreader:\n query = \"INSERT INTO user_and_song (song, \\\n user_id, \\\n first_name, \\\n last_name)\"\n query = query + \" VALUES (%s, %s, %s, %s)\"\n session.execute(query, (line[9], int(line[10]), line[1], line[4]))",
"_____no_output_____"
],
[
"# SELECT statement:\n# To answer Query-3, this CQL query \n# * matches song (=All Hands Against His Own) to \n# * return first_name and last_name (of users) from user_and_song table.\n\nquery = \"SELECT first_name, last_name \\\n FROM user_and_song \\\n WHERE song = 'All Hands Against His Own'\"\ntry:\n users = session.execute(query)\nexcept Exception as e:\n print(e)\n\nfor row in users:\n print (row.first_name, row.last_name)",
"_____no_output_____"
]
],
[
[
"### Drop the tables before closing out the sessions",
"_____no_output_____"
]
],
[
[
"## Drop the table before closing out the sessions\nquery = \"DROP TABLE song_in_session\"\ntry:\n rows = session.execute(query)\nexcept Exception as e:\n print(e)\n\nquery2 = \"DROP TABLE artist_in_session\"\ntry:\n rows = session.execute(query2)\nexcept Exception as e:\n print(e)\n \nquery3 = \"DROP TABLE user_and_song\"\ntry:\n rows = session.execute(query3)\nexcept Exception as e:\n print(e)",
"_____no_output_____"
]
],
[
[
"### Close the session and cluster connection¶",
"_____no_output_____"
]
],
[
[
"session.shutdown()\ncluster.shutdown()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8c338f4a3653fd98d40f0a61e0d8dfd85b476d
| 11,279 |
ipynb
|
Jupyter Notebook
|
docs/source/nb/.ipynb_checkpoints/MNIST_demo-checkpoint.ipynb
|
StatML-dAI/dnn-inference
|
f17b6945dfff80118e25f1b39b56c3e4027b3b92
|
[
"MIT"
] | null | null | null |
docs/source/nb/.ipynb_checkpoints/MNIST_demo-checkpoint.ipynb
|
StatML-dAI/dnn-inference
|
f17b6945dfff80118e25f1b39b56c3e4027b3b92
|
[
"MIT"
] | null | null | null |
docs/source/nb/.ipynb_checkpoints/MNIST_demo-checkpoint.ipynb
|
StatML-dAI/dnn-inference
|
f17b6945dfff80118e25f1b39b56c3e4027b3b92
|
[
"MIT"
] | 2 |
2022-02-17T15:57:42.000Z
|
2022-02-20T11:50:45.000Z
| 60.639785 | 3,288 | 0.660697 |
[
[
[
"## ``dnn-inference`` in MNIST dataset",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom tensorflow import keras\nfrom tensorflow.keras.datasets import mnist\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPooling2D\nfrom tensorflow.python.keras import backend as K\nimport time\nfrom sklearn.model_selection import train_test_split\nfrom tensorflow.keras.optimizers import Adam, SGD",
"_____no_output_____"
],
[
"np.random.seed(0)\nnum_classes = 2\n# input image dimensions\nimg_rows, img_cols = 28, 28\n\n# the data, split between train and test sets\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\nX = np.vstack((x_train, x_test))\ny = np.hstack((y_train, y_test))\nind = (y == 9) + (y == 7)\nX, y = X[ind], y[ind]\nX = X.astype('float32')\nX += .01*abs(np.random.randn(14251, 28, 28))\ny[y==7], y[y==9] = 0, 1\n\nif K.image_data_format() == 'channels_first':\n\tX = X.reshape(x.shape[0], 1, img_rows, img_cols)\n\tinput_shape = (1, img_rows, img_cols)\nelse:\n\tX = X.reshape(X.shape[0], img_rows, img_cols, 1)\n\tinput_shape = (img_rows, img_cols, 1)\n\nX /= 255.\n\n# convert class vectors to binary class matrices\ny = keras.utils.to_categorical(y, num_classes)",
"_____no_output_____"
],
[
"## define the learning models\ndef cnn():\n\tmodel = Sequential()\n\tmodel.add(Conv2D(32, kernel_size=(3, 3), activation='relu', input_shape=input_shape))\n\tmodel.add(Conv2D(64, (3, 3), activation='relu'))\n\tmodel.add(MaxPooling2D(pool_size=(2, 2)))\n\tmodel.add(Dropout(0.25))\n\tmodel.add(Flatten())\n\tmodel.add(Dense(128, activation='relu'))\n\tmodel.add(Dropout(0.5))\n\tmodel.add(Dense(num_classes, activation='softmax'))\n\tmodel.compile(loss=keras.losses.binary_crossentropy, optimizer=keras.optimizers.Adam(0.0005), metrics=['accuracy'])\n\treturn model\n\nmodel, model_mask = cnn(), cnn()",
"2021-12-15 14:51:53.708972: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:53.714047: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:53.714322: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:53.714801: I tensorflow/core/platform/cpu_feature_guard.cc:151] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 AVX512F FMA\nTo enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.\n2021-12-15 14:51:53.715620: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:53.715889: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:53.716137: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:54.025005: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:54.025285: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:54.025520: I tensorflow/stream_executor/cuda/cuda_gpu_executor.cc:939] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero\n2021-12-15 14:51:54.025786: I tensorflow/core/common_runtime/gpu/gpu_device.cc:1525] Created device /job:localhost/replica:0/task:0/device:GPU:0 with 3403 MB memory: -> device: 0, name: NVIDIA GeForce RTX 2060, pci bus id: 0000:01:00.0, compute capability: 7.5\n"
],
[
"## fitting param\nfrom tensorflow.keras.callbacks import EarlyStopping\nes = EarlyStopping(monitor='val_accuracy', mode='max', verbose=0, patience=15, restore_best_weights=True)\n\nfit_params = {'callbacks': [es],\n\t\t\t 'epochs': 5,\n\t\t\t 'batch_size': 32,\n\t\t\t 'validation_split': .2,\n\t\t\t 'verbose': 0}\n\nsplit_params = {'split': 'one-split',\n\t\t\t\t'perturb': None,\n\t\t\t\t'num_perm': 100,\n\t\t\t\t'ratio_grid': [.2, .4, .6, .8],\n\t\t\t\t'perturb_grid': [.001, .005, .01, .05, .1],\n\t\t\t\t'min_inf': 100,\n\t\t\t\t'min_est': 1000,\n\t\t\t\t'ratio_method': 'fuse',\n\t\t\t\t'cv_num': 1,\n\t\t\t\t'cp': 'min',\n\t\t\t\t'verbose': 1}",
"_____no_output_____"
],
[
"## Inference based on dnn_inference\nfrom dnn_inference.BBoxTest import split_test\n## testing based on learning models\ninf_feats = [[np.arange(19,28), np.arange(13,20)], [np.arange(21,28), np.arange(4, 13)],[np.arange(7,16), np.arange(9,16)]]\nshiing = split_test(inf_feats=inf_feats, model=model, model_mask=model_mask, change='mask', eva_metric='zero-one')\np_value_tmp = shiing.testing(X, y, cv_num=3, cp='hommel', fit_params=fit_params, split_params=split_params)\n## visualize testing results\nshiing.visual(X,y)\nprint('P-values: %s' %p_value_tmp)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a8c3c1706ba65fccfce22f053085b54c587723f
| 5,413 |
ipynb
|
Jupyter Notebook
|
README.ipynb
|
flaport/mplppt
|
4838568d99b4ac36ffbe4a46f18d341e2acd7c9a
|
[
"MIT"
] | 4 |
2019-08-17T10:56:58.000Z
|
2020-11-04T11:49:39.000Z
|
README.ipynb
|
flaport/mplppt
|
4838568d99b4ac36ffbe4a46f18d341e2acd7c9a
|
[
"MIT"
] | 1 |
2021-11-24T03:39:00.000Z
|
2021-11-25T08:28:58.000Z
|
README.ipynb
|
flaport/mplppt
|
4838568d99b4ac36ffbe4a46f18d341e2acd7c9a
|
[
"MIT"
] | null | null | null | 23.4329 | 264 | 0.552004 |
[
[
[
"# MPLPPT\n`mplppt` is a simple library made from some hacky scripts I used to use to convert matplotlib figures to powerpoint figures. Which makes this a hacky library, I guess 😀.",
"_____no_output_____"
],
[
"## Goal",
"_____no_output_____"
],
[
"`mplppt` seeks to implement an alternative `savefig` function for `matplotlib` figures. This `savefig` function saves a `matplotlib` figure with a single axis to a powerpoint presentation with a single slide containing the figure. ",
"_____no_output_____"
],
[
"## Installation\n```bash\npip install mplppt\n```",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import mplppt\n%matplotlib inline\nimport numpy as np\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Supported Conversions",
"_____no_output_____"
],
[
"`mplppt` supports [partly] conversion of the following matplotlib objects:\n* Lines [`matplotlib.lines.Line2D`]\n* Rectangles [`matplotlib.patches.Rectangle`]\n* Polygons [`matplotlib.patches.Polygon`]\n* pcolormesh [`matplotlib.collections.QuadMesh`]\n* text [`matplotlib.text.Text`]\n\nso far `mplppt` does not (yet) support (out of many other things):\n* markers (including tick marks)\n* linestyle\n",
"_____no_output_____"
],
[
"## Simple Example\nAn example of all different conversions available for mplppt. Below we give an example of how all these objects can be combined into a single plot, which can then be exported to powerpoint:",
"_____no_output_____"
]
],
[
[
"# plot [Line2D]\nx = np.linspace(-1,5)\ny = np.sin(x)\nplt.plot(x,y,color='C1')\n\n# rectangle\nplt.gca().add_patch(mpl.patches.Rectangle((0, 0), 3, 0.5))\n\n# polygon\nplt.gca().add_patch(mpl.patches.Polygon(np.array([[5.0,1.0],[4.0,-0.2],[2.0,0.6]]), color=\"red\"))\n\n# pcolormesh\nx = np.linspace(0,1, 100)\ny = np.linspace(0,1, 100)\nX, Y = np.meshgrid(x,y)\nZ = X**2 + Y**2\nplt.pcolormesh(X,Y,Z)\n\n# text\ntext = plt.text(0,0,'hello')\n\n# set limits\nplt.ylim(-0.5,1)\n\n# Save figure to pptx\nmplppt.savefig('first_example.pptx')\n\n# show figure\nplt.show()",
"_____no_output_____"
]
],
[
[
"Which results in a powerpoint slide which looks as follows:",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## Cool! What else can I do with this?",
"_____no_output_____"
],
[
"You are not bound to using matplotlib! The `mplppt` repository contains some standard powerpoint shapes that you can use. Try something like:",
"_____no_output_____"
]
],
[
[
"ppt = mplppt.Group() # Create a new group of objects\nppt += mplppt.Rectangle(name='rect', x=0, y=0, cx=100, cy=100, slidesize=(10,5)) # add an object to the group\nppt.save('second_example.pptx') # export the group as a ppt slide",
"_____no_output_____"
]
],
[
[
"## Is any of this documented?",
"_____no_output_____"
],
[
"No.",
"_____no_output_____"
],
[
"## How does this work?",
"_____no_output_____"
],
[
"The repository contains a template folder, which is nothing more than an empty powerpoint presentation which is unzipped. After making a copy of the template folder and adding some `xml` code for the shapes, the modified folder is zipped into a `.pptx` file.",
"_____no_output_____"
],
[
"## Copyright",
"_____no_output_____"
],
[
"© Floris Laporte - MIT License",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a8c6ae12795bbb6cc388ac5ea04e4055bdd13c6
| 44,382 |
ipynb
|
Jupyter Notebook
|
Crime Rate/CrimeData-2018.ipynb
|
RUB-Project-Team/NJ-Housing-2020
|
931630fcf263be2a9cb105ae8419f430c5ed61d1
|
[
"FSFAP"
] | null | null | null |
Crime Rate/CrimeData-2018.ipynb
|
RUB-Project-Team/NJ-Housing-2020
|
931630fcf263be2a9cb105ae8419f430c5ed61d1
|
[
"FSFAP"
] | null | null | null |
Crime Rate/CrimeData-2018.ipynb
|
RUB-Project-Team/NJ-Housing-2020
|
931630fcf263be2a9cb105ae8419f430c5ed61d1
|
[
"FSFAP"
] | null | null | null | 32.924332 | 279 | 0.352958 |
[
[
[
"# Crime Data File- Year 2018",
"_____no_output_____"
]
],
[
[
"#Import libraries\nimport pandas as pd\nfrom pandas import ExcelWriter\nfrom pandas import ExcelFile\nimport os\n\n#Raw data file path\nfilepath = os.path.join(\"\",\"raw_crime_data\",\"20190815_crimetrend_2018.xlsx\")",
"_____no_output_____"
]
],
[
[
"### Get Raw Data for processing",
"_____no_output_____"
]
],
[
[
"#Get raw data into data frames - bring all excel tab data in to single dataframe\nraw_data = pd.concat(pd.read_excel(filepath,sheet_name=None),ignore_index=True)\n\n#Check column heading\nprint(\"Column headings:\",raw_data.columns)",
"Column headings: Index(['CURRENT DATE: 08/13/2019',\n 'INDEX CRIMES BY COUNTY FOR JAN - 2018 TO DEC - 2018', 'PAGE: 1',\n 'PAGE: 10', 'PAGE: 11', 'PAGE: 12', 'PAGE: 13', 'PAGE: 14', 'PAGE: 15',\n 'PAGE: 16', 'PAGE: 17', 'PAGE: 18', 'PAGE: 19', 'PAGE: 2', 'PAGE: 20',\n 'PAGE: 21', 'PAGE: 3', 'PAGE: 4', 'PAGE: 5', 'PAGE: 6', 'PAGE: 7',\n 'PAGE: 8', 'PAGE: 9', 'Unnamed: 11', 'Unnamed: 2', 'Unnamed: 3',\n 'Unnamed: 4', 'Unnamed: 5', 'Unnamed: 6', 'Unnamed: 7', 'Unnamed: 8',\n 'Unnamed: 9'],\n dtype='object')\n"
],
[
"#Raw Data preview\nraw_data.head(5)",
"_____no_output_____"
]
],
[
[
"### Raw Data Cleanup",
"_____no_output_____"
]
],
[
[
"#Drop all columns between column index 2 to 23 (these are page numbers that corresponds to each excel sheet) \nraw_data = raw_data.drop(raw_data.iloc[:, 2:23], axis = 1) ",
"_____no_output_____"
],
[
"#Check raw data sample\nraw_data.head(5)",
"_____no_output_____"
],
[
"#Drop all rows that has NAN records - these are blank lines from excel got converted as df row \nraw_data=raw_data.dropna(how='all')",
"_____no_output_____"
],
[
"#Check sample data\nraw_data.head(5)",
"_____no_output_____"
],
[
"#Drop first row \nraw_data=raw_data.drop(raw_data.index[0])\nraw_data.head(2)",
"_____no_output_____"
],
[
"#Rename the data frame columns\ncrime_data= raw_data.rename(columns={\"CURRENT DATE: 08/13/2019\": \"ORINumber\", \"INDEX CRIMES BY COUNTY FOR JAN - 2018 TO DEC - 2018\": \"Agency\",\n \"Unnamed: 11\":\"Months\",\"Unnamed: 2\":\"Population\",\"Unnamed: 3\":\"Murder\",\"Unnamed: 4\":\"Rape\",\n \"Unnamed: 5\":\"Robbery\",\"Unnamed: 6\":\"Assault\",\"Unnamed: 7\":\"Burglary\",\"Unnamed: 8\":\"Larceny\",\n \"Unnamed: 9\":\"Auto Theft\"})",
"_____no_output_____"
],
[
"#Display fist hand crime data set sample\ncrime_data.head(2)",
"_____no_output_____"
],
[
"#Drop first row which is now column names and view sample data\ncrime_data=crime_data.drop(crime_data.index[0])\ncrime_data.head(5)",
"_____no_output_____"
]
],
[
[
"### Process crime data and capture required data points ",
"_____no_output_____"
]
],
[
[
"#There are only two rows - City and Rate Per 100,00 - Lets ignore County and other and only consider PD rows\norinumber=[]\ncity=[]\npopulation=[]\nmurder=[]\nrape=[]\nrobbery=[]\nassault=[]\nburglary=[]\nlarceny=[]\nautotheft=[]\nicount=1\n\n# iterate over rows with iterrows()\nfor index, row in crime_data.iterrows():\n #First row is always city/County\n if row['Agency'] !=\"Rate Per 100,000\" and icount==1:\n icity=row['Agency']\n \n #Check if its City and if yes process with data capture\n if \"PD\" in str(icity):\n \n #Capture OriNumber, City and polulation\n orinumber.append(row['ORINumber'])\n city.append(icity)\n population.append(row['Population'])\n \n #Increment the counter\n icount+=1\n else:\n continue\n \n #Access data using column names\n elif row['Agency']==\"Rate Per 100,000\" and icount==2:\n \n #reset counter\n icount=1\n \n #Capture crime data\n try:\n murder.append(row['Murder'])\n except:\n murder.append(0)\n try: \n rape.append(row['Rape'])\n except:\n rape.append(0)\n try:\n robbery.append(row['Robbery'])\n except:\n robbery.append(0)\n try:\n assault.append(row['Assault'])\n except:\n assault.append(0)\n try:\n burglary.append(row['Burglary'])\n except:\n burglary.append(0)\n try: \n larceny.append(row['Larceny'])\n except:\n larceny.append(0)\n try:\n autotheft.append(row['Auto Theft'])\n except:\n autotheft.append(0)\n\n#Just check count of all captured data elements to make sure we catpure info correctly \nif (len(orinumber)==len(murder)==len(rape)==len(robbery)==len(assault)==len(burglary)==len(larceny)==len(autotheft)==len(city)==len(population)==len(murder)==len(rape)==len(robbery)==len(assault)==len(burglary)==len(larceny)==len(autotheft)==len(city)==len(population)):\n print(f\"Successfully processed file. All data point count matched. Number of towns data is process is: {len(city)}\")\nelse:\n print(\"Plese check the raw file. Data point count does not match...\")",
"Successfully processed file. All data point count matched. Number of towns data is process is: 490\n"
],
[
"#Create a crime data frame from series of data points\ntown_crime= {\"ORINumber\":orinumber,\"City\":city,\"Population\":population,\"Murder\":murder,\"Rape\":rape,\n \"Robbery\":robbery,\"Assault\":assault,\"Burglary\":burglary,\"Larceny\":larceny,\"Auto Theft\":autotheft}\ncrime_data=pd.DataFrame(town_crime)",
"_____no_output_____"
],
[
"#Verify crime data points\ncrime_data.head(10)",
"_____no_output_____"
],
[
"#Export data to csv file - not required for the project\n#finished crime data file path\nfilepath = os.path.join(\"..\",\"Final Output Data\",\"Crime_Data_2018.csv\")\ncrime_data.to_csv(filepath, index = False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4a8c7ad6f6e8a199be0b60a133f9e607d850afa5
| 509,523 |
ipynb
|
Jupyter Notebook
|
Tweets_Reach&Ego.ipynb
|
bhavyamendiratta/Twitter-Hashtag-Analytics
|
8442cccd36f90243799503d40530534c75159c14
|
[
"MIT"
] | null | null | null |
Tweets_Reach&Ego.ipynb
|
bhavyamendiratta/Twitter-Hashtag-Analytics
|
8442cccd36f90243799503d40530534c75159c14
|
[
"MIT"
] | null | null | null |
Tweets_Reach&Ego.ipynb
|
bhavyamendiratta/Twitter-Hashtag-Analytics
|
8442cccd36f90243799503d40530534c75159c14
|
[
"MIT"
] | 1 |
2020-10-06T07:34:04.000Z
|
2020-10-06T07:34:04.000Z
| 53.414719 | 46,632 | 0.57379 |
[
[
[
"import tweepy\nimport pandas as pd\nimport sys\nimport json\nconsumer_key = 'Q5kScvH4J2CE6d3w8xesxT1bm'\nconsumer_secret = 'mlGrcssaVjN9hQMi6wI6RqWKt2LcHAEyYCGh6WF8yq20qcTb8T'\naccess_token = '944440837739487232-KTdrvr4vARk7RTKvJkRPUF8I4VOvGIr'\naccess_token_secret = 'bfHE0jC5h3B7W3H18TxV7XsofG1xuB6zeINo2DxmZ8K1W'\n\nauth = tweepy.OAuthHandler(consumer_key, consumer_secret)\nauth.set_access_token(access_token, access_token_secret)\napi = tweepy.API(auth,wait_on_rate_limit=True,wait_on_rate_limit_notify=True,compression=True)\nsearch_words = \"#Kashmir\"\ndate_since = \"2015-01-01\"\n\n#..............................................................\n\ntweets = tweepy.Cursor(api.search,\n q=search_words,\n lang=\"en\",\n since=date_since).items(10)\ntweets\n\n#...............................................................\n\n",
"_____no_output_____"
],
[
"date=[]\nus=[]\no=0;\ntext=[]\nimport csv\nfor tweet in tweets:\n us.append(tweet)\n text.append(tweet.text)\n print(o)\n o+=1\nprint(o)\n\n#..................................................................\n\n\n#..................................................................\n\n\n\n\n#....................................................................\n\n\nimport pandas as pd\ndf=pd.DataFrame()\n\nid=[]\nfor i in range(o):\n id.append(us[i]._json['id'])\ndate=[]\nfor i in range(o):\n date.append(us[i]._json['created_at'])\nuser=[]\nfor i in range(o):\n user.append(us[i]._json['user']['screen_name'])\ntext=[]\nfor i in range(o):\n text.append(us[i]._json['text'])\n\ndf['id']=id\ndf['date']=date\ndf['user']=user\ndf['text']=text\n\n\nretweet=[]\nfor i in range(o):\n a=us[i]._json['text']\n if(a[0]==\"R\" and a[1]==\"T\"):\n retweet.append(\"True\")\n else:\n retweet.append(\"False\")\ndf['retweet']=retweet\n\nretweet_count=[]\nfor i in range(o):\n retweet_count.append(us[i]._json['retweet_count'])\ndf[\"retweet_count\"]=retweet_count\n\nfriends_count=[]\nfollowers_count=[]\nfavourites_count=[]\nlocation=[]\nsource=[]\nfor i in range(o):\n friends_count.append(us[i]._json['user']['friends_count'])\n followers_count.append(us[i]._json['user']['followers_count'])\n favourites_count.append(us[i]._json['user']['favourites_count'])\n location.append(us[i]._json['user']['location'])\n source.append(us[i]._json['source'][-11:-4])\n\ndf['location']=location\ndf['source']=source\ndf['followers_count']=followers_count\ndf['friends_count']=friends_count\ndf['favourite_count']=favourites_count\n\npost_by=[]\nfor i in range(o):\n a=us[i]._json['entities']['user_mentions']\n #print(a)\n if(a):\n post_by.append(us[i]._json['entities']['user_mentions'][0]['screen_name'])\n else:\n post_by.append(\"null\")\ndf['post_by']=post_by\n\n#............................................................................\n\n\n\n\n\n\n#............................................................\n\n\n\n#.........................................................\n\ndf\n\n#......................\n",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n"
],
[
"df.head(n=2)",
"_____no_output_____"
],
[
"df['date']",
"_____no_output_____"
],
[
"datetime?",
"_____no_output_____"
],
[
"df['date']",
"_____no_output_____"
],
[
"a=df['date'].values.tolist()",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"type(a[0])",
"_____no_output_____"
],
[
"b=df['followers_count'].values.tolist()",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
],
[
"type(b[0])",
"_____no_output_____"
],
[
"b[0]+b[1]",
"_____no_output_____"
],
[
"c=[]\nc.append(b[0])\nfor i in range(1,len(b)):\n c.append(c[i-1]+b[i])",
"_____no_output_____"
],
[
"c",
"_____no_output_____"
],
[
"a=[]\nfor i in range(len(b)):\n a.append(i)",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"import pandas as pd\ndf=pd.read_csv('Datawarehouse.csv')\n",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"b=df['followers_count'].values.tolist()",
"_____no_output_____"
],
[
"c=[]\nc.append(b[0])\nfor i in range(1,len(b)):\n c.append(c[i-1]+b[i])",
"_____no_output_____"
],
[
"c[len(b)-1]",
"_____no_output_____"
],
[
"a=df['date'].values.tolist()",
"_____no_output_____"
],
[
"df['date']",
"_____no_output_____"
],
[
"date=df['date'].values.tolist()",
"_____no_output_____"
],
[
"date",
"_____no_output_____"
],
[
"date1=date[:1000]\nc1=c[:1000]",
"_____no_output_____"
],
[
"d=[]",
"_____no_output_____"
],
[
"d.append(c[4925])\nd.append(c[5511])\nd.append(c[9279])\nd.append(c[12384])\nd.append(c[15629])\nd.append(c[18381])\nd.append(c[22174])\nd.append(c[23401])\n\n",
"_____no_output_____"
],
[
"d",
"_____no_output_____"
],
[
"date=[\"Thu Feb 20 00:00\",\" Thu 20 12:00\",\"Fri Feb 21 00:00\",\"Fri Feb 21 12:00\",\"Sat Feb 22 00:00\",\"Sat Feb 22 12:00\",\"Sun Feb 23 00:00\",\"Sun Feb 23 12:00\"]",
"_____no_output_____"
],
[
"plt.plot(date,d,linewidth=3)\nplt.plot(date,d,'bo')\nplt.xlabel('Date',size=15)\nplt.ylabel(\"Potential Impact\",size=15)\nplt.tight_layout(pad=0,rect=(1,2,3,3.5))\nplt.show()",
"_____no_output_____"
],
[
"plt.xlabel?",
"_____no_output_____"
],
[
"from collections import Counter",
"_____no_output_____"
],
[
"aa=df['retweet'].values.tolist()",
"_____no_output_____"
],
[
"len(aa)",
"_____no_output_____"
],
[
"aa1=aa[0:4925]",
"_____no_output_____"
],
[
"aa1=Counter(aa1)",
"_____no_output_____"
],
[
"aa1",
"_____no_output_____"
],
[
"tweets1=aa1[0]",
"_____no_output_____"
],
[
"retweets1=aa1[1]",
"_____no_output_____"
],
[
"print(tweets1,retweets1)",
"1167 3758\n"
],
[
"aa2=aa[4926:5511]\naa2=Counter(aa2)\nprint(aa2)\n\n\n",
"Counter({True: 474, False: 111})\n"
],
[
"tweets2=aa2[0]\nretweets2=aa2[1]\nprint(tweets2,retweets2)",
"111 474\n"
],
[
"aa3=aa[5512:9279]\naa3=Counter(aa3)\nprint(aa3)\n\n\n",
"Counter({True: 3070, False: 697})\n"
],
[
"tweets3=aa3[0]\nretweets3=aa3[1]\nprint(tweets3,retweets3)",
"697 3070\n"
],
[
"aa4=aa[9280:12384]\naa4=Counter(aa4)\nprint(aa4)\n\n\n",
"Counter({True: 2597, False: 507})\n"
],
[
"tweets4=aa4[0]\nretweets4=aa4[1]\nprint(tweets4,retweets4)",
"507 2597\n"
],
[
"aa5=aa[12385:15629]\naa5=Counter(aa5)\nprint(aa5)\n\n\n",
"Counter({True: 2720, False: 524})\n"
],
[
"tweets5=aa5[0]\nretweets5=aa5[1]\nprint(tweets5,retweets5)",
"524 2720\n"
],
[
"aa6=aa[15630:18381]\naa6=Counter(aa6)\nprint(aa6)\n\n\n",
"Counter({True: 2028, False: 723})\n"
],
[
"tweets6=aa6[0]\nretweets6=aa6[1]\nprint(tweets6,retweets6)",
"723 2028\n"
],
[
"aa7=aa[18382:22174]\naa7=Counter(aa7)\nprint(aa7)\n\n\n",
"Counter({True: 2974, False: 818})\n"
],
[
"tweets7=aa7[0]\nretweets7=aa7[1]\nprint(tweets7,retweets7)",
"818 2974\n"
],
[
"aa8=aa[22175:23401]\naa8=Counter(aa8)\nprint(aa8)\n\n\n",
"Counter({True: 1013, False: 213})\n"
],
[
"tweets8=aa8[0]\nretweets8=aa8[1]\nprint(tweets8,retweets8)",
"213 1013\n"
],
[
"tweet_list=[tweets1,tweets2,tweets3,tweets4,tweets5,tweets6,tweets7,tweets8]\nretweet_list=[retweets1,retweets2,retweets3,retweets4,retweets5,retweets6,retweets7,retweets8]",
"_____no_output_____"
],
[
"plt.plot(date,tweet_list,label='number of tweets')\nplt.plot(date,tweet_list,'bo')\nplt.plot(date,retweet_list,label='number of retweets')\nplt.plot(date,retweet_list,'ro')\nplt.xlabel('Date',size=15)\nplt.tight_layout(pad=0,rect=(1,2,3,3.5))\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"print('Nice Work')",
"Nice Work\n"
],
[
"c=df['followers_count'].values.tolist()",
"_____no_output_____"
],
[
"cc1=c[0:4925]\ncc2=c[4926:5511]\ncc3=c[5512:9279]\ncc4=c[9280:12384]\ncc5=c[12385:15629]\ncc6=c[15630:18381]\ncc7=c[18382:22174]\ncc8=c[22175:23401]",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"cc11=np.sum(cc1)",
"_____no_output_____"
],
[
"cc11=np.sum(cc1)\ncc22=np.sum(cc2)\ncc33=np.sum(cc3)\ncc44=np.sum(cc4)\ncc55=np.sum(cc5)\ncc66=np.sum(cc6)\ncc77=np.sum(cc7)\ncc88=np.sum(cc8)",
"_____no_output_____"
],
[
"cc11+cc22+cc33+cc44+cc55+cc66+cc77+cc88",
"_____no_output_____"
]
],
[
[
"# New Section",
"_____no_output_____"
]
],
[
[
"impact=[cc11,cc22,cc33,cc44,cc55,cc66,cc77,cc88]",
"_____no_output_____"
],
[
"plt.plot(date,impact,label='potential impact')\nplt.plot(date,impact,'bo')\nplt.xlabel('Date',size=15)\nplt.tight_layout(pad=0,rect=(1,2,3,3.5))\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n",
"_____no_output_____"
],
[
"df=pd.read_csv('Datawarehouse.csv')",
"_____no_output_____"
],
[
"plt.fill_between(date,0,d,label='cummualative impact')\n#plt.plot(date,d,'bo')\nplt.xlabel('Date',size=15)\nplt.ylabel(\"Potential Impact\",size=15)\nplt.tight_layout(pad=0,rect=(1,2,3,4))\nplt.fill_between(date,impact,label='potential impact')\n#plt.plot(date,impact,'ro')\nplt.xlabel('Date',size=15)\nplt.tight_layout(pad=0,rect=(1,2,3,4))\n\nplt.legend()\nplt.show()",
"_____no_output_____"
],
[
"df.head(n=2)",
"_____no_output_____"
],
[
"dff=df.to_dict()",
"_____no_output_____"
],
[
"uni=[]\nuniqq=[]\nfor i in range(23402):\n if dff['user'][i] not in uni :\n uni.append(dff['user'][i])\n uniqq.append(dff['followers_count'][i])",
"_____no_output_____"
],
[
"len(uni)",
"_____no_output_____"
],
[
"newdf=pd.DataFrame()",
"_____no_output_____"
],
[
"newdf['user']=uni",
"_____no_output_____"
],
[
"newdf['follower_count']=uniqq",
"_____no_output_____"
],
[
"newdf.head(n=5)",
"_____no_output_____"
],
[
"newnewdf=newdf.sort_values('follower_count',ascending=False)",
"_____no_output_____"
],
[
"newnewdf",
"_____no_output_____"
],
[
"a=newnewdf['follower_count'].values",
"_____no_output_____"
],
[
"type(a)",
"_____no_output_____"
],
[
"follower_sum=np.sum(a)",
"_____no_output_____"
],
[
"follower_sum",
"_____no_output_____"
],
[
"follower_avg=follower_sum/len(total_followers)",
"_____no_output_____"
],
[
"print(follower_avg)",
"_____no_output_____"
],
[
"df.head(n=2)",
"_____no_output_____"
],
[
"c=df['retweet'].values",
"_____no_output_____"
],
[
"from collections import Counter",
"_____no_output_____"
],
[
"c=Counter(c)",
"_____no_output_____"
],
[
"c",
"_____no_output_____"
],
[
"## kitne tweet and kitne retweet",
"_____no_output_____"
],
[
"fdf=df.sort_values('retweet',ascending=True)",
"_____no_output_____"
],
[
"fdf",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"import pandas as pd\n\n\n",
"_____no_output_____"
],
[
"df=pd.read_csv(\"Datawarehouse (1).csv\")",
"_____no_output_____"
],
[
"df.head(5)",
"_____no_output_____"
],
[
"dff=df.to_dict()",
"_____no_output_____"
],
[
"#unique user\n#reach\nuni=[]\ndate=[]\nsum=0\nfor i in range(23402):\n if dff['user'][i] not in uni:\n uni.append(dff['user'][i])\n sum+=dff['followers_count'][i]\n date.append(dff['date'][i])",
"_____no_output_____"
],
[
"sum",
"_____no_output_____"
],
[
"date",
"_____no_output_____"
],
[
"import tweepy\nimport pandas as pd\naccess_token = \"944440837739487232-KTdrvr4vARk7RTKvJkRPUF8I4VOvGIr\"\naccess_token_secret = \"bfHE0jC5h3B7W3H18TxV7XsofG1xuB6zeINo2DxmZ8K1W\"\nconsumer_key = \"Q5kScvH4J2CE6d3w8xesxT1bm\"\nconsumer_secret = \"mlGrcssaVjN9hQMi6wI6RqWKt2LcHAEyYCGh6WF8yq20qcTb8T\"\ncol1=[]\ncol2=[]\n\nauth = tweepy.OAuthHandler(consumer_key, consumer_secret)\nauth.set_access_token(access_token, access_token_secret)\napi = tweepy.API(auth,wait_on_rate_limit=True,wait_on_rate_limit_notify=True,compression=True)\n\nfor user in tweepy.Cursor(api.friends, screen_name=\"hasanminhaj\").items(400):\n print(user.screen_name)\n col1.append(\"hasanminhaj\")\n col2.append(user.screen_name) ",
"chrissyteigen\nmitchrichmond23\nninaemlemdi\nJon_Favreau\ndrsanjaygupta\nMattGertz\nJasonSCampbell\njkbjournalist\ntomhanks\nHillaryClinton\nanitakumar01\nBoysClubNY\ndmaq1\nChrisEvans\nSarahxAnwer\nCasey\nmartinandanar\nMarkRuffalo\nTheEllenShow\njohnaugust\nKimKap\n55buckets\nSopanDeb\nBetsyHodges\nleeunkrich\nbessbell\nezraklein\nAOC\npetrodraiz\nchamath\nsophchang\npriyankachopra\nvoxdotcom\nTiffanyHaddish\nadriangrenier\njessetyler\njohnlegend\nalyankovic\nperlmutations\njonnysun\nnikolajcw\nGabbySidibe\nadampally\nJasonRitter\nkenjeong\npronounced_ing\nColeEscola\nshresnik\nzach_r1ce\nJayPharoah\nSacramentoKings\nTVietor08\njaboukie\nTheSamhita\ntomsegura\njimmykimmel\ngabegundacker\nTHEKIDMERO\nGenius\nbrandonjinx\nSheilaVee\njackwhitehall\nrussellhoward\ndstfelix\nyogrishiramdev\numxrshk\nkporzee\nEnesKanter\njennyhan\njenflanz\nhodakatebi\npatriotact\ndavidiserson\nTheDweck\nthatchriskelly\nJokoy\nN_C_B\nRonnieFieg\nattell\nKobiLibii\nMB3FIVE\nACLU\nBarackObama\nnytimes\nmaddow\nSethrogen\nbananapeele\nkathygriffin\nknguyen\nGQMagazine\nfranciaraisa\nBlairImani\nAMANI2020\nFullFrontalSamB\nJHarden13\ncolbertlateshow\nStephenAtHome\nvanitaguptaCR\ntannercolby\nzachdilanzo\nelizacossio\nDevDell\nnbcsnl\nColinJost\nseanogallagher\nLastWeekTonight\nAnikKhan_\nSamanth_S\nGuzKhanOfficial\ndissectpodcast\nmattingebretson\nFaizaPatelBCJ\nLefsetz\nrtsimp\nmrmedina\npaulshipper\nbejohnce\nMamoudouNDiaye\nLilfilm\nFelonious_munk\nJuleykaLantigua\nvcunningham\nReddsaidit\neshagupta2811\nbillyeichner\nJimGaffigan\nPreetBharara\nA24\njeremysliew\nMMFlint\nheymichellelee\nmikehofman\neveewing\nfranklinleonard\nelseed\nairfrance\nKaepernick7\naliamjadrizvi\narturodraws\nStephenKing\nSamHeughan\nedgarwright\nSusannaFogel\nBarryJenkins\nMitchyD\nyunamusic\nKinglimaa\nBenSPLATT\nthevirdas\nrakeshsatyal\nblamethelabel\nTonyRevolori\niffykaysar\nStephenRDaw\nAvanJogia\nmichaelsmith\nMalPal711\nmandamanda___\nSheaSerrano\nmallika_rao\nCariChampion\nMikeDrucker\njk_rowling\nava\nditzkoff\nMekkiLeeper\niamledgin\ncharltonbrooker\nludwiggoransson\nRachelFeinstein\nrealDonaldTrump\nEmilyeOberg\nlevie\nVaynerMedia\nEdgeofSports\ntySchmitt5\nJohnLeguizamo\nMarkDuplass\nVanJones68\nDanAmira\najv\nfinkd\ndavidrocknyc\nalexwagner\nComplex\nseanseaevans\nmarquezjesso\nBRANDONWARDELL\nNPR\nRickFamuyiwa\nLewisHowes\nDreamsickJustin\nGladwell\nMsEmmaBowman\niamsrk\ndeclanwalsh\nmholland85\nBKBMG\ngoodreads\nKrewella\namritsingh\nBlazerRamon\nianbremmer\nRogueTerritory\naspiegelnpr\njanellejcomic\nKingOfQueenz\niJesseWilliams\nlildickytweets\nVasu\ningridnilsen\njoshrogin\nsethmeyers\nramy\nsolomongeorgio\nMrGeorgeWallace\nDaveedDiggs\ngaryvee\ndpmeyer\nstevejang\nringer\npaulwdowns\nrobinthede\ntedtremper\nJustinTrudeau\nwfcgreen\namirkingkhan\nfarantahir_\nmehdirhasan\nhopesolo\nSenSanders\nisaiahlester\nBernieSanders\nNateParker\nkatepurchase\ntelfordk\nAnandWrites\nMRPORTERLIVE\nchancetherapper\nbcamp810\nHOUSE_of_WARIS\nLilly\nbrokemogul\nThaboSefolosha\nbomani_jones\nATTACKATHLETICS\nDLeary0us\nUnitedBlackout\nLenaWaithe\ngkhamba\nJeffreyGurian\natifateeq\nkendricklamar\nMatthewModine\nNickKristof\nDreamville\nKillerMike\nryanleslie\nHEIRMJ\nTheNarcicyst\n"
],
[
"col1",
"_____no_output_____"
],
[
"col2",
"_____no_output_____"
],
[
"df = pd.DataFrame(index=None)\ndf[\"source\"]=col1\ndf[\"target\"]=col2\nprint(df)\ndf.to_csv('fs.csv',index=False)",
" source target\n0 hasanminhaj chrissyteigen\n1 hasanminhaj mitchrichmond23\n2 hasanminhaj ninaemlemdi\n3 hasanminhaj Jon_Favreau\n4 hasanminhaj drsanjaygupta\n.. ... ...\n395 hasanminhaj Mowjood\n396 hasanminhaj Trevornoah\n397 hasanminhaj jinajones\n398 hasanminhaj AdamLowitt\n399 hasanminhaj jordanklepper\n\n[400 rows x 2 columns]\n"
],
[
"df",
"_____no_output_____"
],
[
"c=4\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fs.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fs.csv',index=False)",
"Rate limit reached. Sleeping for: 45\n"
],
[
"c=25\n\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fs.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fs.csv',index=False)",
"Rate limit reached. Sleeping for: 7\n"
],
[
"c=30\n\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fs.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fs.csv',index=False)",
"Rate limit reached. Sleeping for: 11\n"
],
[
"c=50\n\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fs.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fs.csv',index=False)",
"Rate limit reached. Sleeping for: 438\n"
],
[
"c=55\n\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fs.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fs.csv',index=False)",
"Rate limit reached. Sleeping for: 713\n"
],
[
"c=70\n\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fs.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fs.csv',index=False)",
"Rate limit reached. Sleeping for: 593\n"
],
[
"c=300\n\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fs.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fs.csv',index=False)",
"Rate limit reached. Sleeping for: 253\n"
],
[
"import tweepy\nimport pandas as pd\naccess_token = \"944440837739487232-KTdrvr4vARk7RTKvJkRPUF8I4VOvGIr\"\naccess_token_secret = \"bfHE0jC5h3B7W3H18TxV7XsofG1xuB6zeINo2DxmZ8K1W\"\nconsumer_key = \"Q5kScvH4J2CE6d3w8xesxT1bm\"\nconsumer_secret = \"mlGrcssaVjN9hQMi6wI6RqWKt2LcHAEyYCGh6WF8yq20qcTb8T\"\ncol1=[]\ncol2=[]\n\nauth = tweepy.OAuthHandler(consumer_key, consumer_secret)\nauth.set_access_token(access_token, access_token_secret)\napi = tweepy.API(auth,wait_on_rate_limit=True,wait_on_rate_limit_notify=True,compression=True)",
"_____no_output_____"
],
[
"col2=['chrissyteigen',\n 'mitchrichmond23',\n 'ninaemlemdi',\n 'Jon_Favreau',\n 'drsanjaygupta',\n 'MattGertz',\n 'JasonSCampbell',\n 'jkbjournalist',\n 'tomhanks',\n 'HillaryClinton',\n 'anitakumar01',\n 'BoysClubNY',\n 'dmaq1',\n 'ChrisEvans',\n 'SarahxAnwer',\n 'Casey',\n 'martinandanar',\n 'MarkRuffalo',\n 'TheEllenShow',\n 'johnaugust',\n 'KimKap',\n '55buckets',\n 'SopanDeb',\n 'BetsyHodges',\n 'leeunkrich',\n 'bessbell',\n 'ezraklein',\n 'AOC',\n 'petrodraiz',\n 'chamath',\n 'sophchang',\n 'priyankachopra',\n 'voxdotcom',\n 'TiffanyHaddish',\n 'adriangrenier',\n 'jessetyler',\n 'johnlegend',\n 'alyankovic',\n 'perlmutations',\n 'jonnysun',\n 'nikolajcw',\n 'GabbySidibe',\n 'adampally',\n 'JasonRitter',\n 'kenjeong',\n 'pronounced_ing',\n 'ColeEscola',\n 'shresnik',\n 'zach_r1ce',\n 'JayPharoah',\n 'SacramentoKings',\n 'TVietor08',\n 'jaboukie',\n 'TheSamhita',\n 'tomsegura',\n 'jimmykimmel',\n 'gabegundacker',\n 'THEKIDMERO',\n 'Genius',\n 'brandonjinx',\n 'SheilaVee',\n 'jackwhitehall',\n 'russellhoward',\n 'dstfelix',\n 'yogrishiramdev',\n 'umxrshk',\n 'kporzee',\n 'EnesKanter',\n 'jennyhan',\n 'jenflanz',\n 'hodakatebi',\n 'patriotact',\n 'davidiserson',\n 'TheDweck',\n 'thatchriskelly',\n 'Jokoy',\n 'N_C_B',\n 'RonnieFieg',\n 'attell',\n 'KobiLibii',\n 'MB3FIVE',\n 'ACLU',\n 'BarackObama',\n 'nytimes',\n 'maddow',\n 'Sethrogen',\n 'bananapeele',\n 'kathygriffin',\n 'knguyen',\n 'GQMagazine',\n 'franciaraisa',\n 'BlairImani',\n 'AMANI2020',\n 'FullFrontalSamB',\n 'JHarden13',\n 'colbertlateshow',\n 'StephenAtHome',\n 'vanitaguptaCR',\n 'tannercolby',\n 'zachdilanzo',\n 'elizacossio',\n 'DevDell',\n 'nbcsnl',\n 'ColinJost',\n 'seanogallagher',\n 'LastWeekTonight',\n 'AnikKhan_',\n 'Samanth_S',\n 'GuzKhanOfficial',\n 'dissectpodcast',\n 'mattingebretson',\n 'FaizaPatelBCJ',\n 'Lefsetz',\n 'rtsimp',\n 'mrmedina',\n 'paulshipper',\n 'bejohnce',\n 'MamoudouNDiaye',\n 'Lilfilm',\n 'Felonious_munk',\n 'JuleykaLantigua',\n 'vcunningham',\n 'Reddsaidit',\n 'eshagupta2811',\n 'billyeichner',\n 'JimGaffigan',\n 'PreetBharara',\n 'A24',\n 'jeremysliew',\n 'MMFlint',\n 'heymichellelee',\n 'mikehofman',\n 'eveewing',\n 'franklinleonard',\n 'elseed',\n 'airfrance',\n 'Kaepernick7',\n 'aliamjadrizvi',\n 'arturodraws',\n 'StephenKing',\n 'SamHeughan',\n 'edgarwright',\n 'SusannaFogel',\n 'BarryJenkins',\n 'MitchyD',\n 'yunamusic',\n 'Kinglimaa',\n 'BenSPLATT',\n 'thevirdas',\n 'rakeshsatyal',\n 'blamethelabel',\n 'TonyRevolori',\n 'iffykaysar',\n 'StephenRDaw',\n 'AvanJogia',\n 'michaelsmith',\n 'MalPal711',\n 'mandamanda___',\n 'SheaSerrano',\n 'mallika_rao',\n 'CariChampion',\n 'MikeDrucker',\n 'jk_rowling',\n 'ava',\n 'ditzkoff',\n 'MekkiLeeper',\n 'iamledgin',\n 'charltonbrooker',\n 'ludwiggoransson',\n 'RachelFeinstein',\n 'realDonaldTrump',\n 'EmilyeOberg',\n 'levie',\n 'VaynerMedia',\n 'EdgeofSports',\n 'tySchmitt5',\n 'JohnLeguizamo',\n 'MarkDuplass',\n 'VanJones68',\n 'DanAmira',\n 'ajv',\n 'finkd',\n 'davidrocknyc',\n 'alexwagner',\n 'Complex',\n 'seanseaevans',\n 'marquezjesso',\n 'BRANDONWARDELL',\n 'NPR',\n 'RickFamuyiwa',\n 'LewisHowes',\n 'DreamsickJustin',\n 'Gladwell',\n 'MsEmmaBowman',\n 'iamsrk',\n 'declanwalsh',\n 'mholland85',\n 'BKBMG',\n 'goodreads',\n 'Krewella',\n 'amritsingh',\n 'BlazerRamon',\n 'ianbremmer',\n 'RogueTerritory',\n 'aspiegelnpr',\n 'janellejcomic',\n 'KingOfQueenz',\n 'iJesseWilliams',\n 'lildickytweets',\n 'Vasu',\n 'ingridnilsen',\n 'joshrogin',\n 'sethmeyers',\n 'ramy',\n 'solomongeorgio',\n 'MrGeorgeWallace',\n 'DaveedDiggs',\n 'garyvee',\n 'dpmeyer',\n 'stevejang',\n 'ringer',\n 'paulwdowns',\n 'robinthede',\n 'tedtremper',\n 'JustinTrudeau',\n 'wfcgreen',\n 'amirkingkhan',\n 'farantahir_',\n 'mehdirhasan',\n 'hopesolo',\n 'SenSanders',\n 'isaiahlester',\n 'BernieSanders',\n 'NateParker',\n 'katepurchase',\n 'telfordk',\n 'AnandWrites',\n 'MRPORTERLIVE',\n 'chancetherapper',\n 'bcamp810',\n 'HOUSE_of_WARIS',\n 'Lilly',\n 'brokemogul',\n 'ThaboSefolosha',\n 'bomani_jones',\n 'ATTACKATHLETICS',\n 'DLeary0us',\n 'UnitedBlackout',\n 'LenaWaithe',\n 'gkhamba',\n 'JeffreyGurian',\n 'atifateeq',\n 'kendricklamar',\n 'MatthewModine',\n 'NickKristof',\n 'Dreamville',\n 'KillerMike',\n 'ryanleslie',\n 'HEIRMJ',\n 'TheNarcicyst',\n 'joshluber',\n 'nathanfielder',\n 'djkhaled',\n 'ayeshacurry',\n 'MazMHussain',\n 'davidfolkenflik',\n 'JensenKarp',\n 'michaelb4jordan',\n 'JordanPeele',\n 'MattHalfhill',\n 'IanMcKellen',\n 'iraglass',\n 'El_Silvero',\n 'JLaPuma',\n 'anildash',\n 'rameswaram',\n 'AllOfItWNYC',\n 'thismyshow',\n 'EugeneMirman',\n 'ShahanR',\n 'NinaDavuluri',\n 'GrantNapearshow',\n 'CarmichaelDave',\n 'EliseCz',\n 'Andrea_Simmons',\n 'showtoones',\n 'electrolemon',\n 'iamcolinquinn',\n 'abrahamjoseph',\n 'bfishbfish',\n 'SpecialRepMC',\n 'fannynordmark',\n 'JRHavlan',\n 'ambarella',\n 'deray',\n 'hugoandmarie',\n 'IStandWithAhmed',\n 'showmetheravi',\n 'DesiLydic',\n 'roywoodjr',\n 'ronnychieng',\n 'fakedansavage',\n 'chrislhayes',\n 'twitney',\n 'humansofny',\n 'Lin_Manuel',\n 'mdotbrown',\n 'PinnapplePower',\n 'ChrisGethard',\n 'EasyPri',\n 'oldmanebro',\n 'marcecko',\n 'eugcordero',\n 'Iam1Cent',\n 'ajjacobs',\n 'melissamccarthy',\n 'TaheraHAhmad',\n 'patthewanderer',\n 'saladinahmed',\n 'SamSpratt',\n 'jamesmiglehart',\n 'AkilahObviously',\n 'tompapa',\n 'phlaimeaux',\n 'GBerlanti',\n 'AngeloLozada66',\n 'rojoperezzz',\n 'prattprattpratt',\n 'dherzog77',\n 'talkhoops',\n 'VRam_21',\n 'heavenrants',\n 'rastphan',\n 'jsmooth995',\n 'ComedyGroupie',\n 'chrizmillr',\n 'YourAnonNews',\n 'LuciaAniello',\n 'Babyballs69',\n 'paulfeig',\n 'GlitterCheese',\n 'mojorojo',\n 'BinaShah',\n 'ismat',\n 'RonanFarrow',\n 'tejucole',\n 'Zeyba',\n 'joncbenson',\n 'iamsambee',\n 'lsarsour',\n 'joshgondelman',\n 'robcorddry',\n 'kristenschaaled',\n 'danielradosh',\n 'timcarvell',\n 'AymanM',\n 'BergerWorld',\n 'iamjohnoliver',\n 'rabiasquared',\n 'Apey',\n 'heyrubes_',\n 'graceofwrath',\n 'raminhedayati',\n 'jonesinforjason',\n 'Variety',\n 'almadrigal',\n 'philiplord',\n 'HallieHaglund',\n 'dopequeenpheebs',\n 'rianjohnson',\n 'zainabjohnson',\n 'billburr',\n 'mileskahn',\n 'MrChadCarter',\n 'hodgman',\n 'sammorril',\n 'AaronCouch',\n 'brennanshroff',\n 'ComedyCellarUSA',\n 'RoryAlbanese',\n 'mattkoff',\n 'tedalexandro',\n 'JenaFriedman',\n 'OnePerfectShot',\n 'GeorgeKiel3',\n 'morninggloria',\n 'JSim07',\n 'TessaThompson_x',\n 'alicewetterlund',\n 'SarahTreem',\n 'baluchx',\n 'BENBALLER',\n 'TheDailyShow',\n 'larrywilmore',\n 'JessicaPilot212',\n 'Mowjood',\n 'Trevornoah',\n 'jinajones',\n 'AdamLowitt',\n 'jordanklepper']",
"_____no_output_____"
],
[
"col1=['hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj',\n 'hasanminhaj']",
"_____no_output_____"
],
[
"df=pd.DataFrame()\ndf['source']=col1\ndf['target']=col2",
"_____no_output_____"
],
[
"import pandas as pd\nc=310\nslp=0\nfriend = col2.copy()\ncol3=[]\n\nfor i in friend[c:]:\n print(c)\n c=c+1\n col3.append(i)\n for j in friend[c:]:\n friendship=api.show_friendship(source_screen_name=i, target_screen_name=j)\n if(friendship[0].followed_by):\n print(j,i) # i followed by j\n df.loc[len(df)]=[j,i]\n df.to_csv('fin.csv',index=False)\n if(friendship[1].followed_by):\n print(i,j) # j followed by i\n df.loc[len(df)]=[i,j]\n df.to_csv('fin.csv',index=False)",
"310\nlsarsour oldmanebro\nVariety oldmanebro\nJSim07 oldmanebro\nBENBALLER oldmanebro\noldmanebro BENBALLER\n311\nVariety marcecko\nBENBALLER marcecko\nmarcecko BENBALLER\nJessicaPilot212 marcecko\n312\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8c8c004b42969617ea351b9e8cf924d00df7b2
| 9,753 |
ipynb
|
Jupyter Notebook
|
my_models/ipynb/resnet_50_skin_cancer.ipynb
|
munachisonwadike/gram-ood
|
0cfd8de737badfbe3cc26c4761173977e8468dea
|
[
"MIT"
] | 10 |
2020-06-16T04:41:54.000Z
|
2022-03-10T03:52:55.000Z
|
my_models/ipynb/resnet_50_skin_cancer.ipynb
|
munachisonwadike/gram-ood
|
0cfd8de737badfbe3cc26c4761173977e8468dea
|
[
"MIT"
] | 2 |
2020-10-30T17:14:41.000Z
|
2021-11-09T16:15:32.000Z
|
my_models/ipynb/resnet_50_skin_cancer.ipynb
|
munachisonwadike/gram-ood
|
0cfd8de737badfbe3cc26c4761173977e8468dea
|
[
"MIT"
] | 1 |
2020-11-24T17:39:22.000Z
|
2020-11-24T17:39:22.000Z
| 37.511538 | 111 | 0.494412 |
[
[
[
"import sys\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"# Releasing the GPU memory\ntorch.cuda.empty_cache()",
"_____no_output_____"
],
[
"def conv3x3(in_planes, out_planes, stride=1, groups=1, dilation=1):\n \"\"\"3x3 convolution with padding\"\"\"\n return nn.Conv2d(in_planes, out_planes, kernel_size=3, stride=stride,\n padding=dilation, groups=groups, bias=False, dilation=dilation)\n\n\ndef conv1x1(in_planes, out_planes, stride=1):\n \"\"\"1x1 convolution\"\"\"\n return nn.Conv2d(in_planes, out_planes, kernel_size=1, stride=stride, bias=False)\n\n\nclass Bottleneck(nn.Module):\n expansion = 4\n\n def __init__(self, inplanes, planes, stride=1, downsample=None, groups=1,\n base_width=64, dilation=1, norm_layer=None):\n super(Bottleneck, self).__init__()\n if norm_layer is None:\n norm_layer = nn.BatchNorm2d\n width = int(planes * (base_width / 64.)) * groups\n # Both self.conv2 and self.downsample layers downsample the input when stride != 1\n self.conv1 = conv1x1(inplanes, width)\n self.bn1 = norm_layer(width)\n self.conv2 = conv3x3(width, width, stride, groups, dilation)\n self.bn2 = norm_layer(width)\n self.conv3 = conv1x1(width, planes * self.expansion)\n self.bn3 = norm_layer(planes * self.expansion)\n self.relu = nn.ReLU(inplace=True)\n self.downsample = downsample\n self.stride = stride\n\n def forward(self, x):\n identity = x\n\n out = self.conv1(x)\n out = self.bn1(out)\n out = self.relu(out)\n \n out = self.conv2(out)\n out = self.bn2(out)\n out = self.relu(out)\n\n out = self.conv3(out)\n out = self.bn3(out)\n\n if self.downsample is not None:\n identity = self.downsample(x)\n\n out += identity\n out = self.relu(out)\n\n return out\n\n\nclass ResNet(nn.Module):\n\n def __init__(self, block, layers, num_classes=1000, zero_init_residual=False,\n groups=1, width_per_group=64, replace_stride_with_dilation=None,\n norm_layer=None):\n super(ResNet, self).__init__()\n if norm_layer is None:\n norm_layer = nn.BatchNorm2d\n self._norm_layer = norm_layer\n\n self.inplanes = 64\n self.dilation = 1\n if replace_stride_with_dilation is None:\n # each element in the tuple indicates if we should replace\n # the 2x2 stride with a dilated convolution instead\n replace_stride_with_dilation = [False, False, False]\n if len(replace_stride_with_dilation) != 3:\n raise ValueError(\"replace_stride_with_dilation should be None \"\n \"or a 3-element tuple, got {}\".format(replace_stride_with_dilation))\n self.groups = groups\n self.base_width = width_per_group\n self.conv1 = nn.Conv2d(3, self.inplanes, kernel_size=7, stride=2, padding=3,\n bias=False)\n self.bn1 = norm_layer(self.inplanes)\n self.relu = nn.ReLU(inplace=True)\n self.maxpool = nn.MaxPool2d(kernel_size=3, stride=2, padding=1)\n self.layer1 = self._make_layer(block, 64, layers[0])\n self.layer2 = self._make_layer(block, 128, layers[1], stride=2,\n dilate=replace_stride_with_dilation[0])\n self.layer3 = self._make_layer(block, 256, layers[2], stride=2,\n dilate=replace_stride_with_dilation[1])\n self.layer4 = self._make_layer(block, 512, layers[3], stride=2,\n dilate=replace_stride_with_dilation[2])\n self.avgpool = nn.AdaptiveAvgPool2d((1, 1))\n self.fc = nn.Linear(512 * block.expansion, num_classes)\n\n for m in self.modules():\n if isinstance(m, nn.Conv2d):\n nn.init.kaiming_normal_(m.weight, mode='fan_out', nonlinearity='relu')\n elif isinstance(m, (nn.BatchNorm2d, nn.GroupNorm)):\n nn.init.constant_(m.weight, 1)\n nn.init.constant_(m.bias, 0)\n\n def _make_layer(self, block, planes, blocks, stride=1, dilate=False):\n norm_layer = self._norm_layer\n downsample = None\n previous_dilation = self.dilation\n if dilate:\n self.dilation *= stride\n stride = 1\n if stride != 1 or self.inplanes != planes * block.expansion:\n downsample = nn.Sequential(\n conv1x1(self.inplanes, planes * block.expansion, stride),\n norm_layer(planes * block.expansion),\n )\n\n layers = []\n layers.append(block(self.inplanes, planes, stride, downsample, self.groups,\n self.base_width, previous_dilation, norm_layer))\n self.inplanes = planes * block.expansion\n for _ in range(1, blocks):\n layers.append(block(self.inplanes, planes, groups=self.groups,\n base_width=self.base_width, dilation=self.dilation,\n norm_layer=norm_layer))\n\n return nn.Sequential(*layers)\n\n def forward(self, x):\n x = self.conv1(x)\n x = self.bn1(x)\n x = self.relu(x)\n x = self.maxpool(x)\n\n x = self.layer1(x)\n x = self.layer2(x)\n x = self.layer3(x)\n x = self.layer4(x)\n\n x = self.avgpool(x)\n x = x.reshape(x.size(0), -1)\n x = self.fc(x)\n\n return x\n\n\nclass Net (nn.Module):\n\n def __init__(self, num_class, freeze_conv=False, n_extra_info=0, p_dropout=0.5, neurons_class=256,\n feat_reducer=None, classifier=None):\n super(Net, self).__init__()\n\n resnet = ResNet(Bottleneck, [3, 4, 6, 3])\n self.features = nn.Sequential(*list(resnet.children())[:-1])\n\n # freezing the convolution layers\n if freeze_conv:\n for param in self.features.parameters():\n param.requires_grad = False\n\n # Feature reducer\n if feat_reducer is None:\n self.feat_reducer = nn.Sequential(\n nn.Linear(2048, neurons_class),\n nn.BatchNorm1d(neurons_class),\n nn.ReLU(),\n nn.Dropout(p=p_dropout)\n )\n else:\n self.feat_reducer = feat_reducer\n\n # Here comes the extra information (if applicable)\n if classifier is None:\n self.classifier = nn.Linear(neurons_class + n_extra_info, num_class)\n else:\n self.classifier = classifier\n \n self.collecting = False\n\n def forward(self, img, extra_info=None):\n\n x = self.features(img)\n # Flatting\n x = x.view(x.size(0), -1)\n x = self.feat_reducer(x)\n res = self.classifier(x)\n return res\n\ntorch_model = Net(8)\nckpt = torch.load(\"checkpoints/resnet-50_checkpoint.pth\")\ntorch_model.load_state_dict(ckpt['model_state_dict'])\ntorch_model.eval()\ntorch_model.cuda()\nprint(\"Done!\")",
"Done!\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
4a8c910c4cbb56222b6019a87e8c91094b02ec1d
| 6,524 |
ipynb
|
Jupyter Notebook
|
notebooks/birdsong/19.0-day-length-bouts.ipynb
|
timsainb/ParallelsBirdsongLanguagePaper
|
712d4d99b716e121f4e344810bb12f8f45ce7036
|
[
"BSD-3-Clause"
] | 2 |
2019-06-12T08:25:22.000Z
|
2020-12-17T03:59:03.000Z
|
notebooks/birdsong/19.0-day-length-bouts.ipynb
|
timsainb/ParallelsBirdsongLanguagePaper
|
712d4d99b716e121f4e344810bb12f8f45ce7036
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/birdsong/19.0-day-length-bouts.ipynb
|
timsainb/ParallelsBirdsongLanguagePaper
|
712d4d99b716e121f4e344810bb12f8f45ce7036
|
[
"BSD-3-Clause"
] | 1 |
2021-07-30T16:11:49.000Z
|
2021-07-30T16:11:49.000Z
| 23.053004 | 85 | 0.551655 |
[
[
[
"# get bouts per day for each bird\n1. load datasets\n...",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom parallelspaper.config.paths import DATA_DIR\nfrom parallelspaper.birdsong_datasets import MI_seqs, compress_seq",
"_____no_output_____"
],
[
"from parallelspaper.quickplots import plot_model_fits\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Load data",
"_____no_output_____"
]
],
[
[
"starling_seq_df = pd.read_pickle(DATA_DIR / 'song_seq_df/starling.pickle')\nCAVI_CATH_seq_df = pd.read_pickle(DATA_DIR / 'song_seq_df/CAVI_CATH.pickle')\nBF_seq_df = pd.read_pickle(DATA_DIR / 'song_seq_df/BF.pickle')",
"_____no_output_____"
],
[
"seq_dfs = pd.concat([starling_seq_df, CAVI_CATH_seq_df, BF_seq_df])",
"_____no_output_____"
],
[
"# sequence lengths\nseq_dfs['sequence_lens'] = [len(i) for i in seq_dfs.syllables]\n# recording number as integer\nseq_dfs['rec_num'] = seq_dfs.rec_num.values.astype('int32')\n# sort sequences\nseq_dfs = seq_dfs.sort_values(by=['species', 'bird', 'rec_num'])\n# get rid of unID'd birds (CAVI, CATH)\nseq_dfs = seq_dfs[seq_dfs.bird != '?']",
"_____no_output_____"
],
[
"seq_dfs[:3]",
"_____no_output_____"
],
[
"# for each bird, for each day, count the number of bouts in that day",
"_____no_output_____"
],
[
"from tqdm.autonotebook import tqdm",
"_____no_output_____"
],
[
"day_bouts = {}\nfor species in (np.unique(seq_dfs.species)):\n day_bouts[species] = []\n species_df = seq_dfs[seq_dfs.species == species]\n for bird in tqdm(np.unique(species_df.bird.values)):\n bird_df = species_df[species_df.bird == bird]\n for day in np.unique(bird_df.day.values):\n day_df = bird_df[bird_df.day==day]\n day_bouts[species].append(len(day_df)) ",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(ncols=4, figsize=(16,4))\nfor si, species in enumerate(np.unique(seq_dfs.species)):\n axs[si].hist(day_bouts[species])",
"_____no_output_____"
],
[
"np.median(day_bouts['BF'])",
"_____no_output_____"
],
[
"np.median(day_bouts['CAVI'])",
"_____no_output_____"
],
[
"np.median(day_bouts['CATH'])",
"_____no_output_____"
],
[
"np.median(day_bouts['Starling'])",
"_____no_output_____"
],
[
"np.median(np.concatenate([day_bouts[i] for i in day_bouts.keys()]))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8c9c4704a1687e06504930e0042cee12cf8b5b
| 4,725 |
ipynb
|
Jupyter Notebook
|
A_Tour_of_Computer_Animation_Table_of_Contents.ipynb
|
BachiLi/A-Tour-of-Computer-Animation
|
c739e333a5519b1ec379b8a867536de65233d0ea
|
[
"MIT"
] | 4 |
2021-06-15T16:07:28.000Z
|
2021-08-12T02:01:30.000Z
|
A_Tour_of_Computer_Animation_Table_of_Contents.ipynb
|
BachiLi/A-Tour-of-Computer-Animation
|
c739e333a5519b1ec379b8a867536de65233d0ea
|
[
"MIT"
] | null | null | null |
A_Tour_of_Computer_Animation_Table_of_Contents.ipynb
|
BachiLi/A-Tour-of-Computer-Animation
|
c739e333a5519b1ec379b8a867536de65233d0ea
|
[
"MIT"
] | null | null | null | 48.71134 | 559 | 0.649947 |
[
[
[
"<a href=\"https://colab.research.google.com/github/BachiLi/A-Tour-of-Computer-Animation/blob/main/A_Tour_of_Computer_Animation_Table_of_Contents.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"**A Tour of Computer Animation** -- [Tzu-Mao Li](https://cseweb.ucsd.edu/~tzli/)",
"_____no_output_____"
],
[
"This is a note that records my journey into computer animation. The structure of this tour is inspired by the books [\"Physically Based Rendering:From Theory To Implementation\"](https://www.pbr-book.org/), [\"Ray Tracing in One Weekend\"](https://raytracing.github.io/books/RayTracingInOneWeekend.html), and [\"Numerical Tours\"](https://www.numerical-tours.com/). Most books and articles about computer animation and physics simulation are mathematic centric and do not contain much code and experiments. This note is an attempt to bridge the gap.",
"_____no_output_____"
],
[
"This is the hub to the chapters of this tour. I do not assume background on computer animation or graphics, but I do assume basic familiarity on calculus and linear algebra. There will be quite a bit of math -- sorry. The code is going to be all written in numpy and visualize with matplotlib. These are going to be unoptimized code. We will focus slightly more on the foundation instead of immediate practical implementations, so it might take a while before we can render fancy animations. Don't be afraid to play with the code.",
"_____no_output_____"
],
[
"**Table of Contents**\n1. [Newtonian Mechanics and Forward Euler Method](https://colab.research.google.com/drive/1K-Ly9vqZbymrAYe6Krg1ZfSMPY6CnAcY)\n2. [Lagrangian Mechanics and Pendulums](https://colab.research.google.com/drive/1L4QJyq8hSlgllSYytYW5UHTPvd6w7Vz9)\n3. [Time Integration and Stability](https://colab.research.google.com/drive/1mXTlYt2nRnXLrXpnP26BgjHKghjGPTCL?usp=sharing)\n4. [Elastic Simulation and Mass Spring Systems](https://colab.research.google.com/drive/1erjL0a_KCVx8p3lDcE747k8wqbEaxYPY?usp=sharing)\n5. Physics as Constraints Solving and Position-based Dynamics\n",
"_____no_output_____"
],
[
"Some useful textbooks and lectures (they are not prerequisite, but instead this note should be used as an accompanying material to these):\n- [David Levin: CSC417 - Physics-based Animation](https://www.youtube.com/playlist?list=PLTkE7n2CwG_PH09_q0Q7ttjqE2F9yGeM3) (the structure of this tour takes huge inspirations from this course)\n- [The Feynman Lectures on Physics](https://www.feynmanlectures.caltech.edu/)\n- [Doug James: CS348C - Computer Graphics: Animation and Simulation](https://graphics.stanford.edu/courses/cs348c/)\n- [Arnold: Mathematical Methods of Classical Mechanics](https://www.amazon.com/Mathematical-Classical-Mechanics-Graduate-Mathematics/dp/0387968903)\n- [Witkin and Baraff: Physics-based Modeling](https://graphics.pixar.com/pbm2001/)\n- [Bargteil and Shinar: An Introduction to Physics-based Animation](https://cal.cs.umbc.edu/Courses/PhysicsBasedAnimation/)\n- [Åström and Akenine-Möller: Immersive Linear Algebra](http://immersivemath.com/ila/index.html)",
"_____no_output_____"
],
[
"The notes are still work in progress and probably contain a lot of errors. Please send email to me ([email protected]) if you have any suggestions and comments.",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a8c9f004021c36c52723760761ac821e0c7a9f1
| 62,590 |
ipynb
|
Jupyter Notebook
|
shenliting/homework4/Untitled9.ipynb
|
nju-teaching/computational-communication
|
b95bca72bcfbe412fef15df9f3f057e398be7e34
|
[
"MIT"
] | 7 |
2016-03-16T12:11:39.000Z
|
2018-05-03T16:42:08.000Z
|
shenliting/homework4/Untitled9.ipynb
|
nju-teaching/computational-communication
|
b95bca72bcfbe412fef15df9f3f057e398be7e34
|
[
"MIT"
] | 5 |
2016-03-18T02:03:35.000Z
|
2016-05-04T10:20:52.000Z
|
shenliting/homework4/Untitled9.ipynb
|
nju-teaching/computational-communication
|
b95bca72bcfbe412fef15df9f3f057e398be7e34
|
[
"MIT"
] | 12 |
2016-03-16T12:12:13.000Z
|
2017-04-03T09:25:39.000Z
| 29.193097 | 650 | 0.458636 |
[
[
[
"import urllib2\nfrom bs4 import BeautifulSoup",
"_____no_output_____"
],
[
"url = 'https://www.baidu.com/'\ncontent = urllib2.urlopen(url).read() \nsoup = BeautifulSoup(content, 'html.parser') \nsoup",
"_____no_output_____"
],
[
"print(soup.prettify())",
"<html>\n <head>\n <script>\n location.replace(location.href.replace(\"https://\",\"http://\"));\n </script>\n </head>\n <body>\n <noscript>\n <meta content=\"0;url=http://www.baidu.com/\" http-equiv=\"refresh\"/>\n </noscript>\n </body>\n</html>\n"
],
[
"for tag in soup.find_all(True):\n print(tag.name)",
"html\nhead\nscript\nbody\nnoscript\nmeta\n"
],
[
"soup('head')# or soup.head",
"_____no_output_____"
],
[
"soup.body",
"_____no_output_____"
],
[
"soup.body.name",
"_____no_output_____"
],
[
"soup.meta.string",
"_____no_output_____"
],
[
"soup.find_all('noscript',content_='0;url=http://www.baidu.com/')",
"_____no_output_____"
],
[
"soup.find_all('noscript')[0]",
"_____no_output_____"
],
[
"soup.find_all([\"head\",\"script\"])",
"_____no_output_____"
],
[
"soup.get_text()",
"_____no_output_____"
],
[
"print(soup.get_text())",
"\n\n\r\n\t\tlocation.replace(location.href.replace(\"https://\",\"http://\"));\r\n\t\n\n\n\n\n\n"
],
[
"from IPython.display import display_html, HTML\nHTML('<iframe src=http://bbs.tianya.cn/list.jsp?item=free&nextid=%d&order=8&k=PX width=1000 height=500></iframe>')\n# the webpage we would like to crawl",
"_____no_output_____"
],
[
"page_num = 0\nurl = \"http://bbs.tianya.cn/list.jsp?item=free&nextid=%d&order=8&k=PX\" % page_num\ncontent = urllib2.urlopen(url).read() #获取网页的html文本\nsoup = BeautifulSoup(content, \"lxml\") \narticles = soup.find_all('tr')",
"_____no_output_____"
],
[
"print articles[0]",
"<tr>\n<th scope=\"col\"> 标题</th>\n<th scope=\"col\">作者</th>\n<th scope=\"col\">点击</th>\n<th scope=\"col\">回复</th>\n<th scope=\"col\">发表时间</th>\n</tr>\n"
],
[
"print articles[1]",
"<tr class=\"bg\">\n<td class=\"td-title \">\n<span class=\"face\" title=\"\">\n</span>\n<a href=\"/post-free-2849477-1.shtml\" target=\"_blank\">\r\n\t\t\t\t\t\t\t【民间语文第161期】宁波px启示:船进港湾人应上岸<span class=\"art-ico art-ico-3\" title=\"内有0张图片\"></span>\n</a>\n</td>\n<td><a class=\"author\" href=\"http://www.tianya.cn/50499450\" target=\"_blank\">贾也</a></td>\n<td>194684</td>\n<td>2703</td>\n<td title=\"2012-10-29 07:59\">10-29 07:59</td>\n</tr>\n"
],
[
"len(articles[1:])",
"_____no_output_____"
],
[
"for t in articles[1].find_all('td'): print t",
"<td class=\"td-title \">\n<span class=\"face\" title=\"\">\n</span>\n<a href=\"/post-free-2849477-1.shtml\" target=\"_blank\">\r\n\t\t\t\t\t\t\t【民间语文第161期】宁波px启示:船进港湾人应上岸<span class=\"art-ico art-ico-3\" title=\"内有0张图片\"></span>\n</a>\n</td>\n<td><a class=\"author\" href=\"http://www.tianya.cn/50499450\" target=\"_blank\">贾也</a></td>\n<td>194684</td>\n<td>2703</td>\n<td title=\"2012-10-29 07:59\">10-29 07:59</td>\n"
],
[
"td = articles[1].find_all('td')",
"_____no_output_____"
],
[
"print td[0]",
"<td class=\"td-title \">\n<span class=\"face\" title=\"\">\n</span>\n<a href=\"/post-free-2849477-1.shtml\" target=\"_blank\">\r\n\t\t\t\t\t\t\t【民间语文第161期】宁波px启示:船进港湾人应上岸<span class=\"art-ico art-ico-3\" title=\"内有0张图片\"></span>\n</a>\n</td>\n"
],
[
"print(td[0].text)",
"\n\n\n\r\n\t\t\t\t\t\t\t【民间语文第161期】宁波px启示:船进港湾人应上岸\n\n\n"
],
[
"print td[0].a['href']",
"/post-free-2849477-1.shtml\n"
],
[
"print td[1]\nprint td[2]\nprint td[3]\nprint td[4]",
"<td><a class=\"author\" href=\"http://www.tianya.cn/50499450\" target=\"_blank\">贾也</a></td>\n<td>194684</td>\n<td>2703</td>\n<td title=\"2012-10-29 07:59\">10-29 07:59</td>\n"
],
[
"records = []\nfor i in articles[1:]:\n td = i.find_all('td')\n title = td[0].text.strip()\n title_url = td[0].a['href']\n author = td[1].text\n author_url = td[1].a['href']\n views = td[2].text\n replies = td[3].text\n date = td[4]['title']\n record = title + '\\t' + title_url+ '\\t' + author + '\\t'+ author_url + '\\t' + views+ '\\t' + replies+ '\\t'+ date\n records.append(record)",
"_____no_output_____"
],
[
"print records[2]",
"宁波准备停止PX项目了,元芳,你怎么看?\t/post-free-2848797-1.shtml\t牧阳光\thttp://www.tianya.cn/36535656\t82860\t625\t2012-10-28 19:11\n"
],
[
"def crawler(page_num, file_name):\n try:\n # open the browser\n url = \"http://bbs.tianya.cn/list.jsp?item=free&nextid=%d&order=8&k=PX\" % page_num\n content = urllib2.urlopen(url).read() #获取网页的html文本\n soup = BeautifulSoup(content, \"lxml\") \n articles = soup.find_all('tr')\n # write down info\n for i in articles[1:]:\n td = i.find_all('td')\n title = td[0].text.strip()\n title_url = td[0].a['href']\n author = td[1].text\n author_url = td[1].a['href']\n views = td[2].text\n replies = td[3].text\n date = td[4]['title']\n record = title + '\\t' + title_url+ '\\t' + author + '\\t'+ \\\n author_url + '\\t' + views+ '\\t' + replies+ '\\t'+ date\n with open(file_name,'a') as p: # '''Note''':Append mode, run only once!\n p.write(record.encode('utf-8')+\"\\n\") ##!!encode here to utf-8 to avoid encoding\n\n except Exception, e:\n print e\n pass",
"_____no_output_____"
],
[
"# crawl all pages\nfor page_num in range(10):\n print (page_num)\n crawler(page_num, 'D:/GitHub/computational-communication-2016/shenliting/homework4/tianya_bbs_threads_list.txt') ",
"0\n1\n2\n3\n4\n5\n6\n7\n8\n9\n"
],
[
"import pandas as pd\n\ndf = pd.read_csv('D:/GitHub/computational-communication-2016/shenliting/homework4/tianya_bbs_threads_list.txt', sep = \"\\t\", header=None)\ndf[: 2]",
"_____no_output_____"
],
[
"len(df)",
"_____no_output_____"
],
[
"df=df.rename(columns = {0:'title', 1:'link', 2:'author',3:'author_page', 4:'click', 5:'reply', 6:'time'})\ndf[:2]",
"_____no_output_____"
],
[
"len(df.link)",
"_____no_output_____"
],
[
"df.author_page[:5]",
"_____no_output_____"
],
[
"def author_crawler(url, file_name):\n try:\n content = urllib2.urlopen(url).read() #获取网页的html文本\n soup = BeautifulSoup(content, \"lxml\")\n link_info = soup.find_all('div', {'class', 'link-box'})\n followed_num, fans_num = [i.a.text for i in link_info]\n try:\n activity = soup.find_all('span', {'class', 'subtitle'})\n post_num, reply_num = [j.text[2:] for i in activity[:1] for j in i('a')]\n except:\n post_num, reply_num = 1, 0\n record = '\\t'.join([url, followed_num, fans_num, post_num, reply_num])\n with open(file_name,'a') as p: # '''Note''':Append mode, run only once!\n p.write(record.encode('utf-8')+\"\\n\") ##!!encode here to utf-8 to avoid encoding\n\n except Exception, e:\n print e, url\n record = '\\t'.join([url, 'na', 'na', 'na', 'na'])\n with open(file_name,'a') as p: # '''Note''':Append mode, run only once!\n p.write(record.encode('utf-8')+\"\\n\") ##!!encode here to utf-8 to avoid encoding\n pass",
"_____no_output_____"
],
[
"for k, url in enumerate(df.author_page):\n if k % 10==0:\n print k\n author_crawler(url, 'D:/GitHub/computational-communication-2016/shenliting/homework4/tianya_bbs_threads_author_info.txt') ",
"0\n10\n20\n30\n40\nneed more than 0 values to unpack http://www.tianya.cn/67896263\nneed more than 0 values to unpack http://www.tianya.cn/42330613\nsequence item 3: expected string or Unicode, int found http://www.tianya.cn/26517664\n50\nneed more than 0 values to unpack http://www.tianya.cn/75591747\n60\nneed more than 0 values to unpack http://www.tianya.cn/24068399\n70\n80\n90\nneed more than 0 values to unpack http://www.tianya.cn/67896263\nsequence item 3: expected string or Unicode, int found http://www.tianya.cn/62237033\n100\n110\n120\n130\n140\n150\n160\n170\n180\n190\nneed more than 0 values to unpack http://www.tianya.cn/67896263\n200\n210\nneed more than 0 values to unpack http://www.tianya.cn/85353911\n220\n230\n240\n250\n260\n270\n280\nneed more than 0 values to unpack http://www.tianya.cn/67896263\n290\nneed more than 0 values to unpack http://www.tianya.cn/67896263\n300\n310\n320\nneed more than 0 values to unpack http://www.tianya.cn/67896263\n330\n340\n350\n360\n370\nneed more than 0 values to unpack http://www.tianya.cn/67896263\n380\n390\n400\n410\n420\n430\n440\n450\n460\n"
],
[
"url = df.author_page[1]\ncontent = urllib2.urlopen(url).read() #获取网页的html文本\nsoup1 = BeautifulSoup(content, \"lxml\") ",
"_____no_output_____"
],
[
"activity = soup1.find_all('span', {'class', 'subtitle'})\npost_num, reply_num = [j.text[2:] for i in activity[:1] for j in i('a')]\nprint post_num, reply_num",
"2 5\n"
],
[
"print activity[0]",
"<span class=\"subtitle\"><a href=\"http://www.tianya.cn/74341835/bbs?t=post\">主帖2</a> <a href=\"http://www.tianya.cn/74341835/bbs?t=reply\">回帖5</a></span>\n"
],
[
"df.link[2]",
"_____no_output_____"
],
[
"url = 'http://bbs.tianya.cn' + df.link[2]\nurl",
"_____no_output_____"
],
[
"from IPython.display import display_html, HTML\nHTML('<iframe src=http://bbs.tianya.cn/post-free-2848797-1.shtml width=1000 height=500></iframe>')\n# the webpage we would like to crawl",
"_____no_output_____"
],
[
"post = urllib2.urlopen(url).read() #获取网页的html文本\npost_soup = BeautifulSoup(post, \"lxml\") \n#articles = soup.find_all('tr')",
"_____no_output_____"
],
[
"print (post_soup.prettify())[:1000]",
"<!DOCTYPE HTML>\n<html>\n <head>\n <meta charset=\"utf-8\"/>\n <title>\n 宁波准备停止PX项目了,元芳,你怎么看?_天涯杂谈_天涯论坛\n </title>\n <meta content=\"宁波准备停止PX项目了,元芳,你怎么看? 从宁波市政府新闻发言人处获悉,宁波市经与项目投资方研究决定:(1)坚决不上PX项目;(2)炼化一体化项目前期工作停止推进,再作科学论证。...\" name=\"description\"/>\n <meta content=\"IE=EmulateIE9\" http-equiv=\"X-UA-Compatible\"/>\n <meta content=\"牧阳光\" name=\"author\"/>\n <meta content=\"format=xhtml; url=http://bbs.tianya.cn/m/post-free-2848797-1.shtml\" http-equiv=\"mobile-agent\"/>\n <link href=\"http://static.tianyaui.com/global/ty/TY.css\" rel=\"stylesheet\" type=\"text/css\"/>\n <link href=\"http://static.tianyaui.com/global/bbs/web/static/css/bbs_article_c55fffc.css\" rel=\"stylesheet\" type=\"text/css\"/>\n <link href=\"http://static.tianyaui.com/favicon.ico\" rel=\"shortcut icon\" type=\"image/vnd.microsoft.icon\"/>\n <link href=\"http://bbs.tianya.cn/post-free-2848797-2.shtml\" rel=\"next\"/>\n <script type=\"text/javascript\">\n var bbsGlobal = {\r\n\tisEhomeItem : false,\r\n\tisNewArticle : false,\r\n\tauthorId : \"36535656\",\r\n\tauthorNa\n"
],
[
"pa = post_soup.find_all('div', {'class', 'atl-item'})\nlen(pa)",
"_____no_output_____"
],
[
"print pa[0]",
"<div _host=\"%E7%89%A7%E9%98%B3%E5%85%89\" class=\"atl-item host-item\">\n<div class=\"atl-content\">\n<div class=\"atl-con-hd clearfix\">\n<div class=\"atl-con-hd-l\"></div>\n<div class=\"atl-con-hd-r\"></div>\n</div>\n<div class=\"atl-con-bd clearfix\">\n<div class=\"bbs-content clearfix\">\n<br/>\n 从宁波市政府新闻发言人处获悉,宁波市经与项目投资方研究决定:(1)坚决不上PX项目;(2)炼化一体化项目前期工作停止推进,再作科学论证。<br/>\n<br/>\n</div>\n<div class=\"clearfix\" id=\"alt_action\"></div>\n<div class=\"clearfix\">\n<div class=\"host-data\">\n<span>楼主发言:11次</span> <span>发图:0张</span>\n</div>\n<div class=\"atl-reply\" id=\"alt_reply\">\n<a author=\"牧阳光\" authorid=\"36535656\" class=\"reportme a-link\" href=\"javascript:void(0);\" replyid=\"0\" replytime=\"2012-10-28 19:11:00\"> 举报</a> | \r\n\t\t\t\t\t\t\t<a class=\"a-link acl-share\" href=\"javascript:void(0);\">分享</a> | \r\n\t\t\t \t<a class=\"a-link acl-more\" href=\"javascript:void(0);\">更多</a> |\r\n\t\t\t\t\t\t\t<span><a class=\"a-link\" name=\"0\">楼主</a></span>\n<a _name=\"牧阳光\" _time=\"2012-10-28 19:11:00\" class=\"a-link2 replytop\" href=\"#fabu_anchor\">回复</a>\n</div>\n</div>\n<div id=\"ds-quick-box\" style=\"display:none;\"></div>\n</div>\n<div class=\"atl-con-ft clearfix\">\n<div class=\"atl-con-ft-l\"></div>\n<div class=\"atl-con-ft-r\"></div>\n<div id=\"niuren_ifm\"></div>\n</div>\n</div>\n</div>\n"
],
[
"print pa[1]",
"<div _host=\"%E6%80%A8%E9%AD%82%E9%AC%BC\" class=\"atl-item\" id=\"1\" js_restime=\"2012-10-28 19:17:56\" js_username=\"%E6%80%A8%E9%AD%82%E9%AC%BC\" replyid=\"92725226\">\n<div class=\"atl-head\" id=\"ea93038aa568ef2bf7a8cf6b6853b744\">\n<div class=\"atl-head-reply\"></div>\n<div class=\"atl-info\">\n<span>作者:<a class=\"js-vip-check\" href=\"http://www.tianya.cn/73157063\" target=\"_blank\" uid=\"73157063\" uname=\"怨魂鬼\">怨魂鬼</a> </span>\n<span>时间:2012-10-28 19:17:56</span>\n</div>\n</div>\n<div class=\"atl-content\">\n<div class=\"atl-con-hd clearfix\">\n<div class=\"atl-con-hd-l\"></div>\n<div class=\"atl-con-hd-r\"></div>\n</div>\n<div class=\"atl-con-bd clearfix\">\n<div class=\"bbs-content\">\r\n\t\t\t\t\t\t\t 图片分享<img original=\"http://img3.laibafile.cn/p/m/122161321.jpg\" src=\"http://static.tianyaui.com/img/static/2011/imgloading.gif\"/><br/><br/> \r\n\t\t\t\t\t\t\t\r\n\t\t\t\t\t\t</div>\n<div class=\"atl-reply\">\r\n\t\t\t\t\t\t\t来自 <a _stat=\"/stat/bbs/post/来自\" class=\"a-link\" href=\"http://www.tianya.cn/mobile/\" rel=\"nofollow\" target=\"_blank\">天涯社区客户端</a> |\r\n\t\t\t\t\t\t\t<a author=\"怨魂鬼\" authorid=\"73157063\" class=\"reportme a-link\" href=\"javascript:void(0);\" replyid=\"92725226\" replytime=\"2012-10-28 19:17:56\">举报</a> |\r\n\t\t\t\t\t\t\t\t\t\t\t\t\t\t\r\n\t\t\t\t\t\t\t<span>1楼</span> |\r\n\t\t\t\t\t\t\t<a class=\"a-link-2 ir-shang\" floor=\"1\" href=\"javascript:void(0);\" title=\"打赏层主\">\r\n\t\t\t\t\t\t\t\t打赏\r\n\t\t\t\t\t\t\t</a> |\r\n\t\t\t\t\t\t\t<a class=\"a-link-2 reply\" href=\"#fabu_anchor\" title=\"引用回复\">回复</a> |\r\n\t\t\t\t\t\t\t<a _stat=\"/stat/bbs/post/评论\" class=\"a-link-2 ir-remark\" floor=\"1\" href=\"javascript:void(0);\" title=\"插入评论\">\r\n\t\t\t\t\t\t\t\t评论\r\n\t\t\t\t\t\t\t</a>\n</div>\n</div>\n<div class=\"atl-con-ft clearfix\">\n<div class=\"atl-con-ft-l\"></div>\n<div class=\"atl-con-ft-r\"></div>\n</div>\n</div>\n</div>\n"
],
[
"print pa[0].find('div', {'class', 'bbs-content'}).text.strip()",
"从宁波市政府新闻发言人处获悉,宁波市经与项目投资方研究决定:(1)坚决不上PX项目;(2)炼化一体化项目前期工作停止推进,再作科学论证。\n"
],
[
"print pa[87].find('div', {'class', 'bbs-content'}).text.strip()",
"@lice5 2012-10-28 20:37:17 作为宁波人 还是说一句:革命尚未成功 同志仍需努力 ----------------------------- 对 现在说成功还太乐观,就怕说一套做一套\n"
],
[
"pa[1].a",
"_____no_output_____"
],
[
"print pa[0].find('a', class_ = 'reportme a-link')",
"<a author=\"牧阳光\" authorid=\"36535656\" class=\"reportme a-link\" href=\"javascript:void(0);\" replyid=\"0\" replytime=\"2012-10-28 19:11:00\"> 举报</a>\n"
],
[
"print pa[0].find('a', class_ = 'reportme a-link')['replytime']",
"2012-10-28 19:11:00\n"
],
[
"print pa[0].find('a', class_ = 'reportme a-link')['author']",
"牧阳光\n"
],
[
"for i in pa[:10]:\n p_info = i.find('a', class_ = 'reportme a-link')\n p_time = p_info['replytime']\n p_author_id = p_info['authorid']\n p_author_name = p_info['author']\n p_content = i.find('div', {'class', 'bbs-content'}).text.strip()\n p_content = p_content.replace('\\t', '')\n print p_time, '--->', p_author_id, '--->', p_author_name,'--->', p_content, '\\n'",
"2012-10-28 19:11:00 ---> 36535656 ---> 牧阳光 ---> 从宁波市政府新闻发言人处获悉,宁波市经与项目投资方研究决定:(1)坚决不上PX项目;(2)炼化一体化项目前期工作停止推进,再作科学论证。 \n\n2012-10-28 19:17:56 ---> 73157063 ---> 怨魂鬼 ---> 图片分享 \n\n2012-10-28 19:18:17 ---> 73157063 ---> 怨魂鬼 ---> @怨魂鬼 2012-10-28 19:17:56 图片分享 [发自掌中天涯客户端 ] ----------------------------- 2楼我的天下! \n\n2012-10-28 19:18:46 ---> 36535656 ---> 牧阳光 ---> 。。。沙发板凳这么快就被坐了~~ \n\n2012-10-28 19:19:04 ---> 41774471 ---> zgh0213 ---> 元芳你怎么看 \n\n2012-10-28 19:19:37 ---> 73157063 ---> 怨魂鬼 ---> @牧阳光 2012-10-28 19:18:46 。。。沙发板凳这么快就被坐了~~ ----------------------------- 运气、 \n\n2012-10-28 19:20:04 ---> 36535656 ---> 牧阳光 ---> @怨魂鬼 5楼 运气、 ----------------------------- 哈哈。。。 \n\n2012-10-28 19:20:07 ---> 54060837 ---> 帆小叶 ---> 八卦的被和谐了。帖个链接http://api.pwnz.org/0/?url=bG10aC4 wOTIyNzQvNzIvMDEvMjEvc3dlbi9tb2MuYW5 paGN0ZXJjZXMud3d3Ly9BMyVwdHRo \n\n2012-10-28 19:20:33 ---> 36535656 ---> 牧阳光 ---> @怨魂鬼 2楼 2楼我的天下! ----------------------------- 。。。还是掌中天涯,NB的~~ \n\n2012-10-28 19:25:22 ---> 36535656 ---> 牧阳光 ---> 消息来源,官方微博@宁波发布 \n\n"
],
[
"post_soup.find('div', {'class', 'atl-pages'})#['onsubmit']",
"_____no_output_____"
],
[
"post_pages = post_soup.find('div', {'class', 'atl-pages'})\npost_pages = post_pages.form['onsubmit'].split(',')[-1].split(')')[0]\npost_pages",
"_____no_output_____"
],
[
"url = 'http://bbs.tianya.cn' + df.link[2]\nurl_base = ''.join(url.split('-')[:-1]) + '-%d.shtml'\nurl_base",
"_____no_output_____"
],
[
"def parsePage(pa):\n records = []\n for i in pa:\n p_info = i.find('a', class_ = 'reportme a-link')\n p_time = p_info['replytime']\n p_author_id = p_info['authorid']\n p_author_name = p_info['author']\n p_content = i.find('div', {'class', 'bbs-content'}).text.strip()\n p_content = p_content.replace('\\t', '').replace('\\n', '')#.replace(' ', '')\n record = p_time + '\\t' + p_author_id+ '\\t' + p_author_name + '\\t'+ p_content\n records.append(record)\n return records\n\nimport sys\ndef flushPrint(s):\n sys.stdout.write('\\r')\n sys.stdout.write('%s' % s)\n sys.stdout.flush()",
"_____no_output_____"
],
[
"url_1 = 'http://bbs.tianya.cn' + df.link[10]\ncontent = urllib2.urlopen(url_1).read() #获取网页的html文本\npost_soup = BeautifulSoup(content, \"lxml\") \npa = post_soup.find_all('div', {'class', 'atl-item'})\nb = post_soup.find('div', class_= 'atl-pages')\nb",
"_____no_output_____"
],
[
"url_1 = 'http://bbs.tianya.cn' + df.link[0]\ncontent = urllib2.urlopen(url_1).read() #获取网页的html文本\npost_soup = BeautifulSoup(content, \"lxml\") \npa = post_soup.find_all('div', {'class', 'atl-item'})\na = post_soup.find('div', {'class', 'atl-pages'})\na",
"_____no_output_____"
],
[
"a.form",
"_____no_output_____"
],
[
"if b.form:\n print 'true'\nelse:\n print 'false'",
"false\n"
],
[
"import random\nimport time\n\ndef crawler(url, file_name):\n try:\n # open the browser\n url_1 = 'http://bbs.tianya.cn' + url\n content = urllib2.urlopen(url_1).read() #获取网页的html文本\n post_soup = BeautifulSoup(content, \"lxml\") \n # how many pages in a post\n post_form = post_soup.find('div', {'class', 'atl-pages'})\n if post_form.form:\n post_pages = post_form.form['onsubmit'].split(',')[-1].split(')')[0]\n post_pages = int(post_pages)\n url_base = '-'.join(url_1.split('-')[:-1]) + '-%d.shtml'\n else:\n post_pages = 1\n # for the first page\n pa = post_soup.find_all('div', {'class', 'atl-item'})\n records = parsePage(pa)\n with open(file_name,'a') as p: # '''Note''':Append mode, run only once!\n for record in records: \n p.write('1'+ '\\t' + url + '\\t' + record.encode('utf-8')+\"\\n\") \n # for the 2nd+ pages\n if post_pages > 1:\n for page_num in range(2, post_pages+1):\n time.sleep(random.random())\n flushPrint(page_num)\n url2 =url_base % page_num\n content = urllib2.urlopen(url2).read() #获取网页的html文本\n post_soup = BeautifulSoup(content, \"lxml\") \n pa = post_soup.find_all('div', {'class', 'atl-item'})\n records = parsePage(pa)\n with open(file_name,'a') as p: # '''Note''':Append mode, run only once!\n for record in records: \n p.write(str(page_num) + '\\t' +url + '\\t' + record.encode('utf-8')+\"\\n\") \n else:\n pass\n except Exception, e:\n print e\n pass",
"_____no_output_____"
],
[
"url = 'http://bbs.tianya.cn' + df.link[2]\nfile_name = 'D:/GitHub/computational-communication-2016/shenliting/homework4/tianya_bbs_threads_test.txt'\ncrawler(url, file_name)",
"HTTP Error 404: Not Found\n"
],
[
"for k, link in enumerate(df.link):\n flushPrint(link)\n if k % 10== 0:\n print 'This it the post of : ' + str(k)\n file_name = 'D:/GitHub/computational-communication-2016/shenliting/homework4/tianya_bbs_threads_network.txt'\n crawler(link, file_name)",
"/post-free-2849477-1.shtmlThis it the post of : 0\n/post-free-2842180-1.shtmlThis it the post of : 10\n/post-free-3316698-1.shtmlThis it the post of : 20\n/post-free-923387-1.shtmlThis it the post of : 30\n/post-free-4236026-1.shtmlThis it the post of : 40\n/post-free-2850721-1.shtmlThis it the post of : 50\n/post-free-5054821-1.shtmlThis it the post of : 60\n/post-free-3326274-1.shtmlThis it the post of : 70\n/post-free-4236793-1.shtmlThis it the post of : 80\n/post-free-4239792-1.shtmlThis it the post of : 90\n/post-free-5042110-1.shtmlThis it the post of : 100\n/post-free-2241144-1.shtmlThis it the post of : 110\n/post-free-3324561-1.shtmlThis it the post of : 120\n/post-free-921701-1.shtmlThis it the post of : 130\n/post-free-5045950-1.shtmlThis it the post of : 140\n/post-free-2848818-1.shtmlThis it the post of : 150\n/post-free-3352554-1.shtmlThis it the post of : 160\n/post-free-949151-1.shtmlThis it the post of : 170\n/post-free-2848839-1.shtmlThis it the post of : 180\n/post-free-3228423-1.shtmlThis it the post of : 190\n/post-free-2852970-1.shtmlThis it the post of : 200\n/post-free-3325388-1.shtmlThis it the post of : 210\n/post-free-3835748-1.shtmlThis it the post of : 220\n/post-free-3833431-1.shtmlThis it the post of : 230\n/post-free-3378998-1.shtmlThis it the post of : 240\n/post-free-3359022-1.shtmlThis it the post of : 250\n/post-free-3838116-1.shtmlThis it the post of : 260\n/post-free-3396378-1.shtmlThis it the post of : 270\n/post-free-3835212-1.shtmlThis it the post of : 280\n/post-free-4248593-1.shtmlThis it the post of : 290\n/post-free-3833373-1.shtmlThis it the post of : 300\n/post-free-3847600-1.shtmlThis it the post of : 310\n/post-free-3832970-1.shtmlThis it the post of : 320\n/post-free-4076130-1.shtmlThis it the post of : 330\n/post-free-3835673-1.shtmlThis it the post of : 340\n/post-free-3835434-1.shtmlThis it the post of : 350\n/post-free-3368554-1.shtmlThis it the post of : 360\n/post-free-3832938-1.shtmlThis it the post of : 370\n/post-free-3835075-1.shtmlThis it the post of : 380\n/post-free-3832963-1.shtmlThis it the post of : 390\n/post-free-4250604-1.shtmlThis it the post of : 400\n/post-free-3834828-1.shtmlThis it the post of : 410\n/post-free-3835007-1.shtmlThis it the post of : 420\n/post-free-3838253-1.shtmlThis it the post of : 430\n/post-free-3835167-1.shtmlThis it the post of : 440\n/post-free-3835898-1.shtmlThis it the post of : 450\n/post-free-3835123-1.shtmlThis it the post of : 460\n/post-free-3835031-1.shtml"
],
[
"dtt = []\nwith open('D:/GitHub/computational-communication-2016/shenliting/homework4/tianya_bbs_threads_network.txt', 'r') as f:\n for line in f:\n pnum, link, time, author_id, author, content = line.replace('\\n', '').split('\\t')\n dtt.append([pnum, link, time, author_id, author, content])\nlen(dtt)",
"_____no_output_____"
],
[
"dt = pd.DataFrame(dtt)\ndt[:5]",
"_____no_output_____"
],
[
"dt=dt.rename(columns = {0:'page_num', 1:'link', 2:'time', 3:'author',4:'author_name', 5:'reply'})\ndt[:5]",
"_____no_output_____"
],
[
"dt.reply[:100]",
"_____no_output_____"
],
[
"18459/50",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8ca24c6b8bafbfc11672215fb3b8734c1fad54
| 356,785 |
ipynb
|
Jupyter Notebook
|
notebook.ipynb
|
tem-ctrl/time_series_analysis_of_naics
|
7c3a053ac6be3a2dbad43fa9bc1b05935c18b468
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
tem-ctrl/time_series_analysis_of_naics
|
7c3a053ac6be3a2dbad43fa9bc1b05935c18b468
|
[
"MIT"
] | null | null | null |
notebook.ipynb
|
tem-ctrl/time_series_analysis_of_naics
|
7c3a053ac6be3a2dbad43fa9bc1b05935c18b468
|
[
"MIT"
] | null | null | null | 136.961612 | 99,738 | 0.84478 |
[
[
[
"# Time Series Analysis of NAICS: North American employment data from 1997 to 2019\n",
"_____no_output_____"
],
[
"Import necessary libraries",
"_____no_output_____"
]
],
[
[
"import os\nimport pandas as pd\nimport numpy as np\nimport datetime as dt\nfrom glob import glob\nimport re\nimport warnings\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom openpyxl import load_workbook\nfrom tqdm import tqdm\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"## I. Data loading and cleaning",
"_____no_output_____"
]
],
[
[
"def create_n_digits_df(data_path, n):\n \"\"\"\n Make pandas dataframe from all n digits code industries, ordered \n from the oldest (Jan 1997) to the newest (Dec 2019)\n\n Parameter:\n ----------\n data_path : str or Pathlike object\n Path to the CSV data files\n\n n :int\n Number of digits in NAICS code\n\n Returns:\n -------\n pandas.core.frame.DataFrame:\n DataFrame with n digits industries sorted in ascending dates\n \"\"\"\n try:\n isinstance (n, int)\n assert n in [2,3,4]\n except:\n print(f'Wrong value of the parameter n!!!\\nExpected an integer 2, 3 or 4 but got {n}.')\n return\n \n list_n = [x for x in os.listdir(data_path) if re.search(f'_{n}NAICS', x)]\n df = pd.read_csv(data_path + list_n[-1])\n for i in range(len(list_n)-1):\n df2 = pd.read_csv(data_path+list_n[i])\n df = df.append(df2)\n return df",
"_____no_output_____"
],
[
"data_path = 'data/'\ndf2 = create_n_digits_df(data_path, 2)\ndf3 = create_n_digits_df(data_path, 3)\ndf4 = create_n_digits_df(data_path, 4)",
"_____no_output_____"
],
[
"df2.head()",
"_____no_output_____"
],
[
"df3.head()",
"_____no_output_____"
],
[
"df4.head()",
"_____no_output_____"
],
[
"def clean_df(df):\n \"\"\"\n Extract NAICS code from NAICS column and add it as new column,\n Remove code from NAICS column\n Create 'DATE' column from SYEAR and SMONTH\n Drop 'SYEAR' and 'SMTH' columns\n \n Parameters:\n ----------\n df : pandas.core.frame.DataFrame\n Dataframe to trandform\n \n Returns:\n --------\n pandas.core.frame.DataFrame\n Dataframe with 'NAICS_CODE' and 'DATE' columns, and NAICS column without code.\n \n \"\"\"\n def extract_code(x):\n if type(x) == int:\n return [x]\n if '[' not in x:\n y = None\n elif '-' in x: \n code_len = len(x.split('[')[1].replace(']', '').split('-')[0])\n x = x.split('[')[1].replace(']', '').split('-')\n if code_len == 2:\n y = [*range(int(x[0]), int(x[1])+1)]\n else:\n y = [int(i) for i in x]\n else:\n x = x.split('[')[1].replace(']', '')\n y = [int(x)]\n return y\n\n df['NAICS_CODE'] = df['NAICS'].apply(extract_code)\n df['NAICS'] = df['NAICS'].astype('str').str.split('[').str.get(0).str.strip()\n df['DATE'] = pd.to_datetime(df['SYEAR'].astype('str') + df['SMTH'].astype('str'), format='%Y%m').dt.strftime('%Y-%m')\n df.drop(columns=['SYEAR', 'SMTH'], inplace=True)\n\n return df",
"_____no_output_____"
],
[
"df2 = clean_df(df2)\ndf3 = clean_df(df3)\ndf4 = clean_df(df4)",
"_____no_output_____"
],
[
"df2.head(20)",
"_____no_output_____"
],
[
"def find_2(df):\n fi = False\n for code in df.NAICS_CODE:\n if code == None:\n pass\n else:\n if len(code) >= 2:\n fi = True\n break \n return fi\nprint(f'Lines with two or more codes in df2?\\t{find_2(df2)}\\n\\\nLines with two or more codes in df3?\\t{find_2(df3)}\\n\\\nLines with two or more codes in df4?\\t{find_2(df4)}')",
"Lines with two or more codes in df2?\tTrue\nLines with two or more codes in df3?\tFalse\nLines with two or more codes in df4?\tFalse\n"
],
[
"# df3.head()",
"_____no_output_____"
],
[
"# df4.head(20)",
"_____no_output_____"
]
],
[
[
