hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
4ae2bef459961c6dd391b0ec5649505a2dfebfab
| 186,788 |
ipynb
|
Jupyter Notebook
|
section_7-5.ipynb
|
jlperla/PerlaTonettiWaugh.jl
|
a130566ac5c061dc42631b87421f8c66b51c9668
|
[
"MIT"
] | 11 |
2019-05-02T22:36:34.000Z
|
2021-09-27T05:11:05.000Z
|
section_7-5.ipynb
|
mwaugh0328/PerlaTonettiWaugh.jl
|
a130566ac5c061dc42631b87421f8c66b51c9668
|
[
"MIT"
] | 24 |
2019-11-28T22:40:18.000Z
|
2020-03-04T17:06:57.000Z
|
section_7-5.ipynb
|
jlperla/PerlaTonettiWaugh.jl
|
a130566ac5c061dc42631b87421f8c66b51c9668
|
[
"MIT"
] | 9 |
2019-05-02T22:37:02.000Z
|
2020-12-29T13:10:20.000Z
| 241.016774 | 89,012 | 0.897226 |
[
[
[
"import pandas as pd\nimport scipy.io\nimport os\nimport matplotlib.pyplot as plt\n\npath = os.getcwd()\n\nmatlab_exe_path = '''matlab'''\n\njulia_path = '''C:\\\\Users\\\\mwaugh\\\\AppData\\\\Local\\\\Programs\\\\Julia\\\\Julia-1.4.0\\\\bin\\\\julia.exe'''\n\npath = \"src\\\\calibration\"\n#fig_path = \"C:\\\\users\\mwaugh\\\\github\\\\perla_tonetti_waugh\\\\Figures\"",
"_____no_output_____"
]
],
[
[
"---\n### [Equilibrium Technology Diffusion, Trade, and Growth](https://christophertonetti.com/files/papers/PerlaTonettiWaugh_DiffusionTradeAndGrowth.pdf) by Perla, Tonetti, and Waugh (AER 2020)\n---\n## 7.5. The Role of Firm Dynamics and Adoption Costs\n\n#### Table of Contents\n\n- [GBM](#gbm)\n\n- [Delta Shock](#detla)\n\n- [Connection to Welfare Decomposition (Section 7.3)](#dcomp)\n\nThe underlying MATLAB code is described (with links to the relevant ``.m`` files) in the [readme file in the calibraiton folder](/src/calibration/README.md).\n\n---\n\n### <a name=\"gbm\"></a> Importance of Firm Productivity Shocks (GBM)\n\nWe uniformly scale up and down the GBM variance and mean for different values of the adoption cost parameter chi. The large value of chi is ten percent larger than the baseline calibrated value. The small value of chi is ten percent smaller than the baseline calibrated value. All other parameter values are fixed, i.e., we do not re-calibrate the model when changing these parameter values.",
"_____no_output_____"
],
[
"##### Step 1. Compute outcomes for different GBM parameter values\n\nFirst, we compute a key input for the figures, saved to [``/output/robust/gbm/closest_chi_params.csv``](/output/robust/gbm/closest_chi_params.csv). Each row in this file contains the parameter values that generate a BGP equilibrium growth rate that matches the baseline aggregate growth when externally fixing a set value for chi.",
"_____no_output_____"
]
],
[
[
"matlab_cmd = '''\"cd('src\\calibration');robust_no_recalibrate_gbm;\"'''",
"_____no_output_____"
],
[
"!{matlab_exe_path} -batch {matlab_cmd}",
"Calbirated values computed on date\n 20-Apr-2020\n\n%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%\nChi Values\n 0.1409\n\nParameters for given chi which deliver growth closest to baseline growth\n Columns 1 through 13\n\n 'g' 'theta' 'kappa' 'chi' 'mu' 'upsilon' 'zeta' 'delta' 'N' 'gamma' 'eta' 'Theta' 'd_0'\n\n Column 14\n\n 'd_T'\n\n Columns 1 through 13\n\n 0.0080 4.9890 0.1042 0.1409 -0.0251 0.0390 1.0000 0.0200 10.0000 1.0000 0 1.0000 3.0225\n\n Columns 14 through 16\n\n 2.8202 0.0203 3.1669\n\nChi Values\n 0.1268\n\nParameters for given chi which deliver growth closest to baseline growth\n Columns 1 through 13\n\n 'g' 'theta' 'kappa' 'chi' 'mu' 'upsilon' 'zeta' 'delta' 'N' 'gamma' 'eta' 'Theta' 'd_0'\n\n Column 14\n\n 'd_T'\n\n Columns 1 through 13\n\n 0.0079 4.9890 0.1042 0.1268 -0.0311 0.0483 1.0000 0.0200 10.0000 1.0000 0 1.0000 3.0225\n\n Columns 14 through 16\n\n 2.8202 0.0203 3.1669\n\nChi Values\n 0.1153\n\nParameters for given chi which deliver growth closest to baseline growth\n Columns 1 through 13\n\n 'g' 'theta' 'kappa' 'chi' 'mu' 'upsilon' 'zeta' 'delta' 'N' 'gamma' 'eta' 'Theta' 'd_0'\n\n Column 14\n\n 'd_T'\n\n Columns 1 through 13\n\n 0.0079 4.9890 0.1042 0.1153 -0.0372 0.0579 1.0000 0.0200 10.0000 1.0000 0 1.0000 3.0225\n\n Columns 14 through 16\n\n 2.8202 0.0203 3.1669\n\n"
]
],
[
[
"##### Step 2. Create Figure 6 in PTW 2020\n\nThe code below reads in the output from matlab and then plots the results to generate Figure 6 of PTW.",
"_____no_output_____"
]
],
[
[
"cnames = ['gold', 'gnew', 'gdiff', \"welfare\", 'upsilon']\n\nmat = scipy.io.loadmat(path + \"\\\\output\\\\robust\\\\gbm\\\\norecalibrate_values_gbm_1.mat\")\n\nnocaldf = pd.DataFrame(mat[\"record_values\"])\n\nnocaldf.columns = cnames\nnocaldf[\"gdiff\"] = -nocaldf[\"gdiff\"] \n\nnocaldf.sort_values([\"upsilon\"], inplace = True)\n\nbase_chi = str(round(mat[\"chi_value\"][0][0],3))",
"_____no_output_____"
],
[
"mat = scipy.io.loadmat(path + \"\\\\output\\\\robust\\\\gbm\\\\norecalibrate_values_gbm_0.9.mat\")\n\nnocaldf_lowchi = pd.DataFrame(mat[\"record_values\"])\n\nnocaldf_lowchi.columns = cnames\nnocaldf_lowchi[\"gdiff\"] = -nocaldf_lowchi[\"gdiff\"] \n\nnocaldf_lowchi.sort_values([\"upsilon\"], inplace = True)\n\nlow_chi = str(round(mat[\"chi_value\"][0][0],3))",
"_____no_output_____"
],
[
"mat = scipy.io.loadmat(path + \"\\\\output\\\\robust\\\\gbm\\\\norecalibrate_values_gbm_1.1.mat\")\n\nnocaldf_higchi = pd.DataFrame(mat[\"record_values\"])\n\nnocaldf_higchi.columns = cnames\nnocaldf_higchi[\"gdiff\"] = -nocaldf_higchi[\"gdiff\"] \nnocaldf_higchi.sort_values([\"upsilon\"], inplace = True)\n\nhig_chi = str(round(mat[\"chi_value\"][0][0],3))",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(2,2,figsize = (12,10))\n\nfig.tight_layout(pad = 6)\n\n\nposition = (0,0)\n\nax[position].plot(nocaldf.upsilon, 100*nocaldf[\"gdiff\"], lw = 4)\n\nax[position].plot(nocaldf_lowchi.upsilon, 100*nocaldf_lowchi[\"gdiff\"], lw = 4, color = 'r')\n\nax[position].plot(nocaldf_higchi.upsilon, 100*nocaldf_higchi[\"gdiff\"], lw = 4, color = 'k')\n\nxticks = [0.01*float(item) for item in list(range(1,11,1)) ]\n\nax[position].set_xticks(xticks)\n\nax[position].set_xlim(0.004,0.07)\nax[position].set_ylim(0.10,0.35)\nax[position].set_ylabel(\"\\n Change in Productivity Growth \\n Percentage Points\", fontsize = 12)\n\nax[position].spines[\"right\"].set_visible(False)\nax[position].spines[\"top\"].set_visible(False)\n\nax[position].vlines(0.048, 0.00, 0.234, \n color='k', \n linestyle='--',\n lw = 3) \n\nax[position].hlines(0.234, -0.001, 0.048 , \n color='k', \n label=\"Calibrated Values\",\n linestyle='--',\n lw = 3) \n\n\n###################################################################################\n\nposition = (0,1)\n\nax[position].plot(nocaldf.upsilon, 100*nocaldf[\"gold\"], lw = 4, label = \"Calibrated χ = \" + base_chi)\n\nax[position].plot(nocaldf_lowchi.upsilon, 100*nocaldf_lowchi[\"gold\"], lw = 4, color = 'red', label = \"Large χ = \" + low_chi)\n\nax[position].plot(nocaldf_higchi.upsilon, 100*nocaldf_higchi[\"gold\"], lw = 4, color = 'k', label = \"Small χ = \" + hig_chi)\n\nxticks = [0.01*float(item) for item in list(range(1,11,1)) ]\n\nax[position].set_xticks(xticks)\n\nax[position].set_xlim(0.004,0.07)\nax[position].set_ylim(0.0,3.10)\n#ax.set_ylim(0,0.40)\nax[position].set_xlabel(\"\\n GBM Variance Parameter\", fontsize = 12)\nax[position].set_ylabel(\"\\n Initial SS Productivity Growth\", fontsize = 12)\n\nax[position].hlines(0.79, -0.001, 0.048,\n color='k', \n label=\"Calibrated Values\",\n linestyle='--',\n lw = 3) \n\nax[position].vlines(0.048, 0, 0.79, \n color='k', \n linestyle='--',\n lw = 3) \n\nax[position].spines[\"right\"].set_visible(False)\nax[position].spines[\"top\"].set_visible(False)\n\nax[position].legend(bbox_to_anchor=(0., -1.25, 1., .102),frameon = False, fontsize = 14, loc = 4)\n\n#########################################################################################\n\nposition = (1,0)\n\nseries = \"welfare\"\n\nax[position].plot(nocaldf.upsilon, 100*nocaldf[series], lw = 4, label = \"Calibrated χ\")\n\nax[position].plot(nocaldf_lowchi.upsilon, 100*nocaldf_lowchi[series], lw = 4, color = 'red', label = \"Large χ\")\n\nax[position].plot(nocaldf_higchi.upsilon, 100*nocaldf_higchi[series], lw = 4, color = 'k', label = \"Small χ\")\n\nxticks = [0.01*float(item) for item in list(range(1,11,1)) ]\n\nax[position].set_xticks(xticks)\n\nax[position].set_xlim(0.004,0.07)\nax[position].set_ylim(5,15)\n#ax.set_ylim(0,0.40)\nax[position].set_xlabel(\"\\n GBM Variance Parameter\", fontsize = 12)\nax[position].set_ylabel(\"\\n Welfare Gain, Percent\", fontsize = 12)\n\nax[position].spines[\"right\"].set_visible(False)\nax[position].spines[\"top\"].set_visible(False)\n\n\nax[position].vlines(0.048, 0, 11.18, \n color='k', \n linestyle='--',\n lw = 3) # thickness of the line\n\nax[position].hlines(11.18, -0.001, 0.048 , \n color='k', \n label=\"Calibrated Values\", \n linestyle='--',\n lw = 3) \n\n#############################################################################################\nposition = (1,1)\n\nax[position].axis('off')\n\n#plt.savefig(fig_path + \"\\\\gbm_chi.pdf\", bbox_inches = \"tight\", dip = 3600)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"##### Discussion of these results from PTW text\nThe first thing to observe from Figure 6 is that the percentage point change in productivity is nearly constant across different values of the variance parameter. In other words, the variance does not much affect the response of growth to a change in trade costs.\n\nThe parameter which does influence the change in growth is the adoption cost parameter. The three\ndifferent lines on the left panel in Figure 6 illustrate this point. A small value of chi (top black line) corresponds to small costs of adoption. When adoption costs are small, growth is more responsive to changes in trade costs. In contrast, a large value of chi (bottom red line) corresponds to large adoption costs and a smaller response of growth to trade costs. The closed form equations available in the non-GBM version of the model deliver some insight. Equation 35 shows that the change in the growth rate for a given change in trade costs is larger when adoption costs are smaller.\n\nEven though the elasticity of growth to trade costs is not sensitive to the value of the GBM variance parameterholding adoption costs constant, the value of the GBM variance parameter—and, thus, the firm dynamics data—strongly influences the calibrated value of the adoption cost. The right panel in Figure 6 illustrates this point by tracing out how the growth rate in the initial steady state varies with the GBM variance parameter. For a given value, there is a near linear decrease in the steady state growth rate as the variance increases. Across chi values, the slope is essentially the same, but the intercept shifts, with smaller chi values leading to higher growth rates. This is intuitive—lower adoption costs lead to more adoption and faster economic growth.\n\nThe implication of these observations is that data on firm dynamics influences the inferred adoption cost and, thus, the elasticity of growth to trade costs. For example, holding fixed our target of an aggregate growth rate of 0.79 percent, if the transition matrix of relative size (Table 3) had pushed for us to find a smaller value for the GBM variance parameter, then the right panel of Figure 6 shows this would have lead us to calibrate a larger value for chi. Combining this observation with the left panel of Figure 6, our calibration strategy would have then led to a smaller increase in the growth rate for the same decrease in trade costs.\n\nThe lower panel of Figure 6 shows that the welfare gains from trade (comparing BGPs) are nearly constant across values of the GBM variance parameter, but sensitive to the value of chi, just like the elasticity of growth to trade costs. Thus, the value of chi is crucial for determining both the change in growth and the welfare gains from trade. Even though the GBM variance parameter does not much affect the welfare gains from trade when holding all other parameters constant, different values of the GBM variance parameter (which are associated with different firm dynamics moments) affect the calibration of chi. It is in this sense that not just firm heterogeneity, but firm dynamics, matter\nfor the welfare gains from trade in our model.",
"_____no_output_____"
],
[
"Our discussion above, which compares our gains from trade to those in Sampson (2016), strongly suggests this point as well. When the GBM process is shut down and the model is re-calibrated, the gains from trade are still larger than what the ACR formula would imply, but they are far more modest and in line with what Sampson finds. Recall from Section 7.3 that much of the welfare gains arise because the equilibrium has an inefficiently low growth rate and that changes in trade costs change the growth rate. Using the decomposition from Section 7.3, we find that the different values of chi associated with different\nvalues of the GBM variance parameteraffect the welfare gains from trade almost completely because of a change in the sensitivity\nof growth to the trade cost and not because of different levels of inefficiency.\n\nThis point is illustrated in the Connection to the Welfare Decomposition section below.",
"_____no_output_____"
],
[
"---\n\n### <a name=\"delta\"></a> Importance of the Exit Shock\n\nWe uniformly scale up and down the exit shock for different values of the adoption cost parameter chi. The large value of chi is ten percent larger than the baseline calibrated value. The small value of chi is ten percent smaller than the baseline calibrated value. All other parameter values are fixed, i.e., we do not re-calibrate the model when changing these parameter values. ",
"_____no_output_____"
],
[
"##### Step 1. Compute outcomes for different Delta parameter values\n\nThis calls the matlab code to perform this operation. The code appendix below describes each of the different components. The line below executes matlab from the command line/terminal",
"_____no_output_____"
]
],
[
[
"matlab_cmd = '''\"cd('src\\calibration');robust_no_recalibrate_delta;\"'''",
"_____no_output_____"
],
[
"!{matlab_exe_path} -batch {matlab_cmd}",
"Calbirated values computed on date\n 20-Apr-2020\n\n%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%\nChi Values\n 0.1409\n\nChi Values\n 0.1268\n\nChi Values\n 0.1153\n\n"
]
],
[
[
"##### Step 2. Create Figure 7 in PTW 2020\n\nThe code below reads in the output from matlab and then plots the results to create Figure 7 of PTW.",
"_____no_output_____"
]
],
[
[
"cnames = ['gold', 'gnew', 'gdiff', \"welfare\", 'delta']\n\nmat = scipy.io.loadmat(path + \"\\\\output\\\\robust\\\\delta\\\\norecalibrate_values_delta_1.mat\")\n\nnocaldf = pd.DataFrame(mat[\"record_values\"])\n\nnocaldf.columns = cnames\nnocaldf[\"gdiff\"] = -nocaldf[\"gdiff\"] \n\nnocaldf.sort_values([\"delta\"], inplace = True)\n\nbase_chi = str(round(mat[\"chi_value\"][0][0],3))",
"_____no_output_____"
],
[
"mat = scipy.io.loadmat(path + \"\\\\output\\\\robust\\\\delta\\\\norecalibrate_values_delta_0.9.mat\")\n\nnocaldf_lowchi = pd.DataFrame(mat[\"record_values\"])\n\nnocaldf_lowchi.columns = cnames\nnocaldf_lowchi[\"gdiff\"] = -nocaldf_lowchi[\"gdiff\"] \n\nnocaldf_lowchi.sort_values([\"delta\"], inplace = True)\n\nlow_chi = str(round(mat[\"chi_value\"][0][0],3))",
"_____no_output_____"
],
[
"mat = scipy.io.loadmat(path + \"\\\\output\\\\robust\\\\delta\\\\norecalibrate_values_delta_1.1.mat\")\n\nnocaldf_higchi = pd.DataFrame(mat[\"record_values\"])\n\nnocaldf_higchi.columns = cnames\nnocaldf_higchi[\"gdiff\"] = -nocaldf_higchi[\"gdiff\"]\n\nnocaldf_higchi.sort_values([\"delta\"], inplace = True)\n\nhig_chi = str(round(mat[\"chi_value\"][0][0],3))",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(2,2,figsize = (12,10))\n\nfig.tight_layout(pad = 6)\n\n\nposition = (0,0)\n\nax[position].plot(nocaldf.delta, 100*nocaldf[\"gdiff\"], lw = 4)\n\nax[position].plot(nocaldf_lowchi.delta, 100*nocaldf_lowchi[\"gdiff\"], lw = 4, color = 'r')\n\nax[position].plot(nocaldf_higchi.delta, 100*nocaldf_higchi[\"gdiff\"], lw = 4, color = 'k')\n\nxticks = [0.01*float(item) for item in list(range(1,11,1)) ]\n\nax[position].set_xticks(xticks)\n\nax[position].set_xlim(0.01,0.04)\nax[position].set_ylim(0.15,0.4)\n#ax[position].set_xlabel(\"\\n GBM Variance Parameter\", fontsize = 12)\nax[position].set_ylabel(\"\\n Change in Productivity Growth \\n Percentage Points\", fontsize = 12)\n\nax[position].spines[\"right\"].set_visible(False)\nax[position].spines[\"top\"].set_visible(False)\n\nax[position].vlines(0.020, 0.00, 0.234, # Set the value equall to the average\n color='k', # make the color red\n #label='Trade Shock', # this is the label (shows up in the legend)\n linestyle='--',\n lw = 3) # thickness of the line\n\nax[position].hlines(0.234, -0.001, 0.020 , # Set the value equall to the average\n color='k', # make the color red\n label=\"Calibrated Values\", # this is the label (shows up in the legend)\n linestyle='--',\n lw = 3) # thickness of the line\n\n\n##########################################################################################\n\nposition = (0,1)\n\nax[position].plot(nocaldf.delta, 100*nocaldf[\"gold\"], lw = 4, label = \"Calibrated χ = \" + base_chi)\n\nax[position].plot(nocaldf_lowchi.delta, 100*nocaldf_lowchi[\"gold\"], \n lw = 4, color = 'red', label = \"Large χ = \" + low_chi)\n\nax[position].plot(nocaldf_higchi.delta, 100*nocaldf_higchi[\"gold\"], \n lw = 4, color = 'k', label = \"Small χ = \" + hig_chi)\n\n#ax[1].plot(nocaldf_bigchi.upsilon, 100*nocaldf_bigchi[\"gold\"], lw = 4, color = 'k', label = \"Large 1/chi\")\n\nxticks = [0.01*float(item) for item in list(range(1,11,1)) ]\n\nax[position].set_xticks(xticks)\n\nax[position].set_xlim(0.01,0.04)\nax[position].set_ylim(0.20,1.4)\n#ax.set_ylim(0,0.40)\nax[position].set_xlabel(\"\\n Exit Shock Parameter\", fontsize = 12)\nax[position].set_ylabel(\"\\n Initial SS Productivity Growth\", fontsize = 12)\n\nax[position].vlines(0.02, 0.00, 0.79, # Set the value equall to the average\n color='k', # make the color red\n #label='Trade Shock', # this is the label (shows up in the legend)\n linestyle='--',\n lw = 3) # thickness of the line\n\nax[position].hlines(0.79, -0.001, 0.020 , # Set the value equall to the average\n color='k', # make the color red\n label=\"Calibrated Values\", # this is the label (shows up in the legend)\n linestyle='--',\n lw = 3) # thickness of the line\n\nax[position].spines[\"right\"].set_visible(False)\nax[position].spines[\"top\"].set_visible(False)\n\nax[position].legend(bbox_to_anchor=(0., -1.25, 1., .102),frameon = False, fontsize = 14, loc = 4)\n\n#########################################################################################\nposition = (1,0)\n\nseries = \"welfare\"\n\nax[position].plot(nocaldf.delta, 100*nocaldf[series], lw = 4, label = \"Calibrated χ\")\n\nax[position].plot(nocaldf_lowchi.delta, 100*nocaldf_lowchi[series], lw = 4, color = 'red', label = \"Large χ\")\n\nax[position].plot(nocaldf_higchi.delta, 100*nocaldf_higchi[series], lw = 4, color = 'k', label = \"Small χ\")\n\n#ax[1].plot(nocaldf_bigchi.upsilon, 100*nocaldf_bigchi[\"gold\"], lw = 4, color = 'k', label = \"Large 1/chi\")\n\nxticks = [0.01*float(item) for item in list(range(1,11,1)) ]\n\nax[position].set_xticks(xticks)\n\nax[position].set_xlim(0.01,0.04)\nax[position].set_ylim(6,20)\n#ax.set_ylim(0,0.40)\nax[position].set_xlabel(\"\\n Exit Shock Parameter\", fontsize = 12)\nax[position].set_ylabel(\"\\n Welfare Gain, Percent\", fontsize = 12)\n\nax[position].spines[\"right\"].set_visible(False)\nax[position].spines[\"top\"].set_visible(False)\n\n\nax[position].vlines(0.02, 0, 11.18, # Set the value equall to the average\n color='k', # make the color red\n #label='Trade Shock', # this is the label (shows up in the legend)\n linestyle='--',\n lw = 3) # thickness of the line\n\nax[position].hlines(11.18, -0.001, 0.02 , # Set the value equall to the average\n color='k', # make the color red\n label=\"Calibrated Values\", # this is the label (shows up in the legend)\n linestyle='--',\n lw = 3) # thickness of the line\n\n###############################################################################################\nposition = (1,1)\n\nax[position].axis('off')\n\n#plt.savefig(fig_path + \"\\\\delta_chi.pdf\", bbox_inches = \"tight\", dip = 3600)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"##### Discussion of Figure from paper\n\nSimilarly to the GBM variance case, the delta parameter interacts with the adoption cost parameter to affect the calibrated value of chi. The right panel in Figure 6 illustrates this point by tracing out how the growth rate in the initial steady state varies with delta. For a given chi value, the steady state growth rate increases with delta; across chi values, smaller chi values (lower adoption costs) lead to higher growth rates. Figure 6 shows that larger delta values (i.e., more entry observed in the data) would induce the calibration to infer larger chi values. But because these two parameters have opposite effects on economic growth, the change\nin parameter values generates offsetting effects and leaves the model’s elasticity of growth to trade costs unchanged\n\nThe welfare gains from trade display a similar pattern. The bottom panel of Figure 7 shows that the welfare gains from trade increase with the value of delta, holding all else fixed. Again, however, larger values of delta generate larger calibrated values of chi, which offset to keep the welfare gains from trade largely unchanged. Re-calibrating the model holding fixed different values for delta verifies this observation—welfare only increases slightly as delta increases.",
"_____no_output_____"
],
[
"### <a name=\"dcomp\"></a> Connection to the Welfare Decomposition",
"_____no_output_____"
],
[
"This calls Julia to perform the same welfare decomposition exercise as that done in ``section_7-3.ipynb``.",
"_____no_output_____"
]
],
[
[
"!jupyter nbconvert --to script ChiUpsilonDelta.ipynb",
"[NbConvertApp] Converting notebook ChiUpsilonDelta.ipynb to script\n[NbConvertApp] Writing 2986 bytes to ChiUpsilonDelta.jl\n"
],
[
"julia_command = '''ChiUpsilonDelta.jl'''",
"_____no_output_____"
],
[
"!{julia_path} {julia_command}",
"-------------------------------------------------------------------------\n\nBaseline\nGrowth in Baseline: 0.79\nBaseline Inefficincy term (CE units, percent of increase): 44.696\nBaseline Semi-Elasticity of Growth: -2.828\n-------------------------------------------------------------------------\n\nLarge Chi case\nGrowth in Large Chi case: 0.8\nLarge Chi Inefficincy term (CE units, percent of increase): 44.125\nLarge Chi Semi-Elasticity of Growth: -2.136\n-------------------------------------------------------------------------\n\nSmall Chi case\nGrowth in Small Chi case: 0.79\nSmall Chi Inefficincy term (CE units, percent of increase): 45.145\nSmall Chi Semi-Elasticity of Growth: -3.63\n"
]
],
[
[
"The decomposition from Section 7.3, shows how different values of $\\chi$s affect the welfare gains from trade. We find that the different values of $\\chi$ associated with different values of $\\upsilon^{2}$ affect the welfare gains from trade almost completely because of a change in the sensitivity of growth to the trade cost $\\left(\\frac{\\mathrm{d} f_{g}}{\\mathrm{d} d}\\right)$ (i.e., the semi-elasticity of growth changes substantially) and not because of different levels of inefficiency $\\left(U_1 \\frac{ \\partial f_{c}}{ \\partial g} + U_2\\right)$ (which are relativly simmilar across different specifications).",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
4ae2cef8b837a733271d23c32601f1705a52c2cb
| 37,245 |
ipynb
|
Jupyter Notebook
|
examples/coord_radar2glob_roipac.ipynb
|
insarwxw/PySAR
|
62126641ec38adad900863d8bc69beaba56e3598
|
[
"MIT"
] | 6 |
2017-10-13T14:27:07.000Z
|
2022-03-28T07:59:15.000Z
|
examples/coord_radar2glob_roipac.ipynb
|
insarwxw/PySAR
|
62126641ec38adad900863d8bc69beaba56e3598
|
[
"MIT"
] | null | null | null |
examples/coord_radar2glob_roipac.ipynb
|
insarwxw/PySAR
|
62126641ec38adad900863d8bc69beaba56e3598
|
[
"MIT"
] | 2 |
2020-02-09T08:27:50.000Z
|
2021-05-09T23:15:22.000Z
| 344.861111 | 34,154 | 0.925842 |
[
[
[
"'''Convert radar coord into geo coord: comparison between using geomapFile and using corner lat/lon'''\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pysar._readfile as readfile\nimport pysar._pysar_utilities as ut\nimport pysar.view as view\n\nwork_dir = '/Users/jeromezhang/Documents/insarlab/Kyushu/KyushuT422F650AlosA/TIMESERIES'\ngeomapFile = work_dir+'/geomap_4rlks_tight.trans'\nrdrFile = work_dir+'/temporal_coherence.h5'\n\nrg=np.array([1000, 1000])\naz=np.array([1500, 1000])\n\nlat0, lon0 = ut.radar2glob(az, rg, geomapFile, rdrFile)[0:2]\nlat1, lon1 = ut.radar2glob(az, rg, None, rdrFile)[0:2]\n\nimg, atr = readfile.read(geomapFile, (), 'azimuth')\nfig = plt.figure()\nax = fig.add_axes([0.1,0.1,0.8,0.8])\nax = view.plot_matrix(ax, img, atr)[0]\n\nax.plot(lon0, lat0, 'r*') #using geomap*.trans, accurate\nax.plot(lon1, lat1, 'b.') #using corner lat/lon, not accurate\n\nplt.show()\n\n\n",
"reading file: /Users/jeromezhang/Documents/insarlab/Kyushu/KyushuT422F650AlosA/TIMESERIES/geomap_4rlks_tight.trans\nResidul - lat: 7.50000000007e-05, lon: 0.001775\ndata coverage in y/x: (0, 0, 3138, 2695)\nsubset coverage in y/x: (0, 0, 3138, 2695)\ndata coverage in lat/lon: (131.02409876, 33.63756779, 132.07008829999998, 32.739243439999996)\nsubset coverage in lat/lon: (131.02409876, 33.63756779, 132.07008829999998, 32.739243439999996)\n------------------------------------------------------------------------\ncolormap: jet\ndata unit: 1\ndisplay unit: 1\ndata range: 0.0 - 2299.49975586\ndisplay range: 0.0 - 2299.49975586\ndisplay data in transparency: 1.0\nplot in Lat/Lon coordinate\nmap projection: cyl\nplotting Data ...\nplot scale bar\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
4ae2d2241b47861bbd499fde9852b5bf46b97f5d
| 1,277 |
ipynb
|
Jupyter Notebook
|
Untitled2.ipynb
|
BrackenW3/Atom-file-icons
|
be5b867619a8b480c35f8a16593eb2f18afd3101
|
[
"MIT"
] | null | null | null |
Untitled2.ipynb
|
BrackenW3/Atom-file-icons
|
be5b867619a8b480c35f8a16593eb2f18afd3101
|
[
"MIT"
] | null | null | null |
Untitled2.ipynb
|
BrackenW3/Atom-file-icons
|
be5b867619a8b480c35f8a16593eb2f18afd3101
|
[
"MIT"
] | null | null | null | 23.648148 | 233 | 0.480031 |
[
[
[
"<a href=\"https://colab.research.google.com/github/BrackenW3/Atom-file-icons/blob/master/Untitled2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"print(\"Hello Google!\")",
"Hello Google!\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
4ae2e3cba77e4d29ad6757276a1c1b5cee6f2346
| 55,615 |
ipynb
|
Jupyter Notebook
|
0420 - Redo Actual Result Calculation.ipynb
|
LaoKpa/reinforcement_trader
|
1465731269e6d58900a28a040346bf45ffb5cf97
|
[
"MIT"
] | 7 |
2020-09-28T23:36:40.000Z
|
2022-02-22T02:00:32.000Z
|
0420 - Redo Actual Result Calculation.ipynb
|
LaoKpa/reinforcement_trader
|
1465731269e6d58900a28a040346bf45ffb5cf97
|
[
"MIT"
] | 4 |
2020-11-13T18:48:52.000Z
|
2022-02-10T01:29:47.000Z
|
0420 - Redo Actual Result Calculation.ipynb
|
lzcaisg/reinforcement_trader
|
1465731269e6d58900a28a040346bf45ffb5cf97
|
[
"MIT"
] | 3 |
2020-11-23T17:31:59.000Z
|
2021-04-08T10:55:03.000Z
| 42.715054 | 118 | 0.421793 |
[
[
[
"import numpy as np\nimport pandas as pd\nfrom CSVUtils import *\nimport pickle\nfrom os import path\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"ROOT_DIR = \"./from github/Stock-Trading-Environment/\"\nfreq_list = [\n {\n \"freq\": 1,\n \"training\": \"10k\",\n \"DIR\": \"./output/200\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training_detailed-ModelNo-10000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 1,\n \"training\": \"50k\",\n \"DIR\": \"./output/201\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training_detailed-ModelNo-50000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 1,\n \"training\": \"100k\",\n \"DIR\": \"./output/201\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training_detailed-ModelNo-100000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 1,\n \"training\": \"500k\",\n \"DIR\": \"./output/202\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training_detailed-ModelNo-500000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n }\n]\nbnh_ratio={\n \"2015-2019\": 3.0693999444726088,\n \"2001-2004\": 1.0730432708521411,\n \"2007-2010\": 1.8942480597911275,\n}",
"_____no_output_____"
],
[
"ROOT_DIR = \"./from github/Stock-Trading-Environment/\"\nfreq_list = [\n {\n \"freq\": 7,\n \"training\": \"50k\",\n \"DIR\": \"./output/204\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training-punish_detailed-ModelNo-50000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 7,\n \"training\": \"100k\",\n \"DIR\": \"./output/205\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training-swap-nopunish-7d_detailed-ModelNo-100000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 7,\n \"training\": \"200k\",\n \"DIR\": \"./output/204\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training-punish_detailed-ModelNo-200000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n]",
"_____no_output_____"
],
[
"freq_list = [\n {\n \"freq\": 20,\n \"training\": \"10k\",\n \"DIR\": \"./output/306\",\n \"prefix\": \"BRZ_TW_NASDAQ-Selected_Trans-withleakage+RSI-_detailed-ModelNo-10000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 20,\n \"training\": \"50k\",\n \"DIR\": \"./output/306\",\n \"prefix\": \"BRZ_TW_NASDAQ-Selected_Trans-withleakage+RSI-_detailed-ModelNo-50000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 20,\n \"training\": \"100k\",\n \"DIR\": \"./output/306\",\n \"prefix\": \"BRZ_TW_NASDAQ-Selected_Trans-withleakage+RSI-_detailed-ModelNo-100000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 20,\n \"training\": \"200k\",\n \"DIR\": \"./output/306\",\n \"prefix\": \"BRZ_TW_NASDAQ-Selected_Trans-withleakage+RSI-_detailed-ModelNo-200000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n]",
"_____no_output_____"
],
[
"freq_list = [\n {\n \"freq\": 20,\n \"training\": \"BRZ+TW+NASDAQ\",\n \"DIR\": \"./output/205\",\n \"prefix\": \"BRZ+TW+NASDAQ-Training-swap-nopunish-7d_detailed-ModelNo-100000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 20,\n \"training\": \"BRZ+TW+NASDAQ\",\n \"DIR\": \"./output/205\",\n \"prefix\": \"NASDA+QBRZ+TW-Training-swap-nopunish_detailed-ModelNo-100000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n {\n \"freq\": 20,\n \"training\": \"BRZ+TW+NASDAQ\",\n \"DIR\": \"./output/205\",\n \"prefix\": \"TW+NASDAQ+BRZ-Training-swap-nopunish_detailed-ModelNo-100000-\",\n \"start_date\": pd.to_datetime(\"2015-01-01\"),\n \"end_date\": pd.to_datetime(\"2019-12-31\"),\n },\n]",
"_____no_output_____"
],
[
"for experiment in freq_list:\n nominal_rate_list = [] # Model/bnh\n nominal_return_list = [] # profit/300k\n actual_return_list = [] # actual profit/bnh\n \n DIR = path.join(\"./from github/Stock-Trading-Environment\",experiment[\"DIR\"])\n for i in range(10):\n record = pickle.load(open(path.join(DIR,experiment[\"prefix\"]+str(i)+\".out\"), \"rb\"))\n df = pd.DataFrame(record)\n final_nominal_profit = df['net_worth'].iloc[-1]-300000\n bnh_profit = df['buyNhold_balance'].iloc[-1]-300000\n nominal_profit_rate = (final_nominal_profit/bnh_profit) # How much better is the model compare to bnh\n nominal_rate_list.append(nominal_profit_rate)\n \n nominal_return_list.append(final_nominal_profit/300000)\n actual_return_list.append(df['actual_profit'].iloc[-1]/df['buyNhold_balance'].iloc[-1])\n nominal_rate_list=np.array(nominal_rate_list)\n print(experiment['freq'], experiment['training'], \n len(nominal_rate_list[(nominal_rate_list>=1)]),\n len(nominal_rate_list[(nominal_rate_list>=0.50) & (nominal_rate_list<1)]),\n len(nominal_rate_list[(nominal_rate_list>=0) & (nominal_rate_list<0.50)]),\n len(nominal_rate_list[nominal_rate_list<0]),\n np.mean(nominal_return_list),\n np.mean(actual_return_list),\n \n )",
"20 BRZ+TW+NASDAQ 2 7 1 0 0.5863616986965738 -0.10448461598959694\n20 BRZ+TW+NASDAQ 3 5 2 0 0.6119059675870048 -0.0900646474644653\n20 BRZ+TW+NASDAQ 1 5 4 0 0.4647040853053587 -0.17316142813359825\n"
],
[
"df",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae2e4ecbcc02796888be59e8ee1354d20ab4ef1
| 10,529 |
ipynb
|
Jupyter Notebook
|
aula_04/python-exercicios-aula_04.ipynb
|
cidfelipe/mentoria_evolution_aula_09
|
518eb33b5feb08da62f0c9576680f32c1809630c
|
[
"MIT"
] | null | null | null |
aula_04/python-exercicios-aula_04.ipynb
|
cidfelipe/mentoria_evolution_aula_09
|
518eb33b5feb08da62f0c9576680f32c1809630c
|
[
"MIT"
] | null | null | null |
aula_04/python-exercicios-aula_04.ipynb
|
cidfelipe/mentoria_evolution_aula_09
|
518eb33b5feb08da62f0c9576680f32c1809630c
|
[
"MIT"
] | null | null | null | 20.326255 | 130 | 0.493494 |
[
[
[
"# Mentoria Evolution - Exercícios Python ",
"_____no_output_____"
],
[
"https://minerandodados.com.br",
"_____no_output_____"
],
[
"* Para executar uma célula digite **Control + enter** ou clique em **Run**.\n* As celulas para rodar script Python devem ser do tipo code.\n* Crie células abaixo das celulas que foram escrito o enunciado das questões com as respostas.",
"_____no_output_____"
],
[
"**Obs**: Caso de dúvidas, volte na aula anterior de Python. Não desista :)",
"_____no_output_____"
],
[
"## Exercícios de Fixação",
"_____no_output_____"
],
[
"1) Responda: É possível ter elementos de tipos difentes em uma mesma lista? Exemplo: strings e números?",
"_____no_output_____"
]
],
[
[
"'Sim é possivel ter elementos de tipos diferentes em uma mesma lista'\nexemplo = ['String', 22]\nexemplo",
"_____no_output_____"
]
],
[
[
"2) Trabalhando com Listas, faça:\n- a) Cria uma lista de valores inteiros com o **nome** e **idades**.\n- b) Imprima apenas segundo elemento da lista.\n- c) Imprima a quantidade de elementos da lista.\n- d) Substitua o valor do segundo elemento da lista e imprima o resultado.\n- e) Imprima apenas os valores do segundo elemento em diante.\n- f) Remova qualquer elemento da lista e imprima o resultado.\n- g) Defina uma lista chamada salarios com os valores : **900,1200,1500,800,12587,10000**.\n- h) Verifique se contém o valor 10000 na lista de salarios.\n- i) Imprima o menor e maior valor da lista.\n- j) Adicione o valor 7000 a lista.\n- l) Extenda a lista com dois novos elementos utilizando apenas um método.\n- m) Imprima o índice do elemento de valor 800 da lista de salarios.\n- n) Faça uma ordenação dos valores da lista de salarios em ordem crescente e decrescente.",
"_____no_output_____"
]
],
[
[
"nomes = ['Felipe', 22, 'Joao', 35, 'Maria', 18]\nnomes",
"_____no_output_____"
],
[
"nomes[1]",
"_____no_output_____"
],
[
"len(nomes)",
"_____no_output_____"
],
[
"nomes[1] = 28\nnomes",
"_____no_output_____"
],
[
"salarios = [900, 1200, 1500, 800, 12587, 10000]\nsalarios",
"_____no_output_____"
],
[
"10000 in salarios",
"_____no_output_____"
],
[
"min(salarios)",
"_____no_output_____"
],
[
"salarios.append(7000)\nsalarios",
"_____no_output_____"
],
[
"salarios = salarios + [12000, 8700]\nsalarios",
"_____no_output_____"
]
],
[
[
"3) Trabalhando com dicionários:\n- a) Crie um dicionário para armazenar o nome e a idade de pessoas, Exemplo:\n pessoas = {'Rodrigo':30, 'Fulana':18}\n- b) Imprima a idade da pessoa \"Fulana\"\n- c) Imprima as Chaves do dicionario criado anteriormente.\n- d) Imprima os valores das chaves do dicionário\n- e) Busque a chave \"Felipe\" se ela não existe insira esta e o valor 30 (obs: use o método setdefault())",
"_____no_output_____"
]
],
[
[
"pessoas = {'Cid': 40, 'Samara': 38, 'Beatriz': 36}\npessoas",
"_____no_output_____"
],
[
"pessoas['Samara']",
"_____no_output_____"
],
[
"pessoas.keys()",
"_____no_output_____"
],
[
"pessoas.values()",
"_____no_output_____"
],
[
"pessoas.setdefault('Felipe',30)\npessoas",
"_____no_output_____"
]
],
[
[
"4) Estruturas condicionais:\n- a) Verifique se 5 é maior que 1, se sim, imprima \"5 é maior que 1\"\n- b) crie as variávies x1 e y1, defina dois valores quaisquer para as duas variáveis. \n Verifique se x1 é maior que y1. Se sim, imprima \"x1 é maior que y1\", senão imprima: \"y1 é maior que x1\"\n- c) Crie uma lista de valores como [2,3,4,5,6,7] e faça um loop para imprimir todos os valores na tela multiplicados por 2.",
"_____no_output_____"
]
],
[
[
"if 5 > 1:\n print('5 é maior que 1')",
"5 é maior que 1\n"
],
[
"x1, y1 = 3, 8\nprint(x1)\nprint(y1)",
"3\n8\n"
],
[
"if x1 > y1:\n print(\"x1 é maior que y1\")\nelse:\n print(\"y1 é maior que x1\")",
"y1 é maior que x1\n"
],
[
"for i in [2,3,4,5,6,7]:\n print (\"Valor: %s\" %i)",
"Valor: 2\nValor: 3\nValor: 4\nValor: 5\nValor: 6\nValor: 7\n"
]
],
[
[
"- Ao concluir, salve seu notebook e envie suas respostas para **[email protected]**",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4ae2e5e285299d0eddcebdb249901f1c88bed9a6
| 5,532 |
ipynb
|
Jupyter Notebook
|
mysite/static/ipython/3.N-Gram.ipynb
|
YongBeomKim/KakaoBot
|
20aff717dbab31e85dfa930da1e23bf62930c4b2
|
[
"MIT"
] | null | null | null |
mysite/static/ipython/3.N-Gram.ipynb
|
YongBeomKim/KakaoBot
|
20aff717dbab31e85dfa930da1e23bf62930c4b2
|
[
"MIT"
] | null | null | null |
mysite/static/ipython/3.N-Gram.ipynb
|
YongBeomKim/KakaoBot
|
20aff717dbab31e85dfa930da1e23bf62930c4b2
|
[
"MIT"
] | 3 |
2018-07-18T05:15:47.000Z
|
2018-07-18T11:50:26.000Z
| 21.779528 | 87 | 0.41504 |
[
[
[
"<br>\n# **N-Gram**\n1. 단어를 분석하는 방법 중 하나\n1. 여기서 N은 단어의 묶음갯수, 또는 글자의 묶음갯수 모두가능\n1. 이번에는 문장이 짧으므로 글자의 묶음갯수를 기준으로 작업을 진행할 예정이다",
"_____no_output_____"
],
[
"<br>\n## 1 **N-gram**\n1. 문장의 단어들을 잘게 나누기\n1. 접두어 접미어가 모두있는 한글의 특성상 앞/뒤 구분없이 잘르는 작업이 효과적\n1. 단 단어사이를 모두 연결시 의도치 않은 단어가 생성 가능하므로, 단어끼리는 나눠서 작업을 한다",
"_____no_output_____"
]
],
[
[
"sentence = '오늘의 날씨는 어떻게 되나요'\ntexts = sentence.split(' ')\ntexts",
"_____no_output_____"
],
[
"# [ list ] 의 Append\nresult = []\nresult.append('장고')\nresult.append('wework')\nresult.append(2018)\nresult",
"_____no_output_____"
],
[
"ngram = 2\nresult = []\n\nfor text in input_list:\n for no in range(len(text) - (ngram-1)):\n result.append(text[no:no + ngram])\nresult",
"_____no_output_____"
],
[
"# 위의 내용을 함수로 만들기\n# 작업을 함수에 하청을 주고, 우리는 return 결과값만 메모리에 저장된다\n# 기타 중간과정들은 자동으로 메모리에서 버려진다\ndef ngram(text, ngram=2):\n result = []\n input_list = text.split(' ')\n for text in input_list:\n for no in range(0, len(text) - (ngram - 1)):\n result.append(text[no:no + ngram])\n return result\n\nsentence = 'wework 수업날은 항상 날씨가 좋다'\nngram(sentence)",
"_____no_output_____"
],
[
" nowtime = '20120'\n content_dict = {\n '카톡': {\n 'message': {\n 'text':'카카오톡 공식문서에서 답을 찾으세요',\n \"message_button\": {\n \"label\": \"Git Hub\",\n \"url\": \"https://github.com/plusfriend/auto_reply\"\n },\n },\n 'keyboard':{'type':'text'}\n },\n\n '장고': {\n 'message': {\n 'text':'장고학습 동영상 Site 에서 답을 찾아보세요',\n \"message_button\": {\n \"label\": \"장고 웹사이트 만들기\",\n \"url\": \"https://programmers.co.kr/learn/courses/6\"\n },\n },\n 'keyboard':{'type':'text'}\n },\n\n '시간': {\n 'message': {\n 'text': '현재 시간은' + nowtime +'입니다'\n },\n 'keyboard':{'type':'text'},\n },\n }",
"_____no_output_____"
],
[
"content_dict['시간'].keys()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae2f127c21dee2993af2b49d34b289fc3acd52d
| 2,281 |
ipynb
|
Jupyter Notebook
|
Day_046_gradient_boosting_machine.ipynb
|
hengbinxu/ML100-Days
|
ed0eb6e32882239599df57486af3dc398f160d4c
|
[
"MIT"
] | 1 |
2019-01-02T01:18:27.000Z
|
2019-01-02T01:18:27.000Z
|
Day_046_gradient_boosting_machine.ipynb
|
hengbinxu/ML100-Days
|
ed0eb6e32882239599df57486af3dc398f160d4c
|
[
"MIT"
] | null | null | null |
Day_046_gradient_boosting_machine.ipynb
|
hengbinxu/ML100-Days
|
ed0eb6e32882239599df57486af3dc398f160d4c
|
[
"MIT"
] | null | null | null | 22.145631 | 234 | 0.562034 |
[
[
[
"from sklearn import datasets, metrics\nfrom sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.model_selection import train_test_split",
"/anaconda3/lib/python3.7/site-packages/sklearn/ensemble/weight_boosting.py:29: DeprecationWarning: numpy.core.umath_tests is an internal NumPy module and should not be imported. It will be removed in a future NumPy release.\n from numpy.core.umath_tests import inner1d\n"
],
[
"# 讀取鳶尾花資料集\niris = datasets.load_iris()\n\n# 切分訓練集/測試集\nx_train, x_test, y_train, y_test = train_test_split(iris.data, iris.target, test_size=0.25, random_state=4)\n\n# 建立模型\nclf = GradientBoostingClassifier()\n\n# 訓練模型\nclf.fit(x_train, y_train)\n\n# 預測測試集\ny_pred = clf.predict(x_test)",
"_____no_output_____"
],
[
"acc = metrics.accuracy_score(y_test, y_pred)\nprint(\"Acuuracy: \", acc)",
"Acuuracy: 0.9736842105263158\n"
]
],
[
[
"### 作業\n目前已經學過許多的模型,相信大家對整體流程應該比較掌握了,這次作業請改用**手寫辨識資料集**,步驟流程都是一樣的,請試著自己撰寫程式碼來完成所有步驟",
"_____no_output_____"
]
],
[
[
"digits = datasets.load_digits()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae31582ab7e992954f3d72acbcbba904d0ce2a5
| 17,769 |
ipynb
|
Jupyter Notebook
|
lectures/L14/L14.ipynb
|
nate-stein/cs207_nate_stein
|
f8ce68f9d839a0bd0ab4a2e1ebaa7ae985f6b5d0
|
[
"MIT"
] | null | null | null |
lectures/L14/L14.ipynb
|
nate-stein/cs207_nate_stein
|
f8ce68f9d839a0bd0ab4a2e1ebaa7ae985f6b5d0
|
[
"MIT"
] | null | null | null |
lectures/L14/L14.ipynb
|
nate-stein/cs207_nate_stein
|
f8ce68f9d839a0bd0ab4a2e1ebaa7ae985f6b5d0
|
[
"MIT"
] | null | null | null | 27.591615 | 760 | 0.553942 |
[
[
[
"# Lecture 14\n\n### Wednesday, October 25th 2017",
"_____no_output_____"
],
[
"## Last time:\n* Iterators and Iterables\n* Trees, Binary trees, and BSTs\n\n## This time:\n* BST Traversal\n* Generators\n* Memory layouts\n* Heaps?",
"_____no_output_____"
],
[
"# BST Traversal\n\n* We've stored our data in a BST\n* This seemed like a good idea at the time because BSTs have some nice properties\n* To be able to access/use our data, we need to be able to traverse the tree",
"_____no_output_____"
],
[
"#### Traversal Choices\n\nThere are three traversal choices based on an implicit ordering of the tree from left to right:\n\n1. In-order: Traverse left-subtree, then current root, then right sub tree\n2. Post-order: Traverse left subtree, then traverse left subtree, and then current root\n3. Pre-order: Current root, then traverse left subtree, then traverse right subtree",
"_____no_output_____"
],
[
"* Traversing a tree means performing some operation\n* In our examples, the operation will be \"displaying the data\"\n* However, an operation could be \"deleting files\"",
"_____no_output_____"
],
[
"## Example\nTraverse the BST below using *in-order*, *post-order*, and *pre-order* traversals. Write the resulting sorted data structure (as a list is fine).\n",
"_____no_output_____"
],
[
"# Heaps\nWe listed several types of data structures at the beginning of our data structures unit.\n\nSo far, we have discussed lists and trees (in particular binary trees and binary search trees).\n\nHeaps are a type of tree, a little different from binary trees.",
"_____no_output_____"
],
[
"## Some Motivation\n\n### Priority Queues\n\n* People may come to your customer service counter in a certain order, but you might want to serve your executive class first! \n* In other words, there is an \"ordering\" on your customers and you want to serve people in the order of the most VIP.\n* This problem requires us to then sort things by importance and then evaluate things in this sorted order.\n* A priority queue is a data structure for this, which allows us to do things more efficiently than simple sorting every time a new thing comes in.\n\nItems are inserted at one end and deleted from the other end of a queue (first in, first out [FIFO] buffer).",
"_____no_output_____"
],
[
"The basic priority queue is defined to be supporting three primary operations:\n1. Insert: insert an item with \"key\" (e.g. an importance) $k$ into priority queue $Q$.\n2. Find Minimum: get the item, or a pointer to the item, whose key value is smaller than any other key in $Q$.\n3. Delete Minimum: Remove the item with minimum $k$ from $Q$.",
"_____no_output_____"
],
[
"### Comments on Implementation of Priorty Queues\n\nOne could use an unsorted array and store a pointer to the minimum index; accessing the minimum is an $O(1)$ operation.\n* It's cheap to update the pointer when new items are inserted into the array because we update it in $O(1)$ only when the new value is less than the current one.\n* Finding a new minimum after deleting the old one requires a scan of the array ($O(n)$ operation) and then resetting the pointer.",
"_____no_output_____"
],
[
"One could alternatively implement the priority queue with a *balanced* binary tree structure. Then we'll get performance of $O(\\log(n))$!",
"_____no_output_____"
],
[
"This leads us to *heaps*. Heaps are a type of balanced binary tree.\n\n* A heap providing access to minimum values is called a *min-heap*\n* A heap providing access to maximum values is called a *max-heap*\n* Note that you can't have a *min-heap* and *max-heap* together",
"_____no_output_____"
],
[
"### Heapsort\n* Implementing a priority queue with `selection sort` takes $O(n^{2})$ operations\n* Using a heap takes $O(n\\log(n))$ operations\n\nImplementing a sorting algorithm using a heap is called `heapsort`.\n\n`Heapsort` is an *in-place* sort and requires no extra memory.\n\nNote that there are many sorting algorithms nowadays. `Python` uses [`Timsort`](https://en.wikipedia.org/wiki/Timsort).",
"_____no_output_____"
],
[
"### Back to Heaps\n\nA heap has two properties:\n\n1. Shape property\n * A leaf node at depth $k>0$ can exist only if all the nodes at the previous depth exist. Nodes at any partially filled level are added \"from left to right\".\n2. Heap property\n * For a *min-heap*, each node in the tree contains a key less than or equal to either of its two children (if they exist).\n - This is also known as the labeling of a \"parent node\" dominating that of its children. \n * For max heaps we use greater-than-or-equal.",
"_____no_output_____"
],
[
"#### Heap Mechanics\n\n* The first element in the array is the root key\n* The next two elements make up the first level of children. This is done from left to right\n* Then the next four and so on.",
"_____no_output_____"
],
[
"#### More Details on Heap Mechanics\n\nTo construct a heap, insert each new element that comes in at the left-most open spot. \n\nThis maintains the shape property but not the heap property.",
"_____no_output_____"
],
[
"#### Restore the Heap Property by \"Bubbling Up\"\nLook at the parent and if the child \"dominates\" we swap parent and child. Repeat this process until we bubble up to the root.\n\nIdentifying the dominant is now easy because it will be at the top of the tree.\n\nThis process is called `heapify` and must also be done at the first construction of the heap.",
"_____no_output_____"
],
[
"#### Deletion\nRemoving the dominant key creates a hole at the top (the first position in the array).\n\n**Fill this hole with the rightmost position in the array**, or the rightmost leaf node.\n\nThis destroys the heap property!\n\nSo we now bubble this key down until it dominates all its children.",
"_____no_output_____"
],
[
"## Example\n1. Construct a *min-heap* for the array $$\\left[1, 8, 5, 9, 23, 2, 45, 6, 7, 99, -5\\right].$$\n2. Delete $-5$ and update the *min-heap*.",
"_____no_output_____"
],
[
"# Iterables/Iterators Again\nWe have been discussing data structures and simultaneously exploring iterators and iterables.",
"_____no_output_____"
]
],
[
[
"class SentenceIterator:\n def __init__(self, words): \n self.words = words \n self.index = 0\n \n def __next__(self): \n try:\n word = self.words[self.index] \n except IndexError:\n raise StopIteration() \n self.index += 1\n return word \n\n def __iter__(self):\n return self\n\nclass Sentence: # An iterable\n def __init__(self, text): \n self.text = text\n self.words = text.split()\n \n def __iter__(self):\n return SentenceIterator(self.words)\n \n def __repr__(self):\n return 'Sentence(%s)' % reprlib.repr(self.text)",
"_____no_output_____"
]
],
[
[
"### Example Usage",
"_____no_output_____"
]
],
[
[
"a = Sentence(\"Dogs will save the world and cats will eat it.\")\n\nfor item in a:\n print(item)\nprint(\"\\n\")\nit = iter(a) # it is an iterator\nwhile True:\n try:\n nextval = next(it)\n print(nextval)\n except StopIteration:\n del it\n break",
"Dogs\nwill\nsave\nthe\nworld\nand\ncats\nwill\neat\nit.\n\n\nDogs\nwill\nsave\nthe\nworld\nand\ncats\nwill\neat\nit.\n"
]
],
[
[
"#### Every collection in Python is iterable.\n\nWe have already seen iterators are used to make for loops. They are also used to make other collections:\n\n* To loop over a file line by line from disk\n* In the making of list, dict, and set comprehensions\n* In unpacking tuples\n* In parameter unpacking in function calls (*args syntax)\n\nAn iterator defines both `__iter__` and a `__next__` (the first one is only required to make sure an iterator is an iterable).\n\n**Recap:** An iterator retrieves items from a collection. The collection must implement `__iter__`.",
"_____no_output_____"
],
[
"## Generators\n\n* A generator function looks like a normal function, but yields values instead of returning them. \n* The syntax is (unfortunately) the same otherwise ([PEP 255 -- Simple Generators](https://www.python.org/dev/peps/pep-0255/)).\n* A generator is a different beast. When the function runs, it creates a generator.\n* The generator is an iterator and gets an internal implementation of `__iter__` and `__next__`.",
"_____no_output_____"
]
],
[
[
"def gen123():\n print(\"A\")\n yield 1\n print(\"B\")\n yield 2\n print(\"C\")\n yield 3\n\ng = gen123()\n\nprint(gen123, \" \", type(gen123), \" \", type(g))",
"<function gen123 at 0x10d8081e0> <class 'function'> <class 'generator'>\n"
],
[
"print(\"A generator is an iterator.\")\nprint(\"It has {} and {}\".format(g.__iter__, g.__next__))",
"A generator is an iterator.\nIt has <method-wrapper '__iter__' of generator object at 0x10d7cefc0> and <method-wrapper '__next__' of generator object at 0x10d7cefc0>\n"
]
],
[
[
"### Some notes on generators\n* When `next` is called on the generator, the function proceeds until the first yield.\n* The function body is now suspended and the value in the yield is then passed to the calling scope as the outcome of the `next`.\n* When next is called again, it gets `__next__` called again (implicitly) in the generator, and the next value is yielded.\n* This continues until we reach the end of the function, the return of which creates a `StopIteration` in next.\n\nAny Python function that has the yield keyword in its body is a generator function.",
"_____no_output_____"
]
],
[
[
"print(next(g))\nprint(next(g))\nprint(next(g))\nprint(next(g))",
"A\n1\nB\n2\nC\n3\n"
]
],
[
[
"### More notes on generators\n* Generators yield one item at a time\n* In this way, they feed the `for` loop one item at a time",
"_____no_output_____"
]
],
[
[
"for i in gen123():\n print(i, \"\\n\")",
"A\n1 \n\nB\n2 \n\nC\n3 \n\n"
]
],
[
[
"## Lecture Exercise\nCreate a `Sentence` iterator class that uses a generator expression. You will write the generator expression in the `__iter__` special method. Note that the generator automatically gets `__next__`.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4ae31944d67178968298bd43fc1025494356faf1
| 50,365 |
ipynb
|
Jupyter Notebook
|
Untitled.ipynb
|
PatMartin7/Unsupervised
|
8545e976a808bd4092d1e35e2bea679dd9871d2b
|
[
"ADSL"
] | null | null | null |
Untitled.ipynb
|
PatMartin7/Unsupervised
|
8545e976a808bd4092d1e35e2bea679dd9871d2b
|
[
"ADSL"
] | null | null | null |
Untitled.ipynb
|
PatMartin7/Unsupervised
|
8545e976a808bd4092d1e35e2bea679dd9871d2b
|
[
"ADSL"
] | null | null | null | 36.951577 | 6,636 | 0.367219 |
[
[
[
"# Initial imports\nimport pandas as pd\nimport hvplot.pandas\nfrom path import Path\nimport plotly.express as px\nfrom sklearn.preprocessing import StandardScaler, MinMaxScaler\nfrom sklearn.decomposition import PCA\nfrom sklearn.cluster import KMeans",
"_____no_output_____"
],
[
"# Load the crypto_data.csv dataset.\nfile_path = \"crypto_data.csv\"\ndf_crypto = pd.read_csv(file_path,index_col = 0)\ndf_crypto.head()",
"_____no_output_____"
],
[
"# Keep all the cryptocurrencies that are being traded.\ndf_crypto = df_crypto[df_crypto.IsTrading.eq(True)]\ndf_crypto.shape",
"_____no_output_____"
],
[
"#Keep all cryptocurrencies where the algorithm is working\npd.isna(df_crypto['Algorithm'])",
"_____no_output_____"
],
[
"# Remove the \"IsTrading\" column. \ndf_crypto = df_crypto.drop([\"IsTrading\"],axis = 1)",
"_____no_output_____"
],
[
"# Remove rows that have at least 1 null value.\ndf_crypto = df_crypto.dropna(how='any',axis=0) \ndf_crypto",
"_____no_output_____"
],
[
"# Keep the rows where coins are mined.\ndf_crypto = df_crypto[df_crypto.TotalCoinsMined > 0]\ndf_crypto",
"_____no_output_____"
],
[
"# Create a new DataFrame that holds only the cryptocurrencies names.\nnames = df_crypto.filter(['CoinName'], axis=1)\n# Drop the 'CoinName' column since it's not going to be used on the clustering algorithm.\ndf_crypto = df_crypto.drop(['CoinName'],axis = 1)\ndf_crypto",
"_____no_output_____"
],
[
"# Use get_dummies() to create variables for text features.\ncrypto = pd.get_dummies(df_crypto['Algorithm'])\ndummy = pd.get_dummies(df_crypto['ProofType'])\ncombined = pd.concat([crypto,dummy],axis =1)\ndf = df_crypto.merge(combined,left_index = True,right_index = True)\ndf = df.drop(['Algorithm','ProofType'],axis = 1)\ndf",
"_____no_output_____"
],
[
"# Standardize the data with StandardScaler().\ndf_scaled = StandardScaler().fit_transform(df)\nprint(df_scaled)",
"[[-0.11710817 -0.1528703 -0.0433963 ... -0.0433963 -0.0433963\n -0.0433963 ]\n [-0.09396955 -0.145009 -0.0433963 ... -0.0433963 -0.0433963\n -0.0433963 ]\n [ 0.52494561 4.48942416 -0.0433963 ... -0.0433963 -0.0433963\n -0.0433963 ]\n ...\n [-0.09561336 -0.13217937 -0.0433963 ... -0.0433963 -0.0433963\n -0.0433963 ]\n [-0.11694817 -0.15255998 -0.0433963 ... -0.0433963 -0.0433963\n -0.0433963 ]\n [-0.11710536 -0.15285552 -0.0433963 ... -0.0433963 -0.0433963\n -0.0433963 ]]\n"
],
[
"# Using PCA to reduce dimension to three principal components.\npca = PCA(n_components=3)\ndf_pca = pca.fit_transform(df_scaled)\ndf_pca",
"_____no_output_____"
],
[
"# Create a DataFrame with the three principal components.\npcs_df = pd.DataFrame(\n data = df_pca, columns = ['PC1','PC2','PC3'], index = df_crypto.index\n)\npcs_df",
"_____no_output_____"
],
[
"# Create an elbow curve to find the best value for K.\ninertia = []\nk = list(range(1, 11))\n# Calculate the inertia for the range of K values\nfor i in k:\n km = KMeans(n_clusters=i, random_state=0)\n km.fit(pcs_df)\n inertia.append(km.inertia_)\n\nelbow_data = {\"k\":k,\"inertia\":inertia}\ndf_elbow = pd.DataFrame(elbow_data)\ndf_elbow.hvplot.line(x=\"k\",y=\"inertia\",xticks=k,title=\"Elbow Curve\")",
"_____no_output_____"
],
[
"#We did identify the classification 0f 531 cryptocurrencies based on similarities of their features The output is unknown, the best method would be unsupervised learning and clustering algorithms to group the currencies This classification report could be used by an investment bank to propose a new cryptocurrency investment portfolio to its clients.",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae324898eadcbb04d2246e40f3d1ffdd0bfca73
| 10,128 |
ipynb
|
Jupyter Notebook
|
notebooks/federated_learning/federated_learning_basic_concepts_pretrained_model.ipynb
|
SSSuperTIan/Sherpa.ai-Federated-Learning-Framework
|
a30d73a018526f1033ee0ec57489c4c6e2f15b0a
|
[
"Apache-2.0"
] | 1 |
2021-03-18T07:31:36.000Z
|
2021-03-18T07:31:36.000Z
|
notebooks/federated_learning/federated_learning_basic_concepts_pretrained_model.ipynb
|
SSSuperTIan/Sherpa.ai-Federated-Learning-Framework
|
a30d73a018526f1033ee0ec57489c4c6e2f15b0a
|
[
"Apache-2.0"
] | null | null | null |
notebooks/federated_learning/federated_learning_basic_concepts_pretrained_model.ipynb
|
SSSuperTIan/Sherpa.ai-Federated-Learning-Framework
|
a30d73a018526f1033ee0ec57489c4c6e2f15b0a
|
[
"Apache-2.0"
] | null | null | null | 40.190476 | 997 | 0.667062 |
[
[
[
"# Federated learning: pretrained model\n\nIn this notebook, we provide a simple example of how to perform an experiment in a federated environment with the help of the Sherpa.ai Federated Learning framework. We are going to use a popular dataset and a pretrained model. \n## The data\nThe framework provides some functions for loading the [Emnist](https://www.nist.gov/itl/products-and-services/emnist-dataset) digits dataset.",
"_____no_output_____"
]
],
[
[
"import shfl\n\ndatabase = shfl.data_base.Emnist()\ntrain_data, train_labels, test_data, test_labels = database.load_data()",
"_____no_output_____"
]
],
[
[
"Let's inspect some properties of the loaded data.",
"_____no_output_____"
]
],
[
[
"print(len(train_data))\nprint(len(test_data))\nprint(type(train_data[0]))\ntrain_data[0].shape",
"_____no_output_____"
]
],
[
[
"So, as we have seen, our dataset is composed of a set of matrices that are 28 by 28. Before starting with the federated scenario, we can take a look at a sample in the training data.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\nplt.imshow(train_data[0])",
"_____no_output_____"
]
],
[
[
"We are going to simulate a federated learning scenario with a set of client nodes containing private data, and a central server that will be responsible for coordinating the different clients. But, first of all, we have to simulate the data contained in every client. In order to do that, we are going to use the previously loaded dataset. The assumption in this example is that the data is distributed as a set of independent and identically distributed random variables, with every node having approximately the same amount of data. There are a set of different possibilities for distributing the data. The distribution of the data is one of the factors that can have the most impact on a federated algorithm. Therefore, the framework has some of the most common distributions implemented, which allows you to easily experiment with different situations. In [Federated Sampling](./federated_learning_sampling.ipynb), you can dig into the options that the framework provides, at the moment.",
"_____no_output_____"
]
],
[
[
"iid_distribution = shfl.data_distribution.IidDataDistribution(database)\nfederated_data, test_data, test_label = iid_distribution.get_federated_data(num_nodes=20, percent=10)",
"_____no_output_____"
]
],
[
[
"That's it! We have created federated data from the Emnist dataset using 20 nodes and 10 percent of the available data. This data is distributed to a set of data nodes in the form of private data. Let's learn a little more about the federated data.",
"_____no_output_____"
]
],
[
[
"print(type(federated_data))\nprint(federated_data.num_nodes())\nfederated_data[0].private_data",
"_____no_output_____"
]
],
[
[
"As we can see, private data in a node is not directly accessible but the framework provides mechanisms to use this data in a machine learning model. \n## The model\nA federated learning algorithm is defined by a machine learning model, locally deployed in each node, that learns from the respective node's private data and an aggregating mechanism to aggregate the different model parameters uploaded by the client nodes to a central node. In this example, we will use a deep learning model using Keras to build it. The framework provides classes on using Tensorflow (see notebook [Federated learning Tensorflow Model](./federated_learning_basic_concepts_tensorflow.ipynb)) and Keras (see notebook [Federated Learning basic concepts](./federated_learning_basic_concepts.ipynb)) models in a federated learning scenario, your only job is to create a function acting as model builder. Moreover, the framework provides classes to allow using pretrained Tensorflow and Keras models. In this example, we will use a pretrained Keras learning model.",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\n#If you want execute in GPU, you must uncomment this two lines.\n# physical_devices = tf.config.experimental.list_physical_devices('GPU')\n# tf.config.experimental.set_memory_growth(physical_devices[0], True)\n\ntrain_data = train_data.reshape(-1,28,28,1)\n\nmodel = tf.keras.models.Sequential()\nmodel.add(tf.keras.layers.Conv2D(32, kernel_size=(3, 3), padding='same', activation='relu', strides=1, input_shape=(28, 28, 1)))\nmodel.add(tf.keras.layers.MaxPooling2D(pool_size=2, strides=2, padding='valid'))\nmodel.add(tf.keras.layers.Dropout(0.4))\nmodel.add(tf.keras.layers.Conv2D(32, kernel_size=(3, 3), padding='same', activation='relu', strides=1))\nmodel.add(tf.keras.layers.MaxPooling2D(pool_size=2, strides=2, padding='valid'))\nmodel.add(tf.keras.layers.Dropout(0.3))\nmodel.add(tf.keras.layers.Flatten())\nmodel.add(tf.keras.layers.Dense(128, activation='relu'))\nmodel.add(tf.keras.layers.Dropout(0.1))\nmodel.add(tf.keras.layers.Dense(64, activation='relu'))\nmodel.add(tf.keras.layers.Dense(10, activation='softmax'))\n\nmodel.compile(optimizer=\"rmsprop\", loss=\"categorical_crossentropy\", metrics=[\"accuracy\"])\n\nmodel.fit(x=train_data, y=train_labels, batch_size=128, epochs=3, validation_split=0.2, \n verbose=1, shuffle=False)",
"_____no_output_____"
],
[
"def model_builder():\n pretrained_model = model\n \n criterion = tf.keras.losses.CategoricalCrossentropy()\n optimizer = tf.keras.optimizers.RMSprop()\n metrics = [tf.keras.metrics.categorical_accuracy]\n \n return shfl.model.DeepLearningModel(model=pretrained_model, criterion=criterion, optimizer=optimizer, metrics=metrics)",
"_____no_output_____"
]
],
[
[
"Now, the only piece missing is the aggregation operator. Nevertheless, the framework provides some aggregation operators that we can use. In the following piece of code, we define the federated aggregation mechanism. Moreover, we define the federated government based on the Keras learning model, the federated data, and the aggregation mechanism.",
"_____no_output_____"
]
],
[
[
"aggregator = shfl.federated_aggregator.FedAvgAggregator()\nfederated_government = shfl.federated_government.FederatedGovernment(model_builder, federated_data, aggregator)",
"_____no_output_____"
]
],
[
[
"If you want to see all the aggregation operators, you can check out the [Aggregation Operators](./federated_learning_basic_concepts_aggregation_operators.ipynb) notebook. Before running the algorithm, we want to apply a transformation to the data. A good practice is to define a federated operation that will ensure that the transformation is applied to the federated data in all the client nodes. We want to reshape the data, so we define the following FederatedTransformation.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nclass Reshape(shfl.private.FederatedTransformation):\n \n def apply(self, labeled_data):\n labeled_data.data = np.reshape(labeled_data.data, (labeled_data.data.shape[0], labeled_data.data.shape[1], labeled_data.data.shape[2],1))\n\nclass CastFloat(shfl.private.FederatedTransformation):\n \n def apply(self, labeled_data):\n labeled_data.data = labeled_data.data.astype(np.float32)\n \nshfl.private.federated_operation.apply_federated_transformation(federated_data, Reshape())\nshfl.private.federated_operation.apply_federated_transformation(federated_data, CastFloat())",
"_____no_output_____"
]
],
[
[
"## Run the federated learning experiment\nWe are now ready to execute our federated learning algorithm.",
"_____no_output_____"
]
],
[
[
"test_data = np.reshape(test_data, (test_data.shape[0], test_data.shape[1], test_data.shape[2],1))\ntest_data = test_data.astype(np.float32)\nfederated_government.run_rounds(2, test_data, test_label)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae32fd711ef19dd37dce2ec854328ca06b3f870
| 4,407 |
ipynb
|
Jupyter Notebook
|
python-tuts/0-beginner/1-Refresher-Course/02 - Conditionals.ipynb
|
AadityaGupta/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials
|
352dd6d9a785e22fde0ce53a6b0c2e56f4964950
|
[
"Apache-2.0"
] | 3,266 |
2017-08-06T16:51:46.000Z
|
2022-03-30T07:34:24.000Z
|
python-tuts/0-beginner/1-Refresher-Course/02 - Conditionals.ipynb
|
AadityaGupta/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials
|
352dd6d9a785e22fde0ce53a6b0c2e56f4964950
|
[
"Apache-2.0"
] | 150 |
2017-08-28T14:59:36.000Z
|
2022-03-11T23:21:35.000Z
|
python-tuts/0-beginner/1-Refresher-Course/02 - Conditionals.ipynb
|
AadityaGupta/Artificial-Intelligence-Deep-Learning-Machine-Learning-Tutorials
|
352dd6d9a785e22fde0ce53a6b0c2e56f4964950
|
[
"Apache-2.0"
] | 1,449 |
2017-08-06T17:40:59.000Z
|
2022-03-31T12:03:24.000Z
| 18.061475 | 116 | 0.449285 |
[
[
[
"### Conditionals",
"_____no_output_____"
],
[
"A conditional is a construct that allows you to branch your code based on conditions being met (or not)",
"_____no_output_____"
],
[
"This is achieved using **if**, **elif** and **else** or the **ternary operator** (aka conditional expression)",
"_____no_output_____"
]
],
[
[
"a = 2\nif a < 3:\n print('a < 3')\nelse:\n print('a >= 3')",
"a < 3\n"
]
],
[
[
"**if** statements can be nested:",
"_____no_output_____"
]
],
[
[
"a = 15\n\nif a < 5:\n print('a < 5')\nelse:\n if a < 10:\n print('5 <= a < 10')\n else:\n print('a >= 10')",
"a >= 10\n"
]
],
[
[
"But the **elif** statement provides far better readability:",
"_____no_output_____"
]
],
[
[
"a = 15\nif a < 5:\n print('a < 5')\nelif a < 10:\n print('5 <= a < 10')\nelse:\n print('a >= 10')",
"a >= 10\n"
]
],
[
[
"In Python, **elif** is the closest you'll find to the switch/case statement available in some other languages.",
"_____no_output_____"
],
[
"Python also provides a conditional expression (ternary operator):\n\nX if (condition) else Y\n\nreturns (and evaluates) X if (condition) is True, otherwise returns (and evaluates) Y",
"_____no_output_____"
]
],
[
[
"a = 5\nres = 'a < 10' if a < 10 else 'a >= 10'\nprint(res)",
"a < 10\n"
],
[
"a = 15\nres = 'a < 10' if a < 10 else 'a >= 10'\nprint(res)",
"a >= 10\n"
]
],
[
[
"Note that **X** and **Y** can be any expression, not just literal values:",
"_____no_output_____"
]
],
[
[
"def say_hello():\n print('Hello!')\n \ndef say_goodbye():\n print('Goodbye!')",
"_____no_output_____"
],
[
"a = 5\nsay_hello() if a < 10 else say_goodbye()",
"Hello!\n"
],
[
"a = 15\nsay_hello() if a < 10 else say_goodbye()",
"Goodbye!\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4ae33355d31cb88b2a7cf80f4d4c2f554a3ac181
| 5,668 |
ipynb
|
Jupyter Notebook
|
heritageconnector/preprocess data.ipynb
|
LinkedPasts/LaNC-workshop
|
a855da569470e94d9c5bd508b51ddb4c704a93e7
|
[
"MIT"
] | 13 |
2020-12-03T10:25:48.000Z
|
2021-03-16T08:41:47.000Z
|
heritageconnector/preprocess data.ipynb
|
LinkedPasts/LaNC-workshop
|
a855da569470e94d9c5bd508b51ddb4c704a93e7
|
[
"MIT"
] | null | null | null |
heritageconnector/preprocess data.ipynb
|
LinkedPasts/LaNC-workshop
|
a855da569470e94d9c5bd508b51ddb4c704a93e7
|
[
"MIT"
] | 2 |
2020-12-07T16:32:33.000Z
|
2021-12-18T07:29:01.000Z
| 22.403162 | 115 | 0.468772 |
[
[
[
"# process data\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport json",
"_____no_output_____"
]
],
[
[
"## process gazetteers",
"_____no_output_____"
]
],
[
[
"canmore_df = pd.read_csv(\"./HES/canmore_text_extract.csv\", low_memory=False)\n\nparishes = canmore_df['PARISH'].str.title().unique().tolist()\ncounties = canmore_df['COUNTY_NAME'].str.title().unique().tolist()\n\ncanmore_text = parishes + counties\n\nprint(len(parishes), len(counties), len(canmore_text))\n\nparishes[0:10], counties[0:10]",
"183 11 194\n"
],
[
"geonames_df = pd.read_csv(\"./BL/Geonames_GBNI_places.csv\", header=None, names=['place_name'])\ngeonames_text = geonames_df['place_name'].unique().tolist()\n\niams_df = pd.read_csv(\"./BL/IAMS_GBNI_places.csv\", header=None)\niams_text = iams_df[0].unique().tolist()\n\nprint(len(geonames_text), len(iams_text))\n\ngeonames_text[0:10], iams_text[0:10]",
"7168 15873\n"
],
[
"bl_places_df = pd.read_csv(\"./BL/BL_Med_places.csv\", header=None)\nbl_places_text = bl_places[0].apply(lambda i: i.split(\";\")[0].split(\",\")[0].strip()).unique().tolist()\n\nprint(len(bl_places_text))\n\nbl_places_text[0:20]",
"925\n"
],
[
"gazetteer = list(set(parishes + counties + iams_text + bl_places_text))\n\nlen(gazetteer)",
"_____no_output_____"
],
[
"gazetteer_dict = [{\"label\": \"GPE\", \"pattern\": place} for place in gazetteer]\n\nwith open(\"gazetteer.jsonl\", \"w\") as f:\n for item in gazetteer_dict:\n json.dump(item, f)\n f.write(\"\\n\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4ae33e6799e150313bf07a33277825cf8d1794df
| 406,569 |
ipynb
|
Jupyter Notebook
|
doc/source/tutorials/tutorial_binary_under_uncertainty.ipynb
|
GabrielSGoncalves/optbinning
|
49c36210baa5ac9d65d5dadb267b3532d509368a
|
[
"Apache-2.0"
] | null | null | null |
doc/source/tutorials/tutorial_binary_under_uncertainty.ipynb
|
GabrielSGoncalves/optbinning
|
49c36210baa5ac9d65d5dadb267b3532d509368a
|
[
"Apache-2.0"
] | null | null | null |
doc/source/tutorials/tutorial_binary_under_uncertainty.ipynb
|
GabrielSGoncalves/optbinning
|
49c36210baa5ac9d65d5dadb267b3532d509368a
|
[
"Apache-2.0"
] | null | null | null | 108.882967 | 25,920 | 0.790609 |
[
[
[
"# Tutorial: optimal binning with binary target under uncertainty",
"_____no_output_____"
],
[
"The drawback of performing optimal binning given only expected event rates is that variability of event rates in different periods is not taken into account. In this tutorial, we show how scenario-based stochastic programming allows incorporating uncertainty without much difficulty.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\nfrom scipy import stats",
"_____no_output_____"
],
[
"from optbinning import OptimalBinning\nfrom optbinning.binning.uncertainty import SBOptimalBinning",
"_____no_output_____"
]
],
[
[
"### Scenario generation",
"_____no_output_____"
],
[
"We generate three scenarios, all equally likely, aiming to represent three economic scenarios severity using the customer's score variable, for instance.",
"_____no_output_____"
],
[
"**Scenario 0 - Normal (Realistic)**: A low customer' score has a higher event rate (default rate, churn, etc) than a high customer's score. The population corresponding to non-event and event are reasonably separated.",
"_____no_output_____"
]
],
[
[
"N0 = int(1e5)\n\nxe = stats.beta(a=4, b=15).rvs(size=N0, random_state=42)\nye = stats.bernoulli(p=0.7).rvs(size=N0, random_state=42)\nxn = stats.beta(a=6, b=8).rvs(size=N0, random_state=42)\nyn = stats.bernoulli(p=0.2).rvs(size=N0, random_state=42)\n\nx0 = np.concatenate((xn, xe), axis=0)\ny0 = np.concatenate((yn, ye), axis=0)",
"_____no_output_____"
],
[
"def plot_distribution(x, y):\n plt.hist(x[y == 0], label=\"n_nonevent\", color=\"b\", alpha=0.5)\n plt.hist(x[y == 1], label=\"n_event\", color=\"r\", alpha=0.5)\n plt.legend()\n plt.show()",
"_____no_output_____"
],
[
"plot_distribution(x0, y0)",
"_____no_output_____"
]
],
[
[
"**Scenario 1: Good (Optimistic)**: A low customer' score has a much higher event rate (default rate, churn, etc) than a high customer's score. The population corresponding to non-event and event rate are very well separated, showing minimum overlap regions.",
"_____no_output_____"
]
],
[
[
"N1 = int(5e4)\n\nxe = stats.beta(a=25, b=50).rvs(size=N1, random_state=42)\nye = stats.bernoulli(p=0.9).rvs(size=N1, random_state=42)\nxn = stats.beta(a=22, b=25).rvs(size=N1, random_state=42)\nyn = stats.bernoulli(p=0.05).rvs(size=N1, random_state=42)\n\nx1 = np.concatenate((xn, xe), axis=0)\ny1 = np.concatenate((yn, ye), axis=0)",
"_____no_output_____"
],
[
"plot_distribution(x1, y1)",
"_____no_output_____"
]
],
[
[
"**Scenario 2: Bad (Pessimistic)**: Customer's behavior cannot be accurately segmented, and a general increase in event rates is exhibited. The populations corresponding to non-event and event are practically overlapped.",
"_____no_output_____"
]
],
[
[
"N2 = int(5e4)\n\nxe = stats.beta(a=4, b=6).rvs(size=N2, random_state=42)\nye = stats.bernoulli(p=0.7).rvs(size=N2, random_state=42)\nxn = stats.beta(a=8, b=10).rvs(size=N2, random_state=42)\nyn = stats.bernoulli(p=0.4).rvs(size=N2, random_state=42)\n\nx2 = np.concatenate((xn, xe), axis=0)\ny2 = np.concatenate((yn, ye), axis=0)",
"_____no_output_____"
],
[
"plot_distribution(x2, y2)",
"_____no_output_____"
]
],
[
[
"### Scenario-based stochastic optimal binning",
"_____no_output_____"
],
[
"Prepare scenarios data and instantiate an ``SBOptimalBinning`` object class. We set a descending monotonicity constraint with respect to event rate and a minimum bin size.",
"_____no_output_____"
]
],
[
[
"X = [x0, x1, x2]\nY = [y0, y1, y2]",
"_____no_output_____"
],
[
"sboptb = SBOptimalBinning(monotonic_trend=\"descending\", min_bin_size=0.05)\nsboptb.fit(X, Y)",
"_____no_output_____"
],
[
"sboptb.status",
"_____no_output_____"
]
],
[
[
"We obtain \"only\" three splits guaranteeing feasibility for each scenario.",
"_____no_output_____"
]
],
[
[
"sboptb.splits",
"_____no_output_____"
],
[
"sboptb.information(print_level=2)",
"optbinning (Version 0.5.0)\nCopyright (c) 2019-2020 Guillermo Navas-Palencia, Apache License 2.0\n\n Begin options\n name * d\n prebinning_method cart * d\n max_n_prebins 20 * d\n min_prebin_size 0.05 * d\n min_n_bins no * d\n max_n_bins no * d\n min_bin_size 0.05 * U\n max_bin_size no * d\n monotonic_trend descending * U\n min_event_rate_diff 0 * d\n max_pvalue no * d\n max_pvalue_policy consecutive * d\n class_weight no * d\n user_splits no * d\n user_splits_fixed no * d\n special_codes no * d\n split_digits no * d\n time_limit 100 * d\n verbose False * d\n End options\n\n Name : UNKNOWN \n Status : OPTIMAL \n\n Pre-binning statistics\n Number of pre-bins 16\n Number of refinements 1\n\n Solver statistics\n Type cp\n Number of booleans 22\n Number of branches 48\n Number of conflicts 1\n Objective value 2676283\n Best objective bound 2676283\n\n Timing\n Total time 1.23 sec\n Pre-processing 0.01 sec ( 0.51%)\n Pre-binning 0.86 sec ( 69.84%)\n Solver 0.36 sec ( 29.60%)\n model generation 0.34 sec ( 93.32%)\n optimizer 0.02 sec ( 6.68%)\n Post-processing 0.00 sec ( 0.02%)\n\n"
]
],
[
[
"#### The binning table",
"_____no_output_____"
],
[
"As other optimal binning algorithms in OptBinning, ``SBOptimalBinning`` also returns a binning table displaying the binned data considering all scenarios.",
"_____no_output_____"
]
],
[
[
"sboptb.binning_table.build()",
"_____no_output_____"
],
[
"sboptb.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
],
[
"sboptb.binning_table.analysis()",
"---------------------------------------------\nOptimalBinning: Binary Binning Table Analysis\n---------------------------------------------\n\n General metrics\n\n Gini index 0.32783732\n IV (Jeffrey) 0.37573525\n JS (Jensen-Shannon) 0.04592372\n HHI 0.27245330\n HHI (normalized) 0.12694396\n Cramer's V 0.29928851\n Quality score 0.87078922\n\n Significance tests\n\n Bin A Bin B t-statistic p-value P[A > B] P[B > A]\n 0 1 285.174852 5.596834e-64 1.0 1.110223e-16\n 1 2 2873.289985 0.000000e+00 1.0 1.110223e-16\n 2 3 6817.466845 0.000000e+00 1.0 1.110223e-16\n\n"
]
],
[
[
"### Expected value solution (EVS)",
"_____no_output_____"
],
[
"The expected value solution is calculated with the normal (expected) scenario.",
"_____no_output_____"
]
],
[
[
"optb = OptimalBinning(monotonic_trend=\"descending\", min_bin_size=0.05)\noptb.fit(x0, y0)",
"_____no_output_____"
],
[
"optb.binning_table.build()",
"_____no_output_____"
],
[
"optb.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
],
[
"optb.binning_table.analysis()",
"---------------------------------------------\nOptimalBinning: Binary Binning Table Analysis\n---------------------------------------------\n\n General metrics\n\n Gini index 0.42141055\n IV (Jeffrey) 0.59497411\n JS (Jensen-Shannon) 0.07160267\n HHI 0.07501900\n HHI (normalized) 0.01335360\n Cramer's V 0.36927482\n Quality score 0.16335319\n\n Significance tests\n\n Bin A Bin B t-statistic p-value P[A > B] P[B > A]\n 0 1 5.139745 2.338408e-02 0.988706 1.129409e-02\n 1 2 11.534993 6.829832e-04 0.999721 2.787731e-04\n 2 3 26.208899 3.064073e-07 1.000000 7.445353e-09\n 3 4 28.661681 8.619251e-08 1.000000 4.436704e-09\n 4 5 110.500800 7.611468e-26 1.000000 1.110223e-16\n 5 6 125.906119 3.223792e-29 1.000000 1.110223e-16\n 6 7 206.865709 6.632897e-47 1.000000 1.110223e-16\n 7 8 194.419542 3.449032e-44 1.000000 1.110223e-16\n 8 9 175.309976 5.122903e-40 1.000000 1.110223e-16\n 9 10 88.957203 4.034468e-21 1.000000 1.110223e-16\n 10 11 7.648694 5.681344e-03 0.997558 2.442113e-03\n 11 12 0.520543 4.706103e-01 0.764881 2.351195e-01\n 12 13 0.278879 5.974371e-01 0.701329 2.986709e-01\n\n"
]
],
[
[
"### Scenario analysis",
"_____no_output_____"
],
[
"#### Scenario 0 - Normal (Realistic)",
"_____no_output_____"
]
],
[
[
"bt0 = sboptb.binning_table_scenario(scenario_id=0)\nbt0.build()",
"_____no_output_____"
],
[
"bt0.plot(metric=\"event_rate\")",
"_____no_output_____"
],
[
"optb0 = OptimalBinning(monotonic_trend=\"descending\", min_bin_size=0.05)\noptb0.fit(x0, y0)",
"_____no_output_____"
],
[
"optb0.binning_table.build()",
"_____no_output_____"
],
[
"optb0.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
]
],
[
[
"Apply expected value solution to scenario 0.",
"_____no_output_____"
]
],
[
[
"evs_optb0 = OptimalBinning(user_splits=optb.splits)\nevs_optb0.fit(x0, y0)",
"_____no_output_____"
],
[
"evs_optb0.binning_table.build()",
"_____no_output_____"
],
[
"evs_optb0.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
]
],
[
[
"The expected value solution applied to scenarion 0 does not satisfy the ``min_bin_size`` constraint, hence the solution is not feasible.",
"_____no_output_____"
]
],
[
[
"EVS_0 = 0.594974",
"_____no_output_____"
]
],
[
[
"**Scenario 1: Good (Optimistic)**",
"_____no_output_____"
]
],
[
[
"bt1 = sboptb.binning_table_scenario(scenario_id=1)\nbt1.build()",
"_____no_output_____"
],
[
"bt1.plot(metric=\"event_rate\")",
"_____no_output_____"
],
[
"optb1 = OptimalBinning(monotonic_trend=\"descending\", min_bin_size=0.05)\noptb1.fit(x1, y1)",
"_____no_output_____"
],
[
"optb1.binning_table.build()",
"_____no_output_____"
],
[
"optb1.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
]
],
[
[
"Apply expected value solution to scenario 1.",
"_____no_output_____"
]
],
[
[
"evs_optb1 = OptimalBinning(user_splits=optb.splits)\nevs_optb1.fit(x1, y1)",
"_____no_output_____"
],
[
"evs_optb1.binning_table.build()",
"_____no_output_____"
],
[
"evs_optb1.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
]
],
[
[
"The expected value solution applied to scenario 1 satisfies neither the ``min_bin_size`` constraint nor the monotonicity constraint, hence the solution is not feasible.",
"_____no_output_____"
]
],
[
[
"EVS_1 = -np.inf",
"_____no_output_____"
]
],
[
[
"**Scenario 2: Bad (Pessimistic)**",
"_____no_output_____"
]
],
[
[
"bt2 = sboptb.binning_table_scenario(scenario_id=2)\nbt2.build()",
"_____no_output_____"
],
[
"bt2.plot(metric=\"event_rate\")",
"_____no_output_____"
],
[
"optb2 = OptimalBinning(monotonic_trend=\"descending\", min_bin_size=0.05)\noptb2.fit(x2, y2)",
"_____no_output_____"
],
[
"optb2.binning_table.build()",
"_____no_output_____"
],
[
"optb2.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
]
],
[
[
"Apply expected value solution to scenario 2.",
"_____no_output_____"
]
],
[
[
"evs_optb2 = OptimalBinning(user_splits=optb.splits)\nevs_optb2.fit(x2, y2)",
"_____no_output_____"
],
[
"evs_optb2.binning_table.build()",
"_____no_output_____"
],
[
"evs_optb2.binning_table.plot(metric=\"event_rate\")",
"_____no_output_____"
]
],
[
[
"The expected value solution applied to scenario 2 satisfies neither the ``min_bin_size`` constraint nor the monotonicity constraint, hence the solution is not feasible.",
"_____no_output_____"
]
],
[
[
"EVS_2 = -np.inf",
"_____no_output_____"
]
],
[
[
"### Expected value of perfect information (EVPI)",
"_____no_output_____"
],
[
"If we have prior information about the incoming economic scenarios, we could take optimal solutions for each scenario, with total IV:",
"_____no_output_____"
]
],
[
[
"DIV0 = optb0.binning_table.iv\nDIV1 = optb1.binning_table.iv\nDIV2 = optb2.binning_table.iv\nDIV = (DIV0 + DIV1 + DIV2) / 3",
"_____no_output_____"
],
[
"DIV",
"_____no_output_____"
]
],
[
[
"However, this information is unlikely to be available in advance, so the best we can do in the long run is to use the stochastic programming, with expected total IV:",
"_____no_output_____"
]
],
[
[
"SIV = sboptb.binning_table.iv",
"_____no_output_____"
],
[
"SIV",
"_____no_output_____"
]
],
[
[
"The difference, in the case of perfect information, is the expected value of perfect information (EVPI) given by:",
"_____no_output_____"
]
],
[
[
"EVPI = DIV - SIV\nEVPI",
"_____no_output_____"
]
],
[
[
"### Value of stochastic solution (VSS)",
"_____no_output_____"
],
[
"The loss in IV by not considering stochasticity is the difference between the application of the expected value solution for each scenario and the stochastic model IV. The application of the EVS to each scenario results in infeasible solutions, thus",
"_____no_output_____"
]
],
[
[
"VSS = SIV - (EVS_0 + EVS_1 + EVS_2)\nVSS",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
4ae34ce82d2af268a30fd58a6ebcca7f81081d0c
| 353,006 |
ipynb
|
Jupyter Notebook
|
notebooks/isolation_forest.ipynb
|
mpky/property_project
|
f22b8dbd829ceb16a556a5abc99da2bd9c20c5ae
|
[
"MIT"
] | 6 |
2020-05-01T11:54:12.000Z
|
2021-05-26T21:05:55.000Z
|
notebooks/isolation_forest.ipynb
|
mpky/property_project
|
f22b8dbd829ceb16a556a5abc99da2bd9c20c5ae
|
[
"MIT"
] | 3 |
2020-02-26T04:22:56.000Z
|
2021-03-19T12:22:08.000Z
|
notebooks/isolation_forest.ipynb
|
mpky/property_project
|
f22b8dbd829ceb16a556a5abc99da2bd9c20c5ae
|
[
"MIT"
] | null | null | null | 370.804622 | 80,776 | 0.934384 |
[
[
[
"## Using Isolation Forest to Detect Criminally-Linked Properties\n\nThe goal of this notebook is to apply the Isolation Forest anomaly detection algorithm to the property data. The algorithm is particularly good at detecting anomalous data points in cases of extreme class imbalance. After normalizing the data and splitting into a training set and test set, I trained the first model.\n\nNext, I manually selected a few features that, based on my experience investigating money-laundering and asset tracing, I thought would be most important and trained a model on just those.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split, GridSearchCV\nfrom sklearn import preprocessing\nfrom sklearn.metrics import classification_report, confusion_matrix, recall_score, roc_auc_score\nfrom sklearn.metrics import make_scorer, precision_score, accuracy_score\nfrom sklearn.ensemble import IsolationForest\nfrom sklearn.decomposition import PCA\nimport seaborn as sns\nimport itertools\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline\nsns.set_style('dark')",
"_____no_output_____"
]
],
[
[
"#### Load Data and Remove Columns",
"_____no_output_____"
]
],
[
[
"# Read in the data\ndf = pd.read_hdf('../data/processed/bexar_true_labels.h5')\nprint(\"Number of properties:\", len(df))",
"Number of properties: 5563\n"
],
[
"# Get criminal property rate\ncrim_prop_rate = 1 - (len(df[df['crim_prop']==0]) / len(df))\nprint(\"Rate is: {:.5%}\".format(crim_prop_rate))",
"Rate is: 0.84487%\n"
],
[
"# Re-label the normal properties with 1 and the criminal ones with -1\ndf['binary_y'] = [1 if x==0 else -1 for x in df.crim_prop]\nprint(df.binary_y.value_counts())",
" 1 5516\n-1 47\nName: binary_y, dtype: int64\n"
],
[
"# Normalize the data\nX = df.iloc[:,1:-2]\nX_norm = preprocessing.normalize(X)\n\ny = df.binary_y\n\n# Split the data into training and test\nX_train_norm, X_test_norm, y_train_norm, y_test_norm = train_test_split(\n X_norm, y, test_size=0.33, random_state=42\n)",
"_____no_output_____"
]
],
[
[
"#### UDFs",
"_____no_output_____"
]
],
[
[
"# Define function to plot resulting confusion matrix\ndef plot_confusion_matrix(conf_matrix, title, classes=['criminally-linked', 'normal'],\n cmap=plt.cm.Oranges):\n \"\"\"Plot confusion matrix with heatmap and classification statistics.\"\"\"\n conf_matrix = conf_matrix.astype('float') / conf_matrix.sum(axis=1)[:, np.newaxis]\n \n plt.figure(figsize=(8,8))\n plt.imshow(conf_matrix, interpolation='nearest', cmap=cmap)\n plt.title(title,fontsize=18)\n plt.colorbar(pad=.12)\n\n tick_marks = np.arange(len(classes))\n plt.xticks(tick_marks, classes, rotation=45,fontsize=11)\n plt.yticks(tick_marks, classes, rotation=45, fontsize=11)\n\n fmt = '.4%'\n thresh = conf_matrix.max() / 2.\n for i, j in itertools.product(range(conf_matrix.shape[0]), range(conf_matrix.shape[1])):\n plt.text(j, i, format(conf_matrix[i, j], fmt),\n horizontalalignment=\"center\",\n verticalalignment=\"top\",\n fontsize=16,\n color=\"white\" if conf_matrix[i, j] > thresh else \"black\")\n plt.ylabel('True label',fontsize=14, rotation=0)\n plt.xlabel('Predicted label',fontsize=14)\n \n# Function for returning the model metrics\ndef metrics_iforest(y_true,y_pred):\n \"\"\"Return model metrics.\"\"\"\n print('Model recall is',recall_score(\n y_true,\n y_pred,\n zero_division=0,\n pos_label=-1\n ))\n print('Model precision is',precision_score(\n y_true,\n y_pred,\n zero_division=0,\n pos_label=-1\n ))\n\n print(\"Model AUC is\", roc_auc_score(y_true, y_pred))\n \n# Function for histograms of anomaly scores\ndef anomaly_plot(anomaly_scores,anomaly_scores_list,title):\n \"\"\"Plot histograms of anomaly scores.\"\"\"\n plt.figure(figsize=[15,9])\n plt.subplot(211)\n plt.hist(anomaly_scores,bins=100,log=False,color='royalblue')\n for xc in anomaly_scores_list:\n plt.axvline(x=xc,color='red',linestyle='--',linewidth=0.5,label='criminally-linked property')\n plt.title(title,fontsize=16)\n\n handles, labels = plt.gca().get_legend_handles_labels()\n by_label = dict(zip(labels, handles))\n plt.legend(by_label.values(), by_label.keys(),fontsize=14)\n plt.ylabel('Number of properties',fontsize=13)\n\n plt.subplot(212)\n plt.hist(anomaly_scores,bins=100,log=True,color='royalblue')\n for xc in anomaly_scores_list:\n plt.axvline(x=xc,color='red',linestyle='--',linewidth=0.5,label='criminally-linked property')\n plt.xlabel('Anomaly score',fontsize=13)\n plt.ylabel('Number of properties',fontsize=13)\n plt.title('{} (Log Scale)'.format(title),fontsize=16)\n\n plt.show()",
"_____no_output_____"
]
],
[
[
"#### Gridsearch\n\nIsolation Forest is fairly robust to parameter changes, but changes in the contamination rate affect performance. I will gridsearch based on a range of contamination from 0.01 to 0.25 in leaps of 0.05. ",
"_____no_output_____"
]
],
[
[
"# Set what metrics to evaluate predictions\nscoring = {\n 'AUC': 'roc_auc',\n 'Recall': make_scorer(recall_score,pos_label=-1),\n 'Precision': make_scorer(precision_score,pos_label=-1)\n}\n\ngs = GridSearchCV(\n IsolationForest(max_samples=0.25, random_state=42,n_estimators=100),\n param_grid={'contamination': np.arange(0.01, 0.25, 0.05)},\n scoring=scoring, \n refit='Recall',\n verbose=0,\n cv=3\n)\n\n# Fit to training data\ngs.fit(X_train_norm,y_train_norm)",
"_____no_output_____"
],
[
"print(gs.best_params_)",
"{'contamination': 0.21000000000000002}\n"
]
],
[
[
"##### Model Performance on Training Data",
"_____no_output_____"
]
],
[
[
"y_pred_train_gs = gs.predict(X_train_norm)\nmetrics_iforest(y_train_norm,y_pred_train_gs)",
"Model recall is 0.8695652173913043\nModel precision is 0.02554278416347382\nModel AUC is 0.831785848436473\n"
],
[
"conf_matrix = confusion_matrix(y_train_norm, y_pred_train_gs)\nprint(conf_matrix)\nplot_confusion_matrix(conf_matrix, title='Isolation Forest Confusion Matrix on Training Data')",
"[[ 20 3]\n [ 763 2941]]\n"
]
],
[
[
"Model recall is decent, but the precision is quite poor; the model is labeling >20% of innocent properties as criminal.",
"_____no_output_____"
],
[
"##### Model Performance on Test Data",
"_____no_output_____"
]
],
[
[
"y_pred_test_gs = gs.predict(X_test_norm)\nmetrics_iforest(y_test_norm,y_pred_test_gs)",
"Model recall is 0.7916666666666666\nModel precision is 0.046798029556650245\nModel AUC is 0.7890452538631346\n"
],
[
"conf_matrix = confusion_matrix(y_test_norm, y_pred_test_gs)\nprint(conf_matrix)\nplot_confusion_matrix(conf_matrix, title='Isolation Forest Confusion Matrix on Test Data')",
"[[ 19 5]\n [ 387 1425]]\n"
]
],
[
[
"Similar to performance on the training data, the model has a tremendous amount of false positives. While better than false negatives, were this model to be implemented to screen properties, it would waste a lot of manual labor on checking falsely-labeled properties.\n\nGiven the context of detecting money-laundering and ill-gotten funds, more false positives are acceptable to reduce false negatives, but the model produces far too many.",
"_____no_output_____"
],
[
"#### Visualize Distribution of Anomaly Scores\n\nSklearn's Isolation Forest provides anomaly scores for each property where the lower the score, the more anomalous the datapoint is.",
"_____no_output_____"
],
[
"##### Training Data",
"_____no_output_____"
]
],
[
[
"# Grab anomaly scores for criminally-linked properties\ntrain_df = pd.DataFrame(X_train_norm)\ny_train_series = y_train_norm.reset_index()\ntrain_df['y_value'] = y_train_series.binary_y\ntrain_df['anomaly_scores'] = gs.decision_function(X_train_norm)\nanomaly_scores_list = train_df[train_df.y_value==-1]['anomaly_scores']",
"_____no_output_____"
],
[
"print(\"Mean score for outlier properties:\",np.mean(anomaly_scores_list))\nprint(\"Mean score for normal properties:\",np.mean(train_df[train_df.y_value==1]['anomaly_scores']))",
"Mean score for outlier properties: -0.052249827254644575\nMean score for normal properties: 0.01628741354777649\n"
],
[
"anomaly_plot(train_df['anomaly_scores'],\n anomaly_scores_list,\n title='Distribution of Anomaly Scores across Training Data')",
"_____no_output_____"
]
],
[
[
"##### Test Data",
"_____no_output_____"
]
],
[
[
"test_df = pd.DataFrame(X_test_norm)\ny_test_series = y_test_norm.reset_index()\ntest_df['y_value'] = y_test_series.binary_y\ntest_df['anomaly_scores'] = gs.decision_function(X_test_norm)\nanomaly_scores_list_test = test_df[test_df.y_value==-1]['anomaly_scores']",
"_____no_output_____"
],
[
"print(\"Mean score for outlier properties:\",np.mean(anomaly_scores_list_test))\nprint(\"Mean score for normal properties:\",np.mean(test_df[test_df.y_value==1]['anomaly_scores']))",
"Mean score for outlier properties: -0.03406306018822521\nMean score for normal properties: 0.016454369969430074\n"
],
[
"anomaly_plot(test_df['anomaly_scores'], \n anomaly_scores_list_test, \n title='Distribution of Anomaly Scores across Test Data'\n)",
"_____no_output_____"
]
],
[
[
"The top plots give a sense of how skewed the distribution is and how relatively lower the anomaly scores for the criminally-linked properties are when compared to the greater population. The log scale histogram highlights just how many properties do have quite low anomaly scores, which are returned as false positives.",
"_____no_output_____"
],
[
"#### Model with Select Features\n\nWith `feature_importances_` not existing for Isolation Forest, I wanted to see if I could use my background in investigating money laundering to select a few features that would be the best indicators of \"abnormal\" properties.",
"_____no_output_____"
]
],
[
[
"# Grab specific columns\nX_trim = X[['partial_owner','just_established_owner',\n 'foreign_based_owner','out_of_state_owner',\n 'owner_legal_person','owner_likely_company',\n 'owner_owns_multiple','two_gto_reqs']]\n\n# Normalize\nX_trim_norm = preprocessing.normalize(X_trim)\n\n# Split the data into train and test\nX_train_trim, X_test_trim, y_train_trim, y_test_trim = train_test_split(\n X_trim_norm, y, test_size=0.33, random_state=42\n)",
"_____no_output_____"
],
[
"scoring = {\n 'AUC': 'roc_auc',\n 'Recall': make_scorer(recall_score, pos_label=-1),\n 'Precision': make_scorer(precision_score, pos_label=-1)\n}\n\ngs_trim = GridSearchCV(\n IsolationForest(max_samples=0.25, random_state=42,n_estimators=100),\n param_grid={'contamination': np.arange(0.01, 0.25, 0.05)},\n scoring=scoring, \n refit='Recall',\n verbose=0,\n cv=3\n)\n\n# Fit to training data\ngs_trim.fit(X_train_trim,y_train_trim)\nprint(gs_trim.best_params_)",
"{'contamination': 0.11}\n"
]
],
[
[
"##### Training Data",
"_____no_output_____"
]
],
[
[
"y_pred_train_gs_trim = gs_trim.predict(X_train_trim)\nmetrics_iforest(y_train_trim,y_pred_train_gs_trim)",
"Model recall is 0.782608695652174\nModel precision is 0.0627177700348432\nModel AUC is 0.8549922527936896\n"
],
[
"conf_matrix = confusion_matrix(y_train_trim, y_pred_train_gs_trim)\nprint(conf_matrix)\nplot_confusion_matrix(conf_matrix, title='Conf Matrix on Training Data with Select Features')",
"[[ 18 5]\n [ 269 3435]]\n"
]
],
[
[
"Reducing the data to select features worsens the model's true positives by two properties, but massively improves the false positive rate (753 down to 269). Overall, model precision is still poor.",
"_____no_output_____"
],
[
"##### Test Data",
"_____no_output_____"
]
],
[
[
"y_pred_test_trim = gs_trim.predict(X_test_trim)\nmetrics_iforest(y_test_trim,y_pred_test_trim)",
"Model recall is 0.875\nModel precision is 0.1510791366906475\nModel AUC is 0.904939293598234\n"
],
[
"conf_matrix = confusion_matrix(y_test_trim, y_pred_test_trim)\nprint(conf_matrix)\nplot_confusion_matrix(conf_matrix, title='Conf Matrix on Test Data with Select Features')",
"[[ 21 3]\n [ 118 1694]]\n"
]
],
[
[
"The model trained on select features performs better than the first on the test data both in terms of correct labels and reducing false positives.",
"_____no_output_____"
],
[
"#### Final Notes\n\n- For both models, recall is strong, indicating the model is able to detect something anomalous about the criminal properties. However, model precision is awful, meaning it does so at the expense of many false positives.\n- Selecting features based on my experience in the field improves model precision.\n- There are many properties that the models find more \"anomalous\" than the true positives. This could indicate the criminals have done a good job of making their properties appear relatively \"innocent\" in the broad spectrum of residential property ownership in Bexar County.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4ae368398ff3ba672a12cdb1cec050bcbf96a9f7
| 81,416 |
ipynb
|
Jupyter Notebook
|
Response_Analysis.ipynb
|
hrai/research-project
|
1b5e1754ea843f6363725bd5f599e3f9828edf86
|
[
"Apache-2.0"
] | null | null | null |
Response_Analysis.ipynb
|
hrai/research-project
|
1b5e1754ea843f6363725bd5f599e3f9828edf86
|
[
"Apache-2.0"
] | null | null | null |
Response_Analysis.ipynb
|
hrai/research-project
|
1b5e1754ea843f6363725bd5f599e3f9828edf86
|
[
"Apache-2.0"
] | null | null | null | 86.890075 | 46,262 | 0.680026 |
[
[
[
"<a href=\"https://colab.research.google.com/github/hrai/research-project/blob/master/Response_Analysis.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nfrom statistics import *\n\nurl='https://raw.githubusercontent.com/hrai/research-project/master/responce-accuracy.csv'\n\ndf = pd.read_csv(url)\ndf.columns",
"_____no_output_____"
],
[
"rows, cols = df.shape\nprint('total rows - {}'.format(rows))\nprint('total cols - {}'.format(cols))\n",
"total rows - 145\ntotal cols - 3\n"
],
[
"df.head(10)",
"_____no_output_____"
],
[
"group_by_question = df.groupby(by=['Question'])\ngroup_by_question.count()\n",
"_____no_output_____"
],
[
"mean_score=group_by_question.mean()\n\nmean_score",
"_____no_output_____"
],
[
"\nsns.set(style=\"whitegrid\")\nax = sns.barplot(x=\"Score\", y=\"Question\", orient='h', estimator=mean, data=df)",
"_____no_output_____"
],
[
"max_score=group_by_question.max()\nmax_score",
"_____no_output_____"
],
[
"avg_max_score=max_score['Score'].mean()\nprint('average max score is {}'.format(avg_max_score))",
"average max score is 0.7311268554333334\n"
],
[
"min_score=group_by_question.min()\nmin_score",
"_____no_output_____"
],
[
"avg_min_score=min_score['Score'].mean()\nprint('average min score is {}'.format(avg_min_score))",
"average min score is 0.38850378447333334\n"
],
[
"avg_score=df['Score'].mean()\nprint('average score is {}'.format(avg_score))",
"average score is 0.5700631354365515\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae3755ae0611b593572e0367f1856a097466ce6
| 12,756 |
ipynb
|
Jupyter Notebook
|
notebooks/WithPFLOTRAN/OUTLINED/.ipynb_checkpoints/BuildCaseDirectory-checkpoint.ipynb
|
edsaac/bioparticle
|
67e191329ef191fc539b290069524b42fbaf7e21
|
[
"MIT"
] | null | null | null |
notebooks/WithPFLOTRAN/OUTLINED/.ipynb_checkpoints/BuildCaseDirectory-checkpoint.ipynb
|
edsaac/bioparticle
|
67e191329ef191fc539b290069524b42fbaf7e21
|
[
"MIT"
] | 1 |
2020-09-25T23:31:21.000Z
|
2020-09-25T23:31:21.000Z
|
notebooks/WithPFLOTRAN/OUTLINED/.ipynb_checkpoints/BuildCaseDirectory-checkpoint.ipynb
|
edsaac/VirusTransport_RxSandbox
|
67e191329ef191fc539b290069524b42fbaf7e21
|
[
"MIT"
] | 1 |
2021-09-30T05:00:58.000Z
|
2021-09-30T05:00:58.000Z
| 43.684932 | 2,062 | 0.524302 |
[
[
[
"%reset -f\n\n## PFLOTRAN\nimport jupypft.model as mo\nimport jupypft.parameter as pm\nimport jupypft.attachmentRateCFT as arCFT\nimport jupypft.plotBTC as plotBTC",
"_____no_output_____"
]
],
[
[
"# Build the Case Directory",
"_____no_output_____"
]
],
[
[
"## Temperatures\nRef,Atm,Tin = pm.Real(tag=\"<initialTemp>\",value=10.,units=\"C\",mathRep=\"$$T_{0}$$\"),\\\n pm.Real(tag=\"<atmosphereTemp>\",value=10,units=\"C\",mathRep=\"$$T_{atm}$$\"),\\\n pm.Real(tag=\"<leakageTemp>\",value=10., units=\"m³/d\",mathRep=\"$$T_{in}$$\")",
"_____no_output_____"
],
[
"LongDisp = pm.Real(tag=\"<longDisp>\",value=0.0,units=\"m\",mathRep=\"$$\\\\alpha_L$$\")",
"_____no_output_____"
],
[
"#Gradients\nGX,GY,GZ = pm.Real(tag=\"<GradientX>\",value=0.,units=\"-\",mathRep=\"$$\\partial_x h$$\"),\\\n pm.Real(tag=\"<GradientY>\",value=0.,units=\"-\",mathRep=\"$$\\partial_y h$$\"),\\\n pm.Real(tag=\"<Gradient>>\",value=0.,units=\"-\",mathRep=\"$$\\partial_z h$$\")",
"_____no_output_____"
],
[
"## Dimensions\nLX,LY,LZ = pm.Real(\"<LenX>\",value=200,units=\"m\",mathRep=\"$$LX$$\"),\\\n pm.Real(\"<LenY>\",value=100,units=\"m\",mathRep=\"$$LY$$\"),\\\n pm.Real(\"<LenZ>\",value=20,units=\"m\",mathRep=\"$$LZ$$\")",
"_____no_output_____"
],
[
"## Permeability\nkX,kY,kZ = pm.Real(tag=\"<PermX>\",value=1.0E-8,units=\"m²\",mathRep=\"$$k_{xx}$$\"),\\\n pm.Real(tag=\"<PermY>\",value=1.0E-8,units=\"m²\",mathRep=\"$$k_{yy}$$\"),\\\n pm.Real(tag=\"<PermZ>\",value=1.0E-8,units=\"m²\",mathRep=\"$$k_{zz}$$\")\n\ntheta = pm.Real(tag=\"<porosity>\",value=0.35,units=\"adim\",mathRep=\"$$\\\\theta$$\")",
"_____no_output_____"
],
[
"## Extraction well\noutX1,outX2 = pm.Real(tag=\"<outX1>\",value=LX.value/2.,units=\"m\",mathRep=\"$$x_{1,Q_{out}}$$\"),\\\n pm.Real(tag=\"<outX2>\",value=LX.value/2.,units=\"m\",mathRep=\"$$x_{2,Q_{out}}$$\")\n\noutY1,outY2 = pm.Real(tag=\"<outY1>\",value=LY.value/2.,units=\"m\",mathRep=\"$$y_{1,Q_{out}}$$\"),\\\n pm.Real(tag=\"<outY2>\",value=LY.value/2.,units=\"m\",mathRep=\"$$y_{2,Q_{out}}$$\")\n\noutZ1,outZ2 = pm.Real(tag=\"<outZ1>\",value=LZ.value/2. ,units=\"m\",mathRep=\"$$z_{1,Q_{out}}$$\"),\\\n pm.Real(tag=\"<outZ2>\",value=LZ.value - 1.0,units=\"m\",mathRep=\"$$z_{2,Q_{out}}$$\")\n\n## Extraction rate\nQout = pm.Real(tag=\"<outRate>\",value=-21.0,units=\"m³/d\",mathRep=\"$$Q_{out}$$\")",
"_____no_output_____"
],
[
"setbackDist = 40.\n\n## Injection point\ninX1,inX2 = pm.Real(tag=\"<inX1>\",value=outX1.value + setbackDist,units=\"m\",mathRep=\"$$x_{1,Q_{in}}$$\"),\\\n pm.Real(tag=\"<inX2>\",value=outX2.value + setbackDist,units=\"m\",mathRep=\"$$x_{2,Q_{in}}$$\")\n\ninY1,inY2 = pm.Real(tag=\"<inY1>\",value=outY1.value + 0.0,units=\"m\",mathRep=\"$$y_{1,Q_{in}}$$\"),\\\n pm.Real(tag=\"<inY2>\",value=outY2.value + 0.0,units=\"m\",mathRep=\"$$y_{2,Q_{in}}$$\")\n\ninZ1,inZ2 = pm.Real(tag=\"<inZ1>\",value=LZ.value - 5.0,units=\"m\",mathRep=\"$$z_{1,Q_{in}}$$\"),\\\n pm.Real(tag=\"<inZ2>\",value=LZ.value - 1.0,units=\"m\",mathRep=\"$$z_{2,Q_{in}}$$\")",
"_____no_output_____"
],
[
"## Concentration\nC0 = pm.Real(\"<initialConcentration>\", value=1.0, units=\"mol/L\")\n\n## Injection rate\nQin = pm.Real(tag=\"<inRate>\",value=0.24, units=\"m³/d\",mathRep=\"$$Q_{in}$$\")",
"_____no_output_____"
],
[
"## Grid\nnX,nY,nZ = pm.Integer(\"<nX>\",value=41,units=\"-\",mathRep=\"$$nX$$\"),\\\n pm.Integer(\"<nY>\",value=21 ,units=\"-\",mathRep=\"$$nY$$\"),\\\n pm.Integer(\"<nZ>\",value=1,units=\"-\",mathRep=\"$$nZ$$\")\n\ndX,dY,dZ = pm.JustText(\"<dX>\"),\\\n pm.JustText(\"<dY>\"),\\\n pm.JustText(\"<dZ>\")\n\nCellRatio = { 'X' : 2.0, 'Y' : 2.0, 'Z' : 0.75 }\n#CellRatio = { 'X' : 1.00, 'Y' : 0.50, 'Z' : 0.75 }\n\ndX.value = mo.buildDXYZ(LX.value,CellRatio['X'],nX.value,hasBump=True)\ndY.value = mo.buildDXYZ(LY.value,CellRatio['Y'],nY.value,hasBump=True)\n\nif nZ == 1:\n dZ.value = LZ.value\nelse:\n dZ.value = mo.buildDXYZ(LZ.value,CellRatio['Z'],nZ.value,hasBump=False)",
"_____no_output_____"
],
[
"# Time config\nendTime = pm.Real(\"<endTime>\",value=100.,units=\"d\")",
"_____no_output_____"
],
[
"## Bioparticle\nkAtt,kDet = pm.Real(tag=\"<katt>\",value=1.0E-30,units=\"1/s\",mathRep=\"$$k_{att}$$\"),\\\n pm.Real(tag=\"<kdet>\",value=1.0E-30,units=\"1/s\",mathRep=\"$$k_{det}$$\")\n\ndecayAq,decayIm = pm.Real(tag=\"<decayAq>\",value=1.0E-30,units=\"1/s\",mathRep=\"$$\\lambda_{aq}$$\"),\\\n pm.Real(tag=\"<decayIm>\",value=1.0E-30,units=\"1/s\",mathRep=\"$$\\lambda_{im}$$\")",
"_____no_output_____"
],
[
"caseDict = {\n \"Temp\":{\n \"Reference\" : Ref,\n \"Atmosphere\": Atm,\n \"Injection\" : Tin },\n \"longDisp\":LongDisp,\n \"Gradient\":{\n \"X\" :GX,\n \"Y\" :GY,\n \"Z\" :GZ },\n \"L\":{\n \"X\" :LX,\n \"Y\" :LY,\n \"Z\" :LZ },\n \"k\":{\n \"X\" :kX,\n \"Y\" :kY,\n \"Z\" :kZ },\n \"theta\":theta,\n \"outCoord\":{\n \"X\" : { 1 : outX1,\n 2 : outX2},\n \"Y\" : { 1 : outY1,\n 2 : outY2},\n \"Z\" : { 1 : outZ1,\n 2 : outZ2}},\n \"inCoord\":{\n \"X\" : { 1 : inX1,\n 2 : inX2},\n \"Y\" : { 1 : inY1,\n 2 : inY2},\n \"Z\" : { 1 : inZ1,\n 2 : inZ2}},\n \"C0\":C0,\n \"Q\":{\"In\":Qin,\n \"Out\":Qout},\n \"nGrid\":{\"X\":nX,\n \"Y\":nY,\n \"Z\":nZ},\n \"dGrid\":{\"X\":dX,\n \"Y\":dY,\n \"Z\":dZ},\n \"endTime\":endTime,\n \"BIOPARTICLE\":{\n \"katt\" : kAtt,\n \"kdet\" : kDet,\n \"decayAq\" : decayAq,\n \"decayIm\" : decayIm}\n }",
"_____no_output_____"
],
[
"import pickle\n\nwith open('caseDict.pkl', 'wb') as f:\n pickle.dump(caseDict,f)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae3900799a02ac67c7bf65b5e00d829f2983940
| 7,966 |
ipynb
|
Jupyter Notebook
|
Pytorch Study/Pytorch GAN.ipynb
|
UPRMG/Study_Programming
|
8bb0848ece48621b253035bee8cfe0101daa9b82
|
[
"MIT"
] | null | null | null |
Pytorch Study/Pytorch GAN.ipynb
|
UPRMG/Study_Programming
|
8bb0848ece48621b253035bee8cfe0101daa9b82
|
[
"MIT"
] | null | null | null |
Pytorch Study/Pytorch GAN.ipynb
|
UPRMG/Study_Programming
|
8bb0848ece48621b253035bee8cfe0101daa9b82
|
[
"MIT"
] | null | null | null | 28.14841 | 165 | 0.481296 |
[
[
[
"## GAN (Pytorch)",
"_____no_output_____"
],
[
"### Terminal : tensorboard --logdir=./GAN",
"_____no_output_____"
],
[
"Reference : https://github.com/aladdinpersson/Machine-Learning-Collection/blob/master/ML/Pytorch/GANs/1.%20SimpleGAN/fc_gan.py",
"_____no_output_____"
],
[
"$$\n\\underset{\\theta_{g}}min \\underset{\\theta_{d}}max[E_{x\\sim P_{data}}logD_{\\theta_{d}}(x) + E_{z\\sim P_{z}}log(1-D_{\\theta_{d}}(G_{\\theta_{g}}(z)))]\n$$\n\n\n- For D, maximize objective by making 𝑫(𝒙) is close to 1 and 𝑫(𝑮(𝒛)) is close to 0\n- For G, minimize objective by making 𝑫(𝑮(𝒛))",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.optim as optim\nimport torchvision\nimport torchvision.datasets as datasets\nfrom torch.utils.data import DataLoader\nimport torchvision.transforms as transforms\nfrom torch.utils.tensorboard import SummaryWriter\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"class Disciminator(nn.Module):\n def __init__(self, img_dim):\n super().__init__()\n self.disc = nn.Sequential(\n nn.Linear(img_dim, 128),\n nn.LeakyReLU(0.1),\n nn.Linear(128,1),\n nn.Sigmoid(),\n )\n \n def forward(self, x):\n return self.disc(x)\n \nclass Generator(nn.Module):\n def __init__(self, z_dim, img_dim):\n super().__init__()\n self.gen = nn.Sequential(\n nn.Linear(z_dim, 256),\n nn.LeakyReLU(0.1),\n nn.Linear(256, img_dim),\n nn.Tanh(),\n )\n \n def forward(self, x):\n return self.gen(x)",
"_____no_output_____"
],
[
"device = 'cuda' if torch.cuda.is_available() else 'cpu'\nlr = 3e-4\nz_dim = 64 #128, 256\nimage_dim = 28*28*1 #784\nbatch_size = 32\nnum_epochs = 10\n\ndisc = Disciminator(image_dim).to(device)\ngen = Generator(z_dim, image_dim).to(device)\n\nfixed_noise = torch.randn((batch_size, z_dim)).to(device)\n\ntransforms = transforms.Compose(\n[transforms.ToTensor(), transforms.Normalize((0.1307,),(0.3081,))]\n)",
"_____no_output_____"
],
[
"dataset = datasets.MNIST(root='./content',\n transform=transforms,\n download=False)",
"_____no_output_____"
],
[
"loader = DataLoader(dataset, batch_size=batch_size, shuffle=True)",
"_____no_output_____"
],
[
"opt_disc = optim.Adam(disc.parameters(),lr=lr)\nopt_gen = optim.Adam(gen.parameters(),lr=lr)",
"_____no_output_____"
],
[
"criterion = nn.BCELoss()",
"_____no_output_____"
],
[
"writer_fake = SummaryWriter(f\"./GAN\")\nwriter_real = SummaryWriter(f\"./GAN\")",
"_____no_output_____"
],
[
"step = 0",
"_____no_output_____"
],
[
"for epoch in range(num_epochs):\n for batch_idx, (real, _) in enumerate(loader):\n real = real.view(-1, 784).to(device)\n batch_size = real.shape[0]\n \n ### Train Disciminator : max log(D(real)) + log(1-D(G(z)))\n \n noise = torch.randn(batch_size, z_dim).to(device)\n fake = gen(noise)\n \n disc_real = disc(real).view(-1)\n lossD_real = criterion(disc_real, torch.ones_like(disc_real))\n \n disc_fake = disc(fake).view(-1)\n lossD_fake = criterion(disc_fake, torch.ones_like(disc_fake))\n \n lossD = (lossD_real + lossD_fake) / 2\n \n disc.zero_grad()\n lossD.backward(retain_graph=True)\n opt_disc.step()\n \n ### Train Generator min log(1-D(G(z))) <-> max log(D(G(z)))\n output = disc(fake).view(-1)\n lossG = criterion(output, torch.ones_like(output))\n gen.zero_grad()\n lossG.backward()\n opt_gen.step()\n \n if batch_idx == 0:\n print(f\"Epoch [{epoch}/{num_epochs}] \\ \"\n f\"Loss D : {lossD : .4f}, Loss G : {lossG : .4f}\"\n )\n \n with torch.no_grad():\n fake = gen(fixed_noise).reshape(-1,1,28,28)\n data = real.reshape(-1,1,28,28)\n img_grid_fake = torchvision.utils.make_grid(fake, normalize=True)\n img_grid_real = torchvision.utils.make_grid(data, normalize=True)\n \n writer_fake.add_image(\n \"MNIST Fake Images\", img_grid_fake, global_step=step)\n \n writer_real.add_image(\n \"MNIST Real Images\", img_grid_real, global_step=step)\n \n step += 1",
"Epoch [0/10] \\ Loss D : 0.6312, Loss G : 0.6577\nEpoch [1/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [2/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [3/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [4/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [5/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [6/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [7/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [8/10] \\ Loss D : 0.0000, Loss G : 0.0000\nEpoch [9/10] \\ Loss D : 0.0000, Loss G : 0.0000\n"
]
],
[
[
"+ cuda device\n+ change learning rate\n+ change Normalization \n+ change batchnorm\n+ architecture change CNN",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4ae3946e7dda78a534afaf1a1682a384a6cb04ae
| 94,481 |
ipynb
|
Jupyter Notebook
|
lessons/01_phugoid/01_03 Full_Phugoid_Model_solutions.ipynb
|
rbds/Numerical_Methods_working_folder
|
d929ed7506054e7aa7ba059623c37ecf7d6ae993
|
[
"CC-BY-3.0"
] | null | null | null |
lessons/01_phugoid/01_03 Full_Phugoid_Model_solutions.ipynb
|
rbds/Numerical_Methods_working_folder
|
d929ed7506054e7aa7ba059623c37ecf7d6ae993
|
[
"CC-BY-3.0"
] | null | null | null |
lessons/01_phugoid/01_03 Full_Phugoid_Model_solutions.ipynb
|
rbds/Numerical_Methods_working_folder
|
d929ed7506054e7aa7ba059623c37ecf7d6ae993
|
[
"CC-BY-3.0"
] | null | null | null | 174.319188 | 34,572 | 0.889682 |
[
[
[
"from math import sin, cos, log, ceil\nimport numpy\nfrom matplotlib import pyplot\n%matplotlib inline\nfrom matplotlib import rcParams\nrcParams['font.family'] = 'serif'\nrcParams['font.size']=16",
"_____no_output_____"
],
[
"# model parameters:\ng= 9.8 #[m/s^2]\nv_t = 20.0 #[m/s] trim velocity\nC_D = 1/40 #drag coef.\nC_L = 1 #coefficient of lift\n\n#ICs\nv0 = v_t\ntheta0 = 0\nx0 = 0\ny0 = 1000",
"_____no_output_____"
],
[
"def f(u):\n \"\"\" Returns RHS of phugoid system of eqns.\n parameters: \n u - array of float with solution at time n\n \n returns:\n dudt - array of float with solution of RHS given u \n \"\"\"\n \n v = u[0]\n theta = u[1]\n x = u[2]\n y = u[3]\n return numpy.array([-g*sin(theta) - C_D/C_L*g/v_t**2*v**2,\n -g*cos(theta)/v +g/v_t**2*v,\n v*cos(theta),\n v*sin(theta)])",
"_____no_output_____"
],
[
"def euler(u,f,dt):\n \"\"\"Euler's method, returns next time step\n \n u: soln. at previous time step\n f: function to compute RHS of system of equations\n dt: dt.\n \n \"\"\"\n return u + dt*f(u)",
"_____no_output_____"
],
[
"T = 100 #t_final\ndt = 0.1\nN = int(T/dt) + 1\nt = numpy.linspace(0,T,N)\n#initialize array\nu = numpy.empty((N,4))\nu[0] = numpy.array([v0, theta0, x0, y0]) #ICs\n\n#Euler's method\nfor n in range(N-1):\n u[n+1] = euler(u[n], f, dt)",
"_____no_output_____"
],
[
"x = u[:,2]\ny = u[:,3]",
"_____no_output_____"
],
[
"pyplot.figure(figsize=(8,6))\npyplot.grid(True)\npyplot.xlabel(r'x', fontsize=18)\npyplot.ylabel(r'y', fontsize=18)\npyplot.title('Glider trajectory, flight time = %.2f' %T, fontsize=18)\npyplot.plot(x,y, lw=2);",
"_____no_output_____"
],
[
"dt_values = numpy.array([0.1, 0.05, 0.01, 0.005, 0.001])\nu_values = numpy.empty_like(dt_values, dtype=numpy.ndarray)\nfor i, dt in enumerate(dt_values):\n N=int(T/dt) + 1\n t = numpy.linspace(0.0, T, N)\n \n #initialize solution array\n u = numpy.empty((N,4))\n u[0] = numpy.array([v0, theta0, x0, y0])\n\n for n in range(N-1):\n u[n+1] = euler(u[n], f, dt)\n \n u_values[i] = u",
"_____no_output_____"
],
[
"def get_diffgrid(u_current, u_fine, dt):\n \"\"\"Returns the difference between one grid and the finest grid using the L1 norm\n parameters: \n u_current: solution on current grid\n u_finest: solution on fine grid\n dt\n \n returns:\n diffgrid: difference computed in L1 norm\n \"\"\"\n N_current = len(u_current[:,0])\n N_fine = len(u_fine[:,0])\n \n grid_size_ratio = ceil(N_fine/N_current)\n diffgrid = dt*numpy.sum(numpy.abs(u_current[:,2]-u_fine[::grid_size_ratio,2]))\n \n return diffgrid",
"_____no_output_____"
],
[
"diffgrid = numpy.empty_like(dt_values)\n\nfor i, dt in enumerate(dt_values):\n print('dt = {}'.format(dt))\n \n diffgrid[i] = get_diffgrid(u_values[i], u_values[-1], dt)",
"dt = 0.1\ndt = 0.05\ndt = 0.01\ndt = 0.005\ndt = 0.001\n"
],
[
"pyplot.figure(figsize=(6,6))\npyplot.grid(True)\npyplot.xlabel('$\\Delta t$', fontsize=18)\npyplot.ylabel('$L_1$-norm of the grid differences', fontsize=18)\npyplot.axis('equal')\npyplot.loglog(dt_values[:-1], diffgrid[:-1], ls='-', marker='o', lw=2);",
"_____no_output_____"
],
[
"\n\nr = 2 #what is r?\nh = 0.001\n\ndt_values2 = numpy.array([h, r*h, r**2*h])\n\nu_values2 = numpy.empty_like(dt_values2, dtype=numpy.ndarray)\n\ndiffgrid2 = numpy.empty(2)\n\nfor i, dt in enumerate(dt_values2):\n \n N = int(T/dt) + 1 # number of time-steps\n \n ### discretize the time t ###\n t = numpy.linspace(0.0, T, N)\n \n # initialize the array containing the solution for each time-step\n u = numpy.empty((N, 4))\n u[0] = numpy.array([v0, theta0, x0, y0])\n\n for n in range(N-1):\n\n u[n+1] = euler(u[n], f, dt) \n \n # store the value of u related to one grid\n u_values2[i] = u\n \n\n#calculate f2 - f1\ndiffgrid2[0] = get_diffgrid(u_values2[1], u_values2[0], dt_values2[1])\n\n#calculate f3 - f2\ndiffgrid2[1] = get_diffgrid(u_values2[2], u_values2[1], dt_values2[2])\n\n# calculate the order of convergence\np = (log(diffgrid2[1]) - log(diffgrid2[0])) / log(r)\n\nprint('The order of convergence is p = {:.3f}'.format(p));",
"The order of convergence is p = 1.029\n"
]
],
[
[
"Paper Airplane Challenge: \n - Find a combination of launch angle and velocity that gives best distance.\n ",
"_____no_output_____"
]
],
[
[
"L_D = 5.0\nC_D = 1/L_D\nv_t = 4.9 #[m/s]\n#ICs\ntheta0 = 0\nx0 = 0\ny0 = 2 #[m] - a realistic height to throw a paper airplane from\nv0 = v_t\n#height = y0\n#t = [0]\n\ndt = 0.001\nT=20\nN = int(T/dt)+1\nt = numpy.linspace(0,T, N)\n\ndef challenge(arg1, arg2):\n u = numpy.empty((N,4))\n u[0] = numpy.array([arg1, arg2, x0, y0])\n #print(numpy.shape(u))\n #print(N)\n for n in range(N-1):\n u[n+1] = euler(u[n], f, dt)\n n_max = n\n if u[n][3] <= 0:\n break\n #print(numpy.shape(u))\n u = u[:n_max]\n #print(numpy.shape(u))\n\n return u\n\n",
"_____no_output_____"
],
[
"u = challenge(5, 0)\nprint(u[-1][2])",
"10.4577277488\n"
],
[
"#iterate over v0, theta0, taking large steps\n#find a best solution\nmax_dist = 0.0\nmax_params = [0, 0]\nfor theta0 in range (-90, 90, 5):\n for v0 in range (1, 10):\n u = numpy.empty((1,4))\n u_final = challenge(v0, theta0)\n if u_final[-1][2] > max_dist:\n max_params_low_res = [v0, theta0]\n max_dist_low_res = u_final[-1][2]\n best_run_low_res = u_final\n\nprint(max_dist_low_res)\nprint(max_params_low_res)",
"0.635345072227\n[9, 80]\n"
],
[
"#iterate at a finer resolution over previous solution\nmax_dist = 0.0 #\nmax_params = [0, 0]\nfor theta0 in range (max_params_low_res[1] -5, max_params_low_res[1] +5):\n for v0 in range (max_params_low_res[0] -5, max_params_low_res[0] + 5):\n u = numpy.empty((1,4))\n u_final = challenge(v0, theta0)\n if u_final[-1][2] > max_dist:\n max_params= [v0, theta0]\n max_dist = u_final[-1][2]\n best_run = u_final\n\nprint(max_dist)\nprint(max_params)",
"17.9958296006\n[10, 75]\n"
],
[
"#print(u_longest)\nx = best_run[:,2]\ny = best_run[:,3]\npyplot.figure(figsize=(8,6))\npyplot.grid(True)\npyplot.xlabel(r'x', fontsize=18)\npyplot.ylabel(r'y', fontsize=18)\npyplot.title('Paper airplane trajectory', fontsize=18)\npyplot.plot(x,y, 'k-', lw=2);\n\nprint(\"max distance: {:.2f} m, v0= {:.2f}, theta0 = {:.2f}\".format(max_dist, max_params[0], max_params[1]))",
"max distance: 18.00 m, v0= 10.00, theta0 = 75.00\n"
],
[
"\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae39c5dd6a22d38f247ca9607f5d51a450de669
| 14,676 |
ipynb
|
Jupyter Notebook
|
notebooks_vacios/013-NumPy-SeccionesArrays.ipynb
|
caorodriguez/aprendealgo-
|
b8e9692214280cc0c1981bbd414b19fd0fa7dcb8
|
[
"CC-BY-4.0"
] | 86 |
2015-03-05T18:57:16.000Z
|
2021-11-18T00:19:22.000Z
|
notebooks_vacios/013-NumPy-SeccionesArrays.ipynb
|
caorodriguez/aprendealgo-
|
b8e9692214280cc0c1981bbd414b19fd0fa7dcb8
|
[
"CC-BY-4.0"
] | 66 |
2015-01-26T19:08:20.000Z
|
2020-05-20T17:09:58.000Z
|
notebooks_vacios/013-NumPy-SeccionesArrays.ipynb
|
caorodriguez/aprendealgo-
|
b8e9692214280cc0c1981bbd414b19fd0fa7dcb8
|
[
"CC-BY-4.0"
] | 63 |
2015-02-18T15:12:45.000Z
|
2022-03-29T06:18:39.000Z
| 27.431776 | 620 | 0.507155 |
[
[
[
"<img src=\"../images/aeropython_logo.png\" alt=\"AeroPython\" style=\"width: 300px;\"/>",
"_____no_output_____"
],
[
"# Secciones de arrays",
"_____no_output_____"
],
[
"_Hasta ahora sabemos cómo crear arrays y realizar algunas operaciones con ellos, sin embargo, todavía no hemos aprendido cómo acceder a elementos concretos del array_",
"_____no_output_____"
],
[
"## Arrays de una dimensión",
"_____no_output_____"
]
],
[
[
"# Accediendo al primer elemento",
"_____no_output_____"
],
[
"# Accediendo al último",
"_____no_output_____"
]
],
[
[
"##### __¡Atención!__ ",
"_____no_output_____"
],
[
"NumPy devuelve __vistas__ de la sección que le pidamos, no __copias__. Esto quiere decir que debemos prestar mucha atención a este comportamiento:",
"_____no_output_____"
],
[
"Lo mismo ocurre al revés:",
"_____no_output_____"
],
[
"`a` apunta a las direcciones de memoria donde están guardados los elementos del array `arr` que hemos seleccionado, no copia sus valores, a menos que explícitamente hagamos:",
"_____no_output_____"
],
[
"## Arrays de dos dimensiones",
"_____no_output_____"
],
[
"## Secciones de arrays",
"_____no_output_____"
],
[
"Hasta ahora hemos visto cómo acceder a elementos aislados del array, pero la potencia de NumPy está en poder acceder a secciones enteras. Para ello se usa la sintaxis `inicio:final:paso`: si alguno de estos valores no se pone toma un valor por defecto. Veamos ejemplos:",
"_____no_output_____"
]
],
[
[
"# De la segunda a la tercera fila, incluida",
"_____no_output_____"
],
[
"# Hasta la tercera fila sin incluir y de la segunda a la quinta columnas saltando dos\n#M[1:2:1, 1:5:2] # Equivalente",
"_____no_output_____"
]
],
[
[
"##### Ejercicio",
"_____no_output_____"
],
[
"Pintar un tablero de ajedrez usando la función `plt.matshow`.",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"___Hemos aprendido:___\n\n* A acceder a elementos de un array\n* Que las secciones no devuelven copias, sino vistas\n\n__¡Quiero más!__Algunos enlaces:\n\nAlgunos enlaces en Pybonacci:\n\n* [Cómo crear matrices en Python con NumPy](http://pybonacci.wordpress.com/2012/06/11/como-crear-matrices-en-python-con-numpy/).\n* [Números aleatorios en Python con NumPy y SciPy](http://pybonacci.wordpress.com/2013/01/11/numeros-aleatorios-en-python-con-numpy-y-scipy/).\n\n\nAlgunos enlaces en otros sitios:\n\n* [100 numpy exercises](http://www.labri.fr/perso/nrougier/teaching/numpy.100/index.html). Es posible que de momento sólo sepas hacer los primeros, pero tranquilo, pronto sabrás más...\n* [NumPy and IPython SciPy 2013 Tutorial](http://conference.scipy.org/scipy2013/tutorial_detail.php?id=100).\n* [NumPy and SciPy documentation](http://docs.scipy.org/doc/).",
"_____no_output_____"
],
[
"---\n<br/>\n#### <h4 align=\"right\">¡Síguenos en Twitter!\n<br/>\n###### <a href=\"https://twitter.com/AeroPython\" class=\"twitter-follow-button\" data-show-count=\"false\">Follow @AeroPython</a> <script>!function(d,s,id){var js,fjs=d.getElementsByTagName(s)[0],p=/^http:/.test(d.location)?'http':'https';if(!d.getElementById(id)){js=d.createElement(s);js.id=id;js.src=p+'://platform.twitter.com/widgets.js';fjs.parentNode.insertBefore(js,fjs);}}(document, 'script', 'twitter-wjs');</script> \n<br/>\n###### Este notebook ha sido realizado por: Juan Luis Cano y Álex Sáez \n<br/>\n##### <a rel=\"license\" href=\"http://creativecommons.org/licenses/by/4.0/deed.es\"><img alt=\"Licencia Creative Commons\" style=\"border-width:0\" src=\"http://i.creativecommons.org/l/by/4.0/88x31.png\" /></a><br /><span xmlns:dct=\"http://purl.org/dc/terms/\" property=\"dct:title\">Curso AeroPython</span> por <span xmlns:cc=\"http://creativecommons.org/ns#\" property=\"cc:attributionName\">Juan Luis Cano Rodriguez y Alejandro Sáez Mollejo</span> se distribuye bajo una <a rel=\"license\" href=\"http://creativecommons.org/licenses/by/4.0/deed.es\">Licencia Creative Commons Atribución 4.0 Internacional</a>.",
"_____no_output_____"
],
[
"---\n_Las siguientes celdas contienen configuración del Notebook_\n\n_Para visualizar y utlizar los enlaces a Twitter el notebook debe ejecutarse como [seguro](http://ipython.org/ipython-doc/dev/notebook/security.html)_\n\n File > Trusted Notebook",
"_____no_output_____"
]
],
[
[
"# Esta celda da el estilo al notebook\nfrom IPython.core.display import HTML\ncss_file = '../styles/aeropython.css'\nHTML(open(css_file, \"r\").read())",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
4ae3ceed2934acec78b4cf7710ff084e36daf4cf
| 271,711 |
ipynb
|
Jupyter Notebook
|
experiments/tl_3/A_killme/cores_wisig-oracle.run1.framed/trials/13/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null |
experiments/tl_3/A_killme/cores_wisig-oracle.run1.framed/trials/13/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null |
experiments/tl_3/A_killme/cores_wisig-oracle.run1.framed/trials/13/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null | 102.804011 | 73,188 | 0.704267 |
[
[
[
"# Transfer Learning Template",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\n \nimport os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom torch.optim import Adam\nfrom easydict import EasyDict\nimport matplotlib.pyplot as plt\n\nfrom steves_models.steves_ptn import Steves_Prototypical_Network\n\nfrom steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper\nfrom steves_utils.iterable_aggregator import Iterable_Aggregator\nfrom steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader\nfrom steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)\nfrom steves_utils.PTN.utils import independent_accuracy_assesment\n\nfrom torch.utils.data import DataLoader\n\nfrom steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory\n\nfrom steves_utils.ptn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.transforms import get_chained_transform",
"_____no_output_____"
]
],
[
[
"# Allowed Parameters\nThese are allowed parameters, not defaults\nEach of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)\n\nPapermill uses the cell tag \"parameters\" to inject the real parameters below this cell.\nEnable tags to see what I mean",
"_____no_output_____"
]
],
[
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"seed\",\n \"dataset_seed\",\n \"n_shot\",\n \"n_query\",\n \"n_way\",\n \"train_k_factor\",\n \"val_k_factor\",\n \"test_k_factor\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"x_net\",\n \"datasets\",\n \"torch_default_dtype\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"x_shape\",\n}",
"_____no_output_____"
],
[
"from steves_utils.CORES.utils import (\n ALL_NODES,\n ALL_NODES_MINIMUM_1000_EXAMPLES,\n ALL_DAYS\n)\n\nfrom steves_utils.ORACLE.utils_v2 import (\n ALL_DISTANCES_FEET_NARROWED,\n ALL_RUNS,\n ALL_SERIAL_NUMBERS,\n)\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"STANDALONE PTN\"\nstandalone_parameters[\"lr\"] = 0.001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"dataset_seed\"] = 1337\n\nstandalone_parameters[\"n_way\"] = 8\nstandalone_parameters[\"n_shot\"] = 3\nstandalone_parameters[\"n_query\"] = 2\nstandalone_parameters[\"train_k_factor\"] = 1\nstandalone_parameters[\"val_k_factor\"] = 2\nstandalone_parameters[\"test_k_factor\"] = 2\n\n\nstandalone_parameters[\"n_epoch\"] = 50\n\nstandalone_parameters[\"patience\"] = 10\nstandalone_parameters[\"criteria_for_best\"] = \"source_loss\"\n\nstandalone_parameters[\"datasets\"] = [\n {\n \"labels\": ALL_SERIAL_NUMBERS,\n \"domains\": ALL_DISTANCES_FEET_NARROWED,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\", \"minus_two\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE_\"\n },\n {\n \"labels\": ALL_NODES,\n \"domains\": ALL_DAYS,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_power\", \"times_zero\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"CORES_\"\n } \n]\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\n\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n]\n\n# Parameters relevant to results\n# These parameters will basically never need to change\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"\n\n\n\n\n",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"tl_3A:cores+wisig -> oracle.run1.framed\",\n \"device\": \"cuda\",\n \"lr\": 0.001,\n \"x_shape\": [2, 200],\n \"n_shot\": 3,\n \"n_query\": 2,\n \"train_k_factor\": 3,\n \"val_k_factor\": 2,\n \"test_k_factor\": 2,\n \"torch_default_dtype\": \"torch.float32\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_loss\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 200]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 16000, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"n_way\": 16,\n \"datasets\": [\n {\n \"labels\": [\n \"1-10.\",\n \"1-11.\",\n \"1-15.\",\n \"1-16.\",\n \"1-17.\",\n \"1-18.\",\n \"1-19.\",\n \"10-4.\",\n \"10-7.\",\n \"11-1.\",\n \"11-14.\",\n \"11-17.\",\n \"11-20.\",\n \"11-7.\",\n \"13-20.\",\n \"13-8.\",\n \"14-10.\",\n \"14-11.\",\n \"14-14.\",\n \"14-7.\",\n \"15-1.\",\n \"15-20.\",\n \"16-1.\",\n \"16-16.\",\n \"17-10.\",\n \"17-11.\",\n \"17-2.\",\n \"19-1.\",\n \"19-16.\",\n \"19-19.\",\n \"19-20.\",\n \"19-3.\",\n \"2-10.\",\n \"2-11.\",\n \"2-17.\",\n \"2-18.\",\n \"2-20.\",\n \"2-3.\",\n \"2-4.\",\n \"2-5.\",\n \"2-6.\",\n \"2-7.\",\n \"2-8.\",\n \"3-13.\",\n \"3-18.\",\n \"3-3.\",\n \"4-1.\",\n \"4-10.\",\n \"4-11.\",\n \"4-19.\",\n \"5-5.\",\n \"6-15.\",\n \"7-10.\",\n \"7-14.\",\n \"8-18.\",\n \"8-20.\",\n \"8-3.\",\n \"8-8.\",\n ],\n \"domains\": [1, 2, 3, 4, 5],\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": \"/mnt/wd500GB/CSC500/csc500-main/datasets/cores.stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\", \"take_200\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"C_A_\",\n },\n {\n \"labels\": [\n \"1-10\",\n \"1-12\",\n \"1-14\",\n \"1-16\",\n \"1-18\",\n \"1-19\",\n \"1-8\",\n \"10-11\",\n \"10-17\",\n \"10-4\",\n \"10-7\",\n \"11-1\",\n \"11-10\",\n \"11-19\",\n \"11-20\",\n \"11-4\",\n \"11-7\",\n \"12-19\",\n \"12-20\",\n \"12-7\",\n \"13-14\",\n \"13-18\",\n \"13-19\",\n \"13-20\",\n \"13-3\",\n \"13-7\",\n \"14-10\",\n \"14-11\",\n \"14-12\",\n \"14-13\",\n \"14-14\",\n \"14-19\",\n \"14-20\",\n \"14-7\",\n \"14-8\",\n \"14-9\",\n \"15-1\",\n \"15-19\",\n \"15-6\",\n \"16-1\",\n \"16-16\",\n \"16-19\",\n \"16-20\",\n \"17-10\",\n \"17-11\",\n \"18-1\",\n \"18-10\",\n \"18-11\",\n \"18-12\",\n \"18-13\",\n \"18-14\",\n \"18-15\",\n \"18-16\",\n \"18-17\",\n \"18-19\",\n \"18-2\",\n \"18-20\",\n \"18-4\",\n \"18-5\",\n \"18-7\",\n \"18-8\",\n \"18-9\",\n \"19-1\",\n \"19-10\",\n \"19-11\",\n \"19-12\",\n \"19-13\",\n \"19-14\",\n \"19-15\",\n \"19-19\",\n \"19-2\",\n \"19-20\",\n \"19-3\",\n \"19-4\",\n \"19-6\",\n \"19-7\",\n \"19-8\",\n \"19-9\",\n \"2-1\",\n \"2-13\",\n \"2-15\",\n \"2-3\",\n \"2-4\",\n \"2-5\",\n \"2-6\",\n \"2-7\",\n \"2-8\",\n \"20-1\",\n \"20-12\",\n \"20-14\",\n \"20-15\",\n \"20-16\",\n \"20-18\",\n \"20-19\",\n \"20-20\",\n \"20-3\",\n \"20-4\",\n \"20-5\",\n \"20-7\",\n \"20-8\",\n \"3-1\",\n \"3-13\",\n \"3-18\",\n \"3-2\",\n \"3-8\",\n \"4-1\",\n \"4-10\",\n \"4-11\",\n \"5-1\",\n \"5-5\",\n \"6-1\",\n \"6-15\",\n \"6-6\",\n \"7-10\",\n \"7-11\",\n \"7-12\",\n \"7-13\",\n \"7-14\",\n \"7-7\",\n \"7-8\",\n \"7-9\",\n \"8-1\",\n \"8-13\",\n \"8-14\",\n \"8-18\",\n \"8-20\",\n \"8-3\",\n \"8-8\",\n \"9-1\",\n \"9-7\",\n ],\n \"domains\": [1, 2, 3, 4],\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": \"/mnt/wd500GB/CSC500/csc500-main/datasets/wisig.node3-19.stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\", \"take_200\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"W_A_\",\n },\n {\n \"labels\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"domains\": [32, 38, 8, 44, 14, 50, 20, 26],\n \"num_examples_per_domain_per_label\": 2000,\n \"pickle_path\": \"/mnt/wd500GB/CSC500/csc500-main/datasets/oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_mag\", \"take_200\", \"resample_20Msps_to_25Msps\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE.run1_\",\n },\n ],\n \"seed\": 500,\n \"dataset_seed\": 500,\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nif \"x_shape\" not in p:\n p.x_shape = [2,256] # Default to this if we dont supply x_shape\n\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"###########################################\n# The stratified datasets honor this\n###########################################\ntorch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"p.domains_source = []\np.domains_target = []\n\n\ntrain_original_source = []\nval_original_source = []\ntest_original_source = []\n\ntrain_original_target = []\nval_original_target = []\ntest_original_target = []",
"_____no_output_____"
],
[
"# global_x_transform_func = lambda x: normalize(x.to(torch.get_default_dtype()), \"unit_power\") # unit_power, unit_mag\n# global_x_transform_func = lambda x: normalize(x, \"unit_power\") # unit_power, unit_mag",
"_____no_output_____"
],
[
"def add_dataset(\n labels,\n domains,\n pickle_path,\n x_transforms,\n episode_transforms,\n domain_prefix,\n num_examples_per_domain_per_label,\n source_or_target_dataset:str,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n):\n \n if x_transforms == []: x_transform = None\n else: x_transform = get_chained_transform(x_transforms)\n \n if episode_transforms == []: episode_transform = None\n else: raise Exception(\"episode_transforms not implemented\")\n \n episode_transform = lambda tup, _prefix=domain_prefix: (_prefix + str(tup[0]), tup[1])\n\n\n eaf = Episodic_Accessor_Factory(\n labels=labels,\n domains=domains,\n num_examples_per_domain_per_label=num_examples_per_domain_per_label,\n iterator_seed=iterator_seed,\n dataset_seed=dataset_seed,\n n_shot=n_shot,\n n_way=n_way,\n n_query=n_query,\n train_val_test_k_factors=train_val_test_k_factors,\n pickle_path=pickle_path,\n x_transform_func=x_transform,\n )\n\n train, val, test = eaf.get_train(), eaf.get_val(), eaf.get_test()\n train = Lazy_Iterable_Wrapper(train, episode_transform)\n val = Lazy_Iterable_Wrapper(val, episode_transform)\n test = Lazy_Iterable_Wrapper(test, episode_transform)\n\n if source_or_target_dataset==\"source\":\n train_original_source.append(train)\n val_original_source.append(val)\n test_original_source.append(test)\n\n p.domains_source.extend(\n [domain_prefix + str(u) for u in domains]\n )\n elif source_or_target_dataset==\"target\":\n train_original_target.append(train)\n val_original_target.append(val)\n test_original_target.append(test)\n p.domains_target.extend(\n [domain_prefix + str(u) for u in domains]\n )\n else:\n raise Exception(f\"invalid source_or_target_dataset: {source_or_target_dataset}\")\n ",
"_____no_output_____"
],
[
"for ds in p.datasets:\n add_dataset(**ds)",
"_____no_output_____"
],
[
"# from steves_utils.CORES.utils import (\n# ALL_NODES,\n# ALL_NODES_MINIMUM_1000_EXAMPLES,\n# ALL_DAYS\n# )\n\n# add_dataset(\n# labels=ALL_NODES,\n# domains = ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"cores_{u}\"\n# )",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle1_{u}\"\n# )\n",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62,56}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle2_{u}\"\n# )",
"_____no_output_____"
],
[
"# add_dataset(\n# labels=list(range(19)),\n# domains = [0,1,2],\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"metehan.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"met_{u}\"\n# )",
"_____no_output_____"
],
[
"# # from steves_utils.wisig.utils import (\n# # ALL_NODES_MINIMUM_100_EXAMPLES,\n# # ALL_NODES_MINIMUM_500_EXAMPLES,\n# # ALL_NODES_MINIMUM_1000_EXAMPLES,\n# # ALL_DAYS\n# # )\n\n# import steves_utils.wisig.utils as wisig\n\n\n# add_dataset(\n# labels=wisig.ALL_NODES_MINIMUM_100_EXAMPLES,\n# domains = wisig.ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"wisig.node3-19.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"wisig_{u}\"\n# )",
"_____no_output_____"
],
[
"###################################\n# Build the dataset\n###################################\ntrain_original_source = Iterable_Aggregator(train_original_source, p.seed)\nval_original_source = Iterable_Aggregator(val_original_source, p.seed)\ntest_original_source = Iterable_Aggregator(test_original_source, p.seed)\n\n\ntrain_original_target = Iterable_Aggregator(train_original_target, p.seed)\nval_original_target = Iterable_Aggregator(val_original_target, p.seed)\ntest_original_target = Iterable_Aggregator(test_original_target, p.seed)\n\n# For CNN We only use X and Y. And we only train on the source.\n# Properly form the data using a transform lambda and Lazy_Iterable_Wrapper. Finally wrap them in a dataloader\n\ntransform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only\n\ntrain_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)\nval_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)\ntest_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)\n\ntrain_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)\nval_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)\ntest_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"from steves_utils.transforms import get_average_magnitude, get_average_power\n\nprint(set([u for u,_ in val_original_source]))\nprint(set([u for u,_ in val_original_target]))\n\ns_x, s_y, q_x, q_y, _ = next(iter(train_processed_source))\nprint(s_x)\n\n# for ds in [\n# train_processed_source,\n# val_processed_source,\n# test_processed_source,\n# train_processed_target,\n# val_processed_target,\n# test_processed_target\n# ]:\n# for s_x, s_y, q_x, q_y, _ in ds:\n# for X in (s_x, q_x):\n# for x in X:\n# assert np.isclose(get_average_magnitude(x.numpy()), 1.0)\n# assert np.isclose(get_average_power(x.numpy()), 1.0)\n ",
"{'C_A_2', 'C_A_3', 'C_A_4', 'W_A_2', 'W_A_1', 'C_A_5', 'W_A_3', 'C_A_1', 'W_A_4'}\n"
],
[
"###################################\n# Build the model\n###################################\n# easfsl only wants a tuple for the shape\nmodel = Steves_Prototypical_Network(x_net, device=p.device, x_shape=tuple(p.x_shape))\noptimizer = Adam(params=model.parameters(), lr=p.lr)",
"(2, 200)\n"
],
[
"###################################\n# train\n###################################\njig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n patience=p.patience,\n optimizer=optimizer,\n criteria_for_best=p.criteria_for_best,\n)",
