hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cbab390bb07e6ae73027d2172baafb6566bd16bd
| 1,907 |
ipynb
|
Jupyter Notebook
|
Python/13. regex and parsing/87. validating uid.ipynb
|
faisalsanto007/Hakcerrank-problem-solving
|
eaf6404e8896fe3448df8a3cb4c86585fd7bebcc
|
[
"MIT"
] | null | null | null |
Python/13. regex and parsing/87. validating uid.ipynb
|
faisalsanto007/Hakcerrank-problem-solving
|
eaf6404e8896fe3448df8a3cb4c86585fd7bebcc
|
[
"MIT"
] | null | null | null |
Python/13. regex and parsing/87. validating uid.ipynb
|
faisalsanto007/Hakcerrank-problem-solving
|
eaf6404e8896fe3448df8a3cb4c86585fd7bebcc
|
[
"MIT"
] | null | null | null | 25.426667 | 137 | 0.507604 |
[
[
[
"A valid UID must follow the rules below:\n\n It must contain at least 2 uppercase English alphabet characters.\n It must contain at least 3 digits (0 - 9).\n It should only contain alphanumeric characters (a - z, A - Z & 0 - 9).\n No character should repeat.\n There must be exactly 10 characters in a valid UID.",
"_____no_output_____"
]
],
[
[
"import re\n\nfor _ in range(int(input())):\n u = ''.join(sorted(input()))\n \n try:\n assert re.search(r'[A-Z]{2}', u) # assert hocche condition check kore..condition vul hoile, except line er code run hobe\n assert re.search(r'\\d\\d\\d', u)\n assert not re.search(r'[^a-zA-Z0-9]', u) # ^ mane e character chara onno character ase kina check kora\n assert not re.search(r'(.)\\1', u) # prottekta character shudhu 1bar e thakte parbe\n assert len(u) == 10\n \n except:\n print('Invalid')\n else:\n print('Valid')",
"2\nB1CD102354\nInvalid\nB1CDEF2354\nValid\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
cbab46c646ecffd71b9224200d3a35ba6119ace5
| 8,096 |
ipynb
|
Jupyter Notebook
|
02-python/PyBank/main.ipynb
|
senalba/python-homework
|
4ec5b48ccbca7a52f7e58061cfe7271245f54d7b
|
[
"MIT"
] | null | null | null |
02-python/PyBank/main.ipynb
|
senalba/python-homework
|
4ec5b48ccbca7a52f7e58061cfe7271245f54d7b
|
[
"MIT"
] | null | null | null |
02-python/PyBank/main.ipynb
|
senalba/python-homework
|
4ec5b48ccbca7a52f7e58061cfe7271245f54d7b
|
[
"MIT"
] | null | null | null | 25.948718 | 119 | 0.552372 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"df_budget = pd.read_csv('Resources/budget_data.csv')",
"_____no_output_____"
],
[
"dates = df_budget.Date.to_list()\nprofits = df_budget['Profit/Losses'].to_list()",
"_____no_output_____"
],
[
"number_months = len(dates)",
"_____no_output_____"
],
[
"total_amount = sum(profits)",
"_____no_output_____"
],
[
"change_weight = 1/(number_months - 1)\naverage_change = sum((profits[i] - profits[i-1]) * change_weight for i in range(1,len(profits)))\naverage_change = round(average_change, 2)",
"_____no_output_____"
],
[
"greatest_increase = profits[0]\nmonth_g_i = dates[0]\n\ngreatest_decrease = profits[0]\nmonth_g_d = dates[0]\n\nfor idx, amount in enumerate(profits):\n \n if amount > greatest_increase:\n greatest_increase = amount\n month_g_i = dates[idx]\n \n if amount < greatest_decrease:\n greatest_decrease = amount\n month_g_d = dates[idx]",
"_____no_output_____"
],
[
"my_string = 'Financial Analysis\\n'\nmy_string += '-'*20 + '\\n'\nmy_string += 'Total Months: ' + str(number_months) +'\\n'\nmy_string += 'Total: ' + '$' + str(total_amount) +'\\n'\nmy_string += '\\tAverage Change: ' + '$' + str(average_change) +'\\n'\nmy_string += 'Greatest Inrease in Profits: ' + month_g_i + ' ($' + str(greatest_increase) + ')\\n'\nmy_string += 'Greatest Decrease in Profits: ' + month_g_d + ' ($' + str(greatest_decrease) + ')'",
"_____no_output_____"
],
[
"print(my_string)",
"Financial Analysis\n--------------------\nTotal Months: 86\nTotal: $38382578\n\tAverage Change: $-2315.12\nGreatest Inrease in Profits: Feb-2012 ($1170593)\nGreatest Decrease in Profits: Sep-2013 ($-1196225)\n"
],
[
"with open('fin_analysis.txt', 'w') as writer:\n \n writer.write(my_string)",
"_____no_output_____"
]
],
[
[
"# Pandas",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"df_budget = pd.read_csv('Resources/budget_data.csv', index_col='Date')",
"_____no_output_____"
],
[
"#The total number of months included in the dataset.\nnumber_months = df_budget.shape[0]",
"_____no_output_____"
],
[
"#The net total amount of Profit/Losses over the entire period.\ntotal_amount = df_budget['Profit/Losses'].sum()",
"_____no_output_____"
],
[