"Since only 2-digit codes dataset contains lines with more than one code, it becomes concevable to drop those lines but further information from *LMO detailed industries by NAICS* is needed. <br>\nWe now load the detailed industries by NAICS data, which serve as bridge between the n-digit dataframes and the output file.",
"_____no_output_____"
]
],
[
[
"lmo = 'LMO_Detailed_Industries_by_NAICS.xlsx'\ndf_lmo = pd.read_excel(data_path+lmo, usecols=[0,1])",
"_____no_output_____"
],
[
"df_lmo.head()",
"_____no_output_____"
],
[
"def format_lmo_naics(x):\n if type(x) == int:\n y = [x]\n else:\n x = x.replace('&', ',').split(',')\n y = [int(i.strip()) for i in x]\n return y",
"_____no_output_____"
],
[
"df_lmo.NAICS = df_lmo.NAICS.apply(format_lmo_naics)",
"_____no_output_____"
],
[
"df_lmo.head()",
"_____no_output_____"
],
[
"df_lmo.isna().any()",
"_____no_output_____"
],
[
"df_lmo['code_len'] = df_lmo.NAICS.apply(lambda x : len(str(x[0])))",
"_____no_output_____"
],
[
"df_lmo.head()",
"_____no_output_____"
],
[
"def check_lmo(df):\n fi = False\n i = 0\n for naics in df.NAICS:\n if len(naics) >= 2 and len(str(naics[0])) == 2:\n fi = True\n i += 1\n # break\n return i\n\ncheck_lmo(df_lmo)",
"_____no_output_____"
]
],
[
[
"There is a single line with 2 or more 2-digit naics codes. If there is not a line in *df2* with the same naics codes we can drop all lines from *df2* with two or more naics codes, thus making it possible and safe to use integer naics codes in n-digit datasets.",
"_____no_output_____"
]
],
[
[
"for i in range(len(df_lmo)):\n if len(df_lmo.NAICS.iloc[i]) >= 2 and df_lmo.code_len.iloc[i] == 2:\n code_check_df2 = df_lmo.iloc[i].NAICS\n print(f'Code to check in df2 : {code_check_df2}')\n break\nsafety = 'Safe to drop lines with multiple codes!!!'\nfor naic in df2.NAICS_CODE:\n if naic == code_check_df2:\n safety = 'Unsafe to drop lines with multiple codes!!!'\n break\nprint(safety)",
"Code to check in df2 : [55, 56]\nSafe to drop lines with multiple codes!!!\n"
]
],
[
[
"We can safely drop lines with more than one naics code from 2-digit dataset and cconvert *NAICS_CODE* column to *int*",
"_____no_output_____"
]
],
[
[
"df2['to_drop'] = df2.NAICS_CODE.apply(lambda x : len(x)>=2)\ndf2 = df2[df2.to_drop == False]\ndf2.drop('to_drop', axis=1, inplace=True)",
"_____no_output_____"
],
[
"df2.head()",
"_____no_output_____"
],
[
"for df in [df2, df3, df4]:\n df.dropna(inplace = True)\n df.NAICS_CODE = df.NAICS_CODE.apply(lambda x: int(x[0]))\n # df = df.reindex(columns= ['DATE', 'NAICS', 'NAICS_CODE', '_EMPLOYMENT_'])",
"_____no_output_____"
],
[
"df4.head()",
"_____no_output_____"
],
[
"# For Github\n# dic = {'x':['Other [326, 327, 334, 335, 337 & 339]',\n# 'Food[311 & 312]']}\n# dfx = pd.DataFrame(dic)",
"_____no_output_____"
]
],
[
[
"## II. Filling out the output file",
"_____no_output_____"
]
],
[
[
"out_file = 'Data_Output_Template.xlsx' \ndf_out = pd.read_excel(data_path+out_file, usecols = [0,1,2,3])",
"_____no_output_____"
],
[
"df_out.head(2)",
"_____no_output_____"
],
[
"df_out['DATE'] = pd.to_datetime(df_out['SYEAR'].astype('str') + df_out['SMTH'].astype('str'), format='%Y%m').dt.strftime('%Y-%m')\ndf_out.drop(columns=['SYEAR', 'SMTH'], inplace=True)",
"_____no_output_____"
],
[
"df_out.head()",
"_____no_output_____"
],
[
"def employment_rate(i):\n global df_lmo, df2, df3, df4, df_out\n\n employment_out = 0\n naics_name = df_out['LMO_Detailed_Industry'].iloc[i]\n sdate = df_out.DATE.iloc[i]\n naics_codes = df_lmo[df_lmo['LMO_Detailed_Industry']==naics_name].NAICS.item()\n # Choose which n-digit dataset to look in\n code_length = df_lmo[df_lmo['LMO_Detailed_Industry']==naics_name].code_len.item()\n if code_length == 2:\n df = df2\n elif code_length == 3:\n df = df3\n else:\n df = df4\n\n dfg = df.groupby(['NAICS_CODE', 'DATE'], sort=False).agg({'_EMPLOYMENT_': sum})\n for code in naics_codes:\n try:\n employment = dfg.loc[(code, sdate)].item()\n except:\n employment = 0\n employment_out += employment\n \n return int(employment_out)",
"_____no_output_____"
],
[
"for i in tqdm(range(len(df_out))):\n df_out.Employment.iloc[i] = employment_rate(i)",
"100%|██████████| 15576/15576 [15:11<00:00, 17.09it/s]\n"
],
[
"df_out.Employment = df_out.Employment.apply(lambda x : int(x))\ndf_out.head(20)",
"_____no_output_____"
]
],
[
[
"We can now copy the values of employment per naics per date in the excel file.",
"_____no_output_____"
]
],
[
[
"wb = load_workbook(data_path+'Data_Output_Template.xlsx')\nws = wb.active\n\nfor i in tqdm(range(len(df_out))):\n cell = f'D{i+2}'\n ws[cell] = df_out.Employment.iloc[i]\n\nwb.save(data_path+'Data_Output.xlsx')\nwb.close()",
"100%|██████████| 15576/15576 [00:01<00:00, 8202.18it/s]\n"
]
],
[
[
"## III. Times Series Analysis: Answer to the questions\n### III.1. How employment in Construction evolved over time and how this compares to the total employment across all industries?\n#### a. Evolution of employment in construction",
"_____no_output_____"
]
],
[
[
"construction = df_out[df_out.LMO_Detailed_Industry == 'Construction']",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,5))\ng = sns.lineplot(x='DATE', y='Employment', data = construction)\ng.set_xticks([*range(0,264,12)])\ng.set_xticklabels([dat for dat in construction.DATE if '-01' in dat], rotation = 90)\ng.set_title('Employment in construction from Jan 1997 to Dec 2018')\nplt.scatter(x=['2004-02', '2008-08', '2016-01'], y=[120000, 232750, 197250], c='r', s=100)\nplt.axvline(x='2004-02', ymax=0.18, linestyle='--', color='r')\nplt.axvline(x='2008-08', ymax=0.88, linestyle='--', color='r')\nplt.axvline(x='2016-01', ymax=0.68, linestyle='--', color='r')\nplt.annotate('2004-02', xy=('2004-02', 120000), xytext=('2006-01', 140000), arrowprops={'arrowstyle':'->'})\nplt.annotate('2016-01', xy=('2016-01', 197250), xytext=('2013-01', 160000), arrowprops={'arrowstyle':'->'})\nplt.annotate('2008-08', xy=('2008-08', 232750), xytext=('2008-01', 240000))\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"There are four different sections in the evolution of employment rate in construction from $1997$ to $2018$. Two sections of global steadiness (**Jan 1997** $-$ **Feb 2004** and **Aug 2008** $-$ **Jan 2016**) during which the employment rate oscillates around a certain constant, and two section of steep increase (**Feb 2004** $-$ **Aug 2008** and **Jan 2016** $-$ **Dec 2018**).\n\n#### b. Comparison of emplyment in construction with overall employment",
"_____no_output_____"
]
],
[
[
"df_total_per_date = df_out.groupby('DATE').agg({'Employment':np.sum})\ndf_total_per_date['Construction_emp(%)'] = (construction.Employment).values * 100 /df_total_per_date.Employment.values\ndf_total_per_date['Construction_emp(%)'] = df_total_per_date['Construction_emp(%)'].apply(lambda x : round(x,2))\ndf_total_per_date.head()",
"_____no_output_____"
],
[
"df_total_per_date[df_total_per_date['Construction_emp(%)'] == df_total_per_date['Construction_emp(%)'].max()]",
"_____no_output_____"
],
[
"df_total_per_date[df_total_per_date['Construction_emp(%)'] == df_total_per_date['Construction_emp(%)'].min()]",
"_____no_output_____"
],
[
"plt.figure(figsize = (10,5))\ng = sns.lineplot(y='Construction_emp(%)', x='DATE', data=df_total_per_date)\ng.set_xticks([*range(0,264,12)])\ng.set_xticklabels([dat for dat in construction.DATE if '-01' in dat], rotation = 90)\ng.set_title('Percentage of Employment in Construction from 1997 to Dec 2018')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"We notice that the portion of employment in construction follows the same fashion like the evolution of employment in condtruction with a maximum value of $10.23\\%$ in **Aug 2008**, just at the end of the first steep increase region of the employment in construction. In contrast, the lowest value was registered in **Jan 2001**, with only $5.18\\%$.\n\n### III.2. When (Year) was the employment rate the highest between the dedicated time frame?",
"_____no_output_____"
]
],
[
[
"df_out['DATE'] = pd.to_datetime(df_out['DATE'])",
"_____no_output_____"
],
[
"emp_year = df_out.groupby(df_out.DATE.dt.year).agg({'Employment':sum})\n# emp_year = df_out.groupby('DATE').agg({'Employment':sum})\nemp_year.reset_index()\nemp_year.head()",
"_____no_output_____"
],
[
"emp_year.query('Employment==Employment.max()')",
"_____no_output_____"
],
[
"plt.figure(figsize=(10,5))\nsns.lineplot(x=emp_year.index, y='Employment', data =emp_year)\nplt.title('Total employment per Year')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"As we would have expected, $2018$ is the year with the largest employment rate with a total of $29922000$ employees.",
"_____no_output_____"
],
[
"### III.3. Which industry sector, subsector or industry group has had the highest number of employees?",
"_____no_output_____"
]
],
[
[
"total_counts = df_out.groupby('LMO_Detailed_Industry')['Employment'].sum().sort_values(ascending=False)\ntotal_df = pd.DataFrame({'Industry':total_counts.index, 'Employments':total_counts.values})\nfig, ax = plt.subplots(figsize=(10,10))\nax = sns.barplot(x='Employments', y='Industry', data = total_df)\nax.tick_params(axis='y', labelsize=8) \nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"Let's find the number of digits in the NAICS code of **Other retail trade (excluding cars and personal care)** and the number of industry subsectors involved.",
"_____no_output_____"
]
],
[
[
"df_lmo[df_lmo['LMO_Detailed_Industry']==total_df.head(1).Industry.item()].code_len.item()",
"_____no_output_____"
],
[
"df_lmo[df_lmo['LMO_Detailed_Industry']==total_df.head(1).Industry.item()].NAICS.item()",
"_____no_output_____"
]
],
[
[
"As shown by the above figure, **Other retail trade (excluding cars and personal care)** is the industry subsector (three digits NAICS) with the largest number of employees. However, this category indludes $11$ different industry subsectors, so **construction** is definitely the industry sector that employs most people.",
"_____no_output_____"
],
[
"### III.4. As a rapidly developping field, if Data Science industry level (Number of digits) in NAICS, is less or equal to 4 then how has Data Science employment evolved over time? Otherwise, What is the lowest industry level above Data Science and how did it evolve from 1997 to 2019? \n\nData Science NAICS code is $518210$ and its lowest industry sector included in our data (four digit NAICS) is $5182$, the name being **Data processing, hosting, and related services**. [[1]](#naics1), [[2]](#naics2)",
"_____no_output_____"
]
],
[
[
"data_science = df4[df4.NAICS_CODE == 5182][['_EMPLOYMENT_', 'DATE']].reset_index()\ndata_science.drop('index', axis=1, inplace=True)\ndata_science.head()",
"_____no_output_____"
],
[
"plt.figure(figsize = (10,5))\ng = sns.lineplot(y='_EMPLOYMENT_', x='DATE', data=data_science)\ng.set_xticks([*range(0,264,12)])\ng.set_xticklabels([dat for dat in construction.DATE if '-01' in dat], rotation = 90)\ng.set_title('Evolution of data related employment from 1997 to Dec 2018')\nplt.grid()\nplt.show()",
"_____no_output_____"
],
[
"data_science.query('_EMPLOYMENT_ ==_EMPLOYMENT_.max()')",
"_____no_output_____"
]
],
[
[
"We observe that the data-related employment remain quite globally constant until $2013$ then has increased until mid $2017$ when it became steady at a relatively low value, before reaching the peak of $6500$ employees in **Aug 2018**. ",
"_____no_output_____"
],
[
"### III.5. Are there industry sectorsn subsectors or industry groups for which the employment rate decreased over time?\nTo answer this question, we will plot the time series evolution of all the $59$ industries included in our data.",
"_____no_output_____"
]
],
[
[
"data_out = pd.DataFrame(df_out.reset_index().groupby(['LMO_Detailed_Industry', 'DATE'], as_index=False)['Employment'].sum())\ndata_out = data_out.pivot('DATE', 'LMO_Detailed_Industry', 'Employment')\ndata_out.index.freq = 'MS'\ndata_out.fillna(0, inplace=True)\ndata_out.plot(subplots=True, figsize=(10, 120))\nplt.show()",
"_____no_output_____"
]
],
[
[
"At first sight, the employment rate in the following industries decreased over time:\n- Wood product manufacturing\n- Telecommunications\n- Support activities for agriculture and forestery\n- Rail trandportation\n- Primary metal manufacturing\n- Paper manufacturing\n- Fishing, hunting and traping\n\nWhat would be the potential factors that caused the employment to decrease in those industries from $1997$ to $2018$? <br>\nAdditionally, these visualizations are very cumbersom, a dashboard for optimieing their look and presentation is under building. ",
"_____no_output_____"
],
[
"## References\n <a id=\"/naics\" ></a> [1] [North American Industry Classification System (NAICS) Canada](www.statcan.gc.ca), 2017 Version 1.0, P. $48$\n\n <a id=\"/naics2\" ></a> [2] [NAICS code description](https://www.naics.com/naics-code-description/?code=518210)\n ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a8ca79566bea456919f6a1d012fa2f9719090a3
| 63,243 |
ipynb
|
Jupyter Notebook
|
notebooks/04b-classification-resp.ipynb
|
mirgee/thesis_project
|
296f292a84fe4756374d87c81e657ac991766a60
|
[
"MIT"
] | null | null | null |
notebooks/04b-classification-resp.ipynb
|
mirgee/thesis_project
|
296f292a84fe4756374d87c81e657ac991766a60
|
[
"MIT"
] | 2 |
2020-03-24T17:03:19.000Z
|
2020-03-31T03:19:19.000Z
|
notebooks/04b-classification-resp.ipynb
|
mirgee/thesis_project
|
296f292a84fe4756374d87c81e657ac991766a60
|
[
"MIT"
] | null | null | null | 49.915549 | 3,044 | 0.576665 |
[
[
[
"%load_ext autoreload\n%autoreload 2\n%load_ext watermark\n%watermark -v -n -m -p numpy,scipy,sklearn,pandas",
"Thu Aug 22 2019 \n\nCPython 3.6.8\nIPython 7.6.0\n\nnumpy 1.16.4\nscipy 1.2.1\nsklearn 0.21.2\npandas 0.24.2\n\ncompiler : GCC 7.3.0\nsystem : Linux\nrelease : 4.15.0-58-generic\nmachine : x86_64\nprocessor : x86_64\nCPU cores : 4\ninterpreter: 64bit\n"
],
[
"%matplotlib inline\nimport sys\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport os\nimport nolds\nimport data\nimport mne\nfrom random import randint\n\nfrom config import *\nfrom data.utils import prepare_dfs, prepare_resp_non, prepare_dep_non, get_metapkl\nfrom data.data_files import files_builder, DataKind\nfrom classification.prediction import predict\nfrom classification.scorers import scorer_factory\n \nmetapkl = get_metapkl()\nmeta_df = pd.read_excel(os.path.join(RAW_ROOT, META_FILE_NAME), index_col='ID', names=META_COLUMN_NAMES)\ndata = np.transpose(files_builder(DataKind('processed')).single_file('1a.fif').df.values)",
"Opening raw data file /home/miroslav/Source/thesis_project/data/processed/1a.fif...\nThis filename (/home/miroslav/Source/thesis_project/data/processed/1a.fif) does not conform to MNE naming conventions. All raw files should end with raw.fif, raw_sss.fif, raw_tsss.fif, raw.fif.gz, raw_sss.fif.gz or raw_tsss.fif.gz\nIsotrak not found\n Range : 0 ... 19104 = 0.000 ... 76.416 secs\nReady.\n"
],
[
"def ff(row, col, t1, t2=0):\n if row[col] <= t1:\n return -1\n elif row[col] <= t2: \n return 0\n else:\n return 1\n \nmetapkl['resp'] = metapkl.apply(lambda row: ff(row, 'change', 1.8), axis=1)\nmetapkl = metapkl.astype({'resp': 'category'}) \nprint(metapkl.loc[(slice(None), 'a'), 'resp'].value_counts())",
"-1 70\n 1 63\nName: resp, dtype: int64\n"
],
[
"from itertools import combinations\nfrom sklearn import svm, datasets, metrics\nfrom sklearn.feature_selection import (SelectFromModel, RFE, RFECV, SelectKBest, mutual_info_classif, chi2)\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import train_test_split, cross_validate, GridSearchCV\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom genetic_selection import GeneticSelectionCV\nfrom sklearn.neural_network import MLPClassifier\nfrom sklearn.naive_bayes import GaussianNB\n\nestimators = {\n svm.SVC(kernel='linear', class_weight='balanced'): {\n 'C': [0.02, 0.5, 1.0, 1.5, 2, 3, 4, 5, 10, 50],\n 'kernel': ('linear', 'poly', 'rbf'),\n 'gamma': ('auto', 'scale'),\n # 'decision_function_shape' : ('ovo', 'ovr'),\n },\n LogisticRegression(class_weight='balanced'): {\n 'C': [0.02, 0.5, 1.0, 2, 3, 4, 5, 10, 20, 50],\n 'penalty': ['l2', 'l1'],\n },\n}\n\ndef get_selectors(estimator):\n return {\n 'RFECV': RFECV(estimator, 5), \n 'SelectFromModel': SelectFromModel(estimator), \n # 'SelectKBest': SelectKBest(chi2, 3),\n 'Genetic': GeneticSelectionCV(estimator,\n cv=5,\n verbose=0,\n scoring=scorer,\n n_population=80,\n crossover_proba=0.8,\n mutation_proba=0.2,\n n_generations=80,\n crossover_independent_proba=0.5,\n mutation_independent_proba=0.05,\n tournament_size=5,\n caching=True,\n n_jobs=1\n ),\n }\n\nscorer = scorer_factory(metrics.roc_auc_score, average='weighted')\nfeatures = ('corr', 'lyap', 'sampen', 'dfa', 'hurst', 'higu')\ncomb_size = 1\ngrid_search_cv = 5\n\nfor estimator, params in estimators.items():\n for cols in combinations(features, comb_size):\n for selector_name, selector in get_selectors(estimator).items():\n print(cols)\n print(selector_name)\n gs = GridSearchCV(estimator, params, iid=False, scoring=scorer, cv=grid_search_cv)\n predict('resp', 'a', cols, estimator, metapkl, gs=gs, show_selected=True, selector=selector)\n print(gs.best_params_)\n print('\\n\\n')",
"/home/miroslav/anaconda3/envs/thesis/lib/python3.6/site-packages/sklearn/externals/joblib/__init__.py:15: DeprecationWarning: sklearn.externals.joblib is deprecated in 0.21 and will be removed in 0.23. Please import this functionality directly from joblib, which can be installed with: pip install joblib. If this warning is raised when loading pickled models, you may need to re-serialize those models with scikit-learn 0.21+.\n warnings.warn(msg, category=DeprecationWarning)\n/home/miroslav/anaconda3/envs/thesis/lib/python3.6/site-packages/sklearn/model_selection/_split.py:1978: FutureWarning: The default value of cv will change from 3 to 5 in version 0.22. Specify it explicitly to silence this warning.\n warnings.warn(CV_WARNING, FutureWarning)\n"
]
],
[
[
"# CROSS-VALIDATED",
"_____no_output_____"
]
],
[
[
"estimators = [\n # LogisticRegression(C=1, penalty='l1', class_weight='balanced'),\n svm.SVC(C=1, class_weight='balanced', kernel='linear'),\n]\n\n# 75\nchannels = [('FP2', 'lyap'), ('F3', 'lyap'), ('O1', 'lyap'), ('T4', 'lyap'), ('T6', 'lyap'), \n ('F3', 'sampen'), ('C3', 'sampen'), ('T6', 'sampen')]\n# channels = [('F3', 'lyap'), ('O2', 'lyap'), ('T5', 'lyap'), ('T6', 'lyap'), ('FP2', 'corr'),\n# ('F4', 'corr'), ('O2', 'corr')]\n\nfor estimator in estimators:\n est = predict('resp', 'a', None, estimator, channels=channels)",
"Class distribution: {-1: 47, 1: 47}\nAccuracy: 0.75 (+/- 0.10)\nPrecision: 0.77 (+/- 0.09)\nRecall: 0.75 (+/- 0.10)\nF1: 0.75 (+/- 0.10)\n0.75 $\\pm$ 0.10 & 0.77 $\\pm$ 0.09 & 0.75 $\\pm$ 0.10 & 0.75 $\\pm$ 0.10 & \\\\ \\hline\n"
],
[
"estimators = [\n # LogisticRegression(class_weight='balanced'),\n svm.SVC(C=1.5, class_weight='balanced', kernel='linear'),\n]\n# 71\n# channels = [('F3', 'lyap'), ('C4', 'lyap'), ('O1', 'lyap'), ('F7', 'lyap'), ('T3', 'lyap'), ('T6', 'lyap'), ]\nchannels = [('F3', 'lyap'), ('F4', 'lyap'), ('T5', 'lyap'), ('T6', 'lyap')]\n\nfor estimator in estimators:\n est = predict('resp', 'a', None, estimator, channels=channels)",
"Class distribution: {-1: 47, 1: 47}\nAccuracy: 0.71 (+/- 0.08)\nPrecision: 0.73 (+/- 0.08)\nRecall: 0.71 (+/- 0.08)\nF1: 0.70 (+/- 0.09)\n0.71 $\\pm$ 0.08 & 0.73 $\\pm$ 0.08 & 0.71 $\\pm$ 0.08 & 0.70 $\\pm$ 0.09 & \\\\ \\hline\n"
],
[
"estimators = [\n LogisticRegression(C=1, penalty='l2', class_weight='balanced'),\n # svm.SVC(C=1, class_weight='balanced', kernel='linear'),\n]\n# 71\n# channels = [('F3', 'sampen'), ('C4', 'sampen'), ('C3', 'sampen'), ('Fz', 'sampen')]\nchannels = [('FP1', 'sampen'), ('F3', 'sampen'), ('P3', 'sampen'), ('Cz', 'sampen')]\n\nfor estimator in estimators:\n est = predict('resp', 'a', None, estimator, channels=channels)",
"Class distribution: {-1: 47, 1: 47}\nAccuracy: 0.66 (+/- 0.09)\nPrecision: 0.66 (+/- 0.09)\nRecall: 0.66 (+/- 0.09)\nF1: 0.65 (+/- 0.10)\n0.66 $\\pm$ 0.09 & 0.66 $\\pm$ 0.09 & 0.66 $\\pm$ 0.09 & 0.65 $\\pm$ 0.10 & \\\\ \\hline\n"
],
[
"estimators = [\n LogisticRegression(C=1, penalty='l2', class_weight='balanced'),\n # svm.SVC(C=1, class_weight='balanced', kernel='linear'),\n]\n\nchannels = [('F3', 'higu'), ('F8', 'higu')]\n\n\nfor estimator in estimators:\n est = predict('resp', 'a', None, estimator, channels=channels)",
"Class distribution: {-1: 47, 1: 47}\nAccuracy: 0.66 (+/- 0.05)\nPrecision: 0.72 (+/- 0.08)\nRecall: 0.66 (+/- 0.05)\nF1: 0.64 (+/- 0.05)\n0.66 $\\pm$ 0.05 & 0.72 $\\pm$ 0.08 & 0.66 $\\pm$ 0.05 & 0.64 $\\pm$ 0.05 & \\\\ \\hline\n"
],
[
"estimators = [\n # LogisticRegression(C=1, penalty='l2', class_weight='balanced'),\n svm.SVC(C=2, class_weight='balanced', kernel='rbf'),\n]\n\nchannels = [('C3', 'hurst'), ('T6', 'hurst')]\n\n\nfor estimator in estimators:\n est = predict('resp', 'a', None, estimator, channels=channels)",
"Class distribution: {-1: 47, 1: 47}\nAccuracy: 0.63 (+/- 0.09)\nPrecision: 0.64 (+/- 0.10)\nRecall: 0.63 (+/- 0.09)\nF1: 0.62 (+/- 0.09)\n0.63 $\\pm$ 0.09 & 0.64 $\\pm$ 0.10 & 0.63 $\\pm$ 0.09 & 0.62 $\\pm$ 0.09 & \\\\ \\hline\n"
],
[
"estimators = [\n LogisticRegression(C=1, penalty='l2', class_weight='balanced'),\n # svm.SVC(C=2, class_weight='balanced', kernel='rbf'),\n]\n\nchannels = [('F3', 'corr'), ('F4', 'corr'), ('O2', 'corr'), ('Pz', 'corr')]\n\n\nfor estimator in estimators:\n est = predict('resp', 'a', None, estimator, channels=channels)",
"Class distribution: {-1: 47, 1: 47}\nAccuracy: 0.67 (+/- 0.09)\nPrecision: 0.70 (+/- 0.11)\nRecall: 0.67 (+/- 0.09)\nF1: 0.65 (+/- 0.10)\n0.67 $\\pm$ 0.09 & 0.70 $\\pm$ 0.11 & 0.67 $\\pm$ 0.09 & 0.65 $\\pm$ 0.10 & \\\\ \\hline\n"
],
[
"estimators = [\n # LogisticRegression(C=1, penalty='l2', class_weight='balanced'),\n svm.SVC(C=10, class_weight='balanced', kernel='linear'),\n]\n\nchannels = [('T3', 'dfa'), ('T4', 'dfa'), ('Cz', 'dfa')]\n\nfor estimator in estimators:\n est = predict('resp', 'a', None, estimator, channels=channels)",
"Class distribution: {-1: 47, 1: 47}\nAccuracy: 0.64 (+/- 0.15)\nPrecision: 0.65 (+/- 0.15)\nRecall: 0.64 (+/- 0.15)\nF1: 0.63 (+/- 0.15)\n0.64 $\\pm$ 0.15 & 0.65 $\\pm$ 0.15 & 0.64 $\\pm$ 0.15 & 0.63 $\\pm$ 0.15 & \\\\ \\hline\n"
],
[
"channels = [('F3', 'lyap'), ('F4', 'lyap'), ('C4', 'lyap'), ('P3', 'lyap'), ('P4', 'lyap'), ('F8', 'lyap'), \n ('T4', 'lyap'), ('T5', 'lyap'), ('T6', 'lyap'), ('Fz', 'lyap'),]\nchannels = [('F3', 'sampen'), ('C4', 'sampen'), ('C3', 'sampen'), ('Fz', 'sampen')]\nchannels = [('F3', 'higu'), ('P4', 'higu'), ('C3', 'higu'), ('Fz', 'higu'), ('F8', 'higu')]\nchannels = [('C4', 'hurst'), ('T5', 'hurst')]\n\nestimators = {\n svm.SVC(kernel='linear', class_weight='balanced'): {\n 'C': [0.02, 0.5, 1.0, 1.5, 2, 3, 4, 5, 10, 50],\n 'kernel': ('linear', 'poly', 'rbf'),\n # 'decision_function_shape' : ('ovo', 'ovr'),\n },\n LogisticRegression(class_weight='balanced'): {\n 'C': [0.02, 0.5, 1.0, 2, 3, 4, 5, 10, 20, 50],\n 'penalty': ['l2', 'l1'],\n },\n}\n\nfor estimator, params in estimators.items():\n gs = grid_search.GridSearchCV(estimator, params, iid=False, scoring=scorer, cv=5)\n selector = GeneticSelectionCV(estimator,\n cv=5,\n verbose=0,\n scoring=scorer,\n n_population=80,\n crossover_proba=0.8,\n mutation_proba=0.2,\n n_generations=80,\n crossover_independent_proba=0.5,\n mutation_independent_proba=0.05,\n tournament_size=5,\n caching=True,\n n_jobs=-1)\n selector = None\n est = predict('resp', 'a', None, estimator, show_selected=True, channels=channels, gs=gs, selector=selector)\n print(gs.best_params_)",
"/home/kovar/miniconda3/envs/thesis/lib/python3.6/site-packages/pandas/core/reshape/merge.py:544: UserWarning: merging between different levels can give an unintended result (2 levels on the left, 1 on the right)\n warnings.warn(msg, UserWarning)\n"
]
],
[
[
"# RESULTS FOR FIRST AND LAST 15% QUANTILE OF CHANGE (34 / 32)",
"_____no_output_____"
],
[
"## Lyapunov exponent",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(C=3.5, penalty='l1', class_weight='balanced')\npredict('resp', None, ('lyap',), estimator,\n channels=('FP2', 'F3', 'F4', 'C3', 'P3', 'P4', 'F7', 'F8', 'T6', 'Fz', 'Cz'))",
"Training distribution: {-1: 24, 1: 22}\nTesting distribution: {-1: 10, 1: 10}\n0.80 & 0.80 & 0.80 & $\\left( \\begin{smallmatrix} 9 & 1 \\\\ 3 & 7 \\end{smallmatrix} \\right)$ & FP2, F3, F4, C3, P3, P4, F7, F8, T6, Fz, Cz \\\\ \\hline\nAccuracy: 0.64 (+/- 0.19)\n"
]
],
[
[
"## DFA",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(C=50, penalty='l2', class_weight='balanced')\npredict('resp', None, ('dfa',), estimator, channels=('FP2', 'F3', 'O1', 'T5', 'T6', 'Cz'))",
"Training distribution: {-1: 24, 1: 22}\nTesting distribution: {-1: 10, 1: 10}\n0.80 & 0.80 & 0.80 & $\\left( \\begin{smallmatrix} 8 & 2 \\\\ 2 & 8 \\end{smallmatrix} \\right)$ & FP2, F3, O1, T5, T6, Cz \\\\ \\hline\nAccuracy: 0.67 (+/- 0.14)\n"
]
],
[
[
"## Hurst exponent",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(C=3, penalty='l2', class_weight='balanced')\npredict('resp', None, ('hurst',), estimator,\n channels=('FP2', 'F3', 'O1', 'T5', 'T6', 'Cz'))",
"Training distribution: {-1: 24, 1: 22}\nTesting distribution: {-1: 10, 1: 10}\n0.70 & 0.70 & 0.70 & $\\left( \\begin{smallmatrix} 7 & 3 \\\\ 3 & 7 \\end{smallmatrix} \\right)$ & FP2, F3, O1, T5, T6, Cz \\\\ \\hline\nAccuracy: 0.60 (+/- 0.18)\n"
]
],
[
[
"## Higuchi dimension\nResults around 55%.",
"_____no_output_____"
],
[
"## Correlation dimension",
"_____no_output_____"
]
],
[
[
"estimator = svm.SVC(C=2, kernel='rbf', class_weight='balanced', decision_function_shape='ovo')\npredict('resp', None, ('corr'), estimator, \n channels=('F4', 'C4', 'O1', 'F7', 'F8', 'T5', 'T6'))",
"Training distribution: {-1: 24, 1: 22}\nTesting distribution: {-1: 10, 1: 10}\n1.00 & 1.00 & 1.00 & $\\left( \\begin{smallmatrix} 10 & 0 \\\\ 0 & 10 \\end{smallmatrix} \\right)$ & F4, C4, O1, F7, F8, T5, T6 \\\\ \\hline\nAccuracy: 0.65 (+/- 0.21)\n"
]
],
[
[
"# RESULTS FOR FIRST AND LAST TERCILE OF CHANGE (74 / 66)",
"_____no_output_____"
],
[
"## Lyapunov exponent",
"_____no_output_____"
]
],
[
[
"estimator = svm.SVC(C=1, kernel='rbf', class_weight='balanced', decision_function_shape='ovo')\npredict('resp', None, ('lyap'), estimator, channels=('F3', 'F4', 'O1', 'O2', 'T6', 'Fz'))",
"Training distribution: {-1: 51, 1: 47}\nTesting distribution: {-1: 23, 1: 19}\n0.76 & 0.76 & 0.77 & $\\left( \\begin{smallmatrix} 16 & 7 \\\\ 3 & 16 \\end{smallmatrix} \\right)$ & F3, F4, O1, O2, T6, Fz \\\\ \\hline\n"
]
],
[
[
"## DFA",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(class_weight='balanced')\nparams = {'C': [0.5, 1.0, 1.5, 2, 2.5, 3, 3.5],\n 'penalty': ['l2', 'l1'],}\nestimator = grid_search.GridSearchCV(estimator, params, iid=False, scoring=scorer)\npredict('resp', None, ('dfa',), estimator, channels=('C3', 'P4', 'F7', 'T6'))\nestimator.best_params_",
"Training distribution: {-1: 51, 1: 47}\nTesting distribution: {-1: 23, 1: 19}\n0.64 & 0.64 & 0.64 & $\\left( \\begin{smallmatrix} 15 & 8 \\\\ 7 & 12 \\end{smallmatrix} \\right)$ & C3, P4, F7, T6 \\\\ \\hline\n"
]
],
[
[
"### Alpha / Theta envelope\nAll estimators fail (50/50). Very similar values between groupds and small variance. There is still chance for depression score, however.\n\nSame for sample entropy.",
"_____no_output_____"
],
[
"## Hurst exponent",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(C=3, penalty='l2', class_weight='balanced')\npredict('resp', None, ('hurst',), estimator, channels=('P3', 'F7', 'T4', 'T6', 'Cz'))",
"Training distribution: {-1: 51, 1: 47}\nTesting distribution: {-1: 23, 1: 19}\n0.71 & 0.71 & 0.73 & $\\left( \\begin{smallmatrix} 14 & 9 \\\\ 3 & 16 \\end{smallmatrix} \\right)$ & P3, F7, T4, T6, Cz \\\\ \\hline\n"
]
],
[
[
"## Higuchi dimension",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(C=3.5, penalty='l1', class_weight='balanced')\npredict('resp', None, ('higu',), estimator, channels=('FP2',))",
"Training distribution: {-1: 51, 1: 47}\nTesting distribution: {-1: 23, 1: 19}\n0.69 & 0.69 & 0.70 & $\\left( \\begin{smallmatrix} 13 & 10 \\\\ 3 & 16 \\end{smallmatrix} \\right)$ & FP2 \\\\ \\hline\n"
]
],
[
[
"## Correlation dimension",
"_____no_output_____"
]
],
[
[
"estimator = LogisticRegression(C=3.5, penalty='l1', class_weight='balanced')\nestimator = predict('resp', None, ('corr',), estimator, channels=('F4', 'C4', 'T5'))",
"Training distribution: {-1: 51, 1: 47}\nTesting distribution: {-1: 23, 1: 19}\n0.69 & 0.69 & 0.70 & $\\left( \\begin{smallmatrix} 14 & 9 \\\\ 4 & 15 \\end{smallmatrix} \\right)$ & F4, C4, T5 \\\\ \\hline\n"
],
[
"estimator = svm.SVC(C=2, kernel='rbf', class_weight='balanced', decision_function_shape='ovo')\npredict('resp', None, ('corr'), estimator, channels=('F4', 'C4', 'O1', 'F7', 'F8', 'T5'))",
"Training distribution: {-1: 51, 1: 47}\nTesting distribution: {-1: 23, 1: 19}\nAccuracy score: 0.67\nConfusion matrix:\n [[13 10]\n [ 4 15]]\nPrecision score: 0.6901960784313724\nRecall score: 0.6666666666666666\nf1 score: 0.6643939393939394\nROC AUC score: 0.6773455377574371\n"
]
],
[
[
"# Manual selection",
"_____no_output_____"
],
[
"## LLE",
"_____no_output_____"
]
],
[
[
"from random import randint\nestimators = {\n svm.SVC(kernel='linear', class_weight='balanced'): {\n 'C': [0.02, 0.5, 1.0, 1.5, 2, 3, 4, 5, 10, 50],\n 'kernel': ('linear', 'poly', 'rbf'),\n },\n LogisticRegression(class_weight='balanced'): {\n 'C': [0.02, 0.5, 1.0, 2, 3, 4, 5, 10, 20, 50],\n 'penalty': ['l2', 'l1'],\n },\n}\nseed = randint(0, 100000)\nprint('Seed: %s' % seed)\nchannels = ['Fz']\nfor estimator, params in estimators.items():\n est = grid_search.GridSearchCV(estimator, params, iid=False, scoring=scorer, cv=4)\n est = predict('resp', None, ('lyap',), est, show_selected=True, channels=channels, seed=seed)",
"Seed: 5870\nTraining distribution: {-1: 29, 1: 27}\nTesting distribution: {-1: 11, 1: 14}\nAccuracy score: 0.68\nConfusion matrix:\n [[9 2]\n [6 8]]\nPrecision score: 0.7120000000000001\nRecall score: 0.68\nf1 score: 0.6779487179487179\nROC AUC score: 0.6948051948051948\nTraining distribution: {-1: 29, 1: 27}\nTesting distribution: {-1: 11, 1: 14}\nAccuracy score: 0.72\nConfusion matrix:\n [[9 2]\n [5 9]]\nPrecision score: 0.741038961038961\nRecall score: 0.72\nf1 score: 0.7200000000000002\nROC AUC score: 0.7305194805194805\n"
],
[
"seed = randint(0, 100000)\nprint('Seed: %s' % seed)\nchannels = ['P3']\nfor estimator, params in estimators.items():\n est = grid_search.GridSearchCV(estimator, params, iid=False, scoring=scorer, cv=4)\n est = predict('resp', None, ('lyap',), est, show_selected=True, channels=channels, seed=seed)",
"Seed: 91461\nTraining distribution: {-1: 25, 1: 31}\nTesting distribution: {-1: 15, 1: 10}\nAccuracy score: 0.72\nConfusion matrix:\n [[9 6]\n [1 9]]\nPrecision score: 0.78\nRecall score: 0.72\nf1 score: 0.72\nROC AUC score: 0.75\nTraining distribution: {-1: 25, 1: 31}\nTesting distribution: {-1: 15, 1: 10}\nAccuracy score: 0.56\nConfusion matrix:\n [[13 2]\n [ 9 1]]\nPrecision score: 0.4878787878787878\nRecall score: 0.56\nf1 score: 0.48316008316008324\nROC AUC score: 0.4833333333333334\n"
]
],
[
[
"## Higuchi fractal dimension",
"_____no_output_____"
]
],
[
[
"estimators = {\n # svm.SVC(kernel='linear', class_weight='balanced'): {\n # 'C': [0.02, 0.5, 1.0, 1.5, 2, 3, 4, 5, 10, 50],\n # 'kernel': ('linear', 'poly', 'rbf'),\n # },\n LogisticRegression(class_weight='balanced'): {\n 'C': [0.02, 0.5, 1.0, 2, 3, 4, 5, 10, 20, 50],\n 'penalty': ['l2', 'l1'],\n },\n}\n# 71875\nseed = randint(0, 100000)\nprint('Seed: %s' % seed)\nchannels = ['FP2', 'Cz']\nfor estimator, params in estimators.items():\n est = grid_search.GridSearchCV(estimator, params, iid=False, scoring=scorer, cv=4)\n est = predict('resp', None, ('higu',), est, show_selected=True, channels=channels, seed=seed)",
"Seed: 95528\nTraining distribution: {-1: 25, 1: 31}\nTesting distribution: {-1: 15, 1: 10}\nAccuracy score: 0.88\nConfusion matrix:\n [[12 3]\n [ 0 10]]\nPrecision score: 0.9076923076923077\nRecall score: 0.88\nf1 score: 0.8811594202898552\nROC AUC score: 0.9\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8cafc8b7dbd3fcf33fde84c7e66beb0c674112
| 14,446 |
ipynb
|
Jupyter Notebook
|
week04_approx_rl/seminar_tf.ipynb
|
mmamedli/Practical_RL
|
e233e084842bd9a1f40885acdffd61af35dfd3e1
|
[
"Unlicense"
] | 1 |
2020-11-30T10:30:50.000Z
|
2020-11-30T10:30:50.000Z
|
week04_approx_rl/seminar_tf.ipynb
|
mmamedli/Practical_RL
|
e233e084842bd9a1f40885acdffd61af35dfd3e1
|
[
"Unlicense"
] | null | null | null |
week04_approx_rl/seminar_tf.ipynb
|
mmamedli/Practical_RL
|
e233e084842bd9a1f40885acdffd61af35dfd3e1
|
[
"Unlicense"
] | 1 |
2021-07-05T10:37:42.000Z
|
2021-07-05T10:37:42.000Z
| 35.757426 | 283 | 0.595874 |
[
[
[
"# Approximate q-learning\n\nIn this notebook you will teach a __tensorflow__ neural network to do Q-learning.",
"_____no_output_____"
],
[
"__Frameworks__ - we'll accept this homework in any deep learning framework. This particular notebook was designed for tensorflow, but you will find it easy to adapt it to almost any python-based deep learning framework.",
"_____no_output_____"
]
],
[
[
"import sys, os\nif 'google.colab' in sys.modules:\n %tensorflow_version 1.x\n \n if not os.path.exists('.setup_complete'):\n !wget -q https://raw.githubusercontent.com/yandexdataschool/Practical_RL/master/setup_colab.sh -O- | bash\n\n !touch .setup_complete\n\n# This code creates a virtual display to draw game images on.\n# It will have no effect if your machine has a monitor.\nif type(os.environ.get(\"DISPLAY\")) is not str or len(os.environ.get(\"DISPLAY\")) == 0:\n !bash ../xvfb start\n os.environ['DISPLAY'] = ':1'",
"_____no_output_____"
],
[
"import gym\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"env = gym.make(\"CartPole-v0\").env\nenv.reset()\nn_actions = env.action_space.n\nstate_dim = env.observation_space.shape\n\nplt.imshow(env.render(\"rgb_array\"))",
"_____no_output_____"
]
],
[
[
"# Approximate (deep) Q-learning: building the network\n\nTo train a neural network policy one must have a neural network policy. Let's build it.\n\n\nSince we're working with a pre-extracted features (cart positions, angles and velocities), we don't need a complicated network yet. In fact, let's build something like this for starters:\n\n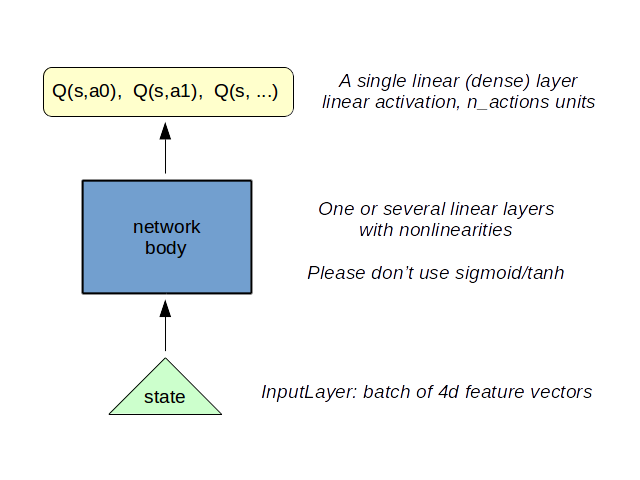\n\nFor your first run, please only use linear layers (`L.Dense`) and activations. Stuff like batch normalization or dropout may ruin everything if used haphazardly. \n\nAlso please avoid using nonlinearities like sigmoid & tanh: since agent's observations are not normalized, sigmoids might be saturated at initialization. Instead, use non-saturating nonlinearities like ReLU.\n\nIdeally you should start small with maybe 1-2 hidden layers with < 200 neurons and then increase network size if agent doesn't beat the target score.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport keras\nimport keras.layers as L\ntf.reset_default_graph()\nsess = tf.InteractiveSession()\nkeras.backend.set_session(sess)",
"_____no_output_____"
],
[
"assert not tf.test.is_gpu_available(), \\\n \"Please complete this assignment without a GPU. If you use a GPU, the code \" \\\n \"will run a lot slower due to a lot of copying to and from GPU memory. \" \\\n \"To disable the GPU in Colab, go to Runtime → Change runtime type → None.\"",
"_____no_output_____"
],
[
"network = keras.models.Sequential()\nnetwork.add(L.InputLayer(state_dim))\n\n<YOUR CODE: stack layers!!!1>",
"_____no_output_____"
],
[
"def get_action(state, epsilon=0):\n \"\"\"\n sample actions with epsilon-greedy policy\n recap: with p = epsilon pick random action, else pick action with highest Q(s,a)\n \"\"\"\n \n q_values = network.predict(state[None])[0]\n \n <YOUR CODE>\n\n return <YOUR CODE: epsilon-greedily selected action>",
"_____no_output_____"
],
[
"assert network.output_shape == (None, n_actions), \"please make sure your model maps state s -> [Q(s,a0), ..., Q(s, a_last)]\"\nassert network.layers[-1].activation == keras.activations.linear, \"please make sure you predict q-values without nonlinearity\"\n\n# test epsilon-greedy exploration\ns = env.reset()\nassert np.shape(get_action(s)) == (), \"please return just one action (integer)\"\nfor eps in [0., 0.1, 0.5, 1.0]:\n state_frequencies = np.bincount([get_action(s, epsilon=eps) for i in range(10000)], minlength=n_actions)\n best_action = state_frequencies.argmax()\n assert abs(state_frequencies[best_action] - 10000 * (1 - eps + eps / n_actions)) < 200\n for other_action in range(n_actions):\n if other_action != best_action:\n assert abs(state_frequencies[other_action] - 10000 * (eps / n_actions)) < 200\n print('e=%.1f tests passed'%eps)",
"_____no_output_____"
]
],
[
[
"### Q-learning via gradient descent\n\nWe shall now train our agent's Q-function by minimizing the TD loss:\n$$ L = { 1 \\over N} \\sum_i (Q_{\\theta}(s,a) - [r(s,a) + \\gamma \\cdot max_{a'} Q_{-}(s', a')]) ^2 $$\n\n\nWhere\n* $s, a, r, s'$ are current state, action, reward and next state respectively\n* $\\gamma$ is a discount factor defined two cells above.\n\nThe tricky part is with $Q_{-}(s',a')$. From an engineering standpoint, it's the same as $Q_{\\theta}$ - the output of your neural network policy. However, when doing gradient descent, __we won't propagate gradients through it__ to make training more stable (see lectures).\n\nTo do so, we shall use `tf.stop_gradient` function which basically says \"consider this thing constant when doingbackprop\".",
"_____no_output_____"
]
],
[
[
"# Create placeholders for the <s, a, r, s'> tuple and a special indicator for game end (is_done = True)\nstates_ph = keras.backend.placeholder(dtype='float32', shape=(None,) + state_dim)\nactions_ph = keras.backend.placeholder(dtype='int32', shape=[None])\nrewards_ph = keras.backend.placeholder(dtype='float32', shape=[None])\nnext_states_ph = keras.backend.placeholder(dtype='float32', shape=(None,) + state_dim)\nis_done_ph = keras.backend.placeholder(dtype='bool', shape=[None])",
"_____no_output_____"
],
[
"#get q-values for all actions in current states\npredicted_qvalues = network(states_ph)\n\n#select q-values for chosen actions\npredicted_qvalues_for_actions = tf.reduce_sum(predicted_qvalues * tf.one_hot(actions_ph, n_actions), axis=1)",
"_____no_output_____"
],
[
"gamma = 0.99\n\n# compute q-values for all actions in next states\npredicted_next_qvalues = <YOUR CODE: apply network to get q-values for next_states_ph>\n\n# compute V*(next_states) using predicted next q-values\nnext_state_values = <YOUR CODE>\n\n# compute \"target q-values\" for loss - it's what's inside square parentheses in the above formula.\ntarget_qvalues_for_actions = <YOUR CODE>\n\n# at the last state we shall use simplified formula: Q(s,a) = r(s,a) since s' doesn't exist\ntarget_qvalues_for_actions = tf.where(is_done_ph, rewards_ph, target_qvalues_for_actions)",
"_____no_output_____"
],
[
"#mean squared error loss to minimize\nloss = (predicted_qvalues_for_actions - tf.stop_gradient(target_qvalues_for_actions)) ** 2\nloss = tf.reduce_mean(loss)\n\n# training function that resembles agent.update(state, action, reward, next_state) from tabular agent\ntrain_step = tf.train.AdamOptimizer(1e-4).minimize(loss)",
"_____no_output_____"
],
[
"assert tf.gradients(loss, [predicted_qvalues_for_actions])[0] is not None, \"make sure you update q-values for chosen actions and not just all actions\"\nassert tf.gradients(loss, [predicted_next_qvalues])[0] is None, \"make sure you don't propagate gradient w.r.t. Q_(s',a')\"\nassert predicted_next_qvalues.shape.ndims == 2, \"make sure you predicted q-values for all actions in next state\"\nassert next_state_values.shape.ndims == 1, \"make sure you computed V(s') as maximum over just the actions axis and not all axes\"\nassert target_qvalues_for_actions.shape.ndims == 1, \"there's something wrong with target q-values, they must be a vector\"",
"_____no_output_____"
]
],
[
[
"### Playing the game",
"_____no_output_____"
]
],
[
[
"sess.run(tf.global_variables_initializer())",
"_____no_output_____"
],
[
"def generate_session(env, t_max=1000, epsilon=0, train=False):\n \"\"\"play env with approximate q-learning agent and train it at the same time\"\"\"\n total_reward = 0\n s = env.reset()\n \n for t in range(t_max):\n a = get_action(s, epsilon=epsilon) \n next_s, r, done, _ = env.step(a)\n \n if train:\n sess.run(train_step,{\n states_ph: [s], actions_ph: [a], rewards_ph: [r], \n next_states_ph: [next_s], is_done_ph: [done]\n })\n\n total_reward += r\n s = next_s\n if done:\n break\n \n return total_reward",
"_____no_output_____"
],
[
"epsilon = 0.5",
"_____no_output_____"
],
[
"for i in range(1000):\n session_rewards = [generate_session(env, epsilon=epsilon, train=True) for _ in range(100)]\n print(\"epoch #{}\\tmean reward = {:.3f}\\tepsilon = {:.3f}\".format(i, np.mean(session_rewards), epsilon))\n \n epsilon *= 0.99\n assert epsilon >= 1e-4, \"Make sure epsilon is always nonzero during training\"\n \n if np.mean(session_rewards) > 300:\n print(\"You Win!\")\n break",
"_____no_output_____"
]
],
[
[
"### How to interpret results\n\n\nWelcome to the f.. world of deep f...n reinforcement learning. Don't expect agent's reward to smoothly go up. Hope for it to go increase eventually. If it deems you worthy.\n\nSeriously though,\n* __ mean reward__ is the average reward per game. For a correct implementation it may stay low for some 10 epochs, then start growing while oscilating insanely and converges by ~50-100 steps depending on the network architecture. \n* If it never reaches target score by the end of for loop, try increasing the number of hidden neurons or look at the epsilon.\n* __ epsilon__ - agent's willingness to explore. If you see that agent's already at < 0.01 epsilon before it's is at least 200, just reset it back to 0.1 - 0.5.",
"_____no_output_____"
],
[
"### Record videos\n\nAs usual, we now use `gym.wrappers.Monitor` to record a video of our agent playing the game. Unlike our previous attempts with state binarization, this time we expect our agent to act ~~(or fail)~~ more smoothly since there's no more binarization error at play.\n\nAs you already did with tabular q-learning, we set epsilon=0 for final evaluation to prevent agent from exploring himself to death.",
"_____no_output_____"
]
],
[
[
"# Record sessions\n\nimport gym.wrappers\n\nwith gym.wrappers.Monitor(gym.make(\"CartPole-v0\"), directory=\"videos\", force=True) as env_monitor:\n sessions = [generate_session(env_monitor, epsilon=0, train=False) for _ in range(100)]",
"_____no_output_____"
],
[
"# Show video. This may not work in some setups. If it doesn't\n# work for you, you can download the videos and view them locally.\n\nfrom pathlib import Path\nfrom IPython.display import HTML\n\nvideo_names = sorted([s for s in Path('videos').iterdir() if s.suffix == '.mp4'])\n\nHTML(\"\"\"\n<video width=\"640\" height=\"480\" controls>\n <source src=\"{}\" type=\"video/mp4\">\n</video>\n\"\"\".format(video_names[-1])) # You can also try other indices",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
4a8cc6bed80a46fd0b9df517157529fa9291410e
| 34,663 |
ipynb
|
Jupyter Notebook
|
site/ja/guide/eager.ipynb
|
PeterJaq/docs-l10n
|
b08620c5c7f2955937c67d3667c82a8ef37ba1ee
|
[
"Apache-2.0"
] | null | null | null |
site/ja/guide/eager.ipynb
|
PeterJaq/docs-l10n
|
b08620c5c7f2955937c67d3667c82a8ef37ba1ee
|
[
"Apache-2.0"
] | null | null | null |
site/ja/guide/eager.ipynb
|
PeterJaq/docs-l10n
|
b08620c5c7f2955937c67d3667c82a8ef37ba1ee
|
[
"Apache-2.0"
] | null | null | null | 27.931507 | 429 | 0.47656 |
[
[
[
"##### Copyright 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Eager Execution の基本",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/guide/eager\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ja/guide/eager.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/ja/guide/eager.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/ja/guide/eager.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: これらのドキュメントは私たちTensorFlowコミュニティが翻訳したものです。コミュニティによる 翻訳は**ベストエフォート**であるため、この翻訳が正確であることや[英語の公式ドキュメント](https://www.tensorflow.org/?hl=en)の 最新の状態を反映したものであることを保証することはできません。 この翻訳の品質を向上させるためのご意見をお持ちの方は、GitHubリポジトリ[tensorflow/docs](https://github.com/tensorflow/docs)にプルリクエストをお送りください。 コミュニティによる翻訳やレビューに参加していただける方は、 [[email protected] メーリングリスト](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-ja)にご連絡ください。",
"_____no_output_____"
],
[
"TensorFlow の Eager Execution は、計算グラフの作成と評価を同時におこなう命令的なプログラミングを行うための環境です:\nオペレーションはあとで実行するための計算グラフでなく、具体的な計算結果の値を返します。\nこの方法を用いることにより、初心者にとって TensorFlow を始めやすくなり、またモデルのデバッグも行いやすくなります。\nさらにコードの記述量も削減されます。\nこのガイドの内容を実行するためには、対話的インタープリタ `python` を起動し、以下のコードサンプルを実行してください。\n\nEager Execution は研究や実験のための柔軟な機械学習環境として、以下を提供します。\n\n* *直感的なインタフェース*— Python のデータ構造を使用して、コードを自然に記述することができます。小規模なモデルとデータに対してすばやく実験を繰り返すことができます。\n* *より簡単なデバッグ*— ops を直接呼び出すことで、実行中のモデルを調査したり、変更をテストすることができます。 Python 標準のデバッグツールを用いて即座にエラーのレポーティングができます。\n* *自然な制御フロー*— TensorFlow のグラフ制御フローの代わりに Python の制御フローを利用するため、動的なモデルの作成をシンプルに行うことができます。\n \nEager Execution は TensorFlow のほとんどのオペレーションとGPUアクセラレーションをサポートします。\n\nNote: いくつかのモデルは Eager Execution を有効化することでオーバヘッドが増える可能性があります。\nパフォーマンス改善を行っていますが、もしも問題を発見したら、バグ報告してベンチマークを共有してください。",
"_____no_output_____"
],
[
"## セットアップと基本的な使い方",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\n\nimport cProfile",
"_____no_output_____"
]
],
[
[
"TensorFlow 2.0 では、 Eager Execution はデフォルトで有効化されます。",
"_____no_output_____"
]
],
[
[
"tf.executing_eagerly()",
"_____no_output_____"
]
],
[
[
"これで TensorFlow のオペレーションを実行してみましょう。結果はすぐに返されます。",
"_____no_output_____"
]
],
[
[
"x = [[2.]]\nm = tf.matmul(x, x)\nprint(\"hello, {}\".format(m))",
"_____no_output_____"
]
],
[
[
"Eager Execution を有効化することで、 TensorFlow の挙動は変わります—TensorFlowは即座に式を評価して結果をPythonに返すようになります。\n`tf.Tensor` オブジェクトは計算グラフのノードへのシンボリックハンドルの代わりに具体的な値を参照します。\nセッションの中で構築して実行する計算グラフが存在しないため、`print()`やデバッガを使って容易に結果を調べることができます。\n勾配計算を遮ることなくテンソル値を評価、出力、およびチェックすることができます。\n\nEager Execution は、[NumPy](http://www.numpy.org/)と一緒に使うことができます。\nNumPy のオペレーションは、`tf.Tensor`を引数として受け取ることができます。\nTensorFlow [math operations](https://www.tensorflow.org/api_guides/python/math_ops) はPython オブジェクトと Numpy array を `tf.Tensor` に変換します。\n`tf.Tensor.numpy` メソッドはオブジェクトの値を NumPy の `ndarray` 形式で返します。",
"_____no_output_____"
]
],
[
[
"a = tf.constant([[1, 2],\n [3, 4]])\nprint(a)",
"_____no_output_____"
],
[
"# ブロードキャストのサポート\nb = tf.add(a, 1)\nprint(b)",
"_____no_output_____"
],
[
"# オペレータのオーバーロードがサポートされている\nprint(a * b)",
"_____no_output_____"
],
[
"# NumPy valueの使用\nimport numpy as np\n\nc = np.multiply(a, b)\nprint(c)",
"_____no_output_____"
],
[
"# Tensor から numpy の値を得る\nprint(a.numpy())\n# => [[1 2]\n# [3 4]]",
"_____no_output_____"
]
],
[
[
"## 動的な制御フロー\n\nEager Execution の主要なメリットは、モデルを実行する際にホスト言語のすべての機能性が利用できることです。\nたとえば、[fizzbuzz](https://en.wikipedia.org/wiki/Fizz_buzz)が簡単に書けます:",
"_____no_output_____"
]
],
[
[
"def fizzbuzz(max_num):\n counter = tf.constant(0)\n max_num = tf.convert_to_tensor(max_num)\n for num in range(1, max_num.numpy()+1):\n num = tf.constant(num)\n if int(num % 3) == 0 and int(num % 5) == 0:\n print('FizzBuzz')\n elif int(num % 3) == 0:\n print('Fizz')\n elif int(num % 5) == 0:\n print('Buzz')\n else:\n print(num.numpy())\n counter += 1",
"_____no_output_____"
],
[
"fizzbuzz(15)",
"_____no_output_____"
]
],
[
[
"この関数はテンソル値に依存する条件式を持ち、実行時にこれらの値を表示します。",
"_____no_output_____"
],
[
"## Eager Execution による学習",
"_____no_output_____"
],
[
"### 勾配の計算\n\n[自動微分](https://en.wikipedia.org/wiki/Automatic_differentiation)はニューラルネットワークの学習で利用される[バックプロパゲーション](https://en.wikipedia.org/wiki/Backpropagation)などの機械学習アルゴリズムの実装を行う上で便利です。\nEager Executionでは、勾配計算をあとで行うためのオペレーションをトレースするために`tf.GradientTape` を利用します。\n\nEager Execution では、学習や勾配計算に, `tf.GradientTape` を利用できます。これは複雑な学習ループを実行するときに特に役立ちます。\n\n各呼び出し中に異なるオペレーションが発生する可能性があるため、すべての forward-pass オペレーションは一つの「テープ」に記録されます。勾配を計算するには、テープを逆方向に再生してから破棄します。特定の `tf.GradientTape`は一つのグラデーションしか計算できません。後続の呼び出しは実行時エラーをスローします。",
"_____no_output_____"
]
],
[
[
"w = tf.Variable([[1.0]])\nwith tf.GradientTape() as tape:\n loss = w * w\n\ngrad = tape.gradient(loss, w)\nprint(grad) # => tf.Tensor([[ 2.]], shape=(1, 1), dtype=float32)",
"_____no_output_____"
]
],
[
[
"### モデル学習\n\n以下の example は MNIST という手書き数字分類を行うマルチレイヤーモデルを作成します。\nEager Execution 環境における学習可能なグラフを構築するためのオプティマイザーとレイヤーAPIを提示します。",
"_____no_output_____"
]
],
[
[
"# mnist データのを取得し、フォーマットする\n(mnist_images, mnist_labels), _ = tf.keras.datasets.mnist.load_data()\n\ndataset = tf.data.Dataset.from_tensor_slices(\n (tf.cast(mnist_images[...,tf.newaxis]/255, tf.float32),\n tf.cast(mnist_labels,tf.int64)))\ndataset = dataset.shuffle(1000).batch(32)",
"_____no_output_____"
],
[
"# モデルを構築する\nmnist_model = tf.keras.Sequential([\n tf.keras.layers.Conv2D(16,[3,3], activation='relu',\n input_shape=(None, None, 1)),\n tf.keras.layers.Conv2D(16,[3,3], activation='relu'),\n tf.keras.layers.GlobalAveragePooling2D(),\n tf.keras.layers.Dense(10)\n])",
"_____no_output_____"
]
],
[
[
"学習を行わずとも、モデルを呼び出して、 Eager Execution により、出力を検査することができます:",
"_____no_output_____"
]
],
[
[
"for images,labels in dataset.take(1):\n print(\"Logits: \", mnist_model(images[0:1]).numpy())",
"_____no_output_____"
]
],
[
[
"keras モデルは組み込みで学習のループを回すメソッド `fit` がありますが、よりカスタマイズが必要な場合もあるでしょう。 Eager Executionを用いて実装された学習ループのサンプルを以下に示します:",
"_____no_output_____"
]
],
[
[
"optimizer = tf.keras.optimizers.Adam()\nloss_object = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)\n\nloss_history = []",
"_____no_output_____"
]
],
[
[
"Note: モデルの状況を確認したいときは、 `tf.debugging` にある assert 機能を利用してください。この機能は Eager Execution と Graph Execution のどちらでも利用できます。",
"_____no_output_____"
]
],
[
[
"def train_step(images, labels):\n with tf.GradientTape() as tape:\n logits = mnist_model(images, training=True)\n \n # assertを入れて出力の型をチェックする。\n tf.debugging.assert_equal(logits.shape, (32, 10))\n \n loss_value = loss_object(labels, logits)\n\n loss_history.append(loss_value.numpy().mean())\n grads = tape.gradient(loss_value, mnist_model.trainable_variables)\n optimizer.apply_gradients(zip(grads, mnist_model.trainable_variables))",
"_____no_output_____"
],
[
"def train():\n for epoch in range(3):\n for (batch, (images, labels)) in enumerate(dataset):\n train_step(images, labels)\n print ('Epoch {} finished'.format(epoch))",
"_____no_output_____"
],
[
"train()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nplt.plot(loss_history)\nplt.xlabel('Batch #')\nplt.ylabel('Loss [entropy]')",
"_____no_output_____"
]
],
[
[
"### Variablesとオプティマイザ\n\n`tf.Variable` オブジェクトは、学習中にアクセスされるミュータブルな `tf.Tensor` 値を格納し、自動微分を容易にします。\nモデルのパラメータは、変数としてクラスにカプセル化できます。\n\n`tf.GradientTape` と共に `tf.Variable` を使うことでモデルパラメータはよりカプセル化されます。たとえば、上の\nの自動微分の例は以下のように書き換えることができます:",
"_____no_output_____"
]
],
[
[
"class Model(tf.keras.Model):\n def __init__(self):\n super(Model, self).__init__()\n self.W = tf.Variable(5., name='weight')\n self.B = tf.Variable(10., name='bias')\n def call(self, inputs):\n return inputs * self.W + self.B\n\n# 3 * x + 2を近似するトイデータセット\nNUM_EXAMPLES = 2000\ntraining_inputs = tf.random.normal([NUM_EXAMPLES])\nnoise = tf.random.normal([NUM_EXAMPLES])\ntraining_outputs = training_inputs * 3 + 2 + noise\n\n# 最適化対象のloss関数\ndef loss(model, inputs, targets):\n error = model(inputs) - targets\n return tf.reduce_mean(tf.square(error))\n\ndef grad(model, inputs, targets):\n with tf.GradientTape() as tape:\n loss_value = loss(model, inputs, targets)\n return tape.gradient(loss_value, [model.W, model.B])\n\n# 定義:\n# 1. モデル\n# 2. モデルパラメータに関する損失関数の導関数\n# 3. 導関数に基づいて変数を更新するストラテジ。\nmodel = Model()\noptimizer = tf.keras.optimizers.SGD(learning_rate=0.01)\n\nprint(\"Initial loss: {:.3f}\".format(loss(model, training_inputs, training_outputs)))\n\n# 学習ループ\nfor i in range(300):\n grads = grad(model, training_inputs, training_outputs)\n optimizer.apply_gradients(zip(grads, [model.W, model.B]))\n if i % 20 == 0:\n print(\"Loss at step {:03d}: {:.3f}\".format(i, loss(model, training_inputs, training_outputs)))\n\nprint(\"Final loss: {:.3f}\".format(loss(model, training_inputs, training_outputs)))\nprint(\"W = {}, B = {}\".format(model.W.numpy(), model.B.numpy()))",
"_____no_output_____"
]
],
[
[
"## Eager Execution の途中でオブジェクトのステータスを使用する\n\nTF 1.x の Graph Execution では、プログラムの状態(Variableなど)は global collection に格納され、それらの存続期間は `tf.Session` オブジェクトによって管理されます。\n対照的に、 Eager Execution の間、状態オブジェクトの存続期間は、対応する Python オブジェクトの存続期間によって決定されます。\n\n### 変数とオブジェクト\n\nEager Execution の間、変数はオブジェクトへの最後の参照が削除され、その後削除されるまで存続します。",
"_____no_output_____"
]
],
[
[
"if tf.test.is_gpu_available():\n with tf.device(\"gpu:0\"):\n v = tf.Variable(tf.random.normal([1000, 1000]))\n v = None # v は GPU メモリを利用しなくなる",
"_____no_output_____"
]
],
[
[
"### オブジェクトベースの保存\n\nこのセクションは、[チェックポイントの学習の手引き](./checkpoint.ipynb) の省略版です。\n\n`tf.train.Checkpoint` はチェックポイントを用いて `tf.Variable` を保存および復元することができます:",
"_____no_output_____"
]
],
[
[
"x = tf.Variable(10.)\ncheckpoint = tf.train.Checkpoint(x=x)",
"_____no_output_____"
],
[
"x.assign(2.) # 新しい値を変数に代入して保存する。\ncheckpoint_path = './ckpt/'\ncheckpoint.save('./ckpt/')",
"_____no_output_____"
],
[
"x.assign(11.) # 保存後に変数の値を変える。\n\n# チェックポイントから変数を復元する\ncheckpoint.restore(tf.train.latest_checkpoint(checkpoint_path))\n\nprint(x) # => 2.0",
"_____no_output_____"
]
],
[
[
"モデルを保存して読み込むために、 `tf.train.Checkpoint` は隠れ変数なしにオブジェクトの内部状態を保存します。 `モデル`、 `オプティマイザ` 、そしてグローバルステップの状態を記録するには、それらを `tf.train.Checkpoint` に渡します。",
"_____no_output_____"
]
],
[
[
"import os\n\nmodel = tf.keras.Sequential([\n tf.keras.layers.Conv2D(16,[3,3], activation='relu'),\n tf.keras.layers.GlobalAveragePooling2D(),\n tf.keras.layers.Dense(10)\n])\noptimizer = tf.keras.optimizers.Adam(learning_rate=0.001)\ncheckpoint_dir = 'path/to/model_dir'\nif not os.path.exists(checkpoint_dir):\n os.makedirs(checkpoint_dir)\ncheckpoint_prefix = os.path.join(checkpoint_dir, \"ckpt\")\nroot = tf.train.Checkpoint(optimizer=optimizer,\n model=model)\n\nroot.save(checkpoint_prefix)\nroot.restore(tf.train.latest_checkpoint(checkpoint_dir))",
"_____no_output_____"
]
],
[
[
"多くの学習ループでは、変数は `tf.train..Checkpoint.restore` が呼ばれたあとに作成されます。これらの変数は作成されてすぐに復元され、チェックポイントがすべてロードされたことを確認するためのアサーションが利用可能になります。詳しくは、 [guide to training checkpoints](./checkpoint.ipynb) を見てください。",
"_____no_output_____"
],
[
"### オブジェクト指向メトリクス\n\n`tfe.keras.metrics`はオブジェクトとして保存されます。新しいデータを呼び出し可能オブジェクトに渡してメトリクスを更新し、 `tfe.keras.metrics.result`メソッドを使って結果を取得します。次に例を示します:",
"_____no_output_____"
]
],
[
[
"m = tf.keras.metrics.Mean(\"loss\")\nm(0)\nm(5)\nm.result() # => 2.5\nm([8, 9])\nm.result() # => 5.5",
"_____no_output_____"
]
],
[
[
"## 高度な自動微分トピック\n\n### 動的なモデル\n\n`tf.GradientTape` は動的モデルでも使うことができます。 \n以下の [バックトラックライン検索](https://wikipedia.org/wiki/Backtracking_line_search)\nアルゴリズムの例は、複雑な制御フローにもかかわらず\n勾配があり、微分可能であることを除いて、通常の NumPy コードのように見えます:",
"_____no_output_____"
]
],
[
[
"def line_search_step(fn, init_x, rate=1.0):\n with tf.GradientTape() as tape:\n # 変数は自動的に記録されるが、Tensorは手動でウォッチする\n tape.watch(init_x)\n value = fn(init_x)\n grad = tape.gradient(value, init_x)\n grad_norm = tf.reduce_sum(grad * grad)\n init_value = value\n while value > init_value - rate * grad_norm:\n x = init_x - rate * grad\n value = fn(x)\n rate /= 2.0\n return x, value",
"_____no_output_____"
]
],
[
[
"### カスタム勾配\n\nカスタム勾配は、勾配を上書きする簡単な方法です。 フォワード関数では、\n入力、出力、または中間結果に関する勾配を定義します。たとえば、逆方向パスにおいて勾配のノルムを制限する簡単な方法は次のとおりです:",
"_____no_output_____"
]
],
[
[
"@tf.custom_gradient\ndef clip_gradient_by_norm(x, norm):\n y = tf.identity(x)\n def grad_fn(dresult):\n return [tf.clip_by_norm(dresult, norm), None]\n return y, grad_fn",
"_____no_output_____"
]
],
[
[
"カスタム勾配は、一連の演算に対して数値的に安定した勾配を提供するために共通的に使用されます。:",
"_____no_output_____"
]
],
[
[
"def log1pexp(x):\n return tf.math.log(1 + tf.exp(x))\n\ndef grad_log1pexp(x):\n with tf.GradientTape() as tape:\n tape.watch(x)\n value = log1pexp(x)\n return tape.gradient(value, x)",
"_____no_output_____"
],
[
"# 勾配計算は x = 0 のときはうまくいく。\ngrad_log1pexp(tf.constant(0.)).numpy()",
"_____no_output_____"
],
[
"# しかし、x = 100のときは数値的不安定により失敗する。\ngrad_log1pexp(tf.constant(100.)).numpy()",
"_____no_output_____"
]
],
[
[
"ここで、 `log1pexp` 関数はカスタム勾配を用いて解析的に単純化することができます。\n以下の実装は、フォワードパスの間に計算された `tf.exp(x)` の値を\n再利用します—冗長な計算を排除することでより効率的になります:",
"_____no_output_____"
]
],
[
[
"@tf.custom_gradient\ndef log1pexp(x):\n e = tf.exp(x)\n def grad(dy):\n return dy * (1 - 1 / (1 + e))\n return tf.math.log(1 + e), grad\n\ndef grad_log1pexp(x):\n with tf.GradientTape() as tape:\n tape.watch(x)\n value = log1pexp(x)\n return tape.gradient(value, x)",
"_____no_output_____"
],
[
"# 上と同様に、勾配計算はx = 0のときにはうまくいきます。\ngrad_log1pexp(tf.constant(0.)).numpy()",
"_____no_output_____"
],
[
"# また、勾配計算はx = 100でも機能します。\ngrad_log1pexp(tf.constant(100.)).numpy()",
"_____no_output_____"
]
],
[
[
"## パフォーマンス\n\nEager Executionの間、計算は自動的にGPUにオフロードされます。計算を実行するデバイスを指定したい場合は、\n`tf.device( '/ gpu:0')` ブロック(もしくはCPUを指定するブロック)で囲むことで指定できます:",
"_____no_output_____"
]
],
[
[
"import time\n\ndef measure(x, steps):\n # TensorFlowはGPUを初めて使用するときに初期化するため、時間計測対象からは除外する。\n tf.matmul(x, x)\n start = time.time()\n for i in range(steps):\n x = tf.matmul(x, x)\n # tf.matmulは、行列乗算が完了する前に戻ることができる。\n # (たとえば、CUDAストリームにオペレーションをエンキューした後に戻すことができる)。\n # 以下のx.numpy()呼び出しは、すべてのキューに入れられたオペレーションが完了したことを確認する。\n # (そして結果をホストメモリにコピーするため、計算時間は単純なmatmulオペレーションよりも多くのことを含む時間になる。)\n _ = x.numpy()\n end = time.time()\n return end - start\n\nshape = (1000, 1000)\nsteps = 200\nprint(\"Time to multiply a {} matrix by itself {} times:\".format(shape, steps))\n\n# CPU上で実行するとき:\nwith tf.device(\"/cpu:0\"):\n print(\"CPU: {} secs\".format(measure(tf.random.normal(shape), steps)))\n\n# GPU上で実行するとき(GPUが利用できれば):\nif tf.test.is_gpu_available():\n with tf.device(\"/gpu:0\"):\n print(\"GPU: {} secs\".format(measure(tf.random.normal(shape), steps)))\nelse:\n print(\"GPU: not found\")",
"_____no_output_____"
]
],
[
[
"`tf.Tensor` オブジェクトはそのオブジェクトに対するオペレーションを実行するために別のデバイスにコピーすることができます:",
"_____no_output_____"
]
],
[
[
"if tf.test.is_gpu_available():\n x = tf.random.normal([10, 10])\n\n x_gpu0 = x.gpu()\n x_cpu = x.cpu()\n\n _ = tf.matmul(x_cpu, x_cpu) # CPU上で実行するとき\n _ = tf.matmul(x_gpu0, x_gpu0) # GPU:0上で実行するとき",
"_____no_output_____"
]
],
[
[
"### ベンチマーク\n\nGPUでの\n[ResNet50](https://github.com/tensorflow/tensorflow/tree/master/tensorflow/contrib/eager/python/examples/resnet50)\nの学習のような、計算量の多いモデルの場合は、Eager Executionのパフォーマンスは `tf.function` のパフォーマンスに匹敵します。\nしかし、この2つの環境下のパフォーマンスの違いは計算量の少ないモデルではより大きくなり、小さなたくさんのオペレーションからなるモデルでホットコードパスを最適化するためにやるべきことがあります。\n\n## functionsの利用\n\nEager Execution は開発とデバッグをより対話的にしますが、\nTensorFlow 1.x スタイルの Graph Execution は分散学習、パフォーマンスの最適化、そしてプロダクション環境へのデプロイの観点で利点があります。\n\n2つの手法のギャップを埋めるために、 TensorFlow 2.0 は `tf.function` という機能を導入しています。\n詳しくは、 [Autograph](./function.ipynb) のガイドを見てください。\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a8cc9c032ce7d27169927857ec50751f9edb8ea
| 742,751 |
ipynb
|
Jupyter Notebook
|
Beginners_guide/.ipynb_checkpoints/03_Loading_data-checkpoint.ipynb
|
mrMustacho/hackathonFACH2020-waterlands
|
b87ce003976fa07cb85de1a9cc2d0d0c107f2087
|
[
"MIT"
] | null | null | null |
Beginners_guide/.ipynb_checkpoints/03_Loading_data-checkpoint.ipynb
|
mrMustacho/hackathonFACH2020-waterlands
|
b87ce003976fa07cb85de1a9cc2d0d0c107f2087
|
[
"MIT"
] | null | null | null |
Beginners_guide/.ipynb_checkpoints/03_Loading_data-checkpoint.ipynb
|
mrMustacho/hackathonFACH2020-waterlands
|
b87ce003976fa07cb85de1a9cc2d0d0c107f2087
|
[
"MIT"
] | null | null | null | 65.87592 | 5,648 | 0.480949 |
[
[
[
"# Loading data from Chile data cube\n\n* **Prerequisites:** Users of this notebook should have a basic understanding of:\n * How to run a [Jupyter notebook](01_Jupyter_notebooks.ipynb)\n * Inspecting available [Products and measurements](02_Products_and_measurements.ipynb)",
"_____no_output_____"
],
[
"## Background\nLoading data from the Chile instance of the [Open Data Cube](https://www.opendatacube.org/) requires the construction of a data query that specifies the what, where, and when of the data request.\nEach query returns a [multi-dimensional xarray object](http://xarray.pydata.org/en/stable/) containing the contents of your query.\nIt is essential to understand the `xarray` data structures as they are fundamental to the structure of data loaded from the datacube.\nManipulations, transformations and visualisation of `xarray` objects provide datacube users with the ability to explore and analyse datasets, as well as pose and answer scientific questions.",
"_____no_output_____"
],
[
"## Description\nThis notebook will introduce how to load data from the Chile datacube through the construction of a query and use of the `dc.load()` function.\nTopics covered include:\n\n* Loading data using `dc.load()`\n* Interpreting the resulting `xarray.Dataset` object\n * Inspecting an individual `xarray.DataArray`\n* Customising parameters passed to the `dc.load()` function\n * Loading specific measurements\n * Loading data for coordinates in a custom coordinate reference system (CRS)\n * Projecting data to a new CRS and spatial resolution \n * Specifying a specific spatial resampling method\n* Loading data using a reusable dictionary query\n* Loading matching data from multiple products using `like`\n* Adding a progress bar to the data load\n\n***",
"_____no_output_____"
],
[
"## Getting started\nTo run this introduction to loading data from the datacube, run all the cells in the notebook starting with the \"Load packages\" cell. For help with running notebook cells, refer back to the [Jupyter Notebooks notebook](01_Jupyter_notebooks.ipynb).",
"_____no_output_____"
],
[
"### Load packages\nFirst we need to load the `datacube` package.\nThis will allow us to query the datacube database and load some data. \nThe `with_ui_cbk` function from `odc.ui` will allow us to show a progress bar when loading large amounts of data.",
"_____no_output_____"
]
],
[
[
"import datacube\nfrom odc.ui import with_ui_cbk",
"_____no_output_____"
]
],
[
[
"### Connect to the datacube\nWe then need to connect to the datacube database.\nWe will then be able to use the `dc` datacube object to load data.\nThe `app` parameter is a unique name used to identify the notebook that does not have any effect on the analysis.",
"_____no_output_____"
]
],
[
[
"dc = datacube.Datacube(app=\"03_Loading_data\")",
"_____no_output_____"
]
],
[
[
"## Loading data using `dc.load()`\n\nLoading data from the datacube uses the [dc.load()](https://datacube-core.readthedocs.io/en/latest/dev/api/generate/datacube.Datacube.load.html) function.\n\nThe function requires the following minimum arguments:\n\n* `product`: A specific product to load (to revise products, see the [Products and measurements](02_Products_and_measurements.ipynb) notebook).\n* `x`: Defines the spatial region in the *x* dimension. By default, the *x* and *y* arguments accept queries in a geographical co-ordinate system WGS84, identified by the EPSG code *4326*.\n* `y`: Defines the spatial region in the *y* dimension. The dimensions ``longitude``/``latitude`` and ``x``/``y`` can be used interchangeably.\n* `time`: Defines the temporal extent. The time dimension can be specified using a tuple of datetime objects or strings in the \"YYYY\", \"YYYY-MM\" or \"YYYY-MM-DD\" format. \n\nLet's run a query to load 2018 data from Landsat 8 over Santiago\n. \nFor this example, we can use the following parameters:\n\n* `product`: `usgs_espa_ls8c1_sr`\n* `x`=`(-71.1, -71.5)`\n* `y`=`(-29.5, -30)`,\n* `time`: `(\"2020-01-01\", \"2020-12-31\")`\n\nRun the following cell to load all datasets from the `usgs_espa_ls8c1_sr` product that match this spatial and temporal extent:",
"_____no_output_____"
]
],
[
[
"ds = dc.load(product=\"usgs_espa_ls8c1_sr\",\n x=(-71.1, -71.5),\n y=(-29.5, -30),\n output_crs = \"EPSG:32719\",\n time = (\"2020-01-01\", \"2020-12-31\"),\n resolution = (-25, 25),\n dask_chunks={\"time\": 1}\n )\nds",
"_____no_output_____"
]
],
[
[
"### Interpreting the resulting `xarray.Dataset`\nThe variable `ds` has returned an `xarray.Dataset` containing all data that matched the spatial and temporal query parameters inputted into `dc.load`.\n\n*Dimensions* \n\n* Identifies the number of timesteps returned in the search (`time: 1`) as well as the number of pixels in the `x` and `y` directions of the data query.\n\n*Coordinates* \n\n* `time` identifies the date attributed to each returned timestep.\n* `x` and `y` are the coordinates for each pixel within the spatial bounds of your query.\n\n*Data variables*\n\n* These are the measurements available for the nominated product. \nFor every date (`time`) returned by the query, the measured value at each pixel (`y`, `x`) is returned as an array for each measurement.\nEach data variable is itself an `xarray.DataArray` object ([see below](#Inspecting-an-individual-xarray.DataArray)). \n\n*Attributes*\n\n* `crs` identifies the coordinate reference system (CRS) of the loaded data. ",
"_____no_output_____"
],
[
"### Inspecting an individual `xarray.DataArray`\nThe `xarray.Dataset` we loaded above is itself a collection of individual `xarray.DataArray` objects that hold the actual data for each data variable/measurement. \nFor example, all measurements listed under _Data variables_ above (e.g. `blue`, `green`, `red`, `nir`, `swir1`, `swir2`) are `xarray.DataArray` objects.\n\nWe can inspect the data in these `xarray.DataArray` objects using either of the following syntaxes:\n```\nds[\"measurement_name\"]\n```\nor:\n```\nds.measurement_name\n```\n\nBeing able to access data from individual data variables/measurements allows us to manipulate and analyse data from individual satellite bands or specific layers in a dataset. \nFor example, we can access data from the near infra-red satellite band (i.e. `nir`):",
"_____no_output_____"
]
],
[
[
"ds.red",
"_____no_output_____"
]
],
[
[
"Note that the object header informs us that it is an `xarray.DataArray` containing data for the `nir` satellite band. \n\nLike an `xarray.Dataset`, the array also includes information about the data's **dimensions** (i.e. `(time: 1, y: 801, x: 644)`), **coordinates** and **attributes**.\nThis particular data variable/measurement contains some additional information that is specific to the `nir` band, including details of array's nodata value (i.e. `nodata: -999`).\n\n> **Note**: For a more in-depth introduction to `xarray` data structures, refer to the [official xarray documentation](http://xarray.pydata.org/en/stable/data-structures.html)",
"_____no_output_____"
],
[
"## Customising the `dc.load()` function\n\nThe `dc.load()` function can be tailored to refine a query.\n\nCustomisation options include:\n\n* `measurements:` This argument is used to provide a list of measurement names to load, as listed in `dc.list_measurements()`. \nFor satellite datasets, measurements contain data for each individual satellite band (e.g. near infrared). \nIf not provided, all measurements for the product will be returned.\n* `crs:` The coordinate reference system (CRS) of the query's `x` and `y` coordinates is assumed to be `WGS84`/`EPSG:4326` unless the `crs` field is supplied, even if the stored data is in another projection or the `output_crs` is specified. \nThe `crs` parameter is required if your query's coordinates are in any other CRS.\n* `group_by:` Satellite datasets based around scenes can have multiple observations per day with slightly different time stamps as the satellite collects data along its path.\nThese observations can be combined by reducing the `time` dimension to the day level using `group_by=solar_day`.\n* `output_crs` and `resolution`: To reproject or change the resolution the data, supply the `output_crs` and `resolution` fields. \n* `resampling`: This argument allows you to specify a custom spatial resampling method to use when data is reprojected into a different CRS. \n\nExample syntax on the use of these options follows in the cells below.\n\n> For help or more customisation options, run `help(dc.load)` in an empty cell or visit the function's [documentation page](https://datacube-core.readthedocs.io/en/latest/dev/api/generate/datacube.Datacube.load.html)\n",
"_____no_output_____"
],
[
"### Specifying measurements\nBy default, `dc.load()` will load *all* measurements in a product.\n\nTo load data from the `red`, `green` and `blue` satellite bands only, we can add `measurements=[\"red\", \"green\", \"blue\"]` to our query:",
"_____no_output_____"
]
],
[
[
"# Note the optional inclusion of the measurements list\nds_rgb = dc.load(product=\"usgs_espa_ls8c1_sr\",\n measurements=[\"red\", \"green\", \"blue\"],\n x=(-71.1, -71.5),\n y=(-29.5, -30),\n output_crs = \"EPSG:32719\",\n time = (\"2020-01-01\", \"2020-12-31\"),\n resolution = (-25, 25),\n dask_chunks={\"time\": 1}\n )\n\nds_rgb",
"_____no_output_____"
]
],
[
[
"Note that the *Data variables* component of the `xarray.Dataset` now includes only the measurements specified in the query (i.e. the `red`, `green` and `blue` satellite bands).",
"_____no_output_____"
],
[
"### Loading data for coordinates in any CRS\nBy default, `dc.load()` assumes that your query `x` and `y` coordinates are provided in degrees in the `WGS84/EPSG:4326` CRS.\nIf your coordinates are in a different coordinate system, you need to specify this using the `crs` parameter.\n\nIn the example below, we load data for a set of `x` and `y` coordinates defined in WGS84 UTM zone 19S (`EPSG:32719`), and ensure that the `dc.load()` function accounts for this by including `crs=\"EPSG:32719\"`:\n",
"_____no_output_____"
]
],
[
[
"# Note the new `x` and `y` coordinates and `crs` parameter\nds_custom_crs = dc.load(product=\"usgs_espa_ls8c1_sr\",\n time=(\"2020-01-01\", \"2020-12-31\"),\n x=(335713, 355713),\n y=(6287592, 6307592),\n crs=\"EPSG:32719\",\n output_crs = \"EPSG:32719\",\n resolution = (-25, 25),\n dask_chunks={\"time\": 1}\n )\n\nds_custom_crs",
"_____no_output_____"
]
],
[
[
"### CRS reprojection\nCertain applications may require that you output your data into a specific CRS.\nYou can reproject your output data by specifying the new `output_crs` and identifying the `resolution` required.\n\nIn this example, we will reproject our data to a new CRS (UTM Zone 34S, `EPSG:32734`) and resolution (250 x 250 m). Note that for most CRSs, the first resolution value is negative (e.g. `(-250, 250)`):",
"_____no_output_____"
]
],
[
[
"ds_reprojected = dc.load(product=\"usgs_espa_ls8c1_sr\",\n measurements=[\"red\", \"green\", \"blue\"],\n x=(-71.1, -71.5),\n y=(-29.5, -30),\n output_crs = \"EPSG:32734\",\n time = (\"2020-01-01\", \"2020-12-31\"),\n resolution = (-250, 250),\n dask_chunks={\"time\": 1}\n )\n\nds_reprojected",
"_____no_output_____"
]
],
[
[
"Note that the `crs` attribute in the *Attributes* section has changed to `EPSG:32734`. \nDue to the larger 250 m resolution, there are also now less pixels on the `x` and `y` dimensions (e.g. `x: 467, y: 344` compared to `x: 801, y: 801` in earlier examples).\n",
"_____no_output_____"
],
[
"### Spatial resampling methods\nWhen a product is re-projected to a different CRS and/or resolution, the new pixel grid may differ from the original input pixels by size, number and alignment.\nIt is therefore necessary to apply a spatial \"resampling\" rule that allocates input pixel values into the new pixel grid.\n\nBy default, `dc.load()` resamples pixel values using \"nearest neighbour\" resampling, which allocates each new pixel with the value of the closest input pixel.\nDepending on the type of data and the analysis being run, this may not be the most appropriate choice (e.g. for continuous data).\n\nThe `resampling` parameter in `dc.load()` allows you to choose a custom resampling method from the following options: \n\n```\n\"nearest\", \"cubic\", \"bilinear\", \"cubic_spline\", \"lanczos\", \n\"average\", \"mode\", \"gauss\", \"max\", \"min\", \"med\", \"q1\", \"q3\"\n```\n\nFor example, we can request that all loaded data is resampled using \"average\" resampling:",
"_____no_output_____"
]
],
[
[
"# Note the additional `resampling` parameter\nds_averageresampling = dc.load(product=\"usgs_espa_ls8c1_sr\",\n measurements=[\"red\", \"green\", \"blue\"],\n x=(-71.1, -71.5),\n y=(-29.5, -30),\n output_crs = \"EPSG:32719\",\n time = (\"2020-01-01\", \"2020-12-31\"),\n resolution = (-250, 250),\n dask_chunks={\"time\": 1},\n resampling=\"average\"\n )\n\nds_averageresampling",
"_____no_output_____"
]
],
[
[
"You can also provide a Python dictionary to request a different sampling method for different measurements. \nThis can be particularly useful when some measurements contain contain categorical data which require resampling methods such as \"nearest\" or \"mode\" that do not modify the input pixel values.\n\nIn the example below, we specify `resampling={\"red\": \"nearest\", \"*\": \"average\"}`, which will use \"nearest\" neighbour resampling for the `red` satellite band only. `\"*\": \"average\"` will apply \"average\" resampling for all other satellite bands:\n",
"_____no_output_____"
]
],
[
[
"ds_customresampling = dc.load(product=\"usgs_espa_ls8c1_sr\",\n measurements=[\"red\", \"green\", \"blue\"],\n x=(-71.1, -71.5),\n y=(-29.5, -30),\n output_crs = \"EPSG:32719\",\n time = (\"2020-01-01\", \"2020-12-31\"),\n resolution = (-250, 250),\n dask_chunks={\"time\": 1},\n resampling={\"red\": \"nearest\", \"*\": \"average\"}\n )\n\nds_customresampling",
"_____no_output_____"
]
],
[
[
"> **Note**: For more information about spatial resampling methods, see the [following guide](https://rasterio.readthedocs.io/en/stable/topics/resampling.html)",
"_____no_output_____"
],
[
"## Loading data using the query dictionary syntax\nIt is often useful to re-use a set of query parameters to load data from multiple products.\nTo achieve this, we can load data using the \"query dictionary\" syntax.\nThis involves placing the query parameters we used to load data above inside a Python dictionary object which we can re-use for multiple data loads:",
"_____no_output_____"
]
],
[
[
"query = {\"x\": (-71.1, -71.5),\n \"y\": (-29.5, -30),\n \"time\": (\"2020-01-01\", \"2020-12-31\"),\n \"output_crs\": \"EPSG:32719\",\n \"time\": (\"2020-01-01\", \"2020-12-31\"),\n \"resolution\": (-250, 250),\n \"dask_chunks\": {\"time\": 1}\n }\n",
"_____no_output_____"
]
],
[
[
"We can then use this query dictionary object as an input to `dc.load()`. \n\n> The `**` syntax below is Python's \"keyword argument unpacking\" operator.\nThis operator takes the named query parameters listed in the dictionary we created (e.g. `\"x\": (153.3, 153.4)`), and \"unpacks\" them into the `dc.load()` function as new arguments. \nFor more information about unpacking operators, refer to the [Python documentation](https://docs.python.org/3/tutorial/controlflow.html#unpacking-argument-lists)",
"_____no_output_____"
]
],
[
[
"ds = dc.load(product=\"usgs_espa_ls8c1_sr\",\n **query)\n\nds",
"_____no_output_____"
]
],
[
[
"Query dictionaries can contain any set of parameters that would usually be provided to `dc.load()`:",
"_____no_output_____"
]
],
[
[
"query = {\"x\": (-71.1, -71.5),\n \"y\": (-29.5, -30),\n \"time\": (\"2020-01-01\", \"2020-12-31\"),\n \"output_crs\": \"EPSG:32719\",\n \"time\": (\"2020-01-01\", \"2020-12-31\"),\n \"resolution\": (-500, 500),\n \"dask_chunks\": {\"time\": 1},\n \"resampling\": {\"red\": \"nearest\", \"*\": \"average\"}\n }\n\nds_ls8 = dc.load(product=\"usgs_espa_ls8c1_sr\",\n **query)\n\nds_ls8\n",
"_____no_output_____"
]
],
[
[
"## Other helpful tricks\n### Loading data \"like\" another dataset\nAnother option for loading matching data from multiple products is to use `dc.load()`'s `like` parameter.\nThis will copy the spatial and temporal extent and the CRS/resolution from an existing dataset, and use these parameters to load a new data from a new product.\n\nIn the example below, we load another WOfS dataset that exactly matches the `ds_ls8` dataset we loaded earlier:\n",
"_____no_output_____"
]
],
[
[
"# THIS WON'T WORK UNTIL WE GET MORE DATA IN THE CHILE DATACUBE\n\n# ds_wofs = dc.load(product=\"ga_ls8c_wofs_2_annual_summary\",\n# like=ds_ls8)\n\n# print(ds_wofs)",
"_____no_output_____"
]
],
[
[
"### Adding a progress bar\nWhen loading large amounts of data, it can be useful to view the progress of the data load. \nThe `progress_cbk` parameter in `dc.load()` allows us to add a progress bar which will indicate how the load is progressing. In this example, we will load 5 years of data (2013, 2014, 2015, 2016 and 2017) from the `ga_ls8c_wofs_2_annual_summary` product with a progress bar:",
"_____no_output_____"
],
[
"This only works when dask chunking is **disabled**. To understand more about Dask, please see [Parallel processing with Dask](08_Parallel_processing_with_dask.ipynb)",
"_____no_output_____"
]
],
[
[
"query = {\"x\": (-71.1, -71.5),\n \"y\": (-29.5, -30),\n \"time\": (\"2020-01-01\", \"2020-12-31\"),\n \"output_crs\": \"EPSG:32719\",\n \"time\": (\"2020-01-01\", \"2020-12-31\"),\n \"resolution\": (-500, 500),\n# \"dask_chunks\": {\"time\": 1},\n \"resampling\": {\"red\": \"nearest\", \"*\": \"average\"}\n }\n\nds_progress = dc.load(product=\"usgs_espa_ls8c1_sr\",\n progress_cbk=with_ui_cbk(),\n **query)\n\nds_progress",
"_____no_output_____"
]
],
[
[
"## Recommended next steps\n\nFor more advanced information about working with Jupyter Notebooks or JupyterLab, you can explore [JupyterLab documentation page](https://jupyterlab.readthedocs.io/en/stable/user/notebook.html).\n\nTo continue working through the notebooks in this beginner's guide, the following notebooks are designed to be worked through in the following order:\n\n1. [Jupyter Notebooks](01_Jupyter_notebooks.ipynb)\n2. [Products and Measurements](02_Products_and_measurements.ipynb)\n3. **Loading data (this notebook)**\n4. [Plotting](04_Plotting.ipynb)\n5. [Performing a basic analysis](05_Basic_analysis.ipynb)\n6. [Introduction to numpy](06_Intro_to_numpy.ipynb)\n7. [Introduction to xarray](07_Intro_to_xarray.ipynb)\n8. [Parallel processing with Dask](08_Parallel_processing_with_dask.ipynb)\n\nOnce you have you have completed the above six tutorials, join advanced users in exploring:\n\n* The \"Datasets\" directory in the repository, where you can explore DE Africa products in depth.\n* The \"Frequently used code\" directory, which contains a recipe book of common techniques and methods for analysing DE Africa data.\n* The \"Real-world examples\" directory, which provides more complex workflows and analysis case studies.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a8ce0d3a5698e9659785817dcbca733df5d8b48
| 6,074 |
ipynb
|
Jupyter Notebook
|
Python/pandas/leoni.ipynb
|
Dataninja/j-r-p
|
b8ff6b9a1a798c0c6581b86df5b7485e0f371bc0
|
[
"MIT"
] | 1 |
2018-11-19T06:40:38.000Z
|
2018-11-19T06:40:38.000Z
|
Python/pandas/leoni.ipynb
|
Dataninja/j-r-p
|
b8ff6b9a1a798c0c6581b86df5b7485e0f371bc0
|
[
"MIT"
] | 7 |
2018-11-18T08:15:55.000Z
|
2018-12-02T22:15:58.000Z
|
Python/pandas/leoni.ipynb
|
Dataninja/j-r-p
|
b8ff6b9a1a798c0c6581b86df5b7485e0f371bc0
|
[
"MIT"
] | 6 |
2018-11-19T18:48:55.000Z
|
2021-04-09T10:37:46.000Z
| 25.20332 | 118 | 0.506915 |
[
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"pageURL = 'https://it.wikipedia.org/wiki/Leone_d%27oro_al_miglior_film'\ntables = pd.read_html(pageURL, match='Anno', header=0)\ndataframe = tables[0]",
"_____no_output_____"
],
[
"dataframe['index_col'] = dataframe.index",
"_____no_output_____"
],
[
"dataframe[14:15]",
"_____no_output_____"
],
[
"# creo un dataframe per i record con le colonne mappate male, in cui l'anno contiene stringhe\ndf2=dataframe[dataframe['Anno'].str.contains(\"[a-zA-Z]\")]\n# creo un dataframe per tutti gli altri valori\ndf1=dataframe[(~dataframe['Anno'].str.contains(\"[a-zA-Z]\")) & (dataframe['Regista'].str.contains(\".+\")) ]",
"_____no_output_____"
],
[
"# aggiungo un colonna con valori nulli e la sposto a inizio dataframe\ndf2.loc[:,'test']=np.nan\ncol = \"test\" \ndf2 = pd.concat([df2[col],df2.drop(col,axis=1)], axis=1)",
"_____no_output_____"
],
[
"# rimuovo da df2 la colonna in più\ncolonna=len(df2.columns)-2\ndf2.drop(df2.columns[colonna], axis = 1,inplace=True)",
"_____no_output_____"
],
[
"# rinomino le colonne dei dataframe con dei numeri crescenti da 0 a salire\ndf2.columns = np.arange(len(df2.columns))\ndf1.columns = np.arange(len(df1.columns))",
"_____no_output_____"
],
[
"# faccio il merge dei dataframe\ndf=df1.append(df2, ignore_index=True)",
"_____no_output_____"
],
[
"# riordino le righe\ndf.sort_values([4],inplace=True)\n# inserisco l'anno nelle celle in cui manca\ndf.fillna(method=\"ffill\",inplace=True)\n# imposto un separatore per le celle con più nazioni\ndf.replace({'\\/.{1}': ':'}, regex=True,inplace=True)\n# rinomino le colonne\ndf.rename(index=str, columns={0: \"Anno\", 1: \"Titolo\",2:\"Regista\",3:\"Nazione\"}, inplace=True)\n# rimuovo una colonna non utile\ndf.drop([4], axis=1)\n# salvo la lista in CSV\ndf.to_csv(\"leoni_pd.csv\",index=False,encoding=\"utf-8\")",
"_____no_output_____"
],
[
"nazioni=df[['Nazione']]",
"_____no_output_____"
],
[
"# splitto le celle con più nazioni associate, traspongo in verticale e creo un dataframe\nnazioni=nazioni['Nazione'].str.split(':', expand=True).stack()\nnazioni=nazioni.to_frame()",
"_____no_output_____"
],
[
"# raggruppo, conteggio e ordino\nnazioni=nazioni.groupby([0]).size().reset_index(name='Conteggio').sort_values(['Conteggio'], ascending=False)\nnazioni.rename(index=str, columns={0: \"Nazione\"}, inplace=True)",
"_____no_output_____"
],
[
"# salvo in CSV\nnazioni.to_csv(\"listaNazioni_pd.csv\",index=False,encoding=\"utf-8\")",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8ce979eed8540ef2ccfa8a4a29eda6436124ee