"epoch: 1, [batch: 1 / 2077], examples_per_second: 126.0577, train_label_loss: 2.7165, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"###################################\n# Evaluate the model\n###################################\nsource_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))\n\nconfusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": dict(p),\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"ptn\"),\n}",
"_____no_output_____"
],
[
"ax = get_loss_curve(experiment)\nplt.show()",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.9674524221453287 Target Test Label Accuracy: 0.19124348958333334\nSource Val Label Accuracy: 0.9636678200692042 Target Val Label Accuracy: 0.18955078125\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae3d4c7b9b2aaef7b38de1fae72f91a66f7e2c3
| 2,387 |
ipynb
|
Jupyter Notebook
|
Coursera/Using Python to Access Web Data/Week-4/Excercise/Urllib-Testing.ipynb
|
manipiradi/Online-Courses-Learning
|
2a4ce7590d1f6d1dfa5cfde632660b562fcff596
|
[
"MIT"
] | 331 |
2019-10-22T09:06:28.000Z
|
2022-03-27T13:36:03.000Z
|
Coursera/Using Python to Access Web Data/Week-4/Excercise/Urllib-Testing.ipynb
|
manipiradi/Online-Courses-Learning
|
2a4ce7590d1f6d1dfa5cfde632660b562fcff596
|
[
"MIT"
] | 8 |
2020-04-10T07:59:06.000Z
|
2022-02-06T11:36:47.000Z
|
Coursera/Using Python to Access Web Data/Week-4/Excercise/Urllib-Testing.ipynb
|
manipiradi/Online-Courses-Learning
|
2a4ce7590d1f6d1dfa5cfde632660b562fcff596
|
[
"MIT"
] | 572 |
2019-07-28T23:43:35.000Z
|
2022-03-27T22:40:08.000Z
| 22.308411 | 307 | 0.495182 |
[
[
[
"import urllib.request, urllib.parse, urllib.error #import url library",
"_____no_output_____"
],
[
"link = urllib.request.urlopen('http://data.pr4e.org/romeo.txt')",
"_____no_output_____"
],
[
"for line in link:\n print(line.decode().strip())",
"But soft what light through yonder window breaks\nIt is the east and Juliet is the sun\nArise fair sun and kill the envious moon\nWho is already sick and pale with grief\n"
],
[
"print(type(link))",
"<class 'http.client.HTTPResponse'>\n"
],
[
"link = urllib.request.urlopen('http://data.pr4e.org/romeo.txt')\n#Word count\nword = dict()\nfor line in link:\n temp = line.decode().split()\n for wd in temp:\n word[wd] = word.get(wd, 0) + 1\n \nprint(word)",
"{'But': 1, 'soft': 1, 'what': 1, 'light': 1, 'through': 1, 'yonder': 1, 'window': 1, 'breaks': 1, 'It': 1, 'is': 3, 'the': 3, 'east': 1, 'and': 3, 'Juliet': 1, 'sun': 2, 'Arise': 1, 'fair': 1, 'kill': 1, 'envious': 1, 'moon': 1, 'Who': 1, 'already': 1, 'sick': 1, 'pale': 1, 'with': 1, 'grief': 1}\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4ae3dbca2e9743df16878b7250f7a4ad58eaa0c7
| 145,724 |
ipynb
|
Jupyter Notebook
|
intro-to-rnns/Anna_KaRNNa.ipynb
|
jellenberger/deep-learning
|
e746bd3fca3503149ca4b5f7fcec316b1c764fba
|
[
"MIT"
] | null | null | null |
intro-to-rnns/Anna_KaRNNa.ipynb
|
jellenberger/deep-learning
|
e746bd3fca3503149ca4b5f7fcec316b1c764fba
|
[
"MIT"
] | null | null | null |
intro-to-rnns/Anna_KaRNNa.ipynb
|
jellenberger/deep-learning
|
e746bd3fca3503149ca4b5f7fcec316b1c764fba
|
[
"MIT"
] | null | null | null | 102.33427 | 5,899 | 0.669389 |
[
[
[
"# Anna KaRNNa\n\nIn this notebook, we'll build a character-wise RNN trained on Anna Karenina, one of my all-time favorite books. It'll be able to generate new text based on the text from the book.\n\nThis network is based off of Andrej Karpathy's [post on RNNs](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) and [implementation in Torch](https://github.com/karpathy/char-rnn). Also, some information [here at r2rt](http://r2rt.com/recurrent-neural-networks-in-tensorflow-ii.html) and from [Sherjil Ozair](https://github.com/sherjilozair/char-rnn-tensorflow) on GitHub. Below is the general architecture of the character-wise RNN.\n\n<img src=\"assets/charseq.jpeg\" width=\"500\">",
"_____no_output_____"
]
],
[
[
"import time\nfrom collections import namedtuple\n\nimport numpy as np\nimport tensorflow as tf",
"_____no_output_____"
]
],
[
[
"First we'll load the text file and convert it into integers for our network to use. Here I'm creating a couple dictionaries to convert the characters to and from integers. Encoding the characters as integers makes it easier to use as input in the network.",
"_____no_output_____"
]
],
[
[
"with open('anna.txt', 'r') as f:\n text=f.read()\nvocab = sorted(set(text))\nvocab_to_int = {c: i for i, c in enumerate(vocab)}\nint_to_vocab = dict(enumerate(vocab))\nencoded = np.array([vocab_to_int[c] for c in text], dtype=np.int32)",
"_____no_output_____"
]
],
[
[
"Let's check out the first 100 characters, make sure everything is peachy. According to the [American Book Review](http://americanbookreview.org/100bestlines.asp), this is the 6th best first line of a book ever.",
"_____no_output_____"
]
],
[
[
"text[:100]",
"_____no_output_____"
]
],
[
[
"And we can see the characters encoded as integers.",
"_____no_output_____"
]
],
[
[
"encoded[:100]",
"_____no_output_____"
]
],
[
[
"Since the network is working with individual characters, it's similar to a classification problem in which we are trying to predict the next character from the previous text. Here's how many 'classes' our network has to pick from.",
"_____no_output_____"
]
],
[
[
"len(vocab)",
"_____no_output_____"
]
],
[
[
"## Making training mini-batches\n\nHere is where we'll make our mini-batches for training. Remember that we want our batches to be multiple sequences of some desired number of sequence steps. Considering a simple example, our batches would look like this:\n\n<img src=\"assets/[email protected]\" width=500px>\n\n\n<br>\n\nWe start with our text encoded as integers in one long array in `encoded`. Let's create a function that will give us an iterator for our batches. I like using [generator functions](https://jeffknupp.com/blog/2013/04/07/improve-your-python-yield-and-generators-explained/) to do this. Then we can pass `encoded` into this function and get our batch generator.\n\nThe first thing we need to do is discard some of the text so we only have completely full batches. Each batch contains $N \\times M$ characters, where $N$ is the batch size (the number of sequences) and $M$ is the number of steps. Then, to get the total number of batches, $K$, we can make from the array `arr`, you divide the length of `arr` by the number of characters per batch. Once you know the number of batches, you can get the total number of characters to keep from `arr`, $N * M * K$.\n\nAfter that, we need to split `arr` into $N$ sequences. You can do this using `arr.reshape(size)` where `size` is a tuple containing the dimensions sizes of the reshaped array. We know we want $N$ sequences (`batch_size` below), let's make that the size of the first dimension. For the second dimension, you can use `-1` as a placeholder in the size, it'll fill up the array with the appropriate data for you. After this, you should have an array that is $N \\times (M * K)$.\n\nNow that we have this array, we can iterate through it to get our batches. The idea is each batch is a $N \\times M$ window on the $N \\times (M * K)$ array. For each subsequent batch, the window moves over by `n_steps`. We also want to create both the input and target arrays. Remember that the targets are the inputs shifted over one character. \n\nThe way I like to do this window is use `range` to take steps of size `n_steps` from $0$ to `arr.shape[1]`, the total number of steps in each sequence. That way, the integers you get from `range` always point to the start of a batch, and each window is `n_steps` wide.\n\n> **Exercise:** Write the code for creating batches in the function below. The exercises in this notebook _will not be easy_. I've provided a notebook with solutions alongside this notebook. If you get stuck, checkout the solutions. The most important thing is that you don't copy and paste the code into here, **type out the solution code yourself.**",
"_____no_output_____"
]
],
[
[
"def get_batches(arr, batch_size, n_steps):\n '''Create a generator that returns batches of size\n batch_size x n_steps from arr.\n \n Arguments\n ---------\n arr: Array you want to make batches from\n batch_size: Batch size, the number of sequences per batch\n n_steps: Number of sequence steps per batch\n '''\n # Get the number of characters per batch and number of batches we can make\n chars_per_batch = batch_size * n_steps\n n_batches = len(arr) // chars_per_batch\n \n # Keep only enough characters to make full batches\n arr = arr[:n_batches * chars_per_batch]\n \n # Reshape into batch_size rows\n arr = arr.reshape((batch_size, -1))\n \n for n in range(0, arr.shape[1], n_steps):\n # The features - grab entire column at n : n + 1\n x = arr[:, n:(n + n_steps)]\n # The targets - same as above, shifted by + 1\n # but for end of last column, sub first chars in x\n y = np.zeros_like(x)\n y[:, :-1], y[:, -1] = x[:, 1:], x[:, 0] \n \n yield x, y",
"_____no_output_____"
]
],
[
[
"Now I'll make my data sets and we can check out what's going on here. Here I'm going to use a batch size of 10 and 50 sequence steps.",
"_____no_output_____"
]
],
[
[
"batches = get_batches(encoded, 10, 50)\nx, y = next(batches)",
"_____no_output_____"
],
[
"print('x\\n', x[:10, :10])\nprint('\\ny\\n', y[:10, :10])",
"x\n [[31 64 57 72 76 61 74 1 16 0]\n [ 1 57 69 1 70 71 76 1 63 71]\n [78 65 70 13 0 0 3 53 61 75]\n [70 1 60 77 74 65 70 63 1 64]\n [ 1 65 76 1 65 75 11 1 75 65]\n [ 1 37 76 1 79 57 75 0 71 70]\n [64 61 70 1 59 71 69 61 1 62]\n [26 1 58 77 76 1 70 71 79 1]\n [76 1 65 75 70 7 76 13 1 48]\n [ 1 75 57 65 60 1 76 71 1 64]]\n\ny\n [[64 57 72 76 61 74 1 16 0 0]\n [57 69 1 70 71 76 1 63 71 65]\n [65 70 13 0 0 3 53 61 75 11]\n [ 1 60 77 74 65 70 63 1 64 65]\n [65 76 1 65 75 11 1 75 65 74]\n [37 76 1 79 57 75 0 71 70 68]\n [61 70 1 59 71 69 61 1 62 71]\n [ 1 58 77 76 1 70 71 79 1 75]\n [ 1 65 75 70 7 76 13 1 48 64]\n [75 57 65 60 1 76 71 1 64 61]]\n"
]
],
[
[
"If you implemented `get_batches` correctly, the above output should look something like \n```\nx\n [[55 63 69 22 6 76 45 5 16 35]\n [ 5 69 1 5 12 52 6 5 56 52]\n [48 29 12 61 35 35 8 64 76 78]\n [12 5 24 39 45 29 12 56 5 63]\n [ 5 29 6 5 29 78 28 5 78 29]\n [ 5 13 6 5 36 69 78 35 52 12]\n [63 76 12 5 18 52 1 76 5 58]\n [34 5 73 39 6 5 12 52 36 5]\n [ 6 5 29 78 12 79 6 61 5 59]\n [ 5 78 69 29 24 5 6 52 5 63]]\n\ny\n [[63 69 22 6 76 45 5 16 35 35]\n [69 1 5 12 52 6 5 56 52 29]\n [29 12 61 35 35 8 64 76 78 28]\n [ 5 24 39 45 29 12 56 5 63 29]\n [29 6 5 29 78 28 5 78 29 45]\n [13 6 5 36 69 78 35 52 12 43]\n [76 12 5 18 52 1 76 5 58 52]\n [ 5 73 39 6 5 12 52 36 5 78]\n [ 5 29 78 12 79 6 61 5 59 63]\n [78 69 29 24 5 6 52 5 63 76]]\n ```\n although the exact numbers will be different. Check to make sure the data is shifted over one step for `y`.",
"_____no_output_____"
],
[
"## Building the model\n\nBelow is where you'll build the network. We'll break it up into parts so it's easier to reason about each bit. Then we can connect them up into the whole network.\n\n<img src=\"assets/charRNN.png\" width=500px>\n\n\n### Inputs\n\nFirst off we'll create our input placeholders. As usual we need placeholders for the training data and the targets. We'll also create a placeholder for dropout layers called `keep_prob`. This will be a scalar, that is a 0-D tensor. To make a scalar, you create a placeholder without giving it a size.\n\n> **Exercise:** Create the input placeholders in the function below.",
"_____no_output_____"
]
],
[
[
"def build_inputs(batch_size, num_steps):\n ''' Define placeholders for inputs, targets, and dropout \n \n Arguments\n ---------\n batch_size: Batch size, number of sequences per batch\n num_steps: Number of sequence steps in a batch\n \n '''\n # Declare placeholders we'll feed into the graph\n inputs = tf.placeholder(tf.int32, shape=[batch_size, num_steps], name='inputs')\n targets = tf.placeholder(tf.int32, shape=[batch_size, num_steps], name='targets')\n \n # Keep probability placeholder for drop out layers\n keep_prob = tf.placeholder(tf.float32, name='keep_prob')\n \n return inputs, targets, keep_prob",
"_____no_output_____"
]
],
[
[
"### LSTM Cell\n\nHere we will create the LSTM cell we'll use in the hidden layer. We'll use this cell as a building block for the RNN. So we aren't actually defining the RNN here, just the type of cell we'll use in the hidden layer.\n\nWe first create a basic LSTM cell with\n\n```python\nlstm = tf.contrib.rnn.BasicLSTMCell(num_units)\n```\n\nwhere `num_units` is the number of units in the hidden layers in the cell. Then we can add dropout by wrapping it with \n\n```python\ntf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)\n```\nYou pass in a cell and it will automatically add dropout to the inputs or outputs. Finally, we can stack up the LSTM cells into layers with [`tf.contrib.rnn.MultiRNNCell`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/rnn/MultiRNNCell). With this, you pass in a list of cells and it will send the output of one cell into the next cell. Previously with TensorFlow 1.0, you could do this\n\n```python\ntf.contrib.rnn.MultiRNNCell([cell]*num_layers)\n```\n\nThis might look a little weird if you know Python well because this will create a list of the same `cell` object. However, TensorFlow 1.0 will create different weight matrices for all `cell` objects. But, starting with TensorFlow 1.1 you actually need to create new cell objects in the list. To get it to work in TensorFlow 1.1, it should look like\n\n```python\ndef build_cell(num_units, keep_prob):\n lstm = tf.contrib.rnn.BasicLSTMCell(num_units)\n drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)\n \n return drop\n \ntf.contrib.rnn.MultiRNNCell([build_cell(num_units, keep_prob) for _ in range(num_layers)])\n```\n\nEven though this is actually multiple LSTM cells stacked on each other, you can treat the multiple layers as one cell.\n\nWe also need to create an initial cell state of all zeros. This can be done like so\n\n```python\ninitial_state = cell.zero_state(batch_size, tf.float32)\n```\n\nBelow, we implement the `build_lstm` function to create these LSTM cells and the initial state.",
"_____no_output_____"
]
],
[
[
"def build_lstm(lstm_size, num_layers, batch_size, keep_prob):\n ''' Build LSTM cell.\n \n Arguments\n ---------\n keep_prob: Scalar tensor (tf.placeholder) for the dropout keep probability\n lstm_size: Size of the hidden layers in the LSTM cells\n num_layers: Number of LSTM layers\n batch_size: Batch size\n\n '''\n ### Build the LSTM Cell\n def build_cell():\n # Use a basic LSTM cell\n lstm = tf.contrib.rnn.BasicLSTMCell(lstm_size)\n \n # Add dropout to the cell outputs\n drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)\n \n return drop\n \n # Stack up multiple LSTM layers, for deep learning\n cell = tf.contrib.rnn.MultiRNNCell([build_cell() for _ in range(num_layers)])\n initial_state = cell.zero_state(batch_size, tf.float32)\n \n return cell, initial_state",
"_____no_output_____"
]
],
[
[
"### RNN Output\n\nHere we'll create the output layer. We need to connect the output of the RNN cells to a full connected layer with a softmax output. The softmax output gives us a probability distribution we can use to predict the next character, so we want this layer to have size $C$, the number of classes/characters we have in our text.\n\nIf our input has batch size $N$, number of steps $M$, and the hidden layer has $L$ hidden units, then the output is a 3D tensor with size $N \\times M \\times L$. The output of each LSTM cell has size $L$, we have $M$ of them, one for each sequence step, and we have $N$ sequences. So the total size is $N \\times M \\times L$. \n\nWe are using the same fully connected layer, the same weights, for each of the outputs. Then, to make things easier, we should reshape the outputs into a 2D tensor with shape $(M * N) \\times L$. That is, one row for each sequence and step, where the values of each row are the output from the LSTM cells. We get the LSTM output as a list, `lstm_output`. First we need to concatenate this whole list into one array with [`tf.concat`](https://www.tensorflow.org/api_docs/python/tf/concat). Then, reshape it (with `tf.reshape`) to size $(M * N) \\times L$.\n\nOne we have the outputs reshaped, we can do the matrix multiplication with the weights. We need to wrap the weight and bias variables in a variable scope with `tf.variable_scope(scope_name)` because there are weights being created in the LSTM cells. TensorFlow will throw an error if the weights created here have the same names as the weights created in the LSTM cells, which they will be default. To avoid this, we wrap the variables in a variable scope so we can give them unique names.\n\n> **Exercise:** Implement the output layer in the function below.",
"_____no_output_____"
]
],
[
[
"def build_output(lstm_output, in_size, out_size):\n ''' Build a softmax layer, return the softmax output and logits.\n \n Arguments\n ---------\n \n lstm_output: List of output tensors from the LSTM layer\n in_size: Size of the input tensor, for example, size of the LSTM cells\n out_size: Size of this softmax layer\n \n '''\n\n # Reshape output so it's a bunch of rows, one row for each step for each sequence.\n # Concatenate lstm_output over axis 1 (the columns)\n seq_output = tf.concat(lstm_output, axis=1)\n # Reshape seq_output to a 2D tensor with lstm_size columns\n x = tf.reshape(seq_output, [-1, in_size])\n \n # Connect the RNN outputs to a softmax layer\n with tf.variable_scope('softmax'):\n # Create the weight and bias variables here\n softmax_w = tf.Variable(tf.truncated_normal((in_size, out_size), stddev=0.1))\n softmax_b = tf.Variable(tf.zeros(out_size))\n \n # Since output is a bunch of rows of RNN cell outputs, logits will be a bunch\n # of rows of logit outputs, one for each step and sequence\n logits = tf.matmul(x, softmax_w) + softmax_b\n \n # Use softmax to get the probabilities for predicted characters\n out = tf.nn.softmax(logits, name='predictions')\n \n return out, logits",
"_____no_output_____"
]
],
[
[
"### Training loss\n\nNext up is the training loss. We get the logits and targets and calculate the softmax cross-entropy loss. First we need to one-hot encode the targets, we're getting them as encoded characters. Then, reshape the one-hot targets so it's a 2D tensor with size $(M*N) \\times C$ where $C$ is the number of classes/characters we have. Remember that we reshaped the LSTM outputs and ran them through a fully connected layer with $C$ units. So our logits will also have size $(M*N) \\times C$.\n\nThen we run the logits and targets through `tf.nn.softmax_cross_entropy_with_logits` and find the mean to get the loss.\n\n>**Exercise:** Implement the loss calculation in the function below.",
"_____no_output_____"
]
],
[
[
"def build_loss(logits, targets, lstm_size, num_classes):\n ''' Calculate the loss from the logits and the targets.\n \n Arguments\n ---------\n logits: Logits from final fully connected layer\n targets: Targets for supervised learning\n lstm_size: Number of LSTM hidden units\n num_classes: Number of classes in targets\n \n '''\n \n # One-hot encode targets and reshape to match logits, one row per batch_size per step\n y_one_hot = tf.one_hot(targets, num_classes)\n y_reshaped = tf.reshape(y_one_hot, logits.get_shape())\n \n # Softmax cross entropy loss\n loss = tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=y_reshaped)\n loss = tf.reduce_mean(loss)\n return loss",
"_____no_output_____"
]
],
[
[
"### Optimizer\n\nHere we build the optimizer. Normal RNNs have have issues gradients exploding and disappearing. LSTMs fix the disappearance problem, but the gradients can still grow without bound. To fix this, we can clip the gradients above some threshold. That is, if a gradient is larger than that threshold, we set it to the threshold. This will ensure the gradients never grow overly large. Then we use an AdamOptimizer for the learning step.",
"_____no_output_____"
]
],
[
[
"def build_optimizer(loss, learning_rate, grad_clip):\n ''' Build optmizer for training, using gradient clipping.\n \n Arguments:\n loss: Network loss\n learning_rate: Learning rate for optimizer\n \n '''\n \n # Optimizer for training, using gradient clipping to control exploding gradients\n tvars = tf.trainable_variables()\n grads, _ = tf.clip_by_global_norm(tf.gradients(loss, tvars), grad_clip)\n train_op = tf.train.AdamOptimizer(learning_rate)\n optimizer = train_op.apply_gradients(zip(grads, tvars))\n \n return optimizer",
"_____no_output_____"
]
],
[
[
"### Build the network\n\nNow we can put all the pieces together and build a class for the network. To actually run data through the LSTM cells, we will use [`tf.nn.dynamic_rnn`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/nn/dynamic_rnn). This function will pass the hidden and cell states across LSTM cells appropriately for us. It returns the outputs for each LSTM cell at each step for each sequence in the mini-batch. It also gives us the final LSTM state. We want to save this state as `final_state` so we can pass it to the first LSTM cell in the the next mini-batch run. For `tf.nn.dynamic_rnn`, we pass in the cell and initial state we get from `build_lstm`, as well as our input sequences. Also, we need to one-hot encode the inputs before going into the RNN. \n\n> **Exercise:** Use the functions you've implemented previously and `tf.nn.dynamic_rnn` to build the network.",
"_____no_output_____"
]
],
[
[
"class CharRNN:\n \n def __init__(self, num_classes, batch_size=64, num_steps=50, \n lstm_size=128, num_layers=2, learning_rate=0.001, \n grad_clip=5, sampling=False):\n \n # When we're using this network for sampling later, we'll be passing in\n # one character at a time, so providing an option for that\n if sampling == True:\n batch_size, num_steps = 1, 1\n else:\n batch_size, num_steps = batch_size, num_steps\n\n tf.reset_default_graph()\n \n # Build the input placeholder tensors\n self.inputs, self.targets, self.keep_prob = build_inputs(batch_size, num_steps)\n\n # Build the LSTM cell\n cell, self.initial_state = build_lstm(lstm_size, num_layers, batch_size, self.keep_prob)\n\n ### Run the data through the RNN layers\n # First, one-hot encode the input tokens\n x_one_hot = tf.one_hot(self.inputs, num_classes)\n \n # Run each sequence step through the RNN and collect the outputs\n outputs, state = tf.nn.dynamic_rnn(cell, x_one_hot, initial_state=self.initial_state)\n self.final_state = state\n \n # Get softmax predictions and logits\n self.prediction, self.logits = build_output(outputs, lstm_size, num_classes)\n \n # Loss and optimizer (with gradient clipping)\n self.loss = build_loss(self.logits, self.targets, lstm_size, num_classes)\n self.optimizer = build_optimizer(self.loss, learning_rate, grad_clip)",
"_____no_output_____"
]
],
[
[
"## Hyperparameters\n\nHere are the hyperparameters for the network.\n\n* `batch_size` - Number of sequences running through the network in one pass.\n* `num_steps` - Number of characters in the sequence the network is trained on. Larger is better typically, the network will learn more long range dependencies. But it takes longer to train. 100 is typically a good number here.\n* `lstm_size` - The number of units in the hidden layers.\n* `num_layers` - Number of hidden LSTM layers to use\n* `learning_rate` - Learning rate for training\n* `keep_prob` - The dropout keep probability when training. If you're network is overfitting, try decreasing this.\n\nHere's some good advice from Andrej Karpathy on training the network. I'm going to copy it in here for your benefit, but also link to [where it originally came from](https://github.com/karpathy/char-rnn#tips-and-tricks).\n\n> ## Tips and Tricks\n\n>### Monitoring Validation Loss vs. Training Loss\n>If you're somewhat new to Machine Learning or Neural Networks it can take a bit of expertise to get good models. The most important quantity to keep track of is the difference between your training loss (printed during training) and the validation loss (printed once in a while when the RNN is run on the validation data (by default every 1000 iterations)). In particular:\n\n> - If your training loss is much lower than validation loss then this means the network might be **overfitting**. Solutions to this are to decrease your network size, or to increase dropout. For example you could try dropout of 0.5 and so on.\n> - If your training/validation loss are about equal then your model is **underfitting**. Increase the size of your model (either number of layers or the raw number of neurons per layer)\n\n> ### Approximate number of parameters\n\n> The two most important parameters that control the model are `lstm_size` and `num_layers`. I would advise that you always use `num_layers` of either 2/3. The `lstm_size` can be adjusted based on how much data you have. The two important quantities to keep track of here are:\n\n> - The number of parameters in your model. This is printed when you start training.\n> - The size of your dataset. 1MB file is approximately 1 million characters.\n\n>These two should be about the same order of magnitude. It's a little tricky to tell. Here are some examples:\n\n> - I have a 100MB dataset and I'm using the default parameter settings (which currently print 150K parameters). My data size is significantly larger (100 mil >> 0.15 mil), so I expect to heavily underfit. I am thinking I can comfortably afford to make `lstm_size` larger.\n> - I have a 10MB dataset and running a 10 million parameter model. I'm slightly nervous and I'm carefully monitoring my validation loss. If it's larger than my training loss then I may want to try to increase dropout a bit and see if that helps the validation loss.\n\n> ### Best models strategy\n\n>The winning strategy to obtaining very good models (if you have the compute time) is to always err on making the network larger (as large as you're willing to wait for it to compute) and then try different dropout values (between 0,1). Whatever model has the best validation performance (the loss, written in the checkpoint filename, low is good) is the one you should use in the end.\n\n>It is very common in deep learning to run many different models with many different hyperparameter settings, and in the end take whatever checkpoint gave the best validation performance.\n\n>By the way, the size of your training and validation splits are also parameters. Make sure you have a decent amount of data in your validation set or otherwise the validation performance will be noisy and not very informative.",
"_____no_output_____"
]
],
[
[
"batch_size = 10 # Sequences per batch\nnum_steps = 50 # Number of sequence steps per batch\nlstm_size = 512 # Size of hidden layers in LSTMs\nnum_layers = 2 # Number of LSTM layers\nlearning_rate = 0.01 # Learning rate\nkeep_prob = 0.5 # Dropout keep probability",
"_____no_output_____"
]
],
[
[
"## Time for training\n\nThis is typical training code, passing inputs and targets into the network, then running the optimizer. Here we also get back the final LSTM state for the mini-batch. Then, we pass that state back into the network so the next batch can continue the state from the previous batch. And every so often (set by `save_every_n`) I save a checkpoint.\n\nHere I'm saving checkpoints with the format\n\n`i{iteration number}_l{# hidden layer units}.ckpt`\n\n> **Exercise:** Set the hyperparameters above to train the network. Watch the training loss, it should be consistently dropping. Also, I highly advise running this on a GPU.",
"_____no_output_____"
]
],
[
[
"epochs = 20\n# Print losses every N interations\nprint_every_n = 50\n\n# Save every N iterations\nsave_every_n = 200\n\nmodel = CharRNN(len(vocab), batch_size=batch_size, num_steps=num_steps,\n lstm_size=lstm_size, num_layers=num_layers, \n learning_rate=learning_rate)\n\nsaver = tf.train.Saver(max_to_keep=100)\nwith tf.Session() as sess:\n sess.run(tf.global_variables_initializer())\n \n # Use the line below to load a checkpoint and resume training\n #saver.restore(sess, 'checkpoints/______.ckpt')\n counter = 0\n for e in range(epochs):\n # Train network\n new_state = sess.run(model.initial_state)\n loss = 0\n for x, y in get_batches(encoded, batch_size, num_steps):\n counter += 1\n start = time.time()\n feed = {model.inputs: x,\n model.targets: y,\n model.keep_prob: keep_prob,\n model.initial_state: new_state}\n batch_loss, new_state, _ = sess.run([model.loss, \n model.final_state, \n model.optimizer], \n feed_dict=feed)\n if (counter % print_every_n == 0):\n end = time.time()\n print('Epoch: {}/{}... '.format(e+1, epochs),\n 'Training Step: {}... '.format(counter),\n 'Training loss: {:.4f}... '.format(batch_loss),\n '{:.4f} sec/batch'.format((end-start)))\n \n if (counter % save_every_n == 0):\n saver.save(sess, \"checkpoints/i{}_l{}.ckpt\".format(counter, lstm_size))\n \n saver.save(sess, \"checkpoints/i{}_l{}.ckpt\".format(counter, lstm_size))",
"Epoch: 1/20... Training Step: 50... Training loss: 3.2373... 0.4576 sec/batch\nEpoch: 1/20... Training Step: 100... Training loss: 3.1583... 0.4646 sec/batch\nEpoch: 1/20... Training Step: 150... Training loss: 3.1531... 0.4599 sec/batch\nEpoch: 1/20... Training Step: 200... Training loss: 3.2320... 0.4538 sec/batch\nEpoch: 1/20... Training Step: 250... Training loss: 3.1975... 0.4711 sec/batch\nEpoch: 1/20... Training Step: 300... Training loss: 3.1303... 0.4576 sec/batch\nEpoch: 1/20... Training Step: 350... Training loss: 3.1254... 0.4642 sec/batch\nEpoch: 1/20... Training Step: 400... Training loss: 3.1819... 0.4629 sec/batch\nEpoch: 1/20... Training Step: 450... Training loss: 3.0995... 0.4530 sec/batch\nEpoch: 1/20... Training Step: 500... Training loss: 3.2739... 0.4785 sec/batch\nEpoch: 1/20... Training Step: 550... Training loss: 3.0376... 0.4589 sec/batch\nEpoch: 1/20... Training Step: 600... Training loss: 3.0934... 0.4574 sec/batch\nEpoch: 1/20... Training Step: 650... Training loss: 3.1416... 0.4801 sec/batch\nEpoch: 1/20... Training Step: 700... Training loss: 3.1047... 0.4669 sec/batch\nEpoch: 1/20... Training Step: 750... Training loss: 3.0679... 0.4570 sec/batch\nEpoch: 1/20... Training Step: 800... Training loss: 3.0859... 0.4513 sec/batch\nEpoch: 1/20... Training Step: 850... Training loss: 3.0900... 0.4465 sec/batch\nEpoch: 1/20... Training Step: 900... Training loss: 3.0075... 0.4431 sec/batch\nEpoch: 1/20... Training Step: 950... Training loss: 3.1291... 0.4542 sec/batch\nEpoch: 1/20... Training Step: 1000... Training loss: 3.0775... 0.4471 sec/batch\nEpoch: 1/20... Training Step: 1050... Training loss: 3.0964... 0.4647 sec/batch\nEpoch: 1/20... Training Step: 1100... Training loss: 3.0604... 0.4463 sec/batch\nEpoch: 1/20... Training Step: 1150... Training loss: 3.0624... 0.4609 sec/batch\nEpoch: 1/20... Training Step: 1200... Training loss: 3.0551... 0.4500 sec/batch\nEpoch: 1/20... Training Step: 1250... Training loss: 3.1015... 0.4621 sec/batch\nEpoch: 1/20... Training Step: 1300... Training loss: 3.1135... 0.4529 sec/batch\nEpoch: 1/20... Training Step: 1350... Training loss: 3.0746... 0.4653 sec/batch\nEpoch: 1/20... Training Step: 1400... Training loss: 3.1666... 0.4532 sec/batch\nEpoch: 1/20... Training Step: 1450... Training loss: 3.2059... 0.4427 sec/batch\nEpoch: 1/20... Training Step: 1500... Training loss: 3.1290... 0.4486 sec/batch\nEpoch: 1/20... Training Step: 1550... Training loss: 3.0154... 0.4534 sec/batch\nEpoch: 1/20... Training Step: 1600... Training loss: 3.0927... 0.4477 sec/batch\nEpoch: 1/20... Training Step: 1650... Training loss: 3.1212... 0.4510 sec/batch\nEpoch: 1/20... Training Step: 1700... Training loss: 3.2449... 0.4592 sec/batch\nEpoch: 1/20... Training Step: 1750... Training loss: 3.1195... 0.4546 sec/batch\nEpoch: 1/20... Training Step: 1800... Training loss: 3.1626... 0.4469 sec/batch\nEpoch: 1/20... Training Step: 1850... Training loss: 3.1343... 0.4518 sec/batch\nEpoch: 1/20... Training Step: 1900... Training loss: 3.0862... 0.4633 sec/batch\nEpoch: 1/20... Training Step: 1950... Training loss: 3.1533... 0.4540 sec/batch\nEpoch: 1/20... Training Step: 2000... Training loss: 3.0689... 0.4482 sec/batch\nEpoch: 1/20... Training Step: 2050... Training loss: 3.1257... 0.4433 sec/batch\nEpoch: 1/20... Training Step: 2100... Training loss: 3.0042... 0.4572 sec/batch\nEpoch: 1/20... Training Step: 2150... Training loss: 3.2172... 0.4541 sec/batch\nEpoch: 1/20... Training Step: 2200... Training loss: 3.0201... 0.4512 sec/batch\nEpoch: 1/20... Training Step: 2250... Training loss: 3.1002... 0.4493 sec/batch\nEpoch: 1/20... Training Step: 2300... Training loss: 3.1027... 0.4529 sec/batch\nEpoch: 1/20... Training Step: 2350... Training loss: 3.1075... 0.4491 sec/batch\nEpoch: 1/20... Training Step: 2400... Training loss: 3.1034... 0.4511 sec/batch\nEpoch: 1/20... Training Step: 2450... Training loss: 3.1817... 0.4469 sec/batch\nEpoch: 1/20... Training Step: 2500... Training loss: 3.1841... 0.4571 sec/batch\nEpoch: 1/20... Training Step: 2550... Training loss: 3.0981... 0.4471 sec/batch\nEpoch: 1/20... Training Step: 2600... Training loss: 3.1383... 0.4629 sec/batch\nEpoch: 1/20... Training Step: 2650... Training loss: 3.1560... 0.4631 sec/batch\nEpoch: 1/20... Training Step: 2700... Training loss: 3.1014... 0.4525 sec/batch\nEpoch: 1/20... Training Step: 2750... Training loss: 3.1540... 0.4486 sec/batch\nEpoch: 1/20... Training Step: 2800... Training loss: 3.2223... 0.4577 sec/batch\nEpoch: 1/20... Training Step: 2850... Training loss: 3.1075... 0.4564 sec/batch\nEpoch: 1/20... Training Step: 2900... Training loss: 3.0366... 0.4583 sec/batch\nEpoch: 1/20... Training Step: 2950... Training loss: 3.0871... 0.4537 sec/batch\nEpoch: 1/20... Training Step: 3000... Training loss: 3.1899... 0.4442 sec/batch\nEpoch: 1/20... Training Step: 3050... Training loss: 3.1101... 0.4447 sec/batch\nEpoch: 1/20... Training Step: 3100... Training loss: 3.1488... 0.4438 sec/batch\nEpoch: 1/20... Training Step: 3150... Training loss: 3.1750... 0.4518 sec/batch\nEpoch: 1/20... Training Step: 3200... Training loss: 3.1264... 0.4434 sec/batch\nEpoch: 1/20... Training Step: 3250... Training loss: 3.0746... 0.4630 sec/batch\nEpoch: 1/20... Training Step: 3300... Training loss: 3.2233... 0.4472 sec/batch\nEpoch: 1/20... Training Step: 3350... Training loss: 3.0536... 0.4536 sec/batch\nEpoch: 1/20... Training Step: 3400... Training loss: 3.0216... 0.4479 sec/batch\nEpoch: 1/20... Training Step: 3450... Training loss: 3.1055... 0.4483 sec/batch\nEpoch: 1/20... Training Step: 3500... Training loss: 3.1200... 0.4613 sec/batch\nEpoch: 1/20... Training Step: 3550... Training loss: 3.1218... 0.4605 sec/batch\nEpoch: 1/20... Training Step: 3600... Training loss: 3.1458... 0.4497 sec/batch\nEpoch: 1/20... Training Step: 3650... Training loss: 3.1743... 0.4546 sec/batch\nEpoch: 1/20... Training Step: 3700... Training loss: 3.1992... 0.4474 sec/batch\nEpoch: 1/20... Training Step: 3750... Training loss: 3.1022... 0.4479 sec/batch\nEpoch: 1/20... Training Step: 3800... Training loss: 3.1365... 0.4566 sec/batch\nEpoch: 1/20... Training Step: 3850... Training loss: 3.0511... 0.4602 sec/batch\nEpoch: 1/20... Training Step: 3900... Training loss: 3.0910... 0.4479 sec/batch\nEpoch: 1/20... Training Step: 3950... Training loss: 3.2298... 0.4723 sec/batch\nEpoch: 2/20... Training Step: 4000... Training loss: 3.1452... 0.4485 sec/batch\nEpoch: 2/20... Training Step: 4050... Training loss: 3.0453... 0.4495 sec/batch\nEpoch: 2/20... Training Step: 4100... Training loss: 3.0328... 0.4446 sec/batch\nEpoch: 2/20... Training Step: 4150... Training loss: 3.0566... 0.4489 sec/batch\nEpoch: 2/20... Training Step: 4200... Training loss: 3.0929... 0.4589 sec/batch\nEpoch: 2/20... Training Step: 4250... Training loss: 3.0745... 0.4583 sec/batch\nEpoch: 2/20... Training Step: 4300... Training loss: 3.0934... 0.4494 sec/batch\nEpoch: 2/20... Training Step: 4350... Training loss: 3.0507... 0.4501 sec/batch\nEpoch: 2/20... Training Step: 4400... Training loss: 3.0531... 0.4485 sec/batch\nEpoch: 2/20... Training Step: 4450... Training loss: 3.1315... 0.4520 sec/batch\nEpoch: 2/20... Training Step: 4500... Training loss: 3.0793... 0.4559 sec/batch\nEpoch: 2/20... Training Step: 4550... Training loss: 3.1483... 0.4536 sec/batch\nEpoch: 2/20... Training Step: 4600... Training loss: 3.2350... 0.4543 sec/batch\nEpoch: 2/20... Training Step: 4650... Training loss: 3.1705... 0.4455 sec/batch\nEpoch: 2/20... Training Step: 4700... Training loss: 3.1777... 0.4485 sec/batch\nEpoch: 2/20... Training Step: 4750... Training loss: 3.1580... 0.4474 sec/batch\nEpoch: 2/20... Training Step: 4800... Training loss: 3.1315... 0.4568 sec/batch\nEpoch: 2/20... Training Step: 4850... Training loss: 3.0613... 0.4456 sec/batch\nEpoch: 2/20... Training Step: 4900... Training loss: 3.1038... 0.4543 sec/batch\nEpoch: 2/20... Training Step: 4950... Training loss: 3.1421... 0.4508 sec/batch\n"
]
],
[
[
"#### Saved checkpoints\n\nRead up on saving and loading checkpoints here: https://www.tensorflow.org/programmers_guide/variables",
"_____no_output_____"
]
],
[
[
"tf.train.get_checkpoint_state('checkpoints')",
"_____no_output_____"
]
],
[
[
"## Sampling\n\nNow that the network is trained, we'll can use it to generate new text. The idea is that we pass in a character, then the network will predict the next character. We can use the new one, to predict the next one. And we keep doing this to generate all new text. I also included some functionality to prime the network with some text by passing in a string and building up a state from that.\n\nThe network gives us predictions for each character. To reduce noise and make things a little less random, I'm going to only choose a new character from the top N most likely characters.\n\n",
"_____no_output_____"
]
],
[
[
"def pick_top_n(preds, vocab_size, top_n=5):\n p = np.squeeze(preds)\n p[np.argsort(p)[:-top_n]] = 0\n p = p / np.sum(p)\n c = np.random.choice(vocab_size, 1, p=p)[0]\n return c",
"_____no_output_____"
],
[
"def sample(checkpoint, n_samples, lstm_size, vocab_size, prime=\"The \"):\n samples = [c for c in prime]\n model = CharRNN(len(vocab), lstm_size=lstm_size, sampling=True)\n saver = tf.train.Saver()\n with tf.Session() as sess:\n saver.restore(sess, checkpoint)\n new_state = sess.run(model.initial_state)\n for c in prime:\n x = np.zeros((1, 1))\n x[0,0] = vocab_to_int[c]\n feed = {model.inputs: x,\n model.keep_prob: 1.,\n model.initial_state: new_state}\n preds, new_state = sess.run([model.prediction, model.final_state], \n feed_dict=feed)\n\n c = pick_top_n(preds, len(vocab))\n samples.append(int_to_vocab[c])\n\n for i in range(n_samples):\n x[0,0] = c\n feed = {model.inputs: x,\n model.keep_prob: 1.,\n model.initial_state: new_state}\n preds, new_state = sess.run([model.prediction, model.final_state], \n feed_dict=feed)\n\n c = pick_top_n(preds, len(vocab))\n samples.append(int_to_vocab[c])\n \n return ''.join(samples)",
"_____no_output_____"
]
],
[
[
"Here, pass in the path to a checkpoint and sample from the network.",
"_____no_output_____"
]
],
[
[
"tf.train.latest_checkpoint('checkpoints')",
"_____no_output_____"
],
[
"checkpoint = tf.train.latest_checkpoint('checkpoints')\nsamp = sample(checkpoint, 2000, lstm_size, len(vocab), prime=\"Far\")\nprint(samp)",
"INFO:tensorflow:Restoring parameters from checkpoints/i79400_l128.ckpt\nFarition, she asked him to his hand about with and teals on, and he saw them, how tried the\nmost call how he saw to as though she stood the\nstill that answered, was not him and with the tone and thousand to\nall her wife. \"I'm bound on\nall and master.\"\n\n\"I'm no or imparicon to home and that if you have been inservone, tean, but\nthen I won't cannot, in such a caming was and to the mustancisals, and and alone on\nthe\npeacation to see.\n\nWhat's to bely one to terrent on their mind. And\nthe point was there only been to her and sand and\nto him, in't artist, and why seeman anything as\nand with his still she was so terry who, they have strange at the peinar that there were not shired.\n\n\"That's harn his husband. He was all the sempt one in the same,\nand at such the bouth and that he say, that's, and a strings is the mergly and were and house to see. I ad that in her artation to be and her his simple, and what to cenfus and help at once.\"\n\n\"What was there is the prince of support, and. I came in to the\nbook,\nit was second and masters., women to the condition,\"\nhe adsteres with him were so of herself and stopping at that sense, but he was believen\nto this same, and he was said on his, as there\nhad steps on the\nprince, with his hand,\nto but her sain her hand of a\nlook to the most said him, whispered on she and an the pase on hasces and horses,\nand she had the same seen\nhes to the children, she had never been had steps in herself; and a lifes that two who had bransher and at his solity, but the\ncould not come alreary to a made the pot and at this same time in the peatance of\nhis house of him to as they had breaked any his\nwife. In this work had believe.\n\n\"I constate what it's a conviction\nand mean as it is not that to see them, and\nit's took her, and there\nwas and and he say, arand it. They's\nbetter how ill on,\" she said.\n\n\"It's become alone of this man, the bears of they said they are to she, if you was that's the master\nwas as she consent his sight and all him in the corsished, and \n"
],
[
"checkpoint = 'checkpoints/i200_l512.ckpt'\nsamp = sample(checkpoint, 1000, lstm_size, len(vocab), prime=\"Far\")\nprint(samp)",
"INFO:tensorflow:Restoring parameters from checkpoints/i200_l512.ckpt\n"
],
[
"checkpoint = 'checkpoints/i600_l512.ckpt'\nsamp = sample(checkpoint, 1000, lstm_size, len(vocab), prime=\"Far\")\nprint(samp)",
"INFO:tensorflow:Restoring parameters from checkpoints/i600_l512.ckpt\n"
],
[
"checkpoint = 'checkpoints/i1200_l512.ckpt'\nsamp = sample(checkpoint, 1000, lstm_size, len(vocab), prime=\"Far\")\nprint(samp)",
"INFO:tensorflow:Restoring parameters from checkpoints/i1200_l512.ckpt\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4ae3e5023849172bb2d7ed1fcb2364c16b880357
| 227,277 |
ipynb
|
Jupyter Notebook
|
KLASTERIZACJA.ipynb
|
MrYokkie/Data-Visualization
|
1111ec7aa9e5f0ddac929eaee069681c5681ded5
|
[
"MIT"
] | null | null | null |
KLASTERIZACJA.ipynb
|
MrYokkie/Data-Visualization
|
1111ec7aa9e5f0ddac929eaee069681c5681ded5
|
[
"MIT"
] | null | null | null |
KLASTERIZACJA.ipynb
|
MrYokkie/Data-Visualization
|
1111ec7aa9e5f0ddac929eaee069681c5681ded5
|
[
"MIT"
] | null | null | null | 119.305512 | 52,308 | 0.766994 |
[
[
[
"import pandas as pd\nfrom scipy.spatial.distance import pdist\nfrom scipy.cluster.hierarchy import *\nfrom matplotlib import pyplot as plt\nfrom matplotlib import rc\nimport numpy as np\nfrom sklearn.cluster import KMeans\nimport seaborn as sns\nfrom scipy.cluster.hierarchy import dendrogram, linkage\nfrom scipy.cluster import hierarchy",
"_____no_output_____"
],
[
"xl1 = pd.ExcelFile('1.xlsx')\nxl2 = pd.ExcelFile('2.xlsx') #!!!!!! измените имя файла и название рабочего листа\nxl1 #эта команда выведет пять случайных строк таблицы, таблица не отобразиться полностью.\nxl2",
"_____no_output_____"
],
[
"xl1.sheet_names",
"_____no_output_____"
],
[
"xl2.sheet_names",
"_____no_output_____"
],
[
"df = xl1.parse('Arkusz1') #wkinut tot sheet w dataframe\ndf.columns",
"_____no_output_____"
],
[
"df1 = xl2.parse('Arkusz2') #wkinut tot sheet w dataframe\ndf1.columns",
"_____no_output_____"
],
[
"# !!!!!! укажите количественные (int, float) столбцы, по которым выполним кластеризацию\ncol1=['połoczenia inhibitor betalaktamazy/ penicylina', 'cefalosporyny 3 generacji','karbapenemy','aminoglikozydy','fluorochinolony', 'sulfonamidy', 'tetracykliny', 'biofilm RT','biofilm 37C', 'swimming RT', 'swimming 37C', 'swarming RT',\n 'swarming 37C']\ncol2=[ 'biofilm RT', 'biofilm 37C', 'swimming RT',\n 'swimming 37C', 'swarming RT', 'swarming 37C', 'AMC', 'TZP',\n 'CXM', 'CTX', 'CAZ', 'FEP', 'IPM', 'MEM', 'ETP', 'AMK', 'CN', 'CIP',\n 'SXT', 'TGC', 'FOX']",
"_____no_output_____"
],
[
"pd.options.mode.chained_assignment = None \ndf[col1].fillna(0, inplace=True) # заменим пропуски данных нулями, в противном случае выдаст ошибку\npd.options.mode.chained_assignment = None \ndf1[col2].fillna(0, inplace=True) # заменим пропуски данных нулями, в противном случае выдаст ошибку",
"_____no_output_____"
],
[
"df[col1].corr() # посмотрим на парные корреляции",
"_____no_output_____"
]
],
[
[
"df[col1].corr() # посмотрим на парные корреляции",
"_____no_output_____"
]
],
[
[
"df1[col2].corr() # посмотрим на парные корреляции",
"_____no_output_____"
],
[
"# загружаем библиотеку препроцесинга данных\n# эта библиотека автоматически приведен данные к нормальным значениям\nfrom sklearn import preprocessing\ndataNorm1 = preprocessing.MinMaxScaler().fit_transform(df[col1].values)\ndataNorm2 = preprocessing.MinMaxScaler().fit_transform(df1[col2].values)",
"_____no_output_____"
],
[
"# Вычислим расстояния между каждым набором данных,\n# т.е. строками массива data_for_clust\n# Вычисляется евклидово расстояние (по умолчанию)\ndata_dist1 = pdist(dataNorm1, 'euclidean')\ndata_dist2 = pdist(dataNorm2, 'euclidean')\n# Главная функция иерархической кластеризии\n# Объедение элементов в кластера и сохранение в \n# специальной переменной (используется ниже для визуализации \n# и выделения количества кластеров\ndata_linkage1 = linkage(data_dist1, method='average')\ndata_linkage2 = linkage(data_dist2, method='average')",
"_____no_output_____"
],
[
"# Метод локтя. Позволячет оценить оптимальное количество сегментов.\n# Показывает сумму внутри групповых вариаций\nlast = data_linkage1[-10:, 2]\nlast_rev = last[::-1]\nidxs = np.arange(1, len(last) + 1)\nplt.plot(idxs, last_rev)\n\nacceleration = np.diff(last, 2) \nacceleration_rev = acceleration[::-1]\nplt.plot(idxs[:-2] + 1, acceleration_rev)\nplt.show()\nk = acceleration_rev.argmax() + 2 \nprint(\"Рекомендованное количество кластеров:\", k)",
"_____no_output_____"
],
[
"last = data_linkage2[-10:, 2]\nlast_rev = last[::-1]\nidxs = np.arange(1, len(last) + 1)\nplt.plot(idxs, last_rev)\n\nacceleration = np.diff(last, 2) \nacceleration_rev = acceleration[::-1]\nplt.plot(idxs[:-2] + 1, acceleration_rev)\nplt.show()\nk = acceleration_rev.argmax() + 2 \nprint(\"Рекомендованное количество кластеров:\", k)",
"_____no_output_____"
],
[
"#функция построения дендрограмм\ndef fancy_dendrogram(*args, **kwargs):\n max_d = kwargs.pop('max_d', None)\n if max_d and 'color_threshold' not in kwargs:\n kwargs['color_threshold'] = max_d\n annotate_above = kwargs.pop('annotate_above', 0)\n\n ddata = dendrogram(*args, **kwargs)\n\n if not kwargs.get('no_plot', False):\n plt.title('Hierarchical Clustering Dendrogram (truncated)')\n plt.xlabel('sample id')\n plt.ylabel('distance')\n for i, d, c in zip(ddata['icoord'], ddata['dcoord'], ddata['color_list']):\n x = 0.5 * sum(i[1:3])\n y = d[1]\n if y > annotate_above:\n plt.plot(x, y, 'o', c=c)\n plt.annotate(\"%.3g\" % y, (x, y), xytext=(0, -5),\n textcoords='offset points',\n va='top', ha='center')\n if max_d:\n plt.axhline(y=max_d, c='k')\n return ddata",
"_____no_output_____"
],
[
"# !!!!!!!!! укажите, какое количество кластеров будете использовать!\nnClust1=34",
"_____no_output_____"
],
[
"nClust2=34",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 35 entries, 0 to 34\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 połoczenia inhibitor betalaktamazy/ penicylina 35 non-null int64 \n 1 cefalosporyny 3 generacji 35 non-null int64 \n 2 karbapenemy 35 non-null int64 \n 3 aminoglikozydy 35 non-null int64 \n 4 fluorochinolony 35 non-null int64 \n 5 sulfonamidy 35 non-null int64 \n 6 tetracykliny 35 non-null int64 \n 7 biofilm RT 35 non-null float64\n 8 biofilm 37C 35 non-null float64\n 9 swimming RT 35 non-null float64\n 10 swimming 37C 35 non-null float64\n 11 swarming RT 35 non-null float64\n 12 swarming 37C 35 non-null float64\ndtypes: float64(6), int64(7)\nmemory usage: 3.7 KB\n"
],
[
"df.describe()",
"_____no_output_____"
],
[
"fancy_dendrogram(\n data_linkage1,\n truncate_mode='level',\n p=nClust1, \n leaf_rotation=90.,\n leaf_font_size=8.,\n show_contracted=True,\n annotate_above=100,\n)\nplt.savefig(\"wykres1.png\",dpi = 300)\nplt.show()",
"_____no_output_____"
],
[
"#строим дендрограмму \nfancy_dendrogram(\n data_linkage2,\n truncate_mode='level',\n p=nClust2, \n leaf_rotation=90.,\n leaf_font_size=8.,\n show_contracted=True,\n annotate_above=100,\n)\nplt.savefig(\"wykres2.png\",dpi = 300)\nplt.show()",
"_____no_output_____"
],
[
"# иерархическая кластеризация\nclusters=fcluster(data_linkage1, nClust1, criterion='maxclust')\nclusters",
"_____no_output_____"
],
[
"clusters=fcluster(data_linkage2, nClust2, criterion='maxclust')\nclusters",
"_____no_output_____"
],
[
"df[df['I']==33] # !!!!! меняйте номер кластера",
"_____no_output_____"
],
[
"# строим кластеризаци методом KMeans\nkm = KMeans(n_clusters=nClust1).fit(dataNorm1)",
"_____no_output_____"
],
[
"# выведем полученное распределение по кластерам\n# так же номер кластера, к котрому относится строка, так как нумерация начинается с нуля, выводим добавляя 1\nkm.labels_ +1",
"_____no_output_____"
],
[
"x=0 # Чтобы построить диаграмму в разных осях, меняйте номера столбцов\ny=2 #\ncentroids = km.cluster_centers_\nplt.figure(figsize=(10, 8))\nplt.scatter(dataNorm1[:,x], dataNorm1[:,y], c=km.labels_, cmap='flag')\nplt.scatter(centroids[:, x], centroids[:, y], marker='*', s=300,\n c='r', label='centroid')\nplt.xlabel(col1[x])\nplt.ylabel(col1[y]);\nplt.show()",
"_____no_output_____"
],
[
"#сохраним результаты в файл\ndf.to_excel('result_claster.xlsx', index=False)",
"_____no_output_____"
],
[
"sns.clustermap(df, metric=\"correlation\", method=\"single\", cmap=\"Blues\", standard_scale=1)\nplt.show()",
"_____no_output_____"
],
[
"sns.clustermap(df1, metric=\"correlation\", figsize=(22, 22), method=\"single\", cmap=\"Blues\", standard_scale=1)\nplt.savefig(\"clustmap2.png\", dpi = 300)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae3eac64737ec7a27567ee517f3c06e9c5261ef
| 6,626 |
ipynb
|
Jupyter Notebook
|
_build/jupyter_execute/ipynb_to_docx.ipynb
|
PyLCARS/Python-and-SPICE-Book
|
0bf02aa16d97115cea955d33a7aab7e02f8d3453
|
[
"MIT"
] | 3 |
2021-01-04T23:56:51.000Z
|
2021-02-22T13:22:30.000Z
|
_build/jupyter_execute/ipynb_to_docx.ipynb
|
PyLCARS/Python-and-SPICE-Book
|
0bf02aa16d97115cea955d33a7aab7e02f8d3453
|
[
"MIT"
] | null | null | null |
_build/jupyter_execute/ipynb_to_docx.ipynb
|
PyLCARS/Python-and-SPICE-Book
|
0bf02aa16d97115cea955d33a7aab7e02f8d3453
|
[
"MIT"
] | null | null | null | 29.061404 | 210 | 0.578328 |
[
[
[
"This is not part of the book, this is a editing tool that is used to create markdownfiles for the jupyter noteboks and then convert them to Microsoft Word in order to use Word and Grammerly to do editing ",
"_____no_output_____"
]
],
[
[
"from pathlib import Path",
"_____no_output_____"
],
[
"path=Path('.')\nnotebooks=[]\nfor p in path.rglob(\"*.ipynb\"):\n if '_build' in str(p.absolute()):\n continue\n \n elif 'ipynb_checkpoints' in str(p.absolute()):\n continue\n else:\n notebooks.append(p)\nnotebooks",
"_____no_output_____"
],
[
"notebooks[2].relative_to(notebooks[2].parent.parent)",
"_____no_output_____"
],
[
"for nb in notebooks:\n #base=str(nb.absolute())\n ipynb='./'+str(nb.relative_to(nb.parent.parent))\n base=ipynb[:-6]\n html=base+'.html'\n docx=base+'.docx'\n print(ipynb)\n \n #uncommment to use\n #!jupyter nbconvert --to html {ipynb}\n \n #testing these but these have yet to yield what I want\n #!jupyter nbconvert --clear-output --to html {ipynb}\n #!jupyter nbconvert --TagRemovePreprocessor.enabled=True --TagRemovePreprocessor.remove_cell_tags=\"['remove_output']\" --to html {ipynb}\n \n #!pandoc -s {html} -o {docx}",
"./intro.ipynb\n./ipynb_to_docx.ipynb\n./needed_libraries.ipynb\n./AC_2/AC_1.ipynb\n./AC_2/AC_1_One-Two-Three_Phase_AC.ipynb\n./AC_2/AC_2_RCL_filters.ipynb\n./AC_2/AC_3_CoupledMag.ipynb\n./AC_2/AC_4_PZ.ipynb\n./AC_2/AC_5_twoports.ipynb\n./AC_2/AC_6_S_transmissionlines.ipynb\n./Appendix/skidl_2_pyspice_check.ipynb\n./DC_1/DC_1.ipynb\n./DC_1/DC_1_op_ohm.ipynb\n./DC_1/DC_2_op_source_transform.ipynb\n./DC_1/DC_3_PracticalSourcs_sweeps_subcirucirts.ipynb\n./DC_1/DC_4_dependent_sources_and_tf.ipynb\n./DC_1/DC_5_Thvenin_Norton.ipynb\n./DC_1/DC_6_DC_MaxPower_MaxEfficiency.ipynb\n./Diodes_7/Diodes_1_Chartiztion.ipynb\n./Diodes_7/Diodes_2_Clipers.ipynb\n./Distortion/Distortion_testing.ipynb\n./Noise_4/Noise_1_noise.ipynb\n./Sensitivity_5/Sensitivity_1_ngspice's.ipynb\n./Sensitivity_5/Sensitivity_2_with_Python.ipynb\n./Trans_3/Untitled.ipynb\n"
]
],
[
[
"# note to self to work building book and github page\nbuild book:\nbe looking at this folder not in it \n```\njupyter-book build Python-and-SPICE-Book/\n```\n\n\nbuild book site:\nbe inside the top repo\n```\nghp-import -n -p -c https://pylcars.github.io/Python-and-SPICE-Book/docs -f _build/html\n```",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4ae408d4f5aa1b6b1d4e636e99810d5ec4b99b73
| 18,808 |
ipynb
|
Jupyter Notebook
|
kaggle/toxic_comments/notebooks/Dense.ipynb
|
radpet/stuff
|
56adcabf712dbefa90c0fd85693acbf49f184189
|
[
"MIT"
] | null | null | null |
kaggle/toxic_comments/notebooks/Dense.ipynb
|
radpet/stuff
|
56adcabf712dbefa90c0fd85693acbf49f184189
|
[
"MIT"
] | 10 |
2020-02-24T17:56:53.000Z
|
2022-03-11T23:18:29.000Z
|
kaggle/toxic_comments/notebooks/Dense.ipynb
|
radpet/stuff
|
56adcabf712dbefa90c0fd85693acbf49f184189
|
[
"MIT"
] | null | null | null | 30.482982 | 194 | 0.446512 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"train = pd.read_csv('./data/train_cleaned.csv')\ntest = pd.read_csv('./data/test_cleaned.csv')",
"_____no_output_____"
],
[
"train[\"comment_text\"].fillna(\"unknown\", inplace=True)\ntest[\"comment_text\"].fillna(\"unknown\", inplace=True)",
"_____no_output_____"
],
[
"from util import labels, RocAucEvaluation",
"Using TensorFlow backend.\n"
],
[
"num_classes = len(labels)\nmax_features = 30000\nhidden = 300\ndropout = 0.4\nbatch_size = 1000",
"_____no_output_____"
],
[
"labels_to_id = {label:key for key,label in enumerate(labels)}",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import TfidfVectorizer",
"_____no_output_____"
],
[
"vectorizer = TfidfVectorizer(max_features=max_features, stop_words='english').fit(train['comment_text'].append(test['comment_text']))",
"_____no_output_____"
],
[
"from keras.layers import Input, Dense, Dropout, BatchNormalization\nfrom keras.models import Model",
"_____no_output_____"
],
[
"def get_model():\n input_comment = Input(shape=(max_features,), sparse=True)\n \n x = Dense(hidden, activation='relu')(input_comment)\n x = BatchNormalization()(x)\n x = Dropout(dropout)(x)\n output_pred = Dense(num_classes, activation='sigmoid')(x)\n \n model = Model(inputs=input_comment, outputs=output_pred)\n return model",
"_____no_output_____"
],
[
"model = get_model()",
"_____no_output_____"
],
[
"model.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) (None, 30000) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 300) 9000300 \n_________________________________________________________________\nbatch_normalization_1 (Batch (None, 300) 1200 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 300) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 6) 1806 \n=================================================================\nTotal params: 9,003,306\nTrainable params: 9,002,706\nNon-trainable params: 600\n_________________________________________________________________\n"
],
[
"model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"train_fold, val_fold = train_test_split(train, train_size=0.95, random_state=123456)",
"/usr/local/lib/python3.5/dist-packages/sklearn/model_selection/_split.py:2010: FutureWarning: From version 0.21, test_size will always complement train_size unless both are specified.\n FutureWarning)\n"
],
[
"train_fold.shape, val_fold.shape",
"_____no_output_____"
],
[
"train_vect = vectorizer.transform(train_fold['comment_text'])",
"_____no_output_____"
],
[
"val_fold_features = vectorizer.transform(val_fold['comment_text'])",
"_____no_output_____"
],
[
"roc_auc = RocAucEvaluation(validation_data=(val_fold_features, val_fold[labels]))",
"_____no_output_____"
],
[
"model.fit(train_vect, train_fold[labels].values, batch_size=batch_size, epochs=4, callbacks=[roc_auc])",
"Epoch 1/4\n151000/151592 [============================>.] - ETA: 0s - loss: 0.2344 - acc: 0.9321\n ROC-AUC - epoch: 1 - score: 0.969580 \n\n151592/151592 [==============================] - 7s 44us/step - loss: 0.2337 - acc: 0.9323\nEpoch 2/4\n151000/151592 [============================>.] - ETA: 0s - loss: 0.0468 - acc: 0.9844- ETA: 0s - loss: 0.0470 - ac\n ROC-AUC - epoch: 2 - score: 0.973836 \n\n151592/151592 [==============================] - 6s 40us/step - loss: 0.0468 - acc: 0.9844\nEpoch 3/4\n151000/151592 [============================>.] - ETA: 0s - loss: 0.0331 - acc: 0.9882- ETA: 2s - loss: 0.0328 - acc - ETA: 1s - loss: 0.0330 - acc: 0 - ETA: 1s - loss: 0.0\n ROC-AUC - epoch: 3 - score: 0.972107 \n\n151592/151592 [==============================] - 6s 41us/step - loss: 0.0331 - acc: 0.9882\nEpoch 4/4\n151000/151592 [============================>.] - ETA: 0s - loss: 0.0258 - acc: 0.9910\n ROC-AUC - epoch: 4 - score: 0.969529 \n\n151592/151592 [==============================] - 6s 41us/step - loss: 0.0258 - acc: 0.9910\n"
],
[
"test_vect = vectorizer.transform(test['comment_text'])",
"_____no_output_____"
],
[
"preds = model.predict(test_vect)",
"_____no_output_____"
],
[
"subm = pd.DataFrame(preds, columns=labels, index=test['id'])",
"_____no_output_____"
],
[
"subm.head(20)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae417d0a0838a7d200c598771bc4c0ff6aaf6bd
| 29,229 |
ipynb
|
Jupyter Notebook
|
tutorials/W1D1_ModelTypes/W1D1_Tutorial2.ipynb
|
DianaMosquera/course-content
|
cc2e0e2e5d9b476e0fb810ead4ed19dc23745152
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 2 |
2021-05-12T02:19:05.000Z
|
2021-05-12T13:49:29.000Z
|
tutorials/W1D1_ModelTypes/W1D1_Tutorial2.ipynb
|
DianaMosquera/course-content
|
cc2e0e2e5d9b476e0fb810ead4ed19dc23745152
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 1 |
2020-09-21T09:58:24.000Z
|
2020-09-21T09:58:24.000Z
|
tutorials/W1D1_ModelTypes/W1D1_Tutorial2.ipynb
|
DianaMosquera/course-content
|
cc2e0e2e5d9b476e0fb810ead4ed19dc23745152
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 1 |
2021-05-02T10:03:07.000Z
|
2021-05-02T10:03:07.000Z
| 38.108214 | 714 | 0.599028 |
[
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W1D1_ModelTypes/W1D1_Tutorial2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Neuromatch Academy: Week 1, Day 1, Tutorial 2\n# Model Types: \"How\" models\n__Content creators:__ Matt Laporte, Byron Galbraith, Konrad Kording\n\n__Content reviewers:__ Dalin Guo, Aishwarya Balwani, Madineh Sarvestani, Maryam Vaziri-Pashkam, Michael Waskom",
"_____no_output_____"
],
[
"___\n# Tutorial Objectives\nThis is tutorial 2 of a 3-part series on different flavors of models used to understand neural data. In this tutorial we will explore models that can potentially explain *how* the spiking data we have observed is produced\n\nTo understand the mechanisms that give rise to the neural data we save in Tutorial 1, we will build simple neuronal models and compare their spiking response to real data. We will:\n- Write code to simulate a simple \"leaky integrate-and-fire\" neuron model \n- Make the model more complicated — but also more realistic — by adding more physiologically-inspired details",
"_____no_output_____"
]
],
[
[
"#@title Video 1: \"How\" models\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='PpnagITsb3E', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"# Setup",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy import stats",
"_____no_output_____"
],
[
"#@title Figure Settings\nimport ipywidgets as widgets #interactive display\n\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/nma.mplstyle\")",
"_____no_output_____"
],
[
"#@title Helper Functions\ndef histogram(counts, bins, vlines=(), ax=None, ax_args=None, **kwargs):\n \"\"\"Plot a step histogram given counts over bins.\"\"\"\n if ax is None:\n _, ax = plt.subplots()\n\n # duplicate the first element of `counts` to match bin edges\n counts = np.insert(counts, 0, counts[0])\n\n ax.fill_between(bins, counts, step=\"pre\", alpha=0.4, **kwargs) # area shading\n ax.plot(bins, counts, drawstyle=\"steps\", **kwargs) # lines\n\n for x in vlines:\n ax.axvline(x, color='r', linestyle='dotted') # vertical line\n\n if ax_args is None:\n ax_args = {}\n\n # heuristically set max y to leave a bit of room\n ymin, ymax = ax_args.get('ylim', [None, None])\n if ymax is None:\n ymax = np.max(counts)\n if ax_args.get('yscale', 'linear') == 'log':\n ymax *= 1.5\n else:\n ymax *= 1.1\n if ymin is None:\n ymin = 0\n\n if ymax == ymin:\n ymax = None\n\n ax_args['ylim'] = [ymin, ymax]\n\n ax.set(**ax_args)\n ax.autoscale(enable=False, axis='x', tight=True)\n\n\ndef plot_neuron_stats(v, spike_times):\n fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(12, 5))\n\n # membrane voltage trace\n ax1.plot(v[0:100])\n ax1.set(xlabel='Time', ylabel='Voltage')\n # plot spike events\n for x in spike_times:\n if x >= 100:\n break\n ax1.axvline(x, color='red')\n\n # ISI distribution\n if len(spike_times)>1:\n isi = np.diff(spike_times)\n n_bins = np.arange(isi.min(), isi.max() + 2) - .5\n counts, bins = np.histogram(isi, n_bins)\n vlines = []\n if len(isi) > 0:\n vlines = [np.mean(isi)]\n xmax = max(20, int(bins[-1])+5)\n histogram(counts, bins, vlines=vlines, ax=ax2, ax_args={\n 'xlabel': 'Inter-spike interval',\n 'ylabel': 'Number of intervals',\n 'xlim': [0, xmax]\n })\n else:\n ax2.set(xlabel='Inter-spike interval',\n ylabel='Number of intervals')\n plt.show()",
"_____no_output_____"
]
],
[
[
"# Section 1: The Linear Integrate-and-Fire Neuron\n\nHow does a neuron spike? \n\nA neuron charges and discharges an electric field across its cell membrane. The state of this electric field can be described by the _membrane potential_. The membrane potential rises due to excitation of the neuron, and when it reaches a threshold a spike occurs. The potential resets, and must rise to a threshold again before the next spike occurs.\n\nOne of the simplest models of spiking neuron behavior is the linear integrate-and-fire model neuron. In this model, the neuron increases its membrane potential $V_m$ over time in response to excitatory input currents $I$ scaled by some factor $\\alpha$:\n\n\\begin{align}\n dV_m = {\\alpha}I\n\\end{align}\n\nOnce $V_m$ reaches a threshold value a spike is produced, $V_m$ is reset to a starting value, and the process continues.\n\nHere, we will take the starting and threshold potentials as $0$ and $1$, respectively. So, for example, if $\\alpha I=0.1$ is constant---that is, the input current is constant---then $dV_m=0.1$, and at each timestep the membrane potential $V_m$ increases by $0.1$ until after $(1-0)/0.1 = 10$ timesteps it reaches the threshold and resets to $V_m=0$, and so on.\n\nNote that we define the membrane potential $V_m$ as a scalar: a single real (or floating point) number. However, a biological neuron's membrane potential will not be exactly constant at all points on its cell membrane at a given time. We could capture this variation with a more complex model (e.g. with more numbers). Do we need to? \n\nThe proposed model is a 1D simplification. There are many details we could add to it, to preserve different parts of the complex structure and dynamics of a real neuron. If we were interested in small or local changes in the membrane potential, our 1D simplification could be a problem. However, we'll assume an idealized \"point\" neuron model for our current purpose.\n\n#### Spiking Inputs\n\nGiven our simplified model for the neuron dynamics, we still need to consider what form the input $I$ will take. How should we specify the firing behavior of the presynaptic neuron(s) providing the inputs to our model neuron? \n\nUnlike in the simple example above, where $\\alpha I=0.1$, the input current is generally not constant. Physical inputs tend to vary with time. We can describe this variation with a distribution.\n\nWe'll assume the input current $I$ over a timestep is due to equal contributions from a non-negative ($\\ge 0$) integer number of input spikes arriving in that timestep. Our model neuron might integrate currents from 3 input spikes in one timestep, and 7 spikes in the next timestep. We should see similar behavior when sampling from our distribution.\n\nGiven no other information about the input neurons, we will also assume that the distribution has a mean (i.e. mean rate, or number of spikes received per timestep), and that the spiking events of the input neuron(s) are independent in time. Are these reasonable assumptions in the context of real neurons?\n\nA suitable distribution given these assumptions is the Poisson distribution, which we'll use to model $I$:\n\n\\begin{align}\n I \\sim \\mathrm{Poisson}(\\lambda)\n\\end{align}\n\nwhere $\\lambda$ is the mean of the distribution: the average rate of spikes received per timestep.",
"_____no_output_____"
],
[
"### Exercise 1: Compute $dV_m$\n\nFor your first exercise, you will write the code to compute the change in voltage $dV_m$ (per timestep) of the linear integrate-and-fire model neuron. The rest of the code to handle numerical integration is provided for you, so you just need to fill in a definition for `dv` in the `lif_neuron` function below. The value of $\\lambda$ for the Poisson random variable is given by the function argument `rate`.\n\n\n\nThe [`scipy.stats`](https://docs.scipy.org/doc/scipy/reference/stats.html) package is a great resource for working with and sampling from various probability distributions. We will use the `scipy.stats.poisson` class and its method `rvs` to produce Poisson-distributed random samples. In this tutorial, we have imported this package with the alias `stats`, so you should refer to it in your code as `stats.poisson`.",
"_____no_output_____"
]
],
[
[
"def lif_neuron(n_steps=1000, alpha=0.01, rate=10):\n \"\"\" Simulate a linear integrate-and-fire neuron.\n\n Args:\n n_steps (int): The number of time steps to simulate the neuron's activity.\n alpha (float): The input scaling factor\n rate (int): The mean rate of incoming spikes\n\n \"\"\"\n # precompute Poisson samples for speed\n exc = stats.poisson(rate).rvs(n_steps)\n\n v = np.zeros(n_steps)\n spike_times = []\n\n ################################################################################\n # Students: compute dv, then comment out or remove the next line\n raise NotImplementedError(\"Excercise: compute the change in membrane potential\")\n ################################################################################\n\n for i in range(1, n_steps):\n\n dv = ...\n\n v[i] = v[i-1] + dv\n if v[i] > 1:\n spike_times.append(i)\n v[i] = 0\n\n return v, spike_times\n\n# Set random seed (for reproducibility)\nnp.random.seed(12)\n\n# Uncomment these lines after completing the lif_neuron function\n# v, spike_times = lif_neuron()\n# plot_neuron_stats(v, spike_times)",
"_____no_output_____"
],
[
"# to_remove solution\ndef lif_neuron(n_steps=1000, alpha=0.01, rate=10):\n \"\"\" Simulate a linear integrate-and-fire neuron.\n\n Args:\n n_steps (int): The number of time steps to simulate the neuron's activity.\n alpha (float): The input scaling factor\n rate (int): The mean rate of incoming spikes\n \"\"\"\n # precompute Poisson samples for speed\n exc = stats.poisson(rate).rvs(n_steps)\n\n v = np.zeros(n_steps)\n spike_times = []\n for i in range(1, n_steps):\n\n dv = alpha * exc[i]\n\n v[i] = v[i-1] + dv\n if v[i] > 1:\n spike_times.append(i)\n v[i] = 0\n\n return v, spike_times\n\n# Set random seed (for reproducibility)\nnp.random.seed(12)\n\nv, spike_times = lif_neuron()\nwith plt.xkcd():\n plot_neuron_stats(v, spike_times)",
"_____no_output_____"
]
],
[
[
"## Interactive Demo: Linear-IF neuron\nLike last time, you can now explore how various parametes of the LIF model influence the ISI distribution.",
"_____no_output_____"
]
],
[
[
"#@title\n\n#@markdown You don't need to worry about how the code works – but you do need to **run the cell** to enable the sliders.\n\ndef _lif_neuron(n_steps=1000, alpha=0.01, rate=10):\n exc = stats.poisson(rate).rvs(n_steps)\n v = np.zeros(n_steps)\n spike_times = []\n for i in range(1, n_steps):\n dv = alpha * exc[i]\n v[i] = v[i-1] + dv\n if v[i] > 1:\n spike_times.append(i)\n v[i] = 0\n return v, spike_times\n\[email protected](\n n_steps=widgets.FloatLogSlider(1000.0, min=2, max=4),\n alpha=widgets.FloatLogSlider(0.01, min=-2, max=-1),\n rate=widgets.IntSlider(10, min=5, max=20)\n)\ndef plot_lif_neuron(n_steps=1000, alpha=0.01, rate=10):\n v, spike_times = _lif_neuron(int(n_steps), alpha, rate)\n plot_neuron_stats(v, spike_times)",
"_____no_output_____"
],
[
"#@title Video 2: Linear-IF models\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='QBD7kulhg4U', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"# Section 2: Inhibitory signals\n",
"_____no_output_____"
],
[
"\nOur linear integrate-and-fire neuron from the previous section was indeed able to produce spikes. However, our ISI histogram doesn't look much like empirical ISI histograms seen in Tutorial 1, which had an exponential-like shape. What is our model neuron missing, given that it doesn't behave like a real neuron?\n\nIn the previous model we only considered excitatory behavior -- the only way the membrane potential could decrease was upon a spike event. We know, however, that there are other factors that can drive $V_m$ down. First is the natural tendency of the neuron to return to some steady state or resting potential. We can update our previous model as follows:\n\n\\begin{align}\n dV_m = -{\\beta}V_m + {\\alpha}I\n\\end{align}\n\nwhere $V_m$ is the current membrane potential and $\\beta$ is some leakage factor. This is a basic form of the popular Leaky Integrate-and-Fire model neuron (for a more detailed discussion of the LIF Neuron, see the Appendix).\n\nWe also know that in addition to excitatory presynaptic neurons, we can have inhibitory presynaptic neurons as well. We can model these inhibitory neurons with another Poisson random variable:\n\n\\begin{align}\nI = I_{exc} - I_{inh} \\\\\nI_{exc} \\sim \\mathrm{Poisson}(\\lambda_{exc}) \\\\\nI_{inh} \\sim \\mathrm{Poisson}(\\lambda_{inh})\n\\end{align}\n\nwhere $\\lambda_{exc}$ and $\\lambda_{inh}$ are the average spike rates (per timestep) of the excitatory and inhibitory presynaptic neurons, respectively.",
"_____no_output_____"
],
[
"### Exercise 2: Compute $dV_m$ with inhibitory signals\n\nFor your second exercise, you will again write the code to compute the change in voltage $dV_m$, though now of the LIF model neuron described above. Like last time, the rest of the code needed to handle the neuron dynamics are provided for you, so you just need to fill in a definition for `dv` below.\n",
"_____no_output_____"
]
],
[
[
"def lif_neuron_inh(n_steps=1000, alpha=0.5, beta=0.1, exc_rate=10, inh_rate=10):\n \"\"\" Simulate a simplified leaky integrate-and-fire neuron with both excitatory\n and inhibitory inputs.\n\n Args:\n n_steps (int): The number of time steps to simulate the neuron's activity.\n alpha (float): The input scaling factor\n beta (float): The membrane potential leakage factor\n exc_rate (int): The mean rate of the incoming excitatory spikes\n inh_rate (int): The mean rate of the incoming inhibitory spikes\n \"\"\"\n\n # precompute Poisson samples for speed\n exc = stats.poisson(exc_rate).rvs(n_steps)\n inh = stats.poisson(inh_rate).rvs(n_steps)\n\n v = np.zeros(n_steps)\n spike_times = []\n\n ###############################################################################\n # Students: compute dv, then comment out or remove the next line\n raise NotImplementedError(\"Excercise: compute the change in membrane potential\")\n ################################################################################\n\n for i in range(1, n_steps):\n\n dv = ...\n\n v[i] = v[i-1] + dv\n if v[i] > 1:\n spike_times.append(i)\n v[i] = 0\n\n return v, spike_times\n\n# Set random seed (for reproducibility)\nnp.random.seed(12)\n\n# Uncomment these lines do make the plot once you've completed the function\n#v, spike_times = lif_neuron_inh()\n#plot_neuron_stats(v, spike_times)",
"_____no_output_____"
],
[
"# to_remove solution\ndef lif_neuron_inh(n_steps=1000, alpha=0.5, beta=0.1, exc_rate=10, inh_rate=10):\n \"\"\" Simulate a simplified leaky integrate-and-fire neuron with both excitatory\n and inhibitory inputs.\n\n Args:\n n_steps (int): The number of time steps to simulate the neuron's activity.\n alpha (float): The input scaling factor\n beta (float): The membrane potential leakage factor\n exc_rate (int): The mean rate of the incoming excitatory spikes\n inh_rate (int): The mean rate of the incoming inhibitory spikes\n \"\"\"\n # precompute Poisson samples for speed\n exc = stats.poisson(exc_rate).rvs(n_steps)\n inh = stats.poisson(inh_rate).rvs(n_steps)\n\n v = np.zeros(n_steps)\n spike_times = []\n for i in range(1, n_steps):\n\n dv = -beta * v[i-1] + alpha * (exc[i] - inh[i])\n\n v[i] = v[i-1] + dv\n if v[i] > 1:\n spike_times.append(i)\n v[i] = 0\n\n return v, spike_times\n\n# Set random seed (for reproducibility)\nnp.random.seed(12)\n\nv, spike_times = lif_neuron_inh()\n\nwith plt.xkcd():\n plot_neuron_stats(v, spike_times)",
"_____no_output_____"
]
],
[
[
"## Interactive Demo: LIF + inhibition neuron",
"_____no_output_____"
]
],
[
[
"#@title\n#@markdown **Run the cell** to enable the sliders.\ndef _lif_neuron_inh(n_steps=1000, alpha=0.5, beta=0.1, exc_rate=10, inh_rate=10):\n \"\"\" Simulate a simplified leaky integrate-and-fire neuron with both excitatory\n and inhibitory inputs.\n\n Args:\n n_steps (int): The number of time steps to simulate the neuron's activity.\n alpha (float): The input scaling factor\n beta (float): The membrane potential leakage factor\n exc_rate (int): The mean rate of the incoming excitatory spikes\n inh_rate (int): The mean rate of the incoming inhibitory spikes\n \"\"\"\n # precompute Poisson samples for speed\n exc = stats.poisson(exc_rate).rvs(n_steps)\n inh = stats.poisson(inh_rate).rvs(n_steps)\n\n v = np.zeros(n_steps)\n spike_times = []\n for i in range(1, n_steps):\n dv = -beta * v[i-1] + alpha * (exc[i] - inh[i])\n v[i] = v[i-1] + dv\n if v[i] > 1:\n spike_times.append(i)\n v[i] = 0\n\n return v, spike_times\n\[email protected](n_steps=widgets.FloatLogSlider(1000.0, min=2.5, max=4),\n alpha=widgets.FloatLogSlider(0.5, min=-1, max=1),\n beta=widgets.FloatLogSlider(0.1, min=-1, max=0),\n exc_rate=widgets.IntSlider(12, min=10, max=20),\n inh_rate=widgets.IntSlider(12, min=10, max=20))\ndef plot_lif_neuron(n_steps=1000, alpha=0.5, beta=0.1, exc_rate=10, inh_rate=10):\n v, spike_times = _lif_neuron_inh(int(n_steps), alpha, beta, exc_rate, inh_rate)\n plot_neuron_stats(v, spike_times)",
"_____no_output_____"
],
[
"#@title Video 3: LIF + inhibition\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id='Aq7JrxRkn2w', width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"#Summary\n\nIn this tutorial we gained some intuition for the mechanisms that produce the observed behavior in our real neural data. First, we built a simple neuron model with excitatory input and saw that it's behavior, measured using the ISI distribution, did not match our real neurons. We then improved our model by adding leakiness and inhibitory input. The behavior of this balanced model was much closer to the real neural data.",
"_____no_output_____"
],
[
"# Bonus",
"_____no_output_____"
],
[
"### Why do neurons spike?\n\nA neuron stores energy in an electric field across its cell membrane, by controlling the distribution of charges (ions) on either side of the membrane. This energy is rapidly discharged to generate a spike when the field potential (or membrane potential) crosses a threshold. The membrane potential may be driven toward or away from this threshold, depending on inputs from other neurons: excitatory or inhibitory, respectively. The membrane potential tends to revert to a resting potential, for example due to the leakage of ions across the membrane, so that reaching the spiking threshold depends not only on the amount of input ever received following the last spike, but also the timing of the inputs.\n\nThe storage of energy by maintaining a field potential across an insulating membrane can be modeled by a capacitor. The leakage of charge across the membrane can be modeled by a resistor. This is the basis for the leaky integrate-and-fire neuron model.",
"_____no_output_____"
],
[
"### The LIF Model Neuron\n\nThe full equation for the LIF neuron is\n\n\\begin{align}\nC_{m}\\frac{dV_m}{dt} = -(V_m - V_{rest})/R_{m} + I\n\\end{align}\n\nwhere $C_m$ is the membrane capacitance, $R_M$ is the membrane resistance, $V_{rest}$ is the resting potential, and $I$ is some input current (from other neurons, an electrode, ...).\n\nIn our above examples we set many of these parameters to convenient values ($C_m = R_m = dt = 1$, $V_{rest} = 0$) to focus more on the general behavior of the model. However, these too can be manipulated to achieve different dynamics, or to ensure the dimensions of the problem are preserved between simulation units and experimental units (e.g. with $V_m$ given in millivolts, $R_m$ in megaohms, $t$ in milliseconds).",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4ae4214200be1be5659fa4ec2f4c303ddf1caa9c
| 9,036 |
ipynb
|
Jupyter Notebook
|
gender_recognition/Gender Recognition Model.ipynb
|
ck090/New_Bechdel
|
0e9b61e974de4716fe5131c0b9010e2b8723d3af
|
[
"MIT"
] | 2 |
2018-10-01T21:46:08.000Z
|
2018-10-02T06:36:21.000Z
|
gender_recognition/Gender Recognition Model.ipynb
|
ck090/New_Bechdel
|
0e9b61e974de4716fe5131c0b9010e2b8723d3af
|
[
"MIT"
] | null | null | null |
gender_recognition/Gender Recognition Model.ipynb
|
ck090/New_Bechdel
|
0e9b61e974de4716fe5131c0b9010e2b8723d3af
|
[
"MIT"
] | null | null | null | 29.337662 | 275 | 0.520142 |
[
[
[
"## A quick Gender Recognition model\nGrabbed from [nlpforhackers](https://nlpforhackers.io/introduction-machine-learning/) webpage.\n1. Firstly convert the dataset into a numpy array to keep only gender and names\n2. Set the feature parameters which takes in different parameters\n3. Vectorize the parametes\n4. Get varied train, test split and test it for validity by checking out the count of the train test split\n5. Transform lists of feature-value mappings to vectors. (When feature values are strings, this transformer will do a binary one-hot (aka one-of-K) coding: one boolean-valued feature is constructed for each of the possible string values that the feature can take on)\n6. Train a decision tree classifier on this and save the model as a pickle file",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.utils import shuffle\nfrom sklearn.feature_extraction import DictVectorizer\nfrom sklearn.tree import DecisionTreeClassifier",
"_____no_output_____"
],
[
"names = pd.read_csv('names_dataset.csv')\nprint(names.head(10))\n \nprint(\"%d names in dataset\" % len(names))",
" index name sex\n0 0 Mary F\n1 1 Anna F\n2 2 Emma F\n3 3 Elizabeth F\n4 4 Minnie F\n5 5 Margaret F\n6 6 Ida F\n7 7 Alice F\n8 8 Bertha F\n9 9 Sarah F\n95025 names in dataset\n"
],
[
"# Get the data out of the dataframe into a numpy matrix and keep only the name and gender columns\nnames = names.as_matrix()[:, 1:]\nprint(names)\n \n# We're using 90% of the data for training\nTRAIN_SPLIT = 0.90",
"[['Mary' 'F']\n ['Anna' 'F']\n ['Emma' 'F']\n ...\n ['Ziyu' 'M']\n ['Zykir' 'M']\n ['Zyus' 'M']]\n"
],
[
"def features(name):\n name = name.lower()\n return {\n 'first-letter': name[0], # First letter\n 'first2-letters': name[0:2], # First 2 letters\n 'first3-letters': name[0:3], # First 3 letters\n 'last-letter': name[-1], # Last letter\n 'last2-letters': name[-2:], # Last 2 letters\n 'last3-letters': name[-3:], # Last 3 letters\n }\n\n# Feature Extraction\nprint(features(\"Alex\"))",
"{'first2-letters': 'al', 'last-letter': 'x', 'first-letter': 'a', 'last2-letters': 'ex', 'last3-letters': 'lex', 'first3-letters': 'ale'}\n"
],
[
"# Vectorize the features function\nfeatures = np.vectorize(features)\nprint(features([\"Anna\", \"Hannah\", \"Paul\"]))\n# [ array({'first2-letters': 'an', 'last-letter': 'a', 'first-letter': 'a', 'last2-letters': 'na', 'last3-letters': 'nna', 'first3-letters': 'ann'}, dtype=object)\n# array({'first2-letters': 'ha', 'last-letter': 'h', 'first-letter': 'h', 'last2-letters': 'ah', 'last3-letters': 'nah', 'first3-letters': 'han'}, dtype=object)\n# array({'first2-letters': 'pa', 'last-letter': 'l', 'first-letter': 'p', 'last2-letters': 'ul', 'last3-letters': 'aul', 'first3-letters': 'pau'}, dtype=object)]\n \n# Extract the features for the whole dataset\nX = features(names[:, 0]) # X contains the features\n \n# Get the gender column\ny = names[:, 1] # y contains the targets\n \n# Test if we built the dataset correctly\nprint(\"\\n\\nName: %s, features=%s, gender=%s\" % (names[0][0], X[0], y[0]))",
"[{'first2-letters': 'an', 'last-letter': 'a', 'first-letter': 'a', 'last2-letters': 'na', 'last3-letters': 'nna', 'first3-letters': 'ann'}\n {'first2-letters': 'ha', 'last-letter': 'h', 'first-letter': 'h', 'last2-letters': 'ah', 'last3-letters': 'nah', 'first3-letters': 'han'}\n {'first2-letters': 'pa', 'last-letter': 'l', 'first-letter': 'p', 'last2-letters': 'ul', 'last3-letters': 'aul', 'first3-letters': 'pau'}]\n\n\nName: Mary, features={'first2-letters': 'ma', 'last-letter': 'y', 'first-letter': 'm', 'last2-letters': 'ry', 'last3-letters': 'ary', 'first3-letters': 'mar'}, gender=F\n"
],
[
"X, y = shuffle(X, y)\nX_train, X_test = X[:int(TRAIN_SPLIT * len(X))], X[int(TRAIN_SPLIT * len(X)):]\ny_train, y_test = y[:int(TRAIN_SPLIT * len(y))], y[int(TRAIN_SPLIT * len(y)):]\n\n# Check to see if the datasets add up\nprint len(X_train), len(X_test), len(y_train), len(y_test)",
"85522 9503 85522 9503\n"
],
[
"# Transforms lists of feature-value mappings to vectors.\nvectorizer = DictVectorizer()\nvectorizer.fit(X_train)\ntransformed = vectorizer.transform(features([\"Mary\", \"John\"]))\nprint transformed\n\nprint type(transformed) # <class 'scipy.sparse.csr.csr_matrix'>\nprint transformed.toarray()[0][12] # 1.0\nprint vectorizer.feature_names_[12] # first-letter=m",
" (0, 12)\t1.0\n (0, 244)\t1.0\n (0, 2766)\t1.0\n (0, 4636)\t1.0\n (0, 4955)\t1.0\n (0, 5290)\t1.0\n (1, 9)\t1.0\n (1, 198)\t1.0\n (1, 2300)\t1.0\n (1, 4625)\t1.0\n (1, 4762)\t1.0\n (1, 7416)\t1.0\n<class 'scipy.sparse.csr.csr_matrix'>\n1.0\nfirst-letter=m\n"
],
[
"clf = DecisionTreeClassifier(criterion = 'gini')\nclf.fit(vectorizer.transform(X_train), y_train)\n\n# Accuracy on training set\nprint clf.score(vectorizer.transform(X_train), y_train) \n \n# Accuracy on test set\nprint clf.score(vectorizer.transform(X_test), y_test)",
"0.9865180889127944\n0.8706724192360308\n"
],
[
"# Therefore, we are getting a decent result from the names\nprint clf.predict(vectorizer.transform(features([\"SMYSLOV\", \"CHASTITY\", \"MISS PERKY\", \"SHARON\", \"ALONSO\", \"SECONDARY OFFICER\"])))",
"['M' 'F' 'F' 'F' 'M' 'M']\n"
],
[
"# Save the model using pickle\nimport pickle",
"_____no_output_____"
],
[
"pickle_out = open(\"gender_recog.pickle\", \"wb\")\npickle.dump(clf, pickle_out)\npickle_out.close()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae442295146074aaef530bf453c89bb82e63c2a
| 3,401 |
ipynb
|
Jupyter Notebook
|
old/03-0_restore_trained_model.ipynb
|
sungjae-cho/arithmetic-neural-nets
|
3a6d4404a66f2a3db912ff00118a8ea8e53ca077
|
[
"MIT"
] | 1 |
2019-03-05T14:07:03.000Z
|
2019-03-05T14:07:03.000Z
|
old/03-0_restore_trained_model.ipynb
|
sungjae-cho/arithmetic-neural-nets
|
3a6d4404a66f2a3db912ff00118a8ea8e53ca077
|
[
"MIT"
] | null | null | null |
old/03-0_restore_trained_model.ipynb
|
sungjae-cho/arithmetic-neural-nets
|
3a6d4404a66f2a3db912ff00118a8ea8e53ca077
|
[
"MIT"
] | 1 |
2019-09-07T16:35:41.000Z
|
2019-09-07T16:35:41.000Z
| 40.011765 | 756 | 0.585416 |
[
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"dir_saved_model = 'saved_models'\nmodel_id = '20180815180530'\nmodel_to_import = '{}/{}-fnn-sigm-24-adam-lr0.001000-bs32-epoch1376-testacc1.000.ckpt'.format(model_id, model_id)\n\nsaver = tf.train.import_meta_graph('{}/{}.meta'.format(dir_saved_model, model_to_import))\n\nconfig = tf.ConfigProto()\nconfig.gpu_options.allow_growth = True\n\nwith tf.Session(config=config) as sess:\n saver.restore(sess, '{}/{}'.format(dir_saved_model, model_to_import))\n \n # End of all epochs \n # Run computing test loss, accuracy\n test_loss, summary, test_accuracy = sess.run(\n [loss, merged_summary_op, accuracy],\n feed_dict={inputs:input_dev, targets:output_dev})\n \n print(\"└ epoch: {}, step: {}, test_loss: {}, test_accuracy: {}\".format(epoch, step, test_loss, test_accuracy))\n model_name = utils.get_fnn_model_name(activation, layer_dims, str_optimizer, \n learning_rate, batch_size, 'test', test_accuracy)\n model_saver.save(sess, '{}/{}.ckpt'.format(dir_saved_models, model_name))\n print(\"Model saved.\")",
"INFO:tensorflow:Restoring parameters from saved_models/20180815180530/20180815180530-fnn-sigm-24-adam-lr0.001000-bs32-epoch1376-testacc1.000.ckpt\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4ae476addbb6f38884e0142e488b8e1c29017421
| 10,335 |
ipynb
|
Jupyter Notebook
|
hunkim_ReinforcementLearning/Lecture04.ipynb
|
Derek-tjhwang/ReinforcementLearning
|
bc9d9604ee1e5d6e0bb84a0170261c07bd048524
|
[
"Apache-2.0"
] | null | null | null |
hunkim_ReinforcementLearning/Lecture04.ipynb
|
Derek-tjhwang/ReinforcementLearning
|
bc9d9604ee1e5d6e0bb84a0170261c07bd048524
|
[
"Apache-2.0"
] | null | null | null |
hunkim_ReinforcementLearning/Lecture04.ipynb
|
Derek-tjhwang/ReinforcementLearning
|
bc9d9604ee1e5d6e0bb84a0170261c07bd048524
|
[
"Apache-2.0"
] | null | null | null | 50.169903 | 5,764 | 0.741945 |
[
[
[
"# Lecture 4. Q-Learning exploit & exploration and discouted reward",
"_____no_output_____"
],
[
"Q-Learning을 완벽하게 하는 방법에 대해서 배운다.\n\nLecture 3. 에서는 Exploit VS Exploration을 하지 않는 다는 문제점이 있었다. \n따라서 이번에는 Exploit and Exploration을 접목해서 학습 시키는 방법에 대해서 해보자.\n\nExploration : E-greedy 정책 \nDecaying E-greedy : 시간이 지날수록 Epsilon 값을 감소 시켜서 Exploration 확률을 줄이는 것 \nRandom Noise : 각각의 Argment에 Random한 값을 더해서 argmax가 다르게 나오도록 하는것.\n\nQ^hat converges to Q.",
"_____no_output_____"
]
],
[
[
"import gym\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom gym.envs.registration import register\n\nregister(\n id='FrozenLake-v3',\n entry_point='gym.envs.toy_text:FrozenLakeEnv',\n kwargs={'map_name': '4x4',\n 'is_slippery': False\n }\n)",
"_____no_output_____"
],
[
"env = gym.make('FrozenLake-v3')",
"_____no_output_____"
],
[
"Q = np.zeros([env.observation_space.n, env.action_space.n])\n\ndis = 0.99\nnum_episodes = 2000\n\nrList = []\n\nfor i in range(num_episodes):\n state = env.reset()\n rAll = 0\n done = False\n \n e = 1 / ((i / 100) +1)\n \n while not done:\n if np.random.rand(1) < e:\n action = env.action_space.sample()\n else:\n action = np.argmax(Q[state, :])\n \n #action = np.argmax(Q[state, :] + np.random.randn(1, env.action_space.n) / (i + 1))\n \n new_state, reward, done, _ = env.step(action)\n \n Q[state, action] = reward + dis * np.max(Q[new_state, :])\n \n rAll += reward\n state = new_state\n \n rList.append(rAll)",
"_____no_output_____"
],
[
"print(\"Success rate: \" + str(sum(rList)/num_episodes))\nprint(\"Final Q-Table Values\")\nprint(Q)\nplt.bar(range(len(rList)), rList, color=\"blue\")\nplt.show()",
"Success rate: 0.0\nFinal Q-Table Values\n[[0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]\n [0. 0. 0. 0.]]\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4ae47d500db3d90fb53af861dfb76358e1278b13
| 846,239 |
ipynb
|
Jupyter Notebook
|
examples/get_started.ipynb
|
Pacman1984/etna
|
9b3ccb980e576d56858f14aca2e06ce2957b0fa9
|
[
"Apache-2.0"
] | 96 |
2021-09-05T06:29:34.000Z
|
2021-11-07T15:22:54.000Z
|
examples/get_started.ipynb
|
Pacman1984/etna
|
9b3ccb980e576d56858f14aca2e06ce2957b0fa9
|
[
"Apache-2.0"
] | 188 |
2021-09-06T15:59:58.000Z
|
2021-11-17T09:34:16.000Z
|
examples/get_started.ipynb
|
Pacman1984/etna
|
9b3ccb980e576d56858f14aca2e06ce2957b0fa9
|
[
"Apache-2.0"
] | 8 |
2021-09-06T09:18:35.000Z
|
2021-11-11T21:18:39.000Z
| 869.72148 | 211,539 | 0.951754 |
[
[
[
"# Get started\n\n<a href=\"https://mybinder.org/v2/gh/tinkoff-ai/etna/master?filepath=examples/get_started.ipynb\">\n <img src=\"https://mybinder.org/badge_logo.svg\" align='left'>\n</a>",
"_____no_output_____"
],
[
"This notebook contains the simple examples of time series forecasting pipeline\nusing ETNA library.\n\n**Table of Contents**\n\n* [Creating TSDataset](#chapter1)\n* [Plotting](#chapter2)\n* [Forecast single time series](#chapter3)\n * [Simple forecast](#section_3_1)\n * [Prophet](#section_3_2)\n * [Catboost](#section_3_3)\n* [Forecast multiple time series](#chapter4)\n* [Pipeline](#chapter5)",
"_____no_output_____"
],
[
"## 1. Creating TSDataset <a class=\"anchor\" id=\"chapter1\"></a>",
"_____no_output_____"
],
[
"Let's load and look at the dataset",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"original_df = pd.read_csv(\"data/monthly-australian-wine-sales.csv\")\noriginal_df.head()",
"_____no_output_____"
]
],
[
[
"etna_ts is strict about data format:\n* column we want to predict should be called `target`\n* column with datatime data should be called `timestamp`\n* because etna is always ready to work with multiple time series, column `segment` is also compulsory\n\nOur library works with the special data structure TSDataset. So, before starting anything, we need to convert the classical DataFrame to TSDataset.\n\nLet's rename first",
"_____no_output_____"
]
],
[
[
"original_df[\"timestamp\"] = pd.to_datetime(original_df[\"month\"])\noriginal_df[\"target\"] = original_df[\"sales\"]\noriginal_df.drop(columns=[\"month\", \"sales\"], inplace=True)\noriginal_df[\"segment\"] = \"main\"\noriginal_df.head()",
"_____no_output_____"
]
],
[
[
"Time to convert to TSDataset!\n\nTo do this, we initially need to convert the classical DataFrame to the special format.",
"_____no_output_____"
]
],
[
[
"from etna.datasets.tsdataset import TSDataset",
"_____no_output_____"
],
[
"df = TSDataset.to_dataset(original_df)\ndf.head()",
"_____no_output_____"
]
],
[
[
"Now we can construct the TSDataset.\n\nAdditionally to passing dataframe we should specify frequency of our data.\nIn this case it is monthly data.",
"_____no_output_____"
]
],
[
[
"ts = TSDataset(df, freq=\"1M\")",
"/Users/y.a.shenshina/repos/etna/etna/datasets/tsdataset.py:102: UserWarning: You probably set wrong freq. Discovered freq in you data is MS, you set 1M\n warnings.warn(\n"
]
],
[
[
"Oups. Let's fix that",
"_____no_output_____"
]
],
[
[
"ts = TSDataset(df, freq=\"MS\")",
"_____no_output_____"
]
],
[
[
"We can look at the basic information about the dataset",
"_____no_output_____"
]
],
[
[
"ts.info()",
"<class 'etna.datasets.TSDataset'>\nnum_segments: 1\nnum_exogs: 0\nnum_regressors: 0\nfreq: MS\n start_timestamp end_timestamp length num_missing\nsegments \nmain 1980-01-01 1994-08-01 176 0\n"
]
],
[
[
"Or in DataFrame format",
"_____no_output_____"
]
],
[
[
"ts.describe()",
"_____no_output_____"
]
],
[
[
"## 2. Plotting <a class=\"anchor\" id=\"chapter2\"></a>\n\nLet's take a look at the time series in the dataset",
"_____no_output_____"
]
],
[
[
"ts.plot()",
"_____no_output_____"
]
],
[
[
"## 3. Forecasting single time series <a class=\"anchor\" id=\"chapter3\"></a>\n\nOur library contains a wide range of different models for time series forecasting. Let's look at some of them.",
"_____no_output_____"
],
[
"### 3.1 Simple forecast<a class=\"anchor\" id=\"section_3_1\"></a>\nLet's predict the monthly values in 1994 in our dataset using the ```NaiveModel```",
"_____no_output_____"
]
],
[
[
"train_ts, test_ts = ts.train_test_split(train_start=\"1980-01-01\",\n train_end=\"1993-12-01\",\n test_start=\"1994-01-01\",\n test_end=\"1994-08-01\")",
"_____no_output_____"
],
[
"HORIZON = 8\nfrom etna.models import NaiveModel\n\n#Fit the model\nmodel = NaiveModel(lag=12)\nmodel.fit(train_ts)\n\n#Make the forecast\nfuture_ts = train_ts.make_future(HORIZON)\nforecast_ts = model.forecast(future_ts)",
"_____no_output_____"
]
],
[
[
"Now let's look at a metric and plot the prediction.\nAll the methods already built-in in etna.",
"_____no_output_____"
]
],
[
[
"from etna.metrics import SMAPE",
"_____no_output_____"
],
[
"smape = SMAPE()\nsmape(y_true=test_ts, y_pred=forecast_ts)",
"_____no_output_____"
],
[
"from etna.analysis import plot_forecast",
"_____no_output_____"
],
[
"plot_forecast(forecast_ts, test_ts, train_ts, n_train_samples=10)",
"_____no_output_____"
]
],
[
[
"### 3.2 Prophet<a class=\"anchor\" id=\"section_3_2\"></a>\n\nNow try to improve the forecast and predict the values with the Facebook Prophet.",
"_____no_output_____"
]
],
[
[
"from etna.models import ProphetModel\n\nmodel = ProphetModel()\nmodel.fit(train_ts)\n\n#Make the forecast\nfuture_ts = train_ts.make_future(HORIZON)\nforecast_ts = model.forecast(future_ts)",
"INFO:prophet:Disabling weekly seasonality. Run prophet with weekly_seasonality=True to override this.\nINFO:prophet:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.\n"
],
[
"smape(y_true=test_ts, y_pred=forecast_ts)\n",
"_____no_output_____"
],
[
"plot_forecast(forecast_ts, test_ts, train_ts, n_train_samples=10)",
"_____no_output_____"
]
],
[
[
"### 3.2 Catboost<a class=\"anchor\" id=\"section_3_3\"></a>\nAnd finally let's try the Catboost model.\n\nAlso etna has wide range of transforms you may apply to your data.\n\nHere how it is done:",
"_____no_output_____"
]
],
[
[
"from etna.transforms import LagTransform\n\nlags = LagTransform(in_column=\"target\", lags=list(range(8, 24, 1)))\ntrain_ts.fit_transform([lags])",
"_____no_output_____"
],
[
"from etna.models import CatBoostModelMultiSegment\n\nmodel = CatBoostModelMultiSegment()\nmodel.fit(train_ts)\nfuture_ts = train_ts.make_future(HORIZON)\nforecast_ts = model.forecast(future_ts)",
"_____no_output_____"
],
[
"from etna.metrics import SMAPE\n\nsmape = SMAPE()\nsmape(y_true=test_ts, y_pred=forecast_ts)",
"_____no_output_____"
],
[
"from etna.analysis import plot_forecast\n\ntrain_ts.inverse_transform()\nplot_forecast(forecast_ts, test_ts, train_ts, n_train_samples=10)",
"_____no_output_____"
]
],
[
[
"## 4. Forecasting multiple time series <a class=\"anchor\" id=\"chapter4\"></a>\n\nIn this section you may see example of how easily etna works\nwith multiple time series and get acquainted with other transforms etna contains.",
"_____no_output_____"
]
],
[
[
"original_df = pd.read_csv(\"data/example_dataset.csv\")\noriginal_df.head()",
"_____no_output_____"
],
[
"df = TSDataset.to_dataset(original_df)\nts = TSDataset(df, freq=\"D\")\nts.plot()",
"_____no_output_____"
],
[
"ts.info()",
"<class 'etna.datasets.TSDataset'>\nnum_segments: 4\nnum_exogs: 0\nnum_regressors: 0\nfreq: D\n start_timestamp end_timestamp length num_missing\nsegments \nsegment_a 2019-01-01 2019-12-31 365 0\nsegment_b 2019-01-01 2019-12-31 365 0\nsegment_c 2019-01-01 2019-12-31 365 0\nsegment_d 2019-01-01 2019-12-31 365 0\n"
],
[
"import warnings\n\nfrom etna.transforms import MeanTransform, LagTransform, LogTransform, \\\n SegmentEncoderTransform, DateFlagsTransform, LinearTrendTransform\n\nwarnings.filterwarnings(\"ignore\")\n\nlog = LogTransform(in_column=\"target\")\ntrend = LinearTrendTransform(in_column=\"target\")\nseg = SegmentEncoderTransform()\n\nlags = LagTransform(in_column=\"target\", lags=list(range(30, 96, 1)))\nd_flags = DateFlagsTransform(day_number_in_week=True,\n day_number_in_month=True,\n week_number_in_month=True,\n week_number_in_year=True,\n month_number_in_year=True,\n year_number=True,\n special_days_in_week=[5, 6])\nmean30 = MeanTransform(in_column=\"target\", window=30)",
"_____no_output_____"
],
[
"HORIZON = 31\ntrain_ts, test_ts = ts.train_test_split(train_start=\"2019-01-01\",\n train_end=\"2019-11-30\",\n test_start=\"2019-12-01\",\n test_end=\"2019-12-31\")\ntrain_ts.fit_transform([log, trend, lags, d_flags, seg, mean30])",
"_____no_output_____"
],
[
"from etna.models import CatBoostModelMultiSegment\n\nmodel = CatBoostModelMultiSegment()\nmodel.fit(train_ts)\nfuture_ts = train_ts.make_future(HORIZON)\nforecast_ts = model.forecast(future_ts)",
"_____no_output_____"
],
[
"smape = SMAPE()\nsmape(y_true=test_ts, y_pred=forecast_ts)",
"_____no_output_____"
],
[
"train_ts.inverse_transform()\nplot_forecast(forecast_ts, test_ts, train_ts, n_train_samples=20)",
"_____no_output_____"
]
],
[
[
"## 5. Pipeline <a class=\"anchor\" id=\"chapter5\"></a>\n\nLet's wrap everything into pipeline to create the end-to-end model from previous section.",
"_____no_output_____"
]
],
[
[
"from etna.pipeline import Pipeline",
"_____no_output_____"
],
[
"train_ts, test_ts = ts.train_test_split(train_start=\"2019-01-01\",\n train_end=\"2019-11-30\",\n test_start=\"2019-12-01\",\n test_end=\"2019-12-31\")",
"_____no_output_____"
]
],
[
[
"We put: **model**, **transforms** and **horizon** in a single object, which has the similar interface with the model(fit/forecast)",
"_____no_output_____"
]
],
[
[
"model = Pipeline(model=CatBoostModelMultiSegment(),\n transforms=[log, trend, lags, d_flags, seg, mean30],\n horizon=HORIZON)\nmodel.fit(train_ts)\nforecast_ts = model.forecast()",
"_____no_output_____"
]
],
[
[
"As in the previous section, let's calculate the metrics and plot the forecast",
"_____no_output_____"
]
],
[
[
"smape = SMAPE()\nsmape(y_true=test_ts, y_pred=forecast_ts)",
"_____no_output_____"
],
[
"plot_forecast(forecast_ts, test_ts, train_ts, n_train_samples=20)\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4ae48497f0c4bb68513b349b6a852fbf115ba471
| 234,407 |
ipynb
|
Jupyter Notebook
|
doc/source/user_guide/numpyro_refitting_xr_lik.ipynb
|
Divyateja04/arviz
|
a911a0f7f3c476058084d129ac4a7202d84c1a22
|
[
"Apache-2.0"
] | null | null | null |
doc/source/user_guide/numpyro_refitting_xr_lik.ipynb
|
Divyateja04/arviz
|
a911a0f7f3c476058084d129ac4a7202d84c1a22
|
[
"Apache-2.0"
] | null | null | null |
doc/source/user_guide/numpyro_refitting_xr_lik.ipynb
|
Divyateja04/arviz
|
a911a0f7f3c476058084d129ac4a7202d84c1a22
|
[
"Apache-2.0"
] | null | null | null | 48.946962 | 19,272 | 0.550193 |
[
[
[
"# Refitting NumPyro models with ArviZ (and xarray)\n\nArviZ is backend agnostic and therefore does not sample directly. In order to take advantage of algorithms that require refitting models several times, ArviZ uses `SamplingWrappers` to convert the API of the sampling backend to a common set of functions. Hence, functions like Leave Future Out Cross Validation can be used in ArviZ independently of the sampling backend used.",
"_____no_output_____"
],
[
"Below there is one example of `SamplingWrapper` usage for NumPyro.",
"_____no_output_____"
]
],
[
[
"import arviz as az\nimport numpyro\nimport numpyro.distributions as dist\nimport jax.random as random\nfrom numpyro.infer import MCMC, NUTS\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport scipy.stats as stats\nimport xarray as xr",
"_____no_output_____"
],
[
"numpyro.set_host_device_count(4)",
"_____no_output_____"
]
],
[
[
"For the example we will use a linear regression.",
"_____no_output_____"
]
],
[
[
"np.random.seed(26)\n\nxdata = np.linspace(0, 50, 100)\nb0, b1, sigma = -2, 1, 3\nydata = np.random.normal(loc=b1 * xdata + b0, scale=sigma)",
"_____no_output_____"
],
[
"plt.plot(xdata, ydata)",
"_____no_output_____"
]
],
[
[
"Now we will write the NumPyro code:",
"_____no_output_____"
]
],
[
[
"def model(N, x, y=None):\n b0 = numpyro.sample(\"b0\", dist.Normal(0, 10))\n b1 = numpyro.sample(\"b1\", dist.Normal(0, 10))\n sigma_e = numpyro.sample(\"sigma_e\", dist.HalfNormal(10))\n numpyro.sample(\"y\", dist.Normal(b0 + b1 * x, sigma_e), obs=y)",
"_____no_output_____"
],
[
"data_dict = {\n \"N\": len(ydata),\n \"y\": ydata,\n \"x\": xdata,\n}\nkernel = NUTS(model)\nsample_kwargs = dict(\n sampler=kernel, \n num_warmup=1000, \n num_samples=1000, \n num_chains=4, \n chain_method=\"parallel\"\n)\nmcmc = MCMC(**sample_kwargs)\nmcmc.run(random.PRNGKey(0), **data_dict)",
"_____no_output_____"
]
],
[
[
"We have defined a dictionary `sample_kwargs` that will be passed to the `SamplingWrapper` in order to make sure that all refits use the same sampler parameters. We follow the same pattern with `az.from_numpyro`.",
"_____no_output_____"
]
],
[
[
"dims = {\"y\": [\"time\"], \"x\": [\"time\"]}\nidata_kwargs = {\n \"dims\": dims,\n \"constant_data\": {\"x\": xdata}\n}\nidata = az.from_numpyro(mcmc, **idata_kwargs)\ndel idata.log_likelihood\nidata",
"_____no_output_____"
]
],
[
[
"We are now missing the `log_likelihood` group because we have not used the `log_likelihood` argument in `idata_kwargs`. We are doing this to ease the job of the sampling wrapper. Instead of going out of our way to get Stan to calculate the pointwise log likelihood values for each refit and for the excluded observation at every refit, we will compromise and manually write a function to calculate the pointwise log likelihood.\n\nEven though it is not ideal to lose part of the straight out of the box capabilities of PyStan-ArviZ integration, this should generally not be a problem. We are basically moving the pointwise log likelihood calculation from the Stan code to the Python code, in both cases we need to manually write the function to calculate the pointwise log likelihood.\n\nMoreover, the Python computation could even be written to be compatible with Dask. Thus it will work even in cases where the large number of observations makes it impossible to store pointwise log likelihood values (with shape `n_samples * n_observations`) in memory.",
"_____no_output_____"
]
],
[
[
"def calculate_log_lik(x, y, b0, b1, sigma_e):\n mu = b0 + b1 * x\n return stats.norm(mu, sigma_e).logpdf(y)",
"_____no_output_____"
]
],
[
[
"This function should work for any shape of the input arrays as long as their shapes are compatible and can broadcast. There is no need to loop over each draw in order to calculate the pointwise log likelihood using scalars.\n\nTherefore, we can use `xr.apply_ufunc` to handle the broadasting and preserve the dimension names:",
"_____no_output_____"
]
],
[
[
"log_lik = xr.apply_ufunc(\n calculate_log_lik,\n idata.constant_data[\"x\"],\n idata.observed_data[\"y\"],\n idata.posterior[\"b0\"],\n idata.posterior[\"b1\"],\n idata.posterior[\"sigma_e\"],\n)\nidata.add_groups(log_likelihood=log_lik)",
"_____no_output_____"
]
],
[
[
"The first argument is the function, followed by as many positional arguments as needed by the function, 5 in our case. As this case does not have many different dimensions nor combinations of these, we do not need to use any extra kwargs passed to [`xr.apply_ufunc`](http://xarray.pydata.org/en/stable/generated/xarray.apply_ufunc.html#xarray.apply_ufunc).\n\nWe are now passing the arguments to `calculate_log_lik` initially as `xr.DataArrays`. What is happening here behind the scenes is that `xr.apply_ufunc` is broadcasting and aligning the dimensions of all the DataArrays involved and afterwards passing numpy arrays to `calculate_log_lik`. Everything works automagically. \n\nNow let's see what happens if we were to pass the arrays directly to `calculate_log_lik` instead:",
"_____no_output_____"
]
],
[
[
"calculate_log_lik(\n idata.constant_data[\"x\"].values,\n idata.observed_data[\"y\"].values,\n idata.posterior[\"b0\"].values,\n idata.posterior[\"b1\"].values,\n idata.posterior[\"sigma_e\"].values\n)",
"_____no_output_____"
]
],
[
[
"If you are still curious about the magic of xarray and `xr.apply_ufunc`, you can also try to modify the `dims` used to generate the InferenceData a couple cells before:\n\n dims = {\"y\": [\"time\"], \"x\": [\"time\"]}\n \nWhat happens to the result if you use a different name for the dimension of `x`?",
"_____no_output_____"
]
],
[
[
"idata",
"_____no_output_____"
]
],
[
[
"We will create a subclass of `az.SamplingWrapper`. Therefore, instead of having to implement all functions required by `az.reloo` we only have to implement `sel_observations` (we are cloning `sample` and `get_inference_data` from the `PyStanSamplingWrapper` in order to use `apply_ufunc` instead of assuming the log likelihood is calculated within Stan). \n\nNote that of the 2 outputs of `sel_observations`, `data__i` is a dictionary because it is an argument of `sample` which will pass it as is to `model.sampling`, whereas `data_ex` is a list because it is an argument to `log_likelihood__i` which will pass it as `*data_ex` to `apply_ufunc`. More on `data_ex` and `apply_ufunc` integration below.",
"_____no_output_____"
]
],
[
[
"class NumPyroSamplingWrapper(az.SamplingWrapper):\n def __init__(self, model, **kwargs): \n self.rng_key = kwargs.pop(\"rng_key\", random.PRNGKey(0))\n \n super(NumPyroSamplingWrapper, self).__init__(model, **kwargs)\n \n def sample(self, modified_observed_data):\n self.rng_key, subkey = random.split(self.rng_key)\n mcmc = MCMC(**self.sample_kwargs)\n mcmc.run(subkey, **modified_observed_data)\n return mcmc\n\n def get_inference_data(self, fit):\n # Cloned from PyStanSamplingWrapper.\n idata = az.from_numpyro(mcmc, **self.idata_kwargs)\n return idata\n \nclass LinRegWrapper(NumPyroSamplingWrapper):\n def sel_observations(self, idx):\n xdata = self.idata_orig.constant_data[\"x\"]\n ydata = self.idata_orig.observed_data[\"y\"]\n mask = np.isin(np.arange(len(xdata)), idx)\n # data__i is passed to numpyro to sample on it -> dict of numpy array\n # data_ex is passed to apply_ufunc -> list of DataArray\n data__i = {\"x\": xdata[~mask].values, \"y\": ydata[~mask].values, \"N\": len(ydata[~mask])}\n data_ex = [xdata[mask], ydata[mask]]\n return data__i, data_ex\n",
"_____no_output_____"
],
[
"loo_orig = az.loo(idata, pointwise=True)\nloo_orig",
"_____no_output_____"
]
],
[
[
"In this case, the Leave-One-Out Cross Validation (LOO-CV) approximation using Pareto Smoothed Importance Sampling (PSIS) works for all observations, so we will use modify `loo_orig` in order to make `az.reloo` believe that PSIS failed for some observations. This will also serve as a validation of our wrapper, as the PSIS LOO-CV already returned the correct value.",
"_____no_output_____"
]
],
[
[
"loo_orig.pareto_k[[13, 42, 56, 73]] = np.array([0.8, 1.2, 2.6, 0.9])",
"_____no_output_____"
]
],
[
[
"We initialize our sampling wrapper. Let's stop and analyze each of the arguments. \n\nWe then use the `log_lik_fun` and `posterior_vars` argument to tell the wrapper how to call `xr.apply_ufunc`. `log_lik_fun` is the function to be called, which is then called with the following positional arguments:\n\n log_lik_fun(*data_ex, *[idata__i.posterior[var_name] for var_name in posterior_vars]\n \nwhere `data_ex` is the second element returned by `sel_observations` and `idata__i` is the InferenceData object result of `get_inference_data` which contains the fit on the subsetted data. We have generated `data_ex` to be a tuple of DataArrays so it plays nicely with this call signature.\n\nWe use `idata_orig` as a starting point, and mostly as a source of observed and constant data which is then subsetted in `sel_observations`.\n\nFinally, `sample_kwargs` and `idata_kwargs` are used to make sure all refits and corresponding InferenceData are generated with the same properties.",
"_____no_output_____"
]
],
[
[
"pystan_wrapper = LinRegWrapper(\n mcmc, \n rng_key=random.PRNGKey(7),\n log_lik_fun=calculate_log_lik, \n posterior_vars=(\"b0\", \"b1\", \"sigma_e\"),\n idata_orig=idata, \n sample_kwargs=sample_kwargs, \n idata_kwargs=idata_kwargs\n)",
"_____no_output_____"
]
],
[
[
"And eventually, we can use this wrapper to call `az.reloo`, and compare the results with the PSIS LOO-CV results.",
"_____no_output_____"
]
],
[
[
"loo_relooed = az.reloo(pystan_wrapper, loo_orig=loo_orig)",
"/home/oriol/miniconda3/envs/arviz/lib/python3.8/site-packages/arviz/stats/stats_refitting.py:99: UserWarning: reloo is an experimental and untested feature\n warnings.warn(\"reloo is an experimental and untested feature\", UserWarning)\narviz.stats.stats_refitting - INFO - Refitting model excluding observation 13\nINFO:arviz.stats.stats_refitting:Refitting model excluding observation 13\narviz.stats.stats_refitting - INFO - Refitting model excluding observation 42\nINFO:arviz.stats.stats_refitting:Refitting model excluding observation 42\narviz.stats.stats_refitting - INFO - Refitting model excluding observation 56\nINFO:arviz.stats.stats_refitting:Refitting model excluding observation 56\narviz.stats.stats_refitting - INFO - Refitting model excluding observation 73\nINFO:arviz.stats.stats_refitting:Refitting model excluding observation 73\n"
],
[
"loo_relooed",
"_____no_output_____"
],
[
"loo_orig",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4ae49198f21fa9493c9cc6f4bb568b38aadc9d1b
| 82,752 |
ipynb
|
Jupyter Notebook
|
docs/Miscellaneous/sigfuncs.ipynb
|
SachsLab/indl
|
531d2e0c2ee765004aedc553af40e258262f86cb
|
[
"Apache-2.0"
] | 1 |
2021-02-22T01:39:50.000Z
|
2021-02-22T01:39:50.000Z
|
docs/Miscellaneous/sigfuncs.ipynb
|
SachsLab/indl
|
531d2e0c2ee765004aedc553af40e258262f86cb
|
[
"Apache-2.0"
] | null | null | null |
docs/Miscellaneous/sigfuncs.ipynb
|
SachsLab/indl
|
531d2e0c2ee765004aedc553af40e258262f86cb
|
[
"Apache-2.0"
] | null | null | null | 544.421053 | 39,224 | 0.94718 |
[
[
[
"# Signal Functions\n\nSome useful signal functions.\n* **sigmoid**\n* **minimum_jerk**",
"_____no_output_____"
]
],
[
[
"from indl.misc.sigfuncs import sigmoid\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nx = np.arange(-6, 6, 0.1)\n\nplt.subplot(1, 2, 1)\nfor B in [0.5, 1, 2, 5, 10]:\n plt.plot(x, sigmoid(x, B=B), label=f\"B={B}\")\nplt.legend()\n\nplt.subplot(1, 2, 2)\nfor x_offset in [-3.0, -1.5, 0, 1.5, 3.0]:\n plt.plot(x, sigmoid(x, x_offset=x_offset), label=f\"{x_offset}\")\nplt.legend()\n\nplt.show()\n",
"_____no_output_____"
],
[
"a = np.array([[0.2, 0.5]]).T\nsigmoid(x, A=a).shape\n",
"_____no_output_____"
],
[
"from indl.misc.sigfuncs import minimum_jerk\nx = np.arange(0, 6.0, 0.1)\nfor degree in [0, 1, 2]:\n plt.plot(x, minimum_jerk(x, degree=degree), label=f\"{degree}\")\nplt.legend()\nplt.show()\n",
"_____no_output_____"
],
[
"a = np.random.rand(5, 2)\nY = minimum_jerk(x, a0=a[:, 0], af=a[:, 1], degree=0)\nplt.plot(x, Y)\nplt.show()\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4ae4b22396411bccbc158029a04a3ab156992aa5
| 8,491 |
ipynb
|
Jupyter Notebook
|
coalescent/.ipynbs/alignment_distance.ipynb
|
kdmurray91/kwip-experiments
|
7a8e8778e0f5ae39d2dd123d9e4216a475d62ba0
|
[
"MIT"
] | 1 |
2020-07-07T04:50:20.000Z
|
2020-07-07T04:50:20.000Z
|
coalescent/.ipynbs/alignment_distance.ipynb
|
kdmurray91/kwip-experiments
|
7a8e8778e0f5ae39d2dd123d9e4216a475d62ba0
|
[
"MIT"
] | null | null | null |
coalescent/.ipynbs/alignment_distance.ipynb
|
kdmurray91/kwip-experiments
|
7a8e8778e0f5ae39d2dd123d9e4216a475d62ba0
|
[
"MIT"
] | null | null | null | 17.800839 | 101 | 0.511365 |
[
[
[
"import numpy as np",
"_____no_output_____"
],
[
"import treedist\nfrom skbio import TreeNode, DistanceMatrix, TabularMSA, DNA\nfrom scipy.spatial.distance import hamming\nimport ete3",
"_____no_output_____"
],
[
"#aln = TabularMSA.read('../runs/run_2016-03-17/all_genomes-0.1.fasta', constructor=DNA)\naln = TabularMSA.read('../runs/run_2016-03-17/ali', constructor=DNA)",
"_____no_output_____"
],
[
"ad = DistanceMatrix.from_iterable([seq.values for seq in aln], metric=hamming, keys=aln.index) ",
"_____no_output_____"
],
[
"t = ete3.Tree('../runs/run_2016-03-17/data/125467289/population.nwk')",
"_____no_output_____"
],
[
"import string\n\nstring.ascii_uppercase",
"_____no_output_____"
],
[
"_, maxdist = t.get_farthest_leaf()\nmaxdist",
"_____no_output_____"
],
[
"list(sorted(t.get_leaf_names()))",
"_____no_output_____"
],
[
"from math import ceil, log",
"_____no_output_____"
],
[
"ceil(log(101)/log(10))",
"_____no_output_____"
],
[
"import itertools as itl\n\nitl.product(string.ascii_uppercase, )\n",
"_____no_output_____"
],
[
"?itl.product",
"_____no_output_____"
],
[
"t = ete3.Tree('../runs/run_2016-03-17/data/125467289/population.nwk')a\n",
"_____no_output_____"
],
[
"dists = []\nfor a, b in itl.combinations(t.get_leaf_names(), 2):\n a = t&a\n dists.append(a.get_distance(b))",
"_____no_output_____"
],
[
"from statistics import median, mean",
"_____no_output_____"
],
[
"mean(dists)",
"_____no_output_____"
],
[
"t.get_farthest_leaf()",
"_____no_output_____"
],
[
"def pwdist()",
"_____no_output_____"
],
[
"t = ete3.Tree('../runs/run_2016-03-17/data/125467289/population.nwk')\n_, maxdist = t.get_farthest_leaf()\nfor ch in t.iter_descendants():\n ch.dist /= maxdist",
"_____no_output_____"
],
[
"t.write(format=5)",
"_____no_output_____"
],
[
"t.write?",
"_____no_output_____"
],
[
"#t.convert_to_ultrametric()\nprint(t.get_ascii())\n\nx = t.describe()",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"l = list(t.iter_descendants())\na = l[0]\na.dist /= t.describe(",
"_____no_output_____"
],
[
"t = TreeNode.read('../runs/run_2016-03-17/data/125467289/population.nwk')",
"_____no_output_____"
],
[
"t.tip_tip_distances()",
"_____no_output_____"
],
[
"print(t.ascii_art())",
"_____no_output_____"
],
[
"d = DistanceMatrix.read('../runs/run_2016-03-17/data/125467289/kwip/100x-0.01-wip.dist')\nd.filter(sorted(d.ids))",
"_____no_output_____"
],
[
"treedist.partition_weighted_distance('../runs/run_2016-03-17/data/125467289/population.nwk')",
"_____no_output_____"
],
[
"from glob import glob",
"_____no_output_____"
],
[
"distfiles = glob(\"../runs/2016-03-15_genomics-nci/125467289/kwip/*.dist\")",
"_____no_output_____"
],
[
"truth = treedist.get_truth(treef, 3)",
"_____no_output_____"
],
[
"truth",
"_____no_output_____"
],
[
"treef = '../runs/2016-03-15_genomics-nci/125467289/population.nwk'",
"_____no_output_____"
],
[
"treedist.get_table(treef, distfiles)k",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae4bba4114119904f3bd1a63aa850662aae4c72
| 2,743 |
ipynb
|
Jupyter Notebook
|
misc/notes/Databases/notes/Nested Routes.ipynb
|
alittlebirdie00/coma
|
4670e4be2f687f48b9c75118788aa7f90b72b5d4
|
[
"MIT"
] | 2 |
2018-09-16T21:39:24.000Z
|
2019-05-03T06:24:18.000Z
|
misc/notes/Databases/notes/Nested Routes.ipynb
|
alittlebirdie00/coma
|
4670e4be2f687f48b9c75118788aa7f90b72b5d4
|
[
"MIT"
] | 46 |
2018-08-28T13:38:04.000Z
|
2019-10-02T18:54:51.000Z
|
misc/notes/Databases/notes/Nested Routes.ipynb
|
alittlebirdie00/coma
|
4670e4be2f687f48b9c75118788aa7f90b72b5d4
|
[
"MIT"
] | 1 |
2018-08-30T11:08:31.000Z
|
2018-08-30T11:08:31.000Z
| 47.293103 | 281 | 0.509661 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4ae4bbf9403b97430477355177705a37969f15f9
| 11,456 |
ipynb
|
Jupyter Notebook
|
helloworld/notebooks/OpIris.ipynb
|
michaelweilsalesforce/TransmogrifAI
|
07fd316d8090a4a321d377baf09471815b44f808
|
[
"BSD-3-Clause"
] | 1 |
2020-04-08T03:50:59.000Z
|
2020-04-08T03:50:59.000Z
|
helloworld/notebooks/OpIris.ipynb
|
michaelweilsalesforce/TransmogrifAI
|
07fd316d8090a4a321d377baf09471815b44f808
|
[
"BSD-3-Clause"
] | null | null | null |
helloworld/notebooks/OpIris.ipynb
|
michaelweilsalesforce/TransmogrifAI
|
07fd316d8090a4a321d377baf09471815b44f808
|
[
"BSD-3-Clause"
] | null | null | null | 33.694118 | 432 | 0.644728 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4ae4bc904c46c2ad0fed7ba39176043b1b313646
| 104,241 |
ipynb
|
Jupyter Notebook
|
create_data.ipynb
|
kaphka/ml-software
|
03c4c0023be3e10fcb9a625796142ed4605e8104
|
[
"Apache-2.0"
] | null | null | null |
create_data.ipynb
|
kaphka/ml-software
|
03c4c0023be3e10fcb9a625796142ed4605e8104
|
[
"Apache-2.0"
] | null | null | null |
create_data.ipynb
|
kaphka/ml-software
|
03c4c0023be3e10fcb9a625796142ed4605e8104
|
[
"Apache-2.0"
] | null | null | null | 209.319277 | 72,842 | 0.888393 |
[
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport pickle\nimport functools as ft",
"_____no_output_____"
],
[
"X = x_train[1]\nX",
"_____no_output_____"
],
[
"not ft.reduce(lambda old, new: old == new,X >= 0)",
"_____no_output_____"
]
],
[
[
"## XOR",
"_____no_output_____"
]
],
[
[
"def xor(X):\n if not ft.reduce(lambda old, new: old == new,X >= 0):\n return 1\n else:\n return 0 \n \nx_train = np.array([(np.random.random_sample(5000) - 0.5) * 2 for dim in range(2)]).transpose()\nx_test = np.array([(np.random.random_sample(100) - 0.5) * 2 for dim in range(2)]).transpose()\ny_train = np.apply_along_axis(xor, 1, x_train)\ny_test = np.apply_along_axis(xor, 1, x_test)\nwith open('data/xor.tuple', 'wb') as xtuple:\n pickle.dump((x_train, y_train, x_test, y_test), xtuple)",
"_____no_output_____"
]
],
[
[
"## Multivariante Regression - Housing Data Set\nhttps://archive.ics.uci.edu/ml/datasets/Housing\n1. CRIM: per capita crime rate by town \n2. ZN: proportion of residential land zoned for lots over 25,000 sq.ft. \n3. INDUS: proportion of non-retail business acres per town \n4. CHAS: Charles River dummy variable (= 1 if tract bounds river; 0 otherwise) \n5. NOX: nitric oxides concentration (parts per 10 million) \n6. RM: average number of rooms per dwelling \n7. AGE: proportion of owner-occupied units built prior to 1940 \n8. DIS: weighted distances to five Boston employment centres \n9. RAD: index of accessibility to radial highways \n10. TAX: full-value property-tax rate per \\$10,000 \n11. PTRATIO: pupil-teacher ratio by town \n12. B: 1000(Bk - 0.63)^2 where Bk is the proportion of blacks by town \n13. LSTAT: \\% lower status of the population \n14. MEDV: Median value of owner-occupied homes in $1000's",
"_____no_output_____"
]
],
[
[
"!wget -P data/ https://archive.ics.uci.edu/ml/machine-learning-databases/housing/housing.data",
"--2016-07-13 19:47:59-- https://archive.ics.uci.edu/ml/machine-learning-databases/housing/housing.data\nAuflösen des Hostnamens »archive.ics.uci.edu (archive.ics.uci.edu)« … 128.195.10.249\nVerbindungsaufbau zu archive.ics.uci.edu (archive.ics.uci.edu)|128.195.10.249|:443 … verbunden.\nHTTP-Anforderung gesendet, auf Antwort wird gewartet … 200 OK\nLänge: 49082 (48K) [text/plain]\nWird in »»data/housing.data«« gespeichert.\n\nhousing.data 100%[===================>] 47.93K 133KB/s in 0.4s \n\n2016-07-13 19:48:00 (133 KB/s) - »data/housing.data« gespeichert [49082/49082]\n\n"
],
[
"housing = pd.read_csv('data/housing.data', delim_whitespace=True, \n names=['CRIM', \n 'ZM', \n 'INDUS', \n 'CHAS', \n 'NOX', \n 'RM', \n 'AGE', \n 'DIS', \n 'RAD',\n 'TAX',\n 'PTRATIO',\n 'B',\n 'LSTAT',\n 'MEDV'])\nhousing.head()\nwith open('data/housing.dframe', 'wb') as dhousing:\n pickle.dump(housing, dhousing)",
"_____no_output_____"
]
],
[
[
"## Binary Classification - Pima Indians Diabetes Data Set\nhttps://archive.ics.uci.edu/ml/datasets/Pima+Indians+Diabetes\n1. Number of times pregnant \n2. Plasma glucose concentration a 2 hours in an oral glucose tolerance test \n3. Diastolic blood pressure (mm Hg) \n4. Triceps skin fold thickness (mm) \n5. 2-Hour serum insulin (mu U/ml) \n6. Body mass index (weight in kg/(height in m)^2) \n7. Diabetes pedigree function \n8. Age (years) \n9. Class variable (0 or 1) ",
"_____no_output_____"
]
],
[
[
"!wget -P data/ https://archive.ics.uci.edu/ml/machine-learning-databases/pima-indians-diabetes/pima-indians-diabetes.data",
"--2016-07-13 19:28:32-- https://archive.ics.uci.edu/ml/machine-learning-databases/pima-indians-diabetes/pima-indians-diabetes.data\nAuflösen des Hostnamens »archive.ics.uci.edu (archive.ics.uci.edu)« … 128.195.10.249\nVerbindungsaufbau zu archive.ics.uci.edu (archive.ics.uci.edu)|128.195.10.249|:443 … verbunden.\nHTTP-Anforderung gesendet, auf Antwort wird gewartet … 200 OK\nLänge: 23279 (23K) [text/plain]\nWird in »»data/pima-indians-diabetes.data«« gespeichert.\n\npima-indians-diabet 100%[===================>] 22.73K 129KB/s in 0.2s \n\n2016-07-13 19:28:33 (129 KB/s) - »data/pima-indians-diabetes.data« gespeichert [23279/23279]\n\n"
],
[
"data = pd.read_csv('data/pima-indians-diabetes.data',\n names=['n_pregnant', \n 'glucose', \n 'mmHg', \n 'triceps', \n 'insulin', \n 'BMI', \n 'pedigree', \n 'age', \n 'class'])\ndata.head()\nx = np.array(data)[:,:-1]\ny = np.array(data)[:,-1]\nn_train = int(len(x) * 0.70)\nx_train = x[:n_train]\nx_test = x[n_train:]\ny_train = y[:n_train]\ny_test = y[n_train:]\nwith open('data/pima-indians-diabetes.tuple', 'wb') as xtuple:\n pickle.dump((x_train, y_train, x_test, y_test), xtuple)",
"_____no_output_____"
]
],
[
[
"## Image Classification - MNIST dataset\nhttp://deeplearning.net/data/mnist/mnist.pkl.gz",
"_____no_output_____"
]
],
[
[
"!wget -P data/ http://deeplearning.net/data/mnist/mnist.pkl.gz",
"--2016-07-13 15:39:19-- http://deeplearning.net/data/mnist/mnist.pkl.gz\nAuflösen des Hostnamens »deeplearning.net (deeplearning.net)« … 132.204.26.28\nVerbindungsaufbau zu deeplearning.net (deeplearning.net)|132.204.26.28|:80 … verbunden.\nHTTP-Anforderung gesendet, auf Antwort wird gewartet … 200 OK\nLänge: 16168813 (15M) [application/x-gzip]\nWird in »»data/mnist.pkl.gz«« gespeichert.\n\nmnist.pkl.gz 100%[===================>] 15.42M 3.95MB/s in 5.7s \n\n2016-07-13 15:39:25 (2.71 MB/s) - »data/mnist.pkl.gz« gespeichert [16168813/16168813]\n\n"
],
[
"import cPickle, gzip, numpy\n\n# Load the dataset\nf = gzip.open('data/mnist.pkl.gz', 'rb')\ntrain_set, valid_set, test_set = cPickle.load(f)\nf.close()",
"_____no_output_____"
],
[
"plt.imshow(train_set[0][0].reshape((28,28)),cmap='gray', interpolation=None)",
"_____no_output_____"
],
[
"!wget -P data/ http://data.dmlc.ml/mxnet/data/mnist.zip\n!unzip -d data/ -u data/mnist.zip",
"--2016-07-14 02:04:00-- http://data.dmlc.ml/mxnet/data/mnist.zip\nAuflösen des Hostnamens »data.dmlc.ml (data.dmlc.ml)« … 128.2.209.42\nVerbindungsaufbau zu data.dmlc.ml (data.dmlc.ml)|128.2.209.42|:80 … verbunden.\nHTTP-Anforderung gesendet, auf Antwort wird gewartet … 200 OK\nLänge: 11595270 (11M) [application/zip]\nWird in »»data/mnist.zip«« gespeichert.\n\nmnist.zip 100%[===================>] 11.06M 2.02MB/s in 8.7s \n\n2016-07-14 02:04:09 (1.26 MB/s) - »data/mnist.zip« gespeichert [11595270/11595270]\n\nArchive: data/mnist.zip\n inflating: data/t10k-images-idx3-ubyte \n inflating: data/t10k-labels-idx1-ubyte \n inflating: data/train-images-idx3-ubyte \n inflating: data/train-labels-idx1-ubyte \n"
]
],
[
[
"## Image Classification - CIFAR-10 dataset\nhttps://www.cs.toronto.edu/~kriz/cifar.html",
"_____no_output_____"
]
],
[
[
"!wget -P data/ https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz\n!tar -xzf data/cifar-10-python.tar.gz -C data/",
"--2016-07-13 15:27:29-- https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz\nAuflösen des Hostnamens »www.cs.toronto.edu (www.cs.toronto.edu)« … 128.100.3.30\nVerbindungsaufbau zu www.cs.toronto.edu (www.cs.toronto.edu)|128.100.3.30|:443 … verbunden.\nHTTP-Anforderung gesendet, auf Antwort wird gewartet … 200 OK\nLänge: 170498071 (163M) [application/x-gzip]\nWird in »»data/cifar-10-python.tar.gz«« gespeichert.\n\ncifar-10-python.tar 100%[===================>] 162.60M 2.12MB/s in 82s \n\n2016-07-13 15:28:51 (1.99 MB/s) - »data/cifar-10-python.tar.gz« gespeichert [170498071/170498071]\n\n"
],
[
"with open('data/cifar-10-batches-py/data_batch_1', 'rb') as batch:\n cifar1 = cPickle.load(batch)",
"_____no_output_____"
],
[
"cifar1.keys()",
"_____no_output_____"
],
[
"img = np.stack([cifar1['data'][0].reshape((3,32,32))[0,:,:],\n cifar1['data'][0].reshape((3,32,32))[1,:,:],\n cifar1['data'][0].reshape((3,32,32))[2,:,:]],axis=2)\nplt.imshow(img, cmap='gray')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4ae4d8877a179cf209985f879be9805601a6c816
| 14,964 |
ipynb
|
Jupyter Notebook
|
Lectures/Addition/To Lecture 1 - 1 What is a tensor.ipynb
|
lev1khachatryan/ASDS_CV
|
c9f0c0412002e929bcb7cc2fc6e5392977a9fa76
|
[
"MIT"
] | 5 |
2019-12-13T16:26:10.000Z
|
2020-01-10T07:44:05.000Z
|
Lectures/Addition/To Lecture 1 - 1 What is a tensor.ipynb
|
lev1khachatryan/ASDS_CV
|
c9f0c0412002e929bcb7cc2fc6e5392977a9fa76
|
[
"MIT"
] | 1 |
2020-01-07T16:48:21.000Z
|
2020-03-18T18:43:37.000Z
|
Lectures/Addition/To Lecture 1 - 1 What is a tensor.ipynb
|
lev1khachatryan/ASDS_CV
|
c9f0c0412002e929bcb7cc2fc6e5392977a9fa76
|
[
"MIT"
] | null | null | null | 38.369231 | 928 | 0.610465 |
[
[
[
"# <div align=\"center\">What is a Tensor</div>\n---------------------------------------------------------------------\n\nyou can Find me on Github:\n> ###### [ GitHub](https://github.com/lev1khachatryan)",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"***Tensors are not generalizations of vectors***. It’s very slightly more understandable to say that tensors are generalizations of matrices, in the same way that it is slightly more accurate to say “vanilla ice cream is a generalization of chocolate ice cream” than it is to say that “vanilla ice cream is a generalization of dessert”, closer, but still false. Vanilla and Chocolate are both ice cream, but chocolate ice cream is not a type of vanilla ice cream, and “dessert” certainly isn’t a type of vanilla ice cream. In fact, technically, ***vectors are generalizations of tensors.*** What we generally think of as vectors are geometrical points in space, and we normally represent them as an array of numbers. That array of numbers is what people are referring to when they say \"Tensors are generalizations of vectors\", but really, even this adjusted claim is fundamentally false and extremely misleading.\n\nAt first let's define what is a vector.",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"## Definition of a vector space",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"The set ***V*** is a vector space with respect to the operations + (which is any operation that maps two elements of the space to another element of the space, not necessarily addition) and * (which is any operation that maps an element in the space and a scalar to another element in the space, not necessarily multiplication) if and only if, for every $x,y,z ∈ V$ and $a,b ∈ R$\n\n* \\+ is commutative, that is x+y=y+x\n\n\n* \\+ is associative, that is (x+y)+z=x+(y+z)\n\n\n* There exists an identity element in the space, that is there exists an element 0 such that x+0=x\n\n\n* Every element has an inverse, that is for every element x there exists an element −x such that x+−x=0\n\n\n* \\* is associative, that is a(b∗x)=(ab)∗x\n\n\n* There is scalar distributivity over +, that is a∗(x+y)=a∗x+a∗y\n\n\n* There is vector distributivity over scalar addition, that is (a+b)∗x=a∗x+b∗x\n\n\n* And finally, 1∗x=x (it can be obtained from above mentioned 7 points)",
"_____no_output_____"
],
[
"***A vector is defined as a member of such a space***. Notice how nothing here is explicitly stated to be numerical. We could be talking about colors, or elephants, or glasses of milk; as long as we meaningfully define these two operations, anything can be a vector. The special case of vectors that we usually think about in physics and geometry satisfy this definition ( i.e. points in space or “arrows”). Thus, “arrows” are special cases of vectors. More formally, every “arrow” v represents the line segment from 0, \"the origin\", which is the identity element of the vector space, to some other point in space. In this view, you can construct a vector space of “arrows” by first picking a point in space, and taking the set of all line segments from that point. (From now on, I will use the term “arrows” to formally distinguish between formal vectors and the type of vectors that have “magnitude and direction”.)\n\nOkay, so anyone trying to understand tensors probably already knows this stuff.\n\nBut here is something you may not have heard about before if you are learning about tensors. When we define a vector space like this, we generally find that it is natural to define an operation that gives us lengths and angles. ***A vector space with lengths and angles is called an inner product space.***",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"## Definition of a Inner product space",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"An inner product space is a vector space V with an additional operation ***⋅*** such that, for all x,y,z ∈ V\n\n* x⋅y ∈ R\n\n\n* x⋅x ≥ 0\n\n\n* x⋅x=0 ⟺ x=0\n\n\n* x⋅(ay)=a(x⋅y)\n\n\n* x⋅y=y⋅x\n\n\n* x⋅(y+z)=x⋅y+x⋅z\n\nWe define the length of a vector x in an inner product space to be $||x|| = \\sqrt[2]{x⋅x}$ , and the angle between two vectors x,y to be $arccos(\\frac{x⋅y}{||x||||y||}).$\n\nThis is the equivalent of the dot product, which is defined to be $||x||||y||cos(θ)$, but note that this is not defined in terms of any sort of \"components\" of the vector, there are no arrays of numbers mentioned. I.e. the dot product is a geometrical operation.\n\nSo I have secretly given you your first glimpse at a tensor. Where was it? Was it x? Was it y? Was it V? Was it the glass of milk???\n\nIt was none of these things; ***it was the operation itself . The dot product itself is an example of a tensor.***\n\nWell, again, ***tensors aren’t generalizations of vectors at all. Vectors, as we defined them above, are generalizations of tensors. And tensors aren’t technically generalizations of matrices. But tensors can certainly be thought of as kind of the same sort of object as a matrix.***\n\nThere are two things that tensors and matrices have in common. The first, and most important thing, is that they are both n-linear maps. This is why tensors are almost generalizations of matrices. The second, and more misleading, thing is that they can be represented as a 2d array of numbers. This second thing is a huge, and I mean HUGE red herring, and has undoubtedly caused an innumerable number of people to be confused.\n\n*Let’s tackle the concept of bilinear maps, and then we can use that knowledge of bilinear maps to help us tackle the concept of representing rank 2 tensors as 2d arrays.*",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"## Bilinear maps",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"The dot product is what the cool kids like to call a bilinear map. This just means that the dot product has the following properties:\n\n* x⋅(y+z)=x⋅y+x⋅z\n\n\n* (y+z)⋅x=y⋅x+z⋅x\n\n\n* x⋅(ay)=a(x⋅y)\n\nWhy is this important? Well if we represent the vector x as $x=x_{1}i+x_{2}j$, and we represent the vector $y=y_{1}i+y_{2}j$, then because ⋅ is linear, the following is true: $x⋅y=y_{1} x_{1} i⋅i + y_{2} x_{2} j⋅j + (x_{1} y_{2} + x_{2} y_{1})i⋅j$\n\nThis means if we know the values of i⋅i, j⋅j, and i⋅j, then we have completely defined the operation ⋅. In other words, knowing just these 3 values allows us to calculate the value of x⋅y for any x and y.\n\nNow we can describe how ⋅ might be represented as a 2d array. If ⋅ is the standard cartesian dot product that you learned about on the first day of your linear algebra or physics class, and i and j are both the standard cartesian unit vectors, then i⋅i=1, j⋅j=1, and j⋅i=i⋅j=0.\n\nTo represent this tensor ⋅ as a 2d array, we would create a table holding these values, i.e.\n\n\\begin{bmatrix}\n ⋅ & i & j \\\\[0.3em]\n i & 1 & 0 \\\\[0.3em]\n j & 0 & 1\n \\end{bmatrix}\n \nOr, more compactly\n\n\\begin{bmatrix}\n 1 & 0 \\\\[0.3em]\n 0 & 1\n \\end{bmatrix}\n \nDO NOT LET THIS SIMILARITY TO THE SIMILAR MATRIX NOTATION FOOL YOU. Multiplying this by a vector will clearly give the wrong answer for many reasons, the most important of which is that the dot product produces a scalar quantity, a matrix produces a vector quantity. This notation is simply a way of neatly writing what the dot product represents, it is not a way of making the dot product into a matrix.\n\nIf we become more general, then we can take arbitrary values for these dot products i⋅i=a, j⋅j=b, and j⋅i=i⋅j=c.\n\nWhich would be represented as\n\n\\begin{bmatrix}\n a & c \\\\[0.3em]\n c & b\n \\end{bmatrix}\n \n***A tensor defined in this way is called the metric tensor. The reason it is called that, and the reason it is so important in general relativity, is that just by changing the values we can change the definition of lengths and angles*** (remember that inner product spaces define length and angles in terms of ⋅), and we can enumerate over all possible definitions of lengths and angles. We call this a rank 2 tensor because it is a 2d array (i.e. it looks like a square), if we had a 3x3 tensor, such as a metric tensor for 3 dimensional space it would still be an example of a rank 2 tensor.\n\n\\begin{bmatrix}\n a & s & d \\\\[0.3em]\n f & g & h \\\\[0.3em]\n z & x & b\n \\end{bmatrix}\n \n(Note: the table is symmetric along the diagonal only because the metric tensor is commutative. A general tensor does not have to be commutative and thus its representation does not have to be symmetric.)\n\nTo get a rank 3 tensor, we would create a cube-like table of values as opposed to a square-like one (I can’t do this in latex so you’ll have to imagine it). A rank 3 tensor would be a trilinear map. A trilinear map m takes 3 vectors from a vector space V, and can be defined in terms of the values it takes when its arguments are the basis vectors of V. E.g. if V has two basis vectors i and j, then m can be defined by defining the values of m(i,i,i), m(i,i,j), m(i,j,i), m(i,j,j), m(j,i,i), m(j,i,j), m(j,j,i), and m(j,j,j) in a 3d array.\n\n\nA rank 4 tensor would be a 4-linear, A.K.A quadrlinear map that would take 4 arguments, and thus be represented as a 4 dimensional array etc.\n\n",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"## Why do people think tensors are generalizations of vectors?",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