"# The average of the changes in Profit/Losses over the entire period.\naverage_change = (df_budget['Profit/Losses'] - df_budget['Profit/Losses'].shift(1)).mean(skipna=True).round(2)\n#average_change = df_budget['Profit/Losses'].mean(skipna=True)",
"_____no_output_____"
],
[
"# The greatest increase in profits (date and amount) over the entire period.\ngreatest_increase = df_budget['Profit/Losses'].max()\nmonth_g_i = df_budget.index[df_budget['Profit/Losses'] == greatest_increase].tolist()[0]",
"_____no_output_____"
],
[
"#The greatest decrease in losses (date and amount) over the entire period.\ngreatest_decrease = df_budget['Profit/Losses'].min()\nmonth_g_d = df_budget.index[df_budget['Profit/Losses'] == greatest_decrease].tolist()[0]",
"_____no_output_____"
],
[
"my_string = 'Financial Analysis\\n'\nmy_string += '-'*20 + '\\n'\nmy_string += 'Total Months: ' + str(number_months) +'\\n'\nmy_string += 'Total: ' + '$' + str(total_amount) +'\\n'\nmy_string += '\\tAverage Change: ' + '$' + str(average_change) +'\\n'\nmy_string += 'Greatest Inrease in Profits: ' + month_g_i + ' ($' + str(greatest_increase) + ')\\n'\nmy_string += 'Greatest Decrease in Profits: ' + month_g_d + ' ($' + str(greatest_decrease) + ')'",
"_____no_output_____"
],
[
"print(my_string)",
"Financial Analysis\n--------------------\nTotal Months: 86\nTotal: $38382578\n\tAverage Change: $-2315.12\nGreatest Inrease in Profits: Feb-2012 ($1170593)\nGreatest Decrease in Profits: Sep-2013 ($-1196225)\n"
],
[
"with open('fin_anaalysis.txt', 'w') as writer:\n \n writer.write(my_string)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbab52a022f6f5ccd99ba67daf7adfa7fb68f435
| 700,325 |
ipynb
|
Jupyter Notebook
|
tutorials/2-Advance/FinRL_Ensemble_StockTrading_ICAIF_2020.ipynb
|
Yaotian-Liu/FinRL
|
5b86578f932eca1a402e4af5a3a3555c437107a5
|
[
"MIT"
] | 1 |
2022-03-20T16:01:09.000Z
|
2022-03-20T16:01:09.000Z
|
tutorials/2-Advance/FinRL_Ensemble_StockTrading_ICAIF_2020.ipynb
|
CancerHenry/FinRL
|
5b86578f932eca1a402e4af5a3a3555c437107a5
|
[
"MIT"
] | null | null | null |
tutorials/2-Advance/FinRL_Ensemble_StockTrading_ICAIF_2020.ipynb
|
CancerHenry/FinRL
|
5b86578f932eca1a402e4af5a3a3555c437107a5
|
[
"MIT"
] | 1 |
2022-03-24T05:38:06.000Z
|
2022-03-24T05:38:06.000Z
| 154.59713 | 445,686 | 0.761901 |
[
[
[
"<a href=\"https://colab.research.google.com/github/AI4Finance-Foundation/FinRL/blob/master/FinRL_Ensemble_StockTrading_ICAIF_2020.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Deep Reinforcement Learning for Stock Trading from Scratch: Multiple Stock Trading Using Ensemble Strategy\n\nTutorials to use OpenAI DRL to trade multiple stocks using ensemble strategy in one Jupyter Notebook | Presented at ICAIF 2020\n\n* This notebook is the reimplementation of our paper: Deep Reinforcement Learning for Automated Stock Trading: An Ensemble Strategy, using FinRL.\n* Check out medium blog for detailed explanations: https://medium.com/@ai4finance/deep-reinforcement-learning-for-automated-stock-trading-f1dad0126a02\n* Please report any issues to our Github: https://github.com/AI4Finance-LLC/FinRL-Library/issues\n* **Pytorch Version** \n\n",
"_____no_output_____"
],
[
"# Content",
"_____no_output_____"
],
[
"* [1. Problem Definition](#0)\n* [2. Getting Started - Load Python packages](#1)\n * [2.1. Install Packages](#1.1) \n * [2.2. Check Additional Packages](#1.2)\n * [2.3. Import Packages](#1.3)\n * [2.4. Create Folders](#1.4)\n* [3. Download Data](#2)\n* [4. Preprocess Data](#3) \n * [4.1. Technical Indicators](#3.1)\n * [4.2. Perform Feature Engineering](#3.2)\n* [5.Build Environment](#4) \n * [5.1. Training & Trade Data Split](#4.1)\n * [5.2. User-defined Environment](#4.2) \n * [5.3. Initialize Environment](#4.3) \n* [6.Implement DRL Algorithms](#5) \n* [7.Backtesting Performance](#6) \n * [7.1. BackTestStats](#6.1)\n * [7.2. BackTestPlot](#6.2) \n * [7.3. Baseline Stats](#6.3) \n * [7.3. Compare to Stock Market Index](#6.4) ",
"_____no_output_____"
],
[
"<a id='0'></a>\n# Part 1. Problem Definition",
"_____no_output_____"
],
[