| 352,009 |
ipynb
|
Jupyter Notebook
|
general/how-to/how_to_developing_advanced_user_interfaces/how_to_developing_advanced_user_interfaces.ipynb
|
transentis/bptk_py_tutorial
|
db622858401fb63f773bc5917414bd42872c5010
|
[
"MIT"
] | 34 |
2020-02-01T04:53:56.000Z
|
2022-03-07T19:28:59.000Z
|
general/how-to/how_to_developing_advanced_user_interfaces/how_to_developing_advanced_user_interfaces.ipynb
|
transentis/bptk_py_tutorial
|
db622858401fb63f773bc5917414bd42872c5010
|
[
"MIT"
] | 3 |
2021-05-04T07:08:26.000Z
|
2022-03-02T11:39:51.000Z
|
general/how-to/how_to_developing_advanced_user_interfaces/how_to_developing_advanced_user_interfaces.ipynb
|
transentis/bptk_py_tutorial
|
db622858401fb63f773bc5917414bd42872c5010
|
[
"MIT"
] | 14 |
2020-03-26T21:08:54.000Z
|
2022-02-04T14:20:01.000Z
| 359.927403 | 71,272 | 0.926925 |
[
[
[
"# Developing Advanced User Interfaces\n*Using Jupyter Widgets, Pandas Dataframes and Matplotlib*",
"_____no_output_____"
],
[
"While BPTK-Py offers a number of high-level functions to quickly plot equations (such as `bptk.plot_scenarios`) or create a dashboard (e.g. `bptk.dashboard`), you may sometimes be in a situation when you want to create more sophisticated plots (e.g. plots with two axes) or a more sophisticated interface dashboard for your simulation. \n\nThis is actually quite easy, because BPTK-Py's high-level functions already utilize some very powerfull open source libraries for data management, plotting and dashboards: Pandas, Matplotlib and Jupyter Widgets.\n\nIn order to harness the full power of these libraries, you only need to understand how to make the data generated by BPTK-Py available to them. This _How To_ illustrates this using a neat little simulation of customer acquisition strategies. You don't need to understand the simulation to follow this document, but if you are interested you can read more about it on our [blog](https://www.transentis.com/an-example-to-illustrate-the-business-prototyping-methodology/). ",
"_____no_output_____"
],
[
"## Advanced Plotting\n\nWe'll start with some advanced plotting of simulation results.",
"_____no_output_____"
]
],
[
[
"## Load the BPTK Package\nfrom BPTK_Py.bptk import bptk \n\nbptk = bptk()",
"_____no_output_____"
]
],
[
[
"BPTK-Py's workhorse for creating plots is the `bptk.plot_scenarios`function. The function generates all the data you would like to plot using the simulation defined by the scenario manager and the settings defined by the scenarios. The data are stored in a Pandas dataframe. When it comes to plotting the results, the framework uses Matplotlib. To illustrate this, we will recreate the plot below directly from the underlying data:",
"_____no_output_____"
]
],
[
[
" bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"base\"], \n equations=['customers'],\n title=\"Base\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"No. of Customers\"\n )",
"_____no_output_____"
]
],
[
[
"You can access the data generated by a scenario by saving it into a dataframe. You can do this by adding the `return_df` flag to `bptk.plot_scenario`:",
"_____no_output_____"
]
],
[
[
" df=bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"base\"], \n equations=['customers'],\n title=\"Base\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"No. of Customers\",\n return_df=True\n )",
"_____no_output_____"
]
],
[
[
"The dataframe is indexed by time and stores the equations (in SD models) or agent properties (in Agent-based models) in the columns",
"_____no_output_____"
]
],
[
[
"df[0:10] # just show the first ten items",
"_____no_output_____"
]
],
[
[
"The frameworks `bptk.plot_scenarios` method first runs the simulation using the setting defined in the scenario and stores the data in a dataframe. It then plots the dataframe using Pandas `df.plot`method.\n\nWe can do the same:",
"_____no_output_____"
]
],
[
[
"subplot=df.plot(None,\"customers\")",
"_____no_output_____"
]
],
[
[
"The plot above doesn't look quite as neat as the plots created by `bptk.plot_scenarios`– this is because the framework applies some styling information. The styling information is stored in BPTK_Py.config, and you can access (and modify) it there.\n\nNow let's apply the config to `df.plot`:",
"_____no_output_____"
]
],
[
[
"import BPTK_Py.config as config\n\nsubplot=df.plot(kind=config.configuration[\"kind\"],\n alpha=config.configuration[\"alpha\"], stacked=config.configuration[\"stacked\"],\n figsize=config.configuration[\"figsize\"],\n title=\"Base\",\n color=config.configuration[\"colors\"],\n lw=config.configuration[\"linewidth\"])",
"_____no_output_____"
]
],
[
[
"Yes! We've recreated the plot from the high level `btpk.plot_scenarios` method using basic plotting functions.\n\nNow let's do something that currently isn't possible using the high-level BPTK-Py methods - let's create a graph that has two axes.\n\nThis is useful when you want to show the results of two equations at the same time, but they have different orders of magnitudes. For instance in the plot below, the number of customers is much smaller than the profit made, so the customer graph looks like a straight line. But it would still be intersting to be able to compare the two graphs.",
"_____no_output_____"
]
],
[
[
" bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"base\"], \n equations=['customers','profit'],\n title=\"Base\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"No. of Customers\"\n )",
"_____no_output_____"
]
],
[
[
"As before, we collect the data in a dataframe.",
"_____no_output_____"
]
],
[
[
" df=bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"base\"], \n equations=['customers','profit'],\n title=\"Base\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"No. of Customers\",\n return_df = True\n )",
"_____no_output_____"
],
[
"df[0:10]",
"_____no_output_____"
]
],
[
[
"Plotting two axes is easy in Pandas (which itself uses the Matplotlib library):",
"_____no_output_____"
]
],
[
[
"ax = df.plot(None,'customers', kind=config.configuration[\"kind\"],\n alpha=config.configuration[\"alpha\"], stacked=config.configuration[\"stacked\"],\n figsize=config.configuration[\"figsize\"],\n title=\"Profit vs. Customers\",\n color=config.configuration[\"colors\"],\n lw=config.configuration[\"linewidth\"])\n# ax is a Matplotlib Axes object\n\nax1 = ax.twinx()\n\n# Matplotlib.axes.Axes.twinx creates a twin y-axis.\n\nplot =df.plot(None,'profit',ax=ax1)",
"_____no_output_____"
]
],
[
[
"Voila! This is actually quite easy one you understand how to access the data (and of course a little knowledge of Pandas and Matplotlib is also useful). If you were writing a document that needed a lot of plots of this kind, you could create your own high-level function to avoide having to copy and paste the code above multiple times.",
"_____no_output_____"
],
[
"## Advanced interactive user interfaces\nNow let's try something a little more challenging: Let's build a dashboard for our simulation that let's you manipulate some of the scenrio settings interactively and plots results in tabs.",
"_____no_output_____"
],
[
"> Note: You need to have widgets enabled in Jupyter for the following to work. Please check the [BPTK-Py installation instructions](https://bptk.transentis-labs.com/en/latest/docs/usage/installation.html) or refer to the [Jupyter Widgets](https://ipywidgets.readthedocs.io/en/latest/user_install.html) documentation ",
"_____no_output_____"
],
[
"First, we need to understand how to create tabs. For this we need to import the `ipywidget` Library and we also need to access Matplotlib's `pyplot`",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nfrom ipywidgets import interact\nimport ipywidgets as widgets",
"_____no_output_____"
]
],
[
[
"Then we can create some tabs that display scenario results as follows:",
"_____no_output_____"
]
],
[
[
"out1 = widgets.Output()\nout2 = widgets.Output()\n\ntab = widgets.Tab(children = [out1, out2])\ntab.set_title(0, 'Customers')\ntab.set_title(1, 'Profit')\ndisplay(tab)\n\nwith out1:\n # turn of pyplot's interactive mode to ensure the plot is not created directly\n plt.ioff() \n # create the plot, but don't show it yet\n bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"hereWeGo\"], \n equations=['customers'],\n title=\"Here We Go\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"No. of Customers\"\n )\n # show the plot\n plt.show()\n # turn interactive mode on again\n plt.ion()\n\nwith out2:\n plt.ioff()\n bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"hereWeGo\"], \n equations=['profit'],\n title=\"Here We Go\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"Euro\"\n )\n plt.show()\n plt.ion()",
"_____no_output_____"
]
],
[
[
"That was easy! The only thing you really need to understand is to turn interactive plotting in `pyplot` off before creating the tabs and then turn it on again to create the plots. If you forget to do that, the plots appear above the tabs (try it and see!). ",
"_____no_output_____"
],
[
"In the next step, we need to add some sliders to manipulate the following scenario settings:\n\n* Referrals\n* Referral Free Months\n* Referral Program Adoption %\n* Advertising Success %",
"_____no_output_____"
],
[
"Creating a slider for the referrals is easy using the integer slider from the `ipywidgets` widget library:",
"_____no_output_____"
]
],
[
[
"widgets.IntSlider(\n value=7,\n min=0,\n max=15,\n step=1,\n description='Referrals:',\n disabled=False,\n continuous_update=False,\n orientation='horizontal',\n readout=True,\n readout_format='d'\n)",
"_____no_output_____"
]
],
[
[
"When manipulating a simulation model, we mostly want to start with a particular scenario and then manipulate some of the scenario settings using interactive widgets. Let's set up a new scenario for this purpose and call it `interactiveScenario`:",
"_____no_output_____"
]
],
[
[
"bptk.register_scenarios(scenario_manager=\"smCustomerAcquisition\", scenarios= \n {\n \"interactiveScenario\":{\n \"constants\":{\n \"referrals\":0,\n \"advertisingSuccessPct\":0.1,\n \"referralFreeMonths\":3,\n \"referralProgamAdoptionPct\":10\n }\n }\n }\n)",
"_____no_output_____"
]
],
[
[
"We can then access the scenario using `bptk.get_scenarios`: ",
"_____no_output_____"
]
],
[
[
"scenario = bptk.get_scenario(\"smCustomerAcquisition\",\"interactiveScenario\")\nscenario.constants",
"_____no_output_____"
],
[
"bptk.plot_scenarios(scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"interactiveScenario\"], \n equations=['profit'],\n title=\"Interactive Scenario\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"Euro\"\n )",
"_____no_output_____"
]
],
[
[
"The scenario constants can be accessed in the constants variable:",
"_____no_output_____"
],
[
"Now we have all the right pieces, we can put them together using the interact function.",
"_____no_output_____"
]
],
[
[
"@interact(advertising_success_pct=widgets.FloatSlider(\n value=0.1,\n min=0,\n max=1,\n step=0.01,\n continuous_update=False,\n description='Advertising Success Pct'\n))\ndef dashboard(advertising_success_pct):\n scenario= bptk.get_scenario(\"smCustomerAcquisition\",\n \"interactiveScenario\")\n \n scenario.constants[\"advertisingSuccessPct\"]=advertising_success_pct\n bptk.reset_scenario_cache(scenario_manager=\"smCustomerAcquisition\",\n scenario=\"interactiveScenario\")\n bptk.plot_scenarios(scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"interactiveScenario\"], \n equations=['profit'],\n title=\"Interactive Scenario\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"Euro\"\n )",
"_____no_output_____"
]
],
[
[
"Now let's combine this with the tabs from above.",
"_____no_output_____"
]
],
[
[
"out1 = widgets.Output()\nout2 = widgets.Output()\n\n\ntab = widgets.Tab(children = [out1, out2])\ntab.set_title(0, 'Customers')\ntab.set_title(1, 'Profit')\ndisplay(tab)\n \n@interact(advertising_success_pct=widgets.FloatSlider(\n value=0.1,\n min=0,\n max=10,\n step=0.01,\n continuous_update=False,\n description='Advertising Success Pct'\n))\ndef dashboardWithTabs(advertising_success_pct):\n scenario= bptk.get_scenario(\"smCustomerAcquisition\",\"interactiveScenario\")\n \n scenario.constants[\"advertisingSuccessPct\"]=advertising_success_pct\n bptk.reset_scenario_cache(scenario_manager=\"smCustomerAcquisition\",\n scenario=\"interactiveScenario\")\n \n \n \n with out1:\n # turn of pyplot's interactive mode to ensure the plot is not created directly\n plt.ioff() \n # clear the widgets output ... otherwise we will end up with a long list of plots, one for each change of settings\n \n # create the plot, but don't show it yet\n bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"interactiveScenario\"], \n equations=['customers'],\n title=\"Interactive Scenario\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"No. of Customers\"\n )\n # show the plot\n out1.clear_output() \n plt.show()\n # turn interactive mode on again\n plt.ion()\n\n with out2:\n plt.ioff()\n out2.clear_output()\n bptk.plot_scenarios(\n scenario_managers=[\"smCustomerAcquisition\"],\n scenarios=[\"interactiveScenario\"], \n equations=['profit'],\n title=\"Interactive Scenario\",\n freq=\"M\",\n x_label=\"Time\",\n y_label=\"Euro\"\n )\n plt.show()\n plt.ion()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8cec51a42abb70dcef925034520184b13b8b7e
| 27,101 |
ipynb
|
Jupyter Notebook
|
Skill-08/cycle-gan/Cycle-GAN-No-Outputs.ipynb
|
vcip2015/The-GAN-Book
|
d0b7f17ff7bd8ac3c7fae124ce4a986f460f1518
|
[
"MIT"
] | 1 |
2022-01-20T06:55:19.000Z
|
2022-01-20T06:55:19.000Z
|
Skill-08/cycle-gan/Cycle-GAN-No-Outputs.ipynb
|
vcip2015/The-GAN-Book
|
d0b7f17ff7bd8ac3c7fae124ce4a986f460f1518
|
[
"MIT"
] | null | null | null |
Skill-08/cycle-gan/Cycle-GAN-No-Outputs.ipynb
|
vcip2015/The-GAN-Book
|
d0b7f17ff7bd8ac3c7fae124ce4a986f460f1518
|
[
"MIT"
] | null | null | null | 35.150454 | 249 | 0.500092 |
[
[
[
"<a href=\"https://colab.research.google.com/github/kartikgill/The-GAN-Book/blob/main/Skill-08/Cycle-GAN-No-Outputs.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Import Useful Libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom tqdm import tqdm_notebook\n%matplotlib inline",
"_____no_output_____"
],
[
"import tensorflow\nprint (tensorflow.__version__)",
"_____no_output_____"
]
],
[
[
"# Download and Unzip Data",
"_____no_output_____"
]
],
[
[
"!wget https://people.eecs.berkeley.edu/~taesung_park/CycleGAN/datasets/horse2zebra.zip",
"_____no_output_____"
],
[
"!unzip horse2zebra.zip",
"_____no_output_____"
],
[
"!ls horse2zebra",
"_____no_output_____"
],
[
"import glob\npath = \"\"\nhorses_train = glob.glob(path + 'horse2zebra/trainA/*.jpg')\nzebras_train = glob.glob(path + 'horse2zebra/trainB/*.jpg')\nhorses_test = glob.glob(path + 'horse2zebra/testA/*.jpg')\nzebras_test = glob.glob(path + 'horse2zebra/testB/*.jpg')",
"_____no_output_____"
],
[
"len(horses_train), len(zebras_train), len(horses_test), len(zebras_test)",
"_____no_output_____"
],
[
"import cv2\nfor file in horses_train[:10]:\n img = cv2.imread(file)\n print (img.shape)",
"_____no_output_____"
]
],
[
[
"# Display few Samples",
"_____no_output_____"
]
],
[
[
"print (\"Horses\")\nfor k in range(2):\n plt.figure(figsize=(15, 15))\n for j in range(6):\n file = np.random.choice(horses_train)\n img = cv2.imread(file)\n img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)\n plt.subplot(660 + 1 + j)\n plt.imshow(img)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()\n\nprint (\"-\"*80)\nprint (\"Zebras\")\nfor k in range(2):\n plt.figure(figsize=(15, 15))\n for j in range(6):\n file = np.random.choice(zebras_train)\n img = cv2.imread(file)\n img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)\n plt.subplot(660 + 1 + j)\n plt.imshow(img)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Define Generator Model (Res-Net Like)",
"_____no_output_____"
]
],
[
[
"#Following function is taken from: https://keras.io/examples/generative/cyclegan/\n\nclass ReflectionPadding2D(tensorflow.keras.layers.Layer):\n \"\"\"Implements Reflection Padding as a layer.\n\n Args:\n padding(tuple): Amount of padding for the\n spatial dimensions.\n\n Returns:\n A padded tensor with the same type as the input tensor.\n \"\"\"\n\n def __init__(self, padding=(1, 1), **kwargs):\n self.padding = tuple(padding)\n super(ReflectionPadding2D, self).__init__(**kwargs)\n\n def call(self, input_tensor, mask=None):\n padding_width, padding_height = self.padding\n padding_tensor = [\n [0, 0],\n [padding_height, padding_height],\n [padding_width, padding_width],\n [0, 0],\n ]\n return tensorflow.pad(input_tensor, padding_tensor, mode=\"REFLECT\")",
"_____no_output_____"
],
[
"import tensorflow_addons as tfa",
"_____no_output_____"
],
[
"# Weights initializer for the layers.\nkernel_init = tensorflow.keras.initializers.RandomNormal(mean=0.0, stddev=0.02)\n# Gamma initializer for instance normalization.\ngamma_init = tensorflow.keras.initializers.RandomNormal(mean=0.0, stddev=0.02)",
"_____no_output_____"
],
[
"def custom_resnet_block(input_data, filters):\n x = ReflectionPadding2D()(input_data)\n x = tensorflow.keras.layers.Conv2D(filters, kernel_size=(3,3), padding='valid', kernel_initializer=kernel_init)(x)\n x = tfa.layers.InstanceNormalization()(x)\n x = tensorflow.keras.layers.Activation('relu')(x)\n\n x = ReflectionPadding2D()(x)\n x = tensorflow.keras.layers.Conv2D(filters, kernel_size=(3,3), padding='valid', kernel_initializer=kernel_init)(x)\n x = tfa.layers.InstanceNormalization()(x)\n\n x = tensorflow.keras.layers.Add()([x, input_data])\n return x",
"_____no_output_____"
],
[
"def make_generator():\n source_image = tensorflow.keras.layers.Input(shape=(256, 256, 3))\n\n x = ReflectionPadding2D(padding=(3, 3))(source_image)\n x = tensorflow.keras.layers.Conv2D(64, kernel_size=(7,7), kernel_initializer=kernel_init, use_bias=False)(x)\n x = tfa.layers.InstanceNormalization()(x)\n x = tensorflow.keras.layers.Activation('relu')(x)\n\n x = tensorflow.keras.layers.Conv2D(128, kernel_size=(3,3), strides=(2,2), padding='same', kernel_initializer=kernel_init)(x)\n x = tfa.layers.InstanceNormalization()(x)\n x = tensorflow.keras.layers.Activation('relu')(x)\n\n x = tensorflow.keras.layers.Conv2D(256, kernel_size=(3,3), strides=(2,2), padding='same', kernel_initializer=kernel_init)(x)\n x = tfa.layers.InstanceNormalization()(x)\n x = tensorflow.keras.layers.Activation('relu')(x)\n\n x = custom_resnet_block(x, 256)\n x = custom_resnet_block(x, 256)\n x = custom_resnet_block(x, 256)\n\n x = custom_resnet_block(x, 256)\n x = custom_resnet_block(x, 256)\n x = custom_resnet_block(x, 256)\n\n x = custom_resnet_block(x, 256)\n x = custom_resnet_block(x, 256)\n x = custom_resnet_block(x, 256)\n\n x = tensorflow.keras.layers.Conv2DTranspose(128, kernel_size=(3,3), strides=(2,2), padding='same', kernel_initializer=kernel_init)(x)\n x = tfa.layers.InstanceNormalization()(x)\n x = tensorflow.keras.layers.Activation('relu')(x)\n\n x = tensorflow.keras.layers.Conv2DTranspose(64, kernel_size=(3,3), strides=(2,2), padding='same', kernel_initializer=kernel_init)(x)\n x = tfa.layers.InstanceNormalization()(x)\n x = tensorflow.keras.layers.Activation('relu')(x)\n\n x = ReflectionPadding2D(padding=(3, 3))(x)\n x = tensorflow.keras.layers.Conv2D(3, kernel_size=(7,7), padding='valid')(x)\n x = tfa.layers.InstanceNormalization()(x)\n\n translated_image = tensorflow.keras.layers.Activation('tanh')(x)\n\n return source_image, translated_image\n\nsource_image, translated_image = make_generator()\ngenerator_network_AB = tensorflow.keras.models.Model(inputs=source_image, outputs=translated_image)\n\nsource_image, translated_image = make_generator()\ngenerator_network_BA = tensorflow.keras.models.Model(inputs=source_image, outputs=translated_image)\n\nprint (generator_network_AB.summary())",
"_____no_output_____"
]
],
[
[
"# Define Discriminator Network",
"_____no_output_____"
]
],
[
[
"def my_conv_layer(input_layer, filters, strides, bn=True):\n x = tensorflow.keras.layers.Conv2D(filters, kernel_size=(4,4), strides=strides, padding='same', kernel_initializer=kernel_init)(input_layer)\n x = tensorflow.keras.layers.LeakyReLU(alpha=0.2)(x)\n if bn:\n x = tfa.layers.InstanceNormalization()(x)\n return x",
"_____no_output_____"
],
[
" def make_discriminator():\n target_image_input = tensorflow.keras.layers.Input(shape=(256, 256, 3))\n\n x = my_conv_layer(target_image_input, 64, (2,2), bn=False)\n x = my_conv_layer(x, 128, (2,2))\n x = my_conv_layer(x, 256, (2,2))\n x = my_conv_layer(x, 512, (1,1))\n\n patch_features = tensorflow.keras.layers.Conv2D(1, kernel_size=(4,4), padding='same')(x)\n return target_image_input, patch_features\n\n\ntarget_image_input, patch_features = make_discriminator()\ndiscriminator_network_A = tensorflow.keras.models.Model(inputs=target_image_input, outputs=patch_features)\n\ntarget_image_input, patch_features = make_discriminator()\ndiscriminator_network_B = tensorflow.keras.models.Model(inputs=target_image_input, outputs=patch_features)\n\nprint (discriminator_network_A.summary())",
"_____no_output_____"
],
[
"adam_optimizer = tensorflow.keras.optimizers.Adam(learning_rate=0.0002, beta_1=0.5)\ndiscriminator_network_A.compile(loss='mse', optimizer=adam_optimizer, metrics=['accuracy'])\ndiscriminator_network_B.compile(loss='mse', optimizer=adam_optimizer, metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"# Define Cycle-GAN",
"_____no_output_____"
]
],
[
[
"source_image_A = tensorflow.keras.layers.Input(shape=(256, 256, 3))\nsource_image_B = tensorflow.keras.layers.Input(shape=(256, 256, 3))\n\n# Domain Transfer\nfake_B = generator_network_AB(source_image_A)\nfake_A = generator_network_BA(source_image_B)\n\n# Restoring original Domain\nget_back_A = generator_network_BA(fake_B)\nget_back_B = generator_network_AB(fake_A)\n\n# Get back Identical/Same Image\nget_same_A = generator_network_BA(source_image_A)\nget_same_B = generator_network_AB(source_image_B)\n \ndiscriminator_network_A.trainable=False\ndiscriminator_network_B.trainable=False\n\n# Tell Real vs Fake, for a given domain\nverify_A = discriminator_network_A(fake_A)\nverify_B = discriminator_network_B(fake_B)\n\ncycle_gan = tensorflow.keras.models.Model(inputs = [source_image_A, source_image_B], \\\n outputs = [verify_A, verify_B, get_back_A, get_back_B, get_same_A, get_same_B])\ncycle_gan.summary()",
"_____no_output_____"
]
],
[
[
"# Compiling Model",
"_____no_output_____"
]
],
[
[
"cycle_gan.compile(loss=['mse', 'mse', 'mae', 'mae', 'mae', 'mae'], loss_weights=[1, 1, 10, 10, 5, 5],\\\n optimizer=adam_optimizer)",
"_____no_output_____"
]
],
[
[
"# Define Data Generators",
"_____no_output_____"
]
],
[
[
"def horses_to_zebras(horses, generator_network):\n generated_samples = generator_network.predict_on_batch(horses)\n return generated_samples\n\ndef zebras_to_horses(zebras, generator_network):\n generated_samples = generator_network.predict_on_batch(zebras)\n return generated_samples\n\ndef get_horse_samples(batch_size):\n random_files = np.random.choice(horses_train, size=batch_size)\n images = []\n for file in random_files:\n img = cv2.imread(file)\n img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)\n images.append((img-127.5)/127.5)\n horse_images = np.array(images)\n return horse_images\n\ndef get_zebra_samples(batch_size):\n random_files = np.random.choice(zebras_train, size=batch_size)\n images = []\n for file in random_files:\n img = cv2.imread(file)\n img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)\n images.append((img-127.5)/127.5)\n zebra_images = np.array(images)\n return zebra_images\n\ndef show_generator_results_horses_to_zebras(generator_network_AB, generator_network_BA):\n images = []\n for j in range(5):\n file = np.random.choice(horses_test)\n img = cv2.imread(file)\n img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)\n images.append(img)\n\n print ('Input Horse Images')\n plt.figure(figsize=(13, 13))\n for j, img in enumerate(images):\n plt.subplot(550 + 1 + j)\n plt.imshow(img)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()\n\n print ('Translated (Horse -> Zebra) Images')\n translated = []\n plt.figure(figsize=(13, 13))\n for j, img in enumerate(images):\n img = (img-127.5)/127.5\n output = horses_to_zebras(np.array([img]), generator_network_AB)[0]\n translated.append(output)\n output = (output+1.0)/2.0\n plt.subplot(550 + 1 + j)\n plt.imshow(output)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()\n\n print ('Translated reverse ( Fake Zebras -> Fake Horses)')\n plt.figure(figsize=(13, 13))\n for j, img in enumerate(translated):\n output = zebras_to_horses(np.array([img]), generator_network_BA)[0]\n output = (output+1.0)/2.0\n plt.subplot(550 + 1 + j)\n plt.imshow(output)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()\n\ndef show_generator_results_zebras_to_horses(generator_network_AB, generator_network_BA):\n images = []\n for j in range(5):\n file = np.random.choice(zebras_test)\n img = cv2.imread(file)\n img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)\n images.append(img)\n\n print ('Input Zebra Images')\n plt.figure(figsize=(13, 13))\n for j, img in enumerate(images):\n plt.subplot(550 + 1 + j)\n plt.imshow(img)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()\n\n print ('Translated (Zebra -> Horse) Images')\n translated = []\n plt.figure(figsize=(13, 13))\n for j, img in enumerate(images):\n img = (img-127.5)/127.5\n output = zebras_to_horses(np.array([img]), generator_network_BA)[0]\n translated.append(output)\n output = (output+1.0)/2.0\n plt.subplot(550 + 1 + j)\n plt.imshow(output)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()\n\n print ('Translated reverse (Fake Horse -> Fake Zebra)')\n plt.figure(figsize=(13, 13))\n for j, img in enumerate(translated):\n output = horses_to_zebras(np.array([img]), generator_network_AB)[0]\n output = (output+1.0)/2.0\n plt.subplot(550 + 1 + j)\n plt.imshow(output)\n plt.axis('off')\n #plt.title(trainY[i])\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Training Cycle-GAN",
"_____no_output_____"
]
],
[
[
"len(horses_train), len(zebras_train)",
"_____no_output_____"
],
[
"epochs = 500\nbatch_size = 1\nsteps = 1067\n\nfor i in range(0, epochs):\n if i%5 == 0:\n show_generator_results_horses_to_zebras(generator_network_AB, generator_network_BA)\n print (\"-\"*100)\n show_generator_results_zebras_to_horses(generator_network_AB, generator_network_BA)\n for j in range(steps): \n # A == Horses\n # B == Zebras\n domain_A_images = get_horse_samples(batch_size)\n domain_B_images = get_zebra_samples(batch_size)\n\n fake_patch = np.zeros((batch_size, 32, 32, 1))\n real_patch = np.ones((batch_size, 32, 32, 1))\n \n fake_B_images = generator_network_AB(domain_A_images)\n fake_A_images = generator_network_BA(domain_B_images)\n \n # Updating Discriminator A weights\n discriminator_network_A.trainable=True\n discriminator_network_B.trainable=False\n loss_d_real_A = discriminator_network_A.train_on_batch(domain_A_images, real_patch)\n loss_d_fake_A = discriminator_network_A.train_on_batch(fake_A_images, fake_patch)\n \n loss_d_A = np.add(loss_d_real_A, loss_d_fake_A)/2.0\n \n # Updating Discriminator B weights\n discriminator_network_B.trainable=True\n discriminator_network_A.trainable=False\n loss_d_real_B = discriminator_network_B.train_on_batch(domain_B_images, real_patch)\n loss_d_fake_B = discriminator_network_B.train_on_batch(fake_B_images, fake_patch)\n \n loss_d_B = np.add(loss_d_real_B, loss_d_fake_B)/2.0\n \n # Make the Discriminator belive that these are real samples and calculate loss to train the generator\n \n discriminator_network_A.trainable=False\n discriminator_network_B.trainable=False\n \n # Updating Generator weights\n loss_g = cycle_gan.train_on_batch([domain_A_images, domain_B_images],\\\n [real_patch, real_patch, domain_A_images, domain_B_images, domain_A_images, domain_B_images])\n \n if j%100 == 0:\n print (\"Epoch:%.0f, Step:%.0f, DA-Loss:%.3f, DA-Acc:%.3f, DB-Loss:%.3f, DB-Acc:%.3f, G-Loss:%.3f\"\\\n %(i,j,loss_d_A[0],loss_d_A[1]*100,loss_d_B[0],loss_d_B[1]*100,loss_g[0]))\n ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a8cf399b37ba5a085e1bba9fc77eb7b3c78c1d7
| 27,002 |
ipynb
|
Jupyter Notebook
|
labs/Work-with-Data.ipynb
|
PeakIndicatorsHub/Getting-Started-On-Azure-ML
|
ee637ddfe7d213cf4759da0ed5f59c7382148870
|
[
"MIT"
] | null | null | null |
labs/Work-with-Data.ipynb
|
PeakIndicatorsHub/Getting-Started-On-Azure-ML
|
ee637ddfe7d213cf4759da0ed5f59c7382148870
|
[
"MIT"
] | null | null | null |
labs/Work-with-Data.ipynb
|
PeakIndicatorsHub/Getting-Started-On-Azure-ML
|
ee637ddfe7d213cf4759da0ed5f59c7382148870
|
[
"MIT"
] | 1 |
2021-02-17T08:54:32.000Z
|
2021-02-17T08:54:32.000Z
| 43.411576 | 570 | 0.631361 |
[
[
[
"# Work with Data\nData is the foundation on which machine learning models are built. Managing data centrally in the cloud, and making it accessible to teams of data scientists who are running experiments and training models on multiple workstations and compute targets is an important part of any professional data science solution.\n\nIn this notebook, you'll explore two Azure Machine Learning objects for working with data: *datastores*, and *datasets*.\n\n## Before you start\nIf you haven't already done so, you must install the latest version of the **azureml-sdk** and **azureml-widgets** packages before running this notebook. To do this, run the cell below and then **restart the kernel** before running the subsequent cells.",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade azureml-sdk azureml-widgets",
"_____no_output_____"
]
],
[
[
"## Connect to your workspace\nWith the latest version of the SDK installed, now you're ready to connect to your workspace.\n\n**Note:** If you haven't already established an authenticated session with your Azure subscription, you'll be prompted to authenticate by clicking a link, entering an authentication code, and signing into Azure.",
"_____no_output_____"
]
],
[
[
"\nimport azureml.core\nfrom azureml.core import Workspace\n\n# Load the workspace from the saved config file\nws = Workspace.from_config()\nprint('Ready to use Azure ML {} to work with {}'.format(azureml.core.VERSION, ws.name))",
"_____no_output_____"
]
],
[
[
"## Work with datastores\nIn Azure ML, *datastores* are references to storage locations, such as Azure Storage blob containers. Every workspace has a default datastore - usually the Azure storage blob container that was created with the workspace. If you need to work with data that is stored in different locations, you can add custom datastores to your workspace and set any of them to be the default.\n\n### View datastores\nRun the following code to determine the datastores in your workspace:",
"_____no_output_____"
]
],
[
[
"# Get the default datastore\ndefault_ds = ws.get_default_datastore()\n\n# Enumerate all datastores, indicating which is the default\nfor ds_name in ws.datastores:\n print(ds_name, \"- Default =\", ds_name == default_ds.name)",
"_____no_output_____"
]
],
[
[
"You can also view and manage datastores in your workspace on the **Datastores** page for your workspace in [Azure Machine Learning studio](https://ml.azure.com/).\n\n### Upload data to a datastore\n\nNow that you have determined the available datastores, you can upload files from your local file system to a datastore so that it will be accessible to experiments running in the workspace, regardless of where the experiment script is actually being run",
"_____no_output_____"
]
],
[
[
"default_ds.upload_files(files=['./data/diabetes.csv', './data/diabetes2.csv'], # Upload the diabetes csv files in /data\n target_path='diabetes-data/', # Put it in a folder path in the datastore\n overwrite=True, # Replace existing files of the same name\n show_progress=True)",
"_____no_output_____"
]
],
[
[
"\n## Work with datasets\nAzure Machine Learning provides an abstraction for data in the form of *datasets*. A dataset is a versioned reference to a specific set of data that you may want to use in an experiment. Datasets can be *tabular* or *file-based*.\n\n### Create a tabular dataset\nLet's create a dataset from the diabetes data you uploaded to the datastore, and view the first 20 records. In this case, the data is in a structured format in a CSV file, so we'll use a tabular dataset.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Dataset\n\n# Get the default datastore\ndefault_ds = ws.get_default_datastore()\n\n#Create a tabular dataset from the path on the datastore (this may take a short while)\ntab_data_set = Dataset.Tabular.from_delimited_files(path=(default_ds, 'diabetes-data/*.csv'))\n\n# Display the first 20 rows as a Pandas dataframe\ntab_data_set.take(20).to_pandas_dataframe()",
"_____no_output_____"
]
],
[
[
"As you can see in the code above, it's easy to convert a tabular dataset to a Pandas dataframe, enabling you to work with the data using common python techniques.",
"_____no_output_____"
],
[
"### Create a file Dataset\n\nThe dataset you created is a *tabular* dataset that can be read as a dataframe containing all of the data in the structured files that are included in the dataset definition. This works well for tabular data, but in some machine learning scenarios you might need to work with data that is unstructured; or you may simply want to handle reading the data from files in your own code. To accomplish this, you can use a file dataset, which creates a list of file paths in a virtual mount point, which you can use to read the data in the files.",
"_____no_output_____"
]
],
[
[
"#Create a file dataset from the path on the datastore (this may take a short while)\nfile_data_set = Dataset.File.from_files(path=(default_ds, 'diabetes-data/*.csv'))\n\n# Get the files in the dataset\nfor file_path in file_data_set.to_path():\n print(file_path)",
"_____no_output_____"
]
],
[
[
"### Register datasets\nNow that you have created datasets that reference the diabetes data, you can register them to make them easily accessible to any experiment being run in the workspace.\n\nWe'll register the tabular dataset as **diabetes dataset**, and the file dataset as **diabetes files**.",
"_____no_output_____"
]
],
[
[
"# Register the tabular dataset\ntry:\n tab_data_set = tab_data_set.register(workspace=ws, \n name='diabetes dataset',\n description='diabetes data',\n tags = {'format':'CSV'},\n create_new_version=True)\nexcept Exception as ex:\n print(ex)\n\n# Register the file dataset\ntry:\n file_data_set = file_data_set.register(workspace=ws,\n name='diabetes file dataset',\n description='diabetes files',\n tags = {'format':'CSV'},\n create_new_version=True)\nexcept Exception as ex:\n print(ex)\n\nprint('Datasets registered')",
"_____no_output_____"
]
],
[
[
"You can view and manage datasets on the **Datasets** page for your workspace in [Azure Machine Learning studio](https://ml.azure.com/). You can also get a list of datasets from the workspace object:",
"_____no_output_____"
]
],
[
[
"print(\"Datasets:\")\nfor dataset_name in list(ws.datasets.keys()):\n dataset = Dataset.get_by_name(ws, dataset_name)\n print(\"\\t\", dataset.name, 'version', dataset.version)",
"_____no_output_____"
]
],
[
[
"\nThe ability to version datasets enables you to redefine datasets without breaking existing experiments or pipelines that rely on previous definitions. By default, the latest version of a named dataset is returned, but you can retrieve a specific version of a dataset by specifying the version number, like this:\n\n`dataset_v1 = Dataset.get_by_name(ws, 'diabetes dataset', version = 1)`",
"_____no_output_____"
],
[
"### Train a model from a tabular dataset\nNow that you have datasets, you're ready to start training models from them. You can pass datasets to scripts as inputs in the estimator being used to run the script.\n\nRun the following two code cells to create:\n\n1. A folder named **diabetes_training_from_tab_dataset**\n2. A script that trains a classification model by using a tabular dataset that is passed to is as an argument.",
"_____no_output_____"
]
],
[
[
"\nimport os\n\n# Create a folder for the experiment files\nexperiment_folder = 'diabetes_training_from_tab_dataset'\nos.makedirs(experiment_folder, exist_ok=True)\nprint(experiment_folder, 'folder created')",
"_____no_output_____"
],
[
"%%writefile $experiment_folder/diabetes_training.py\n# Import libraries\nimport os\nimport argparse\nfrom azureml.core import Run, Dataset\nimport pandas as pd\nimport numpy as np\nimport joblib\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import roc_auc_score\nfrom sklearn.metrics import roc_curve\n\n# Get the script arguments (regularization rate and training dataset ID)\nparser = argparse.ArgumentParser()\nparser.add_argument('--regularization', type=float, dest='reg_rate', default=0.01, help='regularization rate')\nparser.add_argument(\"--input-data\", type=str, dest='training_dataset_id', help='training dataset')\nargs = parser.parse_args()\n\n# Set regularization hyperparameter (passed as an argument to the script)\nreg = args.reg_rate\n\n# Get the experiment run context\nrun = Run.get_context()\n\n# Get the training dataset\nprint(\"Loading Data...\")\ndiabetes = run.input_datasets['training_data'].to_pandas_dataframe()\n\n# Separate features and labels\nX, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values\n\n# Split data into training set and test set\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)\n\n# Train a logistic regression model\nprint('Training a logistic regression model with regularization rate of', reg)\nrun.log('Regularization Rate', np.float(reg))\nmodel = LogisticRegression(C=1/reg, solver=\"liblinear\").fit(X_train, y_train)\n\n# calculate accuracy\ny_hat = model.predict(X_test)\nacc = np.average(y_hat == y_test)\nprint('Accuracy:', acc)\nrun.log('Accuracy', np.float(acc))\n\n# calculate AUC\ny_scores = model.predict_proba(X_test)\nauc = roc_auc_score(y_test,y_scores[:,1])\nprint('AUC: ' + str(auc))\nrun.log('AUC', np.float(auc))\n\nos.makedirs('outputs', exist_ok=True)\n# note file saved in the outputs folder is automatically uploaded into experiment record\njoblib.dump(value=model, filename='outputs/diabetes_model.pkl')\n\nrun.complete()",
"_____no_output_____"
]
],
[
[
"**Note:** In the script, the dataset is passed as a parameter (or argument). In the case of a tabular dataset, this argument will contain the ID of the registered dataset; so you could write code in the script to get the experiment's workspace from the run context, and then get the dataset using its ID; like this:\n\n`run = Run.get_context()`\n\n`ws = run.experiment.workspace`\n\n`dataset = Dataset.get_by_id(ws, id=args.training_dataset_id)`\n\n`diabetes = dataset.to_pandas_dataframe()`\n\nHowever, Azure Machine Learning runs automatically identify arguments that reference named datasets and add them to the run's **input_datasets** collection, so you can also retrieve the dataset from this collection by specifying its \"friendly name\" (which as you'll see shortly, is specified in the argument definition in the script run configuration for the experiment). This is the approach taken in the script above.\n\nNow you can run a script as an experiment, defining an argument for the training dataset, which is read by the script.\n\n**Note:** The **Dataset** class depends on some components in the **azureml-dataprep** package, which includes optional support for **pandas** that is used by the **to_pandas_dataframe()** method. So you need to include this package in the environment where the training experiment will be run.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Experiment, ScriptRunConfig, Environment\nfrom azureml.core.conda_dependencies import CondaDependencies\nfrom azureml.widgets import RunDetails\n\n\n# Create a Python environment for the experiment\nsklearn_env = Environment(\"sklearn-env\")\n\n# Ensure the required packages are installed (we need scikit-learn, Azure ML defaults, and Azure ML dataprep)\npackages = CondaDependencies.create(conda_packages=['scikit-learn','pip'],\n pip_packages=['azureml-defaults','azureml-dataprep[pandas]'])\nsklearn_env.python.conda_dependencies = packages\n\n# Get the training dataset\ndiabetes_ds = ws.datasets.get(\"diabetes dataset\")\n\n# Create a script config\nscript_config = ScriptRunConfig(source_directory=experiment_folder,\n script='diabetes_training.py',\n arguments = ['--regularization', 0.1, # Regularizaton rate parameter\n '--input-data', diabetes_ds.as_named_input('training_data')], # Reference to dataset\n environment=sklearn_env) \n\n# submit the experiment\nexperiment_name = 'mslearn-train-diabetes'\nexperiment = Experiment(workspace=ws, name=experiment_name)\nrun = experiment.submit(config=script_config)\nRunDetails(run).show()\nrun.wait_for_completion()",
"_____no_output_____"
]
],
[
[
"**Note:** The **--input-data** argument passes the dataset as a *named input* that includes a *friendly name* for the dataset, which is used by the script to read it from the **input_datasets** collection in the experiment run. The string value in the **--input-data** argument is actually the registered dataset's ID. As an alternative approach, you could simply pass *diabetes_ds.id*, in which case the script can access the dataset ID from the script arguments and use it to get the dataset from the workspace, but not from the **input_datasets** collection.\n\nThe first time the experiment is run, it may take some time to set up the Python environment - subsequent runs will be quicker.\n\nWhen the experiment has completed, in the widget, view the **azureml-logs/70_driver_log.txt** output log and the metrics generated by the run.\n\n### Register the trained model\nAs with any training experiment, you can retrieve the trained model and register it in your Azure Machine Learning workspace",
"_____no_output_____"
]
],
[
[
"from azureml.core import Model\n\nrun.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',\n tags={'Training context':'Tabular dataset'}, properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})\n\nfor model in Model.list(ws):\n print(model.name, 'version:', model.version)\n for tag_name in model.tags:\n tag = model.tags[tag_name]\n print ('\\t',tag_name, ':', tag)\n for prop_name in model.properties:\n prop = model.properties[prop_name]\n print ('\\t',prop_name, ':', prop)\n print('\\n')",
"_____no_output_____"
]
],
[
[
"\n### Train a model from a file dataset\nYou've seen how to train a model using training data in a tabular dataset; but what about a file dataset?\n\nWhen you're using a file dataset, the dataset argument passed to the script represents a mount point containing file paths. How you read the data from these files depends on the kind of data in the files and what you want to do with it. In the case of the diabetes CSV files, you can use the Python glob module to create a list of files in the virtual mount point defined by the dataset, and read them all into Pandas dataframes that are concatenated into a single dataframe.\n\nRun the following two code cells to create:\n\n1. A folder named **diabetes_training_from_file_dataset**\n\n2. A script that trains a classification model by using a file dataset that is passed to is as an *input*.",
"_____no_output_____"
]
],
[
[
"import os\n\n# Create a folder for the experiment files\nexperiment_folder = 'diabetes_training_from_file_dataset'\nos.makedirs(experiment_folder, exist_ok=True)\nprint(experiment_folder, 'folder created')",
"_____no_output_____"
],
[
"%%writefile $experiment_folder/diabetes_training.py\n# Import libraries\nimport os\nimport argparse\nfrom azureml.core import Dataset, Run\nimport pandas as pd\nimport numpy as np\nimport joblib\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import roc_auc_score\nfrom sklearn.metrics import roc_curve\nimport glob\n\n# Get script arguments (rgularization rate and file dataset mount point)\nparser = argparse.ArgumentParser()\nparser.add_argument('--regularization', type=float, dest='reg_rate', default=0.01, help='regularization rate')\nparser.add_argument('--input-data', type=str, dest='dataset_folder', help='data mount point')\nargs = parser.parse_args()\n\n# Set regularization hyperparameter (passed as an argument to the script)\nreg = args.reg_rate\n\n# Get the experiment run context\nrun = Run.get_context()\n\n# load the diabetes dataset\nprint(\"Loading Data...\")\ndata_path = run.input_datasets['training_files'] # Get the training data path from the input\n# (You could also just use args.data_folder if you don't want to rely on a hard-coded friendly name)\n\n# Read the files\nall_files = glob.glob(data_path + \"/*.csv\")\ndiabetes = pd.concat((pd.read_csv(f) for f in all_files), sort=False)\n\n# Separate features and labels\nX, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values\n\n# Split data into training set and test set\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)\n\n# Train a logistic regression model\nprint('Training a logistic regression model with regularization rate of', reg)\nrun.log('Regularization Rate', np.float(reg))\nmodel = LogisticRegression(C=1/reg, solver=\"liblinear\").fit(X_train, y_train)\n\n# calculate accuracy\ny_hat = model.predict(X_test)\nacc = np.average(y_hat == y_test)\nprint('Accuracy:', acc)\nrun.log('Accuracy', np.float(acc))\n\n# calculate AUC\ny_scores = model.predict_proba(X_test)\nauc = roc_auc_score(y_test,y_scores[:,1])\nprint('AUC: ' + str(auc))\nrun.log('AUC', np.float(auc))\n\nos.makedirs('outputs', exist_ok=True)\n# note file saved in the outputs folder is automatically uploaded into experiment record\njoblib.dump(value=model, filename='outputs/diabetes_model.pkl')\n\nrun.complete()",
"_____no_output_____"
]
],
[
[
"Just as with tabular datasets, you can retrieve a file dataset from the **input_datasets** collection by using its friendly name. You can also retrieve it from the script argument, which in the case of a file dataset contains a mount path to the files (rather than the dataset ID passed for a tabular dataset).\n\nNext we need to change the way we pass the dataset to the script - it needs to define a path from which the script can read the files. You can use either the **as_download** or **as_mount** method to do this. Using **as_download** causes the files in the file dataset to be downloaded to a temporary location on the compute where the script is being run, while **as_mount** creates a mount point from which the files can be streamed directly from the datasetore.\n\nYou can combine the access method with the **as_named_input** method to include the dataset in the **input_datasets** collection in the experiment run (if you omit this, for example by setting the argument to *diabetes_ds.as_mount()*, the script will be able to access the dataset mount point from the script arguments, but not from the **input_datasets** collection).",
"_____no_output_____"
]
],
[
[
"from azureml.core import Experiment\nfrom azureml.widgets import RunDetails\n\n\n# Get the training dataset\ndiabetes_ds = ws.datasets.get(\"diabetes file dataset\")\n\n# Create a script config\nscript_config = ScriptRunConfig(source_directory=experiment_folder,\n script='diabetes_training.py',\n arguments = ['--regularization', 0.1, # Regularizaton rate parameter\n '--input-data', diabetes_ds.as_named_input('training_files').as_download()], # Reference to dataset location\n environment=sklearn_env) # Use the environment created previously\n\n# submit the experiment\nexperiment_name = 'mslearn-train-diabetes'\nexperiment = Experiment(workspace=ws, name=experiment_name)\nrun = experiment.submit(config=script_config)\nRunDetails(run).show()\nrun.wait_for_completion()",
"_____no_output_____"
]
],
[
[
"\nWhen the experiment has completed, in the widget, view the **azureml-logs/70_driver_log.txt** output log to verify that the files in the file dataset were downloaded to a temporary folder to enable the script to read the files.",
"_____no_output_____"
],
[
"### Register the trained model\nOnce again, you can register the model that was trained by the experiment.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Model\n\nrun.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',\n tags={'Training context':'File dataset'}, properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})\n\nfor model in Model.list(ws):\n print(model.name, 'version:', model.version)\n for tag_name in model.tags:\n tag = model.tags[tag_name]\n print ('\\t',tag_name, ':', tag)\n for prop_name in model.properties:\n prop = model.properties[prop_name]\n print ('\\t',prop_name, ':', prop)\n print('\\n')",
"_____no_output_____"
]
],
[
[
"**More Information:** For more information about training with datasets, see [Training with Datasets](https://docs.microsoft.com/azure/machine-learning/how-to-train-with-datasets) in the Azure ML documentation.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a8d0729074000d7c8a75620c8fa19be34861f29
| 105,095 |
ipynb
|
Jupyter Notebook
|
ML/Lectures/02_binomial_polls.ipynb
|
TheFebrin/Machine-Learning
|
3e58b89315960e7d4896e44075a8105fcb78f0c0
|
[
"MIT"
] | null | null | null |
ML/Lectures/02_binomial_polls.ipynb
|
TheFebrin/Machine-Learning
|
3e58b89315960e7d4896e44075a8105fcb78f0c0
|
[
"MIT"
] | null | null | null |
ML/Lectures/02_binomial_polls.ipynb
|
TheFebrin/Machine-Learning
|
3e58b89315960e7d4896e44075a8105fcb78f0c0
|
[
"MIT"
] | null | null | null | 105,095 | 105,095 | 0.907569 |
[
[
[
"%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"import scipy.stats",
"_____no_output_____"
]
],
[
[
"# Introduction\n\nDuring the first lecture we have seen that the goal of machine learning is to train (learn/fit) a \n**model** on a **dataset** such that we will be able to answer several questions about the data using\nthe model. Some useful questions are:\n1. Predict a target $y$ for a new input $x$: predict what is in an image, recognize some audio sample, tell if the stock price will go up or down...\n2. Generate new sample $x$ similar to those form the training dataset. Alternatively, given part of a generate the other part (e.g. given half of an image generate the other half).\n\nHistorically, similar questions were considered by statisticians. In fact, machine learning is very similar to statistics. Some people claim that there is very little difference between the two, and a tongue-in-cheek definition of machine learning is \"statistics without checking for assumptions\", to which ML practitioners reply that they are at least able to solve problems that are too complex for a through and formal statistical analysis. \n\nFor a more in-depth discussion I highly recommend the [\"Two cultures\"](https://projecteuclid.org/euclid.ss/1009213726) essay by Leo Breiman.\n\nDue to the similarity of the two fields we will today explore a few examples of statistical inference. Some of the resulting concepts (maximum likelihood, interpreting the outputs of a model as probabilities) will be used through the semester.",
"_____no_output_____"
],
[
"# Statistical Inference\n\nConsider the polling problem:\n1. There exists **a population** of individuals (e.g. voters).\n2. The individuals have a voting preference (party A or B).\n3. We want the fraction $\\phi$ of voters that prefer A.\n4. But we don't want to ask everyone (which means holding an election)!\n\nInstead we want to conduct a poll (choose a **sample** of people \nand get their mean preference $\\bar\\phi$).\n\nQuestions:\n1. How are $\\phi$ and $\\bar\\phi$ related?\n2. What is our error?\n3. How many persons do we need to ask to achieve a desired error?",
"_____no_output_____"
],
[
"# Polling\n\nSuppose there is a large population of individuals, that support either candidate A or candidate B. We want to establish the fraction of supporters of A in the population $\\phi$.\n\nWe will conduct an opinion poll asking about the support for each party. We will choose randomly a certain number of people, $n$, and ask them about their candidates.\n\nWe want to use the results of the poll to establish:\n1. an estimate of the true population parameter $\\phi$\n2. our confidence about the interval",
"_____no_output_____"
],
[
"## Sampling Model\nFirst, we define a formal model of sampling. We will assume that the population is much bigger than the small sample. Thus we will assume a *sampling with replacement* model: each person is selected independently at random from the full population. We call such a sample IID (Independent Identically Distributed).\n\nHaving the sampling model we establish that the number of supporters of A in the sample follows a *binomial distribution* with:\n* poll size == $n$ == number of samples,\n* fraction of A's supporters == $\\phi$ == success rate.\n\nFor the binomial distribution with $n$ trials and probability of success $\\phi$ the expected number of successes is $n\\phi$ and the variance is $n\\phi(1-\\phi)$. \n\nAlternatively, the *fraction* of successes in the sample has the expected value $\\phi$ and variance $\\frac{\\phi(1-\\phi)}{n}$. \n\nLets plot the PMF (Probability Mass Function) of the number of successes.",
"_____no_output_____"
]
],
[
[
"# Poll variability check: draw samples form the binomial distribution\n\nn = 50\nphi = 0.55\n\n# Simulate a few polls\nfor _ in range(10):\n sample = random.rand(n)<phi\n print (\"Drawn %d samples. Observed success rate: %.2f (true rate: %.2f)\" % \n (n, 1.0*sample.sum()/n, phi))",
"Drawn 50 samples. Observed success rate: 0.68 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.62 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.48 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.56 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.52 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.50 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.56 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.46 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.46 (true rate: 0.55)\nDrawn 50 samples. Observed success rate: 0.60 (true rate: 0.55)\n"
],
[
"# model parameters\nn = 10\nphi = 0.55\n\n# the binomial distribution\nmodel = scipy.stats.binom(n=n, p=phi)\nx = arange(n+1)\n\n# plot the PMF - probability mass function\nstem(x, model.pmf(x), 'b', label='Binomial PMF')\n\n# plot the normal approximation\nmu = phi * n\nstdev = sqrt(phi*(1-phi) * n)\nmodel_norm = scipy.stats.norm(mu, stdev)\nx_cont = linspace(x[0], x[-1], 1000)\nplot(x_cont, model_norm.pdf(x_cont), 'r', label='Norm approx.')\n\naxvline(mu, *xlim(), color='g', label='Mean')\n\nlegend(loc='upper left')",
"_____no_output_____"
]
],
[
[
"## Parameter Estimation\n\nIn Statistics and Machine Learning we only have access to the sample. The goal is to learn something useful about the unknown population. Here we are interested in the true heads probability $\\phi$.\n\nThe MLE (Maximum Likelihood Estimator) for $\\phi$ is just the sample mean $\\bar\\phi$. However, how precise it is? We want the (sample dependent) confidence interval around the sample mean, such that in 95% of experiments (samples taken), the true unknown population parameter $\\phi$ is in the confidence interval.\n\nFormally we want to find $\\bar\\phi$ and $\\epsilon$ such that $P(\\bar\\phi-\\epsilon \\leq \\phi \\leq \\bar\\phi + \\epsilon) > 0.95$ or, equivalently, such that $P(|\\phi-\\bar\\phi| \\leq \\epsilon) > 0.95$.\n\nNote: from the sampling model we know that for a large enough sample (>15 persons) the random variable denoting the sample mean $\\bar\\phi$ is approximately normally distributed with mean $\\phi$ and standard deviation $\\sigma = \\sqrt{(\\phi(1-\\phi)/n)}$. However we do not know $\\phi$. When designing the experiment, we can take the worse value, which is 0.5. Alternatively, we can plug for $\\phi$ the estimated sample mean $\\bar\\phi$. Note: we are being too optimistic here, but the error will be small.\n\nFor a standard normal random variable (mean 0 and standard deviation 1) 96% of samples fall within the range $\\pm 1.96$. \n\nTherefore the confidence interval is approximately $\\bar\\phi \\pm 1.96\\sqrt{\\frac{\\bar\\phi(1-\\bar\\phi)}{n}}$.\n",
"_____no_output_____"
]
],
[
[
"phi=0.55\nn=100\nn_experiments=1000\n\nsamples = rand(n_experiments, n)<phi\nphi_bar = samples.mean(1)\n\nhist(phi_bar, bins=20, label='observed $\\\\bar\\\\phi$')\naxvline([phi], color='r', label='true $\\\\phi$')\ntitle('Histgram of sample means $\\\\bar\\\\phi$ form %d experiments.\\n'\n 'Model: %d trials, %.2f prob of success'%(n_experiments,n,phi))\nlegend()\nxlim(phi-0.15, phi+0.15)\n\nconfidence_intervals = zeros((n_experiments, 2))\nconfidence_intervals[:,0] = phi_bar - 1.96*np.sqrt(phi_bar*(1-phi_bar)/n)\nconfidence_intervals[:,1] = phi_bar + 1.96*np.sqrt(phi_bar*(1-phi_bar)/n)\n\n#note: this also works, can you exmplain how the formula works in numpy?\nconfidence_intervals2 = phi_bar[:,None] + [-1.96, 1.96] * np.sqrt(phi_bar*(1-phi_bar)/n).reshape(-1,1)\nassert np.abs(confidence_intervals-confidence_intervals2).max()==0\n\ngood_experiments = (confidence_intervals[:,0]<phi) & (confidence_intervals[:,1]>phi)\n\nprint (\"Average confidence interval is phi_bar +-%.3f\" \n % ((confidence_intervals[:,1]-confidence_intervals[:,0]).mean()/2.0,))\n\nprint (\"Out of %d experiments, the true phi fell into the confidence interval %d times.\"\n % (n_experiments, good_experiments.sum()))",
"Average confidence interval is phi_bar +-0.097\nOut of 1000 experiments, the true phi fell into the confidence interval 961 times.\n"
]
],
[
[
"## Bootstrap estimation of confidence interval",
"_____no_output_____"
]
],
[
[
"# Here we make a bootstrap analysis of one experiment\nn_bootstraps = 200\nexp_id = 1\nexp0 = samples[exp_id]\n\n# sample answers with replacement\nbootstrap_idx = np.random.randint(low=0, high=n, size=(n_bootstraps, n))\nexp0_bootstraps = exp0[bootstrap_idx]\n\n# compute the mean in each bootstrap sample\nexp0_bootstrap_means = exp0_bootstraps.mean(1)\n\n# Estimate the confidence interval by taking the 2.5 and 97.5 percentile\nsorted_bootstrap_means = np.sort(exp0_bootstrap_means)\nbootstrap_conf_low, bootstrap_conf_high = sorted_bootstrap_means[\n [int(0.025 * n_bootstraps), int(0.975 * n_bootstraps)]]\n\nhist(exp0_bootstrap_means, bins=20, label='bootstrap estims of $\\phi$')\naxvline(phi, 0, 1, label='$\\\\phi$', color='red')\naxvline(phi_bar[exp_id], 0, 1, label='$\\\\bar{\\\\phi}$', color='green')\naxvspan(confidence_intervals[exp_id, 0], confidence_intervals[exp_id, 1], # ymin=0.5, ymax=1.0, \n alpha=0.2, label='theoretical 95% conf int', color='green')\naxvspan(bootstrap_conf_low, bootstrap_conf_high, # ymin=0.0, ymax=0.5, \n alpha=0.2, label='bootsrap 95% conf int', color='blue')\nlegend()\n_ = xlim(phi-0.15, phi+0.15)\ntitle('Theoretical and bootstrap confidence intervals')",
"_____no_output_____"
]
],
[
[
"## Practical conclusions about polls\nPractical outcome: in the worst case ($\\phi=0.5$) the 95% confidence interval is $\\pm 1.96\\sqrt{\\frac{0.5(1-0.5)}{n}} = \\pm \\frac{0.975}{\\sqrt{n}}$. To get the usually acceptable polling error of 3 percentage points, one needs to sample 1056 persons. Polling companies typically ask between 1000-3000 persons.",
"_____no_output_____"
],
[
"Questions:\n1. How critical is the IID sampling assumption?\n2. What do you think is a larger problem: approximating the PDF with a Gaussian distribution, or people lying in the questionnaire?",
"_____no_output_____"
],
[
"# Bayesian reasoning\n\nWe will treat $\\phi$ - the unknown fraction of A supporters in the population as a random variable. Its probability distribution will express *our subjective* uncertainty about its value.\n\nWe will need to start with a *prior* assumption about our belief of $\\phi$. For convenience we will choose a *conjugate prior*, the Beta distribution, because the formula for its PDF is similar to the formula for the likelihood.",
"_____no_output_____"
]
],
[
[
"support = linspace(0,1,512)\n\nA=1\nB=1\n\nplot(support, scipy.stats.beta.pdf(support, A,B))\ntitle(\"Prior: Beta(%.1f, %.1f) distribution\" %(A,B))",
"_____no_output_____"
]
],
[
[
"Then we will collect samples, and after each sample update our belief about $p$.",
"_____no_output_____"
]
],
[
[
"n_successes = 0\nn_failures = 0\nphi = 0.6",
"_____no_output_____"
],
[
"for _ in range(10):\n if rand() < phi:\n n_successes += 1\n else:\n n_failures +=1\n\nplot(support, scipy.stats.beta.pdf(support, A+n_successes, B+n_failures), label='posterior')\naxvline(phi, color='r', label='True $\\\\phi$')\nconf_int_low, conf_int_high = scipy.stats.beta.ppf((0.025,0.975), A+n_successes, B+n_failures)\naxvspan(conf_int_low, conf_int_high, alpha=0.2, label='95% conf int')\ntitle(\"Posterior after seeing %d successes and %d failures\\n\"\n \"Prior pseudo-counts: A=%.1f, B=%.1f\\n\"\n \"MAP estimate: %f, MLE estimate: %f\\n\"\n \"conf_int: (%f, %f)\"% (n_successes, n_failures, A, B, \n 1.0*(A+n_successes-1)/(A+n_successes+B+n_failures-2),\n 1.0*n_successes/(n_successes+n_failures),\n conf_int_low, conf_int_high))\nlegend()",
"_____no_output_____"
]
],
[
[
"Please note: in the Bayesian framework we treat the quantities we want to estimate as random variables. \n\nWe need to define our prior beliefs about them. In the example, the prior was a Beta distribution.\n\nAfter seeing the data we update our belief about the world. In the example, this is vary easy - we keep running counts of the number of failures and successes observed. We update them seeing the data. The prior conveniently can be treated as *pseudo-counts*. \n\nTo summarize the distribution over the parameter, we typically take its mode (the most likely value), calling the approach MAP (Maximum a Posteriori).",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8d0c67b1495596abd4e8e401ef92b1b53fe5ae
| 24,460 |
ipynb
|
Jupyter Notebook
|
notebooks/Determine best dynamic min_samples factor.ipynb
|
CitizenScienceInAstronomyWorkshop/P4_sandbox
|
d04f688945848c0627f67496a55a654d661fb758
|
[
"0BSD"
] | 1 |
2016-09-29T21:12:51.000Z
|
2016-09-29T21:12:51.000Z
|
notebooks/Determine best dynamic min_samples factor.ipynb
|
CitizenScienceInAstronomyWorkshop/P4_sandbox
|
d04f688945848c0627f67496a55a654d661fb758
|
[
"0BSD"
] | 47 |
2015-08-10T05:57:27.000Z
|
2020-06-12T21:21:20.000Z
|
notebooks/Determine best dynamic min_samples factor.ipynb
|
CitizenScienceInAstronomyWorkshop/P4_sandbox
|
d04f688945848c0627f67496a55a654d661fb758
|
[
"0BSD"
] | 1 |
2015-08-16T19:20:48.000Z
|
2015-08-16T19:20:48.000Z
| 161.986755 | 21,432 | 0.902494 |
[
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"from planet4 import io, plotting, markings",
"_____no_output_____"
],
[
"db = io.DBManager()",
"_____no_output_____"
],
[
"ids = db.image_ids",
"_____no_output_____"
],
[
"sample = np.random.choice(ids, 300)",
"_____no_output_____"
],
[
"results = []\nfor s in sample:\n d = {}\n d['image_id'] = s\n data = db.get_image_id_markings(s)\n unique_users = data.user_name.nunique()\n d['n_users'] = unique_users\n not_logged_in_users = []\n logged_in_users = []\n for user in data.user_name.unique():\n val = len(data[data.user_name==user])\n if user.startswith('not-logged-in'):\n not_logged_in_users.append(val)\n else:\n logged_in_users.append(val)\n d['logged_in'] = np.mean(logged_in_users)\n d['not_logged_in'] = np.mean(not_logged_in_users)\n results.append(d)",
"_____no_output_____"
],
[
"pd.DataFrame(results).plot(kind='scatter', x='not_logged_in', y='logged_in')\nplt.plot(np.arange(30), np.arange(30))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8d10c07e3244a87cad029ab29ae47897c445cd
| 54,289 |
ipynb
|
Jupyter Notebook
|
3. Natural Language Processing with Sequence Models/Week 2/C3_W2_Assignment.ipynb
|
aditya-bhat/Natural-Language-Processing-Specialization
|
05d4de852ad631ce027e18f3b1404141a31edce6
|
[
"MIT"
] | null | null | null |
3. Natural Language Processing with Sequence Models/Week 2/C3_W2_Assignment.ipynb
|
aditya-bhat/Natural-Language-Processing-Specialization
|
05d4de852ad631ce027e18f3b1404141a31edce6
|
[
"MIT"
] | null | null | null |
3. Natural Language Processing with Sequence Models/Week 2/C3_W2_Assignment.ipynb
|
aditya-bhat/Natural-Language-Processing-Specialization
|
05d4de852ad631ce027e18f3b1404141a31edce6
|
[
"MIT"
] | null | null | null | 37.990903 | 620 | 0.576415 |
[
[
[
"# Assignment 2: Deep N-grams\n\nWelcome to the second assignment of course 3. In this assignment you will explore Recurrent Neural Networks `RNN`.\n- You will be using the fundamentals of google's [trax](https://github.com/google/trax) package to implement any kind of deeplearning model. \n\nBy completing this assignment, you will learn how to implement models from scratch:\n- How to convert a line of text into a tensor\n- Create an iterator to feed data to the model\n- Define a GRU model using `trax`\n- Train the model using `trax`\n- Compute the accuracy of your model using the perplexity\n- Predict using your own model",
"_____no_output_____"
],
[
"## Outline\n\n- [Overview](#0)\n- [Part 1: Importing the Data](#1)\n - [1.1 Loading in the data](#1.1)\n - [1.2 Convert a line to tensor](#1.2)\n - [Exercise 01](#ex01)\n - [1.3 Batch generator](#1.3)\n - [Exercise 02](#ex02)\n - [1.4 Repeating Batch generator](#1.4) \n- [Part 2: Defining the GRU model](#2)\n - [Exercise 03](#ex03)\n- [Part 3: Training](#3)\n - [3.1 Training the Model](#3.1)\n - [Exercise 04](#ex04)\n- [Part 4: Evaluation](#4)\n - [4.1 Evaluating using the deep nets](#4.1)\n - [Exercise 05](#ex05)\n- [Part 5: Generating the language with your own model](#5) \n- [Summary](#6)\n",
"_____no_output_____"
],
[
"<a name='0'></a>\n### Overview\n\nYour task will be to predict the next set of characters using the previous characters. \n- Although this task sounds simple, it is pretty useful.\n- You will start by converting a line of text into a tensor\n- Then you will create a generator to feed data into the model\n- You will train a neural network in order to predict the new set of characters of defined length. \n- You will use embeddings for each character and feed them as inputs to your model. \n - Many natural language tasks rely on using embeddings for predictions. \n- Your model will convert each character to its embedding, run the embeddings through a Gated Recurrent Unit `GRU`, and run it through a linear layer to predict the next set of characters.\n\n<img src = \"model.png\" style=\"width:600px;height:150px;\"/>\n\nThe figure above gives you a summary of what you are about to implement. \n- You will get the embeddings;\n- Stack the embeddings on top of each other;\n- Run them through two layers with a relu activation in the middle;\n- Finally, you will compute the softmax. \n\nTo predict the next character:\n- Use the softmax output and identify the word with the highest probability.\n- The word with the highest probability is the prediction for the next word.",
"_____no_output_____"
]
],
[
[
"import os\nimport trax\nimport trax.fastmath.numpy as np\nimport pickle\nimport numpy\nimport random as rnd\nfrom trax import fastmath\nfrom trax import layers as tl\n\n# set random seed\ntrax.supervised.trainer_lib.init_random_number_generators(32)\nrnd.seed(32)",
"INFO:tensorflow:tokens_length=568 inputs_length=512 targets_length=114 noise_density=0.15 mean_noise_span_length=3.0 \n"
]
],
[
[
"<a name='1'></a>\n# Part 1: Importing the Data\n\n<a name='1.1'></a>\n### 1.1 Loading in the data\n\n<img src = \"shakespeare.png\" style=\"width:250px;height:250px;\"/>\n\nNow import the dataset and do some processing. \n- The dataset has one sentence per line.\n- You will be doing character generation, so you have to process each sentence by converting each **character** (and not word) to a number. \n- You will use the `ord` function to convert a unique character to a unique integer ID. \n- Store each line in a list.\n- Create a data generator that takes in the `batch_size` and the `max_length`. \n - The `max_length` corresponds to the maximum length of the sentence.",
"_____no_output_____"
]
],
[
[
"dirname = 'data/'\nlines = [] # storing all the lines in a variable. \nfor filename in os.listdir(dirname):\n with open(os.path.join(dirname, filename)) as files:\n for line in files:\n # remove leading and trailing whitespace\n pure_line = line.strip()\n \n # if pure_line is not the empty string,\n if pure_line:\n # append it to the list\n lines.append(pure_line)",
"_____no_output_____"
],
[
"n_lines = len(lines)\nprint(f\"Number of lines: {n_lines}\")\nprint(f\"Sample line at position 0 {lines[0]}\")\nprint(f\"Sample line at position 999 {lines[999]}\")",
"Number of lines: 125097\nSample line at position 0 A LOVER'S COMPLAINT\nSample line at position 999 With this night's revels and expire the term\n"
]
],
[
[
"Notice that the letters are both uppercase and lowercase. In order to reduce the complexity of the task, we will convert all characters to lowercase. This way, the model only needs to predict the likelihood that a letter is 'a' and not decide between uppercase 'A' and lowercase 'a'.",
"_____no_output_____"
]
],
[
[
"# go through each line\nfor i, line in enumerate(lines):\n # convert to all lowercase\n lines[i] = line.lower()\n\nprint(f\"Number of lines: {n_lines}\")\nprint(f\"Sample line at position 0 {lines[0]}\")\nprint(f\"Sample line at position 999 {lines[999]}\")",
"Number of lines: 125097\nSample line at position 0 a lover's complaint\nSample line at position 999 with this night's revels and expire the term\n"
],
[
"eval_lines = lines[-1000:] # Create a holdout validation set\nlines = lines[:-1000] # Leave the rest for training\n\nprint(f\"Number of lines for training: {len(lines)}\")\nprint(f\"Number of lines for validation: {len(eval_lines)}\")",
"Number of lines for training: 124097\nNumber of lines for validation: 1000\n"
]
],
[
[
"<a name='1.2'></a>\n### 1.2 Convert a line to tensor\n\nNow that you have your list of lines, you will convert each character in that list to a number. You can use Python's `ord` function to do it. \n\nGiven a string representing of one Unicode character, the `ord` function return an integer representing the Unicode code point of that character.\n\n",
"_____no_output_____"
]
],
[
[
"# View the unique unicode integer associated with each character\nprint(f\"ord('a'): {ord('a')}\")\nprint(f\"ord('b'): {ord('b')}\")\nprint(f\"ord('c'): {ord('c')}\")\nprint(f\"ord(' '): {ord(' ')}\")\nprint(f\"ord('x'): {ord('x')}\")\nprint(f\"ord('y'): {ord('y')}\")\nprint(f\"ord('z'): {ord('z')}\")\nprint(f\"ord('1'): {ord('1')}\")\nprint(f\"ord('2'): {ord('2')}\")\nprint(f\"ord('3'): {ord('3')}\")",
"ord('a'): 97\nord('b'): 98\nord('c'): 99\nord(' '): 32\nord('x'): 120\nord('y'): 121\nord('z'): 122\nord('1'): 49\nord('2'): 50\nord('3'): 51\n"
]
],
[
[
"<a name='ex01'></a>\n### Exercise 01\n\n**Instructions:** Write a function that takes in a single line and transforms each character into its unicode integer. This returns a list of integers, which we'll refer to as a tensor.\n- Use a special integer to represent the end of the sentence (the end of the line).\n- This will be the EOS_int (end of sentence integer) parameter of the function.\n- Include the EOS_int as the last integer of the \n- For this exercise, you will use the number `1` to represent the end of a sentence.",
"_____no_output_____"
]
],
[
[
"# UNQ_C1 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: line_to_tensor\ndef line_to_tensor(line, EOS_int=1):\n \"\"\"Turns a line of text into a tensor\n\n Args:\n line (str): A single line of text.\n EOS_int (int, optional): End-of-sentence integer. Defaults to 1.\n\n Returns:\n list: a list of integers (unicode values) for the characters in the `line`.\n \"\"\"\n \n # Initialize the tensor as an empty list\n tensor = []\n ### START CODE HERE (Replace instances of 'None' with your code) ###\n # for each character:\n for c in line:\n \n # convert to unicode int\n c_int = ord(c)\n \n # append the unicode integer to the tensor list\n tensor.append(c_int)\n \n # include the end-of-sentence integer\n tensor.append(1)\n ### END CODE HERE ###\n\n return tensor",
"_____no_output_____"
],
[
"# Testing your output\nline_to_tensor('abc xyz')",
"_____no_output_____"
]
],
[
[
"##### Expected Output\n```CPP\n[97, 98, 99, 32, 120, 121, 122, 1]\n```",
"_____no_output_____"
],
[
"<a name='1.3'></a>\n### 1.3 Batch generator \n\nMost of the time in Natural Language Processing, and AI in general we use batches when training our data sets. Here, you will build a data generator that takes in a text and returns a batch of text lines (lines are sentences).\n- The generator converts text lines (sentences) into numpy arrays of integers padded by zeros so that all arrays have the same length, which is the length of the longest sentence in the entire data set.\n\nOnce you create the generator, you can iterate on it like this:\n\n```\nnext(data_generator)\n```\n\nThis generator returns the data in a format that you could directly use in your model when computing the feed-forward of your algorithm. This iterator returns a batch of lines and per token mask. The batch is a tuple of three parts: inputs, targets, mask. The inputs and targets are identical. The second column will be used to evaluate your predictions. Mask is 1 for non-padding tokens.\n\n<a name='ex02'></a>\n### Exercise 02\n**Instructions:** Implement the data generator below. Here are some things you will need. \n\n- While True loop: this will yield one batch at a time.\n- if index >= num_lines, set index to 0. \n- The generator should return shuffled batches of data. To achieve this without modifying the actual lines a list containing the indexes of `data_lines` is created. This list can be shuffled and used to get random batches everytime the index is reset.\n- if len(line) < max_length append line to cur_batch.\n - Note that a line that has length equal to max_length should not be appended to the batch. \n - This is because when converting the characters into a tensor of integers, an additional end of sentence token id will be added. \n - So if max_length is 5, and a line has 4 characters, the tensor representing those 4 characters plus the end of sentence character will be of length 5, which is the max length.\n- if len(cur_batch) == batch_size, go over every line, convert it to an int and store it.\n\n**Remember that when calling np you are really calling trax.fastmath.numpy which is trax’s version of numpy that is compatible with JAX. As a result of this, where you used to encounter the type numpy.ndarray now you will find the type jax.interpreters.xla.DeviceArray.**",
"_____no_output_____"
],
[
"<details> \n<summary>\n <font size=\"3\" color=\"darkgreen\"><b>Hints</b></font>\n</summary>\n<p>\n<ul>\n <li>Use the line_to_tensor function above inside a list comprehension in order to pad lines with zeros.</li>\n <li>Keep in mind that the length of the tensor is always 1 + the length of the original line of characters. Keep this in mind when setting the padding of zeros.</li>\n</ul>\n</p>",
"_____no_output_____"
]
],
[
[
"# UNQ_C2 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: data_generator\ndef data_generator(batch_size, max_length, data_lines, line_to_tensor=line_to_tensor, shuffle=True):\n \"\"\"Generator function that yields batches of data\n\n Args:\n batch_size (int): number of examples (in this case, sentences) per batch.\n max_length (int): maximum length of the output tensor.\n NOTE: max_length includes the end-of-sentence character that will be added\n to the tensor. \n Keep in mind that the length of the tensor is always 1 + the length\n of the original line of characters.\n data_lines (list): list of the sentences to group into batches.\n line_to_tensor (function, optional): function that converts line to tensor. Defaults to line_to_tensor.\n shuffle (bool, optional): True if the generator should generate random batches of data. Defaults to True.\n\n Yields:\n tuple: two copies of the batch (jax.interpreters.xla.DeviceArray) and mask (jax.interpreters.xla.DeviceArray).\n NOTE: jax.interpreters.xla.DeviceArray is trax's version of numpy.ndarray\n \"\"\"\n # initialize the index that points to the current position in the lines index array\n index = 0\n \n # initialize the list that will contain the current batch\n cur_batch = []\n \n # count the number of lines in data_lines\n num_lines = len(data_lines)\n \n # create an array with the indexes of data_lines that can be shuffled\n lines_index = [*range(num_lines)]\n \n # shuffle line indexes if shuffle is set to True\n if shuffle:\n rnd.shuffle(lines_index)\n \n ### START CODE HERE (Replace instances of 'None' with your code) ###\n while True:\n \n # if the index is greater or equal than to the number of lines in data_lines\n if index >= num_lines:\n # then reset the index to 0\n index = 0\n # shuffle line indexes if shuffle is set to True\n if shuffle:\n rnd.shuffle(lines_index)\n \n # get a line at the `lines_index[index]` position in data_lines\n line = data_lines[lines_index[index]]\n \n # if the length of the line is less than max_length\n if len(line) < max_length:\n # append the line to the current batch\n cur_batch.append(line)\n \n # increment the index by one\n index += 1\n \n # if the current batch is now equal to the desired batch size\n if len(cur_batch) == batch_size:\n \n batch = []\n mask = []\n \n # go through each line (li) in cur_batch\n for li in cur_batch:\n # convert the line (li) to a tensor of integers\n tensor = line_to_tensor(li)\n \n # Create a list of zeros to represent the padding\n # so that the tensor plus padding will have length `max_length`\n pad = [0] * (max_length - len(tensor))\n \n # combine the tensor plus pad\n tensor_pad = tensor + pad\n \n # append the padded tensor to the batch\n batch.append(tensor_pad)\n\n # A mask for tensor_pad is 1 wherever tensor_pad is not\n # 0 and 0 wherever tensor_pad is 0, i.e. if tensor_pad is\n # [1, 2, 3, 0, 0, 0] then example_mask should be\n # [1, 1, 1, 0, 0, 0]\n # Hint: Use a list comprehension for this\n example_mask = [0 if val == 0 else 1 for val in tensor_pad]\n mask.append(example_mask)\n \n # convert the batch (data type list) to a trax's numpy array\n batch_np_arr = np.array(batch)\n mask_np_arr = np.array(mask)\n \n ### END CODE HERE ##\n \n # Yield two copies of the batch and mask.\n yield batch_np_arr, batch_np_arr, mask_np_arr\n \n # reset the current batch to an empty list\n cur_batch = []\n ",
"_____no_output_____"
],
[
"# Try out your data generator\ntmp_lines = ['12345678901', #length 11\n '123456789', # length 9\n '234567890', # length 9\n '345678901'] # length 9\n\n# Get a batch size of 2, max length 10\ntmp_data_gen = data_generator(batch_size=2, \n max_length=10, \n data_lines=tmp_lines,\n shuffle=False)\n\n# get one batch\ntmp_batch = next(tmp_data_gen)\n\n# view the batch\ntmp_batch",
"_____no_output_____"
]
],
[
[
"##### Expected output\n\n```CPP\n(DeviceArray([[49, 50, 51, 52, 53, 54, 55, 56, 57, 1],\n [50, 51, 52, 53, 54, 55, 56, 57, 48, 1]], dtype=int32),\n DeviceArray([[49, 50, 51, 52, 53, 54, 55, 56, 57, 1],\n [50, 51, 52, 53, 54, 55, 56, 57, 48, 1]], dtype=int32),\n DeviceArray([[1, 1, 1, 1, 1, 1, 1, 1, 1, 1],\n [1, 1, 1, 1, 1, 1, 1, 1, 1, 1]], dtype=int32))\n```",
"_____no_output_____"
],
[
"Now that you have your generator, you can just call them and they will return tensors which correspond to your lines in Shakespeare. The first column and the second column are identical. Now you can go ahead and start building your neural network. ",
"_____no_output_____"
],
[
"<a name='1.4'></a>\n### 1.4 Repeating Batch generator \n\nThe way the iterator is currently defined, it will keep providing batches forever.\n\nAlthough it is not needed, we want to show you the `itertools.cycle` function which is really useful when the generator eventually stops\n\nNotice that it is expected to use this function within the training function further below\n\nUsually we want to cycle over the dataset multiple times during training (i.e. train for multiple *epochs*).\n\nFor small datasets we can use [`itertools.cycle`](https://docs.python.org/3.8/library/itertools.html#itertools.cycle) to achieve this easily.",
"_____no_output_____"
]
],
[
[
"import itertools\n\ninfinite_data_generator = itertools.cycle(\n data_generator(batch_size=2, max_length=10, data_lines=tmp_lines))",
"_____no_output_____"
]
],
[
[
"You can see that we can get more than the 5 lines in tmp_lines using this.",
"_____no_output_____"
]
],
[
[
"ten_lines = [next(infinite_data_generator) for _ in range(10)]\nprint(len(ten_lines))",
"10\n"
]
],
[
[