"So now we come to why people think tensors are generalizations of vectors. Its because, if we take a function $f(y)=x⋅y$, then f, being the linear scallawag it is, can be defined with only 2 values. $f(y)=y_{1}f(i)+y_{2}f(j)$, so knowing the values of f(i)and f(j) completely define f. And therefore, f is a rank 1 tensor, i.e. a multilinear map with one argument. This would be represented as a 1d array, very much like the common notion of a vector. Furthermore, these values completely define x as well. If ⋅ is specifically the cartesian metric tensor, then the values of the representation of x and the values of the representation of f are exactly the same. This is why people think tensors are generalizations of vectors.\n\nBut if ⋅ is given different values, then the representation of x and the representation of f will have different values. ***Vectors by themselves are not linear maps, they can just be thought of as linear maps***. In order for them to actually be linear maps, they need to be combined with some sort of linear operator such as ⋅.\n\nSo here is the definition: ***A tensor is any multilinear map from a vector space to a scalar field***. (Note: A multilinear map is just a generalization of linear and bilinear maps to maps that have more than 2 arguments. I.e. any map which is distributive over addition and scalar multiplication. Linear maps are considered a type of multilinear map)\n\nThis definition as a multilinear maps is another reason people think tensors are generalization of matrices, because matrices are linear maps just like tensors. But the distinction is that matrices take a vector space to itself, while tensors take a vector space to a scalar field. So a matrix is not strictly speaking a tensor.",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
],
[
" ",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4ae4e9733598a68ed0d06b0d8392c525c60bb2ad
| 6,529 |
ipynb
|
Jupyter Notebook
|
component-library/transform/spark-parquet-to-csv.ipynb
|
ibm-developer-skills-network/dphcz-ETL_ProjectRepo
|
934a02a99057ad0f6c35d07a94cf25ee849ff168
|
[
"Apache-2.0"
] | 1 |
2021-10-17T07:17:51.000Z
|
2021-10-17T07:17:51.000Z
|
transform/spark-parquet-to-csv.ipynb
|
diocom/component-library
|
1728e9c11a18588bd22e7c445d5822ce05659521
|
[
"Apache-2.0"
] | null | null | null |
transform/spark-parquet-to-csv.ipynb
|
diocom/component-library
|
1728e9c11a18588bd22e7c445d5822ce05659521
|
[
"Apache-2.0"
] | null | null | null | 26.116 | 181 | 0.555828 |
[
[
[
"# Converts a parquet file to CSV file with header using ApacheSpark",
"_____no_output_____"
]
],
[
[
"%%bash\nexport version=`python --version |awk '{print $2}' |awk -F\".\" '{print $1$2}'`\n\nif [ $version == '36' ]; then\n pip install pyspark==2.4.8 wget==3.2 pyspark2pmml==0.5.1\nelif [ $version == '38' ]; then\n pip install pyspark==3.1.2 wget==3.2 pyspark2pmml==0.5.1\nelse\n echo 'Currently only python 3.6 and 3.8 is supported, in case you need a different version please open an issue at https://github.com/elyra-ai/component-library/issues'\n exit -1\nfi",
"_____no_output_____"
],
[
"# @param data_dir temporal data storage for local execution\n# @param data_csv csv path and file name (default: data.csv)\n# @param data_parquet path and parquet file name (default: data.parquet)\n# @param master url of master (default: local mode)",
"_____no_output_____"
],
[
"from pyspark import SparkContext, SparkConf\nfrom pyspark.sql import SparkSession\nimport os\nimport shutil\nimport glob",
"_____no_output_____"
],
[
"data_csv = os.environ.get('data_csv', 'data.csv')\ndata_parquet = os.environ.get('data_parquet', 'data.parquet')\nmaster = os.environ.get('master', \"local[*]\")\ndata_dir = os.environ.get('data_dir', '../../data/')",
"_____no_output_____"
],
[
"data_parquet = 'trends.parquet'\ndata_csv = 'trends.csv'",
"_____no_output_____"
],
[
"skip = False\nif os.path.exists(data_dir + data_csv):\n skip = True",
"_____no_output_____"
],
[
"if not skip:\n sc = SparkContext.getOrCreate(SparkConf().setMaster(master))\n spark = SparkSession.builder.getOrCreate()",
"_____no_output_____"
],
[
"if not skip:\n df = spark.read.parquet(data_dir + data_parquet)",
"_____no_output_____"
],
[
"if not skip:\n if os.path.exists(data_dir + data_csv):\n shutil.rmtree(data_dir + data_csv)\n df.coalesce(1).write.option(\"header\", \"true\").csv(data_dir + data_csv)\n file = glob.glob(data_dir + data_csv + '/part-*')\n shutil.move(file[0], data_dir + data_csv + '.tmp')\n shutil.rmtree(data_dir + data_csv)\n shutil.move(data_dir + data_csv + '.tmp', data_dir + data_csv)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae4ef8b9ed3b0f4d823d7212ac024b8e70c4cd1
| 3,396 |
ipynb
|
Jupyter Notebook
|
Create climate gif.ipynb
|
hausfath/scrape_global_temps
|
3cce1cc5ecfc0cfb3e29188d37ec6c18c870f037
|
[
"MIT"
] | 34 |
2018-04-20T14:48:08.000Z
|
2022-01-20T19:56:15.000Z
|
Create climate gif.ipynb
|
hausfath/scrape_global_temps
|
3cce1cc5ecfc0cfb3e29188d37ec6c18c870f037
|
[
"MIT"
] | 2 |
2018-04-20T15:54:10.000Z
|
2018-07-31T18:27:26.000Z
|
Create climate gif.ipynb
|
hausfath/scrape_global_temps
|
3cce1cc5ecfc0cfb3e29188d37ec6c18c870f037
|
[
"MIT"
] | 11 |
2018-04-20T18:08:50.000Z
|
2021-03-06T07:02:02.000Z
| 34.653061 | 104 | 0.534747 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport pylab as plt\nfrom matplotlib.animation import FuncAnimation\n\nfig, ax = plt.subplots()\nfig.set_tight_layout(True)\n\ndata = pd.read_csv('/Users/hausfath/Desktop/Climate Science/GHCN Monthly/model_obs_annual.csv')\nprint(data.head())\n\n#Plot data that persists (isn't redrawn)\nplt.xlim(data['year'].min(),data['year'].max())\nplt.plot(data['year'], data['model_mean'], 'r-', label='Multimodel Mean')\nplt.fill_between(data['year'], data['high_model'], data['low_model'], color='k',alpha=.2)\nplt.plot(data['year'][0], data['berkeley'][0], 'b-', label='Berkeley Earth')\nleg=plt.legend(loc='upper left', numpoints=1, fancybox=True)\nplt.ylabel('Degrees C w.r.t. 1979-2000') \n\ndef update(i):\n #Plot data that is redrawn\n plt.plot(data['year'][0:i], data['berkeley'][0:i], 'b-', label='Berkeley Earth')\n return plt\n\nif __name__ == '__main__':\n # FuncAnimation will call the 'update' function for each frame\n anim = FuncAnimation(fig, update, frames=np.arange(data['year'].count()), interval=100)\n anim.save('line.gif', dpi=100, writer='imagemagick')\n #plt.show()\n\n",
" year gistemp hadcrut4 ncdc cowtanandway berkeley model_mean \\\n0 1880 -0.516905 -0.406484 -0.460254 -0.473833 -0.586254 -0.372227 \n1 1881 -0.431071 -0.385234 -0.413013 -0.417500 -0.495671 -0.354215 \n2 1882 -0.417738 -0.394984 -0.412096 -0.470000 -0.534337 -0.351653 \n3 1883 -0.526071 -0.475317 -0.487329 -0.524083 -0.607837 -0.401751 \n4 1884 -0.596071 -0.589401 -0.549071 -0.684667 -0.761004 -0.643350 \n\n model_sd model_p2 model_p98 high_model low_model med_high_model \\\n0 0.186660 -0.757706 -0.066128 0.001092 -0.745546 -0.185567 \n1 0.169575 -0.670132 -0.040724 -0.015065 -0.693366 -0.184640 \n2 0.183873 -0.650893 0.009126 0.016093 -0.719399 -0.167780 \n3 0.178052 -0.752952 -0.024888 -0.045647 -0.757856 -0.223699 \n4 0.255575 -1.112845 -0.170002 -0.132200 -1.154500 -0.387775 \n\n med_low_model \n0 -0.558887 \n1 -0.523790 \n2 -0.535526 \n3 -0.579804 \n4 -0.898925 \n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
4ae4f8bdf217b4fb663383eec571c29552aa3004
| 21,194 |
ipynb
|
Jupyter Notebook
|
Pytorch Classification Problem - Diabetics with NN.ipynb
|
ajuhz/Udemy-neural-network-boorcamp
|
242bebf9c6e8da843db6da62859e29375ecf2a3e
|
[
"MIT"
] | null | null | null |
Pytorch Classification Problem - Diabetics with NN.ipynb
|
ajuhz/Udemy-neural-network-boorcamp
|
242bebf9c6e8da843db6da62859e29375ecf2a3e
|
[
"MIT"
] | null | null | null |
Pytorch Classification Problem - Diabetics with NN.ipynb
|
ajuhz/Udemy-neural-network-boorcamp
|
242bebf9c6e8da843db6da62859e29375ecf2a3e
|
[
"MIT"
] | null | null | null | 30.062411 | 180 | 0.412334 |
[
[
[
"# Pytorch : Classification Problem - Diabetics with NN",
"_____no_output_____"
]
],
[
[
"#import necessary libraries\n#describe reason for import each libraries \nimport numpy as np # converting data from pandas to torch\nimport torch \nimport torch.nn as nn #main library to define the architecture of the neural network\nimport pandas as pd # to read the data from the csv file \nfrom sklearn.preprocessing import StandardScaler # used for feature normalization\nfrom torch.utils.data import Dataset,DataLoader\nimport matplotlib.pyplot as plt # to plot loss with epochs",
"_____no_output_____"
]
],
[
[
"# Data Preprocessing",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('diabetes.csv')",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"#Extract features X and o/p y from the data\nX = data.iloc[:,:-1]\nX = np.array(X)\ny = data.iloc[:,-1]\ny = np.array(y) # need to convert datatype into float else not possible to convert into tensor",
"_____no_output_____"
],
[
"y[y=='positive']=1.\ny[y=='negative']=0.",
"_____no_output_____"
],
[
"y = np.array(y,dtype=np.float64)",
"_____no_output_____"
],
[
"y = y.reshape(len(y),1)",
"_____no_output_____"
]
],
[
[
"# Feature normalization",
"_____no_output_____"
],
[
"# Formula: $x^{\\prime}=\\frac{x-\\mu}{\\sigma}$ *where $\\mu$ is mean and $\\sigma$ is std",
"_____no_output_____"
]
],
[
[
"mean = X.mean(axis = 0) # taking mean along \nstd = X.std(axis = 0)",
"_____no_output_____"
],
[
"X_norm = (X-mean)/std",
"_____no_output_____"
],
[
"X_norm",
"_____no_output_____"
],
[
"#alternate approach \nsc = StandardScaler()\nX_norm1 = sc.fit_transform(X)",
"_____no_output_____"
],
[
"X_norm1",
"_____no_output_____"
],
[
"#Converting numpy array into tensor\nX_tensor = torch.tensor(X_norm)\ny_tensor = torch.tensor(y)",
"_____no_output_____"
],
[
"print(X_tensor.shape)\nprint(y_tensor.shape)",
"torch.Size([768, 7])\ntorch.Size([768, 1])\n"
],
[
"# We need to create custom dataset class to feed the data into dataloader\n#because as per pytorch standard dataloader accepts dataset class\n#this part can be copy pasted in case of use of custom data in X,y format\nclass Dataset(Dataset):\n def __init__(self,x,y):\n self.x = x\n self.y = y\n \n def __getitem__(self,index):\n # Get one item from the dataset\n return self.x[index], self.y[index]\n \n def __len__(self):\n return len(self.x)",
"_____no_output_____"
],
[
"dataset = Dataset(X_tensor,y_tensor)",
"_____no_output_____"
],
[
"print(dataset.__getitem__(2))\nprint(dataset.__len__())",
"(tensor([ 1.2339, 1.9437, -0.2639, -1.2882, -0.6929, -1.1033, -0.1056],\n dtype=torch.float64), tensor([1.], dtype=torch.float64))\n768\n"
],
[
"#create the dataloader for the model\ndataloader = DataLoader(dataset, batch_size=32, shuffle=True)",
"_____no_output_____"
],
[
"#Let's check the dataloader and iterate through it\nprint('Length of the dataloader:{}'.format(str(len(dataloader)))) # i.e no of batches\nfor (x,y) in dataloader:\n print(\"For one iteration (batch), there is:\")\n print(\"Data: {}\".format(x.shape))\n print(\"Labels: {}\".format(y.shape))\n break ",
"Length of the dataloader:24\nFor one iteration (batch), there is:\nData: torch.Size([32, 7])\nLabels: torch.Size([32, 1])\n"
]
],
[
[
"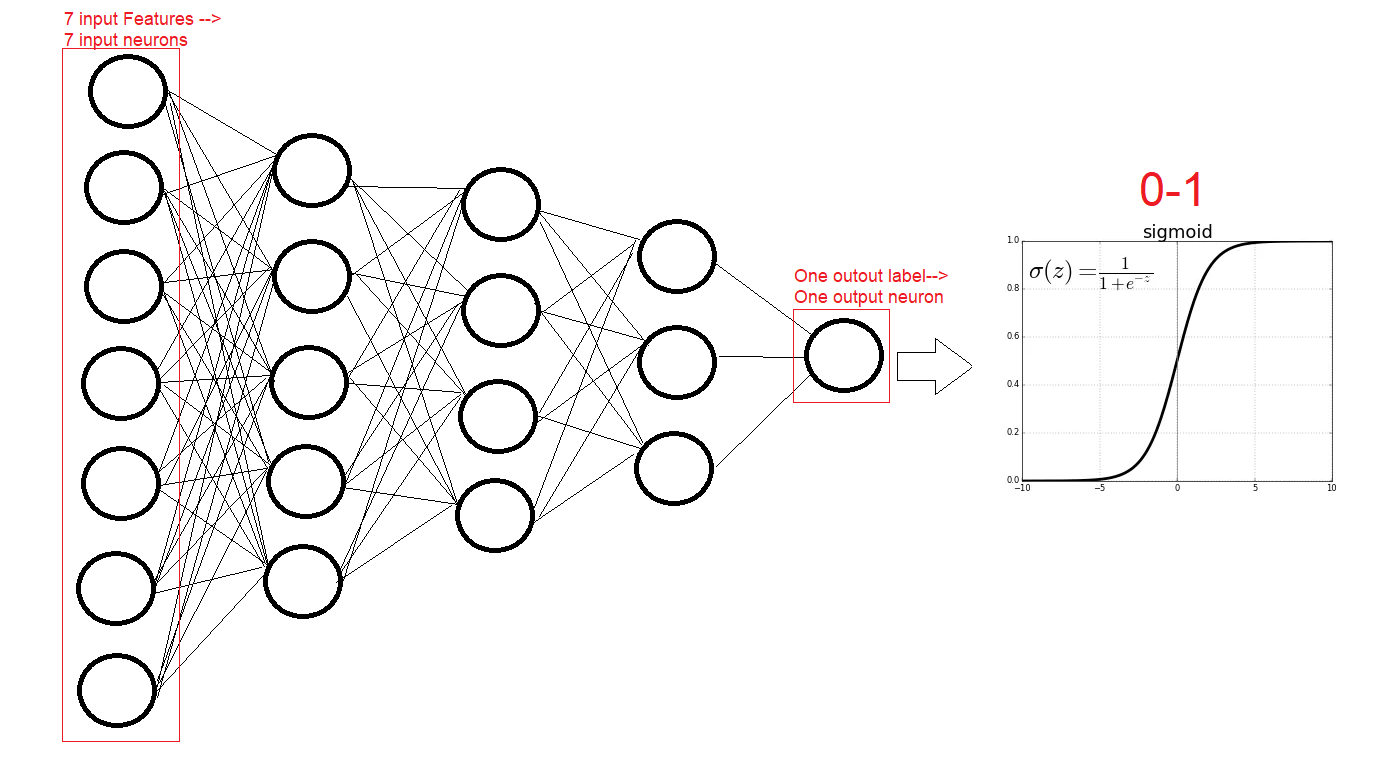",
"_____no_output_____"
]
],
[
[
"#create NN architecture as depicted above\nclass Model(nn.Module):\n def __init__(self,input_features,labels):\n super(Model, self).__init__() # if we dont include this then we will get error cannot assign module before Module.__init__() call\n self.input_features = input_features\n self.fc1 = nn.Linear(input_features,5)\n self.fc2 = nn.Linear(5,4)\n self.fc3 = nn.Linear(4,3)\n self.fc4 = nn.Linear(3,labels)\n self.sigmoid = nn.Sigmoid() # activation fn for the o/p\n self.tanh = nn.Tanh() # activation function for the hidden layers\n \n def forward(self,X):\n output = self.tanh(self.fc1(X))\n output = self.tanh(self.fc2(output))\n output = self.tanh(self.fc3(output))\n output = self.sigmoid(self.fc4(output))\n return output",
"_____no_output_____"
]
],
[
[
"**Reference only\n# Note: we don't need to manually derive this, in pytorch we can use the cost function provided in the library\n\n$H_{p}(q)=-\\frac{1}{N} \\sum_{i=1}^{N} y_{i} \\cdot \\log \\left(p\\left(y_{i}\\right)\\right)+\\left(1-y_{i}\\right) \\cdot \\log \\left(1-p\\left(y_{i}\\right)\\right)$\n\n\ncost = -(Y * torch.log(hypothesis) + (1 - Y) * torch.log(1 - hypothesis)).mean()",
"_____no_output_____"
]
],
[
[
"def train(model,epochs,criterion,optimizer,dataloader):\n average_loss =[]\n \n for epoch in range(epochs):\n minibatch_loss=0\n for x,y in dataloader:\n x = x.float() # converting into float to avoid any error for type casting\n y = y.float()\n #forward propagation\n output = model(x)\n #calculate loss\n loss = criterion(output,y)\n minibatch_loss += loss\n #cleat gradient buffer # check the reason why the optimizer is zero\n optimizer.zero_grad()\n # Backward propagation\n loss.backward()\n # Weight Update: w <-- w - lr * gradient\n optimizer.step()\n #calculate accuracy for one epoch\n average_loss.append(minibatch_loss/len(dataloader)) \n output = (output>0.5).float()\n #how many correct prection taking average of that\n accuracy = (output == y).float().mean()\n if (epoch+1)%50 ==0:\n print(\"Epoch {}/{}, Loss: {:.3f}, Accuracy: {:.3f}\".format(epoch+1,epochs, average_loss[-1], accuracy))\n return [average_loss,model]",
"_____no_output_____"
],
[
"# call train function:\nmodel = Model(X_tensor.shape[1],y_tensor.shape[1])\nepochs = 150\n# Loss function - Binary Cross Entropy \n#In Binary Cross Entropy: the input and output should have the same shape \n#size_average = True --> the losses are averaged over observations for each minibatch\n#criterion = nn.BCELoss(size_average=True)\ncriterion = nn.BCELoss(reduction='mean')\n#We will use SGD with momentum\n# Need to read about torch modules that is getting used here\noptimizer = torch.optim.SGD(model.parameters(), lr=0.1, momentum=0.9)\ndataloader =dataloader\naverage_loss,model = train(model,epochs,criterion,optimizer,dataloader)",
"Epoch 50/150, Loss: 0.432, Accuracy: 0.750\nEpoch 100/150, Loss: 0.418, Accuracy: 0.781\nEpoch 150/150, Loss: 0.405, Accuracy: 0.625\n"
],
[
"def BCELossfn(output,y):\n loss = -(torch.sum(y*torch.log(output)+(1-y)*torch.log(1-output)))\n return loss\n ",
"_____no_output_____"
],
[
"def predict(model,X,y):\n output = model(X)\n if output>0.5:\n output = 1.\n else:\n output = 0.\n print('Predicted output:{}'.format(str(output)))\n print('Ground truth:{}'.format(str(y.item())))",
"_____no_output_____"
],
[
"predict(model,X_tensor[0].float(),y_tensor[0].float())",
"Predicted output:1.0\nGround truth:1.0\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4ae4fbc872c108ab85bd07eb11ec71be91b7b854
| 197,819 |
ipynb
|
Jupyter Notebook
|
4_NN_Classifier02.ipynb
|
Vidhin05/Classifiers-Neural-Networks
|
edeb36b9a7bfb902bfbc0c1c06cde7844ea9d253
|
[
"MIT"
] | null | null | null |
4_NN_Classifier02.ipynb
|
Vidhin05/Classifiers-Neural-Networks
|
edeb36b9a7bfb902bfbc0c1c06cde7844ea9d253
|
[
"MIT"
] | null | null | null |
4_NN_Classifier02.ipynb
|
Vidhin05/Classifiers-Neural-Networks
|
edeb36b9a7bfb902bfbc0c1c06cde7844ea9d253
|
[
"MIT"
] | null | null | null | 322.180782 | 54,972 | 0.918597 |
[
[
[
"# Image Classifier\n## Dataset : 28x28 pixel Low Res Images",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nfrom tensorflow import keras\nimport pandas as pd\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport os\n\ntf.logging.set_verbosity(tf.logging.ERROR)\npd.options.display.max_rows = 7",
"/Users/rutvikshah/anaconda3/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"CATEGORIES = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat','Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']",
"_____no_output_____"
]
],
[
[
"## Getting the Data",
"_____no_output_____"
]
],
[
[
"data = keras.datasets.fashion_mnist\n(train_images, train_labels), (test_images, test_labels) = data.load_data()",
"_____no_output_____"
],
[
"print(\"Train Dataset Dimensions : \",train_images.shape)\nprint(\"Test Dataset Dimensions : \",test_images.shape)",
"Train Dataset Dimensions : (60000, 28, 28)\nTest Dataset Dimensions : (10000, 28, 28)\n"
],
[
"train_labels",
"_____no_output_____"
],
[
"test_labels",
"_____no_output_____"
]
],
[
[
"### Sample Train Data",
"_____no_output_____"
]
],
[
[
"plt.figure()\nplt.imshow(train_images[17])\nplt.colorbar()\nplt.grid(False)",
"_____no_output_____"
]
],
[
[
"### Sample Test Data",
"_____no_output_____"
]
],
[
[
"plt.figure()\nplt.imshow(test_images[17])\nplt.colorbar()\nplt.grid(False)",
"_____no_output_____"
]
],
[
[
"## Preprocessing (Normalisation)",
"_____no_output_____"
]
],
[
[
"train_images = train_images / 255.0\ntest_images = test_images / 255.0",
"_____no_output_____"
]
],
[
[
"### Neural Network\n##### Structure of the NNs:\n\n### Model I\n• Input Feature (Flattened/Linearised)<br>\n• Hidden Dense Layer 1: 128 neurons (AF : RELU)<br>\n• Output Layer : 10 neurons (AF : SOFTMAX)<br>\n\n\n### Model II\n• Input Feature (Flattened/Linearised)<br>\n• Hidden Dense Layer 1: 128 neurons (AF : RELU)<br>\n• Hidden Dense Layer 2: 128 neurons (AF : RELU)<br>\n• Output Layer : 10 neurons (AF : SOFTMAX)<br>\n\n\n### Model III\n• Input Feature (Flattened/Linearised)<br>\n• Hidden Dense Layer 1: 128 neurons (AF : RELU)<br>\n• Hidden Dense Layer 2: 128 neurons (AF : RELU)<br>\n• Hidden Dense Layer 3: 128 neurons (AF : RELU)<br>\n• Output Layer : 10 neurons (AF : SOFTMAX)<br>",
"_____no_output_____"
]
],
[
[
"classifier1 = keras.Sequential([\n keras.layers.Flatten(input_shape=(28, 28)),\n keras.layers.Dense(128, activation=tf.nn.relu),\n keras.layers.Dense(10, activation=tf.nn.softmax)\n ])\n\nclassifier2 = keras.Sequential([\n keras.layers.Flatten(input_shape=(28, 28)),\n keras.layers.Dense(128, activation=tf.nn.relu),\n keras.layers.Dense(128, activation=tf.nn.relu),\n keras.layers.Dense(10, activation=tf.nn.softmax)\n ])\n\nclassifier3 = keras.Sequential([\n keras.layers.Flatten(input_shape=(28, 28)),\n keras.layers.Dense(128, activation=tf.nn.relu),\n keras.layers.Dense(128, activation=tf.nn.relu),\n keras.layers.Dense(128, activation=tf.nn.relu),\n keras.layers.Dense(10, activation=tf.nn.softmax)\n ])",
"_____no_output_____"
],
[
"classifier1.compile(optimizer=tf.train.AdamOptimizer(), loss='sparse_categorical_crossentropy', metrics=['accuracy'])\nclassifier2.compile(optimizer=tf.train.AdamOptimizer(), loss='sparse_categorical_crossentropy', metrics=['accuracy'])\nclassifier3.compile(optimizer=tf.train.AdamOptimizer(), loss='sparse_categorical_crossentropy', metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## Training All Classifiers/Models",
"_____no_output_____"
]
],
[
[
"classifier1.fit(train_images, train_labels, epochs=7)",
"Epoch 1/7\n60000/60000 [==============================] - 6s 108us/step - loss: 0.4932 - acc: 0.8261\nEpoch 2/7\n60000/60000 [==============================] - 6s 94us/step - loss: 0.3721 - acc: 0.8666\nEpoch 3/7\n60000/60000 [==============================] - 6s 94us/step - loss: 0.3353 - acc: 0.8778\nEpoch 4/7\n60000/60000 [==============================] - 6s 95us/step - loss: 0.3121 - acc: 0.8855\nEpoch 5/7\n60000/60000 [==============================] - 6s 97us/step - loss: 0.2926 - acc: 0.8920\nEpoch 6/7\n60000/60000 [==============================] - 6s 97us/step - loss: 0.2795 - acc: 0.8961\nEpoch 7/7\n60000/60000 [==============================] - 6s 97us/step - loss: 0.2659 - acc: 0.9013\n"
],
[
"classifier2.fit(train_images, train_labels, epochs=7)",
"Epoch 1/7\n60000/60000 [==============================] - 7s 123us/step - loss: 0.4839 - acc: 0.8272\nEpoch 2/7\n60000/60000 [==============================] - 7s 116us/step - loss: 0.3622 - acc: 0.8681\nEpoch 3/7\n60000/60000 [==============================] - 7s 116us/step - loss: 0.3292 - acc: 0.8776\nEpoch 4/7\n60000/60000 [==============================] - 7s 116us/step - loss: 0.3029 - acc: 0.8886\nEpoch 5/7\n60000/60000 [==============================] - 7s 117us/step - loss: 0.2867 - acc: 0.8928\nEpoch 6/7\n60000/60000 [==============================] - 7s 116us/step - loss: 0.2723 - acc: 0.8991\nEpoch 7/7\n60000/60000 [==============================] - 7s 115us/step - loss: 0.2585 - acc: 0.9032\n"
],
[
"classifier3.fit(train_images, train_labels, epochs=7)",
"Epoch 1/7\n60000/60000 [==============================] - 8s 137us/step - loss: 0.4870 - acc: 0.8237\nEpoch 2/7\n60000/60000 [==============================] - 8s 129us/step - loss: 0.3661 - acc: 0.8654\nEpoch 3/7\n60000/60000 [==============================] - 8s 129us/step - loss: 0.3311 - acc: 0.8779\nEpoch 4/7\n60000/60000 [==============================] - 8s 129us/step - loss: 0.3067 - acc: 0.8858\nEpoch 5/7\n60000/60000 [==============================] - 8s 131us/step - loss: 0.2909 - acc: 0.8921\nEpoch 6/7\n60000/60000 [==============================] - 8s 130us/step - loss: 0.2769 - acc: 0.8969\nEpoch 7/7\n60000/60000 [==============================] - 8s 129us/step - loss: 0.2640 - acc: 0.8999\n"
]
],
[
[
"## Finding Accuracy Of Each Classifier On Test Set",
"_____no_output_____"
]
],
[
[
"test_loss, test_acc = classifier1.evaluate(test_images, test_labels)\nprint('Test accuracy:', test_acc)",
"10000/10000 [==============================] - 0s 47us/step\nTest accuracy: 0.8766\n"
],
[
"test_loss, test_acc = classifier2.evaluate(test_images, test_labels)\nprint('Test accuracy:', test_acc)",
"10000/10000 [==============================] - 1s 51us/step\nTest accuracy: 0.88\n"
],
[
"test_loss, test_acc = classifier3.evaluate(test_images, test_labels)\nprint('Test accuracy:', test_acc)",
"10000/10000 [==============================] - 1s 52us/step\nTest accuracy: 0.8739\n"
],
[
"class1_predict = classifier1.predict(test_images)\nclass2_predict = classifier2.predict(test_images)\nclass3_predict = classifier3.predict(test_images)",
"_____no_output_____"
],
[
"def plot_image(i, arr, actual_label, img):\n arr, actual_label, img = arr[i], actual_label[i], img[i]\n plt.grid(False)\n plt.xticks([])\n plt.yticks([])\n\n plt.imshow(img, cmap=plt.cm.binary)\n\n predicted_label = np.argmax(arr)\n if predicted_label == actual_label:\n color = 'blue'\n else:\n color = 'red'\n\n plt.xlabel(\"{} {:2.0f}% ({})\".format(CATEGORIES[predicted_label],\n 100*np.max(arr),\n CATEGORIES[actual_label]),\n color=color)\n\ndef plot_value_array(i, arr, actual_label):\n arr, actual_label = arr[i], actual_label[i]\n plt.grid(False)\n plt.xticks([])\n plt.yticks([])\n thisplot = plt.bar(range(10), arr, color=\"#777777\")\n plt.ylim([0, 1]) \n predicted_label = np.argmax(arr)\n\n thisplot[predicted_label].set_color('red')\n thisplot[actual_label].set_color('blue')",
"_____no_output_____"
],
[
"num_rows = 4\nnum_cols = 4\nnum_images = num_rows*num_cols\nplt.figure(figsize=(2*2*num_cols, 2*num_rows))\nfor i in range(num_images):\n plt.subplot(num_rows, 2*num_cols, 2*i+1)\n plot_image(i, class1_predict, test_labels, test_images)\n plt.subplot(num_rows, 2*num_cols, 2*i+2)\n plot_value_array(i, class1_predict, test_labels)",
"_____no_output_____"
],
[
"num_rows = 4\nnum_cols = 4\nnum_images = num_rows*num_cols\nplt.figure(figsize=(2*2*num_cols, 2*num_rows))\nfor i in range(num_images):\n plt.subplot(num_rows, 2*num_cols, 2*i+1)\n plot_image(i, class3_predict, test_labels, test_images)\n plt.subplot(num_rows, 2*num_cols, 2*i+2)\n plot_value_array(i, class3_predict, test_labels)",
"_____no_output_____"
],
[
"num_rows = 4\nnum_cols = 4\nnum_images = num_rows*num_cols\nplt.figure(figsize=(2*2*num_cols, 2*num_rows))\nfor i in range(num_images):\n plt.subplot(num_rows, 2*num_cols, 2*i+1)\n plot_image(i, class2_predict, test_labels, test_images)\n plt.subplot(num_rows, 2*num_cols, 2*i+2)\n plot_value_array(i, class2_predict, test_labels)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae50c6162e64583f6cad3c5ddc0ffe4f3b2e437
| 445,986 |
ipynb
|
Jupyter Notebook
|
Pharmaceuticals_Matplotlib.ipynb
|
rimsha-aziz/Matplotlib-PowerOfPlots
|
4867161db8498ce333811e10eef4e1c1b139dbfe
|
[
"ADSL"
] | null | null | null |
Pharmaceuticals_Matplotlib.ipynb
|
rimsha-aziz/Matplotlib-PowerOfPlots
|
4867161db8498ce333811e10eef4e1c1b139dbfe
|
[
"ADSL"
] | null | null | null |
Pharmaceuticals_Matplotlib.ipynb
|
rimsha-aziz/Matplotlib-PowerOfPlots
|
4867161db8498ce333811e10eef4e1c1b139dbfe
|
[
"ADSL"
] | null | null | null | 76.656239 | 31,432 | 0.735057 |
[
[
[
"Dependencies and starter code",
"_____no_output_____"
],
[
"Observations:\n1. The number of data points per drug regimen group were not equal. Capomulin and Ramicane had the most amount of data points and they were also a part of the top four most promising treatment regimens. Their inclusion in this category may be because there was a larger data set of points thus allowing for greater accuracy when analyzing their efficacy and promising nature. \n2. Capomulin seems to be the most effective drug regimen because it greatly reduced tumor volume as it has the lowest mean tumor volume and the second lowest SEM. \n3. Tumor volume is positively correlated to mouse weight for a mouse that is treated with Capomulin. This is supported by the R value of this relationship being, +0.96. ",
"_____no_output_____"
]
],
[
[
"# Dependencies and Setup\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport scipy.stats as st\nimport numpy as np\nfrom scipy.stats import linregress\n\n# Study data files\nmouse_metadata = \"data/Mouse_metadata.csv\"\nstudy_results = \"data/Study_results.csv\"\n\n# Read the mouse data and the study results\nmouse_metadata = pd.read_csv(mouse_metadata)\nstudy_results = pd.read_csv(study_results)\n\n\n# Combine the data into a single dataset\ncombined_mouse = pd.merge(mouse_metadata, study_results,\n how='outer', on='Mouse ID')\ncombined_mouse",
"_____no_output_____"
]
],
[
[
"Summary statistics",
"_____no_output_____"
]
],
[
[
"# Generate a summary statistics table of mean, median, variance, standard deviation, and SEM of the tumor volume for each regimen\n\nRegimens = combined_mouse.groupby([\"Drug Regimen\"])\nRegimens\n\nregimen_mean = Regimens[\"Tumor Volume (mm3)\"].mean()\nregimen_median = Regimens[\"Tumor Volume (mm3)\"].median()\nregimen_variance = Regimens[\"Tumor Volume (mm3)\"].var()\nregimen_std = Regimens[\"Tumor Volume (mm3)\"].std()\nregimen_sem = Regimens[\"Tumor Volume (mm3)\"].sem()",
"_____no_output_____"
],
[
"summary_stats = pd.DataFrame({\"Mean\": regimen_mean, \"Median\":regimen_median, \"Variance\":regimen_variance, \"Standard Deviation\": regimen_std, \"SEM\": regimen_sem})\n\nsummary_stats",
"_____no_output_____"
]
],
[
[
"Bar plots",
"_____no_output_____"
]
],
[
[
"# Generate a bar plot showing number of data points for each treatment regimen using pandas\nregimen_data_points = Regimens.count()[\"Mouse ID\"]\nregimen_data_points",
"_____no_output_____"
],
[
"# Generate a bar plot showing number of data points for each treatment regimen using pyplot\nbar_regimen = regimen_data_points.plot(kind='bar')\nplt.title(\"Data Points and Drug Regimen\")\nplt.xlabel(\"Drug Regimen\")\nplt.ylabel(\"Data Points\")\nplt.ylim(0, 240)",
"_____no_output_____"
],
[
"# Generate a bar plot showing number of data points for each treatment regimen using pyplot\ndata_points = [230, 178,178,188,186,181,161,228,181,182]\nx_axis = np.arange(len(regimen_data_points))\nplt.bar(x_axis, data_points, color='r', alpha=0.5, align=\"center\")\ntick_locations = [value for value in x_axis]\nplt.xticks(tick_locations, [\"Capomulin\", \"Ceftamin\", \"Infubrinol\", \"Ketapril\",\"Naftisol\", \"Placebo\",\"Propriva\",\"Ramicane\",\"Stekasyn\",\"Zoniferol\"],rotation='vertical')\n\nplt.title(\"Data Points Using Pyplot\")\nplt.xlabel(\"Drug Regimen\")\nplt.ylabel(\"Data Points\")\n\nplt.xlim(-0.75, len(x_axis)-0.25)\n\nplt.ylim(0, 250)\n",
"_____no_output_____"
]
],
[
[
"Pie Plots",
"_____no_output_____"
]
],
[
[
"# Generate a pie plot showing the distribution of female versus male mice using pandas\ngender_count = combined_mouse.groupby(\"Sex\")[\"Mouse ID\"].nunique()\ngender_count.head()",
"_____no_output_____"
],
[
"total_count = len(combined_mouse[\"Mouse ID\"].unique())\ntotal_count",
"_____no_output_____"
],
[
"gender_percent = (gender_count/total_count)*100\ngp= gender_percent.round(2)",
"_____no_output_____"
],
[
"gender_df = pd.DataFrame({\"Sex Count\":gender_count,\n \"Sex Percentage\":gp})\n \ngender_df",
"_____no_output_____"
],
[
"colors = ['pink', 'lightblue']\nexplode = (0.1, 0)\nplot = gender_df.plot.pie(y=\"Sex Count\",figsize=(6,6), colors = colors, startangle=140, explode = explode, shadow = True, autopct=\"%1.1f%%\")\nplt.title(\"Percentage of Female vs. Male Mice Using Pandas\")",
"_____no_output_____"
],
[
"# Generate a pie plot showing the distribution of female versus male mice using pyplot",
"_____no_output_____"
],
[
"sex = [\"Female\",\"Male\"]\nsex_percent = [gp]\ncolors = [\"pink\",\"lightblue\"]\nexplode = (0.1,0)\nplt.pie(sex_percent, explode=explode, labels=sex, colors=colors,\n autopct=\"%1.1f%%\", shadow=True, startangle=140)\nplt.axis(\"equal\")\nplt.title(\"Percentage of Female vs. Male Mice Using Pyplot\")\nplt.show()",
"C:\\Users\\rimsh\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\ipykernel_launcher.py:6: MatplotlibDeprecationWarning: Non-1D inputs to pie() are currently squeeze()d, but this behavior is deprecated since 3.1 and will be removed in 3.3; pass a 1D array instead.\n \n"
]
],
[
[
"Quartiles, outliers and boxplots",
"_____no_output_____"
]
],
[
[
"# Calculate the final tumor volume of each mouse across four of the most promising treatment regimens. Calculate the IQR and quantitatively determine if there are any potential outliers. \n\ntop_4 = combined_mouse[[\"Drug Regimen\", \"Mouse ID\", \"Timepoint\", \"Tumor Volume (mm3)\"]]\ntop_4\ntop_four = top_4.sort_values(\"Timepoint\", ascending=False)\ntop_four.head(4)\n",
"_____no_output_____"
],
[
"tumor_naftisol= top_four.loc[(top_four[\"Drug Regimen\"] == \"Naftisol\") | (top_four[\"Timepoint\"] == \"45\"),:]\ntumor_naftisol",
"C:\\Users\\rimsh\\AppData\\Local\\Continuum\\anaconda3\\lib\\site-packages\\pandas\\core\\ops\\__init__.py:1115: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison\n result = method(y)\n"
],
[
"quartiles = tumor_naftisol['Tumor Volume (mm3)'].quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq-lowerq\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(lower_bound)\nprint(upper_bound) ",
"28.270133771249995\n78.97877414125\n"
],
[
"outliers = []\nfor vol in tumor_naftisol:\n for row in tumor_naftisol[\"Tumor Volume (mm3)\"]:\n if row > upper_bound:\n print (f'{row} is an outlier')\n if row < lower_bound:\n print(f'{row} is an outlier')\n else:\n print(f\"This mouse is not an outlier for Drug Regimen Naftisol\")",
"This mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\nThis mouse is not an outlier for Drug Regimen Naftisol\n"
],
[
"tumor_capomulin= top_four.loc[(top_four[\"Drug Regimen\"] == \"Capomulin\") | (top_four[\"Timepoint\"] == \"45\"),:]\ntumor_capomulin",
"_____no_output_____"
],
[
"quartiles = tumor_capomulin['Tumor Volume (mm3)'].quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq-lowerq\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(lower_bound)\nprint(upper_bound)",
"26.714832162499995\n55.9711007025\n"
],
[
"for vol in tumor_capomulin:\n for row in tumor_capomulin[\"Tumor Volume (mm3)\"]:\n if row < lower_bound:\n print(f'{row} is an outlier')\n if row > upper_bound:\n print(f'{row} is an outlier')\n else:\n print(\"This mouse is not an outlier for Drug Regimen Capomulin\")\n",
"This mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n23.34359787 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n25.47214326 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n23.34359787 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n25.47214326 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n23.34359787 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n25.47214326 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n23.34359787 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n25.47214326 is an outlier\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\nThis mouse is not an outlier for Drug Regimen Capomulin\n"
],
[
"tumor_placebo = top_four.loc[(top_four[\"Drug Regimen\"] == \"Placebo\") | (top_four[\"Timepoint\"] == \"45\"),:]\ntumor_placebo\n",
"_____no_output_____"
],
[
"quartiles = tumor_placebo['Tumor Volume (mm3)'].quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq-lowerq\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(lower_bound)\nprint(upper_bound)",
"28.772230604999994\n78.603756725\n"
],
[
"for vol in tumor_placebo:\n for row in tumor_placebo[\"Tumor Volume (mm3)\"]:\n if row < lower_bound:\n print(f'{row} is an outlier')\n if row > upper_bound:\n print(f'{row} is an outlier')\n else:\n print(\"This mouse is not an outlier for Drug Regimen Placebo\")",
"This mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\n"
],
[
"tumor_ramicane= top_four.loc[(top_four[\"Drug Regimen\"] == \"Ramicane\") | (top_four[\"Timepoint\"] == \"45\"),:]\ntumor_ramicane\n",
"_____no_output_____"
],
[
"quartiles = tumor_ramicane['Tumor Volume (mm3)'].quantile([.25,.5,.75])\nlowerq = quartiles[0.25]\nupperq = quartiles[0.75]\niqr = upperq-lowerq\nlower_bound = lowerq - (1.5*iqr)\nupper_bound = upperq + (1.5*iqr)\nprint(lower_bound)\nprint(upper_bound)",
"24.18658646249998\n57.48804812250001\n"
],
[
"for vol in tumor_ramicane:\n for row in tumor_ramicane[\"Tumor Volume (mm3)\"]:\n if row < lower_bound:\n print(f'{row} is an outlier')\n if row > upper_bound:\n print(f'{row} is an outlier')\n else:\n print(\"This mouse is not an outlier for Drug Regimen Placebo\")",
"This mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\n22.05012627 is an outlier\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\n22.05012627 is an outlier\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\nThis mouse is not an outlier for Drug Regimen Placebo\n"
],
[
"# Generate a box plot of the final tumor volume of each mouse across four regimens of interest\n\nnaftisol_vol = tumor_naftisol[\"Tumor Volume (mm3)\"]\ncapomulin_vol = tumor_capomulin[\"Tumor Volume (mm3)\"]\nplacebo_vol = tumor_placebo[\"Tumor Volume (mm3)\"]\nramicane_vol = tumor_ramicane[\"Tumor Volume (mm3)\"]\nnaf = plt.boxplot(naftisol_vol,positions = [1],widths= 0.5)\ncap = plt.boxplot(capomulin_vol,positions = [2],widths = 0.5)\nplac = plt.boxplot(placebo_vol,positions = [3],widths = 0.5)\nram = plt.boxplot(ramicane_vol,positions = [4],widths =0.5)\nplt.title(\"Final tumor volume of each mouse across four of the most promising treatment regimens\")\nplt.ylabel(\"Tumor Volume\")\nplt.xlabel(\"Treatments\")\nplt.xticks([1, 2, 3,4], ['Naftisol', 'Capomulin', 'Placebo','Ramicane'])\nplt.ylim(10, 80)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Line and scatter plots",
"_____no_output_____"
]
],
[
[
"# Generate a line plot of time point versus tumor volume for a mouse treated with Capomulin\ncap_mouse = combined_mouse.loc[(combined_mouse[\"Mouse ID\"] == \"j119\"),:]\ncap_mouse",
"_____no_output_____"
],
[
"x_axis = cap_mouse[\"Timepoint\"]\ny_axis = cap_mouse[\"Tumor Volume (mm3)\"]\nplt.plot(x_axis,y_axis, marker ='o', color='blue')\nplt.title(\"Time point versus tumor volume for a mouse treated with Capomulin\")\nplt.xlabel(\"Timepoint\")\nplt.ylabel(\"Tumor Volume (mm3)\")\nplt.show()",
"_____no_output_____"
],
[
"# Generate a scatter plot of mouse weight versus average tumor volume for the Capomulin regimen\ncap_df = combined_mouse[[\"Mouse ID\",\"Weight (g)\", \"Tumor Volume (mm3)\",\"Drug Regimen\"]]\ncap_df",
"_____no_output_____"
],
[
"cap_scatter = cap_df.loc[(cap_df[\"Drug Regimen\"] == \"Capomulin\"),:]\ncap_scatter",
"_____no_output_____"
],
[
"cap_weight = cap_scatter.groupby(\"Weight (g)\")[\"Tumor Volume (mm3)\"].mean()\ncap_weight",
"_____no_output_____"
],
[
"cap_weight_df = pd.DataFrame(cap_weight)\ncap_weight_df",
"_____no_output_____"
],
[
"capo_final = pd.DataFrame(cap_weight_df).reset_index()\ncapo_final",
"_____no_output_____"
],
[
"plt.scatter(x=capo_final['Weight (g)'], y=capo_final['Tumor Volume (mm3)'])\nplt.title(\"Mouse weight versus average tumor volume for the Capomulin regimen\")\nplt.xlabel(\"Weight (g)\")\nplt.ylabel(\"Average Tumor Volume (mm3)\")\nplt.show()",
"_____no_output_____"
],
[
"# Calculate the correlation coefficient and linear regression model for mouse weight and average tumor volume for the Capomulin regimen\nx_values = capo_final[\"Weight (g)\"]\ny_values = capo_final[\"Tumor Volume (mm3)\"]\n(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)\nregress_values = x_values * slope + intercept\nline_eq = \"y =\" + str(round(slope,2)) + \"x + \" + str(round(intercept,2))\nplt.scatter(x_values, y_values)\nplt.plot(x_values,regress_values,\"r-\")\nplt.annotate(line_eq,(10,10),fontsize=15,color=\"black\")\nplt.xlabel(\"Weight (g)\")\nplt.ylabel(\"Average Tumor Volume (mm3)\")\nplt.title(\"Mouse weight versus average tumor volume for the Capomulin regimen\")\nprint(f\"The r-squared is: {rvalue}\")\nplt.show()",
"The r-squared is: 0.950524396185527\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae5139291a556bb0d8767cd983e9f82bd10b7e0
| 6,127 |
ipynb
|
Jupyter Notebook
|
jupyter_notebooks/temp/temp_fix_continuation_mpc_to_all.ipynb
|
noambuckman/mpc-multiple-vehicles
|
a20949c335f1af97962569eed112e6cef46174d9
|
[
"MIT"
] | 1 |
2021-11-02T15:16:17.000Z
|
2021-11-02T15:16:17.000Z
|
jupyter_notebooks/temp/temp_fix_continuation_mpc_to_all.ipynb
|
noambuckman/mpc-multiple-vehicles
|
a20949c335f1af97962569eed112e6cef46174d9
|
[
"MIT"
] | 5 |
2021-04-14T17:08:59.000Z
|
2021-05-27T21:41:02.000Z
|
jupyter_notebooks/temp/temp_fix_continuation_mpc_to_all.ipynb
|
noambuckman/mpc-multiple-vehicles
|
a20949c335f1af97962569eed112e6cef46174d9
|
[
"MIT"
] | 2 |
2022-02-07T08:16:05.000Z
|
2022-03-09T23:30:17.000Z
| 40.045752 | 171 | 0.607149 |
[
[
[
"%load_ext autoreload\n%autoreload 2\nimport numpy as np\nnp.set_printoptions(precision=2)\nimport matplotlib.pyplot as plt\nimport copy as cp\nimport sys, json, pickle\nPROJECT_PATHS = ['/home/nbuckman/Dropbox (MIT)/DRL/2020_01_cooperative_mpc/mpc-multiple-vehicles/', '/Users/noambuckman/mpc-multiple-vehicles/']\nfor p in PROJECT_PATHS:\n sys.path.append(p)\nimport src.traffic_world as tw\nimport src.multiagent_mpc as mpc\nimport src.car_plotting_multiple as cmplot\nimport src.solver_helper as helper\nimport src.vehicle as vehicle",
"_____no_output_____"
],
[
"i_mpc_start = 1\ni_mpc = i_mpc_start\nlog_directory = '/home/nbuckman/mpc_results/f509-425f-20200907-153800/'",
"_____no_output_____"
],
[
"with open(log_directory + \"params.json\",'rb') as fp:\n params = json.load(fp)",
"_____no_output_____"
],
[
"n_rounds_mpc = params['n_rounds_mpc']\nnumber_ctrl_pts_executed = params['number_ctrl_pts_executed']\n\n\nxamb_actual, uamb_actual = np.zeros((6, n_rounds_mpc*number_ctrl_pts_executed + 1)), np.zeros((2, n_rounds_mpc*number_ctrl_pts_executed)) \nxothers_actual = [np.zeros((6, n_rounds_mpc*number_ctrl_pts_executed + 1)) for i in range(params['n_other'])]\nuothers_actual = [np.zeros((2, n_rounds_mpc*number_ctrl_pts_executed)) for i in range(params['n_other'])] ",
"_____no_output_____"
],
[
"actual_t = 0\nlast_mpc_i = 104\nfor i_mpc_start in range(1,last_mpc_i+2):\n previous_mpc_file = folder + 'data/mpc_%02d'%(i_mpc_start - 1)\n xamb_executed, uamb_executed, _, all_other_x_executed, all_other_u_executed, _, = mpc.load_state(previous_mpc_file, params['n_other'])\n all_other_u_mpc = all_other_u_executed\n uamb_mpc = uamb_executed\n previous_all_file = folder + 'data/all_%02d'%(i_mpc_start -1)\n# xamb_actual_prev, uamb_actual_prev, _, xothers_actual_prev, uothers_actual_prev, _ = mpc.load_state(previous_all_file, params['n_other'], ignore_des = True)\n t_end = actual_t+number_ctrl_pts_executed+1\n xamb_actual[:, actual_t:t_end] = xamb_executed[:,:number_ctrl_pts_executed+1]\n uamb_actual[:, actual_t:t_end] = uamb_executed[:,:number_ctrl_pts_executed+1]\n for i in range(len(xothers_actual_prev)):\n xothers_actual[i][:, actual_t:t_end] = all_other_x_executed[i][:,:number_ctrl_pts_executed+1]\n uothers_actual[i][:, actual_t:t_end] = all_other_u_executed[i][:,:number_ctrl_pts_executed+1]\n# print(xamb_actual[0,:t_end])\n\n# print(\" \")\n file_name = folder + \"data/\"+'all_%02d'%(i_mpc_start-1) \n mpc.save_state(file_name, xamb_actual, uamb_actual, None, xothers_actual, uothers_actual, None, end_t = actual_t+number_ctrl_pts_executed+1)\n actual_t += number_ctrl_pts_executed\nprint(\"Loaded initial positions from %s\"%(previous_mpc_file))\nprint(xothers_actual[0][0,:t_end])",
"Loaded initial positions from /home/nbuckman/mpc_results/f509-425f-20200907-153800/data/mpc_104\n[ 20. 22.22 24.46 26.69 28.93 31.16 33.4 35.63 37.87 40.1\n 42.34 44.57 46.81 49.04 51.28 53.51 55.75 57.98 60.22 62.45\n 64.69 66.92 69.16 71.39 73.63 75.86 78.1 80.33 82.57 84.8\n 87.04 89.27 91.5 93.74 95.97 98.21 100.44 102.68 104.91 107.15\n 109.38 111.62 113.85 116.09 118.32 120.56 122.79 125.03 127.26 129.5\n 131.73 133.97 136.2 138.44 140.67 142.91 145.14 147.38 149.61 151.85\n 154.08 156.32 158.55 160.79 163.02 165.26 167.49 169.73 171.96 174.2\n 176.43 178.67 180.9 183.13 185.37 187.6 189.84 192.07 194.31 196.54\n 198.78 201.01 203.25 205.48 207.72 209.95 212.19 214.42 216.66 218.89\n 221.13 223.36 225.6 227.83 230.07 232.3 234.54 236.77 239.01 241.24\n 243.48 245.71 247.95 250.18 252.42 254.65 256.89 259.12 261.36 263.59\n 265.83 268.06 270.3 272.53 274.77 277. 279.24 281.47 283.71 285.94\n 288.18 290.41 292.65 294.88 297.12 299.35 301.59 303.82 306.06 308.29\n 310.53 312.76 315. 317.23 319.47 321.7 323.94 326.17 328.41 330.64\n 332.88 335.11 337.35 339.58 341.82 344.05 346.29 348.52 350.76 352.99\n 355.23 357.46 359.7 361.93 364.17 366.4 368.64 370.87 373.11 375.34\n 377.58 379.81 382.05 384.28 386.52 388.75 390.99 393.22 395.46 397.69\n 399.93 402.16 404.4 406.63 408.87 411.1 413.34 415.57 417.81 420.04\n 422.28 424.51 426.75 428.98 431.22 433.45 435.69 437.92 440.16 442.39\n 444.63 446.86 449.1 451.33 453.57 455.8 458.04 460.27 462.51 464.74\n 466.98 469.21 471.45 473.68 475.92 478.15 480.39 482.62 484.86 487.09\n 489.33]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4ae5216aad9485d5478b3154188c2586e662b4d9
| 1,925 |
ipynb
|
Jupyter Notebook
|
notebooks/Lyna/Gff3ToBed.ipynb
|
VCMason/PyGenToolbox
|
3367a9b3df3bdb0223dd9671e9d355b81455fe2f
|
[
"MIT"
] | null | null | null |
notebooks/Lyna/Gff3ToBed.ipynb
|
VCMason/PyGenToolbox
|
3367a9b3df3bdb0223dd9671e9d355b81455fe2f
|
[
"MIT"
] | null | null | null |
notebooks/Lyna/Gff3ToBed.ipynb
|
VCMason/PyGenToolbox
|
3367a9b3df3bdb0223dd9671e9d355b81455fe2f
|
[
"MIT"
] | null | null | null | 23.192771 | 140 | 0.575584 |
[
[
[
"%load_ext autoreload\n%autoreload 2\nimport datetime\nimport os\nimport pandas as pd\nprint(datetime.datetime.now())\n\n#dir(pygentoolbox.Tools)\n%matplotlib inline\nimport matplotlib.pyplot as plt\nfrom pygentoolbox.Gff3ToBed import main",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n2020-07-30 22:52:29.068000\n"
],
[
"gff3file = 'D:\\\\LinuxShare\\\\Ciliates\\\\Genomes\\\\Annotations\\\\internal_eliminated_sequence_PGM_ParTIES.pt_51_with_ies.gff3'\n\nmain(gff3file)",
"Reading input: D:\\LinuxShare\\Ciliates\\Genomes\\Annotations\\internal_eliminated_sequence_PGM_ParTIES.pt_51_with_ies.gff3\noutput file: D:\\LinuxShare\\Ciliates\\Genomes\\Annotations\\internal_eliminated_sequence_PGM_ParTIES.pt_51_with_ies.bed\nfinished converting gff3 to bed file\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4ae525654920e348e4f15dec54c4255124eb9e09
| 182,152 |
ipynb
|
Jupyter Notebook
|
Coding_exercices/Classifying Iris Species.ipynb
|
FelipeChiriboga/s_a_projects
|
98e5432d88bd5fc0e6f1dcd25975f7b567c390aa
|
[
"MIT"
] | null | null | null |
Coding_exercices/Classifying Iris Species.ipynb
|
FelipeChiriboga/s_a_projects
|
98e5432d88bd5fc0e6f1dcd25975f7b567c390aa
|
[
"MIT"
] | null | null | null |
Coding_exercices/Classifying Iris Species.ipynb
|
FelipeChiriboga/s_a_projects
|
98e5432d88bd5fc0e6f1dcd25975f7b567c390aa
|
[
"MIT"
] | null | null | null | 245.819163 | 157,888 | 0.894297 |
[
[
[
"# Meet the data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.datasets import load_iris\nfrom sklearn.neighbors import KNeighborsClassifier",
"_____no_output_____"
],
[
"iris_dataset = load_iris()",
"_____no_output_____"
],
[
"iris_dataset",
"_____no_output_____"
],
[
"print(iris_dataset['DESCR'])",
".. _iris_dataset:\n\nIris plants dataset\n--------------------\n\n**Data Set Characteristics:**\n\n :Number of Instances: 150 (50 in each of three classes)\n :Number of Attributes: 4 numeric, predictive attributes and the class\n :Attribute Information:\n - sepal length in cm\n - sepal width in cm\n - petal length in cm\n - petal width in cm\n - class:\n - Iris-Setosa\n - Iris-Versicolour\n - Iris-Virginica\n \n :Summary Statistics:\n\n ============== ==== ==== ======= ===== ====================\n Min Max Mean SD Class Correlation\n ============== ==== ==== ======= ===== ====================\n sepal length: 4.3 7.9 5.84 0.83 0.7826\n sepal width: 2.0 4.4 3.05 0.43 -0.4194\n petal length: 1.0 6.9 3.76 1.76 0.9490 (high!)\n petal width: 0.1 2.5 1.20 0.76 0.9565 (high!)\n ============== ==== ==== ======= ===== ====================\n\n :Missing Attribute Values: None\n :Class Distribution: 33.3% for each of 3 classes.\n :Creator: R.A. Fisher\n :Donor: Michael Marshall (MARSHALL%[email protected])\n :Date: July, 1988\n\nThe famous Iris database, first used by Sir R.A. Fisher. The dataset is taken\nfrom Fisher's paper. Note that it's the same as in R, but not as in the UCI\nMachine Learning Repository, which has two wrong data points.\n\nThis is perhaps the best known database to be found in the\npattern recognition literature. Fisher's paper is a classic in the field and\nis referenced frequently to this day. (See Duda & Hart, for example.) The\ndata set contains 3 classes of 50 instances each, where each class refers to a\ntype of iris plant. One class is linearly separable from the other 2; the\nlatter are NOT linearly separable from each other.\n\n.. topic:: References\n\n - Fisher, R.A. \"The use of multiple measurements in taxonomic problems\"\n Annual Eugenics, 7, Part II, 179-188 (1936); also in \"Contributions to\n Mathematical Statistics\" (John Wiley, NY, 1950).\n - Duda, R.O., & Hart, P.E. (1973) Pattern Classification and Scene Analysis.\n (Q327.D83) John Wiley & Sons. ISBN 0-471-22361-1. See page 218.\n - Dasarathy, B.V. (1980) \"Nosing Around the Neighborhood: A New System\n Structure and Classification Rule for Recognition in Partially Exposed\n Environments\". IEEE Transactions on Pattern Analysis and Machine\n Intelligence, Vol. PAMI-2, No. 1, 67-71.\n - Gates, G.W. (1972) \"The Reduced Nearest Neighbor Rule\". IEEE Transactions\n on Information Theory, May 1972, 431-433.\n - See also: 1988 MLC Proceedings, 54-64. Cheeseman et al\"s AUTOCLASS II\n conceptual clustering system finds 3 classes in the data.\n - Many, many more ...\n"
],
[
"print('Target names: {}'.format(iris_dataset['target_names']))",
"Target names: ['setosa' 'versicolor' 'virginica']\n"
],
[
"print('Feature names: {}'.format(iris_dataset['feature_names']))",
"Feature names: ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']\n"
],
[
"print('Type of data: {}'.format(type(iris_dataset['data'])))",
"Type of data: <class 'numpy.ndarray'>\n"
],
[
"print('Shape of data: {}'.format(iris_dataset['data'].shape))",
"Shape of data: (150, 4)\n"
],
[
"print('First five columns of data:\\n{}'.format(iris_dataset['data'][:5]))",
"First five columns of data:\n[[5.1 3.5 1.4 0.2]\n [4.9 3. 1.4 0.2]\n [4.7 3.2 1.3 0.2]\n [4.6 3.1 1.5 0.2]\n [5. 3.6 1.4 0.2]]\n"
],
[
"print('Type of target: {}'.format(type(iris_dataset['target'])))",
"Type of target: <class 'numpy.ndarray'>\n"
],
[
"print('Shape of target: {}'.format(iris_dataset['target'].shape))",
"Shape of target: (150,)\n"
],
[
"print('target:\\n{}'.format(iris_dataset['target']))",
"target:\n[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0\n 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1\n 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2\n 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2\n 2 2]\n"
]
],
[
[
"# Training and Testing Data",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X_train, X_test, y_train, y_test = train_test_split(iris_dataset['data'],\n iris_dataset['target'],\n random_state=0)",
"_____no_output_____"
],
[
"print('X_train shape: {}'.format(X_train.shape))\nprint('y_train shape: {}'.format(y_train.shape))",
"X_train shape: (112, 4)\ny_train shape: (112,)\n"
],
[
"print('X_test shape: {}'.format(X_test.shape))\nprint('y_test shape: {}'.format(y_test.shape))",
"X_test shape: (38, 4)\ny_test shape: (38,)\n"
]
],
[
[
"# Inspect the Data!",
"_____no_output_____"
]
],
[
[
"# create dataframe from data in x_train\n# label the columns using the string in iris_dataset.feature_names\n\niris_dataframe = pd.DataFrame(X_train, columns=iris_dataset.feature_names)",
"_____no_output_____"
],
[
"# create a scatter matrix from the dataframe, color by y_train\ngrr = pd.scatter_matrix(iris_dataframe, c=y_train, figsize=(10, 10), marker='o',\n hist_kwds={'bins':20}, s=60, alpha=.8)",
"C:\\Users\\Pippo\\Anaconda3\\envs\\Test\\lib\\site-packages\\ipykernel_launcher.py:3: FutureWarning: pandas.scatter_matrix is deprecated, use pandas.plotting.scatter_matrix instead\n This is separate from the ipykernel package so we can avoid doing imports until\n"
]
],
[
[
"# K-Nearest Neighbors Classifier",
"_____no_output_____"
]
],
[
[
"knn = KNeighborsClassifier(n_neighbors=1)",
"_____no_output_____"
],
[
"knn.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"# Making Predictions",
"_____no_output_____"
]
],
[
[
"X_new = np.array([[5, 2.9, 1, 0.2]])\nprint('X_new.shape: {}'.format(X_new.shape))",
"X_new.shape: (1, 4)\n"
],
[
"prediction = knn.predict(X_new)\nprint('Prediction: {}'.format(prediction))\nprint('Predicted target name: {}'.format(iris_dataset['target_names'][prediction]))",
"Prediction: [0]\nPredicted target name: ['setosa']\n"
]
],
[
[
"# Evaluating the Model",
"_____no_output_____"
]
],
[
[
"y_pred = knn.predict(X_test)\nprint('Test set predictions:\\n {}'.format(y_pred))",
"Test set predictions:\n [2 1 0 2 0 2 0 1 1 1 2 1 1 1 1 0 1 1 0 0 2 1 0 0 2 0 0 1 1 0 2 1 0 2 2 1 0\n 2]\n"
],
[
"print('Test set score: {:.2f}'.format(np.mean(y_pred == y_test)))",
"Test set score: 0.97\n"
],
[
"print('Test set score: {:.2f}'.format(knn.score(X_test, y_test)))",
"Test set score: 0.97\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4ae534b3581f74334fe3def65271e9568da65884
| 6,619 |
ipynb
|
Jupyter Notebook
|
Stock Prediction Research Proposal.ipynb
|
shobes572/DSI_Module-34_Capstone
|
e0933ba5e49f8fe2f409ee1c87eedbcd8d25935f
|
[
"MIT"
] | null | null | null |
Stock Prediction Research Proposal.ipynb
|
shobes572/DSI_Module-34_Capstone
|
e0933ba5e49f8fe2f409ee1c87eedbcd8d25935f
|
[
"MIT"
] | null | null | null |
Stock Prediction Research Proposal.ipynb
|
shobes572/DSI_Module-34_Capstone
|
e0933ba5e49f8fe2f409ee1c87eedbcd8d25935f
|
[
"MIT"
] | null | null | null | 70.414894 | 626 | 0.714005 |
[
[
[
"# Stock Prediction Research Proposal",
"_____no_output_____"
],
[
"### Introduction\n\nThe main purpose of this research project is to create a Stock-prediction application to be used as a day-trading application to support investment decisions for beginners. The Target Audience for the project would be a tech company to whom I would be selling my model my model, or pushing my model to the implementation team who would integrate with a web app or on a mobile device. \n\n### Research Design\n\n[Day Trading](https://www.thestreet.com/investing/how-much-money-do-you-need-to-start-day-trading-15176512) has a few legal definitions outlined by the [SEC](https://www.sec.gov/files/daytrading.pdf), with some possible broader definitions being applied by your broker. For a beginning investor that won't necessarily want to commit $25,000, I will develop a model and application that can accurately predict high-yield trades over a timeframe that doesn't quite qualify as day-trading. For the purposes of this research, we will not be including options and other securities as possible transactions for the model.\n\n>_Executing four or more day trades within five business days_\n>\n> #### _-SEC_\n\nSo with a limit of four buy/sell transactions per week I will begin the model-building process with the assumed structure that we will buy a single stock on Monday Morning at opening and sell on Tuesday Morning at opening, and so on. \n* First, we'd need to define our stock universe\n * **Consideration 1:** Liquidity is one of the primary concerns when trading on short timespans. Just because you want to sell your stocks from what your algorithm is showing, doesn't mean that there are people buying.\n * **Consideration 2:** Volatility is the next factor we'll be considering. A higher volatility offers the opportunity for more gains at an increased risk. This measure is often calculated with the [Sharpe Ratio](https://www.investopedia.com/terms/s/sharperatio.asp) but the [Sortino Ratio](https://www.investopedia.com/terms/s/sortinoratio.asp) will likely be a better metric since it doesn't penalize net positive volatility.\n * Taking both of these consideration, I will be building the model with mid-cap stocks\n* Define the Historic timeframe for or predictions (2 days, 1 week, 2 weeks, etc.)\n* Decompose the time series and build a Deep Learning ML model on the data\n* Validate model accuracy and test live with paper trading on Alpaca\n\nAs we all know, predicting stock values based purely on the time series data is difficult due to the volatile nature of mid-cap stocks. So I will also include some of the following as market indicators and techniques for a multi-input deeplearning model:\n* Text data, vectorized with TFIDF or similar vectorizing techniques in NLP to measure sentiment toward a company in a given time period from one of the following sources:\n * Financial News Headlines\n * [Twitter data](https://developer.twitter.com/en/docs)\n * [stockwits](https://api.stocktwits.com/developers/docs)\n* [Applying CNN techniques](https://arxiv.org/abs/2001.09769) is a relatively new, novel way of modeling stock data that can apparently get better prediction accuracy than traditional time series modeling techniques used for stock data.",
"_____no_output_____"
],
[
"### Data\n\nThe primary focus for data will be acquiring market data from either [Polygon](https://polygon.io/stocks?gclid=Cj0KCQjwudb3BRC9ARIsAEa-vUu_C5pdMe26WRhWcj7nSpezXzIyXIs-Dec_LxrlkweD0nFN0MUlCPMaAo_OEALw_wcB) ([Alpaca](https://alpaca.markets/docs/) uses live Polygon data and can be used for live trading and paper trading to validate the model's efficacy) or [Yahoo Finance](https://rapidapi.com/apidojo/api/yahoo-finance1) and then adding predictors to increase the accuracy of the model, via [Twitter](https://developer.twitter.com/en/docs), [stockwits](https://api.stocktwits.com/developers/docs), News, etc.\n\n### Conclusion\n\nThe final, intended use-case for this model would be an optimized, automated, investor that would be able to perform multiple 'smart' decisions on trading. Once model integrity is established with a goal performance of 15% portfolio growth (average market return for both the S&P500 and Dow Jones Industrial Average in 2019)(**Note:** the projected market return for 2020 is around _6-10%_ , and the running 10-year average stays around _10-12%_ ). The model will also keep a live, ranked list of top stocks to invest in as an additional output for investors that may not be comfortable using algorithmic trading.\n\nThis will all be done without input from the end user. Future features can be added that would allow an investor to define their own stock universe, transaction limit frequency, portfolio diversity, etc. Alpaca is the broker for the trading in this model, so the end user would need to have an account created and linked to their bank account for them to be able to use trading script. The final script would need to be hosted on some sort of cloud system like [Amazon EC2](https://aws.amazon.com/ec2/), [Ubuntu Server](https://ubuntu.com/server), [OVHCloud](https://www.ovh.co.uk/), etc.",
"_____no_output_____"
],
[
"### Additional Resources\n\n* [SimFin](https://github.com/SimFin/simfin-tutorials)\n* [Alpaca Pipeline](https://github.com/alpacahq/pipeline-live)\n* [Quantitative Finance on Udemy](https://www.udemy.com/course/quantitative-finance-algorithmic-trading-in-python/)\n* [Pandas Technical Analysis](https://github.com/twopirllc/pandas-ta)\n* [Stock Universe](https://www.robertbrain.com/share-market/your-stock-universe.html)",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4ae534d16eafb14cbaf8244876bb66986552c4aa
| 502,084 |
ipynb
|
Jupyter Notebook
|
_notebooks/2021-05-30-Classification example 2 using Health Data with PyCaret.ipynb
|
davidrkearney/Kearney_Data_Science
|
25690d87595dfb6e07d0069e248ca8d96e275105
|
[
"Apache-2.0"
] | 1 |
2020-11-06T14:27:55.000Z
|
2020-11-06T14:27:55.000Z
|
_notebooks/2021-05-30-Classification example 2 using Health Data with PyCaret.ipynb
|
davidrkearney/Kearney_Data_Science
|
25690d87595dfb6e07d0069e248ca8d96e275105
|
[
"Apache-2.0"
] | 2 |
2021-05-20T18:01:16.000Z
|
2022-02-26T08:19:01.000Z
|
_notebooks/2021-05-30-Classification example 2 using Health Data with PyCaret.ipynb
|
davidrkearney/Kearney_Data_Science
|
25690d87595dfb6e07d0069e248ca8d96e275105
|
[
"Apache-2.0"
] | null | null | null | 146.039558 | 110,528 | 0.754533 |
[
[
[
"# Classification example 2 using Health Data with PyCaret\n",
"_____no_output_____"
]
],
[
[
"#Code from https://github.com/pycaret/pycaret/",
"_____no_output_____"
],
[
"# check version\nfrom pycaret.utils import version\nversion()",
"_____no_output_____"
]
],
[
[
"# 1. Data Repository",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nurl = 'https://raw.githubusercontent.com/davidrkearney/colab-notebooks/main/datasets/strokes_training.csv'\ndf = pd.read_csv(url, error_bad_lines=False)\ndf",
"_____no_output_____"
],
[
"data=df",
"_____no_output_____"
]
],
[
[
"# 2. Initialize Setup",
"_____no_output_____"
]
],
[
[
"from pycaret.classification import *\nclf1 = setup(df, target = 'stroke', session_id=123, log_experiment=True, experiment_name='health2')",
"_____no_output_____"
]
],
[
[
"# 3. Compare Baseline",
"_____no_output_____"
]
],
[
[
"best_model = compare_models()",
"_____no_output_____"
]
],
[
[
"# 4. Create Model",
"_____no_output_____"
]
],
[
[
"lr = create_model('lr')",
"_____no_output_____"
],
[
"dt = create_model('dt')",
"_____no_output_____"
],
[
"rf = create_model('rf', fold = 5)",
"_____no_output_____"
],
[
"models()",
"_____no_output_____"
],
[
"models(type='ensemble').index.tolist()",
"_____no_output_____"
],
[
"#ensembled_models = compare_models(whitelist = models(type='ensemble').index.tolist(), fold = 3)",
"_____no_output_____"
]
],
[
[
"# 5. Tune Hyperparameters",
"_____no_output_____"
]
],
[
[
"tuned_lr = tune_model(lr)",
"_____no_output_____"
],
[
"tuned_rf = tune_model(rf)",
"_____no_output_____"
]
],
[
[
"# 6. Ensemble Model",
"_____no_output_____"
]
],
[
[
"bagged_dt = ensemble_model(dt)",
"_____no_output_____"
],
[
"boosted_dt = ensemble_model(dt, method = 'Boosting')",
"_____no_output_____"
]
],
[
[
"# 7. Blend Models",
"_____no_output_____"
]
],
[
[
"blender = blend_models(estimator_list = [boosted_dt, bagged_dt, tuned_rf], method = 'soft')",
"_____no_output_____"
]
],
[
[
"# 8. Stack Models",
"_____no_output_____"
]
],
[
[
"stacker = stack_models(estimator_list = [boosted_dt,bagged_dt,tuned_rf], meta_model=rf)",
"_____no_output_____"
]
],
[
[
"# 9. Analyze Model",
"_____no_output_____"
]
],
[
[
"plot_model(rf)",
"_____no_output_____"
],
[
"plot_model(rf, plot = 'confusion_matrix')",
"_____no_output_____"
],
[
"plot_model(rf, plot = 'boundary')",
"_____no_output_____"
],
[
"plot_model(rf, plot = 'feature')",
"_____no_output_____"
],
[
"plot_model(rf, plot = 'pr')",
"_____no_output_____"
],
[
"plot_model(rf, plot = 'class_report')",
"_____no_output_____"
],
[
"evaluate_model(rf)",
"_____no_output_____"
]
],
[
[
"# 10. Interpret Model",
"_____no_output_____"
]
],
[
[
"catboost = create_model('rf', cross_validation=False)",
"_____no_output_____"
],
[
"interpret_model(catboost)",
"_____no_output_____"
],
[
"interpret_model(catboost, plot = 'correlation')",
"_____no_output_____"
],
[
"interpret_model(catboost, plot = 'reason', observation = 12)",
"_____no_output_____"
]
],
[
[
"# 11. AutoML()",
"_____no_output_____"
]
],
[
[
"best = automl(optimize = 'Recall')\nbest",
"_____no_output_____"
]
],
[
[
"# 12. Predict Model",
"_____no_output_____"
]
],
[
[
"pred_holdouts = predict_model(lr)\npred_holdouts.head()",
"_____no_output_____"
],
[
"new_data = data.copy()\nnew_data.drop(['Purchase'], axis=1, inplace=True)\npredict_new = predict_model(best, data=new_data)\npredict_new.head()",
"_____no_output_____"
]
],
[
[
"# 13. Save / Load Model",
"_____no_output_____"
]
],
[
[
"save_model(best, model_name='best-model')",
"_____no_output_____"
],
[
"loaded_bestmodel = load_model('best-model')\nprint(loaded_bestmodel)",
"_____no_output_____"
],
[
"from sklearn import set_config\nset_config(display='diagram')\nloaded_bestmodel[0]",
"_____no_output_____"
],
[
"from sklearn import set_config\nset_config(display='text')",
"_____no_output_____"
]
],
[
[
"# 14. Deploy Model",
"_____no_output_____"
]
],
[
[
"deploy_model(best, model_name = 'best-aws', authentication = {'bucket' : 'pycaret-test'})",
"_____no_output_____"
]
],
[
[
"# 15. Get Config / Set Config",
"_____no_output_____"
]
],
[
[
"X_train = get_config('X_train')\nX_train.head()",
"_____no_output_____"
],
[
"get_config('seed')",
"_____no_output_____"
],
[
"from pycaret.classification import set_config\nset_config('seed', 999)",
"_____no_output_____"
],
[
"get_config('seed')",
"_____no_output_____"
]
],
[
[
"# 16. MLFlow UI",
"_____no_output_____"
]
],
[
[
"# !mlflow ui",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae54ad62cce206578388329542fd3ac26014421
| 84,953 |
ipynb
|
Jupyter Notebook
|
notebooks/tweet_question_generation_corrected_data.ipynb
|
mayaang/question_generation
|
20255d2d3164f93b9fe55e062cbffa3db4d54ce9
|
[
"MIT"
] | null | null | null |
notebooks/tweet_question_generation_corrected_data.ipynb
|
mayaang/question_generation
|
20255d2d3164f93b9fe55e062cbffa3db4d54ce9
|
[
"MIT"
] | null | null | null |
notebooks/tweet_question_generation_corrected_data.ipynb
|
mayaang/question_generation
|
20255d2d3164f93b9fe55e062cbffa3db4d54ce9
|
[
"MIT"
] | null | null | null | 56.409695 | 558 | 0.665533 |
[
[
[
"<a href=\"https://colab.research.google.com/github/patil-suraj/question_generation/blob/master/question_generation.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip install -U transformers==3.0.0",
"Requirement already up-to-date: transformers==3.0.0 in /usr/local/lib/python3.6/dist-packages (3.0.0)\nRequirement already satisfied, skipping upgrade: tqdm>=4.27 in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (4.41.1)\nRequirement already satisfied, skipping upgrade: tokenizers==0.8.0-rc4 in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (0.8.0rc4)\nRequirement already satisfied, skipping upgrade: dataclasses; python_version < \"3.7\" in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (0.7)\nRequirement already satisfied, skipping upgrade: requests in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (2.23.0)\nRequirement already satisfied, skipping upgrade: sacremoses in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (0.0.43)\nRequirement already satisfied, skipping upgrade: regex!=2019.12.17 in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (2019.12.20)\nRequirement already satisfied, skipping upgrade: packaging in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (20.4)\nRequirement already satisfied, skipping upgrade: filelock in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (3.0.12)\nRequirement already satisfied, skipping upgrade: sentencepiece in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (0.1.91)\nRequirement already satisfied, skipping upgrade: numpy in /usr/local/lib/python3.6/dist-packages (from transformers==3.0.0) (1.18.5)\nRequirement already satisfied, skipping upgrade: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->transformers==3.0.0) (2.10)\nRequirement already satisfied, skipping upgrade: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->transformers==3.0.0) (2020.6.20)\nRequirement already satisfied, skipping upgrade: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->transformers==3.0.0) (1.24.3)\nRequirement already satisfied, skipping upgrade: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->transformers==3.0.0) (3.0.4)\nRequirement already satisfied, skipping upgrade: six in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers==3.0.0) (1.15.0)\nRequirement already satisfied, skipping upgrade: joblib in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers==3.0.0) (0.16.0)\nRequirement already satisfied, skipping upgrade: click in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers==3.0.0) (7.1.2)\nRequirement already satisfied, skipping upgrade: pyparsing>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from packaging->transformers==3.0.0) (2.4.7)\n"
],
[
"!python -m nltk.downloader punkt",
"/usr/lib/python3.6/runpy.py:125: RuntimeWarning: 'nltk.downloader' found in sys.modules after import of package 'nltk', but prior to execution of 'nltk.downloader'; this may result in unpredictable behaviour\n warn(RuntimeWarning(msg))\n[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n"
],
[
"!pip3 install nlp",
"Requirement already satisfied: nlp in /usr/local/lib/python3.6/dist-packages (0.4.0)\nRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.6/dist-packages (from nlp) (4.41.1)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from nlp) (1.18.5)\nRequirement already satisfied: dataclasses; python_version < \"3.7\" in /usr/local/lib/python3.6/dist-packages (from nlp) (0.7)\nRequirement already satisfied: filelock in /usr/local/lib/python3.6/dist-packages (from nlp) (3.0.12)\nRequirement already satisfied: requests>=2.19.0 in /usr/local/lib/python3.6/dist-packages (from nlp) (2.23.0)\nRequirement already satisfied: xxhash in /usr/local/lib/python3.6/dist-packages (from nlp) (2.0.0)\nRequirement already satisfied: dill in /usr/local/lib/python3.6/dist-packages (from nlp) (0.3.2)\nRequirement already satisfied: pandas in /usr/local/lib/python3.6/dist-packages (from nlp) (1.0.5)\nRequirement already satisfied: pyarrow>=0.16.0 in /usr/local/lib/python3.6/dist-packages (from nlp) (1.0.1)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19.0->nlp) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19.0->nlp) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19.0->nlp) (2020.6.20)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19.0->nlp) (2.10)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas->nlp) (2018.9)\nRequirement already satisfied: python-dateutil>=2.6.1 in /usr/local/lib/python3.6/dist-packages (from pandas->nlp) (2.8.1)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil>=2.6.1->pandas->nlp) (1.15.0)\n"
],
[
"!pip3 install git+https://github.com/Maluuba/nlg-eval.git@master\n",
"Collecting git+https://github.com/Maluuba/nlg-eval.git@master\n Cloning https://github.com/Maluuba/nlg-eval.git (to revision master) to /tmp/pip-req-build-ql1r64u3\n Running command git clone -q https://github.com/Maluuba/nlg-eval.git /tmp/pip-req-build-ql1r64u3\nRequirement already satisfied (use --upgrade to upgrade): nlg-eval==2.3 from git+https://github.com/Maluuba/nlg-eval.git@master in /usr/local/lib/python3.6/dist-packages\nRequirement already satisfied: click>=6.3 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (7.1.2)\nRequirement already satisfied: nltk>=3.1 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (3.2.5)\nRequirement already satisfied: numpy>=1.11.0 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (1.18.5)\nRequirement already satisfied: psutil>=5.6.2 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (5.7.2)\nRequirement already satisfied: requests>=2.19 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (2.23.0)\nRequirement already satisfied: six>=1.11 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (1.15.0)\nRequirement already satisfied: scipy>=0.17.0 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (1.4.1)\nRequirement already satisfied: scikit-learn>=0.17 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (0.22.2.post1)\nRequirement already satisfied: gensim>=3 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (3.6.0)\nRequirement already satisfied: Theano>=0.8.1 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (1.0.5)\nRequirement already satisfied: tqdm>=4.24 in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (4.41.1)\nRequirement already satisfied: xdg in /usr/local/lib/python3.6/dist-packages (from nlg-eval==2.3) (4.0.1)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19->nlg-eval==2.3) (2.10)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19->nlg-eval==2.3) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19->nlg-eval==2.3) (3.0.4)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests>=2.19->nlg-eval==2.3) (2020.6.20)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.17->nlg-eval==2.3) (0.16.0)\nRequirement already satisfied: smart-open>=1.2.1 in /usr/local/lib/python3.6/dist-packages (from gensim>=3->nlg-eval==2.3) (2.1.0)\nRequirement already satisfied: boto3 in /usr/local/lib/python3.6/dist-packages (from smart-open>=1.2.1->gensim>=3->nlg-eval==2.3) (1.14.37)\nRequirement already satisfied: boto in /usr/local/lib/python3.6/dist-packages (from smart-open>=1.2.1->gensim>=3->nlg-eval==2.3) (2.49.0)\nRequirement already satisfied: s3transfer<0.4.0,>=0.3.0 in /usr/local/lib/python3.6/dist-packages (from boto3->smart-open>=1.2.1->gensim>=3->nlg-eval==2.3) (0.3.3)\nRequirement already satisfied: botocore<1.18.0,>=1.17.37 in /usr/local/lib/python3.6/dist-packages (from boto3->smart-open>=1.2.1->gensim>=3->nlg-eval==2.3) (1.17.37)\nRequirement already satisfied: jmespath<1.0.0,>=0.7.1 in /usr/local/lib/python3.6/dist-packages (from boto3->smart-open>=1.2.1->gensim>=3->nlg-eval==2.3) (0.10.0)\nRequirement already satisfied: docutils<0.16,>=0.10 in /usr/local/lib/python3.6/dist-packages (from botocore<1.18.0,>=1.17.37->boto3->smart-open>=1.2.1->gensim>=3->nlg-eval==2.3) (0.15.2)\nRequirement already satisfied: python-dateutil<3.0.0,>=2.1 in /usr/local/lib/python3.6/dist-packages (from botocore<1.18.0,>=1.17.37->boto3->smart-open>=1.2.1->gensim>=3->nlg-eval==2.3) (2.8.1)\nBuilding wheels for collected packages: nlg-eval\n Building wheel for nlg-eval (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for nlg-eval: filename=nlg_eval-2.3-cp36-none-any.whl size=68175138 sha256=d4bceb33ce44b1062eb98002f95238c29595278e8f514710080644c9785af7d7\n Stored in directory: /tmp/pip-ephem-wheel-cache-_vwnh7yb/wheels/a5/7c/fd/f312beca2adcc3f49cb40570730658dad37bb5709f5d237a56\nSuccessfully built nlg-eval\n"
],
[
"!nlg-eval --setup",
"[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n\u001b[31mInstalling to /root/.cache/nlgeval\u001b[0m\n\u001b[31mIn case of incomplete downloads, delete the directory and run `nlg-eval --setup /root/.cache/nlgeval' again.\u001b[0m\nDownloading https://raw.githubusercontent.com/robmsmt/glove-gensim/4c2224bccd61627b76c50a5e1d6afd1c82699d22/glove2word2vec.py to /usr/local/lib/python3.6/dist-packages/nlgeval/word2vec.\nglove2word2vec.py: 100% 1.00/1.00 [00:00<00:00, 616 chunks/s]\n"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly&response_type=code\n\nEnter your authorization code:\n··········\nMounted at /content/drive\n"
],
[
"%cd drive/My\\ Drive/question_generation/question_generation/",
"/content/drive/My Drive/question_generation/question_generation\n"
],
[
"ls data/tweet_manual_multitask/",
"tweet_dev_squad_manual_edit.json tweet_train_squad_manual_edit.json\ntweet_manual_multitask.py\n"
],
[
"# Already DONE !!!!!\n#!python3 prepare_data.py --task qg --model_type t5 --dataset_path data/tweet_manual_multitask --qg_format highlight_qg_format --max_source_length 512 --max_target_length 32 --train_file_name train_data_qg_hl_t5_tweet_manual.pt --valid_file_name valid_data_qg_hl_t5_tweet_manual.pt",
"2020-08-20 19:05:37.584306: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\n08/20/2020 19:05:39 - INFO - transformers.tokenization_utils_base - loading file https://s3.amazonaws.com/models.huggingface.co/bert/t5-spiece.model from cache at /root/.cache/torch/transformers/68f1b8dbca4350743bb54b8c4169fd38cbabaad564f85a9239337a8d0342af9f.9995af32582a1a7062cb3173c118cb7b4636fa03feb967340f20fc37406f021f\n08/20/2020 19:05:39 - INFO - transformers.tokenization_utils - Adding <sep> to the vocabulary\n08/20/2020 19:05:39 - INFO - transformers.tokenization_utils - Adding <hl> to the vocabulary\n08/20/2020 19:05:39 - INFO - nlp.load - Checking data/tweet_manual_multitask/tweet_manual_multitask.py for additional imports.\n08/20/2020 19:05:39 - INFO - filelock - Lock 140323353226040 acquired on data/tweet_manual_multitask/tweet_manual_multitask.py.lock\n08/20/2020 19:05:39 - INFO - nlp.load - Found main folder for dataset data/tweet_manual_multitask/tweet_manual_multitask.py at /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask\n08/20/2020 19:05:39 - INFO - nlp.load - Creating specific version folder for dataset data/tweet_manual_multitask/tweet_manual_multitask.py at /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d\n08/20/2020 19:05:39 - INFO - nlp.load - Copying script file from data/tweet_manual_multitask/tweet_manual_multitask.py to /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tweet_manual_multitask.py\n08/20/2020 19:05:39 - INFO - nlp.load - Couldn't find dataset infos file at data/tweet_manual_multitask/dataset_infos.json\n08/20/2020 19:05:39 - INFO - nlp.load - Creating metadata file for dataset data/tweet_manual_multitask/tweet_manual_multitask.py at /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tweet_manual_multitask.json\n08/20/2020 19:05:39 - INFO - filelock - Lock 140323353226040 released on data/tweet_manual_multitask/tweet_manual_multitask.py.lock\n[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n08/20/2020 19:05:39 - INFO - nlp.builder - Generating dataset squad_multitask (/root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d)\nDownloading and preparing dataset squad_multitask/highlight_qg_format (download: Unknown size, generated: Unknown size, post-processed: Unknown sizetotal: Unknown size) to /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d...\n08/20/2020 19:05:40 - INFO - nlp.builder - Dataset not on Hf google storage. Downloading and preparing it from source\n08/20/2020 19:05:40 - INFO - nlp.utils.info_utils - Unable to verify checksums.\n08/20/2020 19:05:40 - INFO - nlp.builder - Generating split train\n0 examples [00:00, ? examples/s]08/20/2020 19:05:40 - INFO - root - generating examples from = /content/drive/My Drive/question_generation/question_generation/data/tweet_manual_multitask/tweet_train_squad_manual_edit.json\n08/20/2020 19:05:42 - INFO - nlp.arrow_writer - Done writing 41507 examples in 10296027 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d.incomplete/squad_multitask-train.arrow.\n08/20/2020 19:05:42 - INFO - nlp.builder - Generating split validation\n0 examples [00:00, ? examples/s]08/20/2020 19:05:42 - INFO - root - generating examples from = /content/drive/My Drive/question_generation/question_generation/data/tweet_manual_multitask/tweet_dev_squad_manual_edit.json\n08/20/2020 19:05:42 - INFO - nlp.arrow_writer - Done writing 3469 examples in 880816 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d.incomplete/squad_multitask-validation.arrow.\n08/20/2020 19:05:42 - INFO - nlp.utils.info_utils - Unable to verify splits sizes.\nDataset squad_multitask downloaded and prepared to /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d. Subsequent calls will reuse this data.\n08/20/2020 19:05:42 - INFO - nlp.builder - Constructing Dataset for split train, from /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d\n08/20/2020 19:05:42 - INFO - nlp.utils.info_utils - Unable to verify checksums.\n08/20/2020 19:05:42 - INFO - nlp.load - Checking data/tweet_manual_multitask/tweet_manual_multitask.py for additional imports.\n08/20/2020 19:05:42 - INFO - filelock - Lock 140323353196472 acquired on data/tweet_manual_multitask/tweet_manual_multitask.py.lock\n08/20/2020 19:05:42 - INFO - nlp.load - Found main folder for dataset data/tweet_manual_multitask/tweet_manual_multitask.py at /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask\n08/20/2020 19:05:42 - INFO - nlp.load - Found specific version folder for dataset data/tweet_manual_multitask/tweet_manual_multitask.py at /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d\n08/20/2020 19:05:42 - INFO - nlp.load - Found script file from data/tweet_manual_multitask/tweet_manual_multitask.py to /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tweet_manual_multitask.py\n08/20/2020 19:05:42 - INFO - nlp.load - Couldn't find dataset infos file at data/tweet_manual_multitask/dataset_infos.json\n08/20/2020 19:05:42 - INFO - nlp.load - Found metadata file for dataset data/tweet_manual_multitask/tweet_manual_multitask.py at /usr/local/lib/python3.6/dist-packages/nlp/datasets/tweet_manual_multitask/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tweet_manual_multitask.json\n08/20/2020 19:05:42 - INFO - filelock - Lock 140323353196472 released on data/tweet_manual_multitask/tweet_manual_multitask.py.lock\n08/20/2020 19:05:42 - INFO - nlp.builder - Overwrite dataset info from restored data version.\n08/20/2020 19:05:42 - INFO - nlp.info - Loading Dataset info from /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d\n08/20/2020 19:05:42 - INFO - nlp.builder - Reusing dataset squad_multitask (/root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d)\n08/20/2020 19:05:42 - INFO - nlp.builder - Constructing Dataset for split validation, from /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d\n08/20/2020 19:05:43 - INFO - nlp.utils.info_utils - Unable to verify checksums.\n08/20/2020 19:05:43 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-385f8db752e9b46ea6fe2a445e7b5453.arrow\n100% 42/42 [00:00<00:00, 159.14it/s]\n08/20/2020 19:05:43 - INFO - nlp.arrow_writer - Done writing 10691 examples in 2848933 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmpphximduv.\n08/20/2020 19:05:43 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-c964c7ea09b11bbc7f3cfe6fdba16451.arrow\n100% 4/4 [00:00<00:00, 185.02it/s]\n08/20/2020 19:05:43 - INFO - nlp.arrow_writer - Done writing 1085 examples in 277865 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmp7sre9ob8.\n08/20/2020 19:05:43 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-1bde36e2c6adacfafd4a1912196beb6c.arrow\n100% 10691/10691 [00:00<00:00, 35968.66it/s]\n08/20/2020 19:05:43 - INFO - nlp.arrow_writer - Done writing 10691 examples in 2955471 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmpleb82ip9.\n08/20/2020 19:05:43 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-768a25c198123fc7c827b05d9d581491.arrow\n100% 10691/10691 [00:00<00:00, 35161.27it/s]\n08/20/2020 19:05:43 - INFO - nlp.arrow_writer - Done writing 10691 examples in 2827179 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmpkwzfpmlx.\n08/20/2020 19:05:43 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-3f5feb2034af54d5401b0a93ae99378a.arrow\n100% 11/11 [00:20<00:00, 1.83s/it]\n08/20/2020 19:06:03 - INFO - nlp.arrow_writer - Done writing 10691 examples in 93273171 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmpto1bytbt.\n08/20/2020 19:06:03 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-650cdd2eba7b638d930d545ddc47079c.arrow\n100% 1085/1085 [00:00<00:00, 34153.78it/s]\n08/20/2020 19:06:04 - INFO - nlp.arrow_writer - Done writing 1085 examples in 288691 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmpyepojtld.\n08/20/2020 19:06:04 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-d9c7ed0bbdf0e3593d0ef443dd95d48e.arrow\n100% 1085/1085 [00:00<00:00, 34618.05it/s]\n08/20/2020 19:06:04 - INFO - nlp.arrow_writer - Done writing 1085 examples in 275671 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmpgg1hhp_5.\n08/20/2020 19:06:04 - INFO - nlp.arrow_dataset - Caching processed dataset at /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/cache-d2c57ee0b1d706e7a387ef1fcc90a88a.arrow\n100% 2/2 [00:02<00:00, 1.03s/it]\n08/20/2020 19:06:06 - INFO - nlp.arrow_writer - Done writing 1085 examples in 9454795 bytes /root/.cache/huggingface/datasets/squad_multitask/highlight_qg_format/1.0.0/a5b4dc1174978fb27b04d14e1b35abd5f90bd94bfbbf6f9889e7a84626ebce6d/tmpg5f30ox6.\n08/20/2020 19:06:06 - INFO - nlp.arrow_dataset - Set __getitem__(key) output type to torch for ['source_ids', 'target_ids', 'attention_mask'] columns (when key is int or slice) and don't output other (un-formated) columns.\n08/20/2020 19:06:06 - INFO - nlp.arrow_dataset - Set __getitem__(key) output type to torch for ['source_ids', 'target_ids', 'attention_mask'] columns (when key is int or slice) and don't output other (un-formated) columns.\n08/20/2020 19:06:06 - INFO - __main__ - saved train dataset at data/train_data_qg_hl_t5_tweet_manual.pt\n08/20/2020 19:06:06 - INFO - __main__ - saved validation dataset at data/valid_data_qg_hl_t5_tweet_manual.pt\n08/20/2020 19:06:08 - INFO - __main__ - saved tokenizer at t5_qg_tokenizer\n"
],
[
"###################### \n# valhalla/t5-small-qg-hl\n######################\n#08/15/2020 15:57:21 - INFO - transformers.configuration_utils - Configuration saved in t5-small-qg-hl/config.json\n#08/15/2020 15:57:23 - INFO - transformers.modeling_utils - Model weights saved in t5-small-qg-hl/pytorch_model.bin\n%%capture loggingfine\n!python3 run_qg.py \\\n --model_name_or_path valhalla/t5-small-qg-hl \\\n --model_type t5 \\\n --tokenizer_name_or_path t5_qg_tokenizer \\\n --output_dir t5-small-qg-hl-15-manual \\\n --train_file_path data/train_data_qg_hl_t5_tweet_manual.pt \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --per_device_train_batch_size 16 \\\n --per_device_eval_batch_size 16 \\\n --gradient_accumulation_steps 8 \\\n --learning_rate 1e-4 \\\n --num_train_epochs 15 \\\n --seed 42 \\\n --do_train \\\n --do_eval \\\n --evaluate_during_training \\\n --logging_steps 100\\\n --logging_dir t5-small-qg-15-manual-log\\\n --overwrite_output_dir",
"_____no_output_____"
],
[
"#using originally pretrained model small t5 valhalla\n!python eval.py \\\n --model_name_or_path valhalla/t5-small-qg-hl \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_t5-original-small-qg-hl_tweet_manual_model.txt",
"2020-08-21 19:23:23.546867: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\nDownloading: 100% 792k/792k [00:00<00:00, 3.31MB/s]\nDownloading: 100% 31.0/31.0 [00:00<00:00, 22.4kB/s]\nDownloading: 100% 65.0/65.0 [00:00<00:00, 41.6kB/s]\nDownloading: 100% 90.0/90.0 [00:00<00:00, 60.6kB/s]\n 0% 0/34 [00:00<?, ?it/s]/usr/local/lib/python3.6/dist-packages/nlp/utils/py_utils.py:191: UserWarning: The given NumPy array is not writeable, and PyTorch does not support non-writeable tensors. This means you can write to the underlying (supposedly non-writeable) NumPy array using the tensor. You may want to copy the array to protect its data or make it writeable before converting it to a tensor. This type of warning will be suppressed for the rest of this program. (Triggered internally at /pytorch/torch/csrc/utils/tensor_numpy.cpp:141.)\n return function(data_struct)\n100% 34/34 [01:14<00:00, 2.19s/it]\n"
],
[
", #evaluate valhalla small model\n!nlg-eval --hypothesis=hypothesis_t5-original-small-qg-hl_tweet_manual_model.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"\u001b[32mUsing data from /root/.cache/nlgeval\u001b[0m\n\u001b[32mIn case of broken downloads, remove the directory and run setup again.\u001b[0m\nBleu_1: 0.168221\nBleu_2: 0.101524\nBleu_3: 0.067962\nBleu_4: 0.047070\nMETEOR: 0.200778\nROUGE_L: 0.183403\nCIDEr: 0.552163\n"
],
[
"#using finetuned small model with 15 epochs\n!python eval.py \\\n --model_name_or_path t5-small-qg-hl-15-manual \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_t5-small-fine-tuned-qg-hl_t5_tweet_manual.txt",
"2020-08-21 19:25:48.493103: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\n 0% 0/34 [00:00<?, ?it/s]/usr/local/lib/python3.6/dist-packages/nlp/utils/py_utils.py:191: UserWarning: The given NumPy array is not writeable, and PyTorch does not support non-writeable tensors. This means you can write to the underlying (supposedly non-writeable) NumPy array using the tensor. You may want to copy the array to protect its data or make it writeable before converting it to a tensor. This type of warning will be suppressed for the rest of this program. (Triggered internally at /pytorch/torch/csrc/utils/tensor_numpy.cpp:141.)\n return function(data_struct)\n100% 34/34 [01:08<00:00, 2.03s/it]\n"
],
[
", #evaluate finetuned small model\n!nlg-eval --hypothesis=hypothesis_t5-small-fine-tuned-qg-hl_t5_tweet_manual.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"\u001b[32mUsing data from /root/.cache/nlgeval\u001b[0m\n\u001b[32mIn case of broken downloads, remove the directory and run setup again.\u001b[0m\nBleu_1: 0.351202\nBleu_2: 0.246024\nBleu_3: 0.182722\nBleu_4: 0.141755\nMETEOR: 0.225531\nROUGE_L: 0.371697\nCIDEr: 1.479602\n"
],
[
"#valhalla/t5-base-qg-hl\n###########################\n#08/15/2020 15:57:21 - INFO - transformers.configuration_utils - Configuration saved in t5-small-qg-hl/config.json\n#08/15/2020 15:57:23 - INFO - transformers.modeling_utils - Model weights saved in t5-small-qg-hl/pytorch_model.bin\n%%capture loggingfine\n!python3 run_qg.py \\\n --model_name_or_path valhalla/t5-base-qg-hl \\\n --model_type t5 \\\n --tokenizer_name_or_path t5_qg_tokenizer \\\n --output_dir t5-base-qg-hl-15-manual \\\n --train_file_path data/train_data_qg_hl_t5_tweet_manual.pt \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --per_device_train_batch_size 16 \\\n --per_device_eval_batch_size 16 \\\n --gradient_accumulation_steps 8 \\\n --learning_rate 1e-4 \\\n --num_train_epochs 15 \\\n --seed 42 \\\n --do_train \\\n --do_eval \\\n --evaluate_during_training \\\n --logging_steps 100\\\n --logging_dir t5-base-qg-15-manual-log\\\n --overwrite_output_dir",
"_____no_output_____"
],
[
"#using originally pretrained model base t5 valhalla\n!python eval.py \\\n --model_name_or_path valhalla/t5-base-qg-hl \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_original_base_base-qg-hl_tweet_manual_model.txt",
"2020-08-22 09:08:49.640018: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\nDownloading: 100% 1.02k/1.02k [00:00<00:00, 905kB/s]\nDownloading: 100% 792k/792k [00:00<00:00, 2.42MB/s]\nDownloading: 100% 15.0/15.0 [00:00<00:00, 11.3kB/s]\nDownloading: 100% 1.79k/1.79k [00:00<00:00, 1.41MB/s]\nDownloading: 100% 129/129 [00:00<00:00, 95.3kB/s]\nDownloading: 100% 892M/892M [00:29<00:00, 29.8MB/s]\n 0% 0/34 [00:00<?, ?it/s]/usr/local/lib/python3.6/dist-packages/nlp/utils/py_utils.py:191: UserWarning: The given NumPy array is not writeable, and PyTorch does not support non-writeable tensors. This means you can write to the underlying (supposedly non-writeable) NumPy array using the tensor. You may want to copy the array to protect its data or make it writeable before converting it to a tensor. This type of warning will be suppressed for the rest of this program. (Triggered internally at /pytorch/torch/csrc/utils/tensor_numpy.cpp:141.)\n return function(data_struct)\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [96,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [97,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [98,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [99,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [100,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [101,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [102,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [103,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [104,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [105,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [106,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [107,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [108,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [109,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [110,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [111,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [112,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [113,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [114,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [115,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [116,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [117,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [118,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [119,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [120,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [121,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [122,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [123,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [124,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [125,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [126,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [127,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [32,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [33,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [34,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [35,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [36,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [37,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [38,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [39,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [40,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [41,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [42,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [43,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [44,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [45,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [46,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [47,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [48,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [49,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [50,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [51,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [52,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [53,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [54,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [55,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [56,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [57,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [58,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [59,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [60,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [61,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [62,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [63,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [0,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [1,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [2,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [3,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [4,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [5,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [6,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [7,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [8,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [9,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [10,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [11,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [12,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [13,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [14,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [15,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [16,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [17,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [18,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [19,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [20,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [21,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [22,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [23,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [24,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [25,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [26,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [27,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [28,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [29,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [30,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [31,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [64,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [65,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [66,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [67,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [68,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [69,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [70,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [71,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [72,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [73,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [74,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [75,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [76,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [77,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [78,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [79,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [80,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [81,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [82,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [83,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [84,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [85,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [86,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [87,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [88,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [89,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [90,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [91,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [92,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [93,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [94,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n/pytorch/aten/src/THC/THCTensorIndex.cu:272: indexSelectLargeIndex: block: [78,0,0], thread: [95,0,0] Assertion `srcIndex < srcSelectDimSize` failed.\n 0% 0/34 [00:00<?, ?it/s]\nTraceback (most recent call last):\n File \"eval.py\", line 92, in <module>\n main()\n File \"eval.py\", line 82, in main\n max_length=args.max_decoding_length\n File \"eval.py\", line 52, in get_predictions\n length_penalty=length_penalty,\n File \"/usr/local/lib/python3.6/dist-packages/torch/autograd/grad_mode.py\", line 15, in decorate_context\n return func(*args, **kwargs)\n File \"/usr/local/lib/python3.6/dist-packages/transformers/modeling_utils.py\", line 1159, in generate\n encoder_outputs: tuple = encoder(input_ids, attention_mask=attention_mask)\n File \"/usr/local/lib/python3.6/dist-packages/torch/nn/modules/module.py\", line 722, in _call_impl\n result = self.forward(*input, **kwargs)\n File \"/usr/local/lib/python3.6/dist-packages/transformers/modeling_t5.py\", line 748, in forward\n output_attentions=output_attentions,\n File \"/usr/local/lib/python3.6/dist-packages/torch/nn/modules/module.py\", line 722, in _call_impl\n result = self.forward(*input, **kwargs)\n File \"/usr/local/lib/python3.6/dist-packages/transformers/modeling_t5.py\", line 516, in forward\n output_attentions=output_attentions,\n File \"/usr/local/lib/python3.6/dist-packages/torch/nn/modules/module.py\", line 722, in _call_impl\n result = self.forward(*input, **kwargs)\n File \"/usr/local/lib/python3.6/dist-packages/transformers/modeling_t5.py\", line 423, in forward\n output_attentions=output_attentions,\n File \"/usr/local/lib/python3.6/dist-packages/torch/nn/modules/module.py\", line 722, in _call_impl\n result = self.forward(*input, **kwargs)\n File \"/usr/local/lib/python3.6/dist-packages/transformers/modeling_t5.py\", line 338, in forward\n q = shape(self.q(input)) # (bs, n_heads, qlen, dim_per_head)\n File \"/usr/local/lib/python3.6/dist-packages/torch/nn/modules/module.py\", line 722, in _call_impl\n result = self.forward(*input, **kwargs)\n File \"/usr/local/lib/python3.6/dist-packages/torch/nn/modules/linear.py\", line 91, in forward\n return F.linear(input, self.weight, self.bias)\n File \"/usr/local/lib/python3.6/dist-packages/torch/nn/functional.py\", line 1676, in linear\n output = input.matmul(weight.t())\nRuntimeError: CUDA error: CUBLAS_STATUS_ALLOC_FAILED when calling `cublasCreate(handle)`\n"
],
[
", #evaluate valhalla base model\n!nlg-eval --hypothesis=hypothesis_original_base_base-qg-hl_tweet_manual_model.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"Usage: nlg-eval [OPTIONS]\nTry 'nlg-eval --help' for help.\n\nError: Invalid value for '--hypothesis': Path 'hypothesis_original_base_base-qg-hl_tweet_manual_model.txt' does not exist.\n"
],
[
"#using originally pretrained model base t5 valhalla\n!python eval.py \\\n --model_name_or_path t5-base-qg-hl-15-manual \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_fine-tuned_base_qg-hl_tweet_manual_model.txt",
"python3: can't open file 'eval.py': [Errno 2] No such file or directory\n"
],
[
"#evaluate fine-tuned base model\n!nlg-eval --hypothesis=hypothesis_fine-tuned_base_qg-hl_tweet_manual_model.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"\u001b[32mUsing data from /root/.cache/nlgeval\u001b[0m\n\u001b[32mIn case of broken downloads, remove the directory and run setup again.\u001b[0m\nBleu_1: 0.400382\nBleu_2: 0.293443\nBleu_3: 0.222170\nBleu_4: 0.173488\nMETEOR: 0.252304\nROUGE_L: 0.423129\nCIDEr: 1.833456\n"
],
[
"#valhalla/t5-small-e2e-qg\n###########################\n#08/15/2020 15:57:21 - INFO - transformers.configuration_utils - Configuration saved in t5-small-qg-hl/config.json\n#08/15/2020 15:57:23 - INFO - transformers.modeling_utils - Model weights saved in t5-small-qg-hl/pytorch_model.bin\n%%capture loggingfine\n!python3 run_qg.py \\\n --model_name_or_path valhalla/t5-small-e2e-qg \\\n --model_type t5 \\\n --tokenizer_name_or_path t5_qg_tokenizer \\\n --output_dir t5-small-E2E-qg-15-manual \\\n --train_file_path data/train_data_qg_hl_t5_tweet_manual.pt \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --per_device_train_batch_size 16 \\\n --per_device_eval_batch_size 16 \\\n --gradient_accumulation_steps 8 \\\n --learning_rate 1e-4 \\\n --num_train_epochs 15 \\\n --seed 42 \\\n --do_train \\\n --do_eval \\\n --evaluate_during_training \\\n --logging_steps 100\\\n --logging_dir t5-small-E2E-qg-15-manual-log\\\n --overwrite_output_dir",
"_____no_output_____"
],
[
"#using ORIGINAL small valhalla/t5-small-e2e-qg 15 epochs\n!python eval.py \\\n --model_name_or_path valhalla/t5-small-e2e-qg \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_t5-small-original-E2E-qg-hl_t5_tweet_manual.txt",
"2020-08-21 23:45:32.858092: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\nDownloading: 100% 792k/792k [00:00<00:00, 1.84MB/s]\nDownloading: 100% 31.0/31.0 [00:00<00:00, 36.6kB/s]\nDownloading: 100% 1.79k/1.79k [00:00<00:00, 1.80MB/s]\nDownloading: 100% 124/124 [00:00<00:00, 134kB/s]\n 0% 0/34 [00:00<?, ?it/s]/usr/local/lib/python3.6/dist-packages/nlp/utils/py_utils.py:191: UserWarning: The given NumPy array is not writeable, and PyTorch does not support non-writeable tensors. This means you can write to the underlying (supposedly non-writeable) NumPy array using the tensor. You may want to copy the array to protect its data or make it writeable before converting it to a tensor. This type of warning will be suppressed for the rest of this program. (Triggered internally at /pytorch/torch/csrc/utils/tensor_numpy.cpp:141.)\n return function(data_struct)\n100% 34/34 [00:49<00:00, 1.46s/it]\n"
],
[
"#evaluate ORGIINAL E2E small model\n!nlg-eval --hypothesis=hypothesis_t5-small-original-E2E-qg-hl_t5_tweet_manual.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"\u001b[32mUsing data from /root/.cache/nlgeval\u001b[0m\n\u001b[32mIn case of broken downloads, remove the directory and run setup again.\u001b[0m\nBleu_1: 0.053089\nBleu_2: 0.026385\nBleu_3: 0.015345\nBleu_4: 0.009345\nMETEOR: 0.109112\nROUGE_L: 0.082893\nCIDEr: 0.025654\n"
],
[
"#using FINE-TUNED small valhalla/t5-small-e2e-qg 15 epochs\n!python eval.py \\\n --model_name_or_path t5-small-E2E-qg-15-manual \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_t5-small-fine-tuned-E2E-qg-hl_t5_tweet_manual.txt",
"2020-08-21 23:47:03.634999: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\n 0% 0/34 [00:00<?, ?it/s]/usr/local/lib/python3.6/dist-packages/nlp/utils/py_utils.py:191: UserWarning: The given NumPy array is not writeable, and PyTorch does not support non-writeable tensors. This means you can write to the underlying (supposedly non-writeable) NumPy array using the tensor. You may want to copy the array to protect its data or make it writeable before converting it to a tensor. This type of warning will be suppressed for the rest of this program. (Triggered internally at /pytorch/torch/csrc/utils/tensor_numpy.cpp:141.)\n return function(data_struct)\n100% 34/34 [00:34<00:00, 1.00s/it]\n"
],
[
"#evaluate fine-tuned small model\n!nlg-eval --hypothesis=hypothesis_t5-small-fine-tuned-E2E-qg-hl_t5_tweet_manual.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"\u001b[32mUsing data from /root/.cache/nlgeval\u001b[0m\n\u001b[32mIn case of broken downloads, remove the directory and run setup again.\u001b[0m\nBleu_1: 0.333872\nBleu_2: 0.230703\nBleu_3: 0.169098\nBleu_4: 0.129525\nMETEOR: 0.215522\nROUGE_L: 0.353338\nCIDEr: 1.348371\n"
],
[
"#valhalla/t5-base-e2e-qg\n##########################\n#08/15/2020 15:57:21 - INFO - transformers.configuration_utils - Configuration saved in t5-small-qg-hl/config.json\n#08/15/2020 15:57:23 - INFO - transformers.modeling_utils - Model weights saved in t5-small-qg-hl/pytorch_model.bin\n%%capture loggingfine\n!python3 run_qg.py \\\n --model_name_or_path valhalla/t5-base-e2e-qg \\\n --model_type t5 \\\n --tokenizer_name_or_path t5_qg_tokenizer \\\n --output_dir t5-base-E2E-qg-15-manual \\\n --train_file_path data/train_data_qg_hl_t5_tweet_manual.pt \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --per_device_train_batch_size 16 \\\n --per_device_eval_batch_size 16 \\\n --gradient_accumulation_steps 8 \\\n --learning_rate 1e-4 \\\n --num_train_epochs 15 \\\n --seed 42 \\\n --do_train \\\n --do_eval \\\n --evaluate_during_training \\\n --logging_steps 100\\\n --logging_dir t5-base-E2E-qg-15-manual-log\\\n --overwrite_output_dir",
"_____no_output_____"
],
[
"#using ORIGINAL base valhalla/t5-base-e2e-qg 15 epochs\n!python eval.py \\\n --model_name_or_path valhalla/t5-base-e2e-qg \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_t5-base-original-E2E-qg-hl_t5_tweet_manual.txt",
"2020-08-22 09:13:56.967399: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\nDownloading: 100% 1.35k/1.35k [00:00<00:00, 1.02MB/s]\nDownloading: 100% 792k/792k [00:00<00:00, 2.32MB/s]\nDownloading: 100% 31.0/31.0 [00:00<00:00, 25.9kB/s]\nDownloading: 100% 1.79k/1.79k [00:00<00:00, 1.58MB/s]\nDownloading: 100% 195/195 [00:00<00:00, 168kB/s]\nDownloading: 100% 892M/892M [00:27<00:00, 32.4MB/s]\n 0% 0/34 [00:00<?, ?it/s]/usr/local/lib/python3.6/dist-packages/nlp/utils/py_utils.py:191: UserWarning: The given NumPy array is not writeable, and PyTorch does not support non-writeable tensors. This means you can write to the underlying (supposedly non-writeable) NumPy array using the tensor. You may want to copy the array to protect its data or make it writeable before converting it to a tensor. This type of warning will be suppressed for the rest of this program. (Triggered internally at /pytorch/torch/csrc/utils/tensor_numpy.cpp:141.)\n return function(data_struct)\n100% 34/34 [01:10<00:00, 2.08s/it]\n"
],
[
"#evaluate ORIGINAL E2E base model\n!nlg-eval --hypothesis=hypothesis_t5-base-original-E2E-qg-hl_t5_tweet_manual.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"\u001b[32mUsing data from /root/.cache/nlgeval\u001b[0m\n\u001b[32mIn case of broken downloads, remove the directory and run setup again.\u001b[0m\nBleu_1: 0.065108\nBleu_2: 0.035949\nBleu_3: 0.022479\nBleu_4: 0.014611\nMETEOR: 0.127798\nROUGE_L: 0.101185\nCIDEr: 0.048939\n"
],
[
"#using FINE-TUNED base valhalla/t5-base-e2e-qg 15 epochs\n!python eval.py \\\n --model_name_or_path t5-base-E2E-qg-15-manual \\\n --valid_file_path data/valid_data_qg_hl_t5_tweet_manual.pt \\\n --model_type t5 \\\n --num_beams 4 \\\n --max_decoding_length 32 \\\n --output_path hypothesis_t5-base-fine-tuned-E2E-qg-hl_t5_tweet_manual.txt",
"2020-08-22 09:16:26.929314: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\n 0% 0/34 [00:00<?, ?it/s]/usr/local/lib/python3.6/dist-packages/nlp/utils/py_utils.py:191: UserWarning: The given NumPy array is not writeable, and PyTorch does not support non-writeable tensors. This means you can write to the underlying (supposedly non-writeable) NumPy array using the tensor. You may want to copy the array to protect its data or make it writeable before converting it to a tensor. This type of warning will be suppressed for the rest of this program. (Triggered internally at /pytorch/torch/csrc/utils/tensor_numpy.cpp:141.)\n return function(data_struct)\n100% 34/34 [00:51<00:00, 1.50s/it]\n"
],
[
"#evaluate fine-tuned base model\n!nlg-eval --hypothesis=hypothesis_t5-base-fine-tuned-E2E-qg-hl_t5_tweet_manual.txt --references=data/tweet_dev_automatic_reference.txt --no-skipthoughts --no-glove ",
"\u001b[32mUsing data from /root/.cache/nlgeval\u001b[0m\n\u001b[32mIn case of broken downloads, remove the directory and run setup again.\u001b[0m\nBleu_1: 0.399541\nBleu_2: 0.291154\nBleu_3: 0.219786\nBleu_4: 0.170864\nMETEOR: 0.251499\nROUGE_L: 0.422600\nCIDEr: 1.826197\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae54fa41167156193ae8b4346f3a8cf11a748a1
| 8,985 |
ipynb
|
Jupyter Notebook
|
2.keras_jupyter/MLP/MLP.ipynb
|
Finfra/DeepLearningStudy
|
3c732cfdeb814e30cda3f166b0c722073e266c45
|
[
"MIT"
] | 1 |
2020-02-04T10:27:49.000Z
|
2020-02-04T10:27:49.000Z
|
2.keras_jupyter/MLP/MLP.ipynb
|
Finfra/DeepLearningStudy
|
3c732cfdeb814e30cda3f166b0c722073e266c45
|
[
"MIT"
] | null | null | null |
2.keras_jupyter/MLP/MLP.ipynb
|
Finfra/DeepLearningStudy
|
3c732cfdeb814e30cda3f166b0c722073e266c45
|
[
"MIT"
] | 3 |
2020-02-04T05:03:18.000Z
|
2020-02-04T10:27:51.000Z
| 37.4375 | 291 | 0.590762 |
[
[
[
"Trains a simple deep NN on the MNIST dataset.\n\nGets to 98.40% test accuracy after 20 epochs\n(there is *a lot* of margin for parameter tuning).\n2 seconds per epoch on a K520 GPU.\n",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\n\nfrom tensorflow.python import keras\nfrom tensorflow.python.keras.datasets import mnist\nfrom tensorflow.python.keras.models import Sequential\nfrom tensorflow.python.keras.layers import Dense, Dropout\nfrom tensorflow.python.keras.optimizers import RMSprop",
"_____no_output_____"
],
[
"batch_size = 128\nnum_classes = 10\nepochs = 20",
"_____no_output_____"
],
[
"# # the data, split between train and test sets\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\n# x_train = x_train.reshape(60000, 784)\n# x_test = x_test.reshape(10000, 784)\n# x_train = x_train.astype('float32')\n# x_test = x_test.astype('float32')\n# x_train /= 255\n# x_test /= 255\n# print(x_train.shape[0], 'train samples')\n# print(x_test.shape[0], 'test samples')\n\n# # convert class vectors to binary class matrices\ny_train = keras.utils.to_categorical(y_train, num_classes)\n# y_test = keras.utils.to_categorical(y_test, num_classes)\ny_train\n\n\n# print(x_train.shape,y_train.shape,x_test.shape,y_test.shape)",
"_____no_output_____"
],
[
"from tensorflow.examples.tutorials.mnist import input_data\nm = input_data.read_data_sets('./data', one_hot=True)\nx_train=m.train.images\ny_train=m.train.labels\nx_test=m.test.images\ny_test=m.test.labels\nprint(x_train.shape,y_train.shape,x_test.shape,y_test.shape)",
"WARNING:tensorflow:From <ipython-input-4-451e2c36d0ff>:2: read_data_sets (from tensorflow.contrib.learn.python.learn.datasets.mnist) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use alternatives such as official/mnist/dataset.py from tensorflow/models.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow_core/contrib/learn/python/learn/datasets/mnist.py:260: maybe_download (from tensorflow.contrib.learn.python.learn.datasets.base) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease write your own downloading logic.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow_core/contrib/learn/python/learn/datasets/mnist.py:262: extract_images (from tensorflow.contrib.learn.python.learn.datasets.mnist) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use tf.data to implement this functionality.\nExtracting ./data/train-images-idx3-ubyte.gz\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow_core/contrib/learn/python/learn/datasets/mnist.py:267: extract_labels (from tensorflow.contrib.learn.python.learn.datasets.mnist) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use tf.data to implement this functionality.\nExtracting ./data/train-labels-idx1-ubyte.gz\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow_core/contrib/learn/python/learn/datasets/mnist.py:110: dense_to_one_hot (from tensorflow.contrib.learn.python.learn.datasets.mnist) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use tf.one_hot on tensors.\nExtracting ./data/t10k-images-idx3-ubyte.gz\nExtracting ./data/t10k-labels-idx1-ubyte.gz\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow_core/contrib/learn/python/learn/datasets/mnist.py:290: DataSet.__init__ (from tensorflow.contrib.learn.python.learn.datasets.mnist) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use alternatives such as official/mnist/dataset.py from tensorflow/models.\n(55000, 784) (55000, 10) (10000, 784) (10000, 10)\n"
],
[
"model = Sequential()\nmodel.add(Dense(512, activation='relu', input_shape=(784,)))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(512, activation='relu'))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(num_classes, activation='softmax'))\n\nmodel.summary()\n\nmodel.compile(loss='categorical_crossentropy',\n optimizer=RMSprop(),\n metrics=['accuracy'])",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow_core/python/ops/resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\nModel: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 512) 401920 \n_________________________________________________________________\ndropout (Dropout) (None, 512) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 512) 262656 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 512) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 10) 5130 \n=================================================================\nTotal params: 669,706\nTrainable params: 669,706\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"history = model.fit(x_train, y_train,\n batch_size=batch_size,\n epochs=epochs,\n verbose=1,\n validation_data=(x_test, y_test))\n",
"Train on 55000 samples, validate on 10000 samples\n"
],
[
"score = model.evaluate(x_test, y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])",
"Test loss: 0.12230103779537749\nTest accuracy: 0.9845\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae565b343622544ad55e03f30d43570a987f9ce
| 552,884 |
ipynb
|
Jupyter Notebook
|
notebooks/BiasVariance.ipynb
|
jeremy9959/Math-5800-Spring-2020
|
f5be701e80a4d930439e66b1a49a831aa2e6fb97
|
[
"MIT"
] | null | null | null |
notebooks/BiasVariance.ipynb
|
jeremy9959/Math-5800-Spring-2020
|
f5be701e80a4d930439e66b1a49a831aa2e6fb97
|
[
"MIT"
] | null | null | null |
notebooks/BiasVariance.ipynb
|
jeremy9959/Math-5800-Spring-2020
|
f5be701e80a4d930439e66b1a49a831aa2e6fb97
|
[
"MIT"
] | null | null | null | 2,547.852535 | 220,968 | 0.960847 |
[
[
[
"## Bias and Variance\n\nFor the sake of this discussion, let's assume we are looking at a regression problem. For values of \n$x$ in the interval $[-1,1]$ there is a function $f(x)$ so that \n$$\nY = f(x)+\\epsilon\n$$\nwhere $\\epsilon$ is a noise term -- say, normally distributed with mean zero and variance $\\sigma^2$.\n\nWe have a set $D$ of \"training data\" which is a collection of $N$ points $(x_i,y_i)$ drawn from the model and\nwe want to try to reconstruct the function $f(x)$ so that we can accurately predict $f(x)$ for some new,\ntest value of the function $x$.\n\nGiven $D$, we construct a function $h_{D}$ so as to minimize the mean squared error (or loss)\n$$\nL = \\frac{1}{N}\\sum_{i=1}^{N} (h_{D}(x_i)-y_i)^2.\n$$\nThis value is called the *training loss* or the *training error*.\n\nNow suppose we pick a point $x_0$ and we'd like to understand the error in our prediction $h_{D}(x_0)$.\nWe can ask what the expected value of the squared error $(h_D(x_0)-Y)^2$ where $Y$ is the random value\nobtained from the \"true\" model. First we write\n$$\nh_D(x_0)-Y = h_D(x_0)-E(Y)+E(Y)-Y\n$$\nand use that $E(Y)=f(x_0)$ \nto get\n$$\nE((h_D(x_0)-Y)^2) = E((h_D(x_0)-f(x_0)^2)+E((E(Y)-Y)^2) + 2E((h_D(x_0)-E(Y))(E(Y)-Y)).\n$$\nSince $E(Y)-Y=\\epsilon$ is independent of $h_D(x_0)-E(Y)$ and $E(\\epsilon)=0$ the third term vanishes\nand the second term is $\\sigma^2$. We further split of the first term as\n$$\n(h_D(x_0)-f(x_0))=(h_D(x_0)-Eh_D(x_0)+Eh_D(x_0)-f(x_0)\n$$\nwhere $Eh_D(x_0)$ is the average prediction at $x_0$ as $h$ varies over all possible training sets.\nFrom this we get\n$$\nE((h_D(x_0)-f(x_0))^2) = E((h_D(x_0)-Eh_D(x_0))^2) + E((f(x_0)-Eh_D(x_0))^2)\n$$\nThe first of these terms has nothing do do with the \"true\" function $f$; it measures how much the\npredicted value at $x_0$ varies as the training set varies. This term is called the *variance*.\nIn the second term, the expectation is \nirrelevant because the term inside doesn't depend on the training set; it measures the (square of) the\ndifference between the average predicted value and the value of $f(x_0)$; it is called the *(squared) bias.*\n\nPutting all of this together, the error in prediction at a single point $x_0$ is made up of three terms:\n\n- the underlying variance in the process that generated the data, $\\sigma^2$. \n- the sensitivity of the predictive function to the choice of training set (the variance)\n- the degree to which the predictive function accurately guesses the true value *on average*.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.preprocessing import PolynomialFeatures\nfrom sklearn.linear_model import LinearRegression, Lasso\nfrom sklearn.pipeline import make_pipeline\nfrom numpy.random import normal,uniform\nimport matplotlib.pyplot as plt\nplt.style.use('ggplot')\n\ndef sample(f, sigma=.5, N=5):\n \"\"\"choose N x values as a training set from the interval at random and return f(x)+n(0,sigma^2) as the data at\n that training set\"\"\"\n x = uniform(-1,1,size=(N,1))\n y = f(x)+normal(0,sigma,size=(N,1))\n return np.concatenate([x,y],axis=1)\n\ndef bias_variance_plot(degree=2,samples=20,training_set_size=10,truth=None,sigma=.5): \n plt.figure(figsize=(10,10))\n if not truth:\n truth = lambda x: 0\n pipeline = make_pipeline(PolynomialFeatures(degree=degree),LinearRegression())\n _=plt.title(\"Fitting {} polynomials of degree {} to training sets of size {}\\nsigma={}\".format(samples,degree,training_set_size,sigma))\n x =np.linspace(-1,1,20)\n\n plt.ylim([-2,2])\n avg = np.zeros(x.shape)\n for i in range(samples):\n T= sample(truth,sigma=sigma,N=training_set_size)\n plt.scatter(T[:,0],T[:,1])\n model=pipeline.fit(T[:,0].reshape(-1,1),T[:,1])\n y = model.predict(x.reshape(-1,1))\n avg += y\n _=plt.plot(x,model.predict(x.reshape(-1,1)))\n _=plt.plot(x,avg/samples,color='black',label='mean predictor')\n _=plt.legend()",
"_____no_output_____"
]
],
[
[
"Here are some examples. Suppose that the underlying data comes from the parabola $f(x)=-x+x^2$ with the\nstandard deviation of the underlying error equal to $.5$.\n\nFirst we underfit the data, by using a least squares fit of a linear equation, getting a group of fits that have high bias (the average fit doesn't match the data well) but the solutions don't vary much with the training set.\n",
"_____no_output_____"
]
],
[
[
"bias_variance_plot(degree=1,training_set_size=20,samples=10,truth=lambda x: -x+x**2)\n",
"_____no_output_____"
]
],
[
[
"Next we plot a quadratic fit, which has low bias (in fact the average fit is exactly right) and the variance is also controlled.",
"_____no_output_____"
]
],
[
[
"bias_variance_plot(degree=2,training_set_size=10,samples=20,truth=lambda x: -x+x**2)\n",
"_____no_output_____"
]
],
[
[
"Finally we overfit the training data using a degree 3 polynomial. Again, the bias is good -- in the limit the\ncubic fits average to the quadratic solution -- but now the variance is very high so there is a lot of dependence on the test set.",
"_____no_output_____"
]
],
[
[
"bias_variance_plot(degree=3,training_set_size=10,samples=20,truth=lambda x: -x+x**2)\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae57aa603f59c846417ff854e3b101c6529c5b1
| 45,985 |
ipynb
|
Jupyter Notebook
|
lectures/other/Interval arithmetic.ipynb
|
snowdj/18S096-iap17
|
0a922819405312f4201c82828bc0fe94ef186b24
|
[
"MIT"
] | 107 |
2019-02-20T01:38:18.000Z
|
2022-03-21T03:01:03.000Z
|
lectures/other/Interval arithmetic.ipynb
|
snowdj/18S096-iap17
|
0a922819405312f4201c82828bc0fe94ef186b24
|
[
"MIT"
] | null | null | null |
lectures/other/Interval arithmetic.ipynb
|
snowdj/18S096-iap17
|
0a922819405312f4201c82828bc0fe94ef186b24
|
[
"MIT"
] | 25 |
2019-04-16T20:43:02.000Z
|
2022-03-24T22:18:06.000Z
| 86.114232 | 356 | 0.724301 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4ae59c51558dc03076cd38c0c5884c84e6e56c04
| 673,230 |
ipynb
|
Jupyter Notebook
|
notebook/2020.03.20_GCP_TCGA_CCLE_raw_processing.ipynb
|
wconnell/metrx
|
54296bac5bdd861dc4f43c37a5024de8d285afaa
|
[
"BSD-3-Clause"
] | null | null | null |
notebook/2020.03.20_GCP_TCGA_CCLE_raw_processing.ipynb
|
wconnell/metrx
|
54296bac5bdd861dc4f43c37a5024de8d285afaa
|
[
"BSD-3-Clause"
] | null | null | null |
notebook/2020.03.20_GCP_TCGA_CCLE_raw_processing.ipynb
|
wconnell/metrx
|
54296bac5bdd861dc4f43c37a5024de8d285afaa
|
[
"BSD-3-Clause"
] | null | null | null | 659.382958 | 416,436 | 0.9409 |
[
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import sys\nsys.path.append(\"..\")\n \nimport numpy as np\nimport pandas as pd\npd.set_option('display.max_columns', 100)\n\n# viz\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set(rc={'figure.figsize':(12.7,10.27)})\n\n# notebook settings\nfrom IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = \"all\"\nfrom IPython.display import set_matplotlib_formats\nset_matplotlib_formats('retina')\n\nimport os\nos.environ[\"CUDA_VISIBLE_DEVICES\"]=\"3\"",
"_____no_output_____"
],
[
"ls /srv/nas/mk2/projects/pan-cancer/TCGA_CCLE_GCP/TCGA",
"\u001b[0m\u001b[01;32mgdc_sample_sheet.2020-03-19_1.tsv\u001b[0m* \u001b[01;32mTCGA_LGG_counts.tsv.gz\u001b[0m*\n\u001b[01;32mgdc_sample_sheet.2020-03-19_2.tsv\u001b[0m* \u001b[01;32mTCGA_LIHC_counts.tsv.gz\u001b[0m*\n\u001b[01;32mgdc_sample_sheet.tsv\u001b[0m* \u001b[01;32mTCGA_LUAD_counts.tsv.gz\u001b[0m*\n\u001b[01;32mmap_TCGA_id.py\u001b[0m* \u001b[01;32mTCGA_LUSC_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_ACC_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_MESO_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_BCLA_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_Metadata.csv.gz\u001b[0m*\n\u001b[01;32mTCGA_BRCA_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_OV_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_CESC_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_PAAD_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_CHOL_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_PCPG_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_COAD_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_PRAD_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_DLBC_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_READ_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_ESCA_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_SARC_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_GBM_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_SKCM_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_GDC_ID_MAP.tsv\u001b[0m* \u001b[01;32mTCGA_STAD_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_HNSC_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_TGCT_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_ID_MAP.csv\u001b[0m* \u001b[01;32mTCGA_THCA_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_ID_samples.csv\u001b[0m* \u001b[01;32mTCGA_THYM_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_KICH_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_UCEC_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_KIRC_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_UCS_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_KIRP_counts.tsv.gz\u001b[0m* \u001b[01;32mTCGA_UVM_counts.tsv.gz\u001b[0m*\n\u001b[01;32mTCGA_LAML_counts.tsv.gz\u001b[0m*\n"
],
[
"def getTCGA(disease):\n path = \"/srv/nas/mk2/projects/pan-cancer/TCGA_CCLE_GCP/TCGA/TCGA_{}_counts.tsv.gz\"\n files = [path.format(d) for d in disease]\n return files",
"_____no_output_____"
],
[
"def readGCP(files):\n \"\"\"\n Paths to count matrices.\n \"\"\"\n data_dict = {}\n for f in files:\n key = os.path.basename(f).split(\"_\")[1]\n data = pd.read_csv(f, sep='\\t', index_col=0)\n meta = pd.DataFrame([row[:-1] for row in data.index.str.split(\"|\")],\n columns=['ENST', 'ENSG', 'OTTHUMG', 'OTTHUMT', 'GENE-NUM', 'GENE', 'NUM', 'TYPE'])\n data.index = meta['GENE']\n data_dict[key] = data.T\n return data_dict",
"_____no_output_____"
],
[
"def renameTCGA(data_dict, mapper):\n for key in data_dict.keys():\n data_dict[key] = data_dict[key].rename(mapper)\n return data_dict",
"_____no_output_____"
],
[
"def uq_norm(df, q=0.75):\n \"\"\"\n Upper quartile normalization of GEX for samples.\n \"\"\"\n quantiles = df.quantile(q=q, axis=1)\n norm = df.divide(quantiles, axis=0)\n return norm",
"_____no_output_____"
],
[
"base = \"/srv/nas/mk2/projects/pan-cancer/TCGA_CCLE_GCP\"\ndisease = ['BRCA', 'LUAD', 'KIRC', 'THCA', 'PRAD', 'SKCM']\n\ntcga_files = getTCGA(disease)\ntcga_meta = pd.read_csv(os.path.join(base, \"TCGA/TCGA_GDC_ID_MAP.tsv\"), sep=\"\\t\")\ntcga = readGCP(tcga_files)",
"_____no_output_____"
],
[
"# rename samples to reflect canonical IDs\ntcga = renameTCGA(tcga, mapper=dict(zip(tcga_meta['CGHubAnalysisID'], tcga_meta['Sample ID'])))",
"_____no_output_____"
],
[
"# combine samples\ntcga = pd.concat(tcga.values())",
"_____no_output_____"
]
],
[
[
"## Normalization",
"_____no_output_____"
]
],
[
[
"# Upper quartile normalization\ntcga = uq_norm(tcga)",
"_____no_output_____"
],
[
"# log norm\ntcga = tcga.transform(np.log1p)",
"_____no_output_____"
],
[
"# downsample\ntcga = tcga.sample(n=15000, axis=1)",
"_____no_output_____"
],
[
"train_data.shap",
"_____no_output_____"
],
[
"tcga_meta[tcga_meta['Sample ID'] == 'TCGA-A7-A26F-01B']",
"_____no_output_____"
]
],
[
[
"# Model",
"_____no_output_____"
],
[
"### Experimental Setup",
"_____no_output_____"
]
],
[
[
"from collections import OrderedDict \nhierarchy = OrderedDict({'Disease':['BRCA', 'LUAD', 'KIRC', 'THCA', 'PRAD', 'SKCM'],\n 'Sample Type':['Primary Tumor', 'Solid Tissue Normal', 'Metastatic']})",
"_____no_output_____"
],
[
"class Experiment():\n \"\"\"\n Defines an experimental class hierarchy object.\n \"\"\"\n def __init__(self, meta_data, hierarchy, cases, min_samples):\n self.hierarchy = hierarchy\n self.meta_data = self.categorize(meta_data, self.hierarchy, min_samples)\n self.cases = self.meta_data[cases].unique()\n self.labels = self.meta_data['meta'].cat.codes.values.astype('int')\n self.labels_dict = {key:val for key,val in enumerate(self.meta_data['meta'].cat.categories.values)}\n \n def categorize(self, meta_data, hierarchy, min_samples):\n assert isinstance(hierarchy, OrderedDict), \"Argument of wrong type.\"\n # downsample data\n for key,val in hierarchy.items():\n meta_data = meta_data[meta_data[key].isin(val)]\n # unique meta classes\n meta_data['meta'] = meta_data[list(hierarchy.keys())].apply(lambda row: ':'.join(row.values.astype(str)), axis=1)\n # filter meta classes\n counts = meta_data['meta'].value_counts()\n keep = counts[counts > min_samples].index\n meta_data = meta_data[meta_data['meta'].isin(keep)]\n # generate class categories\n meta_data['meta'] = meta_data['meta'].astype('category')\n return meta_data\n \n def holdout(self, holdout):\n self.holdout = holdout\n self.holdout_samples = self.meta_data[self.meta_data['meta'].isin(holdout)]\n self.meta_data = self.meta_data[~self.meta_data['meta'].isin(holdout)]",
"_____no_output_____"
],
[
"from dutils import train_test_split_case\nexp = Experiment(meta_data=tcga_meta,\n hierarchy=hierarchy,\n cases='Case ID',\n min_samples=20)\nexp.holdout(holdout=['SKCM:Metastatic'])",
"_____no_output_____"
],
[
"exp.meta_data['meta'].value_counts()\nexp.holdout_samples['meta'].value_counts()",
"_____no_output_____"
],
[
"# Define Train / Test sample split\ntarget = 'meta'\n\ntrain, test = train_test_split_case(exp.meta_data, cases='Case ID')\n# stratification is not quite perfect but close\n# in order to preserve matched samples for each case together\n# in train or test set\ncase_counts = exp.meta_data[target].value_counts()\ntrain[target].value_counts()[case_counts.index.to_numpy()] / case_counts\ntest[target].value_counts()[case_counts.index.to_numpy()] / case_counts",
"_____no_output_____"
],
[
"# split data\ntrain_data = tcga[tcga.index.isin(train['Sample ID'])].astype(np.float16)\ntest_data = tcga[tcga.index.isin(test['Sample ID'])].astype(np.float16)",
"_____no_output_____"
],
[
"import torch\nfrom torch.optim import lr_scheduler\nimport torch.optim as optim\nfrom torch.autograd import Variable\n#torch.manual_seed(123)\n\nfrom trainer import fit\nimport visualization as vis\nimport numpy as np\ncuda = torch.cuda.is_available()\nprint(\"Cuda is available: {}\".format(cuda))",
"Cuda is available: True\n"
],
[
"import torch\nfrom torch.utils.data import Dataset\n\nclass SiameseDataset(Dataset):\n \"\"\"\n Train: For each sample creates randomly a positive or a negative pair\n Test: Creates fixed pairs for testing\n \"\"\"\n\n def __init__(self, experiment, data, train=False):\n self.train = train\n self.labels = experiment.meta_data[experiment\n .meta_data['Sample ID']\n .isin(data.index)]['meta'].cat.codes.values.astype('int')\n assert len(data) == len(self.labels)\n\n if self.train:\n self.train_labels = self.labels\n self.train_data = torch.from_numpy(data.values).float()\n self.labels_set = set(self.train_labels)\n self.label_to_indices = {label: np.where(self.train_labels == label)[0]\n for label in self.labels_set}\n else:\n # generate fixed pairs for testing\n self.test_labels = self.labels\n self.test_data = torch.from_numpy(data.values).float()\n self.labels_set = set(self.test_labels)\n self.label_to_indices = {label: np.where(self.test_labels == label)[0]\n for label in self.labels_set}\n\n random_state = np.random.RandomState(29)\n\n positive_pairs = [[i,\n random_state.choice(self.label_to_indices[self.test_labels[i].item()]),\n 1]\n for i in range(0, len(self.test_data), 2)]\n\n negative_pairs = [[i,\n random_state.choice(self.label_to_indices[\n np.random.choice(\n list(self.labels_set - set([self.test_labels[i].item()]))\n )\n ]),\n 0]\n for i in range(1, len(self.test_data), 2)]\n self.test_pairs = positive_pairs + negative_pairs\n\n def __getitem__(self, index):\n if self.train:\n target = np.random.randint(0, 2)\n img1, label1 = self.train_data[index], self.train_labels[index].item()\n if target == 1:\n siamese_index = index\n while siamese_index == index:\n siamese_index = np.random.choice(self.label_to_indices[label1])\n else:\n siamese_label = np.random.choice(list(self.labels_set - set([label1])))\n siamese_index = np.random.choice(self.label_to_indices[siamese_label])\n img2 = self.train_data[siamese_index]\n else:\n img1 = self.test_data[self.test_pairs[index][0]]\n img2 = self.test_data[self.test_pairs[index][1]]\n target = self.test_pairs[index][2]\n \n return (img1, img2), target\n\n def __len__(self):\n if self.train:\n return len(self.train_data)\n else:\n return len(self.test_data)",
"_____no_output_____"
]
],
[
[
"# Siamese Network",
"_____no_output_____"
]
],
[
[
"siamese_train_dataset = SiameseDataset(experiment=exp,\n data=train_data,\n train=True)\nsiamese_test_dataset = SiameseDataset(experiment=exp,\n data=test_data,\n train=False)",
"_____no_output_____"
],
[
"batch_size = 8\nkwargs = {'num_workers': 10, 'pin_memory': True} if cuda else {'num_workers': 10}\nsiamese_train_loader = torch.utils.data.DataLoader(siamese_train_dataset, batch_size=batch_size, shuffle=True, **kwargs)\nsiamese_test_loader = torch.utils.data.DataLoader(siamese_test_dataset, batch_size=batch_size, shuffle=False, **kwargs)\n\n# Set up the network and training parameters\nfrom tcga_networks import EmbeddingNet, SiameseNet\nfrom losses import ContrastiveLoss, TripletLoss\nfrom metrics import AccumulatedAccuracyMetric\n\n# Step 2\nn_samples, n_features = siamese_train_dataset.train_data.shape\nembedding_net = EmbeddingNet(n_features, 2)\n# Step 3\nmodel = SiameseNet(embedding_net)\nif cuda:\n model.cuda()\n \n# Step 4\nmargin = 1.\nloss_fn = ContrastiveLoss(margin)\nlr = 1e-3\noptimizer = optim.Adam(model.parameters(), lr=lr)\nscheduler = lr_scheduler.StepLR(optimizer, 8, gamma=0.1, last_epoch=-1)\nn_epochs = 10\n# print training metrics every log_interval * batch_size\nlog_interval = round(len(siamese_train_dataset)/4/batch_size)",
"_____no_output_____"
],
[
"print('Active CUDA Device: GPU', torch.cuda.current_device())\n\nprint ('Available devices ', torch.cuda.device_count())\nprint ('Current cuda device ', torch.cuda.current_device())",
"Active CUDA Device: GPU 0\nAvailable devices 1\nCurrent cuda device 0\n"
],
[
"train_loss, val_loss = fit(siamese_train_loader, siamese_test_loader, model, loss_fn, optimizer, scheduler, \n n_epochs, cuda, log_interval)",
"Train: [0/2931 (0%)]\tLoss: 0.311600\nTrain: [736/2931 (25%)]\tLoss: 0.206912\nTrain: [1472/2931 (50%)]\tLoss: 0.144015\nTrain: [2208/2931 (75%)]\tLoss: 0.142308\nEpoch: 1/10. Train set: Average loss: 0.1668\nEpoch: 1/10. Validation set: Average loss: 0.1735\nTrain: [0/2931 (0%)]\tLoss: 0.190095\nTrain: [736/2931 (25%)]\tLoss: 0.133940\nTrain: [1472/2931 (50%)]\tLoss: 0.323029\nTrain: [2208/2931 (75%)]\tLoss: 0.155684\nEpoch: 2/10. Train set: Average loss: 0.1887\nEpoch: 2/10. Validation set: Average loss: 0.1408\nTrain: [0/2931 (0%)]\tLoss: 0.105760\nTrain: [736/2931 (25%)]\tLoss: 0.166466\nTrain: [1472/2931 (50%)]\tLoss: 0.250662\nTrain: [2208/2931 (75%)]\tLoss: 0.159311\nEpoch: 3/10. Train set: Average loss: 0.1740\nEpoch: 3/10. Validation set: Average loss: 0.1344\nTrain: [0/2931 (0%)]\tLoss: 0.169786\nTrain: [736/2931 (25%)]\tLoss: 0.133313\nTrain: [1472/2931 (50%)]\tLoss: 4.163010\nTrain: [2208/2931 (75%)]\tLoss: 0.357001\nEpoch: 4/10. Train set: Average loss: 1.2080\nEpoch: 4/10. Validation set: Average loss: 0.1537\nTrain: [0/2931 (0%)]\tLoss: 0.099442\nTrain: [736/2931 (25%)]\tLoss: 0.137219\nTrain: [1472/2931 (50%)]\tLoss: 0.304385\nTrain: [2208/2931 (75%)]\tLoss: 140.985126\nEpoch: 5/10. Train set: Average loss: 35.5313\nEpoch: 5/10. Validation set: Average loss: 0.7785\nTrain: [0/2931 (0%)]\tLoss: 0.907264\nTrain: [736/2931 (25%)]\tLoss: 0.243135\nTrain: [1472/2931 (50%)]\tLoss: 14.033909\nTrain: [2208/2931 (75%)]\tLoss: 0.170947\nEpoch: 6/10. Train set: Average loss: 3.6675\nEpoch: 6/10. Validation set: Average loss: 0.1722\nTrain: [0/2931 (0%)]\tLoss: 0.143987\nTrain: [736/2931 (25%)]\tLoss: 0.165980\nTrain: [1472/2931 (50%)]\tLoss: 1.681781\nTrain: [2208/2931 (75%)]\tLoss: 0.406440\nEpoch: 7/10. Train set: Average loss: 0.6060\nEpoch: 7/10. Validation set: Average loss: 0.1751\nTrain: [0/2931 (0%)]\tLoss: 0.140763\nTrain: [736/2931 (25%)]\tLoss: 0.190577\nTrain: [1472/2931 (50%)]\tLoss: 0.161815\nTrain: [2208/2931 (75%)]\tLoss: 0.163189\nEpoch: 8/10. Train set: Average loss: 0.1692\nEpoch: 8/10. Validation set: Average loss: 0.1710\nTrain: [0/2931 (0%)]\tLoss: 0.177842\nTrain: [736/2931 (25%)]\tLoss: 0.162595\nTrain: [1472/2931 (50%)]\tLoss: 0.164396\nTrain: [2208/2931 (75%)]\tLoss: 0.155691\nEpoch: 9/10. Train set: Average loss: 0.1622\nEpoch: 9/10. Validation set: Average loss: 0.1699\nTrain: [0/2931 (0%)]\tLoss: 0.131774\nTrain: [736/2931 (25%)]\tLoss: 0.160130\nTrain: [1472/2931 (50%)]\tLoss: 1.366779\nTrain: [2208/2931 (75%)]\tLoss: 0.226311\nEpoch: 10/10. Train set: Average loss: 0.4963\nEpoch: 10/10. Validation set: Average loss: 0.2211\n"