"This problem is to design an automated trading solution for single stock trading. We model the stock trading process as a Markov Decision Process (MDP). We then formulate our trading goal as a maximization problem.\n\nThe algorithm is trained using Deep Reinforcement Learning (DRL) algorithms and the components of the reinforcement learning environment are:\n\n\n* Action: The action space describes the allowed actions that the agent interacts with the\nenvironment. Normally, a ∈ A includes three actions: a ∈ {−1, 0, 1}, where −1, 0, 1 represent\nselling, holding, and buying one stock. Also, an action can be carried upon multiple shares. We use\nan action space {−k, ..., −1, 0, 1, ..., k}, where k denotes the number of shares. For example, \"Buy\n10 shares of AAPL\" or \"Sell 10 shares of AAPL\" are 10 or −10, respectively\n\n* Reward function: r(s, a, s′) is the incentive mechanism for an agent to learn a better action. The change of the portfolio value when action a is taken at state s and arriving at new state s', i.e., r(s, a, s′) = v′ − v, where v′ and v represent the portfolio\nvalues at state s′ and s, respectively\n\n* State: The state space describes the observations that the agent receives from the environment. Just as a human trader needs to analyze various information before executing a trade, so\nour trading agent observes many different features to better learn in an interactive environment.\n\n* Environment: Dow 30 consituents\n\n\nThe data of the single stock that we will be using for this case study is obtained from Yahoo Finance API. The data contains Open-High-Low-Close price and volume.\n",
"_____no_output_____"
],
[
"<a id='1'></a>\n# Part 2. Getting Started- Load Python Packages",
"_____no_output_____"
],
[
"<a id='1.1'></a>\n## 2.1. Install all the packages through FinRL library\n",
"_____no_output_____"
]
],
[
[
"# ## install finrl library\n!pip install git+https://github.com/AI4Finance-LLC/FinRL-Library.git",
"Collecting git+https://github.com/AI4Finance-LLC/FinRL-Library.git\n Cloning https://github.com/AI4Finance-LLC/FinRL-Library.git to /tmp/pip-req-build-c3oa36fu\n Running command git clone -q https://github.com/AI4Finance-LLC/FinRL-Library.git /tmp/pip-req-build-c3oa36fu\nCollecting pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2\n Cloning https://github.com/quantopian/pyfolio.git to /tmp/pip-install-ezrt4uw9/pyfolio_d988d0f156764ce5b3d783a589e28c15\n Running command git clone -q https://github.com/quantopian/pyfolio.git /tmp/pip-install-ezrt4uw9/pyfolio_d988d0f156764ce5b3d783a589e28c15\nCollecting elegantrl@ git+https://github.com/AI4Finance-Foundation/ElegantRL.git#egg=elegantrl\n Cloning https://github.com/AI4Finance-Foundation/ElegantRL.git to /tmp/pip-install-ezrt4uw9/elegantrl_55a3f3c3f82a4ab7924333e69dd21b06\n Running command git clone -q https://github.com/AI4Finance-Foundation/ElegantRL.git /tmp/pip-install-ezrt4uw9/elegantrl_55a3f3c3f82a4ab7924333e69dd21b06\nRequirement already satisfied: numpy>=1.17.3 in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (1.19.5)\nRequirement already satisfied: pandas>=1.1.5 in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (1.1.5)\nCollecting stockstats>=0.4.0\n Downloading stockstats-0.4.1-py2.py3-none-any.whl (19 kB)\nCollecting yfinance\n Downloading yfinance-0.1.69-py2.py3-none-any.whl (26 kB)\nCollecting elegantrl\n Downloading elegantrl-0.3.3-py3-none-any.whl (234 kB)\n\u001b[K |████████████████████████████████| 234 kB 10.8 MB/s \n\u001b[?25hRequirement already satisfied: matplotlib in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (3.2.2)\nRequirement already satisfied: scikit-learn>=0.21.0 in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (1.0.2)\nRequirement already satisfied: gym>=0.17 in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (0.17.3)\nCollecting stable-baselines3[extra]\n Downloading stable_baselines3-1.3.0-py3-none-any.whl (174 kB)\n\u001b[K |████████████████████████████████| 174 kB 55.1 MB/s \n\u001b[?25hCollecting ray[default]\n Downloading ray-1.9.2-cp37-cp37m-manylinux2014_x86_64.whl (57.6 MB)\n\u001b[K |████████████████████████████████| 57.6 MB 1.2 MB/s \n\u001b[?25hCollecting lz4\n Downloading lz4-3.1.10-cp37-cp37m-manylinux2010_x86_64.whl (1.8 MB)\n\u001b[K |████████████████████████████████| 1.8 MB 21.0 MB/s \n\u001b[?25hCollecting tensorboardX\n Downloading tensorboardX-2.4.1-py2.py3-none-any.whl (124 kB)\n\u001b[K |████████████████████████████████| 124 kB 52.5 MB/s \n\u001b[?25hCollecting gputil\n Downloading GPUtil-1.4.0.tar.gz (5.5 kB)\nCollecting exchange_calendars\n Downloading exchange_calendars-3.5.tar.gz (147 kB)\n\u001b[K |████████████████████████████████| 147 kB 60.1 MB/s \n\u001b[?25hCollecting alpaca_trade_api\n Downloading alpaca_trade_api-1.4.3-py3-none-any.whl (36 kB)\nCollecting