"<a name='2'></a>\n\n# Part 2: Defining the GRU model\n\nNow that you have the input and output tensors, you will go ahead and initialize your model. You will be implementing the `GRULM`, gated recurrent unit model. To implement this model, you will be using google's `trax` package. Instead of making you implement the `GRU` from scratch, we will give you the necessary methods from a build in package. You can use the following packages when constructing the model: \n\n\n- `tl.Serial`: Combinator that applies layers serially (by function composition). [docs](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Serial) / [source code](https://github.com/google/trax/blob/1372b903bb66b0daccee19fd0b1fdf44f659330b/trax/layers/combinators.py#L26)\n - You can pass in the layers as arguments to `Serial`, separated by commas. \n - For example: `tl.Serial(tl.Embeddings(...), tl.Mean(...), tl.Dense(...), tl.LogSoftmax(...))`\n\n___\n\n- `tl.ShiftRight`: Allows the model to go right in the feed forward. [docs](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.attention.ShiftRight) / [source code](https://github.com/google/trax/blob/1372b903bb66b0daccee19fd0b1fdf44f659330b/trax/layers/attention.py#L297)\n - `ShiftRight(n_shifts=1, mode='train')` layer to shift the tensor to the right n_shift times\n - Here in the exercise you only need to specify the mode and not worry about n_shifts\n\n___\n\n- `tl.Embedding`: Initializes the embedding. In this case it is the size of the vocabulary by the dimension of the model. [docs](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Embedding) / [source code](https://github.com/google/trax/blob/1372b903bb66b0daccee19fd0b1fdf44f659330b/trax/layers/core.py#L113) \n - `tl.Embedding(vocab_size, d_feature)`.\n - `vocab_size` is the number of unique words in the given vocabulary.\n - `d_feature` is the number of elements in the word embedding (some choices for a word embedding size range from 150 to 300, for example).\n___\n\n- `tl.GRU`: `Trax` GRU layer. [docs](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.rnn.GRU) / [source code](https://github.com/google/trax/blob/1372b903bb66b0daccee19fd0b1fdf44f659330b/trax/layers/rnn.py#L143)\n - `GRU(n_units)` Builds a traditional GRU of n_cells with dense internal transformations.\n - `GRU` paper: https://arxiv.org/abs/1412.3555\n___\n\n- `tl.Dense`: A dense layer. [docs](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.Dense) / [source code](https://github.com/google/trax/blob/1372b903bb66b0daccee19fd0b1fdf44f659330b/trax/layers/core.py#L28)\n - `tl.Dense(n_units)`: The parameter `n_units` is the number of units chosen for this dense layer.\n___\n\n- `tl.LogSoftmax`: Log of the output probabilities. [docs](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.core.LogSoftmax) / [source code](https://github.com/google/trax/blob/1372b903bb66b0daccee19fd0b1fdf44f659330b/trax/layers/core.py#L242)\n - Here, you don't need to set any parameters for `LogSoftMax()`.\n___\n\n<a name='ex03'></a>\n### Exercise 03\n**Instructions:** Implement the `GRULM` class below. You should be using all the methods explained above.\n",
"_____no_output_____"
]
],
[
[
"# UNQ_C3 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: GRULM\ndef GRULM(vocab_size=256, d_model=512, n_layers=2, mode='train'):\n \"\"\"Returns a GRU language model.\n\n Args:\n vocab_size (int, optional): Size of the vocabulary. Defaults to 256.\n d_model (int, optional): Depth of embedding (n_units in the GRU cell). Defaults to 512.\n n_layers (int, optional): Number of GRU layers. Defaults to 2.\n mode (str, optional): 'train', 'eval' or 'predict', predict mode is for fast inference. Defaults to \"train\".\n\n Returns:\n trax.layers.combinators.Serial: A GRU language model as a layer that maps from a tensor of tokens to activations over a vocab set.\n \"\"\"\n ### START CODE HERE (Replace instances of 'None' with your code) ###\n model = tl.Serial(\n tl.ShiftRight(mode=mode), # Stack the ShiftRight layer\n tl.Embedding(vocab_size=vocab_size, d_feature=d_model), # Stack the embedding layer\n [tl.GRU(n_units=d_model) for _ in range(n_layers)], # Stack GRU layers of d_model units keeping n_layer parameter in mind (use list comprehension syntax)\n tl.Dense(n_units=vocab_size), # Dense layer\n tl.LogSoftmax() # Log Softmax\n )\n ### END CODE HERE ###\n return model\n",
"_____no_output_____"
],
[
"# testing your model\nmodel = GRULM()\nprint(model)",
"Serial[\n ShiftRight(1)\n Embedding_256_512\n GRU_512\n GRU_512\n Dense_256\n LogSoftmax\n]\n"
]
],
[
[
"##### Expected output\n\n```CPP\nSerial[\n ShiftRight(1)\n Embedding_256_512\n GRU_512\n GRU_512\n Dense_256\n LogSoftmax\n]\n```",
"_____no_output_____"
],
[
"<a name='3'></a>\n# Part 3: Training\n\nNow you are going to train your model. As usual, you have to define the cost function, the optimizer, and decide whether you will be training it on a `gpu` or `cpu`. You also have to feed in a built model. Before, going into the training, we re-introduce the `TrainTask` and `EvalTask` abstractions from the last week's assignment.\n\nTo train a model on a task, Trax defines an abstraction `trax.supervised.training.TrainTask` which packages the train data, loss and optimizer (among other things) together into an object.\n\nSimilarly to evaluate a model, Trax defines an abstraction `trax.supervised.training.EvalTask` which packages the eval data and metrics (among other things) into another object.\n\nThe final piece tying things together is the `trax.supervised.training.Loop` abstraction that is a very simple and flexible way to put everything together and train the model, all the while evaluating it and saving checkpoints.\nUsing `training.Loop` will save you a lot of code compared to always writing the training loop by hand, like you did in courses 1 and 2. More importantly, you are less likely to have a bug in that code that would ruin your training.",
"_____no_output_____"
]
],
[
[
"batch_size = 32\nmax_length = 64",
"_____no_output_____"
]
],
[
[
"An `epoch` is traditionally defined as one pass through the dataset.\n\nSince the dataset was divided in `batches` you need several `steps` (gradient evaluations) in order to complete an `epoch`. So, one `epoch` corresponds to the number of examples in a `batch` times the number of `steps`. In short, in each `epoch` you go over all the dataset. \n\nThe `max_length` variable defines the maximum length of lines to be used in training our data, lines longer that that length are discarded. \n\nBelow is a function and results that indicate how many lines conform to our criteria of maximum length of a sentence in the entire dataset and how many `steps` are required in order to cover the entire dataset which in turn corresponds to an `epoch`.",
"_____no_output_____"
]
],
[
[
"def n_used_lines(lines, max_length):\n '''\n Args: \n lines: all lines of text an array of lines\n max_length - max_length of a line in order to be considered an int\n output_dir - folder to save your file an int\n Return:\n number of efective examples\n '''\n\n n_lines = 0\n for l in lines:\n if len(l) <= max_length:\n n_lines += 1\n return n_lines\n\nnum_used_lines = n_used_lines(lines, 32)\nprint('Number of used lines from the dataset:', num_used_lines)\nprint('Batch size (a power of 2):', int(batch_size))\nsteps_per_epoch = int(num_used_lines/batch_size)\nprint('Number of steps to cover one epoch:', steps_per_epoch)",
"Number of used lines from the dataset: 25881\nBatch size (a power of 2): 32\nNumber of steps to cover one epoch: 808\n"
]
],
[
[
"**Expected output:** \n\nNumber of used lines from the dataset: 25881\n\nBatch size (a power of 2): 32\n\nNumber of steps to cover one epoch: 808",
"_____no_output_____"
],
[
"<a name='3.1'></a>\n### 3.1 Training the model\n\nYou will now write a function that takes in your model and trains it. To train your model you have to decide how many times you want to iterate over the entire data set. \n\n<a name='ex04'></a>\n### Exercise 04\n\n**Instructions:** Implement the `train_model` program below to train the neural network above. Here is a list of things you should do:\n\n- Create a `trax.supervised.trainer.TrainTask` object, this encapsulates the aspects of the dataset and the problem at hand:\n - labeled_data = the labeled data that we want to *train* on.\n - loss_fn = [tl.CrossEntropyLoss()](https://trax-ml.readthedocs.io/en/latest/trax.layers.html?highlight=CrossEntropyLoss#trax.layers.metrics.CrossEntropyLoss)\n - optimizer = [trax.optimizers.Adam()](https://trax-ml.readthedocs.io/en/latest/trax.optimizers.html?highlight=Adam#trax.optimizers.adam.Adam) with learning rate = 0.0005\n\n- Create a `trax.supervised.trainer.EvalTask` object, this encapsulates aspects of evaluating the model:\n - labeled_data = the labeled data that we want to *evaluate* on.\n - metrics = [tl.CrossEntropyLoss()](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.metrics.CrossEntropyLoss) and [tl.Accuracy()](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.metrics.Accuracy)\n - How frequently we want to evaluate and checkpoint the model.\n\n- Create a `trax.supervised.trainer.Loop` object, this encapsulates the following:\n - The previously created `TrainTask` and `EvalTask` objects.\n - the training model = [GRULM](#ex03)\n - optionally the evaluation model, if different from the training model. NOTE: in presence of Dropout etc we usually want the evaluation model to behave slightly differently than the training model.\n\nYou will be using a cross entropy loss, with Adam optimizer. Please read the [trax](https://trax-ml.readthedocs.io/en/latest/index.html) documentation to get a full understanding. Make sure you use the number of steps provided as a parameter to train for the desired number of steps.\n\n**NOTE:** Don't forget to wrap the data generator in `itertools.cycle` to iterate on it for multiple epochs.",
"_____no_output_____"
]
],
[
[
"from trax.supervised import training\n\n# UNQ_C4 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: train_model\ndef train_model(model, data_generator, batch_size=32, max_length=64, lines=lines, eval_lines=eval_lines, n_steps=1, output_dir='model/'): \n \"\"\"Function that trains the model\n\n Args:\n model (trax.layers.combinators.Serial): GRU model.\n data_generator (function): Data generator function.\n batch_size (int, optional): Number of lines per batch. Defaults to 32.\n max_length (int, optional): Maximum length allowed for a line to be processed. Defaults to 64.\n lines (list, optional): List of lines to use for training. Defaults to lines.\n eval_lines (list, optional): List of lines to use for evaluation. Defaults to eval_lines.\n n_steps (int, optional): Number of steps to train. Defaults to 1.\n output_dir (str, optional): Relative path of directory to save model. Defaults to \"model/\".\n\n Returns:\n trax.supervised.training.Loop: Training loop for the model.\n \"\"\"\n \n ### START CODE HERE (Replace instances of 'None' with your code) ###\n bare_train_generator = data_generator(batch_size, max_length, data_lines=lines)\n infinite_train_generator = itertools.cycle(bare_train_generator)\n \n bare_eval_generator = data_generator(batch_size, max_length, data_lines=eval_lines)\n infinite_eval_generator = itertools.cycle(bare_eval_generator)\n \n train_task = training.TrainTask(\n labeled_data=infinite_train_generator, # Use infinite train data generator\n loss_layer=tl.CrossEntropyLoss(), # Don't forget to instantiate this object\n optimizer=trax.optimizers.Adam(0.0005) # Don't forget to add the learning rate parameter\n )\n\n eval_task = training.EvalTask(\n labeled_data=infinite_eval_generator, # Use infinite eval data generator\n metrics=[tl.CrossEntropyLoss(), tl.Accuracy()], # Don't forget to instantiate these objects\n n_eval_batches=3 # For better evaluation accuracy in reasonable time\n )\n \n training_loop = training.Loop(model,\n train_task,\n eval_task=eval_task,\n output_dir=output_dir)\n\n training_loop.run(n_steps=n_steps)\n \n ### END CODE HERE ###\n \n # We return this because it contains a handle to the model, which has the weights etc.\n return training_loop\n",
"_____no_output_____"
],
[
"# Train the model 1 step and keep the `trax.supervised.training.Loop` object.\ntraining_loop = train_model(GRULM(), data_generator)",
"Step 1: train CrossEntropyLoss | 5.54486227\nStep 1: eval CrossEntropyLoss | 5.48863840\nStep 1: eval Accuracy | 0.17598992\n"
]
],
[
[
"The model was only trained for 1 step due to the constraints of this environment. Even on a GPU accelerated environment it will take many hours for it to achieve a good level of accuracy. For the rest of the assignment you will be using a pretrained model but now you should understand how the training can be done using Trax.",
"_____no_output_____"
],
[
"<a name='4'></a>\n# Part 4: Evaluation \n<a name='4.1'></a>\n### 4.1 Evaluating using the deep nets\n\nNow that you have learned how to train a model, you will learn how to evaluate it. To evaluate language models, we usually use perplexity which is a measure of how well a probability model predicts a sample. Note that perplexity is defined as: \n\n$$P(W) = \\sqrt[N]{\\prod_{i=1}^{N} \\frac{1}{P(w_i| w_1,...,w_{n-1})}}$$\n\nAs an implementation hack, you would usually take the log of that formula (to enable us to use the log probabilities we get as output of our `RNN`, convert exponents to products, and products into sums which makes computations less complicated and computationally more efficient). You should also take care of the padding, since you do not want to include the padding when calculating the perplexity (because we do not want to have a perplexity measure artificially good).\n\n\n$$log P(W) = {log\\big(\\sqrt[N]{\\prod_{i=1}^{N} \\frac{1}{P(w_i| w_1,...,w_{n-1})}}\\big)}$$\n\n$$ = {log\\big({\\prod_{i=1}^{N} \\frac{1}{P(w_i| w_1,...,w_{n-1})}}\\big)^{\\frac{1}{N}}}$$ \n\n$$ = {log\\big({\\prod_{i=1}^{N}{P(w_i| w_1,...,w_{n-1})}}\\big)^{-\\frac{1}{N}}} $$\n$$ = -\\frac{1}{N}{log\\big({\\prod_{i=1}^{N}{P(w_i| w_1,...,w_{n-1})}}\\big)} $$\n$$ = -\\frac{1}{N}{\\big({\\sum_{i=1}^{N}{logP(w_i| w_1,...,w_{n-1})}}\\big)} $$\n\n\n<a name='ex05'></a>\n### Exercise 05\n**Instructions:** Write a program that will help evaluate your model. Implementation hack: your program takes in preds and target. Preds is a tensor of log probabilities. You can use [`tl.one_hot`](https://github.com/google/trax/blob/22765bb18608d376d8cd660f9865760e4ff489cd/trax/layers/metrics.py#L154) to transform the target into the same dimension. You then multiply them and sum. \n\nYou also have to create a mask to only get the non-padded probabilities. Good luck! ",
"_____no_output_____"
],
[
"<details> \n<summary>\n <font size=\"3\" color=\"darkgreen\"><b>Hints</b></font>\n</summary>\n<p>\n<ul>\n <li>To convert the target into the same dimension as the predictions tensor use tl.one.hot with target and preds.shape[-1].</li>\n <li>You will also need the np.equal function in order to unpad the data and properly compute perplexity.</li>\n <li>Keep in mind while implementing the formula above that <em> w<sub>i</sub></em> represents a letter from our 256 letter alphabet.</li>\n</ul>\n</p>",
"_____no_output_____"
]
],
[
[
"# UNQ_C5 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# GRADED FUNCTION: test_model\ndef test_model(preds, target):\n \"\"\"Function to test the model.\n\n Args:\n preds (jax.interpreters.xla.DeviceArray): Predictions of a list of batches of tensors corresponding to lines of text.\n target (jax.interpreters.xla.DeviceArray): Actual list of batches of tensors corresponding to lines of text.\n\n Returns:\n float: log_perplexity of the model.\n \"\"\"\n ### START CODE HERE (Replace instances of 'None' with your code) ###\n total_log_ppx = np.sum(preds * tl.one_hot(target, preds.shape[-1]),axis= -1) # HINT: tl.one_hot() should replace one of the Nones\n\n non_pad = 1.0 - np.equal(target, 0) # You should check if the target equals 0\n ppx = total_log_ppx * non_pad # Get rid of the padding\n\n log_ppx = np.sum(ppx) / np.sum(non_pad)\n ### END CODE HERE ###\n \n return -log_ppx",
"_____no_output_____"
],
[
"# UNQ_C6 (UNIQUE CELL IDENTIFIER, DO NOT EDIT)\n# Testing \nmodel = GRULM()\nmodel.init_from_file('model.pkl.gz')\nbatch = next(data_generator(batch_size, max_length, lines, shuffle=False))\npreds = model(batch[0])\nlog_ppx = test_model(preds, batch[1])\nprint('The log perplexity and perplexity of your model are respectively', log_ppx, np.exp(log_ppx))",
"The log perplexity and perplexity of your model are respectively 1.9785146 7.2319922\n"
]
],
[
[
"**Expected Output:** The log perplexity and perplexity of your model are respectively around 1.9 and 7.2.",
"_____no_output_____"
],
[
"<a name='5'></a>\n# Part 5: Generating the language with your own model\n\nWe will now use your own language model to generate new sentences for that we need to make draws from a Gumble distribution.",
"_____no_output_____"
],
[
"The Gumbel Probability Density Function (PDF) is defined as: \n\n$$ f(z) = {1\\over{\\beta}}e^{(-z+e^{(-z)})} $$\n\nwhere: $$ z = {(x - \\mu)\\over{\\beta}}$$\n\nThe maximum value, which is what we choose as the prediction in the last step of a Recursive Neural Network `RNN` we are using for text generation, in a sample of a random variable following an exponential distribution approaches the Gumbel distribution when the sample increases asymptotically. For that reason, the Gumbel distribution is used to sample from a categorical distribution.",
"_____no_output_____"
]
],
[
[
"# Run this cell to generate some news sentence\ndef gumbel_sample(log_probs, temperature=1.0):\n \"\"\"Gumbel sampling from a categorical distribution.\"\"\"\n u = numpy.random.uniform(low=1e-6, high=1.0 - 1e-6, size=log_probs.shape)\n g = -np.log(-np.log(u))\n return np.argmax(log_probs + g * temperature, axis=-1)\n\ndef predict(num_chars, prefix):\n inp = [ord(c) for c in prefix]\n result = [c for c in prefix]\n max_len = len(prefix) + num_chars\n for _ in range(num_chars):\n cur_inp = np.array(inp + [0] * (max_len - len(inp)))\n outp = model(cur_inp[None, :]) # Add batch dim.\n next_char = gumbel_sample(outp[0, len(inp)])\n inp += [int(next_char)]\n \n if inp[-1] == 1:\n break # EOS\n result.append(chr(int(next_char)))\n \n return \"\".join(result)\n\nprint(predict(32, \"\"))",
"And in the shapes of heaven, he \n"
],
[
"print(predict(32, \"\"))\nprint(predict(32, \"\"))\nprint(predict(32, \"\"))\n",
"MARK ANTONY\tTo go, good sir.\nEven with a countenance, exempt \nI'll leave him to; so 'twere so \n"
]
],
[
[
"In the generated text above, you can see that the model generates text that makes sense capturing dependencies between words and without any input. A simple n-gram model would have not been able to capture all of that in one sentence.",
"_____no_output_____"
],
[
"<a name='6'></a>\n### <span style=\"color:blue\"> On statistical methods </span>\n\nUsing a statistical method like the one you implemented in course 2 will not give you results that are as good. Your model will not be able to encode information seen previously in the data set and as a result, the perplexity will increase. Remember from course 2 that the higher the perplexity, the worse your model is. Furthermore, statistical ngram models take up too much space and memory. As a result, it will be inefficient and too slow. Conversely, with deepnets, you can get a better perplexity. Note, learning about n-gram language models is still important and allows you to better understand deepnets.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4a8d42d4bf9aa6779aa5578c782db2528b471fe3
| 4,881 |
ipynb
|
Jupyter Notebook
|
Theoretical_question_hmw3.ipynb
|
Gcorsetti/ADM-HMW3_AnimeList
|
41186a94310898ff437c27aa3910ee4ca4f0a798
|
[
"MIT"
] | 1 |
2021-11-14T21:59:42.000Z
|
2021-11-14T21:59:42.000Z
|
Theoretical_question_hmw3.ipynb
|
FilMastro/ADM-Homework3
|
8989b982f0201433a71b418dec7f9afa96de7bae
|
[
"MIT"
] | null | null | null |
Theoretical_question_hmw3.ipynb
|
FilMastro/ADM-Homework3
|
8989b982f0201433a71b418dec7f9afa96de7bae
|
[
"MIT"
] | null | null | null | 32.54 | 747 | 0.550297 |
[
[
[
"# 5. Algorithmic Question",
"_____no_output_____"
],
[
" You consult for a personal trainer who has a back-to-back sequence of requests for appointments. A sequence of requests is of the form : 30, 40, 25, 50, 30, 20 where each number is the time that the person who makes the appointment wants to spend. You need to accept some requests, however you need a break between them, so you cannot accept two consecutive requests. For example, [30, 50, 20] is an acceptable solution (of duration 100), but [30, 40, 50, 20] is not, because 30 and 40 are two consecutive appointments. Your goal is to provide to the personal trainer a schedule that maximizes the total length of the accepted appointments. For example, in the previous instance, the optimal solution is [40, 50, 20], of total duration 110.",
"_____no_output_____"
],
[
"-----------------------------------------",
"_____no_output_____"
],
[
"1. Write an algorithm that computes the acceptable solution with the longest possible duration.\n2. Implement a program that given in input an instance in the form given above, gives the optimal solution.",
"_____no_output_____"
],
[
"The following algorithm is actually the merge of 2 algorithm :\n--\n- Simple Comparison of the two possible sub lists (taking every other number)\n- Using a greedy Heuristic\n\nThe app_setter function basically checks the input array with this two very simple algorithm and then compares the results. We chose this approach in order to shield the function from the vulnerabilities of the single algorithms since there are specific cases in which is possible to demonstrate the ineffectivness of the two. Nevertheless this cross result solves many problems from this point of view\n\n",
"_____no_output_____"
]
],
[
[
"def app_setter(A):\n l1,l2,l3,B,t = [],[],[],A.copy(),0\n try:\n for i in range(0,len(A),2): #simple comparison of the two everyother lists\n l1.append(A[i])\n for i in range(1,len(A),2):\n l2.append(A[i])\n except IndexError:\n pass\n \n while t < len(B)/2: #greedy\n m = max(B)\n try:\n l3.append(m)\n except:\n pass\n try :\n B[B.index(m)+1] = 0\n B[B.index(m)-1] = 0\n B[B.index(m)] = 0\n B.remove(0)\n except IndexError:\n pass\n t+=1\n \n if sum(l1)>= sum(l2) and sum(l1)>=sum(l3):\n return l1\n if sum(l2)>= sum(l1) and sum(l2)>=sum(l3):\n return l2\n if sum(l3)>= sum(l1) and sum(l3)>=sum(l2):\n return l3",
"_____no_output_____"
],
[
"app_setter([10, 50, 10, 50, 10, 50, 150, 120])",
"_____no_output_____"
],
[
"[10, 50, 10, 50, 10, 50, 150, 120]\n[150, 50, 50] = 250 ---> Algorithm greedy\n[50, 50, 50, 120] = 270 ---> Simple every other i+1\n[10, 10, 10, 150] = 180 ---> Simple every other",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a8d502e5b82303bee61592a880f65c23083af90
| 29,314 |
ipynb
|
Jupyter Notebook
|
Labs/Deep Learning/200 - Machine Learning in Python/3 - Predict/Resources/FlightData.ipynb
|
Mirza-Younus-Baig/computerscience
|
e3cd81f3da00e4703575f812cfbb129e47b4e570
|
[
"MIT"
] | 4 |
2020-01-30T13:12:04.000Z
|
2022-01-21T01:14:43.000Z
|
Labs/Deep Learning/200 - Machine Learning in Python/3 - Predict/Resources/FlightData.ipynb
|
Mirza-Younus-Baig/computerscience
|
e3cd81f3da00e4703575f812cfbb129e47b4e570
|
[
"MIT"
] | null | null | null |
Labs/Deep Learning/200 - Machine Learning in Python/3 - Predict/Resources/FlightData.ipynb
|
Mirza-Younus-Baig/computerscience
|
e3cd81f3da00e4703575f812cfbb129e47b4e570
|
[
"MIT"
] | 4 |
2020-04-11T18:25:02.000Z
|
2020-04-11T19:54:57.000Z
| 30.283058 | 119 | 0.289282 |
[
[
[
"import pandas as pd\n\ndf = pd.read_csv('flightdata.csv')\ndf.head()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.isnull().values.any()",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
],
[
"df = df.drop('Unnamed: 25', axis=1)\ndf.isnull().sum()",
"_____no_output_____"
],
[
"df = df[[\"MONTH\", \"DAY_OF_MONTH\", \"DAY_OF_WEEK\", \"ORIGIN\", \"DEST\", \"CRS_DEP_TIME\", \"ARR_DEL15\"]]\ndf.isnull().sum()",
"_____no_output_____"
],
[
"df[df.isnull().values.any(axis=1)].head()",
"_____no_output_____"
],
[
"df = df.fillna({'ARR_DEL15': 1})\ndf.iloc[177:185]",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"import math\n\nfor index, row in df.iterrows():\n df.loc[index, 'CRS_DEP_TIME'] = math.floor(row['CRS_DEP_TIME'] / 100)\ndf.head()",
"_____no_output_____"
],
[
"df = pd.get_dummies(df, columns=['ORIGIN', 'DEST'])\ndf.head()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8d6eb3f454d0997ec32ac3eec8aea8c9072902
| 65,239 |
ipynb
|
Jupyter Notebook
|
Explained_variance_fits.ipynb
|
zqwei/Spike2Fluroscence_jGCaMP8
|
be7dab9a5de341ec50ed9906b8148eb7ba4a1b85
|
[
"MIT"
] | null | null | null |
Explained_variance_fits.ipynb
|
zqwei/Spike2Fluroscence_jGCaMP8
|
be7dab9a5de341ec50ed9906b8148eb7ba4a1b85
|
[
"MIT"
] | null | null | null |
Explained_variance_fits.ipynb
|
zqwei/Spike2Fluroscence_jGCaMP8
|
be7dab9a5de341ec50ed9906b8148eb7ba4a1b85
|
[
"MIT"
] | null | null | null | 407.74375 | 40,672 | 0.93858 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport warnings\nwarnings.filterwarnings('ignore')\nimport seaborn as sns\nsns.set(font_scale=1.5, style='ticks')\nimport pandas as pd\n%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\nhits_label = ['jGCaMP8f', 'jGCaMP8m', 'jGCaMP8s', 'jGCaMP7f', 'XCaMP-Gf', 'GCaMP6s', 'GCaMP6f', 'TG-GCaMP6s', 'TG-GCaMP6f']\nhits_colors = ['#0000ff', '#ff0000', '#666666', '#00ff00', '#0099ff', '#FFD670', '#FFD670', '#FFD670', '#FFD670']\n\nsensor6_df = pd.read_csv('GCaMP8_exported_ROIs_s2f_full/GCaMP6s_ev.csv')\nsensor8_df = pd.read_csv('GCaMP8_exported_ROIs_s2f_full/GCaMP8s_ev.csv')\nsensor_df = pd.concat([sensor8_df, sensor6_df])\nsensor8_df_sub = sensor8_df[sensor8_df['Cell'].str.contains('jGCaMP8')]",
"_____no_output_____"
]
],
[
[
"## Figure B: Explained variance -- linear vs sigmoid model",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(4, 3))\nidx = sensor8_df_sub['Type']\nplt.scatter(sensor8_df_sub['EV_l'][idx], sensor8_df_sub['EV_s'][idx], c='k')\nplt.scatter(sensor8_df_sub['EV_l'][~idx], sensor8_df_sub['EV_s'][~idx], edgecolors='k', facecolors='none')\nplt.plot([0, 1], [0, 1], '--r')\nsns.despine()\nplt.xlim([0, 1])\nplt.ylim([0, 1])\nplt.xlabel('EV linear model')\nplt.ylabel('EV sigmoid model')\nsns.despine()\nplt.savefig('figures/EV_linear_sigmoid.pdf')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Figure C: Variance explained (sigmoid - linear)",
"_____no_output_____"
]
],
[
[
"sns.swarmplot(data=sensor_df, \n x='Cell', \n y='EV_diff', \n order = hits_label, \n size=3,\n alpha=0.8,\n color='darkgray',\n edgecolor = None,\n linewidth=0)\nax = sns.boxplot(data=sensor_df, \n x='Cell', \n y='EV_diff',\n order = hits_label, \n showfliers=False, \n linewidth=2,\n width=.5,\n palette = hits_colors)\n\nsns.despine()\nax.set_xticklabels(ax.get_xticklabels(),rotation=45)\nplt.ylim([-0.03, 0.42])\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8d7f45b3ef0d14265e27bb6bdd83b3870beaed
| 18,755 |
ipynb
|
Jupyter Notebook
|
{{ cookiecutter.repo_name }}/notebooks/io.ipynb
|
wcbeard/cookiecutter-data-science
|
d56640f567635c03bfca3c7feac01c69d973473a
|
[
"MIT"
] | null | null | null |
{{ cookiecutter.repo_name }}/notebooks/io.ipynb
|
wcbeard/cookiecutter-data-science
|
d56640f567635c03bfca3c7feac01c69d973473a
|
[
"MIT"
] | null | null | null |
{{ cookiecutter.repo_name }}/notebooks/io.ipynb
|
wcbeard/cookiecutter-data-science
|
d56640f567635c03bfca3c7feac01c69d973473a
|
[
"MIT"
] | null | null | null | 36.702544 | 3,599 | 0.589976 |
[
[
[
" # Table of Contents\n<div class=\"toc\" style=\"margin-top: 1em;\"><ul class=\"toc-item\" id=\"toc-level0\"><ul class=\"toc-item\"><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Import\" data-toc-modified-id=\"Import-0.1\"><span class=\"toc-item-num\">0.1 </span>Import</a></span><ul class=\"toc-item\"><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Common\" data-toc-modified-id=\"Common-0.1.1\"><span class=\"toc-item-num\">0.1.1 </span>Common</a></span></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Libs\" data-toc-modified-id=\"Libs-0.1.2\"><span class=\"toc-item-num\">0.1.2 </span>Libs</a></span></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Special\" data-toc-modified-id=\"Special-0.1.3\"><span class=\"toc-item-num\">0.1.3 </span>Special</a></span></li></ul></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Load-longitudinal-retention\" data-toc-modified-id=\"Load-longitudinal-retention-0.2\"><span class=\"toc-item-num\">0.2 </span>Load longitudinal retention</a></span><ul class=\"toc-item\"><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Move\" data-toc-modified-id=\"Move-0.2.1\"><span class=\"toc-item-num\">0.2.1 </span>Move</a></span></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Convert\" data-toc-modified-id=\"Convert-0.2.2\"><span class=\"toc-item-num\">0.2.2 </span>Convert</a></span></li></ul></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Load-longitudinal-seq=1\" data-toc-modified-id=\"Load-longitudinal-seq=1-0.3\"><span class=\"toc-item-num\">0.3 </span>Load longitudinal seq=1</a></span><ul class=\"toc-item\"><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Convert-to-parq\" data-toc-modified-id=\"Convert-to-parq-0.3.1\"><span class=\"toc-item-num\">0.3.1 </span>Convert to parq</a></span></li></ul></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Load-longitudinal-seq=2\" data-toc-modified-id=\"Load-longitudinal-seq=2-0.4\"><span class=\"toc-item-num\">0.4 </span>Load longitudinal seq=2</a></span><ul class=\"toc-item\"><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Move-raw\" data-toc-modified-id=\"Move-raw-0.4.1\"><span class=\"toc-item-num\">0.4.1 </span>Move raw</a></span></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Convert-to-parq\" data-toc-modified-id=\"Convert-to-parq-0.4.2\"><span class=\"toc-item-num\">0.4.2 </span>Convert to parq</a></span><ul class=\"toc-item\"><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#debug\" data-toc-modified-id=\"debug-0.4.2.1\"><span class=\"toc-item-num\">0.4.2.1 </span>debug</a></span></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#debug-end\" data-toc-modified-id=\"debug-end-0.4.2.2\"><span class=\"toc-item-num\">0.4.2.2 </span>debug end</a></span></li></ul></li></ul></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#Load-Onboarding-A/B\" data-toc-modified-id=\"Load-Onboarding-A/B-0.5\"><span class=\"toc-item-num\">0.5 </span>Load Onboarding A/B</a></span></li><li><span><a href=\"http://localhost:8888/notebooks/notebooks/io.ipynb#OOM\" data-toc-modified-id=\"OOM-0.6\"><span class=\"toc-item-num\">0.6 </span>OOM</a></span></li></ul></ul></div>",
"_____no_output_____"
],
[
"## Import",
"_____no_output_____"
],
[
"### Common",
"_____no_output_____"
]
],
[
[
"import boot_utes as bu; bu.reload(bu); from boot_utes import (reload, add_path, path)\nadd_path('../src/', '~/repos/myutils/')",
"_____no_output_____"
],
[
"from collections import defaultdict, Counter, OrderedDict\nfrom functools import partial\nfrom itertools import count\nfrom operator import itemgetter as itg\nfrom pprint import pprint\nimport os, sys, itertools as it, simplejson, time\nfrom os.path import join\nfrom glob import glob",
"_____no_output_____"
]
],
[
[
"### Libs",
"_____no_output_____"
]
],
[
[
"# from gensim.models import Word2Vec\n\nimport feather\nimport numpy as np\nimport numpy.random as nr\nimport pandas as pd\nfrom pandas import DataFrame, Series\nfrom pandas.compat import lmap, lrange, lfilter, lzip\n\nimport toolz.curried as z\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline\npd.options.mode.use_inf_as_na = True\n\n# %mkdir cache\nimport joblib; mem = joblib.Memory(location='cache')\ndbx = '/Users/wbeard/miniconda3/envs/tmpdecomp2/bin/databricks'",
"_____no_output_____"
],
[
"def norm_rows(df):\n return df.div(df.sum(axis=1), axis=0)",
"_____no_output_____"
],
[
"from os.path import join\nimport re\n\ndef get_spk_parq_fn(dir, ext='parquet'):\n s3parq_fs = !aws s3 ls $dir/\n# print(dir)\n# print(s3parq_fs)\n spk_parq_re = re.compile(r'part.+?\\.{}'.format(ext))\n fn_all = spk_parq_re.findall(s3parq_fs[-1])\n if not len(fn_all) == 1:\n print('Uh oh: fn_all=', fn_all)\n [fn] = fn_all\n return fn\n\ndef show_s3_opts(*dir_elems):\n dir = join(*dir_elems)\n if not dir.endswith('/'):\n dir += '/'\n res = !aws s3 ls $dir\n return res\n \ndef mv_s3_pq(from_dir='', to_dir='data/pings', fn='pings_0125.parq', s3base=None, ext='parquet'):\n s3base = s3base or 's3://mozilla-databricks-telemetry-test/wbeard/'\n s3dir = join(s3base, from_dir, fn)\n \n try:\n pqn_ = get_spk_parq_fn(s3dir, ext=ext)\n print('Found pq file', pqn_)\n except IndexError:\n print('ERROR: File {} not found'.format(s3dir))\n opts = show_s3_opts(s3base, from_dir)\n print('\\nPossible options:\\n\\t', '\\n\\t'.join(opts))\n return\n \n \n s3loc = join(s3base, from_dir, fn, pqn_)\n to_loc = join(to_dir, fn)\n \n \n# print(s3loc)\n# print(to_loc)\n !aws s3 cp $s3loc $to_loc\n return to_loc\n \n",
"_____no_output_____"
]
],
[
[
"### Special",
"_____no_output_____"
]
],
[
[
"# import utils.seq_utils as su; reload(su)\n# import utils.events_utils as eu; reload(eu)\n# import utils.feather_counter as fc; reload(fc)\n# import utils.mutes as mt; reload(mt)\n\nimport myutils as mu\nimport functoolz as fz; reload(fz)\nimport numba_utils as nu; reload(nu)\n\nmt.set_mem(mem=mem, json=simplejson)\nsu.set_memo(memoizer=mt.json_memo())\n;;",
"_____no_output_____"
],
[
"def move_new(dbx_dir, outdir=None, outdir_base='../data/raw/'):\n full_dbx_dir = join(\"dbfs:/wbeard/\", dbx_dir)\n outdir = join(outdir_base, dbx_dir) if outdir is None else outdir\n dbx_fs = !$dbx fs ls $full_dbx_dir\n written = os.listdir(outdir)\n tobewritten = [f for f in dbx_fs if f not in written]\n for f in tobewritten:\n dbx_fn = join(full_dbx_dir, f)\n print('{} => {}'.format(dbx_fn, outdir))\n !$dbx fs cp $dbx_fn $outdir\n return tobewritten, written\n \n\ndef keep_moving(dbx_dir, outdir=None, outdir_base='../data/raw/', maxn=5):\n while 1:\n time.sleep(3)\n tobewritten, written = move_new(dbx_dir, outdir=outdir,\n outdir_base=outdir_base)\n\n if maxn and (len(tobewritten) + len(written) >= maxn):\n return\n print('.', end='')\n \n \nvc = lambda x: Series(x).value_counts(normalize=0).sort_index()\n",
"_____no_output_____"
],
[
"!aws s3 sync 's3://mozilla-databricks-telemetry-test/wbeard/crash_pings/p_201801' data/p29_0202",
"download: s3://mozilla-databricks-telemetry-test/wbeard/crash_pings/p_20180126_20180128_eq_42.parq/_SUCCESS to data/p26_8/_SUCCESS\ndownload: s3://mozilla-databricks-telemetry-test/wbeard/crash_pings/p_20180126_20180128_eq_42.parq/_committed_6840194297520358502 to data/p26_8/_committed_6840194297520358502\ndownload: s3://mozilla-databricks-telemetry-test/wbeard/crash_pings/p_20180126_20180128_eq_42.parq/_started_6840194297520358502 to data/p26_8/_started_6840194297520358502\ndownload: s3://mozilla-databricks-telemetry-test/wbeard/crash_pings/p_20180126_20180128_eq_42.parq/part-00002-tid-6840194297520358502-d54968ad-db37-4285-8f8c-e2e58808f762-0-c000.snappy.parquet to data/p26_8/part-00002-tid-6840194297520358502-d54968ad-db37-4285-8f8c-e2e58808f762-0-c000.snappy.parquet\ndownload: s3://mozilla-databricks-telemetry-test/wbeard/crash_pings/p_20180126_20180128_eq_42.parq/part-00000-tid-6840194297520358502-d54968ad-db37-4285-8f8c-e2e58808f762-0-c000.snappy.parquet to data/p26_8/part-00000-tid-6840194297520358502-d54968ad-db37-4285-8f8c-e2e58808f762-0-c000.snappy.parquet\ndownload: s3://mozilla-databricks-telemetry-test/wbeard/crash_pings/p_20180126_20180128_eq_42.parq/part-00001-tid-6840194297520358502-d54968ad-db37-4285-8f8c-e2e58808f762-0-c000.snappy.parquet to data/p26_8/part-00001-tid-6840194297520358502-d54968ad-db37-4285-8f8c-e2e58808f762-0-c000.snappy.parquet\n"
]
],
[
[
"## Win_proc",
"_____no_output_____"
]
],
[
[
"ls ../../win_proc/data/raw/",
"till_0204.pq\n"
],
[
"local_dir = '../../win_proc/data/raw/'\nbase = 's3://net-mozaws-prod-us-west-2-pipeline-analysis/wbeard/wl/'",
"_____no_output_____"
],
[
"# base = 'dbfs:/wbeard/apb/'\npqdir = 'till_0207.pq'\n# pqdir = 'abc'\n\n# , ext='json'\nmv_s3_pq(from_dir='', to_dir=local_dir, fn=pqdir, s3base=base)",
"Found pq file part-00000-tid-6114354128941207063-a497a401-5843-4966-9646-3d2d6bae1b20-2389165-c000.snappy.parquet\ndownload: s3://net-mozaws-prod-us-west-2-pipeline-analysis/wbeard/wl/till_0207.pq/part-00000-tid-6114354128941207063-a497a401-5843-4966-9646-3d2d6bae1b20-2389165-c000.snappy.parquet to ../data/raw/till_0207.pq\n"
],
[
"json_file = join(base, pqdir)\nprint(json_file)\n!aws s3 cp \"$json_file\" \"$local_dir\"",
"s3://net-mozaws-prod-us-west-2-pipeline-analysis/wbeard/ipc/samp.json\nfatal error: An error occurred (404) when calling the HeadObject operation: Key \"wbeard/ipc/samp.json\" does not exist\n"
],
[
"!aws s3 ls \"$json_file/\"",
"2018-11-28 12:23:24 0 _SUCCESS\n2018-11-28 12:23:24 117 _committed_3179433506591924477\n2018-11-28 12:23:23 0 _started_3179433506591924477\n2018-11-28 12:23:24 51818 part-00000-tid-3179433506591924477-7c90a292-abf3-44e3-98b3-7213773c5a1d-10458612-c000.json\n"
]
],
[
[
"_fns = !$dbx fs ls 'dbfs:/wbeard/ipc/'\n_fns = [f for f in _fns if not path.exists(join(local_dir, f))]\nprint(_fns)",
"_____no_output_____"
],
[
"!$dbx fs cp \"dbfs:/wbeard/apb/cid_ret_1122.pq\" $local_dir",
"_____no_output_____"
],
[
"# _fns = 'r23-25_counts.json r23-25_prompt.json r23-25_settings.json'.split()\nfor _fn in _fns:\n fn = join('dbfs:/wbeard/apb/day', _fn)\n print(fn)\n !$dbx fs cp $fn $local_dir",
"_____no_output_____"
]
],
[
[
"\n\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"raw",
"raw",
"raw"
],
[
"code"
]
] |
4a8d809e2c8879085d226201ee297724f61a446b
| 38,566 |
ipynb
|
Jupyter Notebook
|
danceClassifierAppMulticat.ipynb
|
vhpvmx/danceClassifierModelMulticat
|
c2c29ad979f2d10a01d3cdc548eb8a1e29a9f23d
|
[
"MIT"
] | null | null | null |
danceClassifierAppMulticat.ipynb
|
vhpvmx/danceClassifierModelMulticat
|
c2c29ad979f2d10a01d3cdc548eb8a1e29a9f23d
|
[
"MIT"
] | null | null | null |
danceClassifierAppMulticat.ipynb
|
vhpvmx/danceClassifierModelMulticat
|
c2c29ad979f2d10a01d3cdc548eb8a1e29a9f23d
|
[
"MIT"
] | null | null | null | 54.548798 | 18,814 | 0.764508 |
[
[
[
"import fastbook\nfastbook.setup_book()",
"_____no_output_____"
],
[
"from fastai.vision.all import *\nfrom fastai.vision.widgets import *",
"_____no_output_____"
],
[
"def parent_label_multi(o):\n return [Path(o).parent.name]",
"_____no_output_____"
],
[
"path = Path()\nlearn_inf = load_learner(path/'model.pkl', cpu=True)",
"_____no_output_____"
],
[
"btn_upload = widgets.FileUpload()\nout_pl = widgets.Output()\nlbl_pred = widgets.Label()\nbtn_run = widgets.Button(description='Classify')\n\ndef on_click_classify(change):\n img = PILImage.create(btn_upload.data[-1])\n out_pl.clear_output()\n with out_pl: display(img.to_thumb(512,512))\n pred,pred_idx,probs = learn_inf.predict(img)\n #lbl_pred.value = f'Prediction: {pred}; Probability: {probs[pred_idx]:.04f}'\n lbl_pred.value = f'Prediction: {pred}'\n\nbtn_run.on_click(on_click_classify)",
"_____no_output_____"
],
[
"#hide_output\nVBox([widgets.Label('Select your dancer!'), \n btn_upload, btn_run, out_pl, lbl_pred])",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8d89b20da70e33726215a0df231b5c9c90c875
| 181,863 |
ipynb
|
Jupyter Notebook
|
src/SentimentAnalysis2.ipynb
|
voschezang/books
|
5ce6ad693b8995ac43f5e597573a7c71e39933a5
|
[
"MIT"
] | null | null | null |
src/SentimentAnalysis2.ipynb
|
voschezang/books
|
5ce6ad693b8995ac43f5e597573a7c71e39933a5
|
[
"MIT"
] | null | null | null |
src/SentimentAnalysis2.ipynb
|
voschezang/books
|
5ce6ad693b8995ac43f5e597573a7c71e39933a5
|
[
"MIT"
] | null | null | null | 43.300714 | 3,180 | 0.373721 |
[
[
[
"from vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer\nimport os\nfrom nltk.tokenize import sent_tokenize\nimport pandas as pd\nfrom wordcloud import WordCloud\nfrom PIL import Image\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport random\nfrom nltk.corpus import stopwords\nfrom data import reduce_genres",
"Using TensorFlow backend.\n"
],
[
"import config\nimport tfidf2 as tfidf\nos.getcwd()",
"_____no_output_____"
],
[
"os.listdir(config.dataset_dir)",
"_____no_output_____"
],
[
"def make_lex_dict(lexicon_file):\n \"\"\"\n Convert lexicon file to a dictionary\n \"\"\"\n lex_dict = {}\n for line in lexicon_file.split('\\n'):\n (word, measure) = line.strip().split('\\t')[0:2]\n lex_dict[word] = float(measure)\n return lex_dict\n \nsent_dict = make_lex_dict(open('/Users/Tristan/books/src/' +'vader_lexicon.txt', 'r').read())\n",
"_____no_output_____"
]
],
[
[
"Sentiment analysis. Analysis is performed for each sentence and the sentiment scores kept in lists. Sentiment scores are calculated by averaging the sentiment scores for all sentences.",
"_____no_output_____"
]
],
[
[
"def return_sentiment_scores(sentence):\n # return just the sentiment scores\n snt = analyser.polarity_scores(sentence)\n return snt\n\ndef sentiment_analysis(directory):\n analyser = SentimentIntensityAnalyzer() \n # returns the sentiment of every book in the directory\n data = pd.read_csv(config.dataset_dir + 'output/final_data.csv', index_col=0)\n print(len(data.index))\n# max_amt = len(data.index) + 2\n# print(data.index, len(os.listdir(directory)))\n pos_list = []\n neg_list = []\n neu_list = []\n comp_list = []\n \n # for every book\n for filename in data['filename']:#[:max_amt]:\n \n sub_pos_list = []\n sub_neg_list = []\n sub_neu_list = []\n sub_comp_list = []\n \n # if file is a textfile\n if filename.endswith(\".txt\"):\n text = open(os.path.join(directory, filename), 'r', errors='replace')\n # for every line in the text\n for line in text.readlines():\n scores = return_sentiment_scores(line)\n # save sentiment scores \n sub_neg_list.append(scores['neg'])\n sub_neu_list.append(scores['neu'])\n sub_pos_list.append(scores['pos'])\n sub_comp_list.append(scores['compound'])\n \n # then save average sentiment scores for each book\n neg_list.append((sum(sub_neg_list) / float(len(sub_neg_list))))\n pos_list.append((sum(sub_pos_list) / float(len(sub_pos_list))))\n neu_list.append((sum(sub_neu_list) / float(len(sub_neu_list))))\n comp_list.append((sum(sub_comp_list) / float(len(sub_comp_list))))\n \n # convert scores to pandas compatible list\n neg = pd.Series(neg_list)\n pos = pd.Series(pos_list)\n neu = pd.Series(neu_list)\n com = pd.Series(comp_list)\n\n print(len(neg), len(pos), len(neu), len(com))\n # fill the right columns with the right data\n print(type(data),'type')\n print(neg)\n data['neg score'] = neg.values\n data['pos score'] = pos.values\n data['neu score'] = neu.values\n data['comp score'] = com.values\n data.to_csv(config.dataset_dir + 'output/final_data.csv')\n return data",
"_____no_output_____"
],
[
"analyser = SentimentIntensityAnalyzer() \nsentiment_analysis(config.dataset_dir + 'bookdatabase/books/')",
"915\n915 915 915 915\n<class 'pandas.core.frame.DataFrame'> type\n0 0.064028\n1 0.044295\n2 0.049711\n3 0.047641\n4 0.061587\n5 0.048907\n6 0.057010\n7 0.045972\n8 0.054442\n9 0.037464\n10 0.037340\n11 0.055954\n12 0.041392\n13 0.055000\n14 0.068802\n15 0.052302\n16 0.041800\n17 0.043438\n18 0.026211\n19 0.056150\n20 0.044526\n21 0.034635\n22 0.033165\n23 0.040971\n24 0.054841\n25 0.048444\n26 0.058575\n27 0.052053\n28 0.078131\n29 0.045618\n ... \n885 0.040172\n886 0.060173\n887 0.017144\n888 0.038590\n889 0.044522\n890 0.057267\n891 0.061412\n892 0.046271\n893 0.057172\n894 0.054442\n895 0.061912\n896 0.056598\n897 0.051939\n898 0.057441\n899 0.038862\n900 0.032124\n901 0.051378\n902 0.073029\n903 0.043030\n904 0.064964\n905 0.048816\n906 0.043717\n907 0.055604\n908 0.029775\n909 0.035116\n910 0.050853\n911 0.053077\n912 0.051314\n913 0.053722\n914 0.047641\nLength: 915, dtype: float64\n"
]
],
[
[
"We also want to count the amount of positive and negative words as features. We also create a new file for each book with just the sentiment words. As a result, we will be able to do tfidf on these files later and create wordclouds per genre.",
"_____no_output_____"
]
],
[
[
"def count_sentiment_words(directory):\n sent_words_list =[]\n pos_list = []\n neg_list = []\n \n data = pd.read_csv(config.dataset_dir + 'output/final_data.csv', index_col=0)\n\n for filename in data['filename']:\n sent_words_list =[]\n pos_count = 0\n neg_count = 0\n \n if filename.endswith(\".txt\"):\n text = open(os.path.join(directory, filename), 'r', errors='replace')\n sentiment_file = open(config.dataset_dir +'output/sentiment_word_texts/' + filename , 'w')\n\n for line in text.readlines():\n for word in line.split(\" \"):\n if word in sent_dict:\n if sent_dict[word] >= 0:\n pos_count += 1\n sent_words_list.append(word)\n sentiment_file.write(\"%s\" % word)\n sentiment_file.write(\" \")\n else:\n neg_count += 1\n sentiment_file.write(\"%s\" % word)\n sentiment_file.write(\" \")\n\n pos_list.append(pos_count)\n neg_list.append(neg_count)\n \n data['amt pos'] = pos_list \n data['amt neg'] = neg_list\n \n data.to_csv(config.dataset_dir + 'output/final_data.csv')\n return data\n\ncount_sentiment_words(config.dataset_dir + 'bookdatabase/books/')\n",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"def read_unique_genres():\n genres_file = open(config.dataset_dir + 'unique_genres.txt', 'r')\n return[genre.strip('\\n') for genre in genres_file.readlines()]",
"_____no_output_____"
],
[
"def create_wordcloud(scores, genre):\n \n font_path = config.dataset_dir + 'Open_Sans_Condensed/OpenSansCondensed-Light.ttf'\n stopWords = set(stopwords.words('english'))\n\n try:\n w = WordCloud(stopwords = stopWords, background_color='white', min_font_size=14, font_path=font_path, width = 1000, height = 500,relative_scaling=1,normalize_plurals=False)\n wordcloud = w.generate_from_frequencies(scores)\n wordcloud.recolor(color_func=grey_color_func)\n\n \n except ZeroDivisionError:\n print('shit')\n return\n \n plt.figure(figsize=(15,8))\n plt.imshow(wordcloud, interpolation='bilinear')\n plt.axis(\"off\")\n plt.savefig(config.dataset_dir + 'output/wordclouds/' + genre + '.png')\n plt.close()\n \ndef grey_color_func(word, font_size, position, orientation, random_state=None, **kwargs):\n return \"hsl(0, 0%%, %d%%)\" % random.randint(10, 50)\n\n\ndef tfidf_per_genre(plot_wc=False):\n data = pd.read_csv(config.dataset_dir + 'output/final_data.csv')\n genres_file = open(config.dataset_dir + 'unique_genres.txt', 'r')\n pre_genre_list = [genre.strip('\\n') for genre in genres_file.readlines()]\n directory = config.dataset_dir + 'output/sentiment_word_texts/'\n doc_list = []\n genre_list = reduce_genres(pre_genre_list)\n print(pre_genre_list)\n print(genre_list)\n # create a list of lists containing all tokens contained in the text of a certain genre\n for genre in genre_list:\n book_list = []\n genre = genre.replace('/', ' ')\n books_of_genre = data.loc[data['genre'] == genre]\n \n for book in books_of_genre['filename']:\n book_list.append(book)\n \n genre_document = tfidf.genre_document(book_list, directory)\n doc_list.append(genre_document)\n\n# create index\n index = tfidf.create_index(genre_list, doc_list)\n # create tf_matrix\n tf_matrix = tfidf.create_tf_matrix(genre_list, doc_list)\n \n # create scores for each genre\n for genre, document in zip(genre_list, doc_list):\n genre = genre.replace('/', ' ')\n score_dict = {}\n document = set(document)\n try:\n \n for term in document:\n\n score = tfidf.tfidf(term, genre, doc_list, index, tf_matrix)\n score_dict[term] = score\n\n scores_file = open(config.dataset_dir +'output/top200_per_genre/' + genre + '.txt', 'w')\n\n for w in sorted(score_dict, key=score_dict.get, reverse=True):\n\n scores_file.write('%s/n' % w)\n\n scores_file.close()\n\n print('success')\n\n if plot_wc:\n font_path = config.dataset_dir + 'Open_Sans_Condensed/OpenSansCondensed-Light.ttf'\n create_wordcloud(score_dict, genre)\n\n except ZeroDivisionError:\n print('reaallly')\n continue\n except ValueError:\n continue\n \n\ntfidf_dict_per_genre = tfidf_per_genre(plot_wc=True)",
"['Science fiction fandom', 'Diary and Novel', 'Fantasy literature', 'War', 'Epistolary novel', 'Künstlerroman', 'Short stories', 'Legal thriller', 'Bush poetry', 'Fantasy novel', 'Sketch story', 'Romance novel', 'Christian literature', 'Autobiographical novel', 'Business', 'Poem', 'Planetary romance', 'Utopian fiction', 'Romantic novel', 'Young-adult fiction', 'Non-fiction', 'Poetry', 'Historical', 'unknown', 'Nature writing', 'Non-Fiction', 'Christian fiction', 'Tragicomedy', 'Detective stories', \"Children's Literature\", 'Encyclopedia', 'Thriller (genre)', 'Christian novel', 'Fairytale fantasy', 'Satirical', 'Christian Apologetics', 'Crime fiction', 'Prose poetry', 'Young adult fiction', 'Philosophy', 'Humor', 'Historical fantasy', 'Detective fiction', 'Hindi', 'Biography in literature', 'Decadent movement', 'Cookbook', 'Verse novel', 'Apocalyptic and post-apocalyptic fiction', 'Textbook', 'Essay', 'History of Ideas', 'Adventure novel', 'Family saga', 'Mystery novel', 'Short story', 'Novella', \"Children's literature\", 'Dystopian', 'Captivity narrative', 'Biographical novel', 'Comedy', 'Science fantasy', 'Detective novel', 'Lost World (genre)', 'Spiritual autobiography', 'Short story cycle', 'Social commentary', 'Political journalism', 'Adventure fiction', 'Nonfiction', 'Thriller fiction', 'Picture book', 'Historical mystery', 'Comic novel', 'Monograph', 'Alternate history', 'Political philosophy', 'Horror novel', 'Comedy novel', 'Science', 'Photography', 'Science fiction', 'Historical Fiction', 'Travel literature', 'Thriller novel', 'Literary realism', 'Historical fiction', 'Philosophical novel', 'Fiction', 'Ruritanian romance', 'Tragedy', 'Dystopian novel', 'Cryptozoology', \"Children's book\", 'Black comedy', 'Cultural criticism', 'Fantasy/Ethnic novel', 'Crime novel', 'Western fiction', 'Economics', 'Political', 'Gothic fiction', 'Satire', 'Syair', 'Temperance movement', 'Sword and sorcery', 'Wuxia', 'Utopian novel', 'Bildungsroman', 'Fantasy', \"Children's novel\", 'Slave narrative', 'Frame story', 'Non-Profit', 'Novel of manners', 'Canadian literature', 'Interior design', 'Victorian literature', 'Folklore', 'Christian mythology', 'Military science fiction', 'Historical novels', 'Chivalric romance', 'Juvenile literature', 'Reference work', 'Young-Adult Fiction', 'Drama', 'Travel writing', 'Political novel', 'Science Fantasy', 'Romanticism', 'Christian mysticism', 'Utopian and dystopian fiction', 'Mystery (fiction)', 'Western novel', \"Children's fiction\", 'World War II', 'Theodicy', 'Feminist science fiction', 'Fairy tale', 'Horror fiction', 'Sensation novel', 'Psychological novel', 'Military history', 'Western (genre)', 'Autobiography', 'War novel', 'Political thriller', 'Self-help', 'Roman à clef', 'Fantasy fiction', \"Children's books\", 'Historical Novel', 'Biography', 'Psychological Thriller', 'Mystery fiction', 'Young adult novel', 'Historical novel', 'Gaucho literature', 'Contemporary romance', 'Apocalyptic fiction', 'Nonsense poetry', 'Adventure', 'Philosophical literature', 'Spy novel', 'History', 'Philosophical fiction', 'Young adult novels', 'Memoir', 'Dark fantasy', 'Gothic novel', 'Novel']\n{'fiction', 'short stori', 'militariscienc', 'romantic', 'econom', 'satir', 'monograph', 'novel', 'captiv narr', 'frame stori', 'knstlerroman', 'crime', 'altern histori', 'science', 'histori of idea', 'biographiin', 'folklor', 'diariand', 'photographi', 'natur write', 'children', 'novella', 'polit', 'syair', 'roman clef', 'ruritanian romanc', 'bildungsroman', 'chivalr romanc', 'travel write', 'epistolari', 'short stori cycl', 'victorian', 'nonprofit', 'militari histori', 'autobiograph', 'gaucho', 'bush poetri', 'fantasi', 'refer work', 'poem', 'spiritu autobiographi', 'apocalypt', 'detect', 'planetari romanc', 'contemporari romanc', 'comedi', 'detect stori', 'selfhelp', 'biographi', 'fairi tale', 'mysteri', 'feministscienc', 'poetri', 'vers', 'essay', 'interior design', 'famili saga', 'drama', 'nonfict', 'black comedi', 'utopian', 'biograph', 'autobiographi', 'humor', 'temper movement', 'thriller', 'pictur book', 'dystopian', 'tragedi', 'nonsens poetri', 'hindi', 'lost world genr', 'travel', 'war', 'literari realism', 'decad movement', 'textbook', 'utopiananddystopian', 'sword and sorceri', 'slave narr', 'cultur critic', 'christian', 'memoir', 'world war ii', 'theodici', 'busi', 'social commentari', 'wuxia', 'spi', 'juvenil', 'tragicomedi', 'sensat', 'science fiction', 'psycholog', 'western', 'sketch stori', 'cookbook', 'fantasyethn', 'comic', 'adventur', 'encyclopedia', 'cryptozoolog', 'horror', 'histor', 'novel of manner', 'young adult', 'canadian', 'apocalyptandpostapocalypt', 'unknown', 'prose poetri', 'gothic', 'philosophi', 'romance'}\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\nsuccess\n"
],
[
"list(tfidf_dict_per_genre.keys())[:4]",
"_____no_output_____"
],
[
"len(list(tfidf_dict_per_genre.keys()))",
"_____no_output_____"
],
[
"len(tfidf_dict_per_genre['Diary and Novel']) # may differ per genre",
"_____no_output_____"
],
[
"n_words_per_genre = 100",
"_____no_output_____"
],
[
"sample = tfidf_dict_per_genre['War']\ni = list(sample.keys())[-1]\nsample[i]",
"_____no_output_____"
],
[
"max(list(sample.values()))",
"_____no_output_____"
],
[
"sample",
"_____no_output_____"
]
],
[
[
"## Generate labels file",
"_____no_output_____"
]
],
[
[
"import pandas, os\nimport data, config\nfrom utils import io",
"Using TensorFlow backend.\n"
],
[
"info = pandas.read_csv(config.info_file)\nbook_list = os.listdir(config.sentiment_words_dir)[:]\nlabels = data.extract_genres(info, book_list)",
"_____no_output_____"
],
[
"labels",
"_____no_output_____"
],
[
"io.save_dict_to_csv(config.dataset_dir, 'labels', labels)",
"_____no_output_____"
]
],
[
[
"# (oud)\nChoose to most important to be kept in the feature-vector",
"_____no_output_____"
]
],
[
[
"from wordcloud import WordCloud\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"import data, config, tfidf",
"_____no_output_____"
],
[
"directory = config.dataset_dir + 'output/sentiment_word_texts'\nbook_list = os.listdir(directory)\nbook_list = book_list[:20]",
"_____no_output_____"
],
[
"index = tfidf.create_index(directory, book_list)\ntf_matrix = tfidf.create_tf_matrix(directory, book_list)",
"_____no_output_____"
],
[
"tfidf_dict = tfidf.perform_tfidf(directory, book_list, index, tf_matrix)",
"_____no_output_____"
],
[
"# (optional) show the result\nw = WordCloud(background_color='white', width=900, height=500, \n max_words=1628,relative_scaling=1,normalize_plurals=False)\nwordcloud = w.generate_from_frequencies(tfidf_dict)",
"_____no_output_____"
],
[
"plt.imshow(wordcloud, interpolation='bilinear')\nplt.axis(\"off\")\n# plt.savefig(config.dataset_dir + 'output/wordclouds/' + genre + '.png')",
"_____no_output_____"
],
[
"# tfidf_dict_per_genre = wordcloud_per_genre()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8d8a635e0e914e8d04822215a87eb65f4618ff
| 2,805 |
ipynb
|
Jupyter Notebook
|
4_basic_image_recognition/1_download_testdata.ipynb
|
xiabai84/tensorflow_for_engineer
|
8036239050e1142aa708429d5c3febff4cfb9a88
|
[
"MIT"
] | 1 |
2020-08-14T14:54:28.000Z
|
2020-08-14T14:54:28.000Z
|
4_basic_image_recognition/1_download_testdata.ipynb
|
xiabai84/tensorflow_for_enginner
|
8036239050e1142aa708429d5c3febff4cfb9a88
|
[
"MIT"
] | null | null | null |
4_basic_image_recognition/1_download_testdata.ipynb
|
xiabai84/tensorflow_for_enginner
|
8036239050e1142aa708429d5c3febff4cfb9a88
|
[
"MIT"
] | null | null | null | 28.917526 | 129 | 0.505882 |
[
[
[
"destination_folder = 'data/'\n\nfile_names = ['train-images-idx3-ubyte.gz',\n 'train-labels-idx1-ubyte.gz',\n 't10k-images-idx3-ubyte.gz',\n 't10k-labels-idx1-ubyte.gz']\n\ndownload_url = 'http://yann.lecun.com/exdb/mnist/'",
"_____no_output_____"
]
],
[
[
"## Download test data from internet, make sure you have internet access",
"_____no_output_____"
]
],
[
[
"import wget\nfrom sh import gunzip\n\nfor file in file_names:\n print(\"Donwload and extract %s from %s\" % (file, download_url+file))\n wget.download(url=download_url + file, out='data/')\n #gunzip(destination_folder + file)\n ",
"Donwload and extract train-images-idx3-ubyte.gz from http://yann.lecun.com/exdb/mnist/train-images-idx3-ubyte.gz\nDonwload and extract train-labels-idx1-ubyte.gz from http://yann.lecun.com/exdb/mnist/train-labels-idx1-ubyte.gz\nDonwload and extract t10k-images-idx3-ubyte.gz from http://yann.lecun.com/exdb/mnist/t10k-images-idx3-ubyte.gz\nDonwload and extract t10k-labels-idx1-ubyte.gz from http://yann.lecun.com/exdb/mnist/t10k-labels-idx1-ubyte.gz\n"
],
[
"for file in file_names:\n gunzip(destination_folder + file)\n\nprint('finished!')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a8d8ec447cfcf8e8a4e984cf6fa0a5e62aa3f3a
| 29,410 |
ipynb
|
Jupyter Notebook
|
model.ipynb
|
sushant1212/Sentence_Decoding_NLP
|
907db91931141926dffc9394e007a3715d9b0042
|
[
"MIT"
] | 1 |
2021-11-29T10:08:57.000Z
|
2021-11-29T10:08:57.000Z
|
model.ipynb
|
sushant1212/Sentence_Decoding_NLP
|
907db91931141926dffc9394e007a3715d9b0042
|
[
"MIT"
] | null | null | null |
model.ipynb
|
sushant1212/Sentence_Decoding_NLP
|
907db91931141926dffc9394e007a3715d9b0042
|
[
"MIT"
] | null | null | null | 30.795812 | 129 | 0.50765 |
[
[
[
"import pickle\nimport pandas as pd\nimport re\nimport nltk\nfrom nltk.corpus import stopwords\nfrom nltk.stem import WordNetLemmatizer\nimport numpy as np\nimport bcolz\nimport unicodedata\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport time\nimport torch.optim as optim\nimport matplotlib.pyplot as plt\nplt.switch_backend('agg')\nimport matplotlib.ticker as ticker\nimport numpy as np\nimport random",
"_____no_output_____"
]
],
[
[
"# Preprocessing the text data",
"_____no_output_____"
]
],
[
[
"def unicodeToAscii(s):\n return ''.join(\n c for c in unicodedata.normalize('NFD', s)\n if unicodedata.category(c) != 'Mn'\n )\n",
"_____no_output_____"
],
[
"def normalizeString(s):\n s = unicodeToAscii(s.lower().strip())\n s = s.replace(\"'\",\"\")\n s = re.sub(r\"([.!?])\", r\" \\1\", s)\n s = re.sub(r\"[^a-zA-Z.!?]+\", r\" \", s)\n return s",
"_____no_output_____"
],
[
"def preprocess(df):\n nrows = len(df)\n real_preprocess = []\n df['Content_Parsed_1'] = df['transcription']\n for row in range(0, nrows):\n\n # Create an empty list containing preprocessed words\n real_preprocess = []\n\n # Save the text and its words into an object\n text = df.loc[row]['transcription']\n text = normalizeString(text)\n\n\n df.loc[row]['Content_Parsed_1'] = text\n\n df['action'] = df['action'].str.lower()\n df['object'] = df['object'].str.lower()\n df['location'] = df['location'].str.lower()",
"_____no_output_____"
],
[
"nltk.download('wordnet')\n",
"[nltk_data] Downloading package wordnet to\n[nltk_data] C:\\Users\\DELL\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n"
],
[
"def lemmatize(df):\n wordnet_lemmatizer = WordNetLemmatizer()\n # Lemmatizing the content\n nrows = len(df)\n lemmatized_text_list = []\n for row in range(0, nrows):\n\n # Create an empty list containing lemmatized words\n lemmatized_list = []\n\n # Save the text and its words into an object\n text = df.loc[row]['Content_Parsed_1']\n text_words = text.split(\" \")\n\n # Iterate through every word to lemmatize\n for word in text_words:\n lemmatized_list.append(wordnet_lemmatizer.lemmatize(word, pos=\"v\"))\n\n # Join the list\n lemmatized_text = \" \".join(lemmatized_list)\n\n # Append to the list containing the texts\n lemmatized_text_list.append(lemmatized_text)\n df['Content_Parsed_2'] = lemmatized_text_list\n",
"_____no_output_____"
],
[
"path_df = \"E:/saarthi/task_data/train_data.csv\"\nwith open(path_df, 'rb') as data:\n df = pd.read_csv(data)",
"_____no_output_____"
],
[
"path_df_val = \"E:/saarthi/task_data/valid_data.csv\"\nwith open(path_df, 'rb') as data:\n df_val = pd.read_csv(data)",
"_____no_output_____"
],
[
"preprocess(df_val)\nlemmatize(df_val)",
"_____no_output_____"
],
[
"preprocess(df)\nlemmatize(df)",
"_____no_output_____"
]
],
[
[
"# Getting Glove Word embeddings ",
"_____no_output_____"
]
],
[
[
"glove_path = \"E:\"",
"_____no_output_____"
],
[
"vectors = bcolz.open(f'{glove_path}/6B.50.dat')[:]\nwords = pickle.load(open(f'{glove_path}/6B.50_words.pkl', 'rb'))\nword2idx = pickle.load(open(f'{glove_path}/6B.50_idx.pkl', 'rb'))\nglove = {w: vectors[word2idx[w]] for w in words}",
"_____no_output_____"
],
[
"target_vocab = []\nnrows = len(df)\nfor row in range(0, nrows):\n text = df.loc[row]['Content_Parsed_2']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
],
[
"target_vocab = []\nnrows = len(df_val)\nfor row in range(0, nrows):\n text = df.loc[row]['Content_Parsed_2']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
],
[
"nrows = len(df)\nfor row in range(0, nrows):\n text = df.loc[row]['action']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
],
[
"nrows = len(df_val)\nfor row in range(0, nrows):\n text = df.loc[row]['action']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
],
[
"nrows = len(df)\nfor row in range(0, nrows):\n text = df.loc[row]['object']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
],
[
"nrows = len(df_val)\nfor row in range(0, nrows):\n text = df.loc[row]['object']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
],
[
"nrows = len(df)\nfor row in range(0, nrows):\n text = df.loc[row]['location']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
],
[
"nrows = len(df_val)\nfor row in range(0, nrows):\n text = df.loc[row]['location']\n text_words = text.split(\" \")\n for word in text_words:\n if word not in target_vocab:\n target_vocab.append(word)",
"_____no_output_____"
]
],
[
[
"# Creating an embedding matrix",
"_____no_output_____"
]
],
[
[
"vocab_size = len(target_vocab)\ninput_size = 50\n\nembedding_matrix = torch.zeros((vocab_size, input_size))\nfor w in target_vocab:\n i = word_to_idx(w)\n \n embedding_matrix[i, :] = torch.from_numpy(glove[w]).float()",
"_____no_output_____"
]
],
[
[
"# Defining utility functions",
"_____no_output_____"
]
],
[
[
"def word_to_idx(word):\n for i, w in enumerate(target_vocab):\n if w == word:\n return i\n return -1",
"_____no_output_____"
],
[
"def sentence_to_matrix(sentence):\n words = sentence.split(\" \")\n n = len(words)\n m = torch.zeros((n, input_size))\n for i, w in enumerate(words):\n m[i] = embedding_matrix[word_to_idx(w)]\n return m",
"_____no_output_____"
],
[
"def sentence_to_index(sentence):\n w = sentence.split(\" \")\n l = []\n for word in w:\n l.append(word_to_idx(word))\n t = torch.tensor(l, dtype=torch.float32)\n return t",
"_____no_output_____"
],
[
"output_size = len(target_vocab)\ninput_size = 50\nhidden_size = 50",
"_____no_output_____"
],
[
"def showPlot(points):\n plt.figure()\n fig, ax = plt.subplots()\n # this locator puts ticks at regular intervals\n loc = ticker.MultipleLocator(base=0.2)\n ax.yaxis.set_major_locator(loc)\n plt.plot(points)",
"_____no_output_____"
],
[
"import time\nimport math\n\n\ndef asMinutes(s):\n m = math.floor(s / 60)\n s -= m * 60\n return '%dm %ds' % (m, s)\n\n\ndef timeSince(since, percent):\n now = time.time()\n s = now - since\n es = s / (percent)\n rs = es - s\n return '%s (- %s)' % (asMinutes(s), asMinutes(rs))",
"_____no_output_____"
]
],
[
[
"# Creating the Networks",
"_____no_output_____"
]
],
[
[
"class EncoderRNN(nn.Module):\n def __init__(self, input_size, hidden_size):\n super(EncoderRNN, self).__init__()\n self.hidden_size = hidden_size\n self.gru = nn.GRU(input_size, hidden_size)\n\n def forward(self, x, hidden):\n x = x.unsqueeze(0)\n output, hidden = self.gru(x, hidden)\n return output, hidden\n\n def initHidden(self):\n return torch.zeros(1, 1, self.hidden_size, device=device)",
"_____no_output_____"
],
[
"s = \"turn down the bathroom temperature\"\ndevice = 'cuda:0' if torch.cuda.is_available() else 'cpu'\nmatrix = sentence_to_matrix(s)\nprint(matrix[0].unsqueeze(0).shape)\nencoder = EncoderRNN(input_size, hidden_size)\nhidden = encoder.initHidden()\nfor i in range(matrix.shape[0]):\n out, hidden = encoder(matrix[i].unsqueeze(0), hidden)\nprint(out.shape)\nprint(hidden.shape)",
"torch.Size([1, 50])\n5\ntorch.Size([1, 1, 50])\ntorch.Size([1, 1, 50])\n"
],
[
"class DecoderRNN(nn.Module):\n def __init__(self, hidden_size, output_size):\n super(DecoderRNN, self).__init__()\n self.hidden_size = hidden_size\n self.gru = nn.GRU(hidden_size, hidden_size)\n self.out = nn.Linear(hidden_size, output_size)\n self.softmax = nn.LogSoftmax(dim=1)\n\n def forward(self, x, hidden):\n output = F.relu(x)\n output, hidden = self.gru(output, hidden)\n output_softmax = self.softmax(self.out(output[0]))\n return output, hidden, output_softmax\n\n def initHidden(self):\n return torch.zeros(1, 1, self.hidden_size, device=device)",
"_____no_output_____"
],
[
"decoder_hidden = hidden\ndecoder_input = torch.ones((1,1,50))\ndecoder = DecoderRNN(hidden_size, output_size)\noutput_sentence = df.loc[3][\"action\"] + \" \"+ df.loc[3][\"object\"] + \" \" + df.loc[3][\"location\"]\nprint(output_sentence)\ntarget_tensor = sentence_to_index(output_sentence)\ncriterion = nn.NLLLoss()\nloss = 0\nfor i in range(target_tensor.shape[0]):\n decoder_input, decoder_hidden, decoder_output_softmax = decoder(decoder_input, decoder_hidden)\n loss += criterion(decoder_output_softmax, target_tensor[i].unsqueeze(0).long())\n print(torch.argmax(decoder_output_softmax, dim=1))",
"decrease heat washroom\ntensor([25])\ntensor([25])\ntensor([22])\n"
]
],
[
[
"# Training the networks",
"_____no_output_____"
]
],
[
[
"def train(input_tensor, target_tensor, encoder, decoder, encoder_optimizer, decoder_optimizer, criterion):\n \n encoder_hidden = encoder.initHidden()\n encoder_optimizer.zero_grad()\n decoder_optimizer.zero_grad()\n\n input_length = input_tensor.size(0)\n target_length = target_tensor.size(0)\n\n loss = 0\n\n for ei in range(input_length):\n encoder_output, encoder_hidden = encoder(input_tensor[ei].unsqueeze(0), encoder_hidden)\n\n decoder_input = torch.ones((1,1,50))\n\n decoder_hidden = encoder_hidden\n\n \n for i in range(target_tensor.shape[0]):\n decoder_input, decoder_hidden, decoder_output_softmax = decoder(decoder_input, decoder_hidden)\n loss += criterion(decoder_output_softmax, target_tensor[i].unsqueeze(0).long())\n\n loss.backward()\n\n encoder_optimizer.step()\n decoder_optimizer.step()\n\n return loss.item() / target_length",
"_____no_output_____"
],
[
"def trainIters(encoder, decoder, n_iters, df, print_every=1000, plot_every=100, learning_rate=0.01):\n start = time.time()\n plot_losses = []\n print_loss_total = 0 # Reset every print_every\n plot_loss_total = 0 # Reset every plot_every\n\n encoder_optimizer = optim.SGD(encoder.parameters(), lr=learning_rate)\n decoder_optimizer = optim.SGD(decoder.parameters(), lr=learning_rate)\n criterion = nn.NLLLoss()\n \n nrows = len(df)\n\n for iter in range(1, n_iters + 1):\n i = random.randint(0, n_iters)\n i = (i % nrows)\n \n s = df.loc[i][\"Content_Parsed_2\"]\n\n \n input_tensor = sentence_to_matrix(s)\n \n output_sentence = df.loc[i][\"action\"] + \" \"+ df.loc[i][\"object\"] + \" \" + df.loc[i][\"location\"]\n target_tensor = sentence_to_index(output_sentence)\n\n loss = train(input_tensor, target_tensor, encoder, decoder, encoder_optimizer, decoder_optimizer, criterion)\n print_loss_total += loss\n plot_loss_total += loss\n\n if iter % print_every == 0:\n print_loss_avg = print_loss_total / print_every\n print_loss_total = 0\n print('%s (%d %d%%) %.4f' % (timeSince(start, iter / n_iters),\n iter, iter / n_iters * 100, print_loss_avg))\n\n if iter % plot_every == 0:\n plot_loss_avg = plot_loss_total / plot_every\n plot_losses.append(plot_loss_avg)\n plot_loss_total = 0\n\n showPlot(plot_losses)",
"_____no_output_____"
],
[
"def predict(encoder, decoder, input_sentence):\n encoder_hidden = encoder.initHidden()\n input_tensor = sentence_to_matrix(input_sentence)\n decoder_input = torch.ones((1,1,50))\n input_length = input_tensor.size(0)\n \n for ei in range(input_length):\n encoder_output, encoder_hidden = encoder(input_tensor[ei].unsqueeze(0), encoder_hidden)\n \n decoder_hidden = encoder_hidden\n \n for i in range(3):\n decoder_input, decoder_hidden, decoder_output_softmax = decoder(decoder_input, decoder_hidden)\n idx = torch.argmax(decoder_output_softmax)\n print(target_vocab[idx])",
"_____no_output_____"
],
[
"def evaluate(encoder, decoder, input_sentence, target_tensor):\n encoder_hidden = encoder.initHidden()\n input_tensor = sentence_to_matrix(input_sentence)\n decoder_input = torch.ones((1,1,50))\n input_length = input_tensor.size(0)\n \n for ei in range(input_length):\n encoder_output, encoder_hidden = encoder(input_tensor[ei].unsqueeze(0), encoder_hidden)\n \n decoder_hidden = encoder_hidden\n correct = 0\n for i in range(3):\n decoder_input, decoder_hidden, decoder_output_softmax = decoder(decoder_input, decoder_hidden)\n idx = torch.argmax(decoder_output_softmax)\n if(idx == target_tensor[i]):\n correct += 1\n if(correct == 3):\n return 1\n else: \n return 0",
"_____no_output_____"
],
[
"encoder = EncoderRNN(input_size, hidden_size).to(device)\ndecoder = DecoderRNN(hidden_size, output_size)\n\ntrainIters(encoder, decoder, 150000, df)",
"0m 3s (- 9m 23s) (1000 0%) 2.1604\n0m 7s (- 9m 40s) (2000 1%) 1.0210\n0m 11s (- 9m 27s) (3000 2%) 0.5684\n0m 16s (- 9m 47s) (4000 2%) 0.4079\n0m 20s (- 9m 48s) (5000 3%) 0.3162\n0m 24s (- 9m 44s) (6000 4%) 0.2225\n0m 28s (- 9m 43s) (7000 4%) 0.1346\n0m 32s (- 9m 35s) (8000 5%) 0.0895\n0m 36s (- 9m 36s) (9000 6%) 0.0623\n0m 40s (- 9m 33s) (10000 6%) 0.0533\n0m 45s (- 9m 36s) (11000 7%) 0.0448\n0m 49s (- 9m 31s) (12000 8%) 0.0309\n0m 53s (- 9m 23s) (13000 8%) 0.0360\n0m 58s (- 9m 24s) (14000 9%) 0.0314\n1m 2s (- 9m 21s) (15000 10%) 0.0271\n1m 6s (- 9m 15s) (16000 10%) 0.0204\n1m 10s (- 9m 13s) (17000 11%) 0.0163\n1m 14s (- 9m 8s) (18000 12%) 0.0150\n1m 19s (- 9m 7s) (19000 12%) 0.0162\n1m 24s (- 9m 8s) (20000 13%) 0.0127\n1m 29s (- 9m 10s) (21000 14%) 0.0138\n1m 35s (- 9m 14s) (22000 14%) 0.0132\n1m 40s (- 9m 14s) (23000 15%) 0.0171\n1m 46s (- 9m 18s) (24000 16%) 0.0113\n1m 52s (- 9m 23s) (25000 16%) 0.0107\n1m 57s (- 9m 18s) (26000 17%) 0.0094\n2m 2s (- 9m 17s) (27000 18%) 0.0114\n2m 7s (- 9m 16s) (28000 18%) 0.0154\n2m 13s (- 9m 15s) (29000 19%) 0.0077\n2m 19s (- 9m 17s) (30000 20%) 0.0088\n2m 24s (- 9m 15s) (31000 20%) 0.0059\n2m 30s (- 9m 13s) (32000 21%) 0.0065\n2m 36s (- 9m 13s) (33000 22%) 0.0060\n2m 42s (- 9m 12s) (34000 22%) 0.0066\n2m 46s (- 9m 8s) (35000 23%) 0.0066\n2m 52s (- 9m 4s) (36000 24%) 0.0098\n2m 57s (- 9m 3s) (37000 24%) 0.0059\n3m 2s (- 8m 59s) (38000 25%) 0.0065\n3m 8s (- 8m 56s) (39000 26%) 0.0062\n3m 13s (- 8m 51s) (40000 26%) 0.0057\n3m 18s (- 8m 46s) (41000 27%) 0.0103\n3m 24s (- 8m 45s) (42000 28%) 0.0099\n3m 30s (- 8m 42s) (43000 28%) 0.0056\n3m 35s (- 8m 38s) (44000 29%) 0.0044\n3m 41s (- 8m 35s) (45000 30%) 0.0038\n3m 46s (- 8m 31s) (46000 30%) 0.0058\n3m 51s (- 8m 26s) (47000 31%) 0.0040\n3m 56s (- 8m 22s) (48000 32%) 0.0065\n4m 1s (- 8m 17s) (49000 32%) 0.0050\n4m 6s (- 8m 13s) (50000 33%) 0.0034\n4m 12s (- 8m 10s) (51000 34%) 0.0035\n4m 17s (- 8m 4s) (52000 34%) 0.0024\n4m 22s (- 8m 0s) (53000 35%) 0.0028\n4m 28s (- 7m 57s) (54000 36%) 0.0025\n4m 34s (- 7m 53s) (55000 36%) 0.0028\n4m 38s (- 7m 48s) (56000 37%) 0.0022\n4m 44s (- 7m 43s) (57000 38%) 0.0024\n4m 49s (- 7m 39s) (58000 38%) 0.0020\n4m 54s (- 7m 33s) (59000 39%) 0.0022\n5m 0s (- 7m 30s) (60000 40%) 0.0019\n5m 5s (- 7m 26s) (61000 40%) 0.0019\n5m 10s (- 7m 21s) (62000 41%) 0.0020\n5m 16s (- 7m 17s) (63000 42%) 0.0020\n5m 21s (- 7m 12s) (64000 42%) 0.0017\n5m 26s (- 7m 7s) (65000 43%) 0.0018\n5m 31s (- 7m 2s) (66000 44%) 0.0018\n5m 37s (- 6m 58s) (67000 44%) 0.0016\n5m 43s (- 6m 53s) (68000 45%) 0.0017\n5m 48s (- 6m 49s) (69000 46%) 0.0017\n5m 53s (- 6m 44s) (70000 46%) 0.0017\n5m 59s (- 6m 39s) (71000 47%) 0.0017\n6m 4s (- 6m 34s) (72000 48%) 0.0017\n6m 10s (- 6m 30s) (73000 48%) 0.0016\n6m 15s (- 6m 25s) (74000 49%) 0.0015\n6m 21s (- 6m 21s) (75000 50%) 0.0015\n6m 27s (- 6m 16s) (76000 50%) 0.0015\n6m 32s (- 6m 12s) (77000 51%) 0.0013\n6m 38s (- 6m 7s) (78000 52%) 0.0014\n6m 43s (- 6m 2s) (79000 52%) 0.0013\n6m 49s (- 5m 57s) (80000 53%) 0.0013\n6m 54s (- 5m 52s) (81000 54%) 0.0013\n7m 0s (- 5m 48s) (82000 54%) 0.0013\n7m 5s (- 5m 43s) (83000 55%) 0.0012\n7m 11s (- 5m 38s) (84000 56%) 0.0012\n7m 16s (- 5m 33s) (85000 56%) 0.0013\n7m 21s (- 5m 28s) (86000 57%) 0.0012\n7m 26s (- 5m 23s) (87000 57%) 0.0011\n7m 31s (- 5m 17s) (88000 58%) 0.0012\n7m 36s (- 5m 13s) (89000 59%) 0.0011\n7m 42s (- 5m 8s) (90000 60%) 0.0012\n7m 49s (- 5m 4s) (91000 60%) 0.0012\n7m 55s (- 4m 59s) (92000 61%) 0.0011\n8m 1s (- 4m 55s) (93000 62%) 0.0010\n8m 8s (- 4m 50s) (94000 62%) 0.0011\n8m 13s (- 4m 45s) (95000 63%) 0.0010\n8m 18s (- 4m 40s) (96000 64%) 0.0011\n8m 24s (- 4m 35s) (97000 64%) 0.0010\n8m 29s (- 4m 30s) (98000 65%) 0.0011\n8m 34s (- 4m 24s) (99000 66%) 0.0010\n8m 39s (- 4m 19s) (100000 66%) 0.0009\n8m 45s (- 4m 14s) (101000 67%) 0.0010\n8m 50s (- 4m 9s) (102000 68%) 0.0010\n8m 55s (- 4m 4s) (103000 68%) 0.0009\n9m 1s (- 3m 59s) (104000 69%) 0.0009\n9m 7s (- 3m 54s) (105000 70%) 0.0010\n9m 13s (- 3m 49s) (106000 70%) 0.0009\n9m 19s (- 3m 44s) (107000 71%) 0.0009\n9m 25s (- 3m 39s) (108000 72%) 0.0009\n9m 31s (- 3m 34s) (109000 72%) 0.0008\n9m 37s (- 3m 30s) (110000 73%) 0.0009\n9m 43s (- 3m 25s) (111000 74%) 0.0009\n9m 50s (- 3m 20s) (112000 74%) 0.0008\n9m 55s (- 3m 15s) (113000 75%) 0.0008\n10m 2s (- 3m 10s) (114000 76%) 0.0008\n10m 7s (- 3m 5s) (115000 76%) 0.0008\n10m 13s (- 2m 59s) (116000 77%) 0.0008\n10m 19s (- 2m 54s) (117000 78%) 0.0008\n10m 25s (- 2m 49s) (118000 78%) 0.0008\n10m 31s (- 2m 44s) (119000 79%) 0.0008\n10m 37s (- 2m 39s) (120000 80%) 0.0008\n10m 43s (- 2m 34s) (121000 80%) 0.0008\n10m 49s (- 2m 28s) (122000 81%) 0.0008\n10m 54s (- 2m 23s) (123000 82%) 0.0008\n10m 59s (- 2m 18s) (124000 82%) 0.0008\n11m 4s (- 2m 12s) (125000 83%) 0.0007\n11m 9s (- 2m 7s) (126000 84%) 0.0007\n11m 14s (- 2m 2s) (127000 84%) 0.0007\n11m 20s (- 1m 56s) (128000 85%) 0.0007\n11m 27s (- 1m 51s) (129000 86%) 0.0007\n11m 32s (- 1m 46s) (130000 86%) 0.0007\n11m 38s (- 1m 41s) (131000 87%) 0.0008\n11m 44s (- 1m 36s) (132000 88%) 0.0007\n11m 49s (- 1m 30s) (133000 88%) 0.0007\n11m 55s (- 1m 25s) (134000 89%) 0.0007\n12m 1s (- 1m 20s) (135000 90%) 0.0007\n12m 7s (- 1m 14s) (136000 90%) 0.0007\n12m 13s (- 1m 9s) (137000 91%) 0.0007\n12m 18s (- 1m 4s) (138000 92%) 0.0007\n12m 23s (- 0m 58s) (139000 92%) 0.0007\n12m 29s (- 0m 53s) (140000 93%) 0.0007\n12m 35s (- 0m 48s) (141000 94%) 0.0006\n12m 41s (- 0m 42s) (142000 94%) 0.0006\n12m 46s (- 0m 37s) (143000 95%) 0.0006\n12m 52s (- 0m 32s) (144000 96%) 0.0006\n12m 58s (- 0m 26s) (145000 96%) 0.0006\n13m 4s (- 0m 21s) (146000 97%) 0.0006\n13m 10s (- 0m 16s) (147000 98%) 0.0006\n13m 16s (- 0m 10s) (148000 98%) 0.0006\n13m 22s (- 0m 5s) (149000 99%) 0.0006\n13m 27s (- 0m 0s) (150000 100%) 0.0006\n"
]
],
[
[
"# Evaluating the model",
"_____no_output_____"
]
],
[
[
"n = len(df_val)\ntotal = 0\ncorrect = 0\nfor i in range(n):\n output_sentence = df_val.loc[i][\"action\"] + \" \"+ df_val.loc[i][\"object\"] + \" \" + df_val.loc[i][\"location\"]\n target_tensor = sentence_to_index(output_sentence)\n \n input_sentence = df_val.loc[i][\"Content_Parsed_2\"]\n correct += evaluate(encoder, decoder, input_sentence, target_tensor)\n total += 1\nprint(correct)\nprint(total)\nprint(f\"Accuracy on Val test : {(float(correct)/total)*100}\")",
"11566\n11566\nAccuracy on Val test : 100.0\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8d9a35e8dfb6b9785b5b238ba381b1c4733a22
| 14,608 |
ipynb
|
Jupyter Notebook
|
DeOldify_colab.ipynb
|
cclauss/DeOldify
|
61486825398cdd89d5cc17308fcdbe305c7ec030
|
[
"MIT"
] | null | null | null |
DeOldify_colab.ipynb
|
cclauss/DeOldify
|
61486825398cdd89d5cc17308fcdbe305c7ec030
|
[
"MIT"
] | null | null | null |
DeOldify_colab.ipynb
|
cclauss/DeOldify
|
61486825398cdd89d5cc17308fcdbe305c7ec030
|
[
"MIT"
] | null | null | null | 26.80367 | 260 | 0.590156 |
[
[
[
"<a href=\"https://colab.research.google.com/github/jantic/DeOldify/blob/master/DeOldify_colab.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# DeOldify on Colab #\n\nThis notebook shows how to get your own version of [DeOldify](https://github.com/jantic/DeOldify) working on Google Colab. A lot of the initial steps are just installs -- but these are also the steps that can make running the model a tedious exercise.\n\nInitially, one must `pip install` a few dependencies, then `wget` is used to download the appropriate picture data.\n\nNECESSARY PRELIMINARY STEP: Please make sure you have gone up to the \"Runtime\" menu above and \"Change Runtime Type\" to Python3 and GPU.\n\nI hope you have fun, and thanks to Jason Antic for this awesome tool!\n\n-Matt Robinson, <[email protected]>\n\nNEW: You can now load your files from you own Google Drive, check the last cell of the notebook for more information.",
"_____no_output_____"
]
],
[
[
"!git clone https://github.com/jantic/DeOldify.git DeOldify",
"_____no_output_____"
],
[
"from os import path\nfrom wheel.pep425tags import get_abbr_impl, get_impl_ver, get_abi_tag\nplatform = '{}{}-{}'.format(get_abbr_impl(), get_impl_ver(), get_abi_tag())\n\naccelerator = 'cu80' if path.exists('/opt/bin/nvidia-smi') else 'cpu'\n\n!pip install -q http://download.pytorch.org/whl/{accelerator}/torch-0.4.1-{platform}-linux_x86_64.whl torchvision\nimport torch\nprint(torch.__version__)\nprint(torch.cuda.is_available())",
"_____no_output_____"
],
[
"cd DeOldify",
"_____no_output_____"
],
[
"!pip install -e .",
"_____no_output_____"
],
[
"%matplotlib inline\n%reload_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"# Doing work so I can access data from my google drive\n!pip install PyDrive",
"_____no_output_____"
],
[
"# Work around with Pillow being preinstalled on these Colab VMs, causing conflicts otherwise.\n!pip install Pillow==4.1.1",
"_____no_output_____"
],
[
"import os\nfrom pydrive.auth import GoogleAuth\nfrom pydrive.drive import GoogleDrive\nfrom google.colab import auth\nfrom oauth2client.client import GoogleCredentials\nimport multiprocessing\nfrom torch import autograd\nfrom fastai.transforms import TfmType\nfrom fasterai.transforms import *\nfrom fastai.conv_learner import *\nfrom fasterai.images import *\nfrom fasterai.dataset import *\nfrom fasterai.visualize import *\nfrom fasterai.callbacks import *\nfrom fasterai.loss import *\nfrom fasterai.modules import *\nfrom fasterai.training import *\nfrom fasterai.generators import *\nfrom fastai.torch_imports import *\nfrom fasterai.filters import *\nfrom pathlib import Path\nfrom itertools import repeat\nfrom google.colab import drive\nfrom IPython.display import Image\nimport tensorboardX\ntorch.cuda.set_device(0)\nplt.style.use('dark_background')\ntorch.backends.cudnn.benchmark=True",
"_____no_output_____"
],
[
"auth.authenticate_user()\ngauth = GoogleAuth()\ngauth.credentials = GoogleCredentials.get_application_default()\ndrive = GoogleDrive(gauth)",
"_____no_output_____"
]
],
[
[
"Note that the above requires a verification step. It isn't too bad.",
"_____no_output_____"
]
],
[
[
"# Now download the pretrained weights, which I have saved to my google drive\n# note that the id is the ending part of the shareable link url (after open?id=)\n# The pretrained weights can be downloaded from https://www.dropbox.com/s/7r2wu0af6okv280/colorize_gen_192.h5\ndownload = drive.CreateFile({'id': '1mRRvS3WIHPdp36G0yc1jC0XI6i-Narv6'})\ndownload.GetContentFile('pretrained_weights.h5')",
"_____no_output_____"
]
],
[
[
"With access to your Google Drive, the \"deOldifyImages\" directory will be created. Drop there your personal images, and after the full execution of the notebook find the results at its subdirectory \"results\"",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')\n!mkdir \"/content/drive/My Drive/deOldifyImages\"\n!mkdir \"/content/drive/My Drive/deOldifyImages/results\"",
"_____no_output_____"
],
[
"weights_path = 'pretrained_weights.h5'\nresults_dir='/content/drive/My Drive/deOldifyImages/results'\n\n#The higher the render_factor, the more GPU memory will be used and generally images will look better. \n#11GB can take a factor of 42 max. Performance generally gracefully degrades with lower factors, \n#though you may also find that certain images will actually render better at lower numbers. \n#This tends to be the case with the oldest photos.\nrender_factor=42\nfilters = [Colorizer(gpu=0, weights_path=weights_path)]\nvis = ModelImageVisualizer(filters, render_factor=render_factor, results_dir=results_dir)",
"_____no_output_____"
],
[
"# download an example picture to try.\n# NOTE: All the jpg files cloned from the git repo are corrupted. Must download yourself.\n!wget \"https://media.githubusercontent.com/media/jantic/DeOldify/master/test_images/abe.jpg\" -O \"abe2.jpg\"",
"_____no_output_____"
],
[
"# %matplotlib inline\nvis.plot_transformed_image('abe2.jpg', render_factor=25)",
"_____no_output_____"
],
[
"!wget \"https://media.githubusercontent.com/media/jantic/DeOldify/master/test_images/TV1930s.jpg\" -O \"family_TV.jpg\"",
"_____no_output_____"
],
[
"vis.plot_transformed_image('family_TV.jpg', render_factor=41)",
"_____no_output_____"
]
],
[
[
"Let's see how well it does Dorothy before her world turns to color in the Wizard of Oz:",
"_____no_output_____"
]
],
[
[
"!wget \"https://magnoliaforever.files.wordpress.com/2011/09/wizard-of-oz.jpg\" -O \"Dorothy.jpg\"",
"_____no_output_____"
],
[
"vis.plot_transformed_image('Dorothy.jpg', render_factor=30)",
"_____no_output_____"
]
],
[
[
"Let's now try Butch and Sundance. Famously the last scene ends with a black and white still. So we know what the color was beforehand.",
"_____no_output_____"
]
],
[
[
"!wget \"https://i.ebayimg.com/images/g/HqkAAOSwRLZUAwyS/s-l300.jpg\" -O \"butch_and_sundance.jpg\"",
"_____no_output_____"
],
[
"vis.plot_transformed_image('butch_and_sundance.jpg', render_factor=29)",
"_____no_output_____"
]
],
[
[
"Let's get a picture of what they were actually wearing:",
"_____no_output_____"
]
],
[
[
"!wget \"https://bethanytompkins.files.wordpress.com/2015/09/freezeframe.jpg\" -O \"butch_and_sundance_color.jpg\"",
"_____no_output_____"
],
[
"Image('butch_and_sundance_color.jpg')",
"_____no_output_____"
]
],
[
[
"If you want to colorise pictures from your drive, drop them in a directory named deOldifyImages (in the root of your drive) and the next cell will save the colorise pictures in deOldifyImages/results.",