],
[
"plt.plot(range(0, n_epochs), train_loss, 'rx-', label='train')\nplt.plot(range(0, n_epochs), val_loss, 'bx-', label='validation')\nplt.legend()",
"_____no_output_____"
],
[
" \ndef extract_embeddings(samples, target, model):\n cuda = torch.cuda.is_available()\n with torch.no_grad():\n model.eval()\n assert len(samples) == len(target)\n embeddings = np.zeros((len(samples), 2))\n labels = np.zeros(len(target))\n k = 0\n if cuda:\n samples = samples.cuda()\n if isinstance(model, torch.nn.DataParallel):\n embeddings[k:k+len(samples)] = model.module.get_embedding(samples).data.cpu().numpy()\n else:\n embeddings[k:k+len(samples)] = model.get_embedding(samples).data.cpu().numpy()\n labels[k:k+len(samples)] = target\n k += len(samples)\n return embeddings, labels",
"_____no_output_____"
],
[
"train_embeddings_cl, train_labels_cl = extract_embeddings(siamese_train_dataset.train_data, siamese_train_dataset.labels, model)\nvis.sns_plot_embeddings(train_embeddings_cl, train_labels_cl, exp.labels_dict, \n hue='meta', style='Sample Type', alpha=0.5)\nplt.title('PanCancer Train: Siamese')\nplt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)",
"_____no_output_____"
],
[
"val_embeddings_baseline, val_labels_baseline = extract_embeddings(siamese_test_dataset.test_data, siamese_test_dataset.labels, model)\nvis.sns_plot_embeddings(val_embeddings_baseline, val_labels_baseline, exp.labels_dict, \n hue='meta', style='Sample Type', alpha=0.5)\nplt.title('PanCancer Test: Siamese')\nplt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae5a40b2ec63c64acc373644654252c9ad3a22d
| 35,478 |
ipynb
|
Jupyter Notebook
|
finviz.ipynb
|
sahudsonleck/finviz-analytics
|
845fc717ffe08910e16e3a22ee415d297f886658
|
[
"MIT"
] | null | null | null |
finviz.ipynb
|
sahudsonleck/finviz-analytics
|
845fc717ffe08910e16e3a22ee415d297f886658
|
[
"MIT"
] | null | null | null |
finviz.ipynb
|
sahudsonleck/finviz-analytics
|
845fc717ffe08910e16e3a22ee415d297f886658
|
[
"MIT"
] | null | null | null | 30.375 | 298 | 0.54538 |
[
[
[
"# Finviz Analytics\n\n\n### What is Finviz?\nFinViz aims to make market information accessible and provides a lot of data in visual snapshots, allowing traders and investors to quickly find the stock, future or forex pair they are looking for. The site provides advanced screeners, market maps, analysis, comparative tools and charts.\n\n### Why?\nLeverage NRT financial stats to create custom stock screens and perspectives to identify value with in volatile market conditions.",
"_____no_output_____"
],
[
"***\n## Prerequisites \n\n### finviz\nFinviz is a stock screener and trading tool used for creating financial displays. Professional traders frequently use this platform to save time because Finviz allows traders and investors to quickly screen and find stocks based on set criteria.\n\n\n### pandas, pandas_profiling\nPandas needs no introduction. Pandas_profiling creates beautiful html data profiles.\n\n### nest_asyncio\nNest_asyncio supports asynchronous call for use with an interactive broker.\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport logging \nfrom finviz.screener import Screener\n\nlog = logging.getLogger() \nconsole = logging.StreamHandler()\nformat_str = '%(asctime)s\\t%(levelname)s -- %(processName)s -- %(message)s'\nconsole.setFormatter(logging.Formatter(format_str))\nlog.addHandler(console) \nlog.setLevel(logging.INFO) \n",
"_____no_output_____"
]
],
[
[
"***\n## Load environment and runtime variables\n\n",
"_____no_output_____"
]
],
[
[
"'''\nMODULE is used to identify and segment runtime and environment variables from config files.\n'''\nMODULE = 'ldg_finviz'\n\n\n'''\nLoad configuration fiiles from \\config. Instansiate variables with config file names. \n'''\nimport os \nd = os.getcwd()\ndf = d + '\\\\config\\\\'\ntry: \n for i in os.listdir(df):\n k = i[:-4] \n v = open(df + i).read()\n v = eval(v)\n exec(\"%s=%s\" % (k,v)) \n log.info('loaded: ' + k)\nexcept:\n log.error('issue encountered with eval(data): ' + str(v))\n",
"2020-06-30 18:29:39,405\tINFO -- MainProcess -- loaded: api_params\n2020-06-30 18:29:39,408\tINFO -- MainProcess -- loaded: data_var\n2020-06-30 18:29:39,410\tINFO -- MainProcess -- loaded: env_var\n2020-06-30 18:29:39,411\tINFO -- MainProcess -- loaded: err_var\n2020-06-30 18:29:39,412\tINFO -- MainProcess -- loaded: map_generic_fn\n2020-06-30 18:29:39,413\tINFO -- MainProcess -- loaded: map_landing_dataset_code\n2020-06-30 18:29:39,414\tINFO -- MainProcess -- loaded: transform\n"
],
[
"def get_data_finviz(generate_data_profile=False):\n '''\n Download FinViz 15min delayed stock data. \n * filter - Filter stock universe using filters variable. \n * Select datasets to download using the map_api_fv_table_key.config \n * Dataset options include: \n 'Overview': '111',\n 'Valuation': '121',\n 'Ownership': '131',\n 'Performance': '141',\n 'Custom': '152',\n 'Financial': '161',\n 'Technical': '171'\n * Refer to /docs for dataset details.\n \n Output data in .csv format to landing.\n '''\n import nest_asyncio\n nest_asyncio.apply()\n \n #load variables\n ldg_path = env_var.get('ldg_path') \n filters = api_params['filter']\n\n #loop through datasets to download from Screener & write to file.\n for i in api_params['datasets']:\n log.info('downloading:' + i.get('dataset').lower())\n stock_list = Screener(filters=filters, table= i.get('dataset'))\n stock_list.to_csv(ldg_path + 'stock_screener_' + i.get('dataset').lower() + '.csv')\n \n if generate_data_profile == True:\n log.info('begin pandas profile.html generation')\n generate_docs('ldg_path')\n\ndef get_transform(target):\n '''\n Get transform from transform.cfg for target dataset\n #returns a list of dicts with transform logic in format {field:value, fn:value} \n '''\n\n lst = [i.get(target) for i in transform['root'] if i.get(target) != None]\n if lst != []: lst = lst[0]\n\n return lst\n\ndef apply_transform(df, transform, target):\n '''\n apply list of tranformstions to dataframe\n '''\n import numpy as np\n log.info('begin transforms for: ' + target)\n \n try: \n for t in transform: \n\n #get function to apply\n fn = t.get('fn') \n #get reusable function from map_generic_fn if fn starts with $ \n if fn[0] == '$': fn = map_generic_fn.get(fn)\n\n #get field or fields to update\n field = t.get('field')\n if field[0] == '[':field = eval(field) \n #log.info('apply transform: {field, function} ' + str(field) + ' ' + fn)\n\n #apply transformation\n df[field] = eval(fn)\n except: \n log.error('error encountered with table:' + str(target) + ' field:' + str(field) + ' fn:' + str(fn) )\n \n log.info('end transforms for: ' + target)\n return \n\ndef normalize_data():\n '''\n Perform preprocessing and copy data to staging area. \n Preprocessing steps are included in transform.cfg and typically include:\n - metadata validation / data contract\n - preliminary schema normalization\n - data type validation & associated cleansing. \n '''\n \n ldg_path = env_var.get('ldg_path') \n stg_path = env_var.get('stg_path')\n \n try:\n #for each dataset in map_landing_dataset_code\n for i in map_landing_dataset_code.get(MODULE):\n\n #load file for meta contract and data type conversion\n file = ldg_path + i.get('file')\n code = i.get('code')\n df = pd.read_csv(file)\n\n trns = get_transform(code)\n if trns != []:\n #apply transformation to dataframe\n apply_transform(df,trns,code)\n\n df.to_csv(stg_path + i.get('file'))\n #TODO Normalize column names, tolwower() with underscores... \n except:\n log.error('error in normalize_data()')\n \ndef generate_docs(path):\n '''\n generate pandas_profile.html reports\n '''\n from pandas_profiling import ProfileReport\n \n data_path = env_var.get(path)\n \n try: \n for i in map_landing_dataset_code[MODULE]:\n file = i.get('file')\n print(file)\n df = pd.read_csv(data_path + file)\n profile = ProfileReport(df, title= 'Profile: ' + file + ' (Landing)')\n fpdf = data_path + 'profile_' + file[0:-4] + '.pdf'\n \n profile.to_file(fpdf)\n #convert_file_format(fhtml,fpdf)\n \n except:\n log.error('error in generating pandas_profile.html')\n\ndef convert_file_format(fromfile, tofile):\n import pdfkit as pk\n pk.from_file(fromfile,tofile)\n \n\ndef convert_unit(u):\n if len(u) != 1: \n ua = u\n u = str(u[-1])\n val = str(ua[0:-1]).replace('.','')\n else: val = ''\n u=u.lower()\n\n if u == 'm':\n val += '0000'\n elif u == 'b':\n val += '0000000'\n return val ",
"_____no_output_____"
]
],
[
[
"# Download & stage data ",
"_____no_output_____"
]
],
[
[
"log.info('begin downloading finviz')\nget_data_finviz(generate_data_profile=True)\nlog.info('end downloading finviz')\n\n\nlog.info('begin finviz preprocessing')\nnormalize_data()\nlog.info('end finviz preprocessing')",
"2020-06-30 18:29:45,048\tINFO -- MainProcess -- begin downloading finviz\n2020-06-30 18:29:45,051\tINFO -- MainProcess -- downloading:overview\n2020-06-30 18:29:51,582\tINFO -- MainProcess -- downloading:valuation\n2020-06-30 18:29:58,655\tINFO -- MainProcess -- downloading:ownership\n2020-06-30 18:30:06,428\tINFO -- MainProcess -- downloading:performance\n2020-06-30 18:30:13,557\tINFO -- MainProcess -- downloading:custom\n2020-06-30 18:30:19,675\tINFO -- MainProcess -- downloading:financial\n2020-06-30 18:30:26,674\tINFO -- MainProcess -- downloading:technical\n2020-06-30 18:30:33,519\tINFO -- MainProcess -- begin pandas profile.html generation\nC:\\ProgramData\\Anaconda3\\lib\\site-packages\\statsmodels\\tools\\_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"def enrich_data_finviz():\n import numpy as np\n \n ENF = err_var.get('no_param')\n stg_path = env_var.get('stg_path')\n \n fown =stg_path + data_var[MODULE].get('stock_ownership',ENF)\n fovr =stg_path + data_var[MODULE].get('stock_overview',ENF)\n key = data_var[MODULE].get('stock_key',ENF)\n \n if (fown==ENF) or (fovr== ENF) or (key == ENF):\n e = 'missing file or key name'\n log_diagnostics('enrich_data_finviz',e,env_var)\n return\n \n #generate additional attibutes\n df_own = pd.read_csv(fown).reset_index()\n df_own.set_index(key,inplace=True)\n df_view = pd.read_csv(fovr).reset_index()\n df_view.set_index(key,inplace=True)\n df = pd.merge(df_own,df_view,how='inner',left_index = True, right_index=True)\n df.reset_index(inplace=True)\n\n target = 'stg_finviz_summary' \n transform = get_transform(target) \n apply_transform(df,transform,target)\n df = df[['Ticker','eps','earnings','P/E','e/p','Outstanding']]\n\n df.to_csv(stg_path + 'stock_screener_summary.csv')\n\n\nenrich_data_finviz()",
"_____no_output_____"
],
[
"!pip uninstall pdfkit\n!pip uninstall weasyprint",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4ae5a9e00016f7ed750083b23aebda09e3164702
| 6,168 |
ipynb
|
Jupyter Notebook
|
example/textual_language_augmenter.ipynb
|
So-AI-love/nlpaug
|
3aff5754609cb6bf092709d9af2089ccd55ffc93
|
[
"MIT"
] | 3,121 |
2019-04-21T07:02:47.000Z
|
2022-03-31T22:17:36.000Z
|
example/textual_language_augmenter.ipynb
|
So-AI-love/nlpaug
|
3aff5754609cb6bf092709d9af2089ccd55ffc93
|
[
"MIT"
] | 186 |
2019-05-31T18:18:13.000Z
|
2022-03-28T10:11:05.000Z
|
example/textual_language_augmenter.ipynb
|
So-AI-love/nlpaug
|
3aff5754609cb6bf092709d9af2089ccd55ffc93
|
[
"MIT"
] | 371 |
2019-03-17T17:59:56.000Z
|
2022-03-31T01:45:15.000Z
| 23.18797 | 101 | 0.542964 |
[
[
[
"%load_ext autoreload\n%autoreload 2\nimport importlib",
"_____no_output_____"
],
[
"import os\nos.environ[\"MODEL_DIR\"] = '../model'",
"_____no_output_____"
]
],
[
[
"# Config",
"_____no_output_____"
]
],
[
[
"import nlpaug.augmenter.char as nac\nimport nlpaug.augmenter.word as naw\nimport nlpaug.augmenter.sentence as nas\nimport nlpaug.flow as nafc\n\nfrom nlpaug.util import Action",
"_____no_output_____"
]
],
[
[
"# Synonym Augmenter (WordNet, Spanish)",
"_____no_output_____"
]
],
[
[
"text = 'Un rápido zorro marrón salta sobre el perro perezoso'\naug = naw.SynonymAug(aug_src='wordnet', lang='spa')\naugmented_text = aug.augment(text)\nprint(\"Original:\")\nprint(text)\nprint(\"Augmented Text:\")\nprint(augmented_text)",
"Original:\nUn rápido zorro marrón salta sobre el perro perezoso\nAugmented Text:\nUn rápido zorro marrón salta sobre el can perezoso\n"
]
],
[
[
"# Word Embeddings Augmenter (word2vec, French)",
"_____no_output_____"
]
],
[
[
"augmented_text = aug.augment(text)\nprint(\"Original:\")\nprint(text)\nprint(\"Augmented Text:\")\nprint(augmented_text)",
"Original:\nUn rápido zorro marrón salta sobre el perro perezoso\nAugmented Text:\nUn rápido zorro marrón salta sobre el perro moroso\n"
]
],
[
[
"# Word Embeddings Augmenter (fasttext, Japanese)",
"_____no_output_____"
]
],
[
[
"# https://github.com/taishi-i/nagisa\nimport nagisa\ndef tokenizer(x):\n return nagisa.tagging(text).words\n\ntext = '速い茶色の狐が怠惰なな犬を飛び越えます'\naug = naw.WordEmbsAug(model_type='fasttext', tokenizer=tokenizer,\n model_path=os.path.join(os.environ.get(\"MODEL_DIR\"), 'wiki.ja.vec'))\naugmented_text = aug.augment(text)\nprint(\"Original:\")\nprint(text)\nprint(\"Augmented Text:\")\nprint(augmented_text)",
"Original:\n速い茶色の狐が怠惰なな犬を飛び越えます\nAugmented Text:\n速い 茶色 後に 狐 が 苦 な な 犬 から 飛び越え ます\n"
]
],
[
[
"# Contextual Word Embeddings Augmenter (BERT)",
"_____no_output_____"
]
],
[
[
"# Augment French by BERT\naug = naw.ContextualWordEmbsAug(model_path='bert-base-multilingual-uncased', aug_p=0.1)\ntext = \"Bonjour, J'aimerais une attestation de l'employeur certifiant que je suis en CDI.\"\naugmented_text = aug.augment(text)\nprint(\"Original:\")\nprint(text)\nprint(\"Augmented Text:\")\nprint(augmented_text)",
"Original:\nBonjour, J'aimerais une attestation de l'employeur certifiant que je suis en CDI.\nAugmented Text:\nbonjour , j ' aimerais ! attestation de l ' employeur certifiant comment je suis en cdi .\n"
],
[
"# Augment Japanese by BERT\naug = naw.ContextualWordEmbsAug(model_path='bert-base-multilingual-uncased', aug_p=0.1)\ntext = '速い茶色の狐が怠惰なな犬を飛び越えます'\naugmented_text = aug.augment(text)\nprint(\"Original:\")\nprint(text)\nprint(\"Augmented Text:\")\nprint(augmented_text)",
"Original:\n速い茶色の狐が怠惰なな犬を飛び越えます\nAugmented Text:\n速 い 黄 色 の 狐 か 怠 惰 なな 犬 を 飛 ひ 越 えます\n"
],
[
"# Augment Spanish by BERT\naug = naw.ContextualWordEmbsAug(model_path='bert-base-multilingual-uncased', aug_p=0.1)\ntext = 'Un rápido zorro marrón salta sobre el perro perezoso'\naugmented_text = aug.augment(text)\nprint(\"Original:\")\nprint(text)\nprint(\"Augmented Text:\")\nprint(augmented_text)",
"Original:\nUn rápido zorro marrón salta sobre el perro perezoso\nAugmented Text:\nun rapido gato marron salta sobre el perro perezoso\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4ae5ab4bbd256869fd5686c58b138f0e7d78fb08
| 1,170 |
ipynb
|
Jupyter Notebook
|
Capstone Project Course.ipynb
|
Marouane1443/Coursera_Capstone
|
f04700344f710a35dbe88dab1a8464218cca2b4c
|
[
"BSD-3-Clause"
] | null | null | null |
Capstone Project Course.ipynb
|
Marouane1443/Coursera_Capstone
|
f04700344f710a35dbe88dab1a8464218cca2b4c
|
[
"BSD-3-Clause"
] | null | null | null |
Capstone Project Course.ipynb
|
Marouane1443/Coursera_Capstone
|
f04700344f710a35dbe88dab1a8464218cca2b4c
|
[
"BSD-3-Clause"
] | null | null | null | 17.462687 | 64 | 0.513675 |
[
[
[
"This notebook will be mainly used for the capstone project",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"print(\"Hello Capstone Project Course!\")",
"Hello Capstone Project Course!\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
]
] |
4ae5b0072a3dd8b3de37be98e433019945eb2614
| 517,962 |
ipynb
|
Jupyter Notebook
|
Classification/Logistic_Reg_sklearn_statsmodels.ipynb
|
thirupathi-chintu/Machine-Learning-with-Python
|
0bb8753a5140c8e69a24f2ab95c7ef133ac025a6
|
[
"BSD-2-Clause"
] | 1,803 |
2018-11-26T20:53:23.000Z
|
2022-03-31T15:25:29.000Z
|
Classification/Logistic_Reg_sklearn_statsmodels.ipynb
|
thirupathi-chintu/Machine-Learning-with-Python
|
0bb8753a5140c8e69a24f2ab95c7ef133ac025a6
|
[
"BSD-2-Clause"
] | 8 |
2019-02-05T04:09:57.000Z
|
2022-02-19T23:46:27.000Z
|
Classification/Logistic_Reg_sklearn_statsmodels.ipynb
|
thirupathi-chintu/Machine-Learning-with-Python
|
0bb8753a5140c8e69a24f2ab95c7ef133ac025a6
|
[
"BSD-2-Clause"
] | 1,237 |
2018-11-28T19:48:55.000Z
|
2022-03-31T15:25:07.000Z
| 178.238816 | 391,580 | 0.871558 |
[
[
[
"# Logistic regression example\n### Dr. Tirthajyoti Sarkar, Fremont, CA 94536\n\n---\n\nThis notebook demonstrates solving a logistic regression problem of predicting Hypothyrodism with **Scikit-learn** and **Statsmodels** libraries.\n\nThe dataset is taken from UCI ML repository.\n<br>Here is the link: https://archive.ics.uci.edu/ml/datasets/Thyroid+Disease",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"### Read the dataset",
"_____no_output_____"
]
],
[
[
"names = 'response age sex on_thyroxine query_on_thyroxine antithyroid_medication thyroid_surgery query_hypothyroid query_hyperthyroid pregnant \\\nsick tumor lithium goitre TSH_measured TSH T3_measured \\\nT3 TT4_measured TT4 T4U_measured T4U FTI_measured FTI TBG_measured TBG'",
"_____no_output_____"
],
[
"names = names.split(' ')",
"_____no_output_____"
],
[
"#!wget https://raw.githubusercontent.com/tirthajyoti/Machine-Learning-with-Python/master/Datasets/hypothyroid.csv\n\n#!mkdir Data\n#!mv hypothyroid.csv Data/",
"_____no_output_____"
],
[
"df = pd.read_csv('Data/hypothyroid.csv',index_col=False,names=names,na_values=['?'])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"to_drop=[]\nfor c in df.columns:\n if 'measured' in c or 'query' in c:\n to_drop.append(c)",
"_____no_output_____"
],
[
"to_drop",
"_____no_output_____"
],
[
"to_drop.append('TBG')",
"_____no_output_____"
],
[
"df.drop(to_drop,axis=1,inplace=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"### Let us see the basic statistics on the dataset",
"_____no_output_____"
]
],
[
[
"df.describe().T",
"_____no_output_____"
]
],
[
[
"### Are any data points are missing? We can check it using `df.isna()` method\nThe `df.isna()` method gives back a full DataFrame with Boolean values - True for data present, False for missing data. We can use `sum()` on that DataFrame to see and calculate the number of missing values per column.",
"_____no_output_____"
]
],
[
[
"df.isna().sum()",
"_____no_output_____"
]
],
[
[
"### We can use `df.dropna()` method to drop those missing rows",
"_____no_output_____"
]
],
[
[
"df.dropna(inplace=True)",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
]
],
[
[
"### Creating a transformation function to convert `+` or `-` responses to 1 and 0",
"_____no_output_____"
]
],
[
[
"def class_convert(response):\n if response=='hypothyroid':\n return 1\n else:\n return 0",
"_____no_output_____"
],
[
"df['response']=df['response'].apply(class_convert)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
]
],
[
[
"### Exploratory data analysis",
"_____no_output_____"
]
],
[
[
"for var in ['age','TSH','T3','TT4','T4U','FTI']:\n sns.boxplot(x='response',y=var,data=df)\n plt.show()",
"_____no_output_____"
],
[
"sns.pairplot(data=df[df.columns[1:]],diag_kws={'edgecolor':'k','bins':25},plot_kws={'edgecolor':'k'})\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Create dummy variables for the categorical variables",
"_____no_output_____"
]
],
[
[
"df_dummies = pd.get_dummies(data=df)",
"_____no_output_____"
],
[
"df_dummies.shape",
"_____no_output_____"
],
[
"df_dummies.sample(10)",
"_____no_output_____"
]
],
[
[
"### Test/train split",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(df_dummies.drop('response',axis=1), \n df_dummies['response'], test_size=0.30, \n random_state=42)",
"_____no_output_____"
],
[
"print(\"Training set shape\",X_train.shape)\nprint(\"Test set shape\",X_test.shape)",
"Training set shape (1400, 24)\nTest set shape (600, 24)\n"
]
],
[
[
"### Using `LogisticRegression` estimator from Scikit-learn\nWe are using the L2 regularization by default",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"clf1 = LogisticRegression(penalty='l2',solver='newton-cg')",
"_____no_output_____"
],
[
"clf1.fit(X_train,y_train)",
"_____no_output_____"
]
],
[
[
"### Intercept, coefficients, and score",
"_____no_output_____"
]
],
[
[
"clf1.intercept_",
"_____no_output_____"
],
[
"clf1.coef_",
"_____no_output_____"
],
[
"clf1.score(X_test,y_test)",
"_____no_output_____"
]
],
[
[
"### For `LogisticRegression` estimator, there is a special `predict_proba` method which computes the raw probability values",
"_____no_output_____"
]
],
[
[
"prob_threshold = 0.5",
"_____no_output_____"
],
[
"prob_df=pd.DataFrame(clf1.predict_proba(X_test[:10]),columns=['Prob of NO','Prob of YES'])\nprob_df['Decision']=(prob_df['Prob of YES']>prob_threshold).apply(int)\nprob_df",
"_____no_output_____"
],
[
"y_test[:10]",
"_____no_output_____"
]
],
[
[
"### Classification report, and confusion matrix",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report, confusion_matrix",
"_____no_output_____"
],
[
"print(classification_report(y_test, clf1.predict(X_test)))",
" precision recall f1-score support\n\n 0 0.99 0.99 0.99 569\n 1 0.86 0.77 0.81 31\n\n micro avg 0.98 0.98 0.98 600\n macro avg 0.92 0.88 0.90 600\nweighted avg 0.98 0.98 0.98 600\n\n"
],
[
"pd.DataFrame(confusion_matrix(y_test, clf1.predict(X_test)),columns=['Predict-YES','Predict-NO'],index=['YES','NO'])",
"_____no_output_____"
]
],
[
[
"### Using `statsmodels` library",
"_____no_output_____"
]
],
[
[
"import statsmodels.formula.api as smf\nimport statsmodels.api as sm",
"_____no_output_____"
],
[
"df_dummies.columns",
"_____no_output_____"
]
],
[
[
"### Create a 'formula' in the same style as in R language",
"_____no_output_____"
]
],
[
[
"formula = 'response ~ ' + '+'.join(df_dummies.columns[1:])",
"_____no_output_____"
],
[
"formula",
"_____no_output_____"
]
],
[
[
"### Fit a GLM (Generalized Linear model) with this formula and choosing `Binomial` as the family of function",
"_____no_output_____"
]
],
[
[
"model = smf.glm(formula = formula, data=df_dummies, family=sm.families.Binomial())",
"_____no_output_____"
],
[
"result=model.fit()",
"_____no_output_____"
]
],
[
[
"### `summary` method shows a R-style table with all kind of statistical information",
"_____no_output_____"
]
],
[
[
"print(result.summary())",
" Generalized Linear Model Regression Results \n==============================================================================\nDep. Variable: response No. Observations: 2000\nModel: GLM Df Residuals: 1984\nModel Family: Binomial Df Model: 15\nLink Function: logit Scale: 1.0000\nMethod: IRLS Log-Likelihood: -123.59\nDate: Wed, 21 Aug 2019 Deviance: 247.17\nTime: 14:54:43 Pearson chi2: 9.10e+03\nNo. Iterations: 22 Covariance Type: nonrobust\n============================================================================================\n coef std err z P>|z| [0.025 0.975]\n--------------------------------------------------------------------------------------------\nIntercept -3.1161 4981.804 -0.001 1.000 -9767.273 9761.041\nage 0.0219 0.011 2.062 0.039 0.001 0.043\nTSH 0.0214 0.008 2.823 0.005 0.007 0.036\nT3 -0.0233 0.320 -0.073 0.942 -0.651 0.604\nTT4 0.0367 0.027 1.376 0.169 -0.016 0.089\nT4U -1.5693 1.656 -0.947 0.343 -4.816 1.677\nFTI -0.1420 0.030 -4.709 0.000 -0.201 -0.083\nsex_F -1.4005 2490.902 -0.001 1.000 -4883.479 4880.678\nsex_M -1.7156 2490.902 -0.001 0.999 -4883.794 4880.363\non_thyroxine_f -1.2357 2490.902 -0.000 1.000 -4883.314 4880.843\non_thyroxine_t -1.8804 2490.902 -0.001 0.999 -4883.959 4880.198\nantithyroid_medication_f -0.3206 2490.902 -0.000 1.000 -4882.399 4881.758\nantithyroid_medication_t -2.7954 2490.902 -0.001 0.999 -4884.874 4879.283\nthyroid_surgery_f -2.2489 2490.902 -0.001 0.999 -4884.328 4879.830\nthyroid_surgery_t -0.8672 2490.902 -0.000 1.000 -4882.946 4881.211\npregnant_f -0.2489 2490.903 -9.99e-05 1.000 -4882.328 4881.830\npregnant_t -2.8671 2490.903 -0.001 0.999 -4884.947 4879.213\nsick_f -1.1899 2490.902 -0.000 1.000 -4883.269 4880.889\nsick_t -1.9262 2490.902 -0.001 0.999 -4884.005 4880.153\ntumor_f 8.1294 6275.938 0.001 0.999 -1.23e+04 1.23e+04\ntumor_t -11.2454 7357.670 -0.002 0.999 -1.44e+04 1.44e+04\nlithium_f 8.7704 2.42e+04 0.000 1.000 -4.75e+04 4.75e+04\nlithium_t -11.8864 2.91e+04 -0.000 1.000 -5.7e+04 5.7e+04\ngoitre_f -1.9841 2490.902 -0.001 0.999 -4884.063 4880.095\ngoitre_t -1.1320 2490.902 -0.000 1.000 -4883.211 4880.947\n============================================================================================\n"
]
],
[
[
"### The `predict` method computes probability for the test dataset",
"_____no_output_____"
]
],
[
[
"result.predict(X_test[:10])",
"_____no_output_____"
]
],
[
[
"### To create binary predictions, you have to apply a threshold probability and convert the booleans into integers",
"_____no_output_____"
]
],
[
[
"y_pred=(result.predict(X_test)>prob_threshold).apply(int)",
"_____no_output_____"
],
[
"print(classification_report(y_test,y_pred))",
" precision recall f1-score support\n\n 0 0.99 0.99 0.99 569\n 1 0.89 0.77 0.83 31\n\n micro avg 0.98 0.98 0.98 600\n macro avg 0.94 0.88 0.91 600\nweighted avg 0.98 0.98 0.98 600\n\n"
],
[
"pd.DataFrame(confusion_matrix(y_test, y_pred),columns=['Predict-YES','Predict-NO'],index=['YES','NO'])",
"_____no_output_____"
]
],
[
[
"### A smaller model with only first few variables\n\nWe saw that majority of variables in the logistic regression model has p-values very high and therefore they are not statistically significant. We create another smaller model removing those variables.",
"_____no_output_____"
]
],
[
[
"formula = 'response ~ ' + '+'.join(df_dummies.columns[1:7])\nformula",
"_____no_output_____"
],
[
"model = smf.glm(formula = formula, data=df_dummies, family=sm.families.Binomial())\nresult=model.fit()\nprint(result.summary())",
" Generalized Linear Model Regression Results \n==============================================================================\nDep. Variable: response No. Observations: 2000\nModel: GLM Df Residuals: 1993\nModel Family: Binomial Df Model: 6\nLink Function: logit Scale: 1.0000\nMethod: IRLS Log-Likelihood: -131.55\nDate: Wed, 21 Aug 2019 Deviance: 263.09\nTime: 14:54:43 Pearson chi2: 5.65e+03\nNo. Iterations: 9 Covariance Type: nonrobust\n==============================================================================\n coef std err z P>|z| [0.025 0.975]\n------------------------------------------------------------------------------\nIntercept 4.1596 1.995 2.085 0.037 0.250 8.069\nage 0.0248 0.010 2.481 0.013 0.005 0.044\nTSH 0.0212 0.007 2.892 0.004 0.007 0.035\nT3 0.0704 0.297 0.237 0.812 -0.511 0.652\nTT4 0.0296 0.026 1.134 0.257 -0.022 0.081\nT4U -1.2903 1.665 -0.775 0.438 -4.553 1.973\nFTI -0.1307 0.029 -4.449 0.000 -0.188 -0.073\n==============================================================================\n"
],
[
"y_pred=(result.predict(X_test)>prob_threshold).apply(int)\nprint(classification_report(y_pred,y_test))",
" precision recall f1-score support\n\n 0 0.99 0.99 0.99 573\n 1 0.74 0.85 0.79 27\n\n micro avg 0.98 0.98 0.98 600\n macro avg 0.87 0.92 0.89 600\nweighted avg 0.98 0.98 0.98 600\n\n"
],
[
"pd.DataFrame(confusion_matrix(y_test, y_pred),columns=['Predict-YES','Predict-NO'],index=['YES','NO'])",
"_____no_output_____"
]
],
[
[
"### How do the probabilities compare between `Scikit-learn` and `Statsmodels` predictions? ",
"_____no_output_____"
]
],
[
[
"sklearn_prob = clf1.predict_proba(X_test)[...,1][:10]\nstatsmodels_prob = result.predict(X_test[:10])",
"_____no_output_____"
],
[
"prob_comp_df=pd.DataFrame(data={'Scikit-learn Prob':list(sklearn_prob),'Statsmodels Prob':list(statsmodels_prob)})\nprob_comp_df",
"_____no_output_____"
]
],
[
[
"### Coefficient interpretation\n\nWhat is the interpretation of the coefficient value for `age` and `FTI`?\n\n- With every one year of age increase, the log odds of the hypothyrodism **increases** by 0.0248 or the odds of hypothyroidsm increases by a factor of exp(0.0248) = 1.025 i.e. almost 2.5%.\n- With every one unit of FTI increase, the log odds of the hypothyrodism **decreases** by 0.1307 or the odds of hypothyroidsm decreases by a factor of exp(0.1307) = 1.1396 i.e. almost by 12.25%.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4ae5b88427f6caa0f6243ebc9a1a476fa78613f9
| 79,889 |
ipynb
|
Jupyter Notebook
|
Notebooks/H1N1 Modeling.ipynb
|
MikeAnderson89/H1N1_and_Flu_Predictions
|
9fd22b12b59e5042ecfa6af108e6c29f81cea5f0
|
[
"MIT"
] | null | null | null |
Notebooks/H1N1 Modeling.ipynb
|
MikeAnderson89/H1N1_and_Flu_Predictions
|
9fd22b12b59e5042ecfa6af108e6c29f81cea5f0
|
[
"MIT"
] | null | null | null |
Notebooks/H1N1 Modeling.ipynb
|
MikeAnderson89/H1N1_and_Flu_Predictions
|
9fd22b12b59e5042ecfa6af108e6c29f81cea5f0
|
[
"MIT"
] | null | null | null | 50.055764 | 1,711 | 0.514939 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport tensorflow as tf\nfrom datetime import datetime\nimport matplotlib.pyplot as plt\nimport seaborn as sns",
"_____no_output_____"
],
[
"features = pd.read_csv('../Data/training_set_features.csv')\nlabels = pd.read_csv('../Data/training_set_labels.csv')",
"_____no_output_____"
],
[
"df = pd.merge(features, labels, on='respondent_id', how='inner')",
"_____no_output_____"
],
[
"df = df.drop(columns=['employment_occupation', 'employment_industry', 'health_insurance', 'respondent_id'])",
"_____no_output_____"
],
[
"seas_df = df.drop(columns=['h1n1_concern',\n 'h1n1_knowledge',\n 'doctor_recc_h1n1',\n 'opinion_h1n1_vacc_effective',\n 'opinion_h1n1_risk',\n 'opinion_h1n1_sick_from_vacc',\n 'h1n1_vaccine'])\n\nh1n1_df = df.drop(columns=['doctor_recc_seasonal',\n 'opinion_seas_vacc_effective',\n 'opinion_seas_risk',\n 'opinion_seas_sick_from_vacc',\n 'seasonal_vaccine'])",
"_____no_output_____"
],
[
"categorical_columns = [\n 'sex',\n 'hhs_geo_region',\n 'census_msa',\n 'race',\n 'age_group',\n 'behavioral_face_mask',\n 'behavioral_wash_hands',\n 'behavioral_antiviral_meds',\n 'behavioral_outside_home',\n 'behavioral_large_gatherings',\n 'behavioral_touch_face',\n 'behavioral_avoidance',\n 'health_worker',\n 'child_under_6_months',\n 'chronic_med_condition',\n 'education',\n 'marital_status',\n 'employment_status',\n 'rent_or_own',\n 'doctor_recc_h1n1',\n 'doctor_recc_seasonal',\n 'income_poverty'\n]\n\nnumerical_columns = [\n 'household_children',\n 'household_adults',\n 'h1n1_concern',\n 'h1n1_knowledge',\n 'opinion_h1n1_risk',\n 'opinion_h1n1_vacc_effective',\n 'opinion_h1n1_sick_from_vacc',\n 'opinion_seas_vacc_effective',\n 'opinion_seas_risk',\n 'opinion_seas_sick_from_vacc',\n \n]\n\nfor column in categorical_columns:\n curr_col = df[column]\n df.loc[df[column] == 1, column] = 'Yes'\n df.loc[df[column] == 0, column] = 'No'\n\n",
"_____no_output_____"
]
],
[
[
"## Deal with NAs",
"_____no_output_____"
]
],
[
[
"((df.isnull().sum() / len(df)) * 100).sort_values()",
"_____no_output_____"
],
[
"for column in numerical_columns:\n df[column] = df[column].fillna(df[column].mean())\n\ndf = df.dropna()",
"_____no_output_____"
]
],
[
[
"## Initial Run",
"_____no_output_____"
]
],
[
[
"X = df.drop(columns=['h1n1_vaccine', 'seasonal_vaccine'])\ny = df[['h1n1_vaccine', 'seasonal_vaccine']]\ny_h1n1 = df[['h1n1_vaccine']]\ny_seas = df[['seasonal_vaccine']]",
"_____no_output_____"
]
],
[
[
"#### Categorical",
"_____no_output_____"
]
],
[
[
"#Get Binary Data for Categorical Variables\ncat_df = X[categorical_columns]",
"_____no_output_____"
],
[
"recat_df = pd.get_dummies(data=cat_df)",
"_____no_output_____"
]
],
[
[
"#### Numerical",
"_____no_output_____"
]
],
[
[
"num_df = X[numerical_columns]",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\n\n#Scale Numerical Data\nscaler = StandardScaler()\nscaled_num = scaler.fit_transform(num_df)\nscaled_num_df = pd.DataFrame(scaled_num, index=num_df.index, columns=num_df.columns)",
"_____no_output_____"
],
[
"encoded_df = pd.concat([recat_df, scaled_num_df], axis=1)",
"_____no_output_____"
],
[
"encoded_df",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(encoded_df, y, test_size=0.3, random_state=42)",
"_____no_output_____"
],
[
"X_train = np.asarray(X_train)\nX_test = np.asarray(X_test)\ny_train = np.asarray(y_train)\ny_test = np.asarray(y_test)\nX = np.asarray(encoded_df)",
"_____no_output_____"
]
],
[
[
"# Neural Network",
"_____no_output_____"
]
],
[
[
"from tensorflow import keras\n\nmodel = keras.Sequential([\n keras.layers.Dense(60, activation='selu', input_dim=84),\n keras.layers.Dense(100, activation='relu'),\n keras.layers.Dense(200, activation='selu'),\n keras.layers.Dense(32, activation='relu'),\n keras.layers.Dense(5, activation='swish'),\n keras.layers.Dense(2, activation='swish')\n])",
"_____no_output_____"
],
[
"model.compile(optimizer='adam', \n loss=tf.losses.CategoricalCrossentropy(from_logits=True),\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"history = model.fit(\n X_train,\n y_train,\n batch_size=200,\n epochs=5000,\n validation_data=(X_test, y_test)\n)",
"Epoch 1/5000\n94/94 [==============================] - 0s 3ms/step - loss: 0.4581 - accuracy: 0.4116 - val_loss: 0.5046 - val_accuracy: 0.4026\nEpoch 2/5000\n94/94 [==============================] - 0s 2ms/step - loss: 4.9057 - accuracy: 0.5707 - val_loss: 10.5480 - val_accuracy: 0.6942\nEpoch 3/5000\n94/94 [==============================] - 0s 2ms/step - loss: 93.9989 - accuracy: 0.6465 - val_loss: 56.4817 - val_accuracy: 0.7146\nEpoch 4/5000\n94/94 [==============================] - 0s 2ms/step - loss: 728.9608 - accuracy: 0.6463 - val_loss: 2226.2800 - val_accuracy: 0.6235\nEpoch 5/5000\n94/94 [==============================] - 0s 2ms/step - loss: 4726.7036 - accuracy: 0.6437 - val_loss: 5452.4375 - val_accuracy: 0.7118\nEpoch 6/5000\n94/94 [==============================] - 0s 2ms/step - loss: 9564.6445 - accuracy: 0.6428 - val_loss: 5383.6187 - val_accuracy: 0.7146\nEpoch 7/5000\n94/94 [==============================] - 0s 2ms/step - loss: 27876.6602 - accuracy: 0.6423 - val_loss: 17322.0020 - val_accuracy: 0.7146\nEpoch 8/5000\n94/94 [==============================] - 0s 2ms/step - loss: 44396.2930 - accuracy: 0.6469 - val_loss: 32027.7168 - val_accuracy: 0.7002\nEpoch 9/5000\n94/94 [==============================] - 0s 2ms/step - loss: 149833.9688 - accuracy: 0.6418 - val_loss: 80736.3750 - val_accuracy: 0.6889\nEpoch 10/5000\n94/94 [==============================] - 0s 2ms/step - loss: 134787.2500 - accuracy: 0.6495 - val_loss: 325050.9688 - val_accuracy: 0.6216\nEpoch 11/5000\n94/94 [==============================] - 0s 2ms/step - loss: 251180.8281 - accuracy: 0.6439 - val_loss: 11821.4316 - val_accuracy: 0.6231\nEpoch 12/5000\n94/94 [==============================] - 0s 2ms/step - loss: 362681.5625 - accuracy: 0.6436 - val_loss: 1113056.7500 - val_accuracy: 0.7146\nEpoch 13/5000\n94/94 [==============================] - 0s 2ms/step - loss: 1118073.1250 - accuracy: 0.6427 - val_loss: 766207.4375 - val_accuracy: 0.6194\nEpoch 14/5000\n94/94 [==============================] - 0s 2ms/step - loss: 1232748.3750 - accuracy: 0.6426 - val_loss: 2337703.0000 - val_accuracy: 0.6217\nEpoch 15/5000\n94/94 [==============================] - 0s 2ms/step - loss: 1647592.1250 - accuracy: 0.6438 - val_loss: 386363.5000 - val_accuracy: 0.7127\nEpoch 16/5000\n94/94 [==============================] - 0s 2ms/step - loss: 1800699.0000 - accuracy: 0.6442 - val_loss: 3408100.7500 - val_accuracy: 0.6227\nEpoch 17/5000\n94/94 [==============================] - 0s 2ms/step - loss: 3808177.5000 - accuracy: 0.6435 - val_loss: 726046.0000 - val_accuracy: 0.6227\nEpoch 18/5000\n94/94 [==============================] - 0s 2ms/step - loss: 2553061.0000 - accuracy: 0.6409 - val_loss: 3051980.5000 - val_accuracy: 0.6891\nEpoch 19/5000\n94/94 [==============================] - 0s 2ms/step - loss: 4217261.0000 - accuracy: 0.6406 - val_loss: 11172239.0000 - val_accuracy: 0.6215\nEpoch 20/5000\n94/94 [==============================] - 0s 2ms/step - loss: 8031296.5000 - accuracy: 0.6408 - val_loss: 19805110.0000 - val_accuracy: 0.6215\nEpoch 21/5000\n94/94 [==============================] - 0s 2ms/step - loss: 7874280.5000 - accuracy: 0.6420 - val_loss: 12199877.0000 - val_accuracy: 0.6211\nEpoch 22/5000\n94/94 [==============================] - 0s 2ms/step - loss: 12675631.0000 - accuracy: 0.6437 - val_loss: 3006174.0000 - val_accuracy: 0.6210\nEpoch 23/5000\n94/94 [==============================] - 0s 2ms/step - loss: 10422905.0000 - accuracy: 0.6418 - val_loss: 8610656.0000 - val_accuracy: 0.6224\nEpoch 24/5000\n94/94 [==============================] - 0s 2ms/step - loss: 10953854.0000 - accuracy: 0.6421 - val_loss: 21237436.0000 - val_accuracy: 0.6210\nEpoch 25/5000\n94/94 [==============================] - 0s 2ms/step - loss: 14320600.0000 - accuracy: 0.6442 - val_loss: 8601043.0000 - val_accuracy: 0.6224\nEpoch 26/5000\n94/94 [==============================] - 0s 2ms/step - loss: 21852488.0000 - accuracy: 0.6398 - val_loss: 9508753.0000 - val_accuracy: 0.6216\nEpoch 27/5000\n94/94 [==============================] - 0s 2ms/step - loss: 25215360.0000 - accuracy: 0.6411 - val_loss: 1934189.3750 - val_accuracy: 0.6829\nEpoch 28/5000\n94/94 [==============================] - 0s 2ms/step - loss: 26388914.0000 - accuracy: 0.6466 - val_loss: 30157924.0000 - val_accuracy: 0.6215\nEpoch 29/5000\n94/94 [==============================] - 0s 2ms/step - loss: 33147992.0000 - accuracy: 0.6427 - val_loss: 33740512.0000 - val_accuracy: 0.7146\nEpoch 30/5000\n94/94 [==============================] - 0s 2ms/step - loss: 27107508.0000 - accuracy: 0.6455 - val_loss: 35969624.0000 - val_accuracy: 0.6204\nEpoch 31/5000\n94/94 [==============================] - 0s 2ms/step - loss: 45858880.0000 - accuracy: 0.6457 - val_loss: 12904045.0000 - val_accuracy: 0.7091\nEpoch 32/5000\n94/94 [==============================] - 0s 2ms/step - loss: 37509968.0000 - accuracy: 0.6476 - val_loss: 62433776.0000 - val_accuracy: 0.7146\nEpoch 33/5000\n94/94 [==============================] - 0s 2ms/step - loss: 37470476.0000 - accuracy: 0.6475 - val_loss: 12822841.0000 - val_accuracy: 0.6331\nEpoch 34/5000\n94/94 [==============================] - 0s 2ms/step - loss: 45705024.0000 - accuracy: 0.6449 - val_loss: 33301480.0000 - val_accuracy: 0.7066\nEpoch 35/5000\n94/94 [==============================] - 0s 2ms/step - loss: 52550988.0000 - accuracy: 0.6473 - val_loss: 13784739.0000 - val_accuracy: 0.7029\nEpoch 36/5000\n94/94 [==============================] - 0s 2ms/step - loss: 66794836.0000 - accuracy: 0.6411 - val_loss: 183237440.0000 - val_accuracy: 0.7146\nEpoch 37/5000\n94/94 [==============================] - 0s 2ms/step - loss: 62280220.0000 - accuracy: 0.6471 - val_loss: 39697756.0000 - val_accuracy: 0.6878\nEpoch 38/5000\n94/94 [==============================] - 0s 2ms/step - loss: 90092792.0000 - accuracy: 0.6407 - val_loss: 105631896.0000 - val_accuracy: 0.6229\nEpoch 39/5000\n94/94 [==============================] - 0s 2ms/step - loss: 130031096.0000 - accuracy: 0.6459 - val_loss: 47853144.0000 - val_accuracy: 0.6211\nEpoch 40/5000\n94/94 [==============================] - 0s 2ms/step - loss: 95973840.0000 - accuracy: 0.6432 - val_loss: 112537032.0000 - val_accuracy: 0.6202\nEpoch 41/5000\n94/94 [==============================] - 0s 2ms/step - loss: 56965136.0000 - accuracy: 0.6406 - val_loss: 54638008.0000 - val_accuracy: 0.6923\nEpoch 42/5000\n94/94 [==============================] - 0s 2ms/step - loss: 121091432.0000 - accuracy: 0.6422 - val_loss: 28487460.0000 - val_accuracy: 0.6226\nEpoch 43/5000\n94/94 [==============================] - 0s 2ms/step - loss: 164656016.0000 - accuracy: 0.6403 - val_loss: 138217360.0000 - val_accuracy: 0.6220\nEpoch 44/5000\n94/94 [==============================] - 0s 2ms/step - loss: 112128040.0000 - accuracy: 0.6400 - val_loss: 191743680.0000 - val_accuracy: 0.7024\nEpoch 45/5000\n94/94 [==============================] - 0s 2ms/step - loss: 118058280.0000 - accuracy: 0.6465 - val_loss: 99461808.0000 - val_accuracy: 0.7146\nEpoch 46/5000\n94/94 [==============================] - 0s 2ms/step - loss: 113831240.0000 - accuracy: 0.6441 - val_loss: 149199264.0000 - val_accuracy: 0.6215\nEpoch 47/5000\n94/94 [==============================] - 0s 2ms/step - loss: 207023632.0000 - accuracy: 0.6404 - val_loss: 197590640.0000 - val_accuracy: 0.7146\nEpoch 48/5000\n94/94 [==============================] - 0s 2ms/step - loss: 144444128.0000 - accuracy: 0.6431 - val_loss: 207115776.0000 - val_accuracy: 0.6222\nEpoch 49/5000\n94/94 [==============================] - 0s 2ms/step - loss: 228580752.0000 - accuracy: 0.6432 - val_loss: 191666256.0000 - val_accuracy: 0.6229\nEpoch 50/5000\n94/94 [==============================] - 0s 2ms/step - loss: 200370592.0000 - accuracy: 0.6433 - val_loss: 163128752.0000 - val_accuracy: 0.6227\nEpoch 51/5000\n94/94 [==============================] - 0s 2ms/step - loss: 176418736.0000 - accuracy: 0.6428 - val_loss: 159334064.0000 - val_accuracy: 0.6215\nEpoch 52/5000\n94/94 [==============================] - 0s 2ms/step - loss: 208566064.0000 - accuracy: 0.6445 - val_loss: 374760736.0000 - val_accuracy: 0.6229\nEpoch 53/5000\n94/94 [==============================] - 0s 2ms/step - loss: 262661808.0000 - accuracy: 0.6465 - val_loss: 314313344.0000 - val_accuracy: 0.6217\nEpoch 54/5000\n94/94 [==============================] - 0s 2ms/step - loss: 241026192.0000 - accuracy: 0.6475 - val_loss: 394748768.0000 - val_accuracy: 0.6215\nEpoch 55/5000\n94/94 [==============================] - 0s 2ms/step - loss: 367754272.0000 - accuracy: 0.6451 - val_loss: 138436176.0000 - val_accuracy: 0.6911\nEpoch 56/5000\n94/94 [==============================] - 0s 2ms/step - loss: 241240512.0000 - accuracy: 0.6465 - val_loss: 240968560.0000 - val_accuracy: 0.6239\nEpoch 57/5000\n94/94 [==============================] - 0s 2ms/step - loss: 413072256.0000 - accuracy: 0.6429 - val_loss: 1016240128.0000 - val_accuracy: 0.6215\nEpoch 58/5000\n94/94 [==============================] - 0s 2ms/step - loss: 493040544.0000 - accuracy: 0.6423 - val_loss: 44767760.0000 - val_accuracy: 0.6551\nEpoch 59/5000\n94/94 [==============================] - 0s 2ms/step - loss: 195869408.0000 - accuracy: 0.6423 - val_loss: 390878464.0000 - val_accuracy: 0.6227\nEpoch 60/5000\n94/94 [==============================] - 0s 2ms/step - loss: 372289024.0000 - accuracy: 0.6420 - val_loss: 127319368.0000 - val_accuracy: 0.6214\nEpoch 61/5000\n94/94 [==============================] - 0s 2ms/step - loss: 706285440.0000 - accuracy: 0.6486 - val_loss: 721652160.0000 - val_accuracy: 0.6214\nEpoch 62/5000\n94/94 [==============================] - 0s 2ms/step - loss: 408792672.0000 - accuracy: 0.6475 - val_loss: 693527616.0000 - val_accuracy: 0.7146\nEpoch 63/5000\n94/94 [==============================] - 0s 2ms/step - loss: 351692480.0000 - accuracy: 0.6391 - val_loss: 560084032.0000 - val_accuracy: 0.6874\nEpoch 64/5000\n94/94 [==============================] - 0s 3ms/step - loss: 645895296.0000 - accuracy: 0.6422 - val_loss: 596730432.0000 - val_accuracy: 0.6220\nEpoch 65/5000\n94/94 [==============================] - 0s 2ms/step - loss: 551598272.0000 - accuracy: 0.6458 - val_loss: 86612256.0000 - val_accuracy: 0.6215\nEpoch 66/5000\n94/94 [==============================] - 0s 2ms/step - loss: 633274944.0000 - accuracy: 0.6463 - val_loss: 1054557568.0000 - val_accuracy: 0.6214\nEpoch 67/5000\n94/94 [==============================] - 0s 2ms/step - loss: 647588032.0000 - accuracy: 0.6452 - val_loss: 657145792.0000 - val_accuracy: 0.6215\nEpoch 68/5000\n94/94 [==============================] - 0s 2ms/step - loss: 354433952.0000 - accuracy: 0.6433 - val_loss: 675115200.0000 - val_accuracy: 0.7146\nEpoch 69/5000\n94/94 [==============================] - 0s 2ms/step - loss: 636722176.0000 - accuracy: 0.6443 - val_loss: 208832032.0000 - val_accuracy: 0.6440\nEpoch 70/5000\n94/94 [==============================] - 0s 2ms/step - loss: 553934208.0000 - accuracy: 0.6477 - val_loss: 847264896.0000 - val_accuracy: 0.6222\nEpoch 71/5000\n62/94 [==================>...........] - ETA: 0s - loss: 724060672.0000 - accuracy: 0.6450"
],
[
"y_true = y_test\ny_predicted = model.predict(X_test)\ny_predicted_binary = np.where(y_predicted > 0.5, 1, 0)",
"_____no_output_____"
],
[
"from sklearn.metrics import roc_auc_score\n\nroc_auc_score(y_true, y_predicted)",
"_____no_output_____"
]
],
[
[
"# Random Forest",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor\nfrom sklearn.metrics import r2_score\n\nmodel = RandomForestRegressor()\nmodel.fit(X_train, y_train)\n\ny_predicted = model.predict(X_test)\n\nr2_score(y_predicted, y_test)",
"_____no_output_____"
],
[
"from sklearn.model_selection import RandomizedSearchCV\n# Number of trees in random forest\nn_estimators = [int(x) for x in np.linspace(start = 200, stop = 2000, num = 10)]\n# Number of features to consider at every split\nmax_features = ['auto', 'sqrt']\n# Maximum number of levels in tree\nmax_depth = [int(x) for x in np.linspace(10, 110, num = 11)]\nmax_depth.append(None)\n# Minimum number of samples required to split a node\nmin_samples_split = [2, 5, 10]\n# Minimum number of samples required at each leaf node\nmin_samples_leaf = [1, 2, 4]\n# Method of selecting samples for training each tree\nbootstrap = [True, False]\n# Create the random grid\nrandom_grid = {'n_estimators': n_estimators,\n 'max_features': max_features,\n 'max_depth': max_depth,\n 'min_samples_split': min_samples_split,\n 'min_samples_leaf': min_samples_leaf,\n 'bootstrap': bootstrap}\nprint(random_grid)",
"{'n_estimators': [200, 400, 600, 800, 1000, 1200, 1400, 1600, 1800, 2000], 'max_features': ['auto', 'sqrt'], 'max_depth': [10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, None], 'min_samples_split': [2, 5, 10], 'min_samples_leaf': [1, 2, 4], 'bootstrap': [True, False]}\n"
],
[
"rf = RandomForestRegressor()\nrf_random = RandomizedSearchCV(estimator = rf, param_distributions = random_grid, n_iter = 100, cv = 3, verbose=2, random_state=42, n_jobs = -1)",
"_____no_output_____"
],
[
"model = rf_random\nmodel.fit(X_train, y_train)",
"Fitting 3 folds for each of 100 candidates, totalling 300 fits\n"
],
[
"model.best_params_",
"_____no_output_____"
],
[
"def evaluate(model, X_test, y_test):\n predictions = model.predict(X_test)\n errors = abs(predictions - y_test)\n mape = 100 * np.mean(errors / y_test)\n accuracy = 100 - mape\n print('Model Performance')\n print('Average Error: {:0.4f} degrees.'.format(np.mean(errors)))\n print('Accuracy = {:0.2f}%.'.format(accuracy))\n \n return accuracy\n\n\nbase_model = RandomForestRegressor(n_estimators = 10, random_state = 42)\nbase_model.fit(X_train, y_train)\nbase_accuracy = evaluate(base_model, X_test, y_test)\n\n\n\nbest_random = rf_random.best_estimator_\nrandom_accuracy = evaluate(best_random, X_test, y_test)",
"<ipython-input-114-2d379f86ada3>:4: RuntimeWarning: divide by zero encountered in true_divide\n mape = 100 * np.mean(errors / y_test)\n<ipython-input-114-2d379f86ada3>:4: RuntimeWarning: invalid value encountered in true_divide\n mape = 100 * np.mean(errors / y_test)\n"
],
[
"y_predicted = best_random.predict(X_test)\nmodel = best_random\n\nfrom sklearn.metrics import roc_auc_score\n\nroc_auc_score(y_test, y_predicted)",
"_____no_output_____"
]
],
[
[
"## Submission Data",
"_____no_output_____"
]
],
[
[
"test_data = pd.read_csv('../Data/test_set_features.csv')\ndf_full = test_data",
"_____no_output_____"
],
[
"df = df_full.drop(columns=['employment_occupation', 'employment_industry', 'health_insurance', 'respondent_id'])\n\ncategorical_columns = [\n 'sex',\n 'hhs_geo_region',\n 'census_msa',\n 'race',\n 'age_group',\n 'behavioral_face_mask',\n 'behavioral_wash_hands',\n 'behavioral_antiviral_meds',\n 'behavioral_outside_home',\n 'behavioral_large_gatherings',\n 'behavioral_touch_face',\n 'behavioral_avoidance',\n 'health_worker',\n 'child_under_6_months',\n 'chronic_med_condition',\n 'education',\n 'marital_status',\n 'employment_status',\n 'rent_or_own',\n 'doctor_recc_h1n1',\n 'doctor_recc_seasonal',\n 'income_poverty'\n]\n\nnumerical_columns = [\n 'household_children',\n 'household_adults',\n 'h1n1_concern',\n 'h1n1_knowledge',\n 'opinion_h1n1_risk',\n 'opinion_h1n1_vacc_effective',\n 'opinion_h1n1_sick_from_vacc',\n 'opinion_seas_vacc_effective',\n 'opinion_seas_risk',\n 'opinion_seas_sick_from_vacc',\n \n]\n\nfor column in categorical_columns:\n curr_col = df[column]\n df.loc[df[column] == 1, column] = 'Yes'\n df.loc[df[column] == 0, column] = 'No'",
"_____no_output_____"
],
[
"for column in numerical_columns:\n df[column] = df[column].fillna(df[column].mean())\n\ndf['health_worker'] = df['health_worker'].fillna(0)\ndf['behavioral_face_mask'] = df['behavioral_face_mask'].fillna(0)\ndf['behavioral_wash_hands'] = df['behavioral_wash_hands'].fillna(0)\ndf['behavioral_antiviral_meds'] = df['behavioral_antiviral_meds'].fillna(0)\ndf['behavioral_outside_home'] = df['behavioral_outside_home'].fillna(0)\ndf['behavioral_large_gatherings'] = df['behavioral_large_gatherings'].fillna(0)\ndf['behavioral_touch_face'] = df['behavioral_touch_face'].fillna(0)\ndf['behavioral_avoidance'] = df['behavioral_avoidance'].fillna(0)\ndf['child_under_6_months'] = df['child_under_6_months'].fillna(0)\ndf['chronic_med_condition'] = df['chronic_med_condition'].fillna(0)\ndf['marital_status'] = df['marital_status'].fillna('Not Married')\ndf['rent_or_own'] = df['rent_or_own'].fillna('Rent')\ndf['education'] = df['education'].fillna('Some College')\ndf['employment_status'] = df['employment_status'].fillna('Employed')\ndf['doctor_recc_h1n1'] = df['doctor_recc_h1n1'].fillna(1)\ndf['doctor_recc_seasonal'] = df['doctor_recc_seasonal'].fillna(1)\ndf['income_poverty'] = df['income_poverty'].fillna('<= $75,000, Above Poverty')",
"_____no_output_____"
],
[
"X = df\n\n#Get Binary Data for Categorical Variables\ncat_df = X[categorical_columns]\nrecat_df = pd.get_dummies(data=cat_df)\n\nnum_df = X[numerical_columns]\n\nfrom sklearn.preprocessing import StandardScaler\n\n#Scale Numerical Data\nscaler = StandardScaler()\nscaled_num = scaler.fit_transform(num_df)\nscaled_num_df = pd.DataFrame(scaled_num, index=num_df.index, columns=num_df.columns)\n\nencoded_df = pd.concat([recat_df, scaled_num_df], axis=1)\n\nX = np.asarray(encoded_df)",
"_____no_output_____"
],
[
"y = model.predict(X)\ny_df = pd.DataFrame(y, columns=['h1n1_vaccine', 'seasonal_vaccine'])",
"_____no_output_____"
],
[
"results = pd.concat([df_full, y_df], axis=1)",
"_____no_output_____"
],
[
"results = results[['respondent_id', 'h1n1_vaccine', 'seasonal_vaccine']]",
"_____no_output_____"
],
[
"results.to_csv('../Submissions/Submission 6.29.21.csv', index=False)",
"_____no_output_____"
],
[
"from sklearn.feature_selection import SelectFromModel\n\nsel = SelectFromModel(RandomForestRegressor(n_estimators= 800,\n min_samples_split= 2,\n min_samples_leaf= 4,\nmax_features= 'sqrt',\n max_depth= 20,\n bootstrap= False))\nsel.fit(X_train, y_train)",
"_____no_output_____"
],
[
"selected_feat= encoded_df.columns[(sel.get_support())]\nlen(selected_feat)",
"_____no_output_____"
],
[
"selected_feat",
"_____no_output_____"
],
[
"pd.Series(sel.estimator_, feature_importances_.ravel()).hist()",
"_____no_output_____"
],
[
"for x in selected_feat:\n print(x)",
"age_group_18 - 34 Years\nage_group_65+ Years\nhealth_worker_No\nhealth_worker_Yes\ndoctor_recc_h1n1_No\ndoctor_recc_h1n1_Yes\ndoctor_recc_seasonal_No\ndoctor_recc_seasonal_Yes\nhousehold_children\nh1n1_concern\nh1n1_knowledge\nopinion_h1n1_risk\nopinion_h1n1_vacc_effective\nopinion_h1n1_sick_from_vacc\nopinion_seas_vacc_effective\nopinion_seas_risk\nopinion_seas_sick_from_vacc\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae5d45f5707352eb47d8267d355fc9f4f631921
| 657,005 |
ipynb
|
Jupyter Notebook
|
CaseStudies/Capstone Projects/1-NLP-Automatic.Ticket.Assignment/Automatic Ticket Assignment - Milestone 2.ipynb
|
averma111/AIML-PGP
|
c9f61dadcfb2ea8cb2ff5412c0ab583f7e3807ca
|
[
"Apache-2.0"
] | null | null | null |
CaseStudies/Capstone Projects/1-NLP-Automatic.Ticket.Assignment/Automatic Ticket Assignment - Milestone 2.ipynb
|
averma111/AIML-PGP
|
c9f61dadcfb2ea8cb2ff5412c0ab583f7e3807ca
|
[
"Apache-2.0"
] | null | null | null |
CaseStudies/Capstone Projects/1-NLP-Automatic.Ticket.Assignment/Automatic Ticket Assignment - Milestone 2.ipynb
|
averma111/AIML-PGP
|
c9f61dadcfb2ea8cb2ff5412c0ab583f7e3807ca
|
[
"Apache-2.0"
] | null | null | null | 168.074955 | 143,716 | 0.854334 |
[
[
[
"## Automatic Ticket Assignment\nOne of the key activities of any IT function is to ensure there is no\nimpact to the Business operations. <b>IT leverages Incident Management process to achieve the\nabove Objective.</b> An incident is something that is unplanned interruption to an IT service or\nreduction in the quality of an IT service that affects the Users and the Business. <b><i>The main goal\nof Incident Management process is to provide a quick fix / workarounds or solutions that resolves the interruption and restores the service to its full capacity to ensure no business impact.</i></b>\n\nIn most of the organizations, incidents are created by various Business and IT Users, End Users/ Vendors if they have access to ticketing systems, and from the integrated monitoring\nsystems and tools. <b>Assigning the incidents to the appropriate person or unit in the support team has critical importance to provide improved user satisfaction while ensuring better allocation of support resources.</b>\n\n<i> Manual assignment of incidents is time consuming and requires human efforts. There may be mistakes due to human errors and resource consumption is carried out ineffectively because of the misaddressing. On the other hand, manual assignment increases the response and resolution times which result in user satisfaction deterioration / poor customer service.</i>\n#### <b>Business Domain Value:</b> \nIn the support process, incoming incidents are analyzed and assessed by organization’s support teams to fulfill the request. In many organizations, better allocation and effective usage of the valuable support resources will directly result in substantial cost savings.\n\nCurrently the incidents are created by various stakeholders (Business Users, IT Users and Monitoring Tools) within IT Service Management Tool and are assigned to Service Desk teams (L1 / L2 teams). This team will review the incidents for right ticket categorization, priorities and then carry out initial diagnosis to see if they can resolve. Around ~54% of the incidents are resolved by L1 / L2 teams. Incase L1 / L2 is unable to resolve, they will then escalate / assign the tickets to Functional teams from Applications and Infrastructure (L3 teams). Some portions of incidents are directly assigned to L3 teams by either Monitoring tools or Callers / Requestors. L3 teams will carry out detailed diagnosis and resolve the incidents. Around ~56%\nof incidents are resolved by Functional / L3 teams. Incase if vendor support is needed, they will reach out for their support towards incident closure.\n\nL1 / L2 needs to spend time reviewing Standard Operating Procedures (SOPs) before assigning to Functional teams (Minimum ~25-30% of incidents needs to be reviewed for SOPs before ticket assignment). 15 min is being spent for SOP review for each incident. Minimum of ~1 FTE effort needed only for incident assignment to L3 teams. During the process of incident assignments by L1 / L2 teams to functional groups, there were multiple instances of incidents getting assigned to wrong functional groups. Around ~25% of Incidents are wrongly assigned to functional teams. Additional effort needed for Functional teams to re-assign to right functional groups. During this process, some of the incidents are in queue and not addressed timely resulting in poor customer service.\n\n## Objective:\n### Build Multi-Class classifier that can classify the tickets by analysing text.###\nGuided by powerful AI techniques that can classify incidents to right functional groups can help organizations to reduce the resolving time of the issue and can focus on more productive tasks. In the previous milestone we've already covered Data cleaning, preprocessing, Exploratory Data Analysis\n\nMilestone 2: Test the Model, Fine-tuning and Repeat\n 1. Test the model and report as per evaluation metrics\n 2. Try different models\n 3. Try different evaluation metrics\n 4. Set different hyper parameters, by trying different optimizers, loss functions, epochs, learning rate, batch size, checkpointing, early stopping etc..for these models to fine-tune them\n 5. Report evaluation metrics for these models along with your observation on how changing different hyper parameters leads to change in the final evaluation metric.\n---\n\n### <u>Imports and Configurations</u>\nSection to import all necessary packages. Install the libraries which are not included in Anaconda distribution by default using pypi channel or conda forge\n**``!pip install ftfy wordcloud goslate spacy plotly cufflinks gensim pyLDAvis``**<br/>\n**``conda install -c conda-forge ftfy wordcloud goslate spacy plotly cufflinks gensim pyLDAvis``**",
"_____no_output_____"
]
],
[
[
"# Utilities\nfrom time import time\nfrom PIL import Image\nfrom pprint import pprint\nfrom zipfile import ZipFile\nimport os, sys, itertools, re, calendar\nimport warnings, pickle, string, timestring\nfrom IPython.display import IFrame\nfrom ftfy import fix_encoding, fix_text, badness\n\n# Translation APIs\nfrom goslate import Goslate # Provided by Google\n\n# Numerical calculation\nimport numpy as np\n\n# Data Handling\nimport pandas as pd\n\n# Data Visualization\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport cufflinks as cf\nimport plotly as py\nimport plotly.graph_objs as go\nfrom plotly.offline import download_plotlyjs,init_notebook_mode,plot,iplot\nimport pyLDAvis\nimport pyLDAvis.gensim\n\n# Sequential Modeling\nimport keras.backend as K\nfrom keras.datasets import imdb\nfrom keras.models import Sequential, Model\nfrom keras.layers.merge import Concatenate\nfrom keras.layers import Input, Dropout, Flatten, Dense, Embedding, LSTM, GRU\nfrom keras.layers import BatchNormalization, TimeDistributed, Conv1D, MaxPooling1D\nfrom keras.constraints import max_norm, unit_norm\nfrom keras.preprocessing.text import Tokenizer, text_to_word_sequence\nfrom keras.preprocessing.sequence import pad_sequences\nfrom keras.callbacks import EarlyStopping, ModelCheckpoint\n\n# Traditional Modeling\nfrom sklearn.naive_bayes import MultinomialNB\nfrom sklearn.neighbors import KNeighborsClassifier\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer, TfidfTransformer\nfrom sklearn.svm import SVC, LinearSVC\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier\n\n# Topic Modeling\nimport gensim\nimport gensim.corpora as corpora\nfrom gensim.utils import simple_preprocess\nfrom gensim.parsing import preprocessing\nfrom gensim.test.utils import common_texts\nfrom gensim.models.doc2vec import Doc2Vec, TaggedDocument\nfrom gensim.models.phrases import Phraser\nfrom gensim.models import Phrases, CoherenceModel\n\n# Tools & Evaluation metrics\nfrom sklearn.metrics import confusion_matrix, classification_report, auc\nfrom sklearn.metrics import roc_curve, accuracy_score, precision_recall_curve\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.model_selection import train_test_split, GridSearchCV\nfrom collections import Counter\nfrom imblearn.under_sampling import RandomUnderSampler\n\n# NLP toolkits\nimport spacy\nimport nltk\nfrom nltk import tokenize\nfrom nltk.corpus import stopwords",
"_____no_output_____"
],
[
"# Configure for any default setting of any library\nnltk.download('stopwords')\nwarnings.filterwarnings('ignore')\nget_ipython().magic(u'matplotlib inline')\nplt.style.use('ggplot')\npyLDAvis.enable_notebook()\ninit_notebook_mode(connected=True)\ncf.go_offline()\n%matplotlib inline",
"[nltk_data] Downloading package stopwords to\n[nltk_data] C:\\Users\\aksha\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
]
],
[
[
"### <u>Set the working directory</u>\nMount the drive and set the project path to cureent working directory, when running in Google Colab. No changes are required in case of running in Local PC.",
"_____no_output_____"
]
],
[
[
"# Block which runs on both Google Colab and Local PC without any modification\nif 'google.colab' in sys.modules: \n project_path = \"/content/drive/My Drive/Colab Notebooks/DLCP/Capstone-NLP/\"\n # Google Colab lib\n from google.colab import drive\n # Mount the drive\n drive.mount('/content/drive/', force_remount=True)\n sys.path.append(project_path)\n %cd $project_path\n\n# Let's look at the sys path\nprint('Current working directory', os.getcwd())",
"Current working directory D:\\Hands-On\\PGP-AIML\\CaseStudies\\Capstone Projects\\1-NLP-Automatic.Ticket.Assignment\n"
]
],
[
[
"### <u>Create Word Embbeddings</u>\nWe've observed poor performance in the 1st milestone, which enables us to create our own word embbeddings. Let's load the preprocessed dataset and use Gensim model to create Word2Vec embbeddings.\n\nWord embedding is one of the most important techniques in natural language processing(NLP), where words are mapped to vectors of real numbers. Word embedding is capable of capturing the meaning of a word in a document, semantic and syntactic similarity, relation with other words.\n\nThe word2vec algorithms include skip-gram and CBOW models, using either hierarchical softmax or negative sampling.\n\n",
"_____no_output_____"
]
],
[
[
"# Load the preprocessed pickle dataset\nwith open('preprocessed_ticket.pkl','rb') as f:\n ticket = pickle.load(f)\n \n# Function to create the tokenized sentence\ndef tokenize_sentences(sentence):\n doc = nlp(sentence)\n return [token.lemma_ for token in doc if token.lemma_ !='-PRON-' and not token.is_stop]\n\nsentence_stream=[]\nfor sent in ticket.Summary.values.tolist():\n sentence_stream.append(tokenize_sentences(sent))",
"_____no_output_____"
],
[
"# Create the Bigram and Trigram models\nbigram = Phrases(sentence_stream, min_count=2, threshold=2)\ntrigram = Phrases(bigram[sentence_stream], min_count=2, threshold=1)\nbigram_phraser = Phraser(bigram)\ntrigram_phraser = Phraser(trigram)\nngram_sentences=[]\nfor sent in sentence_stream:\n tokens_ = bigram_phraser[sent]\n #print(\"Bigrams Tokens:\\t\", tokens_)\n tokens_ = trigram_phraser[tokens_]\n ngram_sentences.append(tokens_)\n \n#Serialize bigram and trigram for future\nbigram_phraser.save('bigram_mdl_14_03_2020.pkl')\ntrigram_phraser.save('trigram_mdl_14_03_2020.pkl')",
"_____no_output_____"
],
[
"# Create the tagged documents\ndocuments = [TaggedDocument(words=doc, tags=[i]) for i, doc in enumerate(ngram_sentences)]\nprint(\"Length of Tagged Documents:\",len(documents))\nprint(\"Tagged Documents[345]:\",documents[345])",
"Length of Tagged Documents: 27101\nTagged Documents[345]: TaggedDocument(['monitor', 'video', 'measure_machine', 'supply', 'oziflwma', 'nhgvmqdl', 'monitor', 'video', 'measure_machine', 'supply', 'oziflwma', 'nhgvmqdl', 'general_lines1'], [345])\n"
],
[
"# Build the Word2Vec model\nmax_epochs = 100\nvec_size = 300\nalpha = 0.025\nmodel = Doc2Vec(vector_size=vec_size,window=2,\n alpha=alpha, \n min_alpha=0.00025,\n min_count=2,\n dm =1)\nmodel.build_vocab(documents)\n\nfor epoch in range(max_epochs):\n model.train(documents, \n total_examples=model.corpus_count,\n epochs=model.iter)\n # decrease the learning rate\n model.alpha -= 0.0002\n # fix the learning rate, no decay\n model.min_alpha = model.alpha\n\nmodel.save(\"d2v_inc_model.mdl\")\nprint(\"Model Saved\")",
"Model Saved\n"
]
],
[
[
"**Comments**:\nWord Embbeddings are generated from the corpus of our tickets dataset and serialized for further use.\n\n### <u>Load the dataset</u>\nWe've observed poor performance in the 1st milestone, which enables us to introduce 2 more attributes, such as:\n\n- **Shift**: Working shift of the support associate in which the ticket was recieved OR failure occured\n- **Lines**: Lines of text present in the ticket description column\nLoad the searialized dataset stored after 1st milestone's EDA and append the above attributes to them. Also drop sd_len, sd_word_count, desc_len, desc_word_count columns",
"_____no_output_____"
]
],
[
[
"# Function to determine the Part of the Day (POD)\ndef get_POD(tkt):\n dt1 = r\"(?:\\d{1,2}[\\/-]){2}\\d{4} (?:\\d{2}:?){3}\"\n dt2 = r\"\\d{4}(?:[\\/-]\\d{1,2}){2} (?:\\d{2}:?){3}\"\n months = '|'.join(calendar.month_name[1:])\n dt3 = fr'[a-zA-Z]+day, (?i:{months}) \\d{{1,2}}, \\d{{4}} \\d{{1,2}}:\\d{{1,2}} (?i:am|pm)'\n matches = set(re.findall('|'.join([dt1,dt2,dt3]), tkt))\n if len(matches):\n try:\n hr = timestring.Date(list(matches)[0]).hour\n return 'Morning' if (hr >= 6) and (hr < 18) else 'Night'\n except:\n pass\n return 'General'",
"_____no_output_____"
],
[
"# Get POD and lines of Desc from the unprocessed pickle\nwith open('translated_ticket.pkl','rb') as f:\n ticket = pickle.load(f)\n\nlines = ticket.Description.apply(lambda x: len(str(x).split('\\n')))\nshifts = ticket[['Short description', 'Description']].agg(lambda x: get_POD(str(x[0]) + str(x[1])), axis=1)\nshifts.value_counts()",
"_____no_output_____"
],
[
"# Load the serialized dataset after milestone-1\nwith open('model_ready.pkl','rb') as handle:\n ticket = pickle.load(handle)\n\n# Drop the unwanted columns\nticket.drop(['sd_len','sd_word_count','desc_len','desc_word_count','Caller'], axis=1, inplace=True)\n# Insert the new attributes\nticket.insert(loc=ticket.shape[1]-1, column='Shift', value=shifts)\nticket.insert(loc=ticket.shape[1]-1, column='Lines', value=lines)\n\n# Check the head of the dataset\nticket.head()",
"_____no_output_____"
]
],
[
[
"#### Observation from Milestone-1\nOut of all the models we've tried in Milestone-1, Support Vector Machine (SVM) under statistical ML algorithms and Neural Networks are performing better than all others. The models were highly overfitted and one of the obvious reason was the dataset was highly imbalanced. Ratio of GRP_0 to all others is 47:53 and there are 40 groups having less than or equal to 30 tickets assigned each.\n\nLet's address this problem to fine tune the model accuracy by implementing\n- Dealing with imbalanced dataset. \n - Creating distinctive clusters under GRP_0 and downsampling top clusters\n - Clubbing together all those groups into one which has 30 or less tickets assigned\n- Replacing TF-IDF vectorizer technique with word embeddings for statistical ML algorithms.\n\n### <u>Resampling the Imbalanced dataset</u>\nA widely adopted technique for dealing with highly unbalanced datasets is called resampling. It consists of removing samples from the majority class (under-sampling) and / or adding more examples from the minority class (over-sampling).\n\n\n\n### Topic Modeling\nTopic Modeling is a technique to extract the hidden topics from large volumes of text. **Latent Dirichlet Allocation(LDA)** is a popular algorithm for topic modeling with excellent implementations in the Python’s Gensim package.\n\nLet's first use gensim to implement LDA and find out any distinctive topics among GRP_0, followed by down-sampling the top 3 topics to contain maximum number of tickets created for.\n\nInstallation:<br/>\nusing pypi: **`!pip install gensim`**<br/>\nusing conda: **`conda install -c conda-forge gensim`**\n\n#### 1. Prepare Stopwords\nUsed english stopwords from NLTK and extended it to include domain specific frequent words",
"_____no_output_____"
]
],
[
[
"# Records assigned to only GRP_0\ngrp0_tickets = ticket[ticket['Assignment group'] == 'GRP_0']\n\n# Prepare NLTK STOPWORDS\nSTOP_WORDS = stopwords.words('english')\nSTOP_WORDS.extend(['yes','na','hi','receive','hello','regards','thanks','see','help',\n 'from','greeting','forward','reply','will','please','able'])",
"_____no_output_____"
]
],
[
[
"#### 2. Tokenize words and Clean-up text\nTokenize each sentence into a list of words, removing punctuations and unnecessary characters altogether.",
"_____no_output_____"
]
],
[
[
"# Vectorizations\ndef sent_to_words(sentences):\n for sentence in sentences:\n yield(gensim.utils.simple_preprocess(str(sentence), deacc=True)) # deacc=True removes punctuations\n\n# Tokenize the Summary attribute of GRP_0 records\ndata_words = list(sent_to_words(grp0_tickets['Summary'].values.tolist()))\ndata_words_nostops = [[word for word in simple_preprocess(str(doc)) if word not in STOP_WORDS] for doc in data_words]",
"_____no_output_____"
]
],
[
[
"#### 3. Bigram and Trigram Models\nBigrams and Trigrams are two and three words frequently occurring together respectively in a document.",
"_____no_output_____"
]
],
[
[
"# Build the bigram and trigram models\nbigram = gensim.models.Phrases(data_words, min_count=5, threshold=100) # higher threshold fewer phrases.\ntrigram = gensim.models.Phrases(bigram[data_words], threshold=100) \n\n# Faster way to get a sentence clubbed as a trigram/bigram\nbigram_mod = gensim.models.phrases.Phraser(bigram)\ndata_words_bigrams = [bigram_mod[doc] for doc in data_words_nostops]\n\ntrigram_mod = gensim.models.phrases.Phraser(trigram)\ndata_words_trigrams = [trigram_mod[doc] for doc in data_words_nostops]",
"_____no_output_____"
]
],
[
[
"#### 4. Dictionary and Corpus needed for Topic Modeling\nCreare the two main inputs to the LDA topic model are the dictionary(id2word) and the corpus.",
"_____no_output_____"
]
],
[
[
"# Create Dictionary\nid2word = corpora.Dictionary(data_words_bigrams)\n\n# Term Document Frequency\ncorpus = [id2word.doc2bow(text) for text in data_words_bigrams]",
"_____no_output_____"
]
],
[
[
"#### 5. Building the Topic Model\nBuild a Topic Model with top 3 different topics where each topic is a combination of keywords and each keyword contributes a certain weightage to the topic.",
"_____no_output_____"
]
],
[
[
"# Build LDA model\nlda_model = gensim.models.ldamodel.LdaModel(corpus=corpus,\n id2word=id2word,\n num_topics=3, \n random_state=100,\n update_every=1,\n chunksize=100,\n passes=10,\n alpha='auto',\n per_word_topics=True)\n\nfor idx, topic in lda_model.print_topics():\n print('Topic: {} \\nWords: {}'.format(idx+1, topic))\n print()",
"\nTopic: 1 \nWords: 0.050*\"unable\" + 0.034*\"outlook\" + 0.031*\"issue\" + 0.023*\"work\" + 0.020*\"error\" + 0.019*\"connect\" + 0.018*\"open\" + 0.016*\"get\" + 0.016*\"skype\" + 0.011*\"vpn\"\n\nTopic: 2 \nWords: 0.098*\"password\" + 0.066*\"erp\" + 0.061*\"reset\" + 0.054*\"account\" + 0.048*\"user\" + 0.037*\"login\" + 0.037*\"sid\" + 0.034*\"lock\" + 0.017*\"unlock\" + 0.016*\"request\"\n\nTopic: 3 \nWords: 0.043*\"gmail\" + 0.042*\"com\" + 0.026*\"access\" + 0.020*\"company\" + 0.020*\"update\" + 0.015*\"email\" + 0.013*\"ticket\" + 0.011*\"device\" + 0.009*\"mobile\" + 0.008*\"send\"\n\n"
]
],
[
[
"**How to interpret this?**\n\nTopic 1 is a represented as `0.060*\"company\" + 0.028*\"windows\" + 0.026*\"device\" + 0.021*\"vpn\" + 0.021*\"connect\" + 0.018*\"message\" + 0.014*\"link\" + 0.013*\"window\" + 0.011*\"follow\" + 0.011*\"use\"`\n\nIt means the top 10 keywords that contribute to this topic are: ‘company’, ‘windows’, ‘device’.. and so on and the weight of ‘windows’ on topic 1 is 0.028.\n\nThe weights reflect how important a keyword is to that topic.\n\n\n\n#### 6. Model Perplexity and Coherence Score\nModel perplexity and topic coherence provide a convenient measure to judge how good a given topic model is.",
"_____no_output_____"
]
],
[
[
"# Compute Perplexity\nprint('\\nPerplexity: ', lda_model.log_perplexity(corpus)) # a measure of how good the model is. lower the better.\n\n# Compute Coherence Score\ncoherence_model_lda = CoherenceModel(model=lda_model, texts=data_words_bigrams, dictionary=id2word, coherence='c_v')\ncoherence_lda = coherence_model_lda.get_coherence()\nprint('\\nCoherence Score: ', coherence_lda)",
"\nPerplexity: -6.800106460957963\n\nCoherence Score: 0.4568991626530355\n"
]
],
[
[
"#### 7. Visualize the topics-keywords\nExamine the produced topics and the associated keywords using pyLDAvis.",
"_____no_output_____"
]
],
[
[
"# Visualize the topics\npyLDAvis.save_html(pyLDAvis.gensim.prepare(lda_model, corpus, id2word), 'lda.html')\nIFrame(src='./lda.html', width=1220, height=858)",
"_____no_output_____"
]
],
[
[
"\n#### 8. Topic assignment for GRP_0 tickets\nRun LDA for each record of GRP_0 to find the associated topic based on the LDA score. As the topic modeling has been trained to accomodate only top 3 topics for entire GRP_0 data, any record scoring less than 50%, we categorize them into 4th(other) topic and such tickets are not the candidates for resampling.",
"_____no_output_____"
]
],
[
[
"# Function to Determine topic\nTOPICS = {1:\"Communication Issue\", 2:\"Account/Password Reset\", 3:\"Access Issue\", 4:\"Other Issues\"}\ndef get_groups(text):\n bow_vector = id2word.doc2bow([word for word in simple_preprocess(text) if word not in STOP_WORDS])\n index, score = sorted(lda_model[bow_vector][0], key=lambda tup: tup[1], reverse=True)[0]\n return TOPICS[index+1 if score > 0.5 else 4], round(score, 2)\n\n# Check for a Random record\ntext = grp0_tickets.reset_index().loc[np.random.randint(0, grp0_tickets.shape[1]),'Summary']\ntopic, score = get_groups(text)\nprint(\"\\033[1mText:\\033[0m {}\\n\\033[1mTopic:\\033[0m {}\\n\\033[1mScore:\\033[0m {}\".format(text, topic, score))",
"\u001b[1mText:\u001b[0m outlook receive from [email protected] hello team meetingsskype meeting etc be not appear in outlook calendar can somebody please advise how to correct this kind\n\u001b[1mTopic:\u001b[0m Communication Issue\n\u001b[1mScore:\u001b[0m 0.5099999904632568\n"
],
[
"# Apply the function to the dataset\ngrp0_tickets.insert(loc=grp0_tickets.shape[1]-1, \n column='Topic', \n value=[get_groups(text)[0] for text in grp0_tickets.Summary])\ngrp0_tickets.head()",
"_____no_output_____"
],
[
"# Count the records based on Topics\ngrp0_tickets.Topic.value_counts()",
"_____no_output_____"
]
],
[
[
"**Observations**:\n- From the above analysis, it's evident that the tope 3 topics are present in maximum numbers. The ratio of top 3 topics and other topic is $33:33:26:8$\n- Except for the Other Issues, rest 3 categories of records can be down sampled to balance the dataset\n\n#### 9. Down-sampling the majority topics under GRP_0\nUnder-sample the majority class(es) by randomly picking samples with or without replacement. We're using RandomUnderSampler class from imblearn.",
"_____no_output_____"
]
],
[
[
"# Instantiate the UnderSampler class\nsampler = RandomUnderSampler(sampling_strategy='auto', random_state=0)\n# Fit the data\nX_res, y_res = sampler.fit_resample(grp0_tickets.drop(['Assignment group','Topic'], axis=1), grp0_tickets.Topic)\n# Check the ratio of output topics\ny_res.value_counts()",
"_____no_output_____"
]
],
[
[
"**Observation:**<br/>\nThe output of the UnderSampling technique shows that all the 4 distinct topics are resampled to exactly match the records in each topic making them a perfectly balanced distribution under GRP_0.\n\nLet's combine the Topic and Assignment group columns to maintain a single target attribute.",
"_____no_output_____"
]
],
[
[
"# Combine Topic and Assignment Group columns\ngrp0_tickets = pd.concat([X_res, y_res], axis=1)\ngrp0_tickets['Assignment group'] = grp0_tickets['Topic'].apply(lambda x: f'GRP_0 ({x})')\n# Drop the Topic column\ngrp0_tickets.drop(['Topic'], axis=1, inplace=True)\nprint(f\"\\033[1mNew size of GRP_0 tickets:\\033[0m {grp0_tickets.shape}\")\ngrp0_tickets.head()",
"\u001b[1mNew size of GRP_0 tickets:\u001b[0m (1368, 7)\n"
]
],
[
[
"#### 10. Club groups with lesser tickets assigned\nCombine all groups with less than 25 tickets assigned into one separate group named ***Miscellaneous***",
"_____no_output_____"
]
],
[
[
"# Find out the Assignment Groups with less than equal to 25 tickets assigned\nrare_tickets = ticket.groupby(['Assignment group']).filter(lambda x: len(x) <= 25)\nprint('\\033[1m#Groups with less than equal to 25 tickets assigned:\\033[0m', rare_tickets['Assignment group'].nunique())\n\n# Visualize the distribution\nrare_tickets['Assignment group'].iplot(\n kind='hist',\n xTitle='Assignment Group',\n yTitle='count',\n colorscale='-orrd',\n title='#Records by rare Assignment Groups- Histogram')\n\n# Rename the Assignment group attribute\nrare_tickets['Assignment group'] = 'Miscellaneous'",
"\u001b[1m#Groups with less than equal to 25 tickets assigned:\u001b[0m 37\n"
]
],
[
[
"#### 11. Join and prepare the balanced dataset\nLet's club together resampled topics under GRP_0 with Miscellaneous group with less than 25 tickets with all others",
"_____no_output_____"
]
],
[
[
"# Find tickets with good number of tickets assigned\ngood_tickets = ticket.iloc[[idx for idx in ticket.index if idx not in rare_tickets.index]]\ngood_tickets = good_tickets[good_tickets['Assignment group'] != 'GRP_0']\n\n# Join all the 3 datasets\nticket = pd.concat([grp0_tickets, good_tickets, rare_tickets]).reset_index(drop=True)\n\n# Serialize the balanced dataset\nwith open('balanced_ticket.pkl','wb') as f:\n pickle.dump(ticket[['Summary','Assignment group']], f, pickle.HIGHEST_PROTOCOL)\n\nticket.head()",
"_____no_output_____"
],
[
"# Visualize the assignment groups distribution\nprint('\\033[1m#Unique groups remaining:\\033[0m', ticket['Assignment group'].nunique())\npd.DataFrame(ticket.groupby('Assignment group').size(),columns = ['Count']).reset_index().iplot(\n kind='pie', \n labels='Assignment group', \n values='Count', \n title='#Records by Assignment groups',\n pull=np.linspace(0,0.3,ticket['Assignment group'].nunique()))",
"\u001b[1m#Unique groups remaining:\u001b[0m 41\n"
]
],
[
[
"**Comments:**\n- It's evident from the pie chart above the dataset is nearly balanced which can be considered for model building.\n\n## <u>Model Building</u>\nLet's load the balanced dataset and Word2Vec model to generate word embbeddings and feed it into LSTM.\n\n### <u>RNN with LSTM networks</u>\nLong Short-Term Memory~(LSTM) was introduced by S. Hochreiter and J. Schmidhuber and developed by many research scientists.\n\nTo deal with these problems Long Short-Term Memory (LSTM) is a special type of RNN that preserves long term dependency in a more effective way compared to the basic RNNs. This is particularly useful to overcome vanishing gradient problem as LSTM uses multiple gates to carefully regulate the amount of information that will be allowed into each node state. The figure shows the basic cell of a LSTM model.\n\n\n\nLet's create another column of categorical datatype from Assignment groups. Let's write some generic methods for utilities and to plot evaluation metrics.",
"_____no_output_____"
]
],
[
[
"# A class that logs the time\nclass Timer():\n '''\n A generic class to log the time\n '''\n def __init__(self):\n self.start_ts = None\n def start(self):\n self.start_ts = time()\n def stop(self):\n return 'Time taken: %2fs' % (time()-self.start_ts)\n \ntimer = Timer()\n\n# A method that plots the Precision-Recall curve\ndef plot_prec_recall_vs_thresh(precisions, recalls, thresholds):\n plt.figure(figsize=(10,5))\n plt.plot(thresholds, precisions[:-1], 'b--', label='precision')\n plt.plot(thresholds, recalls[:-1], 'g--', label = 'recall')\n plt.xlabel('Threshold')\n plt.legend()\n\n# A method to train and test the model\ndef run_classification(estimator, X_train, X_test, y_train, y_test, arch_name=None, pipelineRequired=True, isDeepModel=False):\n timer.start()\n # train the model\n clf = estimator\n\n if pipelineRequired :\n clf = Pipeline([('vect', CountVectorizer()),\n ('tfidf', TfidfTransformer()),\n ('clf', estimator),\n ])\n \n if isDeepModel :\n clf.fit(X_train, y_train, validation_data=(X_test, y_test),epochs=10, batch_size=128,verbose=1,callbacks=call_backs(arch_name))\n # predict from the claffier\n y_pred = clf.predict(X_test)\n y_pred = np.argmax(y_pred, axis=1)\n y_train_pred = clf.predict(X_train)\n y_train_pred = np.argmax(y_train_pred, axis=1)\n else :\n clf.fit(X_train, y_train)\n # predict from the claffier\n y_pred = clf.predict(X_test)\n y_train_pred = clf.predict(X_train)\n \n print('Estimator:', clf)\n print('='*80)\n print('Training accuracy: %.2f%%' % (accuracy_score(y_train,y_train_pred) * 100))\n print('Testing accuracy: %.2f%%' % (accuracy_score(y_test, y_pred) * 100))\n print('='*80)\n print('Confusion matrix:\\n %s' % (confusion_matrix(y_test, y_pred)))\n print('='*80)\n print('Classification report:\\n %s' % (classification_report(y_test, y_pred)))\n print(timer.stop(), 'to run the model')",
"_____no_output_____"
],
[
"# Load the balanced dataset\nwith open('balanced_ticket.pkl','rb') as f:\n ticket = pickle.load(f)\n# Load the Word2Vec model\nwmodel = Doc2Vec.load('d2v_inc_model.mdl')",
"_____no_output_____"
],
[
"w2v_weights = wmodel.wv.vectors\nvocab_size, embedding_size = w2v_weights.shape\nprint(\"Vocabulary Size: {} - Embedding Dim: {}\".format(vocab_size, embedding_size))",
"Vocabulary Size: 19400 - Embedding Dim: 300\n"
],
[
"# Sequences will be padded or truncated to this length\nMAX_SEQUENCE_LENGTH = 75\n\n# Prepare the embbedings with 0's padding to max sequence length\nX = ticket.Summary.values.tolist()\nset_X=[]\nfor sent in X:\n #print(sent[0])\n set_X.append(np.array([word2token(w) for w in tokenize_sentences(sent[0])[:MAX_SEQUENCE_LENGTH] if w != '']))\n\nset_X = pad_sequences(set_X, maxlen=MAX_SEQUENCE_LENGTH, padding='pre', value=0)\ny = pd.get_dummies(upsmpl_dset['group']).values\nprint('Shape of label Y:', (27470, 41))\nprint('Shape of label X:', (27470, 75))",
"Shape of label Y: (27470, 41)\nShape of label X: (27470, 75)\n"
],
[
"# Divide the original dataset into train and test split\nX_train, X_test, y_train, y_test = train_test_split(set_X, y, test_size=0.3, random_state=47)\nX_train.shape, X_test.shape, y_train.shape, y_test.shape",
"(19229, 75) (8241, 75) (19229, 41) (8241, 41)\n"
],
[
"# Visualize a random training sample\nX_train[67]",
"_____no_output_____"
],
[
"# CREATE the MODEL\n\n# Samples of categories with less than this number of samples will be ignored\nDROP_THRESHOLD = 10000\nmodel_seq = Sequential()\nmodel_seq.add(Embedding(input_dim=vocab_size, \n output_dim=embedding_size, \n weights=[w2v_weights], \n input_length=MAX_SEQUENCE_LENGTH, \n mask_zero=True,\n trainable=False))\nmodel_seq.add(SpatialDropout1D(0.2))\nmodel_seq.add(LSTM(100, dropout=0.2, recurrent_dropout=0.2))\nmodel_seq.add(Dense(41, activation='softmax'))\nmodel_seq.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nepochs = 20\nbatch_size = 64\n\nhistory = model_seq.fit(X_train, \n Y_train, \n epochs=epochs, \n batch_size=batch_size,\n validation_split=0.1,\n callbacks=[EarlyStopping(monitor='val_loss', patience=3, min_delta=0.0001)])\n",
"WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:541: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:4432: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:190: The name tf.get_default_session is deprecated. Please use tf.compat.v1.get_default_session instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:197: The name tf.ConfigProto is deprecated. Please use tf.compat.v1.ConfigProto instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:203: The name tf.Session is deprecated. Please use tf.compat.v1.Session instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:207: The name tf.global_variables is deprecated. Please use tf.compat.v1.global_variables instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:216: The name tf.is_variable_initialized is deprecated. Please use tf.compat.v1.is_variable_initialized instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:223: The name tf.variables_initializer is deprecated. Please use tf.compat.v1.variables_initializer instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:148: The name tf.placeholder_with_default is deprecated. Please use tf.compat.v1.placeholder_with_default instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:3733: calling dropout (from tensorflow.python.ops.nn_ops) with keep_prob is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:3239: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/optimizers.py:793: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:3576: The name tf.log is deprecated. Please use tf.math.log instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:1033: The name tf.assign_add is deprecated. Please use tf.compat.v1.assign_add instead.\n\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/keras/backend/tensorflow_backend.py:1020: The name tf.assign is deprecated. Please use tf.compat.v1.assign instead.\n\nTrain on 17306 samples, validate on 1923 samples\nEpoch 1/20\n17306/17306 [==============================] - 71s 4ms/step - loss: 2.7391 - acc: 0.2600 - val_loss: 1.8067 - val_acc: 0.4893\nEpoch 2/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 1.6989 - acc: 0.5259 - val_loss: 1.2630 - val_acc: 0.6453\nEpoch 3/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 1.2872 - acc: 0.6424 - val_loss: 1.0008 - val_acc: 0.7150\nEpoch 4/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 1.0489 - acc: 0.7076 - val_loss: 0.8288 - val_acc: 0.7577\nEpoch 5/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.8877 - acc: 0.7556 - val_loss: 0.7168 - val_acc: 0.7894\nEpoch 6/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.7869 - acc: 0.7783 - val_loss: 0.6305 - val_acc: 0.8216\nEpoch 7/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.7030 - acc: 0.8004 - val_loss: 0.5709 - val_acc: 0.8263\nEpoch 8/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.6372 - acc: 0.8160 - val_loss: 0.5208 - val_acc: 0.8461\nEpoch 9/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.5823 - acc: 0.8320 - val_loss: 0.4781 - val_acc: 0.8549\nEpoch 10/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.5367 - acc: 0.8436 - val_loss: 0.4431 - val_acc: 0.8684\nEpoch 11/20\n17306/17306 [==============================] - 66s 4ms/step - loss: 0.5025 - acc: 0.8521 - val_loss: 0.4179 - val_acc: 0.8783\nEpoch 12/20\n17306/17306 [==============================] - 66s 4ms/step - loss: 0.4677 - acc: 0.8601 - val_loss: 0.3887 - val_acc: 0.8856\nEpoch 13/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.4353 - acc: 0.8709 - val_loss: 0.3816 - val_acc: 0.8747\nEpoch 14/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.4210 - acc: 0.8737 - val_loss: 0.3483 - val_acc: 0.8965\nEpoch 15/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.4004 - acc: 0.8802 - val_loss: 0.3453 - val_acc: 0.9012\nEpoch 16/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.3892 - acc: 0.8844 - val_loss: 0.3309 - val_acc: 0.8981\nEpoch 17/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.3626 - acc: 0.8892 - val_loss: 0.3243 - val_acc: 0.9028\nEpoch 18/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.3525 - acc: 0.8916 - val_loss: 0.3096 - val_acc: 0.9100\nEpoch 19/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.3394 - acc: 0.8948 - val_loss: 0.3087 - val_acc: 0.9085\nEpoch 20/20\n17306/17306 [==============================] - 66s 4ms/step - loss: 0.3217 - acc: 0.9036 - val_loss: 0.3074 - val_acc: 0.9043\n"
],
[
"## Iteration 1 ...changing the dropout value\n\nmodel_seq = Sequential()\nmodel_seq.add(Embedding(input_dim=vocab_size, \n output_dim=embedding_size, \n weights=[w2v_weights], \n input_length=MAX_SEQUENCE_LENGTH, \n mask_zero=True,\n trainable=False))\nmodel_seq.add(SpatialDropout1D(0.1))\nmodel_seq.add(LSTM(100, dropout=0.1, recurrent_dropout=0.1))\nmodel_seq.add(Dense(41, activation='softmax'))\nmodel_seq.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nhistory = model_seq.fit(X_train, \n Y_train, \n epochs=epochs, \n batch_size=batch_size,\n validation_split=0.1,\n callbacks=[EarlyStopping(monitor='val_loss', patience=3, min_delta=0.0001)])",
"Train on 17306 samples, validate on 1923 samples\nEpoch 1/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 2.4506 - acc: 0.3459 - val_loss: 1.5717 - val_acc: 0.5757\nEpoch 2/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 1.3551 - acc: 0.6298 - val_loss: 1.0543 - val_acc: 0.7150\nEpoch 3/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.9840 - acc: 0.7384 - val_loss: 0.7932 - val_acc: 0.7920\nEpoch 4/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.7751 - acc: 0.7847 - val_loss: 0.6645 - val_acc: 0.8185\nEpoch 5/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.6358 - acc: 0.8242 - val_loss: 0.5489 - val_acc: 0.8393\nEpoch 6/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.5429 - acc: 0.8473 - val_loss: 0.5011 - val_acc: 0.8601\nEpoch 7/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.4804 - acc: 0.8630 - val_loss: 0.4168 - val_acc: 0.8799\nEpoch 8/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.4222 - acc: 0.8803 - val_loss: 0.3849 - val_acc: 0.8856\nEpoch 9/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.3868 - acc: 0.8877 - val_loss: 0.3563 - val_acc: 0.8913\nEpoch 10/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.3547 - acc: 0.8960 - val_loss: 0.3427 - val_acc: 0.8991\nEpoch 11/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.3214 - acc: 0.9077 - val_loss: 0.3220 - val_acc: 0.9048\nEpoch 12/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.3049 - acc: 0.9098 - val_loss: 0.2978 - val_acc: 0.9059\nEpoch 13/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.2796 - acc: 0.9177 - val_loss: 0.2876 - val_acc: 0.9137\nEpoch 14/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.2648 - acc: 0.9236 - val_loss: 0.2853 - val_acc: 0.9059\nEpoch 15/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.2531 - acc: 0.9246 - val_loss: 0.2784 - val_acc: 0.9069\nEpoch 16/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.2337 - acc: 0.9291 - val_loss: 0.2719 - val_acc: 0.9100\nEpoch 17/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.2279 - acc: 0.9298 - val_loss: 0.2550 - val_acc: 0.9210\nEpoch 18/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.2209 - acc: 0.9332 - val_loss: 0.2649 - val_acc: 0.9210\nEpoch 19/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 0.2115 - acc: 0.9341 - val_loss: 0.2522 - val_acc: 0.9256\nEpoch 20/20\n17306/17306 [==============================] - 66s 4ms/step - loss: 0.2023 - acc: 0.9350 - val_loss: 0.2434 - val_acc: 0.9308\n"
],
[
"## Iteration 2 ..adding more core to LTSM\n\nmodel_seq = Sequential()\nmodel_seq.add(Embedding(input_dim=vocab_size, \n output_dim=embedding_size, \n weights=[w2v_weights], \n input_length=MAX_SEQUENCE_LENGTH, \n mask_zero=True,\n trainable=False))\nmodel_seq.add(SpatialDropout1D(0.1))\nmodel_seq.add(LSTM(150, dropout=0.1, recurrent_dropout=0.1))\nmodel_seq.add(Dense(41, activation='softmax'))\nmodel_seq.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nhistory = model_seq.fit(X_train, \n Y_train, \n epochs=epochs, \n batch_size=batch_size,\n validation_split=0.1,\n callbacks=[EarlyStopping(monitor='val_loss', patience=3, min_delta=0.0001)])",
"Train on 17306 samples, validate on 1923 samples\nEpoch 1/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 2.2320 - acc: 0.3925 - val_loss: 1.3308 - val_acc: 0.6365\nEpoch 2/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 1.1154 - acc: 0.6917 - val_loss: 0.7884 - val_acc: 0.7826\nEpoch 3/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.7616 - acc: 0.7887 - val_loss: 0.5929 - val_acc: 0.8310\nEpoch 4/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.5980 - acc: 0.8329 - val_loss: 0.4852 - val_acc: 0.8492\nEpoch 5/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.4662 - acc: 0.8706 - val_loss: 0.4057 - val_acc: 0.8721\nEpoch 6/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.3935 - acc: 0.8880 - val_loss: 0.3604 - val_acc: 0.8970\nEpoch 7/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.3435 - acc: 0.9000 - val_loss: 0.3185 - val_acc: 0.8965\nEpoch 8/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.3005 - acc: 0.9125 - val_loss: 0.3007 - val_acc: 0.9038\nEpoch 9/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.2659 - acc: 0.9227 - val_loss: 0.2854 - val_acc: 0.9090\nEpoch 10/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.2436 - acc: 0.9284 - val_loss: 0.2743 - val_acc: 0.9095\nEpoch 11/20\n17306/17306 [==============================] - 71s 4ms/step - loss: 0.2261 - acc: 0.9303 - val_loss: 0.2500 - val_acc: 0.9256\nEpoch 12/20\n17306/17306 [==============================] - 72s 4ms/step - loss: 0.2108 - acc: 0.9365 - val_loss: 0.2508 - val_acc: 0.9272\nEpoch 13/20\n17306/17306 [==============================] - 71s 4ms/step - loss: 0.1933 - acc: 0.9410 - val_loss: 0.2474 - val_acc: 0.9225\nEpoch 14/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1887 - acc: 0.9401 - val_loss: 0.2359 - val_acc: 0.9288\nEpoch 15/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1724 - acc: 0.9449 - val_loss: 0.2413 - val_acc: 0.9324\nEpoch 16/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.1670 - acc: 0.9473 - val_loss: 0.2311 - val_acc: 0.9324\nEpoch 17/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.1642 - acc: 0.9466 - val_loss: 0.2288 - val_acc: 0.9303\nEpoch 18/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.1619 - acc: 0.9476 - val_loss: 0.2313 - val_acc: 0.9314\nEpoch 19/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1528 - acc: 0.9497 - val_loss: 0.2259 - val_acc: 0.9314\nEpoch 20/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1509 - acc: 0.9493 - val_loss: 0.2269 - val_acc: 0.9246\n"
]
],
[
[
"#### Finding Confidence Interval\nAs this iteration is having more accuracy and no overfitting, let's find out the confidence interval.",
"_____no_output_____"
]
],
[
[
"acc = history.history['acc']\nplt.figure(figsize=(10,7), dpi= 80)\nsns.distplot(acc, color=\"dodgerblue\", label=\"Compact\")\naccr = model_seq.evaluate(X_test,Y_test)\nprint('Test set\\n Loss: {:0.3f}\\n Accuracy: {:0.3f}'.format(accr[0],accr[1]*100))",
"8241/8241 [==============================] - 20s 2ms/step\n Test set\n Loss: 0.265\n Accuracy: 92.768\n"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"accuracy=0.9276\nn = 8241\ninterval = 1.96 * np.sqrt( (accuracy * (1 - accuracy)) / n)\nprint(interval*100)",
"0.5595201963469515\n"
]
],
[
[
"**Observation**:\n- There is a 95% likelihood that the confidence interval [92.21, 93.31] covers the true classification of the model on unseen data.",
"_____no_output_____"
]
],
[
[
"## Iteration 3 ....adding a dense and dropout and batchNormalistaion layer\n\nmodel_seq = Sequential()\nmodel_seq.add(Embedding(input_dim=vocab_size, \n output_dim=embedding_size, \n weights=[w2v_weights], \n input_length=MAX_SEQUENCE_LENGTH, \n mask_zero=True,\n trainable=False))\nmodel_seq.add(SpatialDropout1D(0.1))\nmodel_seq.add(LSTM(150, dropout=0.1, recurrent_dropout=0.1))\nmodel_seq.add(Dense(150, activation='relu'))\nmodel_seq.add(BatchNormalization(momentum=0.9,epsilon=0.02))\nmodel_seq.add(Dropout(0.1))\nmodel_seq.add(Dense(41, activation='softmax'))\nmodel_seq.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])\n\nhistory = model_seq.fit(X_train, \n Y_train, \n epochs=epochs, \n batch_size=batch_size,\n validation_split=0.1,\n callbacks=[EarlyStopping(monitor='val_loss', patience=3, min_delta=0.0001)])",
"Train on 17306 samples, validate on 1923 samples\nEpoch 1/20\n17306/17306 [==============================] - 71s 4ms/step - loss: 2.3474 - acc: 0.3646 - val_loss: 1.3097 - val_acc: 0.6495\nEpoch 2/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 1.1205 - acc: 0.6874 - val_loss: 0.7741 - val_acc: 0.7780\nEpoch 3/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.7102 - acc: 0.7980 - val_loss: 0.5141 - val_acc: 0.8461\nEpoch 4/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.5083 - acc: 0.8520 - val_loss: 0.4037 - val_acc: 0.8820\nEpoch 5/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.3922 - acc: 0.8822 - val_loss: 0.3437 - val_acc: 0.9017\nEpoch 6/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.3192 - acc: 0.9032 - val_loss: 0.3259 - val_acc: 0.8970\nEpoch 7/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.2799 - acc: 0.9118 - val_loss: 0.2821 - val_acc: 0.9100\nEpoch 8/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.2446 - acc: 0.9223 - val_loss: 0.2820 - val_acc: 0.9111\nEpoch 9/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.2292 - acc: 0.9255 - val_loss: 0.2788 - val_acc: 0.9225\nEpoch 10/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.2104 - acc: 0.9309 - val_loss: 0.2573 - val_acc: 0.9262\nEpoch 11/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1971 - acc: 0.9352 - val_loss: 0.2676 - val_acc: 0.9225\nEpoch 12/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1875 - acc: 0.9379 - val_loss: 0.2547 - val_acc: 0.9204\nEpoch 13/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.1803 - acc: 0.9411 - val_loss: 0.2624 - val_acc: 0.9272\nEpoch 14/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1726 - acc: 0.9444 - val_loss: 0.2500 - val_acc: 0.9272\nEpoch 15/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.1689 - acc: 0.9434 - val_loss: 0.2570 - val_acc: 0.9288\nEpoch 16/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.1643 - acc: 0.9444 - val_loss: 0.2843 - val_acc: 0.9199\nEpoch 17/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1722 - acc: 0.9422 - val_loss: 0.2800 - val_acc: 0.9241\n"
],
[
"## iteration 4 ...optimizing adam\nfrom keras.optimizers import Adam\n\nadam = Adam(lr=0.001, beta_1=0.9, beta_2=0.999, epsilon=1e-08, decay=0.0, amsgrad=False)\n\nmodel_seq = Sequential()\nmodel_seq.add(Embedding(input_dim=vocab_size, \n output_dim=embedding_size, \n weights=[w2v_weights], \n input_length=MAX_SEQUENCE_LENGTH, \n mask_zero=True,\n trainable=False))\nmodel_seq.add(SpatialDropout1D(0.1))\nmodel_seq.add(LSTM(150, dropout=0.1, recurrent_dropout=0.1))\nmodel_seq.add(Dense(150, activation='relu'))\nmodel_seq.add(BatchNormalization(momentum=0.9,epsilon=0.02))\nmodel_seq.add(Dropout(0.1))\nmodel_seq.add(Dense(41, activation='softmax'))\nmodel_seq.compile(loss='categorical_crossentropy', optimizer=adam, metrics=['accuracy'])\n\nhistory = model_seq.fit(X_train, \n Y_train, \n epochs=epochs, \n batch_size=batch_size,\n validation_split=0.1,\n callbacks=[EarlyStopping(monitor='val_loss', patience=3, min_delta=0.0001)])",
"Train on 17306 samples, validate on 1923 samples\nEpoch 1/20\n17306/17306 [==============================] - 72s 4ms/step - loss: 2.3593 - acc: 0.3639 - val_loss: 1.3768 - val_acc: 0.6355\nEpoch 2/20\n17306/17306 [==============================] - 67s 4ms/step - loss: 1.1354 - acc: 0.6848 - val_loss: 0.7751 - val_acc: 0.7842\nEpoch 3/20\n17306/17306 [==============================] - 68s 4ms/step - loss: 0.7054 - acc: 0.7992 - val_loss: 0.5247 - val_acc: 0.8482\nEpoch 4/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.5034 - acc: 0.8542 - val_loss: 0.4234 - val_acc: 0.8762\nEpoch 5/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.3971 - acc: 0.8824 - val_loss: 0.3549 - val_acc: 0.8934\nEpoch 6/20\n17306/17306 [==============================] - 71s 4ms/step - loss: 0.3281 - acc: 0.8978 - val_loss: 0.3206 - val_acc: 0.9100\nEpoch 7/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.2791 - acc: 0.9123 - val_loss: 0.2898 - val_acc: 0.9126\nEpoch 8/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.2475 - acc: 0.9207 - val_loss: 0.3115 - val_acc: 0.9142\nEpoch 9/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.2281 - acc: 0.9264 - val_loss: 0.2908 - val_acc: 0.9210\nEpoch 10/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.2093 - acc: 0.9316 - val_loss: 0.2688 - val_acc: 0.9262\nEpoch 11/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1924 - acc: 0.9361 - val_loss: 0.2657 - val_acc: 0.9236\nEpoch 12/20\n17306/17306 [==============================] - 70s 4ms/step - loss: 0.1948 - acc: 0.9356 - val_loss: 0.2895 - val_acc: 0.9173\nEpoch 13/20\n17306/17306 [==============================] - 71s 4ms/step - loss: 0.1829 - acc: 0.9401 - val_loss: 0.2876 - val_acc: 0.9298\nEpoch 14/20\n17306/17306 [==============================] - 69s 4ms/step - loss: 0.1766 - acc: 0.9415 - val_loss: 0.2804 - val_acc: 0.9288\n"
],
[
"accr = model_seq.evaluate(X_test,Y_test)\nprint('Test set\\n Loss: {:0.3f}\\n Accuracy: {:0.3f}'.format(accr[0],accr[1]*100))",
"8241/8241 [==============================] - 23s 3ms/step\nTest set\n Loss: 0.318\n Accuracy: 92.549\n"
],
[
"# Data Visualization\nimport matplotlib.pyplot as plt\n\nplt.title('Loss')\nplt.plot(history.history['loss'], label='train')\nplt.plot(history.history['val_loss'], label='validation')\nplt.legend()\nplt.show();",
"_____no_output_____"
],
[
"plt.title('Accuracy')\nplt.plot(history.history['acc'], label='train')\nplt.plot(history.history['val_acc'], label='validation')\nplt.legend()\nplt.show();",
"_____no_output_____"
]
],
[
[
"### Summary\nThe accuracy of each flavors of LSTM model is as follows in the table. This is clear indicative of how LSTM, in the family of RNN is efficient of dealing with textual data.\n- We've been able to bump up the model performance upto the range 92.21 to 93.31 with 95% confidence level.\n- Making the dataset balanced, helped the model to be trained more accurately.\n- Creating our own word embbeddings helped finding better representation of keywords of our corpus.\n- Hyperparameter tuning resulted in finding the model with more accuracy without overfitting, which is evident from the train vs. validation accuracy curve.\n\n\n### Automation of Ticket Assignment has following benefits: -\n1.\tIncrease in Customer Satisfaction.\n2.\tDecrease in the response and resolution time.\n3.\tEliminate human error in Ticket Assignment. (Which was ~25% Incidents)\n4.\tAvoid missing SLAs due to error in Ticket Assignment.\n5.\tEliminate any Financial penalty associated with missed SLAs.\n6.\tExcellent Customer Service.\n7.\tReallocate (~1 FTE) requirement for Productive Work. \n8.\tIncrease in morale of L1 / L2 Team.\n9.\tEradicate 15 mins Effort spent for SOP review (~25-30% of Incidents OR 531.25-637.5 Person Hours).\n10.\tDecrease in associated Expense.\n11.\tL1 / L2 Team can focus on resolving ~54% of the incidents\n12.\tFunctional / L3 teams can focus on resolving ~56% of incidents\n\n**~1 FTE from L1 / L2 Team saved through automating Ticket Assignment can focus on Continuous Improvement activities.\n~25% of Incidents which is 2125 additional Incidents will now get resolved within SLA.**\n\n### Additional Business Insights\n1. Root cause analysis (RCA) need to be performed on job_scheduler, to understand the cause of failure.\nNo. of Incident Ticket reduction expected by performing RCA:- 1928. \n22.68% of Total Incident volume of 8500.\nHence, we can reduce the Resource / FTE allocation also by approximately 22.68%.\n2. Password Rest process need to be automated.\nNo. of Incident Ticket reduction expected by automating password reset process:- 1246\n14.66% of Total Incident volume of 8500. \nHence, we can reduce the Resource / FTE allocation also by approximately 14.66%.\nHence a cumulative reduction of 3174 Incidents means 37.34% reduction in Total Incident volume of 8500. \nHence, cumulative Resource / FTE allocation reduction by approximately 37.34%.\nBusiness can operate at ~62.66% of original Estimates.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4ae5d9261bd9bd96809eb1baa4383685942e879a
| 234,422 |
ipynb
|
Jupyter Notebook
|
Q3/batch1/DeepLearning Part -2/Chapter7/7_1_Keras_functional_API.ipynb
|
aiwithqasim/PIAIC-Artificial-Intelligence
|
c4350727fe42c834eb119faf96543f419b098ac6
|
[
"MIT"
] | 1 |
2022-03-25T16:47:09.000Z
|
2022-03-25T16:47:09.000Z
|
Q3/batch1/DeepLearning Part -2/Chapter7/7_1_Keras_functional_API.ipynb
|
aiwithqasim/PIAIC-Artificial-Intelligence
|
c4350727fe42c834eb119faf96543f419b098ac6
|
[
"MIT"
] | null | null | null |
Q3/batch1/DeepLearning Part -2/Chapter7/7_1_Keras_functional_API.ipynb
|
aiwithqasim/PIAIC-Artificial-Intelligence
|
c4350727fe42c834eb119faf96543f419b098ac6
|
[
"MIT"
] | null | null | null | 118.096725 | 23,404 | 0.584945 |
[
[
[
"# Keras Functional API",
"_____no_output_____"
]
],
[
[
"# sudo pip3 install --ignore-installed --upgrade tensorflow\nimport keras\nimport tensorflow as tf\nprint(keras.__version__)\nprint(tf.__version__)\n# To ignore keep_dims warning\ntf.logging.set_verbosity(tf.logging.ERROR)",
"2.0.9\n1.8.0\n"
]
],
[
[
"Let’s start with a minimal example that shows side by side a simple Sequential model and its equivalent in the functional API:",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential, Model \nfrom keras import layers \nfrom keras import Input \n\nseq_model = Sequential()\nseq_model.add(layers.Dense(32, activation='relu', input_shape=(64,))) \nseq_model.add(layers.Dense(32, activation='relu'))\nseq_model.add(layers.Dense(10, activation='softmax'))\n\ninput_tensor = Input(shape=(64,))\nx = layers.Dense(32, activation='relu')(input_tensor)\nx = layers.Dense(32, activation='relu')(x)\noutput_tensor = layers.Dense(10, activation='softmax')(x)\n\nmodel = Model(input_tensor, output_tensor)",
"_____no_output_____"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_2 (InputLayer) (None, 64) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 32) 2080 \n_________________________________________________________________\ndense_6 (Dense) (None, 32) 1056 \n_________________________________________________________________\ndense_7 (Dense) (None, 10) 330 \n=================================================================\nTotal params: 3,466\nTrainable params: 3,466\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"The only part that may seem a bit magical at this point is instantiating a Model object using only an input tensor and an output tensor. Behind the scenes, Keras retrieves every layer involved in going from input_tensor to output_tensor, bringing them together into a graph-like data structure—a Model. Of course, the reason it works is that output_tensor was obtained by repeatedly transforming input_tensor. \n\nIf you tried to build a model from **inputs and outputs that weren’t related**, you’d get a RuntimeError:",
"_____no_output_____"
]
],
[
[
"unrelated_input = Input(shape=(32,))\nbad_model = Model(unrelated_input, output_tensor)",
"_____no_output_____"
]
],
[
[
"This error tells you, in essence, that Keras couldn’t reach input_2 from the provided output tensor. \n\nWhen it comes to compiling, training, or evaluating such an instance of Model, the API is *the same as that of Sequential*:",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='rmsprop', loss='categorical_crossentropy')\n\nimport numpy as np\nx_train = np.random.random((1000, 64))\ny_train = np.random.random((1000, 10)) \n\nmodel.fit(x_train, y_train, epochs=10, batch_size=128)\n\nscore = model.evaluate(x_train, y_train)",
"WARNING:tensorflow:From /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/keras/backend/tensorflow_backend.py:2857: calling reduce_sum (from tensorflow.python.ops.math_ops) with keep_dims is deprecated and will be removed in a future version.\nInstructions for updating:\nkeep_dims is deprecated, use keepdims instead\nWARNING:tensorflow:From /Library/Frameworks/Python.framework/Versions/3.6/lib/python3.6/site-packages/keras/backend/tensorflow_backend.py:1340: calling reduce_mean (from tensorflow.python.ops.math_ops) with keep_dims is deprecated and will be removed in a future version.\nInstructions for updating:\nkeep_dims is deprecated, use keepdims instead\nEpoch 1/10\n1000/1000 [==============================] - 0s 222us/step - loss: 11.6923\nEpoch 2/10\n1000/1000 [==============================] - 0s 18us/step - loss: 11.5943\nEpoch 3/10\n1000/1000 [==============================] - 0s 17us/step - loss: 11.5780\nEpoch 4/10\n1000/1000 [==============================] - 0s 16us/step - loss: 11.5710\nEpoch 5/10\n1000/1000 [==============================] - 0s 18us/step - loss: 11.5663\nEpoch 6/10\n1000/1000 [==============================] - 0s 19us/step - loss: 11.5621\nEpoch 7/10\n1000/1000 [==============================] - 0s 18us/step - loss: 11.5598\nEpoch 8/10\n1000/1000 [==============================] - 0s 18us/step - loss: 11.5562\nEpoch 9/10\n1000/1000 [==============================] - 0s 18us/step - loss: 11.5546\nEpoch 10/10\n1000/1000 [==============================] - 0s 18us/step - loss: 11.5529\n1000/1000 [==============================] - 0s 78us/step\n"
]
],
[
[
"## Multi-input models\n\n\n#### A question-answering model example\n\nFollowing is an example of how you can build such a model with the functional API. You set up two independent branches, encoding the text input and the question input as representation vectors; then, concatenate these vectors; and finally, add a softmax classifier on top of the concatenated representations.",
"_____no_output_____"
]
],
[
[
"from keras.models import Model\nfrom keras import layers\nfrom keras import Input\n\ntext_vocabulary_size = 10000\nquestion_vocabulary_size = 10000\nanswer_vocabulary_size = 500\n\n# The text input is a variable-length sequence of integers. \n# Note that you can optionally name the inputs.\ntext_input = Input(shape=(None,), dtype='int32', name='text')\n\n# Embeds the inputs into a sequence of vectors of size 64\n# embedded_text = layers.Embedding(64, text_vocabulary_size)(text_input)\n# embedded_text = layers.Embedding(output_dim=64, input_dim=text_vocabulary_size)(text_input)\nembedded_text = layers.Embedding(text_vocabulary_size,64)(text_input)\n\n# Encodes the vectors in a single vector via an LSTM\nencoded_text = layers.LSTM(32)(embedded_text)\n\n\n# Same process (with different layer instances) for the question\nquestion_input = Input(shape=(None,),dtype='int32',name='question')\n\n# embedded_question = layers.Embedding(32, question_vocabulary_size)(question_input)\n# embedded_question = layers.Embedding(output_dim=32, input_dim=question_vocabulary_size)(question_input)\nembedded_question = layers.Embedding(question_vocabulary_size,32)(question_input)\n\nencoded_question = layers.LSTM(16)(embedded_question) \n\n# Concatenates the encoded question and encoded text\nconcatenated = layers.concatenate([encoded_text, encoded_question],axis=-1)\n\n# Adds a softmax classifier on top\nanswer = layers.Dense(answer_vocabulary_size, activation='softmax')(concatenated)\n\n# At model instantiation, you specify the two inputs and the output.\nmodel = Model([text_input, question_input], answer)\nmodel.compile(optimizer='rmsprop',loss='categorical_crossentropy',metrics=['acc'])",
"_____no_output_____"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ntext (InputLayer) (None, None) 0 \n__________________________________________________________________________________________________\nquestion (InputLayer) (None, None) 0 \n__________________________________________________________________________________________________\nembedding_3 (Embedding) (None, None, 64) 640000 text[0][0] \n__________________________________________________________________________________________________\nembedding_4 (Embedding) (None, None, 32) 320000 question[0][0] \n__________________________________________________________________________________________________\nlstm_3 (LSTM) (None, 32) 12416 embedding_3[0][0] \n__________________________________________________________________________________________________\nlstm_4 (LSTM) (None, 16) 3136 embedding_4[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 48) 0 lstm_3[0][0] \n lstm_4[0][0] \n__________________________________________________________________________________________________\ndense_9 (Dense) (None, 500) 24500 concatenate_2[0][0] \n==================================================================================================\nTotal params: 1,000,052\nTrainable params: 1,000,052\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"Now, how do you **train** this two-input model? \n\nThere are two possible APIs: \n* you can feed the model a list of Numpy arrays as inputs\n* you can feed it a dictionary that maps input names to Numpy arrays. \n\nNaturally, the latter option is available only if you give names to your inputs. \n\n#### Training the multi-input model",
"_____no_output_____"
]
],
[
[
"import numpy as np\nnum_samples = 1000 \nmax_length = 100\n\n# Generates dummy Numpy data\ntext = np.random.randint(1, text_vocabulary_size,size=(num_samples, max_length))\nquestion = np.random.randint(1, question_vocabulary_size,size=(num_samples, max_length)) \n\n# Answers are one-hot encoded, not integers\n# answers = np.random.randint(0, 1,size=(num_samples, answer_vocabulary_size))\nanswers = np.random.randint(answer_vocabulary_size, size=(num_samples))\nanswers = keras.utils.to_categorical(answers, answer_vocabulary_size)\n\n# Fitting using a list of inputs\nprint('-'*10,\"First training run with list of NumPy arrays\",'-'*60)\n\nmodel.fit([text, question], answers, epochs=10, batch_size=128)\n\nprint()\n\n# Fitting using a dictionary of inputs (only if inputs are named)\nprint('-'*10,\"Second training run with dictionary and named inputs\",'-'*60)\n\nmodel.fit({'text': text, 'question': question}, answers,epochs=10, batch_size=128)",
"_____no_output_____"
]
],
[
[
"## Multi-output models\nYou can also use the functional API to build models with multiple outputs (or multiple *heads*). \n\n#### Example - prediction of Age, Gender and Income from social media posts\nA simple example is a network that attempts to simultaneously predict different properties of the data, such as a network that takes as input a series of social media posts from a single anonymous person and tries to predict attributes of that person, such as age, gender, and income level.\n",
"_____no_output_____"
]
],
[
[
"from keras import layers\nfrom keras import Input \nfrom keras.models import Model \n\nvocabulary_size = 50000 \nnum_income_groups = 10 \n\nposts_input = Input(shape=(None,), dtype='int32', name='posts')\n\n#embedded_posts = layers.Embedding(256, vocabulary_size)(posts_input) \nembedded_posts = layers.Embedding(vocabulary_size,256)(posts_input)\n\nx = layers.Conv1D(128, 5, activation='relu', padding='same')(embedded_posts)\nx = layers.MaxPooling1D(5)(x)\nx = layers.Conv1D(256, 5, activation='relu', padding='same')(x)\nx = layers.Conv1D(256, 5, activation='relu', padding='same')(x)\nx = layers.MaxPooling1D(5)(x)\nx = layers.Conv1D(256, 5, activation='relu', padding='same')(x)\nx = layers.Conv1D(256, 5, activation='relu', padding='same')(x) \nx = layers.GlobalMaxPooling1D()(x)\nx = layers.Dense(128, activation='relu')(x) \n\n# Note that the output layers are given names.\n\nage_prediction = layers.Dense(1, name='age')(x)\n\nincome_prediction = layers.Dense(num_income_groups, activation='softmax',name='income')(x)\n\ngender_prediction = layers.Dense(1, activation='sigmoid', name='gender')(x)\n\nmodel = Model(posts_input,[age_prediction, income_prediction, gender_prediction])\n\nprint(\"Model is ready!\")",
"Model is ready!\n"
]
],
[
[
"#### Compilation options of a multi-output model: multiple losses",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='rmsprop', loss=['mse', 'categorical_crossentropy', 'binary_crossentropy'])\n\n# Equivalent (possible only if you give names to the output layers)\nmodel.compile(optimizer='rmsprop',loss={'age': 'mse',\n 'income': 'categorical_crossentropy',\n 'gender': 'binary_crossentropy'})",
"_____no_output_____"
],
[
"model.compile(optimizer='rmsprop',\n loss=['mse', 'categorical_crossentropy', 'binary_crossentropy'],\n loss_weights=[0.25, 1., 10.]) \n\n# Equivalent (possible only if you give names to the output layers)\nmodel.compile(optimizer='rmsprop',\n loss={'age': 'mse','income': 'categorical_crossentropy','gender': 'binary_crossentropy'},\n loss_weights={'age': 0.25,\n 'income': 1.,\n 'gender': 10.})",
"_____no_output_____"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nposts (InputLayer) (None, None) 0 \n__________________________________________________________________________________________________\nembedding_5 (Embedding) (None, None, 256) 12800000 posts[0][0] \n__________________________________________________________________________________________________\nconv1d_1 (Conv1D) (None, None, 128) 163968 embedding_5[0][0] \n__________________________________________________________________________________________________\nmax_pooling1d_1 (MaxPooling1D) (None, None, 128) 0 conv1d_1[0][0] \n__________________________________________________________________________________________________\nconv1d_2 (Conv1D) (None, None, 256) 164096 max_pooling1d_1[0][0] \n__________________________________________________________________________________________________\nconv1d_3 (Conv1D) (None, None, 256) 327936 conv1d_2[0][0] \n__________________________________________________________________________________________________\nmax_pooling1d_2 (MaxPooling1D) (None, None, 256) 0 conv1d_3[0][0] \n__________________________________________________________________________________________________\nconv1d_4 (Conv1D) (None, None, 256) 327936 max_pooling1d_2[0][0] \n__________________________________________________________________________________________________\nconv1d_5 (Conv1D) (None, None, 256) 327936 conv1d_4[0][0] \n__________________________________________________________________________________________________\nglobal_max_pooling1d_1 (GlobalM (None, 256) 0 conv1d_5[0][0] \n__________________________________________________________________________________________________\ndense_10 (Dense) (None, 128) 32896 global_max_pooling1d_1[0][0] \n__________________________________________________________________________________________________\nage (Dense) (None, 1) 129 dense_10[0][0] \n__________________________________________________________________________________________________\nincome (Dense) (None, 10) 1290 dense_10[0][0] \n__________________________________________________________________________________________________\ngender (Dense) (None, 1) 129 dense_10[0][0] \n==================================================================================================\nTotal params: 14,146,316\nTrainable params: 14,146,316\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"#### Feeding data to a multi-output model\n\nMuch as in the case of multi-input models, you can pass Numpy data to the model for training either via a list of arrays or via a dictionary of arrays.",
"_____no_output_____"
],
[
"#### Training a multi-output model",
"_____no_output_____"
]
],
[
[
"import numpy as np \n\nTRACE = False\n\nnum_samples = 1000 \nmax_length = 100 \n\nposts = np.random.randint(1, vocabulary_size, size=(num_samples, max_length))\nif TRACE:\n print(\"*** POSTS ***\")\n print(posts.shape)\n print(posts[:10])\n print()\n\nage_targets = np.random.randint(0, 100, size=(num_samples,1))\nif TRACE:\n print(\"*** AGE ***\")\n print(age_targets.shape)\n print(age_targets[:10])\n print()\n\nincome_targets = np.random.randint(1, num_income_groups, size=(num_samples,1))\nincome_targets = keras.utils.to_categorical(income_targets,num_income_groups)\nif TRACE:\n print(\"*** INCOME ***\")\n print(income_targets.shape)\n print(income_targets[:10])\n print()\n\ngender_targets = np.random.randint(0, 2, size=(num_samples,1))\nif TRACE:\n print(\"*** GENDER ***\")\n print(gender_targets.shape)\n print(gender_targets[:10])\n print()\n\nprint('-'*10, \"First training run with NumPy arrays\", '-'*60)\n\n# age_targets, income_targets, and gender_targets are assumed to be Numpy arrays.\n\nmodel.fit(posts, [age_targets, income_targets, gender_targets], epochs=10, batch_size=64)\n\nprint('-'*10,\"Second training run with dictionary and named outputs\",'-'*60)\n\n# Equivalent (possible only if you give names to the output layers)\n\nmodel.fit(posts, {'age': age_targets,\n 'income': income_targets,\n 'gender': gender_targets},\n epochs=10, batch_size=64)",
"---------- First training run with NumPy arrays ------------------------------------------------------------\nEpoch 1/10\n1000/1000 [==============================] - 9s 9ms/step - loss: 2034.5990 - age_loss: 8066.3788 - income_loss: 4.0558 - gender_loss: 1.3948\nEpoch 2/10\n1000/1000 [==============================] - 9s 9ms/step - loss: 150.4753 - age_loss: 542.2556 - income_loss: 2.4038 - gender_loss: 1.2508\nEpoch 3/10\n1000/1000 [==============================] - 10s 10ms/step - loss: 163.5628 - age_loss: 575.2898 - income_loss: 2.6241 - gender_loss: 1.7116\nEpoch 4/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 402.7297 - age_loss: 1402.9028 - income_loss: 9.0451 - gender_loss: 4.2959\nEpoch 5/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 17855.3646 - age_loss: 71078.0439 - income_loss: 13.6580 - gender_loss: 7.2195\nEpoch 6/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 585917.7878 - age_loss: 2343302.9990 - income_loss: 14.3116 - gender_loss: 7.7740\nEpoch 7/10\n1000/1000 [==============================] - 10s 10ms/step - loss: 81165.1498 - age_loss: 324283.9278 - income_loss: 14.2973 - gender_loss: 7.9871\nEpoch 8/10\n1000/1000 [==============================] - 9s 9ms/step - loss: 43909.6058 - age_loss: 175262.1013 - income_loss: 14.2088 - gender_loss: 7.9871\nEpoch 9/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 1022139.5908 - age_loss: 4088181.4002 - income_loss: 14.3887 - gender_loss: 7.9871\nEpoch 10/10\n1000/1000 [==============================] - 9s 9ms/step - loss: 119479.2284 - age_loss: 477539.7231 - income_loss: 14.4289 - gender_loss: 7.9871\n---------- Second training run with dictionary and named outputs ------------------------------------------------------------\nEpoch 1/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 124047.4452 - age_loss: 495812.8410 - income_loss: 14.3610 - gender_loss: 7.9871\nEpoch 2/10\n1000/1000 [==============================] - 7s 7ms/step - loss: 40409.4534 - age_loss: 161260.9529 - income_loss: 14.3451 - gender_loss: 7.9871\nEpoch 3/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 2701707.6878 - age_loss: 10806453.8409 - income_loss: 14.4096 - gender_loss: 7.9871\nEpoch 4/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 326507.1074 - age_loss: 1305652.0114 - income_loss: 14.2323 - gender_loss: 7.9871\nEpoch 5/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 960.2075 - age_loss: 3463.2557 - income_loss: 14.5223 - gender_loss: 7.9871\nEpoch 6/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 1366666.2853 - age_loss: 5466288.3055 - income_loss: 14.3686 - gender_loss: 7.9871\nEpoch 7/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 936314.2207 - age_loss: 3744880.3891 - income_loss: 14.2731 - gender_loss: 7.9871\nEpoch 8/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 30479.6221 - age_loss: 121541.4295 - income_loss: 14.3935 - gender_loss: 7.9871\nEpoch 9/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 3721.0663 - age_loss: 14507.2060 - income_loss: 14.3935 - gender_loss: 7.9871\nEpoch 10/10\n1000/1000 [==============================] - 8s 8ms/step - loss: 6087.8961 - age_loss: 23974.5251 - income_loss: 14.3935 - gender_loss: 7.9871\n"
]
],
[
[
"### 7.1.4 Directed acyclic graphs of layers \n\nWith the functional API, not only can you build models with multiple inputs and multiple outputs, but you can also implement networks with a complex internal topology. \n\nNeural networks in Keras are allowed to be arbitrary directed acyclic graphs of layers (the only processing loops that are allowed are those internal to recurrent layers).\n\nSeveral common neural-network components are implemented as graphs. Two notable ones are <i>Inception modules</i> and <i>residual connections</i>. To better understand how the functional API can be used to build graphs of layers, let’s take a look at how you can implement both of them in Keras.",
"_____no_output_____"
],
[
"#### Inception modules \n\nInception [3] is a popular type of network architecture for convolutional neural networks. It consists of a stack of modules that themselves look like small independent networks, split into several parallel branches.\n\n##### The purpose of 1 × 1 convolutions \n\n1 × 1 convolutions (also called pointwise convolutions) are featured in Inception modules, where they contribute to factoring out channel-wise feature learning and space-wise feature learning.\n ",
"_____no_output_____"
]
],
[
[
"from keras import layers \nfrom keras.layers import Input\n\n# This example assumes the existence of a 4D input tensor x:\n# This returns a typical image tensor like those of MNIST dataset \nx = Input(shape=(28, 28, 1), dtype='float32', name='images')\nprint(\"x.shape:\",x.shape)\n\n# Every branch has the same stride value (2), which is necessary to \n# keep all branch outputs the same size so you can concatenate them\nbranch_a = layers.Conv2D(128, 1, padding='same', activation='relu', strides=2)(x)\n\n# In this branch, the striding occurs in the spatial convolution layer.\nbranch_b = layers.Conv2D(128, 1, padding='same', activation='relu')(x)\nbranch_b = layers.Conv2D(128, 3, padding='same', activation='relu', strides=2)(branch_b)\n\n# In this branch, the striding occurs in the average pooling layer.\nbranch_c = layers.AveragePooling2D(3, padding='same', strides=2)(x)\nbranch_c = layers.Conv2D(128, 3, padding='same', activation='relu')(branch_c)\n\nbranch_d = layers.Conv2D(128, 1, padding='same', activation='relu')(x) \nbranch_d = layers.Conv2D(128, 3, padding='same', activation='relu')(branch_d)\nbranch_d = layers.Conv2D(128, 3, padding='same', activation='relu', strides=2)(branch_d)\n\n# Concatenates the branch outputs to obtain the module output\noutput = layers.concatenate([branch_a, branch_b, branch_c, branch_d], axis=-1)\n\n# Adding a classifier on top of the convnet\noutput = layers.Flatten()(output)\noutput = layers.Dense(512, activation='relu')(output)\npredictions = layers.Dense(10, activation='softmax')(output)\n\nmodel = keras.models.Model(inputs=x, outputs=predictions)",
"x.shape: (?, 28, 28, 1)\n"
]
],
[
[
"#### Train the Inception model using the Dataset API and the MNIST data\n\nInspired by: https://github.com/keras-team/keras/blob/master/examples/mnist_dataset_api.py",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport os\nimport tempfile\n\nimport keras\nfrom keras import backend as K\nfrom keras import layers\nfrom keras.datasets import mnist\n\nimport tensorflow as tf\n\nif K.backend() != 'tensorflow':\n raise RuntimeError('This example can only run with the TensorFlow backend,'\n ' because it requires the Dataset API, which is not'\n ' supported on other platforms.')\n\nbatch_size = 128\nbuffer_size = 10000\nsteps_per_epoch = int(np.ceil(60000 / float(batch_size))) # = 469\nepochs = 5\nnum_classes = 10\n\ndef cnn_layers(x):\n \n # This example assumes the existence of a 4D input tensor x:\n # This returns a typical image tensor like those of MNIST dataset \n print(\"x.shape:\",x.shape)\n\n # Every branch has the same stride value (2), which is necessary to \n # keep all branch outputs the same size so you can concatenate them\n branch_a = layers.Conv2D(128, 1, padding='same', activation='relu', strides=2)(x)\n\n # In this branch, the striding occurs in the spatial convolution layer.\n branch_b = layers.Conv2D(128, 1, padding='same', activation='relu')(x)\n branch_b = layers.Conv2D(128, 3, padding='same', activation='relu', strides=2)(branch_b)\n\n # In this branch, the striding occurs in the average pooling layer.\n branch_c = layers.AveragePooling2D(3, padding='same', strides=2)(x)\n branch_c = layers.Conv2D(128, 3, padding='same', activation='relu')(branch_c)\n\n branch_d = layers.Conv2D(128, 1, padding='same', activation='relu')(x) \n branch_d = layers.Conv2D(128, 3, padding='same', activation='relu')(branch_d)\n branch_d = layers.Conv2D(128, 3, padding='same', activation='relu', strides=2)(branch_d)\n\n # Concatenates the branch outputs to obtain the module output\n output = layers.concatenate([branch_a, branch_b, branch_c, branch_d], axis=-1)\n\n # Adding a classifier on top of the convnet\n output = layers.Flatten()(output)\n output = layers.Dense(512, activation='relu')(output)\n predictions = layers.Dense(num_classes, activation='softmax')(output)\n \n return predictions\n\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\nx_train = x_train.astype(np.float32) / 255\nx_train = np.expand_dims(x_train, -1)\ny_train = tf.one_hot(y_train, num_classes)\n\n# Create the dataset and its associated one-shot iterator.\ndataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\ndataset = dataset.repeat()\ndataset = dataset.shuffle(buffer_size)\ndataset = dataset.batch(batch_size)\niterator = dataset.make_one_shot_iterator()\n\n# Model creation using tensors from the get_next() graph node.\ninputs, targets = iterator.get_next()\n\nprint(\"inputs.shape:\",inputs.shape)\nprint(\"targets.shape:\",targets.shape)\n\nmodel_input = layers.Input(tensor=inputs)\nmodel_output = cnn_layers(model_input)\n\nmodel = keras.models.Model(inputs=model_input, outputs=model_output)\n\nmodel.compile(optimizer=keras.optimizers.RMSprop(lr=2e-3, decay=1e-5),\n loss='categorical_crossentropy',\n metrics=['accuracy'],\n target_tensors=[targets])",
"inputs.shape: (?, 28, 28, 1)\ntargets.shape: (?, 10)\nx.shape: (?, 28, 28, 1)\n"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_6 (InputLayer) (None, 28, 28, 1) 0 \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 28, 28, 128) 256 input_6[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 28, 28, 128) 256 input_6[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_4 (AveragePoo (None, 14, 14, 1) 0 input_6[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 28, 28, 128) 147584 conv2d_26[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 14, 14, 128) 256 input_6[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 14, 14, 128) 147584 conv2d_23[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 14, 14, 128) 1280 average_pooling2d_4[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 14, 14, 128) 147584 conv2d_27[0][0] \n__________________________________________________________________________________________________\nconcatenate_6 (Concatenate) (None, 14, 14, 512) 0 conv2d_22[0][0] \n conv2d_24[0][0] \n conv2d_25[0][0] \n conv2d_28[0][0] \n__________________________________________________________________________________________________\nflatten_4 (Flatten) (None, 100352) 0 concatenate_6[0][0] \n__________________________________________________________________________________________________\ndense_17 (Dense) (None, 512) 51380736 flatten_4[0][0] \n__________________________________________________________________________________________________\ndense_18 (Dense) (None, 10) 5130 dense_17[0][0] \n==================================================================================================\nTotal params: 51,830,666\nTrainable params: 51,830,666\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"#### Train Inception model",
"_____no_output_____"
]
],
[
[
"model.fit(epochs=epochs,\n steps_per_epoch=steps_per_epoch)\n\n# Save the model weights.\nweight_path = os.path.join(tempfile.gettempdir(), 'saved_Inception_wt.h5')\nmodel.save_weights(weight_path)",
"Epoch 1/5\n469/469 [==============================] - 2079s 4s/step - loss: 0.0478 - acc: 0.9861\nEpoch 2/5\n469/469 [==============================] - 1916s 4s/step - loss: 0.0264 - acc: 0.9920\nEpoch 3/5\n469/469 [==============================] - 1843s 4s/step - loss: 0.0182 - acc: 0.9946\nEpoch 4/5\n469/469 [==============================] - 1825s 4s/step - loss: 0.0122 - acc: 0.9968\nEpoch 5/5\n469/469 [==============================] - 1828s 4s/step - loss: 0.0101 - acc: 0.9974\n"
]
],
[
[
"#### Test the Inception model\n\nSecond session to test loading trained model without tensors.",
"_____no_output_____"
]
],
[
[
"# Clean up the TF session.\nK.clear_session()\n\n# Second session to test loading trained model without tensors.\nx_test = x_test.astype(np.float32)\nx_test = np.expand_dims(x_test, -1)\n\nx_test_inp = layers.Input(shape=x_test.shape[1:])\ntest_out = cnn_layers(x_test_inp)\ntest_model = keras.models.Model(inputs=x_test_inp, outputs=test_out)\n\nweight_path = os.path.join(tempfile.gettempdir(), 'saved_Inception_wt.h5')\ntest_model.load_weights(weight_path)\n\ntest_model.compile(optimizer='rmsprop',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\ntest_model.summary()\n\nSVG(model_to_dot(test_model).create(prog='dot', format='svg'))\n\nloss, acc = test_model.evaluate(x_test, y_test, num_classes)\nprint('\\nTest accuracy: {0}'.format(acc))",