ccxt>=1.66.32\n Downloading ccxt-1.67.31-py2.py3-none-any.whl (2.3 MB)\n\u001b[K |████████████████████████████████| 2.3 MB 64.8 MB/s \n\u001b[?25hCollecting jqdatasdk\n Downloading jqdatasdk-1.8.10-py3-none-any.whl (153 kB)\n\u001b[K |████████████████████████████████| 153 kB 41.8 MB/s \n\u001b[?25hCollecting wrds\n Downloading wrds-3.1.1-py3-none-any.whl (12 kB)\nRequirement already satisfied: pytest in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (3.6.4)\nRequirement already satisfied: setuptools>=41.4.0 in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (57.4.0)\nRequirement already satisfied: wheel>=0.33.6 in /usr/local/lib/python3.7/dist-packages (from finrl==0.3.4) (0.37.1)\nCollecting pre-commit\n Downloading pre_commit-2.16.0-py2.py3-none-any.whl (191 kB)\n\u001b[K |████████████████████████████████| 191 kB 35.1 MB/s \n\u001b[?25hCollecting pybullet\n Downloading pybullet-3.2.1-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.whl (90.8 MB)\n\u001b[K |████████████████████████████████| 90.8 MB 315 bytes/s \n\u001b[?25hRequirement already satisfied: torch in /usr/local/lib/python3.7/dist-packages (from elegantrl@ git+https://github.com/AI4Finance-Foundation/ElegantRL.git#egg=elegantrl->finrl==0.3.4) (1.10.0+cu111)\nRequirement already satisfied: opencv-python in /usr/local/lib/python3.7/dist-packages (from elegantrl@ git+https://github.com/AI4Finance-Foundation/ElegantRL.git#egg=elegantrl->finrl==0.3.4) (4.1.2.30)\nCollecting box2d-py\n Downloading box2d_py-2.3.8-cp37-cp37m-manylinux1_x86_64.whl (448 kB)\n\u001b[K |████████████████████████████████| 448 kB 61.8 MB/s \n\u001b[?25hRequirement already satisfied: ipython>=3.2.3 in /usr/local/lib/python3.7/dist-packages (from pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (5.5.0)\nRequirement already satisfied: pytz>=2014.10 in /usr/local/lib/python3.7/dist-packages (from pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (2018.9)\nRequirement already satisfied: scipy>=0.14.0 in /usr/local/lib/python3.7/dist-packages (from pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (1.4.1)\nRequirement already satisfied: seaborn>=0.7.1 in /usr/local/lib/python3.7/dist-packages (from pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (0.11.2)\nCollecting empyrical>=0.5.0\n Downloading empyrical-0.5.5.tar.gz (52 kB)\n\u001b[K |████████████████████████████████| 52 kB 1.3 MB/s \n\u001b[?25hCollecting aiohttp>=3.8\n Downloading aiohttp-3.8.1-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (1.1 MB)\n\u001b[K |████████████████████████████████| 1.1 MB 51.5 MB/s \n\u001b[?25hCollecting cryptography>=2.6.1\n Downloading cryptography-36.0.1-cp36-abi3-manylinux_2_24_x86_64.whl (3.6 MB)\n\u001b[K |████████████████████████████████| 3.6 MB 61.2 MB/s \n\u001b[?25hRequirement already satisfied: certifi>=2018.1.18 in /usr/local/lib/python3.7/dist-packages (from ccxt>=1.66.32->finrl==0.3.4) (2021.10.8)\nCollecting yarl==1.7.2\n Downloading yarl-1.7.2-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (271 kB)\n\u001b[K |████████████████████████████████| 271 kB 60.1 MB/s \n\u001b[?25hRequirement already satisfied: requests>=2.18.4 in /usr/local/lib/python3.7/dist-packages (from ccxt>=1.66.32->finrl==0.3.4) (2.23.0)\nCollecting aiodns>=1.1.1\n Downloading aiodns-3.0.0-py3-none-any.whl (5.0 kB)\nCollecting multidict>=4.0\n Downloading multidict-5.2.0-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (160 kB)\n\u001b[K |████████████████████████████████| 160 kB 60.0 MB/s \n\u001b[?25hRequirement already satisfied: idna>=2.0 in /usr/local/lib/python3.7/dist-packages (from yarl==1.7.2->ccxt>=1.66.32->finrl==0.3.4) (2.10)\nRequirement already satisfied: typing-extensions>=3.7.4 in /usr/local/lib/python3.7/dist-packages (from yarl==1.7.2->ccxt>=1.66.32->finrl==0.3.4) (3.10.0.2)\nCollecting pycares>=4.0.0\n Downloading pycares-4.1.2-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (291 kB)\n\u001b[K |████████████████████████████████| 291 kB 57.3 MB/s \n\u001b[?25hRequirement already satisfied: charset-normalizer<3.0,>=2.0 in /usr/local/lib/python3.7/dist-packages (from aiohttp>=3.8->ccxt>=1.66.32->finrl==0.3.4) (2.0.10)\nCollecting asynctest==0.13.0\n Downloading asynctest-0.13.0-py3-none-any.whl (26 kB)\nCollecting aiosignal>=1.1.2\n Downloading aiosignal-1.2.0-py3-none-any.whl (8.2 kB)\nRequirement already satisfied: attrs>=17.3.0 in /usr/local/lib/python3.7/dist-packages (from aiohttp>=3.8->ccxt>=1.66.32->finrl==0.3.4) (21.4.0)\nCollecting frozenlist>=1.1.1\n Downloading frozenlist-1.2.0-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (192 kB)\n\u001b[K |████████████████████████████████| 192 kB 30.1 MB/s \n\u001b[?25hCollecting async-timeout<5.0,>=4.0.0a3\n Downloading async_timeout-4.0.2-py3-none-any.whl (5.8 kB)\nRequirement already satisfied: cffi>=1.12 in /usr/local/lib/python3.7/dist-packages (from cryptography>=2.6.1->ccxt>=1.66.32->finrl==0.3.4) (1.15.0)\nRequirement already satisfied: pycparser in /usr/local/lib/python3.7/dist-packages (from cffi>=1.12->cryptography>=2.6.1->ccxt>=1.66.32->finrl==0.3.4) (2.21)\nRequirement already satisfied: pandas-datareader>=0.2 in /usr/local/lib/python3.7/dist-packages (from empyrical>=0.5.0->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (0.9.0)\nRequirement already satisfied: pyglet<=1.5.0,>=1.4.0 in /usr/local/lib/python3.7/dist-packages (from gym>=0.17->finrl==0.3.4) (1.5.0)\nRequirement already satisfied: cloudpickle<1.7.0,>=1.2.0 in /usr/local/lib/python3.7/dist-packages (from gym>=0.17->finrl==0.3.4) (1.3.0)\nRequirement already satisfied: traitlets>=4.2 in /usr/local/lib/python3.7/dist-packages (from ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (5.1.1)\nRequirement already satisfied: pexpect in /usr/local/lib/python3.7/dist-packages (from ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (4.8.0)\nRequirement already satisfied: prompt-toolkit<2.0.0,>=1.0.4 in /usr/local/lib/python3.7/dist-packages (from ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (1.0.18)\nRequirement already satisfied: decorator in /usr/local/lib/python3.7/dist-packages (from ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (4.4.2)\nRequirement already satisfied: pickleshare in /usr/local/lib/python3.7/dist-packages (from ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (0.7.5)\nRequirement already satisfied: simplegeneric>0.8 in /usr/local/lib/python3.7/dist-packages (from ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (0.8.1)\nRequirement already satisfied: pygments in /usr/local/lib/python3.7/dist-packages (from ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (2.6.1)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.7/dist-packages (from matplotlib->finrl==0.3.4) (0.11.0)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->finrl==0.3.4) (1.3.2)\nRequirement already satisfied: python-dateutil>=2.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->finrl==0.3.4) (2.8.2)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.7/dist-packages (from matplotlib->finrl==0.3.4) (3.0.6)\nRequirement already satisfied: lxml in /usr/local/lib/python3.7/dist-packages (from pandas-datareader>=0.2->empyrical>=0.5.0->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (4.2.6)\nRequirement already satisfied: six>=1.9.0 in /usr/local/lib/python3.7/dist-packages (from prompt-toolkit<2.0.0,>=1.0.4->ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (1.15.0)\nRequirement already satisfied: wcwidth in /usr/local/lib/python3.7/dist-packages (from prompt-toolkit<2.0.0,>=1.0.4->ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (0.2.5)\nRequirement already satisfied: future in /usr/local/lib/python3.7/dist-packages (from pyglet<=1.5.0,>=1.4.0->gym>=0.17->finrl==0.3.4) (0.16.0)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests>=2.18.4->ccxt>=1.66.32->finrl==0.3.4) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests>=2.18.4->ccxt>=1.66.32->finrl==0.3.4) (1.24.3)\nRequirement already satisfied: threadpoolctl>=2.0.0 in /usr/local/lib/python3.7/dist-packages (from scikit-learn>=0.21.0->finrl==0.3.4) (3.0.0)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.7/dist-packages (from scikit-learn>=0.21.0->finrl==0.3.4) (1.1.0)\nCollecting websockets<10,>=8.0\n Downloading websockets-9.1-cp37-cp37m-manylinux2010_x86_64.whl (103 kB)\n\u001b[K |████████████████████████████████| 103 kB 70.9 MB/s \n\u001b[?25hCollecting websocket-client<2,>=0.56.0\n Downloading websocket_client-1.2.3-py3-none-any.whl (53 kB)\n\u001b[K |████████████████████████████████| 53 kB 2.0 MB/s \n\u001b[?25hCollecting msgpack==1.0.2\n Downloading msgpack-1.0.2-cp37-cp37m-manylinux1_x86_64.whl (273 kB)\n\u001b[K |████████████████████████████████| 273 kB 72.2 MB/s \n\u001b[?25hCollecting PyYAML==5.4.1\n Downloading PyYAML-5.4.1-cp37-cp37m-manylinux1_x86_64.whl (636 