"_____no_output_____"
]
],
[
[
"for img in os.listdir(\"/content/drive/My Drive/deOldifyImages/\"):\n img_path = str(\"/content/drive/My Drive/deOldifyImages/\") + img\n if os.path.isfile(img_path):\n vis.plot_transformed_image(img_path)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8d9d19a1a926f371c6c6663c61c66bd0db20e4
| 67,020 |
ipynb
|
Jupyter Notebook
|
Embedding/PyTorch/Advanced/ProtElectra.ipynb
|
taneishi/ProtTrans
|
9c613d5431c343429e693ccda91e8781aaa052c8
|
[
"AFL-3.0"
] | 401 |
2020-07-13T18:41:22.000Z
|
2022-03-31T07:19:26.000Z
|
Embedding/PyTorch/Advanced/ProtElectra.ipynb
|
taneishi/ProtTrans
|
9c613d5431c343429e693ccda91e8781aaa052c8
|
[
"AFL-3.0"
] | 71 |
2020-07-14T14:51:43.000Z
|
2022-03-17T09:28:45.000Z
|
Embedding/PyTorch/Advanced/ProtElectra.ipynb
|
taneishi/ProtTrans
|
9c613d5431c343429e693ccda91e8781aaa052c8
|
[
"AFL-3.0"
] | 68 |
2020-07-14T08:32:17.000Z
|
2022-03-27T07:07:26.000Z
| 33.543544 | 254 | 0.489421 |
[
[
[
"<a href=\"https://colab.research.google.com/github/agemagician/Prot-Transformers/blob/master/Embedding/Advanced/Electra.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"<h3> Extracting protein sequences' features using ProtElectra pretrained-model <h3>",
"_____no_output_____"
],
[
"<b>1. Load necessry libraries including huggingface transformers<b>",
"_____no_output_____"
]
],
[
[
"!pip install -q transformers",
"\u001b[K |████████████████████████████████| 675kB 7.1MB/s \n\u001b[K |████████████████████████████████| 1.1MB 33.0MB/s \n\u001b[K |████████████████████████████████| 3.8MB 45.5MB/s \n\u001b[K |████████████████████████████████| 890kB 34.7MB/s \n\u001b[?25h Building wheel for sacremoses (setup.py) ... \u001b[?25l\u001b[?25hdone\n"
],
[
"import torch\nfrom transformers import ElectraTokenizer, ElectraForPreTraining, ElectraForMaskedLM, ElectraModel\nimport re\nimport os\nimport requests\nfrom tqdm.auto import tqdm",
"_____no_output_____"
]
],
[
[
"<b>2. Set the url location of ProtElectra and the vocabulary file<b>",
"_____no_output_____"
]
],
[
[
"generatorModelUrl = 'https://www.dropbox.com/s/5x5et5q84y3r01m/pytorch_model.bin?dl=1'\ndiscriminatorModelUrl = 'https://www.dropbox.com/s/9ptrgtc8ranf0pa/pytorch_model.bin?dl=1'\n\ngeneratorConfigUrl = 'https://www.dropbox.com/s/9059fvix18i6why/config.json?dl=1'\ndiscriminatorConfigUrl = 'https://www.dropbox.com/s/jq568evzexyla0p/config.json?dl=1'\n\nvocabUrl = 'https://www.dropbox.com/s/wck3w1q15bc53s0/vocab.txt?dl=1'",
"_____no_output_____"
]
],
[
[
"<b>3. Download ProtElectra models and vocabulary files<b>",
"_____no_output_____"
]
],
[
[
"downloadFolderPath = 'models/electra/'",
"_____no_output_____"
],
[
"discriminatorFolderPath = os.path.join(downloadFolderPath, 'discriminator')\ngeneratorFolderPath = os.path.join(downloadFolderPath, 'generator')\n\ndiscriminatorModelFilePath = os.path.join(discriminatorFolderPath, 'pytorch_model.bin')\ngeneratorModelFilePath = os.path.join(generatorFolderPath, 'pytorch_model.bin')\n\ndiscriminatorConfigFilePath = os.path.join(discriminatorFolderPath, 'config.json')\ngeneratorConfigFilePath = os.path.join(generatorFolderPath, 'config.json')\n\nvocabFilePath = os.path.join(downloadFolderPath, 'vocab.txt')",
"_____no_output_____"
],
[
"if not os.path.exists(discriminatorFolderPath):\n os.makedirs(discriminatorFolderPath)\nif not os.path.exists(generatorFolderPath):\n os.makedirs(generatorFolderPath)",
"_____no_output_____"
],
[
"def download_file(url, filename):\n response = requests.get(url, stream=True)\n with tqdm.wrapattr(open(filename, \"wb\"), \"write\", miniters=1,\n total=int(response.headers.get('content-length', 0)),\n desc=filename) as fout:\n for chunk in response.iter_content(chunk_size=4096):\n fout.write(chunk)",
"_____no_output_____"
],
[
"if not os.path.exists(generatorModelFilePath):\n download_file(generatorModelUrl, generatorModelFilePath)\n\nif not os.path.exists(discriminatorModelFilePath):\n download_file(discriminatorModelUrl, discriminatorModelFilePath)\n \nif not os.path.exists(generatorConfigFilePath):\n download_file(generatorConfigUrl, generatorConfigFilePath)\n\nif not os.path.exists(discriminatorConfigFilePath):\n download_file(discriminatorConfigUrl, discriminatorConfigFilePath)\n \nif not os.path.exists(vocabFilePath):\n download_file(vocabUrl, vocabFilePath)",
"_____no_output_____"
]
],
[
[
"<b>4. Load the vocabulary and ProtElectra discriminator and generator Models<b>",
"_____no_output_____"
]
],
[
[
"tokenizer = ElectraTokenizer(vocabFilePath, do_lower_case=False )",
"_____no_output_____"
],
[
"discriminator = ElectraForPreTraining.from_pretrained(discriminatorFolderPath)",
"_____no_output_____"
],
[
"generator = ElectraForMaskedLM.from_pretrained(generatorFolderPath)",
"_____no_output_____"
],
[
"electra = ElectraModel.from_pretrained(discriminatorFolderPath)",
"_____no_output_____"
]
],
[
[
"<b>5. Load the model into the GPU if avilabile and switch to inference mode<b>",
"_____no_output_____"
]
],
[
[
"device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')",
"_____no_output_____"
],
[
"discriminator = discriminator.to(device)\ndiscriminator = discriminator.eval()",
"_____no_output_____"
],
[
"generator = generator.to(device)\ngenerator = generator.eval()",
"_____no_output_____"
],
[
"electra = electra.to(device)\nelectra = electra.eval()",
"_____no_output_____"
]
],
[
[
"<b>6. Create or load sequences and map rarely occured amino acids (U,Z,O,B) to (X)<b>",
"_____no_output_____"
]
],
[
[
"sequences_Example = [\"A E T C Z A O\",\"S K T Z P\"]",
"_____no_output_____"
],
[
"sequences_Example = [re.sub(r\"[UZOB]\", \"X\", sequence) for sequence in sequences_Example]",
"_____no_output_____"
]
],
[
[
"<b>7. Tokenize, encode sequences and load it into the GPU if possibile<b>",
"_____no_output_____"
]
],
[
[
"ids = tokenizer.batch_encode_plus(sequences_Example, add_special_tokens=True, pad_to_max_length=True)",
"_____no_output_____"
],
[
"input_ids = torch.tensor(ids['input_ids']).to(device)\nattention_mask = torch.tensor(ids['attention_mask']).to(device)",
"_____no_output_____"
]
],
[
[
"<b>8. Extracting sequences' features and load it into the CPU if needed<b>",
"_____no_output_____"
]
],
[
[
"with torch.no_grad():\n discriminator_embedding = discriminator(input_ids=input_ids,attention_mask=attention_mask)[0]",
"_____no_output_____"
],
[
"discriminator_embedding = discriminator_embedding.cpu().numpy()",
"_____no_output_____"
],
[
"with torch.no_grad():\n generator_embedding = generator(input_ids=input_ids,attention_mask=attention_mask)[0]",
"_____no_output_____"
],
[
"generator_embedding = generator_embedding.cpu().numpy()",
"_____no_output_____"
],
[
"with torch.no_grad():\n electra_embedding = electra(input_ids=input_ids,attention_mask=attention_mask)[0]",
"_____no_output_____"
],
[
"electra_embedding = electra_embedding.cpu().numpy()",
"_____no_output_____"
]
],
[
[
"<b>9. Remove padding ([PAD]) and special tokens ([CLS],[SEP]) that is added by Electra model<b>",
"_____no_output_____"
]
],
[
[
"features = [] \nfor seq_num in range(len(electra_embedding)):\n seq_len = (attention_mask[seq_num] == 1).sum()\n seq_emd = electra_embedding[seq_num][1:seq_len-1]\n features.append(seq_emd)",
"_____no_output_____"
],
[
"print(features)",
"[array([[-3.11754458e-02, -1.18080616e-01, -1.51422679e-01, ...,\n -8.80782455e-02, -2.03649044e-01, 2.34545898e-02],\n [-6.92143589e-02, -7.63380080e-02, -1.78088211e-02, ...,\n -4.15132381e-02, -3.08615528e-02, -8.58288854e-02],\n [ 3.80904488e-02, -1.71692267e-01, -5.64219430e-02, ...,\n -1.18378937e-01, -9.77956504e-02, 2.44725216e-02],\n ...,\n [ 1.27263516e-01, -1.34989679e-01, -3.06518644e-01, ...,\n 3.99149172e-02, -4.54527065e-02, -3.57910693e-01],\n [-5.05245999e-02, -9.02514085e-02, 6.78477362e-02, ...,\n -4.76730466e-02, -9.57428291e-02, -1.68221351e-02],\n [ 3.07775717e-02, 7.57525049e-05, -5.32222912e-02, ...,\n -1.47995083e-02, -1.57044619e-01, -9.64660496e-02]], dtype=float32), array([[-6.04737513e-02, -1.60797983e-01, -1.63700715e-01, ...,\n -7.67330825e-02, -1.51252389e-01, -4.52133343e-02],\n [-9.30745900e-02, -5.02012298e-02, -1.62957162e-02, ...,\n -2.65192648e-04, -2.70886812e-03, -2.37740427e-02],\n [-4.23046909e-02, -1.51860267e-01, -6.50829077e-02, ...,\n -1.45550948e-02, -7.37645999e-02, -5.66908680e-02],\n [ 4.75764386e-02, -7.08769858e-02, -2.75032073e-01, ...,\n -1.02747001e-01, -1.06809154e-01, -2.18676060e-01],\n [-4.99716476e-02, 1.03257410e-02, -9.70151871e-02, ...,\n -1.18601866e-01, 7.05851475e-03, -1.71629295e-01]], dtype=float32)]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a8da2a0569ce350e48acbbcc3a55ba990553853
| 245,364 |
ipynb
|
Jupyter Notebook
|
Python_Stock/Technical_Indicators/PMO.ipynb
|
chunsj/Stock_Analysis_For_Quant
|
5f28ef9537885a695245d26f3010592a29d45a34
|
[
"MIT"
] | 962 |
2019-07-17T09:57:41.000Z
|
2022-03-29T01:55:20.000Z
|
Python_Stock/Technical_Indicators/PMO.ipynb
|
chunsj/Stock_Analysis_For_Quant
|
5f28ef9537885a695245d26f3010592a29d45a34
|
[
"MIT"
] | 5 |
2020-04-29T16:54:30.000Z
|
2022-02-10T02:57:30.000Z
|
Python_Stock/Technical_Indicators/PMO.ipynb
|
chunsj/Stock_Analysis_For_Quant
|
5f28ef9537885a695245d26f3010592a29d45a34
|
[
"MIT"
] | 286 |
2019-08-04T10:37:58.000Z
|
2022-03-28T06:31:56.000Z
| 141.50173 | 90,310 | 0.785779 |
[
[
[
"# Decision Point Price Momentum Oscillator (PMO)",
"_____no_output_____"
],
[
"https://stockcharts.com/school/doku.php?id=chart_school:technical_indicators:dppmo",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\n# fix_yahoo_finance is used to fetch data \nimport fix_yahoo_finance as yf\nyf.pdr_override()",
"_____no_output_____"
],
[
"# input\nsymbol = 'AAPL'\nstart = '2017-01-01'\nend = '2019-01-01'\n\n# Read data \ndf = yf.download(symbol,start,end)\n\n# View Columns\ndf.head()",
"[*********************100%***********************] 1 of 1 downloaded\n"
],
[
"df.tail()",
"_____no_output_____"
],
[
"df['ROC'] = ((df['Adj Close'] - df['Adj Close'].shift(1))/df['Adj Close'].shift(1)) * 100\ndf = df.dropna()\ndf.head()",
"_____no_output_____"
],
[
"df['35_Custom_EMA_ROC'] = df['ROC'].ewm(ignore_na=False,span=35,min_periods=0,adjust=True).mean()\ndf.head()",
"_____no_output_____"
],
[
"df['35_Custom_EMA_ROC_10'] = df['35_Custom_EMA_ROC']*10\ndf.head()",
"_____no_output_____"
],
[
"df = df.dropna()\ndf.head(20)",
"_____no_output_____"
],
[
"df['PMO_Line'] = df['35_Custom_EMA_ROC_10'].ewm(ignore_na=False,span=20,min_periods=0,adjust=True).mean()\ndf.head()",
"_____no_output_____"
],
[
"df['PMO_Signal_Line'] = df['PMO_Line'].ewm(ignore_na=False,span=10,min_periods=0,adjust=True).mean()",
"_____no_output_____"
],
[
"df = df.dropna()\ndf.head()",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(14,10))\nax1 = plt.subplot(2, 1, 1)\nax1.plot(df['Adj Close'])\nax1.set_title('Stock '+ symbol +' Closing Price')\nax1.set_ylabel('Price')\nax1.legend(loc='best')\n\nax2 = plt.subplot(2, 1, 2)\nax2.plot(df['PMO_Line'], label='PMO Line')\nax2.plot(df['PMO_Signal_Line'], label='PMO Signal Line')\nax2.axhline(y=0, color='red')\nax2.grid()\nax2.legend(loc='best')\nax2.set_ylabel('PMO')\nax2.set_xlabel('Date')",
"_____no_output_____"
]
],
[
[
"## Candlestick with PMO",
"_____no_output_____"
]
],
[
[
"from matplotlib import dates as mdates\nimport datetime as dt\n\ndfc = df.copy()\ndfc['VolumePositive'] = dfc['Open'] < dfc['Adj Close']\n#dfc = dfc.dropna()\ndfc = dfc.reset_index()\ndfc['Date'] = mdates.date2num(dfc['Date'].astype(dt.date))\ndfc.head()",
"_____no_output_____"
],
[
"from mpl_finance import candlestick_ohlc\n\nfig = plt.figure(figsize=(14,10))\nax1 = plt.subplot(2, 1, 1)\ncandlestick_ohlc(ax1,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)\nax1.xaxis_date()\nax1.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))\nax1.grid(True, which='both')\nax1.minorticks_on()\nax1v = ax1.twinx()\ncolors = dfc.VolumePositive.map({True: 'g', False: 'r'})\nax1v.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)\nax1v.axes.yaxis.set_ticklabels([])\nax1v.set_ylim(0, 3*df.Volume.max())\nax1.set_title('Stock '+ symbol +' Closing Price')\nax1.set_ylabel('Price')\n\nax2 = plt.subplot(2, 1, 2)\nax2.plot(df['PMO_Line'], label='PMO_Line')\nax2.plot(df['PMO_Signal_Line'], label='PMO_Signal_Line')\nax2.axhline(y=0, color='red')\nax2.grid()\nax2.set_ylabel('PMO')\nax2.set_xlabel('Date')\nax2.legend(loc='best')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a8da5c6e2a8ea9b4f951326184a9604140d5ecf
| 28,415 |
ipynb
|
Jupyter Notebook
|
Datascrape/Datascrape/spiders/Hipflat Bankok Rentals.ipynb
|
poomkhor/Scrapy
|
e245ac24df4ea9ee11b94cf0168a3faccef1cfa1
|
[
"MIT"
] | null | null | null |
Datascrape/Datascrape/spiders/Hipflat Bankok Rentals.ipynb
|
poomkhor/Scrapy
|
e245ac24df4ea9ee11b94cf0168a3faccef1cfa1
|
[
"MIT"
] | null | null | null |
Datascrape/Datascrape/spiders/Hipflat Bankok Rentals.ipynb
|
poomkhor/Scrapy
|
e245ac24df4ea9ee11b94cf0168a3faccef1cfa1
|
[
"MIT"
] | null | null | null | 31.156798 | 145 | 0.351258 |
[
[
[
"import pandas as pd\r\nimport numpy as np\r\ndf = pd.read_csv(\"hipflat_rent.csv\")\r\nprint (df)",
" condo district room_area rental_rate\n0 Tubtim Mansion Chom Thong 170 50000.0\n1 Tubtim Mansion Chom Thong 250 58000.0\n2 Tubtim Mansion Chom Thong 170 50000.0\n3 Tubtim Mansion Chom Thong 250 58000.0\n4 Tubtim Mansion Chom Thong 175 45000.0\n... ... ... ... ...\n27623 Esta Bliss Min Buri 22.99 8000.0\n27624 Esta Bliss Min Buri 36 12000.0\n27625 Esta Bliss Min Buri 46 15000.0\n27626 Esta Bliss Min Buri 23 7500.0\n27627 Esta Bliss Min Buri 23 7000.0\n\n[27628 rows x 4 columns]\n"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df['room_area'] = pd.to_numeric(df['room_area'], errors = 'coerce')\r\ndf['rental_rate'] = pd.to_numeric(df['rental_rate'])\r\ndf",
"_____no_output_____"
],
[
"df['rental_rate'][df['district']=='Min Buri'].sum()",
"_____no_output_____"
],
[
"table = pd.pivot_table(df, index = 'district',values=['rental_rate','room_area'], aggfunc={'room_area':np.mean,'rental_rate':np.mean})\r\ntable = table.round(decimals=2)\r\ntable.head()",
"_____no_output_____"
]
],
[
[
"### Return the district with maximum rental rate per square meter",
"_____no_output_____"
]
],
[
[
"table['rate_per_sqm'] = table['rental_rate']/table['room_area']",
"_____no_output_____"
],
[
"table.head()",
"_____no_output_____"
],
[
"max_rate = table.idxmax()\r\nmax_rate",
"_____no_output_____"
],
[
"table.sort_values('rate_per_sqm', ascending=False)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4a8daa270d023383ac8e7725e03654837c2cfbb3
| 3,880 |
ipynb
|
Jupyter Notebook
|
virtualize-localization.ipynb
|
oldshuren/Hierarchical-Localization
|
334327956138c98af209e5ae9e13a0e222e97c2c
|
[
"Apache-2.0"
] | null | null | null |
virtualize-localization.ipynb
|
oldshuren/Hierarchical-Localization
|
334327956138c98af209e5ae9e13a0e222e97c2c
|
[
"Apache-2.0"
] | null | null | null |
virtualize-localization.ipynb
|
oldshuren/Hierarchical-Localization
|
334327956138c98af209e5ae9e13a0e222e97c2c
|
[
"Apache-2.0"
] | null | null | null | 27.913669 | 129 | 0.603093 |
[
[
[
"%load_ext autoreload\n%autoreload 2\n\nfrom pathlib import Path\nfrom pprint import pformat\n\nfrom hloc import extract_features, match_features, pairs_from_covisibility\nfrom hloc import colmap_from_nvm, triangulation, localize_sfm, visualization",
"_____no_output_____"
]
],
[
[
"# Visualize localization",
"_____no_output_____"
]
],
[
[
"results='/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr5s10_netvlad3_SfM/localized/localize_from_sfm_results.txt'\nquery_images=Path('/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr3')\ndb_images=Path('/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr5s10') \nreference_sfm=Path('/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr5s10_netvlad3_SfM/sfm_superpoint_inloc+superglue') ",
"_____no_output_____"
]
],
[
[
"## Visualizing the localization\nWe parse the localization logs and for each query image plot matches and inliers with a few database images.",
"_____no_output_____"
]
],
[
[
"visualization.visualize_loc(\n results, query_images, db_image_dir=db_images, sfm_model=reference_sfm / 'models/0', n=10, top_k_db=1, seed=2)",
"_____no_output_____"
],
[
"visualization.visualize_loc(\n results, query_images, db_image_dir=db_images, sfm_model=reference_sfm / 'models/0', n=10, top_k_db=1, seed=32)",
"_____no_output_____"
],
[
"visualization.visualize_loc(\n results, query_images, db_image_dir=db_images, sfm_model=reference_sfm / 'models/0', selected=['frame-001196.png'])",
"_____no_output_____"
],
[
"# d2net + NN\nresults='/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr5s10_SfM-3/localized/localize_from_sfm_results.txt'\nquery_images=Path('/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr3')\ndb_images=Path('/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr5s10') \nreference_sfm=Path('/nas4/dliu/wrk/robotics/datasets/RobotLab/rlab_tr5s10_SfM-3/sfm_d2net-ss+NN') ",
"_____no_output_____"
],
[
"visualization.visualize_loc(\n results, query_images, db_image_dir=db_images, sfm_model=reference_sfm / 'models/0', n=10, top_k_db=1, seed=32)",
"_____no_output_____"
],
[
"visualization.visualize_loc(\n results, query_images, db_image_dir=db_images, sfm_model=reference_sfm / 'models/0', selected=['frame-001196.png'])",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8db1efb44922a4ce01b4013b85fb68409e6846
| 36,181 |
ipynb
|
Jupyter Notebook
|
tutorial-reference/Day 18/notebooks/3 - Parse URL with Regex.ipynb
|
Rohianon/30-Days-of-Python
|
d82c5ae622d8079545eca940f9fa5992103709c2
|
[
"MIT"
] | 2,044 |
2016-03-11T02:56:41.000Z
|
2022-03-29T16:34:58.000Z
|
tutorial-reference/Day 18/notebooks/3 - Parse URL with Regex.ipynb
|
Yasin-Shah/30-Days-of-Python
|
161375734d7added6695b8cea6f4e730dc90505c
|
[
"MIT"
] | 28 |
2016-12-26T10:51:08.000Z
|
2021-12-13T20:49:47.000Z
|
tutorial-reference/Day 18/notebooks/3 - Parse URL with Regex.ipynb
|
Yasin-Shah/30-Days-of-Python
|
161375734d7added6695b8cea6f4e730dc90505c
|
[
"MIT"
] | 1,448 |
2016-03-12T04:44:06.000Z
|
2022-03-28T11:43:45.000Z
| 87.39372 | 15,934 | 0.715624 |
[
[
[
"import re\nimport requests\nimport time\nfrom requests_html import HTML\nfrom selenium import webdriver\nfrom selenium.webdriver.chrome.options import Options",
"_____no_output_____"
],
[
"options = Options()\noptions.add_argument(\"--headless\")\n\ndriver = webdriver.Chrome(options=options)",
"_____no_output_____"
],
[
"categories = [\n \"https://www.amazon.com/Best-Sellers-Toys-Games/zgbs/toys-and-games/\",\n \"https://www.amazon.com/Best-Sellers-Electronics/zgbs/electronics/\",\n \"https://www.amazon.com/Best-Sellers/zgbs/fashion/\"\n]",
"_____no_output_____"
],
[
"# categories",
"_____no_output_____"
],
[
"first_url = categories[0]",
"_____no_output_____"
],
[
"driver.get(first_url)",
"_____no_output_____"
],
[
"body_el = driver.find_element_by_css_selector(\"body\")\nhtml_str = body_el.get_attribute(\"innerHTML\")",
"_____no_output_____"
],
[
"html_obj = HTML(html=html_str)",
"_____no_output_____"
],
[
"page_links = [f\"https://www.amazon.com{x}\" for x in html_obj.links if x.startswith(\"/\")]\n# new_links = [x for x in new_links if \"product-reviews/\" not in x]",
"_____no_output_____"
],
[
"# page_links",
"_____no_output_____"
],
[
"def scrape_product_page(url, title_lookup = \"#productTitle\", price_lookup = \"#priceblock_ourprice\"):\n driver.get(url)\n time.sleep(0.5)\n body_el = driver.find_element_by_css_selector(\"body\")\n html_str = body_el.get_attribute(\"innerHTML\")\n html_obj = HTML(html=html_str)\n product_title = html_obj.find(title_lookup, first=True).text\n product_price = html_obj.find(price_lookup, first=True).text\n return product_title, product_price",
"_____no_output_____"
],
[
"# https://www.amazon.com/LEGO-Classic-Medium-Creative-Brick/dp/B00NHQFA1I/\n# https://www.amazon.com/Crayola-Washable-Watercolors-8-ea/dp/B000HHKAE2/\n\n# <base-url>/<slug>/dp/<product_id>/",
"_____no_output_____"
],
[
"# my_regex_pattern = r\"https://www.amazon.com/(?P<slug>[\\w-]+)/dp/(?P<product_id>[\\w-]+)/\"\n# my_url = 'https://www.amazon.com/Crayola-Washable-Watercolors-8-ea/dp/B000HHKAE2/'",
"_____no_output_____"
],
[
"# regex = re.compile(my_regex_pattern)",
"_____no_output_____"
],
[
"# my_match = regex.match(my_url)\n# print(my_match)\n# my_match['product_id']",
"_____no_output_____"
],
[
"# my_match['slug']",
"_____no_output_____"
],
[
"regex_options = [\n r\"https://www.amazon.com/gp/product/(?P<product_id>[\\w-]+)/\",\n r\"https://www.amazon.com/dp/(?P<product_id>[\\w-]+)/\",\n r\"https://www.amazon.com/(?P<slug>[\\w-]+)/dp/(?P<product_id>[\\w-]+)/\",\n]\n\ndef extract_product_id_from_url(url):\n product_id = None\n for regex_str in regex_options:\n regex = re.compile(regex_str)\n match = regex.match(url)\n if match != None:\n try:\n product_id = match['product_id']\n except:\n pass\n return product_id",
"_____no_output_____"
],
[
"\n# page_links = [x for x in page_links if extract_product_id_from_url(x) != None]\ndef clean_page_links(page_links=[]):\n final_page_links = []\n for url in page_links:\n product_id = extract_product_id_from_url(url)\n if product_id != None:\n final_page_links.append({\"url\": url, \"product_id\": product_id})\n return final_page_links\n\ncleaned_links = clean_page_links(page_links)",
"_____no_output_____"
],
[
"len(page_links) # == len(cleaned_links)",
"_____no_output_____"
],
[
"len(cleaned_links)",
"_____no_output_____"
],
[
"def perform_scrape(cleaned_items=[]):\n data_extracted = []\n for obj in cleaned_items:\n link = obj['url']\n product_id = obj['product_id']\n title, price = (None, None)\n try:\n title, price = scrape_product_page(link)\n except:\n pass\n if title != None and price != None:\n print(link, title, price)\n product_data = {\n \"url\": link,\n \"product_id\": product_id,\n \"title\": title,\n \"price\": price\n }\n data_extracted.append(product_data)\n return data_extracted",
"_____no_output_____"
],
[
"extracted_data = perform_scrape(cleaned_items=cleaned_links)",
"https://www.amazon.com/Crayola-Colored-Pre-sharpened-Coloring-Stocking/dp/B018HB2QFU/ref=zg_bs_toys-and-games_10/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Crayola Colored Pencils, Adult Coloring, Fun At Home Activities, 50 Count $19.38\nhttps://www.amazon.com/Little-Tikes-Spiralin-Waterpark-Table/dp/B004INDQWY/ref=zg_bs_toys-and-games_37/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Little Tikes Spiralin’ Seas Waterpark Water Table $44.99\nhttps://www.amazon.com/Crayola-Washable-Watercolors-8-ea/dp/B000HHKAE2/ref=zg_bs_toys-and-games_21/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Crayola Washable Watercolors, Paint Set For Kids, 8Count $1.99\nhttps://www.amazon.com/Munchkin-17040-Fishin-Bath-Toy/dp/B01N52DUNK/ref=zg_bs_toys-and-games_48/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Munchkin Fishin' Bath Toy $7.73\nhttps://www.amazon.com/Hasbro-B0965-Monopoly-Deal-Card/dp/B00NQQTZCO/ref=zg_bs_toys-and-games_18/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Monopoly Deal Card Game $7.99\nhttps://www.amazon.com/LEGO-Classic-Medium-Creative-Brick/dp/B00NHQFA1I/ref=zg_bs_toys-and-games_19/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD LEGO Classic Medium Creative Brick Box 10696 Building Toys for Creative Play; Kids Creative Kit (484 Pieces) $27.99\nhttps://www.amazon.com/Taco-Cat-Goat-Cheese-Pizza/dp/B077Z1R28P/ref=zg_bs_toys-and-games_30/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Taco Cat Goat Cheese Pizza $9.99\nhttps://www.amazon.com/Crayola-Twistables-Colored-Exclusive-Stocking/dp/B07D4RN9NH/ref=zg_bs_toys-and-games_36/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Crayola Twistables Colored Pencils Coloring Set, Kids Indoor Activities At Home, Gift Age 3+ - 50 Count $12.99\nhttps://www.amazon.com/SunWorks-Construction-Assorted-Colors-Sheets/dp/B0017OJKLI/ref=zg_bs_toys-and-games_45/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD SunWorks Construction Paper, 10 Assorted Colors, 9\" x 12\", 100 Sheets $8.29\nhttps://www.amazon.com/Crayola-Erasable-Non-Toxic-Pre-Sharpened-Gradation/dp/B000PCWKBA/ref=zg_bs_toys-and-games_20/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Crayola Erasable Colored Pencils, Kids At Home Activities, 24 Count $5.97\nhttps://www.amazon.com/Play-Doh-A5417-Sparkle-Compound-Collection/dp/B00IGNWYNE/ref=zg_bs_toys-and-games_5/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Play-Doh Sparkle Compound Collection $4.94\nhttps://www.amazon.com/Hasbro-A5640-Connect-4-Game/dp/B00D8STBHY/ref=zg_bs_toys-and-games_2/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Hasbro Connect 4 Game $12.99\nhttps://www.amazon.com/Crayola-Crayons-Assorted-Toddler-16Count/dp/B07L6VQDSC/ref=zg_bs_toys-and-games_29/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Crayola Jumbo Crayons, Assorted Colors, Great Toddler Crayons, 16Count $6.25\nhttps://www.amazon.com/Postage-Stamps-envelopes-Wayne-Global/dp/B081J497K2/ref=zg_bs_toys-and-games_44/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD 20 Postage Stamps with 20 (#10) envelopes Bundle by Wayne Global- Stamp Design May Vary $16.90\nhttps://www.amazon.com/ThinkFun-Zingo-Winning-Pre-Readers-Readers/dp/B01DY818JG/ref=zg_bs_toys-and-games_46/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD ThinkFun Zingo Bingo Award Winning Preschool Game for Pre-Readers and Early Readers Age 4 and Up - One of the Most Popular Board Games for Boys and Girls and their Parents, Amazon Exclusive Version $19.99\nhttps://www.amazon.com/Disney-Princess-Necklace-Activity-Set/dp/B0747ZGTTG/ref=zg_bs_toys-and-games_25/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Tara Toy Disney Princess Necklace Activity Set $12.79\nhttps://www.amazon.com/Learning-Resources-Spike-Hedgehog-Sensory/dp/B078WM314M/ref=zg_bs_toys-and-games_33/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Learning Resources Spike The Fine Motor Hedgehog, Sensory, Fine Motor Toy, Easter Basket Toy, Ages 18 months+ $10.99\nhttps://www.amazon.com/Cards-Against-Humanity-LLC-CAHUS/dp/B004S8F7QM/ref=zg_bs_toys-and-games_13/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Cards Against Humanity $25.00\nhttps://www.amazon.com/L-L-Surprise-Surprises-Including/dp/B07XSPCBVP/ref=zg_bs_toys-and-games_24/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD L.O.L. Surprise! Lights Glitter Doll with 8 Surprises Including Black Light Surprises $10.88\nhttps://www.amazon.com/Crayola-Washable-Special-Sidewalk-Anti-Roll/dp/B00PY47LHW/ref=zg_bs_toys-and-games_42/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Crayola Washable Sidewalk Chalk, Neon Chalk $4.99\nhttps://www.amazon.com/L-L-Surprise-Candylicious-Multicolor/dp/B07XSQ3BYJ/ref=zg_bs_toys-and-games_47/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD L.O.L. Surprise! O.M.G. Candylicious Fashion Doll with 20 Surprises,Multicolor $26.88\nhttps://www.amazon.com/Elmers-Liquid-Glitter-Washable-Assorted/dp/B008M56YZU/ref=zg_bs_toys-and-games_39/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Elmer's Liquid Glitter Glue, Great For Making Slime, Washable, Assorted Colors, 6 Ounces Each, 3 Count $11.95\nhttps://www.amazon.com/Melissa-Doug-Pretend-Frustration-Free-Packaging/dp/B07PBXRJ7G/ref=zg_bs_toys-and-games_43/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Melissa & Doug Dust! Sweep! Mop! (Frustration Free Packaging) $25.09\nhttps://www.amazon.com/Chalk-Jumbo-Sidewalk-Count-colors/dp/B079WL9MXC/ref=zg_bs_toys-and-games_31/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Chalk Jumbo Sidewalk Chalk 20 Count- 5 colors $11.54\nhttps://www.amazon.com/Melissa-Doug-Activity-Child-Safe-Scissors/dp/B00EJAEUBC/ref=zg_bs_toys-and-games_15/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Melissa & Doug Scissor Skills Activity Pad $4.99\nhttps://www.amazon.com/Nuby-Floating-Purple-Octopus-Interactive/dp/B083ZZSGLR/ref=zg_bs_toys-and-games_49/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Nuby Floating Purple Octopus with 3 Hoopla Rings Interactive Bath Toy $6.89\nhttps://www.amazon.com/Crayola-Twistables-Classrooms-Preschools-Self-Sharpening/dp/B00062J99K/ref=zg_bs_toys-and-games_8/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Crayola Twistables Crayons Coloring Set, Kids Indoor Activities at Home, 24 Count $4.49\nhttps://www.amazon.com/Creativity-Kids-Grow-Glow-Terrarium/dp/B00I9KDFK0/ref=zg_bs_toys-and-games_9/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Creativity For Kids Grow 'N Glow Terrarium Science Kits for Kids - Create Your Own Mini Ecosystem, Educational Toys $12.99\nhttps://www.amazon.com/Regal-Games-Chalk-City-Washable/dp/B071CKSMS7/ref=zg_bs_toys-and-games_4/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Regal Games Chalk City - 20 Piece Jumbo Washable Sidewalk Chalk $12.99\nhttps://www.amazon.com/First-Years-Stack-Up-Cups/dp/B00005C5H4/ref=zg_bs_toys-and-games_27/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD The First Years Stack Up Cup Toys $6.00\nhttps://www.amazon.com/Melissa-Doug-Decorate-Your-Own-Butterfly-Magnets/dp/B00Y8YOVOA/ref=zg_bs_toys-and-games_12/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Melissa & Doug Created by Me! Butterfly Magnets - The Original (Kids Craft Kit, Supplies for 4 Projects, Great Gift for Girls and Boys – Best for 4, 5, 6, 7 and 8 Year Olds), Mix $4.99\nhttps://www.amazon.com/Insect-Lore-Butterfly-Growing-Kit/dp/B00000ISC5/ref=zg_bs_toys-and-games_3/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD Insect Lore Butterfly Growing Kit - With Voucher to Redeem Caterpillars Later $22.99\nhttps://www.amazon.com/L-L-Surprise-Sparkle-Multicolor/dp/B07PQVRPR1/ref=zg_bs_toys-and-games_11/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD L.O.L. Surprise Dolls Sparkle Series A, Multicolor $10.88\n"
],
[
"print(extracted_data)",
"[{'url': 'https://www.amazon.com/Crayola-Colored-Pre-sharpened-Coloring-Stocking/dp/B018HB2QFU/ref=zg_bs_toys-and-games_10/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B018HB2QFU', 'title': 'Crayola Colored Pencils, Adult Coloring, Fun At Home Activities, 50 Count', 'price': '$19.38'}, {'url': 'https://www.amazon.com/Little-Tikes-Spiralin-Waterpark-Table/dp/B004INDQWY/ref=zg_bs_toys-and-games_37/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B004INDQWY', 'title': 'Little Tikes Spiralin’ Seas Waterpark Water Table', 'price': '$44.99'}, {'url': 'https://www.amazon.com/Crayola-Washable-Watercolors-8-ea/dp/B000HHKAE2/ref=zg_bs_toys-and-games_21/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B000HHKAE2', 'title': 'Crayola Washable Watercolors, Paint Set For Kids, 8Count', 'price': '$1.99'}, {'url': 'https://www.amazon.com/Munchkin-17040-Fishin-Bath-Toy/dp/B01N52DUNK/ref=zg_bs_toys-and-games_48/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B01N52DUNK', 'title': \"Munchkin Fishin' Bath Toy\", 'price': '$7.73'}, {'url': 'https://www.amazon.com/Hasbro-B0965-Monopoly-Deal-Card/dp/B00NQQTZCO/ref=zg_bs_toys-and-games_18/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00NQQTZCO', 'title': 'Monopoly Deal Card Game', 'price': '$7.99'}, {'url': 'https://www.amazon.com/LEGO-Classic-Medium-Creative-Brick/dp/B00NHQFA1I/ref=zg_bs_toys-and-games_19/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00NHQFA1I', 'title': 'LEGO Classic Medium Creative Brick Box 10696 Building Toys for Creative Play; Kids Creative Kit (484 Pieces)', 'price': '$27.99'}, {'url': 'https://www.amazon.com/Taco-Cat-Goat-Cheese-Pizza/dp/B077Z1R28P/ref=zg_bs_toys-and-games_30/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B077Z1R28P', 'title': 'Taco Cat Goat Cheese Pizza', 'price': '$9.99'}, {'url': 'https://www.amazon.com/Crayola-Twistables-Colored-Exclusive-Stocking/dp/B07D4RN9NH/ref=zg_bs_toys-and-games_36/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07D4RN9NH', 'title': 'Crayola Twistables Colored Pencils Coloring Set, Kids Indoor Activities At Home, Gift Age 3+ - 50 Count', 'price': '$12.99'}, {'url': 'https://www.amazon.com/SunWorks-Construction-Assorted-Colors-Sheets/dp/B0017OJKLI/ref=zg_bs_toys-and-games_45/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B0017OJKLI', 'title': 'SunWorks Construction Paper, 10 Assorted Colors, 9\" x 12\", 100 Sheets', 'price': '$8.29'}, {'url': 'https://www.amazon.com/Crayola-Erasable-Non-Toxic-Pre-Sharpened-Gradation/dp/B000PCWKBA/ref=zg_bs_toys-and-games_20/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B000PCWKBA', 'title': 'Crayola Erasable Colored Pencils, Kids At Home Activities, 24 Count', 'price': '$5.97'}, {'url': 'https://www.amazon.com/Play-Doh-A5417-Sparkle-Compound-Collection/dp/B00IGNWYNE/ref=zg_bs_toys-and-games_5/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00IGNWYNE', 'title': 'Play-Doh Sparkle Compound Collection', 'price': '$4.94'}, {'url': 'https://www.amazon.com/Hasbro-A5640-Connect-4-Game/dp/B00D8STBHY/ref=zg_bs_toys-and-games_2/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00D8STBHY', 'title': 'Hasbro Connect 4 Game', 'price': '$12.99'}, {'url': 'https://www.amazon.com/Crayola-Crayons-Assorted-Toddler-16Count/dp/B07L6VQDSC/ref=zg_bs_toys-and-games_29/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07L6VQDSC', 'title': 'Crayola Jumbo Crayons, Assorted Colors, Great Toddler Crayons, 16Count', 'price': '$6.25'}, {'url': 'https://www.amazon.com/Postage-Stamps-envelopes-Wayne-Global/dp/B081J497K2/ref=zg_bs_toys-and-games_44/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B081J497K2', 'title': '20 Postage Stamps with 20 (#10) envelopes Bundle by Wayne Global- Stamp Design May Vary', 'price': '$16.90'}, {'url': 'https://www.amazon.com/ThinkFun-Zingo-Winning-Pre-Readers-Readers/dp/B01DY818JG/ref=zg_bs_toys-and-games_46/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B01DY818JG', 'title': 'ThinkFun Zingo Bingo Award Winning Preschool Game for Pre-Readers and Early Readers Age 4 and Up - One of the Most Popular Board Games for Boys and Girls and their Parents, Amazon Exclusive Version', 'price': '$19.99'}, {'url': 'https://www.amazon.com/Disney-Princess-Necklace-Activity-Set/dp/B0747ZGTTG/ref=zg_bs_toys-and-games_25/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B0747ZGTTG', 'title': 'Tara Toy Disney Princess Necklace Activity Set', 'price': '$12.79'}, {'url': 'https://www.amazon.com/Learning-Resources-Spike-Hedgehog-Sensory/dp/B078WM314M/ref=zg_bs_toys-and-games_33/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B078WM314M', 'title': 'Learning Resources Spike The Fine Motor Hedgehog, Sensory, Fine Motor Toy, Easter Basket Toy, Ages 18 months+', 'price': '$10.99'}, {'url': 'https://www.amazon.com/Cards-Against-Humanity-LLC-CAHUS/dp/B004S8F7QM/ref=zg_bs_toys-and-games_13/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B004S8F7QM', 'title': 'Cards Against Humanity', 'price': '$25.00'}, {'url': 'https://www.amazon.com/L-L-Surprise-Surprises-Including/dp/B07XSPCBVP/ref=zg_bs_toys-and-games_24/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07XSPCBVP', 'title': 'L.O.L. Surprise! Lights Glitter Doll with 8 Surprises Including Black Light Surprises', 'price': '$10.88'}, {'url': 'https://www.amazon.com/Crayola-Washable-Special-Sidewalk-Anti-Roll/dp/B00PY47LHW/ref=zg_bs_toys-and-games_42/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00PY47LHW', 'title': 'Crayola Washable Sidewalk Chalk, Neon Chalk', 'price': '$4.99'}, {'url': 'https://www.amazon.com/L-L-Surprise-Candylicious-Multicolor/dp/B07XSQ3BYJ/ref=zg_bs_toys-and-games_47/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07XSQ3BYJ', 'title': 'L.O.L. Surprise! O.M.G. Candylicious Fashion Doll with 20 Surprises,Multicolor', 'price': '$26.88'}, {'url': 'https://www.amazon.com/SplashEZ-Sprinkler-Splash-Wading-Learning/dp/B07MNMT3M7/ref=zg_bs_toys-and-games_23/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07MNMT3M7', 'title': None, 'price': None}, {'url': 'https://www.amazon.com/Elmers-Liquid-Glitter-Washable-Assorted/dp/B008M56YZU/ref=zg_bs_toys-and-games_39/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B008M56YZU', 'title': \"Elmer's Liquid Glitter Glue, Great For Making Slime, Washable, Assorted Colors, 6 Ounces Each, 3 Count\", 'price': '$11.95'}, {'url': 'https://www.amazon.com/Crayola-Washable-Sidewalk-Outdoor-Exclusive/dp/B00LH1WN4W/ref=zg_bs_toys-and-games_38/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00LH1WN4W', 'title': None, 'price': None}, {'url': 'https://www.amazon.com/Melissa-Doug-Pretend-Frustration-Free-Packaging/dp/B07PBXRJ7G/ref=zg_bs_toys-and-games_43/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07PBXRJ7G', 'title': 'Melissa & Doug Dust! Sweep! Mop! (Frustration Free Packaging)', 'price': '$25.09'}, {'url': 'https://www.amazon.com/Chalk-Jumbo-Sidewalk-Count-colors/dp/B079WL9MXC/ref=zg_bs_toys-and-games_31/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B079WL9MXC', 'title': 'Chalk Jumbo Sidewalk Chalk 20 Count- 5 colors', 'price': '$11.54'}, {'url': 'https://www.amazon.com/Melissa-Doug-Activity-Child-Safe-Scissors/dp/B00EJAEUBC/ref=zg_bs_toys-and-games_15/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00EJAEUBC', 'title': 'Melissa & Doug Scissor Skills Activity Pad', 'price': '$4.99'}, {'url': 'https://www.amazon.com/Jenga-A2120EU4-Classic-Game/dp/B00ABA0ZOA/ref=zg_bs_toys-and-games_1/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00ABA0ZOA', 'title': None, 'price': None}, {'url': 'https://www.amazon.com/Nuby-Floating-Purple-Octopus-Interactive/dp/B083ZZSGLR/ref=zg_bs_toys-and-games_49/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B083ZZSGLR', 'title': 'Nuby Floating Purple Octopus with 3 Hoopla Rings Interactive Bath Toy', 'price': '$6.89'}, {'url': 'https://www.amazon.com/Crayola-Twistables-Classrooms-Preschools-Self-Sharpening/dp/B00062J99K/ref=zg_bs_toys-and-games_8/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00062J99K', 'title': 'Crayola Twistables Crayons Coloring Set, Kids Indoor Activities at Home, 24 Count', 'price': '$4.49'}, {'url': 'https://www.amazon.com/Creativity-Kids-Grow-Glow-Terrarium/dp/B00I9KDFK0/ref=zg_bs_toys-and-games_9/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00I9KDFK0', 'title': \"Creativity For Kids Grow 'N Glow Terrarium Science Kits for Kids - Create Your Own Mini Ecosystem, Educational Toys\", 'price': '$12.99'}, {'url': 'https://www.amazon.com/Regal-Games-Chalk-City-Washable/dp/B071CKSMS7/ref=zg_bs_toys-and-games_4/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B071CKSMS7', 'title': 'Regal Games Chalk City - 20 Piece Jumbo Washable Sidewalk Chalk', 'price': '$12.99'}, {'url': 'https://www.amazon.com/Step2-874600-Showers-Playset-Multi-Colored/dp/B01K1K0K6M/ref=zg_bs_toys-and-games_6/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B01K1K0K6M', 'title': None, 'price': None}, {'url': 'https://www.amazon.com/First-Years-Stack-Up-Cups/dp/B00005C5H4/ref=zg_bs_toys-and-games_27/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00005C5H4', 'title': 'The First Years Stack Up Cup Toys', 'price': '$6.00'}, {'url': 'https://www.amazon.com/Crayola-Washable-Glitter-Exclusive-Stocking/dp/B07BYWS5XW/ref=zg_bs_toys-and-games_32/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07BYWS5XW', 'title': None, 'price': None}, {'url': 'https://www.amazon.com/Melissa-Doug-Decorate-Your-Own-Butterfly-Magnets/dp/B00Y8YOVOA/ref=zg_bs_toys-and-games_12/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00Y8YOVOA', 'title': 'Melissa & Doug Created by Me! Butterfly Magnets - The Original (Kids Craft Kit, Supplies for 4 Projects, Great Gift for Girls and Boys – Best for 4, 5, 6, 7 and 8 Year Olds), Mix', 'price': '$4.99'}, {'url': 'https://www.amazon.com/Insect-Lore-Butterfly-Growing-Kit/dp/B00000ISC5/ref=zg_bs_toys-and-games_3/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00000ISC5', 'title': 'Insect Lore Butterfly Growing Kit - With Voucher to Redeem Caterpillars Later', 'price': '$22.99'}, {'url': 'https://www.amazon.com/L-L-Surprise-Sparkle-Multicolor/dp/B07PQVRPR1/ref=zg_bs_toys-and-games_11/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07PQVRPR1', 'title': 'L.O.L. Surprise Dolls Sparkle Series A, Multicolor', 'price': '$10.88'}, {'url': 'https://www.amazon.com/Crayola-Outdoor-Glitter-Sidewalk-Summer/dp/B00PY47F8M/ref=zg_bs_toys-and-games_16/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00PY47F8M', 'title': 'Crayola Outdoor Chalk, Glitter Sidewalk Chalk, Summer Toys, 5 Count', 'price': '$8.73'}, {'url': 'https://www.amazon.com/Jenga-A2120EU4-Classic-Game/dp/B00ABA0ZOA/ref=zg_bs_toys-and-games_map_1/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00ABA0ZOA', 'title': None, 'price': None}, {'url': 'https://www.amazon.com/Creative-Roots-Stepping-Horizon-Assorted/dp/B07HSJ5R7V/ref=zg_bs_toys-and-games_50/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07HSJ5R7V', 'title': 'CREATIVE ROOTS Paint Your Own Flower Stepping Stone by Horizon Group USA Toy, Assorted', 'price': '$7.99'}, {'url': 'https://www.amazon.com/Kinetic-Sand-Beach-Kingdom-Playset/dp/B079P87RVB/ref=zg_bs_toys-and-games_41/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B079P87RVB', 'title': 'Kinetic Sand Beach Sand Kingdom Playset with 3lbs of Beach Sand, for Ages 3 and Up', 'price': '$19.97'}, {'url': 'https://www.amazon.com/Bunch-Balloons-Pack-Amazon-Exclusive/dp/B07GW2QQWN/ref=zg_bs_toys-and-games_28/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07GW2QQWN', 'title': 'Bunch O Balloons - 350 Rapid-Fill Water Balloons (10 Pack) Amazon Exclusive', 'price': '$27.74'}, {'url': 'https://www.amazon.com/Crayola-Markers-Assorted-Colors-Coloring/dp/B01C64BASI/ref=zg_bs_toys-and-games_40/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B01C64BASI', 'title': 'Crayola Fine Line Markers Adult Coloring Set, Gift Age 12+ - 40 Count', 'price': '$12.99'}, {'url': 'https://www.amazon.com/LEGO-Classic-Green-Baseplate-Supplement/dp/B00NHQF65S/ref=zg_bs_toys-and-games_14/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00NHQF65S', 'title': 'LEGO Classic Green Baseplate 2304 Supplement for Building, Playing, and Displaying LEGO Creations, 10cm x 10cm, Large Building Base Accessory for Kids and Adults (1 Piece)', 'price': '$4.99'}, {'url': 'https://www.amazon.com/Aqua-Monterey-Multi-Purpose-Inflatable-Portable/dp/B073WMYP6M/ref=zg_bs_toys-and-games_26/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B073WMYP6M', 'title': 'Aqua 4-in-1 Monterey Hammock Inflatable Pool Float, Multi-Purpose Pool Hammock (Saddle, Lounge Chair, Hammock, Drifter) Pool Chair, Portable Water Hammock, Navy/White Stripe', 'price': '$15.07'}, {'url': 'https://www.amazon.com/Play-Doh-Modeling-Compound-Non-Toxic-Exclusive/dp/B00JM5GZGW/ref=zg_bs_toys-and-games_17/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B00JM5GZGW', 'title': 'Play-Doh Modeling Compound 36-Pack Case of Colors, Non-Toxic, Assorted Colors, 3-Ounce Cans (Amazon Exclusive)', 'price': '$24.99'}, {'url': 'https://www.amazon.com/Darice-Solution-Top-Works-Machines-Birthdays/dp/B07RYBB4NZ/ref=zg_bs_toys-and-games_7/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B07RYBB4NZ', 'title': 'Darice 64-Ounce Bubble Solution-Includes Wand and Easy Pour Funnel Top-Works with Bubble Machines-for Weddings, Birthdays and Outdoor Events', 'price': '$9.00'}, {'url': 'https://www.amazon.com/FoxPrint-Princess-conveniently-Carrying-Foldable/dp/B0120XRWLE/ref=zg_bs_toys-and-games_34/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B0120XRWLE', 'title': None, 'price': None}, {'url': 'https://www.amazon.com/SunWorks-Construction-Paper-White-Sheets/dp/B0017OHG1O/ref=zg_bs_toys-and-games_35/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B0017OHG1O', 'title': 'SunWorks Heavyweight Construction Paper, 9 x 12 Inches, White, 100 Sheets', 'price': '$3.69'}, {'url': 'https://www.amazon.com/Battleship-Planes-Strategy-Amazon-Exclusive/dp/B06Y1N3PTX/ref=zg_bs_toys-and-games_22/138-0291413-2906220?_encoding=UTF8&psc=1&refRID=PZN4KF19MNZDRXDYK2WD', 'product_id': 'B06Y1N3PTX', 'title': 'Battleship With Planes Strategy Board Game For Ages 7 and Up (Amazon Exclusive)', 'price': '$14.99'}]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8dc4be5e4a72d2d11f4d908001988ebc9a8837
| 72,265 |
ipynb
|
Jupyter Notebook
|
casestudies/statelevel/patients/patient_allocation_county.ipynb
|
flixpar/covid-resource-allocation
|
bda248346d8b1477a8ce209e9c37a691c4a6bdba
|
[
"MIT"
] | 4 |
2020-11-12T19:22:05.000Z
|
2021-08-09T09:11:21.000Z
|
casestudies/statelevel/patients/patient_allocation_county.ipynb
|
flixpar/covid-resource-allocation
|
bda248346d8b1477a8ce209e9c37a691c4a6bdba
|
[
"MIT"
] | null | null | null |
casestudies/statelevel/patients/patient_allocation_county.ipynb
|
flixpar/covid-resource-allocation
|
bda248346d8b1477a8ce209e9c37a691c4a6bdba
|
[
"MIT"
] | 1 |
2021-07-31T17:59:16.000Z
|
2021-07-31T17:59:16.000Z
| 86.648681 | 7,669 | 0.446399 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a8dc6e1fb1ebd1f174ea5b44f13d53774df7615
| 51,458 |
ipynb
|
Jupyter Notebook
|
house_prices/01.ipynb
|
yunshuipiao/sw_kaggle
|
2bd32670a74fcb407486bd382b414937c109160f
|
[
"Apache-2.0"
] | 6 |
2018-05-19T06:35:58.000Z
|
2021-02-16T07:28:24.000Z
|
house_prices/01.ipynb
|
yunshuipiao/sw_kaggle
|
2bd32670a74fcb407486bd382b414937c109160f
|
[
"Apache-2.0"
] | null | null | null |
house_prices/01.ipynb
|
yunshuipiao/sw_kaggle
|
2bd32670a74fcb407486bd382b414937c109160f
|
[
"Apache-2.0"
] | 5 |
2019-02-19T03:06:45.000Z
|
2021-02-16T07:29:29.000Z
| 36.238028 | 163 | 0.440728 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import Imputer",
"/anaconda3/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n return f(*args, **kwds)\n/anaconda3/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88\n return f(*args, **kwds)\n"
],
[
"# 上述函数,其输入是包含1个多个枚举类别的2D数组,需要reshape成为这种数组\n# from sklearn.preprocessing import CategoricalEncoder #后面会添加这个方法\n\nfrom sklearn.base import BaseEstimator, TransformerMixin\nfrom sklearn.utils import check_array\nfrom sklearn.preprocessing import LabelEncoder\n\nfrom scipy import sparse\n\n# 后面再去理解\nclass CategoricalEncoder(BaseEstimator, TransformerMixin):\n \"\"\"Encode categorical features as a numeric array.\n The input to this transformer should be a matrix of integers or strings,\n denoting the values taken on by categorical (discrete) features.\n The features can be encoded using a one-hot aka one-of-K scheme\n (``encoding='onehot'``, the default) or converted to ordinal integers\n (``encoding='ordinal'``).\n This encoding is needed for feeding categorical data to many scikit-learn\n estimators, notably linear models and SVMs with the standard kernels.\n Read more in the :ref:`User Guide <preprocessing_categorical_features>`.\n Parameters\n ----------\n encoding : str, 'onehot', 'onehot-dense' or 'ordinal'\n The type of encoding to use (default is 'onehot'):\n - 'onehot': encode the features using a one-hot aka one-of-K scheme\n (or also called 'dummy' encoding). This creates a binary column for\n each category and returns a sparse matrix.\n - 'onehot-dense': the same as 'onehot' but returns a dense array\n instead of a sparse matrix.\n - 'ordinal': encode the features as ordinal integers. This results in\n a single column of integers (0 to n_categories - 1) per feature.\n categories : 'auto' or a list of lists/arrays of values.\n Categories (unique values) per feature:\n - 'auto' : Determine categories automatically from the training data.\n - list : ``categories[i]`` holds the categories expected in the ith\n column. The passed categories are sorted before encoding the data\n (used categories can be found in the ``categories_`` attribute).\n dtype : number type, default np.float64\n Desired dtype of output.\n handle_unknown : 'error' (default) or 'ignore'\n Whether to raise an error or ignore if a unknown categorical feature is\n present during transform (default is to raise). When this is parameter\n is set to 'ignore' and an unknown category is encountered during\n transform, the resulting one-hot encoded columns for this feature\n will be all zeros.\n Ignoring unknown categories is not supported for\n ``encoding='ordinal'``.\n Attributes\n ----------\n categories_ : list of arrays\n The categories of each feature determined during fitting. When\n categories were specified manually, this holds the sorted categories\n (in order corresponding with output of `transform`).\n Examples\n --------\n Given a dataset with three features and two samples, we let the encoder\n find the maximum value per feature and transform the data to a binary\n one-hot encoding.\n >>> from sklearn.preprocessing import CategoricalEncoder\n >>> enc = CategoricalEncoder(handle_unknown='ignore')\n >>> enc.fit([[0, 0, 3], [1, 1, 0], [0, 2, 1], [1, 0, 2]])\n ... # doctest: +ELLIPSIS\n CategoricalEncoder(categories='auto', dtype=<... 'numpy.float64'>,\n encoding='onehot', handle_unknown='ignore')\n >>> enc.transform([[0, 1, 1], [1, 0, 4]]).toarray()\n array([[ 1., 0., 0., 1., 0., 0., 1., 0., 0.],\n [ 0., 1., 1., 0., 0., 0., 0., 0., 0.]])\n See also\n --------\n sklearn.preprocessing.OneHotEncoder : performs a one-hot encoding of\n integer ordinal features. The ``OneHotEncoder assumes`` that input\n features take on values in the range ``[0, max(feature)]`` instead of\n using the unique values.\n sklearn.feature_extraction.DictVectorizer : performs a one-hot encoding of\n dictionary items (also handles string-valued features).\n sklearn.feature_extraction.FeatureHasher : performs an approximate one-hot\n encoding of dictionary items or strings.\n \"\"\"\n\n def __init__(self, encoding='onehot', categories='auto', dtype=np.float64,\n handle_unknown='error'):\n self.encoding = encoding\n self.categories = categories\n self.dtype = dtype\n self.handle_unknown = handle_unknown\n\n def fit(self, X, y=None):\n \"\"\"Fit the CategoricalEncoder to X.\n Parameters\n ----------\n X : array-like, shape [n_samples, n_feature]\n The data to determine the categories of each feature.\n Returns\n -------\n self\n \"\"\"\n\n if self.encoding not in ['onehot', 'onehot-dense', 'ordinal']:\n template = (\"encoding should be either 'onehot', 'onehot-dense' \"\n \"or 'ordinal', got %s\")\n raise ValueError(template % self.handle_unknown)\n\n if self.handle_unknown not in ['error', 'ignore']:\n template = (\"handle_unknown should be either 'error' or \"\n \"'ignore', got %s\")\n raise ValueError(template % self.handle_unknown)\n\n if self.encoding == 'ordinal' and self.handle_unknown == 'ignore':\n raise ValueError(\"handle_unknown='ignore' is not supported for\"\n \" encoding='ordinal'\")\n\n X = check_array(X, dtype=np.object, accept_sparse='csc', copy=True)\n n_samples, n_features = X.shape\n\n self._label_encoders_ = [LabelEncoder() for _ in range(n_features)]\n\n for i in range(n_features):\n le = self._label_encoders_[i]\n Xi = X[:, i]\n if self.categories == 'auto':\n le.fit(Xi)\n else:\n valid_mask = np.in1d(Xi, self.categories[i])\n if not np.all(valid_mask):\n if self.handle_unknown == 'error':\n diff = np.unique(Xi[~valid_mask])\n msg = (\"Found unknown categories {0} in column {1}\"\n \" during fit\".format(diff, i))\n raise ValueError(msg)\n le.classes_ = np.array(np.sort(self.categories[i]))\n\n self.categories_ = [le.classes_ for le in self._label_encoders_]\n\n return self\n\n def transform(self, X):\n \"\"\"Transform X using one-hot encoding.\n Parameters\n ----------\n X : array-like, shape [n_samples, n_features]\n The data to encode.\n Returns\n -------\n X_out : sparse matrix or a 2-d array\n Transformed input.\n \"\"\"\n X = check_array(X, accept_sparse='csc', dtype=np.object, copy=True)\n n_samples, n_features = X.shape\n X_int = np.zeros_like(X, dtype=np.int)\n X_mask = np.ones_like(X, dtype=np.bool)\n\n for i in range(n_features):\n valid_mask = np.in1d(X[:, i], self.categories_[i])\n\n if not np.all(valid_mask):\n if self.handle_unknown == 'error':\n diff = np.unique(X[~valid_mask, i])\n msg = (\"Found unknown categories {0} in column {1}\"\n \" during transform\".format(diff, i))\n raise ValueError(msg)\n else:\n # Set the problematic rows to an acceptable value and\n # continue `The rows are marked `X_mask` and will be\n # removed later.\n X_mask[:, i] = valid_mask\n X[:, i][~valid_mask] = self.categories_[i][0]\n X_int[:, i] = self._label_encoders_[i].transform(X[:, i])\n\n if self.encoding == 'ordinal':\n return X_int.astype(self.dtype, copy=False)\n\n mask = X_mask.ravel()\n n_values = [cats.shape[0] for cats in self.categories_]\n n_values = np.array([0] + n_values)\n indices = np.cumsum(n_values)\n\n column_indices = (X_int + indices[:-1]).ravel()[mask]\n row_indices = np.repeat(np.arange(n_samples, dtype=np.int32),\n n_features)[mask]\n data = np.ones(n_samples * n_features)[mask]\n\n out = sparse.csc_matrix((data, (row_indices, column_indices)),\n shape=(n_samples, indices[-1]),\n dtype=self.dtype).tocsr()\n if self.encoding == 'onehot-dense':\n return out.toarray()\n else:\n return out\n",
"_____no_output_____"
],
[
"# 另一个转换器:用于选择子集\nfrom sklearn.base import BaseEstimator, TransformerMixin\n\nclass DataFrameSelector(BaseEstimator, TransformerMixin):\n def __init__(self, attribute_names):\n self.attribute_names = attribute_names\n def fit(self, X, y=None):\n return self\n def transform(self, X):\n return X[self.attribute_names]\n\nclass DataFrameFillCat(BaseEstimator, TransformerMixin):\n def __init__(self, arrtibute_names):\n self.attribute_names = arrtibute_names\n def fit(self, X):\n return self\n def transform(self, X):\n print(type(X))\n for attributename in self.attribute_names:\n # print(X[attributename])\n freq_cat = X[attributename].dropna().mode()[0]\n# print(freq_cat)\n X[attributename] = X[attributename].fillna(freq_cat)\n return X.values",
"_____no_output_____"
],
[
"# 加载数据\ntrain_df = pd.read_csv(\"./datasets/train.csv\")\ntest_df = pd.read_csv(\"./datasets/test.csv\")\ncombine = [train_df, test_df]",
"_____no_output_____"
],
[
"train_df.head()",
"_____no_output_____"
],
[
"train_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1460 entries, 0 to 1459\nData columns (total 81 columns):\nId 1460 non-null int64\nMSSubClass 1460 non-null int64\nMSZoning 1460 non-null object\nLotFrontage 1201 non-null float64\nLotArea 1460 non-null int64\nStreet 1460 non-null object\nAlley 91 non-null object\nLotShape 1460 non-null object\nLandContour 1460 non-null object\nUtilities 1460 non-null object\nLotConfig 1460 non-null object\nLandSlope 1460 non-null object\nNeighborhood 1460 non-null object\nCondition1 1460 non-null object\nCondition2 1460 non-null object\nBldgType 1460 non-null object\nHouseStyle 1460 non-null object\nOverallQual 1460 non-null int64\nOverallCond 1460 non-null int64\nYearBuilt 1460 non-null int64\nYearRemodAdd 1460 non-null int64\nRoofStyle 1460 non-null object\nRoofMatl 1460 non-null object\nExterior1st 1460 non-null object\nExterior2nd 1460 non-null object\nMasVnrType 1452 non-null object\nMasVnrArea 1452 non-null float64\nExterQual 1460 non-null object\nExterCond 1460 non-null object\nFoundation 1460 non-null object\nBsmtQual 1423 non-null object\nBsmtCond 1423 non-null object\nBsmtExposure 1422 non-null object\nBsmtFinType1 1423 non-null object\nBsmtFinSF1 1460 non-null int64\nBsmtFinType2 1422 non-null object\nBsmtFinSF2 1460 non-null int64\nBsmtUnfSF 1460 non-null int64\nTotalBsmtSF 1460 non-null int64\nHeating 1460 non-null object\nHeatingQC 1460 non-null object\nCentralAir 1460 non-null object\nElectrical 1459 non-null object\n1stFlrSF 1460 non-null int64\n2ndFlrSF 1460 non-null int64\nLowQualFinSF 1460 non-null int64\nGrLivArea 1460 non-null int64\nBsmtFullBath 1460 non-null int64\nBsmtHalfBath 1460 non-null int64\nFullBath 1460 non-null int64\nHalfBath 1460 non-null int64\nBedroomAbvGr 1460 non-null int64\nKitchenAbvGr 1460 non-null int64\nKitchenQual 1460 non-null object\nTotRmsAbvGrd 1460 non-null int64\nFunctional 1460 non-null object\nFireplaces 1460 non-null int64\nFireplaceQu 770 non-null object\nGarageType 1379 non-null object\nGarageYrBlt 1379 non-null float64\nGarageFinish 1379 non-null object\nGarageCars 1460 non-null int64\nGarageArea 1460 non-null int64\nGarageQual 1379 non-null object\nGarageCond 1379 non-null object\nPavedDrive 1460 non-null object\nWoodDeckSF 1460 non-null int64\nOpenPorchSF 1460 non-null int64\nEnclosedPorch 1460 non-null int64\n3SsnPorch 1460 non-null int64\nScreenPorch 1460 non-null int64\nPoolArea 1460 non-null int64\nPoolQC 7 non-null object\nFence 281 non-null object\nMiscFeature 54 non-null object\nMiscVal 1460 non-null int64\nMoSold 1460 non-null int64\nYrSold 1460 non-null int64\nSaleType 1460 non-null object\nSaleCondition 1460 non-null object\nSalePrice 1460 non-null int64\ndtypes: float64(3), int64(35), object(43)\nmemory usage: 924.0+ KB\n"
],
[
"train_df.describe()",
"_____no_output_____"
],
[
"train_df.describe(include=np.object)",
"_____no_output_____"
],
[
"num_attribute = ['MSSubClass', 'LotArea', 'OverallQual',\n 'OverallCond', 'YearBuilt', 'YearRemodAdd', 'MasVnrArea', 'BsmtFinSF1',\n 'BsmtFinSF2', 'BsmtUnfSF', 'TotalBsmtSF', '1stFlrSF', '2ndFlrSF',\n 'LowQualFinSF', 'GrLivArea', 'BsmtFullBath', 'BsmtHalfBath', 'FullBath',\n 'HalfBath', 'BedroomAbvGr', 'KitchenAbvGr', 'TotRmsAbvGrd',\n 'Fireplaces', 'GarageYrBlt', 'GarageCars', 'GarageArea', 'WoodDeckSF',\n 'OpenPorchSF', 'EnclosedPorch', '3SsnPorch', 'ScreenPorch', 'PoolArea',\n 'MiscVal', 'MoSold', 'YrSold',]\ncat_attribute = ['MSZoning', 'Street', 'LotShape', 'LandContour', 'Utilities',\n 'LotConfig', 'LandSlope', 'Neighborhood', 'Condition1', 'Condition2',\n 'BldgType', 'HouseStyle', 'RoofStyle', 'RoofMatl', 'Exterior1st',\n 'Exterior2nd', 'MasVnrType', 'ExterQual', 'ExterCond', 'Foundation',\n 'BsmtQual', 'BsmtCond', 'BsmtExposure', 'BsmtFinType1', 'BsmtFinType2',\n 'Heating', 'HeatingQC', 'CentralAir', 'Electrical', 'KitchenQual',\n 'Functional', 'GarageType', 'GarageFinish', 'GarageQual',\n 'GarageCond', 'PavedDrive',\n 'SaleType', 'SaleCondition']",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\nnum_pipeline = Pipeline([\n (\"selector\", DataFrameSelector(num_attribute)),\n (\"imputer\", Imputer(strategy=\"median\")),\n (\"std_scaler\", StandardScaler())\n])",
"_____no_output_____"
],
[
"cat_pipeline = Pipeline([\n (\"selector\", DataFrameSelector(cat_attribute)),\n (\"fillna\", DataFrameFillCat(cat_attribute)),\n (\"cat_encoder\", CategoricalEncoder(encoding=\"onehot-dense\"))\n])",
"_____no_output_____"
],
[
"X_train = train_df\nX_train_cat_pipeline = num_pipeline.fit_transform(X_train)",
"_____no_output_____"
],
[
"from sklearn.pipeline import FeatureUnion\nfull_pipeline = FeatureUnion(transformer_list=[\n (\"num_pipeline\", num_pipeline),\n (\"cat_pipeline\", cat_pipeline),\n])",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train = train_df.drop([\"Id\", \"SalePrice\"], axis = 1)\ny_train = train_df[\"SalePrice\"]\n# X_train.info()",
"_____no_output_____"
],
[
"X_train_pipeline = full_pipeline.fit_transform(X_train)",
"<class 'pandas.core.frame.DataFrame'>\n"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X_train_pipeline, y_train, test_size=0.1)",
"_____no_output_____"
],
[
"X_train.shape, X_test.shape, y_train.shape",
"_____no_output_____"
],
[
"# X_test_pipeline = full_pipeline.transform(X_test)",
"_____no_output_____"
],
[
"from sklearn.ensemble import RandomForestRegressor\nrdf_reg = RandomForestRegressor()\nrdf_reg.fit(X_train, y_train)",
"_____no_output_____"
],
[
"y_pred = rdf_reg.predict(X_test)\n# y_pred = rdf_reg.predict(X_test_pipeline)",
"_____no_output_____"
],
[
"from sklearn.metrics import mean_squared_error\nscores_mse = mean_squared_error(y_pred, y_test)\nscores_mse",
"_____no_output_____"
],
[
"from sklearn.ensemble import GradientBoostingRegressor\ngbr_reg = GradientBoostingRegressor(n_estimators=1000, max_depth=2)\ngbr_reg.fit(X_train, y_train)\ny_pred = gbr_reg.predict(X_test)\nscores_mse = mean_squared_error(y_pred, y_test)\nscores_mse",
"_____no_output_____"
],
[
"test_df_data = test_df.drop([\"Id\"], axis=1)",
"_____no_output_____"
],
[
"X_test_pipeline = full_pipeline.transform(test_df_data)\n# test_df_data.info()\n# test_df_data.info()",
"<class 'pandas.core.frame.DataFrame'>\n"
],
[
"y_pred = gbr_reg.predict(X_test_pipeline)",
"_____no_output_____"
],
[
"result =pd.DataFrame({\n \"Id\": test_df[\"Id\"],\n \"SalePrice\": y_pred\n })\nresult.to_csv(\"result.csv\", index=False)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8dec628bd6eb73d1c0f772b8a65445bbb792c5
| 11,444 |
ipynb
|
Jupyter Notebook
|
notebooks/experiment2_test_w2v.ipynb
|
Sapphirine/202112-20-Sentiment-Prediction-of-Game-Reviews
|
61665913578209723472d87ac9c79347b8f69842
|
[
"MIT"
] | null | null | null |
notebooks/experiment2_test_w2v.ipynb
|
Sapphirine/202112-20-Sentiment-Prediction-of-Game-Reviews
|
61665913578209723472d87ac9c79347b8f69842
|
[
"MIT"
] | null | null | null |
notebooks/experiment2_test_w2v.ipynb
|
Sapphirine/202112-20-Sentiment-Prediction-of-Game-Reviews
|
61665913578209723472d87ac9c79347b8f69842
|
[
"MIT"
] | null | null | null | 11,444 | 11,444 | 0.687522 |
[
[
[
"from google.colab import drive\ndrive.mount(\"/content/gdrive\")",
"Drive already mounted at /content/gdrive; to attempt to forcibly remount, call drive.mount(\"/content/gdrive\", force_remount=True).\n"
],
[
"import numpy as np\nimport pandas as pd\nimport numpy as np\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\nimport re\nimport torch\ndf = pd.read_csv(\"/content/gdrive/MyDrive/tidydata.csv\")\ndf['label'] = df['recommended'].apply(lambda x: 0 if x== True else 1)\nX = df[['review']]\ny = df['label']\nX_train, X_test, y_train, y_test = train_test_split(df.index.values, df.label.values, test_size=0.2, random_state=40, stratify=df.label.values)\ndf['type'] = ['tmp']*df.shape[0]\ndf.loc[X_train, 'type'] = 'train'\ndf.loc[X_test, 'type'] = 'test'\nX_train_list = list(df[df.type=='train'].review.values)\nY_train_list = list(df[df.type=='train'].label.values)\ntmp1 = []\ntmp2 = []\nfor i in range(len(X_train_list)):\n if X_train_list[i]==X_train_list[i]:\n tmp1.append(X_train_list[i])\n tmp2.append(Y_train_list[i])\nX_train_list = tmp1\nY_train_list = tmp2\n\nX_test_list = list(df[df.type=='test'].review.values)\nY_test_list = list(df[df.type=='test'].label.values)\ntmp1 = []\ntmp2 = []\nfor i in range(len(X_test_list)):\n if X_test_list[i]==X_test_list[i]:\n tmp1.append(X_test_list[i])\n tmp2.append(Y_test_list[i])\nX_test_list = tmp1\nY_test_list = tmp2",
"_____no_output_____"
],
[
"import json\nwith open(\"/content/gdrive/MyDrive/df_train_w2v.json\", \"r\") as file:\n train_emb = json.load(file)\nwith open(\"/content/gdrive/MyDrive/df_test_w2v.json\", \"r\") as file:\n test_emb = json.load(file)",
"_____no_output_____"
],
[
"train_emb[0][0]",
"_____no_output_____"
],
[
"from sklearn.utils import shuffle\nX_train, Y_train = shuffle(train_emb, Y_train_list)\nfrom sklearn.neural_network import MLPClassifier\nfrom sklearn.linear_model import LogisticRegression\n#NN = MLPClassifier(solver='lbfgs', alpha=1e-5, hidden_layer_sizes=(128, 2))\nLR = LogisticRegression(random_state=0).fit(X_train, Y_train)\n#NN.fit(X_train, Y_train)\nLR.fit(X_train, Y_train)",
"/usr/local/lib/python3.7/dist-packages/sklearn/linear_model/_logistic.py:818: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG,\n/usr/local/lib/python3.7/dist-packages/sklearn/linear_model/_logistic.py:818: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG,\n"
],
[
"from sklearn.metrics import classification_report\ny_pred = LR.predict(test_emb)\nacc = 0\nfor i in range(len(y_pred)):\n if y_pred[i]==Y_test_list[i]:\n acc += 1\nacc = acc/len(y_pred)\nprint(acc)\nprint(classification_report(y_pred, Y_test_list, digits=3))",
"0.8325318586198606\n precision recall f1-score support\n\n 0 0.822 0.841 0.831 20373\n 1 0.843 0.825 0.834 21217\n\n accuracy 0.833 41590\n macro avg 0.833 0.833 0.833 41590\nweighted avg 0.833 0.833 0.833 41590\n\n"
],
[
"sum(Y_test_list)",
"_____no_output_____"
],
[
"import numpy as np\nfrom sklearn import metrics\nfpr, tpr, thresholds = metrics.roc_curve(Y_test_list, y_pred, pos_label=1)\nmetrics.auc(fpr, tpr)",
"_____no_output_____"
],
[
"from sklearn.metrics import classification_report\nfrom sklearn.naive_bayes import GaussianNB\ngnb = GaussianNB()\ngnb.fit(X_train, Y_train)",
"_____no_output_____"
],
[
"y_pred = gnb.predict(test_emb)\nacc = 0\nfor i in range(len(y_pred)):\n if y_pred[i]==Y_test_list[i]:\n acc += 1\nacc = acc/len(y_pred)\nprint(classification_report(y_pred, Y_test_list, digits=3))",
" precision recall f1-score support\n\n 0 0.500 0.747 0.599 13948\n 1 0.830 0.623 0.712 27642\n\n accuracy 0.665 41590\n macro avg 0.665 0.685 0.655 41590\nweighted avg 0.719 0.665 0.674 41590\n\n"
],
[
"from sklearn.ensemble import RandomForestClassifier\nfrom sklearn.datasets import make_classification\nX_train, Y_train = shuffle(train_emb, Y_train_list)\nclf = RandomForestClassifier(max_depth=10, random_state=0)\nclf.fit(X_train, Y_train)\n#df_tfidf_test\nprint(classification_report(clf.predict(test_emb), Y_test_list, digits=3))",
" precision recall f1-score support\n\n 0 0.804 0.824 0.814 20336\n 1 0.827 0.808 0.817 21254\n\n accuracy 0.816 41590\n macro avg 0.816 0.816 0.816 41590\nweighted avg 0.816 0.816 0.816 41590\n\n"
],
[
"y_pred = clf.predict(test_emb)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8dfd660ecde4bef30808f06a0d57bde7b88730
| 53,410 |
ipynb
|
Jupyter Notebook
|
SNRtrial.ipynb
|
xianglight/MC_DNN
|
8dad8ef738fa7c8a8082fa43c87db4719d356dfb
|
[
"MIT"
] | null | null | null |
SNRtrial.ipynb
|
xianglight/MC_DNN
|
8dad8ef738fa7c8a8082fa43c87db4719d356dfb
|
[
"MIT"
] | null | null | null |
SNRtrial.ipynb
|
xianglight/MC_DNN
|
8dad8ef738fa7c8a8082fa43c87db4719d356dfb
|
[
"MIT"
] | null | null | null | 220.702479 | 21,638 | 0.70719 |
[
[
[
"import pandas as pd\n\n# Read in white wine data \nMCdata = pd.read_csv(r\"C:\\Users\\soari\\Documents\\GitHub\\MC_SNR_DNN/SNRdata.csv\",header=None)\n\n# Read in red wine data \nMClabel = pd.read_csv(r\"C:\\Users\\soari\\Documents\\GitHub\\MC_SNR_DNN/SNRlabel.csv\",header=None)",
"_____no_output_____"
],
[
"print(MCdata.info())\nprint(MClabel.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 12000 entries, 0 to 11999\nData columns (total 100 columns):\n0 12000 non-null int64\n1 12000 non-null float64\n2 12000 non-null float64\n3 12000 non-null float64\n4 12000 non-null float64\n5 12000 non-null float64\n6 12000 non-null float64\n7 12000 non-null float64\n8 12000 non-null float64\n9 12000 non-null float64\n10 12000 non-null float64\n11 12000 non-null float64\n12 12000 non-null float64\n13 12000 non-null float64\n14 12000 non-null float64\n15 12000 non-null float64\n16 12000 non-null float64\n17 12000 non-null float64\n18 12000 non-null float64\n19 12000 non-null float64\n20 12000 non-null float64\n21 12000 non-null float64\n22 12000 non-null float64\n23 12000 non-null float64\n24 12000 non-null float64\n25 12000 non-null float64\n26 12000 non-null float64\n27 12000 non-null float64\n28 12000 non-null float64\n29 12000 non-null float64\n30 12000 non-null float64\n31 12000 non-null float64\n32 12000 non-null float64\n33 12000 non-null float64\n34 12000 non-null float64\n35 12000 non-null float64\n36 12000 non-null float64\n37 12000 non-null float64\n38 12000 non-null float64\n39 12000 non-null float64\n40 12000 non-null float64\n41 12000 non-null float64\n42 12000 non-null float64\n43 12000 non-null float64\n44 12000 non-null float64\n45 12000 non-null float64\n46 12000 non-null float64\n47 12000 non-null float64\n48 12000 non-null float64\n49 12000 non-null float64\n50 12000 non-null float64\n51 12000 non-null float64\n52 12000 non-null float64\n53 12000 non-null float64\n54 12000 non-null float64\n55 12000 non-null float64\n56 12000 non-null float64\n57 12000 non-null float64\n58 12000 non-null float64\n59 12000 non-null float64\n60 12000 non-null float64\n61 12000 non-null float64\n62 12000 non-null float64\n63 12000 non-null float64\n64 12000 non-null float64\n65 12000 non-null float64\n66 12000 non-null float64\n67 12000 non-null float64\n68 12000 non-null float64\n69 12000 non-null float64\n70 12000 non-null float64\n71 12000 non-null float64\n72 12000 non-null float64\n73 12000 non-null float64\n74 12000 non-null float64\n75 12000 non-null float64\n76 12000 non-null float64\n77 12000 non-null float64\n78 12000 non-null float64\n79 12000 non-null float64\n80 12000 non-null float64\n81 12000 non-null float64\n82 12000 non-null float64\n83 12000 non-null float64\n84 12000 non-null float64\n85 12000 non-null float64\n86 12000 non-null float64\n87 12000 non-null float64\n88 12000 non-null float64\n89 12000 non-null float64\n90 12000 non-null float64\n91 12000 non-null float64\n92 12000 non-null float64\n93 12000 non-null float64\n94 12000 non-null float64\n95 12000 non-null float64\n96 12000 non-null float64\n97 12000 non-null float64\n98 12000 non-null float64\n99 12000 non-null float64\ndtypes: float64(99), int64(1)\nmemory usage: 9.2 MB\nNone\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 12000 entries, 0 to 11999\nData columns (total 1 columns):\n0 12000 non-null int64\ndtypes: int64(1)\nmemory usage: 93.9 KB\nNone\n"
],
[
"import matplotlib.pyplot as plt\npm = MCdata[100:101]\n# pm.plot(kind='bar',legend=False)\nax = pm.transpose().plot(kind='line', title =\"CDF\", figsize=(6, 4), legend=False, fontsize=12) \nax.set_xlabel(\"Bins\", fontsize=12)\nax.set_ylabel(\"CDF\", fontsize=12)\nplt.show()\n # plt.savefig('filename.png', dpi=600)\n",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nimport numpy as np\n\n# Split the data up in train and test sets\nX_train, X_test, y_train, y_test = train_test_split(MCdata, MClabel, test_size=0.33, random_state=42)\n\n",
"_____no_output_____"
],
[
"# Import `Sequential` from `keras.models`\nfrom keras.models import Sequential\n\n# Import `Dense` from `keras.layers`\nfrom keras.layers import Dense\nfrom tensorflow.keras import layers\nfrom tensorflow.keras import regularizers\n\n# Initialize the constructor\nmodel = Sequential()\n\n# # Add an input layer \n# model.add(Dense(100, activation='relu', input_shape=(100,)))\n\n# # Add one hidden layer \n# model.add(Dense(20, activation='relu'))\n\n# # Add an output layer \n# model.add(Dense(3, activation='sigmoid'))\n\n\n# # Strategy 1: add weight regulation to avoid overfitting \n # # Add an input layer \n # model.add(Dense(100, activation='relu', kernel_regularizer=regularizers.l2(0.001), input_shape=(100,)))\n\n # # Add one hidden layer \n # model.add(Dense(20, kernel_regularizer=regularizers.l2(0.001),activation='relu'))\n # # l2(0.001) means that every coefficient in the weight matrix of the layer will add 0.001 * weight_coefficient_value**2 to the total loss of the network.\n\n # Strategy 2: Dropout \nmodel.add(Dense(100, activation='relu', input_shape=(100,)))\nlayers.Dropout(0.5),\nmodel.add(Dense(20, activation='relu'))\nlayers.Dropout(0.5),\nmodel.add(Dense(1)\n\n\n# Model output shape\nmodel.output_shape\n\n# Model summary\nmodel.summary()\n\n# Model config\nmodel.get_config()\n\n# List all weight tensors \nmodel.get_weights()",
"Using TensorFlow backend.\nWARNING:tensorflow:From C:\\Users\\soari\\Anaconda3\\envs\\tf1\\lib\\site-packages\\tensorflow_core\\python\\ops\\resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\nModel: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_1 (Dense) (None, 100) 10100 \n_________________________________________________________________\ndense_2 (Dense) (None, 20) 2020 \n_________________________________________________________________\ndense_3 (Dense) (None, 3) 63 \n=================================================================\nTotal params: 12,183\nTrainable params: 12,183\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"X_train=X_train.transpose()\nX_test=X_test.transpose()\ny_train=y_train.transpose()\ny_test=y_test.transpose()\n\n\nfrom keras import optimizers\nsgd = optimizers.SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)\nmodel.compile(loss='mse',optimizer='adam',metrics=['mae','mse'])\nhistory=model.fit(X_train,y_train,epochs=5,batch_size=1,verbose=1,validation_split=0.2)\ny_pred = model.predict(X_test)\n\nscore = model.evaluate(X_test, y_test,verbose=1)\n# print(\"Test Score:\", score[0])\n# print(\"Test Accuracy:\", score[1])",
"_____no_output_____"
],
[
"# Plot training & validation accuracy values\nplt.plot(history.history['acc'])\nplt.plot(history.history['val_acc'])\n\nplt.title('model accuracy')\nplt.ylabel('accuracy')\nplt.xlabel('epoch')\nplt.legend(['train','test'], loc='upper left')\nplt.show()\n\n# Plot training & validation loss values\nplt.plot(history.history['loss'])\nplt.plot(history.history['val_loss'])\n\nplt.title('model loss')\nplt.ylabel('loss')\nplt.xlabel('epoch')\nplt.legend(['train','test'], loc='upper left')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8e09c9a973e80f769105ac5bb465d68f37ea51
| 802,126 |
ipynb
|
Jupyter Notebook
|
Chapter01/runs/2019-11-13T07-40-54Z/Train Your First Deep Neural Network.ipynb
|
PacktPublishing/Deep-Learning-with-R-Cookbook
|
65a1b7d18de00a3f47b674e958049037ed636b91
|
[
"MIT"
] | 4 |
2021-03-21T01:46:32.000Z
|
2022-01-08T07:58:19.000Z
|
Chapter01/runs/2019-11-13T07-40-54Z/Train Your First Deep Neural Network.ipynb
|
PacktPublishing/Deep-Learning-with-R-Cookbook
|
65a1b7d18de00a3f47b674e958049037ed636b91
|
[
"MIT"
] | null | null | null |
Chapter01/runs/2019-11-13T07-40-54Z/Train Your First Deep Neural Network.ipynb
|
PacktPublishing/Deep-Learning-with-R-Cookbook
|
65a1b7d18de00a3f47b674e958049037ed636b91
|
[
"MIT"
] | 1 |
2021-03-21T01:46:35.000Z
|
2021-03-21T01:46:35.000Z
| 24.405951 | 18,242 | 0.341682 |
[
[
[
"library(keras)",
"_____no_output_____"
]
],
[
[
"**Loading MNIST dataset from the library datasets**",
"_____no_output_____"
]
],
[
[
"mnist <- dataset_mnist()\nx_train <- mnist$train$x\ny_train <- mnist$train$y\nx_test <- mnist$test$x\ny_test <- mnist$test$y",
"_____no_output_____"
]
],
[
[
"**Data Preprocessing**",
"_____no_output_____"
]
],
[
[
"# reshape\nx_train <- array_reshape(x_train, c(nrow(x_train), 784))\nx_test <- array_reshape(x_test, c(nrow(x_test), 784))\n# rescale\nx_train <- x_train / 255\nx_test <- x_test / 255",
"_____no_output_____"
]
],
[
[
"The y data is an integer vector with values ranging from 0 to 9.\nTo prepare this data for training we one-hot encode the vectors into binary class matrices using the Keras to_categorical() function:",
"_____no_output_____"
]
],
[
[
"y_train <- to_categorical(y_train, 10)\ny_test <- to_categorical(y_test, 10)",
"_____no_output_____"
]
],
[
[
"**Building model**",
"_____no_output_____"
]
],
[
[
"model <- keras_model_sequential() \nmodel %>% \n layer_dense(units = 256, activation = 'relu', input_shape = c(784)) %>% \n layer_dropout(rate = 0.4) %>% \n layer_dense(units = 128, activation = 'relu') %>%\n layer_dropout(rate = 0.3) %>%\n layer_dense(units = 10, activation = 'softmax')",
"_____no_output_____"
],
[
"# Use the summary() function to print the details of the model:\nsummary(model)",
"________________________________________________________________________________\nLayer (type) Output Shape Param # \n================================================================================\ndense_1 (Dense) (None, 256) 200960 \n________________________________________________________________________________\ndropout_1 (Dropout) (None, 256) 0 \n________________________________________________________________________________\ndense_2 (Dense) (None, 128) 32896 \n________________________________________________________________________________\ndropout_2 (Dropout) (None, 128) 0 \n________________________________________________________________________________\ndense_3 (Dense) (None, 10) 1290 \n================================================================================\nTotal params: 235,146\nTrainable params: 235,146\nNon-trainable params: 0\n________________________________________________________________________________\n"
]
],
[
[
"**Compiling the model**",
"_____no_output_____"
]
],
[
[
"model %>% compile(\n loss = 'categorical_crossentropy',\n optimizer = optimizer_rmsprop(),\n metrics = c('accuracy')\n)",
"_____no_output_____"
]
],
[
[
"**Training and Evaluation**",
"_____no_output_____"
]
],
[
[
"history <- model %>% fit(\n x_train, y_train, \n epochs = 30, batch_size = 128, \n validation_split = 0.2\n)",
"_____no_output_____"
],
[
"plot(history)",
"_____no_output_____"
],
[
"# Plot the accuracy of the training data \nplot(history$metrics$acc, main=\"Model Accuracy\", xlab = \"epoch\", ylab=\"accuracy\", col=\"blue\", type=\"l\")\n\n# Plot the accuracy of the validation data\nlines(history$metrics$val_acc, col=\"green\")\n\n# Add Legend\nlegend(\"bottomright\", c(\"train\",\"test\"), col=c(\"blue\", \"green\"), lty=c(1,1))",
"_____no_output_____"
],
[
"# Plot the model loss of the training data\nplot(history$metrics$loss, main=\"Model Loss\", xlab = \"epoch\", ylab=\"loss\", col=\"blue\", type=\"l\")\n\n# Plot the model loss of the test data\nlines(history$metrics$val_loss, col=\"green\")\n\n# Add legend\nlegend(\"topright\", c(\"train\",\"test\"), col=c(\"blue\", \"green\"), lty=c(1,1))",
"_____no_output_____"
]
],
[
[
"**Predicting for the test data**",
"_____no_output_____"
]
],
[
[
"model %>% predict_classes(x_test)",
"_____no_output_____"
],
[
"# Evaluate on test data and labels\nscore <- model %>% evaluate(x_test, y_test, batch_size = 128)\n\n# Print the score\nprint(score)",
"$loss\n[1] 0.1062629\n\n$acc\n[1] 0.9793\n\n"
]
],
[
[
"## Hyperparameter tuning",
"_____no_output_____"
]
],
[
[
"# install.packages(\"tfruns\")",
"_____no_output_____"
],
[
"library(tfruns)",
"_____no_output_____"
],
[
"runs <- tuning_run(file = \"hyperparameter_tuning_model.r\", flags = list(\n dense_units1 = c(8,16),\n dropout1 = c(0.2, 0.3, 0.4),\n dense_units2 = c(8,16),\n dropout2 = c(0.2, 0.3, 0.4)\n))",
"36 total combinations of flags (use sample parameter to run a random subset)\n"
],
[
"runs",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4a8e0bb1011fd4fff69728bb76d3c41f484217b6
| 5,267 |
ipynb
|
Jupyter Notebook
|
libgeohash.ipynb
|
jclosure/libgeohash
|
9d47e2d658d60fb052ed86daab94ba735706d368
|
[
"BSD-3-Clause"
] | null | null | null |
libgeohash.ipynb
|
jclosure/libgeohash
|
9d47e2d658d60fb052ed86daab94ba735706d368
|
[
"BSD-3-Clause"
] | null | null | null |
libgeohash.ipynb
|
jclosure/libgeohash
|
9d47e2d658d60fb052ed86daab94ba735706d368
|
[
"BSD-3-Clause"
] | null | null | null | 21.323887 | 116 | 0.494209 |
[
[
[
"## Python wrapper library for libgeohash\n\nhttps://github.com/simplegeo/libgeohash",
"_____no_output_____"
]
],
[
[
"import ctypes",
"_____no_output_____"
],
[
"# compile .so with: gcc -fPIC -shared -o geohash.so geohash.c\n\n_geohash = ctypes.CDLL('./geohash_macos.so')",
"_____no_output_____"
],
[
"# convenience function for wrapping c functions\n\ndef wrap_function(lib, funcname, restype, argtypes):\n \"\"\"Simplify wrapping ctypes functions\"\"\"\n func = lib.__getattr__(funcname)\n func.restype = restype\n func.argtypes = argtypes\n return func",
"_____no_output_____"
]
],
[
[
"### Define classes to represent structs used in the c api",
"_____no_output_____"
]
],
[
[
"class GeoBoxDimension(ctypes.Structure):\n _fields_ = [('height', ctypes.c_double), ('width', ctypes.c_double)]\n\n def __repr__(self):\n return '({0}, {1})'.format(self.height, self.width)\n \n \nclass GeoCoord(ctypes.Structure):\n _fields_ = [('latitude', ctypes.c_double), ('longitude', ctypes.c_double), ('north', ctypes.c_double), \n ('east', ctypes.c_double), ('south', ctypes.c_double), ('west', ctypes.c_double), \n ('dimension', GeoBoxDimension)]\n\n def __repr__(self):\n return '({0}, {1})'.format(self.latitude, self.longitude)",
"_____no_output_____"
]
],
[
[
"### Wrap the c api in python functions",
"_____no_output_____"
]
],
[
[
"geohash_encode = wrap_function(\n _geohash, \n 'geohash_encode', \n ctypes.c_char_p, \n (ctypes.c_double, ctypes.c_double, ctypes.c_int)\n)\n\n\ngeohash_decode = wrap_function(\n _geohash,\n 'geohash_decode',\n GeoCoord,\n [ctypes.c_char_p]\n)\n\n\ngeohash_neighbors = wrap_function(\n _geohash,\n 'geohash_neighbors',\n ctypes.POINTER(ctypes.c_char_p),\n [ctypes.c_char_p]\n)\n\n\ngeohash_dimensions_for_precision = wrap_function(\n _geohash, \n 'geohash_dimensions_for_precision', \n GeoBoxDimension, \n [ctypes.c_int]\n)",
"_____no_output_____"
]
],
[
[
"### exercise the api",
"_____no_output_____"
]
],
[
[
"geohash_encode(41.41845703125, 2.17529296875, 5)",
"_____no_output_____"
],
[
"geohash_decode(b'sp3e9')",
"_____no_output_____"
],
[
"a = geohash_neighbors(b'sp3e9')\n[a[i] for i in range(0,8)] # there are 8 neighbors",
"_____no_output_____"
],
[
"geohash_dimensions_for_precision(6)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4a8e0e01cb277453c79e09144de584298fdbf7c1
| 3,701 |
ipynb
|
Jupyter Notebook
|
Create_Job_Submission_Script.ipynb
|
aaleksopoulos/Udacity_Intel_Edge_AI_project2_smart-queue-system
|
2760d71f7ec5937bec013f6b30780e3fb9c65e6c
|
[
"MIT"
] | null | null | null |
Create_Job_Submission_Script.ipynb
|
aaleksopoulos/Udacity_Intel_Edge_AI_project2_smart-queue-system
|
2760d71f7ec5937bec013f6b30780e3fb9c65e6c
|
[
"MIT"
] | null | null | null |
Create_Job_Submission_Script.ipynb
|
aaleksopoulos/Udacity_Intel_Edge_AI_project2_smart-queue-system
|
2760d71f7ec5937bec013f6b30780e3fb9c65e6c
|
[
"MIT"
] | null | null | null | 35.247619 | 308 | 0.597947 |
[
[
[