"x.shape: (?, 28, 28, 1)\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 28, 28, 1) 0 \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 28, 28, 128) 256 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 28, 28, 128) 256 input_1[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_1 (AveragePoo (None, 14, 14, 1) 0 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 28, 28, 128) 147584 conv2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 14, 14, 128) 256 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 14, 14, 128) 147584 conv2d_2[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 14, 14, 128) 1280 average_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 14, 14, 128) 147584 conv2d_6[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 14, 14, 512) 0 conv2d_1[0][0] \n conv2d_3[0][0] \n conv2d_4[0][0] \n conv2d_7[0][0] \n__________________________________________________________________________________________________\nflatten_1 (Flatten) (None, 100352) 0 concatenate_1[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 512) 51380736 flatten_1[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 10) 5130 dense_1[0][0] \n==================================================================================================\nTotal params: 51,830,666\nTrainable params: 51,830,666\nNon-trainable params: 0\n__________________________________________________________________________________________________\n10000/10000 [==============================] - 124s 12ms/step\n\nTest accuracy: 0.9828999968171119\n"
]
],
[
[
"#### Residual connections - ResNET\n\nResidual connections or ResNET are a common graph-like network component found in many post-2015 network architectures, including Xception. They were introduced by He et al. from Microsoft and are figthing two common problems with large-scale deep-learning model: vanishing gradients and representational bottlenecks. \n\nA residual connection consists of making the output of an earlier layer available as input to a later layer, effectively creating a shortcut in a sequential network. Rather than being concatenated to the later activation, the earlier output is summed with the later activation, which assumes that both activations are the same size. If they’re different sizes, you can use a linear transformation to reshape the earlier activation into the target shape (for example, a Dense layer without an activation or, for convolutional feature maps, a 1 × 1 convolution without an activation). \n\n###### ResNET implementation when the feature-map sizes are the same\n\nHere’s how to implement a residual connection in Keras when the feature-map sizes are the same, using identity residual connections. This example assumes the existence of a 4D input tensor x:",
"_____no_output_____"
]
],
[
[
"from keras import layers \nfrom keras.layers import Input\n\n# This example assumes the existence of a 4D input tensor x:\n# This returns a typical image tensor like those of MNIST dataset \nx = Input(shape=(28, 28, 1), dtype='float32', name='images')\nprint(\"x.shape:\",x.shape)\n\n# Applies a transformation to x\ny = layers.Conv2D(128, 3, activation='relu', padding='same')(x)\ny = layers.Conv2D(128, 3, activation='relu', padding='same')(y)\ny = layers.Conv2D(128, 3, activation='relu', padding='same')(y)\n\n# Adds the original x back to the output features\noutput = layers.add([y, x])\n\n# Adding a classifier on top of the convnet\noutput = layers.Flatten()(output)\noutput = layers.Dense(512, activation='relu')(output)\npredictions = layers.Dense(10, activation='softmax')(output)",
"x.shape: (?, 28, 28, 1)\n"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_6 (InputLayer) (None, 28, 28, 1) 0 \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 28, 28, 128) 256 input_6[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 28, 28, 128) 256 input_6[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d_4 (AveragePoo (None, 14, 14, 1) 0 input_6[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 28, 28, 128) 147584 conv2d_26[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 14, 14, 128) 256 input_6[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 14, 14, 128) 147584 conv2d_23[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 14, 14, 128) 1280 average_pooling2d_4[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 14, 14, 128) 147584 conv2d_27[0][0] \n__________________________________________________________________________________________________\nconcatenate_6 (Concatenate) (None, 14, 14, 512) 0 conv2d_22[0][0] \n conv2d_24[0][0] \n conv2d_25[0][0] \n conv2d_28[0][0] \n__________________________________________________________________________________________________\nflatten_4 (Flatten) (None, 100352) 0 concatenate_6[0][0] \n__________________________________________________________________________________________________\ndense_17 (Dense) (None, 512) 51380736 flatten_4[0][0] \n__________________________________________________________________________________________________\ndense_18 (Dense) (None, 10) 5130 dense_17[0][0] \n==================================================================================================\nTotal params: 51,830,666\nTrainable params: 51,830,666\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"###### ResNET implementation when the feature-map sizes differ\n\nAnd the following implements a residual connection when the feature-map sizes differ, using a linear residual connection (again, assuming the existence of a 4D input tensor x):",
"_____no_output_____"
]
],
[
[
"from keras import layers \nfrom keras.layers import Input\n\n# This example assumes the existence of a 4D input tensor x:\n# This returns a typical image tensor like those of MNIST dataset \nx = Input(shape=(28, 28, 1), dtype='float32', name='images')\nprint(\"x.shape:\",x.shape)\n\n# Applies a transformation to x\ny = layers.Conv2D(128, 3, activation='relu', padding='same')(x)\ny = layers.Conv2D(128, 3, activation='relu', padding='same')(y)\ny = layers.MaxPooling2D(2, strides=2)(y)\n\n# Uses a 1 × 1 convolution to linearly downsample the original x tensor to the same shape as y\nresidual = layers.Conv2D(128, 1, strides=2, padding='same')(x)\n\n# Adds the residual tensor back to the output features\noutput = layers.add([y, residual])\n\n# Adding a classifier on top of the convnet\noutput = layers.Flatten()(output)\noutput = layers.Dense(512, activation='relu')(output)\npredictions = layers.Dense(10, activation='softmax')(output)\nmodel = keras.models.Model(inputs=x, outputs=predictions)",
"x.shape: (?, 28, 28, 1)\n"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nimages (InputLayer) (None, 28, 28, 1) 0 \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 28, 28, 128) 1280 images[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 28, 28, 128) 147584 conv2d_11[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_1 (MaxPooling2D) (None, 14, 14, 128) 0 conv2d_12[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 14, 14, 128) 256 images[0][0] \n__________________________________________________________________________________________________\nadd_2 (Add) (None, 14, 14, 128) 0 max_pooling2d_1[0][0] \n conv2d_13[0][0] \n__________________________________________________________________________________________________\nflatten_3 (Flatten) (None, 25088) 0 add_2[0][0] \n__________________________________________________________________________________________________\ndense_5 (Dense) (None, 512) 12845568 flatten_3[0][0] \n__________________________________________________________________________________________________\ndense_6 (Dense) (None, 10) 5130 dense_5[0][0] \n==================================================================================================\nTotal params: 12,999,818\nTrainable params: 12,999,818\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"#### Train the ResNET model using the Dataset API and the MNIST data\n(when the feature-map sizes are the same)",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport os\nimport tempfile\n\nimport keras\nfrom keras import backend as K\nfrom keras import layers\nfrom keras.datasets import mnist\n\nimport tensorflow as tf\n\nif K.backend() != 'tensorflow':\n raise RuntimeError('This example can only run with the TensorFlow backend,'\n ' because it requires the Dataset API, which is not'\n ' supported on other platforms.')\n\nbatch_size = 128\nbuffer_size = 10000\nsteps_per_epoch = int(np.ceil(60000 / float(batch_size))) # = 469\nepochs = 5\nnum_classes = 10\n\ndef cnn_layers(x):\n # This example assumes the existence of a 4D input tensor x:\n # This returns a typical image tensor like those of MNIST dataset \n print(\"x.shape:\",x.shape)\n # Applies a transformation to x\n y = layers.Conv2D(128, 3, activation='relu', padding='same')(x)\n y = layers.Conv2D(128, 3, activation='relu', padding='same')(y)\n y = layers.Conv2D(128, 3, activation='relu', padding='same')(y)\n # Adds the original x back to the output features\n output = layers.add([y, x])\n\n # Adding a classifier on top of the convnet\n output = layers.Flatten()(output)\n output = layers.Dense(512, activation='relu')(output)\n predictions = layers.Dense(10, activation='softmax')(output)\n return predictions\n\n(x_train, y_train), (x_test, y_test) = mnist.load_data()\nx_train = x_train.astype(np.float32) / 255\nx_train = np.expand_dims(x_train, -1)\ny_train = tf.one_hot(y_train, num_classes)\n\n# Create the dataset and its associated one-shot iterator.\ndataset = tf.data.Dataset.from_tensor_slices((x_train, y_train))\ndataset = dataset.repeat()\ndataset = dataset.shuffle(buffer_size)\ndataset = dataset.batch(batch_size)\niterator = dataset.make_one_shot_iterator()\n\n# Model creation using tensors from the get_next() graph node.\ninputs, targets = iterator.get_next()\n\nprint(\"inputs.shape:\",inputs.shape)\nprint(\"targets.shape:\",targets.shape)\n\nmodel_input = layers.Input(tensor=inputs)\nmodel_output = cnn_layers(model_input)\n\nmodel = keras.models.Model(inputs=model_input, outputs=model_output)\n\nmodel.compile(optimizer=keras.optimizers.RMSprop(lr=2e-3, decay=1e-5),\n loss='categorical_crossentropy',\n metrics=['accuracy'],\n target_tensors=[targets])",
"inputs.shape: (?, 28, 28, 1)\ntargets.shape: (?, 10)\nx.shape: (?, 28, 28, 1)\n"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_2 (InputLayer) (None, 28, 28, 1) 0 \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 28, 28, 128) 1280 input_2[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 28, 28, 128) 147584 conv2d_14[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 28, 28, 128) 147584 conv2d_15[0][0] \n__________________________________________________________________________________________________\nadd_3 (Add) (None, 28, 28, 128) 0 conv2d_16[0][0] \n input_2[0][0] \n__________________________________________________________________________________________________\nflatten_4 (Flatten) (None, 100352) 0 add_3[0][0] \n__________________________________________________________________________________________________\ndense_7 (Dense) (None, 512) 51380736 flatten_4[0][0] \n__________________________________________________________________________________________________\ndense_8 (Dense) (None, 10) 5130 dense_7[0][0] \n==================================================================================================\nTotal params: 51,682,314\nTrainable params: 51,682,314\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"#### Train and Save the ResNet model",
"_____no_output_____"
]
],
[
[
"model.fit(epochs=epochs,\n steps_per_epoch=steps_per_epoch)\n\n# Save the model weights.\nweight_path = os.path.join(tempfile.gettempdir(), 'saved_ResNet_wt.h5')\nmodel.save_weights(weight_path)",
"Epoch 1/5\n469/469 [==============================] - 2159s 5s/step - loss: 7.3090 - acc: 0.5429\nEpoch 2/5\n469/469 [==============================] - 2127s 5s/step - loss: 5.7656 - acc: 0.6412\nEpoch 3/5\n469/469 [==============================] - 2129s 5s/step - loss: 5.3445 - acc: 0.6674\nEpoch 4/5\n469/469 [==============================] - 2127s 5s/step - loss: 4.4341 - acc: 0.7239\nEpoch 5/5\n469/469 [==============================] - 2126s 5s/step - loss: 4.3280 - acc: 0.7306\n"
]
],
[
[
"#### Second session to test loading trained model without tensors.",
"_____no_output_____"
]
],
[
[
"# Clean up the TF session.\nK.clear_session()\n\n# Second session to test loading trained model without tensors.\nx_test = x_test.astype(np.float32)\nx_test = np.expand_dims(x_test, -1)\n\nx_test_inp = layers.Input(shape=x_test.shape[1:])\ntest_out = cnn_layers(x_test_inp)\ntest_model = keras.models.Model(inputs=x_test_inp, outputs=test_out)\n\nweight_path = os.path.join(tempfile.gettempdir(), 'saved_ResNet_wt.h5')\ntest_model.load_weights(weight_path)\ntest_model.compile(optimizer='rmsprop',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])\ntest_model.summary()\n\nloss, acc = test_model.evaluate(x_test, y_test, num_classes)\nprint('\\nTest accuracy: {0}'.format(acc))",
"x.shape: (?, 28, 28, 1)\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 28, 28, 1) 0 \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 28, 28, 128) 1280 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 28, 28, 128) 147584 conv2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 28, 28, 128) 147584 conv2d_2[0][0] \n__________________________________________________________________________________________________\nadd_1 (Add) (None, 28, 28, 128) 0 conv2d_3[0][0] \n input_1[0][0] \n__________________________________________________________________________________________________\nflatten_1 (Flatten) (None, 100352) 0 add_1[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 512) 51380736 flatten_1[0][0] \n__________________________________________________________________________________________________\ndense_2 (Dense) (None, 10) 5130 dense_1[0][0] \n==================================================================================================\nTotal params: 51,682,314\nTrainable params: 51,682,314\nNon-trainable params: 0\n__________________________________________________________________________________________________\n10000/10000 [==============================] - 148s 15ms/step\n\nTest accuracy: 0.7399000000655651\n"
]
],
[
[
"Not very good... probably normal since residual connection are good with very deep network but here we have only 2 hidden layers. ",
"_____no_output_____"
],
[
"### 7.1.5. Layer weights sharing\n\nOne more important feature of the functional API is the ability to reuse a layer instance several times where instead of instantiating a new layer for each call, you reuse the same weights with every call. This allows you to build models that have shared branches—several branches that all share the same knowledge and perform the same operations. \n\n#### Example - semantic similarity between two sentences\n\nFor example, consider a model that attempts to assess the semantic similarity between two sentences. The model has two inputs (the two sentences to compare) and outputs a score between 0 and 1, where 0 means unrelated sentences and 1 means sentences that are either identical or reformulations of each other. Such a model could be useful in many applications, including deduplicating natural-language queries in a dialog system. \n\nIn this setup, the two input sentences are interchangeable, because semantic similarity is a symmetrical relationship: the similarity of A to B is identical to the similarity of B to A. For this reason, it wouldn’t make sense to learn two independent models for processing each input sentence. Rather, you want to process both with a single LSTM layer. The representations of this LSTM layer (its weights) are learned based on both inputs simultaneously. This is what we call a Siamese LSTM model or a shared LSTM.\n\n Note: Siamese network is a special type of neural network architecture. Instead of learning to classify its\n inputs, the Siamese neural network learns to differentiate between two inputs. It learns the similarity.\n\nHere’s how to implement such a model using layer sharing (layer reuse) in the Keras functional API:",
"_____no_output_____"
]
],
[
[
"from keras import layers\nfrom keras import Input\nfrom keras.models import Model\n\n# Instantiates a single LSTM layer, once\nlstm = layers.LSTM(32)\n\n# Building the left branch of the model: \n# inputs are variable-length sequences of vectors of size 128.\nleft_input = Input(shape=(None, 128))\nleft_output = lstm(left_input)\n\n# Building the right branch of the model:\n# when you call an existing layer instance, you reuse its weights.\nright_input = Input(shape=(None, 128))\nright_output = lstm(right_input)\n\n# Builds the classifier on top\nmerged = layers.concatenate([left_output, right_output], axis=-1)\npredictions = layers.Dense(1, activation='sigmoid')(merged)\n\n# Instantiating the model\nmodel = Model([left_input, right_input], predictions)",
"_____no_output_____"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_12 (InputLayer) (None, None, 128) 0 \n__________________________________________________________________________________________________\ninput_13 (InputLayer) (None, None, 128) 0 \n__________________________________________________________________________________________________\nlstm_3 (LSTM) (None, 32) 20608 input_12[0][0] \n input_13[0][0] \n__________________________________________________________________________________________________\nconcatenate_5 (Concatenate) (None, 64) 0 lstm_3[0][0] \n lstm_3[1][0] \n__________________________________________________________________________________________________\ndense_7 (Dense) (None, 1) 65 concatenate_5[0][0] \n==================================================================================================\nTotal params: 20,673\nTrainable params: 20,673\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
],
[
"import numpy as np\n\nnum_samples = 100\nnum_symbols = 2\n\nTRACE = False\n\nleft_data = np.random.randint(0,num_symbols, size=(num_samples,1,128))\nif TRACE:\n print(type(left_data))\n print(left_data.shape)\n print(left_data)\n print('-'*50)\n\nright_data = np.random.randint(0,num_symbols, size=(num_samples,1,128))\nif TRACE:\n print(type(right_data))\n print(right_data.shape)\n print(right_data)\n print('-'*50)\n\nmatching_list = [np.random.randint(0,num_symbols) for _ in range(num_samples)]\ntargets = np.array(matching_list)\nif TRACE:\n print(type(targets))\n print(targets.shape)\n print(targets)\n print('-'*50)\n\n# We must compile a model before training/testing.\nmodel.compile(optimizer='rmsprop',loss='binary_crossentropy',metrics=['acc'])\n\n# Training the model: when you train such a model,\n# the weights of the LSTM layer are updated based on both inputs.\nmodel.fit([left_data, right_data],targets)",
"Epoch 1/1\n100/100 [==============================] - 4s 37ms/step - loss: 0.7013 - acc: 0.4800\n"
]
],
[
[
"### 7.1.6. Models as layers\n\nImportantly, in the functional API, models can be used as you’d use layers—effectively, you can think of a model as a “bigger layer.” This is true of both the Sequential and Model classes. This means you can call a model on an input tensor and retrieve an output tensor: \n\n y = model(x)\n\nIf the model has multiple input tensors and multiple output tensors, it should be called with a list of tensors: \n\n y1, y2 = model([x1, x2])\n\nWhen you call a model instance, you’re reusing the weights of the model—exactly like what happens when you call a layer instance. Calling an instance, whether it’s a layer instance or a model instance, will always reuse the existing learned representations of the instance—which is intuitive.",
"_____no_output_____"
]
],
[
[
"from keras import layers\nfrom keras import applications \nfrom keras import Input\n\nnbr_classes = 10\n\n# The base image-processing model is the Xception network (convolutional base only).\nxception_base = applications.Xception(weights=None,include_top=False)\n\n# The inputs are 250 × 250 RGB images.\nleft_input = Input(shape=(250, 250, 3))\nright_input = Input(shape=(250, 250, 3))\n\nleft_features = xception_base(left_input)\n# right_input = xception_base(right_input)\nright_features = xception_base(right_input)\n\nmerged_features = layers.concatenate([left_features, right_features], axis=-1)\n\npredictions = layers.Dense(nbr_classes, activation='softmax')(merged_features)\n\n# Instantiating the model\nmodel = Model([left_input, right_input], predictions)",
"_____no_output_____"
],
[
"model.summary()\n\nfrom IPython.display import SVG\nfrom keras.utils.vis_utils import model_to_dot\n\nSVG(model_to_dot(model,show_shapes=True).create(prog='dot', format='svg'))",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_15 (InputLayer) (None, 250, 250, 3) 0 \n__________________________________________________________________________________________________\ninput_16 (InputLayer) (None, 250, 250, 3) 0 \n__________________________________________________________________________________________________\nxception (Model) multiple 20861480 input_15[0][0] \n input_16[0][0] \n__________________________________________________________________________________________________\nconcatenate_6 (Concatenate) (None, 8, 8, 4096) 0 xception[1][0] \n xception[2][0] \n__________________________________________________________________________________________________\ndense_8 (Dense) (None, 8, 8, 10) 40970 concatenate_6[0][0] \n==================================================================================================\nTotal params: 20,902,450\nTrainable params: 20,847,922\nNon-trainable params: 54,528\n__________________________________________________________________________________________________\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4ae5fb13a80ef66f0646e5b30296214afb27e1a5
| 3,690 |
ipynb
|
Jupyter Notebook
|
demos/gpu/rapids/python-agg.ipynb
|
omesser/tutorials
|
4ffa0cc474ffe3bb6c673e89aa1361990fdf5bd7
|
[
"Apache-2.0"
] | null | null | null |
demos/gpu/rapids/python-agg.ipynb
|
omesser/tutorials
|
4ffa0cc474ffe3bb6c673e89aa1361990fdf5bd7
|
[
"Apache-2.0"
] | null | null | null |
demos/gpu/rapids/python-agg.ipynb
|
omesser/tutorials
|
4ffa0cc474ffe3bb6c673e89aa1361990fdf5bd7
|
[
"Apache-2.0"
] | null | null | null | 26.546763 | 183 | 0.511924 |
[
[
[
"# Standalone Python Function for Unified Data Batching and Aggregation with RAPIDS cuDF",
"_____no_output_____"
],
[
"## Installation",
"_____no_output_____"
]
],
[
[
"!pip install kafka",
"Collecting kafka\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/21/71/73286e748ac5045b6a669c2fe44b03ac4c5d3d2af9291c4c6fc76438a9a9/kafka-1.3.5-py2.py3-none-any.whl (207kB)\n\u001b[K 100% |████████████████████████████████| 215kB 20.1MB/s ta 0:00:01\n\u001b[?25hInstalling collected packages: kafka\nSuccessfully installed kafka-1.3.5\n"
]
],
[
[
"## Script",
"_____no_output_____"
]
],
[
[
"from kafka import KafkaConsumer\nimport os\nimport glob\nfrom datetime import datetime, timedelta\nimport time\nimport itertools\nimport json\n\n# Select the DataFrame factory\nimport cudf as pd\n# Import pandas as pd\n\n# Basic configuration\nmetric_names = ['cpu_utilization', 'latency', 'packet_loss', 'throughput']\nbatch_len = 100\nbatch = list()\n\n# Kafka configuration\ntopic = ''\nservers = []\noffset = 'earliest'\n\ndef handler(event):\n '''\n Processing function\n '''\n global batch\n global metric_names\n \n # Aggregate event JSON objects\n batch.append(event.body)\n \n # Verify that there are enough events to perform aggregations\n if len(batch) >= interval:\n \n # Create a DataFrame from the batch of event JSON objects\n df = cudf.read_json('\\n'.join(batch), lines=True)\n df = df.reset_index(drop=True)\n \n # Perform aggregations\n df = df.groupby(['company']).\\\n agg({k: ['min', 'max', 'mean'] for k in metric_names})\n \n # Save to Parquet\n filename = f'{time.time()}.parquet'\n filepath = os.path.join(sink, filename)\n new_index = [f'{e[0]}_{e[1]}' for e in list(df.columns)]\n df.columns = new_index\n df.to_parquet(filepath)\n \n # Reset the batch\n batch = list()\n\n# Kafka handling\nconsumer = KafkaConsumer(\n topic,\n bootstrap_servers=servers,\n auto_offset_reset='offset',\n value_deserializer=lambda x: x.decode('utf-8'))\n\nfor message in consumer:\n message = message.value\n handler(message)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae5fbecaa01b2a3f796db5d62df60b3b9aaed76
| 4,052 |
ipynb
|
Jupyter Notebook
|
Python Notebooks/_args _ _kwargs - Bootcamp.ipynb
|
Isaquehg/Scripts
|
d6d94e2d32171262b8286bae82ccd83b3baf30a5
|
[
"MIT"
] | null | null | null |
Python Notebooks/_args _ _kwargs - Bootcamp.ipynb
|
Isaquehg/Scripts
|
d6d94e2d32171262b8286bae82ccd83b3baf30a5
|
[
"MIT"
] | null | null | null |
Python Notebooks/_args _ _kwargs - Bootcamp.ipynb
|
Isaquehg/Scripts
|
d6d94e2d32171262b8286bae82ccd83b3baf30a5
|
[
"MIT"
] | null | null | null | 4,052 | 4,052 | 0.651283 |
[
[
[
"#Function to find 5% of a number sum\r\n\r\ndef myfunc(a,b):\r\n return sum((a,b)) * 0.05\r\n\r\nmyfunc(40,60) #5% of 100",
"_____no_output_____"
],
[
"#*ARGS\r\n#The name mustn´t be ARGS. We can put any name like *abc...\r\ndef myfunc(*args):\r\n print(args)#*arg creates a TUPLE!\r\n return sum(args) * 0.05\r\n\r\n#*args let us to add infinite arguments into the func!\r\nmyfunc(40,60,90,100)",
"(40, 60, 90, 100)\n"
],
[
"#**KWARGS\r\ndef myfunc(**kwargs):\r\n print(kwargs)#NOTE that ** creates a DICTIONARY and * creates a tuple.\r\n\r\n if \"fruit\" in kwargs:\r\n print(\"My favorite fruit is {}\".format(kwargs[\"fruit\"]))\r\n else:\r\n print(\"There´s no fruit here!\")\r\n\r\nmyfunc(fruit=\"orange\")",
"{'fruit': 'orange'}\nMy favorite fruit is orange\n"
],
[
"#Bringing them together\r\ndef somefunc(*arg,**kwarg):\r\n print(arg)\r\n print(kwarg)\r\n print(\"I´d like {} {}\".format(arg[0],kwarg[\"food\"]))\r\n\r\nsomefunc(10,20,30,food=\"eggs\",animal=\"dogs\",auto=\"car\")",
"(10, 20, 30)\n{'food': 'eggs', 'animal': 'dogs', 'auto': 'car'}\nI´d like 10 eggs\n"
],
[
"#Creating a list with even number using random args\r\nlist1 = []\r\ndef myfunc(*args):\r\n for i in args:\r\n if i % 2 == 0:\r\n list1.append(i)\r\n else:\r\n pass\r\n return list1\r\n\r\nmyfunc(0,1,2,3,4,5,6,7)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4ae5fdb1d64b5220262706f6402fa57f7e092616
| 330,890 |
ipynb
|
Jupyter Notebook
|
jupyter-notebooks/Mri_Autoencoder.ipynb
|
sprtkd/mri-msdetect
|
d752f857cc9e6f61b3f2695bdc5fcc5fdaa67575
|
[
"MIT"
] | null | null | null |
jupyter-notebooks/Mri_Autoencoder.ipynb
|
sprtkd/mri-msdetect
|
d752f857cc9e6f61b3f2695bdc5fcc5fdaa67575
|
[
"MIT"
] | null | null | null |
jupyter-notebooks/Mri_Autoencoder.ipynb
|
sprtkd/mri-msdetect
|
d752f857cc9e6f61b3f2695bdc5fcc5fdaa67575
|
[
"MIT"
] | null | null | null | 330,890 | 330,890 | 0.909728 |
[
[
[
"# Basic Init",
"_____no_output_____"
],
[
" **Imports**",
"_____no_output_____"
]
],
[
[
"import nibabel as nib\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom random import randint\nimport tensorflow as tf\nimport glob\nimport pickle\nimport os\nfrom keras.layers import Input, Dense, Conv3D, MaxPooling3D, UpSampling3D, Conv3DTranspose\nfrom keras.models import Model, load_model\nfrom keras import backend as K\nfrom google.colab import drive\nfrom keras import optimizers",
"Using TensorFlow backend.\n"
]
],
[
[
"**Connection to gdrive and file listing**",
"_____no_output_____"
]
],
[
[
"drive.mount('/content/gdrive')\n!ls",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&scope=email%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdocs.test%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive.photos.readonly%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fpeopleapi.readonly&response_type=code\n\nEnter your authorization code:\n··········\nMounted at /content/gdrive\ngdrive\tsample_data\n"
],
[
"base_path = 'gdrive/My Drive/projects/Brain MRI BTech Project/PreprocData/'",
"_____no_output_____"
]
],
[
[
"**List Resource**",
"_____no_output_____"
]
],
[
[
"# memory footprint support libraries/code\n!ln -sf /opt/bin/nvidia-smi /usr/bin/nvidia-smi\n!pip install gputil\n!pip install psutil\n!pip install humanize\nimport psutil\nimport humanize\nimport os\nimport GPUtil as GPU",
"Requirement already satisfied: gputil in /usr/local/lib/python3.6/dist-packages (1.4.0)\nRequirement already satisfied: psutil in /usr/local/lib/python3.6/dist-packages (5.4.8)\nRequirement already satisfied: humanize in /usr/local/lib/python3.6/dist-packages (0.5.1)\n"
],
[
"GPUs = GPU.getGPUs()\n# XXX: only one GPU on Colab and isn’t guaranteed\ngpu = GPUs[0]\ndef printm():\n process = psutil.Process(os.getpid())\n print(\"Gen RAM Free: \" + humanize.naturalsize( psutil.virtual_memory().available ), \" | Proc size: \" + humanize.naturalsize( process.memory_info().rss))\n print(\"GPU RAM Free: {0:.0f}MB | Used: {1:.0f}MB | Util {2:3.0f}% | Total {3:.0f}MB\".format(gpu.memoryFree, gpu.memoryUsed, gpu.memoryUtil*100, gpu.memoryTotal))\nprintm()",
"Gen RAM Free: 12.9 GB | Proc size: 299.1 MB\nGPU RAM Free: 15079MB | Used: 0MB | Util 0% | Total 15079MB\n"
]
],
[
[
"# Dataset Handling",
"_____no_output_____"
],
[
"**MRI handling functions**",
"_____no_output_____"
]
],
[
[
"def get_rand_slice_list(data_shape):\n x_max, y_max, z_max = data_shape\n x_curr = randint((x_max/2)-(x_max/4), (x_max/2)+(x_max/4))\n y_curr = randint((y_max/2)-(y_max/4), (y_max/2)+(y_max/4))\n z_curr = randint((z_max/2)-(z_max/4), (z_max/2)+(z_max/4))\n return x_curr, y_curr, z_curr\n\n\n\ndef show_mri_slices_random(mri_data, explicit_pos=None):\n \"\"\" Function to display random image slices \"\"\"\n '''Provision to give exact slice numbers'''\n '''Random numbers biased towards middle'''\n\n print('Data Shape = ',mri_data.shape)\n if explicit_pos==None:\n x_curr, y_curr, z_curr = get_rand_slice_list(mri_data.shape)\n else:\n x_curr, y_curr, z_curr = explicit_pos\n print('Data Positions = ',x_curr, y_curr, z_curr)\n slice_0 = mri_data[x_curr, :, :]\n slice_1 = mri_data[:, y_curr, :]\n slice_2 = mri_data[:, :, z_curr]\n print('Slice 1: value: ',x_curr)\n plt.imshow(slice_0.T, cmap='gray', origin=0)\n plt.show()\n print('Slice 2: value: ',y_curr)\n plt.imshow(slice_1.T, cmap='gray', aspect=0.5, origin=0)\n plt.show()\n print('Slice 3: value: ',z_curr)\n plt.imshow(slice_2.T, cmap='gray', aspect=0.5, origin=0)\n plt.show()\n \n \ndef get_mri_data(path):\n img_obj = nib.load(path)\n return img_obj.get_fdata()\n\n\ndef get_mri_data_scaler(path,scale_vals,type_mri):\n img_obj = nib.load(path)\n smax,smin = scale_vals[type_mri]\n curr_data = img_obj.get_fdata()\n curr_data = ((curr_data - smin)/(smax-smin))*smax\n return curr_data\n\ndef id_extract(stringpath):\n name_parts = stringpath.split(os.sep)\n name_parts.pop()\n dataset_name = name_parts.pop()\n return int(dataset_name[-2:])\n \ndef print_Details(dat_paths):\n for dat in dat_paths:\n print(dat['id'])\n for key,val in dat.items():\n if key != 'id':\n aaa = get_mri_data(val)\n print(key,aaa.shape,aaa.max(),aaa.min())\n ",
"_____no_output_____"
]
],
[
[
"**Dataset Loading Function**",
"_____no_output_____"
]
],
[
[
"def load_MS_dataset(base_dataset_path):\n total_dataset = []\n patient_folders =glob.glob(base_dataset_path+'*/')\n patient_folders.sort()\n for curr_data_path in patient_folders:\n curr_dataset={}\n curr_dataset['id'] = id_extract(curr_data_path)\n curr_dataset['flair'] = glob.glob(curr_data_path+'/*flair.nii.gz')[-1]\n curr_dataset['t1'] =glob.glob(curr_data_path+'/*t1.nii.gz')[-1]\n curr_dataset['t2'] = glob.glob(curr_data_path+'/*t2.nii.gz')[-1]\n curr_dataset['label'] = glob.glob(curr_data_path+'/*label.nii.gz')[-1]\n total_dataset.append(curr_dataset)\n print(curr_dataset['id'])\n \n with open(base_dataset_path+'data_details.pickle', \"rb\") as data_details_file:\n dataset_details = pickle.load(data_details_file)\n \n return total_dataset, dataset_details\n ",
"_____no_output_____"
]
],
[
[
"**Model Helper functions**",
"_____no_output_____"
]
],
[
[
"def dice_coef_modified(y_true, y_pred):\n y_true_f = K.flatten(y_true)\n y_pred_f = K.flatten(y_pred)\n intersection = K.sum(y_true_f * y_pred_f)\n return (2. * intersection + K.epsilon()) / (K.sum(y_true_f) + K.sum(y_pred_f) + K.epsilon())\n \n \ndef dice_coefficient(y_true, y_pred, smooth=1.):\n y_true_f = K.flatten(y_true)\n y_pred_f = K.flatten(y_pred)\n intersection = K.sum(y_true_f * y_pred_f)\n return (2. * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)\n\n\ndef dice_coefficient_loss(y_true, y_pred):\n return 1.0-dice_coef_modified(y_true, y_pred)\n\ndef binarise_lesion(lesion_data):\n lesion_data[lesion_data <= 0] = 0\n lesion_data[lesion_data > 0] = 1\n return lesion_data\n\n\ndef Mean_IOU(y_true, y_pred):\n nb_classes = K.int_shape(y_pred)[-1]\n iou = []\n true_pixels = K.argmax(y_true, axis=-1)\n pred_pixels = K.argmax(y_pred, axis=-1)\n void_labels = K.equal(K.sum(y_true, axis=-1), 0)\n for i in range(0, nb_classes): # exclude first label (background) and last label (void)\n true_labels = K.equal(true_pixels, i) & ~void_labels\n pred_labels = K.equal(pred_pixels, i) & ~void_labels\n inter = tf.to_int32(true_labels & pred_labels)\n union = tf.to_int32(true_labels | pred_labels)\n legal_batches = K.sum(tf.to_int32(true_labels), axis=1)>0\n ious = K.sum(inter, axis=1)/K.sum(union, axis=1)\n iou.append(K.mean(tf.gather(ious, indices=tf.where(legal_batches)))) # returns average IoU of the same objects\n iou = tf.stack(iou)\n legal_labels = ~tf.debugging.is_nan(iou)\n iou = tf.gather(iou, indices=tf.where(legal_labels))\n return K.mean(iou)\n",
"_____no_output_____"
]
],
[
[
"**Find Scaling Constants**",
"_____no_output_____"
]
],
[
[
"def scalervals(img_paths):\n flair_max=float('-inf')\n flair_min=float('inf')\n t1_max=float('-inf')\n t1_min=float('inf')\n t2_max=float('-inf')\n t2_min=float('inf')\n for dat in img_paths:\n print(dat['id'])\n curr_flair_data = get_mri_data(dat['flair'])\n curr_t1_data = get_mri_data(dat['t1'])\n curr_t2_data = get_mri_data(dat['t2'])\n \n \n curr_flair_max = np.max(curr_flair_data)\n curr_flair_min = np.min(curr_flair_data)\n if(curr_flair_max>flair_max):\n flair_max = curr_flair_max\n if(curr_flair_min<flair_min):\n flair_min = curr_flair_min\n \n curr_t1_max = np.max(curr_t1_data)\n curr_t1_min = np.min(curr_t1_data)\n if(curr_t1_max>t1_max):\n t1_max = curr_t1_max\n if(curr_t1_min<t1_min): \n t1_min = curr_t1_min\n \n curr_t2_max = np.max(curr_t2_data)\n curr_t2_min = np.min(curr_t2_data)\n if(curr_t2_max>t2_max):\n t2_max = curr_t2_max\n if(curr_t2_min<t2_min):\n t2_min = curr_t2_min\n \n return {'flair': [flair_max, flair_min],'t1': [t1_max,t1_min],'t2': [t2_max,t2_min]}\n",
"_____no_output_____"
]
],
[
[
"# Autoencoder Architecture",
"_____no_output_____"
],
[
"**Main Model Function**\n\nModel Details:\n3D autoencoder\n\n",
"_____no_output_____"
]
],
[
[
"#Model Constants\n\nmodel_input_size = (192, 512, 512, 1) #channels last\ntotal_epochs=400\nepochs_per_item=10\nlearning_rate = 0.0001",
"_____no_output_____"
],
[
"def build_model_3dautoencoder():\n model_Input = Input(shape=model_input_size)\n #Encoder\n Conv3D_layer = Conv3D(filters = 8, kernel_size = (3, 3, 3), activation='relu', padding='same')(model_Input)\n MaxPooling3D_layer = MaxPooling3D(pool_size=(2, 2, 2), padding='same')(Conv3D_layer)\n Conv3D_layer = Conv3D(filters = 16, kernel_size = (3, 3, 3), activation='relu', padding='same')(MaxPooling3D_layer)\n MaxPooling3D_layer = MaxPooling3D(pool_size=(2, 2, 2), padding='same')(Conv3D_layer)\n Conv3D_layer = Conv3D(filters = 32, kernel_size = (3, 3, 3), activation='relu', padding='same')(MaxPooling3D_layer)\n encoding_layer = MaxPooling3D(pool_size=(2, 2, 2), padding='same')(Conv3D_layer)\n \n #decoder\n Conv3D_layer = Conv3D(filters = 32, kernel_size = (3, 3, 3), activation='relu', padding='same')(encoding_layer)\n UpSampling3D_layer = UpSampling3D(size=(2, 2, 2))(Conv3D_layer)\n Conv3D_layer = Conv3D(filters = 16, kernel_size = (3, 3, 3), activation='relu', padding='same')(UpSampling3D_layer)\n UpSampling3D_layer = UpSampling3D(size=(2, 2, 2))(Conv3D_layer)\n Conv3D_layer = Conv3D(filters = 8, kernel_size = (3, 3, 3), activation='relu', padding='same')(UpSampling3D_layer)\n UpSampling3D_layer = UpSampling3D(size=(2, 2, 2))(Conv3D_layer)\n decoding_layer = Conv3D(filters = 1, kernel_size = (3, 3, 3), activation='relu', padding='same')(UpSampling3D_layer)\n \n model_autoencoder_3d = Model(model_Input, decoding_layer)\n model_autoencoder_3d.compile(loss=[dice_coefficient_loss], optimizer=optimizers.Adam(lr=learning_rate))\n model_autoencoder_3d.summary()\n return model_autoencoder_3d\n",
"_____no_output_____"
]
],
[
[
"**Train and Test**",
"_____no_output_____"
]
],
[
[
"def train_lesion_Flair(model, dat_paths, tot_epochs, epoch_per_item):\n loopval=True\n while(loopval):\n for dat in dat_paths:\n if tot_epochs<0:\n loopval=False\n break\n print(\"Epochs left: \",tot_epochs)\n print(\"ID: \",dat['id'])\n curr_flair_data = get_mri_data(dat['flair'])\n curr_label_lesion_data = get_mri_data(dat['label'])\n if np.array_equal(curr_flair_data.shape,( 192, 512, 512)):\n curr_flair_data = np.reshape(curr_flair_data.astype('float32'), (1, 192, 512, 512, 1))\n curr_label_lesion_data = np.reshape(curr_label_lesion_data.astype('float32'), (1, 192, 512, 512, 1)) \n model.fit(curr_flair_data, curr_label_lesion_data, epochs=epoch_per_item)\n tot_epochs = tot_epochs - epoch_per_item\n else:\n print(\"size does not match.. Skipping..\")\n \ndef test_lesion_Flair_show(model,dat_paths,num):\n num=num-1\n curr_flair_data = get_mri_data(dat_paths[num]['flair'])\n curr_flair_data_reshaped = np.reshape(curr_flair_data.astype('float32'), (1, 192, 512, 512, 1))\n curr_label_lesion_data = get_mri_data(dat_paths[num]['label'])\n curr_label_lesion_data_reshaped = np.reshape(curr_label_lesion_data.astype('float32'), (1, 192, 512, 512, 1))\n \n predict_lesion_data = model.predict(curr_flair_data_reshaped)\n predict_lesion_data = (predict_lesion_data.reshape((192, 512, 512))).astype(int)\n curr_slice_list = get_rand_slice_list((192, 512, 512))\n print('MRI')\n show_mri_slices_random(curr_flair_data,curr_slice_list)\n print('Label')\n show_mri_slices_random(curr_label_lesion_data,curr_slice_list)\n print('Predicted')\n predict_lesion_data = binarise_lesion(predict_lesion_data)\n show_mri_slices_random(predict_lesion_data,curr_slice_list)\n print(\"Max=\",predict_lesion_data.max(),\"Min=\",predict_lesion_data.min())\n scores = model.evaluate(curr_flair_data_reshaped,curr_label_lesion_data_reshaped)\n print(\"Scores: \",scores)\n \n return predict_lesion_data\n",
"_____no_output_____"
],
[
"def save_output_mri(model,dat_paths,num):\n num=num-1\n curr_flair_data = get_mri_data(dat_paths[num]['flair'])\n curr_flair_data_reshaped = np.reshape(curr_flair_data.astype('float32'), (1, 192, 512, 512, 1))\n curr_label_lesion_data = get_mri_data(dat_paths[num]['label'])\n curr_label_lesion_data_reshaped = np.reshape(curr_label_lesion_data.astype('float32'), (1, 192, 512, 512, 1))\n predict_lesion_data = model.predict(curr_flair_data_reshaped)\n predict_lesion_data = (predict_lesion_data.reshape((192, 512, 512))).astype(int)\n curr_slice_list = get_rand_slice_list((192, 512, 512))\n predict_lesion_data = binarise_lesion(predict_lesion_data).astype(float)\n \n label_obj = nib.load(dat_paths[num]['label'])\n print(label_obj.get_data().shape)\n print(predict_lesion_data.shape)\n output_obj = nib.Nifti1Image(predict_lesion_data, label_obj.affine)\n nib.save(output_obj, 'output.nii.gz')\n \n",
"_____no_output_____"
]
],
[
[
"**Batching Support added**",
"_____no_output_____"
]
],
[
[
"def create_batch_flair(dat_paths, batch_size, curr_offset):\n total_size = len(dat_paths)\n curr_batch_flair=[]\n curr_batch_lesion=[]\n for curr_iter in range(batch_size):\n curr_id = (curr_offset + curr_iter) % total_size\n print('id: ',curr_id)\n curr_flair_data = get_mri_data(dat_paths[curr_id]['flair'])\n curr_label_lesion_data = curr_flair_data\n if np.array_equal(curr_flair_data.shape,( 192, 512, 512)):\n curr_flair_data = np.reshape(curr_flair_data.astype('float32'), (192, 512, 512, 1))\n curr_label_lesion_data = np.reshape(curr_label_lesion_data.astype('float32'), (192, 512, 512, 1))\n curr_batch_flair.append(curr_flair_data)\n curr_batch_lesion.append(curr_label_lesion_data)\n else:\n print(\"size does not match.. Skipping..\")\n curr_batch_flair = np.array(curr_batch_flair)\n curr_batch_lesion = np.array(curr_batch_lesion)\n print('flair batch:',curr_batch_flair.shape)\n print('lesion batch',curr_batch_lesion.shape)\n new_offset = (curr_offset + batch_size) % total_size\n \n return new_offset, curr_batch_flair, curr_batch_lesion\n\ndef train_lesion_Flair_in_batches(model, dat_paths, tot_epochs, epoch_per_batch, batch_size):\n curr_offset = 0\n while(tot_epochs>0):\n print(\"Epochs left: \",tot_epochs)\n print('seed offset: ', curr_offset)\n curr_offset, curr_batch_flair, curr_batch_lesion = create_batch_flair(dat_paths, batch_size, curr_offset)\n model.fit(curr_batch_flair, curr_batch_lesion, epochs=epoch_per_batch)\n tot_epochs = tot_epochs - epoch_per_batch\n ",
"_____no_output_____"
]
],
[
[
"# **Main PipeLine**",
"_____no_output_____"
],
[
"**Load Dataset and Generate Model**",
"_____no_output_____"
]
],
[
[
"dataset_paths,dataset_details = load_MS_dataset(base_path)\nlesion3dAutoencoder = build_model_3dautoencoder()",
"1\n2\n3\n4\n5\n6\n7\n8\n9\n10\n11\n12\n13\n14\n15\n16\n17\n18\n19\n20\n21\n22\n23\n24\n25\n26\n27\n28\n29\n30\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nColocations handled automatically by placer.\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) (None, 192, 512, 512, 1) 0 \n_________________________________________________________________\nconv3d_1 (Conv3D) (None, 192, 512, 512, 8) 224 \n_________________________________________________________________\nmax_pooling3d_1 (MaxPooling3 (None, 96, 256, 256, 8) 0 \n_________________________________________________________________\nconv3d_2 (Conv3D) (None, 96, 256, 256, 16) 3472 \n_________________________________________________________________\nmax_pooling3d_2 (MaxPooling3 (None, 48, 128, 128, 16) 0 \n_________________________________________________________________\nconv3d_3 (Conv3D) (None, 48, 128, 128, 32) 13856 \n_________________________________________________________________\nmax_pooling3d_3 (MaxPooling3 (None, 24, 64, 64, 32) 0 \n_________________________________________________________________\nconv3d_4 (Conv3D) (None, 24, 64, 64, 32) 27680 \n_________________________________________________________________\nup_sampling3d_1 (UpSampling3 (None, 48, 128, 128, 32) 0 \n_________________________________________________________________\nconv3d_5 (Conv3D) (None, 48, 128, 128, 16) 13840 \n_________________________________________________________________\nup_sampling3d_2 (UpSampling3 (None, 96, 256, 256, 16) 0 \n_________________________________________________________________\nconv3d_6 (Conv3D) (None, 96, 256, 256, 8) 3464 \n_________________________________________________________________\nup_sampling3d_3 (UpSampling3 (None, 192, 512, 512, 8) 0 \n_________________________________________________________________\nconv3d_7 (Conv3D) (None, 192, 512, 512, 1) 217 \n=================================================================\nTotal params: 62,753\nTrainable params: 62,753\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"**Model Training and Save**",
"_____no_output_____"
]
],
[
[
"train_lesion_Flair(lesion3dAutoencoder,dataset_paths,total_epochs,epochs_per_item)\nlesion3dAutoencoder.save('my_model.h5')",
"Epochs left: 400\nID: 1\nEpoch 1/10\n1/1 [==============================] - 7s 7s/step - loss: 0.9946\nEpoch 2/10\n1/1 [==============================] - 4s 4s/step - loss: 0.9619\nEpoch 3/10\n1/1 [==============================] - 4s 4s/step - loss: 0.9537\nEpoch 4/10\n1/1 [==============================] - 4s 4s/step - loss: 0.9509\nEpoch 5/10\n1/1 [==============================] - 4s 4s/step - loss: 0.9491\nEpoch 6/10\n1/1 [==============================] - 5s 5s/step - loss: 0.9473\nEpoch 7/10\n1/1 [==============================] - 5s 5s/step - loss: 0.9453\nEpoch 8/10\n1/1 [==============================] - 5s 5s/step - loss: 0.9432\nEpoch 9/10\n1/1 [==============================] - 5s 5s/step - loss: 0.9412\nEpoch 10/10\n1/1 [==============================] - 5s 5s/step - loss: 0.9393\nEpochs left: 390\nID: 2\nEpoch 1/10\n1/1 [==============================] - 5s 5s/step - loss: 0.9977\nEpoch 2/10\n"
]
],
[
[
"**Model Test**",
"_____no_output_____"
]
],
[
[
"hhh=test_lesion_Flair_show(lesion3dAutoencoder,dataset_paths,6)",
"MRI\nData Shape = (192, 512, 512)\nData Positions = 53 260 250\nSlice 1: value: 53\n"
]
],
[
[
"**Save ouput**",
"_____no_output_____"
]
],
[
[
"save_output_mri(lesion3dAutoencoder,dataset_paths,6)",
"_____no_output_____"
]
],
[
[
"**Load Model**",
"_____no_output_____"
]
],
[
[
"autoencoder_model = load_model('my_model.h5')\nhhh=test_lesion_Flair_show(autoencoder_model,dataset_paths,6)",
"/usr/local/lib/python3.6/dist-packages/keras/engine/saving.py:292: UserWarning: No training configuration found in save file: the model was *not* compiled. Compile it manually.\n warnings.warn('No training configuration found in save file: '\n"
]
],
[
[
"# Generating output for each layer",
"_____no_output_____"
]
],
[
[
"def generate_all_layer_ouputs(model,dat_paths,num):\n num=num-1\n curr_flair_data = get_mri_data(dat_paths[num]['flair'])\n curr_flair_data_reshaped = np.reshape(curr_flair_data.astype('float32'), (1, 192, 512, 512, 1))\n curr_label_lesion_data = get_mri_data(dat_paths[num]['label'])\n curr_label_lesion_data_reshaped = np.reshape(curr_label_lesion_data.astype('float32'), (1, 192, 512, 512, 1))\n inp = model.input # input placeholder\n outputs = [layer.output for layer in model.layers] # all layer outputs\n functor = K.function([inp, K.learning_phase()], outputs ) # evaluation function\n # Testing\n layer_outs = functor([curr_label_lesion_data_reshaped, 1.])\n print(layer_outs)\n \ngenerate_all_layer_ouputs(autoencoder_model,dataset_paths,6)",
"_____no_output_____"
]
],
[
[
"# Try out Batching... Requires huge amt of VRAM",
"_____no_output_____"
]
],
[
[
"train_lesion_Flair_in_batches(lesion3dAutoencoder,dataset_paths,total_epochs,epochs_per_item, 2)",
"_____no_output_____"
]
],
[
[
"# **Kill session**",
"_____no_output_____"
]
],
[
[
"!pkill -9 -f ipykernel_launcher",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae601934fd1f45de1cb85dfbd0c07c3825a0d91
| 4,535 |
ipynb
|
Jupyter Notebook
|
image_augmentaion/lowres/transform_lowres_images_1100_valid_Normal.ipynb
|
UVA-DSI-2019-Capstones/CHRC
|
3b89fb6039e435f383754f933537201402391a07
|
[
"MIT"
] | null | null | null |
image_augmentaion/lowres/transform_lowres_images_1100_valid_Normal.ipynb
|
UVA-DSI-2019-Capstones/CHRC
|
3b89fb6039e435f383754f933537201402391a07
|
[
"MIT"
] | null | null | null |
image_augmentaion/lowres/transform_lowres_images_1100_valid_Normal.ipynb
|
UVA-DSI-2019-Capstones/CHRC
|
3b89fb6039e435f383754f933537201402391a07
|
[
"MIT"
] | 1 |
2019-09-07T14:01:14.000Z
|
2019-09-07T14:01:14.000Z
| 26.676471 | 133 | 0.54333 |
[
[
[
"import staintools\nimport csv\nimport os\nimport glob\nimport re\nfrom pandas import DataFrame, Series\nfrom PIL import Image\nimport timeit\nimport time\nimport cv2\nfrom matplotlib import pyplot as plt\nimport numpy as np",
"_____no_output_____"
],
[
"train_paths = [\"/scratch/kk4ze/data_lowres_1100x1100/valid/Normal/\"]",
"_____no_output_____"
],
[
"# get images\nimages = {}\nimages_by_folder = {}\nfor train_path in train_paths:\n images_by_folder[str(train_path)] = []\n files = glob.glob(os.path.join(train_path, '*.jpg'))\n for fl in files:\n flbase = os.path.basename(fl)\n flbase_noext = os.path.splitext(flbase)[0]\n images[flbase_noext]=fl\n images_by_folder[str(train_path)].append(flbase_noext)",
"_____no_output_____"
],
[
"# initialize stain and brightness normalizer\nstain_normalizer = staintools.StainNormalizer(method='vahadane')\nstandardizer = staintools.BrightnessStandardizer()",
"_____no_output_____"
],
[
"# choose target image\ntarget_image = staintools.read_image(\"/scratch/kk4ze/data_lowres_1100x1100/train/Celiac/C03-05_03_5901_4803_horiz__0.jpg\")\nstandard_target_image = standardizer.transform(target_image)\nstain_normalizer.fit(standard_target_image)",
"_____no_output_____"
],
[
"# get destination path\npath_change_map = {}\n\nfor key in list(images_by_folder.keys()):\n temp = key.replace('data_lowres_1100x1100', 'data_lowres_1100x1100_augmented')\n path_change_map[key] = temp",
"_____no_output_____"
],
[
"for key in images_by_folder.keys():\n for value in list(images_by_folder[key]):\n# print(key)\n# print(value)\n# print (str(count) + ' ' + str(value))\n source_img_path = str(key) + str(value) + '.jpg'\n dest_img_path = str(path_change_map[key]) + str(value) + '.jpg'\n# print(source_img_path)\n img = staintools.read_image(source_img_path)\n if (np.mean(img) > 240) or (np.mean(img) < 10):\n continue\n # standardize brightness\n img_standard = standardizer.transform(img)\n # transform the images\n img_normalized = stain_normalizer.transform(img_standard)\n # write image to path\n\n# plt.imshow(img)\n# plt.title('my picture')\n# plt.show()\n# plt.imshow(img_normalized)\n# plt.title('my picture')\n# plt.show()\n cv2.imwrite(os.path.normpath(dest_img_path), img_normalized)",
"_____no_output_____"
],
[
"path_change_map",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae608c56bcadd0f81cf06d2cc000c36eae62cd2
| 61,751 |
ipynb
|
Jupyter Notebook
|
module2-regression-2/212_guided_project_notes.ipynb
|
LycheeCocoa/DS-Unit-2-Linear-Models
|
00d351cd03a2cd606efdbf9e439b305ca4acc054
|
[
"MIT"
] | 4 |
2019-11-20T05:10:43.000Z
|
2021-04-12T04:59:00.000Z
|
module2-regression-2/212_guided_project_notes.ipynb
|
LycheeCocoa/DS-Unit-2-Linear-Models
|
00d351cd03a2cd606efdbf9e439b305ca4acc054
|
[
"MIT"
] | 30 |
2019-07-20T03:27:01.000Z
|
2021-09-08T03:08:25.000Z
|
module2-regression-2/212_guided_project_notes.ipynb
|
LycheeCocoa/DS-Unit-2-Linear-Models
|
00d351cd03a2cd606efdbf9e439b305ca4acc054
|
[
"MIT"
] | 712 |
2019-07-08T15:50:08.000Z
|
2021-11-10T15:18:57.000Z
| 84.129428 | 38,762 | 0.779939 |
[
[
[
"# Libraries for R^2 visualization\nfrom ipywidgets import interactive, IntSlider, FloatSlider\nfrom math import floor, ceil\nfrom sklearn.base import BaseEstimator, RegressorMixin\n\n# Libraries for model building\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\n# Library for working locally or Colab\nimport sys",
"_____no_output_____"
],
[
"# If you're on Colab:\nif 'google.colab' in sys.modules:\n DATA_PATH = 'https://raw.githubusercontent.com/LambdaSchool/DS-Unit-2-Applied-Modeling/master/data/'\n\n# If you're working locally:\nelse:\n DATA_PATH = '../data/'",
"_____no_output_____"
]
],
[
[
"# I. Wrangle Data",
"_____no_output_____"
]
],
[
[
"df = wrangle(DATA_PATH + 'elections/bread_peace_voting.csv')",
"_____no_output_____"
]
],
[
[
"# II. Split Data\n\n**First** we need to split our **target vector** from our **feature matrix**.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"**Second** we need to split our dataset into **training** and **test** sets.\n\nTwo strategies:\n\n- Random train-test split using [`train_test_split`](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html). Generally we use 80% of the data for training, and 20% of the data for testing.\n- If you have **timeseries**, then you need to do a \"cutoff\" split.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# III. Establish Baseline",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# IV. Build Model",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# V. Check Metrics\n\n## Mean Absolute Error\n\nThe unit of measurement is the same as the unit of measurment for your target (in this case, vote share [%]).",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"## Root Mean Squared Error\n\nThe unit of measurement is the same as the unit of measurment for your target (in this case, vote share [%]).",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"## $R^2$ Score\n\nTL;DR: Usually ranges between 0 (bad) and 1 (good).",
"_____no_output_____"
]
],
[
[
"class BruteForceRegressor(BaseEstimator, RegressorMixin):\n def __init__(self, m=0, b=0):\n self.m = m\n self.b = b\n self.mean = 0\n \n def fit(self, X, y):\n self.mean = np.mean(y)\n return self\n \n def predict(self, X, return_mean=True):\n if return_mean:\n return [self.mean] * len(X)\n else:\n return X * self.m + self.b\n\ndef plot(slope, intercept):\n # Assign data to variables\n x = df['income']\n y = df['incumbent_vote_share']\n \n # Create figure\n fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20,6))\n \n # Set ax limits\n mar = 0.2\n x_lim = floor(x.min() - x.min()*mar), ceil(x.max() + x.min()*mar)\n y_lim = floor(y.min() - y.min()*mar), ceil(y.max() + y.min()*mar)\n \n # Instantiate and train model\n bfr = BruteForceRegressor(slope, intercept)\n bfr.fit(x, y)\n \n # ax1 \n ## Plot data\n ax1.set_xlim(x_lim)\n ax1.set_ylim(y_lim)\n ax1.scatter(x, y)\n \n ## Plot base model\n ax1.axhline(bfr.mean, color='orange', label='baseline model')\n \n ## Plot residual lines\n y_base_pred = bfr.predict(x)\n ss_base = mean_squared_error(y, y_base_pred) * len(y)\n for x_i, y_i, yp_i in zip(x, y, y_base_pred):\n ax1.plot([x_i, x_i], [y_i, yp_i], \n color='gray', linestyle='--', alpha=0.75)\n \n ## Formatting\n ax1.legend()\n ax1.set_title(f'Sum of Squares: {np.round(ss_base, 2)}')\n ax1.set_xlabel('Growth in Personal Incomes')\n ax1.set_ylabel('Incumbent Party Vote Share [%]')\n\n # ax2\n\n ax2.set_xlim(x_lim)\n ax2.set_ylim(y_lim)\n ## Plot data\n ax2.scatter(x, y)\n \n ## Plot model\n x_model = np.linspace(*ax2.get_xlim(), 10)\n y_model = bfr.predict(x_model, return_mean=False)\n ax2.plot(x_model, y_model, color='green', label='our model')\n for x_coord, y_coord in zip(x, y):\n ax2.plot([x_coord, x_coord], [y_coord, x_coord * slope + intercept], \n color='gray', linestyle='--', alpha=0.75) \n \n ss_ours = mean_squared_error(y, bfr.predict(x, return_mean=False)) * len(y)\n \n ## Formatting\n ax2.legend()\n ax2.set_title(f'Sum of Squares: {np.round(ss_ours, 2)}')\n ax2.set_xlabel('Growth in Personal Incomes')\n ax2.set_ylabel('Incumbent Party Vote Share [%]')\n\ny = df['incumbent_vote_share']\nslope_slider = FloatSlider(min=-5, max=5, step=0.5, value=0)\nintercept_slider = FloatSlider(min=int(y.min()), max=y.max(), step=2, value=y.mean())\n \ninteractive(plot, slope=slope_slider, intercept=intercept_slider)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"# VI. Communicate Results\n\n**Challenge:** How can we find the coefficients and intercept for our `model`?",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae610f04d8393884f6337d99f75d73c5a054681
| 174,876 |
ipynb
|
Jupyter Notebook
|
Week-5/ET5003_NLP_SpamClasiffier_2.ipynb
|
davidnol/ET5003_SEM1_2021-2
|
07b13b51b368e8dc22ddf3028203e5675ee7f96a
|
[
"BSD-3-Clause"
] | 1 |
2021-09-06T17:47:57.000Z
|
2021-09-06T17:47:57.000Z
|
Week-5/ET5003_NLP_SpamClasiffier_2.ipynb
|
davidnol/ET5003_SEM1_2021-2
|
07b13b51b368e8dc22ddf3028203e5675ee7f96a
|
[
"BSD-3-Clause"
] | null | null | null |
Week-5/ET5003_NLP_SpamClasiffier_2.ipynb
|
davidnol/ET5003_SEM1_2021-2
|
07b13b51b368e8dc22ddf3028203e5675ee7f96a
|
[
"BSD-3-Clause"
] | null | null | null | 77.930481 | 11,262 | 0.629778 |
[
[
[
"<div>\n<img src=\"https://drive.google.com/uc?export=view&id=1vK33e_EqaHgBHcbRV_m38hx6IkG0blK_\" width=\"350\"/>\n</div> \n\n#**Artificial Intelligence - MSc**\n##ET5003 - MACHINE LEARNING APPLICATIONS \n\n###Instructor: Enrique Naredo\n###ET5003_NLP_SpamClasiffier-2",
"_____no_output_____"
],
[
"\n### Spam Classification",
"_____no_output_____"
],
[
"[Spamming](https://en.wikipedia.org/wiki/Spamming) is the use of messaging systems to send multiple unsolicited messages (spam) to large numbers of recipients for the purpose of commercial advertising, for the purpose of non-commercial proselytizing, for any prohibited purpose (especially the fraudulent purpose of phishing), or simply sending the same message over and over to the same user. ",
"_____no_output_____"
],
[
"Spam Classification: Deciding whether an email is spam or not.\n\n",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"# standard libraries\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Scikit-learn is an open source machine learning library \n# that supports supervised and unsupervised learning\n# https://scikit-learn.org/stable/\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.naive_bayes import MultinomialNB\nfrom sklearn.metrics import accuracy_score, confusion_matrix",
"_____no_output_____"
],
[
"# Regular expression operations\n#https://docs.python.org/3/library/re.html\nimport re \n\n# Natural Language Toolkit\n# https://www.nltk.org/install.html\nimport nltk\n\n# Stemming maps different forms of the same word to a common “stem” \n# https://pypi.org/project/snowballstemmer/\nfrom nltk.stem import SnowballStemmer\n\n# https://www.nltk.org/book/ch02.html\nfrom nltk.corpus import stopwords",
"_____no_output_____"
]
],
[
[
"## Step 1: Load dataset",
"_____no_output_____"
]
],
[
[
"# Mount Google Drive\nfrom google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"# path to your (local/cloud) drive \npath = '/content/drive/MyDrive/Colab Notebooks/Enrique/Data/spam/'\n\n# load dataset\ndf = pd.read_csv(path+'spam.csv', encoding='latin-1')\ndf.rename(columns = {'v1':'class_label', 'v2':'message'}, inplace = True)\ndf.drop(['Unnamed: 2', 'Unnamed: 3', 'Unnamed: 4'], axis = 1, inplace = True)",
"_____no_output_____"
],
[
"# original dataset\ndf.head()",
"_____no_output_____"
]
],
[
[
"The dataset has 4825 ham messages and 747 spam messages. ",
"_____no_output_____"
]
],
[
[
"# histogram\nimport seaborn as sns\nsns.countplot(df['class_label'])",
"/usr/local/lib/python3.7/dist-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variable as a keyword arg: x. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
],
[
"# explore dataset\nvc = df['class_label'].value_counts()\nprint(vc)",
"ham 4825\nspam 747\nName: class_label, dtype: int64\n"
]
],
[
[
"This is an imbalanced dataset\n* The number of ham messages is much higher than those of spam.\n* This can potentially cause our model to be biased.\n* To fix this, we could resample our data to get an equal number of spam/ham messages.",
"_____no_output_____"
]
],
[
[
"# convert class label to numeric\nfrom sklearn import preprocessing\nle = preprocessing.LabelEncoder()\nle.fit(df.class_label)\ndf2 = df\ndf2['class_label'] = le.transform(df.class_label)\ndf2.head()",
"_____no_output_____"
],
[
"# another histogram\ndf2.hist()",
"_____no_output_____"
]
],
[
[
"## Step 2: Pre-processing",
"_____no_output_____"
],
[
"Next, we’ll convert our DataFrame to a list, where every element of that list will be a spam message. Then, we’ll join each element of our list into one big string of spam messages. The lowercase form of that string is the required format needed for our word cloud creation.",
"_____no_output_____"
]
],
[
[
"spam_list = df['message'].tolist()\nspam_list",
"_____no_output_____"
],
[
"new_df = pd.DataFrame({'message':spam_list})",
"_____no_output_____"
],
[
"# removing everything except alphabets\nnew_df['clean_message'] = new_df['message'].str.replace(\"[^a-zA-Z#]\", \" \")",
"_____no_output_____"
],
[
"# removing short words\nshort_word = 4\nnew_df['clean_message'] = new_df['clean_message'].apply(lambda x: ' '.join([w for w in x.split() if len(w)>short_word]))",
"_____no_output_____"
],
[
"# make all text lowercase\nnew_df['clean_message'] = new_df['clean_message'].apply(lambda x: x.lower())",
"_____no_output_____"
],
[
"import nltk\nfrom nltk.corpus import stopwords\nnltk.download('stopwords')\n\nswords = stopwords.words('english')",
"[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Unzipping corpora/stopwords.zip.\n"
],
[
"# tokenization\ntokenized_doc = new_df['clean_message'].apply(lambda x: x.split())",
"_____no_output_____"
],
[
"# remove stop-words\ntokenized_doc = tokenized_doc.apply(lambda x: [item for item in x if item not in swords])",
"_____no_output_____"
],
[
"# de-tokenization\ndetokenized_doc = []\nfor i in range(len(new_df)):\n t = ' '.join(tokenized_doc[i])\n detokenized_doc.append(t)",
"_____no_output_____"
],
[
"new_df['clean_message'] = detokenized_doc",
"_____no_output_____"
],
[
"new_df.head()",
"_____no_output_____"
]
],
[
[
"## Step 3: TfidfVectorizer",
"_____no_output_____"
],
[
"**[TfidfVectorizer](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.TfidfVectorizer.html)**\n\nConvert a collection of raw documents to a matrix of TF-IDF features.",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import TfidfVectorizer\n\nvectorizer = TfidfVectorizer(stop_words='english', max_features= 300, max_df=0.5, smooth_idf=True)\nprint(vectorizer)",
"TfidfVectorizer(analyzer='word', binary=False, decode_error='strict',\n dtype=<class 'numpy.float64'>, encoding='utf-8',\n input='content', lowercase=True, max_df=0.5, max_features=300,\n min_df=1, ngram_range=(1, 1), norm='l2', preprocessor=None,\n smooth_idf=True, stop_words='english', strip_accents=None,\n sublinear_tf=False, token_pattern='(?u)\\\\b\\\\w\\\\w+\\\\b',\n tokenizer=None, use_idf=True, vocabulary=None)\n"
],
[
"X = vectorizer.fit_transform(new_df['clean_message'])\nX.shape",
"_____no_output_____"
],
[
"y = df['class_label']\ny.shape",
"_____no_output_____"
]
],
[
[
"Handle imbalance data through SMOTE ",
"_____no_output_____"
]
],
[
[
"from imblearn.combine import SMOTETomek \nsmk= SMOTETomek()\n\nX_bal, y_bal = smk.fit_sample(X, y)",
"/usr/local/lib/python3.7/dist-packages/sklearn/externals/six.py:31: FutureWarning: The module is deprecated in version 0.21 and will be removed in version 0.23 since we've dropped support for Python 2.7. Please rely on the official version of six (https://pypi.org/project/six/).\n \"(https://pypi.org/project/six/).\", FutureWarning)\n/usr/local/lib/python3.7/dist-packages/sklearn/utils/deprecation.py:144: FutureWarning: The sklearn.neighbors.base module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.neighbors. Anything that cannot be imported from sklearn.neighbors is now part of the private API.\n warnings.warn(message, FutureWarning)\n/usr/local/lib/python3.7/dist-packages/sklearn/utils/deprecation.py:87: FutureWarning: Function safe_indexing is deprecated; safe_indexing is deprecated in version 0.22 and will be removed in version 0.24.\n warnings.warn(msg, category=FutureWarning)\n"
],
[
"# histogram\nimport seaborn as sns\nsns.countplot(y_bal)",
"/usr/local/lib/python3.7/dist-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variable as a keyword arg: x. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.\n FutureWarning\n"
],
[
"from sklearn.model_selection import train_test_split\nX_train, X_test, y_train, y_test = train_test_split(X_bal, y_bal, test_size = 0.20, random_state = 0)",
"_____no_output_____"
],
[
"X_train.todense()",
"_____no_output_____"
]
],
[
[
"## Step 4: Learning",
"_____no_output_____"
],
[
"Training the classifier and making predictions on the test set",
"_____no_output_____"
]
],
[
[
"# create a model\nMNB = MultinomialNB()\n\n# fit to data\nMNB.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# testing the model\n\nprediction_train = MNB.predict(X_train)\nprint('training prediction\\t', prediction_train)\n\nprediction_test = MNB.predict(X_test)\nprint('test prediction\\t\\t', prediction_test)",
"training prediction\t [0 0 0 ... 1 0 0]\ntest prediction\t\t [1 1 1 ... 0 0 1]\n"
],
[
"np.set_printoptions(suppress=True)\n\n# Ham and Spam probabilities in test\nclass_prob = MNB.predict_proba(X_test)\nprint(class_prob)",
"[[0.0172931 0.9827069 ]\n [0.45054822 0.54945178]\n [0.0025568 0.9974432 ]\n ...\n [0.77954955 0.22045045]\n [0.50323918 0.49676082]\n [0.28620661 0.71379339]]\n"
],
[
"# show emails classified as 'spam'\nthreshold = 0.5\nspam_ind = np.where(class_prob[:,1]>threshold)[0]",
"_____no_output_____"
]
],
[
[
"## Step 5: Accuracy",
"_____no_output_____"
]
],
[
[
"# accuracy in training set\ny_pred_train = prediction_train\nprint(\"Train Accuracy: \"+str(accuracy_score(y_train, y_pred_train)))",
"Train Accuracy: 0.9182430681523711\n"
],
[
"# accuracy in test set (unseen data)\ny_true = y_test\ny_pred_test = prediction_test\nprint(\"Test Accuracy: \"+str(accuracy_score(y_true, y_pred_test)))",
"Test Accuracy: 0.9139896373056995\n"
],
[
"# confusion matrix\nconf_mat = confusion_matrix(y_true, y_pred_test)\nprint(\"Confusion Matrix\\n\", conf_mat)",
"Confusion Matrix\n [[894 46]\n [120 870]]\n"
],
[
"import matplotlib.pyplot as plt\nfrom sklearn.metrics import ConfusionMatrixDisplay\n\nlabels = ['Ham','Spam']\n\nfig = plt.figure()\nax = fig.add_subplot(111)\ncax = ax.matshow(conf_mat)\nplt.title('Confusion matrix of the classifier\\n')\nfig.colorbar(cax)\nax.set_xticklabels([''] + labels)\nax.set_yticklabels([''] + labels)\nplt.xlabel('Predicted')\nplt.ylabel('True')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
4ae61327ca0eeb439ec423d8badc6e56b1e59329
| 859 |
ipynb
|
Jupyter Notebook
|
notebooks/graph_deduplication_drills.ipynb
|
cribbslab/mclumi
|
3c38f1dd4de11f0f20121a4027f7e104c4059bbe
|
[
"MIT"
] | 3 |
2021-12-18T09:55:09.000Z
|
2022-01-12T10:25:28.000Z
|
notebooks/graph_deduplication_drills.ipynb
|
cribbslab/mclumi
|
3c38f1dd4de11f0f20121a4027f7e104c4059bbe
|
[
"MIT"
] | null | null | null |
notebooks/graph_deduplication_drills.ipynb
|
cribbslab/mclumi
|
3c38f1dd4de11f0f20121a4027f7e104c4059bbe
|
[
"MIT"
] | null | null | null | 17.18 | 50 | 0.513388 |
[
[
[
"# graph-based UMI deduplication drills",
"_____no_output_____"
],
[
"...",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown"
]
] |
4ae6151161f0d3e988bd76d889830d097dc5c04d
| 29,552 |
ipynb
|
Jupyter Notebook
|
logistic regression example.ipynb
|
jae-finger/DS-Unit-2-Linear-Models
|
3d2269d26133dec7996c9a1d3906a0f928598695
|
[
"MIT"
] | null | null | null |
logistic regression example.ipynb
|
jae-finger/DS-Unit-2-Linear-Models
|
3d2269d26133dec7996c9a1d3906a0f928598695
|
[
"MIT"
] | null | null | null |
logistic regression example.ipynb
|
jae-finger/DS-Unit-2-Linear-Models
|
3d2269d26133dec7996c9a1d3906a0f928598695
|
[
"MIT"
] | null | null | null | 29,552 | 29,552 | 0.809556 |
[
[
[
"Lambda School Data Science\n\n*Unit 2, Sprint 1, Module 4*\n\n---\n\n# Logistic Regression\n",
"_____no_output_____"
],
[
"## Overview",
"_____no_output_____"
],
[
"We'll begin with the **majority class baseline.**\n\n[Will Koehrsen](https://twitter.com/koehrsen_will/status/1088863527778111488)\n\n> A baseline for classification can be the most common class in the training dataset.\n\n[*Data Science for Business*](https://books.google.com/books?id=4ZctAAAAQBAJ&pg=PT276), Chapter 7.3: Evaluation, Baseline Performance, and Implications for Investments in Data\n\n> For classification tasks, one good baseline is the _majority classifier,_ a naive classifier that always chooses the majority class of the training dataset (see Note: Base rate in Holdout Data and Fitting Graphs). This may seem like advice so obvious it can be passed over quickly, but it is worth spending an extra moment here. There are many cases where smart, analytical people have been tripped up in skipping over this basic comparison. For example, an analyst may see a classification accuracy of 94% from her classifier and conclude that it is doing fairly well—when in fact only 6% of the instances are positive. So, the simple majority prediction classifier also would have an accuracy of 94%. ",
"_____no_output_____"
],
[
"## Follow Along",
"_____no_output_____"
],
[
"Determine majority class",
"_____no_output_____"
]
],
[
[
"y_train.value_counts(normalize=True)",
"_____no_output_____"
]
],
[
[
"What if we guessed the majority class for every prediction?",
"_____no_output_____"
]
],
[
[
"majority_class = y_train.mode()[0]\ny_pred = [majority_class] * len(y_train)",
"_____no_output_____"
]
],
[
[
"#### Use a classification metric: accuracy\n\n[Classification metrics are different from regression metrics!](https://scikit-learn.org/stable/modules/model_evaluation.html)\n- Don't use _regression_ metrics to evaluate _classification_ tasks.\n- Don't use _classification_ metrics to evaluate _regression_ tasks.\n\n[Accuracy](https://scikit-learn.org/stable/modules/model_evaluation.html#accuracy-score) is a common metric for classification. Accuracy is the [\"proportion of correct classifications\"](https://en.wikipedia.org/wiki/Confusion_matrix): the number of correct predictions divided by the total number of predictions.",
"_____no_output_____"
],
[
"What is the baseline accuracy if we guessed the majority class for every prediction?",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import accuracy_score\naccuracy_score(y_train, y_pred)",
"_____no_output_____"
],
[
"y_pred = [majority_class] * len(y_val)\naccuracy_score(y_val, y_pred)",
"_____no_output_____"
],
[
"# Using Sklearn DummyClassifier\nfrom sklearn.dummy import DummyClassifier\n\n# Fit the DummyClassifier\nbaseline = DummyClassifier(strategy='most_frequent')\nbaseline.fit(X_train, y_train)\n\n# Make predictions on validation data\ny_pred = baseline.predict(X_val)\naccuracy_score(y_val, y_pred)",
"_____no_output_____"
]
],
[
[
"## Overview\n\nTo help us get an intuition for *Logistic* Regression, let's start by trying *Linear* Regression instead, and see what happens...",
"_____no_output_____"
],
[
"### Logistic Regression!",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\nlog_reg = LogisticRegression(solver='lbfgs')\nlog_reg.fit(X_train_imputed, y_train)\nprint('Validation Accuracy', log_reg.score(X_val_imputed, y_val))",
"Validation Accuracy 0.7354260089686099\n"
],
[
"# The predictions look like this\nlog_reg.predict(X_val_imputed)",
"_____no_output_____"
],
[
"log_reg.predict(test_case)",
"_____no_output_____"
],
[
"log_reg.predict_proba(test_case)",
"_____no_output_____"
],
[
"# What's the math?\nlog_reg.coef_",
"_____no_output_____"
],
[
"log_reg.intercept_",
"_____no_output_____"
],
[
"# The logistic sigmoid \"squishing\" function, implemented to accept numpy arrays\nimport numpy as np\n\ndef sigmoid(x):\n return 1 / (1 + np.e**(-x))",
"_____no_output_____"
],
[
"sigmoid(log_reg.intercept_ + np.dot(log_reg.coef_, np.transpose(test_case)))",
"_____no_output_____"
]
],
[
[
"So, clearly a more appropriate model in this situation! For more on the math, [see this Wikipedia example](https://en.wikipedia.org/wiki/Logistic_regression#Probability_of_passing_an_exam_versus_hours_of_study).",
"_____no_output_____"
],
[
"# Use sklearn.linear_model.LogisticRegression to fit and interpret Logistic Regression models",
"_____no_output_____"
],
[
"## Overview\n\nNow that we have more intuition and interpretation of Logistic Regression, let's use it within a realistic, complete scikit-learn workflow, with more features and transformations.",
"_____no_output_____"
],
[
"## Follow Along\n\nSelect these features: `['Pclass', 'Sex', 'Age', 'SibSp', 'Parch', 'Fare', 'Embarked']`\n\n(Why shouldn't we include the `Name` or `Ticket` features? What would happen here?) \n\nFit this sequence of transformers & estimator:\n\n- [category_encoders.one_hot.OneHotEncoder](https://contrib.scikit-learn.org/categorical-encoding/onehot.html)\n- [sklearn.impute.SimpleImputer](https://scikit-learn.org/stable/modules/generated/sklearn.impute.SimpleImputer.html)\n- [sklearn.preprocessing.StandardScaler](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html)\n- [sklearn.linear_model.LogisticRegressionCV](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegressionCV.html)\n\nGet validation accuracy.",
"_____no_output_____"
]
],
[
[
"import category_encoders as ce\nfrom sklearn.linear_model import LogisticRegressionCV\nfrom sklearn.preprocessing import StandardScaler\n\ntarget = 'Survived'\nfeatures = ['Pclass', 'Sex', 'Age', 'SibSp', 'Parch', 'Fare', 'Embarked']\nX_train = train[features]\ny_train = train[target]\nX_val = val[features]\ny_val = val[target]\nprint(X_train.shape, X_val.shape)\n\nencoder = ce.OneHotEncoder(use_cat_names=True)\nX_train_encoded = encoder.fit_transform(X_train)\nX_val_encoded = encoder.transform(X_val)\nprint(X_train_encoded.shape, X_val_encoded.shape)\n\nimputer = SimpleImputer(strategy='mean')\nX_train_imputed = imputer.fit_transform(X_train_encoded)\nX_val_imputed = imputer.transform(X_val_encoded)\n\nscaler = StandardScaler()\nX_train_scaled = scaler.fit_transform(X_train_imputed)\nX_val_scaled = scaler.transform(X_val_imputed)\n\nmodel = LogisticRegressionCV(cv=5, n_jobs=-1, random_state=42)\nmodel.fit(X_train_scaled, y_train)\nprint('Validation Accuracy', model.score(X_val_scaled, y_val))",
"(668, 7) (223, 7)\n(668, 11) (223, 11)\nValidation Accuracy 0.8071748878923767\n"
]
],
[
[
"Plot coefficients:",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\ncoefficients = pd.Series(model.coef_[0], X_train_encoded.columns)\ncoefficients.sort_values().plot.barh();",
"_____no_output_____"
]
],
[
[
"Generate [Kaggle](https://www.kaggle.com/c/titanic) submission:",
"_____no_output_____"
]
],
[
[
"X_test = test[features]\nX_test_encoded = encoder.transform(X_test)\nX_test_imputed = imputer.transform(X_test_encoded)\nX_test_scaled = scaler.transform(X_test_imputed)\ny_pred = model.predict(X_test_scaled)\nsubmission = test[['PassengerId']].copy()\nsubmission['Survived'] = y_pred\nsubmission.to_csv('titanic-submission-01.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae62e1abc9e927c34ceff300e9bf3f8e3634901
| 43,828 |
ipynb
|
Jupyter Notebook
|
vsm_02_dimreduce.ipynb
|
abgoswam/cs224u
|
33e1a22d1c9586b473f43b388163a74264e9258a
|
[
"Apache-2.0"
] | null | null | null |
vsm_02_dimreduce.ipynb
|
abgoswam/cs224u
|
33e1a22d1c9586b473f43b388163a74264e9258a
|
[
"Apache-2.0"
] | null | null | null |
vsm_02_dimreduce.ipynb
|
abgoswam/cs224u
|
33e1a22d1c9586b473f43b388163a74264e9258a
|
[
"Apache-2.0"
] | null | null | null | 33.817901 | 583 | 0.579812 |
[
[
[
"# Vector-space models: dimensionality reduction",
"_____no_output_____"
]
],
[
[
"__author__ = \"Christopher Potts\"\n__version__ = \"CS224u, Stanford, Spring 2020\"",
"_____no_output_____"
]
],
[
[
"## Contents\n\n1. [Overview](#Overview)\n1. [Set-up](#Set-up)\n1. [Latent Semantic Analysis](#Latent-Semantic-Analysis)\n 1. [Overview of the LSA method](#Overview-of-the-LSA-method)\n 1. [Motivating example for LSA](#Motivating-example-for-LSA)\n 1. [Applying LSA to real VSMs](#Applying-LSA-to-real-VSMs)\n 1. [Other resources for matrix factorization](#Other-resources-for-matrix-factorization)\n1. [GloVe](#GloVe)\n 1. [Overview of the GloVe method](#Overview-of-the-GloVe-method)\n 1. [GloVe implementation notes](#GloVe-implementation-notes)\n 1. [Applying GloVe to our motivating example](#Applying-GloVe-to-our-motivating-example)\n 1. [Testing the GloVe implementation](#Testing-the-GloVe-implementation)\n 1. [Applying GloVe to real VSMs](#Applying-GloVe-to-real-VSMs)\n1. [Autoencoders](#Autoencoders)\n 1. [Overview of the autoencoder method](#Overview-of-the-autoencoder-method)\n 1. [Testing the autoencoder implementation](#Testing-the-autoencoder-implementation)\n 1. [Applying autoencoders to real VSMs](#Applying-autoencoders-to-real-VSMs)\n1. [word2vec](#word2vec)\n 1. [Training data](#Training-data)\n 1. [Basic skip-gram](#Basic-skip-gram)\n 1. [Skip-gram with noise contrastive estimation ](#Skip-gram-with-noise-contrastive-estimation-)\n 1. [word2vec resources](#word2vec-resources)\n1. [Other methods](#Other-methods)\n1. [Exploratory exercises](#Exploratory-exercises)",
"_____no_output_____"
],
[
"## Overview\n\nThe matrix weighting schemes reviewed in the first notebook for this unit deliver solid results. However, they are not capable of capturing higher-order associations in the data. \n\nWith dimensionality reduction, the goal is to eliminate correlations in the input VSM and capture such higher-order notions of co-occurrence, thereby improving the overall space.\n\nAs a motivating example, consider the adjectives _gnarly_ and _wicked_ used as slang positive adjectives. Since both are positive, we expect them to be similar in a good VSM. However, at least stereotypically, _gnarly_ is Californian and _wicked_ is Bostonian. Thus, they are unlikely to occur often in the same texts, and so the methods we've reviewed so far will not be able to model their similarity. \n\nDimensionality reduction techniques are often capable of capturing such semantic similarities (and have the added advantage of shrinking the size of our data structures).",
"_____no_output_____"
],
[
"## Set-up\n\n* Make sure your environment meets all the requirements for [the cs224u repository](https://github.com/cgpotts/cs224u/). For help getting set-up, see [setup.ipynb](setup.ipynb).\n\n* Make sure you've downloaded [the data distribution for this course](http://web.stanford.edu/class/cs224u/data/data.tgz), unpacked it, and placed it in the current directory (or wherever you point `DATA_HOME` to below).",
"_____no_output_____"
]
],
[
[
"from mittens import GloVe\nimport numpy as np\nimport os\nimport pandas as pd\nimport scipy.stats\nfrom torch_autoencoder import TorchAutoencoder\nimport utils\nimport vsm",
"_____no_output_____"
],
[
"# Set all the random seeds for reproducibility:\n\nutils.fix_random_seeds()",
"_____no_output_____"
],
[
"DATA_HOME = os.path.join('data', 'vsmdata')",
"_____no_output_____"
],
[
"imdb5 = pd.read_csv(\n os.path.join(DATA_HOME, 'imdb_window5-scaled.csv.gz'), index_col=0)",
"_____no_output_____"
],
[
"imdb20 = pd.read_csv(\n os.path.join(DATA_HOME, 'imdb_window20-flat.csv.gz'), index_col=0)",
"_____no_output_____"
],
[
"giga5 = pd.read_csv(\n os.path.join(DATA_HOME, 'giga_window5-scaled.csv.gz'), index_col=0)",
"_____no_output_____"
],
[
"giga20 = pd.read_csv(\n os.path.join(DATA_HOME, 'giga_window20-flat.csv.gz'), index_col=0)",
"_____no_output_____"
]
],
[
[
"## Latent Semantic Analysis\n\nLatent Semantic Analysis (LSA) is a prominent dimensionality reduction technique. It is an application of __truncated singular value decomposition__ (SVD) and so uses only techniques from linear algebra (no machine learning needed).",
"_____no_output_____"
],
[
"### Overview of the LSA method\n\nThe central mathematical result is that, for any matrix of real numbers $X$ of dimension $m \\times n$, there is a factorization of $X$ into matrices $T$, $S$, and $D$ such that\n\n$$X_{m \\times n} = T_{m \\times m}S_{m\\times m}D_{n \\times m}^{\\top}$$\n\nThe matrices $T$ and $D$ are __orthonormal__ – their columns are length-normalized and orthogonal to one another (that is, they each have cosine distance of $1$ from each other). The singular-value matrix $S$ is a diagonal matrix arranged by size, so that the first dimension corresponds to the greatest source of variability in the data, followed by the second, and so on.\n\nOf course, we don't want to factorize and rebuild the original matrix, as that wouldn't get us anywhere. The __truncation__ part means that we include only the top $k$ dimensions of $S$. Given our row-oriented perspective on these matrices, this means using\n\n$$T[1{:}m, 1{:}k]S[1{:}k, 1{:}k]$$\n\nwhich gives us a version of $T$ that includes only the top $k$ dimensions of variation. \n\nTo build up intuitions, imagine that everyone on the Stanford campus is associated with a 3d point representing their position: $x$ is east–west, $y$ is north–south, and $z$ is zenith–nadir. Since the campus is spread out and has relatively few deep basements and tall buildings, the top two dimensions of variation will be $x$ and $y$, and the 2d truncated SVD of this space will leave $z$ out. This will, for example, capture the sense in which someone at the top of Hoover Tower is close to someone at its base.",
"_____no_output_____"
],
[
"### Motivating example for LSA\n\nWe can also return to our original motivating example of _wicked_ and _gnarly_. Here is a matrix reflecting those assumptions:",
"_____no_output_____"
]
],
[
[
"gnarly_df = pd.DataFrame(\n np.array([\n [1,0,1,0,0,0],\n [0,1,0,1,0,0],\n [1,1,1,1,0,0],\n [0,0,0,0,1,1],\n [0,0,0,0,0,1]], dtype='float64'),\n index=['gnarly', 'wicked', 'awesome', 'lame', 'terrible'])\n\ngnarly_df",
"_____no_output_____"
]
],
[
[
"No column context includes both _gnarly_ and _wicked_ together so our count matrix places them far apart:",
"_____no_output_____"
]
],
[
[
"vsm.neighbors('gnarly', gnarly_df)",
"_____no_output_____"
]
],
[
[
"Reweighting doesn't help. For example, here is the attempt with Positive PMI:",
"_____no_output_____"
]
],
[
[
"vsm.neighbors('gnarly', vsm.pmi(gnarly_df))",
"_____no_output_____"
]
],
[
[
"However, both words tend to occur with _awesome_ and not with _lame_ or _terrible_, so there is an important sense in which they are similar. LSA to the rescue:",
"_____no_output_____"
]
],
[
[
"gnarly_lsa_df = vsm.lsa(gnarly_df, k=2)",
"_____no_output_____"
],
[
"vsm.neighbors('gnarly', gnarly_lsa_df)",
"_____no_output_____"
]
],
[
[
"### Applying LSA to real VSMs\n\nHere's an example that begins to convey the effect that this can have empirically.\n\nFirst, the original count matrix:",
"_____no_output_____"
]
],
[
[
"vsm.neighbors('superb', imdb5).head()",
"_____no_output_____"
]
],
[
[
"And then LSA with $k=100$:",
"_____no_output_____"
]
],
[
[
"imdb5_svd = vsm.lsa(imdb5, k=100)",
"_____no_output_____"
],
[
"vsm.neighbors('superb', imdb5_svd).head()",
"_____no_output_____"
]
],
[
[
"A common pattern in the literature is to apply PMI first. The PMI values tend to give the count matrix a normal (Gaussian) distribution that better satisfies the assumptions underlying SVD:",
"_____no_output_____"
]
],
[
[
"imdb5_pmi = vsm.pmi(imdb5, positive=False)",
"_____no_output_____"
],
[
"imdb5_pmi_svd = vsm.lsa(imdb5_pmi, k=100)",
"_____no_output_____"
],
[
"vsm.neighbors('superb', imdb5_pmi_svd).head()",
"_____no_output_____"
]
],
[
[
"### Other resources for matrix factorization\n\nThe [sklearn.decomposition](http://scikit-learn.org/stable/modules/classes.html#module-sklearn.decomposition) module contains an implementation of LSA ([TruncatedSVD](http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.TruncatedSVD.html#sklearn.decomposition.TruncatedSVD)) that you might want to switch to for real experiments:\n\n* The `sklearn` version is more flexible than the above in that it can operate on both dense matrices (Numpy arrays) and sparse matrices (from Scipy).\n\n* The `sklearn` version will make it easy to try out other dimensionality reduction methods in your own code; [Principal Component Analysis (PCA)](http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html#sklearn.decomposition.PCA) and [Non-Negative Matrix Factorization (NMF)](http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.NMF.html#sklearn.decomposition.NMF) are closely related methods that are worth a look.",
"_____no_output_____"
],
[
"## GloVe\n\n### Overview of the GloVe method\n\n[Pennington et al. (2014)](http://www.aclweb.org/anthology/D/D14/D14-1162.pdf) introduce an objective function for semantic word representations. Roughly speaking, the objective is to learn vectors for words $w_{i}$ and $w_{j}$ such that their dot product is proportional to their probability of co-occurrence:\n\n$$w_{i}^{\\top}\\widetilde{w}_{k} + b_{i} + \\widetilde{b}_{k} = \\log(X_{ik})$$\n\nThe paper is exceptionally good at motivating this objective from first principles. In their equation (6), they define \n\n$$w_{i}^{\\top}\\widetilde{w}_{k} = \\log(P_{ik}) = \\log(X_{ik}) - \\log(X_{i})$$\n\nIf we allow that the rows and columns can be different, then we would do\n\n$$w_{i}^{\\top}\\widetilde{w}_{k} = \\log(P_{ik}) = \\log(X_{ik}) - \\log(X_{i} \\cdot X_{*k})$$\n\nwhere, as in the paper, $X_{i}$ is the sum of the values in row $i$, and $X_{*k}$ is the sum of the values in column $k$.\n\nThe rightmost expression is PMI by the equivalence $\\log(\\frac{x}{y}) = \\log(x) - \\log(y)$, and hence we can see GloVe as aiming to make the dot product of two learned vectors equal to the PMI!\n\nThe full model is a weighting of this objective:\n\n$$\\sum_{i, j=1}^{|V|} f\\left(X_{ij}\\right)\n \\left(w_i^\\top \\widetilde{w}_j + b_i + \\widetilde{b}_j - \\log X_{ij}\\right)^2$$\n\nwhere $V$ is the vocabulary and $f$ is a scaling factor designed to diminish the impact of very large co-occurrence counts:\n\n$$f(x) \n\\begin{cases}\n(x/x_{\\max})^{\\alpha} & \\textrm{if } x < x_{\\max} \\\\\n1 & \\textrm{otherwise}\n\\end{cases}$$\n\nTypically, $\\alpha$ is set to $0.75$ and $x_{\\max}$ to $100$ (though it is worth assessing how many of your non-zero counts are above this; in dense word $\\times$ word matrices, you could be flattening more than you want to).",
"_____no_output_____"
],
[
"### GloVe implementation notes\n\n* The implementation in `vsm.glove` is the most stripped-down, bare-bones version of the GloVe method I could think of. As such, it is quite slow. \n\n* The required [mittens](https://github.com/roamanalytics/mittens) package includes a vectorized implementation that is much, much faster, so we'll mainly use that. \n\n* For really large jobs, [the official C implementation released by the GloVe team](http://nlp.stanford.edu/projects/glove/) is probably the best choice.",
"_____no_output_____"
],
[
"### Applying GloVe to our motivating example\n\nGloVe should do well on our _gnarly/wicked_ evaluation, though you will see a lot variation due to the small size of this VSM:",
"_____no_output_____"
]
],
[
[
"gnarly_glove = vsm.glove(gnarly_df, n=5, max_iter=1000)",
"Stopping at iteration 592 with error 9.987450681981012e-05\n"
],
[
"vsm.neighbors('gnarly', gnarly_glove)",
"_____no_output_____"
]
],
[
[
"### Testing the GloVe implementation\n\nIt is not easy analyze GloVe values derived from real data, but the following little simulation suggests that `vsm.glove` is working as advertised: it does seem to reliably deliver vectors whose dot products are proportional to the log co-occurrence probability:",
"_____no_output_____"
]
],
[
[
"glove_test_count_df = pd.DataFrame(\n np.array([\n [10.0, 2.0, 3.0, 4.0],\n [ 2.0, 10.0, 4.0, 1.0],\n [ 3.0, 4.0, 10.0, 2.0],\n [ 4.0, 1.0, 2.0, 10.0]]),\n index=['A', 'B', 'C', 'D'],\n columns=['A', 'B', 'C', 'D'])",
"_____no_output_____"
],
[
"glove_test_df = vsm.glove(glove_test_count_df, max_iter=1000, n=4)",
"Stopping at iteration 531 with error 9.969189369208185e-05\n"
],
[
"def correlation_test(true, pred): \n mask = true > 0\n M = pred.dot(pred.T)\n with np.errstate(divide='ignore'):\n log_cooccur = np.log(true)\n log_cooccur[np.isinf(log_cooccur)] = 0.0\n row_log_prob = np.log(true.sum(axis=1))\n row_log_prob = np.outer(row_log_prob, np.ones(true.shape[1]))\n prob = log_cooccur - row_log_prob\n return np.corrcoef(prob[mask], M[mask])[0, 1]",
"_____no_output_____"
],
[
"correlation_test(glove_test_count_df.values, glove_test_df.values)",
"_____no_output_____"
]
],
[
[
"### Applying GloVe to real VSMs",
"_____no_output_____"
],
[
"The `vsm.glove` implementation is too slow to use on real matrices. The distribution in the `mittens` package is significantly faster, making its use possible even without a GPU (and it will be very fast indeed on a GPU machine):",
"_____no_output_____"
]
],
[
[
"glove_model = GloVe()\n\nimdb5_glv = glove_model.fit(imdb5.values)\n\nimdb5_glv = pd.DataFrame(imdb5_glv, index=imdb5.index)",
"WARNING:tensorflow:From C:\\Users\\agoswami\\AppData\\Local\\Continuum\\anaconda3\\envs\\nlu\\lib\\site-packages\\tensorflow_core\\python\\ops\\resource_variable_ops.py:1635: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.\nInstructions for updating:\nIf using Keras pass *_constraint arguments to layers.\nWARNING:tensorflow:From C:\\Users\\agoswami\\AppData\\Local\\Continuum\\anaconda3\\envs\\nlu\\lib\\site-packages\\tensorflow_core\\python\\training\\adagrad.py:76: calling Constant.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\n"
],
[
"vsm.neighbors('superb', imdb5_glv).head()",
"_____no_output_____"
]
],
[
[
"## Autoencoders\n\nAn autoencoder is a machine learning model that seeks to learn parameters that predict its own input. This is meaningful when there are intermediate representations that have lower dimensionality than the inputs. These provide a reduced-dimensional view of the data akin to those learned by LSA, but now we have a lot more design choices and a lot more potential to learn higher-order associations in the underyling data.",
"_____no_output_____"
],
[
"### Overview of the autoencoder method\n\nThe module `torch_autoencoder` uses PyToch to implement a simple one-layer autoencoder:\n\n$$\n\\begin{align}\nh &= \\mathbf{f}(xW + b_{h}) \\\\\n\\widehat{x} &= hW^{\\top} + b_{x}\n\\end{align}$$\n\nHere, we assume that the hidden representation $h$ has a low dimensionality like 100, and that $\\mathbf{f}$ is a non-linear activation function (the default for `TorchAutoencoder` is `tanh`). These are the major design choices internal to the network. It might also be meaningful to assume that there are two matrices of weights $W_{xh}$ and $W_{hx}$, rather than using $W^{\\top}$ for the output step.\n\nThe objective function for autoencoders will implement some kind of assessment of the distance between the inputs and their predicted outputs. For example, one could use the one-half mean squared error:\n\n$$\\frac{1}{m}\\sum_{i=1}^{m} \\frac{1}{2}(\\widehat{X[i]} - X[i])^{2}$$\n\nwhere $X$ is the input matrix of examples (dimension $m \\times n$) and $X[i]$ corresponds to the $i$th example.\n\nWhen you call the `fit` method of `TorchAutoencoder`, it returns the matrix of hidden representations $h$, which is the new embedding space: same row count as the input, but with the column count set by the `hidden_dim` parameter.\n\nFor much more on autoencoders, see the 'Autoencoders' chapter of [Goodfellow et al. 2016](http://www.deeplearningbook.org).",
"_____no_output_____"
],
[
"### Testing the autoencoder implementation\n\nHere's an evaluation that is meant to test the autoencoder implementation – we expect it to be able to full encode the input matrix because we know its rank is equal to the dimensionality of the hidden representation.",
"_____no_output_____"
]
],
[
[
"def randmatrix(m, n, sigma=0.1, mu=0):\n return sigma * np.random.randn(m, n) + mu\n\ndef autoencoder_evaluation(nrow=1000, ncol=100, rank=20, max_iter=20000):\n \"\"\"This an evaluation in which `TfAutoencoder` should be able\n to perfectly reconstruct the input data, because the\n hidden representations have the same dimensionality as\n the rank of the input matrix.\n \"\"\"\n X = randmatrix(nrow, rank).dot(randmatrix(rank, ncol))\n ae = TorchAutoencoder(hidden_dim=rank, max_iter=max_iter)\n ae.fit(X)\n X_pred = ae.predict(X)\n mse = (0.5 * (X_pred - X)**2).mean()\n return(X, X_pred, mse)",
"_____no_output_____"
],
[
"ae_max_iter = 100\n\n_, _, ae = autoencoder_evaluation(max_iter=ae_max_iter)\n\nprint(\"Autoencoder evaluation MSE after {0} evaluations: {1:0.04f}\".format(ae_max_iter, ae))",
"Finished epoch 100 of 100; error is 0.00022285695013124496"
]
],
[
[
"### Applying autoencoders to real VSMs\n\nYou can apply the autoencoder directly to the count matrix, but this could interact very badly with the internal activation function: if the counts are all very high or very low, then everything might get pushed irrevocably towards the extreme values of the activation.\n\nThus, it's a good idea to first normalize the values somehow. Here, I use `vsm.length_norm`:",
"_____no_output_____"
]
],
[
[
"imdb5_l2 = imdb5.apply(vsm.length_norm, axis=1)",
"_____no_output_____"
],
[
"imdb5_l2_ae = TorchAutoencoder(\n max_iter=100, hidden_dim=50, eta=0.001).fit(imdb5_l2)",
"Finished epoch 100 of 100; error is 0.00019485459779389203"
],
[
"vsm.neighbors('superb', imdb5_l2_ae).head()",
"_____no_output_____"
]
],
[
[
"This is very slow and seems not to work all that well. To speed things up, one can first apply LSA or similar:",
"_____no_output_____"
]
],
[
[
"imdb5_l2_svd100 = vsm.lsa(imdb5_l2, k=100)",
"_____no_output_____"
],
[
"imdb_l2_svd100_ae = TorchAutoencoder(\n max_iter=1000, hidden_dim=50, eta=0.01).fit(imdb5_l2_svd100)",
"Finished epoch 1000 of 1000; error is 0.00010619893873808905"
],
[
"vsm.neighbors('superb', imdb_l2_svd100_ae).head()",
"_____no_output_____"
]
],
[
[
"## word2vec\n\nThe label __word2vec__ picks out a family of models in which the embedding for a word $w$ is trained to predict the words that co-occur with $w$. This intuition can be cashed out in numerous ways. Here, we review just the __skip-gram model__, due to [Mikolov et al. 2013](https://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality).",
"_____no_output_____"
],
[
"### Training data\n\nThe most natural starting point is to transform a corpus into a supervised data set by mapping each word to a subset (maybe all) of the words that it occurs with in a given window. Schematically:\n\n__Corpus__: `it was the best of times, it was the worst of times, ...`\n\nWith window size 2:\n\n```\n(it, was)\n(it, the)\n(was, it)\n(was, the)\n(was, best)\n(the, was)\n(the, it)\n(the, best)\n(the, of)\n...\n```",
"_____no_output_____"
],
[
"### Basic skip-gram\n\nThe basic skip-gram model estimates the probability of an input–output pair $(a, b)$ as\n\n$$P(b \\mid a) = \\frac{\\exp(x_{a}w_{b})}{\\sum_{b'\\in V}\\exp(x_{a}w_{b'})}$$\n\nwhere $x_{a}$ is the row (word) vector representation of word $a$ and $w_{b}$ is the column (context) vector representation of word $b$. The objective is to minimize the following quantity:\n\n$$\n-\\sum_{i=1}^{m}\\sum_{k=1}^{|V|}\n\\textbf{1}\\{c_{i}=k\\} \n\\log\n\\frac{\n \\exp(x_{i}w_{k})\n}{\n \\sum_{j=1}^{|V|}\\exp(x_{i}w_{j})\n}$$\n\nwhere $V$ is the vocabulary.\n\nThe inputs $x_{i}$ are the word representations, which get updated during training, and the outputs are one-hot vectors $c$. For example, if `was` is the 560th element in the vocab, then the output $c$ for the first example in the corpus above would be a vector of all $0$s except for a $1$ in the 560th position. $x$ would be the representation of `it` in the embedding space. \n\nThe distribution over the entire output space for a given input word $a$ is thus a standard softmax classifier; here we add a bias term for good measure:\n\n$$c = \\textbf{softmax}(x_{a}W + b)$$\n\nIf we think of this model as taking the entire matrix $X$ as input all at once, then it becomes\n\n$$c = \\textbf{softmax}(XW + b)$$\n\nand it is now very clear that we are back to the core insight that runs through all of our reweighting and dimensionality reduction methods: we have a word matrix $X$ and a context matrix $W$, and we are trying to push the dot products of these two embeddings in a specific direction: here, to maximize the likelihood of the observed co-occurrences in the corpus.",
"_____no_output_____"
],
[
"### Skip-gram with noise contrastive estimation \n\nTraining the basic skip-gram model directly is extremely expensive for large vocabularies, because $W$, $b$, and the outputs $c$ get so large. \n\nA straightforward way to address this is to change the objective to use __noise contrastive estimation__ (negative sampling). Where $\\mathcal{D}$ is the original training corpus and $\\mathcal{D}'$ is a sample of pairs not in the corpus, we minimize\n\n$$\\sum_{a, b \\in \\mathcal{D}}-\\log\\sigma(x_{a}w_{b}) + \\sum_{a, b \\in \\mathcal{D}'}\\log\\sigma(x_{a}w_{b})$$\n\nwith $\\sigma$ the sigmoid activation function $\\frac{1}{1 + \\exp(-x)}$.\n\nThe advice of Mikolov et al. is to sample $\\mathcal{D}'$ proportional to a scaling of the frequency distribution of the underlying vocabulary in the corpus:\n\n$$P(w) = \\frac{\\textbf{count}(w)^{0.75}}{\\sum_{w'\\in V} \\textbf{count}(w')}$$\n\nwhere $V$ is the vocabulary.\n\nAlthough this new objective function is a substantively different objective than the previous one, Mikolov et al. (2013) say that it should approximate it, and it is building on the same insight about words and their contexts. See [Levy and Golberg 2014](http://papers.nips.cc/paper/5477-neural-word-embedding-as-implicit-matrix-factorization) for a proof that this objective reduces to PMI shifted by a constant value. See also [Cotterell et al. 2017](https://aclanthology.coli.uni-saarland.de/papers/E17-2028/e17-2028) for an interpretation of this model as a variant of PCA.",
"_____no_output_____"
],
[
"### word2vec resources\n\n* In the usual presentation, word2vec training involves looping repeatedly over the sequence of tokens in the corpus, sampling from the context window from each word to create the positive training pairs. I assume that this same process could be modeled by sampling (row, column) index pairs from our count matrices proportional to their cell values. However, I couldn't get this to work well. I'd be grateful if someone got it work or figured out why it won't!\n\n* Luckily, there are numerous excellent resources for word2vec. [The TensorFlow tutorial Vector representations of words](https://www.tensorflow.org/tutorials/word2vec) is very clear and links to code that is easy to work with. Because TensorFlow has a built in loss function called `tf.nn.nce_loss`, it is especially simple to define these models – one pretty much just sets up an embedding $X$, a context matrix $W$, and a bias $b$, and then feeds them plus a training batch to the loss function.\n\n* The excellent [Gensim package](https://radimrehurek.com/gensim/) has an implementation that handles the scalability issues related to word2vec.",
"_____no_output_____"
],
[
"## Other methods\n\nLearning word representations is one of the most active areas in NLP right now, so I can't hope to offer a comprehensive summary. I'll settle instead for identifying some overall trends and methods:\n\n* The LexVec model of [Salle et al. 2016](https://aclanthology.coli.uni-saarland.de/papers/P16-2068/p16-2068) combines the core insight of GloVe (learn vectors that approximate PMI) with the insight from word2vec that we should additionally try to push words that don't appear together farther apart in the VSM. (GloVe simply ignores 0 count cells and so can't do this.)\n\n* There is growing awareness that many apparently diverse models can be expressed as matrix factorization methods like SVD/LSA. See especially \n[Singh and Gordon 2008](http://www.cs.cmu.edu/~ggordon/singh-gordon-unified-factorization-ecml.pdf),\n[Levy and Golberg 2014](http://papers.nips.cc/paper/5477-neural-word-embedding-as-implicit-matrix-factorization), [Cotterell et al. 2017](https://www.aclweb.org/anthology/E17-2028/).\n\n* Subword modeling ([reviewed briefly in the previous notebook](vsm_01_distributional.ipynb#Subword-information)) is increasingly yielding dividends. (It would already be central if most of NLP focused on languages with complex morphology!) Check out the papers at the Subword and Character-Level Models for NLP Workshops: [SCLeM 2017](https://sites.google.com/view/sclem2017/home), [SCLeM 2018](https://sites.google.com/view/sclem2018/home).\n\n* Contextualized word representations have proven valuable in many contexts. These methods do not provide representations for individual words, but rather represent them in their linguistic context. This creates space for modeling how word senses vary depending on their context of use. We will study these methods later in the quarter, mainly in the context of identifying ways that might achieve better results on your projects.",
"_____no_output_____"
],
[
"## Exploratory exercises\n\nThese are largely meant to give you a feel for the material, but some of them could lead to projects and help you with future work for the course. These are not for credit.\n\n1. Try out some pipelines of reweighting, `vsm.lsa` at various dimensions, and `TorchAutoencoder` to see which seems best according to your sampling around with `vsm.neighbors` and high-level visualization with `vsm.tsne_viz`. Feel free to use other factorization methods defined in [sklearn.decomposition](http://scikit-learn.org/stable/modules/classes.html#module-sklearn.decomposition) as well.\n\n1. What happens if you set `k=1` using `vsm.lsa`? What do the results look like then? What do you think this first (and now only) dimension is capturing?\n\n1. Modify `vsm.glove` so that it uses [the AdaGrad optimization method](http://www.jmlr.org/papers/volume12/duchi11a/duchi11a.pdf) as in the original paper. It's fine to use [the authors' implementation](http://nlp.stanford.edu/projects/glove/), [Jon Gauthier's implementation](http://www.foldl.me/2014/glove-python/), or the [mittens Numpy implementation](https://github.com/roamanalytics/mittens/blob/master/mittens/np_mittens.py) as references, but you might enjoy the challenge of doing this with no peeking at their code.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4ae63a44dc3adf8aebab040c72b93d63da0a9190
| 203,352 |
ipynb
|
Jupyter Notebook
|
ecg_classification.ipynb
|
jarek-pawlowski/machine-learning-applications
|
3181508a5e70041daf10af42cfb82d94e122defd
|
[
"MIT"
] | null | null | null |
ecg_classification.ipynb
|
jarek-pawlowski/machine-learning-applications
|
3181508a5e70041daf10af42cfb82d94e122defd
|
[
"MIT"
] | null | null | null |
ecg_classification.ipynb
|
jarek-pawlowski/machine-learning-applications
|
3181508a5e70041daf10af42cfb82d94e122defd
|
[
"MIT"
] | null | null | null | 156.184332 | 58,969 | 0.81949 |
[
[
[
"<a href=\"https://colab.research.google.com/github/jarek-pawlowski/machine-learning-applications/blob/main/ecg_classification.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Heart beats classification problem\nA typical task for applied Machine Learning in medicine is an automatic classification of signals from diagnostic devices such as ECG or EEG\n\nTypical pipeline:\n- detect QRS compexes (beats)\n- classify them:\n> - normal beat N\n> - arrhytmia, e.g. *venticular* V, *supraventicular* S arrytmia, or *artial fibrillation* AF\n\n\n\n\n\na couple of links:\n- [exemplary challenge from Physionet](https://physionet.org/content/challenge-2017/1.0.0/)\n- [some recent paper on ECG classification](https://doi.org/10.1016/j.knosys.2020.106589)\n\n## our challenge: classify beats as normal or abnormal (arrhytmia)\n- we will utilize signals from **svdb** database, and grab subsequent beats (data preprocessing)\n- then construct binary classifier using NN, decision trees, ensemble metchods, and SVM or NaiveBayes",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Dataset preparation\n1. Download ecg waves from **svdb** database provided by *PhysioNet*\n2. Divide signals into samples, each containing single heartbeat (with window size of 96 points, *sampling ratio* = 128 points/s)\n3. Take only samples annotated as 'N' (normal beat), or 'S' and 'V' (arrhythmias)",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\n\n# install PhysioNet ecg data package \n!pip install wfdb\nimport wfdb\n\n# list of available datasets\ndbs = wfdb.get_dbs()\ndisplay(dbs)\n\n# we choose svdb\nsvdb_dir = os.path.join(os.getcwd(), 'svdb_dir')\nwfdb.dl_database('svdb', dl_dir=svdb_dir)\n\n# Display the downloaded content\nsvdb_in_files = [os.path.splitext(f)[0] for f in os.listdir(svdb_dir) if f.endswith('.dat')]\nprint(svdb_in_files)",
"Requirement already satisfied: wfdb in /usr/local/lib/python3.7/dist-packages (3.4.1)\nRequirement already satisfied: numpy>=1.10.1 in /usr/local/lib/python3.7/dist-packages (from wfdb) (1.21.5)\nRequirement already satisfied: pandas>=0.17.0 in /usr/local/lib/python3.7/dist-packages (from wfdb) (1.3.5)\nRequirement already satisfied: matplotlib>=3.3.4 in /usr/local/lib/python3.7/dist-packages (from wfdb) (3.5.1)\nRequirement already satisfied: scipy>=0.17.0 in /usr/local/lib/python3.7/dist-packages (from wfdb) (1.4.1)\nRequirement already satisfied: requests>=2.8.1 in /usr/local/lib/python3.7/dist-packages (from wfdb) (2.23.0)\nRequirement already satisfied: python-dateutil>=2.7 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.3.4->wfdb) (2.8.2)\nRequirement already satisfied: pyparsing>=2.2.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.3.4->wfdb) (3.0.7)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.3.4->wfdb) (0.11.0)\nRequirement already satisfied: pillow>=6.2.0 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.3.4->wfdb) (7.1.2)\nRequirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.3.4->wfdb) (4.31.1)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.3.4->wfdb) (1.3.2)\nRequirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.7/dist-packages (from matplotlib>=3.3.4->wfdb) (21.3)\nRequirement already satisfied: pytz>=2017.3 in /usr/local/lib/python3.7/dist-packages (from pandas>=0.17.0->wfdb) (2018.9)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/dist-packages (from python-dateutil>=2.7->matplotlib>=3.3.4->wfdb) (1.15.0)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests>=2.8.1->wfdb) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests>=2.8.1->wfdb) (2021.10.8)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests>=2.8.1->wfdb) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests>=2.8.1->wfdb) (2.10)\n"
],
[
"time_window = 48\nall_beats = []\nall_annotations = []\nfor in_file in svdb_in_files:\n print('...processing...' + in_file + '...file')\n signal, fields = wfdb.rdsamp(os.path.join(svdb_dir,in_file), channels=[0])\n annotations = wfdb.rdann(os.path.join(svdb_dir,in_file), 'atr')\n signal=np.array(signal).flatten()\n # grab subsequent heartbeats within [position-48,position+48] window\n beats = np.zeros((len(annotations.sample[5:-5]), time_window*2))\n # note that we remove first and last few beats to ensure that all beats have equal lengths\n for i, ann_position in enumerate(annotations.sample[5:-5]):\n beats[i] = signal[ann_position-time_window:ann_position+time_window]\n all_beats.append(beats)\n # consequently, we remove first and last few annotations\n all_annotations.append(annotations.symbol[5:-5])\n\nall_beats = np.concatenate(all_beats)\nall_annotations = np.concatenate(all_annotations)\n\n# check which annotations are usable for us, are of N or S or V class\nindices = [i for i, ann in enumerate(all_annotations) if ann in {'N','S','V'}]\n# and get only these\nall_beats = all_beats[indices]\nall_annotations = np.array([all_annotations[i] for i in indices])\n\n# print data statistics\nprint(all_beats.shape, all_annotations.shape)\nprint('no of N beats: ' + str(np.count_nonzero(all_annotations == 'N')))\nprint('no of S beats: ' + str(np.count_nonzero(all_annotations == 'S')))\nprint('no of V beats: ' + str(np.count_nonzero(all_annotations == 'V')))",
"...processing...888...file\n...processing...811...file\n...processing...824...file\n...processing...859...file\n...processing...868...file\n...processing...846...file\n...processing...878...file\n...processing...864...file\n...processing...887...file\n...processing...851...file\n...processing...823...file\n...processing...805...file\n...processing...870...file\n...processing...871...file\n...processing...829...file\n...processing...826...file\n...processing...842...file\n...processing...863...file\n...processing...800...file\n...processing...812...file\n...processing...807...file\n...processing...849...file\n...processing...883...file\n...processing...848...file\n...processing...809...file\n...processing...858...file\n...processing...840...file\n...processing...845...file\n...processing...857...file\n...processing...821...file\n...processing...810...file\n...processing...894...file\n...processing...867...file\n...processing...889...file\n...processing...853...file\n...processing...808...file\n...processing...866...file\n...processing...844...file\n...processing...855...file\n...processing...876...file\n...processing...802...file\n...processing...890...file\n...processing...843...file\n...processing...820...file\n...processing...874...file\n...processing...877...file\n...processing...854...file\n...processing...841...file\n...processing...860...file\n...processing...852...file\n...processing...822...file\n...processing...803...file\n...processing...875...file\n...processing...873...file\n...processing...828...file\n...processing...881...file\n...processing...827...file\n...processing...893...file\n...processing...884...file\n...processing...850...file\n...processing...861...file\n...processing...801...file\n...processing...806...file\n...processing...865...file\n...processing...892...file\n...processing...879...file\n...processing...825...file\n...processing...804...file\n...processing...882...file\n...processing...869...file\n...processing...856...file\n...processing...847...file\n...processing...862...file\n...processing...891...file\n...processing...880...file\n...processing...886...file\n...processing...872...file\n...processing...885...file\n(183707, 96) (183707,)\nno of N beats: 161653\nno of S beats: 12146\nno of V beats: 9908\n"
],
[
"# show example samples\n!pip install matplotlib==3.1.3\nimport matplotlib.pyplot as plt\n\nfig, ax = plt.subplots(1,3)\nfig.set_size_inches(15, 3)\nplt.subplots_adjust(wspace=0.2)\nprint(all_annotations[:100])\nsample_number = [0,6,8]\nfor i, sn in enumerate(sample_number):\n ax[i].plot(all_beats[sn])\n ax[i].set(xlabel='time', ylabel='ecg signal', title='beat type ' + all_annotations[sn])\n ax[i].grid()\nplt.show()",
"['N' 'N' 'N' 'N' 'N' 'N' 'V' 'N' 'S' 'N' 'N' 'N' 'V' 'N' 'N' 'N' 'N' 'N'\n 'S' 'N' 'N' 'N' 'S' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'V'\n 'N' 'N' 'N' 'N' 'S' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N'\n 'S' 'N' 'N' 'N' 'N' 'N' 'N' 'S' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N'\n 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N'\n 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N' 'N']\n"
]
],
[
[
"# Experiments\n\n0. Preliminaries\n> - Divide dataset into train/validation/test subset, and normalize each of them. \n> - Define classification accuracy metrics (dataset is imbalanced)\n>>Confusion matrix\n```\n____Prediction\nT | n s v\nr |N Nn Ns Nv\nu |S Sn Ss Sv\nt |V Vn Vs Vv\nh | \n```\n>> - Total accuracy\n$Acc_T = \\frac{Nn+Ss+Vv}{\\Sigma_N+\\Sigma_S+\\Sigma_V}$,\n>> - Arrhythmia accuracy (S or V cases are more important to be detected):\n$Acc_A = \\frac{Ss+Vv}{\\Sigma_S+\\Sigma_V}$,\n>> - $\\Sigma_N=Nn+Ns+Nv$, $\\Sigma_S=Sn+Ss+Sv$,\n$\\Sigma_V=Vn+Vs+Vv$\n\n1. Standard classifiers: *naive Bayes* and *SVM*\n2. Decision Tree with optimized max_depth\n3. Random Forest with vector of features",
"_____no_output_____"
]
],
[
[
"# prepare datasets and define error metrics\nfrom sklearn import preprocessing\nfrom sklearn.model_selection import train_test_split\n\n# to simplify experiments and speedup training \n# we take only some part of the whole dataset\nX, y = all_beats[::10], all_annotations[::10]\n\n# train/validation/test set splitting \nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.15, random_state=0)\nX_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=0.15/0.85, random_state=0)\nprint(len(y_train), len(y_val), len(y_test))\n\n# perform data normalization: z = (x - u)/s\nscaler = preprocessing.StandardScaler().fit(X_train)\nX_train = scaler.transform(X_train)\n# same for the validation subset\nX_val = preprocessing.StandardScaler().fit_transform(X_val)\n# and for the test subset\nX_test = preprocessing.StandardScaler().fit_transform(X_test)\n\n# define accuracy\ndef calculate_accuracy(y_pred, y_gt, comment='', printout=True):\n acc_t = np.count_nonzero(y_pred == y_gt)/len(y_gt)\n acc_a = np.count_nonzero(\n np.logical_and(y_pred == y_gt, y_gt != 'N'))/np.count_nonzero(y_gt != 'N')\n if printout is True:\n print('-----------------------------------') \n print(comment)\n print('Total accuracy, Acc_T = {:.4f}'.format(acc_t))\n print('Arrhythmia accuracy, Acc_A = {:.4f}'.format(acc_a))\n print('-----------------------------------')\n else: return acc_t, acc_a ",
"12859 2756 2756\n"
],
[
"from sklearn.naive_bayes import GaussianNB\nfrom sklearn.svm import SVC\n\ngnb = GaussianNB()\ny_pred = gnb.fit(X_train, y_train).predict(X_test)\ncalculate_accuracy(y_pred, y_test, comment='naive Bayes classifier')\n\nsvc = SVC()\ny_pred = svc.fit(X_train, y_train).predict(X_test)\ncalculate_accuracy(y_pred, y_test, comment='SVM classifier')\n\nsvc = SVC(class_weight='balanced')\ny_pred = svc.fit(X_train, y_train).predict(X_test)\ncalculate_accuracy(y_pred, y_test, comment='balanced SVM classifier')",
"-----------------------------------\nnaive Bayes classifier\nTotal accuracy, Acc_T = 0.8251\nArrhythmia accuracy, Acc_A = 0.3919\n-----------------------------------\n-----------------------------------\nSVM classifier\nTotal accuracy, Acc_T = 0.9383\nArrhythmia accuracy, Acc_A = 0.5303\n-----------------------------------\n-----------------------------------\nbalanced SVM classifier\nTotal accuracy, Acc_T = 0.9224\nArrhythmia accuracy, Acc_A = 0.7954\n-----------------------------------\n"
]
],
[
[
"Summary of this part:\n1. The goal is to maximize both metrics Acc_T and Acc_A at the same time\n1. naive Bayes performs rather poorly\n> - problem with data imbalace\n2. SVM has simillar problem, but after data balacing works quite good",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier\n\ndtc = DecisionTreeClassifier(criterion='entropy', \n class_weight='balanced', \n min_samples_leaf=10)\ny_pred = dtc.fit(X_train, y_train).predict(X_test)\ncalculate_accuracy(y_pred, y_test, comment='balanced DT')\n\n# tunning max_dept hyperparameter (DT likes to overfit)\ntrain_acc_t = []\ntrain_acc_a = []\nval_acc_t = []\nval_acc_a = []\ndepth_range = range(1,26)\nfor max_depth in depth_range:\n dtc = DecisionTreeClassifier(criterion='entropy', \n class_weight='balanced', \n min_samples_leaf=10,\n max_depth=max_depth)\n dt_fit = dtc.fit(X_train, y_train)\n y_pred_train = dt_fit.predict(X_train)\n y_pred_val = dt_fit.predict(X_val) \n acc_t_train, acc_a_train = calculate_accuracy(y_pred_train, y_train, printout=False)\n acc_t_val, acc_a_val = calculate_accuracy(y_pred_val, y_val, printout=False)\n train_acc_t.append(acc_t_train)\n train_acc_a.append(acc_a_train)\n val_acc_t.append(acc_t_val)\n val_acc_a.append(acc_a_val)\n print('{0:d} {1:.4f} {2:4.4f}'.format(max_depth, acc_t_val, acc_a_val))\n\nimport matplotlib.pyplot as plt\n_, ax = plt.subplots()\nax.plot(depth_range, train_acc_t, label='train acc_t')\nax.plot(depth_range, train_acc_a, label='train acc_a')\nax.plot(depth_range, val_acc_t, label='validation acc_t')\nax.plot(depth_range, val_acc_a , label='validation acc_a')\nax.set(xlabel='max_depth', ylabel='accuracy')\nax.xaxis.set_ticks([1, 5, 10, 15, 20, 25])\nax.legend()\nplt.show()",
"-----------------------------------\nbalanced DT\nTotal accuracy, Acc_T = 0.8512\nArrhythmia accuracy, Acc_A = 0.7291\n-----------------------------------\n1 0.1001 0.7562\n2 0.1121 0.8466\n3 0.8875 0.4493\n4 0.8345 0.4822\n5 0.8110 0.5945\n6 0.7812 0.6466\n7 0.8237 0.6247\n8 0.8618 0.6466\n9 0.8650 0.6438\n10 0.8759 0.6466\n11 0.7656 0.7178\n12 0.7805 0.7288\n13 0.7972 0.7205\n14 0.7605 0.7315\n15 0.8153 0.7178\n16 0.8091 0.7205\n17 0.8320 0.7233\n18 0.8208 0.7178\n19 0.8335 0.7068\n20 0.8364 0.7096\n21 0.8345 0.7096\n22 0.8389 0.7123\n23 0.8371 0.7068\n24 0.8411 0.7068\n25 0.8403 0.7123\n"
],
[
"# optimum acc_a max_depth\ndtc = DecisionTreeClassifier(criterion='entropy', \n class_weight='balanced',\n min_samples_leaf=10,\n max_depth=10)\ny_pred = dtc.fit(X_train, y_train).predict(X_test)\ncalculate_accuracy(y_pred, y_test, comment='DT: Acc_A maximized')\n\n# optimum acc_t & acc_a max_depth\ndtc = DecisionTreeClassifier(criterion='entropy', \n class_weight='balanced',\n min_samples_leaf=10,\n max_depth=14)\ny_pred = dtc.fit(X_train, y_train).predict(X_test)\ncalculate_accuracy(y_pred, y_test, comment='DT: Acc_T + Acc_A maximized')",
"-----------------------------------\nDT: Acc_A maximized\nTotal accuracy, Acc_T = 0.8813\nArrhythmia accuracy, Acc_A = 0.6455\n-----------------------------------\n-----------------------------------\nDT: Acc_T + Acc_A maximized\nTotal accuracy, Acc_T = 0.7587\nArrhythmia accuracy, Acc_A = 0.7579\n-----------------------------------\n"
],
[
"# feature vector via PCA (dimensionlality reduction) works poorly\nfrom sklearn.decomposition import PCA\npca = PCA(n_components=15)\nX_train_ = pca.fit_transform(X_train)\nX_test_ = pca.transform(X_test)\n\ndtc = DecisionTreeClassifier(criterion='entropy', \n class_weight='balanced', \n min_samples_leaf=10,\n max_depth=10)\ny_pred = dtc.fit(X_train_, y_train).predict(X_test_)\ncalculate_accuracy(y_pred, y_test, comment='DT with PCA')",
"-----------------------------------\nDT with PCA\nTotal accuracy, Acc_T = 0.5439\nArrhythmia accuracy, Acc_A = 0.4092\n-----------------------------------\n"
]
],
[
[
"Summary:\n1. Decision Tree works a bit worse (than SVM) and has tendency to overfit. We consider two types of hyperparameters:\n> - *max_depth*\n> - *min_samples_leaf*\n2. Tunning *max_depth* gives Acc_A (*max_depth*=8), or Acc_T & Acc_A (*max_depth*=13) maximum value\n3. Simple dimensionality reduction using PCA works rather poorly",
"_____no_output_____"
]
],
[
[
"import pywt\n\n# extract features using different wavelets and simple differences\ndef extract_features(input_sample):\n out = np.array([])\n# sym8\n cA = pywt.downcoef('a', input_sample, 'sym8', level=4, mode='per')\n out = np.append(out,cA)\n cD = pywt.downcoef('d', input_sample, 'sym8', level=4, mode='per')\n out = np.append(out,cD)\n# db6/9\n cA = pywt.downcoef('a', input_sample, 'db6', level=4, mode='per')\n out = np.append(out,cA)\n cD = pywt.downcoef('d', input_sample, 'db6', level=4, mode='per')\n out = np.append(out,cD)\n cA = pywt.downcoef('a', input_sample, 'db9', level=4, mode='per')\n out = np.append(out,cA)\n cD = pywt.downcoef('d', input_sample, 'db9', level=4, mode='per')\n out = np.append(out,cD)\n# dmey\n cA = pywt.downcoef('a', input_sample, 'dmey', level=4, mode='per')\n out = np.append(out,cA)\n cD = pywt.downcoef('d', input_sample, 'dmey', level=4, mode='per')\n out = np.append(out,cD)\n\n# differences\n differences = np.zeros(16)\n for i, t in enumerate(range(40, 56)):\n differences[i] = input_sample[t+1]-input_sample[t]\n out = np.append(out,differences)\n return out\n\n# collect vector of features for all samples\ndef data_features(input_data):\n return np.array([extract_features(sample) for sample in input_data])\n\nX_train_ = data_features(X_train)\nprint(X_train_.shape)\nX_test_ = data_features(X_test)\nprint(X_test_.shape)\n\ndtc = DecisionTreeClassifier(criterion='entropy', \n class_weight='balanced', \n min_samples_leaf=10,\n max_depth=15)\ny_pred = dtc.fit(X_train_, y_train).predict(X_test_)\ncalculate_accuracy(y_pred, y_test, comment='DT with wavelets')\n\nfrom sklearn.ensemble import RandomForestClassifier\n\nrfc = RandomForestClassifier(criterion='entropy', \n n_estimators=500, \n max_depth=10, \n class_weight='balanced')\ny_pred = rfc.fit(X_train_, y_train).predict(X_test_)\ncalculate_accuracy(y_pred, y_test, comment='RF with wavelets')\n\nfrom sklearn.ensemble import AdaBoostClassifier\n\nabc = AdaBoostClassifier(n_estimators=200)\ny_pred = abc.fit(X_train_, y_train).predict(X_test_)\ncalculate_accuracy(y_pred, y_test, comment='Ada with wavelets')",
"(12859, 64)\n(2756, 64)\n-----------------------------------\nDT with wavelets\nTotal accuracy, Acc_T = 0.8545\nArrhythmia accuracy, Acc_A = 0.7435\n-----------------------------------\n-----------------------------------\nRF with wavelets\nTotal accuracy, Acc_T = 0.9445\nArrhythmia accuracy, Acc_A = 0.6801\n-----------------------------------\n-----------------------------------\nAda with wavelets\nTotal accuracy, Acc_T = 0.8984\nArrhythmia accuracy, Acc_A = 0.4755\n-----------------------------------\n"
]
],
[
[
"# Tasks to do \nPlease choose and complete just **one** of them:\n1. Modify classifier to get **accuracy > 0.81** for both Acc_T *and* Acc_A\n> - play with classifier hyperparameters\n> - add some other features, e.g:\n>> - [mean of absolute value (MAV) of signal](https://www.researchgate.net/publication/46147272_Sequential_algorithm_for_life_threatening_cardiac_pathologies_detection_based_on_mean_signal_strength_and_EMD_functions)\n>> - some other signal features from [scipy signal](https://docs.scipy.org/doc/scipy/reference/signal.html#peak-finding), \n>> - distances between previous and next heartbeats are strong features, see e.g. [here](https://link.springer.com/article/10.1007/s11760-009-0136-1),\n>> - it may be also usefull to perform some feature selection, e.g. choose these with variance higher than some assumed threshold (*intuition*: variance measures amount of information in a given feature), or use *model.feature_importances_* attribute (for more see [here](https://scikit-learn.org/stable/modules/feature_selection.html))\n> - balance dataset by yourself: equalize the size of each of 3 groups (hint: take the whole dataset)\n> - or build your own classifier using [MLP](https://scikit-learn.org/stable/modules/neural_networks_supervised.html#classification) \n2. Compare results for Random Forest with AdaBoost classifier\n> - try to figure out why the default Ada setup won't work good\n> - and fix this problem (hint: resampling)\n> - try Ada with different *base_estimators*\n3. Add deep-neural classifier (like one in previous lab) and compare its preformance with today's best classifier\n> - at first you should create *torch.utils.data.DataLoader* object, see [here](https://stanford.edu/~shervine/blog/pytorch-how-to-generate-data-parallel)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4ae63c5d104b6742d911254e9488b39d1f7d3fdd
| 2,085 |
ipynb
|
Jupyter Notebook
|
gps/09-ExperimentingWithMaps-ipyleaflet.ipynb
|
BLannoo/experimental
|
44e09f4a4a9ff717d7c1b8d3de461d2fa731b177
|
[
"MIT"
] | null | null | null |
gps/09-ExperimentingWithMaps-ipyleaflet.ipynb
|
BLannoo/experimental
|
44e09f4a4a9ff717d7c1b8d3de461d2fa731b177
|
[
"MIT"
] | null | null | null |
gps/09-ExperimentingWithMaps-ipyleaflet.ipynb
|
BLannoo/experimental
|
44e09f4a4a9ff717d7c1b8d3de461d2fa731b177
|
[
"MIT"
] | null | null | null | 22.180851 | 120 | 0.51223 |
[
[
[
"from ipyleaflet import Map, basemaps, basemap_to_tiles\n\nm = Map(\n center=(50.8798, 4.7005),\n zoom=13\n)\n\nm",
"_____no_output_____"
],
[
"from ipyleaflet import Map, Marker, MarkerCluster\n\nm = Map(center=(50.8798, 4.7005), zoom=15)\n\nmarker1 = Marker(location=(50.878, 4.701))\nmarker2 = Marker(location=(50.88, 4.700))\n\nmarker_cluster = MarkerCluster(\n markers=(marker1, marker2)\n)\n\nm.add_layer(marker_cluster);\n\nm",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4ae668b0a66ee6e7c06c8a339b6feba17deddfa1
| 26,072 |
ipynb
|
Jupyter Notebook
|
texel_test_task.ipynb
|
Factotum8/texel_categorical_features
|
d867573896f93b9fc9ddbb140c38ed76930feb08
|
[
"MIT"
] | null | null | null |
texel_test_task.ipynb
|
Factotum8/texel_categorical_features
|
d867573896f93b9fc9ddbb140c38ed76930feb08
|
[
"MIT"
] | null | null | null |
texel_test_task.ipynb
|
Factotum8/texel_categorical_features
|
d867573896f93b9fc9ddbb140c38ed76930feb08
|
[
"MIT"
] | null | null | null | 44.491468 | 9,216 | 0.652961 |
[
[
[
"Младшему исследователю из Вашего отдела дали выполнить задание по дизайну признаков (фич) в новом проекте.\nОпределите слабые стороны этого решения и попробуйте улучшить результат (RMSE).\n\nЗадача была сформулирована так: придумать такие дискретные фичи (не более 5 фичей, каждая фича имеет не более 5 значений), чтобы максимально точно предсказывать по ним значения c3, c4, c5 из имеющегося датасета.\n\nПомимо новых дискретных фич допускается использование c1, c2 и d1 в качестве входных переменных (X).",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"df = pd.read_csv('test_data.csv', header=0, index_col=0)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"def get_discrete_feature1(df):\n if pd.isna(df['c3']) or pd.isna(df['c5']) or pd.isna(df['c4']):\n return None\n\n c5 = df['c5']\n c3 = df['c3']\n c4 = df['c4']\n\n if c3 - c5 >= 92 and c3 - c4 < 230:\n return 'cl1'\n\n if c5 - c3 < 92 and c3 - c5 < 92 and c3 - c4 < 230 and c5 - c4 < 250:\n return 'cl2'\n\n if c3 - c5 <= 25 and c5 - c3 < 92 and c3 - c4 >= 230:\n return 'cl3'\n\n if c5 - c3 >= 92 and c5 - c4 < 230:\n return 'cl4'\n\n else:\n return 'cl5'",
"_____no_output_____"
],
[
"# add categorial features\nfor ind in df.index:\n row = df.loc[ind]\n df.loc[ind, 'd2'] = get_discrete_feature1(row)\ndf = df.dropna() ",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"ax = df['d2'].value_counts().plot(kind='bar', figsize=(14,8), title=\"Number of each value\")\n",
"_____no_output_____"
],
[
"x_features = ['c1', 'c2', 'd1', 'd2']\ny_features = ['c3', 'c4', 'c5']",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\ntrain_df, test_df = train_test_split(df, test_size=0.2, random_state=42, shuffle=True)",
"_____no_output_____"
],
[
"!pip install catboost",
"Collecting catboost\n Downloading catboost-0.24.4-cp38-none-win_amd64.whl (65.4 MB)\nCollecting graphviz\n Downloading graphviz-0.16-py2.py3-none-any.whl (19 kB)\nRequirement already satisfied: numpy>=1.16.0 in c:\\users\\a\\anaconda3\\lib\\site-packages (from catboost) (1.19.2)\nRequirement already satisfied: scipy in c:\\users\\a\\anaconda3\\lib\\site-packages (from catboost) (1.5.2)\nRequirement already satisfied: pandas>=0.24.0 in c:\\users\\a\\anaconda3\\lib\\site-packages (from catboost) (1.1.3)\nCollecting plotly\n Downloading plotly-4.14.1-py2.py3-none-any.whl (13.2 MB)\nRequirement already satisfied: matplotlib in c:\\users\\a\\anaconda3\\lib\\site-packages (from catboost) (3.3.2)\nRequirement already satisfied: six in c:\\users\\a\\anaconda3\\lib\\site-packages (from catboost) (1.15.0)\nRequirement already satisfied: python-dateutil>=2.7.3 in c:\\users\\a\\anaconda3\\lib\\site-packages (from pandas>=0.24.0->catboost) (2.8.1)\nRequirement already satisfied: pytz>=2017.2 in c:\\users\\a\\anaconda3\\lib\\site-packages (from pandas>=0.24.0->catboost) (2020.1)\nCollecting retrying>=1.3.3\n Downloading retrying-1.3.3.tar.gz (10 kB)\nRequirement already satisfied: cycler>=0.10 in c:\\users\\a\\anaconda3\\lib\\site-packages (from matplotlib->catboost) (0.10.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in c:\\users\\a\\anaconda3\\lib\\site-packages (from matplotlib->catboost) (1.3.0)\nRequirement already satisfied: certifi>=2020.06.20 in c:\\users\\a\\anaconda3\\lib\\site-packages (from matplotlib->catboost) (2020.6.20)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.3 in c:\\users\\a\\anaconda3\\lib\\site-packages (from matplotlib->catboost) (2.4.7)\nRequirement already satisfied: pillow>=6.2.0 in c:\\users\\a\\anaconda3\\lib\\site-packages (from matplotlib->catboost) (8.0.1)\nBuilding wheels for collected packages: retrying\n Building wheel for retrying (setup.py): started\n Building wheel for retrying (setup.py): finished with status 'done'\n Created wheel for retrying: filename=retrying-1.3.3-py3-none-any.whl size=11434 sha256=f032a51bc6a9e2a8b0ac0bd3777303565a44a018082ee2b7d23a7ad980e1d806\n Stored in directory: c:\\users\\a\\appdata\\local\\pip\\cache\\wheels\\c4\\a7\\48\\0a434133f6d56e878ca511c0e6c38326907c0792f67b476e56\nSuccessfully built retrying\nInstalling collected packages: graphviz, retrying, plotly, catboost\nSuccessfully installed catboost-0.24.4 graphviz-0.16 plotly-4.14.1 retrying-1.3.3\n"
],
[
"from catboost import CatBoostRegressor\n\nmodel = CatBoostRegressor(silent=True, loss_function='MultiRMSE').fit(train_df[x_features], train_df[y_features], cat_features=[3])",
"_____no_output_____"
],
[
"def rmse(a1, a2):\n diff = (a1 - a2)\n return float(np.mean(np.sqrt(np.mean(diff ** 2, axis=0))))\n",
"_____no_output_____"
],
[
"pred = np.array(model.predict(test_df[x_features]))\n\nprint(rmse(pred, test_df[y_features].to_numpy()))",
"47.61463911502748\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae68a89c37fadd329587b4b6f291b8f4be7f02a
| 113,659 |
ipynb
|
Jupyter Notebook
|
extra/Old Content-Based Model (Terrible Efficiency).ipynb
|
jpfairbanks/NewsGraphing
|
12499f184d4ff2b8e76725aaac47bcdf7f7add0c
|
[
"MIT"
] | null | null | null |
extra/Old Content-Based Model (Terrible Efficiency).ipynb
|
jpfairbanks/NewsGraphing
|
12499f184d4ff2b8e76725aaac47bcdf7f7add0c
|
[
"MIT"
] | null | null | null |
extra/Old Content-Based Model (Terrible Efficiency).ipynb
|
jpfairbanks/NewsGraphing
|
12499f184d4ff2b8e76725aaac47bcdf7f7add0c
|
[
"MIT"
] | null | null | null | 161.447443 | 86,172 | 0.859325 |
[
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nfrom pymongo import MongoClient\nimport tldextract\nimport math\nimport re\nimport pickle\nfrom tqdm import tqdm_notebook as tqdm\nimport spacy\nfrom numpy import dot\nfrom numpy.linalg import norm\nimport csv\nimport random\nimport statistics\nimport copy\nimport itertools\nfrom vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer as SIA\nfrom sklearn import svm\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import KFold\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.feature_extraction.text import TfidfTransformer\nimport scipy",
"_____no_output_____"
],
[
"nlp = spacy.load('en')",
"_____no_output_____"
],
[
"client = MongoClient('mongodb://gdelt:meidnocEf1@gdeltmongo1:27017/')\ndb = client.gdelt.metadata\n\ndef valid(s, d):\n if len(d) > 0 and d[0] not in [\"/\", \"#\", \"{\"] and s not in d :\n return True\n else:\n return False",
"_____no_output_____"
],
[
"re_3986 = re.compile(r\"^(([^:/?#]+):)?(//([^/?#]*))?([^?#]*)(\\?([^#]*))?(#(.*))?\")\nwgo = re.compile(\"www.\")\nwhitelist = [\"NOUN\", \"PROPN\", \"ADJ\", \"ADV\"]",
"_____no_output_____"
],
[
"bias = []\nbiasnames = []\npol = ['L', 'LC', 'C', 'RC', 'R']\nrep = ['VERY LOW', 'LOW', 'MIXED', 'HIGH', 'VERY HIGH']\nflag = ['F', 'X', 'S']\ncats = pol\ns2l = {}\nwith open('bias.csv', 'r') as csvfile:\n reader = csv.reader(csvfile)\n for row in reader:\n name = re_3986.match(row[4]).group(4)\n p = -1\n r = -1\n f = -1\n if row[1] in pol:\n p = pol.index(row[1])\n s2l[name] = row[1]\n if row[2] in rep:\n r = rep.index(row[2])\n if row[3] in flag:\n f = flag.index(row[3])\n s2l[name] = row[3]\n bias.append(row + [name, p, r, f, 1 if p == -1 else 0])\n biasnames.append(name)",
"_____no_output_____"
],
[
"sample = 1000000\nstuff = db.find({},{'text':1,'sourceurl':1}).sort(\"_id\",-1).limit(sample)\n\narts = []\n\nfor obj in tqdm(stuff):\n if 'text' in obj:\n sdom = wgo.sub(\"\", re_3986.match(obj['sourceurl']).group(4))\n if sdom in biasnames:\n doc = nlp.tokenizer(obj['text'][:100*8])\n nlp.tagger(doc)\n arts.append((sdom, doc))\nN = len(arts)",
"_____no_output_____"
],
[
"doc_tdf = {}\ndoc_bgdf = {}\ndoc_tf = {}\ndoc_bgf = {}\ndoc_ts = {}\ndoc_bgs = {}\nsite_tf = {}\nsite_bgf = {}\nsite_ts = {}\nsite_bgs = {}\ncat_tf = {cat : {} for cat in cats}\ncat_bgf = {cat : {} for cat in cats}\ncat_ts = {cat : {} for cat in cats}\ncat_bgs = {cat : {} for cat in cats}\nsa = SIA()\nfor (sdom, obj) in tqdm(leads):\n if sdom not in site_tf:\n site_tf[sdom] = {}\n site_bgf[sdom] = {}\n site_ts[sdom] = {}\n site_bgs[sdom] = {}\n \n \nfor (sdom, doc) in tqdm(arts):\n \n #seen = {}\n mycat = s2l[sdom]\n if mycat in cats:\n c = sa.polarity_scores(doc.text)['compound']\n for word in doc[:-1]:\n if not word.is_stop and word.is_alpha and word.pos_ in whitelist:\n \n # Save the sentiments in a list \n # To be averaged into means later\n\n if word.lemma_ not in doc_ts:\n doc_ts[word.lemma_] = []\n doc_ts[word.lemma_].append(c)\n\n if word.lemma_ not in site_ts[sdom]:\n site_ts[sdom][word.lemma_] = []\n site_ts[sdom][word.lemma_].append(c)\n\n if word.lemma_ not in cat_ts[mycat]:\n cat_ts[mycat][word.lemma_] = []\n cat_ts[mycat][word.lemma_].append(c)\n\n # Record counts of this term\n # To be divided by total to make term frequency later\n\n if word.lemma_ not in doc_tf:\n doc_tf[word.lemma_] = 0\n doc_tf[word.lemma_] += 1\n \n if word.lemma_ not in site_tf[sdom]:\n site_tf[sdom][word.lemma_] = 0\n site_tf[sdom][word.lemma_] += 1\n\n if word.lemma_ not in cat_tf[mycat]:\n cat_tf[mycat][word.lemma_] = 0\n cat_tf[mycat][word.lemma_] += 1\n\n\n # # Record number of documents it appears in\n\n # if word.lemma not in seen:\n # seen[word.lemma] = 1\n # if word.lemma_ not in doc_tf:\n # doc_tf[word.lemma_] = 0\n # doc_tf[word.lemma_] += 1\n\n\n neigh = word.nbor()\n if not neigh.is_stop and neigh.pos_ in whitelist:\n bigram = word.lemma_+\" \"+neigh.lemma_\n\n# # Save the sentiments in a list \n# # To be averaged into means later\n\n if bigram not in doc_bgs:\n doc_bgs[bigram] = []\n doc_bgs[bigram].append(c)\n\n if bigram not in site_bgs[sdom]:\n site_bgs[sdom][bigram] = []\n site_bgs[sdom][bigram].append(c)\n\n if bigram not in cat_bgs[mycat]:\n cat_bgs[mycat][bigram] = []\n cat_bgs[mycat][bigram].append(c)\n\n# # Record counts of this bigram\n# # To be divided by total to make term frequency later\n\n\n if bigram not in doc_bgf:\n doc_bgf[bigram] = 0\n doc_bgf[bigram] += 1\n\n if bigram not in site_bgf[sdom]:\n site_bgf[sdom][bigram] = 0\n site_bgf[sdom][bigram] += 1\n\n if bigram not in cat_bgf[mycat]:\n cat_bgf[mycat][bigram] = 0\n cat_bgf[mycat][bigram] += 1\n\n# # if bigram not in seen:\n# # seen[bigram] = 1\n# # if bigram not in doc_bgf:\n# # doc_bgf[bigram] = 0\n# # doc_bgf[bigram] += 1\n\ndoc_tls = copy.deepcopy(doc_ts)\ndoc_bgls = copy.deepcopy(doc_bgs)\nsite_tls = copy.deepcopy(site_ts)\nsite_bgls = copy.deepcopy(site_bgs)\ncat_tls = copy.deepcopy(cat_ts)\ncat_bgls = copy.deepcopy(cat_bgs)\n\n\n\nfor word in tqdm(doc_ts):\n doc_ts[word] = sum(doc_ts[word])/len(doc_ts[word])\nfor word in tqdm(doc_bgs):\n doc_bgs[word] = sum(doc_bgs[word])/len(doc_bgs[word])\nfor site in tqdm(site_bgs):\n for word in site_ts[site]:\n site_ts[site][word] = sum(site_ts[site][word])/len(site_ts[site][word])\n for word in site_bgs[site]:\n site_bgs[site][word] = sum(site_bgs[site][word])/len(site_bgs[site][word])\nfor cat in tqdm(cats):\n for word in cat_ts[cat]:\n cat_ts[cat][word] = sum(cat_ts[cat][word])/len(cat_ts[cat][word])\n for word in cat_bgs[cat]:\n cat_bgs[cat][word] = sum(cat_bgs[cat][word])/len(cat_bgs[cat][word])\n\ndoc_tc = copy.deepcopy(doc_tf)\ndoc_bgc = copy.deepcopy(doc_bgf)\nsite_tc = copy.deepcopy(site_tf)\nsite_bgc = copy.deepcopy(site_bgf)\ncat_tc = copy.deepcopy(cat_tf)\ncat_bgc = copy.deepcopy(cat_bgf)\n \n \ntot = sum(doc_tf.values())\nfor word in tqdm(doc_tf):\n doc_tf[word] = doc_tf[word]/tot\ntot = sum(doc_bgf.values())\nfor word in tqdm(doc_bgf):\n doc_bgf[word] = doc_bgf[word]/tot\nfor site in tqdm(site_tf):\n tot = sum(site_tf[site].values())\n for word in site_tf[site]:\n site_tf[site][word] = site_tf[site][word]/tot\n tot = sum(site_bgf[site].values())\n for word in site_bgf[site]:\n site_bgf[site][word] = site_bgf[site][word]/tot\nfor cat in tqdm(cats):\n tot = sum(cat_tf[cat].values())\n for word in cat_tf[cat]:\n cat_tf[cat][word] = cat_tf[cat][word]/tot\n tot = sum(cat_bgf[cat].values())\n for word in cat_bgf[cat]:\n cat_bgf[cat][word] = cat_bgf[cat][word]/tot",