"# Step 2: Create Job Submission Script\n\nThe next step is to create our job submission script. In the cell below, you will need to complete the job submission script and run the cell to generate the file using the magic `%%writefile` command. Your main task is to complete the following items of the script:\n\n* Create a variable `MODEL` and assign it the value of the first argument passed to the job submission script.\n* Create a variable `DEVICE` and assign it the value of the second argument passed to the job submission script.\n* Create a variable `VIDEO` and assign it the value of the third argument passed to the job submission script.\n* Create a variable `PEOPLE` and assign it the value of the sixth argument passed to the job submission script.",
"_____no_output_____"
]
],
[
[
"%%writefile queue_job.sh\n#!/bin/bash\n\nexec 1>/output/stdout.log 2>/output/stderr.log\n\n# TODO: Create MODEL variable\nMODEL=$1\n# TODO: Create DEVICE variable\nDEVICE=$2\n# TODO: Create VIDEO variable\nVIDEO=$3\nQUEUE=$4\nOUTPUT=$5\n# TODO: Create PEOPLE variable\nPEOPLE=$6\n\nmkdir -p $5\n\nif echo \"$DEVICE\" | grep -q \"FPGA\"; then # if device passed in is FPGA, load bitstream to program FPGA\n #Environment variables and compilation for edge compute nodes with FPGAs\n export AOCL_BOARD_PACKAGE_ROOT=/opt/intel/openvino/bitstreams/a10_vision_design_sg2_bitstreams/BSP/a10_1150_sg2\n\n source /opt/altera/aocl-pro-rte/aclrte-linux64/init_opencl.sh\n aocl program acl0 /opt/intel/openvino/bitstreams/a10_vision_design_sg2_bitstreams/2020-2_PL2_FP16_MobileNet_Clamp.aocx\n\n export CL_CONTEXT_COMPILER_MODE_INTELFPGA=3\nfi\n\npython3 person_detect.py --model ${MODEL} \\\n --device ${DEVICE} \\\n --video ${VIDEO} \\\n --queue_param ${QUEUE} \\\n --output_path ${OUTPUT}\\\n --max_people ${PEOPLE} \\\n\ncd /output\n\ntar zcvf output.tgz *",
"Overwriting queue_job.sh\n"
]
],
[
[
"# Next Step\n\nNow that you've run the above cell and created your job submission script, you will work through each scenarios notebook in the next three workspaces. In each of these notebooks, you will submit jobs to Intel's DevCloud to load and run inference on each type of hardware and then review the results.\n\n**Note**: As a reminder, if you need to make any changes to the job submission script, you can come back to this workspace to edit and run the above cell to overwrite the file with your changes.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a8e1cda49300afa6f10393268d3ebabe07eedac
| 14,731 |
ipynb
|
Jupyter Notebook
|
COMPAS.ipynb
|
sato9hara/stealthily-biased-sampling
|
1eb69998ae4db2fbfa4f0cf2e4ec9e1867884b32
|
[
"MIT"
] | 2 |
2019-11-18T03:46:47.000Z
|
2021-07-23T17:56:46.000Z
|
COMPAS.ipynb
|
sato9hara/stealthily-biased-sampling
|
1eb69998ae4db2fbfa4f0cf2e4ec9e1867884b32
|
[
"MIT"
] | null | null | null |
COMPAS.ipynb
|
sato9hara/stealthily-biased-sampling
|
1eb69998ae4db2fbfa4f0cf2e4ec9e1867884b32
|
[
"MIT"
] | null | null | null | 33.786697 | 153 | 0.484828 |
[
[
[
"## This notebook contains a sample code for the COMPAS data experiment in Section 5.2.\n\nBefore running the code, please check README.md and install LEMON.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom matplotlib import pyplot as plt\nfrom sklearn import feature_extraction\nfrom sklearn import preprocessing\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nimport stealth_sampling",
"_____no_output_____"
]
],
[
[
"### Functions",
"_____no_output_____"
]
],
[
[
"# split data to bins (s, y) = (1, 1), (1, 0), (0, 1), (0, 0)\ndef split_to_four(X, S, Y):\n Z = np.c_[X, S, Y]\n Z_pos_pos = Z[np.logical_and(S, Y), :]\n Z_pos_neg = Z[np.logical_and(S, np.logical_not(Y)), :]\n Z_neg_pos = Z[np.logical_and(np.logical_not(S), Y), :]\n Z_neg_neg = Z[np.logical_and(np.logical_not(S), np.logical_not(Y)), :]\n Z = [Z_pos_pos, Z_pos_neg, Z_neg_pos, Z_neg_neg]\n return Z\n\n# compute demographic parity\ndef demographic_parity(W):\n p_pos = np.mean(np.concatenate(W[:2]))\n p_neg = np.mean(np.concatenate(W[2:]))\n return np.abs(p_pos - p_neg)\n\n# compute the sampling size from each bin\ndef computeK(Z, Nsample, sampled_spos, sampled_ypos):\n Kpp = Nsample*sampled_spos*sampled_ypos[0]\n Kpn = Nsample*sampled_spos*(1-sampled_ypos[0])\n Knp = Nsample*(1-sampled_spos)*sampled_ypos[1]\n Knn = Nsample*(1-sampled_spos)*(1-sampled_ypos[1])\n K = [Kpp, Kpn, Knp, Knn]\n kratio = min([min(1, z.shape[0]/k) for (z, k) in zip(Z, K)])\n Kpp = int(np.floor(Nsample*kratio*sampled_spos*sampled_ypos[0]))\n Kpn = int(np.floor(Nsample*kratio*sampled_spos*(1-sampled_ypos[0])))\n Knp = int(np.floor(Nsample*kratio*(1-sampled_spos)*sampled_ypos[1]))\n Knn = int(np.floor(Nsample*kratio*(1-sampled_spos)*(1-sampled_ypos[1])))\n K = [max([k, 1]) for k in [Kpp, Kpn, Knp, Knn]]\n return K\n\n# case-contrl sampling\ndef case_control_sampling(X, K):\n q = [(K[i]/sum(K)) * np.ones(x.shape[0]) / x.shape[0] for i, x in enumerate(X)]\n return q\n\n# compute wasserstein distance\ndef compute_wasserstein(X1, S1, X2, S2, timeout=10.0):\n dx = stealth_sampling.compute_wasserstein(X1, X2, path='./', prefix='compas', timeout=timeout)\n dx_s1 = stealth_sampling.compute_wasserstein(X1[S1>0.5, :], X2[S2>0.5, :], path='./', prefix='compas', timeout=timeout)\n dx_s0 = stealth_sampling.compute_wasserstein(X1[S1<0.5, :], X2[S2<0.5, :], path='./', prefix='compas', timeout=timeout)\n return dx, dx_s1, dx_s0",
"_____no_output_____"
]
],
[
[
"### Fetch data and preprocess\nWe modified [https://github.com/mbilalzafar/fair-classification/blob/master/disparate_mistreatment/propublica_compas_data_demo/load_compas_data.py]",
"_____no_output_____"
]
],
[
[
"url = 'https://raw.githubusercontent.com/propublica/compas-analysis/master/compas-scores-two-years.csv'\nfeature_list = ['age_cat', 'race', 'sex', 'priors_count', 'c_charge_degree', 'two_year_recid']\nsensitive = 'race'\nlabel = 'score_text'\n\n# fetch data\ndf = pd.read_table(url, sep=',')\ndf = df.dropna(subset=['days_b_screening_arrest'])\n\n# convert to np array\ndata = df.to_dict('list')\nfor k in data.keys():\n data[k] = np.array(data[k])\n\n# filtering records\nidx = np.logical_and(data['days_b_screening_arrest']<=30, data['days_b_screening_arrest']>=-30)\nidx = np.logical_and(idx, data['is_recid'] != -1)\nidx = np.logical_and(idx, data['c_charge_degree'] != 'O')\nidx = np.logical_and(idx, data['score_text'] != 'NA')\nidx = np.logical_and(idx, np.logical_or(data['race'] == 'African-American', data['race'] == 'Caucasian'))\nfor k in data.keys():\n data[k] = data[k][idx]\n \n# label Y\nY = 1 - np.logical_not(data[label]=='Low').astype(np.int32)\n\n# feature X, sensitive feature S\nX = []\nfor feature in feature_list:\n vals = data[feature]\n if feature == 'priors_count':\n vals = [float(v) for v in vals]\n vals = preprocessing.scale(vals)\n vals = np.reshape(vals, (Y.size, -1))\n else:\n lb = preprocessing.LabelBinarizer()\n lb.fit(vals)\n vals = lb.transform(vals)\n if feature == sensitive:\n S = vals[:, 0]\n X.append(vals)\nX = np.concatenate(X, axis=1)",
"_____no_output_____"
]
],
[
[
"### Experiment",
"_____no_output_____"
]
],
[
[
"# parameter settings\nseed = 0 # random seed\n\n# parameter settings for sampling\nNsample = 2000 # number of data to sample\nsampled_ypos = [0.5, 0.5] # the ratio of positive decisions '\\alpha' in sampling\n\n# parameter settings for complainer\nNref = 1278 # number of referential data",
"_____no_output_____"
],
[
"def sample_and_evaluate(X, S, Y, Nref=1278, Nsample=2000, sampled_ypos=[0.5, 0.5], seed=0):\n \n # load data\n Xbase, Xref, Sbase, Sref, Ybase, Yref = train_test_split(X, S, Y, test_size=Nref, random_state=seed)\n N = Xbase.shape[0]\n scaler = StandardScaler()\n scaler.fit(Xbase)\n Xbase = scaler.transform(Xbase)\n Xref = scaler.transform(Xref)\n\n # wasserstein distance between base and ref\n np.random.seed(seed)\n idx = np.random.permutation(Xbase.shape[0])[:Nsample]\n dx, dx_s1, dx_s0 = compute_wasserstein(Xbase[idx, :], Sbase[idx], Xref, Sref, timeout=10.0)\n\n # demographic parity\n Z = split_to_four(Xbase, Sbase, Ybase)\n parity = demographic_parity([z[:, -1] for z in Z])\n \n # sampling\n results = [[parity, dx, dx_s1, dx_s0]]\n sampled_spos = np.mean(Sbase)\n K = computeK(Z, Nsample, sampled_spos, sampled_ypos)\n for i, sampling in enumerate(['case-control', 'stealth']):\n #print('%s: sampling ...' % (sampling,), end='')\n np.random.seed(seed+i)\n if sampling == 'case-control':\n p = case_control_sampling([z[:, :-1] for z in Z], K)\n elif sampling == 'stealth':\n p = stealth_sampling.stealth_sampling([z[:, :-1] for z in Z], K, path='./', prefix='compas', timeout=30.0)\n idx = np.random.choice(N, sum(K), p=np.concatenate(p), replace=False)\n Xs = np.concatenate([z[:, :-2] for z in Z], axis=0)[idx, :]\n Ss = np.concatenate([z[:, -2] for z in Z], axis=0)[idx]\n Ts = np.concatenate([z[:, -1] for z in Z], axis=0)[idx]\n #print('done.')\n \n # demographic parity of the sampled data\n #print('%s: evaluating ...' % (sampling,), end='')\n Zs = split_to_four(Xs, Ss, Ts)\n parity = demographic_parity([z[:, -1] for z in Zs])\n \n # wasserstein disttance\n dx, dx_s1, dx_s0 = compute_wasserstein(Xs, Ss, Xref, Sref, timeout=10.0)\n #print('done.')\n \n results.append([parity, dx, dx_s1, dx_s0])\n return results",
"_____no_output_____"
]
],
[
[
"#### Experiment (One Run)",
"_____no_output_____"
]
],
[
[
"result = sample_and_evaluate(X, S, Y, Nref=Nref, Nsample=Nsample, sampled_ypos=sampled_ypos, seed=seed)\ndf = pd.DataFrame(result)\ndf.index = ['Baseline', 'Case-control', 'Stealth']\ndf.columns = ['DP', 'WD on Pr[x]', 'WD on Pr[x|s=1]', 'WD on Pr[x|s=0]']\nprint('Result (alpha = %.2f, seed=%d)' % (sampled_ypos[0], seed))\ndf",
"Result (alpha = 0.50, seed=0)\n"
]
],
[
[
"#### Experiment (10 Runs)",
"_____no_output_____"
]
],
[
[
"num_itr = 10\nresult_all = []\nfor i in range(num_itr):\n result_i = sample_and_evaluate(X, S, Y, Nref=Nref, Nsample=Nsample, sampled_ypos=sampled_ypos, seed=i)\n result_all.append(result_i)\nresult_all = np.array(result_all)\ndf = pd.DataFrame(np.mean(result_all, axis=0))\ndf.index = ['Baseline', 'Case-control', 'Stealth']\ndf.columns = ['DP', 'WD on Pr[x]', 'WD on Pr[x|s=1]', 'WD on Pr[x|s=0]']\nprint('Average Result of %d runs (alpha = %.2f)' % (num_itr, sampled_ypos[0]))\ndf",
"Average Result of 10 runs (alpha = 0.50)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8e2773f171e3f9a09d63b0999b49bda969c9c0
| 62,284 |
ipynb
|
Jupyter Notebook
|
IUCN_MM_DistMaps.ipynb
|
ArturoBell/IUCN-Dist_py
|
3bc29b84d9d212ae4ad776a86fec1fa6fcecc04e
|
[
"MIT"
] | null | null | null |
IUCN_MM_DistMaps.ipynb
|
ArturoBell/IUCN-Dist_py
|
3bc29b84d9d212ae4ad776a86fec1fa6fcecc04e
|
[
"MIT"
] | null | null | null |
IUCN_MM_DistMaps.ipynb
|
ArturoBell/IUCN-Dist_py
|
3bc29b84d9d212ae4ad776a86fec1fa6fcecc04e
|
[
"MIT"
] | null | null | null | 182.116959 | 51,588 | 0.890566 |
[
[
[
"# Plotting species distribution areas (IUCN spatial data)",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
],
[
"### Libraries",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport geopandas as gpd\nfrom matplotlib import pyplot as plt\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Data\nFirst, let's load the shapefile containing the distribution data for marine mammals (IUCN data).",
"_____no_output_____"
]
],
[
[
"shp = gpd.read_file('./MARINE_MAMMALS/MARINE_MAMMALS.shp')\n#shore = gpd.read_file('shapefile.shp') # In case you want a custom coastline\n#shp.head()",
"_____no_output_____"
]
],
[
[
"## Custom Functions",
"_____no_output_____"
]
],
[
[
"def RGB(rgb_code):\n \"\"\"Transforms 0-255 rgb colors to 0-1 scale\"\"\"\n col = tuple(x/255 for x in rgb_code) # I'm shure there is a more efficient way; however, it was faster to code a simple function\n return col",
"_____no_output_____"
],
[
"def get_ranges(species, shp, drop_columns = None):\n \"\"\"Returns a list of simplifyied distribution data from a shape downloaded from the IUCN. \n Arguments:\n species: a list of binomial species names to get the distributions from\n shp: the shapefile variable containing the distribution ranges\n drop_columns: Columns with attributes to drop from the original shapefile, if None, no columns are dropped\"\"\"\n\n if drop_columns == None:\n for sp in species:\n if sp == species[0]:\n ranges = shp[shp.binomial == sp]\n else:\n ranges = ranges.append(shp[shp.binomial == sp])\n else:\n shp = shp.drop(columns = drop_columns)\n for sp in species:\n if sp == species[0]:\n ranges = shp[shp.binomial == sp]\n else:\n ranges = ranges.append(shp[shp.binomial == sp]) \n return ranges",
"_____no_output_____"
],
[
"def plot_dist_ranges(ranges, cmap = 'winter', font = {'family': 'sans-serif', 'weight': 500}, shore = None, extent = None):\n \"\"\"Plots the distribution ranges of the species contained in the ranges variable.\n Arguments:\n ranges: A list in which each element corresponds to a distribution range (geometry) and attributes from distribution data downloadaded from the IUCN\n cmap: A string defining the colormap to use. If empty, defaults to 'winter'\n font: Dict. containing the properties of the font to use in the plot.\n shore: Variable storing the coastline. If None, defaults to 'naturalearth_lowres' from the geopandas datasets\n extent: A list with the limits of the plot, in the form: [x_inf, x_sup, y_inf, y_sup]. If none, the whole globe is plotted.\"\"\"\n # Setting the font of the plot. \n plt.rcParams['font.family'] = font['family']\n plt.rcParams['font.weight'] = font['weight']\n \n # Plot environment\n fig, ax = plt.subplots(1,1)\n \n # Plotting the distributions\n sp = list(ranges.binomial.values)\n ranges.plot(column = 'binomial', ax = ax, alpha = 0.35, cmap = cmap,\n legend = True,\n legend_kwds = {'prop': {'style': 'italic', 'size': 8}})\n \n # Plotting the coastline\n if shore == None:\n world = gpd.read_file(gpd.datasets.get_path('naturalearth_lowres'))\n world.plot(color = 'lightslategray', alpha = 0.5, ax = ax)\n else:\n shore.plot(color = 'lightslategray', alpha = 0.5, ax = ax)\n \n # Cleaning the plot\n plt.tight_layout()\n for spine in ax.spines.values():\n spine.set_visible(False)\n\n ax.tick_params(axis = 'both',\n which = 'both',\n bottom = False,\n top = False,\n left = False,\n labelleft = True,\n labelbottom = True)\n \n # Changing the tickmarks of the y axis\n if extent == None:\n extent = [-180, 180, -90, 90]\n plt.axis(extent)\n plt.yticks(np.arange(-80,120,40))\n else:\n long_range = abs(extent[0] - extent[1])/4\n plt.yticks(np.linspace(extent[0], extent[1], 4))\n \n # Changing the axes labels\n plt.xlabel('Longitude (º)')\n plt.ylabel('Latitude (º)')\n #plt.labelweight = font['weight']",
"_____no_output_____"
]
],
[
[
"## Extracting and plotting the data\nAs an example, the distributions of *Stenella attenuata* and *Stenella longirostris* are plotted, and most unused columns are dropped. Also, a custom color map is created to pass to the plotting function.",
"_____no_output_____"
]
],
[
[
"from matplotlib.colors import ListedColormap\nspecies = ['Stenella attenuata', 'Stenella longirostris']\ndrop_columns = ['id_no','presence', 'origin', 'source', 'seasonal', 'compiler', 'yrcompiled',\n 'citation', 'dist_comm', 'island', 'subspecies', 'subpop', 'tax_comm', 'kingdom',\n 'phylum', 'class', 'order_', 'family', 'genus', 'category', 'marine', 'terrestial', 'freshwater']\nranges = get_ranges(species, shp, drop_columns)\ncmap = ListedColormap([RGB((121,227,249)), RGB((27,109,183))])",
"_____no_output_____"
],
[
"plot_dist_ranges(ranges, cmap = cmap, font = {'family': 'Montserrat', 'weight': 500})",
"_____no_output_____"
],
[
"#plt.savefig('Dist.jpg', bbox_inches = 'tight', pad_inches = 0.1, transparent = False, dpi = 300)",
"_____no_output_____"
],
[
"import types\ndef imports():\n for name, val in globals().items():\n if isinstance(val, types.ModuleType):\n yield val.__name__\nlist(imports())",
"_____no_output_____"
],
[
"import pkg_resources\nimport types\ndef get_imports():\n for name, val in globals().items():\n if isinstance(val, types.ModuleType):\n # Split ensures you get root package, \n # not just imported function\n name = val.__name__.split(\".\")[0]\n\n elif isinstance(val, type):\n name = val.__module__.split(\".\")[0]\n\n # Some packages are weird and have different\n # imported names vs. system/pip names. Unfortunately,\n # there is no systematic way to get pip names from\n # a package's imported name. You'll have to add\n # exceptions to this list manually!\n poorly_named_packages = {\n \"PIL\": \"Pillow\",\n \"sklearn\": \"scikit-learn\"\n }\n if name in poorly_named_packages.keys():\n name = poorly_named_packages[name]\n\n yield name\nimports = list(set(get_imports()))\n\n# The only way I found to get the version of the root package\n# from only the name of the package is to cross-check the names \n# of installed packages vs. imported packages\nrequirements = []\nfor m in pkg_resources.working_set:\n if m.project_name in imports and m.project_name!=\"pip\":\n requirements.append((m.project_name, m.version))\n\nfor r in requirements:\n print(\"{}=={}\".format(*r))",
"pandas==1.0.3\nnumpy==1.18.4\nmatplotlib==3.2.1\nipywidgets==7.5.1\ngeopandas==0.8.0\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a8e2c416243a3780894a499e2e6e61352f7b4c2
| 188,778 |
ipynb
|
Jupyter Notebook
|
Ekram_LS_DS_234_assignment.ipynb
|
Ekram49/DS-Unit-2-Applied-Modeling
|
77b133dc891a4a3a274402672f3b81bfb15010c5
|
[
"MIT"
] | null | null | null |
Ekram_LS_DS_234_assignment.ipynb
|
Ekram49/DS-Unit-2-Applied-Modeling
|
77b133dc891a4a3a274402672f3b81bfb15010c5
|
[
"MIT"
] | null | null | null |
Ekram_LS_DS_234_assignment.ipynb
|
Ekram49/DS-Unit-2-Applied-Modeling
|
77b133dc891a4a3a274402672f3b81bfb15010c5
|
[
"MIT"
] | null | null | null | 120.547893 | 78,800 | 0.790775 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Ekram49/DS-Unit-2-Applied-Modeling/blob/master/Ekram_LS_DS_234_assignment.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"\nLambda School Data Science\n\n*Unit 2, Sprint 3, Module 4*\n\n---",
"_____no_output_____"
],
[
"# Model Interpretation\n\nYou will use your portfolio project dataset for all assignments this sprint.\n\n## Assignment\n\nComplete these tasks for your project, and document your work.\n\n- [ ] Continue to iterate on your project: data cleaning, exploratory visualization, feature engineering, modeling.\n- [ ] Make at least 1 partial dependence plot to explain your model.\n- [ ] Make at least 1 Shapley force plot to explain an individual prediction.\n- [ ] **Share at least 1 visualization (of any type) on Slack!**\n\nIf you aren't ready to make these plots with your own dataset, you can practice these objectives with any dataset you've worked with previously. Example solutions are available for Partial Dependence Plots with the Tanzania Waterpumps dataset, and Shapley force plots with the Titanic dataset. (These datasets are available in the data directory of this repository.)\n\nPlease be aware that **multi-class classification** will result in multiple Partial Dependence Plots (one for each class), and multiple sets of Shapley Values (one for each class).",
"_____no_output_____"
],
[
"## Stretch Goals\n\n#### Partial Dependence Plots\n- [ ] Make multiple PDPs with 1 feature in isolation.\n- [ ] Make multiple PDPs with 2 features in interaction. \n- [ ] Use Plotly to make a 3D PDP.\n- [ ] Make PDPs with categorical feature(s). Use Ordinal Encoder, outside of a pipeline, to encode your data first. If there is a natural ordering, then take the time to encode it that way, instead of random integers. Then use the encoded data with pdpbox. Get readable category names on your plot, instead of integer category codes.\n\n#### Shap Values\n- [ ] Make Shapley force plots to explain at least 4 individual predictions.\n - If your project is Binary Classification, you can do a True Positive, True Negative, False Positive, False Negative.\n - If your project is Regression, you can do a high prediction with low error, a low prediction with low error, a high prediction with high error, and a low prediction with high error.\n- [ ] Use Shapley values to display verbal explanations of individual predictions.\n- [ ] Use the SHAP library for other visualization types.\n\nThe [SHAP repo](https://github.com/slundberg/shap) has examples for many visualization types, including:\n\n- Force Plot, individual predictions\n- Force Plot, multiple predictions\n- Dependence Plot\n- Summary Plot\n- Summary Plot, Bar\n- Interaction Values\n- Decision Plots\n\nWe just did the first type during the lesson. The [Kaggle microcourse](https://www.kaggle.com/dansbecker/advanced-uses-of-shap-values) shows two more. Experiment and see what you can learn!",
"_____no_output_____"
],
[
"### Links\n\n#### Partial Dependence Plots\n- [Kaggle / Dan Becker: Machine Learning Explainability — Partial Dependence Plots](https://www.kaggle.com/dansbecker/partial-plots)\n- [Christoph Molnar: Interpretable Machine Learning — Partial Dependence Plots](https://christophm.github.io/interpretable-ml-book/pdp.html) + [animated explanation](https://twitter.com/ChristophMolnar/status/1066398522608635904)\n- [pdpbox repo](https://github.com/SauceCat/PDPbox) & [docs](https://pdpbox.readthedocs.io/en/latest/)\n- [Plotly: 3D PDP example](https://plot.ly/scikit-learn/plot-partial-dependence/#partial-dependence-of-house-value-on-median-age-and-average-occupancy)\n\n#### Shapley Values\n- [Kaggle / Dan Becker: Machine Learning Explainability — SHAP Values](https://www.kaggle.com/learn/machine-learning-explainability)\n- [Christoph Molnar: Interpretable Machine Learning — Shapley Values](https://christophm.github.io/interpretable-ml-book/shapley.html)\n- [SHAP repo](https://github.com/slundberg/shap) & [docs](https://shap.readthedocs.io/en/latest/)",
"_____no_output_____"
]
],
[
[
"%%capture\nimport sys\n\n# If you're on Colab:\nif 'google.colab' in sys.modules:\n DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'\n !pip install category_encoders==2.*\n !pip install eli5\n !pip install pdpbox\n !pip install shap\n\n# If you're working locally:\nelse:\n DATA_PATH = '../data/'",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"df = pd.read_csv('https://raw.githubusercontent.com/Ekram49/DS-Unit-1-Build/master/ContinousDataset.csv')\ndf.head()",
"_____no_output_____"
],
[
"df = df.rename(columns={\"Team 1\": \"Team_1\", \"Team 2\": \"Team_2\",\n \"Team 1\": \"Team_1\",\"Match Date\":\"Match_Date\"})\ndf.head()",
"_____no_output_____"
],
[
"df = df[(((df['Team_1'] == 'India') | (df['Team_2'] == 'India'))) & (((df['Team_1'] == 'Pakistan') | (df['Team_2'] == 'Pakistan'))) ]\n\ndf.head()",
"_____no_output_____"
]
],
[
[
"# Baseline",
"_____no_output_____"
]
],
[
[
"df['Winner'].value_counts(normalize = True)",
"_____no_output_____"
],
[
"import seaborn as sns\nsns.countplot(df['Winner'])",
"_____no_output_____"
],
[
"df.isna().sum().sort_values()",
"_____no_output_____"
],
[
"df = df.fillna('Missing')",
"_____no_output_____"
],
[
"df.isna().sum().sort_values()",
"_____no_output_____"
]
],
[
[
"# New Features",
"_____no_output_____"
]
],
[
[
"df['played_at_home'] = (df['Host_Country'] == 'India')",
"_____no_output_____"
],
[
"df['played_at_Pakistan'] = (df['Host_Country'] == 'Pakistan')",
"_____no_output_____"
],
[
"df['Played_in_neutral'] = (df['Host_Country'] != 'India') & (df['Host_Country'] != 'Pakistan')",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"train, test = train_test_split(df, train_size = .8, test_size = .2,\n stratify = df['Winner'], random_state =42)",
"_____no_output_____"
],
[
"train, val = train_test_split(train, train_size = .8, test_size = .2,\n stratify = train['Winner'], random_state =42)",
"_____no_output_____"
]
],
[
[
"# Feature selection",
"_____no_output_____"
]
],
[
[
"target = 'Winner'",
"_____no_output_____"
],
[
"train.describe(exclude = 'number').T.sort_values(by = 'unique', ascending = False)",
"_____no_output_____"
],
[
"# Removing columns with high cordinality\nhigh_cardinality = 'Scorecard', 'Match_Date'\n# Margin will cause data leakage\nfeatures = train.columns.drop(['Unnamed: 0', 'Winner', 'Scorecard', 'Match_Date', 'Margin'])",
"_____no_output_____"
],
[
"X_train = train[features]\ny_train = train[target]\nX_val = val[features]\ny_val = val[target]\nX_test = test[features]\ny_test = test[target]",
"_____no_output_____"
],
[
"!pip install --upgrade category_encoders",
"ERROR! Session/line number was not unique in database. History logging moved to new session 60\nRequirement already up-to-date: category_encoders in /usr/local/lib/python3.6/dist-packages (2.1.0)\nRequirement already satisfied, skipping upgrade: statsmodels>=0.6.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.10.2)\nRequirement already satisfied, skipping upgrade: numpy>=1.11.3 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (1.18.2)\nRequirement already satisfied, skipping upgrade: scikit-learn>=0.20.0 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.22.2.post1)\nRequirement already satisfied, skipping upgrade: patsy>=0.4.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.5.1)\nRequirement already satisfied, skipping upgrade: scipy>=0.19.0 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (1.4.1)\nRequirement already satisfied, skipping upgrade: pandas>=0.21.1 in /usr/local/lib/python3.6/dist-packages (from category_encoders) (0.25.3)\nRequirement already satisfied, skipping upgrade: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.20.0->category_encoders) (0.14.1)\nRequirement already satisfied, skipping upgrade: six in /usr/local/lib/python3.6/dist-packages (from patsy>=0.4.1->category_encoders) (1.12.0)\nRequirement already satisfied, skipping upgrade: python-dateutil>=2.6.1 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.21.1->category_encoders) (2.8.1)\nRequirement already satisfied, skipping upgrade: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas>=0.21.1->category_encoders) (2018.9)\n"
],
[
"import category_encoders as ce\nfrom sklearn.pipeline import make_pipeline\nfrom xgboost import XGBClassifier",
"_____no_output_____"
],
[
"pipeline = make_pipeline(\n ce.OneHotEncoder(use_cat_names= True),\n XGBClassifier(n_estimators=100, random_state=42, n_jobs=-1)\n\n)\npipeline.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"# Validation accuracy",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"y_pred_val = pipeline.predict(X_val)",
"_____no_output_____"
],
[
"accuracy_score(y_val, y_pred_val)",
"_____no_output_____"
]
],
[
[
"# Test accuracy",
"_____no_output_____"
]
],
[
[
"y_pred_test = pipeline.predict(X_test)",
"_____no_output_____"
],
[
"accuracy_score(y_test, y_pred_test)",
"_____no_output_____"
]
],
[
[
"# PDP",
"_____no_output_____"
]
],
[
[
"import category_encoders as ce\nimport seaborn as sns\nfrom sklearn.ensemble import RandomForestClassifier",
"_____no_output_____"
],
[
"encoder = ce.OrdinalEncoder()\nX_train_encoded = encoder.fit_transform(X_train)\n\nmodel = RandomForestClassifier(n_estimators=100, random_state=42, n_jobs=-1)\nmodel.fit(X_train_encoded, y_train)",
"_____no_output_____"
],
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nfrom pdpbox import pdp\nfeature = 'Host_Country'\npdp_dist = pdp.pdp_isolate(model=model, dataset=X_train_encoded, model_features=features, feature=feature)\npdp.pdp_plot(pdp_dist, feature);",
"findfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.\nfindfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.\nfindfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.\nfindfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.\n"
]
],
[
[
"# With two features",
"_____no_output_____"
]
],
[
[
"from pdpbox.pdp import pdp_interact, pdp_interact_plot",
"_____no_output_____"
],
[
"features = ['Host_Country', 'Ground']\n\ninteraction = pdp_interact(\n model=model, \n dataset=X_train_encoded, \n model_features=X_train_encoded.columns, \n features=features\n)\n\npdp_interact_plot(interaction, plot_type='grid', feature_names=features);",
"findfont: Font family ['Arial'] not found. Falling back to DejaVu Sans.\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a8e2e051eba61b37a6c9436eb970d50b3432595
| 17,600 |
ipynb
|
Jupyter Notebook
|
3-applications/Matrix Multiplication.ipynb
|
YosephKS/Introduction-to-Programming
|
d92070084d611b2b68bc56f20a80095aba099a94
|
[
"MIT"
] | null | null | null |
3-applications/Matrix Multiplication.ipynb
|
YosephKS/Introduction-to-Programming
|
d92070084d611b2b68bc56f20a80095aba099a94
|
[
"MIT"
] | null | null | null |
3-applications/Matrix Multiplication.ipynb
|
YosephKS/Introduction-to-Programming
|
d92070084d611b2b68bc56f20a80095aba099a94
|
[
"MIT"
] | null | null | null | 29.62963 | 443 | 0.502557 |
[
[
[
"# Matrix Multiplication",
"_____no_output_____"
],
[
"Author: Yoseph K. Soenggoro",
"_____no_output_____"
]
],
[
[
"from random import random\nfrom itertools import product\nimport numpy as np",
"_____no_output_____"
],
[
"# Check you NumPy Version (I used 1.16.4).\n# If the program is incompatible with your NumPy version, use pip or conda to set the appropriate version\nnp.__version__",
"_____no_output_____"
],
[
"# Choose the value of n, the dimension for Matrix X and Y\nn = 3\n\n# Choose d as the range of random value for Matrix X and Y.\n# By choosing value d, the element of Matrix X and Y will be any real number between 0 and d, but never d.\nd = 10",
"_____no_output_____"
]
],
[
[
"Before starting to multiply any two matrices, first define two different matrices $X$ and $Y$ using the `random` library.",
"_____no_output_____"
]
],
[
[
"# Define Matrix X and Matrix Y\nX = []\nY = []\n\nfor i in range(0, n):\n x_row = []\n for j in range(0, n):\n x_val = random() * d\n x_row.append(x_val)\n X.append(x_row)\n\nfor i in range(0, n):\n y_row = []\n for j in range(0, n):\n y_val = random() * d\n y_row.append(y_val)\n Y.append(y_row)",
"_____no_output_____"
],
[
"# Function to print the matrices\ndef print_matrix(X):\n matrix_string = ''\n for i, j in product(range(0, n), range(0, n)):\n matrix_string += f'{X[i][j]}' + ('\\t' if j != n - 1 else '\\n')\n print(matrix_string)",
"_____no_output_____"
],
[
"# Print X to Check\nprint_matrix(X)",
"1.411243489014371\t4.823307914767269\t2.9619965649396427\n8.632221076600207\t6.909318166288182\t4.297076325685831\n6.741265901114342\t9.161915445688102\t6.166996253979611\n\n"
],
[
"# Print Y to Check\nprint_matrix(Y)",
"7.627644148989169\t4.412768013974525\t4.55820479886072\n7.864788107651906\t2.491004876229417\t1.2387049762153457\n1.6601811042693615\t6.587244192044375\t9.966139995991005\n\n"
]
],
[
[
"### Matrix Multiplication Formula (Linear Algebra)\n\nGiven a $n \\times n$ matrices $X$ and $Y$, as follows:\n\n\\begin{align}\nX =\n\\begin{bmatrix}\n x_{1, 1} & x_{1, 2} & \\dots & x_{1, n} \\\\\n x_{2, 1} & x_{2, 2} & \\dots & x_{2, n} \\\\\n \\vdots & \\vdots & \\ddots & \\vdots \\\\\n x_{n, 1} & x_{n, 2} & \\dots & x_{n, n}\n\\end{bmatrix}\n, \\quad\nY =\n\\begin{bmatrix}\n y_{1, 1} & y_{1, 2} & \\dots & y_{1, n} \\\\\n y_{2, 1} & y_{2, 2} & \\dots & y_{2, n} \\\\\n \\vdots & \\vdots & \\ddots & \\vdots \\\\\n y_{n, 1} & y_{n, 2} & \\dots & y_{n, n}\n\\end{bmatrix}\n\\end{align}\n\nthen the multiplication is defined by the following formula:\n\\begin{align}\n X \\cdot Y = \\left[\\sum_{k = 1}^n x_{i, k} \\cdot y_{j, k}\\right]_{i, j = 1}^n\n\\end{align}",
"_____no_output_____"
],
[
"### Implementation \\#1: Functional Paradigm",
"_____no_output_____"
],
[
"The simplest way to implement Matrix Multiplication is by using modular functions, that can be used and reused multiple times within a program. Given the formula above, the Python implementation will be as follows.",
"_____no_output_____"
]
],
[
[
"# Function to implement Matrix Multiplication of Matrix X and Y\ndef matrix_mul(X, Y):\n Z = []\n\n for i in range(0, n):\n z_row = []\n for j in range(0, n):\n z_val = 0\n for k in range(0, n):\n z_val += X[i][k] * Y[k][j]\n z_row.append(z_val)\n Z.append(z_row)\n return Z",
"_____no_output_____"
]
],
[
[
"For the multiplication between $X$ and $Y$, the result will be kept in variable $Z$.",
"_____no_output_____"
]
],
[
[
"Z = matrix_mul(X, Y)\nprint_matrix(Z)",
"53.61620859740801\t37.75376833274768\t41.92706479366027\n127.31775885342397\t83.60902536916785\t90.7313025662656\n133.714831698932\t93.1935288493318\t103.53812885700651\n\n"
]
],
[
[
"### Check Validity on Matrix Multiplication Function",
"_____no_output_____"
],
[
"Despite having a working matrix multiplication implementation in functional form, we still have no idea whether the result from our implementation is right or wrong. Therefore, one method to validate the result will be doing a comparison with `NumPy`'s implementation of `matmul` API.",
"_____no_output_____"
]
],
[
[
"# Function to compare the Matrix Multiplication Function to NumPy's matmul\ndef check_matrix_mul(X, Y):\n print('Starting Validation Process...\\n\\n\\n')\n x = np.array(X)\n y = np.array(Y)\n z = np.matmul(x, y)\n Z = matrix_mul(X, Y)\n for i, j in product(range(0, n), range(0, n)): \n print(f'Checking index {(i, j)}... \\t\\t\\t {round(z[i][j], 2) == round(Z[i][j], 2)}')\n \n print('\\n')\n print('Validation Process Completed')",
"_____no_output_____"
],
[
"a = check_matrix_mul(X, Y)",
"Starting Validation Process...\n\n\n\nChecking index (0, 0)... \t\t\t True\nChecking index (0, 1)... \t\t\t True\nChecking index (0, 2)... \t\t\t True\nChecking index (1, 0)... \t\t\t True\nChecking index (1, 1)... \t\t\t True\nChecking index (1, 2)... \t\t\t True\nChecking index (2, 0)... \t\t\t True\nChecking index (2, 1)... \t\t\t True\nChecking index (2, 2)... \t\t\t True\n\n\nValidation Process Completed\n"
]
],
[
[
"Since after checking all the results are True, then it can be confirmed that the implentation works sucessfully.",
"_____no_output_____"
],
[
"### Implementation \\#2: Object-Oriented Paradigm",
"_____no_output_____"
],
[
"Another paradigm that can be used is OOP or Object-Oriented Programming, which represents a program as a set of Objects with various fields and methods to interact with the defined Object. In this case, first defined a generalized form of matrices, which is known as Tensors. The implementation of `Tensor` will be as follows: ",
"_____no_output_____"
]
],
[
[
"class Tensor:\n def __init__(self, X):\n validation = self.__checking_validity(X)\n \n self.__dim = 2\n self.tensor = X if validation else []\n self.__dimension = self.__get_dimension_private(X) if validation else -1\n \n def __get_dimension_private(self, X):\n if not check_child(X):\n return 1\n else:\n # Check whether the size of each child are the same\n for i in range(0, len(X)):\n if not check_child(X[i]):\n return self.__dim\n else:\n get_dimension(X[i])\n self.__dim += 1\n return self.__dim\n \n def __checking_validity(self, X):\n self.__dim = 2\n valid = True\n if not check_child(X):\n return valid\n else:\n dim_0 = get_dimension(X[0])\n # Check whether the size of each child are the same\n for i in range(1, len(X)):\n self.__dim = 2\n if get_dimension(X[i]) != dim_0:\n valid &= False\n break\n return valid\n \n # Getting the Value of Tensor Rank/Dimension (Not to be confused with Matrix Dimension)\n def get_dimension(self):\n return self.__dimension",
"_____no_output_____"
]
],
[
[
"Since Tensors are generalized form of matrices, it implies that it is possible to define `Matrix` class as a child class of `Tensor` with additional methods (some overrides the `Tensor`'s original methods). For operators, I only managed to override the multiplication operator for the sake of implementing Matrix Multiplication. Thus, other operator such as `+`, `-`, `/`, and others will not be available for the current implementation.",
"_____no_output_____"
]
],
[
[
"class Matrix(Tensor):\n def __init__(self, X):\n super().__init__(X)\n self.__matrix_string = ''\n \n def __str__(self):\n return self.__matrix_string if self.__check_matrix_validation() else ''\n \n # Check whether the given input X is a valid Matrix\n def __check_matrix_validation(self):\n valid = True\n try:\n for i, j in product(range(0, n), range(0, n)):\n self.__matrix_string += f'{self.tensor[i][j]}' + ('\\t' if j != n - 1 else '\\n')\n except:\n valid = False\n print('Matrix is Invalid. Create New Instance with appropriate inputs.')\n \n return valid\n \n # Get Matrix Dimension: Number of Columns and Rows\n def get_dimension(self):\n print(f'Matrix Dimension: ({len(self.tensor)}, {len(self.tensor[0])})' if self.__check_matrix_validation() else -1)\n return [len(self.tensor), len(self.tensor[0])]\n \n # Overriding Multiplication Operator for Matrix Multiplication\n # and Integer-Matrix Multiplication\n def __mul__(self, other):\n if isinstance(other, Matrix):\n Z = []\n for i in range(0, n):\n z_row = []\n for j in range(0, n):\n z_val = 0\n for k in range(0, n):\n z_val += self.tensor[i][k] * other.tensor[k][j]\n z_row.append(z_val)\n Z.append(z_row)\n return Matrix(Z)\n elif isinstance(other, int):\n Z = []\n for i in range(0, n):\n z_row = []\n for j in range(0, n):\n z_row.append(self.tensor[i][j] * other)\n Z.append(z_row)\n return Matrix(Z)\n else:\n return NotImplemented\n \n # Overriding Reverse Multiplication to support Matrix-Integer Multiplication\n def __rmul__(self, other):\n if isinstance(other, int):\n Z = []\n for i in range(0, n):\n z_row = []\n for j in range(0, n):\n z_row.append(self.tensor[i][j] * other)\n Z.append(z_row)\n return Matrix(Z)\n else:\n return NotImplemented",
"_____no_output_____"
],
[
"# Transform X and Y to Matrix Object\nx_obj = Matrix(X)\ny_obj = Matrix(Y)\n\n# Implement Matrix Multiplication as follows\nz_obj = x_obj * y_obj\nprint(z_obj)",
"53.61620859740801\t37.75376833274768\t41.92706479366027\n127.31775885342397\t83.60902536916785\t90.7313025662656\n133.714831698932\t93.1935288493318\t103.53812885700651\n\n"
]
],
[
[
"### Check Validity on Matrix Multiplication using OOP",
"_____no_output_____"
],
[
"Similar to the previous section, we still have no idea whether the result from our implementation is right or wrong. Hence, validation is highly important. Therefore, one method to validate the result will be again doing a comparison with `NumPy`'s implementation of `matmul` API.",
"_____no_output_____"
]
],
[
[
"# Function to compare the Matrix Multiplication Function to Numpy's matmul\ndef check_matrix_mul_oop(X, Y):\n print('Starting Validation Process...\\n\\n\\n')\n x = np.array(X)\n y = np.array(Y)\n z = np.matmul(x, y)\n Z = Matrix(X) * Matrix(Y)\n for i, j in product(range(0, n), range(0, n)): \n print(f'Checking index {(i, j)}... \\t\\t\\t {round(z[i][j], 2) == round(Z.tensor[i][j], 2)}')\n \n print('\\n')\n print('Validation Process Completed')",
"_____no_output_____"
],
[
"a = check_matrix_mul_oop(X, Y)",
"Starting Validation Process...\n\n\n\nChecking index (0, 0)... \t\t\t True\nChecking index (0, 1)... \t\t\t True\nChecking index (0, 2)... \t\t\t True\nChecking index (1, 0)... \t\t\t True\nChecking index (1, 1)... \t\t\t True\nChecking index (1, 2)... \t\t\t True\nChecking index (2, 0)... \t\t\t True\nChecking index (2, 1)... \t\t\t True\nChecking index (2, 2)... \t\t\t True\n\n\nValidation Process Completed\n"
]
],
[
[
"Since after checking all the results are True, then it can be confirmed that the implentation works sucessfully.",
"_____no_output_____"
],
[
"# Python Libraries",
"_____no_output_____"
],
[
"- [NumPy](https://numpy.org/)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4a8e38a79bca38809fbdb5ff0863a3629e62c52c
| 3,142 |
ipynb
|
Jupyter Notebook
|
Concept02_evaluating_ops.ipynb
|
kpmonaghan/WK-Machine-Learning
|
f76b3a52c0d23196d83100da756568faf59a1d33
|
[
"Apache-2.0"
] | 4,893 |
2016-12-14T06:29:28.000Z
|
2022-03-24T09:05:57.000Z
|
ch02_basics/Concept02_evaluating_ops.ipynb
|
turgunyusuf/TensorFlow-Book
|
446195326ebf9aaaf4ecebfaf4d1df4b80565fb1
|
[
"MIT"
] | 36 |
2016-12-14T09:50:36.000Z
|
2021-04-02T11:21:47.000Z
|
ch02_basics/Concept02_evaluating_ops.ipynb
|
turgunyusuf/TensorFlow-Book
|
446195326ebf9aaaf4ecebfaf4d1df4b80565fb1
|
[
"MIT"
] | 1,392 |
2016-12-14T06:54:28.000Z
|
2022-03-29T17:13:44.000Z
| 17.455556 | 155 | 0.500318 |
[
[
[
"# Ch `02`: Concept `02`",
"_____no_output_____"
],
[
"## Evaluating ops",
"_____no_output_____"
],
[
"Import TensorFlow:",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
]
],
[
[
"Start with a 1x2 matrix:",
"_____no_output_____"
]
],
[
[
"x = tf.constant([[1, 2]])",
"_____no_output_____"
]
],
[
[
"Let's negate it. Define the negation op to be run on the matrix:",
"_____no_output_____"
]
],
[
[
"neg_x = tf.negative(x)",
"_____no_output_____"
]
],
[
[
"It's nothing special if you print it out. In fact, it doesn't even perform the negation computation. Check out what happens when you simply print it:",
"_____no_output_____"
]
],
[
[
"print(neg_x)",
"Tensor(\"Neg_3:0\", shape=(1, 2), dtype=int32)\n"
]
],
[
[
"You need to summon a session so you can launch the negation op:",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n result = sess.run(neg_x)\n print(result)",
"[[-1 -2]]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8e3a4ad4ee47108b003a8e86c9f85ecfddca44
| 71,279 |
ipynb
|
Jupyter Notebook
|
docs-static/SIRS/Python SIRS Modeling.ipynb
|
uncharted-aske/AMIDOL
|
de01c3aa9914b273b6a5c3c8606a0616ceab24bb
|
[
"BSD-3-Clause"
] | 9 |
2019-05-20T17:23:55.000Z
|
2020-12-03T05:57:42.000Z
|
docs-static/SIRS/Python SIRS Modeling.ipynb
|
uncharted-aske/AMIDOL
|
de01c3aa9914b273b6a5c3c8606a0616ceab24bb
|
[
"BSD-3-Clause"
] | 11 |
2019-06-05T23:19:13.000Z
|
2021-09-27T22:09:23.000Z
|
docs-static/SIRS/Python SIRS Modeling.ipynb
|
uncharted-aske/AMIDOL
|
de01c3aa9914b273b6a5c3c8606a0616ceab24bb
|
[
"BSD-3-Clause"
] | 4 |
2019-10-12T21:49:50.000Z
|
2020-09-14T22:07:48.000Z
| 309.908696 | 54,252 | 0.885099 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport re\nfrom scipy.integrate import odeint\n\n# Read the data in, then select the relevant columns, and adjust the week so it is easier to realize\n# as a time series.\nvirii = [\"A (H1)\", \"A (H3)\", \"A (2009 H1N1)\", \"A (Subtyping not Performed)\", \"B\"]\nvirus = \"B\"\nfile = \"data/2007-2008_Region-5_WHO-NREVSS.csv\"\nfluData = pd.read_csv(file)[[\"YEAR\", \"WEEK\", \"TOTAL SPECIMENS\"] + virii]\nfirstWeek = fluData[\"WEEK\"][0]\nfluData[\"T\"] = fluData[\"WEEK\"] + 52 * (fluData[\"WEEK\"] < firstWeek)\nfluData = fluData.drop([\"YEAR\", \"WEEK\"], axis=1)\n\nmatch = re.match(\"^data/(\\d+-\\d+)_Region-(\\d+)_.*\", file)\ntitle = \"Flu Season \" + match.groups()[0] + \" for HHS Region \" + match.groups()[1]\nregion = \"HHS \" + match.groups()[1]\nmatch = re.match(\"^(\\d+)-\\d+.*\", match.groups()[0])\npopYear = match.groups()[0]",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n%matplotlib inline\n#plt.xkcd()\nplt.style.use('ggplot')\ntableau20 = [(31, 119, 180), (174, 199, 232), (255, 127, 14), (255, 187, 120), \n (44, 160, 44), (152, 223, 138), (214, 39, 40), (255, 152, 150), \n (148, 103, 189), (197, 176, 213), (140, 86, 75), (196, 156, 148), \n (227, 119, 194), (247, 182, 210), (127, 127, 127), (199, 199, 199), \n (188, 189, 34), (219, 219, 141), (23, 190, 207), (158, 218, 229)] \n \n# Scale the RGB values to the [0, 1] range, which is the format matplotlib accepts. \nfor i in range(len(tableau20)): \n r, g, b = tableau20[i] \n tableau20[i] = (r / 255., g / 255., b / 255.) \n\nplt.figure(figsize=(12,6)) \n\nfor idx in [0, 1, 2, 3]:\n plt.plot(fluData['T'], fluData[virii[idx]], ls=\"--\", lw=2.5, color=tableau20[idx*2], alpha=1)\n plt.scatter(fluData['T'], fluData[virii[idx]], color=tableau20[idx*2])\n y_pos = 200 + idx*50\n plt.text(40, y_pos, \"Virus Strain:\" + virii[idx], fontsize=8, color=tableau20[idx*2]) \n \nplt.title(title, fontsize=12)\nplt.xlabel(\"Week of Flu Season\", fontsize=10)\nplt.ylabel(\"Infected Individuals\", fontsize=10)",
"_____no_output_____"
],
[
"# Initial values of our states\npopData = pd.read_csv('data/population_data.csv', index_col=0)\n# N - total population of the region\n# I0 - initial infected -- we assume 1.\n# R0 - initial recovered -- we assume none.\n# S0 - initial susceptible -- S0 = N - I0 - R0\n# N - total population of the region\n# I0 - initial infected -- we assume 1.\n# R0 - initial recovered -- we assume none.\n# S0 - initial susceptible -- S0 = N - I0 - R0\nN = 52000000#int(popData[popData['Year'] == int(popYear)]['HHS 5']) #\nI0 = 1\nR0 = 0\nS0 = N - R0 - I0\nprint(\"S0, \", S0)\n\ngamma = 1/3\nrho = 1.24\nbeta = rho*gamma\n\ndef deriv(y, t, N, beta, gamma):\n S, I, R = y\n dSdt = -beta * S * I / N\n dIdt = beta * S * I / N - gamma * I\n dRdt = gamma * I\n return dSdt, dIdt, dRdt\n\ny0 = S0, I0, R0\n\nmin = 40\nmax = fluData['T'].max()\nt = list(range(min*7, max*7))\nw = [x/7 for x in t]\n\nret = odeint(deriv, y0, t, args=(N, beta, gamma))\nS, I, R = ret.T\n\nincidence_predicted = -np.diff(S[0:len(S)-1:7])\nincidence_observed = fluData['B']\nfraction_confirmed = incidence_observed.sum()/incidence_predicted.sum()\n\n# Correct for the week of missed incidence\nplotT = fluData['T'] - 7\n\nplt.figure(figsize=(6,3))\nplt.plot(plotT[2:], incidence_predicted*fraction_confirmed, color=tableau20[2])\nplt.text(40, 100, \"CDC Data for Influenza B\", fontsize=12, color=tableau20[0])\nplt.text(40, 150, \"SIRS Model Result\", fontsize=12, color=tableau20[2])\nplt.title(title, fontsize=12)\nplt.xlabel(\"Week of Flu Season\", fontsize=10)\nplt.ylabel(\"Infected Individuals\", fontsize=10)",
"('S0, ', 51999999)\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
4a8e3c3d38f480ee02d05c703e30093907298eb5
| 51,470 |
ipynb
|
Jupyter Notebook
|
benchmark/example-notebooks/0/before.ipynb
|
tarmstrong/nbdiff
|
3fdfb89f94fc0f4821bc04999ddf53b34d882ab9
|
[
"MIT"
] | 100 |
2015-01-19T07:35:26.000Z
|
2022-03-29T08:38:38.000Z
|
benchmark/example-notebooks/0/before.ipynb
|
tarmstrong/nbdiff
|
3fdfb89f94fc0f4821bc04999ddf53b34d882ab9
|
[
"MIT"
] | 8 |
2015-06-11T00:21:17.000Z
|
2018-08-09T14:04:32.000Z
|
benchmark/example-notebooks/0/before.ipynb
|
tarmstrong/nbdiff
|
3fdfb89f94fc0f4821bc04999ddf53b34d882ab9
|
[
"MIT"
] | 17 |
2015-01-18T21:08:48.000Z
|
2022-02-23T00:20:24.000Z
| 21.383465 | 26 | 0.498213 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a8e48090e9c34641d30a6bb3d3121eb4439a0c4
| 9,273 |
ipynb
|
Jupyter Notebook
|
SceneClassification2017/backup/doc20171030/1. Preprocess-KerasFolderClasses-Test.ipynb
|
StudyExchange/AIChallenger
|
e06cbccd6762bc4f7438cd8bdf2ca1fa54ab03ff
|
[
"MIT"
] | 1 |
2017-12-20T05:47:40.000Z
|
2017-12-20T05:47:40.000Z
|
SceneClassification2017/backup/doc20171030/1. Preprocess-KerasFolderClasses-Test.ipynb
|
StudyExchange/AIChallenger
|
e06cbccd6762bc4f7438cd8bdf2ca1fa54ab03ff
|
[
"MIT"
] | null | null | null |
SceneClassification2017/backup/doc20171030/1. Preprocess-KerasFolderClasses-Test.ipynb
|
StudyExchange/AIChallenger
|
e06cbccd6762bc4f7438cd8bdf2ca1fa54ab03ff
|
[
"MIT"
] | null | null | null | 24.86059 | 169 | 0.514504 |
[
[
[
"# Scene Classification-Test\n## 1. Preprocess-KerasFolderClasses\n- Import pkg\n- Extract zip file\n- Preview \"scene_classes.csv\"\n- Preview \"scene_{0}_annotations_20170922.json\"\n- Test the image and pickle function\n- Split data into serval pickle file",
"_____no_output_____"
],
[
"This part need jupyter notebook start with \"jupyter notebook --NotebookApp.iopub_data_rate_limit=1000000000\" (https://github.com/jupyter/notebook/issues/2287)\n\nReference:\n- https://challenger.ai/competitions\n- https://github.com/jupyter/notebook/issues/2287",
"_____no_output_____"
],
[
"### Import pkg",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n# import matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport seaborn as sns\n%matplotlib inline\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import confusion_matrix",
"_____no_output_____"
],
[
"from keras.utils.np_utils import to_categorical # convert to one-hot-encoding\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D, BatchNormalization\nfrom keras.optimizers import Adam\nfrom keras.preprocessing.image import ImageDataGenerator\nfrom keras.callbacks import LearningRateScheduler, TensorBoard",
"Using TensorFlow backend.\n"
],
[
"# import zipfile\nimport os\nimport zipfile\nimport math\nfrom time import time\nfrom IPython.display import display\nimport pdb\nimport json\nfrom PIL import Image\nimport glob\nimport pickle",
"_____no_output_____"
]
],
[
[
"### Extract zip file",
"_____no_output_____"
]
],
[
[
"input_path = 'input'\ndatasetName = 'test_a'\ndate = '20170922'\n\ndatasetFolder = input_path + '\\\\data_{0}'.format(datasetName)\nzip_path = input_path + '\\\\ai_challenger_scene_{0}_{1}.zip'.format(datasetName, date)\nextract_path = input_path + '\\\\ai_challenger_scene_{0}_{1}'.format(datasetName, date)\nimage_path = extract_path + '\\\\scene_{0}_images_{1}'.format(datasetName, date)\nscene_classes_path = extract_path + '\\\\scene_classes.csv'\nscene_annotations_path = extract_path + '\\\\scene_{0}_annotations_{1}.json'.format(datasetName, date)\n\nprint(input_path)\nprint(datasetFolder)\nprint(zip_path)\nprint(extract_path)\nprint(image_path)\nprint(scene_classes_path)\nprint(scene_annotations_path)",
"input\ninput\\data_test_a\ninput\\ai_challenger_scene_test_a_20170922.zip\ninput\\ai_challenger_scene_test_a_20170922\ninput\\ai_challenger_scene_test_a_20170922\\scene_test_a_images_20170922\ninput\\ai_challenger_scene_test_a_20170922\\scene_classes.csv\ninput\\ai_challenger_scene_test_a_20170922\\scene_test_a_annotations_20170922.json\n"
],
[
"if not os.path.isdir(extract_path):\n with zipfile.ZipFile(zip_path) as file:\n for name in file.namelist():\n file.extract(name, input_path)",
"_____no_output_____"
]
],
[
[
"### Preview \"scene_classes.csv\"",
"_____no_output_____"
]
],
[
[
"scene_classes = pd.read_csv(scene_classes_path, header=None)\ndisplay(scene_classes.head())",
"_____no_output_____"
],
[
"def get_scene_name(lable_number, scene_classes_path):\n scene_classes = pd.read_csv(scene_classes_path, header=None)\n return scene_classes.loc[lable_number, 2]\nprint(get_scene_name(0, scene_classes_path))",
"airport_terminal\n"
]
],
[
[
"### Copy images to ./input/data_test_a/test",
"_____no_output_____"
]
],
[
[
"from shutil import copy2",
"_____no_output_____"
],
[
"cwd = os.getcwd()\ntest_folder = os.path.join(cwd, datasetFolder)\ntest_sub_folder = os.path.join(test_folder, 'test')\nif not os.path.isdir(test_folder):\n os.mkdir(test_folder)\n os.mkdir(test_sub_folder)\nprint(test_folder)\nprint(test_sub_folder)",
"E:\\SceneClassification\\input\\data_test_a\nE:\\SceneClassification\\input\\data_test_a\\test\n"
],
[
"trainDir = test_sub_folder\nfor image_id in os.listdir(os.path.join(cwd, image_path)):\n fileName = image_path + '/' + image_id\n# print(fileName)\n# print(trainDir)\n copy2(fileName, trainDir)",
"_____no_output_____"
],
[
"print('Done!')",
"Done!\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4a8e6162579beb0c141a1e621c15bf2864b3890c
| 3,358 |
ipynb
|
Jupyter Notebook
|
test/ry/2-cameraView.ipynb
|
svetlanalevit/rai
|
aa59bef2b728fcdb7fc6e029639e26993f9b6c50
|
[
"MIT"
] | null | null | null |
test/ry/2-cameraView.ipynb
|
svetlanalevit/rai
|
aa59bef2b728fcdb7fc6e029639e26993f9b6c50
|
[
"MIT"
] | null | null | null |
test/ry/2-cameraView.ipynb
|
svetlanalevit/rai
|
aa59bef2b728fcdb7fc6e029639e26993f9b6c50
|
[
"MIT"
] | null | null | null | 22.843537 | 145 | 0.534842 |
[
[
[
"import sys\nsys.path.append('../../lib')\nimport numpy as np\nimport libry as ry\n\nC = ry.Config()\nC.addFile('../../../rai-robotModels/pr2/pr2.g')\nC.addFile('../../../rai-robotModels/objects/kitchen.g')\nC.view()",
"_____no_output_____"
]
],
[
[
"## Camera views\n\nWe can also add a frame, attached to the head, which has no shape associated to it, but create a view is associated with that frame:",
"_____no_output_____"
]
],
[
[
"C.addFrame(name='camera', parent='head_tilt_link', args='Q:<d(-90 1 0 0) d(180 0 0 1)> focalLength:.3')\nV = C.cameraView()",
"_____no_output_____"
],
[
"Vimg = V.imageViewer()\nVseg = V.segmentationViewer()\nVpcl = V.pointCloudViewer()",
"_____no_output_____"
],
[
"V.addSensor('kinect', 'endeffKinect', 640, 480, 580./480., -1., [.1, 50.] )\n#V.addSensor(name='camera', frameAttached='camera', width=600, height=400)\nV.selectSensor('kinect')\n[image,depth] = V.computeImageAndDepth()\nseg = V.computeSegmentation()\npcl = V.computePointCloud(depth)\nprint('image shape:', image.shape)\nprint('depth shape:', depth.shape)\nprint('segmentation shape:', seg.shape)\nprint('pcl shape:', pcl.shape)",
"_____no_output_____"
]
],
[
[
"When we move the robot, that view moves with it:",
"_____no_output_____"
]
],
[
[
"C.setJointState([0.5], ['head_pan_joint'])\nC.setJointState([1.], ['head_tilt_joint'])\nV.updateConfig(C)\n[image,depth] = V.computeImageAndDepth()\npcl = V.computePointCloud(depth)",
"_____no_output_____"
]
],
[
[
"To close a view (or destroy a handle to a computational module), we reassign it to zero. We can also remove a frame from the configuration.",
"_____no_output_____"
]
],
[
[
"Vimg = 0\nVseg = 0\nVpcl = 0\nV = 0\nC.delFrame('camera')",
"_____no_output_____"
],
[
"C.view_close()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a8e6467c8f15ee1cb9a9dbe7b881daa3b47a3f7
| 418,977 |
ipynb
|
Jupyter Notebook
|
Exercise 05—Limits of Binary Classification Using a Simple Linear Function.ipynb
|
WeiTing1991/machineintelligence
|
4531f429e2e4eb6cfe8e2b109dcf377fb89378c8
|
[
"Apache-2.0"
] | null | null | null |
Exercise 05—Limits of Binary Classification Using a Simple Linear Function.ipynb
|
WeiTing1991/machineintelligence
|
4531f429e2e4eb6cfe8e2b109dcf377fb89378c8
|
[
"Apache-2.0"
] | null | null | null |
Exercise 05—Limits of Binary Classification Using a Simple Linear Function.ipynb
|
WeiTing1991/machineintelligence
|
4531f429e2e4eb6cfe8e2b109dcf377fb89378c8
|
[
"Apache-2.0"
] | null | null | null | 658.768868 | 35,028 | 0.946794 |
[
[
[
"%config InlineBackend.figure_format = 'retina'\nimport matplotlib.pyplot as plt\nimport numpy as np\nnp.set_printoptions(precision=3)\nnp.set_printoptions(suppress=True)",
"_____no_output_____"
]
],
[
[
"### Simple linear classification algorithm",
"_____no_output_____"
],
[
"| vector [x,y] \t| label \t|\n|--------------\t|-------\t|\n| [ 0.0, 0.7] \t| +1 \t|\n| [-0.3, -0.5] \t| -1 \t|\n| [3.0, 0.1] \t| +1 \t|\n| [-0.1, -1.0] \t| -1 \t|\n| [-1.0, 1.1] \t| -1 \t|\n| [2.1, -3.0] \t| +1 \t|",
"_____no_output_____"
],
[
"We can represent the data as a 2-dimensional numpy array",
"_____no_output_____"
]
],
[
[
"data = np.array([[ 0.0, 0.7],\n [-0.3,-0.5],\n [ 3.0, 0.1],\n [-0.1,-1.0],\n [-1.0, 1.1],\n [ 2.1,-3.0]])",
"_____no_output_____"
]
],
[
[
"We can represent the labels as a simple numpy array of numbers",
"_____no_output_____"
]
],
[
[
"labels = np.array([ 1,\n -1,\n 1,\n -1,\n -1,\n +1])",
"_____no_output_____"
]
],
[
[
"We can plot the data using the following function:",
"_____no_output_____"
]
],
[
[
"def plot_data(data, labels):\n fig = plt.figure(figsize=(5,5))\n ax = fig.add_subplot(111)\n ax.scatter(data[:,0], data[:,1], c=labels, s=50, cmap=plt.cm.bwr,zorder=50)\n nudge = 0.08\n for i in range(data.shape[0]):\n d = data[i]\n ax.annotate(f'{i}',(d[0]+nudge,d[1]+nudge))\n ax.set_aspect('equal', 'datalim')\n plt.show()",
"_____no_output_____"
],
[
"plot_data(data,labels)",
"_____no_output_____"
]
],
[
[
"This is a function to evaluate the accuracy of the training",
"_____no_output_____"
]
],
[
[
"def eval_accuracy(data,labels, A,B,C):\n num_correct = 0;\n data_len = data.shape[0]\n \n for i in range(data_len):\n X,Y = data[i]\n current_label = labels[i] \n output = A*X + B*Y + C\n predicted_label = 1 if output >= 1 else -1 if output <= -1 else 0\n if (predicted_label == current_label):\n num_correct += 1\n return np.round(num_correct / data_len,3)\n\ndef create_meshgrid(data):\n h = 0.02\n x_min, x_max = data[:, 0].min() - 1, data[:, 0].max() + 1\n y_min, y_max = data[:, 1].min() - 1, data[:, 1].max() + 1\n xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))\n return (xx,yy,np.ones(xx.shape))\n\ndef plot_learning_simple(grid,data,labels,A,B,C,iteration, accuracy):\n xx,yy,Z = grid\n \n for i in range(xx.shape[0]): # row\n for j in range(yy.shape[1]): #column\n X, Y = xx[i][j],yy[i][j]\n output = A*X + B*Y + C\n predicted_label = 1 if output >= 1 else -1 if output <= -1 else 0\n Z[i][j] = predicted_label\n\n fig = plt.figure(figsize=(5,5))\n ax = fig.add_subplot(111)\n plt.title(f'accuracy at the iteration {iteration}: {accuracy}')\n ax.contourf(xx, yy, Z, cmap=plt.cm.binary, alpha=0.1, zorder=15)\n ax.scatter(data[:, 0], data[:, 1], c=labels, s=50, cmap=plt.cm.bwr,zorder=50)\n ax.set_aspect('equal')\n nudge = 0.08\n for i in range(data.shape[0]):\n d = data[i]\n ax.annotate(f'{i}',(d[0]+nudge,d[1]+nudge))\n plt.xlim(xx.min(), xx.max())\n plt.ylim(yy.min(), yy.max())\n plt.show()",
"_____no_output_____"
]
],
[
[
"Here is the main algorithm.",
"_____no_output_____"
]
],
[
[
"def train_neural_network(data, labels, step_size, no_loops, iter_info):\n #A, B, and C are parameters of the function F. Here, they are set to 1, -2, -1\n A, B, C = 1, -2, -1\n # this function is used for plotting, it can be ignored\n grid = create_meshgrid(data)\n \n # the main training loop\n for i in range(no_loops):\n # we randomly select the data point, and store its info into: x,y,label\n index = np.random.randint(data.shape[0])\n X,Y = data[index]\n label = labels[index]\n # we calculate the output of the function\n output = A*X + B*Y + C\n # We need to define how to affect the output of our function.\n # If the label is 1 but the output is smaller than 1, we want to maximise.\n # If the label is -1 but the output is larger than -1, we want to minimise.\n sign = 1 if (label == 1 and output < 1) else -1 if (label == -1 and output > -1) else 0\n # partial derivative of dF/dA is X, dF/dB is Y, and of dF/dC is 1. \n dA, dB, dC = X, Y, 1\n # here we update the parameter values using partial derivatives\n A += dA * sign * step_size\n B += dB * sign * step_size\n C += dC * sign * step_size;\n \n # after a number of iterations, show training accuracy and plot it\n if (i%iter_info==0):\n accuracy = eval_accuracy(data, labels, A,B,C)\n plot_learning_simple(grid,data,labels,A,B,C,i,accuracy)\n # the algorithm returns the learned parameters A, B, and C\n return (A,B,C)",
"_____no_output_____"
],
[
"train_1 = train_neural_network(data, labels, 0.01, 2501, 500)",
"_____no_output_____"
]
],
[
[
"We can inspect the result by comparing the real label of a data point and the predicted label:",
"_____no_output_____"
]
],
[
[
"def show_prediction(train, data, labels):\n A, B, C = train\n for i in range(data.shape[0]):\n X,Y = data[i]\n label = labels[i]\n output = A*X + B*Y + C\n predicted_label = 1 if output >= 1 else -1 if output <= -1 else 0\n print (f'data point {i}: real label : {label}, pred. label: {predicted_label}, {(label==predicted_label)}')",
"_____no_output_____"
],
[
"show_prediction(train_1,data,labels)",
"data point 0: real label : 1, pred. label: 1, True\ndata point 1: real label : -1, pred. label: -1, True\ndata point 2: real label : 1, pred. label: 1, True\ndata point 3: real label : -1, pred. label: -1, True\ndata point 4: real label : -1, pred. label: -1, True\ndata point 5: real label : 1, pred. label: 1, True\n"
]
],
[
[
"---",
"_____no_output_____"
],
[
"#### Let's try with a different data set",
"_____no_output_____"
]
],
[
[
"data2 = np.array([[ 1.2, 0.7],\n [-0.3,-0.5],\n [ 3.0, 0.1],\n [-0.1,-1.0],\n [-0.0, 1.1],\n [ 2.1,-1.3],\n [ 3.1,-1.8],\n [ 1.1,-0.1],\n [ 1.5,-2.2],\n [ 4.0,-1.0]])",
"_____no_output_____"
],
[
"labels2 = np.array([ 1,\n -1,\n 1,\n -1,\n -1,\n 1,\n -1,\n 1,\n -1,\n -1])",
"_____no_output_____"
],
[
"plot_data(data2,labels2)",
"_____no_output_____"
],
[
"train_2 = train_neural_network(data2, labels2, 0.01, 2501, 500)",
"_____no_output_____"
],
[
"show_prediction(train_1,data2,labels2)",
"data point 0: real label : 1, pred. label: 1, True\ndata point 1: real label : -1, pred. label: -1, True\ndata point 2: real label : 1, pred. label: 1, True\ndata point 3: real label : -1, pred. label: -1, True\ndata point 4: real label : -1, pred. label: 1, False\ndata point 5: real label : 1, pred. label: 1, True\ndata point 6: real label : -1, pred. label: 1, False\ndata point 7: real label : 1, pred. label: 1, True\ndata point 8: real label : -1, pred. label: 1, False\ndata point 9: real label : -1, pred. label: 1, False\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a8e6803746647418e344be0ab1391afeb41f27e
| 230,272 |
ipynb
|
Jupyter Notebook
|
Rethinking/Chp_11.ipynb
|
rserran/resources
|
2996d387d7a0fe10d60fac3431244e524a124c88
|
[
"MIT"
] | 3 |
2022-03-28T00:59:22.000Z
|
2022-03-31T09:47:42.000Z
|
Rethinking/Chp_11.ipynb
|
rserran/resources
|
2996d387d7a0fe10d60fac3431244e524a124c88
|
[
"MIT"
] | 3 |
2022-03-27T19:12:53.000Z
|
2022-03-30T03:54:10.000Z
|
Rethinking/Chp_11.ipynb
|
rserran/resources
|
2996d387d7a0fe10d60fac3431244e524a124c88
|
[
"MIT"
] | 3 |
2022-03-27T20:44:16.000Z
|
2022-03-31T09:47:44.000Z
| 178.505426 | 77,000 | 0.894121 |
[
[
[
"from collections import OrderedDict\n\nimport arviz as az\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport pymc as pm\nimport scipy as sp\n\nfrom theano import shared",
"_____no_output_____"
],
[
"%config InlineBackend.figure_format = 'retina'\naz.style.use('arviz-darkgrid')",
"_____no_output_____"
]
],
[
[
"#### Code 11.1",
"_____no_output_____"
]
],
[
[
"trolley_df = pd.read_csv('Data/Trolley.csv', sep=';')\ntrolley_df.head()",
"_____no_output_____"
]
],
[
[
"#### Code 11.2",
"_____no_output_____"
]
],
[
[
"ax = (trolley_df.response\n .value_counts()\n .sort_index()\n .plot(kind='bar'))\n\nax.set_xlabel(\"response\", fontsize=14);\nax.set_ylabel(\"Frequency\", fontsize=14);",
"_____no_output_____"
]
],
[
[
"#### Code 11.3",
"_____no_output_____"
]
],
[
[
"ax = (trolley_df.response\n .value_counts()\n .sort_index()\n .cumsum()\n .div(trolley_df.shape[0])\n .plot(marker='o'))\n\nax.set_xlim(0.9, 7.1);\nax.set_xlabel(\"response\", fontsize=14)\nax.set_ylabel(\"cumulative proportion\", fontsize=14);",
"_____no_output_____"
]
],
[
[
"#### Code 11.4",
"_____no_output_____"
]
],
[
[
"resp_lco = (trolley_df.response\n .value_counts()\n .sort_index()\n .cumsum()\n .iloc[:-1]\n .div(trolley_df.shape[0])\n .apply(lambda p: np.log(p / (1. - p))))",
"_____no_output_____"
],
[
"ax = resp_lco.plot(marker='o')\n\nax.set_xlim(0.9, 7);\nax.set_xlabel(\"response\", fontsize=14)\nax.set_ylabel(\"log-cumulative-odds\", fontsize=14);",
"_____no_output_____"
]
],
[
[
"#### Code 11.5",
"_____no_output_____"
]
],
[
[
"with pm.Model() as m11_1:\n a = pm.Normal(\n 'a', 0., 10.,\n transform=pm.distributions.transforms.ordered,\n shape=6, testval=np.arange(6) - 2.5)\n \n resp_obs = pm.OrderedLogistic(\n 'resp_obs', 0., a,\n observed=trolley_df.response.values - 1\n )",
"/home/osvaldo/anaconda3/lib/python3.7/site-packages/theano/tensor/subtensor.py:2197: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n rval = inputs[0].__getitem__(inputs[1:])\n/home/osvaldo/anaconda3/lib/python3.7/site-packages/theano/tensor/subtensor.py:2197: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.\n rval = inputs[0].__getitem__(inputs[1:])\n"
],
[
"with m11_1:\n map_11_1 = pm.find_MAP()",
"/home/osvaldo/proyectos/00_PyMC/pymc/pymc/tuning/starting.py:61: UserWarning: find_MAP should not be used to initialize the NUTS sampler, simply call pymc.sample() and it will automatically initialize NUTS in a better way.\n warnings.warn('find_MAP should not be used to initialize the NUTS sampler, simply call pymc.sample() and it will automatically initialize NUTS in a better way.')\n"
]
],
[
[
"#### Code 11.6",
"_____no_output_____"
]
],
[
[
"map_11_1['a']",
"_____no_output_____"
],
[
"daf",
"_____no_output_____"
]
],
[
[
"#### Code 11.7",
"_____no_output_____"
]
],
[
[
"sp.special.expit(map_11_1['a'])",
"_____no_output_____"
]
],
[
[
"#### Code 11.8",
"_____no_output_____"
]
],
[
[
"with m11_1:\n trace_11_1 = pm.sample(1000, tune=1000)",
"_____no_output_____"
],
[
"az.summary(trace_11_1, var_names=['a'], credible_interval=.89, rount_to=2)",
"_____no_output_____"
]
],
[
[
"#### Code 11.9",
"_____no_output_____"
]
],
[
[
"def ordered_logistic_proba(a):\n pa = sp.special.expit(a)\n p_cum = np.concatenate(([0.], pa, [1.]))\n \n return p_cum[1:] - p_cum[:-1]",
"_____no_output_____"
],
[
"ordered_logistic_proba(trace_11_1['a'].mean(axis=0))",
"_____no_output_____"
]
],
[
[
"#### Code 11.10",
"_____no_output_____"
]
],
[
[
"(ordered_logistic_proba(trace_11_1['a'].mean(axis=0)) \\\n * (1 + np.arange(7))).sum()",
"_____no_output_____"
]
],
[
[
"#### Code 11.11",
"_____no_output_____"
]
],
[
[
"ordered_logistic_proba(trace_11_1['a'].mean(axis=0) - 0.5)",
"_____no_output_____"
]
],
[
[
"#### Code 11.12",
"_____no_output_____"
]
],
[
[
"(ordered_logistic_proba(trace_11_1['a'].mean(axis=0) - 0.5) \\\n * (1 + np.arange(7))).sum()",
"_____no_output_____"
]
],
[
[
"#### Code 11.13",
"_____no_output_____"
]
],
[
[
"action = shared(trolley_df.action.values)\nintention = shared(trolley_df.intention.values)\ncontact = shared(trolley_df.contact.values)\n\nwith pm.Model() as m11_2:\n a = pm.Normal(\n 'a', 0., 10.,\n transform=pm.distributions.transforms.ordered,\n shape=6,\n testval=trace_11_1['a'].mean(axis=0)\n )\n \n bA = pm.Normal('bA', 0., 10.)\n bI = pm.Normal('bI', 0., 10.)\n bC = pm.Normal('bC', 0., 10.)\n phi = bA * action + bI * intention + bC * contact\n\n resp_obs = pm.OrderedLogistic(\n 'resp_obs', phi, a,\n observed=trolley_df.response.values - 1\n )",
"_____no_output_____"
],
[
"with m11_2:\n map_11_2 = pm.find_MAP()",
"_____no_output_____"
]
],
[
[
"#### Code 11.14",
"_____no_output_____"
]
],
[
[
"with pm.Model() as m11_3:\n a = pm.Normal(\n 'a', 0., 10.,\n transform=pm.distributions.transforms.ordered,\n shape=6,\n testval=trace_11_1['a'].mean(axis=0)\n )\n \n bA = pm.Normal('bA', 0., 10.)\n bI = pm.Normal('bI', 0., 10.)\n bC = pm.Normal('bC', 0., 10.)\n bAI = pm.Normal('bAI', 0., 10.)\n bCI = pm.Normal('bCI', 0., 10.)\n phi = bA * action + bI * intention + bC * contact \\\n + bAI * action * intention \\\n + bCI * contact * intention\n\n resp_obs = pm.OrderedLogistic(\n 'resp_obs', phi, a,\n observed=trolley_df.response - 1\n )",
"_____no_output_____"
],
[
"with m11_3:\n map_11_3 = pm.find_MAP()",
"_____no_output_____"
]
],
[
[
"#### Code 11.15",
"_____no_output_____"
]
],
[
[
"def get_coefs(map_est):\n coefs = OrderedDict()\n \n for i, ai in enumerate(map_est['a']):\n coefs[f'a_{i}'] = ai\n \n coefs['bA'] = map_est.get('bA', np.nan)\n coefs['bI'] = map_est.get('bI', np.nan)\n coefs['bC'] = map_est.get('bC', np.nan)\n coefs['bAI'] = map_est.get('bAI', np.nan)\n coefs['bCI'] = map_est.get('bCI', np.nan)\n \n return coefs",
"_____no_output_____"
],
[
"(pd.DataFrame.from_dict(\n OrderedDict([\n ('m11_1', get_coefs(map_11_1)),\n ('m11_2', get_coefs(map_11_2)),\n ('m11_3', get_coefs(map_11_3))\n ]))\n .astype(np.float64)\n .round(2))",
"_____no_output_____"
]
],
[
[
"#### Code 11.16",
"_____no_output_____"
]
],
[
[
"with m11_2:\n trace_11_2 = pm.sample(1000, tune=1000)",
"_____no_output_____"
],
[
"with m11_3:\n trace_11_3 = pm.sample(1000, tune=1000)",
"_____no_output_____"
],
[
"comp_df = pm.compare({m11_1:trace_11_1,\n m11_2:trace_11_2,\n m11_3:trace_11_3})\n\ncomp_df.loc[:,'model'] = pd.Series(['m11.1', 'm11.2', 'm11.3'])\ncomp_df = comp_df.set_index('model')\ncomp_df",
"_____no_output_____"
]
],
[
[
"#### Code 11.17-19",
"_____no_output_____"
]
],
[
[
"pp_df = pd.DataFrame(np.array([[0, 0, 0],\n [0, 0, 1],\n [1, 0, 0],\n [1, 0, 1],\n [0, 1, 0],\n [0, 1, 1]]),\n columns=['action', 'contact', 'intention'])",
"_____no_output_____"
],
[
"pp_df",
"_____no_output_____"
],
[
"action.set_value(pp_df.action.values)\ncontact.set_value(pp_df.contact.values)\nintention.set_value(pp_df.intention.values)\n\nwith m11_3:\n pp_trace_11_3 = pm.sample_ppc(trace_11_3, samples=1500)",
"_____no_output_____"
],
[
"PP_COLS = [f'pp_{i}' for i, _ in enumerate(pp_trace_11_3['resp_obs'])]\n\npp_df = pd.concat((pp_df,\n pd.DataFrame(pp_trace_11_3['resp_obs'].T, columns=PP_COLS)),\n axis=1)",
"_____no_output_____"
],
[
"pp_cum_df = (pd.melt(\n pp_df,\n id_vars=['action', 'contact', 'intention'],\n value_vars=PP_COLS, value_name='resp'\n )\n .groupby(['action', 'contact', 'intention', 'resp'])\n .size()\n .div(1500)\n .rename('proba')\n .reset_index()\n .pivot_table(\n index=['action', 'contact', 'intention'],\n values='proba',\n columns='resp'\n )\n .cumsum(axis=1)\n .iloc[:, :-1])",
"_____no_output_____"
],
[
"pp_cum_df",
"_____no_output_____"
],
[
"for (plot_action, plot_contact), plot_df in pp_cum_df.groupby(level=['action', 'contact']):\n fig, ax = plt.subplots(figsize=(8, 6))\n \n ax.plot([0, 1], plot_df, c='C0');\n ax.plot([0, 1], [0, 0], '--', c='C0');\n ax.plot([0, 1], [1, 1], '--', c='C0');\n \n ax.set_xlim(0, 1);\n ax.set_xlabel(\"intention\");\n \n ax.set_ylim(-0.05, 1.05);\n ax.set_ylabel(\"probability\");\n \n ax.set_title(\n \"action = {action}, contact = {contact}\".format(\n action=plot_action, contact=plot_contact\n )\n );",
"_____no_output_____"
]
],
[
[
"#### Code 11.20",
"_____no_output_____"
]
],
[
[
"# define parameters\nPROB_DRINK = 0.2 # 20% of days\nRATE_WORK = 1. # average 1 manuscript per day\n\n# sample one year of production\nN = 365",
"_____no_output_____"
],
[
"drink = np.random.binomial(1, PROB_DRINK, size=N)\ny = (1 - drink) * np.random.poisson(RATE_WORK, size=N)",
"_____no_output_____"
]
],
[
[
"#### Code 11.21",
"_____no_output_____"
]
],
[
[
"drink_zeros = drink.sum()\nwork_zeros = (y == 0).sum() - drink_zeros",
"_____no_output_____"
],
[
"bins = np.arange(y.max() + 1) - 0.5\n\nplt.hist(y, bins=bins);\nplt.bar(0., drink_zeros, width=1., bottom=work_zeros, color='C1', alpha=.5);\n\nplt.xticks(bins + 0.5);\nplt.xlabel(\"manuscripts completed\");\n\nplt.ylabel(\"Frequency\");",
"_____no_output_____"
]
],
[
[
"#### Code 11.22",
"_____no_output_____"
]
],
[
[
"with pm.Model() as m11_4:\n ap = pm.Normal('ap', 0., 1.)\n p = pm.math.sigmoid(ap)\n \n al = pm.Normal('al', 0., 10.)\n lambda_ = pm.math.exp(al)\n \n y_obs = pm.ZeroInflatedPoisson('y_obs', 1. - p, lambda_, observed=y)",
"_____no_output_____"
],
[
"with m11_4:\n map_11_4 = pm.find_MAP()",
"_____no_output_____"
],
[
"map_11_4",
"_____no_output_____"
]
],
[
[
"#### Code 11.23",
"_____no_output_____"
]
],
[
[
"sp.special.expit(map_11_4['ap']) # probability drink",
"_____no_output_____"
],
[
"np.exp(map_11_4['al']) # rate finish manuscripts, when not drinking",
"_____no_output_____"
]
],
[
[
"#### Code 11.24",
"_____no_output_____"
]
],
[
[
"def dzip(x, p, lambda_, log=True):\n like = p**(x == 0) + (1 - p) * sp.stats.poisson.pmf(x, lambda_)\n \n return np.log(like) if log else like",
"_____no_output_____"
]
],
[
[
"#### Code 11.25",
"_____no_output_____"
]
],
[
[
"PBAR = 0.5\nTHETA = 5.",
"_____no_output_____"
],
[
"a = PBAR * THETA\nb = (1 - PBAR) * THETA",
"_____no_output_____"
],
[
"p = np.linspace(0, 1, 100)\n\nplt.plot(p, sp.stats.beta.pdf(p, a, b));\n\nplt.xlim(0, 1);\nplt.xlabel(\"probability\");\n\nplt.ylabel(\"Density\");",
"_____no_output_____"
]
],
[
[
"#### Code 11.26",
"_____no_output_____"
]
],
[
[
"admit_df = pd.read_csv('Data/UCBadmit.csv', sep=';')\nadmit_df.head()",
"_____no_output_____"
],
[
"with pm.Model() as m11_5:\n a = pm.Normal('a', 0., 2.)\n pbar = pm.Deterministic('pbar', pm.math.sigmoid(a))\n\n theta = pm.Exponential('theta', 1.)\n \n admit_obs = pm.BetaBinomial(\n 'admit_obs',\n pbar * theta, (1. - pbar) * theta,\n admit_df.applications.values,\n observed=admit_df.admit.values\n )",
"_____no_output_____"
],
[
"with m11_5:\n trace_11_5 = pm.sample(1000, tune=1000)",
"_____no_output_____"
]
],
[
[
"#### Code 11.27",
"_____no_output_____"
]
],
[
[
"pm.summary(trace_11_5, alpha=.11).round(2)",
"_____no_output_____"
]
],
[
[
"#### Code 11.28",
"_____no_output_____"
]
],
[
[
"np.percentile(trace_11_5['pbar'], [2.5, 50., 97.5])",
"_____no_output_____"
]
],
[
[
"#### Code 11.29",
"_____no_output_____"
]
],
[
[
"pbar_hat = trace_11_5['pbar'].mean()\ntheta_hat = trace_11_5['theta'].mean()\n\np_plot = np.linspace(0, 1, 100)\n\nplt.plot(\n p_plot,\n sp.stats.beta.pdf(p_plot, pbar_hat * theta_hat, (1. - pbar_hat) * theta_hat)\n);\nplt.plot(\n p_plot,\n sp.stats.beta.pdf(\n p_plot[:, np.newaxis],\n trace_11_5['pbar'][:100] * trace_11_5['theta'][:100],\n (1. - trace_11_5['pbar'][:100]) * trace_11_5['theta'][:100]\n ),\n c='C0', alpha=0.1\n);\n\nplt.xlim(0., 1.);\nplt.xlabel(\"probability admit\");\n\nplt.ylim(0., 3.);\nplt.ylabel(\"Density\");",
"_____no_output_____"
]
],
[
[
"#### Code 11.30",
"_____no_output_____"
]
],
[
[
"with m11_5:\n pp_trace_11_5 = pm.sample_ppc(trace_11_5)",
"_____no_output_____"
],
[
"x_case = np.arange(admit_df.shape[0])\n\nplt.scatter(\n x_case,\n pp_trace_11_5['admit_obs'].mean(axis=0) \\\n / admit_df.applications.values\n);\nplt.scatter(x_case, admit_df.admit / admit_df.applications);\n\nhigh = np.percentile(pp_trace_11_5['admit_obs'], 95, axis=0) \\\n / admit_df.applications.values\nplt.scatter(x_case, high, marker='x', c='k');\n\nlow = np.percentile(pp_trace_11_5['admit_obs'], 5, axis=0) \\\n / admit_df.applications.values\nplt.scatter(x_case, low, marker='x', c='k');",
"_____no_output_____"
]
],
[
[
"#### Code 11.31",
"_____no_output_____"
]
],
[
[
"mu = 3.\ntheta = 1.\n\nx = np.linspace(0, 10, 100)\nplt.plot(x, sp.stats.gamma.pdf(x, mu / theta, scale=theta));",
"_____no_output_____"
],
[
"import platform\nimport sys\n\nimport IPython\nimport matplotlib\nimport scipy\n\nprint(\"This notebook was createad on a computer {} running {} and using:\\nPython {}\\nIPython {}\\nPyMC {}\\nNumPy {}\\nPandas {}\\nSciPy {}\\nMatplotlib {}\\n\".format(platform.machine(), ' '.join(platform.linux_distribution()[:2]), sys.version[:5], IPython.__version__, pm.__version__, np.__version__, pd.__version__, scipy.__version__, matplotlib.__version__))",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a8e6dc0eb94d0c844978a2867ae9293521490f6
| 147,243 |
ipynb
|
Jupyter Notebook
|
module4/BuildWeekProject.ipynb
|
iesous-kurios/DS-Unit-2-Applied-Modeling
|
5cec246e1220cb5e81b8abed77d8d5b3ddf28742
|
[
"MIT"
] | 1 |
2019-12-05T03:08:57.000Z
|
2019-12-05T03:08:57.000Z
|
module4/BuildWeekProject.ipynb
|
iesous-kurios/DS-Unit-2-Applied-Modeling
|
5cec246e1220cb5e81b8abed77d8d5b3ddf28742
|
[
"MIT"
] | null | null | null |
module4/BuildWeekProject.ipynb
|
iesous-kurios/DS-Unit-2-Applied-Modeling
|
5cec246e1220cb5e81b8abed77d8d5b3ddf28742
|
[
"MIT"
] | null | null | null | 113.00307 | 44,854 | 0.808154 |
[
[
[
"<a href=\"https://colab.research.google.com/github/iesous-kurios/DS-Unit-2-Applied-Modeling/blob/master/module4/BuildWeekProject.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"%%capture\nimport sys\n\n# If you're on Colab:\nif 'google.colab' in sys.modules:\n DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'\n !pip install category_encoders==2.*\n !pip install eli5\n\n# If you're working locally:\nelse:\n DATA_PATH = '../data/'\n",
"_____no_output_____"
],
[
"# all imports needed for this sheet\n\nimport numpy as np\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\nimport category_encoders as ce\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.feature_selection import f_regression, SelectKBest\nfrom sklearn.linear_model import Ridge\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.preprocessing import StandardScaler\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import validation_curve\nfrom sklearn.tree import DecisionTreeRegressor\nimport xgboost as xgb\n\n%matplotlib inline\nimport seaborn as sns\nfrom sklearn.metrics import accuracy_score\n\nfrom sklearn.model_selection import GridSearchCV, RandomizedSearchCV\n",
"_____no_output_____"
],
[
"df = pd.read_excel('/content/pipeline_pickle.xlsx')",
"_____no_output_____"
]
],
[
[
"I chose \"exit to permanent\" housing as my target due to my belief that accurately predicting this feature would have the largest impact on actual people experiencing homelessness in my county. Developing and fine tuning an accurate model with our data could also lead to major improvements in our county's efforts at addressing the homelessness problem among singles as well (as our shelter only serves families)",
"_____no_output_____"
]
],
[
[
"exit_reasons = ['Rental by client with RRH or equivalent subsidy', \n 'Rental by client, no ongoing housing subsidy', \n 'Staying or living with family, permanent tenure', \n 'Rental by client, other ongoing housing subsidy',\n 'Permanent housing (other than RRH) for formerly homeless persons', \n 'Staying or living with friends, permanent tenure', \n 'Owned by client, with ongoing housing subsidy', \n 'Rental by client, VASH housing Subsidy'\n ]",
"_____no_output_____"
],
[
"# pull all exit destinations from main data file and sum up the totals of each destination, \n# placing them into new df for calculations\nexits = df['3.12 Exit Destination'].value_counts()",
"_____no_output_____"
],
[
" # create target column (multiple types of exits to perm)\ndf['perm_leaver'] = df['3.12 Exit Destination'].isin(exit_reasons)",
"_____no_output_____"
],
[
"# replace spaces with underscore\ndf.columns = df.columns.str.replace(' ', '_')",
"_____no_output_____"
],
[
"df = df.rename(columns = {'Length_of_Time_Homeless_(3.917_Approximate_Start)':'length_homeless', '4.2_Income_Total_at_Entry':'entry_income' \n })",
"_____no_output_____"
]
],
[
[
"If a person were to guess \"did not exit to permanent\" housing every single time, they would be correct approximately 63 percent of the time. I am hoping that through this project, we will be able to provide more focused case management services to guests that displayed features which my model predicted as contributing negatively toward their chances of having an exit to permanent housing. It is my hope that a year from now, the base case will be flipped, and you would need to guess \"did exit to permanent housing\" to be correct approximately 63 percent of the time. ",
"_____no_output_____"
]
],
[
[
"# base case\ndf['perm_leaver'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"# see size of df prior to dropping empties\ndf.shape",
"_____no_output_____"
],
[
"# drop rows with no exit destination (current guests at time of report)\ndf = df.dropna(subset=['3.12_Exit_Destination'])",
"_____no_output_____"
],
[
"# shape of df after dropping current guests\ndf.shape",
"_____no_output_____"
],
[
"df.to_csv('/content/n_alltime.csv')",
"_____no_output_____"
],
[
"# verify no NaN in exit destination feature\ndf['3.12_Exit_Destination'].isna().value_counts()",
"_____no_output_____"
],
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\n\ntrain = df\n\n# Split train into train & val\n#train, val = train_test_split(train, train_size=0.80, test_size=0.20, \n # stratify=train['perm_leaver'], random_state=42)\n\n# Do train/test split\n# Use data from Jan -March 2019 to train\n# Use data from April 2019 to test\ndf['enroll_date'] = pd.to_datetime(df['3.10_Enroll_Date'], infer_datetime_format=True)\ncutoff = pd.to_datetime('2019-01-01')\ntrain = df[df.enroll_date < cutoff]\ntest = df[df.enroll_date >= cutoff]\n\n\n\n\ndef wrangle(X):\n \"\"\"Wrangle train, validate, and test sets in the same way\"\"\"\n \n # Prevent SettingWithCopyWarning\n X = X.copy()\n \n # drop any private information\n X = X.drop(columns=['3.1_FirstName', '3.1_LastName', '3.2_SocSecNo', \n '3.3_Birthdate', 'V5_Prior_Address'])\n \n # drop unusable columns\n X = X.drop(columns=['2.1_Organization_Name', '2.4_ProjectType',\n 'WorkSource_Referral_Most_Recent', 'YAHP_Referral_Most_Recent',\n 'SOAR_Enrollment_Determination_(Most_Recent)',\n 'R7_General_Health_Status', 'R8_Dental_Health_Status',\n 'R9_Mental_Health_Status', 'RRH_Date_Of_Move-In',\n 'RRH_In_Permanent_Housing', 'R10_Pregnancy_Due_Date',\n 'R10_Pregnancy_Status', 'R1_Referral_Source',\n 'R2_Date_Status_Determined', 'R2_Enroll_Status',\n 'R2_Reason_Why_No_Services_Funded', 'R2_Runaway_Youth',\n 'R3_Sexual_Orientation', '2.5_Utilization_Tracking_Method_(Invalid)',\n '2.2_Project_Name', '2.6_Federal_Grant_Programs', '3.16_Client_Location',\n '3.917_Stayed_Less_Than_90_Days', \n '3.917b_Stayed_in_Streets,_ES_or_SH_Night_Before', \n '3.917b_Stayed_Less_Than_7_Nights', '4.24_In_School_(Retired_Data_Element)',\n 'CaseChildren', 'ClientID', 'HEN-HP_Referral_Most_Recent',\n 'HEN-RRH_Referral_Most_Recent', 'Emergency_Shelter_|_Most_Recent_Enrollment',\n 'ProgramType', 'Days_Enrolled_Until_RRH_Date_of_Move-in',\n 'CurrentDate', 'Current_Age', 'Count_of_Bed_Nights_-_Entire_Episode',\n 'Bed_Nights_During_Report_Period'])\n \n # drop rows with no exit destination (current guests at time of report)\n X = X.dropna(subset=['3.12_Exit_Destination'])\n \n # remove columns to avoid data leakage\n X = X.drop(columns=['3.12_Exit_Destination', '5.9_Household_ID', '5.8_Personal_ID',\n '4.2_Income_Total_at_Exit', '4.3_Non-Cash_Benefit_Count_at_Exit'])\n \n # Drop needless feature\n unusable_variance = ['Enrollment_Created_By', '4.24_Current_Status_(Retired_Data_Element)']\n X = X.drop(columns=unusable_variance)\n\n # Drop columns with timestamp\n timestamp_columns = ['3.10_Enroll_Date', '3.11_Exit_Date', \n 'Date_of_Last_ES_Stay_(Beta)', 'Date_of_First_ES_Stay_(Beta)', \n 'Prevention_|_Most_Recent_Enrollment', 'PSH_|_Most_Recent_Enrollment', \n 'Transitional_Housing_|_Most_Recent_Enrollment', 'Coordinated_Entry_|_Most_Recent_Enrollment', \n 'Street_Outreach_|_Most_Recent_Enrollment', 'RRH_|_Most_Recent_Enrollment', \n 'SOAR_Eligibility_Determination_(Most_Recent)', 'Date_of_First_Contact_(Beta)',\n 'Date_of_Last_Contact_(Beta)', '4.13_Engagement_Date', '4.11_Domestic_Violence_-_When_it_Occurred',\n '3.917_Homeless_Start_Date']\n X = X.drop(columns=timestamp_columns)\n \n # return the wrangled dataframe\n return X\n\n",