"_____no_output_____"
],
[
"def cos_sim(a, b):\n a = site_v[a]\n b = site_v[b]\n return dot(a, b)/(norm(a)*norm(b))",
"_____no_output_____"
],
[
"def isReal(site):\n if s2l[site] in pol:\n return True\n return False\n\nsites = [site for site in site_ts.keys() if site in biasnames]\nα = 0.001\n\ntp = {}\nt_exp = [sum(cat_tc[cat].values()) for cat in cats]\nt_exp = [t/sum(t_exp) for t in t_exp]\nsig_terms = []\nfor term in tqdm(doc_ts.keys()):\n ds = [0]*len(cats)\n df = [0]*len(cats)\n #f = False\n \n for i, cat in enumerate(cats):\n if term in cat_ts[cat]:\n ds[i] = cat_ts[cat][term]-doc_ts[term]\n df[i] = cat_tc[cat][term]\n \n χ, p1 = scipy.stats.chisquare(df, f_exp=[t*sum(df) for t in t_exp])\n if p1 < α or scipy.stats.chisquare(ds)[1] < α:\n sig_terms.append(term)\n tp[term] = p\n \n #print(term + \" \" + str(p))\nsig_terms = sorted(sig_terms, key=lambda x:tp[x])\nprint(len(sig_terms))\nprint(sig_terms[:10])\n\nbgp = {}\nt_exp = [sum(cat_bgc[cat].values()) for cat in cats]\nt_exp = [t/sum(t_exp) for t in t_exp]\nsig_bigrams = []\nfor bigram in tqdm(doc_bgs.keys()):\n ds = [0]*len(cats)\n df = [0]*len(cats)\n \n for i, cat in enumerate(cats):\n if bigram in cat_bgs[cat]:\n ds[i] = cat_bgs[cat][bigram]-doc_bgs[bigram]\n df[i] = cat_bgc[cat][bigram]\n \n χ, p1 = scipy.stats.chisquare(df, f_exp=[t*sum(df) for t in t_exp])\n if p1 < α or scipy.stats.chisquare(ds)[1] < α:\n sig_bigrams.append(bigram)\n bgp[bigram] = p\n \n \n \nsig_bigrams = sorted(sig_bigrams, key=lambda x:bgp[x])\nprint(len(sig_bigrams))\nprint(sig_bigrams[:10])",
"_____no_output_____"
],
[
"site_v = {}\nfor site in tqdm(site_ts.keys()):\n if site in site_bgs:\n v = [0]*(len(sig_terms)+len(sig_bigrams))*2\n #tot_term = sum(site_ts[site].values())\n for i, term in enumerate(sig_terms):\n if term in site_ts[site]:\n v[2*i] = site_ts[site][term]-doc_ts[term]\n if term in site_tf[site]:\n v[2*i+1] = site_tf[site][term]-doc_tf[term]\n for j, bigram in enumerate(sig_bigrams):\n if bigram in site_bgs[site]:\n v[2*i+2*j+2] = site_bgs[site][bigram]-doc_bgs[bigram]\n if bigram in site_bgf[site]:\n v[2*i+2*j+3] = site_bgf[site][bigram]-doc_bgf[bigram]\n site_v[site] = v\nprint(len(site_v))",
"_____no_output_____"
],
[
"\n\nclf = RandomForestClassifier(random_state=42)\n#clf = svm.SVC(random_state=42)\n\nsites = [s for s in s2l if s in site_ts.keys()]\nX = [site_v[s] for s in sites if s2l[s] in cats]\ny = [cats.index(s2l[s]) for s in sites if s2l[s] in cats]\n#y = [1 if s2l[s] in [\"L\", \"LC\", \"C\"] else -1 for s in sites]\n\nX = np.asarray(X)\ny = np.asarray(y)\n\nvn = sig_terms+sig_bigrams\nvn = list(itertools.chain(*zip(vn,vn)))\n\ncscore = cross_val_score(clf, X, y, cv=3)\nprint(cscore)\nprint(sum(cscore)/3)\nclf.fit(X, y)\nmask = [i for i, x in enumerate(clf.feature_importances_) if x > 0.0001]\ncscore = cross_val_score(clf, [x[mask] for x in X], y, cv=3)\nprint(cscore)\nprint(sum(cscore)/3)\n\nfi = clf.feature_importances_\nplt.figure(figsize=(10,10))\nplt.plot(sorted(fi[mask]))\nplt.xticks(range(0, len(mask)), sorted([vn[m] for m in mask], key=lambda x:fi[vn.index(x)]), rotation=90)\nplt.show()\n\ncms = []\nfor train, test in KFold(n_splits=3).split(X):\n clf.fit([x[mask] for x in X[train]], y[train])\n cms.append(confusion_matrix(y[test], clf.predict([x[mask] for x in X[test]])))\n# clf.fit(X[train], y[train])\n# cms.append(confusion_matrix(y[test], clf.predict(X[test])))\nprint(sum(cms))\nplt.imshow(sum(cms))\nplt.show()\nprint(sum(sum(sum(cms))))",
"[ 0.30882353 0.26865672 0.3030303 ]\n0.293503516287\n[ 0.32352941 0.31343284 0.37878788]\n0.338583375458\n"
],
[
"sorted(site_v.keys(), key=lambda x:cos_sim(\"breitbart.com\", x), reverse=False)",
"_____no_output_____"
],
[
"site_id = {}\nfor site in site_v:\n site_id[site] = cos_sim(\"breitbart.com\", site) - cos_sim(\"huffingtonpost.com\", site)\n#print(site_id)\nl = sorted(site_id.keys(), key = lambda x : site_id[x])\nprint(l)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae6a56384f2eb657233a874ae392f547ddeeebb
| 223,576 |
ipynb
|
Jupyter Notebook
|
Notebooks/Speech_recognition_project.ipynb
|
amathsow/wolof_speech_recognition
|
537cb67b67e6df0e77437deeee176c8ec582ce8b
|
[
"MIT"
] | 1 |
2020-07-06T23:26:45.000Z
|
2020-07-06T23:26:45.000Z
|
Notebooks/Speech_recognition_project.ipynb
|
amathsow/wolof_speech_recognition
|
537cb67b67e6df0e77437deeee176c8ec582ce8b
|
[
"MIT"
] | null | null | null |
Notebooks/Speech_recognition_project.ipynb
|
amathsow/wolof_speech_recognition
|
537cb67b67e6df0e77437deeee176c8ec582ce8b
|
[
"MIT"
] | null | null | null | 86.994553 | 24,506 | 0.754048 |
[
[
[
"<a href=\"https://colab.research.google.com/github/amathsow/wolof_speech_recognition/blob/master/Speech_recognition_project.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip3 install torch\n!pip3 install torchvision\n!pip3 install torchaudio\n!pip install comet_ml\n",
"Requirement already satisfied: torch in /usr/local/lib/python3.6/dist-packages (1.5.1+cu101)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torch) (1.18.5)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from torch) (0.16.0)\nRequirement already satisfied: torchvision in /usr/local/lib/python3.6/dist-packages (0.6.1+cu101)\nRequirement already satisfied: torch==1.5.1 in /usr/local/lib/python3.6/dist-packages (from torchvision) (1.5.1+cu101)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torchvision) (1.18.5)\nRequirement already satisfied: pillow>=4.1.1 in /usr/local/lib/python3.6/dist-packages (from torchvision) (7.0.0)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from torch==1.5.1->torchvision) (0.16.0)\nCollecting torchaudio\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/e9/0a/40e53c686c2af65b2a4e818d11d9b76fa79178440caf99f3ceb2a32c3b04/torchaudio-0.5.1-cp36-cp36m-manylinux1_x86_64.whl (3.2MB)\n\u001b[K |████████████████████████████████| 3.2MB 4.5MB/s \n\u001b[?25hRequirement already satisfied: torch==1.5.1 in /usr/local/lib/python3.6/dist-packages (from torchaudio) (1.5.1+cu101)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torch==1.5.1->torchaudio) (1.18.5)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from torch==1.5.1->torchaudio) (0.16.0)\nInstalling collected packages: torchaudio\nSuccessfully installed torchaudio-0.5.1\nCollecting comet_ml\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/ac/d8/99564b9b0d68a8acce733a5b053497a4a51e3ffdc54e4b117808bd6c4c94/comet_ml-3.1.12-py2.py3-none-any.whl (214kB)\n\u001b[K |████████████████████████████████| 215kB 4.5MB/s \n\u001b[?25hCollecting wurlitzer>=1.0.2\n Downloading https://files.pythonhosted.org/packages/0c/1e/52f4effa64a447c4ec0fb71222799e2ac32c55b4b6c1725fccdf6123146e/wurlitzer-2.0.1-py2.py3-none-any.whl\nRequirement already satisfied: requests>=2.18.4 in /usr/local/lib/python3.6/dist-packages (from comet_ml) (2.23.0)\nCollecting everett[ini]>=1.0.1; python_version >= \"3.0\"\n Downloading https://files.pythonhosted.org/packages/12/34/de70a3d913411e40ce84966f085b5da0c6df741e28c86721114dd290aaa0/everett-1.0.2-py2.py3-none-any.whl\nRequirement already satisfied: jsonschema!=3.1.0,>=2.6.0 in /usr/local/lib/python3.6/dist-packages (from comet_ml) (2.6.0)\nCollecting netifaces>=0.10.7\n Downloading https://files.pythonhosted.org/packages/0c/9b/c4c7eb09189548d45939a3d3a6b3d53979c67d124459b27a094c365c347f/netifaces-0.10.9-cp36-cp36m-manylinux1_x86_64.whl\nCollecting comet-git-pure>=0.19.11\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/72/7a/483413046e48908986a0f9a1d8a917e1da46ae58e6ba16b2ac71b3adf8d7/comet_git_pure-0.19.16-py3-none-any.whl (409kB)\n\u001b[K |████████████████████████████████| 419kB 12.4MB/s \n\u001b[?25hRequirement already satisfied: nvidia-ml-py3>=7.352.0 in /usr/local/lib/python3.6/dist-packages (from comet_ml) (7.352.0)\nCollecting websocket-client>=0.55.0\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/4c/5f/f61b420143ed1c8dc69f9eaec5ff1ac36109d52c80de49d66e0c36c3dfdf/websocket_client-0.57.0-py2.py3-none-any.whl (200kB)\n\u001b[K |████████████████████████████████| 204kB 19.7MB/s \n\u001b[?25hRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from comet_ml) (1.12.0)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests>=2.18.4->comet_ml) (2020.6.20)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests>=2.18.4->comet_ml) (2.9)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests>=2.18.4->comet_ml) (1.24.3)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests>=2.18.4->comet_ml) (3.0.4)\nCollecting configobj; extra == \"ini\"\n Downloading https://files.pythonhosted.org/packages/64/61/079eb60459c44929e684fa7d9e2fdca403f67d64dd9dbac27296be2e0fab/configobj-5.0.6.tar.gz\nBuilding wheels for collected packages: configobj\n Building wheel for configobj (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for configobj: filename=configobj-5.0.6-cp36-none-any.whl size=34546 sha256=722443d4f7b03fd6ba7b751f8fbc93daec0b62e7e4368f8597a68189a6f77cb4\n Stored in directory: /root/.cache/pip/wheels/f1/e4/16/4981ca97c2d65106b49861e0b35e2660695be7219a2d351ee0\nSuccessfully built configobj\nInstalling collected packages: wurlitzer, configobj, everett, netifaces, comet-git-pure, websocket-client, comet-ml\nSuccessfully installed comet-git-pure-0.19.16 comet-ml-3.1.12 configobj-5.0.6 everett-1.0.2 netifaces-0.10.9 websocket-client-0.57.0 wurlitzer-2.0.1\n"
],
[
"import os\nfrom comet_ml import Experiment\nimport torch\nimport torch.nn as nn\nimport torch.utils.data as data\nimport torch.optim as optim\nimport torch.nn.functional as F\nimport torchaudio\nimport numpy as np\nimport pandas as pd\nimport librosa",
"_____no_output_____"
]
],
[
[
"## ETL process",
"_____no_output_____"
]
],
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive\n"
],
[
"path_audio= 'drive/My Drive/Speech Recognition project/recordings/'\npath_text = 'drive/My Drive/Speech Recognition project/wolof_text/'\nwav_text = 'drive/My Drive/Speech Recognition project/Wavtext_dataset2.csv'",
"_____no_output_____"
]
],
[
[
"## Data preparation for created the char.txt file from my dataset.",
"_____no_output_____"
]
],
[
[
"datapath = 'drive/My Drive/Speech Recognition project/data/records'",
"_____no_output_____"
],
[
"trainpath = '../drive/My Drive/Speech Recognition project/data/records/train/'\nvalpath = '../drive/My Drive/Speech Recognition project/data/records/val/'\ntestpath = '../drive/My Drive/Speech Recognition project/data/records/test/'\n",
"_____no_output_____"
]
],
[
[
"## Let's create the dataset",
"_____no_output_____"
]
],
[
[
"! git clone https://github.com/facebookresearch/CPC_audio.git",
"Cloning into 'CPC_audio'...\nremote: Enumerating objects: 84, done.\u001b[K\nremote: Counting objects: 100% (84/84), done.\u001b[K\nremote: Compressing objects: 100% (65/65), done.\u001b[K\nremote: Total 84 (delta 13), reused 75 (delta 6), pack-reused 0\u001b[K\nUnpacking objects: 100% (84/84), done.\n"
],
[
"!pip install soundfile\n!pip install torchaudio",
"Collecting soundfile\n Downloading https://files.pythonhosted.org/packages/eb/f2/3cbbbf3b96fb9fa91582c438b574cff3f45b29c772f94c400e2c99ef5db9/SoundFile-0.10.3.post1-py2.py3-none-any.whl\nRequirement already satisfied: cffi>=1.0 in /usr/local/lib/python3.6/dist-packages (from soundfile) (1.14.0)\nRequirement already satisfied: pycparser in /usr/local/lib/python3.6/dist-packages (from cffi>=1.0->soundfile) (2.20)\nInstalling collected packages: soundfile\nSuccessfully installed soundfile-0.10.3.post1\nRequirement already satisfied: torchaudio in /usr/local/lib/python3.6/dist-packages (0.5.1)\nRequirement already satisfied: torch==1.5.1 in /usr/local/lib/python3.6/dist-packages (from torchaudio) (1.5.1+cu101)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from torch==1.5.1->torchaudio) (0.16.0)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torch==1.5.1->torchaudio) (1.18.5)\n"
],
[
"!mkdir checkpoint_data\n!wget https://dl.fbaipublicfiles.com/librilight/CPC_checkpoints/not_hub/2levels_6k_top_ctc/checkpoint_30.pt -P checkpoint_data\n!wget https://dl.fbaipublicfiles.com/librilight/CPC_checkpoints/not_hub/2levels_6k_top_ctc/checkpoint_logs.json -P checkpoint_data\n!wget https://dl.fbaipublicfiles.com/librilight/CPC_checkpoints/not_hub/2levels_6k_top_ctc/checkpoint_args.json -P checkpoint_data\n!ls checkpoint_data",
"--2020-07-06 17:44:26-- https://dl.fbaipublicfiles.com/librilight/CPC_checkpoints/not_hub/2levels_6k_top_ctc/checkpoint_30.pt\nResolving dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)... 104.22.75.142, 172.67.9.4, 104.22.74.142, ...\nConnecting to dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)|104.22.75.142|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 113599715 (108M) [application/octet-stream]\nSaving to: ‘checkpoint_data/checkpoint_30.pt’\n\ncheckpoint_30.pt 100%[===================>] 108.34M 86.0MB/s in 1.3s \n\n2020-07-06 17:44:27 (86.0 MB/s) - ‘checkpoint_data/checkpoint_30.pt’ saved [113599715/113599715]\n\n--2020-07-06 17:44:29-- https://dl.fbaipublicfiles.com/librilight/CPC_checkpoints/not_hub/2levels_6k_top_ctc/checkpoint_logs.json\nResolving dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)... 172.67.9.4, 104.22.75.142, 104.22.74.142, ...\nConnecting to dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)|172.67.9.4|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 20786 (20K) [text/plain]\nSaving to: ‘checkpoint_data/checkpoint_logs.json’\n\ncheckpoint_logs.jso 100%[===================>] 20.30K --.-KB/s in 0.007s \n\n2020-07-06 17:44:29 (2.95 MB/s) - ‘checkpoint_data/checkpoint_logs.json’ saved [20786/20786]\n\n--2020-07-06 17:44:30-- https://dl.fbaipublicfiles.com/librilight/CPC_checkpoints/not_hub/2levels_6k_top_ctc/checkpoint_args.json\nResolving dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)... 104.22.74.142, 104.22.75.142, 172.67.9.4, ...\nConnecting to dl.fbaipublicfiles.com (dl.fbaipublicfiles.com)|104.22.74.142|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 2063 (2.0K) [text/plain]\nSaving to: ‘checkpoint_data/checkpoint_args.json’\n\ncheckpoint_args.jso 100%[===================>] 2.01K --.-KB/s in 0s \n\n2020-07-06 17:44:30 (43.9 MB/s) - ‘checkpoint_data/checkpoint_args.json’ saved [2063/2063]\n\ncheckpoint_30.pt checkpoint_args.json\tcheckpoint_logs.json\n"
],
[
"import torch\nimport torchaudio",
"_____no_output_____"
],
[
"%cd CPC_audio/\nfrom cpc.model import CPCEncoder, CPCAR\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\nDIM_ENCODER=256\nDIM_CONTEXT=256\nKEEP_HIDDEN_VECTOR=False\nN_LEVELS_CONTEXT=1\nCONTEXT_RNN=\"LSTM\"\nN_PREDICTIONS=12\nLEARNING_RATE=2e-4\nN_NEGATIVE_SAMPLE =128",
"/content/CPC_audio\n"
],
[
"encoder = CPCEncoder(DIM_ENCODER).to(device)\ncontext = CPCAR(DIM_ENCODER, DIM_CONTEXT, KEEP_HIDDEN_VECTOR, 1, mode=CONTEXT_RNN).to(device)",
"_____no_output_____"
],
[
"# Several functions that will be necessary to load the data later\nfrom cpc.dataset import findAllSeqs, AudioBatchData, parseSeqLabels\nSIZE_WINDOW = 20480\nBATCH_SIZE=8\ndef load_dataset(path_dataset, file_extension='.flac', phone_label_dict=None):\n data_list, speakers = findAllSeqs(path_dataset, extension=file_extension)\n dataset = AudioBatchData(path_dataset, SIZE_WINDOW, data_list, phone_label_dict, len(speakers))\n return dataset",
"_____no_output_____"
],
[
"class CPCModel(torch.nn.Module):\n\n def __init__(self,\n encoder,\n AR):\n\n super(CPCModel, self).__init__()\n self.gEncoder = encoder\n self.gAR = AR\n\n def forward(self, batch_data):\n \n\n encoder_output = self.gEncoder(batch_data)\n #print(encoder_output.shape)\n # The output of the encoder data does not have the good format \n # indeed it is Batch_size x Hidden_size x temp size\n # while the context requires Batch_size x temp size x Hidden_size\n # thus you need to permute\n context_input = encoder_output.permute(0, 2, 1)\n\n context_output = self.gAR(context_input)\n #print(context_output.shape)\n return context_output, encoder_output",
"_____no_output_____"
],
[
"datapath ='../drive/My Drive/Speech Recognition project/data/records/'\ndatapath2 ='../drive/My Drive/Speech Recognition project/data/'",
"_____no_output_____"
],
[
"!ls .. /checkpoint_data/checkpoint_30.pt",
"ls: cannot access '/checkpoint_data/checkpoint_30.pt': No such file or directory\n..:\ncheckpoint_data CPC_audio drive sample_data\n"
],
[
"%cd CPC_audio/\nfrom cpc.dataset import parseSeqLabels\nfrom cpc.feature_loader import loadModel\n\ncheckpoint_path = '../checkpoint_data/checkpoint_30.pt'\ncpc_model, HIDDEN_CONTEXT_MODEL, HIDDEN_ENCODER_MODEL = loadModel([checkpoint_path])\ncpc_model = cpc_model.cuda()\nlabel_dict, N_PHONES = parseSeqLabels(datapath2+'chars2.txt')\ndataset_train = load_dataset(datapath+'train', file_extension='.wav', phone_label_dict=label_dict)\ndataset_val = load_dataset(datapath+'val', file_extension='.wav', phone_label_dict=label_dict)\ndataset_test = load_dataset(datapath+'test', file_extension='.wav', phone_label_dict=label_dict)\ndata_loader_train = dataset_train.getDataLoader(BATCH_SIZE, \"speaker\", True)\ndata_loader_val = dataset_val.getDataLoader(BATCH_SIZE, \"sequence\", False)\ndata_loader_test = dataset_test.getDataLoader(BATCH_SIZE, \"sequence\", False)",
"[Errno 2] No such file or directory: 'CPC_audio/'\n/content/CPC_audio\nLoading checkpoint ../checkpoint_data/checkpoint_30.pt\nLoading the state dict at ../checkpoint_data/checkpoint_30.pt\n"
]
],
[
[
"## Create Model",
"_____no_output_____"
]
],
[
[
"class PhoneClassifier(torch.nn.Module):\n\n def __init__(self,\n input_dim : int,\n n_phones : int):\n super(PhoneClassifier, self).__init__()\n self.linear = torch.nn.Linear(input_dim, n_phones)\n \n\n def forward(self, x):\n return self.linear(x)",
"_____no_output_____"
],
[
"phone_classifier = PhoneClassifier(HIDDEN_CONTEXT_MODEL, N_PHONES).to(device)",
"_____no_output_____"
],
[
"loss_criterion = torch.nn.CrossEntropyLoss()",
"_____no_output_____"
],
[
"parameters = list(phone_classifier.parameters()) + list(cpc_model.parameters())\nLEARNING_RATE = 2e-4\noptimizer = torch.optim.Adam(parameters, lr=LEARNING_RATE)",
"_____no_output_____"
],
[
"optimizer_frozen = torch.optim.Adam(list(phone_classifier.parameters()), lr=LEARNING_RATE)",
"_____no_output_____"
],
[
"def train_one_epoch(cpc_model, \n phone_classifier, \n loss_criterion, \n data_loader, \n optimizer):\n\n cpc_model.train()\n loss_criterion.train()\n\n avg_loss = 0\n avg_accuracy = 0\n n_items = 0\n for step, full_data in enumerate(data_loader):\n # Each batch is represented by a Tuple of vectors:\n # sequence of size : N x 1 x T\n # label of size : N x T\n # \n # With :\n # - N number of sequence in the batch\n # - T size of each sequence\n sequence, label = full_data\n \n \n\n bs = len(sequence)\n seq_len = label.size(1)\n optimizer.zero_grad()\n context_out, enc_out, _ = cpc_model(sequence.to(device),label.to(device))\n\n scores = phone_classifier(context_out)\n\n scores = scores.permute(0,2,1)\n loss = loss_criterion(scores,label.to(device))\n loss.backward()\n optimizer.step()\n avg_loss+=loss.item()*bs\n n_items+=bs\n correct_labels = scores.argmax(1)\n avg_accuracy += ((label==correct_labels.cpu()).float()).mean(1).sum().item()\n avg_loss/=n_items\n avg_accuracy/=n_items\n return avg_loss, avg_accuracy",
"_____no_output_____"
],
[
"avg_loss, avg_accuracy = train_one_epoch(cpc_model, phone_classifier, loss_criterion, data_loader_train, optimizer_frozen)",
"_____no_output_____"
],
[
"avg_loss, avg_accuracy",
"_____no_output_____"
],
[
"def validation_step(cpc_model, \n phone_classifier, \n loss_criterion, \n data_loader):\n \n cpc_model.eval()\n phone_classifier.eval()\n\n avg_loss = 0\n avg_accuracy = 0\n n_items = 0\n with torch.no_grad():\n for step, full_data in enumerate(data_loader):\n # Each batch is represented by a Tuple of vectors:\n # sequence of size : N x 1 x T\n # label of size : N x T\n # \n # With :\n # - N number of sequence in the batch\n # - T size of each sequence\n sequence, label = full_data\n bs = len(sequence)\n seq_len = label.size(1)\n context_out, enc_out, _ = cpc_model(sequence.to(device),label.to(device))\n scores = phone_classifier(context_out)\n scores = scores.permute(0,2,1)\n loss = loss_criterion(scores,label.to(device))\n avg_loss+=loss.item()*bs\n n_items+=bs\n correct_labels = scores.argmax(1)\n avg_accuracy += ((label==correct_labels.cpu()).float()).mean(1).sum().item()\n avg_loss/=n_items\n avg_accuracy/=n_items\n return avg_loss, avg_accuracy",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom google.colab import files\n\ndef run(cpc_model, \n phone_classifier, \n loss_criterion, \n data_loader_train, \n data_loader_val, \n optimizer,\n n_epoch):\n epoches = []\n train_losses = []\n train_accuracies = []\n val_losses = []\n val_accuracies = []\n for epoch in range(n_epoch):\n\n epoches.append(epoch)\n\n print(f\"Running epoch {epoch + 1} / {n_epoch}\")\n loss_train, acc_train = train_one_epoch(cpc_model, phone_classifier, loss_criterion, data_loader_train, optimizer)\n print(\"-------------------\")\n print(f\"Training dataset :\")\n print(f\"Average loss : {loss_train}. Average accuracy {acc_train}\")\n train_losses.append(loss_train)\n train_accuracies.append(acc_train)\n\n print(\"-------------------\")\n print(\"Validation dataset\")\n loss_val, acc_val = validation_step(cpc_model, phone_classifier, loss_criterion, data_loader_val)\n print(f\"Average loss : {loss_val}. Average accuracy {acc_val}\")\n print(\"-------------------\")\n print()\n\n val_losses.append(loss_val)\n val_accuracies.append(acc_val)\n\n plt.plot(epoches, train_losses, label = \"train loss\")\n plt.plot(epoches, val_losses, label = \"val loss\")\n plt.xlabel('epoches')\n plt.ylabel('loss')\n plt.title('train and validation loss')\n plt.legend()\n # Display a figure.\n plt.savefig(\"loss1.png\")\n files.download(\"loss1.png\") \n plt.show()\n\n plt.plot(epoches, train_accuracies, label = \"train accuracy\")\n plt.plot(epoches, val_accuracies, label = \"vali accuracy\")\n plt.xlabel('epoches')\n plt.ylabel('accuracy')\n plt.title('train and validation accuracy')\n plt.legend()\n plt.savefig(\"val1.png\")\n files.download(\"val1.png\") \n # Display a figure.\n plt.show()\n\n",
"_____no_output_____"
]
],
[
[
"## The Training and Evaluating Script",
"_____no_output_____"
]
],
[
[
"run(cpc_model,phone_classifier,loss_criterion,data_loader_train,data_loader_val,optimizer_frozen,n_epoch=10)",
"Running epoch 1 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.9382019115216806. Average accuracy 0.1054983428030303\n-------------------\nValidation dataset\nAverage loss : 3.846985399723053. Average accuracy 0.119873046875\n-------------------\n\nRunning epoch 2 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.7741444472110635. Average accuracy 0.13301964962121213\n-------------------\nValidation dataset\nAverage loss : 3.69957634806633. Average accuracy 0.145751953125\n-------------------\n\nRunning epoch 3 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.6394398140184805. Average accuracy 0.15589488636363635\n-------------------\nValidation dataset\nAverage loss : 3.5785878598690033. Average accuracy 0.162841796875\n-------------------\n\nRunning epoch 4 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.5329696120637837. Average accuracy 0.16332267992424243\n-------------------\nValidation dataset\nAverage loss : 3.4796801805496216. Average accuracy 0.16943359375\n-------------------\n\nRunning epoch 5 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.4407107974543716. Average accuracy 0.17199337121212122\n-------------------\nValidation dataset\nAverage loss : 3.3975033164024353. Average accuracy 0.1744384765625\n-------------------\n\nRunning epoch 6 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.3651765765565815. Average accuracy 0.17717211174242425\n-------------------\nValidation dataset\nAverage loss : 3.3298254013061523. Average accuracy 0.18408203125\n-------------------\n\nRunning epoch 7 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.3034892949191006. Average accuracy 0.1835049715909091\n-------------------\nValidation dataset\nAverage loss : 3.2736769318580627. Average accuracy 0.19091796875\n-------------------\n\nRunning epoch 8 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.2533017866539233. Average accuracy 0.19202769886363635\n-------------------\nValidation dataset\nAverage loss : 3.226604402065277. Average accuracy 0.194091796875\n-------------------\n\nRunning epoch 9 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.21110581629204. Average accuracy 0.19714725378787878\n-------------------\nValidation dataset\nAverage loss : 3.187675654888153. Average accuracy 0.20068359375\n-------------------\n\nRunning epoch 10 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.172546148300171. Average accuracy 0.2001361268939394\n-------------------\nValidation dataset\nAverage loss : 3.155732214450836. Average accuracy 0.2037353515625\n-------------------\n\n"
],
[
"loss_ctc = torch.nn.CTCLoss(zero_infinity=True)",
"_____no_output_____"
],
[
"%cd CPC_audio/\nfrom cpc.eval.common_voices_eval import SingleSequenceDataset, parseSeqLabels, findAllSeqs\npath_train_data_per = datapath+'train'\npath_val_data_per = datapath+'val'\npath_phone_data_per = datapath2+'chars2.txt'\nBATCH_SIZE=8\n\nphone_labels, N_PHONES = parseSeqLabels(path_phone_data_per)\ndata_train_per, _ = findAllSeqs(path_train_data_per, extension='.wav')\ndataset_train_non_aligned = SingleSequenceDataset(path_train_data_per, data_train_per, phone_labels)\ndata_loader_train = torch.utils.data.DataLoader(dataset_train_non_aligned, batch_size=BATCH_SIZE,\n shuffle=True)\n\ndata_val_per, _ = findAllSeqs(path_val_data_per, extension='.wav')\ndataset_val_non_aligned = SingleSequenceDataset(path_val_data_per, data_val_per, phone_labels)\ndata_loader_val = torch.utils.data.DataLoader(dataset_val_non_aligned, batch_size=BATCH_SIZE,\n shuffle=True)",
"1it [00:00, 76.29it/s]"
],
[
"from cpc.feature_loader import loadModel\n\ncheckpoint_path = '../checkpoint_data/checkpoint_30.pt'\ncpc_model, HIDDEN_CONTEXT_MODEL, HIDDEN_ENCODER_MODEL = loadModel([checkpoint_path])\ncpc_model = cpc_model.cuda()\nphone_classifier = PhoneClassifier(HIDDEN_CONTEXT_MODEL, N_PHONES).to(device)",
"Loading checkpoint ../checkpoint_data/checkpoint_30.pt\nLoading the state dict at ../checkpoint_data/checkpoint_30.pt\n"
],
[
"parameters = list(phone_classifier.parameters()) + list(cpc_model.parameters())\nLEARNING_RATE = 2e-4\noptimizer = torch.optim.Adam(parameters, lr=LEARNING_RATE)\n\noptimizer_frozen = torch.optim.Adam(list(phone_classifier.parameters()), lr=LEARNING_RATE)",
"_____no_output_____"
],
[
"import torch.nn.functional as F\n\ndef train_one_epoch_ctc(cpc_model, \n phone_classifier, \n loss_criterion, \n data_loader, \n optimizer):\n \n cpc_model.train()\n loss_criterion.train()\n\n avg_loss = 0\n avg_accuracy = 0\n n_items = 0\n for step, full_data in enumerate(data_loader):\n\n x, x_len, y, y_len = full_data\n\n x_batch_len = x.shape[-1]\n x, y = x.to(device), y.to(device)\n\n bs=x.size(0)\n optimizer.zero_grad()\n context_out, enc_out, _ = cpc_model(x.to(device),y.to(device))\n \n scores = phone_classifier(context_out)\n scores = scores.permute(1,0,2)\n scores = F.log_softmax(scores,2)\n yhat_len = torch.tensor([int(scores.shape[0]*x_len[i]/x_batch_len) for i in range(scores.shape[1])]) # this is an approximation, should be good enough\n\n loss = loss_criterion(scores.float(),y.float().to(device),yhat_len,y_len)\n loss.backward()\n optimizer.step()\n avg_loss+=loss.item()*bs\n n_items+=bs\n avg_loss/=n_items\n\n return avg_loss\n\ndef validation_step(cpc_model, \n phone_classifier, \n loss_criterion, \n data_loader):\n\n cpc_model.eval()\n phone_classifier.eval()\n avg_loss = 0\n avg_accuracy = 0\n n_items = 0\n with torch.no_grad():\n for step, full_data in enumerate(data_loader):\n\n x, x_len, y, y_len = full_data\n\n x_batch_len = x.shape[-1]\n x, y = x.to(device), y.to(device)\n\n bs=x.size(0)\n context_out, enc_out, _ = cpc_model(x.to(device),y.to(device))\n \n scores = phone_classifier(context_out)\n scores = scores.permute(1,0,2)\n scores = F.log_softmax(scores,2)\n yhat_len = torch.tensor([int(scores.shape[0]*x_len[i]/x_batch_len) for i in range(scores.shape[1])]) # this is an approximation, should be good enough\n\n loss = loss_criterion(scores,y.to(device),yhat_len,y_len)\n avg_loss+=loss.item()*bs\n n_items+=bs\n avg_loss/=n_items\n #print(loss)\n return avg_loss\n\ndef run_ctc(cpc_model, \n phone_classifier, \n loss_criterion, \n data_loader_train, \n data_loader_val, \n optimizer,\n n_epoch):\n epoches = []\n train_losses = []\n val_losses = []\n\n for epoch in range(n_epoch):\n\n print(f\"Running epoch {epoch + 1} / {n_epoch}\")\n loss_train = train_one_epoch_ctc(cpc_model, phone_classifier, loss_criterion, data_loader_train, optimizer)\n print(\"-------------------\")\n print(f\"Training dataset :\")\n print(f\"Average loss : {loss_train}.\")\n\n print(\"-------------------\")\n print(\"Validation dataset\")\n loss_val = validation_step(cpc_model, phone_classifier, loss_criterion, data_loader_val)\n print(f\"Average loss : {loss_val}\")\n print(\"-------------------\")\n print()\n epoches.append(epoch)\n train_losses.append(loss_train)\n val_losses.append(loss_val)\n\n plt.plot(epoches, train_losses, label = \"ctc_train loss\")\n plt.plot(epoches, val_losses, label = \"ctc_val loss\")\n plt.xlabel('epoches')\n plt.ylabel('loss')\n plt.title('train and validation ctc loss')\n plt.legend()\n # Display and save a figure.\n plt.savefig(\"ctc_loss.png\")\n files.download(\"ctc_loss.png\") \n plt.show()\n\n ",
"_____no_output_____"
],
[
"run_ctc(cpc_model,phone_classifier,loss_ctc,data_loader_train,data_loader_val,optimizer_frozen,n_epoch=10)",
"Running epoch 1 / 10\n"
],
[
"import numpy as np\n\ndef get_PER_sequence(ref_seq, target_seq):\n\n # re = g.split()\n # h = h.split()\n n = len(ref_seq)\n m = len(target_seq)\n\n D = np.zeros((n+1,m+1))\n for i in range(1,n+1):\n D[i,0] = D[i-1,0]+1\n for j in range(1,m+1):\n D[0,j] = D[0,j-1]+1\n \n ### TODO compute the alignment\n\n for i in range(1,n+1):\n for j in range(1,m+1):\n D[i,j] = min(\n D[i-1,j]+1,\n D[i-1,j-1]+1,\n D[i,j-1]+1,\n D[i-1,j-1]+ 0 if ref_seq[i-1]==target_seq[j-1] else float(\"inf\")\n )\n return D[n,m]/len(ref_seq)\n \n\n #return PER",
"_____no_output_____"
],
[
"ref_seq = [0, 1, 1, 2, 0, 2, 2]\npred_seq = [1, 1, 2, 2, 0, 0]\n\nexpected_PER = 4. / 7.\nprint(get_PER_sequence(ref_seq, pred_seq) == expected_PER)",
"True\n"
],
[
"import progressbar\nfrom multiprocessing import Pool\n\ndef cut_data(seq, sizeSeq):\n maxSeq = sizeSeq.max()\n return seq[:, :maxSeq]\n\n\ndef prepare_data(data):\n seq, sizeSeq, phone, sizePhone = data\n seq = seq.cuda()\n phone = phone.cuda()\n sizeSeq = sizeSeq.cuda().view(-1)\n sizePhone = sizePhone.cuda().view(-1)\n\n seq = cut_data(seq.permute(0, 2, 1), sizeSeq).permute(0, 2, 1)\n return seq, sizeSeq, phone, sizePhone\n\n\ndef get_per(test_dataloader,\n cpc_model,\n phone_classifier):\n\n downsampling_factor = 160\n cpc_model.eval()\n phone_classifier.eval()\n\n avgPER = 0\n nItems = 0 \n \n per = []\n Item = []\n\n print(\"Starting the PER computation through beam search\")\n bar = progressbar.ProgressBar(maxval=len(test_dataloader))\n bar.start()\n\n for index, data in enumerate(test_dataloader):\n\n bar.update(index)\n\n with torch.no_grad():\n \n seq, sizeSeq, phone, sizePhone = prepare_data(data)\n c_feature, _, _ = cpc_model(seq.to(device),phone.to(device))\n sizeSeq = sizeSeq / downsampling_factor\n predictions = torch.nn.functional.softmax(\n phone_classifier(c_feature), dim=2).cpu()\n phone = phone.cpu()\n sizeSeq = sizeSeq.cpu()\n sizePhone = sizePhone.cpu()\n\n bs = c_feature.size(0)\n data_per = [(predictions[b].argmax(1), phone[b]) for b in range(bs)]\n # data_per = [(predictions[b], sizeSeq[b], phone[b], sizePhone[b],\n # \"criterion.module.BLANK_LABEL\") for b in range(bs)]\n\n with Pool(bs) as p:\n poolData = p.starmap(get_PER_sequence, data_per)\n avgPER += sum([x for x in poolData])\n nItems += len(poolData)\n per.append(sum([x for x in poolData]))\n Item.append(index)\n\n bar.finish()\n\n avgPER /= nItems\n\n print(f\"Average CER {avgPER}\")\n\n plt.plot(Item, per, label = \"Per by item\")\n plt.xlabel('Items')\n plt.ylabel('PER')\n plt.title('trends of the PER')\n plt.legend()\n # Display and save a figure.\n plt.savefig(\"Per.png\")\n files.download(\"Per.png\") \n plt.show()\n return avgPER",
"_____no_output_____"
],
[
"get_per(data_loader_val,cpc_model,phone_classifier)",
"\r \r\rN/A% (0 of 32) | | Elapsed Time: 0:00:00 ETA: --:--:--"
],
[
"# Load a dataset labelled with the letters of each sequence.\n%cd /content/CPC_audio\nfrom cpc.eval.common_voices_eval import SingleSequenceDataset, parseSeqLabels, findAllSeqs\npath_train_data_cer = datapath+'train'\npath_val_data_cer = datapath+'val'\npath_letter_data_cer = datapath2+'chars2.txt'\nBATCH_SIZE=8\n\nletters_labels, N_LETTERS = parseSeqLabels(path_letter_data_cer)\ndata_train_cer, _ = findAllSeqs(path_train_data_cer, extension='.wav')\ndataset_train_non_aligned = SingleSequenceDataset(path_train_data_cer, data_train_cer, letters_labels)\n\n\ndata_val_cer, _ = findAllSeqs(path_val_data_cer, extension='.wav')\ndataset_val_non_aligned = SingleSequenceDataset(path_val_data_cer, data_val_cer, letters_labels)\n\n\n# The data loader will generate a tuple of tensors data, labels for each batch\n# data : size N x T1 x 1 : the audio sequence\n# label : size N x T2 the sequence of letters corresponding to the audio data\n# IMPORTANT NOTE: just like the PER the CER is computed with non-aligned phone data.\ndata_loader_train_letters = torch.utils.data.DataLoader(dataset_train_non_aligned, batch_size=BATCH_SIZE,\n shuffle=True)\ndata_loader_val_letters = torch.utils.data.DataLoader(dataset_val_non_aligned, batch_size=BATCH_SIZE,\n shuffle=True)",
"1it [00:00, 77.80it/s]"
],
[
"from cpc.feature_loader import loadModel\n\ncheckpoint_path = '../checkpoint_data/checkpoint_30.pt'\ncpc_model, HIDDEN_CONTEXT_MODEL, HIDDEN_ENCODER_MODEL = loadModel([checkpoint_path])\ncpc_model = cpc_model.cuda()\ncharacter_classifier = PhoneClassifier(HIDDEN_CONTEXT_MODEL, N_LETTERS).to(device)",
"Loading checkpoint ../checkpoint_data/checkpoint_30.pt\nLoading the state dict at ../checkpoint_data/checkpoint_30.pt\n"
],
[
"parameters = list(character_classifier.parameters()) + list(cpc_model.parameters())\nLEARNING_RATE = 2e-4\noptimizer = torch.optim.Adam(parameters, lr=LEARNING_RATE)\n\noptimizer_frozen = torch.optim.Adam(list(character_classifier.parameters()), lr=LEARNING_RATE)",
"_____no_output_____"
],
[
"loss_ctc = torch.nn.CTCLoss(zero_infinity=True)",
"_____no_output_____"
],
[
"run_ctc(cpc_model,character_classifier,loss_ctc,data_loader_train_letters,data_loader_val_letters,optimizer_frozen,n_epoch=10)",
"Running epoch 1 / 10\n-------------------\nTraining dataset :\nAverage loss : 34.74688169776495.\n-------------------\nValidation dataset\nAverage loss : 29.79711248779297\n-------------------\n\nRunning epoch 2 / 10\n-------------------\nTraining dataset :\nAverage loss : 24.83258881113637.\n-------------------\nValidation dataset\nAverage loss : 20.162071548461913\n-------------------\n\nRunning epoch 3 / 10\n-------------------\nTraining dataset :\nAverage loss : 16.316355175229173.\n-------------------\nValidation dataset\nAverage loss : 12.900087188720702\n-------------------\n\nRunning epoch 4 / 10\n-------------------\nTraining dataset :\nAverage loss : 10.577213947737038.\n-------------------\nValidation dataset\nAverage loss : 8.67247957611084\n-------------------\n\nRunning epoch 5 / 10\n-------------------\nTraining dataset :\nAverage loss : 7.440222170364916.\n-------------------\nValidation dataset\nAverage loss : 6.443285507202148\n-------------------\n\nRunning epoch 6 / 10\n-------------------\nTraining dataset :\nAverage loss : 5.748367740880305.\n-------------------\nValidation dataset\nAverage loss : 5.191079601287842\n-------------------\n\nRunning epoch 7 / 10\n-------------------\nTraining dataset :\nAverage loss : 4.789907754725547.\n-------------------\nValidation dataset\nAverage loss : 4.464350517272949\n-------------------\n\nRunning epoch 8 / 10\n-------------------\nTraining dataset :\nAverage loss : 4.236433330612566.\n-------------------\nValidation dataset\nAverage loss : 4.036692462921143\n-------------------\n\nRunning epoch 9 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.9030140416706027.\n-------------------\nValidation dataset\nAverage loss : 3.7723377685546877\n-------------------\n\nRunning epoch 10 / 10\n-------------------\nTraining dataset :\nAverage loss : 3.6891175634297895.\n-------------------\nValidation dataset\nAverage loss : 3.596484752655029\n-------------------\n\n"
],
[
"get_per(data_loader_val_letters,cpc_model,character_classifier)",
"\r \r\rN/A% (0 of 32) | | Elapsed Time: 0:00:00 ETA: --:--:--"
],
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae6ace236a0565ff1b28a12a07e8a0358fcd9b6
| 26,823 |
ipynb
|
Jupyter Notebook
|
lectures/nx/data-science-live.ipynb
|
malvin0704/PIC16B
|
a88b9e1264dc2f842ef22a7742a2e43c16383c8f
|
[
"MIT"
] | null | null | null |
lectures/nx/data-science-live.ipynb
|
malvin0704/PIC16B
|
a88b9e1264dc2f842ef22a7742a2e43c16383c8f
|
[
"MIT"
] | null | null | null |
lectures/nx/data-science-live.ipynb
|
malvin0704/PIC16B
|
a88b9e1264dc2f842ef22a7742a2e43c16383c8f
|
[
"MIT"
] | null | null | null | 60.006711 | 6,504 | 0.719457 |
[
[
[
"# A Brief Overview of Network Data Science\n\nNetworks are extremely rich data structures which admit a wide variety of insightful data analysis tasks. In this set of notes, we'll consider two of the fundamental tasks in network data science: centrality and clustering. We'll also get a bit more practice with network visualization. ",
"_____no_output_____"
]
],
[
[
"import networkx as nx\nimport numpy as np\nfrom matplotlib import pyplot as plt\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"We'll mostly stick with the Karate Club network for today, as this is a very good network for visualization. ",
"_____no_output_____"
]
],
[
[
"G = nx.karate_club_graph()\n\npos = {i : (0, 0) for i in G.nodes()}\n\n# layout = nx.drawing.layout.spring_layout(G)\n\nnx.draw(G, \n with_labels = True, \n node_color = \"steelblue\")",
"_____no_output_____"
]
],
[
[
"### Centrality in Networks\n\nGiven a system, how can we determine *important* components in that system? In networks, the idea of importance is often cashed out in terms of *centrality*: important nodes are the nodes that are most \"central\" to the network. But how should we define or measure this? \n\nOne good way is by computing the degree (i.e. the number of friends possessed by each node). ",
"_____no_output_____"
],
[
"The degree is a direct measure of popularity. But what if it matters not only *how many* friends you have, but *who* those friends are? Maybe we'd like to measure importance using the following, apparently circular idea: \n\n> Central nodes tend to be connected to other central nodes. \n\nAs it turns out, one way to cash out this idea is in terms of...linear algebra! In particular, let's suppose that *my* importance should be proportional to the sum of the importances of my friends. So, if $v_i$ is the importance of node $i$, then we can write \n\n$$ v_i = \\alpha\\sum_{j \\;\\text{friends with} \\;i} v_j\\;, $$\n\nwhere $\\alpha$ is some constant of proportionality. Let's write this up a little more concisely. Let $\\mathbf{A} \\in \\mathbb{R}^{n \\times n}$ be the *adjacency* matrix, with entries \n\n$$ a_{ij} = \\begin{cases}1 &\\quad i \\;\\text{is friends with} \\;j \\\\ 0 &\\quad \\text{otherwise.}\\end{cases}$$\n\nNow, our equation above can be written in matrix-vector form as: \n\n$$\\mathbf{v} = \\alpha \\mathbf{A} \\mathbf{v}$$. \n\nWait! This says that $\\mathbf{v}$ is an eigenvector of $\\mathbf{A}$ with eigenvalue $\\frac{1}{\\alpha}$! So, we can compute centralities by finding eigenvectors of $\\mathbf{A}$. Usually, we just take the largest one. \n\nLet's try it out! Our first step is to obtain the adjacency matrix $\\mathbf{A}$. ",
"_____no_output_____"
],
[
"Now let's find the eigenvector corresponding to the largest eigenvalue. ",
"_____no_output_____"
],
[
"Now let's use this to create a plot: ",
"_____no_output_____"
],
[
"Compared to our calculation of degrees, this *eigenvector centrality* views nodes such as 17 as highly important, not because 17 has many neighbors but rather because 17 is connected to other important nodes. \n\n## PageRank (Again)",
"_____no_output_____"
],
[
"Did somebody say...PageRank? \n\nAs you may remember from either PIC16A or our lectures on linear algebra, PageRank is an algorithm for finding important entities in a complex, relational system. In fact, it's a form of centrality! While we could obtain the adjacency matrix and do the linear algebra manipulations to compute PageRank, an easier way is to use one of the many centrality measures built in to NetworkX. ",
"_____no_output_____"
],
[
"*Yes, we could have done this for eigenvector centrality as well.* ",
"_____no_output_____"
],
[
"Different centrality measures have different mathematical definitions and properties, which means that appropriately interpreting a given measure can be somewhat tricky. One should be cautious before leaping to conclusions about \"the most important node in the network.\" For example, the results look noticeably different when we use betweenness centrality, a popular heuristic that considers nodes to be more important if they are \"between\" lots of pairs of other nodes. ",
"_____no_output_____"
]
],
[
[
"bc = nx.algorithms.centrality.betweenness_centrality(G)\n\nnx.draw(G, layout,\n with_labels=True, \n node_color = \"steelblue\", \n node_size = [5000*bc[i] for i in G.nodes()],\n edgecolors = \"black\")",
"_____no_output_____"
]
],
[
[
"## Graph Clustering\n\nGraph clustering refers to the problem of finding collections of related nodes in the graph. It is one form of unsupervised machine learning, and is similar to problems that you may have seen like k-means and spectral clustering. Indeed, spectral clustering works well on graphs! \n\nAs mentioned above, a common benchmark for graph clustering algorithms is to attempt to reproduce the observed division of the Karate Club graph. Recall that that looks like this: ",
"_____no_output_____"
],
[
"The core idea of most clustering algorithms is that densely-connected sets of nodes are more likely to be members of the same cluster. There are *many* algorithms for graph clustering, which can lead to very different results. \n\nHere's one example. ",
"_____no_output_____"
],
[
"The variable `comms` is now a list of sets. Nodes in the same set are viewed as belonging to the same cluster. Let's visualize these: ",
"_____no_output_____"
],
[
"The result is clearly related to the observed partition, but we haven't recovered it exactly. Indeed, the algorithm picked up 3 clusters, when the clut only divided into two components! Some algorithms allow you to specify the desired number of clusters in advance, while others don't. \n\nWhat about our good friend, spectral clustering? The adjacency matrix of the graph actually can serve as the affinity or similarity matrix we used when studying point blogs. In fact, spectral clustering is often presented as an algorithm for graphs. \n\nThere's no implementation of spectral clustering within NetworkX, but it's easy enough to obtain the adjacency matrix and use the implementation in Scikit-learn`: ",
"_____no_output_____"
],
[
"The resulting clusters are fairly similar to the \"real\" clusters observed in the fracturing of the club. However, fundamentally, graph clustering is an *unsupervised* machine learning task, which means that the problem of defining what makes a \"good\" set of clusters is quite subtle and depends strongly on the data domain. ",
"_____no_output_____"
],
[
"## Graphs From Data\n\nThe easiest way to construct a graph from data is by converting from a Pandas data frame. When constructing a graph from data, using this or any other method, it's often necessary to do a bit of cleaning in order to produce a reasonable result. \n\nFor example, let's revisit the Hamilton mentions network: ",
"_____no_output_____"
]
],
[
[
"url = \"https://philchodrow.github.io/PIC16A/homework/HW3-hamilton-data.csv\"\ndf = pd.read_csv(url, names = [\"source\", \"target\"])\ndf.head()",
"_____no_output_____"
]
],
[
[
"We can think of this dataframe as a list of *directed* edges. ",
"_____no_output_____"
],
[
"Let's visualize? ",
"_____no_output_____"
],
[
"Well, this isn't very legible. Let's filter out all the characters who are only mentioned, but never mention anyone themselves. The number of outgoing edges from a node is often called the *out-degree*.",
"_____no_output_____"
],
[
"The `subgraph()` method can be used to filter down a network to just a desired set of nodes. ",
"_____no_output_____"
],
[
"Now the plot is a bit easier to read, especially if we add a pleasant layout. ",
"_____no_output_____"
],
[
"Great! Having performed our cleaning steps, we could proceed to analyze this graph. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4ae6c527873e1a5b4dece1bec62c53e1a4829977
| 11,772 |
ipynb
|
Jupyter Notebook
|
mnist_dense_neural_network/mnist_dense_neural_network.ipynb
|
gyan-krishna/exploring-ml-and-dl
|
1230b4ac051cc349c1097f3176c5434cfd0db849
|
[
"MIT"
] | null | null | null |
mnist_dense_neural_network/mnist_dense_neural_network.ipynb
|
gyan-krishna/exploring-ml-and-dl
|
1230b4ac051cc349c1097f3176c5434cfd0db849
|
[
"MIT"
] | null | null | null |
mnist_dense_neural_network/mnist_dense_neural_network.ipynb
|
gyan-krishna/exploring-ml-and-dl
|
1230b4ac051cc349c1097f3176c5434cfd0db849
|
[
"MIT"
] | null | null | null | 48.245902 | 5,128 | 0.712878 |
[
[
[
"#!pip install pydot\n#!pip install graphviz \n#!pip install pydotplus\n!pip3 install keras",
"Requirement already satisfied: keras in c:\\python39\\lib\\site-packages (2.4.3)\nRequirement already satisfied: scipy>=0.14 in c:\\python39\\lib\\site-packages (from keras) (1.6.2)\nRequirement already satisfied: numpy>=1.9.1 in c:\\python39\\lib\\site-packages (from keras) (1.20.2)\nRequirement already satisfied: pyyaml in c:\\python39\\lib\\site-packages (from keras) (5.4.1)\nRequirement already satisfied: h5py in c:\\python39\\lib\\site-packages (from keras) (3.2.1)\n"
],
[
"\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Flatten, Dropout, Activation\nfrom keras.utils import np_utils, plot_model\n\nfrom keras.datasets import mnist\nfrom numpy import argmax\nimport pydot\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"(x_train, y_train), (x_test, y_test) = mnist.load_data()\nplt.imshow(x_train[0])\nprint(x_train.shape)\nprint(y_train.shape)\nprint(y_train[0])",
"(60000, 28, 28)\n(60000,)\n5\n"
],
[
"#normalising the data set\nx_train = x_train.astype('float32')\nx_test = x_test.astype('float32')\nx_train /= 255\nx_test /= 255",
"_____no_output_____"
],
[
"#creating the output matrix as a categorical dataset\nY_train = np_utils.to_categorical(y_train, 10)\nY_test = np_utils.to_categorical(y_test, 10)\nprint(Y_train[0])",
"[0. 0. 0. 0. 0. 1. 0. 0. 0. 0.]\n"
],
[
"#creating the model\n\nmodel = Sequential()\nmodel.add(Flatten(input_shape=(28, 28)))\nmodel.add(Dense(128, activation = 'relu'))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(10))\nmodel.add(Activation('softmax'))\n\nmodel.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nflatten (Flatten) (None, 784) 0 \n_________________________________________________________________\ndense (Dense) (None, 128) 100480 \n_________________________________________________________________\ndropout (Dropout) (None, 128) 0 \n_________________________________________________________________\ndense_1 (Dense) (None, 10) 1290 \n_________________________________________________________________\nactivation (Activation) (None, 10) 0 \n=================================================================\nTotal params: 101,770\nTrainable params: 101,770\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"plot_model(model,\n to_file = \"model.jpg\",\n show_shapes = True,\n show_layer_names = True,\n rankdir = \"TB\",\n expand_nested = False,\n dpi = 96)",
"_____no_output_____"
],
[
"model.compile(loss = 'categorical_crossentropy',\n optimizer='adam',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.fit(x_train, Y_train, batch_size=32, epochs=10, verbose=1)",
"_____no_output_____"
],
[
"score = model.evaluate(x_test, Y_test, verbose = 1)\n#5 epochs = 725us/step - loss: 0.1464 - accuracy: 0.9808\n#10 epochs = 687us/step - loss: 0.0634 - accuracy: 0.9818\n#20 epochs = 671us/step - loss: 0.0972 - accuracy: 0.9787\n#30 epochs = 668us/step - loss: 0.1243 - accuracy: 0.9805\n#40 epochs = 808us/step - loss: 0.1477 - accuracy: 0.9821",
"_____no_output_____"
],
[
"plt.imshow(x_test[0])\nerror_count = 0\nfor i in range(len(x_test)):\n x_test0 = x_test[i].reshape(-1,28,28)\n prediction = model.predict(x_test0)\n predicted_class = argmax(prediction)\n if(predicted_class != y_test[i]):\n error_count+=1\nprint(error_count)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae6ce5ab410cfb45fa2de054f30b3eb73e3fdca
| 41,157 |
ipynb
|
Jupyter Notebook
|
05_03_UCS_dijkstra.ipynb
|
u20181a993/algorithmic_complexity
|
c0040e133d1e46073e82e0678c18f6fdee7d4362
|
[
"CC0-1.0"
] | 17 |
2021-04-11T04:25:02.000Z
|
2022-03-28T22:20:58.000Z
|
05_03_UCS_dijkstra.ipynb
|
u20181a993/algorithmic_complexity
|
c0040e133d1e46073e82e0678c18f6fdee7d4362
|
[
"CC0-1.0"
] | 7 |
2021-10-01T22:27:37.000Z
|
2021-11-27T01:32:03.000Z
|
05_03_UCS_dijkstra.ipynb
|
u20181a993/algorithmic_complexity
|
c0040e133d1e46073e82e0678c18f6fdee7d4362
|
[
"CC0-1.0"
] | 30 |
2021-04-12T15:43:14.000Z
|
2022-03-15T18:07:19.000Z
| 131.913462 | 17,469 | 0.550186 |
[
[
[
"<a href=\"https://colab.research.google.com/github/lmcanavals/algorithmic_complexity/blob/main/05_01_UCS_dijkstra.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Breadth First Search\n\nBFS para los amigos",
"_____no_output_____"
]
],
[
[
"import graphviz as gv\nimport numpy as np\nimport pandas as pd\nimport heapq as hq\nimport math",
"_____no_output_____"
],
[
"def readAdjl(fn, haslabels=False, weighted=False, sep=\"|\"):\n with open(fn) as f:\n labels = None\n if haslabels:\n labels = f.readline().strip().split()\n L = []\n for line in f:\n if weighted:\n L.append([tuple(map(int, p.split(sep))) for p in line.strip().split()])\n # line => \"1|3 2|5 4|4\" ==> [(1, 3), (2, 5), (4, 4)]\n else: \n L.append(list(map(int, line.strip().split()))) # \"1 3 5\" => [1, 3, 5]\n # L.append([int(x) for x in line.strip().split()])\n return L, labels\n\ndef adjlShow(L, labels=None, directed=False, weighted=False, path=[],\n layout=\"sfdp\"):\n g = gv.Digraph(\"G\") if directed else gv.Graph(\"G\")\n g.graph_attr[\"layout\"] = layout\n g.edge_attr[\"color\"] = \"gray\"\n g.node_attr[\"color\"] = \"orangered\"\n g.node_attr[\"width\"] = \"0.1\"\n g.node_attr[\"height\"] = \"0.1\"\n g.node_attr[\"fontsize\"] = \"8\"\n g.node_attr[\"fontcolor\"] = \"mediumslateblue\"\n g.node_attr[\"fontname\"] = \"monospace\"\n g.edge_attr[\"fontsize\"] = \"8\"\n g.edge_attr[\"fontname\"] = \"monospace\"\n n = len(L)\n for u in range(n):\n g.node(str(u), labels[u] if labels else str(u))\n added = set()\n for v, u in enumerate(path):\n if u != None:\n if weighted:\n for vi, w in G[u]:\n if vi == v:\n break\n g.edge(str(u), str(v), str(w), dir=\"forward\", penwidth=\"2\", color=\"orange\")\n else:\n g.edge(str(u), str(v), dir=\"forward\", penwidth=\"2\", color=\"orange\")\n added.add(f\"{u},{v}\")\n added.add(f\"{v},{u}\")\n if weighted:\n for u in range(n):\n for v, w in L[u]:\n if not directed and not f\"{u},{v}\" in added:\n added.add(f\"{u},{v}\")\n added.add(f\"{v},{u}\")\n g.edge(str(u), str(v), str(w))\n elif directed:\n g.edge(str(u), str(v), str(w))\n else:\n for u in range(n):\n for v in L[u]:\n if not directed and not f\"{u},{v}\" in added:\n added.add(f\"{u},{v}\")\n added.add(f\"{v},{u}\")\n g.edge(str(u), str(v))\n elif directed:\n g.edge(str(u), str(v))\n return g",
"_____no_output_____"
]
],
[
[
"## Dijkstra",
"_____no_output_____"
]
],
[
[
"def dijkstra(G, s):\n n = len(G)\n visited = [False]*n\n path = [None]*n\n cost = [math.inf]*n\n cost[s] = 0\n queue = [(0, s)]\n while queue:\n g_u, u = hq.heappop(queue)\n if not visite[u]:\n visited[u] = True\n for v, w in G[u]:\n f = g_u + w\n if f < cost[v]:\n cost[v] = f\n path[v] = u\n hq.heappush(queue, (f, v))\n\n return path, cost",
"_____no_output_____"
],
[
"%%file 1.in\n2|4 7|8 14|3\n2|7 5|7\n0|4 1|7 3|5 6|1\n2|5\n7|7\n1|7 6|1 8|5\n2|1 5|1\n0|8 4|7 8|8\n5|5 7|8 9|8 11|9 12|6\n8|8 10|8 12|9 13|7\n9|8 13|3\n8|9\n8|6 9|9 13|2 15|5\n9|7 10|13 12|2 16|9\n0|3 15|9\n12|5 14|9 17|7\n13|9 17|8\n15|7 16|8",
"Writing 1.in\n"
],
[
"G, _ = readAdjl(\"1.in\", weighted=True)\nfor i, edges in enumerate(G):\n print(f\"{i:2}: {edges}\")\nadjlShow(G, weighted=True)",
" 0: [(2, 4), (7, 8), (14, 3)]\n 1: [(2, 7), (5, 7)]\n 2: [(0, 4), (1, 7), (3, 5), (6, 1)]\n 3: [(2, 5)]\n 4: [(7, 7)]\n 5: [(1, 7), (6, 1), (8, 5)]\n 6: [(2, 1), (5, 1)]\n 7: [(0, 8), (4, 7), (8, 8)]\n 8: [(5, 5), (7, 8), (9, 8), (11, 9), (12, 6)]\n 9: [(8, 8), (10, 8), (12, 9), (13, 7)]\n10: [(9, 8), (13, 3)]\n11: [(8, 9)]\n12: [(8, 6), (9, 9), (13, 2), (15, 5)]\n13: [(9, 7), (10, 13), (12, 2), (16, 9)]\n14: [(0, 3), (15, 9)]\n15: [(12, 5), (14, 9), (17, 7)]\n16: [(13, 9), (17, 8)]\n17: [(15, 7), (16, 8)]\n"
],
[
"path, cost = dijkstra(G, 8)\nprint(path)\nadjlShow(G, weighted=True, path=path)",
"[2, 5, 6, 2, 7, 8, 5, 8, None, 8, 9, 8, 8, 12, 0, 12, 13, 15]\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4ae6d1fe570083385a65d3b3871dbb5642684d33
| 7,550 |
ipynb
|
Jupyter Notebook
|
__site/__generated/A-fit-predict/tutorial.ipynb
|
JuliaAI/DataScienceTutorials.jl
|
76b1d3d1a5045c62b104a8358e4c37049f15ccb6
|
[
"MIT"
] | 29 |
2021-08-09T11:35:53.000Z
|
2022-03-07T06:20:43.000Z
|
__site/__generated/A-fit-predict/tutorial.ipynb
|
JuliaAI/DataScienceTutorials.jl
|
76b1d3d1a5045c62b104a8358e4c37049f15ccb6
|
[
"MIT"
] | 24 |
2021-08-09T09:44:23.000Z
|
2022-01-14T10:02:11.000Z
|
__site/__generated/A-fit-predict/tutorial.ipynb
|
JuliaAI/DataScienceTutorials.jl
|
76b1d3d1a5045c62b104a8358e4c37049f15ccb6
|
[
"MIT"
] | 1 |
2021-11-12T10:52:05.000Z
|
2021-11-12T10:52:05.000Z
| 26.491228 | 264 | 0.58543 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4ae6ecc12c9283ac0bec5c536ae310fa5d90f195
| 4,958 |
ipynb
|
Jupyter Notebook
|
AlphaVantage/AlphaVantage_Get_fundamental_data.ipynb
|
stjordanis/awesome-notebooks
|
aeba5bcf5dda6795b8739c66c0b32ad857ca08d6
|
[
"BSD-3-Clause"
] | 1 |
2021-07-06T21:07:48.000Z
|
2021-07-06T21:07:48.000Z
|
AlphaVantage/AlphaVantage_Get_fundamental_data.ipynb
|
girilv/awesome-notebooks
|
0a0be50b5184f220f4a4ff5dea85107af85036c5
|
[
"BSD-3-Clause"
] | null | null | null |
AlphaVantage/AlphaVantage_Get_fundamental_data.ipynb
|
girilv/awesome-notebooks
|
0a0be50b5184f220f4a4ff5dea85107af85036c5
|
[
"BSD-3-Clause"
] | null | null | null | 26.8 | 800 | 0.612949 |
[
[
[
"<img width=\"10%\" alt=\"Naas\" src=\"https://landen.imgix.net/jtci2pxwjczr/assets/5ice39g4.png?w=160\"/>",
"_____no_output_____"
],
[
"# AlphaVantage - Get fundamental data\n<a href=\"https://app.naas.ai/user-redirect/naas/downloader?url=https://raw.githubusercontent.com/jupyter-naas/awesome-notebooks/master/AlphaVantage/AlphaVantage_Get_fundamental_data.ipynb\" target=\"_parent\">\n<img src=\"https://img.shields.io/badge/-Open%20in%20Naas-success?labelColor=000000&logo=data:image/svg+xml;base64,PD94bWwgdmVyc2lvbj0iMS4wIiBlbmNvZGluZz0iVVRGLTgiPz4KPHN2ZyB3aWR0aD0iMTAyNHB4IiBoZWlnaHQ9IjEwMjRweCIgdmlld0JveD0iMCAwIDEwMjQgMTAyNCIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIiB4bWxuczp4bGluaz0iaHR0cDovL3d3dy53My5vcmcvMTk5OS94bGluayIgdmVyc2lvbj0iMS4xIj4KIDwhLS0gR2VuZXJhdGVkIGJ5IFBpeGVsbWF0b3IgUHJvIDIuMC41IC0tPgogPGRlZnM+CiAgPHRleHQgaWQ9InN0cmluZyIgdHJhbnNmb3JtPSJtYXRyaXgoMS4wIDAuMCAwLjAgMS4wIDIyOC4wIDU0LjUpIiBmb250LWZhbWlseT0iQ29tZm9ydGFhLVJlZ3VsYXIsIENvbWZvcnRhYSIgZm9udC1zaXplPSI4MDAiIHRleHQtZGVjb3JhdGlvbj0ibm9uZSIgZmlsbD0iI2ZmZmZmZiIgeD0iMS4xOTk5OTk5OTk5OTk5ODg2IiB5PSI3MDUuMCI+bjwvdGV4dD4KIDwvZGVmcz4KIDx1c2UgaWQ9Im4iIHhsaW5rOmhyZWY9IiNzdHJpbmciLz4KPC9zdmc+Cg==\"/>\n</a>",
"_____no_output_____"
],
[
"## Company overview",
"_____no_output_____"
]
],
[
[
"import requests\nimport pandas as pd\n\nresponse = requests.get('https://www.alphavantage.co/query?function=OVERVIEW&symbol=IBM&apikey=demo')\ndata = response.json()\ndata",
"_____no_output_____"
]
],
[
[
"## Income statement ",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('https://www.alphavantage.co/query?function=INCOME_STATEMENT&symbol=IBM&apikey=demo')\ndata = response.json()\ndata",
"_____no_output_____"
]
],
[
[
"## Balance sheet ",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('https://www.alphavantage.co/query?function=BALANCE_SHEET&symbol=IBM&apikey=demo')\n\ndata = response.json()\ndata",
"_____no_output_____"
]
],
[
[
"## Cashflow statement",
"_____no_output_____"
]
],
[
[
"import requests\n\nresponse = requests.get('https://www.alphavantage.co/query?function=CASH_FLOW&symbol=IBM&apikey=demo')\n\ndata = response.json()\ndata",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae6eefe8867f2de4c809cc334bddc63bf37be35
| 29,562 |
ipynb
|
Jupyter Notebook
|
reinforcement_learning/rl_deepracer_robomaker_coach_gazebo/rl_deepracer_coach_robomaker.ipynb
|
gwulfs/amazon-sagemaker-examples
|
a8fecc389abc417f6af96f94856caa2a8ebd1c56
|
[
"Apache-2.0"
] | null | null | null |
reinforcement_learning/rl_deepracer_robomaker_coach_gazebo/rl_deepracer_coach_robomaker.ipynb
|
gwulfs/amazon-sagemaker-examples
|
a8fecc389abc417f6af96f94856caa2a8ebd1c56
|
[
"Apache-2.0"
] | null | null | null |
reinforcement_learning/rl_deepracer_robomaker_coach_gazebo/rl_deepracer_coach_robomaker.ipynb
|
gwulfs/amazon-sagemaker-examples
|
a8fecc389abc417f6af96f94856caa2a8ebd1c56
|
[
"Apache-2.0"
] | 2 |
2019-07-09T18:32:20.000Z
|
2020-09-11T19:07:55.000Z
| 38.243208 | 622 | 0.58034 |
[
[
[
"# Distributed DeepRacer RL training with SageMaker and RoboMaker\n\n---\n## Introduction\n\n\nIn this notebook, we will train a fully autonomous 1/18th scale race car using reinforcement learning using Amazon SageMaker RL and AWS RoboMaker's 3D driving simulator. [AWS RoboMaker](https://console.aws.amazon.com/robomaker/home#welcome) is a service that makes it easy for developers to develop, test, and deploy robotics applications. \n\nThis notebook provides a jailbreak experience of [AWS DeepRacer](https://console.aws.amazon.com/deepracer/home#welcome), giving us more control over the training/simulation process and RL algorithm tuning.\n\n\n\n\n---\n## How it works? \n\n\n\nThe reinforcement learning agent (i.e. our autonomous car) learns to drive by interacting with its environment, e.g., the track, by taking an action in a given state to maximize the expected reward. The agent learns the optimal plan of actions in training by trial-and-error through repeated episodes. \n \nThe figure above shows an example of distributed RL training across SageMaker and two RoboMaker simulation envrionments that perform the **rollouts** - execute a fixed number of episodes using the current model or policy. The rollouts collect agent experiences (state-transition tuples) and share this data with SageMaker for training. SageMaker updates the model policy which is then used to execute the next sequence of rollouts. This training loop continues until the model converges, i.e. the car learns to drive and stops going off-track. More formally, we can define the problem in terms of the following: \n\n1. **Objective**: Learn to drive autonomously by staying close to the center of the track.\n2. **Environment**: A 3D driving simulator hosted on AWS RoboMaker.\n3. **State**: The driving POV image captured by the car's head camera, as shown in the illustration above.\n4. **Action**: Six discrete steering wheel positions at different angles (configurable)\n5. **Reward**: Positive reward for staying close to the center line; High penalty for going off-track. This is configurable and can be made more complex (for e.g. steering penalty can be added).",
"_____no_output_____"
],
[
"## Prequisites",
"_____no_output_____"
],
[
"### Imports",
"_____no_output_____"
],
[
"To get started, we'll import the Python libraries we need, set up the environment with a few prerequisites for permissions and configurations.",
"_____no_output_____"
],
[
"You can run this notebook from your local machine or from a SageMaker notebook instance. In both of these scenarios, you can run the following to launch a training job on `SageMaker` and a simulation job on `RoboMaker`.",
"_____no_output_____"
]
],
[
[
"import sagemaker\nimport boto3\nimport sys\nimport os\nimport glob\nimport re\nimport subprocess\nfrom IPython.display import Markdown\nfrom time import gmtime, strftime\nsys.path.append(\"common\")\nfrom misc import get_execution_role, wait_for_s3_object\nfrom sagemaker.rl import RLEstimator, RLToolkit, RLFramework\nfrom markdown_helper import *",
"_____no_output_____"
]
],
[
[
"### Setup S3 bucket",
"_____no_output_____"
],
[
"Set up the linkage and authentication to the S3 bucket that we want to use for checkpoint and metadata.",
"_____no_output_____"
]
],
[
[
"# S3 bucket\nsage_session = sagemaker.session.Session()\ns3_bucket = sage_session.default_bucket() \ns3_output_path = 's3://{}/'.format(s3_bucket) # SDK appends the job name and output folder",
"_____no_output_____"
]
],
[
[
"### Define Variables",
"_____no_output_____"
],
[
"We define variables such as the job prefix for the training jobs and s3_prefix for storing metadata required for synchronization between the training and simulation jobs",
"_____no_output_____"
]
],
[
[
"job_name_prefix = 'rl-deepracer'\n\n# create unique job name\njob_name = s3_prefix = job_name_prefix + \"-sagemaker-\" + strftime(\"%y%m%d-%H%M%S\", gmtime())\n\n# Duration of job in seconds (5 hours)\njob_duration_in_seconds = 3600 * 5\n\naws_region = sage_session.boto_region_name\n\nif aws_region not in [\"us-west-2\", \"us-east-1\", \"eu-west-1\"]:\n raise Exception(\"This notebook uses RoboMaker which is available only in US East (N. Virginia), US West (Oregon) and EU (Ireland). Please switch to one of these regions.\")\nprint(\"Model checkpoints and other metadata will be stored at: {}{}\".format(s3_output_path, job_name))",
"_____no_output_____"
]
],
[
[
"### Create an IAM role\nEither get the execution role when running from a SageMaker notebook `role = sagemaker.get_execution_role()` or, when running from local machine, use utils method `role = get_execution_role('role_name')` to create an execution role.",
"_____no_output_____"
]
],
[
[
"try:\n role = sagemaker.get_execution_role()\nexcept:\n role = get_execution_role('sagemaker')\n\nprint(\"Using IAM role arn: {}\".format(role))",
"_____no_output_____"
]
],
[
[
"> Please note that this notebook cannot be run in `SageMaker local mode` as the simulator is based on AWS RoboMaker service.",
"_____no_output_____"
],
[
"### Permission setup for invoking AWS RoboMaker from this notebook",
"_____no_output_____"
],
[
"In order to enable this notebook to be able to execute AWS RoboMaker jobs, we need to add one trust relationship to the default execution role of this notebook.\n",
"_____no_output_____"
]
],
[
[
"display(Markdown(generate_help_for_robomaker_trust_relationship(role)))",
"_____no_output_____"
]
],
[
[
"### Configure VPC",
"_____no_output_____"
],
[
"Since SageMaker and RoboMaker have to communicate with each other over the network, both of these services need to run in VPC mode. This can be done by supplying subnets and security groups to the job launching scripts. \nWe will use the default VPC configuration for this example.",
"_____no_output_____"
]
],
[
[
"ec2 = boto3.client('ec2')\ndefault_vpc = [vpc['VpcId'] for vpc in ec2.describe_vpcs()['Vpcs'] if vpc[\"IsDefault\"] == True][0]\n\ndefault_security_groups = [group[\"GroupId\"] for group in ec2.describe_security_groups()['SecurityGroups'] \\\n if group[\"GroupName\"] == \"default\" and group[\"VpcId\"] == default_vpc]\n\ndefault_subnets = [subnet[\"SubnetId\"] for subnet in ec2.describe_subnets()[\"Subnets\"] \\\n if subnet[\"VpcId\"] == default_vpc and subnet['DefaultForAz']==True]\n\nprint(\"Using default VPC:\", default_vpc)\nprint(\"Using default security group:\", default_security_groups)\nprint(\"Using default subnets:\", default_subnets)",
"_____no_output_____"
]
],
[
[
"A SageMaker job running in VPC mode cannot access S3 resourcs. So, we need to create a VPC S3 endpoint to allow S3 access from SageMaker container. To learn more about the VPC mode, please visit [this link.](https://docs.aws.amazon.com/sagemaker/latest/dg/train-vpc.html)",
"_____no_output_____"
]
],
[
[
"try:\n route_tables = [route_table[\"RouteTableId\"] for route_table in ec2.describe_route_tables()['RouteTables']\\\n if route_table['VpcId'] == default_vpc]\nexcept Exception as e:\n if \"UnauthorizedOperation\" in str(e):\n display(Markdown(generate_help_for_s3_endpoint_permissions(role)))\n else:\n display(Markdown(create_s3_endpoint_manually(aws_region, default_vpc)))\n raise e\n\nprint(\"Trying to attach S3 endpoints to the following route tables:\", route_tables)\n\nassert len(route_tables) >= 1, \"No route tables were found. Please follow the VPC S3 endpoint creation \"\\\n \"guide by clicking the above link.\"\n\ntry:\n ec2.create_vpc_endpoint(DryRun=False,\n VpcEndpointType=\"Gateway\",\n VpcId=default_vpc,\n ServiceName=\"com.amazonaws.{}.s3\".format(aws_region),\n RouteTableIds=route_tables)\n print(\"S3 endpoint created successfully!\")\nexcept Exception as e:\n if \"RouteAlreadyExists\" in str(e):\n print(\"S3 endpoint already exists.\")\n elif \"UnauthorizedOperation\" in str(e):\n display(Markdown(generate_help_for_s3_endpoint_permissions(role)))\n raise e\n else:\n display(Markdown(create_s3_endpoint_manually(aws_region, default_vpc)))\n raise e",
"_____no_output_____"
]
],
[
[
"## Setup the environment\n",
"_____no_output_____"
],
[
"The environment is defined in a Python file called “deepracer_env.py” and the file can be found at `src/robomaker/environments/`. This file implements the gym interface for our Gazebo based RoboMakersimulator. This is a common environment file used by both SageMaker and RoboMaker. The environment variable - `NODE_TYPE` defines which node the code is running on. So, the expressions that have `rospy` dependencies are executed on RoboMaker only. \n\nWe can experiment with different reward functions by modifying `reward_function` in this file. Action space and steering angles can be changed by modifying the step method in `DeepRacerDiscreteEnv` class.",
"_____no_output_____"
],
[
"### Configure the preset for RL algorithm\nThe parameters that configure the RL training job are defined in `src/robomaker/presets/deepracer.py`. Using the preset file, you can define agent parameters to select the specific agent algorithm. We suggest using Clipped PPO for this example. \nYou can edit this file to modify algorithm parameters like learning_rate, neural network structure, batch_size, discount factor etc.",
"_____no_output_____"
]
],
[
[
"!pygmentize src/robomaker/presets/deepracer.py",
"_____no_output_____"
]
],
[
[
"### Training Entrypoint\nThe training code is written in the file “training_worker.py” which is uploaded in the /src directory. At a high level, it does the following:\n- Uploads SageMaker node's IP address.\n- Starts a Redis server which receives agent experiences sent by rollout worker[s] (RoboMaker simulator).\n- Trains the model everytime after a certain number of episodes are received.\n- Uploads the new model weights on S3. The rollout workers then update their model to execute the next set of episodes.",
"_____no_output_____"
]
],
[
[
"# Uncomment the line below to see the training code\n#!pygmentize src/training_worker.py",
"_____no_output_____"
]
],
[
[
"### Train the RL model using the Python SDK Script mode¶\n",
"_____no_output_____"
],
[
"First, we upload the preset and envrionment file to a particular location on S3, as expected by RoboMaker.",
"_____no_output_____"
]
],
[
[
"s3_location = \"s3://%s/%s\" % (s3_bucket, s3_prefix)\n\n# Make sure nothing exists at this S3 prefix\n!aws s3 rm --recursive {s3_location}\n\n# Make any changes to the envrironment and preset files below and upload these files\n!aws s3 cp src/robomaker/environments/ {s3_location}/environments/ --recursive --exclude \".ipynb_checkpoints*\" --exclude \"*.pyc\"\n!aws s3 cp src/robomaker/presets/ {s3_location}/presets/ --recursive --exclude \".ipynb_checkpoints*\" --exclude \"*.pyc\"",
"_____no_output_____"
]
],
[
[
"Next, we define the following algorithm metrics that we want to capture from cloudwatch logs to monitor the training progress. These are algorithm specific parameters and might change for different algorithm. We use [Clipped PPO](https://coach.nervanasys.com/algorithms/policy_optimization/cppo/index.html) for this example.",
"_____no_output_____"
]
],
[
[
"metric_definitions = [\n # Training> Name=main_level/agent, Worker=0, Episode=19, Total reward=-102.88, Steps=19019, Training iteration=1\n {'Name': 'reward-training',\n 'Regex': '^Training>.*Total reward=(.*?),'},\n \n # Policy training> Surrogate loss=-0.32664725184440613, KL divergence=7.255815035023261e-06, Entropy=2.83156156539917, training epoch=0, learning_rate=0.00025\n {'Name': 'ppo-surrogate-loss',\n 'Regex': '^Policy training>.*Surrogate loss=(.*?),'},\n {'Name': 'ppo-entropy',\n 'Regex': '^Policy training>.*Entropy=(.*?),'},\n \n # Testing> Name=main_level/agent, Worker=0, Episode=19, Total reward=1359.12, Steps=20015, Training iteration=2\n {'Name': 'reward-testing',\n 'Regex': '^Testing>.*Total reward=(.*?),'},\n]",
"_____no_output_____"
]
],
[
[
"We use the RLEstimator for training RL jobs.\n\n1. Specify the source directory which has the environment file, preset and training code.\n2. Specify the entry point as the training code\n3. Specify the choice of RL toolkit and framework. This automatically resolves to the ECR path for the RL Container.\n4. Define the training parameters such as the instance count, instance type, job name, s3_bucket and s3_prefix for storing model checkpoints and metadata. **Only 1 training instance is supported for now.**\n4. Set the RLCOACH_PRESET as \"deepracer\" for this example.\n5. Define the metrics definitions that you are interested in capturing in your logs. These can also be visualized in CloudWatch and SageMaker Notebooks.",
"_____no_output_____"
]
],
[
[
"RLCOACH_PRESET = \"deepracer\"\n\ninstance_type = \"ml.c5.4xlarge\"\n\nestimator = RLEstimator(entry_point=\"training_worker.py\",\n source_dir='src',\n dependencies=[\"common/sagemaker_rl\"],\n toolkit=RLToolkit.COACH,\n toolkit_version='0.10.1',\n framework=RLFramework.TENSORFLOW,\n role=role,\n train_instance_type=instance_type,\n train_instance_count=1,\n output_path=s3_output_path,\n base_job_name=job_name_prefix,\n train_max_run=job_duration_in_seconds, # Maximum runtime in seconds\n hyperparameters={\"s3_bucket\": s3_bucket,\n \"s3_prefix\": s3_prefix,\n \"aws_region\": aws_region,\n \"RLCOACH_PRESET\": RLCOACH_PRESET,\n },\n metric_definitions = metric_definitions,\n subnets=default_subnets, # Required for VPC mode\n security_group_ids=default_security_groups, # Required for VPC mode\n )\n\nestimator.fit(job_name=job_name, wait=False)",
"_____no_output_____"
]
],
[
[
"### Start the Robomaker job",
"_____no_output_____"
]
],
[
[
"from botocore.exceptions import UnknownServiceError\n\nrobomaker = boto3.client(\"robomaker\")",
"_____no_output_____"
]
],
[
[
"### Create Simulation Application",
"_____no_output_____"
],
[
"We first create a RoboMaker simulation application using the `DeepRacer public bundle`. Please refer to [RoboMaker Sample Application Github Repository](https://github.com/aws-robotics/aws-robomaker-sample-application-deepracer) if you want to learn more about this bundle or modify it.",
"_____no_output_____"
]
],
[
[
"bundle_s3_key = 'deepracer/simulation_ws.tar.gz'\nbundle_source = {'s3Bucket': s3_bucket,\n 's3Key': bundle_s3_key,\n 'architecture': \"X86_64\"}\nsimulation_software_suite={'name': 'Gazebo',\n 'version': '7'}\nrobot_software_suite={'name': 'ROS',\n 'version': 'Kinetic'}\nrendering_engine={'name': 'OGRE', 'version': '1.x'}",
"_____no_output_____"
]
],
[
[
"Download the public DeepRacer bundle provided by RoboMaker and upload it in our S3 bucket to create a RoboMaker Simulation Application",
"_____no_output_____"
]
],
[
[
"simulation_application_bundle_location = \"https://s3-us-west-2.amazonaws.com/robomaker-applications-us-west-2-11d8d0439f6a/deep-racer/deep-racer-1.0.57.0.1.0.66.0/simulation_ws.tar.gz\"\n\n!wget {simulation_application_bundle_location}\n!aws s3 cp simulation_ws.tar.gz s3://{s3_bucket}/{bundle_s3_key}\n!rm simulation_ws.tar.gz",
"_____no_output_____"
],
[
"app_name = \"deepracer-sample-application\" + strftime(\"%y%m%d-%H%M%S\", gmtime())\n\ntry:\n response = robomaker.create_simulation_application(name=app_name,\n sources=[bundle_source],\n simulationSoftwareSuite=simulation_software_suite,\n robotSoftwareSuite=robot_software_suite,\n renderingEngine=rendering_engine\n )\n simulation_app_arn = response[\"arn\"]\n print(\"Created a new simulation app with ARN:\", simulation_app_arn)\nexcept Exception as e:\n if \"AccessDeniedException\" in str(e):\n display(Markdown(generate_help_for_robomaker_all_permissions(role)))\n raise e\n else:\n raise e",
"_____no_output_____"
]
],
[
[
"### Launch the Simulation job on RoboMaker\n\nWe create [AWS RoboMaker](https://console.aws.amazon.com/robomaker/home#welcome) Simulation Jobs that simulates the environment and shares this data with SageMaker for training. ",
"_____no_output_____"
]
],
[
[
"# Use more rollout workers for faster convergence\nnum_simulation_workers = 1\n\nenvriron_vars = {\n \"MODEL_S3_BUCKET\": s3_bucket,\n \"MODEL_S3_PREFIX\": s3_prefix,\n \"ROS_AWS_REGION\": aws_region,\n \"WORLD_NAME\": \"hard_track\", # Can be one of \"easy_track\", \"medium_track\", \"hard_track\"\n \"MARKOV_PRESET_FILE\": \"%s.py\" % RLCOACH_PRESET,\n \"NUMBER_OF_ROLLOUT_WORKERS\": str(num_simulation_workers)}\n\nsimulation_application = {\"application\": simulation_app_arn,\n \"launchConfig\": {\"packageName\": \"deepracer_simulation\",\n \"launchFile\": \"distributed_training.launch\",\n \"environmentVariables\": envriron_vars}\n }\n \nvpcConfig = {\"subnets\": default_subnets,\n \"securityGroups\": default_security_groups,\n \"assignPublicIp\": True}\n\nresponses = []\nfor job_no in range(num_simulation_workers):\n response = robomaker.create_simulation_job(iamRole=role,\n clientRequestToken=strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime()),\n maxJobDurationInSeconds=job_duration_in_seconds,\n failureBehavior=\"Continue\",\n simulationApplications=[simulation_application],\n vpcConfig=vpcConfig\n )\n responses.append(response)\n\nprint(\"Created the following jobs:\")\njob_arns = [response[\"arn\"] for response in responses]\nfor job_arn in job_arns:\n print(\"Job ARN\", job_arn) ",
"_____no_output_____"
]
],
[
[
"### Visualizing the simulations in RoboMaker",
"_____no_output_____"
],
[
"You can visit the RoboMaker console to visualize the simulations or run the following cell to generate the hyperlinks.",
"_____no_output_____"
]
],
[
[
"display(Markdown(generate_robomaker_links(job_arns, aws_region)))",
"_____no_output_____"
]
],
[
[
"### Plot metrics for training job",
"_____no_output_____"
]
],
[
[
"tmp_dir = \"/tmp/{}\".format(job_name)\nos.system(\"mkdir {}\".format(tmp_dir))\nprint(\"Create local folder {}\".format(tmp_dir))\nintermediate_folder_key = \"{}/output/intermediate\".format(job_name)",
"_____no_output_____"
],
[
"%matplotlib inline\nimport pandas as pd\n\ncsv_file_name = \"worker_0.simple_rl_graph.main_level.main_level.agent_0.csv\"\nkey = intermediate_folder_key + \"/\" + csv_file_name\nwait_for_s3_object(s3_bucket, key, tmp_dir)\n\ncsv_file = \"{}/{}\".format(tmp_dir, csv_file_name)\ndf = pd.read_csv(csv_file)\ndf = df.dropna(subset=['Training Reward'])\nx_axis = 'Episode #'\ny_axis = 'Training Reward'\n\nplt = df.plot(x=x_axis,y=y_axis, figsize=(12,5), legend=True, style='b-')\nplt.set_ylabel(y_axis);\nplt.set_xlabel(x_axis);",
"_____no_output_____"
]
],
[
[
"### Clean Up",
"_____no_output_____"
],
[
"Execute the cells below if you want to kill RoboMaker and SageMaker job.",
"_____no_output_____"
]
],
[
[
"for job_arn in job_arns:\n robomaker.cancel_simulation_job(job=job_arn)",
"_____no_output_____"
],
[
"sage_session.sagemaker_client.stop_training_job(TrainingJobName=estimator._current_job_name)",
"_____no_output_____"
]
],
[
[
"### Evaluation",
"_____no_output_____"
]
],
[
[
"envriron_vars = {\"MODEL_S3_BUCKET\": s3_bucket,\n \"MODEL_S3_PREFIX\": s3_prefix,\n \"ROS_AWS_REGION\": aws_region,\n \"NUMBER_OF_TRIALS\": str(20),\n \"MARKOV_PRESET_FILE\": \"%s.py\" % RLCOACH_PRESET,\n \"WORLD_NAME\": \"hard_track\",\n }\n\nsimulation_application = {\"application\":simulation_app_arn,\n \"launchConfig\": {\"packageName\": \"deepracer_simulation\",\n \"launchFile\": \"evaluation.launch\",\n \"environmentVariables\": envriron_vars}\n }\n \nvpcConfig = {\"subnets\": default_subnets,\n \"securityGroups\": default_security_groups,\n \"assignPublicIp\": True}\n\nresponse = robomaker.create_simulation_job(iamRole=role,\n clientRequestToken=strftime(\"%Y-%m-%d-%H-%M-%S\", gmtime()),\n maxJobDurationInSeconds=job_duration_in_seconds,\n failureBehavior=\"Continue\",\n simulationApplications=[simulation_application],\n vpcConfig=vpcConfig\n )\nprint(\"Created the following job:\")\nprint(\"Job ARN\", response[\"arn\"])",
"_____no_output_____"
]
],
[
[
"### Clean Up Simulation Application Resource",
"_____no_output_____"
]
],
[
[
"robomaker.delete_simulation_application(application=simulation_app_arn)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae6f9ed1654077c3bca10e94d71d225a3062866
| 11,835 |
ipynb
|
Jupyter Notebook
|
Jupyter/stockcompany.ipynb
|
minplemon/stockThird
|
e32c202c95ba19fe2db97f6e5dd175ac64ee1996
|
[
"MIT"
] | 6 |
2020-03-10T14:54:22.000Z
|
2021-11-28T11:49:06.000Z
|
Jupyter/stockcompany.ipynb
|
minplemon/stockThird
|
e32c202c95ba19fe2db97f6e5dd175ac64ee1996
|
[
"MIT"
] | null | null | null |
Jupyter/stockcompany.ipynb
|
minplemon/stockThird
|
e32c202c95ba19fe2db97f6e5dd175ac64ee1996
|
[
"MIT"
] | 5 |
2019-06-25T09:49:53.000Z
|
2020-03-01T11:56:32.000Z
| 40.255102 | 158 | 0.459569 |
[
[
[
"#公司基本信息\nfrom jqdatasdk import *\nauth('18620668927', 'minpeng123')",
"auth success (JQData现有流量增加活动,详情请咨询JQData管理员,微信号:JQData02)\n"
],
[
"#上市公司基本信息\nq=query(finance.STK_COMPANY_INFO).filter(finance.STK_COMPANY_INFO.code=='000001.XSHE').limit(10)\ndf=finance.run_query(q)\nprint(df)",
" id company_id code full_name short_name a_code b_code h_code \\\n0 3082 430000001 000001.XSHE 平安银行股份有限公司 平安银行 000001 None None \n\n fullname_en shortname_en ... province city_id city \\\n0 Ping An Bank Co., Ltd. Ping An Bank ... 广东 440300 深圳市 \n\n industry_id industry_1 industry_2 cpafirm lawfirm ceo \\\n0 J66 金融业 货币金融服务 普华永道中天会计师事务所(特殊普通合伙) 竞天公诚律师事务所 胡跃飞 \n\n comments \n0 深发展的核心竞争力更在于其基于市场需求基础上的雄厚的创新实力。迄今深发展已推出双周供、存抵贷... \n\n[1 rows x 45 columns]\n"
],
[
"#上市公司员工情况\nq=query(finance.STK_EMPLOYEE_INFO).filter(finance.STK_EMPLOYEE_INFO.code=='000001.XSHE',finance.STK_EMPLOYEE_INFO.pub_date>='2015-01-01').limit(10)\ndf=finance.run_query(q)\nprint(df)",
" id company_id code name end_date pub_date employee \\\n0 26789 430000001 000001.XSHE 平安银行 2014-12-31 2015-03-13 29860 \n1 26790 430000001 000001.XSHE 平安银行 2015-06-30 2015-08-14 30526 \n2 26791 430000001 000001.XSHE 平安银行 2015-12-31 2016-03-10 32299 \n3 26792 430000001 000001.XSHE 平安银行 2016-06-30 2016-08-12 38600 \n4 26793 430000001 000001.XSHE 平安银行 2016-12-31 2017-03-17 36885 \n5 35492 430000001 000001.XSHE 平安银行 2017-06-30 2017-08-11 30292 \n6 35493 430000001 000001.XSHE 平安银行 2017-12-31 2018-03-15 31168 \n7 35494 430000001 000001.XSHE 平安银行 2018-06-30 2018-08-16 32744 \n8 58324 430000001 000001.XSHE 平安银行 2018-12-31 2019-03-07 33708 \n\n retirement graduate_rate college_rate middle_rate \n0 None None None 23237.0 \n1 None None None 23892.0 \n2 None None None 25652.0 \n3 None None None 25652.0 \n4 None None None 29884.0 \n5 None None None NaN \n6 None None None 25720.0 \n7 None None None 27194.0 \n8 None None None 28348.0 \n"
],
[
"#上市公司状态变动\nq=query(finance.STK_STATUS_CHANGE).filter(finance.STK_STATUS_CHANGE.code=='000001.XSHE').limit(10)\ndf=finance.run_query(q)\nprint(df)",
" id company_id code name pub_date change_date change_reason \\\n0 16712 430000001 000001.XSHE 深发展A 1991-04-03 1991-04-03 None \n\n change_type_id change_type comments public_status_id public_status \n0 303009 新股上市 None 301001 正常上市 \n"
],
[
"#股票上市信息\nq=query(finance.STK_LIST).filter(finance.STK_LIST.code=='000001.XSHE').limit(10)\ndf=finance.run_query(q)\nprint(df)",
" id code name short_name category exchange start_date end_date \\\n0 4352 000001.XSHE 平安银行 PAYH A XSHE 1991-04-03 None \n\n company_id company_name ipo_shares book_price par_value state_id state \n0 430000001 平安银行股份有限公司 16657580.0 40.0 1.0 301001 正常上市 \n"
],
[
"#股票简称变更\nq=query(finance.STK_NAME_HISTORY).filter(finance.STK_NAME_HISTORY.code=='000001.XSHE').limit(10)\ndf=finance.run_query(q)\nprint(df)",
" id code company_id new_name new_spelling org_name org_spelling \\\n0 1 000001.XSHE 430000001 深发展A SFZA None None \n1 4206 000001.XSHE 430000001 S深发展A SSFZA None None \n2 5600 000001.XSHE 430000001 深发展A SFZA S深发展A SSFZA \n3 7509 000001.XSHE 430000001 平安银行 PAYX 深发展A SFZA \n\n start_date pub_date reason \n0 1991-04-03 1991-04-03 None \n1 2006-10-09 2006-09-28 None \n2 2007-06-20 2007-06-14 None \n3 2012-08-02 2012-08-02 None \n"
],
[
"#公司管理人员任职情况\nq=query(finance.STK_MANAGEMENT_INFO).filter(finance.STK_MANAGEMENT_INFO.code=='000001.XSHE').order_by(finance.STK_MANAGEMENT_INFO.pub_date).limit(10)\ndf=finance.run_query(q)\nprint(df)",
" id company_id company_name code pub_date person_id \\\n0 138262 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201309346 \n1 138263 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201309346 \n2 138264 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201309346 \n3 138265 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201313341 \n4 138266 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201313342 \n5 138267 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201314655 \n6 138268 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201314656 \n7 138269 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201317398 \n8 138270 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201317399 \n9 138271 430000001 平安银行股份有限公司 000001.XSHE 2014-10-10 201317400 \n\n name title_class_id title_class title ... highest_degree_id \\\n0 肖遂宁 314001 董事会成员 董事 ... 316004.0 \n1 肖遂宁 314003 高管成员 行长 ... 316004.0 \n2 肖遂宁 314001 董事会成员 董事长 ... 316004.0 \n3 罗伯特·巴内姆 314001 董事会成员 独立董事 ... 316002.0 \n4 孙昌基 314001 董事会成员 独立董事 ... NaN \n5 张桐以 314001 董事会成员 董事 ... 316002.0 \n6 王开国 314001 董事会成员 董事 ... 316001.0 \n7 陈武朝 314001 董事会成员 独立董事 ... 316001.0 \n8 谢国忠 314001 董事会成员 独立董事 ... 316001.0 \n9 马雪征 314001 董事会成员 董事 ... 316003.0 \n\n highest_degree title_level_id title_level profession_certificate_id \\\n0 大专及其他 317003.0 高级 None \n1 大专及其他 317003.0 高级 None \n2 大专及其他 317003.0 高级 None \n3 硕士及研究生 NaN None None \n4 None 317003.0 高级 None \n5 硕士及研究生 NaN None None \n6 博士研究生 NaN None None \n7 博士研究生 317003.0 高级 None \n8 博士研究生 NaN None None \n9 本科 NaN None None \n\n profession_certificate nationality_id nationality \\\n0 None None None \n1 None None None \n2 None None None \n3 None None None \n4 None None None \n5 None None None \n6 None None None \n7 注册会计师 None None \n8 None None None \n9 None None None \n\n security_career_start_year \\\n0 None \n1 None \n2 None \n3 None \n4 None \n5 None \n6 None \n7 None \n8 None \n9 None \n\n resume \n0 肖遂宁先生:出生于1948年2月,高级经济师。曾任深圳发展银行总行行长、董事长,平安银行股份... \n1 肖遂宁先生:出生于1948年2月,高级经济师。曾任深圳发展银行总行行长、董事长,平安银行股份... \n2 肖遂宁先生:出生于1948年2月,高级经济师。曾任深圳发展银行总行行长、董事长,平安银行股份... \n3 罗伯特巴内姆先生(RobertT.Barnum)自1997年至今在美国加州的PokerFla... \n4 孙昌基,男,研究员级高级工程师。1942年8月20日出生于上海,1966年9月毕业于清华大学... \n5 张桐以(JustinChang)先生,现居香港,是德州太平洋投资集团和新桥投资的合伙人之一。... \n6 王开国先生:1958年出生,获得经济学博士学位。现任海通证券股份有限公司董事长、党委书记。2... \n7 陈武朝先生:1970年出生,中国国籍,无境外永久居留权。毕业于清华大学会计学专业,博士学历,... \n8 谢国忠先生,独立董事候选人,1960年出生,毕业于同济大学路桥系,于1987年获麻省理工学院... \n9 马雪征:中国香港居民,女,1952年出生,大学学历。1978年至1990年任职于中国科学院;... \n\n[10 rows x 26 columns]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae71125e29f033826ee2cbade1c79a47c58345c
| 123,376 |
ipynb
|
Jupyter Notebook
|
tobias/quick_olhada.ipynb
|
joaopedromattos/DMC2020
|
85838edfc5c0a8d28e2d7863aaa8225d00173166
|
[
"MIT"
] | null | null | null |
tobias/quick_olhada.ipynb
|
joaopedromattos/DMC2020
|
85838edfc5c0a8d28e2d7863aaa8225d00173166
|
[
"MIT"
] | null | null | null |
tobias/quick_olhada.ipynb
|
joaopedromattos/DMC2020
|
85838edfc5c0a8d28e2d7863aaa8225d00173166
|
[
"MIT"
] | 1 |
2020-08-24T22:47:36.000Z
|
2020-08-24T22:47:36.000Z
| 32.553034 | 6,496 | 0.368921 |
[
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"# EDA\n<hr>",
"_____no_output_____"
],
[
"## Table infos",
"_____no_output_____"
]
],
[
[
"infos = pd.read_csv('infos.csv', sep = '|')",
"_____no_output_____"
],
[
"infos.head()",
"_____no_output_____"
],
[
"infos.dtypes",
"_____no_output_____"
],
[
"infos.shape",
"_____no_output_____"
],
[
"len(infos) - infos.count()",
"_____no_output_____"
],
[
"infos['promotion'].unique()",
"_____no_output_____"
]
],
[
[
"## Table items",
"_____no_output_____"
]
],
[
[
"items = pd.read_csv('items.csv', sep = '|')",
"_____no_output_____"
],
[
"items.head()",
"_____no_output_____"
],
[
"items.shape",
"_____no_output_____"
],
[
"items.count()",
"_____no_output_____"
],
[
"items.nunique()",
"_____no_output_____"
]
],
[
[
"## Table orders",
"_____no_output_____"
]
],
[
[
"orders = pd.read_csv('orders.csv', sep = '|', parse_dates=['time'])",
"_____no_output_____"
],
[
"orders.head()",
"_____no_output_____"
],
[
"orders.shape",
"_____no_output_____"
],
[
"orders.count()",
"_____no_output_____"
],
[
"orders.dtypes",
"_____no_output_____"
],
[
"orders.time",
"_____no_output_____"
],
[
"orders.time.dt.week",
"_____no_output_____"
],
[
"orders.groupby('itemID')['salesPrice'].nunique().max()",
"_____no_output_____"
]
],
[
[
"# Other things\n<hr>",
"_____no_output_____"
],
[
"## Evalutation function",
"_____no_output_____"
]
],
[
[
"# custo \n# np.sum((prediction - np.maximum(prediction - target, 0) * 1.6) * simulatedPrice)",
"_____no_output_____"
]
],
[
[
"## Submission structure",
"_____no_output_____"
]
],
[
[
"# submission = items[['itemID']]\n# submission['demandPrediction'] = 0 # prediction here\n# submission.to_csv('submission.csv', sep = '|', index=False)",
"_____no_output_____"
]
],
[
[
"# First Model (aggregating by every two weeks before target)",
"_____no_output_____"
],
[
"## - Creating the structure",
"_____no_output_____"
]
],
[
[
"df = orders.copy()",
"_____no_output_____"
],
[
"df.tail()",
"_____no_output_____"
],
[
"df.tail().time.dt.dayofweek",
"_____no_output_____"
],
[
"# We want the last dayofweek from training to be 6",
"_____no_output_____"
],
[
"(df.tail().time.dt.dayofyear + 2) // 7",
"_____no_output_____"
],
[
"(df.head().time.dt.dayofyear + 2) // 7",
"_____no_output_____"
],
[
"df['week'] = (df.time.dt.dayofyear + 2 + 7) // 14\n# + 7 because we want weeks 25 and 26 to be together, week 0 will be discarded",
"_____no_output_____"
],
[
"maxx = df.week.max()\nminn = df.week.min()\nminn, maxx",
"_____no_output_____"
],
[
"n_items = items['itemID'].nunique()\nprint('total number of items:', n_items)\nprint('expected number of instances:', n_items * (maxx + 1))",
"total number of items: 10463\nexpected number of instances: 146482\n"
],
[
"mi = pd.MultiIndex.from_product([range(0, maxx + 1), items['itemID']], names=['week', 'itemID'])\ndata = pd.DataFrame(index = mi)",
"_____no_output_____"
],
[
"data = data.join(df.groupby(['week', 'itemID'])[['order']].sum(), how = 'left')",
"_____no_output_____"
],
[
"data.fillna(0, inplace = True)",
"_____no_output_____"
],
[
"data.groupby('itemID').count().min()",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.groupby('itemID')['salesPrice'].nunique().describe()",
"_____no_output_____"
],
[
"df.groupby('itemID')['salesPrice'].median()",
"_____no_output_____"
]
],
[
[
"## - Creating features",
"_____no_output_____"
]
],
[
[
"# rolling window example with shift\nrandom_df = pd.DataFrame({'B': [0, 1, 2, 3, 4]})\nrandom_df.shift(1).rolling(2).sum()",
"_____no_output_____"
],
[
"data.reset_index(inplace = True)",
"_____no_output_____"
],
[
"data = pd.merge(data, items[['itemID', 'manufacturer', 'category1', 'category2', 'category3']], on = 'itemID')",
"_____no_output_____"
],
[
"# I am going to create three features: the mean of the orders of the last [1, 2, 4] weeks for each item ",
"_____no_output_____"
],
[
"# TODO:\n# longer windows\n# aggregating by other features\n# week pairs since last peek\n# week pairs from 2nd last to last peek\n# ",
"_____no_output_____"
],
[
"data.sort_values('week', inplace = True)",
"_____no_output_____"
],
[
"data",
"_____no_output_____"
],
[
"features = [\n ('itemID', 'item'),\n ('manufacturer', 'manuf'),\n ('category1', 'cat1'),\n ('category2', 'cat2'),\n ('category3', 'cat3')\n]\nfor f, n in features:\n if f not in data.columns:\n print('ops', f)",
"_____no_output_____"
],
[
"# f, name = ('manufacturer', 'manuf')\nfor f, name in features:\n print(f)\n temp = data.groupby([f, 'week'])[['order']].sum()\n shifted = temp.groupby(f)[['order']].shift(1)\n new_feature_block = pd.DataFrame()\n for n in range(3):\n rolled = shifted.groupby(f, as_index = False)['order'].rolling(2 ** n).mean()\n new_feature_block['%s_%d' % (name, 2 ** n)] = rolled.reset_index(0, drop = True) # rolling has a weird index behavior...\n data = pd.merge(data, new_feature_block.reset_index(), on = [f, 'week'])",
"itemID\nmanufacturer\ncategory1\ncategory2\ncategory3\n"
],
[
"data.count() # the larger the window, more NaN are expected",
"_____no_output_____"
],
[
"data.fillna(-1, inplace=True)",
"_____no_output_____"
],
[
"# checking if we got what we wanted\ndata.query('itemID == 1')",
"_____no_output_____"
]
],
[
[
"## - fit, predict",
"_____no_output_____"
]
],
[
[
"# max expected rmse\nfrom sklearn.metrics import mean_squared_error as mse\n# pred = data.loc[1:12].groupby('itemID')['order'].mean().sort_index()\n# target_week = data.loc[13:, 'order'].reset_index(level = 0, drop = True).sort_index()\n# mse(target_week, pred) ** .5",
"_____no_output_____"
],
[
"train = data.query('1 <= week <= 12').reset_index()\ntest = data.query('week == 13').reset_index()",
"_____no_output_____"
],
[
"y_train = train.pop('order').values\ny_test = test.pop('order').values\n\nX_train = train.values\nX_test = test.values",
"_____no_output_____"
],
[
"import xgboost as xgb",
"_____no_output_____"
],
[
"dtrain = xgb.DMatrix(X_train, y_train, missing = -1)\ndtest = xgb.DMatrix(X_test, y_test, missing = -1)\n# specify parameters via map\nparam = {'max_depth':6, 'eta':0.01, 'objective':'reg:squarederror'}\nnum_round = 200\nbst = xgb.train(param, dtrain,\n num_round, early_stopping_rounds = 5,\n evals = [(dtrain, 'train'), (dtest, 'test')])",
"[0]\ttrain-rmse:105.70586\ttest-rmse:86.53361\nMultiple eval metrics have been passed: 'test-rmse' will be used for early stopping.\n\nWill train until test-rmse hasn't improved in 5 rounds.\n[1]\ttrain-rmse:105.55459\ttest-rmse:86.40955\n[2]\ttrain-rmse:105.40828\ttest-rmse:86.29902\n[3]\ttrain-rmse:105.26123\ttest-rmse:86.18501\n[4]\ttrain-rmse:105.12257\ttest-rmse:86.09409\n[5]\ttrain-rmse:104.98622\ttest-rmse:85.98164\n[6]\ttrain-rmse:104.84803\ttest-rmse:85.90354\n[7]\ttrain-rmse:104.71547\ttest-rmse:85.82388\n[8]\ttrain-rmse:104.58319\ttest-rmse:85.74725\n[9]\ttrain-rmse:104.45547\ttest-rmse:85.68170\n[10]\ttrain-rmse:104.33083\ttest-rmse:85.60940\n[11]\ttrain-rmse:104.20426\ttest-rmse:85.57921\n[12]\ttrain-rmse:104.07585\ttest-rmse:85.51643\n[13]\ttrain-rmse:103.95721\ttest-rmse:85.48312\n[14]\ttrain-rmse:103.83033\ttest-rmse:85.43800\n[15]\ttrain-rmse:103.70905\ttest-rmse:85.41409\n[16]\ttrain-rmse:103.58691\ttest-rmse:85.37125\n[17]\ttrain-rmse:103.46766\ttest-rmse:85.31898\n[18]\ttrain-rmse:103.35364\ttest-rmse:85.29717\n[19]\ttrain-rmse:103.24358\ttest-rmse:85.24615\n[20]\ttrain-rmse:103.13451\ttest-rmse:85.19955\n[21]\ttrain-rmse:103.02550\ttest-rmse:85.19665\n[22]\ttrain-rmse:102.92084\ttest-rmse:85.14963\n[23]\ttrain-rmse:102.81574\ttest-rmse:85.09335\n[24]\ttrain-rmse:102.71040\ttest-rmse:85.08337\n[25]\ttrain-rmse:102.60987\ttest-rmse:85.04078\n[26]\ttrain-rmse:102.50923\ttest-rmse:84.95946\n[27]\ttrain-rmse:102.41042\ttest-rmse:84.96864\n[28]\ttrain-rmse:102.31426\ttest-rmse:84.92600\n[29]\ttrain-rmse:102.21632\ttest-rmse:84.86964\n[30]\ttrain-rmse:102.12246\ttest-rmse:84.85836\n[31]\ttrain-rmse:102.03191\ttest-rmse:84.81554\n[32]\ttrain-rmse:101.93928\ttest-rmse:84.80864\n[33]\ttrain-rmse:101.83908\ttest-rmse:84.81090\n[34]\ttrain-rmse:101.74805\ttest-rmse:84.76908\n[35]\ttrain-rmse:101.65283\ttest-rmse:84.75527\n[36]\ttrain-rmse:101.56637\ttest-rmse:84.72517\n[37]\ttrain-rmse:101.47735\ttest-rmse:84.73549\n[38]\ttrain-rmse:101.39262\ttest-rmse:84.72534\n[39]\ttrain-rmse:101.30880\ttest-rmse:84.69257\n[40]\ttrain-rmse:101.22580\ttest-rmse:84.77545\n[41]\ttrain-rmse:101.14490\ttest-rmse:84.77866\n[42]\ttrain-rmse:101.05984\ttest-rmse:84.75678\n[43]\ttrain-rmse:100.98253\ttest-rmse:84.74509\n[44]\ttrain-rmse:100.90263\ttest-rmse:85.06732\nStopping. Best iteration:\n[39]\ttrain-rmse:101.30880\ttest-rmse:84.69257\n\n"
],
[
"# wtf is happening?",
"_____no_output_____"
],
[
"data.query('itemID == 10')",
"_____no_output_____"
],
[
"data.query('itemID == 100')",
"_____no_output_____"
],
[
"data.query('itemID == 1000')",
"_____no_output_____"
],
[
"zeros = data.groupby('itemID')['order'].apply(lambda x : (x == 0).mean())",
"_____no_output_____"
],
[
"plt.hist(zeros, bins = 60);",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae7188233d1c240f05a09c26d3052bcb60fadcc
| 6,879 |
ipynb
|
Jupyter Notebook
|
A_learning_notes/generate_process.ipynb
|
dywlavender/multi-label-classification
|
ec8567169b3f1b08c7dafeb71f007275621a5a95
|
[
"MIT"
] | 17 |
2019-10-16T04:39:27.000Z
|
2022-01-24T03:21:58.000Z
|
A_learning_notes/generate_process.ipynb
|
dywlavender/multi-label-classification
|
ec8567169b3f1b08c7dafeb71f007275621a5a95
|
[
"MIT"
] | null | null | null |
A_learning_notes/generate_process.ipynb
|
dywlavender/multi-label-classification
|
ec8567169b3f1b08c7dafeb71f007275621a5a95
|
[
"MIT"
] | 5 |
2020-03-04T09:08:02.000Z
|
2021-07-20T02:28:57.000Z
| 29.025316 | 1,071 | 0.513592 |
[
[
[
"# 深度学习模型编程一般框架\n1. 建立keras模型;\n1. 定义loss函数;\n1. 编译模型;\n1. 构造训练数据集;\n1. 开始模型训练;\n",
"_____no_output_____"
],
[
"## 建立keras模型\n1. 定义骨干网络;\n1. 组合输入、输出写成Model对象;",
"_____no_output_____"
]
],
[
[
"from tensorflow import keras\ndef build_net(input_tensor):\n out1 = keras.layers.Dense(1, kernel_initializer='glorot_normal', activation='linear',\n kernel_regularizer=keras.regularizers.l2(10))(input_tensor)\n out2 = keras.layers.Dense(1, kernel_initializer='glorot_normal', activation='linear',\n kernel_regularizer=keras.regularizers.l2(10))(input_tensor)\n return [out1, out2]\n\n\nfeature_input = keras.layers.Input(shape=(2,), name='feature_input')\noutputs = build_net(feature_input)\nmodel = keras.models.Model(feature_input, outputs)",
"_____no_output_____"
]
],
[
[
"## 定义loss函数",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\ndef my_loss(y_dummy, pred):\n loss = tf.keras.losses.mean_absolute_error(y_dummy, pred)\n return loss",
"_____no_output_____"
]
],
[
[
"## 编译模型",
"_____no_output_____"
]
],
[
[
"model.compile(loss=my_loss, optimizer='adam', loss_weights=[0.5, 0.5])\nmodel.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nfeature_input (InputLayer) (None, 2) 0 \n__________________________________________________________________________________________________\ndense_4 (Dense) (None, 1) 3 feature_input[0][0] \n__________________________________________________________________________________________________\ndense_5 (Dense) (None, 1) 3 feature_input[0][0] \n==================================================================================================\nTotal params: 6\nTrainable params: 6\nNon-trainable params: 0\n__________________________________________________________________________________________________\n"
]
],
[
[
"## 构造训练数据集",
"_____no_output_____"
]
],
[
[
"import numpy as np\ninput = np.random.normal(0, 1, [4, 2])\nout_1 = np.random.normal(0, 1, [4, 1])\nout_2 = np.random.normal(0, 1, [4, 1])\ndataset = tf.data.Dataset.from_tensor_slices((input, (out_1, out_2)))\ndataset = dataset.repeat().batch(2).prefetch(buffer_size=4)",
"_____no_output_____"
]
],
[
[
"## 开始模型训练",
"_____no_output_____"
]
],
[
[
"model.fit(dataset, epochs=2, steps_per_epoch=2, verbose=1)\n",
"WARNING:tensorflow:From D:\\software\\Anaconda\\install\\envs\\tf-cpu\\lib\\site-packages\\tensorflow\\python\\ops\\math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.cast instead.\nEpoch 1/2\n\r1/2 [==============>...............] - ETA: 0s - loss: 21.9610 - dense_4_loss: 1.3583 - dense_5_loss: 1.5867\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\r2/2 [==============================] - 0s 140ms/step - loss: 22.1960 - dense_4_loss: 1.9502 - dense_5_loss: 1.5183\nEpoch 2/2\n\r1/2 [==============>...............] - ETA: 0s - loss: 21.8526 - dense_4_loss: 1.3555 - dense_5_loss: 1.5861\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\b\r2/2 [==============================] - 0s 1ms/step - loss: 22.0872 - dense_4_loss: 1.9470 - dense_5_loss: 1.5171\n"
]
],
[
[
"更细致的debug(查看梯度、打印操作等),可看详细查看本工程。\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4ae71fdf9e59727d26ad8d9ba9ae72a3d60dc18d
| 10,951 |
ipynb
|
Jupyter Notebook
|
example/example_commandline.ipynb
|
maxipi/intensity_duration_frequency_analysis
|
c3b58518b04a9d5b804cbd17cc67dcedf9970fb1
|
[
"MIT"
] | 20 |
2018-11-09T17:35:12.000Z
|
2022-03-30T18:27:03.000Z
|
example/example_commandline.ipynb
|
maxipi/intensity_duration_frequency_analysis
|
c3b58518b04a9d5b804cbd17cc67dcedf9970fb1
|
[
"MIT"
] | 3 |
2021-03-07T13:09:00.000Z
|
2021-10-15T13:26:48.000Z
|
example/example_commandline.ipynb
|
maxipi/intensity_duration_frequency_analysis
|
c3b58518b04a9d5b804cbd17cc67dcedf9970fb1
|
[
"MIT"
] | 8 |
2018-11-13T07:14:13.000Z
|
2021-12-03T06:48:20.000Z
| 42.945098 | 123 | 0.514382 |
[
[
[
"# Example Commandline Use",
"_____no_output_____"
]
],
[
[
"! python -m idf_analysis -h",
"usage: __main__.py [-h] -i INPUT\n [-ws {ATV-A_121,DWA-A_531,DWA-A_531_advektiv}]\n [-kind {partial,annual}] [-t {>= 0.5 a and <= 100 a}]\n [-d {>= 5 min and <= 8640 min}] [-r {>= 0 L/s*ha}]\n [-h_N {>= 0 mm}] [--r_720_1] [--plot] [--export_table]\n\nheavy rain as a function of the duration and the return period acc. to DWA-A\n531 (2012) All files will be saved in the same directory of the input file but\nin a subfolder called like the inputfile + \"_idf_data\". Inside this folder a\nfile called \"idf_parameter.yaml\"-file will be saved and contains interim-\ncalculation-results and will be automatically reloaded on the next call.\n\noptional arguments:\n -h, --help show this help message and exit\n -i INPUT, --input INPUT\n input file with the rain time-series (csv or parquet)\n -ws {ATV-A_121,DWA-A_531,DWA-A_531_advektiv}, --worksheet {ATV-A_121,DWA-A_531,DWA-A_531_advektiv}\n From which worksheet the recommendations for\n calculating the parameters should be taken.\n -kind {partial,annual}, --series_kind {partial,annual}\n The kind of series used for the calculation.\n (Calculation with partial series is more precise and\n recommended.)\n -t {>= 0.5 a and <= 100 a}, --return_period {>= 0.5 a and <= 100 a}\n return period in years (If two of the three variables\n (rainfall (height or flow-rate), duration, return\n period) are given, the third variable is calculated.)\n -d {>= 5 min and <= 8640 min}, --duration {>= 5 min and <= 8640 min}\n duration in minutes (If two of the three variables\n (rainfall (height or flow-rate), duration, return\n period) are given, the third variable is calculated.)\n -r {>= 0 L/(s*ha)}, --flow_rate_of_rainfall {>= 0 L/(s*ha)}\n rainfall in Liter/(s * ha) (If two of the three\n variables (rainfall (height or flow-rate), duration,\n return period) are given, the third variable is\n calculated.)\n -h_N {>= 0 mm}, --height_of_rainfall {>= 0 mm}\n rainfall in mm or Liter/m^2 (If two of the three\n variables (rainfall (height or flow-rate), duration,\n return period) are given, the third variable is\n calculated.)\n --r_720_1 design rainfall with a duration of 720 minutes (=12 h)\n and a return period of 1 year\n --plot get a plot of the idf relationship\n --export_table get a table of the most frequent used values\n"
]
],
[
[
"I used the rain-time-series from ehyd.gv.at with the ID 112086 (Graz-Andritz)",
"_____no_output_____"
]
],
[
[
"! python -m idf_analysis -i ehyd_112086.parquet",
"Using the subfolder \"ehyd_112086_idf_data\" for the interim- and final-results.\nFound existing interim-results in \"ehyd_112086_idf_data\\idf_parameters.yaml\" and using them for calculations.\n"
],
[
"! python -m idf_analysis -i ehyd_112086.parquet --r_720_1",
"Using the subfolder \"ehyd_112086_idf_data\" for the interim- and final-results.\nFound existing interim-results in \"ehyd_112086_idf_data\\idf_parameters.yaml\" and using them for calculations.\nResultierende Regenhöhe h_N(T_n=1.0a, D=720.0min) = 49.29 mm\nResultierende Regenspende r_N(T_n=1.0a, D=720.0min) = 11.41 L/(s*ha)\n"
],
[
"! python -m idf_analysis -i ehyd_112086.parquet -d 720 -t 1",
"Using the subfolder \"ehyd_112086_idf_data\" for the interim- and final-results.\nFound existing interim-results in \"ehyd_112086_idf_data\\idf_parameters.yaml\" and using them for calculations.\nResultierende Regenhöhe h_N(T_n=1.0a, D=720.0min) = 49.29 mm\nResultierende Regenspende r_N(T_n=1.0a, D=720.0min) = 11.41 L/(s*ha)\n"
],
[
"! python -m idf_analysis -i ehyd_112086.parquet -d 720 -h_N 60",
"Using the subfolder \"ehyd_112086_idf_data\" for the interim- and final-results.\nFound existing interim-results in \"ehyd_112086_idf_data\\idf_parameters.yaml\" and using them for calculations.\nThe return period is 2.0 years.\nResultierende Regenhöhe h_N(T_n=2.0a, D=720.0min) = 60.00 mm\nResultierende Regenspende r_N(T_n=2.0a, D=720.0min) = 13.89 L/(s*ha)\n"
],
[
"! python -m idf_analysis -i ehyd_112086.parquet -t 5 -t 15",
"_____no_output_____"
],
[
"! python -m idf_analysis -i ehyd_112086.parquet --plot",
"Using the subfolder \"ehyd_112086_idf_data\" for the interim- and final-results.\nFound existing interim-results in \"ehyd_112086_idf_data\\idf_parameters.yaml\" and using them for calculations.\nCreated the IDF-curves-plot and saved the file as \"ehyd_112086_idf_data\\idf__curves_plot.png\".\n"
],
[
"! python -m idf_analysis -i ehyd_112086.parquet --export_table",
"Using the subfolder \"ehyd_112086_idf_data\" for the interim- and final-results.\nFound existing interim-results in \"ehyd_112086_idf_data\\idf_parameters.yaml\" and using them for calculations.\nreturn period (a) 1 2 3 5 10 20 25 30 50 75 100\nfrequency (1/a) 1.000 0.500 0.333 0.200 0.100 0.050 0.040 0.033 0.020 0.013 0.010\nduration (min) \n5.0 9.39 10.97 11.89 13.04 14.61 16.19 16.69 17.11 18.26 19.18 19.83\n10.0 15.15 17.62 19.06 20.88 23.35 25.82 26.62 27.27 29.09 30.54 31.56\n15.0 19.03 22.25 24.13 26.51 29.72 32.94 33.98 34.83 37.20 39.08 40.42\n20.0 21.83 25.71 27.99 30.85 34.73 38.62 39.87 40.89 43.75 46.02 47.63\n30.0 25.60 30.66 33.62 37.35 42.41 47.47 49.10 50.43 54.16 57.12 59.22\n45.0 28.92 35.51 39.37 44.23 50.83 57.42 59.54 61.28 66.14 69.99 72.73\n60.0 30.93 38.89 43.54 49.40 57.36 65.31 67.88 69.97 75.83 80.49 83.79\n90.0 33.37 41.74 46.64 52.80 61.17 69.54 72.23 74.43 80.60 85.49 88.96\n180.0 38.01 47.13 52.46 59.18 68.30 77.42 80.36 82.76 89.48 94.81 98.60\n270.0 41.01 50.60 56.21 63.28 72.87 82.46 85.55 88.07 95.14 100.75 104.73\n360.0 43.29 53.23 59.04 66.37 76.31 86.25 89.45 92.06 99.39 105.20 109.33\n450.0 45.14 55.36 61.33 68.87 79.08 89.30 92.59 95.28 102.81 108.79 113.03\n600.0 47.64 58.23 64.43 72.23 82.82 93.41 96.82 99.61 107.42 113.61 118.01\n720.0 49.29 60.13 66.47 74.45 85.29 96.12 99.61 102.46 110.44 116.78 121.28\n1080.0 54.41 64.97 71.15 78.94 89.50 100.06 103.46 106.24 114.02 120.20 124.58\n1440.0 58.02 67.72 73.39 80.54 90.24 99.93 103.05 105.61 112.75 118.42 122.45\n2880.0 66.70 77.41 83.68 91.57 102.29 113.00 116.45 119.26 127.16 133.42 137.87\n4320.0 71.93 85.72 93.78 103.95 117.73 131.52 135.96 139.58 149.75 157.81 163.53\n5760.0 78.95 95.65 105.42 117.72 134.43 151.13 156.50 160.89 173.20 182.97 189.90\n7200.0 83.53 101.38 111.82 124.98 142.83 160.68 166.43 171.12 184.28 194.72 202.13\n8640.0 85.38 104.95 116.40 130.82 150.38 169.95 176.25 181.40 195.82 207.27 215.39\nCreated the IDF-curves-plot and saved the file as \"ehyd_112086_idf_data\\idf_table.csv\".\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae726d88197bc5a8d6298a1d865c5f2ad30a4ba
| 237,339 |
ipynb
|
Jupyter Notebook
|
_in/2019/jp/08.ipynb
|
r-e-x-a-g-o-n/scalable-data-science
|
a97451a768cf12eec9a20fbe5552bbcaf215d662
|
[
"Unlicense"
] | 138 |
2017-07-25T06:48:28.000Z
|
2022-03-31T12:23:36.000Z
|
_in/2019/jp/08.ipynb
|
r-e-x-a-g-o-n/scalable-data-science
|
a97451a768cf12eec9a20fbe5552bbcaf215d662
|
[
"Unlicense"
] | 11 |
2017-08-17T13:45:54.000Z
|
2021-06-04T09:06:53.000Z
|
_in/2019/jp/08.ipynb
|
r-e-x-a-g-o-n/scalable-data-science
|
a97451a768cf12eec9a20fbe5552bbcaf215d662
|
[
"Unlicense"
] | 74 |
2017-08-18T17:04:46.000Z
|
2022-03-21T14:30:51.000Z
| 75.226307 | 52,988 | 0.782311 |
[
[
[
"# [Introduction to Data Science: A Comp-Math-Stat Approach](https://lamastex.github.io/scalable-data-science/as/2019/)\n## YOIYUI001, Summer 2019 \n©2019 Raazesh Sainudiin. [Attribution 4.0 International (CC BY 4.0)](https://creativecommons.org/licenses/by/4.0/)",
"_____no_output_____"
],
[
"# 08. Pseudo-Random Numbers, Simulating from Some Discrete and Continuous Random Variables\n\n\n- The $Uniform(0,1)$ RV\n- The $Bernoulli(\\theta)$ RV\n- Simulating from the $Bernoulli(\\theta)$ RV\n- The Equi-Probable $de\\,Moivre(k)$ RV\n- Simulating from the Equi-Probable $de\\,Moivre(k)$ RV\n- The $Uniform(\\theta_1, \\theta_2)$ RV\n- Simulating from the $Uniform(\\theta_1, \\theta_2)$ RV\n- The $Exponential(\\lambda)$ RV\n- Simulating from the $Exponential(\\lambda)$ RV\n- The standard $Cauchy$ RV\n- Simulating from the standard $Cauchy$ RV\n- Investigating running means\n- Replicable samples\n- A simple simulation\n \n\nIn the last notebook, we started to look at how we can produce realisations from the most elementary $Uniform(0,1)$ random variable.\n\ni.e., how can we produce samples $(x_1, x_2, \\ldots, x_n)$ from $X_1, X_2, \\ldots, X_n$ $\\overset{IID}{\\thicksim}$ $Uniform(0,1)$?\n\nWhat is SageMath doing when we ask for random()?",
"_____no_output_____"
]
],
[
[
"random()",
"_____no_output_____"
]
],
[
[
"We looked at how Modular arithmetic and number theory gives us pseudo-random number generators.\n\nWe used linear congruential generators (LCG) as simple pseudo-random number generators.\n\nRemember that \"pseudo-random\" means that the numbers are not really random. We saw that some linear congruential generators (LCG) have much shorter, more predictable, patterns than others and we learned what makes a good LCG.\n\nWe introduced the pseudo-random number generator (PRNG) called the Mersenne Twister that we will use for simulation purposes in this course. It is based on more sophisticated theory than that of LCG but the basic principles of recurrence relations are the same. \n\n# The $Uniform(0,1)$ Random Variable\n\nRecall that the $Uniform(0,1)$ random variable is the fundamental model as we can transform it to any other random variable, random vector or random structure. The PDF $f$ and DF $F$ of $X \\sim Uniform(0,1)$ are:\n\n$f(x) = \\begin{cases} 0 & \\text{if} \\ x \\notin [0,1] \\\\ 1 & \\text{if} \\ x \\in [0,1] \\end{cases}$\n\n$F(x) = \\begin{cases} 0 & \\text{if} \\ x < 0 \\\\ 1 & \\text{if} \\ x > 1 \\\\ x & \\text{if} \\ x \\in [0,1] \\end{cases}$\n\nWe use the Mersenne twister pseudo-random number generator to mimic independent and identically distributed draws from the $uniform(0,1)$ RV. \n\nIn Sage, we use the python random module to generate pseudo-random numbers for us. (We have already used it: remember randint?)\n\nrandom() will give us one simulation from the $Uniform(0,1)$ RV:",
"_____no_output_____"
]
],
[
[
"random()",
"_____no_output_____"
]
],
[
[
"If we want a whole simulated sample we can use a list comprehension. We will be using this technique frequently so make sure you understand what is going on. \"for i in range(3)\" is acting like a counter to give us 3 simulated values in the list we are making",
"_____no_output_____"
]
],
[
[
"[random() for i in range(3)]",
"_____no_output_____"
],
[
"listOfUniformSamples = [random() for i in range(3) ]\nlistOfUniformSamples",
"_____no_output_____"
]
],
[
[
"If we do this again, we will get a different sample:",
"_____no_output_____"
]
],
[
[
"listOfUniformSamples2 = [random() for i in range(3) ]\nlistOfUniformSamples2",
"_____no_output_____"
]
],
[
[
"Often is it useful to be able to replicate the same random sample. For example, if we were writing some code to do some simulations using samples from a PRNG, and we \"improved\" the way that we were doing it, how would we want to test our improvement? If we could replicate the same samples then we could show that our new code was equivalent to our old code, just more efficient. \n\nRemember when we were using the LCGs, and we could set the seed $x_0$? More sophisticated PRNGs like the Mersenne Twister also have a seed. By setting this seed to a specified value we can make sure that we can replicate samples. ",
"_____no_output_____"
]
],
[
[
"?set_random_seed",
"_____no_output_____"
],
[
"set_random_seed(256526)",
"_____no_output_____"
],
[
"listOfUniformSamples = [random() for i in range(3) ]\nlistOfUniformSamples",
"_____no_output_____"
],
[
"initial_seed()",
"_____no_output_____"
]
],
[
[
"Now we can replicate the same sample again by setting the seed to the same value:",
"_____no_output_____"
]
],
[
[
"set_random_seed(256526)\nlistOfUniformSamples2 = [random() for i in range(3) ]\nlistOfUniformSamples2",
"_____no_output_____"
],
[
"initial_seed()",
"_____no_output_____"
],
[
"set_random_seed(2676676766)\nlistOfUniformSamples2 = [random() for i in range(3) ]\nlistOfUniformSamples2",
"_____no_output_____"
],
[
"initial_seed()",
"_____no_output_____"
]
],
[
[
"We can compare some samples visually by plotting them:",
"_____no_output_____"
]
],
[
[
"set_random_seed(256526)\nlistOfUniformSamples = [(i,random()) for i in range(100)]\nplotsSeed1 = points(listOfUniformSamples)\nt1 = text('Seed 1 = 256626', (60,1.2), rgbcolor='blue',fontsize=10) \nset_random_seed(2676676766)\nplotsSeed2 = points([(i,random()) for i in range(100)],rgbcolor=\"red\")\nt2 = text('Seed 2 = 2676676766', (60,1.2), rgbcolor='red',fontsize=10) \nbothSeeds = plotsSeed1 + plotsSeed2\nt31 = text('Seed 1 and', (30,1.2), rgbcolor='blue',fontsize=10) \nt32 = text('Seed 2', (65,1.2), rgbcolor='red',fontsize=10)\nshow(graphics_array( (plotsSeed1+t1,plotsSeed2+t2, bothSeeds+t31+t32)),figsize=[9,3])",
"_____no_output_____"
]
],
[
[
"### YouTry\n\nTry looking at the more advanced documentation and play a bit.",
"_____no_output_____"
]
],
[
[
"#?sage.misc.randstate",
"_____no_output_____"
]
],
[
[
"(end of You Try)\n\n---\n\n---\n\n### Question:\n\nWhat can we do with samples from a $Uniform(0,1)$ RV? Why bother?\n\n### Answer:\n\nWe can use them to sample or simulate from other, more complex, random variables. \n\n \n\n# The $Bernoulli(\\theta)$ Random Variable\n\nThe $Bernoulli(\\theta)$ RV $X$ with PMF $f(x;\\theta)$ and DF $F(x;\\theta)$ parameterised by some real $\\theta\\in [0,1]$ is a discrete random variable with only two possible outcomes. \n\n$f(x;\\theta)= \\theta^x (1-\\theta)^{1-x} \\mathbf{1}_{\\{0,1\\}}(x) =\n\\begin{cases}\n\\theta & \\text{if} \\ x=1,\\\\\n1-\\theta & \\text{if} \\ x=0,\\\\\n0 & \\text{otherwise}\n\\end{cases}$\n\n$F(x;\\theta) =\n\\begin{cases}\n1 & \\text{if} \\ 1 \\leq x,\\\\\n1-\\theta & \\text{if} \\ 0 \\leq x < 1,\\\\\n0 & \\text{otherwise}\n\\end{cases}$\n\nHere are some functions for the PMF and DF for a $Bernoulli$ RV along with various useful functions for us in the sequel. Let's take a quick look at them.",
"_____no_output_____"
]
],
[
[
"def bernoulliPMF(x, theta):\n '''Probability mass function for Bernoulli(theta).\n \n Param x is the value to find the Bernoulli probability mass of.\n Param theta is the theta parameterising this Bernoulli RV.'''\n \n retValue = 0\n if x == 1:\n retValue = theta\n elif x == 0:\n retValue = 1 - theta\n return retValue\n \ndef bernoulliCDF(x, theta):\n '''DF for Bernoulli(theta).\n \n Param x is the value to find the Bernoulli cumulative density function of.\n Param theta is the theta parameterising this Bernoulli RV.'''\n \n retValue = 0\n if x >= 1:\n retValue = 1\n elif x >= 0:\n retValue = 1 - theta\n # in the case where x < 0, retValue is the default of 0\n return retValue\n\n# PFM plot\ndef pmfPlot(outcomes, pmf_values):\n '''Returns a pmf plot for a discrete distribution.'''\n \n pmf = points(zip(outcomes,pmf_values), rgbcolor=\"blue\", pointsize='20')\n for i in range(len(outcomes)):\n pmf += line([(outcomes[i], 0),(outcomes[i], pmf_values[i])], rgbcolor=\"blue\", linestyle=\":\")\n # padding\n pmf += point((0,1), rgbcolor=\"black\", pointsize=\"0\")\n return pmf\n\n# CDF plot\ndef cdfPlot(outcomes, cdf_values):\n '''Returns a DF plot for a discrete distribution.'''\n \n cdf_pairs = zip(outcomes, cdf_values)\n cdf = point(cdf_pairs, rgbcolor = \"red\", faceted = false, pointsize=\"20\")\n for k in range(len(cdf_pairs)):\n x, kheight = cdf_pairs[k] # unpack tuple\n previous_x = 0\n previous_height = 0\n if k > 0:\n previous_x, previous_height = cdf_pairs[k-1] # unpack previous tuple\n cdf += line([(previous_x, previous_height),(x, previous_height)], rgbcolor=\"grey\")\n cdf += points((x, previous_height),rgbcolor = \"white\", faceted = true, pointsize=\"20\")\n cdf += line([(x, previous_height),(x, kheight)], rgbcolor=\"blue\", linestyle=\":\")\n \n # padding\n max_index = len(outcomes)-1\n cdf += line([(outcomes[0]-0.2, 0),(outcomes[0], 0)], rgbcolor=\"grey\")\n cdf += line([(outcomes[max_index],cdf_values[max_index]),(outcomes[max_index]+0.2, cdf_values[max_index])], \\\n rgbcolor=\"grey\")\n return cdf\n \ndef makeFreqDictHidden(myDataList):\n '''Make a frequency mapping out of a list of data.\n \n Param myDataList, a list of data.\n Return a dictionary mapping each data value from min to max in steps of 1 to its frequency count.'''\n \n freqDict = {} # start with an empty dictionary\n sortedMyDataList = sorted(myDataList)\n for k in sortedMyDataList:\n freqDict[k] = myDataList.count(k)\n \n return freqDict # return the dictionary created\n\ndef makeEMFHidden(myDataList):\n '''Make an empirical mass function from a data list.\n \n Param myDataList, list of data to make emf from.\n Return list of tuples comprising (data value, relative frequency) ordered by data value.'''\n \n freqs = makeFreqDictHidden(myDataList) # make the frequency counts mapping\n totalCounts = sum(freqs.values())\n relFreqs = [fr/(1.0*totalCounts) for fr in freqs.values()] # use a list comprehension\n numRelFreqPairs = zip(freqs.keys(), relFreqs) # zip the keys and relative frequencies together\n numRelFreqPairs.sort() # sort the list of tuples\n\n return numRelFreqPairs\n\nfrom pylab import array\n\ndef makeEDFHidden(myDataList):\n '''Make an empirical distribution function from a data list.\n \n Param myDataList, list of data to make emf from.\n Return list of tuples comprising (data value, cumulative relative frequency) ordered by data value.'''\n \n freqs = makeFreqDictHidden(myDataList) # make the frequency counts mapping\n totalCounts = sum(freqs.values())\n relFreqs = [fr/(1.0*totalCounts) for fr in freqs.values()] # use a list comprehension\n relFreqsArray = array(relFreqs)\n cumFreqs = list(relFreqsArray.cumsum())\n numCumFreqPairs = zip(freqs.keys(), cumFreqs) # zip the keys and culm relative frequencies together\n numCumFreqPairs.sort() # sort the list of tuples\n \n return numCumFreqPairs\n \n# EPMF plot\ndef epmfPlot(samples):\n '''Returns an empirical probability mass function plot from samples data.'''\n \n epmf_pairs = makeEMFHidden(samples)\n epmf = point(epmf_pairs, rgbcolor = \"blue\", pointsize=\"20\")\n for k in epmf_pairs: # for each tuple in the list\n kkey, kheight = k # unpack tuple\n epmf += line([(kkey, 0),(kkey, kheight)], rgbcolor=\"blue\", linestyle=\":\")\n # padding\n epmf += point((0,1), rgbcolor=\"black\", pointsize=\"0\")\n return epmf\n \n\n# ECDF plot\ndef ecdfPlot(samples):\n '''Returns an empirical probability mass function plot from samples data.'''\n ecdf_pairs = makeEDFHidden(samples)\n ecdf = point(ecdf_pairs, rgbcolor = \"red\", faceted = false, pointsize=\"20\")\n for k in range(len(ecdf_pairs)):\n x, kheight = ecdf_pairs[k] # unpack tuple\n previous_x = 0\n previous_height = 0\n if k > 0:\n previous_x, previous_height = ecdf_pairs[k-1] # unpack previous tuple\n ecdf += line([(previous_x, previous_height),(x, previous_height)], rgbcolor=\"grey\")\n ecdf += points((x, previous_height),rgbcolor = \"white\", faceted = true, pointsize=\"20\")\n ecdf += line([(x, previous_height),(x, kheight)], rgbcolor=\"blue\", linestyle=\":\")\n # padding\n ecdf += line([(ecdf_pairs[0][0]-0.2, 0),(ecdf_pairs[0][0], 0)], rgbcolor=\"grey\")\n max_index = len(ecdf_pairs)-1\n ecdf += line([(ecdf_pairs[max_index][0], ecdf_pairs[max_index][1]),(ecdf_pairs[max_index][0]+0.2, \\\n ecdf_pairs[max_index][1])],rgbcolor=\"grey\")\n return ecdf",
"_____no_output_____"
]
],
[
[
"We can see the effect of varying $\\theta$ interactively:",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(theta=(0.5)):\n '''Interactive function to plot the bernoulli pmf and cdf.'''\n if theta <=1 and theta >= 0:\n outcomes = (0, 1) # define the bernoulli outcomes\n print \"Bernoulli (\", RR(theta).n(digits=2), \") pmf and cdf\"\n # pmf plot\n pmf_values = [bernoulliPMF(x, theta) for x in outcomes]\n pmf = pmfPlot(outcomes, pmf_values) # this is one of our own, hidden, functions\n # cdf plot\n cdf_values = [bernoulliCDF(x, theta) for x in outcomes]\n cdf = cdfPlot(outcomes, cdf_values) # this is one of our own, hidden, functions\n show(graphics_array([pmf, cdf]),figsize=[8,3])\n else:\n print \"0 <= theta <= 1\"",
"_____no_output_____"
]
],
[
[
"Don't worry about how these plots are done: you are not expected to be able to understand all of these details now. \n\nJust use them to see the effect of varying $\\theta$.\n\n## Simulating a sample from the $Bernoulli(\\theta)$ RV\n\nWe can simulate a sample from a $Bernoulli$ distribution by transforming input from a $Uniform(0,1)$ distribution using the floor() function in Sage. In maths, $\\lfloor x \\rfloor$, the 'floor of $x$' is the largest integer that is smaller than or equal to $x$. For example, $\\lfloor 3.8 \\rfloor = 3$.",
"_____no_output_____"
]
],
[
[
"z=3.8\nfloor(z)",
"_____no_output_____"
]
],
[
[
"Using floor, we can do inversion sampling from the $Bernoulli(\\theta)$ RV using the the $Uniform(0,1)$ random variable that we said is the fundamental model.\n\nWe will introduce inversion sampling more formally later. In general, inversion sampling means using the inverse of the CDF $F$, $F^{[-1]}$, to transform input from a $Uniform(0,1)$ distribution. \n\nTo simulate from the $Bernoulli(\\theta)$, we can use the following algorithm:\n\n### Input:\n\n- $u \\thicksim Uniform(0,1)$ from a PRNG, $\\qquad \\qquad \\text{where, } \\sim$ means \"sample from\"\n- $\\theta$, the parameter\n\n### Output:\n\n$x \\thicksim Bernoulli(\\theta)$\n\n### Steps:\n\n- $u \\leftarrow Uniform(0,1)$\n- $x \\leftarrow \\lfloor u + \\theta \\rfloor$\n- Return $x$\n\n We can illustrate this with SageMath:",
"_____no_output_____"
]
],
[
[
"theta = 0.5 # theta must be such that 0 <= theta <= 1\nu = random()\nx = floor(u + theta)\nx",
"_____no_output_____"
]
],
[
[
"To make a number of simulations, we can use list comprehensions again:",
"_____no_output_____"
]
],
[
[
"theta = 0.5\nn = 20\nrandomUs = [random() for i in range(n)]\nsimulatedBs = [floor(u + theta) for u in randomUs]\nsimulatedBs",
"_____no_output_____"
]
],
[
[
"To make modular reusable code we can package up what we have done as functions. \n\nThe function `bernoulliFInverse(u, theta)` codes the inverse of the CDF of a Bernoulli distribution parameterised by `theta`. The function `bernoulliSample(n, theta)` uses `bernoulliFInverse(...)` in a list comprehension to simulate n samples from a Bernoulli distribution parameterised by theta, i.e., the distribution of our $Bernoulli(\\theta)$ RV.",
"_____no_output_____"
]
],
[
[
"def bernoulliFInverse(u, theta):\n '''A function to evaluate the inverse CDF of a bernoulli.\n \n Param u is the value to evaluate the inverse CDF at.\n Param theta is the distribution parameters.\n Returns inverse CDF under theta evaluated at u'''\n \n return floor(u + theta)\n \ndef bernoulliSample(n, theta):\n '''A function to simulate samples from a bernoulli distribution.\n \n Param n is the number of samples to simulate.\n Param theta is the bernoulli distribution parameter.\n Returns a simulated Bernoulli sample as a list'''\n \n us = [random() for i in range(n)]\n # use bernoulliFInverse in a list comprehension\n return [bernoulliFInverse(u, theta) for u in us] \n",
"_____no_output_____"
]
],
[
[
"Note that we are using a list comprehension and the built-in SageMath `random()` function to make a list of pseudo-random simulations from the $Uniform(0,1)$. The length of the list is determined by the value of n. Inside the body of the function we assign this list to a variable named `us` (i.e., u plural). We then use another list comprehension to make our simulated sample. This list comprehension works by calling our function `bernoulliFInverse(...)` and passing in values for theta together with each u in us in turn.\n\nLet's try a small number of samples:",
"_____no_output_____"
]
],
[
[
"theta = 0.2\nn = 10\nsamples = bernoulliSample(n, theta)\nsamples",
"_____no_output_____"
]
],
[
[
"Now lets explore the effect of interactively varying n and $\\theta$:",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(theta=(0.5), n=(10,(0..1000))):\n '''Interactive function to plot samples from bernoulli distribution.'''\n if theta >= 0 and theta <= 1:\n print \"epmf and ecdf for \", n, \" samples from Bernoulli (\", theta, \")\"\n samples = bernoulliSample(n, theta)\n # epmf plot\n epmf = epmfPlot(samples) # this is one of our hidden functions\n # ecdf plot\n ecdf = ecdfPlot(samples) # this is one of our hidden functions\n show(graphics_array([epmf, ecdf]),figsize=[8,3])\n else:\n print \"0 <= theta <=1, n>0\"",
"_____no_output_____"
]
],
[
[
"You can vary $\\theta$ and $n$ on the interactive plot. You should be able to see that as $n$ increases, the empirical plots should get closer to the theoretical $f$ and $F$. \n\n\n\n### YouTry\n\nCheck that you understand what `floor` is doing. We have put some extra print statements into our demonstration of floor so that you can see what is going on in each step. Try evaluating this cell several times so that you see what happens with different values of `u`.",
"_____no_output_____"
]
],
[
[
"theta = 0.5 # theta must be such that 0 <= theta <= 1\nu = random()\nprint \"u is\", u\nprint \"u + theta is\", (u + theta)\nprint \"floor(u + theta) is\", floor(u + theta)",
"u is 0.634132126453\nu + theta is 1.13413212645321\nfloor(u + theta) is 1\n"
]
],
[
[
"In the cell below we use floor to get 1's and 0's from the pseudo-random u's given by random(). It is effectively doing exactly the same thing as the functions above that we use to simulate a specified number of $Bernoulli(\\theta)$ RVs, but the why that it is written may be easier to understand. If `floor` is doing what we want it to, then when `n` is sufficiently large, we'd expect our proportion of `1`s to be close to `theta` (remember Kolmogorov's axiomatic motivations for probability!). Try changing the value assigned to the variable `theta` and re-evaluting the cell to check this. ",
"_____no_output_____"
]
],
[
[
"theta = 0.7 # theta must be such that 0 <= theta <= 1\nlistFloorResults = [] # an empty list to store results in\nn = 100000 # how many iterations to do\nfor i in range(n): # a for loop to do something n times\n u = random() # generate u\n x = floor(u + theta) # use floor\n listFloorResults.append(x) # add x to the list of results\nlistFloorResults.count(1)*1.0/len(listFloorResults) # proportion of 1s in the results",
"_____no_output_____"
]
],
[
[
"# The equi-probable $de~Moivre(\\theta)$ Random Variable\n\nThe $de~Moivre(\\theta_1,\\theta_2,\\ldots,\\theta_k)$ RV is the natural generalisation of the $Bernoulli (\\theta)$ RV to more than two outcomes. Take a die (i.e. one of a pair of dice): there are 6 possible outcomes from tossing a die if the die is a normal six-sided one (the outcome is which face is the on the top). To start with we can allow the possibility that the different faces could be loaded so that they have different probabilities of being the face on the top if we throw the die. In this case, k=6 and the parameters $\\theta_1$, $\\theta_2$, ...$\\theta_6$ specify how the die is loaded, and the number on the upper-most face if the die is tossed is a $de\\,Moivre$ random variable parameterised by $\\theta_1,\\theta_2,\\ldots,\\theta_6$. \n\nIf $\\theta_1=\\theta_2=\\ldots=\\theta_6= \\frac{1}{6}$ then we have a fair die.\n\nHere are some functions for the equi-probable $de\\, Moivre$ PMF and CDF where we code the possible outcomes as the numbers on the faces of a k-sided die, i.e, 1,2,...k.",
"_____no_output_____"
]
],
[
[
"def deMoivrePMF(x, k):\n '''Probability mass function for equi-probable de Moivre(k).\n \n Param x is the value to evaluate the deMoirve pmf at.\n Param k is the k parameter for an equi-probable deMoivre.\n Returns the evaluation of the deMoivre(k) pmf at x.'''\n \n if (int(x)==x) & (x > 0) & (x <= k):\n return 1.0/k\n else:\n return 0\n \ndef deMoivreCDF(x, k):\n '''DF for equi-probable de Moivre(k).\n \n Param x is the value to evaluate the deMoirve cdf at.\n Param k is the k parameter for an equi-probable deMoivre.\n Returns the evaluation of the deMoivre(k) cdf at x.'''\n \n return 1.0*x/k",
"_____no_output_____"
],
[
"@interact\ndef _(k=(6)):\n '''Interactive function to plot the de Moivre pmf and cdf.'''\n if (int(k) == k) and (k >= 1):\n outcomes = range(1,k+1,1) # define the outcomes\n pmf_values = [deMoivrePMF(x, k) for x in outcomes]\n print \"equi-probable de Moivre (\", k, \") pmf and cdf\"\n # pmf plot\n pmf = pmfPlot(outcomes, pmf_values) # this is one of our hidden functions\n \n # cdf plot\n cdf_values = [deMoivreCDF(x, k) for x in outcomes]\n cdf = cdfPlot(outcomes, cdf_values) # this is one of our hidden functions\n \n show(graphics_array([pmf, cdf]),figsize=[8,3])\n else:\n print \"k must be an integer, k>0\"",
"_____no_output_____"
]
],
[
[
"### YouTry\nTry changing the value of k in the above interact.",
"_____no_output_____"
],
[
"## Simulating a sample from the equi-probable $de\\,Moivre(k)$ random variable\n\nWe use floor ($\\lfloor \\, \\rfloor$) again for simulating from the equi-probable $de \\, Moivre(k)$ RV, but because we are defining our outcomes as 1, 2, ... k, we just add 1 to the result. ",
"_____no_output_____"
]
],
[
[
"k = 6\nu = random()\nx = floor(u*k)+1\nx",
"_____no_output_____"
]
],
[
[
"To simulate from the equi-probable $de\\,Moivre(k)$, we can use the following algorithm:\n\n#### Input:\n\n- $u \\thicksim Uniform(0,1)$ from a PRNG\n- $k$, the parameter\n\n#### Output:\n\n- $x \\thicksim \\text{equi-probable } de \\, Moivre(k)$\n\n#### Steps:\n\n- $u \\leftarrow Uniform(0,1)$\n- $x \\leftarrow \\lfloor uk \\rfloor + 1$\n- return $x$\n\nWe can illustrate this with SageMath:",
"_____no_output_____"
]
],
[
[
"def deMoivreFInverse(u, k):\n '''A function to evaluate the inverse CDF of an equi-probable de Moivre.\n \n Param u is the value to evaluate the inverse CDF at.\n Param k is the distribution parameter.\n Returns the inverse CDF for a de Moivre(k) distribution evaluated at u.'''\n \n return floor(k*u) + 1\n\ndef deMoivreSample(n, k):\n '''A function to simulate samples from an equi-probable de Moivre.\n \n Param n is the number of samples to simulate.\n Param k is the bernoulli distribution parameter.\n Returns a simulated sample of size n from an equi-probable de Moivre(k) distribution as a list.'''\n \n us = [random() for i in range(n)]\n \n return [deMoivreFInverse(u, k) for u in us]",
"_____no_output_____"
]
],
[
[
"A small sample:",
"_____no_output_____"
]
],
[
[
"deMoivreSample(15,6)",
"_____no_output_____"
]
],
[
[
"You should understand the `deMoivreFInverse` and `deMoivreSample` functions and be able to write something like them if you were asked to. \n\nYou are not expected to be to make the interactive plots below (but this is not too hard to do by syntactic mimicry and google searches!). \n\nNow let's do some interactive sampling where you can vary $k$ and the sample size $n$:",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(k=(6), n=(10,(0..500))):\n '''Interactive function to plot samples from equi-probable de Moivre distribution.'''\n if n > 0 and k >= 0 and int(k) == k:\n print \"epmf and ecdf for \", n, \" samples from equi-probable de Moivre (\", k, \")\"\n outcomes = range(1,k+1,1) # define the outcomes\n samples = deMoivreSample(n, k) # get the samples\n epmf = epmfPlot(samples) # this is one of our hidden functions\n \n ecdf = ecdfPlot(samples) # this is one of our hidden functions\n \n show(graphics_array([epmf, ecdf]),figsize=[10,3])\n else:\n print \"k>0 must be an integer, n>0\"",
"_____no_output_____"
]
],
[
[
"Try changing $n$ and/or $k$. With $k = 40$ for example, you could be simulating the number on the first ball for $n$ Lotto draws.",
"_____no_output_____"
],
[
"### YouTry\n\nA useful counterpart to the floor of a number is the ceiling, denoted $\\lceil \\, \\rceil$. In maths, $\\lceil x \\rceil$, the 'ceiling of $x$' is the smallest integer that is larger than or equal to $x$. For example, $\\lceil 3.8 \\rceil = 4$. We can use the ceil function to do this in Sage:",
"_____no_output_____"
]
],
[
[
"ceil(3.8)",
"_____no_output_____"
]
],
[
[
"Try using `ceil` to check that you understand what it is doing. What would `ceil(0)` be? ",
"_____no_output_____"
],
[