"_____no_output_____"
],
[
"train.shape",
"_____no_output_____"
],
[
"test.shape",
"_____no_output_____"
],
[
"train = wrangle(train)\ntest = wrangle(test)",
"_____no_output_____"
],
[
"# Hand pick features only known at entry to avoid data leakage\nfeatures = ['CaseMembers',\n '3.2_Social_Security_Quality', '3.3_Birthdate_Quality',\n 'Age_at_Enrollment', '3.4_Race', '3.5_Ethnicity', '3.6_Gender',\n '3.7_Veteran_Status', '3.8_Disabling_Condition_at_Entry',\n '3.917_Living_Situation', 'length_homeless',\n '3.917_Times_Homeless_Last_3_Years', '3.917_Total_Months_Homeless_Last_3_Years', \n 'V5_Last_Permanent_Address', 'V5_State', 'V5_Zip', 'Municipality_(City_or_County)',\n '4.1_Housing_Status', '4.4_Covered_by_Health_Insurance', '4.11_Domestic_Violence',\n '4.11_Domestic_Violence_-_Currently_Fleeing_DV?', 'Household_Type', \n 'R4_Last_Grade_Completed', 'R5_School_Status',\n 'R6_Employed_Status', 'R6_Why_Not_Employed', 'R6_Type_of_Employment',\n 'R6_Looking_for_Work', 'entry_income',\n '4.3_Non-Cash_Benefit_Count', 'Barrier_Count_at_Entry',\n 'Chronic_Homeless_Status', 'Under_25_Years_Old',\n '4.10_Alcohol_Abuse_(Substance_Abuse)', '4.07_Chronic_Health_Condition',\n '4.06_Developmental_Disability', '4.10_Drug_Abuse_(Substance_Abuse)',\n '4.08_HIV/AIDS', '4.09_Mental_Health_Problem',\n '4.05_Physical_Disability'\n ]\n",
"_____no_output_____"
],
[
"target = 'perm_leaver'\nX_train = train[features]\ny_train = train[target]\nX_test = test[features]\ny_test = test[target]",
"_____no_output_____"
],
[
"# base case\ndf['perm_leaver'].value_counts(normalize=True)",
"_____no_output_____"
],
[
"# fit linear model to get a 3 on Sprint\n\nfrom sklearn.linear_model import LogisticRegression\n\nencoder = ce.OneHotEncoder(use_cat_names=True)\nX_train_encoded = encoder.fit_transform(X_train)\nX_test_encoded = encoder.transform(X_test)\n\n\nimputer = SimpleImputer()\nX_train_imputed = imputer.fit_transform(X_train_encoded)\nX_test_imputed = imputer.transform(X_test_encoded)\n\nscaler = StandardScaler()\nX_train_scaled = scaler.fit_transform(X_train_imputed)\nX_test_scaled = scaler.transform(X_test_imputed)\n\nmodel = LogisticRegression(random_state=42, max_iter=5000)\nmodel.fit(X_train_scaled, y_train)\nprint ('Validation Accuracy', model.score(X_test_scaled,y_test))",
"Validation Accuracy 0.6315028901734104\n"
]
],
[
[
"Linear model above beat the baseline model, now let's see if we can get even more accurate with a tree-based model",
"_____no_output_____"
]
],
[
[
"import category_encoders as ce\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.ensemble import GradientBoostingClassifier\n\n# Make pipeline!\npipeline = make_pipeline(\n ce.OrdinalEncoder(), \n SimpleImputer(strategy='most_frequent'), \n RandomForestClassifier(n_estimators=100, n_jobs=-1, \n random_state=42, \n )\n)\n\n# Fit on train, score on val\npipeline.fit(X_train, y_train)\ny_pred = pipeline.predict(X_test)\nprint('Validation Accuracy', accuracy_score(y_test, y_pred))",
"Validation Accuracy 0.75\n"
],
[
"from joblib import dump\ndump(pipeline, 'pipeline.joblib', compress=True)",
"_____no_output_____"
],
[
"# get and plot feature importances\n\n# Linear models have coefficients whereas decision trees have \"Feature Importances\"\nimport matplotlib.pyplot as plt\n\nmodel = pipeline.named_steps['randomforestclassifier']\nencoder = pipeline.named_steps['ordinalencoder']\nencoded_columns = encoder.transform(X_test).columns\nimportances = pd.Series(model.feature_importances_, encoded_columns)\nn = 20\nplt.figure(figsize=(10,n/2))\nplt.title(f'Top {n} features')\nimportances.sort_values()[-n:].plot.barh(color='grey');",
"_____no_output_____"
],
[
"# cross validation \n\nk = 3\nscores = cross_val_score(pipeline, X_train, y_train, cv=k, \n scoring='accuracy')\nprint(f'MAE for {k} folds:', -scores)",
"MAE for 3 folds: [-0.61710037 -0.63432836 -0.64552239]\n"
],
[
"-scores.mean()",
"_____no_output_____"
]
],
[
[
"Now that we have beaten the linear model with a tree based model, let us see if xgboost does a better job at predicting exit destination",
"_____no_output_____"
]
],
[
[
"\n\nfrom xgboost import XGBClassifier\n\npipeline = make_pipeline(\n ce.OrdinalEncoder(),\n XGBClassifier(n_estimators=100, random_state=42, n_jobs=-1)\n)\n\n# Fit on train, score on val\npipeline.fit(X_train, y_train)\nprint('Validation Accuracy:', pipeline.score(X_test, y_test))",
"Validation Accuracy: 0.703757225433526\n"
]
],
[
[
"xgboost failed to beat my tree-based model, so the tree-based model is what I will use for my prediction on my web-app",
"_____no_output_____"
]
],
[
[
"# get and plot feature importances\n\n# Linear models have coefficients whereas decision trees have \"Feature Importances\"\nimport matplotlib.pyplot as plt\n\nmodel = pipeline.named_steps['xgbclassifier']\nencoder = pipeline.named_steps['ordinalencoder']\nencoded_columns = encoder.transform(X_test).columns\nimportances = pd.Series(model.feature_importances_, encoded_columns)\nn = 20\nplt.figure(figsize=(10,n/2))\nplt.title(f'Top {n} features')\nimportances.sort_values()[-n:].plot.barh(color='grey');\n\n",
"_____no_output_____"
],
[
"history = pd.read_csv('/content/n_alltime.csv')\nfrom plotly.tools import mpl_to_plotly\nimport seaborn as sns\nfrom sklearn.metrics import accuracy_score\n\nfrom sklearn.model_selection import GridSearchCV, RandomizedSearchCV\n\n\n\n\n# Assign to X, y to avoid data leakage\nfeatures = ['CaseMembers',\n '3.2_Social_Security_Quality', '3.3_Birthdate_Quality',\n 'Age_at_Enrollment', '3.4_Race', '3.5_Ethnicity', '3.6_Gender',\n '3.7_Veteran_Status', '3.8_Disabling_Condition_at_Entry',\n '3.917_Living_Situation', 'length_homeless',\n '3.917_Times_Homeless_Last_3_Years', '3.917_Total_Months_Homeless_Last_3_Years', \n 'V5_Last_Permanent_Address', 'V5_State', 'V5_Zip', 'Municipality_(City_or_County)',\n '4.1_Housing_Status', '4.4_Covered_by_Health_Insurance', '4.11_Domestic_Violence',\n '4.11_Domestic_Violence_-_Currently_Fleeing_DV?', 'Household_Type', \n 'R4_Last_Grade_Completed', 'R5_School_Status',\n 'R6_Employed_Status', 'R6_Why_Not_Employed', 'R6_Type_of_Employment',\n 'R6_Looking_for_Work', 'entry_income',\n '4.3_Non-Cash_Benefit_Count', 'Barrier_Count_at_Entry',\n 'Chronic_Homeless_Status', 'Under_25_Years_Old',\n '4.10_Alcohol_Abuse_(Substance_Abuse)', '4.07_Chronic_Health_Condition',\n '4.06_Developmental_Disability', '4.10_Drug_Abuse_(Substance_Abuse)',\n '4.08_HIV/AIDS', '4.09_Mental_Health_Problem',\n '4.05_Physical_Disability', 'perm_leaver'\n ]\n\n\nX = history[features]\nX = X.drop(columns='perm_leaver')\n\n\n\ny_pred = pipeline.predict(X)\n\n\nfig, ax = plt.subplots()\nsns.distplot(test['perm_leaver'], hist=False, kde=True, ax=ax, label='Actual')\nsns.distplot(y_pred, hist=False, kde=True, ax=ax, label='Predicted')\nax.set_title('Distribution of Actual Exit compared to prediction')\nax.legend().set_visible(True)\n",
"_____no_output_____"
],
[
"pipeline = make_pipeline(\n ce.OrdinalEncoder(), \n SimpleImputer(), \n RandomForestClassifier(random_state=42)\n)\n\nparam_distributions = {\n \n \n 'simpleimputer__strategy': ['most_frequent', 'mean', 'median'], \n 'randomforestclassifier__bootstrap': [True, False],\n 'randomforestclassifier__max_depth': [10, 20, 30, 40, 50, 60, 70, 80, 90, 100, None],\n 'randomforestclassifier__max_features': ['auto', 'sqrt'],\n 'randomforestclassifier__min_samples_leaf': [1, 2, 4],\n 'randomforestclassifier__min_samples_split': [2, 5, 10],\n 'randomforestclassifier__n_estimators': [200, 400, 600, 800, 1000, 1200, 1400, 1600, 1800, 2000]}\n\n# If you're on Colab, decrease n_iter & cv parameters\nsearch = RandomizedSearchCV(\n pipeline, \n param_distributions=param_distributions, \n n_iter=1, \n cv=3, \n scoring='accuracy', \n verbose=10, \n return_train_score=True, \n n_jobs=-1\n)\n\n\n",
"_____no_output_____"
],
[
"# Fit on train, score on val\nsearch.fit(X_train, y_train)",
"Fitting 3 folds for each of 1 candidates, totalling 3 fits\n"
],
[
"print('Best hyperparameters', search.best_params_)\nprint('Cross-validation accuracy score', -search.best_score_)",
"Best hyperparameters {'simpleimputer__strategy': 'most_frequent', 'randomforestclassifier__n_estimators': 1000, 'randomforestclassifier__min_samples_split': 2, 'randomforestclassifier__min_samples_leaf': 4, 'randomforestclassifier__max_features': 'sqrt', 'randomforestclassifier__max_depth': None, 'randomforestclassifier__bootstrap': False}\nCross-validation accuracy score -0.622389909190109\n"
],
[
"\nprint('Validation Accuracy', search.score(X_test, y_test))",
"Validation Accuracy 0.7355491329479769\n"
],
[
"y_pred.shape",
"_____no_output_____"
],
[
"history['perm_leaver'].value_counts()",
"_____no_output_____"
],
[
"1282+478",
"_____no_output_____"
],
[
"from joblib import dump\ndump(pipeline, 'pipeline2.joblib', compress=True)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8e79cfe344785b47ba4e5e50a5bded7f263fa6
| 1,230 |
ipynb
|
Jupyter Notebook
|
Untitled0.ipynb
|
PetricaR/AutoSeries
|
d60b583ad7fa5227e36e0c5cbfca40156f478e25
|
[
"MIT"
] | null | null | null |
Untitled0.ipynb
|
PetricaR/AutoSeries
|
d60b583ad7fa5227e36e0c5cbfca40156f478e25
|
[
"MIT"
] | null | null | null |
Untitled0.ipynb
|
PetricaR/AutoSeries
|
d60b583ad7fa5227e36e0c5cbfca40156f478e25
|
[
"MIT"
] | null | null | null | 22.363636 | 227 | 0.473984 |
[
[
[
"<a href=\"https://colab.research.google.com/github/PetricaR/AutoSeries/blob/master/Untitled0.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"\"radan\"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
]
] |
4a8e7cc871c514c8cabc7af7603087d65304eb12
| 8,061 |
ipynb
|
Jupyter Notebook
|
Notebooks/Spark-Example-02.ipynb
|
Miked387/big_data
|
405aca20c8db4350a7a0d04607ff05f3c4e97c2f
|
[
"BSD-3-Clause"
] | 32 |
2020-07-02T00:51:13.000Z
|
2022-03-31T22:31:30.000Z
|
Notebooks/Spark-Example-1.ipynb
|
ivan-kunfei/BigDataAnalytics
|
57842067043cbcbf291cd704666527c596e4ca69
|
[
"BSD-3-Clause"
] | 2 |
2021-02-17T00:42:23.000Z
|
2021-02-18T17:28:38.000Z
|
Notebooks/Spark-Example-1.ipynb
|
ivan-kunfei/BigDataAnalytics
|
57842067043cbcbf291cd704666527c596e4ca69
|
[
"BSD-3-Clause"
] | 47 |
2020-07-04T15:29:15.000Z
|
2022-03-25T05:08:17.000Z
| 19.424096 | 215 | 0.434686 |
[
[
[
"# Spark Example\n\nThis is a first tutorial on apache spark\n\nsc is here Spark Context object. You do not need to create the \"sc\" object, it is already loaded into memory. ",
"_____no_output_____"
]
],
[
[
"print(sc.version)\nprint(sc.pythonVer)\nprint(sc.master)",
"3.0.0\n3.8\nlocal[*]\n"
],
[
"# We create here an example text data. \na = [\"What Will It Take for BU Commuters to Leave Their Cars for the MBTA? University boosts T pass subsidies to cover half the cost, raises parking fees, all part of broader strategy to build a greener BU\"]",
"_____no_output_____"
],
[
"print(a)",
"['What Will It Take for BU Commuters to Leave Their Cars for the MBTA? University boosts T pass subsidies to cover half the cost, raises parking fees, all part of broader strategy to build a greener BU']\n"
],
[
"# Now, we can go ahead and parallize it, i.e., load it in a distributed data structure as RDD\nrdd = sc.parallelize(a)\n\n# sc is the spark context and it is already loaded \n# When you write spark pyscripts you need to create it. ",
"_____no_output_____"
],
[
"# lines = sc.textFile(sys.argv[1], 1)\nwords = rdd.flatMap(lambda x: x.split(' '))\n\n",
"_____no_output_____"
],
[
"words.take(10)",
"_____no_output_____"
],
[
"counts = rdd.flatMap(lambda x: x.split(' ')) \\\n.map(lambda x: (x, 1)) \\\n.reduceByKey(lambda x, y: x+y)\n\ncounts.collect()",
"_____no_output_____"
],
[
"counts.top(1)",
"_____no_output_____"
],
[
"# top ()\n\n# top(num, key=None)[source]\n# Get the top N elements from an RDD.",
"_____no_output_____"
],
[
"sc.parallelize([10, 4, 2, 12, 3]).top(1)",
"_____no_output_____"
],
[
"sc.parallelize([2, 3, 4, 5, 6], 2).top(2)",
"_____no_output_____"
],
[
"sc.parallelize([10, 4, 2, 12, 3]).top(3, key=str)",
"_____no_output_____"
],
[
"rdd1=sc.parallelize([('a', 10), ('c',4), ('z', 2), ('d', 12)])",
"_____no_output_____"
],
[
"rdd1.top(3, key=lambda x: x[1])",
"_____no_output_____"
],
[
"rdd1.top(3, key=lambda x: x[0])",
"_____no_output_____"
],
[
"rdd1.top(3, lambda x: x[0])",
"_____no_output_____"
],
[
"rdd1.top(3)",
"_____no_output_____"
],
[
"rdd1.top(3, str)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8e873c291e22f0330c54a55a1262b3d7bf86f7
| 97,359 |
ipynb
|
Jupyter Notebook
|
lab13.ipynb
|
hannaht0808/ia241
|
9e87ba6c65e1be3449e0aabe0cc18de23231a434
|
[
"MIT"
] | null | null | null |
lab13.ipynb
|
hannaht0808/ia241
|
9e87ba6c65e1be3449e0aabe0cc18de23231a434
|
[
"MIT"
] | null | null | null |
lab13.ipynb
|
hannaht0808/ia241
|
9e87ba6c65e1be3449e0aabe0cc18de23231a434
|
[
"MIT"
] | null | null | null | 122.618388 | 27,888 | 0.80992 |
[
[
[
"# Visualize Covid19 Data in Python",
"_____no_output_____"
],
[
"## data source",
"_____no_output_____"
],
[
"the data is from [European Centre for Disease Prevention and Control](https://www.ecdc.europa.eu/en/publications-data/download-todays-data-geographic-distribution-covid-19-cases-worldwide)",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pandas",
"_____no_output_____"
]
],
[
[
"## a quick view of the data",
"_____no_output_____"
]
],
[
[
"df = pandas.read_excel('s3://taylor-ia241-2021spring/COVID-19-geographic-disbtribution-worldwide-2020-12-14.xls')\ndf[:10]",
"_____no_output_____"
]
],
[
[
"## trend of the number of cases",
"_____no_output_____"
]
],
[
[
"sum_cases_per_day=df.groupby('dateRep').sum()['cases']",
"_____no_output_____"
],
[
"sum_cases_per_day.plot()",
"_____no_output_____"
]
],
[
[
"## the top 10 countries with the highest deaths",
"_____no_output_____"
]
],
[
[
"sum_of_death_per_country=df.groupby('countriesAndTerritories').sum()['deaths']",
"_____no_output_____"
],
[
"sum_of_death_per_country.nlargest(10).plot.bar()",
"_____no_output_____"
]
],
[
[
"## list of all countries",
"_____no_output_____"
]
],
[
[
"pandas.unique(df['countriesAndTerritories'])",
"_____no_output_____"
]
],
[
[
"## The USA data",
"_____no_output_____"
]
],
[
[
"usa_data = df.loc[ df['countriesAndTerritories']=='United_States_of_America']\n\nusa_data[:10]",
"_____no_output_____"
]
],
[
[
"## how the # death is related to the # case in the USA",
"_____no_output_____"
]
],
[
[
"usa_data.plot.scatter(x='cases',y='deaths',c='month')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8e8d0436513b0187a7f960f0200c4a63f89d79
| 51,832 |
ipynb
|
Jupyter Notebook
|
chap3/chapter_3_examples.ipynb
|
chicolucio-python-learning/introduction-to-python-for-science
|
fe3aebd1342253615bf47c4fa4f01cad1db36d79
|
[
"MIT"
] | 2 |
2019-12-12T20:49:35.000Z
|
2021-09-13T19:07:24.000Z
|
chap3/chapter_3_examples.ipynb
|
chicolucio-python-learning/introduction-to-python-for-science
|
fe3aebd1342253615bf47c4fa4f01cad1db36d79
|
[
"MIT"
] | null | null | null |
chap3/chapter_3_examples.ipynb
|
chicolucio-python-learning/introduction-to-python-for-science
|
fe3aebd1342253615bf47c4fa4f01cad1db36d79
|
[
"MIT"
] | null | null | null | 19.456456 | 474 | 0.453465 |
[
[
[
"# Chapter 3\n\n***Ver como criar uma tabela de conteúdo TOC**",
"_____no_output_____"
],
[
"## Strings",
"_____no_output_____"
]
],
[
[
"a = \"My dog's name is\"\nb = \"Bingo\"",
"_____no_output_____"
],
[
"c = a + \" \" + b",
"_____no_output_____"
],
[
"c",
"_____no_output_____"
],
[
"#trying to add string and integer\nd = \"927\"\ne = 927",
"_____no_output_____"
],
[
"d + e",
"_____no_output_____"
]
],
[
[
"## Lists",
"_____no_output_____"
]
],
[
[
"a = [0, 1, 1, 2, 3, 5, 8, 13]\nb = [5., \"girl\", 2+0j, \"horse\", 21]",
"_____no_output_____"
],
[
"b[0]",
"_____no_output_____"
],
[
"b[1]",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-warning\">\n<big><center>Lists are <span style=\"color:red\"> *zero-indexed*</span> </center></big>\n</div>\n\n<div class=\"alert alert-block alert-success\">\n$$\n\\begin{align}\nlist = &[a, b, c, d, e]\\\\\n&\\color{red}\\Downarrow\n\\hspace{2.2pc}\\color{red}\\Downarrow\\\\\n&\\color{purple}{list[0]}\n\\hspace{1.2pc}\n\\color{purple}{list[4]}\\\\\n&\\color{brown}{list[-5]}\n\\hspace{0.7pc}\n\\color{brown}{list[-1]}\n\\end{align}\n$$\n</div>",
"_____no_output_____"
]
],
[
[
"b[-1]",
"_____no_output_____"
],
[
"b[-5]",
"_____no_output_____"
],
[
"b[4]",
"_____no_output_____"
],
[
"b = [5., \"girl\", 2+0j, \"horse\", 21]\nb[0] = b[0]+2\nimport numpy as np\nb[3] = np.pi",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-warning\">\n<big><center>Adding lists <span style=\"color:red\"> *concatenates*</span> them, just as the **+** operator concatenates strings. </center></big>\n</div>",
"_____no_output_____"
]
],
[
[
"a+a",
"_____no_output_____"
]
],
[
[
"### Slicing lists\n\n<div class=\"alert alert-block alert-warning\">\n<big>Reparar que <span style=\"color:red\"> *não*</span> se inclui o último elemento.</big>\n</div>",
"_____no_output_____"
]
],
[
[
"b",
"_____no_output_____"
],
[
"b[1:4]",
"_____no_output_____"
],
[
"b[3:5]",
"_____no_output_____"
],
[
"b[2:]",
"_____no_output_____"
],
[
"b[:3]",
"_____no_output_____"
],
[
"b[:]",
"_____no_output_____"
],
[
"b[1:-1]",
"_____no_output_____"
],
[
"len(b) #len --> length",
"_____no_output_____"
],
[
"? range",
"_____no_output_____"
]
],
[
[
"### Creating and modifying lists\n\n<div class=\"alert alert-block alert-info\">\nrange(stop) -> range object <br>\nrange(start, stop[, step]) -> range object\n</div>\n\nÚtil para criar *PAs (progressões aritméticas)*",
"_____no_output_____"
]
],
[
[
"range(10) #começa de zero por padrão, armazena apenas início, fim e step. Útil para economizar memória",
"_____no_output_____"
],
[
"print(range(10))",
"range(0, 10)\n"
],
[
"list(range(10)) #para explicitar todos os integrantes",
"_____no_output_____"
],
[
"list(range(3,10))",
"_____no_output_____"
],
[
"list(range(0,10,2))",
"_____no_output_____"
],
[
"a = range(1,10,3)",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"list(a)",
"_____no_output_____"
],
[
"a += [16, 31, 64, 127]",
"_____no_output_____"
],
[
"a = a + [16, 31, 64,127]",
"_____no_output_____"
],
[
"a = list(a) + [16, 31, 64,127]",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a = [0, 0] + a",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"b = a[:5] + [101, 102] + a[5:]",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
]
],
[
[
"### Tuples\n\n<div class=\"alert alert-block alert-warning\">\n<big><center>**Tuples** are lists that are <span style=\"color:red\"> *immutable*</span></center></big>\n</div>\n\nLogo, pode ser usado para armazenar constantes, por exemplo.",
"_____no_output_____"
]
],
[
[
"c = (1, 1, 2, 3, 5, 8, 13)",
"_____no_output_____"
],
[
"c[4]",
"_____no_output_____"
],
[
"c[4] = 7",
"_____no_output_____"
]
],
[
[
"### Multidimensional lists and tuples\n\nUseful in making tables and other structures.",
"_____no_output_____"
]
],
[
[
"a = [[3,9], [8,5], [11,1]] #list",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a[0]",
"_____no_output_____"
],
[
"a[1][0]",
"_____no_output_____"
],
[
"a[1][0] = 10",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a = ([3,9], [8,5], [11,1]) #? Não forma tuple assim... tudo deve ser parêntese. Ver abaixo",
"_____no_output_____"
],
[
"a[1][0]",
"_____no_output_____"
],
[
"a[1][0] = 10",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a = ((3,9), (8,5), (11,1)) #tuple",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a[1][0]",
"_____no_output_____"
],
[
"a[1][0] = 10",
"_____no_output_____"
]
],
[
[
"## NumPy arrays\n\n- all the elements are of the same type.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\na = [0, 0, 1, 4, 7, 16, 31, 64,127]",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"b = np.array(a) #converts a list to an array",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
]
],
[
[
"- the `array` function promotes all of the numbers to the type of the most general entry in the list.",
"_____no_output_____"
]
],
[
[
"c = np.array([1, 4., -2,7]) #todos se tornarão float",
"_____no_output_____"
],
[
"c",
"_____no_output_____"
],
[
"? np.linspace",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\nnp.linspace(start, stop, num=50, endpoint=True, retstep=False, dtype=None)\n</div>\nReturn evenly spaced numbers over a specified interval.\n\nReturns `num` evenly spaced samples, calculated over the interval [`start`, `stop`].\n\nThe endpoint of the interval can optionally be excluded.",
"_____no_output_____"
]
],
[
[
"np.linspace(0, 10, 5)",
"_____no_output_____"
],
[
"np.linspace(0, 10, 5, endpoint=False)",
"_____no_output_____"
],
[
"np.linspace(0, 10, 5, retstep=True)",
"_____no_output_____"
],
[
"? np.logspace",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\nnp.logspace(start, stop, num=50, endpoint=True, base=10.0, dtype=None)\n</div>\n\nReturn numbers spaced evenly on a log scale.\n\nIn linear space, the sequence starts at ``base**start`` (`base` to the power of `start`) and ends with ``base**stop``.",
"_____no_output_____"
]
],
[
[
"np.logspace(1,3,5)",
"_____no_output_____"
],
[
"%precision 1\nnp.logspace(1,3,5)",
"_____no_output_____"
],
[
"? np.arange",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\narange([start,] stop[, step,], dtype=None)\n</div>\nReturn evenly spaced values within a given interval.\n\nValues are generated within the half-open interval ``[start, stop)`` (in other words, the interval including `start` but excluding `stop`). For integer arguments the function is equivalent to the Python built-in\n`range <http://docs.python.org/lib/built-in-funcs.html>`_ function, but returns an ndarray rather than a list.",
"_____no_output_____"
]
],
[
[
"np.arange(0, 10, 2)",
"_____no_output_____"
],
[
"np.arange(0., 10, 2) #todos serão float",
"_____no_output_____"
],
[
"np.arange(0, 10, 1.5)",
"_____no_output_____"
]
],
[
[
"### Criação de arrays de zeros e uns.",
"_____no_output_____"
]
],
[
[
"np.zeros(6)",
"_____no_output_____"
],
[
"np.ones(8)",
"_____no_output_____"
],
[
"np.ones(8, dtype=int)",
"_____no_output_____"
]
],
[
[
"### Mathematical operations with arrays",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\na = np.linspace(-1, 5, 7)",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"a*6",
"_____no_output_____"
],
[
"np.sin(a)",
"_____no_output_____"
],
[
"x = np.linspace(-3.14, 3.14, 21)",
"_____no_output_____"
],
[
"y = np.cos(x)",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"y #fazer o plot disto futuramente",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"np.log(a)",
"/home/chico/anaconda3/lib/python3.6/site-packages/ipykernel/__main__.py:1: RuntimeWarning: divide by zero encountered in log\n if __name__ == '__main__':\n/home/chico/anaconda3/lib/python3.6/site-packages/ipykernel/__main__.py:1: RuntimeWarning: invalid value encountered in log\n if __name__ == '__main__':\n"
],
[
"a = np.array([34., -12, 5.])",
"_____no_output_____"
],
[
"b = np.array([68., 5., 20.])",
"_____no_output_____"
],
[
"a+b #vectorized operations",
"_____no_output_____"
]
],
[
[
"### Slicing and addressing arrays\n\nFórmula para a velocidade média em um intervalo de tempo *i*:\n\n$$\nv_i = \\frac{y_i - y_{i-1}}{t_i - t_{i-1}}\n$$",
"_____no_output_____"
]
],
[
[
"y = np.array([0., 1.3, 5., 10.9, 18.9, 28.7, 40.])\nt = np.array([0., 0.49, 1., 1.5, 2.08, 2.55, 3.2])",
"_____no_output_____"
],
[
"y[:-1]",
"_____no_output_____"
],
[
"y[1:]",
"_____no_output_____"
],
[
"v = (y[1:]-y[:-1])/(t[1:]-t[:-1])",
"_____no_output_____"
],
[
"v",
"_____no_output_____"
]
],
[
[
"### Multi-dimensional arrays and matrices",
"_____no_output_____"
]
],
[
[
"b = np.array([[1., 4, 5], [9, 7, 4]])",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
],
[
"#all elements of a MunPy array must be of the same data type: floats, integers, complex numbers, etc.",
"_____no_output_____"
],
[
"a = np.ones((3,4), dtype=float)",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"np.eye(4)",
"_____no_output_____"
],
[
"c = np.arange(6)",
"_____no_output_____"
],
[
"c",
"_____no_output_____"
],
[
"c = np.reshape(c, (2,3))",
"_____no_output_____"
],
[
"c",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
],
[
"b[0][2]",
"_____no_output_____"
],
[
"b[0,2] #0 indexed",
"_____no_output_____"
],
[
"b[1,2]",
"_____no_output_____"
],
[
"2*b",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-block alert-warning\">\n*Beware*: array multiplication, done on an element-by-element basis, <span, style=\"color:red\">*is not the same as **matrix** multiplication*</span> as defined in linear algebra. Therefore, we distinguish between *array* multiplication and *matrix* multiplication in Python.\n</div>",
"_____no_output_____"
]
],
[
[
"b*c",
"_____no_output_____"
],
[
"d = c.T #cria matriz transposta",
"_____no_output_____"
],
[
"d",
"_____no_output_____"
],
[
"np.dot(b,d) #faz multiplicação matricial",
"_____no_output_____"
]
],
[
[
"## Dictionaries\n\n\\* Também chamados de *hashmaps* ou *associative arrays* em outras linguagens de programação.",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-success\">\n$$\n\\begin{align}\nroom =&\\text{{\"Emma\":309, \"Jacob\":582, \"Olivia\":764}}\\\\\n&\\hspace{1.0pc}\\color{red}\\Downarrow\n\\hspace{1.5pc}\\color{red}\\Downarrow\\\\\n&\\hspace{0.7pc}\\color{purple}{key} \\hspace{1.5pc}\\color{purple}{value}\n\\end{align}\n$$\n</div>",
"_____no_output_____"
]
],
[
[
"room = {\"Emma\":309, \"Jacob\":582, \"Olivia\":764}",
"_____no_output_____"
],
[
"room[\"Olivia\"]",
"_____no_output_____"
],
[
"weird = {\"tank\":52, 846:\"horse\", \"bones\":[23, \"fox\", \"grass\"], \"phrase\":\"I am here\"}",
"_____no_output_____"
],
[
"weird[\"tank\"]",
"_____no_output_____"
],
[
"weird[846]",
"_____no_output_____"
],
[
"weird[\"bones\"]",
"_____no_output_____"
],
[
"weird[\"phrase\"]",
"_____no_output_____"
],
[
"d = {}",
"_____no_output_____"
],
[
"d[\"last name\"] = \"Alberts\"\nd[\"first name\"] = \"Marie\"\nd[\"birthday\"] = \"January 27\"",
"_____no_output_____"
],
[
"d",
"_____no_output_____"
],
[
"d.keys()",
"_____no_output_____"
],
[
"d.values()",
"_____no_output_____"
]
],
[
[
"## Random numbers\n\n`np.random.rand(num)` creates an array of `num` floats **uniformly** distributed on the interval from 0 to 1.\n\n`np.random.randn(num)` produces a **normal (Gaussian)** distribution of `num` random numbers with a mean of 0 and a standard deviation of 1. They are distributed according to\n$$\nP(x)=\\frac{1}{\\sqrt{2\\pi}}e^{-\\frac{1}{2}x²}\n$$\n\n`np.random.randint(low, high, num)` produces a **uniform** random distribution of `num` integers between `low` (inclusive) and `high` (exclusive).",
"_____no_output_____"
]
],
[
[
"np.random.rand()",
"_____no_output_____"
],
[
"np.random.rand(5)",
"_____no_output_____"
],
[
"a, b = 10, 20\n(b-a)*np.random.rand(20) + a #setting interval",
"_____no_output_____"
],
[
"x0, sigma = 15, 10\nsigma*np.random.randn(20) + x0 #setting width and center of normal distribution",
"_____no_output_____"
],
[
"np.random.randint(1, 7, 12) #simutaling a dozen rolls of a single die",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a8e8f8f8c0c37cc92364f71bf121107f57f78be
| 1,892 |
ipynb
|
Jupyter Notebook
|
ipydn/imeshell.ipynb
|
nufeng1999/MyWSL_shell
|
8c3c857001cc91f23dcf6e6e29f9dd4d27a0c8f1
|
[
"Apache-2.0"
] | null | null | null |
ipydn/imeshell.ipynb
|
nufeng1999/MyWSL_shell
|
8c3c857001cc91f23dcf6e6e29f9dd4d27a0c8f1
|
[
"Apache-2.0"
] | null | null | null |
ipydn/imeshell.ipynb
|
nufeng1999/MyWSL_shell
|
8c3c857001cc91f23dcf6e6e29f9dd4d27a0c8f1
|
[
"Apache-2.0"
] | null | null | null | 22.52381 | 102 | 0.515856 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a8e94f20bf2f62ee85ea059032f71095a58cfb4
| 8,460 |
ipynb
|
Jupyter Notebook
|
python/udemy-data-analysis-and-visualization/lecture32_data_combine_dataframes.ipynb
|
juancarlosqr/datascience
|
2e4d78365b059a3e501e988bee53970ac0d718fc
|
[
"MIT"
] | null | null | null |
python/udemy-data-analysis-and-visualization/lecture32_data_combine_dataframes.ipynb
|
juancarlosqr/datascience
|
2e4d78365b059a3e501e988bee53970ac0d718fc
|
[
"MIT"
] | null | null | null |
python/udemy-data-analysis-and-visualization/lecture32_data_combine_dataframes.ipynb
|
juancarlosqr/datascience
|
2e4d78365b059a3e501e988bee53970ac0d718fc
|
[
"MIT"
] | null | null | null | 21.097257 | 66 | 0.341253 |
[
[
[
"import numpy as np\nimport pandas as pd\nfrom pandas import Series,DataFrame",
"_____no_output_____"
],
[
"ser1 = Series([2,np.nan,4,np.nan,6,np.nan],\n index=['Q','R','S','T','U','V'])\n\nser1",
"_____no_output_____"
],
[
"ser2 = Series(np.arange(len(ser1)),\n index=['Q','R','S','T','U','V'],\n dtype=np.float64)\nser2",
"_____no_output_____"
],
[
"Series(np.where(pd.isnull(ser1),ser2,ser1),index=ser1.index)",
"_____no_output_____"
],
[
"# shorcut to previous command\nser1.combine_first(ser2)",
"_____no_output_____"
],
[
"# dataframes\nnan = np.nan\ndf_odds = DataFrame({'X':[1.,nan,3.,nan],\n 'Y':[nan,5.,nan,7.],\n 'Z':[nan,9.,nan,11.]})",
"_____no_output_____"
],
[
"df_evens = DataFrame({'X':[2.,4.,nan,6.,8.],\n 'Y':[nan,10.,12.,14.,16.]})",
"_____no_output_____"
],
[
"df_odds",
"_____no_output_____"
],
[
"df_evens",
"_____no_output_____"
],
[
"df_odds.combine_first(df_evens)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8ea993db06e6d8f5ea94e3f498a623b00f461a
| 645 |
ipynb
|
Jupyter Notebook
|
notebooks/moment_curvature.ipynb
|
bmcs-group/bmcs_tutorial
|
4e008e72839fad8820a6b663a20d3f188610525d
|
[
"MIT"
] | null | null | null |
notebooks/moment_curvature.ipynb
|
bmcs-group/bmcs_tutorial
|
4e008e72839fad8820a6b663a20d3f188610525d
|
[
"MIT"
] | null | null | null |
notebooks/moment_curvature.ipynb
|
bmcs-group/bmcs_tutorial
|
4e008e72839fad8820a6b663a20d3f188610525d
|
[
"MIT"
] | null | null | null | 16.973684 | 42 | 0.525581 |
[] |
[] |
[] |
4a8eade7a8d064192cbc97826cec5dd9b34eed7d
| 15,914 |
ipynb
|
Jupyter Notebook
|
PythonCodes/Untitled.ipynb
|
Nicolucas/C-Scripts
|
2608df5c2e635ad16f422877ff440af69f98f960
|
[
"MIT"
] | null | null | null |
PythonCodes/Untitled.ipynb
|
Nicolucas/C-Scripts
|
2608df5c2e635ad16f422877ff440af69f98f960
|
[
"MIT"
] | null | null | null |
PythonCodes/Untitled.ipynb
|
Nicolucas/C-Scripts
|
2608df5c2e635ad16f422877ff440af69f98f960
|
[
"MIT"
] | null | null | null | 133.731092 | 13,676 | 0.902853 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"x = np.linspace(-10,10,100)",
"_____no_output_____"
],
[
"y = np.exp((x*x)/(x*x-10.1*10.1)) ",
"_____no_output_____"
],
[
"plt.xlim(-12,12)\nplt.plot(x,y)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a8eb696836ccc31902e30df0fe1b9cf16706789
| 18,352 |
ipynb
|
Jupyter Notebook
|
uhecr_model/notebooks/fit_to_simulation/run_simulation.ipynb
|
uhecr-project/uhecr_model
|
8a2e8ab6f11cd2700f6455dff54c746cf3ffb143
|
[
"MIT"
] | null | null | null |
uhecr_model/notebooks/fit_to_simulation/run_simulation.ipynb
|
uhecr-project/uhecr_model
|
8a2e8ab6f11cd2700f6455dff54c746cf3ffb143
|
[
"MIT"
] | 2 |
2021-07-12T08:01:11.000Z
|
2021-08-04T03:01:30.000Z
|
uhecr_model/notebooks/fit_to_simulation/run_simulation.ipynb
|
uhecr-project/uhecr_model
|
8a2e8ab6f11cd2700f6455dff54c746cf3ffb143
|
[
"MIT"
] | null | null | null | 33.007194 | 255 | 0.536781 |
[
[
[
"# Comparison of arrival direction and joint models\n\nIn order to verify the model is working, we fit simulations made under the assumptions of the model. We also compare the differences between a model for only the UHECR arrival directions and one for both the UHECR arrival directions and energies.\n<br>\n<br>\n*This code is used to produce the data shown in Figures 6, 7 and 8 (left panel) in Capel & Mortlock (2019).* \n*See the separate notebook in this directory for the actual plotting of figures.*",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport os\nimport h5py\nfrom matplotlib import pyplot as plt\nfrom pandas import DataFrame\n\nfrom fancy import Data, Model, Analysis\nfrom fancy.interfaces.stan import get_simulation_input",
"_____no_output_____"
],
[
"'''Setting up'''\n\n# Define location of Stan files\nstan_path = '../../stan/'\n\n# Define file containing source catalogue information\nsource_file = '../../data/sourcedata.h5'\n\n# make output directory if it doesnt exist\nif not os.path.isdir(\"output\"):\n os.mkdir(\"output\")\n\n# source_types = [\"SBG_23\", \"2FHL_250Mpc\", \"swift_BAT_213\"]\nsource_types = [\"SBG_23\"]\n\n# detector_types = [\"auger2010\", \"auger2014\", \"TA2015\"]\n# detector_type = \"auger2014\"\ndetector_type = \"TA2015\"\n\n# set random seed\n# random_seed = 19990308\nrandom_seeds = [980, 546, 7984, 333, 2]\n\n# flag to control showing plots or not\nshow_plot = True",
"_____no_output_____"
],
[
"'''set detector and detector properties'''\nif detector_type == \"TA2015\":\n from fancy.detector.TA2015 import detector_properties, alpha_T, M, Eth\nelif detector_type == \"auger2014\":\n from fancy.detector.auger2014 import detector_properties, alpha_T, M, Eth\nelif detector_type == \"auger2010\":\n from fancy.detector.auger2010 import detector_properties, alpha_T, M, Eth\nelse:\n raise Exception(\"Undefined detector type!\")\n",
"_____no_output_____"
],
[
"'''Create joint simulated dataset'''\n\n# Define a Stan simulation to run\nsim_name = stan_path + 'joint_model_sim.stan' # simulate all processes\n\n# Define simulation using Model object and compile Stan code if necessary\nsimulation = Model(sim_filename = sim_name, include_paths = stan_path)\nsimulation.compile(reset=False)\n\nfor random_seed in random_seeds:\n for source_type in source_types:\n print(\"Current Source: {0}\".format(source_type))\n # define separate files\n table_file = '../tables/tables_{0}_{1}.h5'.format(source_type, detector_type)\n sim_output_file = 'output/joint_model_simulation_{0}_{1}_{2}.h5'.format(source_type, detector_type, random_seed)\n\n # Define a source catalogue and detector exposure\n # In the paper we use the SBG catalogue\n data = Data()\n data.add_source(source_file, source_type)\n data.add_detector(detector_properties)\n\n # Plot the sources in Galactic coordinates\n # if show_plot:\n # data.show();\n\n # Define associated fraction\n f = 0.5 \n\n # Simulation input\n B = 20 # nG\n alpha = 3.0\n Eth = Eth \n Eth_sim = 20 # EeV\n ptype = \"p\" # assume proton\n\n # number of simulated inputs\n # changes the background flux linearly\n # should choose Nsim such that FT is the same for\n # each observatory\n # this ensures that L, F0 are the same\n # \n # for PAO, we saw that FT, detector_type = 0.3601\n # FT_PAO = 0.3601 # total, detector_type flux using {1} data with Nsim = 2500, detector_type\n # Nsim_expected = FT_PAO / (M / alpha_T)\n # Nsim = int(np.round(Nsim_expected))\n Nsim = 200\n\n # check value for Nsim\n print(\"Simulated events: {0}\".format(Nsim))\n\n\n # L in yr^-1, F in km^-2 yr^-1\n L, F0 = get_simulation_input(Nsim, f, data.source.distance, M, alpha_T)\n\n # To scale between definition of flux in simulations and fits\n flux_scale = (Eth / Eth_sim)**(1 - alpha)\n\n simulation.input(B = B, L = L, F0 = F0,\n alpha = alpha, Eth = Eth, ptype=ptype)\n\n # check luminosity and isotropic flux values\n # L ~ O(10^39), F0 ~ 0.18\n # same luminosity so only need to check one value\n print(\"Simulated Luminosity: {0:.3e}\".format(L[0]))\n print(\"Simulated isotropic flux: {0:.3f}\".format(F0))\n\n\n # What is happening \n summary = b'Simulation using the joint model and SBG catalogue' # must be a byte str\n \n # Define an Analysis object to bring together Data and Model objects\n sim_analysis = Analysis(data, simulation, analysis_type = 'joint', \n filename = sim_output_file, summary = summary)\n\n print(\"Building tables...\")\n\n # Build pre-computed values for the simulation as you go\n # So that you can try out different parameters\n sim_analysis.build_tables(sim_only = True)\n\n print(\"Running simulation...\")\n # Run simulation\n sim_analysis.simulate(seed = random_seed, Eth_sim = Eth_sim)\n\n # Save to file \n sim_analysis.save()\n\n # print resulting UHECR observed after propagation and Elosses\n print(\"Observed simulated UHECRs: {0}\\n\".format(len(sim_analysis.source_labels)))\n\n\n # print plots if flag is set to true\n # if show_plot:\n # sim_analysis.plot(\"arrival_direction\");\n # sim_analysis.plot(\"energy\");",
"Using cached StanModel\nCurrent Source: SBG_23\nSimulated events: 200\nSimulated Luminosity: 1.486e+39\nSimulated isotropic flux: 0.080\nBuilding tables...\n"
],
[
"'''Fit using arrival direction model'''\nfor random_seed in random_seeds:\n for source_type in source_types:\n print(\"Current Source: {0}\".format(source_type))\n # define separate files\n table_file = '../../tables/tables_{0}_{1}.h5'.format(source_type, detector_type)\n sim_output_file = 'output/joint_model_simulation_{0}_{1}_{2}.h5'.format(source_type, detector_type, random_seed)\n arrival_output_file = 'output/arrival_direction_fit_{0}_{1}_{2}.h5'.format(source_type, detector_type, random_seed)\n # joint_output_file = 'output/joint_fit_{0}_PAO.h5'.format(source_type)\n\n # Define data from simulation\n data = Data()\n data.from_file(sim_output_file)\n\n # if show_plot:\n # data.show()\n\n # Arrival direction model\n model_name = stan_path + 'arrival_direction_model.stan'\n\n # Compile\n model = Model(model_filename = model_name, include_paths = stan_path)\n model.compile(reset=False)\n\n # Define threshold energy in EeV\n model.input(Eth = Eth)\n\n # What is happening \n summary = b'Fit of the arrival direction model to the joint simulation' \n \n # Define an Analysis object to bring together Data and Model objects\n analysis = Analysis(data, model, analysis_type = 'joint', \n filename = arrival_output_file, summary = summary)\n\n # Define location of pre-computed values used in fits \n # (see relevant notebook for how to make these files) \n # Each catalogue has a file of pre-computed values\n analysis.use_tables(table_file)\n\n # Fit the Stan model\n fit = analysis.fit_model(chains = 16, iterations = 500, seed = random_seed)\n\n # Save to analysis file\n analysis.save()\n",
"Current Source: SBG_23\nUsing cached StanModel\nPerforming fitting...\nChecking all diagnostics...\nDone!\nCurrent Source: SBG_23\nUsing cached StanModel\nPerforming fitting...\nChecking all diagnostics...\nDone!\nCurrent Source: SBG_23\nUsing cached StanModel\nPerforming fitting...\nChecking all diagnostics...\nDone!\nCurrent Source: SBG_23\nUsing cached StanModel\nPerforming fitting...\nChecking all diagnostics...\nDone!\nCurrent Source: SBG_23\nUsing cached StanModel\nPerforming fitting...\n"
],
[
"'''Fit using joint model'''\nfor random_seed in random_seeds:\n for source_type in source_types:\n print(\"Current Source: {0}\".format(source_type))\n # define separate files\n table_file = '../../tables/tables_{0}_{1}.h5'.format(source_type, detector_type)\n sim_output_file = 'output/joint_model_simulation_{0}_{1}_{2}.h5'.format(source_type, detector_type, random_seed)\n # arrival_output_file = 'output/arrival_direction_fit_{0}_{1}.h5'.format(source_type, detector_type)\n joint_output_file = 'output/joint_fit_{0}_{1}_{2}.h5'.format(source_type, detector_type, random_seed)\n\n # Define data from simulation\n data = Data()\n data.from_file(sim_output_file)\n\n # create Model and compile \n model_name = stan_path + 'joint_model.stan'\n model = Model(model_filename = model_name, include_paths = stan_path)\n model.compile(reset=False)\n model.input(Eth = Eth)\n\n # create Analysis object\n summary = b'Fit of the joint model to the joint simulation' \n analysis = Analysis(data, model, analysis_type = 'joint', \n filename = joint_output_file, summary = summary)\n analysis.use_tables(table_file)\n\n # Fit the Stan model\n fit = analysis.fit_model(chains = 16, iterations = 500, seed = random_seed)\n\n # Save to analysis file\n analysis.save()\n",
"Current Source: SBG_23\nUsing cached StanModel\nPerforming fitting...\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8eb8076a91e977eb5c4b6d5b4d3c440a60e4f6
| 460,381 |
ipynb
|
Jupyter Notebook
|
Topics_Master/13-Logistic-Regression/03-Logistic Regression Project - Solutions.ipynb
|
meyash/ml_master
|
871ea710f06d4e0052f8c61aa1fbf6c1e39257d4
|
[
"MIT"
] | null | null | null |
Topics_Master/13-Logistic-Regression/03-Logistic Regression Project - Solutions.ipynb
|
meyash/ml_master
|
871ea710f06d4e0052f8c61aa1fbf6c1e39257d4
|
[
"MIT"
] | null | null | null |
Topics_Master/13-Logistic-Regression/03-Logistic Regression Project - Solutions.ipynb
|
meyash/ml_master
|
871ea710f06d4e0052f8c61aa1fbf6c1e39257d4
|
[
"MIT"
] | null | null | null | 630.658904 | 316,748 | 0.932467 |
[
[
[
"___\n\n___\n# Logistic Regression Project - Solutions\n\nIn this project we will be working with a fake advertising data set, indicating whether or not a particular internet user clicked on an Advertisement on a company website. We will try to create a model that will predict whether or not they will click on an ad based off the features of that user.\n\nThis data set contains the following features:\n\n* 'Daily Time Spent on Site': consumer time on site in minutes\n* 'Age': cutomer age in years\n* 'Area Income': Avg. Income of geographical area of consumer\n* 'Daily Internet Usage': Avg. minutes a day consumer is on the internet\n* 'Ad Topic Line': Headline of the advertisement\n* 'City': City of consumer\n* 'Male': Whether or not consumer was male\n* 'Country': Country of consumer\n* 'Timestamp': Time at which consumer clicked on Ad or closed window\n* 'Clicked on Ad': 0 or 1 indicated clicking on Ad\n\n## Import Libraries\n\n**Import a few libraries you think you'll need (Or just import them as you go along!)**",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Get the Data\n**Read in the advertising.csv file and set it to a data frame called ad_data.**",
"_____no_output_____"
]
],
[
[
"ad_data = pd.read_csv('advertising.csv')",
"_____no_output_____"
]
],
[
[
"**Check the head of ad_data**",
"_____no_output_____"
]
],
[
[
"ad_data.head()",
"_____no_output_____"
]
],
[
[
"** Use info and describe() on ad_data**",
"_____no_output_____"
]
],
[
[
"ad_data.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1000 entries, 0 to 999\nData columns (total 10 columns):\nDaily Time Spent on Site 1000 non-null float64\nAge 1000 non-null int64\nArea Income 1000 non-null float64\nDaily Internet Usage 1000 non-null float64\nAd Topic Line 1000 non-null object\nCity 1000 non-null object\nMale 1000 non-null int64\nCountry 1000 non-null object\nTimestamp 1000 non-null object\nClicked on Ad 1000 non-null int64\ndtypes: float64(3), int64(3), object(4)\nmemory usage: 78.2+ KB\n"
],
[
"ad_data.describe()",
"_____no_output_____"
]
],
[
[
"## Exploratory Data Analysis\n\nLet's use seaborn to explore the data!\n\nTry recreating the plots shown below!\n\n** Create a histogram of the Age**",
"_____no_output_____"
]
],
[
[
"sns.set_style('whitegrid')\nad_data['Age'].hist(bins=30)\nplt.xlabel('Age')",
"_____no_output_____"
]
],
[
[
"**Create a jointplot showing Area Income versus Age.**",
"_____no_output_____"
]
],
[
[
"sns.jointplot(x='Age',y='Area Income',data=ad_data)",
"_____no_output_____"
]
],
[
[
"**Create a jointplot showing the kde distributions of Daily Time spent on site vs. Age.**",
"_____no_output_____"
]
],
[
[
"sns.jointplot(x='Age',y='Daily Time Spent on Site',data=ad_data,color='red',kind='kde');",
"_____no_output_____"
]
],
[
[
"** Create a jointplot of 'Daily Time Spent on Site' vs. 'Daily Internet Usage'**",
"_____no_output_____"
]
],
[
[
"sns.jointplot(x='Daily Time Spent on Site',y='Daily Internet Usage',data=ad_data,color='green')",
"_____no_output_____"
]
],
[
[
"** Finally, create a pairplot with the hue defined by the 'Clicked on Ad' column feature.**",
"_____no_output_____"
]
],
[
[
"sns.pairplot(ad_data,hue='Clicked on Ad',palette='bwr')",
"_____no_output_____"
]
],
[
[
"# Logistic Regression\n\nNow it's time to do a train test split, and train our model!\n\nYou'll have the freedom here to choose columns that you want to train on!",
"_____no_output_____"
],
[
"** Split the data into training set and testing set using train_test_split**",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X = ad_data[['Daily Time Spent on Site', 'Age', 'Area Income','Daily Internet Usage', 'Male']]\ny = ad_data['Clicked on Ad']",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=42)",
"_____no_output_____"
]
],
[
[
"** Train and fit a logistic regression model on the training set.**",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"logmodel = LogisticRegression()\nlogmodel.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"## Predictions and Evaluations\n** Now predict values for the testing data.**",
"_____no_output_____"
]
],
[
[
"predictions = logmodel.predict(X_test)",
"_____no_output_____"
]
],
[
[
"** Create a classification report for the model.**",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report",
"_____no_output_____"
],
[
"print(classification_report(y_test,predictions))",
" precision recall f1-score support\n\n 0 0.87 0.96 0.91 162\n 1 0.96 0.86 0.91 168\n\navg / total 0.91 0.91 0.91 330\n\n"
]
],
[
[
"## Great Job!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a8ec192416dab28b1871856939e6bb3894abcf9
| 54,607 |
ipynb
|
Jupyter Notebook
|
Lesson06/Activity01.ipynb
|
bsitati/Big-Data-Analysis-with-Python
|
8d1fd97602c9033d1bdee833945c76beb5f8300c
|
[
"MIT"
] | 17 |
2019-06-08T14:46:11.000Z
|
2022-02-28T09:48:30.000Z
|
Lesson06/Activity01.ipynb
|
bsitati/Big-Data-Analysis-with-Python
|
8d1fd97602c9033d1bdee833945c76beb5f8300c
|
[
"MIT"
] | null | null | null |
Lesson06/Activity01.ipynb
|
bsitati/Big-Data-Analysis-with-Python
|
8d1fd97602c9033d1bdee833945c76beb5f8300c
|
[
"MIT"
] | 39 |
2018-11-16T10:50:28.000Z
|
2022-03-08T05:54:34.000Z
| 161.082596 | 25,054 | 0.844416 |
[
[
[
"## Activity 1: Carry Out Mapping to Gaussian Distribution of Numeric Features from the Given Data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport seaborn as sns\nimport time\nimport re\nimport os\nimport matplotlib.pyplot as plt\nsns.set(style=\"ticks\")\n\nimport sklearn as sk\nfrom scipy import stats\nfrom sklearn import preprocessing\n\n\n# read the downloaded input data (marketing data)\ndf = pd.read_csv('https://raw.githubusercontent.com/TrainingByPackt/Big-Data-Analysis-with-Python/master/Lesson07/Dataset/bank.csv', sep=';')\n",
"_____no_output_____"
],
[
"numeric_df = df._get_numeric_data()\nnumeric_df.head()\n",
"_____no_output_____"
],
[
"numeric_df_array = np.array(numeric_df) # converting to numpy arrays for more efficient computation\n\nloop_c = -1\ncol_for_normalization = list()\n\nfor column in numeric_df_array.T:\n loop_c+=1\n x = column\n k2, p = stats.normaltest(x) \n alpha = 0.001\n print(\"p = {:g}\".format(p))\n \n # rules for printing the normality output\n if p < alpha:\n test_result = \"non_normal_distr\"\n col_for_normalization.append((loop_c)) # applicable if yeo-johnson is used\n \n #if min(x) > 0: # applicable if box-cox is used\n #col_for_normalization.append((loop_c)) # applicable if box-cox is used\n print(\"The null hypothesis can be rejected: non-normal distribution\")\n \n else:\n test_result = \"normal_distr\"\n print(\"The null hypothesis cannot be rejected: normal distribution\")\n",
"p = 1.98749e-70\nThe null hypothesis can be rejected: non-normal distribution\np = 0\nThe null hypothesis can be rejected: non-normal distribution\np = 3.08647e-278\nThe null hypothesis can be rejected: non-normal distribution\np = 0\nThe null hypothesis can be rejected: non-normal distribution\np = 0\nThe null hypothesis can be rejected: non-normal distribution\np = 0\nThe null hypothesis can be rejected: non-normal distribution\np = 0\nThe null hypothesis can be rejected: non-normal distribution\n"
],
[
"columns_to_normalize = numeric_df[numeric_df.columns[col_for_normalization]]\nnames_col = list(columns_to_normalize)\n\n# density plots of the features to check the normality\ncolumns_to_normalize.plot.kde(bw_method=3)\n",
"_____no_output_____"
],
[
"pt = preprocessing.PowerTransformer(method='yeo-johnson', standardize=True, copy=True)\nnormalized_columns = pt.fit_transform(columns_to_normalize)\nnormalized_columns = pd.DataFrame(normalized_columns, columns=names_col)\n",
"_____no_output_____"
],
[
"normalized_columns.plot.kde(bw_method=3)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8ec5b7a40bd7ffb6eee6e5878f1889cd3bafaa
| 12,170 |
ipynb
|
Jupyter Notebook
|
docs/_downloads/80c4696c772e675d3973ed5c145a67b0/Captum_Recipe.ipynb
|
YonghyunRyu/PyTorch-tutorials-kr-exercise
|
bb527494f304f76a4d2cf5a689f00039336fc0c1
|
[
"BSD-3-Clause"
] | 221 |
2018-04-06T01:42:58.000Z
|
2021-11-28T10:12:45.000Z
|
docs/_downloads/80c4696c772e675d3973ed5c145a67b0/Captum_Recipe.ipynb
|
YonghyunRyu/PyTorch-tutorials-kr-exercise
|
bb527494f304f76a4d2cf5a689f00039336fc0c1
|
[
"BSD-3-Clause"
] | 280 |
2018-05-25T08:53:21.000Z
|
2021-12-02T05:37:25.000Z
|
docs/_downloads/80c4696c772e675d3973ed5c145a67b0/Captum_Recipe.ipynb
|
YonghyunRyu/PyTorch-tutorials-kr-exercise
|
bb527494f304f76a4d2cf5a689f00039336fc0c1
|
[
"BSD-3-Clause"
] | 181 |
2018-05-25T02:00:28.000Z
|
2021-11-19T11:56:39.000Z
| 76.0625 | 1,825 | 0.617831 |
[
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\nCaptum을 사용하여 모델 해석하기\n===================================\n\n**번역**: `정재민 <https://github.com/jjeamin>`_\n",
"_____no_output_____"
],
[
"Captum을 사용하면 데이터 특징(features)이 모델의 예측 또는 뉴런 활성화에\n미치는 영향을 이해하고, 모델의 동작 방식을 알 수 있습니다.\n\n그리고 \\ ``Integrated Gradients``\\ 와 \\ ``Guided GradCam``\\ 과 같은\n최첨단의 feature attribution 알고리즘을 적용할 수 있습니다.\n\n이 레시피에서는 Captum을 사용하여 다음을 수행하는 방법을 배웁니다.\n\\* 이미지 분류기(classifier)의 예측을 해당 이미지의 특징(features)에 표시하기\n\\* 속성(attribution) 결과를 시각화 하기\n\n",
"_____no_output_____"
],
[
"시작하기 전에\n----------------\n\n\n",
"_____no_output_____"
],
[
"Captum이 Python 환경에 설치되어 있는지 확인해야 합니다.\nCaptum은 Github에서 ``pip`` 패키지 또는 ``conda`` 패키지로 제공됩니다.\n자세한 지침은 https://captum.ai/ 의 설치 안내서를 참조하면 됩니다.\n\n",
"_____no_output_____"
],
[
"모델의 경우, PyTorch에 내장 된 이미지 분류기(classifier)를 사용합니다.\nCaptum은 샘플 이미지의 어떤 부분이 모델에 의해 만들어진\n특정한 예측에 도움을 주는지 보여줍니다.\n\n",
"_____no_output_____"
]
],
[
[
"import torchvision\nfrom torchvision import transforms\nfrom PIL import Image\nimport requests\nfrom io import BytesIO\n\nmodel = torchvision.models.resnet18(pretrained=True).eval()\n\nresponse = requests.get(\"https://image.freepik.com/free-photo/two-beautiful-puppies-cat-dog_58409-6024.jpg\")\nimg = Image.open(BytesIO(response.content))\n\ncenter_crop = transforms.Compose([\n transforms.Resize(256),\n transforms.CenterCrop(224),\n])\n\nnormalize = transforms.Compose([\n transforms.ToTensor(), # 이미지를 0에서 1사이의 값을 가진 Tensor로 변환\n transforms.Normalize( # 0을 중심으로 하는 imagenet 픽셀의 rgb 분포를 따르는 정규화\n mean=[0.485, 0.456, 0.406],\n std=[0.229, 0.224, 0.225]\n )\n])\ninput_img = normalize(center_crop(img)).unsqueeze(0)",
"_____no_output_____"
]
],
[
[
"속성(attribution) 계산하기\n---------------------\n\n",
"_____no_output_____"
],
[
"모델의 top-3 예측 중에는 개와 고양이에 해당하는 클래스 208과 283이 있습니다.\n\nCaptum의 \\ ``Occlusion``\\ 알고리즘을 사용하여 각 예측을 입력의 해당 부분에 표시합니다.\n\n",
"_____no_output_____"
]
],
[
[
"from captum.attr import Occlusion\n\nocclusion = Occlusion(model)\n\nstrides = (3, 9, 9) # 작을수록 = 세부적인 속성이지만 느림\ntarget=208, # ImageNet에서 Labrador의 인덱스\nsliding_window_shapes=(3,45, 45) # 객체의 모양을 변화시키기에 충분한 크기를 선택\nbaselines = 0 # 이미지를 가릴 값, 0은 회색\n\nattribution_dog = occlusion.attribute(input_img,\n strides = strides,\n target=target,\n sliding_window_shapes=sliding_window_shapes,\n baselines=baselines)\n\n\ntarget=283, # ImageNet에서 Persian cat의 인덱스\nattribution_cat = occlusion.attribute(input_img,\n strides = strides,\n target=target,\n sliding_window_shapes=sliding_window_shapes,\n baselines=0)",
"_____no_output_____"
]
],
[
[
"Captum은 ``Occlusion`` 외에도 \\ ``Integrated Gradients``\\ , \\ ``Deconvolution``\\ ,\n\\ ``GuidedBackprop``\\ , \\ ``Guided GradCam``\\ , \\ ``DeepLift``\\ ,\n그리고 \\ ``GradientShap``\\과 같은 많은 알고리즘을 제공합니다.\n이러한 모든 알고리즘은 초기화할 때 모델을 호출 가능한 \\ ``forward_func``\\ 으로 기대하며\n속성(attribution) 결과를 통합해서 반환하는 ``attribute(...)`` 메소드를 가지는\n``Attribution`` 의 서브클래스 입니다.\n\n이미지인 경우 속성(attribution) 결과를 시각화 해보겠습니다.\n\n\n",
"_____no_output_____"
],
[
"결과 시각화하기\n-----------------------\n\n\n",
"_____no_output_____"
],
[
"Captum의 \\ ``visualization``\\ 유틸리티는 그림과 텍스트 입력 모두에 대한\n속성(attribution) 결과를 시각화 할 수 있는 즉시 사용가능한 방법을 제공합니다.\n\n\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom captum.attr import visualization as viz\n\n# 계산 속성 Tensor를 이미지 같은 numpy 배열로 변환합니다.\nattribution_dog = np.transpose(attribution_dog.squeeze().cpu().detach().numpy(), (1,2,0))\n\nvis_types = [\"heat_map\", \"original_image\"]\nvis_signs = [\"all\", \"all\"] # \"positive\", \"negative\", 또는 모두 표시하는 \"all\"\n# positive 속성은 해당 영역의 존재가 예측 점수를 증가시킨다는 것을 의미합니다.\n# negative 속성은 해당 영역의 존재가 예측 점수를 낮추는 오답 영역을 의미합니다.\n\n_ = viz.visualize_image_attr_multiple(attribution_dog,\n center_crop(img),\n vis_types,\n vis_signs,\n [\"attribution for dog\", \"image\"],\n show_colorbar = True\n )\n\n\nattribution_cat = np.transpose(attribution_cat.squeeze().cpu().detach().numpy(), (1,2,0))\n\n_ = viz.visualize_image_attr_multiple(attribution_cat,\n center_crop(img),\n [\"heat_map\", \"original_image\"],\n [\"all\", \"all\"], # positive/negative 속성 또는 all\n [\"attribution for cat\", \"image\"],\n show_colorbar = True\n )",
"_____no_output_____"
]
],
[
[
"만약 데이터가 텍스트인 경우 ``visualization.visualize_text()`` 는\n입력 텍스트 위에 속성(attribution)을 탐색할 수 있는 전용 뷰(view)를 제공합니다.\nhttp://captum.ai/tutorials/IMDB_TorchText_Interpret 에서 자세한 내용을 확인하세요.\n\n\n",
"_____no_output_____"
],
[
"마지막 노트\n-----------\n\n\n",
"_____no_output_____"
],
[
"Captum은 이미지, 텍스트 등을 포함하여 다양한 방식으로 PyTorch에서 대부분의 모델 타입을 처리할 수 있습니다.\nCaptum을 사용하면 다음을 수행할 수 있습니다.\n\\* 위에서 설명한 것처럼 특정한 출력을 모델 입력에 표시하기\n\\* 특정한 출력을 은닉층의 뉴런에 표시하기 (Captum API reference를 보세요).\n\\* 모델 입력에 대한 은닉층 뉴런의 반응을 표시하기 (Captum API reference를 보세요).\n\n지원되는 메소드의 전체 API와 튜토리얼의 목록은 http://captum.ai 를 참조하세요.\n\nGilbert Tanner의 또 다른 유용한 게시물 :\nhttps://gilberttanner.com/blog/interpreting-pytorch-models-with-captum\n\n\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4a8ecd8412d3d7bcd47d1b3d3c4083396b9576ae
| 565,878 |
ipynb
|
Jupyter Notebook
|
notebooks/SDn.ipynb
|
ScottHaselschwardt/NeutrinoFog
|
6fa09a9ac65f96862b2b764b89b1c11971426291
|
[
"MIT"
] | 8 |
2021-09-07T13:16:07.000Z
|
2021-12-20T13:53:06.000Z
|
notebooks/SDn.ipynb
|
ScottHaselschwardt/NeutrinoFog
|
6fa09a9ac65f96862b2b764b89b1c11971426291
|
[
"MIT"
] | null | null | null |
notebooks/SDn.ipynb
|
ScottHaselschwardt/NeutrinoFog
|
6fa09a9ac65f96862b2b764b89b1c11971426291
|
[
"MIT"
] | 5 |
2021-09-08T10:06:09.000Z
|
2022-02-23T23:17:07.000Z
| 2,272.60241 | 233,296 | 0.961534 |
[
[
[
"import sys\nsys.path.append('../src')\nfrom numpy import *\nimport matplotlib.pyplot as plt\nfrom Like import *\nfrom PlotFuncs import *\nimport WIMPFuncs\n\npek = line_background(6,'k')\n\nfig,ax = MakeLimitPlot_SDn()\n\nalph = 0.25\ncols = cm.bone(linspace(0.3,0.7,4))\nnucs = ['Xe','Ge','NaI']\nzos = [0,-50,-100,-50]\nC_Si = WIMPFuncs.C_SDp(Si29)/WIMPFuncs.C_SDn(Si29)\nC_Ge = WIMPFuncs.C_SDp(Ge73)/WIMPFuncs.C_SDn(Ge73)\nCs = [1.0,C_Ge,1.0]\nfroots = ['SDn','SDp','SDn']\n\nfor nuc,zo,col,C,froot in zip(nucs,zos,cols,Cs,froots):\n data = loadtxt('../data/WIMPLimits/mylimits/DLNuFloor'+nuc+'_detailed_'+froot+'.txt')\n m,sig,NUFLOOR,DY = Floor_2D(data)\n plt.plot(m,NUFLOOR*C,'-',color=col,lw=3,path_effects=pek,zorder=zo)\n plt.fill_between(m,NUFLOOR*C,y2=1e-99,color=col,zorder=zo,alpha=alph)\n\n#plt.text(0.12,0.2e-35,r'{\\bf Silicon}',rotation=45,color='k')\nplt.text(0.23,1.5e-38,r'{\\bf Ge}',rotation=25,color='k')\nplt.text(0.18,5e-38,r'{\\bf NaI}',rotation=26,color='k')\nplt.text(0.175,5e-40,r'{\\bf Xenon}',rotation=31,color='k')\n\nMySaveFig(fig,'NuFloor_Targets_SDn')",
"../src/Like.py:33: RuntimeWarning: invalid value encountered in greater\n Exmin = amin(Ex[Ex>0])\n../src/Like.py:58: RuntimeWarning: invalid value encountered in less\n DY[DY<2] = 2\n../src/Like.py:33: RuntimeWarning: invalid value encountered in greater\n Exmin = amin(Ex[Ex>0])\n../src/Like.py:58: RuntimeWarning: invalid value encountered in less\n DY[DY<2] = 2\n../src/Like.py:33: RuntimeWarning: invalid value encountered in greater\n Exmin = amin(Ex[Ex>0])\n../src/Like.py:58: RuntimeWarning: invalid value encountered in less\n DY[DY<2] = 2\n"
],
[
"pek = line_background(6,'k')\ncmap = cm.terrain_r\n\nfig,ax = MakeLimitPlot_SDn(Collected=True,alph=1,edgecolor=col_alpha('gray',0.75),facecolor=col_alpha('gray',0.5))\n\ndata = loadtxt('../data/WIMPLimits/mylimits/DLNuFloorXe_detailed_SDn.txt')\nm,sig,NUFLOOR,DY = Floor_2D(data,filt=True,filt_width=2,Ex_crit=1e10)\ncnt = plt.contourf(m,sig,DY,levels=linspace(2,15,100),vmax=8,vmin=2.2,cmap=cmap)\nfor c in cnt.collections:\n c.set_edgecolor(\"face\")\nplt.plot(m,NUFLOOR,'-',color='brown',lw=3,path_effects=pek,zorder=100)\n\nim = plt.pcolormesh(-m,sig,DY,vmax=6,vmin=2.2,cmap=cmap,rasterized=True)\ncbar(im,extend='min')\nplt.gcf().text(0.82,0.9,r'$\\left(\\frac{{\\rm d}\\ln\\sigma}{{\\rm d}\\ln N}\\right)^{-1}$',fontsize=35)\n\nplt.gcf().text(0.15*(1-0.01),0.16*(1+0.01),r'{\\bf Xenon}',color='k',fontsize=50,alpha=0.2)\nplt.gcf().text(0.15,0.16,r'{\\bf Xenon}',color='brown',fontsize=50)\n\nMySaveFig(fig,'NuFloorDetailed_Xe_SDn')",
"../src/Like.py:33: RuntimeWarning: invalid value encountered in greater\n Exmin = amin(Ex[Ex>0])\n../src/Like.py:58: RuntimeWarning: invalid value encountered in less\n DY[DY<2] = 2\n"
],
[
"fig,ax = MakeLimitPlot_SDn(Collected=True,alph=1,edgecolor=col_alpha('gray',0.75),facecolor=col_alpha('gray',0.5))\n\ndata = loadtxt('../data/WIMPLimits/mylimits/DLNuFloorNaI_detailed_SDn.txt')\nm,sig,NUFLOOR,DY = Floor_2D(data,filt=True,filt_width=2,Ex_crit=1e11)\ncnt = plt.contourf(m,sig,DY,levels=linspace(2,15,100),vmax=6,vmin=2.2,cmap=cmap)\nfor c in cnt.collections:\n c.set_edgecolor(\"face\")\nplt.plot(m,NUFLOOR,'-',color='brown',lw=3,path_effects=pek,zorder=100)\n\nim = plt.pcolormesh(-m,sig,DY,vmax=6,vmin=2.2,cmap=cmap,rasterized=True)\ncbar(im,extend='min')\nplt.gcf().text(0.82,0.9,r'$\\left(\\frac{{\\rm d}\\ln\\sigma}{{\\rm d}\\ln N}\\right)^{-1}$',fontsize=35)\n\nplt.gcf().text(0.15*(1-0.01),0.16*(1+0.01),r'{\\bf NaI}',color='k',fontsize=50,alpha=0.2)\nplt.gcf().text(0.15,0.16,r'{\\bf NaI}',color='brown',fontsize=50)\n\nMySaveFig(fig,'NuFloorDetailed_NaI_SDn')",
"../src/Like.py:33: RuntimeWarning: invalid value encountered in greater\n Exmin = amin(Ex[Ex>0])\n../src/Like.py:58: RuntimeWarning: invalid value encountered in less\n DY[DY<2] = 2\n"
],
[
"dat1 = loadtxt(\"../data/WIMPLimits/SDn/XENON1T.txt\")\ndat2 = loadtxt(\"../data/WIMPLimits/SDn/PandaX.txt\")\ndat3 = loadtxt(\"../data/WIMPLimits/SDn/CDMSlite.txt\")\ndat4 = loadtxt(\"../data/WIMPLimits/SDn/CRESST.txt\")\n\ndats = [dat1,dat2,dat3,dat4]\n\nmmin = amin(dat4[:,0])\nmmax = 1e4\nmvals = logspace(log10(mmin),log10(mmax),1000)\n\nsig = zeros(shape=1000)\nfor dat in dats:\n sig1 = 10**interp(log10(mvals),log10(dat[:,0]),log10(dat[:,1]))\n sig1[mvals<amin(dat[:,0])] = inf\n sig1[mvals>amax(dat[:,0])] = inf\n sig = column_stack((sig,sig1))\nsig = sig[:,1:]\n\nsig = amin(sig,1)\n\nplt.loglog(mvals,sig,color='r',alpha=1,zorder=0.5,lw=2)\n\nsavetxt('../data/WIMPLimits/SDn/AllLimits-2021.txt',column_stack((mvals,sig)))\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a8ecfa65cd1bf2bac7aa2de733f89a495ae0432
| 28,826 |
ipynb
|
Jupyter Notebook
|
doc/hu_spp.ipynb
|
kingablgh/PySprint
|
c3e76fbf1287d18d78699145f5301593aff47ba0
|
[
"MIT"
] | null | null | null |
doc/hu_spp.ipynb
|
kingablgh/PySprint
|
c3e76fbf1287d18d78699145f5301593aff47ba0
|
[
"MIT"
] | null | null | null |
doc/hu_spp.ipynb
|
kingablgh/PySprint
|
c3e76fbf1287d18d78699145f5301593aff47ba0
|
[
"MIT"
] | null | null | null | 31.746696 | 844 | 0.556199 |
[
[
[
"## 5. Állandó fázisú pont módszere, SPPMethod\n\nEz a módszer alapjaiban kissé különbözik a többitől. Az előzőleg leírt globális metódusok, mint domain átváltás, kivágás, stb. itt is működnek, de másképpen kell kezelni őket. *Megjegyezném, hogy mivel ez a módszer interaktív elemet tartalmaz egyelőre csak Jupyter Notebook-ban stabil.*",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pysprint as ps",
"_____no_output_____"
]
],
[
[
"Példaként a korábban már bemutatott `ps.Generator` segítségével generálni fogok egy sorozat interferogramot, majd azon bemutatom a kiértékelés menetét. Valós méréseknél teljesen hasonlóképpen végezhető a kiértékelés. A legegyszerűbb módszer, hogy különböző karok közti időbeli késleltetésnél generáljunk és elmentsük azokat az alábbi cellában látható. A megkülönböztethetőség miatt minden fájlt a hozzá tartozó karok közti időbeli késleltetésnek megfelelően nevezem el.",
"_____no_output_____"
]
],
[
[
"for delay in range(-200, 201, 50):\n g = ps.Generator(1, 3, 2, delay, GDD=400, TOD=-500, normalize=True)\n g.generate_freq()\n np.savetxt(f'{delay}.txt', np.transpose([g.x, g.y]), delimiter=',')",
"_____no_output_____"
]
],
[
[
"A kód lefuttatásával a munkafüzet környtárában megjelent 7 új txt fájl. \n\nEhhez a kiértékelési módszerhez először fel kell építeni egy listát a felhasználandó interferogramok fájlneveivel. Ezt manuálisan is megtehetjük, itt ezt elkerülve egy rövidítést fogok használni.",
"_____no_output_____"
]
],
[
[
"ifg_files = [f\"{delay}.txt\" for delay in range(-200, 201, 50)]",
"_____no_output_____"
],
[
"print(ifg_files)",
"['-200.txt', '-150.txt', '-100.txt', '-50.txt', '0.txt', '50.txt', '100.txt', '150.txt', '200.txt']\n"
]
],
[
[
"Ha nem hasonló sémára épülnek a felhasználandó fájlok nevei, akkor természetesen a fenti trükk nem működik és egyenként kell beírnunk őket. Miután definiáltuk a fájlneveket a következő lépés a\n```python\nps.SPPMethod(ifg_names, sam_names=None, ref_names=None, **kwargs)\n```\nmeghívása:",
"_____no_output_____"
]
],
[
[
"myspp = ps.SPPMethod(ifg_files, decimal=\".\", sep=\",\", skiprows=0, meta_len=0)",
"_____no_output_____"
]
],
[
[
"A `**kwargs` keyword argumentumok itt elfogadják a korábban már bemutatott `parse_raw` funkció argumentumait (a kódban belül azt is hívja meg egyesével minden interferogramon), hiszen a fájlok sémáját itt is fontos megadni a helyes betöltéshez. A tárgy- és referencianyaláb spektrumai természetesen opcionális argumentumok, mi dönthetjük el, hogy normáljuk-e az interferogramokat.\n\nAz `SPPMethod` objektum először ellenőrzi, hogy a listában lévő fájlnevek valóban léteznek-e, és ha nem, akkor hibával tér vissza. Az `SPPMethod`-nak vannak további metódusai, ilyen pl. a `len(..)`, vagy az `SPPMethod.info`. Az első visszaadja, hogy hány interferogram van jelenleg az objektumban (ez jelen esetben 9), a második pedig a kiértékelés során mutatja majd, hogy hány interferogramból rögzítettünk információt (ez jelenleg 0/9). Később talán `append` (ilyen már van a 0.12.5 verzióban), `insert` és `delete` metódusokat is beépítek.",
"_____no_output_____"
]
],
[
[
"print(len(myspp))",
"9\n"
],
[
"print(myspp.info)",
"Progress: 0/9\n"
]
],
[
[
"Az SPPMethod objektum listaszerűen viselkedik: lehet indexelni is. Mivel benne 9 darab interferogram van, ezért egy ilyen indexelés egy `ps.Dataset` objektumot ad vissza. Ez az alapja minden kiértékelési módszernek, így ez ismeri a korábban bemutatott metódusokat. Tegyük fel, hogy a 3. interferogram adatait ki szeretnénk iratni, és szeretnénk megkapni az y értékeit `np.ndarray`-ként. Ekkor a 2 indexet használva (mivel itt is 0-tól indul a számozás):",
"_____no_output_____"
]
],
[
[
"# a harmadik interferogram adatainak kiíratása\nprint(myspp[2])",
"Dataset\n----------\nParameters\n----------\nDatapoints: 12559\nPredicted domain: frequency\nRange: from 0.99998 to 3.00000 PHz\nNormalized: False\nDelay value: Not given\nSPP position(s): Not given\n----------------------------\nMetadata extracted from file\n----------------------------\n{}\n"
],
[
"# a harmadik interferogram y értékeinek kinyerése, mint np.array\ny_ertekek = myspp[2].data.y.values\nprint(y_ertekek)\nprint(type(y_ertekek))",
"[4.22567237e-08 4.23424876e-08 4.24102757e-08 ... 4.41394357e-08\n 4.30227521e-08 4.18888151e-08]\n<class 'numpy.ndarray'>\n"
]
],
[
[
"Újra hangsúlyozom, minden eddig bemutatott metódus ezeken a kvázi listaelemeken is működik, köztük a `chdomain`, vagy `slice` is. Ezt használjuk ki a kiértékeléshez egy *for* ciklusban. A kiértékeléshez a definiált `SPPMethod`-on meg kell hívni egy for ciklust. Ez végigfut a benne lévő összes interferogramon. Azt, hogy mit akarunk csinálni adott interferogrammal, azt a cikluson belül tudjuk megadni. Az alapvető séma a következő:\n<pre>\nfor ifg in myspp:\n - előfeldolgozása az adott interferogramnak\n - az interaktív SPP Panel megnyitása és adatok rögzítése\n \n- a calculate metódus meghívása a cikluson kívül(!)\n</pre>\n\nEz kód formájában az alábbi cellában látható. Itt külön jelöltem, hogy melyik rész meddig tart.",
"_____no_output_____"
]
],
[
[
"# az interaktív számításokat fontos a with blokkon belülre írni\n\nwith ps.interactive():\n\n for ifg in myspp:\n\n # -----------------------------------Előfeldolgozás-----------------------------------------\n # Ha valós mérésünk van, érdemes valamilyen módon kiíratni a kommentet,\n # ami az interferogram fájlban van, hogy meg tudjuk állapítani milyen késleltetésnél készült.\n # Jelen esetben ennek nincs értelme, mivel a szimulált fájlokkal dolgozom.\n # Ezt legegyszerűbben az alábbi sorral tehetnénk meg: \n # print(ifg.meta['comment'])\n # vagy esetleg a teljes metaadatok kiíratása:\n # print(ifg.meta)\n\n # Ha hullámhossztartományban vagyunk, először át kell váltani.\n # Én frekvenciatartományban szimuláltam, ezért itt kihagyom. Ha szükség van rá a\n # következő sort kell használni.\n # ifg.chdomain() \n\n # Pl. 1.2 PHz alatti körfrekvenciaértékek kivágása. Mivel nem adtam meg stop értéket, így a felső\n # határt érintetlenül hagyná, ha futtatnám. Nyilván ez is opcionális.\n # ifg.slice(start=1.2)\n\n # -----------------------------Az interaktív panel megnyitása-------------------------------\n ifg.open_SPP_panel()\n\n# ---------------------------------A ciklus utáni rész------------------------------------------\n# A cikluson kívül a save_data metódus meghívása, hogy elmentsük a beírt adatainkat fájlba is.\n# Ez természetesen opcionális, de annak érdekében, hogy ne veszítsünk adatot érdemes ezt is elvégezni.\nmyspp.save_data('spp.txt')\n\n\n# a cikluson kívül meghívjuk a calculate függvényt\nmyspp.calculate(reference_point=2, order=3);",
"_____no_output_____"
]
],
[
[
"A magyarázatok nélkül szimulált esetben az egész kód az alábbi, összesen 8 sorra egyszerűsödik. Valós mérés esetén néhány előfeldolgozási lépés és kiíratás természetesen még hozzáadódhat ehhez.\n\n```python\nimport pysprint as ps\n\nifg_files = [f\"{delay}.txt\" for delay in range(-200, 201, 50)]\n\ns = ps.SPPMethod(ifg_files, decimal=\".\", sep=\",\", skiprows=0, meta_len=0)\n\nwith ps.interactive():\n for ifg in s:\n ifg.open_SPP_panel()\n \ns.save_data('spp.txt')\ns.calculate(reference_point=2, order=2, show_graph=True)\n```\nMiután a számolást már elvégezte a program, akkor elérhetővé válik rajta a `GD` property. Ez az illesztett görbét reprezentálja, típusa `ps.core.phase.Phase`. Bővebben erről a `Phase` leírásában.",
"_____no_output_____"
]
],
[
[
"myspp.GD",
"_____no_output_____"
]
],
[
[
"#### 5.1 Számolás nyers adatokból\nMivel az `spp.txt` fájlba elmentettük az bevitt adatokat, azokból egyszerűen lehet újraszámolni az illesztést. Töltsük be `np.loadtxt` segítségével, majd használjuk a `ps.SPPMethod.calculate_from_raw` függvényt.",
"_____no_output_____"
]
],
[
[
"delay, position = np.loadtxt('spp.txt', delimiter=',', unpack=True)\n\nmyspp.calculate_from_raw(delay, position, reference_point=2, order=3);",
"_____no_output_____"
]
],
[
[
"Az előbbi esetben látható, hogy ugyan azt az eredményt kaptuk, mint előzőleg. Ez akkor is hasznos lehet, ha már megvannak a leolvasott SPP pozícióink a hozzá tartozó késleltetésekkel és csak a számolást akarjuk elvégezni. Ekkor még létre sem kell hozni egy új objektumot, csak meghívhatjuk a függvényt következő módon:\n",
"_____no_output_____"
]
],
[
[
"# ehhez beírtam egy teljesen véletlenszerű adatsort\ndelay_minta = [-100, 200, 500, 700, 900]\nposition_minta = [2, 2.1, 2.3, 2.45, 2.6]\n\nps.SPPMethod.calculate_from_raw(delay_minta, position_minta, reference_point=2, order=3);",
"_____no_output_____"
]
],
[
[
"**FONTOS MEGJEGYZÉS:**\n\nAz `order` argumentum a program során mindig a keresett diszperzió rendjét adja meg.",
"_____no_output_____"
],
[
"#### 5.2 Számolás egy további módon",
"_____no_output_____"
],
[
"Mivel továbbra is ugyan ezekkel az adatsorokkal és a `myspp` objektummal dolgozom, most törlöm az összes rögzített adatot belőlük. Ehhez a `SPPMethod.flush` függvényt használom. (Valószínűleg ez a felhasználónak kevésszer szükséges, de elérhető.)",
"_____no_output_____"
]
],
[
[
"myspp.flush()",
"_____no_output_____"
]
],
[
[
"Korábban már észrevehettük, hogy a kiíratás során - legyen bármilyen módszerről is szó - megjelentek olyan sorok is, hogy `Delay value: Not given` és `SPP position(s): Not given`. Például a `myspp` első interferogramja esetén most ez a helyzet:",
"_____no_output_____"
]
],
[
[
"print(myspp[0])",
"Dataset\n----------\nParameters\n----------\nDatapoints: 12559\nPredicted domain: frequency\nRange: from 0.99998 to 3.00000 PHz\nNormalized: False\nDelay value: Not given\nSPP position(s): Not given\n----------------------------\nMetadata extracted from file\n----------------------------\n{}\n"
]
],
[
[
"Ahogyan a `Dataset` leírásában már szerepelt, lehetőségünk van megadni a betöltött interferogramokon az SPP módszerhez szükséges adatokat. Ekkor a `ps.SPPMethod.calculate_from_ifg(ifgs, reference_point, order)` függvénnyel kiértékelhetjük a benne lévő interferogramokat a következő módon:",
"_____no_output_____"
]
],
[
[
"# kicsomagolok öt interferogramot a generált 7 közül\n\nelso_ifg = myspp[0]\nmasodik_ifg = myspp[1]\nharmadik_ifg = myspp[2]\nnegyedik_ifg = myspp[3]\notodik_ifg = myspp[4]",
"_____no_output_____"
],
[
"# beállítok rájuk véletlenszerűen SPP adatokat\n\nelso_ifg.delay = 0\nelso_ifg.positions = 2\n\nmasodik_ifg.delay = 100\nmasodik_ifg.positions = 2\n\nharmadik_ifg.delay = 150\nharmadik_ifg.positions = 1.6\n\nnegyedik_ifg.delay = 200\nnegyedik_ifg.positions = 1.2\n\notodik_ifg.delay = 250\notodik_ifg.positions = 1, 3, 1.2\n\n\n# listába teszem őket\nifgs = [elso_ifg, masodik_ifg, harmadik_ifg, negyedik_ifg, otodik_ifg]",
"_____no_output_____"
],
[
"# meghívom a calculate_from_ifg függvényt\nps.SPPMethod.calculate_from_ifg(ifgs, reference_point=2, order=3);",
"_____no_output_____"
]
],
[
[
"Ez úgy lehet hasznos, hogy amikor más módszerrel több interferogramot is kiértékelünk egymás után, csak rögzítjük az SPP adatokat is, aztán a program ezekből egyenként összegyűjti a szükséges információt a kiértékeléshez, majd abból számol.",
"_____no_output_____"
],
[
"#### 5.3 Az SPPMethod működéséről mélyebben, cache, callbacks\n\n\nAz `SPPMethod` alapvető működését az adatok rögzítése közben az alábbi ábra mutatja.\n\n\n\nA hurok az `SPPMethod`-ból indul, ahol a használandó fájlok neveit, betöltési adatokat, stb. adunk meg. Ezen a ponton még semmilyen számolás és betöltés nem történik. Ezután az `SPPMethod` bármely elemének hívására egy `Dataset` objektum jön létre. Ezen megnyitható az `SPPEditor`, amiben az állandó fázisú pont(ok) helyét és a karok közti késleltetést lehet megadni. Hitelesítés után az SPP-vel kapcsolatos információk az interaktív szerkesztőből visszakerülnek a létrehozott `Dataset` objektumba és ott rögzítődnek. Minden így létrejött `Dataset` objektum kapcsolva van az `SPPMethod`-hoz, amiből felépült, így amikor megváltozik egy SPP-vel kapcsolatos adat, az egyből megváltozik az `SPPMetod`-ban is. A `Registry` gondoskodik arról, hogy minden objektum ami a memóriában van az rögzítődjön, illetve szükség esetén elérhető legyen.\n\n\n**Cache**\n\nHa próbálunk elérni egy adott elemet (akár a `for` ciklussal, akár indexelve, vagy egyéb módon), létrejön egy `Dataset` objektum. Ez a `Dataset` objektum miután már egyszer elértük a memóriában marad és megtart minden rajta végrehajtott változtatást, beállítást. Alapértelmezetten *128 db* interferogram marad a memóriában egyszerre, de ez a határ szükség esetén megváltoztatható. Az éppen aktuálisan a memóriában lévő interferogramok száma (az adott `SPPMethod`-hoz tartozó) a kiíratás során a `Interferograms cached` cellában látható.\n\n**Callbacks**\n\nA fenti ábrán a ciklus utolsó lépése során (ahol a `Dataset` átadja az SPP-vel kapcsolatos adatait a `SPPMethod`-nak) lehetőség van további ún. *callback* függvények meghívására. Egy ilyen beépített callback függvény a `pysprint.eager_executor`. Ez arra használható, hogy minden egyes SPP-vel kapcsolatos adat rögzítése/változtatása után a program azonnal kiszámolja az éppen meglévő adatokból a diszperziót. A korábbiakhoz teljesen hasonlóan kell eljárnunk, csupán a `callback` argumentumot kell megadnunk kiegészítésként. Itt a kötelező argumentumokon túl megadtam a `logfile` és `verbosity` értékeit is: ez minden lépés során a `\"mylog.log\"` fájlba el fogja menteni az adott illesztés eredményeit és egyéb információkat, továbbá a `verbosity=1` miatt a rögzített adatsort is. Ezzel akár könnyen nyomon köthető a kiértékelés menete.",
"_____no_output_____"
]
],
[
[
"# a folyamatos kiértékeléshez szükséges callback függvény importálása\nfrom pysprint import eager_executor\n\nmyspp2 = ps.SPPMethod(\n ifg_files,\n decimal=\".\",\n sep=\",\",\n skiprows=0,\n meta_len=0,\n callback=eager_executor(reference_point=2, order=3, logfile=\"mylog.log\", verbosity=1)\n)",
"_____no_output_____"
],
[
"myspp2",
"_____no_output_____"
]
],
[
[
"Ekkor láthatjuk, hogy az `Eagerly calculating` már `True` értékre változik.",
"_____no_output_____"
],
[
"Természetesen a program csak értelmes esetekben fogja elvégezni a számolást (pl. szükséges, hogy az adatpontok száma nagyobb legyen, mint az illesztés rendje ). A teljesség kedvéért megemlítendő, hogy könnyen írható akár saját callback függvény is. Futtassuk le az újonnan létrehozott `myspp2`-n a már megismert *for* ciklust:",
"_____no_output_____"
]
],
[
[
"with ps.interactive():\n for ifg in myspp2:\n ifg.open_SPP_panel()",
"_____no_output_____"
]
],
[
[
"A fenti cella futtatása közben - miután rögzítettünk elég adatot - megjelentek az eredmények, és minden új adatpont hozzáadása esetén frissültek is. Az adatok rögzítését itt ugyan az interaktív felületet használva végeztem, de akár kódban is megtehető: a `myspp` elemein kell az `delay` és `positions` argumentumokat beállítani, és minden új adat hozzáadásánál újra fogja számolni a program. Az előző számolásom közben készült logfile a következő:",
"_____no_output_____"
]
],
[
[
"!type mylog.log",
"\n---------------------------------------------------------------------------------------\nDate: 2020-10-05 15:43:11.098910\n\nDatapoints used: 3\n\nR^2: 1.00000\n\nResults:\nGD = -0.00000 fs^1\nGDD = -357.48602 fs^2\nTOD = 66.15894 fs^3\n\nValues used:\nx: [2.29569 2.14981 2.00788]\n\ny: [-100. -50. 0.]\n\n---------------------------------------------------------------------------------------\nDate: 2020-10-05 15:43:17.277431\n\nDatapoints used: 4\n\nR^2: 0.99981\n\nResults:\nGD = 1.07604 fs^1\nGDD = -384.79434 fs^2\nTOD = 228.58592 fs^3\n\nValues used:\nx: [2.29569 2.14981 2.00788 1.88565]\n\ny: [-100. -50. 0. 50.]\n\n---------------------------------------------------------------------------------------\nDate: 2020-10-05 15:43:21.772308\n\nDatapoints used: 5\n\nR^2: 0.99946\n\nResults:\nGD = 1.78742 fs^1\nGDD = -366.69986 fs^2\nTOD = 73.95953 fs^3\n\nValues used:\nx: [2.29569 2.14981 2.00788 1.88565 1.74372]\n\ny: [-100. -50. 0. 50. 100.]\n\n---------------------------------------------------------------------------------------\nDate: 2020-10-05 15:43:25.562731\n\nDatapoints used: 6\n\nR^2: 0.99351\n\nResults:\nGD = -1.25001 fs^1\nGDD = -389.64917 fs^2\nTOD = 350.18415 fs^3\n\nValues used:\nx: [2.29569 2.14981 2.00788 1.88565 1.74372 1.68852]\n\ny: [-100. -50. 0. 50. 100. 150.]\n\n---------------------------------------------------------------------------------------\nDate: 2020-10-05 15:43:26.518332\n\nDatapoints used: 6\n\nR^2: 0.99351\n\nResults:\nGD = -1.25001 fs^1\nGDD = -389.64917 fs^2\nTOD = 350.18415 fs^3\n\nValues used:\nx: [2.29569 2.14981 2.00788 1.88565 1.74372 1.68852]\n\ny: [-100. -50. 0. 50. 100. 150.]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a8ed2b501ee9e2f0326ae2d6cb581424dad1424
| 7,789 |
ipynb
|
Jupyter Notebook
|
notebooks/chapter08_ml/04_text.ipynb
|
svaksha/cookbook-code
|
960becec4cc48f14991ed9d8525d5bcd21bc42a7
|
[
"BSD-2-Clause"
] | 5 |
2015-11-26T14:18:23.000Z
|
2018-06-08T00:46:35.000Z
|
notebooks/chapter08_ml/04_text.ipynb
|
kunalj101/cookbook-code
|
adcbdeb6b92e448350ce2643003a2a0719e574ca
|
[
"BSD-2-Clause"
] | null | null | null |
notebooks/chapter08_ml/04_text.ipynb
|
kunalj101/cookbook-code
|
adcbdeb6b92e448350ce2643003a2a0719e574ca
|
[
"BSD-2-Clause"
] | 8 |
2015-11-14T23:18:50.000Z
|
2019-08-20T22:47:07.000Z
| 29.503788 | 492 | 0.520221 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a8edb83752b8ada0a0a4f17214b5bdbfe2b6bcd
| 66,656 |
ipynb
|
Jupyter Notebook
|
chapter_natural-language-processing/word2vec-data-set.ipynb
|
pedro-abundio-wang/d2l-numpy
|
59e3e536f81d355f10a99a4e936d2b3e68201f1d
|
[
"Apache-2.0"
] | null | null | null |
chapter_natural-language-processing/word2vec-data-set.ipynb
|
pedro-abundio-wang/d2l-numpy
|
59e3e536f81d355f10a99a4e936d2b3e68201f1d
|
[
"Apache-2.0"
] | 15 |
2019-10-10T13:01:15.000Z
|
2022-02-10T00:21:14.000Z
|
chapter_natural-language-processing/word2vec-data-set.ipynb
|
pedro-abundio-wang/d2l-numpy
|
59e3e536f81d355f10a99a4e936d2b3e68201f1d
|
[
"Apache-2.0"
] | null | null | null | 37.765439 | 1,352 | 0.503226 |
[
[
[
"# Data Sets for Word2vec\n\n:label:`chapter_word2vec_data`\n\n\nIn this section, we will introduce how to preprocess a data set with\nnegative sampling :numref:`chapter_approx_train` and load into mini-batches for\nword2vec training. The data set we use is [Penn Tree Bank (PTB)]( https://catalog.ldc.upenn.edu/LDC99T42), which is a small but commonly-used corpus. It takes samples from Wall Street Journal articles and includes training sets, validation sets, and test sets. \n\nFirst, import the packages and modules required for the experiment.",
"_____no_output_____"
]
],
[
[
"import collections\nimport d2l\nimport math\nfrom mxnet import np, gluon\nimport random\nimport zipfile",
"_____no_output_____"
]
],
[
[
"## Read and Preprocessing\n\nThis data set has already been preprocessed. Each line of the data set acts as a sentence. All the words in a sentence are separated by spaces. In the word embedding task, each word is a token.",
"_____no_output_____"
]
],
[
[
"# Save to the d2l package.\ndef read_ptb():\n with zipfile.ZipFile('../data/ptb.zip', 'r') as f:\n raw_text = f.read('ptb/ptb.train.txt').decode(\"utf-8\")\n return [line.split() for line in raw_text.split('\\n')]\n\nsentences = read_ptb()\n'# sentences: %d' % len(sentences)",
"_____no_output_____"
]
],
[
[
"Next we build a vocabulary with words appeared not greater than 10 times mapped into a \"<unk>\" token. Note that the preprocessed PTB data also contains \"<unk>\" tokens presenting rare words.",
"_____no_output_____"
]
],
[
[
"vocab = d2l.Vocab(sentences, min_freq=10)\n'vocab size: %d' % len(vocab)",
"_____no_output_____"
]
],
[
[
"## Subsampling\n\nIn text data, there are generally some words that appear at high frequencies, such \"the\", \"a\", and \"in\" in English. Generally speaking, in a context window, it is better to train the word embedding model when a word (such as \"chip\") and a lower-frequency word (such as \"microprocessor\") appear at the same time, rather than when a word appears with a higher-frequency word (such as \"the\"). Therefore, when training the word embedding model, we can perform subsampling[2] on the words. Specifically, each indexed word $w_i$ in the data set will drop out at a certain probability. The dropout probability is given as:\n\n$$ \\mathbb{P}(w_i) = \\max\\left(1 - \\sqrt{\\frac{t}{f(w_i)}}, 0\\right),$$\n\nHere, $f(w_i)$ is the ratio of the instances of word $w_i$ to the total number of words in the data set, and the constant $t$ is a hyper-parameter (set to $10^{-4}$ in this experiment). As we can see, it is only possible to drop out the word $w_i$ in subsampling when $f(w_i) > t$. The higher the word's frequency, the higher its dropout probability.",
"_____no_output_____"
]
],
[
[
"# Save to the d2l package.\ndef subsampling(sentences, vocab):\n # Map low frequency words into <unk>\n sentences = [[vocab.idx_to_token[vocab[tk]] for tk in line]\n for line in sentences]\n # Count the frequency for each word\n counter = d2l.count_corpus(sentences)\n num_tokens = sum(counter.values())\n # Return True if to keep this token during subsampling\n keep = lambda token: (\n random.uniform(0, 1) < math.sqrt(1e-4 / counter[token] * num_tokens))\n # Now do the subsampling.\n return [[tk for tk in line if keep(tk)] for line in sentences]\n\nsubsampled = subsampling(sentences, vocab)",
"_____no_output_____"
]
],
[
[
"Compare the sequence lengths before and after sampling, we can see subsampling significantly reduced the sequence length.",
"_____no_output_____"
]
],
[
[
"d2l.set_figsize((3.5, 2.5))\nd2l.plt.hist([[len(line) for line in sentences],\n [len(line) for line in subsampled]] )\nd2l.plt.xlabel('# tokens per sentence')\nd2l.plt.ylabel('count')\nd2l.plt.legend(['origin', 'subsampled']);",
"_____no_output_____"
]
],
[
[
"For individual tokens, the sampling rate of the high-frequency word \"the\" is less than 1/20.",