"# Inversion Sampler for Continuous Random Variables\n\nWhen we simulated from the discrete RVs above, the $Bernoulli(\\theta)$ and the equi-probable $de\\,Moivre(k)$, we transformed some $u \\thicksim Uniform(0,1)$ into some value for the RV. \n\nNow we will look at the formal idea of an inversion sampler for continuous random variables. Inversion sampling for continuous random variables is a way to simulate values for a continuous random variable $X$ using $u \\thicksim Uniform(0,1)$. \n\nThe idea of the inversion sampler is to treat $u \\thicksim Uniform(0,1)$ as some value taken by the CDF $F$ and find the value $x$ at which $F(X \\le x) = u$.\n\nTo find x where $F(X \\le x) = u$ we need to use the inverse of $F$, $F^{[-1]}$. This is why it is called an **inversion sampler**.\n\nFormalising this,\n\n### Proposition\n\nLet $F(x) := \\int_{- \\infty}^{x} f(y) \\,d y : \\mathbb{R} \\rightarrow [0,1]$ be a continuous DF with density $f$, and let its inverse $F^{[-1]} $ be:\n\n$$ F^{[-1]}(u) := \\inf \\{ x : F(x) = u \\} : [0,1] \\rightarrow \\mathbb{R} $$\n\nThen, $F^{[-1]}(U)$ has the distribution function $F$, provided $U \\thicksim Uniform(0,1)$ ($U$ is a $Uniform(0,1)$ RV).\n\nNote:\n\nThe infimum of a set A of real numbers, denoted by $\\inf(A)$, is the greatest lower bound of every element of $A$.\n\nProof\n\nThe \"one-line proof\" of the proposition is due to the following equalities: \n\n$$P(F^{[-1]}(U) \\leq x) = P(\\inf \\{ y : F(y) = U)\\} \\leq x ) = P(U \\leq F(x)) = F(x), \\quad \\text{for all } x \\in \\mathbb{R} . $$\n\n# Algorithm for Inversion Sampler\n\n#### Input:\n\n- A PRNG for $Uniform(0,1)$ samples\n- A procedure to give us $F^{[-1]}(u)$, inverse of the DF of the target RV $X$ evaluated at $u$\n\n#### Output:\n\n- A sample $x$ from $X$ distributed according to $F$\n\n#### Algorithm steps:\n\n- Draw $u \\sim Uniform(0,1)$\n- Calculate $x = F^{[-1]}(u)$\n\n# The $Uniform(\\theta_1, \\theta_2)$RV\n\nWe have already met the$Uniform(\\theta_1, \\theta_2)$ RV.\n\nGiven two real parameters $\\theta_1,\\theta_2 \\in \\mathbb{R}$, such that $\\theta_1 < \\theta_2$, the PDF of the $Uniform(\\theta_1,\\theta_2)$ RV $X$ is:\n\n$$f(x;\\theta_1,\\theta_2) =\n\\begin{cases}\n\\frac{1}{\\theta_2 - \\theta_1} & \\text{if }\\theta_1 \\leq x \\leq \\theta_2\\text{,}\\\\\n0 & \\text{otherwise}\n\\end{cases}\n$$\n\nand its DF given by $F(x;\\theta_1,\\theta_2) = \\int_{- \\infty}^x f(y; \\theta_1,\\theta_2) \\, dy$ is:\n\n$$\nF(x; \\theta_1,\\theta_2) =\n\\begin{cases}\n0 & \\text{if }x < \\theta_1 \\\\\n\\frac{x-\\theta_1}{\\theta_2-\\theta_1} & \\text{if}~\\theta_1 \\leq x \\leq \\theta_2,\\\\\n1 & \\text{if} x > \\theta_2 \n\\end{cases}\n$$\n\nFor example, here are the PDF, CDF and inverse CDF for the $Uniform(-1,1)$:\n\n<img src=\"images/UniformMinus11ThreeCharts.png\" width=800>\n\nAs usual, we can make some SageMath functions for the PDF and CDF:",
"_____no_output_____"
]
],
[
[
"# uniform pdf\ndef uniformPDF(x, theta1, theta2):\n '''Uniform(theta1, theta2) pdf function f(x; theta1, theta2).\n \n x is the value to evaluate the pdf at.\n theta1, theta2 are the distribution parameters.'''\n \n retvalue = 0 # default return value\n if x >= theta1 and x <= theta2:\n retvalue = 1.0/(theta2-theta1)\n return retvalue\n\n# uniform cdf \ndef uniformCDF(x, theta1, theta2):\n '''Uniform(theta1, theta2) CDF or DF function F(x; theta1, theta2).\n \n x is the value to evaluate the cdf at.\n theta1, theta2 are the distribution parameters.'''\n \n retvalue = 0 # default return value\n if (x > theta2):\n retvalue = 1\n elif (x > theta1): # else-if\n retvalue = (x - theta1) / (theta2-theta1) \n # if (x < theta1), retvalue will be 0\n return retvalue",
"_____no_output_____"
]
],
[
[
"Using these functions in an interactive plot, we can see the effect of changing the distribution parameters $\\theta_1$ and $\\theta_2$.",
"_____no_output_____"
]
],
[
[
"@interact\ndef InteractiveUniformPDFCDFPlots(theta1=0,theta2=1):\n if theta2 > theta1:\n print \"Uniform(\", + RR(theta1).n(digits=2), \",\", RR(theta2).n(digits=2), \") pdf and cdf\"\n p1 = line([(theta1-1,0), (theta1,0)], rgbcolor='blue')\n p1 += line([(theta1,1/(theta2-theta1)), (theta2,1/(theta2-theta1))], rgbcolor='blue')\n p1 += line([(theta2,0), (theta2+1,0)], rgbcolor='blue')\n \n p2 = line([(theta1-1,0), (theta1,0)], rgbcolor='red')\n p2 += line([(theta1,0), (theta2,1)], rgbcolor='red')\n p2 += line([(theta2,1), (theta2+1,1)], rgbcolor='red')\n show(graphics_array([p1, p2]),figsize=[8,3])\n else:\n print \"theta2 must be greater than theta1\"",
"_____no_output_____"
]
],
[
[
"# Simulating from the $Uniform(\\theta_1, \\theta_2)$ RV\n\nWe can simulate from the $Uniform(\\theta_1,\\theta_2)$ using the inversion sampler, provided that we can get an expression for $F^{[-1]}$ that can be implemented as a procedure. \n\nWe can get this by solving for $x$ in terms of $u=F(x;\\theta_1,\\theta_2)$:\n\n$$\nu = \\frac{x-\\theta_1}{\\theta_2-\\theta_1} \\quad \\iff \\quad x = (\\theta_2-\\theta_1)u+\\theta_1 \\quad \\iff \\quad F^{[-1]}(u;\\theta_1,\\theta_2) = \\theta_1+(\\theta_2-\\theta_1)u \n$$\n\n<img src=\"images/Week7InverseUniformSampler.png\" width=600>\n\n## Algorithm for Inversion Sampler for the $Uniform(\\theta_1, \\theta_2)$ RV\n\n#### Input:\n\n- $u \\thicksim Uniform(0,1)$\n- $F^{[-1]}(u)$\n- $\\theta_1$, $\\theta_2$\n\n#### Output:\n\n- A sample $x \\thicksim Uniform(\\theta_1, \\theta_2)$\n\n#### Algorithm steps:\n\n- Draw $u \\sim Uniform(0,1)$\n- Calculate $x = F^{[-1]}(u) = (\\theta_1 + u(\\theta_2 - \\theta_1))$\n- Return $x$\n\nWe can illustrate this with SageMath by writing a function to calculate the inverse of the CDF of a uniform distribution parameterised by theta1 and theta2. Given a value between 0 and 1 for the parameter u, it returns the height of the inverse CDF at this point, i.e. the value in the range theta1 to theta2 where the CDF evaluates to u.\n",
"_____no_output_____"
]
],
[
[
"def uniformFInverse(u, theta1, theta2):\n '''A function to evaluate the inverse CDF of a uniform(theta1, theta2) distribution.\n \n u, u should be 0 <= u <= 1, is the value to evaluate the inverse CDF at.\n theta1, theta2, theta2 > theta1, are the uniform distribution parameters.'''\n \n return theta1 + (theta2 - theta1)*u",
"_____no_output_____"
]
],
[
[
"This function transforms a single $u$ into a single simulated value from the $Uniform(\\theta_1, \\theta_2)$, for example:",
"_____no_output_____"
]
],
[
[
"u = random()\ntheta1, theta2 = 3, 6\nuniformFInverse(u, theta1, theta2)",
"_____no_output_____"
]
],
[
[
"Then we can use this function inside another function to generate a number of samples:",
"_____no_output_____"
]
],
[
[
"def uniformSample(n, theta1, theta2):\n '''A function to simulate samples from a uniform distribution.\n \n n > 0 is the number of samples to simulate.\n theta1, theta2 (theta2 > theta1) are the uniform distribution parameters.'''\n \n us = [random() for i in range(n)]\n \n return [uniformFInverse(u, theta1, theta2) for u in us]",
"_____no_output_____"
]
],
[
[
"The basic strategy is the same as for simulating $Bernoulli$ and $de \\, Moirve$ samples: we are using a list comprehension and the built-in SAGE random() function to make a list of pseudo-random simulations from the $Uniform(0,1)$. The length of the list is determined by the value of n. Inside the body of the function we assign this list to a variable named us (i.e., u plural). We then use another list comprehension to make our simulated sample. This list comprehension works by calling our function uniformFInverse(...) and passing in values for theta1 and theta2 together with each u in us in turn.\n\nYou should be able to write simple functions like uniformFinverse and uniformSample yourself.\n\nTry this for a small sample:",
"_____no_output_____"
]
],
[
[
"param1 = -5\nparam2 = 5\nnToGenerate = 30\nmyUniformSample = uniformSample(nToGenerate, param1, param2)\nprint(myUniformSample)",
"[2.8957510482187026, 0.03128423174056394, 2.5419773139045034, 4.587000660522651, 3.017723728004537, 0.05305196079818053, 3.5570768970634745, 3.098468698217623, -4.156826181220163, 3.8334293816113494, 0.11277226038258892, 3.678064489253778, -2.49096599755666, -4.9393678427190055, -0.7536381266056988, 0.4943773601439805, 4.152171454112787, -3.176695740118023, -1.6627741539117071, 2.411177478936607, -3.6743958858960735, 4.2637761390893605, -3.3123332132398167, 1.1147343318388945, 0.857106205419214, -4.408559113596703, 1.5718140253264865, 3.6437636999952563, -4.768100573981843, 0.22751787199277906]\n"
]
],
[
[
"Much more fun, we can make an interactive plot which uses the uniformSample(...) function to generate and plot while you choose the parameters and number to generate (you are not expected to be able to make interactive plots like this):",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(theta1=-1, theta2=1, n=(1..5000)):\n '''Interactive function to plot samples from uniform distribution.'''\n if theta2 > theta1:\n if n == 1:\n print n, \"uniform(\", + RR(theta1).n(digits=2), \",\", RR(theta2).n(digits=2), \") sample\"\n else:\n print n, \"uniform(\", + RR(theta1).n(digits=2), \",\", RR(theta2).n(digits=2), \") samples\" \n sample = uniformSample(n, theta1, theta2)\n pts = zip(range(1,n+1,1),sample) # plot so that first sample is at x=1\n p=points(pts)\n p+= text(str(theta1), (0, theta1), fontsize=10, color='black') # add labels manually\n p+= text(str(theta2), (0, theta2), fontsize=10, color='black')\n p.show(xmin=0, xmax = n+1, ymin=theta1, ymax = theta2, axes=false, gridlines=[[0,n+1],[theta1,theta2]], \\\n figsize=[7,3])\n \n else:\n print \"Theta1 must be less than theta2\"",
"_____no_output_____"
]
],
[
[
"We can get a better idea of the distribution of our sample using a histogram (the minimum sample size has been set to 50 here because the automatic histogram generation does not do a very good job with small samples).",
"_____no_output_____"
]
],
[
[
"import pylab\n@interact\ndef _(theta1=0, theta2=1, n=(50..5000), Bins=5):\n '''Interactive function to plot samples from uniform distribution as a histogram.'''\n if theta2 > theta1:\n sample = uniformSample(n, theta1, theta2)\n pylab.clf() # clear current figure\n n, bins, patches = pylab.hist(sample, Bins, density=true) \n pylab.ylabel('normalised count')\n pylab.title('Normalised histogram')\n pylab.savefig('myHist') # to actually display the figure\n pylab.show()\n else:\n print \"Theta1 must be less than theta2\"",
"_____no_output_____"
]
],
[
[
"# The $Exponential(\\lambda)$ Random Variable\n\nFor a given $\\lambda$ > 0, an $Exponential(\\lambda)$ Random Variable has the following PDF $f$ and DF $F$:\n\n \n\n$$\nf(x;\\lambda) =\\begin{cases}\\lambda e^{-\\lambda x} & \\text{if }x \\ge 0\\text{,}\\\\ 0 & \\text{otherwise}\\end{cases}\n$$\n\n$$\nF(x;\\lambda) =\\begin{cases}1 - e^{-\\lambda x} & \\text{if }x \\ge 0\\text{,}\\\\ 0 & \\text{otherwise}\\end{cases}\n$$\n\nAn exponential distribution is useful because is can often be used to model inter-arrival times or making inter-event measurements (if you are familiar with the $Poisson$ distribution, a discrete distribution, you may have also met the $Exponential$ distribution as the time between $Poisson$ events). Here are some examples of random variables which are sometimes modelled with an exponential distribution:\n\ntime between the arrival of buses at a bus-stop\ndistance between roadkills on a stretch of highway\nIn SageMath, the we can use `exp(x)` to calculate $e^x$, for example:",
"_____no_output_____"
]
],
[
[
"x = 3.0\nexp(x)",
"_____no_output_____"
]
],
[
[
"We can code some functions for the PDF and DF of an $Exponential$ parameterised by lambda like this $\\lambda$. \n\n**Note** that we cannot or should not use the name `lambda` for the parameter because in SageMath (and Python), the term `lambda` has a special meaning. Do you recall lambda expressions?",
"_____no_output_____"
]
],
[
[
"def exponentialPDF(x, lam):\n '''Exponential pdf function.\n \n x is the value we want to evaluate the pdf at.\n lam is the exponential distribution parameter.'''\n \n return lam*exp(-lam*x)\n \ndef exponentialCDF(x, lam):\n '''Exponential cdf or df function.\n \n x is the value we want to evaluate the cdf at.\n lam is the exponential distribution parameter.'''\n \n return 1 - exp(-lam*x)",
"_____no_output_____"
]
],
[
[
"You should be able to write simple functions like `exponentialPDF` and `exponentialCDF` yourself, but you are not expected to be able to make the interactive plots.\n\nYou can see the shapes of the PDF and CDF for different values of $\\lambda$ using the interactive plot below.",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(lam=('lambda',0.5),Xmax=(5..100)):\n '''Interactive function to plot the exponential pdf and cdf.'''\n if lam > 0:\n print \"Exponential(\", RR(lam).n(digits=2), \") pdf and cdf\"\n from pylab import arange\n xvalues = list(arange(0.1, Xmax, 0.1))\n p1 = line(zip(xvalues, [exponentialPDF(y, lam) for y in xvalues]), rgbcolor='blue')\n p2 = line(zip(xvalues, [exponentialCDF(y, lam) for y in xvalues]), rgbcolor='red')\n show(graphics_array([p1, p2]),figsize=[8,3])\n \n else:\n print \"Lambda must be greater than 0\"",
"_____no_output_____"
]
],
[
[
"We are going to write some functions to help us to do inversion sampling from the $Exponential(\\lambda)$ RV.\n\nAs before, we need an expression for $F^{[-1]}$ that can be implemented as a procedure.\n\nWe can get this by solving for $x$ in terms of $u=F(x;\\lambda)$\n\n\n### YouTry later\n\nShow that\n\n$$\nF^{[-1]}(u;\\lambda) =\\frac{-1}{\\lambda} \\ln(1-u)\n$$\n\n$\\ln = \\log_e$ is the natural logarthim.\n\n(end of You try)\n\n---\n\n---\n \n\n\n\n# Simulating from the $Exponential(\\lambda)$ RV\n\nAlgorithm for Inversion Sampler for the $Exponential(\\lambda)$ RV\n\n#### Input:\n\n- $u \\thicksim Uniform(0,1)$\n- $F^{[-1]}(u)$\n- $\\lambda$\n\n### Output:\n\n- sample $x \\thicksim Exponential(\\lambda)$\n\n#### Algorithm steps:\n\n- Draw $u \\sim Uniform(0,1)$\n- Calculate $x = F^{[-1]}(u) = \\frac{-1}{\\lambda}\\ln(1-u)$\n- Return $x$\n\nThe function `exponentialFInverse(u, lam)` codes the inverse of the CDF of an exponential distribution parameterised by `lam`. Given a value between 0 and 1 for the parameter `u`, it returns the height of the inverse CDF of the exponential distribution at this point, i.e. the value where the CDF evaluates to `u`. The function `exponentialSample(n, lam)` uses `exponentialFInverse(...)` to simulate `n` samples from an exponential distribution parameterised by `lam`. ",
"_____no_output_____"
]
],
[
[
"def exponentialFInverse(u, lam):\n '''A function to evaluate the inverse CDF of a exponential distribution.\n \n u is the value to evaluate the inverse CDF at.\n lam is the exponential distribution parameter.'''\n \n # log without a base is the natural logarithm\n return (-1.0/lam)*log(1 - u)\n \ndef exponentialSample(n, lam):\n '''A function to simulate samples from an exponential distribution.\n \n n is the number of samples to simulate.\n lam is the exponential distribution parameter.'''\n \n us = [random() for i in range(n)]\n \n return [exponentialFInverse(u, lam) for u in us]",
"_____no_output_____"
]
],
[
[
"We can have a look at a small sample:",
"_____no_output_____"
]
],
[
[
"lam = 0.5\nnToGenerate = 30\nsample = exponentialSample(nToGenerate, lam)\nprint(sorted(sample)) # recall that sorted makes a new sorted list",
"[0.0125113588205858, 0.0446106177884152, 0.168104035485009, 0.197652284327824, 0.223456265784130, 0.230155647487195, 0.289451334866385, 0.330234991767644, 0.524450122724420, 0.615868006331397, 0.855682886147430, 0.861598562693223, 0.899938425232603, 1.12601683541338, 1.18879810930181, 1.50404136083550, 1.67626340919313, 1.80407870498492, 1.85186057920650, 1.92968601068065, 2.05984179746552, 2.35359507332389, 4.17665957500900, 4.20066826966049, 4.53052080151337, 4.69313988593758, 5.17968272452207, 5.42884373902885, 8.84366900758729, 17.0836471409921]\n"
]
],
[
[
"You should be able to write simple functions like `exponentialFinverse` and `exponentialSample` yourself by now.\n\nThe best way to visualise the results is to use a histogram. With this interactive plot you can explore the effect of varying lambda and n:",
"_____no_output_____"
]
],
[
[
"import pylab\n@interact\ndef _(lam=('lambda',0.5), n=(50,(10..10000)), Bins=(5,(1,1000))):\n '''Interactive function to plot samples from exponential distribution.'''\n if lam > 0:\n pylab.clf() # clear current figure\n n, bins, patches = pylab.hist(exponentialSample(n, lam), Bins, density=true) \n pylab.ylabel('normalised count')\n pylab.title('Normalised histogram')\n pylab.savefig('myHist') # to actually display the figure\n pylab.show()\n\n\n else:\n print \"Lambda must be greater than 0\"",
"_____no_output_____"
]
],
[
[
"# The Standard $Cauchy$ Random Variable\n\nA standard $Cauchy$ Random Variable has the following PDF $f$ and DF $F$:\n\n$$\nf(x) =\\frac{1}{\\pi(1+x^2)}\\text{,}\\,\\, -\\infty < x < \\infty\n$$\n\n$$\nF(x) = \\frac{1}{\\pi}\\tan^{-1}(x) + 0.5\n$$\n\n \n\nThe $Cauchy$ distribution is an interesting distribution because the expectation does not exist:\n\n$$\n\\int \\left|x\\right|\\,dF(x) = \\frac{2}{\\pi} \\int_0^{\\infty} \\frac{x}{1+x^2}\\,dx = \\left(x \\tan^{-1}(x) \\right]_0^{\\infty} - \\int_0^{\\infty} \\tan^{-1}(x)\\, dx = \\infty \\ .\n$$\n\nIn SageMath, we can use the `arctan` function for $tan^{-1}$, and `pi` for $\\pi$ and code some functions for the PDF and DF of the standard Cauchy as follows.",
"_____no_output_____"
]
],
[
[
"def cauchyPDF(x):\n '''Standard Cauchy pdf function.\n \n x is the value to evaluate the pdf at.'''\n \n return 1.0/(pi.n()*(1+x^2))\n \ndef cauchyCDF(x):\n '''Standard Cauchy cdf function.\n \n x is the value to evaluate the cdf at.'''\n \n return (1.0/pi.n())*arctan(x) + 0.5",
"_____no_output_____"
]
],
[
[
"\nYou can see the shapes of the PDF and CDF using the plot below. Note from the PDF $f$ above is defined for $-\\infty < x < \\infty$. This means we should set some arbitrary limits on the minimum and maximum values to use for the x-axis on the plots. You can change these limits interactively.",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(lower=(-4), upper=(4)):\n '''Interactive function to plot the Cauchy pdf and cdf.'''\n if lower < upper:\n print \"Standard Cauchy pdf and cdf\" \n p1 = plot(cauchyPDF, lower,upper, rgbcolor='blue')\n p2 = plot(cauchyCDF, lower,upper, rgbcolor='red')\n show(graphics_array([p1, p2]),figsize=[8,3])\n else:\n print \"Upper must be greater than lower\"",
"_____no_output_____"
]
],
[
[
"#### Constructing a standard $Cauchy$ RVs\n\n- Place a double light sabre (i.e., one that can shoot its lazer beam from both ends, like that of Darth Mole in Star Wars) on a cartesian axis so that it is centred on $(0, 1)$. \n- Randomly spin it (so that its spin angle to the x-axis is $\\theta \\thicksim Uniform (0, 2\\pi)$). \n- Let it come to rest.\n- The y-coordinate of the point of intersection with the y-axis is a standard Cauchy RV. \n\nYou can see that we are equally likely to get positive and negative values (the density function of the standard $Cauchy$ RV is symmetrical about 0) and whenever the spin angle is close to $\\frac{\\pi}{4}$ ($90^{\\circ}$) or $\\frac{3\\pi}{2}$ ($270^{\\circ}$), the intersections will be a long way out up or down the y-axis, i.e. very negative or very positive values. If the light sabre is exactly parallel to the y-axis there will be no intersection: a $Cauchy$ RV $X$ can take values $-\\infty < x < \\infty$\n\n \n\n<img src=\"images/Week7CauchyLightSabre.png\" width=300>\n\n \n\n## Simulating from the standard $Cauchy$\n\nWe can perform inversion sampling on the $Cauchy$ RV by transforming a $Uniform(0,1)$ random variable into a $Cauchy$ random variable using the inverse CDF.\n\nWe can get this by replacing $F(x)$ by $u$ in the expression for $F(x)$:\n\n$$\n\\frac{1}{\\pi}tan^{-1}(x) + 0.5 = u\n$$\n\nand solving for $x$:\n\n$$\n\\begin{array}{lcl} \\frac{1}{\\pi}tan^{-1}(x) + 0.5 = u & \\iff & \\frac{1}{\\pi} tan^{-1}(x) = u - \\frac{1}{2}\\\\ & \\iff & tan^{-1}(x) = (u - \\frac{1}{2})\\pi\\\\ & \\iff & tan(tan^{-1}(x)) = tan((u - \\frac{1}{2})\\pi)\\\\ & \\iff & x = tan((u - \\frac{1}{2})\\pi) \\end{array}\n$$\n\n## Inversion Sampler for the standard $Cauchy$ RV\n\n#### Input:\n\n- $u \\thicksim Uniform(0,1)$\n- $F^{[-1]}(u)$\n\n#### Output:\n\n- A sample $x \\thicksim \\text{standard } Cauchy$\n\n#### Algorithm steps:\n\n- Draw $u \\sim Uniform(0,1)$\n- Calculate $x = F^{[-1]}(u) = tan((u - \\frac{1}{2})\\pi)$\n- Return $x$\n\nThe function `cauchyFInverse(u)` codes the inverse of the CDF of the standard Cauchy distribution. Given a value between 0 and 1 for the parameter u, it returns the height of the inverse CDF of the standard $Cauchy$ at this point, i.e. the value where the CDF evaluates to u. The function `cauchySample(n`) uses `cauchyFInverse(...)` to simulate `n` samples from a standard Cauchy distribution.",
"_____no_output_____"
]
],
[
[
"def cauchyFInverse(u):\n '''A function to evaluate the inverse CDF of a standard Cauchy distribution.\n \n u is the value to evaluate the inverse CDF at.'''\n \n return RR(tan(pi*(u-0.5)))\n \ndef cauchySample(n):\n '''A function to simulate samples from a standard Cauchy distribution.\n \n n is the number of samples to simulate.'''\n \n us = [random() for i in range(n)]\n return [cauchyFInverse(u) for u in us]",
"_____no_output_____"
]
],
[
[
"And we can visualise these simulated samples with an interactive plot:",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(n=(50,(0..5000))):\n '''Interactive function to plot samples from standard Cauchy distribution.'''\n if n == 1:\n print n, \"Standard Cauchy sample\"\n else:\n print n, \"Standard Cauchy samples\"\n sample = cauchySample(n)\n pts = zip(range(1,n+1,1),sample)\n p=points(pts)\n p+= text(str(floor(min(sample))), (0, floor(min(sample))), \\\n fontsize=10, color='black') # add labels manually\n p+= text(str(ceil(max(sample))), (0, ceil(max(sample))), \\\n fontsize=10, color='black')\n p.show(xmin=0, xmax = n+1, ymin=floor(min(sample)), \\\n ymax = ceil(max(sample)), axes=false, \\\n gridlines=[[0,n+1],[floor(min(sample)),ceil(max(sample))]],\\\n figsize=[7,3])",
"_____no_output_____"
]
],
[
[
"Notice how we can get some very extreme values This is because of the 'thick tails' of the density function of the $Cauchy$ RV. Think about this in relation to the double light sabre visualisation. We can see effect of the extreme values with a histogram visualisation as well. The interactive plot below will only use values between lower and upper in the histogram. Try increasing the sample size to something like 1000 and then gradually widening the limits:",
"_____no_output_____"
]
],
[
[
"import pylab\n@interact\ndef _(n=(50,(0..5000)), lower=(-4), upper=(4), Bins=(5,(1,100))):\n '''Interactive function to plot samples from \n standard Cauchy distribution.'''\n if lower < upper:\n if n == 1:\n print n, \"Standard Cauchy sample\"\n else:\n print n, \"Standard Cauchy samples\"\n sample = cauchySample(n) # the whole sample\n sampleToShow=[c for c in sample if (c >= lower and c <= upper)]\n pylab.clf() # clear current figure\n n, bins, patches = pylab.hist(sampleToShow, Bins, density=true) \n pylab.ylabel('normalised count')\n pylab.title('Normalised histogram, values between ' \\\n + str(floor(lower)) + ' and ' + str(ceil(upper)))\n pylab.savefig('myHist') # to actually display the figure\n pylab.show()\n else:\n print \"lower must be less than upper\"",
"_____no_output_____"
]
],
[
[
"# Running means\n\nWhen we introduced the $Cauchy$ distribution, we noted that the expectation of the $Cauchy$ RV does not exist. This means that attempts to estimate the mean of a $Cauchy$ RV by looking at a sample mean will not be successful: as you take larger and larger samples, the effect of the extreme values will still cause the sample mean to swing around wildly (we will cover estimation properly soon). You are going to investigate the sample mean of simulated $Cauchy$ samples of steadily increasing size and show how unstable this is. A convenient way of doing this is to look at a running mean. We will start by working through the process of calculating some running means for the $Uniform(0,10)$, which do stabilise. You will then do the same thing for the $Cauchy$ and be able to see the instability.\n\nWe will be using the pylab.cumsum function, so we make sure that we have it available. We then generate a sample from the $Uniform(0,10)$",
"_____no_output_____"
]
],
[
[
"from pylab import cumsum\nnToGenerate = 10 # sample size to generate\ntheta1, theta2 = 0, 10 # uniform parameters\nuSample = uniformSample(nToGenerate, theta1, theta2)\nprint(uSample)",
"[5.234123372317886, 8.494432455891982, 9.693289516301245, 5.025095534949817, 6.860022865668135, 5.194665625019904, 3.236702879672173, 1.3642271829174146, 4.653495098823827, 0.5932379933902066]\n"
]
],
[
[
"We are going to treat this sample as though it is actually 10 samples of increasing size:\n\n- sample 1 is the first element in uSample\n- sample 2 contains the first 2 elements in uSample\n- sample 3 contains the first 3 elements in uSample\n- ...\n- sample10 contains the first 10 elements in uSample\n\nWe know that a sample mean is the sum of the elements in the sample divided by the number of elements in the sample $n$:\n\n$$\n\\bar{x} = \\frac{1}{n} \\sum_{i=1}^n x_i\n$$\n\nWe can get the sum of the elements in each of our 10 samples with the cumulative sum of `uSample`. \n\nWe use `cumsum` to get the cumulative sum. This will be a `pylab.array` (or `numpy.arrat`) type, so we use the `list` function to turn it back into a list:",
"_____no_output_____"
]
],
[
[
"csUSample = list(cumsum(uSample))\nprint(csUSample)",
"[5.234123372317886, 13.728555828209867, 23.421845344511112, 28.44694087946093, 35.30696374512907, 40.50162937014897, 43.73833224982114, 45.10255943273856, 49.756054531562384, 50.34929252495259]\n"
]
],
[
[
"What we have now is effectively a list\n\n$$\\left[\\displaystyle\\sum_{i=1}^1x_i, \\sum_{i=1}^2x_i, \\sum_{i=1}^3x_i, \\ldots, \\sum_{i=1}^{10}x_i\\right]$$\n\nSo all we have to do is divide each element in `csUSample` by the number of elements that were summed to make it, and we have a list of running means \n\n$$\\left[\\frac{1}{1}\\displaystyle\\sum_{i=1}^1x_i, \\frac{1}{2}\\sum_{i=1}^2x_i, \\frac{1}{3}\\sum_{i=1}^3x_i, \\ldots, \\frac{1}{10}\\sum_{i=1}^{10}x_i\\right]$$\n\nWe can get the running sample sizes using the `range` function:",
"_____no_output_____"
]
],
[
[
"samplesizes = range(1, len(uSample)+1,1)\nsamplesizes",
"_____no_output_____"
]
],
[
[
"And we can do the division with list comprehension:",
"_____no_output_____"
]
],
[
[
"uniformRunningMeans = [csUSample[i]/samplesizes[i] for i in range(nToGenerate)]\nprint(uniformRunningMeans)",
"[5.234123372317886, 6.8642779141049335, 7.807281781503704, 7.111735219865232, 7.0613927490258135, 6.750271561691495, 6.248333178545877, 5.63781992909232, 5.528450503506932, 5.0349292524952585]\n"
]
],
[
[
"We could pull all of this together into a function which produced a list of running means for sample sizes 1 to $n$.",
"_____no_output_____"
]
],
[
[
"def uniformRunningMeans(n, theta1, theta2):\n '''Function to give a list of n running means from uniform(theta1, theta2).\n \n n is the number of running means to generate.\n theta1, theta2 are the uniform distribution parameters.\n return a list of n running means.'''\n \n sample = uniformSample(n, theta1, theta2)\n from pylab import cumsum # we can import in the middle of code!\n csSample = list(cumsum(sample))\n samplesizes = range(1, n+1,1)\n return [csSample[i]/samplesizes[i] for i in range(n)]",
"_____no_output_____"
]
],
[
[
"Have a look at the running means of 10 incrementally-sized samples:",
"_____no_output_____"
]
],
[
[
"nToGenerate = 10\ntheta1, theta2 = 0, 10\nuRunningMeans = uniformRunningMeans(nToGenerate, theta1, theta2)\npts = zip(range(1, len(uRunningMeans)+1,1),uRunningMeans)\np = points(pts)\nshow(p, figsize=[5,3])",
"_____no_output_____"
]
],
[
[
"Recall that the expectation $E_{(\\theta_1, \\theta_2)}(X)$ of a $X \\thicksim Uniform(\\theta_1, \\theta_2) = \\frac{(\\theta_1 +\\theta_2)}{2}$\n\nIn our simulations we are using $\\theta_1 = 0$, $\\theta_2 = 10$, so if $X \\thicksim Uniform(0,10)$, $E(X) = 5$\n\nTo show that the running means of different simulations from a $Uniform$ distribution settle down to be close to the expectation, we can plot say 5 different groups of running means for sample sizes $1, \\ldots, 1000$. We will use a line plot rather than plotting individual points. ",
"_____no_output_____"
]
],
[
[
"nToGenerate = 1000\ntheta1, theta2 = 0, 10\niterations = 5\nxvalues = range(1, nToGenerate+1,1)\nfor i in range(iterations):\n redshade = 0.5*(iterations - 1 - i)/iterations # to get different colours for the lines\n uRunningMeans = uniformRunningMeans(nToGenerate, theta1, theta2)\n pts = zip(xvalues,uRunningMeans)\n if (i == 0):\n p = line(pts, rgbcolor = (redshade,0,1))\n else:\n p += line(pts, rgbcolor = (redshade,0,1))\nshow(p, figsize=[5,3])",
"_____no_output_____"
]
],
[
[
"### YouTry!\n\nYour task is to now do the same thing for some standard Cauchy running means.\n\nTo start with, do not put everything into a function, just put statements into the cell(s) below to:\n\nMake variable for the number of running means to generate; assign it a small value like 10 at this stage\nUse the cauchySample function to generate the sample from the standard $Cauchy$; have a look at your sample\nMake a named list of cumulative sums of your $Cauchy$ sample using list and cumsum, as we did above; have a look at your cumulative sums\nMake a named list of sample sizes, as we did above\nUse a list comprehension to turn the cumulative sums and sample sizes into a list of running means, as we did above\nHave a look at your running means; do they make sense to you given the individual sample values?\nAdd more cells as you need them.",
"_____no_output_____"
],
[
"When you are happy that you are doing the right things, **write a function**, parameterised by the number of running means to do, that returns a list of running means. Try to make your own function rather than copying and changing the one we used for the $Uniform$: you will learn more by trying to do it yourself. Please call your function `cauchyRunningMeans`, so that (if you have done everything else right), you'll be able to use some code we will supply you with to plot the results. ",
"_____no_output_____"
],
[
"Try checking your function by using it to create a small list of running means. Check that the function does not report an error and gives you the kind of list you expect.",
"_____no_output_____"
],
[
"When you think that your function is working correctly, try evaluating the cell below: this will put the plot of 5 groups of $Uniform(0,10)$ running means beside a plot of 5 groups of standard $Cauchy$ running means produced by your function. ",
"_____no_output_____"
]
],
[
[
"nToGenerate = 10000\ntheta1, theta2 = 0, 10\niterations = 5\nxvalues = range(1, nToGenerate+1,1)\nfor i in range(iterations):\n shade = 0.5*(iterations - 1 - i)/iterations # to get different colours for the lines\n uRunningMeans = uniformRunningMeans(nToGenerate, theta1, theta2)\n problemStr=\"\" # an empty string\n # use try to catch problems with cauchyRunningMeans functions\n try:\n cRunningMeans = cauchyRunningMeans(nToGenerate)\n ##cRunningMeans = hiddenCauchyRunningMeans(nToGenerate)\n cPts = zip(xvalues, cRunningMeans)\n except NameError, e:\n # cauchyRunningMeans is not defined\n cRunningMeans = [1 for c in range(nToGenerate)] # default value\n problemStr = \"No \" \n except Exception, e:\n # some other problem with cauchyRunningMeans\n cRunningMeans = [1 for c in range(nToGenerate)]\n problemStr = \"Problem with \" \n uPts = zip(xvalues, uRunningMeans)\n cPts = zip(xvalues, cRunningMeans)\n if (i < 1):\n p1 = line(uPts, rgbcolor = (shade, 0, 1))\n p2 = line(cPts, rgbcolor = (1-shade, 0, shade))\n cauchyTitleMax = max(cRunningMeans) # for placement of cauchy title\n else:\n p1 += line(uPts, rgbcolor = (shade, 0, 1))\n p2 += line(cPts, rgbcolor = (1-shade, 0, shade))\n if max(cRunningMeans) > cauchyTitleMax:\n cauchyTitleMax = max(cRunningMeans)\ntitleText1 = \"Uniform(\" + str(theta1) + \",\" + str(theta2) + \") running means\" # make title text\nt1 = text(titleText1, (nToGenerate/2,theta2), rgbcolor='blue',fontsize=10) \ntitleText2 = problemStr + \"standard Cauchy running means\" # make title text\nt2 = text(titleText2, (nToGenerate/2,ceil(cauchyTitleMax)+1), rgbcolor='red',fontsize=10) \nshow(graphics_array((p1+t1,p2+t2)),figsize=[10,5])",
"_____no_output_____"
]
],
[
[
"# Replicable samples\n\nRemember that we know how to set the seed of the PRNG used by `random()` with `set_random_seed`? If we wanted our sampling functions to give repeatable samples, we could also pass the functions the seed to use. Try making a new version of `uniformSample` which has a parameter for a value to use as the random number generator seed. Call your new version `uniformSampleSeeded` to distinguish it from the original one. ",
"_____no_output_____"
],
[
"Try out your new `uniformSampleSeeded` function: if you generate two samples using the same seed they should be exactly the same. You could try using a large sample and checking on sample statistics such as the mean, min, max, variance etc, rather than comparing small samples by eye. ",
"_____no_output_____"
],
[
"Recall that you can also give parameters default values in SageMath. Using a default value means that if no value is passed to the function for that parameter, the default value is used. Here is an example with a very simple function:",
"_____no_output_____"
]
],
[
[
"# we already saw default parameters in use - here's a careful walkthrough of how it works \ndef simpleDefaultExample(x, y=0):\n '''A simple function to demonstrate default parameter values.\n \n x is the first parameter, with no default value.\n y is the second parameter, defaulting to 0.'''\n \n return x + y",
"_____no_output_____"
]
],
[
[
"Note that parameters with default values need to come after parameters without default values when we define the function. \n\nNow you can try the function - evaluate the following cells to see what you get:",
"_____no_output_____"
]
],
[
[
"simpleDefaultExample (1,3) # specifying two arguments for the function",
"_____no_output_____"
],
[
"simpleDefaultExample (1) # specifying one argument for the function",
"_____no_output_____"
],
[
"# another way to specify one argument for the function\nsimpleDefaultExample (x=6) ",
"_____no_output_____"
],
[
"# uncomment next line and evaluate - but this will give an error because x has no default value\n#simpleDefaultExample() ",
"_____no_output_____"
],
[
"# uncomment next line and evaluate - but this will also give an error because x has no default value\n# simpleDefaultExample (y=9) ",
"_____no_output_____"
]
],
[
[
"Try making yet another version of the uniform sampler which takes a value to be used as a random number generator seed, but defaults to `None` if no value is supplied for that parameter. `None` is a special Python type.",
"_____no_output_____"
]
],
[
[
"x = None\ntype(x)",
"_____no_output_____"
]
],
[
[
"Using `set_random_seed(None)` will mean that the random seed is actually reset to a new ('random') value. You can see this by testing what happens when you do this twice in succession and then check what seed is being used with `initial_seed`:",
"_____no_output_____"
]
],
[
[
"set_random_seed(None)\ninitial_seed()",
"_____no_output_____"
],
[
"set_random_seed(None)\ninitial_seed()",
"_____no_output_____"
]
],
[
[
"Do another version of the `uniformSampleSeeded` function with a default value for the seed of `None`.",
"_____no_output_____"
],
[
"Check your function again by testing with both when you supply a value for the seed and when you don't.",
"_____no_output_____"
],
[
"---\n## Assignment 2, PROBLEM 4\nMaximum Points = 1",
"_____no_output_____"
],
[
"\nFirst read and understand the following simple simulation (originally written by Jenny Harlow). Then you will modify the simulation to find the solution to this problem.\n\n### A Simple Simulation\n\nWe could use the samplers we have made to do a very simple simulation. Suppose the inter-arrival times, in minutes, of Orbiter buses at an Orbiter stop in Christchurch follows an $Exponential(\\lambda = 0.1)$ distribution. Also suppose that this is quite a popular bus stop, and the arrival of people is very predictable: one new person will arrive in each whole minute. This means that the longer another bus takes in coming, the more people arrive to join the queue. Also suppose that the number of free seats available on any bus follows a $de\\, Moivre(k=40)$ distribution, i.e, there are equally like to to be 1, or 2, or 3 ... or 40 spare seats. If there are more spare seats than people in the queue, everyone can get onto the bus and nobody is left waiting, but if there are not enough spare seats some people will be left waiting for the next bus. As they wait, more people arrive to join the queue....\n\nThis is not very realistic - we would want a better model for how many people arrive at the stop at least, and for the number of spare seats there will be on the bus. However, we are just using this as a simple example that you can do using the random variables you already know how to simulate samples from.\n\nTry to code this example yourself, using our suggested steps. We have put our version the code into a cell below, but you will get more out of this example by trying to do it yourself first.\n\n#### Suggested steps:\n\n- Get a list of 100 $Exponential(\\lambda = 0.1)$ samples using the `exponentialSamples` function. Assign the list to a variable named something like `busTime`s. These are your 100 simulated bus inter-arrival times. \n- Choose a value for the number of people who will be waiting at the busstop when you start the simulation. Call this something like `waiting`. \n- Make a list called something like `leftWaiting`, which to begin with contains just the value assigned to `waiting`. \n- Make an empty list called something like `boardBus`. \n- Start a for loop which takes each element in `busTimes` in turn, i.e. each bus inter-arrival time, and within the for loop:\n - Calculate the number of people arriving at the stop as the floor of the time taken for that bus to arrive (i.e., one person for each whole minute until the bus arrives)\n - Add this to the number of people waiting (e.g., if the number of arrivals is assigned to a variable arrivals, then waiting = waiting + arrivals will increment the value assigned to the waiting variable by the value of arrivals).\n - Simulate a value for the number of seats available on the bus as one simulation from a $de \\, Moirve(k=40)$ RV (it may be easier to use `deMoivreFInverse` rather than `deMoivreSample` because you only need one value - remember that you will have to pass a simulated $u \\thicksim Uniform(0,1)$ to `deMoivreFInverse` as well as the value of the parameter $k$).\n - The number of people who can get on the bus is the minimum of the number of people waiting in the queue and the number of seats on the bus. Calculate this value and assign it to a variable called something like `getOnBus`.\n - Append `getOnBus` to the list `boardBus`.\n - Subtract `getOnBus` from the number of people waiting, waiting (e.g., `waiting = waiting - getOnBus` will decrement waiting by the number of people who get on the bus).\n - Append the new value of `waiting` to the list `leftWaiting`. \n- That is the end of the for loop: you now have two lists, one for the number of people waiting at the stop and one for the number of people who can board each bus as it arrives.",
"_____no_output_____"
],
[
"## YouTry",
"_____no_output_____"
],
[
"\nHere is our code to do the bus stop simulation. \nYours may be different - maybe it will be better!\n\n*You are expected to find the needed functions from the latest notebook this assignment came from and be able to answer this question. Unless you can do it in your head.*",
"_____no_output_____"
]
],
[
[
"def busStopSimulation(buses, lam, seats):\n '''A Simple Simulation - see description above!'''\n BusTimes = exponentialSample(buses,lam)\n waiting = 0 # how many people are waiting at the start of the simulation\n BoardBus = [] # empty list\n LeftWaiting = [waiting] # list with just waiting in it\n for time in BusTimes: # for each bus inter-arrival time\n arrivals = floor(time) # people who arrive at the stop before the bus gets there\n waiting = waiting + arrivals # add them to the queue\n busSeats = deMoivreFInverse(random(), seats) # how many seats available on the bus\n getOnBus = min(waiting, busSeats) # how many people can get on the bus\n BoardBus.append(getOnBus) # add to the list\n waiting = waiting - getOnBus # take the people who board the bus out of the queue\n LeftWaiting.append(waiting) # add to the list\n return [LeftWaiting, BoardBus, BusTimes]",
"_____no_output_____"
],
[
"# let's simulate the people left waiting at the bus stop\nset_random_seed(None) # replace None by a integer to fix seed and output of simulation\nbuses = 100\nlam = 0.1\nseats = 40\nleftWaiting, boardBus, busTimes = busStopSimulation(buses, lam, seats)\n\nprint(leftWaiting) # look at the leftWaiting list\n\nprint(boardBus) # boad bus\n\nprint(busTimes)",
"[0, 0, 0, 0, 0, 0, 0, 3, 0, 0, 0, 0, 0, 11, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 29, 10, 0, 0, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 6, 0, 0, 4, 0, 5, 0, 0, 0, 0, 0, 8, 0, 0, 0, 0, 0, 0, 13, 7, 0, 0, 0, 0, 0, 0, 0, 0, 0, 37, 4, 3, 0, 0, 0, 0, 0, 0, 8, 0, 0, 0, 0, 0, 0, 0, 6, 0, 0, 0, 0, 16]\n[6, 9, 7, 4, 9, 4, 11, 20, 0, 1, 5, 2, 29, 12, 0, 2, 10, 2, 2, 4, 4, 7, 2, 0, 1, 20, 1, 1, 0, 6, 0, 1, 21, 21, 20, 0, 9, 2, 12, 5, 2, 12, 6, 0, 6, 14, 11, 6, 11, 7, 5, 9, 10, 6, 6, 19, 0, 3, 0, 15, 15, 6, 10, 3, 9, 10, 21, 14, 8, 7, 1, 15, 7, 3, 16, 5, 0, 19, 37, 6, 7, 1, 2, 17, 0, 1, 3, 21, 7, 4, 0, 2, 3, 1, 10, 6, 2, 12, 4, 4]\n[6.48518705515480, 9.18039802685999, 7.07762986049353, 4.93015320786531, 9.07759248615945, 4.30282281953945, 14.8981154360975, 17.4180720565381, 0.754776218417572, 1.11215113917905, 5.09901931444744, 2.93787286934829, 40.4810051956517, 1.24993523913344, 0.430252482162418, 2.51514718931127, 10.5337864200131, 2.29517394759063, 2.63875173590370, 4.52676112167308, 7.38748670945393, 4.91187079095663, 2.84839579995237, 0.753361138824570, 1.27555413498616, 20.9895756110382, 1.39386557435748, 1.79815852719863, 0.229253572636612, 6.72226120904483, 0.398741918095855, 1.23603939111658, 50.9906945876376, 2.44480341943560, 10.3582448845091, 0.530410089006438, 9.68779852398168, 2.24821906573067, 12.5152133665588, 5.47540162751441, 2.77486351339220, 14.4279436283841, 4.73928352145235, 0.0464266496064611, 6.53778445312108, 14.9434744629101, 11.0903512124119, 6.01400927515197, 17.7029630031255, 1.98422050518540, 5.12658282075989, 13.5590398823156, 6.72276333192598, 11.8379685793199, 1.84702578444348, 19.1196559717640, 0.888964086803038, 3.87853598195318, 0.117246321280377, 23.3035391183737, 7.19161616333384, 6.99355959059122, 10.0963223428414, 3.28422586108564, 9.34529914853560, 10.6415145844164, 34.7894979587235, 8.98315494599303, 1.58917281390920, 7.78975699006630, 1.81173445799809, 15.0243121919514, 7.76878221610191, 3.92328481043838, 16.2578186553004, 5.52551904321898, 0.867800140837969, 56.4817146729685, 4.30516345740088, 5.64554807961943, 4.82579249465213, 1.07717718215008, 2.94114785110358, 17.7143322043820, 0.733212005579067, 1.81341900874206, 11.8281274454670, 13.2460906218122, 7.40162283611479, 4.98010701724401, 0.747383407699742, 2.25879129051231, 3.21073518501013, 1.84093552508158, 16.5679701762106, 0.743593171058093, 2.19910023217475, 12.1292252091886, 4.48024682996023, 20.4460482087548]\n"
]
],
[
[
"We could do an interactive visualisation of this by evaluating the next cell. This will be showing the number of people able to board the bus and the number of people left waiting at the bus stop by the height of lines on the plot.",
"_____no_output_____"
]
],
[
[
"@interact\ndef _(seed=[0,123,456], lam=[0.1,0.01], seats=[40,10,1000]):\n set_random_seed(seed)\n buses=100\n leftWaiting, boardBus, busTimes = busStopSimulation(buses, lam,seats)\n p1 = line([(0.5,0),(0.5,leftWaiting[0])])\n from pylab import cumsum\n csBusTimes=list(cumsum(busTimes))\n for i in range(1, len(leftWaiting), 1):\n \n p1+= line([(csBusTimes[i-1],0),(csBusTimes[i-1],boardBus[i-1])], rgbcolor='green')\n p1+= line([(csBusTimes[i-1]+.01,0),(csBusTimes[i-1]+.01,leftWaiting[i])], rgbcolor='red')\n\n t1 = text(\"Boarding the bus\", (csBusTimes[len(busTimes)-1]/3,max(max(boardBus),max(leftWaiting))+1), \\\n rgbcolor='green',fontsize=10) \n t2 = text(\"Waiting\", (csBusTimes[len(busTimes)-1]*(2/3),max(max(boardBus),max(leftWaiting))+1), \\\n rgbcolor='red',fontsize=10) \n xaxislabel = text(\"Time\", (csBusTimes[len(busTimes)-1],-10),fontsize=10,color='black')\n yaxislabel = text(\"People\", (-50,max(max(boardBus),max(leftWaiting))+1),fontsize=10,color='black')\n show(p1+t1+t2+xaxislabel+yaxislabel,figsize=[8,5])",
"_____no_output_____"
]
],
[
[
"\nVery briefly explain the effect of varying one of the three parameters:\n\n- `seed`\n- `lam`\n- `seats`\n\nwhile holding the other two parameters fixed on:\n\n- the number of people waiting at the bus stop and\n- the number of people boarding the bus \n\nby using the dropdown menus in the `@interact` above. Think if the simulation makes sense and explain why. You can write down your answers using keyboard by double-clicking this cell and writing between `---` and `---`.\n\n---\n\n\n---\n",
"_____no_output_____"
],
[
"#### Solution for CauchyRunningMeans",
"_____no_output_____"
]
],
[
[
"def hiddenCauchyRunningMeans(n):\n '''Function to give a list of n running means from standardCauchy.\n \n n is the number of running means to generate.'''\n \n sample = cauchySample(n)\n from pylab import cumsum\n csSample = list(cumsum(sample))\n samplesizes = range(1, n+1,1)\n return [csSample[i]/samplesizes[i] for i in range(n)]",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
4ae72c42328b0441e4cba6da92f9a12e4be38656
| 324,892 |
ipynb
|
Jupyter Notebook
|
nn-example.ipynb
|
BNL-ML-Group/example-project
|
ccdab577066a54ccd82d5f1c76f3bde5a635f3ab
|
[
"MIT"
] | null | null | null |
nn-example.ipynb
|
BNL-ML-Group/example-project
|
ccdab577066a54ccd82d5f1c76f3bde5a635f3ab
|
[
"MIT"
] | null | null | null |
nn-example.ipynb
|
BNL-ML-Group/example-project
|
ccdab577066a54ccd82d5f1c76f3bde5a635f3ab
|
[
"MIT"
] | null | null | null | 428.617414 | 43,392 | 0.721335 |
[
[
[
"Jeremy Thaller - Aug. 2021 \n\n*Write a quick summary of the project here. For example: CNN to predict MSD values from XANES spectra.*",
"_____no_output_____"
]
],
[
[
"import numpy as np\r\nimport pandas as pd\r\nimport datetime\r\nimport seaborn as sns\r\nsns.set_style('whitegrid')\r\nimport matplotlib.pyplot as plt\r\nimport tensorflow as tf\r\nfrom tensorflow.keras.layers.experimental import preprocessing\r\nfrom tensorflow import keras\r\nfrom tensorflow.keras import layers\r\n# often I re-use code I've already written, or hide boiler plate code in a \r\n# separate script to keep the main notebook cleaner\r\nfrom scripts.nn_buddy import * \r\n\r\n# Make numpy values easier to read.\r\nnp.set_printoptions(precision=6, suppress=True)\r\n#fix blas GEMM error\r\nphysical_devices = tf.config.list_physical_devices('GPU') \r\n# un-comment the line below when using a GPU\r\n# tf.config.experimental.set_memory_growth(physical_devices[0], True)\r\nprint(\"GPU power activated 🚀🚀\" if len(physical_devices) > 0 else \"No GPU found\") ",
"No GPU found\n"
]
],
[
[
"# EDA and Dataloading\nWith any notebook, the first thing to do after importing everything is to load the data and do some basic data exploratory analysis. Even if you have done some in-depth EDA in another notebook, it's worth printing the dataframe and maybe a plot to double check everything loaded correctly.",
"_____no_output_____"
],
[
"Note, I'm calling `load_all_spectra`, a function I wrote in nn_buddy.py. The first time you run it, it will load all the CSV files into one large dataframe. The columns are the energy values (your features), and each row is an absorption spectrum. After it does this the first time, it saves the dataframe as a HDF file, which it will import next time instead. If you have 1000's of csv files and have to do lots of operations (mainly transposing the data can be time-intensive), loading all the spectra can become slow. Saving the dataframe as an HDF might save you time in the long-run.",
"_____no_output_____"
]
],
[
[
"DATA_PATH = 'DATA'\r\ndataset = load_all_spectra(path_=DATA_PATH, header=3)\r\ndataset.head()",
"hdf file found\ndataset shape: (11, 83)\n"
],
[
"energy_grid = dataset.columns[:-1].to_numpy()\r\nsns.lineplot(x=energy_grid, y=dataset.iloc[0,:-1])\r\nplt.ylabel('$\\mu$'), plt.xlabel('E (eV)')\r\nplt.title('Example Plot');",
"_____no_output_____"
]
],
[
[
"Or, if you know you can write a plotting function in `nn_buddy.py` to make repetitive plotting easier",
"_____no_output_____"
]
],
[
[
"plot_spectrum(dataset, index=1, title='Example Spectrum', save_as='Figures/example.pdf');",
"_____no_output_____"
]
],
[
[
"Split your testing set into a training set and testing set. Don't touch the testing set until the very end (i.e. data leakage). If you have a sparse dataset, you might consider using k-fold cross validation instead",
"_____no_output_____"
]
],
[
[
"# train-test split\r\ntrain_dataset = dataset.sample(frac=0.8, random_state=0)\r\ntest_dataset = dataset.drop(train_dataset.index)\r\n# features dataframe \r\ntrain_features = train_dataset.copy()\r\ntest_features = test_dataset.copy()\r\n# labels dataframe\r\ntrain_labels = train_features.pop('MSD')\r\ntest_labels = test_features.pop('MSD')",
"_____no_output_____"
]
],
[
[
"# Preprocessing\n## Normalization\nFrom TF docs: *This layer will coerce its inputs into a distribution centered around 0 with standard deviation 1. It accomplishes this by precomputing the mean and variance of the data, and calling $\\frac{(input-mean)}{\\sqrt{variance}}$ at runtime.*\n\nWhat happens in adapt: *Compute mean and variance of the data and store them as the layer's weights. adapt should be called before fit, evaluate, or predict.*",
"_____no_output_____"
]
],
[
[
"# I'm using keras' version, but you can define your own noramlization\r\nnormalizer = preprocessing.Normalization()\r\nnormalizer.adapt(np.array(train_features))",
"_____no_output_____"
]
],
[
[
"We need to scale the training labels as well, because they have a very limit range and will restrict the ability of the network to learn. A network that just guesses the mean of the labels will have a very small loss if the range is also very small. This will scale the labels from 0-1 (for the training data, and something close to that for the validation data.)\n\nWe will rescale via $ z_i = \\frac{x_i - \\text{min}(x)}{\\text{max}(x) - \\text{min}(x)}$, but fix the min, and max values so that we can reliably \"unscale\" the data afterwords to retrieve the correct NN predictions. Note, the min, and max are of just the training data. This is on purpose to prevent data leakage. The testing data needs to be scaled with this same value.\n\nTo \"unscale\" or \"denormalize\", we use $x_i = z_i (\\text{max}(x) - \\text{min}(x)) + \\text{min}(x)$",
"_____no_output_____"
]
],
[
[
"# given a np.array\r\ndef normalize_labels(labels):\r\n min, max = np.min(train_labels), np.max(train_labels)\r\n return labels.apply(lambda x: (x - min)/(max-min))\r\n\r\ndef unnormalize_labels(labels):\r\n min, max = np.min(train_labels), np.max(train_labels)\r\n if isinstance(labels, np.ndarray):\r\n labels = labels.flatten()\r\n return [x*(max - min) + min for x in labels]\r\n else:\r\n return labels.apply(lambda x: x*(max - min) + min)",
"_____no_output_____"
],
[
"normalized_train_labels = normalize_labels(train_labels)\r\nnormalized_train_labels.describe()",
"_____no_output_____"
]
],
[
[
"# Model Building and Testing\nNow the fun part. You can try out different architectures and models here. I'd suggest putting each unique type of model in a subheading so you can minimize the section when working on somethign else. Note nothing here should work well in this examle notebook because I'm only including a few spectra in the dataset",
"_____no_output_____"
],
[
"## Simple Neural Network\r\nFor a nice procedure on how to build a neural network, see check out the end of Chapter 3 of [my thesis](https://github.com/jthaller/BNL_Thesis/blob/main/MainDraft.pdf)\r\n\r\nFor hyperparameter tuning, check out the `Optuna package` and utilize `tensorboard` to compare you models. You can find specifc examples of how I used it in the `nn-rdf.ipynb` script in [this repository](https://github.com/BNL-ML-Group/xanes-disorder-nn). This also is a good example for how to write a useful `README.md` for a project",
"_____no_output_____"
]
],
[
[
"# norm = normalizer\r\ndef build_and_compile_model(norm):\r\n model = tf.keras.Sequential([\r\n norm,\r\n layers.Dense(64, activation='relu'), #kernal_initializer = tf.keras.initializers.LecunNormal()\r\n layers.Dense(32, activation='relu'),\r\n layers.Dense(64, activation='relu'),\r\n layers.Dense(1)\r\n ])\r\n\r\n model.compile(loss='mean_absolute_error', # L1 = Lass0 = 'mean_absolute_error'; L2 = ridge = 'mean_squared_error'\r\n optimizer=tf.keras.optimizers.Adam(0.001),\r\n metrics=[tf.keras.metrics.MeanAbsolutePercentageError()]\r\n )\r\n return model\r\n\r\nnn_model = build_and_compile_model(normalizer)",
"_____no_output_____"
],
[
"log_dir = \"./logs/fit/\" + datetime.datetime.now().strftime(\"%Y%m%d-%H%M%S\")\r\ntensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=log_dir)\r\nhistory = linear_model.fit(train_features, normalized_train_labels, \r\n epochs=5, callbacks=[tensorboard_callback],\r\n verbose=0, validation_split=.2) # histogram_frequency=1\r\nnn_model.save('./Models/nn_model')\r\nhist = pd.DataFrame(history.history)\r\nhist['epoch'] = history.epoch\r\nplot_loss(history)",
"INFO:tensorflow:Assets written to: ./Models/saved_linear_model/assets\n"
]
],
[
[
"## XG-Boost",
"_____no_output_____"
]
],
[
[
"import xgboost as xgb",
"_____no_output_____"
],
[
"regressor = xgb.XGBRegressor(\r\n n_estimators=100,\r\n reg_lambda=0,\r\n gamma=0,\r\n max_depth=10\r\n)\r\n# note I'm not normalizing the features for xgboost \r\n# https://datascience.stackexchange.com/questions/60950/is-it-necessary-to-normalise-data-for-xgboost/60954\r\nregressor.fit(train_features, normalized_train_labels)",
"_____no_output_____"
]
],
[
[
"# Make Predictions",
"_____no_output_____"
],
[
"## NN - linear model",
"_____no_output_____"
]
],
[
[
"nn_model = keras.models.load_model('./Models/nn_model', compile = True)\r\nnn_model.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nnormalization_1 (Normalizati (None, 82) 165 \n_________________________________________________________________\ndense_4 (Dense) (None, 64) 5312 \n_________________________________________________________________\ndense_5 (Dense) (None, 32) 2080 \n_________________________________________________________________\ndense_6 (Dense) (None, 64) 2112 \n_________________________________________________________________\ndense_7 (Dense) (None, 1) 65 \n=================================================================\nTotal params: 9,734\nTrainable params: 9,569\nNon-trainable params: 165\n_________________________________________________________________\n"
],
[
"preds = unnormalize_labels(linear_model.predict(test_features))\r\nplot_true_vs_pred(test_labels, preds, limit=.001)",
"_____no_output_____"
]
],
[
[
"## XG-Boost",
"_____no_output_____"
]
],
[
[
"preds = regressor.predict(test_features)\r\nplot_true_vs_pred(test_labels, preds, limit=.15)",
"_____no_output_____"
],
[
"preds",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4ae72caafff0f20e73996986fc1685d74a58924e
| 363,847 |
ipynb
|
Jupyter Notebook
|
Hurst_Exponent/HS_blogPost.ipynb
|
delectaGit/BlogPosts
|
209f22e30405ff43665a8d898ed963e66f58e10a
|
[
"MIT"
] | null | null | null |
Hurst_Exponent/HS_blogPost.ipynb
|
delectaGit/BlogPosts
|
209f22e30405ff43665a8d898ed963e66f58e10a
|
[
"MIT"
] | null | null | null |
Hurst_Exponent/HS_blogPost.ipynb
|
delectaGit/BlogPosts
|
209f22e30405ff43665a8d898ed963e66f58e10a
|
[
"MIT"
] | null | null | null | 318.884312 | 50,168 | 0.91298 |
[
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np\nimport glob\nimport pickle as pkl\nfrom scipy import stats\nimport random \nimport time",
"_____no_output_____"
],
[
"import utility_funcs as uf",
"_____no_output_____"
]
],
[
[
"### The following Hurst function was taken in part from <a href = \"https://www.quantstart.com/articles/Basics-of-Statistical-Mean-Reversion-Testing\">here</a>",
"_____no_output_____"
]
],
[
[
"def hurst(p):\n '''\n Description:\n Given an iterable (p), this functions calculates the Hurst exponent\n by sampling from the linear space\n Inputs:\n p: an iterable\n Outputs:\n the Hurst exponent\n '''\n # find variances for different sets of price differences:\n p = np.array(p)\n tau = np.arange(2,100)\n variancetau = [np.var(np.subtract(p[lag:], p[:-lag])) for lag in tau]\n\n # find the slope of the fitting line in the log-log plane:\n tau = np.log(tau)\n variancetau = np.log(variancetau)\n # find and remove mean:\n xb = np.mean(tau)\n yb = np.mean(variancetau)\n tau -= xb\n variancetau -= yb\n # find the slope:\n m = np.dot(tau, variancetau) / np.dot(tau, tau)\n\n return m / 2",
"_____no_output_____"
],
[
"def add_cur_name(df,cur_name):\n df[\"cur_name\"] = cur_name\n print(cur_name,\"done!\")\n\ndef remove_old_days(df,yr='2018'):\n cond = df.Date > yr+\"-01-01\"\n df = df[cond].copy()\n return df\ndef func_collection(df,cur_name,yr=\"2018\"):\n df = remove_old_days(df,yr)\n add_cur_name(df,cur_name)\n \n return df",
"_____no_output_____"
],
[
"def gaussian(x, mu, sig):\n return np.exp(-np.power(x - mu, 2.) / (2 * np.power(sig, 2.)))",
"_____no_output_____"
],
[
"def rnd_walk_simulator(sigma = 1, candle_bundle = 100, num_bundles = 200, initial = 1,\\\n generator = 'normal', seed = None):\n '''\n Description:\n Generates random-walks of various size, and puts them in a pandas dataframe, in a column\n named 'close'\n Inputs:\n sigma: the scale to be used for each step\n candle_bundle: the number of samples to bundle together\n num_bundles: the total random-walk length\n initial: the initial value to use, first element of the random-walk\n '''\n df = pd.DataFrame()\n close_var = initial \n close_list = []\n np.random.seed(seed)\n for x in range(num_bundles):\n tick_data = []\n if generator == 'normal':\n rnd = np.random.normal(loc=0.0, scale=sigma, size = candle_bundle)\n close_var += np.sum(rnd)\n elif generator == 'uniform':\n rnd = np.random.uniform(low=0, high= 1, size = candle_bundle)\n close_var += np.sum((rnd - 0.5)*sigma)\n elif generator == 'poisson':\n rnd = np.random.poisson(lam = 1, size = candle_bundle)\n close_var += np.sum((rnd - 1)*sigma)\n \n close_list.append(close_var) \n \n df[\"close\"] = close_list\n \n return df",
"_____no_output_____"
],
[
"file_list = glob.glob(\"./data/*\")\nfile_dict = {f:f.split(\"/\")[-1][:-4] for f in file_list}\n \nprint(file_list)",
"['./data/NZD_CHF.csv', './data/GBP_USD.csv']\n"
],
[
"df = uf.read_many_files(file_list,add_to_each_df_func=lambda df,x: func_collection(df,x,yr=\"2017\"),\\\n func_args=file_dict)\n\ndf = df.dropna(axis = 0)\ndf.head()",
"NZD_CHF done!\nGBP_USD done!\n"
],
[
"cond = df.cur_name == \"GBP_USD\"\nprint(hurst(df[cond].close ))\ndf[cond].close.plot()\n\nframe = plt.gca()\nframe.axes.get_xaxis().set_ticks([])\nframe.axes.get_yaxis().set_visible(False)\nplt.xlabel('Example A',fontsize = 14)",
"0.4919592117920463\n"
],
[
"cond = df.cur_name == \"NZD_CHF\"\nprint(hurst(df[cond].close))\ndf[cond].close.plot()\n\nframe = plt.gca()\nframe.axes.get_xaxis().set_ticks([])\nframe.axes.get_yaxis().set_visible(False)\nplt.xlabel('Example B',fontsize = 14)",
"0.466222952573976\n"
],
[
"df_rnd1 = rnd_walk_simulator(seed=10, sigma= 0.00005, num_bundles=300000)\nprint(hurst(df_rnd1.close))\ndf_rnd1.close.plot()\n\nframe = plt.gca()\nframe.axes.get_xaxis().set_ticks([])\nframe.axes.get_yaxis().set_visible(False)\nplt.xlabel('Example C',fontsize = 14)",
"0.49815925500399966\n"
],
[
"df_rnd2 = rnd_walk_simulator(seed=100, sigma= 0.00005, num_bundles=300000)\nprint(hurst(df_rnd2.close))\ndf_rnd2.close.plot()\n\nframe = plt.gca()\nframe.axes.get_xaxis().set_ticks([])\nframe.axes.get_yaxis().set_visible(False)\nplt.xlabel('Example D',fontsize = 14)",
"0.49433492766978543\n"
]
],
[
[
"# Hurst exponent for Forex market:\n### For a nice post on Hurst exponent and its indications look at <a href = \"http://epchan.blogspot.com/2016/04/mean-reversion-momentum-and-volatility.html\">here</a>.",
"_____no_output_____"
],
[
"## all data:",
"_____no_output_____"
]
],
[
[
"for pair in df.cur_name.unique():\n cond = df.cur_name == pair\n hs = hurst(df[cond].close)\n print(\"Hurst for %s is %.5f\"%(pair,hs),end = ' , ')\n print(\"total len of the df is:\",len(df[cond]))",
"Hurst for NZD_CHF is 0.46622 , total len of the df is: 347646\nHurst for GBP_USD is 0.49196 , total len of the df is: 343709\n"
]
],
[
[
"# Random Walks:",
"_____no_output_____"
],
[
"### rnd_steps = 10000:",
"_____no_output_____"
],
[
"### normal:",
"_____no_output_____"
]
],
[
[
"hurst_li10n = []\nst = time.time()\nfor ii in range(10000):\n df_norm = rnd_walk_simulator(sigma = 0.002,\\\n candle_bundle=1,\\\n num_bundles = 10000,\\\n seed = ii,\\\n generator='normal')\n \n hs = hurst(df_norm.close.values)\n hurst_li10n.append(hs)\n if ii%500 == 0:\n print(\"%d done, time= %.4f\"%(ii,time.time()-st),end=\", \")\n st = time.time()\n",
"0 done, time= 0.0962, 500 done, time= 34.9130, 1000 done, time= 33.4250, 1500 done, time= 33.4468, 2000 done, time= 33.3330, 2500 done, time= 33.3497, 3000 done, time= 33.4137, 3500 done, time= 33.3155, 4000 done, time= 33.4977, 4500 done, time= 33.4594, 5000 done, time= 33.7591, 5500 done, time= 33.3843, 6000 done, time= 33.4233, 6500 done, time= 33.3005, 7000 done, time= 33.3244, 7500 done, time= 34.0827, 8000 done, time= 33.7646, 8500 done, time= 34.2802, 9000 done, time= 33.2515, 9500 done, time= 33.5301, "
],
[
"pkl.dump(hurst_li10n,open(\"./hurst_li10_n.pkl\",\"wb\"))",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,8))\nprint(np.mean(hurst_li10n),np.std(hurst_li10n) )\na = plt.hist(hurst_li10n,bins=30,normed=True)\n\nx_range = np.arange(0.42,0.58,0.002)\namp = np.max(a[0])\nplt.plot(x_range, amp*gaussian(x_range,np.mean(hurst_li10n),np.std(hurst_li10n)),'r')\n\nplt.text(0.412,amp,\"Random-Walk Length = 10000\",fontsize = 20)\nplt.text(0.412,amp-1.5,\"mean = \"+'{0:.4f}'.format(np.mean(hurst_li10n)),fontsize = 20)\nplt.text(0.412,amp-3,\"std = \"+'{0:.4f}'.format(np.std(hurst_li10n)),fontsize = 20)\nplt.xlabel(\"Hurst Exponent\",fontsize=18)\nplt.ylabel(\"frequency\",fontsize=18)\nplt.xticks(fontsize=16)\nplt.yticks(fontsize=16)\nplt.xlim(0.41,0.58)",
"0.4973285518801711 0.019287160134145304\n"
]
],
[
[
"### uniform:",
"_____no_output_____"
]
],
[
[
"hurst_li10u = []\nst = time.time()\nfor ii in range(10000):\n df_norm = rnd_walk_simulator(sigma = 0.002,\\\n candle_bundle=1,\\\n num_bundles = 10000,\\\n seed = ii,\\\n generator='uniform')\n \n hs = hurst(df_norm.close.values)\n hurst_li10u.append(hs)\n if ii%500 == 0:\n print(\"%d done, time= %.4f\"%(ii,time.time()-st),end=\", \")\n st = time.time()",
"0 done, time= 0.1095, 500 done, time= 42.9416, 1000 done, time= 43.0851, 1500 done, time= 39.1965, 2000 done, time= 39.0544, 2500 done, time= 39.1885, 3000 done, time= 38.9128, 3500 done, time= 38.9099, 4000 done, time= 38.9808, 4500 done, time= 38.8322, 5000 done, time= 39.4153, 5500 done, time= 38.7849, 6000 done, time= 38.5917, 6500 done, time= 38.7031, 7000 done, time= 38.5378, 7500 done, time= 38.5613, 8000 done, time= 38.5767, 8500 done, time= 38.5169, 9000 done, time= 38.4373, 9500 done, time= 38.6781, "
],
[
"pkl.dump(hurst_li10u,open(\"./hurst_li10_u.pkl\",\"wb\"))",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,8))\nprint(np.mean(hurst_li10u),np.std(hurst_li10u) )\na = plt.hist(hurst_li10u,bins=25,normed=True)\n\nx_range = np.arange(0.42,0.58,0.002)\namp = np.max(a[0])\nplt.plot(x_range, amp*gaussian(x_range,np.mean(hurst_li10u),np.std(hurst_li10u)),'r')\n\nplt.text(0.412,amp,\"Random-Walk Length = 10000\",fontsize = 19)\nplt.text(0.412,amp-1.5,\"mean = \"+'{0:.4f}'.format(np.mean(hurst_li10u)),fontsize = 19)\nplt.text(0.412,amp-3,\"std = \"+'{0:.4f}'.format(np.std(hurst_li10u)),fontsize = 19)\nplt.xlabel(\"Hurst Exponent\",fontsize=18)\nplt.ylabel(\"frequency\",fontsize=18)\nplt.xticks(fontsize=16)\nplt.yticks(fontsize=16)\nplt.xlim(0.41,0.58)",
"0.4975865490974271 0.01920499738172972\n"
]
],
[
[
"### Poisson:",
"_____no_output_____"
]
],
[
[
"hurst_li10p = []\nst = time.time()\nfor ii in range(10000):\n df_norm = rnd_walk_simulator(sigma = 0.002,\\\n candle_bundle=1,\\\n num_bundles = 10000,\\\n seed = ii,\\\n generator='poisson')\n \n hs = hurst(df_norm.close.values)\n hurst_li10p.append(hs)\n if ii%500 == 0:\n print(\"%d done, time= %.4f\"%(ii,time.time()-st),end=\", \")\n st = time.time()",
"0 done, time= 0.1370, 500 done, time= 42.1722, 1000 done, time= 43.8220, 1500 done, time= 42.4492, 2000 done, time= 42.4437, 2500 done, time= 42.3429, 3000 done, time= 43.9027, 3500 done, time= 42.2558, 4000 done, time= 42.3798, 4500 done, time= 42.1131, 5000 done, time= 42.0931, 5500 done, time= 42.0448, 6000 done, time= 42.1390, 6500 done, time= 42.2498, 7000 done, time= 42.1194, 7500 done, time= 42.1572, 8000 done, time= 42.3313, 8500 done, time= 42.5118, 9000 done, time= 42.0514, 9500 done, time= 42.1852, "
],
[
"pkl.dump(hurst_li10p,open(\"./hurst_li10_p.pkl\",\"wb\"))",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,8))\nprint(np.mean(hurst_li10p),np.std(hurst_li10p) )\na = plt.hist(hurst_li10p,bins=25,normed=True)\n\nx_range = np.arange(0.42,0.58,0.002)\namp = np.max(a[0])\nplt.plot(x_range, amp*gaussian(x_range,np.mean(hurst_li10p),np.std(hurst_li10p)),'r')\n\nplt.text(0.412,amp,\"Random-Walk Length = 10000\",fontsize = 19)\nplt.text(0.412,amp-1.5,\"mean = \"+'{0:.4f}'.format(np.mean(hurst_li10p)),fontsize = 19)\nplt.text(0.412,amp-3,\"std = \"+'{0:.4f}'.format(np.std(hurst_li10p)),fontsize = 19)\nplt.xlabel(\"Hurst Exponent\",fontsize=18)\nplt.ylabel(\"frequency\",fontsize=18)\nplt.xticks(fontsize=16)\nplt.yticks(fontsize=16)\nplt.xlim(0.41,0.58)",
"0.49761077945781906 0.019397083706689088\n"
]
],
[
[
"### rnd_steps = 100000:",
"_____no_output_____"
],
[
"### normal:",
"_____no_output_____"
]
],
[
[
"hurst_li100 = []\nst = time.time()\nfor ii in range(10000):\n df_norm = rnd_walk_simulator(sigma = 0.002,\\\n candle_bundle=1,\\\n num_bundles = 100000,\\\n seed = ii,\\\n generator='normal')\n \n hs = hurst(df_norm.close.values)\n hurst_li100.append(hs)\n if ii%500 == 0:\n print(\"%d done, time= %.4f\"%(ii,time.time()-st),end=\", \")\n st = time.time()",
"0 done, time= 0.6589, 500 done, time= 360.5264, 1000 done, time= 369.0569, 1500 done, time= 381.9417, 2000 done, time= 375.6941, 2500 done, time= 352.2228, 3000 done, time= 360.5117, 3500 done, time= 377.1919, 4000 done, time= 382.5946, 4500 done, time= 347.6013, 5000 done, time= 344.6007, 5500 done, time= 346.9138, 6000 done, time= 352.7429, 6500 done, time= 353.7730, 7000 done, time= 350.1097, 7500 done, time= 351.7248, 8000 done, time= 348.2312, 8500 done, time= 349.3438, 9000 done, time= 350.8850, 9500 done, time= 351.5927, "
],
[
"hurst_li100 = pkl.load(open(\"./hurst_li100.pkl\",\"rb\"))",
"_____no_output_____"
],
[
"plt.figure(figsize=(12,8))\nprint(np.mean(hurst_li100),np.std(hurst_li100) )\na = plt.hist(hurst_li100,bins=30,normed=True)\n\nx_range = np.arange(0.47,0.53,0.0005)\namp = np.max(a[0])\nplt.plot(x_range, amp*gaussian(x_range,np.mean(hurst_li100),np.std(hurst_li100)),'r')\n\nplt.text(0.4755,amp,\"Random-Walk Length = 100000\",fontsize = 20)\nplt.text(0.4755,amp-5,\"mean = \"+'{0:.4f}'.format(np.mean(hurst_li100)),fontsize = 20)\nplt.text(0.4755,amp-10,\"std = \"+'{0:.4f}'.format(np.std(hurst_li100)),fontsize = 20)\nplt.xlabel(\"Hurst Exponent\",fontsize=18)\nplt.ylabel(\"frequency\",fontsize=18)\nplt.xticks(fontsize=16)\nplt.yticks(fontsize=16)\nplt.xlim(0.475,0.525)",
"0.4996890151279655 0.0060831918456582465\n"
],
[
"print(stats.skew(hurst_li100))\nprint(stats.kurtosis(hurst_li100))",
"-0.023029401278034163\n0.0022576095483266556\n"
],
[
"pkl.dump(hurst_li100,open(\"./hurst_li100.pkl\",\"wb\"))",
"_____no_output_____"
]
],
[
[
"### uniform:",
"_____no_output_____"
]
],
[
[
"hurst_li100u = []\nst = time.time()\nfor ii in range(10000):\n df_norm = rnd_walk_simulator(sigma = 0.002,\\\n candle_bundle=1,\\\n num_bundles = 100000,\\\n seed = ii,\\\n generator='uniform')\n \n hs = hurst(df_norm.close.values)\n hurst_li100u.append(hs)\n if ii%500 == 0:\n print(\"%d done, time= %.4f\"%(ii,time.time()-st),end=\", \")\n st = time.time()",
"0 done, time= 0.7648, 500 done, time= 3415.9947, 1000 done, time= 367.2228, 1500 done, time= 366.4329, 2000 done, time= 367.1245, 2500 done, time= 368.8297, 3000 done, time= 370.5546, 3500 done, time= 369.9341, 4000 done, time= 371.6526, 4500 done, time= 416.5966, 5000 done, time= 465.9186, 5500 done, time= 436.6161, 6000 done, time= 445.7589, 6500 done, time= 445.5698, 7000 done, time= 443.8797, 7500 done, time= 425.7276, 8000 done, time= 427.9445, 8500 done, time= 419.9981, 9000 done, time= 388.9699, 9500 done, time= 384.5860, "
],
[
"plt.figure(figsize=(12,8))\nprint(np.mean(hurst_li100u),np.std(hurst_li100u) )\na = plt.hist(hurst_li100u,bins=30,normed=True)\n\nx_range = np.arange(0.47,0.53,0.0005)\namp = np.max(a[0])\nplt.plot(x_range, amp*gaussian(x_range,np.mean(hurst_li100u),np.std(hurst_li100u)),'r')\n\nplt.text(0.4755,amp,\"Random-Walk Length = 10000\",fontsize = 20)\nplt.text(0.4755,amp-5,\"mean = \"+'{0:.4f}'.format(np.mean(hurst_li100u)),fontsize = 20)\nplt.text(0.4755,amp-10,\"std = \"+'{0:.4f}'.format(np.std(hurst_li100u)),fontsize = 20)\nplt.xlabel(\"Hurst Exponent\",fontsize=18)\nplt.ylabel(\"frequency\",fontsize=18)\nplt.xticks(fontsize=16)\nplt.yticks(fontsize=16)\nplt.xlim(0.475,0.525)",
"0.49970485614083465 0.006022231078746345\n"
],
[
"pkl.dump(hurst_li100u,open(\"./hurst_li100u.pkl\",\"wb\"))",
"_____no_output_____"
]
],
[
[
"### rnd_steps = 300000:",
"_____no_output_____"
]
],
[
[
"hurst_li300 = []\nst = time.time()\nfor ii in range(1000):\n df_norm = rnd_walk_simulator(sigma = 0.002,\\\n num_bundles = 300000,\\\n seed = ii)\n \n hs = hurst(df_norm.close.values)\n hurst_li300.append(hs)\n if ii%100 == 0:\n print(\"%d done, time= %.4f\"%(ii,time.time()-st),end=\", \")\n st = time.time()",
"0 done, time= 3.3026, 100 done, time= 363.5886, 200 done, time= 370.8800, 300 done, time= 344.2197, 400 done, time= 349.9841, 500 done, time= 354.0914, 600 done, time= 339.4433, 700 done, time= 325.0656, 800 done, time= 344.1690, 900 done, time= 332.2664, "
],
[
"plt.figure(figsize=(12,8))\nprint(np.mean(hurst_li300),np.std(hurst_li300) )\n_ = plt.hist(hurst_li300,bins=12)\n\nplt.text(0.486,200,\"Random-Walk Length = 300000\",fontsize = 15)\nplt.text(0.486,180,\"mean = \"+'{0:.4f}'.format(np.mean(hurst_li300)),fontsize = 15)\nplt.text(0.486,160,\"std = \"+'{0:.4f}'.format(np.std(hurst_li300)),fontsize = 15)\nplt.xlabel(\"Hurst Exponent\",fontsize=14)\nplt.ylabel(\"frequency\",fontsize=14)\nplt.xticks(fontsize=13)\nplt.yticks(fontsize=13)",
"0.4996838765361534 0.003520648542656769\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4ae742a95bebb317ed32ff6f503192386b481a9f
| 20,424 |
ipynb
|
Jupyter Notebook
|
code/processing/2021-02-12_22-13-24/_run_jnb/.ipynb_checkpoints/2021-02-12_22-13-24_Or179_Or177_overnight-output-checkpoint.ipynb
|
hamdanspam/LoisLFP
|
c328f3734bf8b700961e9c48e2f9c172b5df86f0
|
[
"MIT"
] | null | null | null |
code/processing/2021-02-12_22-13-24/_run_jnb/.ipynb_checkpoints/2021-02-12_22-13-24_Or179_Or177_overnight-output-checkpoint.ipynb
|
hamdanspam/LoisLFP
|
c328f3734bf8b700961e9c48e2f9c172b5df86f0
|
[
"MIT"
] | null | null | null |
code/processing/2021-02-12_22-13-24/_run_jnb/.ipynb_checkpoints/2021-02-12_22-13-24_Or179_Or177_overnight-output-checkpoint.ipynb
|
hamdanspam/LoisLFP
|
c328f3734bf8b700961e9c48e2f9c172b5df86f0
|
[
"MIT"
] | null | null | null | 33.155844 | 157 | 0.534763 |
[
[
[
"import os\n\nimport numpy as np\nimport pandas as pd\nimport spikeextractors as se\nimport spiketoolkit as st\nimport spikewidgets as sw\n\nimport tqdm.notebook as tqdm\nfrom scipy.signal import periodogram, spectrogram\n\nimport matplotlib.pyplot as plt\n# %matplotlib inline\n# %config InlineBackend.figure_format='retina'\n\nimport holoviews as hv\nimport holoviews.operation.datashader\nimport holoviews.operation.timeseries\n\n\nhv.extension(\"bokeh\")\n\nimport panel as pn\nimport panel.widgets as pnw\n\npn.extension()\n\nfrom LoisLFPutils.utils import *",
"_____no_output_____"
],
[
"# Path to the data folder in the repo\n\ndata_path = r\"\"\n\n\n\n\n# !!! start assign jupyter notebook parameter(s) !!!\n\ndata_path = '2021-02-12_22-13-24_Or179_Or177_overnight'\n\n# !!! end assign jupyter notebook parameter(s) !!!",
"_____no_output_____"
],
[
"data_path = os.path.join('../../../../data/',data_path)\n\n# Path to the raw data in the hard drive\nwith open(os.path.normpath(os.path.join(data_path, 'LFP_location.txt'))) as f: \n OE_data_path = f.read()",
"_____no_output_____"
]
],
[
[
"### Get each bird's recording, and their microphone channels",
"_____no_output_____"
]
],
[
[
"# This needs to be less repetitive\nif 'Or177' in data_path:\n # Whole recording from the hard drive\n recording = se.BinDatRecordingExtractor(OE_data_path,30000,40, dtype='int16')\n\n # Note I am adding relevant ADC channels\n # First bird\n Or179_recording = se.SubRecordingExtractor(\n recording,\n channel_ids=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,11,12,13,14,15, 32])\n\n # Second bird\n Or177_recording = se.SubRecordingExtractor(\n recording,\n channel_ids=[16, 17,18,19,20,21,22,23,24,25,26,27,28,29,30,31, 33])\n\n # Bandpass fiter microphone recoridngs\n mic_recording = st.preprocessing.bandpass_filter(\n se.SubRecordingExtractor(recording,channel_ids=[32,33]),\n freq_min=500,\n freq_max=1400\n )\nelse:\n # Whole recording from the hard drive\n recording = se.BinDatRecordingExtractor(OE_data_path, 30000, 24, dtype='int16')\n\n # Note I am adding relevant ADC channels\n # First bird\n Or179_recording = se.SubRecordingExtractor(\n recording,\n channel_ids=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10,11,12,13,14,15,16])\n\n # Bandpass fiter microphone recoridngs\n mic_recording = st.preprocessing.bandpass_filter(\n se.SubRecordingExtractor(recording,channel_ids=[16]),\n freq_min=500,\n freq_max=1400\n )",
"_____no_output_____"
],
[
"# Get wav files\nwav_names = [file_name for file_name in os.listdir(data_path) if file_name.endswith('.wav')]\nwav_paths = [os.path.join(data_path,wav_name) for wav_name in wav_names]\n\n# Get tranges for wav files in the actual recording\n# OE_data_path actually contains the path all the way to the .bin. We just need the parent directory\n# with the timestamp.\n\n# Split up the path\nOE_data_path_split= OE_data_path.split(os.sep)\n\n# Take only the first three. os.path is weird so we manually add the separator after the \n# drive name.\nOE_parent_path = os.path.join(OE_data_path_split[0] + os.sep, *OE_data_path_split[1:3])\n\n# Get all time ranges given the custom offset.\ntranges=np.array([\n get_trange(OE_parent_path, path, offset=datetime.timedelta(seconds=0), duration=3) \n for path in wav_paths])\n",
"_____no_output_____"
],
[
"wav_df = pd.DataFrame({'wav_paths':wav_paths, 'wav_names':wav_names, 'trange0':tranges[:, 0], 'trange1':tranges[:, 1]})\nwav_df.head()",
"_____no_output_____"
]
],
[
[
"Connect the wav files to the recording. Manually input to gut check yourself. If it is before 2021 02 21 at 11:00 am PST, you need to add a time delay.",
"_____no_output_____"
]
],
[
[
"wav_f,_,_,_=wav_df.loc[0,:]\nwav_f, data_path\ndatetime.datetime(2021,2,23,8,11,1) - datetime.datetime(2021, 2, 22,22,0,20) ",
"_____no_output_____"
],
[
"paths, name, tr0, tr1 = wav_df.loc[0,:]\nsw.plot_spectrogram(mic_recording, trange= [tr0,tr1+10], freqrange=[300,4000], nfft=2**10, channel=32)",
"_____no_output_____"
],
[
"np.linspace(0,130,14)",
"_____no_output_____"
],
[
"# Set up widgets\nwav_selector = pnw.Select(options=[(i, name) for i, name in enumerate(wav_df.wav_names.values)], name=\"Select song file\")\n# offset_selector = pnw.Select(options=np.linspace(-10,10,21).tolist(), name=\"Select offset\")\nwindow_radius_selector = pnw.Select(options=[10,20,30,40,60], name=\"Select window radius\")\nspect_chan_selector = pnw.Select(options=list(range(16)), name=\"Spectrogram channel\")\nspect_freq_lo = pnw.Select(options=np.linspace(0,130,14).tolist(), name=\"Low frequency for spectrogram (Hz)\")\nspect_freq_hi = pnw.Select(options=np.linspace(130,0,14).tolist(), name=\"Hi frequency for spectrogram (Hz)\")\nlog_nfft_selector = pnw.Select(options=np.linspace(10,16,7).tolist(), value=14, name=\"magnitude of nfft (starts at 256)\")\n\[email protected](\n wav_selector=wav_selector.param.value,\n# offset=offset_selector.param.value,\n window_radius=window_radius_selector.param.value,\n spect_chan=spect_chan_selector.param.value,\n spect_freq_lo=spect_freq_lo.param.value,\n spect_freq_hi=spect_freq_hi.param.value,\n log_nfft=log_nfft_selector.param.value\n)\ndef create_figure(wav_selector, \n# offset,\n window_radius, spect_chan,\n spect_freq_lo, spect_freq_hi, log_nfft):\n \n # Each column in each row to a tuple that we unpack\n wav_file_path, wav_file_name, tr0, tr1 = wav_df.loc[wav_selector[0],:]\n \n # Set up figure\n fig,axes = plt.subplots(4,1, figsize=(16,12))\n \n # Get wav file numpy recording object\n wav_recording = get_wav_recording(wav_file_path)\n \n # Apply offset and apply window radius\n offset = 0\n tr0 = tr0+ offset-window_radius\n # Add duration of wav file\n tr1 = tr1+ offset+window_radius+wav_recording.get_num_frames()/wav_recording.get_sampling_frequency()\n \n \n '''Plot sound spectrogram (Hi fi mic)'''\n sw.plot_spectrogram(wav_recording, channel=0, freqrange=[300,14000],ax=axes[0])\n axes[0].set_title('Hi fi mic spectrogram')\n \n '''Plot sound spectrogram (Lo fi mic)''' \n if 'Or179' in wav_file_name:\n LFP_recording = Or179_recording\n elif 'Or177' in wav_file_name:\n LFP_recording = Or177_recording\n \n mic_channel = LFP_recording.get_channel_ids()[-1]\n \n sw.plot_spectrogram(\n mic_recording,\n mic_channel,\n trange=[tr0, tr1],\n freqrange=[600,4000],\n ax=axes[1]\n )\n \n axes[1].set_title('Lo fi mic spectrogram')\n \n '''Plot LFP timeseries'''\n chan_ids = np.array([LFP_recording.get_channel_ids()]).flatten()\n sw.plot_timeseries(\n LFP_recording,\n channel_ids=[chan_ids[spect_chan]],\n trange=[tr0, tr1],\n ax=axes[2]\n )\n axes[2].set_title('Raw LFP')\n \n # Clean lines\n for line in plt.gca().lines:\n line.set_linewidth(0.5) \n \n '''Plot LFP spectrogram'''\n sw.plot_spectrogram(\n LFP_recording,\n channel=chan_ids[spect_chan],\n freqrange=[spect_freq_lo,spect_freq_hi],\n trange=[tr0, tr1],\n ax=axes[3],\n nfft=int(2**log_nfft)\n )\n axes[3].set_title('LFP')\n \n for i, ax in enumerate(axes):\n ax.set_yticks([ax.get_ylim()[1]])\n ax.set_yticklabels([ax.get_ylim()[1]])\n ax.set_xlabel('')\n \n # Show 30 Hz\n ax.set_yticks([30, ax.get_ylim()[1]])\n ax.set_yticklabels([30, ax.get_ylim()[1]])\n \n \n return fig\n\ndash = pn.Column(\n pn.Row(wav_selector, window_radius_selector,spect_chan_selector),\n pn.Row(spect_freq_lo,spect_freq_hi,log_nfft_selector),\n create_figure\n);\n\ndash",
"_____no_output_____"
]
],
[
[
"## Looking at all channels at a time:",
"_____no_output_____"
]
],
[
[
"# Make chanmap\nchanmap=np.array([[3, 7, 11, 15],[2, 4, 10, 14],[4, 8, 12, 16],[1, 5, 9, 13]])\n\n# Set up widgets\nwav_selector = pnw.Select(options=[(i, name) for i, name in enumerate(wav_df.wav_names.values)], name=\"Select song file\")\nwindow_radius_selector = pnw.Select(options=[10,20,30,40,60], name=\"Select window radius\")\nspect_freq_lo = pnw.Select(options=np.linspace(0,130,14).tolist(), name=\"Low frequency for spectrogram (Hz)\")\nspect_freq_hi = pnw.Select(options=np.linspace(130,0,14).tolist(), name=\"Hi frequency for spectrogram (Hz)\")\nlog_nfft_selector = pnw.Select(options=np.linspace(10,16,7).tolist(),value=14, name=\"magnitude of nfft (starts at 256)\")\n\ndef housekeeping(wav_selector, window_radius):\n # Each column in each row to a tuple that we unpack\n wav_file_path, wav_file_name, tr0, tr1 = wav_df.loc[wav_selector[0],:]\n \n # Get wav file numpy recording object\n wav_recording = get_wav_recording(wav_file_path)\n \n # Apply offset and apply window radius\n offset = 0\n tr0 = tr0+ offset-window_radius\n \n # Add duration of wav file\n tr1 = tr1+ offset+window_radius+wav_recording.get_num_frames()/wav_recording.get_sampling_frequency()\n \n return wav_recording, tr0, tr1\n \[email protected](\n wav_selector=wav_selector.param.value,\n window_radius=window_radius_selector.param.value)\ndef create_sound_figure(wav_selector, window_radius):\n \n # Housekeeping\n wav_recording, tr0, tr1 = housekeeping(wav_selector, window_radius)\n\n # Set up figure for sound\n fig,axes = plt.subplots(1,2, figsize=(16,2))\n \n '''Plot sound spectrogram (Hi fi mic)'''\n sw.plot_spectrogram(wav_recording, channel=0, freqrange=[300,14000], ax=axes[0])\n axes[0].set_title('Hi fi mic spectrogram')\n \n '''Plot sound spectrogram (Lo fi mic)''' \n if 'Or179' in wav_file_name:\n LFP_recording = Or179_recording\n elif 'Or177' in wav_file_name:\n LFP_recording = Or177_recording\n \n mic_channel = LFP_recording.get_channel_ids()[-1]\n \n sw.plot_spectrogram(\n mic_recording,\n mic_channel,\n trange=[tr0, tr1],\n freqrange=[600,4000],\n ax=axes[1]\n )\n \n axes[1].set_title('Lo fi mic spectrogram')\n for ax in axes:\n ax.axis('off')\n \n return fig\n\[email protected](\n wav_selector=wav_selector.param.value,\n window_radius=window_radius_selector.param.value,\n spect_freq_lo=spect_freq_lo.param.value,\n spect_freq_hi=spect_freq_hi.param.value,\n log_nfft=log_nfft_selector.param.value\n)\ndef create_LFP_figure(wav_selector, window_radius,\n spect_freq_lo, spect_freq_hi, log_nfft):\n # Housekeeping\n wav_recording, tr0, tr1 = housekeeping(wav_selector, window_radius)\n \n fig,axes=plt.subplots(4,4,figsize=(16,8))\n \n '''Plot LFP'''\n for i in range(axes.shape[0]):\n for j in range(axes.shape[1]):\n ax = axes[i][j]\n sw.plot_spectrogram(recording, chanmap[i][j], trange=[tr0, tr1],\n freqrange=[spect_freq_lo,spect_freq_hi],\n nfft=int(2**log_nfft), ax=ax, cmap='magma')\n ax.axis('off')\n # Set channel as title\n ax.set_title(chanmap[i][j])\n \n # Clean up\n for i in range(axes.shape[0]):\n for j in range(axes.shape[1]):\n ax=axes[i][j]\n \n ax.set_yticks([ax.get_ylim()[1]])\n ax.set_yticklabels([ax.get_ylim()[1]])\n ax.set_xlabel('')\n\n # Show 30 Hz\n ax.set_yticks([30, ax.get_ylim()[1]])\n ax.set_yticklabels([30, ax.get_ylim()[1]])\n \n \n return fig\n\ndash = pn.Column(\n pn.Row(wav_selector,window_radius_selector),\n pn.Row(spect_freq_lo,spect_freq_hi,log_nfft_selector),\n create_sound_figure, create_LFP_figure\n);",
"_____no_output_____"
]
],
[
[
"# Sleep data analysis!",
"_____no_output_____"
]
],
[
[
"csvs = [os.path.normpath(os.path.join(data_path,file)) for file in os.listdir(data_path) if file.endswith('.csv')] \ncsvs",
"_____no_output_____"
],
[
"csv = csvs[0]\ndf = pd.read_csv(csv)\ndel df['Unnamed: 0']\ndf.head()",
"_____no_output_____"
],
[
"csv_name = csv.split(os.sep)[-1]\nrec=None\nif 'Or179' in csv_name:\n rec = Or179_recording\nelif 'Or177' in csv_name:\n rec = Or177_recording\n\n# Get second to last element in split\nchannel = int(csv_name.split('_')[-2])",
"_____no_output_____"
],
[
"window_slider = pn.widgets.DiscreteSlider(\n name='window size',\n options=[*range(1,1000)],\n value=1\n)\n\nfreq_slider_1 = pn.widgets.DiscreteSlider(\n name='f (Hz)',\n options=[*range(1,200)],\n value=30\n)\n\nfreq_slider_2 = pn.widgets.DiscreteSlider(\n name='f (Hz)',\n options=[*range(1,200)],\n value=10\n)\n\nfreq_slider_3 = pn.widgets.DiscreteSlider(\n name='f (Hz)',\n options=[*range(1,200)],\n value=4\n)\n\nrange_slider = pn.widgets.RangeSlider(\n start=0,\n end=df.t.max(),\n step=10,\n value=(0, 500),\n name=\"Time range\",\n value_throttled=(0,500)\n)\n\[email protected](window=window_slider.param.value,\n freq_1=freq_slider_1.param.value,\n freq_2=freq_slider_2.param.value,\n freq_3=freq_slider_3.param.value,\n rang=range_slider.param.value_throttled)\ndef plot_ts(window, freq_1, freq_2, freq_3, rang):\n# subdf = df.loc[\n# ((df['f']==freq_1)|(df['f']==freq_2)|(df['f']==freq_3)) & (df['t'] < 37800),:]\n subdf = df.loc[\n ((df['f']==freq_1)|(df['f']==freq_2)|(df['f']==freq_3)) \n & ((df['t'] > rang[0]) & (df['t'] < rang[1])),:]\n \n return hv.operation.timeseries.rolling(\n hv.Curve(\n data = subdf,\n kdims=[\"t\", \"f\"],\n vdims=\"logpower\"\n ).groupby(\"f\").overlay().opts(width=1200, height=300),\n rolling_window=window\n )\n\[email protected](rang=range_slider.param.value_throttled)\ndef plot_raw_ts(rang):\n sr = rec.get_sampling_frequency()\n \n return hv.operation.datashader.datashade(\n hv.Curve(\n rec.get_traces(channel_ids=[channel], start_frame=sr*rang[0], end_frame=sr*rang[1]).flatten()\n ),\n aggregator=\"any\"\n ).opts(width=1200, height=300)\n \npn.Column(\n window_slider,freq_slider_1, freq_slider_2, freq_slider_3,range_slider,\n plot_ts,\n plot_raw_ts\n)",
"_____no_output_____"
]
],
[
[
"# TODOs:\n- Does phase vary systematically with frequency???\n- Does the log power increase with time over the nzight??\n- Observation: these birds start singing around 6, before the lights turn on.\n- Possibly add spikes for when song occurs \n- Possibly add timerange slider",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4ae749627dc36a3a356588d9956d55fd3cdf937b
| 69,896 |
ipynb
|
Jupyter Notebook
|
2_autompg_linearregression.ipynb
|
ginttone/test_visuallization
|
bd73af65bec070a42f89728dda4f1011b8130177
|
[
"Apache-2.0"
] | null | null | null |
2_autompg_linearregression.ipynb
|
ginttone/test_visuallization
|
bd73af65bec070a42f89728dda4f1011b8130177
|
[
"Apache-2.0"
] | null | null | null |
2_autompg_linearregression.ipynb
|
ginttone/test_visuallization
|
bd73af65bec070a42f89728dda4f1011b8130177
|
[
"Apache-2.0"
] | null | null | null | 31.857794 | 253 | 0.363826 |
[
[
[
"<a href=\"https://colab.research.google.com/github/ginttone/test_visuallization/blob/master/2_autompg_linearregression.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# 머신러닝\n\n- 정보(데이터)단계<br>\ndropna:info(), describe()<br>\nfillna, replace:describe(), value_counts()<br>\n - 시각화 :통계 선택<br>\nstandard scaler\n혹은 one hot encoding<br> 뭐로 할지 정하기\n\n- 교육단계<br>\nstadard scaler, get_dummies(one hot encoding)<br>\nmodel learning<br>\ncheck score<br>\n\n- 서비스단계 <br>\npickle: dump,load<br>\nrecive data <br>\napply prediction<br>",
"_____no_output_____"
],
[
"## 데이터 로딩\n\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf= pd.read_csv('./auto-mpg.csv', header=None)\ndf.columns=['mpg','cylinders','displacement','horsepower','weight',\n 'acceleration','model year','origin','name']\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 398 entries, 0 to 397\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 mpg 398 non-null float64\n 1 cylinders 398 non-null int64 \n 2 displacement 398 non-null float64\n 3 horsepower 398 non-null object \n 4 weight 398 non-null float64\n 5 acceleration 398 non-null float64\n 6 model year 398 non-null int64 \n 7 origin 398 non-null int64 \n 8 name 398 non-null object \ndtypes: float64(4), int64(3), object(2)\nmemory usage: 28.1+ KB\n"
],
[
"df[['horsepower','name']].describe(include='all')",
"_____no_output_____"
]
],
[
[
"## replace",
"_____no_output_____"
]
],
[
[
"df['horsepower'].value_counts()",
"_____no_output_____"
],
[
"df['horsepower'].unique()",
"_____no_output_____"
],
[
"df_horsepower=df['horsepower'].replace(to_replace='?',value=None,inplace=False)\ndf_horsepower.unique()",
"_____no_output_____"
],
[
"df_horsepower=df_horsepower.astype('float')",
"_____no_output_____"
],
[
"df_horsepower.mean()",
"_____no_output_____"
],
[
"df['horsepower']=df_horsepower.fillna(104)\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 398 entries, 0 to 397\nData columns (total 9 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 mpg 398 non-null float64\n 1 cylinders 398 non-null int64 \n 2 displacement 398 non-null float64\n 3 horsepower 398 non-null float64\n 4 weight 398 non-null float64\n 5 acceleration 398 non-null float64\n 6 model year 398 non-null int64 \n 7 origin 398 non-null int64 \n 8 name 398 non-null object \ndtypes: float64(5), int64(3), object(1)\nmemory usage: 28.1+ KB\n"
],
[
"\ndf['name'].unique()",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"## 분류와 연속 컬럼 구분",
"_____no_output_____"
]
],
[
[
"df.head(8)",
"_____no_output_____"
]
],
[
[
"### 컬럼 형태 분류(check columns)\n* 연속형: displacement, horsepower, weigth,acceleration, \n* 중립형: mpg,cylinders,origin\n* 분류형: model year, name\n\n(대게 카테고리형은 소숫점 및 값이 존재하지 않는다.)\n\nstandard scaler : 연속형<br>\none hot incoding : 분류형 \n\n하나의 데이터에 300개는 있어야 교육이된다\n그래서 name은 교육시키기에 카운터 수가 작아서 빼고 진행",
"_____no_output_____"
],
[
"#### 중립형을 연속형인지 분륳형인지 판단",
"_____no_output_____"
]
],
[
[
"df['name'].value_counts()",
"_____no_output_____"
],
[
"df['mpg'].describe(include='all')",
"_____no_output_____"
]
],
[
[
"예) count 398인데 mpg length가 129 값이 나왔다. 이때 분류형일까 연속형일까?\n\n* 왼쪽카테고리에는 소숫점 이하 값 있어 이럴땐 연속형으로 분류할수 있어/ 오른쪽 반복된카운트값",
"_____no_output_____"
]
],
[
[
"df['mpg'].value_counts()",
"_____no_output_____"
],
[
"df['cylinders'].describe()",
"_____no_output_____"
],
[
"df['cylinders'].value_counts()",
"_____no_output_____"
],
[
"df['origin'].describe()",
"_____no_output_____"
],
[
"df['origin'].value_counts()",
"_____no_output_____"
]
],
[
[
"* 연속형: displacement, horsepower, weigth,acceleration, mpg\n\n* 분류형: model year, name,cylinders,origin\n\nlabel:mpg<br>\nfeature: others without name",
"_____no_output_____"
],
[
"## 시각화: 통계",
"_____no_output_____"
],
[
"### 정규화 단계:\n * standard scaler<br>\nz = (x-u) / s <br>\nz(모집단)<br>\nx(샘플의 표준 점수)<br>\nu(훈련 샘플의 평균)<br>\ns(훈련 샘플의 표준 편차)<br>\n\n fit(X [, y, 샘플 _ 가중치])",
"_____no_output_____"
]
],
[
[
"Y = df['mpg']\nX_continus = df[['displacement','horsepower', 'weight','acceleration']]\nX_category = df[['model year','cylinders','origin']]",
"_____no_output_____"
],
[
"from sklearn import preprocessing",
"_____no_output_____"
],
[
"scaler = preprocessing.StandardScaler()\ntype(scaler)",
"_____no_output_____"
],
[
"#연속형만 적용해서 교육시킨다(패턴만 갖게 됨)\nscaler.fit(X_continus)",
"_____no_output_____"
],
[
"#패턴에 의해 값을 변환해서 X에 담는다\nX = scaler.transform(X_continus)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression",
"_____no_output_____"
],
[
"#리니어 레그래이션 쌍둥이 만들기 lr\nlr = LinearRegression()\n\ntype(lr)",
"_____no_output_____"
],
[
"#해당 내용을 fitting한다, 교육끝\n#fit은 하면 패턴만 갖게된다~ 1차방정식\n\nlr.fit(X,Y)",
"_____no_output_____"
],
[
"lr.score(X,Y)",
"_____no_output_____"
]
],
[
[
"## predict로 전달 하기\nX_continus = df[['displacement','horsepower', 'weight','acceleration']]\n\n훈련시킬때 넣은 순서로 넣어줘야함",
"_____no_output_____"
]
],
[
[
"df.head(1)",
"_____no_output_____"
],
[
"#소비자에게 알려줘야 할 값이 결과로 나온다\n\n#lr.predict([[307.0,130.0,3504.0,12.0]])",
"_____no_output_____"
],
[
"#scaler를 이미 해서 패턴을 만들어놨기 때문에 transform을 사용한다.\nx_cusmter = scaler.transform([[307.0,130.0,3504.0,12.0]])\nx_cusmter.shape",
"_____no_output_____"
],
[
"#lr.predict에 넣어주기\nlr.predict(x_cusmter)",
"_____no_output_____"
]
],
[
[
"## 서비스 단계\n\npickle — Python object serialization\n\npickle은 바이너리 방식(wb) 파일로 저장이 된다. <br>\n바이너리 사람이 익숙한 형태가 아니다.<br>\nclass만 담을수 있다.<br>\n",
"_____no_output_____"
]
],
[
[
"import pickle",
"_____no_output_____"
],
[
"pickle.dump(lr, open('./autompg_lr.pkl', 'wb'))",
"_____no_output_____"
]
],
[
[
"pickle이 저장 된 상태에서 저장된 autompg_lr.pkl을 다운로드한다\n\nsaves 폴더를 만들고 다운로드 했던 autompg_lr.pkl 을 끌어온다\n\npickle은 바이너리 방식(rb) 파일 불러온다.\n",
"_____no_output_____"
]
],
[
[
"!ls -l ./autompg_lr.pkl",
"-rw-r--r-- 1 root root 567 Jul 6 06:28 ./autompg_lr.pkl\n"
],
[
"pickle.load(open('./autompg_lr.pkl', 'rb'))",
"_____no_output_____"
]
],
[
[
"* pickle로 상대 서비스개발자에게 scaler를 넘겨줄때 쓰는 방법",
"_____no_output_____"
]
],
[
[
"pickle.dump(scaler, open('./autompt_standardscaler.pkl', 'wb'))",
"_____no_output_____"
]
],
[
[
"서비스 개발자는 저장한 autompt_standardscaler.pkl를 다운로드해서 본인 일하는곳에 로드하기\n",
"_____no_output_____"
],
[
"## one hot encoding\n",
"_____no_output_____"
]
],
[
[
"X_category",
"_____no_output_____"
],
[
"X_category['origin'].value_counts()\n\n#카테고리\n#1 , 2 , 3\n#? | ? | ?\n#1 | 0 | 0\n#0 | 1 | 0\n#0 | 0 | 1",
"_____no_output_____"
],
[
"df_origin=pd.get_dummies(X_category['origin'], prefix='origin')",
"_____no_output_____"
],
[
"df_cylinders=pd.get_dummies(X_category['cylinders'],prefix='cylinders')",
"_____no_output_____"
],
[
"df_origin.shape, df_cylinders.shape",
"_____no_output_____"
],
[
"# X_continus + df_cylinders + df_origin\nX_continus.head(3)",
"_____no_output_____"
]
],
[
[
"컬럼명 잘 봐야 함, 서비스하려면 작업을해줘야함",
"_____no_output_____"
]
],
[
[
"#원한인코딩응로 붙히는 작업:pd.concat([X_continus, df_cylinders, df_origin], axis='columns')\nX= pd.concat([X_continus, df_cylinders, df_origin], axis='columns')",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nX_train,X_test,Y_train,Y_test = train_test_split(X,Y)\nX_train.shape,X_test.shape,Y_train.shape,Y_test.shape",
"_____no_output_____"
],
[
"#좋은 스코어를 뽑아내기 위한 모델중의 하나가 xgboot(dicision tree 태생)\nimport xgboost",
"_____no_output_____"
],
[
"xgb= xgboost.XGBRegressor()\nxgb",
"_____no_output_____"
],
[
"xgb.fit(X_train,Y_train)",
"[07:23:34] WARNING: /workspace/src/objective/regression_obj.cu:152: reg:linear is now deprecated in favor of reg:squarederror.\n"
],
[
"#교육시킨 데이터 train으로 스코어 확인\nxgb.score(X_train,Y_train)",
"_____no_output_____"
],
[
"#교육시키지 않은 데이터 test로 스코어 확인\nxgb.score(X_test,Y_test)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae74aae24f32ba0826d59e874bb7e778f1c2f0e
| 94,299 |
ipynb
|
Jupyter Notebook
|
Convolution_model_practical_scratch.ipynb
|
shamiulshifat/Machine-Learning-with-Scratch
|
b57bd285223d11a8899625f51323fd4b50c3a204
|
[
"Apache-2.0"
] | 1 |
2020-06-22T15:00:04.000Z
|
2020-06-22T15:00:04.000Z
|
Convolution_model_practical_scratch.ipynb
|
shamiulshifat/Machine-Learning-with-Scratch
|
b57bd285223d11a8899625f51323fd4b50c3a204
|
[
"Apache-2.0"
] | null | null | null |
Convolution_model_practical_scratch.ipynb
|
shamiulshifat/Machine-Learning-with-Scratch
|
b57bd285223d11a8899625f51323fd4b50c3a204
|
[
"Apache-2.0"
] | null | null | null | 95.347826 | 18,850 | 0.793444 |
[
[
[
"# Convolutional Neural Networks: Application\n\nWelcome to Course 4's second assignment! In this notebook, you will:\n\n- Implement helper functions that you will use when implementing a TensorFlow model\n- Implement a fully functioning ConvNet using TensorFlow \n\n**After this assignment you will be able to:**\n\n- Build and train a ConvNet in TensorFlow for a classification problem \n\nWe assume here that you are already familiar with TensorFlow. If you are not, please refer the *TensorFlow Tutorial* of the third week of Course 2 (\"*Improving deep neural networks*\").",
"_____no_output_____"
],
[
"### <font color='darkblue'> Updates to Assignment <font>\n\n#### If you were working on a previous version\n* The current notebook filename is version \"1a\". \n* You can find your work in the file directory as version \"1\".\n* To view the file directory, go to the menu \"File->Open\", and this will open a new tab that shows the file directory.\n\n#### List of Updates\n* `initialize_parameters`: added details about tf.get_variable, `eval`. Clarified test case.\n* Added explanations for the kernel (filter) stride values, max pooling, and flatten functions.\n* Added details about softmax cross entropy with logits.\n* Added instructions for creating the Adam Optimizer.\n* Added explanation of how to evaluate tensors (optimizer and cost).\n* `forward_propagation`: clarified instructions, use \"F\" to store \"flatten\" layer.\n* Updated print statements and 'expected output' for easier visual comparisons.\n* Many thanks to Kevin P. Brown (mentor for the deep learning specialization) for his suggestions on the assignments in this course!",
"_____no_output_____"
],
[
"## 1.0 - TensorFlow model\n\nIn the previous assignment, you built helper functions using numpy to understand the mechanics behind convolutional neural networks. Most practical applications of deep learning today are built using programming frameworks, which have many built-in functions you can simply call. \n\nAs usual, we will start by loading in the packages. ",
"_____no_output_____"
]
],
[
[
"import math\nimport numpy as np\nimport h5py\nimport matplotlib.pyplot as plt\nimport scipy\nfrom PIL import Image\nfrom scipy import ndimage\nimport tensorflow as tf\nfrom tensorflow.python.framework import ops\nfrom cnn_utils import *\n\n%matplotlib inline\nnp.random.seed(1)",
"_____no_output_____"
]
],
[
[
"Run the next cell to load the \"SIGNS\" dataset you are going to use.",
"_____no_output_____"
]
],
[
[
"# Loading the data (signs)\nX_train_orig, Y_train_orig, X_test_orig, Y_test_orig, classes = load_dataset()",
"_____no_output_____"
]
],
[
[
"As a reminder, the SIGNS dataset is a collection of 6 signs representing numbers from 0 to 5.\n\n<img src=\"images/SIGNS.png\" style=\"width:800px;height:300px;\">\n\nThe next cell will show you an example of a labelled image in the dataset. Feel free to change the value of `index` below and re-run to see different examples. ",
"_____no_output_____"
]
],
[
[
"# Example of a picture\nindex = 6\nplt.imshow(X_train_orig[index])\nprint (\"y = \" + str(np.squeeze(Y_train_orig[:, index])))",
"y = 2\n"
]
],
[
[
"In Course 2, you had built a fully-connected network for this dataset. But since this is an image dataset, it is more natural to apply a ConvNet to it.\n\nTo get started, let's examine the shapes of your data. ",
"_____no_output_____"
]
],
[
[
"X_train = X_train_orig/255.\nX_test = X_test_orig/255.\nY_train = convert_to_one_hot(Y_train_orig, 6).T\nY_test = convert_to_one_hot(Y_test_orig, 6).T\nprint (\"number of training examples = \" + str(X_train.shape[0]))\nprint (\"number of test examples = \" + str(X_test.shape[0]))\nprint (\"X_train shape: \" + str(X_train.shape))\nprint (\"Y_train shape: \" + str(Y_train.shape))\nprint (\"X_test shape: \" + str(X_test.shape))\nprint (\"Y_test shape: \" + str(Y_test.shape))\nconv_layers = {}",
"number of training examples = 1080\nnumber of test examples = 120\nX_train shape: (1080, 64, 64, 3)\nY_train shape: (1080, 6)\nX_test shape: (120, 64, 64, 3)\nY_test shape: (120, 6)\n"
]
],
[
[
"### 1.1 - Create placeholders\n\nTensorFlow requires that you create placeholders for the input data that will be fed into the model when running the session.\n\n**Exercise**: Implement the function below to create placeholders for the input image X and the output Y. You should not define the number of training examples for the moment. To do so, you could use \"None\" as the batch size, it will give you the flexibility to choose it later. Hence X should be of dimension **[None, n_H0, n_W0, n_C0]** and Y should be of dimension **[None, n_y]**. [Hint: search for the tf.placeholder documentation\"](https://www.tensorflow.org/api_docs/python/tf/placeholder).",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: create_placeholders\n\ndef create_placeholders(n_H0, n_W0, n_C0, n_y):\n \"\"\"\n Creates the placeholders for the tensorflow session.\n \n Arguments:\n n_H0 -- scalar, height of an input image\n n_W0 -- scalar, width of an input image\n n_C0 -- scalar, number of channels of the input\n n_y -- scalar, number of classes\n \n Returns:\n X -- placeholder for the data input, of shape [None, n_H0, n_W0, n_C0] and dtype \"float\"\n Y -- placeholder for the input labels, of shape [None, n_y] and dtype \"float\"\n \"\"\"\n\n ### START CODE HERE ### (≈2 lines)\n X = tf.placeholder(tf.float32, shape=(None, n_H0, n_W0, n_C0), name='X')\n Y = tf.placeholder(tf.float32, shape=(None, n_y), name='Y')\n ### END CODE HERE ###\n \n return X, Y",
"_____no_output_____"
],
[
"X, Y = create_placeholders(64, 64, 3, 6)\nprint (\"X = \" + str(X))\nprint (\"Y = \" + str(Y))",
"X = Tensor(\"X:0\", shape=(?, 64, 64, 3), dtype=float32)\nY = Tensor(\"Y:0\", shape=(?, 6), dtype=float32)\n"
]
],
[
[
"**Expected Output**\n\n<table> \n<tr>\n<td>\n X = Tensor(\"Placeholder:0\", shape=(?, 64, 64, 3), dtype=float32)\n\n</td>\n</tr>\n<tr>\n<td>\n Y = Tensor(\"Placeholder_1:0\", shape=(?, 6), dtype=float32)\n\n</td>\n</tr>\n</table>",
"_____no_output_____"
],
[
"### 1.2 - Initialize parameters\n\nYou will initialize weights/filters $W1$ and $W2$ using `tf.contrib.layers.xavier_initializer(seed = 0)`. You don't need to worry about bias variables as you will soon see that TensorFlow functions take care of the bias. Note also that you will only initialize the weights/filters for the conv2d functions. TensorFlow initializes the layers for the fully connected part automatically. We will talk more about that later in this assignment.\n\n**Exercise:** Implement initialize_parameters(). The dimensions for each group of filters are provided below. Reminder - to initialize a parameter $W$ of shape [1,2,3,4] in Tensorflow, use:\n```python\nW = tf.get_variable(\"W\", [1,2,3,4], initializer = ...)\n```\n#### tf.get_variable()\n[Search for the tf.get_variable documentation](https://www.tensorflow.org/api_docs/python/tf/get_variable). Notice that the documentation says:\n```\nGets an existing variable with these parameters or create a new one.\n```\nSo we can use this function to create a tensorflow variable with the specified name, but if the variables already exist, it will get the existing variable with that same name.\n",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: initialize_parameters\n\ndef initialize_parameters():\n \"\"\"\n Initializes weight parameters to build a neural network with tensorflow. The shapes are:\n W1 : [4, 4, 3, 8]\n W2 : [2, 2, 8, 16]\n Note that we will hard code the shape values in the function to make the grading simpler.\n Normally, functions should take values as inputs rather than hard coding.\n Returns:\n parameters -- a dictionary of tensors containing W1, W2\n \"\"\"\n \n tf.set_random_seed(1) # so that your \"random\" numbers match ours\n \n ### START CODE HERE ### (approx. 2 lines of code)\n W1 = tf.get_variable('W1', shape=(4,4,3,8), initializer=tf.contrib.layers.xavier_initializer(seed=0))\n W2 = tf.get_variable('W2', shape=(2,2,8,16), initializer=tf.contrib.layers.xavier_initializer(seed=0))\n ### END CODE HERE ###\n\n parameters = {\"W1\": W1,\n \"W2\": W2}\n \n return parameters",
"_____no_output_____"
],
[
"tf.reset_default_graph()\nwith tf.Session() as sess_test:\n parameters = initialize_parameters()\n init = tf.global_variables_initializer()\n sess_test.run(init)\n print(\"W1[1,1,1] = \\n\" + str(parameters[\"W1\"].eval()[1,1,1]))\n print(\"W1.shape: \" + str(parameters[\"W1\"].shape))\n print(\"\\n\")\n print(\"W2[1,1,1] = \\n\" + str(parameters[\"W2\"].eval()[1,1,1]))\n print(\"W2.shape: \" + str(parameters[\"W2\"].shape))",
"W1[1,1,1] = \n[ 0.00131723 0.14176141 -0.04434952 0.09197326 0.14984085 -0.03514394\n -0.06847463 0.05245192]\nW1.shape: (4, 4, 3, 8)\n\n\nW2[1,1,1] = \n[-0.08566415 0.17750949 0.11974221 0.16773748 -0.0830943 -0.08058\n -0.00577033 -0.14643836 0.24162132 -0.05857408 -0.19055021 0.1345228\n -0.22779644 -0.1601823 -0.16117483 -0.10286498]\nW2.shape: (2, 2, 8, 16)\n"
]
],
[
[
"** Expected Output:**\n\n```\nW1[1,1,1] = \n[ 0.00131723 0.14176141 -0.04434952 0.09197326 0.14984085 -0.03514394\n -0.06847463 0.05245192]\nW1.shape: (4, 4, 3, 8)\n\n\nW2[1,1,1] = \n[-0.08566415 0.17750949 0.11974221 0.16773748 -0.0830943 -0.08058\n -0.00577033 -0.14643836 0.24162132 -0.05857408 -0.19055021 0.1345228\n -0.22779644 -0.1601823 -0.16117483 -0.10286498]\nW2.shape: (2, 2, 8, 16)\n```",
"_____no_output_____"
],
[
"### 1.3 - Forward propagation\n\nIn TensorFlow, there are built-in functions that implement the convolution steps for you.\n\n- **tf.nn.conv2d(X,W, strides = [1,s,s,1], padding = 'SAME'):** given an input $X$ and a group of filters $W$, this function convolves $W$'s filters on X. The third parameter ([1,s,s,1]) represents the strides for each dimension of the input (m, n_H_prev, n_W_prev, n_C_prev). Normally, you'll choose a stride of 1 for the number of examples (the first value) and for the channels (the fourth value), which is why we wrote the value as `[1,s,s,1]`. You can read the full documentation on [conv2d](https://www.tensorflow.org/api_docs/python/tf/nn/conv2d).\n\n- **tf.nn.max_pool(A, ksize = [1,f,f,1], strides = [1,s,s,1], padding = 'SAME'):** given an input A, this function uses a window of size (f, f) and strides of size (s, s) to carry out max pooling over each window. For max pooling, we usually operate on a single example at a time and a single channel at a time. So the first and fourth value in `[1,f,f,1]` are both 1. You can read the full documentation on [max_pool](https://www.tensorflow.org/api_docs/python/tf/nn/max_pool).\n\n- **tf.nn.relu(Z):** computes the elementwise ReLU of Z (which can be any shape). You can read the full documentation on [relu](https://www.tensorflow.org/api_docs/python/tf/nn/relu).\n\n- **tf.contrib.layers.flatten(P)**: given a tensor \"P\", this function takes each training (or test) example in the batch and flattens it into a 1D vector. \n * If a tensor P has the shape (m,h,w,c), where m is the number of examples (the batch size), it returns a flattened tensor with shape (batch_size, k), where $k=h \\times w \\times c$. \"k\" equals the product of all the dimension sizes other than the first dimension.\n * For example, given a tensor with dimensions [100,2,3,4], it flattens the tensor to be of shape [100, 24], where 24 = 2 * 3 * 4. You can read the full documentation on [flatten](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/flatten).\n\n- **tf.contrib.layers.fully_connected(F, num_outputs):** given the flattened input F, it returns the output computed using a fully connected layer. You can read the full documentation on [full_connected](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/fully_connected).\n\nIn the last function above (`tf.contrib.layers.fully_connected`), the fully connected layer automatically initializes weights in the graph and keeps on training them as you train the model. Hence, you did not need to initialize those weights when initializing the parameters.\n\n\n#### Window, kernel, filter\nThe words \"window\", \"kernel\", and \"filter\" are used to refer to the same thing. This is why the parameter `ksize` refers to \"kernel size\", and we use `(f,f)` to refer to the filter size. Both \"kernel\" and \"filter\" refer to the \"window.\"",
"_____no_output_____"
],
[
"**Exercise**\n\nImplement the `forward_propagation` function below to build the following model: `CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED`. You should use the functions above. \n\nIn detail, we will use the following parameters for all the steps:\n - Conv2D: stride 1, padding is \"SAME\"\n - ReLU\n - Max pool: Use an 8 by 8 filter size and an 8 by 8 stride, padding is \"SAME\"\n - Conv2D: stride 1, padding is \"SAME\"\n - ReLU\n - Max pool: Use a 4 by 4 filter size and a 4 by 4 stride, padding is \"SAME\"\n - Flatten the previous output.\n - FULLYCONNECTED (FC) layer: Apply a fully connected layer without an non-linear activation function. Do not call the softmax here. This will result in 6 neurons in the output layer, which then get passed later to a softmax. In TensorFlow, the softmax and cost function are lumped together into a single function, which you'll call in a different function when computing the cost. ",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: forward_propagation\n\ndef forward_propagation(X, parameters):\n \"\"\"\n Implements the forward propagation for the model:\n CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED\n \n Note that for simplicity and grading purposes, we'll hard-code some values\n such as the stride and kernel (filter) sizes. \n Normally, functions should take these values as function parameters.\n \n Arguments:\n X -- input dataset placeholder, of shape (input size, number of examples)\n parameters -- python dictionary containing your parameters \"W1\", \"W2\"\n the shapes are given in initialize_parameters\n\n Returns:\n Z3 -- the output of the last LINEAR unit\n \"\"\"\n \n # Retrieve the parameters from the dictionary \"parameters\" \n W1 = parameters['W1']\n W2 = parameters['W2']\n \n ### START CODE HERE ###\n # CONV2D: stride of 1, padding 'SAME'\n Z1 = tf.nn.conv2d(X, W1, strides=[1, 1, 1, 1], padding='SAME')\n # RELU\n A1 = tf.nn.relu(Z1)\n # MAXPOOL: window 8x8, sride 8, padding 'SAME'\n P1 = tf.nn.max_pool(A1, ksize=[1, 8, 8, 1], strides=[1, 8, 8, 1], padding='SAME')\n # CONV2D: filters W2, stride 1, padding 'SAME'\n Z2 = tf.nn.conv2d(P1, W2, strides=[1, 1, 1, 1], padding='SAME')\n # RELU\n A2 = tf.nn.relu(Z2)\n # MAXPOOL: window 4x4, stride 4, padding 'SAME'\n P2 = tf.nn.max_pool(A2, ksize=[1, 4, 4, 1], strides=[1, 4, 4, 1], padding='SAME')\n # FLATTEN\n P2 = tf.contrib.layers.flatten(P2)\n # FULLY-CONNECTED without non-linear activation function (not not call softmax).\n # 6 neurons in output layer. Hint: one of the arguments should be \"activation_fn=None\" \n Z3 = tf.contrib.layers.fully_connected(P2, 6, activation_fn=None)\n ### END CODE HERE ###\n\n return Z3",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as sess:\n np.random.seed(1)\n X, Y = create_placeholders(64, 64, 3, 6)\n parameters = initialize_parameters()\n Z3 = forward_propagation(X, parameters)\n init = tf.global_variables_initializer()\n sess.run(init)\n a = sess.run(Z3, {X: np.random.randn(2,64,64,3), Y: np.random.randn(2,6)})\n print(\"Z3 = \\n\" + str(a))",
"Z3 = \n[[-0.44670227 -1.57208765 -1.53049231 -2.31013036 -1.29104376 0.46852064]\n [-0.17601591 -1.57972014 -1.4737016 -2.61672091 -1.00810647 0.5747785 ]]\n"
]
],
[
[
"**Expected Output**:\n\n```\nZ3 = \n[[-0.44670227 -1.57208765 -1.53049231 -2.31013036 -1.29104376 0.46852064]\n [-0.17601591 -1.57972014 -1.4737016 -2.61672091 -1.00810647 0.5747785 ]]\n```",
"_____no_output_____"
],
[
"### 1.4 - Compute cost\n\nImplement the compute cost function below. Remember that the cost function helps the neural network see how much the model's predictions differ from the correct labels. By adjusting the weights of the network to reduce the cost, the neural network can improve its predictions.\n\nYou might find these two functions helpful: \n\n- **tf.nn.softmax_cross_entropy_with_logits(logits = Z, labels = Y):** computes the softmax entropy loss. This function both computes the softmax activation function as well as the resulting loss. You can check the full documentation [softmax_cross_entropy_with_logits](https://www.tensorflow.org/api_docs/python/tf/nn/softmax_cross_entropy_with_logits).\n- **tf.reduce_mean:** computes the mean of elements across dimensions of a tensor. Use this to calculate the sum of the losses over all the examples to get the overall cost. You can check the full documentation [reduce_mean](https://www.tensorflow.org/api_docs/python/tf/reduce_mean).\n\n#### Details on softmax_cross_entropy_with_logits (optional reading)\n* Softmax is used to format outputs so that they can be used for classification. It assigns a value between 0 and 1 for each category, where the sum of all prediction values (across all possible categories) equals 1.\n* Cross Entropy is compares the model's predicted classifications with the actual labels and results in a numerical value representing the \"loss\" of the model's predictions.\n* \"Logits\" are the result of multiplying the weights and adding the biases. Logits are passed through an activation function (such as a relu), and the result is called the \"activation.\"\n* The function is named `softmax_cross_entropy_with_logits` takes logits as input (and not activations); then uses the model to predict using softmax, and then compares the predictions with the true labels using cross entropy. These are done with a single function to optimize the calculations.\n\n** Exercise**: Compute the cost below using the function above.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: compute_cost \n\ndef compute_cost(Z3, Y):\n \"\"\"\n Computes the cost\n \n Arguments:\n Z3 -- output of forward propagation (output of the last LINEAR unit), of shape (number of examples, 6)\n Y -- \"true\" labels vector placeholder, same shape as Z3\n \n Returns:\n cost - Tensor of the cost function\n \"\"\"\n \n ### START CODE HERE ### (1 line of code)\n cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=Z3, labels=Y))\n ### END CODE HERE ###\n \n return cost",
"_____no_output_____"
],
[
"tf.reset_default_graph()\n\nwith tf.Session() as sess:\n np.random.seed(1)\n X, Y = create_placeholders(64, 64, 3, 6)\n parameters = initialize_parameters()\n Z3 = forward_propagation(X, parameters)\n cost = compute_cost(Z3, Y)\n init = tf.global_variables_initializer()\n sess.run(init)\n a = sess.run(cost, {X: np.random.randn(4,64,64,3), Y: np.random.randn(4,6)})\n print(\"cost = \" + str(a))",
"cost = 2.91034\n"
]
],
[
[
"**Expected Output**: \n```\ncost = 2.91034\n```",
"_____no_output_____"
],
[
"## 1.5 Model \n\nFinally you will merge the helper functions you implemented above to build a model. You will train it on the SIGNS dataset. \n\n**Exercise**: Complete the function below. \n\nThe model below should:\n\n- create placeholders\n- initialize parameters\n- forward propagate\n- compute the cost\n- create an optimizer\n\nFinally you will create a session and run a for loop for num_epochs, get the mini-batches, and then for each mini-batch you will optimize the function. [Hint for initializing the variables](https://www.tensorflow.org/api_docs/python/tf/global_variables_initializer)",
"_____no_output_____"
],
[
"#### Adam Optimizer\nYou can use `tf.train.AdamOptimizer(learning_rate = ...)` to create the optimizer. The optimizer has a `minimize(loss=...)` function that you'll call to set the cost function that the optimizer will minimize.\n\nFor details, check out the documentation for [Adam Optimizer](https://www.tensorflow.org/api_docs/python/tf/train/AdamOptimizer)",
"_____no_output_____"
],
[
"#### Random mini batches\nIf you took course 2 of the deep learning specialization, you implemented `random_mini_batches()` in the \"Optimization\" programming assignment. This function returns a list of mini-batches. It is already implemented in the `cnn_utils.py` file and imported here, so you can call it like this:\n```Python\nminibatches = random_mini_batches(X, Y, mini_batch_size = 64, seed = 0)\n```\n(You will want to choose the correct variable names when you use it in your code).",
"_____no_output_____"
],
[
"#### Evaluating the optimizer and cost\n\nWithin a loop, for each mini-batch, you'll use the `tf.Session` object (named `sess`) to feed a mini-batch of inputs and labels into the neural network and evaluate the tensors for the optimizer as well as the cost. Remember that we built a graph data structure and need to feed it inputs and labels and use `sess.run()` in order to get values for the optimizer and cost.\n\nYou'll use this kind of syntax:\n```\noutput_for_var1, output_for_var2 = sess.run(\n fetches=[var1, var2],\n feed_dict={var_inputs: the_batch_of_inputs,\n var_labels: the_batch_of_labels}\n )\n```\n* Notice that `sess.run` takes its first argument `fetches` as a list of objects that you want it to evaluate (in this case, we want to evaluate the optimizer and the cost). \n* It also takes a dictionary for the `feed_dict` parameter. \n* The keys are the `tf.placeholder` variables that we created in the `create_placeholders` function above. \n* The values are the variables holding the actual numpy arrays for each mini-batch. \n* The sess.run outputs a tuple of the evaluated tensors, in the same order as the list given to `fetches`. \n\nFor more information on how to use sess.run, see the documentation [tf.Sesssion#run](https://www.tensorflow.org/api_docs/python/tf/Session#run) documentation.",
"_____no_output_____"
]
],
[
[
"# GRADED FUNCTION: model\n\ndef model(X_train, Y_train, X_test, Y_test, learning_rate = 0.009,\n num_epochs = 100, minibatch_size = 64, print_cost = True):\n \"\"\"\n Implements a three-layer ConvNet in Tensorflow:\n CONV2D -> RELU -> MAXPOOL -> CONV2D -> RELU -> MAXPOOL -> FLATTEN -> FULLYCONNECTED\n \n Arguments:\n X_train -- training set, of shape (None, 64, 64, 3)\n Y_train -- test set, of shape (None, n_y = 6)\n X_test -- training set, of shape (None, 64, 64, 3)\n Y_test -- test set, of shape (None, n_y = 6)\n learning_rate -- learning rate of the optimization\n num_epochs -- number of epochs of the optimization loop\n minibatch_size -- size of a minibatch\n print_cost -- True to print the cost every 100 epochs\n \n Returns:\n train_accuracy -- real number, accuracy on the train set (X_train)\n test_accuracy -- real number, testing accuracy on the test set (X_test)\n parameters -- parameters learnt by the model. They can then be used to predict.\n \"\"\"\n \n ops.reset_default_graph() # to be able to rerun the model without overwriting tf variables\n tf.set_random_seed(1) # to keep results consistent (tensorflow seed)\n seed = 3 # to keep results consistent (numpy seed)\n (m, n_H0, n_W0, n_C0) = X_train.shape \n n_y = Y_train.shape[1] \n costs = [] # To keep track of the cost\n \n # Create Placeholders of the correct shape\n ### START CODE HERE ### (1 line)\n X, Y = create_placeholders(n_H0, n_W0, n_C0, n_y)\n ### END CODE HERE ###\n\n # Initialize parameters\n ### START CODE HERE ### (1 line)\n parameters = initialize_parameters()\n ### END CODE HERE ###\n \n # Forward propagation: Build the forward propagation in the tensorflow graph\n ### START CODE HERE ### (1 line)\n Z3 = forward_propagation(X, parameters)\n ### END CODE HERE ###\n \n # Cost function: Add cost function to tensorflow graph\n ### START CODE HERE ### (1 line)\n cost = compute_cost(Z3, Y)\n ### END CODE HERE ###\n \n # Backpropagation: Define the tensorflow optimizer. Use an AdamOptimizer that minimizes the cost.\n ### START CODE HERE ### (1 line)\n optimizer = tf.train.AdamOptimizer(learning_rate).minimize(cost)\n ### END CODE HERE ###\n \n # Initialize all the variables globally\n init = tf.global_variables_initializer()\n \n # Start the session to compute the tensorflow graph\n with tf.Session() as sess:\n \n # Run the initialization\n sess.run(init)\n \n # Do the training loop\n for epoch in range(num_epochs):\n\n minibatch_cost = 0.\n num_minibatches = int(m / minibatch_size) # number of minibatches of size minibatch_size in the train set\n seed = seed + 1\n minibatches = random_mini_batches(X_train, Y_train, minibatch_size, seed)\n\n for minibatch in minibatches:\n\n # Select a minibatch\n (minibatch_X, minibatch_Y) = minibatch\n \"\"\"\n # IMPORTANT: The line that runs the graph on a minibatch.\n # Run the session to execute the optimizer and the cost.\n # The feedict should contain a minibatch for (X,Y).\n \"\"\"\n ### START CODE HERE ### (1 line)\n _ , temp_cost =sess.run([optimizer, cost], {X: minibatch_X, Y: minibatch_Y})\n ### END CODE HERE ###\n \n minibatch_cost += temp_cost / num_minibatches\n \n\n # Print the cost every epoch\n if print_cost == True and epoch % 5 == 0:\n print (\"Cost after epoch %i: %f\" % (epoch, minibatch_cost))\n if print_cost == True and epoch % 1 == 0:\n costs.append(minibatch_cost)\n \n \n # plot the cost\n plt.plot(np.squeeze(costs))\n plt.ylabel('cost')\n plt.xlabel('iterations (per tens)')\n plt.title(\"Learning rate =\" + str(learning_rate))\n plt.show()\n\n # Calculate the correct predictions\n predict_op = tf.argmax(Z3, 1)\n correct_prediction = tf.equal(predict_op, tf.argmax(Y, 1))\n \n # Calculate accuracy on the test set\n accuracy = tf.reduce_mean(tf.cast(correct_prediction, \"float\"))\n print(accuracy)\n train_accuracy = accuracy.eval({X: X_train, Y: Y_train})\n test_accuracy = accuracy.eval({X: X_test, Y: Y_test})\n print(\"Train Accuracy:\", train_accuracy)\n print(\"Test Accuracy:\", test_accuracy)\n \n return train_accuracy, test_accuracy, parameters",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your model for 100 epochs. Check if your cost after epoch 0 and 5 matches our output. If not, stop the cell and go back to your code!",
"_____no_output_____"
]
],
[
[
"_, _, parameters = model(X_train, Y_train, X_test, Y_test)",
"Cost after epoch 0: 1.917929\nCost after epoch 5: 1.506757\nCost after epoch 10: 0.955359\nCost after epoch 15: 0.845802\nCost after epoch 20: 0.701174\nCost after epoch 25: 0.571977\nCost after epoch 30: 0.518435\nCost after epoch 35: 0.495806\nCost after epoch 40: 0.429827\nCost after epoch 45: 0.407291\nCost after epoch 50: 0.366394\nCost after epoch 55: 0.376922\nCost after epoch 60: 0.299491\nCost after epoch 65: 0.338870\nCost after epoch 70: 0.316400\nCost after epoch 75: 0.310413\nCost after epoch 80: 0.249549\nCost after epoch 85: 0.243457\nCost after epoch 90: 0.200031\nCost after epoch 95: 0.175452\n"
]
],
[
[
"**Expected output**: although it may not match perfectly, your expected output should be close to ours and your cost value should decrease.\n\n<table> \n<tr>\n <td> \n **Cost after epoch 0 =**\n </td>\n\n <td> \n 1.917929\n </td> \n</tr>\n<tr>\n <td> \n **Cost after epoch 5 =**\n </td>\n\n <td> \n 1.506757\n </td> \n</tr>\n<tr>\n <td> \n **Train Accuracy =**\n </td>\n\n <td> \n 0.940741\n </td> \n</tr> \n\n<tr>\n <td> \n **Test Accuracy =**\n </td>\n\n <td> \n 0.783333\n </td> \n</tr> \n</table>",
"_____no_output_____"
],
[
"Congratulations! You have finished the assignment and built a model that recognizes SIGN language with almost 80% accuracy on the test set. If you wish, feel free to play around with this dataset further. You can actually improve its accuracy by spending more time tuning the hyperparameters, or using regularization (as this model clearly has a high variance). \n\nOnce again, here's a thumbs up for your work! ",
"_____no_output_____"
]
],
[
[
"fname = \"images/thumbs_up.jpg\"\nimage = np.array(ndimage.imread(fname, flatten=False))\nmy_image = scipy.misc.imresize(image, size=(64,64))\nplt.imshow(my_image)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
4ae75893fb243d137a9e2c98a8b463b5a0d77c41
| 158,776 |
ipynb
|
Jupyter Notebook
|
ML-MIPT-basic/homework_basic/assignment0_01_kNN/kNN_practice_0_01.ipynb
|
kryvokhyzha/examples-and-courses
|
477e82ee24e6abba8a6b6d92555f2ed549ca682c
|
[
"MIT"
] | 1 |
2021-12-13T15:41:48.000Z
|
2021-12-13T15:41:48.000Z
|
ML-MIPT-basic/homework_basic/assignment0_01_kNN/kNN_practice_0_01.ipynb
|
kryvokhyzha/examples-and-courses
|
477e82ee24e6abba8a6b6d92555f2ed549ca682c
|
[
"MIT"
] | 15 |
2021-09-12T15:06:13.000Z
|
2022-03-31T19:02:08.000Z
|
ML-MIPT-basic/homework_basic/assignment0_01_kNN/kNN_practice_0_01.ipynb
|
kryvokhyzha/examples-and-courses
|
477e82ee24e6abba8a6b6d92555f2ed549ca682c
|
[
"MIT"
] | 1 |
2022-01-29T00:37:52.000Z
|
2022-01-29T00:37:52.000Z
| 216.021769 | 112,860 | 0.907284 |
[
[
[
"# k-Nearest Neighbor (kNN) implementation\n\n*Credits: this notebook is deeply based on Stanford CS231n course assignment 1. Source link: http://cs231n.github.io/assignments2019/assignment1/*\n\nThe kNN classifier consists of two stages:\n\n- During training, the classifier takes the training data and simply remembers it\n- During testing, kNN classifies every test image by comparing to all training images and transfering the labels of the k most similar training examples\n- The value of k is cross-validated\n\nIn this exercise you will implement these steps and understand the basic Image Classification pipeline and gain proficiency in writing efficient, vectorized code.\n\nWe will work with the handwritten digits dataset. Images will be flattened (8x8 sized image -> 64 sized vector) and treated as vectors.",
"_____no_output_____"
]
],
[
[
"'''\nIf you are using Google Colab, uncomment the next line to download `k_nearest_neighbor.py`. \nYou can open and change it in Colab using the \"Files\" sidebar on the left.\n'''\n# !wget https://raw.githubusercontent.com/girafe-ai/ml-mipt/basic_s20/homeworks_basic/assignment0_01_kNN/k_nearest_neighbor.py",
"_____no_output_____"
],
[
"from sklearn import datasets\n\ndataset = datasets.load_digits()\nprint(dataset.DESCR)",
".. _digits_dataset:\n\nOptical recognition of handwritten digits dataset\n--------------------------------------------------\n\n**Data Set Characteristics:**\n\n :Number of Instances: 5620\n :Number of Attributes: 64\n :Attribute Information: 8x8 image of integer pixels in the range 0..16.\n :Missing Attribute Values: None\n :Creator: E. Alpaydin (alpaydin '@' boun.edu.tr)\n :Date: July; 1998\n\nThis is a copy of the test set of the UCI ML hand-written digits datasets\nhttps://archive.ics.uci.edu/ml/datasets/Optical+Recognition+of+Handwritten+Digits\n\nThe data set contains images of hand-written digits: 10 classes where\neach class refers to a digit.\n\nPreprocessing programs made available by NIST were used to extract\nnormalized bitmaps of handwritten digits from a preprinted form. From a\ntotal of 43 people, 30 contributed to the training set and different 13\nto the test set. 32x32 bitmaps are divided into nonoverlapping blocks of\n4x4 and the number of on pixels are counted in each block. This generates\nan input matrix of 8x8 where each element is an integer in the range\n0..16. This reduces dimensionality and gives invariance to small\ndistortions.\n\nFor info on NIST preprocessing routines, see M. D. Garris, J. L. Blue, G.\nT. Candela, D. L. Dimmick, J. Geist, P. J. Grother, S. A. Janet, and C.\nL. Wilson, NIST Form-Based Handprint Recognition System, NISTIR 5469,\n1994.\n\n.. topic:: References\n\n - C. Kaynak (1995) Methods of Combining Multiple Classifiers and Their\n Applications to Handwritten Digit Recognition, MSc Thesis, Institute of\n Graduate Studies in Science and Engineering, Bogazici University.\n - E. Alpaydin, C. Kaynak (1998) Cascading Classifiers, Kybernetika.\n - Ken Tang and Ponnuthurai N. Suganthan and Xi Yao and A. Kai Qin.\n Linear dimensionalityreduction using relevance weighted LDA. School of\n Electrical and Electronic Engineering Nanyang Technological University.\n 2005.\n - Claudio Gentile. A New Approximate Maximal Margin Classification\n Algorithm. NIPS. 2000.\n"
],
[
"# First 100 images will be used for testing. This dataset is not sorted by the labels, so it's ok\n# to do the split this way.\n# Please be careful when you split your data into train and test in general.\ntest_border = 100\nX_train, y_train = dataset.data[test_border:], dataset.target[test_border:]\nX_test, y_test = dataset.data[:test_border], dataset.target[:test_border]\n\nprint('Training data shape: ', X_train.shape)\nprint('Training labels shape: ', y_train.shape)\nprint('Test data shape: ', X_test.shape)\nprint('Test labels shape: ', y_test.shape)\nnum_test = X_test.shape[0]",
"Training data shape: (1697, 64)\nTraining labels shape: (1697,)\nTest data shape: (100, 64)\nTest labels shape: (100,)\n"
],
[
"# Run some setup code for this notebook.\nimport random\nimport numpy as np\nimport matplotlib.pyplot as plt\n\n# This is a bit of magic to make matplotlib figures appear inline in the notebook\n# rather than in a new window.\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (14.0, 12.0) # set default size of plots\nplt.rcParams['image.interpolation'] = 'nearest'\nplt.rcParams['image.cmap'] = 'gray'\n\n# Some more magic so that the notebook will reload external python modules;\n# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython\n%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"# Visualize some examples from the dataset.\n# We show a few examples of training images from each class.\nclasses = list(np.arange(10))\nnum_classes = len(classes)\nsamples_per_class = 7\nfor y, cls in enumerate(classes):\n idxs = np.flatnonzero(y_train == y)\n idxs = np.random.choice(idxs, samples_per_class, replace=False)\n for i, idx in enumerate(idxs):\n plt_idx = i * num_classes + y + 1\n plt.subplot(samples_per_class, num_classes, plt_idx)\n plt.imshow(X_train[idx].reshape((8, 8)).astype('uint8'))\n plt.axis('off')\n if i == 0:\n plt.title(cls)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Autoreload is a great stuff, but sometimes it does not work as intended. The code below aims to fix than. __Do not forget to save your changes in the `.py` file before reloading the `KNearestNeighbor` class.__",
"_____no_output_____"
]
],
[
[
"# This dirty hack might help if the autoreload has failed for some reason\ntry:\n del KNearestNeighbor\nexcept:\n pass\n\nfrom k_nearest_neighbor import KNearestNeighbor\n\n# Create a kNN classifier instance. \n# Remember that training a kNN classifier is a noop: \n# the Classifier simply remembers the data and does no further processing \nclassifier = KNearestNeighbor()\nclassifier.fit(X_train, y_train)",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
]
],
[
[
"We would now like to classify the test data with the kNN classifier. Recall that we can break down this process into two steps: \n\n1. First we must compute the distances between all test examples and all train examples. \n2. Given these distances, for each test example we find the k nearest examples and have them vote for the label\n\nLets begin with computing the distance matrix between all training and test examples. For example, if there are **Ntr** training examples and **Nte** test examples, this stage should result in a **Nte x Ntr** matrix where each element (i,j) is the distance between the i-th test and j-th train example.\n\n**Note: For the three distance computations that we require you to implement in this notebook, you may not use the np.linalg.norm() function that numpy provides.**\n\nFirst, open `k_nearest_neighbor.py` and implement the function `compute_distances_two_loops` that uses a (very inefficient) double loop over all pairs of (test, train) examples and computes the distance matrix one element at a time.",
"_____no_output_____"
]
],
[
[
"# Open k_nearest_neighbor.py and implement\n# compute_distances_two_loops.\n\n# Test your implementation:\ndists = classifier.compute_distances_two_loops(X_test)\nprint(dists.shape)",
"(100, 1697)\n"
],
[
"# We can visualize the distance matrix: each row is a single test example and\n# its distances to training examples\nplt.imshow(dists, interpolation='none')\nplt.show()",
"_____no_output_____"
]
],
[
[
"**Inline Question 1** \n\nNotice the structured patterns in the distance matrix, where some rows or columns are visible brighter. (Note that with the default color scheme black indicates low distances while white indicates high distances.)\n\n- What in the data is the cause behind the distinctly bright rows?\n- What causes the columns?\n\n$\\color{blue}{\\textit Your Answer:}$ *To my mind, if some point in the test data is noisy (we can't recognize it), this corresponding row will be brighter. For columns, we have the same situation: if some noisy point exists in train data, we will have a brighter column.*\n\n",
"_____no_output_____"
]
],
[
[
"# Now implement the function predict_labels and run the code below:\n# We use k = 1 (which is Nearest Neighbor).\ny_test_pred = classifier.predict_labels(dists, k=1)\n\n# Compute and print the fraction of correctly predicted examples\nnum_correct = np.sum(y_test_pred == y_test)\naccuracy = float(num_correct) / num_test\nprint('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy))",
"Got 95 / 100 correct => accuracy: 0.950000\n"
]
],
[
[
"You should expect to see approximately `95%` accuracy. Now lets try out a larger `k`, say `k = 5`:",
"_____no_output_____"
]
],
[
[
"y_test_pred = classifier.predict_labels(dists, k=5)\nnum_correct = np.sum(y_test_pred == y_test)\naccuracy = float(num_correct) / num_test\nprint('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy))",
"Got 93 / 100 correct => accuracy: 0.930000\n"
]
],
[
[
"Accuracy should slightly decrease with `k = 5` compared to `k = 1`.",
"_____no_output_____"
],
[
"**Inline Question 2**\n\nWe can also use other distance metrics such as L1 distance.\nFor pixel values $p_{ij}^{(k)}$ at location $(i,j)$ of some image $I_k$, \n\nthe mean $\\mu$ across all pixels over all images is $$\\mu=\\frac{1}{nhw}\\sum_{k=1}^n\\sum_{i=1}^{h}\\sum_{j=1}^{w}p_{ij}^{(k)}$$\nAnd the pixel-wise mean $\\mu_{ij}$ across all images is \n$$\\mu_{ij}=\\frac{1}{n}\\sum_{k=1}^np_{ij}^{(k)}.$$\nThe general standard deviation $\\sigma$ and pixel-wise standard deviation $\\sigma_{ij}$ is defined similarly.\n\nWhich of the following preprocessing steps will not change the performance of a Nearest Neighbor classifier that uses L1 distance? Select all that apply.\n1. Subtracting the mean $\\mu$ ($\\tilde{p}_{ij}^{(k)}=p_{ij}^{(k)}-\\mu$.)\n2. Subtracting the per pixel mean $\\mu_{ij}$ ($\\tilde{p}_{ij}^{(k)}=p_{ij}^{(k)}-\\mu_{ij}$.)\n3. Subtracting the mean $\\mu$ and dividing by the standard deviation $\\sigma$.\n4. Subtracting the pixel-wise mean $\\mu_{ij}$ and dividing by the pixel-wise standard deviation $\\sigma_{ij}$.\n5. Rotating the coordinate axes of the data.\n\n$\\color{blue}{\\textit Your Answer:}$ 1, 2, 3, 4\n\n\n$\\color{blue}{\\textit Your Explanation:}$\n1. We just substruct some value from all points. It means that all points will move along the axis.\n2. The same.\n3. We just scale the distances between points, but it doesn't change the performance.\n4. The same\n5. Rotating the coordinate changed the performance of kNN. It relates to projection, which can change distances between points.\n",
"_____no_output_____"
]
],
[
[
"# Now lets speed up distance matrix computation by using partial vectorization\n# with one loop. Implement the function compute_distances_one_loop and run the\n# code below:\ndists_one = classifier.compute_distances_one_loop(X_test)\n\n# To ensure that our vectorized implementation is correct, we make sure that it\n# agrees with the naive implementation. There are many ways to decide whether\n# two matrices are similar; one of the simplest is the Frobenius norm. In case\n# you haven't seen it before, the Frobenius norm of two matrices is the square\n# root of the squared sum of differences of all elements; in other words, reshape\n# the matrices into vectors and compute the Euclidean distance between them.\ndifference = np.linalg.norm(dists - dists_one, ord='fro')\nprint('One loop difference was: %f' % (difference, ))\nif difference < 0.001:\n print('Good! The distance matrices are the same')\nelse:\n print('Uh-oh! The distance matrices are different')",
"One loop difference was: 0.000000\nGood! The distance matrices are the same\n"
],
[
"# Now implement the fully vectorized version inside compute_distances_no_loops\n# and run the code\ndists_two = classifier.compute_distances_no_loops(X_test)\n\n# check that the distance matrix agrees with the one we computed before:\ndifference = np.linalg.norm(dists - dists_two, ord='fro')\nprint('No loop difference was: %f' % (difference, ))\nif difference < 0.001:\n print('Good! The distance matrices are the same')\nelse:\n print('Uh-oh! The distance matrices are different')",
"No loop difference was: 0.000000\nGood! The distance matrices are the same\n"
]
],
[
[
"### Comparing handcrafted and `sklearn` implementations\nIn this section we will just compare the performance of handcrafted and `sklearn` kNN algorithms. The predictions should be the same. No need to write any code in this section.",
"_____no_output_____"
]
],
[
[
"from sklearn import neighbors",
"_____no_output_____"
],
[
"implemented_knn = KNearestNeighbor()\nimplemented_knn.fit(X_train, y_train)",
"_____no_output_____"
],
[
"n_neighbors = 1\nexternal_knn = neighbors.KNeighborsClassifier(n_neighbors=n_neighbors)\nexternal_knn.fit(X_train, y_train)\nprint('sklearn kNN (k=1) implementation achieves: {} accuracy on the test set'.format(\n external_knn.score(X_test, y_test)\n))\ny_predicted = implemented_knn.predict(X_test, k=n_neighbors).astype(int)\naccuracy_score = sum((y_predicted==y_test).astype(float)) / num_test\nprint('Handcrafted kNN (k=1) implementation achieves: {} accuracy on the test set'.format(accuracy_score))\nassert np.array_equal(\n external_knn.predict(X_test),\n y_predicted\n), 'Labels predicted by handcrafted and sklearn kNN implementations are different!'\nprint('\\nsklearn and handcrafted kNN implementations provide same predictions')\nprint('_'*76)\n\n\nn_neighbors = 5\nexternal_knn = neighbors.KNeighborsClassifier(n_neighbors=n_neighbors)\nexternal_knn.fit(X_train, y_train)\nprint('sklearn kNN (k=5) implementation achieves: {} accuracy on the test set'.format(\n external_knn.score(X_test, y_test)\n))\ny_predicted = implemented_knn.predict(X_test, k=n_neighbors).astype(int)\naccuracy_score = sum((y_predicted==y_test).astype(float)) / num_test\nprint('Handcrafted kNN (k=5) implementation achieves: {} accuracy on the test set'.format(accuracy_score))\nassert np.array_equal(\n external_knn.predict(X_test),\n y_predicted\n), 'Labels predicted by handcrafted and sklearn kNN implementations are different!'\nprint('\\nsklearn and handcrafted kNN implementations provide same predictions')\nprint('_'*76)\n\n",
"sklearn kNN (k=1) implementation achieves: 0.95 accuracy on the test set\nHandcrafted kNN (k=1) implementation achieves: 0.95 accuracy on the test set\n\nsklearn and handcrafted kNN implementations provide same predictions\n____________________________________________________________________________\nsklearn kNN (k=5) implementation achieves: 0.93 accuracy on the test set\nHandcrafted kNN (k=5) implementation achieves: 0.93 accuracy on the test set\n\nsklearn and handcrafted kNN implementations provide same predictions\n____________________________________________________________________________\n"
]
],
[
[
"### Measuring the time\nFinally let's compare how fast the implementations are.\n\nTo make the difference more noticable, let's repeat the train and test objects (there is no point but to compute the distance between more pairs).",
"_____no_output_____"
]
],
[
[
"X_train_big = np.vstack([X_train]*5)\nX_test_big = np.vstack([X_test]*5)\ny_train_big = np.hstack([y_train]*5)\ny_test_big = np.hstack([y_test]*5)",
"_____no_output_____"
],
[
"classifier_big = KNearestNeighbor()\nclassifier_big.fit(X_train_big, y_train_big)\n# Let's compare how fast the implementations are\ndef time_function(f, *args):\n \"\"\"\n Call a function f with args and return the time (in seconds) that it took to execute.\n \"\"\"\n import time\n tic = time.time()\n f(*args)\n toc = time.time()\n return toc - tic\n\ntwo_loop_time = time_function(classifier_big.compute_distances_two_loops, X_test_big)\nprint('Two loop version took %f seconds' % two_loop_time)\n\none_loop_time = time_function(classifier_big.compute_distances_one_loop, X_test_big)\nprint('One loop version took %f seconds' % one_loop_time)\n\nno_loop_time = time_function(classifier_big.compute_distances_no_loops, X_test_big)\nprint('No loop version took %f seconds' % no_loop_time)\n\n# You should see significantly faster performance with the fully vectorized implementation!\n\n# NOTE: depending on what machine you're using, \n# you might not see a speedup when you go from two loops to one loop, \n# and might even see a slow-down.",
"Two loop version took 39.726853 seconds\nOne loop version took 2.033814 seconds\nNo loop version took 0.130528 seconds\n"
]
],
[
[
"The improvement seems significant. (On some hardware one loop version may take even more time, than two loop, but no loop should definitely be the fastest. ",
"_____no_output_____"
],
[
"**Inline Question 3**\n\nWhich of the following statements about $k$-Nearest Neighbor ($k$-NN) are true in a classification setting, and for all $k$? Select all that apply.\n1. The decision boundary (hyperplane between classes in feature space) of the k-NN classifier is linear.\n2. The training error of a 1-NN will always be lower than that of 5-NN.\n3. The test error of a 1-NN will always be lower than that of a 5-NN.\n4. The time needed to classify a test example with the k-NN classifier grows with the size of the training set.\n5. None of the above.\n\n$\\color{blue}{\\textit Your Answer:}$ 2, 4\n\n\n$\\color{blue}{\\textit Your Explanation:}$\n1. Decision boundary depends on distances to closest k points. But we don't have any supposes about linearity.\n2. Yes, because of overfitting on one closest point.\n3. Not always, but in most cases test error of a 1-NN will be higher than test error of a 5-NN because of using just the one-point to predict the label for test point.\n4. Yes, it is true because of computing distances to all points.\n",
"_____no_output_____"
],
[
"### Submitting your work\nTo submit your work you need to log into Yandex contest (link will be provided later) and upload the `k_nearest_neighbor.py` file for the corresponding problem",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4ae76f07338aa58dae6b365e9c395d7b2f797ef1
| 6,998 |
ipynb
|
Jupyter Notebook
|
examples/helmholtz.ipynb
|
UnofficialJuliaMirror/JuAFEM.jl-30d91d44-8115-11e8-1d28-c19a5ac16de8
|
5925f7ca1cf66b7b46db7a65e526e3c8891ec409
|
[
"MIT"
] | null | null | null |
examples/helmholtz.ipynb
|
UnofficialJuliaMirror/JuAFEM.jl-30d91d44-8115-11e8-1d28-c19a5ac16de8
|
5925f7ca1cf66b7b46db7a65e526e3c8891ec409
|
[
"MIT"
] | null | null | null |
examples/helmholtz.ipynb
|
UnofficialJuliaMirror/JuAFEM.jl-30d91d44-8115-11e8-1d28-c19a5ac16de8
|
5925f7ca1cf66b7b46db7a65e526e3c8891ec409
|
[
"MIT"
] | null | null | null | 27.124031 | 110 | 0.472278 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4ae776afba691b09c5b8dd495a49ed106324a092
| 929,376 |
ipynb
|
Jupyter Notebook
|
notebooks/Run-Modes.ipynb
|
gabriel-abrahao/pymagicc
|
c9bdd610f97aa832dd879796aa6b9bfe11fa0f07
|
[
"BSD-3-Clause"
] | 25 |
2017-02-03T10:47:13.000Z
|
2020-01-21T21:58:22.000Z
|
notebooks/Run-Modes.ipynb
|
gabriel-abrahao/pymagicc
|
c9bdd610f97aa832dd879796aa6b9bfe11fa0f07
|
[
"BSD-3-Clause"
] | 248 |
2017-02-15T19:47:31.000Z
|
2020-03-05T04:15:05.000Z
|
notebooks/Run-Modes.ipynb
|
gabriel-abrahao/pymagicc
|
c9bdd610f97aa832dd879796aa6b9bfe11fa0f07
|
[
"BSD-3-Clause"
] | 8 |
2020-05-08T04:02:04.000Z
|
2021-12-06T16:10:51.000Z
| 285.698125 | 40,744 | 0.81783 |
[
[
[
"# Run Modes\n\nRunning MAGICC in different modes can be non-trivial. In this notebook we show how to set MAGICC's config flags so that it will run as desired for a few different cases.",
"_____no_output_____"
]
],
[
[
"# NBVAL_IGNORE_OUTPUT\nfrom os.path import join\nimport datetime\nimport dateutil\nfrom copy import deepcopy\n\n\nimport numpy as np\nimport pandas as pd\n\nfrom pymagicc import MAGICC6, rcp26, zero_emissions\nfrom pymagicc.io import MAGICCData",
"_____no_output_____"
],
[
"%matplotlib inline\nfrom matplotlib import pyplot as plt\n\nplt.style.use(\"ggplot\")\nplt.rcParams[\"figure.figsize\"] = (12, 6)",
"_____no_output_____"
]
],
[
[
"## Concentration to emissions hybrid\n\nThis is MAGICC's default run mode. In this run mode, MAGICC will run with prescribed concentrations (or a quantity which scales linearly with radiative forcing for aerosol species) until a given point in time and will then switch to running in emissions driven mode.",
"_____no_output_____"
]
],
[
[
"with MAGICC6() as magicc:\n res = magicc.run(rcp26)",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nres.head()",
"_____no_output_____"
],
[
"plt.figure()\nres.filter(variable=\"Emis*CO2*\", region=\"World\").line_plot(hue=\"variable\")\nplt.figure()\nres.filter(variable=\"Atmos*Conc*CO2\", region=\"World\").line_plot(hue=\"variable\");",
"_____no_output_____"
]
],
[
[
"The switches which control the time at which MAGICC switches from concentrations driven to emissions driven are all in the form `GAS_SWITCHFROMXXX2EMIS_YEAR` e.g. `CO2_SWITCHFROMCONC2EMIS_YEAR` and `BCOC_SWITCHFROMRF2EMIS_YEAR`. \n\nChanging the value of these switches will alter how MAGICC runs.",
"_____no_output_____"
]
],
[
[
"# NBVAL_IGNORE_OUTPUT\ndf = deepcopy(rcp26)\ndf[\"scenario\"] = \"RCP26_altered_co2_switch\"\nwith MAGICC6() as magicc:\n res = res.append(magicc.run(df, co2_switchfromconc2emis_year=1850))",
"_____no_output_____"
],
[
"plt.figure()\nres.filter(variable=\"Emis*CO2*\", region=\"World\").line_plot(hue=\"variable\")\nplt.figure()\nres.filter(variable=\"Atmos*Conc*CO2\", region=\"World\").line_plot(hue=\"variable\");",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nres.timeseries()",
"_____no_output_____"
]
],
[
[
"As we can see, the emissions remain unchanged but the concentrations are altered as MAGICC is now running emissions driven from 1850 rather than 2005 (the default).\n\nTo get a fully emissions driven run, you need to change all of the relevant `GAS_SWITCHXXX2EMIS_YEAR` flags.",
"_____no_output_____"
],
[
"## CO$_2$ Emissions Driven Only\n\nWe can get a CO$_2$ emissions only driven run like shown.",
"_____no_output_____"
]
],
[
[
"df = zero_emissions.timeseries()\n\ntime = zero_emissions[\"time\"]\ndf.loc[\n (\n df.index.get_level_values(\"variable\")\n == \"Emissions|CO2|MAGICC Fossil and Industrial\"\n ),\n :,\n] = np.linspace(0, 30, len(time))\n\nscen = MAGICCData(df)\nscen.filter(variable=\"Em*CO2*Fossil*\").line_plot(\n x=\"time\", label=\"CO2 Fossil\", hue=None\n)\nscen.filter(variable=\"Em*CO2*Fossil*\", keep=False).line_plot(\n x=\"time\", label=\"Everything else\", hue=None\n);",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nwith MAGICC6() as magicc:\n co2_only_res = magicc.run(\n scen,\n endyear=scen[\"time\"].max().year,\n rf_total_constantafteryr=5000,\n rf_total_runmodus=\"CO2\",\n co2_switchfromconc2emis_year=min(scen[\"time\"]).year,\n )",
"/home/jared/code/uom/pymagicc/pymagicc/io/scen.py:254: UserWarning: Ignoring input data which is not required for .SCEN file\n warnings.warn(\"Ignoring input data which is not required for .SCEN file\")\n"
],
[
"for v in [\n \"Emis*CO2*\",\n \"Atmos*Conc*CO2\",\n \"Radiative Forcing\",\n \"Surface Temperature\",\n]:\n plt.figure()\n co2_only_res.filter(variable=v, region=\"World\").line_plot(hue=\"variable\")",
"_____no_output_____"
]
],
[
[
"## Prescribed Forcing Driven Only\n\nIt is also possible to examine MAGICC's response to a prescribed radiative forcing only.",
"_____no_output_____"
]
],
[
[
"time = zero_emissions[\"time\"]\n\nforcing_external = 2.0 * np.arange(0, len(time)) / len(time)\nforcing_ext = MAGICCData(\n forcing_external,\n index=time,\n columns={\n \"scenario\": [\"idealised\"],\n \"model\": [\"unspecified\"],\n \"climate_model\": [\"unspecified\"],\n \"variable\": [\"Radiative Forcing|Extra\"],\n \"unit\": [\"W / m^2\"],\n \"todo\": [\"SET\"],\n \"region\": [\"World\"],\n },\n)\nforcing_ext.metadata = {\n \"header\": \"External radiative forcing with linear increase\"\n}\nforcing_ext.line_plot(x=\"time\");",
"_____no_output_____"
],
[
"with MAGICC6() as magicc:\n forcing_ext_filename = \"CUSTOM_EXTRA_RF.IN\"\n forcing_ext.write(\n join(magicc.run_dir, forcing_ext_filename), magicc.version\n )\n ext_forc_only_res = magicc.run(\n rf_extra_read=1,\n file_extra_rf=forcing_ext_filename,\n rf_total_runmodus=\"QEXTRA\",\n endyear=max(time).year,\n rf_initialization_method=\"ZEROSTARTSHIFT\", # this is default but to be sure\n rf_total_constantafteryr=5000,\n )",
"_____no_output_____"
],
[
"ext_forc_only_res.filter(\n variable=[\"Radiative Forcing\", \"Surface Temperature\"], region=\"World\"\n).line_plot(hue=\"variable\")",
"_____no_output_____"
]
],
[
[
"## Zero Temperature Output\n\nGetting MAGICC to return zero for its temperature output is surprisingly difficult. To help address this, we add the `set_zero_config` method to our MAGICC classes.",
"_____no_output_____"
]
],
[
[
"print(MAGICC6.set_zero_config.__doc__)",
"Set config such that radiative forcing and temperature output will be zero\n\n This method is intended as a convenience only, it does not handle everything in\n an obvious way. Adjusting the parameter settings still requires great care and\n may behave unepexctedly.\n \n"
],
[
"# NBVAL_IGNORE_OUTPUT\nwith MAGICC6() as magicc:\n magicc.set_zero_config()\n res_zero = magicc.run()",
"/home/jared/code/uom/pymagicc/pymagicc/io/scen.py:254: UserWarning: Ignoring input data which is not required for .SCEN file\n warnings.warn(\"Ignoring input data which is not required for .SCEN file\")\n"
],
[
"res_zero.filter(\n variable=[\"Surface Temperature\", \"Radiative Forcing\"], region=\"World\"\n).line_plot(x=\"time\");",
"_____no_output_____"
]
],
[
[
"## CO$_2$ Emissions and Prescribed Forcing\n\nIt is also possible to run MAGICC in a mode which is CO$_2$ emissions driven but also includes a prescribed external forcing.",
"_____no_output_____"
]
],
[
[
"df = zero_emissions.timeseries()\n\ntime = zero_emissions[\"time\"]\nemms_fossil_co2 = (\n np.linspace(0, 3, len(time))\n - (1 + (np.arange(len(time)) - 500) / 500) ** 2\n)\ndf.loc[\n (\n df.index.get_level_values(\"variable\")\n == \"Emissions|CO2|MAGICC Fossil and Industrial\"\n ),\n :,\n] = emms_fossil_co2\n\nscen = MAGICCData(df)\nscen.filter(variable=\"Em*CO2*Fossil*\").line_plot(x=\"time\", hue=\"variable\")\nscen.filter(variable=\"Em*CO2*Fossil*\", keep=False).line_plot(\n x=\"time\", label=\"Everything Else\"\n)\n\nforcing_external = 3.0 * np.arange(0, len(time)) / len(time)\nforcing_ext = MAGICCData(\n forcing_external,\n index=time,\n columns={\n \"scenario\": [\"idealised\"],\n \"model\": [\"unspecified\"],\n \"climate_model\": [\"unspecified\"],\n \"variable\": [\"Radiative Forcing|Extra\"],\n \"unit\": [\"W / m^2\"],\n \"todo\": [\"SET\"],\n \"region\": [\"World\"],\n },\n)\nforcing_ext.metadata = {\n \"header\": \"External radiative forcing with linear increase\"\n}\nforcing_ext.line_plot(x=\"time\", hue=\"variable\");",
"_____no_output_____"
],
[
"# NBVAL_IGNORE_OUTPUT\nscen.timeseries()",
"_____no_output_____"
],
[
"with MAGICC6() as magicc:\n magicc.set_zero_config() # very important, try commenting this out and see what happens\n forcing_ext_filename = \"CUSTOM_EXTRA_RF.IN\"\n forcing_ext.write(\n join(magicc.run_dir, forcing_ext_filename), magicc.version\n )\n co2_emms_ext_forc_res = magicc.run(\n scen,\n endyear=scen[\"time\"].max().year,\n co2_switchfromconc2emis_year=min(scen[\"time\"]).year,\n rf_extra_read=1,\n file_extra_rf=forcing_ext_filename,\n rf_total_runmodus=\"ALL\", # default but just in case\n rf_initialization_method=\"ZEROSTARTSHIFT\", # this is default but to be sure\n rf_total_constantafteryr=5000,\n )",
"_____no_output_____"
],
[
"plt.figure()\nco2_emms_ext_forc_res.filter(variable=\"Emis*CO2*\", region=\"World\").line_plot(\n x=\"time\", hue=\"variable\"\n)\nplt.figure()\nco2_emms_ext_forc_res.filter(\n variable=\"Atmos*Conc*CO2\", region=\"World\"\n).line_plot(x=\"time\")\nplt.figure()\nco2_emms_ext_forc_res.filter(\n variable=\"Radiative Forcing\", region=\"World\"\n).line_plot(x=\"time\")\nplt.figure()\nco2_emms_ext_forc_res.filter(\n variable=\"Surface Temperature\", region=\"World\"\n).line_plot(x=\"time\");",
"_____no_output_____"
]
],
[
[
"If we adjust MAGICC's CO$_2$ temperature feedback start year, it is easier to see what is going on.",
"_____no_output_____"
]
],
[
[
"with MAGICC6() as magicc:\n magicc.set_zero_config()\n\n forcing_ext_filename = \"CUSTOM_EXTRA_RF.IN\"\n forcing_ext.write(\n join(magicc.run_dir, forcing_ext_filename), magicc.version\n )\n for temp_feedback_year in [2000, 2100, 3000]:\n scen[\"scenario\"] = \"idealised_{}_CO2_temperature_feedback\".format(\n temp_feedback_year\n )\n\n co2_emms_ext_forc_res.append(\n magicc.run(\n scen,\n endyear=scen[\"time\"].max().year,\n co2_switchfromconc2emis_year=min(scen[\"time\"]).year,\n rf_extra_read=1,\n file_extra_rf=forcing_ext_filename,\n rf_total_runmodus=\"ALL\",\n rf_initialization_method=\"ZEROSTARTSHIFT\",\n rf_total_constantafteryr=5000,\n co2_tempfeedback_yrstart=temp_feedback_year,\n )\n )",
"_____no_output_____"
],
[
"co2_emms_ext_forc_res.filter(variable=\"Emis*CO2*\", region=\"World\").line_plot(\n x=\"time\", hue=\"variable\"\n)\nplt.figure()\nco2_emms_ext_forc_res.filter(\n variable=\"Atmos*Conc*CO2\", region=\"World\"\n).line_plot(x=\"time\")\nplt.figure()\nco2_emms_ext_forc_res.filter(\n variable=\"Radiative Forcing\", region=\"World\"\n).line_plot(x=\"time\")\nplt.figure()\nco2_emms_ext_forc_res.filter(\n variable=\"Surface Temperature\", region=\"World\"\n).line_plot(x=\"time\");",
"_____no_output_____"
]
],
[
[
"## CO$_2$ Concentrations Driven",
"_____no_output_____"
]
],
[
[
"time = zero_emissions[\"time\"]\n\nco2_concs = 278 * np.ones_like(time)\nco2_concs[105:] = 278 * 1.01 ** (np.arange(0, len(time[105:])))\n\nco2_concs = MAGICCData(\n co2_concs,\n index=time,\n columns={\n \"scenario\": [\"1%/yr CO2\"],\n \"model\": [\"unspecified\"],\n \"climate_model\": [\"unspecified\"],\n \"variable\": [\"Atmospheric Concentrations|CO2\"],\n \"unit\": [\"ppm\"],\n \"todo\": [\"SET\"],\n \"region\": [\"World\"],\n },\n)\nco2_concs = co2_concs.filter(year=range(1700, 2001))\ntime = co2_concs[\"time\"]\nco2_concs.metadata = {\"header\": \"1%/yr atmospheric CO2 concentration increase\"}\nco2_concs.line_plot(x=\"time\");",
"_____no_output_____"
],
[
"with MAGICC6() as magicc:\n co2_conc_filename = \"1PCT_CO2_CONC.IN\"\n co2_concs.write(join(magicc.run_dir, co2_conc_filename), magicc.version)\n co2_conc_driven_res = magicc.run(\n file_co2_conc=co2_conc_filename,\n co2_switchfromconc2emis_year=max(time).year,\n co2_tempfeedback_switch=1,\n co2_tempfeedback_yrstart=1870,\n co2_fertilization_yrstart=1870,\n rf_total_runmodus=\"CO2\",\n rf_total_constantafteryr=max(time).year,\n endyear=max(time).year,\n out_inverseemis=1,\n )",
"_____no_output_____"
],
[
"plt.figure()\nco2_conc_driven_res.filter(\n variable=\"Inverse Emis*CO2*\", region=\"World\"\n).line_plot()\nplt.figure()\nco2_conc_driven_res.filter(\n variable=\"Atmos*Conc*CO2\", region=\"World\"\n).line_plot()\nplt.figure()\nco2_conc_driven_res.filter(\n variable=\"Radiative Forcing\", region=\"World\"\n).line_plot()\nplt.figure()\nco2_conc_driven_res.filter(\n variable=\"Surface Temperature\", region=\"World\"\n).line_plot();",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4ae78011d5e4b72445b0bd7d92ae91e9c92df9e6
| 963 |
ipynb
|
Jupyter Notebook
|
Untitled1.ipynb
|
OHincapie/DataScience300
|
7a2c40061d9a6764443f0c49e53ba355147c3305
|
[
"MIT"
] | null | null | null |
Untitled1.ipynb
|
OHincapie/DataScience300
|
7a2c40061d9a6764443f0c49e53ba355147c3305
|
[
"MIT"
] | null | null | null |
Untitled1.ipynb
|
OHincapie/DataScience300
|
7a2c40061d9a6764443f0c49e53ba355147c3305
|
[
"MIT"
] | null | null | null | 22.928571 | 230 | 0.504673 |
[
[
[
"<a href=\"https://colab.research.google.com/github/OHincapie/DataScience300/blob/main/Untitled1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
4ae781589ab987070b3175525e95e7dc811930d9
| 122,833 |
ipynb
|
Jupyter Notebook
|
download_code/bond-calculaltion-lb-0-82.ipynb
|
fkubota/kaggle-Predicting-Molecular-Properties
|
ceaf401a2bfab10a3314f3122b12cf07b7c6bf2c
|
[
"MIT"
] | null | null | null |
download_code/bond-calculaltion-lb-0-82.ipynb
|
fkubota/kaggle-Predicting-Molecular-Properties
|
ceaf401a2bfab10a3314f3122b12cf07b7c6bf2c
|
[
"MIT"
] | null | null | null |
download_code/bond-calculaltion-lb-0-82.ipynb
|
fkubota/kaggle-Predicting-Molecular-Properties
|
ceaf401a2bfab10a3314f3122b12cf07b7c6bf2c
|
[
"MIT"
] | 2 |
2020-09-26T08:38:36.000Z
|
2021-01-10T10:56:57.000Z
| 40.102187 | 851 | 0.503993 |
[
[
[
"<img src=\"https://raw.githubusercontent.com/EdsonAvelar/auc/master/molecular_banner.png\" width=1900px height=400px />",
"_____no_output_____"
],
[
"# Predicting Molecular Properties\n<h3 style=\"color:red\">If this kernel helps you, up vote to keep me motivated 😁<br>Thanks!</h3>\n\n\n<h3> Can you measure the magnetic interactions between a pair of atoms? </h3>\n\nThis kernel is a combination of multiple kernels. The goal is to organize and explain the code to beginner competitors like me.<br>\nThis Kernels creates lots of new features and uses lightgbm as model.\n> Update: Using Bond Calculation",
"_____no_output_____"
],
[
"# Table of Contents:\n\n**1. [Problem Definition](#id1)** <br>\n**2. [Get the Data (Collect / Obtain)](#id2)** <br>\n**3. [Load the Dataset](#id3)** <br>\n**4. [Data Pre-processing](#id4)** <br>\n**5. [Model](#id5)** <br>\n**6. [Visualization and Analysis of Results](#id6)** <br>\n**7. [Submittion](#id7)** <br>\n**8. [References](#ref)** <br>",
"_____no_output_____"
],
[
"<a id=\"id1\"></a> <br> \n# **1. Problem Definition:** \n\nThis challenge aims to predict interactions between atoms. The main task is develop an algorithm that can predict the magnetic interaction between two atoms in a molecule (i.e., the scalar coupling constant)<br>\n\nIn this competition, you will be predicting the scalar_coupling_constant between atom pairs in molecules, given the two atom types (e.g., C and H), the coupling type (e.g., 2JHC), and any features you are able to create from the molecule structure (xyz) files.\n\n**Data**\n* **train.csv** - the training set, where the first column (molecule_name) is the name of the molecule where the coupling constant originates, the second (atom_index_0) and third column (atom_index_1) is the atom indices of the atom-pair creating the coupling and the fourth column (**scalar_coupling_constant**) is the scalar coupling constant that we want to be able to predict\n* **test.csv** - the test set; same info as train, without the target variable\n* **sample_submission.csv** - a sample submission file in the correct format\n* **structures.csv** - this file contains the same information as the individual xyz structure files, but in a single file\n\n**Additional Data**<br>\n*NOTE: additional data is provided for the molecules in Train only!*\n* **scalar_coupling_contributions.csv** - The scalar coupling constants in train.csv are a sum of four terms. The first column (**molecule_name**) are the name of the molecule, the second (**atom_index_0**) and third column (**atom_index_1**) are the atom indices of the atom-pair, the fourth column indicates the **type** of coupling, the fifth column (**fc**) is the Fermi Contact contribution, the sixth column (**sd**) is the Spin-dipolar contribution, the seventh column (**pso**) is the Paramagnetic spin-orbit contribution and the eighth column (**dso**) is the Diamagnetic spin-orbit contribution.\n\n",
"_____no_output_____"
],
[
"<a id=\"id2\"></a> <br> \n# **2. Get the Data (Collect / Obtain):** ",
"_____no_output_____"
],
[
"## All imports used in this kernel",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport os\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom tqdm import tqdm_notebook\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.svm import NuSVR, SVR\nfrom sklearn.metrics import mean_absolute_error\npd.options.display.precision = 15\n\nimport lightgbm as lgb\nimport xgboost as xgb\nimport time\nimport datetime\nfrom catboost import CatBoostRegressor\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.model_selection import StratifiedKFold, KFold, RepeatedKFold\nfrom sklearn import metrics\nfrom sklearn import linear_model\nimport gc\nimport seaborn as sns\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\nfrom IPython.display import HTML\nimport json\nimport altair as alt\n\nimport networkx as nx\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport os\nimport time\nimport datetime\nimport json\nimport gc\nfrom numba import jit\n\nimport numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom tqdm import tqdm_notebook\n\nimport lightgbm as lgb\nimport xgboost as xgb\nfrom catboost import CatBoostRegressor, CatBoostClassifier\nfrom sklearn import metrics\n\nfrom itertools import product\n\nimport altair as alt\nfrom altair.vega import v3\nfrom IPython.display import HTML\nalt.renderers.enable('notebook')",
"_____no_output_____"
]
],
[
[
"## All function used in this kernel",
"_____no_output_____"
]
],
[
[
"# using ideas from this kernel: https://www.kaggle.com/notslush/altair-visualization-2018-stackoverflow-survey\ndef prepare_altair():\n \"\"\"\n Helper function to prepare altair for working.\n \"\"\"\n vega_url = 'https://cdn.jsdelivr.net/npm/vega@' + v3.SCHEMA_VERSION\n vega_lib_url = 'https://cdn.jsdelivr.net/npm/vega-lib'\n vega_lite_url = 'https://cdn.jsdelivr.net/npm/vega-lite@' + alt.SCHEMA_VERSION\n vega_embed_url = 'https://cdn.jsdelivr.net/npm/vega-embed@3'\n noext = \"?noext\"\n \n paths = {\n 'vega': vega_url + noext,\n 'vega-lib': vega_lib_url + noext,\n 'vega-lite': vega_lite_url + noext,\n 'vega-embed': vega_embed_url + noext\n }\n \n workaround = f\"\"\" requirejs.config({{\n baseUrl: 'https://cdn.jsdelivr.net/npm/',\n paths: {paths}\n }});\n \"\"\"\n \n return workaround\n \n\ndef add_autoincrement(render_func):\n # Keep track of unique <div/> IDs\n cache = {}\n def wrapped(chart, id=\"vega-chart\", autoincrement=True):\n if autoincrement:\n if id in cache:\n counter = 1 + cache[id]\n cache[id] = counter\n else:\n cache[id] = 0\n actual_id = id if cache[id] == 0 else id + '-' + str(cache[id])\n else:\n if id not in cache:\n cache[id] = 0\n actual_id = id\n return render_func(chart, id=actual_id)\n # Cache will stay outside and \n return wrapped\n \n\n@add_autoincrement\ndef render(chart, id=\"vega-chart\"):\n \"\"\"\n Helper function to plot altair visualizations.\n \"\"\"\n chart_str = \"\"\"\n <div id=\"{id}\"></div><script>\n require([\"vega-embed\"], function(vg_embed) {{\n const spec = {chart}; \n vg_embed(\"#{id}\", spec, {{defaultStyle: true}}).catch(console.warn);\n console.log(\"anything?\");\n }});\n console.log(\"really...anything?\");\n </script>\n \"\"\"\n return HTML(\n chart_str.format(\n id=id,\n chart=json.dumps(chart) if isinstance(chart, dict) else chart.to_json(indent=None)\n )\n )\n \n\ndef reduce_mem_usage(df, verbose=True):\n numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']\n start_mem = df.memory_usage().sum() / 1024**2 \n for col in df.columns:\n col_type = df[col].dtypes\n if col_type in numerics:\n c_min = df[col].min()\n c_max = df[col].max()\n if str(col_type)[:3] == 'int':\n if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:\n df[col] = df[col].astype(np.int8)\n elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:\n df[col] = df[col].astype(np.int16)\n elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:\n df[col] = df[col].astype(np.int32)\n elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:\n df[col] = df[col].astype(np.int64) \n else:\n if c_min > np.finfo(np.float16).min and c_max < np.finfo(np.float16).max:\n df[col] = df[col].astype(np.float16)\n elif c_min > np.finfo(np.float32).min and c_max < np.finfo(np.float32).max:\n df[col] = df[col].astype(np.float32)\n else:\n df[col] = df[col].astype(np.float64) \n end_mem = df.memory_usage().sum() / 1024**2\n if verbose: print('Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)'.format(end_mem, 100 * (start_mem - end_mem) / start_mem))\n return df\n \n\n@jit\ndef fast_auc(y_true, y_prob):\n \"\"\"\n fast roc_auc computation: https://www.kaggle.com/c/microsoft-malware-prediction/discussion/76013\n \"\"\"\n y_true = np.asarray(y_true)\n y_true = y_true[np.argsort(y_prob)]\n nfalse = 0\n auc = 0\n n = len(y_true)\n for i in range(n):\n y_i = y_true[i]\n nfalse += (1 - y_i)\n auc += y_i * nfalse\n auc /= (nfalse * (n - nfalse))\n return auc\n\n\ndef eval_auc(y_true, y_pred):\n \"\"\"\n Fast auc eval function for lgb.\n \"\"\"\n return 'auc', fast_auc(y_true, y_pred), True\n\n\ndef group_mean_log_mae(y_true, y_pred, types, floor=1e-9):\n \"\"\"\n Fast metric computation for this competition: https://www.kaggle.com/c/champs-scalar-coupling\n Code is from this kernel: https://www.kaggle.com/uberkinder/efficient-metric\n \"\"\"\n maes = (y_true-y_pred).abs().groupby(types).mean()\n return np.log(maes.map(lambda x: max(x, floor))).mean()\n \n\ndef train_model_regression(X, X_test, y, params, folds, model_type='lgb', eval_metric='mae', columns=None, plot_feature_importance=False, model=None,\n verbose=10000, early_stopping_rounds=200, n_estimators=50000):\n \"\"\"\n A function to train a variety of regression models.\n Returns dictionary with oof predictions, test predictions, scores and, if necessary, feature importances.\n \n :params: X - training data, can be pd.DataFrame or np.ndarray (after normalizing)\n :params: X_test - test data, can be pd.DataFrame or np.ndarray (after normalizing)\n :params: y - target\n :params: folds - folds to split data\n :params: model_type - type of model to use\n :params: eval_metric - metric to use\n :params: columns - columns to use. If None - use all columns\n :params: plot_feature_importance - whether to plot feature importance of LGB\n :params: model - sklearn model, works only for \"sklearn\" model type\n \n \"\"\"\n columns = X.columns if columns is None else columns\n X_test = X_test[columns]\n \n # to set up scoring parameters\n metrics_dict = {'mae': {'lgb_metric_name': 'mae',\n 'catboost_metric_name': 'MAE',\n 'sklearn_scoring_function': metrics.mean_absolute_error},\n 'group_mae': {'lgb_metric_name': 'mae',\n 'catboost_metric_name': 'MAE',\n 'scoring_function': group_mean_log_mae},\n 'mse': {'lgb_metric_name': 'mse',\n 'catboost_metric_name': 'MSE',\n 'sklearn_scoring_function': metrics.mean_squared_error}\n }\n\n \n result_dict = {}\n \n # out-of-fold predictions on train data\n oof = np.zeros(len(X))\n \n # averaged predictions on train data\n prediction = np.zeros(len(X_test))\n \n # list of scores on folds\n scores = []\n feature_importance = pd.DataFrame()\n \n # split and train on folds\n for fold_n, (train_index, valid_index) in enumerate(folds.split(X)):\n print(f'Fold {fold_n + 1} started at {time.ctime()}')\n if type(X) == np.ndarray:\n X_train, X_valid = X[columns][train_index], X[columns][valid_index]\n y_train, y_valid = y[train_index], y[valid_index]\n else:\n X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]\n y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]\n \n if model_type == 'lgb':\n model = lgb.LGBMRegressor(**params, n_estimators = n_estimators, n_jobs = -1)\n model.fit(X_train, y_train, \n eval_set=[(X_train, y_train), (X_valid, y_valid)], eval_metric=metrics_dict[eval_metric]['lgb_metric_name'],\n verbose=verbose, early_stopping_rounds=early_stopping_rounds)\n \n y_pred_valid = model.predict(X_valid)\n y_pred = model.predict(X_test, num_iteration=model.best_iteration_)\n \n if model_type == 'xgb':\n train_data = xgb.DMatrix(data=X_train, label=y_train, feature_names=X.columns)\n valid_data = xgb.DMatrix(data=X_valid, label=y_valid, feature_names=X.columns)\n\n watchlist = [(train_data, 'train'), (valid_data, 'valid_data')]\n model = xgb.train(dtrain=train_data, num_boost_round=20000, evals=watchlist, early_stopping_rounds=200, verbose_eval=verbose, params=params)\n y_pred_valid = model.predict(xgb.DMatrix(X_valid, feature_names=X.columns), ntree_limit=model.best_ntree_limit)\n y_pred = model.predict(xgb.DMatrix(X_test, feature_names=X.columns), ntree_limit=model.best_ntree_limit)\n \n if model_type == 'sklearn':\n model = model\n model.fit(X_train, y_train)\n \n y_pred_valid = model.predict(X_valid).reshape(-1,)\n score = metrics_dict[eval_metric]['sklearn_scoring_function'](y_valid, y_pred_valid)\n print(f'Fold {fold_n}. {eval_metric}: {score:.4f}.')\n print('')\n \n y_pred = model.predict(X_test).reshape(-1,)\n \n if model_type == 'cat':\n model = CatBoostRegressor(iterations=20000, eval_metric=metrics_dict[eval_metric]['catboost_metric_name'], **params,\n loss_function=metrics_dict[eval_metric]['catboost_metric_name'])\n model.fit(X_train, y_train, eval_set=(X_valid, y_valid), cat_features=[], use_best_model=True, verbose=False)\n\n y_pred_valid = model.predict(X_valid)\n y_pred = model.predict(X_test)\n \n oof[valid_index] = y_pred_valid.reshape(-1,)\n if eval_metric != 'group_mae':\n scores.append(metrics_dict[eval_metric]['sklearn_scoring_function'](y_valid, y_pred_valid))\n else:\n scores.append(metrics_dict[eval_metric]['scoring_function'](y_valid, y_pred_valid, X_valid['type']))\n\n prediction += y_pred \n \n if model_type == 'lgb' and plot_feature_importance:\n # feature importance\n fold_importance = pd.DataFrame()\n fold_importance[\"feature\"] = columns\n fold_importance[\"importance\"] = model.feature_importances_\n fold_importance[\"fold\"] = fold_n + 1\n feature_importance = pd.concat([feature_importance, fold_importance], axis=0)\n\n prediction /= folds.n_splits\n \n print('CV mean score: {0:.4f}, std: {1:.4f}.'.format(np.mean(scores), np.std(scores)))\n \n result_dict['oof'] = oof\n result_dict['prediction'] = prediction\n result_dict['scores'] = scores\n \n if model_type == 'lgb':\n if plot_feature_importance:\n feature_importance[\"importance\"] /= folds.n_splits\n cols = feature_importance[[\"feature\", \"importance\"]].groupby(\"feature\").mean().sort_values(\n by=\"importance\", ascending=False)[:50].index\n\n best_features = feature_importance.loc[feature_importance.feature.isin(cols)]\n\n plt.figure(figsize=(16, 12));\n sns.barplot(x=\"importance\", y=\"feature\", data=best_features.sort_values(by=\"importance\", ascending=False));\n plt.title('LGB Features (avg over folds)');\n \n result_dict['feature_importance'] = feature_importance\n \n return result_dict\n \n\n\ndef train_model_classification(X, X_test, y, params, folds, model_type='lgb', eval_metric='auc', columns=None, plot_feature_importance=False, model=None,\n verbose=10000, early_stopping_rounds=200, n_estimators=50000):\n \"\"\"\n A function to train a variety of regression models.\n Returns dictionary with oof predictions, test predictions, scores and, if necessary, feature importances.\n \n :params: X - training data, can be pd.DataFrame or np.ndarray (after normalizing)\n :params: X_test - test data, can be pd.DataFrame or np.ndarray (after normalizing)\n :params: y - target\n :params: folds - folds to split data\n :params: model_type - type of model to use\n :params: eval_metric - metric to use\n :params: columns - columns to use. If None - use all columns\n :params: plot_feature_importance - whether to plot feature importance of LGB\n :params: model - sklearn model, works only for \"sklearn\" model type\n \n \"\"\"\n columns = X.columns if columns == None else columns\n X_test = X_test[columns]\n \n # to set up scoring parameters\n metrics_dict = {'auc': {'lgb_metric_name': eval_auc,\n 'catboost_metric_name': 'AUC',\n 'sklearn_scoring_function': metrics.roc_auc_score},\n }\n \n result_dict = {}\n \n # out-of-fold predictions on train data\n oof = np.zeros((len(X), len(set(y.values))))\n \n # averaged predictions on train data\n prediction = np.zeros((len(X_test), oof.shape[1]))\n \n # list of scores on folds\n scores = []\n feature_importance = pd.DataFrame()\n \n # split and train on folds\n for fold_n, (train_index, valid_index) in enumerate(folds.split(X)):\n print(f'Fold {fold_n + 1} started at {time.ctime()}')\n if type(X) == np.ndarray:\n X_train, X_valid = X[columns][train_index], X[columns][valid_index]\n y_train, y_valid = y[train_index], y[valid_index]\n else:\n X_train, X_valid = X[columns].iloc[train_index], X[columns].iloc[valid_index]\n y_train, y_valid = y.iloc[train_index], y.iloc[valid_index]\n \n if model_type == 'lgb':\n model = lgb.LGBMClassifier(**params, n_estimators=n_estimators, n_jobs = -1)\n model.fit(X_train, y_train, \n eval_set=[(X_train, y_train), (X_valid, y_valid)], eval_metric=metrics_dict[eval_metric]['lgb_metric_name'],\n verbose=verbose, early_stopping_rounds=early_stopping_rounds)\n \n y_pred_valid = model.predict_proba(X_valid)\n y_pred = model.predict_proba(X_test, num_iteration=model.best_iteration_)\n \n if model_type == 'xgb':\n train_data = xgb.DMatrix(data=X_train, label=y_train, feature_names=X.columns)\n valid_data = xgb.DMatrix(data=X_valid, label=y_valid, feature_names=X.columns)\n\n watchlist = [(train_data, 'train'), (valid_data, 'valid_data')]\n model = xgb.train(dtrain=train_data, num_boost_round=n_estimators, evals=watchlist, early_stopping_rounds=early_stopping_rounds, verbose_eval=verbose, params=params)\n y_pred_valid = model.predict(xgb.DMatrix(X_valid, feature_names=X.columns), ntree_limit=model.best_ntree_limit)\n y_pred = model.predict(xgb.DMatrix(X_test, feature_names=X.columns), ntree_limit=model.best_ntree_limit)\n \n if model_type == 'sklearn':\n model = model\n model.fit(X_train, y_train)\n \n y_pred_valid = model.predict(X_valid).reshape(-1,)\n score = metrics_dict[eval_metric]['sklearn_scoring_function'](y_valid, y_pred_valid)\n print(f'Fold {fold_n}. {eval_metric}: {score:.4f}.')\n print('')\n \n y_pred = model.predict_proba(X_test)\n \n if model_type == 'cat':\n model = CatBoostClassifier(iterations=n_estimators, eval_metric=metrics_dict[eval_metric]['catboost_metric_name'], **params,\n loss_function=metrics_dict[eval_metric]['catboost_metric_name'])\n model.fit(X_train, y_train, eval_set=(X_valid, y_valid), cat_features=[], use_best_model=True, verbose=False)\n\n y_pred_valid = model.predict(X_valid)\n y_pred = model.predict(X_test)\n \n oof[valid_index] = y_pred_valid\n scores.append(metrics_dict[eval_metric]['sklearn_scoring_function'](y_valid, y_pred_valid[:, 1]))\n\n prediction += y_pred \n \n if model_type == 'lgb' and plot_feature_importance:\n # feature importance\n fold_importance = pd.DataFrame()\n fold_importance[\"feature\"] = columns\n fold_importance[\"importance\"] = model.feature_importances_\n fold_importance[\"fold\"] = fold_n + 1\n feature_importance = pd.concat([feature_importance, fold_importance], axis=0)\n\n prediction /= folds.n_splits\n \n print('CV mean score: {0:.4f}, std: {1:.4f}.'.format(np.mean(scores), np.std(scores)))\n \n result_dict['oof'] = oof\n result_dict['prediction'] = prediction\n result_dict['scores'] = scores\n \n if model_type == 'lgb':\n if plot_feature_importance:\n feature_importance[\"importance\"] /= folds.n_splits\n cols = feature_importance[[\"feature\", \"importance\"]].groupby(\"feature\").mean().sort_values(\n by=\"importance\", ascending=False)[:50].index\n\n best_features = feature_importance.loc[feature_importance.feature.isin(cols)]\n\n plt.figure(figsize=(16, 12));\n sns.barplot(x=\"importance\", y=\"feature\", data=best_features.sort_values(by=\"importance\", ascending=False));\n plt.title('LGB Features (avg over folds)');\n \n result_dict['feature_importance'] = feature_importance\n \n return result_dict\n\n# setting up altair\nworkaround = prepare_altair()\nHTML(\"\".join((\n \"<script>\",\n workaround,\n \"</script>\",\n)))",
"_____no_output_____"
]
],
[
[
"<a id=\"id3\"></a> <br> \n# **3. Load the Dataset** \n\nLet's load all necessary datasets",
"_____no_output_____"
]
],
[
[
"train = pd.read_csv('../input/train.csv')\ntest = pd.read_csv('../input/test.csv')\nsub = pd.read_csv('../input/sample_submission.csv')\nstructures = pd.read_csv('../input/structures.csv')\nscalar_coupling_contributions = pd.read_csv('../input/scalar_coupling_contributions.csv')\n\nprint('Train dataset shape is -> rows: {} cols:{}'.format(train.shape[0],train.shape[1]))\nprint('Test dataset shape is -> rows: {} cols:{}'.format(test.shape[0],test.shape[1]))\nprint('Sub dataset shape is -> rows: {} cols:{}'.format(sub.shape[0],sub.shape[1]))\nprint('Structures dataset shape is -> rows: {} cols:{}'.format(structures.shape[0],structures.shape[1]))\nprint('Scalar_coupling_contributions dataset shape is -> rows: {} cols:{}'.format(scalar_coupling_contributions.shape[0],\n scalar_coupling_contributions.shape[1]))",
"Train dataset shape is -> rows: 4658147 cols:6\nTest dataset shape is -> rows: 2505542 cols:5\nSub dataset shape is -> rows: 2505542 cols:2\nStructures dataset shape is -> rows: 2358657 cols:6\nScalar_coupling_contributions dataset shape is -> rows: 4658147 cols:8\n"
]
],
[
[
"For an fast model/feature evaluation, get only 10% of dataset. Final submission must remove/coments this code",
"_____no_output_____"
]
],
[
[
"n_estimators_default = 3000",
"_____no_output_____"
],
[
"'''\nsize = round(0.10*train.shape[0])\ntrain = train[:size]\ntest = test[:size]\nsub = sub[:size]\nstructures = structures[:size]\nscalar_coupling_contributions = scalar_coupling_contributions[:size]\n\nprint('Train dataset shape is now rows: {} cols:{}'.format(train.shape[0],train.shape[1]))\nprint('Test dataset shape is now rows: {} cols:{}'.format(test.shape[0],test.shape[1]))\nprint('Sub dataset shape is now rows: {} cols:{}'.format(sub.shape[0],sub.shape[1]))\nprint('Structures dataset shape is now rows: {} cols:{}'.format(structures.shape[0],structures.shape[1]))\nprint('Scalar_coupling_contributions dataset shape is now rows: {} cols:{}'.format(scalar_coupling_contributions.shape[0],\n scalar_coupling_contributions.shape[1]))\n'''",
"_____no_output_____"
]
],
[
[
"The importante things to know is that the scalar coupling constants in train.csv are a sum of four terms. \n```\n* fc is the Fermi Contact contribution\n* sd is the Spin-dipolar contribution\n* pso is the Paramagnetic spin-orbit contribution\n* dso is the Diamagnetic spin-orbit contribution. \n```\nLet's merge this into train",
"_____no_output_____"
]
],
[
[
"train = pd.merge(train, scalar_coupling_contributions, how = 'left',\n left_on = ['molecule_name', 'atom_index_0', 'atom_index_1', 'type'],\n right_on = ['molecule_name', 'atom_index_0', 'atom_index_1', 'type'])",
"_____no_output_____"
],
[
"train.head(10)",
"_____no_output_____"
],
[
"test.head(10)",
"_____no_output_____"
],
[
"scalar_coupling_contributions.head(5)",
"_____no_output_____"
]
],
[
[
"`train['scalar_coupling_constant'] and scalar_coupling_contributions['fc']` quite similar",
"_____no_output_____"
]
],
[
[
"pd.concat(objs=[train['scalar_coupling_constant'],scalar_coupling_contributions['fc'] ],axis=1)[:10]",
"_____no_output_____"
]
],
[
[
"Based in others ideais we can:<br>\n\n- train a model to predict `fc` feature;\n- add this feature to train and test and train the same model to compare performance;\n- train a better model;",
"_____no_output_____"
],
[
"<a id=\"id4\"></a> <br> \n# **4. Data Pre-processing** ",
"_____no_output_____"
],
[
"## Feature generation",
"_____no_output_____"
],
[
"I use this great kernel to get x,y,z position. https://www.kaggle.com/seriousran/just-speed-up-calculate-distance-from-benchmark",
"_____no_output_____"
]
],
[
[
"from tqdm import tqdm_notebook as tqdm\natomic_radius = {'H':0.38, 'C':0.77, 'N':0.75, 'O':0.73, 'F':0.71} # Without fudge factor\n\nfudge_factor = 0.05\natomic_radius = {k:v + fudge_factor for k,v in atomic_radius.items()}\nprint(atomic_radius)\n\nelectronegativity = {'H':2.2, 'C':2.55, 'N':3.04, 'O':3.44, 'F':3.98}\n\n#structures = pd.read_csv(structures, dtype={'atom_index':np.int8})\n\natoms = structures['atom'].values\natoms_en = [electronegativity[x] for x in tqdm(atoms)]\natoms_rad = [atomic_radius[x] for x in tqdm(atoms)]\n\nstructures['EN'] = atoms_en\nstructures['rad'] = atoms_rad\n\ndisplay(structures.head())",
"{'H': 0.43, 'C': 0.8200000000000001, 'N': 0.8, 'O': 0.78, 'F': 0.76}\n"
]
],
[
[
"### Chemical Bond Calculation",
"_____no_output_____"
]
],
[
[
"i_atom = structures['atom_index'].values\np = structures[['x', 'y', 'z']].values\np_compare = p\nm = structures['molecule_name'].values\nm_compare = m\nr = structures['rad'].values\nr_compare = r\n\nsource_row = np.arange(len(structures))\nmax_atoms = 28\n\nbonds = np.zeros((len(structures)+1, max_atoms+1), dtype=np.int8)\nbond_dists = np.zeros((len(structures)+1, max_atoms+1), dtype=np.float32)\n\nprint('Calculating bonds')\n\nfor i in tqdm(range(max_atoms-1)):\n p_compare = np.roll(p_compare, -1, axis=0)\n m_compare = np.roll(m_compare, -1, axis=0)\n r_compare = np.roll(r_compare, -1, axis=0)\n \n mask = np.where(m == m_compare, 1, 0) #Are we still comparing atoms in the same molecule?\n dists = np.linalg.norm(p - p_compare, axis=1) * mask\n r_bond = r + r_compare\n \n bond = np.where(np.logical_and(dists > 0.0001, dists < r_bond), 1, 0)\n \n source_row = source_row\n target_row = source_row + i + 1 #Note: Will be out of bounds of bonds array for some values of i\n target_row = np.where(np.logical_or(target_row > len(structures), mask==0), len(structures), target_row) #If invalid target, write to dummy row\n \n source_atom = i_atom\n target_atom = i_atom + i + 1 #Note: Will be out of bounds of bonds array for some values of i\n target_atom = np.where(np.logical_or(target_atom > max_atoms, mask==0), max_atoms, target_atom) #If invalid target, write to dummy col\n \n bonds[(source_row, target_atom)] = bond\n bonds[(target_row, source_atom)] = bond\n bond_dists[(source_row, target_atom)] = dists\n bond_dists[(target_row, source_atom)] = dists\n\nbonds = np.delete(bonds, axis=0, obj=-1) #Delete dummy row\nbonds = np.delete(bonds, axis=1, obj=-1) #Delete dummy col\nbond_dists = np.delete(bond_dists, axis=0, obj=-1) #Delete dummy row\nbond_dists = np.delete(bond_dists, axis=1, obj=-1) #Delete dummy col\n\nprint('Counting and condensing bonds')\n\nbonds_numeric = [[i for i,x in enumerate(row) if x] for row in tqdm(bonds)]\nbond_lengths = [[dist for i,dist in enumerate(row) if i in bonds_numeric[j]] for j,row in enumerate(tqdm(bond_dists))]\nbond_lengths_mean = [ np.mean(x) for x in bond_lengths]\nn_bonds = [len(x) for x in bonds_numeric]\n\n#bond_data = {'bond_' + str(i):col for i, col in enumerate(np.transpose(bonds))}\n#bond_data.update({'bonds_numeric':bonds_numeric, 'n_bonds':n_bonds})\n\nbond_data = {'n_bonds':n_bonds, 'bond_lengths_mean': bond_lengths_mean }\nbond_df = pd.DataFrame(bond_data)\nstructures = structures.join(bond_df)\ndisplay(structures.head(20))",
"Calculating bonds\n"
],
[
"def map_atom_info(df, atom_idx):\n df = pd.merge(df, structures, how = 'left',\n left_on = ['molecule_name', f'atom_index_{atom_idx}'],\n right_on = ['molecule_name', 'atom_index'])\n \n #df = df.drop('atom_index', axis=1)\n df = df.rename(columns={'atom': f'atom_{atom_idx}',\n 'x': f'x_{atom_idx}',\n 'y': f'y_{atom_idx}',\n 'z': f'z_{atom_idx}'})\n return df\n\ntrain = map_atom_info(train, 0)\ntrain = map_atom_info(train, 1)\n\ntest = map_atom_info(test, 0)\ntest = map_atom_info(test, 1)",
"_____no_output_____"
]
],
[
[
"Let's get the distance between atoms first.",
"_____no_output_____"
]
],
[
[
"train_p_0 = train[['x_0', 'y_0', 'z_0']].values\ntrain_p_1 = train[['x_1', 'y_1', 'z_1']].values\ntest_p_0 = test[['x_0', 'y_0', 'z_0']].values\ntest_p_1 = test[['x_1', 'y_1', 'z_1']].values\n\ntrain['dist'] = np.linalg.norm(train_p_0 - train_p_1, axis=1)\ntest['dist'] = np.linalg.norm(test_p_0 - test_p_1, axis=1)\ntrain['dist_x'] = (train['x_0'] - train['x_1']) ** 2\ntest['dist_x'] = (test['x_0'] - test['x_1']) ** 2\ntrain['dist_y'] = (train['y_0'] - train['y_1']) ** 2\ntest['dist_y'] = (test['y_0'] - test['y_1']) ** 2\ntrain['dist_z'] = (train['z_0'] - train['z_1']) ** 2\ntest['dist_z'] = (test['z_0'] - test['z_1']) ** 2\n\ntrain['type_0'] = train['type'].apply(lambda x: x[0])\ntest['type_0'] = test['type'].apply(lambda x: x[0])\n",
"_____no_output_____"
],
[
"def create_features(df):\n df['molecule_couples'] = df.groupby('molecule_name')['id'].transform('count')\n df['molecule_dist_mean'] = df.groupby('molecule_name')['dist'].transform('mean')\n df['molecule_dist_min'] = df.groupby('molecule_name')['dist'].transform('min')\n df['molecule_dist_max'] = df.groupby('molecule_name')['dist'].transform('max')\n df['atom_0_couples_count'] = df.groupby(['molecule_name', 'atom_index_0'])['id'].transform('count')\n df['atom_1_couples_count'] = df.groupby(['molecule_name', 'atom_index_1'])['id'].transform('count')\n df[f'molecule_atom_index_0_x_1_std'] = df.groupby(['molecule_name', 'atom_index_0'])['x_1'].transform('std')\n df[f'molecule_atom_index_0_y_1_mean'] = df.groupby(['molecule_name', 'atom_index_0'])['y_1'].transform('mean')\n df[f'molecule_atom_index_0_y_1_mean_diff'] = df[f'molecule_atom_index_0_y_1_mean'] - df['y_1']\n df[f'molecule_atom_index_0_y_1_mean_div'] = df[f'molecule_atom_index_0_y_1_mean'] / df['y_1']\n df[f'molecule_atom_index_0_y_1_max'] = df.groupby(['molecule_name', 'atom_index_0'])['y_1'].transform('max')\n df[f'molecule_atom_index_0_y_1_max_diff'] = df[f'molecule_atom_index_0_y_1_max'] - df['y_1']\n df[f'molecule_atom_index_0_y_1_std'] = df.groupby(['molecule_name', 'atom_index_0'])['y_1'].transform('std')\n df[f'molecule_atom_index_0_z_1_std'] = df.groupby(['molecule_name', 'atom_index_0'])['z_1'].transform('std')\n df[f'molecule_atom_index_0_dist_mean'] = df.groupby(['molecule_name', 'atom_index_0'])['dist'].transform('mean')\n df[f'molecule_atom_index_0_dist_mean_diff'] = df[f'molecule_atom_index_0_dist_mean'] - df['dist']\n df[f'molecule_atom_index_0_dist_mean_div'] = df[f'molecule_atom_index_0_dist_mean'] / df['dist']\n df[f'molecule_atom_index_0_dist_max'] = df.groupby(['molecule_name', 'atom_index_0'])['dist'].transform('max')\n df[f'molecule_atom_index_0_dist_max_diff'] = df[f'molecule_atom_index_0_dist_max'] - df['dist']\n df[f'molecule_atom_index_0_dist_max_div'] = df[f'molecule_atom_index_0_dist_max'] / df['dist']\n df[f'molecule_atom_index_0_dist_min'] = df.groupby(['molecule_name', 'atom_index_0'])['dist'].transform('min')\n df[f'molecule_atom_index_0_dist_min_diff'] = df[f'molecule_atom_index_0_dist_min'] - df['dist']\n df[f'molecule_atom_index_0_dist_min_div'] = df[f'molecule_atom_index_0_dist_min'] / df['dist']\n df[f'molecule_atom_index_0_dist_std'] = df.groupby(['molecule_name', 'atom_index_0'])['dist'].transform('std')\n df[f'molecule_atom_index_0_dist_std_diff'] = df[f'molecule_atom_index_0_dist_std'] - df['dist']\n df[f'molecule_atom_index_0_dist_std_div'] = df[f'molecule_atom_index_0_dist_std'] / df['dist']\n df[f'molecule_atom_index_1_dist_mean'] = df.groupby(['molecule_name', 'atom_index_1'])['dist'].transform('mean')\n df[f'molecule_atom_index_1_dist_mean_diff'] = df[f'molecule_atom_index_1_dist_mean'] - df['dist']\n df[f'molecule_atom_index_1_dist_mean_div'] = df[f'molecule_atom_index_1_dist_mean'] / df['dist']\n df[f'molecule_atom_index_1_dist_max'] = df.groupby(['molecule_name', 'atom_index_1'])['dist'].transform('max')\n df[f'molecule_atom_index_1_dist_max_diff'] = df[f'molecule_atom_index_1_dist_max'] - df['dist']\n df[f'molecule_atom_index_1_dist_max_div'] = df[f'molecule_atom_index_1_dist_max'] / df['dist']\n df[f'molecule_atom_index_1_dist_min'] = df.groupby(['molecule_name', 'atom_index_1'])['dist'].transform('min')\n df[f'molecule_atom_index_1_dist_min_diff'] = df[f'molecule_atom_index_1_dist_min'] - df['dist']\n df[f'molecule_atom_index_1_dist_min_div'] = df[f'molecule_atom_index_1_dist_min'] / df['dist']\n df[f'molecule_atom_index_1_dist_std'] = df.groupby(['molecule_name', 'atom_index_1'])['dist'].transform('std')\n df[f'molecule_atom_index_1_dist_std_diff'] = df[f'molecule_atom_index_1_dist_std'] - df['dist']\n df[f'molecule_atom_index_1_dist_std_div'] = df[f'molecule_atom_index_1_dist_std'] / df['dist']\n df[f'molecule_atom_1_dist_mean'] = df.groupby(['molecule_name', 'atom_1'])['dist'].transform('mean')\n df[f'molecule_atom_1_dist_min'] = df.groupby(['molecule_name', 'atom_1'])['dist'].transform('min')\n df[f'molecule_atom_1_dist_min_diff'] = df[f'molecule_atom_1_dist_min'] - df['dist']\n df[f'molecule_atom_1_dist_min_div'] = df[f'molecule_atom_1_dist_min'] / df['dist']\n df[f'molecule_atom_1_dist_std'] = df.groupby(['molecule_name', 'atom_1'])['dist'].transform('std')\n df[f'molecule_atom_1_dist_std_diff'] = df[f'molecule_atom_1_dist_std'] - df['dist']\n df[f'molecule_type_0_dist_std'] = df.groupby(['molecule_name', 'type_0'])['dist'].transform('std')\n df[f'molecule_type_0_dist_std_diff'] = df[f'molecule_type_0_dist_std'] - df['dist']\n df[f'molecule_type_dist_mean'] = df.groupby(['molecule_name', 'type'])['dist'].transform('mean')\n df[f'molecule_type_dist_mean_diff'] = df[f'molecule_type_dist_mean'] - df['dist']\n df[f'molecule_type_dist_mean_div'] = df[f'molecule_type_dist_mean'] / df['dist']\n df[f'molecule_type_dist_max'] = df.groupby(['molecule_name', 'type'])['dist'].transform('max')\n df[f'molecule_type_dist_min'] = df.groupby(['molecule_name', 'type'])['dist'].transform('min')\n df[f'molecule_type_dist_std'] = df.groupby(['molecule_name', 'type'])['dist'].transform('std')\n df[f'molecule_type_dist_std_diff'] = df[f'molecule_type_dist_std'] - df['dist']\n df = reduce_mem_usage(df)\n return df\n\ntrain = create_features(train)\ntest = create_features(test)",
"Mem. usage decreased to 932.89 Mb (69.8% reduction)\nMem. usage decreased to 468.34 Mb (70.1% reduction)\n"
],
[
"def map_atom_info(df_1,df_2, atom_idx):\n df = pd.merge(df_1, df_2, how = 'left',\n left_on = ['molecule_name', f'atom_index_{atom_idx}'],\n right_on = ['molecule_name', 'atom_index'])\n df = df.drop('atom_index', axis=1)\n\n return df\n\ndef create_closest(df_train):\n #I apologize for my poor coding skill. Please make the better one.\n df_temp=df_train.loc[:,[\"molecule_name\",\"atom_index_0\",\"atom_index_1\",\"dist\",\"x_0\",\"y_0\",\"z_0\",\"x_1\",\"y_1\",\"z_1\"]].copy()\n df_temp_=df_temp.copy()\n df_temp_= df_temp_.rename(columns={'atom_index_0': 'atom_index_1',\n 'atom_index_1': 'atom_index_0',\n 'x_0': 'x_1',\n 'y_0': 'y_1',\n 'z_0': 'z_1',\n 'x_1': 'x_0',\n 'y_1': 'y_0',\n 'z_1': 'z_0'})\n df_temp=pd.concat(objs=[df_temp,df_temp_],axis=0)\n\n df_temp[\"min_distance\"]=df_temp.groupby(['molecule_name', 'atom_index_0'])['dist'].transform('min')\n df_temp= df_temp[df_temp[\"min_distance\"]==df_temp[\"dist\"]]\n\n df_temp=df_temp.drop(['x_0','y_0','z_0','min_distance'], axis=1)\n df_temp= df_temp.rename(columns={'atom_index_0': 'atom_index',\n 'atom_index_1': 'atom_index_closest',\n 'distance': 'distance_closest',\n 'x_1': 'x_closest',\n 'y_1': 'y_closest',\n 'z_1': 'z_closest'})\n\n for atom_idx in [0,1]:\n df_train = map_atom_info(df_train,df_temp, atom_idx)\n df_train = df_train.rename(columns={'atom_index_closest': f'atom_index_closest_{atom_idx}',\n 'distance_closest': f'distance_closest_{atom_idx}',\n 'x_closest': f'x_closest_{atom_idx}',\n 'y_closest': f'y_closest_{atom_idx}',\n 'z_closest': f'z_closest_{atom_idx}'})\n return df_train\n\n#dtrain = create_closest(train)\n#dtest = create_closest(test)\n#print('dtrain size',dtrain.shape)\n#print('dtest size',dtest.shape)",
"_____no_output_____"
]
],
[
[
"### cosine angles calculation",
"_____no_output_____"
]
],
[
[
"def add_cos_features(df):\n df[\"distance_0\"]=((df['x_0']-df['x_closest_0'])**2+(df['y_0']-df['y_closest_0'])**2+(df['z_0']-df['z_closest_0'])**2)**(1/2)\n df[\"distance_1\"]=((df['x_1']-df['x_closest_1'])**2+(df['y_1']-df['y_closest_1'])**2+(df['z_1']-df['z_closest_1'])**2)**(1/2)\n df[\"vec_0_x\"]=(df['x_0']-df['x_closest_0'])/df[\"distance_0\"]\n df[\"vec_0_y\"]=(df['y_0']-df['y_closest_0'])/df[\"distance_0\"]\n df[\"vec_0_z\"]=(df['z_0']-df['z_closest_0'])/df[\"distance_0\"]\n df[\"vec_1_x\"]=(df['x_1']-df['x_closest_1'])/df[\"distance_1\"]\n df[\"vec_1_y\"]=(df['y_1']-df['y_closest_1'])/df[\"distance_1\"]\n df[\"vec_1_z\"]=(df['z_1']-df['z_closest_1'])/df[\"distance_1\"]\n df[\"vec_x\"]=(df['x_1']-df['x_0'])/df[\"dist\"]\n df[\"vec_y\"]=(df['y_1']-df['y_0'])/df[\"dist\"]\n df[\"vec_z\"]=(df['z_1']-df['z_0'])/df[\"dist\"]\n df[\"cos_0_1\"]=df[\"vec_0_x\"]*df[\"vec_1_x\"]+df[\"vec_0_y\"]*df[\"vec_1_y\"]+df[\"vec_0_z\"]*df[\"vec_1_z\"]\n df[\"cos_0\"]=df[\"vec_0_x\"]*df[\"vec_x\"]+df[\"vec_0_y\"]*df[\"vec_y\"]+df[\"vec_0_z\"]*df[\"vec_z\"]\n df[\"cos_1\"]=df[\"vec_1_x\"]*df[\"vec_x\"]+df[\"vec_1_y\"]*df[\"vec_y\"]+df[\"vec_1_z\"]*df[\"vec_z\"]\n df=df.drop(['vec_0_x','vec_0_y','vec_0_z','vec_1_x','vec_1_y','vec_1_z','vec_x','vec_y','vec_z'], axis=1)\n return df\n \n#train = add_cos_features(train)\n#test = add_cos_features(test)\n\n#print('train size',train.shape)\n#print('test size',test.shape)",
"_____no_output_____"
]
],
[
[
"Dropping molecule_name and encode atom_0, atom_1 and type_0.<br>\n**@TODO:** Try others encoders ",
"_____no_output_____"
]
],
[
[
"del_cols_list = ['id','molecule_name','sd','pso','dso']\ndef del_cols(df, cols):\n del_cols_list_ = [l for l in del_cols_list if l in df]\n df = df.drop(del_cols_list_,axis=1)\n return df\n\ntrain = del_cols(train,del_cols_list)\ntest = del_cols(test,del_cols_list)",
"_____no_output_____"
],
[
"def encode_categoric_single(df):\n lbl = LabelEncoder()\n cat_cols=[]\n try:\n cat_cols = df.describe(include=['O']).columns.tolist()\n for cat in cat_cols:\n df[cat] = lbl.fit_transform(list(df[cat].values))\n except Exception as e:\n print('error: ', str(e) )\n\n return df",
"_____no_output_____"
],
[
"def encode_categoric(dtrain,dtest):\n lbl = LabelEncoder()\n objs_n = len(dtrain)\n dfmerge = pd.concat(objs=[dtrain,dtest],axis=0)\n cat_cols=[]\n try:\n cat_cols = dfmerge.describe(include=['O']).columns.tolist()\n for cat in cat_cols:\n dfmerge[cat] = lbl.fit_transform(list(dfmerge[cat].values))\n except Exception as e:\n print('error: ', str(e) )\n\n dtrain = dfmerge[:objs_n]\n dtest = dfmerge[objs_n:]\n return dtrain,dtest\n\n",
"_____no_output_____"
],
[
"train = encode_categoric_single(train)\ntest = encode_categoric_single(test)",
"_____no_output_____"
],
[
"y_fc = train['fc']\nX = train.drop(['scalar_coupling_constant','fc'],axis=1)\ny = train['scalar_coupling_constant']\n\nX_test = test.copy()",
"_____no_output_____"
],
[
"print('X size',X.shape)\nprint('X_test size',X_test.shape)\nprint('dtest size',test.shape)\nprint('y_fc size',y_fc.shape)\n\ndel train, test\ngc.collect()\n",
"X size (4658147, 79)\nX_test size (2505542, 79)\ndtest size (2505542, 79)\ny_fc size (4658147,)\n"
],
[
"good_columns = ['bond_lengths_mean_y',\n 'molecule_atom_index_0_dist_max',\n 'bond_lengths_mean_x',\n 'molecule_atom_index_0_dist_mean',\n 'molecule_atom_index_0_dist_std',\n 'molecule_couples',\n 'molecule_atom_index_0_y_1_std',\n 'molecule_dist_mean',\n 'molecule_dist_max',\n 'dist_y',\n 'molecule_atom_index_0_z_1_std',\n 'molecule_atom_index_1_dist_max',\n 'molecule_atom_index_1_dist_min',\n 'molecule_atom_index_0_x_1_std',\n 'molecule_atom_index_1_dist_std',\n 'molecule_atom_index_0_y_1_mean_div',\n 'y_0',\n 'molecule_atom_index_1_dist_mean',\n 'molecule_atom_1_dist_mean',\n 'x_0',\n 'dist_x',\n 'molecule_type_dist_std',\n 'dist_z',\n 'molecule_atom_index_1_dist_std_diff',\n 'molecule_type_dist_mean_diff',\n 'molecule_atom_index_0_dist_max_div',\n 'molecule_atom_1_dist_std',\n 'molecule_type_0_dist_std',\n 'z_0',\n 'molecule_type_dist_std_diff',\n 'molecule_atom_index_0_y_1_mean_diff',\n 'molecule_atom_index_0_dist_std_diff',\n 'molecule_atom_index_0_dist_mean_div',\n 'molecule_atom_index_0_dist_max_diff',\n 'x_1',\n 'molecule_type_dist_max',\n 'molecule_atom_index_0_dist_std_div',\n 'molecule_atom_index_0_dist_mean_diff',\n 'molecule_atom_1_dist_std_diff',\n 'molecule_atom_index_0_y_1_max_diff',\n 'z_1',\n 'molecule_atom_index_0_y_1_max',\n 'molecule_atom_index_0_y_1_mean',\n 'y_1',\n 'molecule_type_0_dist_std_diff',\n 'molecule_dist_min',\n 'molecule_atom_index_1_dist_std_div',\n 'molecule_atom_1_dist_min',\n 'molecule_atom_index_1_dist_max_diff','type']\n\nX = X[good_columns].copy()\nX_test = X_test[good_columns].copy()",
"_____no_output_____"
]
],
[
[
"<a id=\"id5\"></a> <br> \n# **5. Model** \n",
"_____no_output_____"
]
],
[
[
"n_fold = 3\nfolds = KFold(n_splits=n_fold, shuffle=True, random_state=11)",
"_____no_output_____"
]
],
[
[
"## Create out of fold feature",
"_____no_output_____"
]
],
[
[
"params = {'num_leaves': 50,\n 'min_child_samples': 79,\n 'min_data_in_leaf' : 100,\n 'objective': 'regression',\n 'max_depth': 9,\n 'learning_rate': 0.2,\n \"boosting_type\": \"gbdt\",\n \"subsample_freq\": 1,\n \"subsample\": 0.9,\n \"bagging_seed\": 11,\n \"metric\": 'mae',\n \"verbosity\": -1,\n 'reg_alpha': 0.1,\n 'reg_lambda': 0.3,\n 'colsample_bytree': 1.0\n }\nresult_dict_lgb_oof = train_model_regression(X=X, X_test=X_test, y=y_fc, params=params, folds=folds, model_type='lgb', eval_metric='group_mae', plot_feature_importance=False,\n verbose=500, early_stopping_rounds=200, n_estimators=n_estimators_default)\n",
"Fold 1 started at Thu Jun 20 01:45:26 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 1.23537\tvalid_1's l1: 1.25803\n[1000]\ttraining's l1: 1.10539\tvalid_1's l1: 1.14623\n[1500]\ttraining's l1: 1.03041\tvalid_1's l1: 1.08684\n[2000]\ttraining's l1: 0.975876\tvalid_1's l1: 1.04676\n[2500]\ttraining's l1: 0.933263\tvalid_1's l1: 1.01742\n[3000]\ttraining's l1: 0.897684\tvalid_1's l1: 0.994291\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.897684\tvalid_1's l1: 0.994291\nFold 2 started at Thu Jun 20 02:05:23 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 1.23369\tvalid_1's l1: 1.25801\n[1000]\ttraining's l1: 1.10276\tvalid_1's l1: 1.14434\n[1500]\ttraining's l1: 1.02772\tvalid_1's l1: 1.08488\n[2000]\ttraining's l1: 0.974929\tvalid_1's l1: 1.04635\n[2500]\ttraining's l1: 0.931313\tvalid_1's l1: 1.01582\n[3000]\ttraining's l1: 0.89601\tvalid_1's l1: 0.993103\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.89601\tvalid_1's l1: 0.993103\nFold 3 started at Thu Jun 20 02:25:18 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 1.2321\tvalid_1's l1: 1.25659\n[1000]\ttraining's l1: 1.10212\tvalid_1's l1: 1.14408\n[1500]\ttraining's l1: 1.02926\tvalid_1's l1: 1.0874\n[2000]\ttraining's l1: 0.974672\tvalid_1's l1: 1.04699\n[2500]\ttraining's l1: 0.931167\tvalid_1's l1: 1.01644\n[3000]\ttraining's l1: 0.895356\tvalid_1's l1: 0.993243\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.895356\tvalid_1's l1: 0.993243\nCV mean score: -0.1593, std: 0.0012.\n"
],
[
"X['oof_fc'] = result_dict_lgb_oof['oof']\nX_test['oof_fc'] = result_dict_lgb_oof['prediction']",
"_____no_output_____"
],
[
"good_columns = ['oof_fc',\n 'bond_lengths_mean_y',\n 'molecule_atom_index_0_dist_max',\n 'bond_lengths_mean_x',\n 'molecule_atom_index_0_dist_mean',\n 'molecule_atom_index_0_dist_std',\n 'molecule_couples',\n 'molecule_atom_index_0_y_1_std',\n 'molecule_dist_mean',\n 'molecule_dist_max',\n 'dist_y',\n 'molecule_atom_index_0_z_1_std',\n 'molecule_atom_index_1_dist_max',\n 'molecule_atom_index_1_dist_min',\n 'molecule_atom_index_0_x_1_std',\n 'molecule_atom_index_1_dist_std',\n 'molecule_atom_index_0_y_1_mean_div',\n 'y_0',\n 'molecule_atom_index_1_dist_mean',\n 'molecule_atom_1_dist_mean',\n 'x_0',\n 'dist_x',\n 'molecule_type_dist_std',\n 'dist_z',\n 'molecule_atom_index_1_dist_std_diff',\n 'molecule_type_dist_mean_diff',\n 'molecule_atom_index_0_dist_max_div',\n 'molecule_atom_1_dist_std',\n 'molecule_type_0_dist_std',\n 'z_0',\n 'molecule_type_dist_std_diff',\n 'molecule_atom_index_0_y_1_mean_diff',\n 'molecule_atom_index_0_dist_std_diff',\n 'molecule_atom_index_0_dist_mean_div',\n 'molecule_atom_index_0_dist_max_diff',\n 'x_1',\n 'molecule_type_dist_max',\n 'molecule_atom_index_0_dist_std_div',\n 'molecule_atom_index_0_dist_mean_diff',\n 'molecule_atom_1_dist_std_diff',\n 'molecule_atom_index_0_y_1_max_diff',\n 'z_1',\n 'molecule_atom_index_0_y_1_max',\n 'molecule_atom_index_0_y_1_mean',\n 'y_1',\n 'molecule_type_0_dist_std_diff',\n 'molecule_dist_min',\n 'molecule_atom_index_1_dist_std_div',\n 'molecule_atom_1_dist_min',\n 'molecule_atom_index_1_dist_max_diff','type']\n\nX = X[good_columns].copy()\nX_test = X_test[good_columns].copy()",
"_____no_output_____"
],
[
"def create_bunch_of_features(dtrain,dtest,cat_features):\n n_new_features = 0\n train_objs_num = len(dtrain)\n df_merge = pd.concat(objs=[dtrain, dtest], axis=0)\n \n for feature in cat_features:\n #Log Transform\n df_merge[feature+'_log'] = np.log (1 + df_merge[feature])\n n_new_features = n_new_features +1\n\n dtrain = df_merge[:train_objs_num]\n dtest = df_merge[train_objs_num:]\n del df_merge\n gc.collect()\n print('Features Created: {} \\nTotal Features {}'.format(n_new_features,len(dtrain.columns)))\n return dtrain,dtest\n\n#features = list(X.columns)\n#X, X_test = create_bunch_of_features(X,X_test,features)",
"_____no_output_____"
]
],
[
[
"# Checking Best Feature for Final Model",
"_____no_output_____"
]
],
[
[
"params = {'num_leaves': 128,\n 'min_child_samples': 79,\n 'objective': 'regression',\n 'max_depth': 9,\n 'learning_rate': 0.2,\n \"boosting_type\": \"gbdt\",\n \"subsample_freq\": 1,\n \"subsample\": 0.9,\n \"bagging_seed\": 11,\n \"metric\": 'mae',\n \"verbosity\": -1,\n 'reg_alpha': 0.1,\n 'reg_lambda': 0.3,\n 'colsample_bytree': 1.0\n }\n#result_dict_lgb2 = train_model_regression(X=X, X_test=X_test, y=y, params=params, folds=folds, model_type='lgb', eval_metric='group_mae', plot_feature_importance=True,\n# verbose=500, early_stopping_rounds=200, n_estimators=n_estimators_default)\n",
"_____no_output_____"
],
[
"#Best Features? \n''' \nfeature_importance = result_dict_lgb2['feature_importance']\nbest_features = feature_importance[['feature','importance']].groupby(['feature']).mean().sort_values(\n by='importance',ascending=False).iloc[:50,0:0].index.tolist()\nbest_features'''",
"_____no_output_____"
]
],
[
[
"<a id=\"id6\"></a> <br> \n# **6. Final Model** ",
"_____no_output_____"
],
[
"## Training models for each type",
"_____no_output_____"
]
],
[
[
"X_short = pd.DataFrame({'ind': list(X.index), 'type': X['type'].values, 'oof': [0] * len(X), 'target': y.values})\nX_short_test = pd.DataFrame({'ind': list(X_test.index), 'type': X_test['type'].values, 'prediction': [0] * len(X_test)})\nfor t in X['type'].unique():\n print(f'Training of type {t}')\n X_t = X.loc[X['type'] == t]\n X_test_t = X_test.loc[X_test['type'] == t]\n y_t = X_short.loc[X_short['type'] == t, 'target']\n result_dict_lgb3 = train_model_regression(X=X_t, X_test=X_test_t, y=y_t, params=params, folds=folds, model_type='lgb', eval_metric='group_mae', plot_feature_importance=False,\n verbose=500, early_stopping_rounds=200, n_estimators=n_estimators_default)\n X_short.loc[X_short['type'] == t, 'oof'] = result_dict_lgb3['oof']\n X_short_test.loc[X_short_test['type'] == t, 'prediction'] = result_dict_lgb3['prediction']",
"Training of type 0\nFold 1 started at Thu Jun 20 02:45:33 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 1.29748\tvalid_1's l1: 1.59058\n[1000]\ttraining's l1: 1.02789\tvalid_1's l1: 1.5289\n[1500]\ttraining's l1: 0.838422\tvalid_1's l1: 1.48906\n[2000]\ttraining's l1: 0.696969\tvalid_1's l1: 1.46429\n[2500]\ttraining's l1: 0.587396\tvalid_1's l1: 1.44756\n[3000]\ttraining's l1: 0.500955\tvalid_1's l1: 1.43526\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.500955\tvalid_1's l1: 1.43526\nFold 2 started at Thu Jun 20 02:51:52 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 1.28646\tvalid_1's l1: 1.59573\n[1000]\ttraining's l1: 1.01913\tvalid_1's l1: 1.53281\n[1500]\ttraining's l1: 0.831583\tvalid_1's l1: 1.49459\n[2000]\ttraining's l1: 0.692051\tvalid_1's l1: 1.47032\n[2500]\ttraining's l1: 0.582964\tvalid_1's l1: 1.45358\n[3000]\ttraining's l1: 0.496579\tvalid_1's l1: 1.44245\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.496579\tvalid_1's l1: 1.44245\nFold 3 started at Thu Jun 20 02:58:13 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 1.29479\tvalid_1's l1: 1.59866\n[1000]\ttraining's l1: 1.02568\tvalid_1's l1: 1.53532\n[1500]\ttraining's l1: 0.835688\tvalid_1's l1: 1.495\n[2000]\ttraining's l1: 0.693757\tvalid_1's l1: 1.46931\n[2500]\ttraining's l1: 0.585096\tvalid_1's l1: 1.45281\n[3000]\ttraining's l1: 0.498858\tvalid_1's l1: 1.44174\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.498858\tvalid_1's l1: 1.44174\nCV mean score: 0.3645, std: 0.0022.\nTraining of type 3\nFold 1 started at Thu Jun 20 03:04:23 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.248579\tvalid_1's l1: 0.370889\n[1000]\ttraining's l1: 0.171579\tvalid_1's l1: 0.351225\n[1500]\ttraining's l1: 0.126701\tvalid_1's l1: 0.344196\n[2000]\ttraining's l1: 0.0966678\tvalid_1's l1: 0.340717\n[2500]\ttraining's l1: 0.075151\tvalid_1's l1: 0.338827\n[3000]\ttraining's l1: 0.0587571\tvalid_1's l1: 0.337613\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.0587571\tvalid_1's l1: 0.337613\nFold 2 started at Thu Jun 20 03:08:17 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.246318\tvalid_1's l1: 0.372075\n[1000]\ttraining's l1: 0.170408\tvalid_1's l1: 0.352281\n[1500]\ttraining's l1: 0.125565\tvalid_1's l1: 0.344637\n[2000]\ttraining's l1: 0.09552\tvalid_1's l1: 0.340631\n[2500]\ttraining's l1: 0.0738895\tvalid_1's l1: 0.338508\n[3000]\ttraining's l1: 0.0581285\tvalid_1's l1: 0.337256\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.0581285\tvalid_1's l1: 0.337256\nFold 3 started at Thu Jun 20 03:12:11 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.248881\tvalid_1's l1: 0.374139\n[1000]\ttraining's l1: 0.171995\tvalid_1's l1: 0.354095\n[1500]\ttraining's l1: 0.127863\tvalid_1's l1: 0.346715\n[2000]\ttraining's l1: 0.0974023\tvalid_1's l1: 0.343148\n[2500]\ttraining's l1: 0.0755893\tvalid_1's l1: 0.341035\n[3000]\ttraining's l1: 0.0592109\tvalid_1's l1: 0.339608\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.0592109\tvalid_1's l1: 0.339608\nCV mean score: -1.0842, std: 0.0031.\nTraining of type 1\nFold 1 started at Thu Jun 20 03:16:10 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.241771\tvalid_1's l1: 0.646707\n[1000]\ttraining's l1: 0.102734\tvalid_1's l1: 0.631295\n[1500]\ttraining's l1: 0.0444509\tvalid_1's l1: 0.627393\n[2000]\ttraining's l1: 0.0198237\tvalid_1's l1: 0.626197\n[2500]\ttraining's l1: 0.00848043\tvalid_1's l1: 0.625763\n[3000]\ttraining's l1: 0.004035\tvalid_1's l1: 0.625594\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.004035\tvalid_1's l1: 0.625594\nFold 2 started at Thu Jun 20 03:17:08 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.242108\tvalid_1's l1: 0.651558\n[1000]\ttraining's l1: 0.103092\tvalid_1's l1: 0.635512\n[1500]\ttraining's l1: 0.0451569\tvalid_1's l1: 0.632018\n[2000]\ttraining's l1: 0.0201199\tvalid_1's l1: 0.631067\n[2500]\ttraining's l1: 0.00879591\tvalid_1's l1: 0.630561\n[3000]\ttraining's l1: 0.0042111\tvalid_1's l1: 0.630444\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.0042111\tvalid_1's l1: 0.630444\nFold 3 started at Thu Jun 20 03:18:03 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.253697\tvalid_1's l1: 0.654475\n[1000]\ttraining's l1: 0.105285\tvalid_1's l1: 0.636052\n[1500]\ttraining's l1: 0.0468043\tvalid_1's l1: 0.632225\n[2000]\ttraining's l1: 0.0208471\tvalid_1's l1: 0.631054\n[2500]\ttraining's l1: 0.00907399\tvalid_1's l1: 0.630626\n[3000]\ttraining's l1: 0.00432031\tvalid_1's l1: 0.630484\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.00432031\tvalid_1's l1: 0.630484\nCV mean score: -0.4639, std: 0.0037.\nTraining of type 4\nFold 1 started at Thu Jun 20 03:19:00 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.168725\tvalid_1's l1: 0.397807\n[1000]\ttraining's l1: 0.0837682\tvalid_1's l1: 0.387487\n[1500]\ttraining's l1: 0.0450821\tvalid_1's l1: 0.384592\n[2000]\ttraining's l1: 0.0250795\tvalid_1's l1: 0.383465\n[2500]\ttraining's l1: 0.0143791\tvalid_1's l1: 0.382916\n[3000]\ttraining's l1: 0.00854199\tvalid_1's l1: 0.382715\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.00854199\tvalid_1's l1: 0.382715\nFold 2 started at Thu Jun 20 03:20:50 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.174663\tvalid_1's l1: 0.403034\n[1000]\ttraining's l1: 0.086048\tvalid_1's l1: 0.390925\n[1500]\ttraining's l1: 0.0454008\tvalid_1's l1: 0.387586\n[2000]\ttraining's l1: 0.0251721\tvalid_1's l1: 0.386366\n[2500]\ttraining's l1: 0.0142851\tvalid_1's l1: 0.385912\n[3000]\ttraining's l1: 0.00852016\tvalid_1's l1: 0.385732\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.00852016\tvalid_1's l1: 0.385732\nFold 3 started at Thu Jun 20 03:22:42 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.167607\tvalid_1's l1: 0.395742\n[1000]\ttraining's l1: 0.0832846\tvalid_1's l1: 0.386114\n[1500]\ttraining's l1: 0.0447508\tvalid_1's l1: 0.383246\n[2000]\ttraining's l1: 0.0249328\tvalid_1's l1: 0.382155\n[2500]\ttraining's l1: 0.0143471\tvalid_1's l1: 0.381628\n[3000]\ttraining's l1: 0.00857127\tvalid_1's l1: 0.381386\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.00857127\tvalid_1's l1: 0.381386\nCV mean score: -0.9590, std: 0.0047.\nTraining of type 2\nFold 1 started at Thu Jun 20 03:24:34 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.658441\tvalid_1's l1: 0.76777\n[1000]\ttraining's l1: 0.534774\tvalid_1's l1: 0.716643\n[1500]\ttraining's l1: 0.452647\tvalid_1's l1: 0.689575\n[2000]\ttraining's l1: 0.391384\tvalid_1's l1: 0.673315\n[2500]\ttraining's l1: 0.34279\tvalid_1's l1: 0.662028\n[3000]\ttraining's l1: 0.303332\tvalid_1's l1: 0.654806\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.303332\tvalid_1's l1: 0.654806\nFold 2 started at Thu Jun 20 03:33:52 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.657635\tvalid_1's l1: 0.769486\n[1000]\ttraining's l1: 0.535111\tvalid_1's l1: 0.718452\n[1500]\ttraining's l1: 0.452732\tvalid_1's l1: 0.691325\n[2000]\ttraining's l1: 0.390796\tvalid_1's l1: 0.673658\n[2500]\ttraining's l1: 0.34257\tvalid_1's l1: 0.662388\n[3000]\ttraining's l1: 0.303085\tvalid_1's l1: 0.654742\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.303085\tvalid_1's l1: 0.654742\nFold 3 started at Thu Jun 20 03:43:15 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.662275\tvalid_1's l1: 0.7711\n[1000]\ttraining's l1: 0.537656\tvalid_1's l1: 0.717856\n[1500]\ttraining's l1: 0.454699\tvalid_1's l1: 0.689628\n[2000]\ttraining's l1: 0.392534\tvalid_1's l1: 0.672304\n[2500]\ttraining's l1: 0.343887\tvalid_1's l1: 0.661054\n[3000]\ttraining's l1: 0.303548\tvalid_1's l1: 0.65291\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.303548\tvalid_1's l1: 0.65291\nCV mean score: -0.4244, std: 0.0013.\nTraining of type 6\nFold 1 started at Thu Jun 20 03:52:34 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.316433\tvalid_1's l1: 0.416042\n[1000]\ttraining's l1: 0.234234\tvalid_1's l1: 0.388292\n[1500]\ttraining's l1: 0.18352\tvalid_1's l1: 0.376302\n[2000]\ttraining's l1: 0.148321\tvalid_1's l1: 0.370256\n[2500]\ttraining's l1: 0.121773\tvalid_1's l1: 0.366672\n[3000]\ttraining's l1: 0.101318\tvalid_1's l1: 0.36411\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.101318\tvalid_1's l1: 0.36411\nFold 2 started at Thu Jun 20 03:58:06 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.316116\tvalid_1's l1: 0.417274\n[1000]\ttraining's l1: 0.233325\tvalid_1's l1: 0.389466\n[1500]\ttraining's l1: 0.182957\tvalid_1's l1: 0.378067\n[2000]\ttraining's l1: 0.147678\tvalid_1's l1: 0.371791\n[2500]\ttraining's l1: 0.121385\tvalid_1's l1: 0.367939\n[3000]\ttraining's l1: 0.10088\tvalid_1's l1: 0.365497\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.10088\tvalid_1's l1: 0.365497\nFold 3 started at Thu Jun 20 04:03:40 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.317269\tvalid_1's l1: 0.416084\n[1000]\ttraining's l1: 0.234162\tvalid_1's l1: 0.38746\n[1500]\ttraining's l1: 0.184096\tvalid_1's l1: 0.376038\n[2000]\ttraining's l1: 0.148821\tvalid_1's l1: 0.369749\n[2500]\ttraining's l1: 0.122245\tvalid_1's l1: 0.365847\n[3000]\ttraining's l1: 0.101748\tvalid_1's l1: 0.363281\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.101748\tvalid_1's l1: 0.363281\nCV mean score: -1.0098, std: 0.0025.\nTraining of type 5\nFold 1 started at Thu Jun 20 04:09:20 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.709225\tvalid_1's l1: 0.796514\n[1000]\ttraining's l1: 0.602832\tvalid_1's l1: 0.751339\n[1500]\ttraining's l1: 0.525412\tvalid_1's l1: 0.723942\n[2000]\ttraining's l1: 0.46475\tvalid_1's l1: 0.704196\n[2500]\ttraining's l1: 0.415769\tvalid_1's l1: 0.690394\n[3000]\ttraining's l1: 0.374926\tvalid_1's l1: 0.679877\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.374926\tvalid_1's l1: 0.679877\nFold 2 started at Thu Jun 20 04:20:27 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.70802\tvalid_1's l1: 0.795716\n[1000]\ttraining's l1: 0.60196\tvalid_1's l1: 0.750761\n[1500]\ttraining's l1: 0.525244\tvalid_1's l1: 0.722729\n[2000]\ttraining's l1: 0.465151\tvalid_1's l1: 0.703545\n[2500]\ttraining's l1: 0.415711\tvalid_1's l1: 0.689358\n[3000]\ttraining's l1: 0.374916\tvalid_1's l1: 0.679016\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.374916\tvalid_1's l1: 0.679016\nFold 3 started at Thu Jun 20 04:31:48 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.706508\tvalid_1's l1: 0.791906\n[1000]\ttraining's l1: 0.601128\tvalid_1's l1: 0.748282\n[1500]\ttraining's l1: 0.524545\tvalid_1's l1: 0.721572\n[2000]\ttraining's l1: 0.46473\tvalid_1's l1: 0.702281\n[2500]\ttraining's l1: 0.415166\tvalid_1's l1: 0.687838\n[3000]\ttraining's l1: 0.374753\tvalid_1's l1: 0.677945\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.374753\tvalid_1's l1: 0.677945\nCV mean score: -0.3872, std: 0.0012.\nTraining of type 7\nFold 1 started at Thu Jun 20 04:43:13 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.135772\tvalid_1's l1: 0.283488\n[1000]\ttraining's l1: 0.073515\tvalid_1's l1: 0.273716\n[1500]\ttraining's l1: 0.0426127\tvalid_1's l1: 0.270277\n[2000]\ttraining's l1: 0.0257542\tvalid_1's l1: 0.268876\n[2500]\ttraining's l1: 0.0160195\tvalid_1's l1: 0.268179\n[3000]\ttraining's l1: 0.0103484\tvalid_1's l1: 0.267788\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.0103484\tvalid_1's l1: 0.267788\nFold 2 started at Thu Jun 20 04:45:29 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.136714\tvalid_1's l1: 0.284597\n[1000]\ttraining's l1: 0.0742557\tvalid_1's l1: 0.274226\n[1500]\ttraining's l1: 0.0435549\tvalid_1's l1: 0.27091\n[2000]\ttraining's l1: 0.0262644\tvalid_1's l1: 0.269316\n[2500]\ttraining's l1: 0.0164911\tvalid_1's l1: 0.268607\n[3000]\ttraining's l1: 0.0106238\tvalid_1's l1: 0.268256\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.0106238\tvalid_1's l1: 0.268256\nFold 3 started at Thu Jun 20 04:47:43 2019\nTraining until validation scores don't improve for 200 rounds.\n[500]\ttraining's l1: 0.137495\tvalid_1's l1: 0.284328\n[1000]\ttraining's l1: 0.0735836\tvalid_1's l1: 0.274279\n[1500]\ttraining's l1: 0.0427938\tvalid_1's l1: 0.271087\n[2000]\ttraining's l1: 0.0257932\tvalid_1's l1: 0.269622\n[2500]\ttraining's l1: 0.0160693\tvalid_1's l1: 0.268888\n[3000]\ttraining's l1: 0.0103497\tvalid_1's l1: 0.268518\nDid not meet early stopping. Best iteration is:\n[3000]\ttraining's l1: 0.0103497\tvalid_1's l1: 0.268518\nCV mean score: -1.3161, std: 0.0011.\n"
]
],
[
[
"\n",
"_____no_output_____"
],
[
"<a id=\"id7\"></a> <br> \n# **7. Submittion** ",
"_____no_output_____"
]
],
[
[
"#Training models for type\nsub['scalar_coupling_constant'] = X_short_test['prediction']\nsub.to_csv('submission_type.csv', index=False)\nsub.head()\n",
"_____no_output_____"
]
],
[
[
"<a id=\"ref\"></a> <br> \n# **8. References** \n\n[1] OOF Model: https://www.kaggle.com/adarshchavakula/out-of-fold-oof-model-cross-validation<br>\n[2] Using Meta Features: https://www.kaggle.com/artgor/using-meta-features-to-improve-model<br>\n[3] Lot of Features: https://towardsdatascience.com/understanding-feature-engineering-part-1-continuous-numeric-data-da4e47099a7b <br>\n[4] Angle Feature: https://www.kaggle.com/kmat2019/effective-feature <br>\n[5] Recovering bonds from structure: https://www.kaggle.com/aekoch95/bonds-from-structure-data <br>\n\n<h3 style=\"color:red\">If this kernel helps you, up vote to keep me motivated 😁<br>Thanks!</h3>",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4ae782fae1c67b759f405cb7df4fc48be4fdf585
| 8,239 |
ipynb
|
Jupyter Notebook
|
d2l-en/pytorch/chapter_preliminaries/lookup-api.ipynb
|
gr8khan/d2lai
|
7c10432f38c80e86978cd075d0024902b47842a0
|
[
"MIT"
] | null | null | null |
d2l-en/pytorch/chapter_preliminaries/lookup-api.ipynb
|
gr8khan/d2lai
|
7c10432f38c80e86978cd075d0024902b47842a0
|
[
"MIT"
] | null | null | null |
d2l-en/pytorch/chapter_preliminaries/lookup-api.ipynb
|
gr8khan/d2lai
|
7c10432f38c80e86978cd075d0024902b47842a0
|
[
"MIT"
] | null | null | null | 39.233333 | 1,798 | 0.604078 |
[
[
[
"# Documentation\n",
"_____no_output_____"
],
[
"Due to constraints on the length of this book, we cannot possibly introduce every single PyTorch function and class (and you probably would not want us to). The API documentation and additional tutorials and examples provide plenty of documentation beyond the book. In this section we provide you with some guidance to exploring the PyTorch API.\n",
"_____no_output_____"
],
[
"## Finding All the Functions and Classes in a Module\n\nIn order to know which functions and classes can be called in a module, we\ninvoke the `dir` function. For instance, we can query all properties in the\nmodule for generating random numbers:\n",
"_____no_output_____"
]
],
[
[
"import torch\nprint(dir(torch.distributions))",
"['AbsTransform', 'AffineTransform', 'Bernoulli', 'Beta', 'Binomial', 'CatTransform', 'Categorical', 'Cauchy', 'Chi2', 'ComposeTransform', 'ContinuousBernoulli', 'CorrCholeskyTransform', 'Dirichlet', 'Distribution', 'ExpTransform', 'Exponential', 'ExponentialFamily', 'FisherSnedecor', 'Gamma', 'Geometric', 'Gumbel', 'HalfCauchy', 'HalfNormal', 'Independent', 'Kumaraswamy', 'LKJCholesky', 'Laplace', 'LogNormal', 'LogisticNormal', 'LowRankMultivariateNormal', 'LowerCholeskyTransform', 'MixtureSameFamily', 'Multinomial', 'MultivariateNormal', 'NegativeBinomial', 'Normal', 'OneHotCategorical', 'OneHotCategoricalStraightThrough', 'Pareto', 'Poisson', 'PowerTransform', 'RelaxedBernoulli', 'RelaxedOneHotCategorical', 'SigmoidTransform', 'SoftmaxTransform', 'StackTransform', 'StickBreakingTransform', 'StudentT', 'TanhTransform', 'Transform', 'TransformedDistribution', 'Uniform', 'VonMises', 'Weibull', '__all__', '__builtins__', '__cached__', '__doc__', '__file__', '__loader__', '__name__', '__package__', '__path__', '__spec__', 'bernoulli', 'beta', 'biject_to', 'binomial', 'categorical', 'cauchy', 'chi2', 'constraint_registry', 'constraints', 'continuous_bernoulli', 'dirichlet', 'distribution', 'exp_family', 'exponential', 'fishersnedecor', 'gamma', 'geometric', 'gumbel', 'half_cauchy', 'half_normal', 'identity_transform', 'independent', 'kl', 'kl_divergence', 'kumaraswamy', 'laplace', 'lkj_cholesky', 'log_normal', 'logistic_normal', 'lowrank_multivariate_normal', 'mixture_same_family', 'multinomial', 'multivariate_normal', 'negative_binomial', 'normal', 'one_hot_categorical', 'pareto', 'poisson', 'register_kl', 'relaxed_bernoulli', 'relaxed_categorical', 'studentT', 'transform_to', 'transformed_distribution', 'transforms', 'uniform', 'utils', 'von_mises', 'weibull']\n"
]
],
[
[
"Generally, we can ignore functions that start and end with `__` (special objects in Python) or functions that start with a single `_`(usually internal functions). Based on the remaining function or attribute names, we might hazard a guess that this module offers various methods for generating random numbers, including sampling from the uniform distribution (`uniform`), normal distribution (`normal`), and multinomial distribution (`multinomial`).\n\n## Finding the Usage of Specific Functions and Classes\n\nFor more specific instructions on how to use a given function or class, we can invoke the `help` function. As an example, let us explore the usage instructions for tensors' `ones` function.\n",
"_____no_output_____"
]
],
[
[
"help(torch.ones)",
"Help on built-in function ones:\n\nones(...)\n ones(*size, *, out=None, dtype=None, layout=torch.strided, device=None, requires_grad=False) -> Tensor\n \n Returns a tensor filled with the scalar value `1`, with the shape defined\n by the variable argument :attr:`size`.\n \n Args:\n size (int...): a sequence of integers defining the shape of the output tensor.\n Can be a variable number of arguments or a collection like a list or tuple.\n out (Tensor, optional): the output tensor.\n dtype (:class:`torch.dtype`, optional): the desired data type of returned tensor.\n Default: if ``None``, uses a global default (see :func:`torch.set_default_tensor_type`).\n layout (:class:`torch.layout`, optional): the desired layout of returned Tensor.\n Default: ``torch.strided``.\n device (:class:`torch.device`, optional): the desired device of returned tensor.\n Default: if ``None``, uses the current device for the default tensor type\n (see :func:`torch.set_default_tensor_type`). :attr:`device` will be the CPU\n for CPU tensor types and the current CUDA device for CUDA tensor types.\n requires_grad (bool, optional): If autograd should record operations on the\n returned tensor. Default: ``False``.\n \n Example::\n \n >>> torch.ones(2, 3)\n tensor([[ 1., 1., 1.],\n [ 1., 1., 1.]])\n \n >>> torch.ones(5)\n tensor([ 1., 1., 1., 1., 1.])\n\n"
]
],
[
[
"From the documentation, we can see that the `ones` function creates a new tensor with the specified shape and sets all the elements to the value of 1. Whenever possible, you should run a quick test to confirm your interpretation:\n",
"_____no_output_____"
]
],
[
[
"torch.ones(4)",
"_____no_output_____"
]
],
[
[
"In the Jupyter notebook, we can use `?` to display the document in another\nwindow. For example, `list?` will create content that is almost\nidentical to `help(list)`, displaying it in a new browser\nwindow. In addition, if we use two question marks, such as\n`list??`, the Python code implementing the function will also be\ndisplayed.\n\n\n## Summary\n\n* The official documentation provides plenty of descriptions and examples that are beyond this book.\n* We can look up documentation for the usage of an API by calling the `dir` and `help` functions, or `?` and `??` in Jupyter notebooks.\n\n\n## Exercises\n\n1. Look up the documentation for any function or class in the deep learning framework. Can you also find the documentation on the official website of the framework?\n",
"_____no_output_____"
],
[
"[Discussions](https://discuss.d2l.ai/t/39)\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4ae799f56e68caa4e872d44a7ef40da0b2f7f1e3
| 424,967 |
ipynb
|
Jupyter Notebook
|
plotting_notebooks/Figure_3/iPSC_Sarkar2019_overview.ipynb
|
single-cell-genetics/singlecell_neuroseq_paper
|
83e6f36b1b4c1baf9b64cacee8ff85c9cf3bc076
|
[
"Apache-2.0"
] | 12 |
2020-05-28T11:39:48.000Z
|
2021-11-04T04:37:31.000Z
|
plotting_notebooks/Figure_3/iPSC_Sarkar2019_overview.ipynb
|
single-cell-genetics/singlecell_neuroseq_paper
|
83e6f36b1b4c1baf9b64cacee8ff85c9cf3bc076
|
[
"Apache-2.0"
] | 1 |
2021-05-24T21:13:06.000Z
|
2021-05-24T21:13:06.000Z
|
plotting_notebooks/Figure_3/iPSC_Sarkar2019_overview.ipynb
|
single-cell-genetics/singlecell_neuroseq_paper
|
83e6f36b1b4c1baf9b64cacee8ff85c9cf3bc076
|
[
"Apache-2.0"
] | 4 |
2021-03-06T14:54:18.000Z
|
2021-12-20T17:54:02.000Z
| 512.008434 | 307,252 | 0.932988 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport os\nfrom matplotlib.pyplot import *\nfrom IPython.display import display, HTML\nimport glob\nimport scanpy as sc\nimport pandas as pd\nimport seaborn as sns\nimport scipy.stats\n\n%matplotlib inline\n",
"/nfs/software/stegle/users/acuomo/conda-envs/myscanpy/lib/python3.7/site-packages/anndata/_core/anndata.py:21: FutureWarning: pandas.core.index is deprecated and will be removed in a future version. The public classes are available in the top-level namespace.\n from pandas.core.index import RangeIndex\n"
],
[
"file = '/nfs/leia/research/stegle/dseaton/hipsci/singlecell_neuroseq/data/ipsc_singlecell_analysis/sarkar2019_yoruba_ipsc/version0/sarkar2019_yoruba_ipsc.scanpy.dimreduction.harmonyPCA.clustered.h5'\nadata_clustered = sc.read(file)\n\nfile = '/nfs/leia/research/stegle/dseaton/hipsci/singlecell_neuroseq/data/ipsc_singlecell_analysis/sarkar2019_yoruba_ipsc/version0/sarkar2019_yoruba_ipsc.scanpy.h5'\nadatafull = sc.read(file)\n\nin_dir = os.path.dirname(file)\n\nadatafull.obs['cluster_id'] = adata_clustered.obs['louvain'].astype(str)\nadatafull.obsm['X_umap'] = adata_clustered.obsm['X_umap']",
"WARNING: Your filename has more than two extensions: ['.scanpy', '.dimreduction', '.harmonyPCA', '.clustered', '.h5'].\nOnly considering the two last: ['.clustered', '.h5'].\nWARNING: Your filename has more than two extensions: ['.scanpy', '.dimreduction', '.harmonyPCA', '.clustered', '.h5'].\nOnly considering the two last: ['.clustered', '.h5'].\n"
],
[
"adatafull.obs['day'] = 'day0'\nadatafull.obs['donor_long_id'] = adatafull.obs['chip_id']",
"_____no_output_____"
],
[
"adatafull.obs.head()",
"_____no_output_____"
],
[
"#subsample\n\nfraction = 1.0\nadata = sc.pp.subsample(adatafull, fraction, copy=True)\n\nadata.raw = adata",
"_____no_output_____"
],
[
"fig_format = 'png'\n# fig_format = 'pdf'\nsc.set_figure_params(dpi_save=200,format=fig_format)\n#rcParams['figure.figsize'] = 5,4\nrcParams['figure.figsize'] = 5,4",
"_____no_output_____"
],
[
"plotting_fcn = sc.pl.umap",
"_____no_output_____"
],
[
"plotting_fcn(adata, color='cluster_id',size=10)",
"... storing 'cluster_id' as categorical\n... storing 'day' as categorical\n"
],
[
"adata.var",
"_____no_output_____"
],
[
"# gene_list = ['NANOG','SOX2','POU5F1','UTF1','SP8']\n# ensembl gene ids correspoinding\n# gene_list = ['ENSG00000111704','ENSG00000181449','ENSG00000204531','ENSG00000171794','ENSG00000164651']\ngene_list = ['ENSG00000111704','ENSG00000181449','ENSG00000204531','ENSG00000171794','ENSG00000166863']\nsc.pl.stacked_violin(adata, gene_list, groupby='cluster_id', figsize=(5,4))",
"_____no_output_____"
],
[
"df = adata.obs.groupby(['donor_long_id','experiment','cluster_id'])[['day']].count().fillna(0.0).rename(columns={'day':'count'})\ntotal_counts = adata.obs.groupby(['donor_long_id','experiment'])[['day']].count().rename(columns={'day':'total_count'})\n\ndf = df.reset_index()\n\n#.join(donor_total_counts)\n\ndf['f_cells'] = df.apply(lambda x: x['count']/total_counts.loc[(x['donor_long_id'],x['experiment']),'total_count'], axis=1)\n\ndf = df.dropna()\ndf.head()",
"_____no_output_____"
],
[
"mydir = \"/hps/nobackup/stegle/users/acuomo/all_scripts/sc_neuroseq/iPSC_scanpy/\"\n\nfilename = mydir + 'Sarkar_cluster_cell_fractions_by_donor_experiment.csv'\ndf.to_csv(filename)",
"_____no_output_____"
],
[
"sc.tl.rank_genes_groups(adata, groupby='cluster_id', n_genes=1e6)",
"_____no_output_____"
],
[
"# group_names = pval_df.columns\ngroup_names = [str(x) for x in range(4)]\ndf_list = []\nfor group_name in group_names:\n column_names = ['names','pvals','pvals_adj','logfoldchanges','scores']\n data = [pd.DataFrame(adata.uns['rank_genes_groups'][col])[group_name] for col in column_names]\n temp_df = pd.DataFrame(data, index=column_names).transpose()\n temp_df['cluster_id'] = group_name\n df_list.append(temp_df)\n \ndiff_expression_df = pd.concat(df_list)\n\ndiff_expression_df.head()",
"_____no_output_____"
],
[
"diff_exp_file = mydir + 'Sarkar2019' + '.cluster_expression_markers.tsv'\n\ndiff_expression_df.to_csv(diff_exp_file, sep='\\t', index=False)\ndiff_expression_df.query('cluster_id==\"0\"').to_csv(diff_exp_file.replace('.tsv','.cluster0.tsv'), sep='\\t', index=False)\ndiff_expression_df.query('cluster_id==\"1\"').to_csv(diff_exp_file.replace('.tsv','.cluster1.tsv'), sep='\\t', index=False)\ndiff_expression_df.query('cluster_id==\"2\"').to_csv(diff_exp_file.replace('.tsv','.cluster2.tsv'), sep='\\t', index=False)\ndiff_expression_df.query('cluster_id==\"3\"').to_csv(diff_exp_file.replace('.tsv','.cluster3.tsv'), sep='\\t', index=False)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4ae7aef47305234ab64eab4eea2ff36da34b0e29
| 11,179 |
ipynb
|
Jupyter Notebook
|
examples/heatmap_slicer.ipynb
|
jrussell25/mpl-interactions
|
fd41f510a4c813befa984df76a35e240c382963a
|
[
"BSD-3-Clause"
] | null | null | null |
examples/heatmap_slicer.ipynb
|
jrussell25/mpl-interactions
|
fd41f510a4c813befa984df76a35e240c382963a
|
[
"BSD-3-Clause"
] | null | null | null |
examples/heatmap_slicer.ipynb
|
jrussell25/mpl-interactions
|
fd41f510a4c813befa984df76a35e240c382963a
|
[
"BSD-3-Clause"
] | null | null | null | 25.291855 | 278 | 0.508274 |
[
[
[
"%matplotlib widget\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom mpl_interactions import heatmap_slicer",
"_____no_output_____"
]
],
[
[
"## Comparing heatmaps\n\nSometimes I find myself wanting to compare horizontal or vertical slices across two different heatmaps with the same shape. The function `heatmap_slicer` makes this easy and should work for any number of heatmaps from 1 to many (likely not all the way $\\inf$ though). \n\nThe most important options to play with are `slices = {'both', 'vertical', 'horizontal'}`, and `interaction_type = {'move', 'click'}`",
"_____no_output_____"
]
],
[
[
"x = np.linspace(0,np.pi,100)\ny = np.linspace(0,10,200)\nX,Y = np.meshgrid(x,y)\ndata1 = np.sin(X)+np.exp(np.cos(Y))\ndata2 = np.cos(X)+np.exp(np.sin(Y))\nfig,axes = heatmap_slicer(x,y,(data1,data2),slices='both',heatmap_names=('dataset 1','dataset 2'),labels=('Some wild X variable','Y axis'),interaction_type='move')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4ae7b0b5ebbce99bef8d6c66a90647381f94f691
| 399,891 |
ipynb
|
Jupyter Notebook
|
jupyterbook_hnishi/data-analysis/pca.ipynb
|
hnishi/jupyterbook-hnishi
|
640460894973fc3a5b5cd0a46152f8db5d8b2dea
|
[
"MIT"
] | null | null | null |
jupyterbook_hnishi/data-analysis/pca.ipynb
|
hnishi/jupyterbook-hnishi
|
640460894973fc3a5b5cd0a46152f8db5d8b2dea
|
[
"MIT"
] | null | null | null |
jupyterbook_hnishi/data-analysis/pca.ipynb
|
hnishi/jupyterbook-hnishi
|
640460894973fc3a5b5cd0a46152f8db5d8b2dea
|
[
"MIT"
] | null | null | null | 141.855623 | 52,068 | 0.87081 |
[
[
[
"<a href=\"https://colab.research.google.com/github/hnishi/jupyterbook-hnishi/blob/colab-dev/pca.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# 主成分分析 (主成分解析、Principal component analysis : PCA)",
"_____no_output_____"
],
[
"## 概要\n\n- 主成分分析は、教師なし線形変換法の1つ\n - データセットの座標軸を、データの分散が最大になる方向に変換し、元の次元と同じもしくは、元の次元数より低い新しい特徴部分空間を作成する手法\n- 主なタスク\n - 次元削減\n- 次元削減を行うことで以下の目的を達成できる\n - 特徴抽出\n - データの可視化\n- 次元削減を行うメリット\n - 計算コスト(計算時間、メモリ使用量)を削減できる\n - 特徴量を削減したことによる情報の喪失をできるだけ小さくする\n - モデルを簡素化できる(パラメータが減る)ため、オーバーフィッティングを防げる\n - 人間が理解可能な空間にデータを投影することができる(非常に高次元な空間を、身近な3次元、2次元に落とし込むことができる)",
"_____no_output_____"
],
[
"## 応用例\n\n- タンパク質分子の立体構造モデルの構造空間の次元削減と可視化\n- タンパク質の全原子モデルの立体構造は、分子内に含まれる原子の座標情報で表すことができる (原子数 × 3 (x, y, z) 次元のベクトル)\n\n以下は、タンパク質の分子シミュレーションで使われるモデルの1例。 \n(紫色とオレンジ色で表されたリボンモデルがタンパク質で、周りに水とイオンが表示されている) \n(この場合、3547 個の原子 --> 10641 次元)\n\n<img src=\"https://github.com/hnishi/hnishi_da_handson/blob/master/images/cdr-h3-pbc.png?raw=true\" width=\"50%\">\n\n主成分分析により、この立体構造空間を、2次元空間に投影することができる。 \n以下は、その投影に対して自由エネルギーを計算した図。\n\n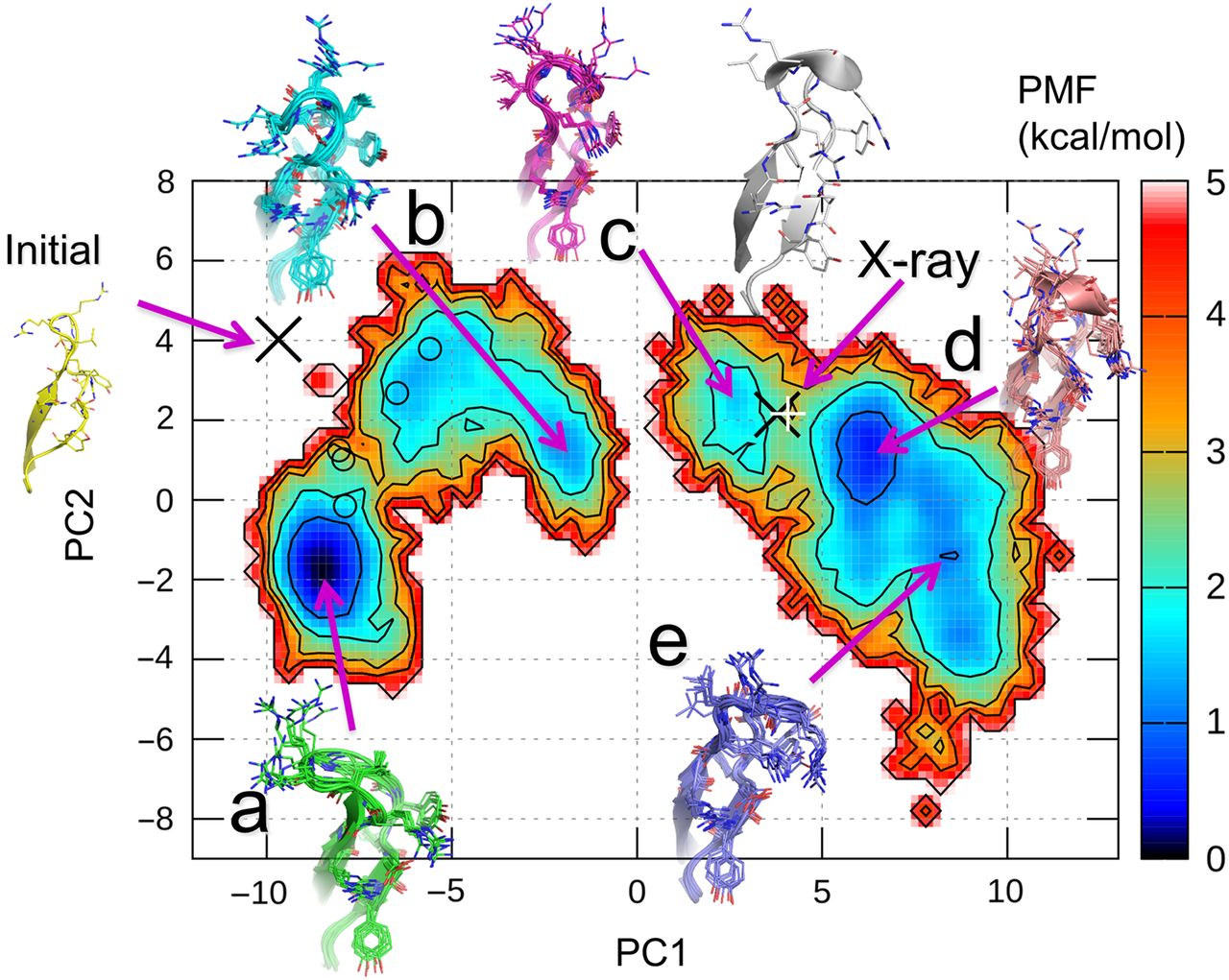\n\n2次元空間上の1点が、1つの立体構造を表している。 \nつまり、この例では、もともと10641次元あった空間を2次元にまで削減している。\n\nRef) [Nishigami, H., Kamiya, N., & Nakamura, H. (2016). Revisiting antibody modeling assessment for CDR-H3 loop. Protein Engineering, Design and Selection, 29(11), 477-484.](https://academic.oup.com/peds/article/29/11/477/2462452)",
"_____no_output_____"
],
[
"## 主成分分析 (PCA) が行う座標変換のイメージ\n\n以下は、PCAが行う座標変換の例\n\n$x_1$ , $x_2$ は、データセットの元々の座標軸であり、 \nPC1, PC2 は座標変換後に得られる新しい座標軸、主成分1、主成分2 である (Principal Components)。 \n\n",
"_____no_output_____"
],
[
"<img src=\"https://github.com/rasbt/python-machine-learning-book-2nd-edition/blob/master/code/ch05/images/05_01.png?raw=true\" width=\"50%\">",
"_____no_output_____"
],
[
"\n- PCA は、高次元データにおいて分散が最大となる方向を見つけ出し、座標を変換する (これはつまり、すべての主成分が、他の主成分と相関がない(直交する) ように座標変換している)\n- 最初の主成分 (PC1) の分散が最大となる",
"_____no_output_____"
],
[
"## 主成分分析の主要な手順\n\nd 次元のデータを k 次元に削減する場合\n\n1. d 次元のデータの標準化(特徴量間のスケールが異なる場合のみ)\n1. 分散共分散行列の作成\n1. 分散共分散行列の固有値と固有ベクトルを求める\n1. 固有値を降順にソートして、固有ベクトルをランク付けする\n1. 最も大きい k 個の固有値に対応する k 個の固有ベクトルを選択 (k ≦ d)\n1. k 個の固有ベクトルから射影(変換)行列 W を作成\n1. 射影(変換)行列を使って d 次元の入力データセットを新しい k 次元の特徴部分空間を取得する\n\n---\n\n固有値問題を解くことで、線形独立な基底ベクトルを得ることができる。 \n詳細は、線形代数の書籍等を参考にする(ここでは詳細な解説をしない)。\n\n参考) \n\nhttps://dora.bk.tsukuba.ac.jp/~takeuchi/?%E7%B7%9A%E5%BD%A2%E4%BB%A3%E6%95%B0II%2F%E5%9B%BA%E6%9C%89%E5%80%A4%E5%95%8F%E9%A1%8C%E3%83%BB%E5%9B%BA%E6%9C%89%E7%A9%BA%E9%96%93%E3%83%BB%E3%82%B9%E3%83%9A%E3%82%AF%E3%83%88%E3%83%AB%E5%88%86%E8%A7%A3",
"_____no_output_____"
],
[
"## python による PCA の実行\n\n以下、Python を使った PCA の実行を順番に見ていく。 \nその後、scikit-learn ライブラリを使った PCA の簡単で効率のよい実装を見る。 \n\n### データセット\n\n- データセットは、 [Wine](https://archive.ics.uci.edu/ml/datasets/Wine) というオープンソースのデータセットを使う。 \n- 178 行のワインサンプルと、それらの化学的性質を表す 13 列の特徴量で構成されている。\n- それぞれのサンプルに、クラス 1, 2, 3 のいずれかがラベルされており、 \nイタリアの同じ地域で栽培されている異なる品種のブドウを表している \n(PCA は教師なし学習なので、学習時にラベルは使わない)。\n",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\n%matplotlib inline",
"_____no_output_____"
],
[
"import pandas as pd\n\n# df_wine = pd.read_csv('https://archive.ics.uci.edu/ml/'\n# 'machine-learning-databases/wine/wine.data',\n# header=None)\n\n# if the Wine dataset is temporarily unavailable from the\n# UCI machine learning repository, un-comment the following line\n# of code to load the dataset from a local path:\n\ndf_wine = pd.read_csv('https://github.com/rasbt/python-machine-learning-book-2nd-edition'\n '/raw/master/code/ch05/wine.data',\n header=None)\n\ndf_wine.columns = ['Class label', 'Alcohol', 'Malic acid', 'Ash',\n 'Alcalinity of ash', 'Magnesium', 'Total phenols',\n 'Flavanoids', 'Nonflavanoid phenols', 'Proanthocyanins',\n 'Color intensity', 'Hue',\n 'OD280/OD315 of diluted wines', 'Proline']\n\ndf_wine.head()",
"_____no_output_____"
]
],
[
[
"Wine データセットの先頭 5 行のデータは上記。",
"_____no_output_____"
]
],
[
[
"for i_label in df_wine['Class label'].unique():\n print('label:', i_label)\n print('shape:', df_wine[df_wine['Class label'] == i_label].shape)",
"label: 1\nshape: (59, 14)\nlabel: 2\nshape: (71, 14)\nlabel: 3\nshape: (48, 14)\n"
]
],
[
[
"ラベルの数はおおよそ揃っている。 \n次に、ラベルごとにデータの分布を見てみる。",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n\nfor i_feature in df_wine.columns:\n if i_feature == 'Class label': continue\n print('feature: ' + str(i_feature))\n # ヒストグラムの描画\n plt.hist(df_wine[df_wine['Class label'] == 1][i_feature], alpha=0.5, bins=20, label=\"1\") \n plt.hist(df_wine[df_wine['Class label'] == 2][i_feature], alpha=0.3, bins=20, label=\"2\", color='r')\n plt.hist(df_wine[df_wine['Class label'] == 3][i_feature], alpha=0.1, bins=20, label=\"3\", color='g') \n plt.legend(loc=\"upper left\", fontsize=13) # 凡例表示\n plt.show()",
"feature: Alcohol\n"
]
],
[
[
"データを70%のトレーニングと30%のテストサブセットに分割する。",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\nX, y = df_wine.iloc[:, 1:].values, df_wine.iloc[:, 0].values\n\nX_train, X_test, y_train, y_test = \\\n train_test_split(X, y, test_size=0.3, \n stratify=y,\n random_state=0)",
"_____no_output_____"
]
],
[
[
"データの標準化を行う。",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import StandardScaler\n\nsc = StandardScaler()\nX_train_std = sc.fit_transform(X_train) # トレーニングセットの標準偏差と平均値を使って、標準化を行う\nX_test_std = sc.transform(X_test) # \"トレーニングセット\"の標準偏差と平均値を使って、標準化を行う\n\n# いずれの特徴量も、値がおおよそ、-1 から +1 の範囲にあることを確認する。\nprint('standardize train', X_train_std[0:2])\nprint('standardize test', X_test_std[0:2])",
"standardize train [[ 0.71225893 2.22048673 -0.13025864 0.05962872 -0.50432733 -0.52831584\n -1.24000033 0.84118003 -1.05215112 -0.29218864 -0.20017028 -0.82164144\n -0.62946362]\n [ 0.88229214 -0.70457155 1.17533605 -0.09065504 2.34147876 1.01675879\n 0.66299475 1.0887425 -0.49293533 0.13152077 1.33982592 0.54931269\n 1.47568796]]\nstandardize test [[ 0.89443737 -0.38811788 1.10073064 -0.81201711 1.13201117 1.09807851\n 0.71204102 0.18101342 0.06628046 0.51285923 0.79629785 0.44829502\n 1.90593792]\n [-1.04879931 -0.77299397 0.54119006 -0.24093881 0.3494145 -0.70721922\n -0.30812129 0.67613838 -1.03520519 -0.90656727 2.24570604 -0.56188171\n -1.22874035]]\n"
]
],
[
[
"---\n\n**注意**\n\nテストデータの標準化の際に、テストデータの標準偏差と平均値を用いてはいけない(トレーニングデータの標準偏差と平均値を用いること)。 \nまた、ここで求めた標準偏差と平均値は、未知のデータを標準化する際にも再使用するので、記録しておくこと。 \n(今回は、ノートブックだけで完結するので、外部ファイル等に記録しなくても問題ない) ",
"_____no_output_____"
],
[
"- 分散共分散行列を作成\n- 固有値問題を解いて、固有値と固有ベクトルを求める\n\n固有値問題とは、以下の条件を満たす、固有ベクトル $v$ と、スカラー値である固有値 $\\lambda$ を求める問題のことである \n(詳細は線形代数の書籍等を参考)。\n\n$$\\Sigma v=\\lambda v$$\n\n$\\Sigma$ は分散共分散行列である(総和記号ではないことに注意)。 \n \n分散共分散行列に関しては、 [前回の資料](https://github.com/hnishi/hnishi_da_handson/blob/master/da_handson_basic_statistic_values.ipynb) を参照。\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport seaborn as sns\n\ncov_mat = np.cov(X_train_std.T)\n\n# 共分散行列のヒートマップ\ndf = pd.DataFrame(cov_mat, index=df_wine.columns[1:], columns=df_wine.columns[1:])\nax = sns.heatmap(df, cmap=\"YlGnBu\") ",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"# 固有値問題を解く(固有値分解)\neigen_vals, eigen_vecs = np.linalg.eigh(cov_mat)\n\nprint('\\nEigenvalues \\n%s' % eigen_vals)\nprint('\\nShape of eigen vectors\\n', eigen_vecs.shape)",
"\nEigenvalues \n[0.10754642 0.15362835 0.1808613 0.21357215 0.3131368 0.34650377\n 0.51828472 0.6620634 0.84166161 0.96120438 1.54845825 2.41602459\n 4.84274532]\n\nShape of eigen vectors\n (13, 13)\n"
]
],
[
[
"**注意**: \n\n固有値分解(固有分解とも呼ばれる)する numpy の関数は、\n\n- [`numpy.linalg.eig`](http://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.eig.html)\n- [`numpy.linalg.eigh`](http://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.eigh.html) \n\nがある。 \n`numpy.linalg.eig` は対称正方行列と非対称正方行列を固有値分解する関数。複素数の固有値を返すことがある。 \n`numpy.linalg.eigh` はエルミート行列(各成分が複素数で、転置させた各成分の虚部の値の正負を反転させたものがもとの行列と等しくなる行列)を固有値分解する関数。常に実数の固有値を返す。 \n\n分散共分散行列は、対称正方行列であり、虚数部が 0 のエルミート行列でもある。 \n対称正方行列の操作では、`numpy.linalg.eigh` の方が数値的に安定しているらしい。\n\nRef) *Python Machine Learning 2nd Edition* by [Sebastian Raschka](https://sebastianraschka.com), Packt Publishing Ltd. 2017.\n",
"_____no_output_____"
],
[
"## 全分散と説明分散(Total and explained variance)\n\n- 固有値の大きさは、データに含まれる情報(分散)の大きさに対応している\n- 主成分j (PCj: j-th principal component) に対応する固有値 $\\lambda_j$ の分散説明率(寄与率、contribution ratio/propotion とも呼ばれる)は以下のように定義される。\n\n$$\\dfrac {\\lambda _{j}}{\\sum ^{d}_{j=1}\\lambda j}$$ \n\n$\\lambda_j$ は、j 番目の固有値、d は全固有値の数(元々の特徴量の数/次元数)。 \n\n分散説明率を見ることで、その主成分が特徴量全体がもつ情報のうち、どれぐらいの情報を表すことができているかを確認できる。 \n以下に、分散説明率と、その累積和をプロットする。",
"_____no_output_____"
]
],
[
[
"# 固有値の合計\ntot = sum(eigen_vals)\n# 分散説明率の配列を作成\nvar_exp = [(i / tot) for i in sorted(eigen_vals, reverse=True)]\n# 分散説明率の累積和を作成\ncum_var_exp = np.cumsum(var_exp)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\n\nplt.bar(range(1, 14), var_exp, alpha=0.5, align='center',\n label='individual explained variance')\nplt.step(range(1, 14), cum_var_exp, where='mid',\n label='cumulative explained variance')\nplt.ylabel('Explained variance ratio')\nplt.xlabel('Principal component index')\nplt.legend(loc='best')\nplt.tight_layout()\n# plt.savefig('images/05_02.png', dpi=300)\nplt.show()",
"_____no_output_____"
]
],
[
[
"グラフから以下のことがわかる。\n\n- 最初の主成分だけで、全体の約 4 割の分散を占めている\n- 2 つの主成分も用いるだけで、もともとあった特徴量全体の約 6 割を説明できている",
"_____no_output_____"
],
[
"## 因子負荷量 (Factor loading)\n\n各主成分軸の意味を知るためには、因子負荷量を見れば良い。\n因子負荷量とは、主成分軸における値と、変換前の軸における値との間の相関値のことである。\n\nこの相関が大きくなるほど、その特徴量が、その主成分に寄与する程度が大きいということを意味している。\n\nRef: https://statistics.co.jp/reference/software_R/statR_9_principal.pdf",
"_____no_output_____"
],
[
"## 特徴変換 (Feature transformation)\n\n射影(変換)行列を取得し、適用して特徴変換を行う。\n\n---\n\n$X' = XW$ \n\n$X'$ : 射影(変換)後の座標(行列) \n$X$ : もともとの座標(行列) \n$W$ : 射影(変換)行列 \n \n$W$ は、次元削減後の次元数の固有ベクトルから構成される。 \n\n$W = [v_1 v_2 ... v_k] \\in \\mathbb{R} ^{n\\times k}$ ",
"_____no_output_____"
]
],
[
[
"# Make a list of (eigenvalue, eigenvector) tuples\neigen_pairs = [(np.abs(eigen_vals[i]), eigen_vecs[:, i])\n for i in range(len(eigen_vals))]\n\n# Sort the (eigenvalue, eigenvector) tuples from high to low\neigen_pairs.sort(key=lambda k: k[0], reverse=True)",
"_____no_output_____"
]
],
[
[
"### まずは、次元削減を行わずに、13 次元 --> 13 次元の座標変換を見てみる\n\n$X' = XW$ \n\n$W = [v_1 v_2 ... v_13] \\in \\mathbb{R} ^{13\\times 13}$ \n$x \\in \\mathbb{R} ^{13}$ \n$x' \\in \\mathbb{R} ^{13}$ ",
"_____no_output_____"
]
],
[
[
"# 変換行列 w の作成\nw = eigen_pairs[0][1][:, np.newaxis]\nfor i in range(1, len(eigen_pairs)):\n # print(i)\n w = np.hstack((w, eigen_pairs[i][1][:, np.newaxis]))\nw.shape",
"_____no_output_____"
],
[
"# 座標変換\nX_train_pca = X_train_std.dot(w)\n# print(X_train_pca.shape)\n\ncov_mat = np.cov(X_train_pca.T)\n\n# 共分散行列のヒートマップ\ndf = pd.DataFrame(cov_mat)\nax = sns.heatmap(df, cmap=\"YlGnBu\") ",
"_____no_output_____"
]
],
[
[
"主成分空間に変換後の各特徴量は、互いに相関が全くないことがわかる(互いに線形独立)。 \n対角成分は分散値であり、第1主成分から大きい順に並んでいることがわかる。",
"_____no_output_____"
],
[
"### 座標変換された空間から元の空間への復元",
"_____no_output_____"
],
[
"## 座標変換された空間から元の空間への復元\n\n$X = X'W^T$ \n\n$X'$ : 座標変換後の座標(行列) \n$X$ : もともとの空間に復元された座標(行列) \n$W^T \\in \\mathbb{R} ^{n\\times n}$ : 転置された変)行列 \n \n$x' \\in \\mathbb{R} ^{n}$ \n$x_{approx} \\in \\mathbb{R} ^{n}$ \n",
"_____no_output_____"
]
],
[
[
"# 1つ目のサンプルに射影行列を適用(内積を作用させる)\nx0 = X_train_std[0]\nprint('もともとの特徴量:', x0)\nz0 = x0.dot(w)\nprint('変換後の特徴量:', z0)\nx0_reconstructed = z0.dot(w.T)\nprint('復元された特徴量:', x0_reconstructed)",
"もともとの特徴量: [ 0.71225893 2.22048673 -0.13025864 0.05962872 -0.50432733 -0.52831584\n -1.24000033 0.84118003 -1.05215112 -0.29218864 -0.20017028 -0.82164144\n -0.62946362]\n変換後の特徴量: [ 2.38299011 0.45458499 0.22703207 0.57988399 -0.57994169 -1.73317476\n -0.70180475 -0.21617248 0.23666876 0.16548767 -0.29726982 -0.23489704\n 0.40161994]\n復元された特徴量: [ 0.71225893 2.22048673 -0.13025864 0.05962872 -0.50432733 -0.52831584\n -1.24000033 0.84118003 -1.05215112 -0.29218864 -0.20017028 -0.82164144\n -0.62946362]\n"
]
],
[
[
"完全に復元できていることがわかる。",
"_____no_output_____"
],
[
"### 13 次元 --> 2 次元に次元削減する\n\n$X' = XW$ \n\n$W = [v_1 v_2] \\in \\mathbb{R} ^{13\\times 2}$ \n$x \\in \\mathbb{R} ^{13}$ \n$x' \\in \\mathbb{R} ^{2}$ ",
"_____no_output_____"
]
],
[
[
"w = np.hstack((eigen_pairs[0][1][:, np.newaxis],\n eigen_pairs[1][1][:, np.newaxis]))\nprint('Matrix W:\\n', w)",
"Matrix W:\n [[-0.13724218 0.50303478]\n [ 0.24724326 0.16487119]\n [-0.02545159 0.24456476]\n [ 0.20694508 -0.11352904]\n [-0.15436582 0.28974518]\n [-0.39376952 0.05080104]\n [-0.41735106 -0.02287338]\n [ 0.30572896 0.09048885]\n [-0.30668347 0.00835233]\n [ 0.07554066 0.54977581]\n [-0.32613263 -0.20716433]\n [-0.36861022 -0.24902536]\n [-0.29669651 0.38022942]]\n"
]
],
[
[
"**注意**\n\nNumPy と LAPACK のバージョンによっては、上記の例とは符号が反転した射影行列 w が作成されることがあるが、問題はない。 \n以下の式が成り立つからである。 \n\n行列 $\\Sigma$ に対して、 $v$ が固有ベクトル、$\\lambda$ が固有値のとき、 \n$$\\Sigma v = \\lambda v,$$\n\nここで $-v$ もまた同じ固有値をもつ固有ベクトルとなる。 \n$$\\Sigma \\cdot (-v) = -\\Sigma v = -\\lambda v = \\lambda \\cdot (-v).$$\n\n(主成分軸のベクトルの向きの違い)",
"_____no_output_____"
]
],
[
[
"# 各サンプルに射影行列を適用(内積を作用)させることで、変換後の座標(特徴量)を得ることができる。\nX_train_std[0].dot(w)",
"_____no_output_____"
]
],
[
[
"### 2次元に射影後の全データを、ラベルごとに色付けしてプロットする",
"_____no_output_____"
]
],
[
[
"X_train_pca = X_train_std.dot(w)\ncolors = ['r', 'b', 'g']\nmarkers = ['s', 'x', 'o']\n\nfor l, c, m in zip(np.unique(y_train), colors, markers):\n plt.scatter(X_train_pca[y_train == l, 0], \n X_train_pca[y_train == l, 1], \n c=c, label=l, marker=m)\n\nplt.xlabel('PC 1')\nplt.ylabel('PC 2')\nplt.legend(loc='lower left')\nplt.tight_layout()\n# plt.savefig('images/05_03.png', dpi=300)\nplt.show()",
"_____no_output_____"
]
],
[
[
"PC1 軸方向をみると、PC2 軸方向よりもよりもデータが広く分布しており、データをよりよく区別できていることがわかる。",
"_____no_output_____"
],
[
"## 次元削減された空間から元の空間への復元\n\n$X_{approx} = X'W^T$ \n\n$X'$ : 射影後の座標(行列) \n$X_{approx}$ : もともとの空間に、近似的に、復元された座標(行列) \n$W^T \\in \\mathbb{R} ^{n\\times k}$ : 転置された射影(変換)行列 \n \n$x' \\in \\mathbb{R} ^{k}$ \n$x_{approx} \\in \\mathbb{R} ^{n}$ \n \n$k = n$ のとき、$X = X_{approx}$ が成り立つ(上述)。",
"_____no_output_____"
]
],
[
[
"# 1つ目のサンプルに射影行列を適用(内積を作用させる)\nx0 = X_train_std[0]\nprint('もともとの特徴量:', x0)\nz0 = x0.dot(w)\nprint('変換後の特徴量:', z0)\nx0_reconstructed = z0.dot(w.T)\nprint('復元された特徴量:', x0_reconstructed)",
"もともとの特徴量: [ 0.71225893 2.22048673 -0.13025864 0.05962872 -0.50432733 -0.52831584\n -1.24000033 0.84118003 -1.05215112 -0.29218864 -0.20017028 -0.82164144\n -0.62946362]\n変換後の特徴量: [2.38299011 0.45458499]\n復元された特徴量: [-0.09837469 0.66412622 0.05052458 0.44153949 -0.23613841 -0.91525549\n -1.00494135 0.76968396 -0.72702683 0.42993247 -0.87134462 -0.9915977\n -0.53417827]\n"
]
],
[
[
"完全には復元できていないことがわかる(近似値に復元される)。",
"_____no_output_____"
],
[
"## Principal component analysis in scikit-learn",
"_____no_output_____"
],
[
"上記で行った PCA の実装は、scikit-learn を使うことで簡単に実装できる。\n以下にその実装を示す。",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\n\npca = PCA()\n# 主成分分析の実行\nX_train_pca = pca.fit_transform(X_train_std)\n# 分散説明率の表示\npca.explained_variance_ratio_",
"_____no_output_____"
],
[
"# 分散説明率とその累積和のプロット\nplt.bar(range(1, 14), pca.explained_variance_ratio_, alpha=0.5, align='center')\nplt.step(range(1, 14), np.cumsum(pca.explained_variance_ratio_), where='mid')\nplt.ylabel('Explained variance ratio')\nplt.xlabel('Principal components')\n\nplt.show()",
"_____no_output_____"
],
[
"# 2次元に削減\npca = PCA(n_components=2)\nX_train_pca = pca.fit_transform(X_train_std)\nX_test_pca = pca.transform(X_test_std)",
"_____no_output_____"
],
[
"# 2次元空間にプロット\nplt.scatter(X_train_pca[:, 0], X_train_pca[:, 1])\nplt.xlabel('PC 1')\nplt.ylabel('PC 2')\nplt.show()",
"_____no_output_____"
]
],
[
[
"### 因子負荷量の確認\n\n以下のように、`pca.components_` を見ることで、因子負荷量を確認することができる。",
"_____no_output_____"
]
],
[
[
"pd.DataFrame(pca.components_.T,index=df_wine.columns[1:],columns=['PC1','PC2']).sort_values('PC1')",
"_____no_output_____"
]
],
[
[
"値の絶対値が大きい特徴量を見れば良い。\n\nつまり、第1主成分 (PC1) でよく表されている特徴量は、 \"Flavanoids\" と \"Total phenols\" である。\n一方、第2主成分 (PC2) でよく表されている特徴量は、 \"Color intensity\" と \"Alcohol\" である。",
"_____no_output_____"
],
[
"## 2次元に次元削減された特徴量を用いてロジスティック回帰を行ってみる\n",
"_____no_output_____"
]
],
[
[
"from matplotlib.colors import ListedColormap\n\ndef plot_decision_regions(X, y, classifier, resolution=0.02):\n\n # setup marker generator and color map\n markers = ('s', 'x', 'o', '^', 'v')\n colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')\n cmap = ListedColormap(colors[:len(np.unique(y))])\n\n # plot the decision surface\n x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1\n x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1\n xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),\n np.arange(x2_min, x2_max, resolution))\n Z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)\n Z = Z.reshape(xx1.shape)\n plt.contourf(xx1, xx2, Z, alpha=0.4, cmap=cmap)\n plt.xlim(xx1.min(), xx1.max())\n plt.ylim(xx2.min(), xx2.max())\n\n # plot class samples\n for idx, cl in enumerate(np.unique(y)):\n plt.scatter(x=X[y == cl, 0], \n y=X[y == cl, 1],\n alpha=0.6, \n c=cmap(idx),\n edgecolor='black',\n marker=markers[idx], \n label=cl)",
"_____no_output_____"
]
],
[
[
"Training logistic regression classifier using the first 2 principal components.",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\npca = PCA(n_components=2)\nX_train_pca = pca.fit_transform(X_train_std)\nX_test_pca = pca.transform(X_test_std)\n\nlr = LogisticRegression(penalty='l2', C=1.0)\n# lr = LogisticRegression(penalty='none')\nlr = lr.fit(X_train_pca, y_train)",
"_____no_output_____"
],
[
"print(X_train_pca.shape)",
"(124, 2)\n"
],
[
"print('Cumulative explained variance ratio:', sum(pca.explained_variance_ratio_))",
"Cumulative explained variance ratio: 0.5538639565949177\n"
]
],
[
[
"### 学習時間の計測",
"_____no_output_____"
]
],
[
[
"%timeit lr.fit(X_train_pca, y_train)",
"100 loops, best of 3: 5.08 ms per loop\n"
],
[
"from sklearn.metrics import plot_confusion_matrix\n\n# 精度\nprint('accuracy', lr.score(X_train_pca, y_train))\n# confusion matrix\nplot_confusion_matrix(lr, X_train_pca, y_train)",
"accuracy 0.9838709677419355\n"
]
],
[
[
"### トレーニングデータセットの予測結果",
"_____no_output_____"
]
],
[
[
"plot_decision_regions(X_train_pca, y_train, classifier=lr)\nplt.xlabel('PC 1')\nplt.ylabel('PC 2')\nplt.legend(loc='lower left')\nplt.tight_layout()\n# plt.savefig('images/05_04.png', dpi=300)\nplt.show()",
"'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.\n'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.\n'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.\n"
]
],
[
[
"### テストデータに対する予測結果",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import plot_confusion_matrix\n\n# 精度\nprint('accuracy', lr.score(X_test_pca, y_test))\n# confusion matrix\nplot_confusion_matrix(lr, X_test_pca, y_test)",
"accuracy 0.9259259259259259\n"
],
[
"plot_decision_regions(X_test_pca, y_test, classifier=lr)\nplt.xlabel('PC 1')\nplt.ylabel('PC 2')\nplt.legend(loc='lower left')\nplt.tight_layout()\n# plt.savefig('images/05_05.png', dpi=300)\nplt.show()",
"'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.\n'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.\n'c' argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with 'x' & 'y'. Please use a 2-D array with a single row if you really want to specify the same RGB or RGBA value for all points.\n"
]
],
[
[
"次元削減せずに全てのの主成分を取得したい場合は、 `n_components=None` にする。",
"_____no_output_____"
]
],
[
[
"pca = PCA(n_components=None)\nX_train_pca = pca.fit_transform(X_train_std)\npca.explained_variance_ratio_",
"_____no_output_____"
]
],
[
[
"## 3 次元に次元削減された特徴量を用いてロジスティック回帰を行ってみる",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\nk = 3\n\npca = PCA(n_components=3)\nX_train_pca = pca.fit_transform(X_train_std)\nX_test_pca = pca.transform(X_test_std)\n\nlr = LogisticRegression(penalty='l2', C=1.0)\n# lr = LogisticRegression(penalty='none')\nlr = lr.fit(X_train_pca, y_train)",
"_____no_output_____"
],
[
"print(X_train_pca.shape)",
"(124, 3)\n"
],
[
"print('Cumulative explained variance ratio:', sum(pca.explained_variance_ratio_))",
"Cumulative explained variance ratio: 0.6720155475408875\n"
],
[
"%timeit lr.fit(X_train_pca, y_train)",
"100 loops, best of 3: 6.02 ms per loop\n"
],
[
"from sklearn.metrics import plot_confusion_matrix\n\n# 精度\nprint('accuracy', lr.score(X_train_pca, y_train))\n# confusion matrix\nplot_confusion_matrix(lr, X_train_pca, y_train)",
"accuracy 0.9838709677419355\n"
],
[
"from mpl_toolkits.mplot3d import Axes3D\nimport matplotlib.pyplot as plt\n\nfig = plt.figure()\nax = fig.add_subplot(111, projection='3d')\n\n# ax.scatter(X_train_pca[:,0], X_train_pca[:,1], X_train_pca[:,2], c='r', marker='o')\nax.scatter(X_train_pca[:,0], X_train_pca[:,1], X_train_pca[:,2], c=y_train, marker='o')\n\nax.set_xlabel('PC1')\nax.set_ylabel('PC2')\nax.set_zlabel('PC3')\n\nplt.show()",
"_____no_output_____"
],
[
"# plotly を使った interactive な 3D 散布図\nimport plotly.express as px\n\ndf = pd.DataFrame(X_train_pca, columns=['PC1', 'PC2', 'PC3'])\ndf['label'] = y_train\n\nfig = px.scatter_3d(df, x='PC1', y='PC2', z='PC3',\n color='label', opacity=0.7, )\nfig.show()",
"_____no_output_____"
]
],
[
[
"人間の目で確認できるのは 3 次元が限界。",
"_____no_output_____"
],
[
"## 次元削減せずにロジスティック回帰を行ってみる\n\n",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\nlr = LogisticRegression(penalty='l2', C=1.0)\n# lr = LogisticRegression(penalty='none')\nlr = lr.fit(X_train_std, y_train)",
"_____no_output_____"
],
[
"# 学習時間\n%timeit lr.fit(X_train_std, y_train)",
"100 loops, best of 3: 7.24 ms per loop\n"
],
[
"from sklearn.metrics import plot_confusion_matrix\n\nprint('Evaluation of training dataset')\n# 精度\nprint('accuracy', lr.score(X_train_std, y_train))\n# confusion matrix\nplot_confusion_matrix(lr, X_train_std, y_train)\n\nprint('Evaluation of test dataset')\n# 精度\nprint('accuracy', lr.score(X_test_std, y_test))\n# confusion matrix\nplot_confusion_matrix(lr, X_test_std, y_test)",
"Evaluation of training dataset\naccuracy 1.0\nEvaluation of test dataset\naccuracy 1.0\n"
]
],
[
[
"元々の全ての特徴量を使って学習させた方が精度が高くなった。 \n学習時間は、次元削減したほうがわずかに早くなっている。\n(主成分 2 つで学習した場合 4.9 ms に対し、元々の特徴量全て使った場合 5.64 ms) \n結論として、今回のタスクでは、PCA を適用するべきではなく、すべての特徴量を使用したほうが良い。 \n \nもっとデータ数が大きい場合や、モデルのパラメータ数が多い場合には、次元削減が効果的となる。",
"_____no_output_____"
],
[
"### 2つの特徴量だけでロジスティック回帰を行ってみる\n\n",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\n\nlr = LogisticRegression(penalty='l2', C=1.0)\n# lr = LogisticRegression(penalty='none')\nlr = lr.fit(X_train_std[:,:2], y_train)",
"_____no_output_____"
],
[
"%timeit lr.fit(X_train_std[:,:2], y_train)",
"100 loops, best of 3: 3.6 ms per loop\n"
],
[
"from sklearn.metrics import plot_confusion_matrix\n\nprint('Evaluation of training dataset')\n# 精度\nprint('accuracy', lr.score(X_train_std[:,:2], y_train))\n# confusion matrix\nplot_confusion_matrix(lr, X_train_std[:,:2], y_train)\n\nprint('Evaluation of test dataset')\n# 精度\nprint('accuracy', lr.score(X_test_std[:,:2], y_test))\n# confusion matrix\nplot_confusion_matrix(lr, X_test_std[:,:2], y_test)",
"Evaluation of training dataset\naccuracy 0.7580645161290323\nEvaluation of test dataset\naccuracy 0.7777777777777778\n"
]
],
[
[
"もともとの特徴量を 2 つだけ使った場合、精度はかなり下がる。\nこれと比べると、PCA によって特徴抽出した 2 つの主成分を使った場合には、精度がかなり高いことがわかる。",
"_____no_output_____"
],
[
"## まとめ\n\n主成分分析により以下のタスクを行うことができる。\n\n- 次元削減\n - データを格納するためのメモリやディスク使用量を削減できる\n - 学習アルゴリズムを高速化できる\n- 可視化\n - 多数の特徴量(次元)をもつデータを2次元などの理解しやすい空間に落とし込んで議論、解釈することができる。\n \nしかし、機械学習の前処理として利用する場合には、以下のことに注意する必要がある。\n\n- 次元削減を行うことによって、多少なりとも情報が失われている\n- まずは、すべての特徴量を使ってトレーニングを試すことが大事 \n- 次元削減によってオーバーフィッティングを防ぐことができるが、次元削減を使う前に正則化を使うべし\n- 上記を試してから、それでも望む結果を得られない場合、次元削減を使う\n- 機械学習のトレーニングでは、通常は、99% の累積寄与率が得られるように削減後の次元数を選ぶことが多い \n\n参考) [Andrew Ng先生の講義](https://www.coursera.org/learn/machine-learning)",
"_____no_output_____"
],
[
"## References",
"_____no_output_____"
],
[
"1. *Python Machine Learning 2nd Edition* by [Sebastian Raschka](https://sebastianraschka.com), Packt Publishing Ltd. 2017. Code Repository: https://github.com/rasbt/python-machine-learning-book-2nd-edition\n1. [Andrew Ng先生の講義](https://www.coursera.org/learn/machine-learning)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4ae7c889db16d34d47332c9b96ccacdb6674e3bb
| 2,799 |
ipynb
|
Jupyter Notebook
|
models/5-Deepflash2/input/deepflash2-lfs/nbs/model_library.ipynb
|
cns-iu/HuBMAP---Hacking-the-Kidney
|
1a41c887f8edb0b52f5afade384a17dc3d3efec4
|
[
"MIT"
] | 33 |
2020-03-23T13:00:27.000Z
|
2022-03-11T21:16:48.000Z
|
nbs/model_library.ipynb
|
matjesg/DeepFLaSH2
|
21fed72cae95aecabee574ab18288ccda8043475
|
[
"Apache-2.0"
] | 25 |
2020-11-08T13:09:03.000Z
|
2022-03-04T15:25:36.000Z
|
nbs/model_library.ipynb
|
matjesg/DeepFLaSH2
|
21fed72cae95aecabee574ab18288ccda8043475
|
[
"Apache-2.0"
] | 8 |
2021-02-16T15:43:49.000Z
|
2022-03-02T16:15:38.000Z
| 28.561224 | 320 | 0.590568 |
[
[
[
"# Model Library\n> Information on the datasets of the pretrained models.",
"_____no_output_____"
],
[
"## wue-cFOS\n\n**Dataset Description**:\nTrained on 36 image-mask pairs of cFOS labels in the dorsal hippocampus (including 12 images of each sub-region: dentate gyrus, CA3 and CA1). Masks for training were prepared by five independent experts. Images were acquired using laser-scanning confocal microscopy with a resolution of 1.6 pixel per µm.\n\n- Images: 36 (laser-scanning confocal microscopy with a resolution of 1.6 pixel per µm)\n- Experts: 5 (Ground Truth Estimation: STAPLE)\n- Training: tbd.",
"_____no_output_____"
],
[
"<p align=\"center\">\n <img src=\"https://raw.githubusercontent.com/matjesg/DeepFLaSH/master/assets/cFOS_Wue.png\" width=\"45%\" alt=\"cFOS_Wue\"> \n \n <img src=\"https://raw.githubusercontent.com/matjesg/DeepFLaSH/master/assets/cFOS_Wue_mask.png\" width=\"45%\" alt=\"cFOS_Wue\"> \n</p>\n",
"_____no_output_____"
],
[
"## wue-Parv\nTrained on 36 image-mask pairs of Parvalbumin-labels in the dorsal hippocampus (including 12 images of each sub-region: dentate gyrus, CA3 and CA1). Masks for training were prepared by five independent experts. Images were acquired using laser-scanning confocal microscopy with a resolution of 1.6 pixel per µm.\n\n- Images: 36 (laser-scanning confocal microscopy with a resolution of 1.6 pixel per µm)\n- Experts: 5 (Ground Truth Estimation: STAPLE)\n- Training: tbd.",
"_____no_output_____"
],
[
"<p align=\"center\">\n <img src=\"https://raw.githubusercontent.com/matjesg/DeepFLaSH/master/assets/Parv.png\" width=\"45%\" alt=\"cFOS_Wue\"> \n \n <img src=\"https://raw.githubusercontent.com/matjesg/DeepFLaSH/master/assets/Parv_mask.png\" width=\"45%\" alt=\"cFOS_Wue\"> \n</p>",
"_____no_output_____"
],
[
"## wue-OPN3",
"_____no_output_____"
],
[
"tbd.",
"_____no_output_____"
],
[
"## wue-GFP",
"_____no_output_____"
],
[
"tbd.",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4ae7cf301addb337164f763e1761ca47c83bcbdf
| 957,427 |
ipynb
|
Jupyter Notebook
|
notebooks/generate_images.ipynb
|
H00N24/visual-analysis-of-big-time-series-datasets
|
8c9c14ca5d16f5d9ef8b623c84f92fe62eee1f86
|
[
"MIT"
] | 2 |
2021-07-30T04:07:43.000Z
|
2022-01-24T07:28:44.000Z
|
notebooks/generate_images.ipynb
|
H00N24/visual-analysis-of-big-time-series-datasets
|
8c9c14ca5d16f5d9ef8b623c84f92fe62eee1f86
|
[
"MIT"
] | null | null | null |
notebooks/generate_images.ipynb
|
H00N24/visual-analysis-of-big-time-series-datasets
|
8c9c14ca5d16f5d9ef8b623c84f92fe62eee1f86
|
[
"MIT"
] | null | null | null | 1,331.609179 | 247,488 | 0.956833 |
[
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
]
],
[
[
"# Generate images",
"_____no_output_____"
]
],
[
[
"from pathlib import Path\n\nimport numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nSMALL_SIZE = 15\nMEDIUM_SIZE = 20\nBIGGER_SIZE = 25\n\nplt.rc(\"font\", size=SMALL_SIZE)\nplt.rc(\"axes\", titlesize=SMALL_SIZE)\nplt.rc(\"axes\", labelsize=MEDIUM_SIZE)\nplt.rc(\"xtick\", labelsize=SMALL_SIZE)\nplt.rc(\"ytick\", labelsize=SMALL_SIZE)\nplt.rc(\"legend\", fontsize=SMALL_SIZE)\nplt.rc(\"figure\", titlesize=BIGGER_SIZE)",
"_____no_output_____"
],
[
"DATA_PATH = Path(\"../thesis/img/\")",
"_____no_output_____"
]
],
[
[
"# DTW",
"_____no_output_____"
]
],
[
[
"from fastdtw import fastdtw",
"_____no_output_____"
],
[
"ts_0 = np.sin(np.logspace(0, np.log10(2 * np.pi), 30))\nts_1 = np.sin(np.linspace(1, 2 * np.pi, 30))",
"_____no_output_____"
],
[
"distance, warping_path = fastdtw(ts_0, ts_1)",
"_____no_output_____"
],
[
"fig, axs = plt.subplots(2, 1, figsize=(8, 8), sharex=True)\n\nfor name, ax in zip([\"Euclidian distance\", \"Dynamic Time Warping\"], axs):\n ax.plot(ts_0 + 1, \"o-\", linewidth=3)\n ax.plot(ts_1, \"o-\", linewidth=3)\n ax.set_yticks([])\n ax.set_xticks([])\n ax.set_title(name)\n\nfor x, y in zip(zip(np.arange(30), np.arange(30)), zip(ts_0 + 1, ts_1)):\n axs[0].plot(x, y, \"r--\", linewidth=2, alpha=0.5)\n\nfor x_0, x_1 in warping_path:\n axs[1].plot([x_0, x_1], [ts_0[x_0] + 1, ts_1[x_1]], \"r--\", linewidth=2, alpha=0.5)\n\nplt.savefig(DATA_PATH / \"dtw_vs_euclid.svg\")\n\nplt.tight_layout()\nplt.show()",
"_____no_output_____"
],
[
"matrix = (ts_0.reshape(-1, 1) - ts_1) ** 2\n\nx = [x for x, _ in warping_path]\ny = [y for _, y in warping_path]\n\n# plt.close('all')\nfig = plt.figure(figsize=(8, 8))\n\n\ngs = fig.add_gridspec(\n 2,\n 2,\n width_ratios=(1, 8),\n height_ratios=(8, 1),\n left=0.1,\n right=0.9,\n bottom=0.1,\n top=0.9,\n wspace=0.01,\n hspace=0.01,\n)\n\nfig.tight_layout()\n\nax_ts_x = fig.add_subplot(gs[0, 0])\nax_ts_y = fig.add_subplot(gs[1, 1])\nax = fig.add_subplot(gs[0, 1], sharex=ax_ts_y, sharey=ax_ts_x)\n\nax.set_xticks([])\nax.set_yticks([])\nax.tick_params(axis=\"x\", labelbottom=False)\nax.tick_params(axis=\"y\", labelleft=False)\nfig.suptitle(\"DTW calculated optimal warping path\")\n\nim = ax.imshow(np.log1p(matrix), origin=\"lower\", cmap=\"bone_r\")\n\n\nax.plot(y, x, \"r\", linewidth=4, label=\"Optimal warping path\")\nax.plot(\n [0, 29], [0, 29], \"--\", linewidth=3, color=\"black\", label=\"Default warping path\"\n)\n\nax.legend()\n\nax_ts_x.plot(ts_0 * -1, np.arange(30), linewidth=4, color=\"#1f77b4\")\n# ax_ts_x.set_yticks(np.arange(30))\nax_ts_x.set_ylim(-0.5, 29.5)\nax_ts_x.set_xlim(-1.5, 1.5)\nax_ts_x.set_xticks([])\n\n\nax_ts_y.plot(ts_1, linewidth=4, color=\"#ff7f0e\")\n# ax_ts_y.set_xticks(np.arange(30))\nax_ts_y.set_xlim(-0.5, 29.5)\nax_ts_y.set_ylim(-1.5, 1.5)\nax_ts_y.set_yticks([])\n\n\n# cbar = plt.colorbar(im, ax=ax, use_gridspec=False, panchor=False)\n\n\nplt.savefig(DATA_PATH / \"dtw_warping_path.svg\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# TSNE",
"_____no_output_____"
]
],
[
[
"import mpl_toolkits.mplot3d.axes3d as p3\nfrom sklearn.datasets import make_s_curve, make_swiss_roll\nfrom sklearn.decomposition import PCA\nfrom sklearn.manifold import TSNE",
"_____no_output_____"
],
[
"n_samples = 1500\nX, y = make_swiss_roll(n_samples, noise=0.1)\nX, y = make_s_curve(n_samples, random_state=42)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 10))\nax = fig.gca(projection=\"3d\")\nax.view_init(20, -60)\n# ax.set_title(\"S curve dataset\", fontsize=18)\n\nax.scatter(X[:, 0], X[:, 1], X[:, 2], c=y)\nax.set_yticklabels([])\nax.set_xticklabels([])\nax.set_zticklabels([])\nfig.tight_layout()\n\nplt.savefig(DATA_PATH / \"s_dataset.svg\", bbox_inches=0)\n\nplt.show()",
"<ipython-input-12-849cbf660a66>:2: MatplotlibDeprecationWarning: Calling gca() with keyword arguments was deprecated in Matplotlib 3.4. Starting two minor releases later, gca() will take no keyword arguments. The gca() function should only be used to get the current axes, or if no axes exist, create new axes with default keyword arguments. To create a new axes with non-default arguments, use plt.axes() or plt.subplot().\n ax = fig.gca(projection=\"3d\")\n<ipython-input-12-849cbf660a66>:10: UserWarning: Tight layout not applied. The left and right margins cannot be made large enough to accommodate all axes decorations. \n fig.tight_layout()\n"
],
[
"X_pca = PCA(n_components=2, random_state=42).fit_transform(X)\nX_tsne = TSNE(n_components=2, perplexity=30, init=\"pca\", random_state=42).fit_transform(\n X\n)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 10))\n\n# plt.title(\"PCA transformation\", fontsize=18)\nplt.scatter(X_pca[:, 0], X_pca[:, 1], c=y)\nplt.xticks([])\nplt.yticks([])\n\nplt.savefig(DATA_PATH / \"s_dataset_pca.svg\")\n\nplt.show()",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(10, 10))\n\n# plt.title(\"t-SNE transformation\", fontsize=18)\n\nplt.scatter(X_tsne[:, 0], X_tsne[:, 1], c=y)\n\nplt.xticks([])\nplt.yticks([])\n\n\nplt.savefig(DATA_PATH / \"s_dataset_tsne.svg\")\n\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Datashader",
"_____no_output_____"
]
],
[
[
"import datashader as ds\nimport datashader.transfer_functions as tf\nimport matplotlib.patches as mpatches\nfrom lttb import downsample",
"_____no_output_____"
],
[
"np.random.seed(42)\nsignal = np.random.normal(0, 10, size=10 ** 6).cumsum() + np.sin(\n np.linspace(0, 100 * np.pi, 10 ** 6)\n) * np.random.normal(0, 1, size=10 ** 6)\n\ns_frame = pd.DataFrame(signal, columns=[\"signal\"]).reset_index()",
"_____no_output_____"
],
[
"x = 1500\ny = 500\n\ncvs = ds.Canvas(plot_height=y, plot_width=x)\nline = cvs.line(s_frame, \"index\", \"signal\")\n\nimg = tf.shade(line).to_pil()",
"_____no_output_____"
],
[
"trans = downsample(s_frame.values, 100)\ntrans[:, 0] /= trans[:, 0].max()\ntrans[:, 0] *= x\ntrans[:, 1] *= -1\ntrans[:, 1] -= trans[:, 1].min()\ntrans[:, 1] /= trans[:, 1].max()\ntrans[:, 1] *= y",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(x / 60, y / 60))\n\nplt.imshow(img, origin=\"upper\")\n\nplt.plot(*trans.T, \"r\", alpha=0.6, linewidth=2)\nplt.legend(\n handles=[\n mpatches.Patch(color=\"blue\", label=\"Datashader (10^6 points)\"),\n mpatches.Patch(color=\"red\", label=\"LTTB (10^3 points)\"),\n ],\n prop={\"size\": 25},\n)\nax.set_xticklabels([])\nax.set_yticklabels([])\nplt.ylabel(\"Value\", fontsize=25)\nplt.xlabel(\"Time\", fontsize=25)\nplt.tight_layout()\nplt.savefig(DATA_PATH / \"datashader.png\")\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"# LTTB",
"_____no_output_____"
]
],
[
[
"from matplotlib import cm\nfrom matplotlib.colors import Normalize\nfrom matplotlib.patches import Polygon",
"_____no_output_____"
],
[
"np.random.seed(42)\nns = np.random.normal(0, 1, size=26).cumsum()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(15, 5))\n\nplt.plot(ns, \"-o\", linewidth=2)\n\nmapper = cm.ScalarMappable(Normalize(vmin=0, vmax=15, clip=True), cmap=\"autumn_r\")\n\nareas = []\n\nfor i, data in enumerate(ns[:-2], 1):\n\n cors = [[i + ui, ns[i + ui]] for ui in range(-1, 2)]\n x = [m[0] for m in cors]\n y = [m[1] for m in cors]\n\n ea = 0.5 * np.abs(np.dot(x, np.roll(y, 1)) - np.dot(y, np.roll(x, 1))) * 10\n\n areas.append(ea)\n\n color = mapper.to_rgba(ea)\n\n plt.plot([i], [ns[i]], \"o\", color=color)\n\n ax.add_patch(\n Polygon(\n cors,\n closed=True,\n fill=True,\n alpha=0.3,\n color=color,\n )\n )\n\n\ncbar = plt.colorbar(mapper, alpha=0.3)\ncbar.set_label(\"Effective Area Size\")\n\nfig.suptitle(\"Effective Area of Data Points\")\nplt.ylabel(\"Value\")\nplt.xlabel(\"Time\")\n\nplt.tight_layout()\nplt.savefig(DATA_PATH / \"effective-area.svg\")\nplt.savefig(DATA_PATH / \"effective-area.png\")\n\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(15, 5))\n\nplt.plot(ns, \"--o\", linewidth=2, label=\"Original time series\")\n\nmapper = cm.ScalarMappable(Normalize(vmin=0, vmax=15, clip=True), cmap=\"autumn_r\")\n\n\nlotb = np.concatenate(\n [[0], np.arange(1, 25, 3) + np.array(areas).reshape(-1, 3).argmax(axis=1), [25]]\n)\n\nfor i, data in enumerate(ns[:-2], 1):\n\n cors = [[i + ui, ns[i + ui]] for ui in range(-1, 2)]\n\n x = [m[0] for m in cors]\n y = [m[1] for m in cors]\n\n ea = 0.5 * np.abs(np.dot(x, np.roll(y, 1)) - np.dot(y, np.roll(x, 1))) * 10\n\n color = mapper.to_rgba(ea) # cm.tab10.colors[i % 5 + 1]\n\n plt.plot([i], [ns[i]], \"o\", color=color)\n\n ax.add_patch(\n Polygon(\n cors,\n closed=True,\n fill=True,\n alpha=0.3,\n color=color,\n )\n )\n\nplt.plot(\n lotb, ns[lotb], \"-x\", linewidth=2, color=\"tab:purple\", label=\"LTOB approximation\"\n)\n\ncbar = plt.colorbar(mapper, alpha=0.3)\ncbar.set_label(\"Effective Area Size\")\n\nplt.vlines(np.linspace(0.5, 24.5, 9), ns.min(), ns.max(), \"black\", \"--\", alpha=0.5)\nplt.ylabel(\"Value\")\nplt.xlabel(\"Time\")\n\nfig.suptitle(\"LTOB downsampling\")\nplt.legend()\nplt.tight_layout()\n\n\nplt.savefig(DATA_PATH / \"ltob.svg\")\nplt.savefig(DATA_PATH / \"ltob.png\")\nplt.show()",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 1, figsize=(15, 5))\n\nplt.plot(ns, \"--o\", linewidth=2, label=\"Original time series\")\n\nds = downsample(np.vstack([np.arange(26), ns]).T, 10)\n\n\nplt.plot(*ds.T, \"-x\", linewidth=2, label=\"LTTB approximation\")\n# plt.plot(ns, \"x\")\n\nplt.vlines(np.linspace(0.5, 24.5, 9), ns.min(), ns.max(), \"black\", \"--\", alpha=0.5)\nplt.ylabel(\"Value\")\nplt.xlabel(\"Time\")\n\nfig.suptitle(\"LTTB downsampling\")\nplt.legend()\nplt.tight_layout()\n\nplt.savefig(DATA_PATH / \"lttb.svg\")\nplt.savefig(DATA_PATH / \"lttb.png\")\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.