"_____no_output_____"
]
],
[
[
"def compare_counts(token):\n return '# of \"%s\": before=%d, after=%d' % (token, sum(\n [line.count(token) for line in sentences]), sum(\n [line.count(token) for line in subsampled]))\n\ncompare_counts('the')",
"_____no_output_____"
]
],
[
[
"But the low-frequency word \"join\" is completely preserved.",
"_____no_output_____"
]
],
[
[
"compare_counts('join')",
"_____no_output_____"
]
],
[
[
"Lastly, we map each token into an index to construct the corpus.",
"_____no_output_____"
]
],
[
[
"corpus = [vocab[line] for line in subsampled]\ncorpus[0:3]",
"_____no_output_____"
]
],
[
[
"## Load the Data Set\n\nNext we read the corpus with token indicies into data batches for training.\n\n### Extract Central Target Words and Context Words\n\nWe use words with a distance from the central target word not exceeding the context window size as the context words of the given center target word. The following definition function extracts all the central target words and their context words. It uniformly and randomly samples an integer to be used as the context window size between integer 1 and the `max_window_size` (maximum context window).",
"_____no_output_____"
]
],
[
[
"# Save to the d2l package.\ndef get_centers_and_contexts(corpus, max_window_size):\n centers, contexts = [], []\n for line in corpus:\n # Each sentence needs at least 2 words to form a\n # \"central target word - context word\" pair\n if len(line) < 2: continue\n centers += line\n for i in range(len(line)): # Context window centered at i\n window_size = random.randint(1, max_window_size)\n indices = list(range(max(0, i - window_size),\n min(len(line), i + 1 + window_size)))\n # Exclude the central target word from the context words\n indices.remove(i)\n contexts.append([line[idx] for idx in indices])\n return centers, contexts",
"_____no_output_____"
]
],
[
[
"Next, we create an artificial data set containing two sentences of 7 and 3 words, respectively. Assume the maximum context window is 2 and print all the central target words and their context words.",
"_____no_output_____"
]
],
[
[
"tiny_dataset = [list(range(7)), list(range(7, 10))]\nprint('dataset', tiny_dataset)\nfor center, context in zip(*get_centers_and_contexts(tiny_dataset, 2)):\n print('center', center, 'has contexts', context)",
"dataset [[0, 1, 2, 3, 4, 5, 6], [7, 8, 9]]\ncenter 0 has contexts [1, 2]\ncenter 1 has contexts [0, 2]\ncenter 2 has contexts [0, 1, 3, 4]\ncenter 3 has contexts [2, 4]\ncenter 4 has contexts [3, 5]\ncenter 5 has contexts [4, 6]\ncenter 6 has contexts [4, 5]\ncenter 7 has contexts [8, 9]\ncenter 8 has contexts [7, 9]\ncenter 9 has contexts [7, 8]\n"
]
],
[
[
"We set the maximum context window size to 5. The following extracts all the central target words and their context words in the data set.",
"_____no_output_____"
]
],
[
[
"all_centers, all_contexts = get_centers_and_contexts(corpus, 5)\n'# center-context pairs: %d' % len(all_centers)",
"_____no_output_____"
]
],
[
[
"### Negative Sampling\n\nWe use negative sampling for approximate training. For a central and context word pair, we randomly sample $K$ noise words ($K=5$ in the experiment). According to the suggestion in the Word2vec paper, the noise word sampling probability $\\mathbb{P}(w)$ is the ratio of the word frequency of $w$ to the total word frequency raised to the power of 0.75 [2].\n\nWe first define a class to draw a candidate according to the sampling weights. It caches a 10000 size random number bank instead of calling `random.choices` every time.",
"_____no_output_____"
]
],
[
[
"# Save to the d2l package.\nclass RandomGenerator(object):\n \"\"\"Draw a random int in [0, n] according to n sampling weights\"\"\"\n def __init__(self, sampling_weights):\n self.population = list(range(len(sampling_weights)))\n self.sampling_weights = sampling_weights\n self.candidates = []\n self.i = 0\n\n def draw(self):\n if self.i == len(self.candidates):\n self.candidates = random.choices(\n self.population, self.sampling_weights, k=10000)\n self.i = 0\n self.i += 1\n return self.candidates[self.i-1]\n\ngenerator = RandomGenerator([2,3,4])\n[generator.draw() for _ in range(10)]",
"_____no_output_____"
],
[
"# Save to the d2l package.\ndef get_negatives(all_contexts, corpus, K):\n counter = d2l.count_corpus(corpus)\n sampling_weights = [counter[i]**0.75 for i in range(len(counter))]\n all_negatives, generator = [], RandomGenerator(sampling_weights)\n for contexts in all_contexts:\n negatives = []\n while len(negatives) < len(contexts) * K:\n neg = generator.draw()\n # Noise words cannot be context words\n if neg not in contexts:\n negatives.append(neg)\n all_negatives.append(negatives)\n return all_negatives\n\nall_negatives = get_negatives(all_contexts, corpus, 5)",
"_____no_output_____"
]
],
[
[
"### Read into Batches\n\nWe extract all central target words `all_centers`, and the context words `all_contexts` and noise words `all_negatives` of each central target word from the data set. We will read them in random mini-batches.\n\nIn a mini-batch of data, the $i$-th example includes a central word and its corresponding $n_i$ context words and $m_i$ noise words. Since the context window size of each example may be different, the sum of context words and noise words, $n_i+m_i$, will be different. When constructing a mini-batch, we concatenate the context words and noise words of each example, and add 0s for padding until the length of the concatenations are the same, that is, the length of all concatenations is $\\max_i n_i+m_i$(`max_len`). In order to avoid the effect of padding on the loss function calculation, we construct the mask variable `masks`, each element of which corresponds to an element in the concatenation of context and noise words, `contexts_negatives`. When an element in the variable `contexts_negatives` is a padding, the element in the mask variable `masks` at the same position will be 0. Otherwise, it takes the value 1. In order to distinguish between positive and negative examples, we also need to distinguish the context words from the noise words in the `contexts_negatives` variable. Based on the construction of the mask variable, we only need to create a label variable `labels` with the same shape as the `contexts_negatives` variable and set the elements corresponding to context words (positive examples) to 1, and the rest to 0.\n\nNext, we will implement the mini-batch reading function `batchify`. Its mini-batch input `data` is a list whose length is the batch size, each element of which contains central target words `center`, context words `context`, and noise words `negative`. The mini-batch data returned by this function conforms to the format we need, for example, it includes the mask variable.",
"_____no_output_____"
]
],
[
[
"# Save to the d2l package.\ndef batchify(data):\n max_len = max(len(c) + len(n) for _, c, n in data)\n centers, contexts_negatives, masks, labels = [], [], [], []\n for center, context, negative in data:\n cur_len = len(context) + len(negative)\n centers += [center]\n contexts_negatives += [context + negative + [0] * (max_len - cur_len)]\n masks += [[1] * cur_len + [0] * (max_len - cur_len)]\n labels += [[1] * len(context) + [0] * (max_len - len(context))]\n return (np.array(centers).reshape(-1, 1), np.array(contexts_negatives),\n np.array(masks), np.array(labels))",
"_____no_output_____"
]
],
[
[
"Construct two simple examples:",
"_____no_output_____"
]
],
[
[
"x_1 = (1, [2,2], [3,3,3,3])\nx_2 = (1, [2,2,2], [3,3])\nbatch = batchify((x_1, x_2))\n\nnames = ['centers', 'contexts_negatives', 'masks', 'labels']\nfor name, data in zip(names, batch):\n print(name, '=', data)",
"centers = [[1.]\n [1.]]\ncontexts_negatives = [[2. 2. 3. 3. 3. 3.]\n [2. 2. 2. 3. 3. 0.]]\nmasks = [[1. 1. 1. 1. 1. 1.]\n [1. 1. 1. 1. 1. 0.]]\nlabels = [[1. 1. 0. 0. 0. 0.]\n [1. 1. 1. 0. 0. 0.]]\n"
]
],
[
[
"We use the `batchify` function just defined to specify the mini-batch reading method in the `DataLoader` instance. \n\n## Put All Things Together\n\nLastly, we define the `load_data_ptb` function that read the PTB data set and return the data loader.",
"_____no_output_____"
]
],
[
[
"# Save to the d2l package.\ndef load_data_ptb(batch_size, max_window_size, num_noise_words):\n sentences = read_ptb()\n vocab = d2l.Vocab(sentences, min_freq=10)\n subsampled = subsampling(sentences, vocab)\n corpus = [vocab[line] for line in subsampled]\n all_centers, all_contexts = get_centers_and_contexts(\n corpus, max_window_size)\n all_negatives = get_negatives(all_contexts, corpus, num_noise_words)\n dataset = gluon.data.ArrayDataset(\n all_centers, all_contexts, all_negatives)\n data_iter = gluon.data.DataLoader(dataset, batch_size, shuffle=True,\n batchify_fn=batchify)\n return data_iter, vocab",
"_____no_output_____"
]
],
[
[
"Let's print the first mini-batch of the data iterator.",
"_____no_output_____"
]
],
[
[
"data_iter, vocab = load_data_ptb(512, 5, 5)\nfor batch in data_iter:\n for name, data in zip(names, batch):\n print(name, 'shape:', data.shape)\n break",
"centers shape: (512, 1)\ncontexts_negatives shape: (512, 60)\nmasks shape: (512, 60)\nlabels shape: (512, 60)\n"
]
],
[
[
"## Summary\n\n* Subsampling attempts to minimize the impact of high-frequency words on the training of a word embedding model.\n* We can pad examples of different lengths to create mini-batches with examples of all the same length and use mask variables to distinguish between padding and non-padding elements, so that only non-padding elements participate in the calculation of the loss function.\n\n## Exercises\n\n* We use the `batchify` function to specify the mini-batch reading method in the `DataLoader` instance and print the shape of each variable in the first batch read. How should these shapes be calculated?\n\n## Scan the QR Code to [Discuss](https://discuss.mxnet.io/t/4356)\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a8ee772af48d424add55b249a18be6cf37bf41b
| 119,790 |
ipynb
|
Jupyter Notebook
|
notebooks/book1/13/linregRbfDemo.ipynb
|
karm-patel/pyprobml
|
af8230a0bc0d01bb0f779582d87e5856d25e6211
|
[
"MIT"
] | null | null | null |
notebooks/book1/13/linregRbfDemo.ipynb
|
karm-patel/pyprobml
|
af8230a0bc0d01bb0f779582d87e5856d25e6211
|
[
"MIT"
] | null | null | null |
notebooks/book1/13/linregRbfDemo.ipynb
|
karm-patel/pyprobml
|
af8230a0bc0d01bb0f779582d87e5856d25e6211
|
[
"MIT"
] | null | null | null | 777.857143 | 115,088 | 0.949612 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\ntry:\n from cycler import cycler\nexcept ModuleNotFoundError:\n %pip install cycler\n from cycler import cycler\nfrom scipy.spatial.distance import cdist\n\ntry:\n import probml_utils as pml\nexcept ModuleNotFoundError:\n %pip install git+https://github.com/probml/probml-utils.git\n import probml_utils as pml\n\nnp.random.seed(0)\nCB_color = [\"#377eb8\", \"#ff7f00\"]\n\ncb_cycler = cycler(linestyle=[\"-\", \"--\", \"-.\"]) * cycler(color=CB_color)\nplt.rc(\"axes\", prop_cycle=cb_cycler)\n\n\ndef fun(x, w):\n return w[0] * x + w[1] * np.square(x)\n\n\n# 'Data as mentioned in the matlab code'\ndef polydatemake():\n n = 21\n sigma = 2\n xtrain = np.linspace(0, 20, n)\n xtest = np.arange(0, 20.1, 0.1)\n w = np.array([-1.5, 1 / 9])\n ytrain = fun(xtrain, w).reshape(-1, 1) + np.random.randn(xtrain.shape[0], 1)\n ytestNoisefree = fun(xtest, w)\n ytestNoisy = ytestNoisefree + sigma * np.random.randn(xtest.shape[0], 1) * sigma\n\n return xtrain, ytrain, xtest, ytestNoisefree, ytestNoisy\n\n\n[xtrain, ytrain, xtest, ytestNoisefree, ytestNoisy] = polydatemake()\n\nsigmas = [0.5, 10, 50]\nK = 10\ncenters = np.linspace(np.min(xtrain), np.max(xtrain), K)\n\n\ndef addones(x):\n # x is of shape (s,)\n return np.insert(x[:, np.newaxis], 0, [[1]], axis=1)\n\n\ndef rbf_features(X, centers, sigma):\n dist_mat = cdist(X, centers, \"minkowski\", p=2.0)\n return np.exp((-0.5 / (sigma**2)) * (dist_mat**2))\n\n\n# using matrix inversion for ridge regression\ndef ridgeReg(X, y, lambd): # returns weight vectors.\n D = X.shape[1]\n w = np.linalg.inv(X.T @ X + lambd * np.eye(D, D)) @ X.T @ y\n\n return w\n\n\nfig, ax = plt.subplots(3, 3, figsize=(10, 10))\nplt.tight_layout()\n\nfor (i, s) in enumerate(sigmas):\n rbf_train = rbf_features(addones(xtrain), addones(centers), s)\n rbf_test = rbf_features(addones(xtest), addones(centers), s)\n reg_w = ridgeReg(rbf_train, ytrain, 0.3)\n ypred = rbf_test @ reg_w\n\n ax[i, 0].plot(xtrain, ytrain, \".\", markersize=8)\n ax[i, 0].plot(xtest, ypred)\n ax[i, 0].set_ylim([-10, 20])\n ax[i, 0].set_xticks(np.arange(0, 21, 5))\n\n for j in range(K):\n ax[i, 1].plot(xtest, rbf_test[:, j], \"b-\")\n ax[i, 1].set_xticks(np.arange(0, 21, 5))\n ax[i, 1].ticklabel_format(style=\"sci\", scilimits=(-2, 2))\n\n ax[i, 2].imshow(rbf_train, interpolation=\"nearest\", aspect=\"auto\", cmap=plt.get_cmap(\"viridis\"))\n ax[i, 2].set_yticks(np.arange(20, 4, -5))\n ax[i, 2].set_xticks(np.arange(2, 10, 2))\npml.savefig(\"rbfDemoALL.pdf\", dpi=300)\nplt.show()",
"/home/patel_zeel/miniconda3/envs/probml_py3912/lib/python3.9/site-packages/probml_utils/plotting.py:74: UserWarning: set FIG_DIR environment variable to save figures\n warnings.warn(\"set FIG_DIR environment variable to save figures\")\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
4a8eef1cc6421c74591bc2061beec533740daef6
| 123,673 |
ipynb
|
Jupyter Notebook
|
jupyter-notebooks/Tox and Adme dataset similarity.ipynb
|
cdd/os-models
|
443536657a685de9e4e769b11714b7432307af17
|
[
"Apache-2.0"
] | 1 |
2019-10-28T09:43:28.000Z
|
2019-10-28T09:43:28.000Z
|
jupyter-notebooks/Tox and Adme dataset similarity.ipynb
|
cdd/os-models
|
443536657a685de9e4e769b11714b7432307af17
|
[
"Apache-2.0"
] | null | null | null |
jupyter-notebooks/Tox and Adme dataset similarity.ipynb
|
cdd/os-models
|
443536657a685de9e4e769b11714b7432307af17
|
[
"Apache-2.0"
] | 1 |
2018-12-05T02:39:37.000Z
|
2018-12-05T02:39:37.000Z
| 92.155738 | 69,218 | 0.680108 |
[
[
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n\nimport seaborn as sns\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as n\n\nimport sys\nimport re",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"\n#df = pd.read_csv('/home/gareth/src/os-models/data/vault/adme_tox_cmc.csv.gz')\ndf = pd.read_csv('/home/gareth/src/os-models/data/vault/adme_tox_nci.csv.gz')",
"_____no_output_____"
],
[
"def rename_columns(df):\n def rename_col(no, n):\n new_n = re.sub(r\" \\[.*\\]$\", \"\", n)\n return new_n\n columns_map = {n:rename_col(i, n) for i, n in enumerate(df.columns)}\n df.rename(columns=columns_map, inplace=True)\n return df\n\ndf = rename_columns(df)\ndf = df.drop(['ID'], axis=1)\n",
"_____no_output_____"
],
[
"df\n",
"_____no_output_____"
],
[
"\nfig, axes = plt.subplots(nrows=1, ncols=1, figsize=(20, 12))\n#sns.set(font_scale=1.2)\nsns.boxplot(data=df, ax=axes)\n#sns.swarmplot(data=df, ax=axes, color=\".0\")\nfor tick in axes.get_xticklabels():\n tick.set_rotation(80)\naxes.tick_params(axis='both', labelsize=15)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a8eefb951975826803c48acddb29867ffa2faf7
| 416,043 |
ipynb
|
Jupyter Notebook
|
soln/chap09.ipynb
|
sonvt1710/ThinkBayes2
|
704af5f2520db2f1320e5368d8dce4bf36a8fe02
|
[
"MIT"
] | null | null | null |
soln/chap09.ipynb
|
sonvt1710/ThinkBayes2
|
704af5f2520db2f1320e5368d8dce4bf36a8fe02
|
[
"MIT"
] | null | null | null |
soln/chap09.ipynb
|
sonvt1710/ThinkBayes2
|
704af5f2520db2f1320e5368d8dce4bf36a8fe02
|
[
"MIT"
] | null | null | null | 114.612397 | 28,580 | 0.87061 |
[
[
[
"# Decision Analysis",
"_____no_output_____"
],
[
"Think Bayes, Second Edition\n\nCopyright 2020 Allen B. Downey\n\nLicense: [Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0)](https://creativecommons.org/licenses/by-nc-sa/4.0/)",
"_____no_output_____"
]
],
[
[
"# If we're running on Colab, install empiricaldist\n# https://pypi.org/project/empiricaldist/\n\nimport sys\nIN_COLAB = 'google.colab' in sys.modules\n\nif IN_COLAB:\n !pip install empiricaldist",
"_____no_output_____"
],
[
"# Get utils.py\n\nfrom os.path import basename, exists\n\ndef download(url):\n filename = basename(url)\n if not exists(filename):\n from urllib.request import urlretrieve\n local, _ = urlretrieve(url, filename)\n print('Downloaded ' + local)\n \ndownload('https://github.com/AllenDowney/ThinkBayes2/raw/master/soln/utils.py')",
"_____no_output_____"
],
[
"from utils import set_pyplot_params\nset_pyplot_params()",
"_____no_output_____"
]
],
[
[
"This chapter presents a problem inspired by the game show *The Price is Right*.\nIt is a silly example, but it demonstrates a useful process called Bayesian [decision analysis](https://en.wikipedia.org/wiki/Decision_analysis).\n\nAs in previous examples, we'll use data and prior distribution to compute a posterior distribution; then we'll use the posterior distribution to choose an optimal strategy in a game that involves bidding.\n\nAs part of the solution, we will use kernel density estimation (KDE) to estimate the prior distribution, and a normal distribution to compute the likelihood of the data.\n\nAnd at the end of the chapter, I pose a related problem you can solve as an exercise.",
"_____no_output_____"
],
[
"## The Price Is Right Problem\n\nOn November 1, 2007, contestants named Letia and Nathaniel appeared on *The Price is Right*, an American television game show. They competed in a game called \"The Showcase\", where the objective is to guess the price of a collection of prizes. The contestant who comes closest to the actual price, without going over, wins the prizes.\n\nNathaniel went first. His showcase included a dishwasher, a wine cabinet, a laptop computer, and a car. He bid \\\\$26,000.\n\nLetia's showcase included a pinball machine, a video arcade game, a pool table, and a cruise of the Bahamas. She bid \\\\$21,500.\n\nThe actual price of Nathaniel's showcase was \\\\$25,347. His bid was too high, so he lost.\n\nThe actual price of Letia's showcase was \\\\$21,578. \n\nShe was only off by \\\\$78, so she won her showcase and, because her bid was off by less than 250, she also won Nathaniel's showcase.",
"_____no_output_____"
],
[
"For a Bayesian thinker, this scenario suggests several questions:\n\n1. Before seeing the prizes, what prior beliefs should the contestants have about the price of the showcase?\n\n2. After seeing the prizes, how should the contestants update those beliefs?\n\n3. Based on the posterior distribution, what should the contestants bid?\n\nThe third question demonstrates a common use of Bayesian methods: decision analysis.\n\nThis problem is inspired by [an example](https://nbviewer.jupyter.org/github/CamDavidsonPilon/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers/blob/master/Chapter5_LossFunctions/Ch5_LossFunctions_PyMC3.ipynb) in Cameron Davidson-Pilon's book, [*Probablistic Programming and Bayesian Methods for Hackers*](http://camdavidsonpilon.github.io/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers).",
"_____no_output_____"
],
[
"## The Prior\n\nTo choose a prior distribution of prices, we can take advantage of data from previous episodes. Fortunately, [fans of the show keep detailed records](https://web.archive.org/web/20121107204942/http://www.tpirsummaries.8m.com/). \n\nFor this example, I downloaded files containing the price of each showcase from the 2011 and 2012 seasons and the bids offered by the contestants.",
"_____no_output_____"
],
[
"The following cells load the data files.",
"_____no_output_____"
]
],
[
[
"# Load the data files\n\ndownload('https://raw.githubusercontent.com/AllenDowney/ThinkBayes2/master/data/showcases.2011.csv')\ndownload('https://raw.githubusercontent.com/AllenDowney/ThinkBayes2/master/data/showcases.2012.csv')",
"_____no_output_____"
]
],
[
[
"The following function reads the data and cleans it up a little.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ndef read_data(filename):\n \"\"\"Read the showcase price data.\"\"\"\n df = pd.read_csv(filename, index_col=0, skiprows=[1])\n return df.dropna().transpose()",
"_____no_output_____"
]
],
[
[
"I'll read both files and concatenate them.",
"_____no_output_____"
]
],
[
[
"df2011 = read_data('showcases.2011.csv')\ndf2012 = read_data('showcases.2012.csv')\n\ndf = pd.concat([df2011, df2012], ignore_index=True)",
"_____no_output_____"
],
[
"print(df2011.shape, df2012.shape, df.shape)",
"(191, 6) (122, 6) (313, 6)\n"
]
],
[
[
"Here's what the dataset looks like:",
"_____no_output_____"
]
],
[
[
"df.head(3)",
"_____no_output_____"
]
],
[
[
"The first two columns, `Showcase 1` and `Showcase 2`, are the values of the showcases in dollars.\nThe next two columns are the bids the contestants made.\nThe last two columns are the differences between the actual values and the bids.",
"_____no_output_____"
],
[
"## Kernel Density Estimation\n\nThis dataset contains the prices for 313 previous showcases, which we can think of as a sample from the population of possible prices.\n\nWe can use this sample to estimate the prior distribution of showcase prices. One way to do that is kernel density estimation (KDE), which uses the sample to estimate a smooth distribution. If you are not familiar with KDE, you can [read about it here](https://mathisonian.github.io/kde).\n\nSciPy provides `gaussian_kde`, which takes a sample and returns an object that represents the estimated distribution.\n\nThe following function takes `sample`, makes a KDE, evaluates it at a given sequence of quantities, `qs`, and returns the result as a normalized PMF.",
"_____no_output_____"
]
],
[
[
"from scipy.stats import gaussian_kde\nfrom empiricaldist import Pmf\n\ndef kde_from_sample(sample, qs):\n \"\"\"Make a kernel density estimate from a sample.\"\"\"\n kde = gaussian_kde(sample)\n ps = kde(qs)\n pmf = Pmf(ps, qs)\n pmf.normalize()\n return pmf",
"_____no_output_____"
]
],
[
[
"We can use it to estimate the distribution of values for Showcase 1:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nqs = np.linspace(0, 80000, 81)\nprior1 = kde_from_sample(df['Showcase 1'], qs)",
"_____no_output_____"
]
],
[
[
"Here's what it looks like:",
"_____no_output_____"
]
],
[
[
"from utils import decorate\n\ndef decorate_value(title=''):\n decorate(xlabel='Showcase value ($)',\n ylabel='PMF',\n title=title)",
"_____no_output_____"
],
[
"prior1.plot(label='Prior 1')\ndecorate_value('Prior distribution of showcase value')",
"_____no_output_____"
]
],
[
[
"**Exercise:** Use this function to make a `Pmf` that represents the prior distribution for Showcase 2, and plot it.",
"_____no_output_____"
]
],
[
[
"# Solution\n\nqs = np.linspace(0, 80000, 81)\nprior2 = kde_from_sample(df['Showcase 2'], qs)",
"_____no_output_____"
],
[
"# Solution\n\nprior1.plot(label='Prior 1')\nprior2.plot(label='Prior 2')\n\ndecorate_value('Prior distributions of showcase value')",
"_____no_output_____"
]
],
[
[
"## Distribution of Error\n\nTo update these priors, we have to answer these questions:\n\n* What data should we consider and how should we quantify it?\n\n* Can we compute a likelihood function; that is, for each hypothetical price, can we compute the conditional likelihood of the data?\n\nTo answer these questions, I will model each contestant as a price-guessing instrument with known error characteristics. \nIn this model, when the contestant sees the prizes, they guess the price of each prize and add up the prices.\nLet's call this total `guess`.\n\nNow the question we have to answer is, \"If the actual price is `price`, what is the likelihood that the contestant's guess would be `guess`?\"\n\nEquivalently, if we define `error = guess - price`, we can ask, \"What is the likelihood that the contestant's guess is off by `error`?\"\n\nTo answer this question, I'll use the historical data again. \nFor each showcase in the dataset, let's look at the difference between the contestant's bid and the actual price:",
"_____no_output_____"
]
],
[
[
"sample_diff1 = df['Bid 1'] - df['Showcase 1']\nsample_diff2 = df['Bid 2'] - df['Showcase 2']",
"_____no_output_____"
]
],
[
[
"To visualize the distribution of these differences, we can use KDE again.",
"_____no_output_____"
]
],
[
[
"qs = np.linspace(-40000, 20000, 61)\nkde_diff1 = kde_from_sample(sample_diff1, qs)\nkde_diff2 = kde_from_sample(sample_diff2, qs)",
"_____no_output_____"
]
],
[
[
"Here's what these distributions look like:",
"_____no_output_____"
]
],
[
[
"kde_diff1.plot(label='Diff 1', color='C8')\nkde_diff2.plot(label='Diff 2', color='C4')\n\ndecorate(xlabel='Difference in value ($)',\n ylabel='PMF',\n title='Difference between bid and actual value')",
"_____no_output_____"
]
],
[
[
"It looks like the bids are too low more often than too high, which makes sense. Remember that under the rules of the game, you lose if you overbid, so contestants probably underbid to some degree deliberately.\n\nFor example, if they guess that the value of the showcase is \\\\$40,000, they might bid \\\\$36,000 to avoid going over.",
"_____no_output_____"
],
[
"It looks like these distributions are well modeled by a normal distribution, so we can summarize them with their mean and standard deviation.\n\nFor example, here is the mean and standard deviation of `Diff` for Player 1.",
"_____no_output_____"
]
],
[
[
"mean_diff1 = sample_diff1.mean()\nstd_diff1 = sample_diff1.std()\n\nprint(mean_diff1, std_diff1)",
"-4116.3961661341855 6899.909806377117\n"
]
],
[
[
"Now we can use these differences to model the contestant's distribution of errors.\nThis step is a little tricky because we don't actually know the contestant's guesses; we only know what they bid.\n\nSo we have to make some assumptions:\n\n* I'll assume that contestants underbid because they are being strategic, and that on average their guesses are accurate. In other words, the mean of their errors is 0.\n\n* But I'll assume that the spread of the differences reflects the actual spread of their errors. So, I'll use the standard deviation of the differences as the standard deviation of their errors.\n\nBased on these assumptions, I'll make a normal distribution with parameters 0 and `std_diff1`.\n\nSciPy provides an object called `norm` that represents a normal distribution with the given mean and standard deviation.",
"_____no_output_____"
]
],
[
[
"from scipy.stats import norm\n\nerror_dist1 = norm(0, std_diff1)",
"_____no_output_____"
]
],
[
[
"The result is an object that provides `pdf`, which evaluates the probability density function of the normal distribution.\n\nFor example, here is the probability density of `error=-100`, based on the distribution of errors for Player 1.",
"_____no_output_____"
]
],
[
[
"error = -100\nerror_dist1.pdf(error)",
"_____no_output_____"
]
],
[
[
"By itself, this number doesn't mean very much, because probability densities are not probabilities. But they are proportional to probabilities, so we can use them as likelihoods in a Bayesian update, as we'll see in the next section.",
"_____no_output_____"
],
[
"## Update\n\nSuppose you are Player 1. You see the prizes in your showcase and your guess for the total price is \\\\$23,000.\n\nFrom your guess I will subtract away each hypothetical price in the prior distribution; the result is your error under each hypothesis.",
"_____no_output_____"
]
],
[
[
"guess1 = 23000\nerror1 = guess1 - prior1.qs",
"_____no_output_____"
]
],
[
[
"Now suppose we know, based on past performance, that your estimation error is well modeled by `error_dist1`.\nUnder that assumption we can compute the likelihood of your error under each hypothesis.",
"_____no_output_____"
]
],
[
[
"likelihood1 = error_dist1.pdf(error1)",
"_____no_output_____"
]
],
[
[
"The result is an array of likelihoods, which we can use to update the prior.",
"_____no_output_____"
]
],
[
[
"posterior1 = prior1 * likelihood1\nposterior1.normalize()",
"_____no_output_____"
]
],
[
[
"Here's what the posterior distribution looks like:",
"_____no_output_____"
]
],
[
[
"prior1.plot(color='C5', label='Prior 1')\nposterior1.plot(color='C4', label='Posterior 1')\n\ndecorate_value('Prior and posterior distribution of showcase value')",
"_____no_output_____"
]
],
[
[
"Because your initial guess is in the lower end of the range, the posterior distribution has shifted to the left. We can compute the posterior mean to see by how much.",
"_____no_output_____"
]
],
[
[
"prior1.mean(), posterior1.mean()",
"_____no_output_____"
]
],
[
[
"Before you saw the prizes, you expected to see a showcase with a value close to \\\\$30,000.\nAfter making a guess of \\\\$23,000, you updated the prior distribution.\nBased on the combination of the prior and your guess, you now expect the actual price to be about \\\\$26,000.",
"_____no_output_____"
],
[
"**Exercise:** Now suppose you are Player 2. When you see your showcase, you guess that the total price is \\\\$38,000.\n\nUse `diff2` to construct a normal distribution that represents the distribution of your estimation errors.\n\nCompute the likelihood of your guess for each actual price and use it to update `prior2`.\n\nPlot the posterior distribution and compute the posterior mean. Based on the prior and your guess, what do you expect the actual price of the showcase to be?",
"_____no_output_____"
]
],
[
[
"# Solution\n\nmean_diff2 = sample_diff2.mean()\nstd_diff2 = sample_diff2.std()\n\nprint(mean_diff2, std_diff2)",
"-3675.891373801917 6886.260711323408\n"
],
[
"# Solution\n\nerror_dist2 = norm(0, std_diff2)",
"_____no_output_____"
],
[
"# Solution\n\nguess2 = 38000\nerror2 = guess2 - prior2.qs\n\nlikelihood2 = error_dist2.pdf(error2)",
"_____no_output_____"
],
[
"# Solution\n\nposterior2 = prior2 * likelihood2\nposterior2.normalize()",
"_____no_output_____"
],
[
"# Solution\n\nprior2.plot(color='C5', label='Prior 2')\nposterior2.plot(color='C15', label='Posterior 2')\n\ndecorate_value('Prior and posterior distribution of showcase value')",
"_____no_output_____"
],
[
"# Solution\n\nprint(prior2.mean(), posterior2.mean())",
"31047.62371912252 34305.20161642468\n"
]
],
[
[
"## Probability of Winning\n\nNow that we have a posterior distribution for each player, let's think about strategy.\n\nFirst, from the point of view of Player 1, let's compute the probability that Player 2 overbids. To keep it simple, I'll use only the performance of past players, ignoring the value of the showcase. \n\nThe following function takes a sequence of past bids and returns the fraction that overbid.",
"_____no_output_____"
]
],
[
[
"def prob_overbid(sample_diff):\n \"\"\"Compute the probability of an overbid.\"\"\"\n return np.mean(sample_diff > 0)",
"_____no_output_____"
]
],
[
[
"Here's an estimate for the probability that Player 2 overbids.",
"_____no_output_____"
]
],
[
[
"prob_overbid(sample_diff2)",
"_____no_output_____"
]
],
[
[
"Now suppose Player 1 underbids by \\\\$5000.\nWhat is the probability that Player 2 underbids by more?\n\nThe following function uses past performance to estimate the probability that a player underbids by more than a given amount, `diff`:",
"_____no_output_____"
]
],
[
[
"def prob_worse_than(diff, sample_diff):\n \"\"\"Probability opponent diff is worse than given diff.\"\"\"\n return np.mean(sample_diff < diff)",
"_____no_output_____"
]
],
[
[
"Here's the probability that Player 2 underbids by more than \\\\$5000.",
"_____no_output_____"
]
],
[
[
"prob_worse_than(-5000, sample_diff2)",
"_____no_output_____"
]
],
[
[
"And here's the probability they underbid by more than \\\\$10,000.",
"_____no_output_____"
]
],
[
[
"prob_worse_than(-10000, sample_diff2)",
"_____no_output_____"
]
],
[
[
"We can combine these functions to compute the probability that Player 1 wins, given the difference between their bid and the actual price:",
"_____no_output_____"
]
],
[
[
"def compute_prob_win(diff, sample_diff):\n \"\"\"Probability of winning for a given diff.\"\"\"\n # if you overbid you lose\n if diff > 0:\n return 0\n \n # if the opponent overbids, you win\n p1 = prob_overbid(sample_diff)\n \n # or of their bid is worse than yours, you win\n p2 = prob_worse_than(diff, sample_diff)\n \n # p1 and p2 are mutually exclusive, so we can add them\n return p1 + p2",
"_____no_output_____"
]
],
[
[
"Here's the probability that you win, given that you underbid by \\\\$5000.",
"_____no_output_____"
]
],
[
[
"compute_prob_win(-5000, sample_diff2)",
"_____no_output_____"
]
],
[
[
"Now let's look at the probability of winning for a range of possible differences.",
"_____no_output_____"
]
],
[
[
"xs = np.linspace(-30000, 5000, 121)\nys = [compute_prob_win(x, sample_diff2) \n for x in xs]",
"_____no_output_____"
]
],
[
[
"Here's what it looks like:",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nplt.plot(xs, ys)\n\ndecorate(xlabel='Difference between bid and actual price ($)',\n ylabel='Probability of winning',\n title='Player 1')",
"_____no_output_____"
]
],
[
[
"If you underbid by \\\\$30,000, the chance of winning is about 30%, which is mostly the chance your opponent overbids.\n\nAs your bids gets closer to the actual price, your chance of winning approaches 1.\n\nAnd, of course, if you overbid, you lose (even if your opponent also overbids).",
"_____no_output_____"
],
[
"**Exercise:** Run the same analysis from the point of view of Player 2. Using the sample of differences from Player 1, compute:\n\n1. The probability that Player 1 overbids.\n\n2. The probability that Player 1 underbids by more than \\\\$5000.\n\n3. The probability that Player 2 wins, given that they underbid by \\\\$5000.\n\nThen plot the probability that Player 2 wins for a range of possible differences between their bid and the actual price.",
"_____no_output_____"
]
],
[
[
"# Solution\n\nprob_overbid(sample_diff1)",
"_____no_output_____"
],
[
"# Solution\n\nprob_worse_than(-5000, sample_diff1)",
"_____no_output_____"
],
[
"# Solution\n\ncompute_prob_win(-5000, sample_diff1)",
"_____no_output_____"
],
[
"# Solution\n\nxs = np.linspace(-30000, 5000, 121)\nys = [compute_prob_win(x, sample_diff1) for x in xs]",
"_____no_output_____"
],
[
"# Solution\n\nplt.plot(xs, ys)\n\ndecorate(xlabel='Difference between bid and actual price ($)',\n ylabel='Probability of winning',\n title='Player 2')",
"_____no_output_____"
]
],
[
[
"## Decision Analysis\n\nIn the previous section we computed the probability of winning, given that we have underbid by a particular amount.\n\nIn reality the contestants don't know how much they have underbid by, because they don't know the actual price.\n\nBut they do have a posterior distribution that represents their beliefs about the actual price, and they can use that to estimate their probability of winning with a given bid.\n\nThe following function takes a possible bid, a posterior distribution of actual prices, and a sample of differences for the opponent.\n\nIt loops through the hypothetical prices in the posterior distribution and, for each price,\n\n1. Computes the difference between the bid and the hypothetical price,\n\n2. Computes the probability that the player wins, given that difference, and\n\n3. Adds up the weighted sum of the probabilities, where the weights are the probabilities in the posterior distribution. ",
"_____no_output_____"
]
],
[
[
"def total_prob_win(bid, posterior, sample_diff):\n \"\"\"Computes the total probability of winning with a given bid.\n\n bid: your bid\n posterior: Pmf of showcase value\n sample_diff: sequence of differences for the opponent\n \n returns: probability of winning\n \"\"\"\n total = 0\n for price, prob in posterior.items():\n diff = bid - price\n total += prob * compute_prob_win(diff, sample_diff)\n return total",
"_____no_output_____"
]
],
[
[
"This loop implements the law of total probability:\n\n$$P(win) = \\sum_{price} P(price) ~ P(win ~|~ price)$$\n\nHere's the probability that Player 1 wins, based on a bid of \\\\$25,000 and the posterior distribution `posterior1`.",
"_____no_output_____"
]
],
[
[
"total_prob_win(25000, posterior1, sample_diff2)",
"_____no_output_____"
]
],
[
[
"Now we can loop through a series of possible bids and compute the probability of winning for each one.",
"_____no_output_____"
]
],
[
[
"bids = posterior1.qs\n\nprobs = [total_prob_win(bid, posterior1, sample_diff2) \n for bid in bids]\n\nprob_win_series = pd.Series(probs, index=bids)",
"_____no_output_____"
]
],
[
[
"Here are the results.",
"_____no_output_____"
]
],
[
[
"prob_win_series.plot(label='Player 1', color='C1')\n\ndecorate(xlabel='Bid ($)',\n ylabel='Probability of winning',\n title='Optimal bid: probability of winning')",
"_____no_output_____"
]
],
[
[
"And here's the bid that maximizes Player 1's chance of winning.",
"_____no_output_____"
]
],
[
[
"prob_win_series.idxmax()",
"_____no_output_____"
],
[
"prob_win_series.max()",
"_____no_output_____"
]
],
[
[
"Recall that your guess was \\\\$23,000.\nUsing your guess to compute the posterior distribution, the posterior mean is about \\\\$26,000.\nBut the bid that maximizes your chance of winning is \\\\$21,000.",
"_____no_output_____"
],
[
"**Exercise:** Do the same analysis for Player 2.",
"_____no_output_____"
]
],
[
[
"# Solution\n\nbids = posterior2.qs\n\nprobs = [total_prob_win(bid, posterior2, sample_diff1) \n for bid in bids]\n\nprob_win_series = pd.Series(probs, index=bids)",
"_____no_output_____"
],
[
"# Solution\n\nprob_win_series.plot(label='Player 2', color='C1')\n\ndecorate(xlabel='Bid ($)',\n ylabel='Probability of winning',\n title='Optimal bid: probability of winning')",
"_____no_output_____"
],
[
"# Solution\n\nprob_win_series.idxmax()",
"_____no_output_____"
],
[
"# Solution\n\nprob_win_series.max()",
"_____no_output_____"
]
],
[
[
"## Maximizing Expected Gain\n\nIn the previous section we computed the bid that maximizes your chance of winning.\nAnd if that's your goal, the bid we computed is optimal.\n\nBut winning isn't everything.\nRemember that if your bid is off by \\\\$250 or less, you win both showcases.\nSo it might be a good idea to increase your bid a little: it increases the chance you overbid and lose, but it also increases the chance of winning both showcases.\n\nLet's see how that works out.\nThe following function computes how much you will win, on average, given your bid, the actual price, and a sample of errors for your opponent.",
"_____no_output_____"
]
],
[
[
"def compute_gain(bid, price, sample_diff):\n \"\"\"Compute expected gain given a bid and actual price.\"\"\"\n diff = bid - price\n prob = compute_prob_win(diff, sample_diff)\n\n # if you are within 250 dollars, you win both showcases\n if -250 <= diff <= 0:\n return 2 * price * prob\n else:\n return price * prob",
"_____no_output_____"
]
],
[
[
"For example, if the actual price is \\\\$35000 \nand you bid \\\\$30000, \nyou will win about \\\\$23,600 worth of prizes on average, taking into account your probability of losing, winning one showcase, or winning both.",
"_____no_output_____"
]
],
[
[
"compute_gain(30000, 35000, sample_diff2)",
"_____no_output_____"
]
],
[
[
"In reality we don't know the actual price, but we have a posterior distribution that represents what we know about it.\nBy averaging over the prices and probabilities in the posterior distribution, we can compute the expected gain for a particular bid.\n\nIn this context, \"expected\" means the average over the possible showcase values, weighted by their probabilities.",
"_____no_output_____"
]
],
[
[
"def expected_gain(bid, posterior, sample_diff):\n \"\"\"Compute the expected gain of a given bid.\"\"\"\n total = 0\n for price, prob in posterior.items():\n total += prob * compute_gain(bid, price, sample_diff)\n return total",
"_____no_output_____"
]
],
[
[
"For the posterior we computed earlier, based on a guess of \\\\$23,000, the expected gain for a bid of \\\\$21,000 is about \\\\$16,900.",
"_____no_output_____"
]
],
[
[
"expected_gain(21000, posterior1, sample_diff2)",
"_____no_output_____"
]
],
[
[
"But can we do any better? \n\nTo find out, we can loop through a range of bids and find the one that maximizes expected gain.",
"_____no_output_____"
]
],
[
[
"bids = posterior1.qs\n\ngains = [expected_gain(bid, posterior1, sample_diff2) for bid in bids]\n\nexpected_gain_series = pd.Series(gains, index=bids)",
"_____no_output_____"
]
],
[
[
"Here are the results.",
"_____no_output_____"
]
],
[
[
"expected_gain_series.plot(label='Player 1', color='C2')\n\ndecorate(xlabel='Bid ($)',\n ylabel='Expected gain ($)',\n title='Optimal bid: expected gain')",
"_____no_output_____"
]
],
[
[
"Here is the optimal bid.",
"_____no_output_____"
]
],
[
[
"expected_gain_series.idxmax()",
"_____no_output_____"
]
],
[
[
"With that bid, the expected gain is about \\\\$17,400.",
"_____no_output_____"
]
],
[
[
"expected_gain_series.max()",
"_____no_output_____"
]
],
[
[
"Recall that your initial guess was \\\\$23,000.\nThe bid that maximizes the chance of winning is \\\\$21,000.\nAnd the bid that maximizes your expected gain is \\\\$22,000.",
"_____no_output_____"
],
[
"**Exercise:** Do the same analysis for Player 2.",
"_____no_output_____"
]
],
[
[
"# Solution\n\nbids = posterior2.qs\n\ngains = [expected_gain(bid, posterior2, sample_diff1) for bid in bids]\n\nexpected_gain_series = pd.Series(gains, index=bids)",
"_____no_output_____"
],
[
"# Solution\n\nexpected_gain_series.plot(label='Player 2', color='C2')\n\ndecorate(xlabel='Bid ($)',\n ylabel='Expected gain ($)',\n title='Optimal bid: expected gain')",
"_____no_output_____"
],
[
"# Solution\n\nexpected_gain_series.idxmax()",
"_____no_output_____"
],
[
"# Solution\n\nexpected_gain_series.max()",
"_____no_output_____"
]
],
[
[
"## Summary\n\nThere's a lot going on this this chapter, so let's review the steps:\n\n1. First we used KDE and data from past shows to estimate prior distributions for the values of the showcases.\n\n2. Then we used bids from past shows to model the distribution of errors as a normal distribution.\n\n3. We did a Bayesian update using the distribution of errors to compute the likelihood of the data.\n\n4. We used the posterior distribution for the value of the showcase to compute the probability of winning for each possible bid, and identified the bid that maximizes the chance of winning.\n\n5. Finally, we used probability of winning to compute the expected gain for each possible bid, and identified the bid that maximizes expected gain.\n\nIncidentally, this example demonstrates the hazard of using the word \"optimal\" without specifying what you are optimizing.\nThe bid that maximizes the chance of winning is not generally the same as the bid that maximizes expected gain.",
"_____no_output_____"
],
[
"## Discussion\n\nWhen people discuss the pros and cons of Bayesian estimation, as contrasted with classical methods sometimes called \"frequentist\", they often claim that in many cases Bayesian methods and frequentist methods produce the same results.\n\nIn my opinion, this claim is mistaken because Bayesian and frequentist method produce different *kinds* of results:\n\n* The result of frequentist methods is usually a single value that is considered to be the best estimate (by one of several criteria) or an interval that quantifies the precision of the estimate.\n\n* The result of Bayesian methods is a posterior distribution that represents all possible outcomes and their probabilities.",
"_____no_output_____"
],
[
"Granted, you can use the posterior distribution to choose a \"best\" estimate or compute an interval.\nAnd in that case the result might be the same as the frequentist estimate.\n\nBut doing so discards useful information and, in my opinion, eliminates the primary benefit of Bayesian methods: the posterior distribution is more useful than a single estimate, or even an interval.",
"_____no_output_____"
],
[
"The example in this chapter demonstrates the point.\nUsing the entire posterior distribution, we can compute the bid that maximizes the probability of winning, or the bid that maximizes expected gain, even if the rules for computing the gain are complicated (and nonlinear).\n\nWith a single estimate or an interval, we can't do that, even if they are \"optimal\" in some sense.\nIn general, frequentist estimation provides little guidance for decision-making.\n\nIf you hear someone say that Bayesian and frequentist methods produce the same results, you can be confident that they don't understand Bayesian methods.",
"_____no_output_____"
],
[
"## Exercises",
"_____no_output_____"
],
[
"**Exercise:** When I worked in Cambridge, Massachusetts, I usually took the subway to South Station and then a commuter train home to Needham. Because the subway was unpredictable, I left the office early enough that I could wait up to 15 minutes and still catch the commuter train.\n\nWhen I got to the subway stop, there were usually about 10 people waiting on the platform. If there were fewer than that, I figured I just missed a train, so I expected to wait a little longer than usual. And if there there more than that, I expected another train soon.\n\nBut if there were a *lot* more than 10 passengers waiting, I inferred that something was wrong, and I expected a long wait. In that case, I might leave and take a taxi.\n\nWe can use Bayesian decision analysis to quantify the analysis I did intuitively. Given the number of passengers on the platform, how long should we expect to wait? And when should we give up and take a taxi?\n\nMy analysis of this problem is in `redline.ipynb`, which is in the repository for this book. [Click here to run this notebook on Colab](https://colab.research.google.com/github/AllenDowney/ThinkBayes2/blob/master/notebooks/redline.ipynb).",
"_____no_output_____"
],
[
"**Exercise:** This exercise is inspired by a true story. In 2001 I created [Green Tea Press](https://greenteapress.com) to publish my books, starting with *Think Python*. I ordered 100 copies from a short run printer and made the book available for sale through a distributor. \n\nAfter the first week, the distributor reported that 12 copies were sold. Based that report, I thought I would run out of copies in about 8 weeks, so I got ready to order more. My printer offered me a discount if I ordered more than 1000 copies, so I went a little crazy and ordered 2000. \n\nA few days later, my mother called to tell me that her *copies* of the book had arrived. Surprised, I asked how many. She said ten.\n\nIt turned out I had sold only two books to non-relatives. And it took a lot longer than I expected to sell 2000 copies.",
"_____no_output_____"
],
[
"The details of this story are unique, but the general problem is something almost every retailer has to figure out. Based on past sales, how do you predict future sales? And based on those predictions, how do you decide how much to order and when?\n\nOften the cost of a bad decision is complicated. If you place a lot of small orders rather than one big one, your costs are likely to be higher. If you run out of inventory, you might lose customers. And if you order too much, you have to pay the various costs of holding inventory.\n\nSo, let's solve a version of the problem I faced. It will take some work to set up the problem; the details are in the notebook for this chapter.",
"_____no_output_____"
],
[
"Suppose you start selling books online. During the first week you sell 10 copies (and let's assume that none of the customers are your mother). During the second week you sell 9 copies.\n\nAssuming that the arrival of orders is a Poisson process, we can think of the weekly orders as samples from a Poisson distribution with an unknown rate.\nWe can use orders from past weeks to estimate the parameter of this distribution, generate a predictive distribution for future weeks, and compute the order size that maximized expected profit.\n\n* Suppose the cost of printing the book is \\\\$5 per copy, \n\n* But if you order 100 or more, it's \\\\$4.50 per copy.\n\n* For every book you sell, you get \\\\$10.\n\n* But if you run out of books before the end of 8 weeks, you lose \\\\$50 in future sales for every week you are out of stock.\n\n* If you have books left over at the end of 8 weeks, you lose \\\\$2 in inventory costs per extra book.\n\nFor example, suppose you get orders for 10 books per week, every week. If you order 60 books, \n\n* The total cost is \\\\$300. \n\n* You sell all 60 books, so you make \\\\$600. \n\n* But the book is out of stock for two weeks, so you lose \\\\$100 in future sales.\n\nIn total, your profit is \\\\$200.\n\nIf you order 100 books,\n\n* The total cost is \\\\$450.\n\n* You sell 80 books, so you make \\\\$800.\n\n* But you have 20 books left over at the end, so you lose \\\\$40.\n\nIn total, your profit is \\\\$310.\n\nCombining these costs with your predictive distribution, how many books should you order to maximize your expected profit?",
"_____no_output_____"
],
[
"To get you started, the following functions compute profits and costs according to the specification of the problem:",
"_____no_output_____"
]
],
[
[
"def print_cost(printed):\n \"\"\"Compute print costs.\n \n printed: integer number printed\n \"\"\"\n if printed < 100:\n return printed * 5\n else:\n return printed * 4.5",
"_____no_output_____"
],
[
"def total_income(printed, orders):\n \"\"\"Compute income.\n \n printed: integer number printed\n orders: sequence of integer number of books ordered\n \"\"\"\n sold = min(printed, np.sum(orders))\n return sold * 10",
"_____no_output_____"
],
[
"def inventory_cost(printed, orders):\n \"\"\"Compute inventory costs.\n \n printed: integer number printed\n orders: sequence of integer number of books ordered\n \"\"\"\n excess = printed - np.sum(orders)\n if excess > 0:\n return excess * 2\n else:\n return 0",
"_____no_output_____"
],
[
"def out_of_stock_cost(printed, orders):\n \"\"\"Compute out of stock costs.\n \n printed: integer number printed\n orders: sequence of integer number of books ordered\n \"\"\"\n weeks = len(orders)\n total_orders = np.cumsum(orders)\n for i, total in enumerate(total_orders):\n if total > printed:\n return (weeks-i) * 50\n return 0",
"_____no_output_____"
],
[
"def compute_profit(printed, orders):\n \"\"\"Compute profit.\n \n printed: integer number printed\n orders: sequence of integer number of books ordered\n \"\"\"\n return (total_income(printed, orders) -\n print_cost(printed)-\n out_of_stock_cost(printed, orders) -\n inventory_cost(printed, orders))",
"_____no_output_____"
]
],
[
[
"To test these functions, suppose we get exactly 10 orders per week for eight weeks:",
"_____no_output_____"
]
],
[
[
"always_10 = [10] * 8\nalways_10",
"_____no_output_____"
]
],
[
[
"If you print 60 books, your net profit is \\\\$200, as in the example.",
"_____no_output_____"
]
],
[
[
"compute_profit(60, always_10)",
"_____no_output_____"
]
],
[
[
"If you print 100 books, your net profit is \\\\$310.",
"_____no_output_____"
]
],
[
[
"compute_profit(100, always_10)",
"_____no_output_____"
]
],
[
[
"Of course, in the context of the problem you don't know how many books will be ordered in any given week. You don't even know the average rate of orders. However, given the data and some assumptions about the prior, you can compute the distribution of the rate of orders.\n\nYou'll have a chance to do that, but to demonstrate the decision analysis part of the problem, I'll start with the arbitrary assumption that order rates come from a gamma distribution with mean 9.\n\nHere's a `Pmf` that represents this distribution.",
"_____no_output_____"
]
],
[
[
"from scipy.stats import gamma\n\nalpha = 9\nqs = np.linspace(0, 25, 101)\nps = gamma.pdf(qs, alpha)\npmf = Pmf(ps, qs)\npmf.normalize()\npmf.mean()",
"_____no_output_____"
]
],
[
[
"And here's what it looks like:",
"_____no_output_____"
]
],
[
[
"pmf.plot(color='C1')\ndecorate(xlabel=r'Book ordering rate ($\\lambda$)',\n ylabel='PMF')",
"_____no_output_____"
]
],
[
[
"Now, we *could* generate a predictive distribution for the number of books ordered in a given week, but in this example we have to deal with a complicated cost function. In particular, `out_of_stock_cost` depends on the sequence of orders.\n\nSo, rather than generate a predictive distribution, I suggest we run simulations. I'll demonstrate the steps.\n\nFirst, from our hypothetical distribution of rates, we can draw a random sample of 1000 values. ",
"_____no_output_____"
]
],
[
[
"rates = pmf.choice(1000)\nnp.mean(rates)",
"_____no_output_____"
]
],
[
[
"For each possible rate, we can generate a sequence of 8 orders.",
"_____no_output_____"
]
],
[
[
"np.random.seed(17)\norder_array = np.random.poisson(rates, size=(8, 1000)).transpose()\norder_array[:5, :]",
"_____no_output_____"
]
],
[
[
"Each row of this array is a hypothetical sequence of orders based on a different hypothetical order rate.\n\nNow, if you tell me how many books you printed, I can compute your expected profits, averaged over these 1000 possible sequences.",
"_____no_output_____"
]
],
[
[
"def compute_expected_profits(printed, order_array):\n \"\"\"Compute profits averaged over a sample of orders.\n \n printed: number printed\n order_array: one row per sample, one column per week\n \"\"\"\n profits = [compute_profit(printed, orders)\n for orders in order_array]\n return np.mean(profits)",
"_____no_output_____"
]
],
[
[
"For example, here are the expected profits if you order 70, 80, or 90 books.",
"_____no_output_____"
]
],
[
[
"compute_expected_profits(70, order_array)",
"_____no_output_____"
],
[
"compute_expected_profits(80, order_array)",
"_____no_output_____"
],
[
"compute_expected_profits(90, order_array)",
"_____no_output_____"
]
],
[
[
"Now, let's sweep through a range of values and compute expected profits as a function of the number of books you print.",
"_____no_output_____"
]
],
[
[
"printed_array = np.arange(70, 110)\nt = [compute_expected_profits(printed, order_array)\n for printed in printed_array]\nexpected_profits = pd.Series(t, printed_array)",
"_____no_output_____"
],
[
"expected_profits.plot(label='')\n\ndecorate(xlabel='Number of books printed',\n ylabel='Expected profit ($)')",
"_____no_output_____"
]
],
[
[
"Here is the optimal order and the expected profit.",
"_____no_output_____"
]
],
[
[
"expected_profits.idxmax(), expected_profits.max()",
"_____no_output_____"
]
],
[
[
"Now it's your turn. Choose a prior that you think is reasonable, update it with the data you are given, and then use the posterior distribution to do the analysis I just demonstrated.",
"_____no_output_____"
]
],
[
[
"# Solution\n\n# For a prior I chose a log-uniform distribution; \n# that is, a distribution that is uniform in log-space\n# from 1 to 100 books per week.\n\nqs = np.logspace(0, 2, 101)\nprior = Pmf(1, qs)\nprior.normalize()",
"_____no_output_____"
],
[
"# Solution\n\n# Here's the CDF of the prior\n\nprior.make_cdf().plot(color='C1')\ndecorate(xlabel=r'Book ordering rate ($\\lambda$)',\n ylabel='CDF')",
"_____no_output_____"
],
[
"# Solution\n\n# Here's a function that updates the distribution of lambda\n# based on one week of orders\n\nfrom scipy.stats import poisson\n\ndef update_book(pmf, data):\n \"\"\"Update book ordering rate.\n \n pmf: Pmf of book ordering rates\n data: observed number of orders in one week\n \"\"\"\n k = data\n lams = pmf.index\n likelihood = poisson.pmf(k, lams)\n pmf *= likelihood\n pmf.normalize()",
"_____no_output_____"
],
[
"# Solution\n\n# Here's the update after week 1.\n\nposterior1 = prior.copy()\nupdate_book(posterior1, 10)",
"_____no_output_____"
],
[
"# Solution\n\n# And the update after week 2.\n\nposterior2 = posterior1.copy()\nupdate_book(posterior2, 9)",
"_____no_output_____"
],
[
"# Solution\n\nprior.mean(), posterior1.mean(), posterior2.mean()",
"_____no_output_____"
],
[
"# Solution\n\n# Now we can generate a sample of 1000 values from the posterior\n\nrates = posterior2.choice(1000)\nnp.mean(rates)",
"_____no_output_____"
],
[
"# Solution\n\n# And we can generate a sequence of 8 weeks for each value\n\norder_array = np.random.poisson(rates, size=(8, 1000)).transpose()\norder_array[:5, :]",
"_____no_output_____"
],
[
"# Solution\n\n# Here are the expected profits for each possible order\n\nprinted_array = np.arange(70, 110)\nt = [compute_expected_profits(printed, order_array)\n for printed in printed_array]\nexpected_profits = pd.Series(t, printed_array)",
"_____no_output_____"
],
[
"# Solution\n\n# And here's what they look like.\n\nexpected_profits.plot(label='')\n\ndecorate(xlabel='Number of books printed',\n ylabel='Expected profit ($)')",
"_____no_output_____"
],
[
"# Solution\n\n# Here's the optimal order.\n\nexpected_profits.idxmax()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8efd86b8a6f3d315bc6606a36dc8dfce7d564c
| 550,506 |
ipynb
|
Jupyter Notebook
|
hw1-geomviz/Special_Euclidean/Special_EUC.ipynb
|
bioshape-lab/ece594n
|
01ab65108ee6bdff0e88fd2a0b4dcc6e0d2cf580
|
[
"MIT"
] | 2 |
2022-03-30T00:47:45.000Z
|
2022-03-31T18:22:16.000Z
|
hw1-geomviz/Special_Euclidean/Special_EUC.ipynb
|
bioshape-lab/ece594n
|
01ab65108ee6bdff0e88fd2a0b4dcc6e0d2cf580
|
[
"MIT"
] | null | null | null |
hw1-geomviz/Special_Euclidean/Special_EUC.ipynb
|
bioshape-lab/ece594n
|
01ab65108ee6bdff0e88fd2a0b4dcc6e0d2cf580
|
[
"MIT"
] | null | null | null | 1,118.914634 | 129,912 | 0.960148 |
[
[
[
"# <center>HW 01: Geomviz: Visualizing Differential Geometry<center>\n\n## <center>Special Euclidean Group SE(n)<center>\n\n<center>$\\color{#003660}{\\text{Swetha Pillai, Ryan Guajardo}}$<center>\n",
"_____no_output_____"
],
[
"# <center> 1.) Mathematical Definition of Special Euclidean SE(n)<center>\n### <center> This group is defined as the set of direct isometries - or rigid-body transformations - of $R^n$.<center>\n<center>i.e. the linear transformations of the affine space $R^n$ that preserve its canonical inner-product, or euclidean distance between points.<center>\n \n \n \n \n\n***\n$$\n\\rho(x) = Rx + u\n$$\n***\n\n\n<center>$\\rho$ is comprised of a rotational part $R$ and a translational part $u$.<center>\n\n$$\n \\newline\n$$\n\n \n \n$$\n\\newline\nSE(n) = \\{(R,u)\\ \\vert\\ R \\in SO(n), u \\in R^n\\}\n\\newline\n$$\n \n<center>Where SO(n) is the special orthogonal group.<center>",
"_____no_output_____"
],
[
"# <center> 2.) Uses of Special Euclidean SE(n) in real-world applications<center>\n\n\n## <center>Rigid Body Kinematics<center>\n Can represent linear and angular displacements in rigid bodies, commonly in SE(3)\n \n\n<center><img src=\"rigid.png\" width=\"500\"/></center>\n ",
"_____no_output_____"
],
[
"\n\n## <center> Autonomous Quadcoptor Path Planning!<center>\n If we want to make a quadcopter autonomous a predefined \n path must be computed by finding collision free paths throughout\n a space whose topological structure is SE(3)\n \n<center><img src=\"quadcopterpic.jpeg\" width=\"500\"/></center>\n \n\n \n \n ",
"_____no_output_____"
],
[
"## <center> Optimal Paths for Polygonal Robots SE(2) <center>\n Similar to Autonomous Quadcopter but we are now in a 2 dimensional plane, hence SE(2)...\n \n<center><img src=\"polygonal.png\" width=\"500\"/></center>\n\n\n \n\n \n\n \n \n \n \n \n ",
"_____no_output_____"
],
[
"## <center>Projective Model of Ideal Pinhole Camera <center>\n Camera coordinate system to World coordinate system transform.\n\n<center><img src=\"pinhole.png\" width=\"500\"/></center>\n \n \n\n \n \n ",
"_____no_output_____"
],
[
"## <center>Pose Estimation <center>\n \n\n \n \n \n \n ",
"_____no_output_____"
],
[
"# 3.) Visualization of Elementary Operation on SE(3)\nShowcase your visualization can be used by plotting the inputs and outputs of operations such as exp, log, geodesics.\n",
"_____no_output_____"
]
],
[
[
"%pip install geomstats\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"Requirement already satisfied: geomstats in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (2.4.2)\nRequirement already satisfied: scikit-learn>=0.22.1 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from geomstats) (1.0.2)\nRequirement already satisfied: scipy>=1.4.1 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from geomstats) (1.7.3)\nRequirement already satisfied: matplotlib>=3.3.4 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from geomstats) (3.5.1)\nRequirement already satisfied: joblib>=0.14.1 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from geomstats) (1.1.0)\nRequirement already satisfied: pandas>=1.1.5 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from geomstats) (1.3.4)\nRequirement already satisfied: numpy>=1.18.1 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from geomstats) (1.21.5)\nRequirement already satisfied: fonttools>=4.22.0 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from matplotlib>=3.3.4->geomstats) (4.32.0)\nRequirement already satisfied: packaging>=20.0 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from matplotlib>=3.3.4->geomstats) (21.3)\nRequirement already satisfied: cycler>=0.10 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from matplotlib>=3.3.4->geomstats) (0.11.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from matplotlib>=3.3.4->geomstats) (1.3.2)\nRequirement already satisfied: python-dateutil>=2.7 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from matplotlib>=3.3.4->geomstats) (2.8.2)\nRequirement already satisfied: pyparsing>=2.2.1 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from matplotlib>=3.3.4->geomstats) (3.0.8)\nRequirement already satisfied: pillow>=6.2.0 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from matplotlib>=3.3.4->geomstats) (9.0.1)\nRequirement already satisfied: pytz>=2017.3 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from pandas>=1.1.5->geomstats) (2021.3)\nRequirement already satisfied: six>=1.5 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from python-dateutil>=2.7->matplotlib>=3.3.4->geomstats) (1.16.0)\nRequirement already satisfied: threadpoolctl>=2.0.0 in /Users/spillai/opt/anaconda3/lib/python3.9/site-packages (from scikit-learn>=0.22.1->geomstats) (3.1.0)\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"from Special_Euclidean import *\nmanifold = Special_Euclidean()",
"INFO: Using numpy backend\n"
],
[
"point = manifold.random_point()\n# point = np.array([1,1,1,1,1,1])\nmanifold.plot(point)",
"Point: [ 0.12519032 0.5730147 -0.35738215 -0.60265131 -0.87291524 -0.09512372]\nRotation: [ 0.12519032 0.5730147 -0.35738215]\nTranslation: [-0.60265131 -0.87291524 -0.09512372]\n"
],
[
"manifold.scatter(5)",
"_____no_output_____"
],
[
"random_points = manifold.random_point(2)\nmanifold.plot_exp(random_points[0], random_points[1])",
"the initial point is: [ 0.94922812 -0.73860135 -0.68533513 0.08603819 -0.64430182 -0.29405483] \n\nthe end point is: [ 0.8051104 0.5948441 -0.74687418 -0.94933289 -0.81885875 0.26294222] \n\nthe exp of the two points is: [ 1.68618658 -0.00376562 -1.39479492 -0.8632947 -1.46316058 -0.0311126 ] \n\n"
],
[
"manifold.plot_log(random_points[0], random_points[1])",
"the initial point is: [ 0.94922812 -0.73860135 -0.68533513 0.08603819 -0.64430182 -0.29405483] \n\nthe end point is: [ 0.8051104 0.5948441 -0.74687418 -0.94933289 -0.81885875 0.26294222] \n\nthe log of the two points is: [-0.01567181 1.33971892 -0.16867757 -1.03537108 -0.17455693 0.55699705] \n\n"
],
[
"# point = np.eye(6)\npoint = np.array([0,0,0,0,0,0])\n\n# all rotations and vectors equal\nvector = np.array([0.5, 0.5, 0.5, 0.5, 0.5, 0.5])\n\n#rotation in one dimension, no translation\n# vector = np.array([0,0,.9,0,0,0])\n\n#rotation in one dimension, translation in one direction\n# vector = np.array([0,0,.9,0,0,.5])\n\nN_STEPS = 10\nmanifold.plot_geodesic(point, vector, N_STEPS)",
"_____no_output_____"
]
],
[
[
"# 4.) Conclusion\n\n## SE(n) is very useful.\n\nGeomstats: https://github.com/geomstats/geomstats\n\nhttp://ingmec.ual.es/~jlblanco/papers/jlblanco2010geometry3D_techrep.pdf\n\nhttps://ieeexplore.ieee.org/document/7425231\n\nhttps://arm.stanford.edu/publications/optimal-paths-polygonal-robots-se2\n\nhttps://mappingignorance.org/2015/10/14/shortcuts-for-efficiently-moving-a-quadrotor-throughout-the-special-euclidean-group-se3-and-2/\n\nhttps://arxiv.org/abs/2111.00190\n\nhttps://www.seas.upenn.edu/~meam620/slides/kinematicsI.pdf",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a8f090cc62118ec640726ce32930bec4dcd661b
| 4,893 |
ipynb
|
Jupyter Notebook
|
copy_files.ipynb
|
Adrian-St/ba-evaluation
|
89132f6ec6748f76ad413cf0359cfd7bb4d72fcf
|
[
"MIT"
] | null | null | null |
copy_files.ipynb
|
Adrian-St/ba-evaluation
|
89132f6ec6748f76ad413cf0359cfd7bb4d72fcf
|
[
"MIT"
] | null | null | null |
copy_files.ipynb
|
Adrian-St/ba-evaluation
|
89132f6ec6748f76ad413cf0359cfd7bb4d72fcf
|
[
"MIT"
] | null | null | null | 42.181034 | 94 | 0.690578 |
[
[
[
"import os\nimport json\nfrom os.path import expanduser\n\nlabels_dir = os.path.join('labeled_data', 'evaluation', 'labels')\nhome = expanduser(\"~\")\nimages_dir = os.path.join(home, 'Pictures', 'Photos', 'Photos')\ntarget_path = os.path.join('labeled_data', 'evaluation', 'groundtruths')\n\nfor file in os.listdir(labels_dir):\n f_path = os.path.join(labels_dir, file)\n print(f_path)\n if os.path.isfile(f_path):\n label_json = json.load(open(f_path))\n image_path = os.path.join(images_dir, label_json['imagePath'])\n if os.path.isfile(image_path):\n os.rename(image_path, os.path.join(target_path, label_json['imagePath']))\n else:\n raise RuntimeError(\"Copying failed\")\n else:\n raise RuntimeError(\"Something went wrong\")\n",
"labeled_data/evaluation/labels/IMG_20181214_155028.json\nlabeled_data/evaluation/labels/IMG_20190118_154316.json\nlabeled_data/evaluation/labels/IMG_20181218_112713.json\nlabeled_data/evaluation/labels/IMG_20190122_153837.json\nlabeled_data/evaluation/labels/IMG_20181211_161943.json\nlabeled_data/evaluation/labels/IMG_20181211_161911.json\nlabeled_data/evaluation/labels/IMG_20190118_142459.json\nlabeled_data/evaluation/labels/IMG_20190118_154330.json\nlabeled_data/evaluation/labels/IMG_20181211_153048.json\nlabeled_data/evaluation/labels/IMG_20181211_161931.json\nlabeled_data/evaluation/labels/IMG_20190122_161516.json\nlabeled_data/evaluation/labels/IMG_20181211_161917.json\nlabeled_data/evaluation/labels/IMG_20190122_125829.json\nlabeled_data/evaluation/labels/IMG_20181218_123700.json\nlabeled_data/evaluation/labels/IMG_20190118_154254.json\nlabeled_data/evaluation/labels/IMG_20190122_153840.json\nlabeled_data/evaluation/labels/IMG_20181214_115346.json\nlabeled_data/evaluation/labels/IMG_20181214_155025.json\nlabeled_data/evaluation/labels/IMG_20181204_103102.json\nlabeled_data/evaluation/labels/IMG_20190118_111909.json\nlabeled_data/evaluation/labels/IMG_20190118_154309.json\nlabeled_data/evaluation/labels/IMG_20181211_161850.json\nlabeled_data/evaluation/labels/IMG_20181211_161937.json\nlabeled_data/evaluation/labels/20181130_FFwd_5_prototype.json\nlabeled_data/evaluation/labels/IMG_20181218_123718.json\nlabeled_data/evaluation/labels/IMG_20190122_161035.json\nlabeled_data/evaluation/labels/IMG_20190122_114813.json\nlabeled_data/evaluation/labels/IMG_20181211_163037.json\nlabeled_data/evaluation/labels/IMG_20190115_171503.json\nlabeled_data/evaluation/labels/IMG_20190122_125824.json\nlabeled_data/evaluation/labels/IMG_20190118_142510.json\nlabeled_data/evaluation/labels/IMG_20181214_114154.json\nlabeled_data/evaluation/labels/20181130_FFwd_1_stakeholders.json\nlabeled_data/evaluation/labels/IMG_20190118_154320.json\nlabeled_data/evaluation/labels/IMG_20181214_115253.json\nlabeled_data/evaluation/labels/IMG_20181211_161925.json\nlabeled_data/evaluation/labels/IMG_20190118_154312.json\nlabeled_data/evaluation/labels/IMG_20190115_113502.json\nlabeled_data/evaluation/labels/IMG_20190122_125836.json\nlabeled_data/evaluation/labels/IMG_20181214_114152.json\nlabeled_data/evaluation/labels/IMG_20181218_173238.json\nlabeled_data/evaluation/labels/IMG_20181214_133954.json\nlabeled_data/evaluation/labels/IMG_20190115_171450.json\nlabeled_data/evaluation/labels/IMG_20181211_161854.json\nlabeled_data/evaluation/labels/IMG_20181214_102412.json\nlabeled_data/evaluation/labels/IMG_20181211_161858.json\nlabeled_data/evaluation/labels/IMG_20181214_111206.json\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
4a8f1114e395f4f839e3d786c240ef6faa13fa04
| 71,792 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Intervar_data_checking-checkpoint.ipynb
|
krishdb38/Bio_python
|
e20ba5579e1e6b4ecf9fe72ef9e21a5960416ae4
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Intervar_data_checking-checkpoint.ipynb
|
krishdb38/Bio_python
|
e20ba5579e1e6b4ecf9fe72ef9e21a5960416ae4
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Intervar_data_checking-checkpoint.ipynb
|
krishdb38/Bio_python
|
e20ba5579e1e6b4ecf9fe72ef9e21a5960416ae4
|
[
"MIT"
] | null | null | null | 51.024876 | 11,160 | 0.451931 |
[
[
[
"import pandas as pd\nfrom IPython.display import display\npd.options.display.max_columns = 20\npd.set_option('display.max_colwidth',500)\n# Since Our Data is Big so Lets see in small\n\nimport numpy as np\n\n# For Pyspark \nfrom pyspark.sql import SparkSession",
"_____no_output_____"
]
],
[
[
"## df = Panda DataFrame ds = Spark Data Frame",
"_____no_output_____"
]
],
[
[
"#df = pd.read_csv(\"../datas_not_to_upload/989.hg19_multianno.txt.intervar\", sep=\"\\t\", low_memory=False)\ndf = pd.read_csv(\"../datas_not_to_upload/1428/1428.hg19_multianno.txt.intervar\", sep = \"\\t\", low_memory=False)",
"_____no_output_____"
],
[
"df.style.set_table_attributes('style=\"font-size:10px\"')\ndf.head()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"new_col = ['Chr', 'Start', 'End', 'Ref', 'Alt', 'Ref_Gene', 'Func_refGene',\n 'ExonicFunc_refGene', 'Gene_ensGene', 'avsnp147', 'AAChange_ensGene',\n 'AAChange_refGene', 'Clinvar','InterVar_Evidence ', 'Freq_gnomAD_genome_ALL',\n 'Freq_esp6500siv2_all', 'Freq_1000g2015aug_all', 'CADD_raw',\n 'CADD_phred', 'SIFT_score', 'GERP++_RS', 'phyloP46way_placental',\n 'dbscSNV_ADA_SCORE', 'dbscSNV_RF_SCORE', 'Interpro_domain',\n 'AAChange.knownGene', 'rmsk', 'MetaSVM_score',\n 'Freq_gnomAD_genome_POPs', 'OMIM', 'Phenotype_MIM', 'OrphaNumber',\n 'Orpha', 'Otherinfo']",
"_____no_output_____"
],
[
"df.dtypes # Columns name with Data Type ",
"_____no_output_____"
]
],
[
[
"## Loading By Using Pyspark",
"_____no_output_____"
]
],
[
[
"spark = SparkSession.builder.appName(\"Inter Var data analysis\")\\\n .config(\"spark.some.config.option\", \"some-value\")\\\n .getOrCreate()",
"_____no_output_____"
],
[
"#df = spark.read.format(\"com.databricks.spark.csv\").\\\n# options(header = \"true\", inferschema = \"true\").\\\n# load(\"../datas_not_to_upload/989.hg19_multianno.txt.intervar\")\n\n# Load Data Frame from CSV\nds = spark.read.csv(\"../datas_not_to_upload/989.hg19_multianno.txt.intervar\", header= True, inferSchema= True,sep=\"\\t\")",
"_____no_output_____"
],
[
"ds.show() ",
"+----+-----+-----+---+---+------------+--------------+------------------+--------------------+-----------+----------------+----------------+-----------------+---------------------------------+----------------------+--------------------+---------------------+--------+----------+----------+---------+---------------------+-----------------+----------------+---------------+------------------+----------------+-------------+-----------------------+----+-------------+-----------+-----+---------+\n|#Chr|Start| End|Ref|Alt| Ref.Gene| Func.refGene|ExonicFunc.refGene| Gene.ensGene| avsnp147|AAChange.ensGene|AAChange.refGene|clinvar: Clinvar | InterVar: InterVar and Evidence |Freq_gnomAD_genome_ALL|Freq_esp6500siv2_all|Freq_1000g2015aug_all|CADD_raw|CADD_phred|SIFT_score|GERP++_RS|phyloP46way_placental|dbscSNV_ADA_SCORE|dbscSNV_RF_SCORE|Interpro_domain|AAChange.knownGene| rmsk|MetaSVM_score|Freq_gnomAD_genome_POPs|OMIM|Phenotype_MIM|OrphaNumber|Orpha|Otherinfo|\n+----+-----+-----+---+---+------------+--------------+------------------+--------------------+-----------+----------------+----------------+-----------------+---------------------------------+----------------------+--------------------+---------------------+--------+----------+----------+---------+---------------------+-----------------+----------------+---------------+------------------+----------------+-------------+-----------------------+----+-------------+-----------+-----+---------+\n| 1|13868|13868| A| G| DDX11L1| ncRNA_exonic| .| ENSG00000223972|rs796086906| .| .| clinvar: UNK | InterVar: Benign...| 0.1244| .| .| .| .| .| .| .| .| .| .| .| .| .| AFR:0.0225,AMR:0....| .| .| null| null| 1|\n| 1|13868|13868| A| G|LOC102725121| ncRNA_exonic| .| ENSG00000223972|rs796086906| .| .| clinvar: UNK | InterVar: Benign...| 0.1244| .| .| .| .| .| .| .| .| .| .| .| .| .| AFR:0.0225,AMR:0....| .| .| null| null| 1|\n| 1|15274|15274| A| G| WASH7P|ncRNA_intronic| .| ENSG00000227232| rs62636497| .| .| clinvar: UNK | InterVar: Benign...| 0.3717| .| 0.347244| .| .| .| .| .| .| .| .| .| Name=MIR3| .| AFR:0.3824,AMR:0....| .| .| null| null| 1|\n| 1|15820|15820| G| T| WASH7P| ncRNA_exonic| .| ENSG00000227232| rs2691315| .| .| clinvar: UNK | InterVar: Benign...| 0.2286| .| 0.410543| .| .| .| .| .| .| .| .| .| .| .| AFR:0.4290,AMR:0....| .| .| null| null| 1|\n| 1|15903|15903| -| C| WASH7P| ncRNA_exonic| .|ENSG00000227232;E...|rs557514207| .| .| clinvar: UNK | InterVar: Benign...| 0.5042| .| 0.441094| .| .| .| .| .| .| .| .| .| .| .| AFR:0.2031,AMR:0....| .| .| null| null| 1|\n| 1|16921|16922| GG| -| WASH7P| ncRNA_exonic| .| ENSG00000227232| .| .| .| clinvar: UNK | InterVar: Uncert...| .| .| .| .| .| .| .| .| .| .| .| .| .| .| AFR:.,AMR:.,EAS:....| .| .| null| null| 1|\n| 1|19004|19004| A| G| WASH7P|ncRNA_intronic| .| ENSG00000227232| rs62101625| .| .| clinvar: UNK | InterVar: Benign...| 0.4806| .| .| .| .| .| .| .| .| .| .| .| Name=L2a| .| AFR:0.4218,AMR:0....| .| .| null| null| 1|\n| 1|20304|20304| G| C| WASH7P|ncRNA_intronic| .| ENSG00000227232| rs6682950| .| .| clinvar: UNK | InterVar: Benign...| 0.4988| .| .| .| .| .| .| .| .| .| .| .| Name=L3| .| AFR:0.4909,AMR:0....| .| .| null| null| 1|\n| 1|20321|20321| A| C| WASH7P|ncRNA_intronic| .| ENSG00000227232| rs6674403| .| .| clinvar: UNK | InterVar: Benign...| 0.5281| .| .| .| .| .| .| .| .| .| .| .| Name=L3| .| AFR:0.5059,AMR:0....| .| .| null| null| 1|\n| 1|20467|20467| C| T| WASH7P|ncRNA_intronic| .| ENSG00000227232| rs6661256| .| .| clinvar: UNK | InterVar: Benign...| 0.4989| .| .| .| .| .| .| .| .| .| .| .| .| .| AFR:0.4940,AMR:0....| .| .| null| null| 1|\n| 1|39255|39255| A| C| FAM138A| intergenic| .|ENSG00000237613;E...| rs62637793| .| .| clinvar: UNK | InterVar: Benign...| 0.3953| .| .| .| .| .| .| .| .| .| .| .|Name=MLT1E1A-int| .| AFR:0.3983,AMR:0....| .| .| null| null| 1|\n| 1|39255|39255| A| C| OR4F5| intergenic| .|ENSG00000237613;E...| rs62637793| .| .| clinvar: UNK | InterVar: Benign...| 0.3953| .| .| .| .| .| .| .| .| .| .| .|Name=MLT1E1A-int| .| AFR:0.3983,AMR:0....| .| .| null| null| 1|\n| 1|39261|39261| T| C| FAM138A| intergenic| .|ENSG00000237613;E...|rs200677948| .| .| clinvar: UNK | InterVar: Benign...| 0.5604| .| .| .| .| .| .| .| .| .| .| .|Name=MLT1E1A-int| .| AFR:0.5066,AMR:0....| .| .| null| null| 1|\n| 1|39261|39261| T| C| OR4F5| intergenic| .|ENSG00000237613;E...|rs200677948| .| .| clinvar: UNK | InterVar: Benign...| 0.5604| .| .| .| .| .| .| .| .| .| .| .|Name=MLT1E1A-int| .| AFR:0.5066,AMR:0....| .| .| null| null| 1|\n| 1|40706|40706| G| A| FAM138A| intergenic| .|ENSG00000237613;E...| .| .| .| clinvar: UNK | InterVar: Uncert...| 0| .| .| .| .| .| .| .| .| .| .| .| Name=AluSz6| .| AFR:0,AMR:0,EAS:....| .| .| null| null| 1|\n| 1|40706|40706| G| A| OR4F5| intergenic| .|ENSG00000237613;E...| .| .| .| clinvar: UNK | InterVar: Uncert...| 0| .| .| .| .| .| .| .| .| .| .| .| Name=AluSz6| .| AFR:0,AMR:0,EAS:....| .| .| null| null| 1|\n| 1|46873|46873| A| T| FAM138A| intergenic| .|ENSG00000237613;E...|rs768965109| .| .| clinvar: UNK | InterVar: Benign...| 0.1786| .| .| .| .| .| .| .| .| .| .| .| .| .| AFR:0.1232,AMR:0....| .| .| null| null| 1|\n| 1|46873|46873| A| T| OR4F5| intergenic| .|ENSG00000237613;E...|rs768965109| .| .| clinvar: UNK | InterVar: Benign...| 0.1786| .| .| .| .| .| .| .| .| .| .| .| .| .| AFR:0.1232,AMR:0....| .| .| null| null| 1|\n| 1|49298|49298| T| C| FAM138A| intergenic| .|ENSG00000237613;E...| rs10399793| .| .| clinvar: UNK | InterVar: Benign...| 0.6127| .| 0.782149| .| .| .| .| .| .| .| .| .| Name=L1PA14| .| AFR:0.3537,AMR:0....| .| .| null| null| 1|\n| 1|49298|49298| T| C| OR4F5| intergenic| .|ENSG00000237613;E...| rs10399793| .| .| clinvar: UNK | InterVar: Benign...| 0.6127| .| 0.782149| .| .| .| .| .| .| .| .| .| Name=L1PA14| .| AFR:0.3537,AMR:0....| .| .| null| null| 1|\n+----+-----+-----+---+---+------------+--------------+------------------+--------------------+-----------+----------------+----------------+-----------------+---------------------------------+----------------------+--------------------+---------------------+--------+----------+----------+---------+---------------------+-----------------+----------------+---------------+------------------+----------------+-------------+-----------------------+----+-------------+-----------+-----+---------+\nonly showing top 20 rows\n\n"
],
[
"ds.printSchema() # Column Name",
"root\n |-- #Chr: string (nullable = true)\n |-- Start: integer (nullable = true)\n |-- End: integer (nullable = true)\n |-- Ref: string (nullable = true)\n |-- Alt: string (nullable = true)\n |-- Ref.Gene: string (nullable = true)\n |-- Func.refGene: string (nullable = true)\n |-- ExonicFunc.refGene: string (nullable = true)\n |-- Gene.ensGene: string (nullable = true)\n |-- avsnp147: string (nullable = true)\n |-- AAChange.ensGene: string (nullable = true)\n |-- AAChange.refGene: string (nullable = true)\n |-- clinvar: Clinvar : string (nullable = true)\n |-- InterVar: InterVar and Evidence : string (nullable = true)\n |-- Freq_gnomAD_genome_ALL: string (nullable = true)\n |-- Freq_esp6500siv2_all: string (nullable = true)\n |-- Freq_1000g2015aug_all: string (nullable = true)\n |-- CADD_raw: string (nullable = true)\n |-- CADD_phred: string (nullable = true)\n |-- SIFT_score: string (nullable = true)\n |-- GERP++_RS: string (nullable = true)\n |-- phyloP46way_placental: string (nullable = true)\n |-- dbscSNV_ADA_SCORE: string (nullable = true)\n |-- dbscSNV_RF_SCORE: string (nullable = true)\n |-- Interpro_domain: string (nullable = true)\n |-- AAChange.knownGene: string (nullable = true)\n |-- rmsk: string (nullable = true)\n |-- MetaSVM_score: string (nullable = true)\n |-- Freq_gnomAD_genome_POPs: string (nullable = true)\n |-- OMIM: string (nullable = true)\n |-- Phenotype_MIM: string (nullable = true)\n |-- OrphaNumber: string (nullable = true)\n |-- Orpha: string (nullable = true)\n |-- Otherinfo: string (nullable = true)\n\n"
]
],
[
[
"## Checking Missing Value Using PySpark",
"_____no_output_____"
]
],
[
[
"from pyspark.sql.functions import count\n\ndef my_count(ds):\n \" Spark data Frame\"\n ds.agg(*[count(c).alias(c) for c in ds.columns]).show()",
"_____no_output_____"
],
[
"# fill na value in spark Data Frame\n# df.na.fill() # Replace Null Values\n# df.na.drop() # Dropping any rows with null Valus\n#df.where() # Filter rows using the given condition\n# df.filter() # Filters rows using the given Condition\n# df.distinct() # Returns distinct rows in this DataFrame\n# df.sample() # Returns a sampld subset of this DataDrame\n# df.sampleBay() # Returns a stratified sample without replacement\n",
"_____no_output_____"
]
],
[
[
"#### Joining the data Using Pyspark",
"_____no_output_____"
]
],
[
[
"# Data Join\n# left.join(right, key, how = \"*\") # * = left, right, inner, full\n",
"_____no_output_____"
],
[
"ds.describe() # Describe the data Frame Using Spark \n# Our data set is not feasible RIght Now\n",
"_____no_output_____"
],
[
"len(ds.columns)",
"_____no_output_____"
],
[
"ds.sample(fraction = 0.001).limit(10).toPandas()",
"_____no_output_____"
]
],
[
[
"### Pandas",
"_____no_output_____"
]
],
[
[
"df.head()",
"_____no_output_____"
],
[
"import pandas_profiling as pp\nimport seaborn as sns",
"_____no_output_____"
],
[
"#pp.ProfileReport(df) # Since Our Data Set is Too Big It will Take long Time ",
"_____no_output_____"
],
[
"genes_gestational = [\"EBF1\", \"EEFSEC\", \"AGTR2\", \"WNT4\", \"ADCY5\", \"RAP2C\"]\ngenes_premature = [\"EB1\", \"EEFSEC\", \"AGTR2\"]",
"_____no_output_____"
],
[
"sns.countplot(df[df[\"Ref.Gene\"].isin(genes_gestational)][\"Ref.Gene\"])",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"#df[\"clinvar: Clinvar \"].unique()\n#df['ExonicFunc.refGene'].unique()\n#df['Func.refGene'].unique()\n#df[' InterVar: InterVar and Evidence '].nunique()\n#df['Freq_gnomAD_genome_ALL'].nunique() # More than 13470\n#df['Freq_esp6500siv2_all']\n#df['CADD_raw'].nunique() # 5805\n#df[' InterVar: InterVar and Evidence '].unique()",
"_____no_output_____"
],
[
"#df.head().T\n#df[\"clinvar: Clinvar \"]",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a8f1308fcf1c60c76edc9fff5037353c5fed504
| 645 |
ipynb
|
Jupyter Notebook
|
prospect-guide/_build/html/_sources/survey/Survey.ipynb
|
deppen8/prospect-guide
|
70466a76343686f693a40af8e854c3086f7689cb
|
[
"MIT"
] | null | null | null |
prospect-guide/_build/html/_sources/survey/Survey.ipynb
|
deppen8/prospect-guide
|
70466a76343686f693a40af8e854c3086f7689cb
|
[
"MIT"
] | 9 |
2021-02-02T03:40:32.000Z
|
2021-09-10T13:35:05.000Z
|
docs/prospect-guide/survey/Survey.ipynb
|
deppen8/prospect
|
fbde75e57fef967643ca0f4c43fe53004d11da70
|
[
"MIT"
] | null | null | null | 16.538462 | 34 | 0.510078 |
[
[
[
"# Survey",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.