kB)\n\u001b[K |████████████████████████████████| 636 kB 72.5 MB/s \n\u001b[?25hCollecting alpaca_trade_api\n Downloading alpaca_trade_api-1.4.2-py3-none-any.whl (36 kB)\n Downloading alpaca_trade_api-1.4.1-py3-none-any.whl (36 kB)\n Downloading alpaca_trade_api-1.4.0-py3-none-any.whl (34 kB)\n Downloading alpaca_trade_api-1.3.0-py3-none-any.whl (43 kB)\n\u001b[K |████████████████████████████████| 43 kB 1.6 MB/s \n\u001b[?25h Downloading alpaca_trade_api-1.2.3-py3-none-any.whl (40 kB)\n\u001b[K |████████████████████████████████| 40 kB 5.4 MB/s \n\u001b[?25hCollecting pyluach\n Downloading pyluach-1.3.0-py3-none-any.whl (17 kB)\nRequirement already satisfied: toolz in /usr/local/lib/python3.7/dist-packages (from exchange_calendars->finrl==0.3.4) (0.11.2)\nRequirement already satisfied: korean_lunar_calendar in /usr/local/lib/python3.7/dist-packages (from exchange_calendars->finrl==0.3.4) (0.2.1)\nRequirement already satisfied: SQLAlchemy>=1.2.8 in /usr/local/lib/python3.7/dist-packages (from jqdatasdk->finrl==0.3.4) (1.4.29)\nCollecting pymysql>=0.7.6\n Downloading PyMySQL-1.0.2-py3-none-any.whl (43 kB)\n\u001b[K |████████████████████████████████| 43 kB 2.0 MB/s \n\u001b[?25hCollecting thriftpy2>=0.3.9\n Downloading thriftpy2-0.4.14.tar.gz (361 kB)\n\u001b[K |████████████████████████████████| 361 kB 72.3 MB/s \n\u001b[?25hRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from SQLAlchemy>=1.2.8->jqdatasdk->finrl==0.3.4) (4.10.0)\nRequirement already satisfied: greenlet!=0.4.17 in /usr/local/lib/python3.7/dist-packages (from SQLAlchemy>=1.2.8->jqdatasdk->finrl==0.3.4) (1.1.2)\nCollecting ply<4.0,>=3.4\n Downloading ply-3.11-py2.py3-none-any.whl (49 kB)\n\u001b[K |████████████████████████████████| 49 kB 5.4 MB/s \n\u001b[?25hRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata->SQLAlchemy>=1.2.8->jqdatasdk->finrl==0.3.4) (3.7.0)\nRequirement already satisfied: ptyprocess>=0.5 in /usr/local/lib/python3.7/dist-packages (from pexpect->ipython>=3.2.3->pyfolio@ git+https://github.com/quantopian/pyfolio.git#egg=pyfolio-0.9.2->finrl==0.3.4) (0.7.0)\nCollecting nodeenv>=0.11.1\n Downloading nodeenv-1.6.0-py2.py3-none-any.whl (21 kB)\nCollecting identify>=1.0.0\n Downloading identify-2.4.3-py2.py3-none-any.whl (98 kB)\n\u001b[K |████████████████████████████████| 98 kB 7.3 MB/s \n\u001b[?25hCollecting virtualenv>=20.0.8\n Downloading virtualenv-20.13.0-py2.py3-none-any.whl (6.5 MB)\n\u001b[K |████████████████████████████████| 6.5 MB 50.3 MB/s \n\u001b[?25hRequirement already satisfied: toml in /usr/local/lib/python3.7/dist-packages (from pre-commit->finrl==0.3.4) (0.10.2)\nCollecting pyyaml>=5.1\n Downloading PyYAML-6.0-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (596 kB)\n\u001b[K |████████████████████████████████| 596 kB 54.8 MB/s \n\u001b[?25hCollecting cfgv>=2.0.0\n Downloading cfgv-3.3.1-py2.py3-none-any.whl (7.3 kB)\nRequirement already satisfied: filelock<4,>=3.2 in /usr/local/lib/python3.7/dist-packages (from virtualenv>=20.0.8->pre-commit->finrl==0.3.4) (3.4.2)\nCollecting distlib<1,>=0.3.1\n Downloading distlib-0.3.4-py2.py3-none-any.whl (461 kB)\n\u001b[K |████████████████████████████████| 461 kB 48.2 MB/s \n\u001b[?25hCollecting platformdirs<3,>=2\n Downloading platformdirs-2.4.1-py3-none-any.whl (14 kB)\nRequirement already satisfied: more-itertools>=4.0.0 in /usr/local/lib/python3.7/dist-packages (from pytest->finrl==0.3.4) (8.12.0)\nRequirement already satisfied: py>=1.5.0 in /usr/local/lib/python3.7/dist-packages (from pytest->finrl==0.3.4) (1.11.0)\nRequirement already satisfied: pluggy<0.8,>=0.5 in /usr/local/lib/python3.7/dist-packages (from pytest->finrl==0.3.4) (0.7.1)\nRequirement already satisfied: atomicwrites>=1.0 in /usr/local/lib/python3.7/dist-packages (from pytest->finrl==0.3.4) (1.4.0)\nRequirement already satisfied: click>=7.0 in /usr/local/lib/python3.7/dist-packages (from ray[default]->finrl==0.3.4) (7.1.2)\nRequirement already satisfied: protobuf>=3.15.3 in /usr/local/lib/python3.7/dist-packages (from ray[default]->finrl==0.3.4) (3.17.3)\nRequirement already satisfied: jsonschema in /usr/local/lib/python3.7/dist-packages (from ray[default]->finrl==0.3.4) (4.3.3)\nCollecting redis>=3.5.0\n Downloading redis-4.1.0-py3-none-any.whl (171 kB)\n\u001b[K |████████████████████████████████| 171 kB 70.6 MB/s \n\u001b[?25hRequirement already satisfied: grpcio>=1.28.1 in /usr/local/lib/python3.7/dist-packages (from ray[default]->finrl==0.3.4) (1.43.0)\nCollecting opencensus\n Downloading opencensus-0.8.0-py2.py3-none-any.whl (128 kB)\n\u001b[K |████████████████████████████████| 128 kB 63.0 MB/s \n\u001b[?25hRequirement already satisfied: smart-open in /usr/local/lib/python3.7/dist-packages (from ray[default]->finrl==0.3.4) (5.2.1)\nCollecting colorful\n Downloading colorful-0.5.4-py2.py3-none-any.whl (201 kB)\n\u001b[K |████████████████████████████████| 201 kB 59.3 MB/s \n\u001b[?25hCollecting aiohttp-cors\n Downloading aiohttp_cors-0.7.0-py3-none-any.whl (27 kB)\nCollecting aioredis<2\n Downloading aioredis-1.3.1-py3-none-any.whl (65 kB)\n\u001b[K |████████████████████████████████| 65 kB 3.4 MB/s \n\u001b[?25hRequirement already satisfied: prometheus-client>=0.7.1 in /usr/local/lib/python3.7/dist-packages (from ray[default]->finrl==0.3.4) (0.12.0)\nCollecting gpustat>=1.0.0b1\n Downloading gpustat-1.0.0b1.tar.gz (82 kB)\n\u001b[K |████████████████████████████████| 82 kB 212 kB/s \n\u001b[?25hCollecting py-spy>=0.2.0\n Downloading py_spy-0.3.11-py2.py3-none-manylinux_2_5_x86_64.manylinux1_x86_64.whl (3.0 MB)\n\u001b[K |████████████████████████████████| 3.0 MB 55.2 MB/s \n\u001b[?25hCollecting hiredis\n Downloading hiredis-2.0.0-cp37-cp37m-manylinux2010_x86_64.whl (85 kB)\n\u001b[K |████████████████████████████████| 85 kB 3.7 MB/s \n\u001b[?25hRequirement already satisfied: nvidia-ml-py3>=7.352.0 in /usr/local/lib/python3.7/dist-packages (from gpustat>=1.0.0b1->ray[default]->finrl==0.3.4) (7.352.0)\nRequirement already satisfied: psutil in /usr/local/lib/python3.7/dist-packages (from gpustat>=1.0.0b1->ray[default]->finrl==0.3.4) (5.4.8)\nCollecting blessed>=1.17.1\n Downloading blessed-1.19.0-py2.py3-none-any.whl (57 kB)\n\u001b[K |████████████████████████████████| 57 kB 5.2 MB/s \n\u001b[?25hCollecting deprecated>=1.2.3\n Downloading Deprecated-1.2.13-py2.py3-none-any.whl (9.6 kB)\nRequirement already satisfied: packaging>=21.3 in /usr/local/lib/python3.7/dist-packages (from redis>=3.5.0->ray[default]->finrl==0.3.4) (21.3)\nRequirement already satisfied: wrapt<2,>=1.10 in /usr/local/lib/python3.7/dist-packages (from deprecated>=1.2.3->redis>=3.5.0->ray[default]->finrl==0.3.4) (1.13.3)\nRequirement already satisfied: importlib-resources>=1.4.0 in /usr/local/lib/python3.7/dist-packages (from jsonschema->ray[default]->finrl==0.3.4) (5.4.0)\nRequirement already satisfied: pyrsistent!=0.17.0,!=0.17.1,!=0.17.2,>=0.14.0 in /usr/local/lib/python3.7/dist-packages (from jsonschema->ray[default]->finrl==0.3.4) (0.18.0)\nCollecting opencensus-context==0.1.2\n Downloading opencensus_context-0.1.2-py2.py3-none-any.whl (4.4 kB)\nRequirement already satisfied: google-api-core<3.0.0,>=1.0.0 in /usr/local/lib/python3.7/dist-packages (from opencensus->ray[default]->finrl==0.3.4) (1.26.3)\nRequirement already satisfied: googleapis-common-protos<2.0dev,>=1.6.0 in /usr/local/lib/python3.7/dist-packages (from google-api-core<3.0.0,>=1.0.0->opencensus->ray[default]->finrl==0.3.4) (1.54.0)\nRequirement already satisfied: google-auth<2.0dev,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from google-api-core<3.0.0,>=1.0.0->opencensus->ray[default]->finrl==0.3.4) (1.35.0)\nRequirement already satisfied: cachetools<5.0,>=2.0.0 in /usr/local/lib/python3.7/dist-packages (from google-auth<2.0dev,>=1.21.1->google-api-core<3.0.0,>=1.0.0->opencensus->ray[default]->finrl==0.3.4) (4.2.4)\nRequirement already satisfied: rsa<5,>=3.1.4 in /usr/local/lib/python3.7/dist-packages (from google-auth<2.0dev,>=1.21.1->google-api-core<3.0.0,>=1.0.0->opencensus->ray[default]->finrl==0.3.4) (4.8)\nRequirement already satisfied: pyasn1-modules>=0.2.1 in /usr/local/lib/python3.7/dist-packages (from google-auth<2.0dev,>=1.21.1->google-api-core<3.0.0,>=1.0.0->opencensus->ray[default]->finrl==0.3.4) (0.2.8)\nRequirement already satisfied: pyasn1<0.5.0,>=0.4.6 in /usr/local/lib/python3.7/dist-packages (from pyasn1-modules>=0.2.1->google-auth<2.0dev,>=1.21.1->google-api-core<3.0.0,>=1.0.0->opencensus->ray[default]->finrl==0.3.4) (0.4.8)\nRequirement already satisfied: tabulate in /usr/local/lib/python3.7/dist-packages (from ray[default]->finrl==0.3.4) (0.8.9)\nRequirement already satisfied: atari-py~=0.2.0 in /usr/local/lib/python3.7/dist-packages (from stable-baselines3[extra]->finrl==0.3.4) (0.2.9)\nRequirement already satisfied: tensorboard>=2.2.0 in /usr/local/lib/python3.7/dist-packages (from stable-baselines3[extra]->finrl==0.3.4) (2.7.0)\nRequirement already satisfied: pillow in /usr/local/lib/python3.7/dist-packages (from stable-baselines3[extra]->finrl==0.3.4) (7.1.2)\nRequirement already satisfied: absl-py>=0.4 in /usr/local/lib/python3.7/dist-packages (from tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (0.12.0)\nRequirement already satisfied: werkzeug>=0.11.15 in /usr/local/lib/python3.7/dist-packages (from tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (1.0.1)\nRequirement already satisfied: tensorboard-plugin-wit>=1.6.0 in /usr/local/lib/python3.7/dist-packages (from tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (1.8.1)\nRequirement already satisfied: tensorboard-data-server<0.7.0,>=0.6.0 in /usr/local/lib/python3.7/dist-packages (from tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (0.6.1)\nRequirement already satisfied: markdown>=2.6.8 in /usr/local/lib/python3.7/dist-packages (from tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (3.3.6)\nRequirement already satisfied: google-auth-oauthlib<0.5,>=0.4.1 in /usr/local/lib/python3.7/dist-packages (from tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (0.4.6)\nRequirement already satisfied: requests-oauthlib>=0.7.0 in /usr/local/lib/python3.7/dist-packages (from google-auth-oauthlib<0.5,>=0.4.1->tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (1.3.0)\nRequirement already satisfied: oauthlib>=3.0.0 in /usr/local/lib/python3.7/dist-packages (from requests-oauthlib>=0.7.0->google-auth-oauthlib<0.5,>=0.4.1->tensorboard>=2.2.0->stable-baselines3[extra]->finrl==0.3.4) (3.1.1)\nCollecting mock\n Downloading mock-4.0.3-py3-none-any.whl (28 kB)\nCollecting psycopg2-binary\n Downloading psycopg2_binary-2.9.3-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (3.0 MB)\n\u001b[K |████████████████████████████████| 3.0 MB 48.5 MB/s \n\u001b[?25hCollecting requests>=2.18.4\n Downloading requests-2.27.1-py2.py3-none-any.whl (63 kB)\n\u001b[K |████████████████████████████████| 63 kB 1.8 MB/s \n\u001b[?25hCollecting lxml\n Downloading lxml-4.7.1-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.manylinux_2_24_x86_64.whl (6.4 MB)\n\u001b[K |████████████████████████████████| 6.4 MB 53.5 MB/s \n\u001b[?25hRequirement already satisfied: multitasking>=0.0.7 in /usr/local/lib/python3.7/dist-packages (from yfinance->finrl==0.3.4) (0.0.10)\nBuilding wheels for collected packages: finrl, elegantrl, pyfolio, empyrical, exchange-calendars, gputil, thriftpy2, gpustat\n Building wheel for finrl (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for finrl: filename=finrl-0.3.4-py3-none-any.whl size=3885434 sha256=6b8baae9760d09314c0a59fa9ef62e9f037664bec2f04d47d8b0d3c5374a8510\n Stored in directory: /tmp/pip-ephem-wheel-cache-gr647fxc/wheels/17/ff/bd/1bc602a0352762b0b24041b88536d803ae343ed0a711fcf55e\n Building wheel for elegantrl (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for elegantrl: filename=elegantrl-0.3.3-py3-none-any.whl size=188472 sha256=2c5b9ab71f0f4482c6a5a98f0572a436a43bb607408a90c30fc0afaf43959351\n Stored in directory: /tmp/pip-ephem-wheel-cache-gr647fxc/wheels/99/85/5e/86cb3a9f47adfca5e248295e93113e1b298d60883126d62c84\n Building wheel for pyfolio (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for pyfolio: filename=pyfolio-0.9.2+75.g4b901f6-py3-none-any.whl size=75774 sha256=9fde427dbea0dfb404cc00e1e8a8192579b8a3003862be8db37698dc7c2e3a25\n Stored in directory: /tmp/pip-ephem-wheel-cache-gr647fxc/wheels/ef/09/e5/2c1bf37c050d22557c080deb1be986d06424627c04aeca19b9\n Building wheel for empyrical (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for empyrical: filename=empyrical-0.5.5-py3-none-any.whl size=39780 sha256=6214337999e9c719132d608b0a6401b8357b3dd8e4993a3be6ae6d364ece96c0\n Stored in directory: /root/.cache/pip/wheels/d9/91/4b/654fcff57477efcf149eaca236da2fce991526cbab431bf312\n Building wheel for exchange-calendars (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for exchange-calendars: filename=exchange_calendars-3.5-py3-none-any.whl size=179486 sha256=e9b623cfad94cfdf693e78b266c1edaa44deda176197fe6d01ca6b825a5de66f\n Stored in directory: /root/.cache/pip/wheels/69/21/43/b6ae2605dd767f6cd5a5b0b70c93a9a75823e44b3ccb92bce7\n Building wheel for gputil (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for gputil: filename=GPUtil-1.4.0-py3-none-any.whl size=7411 sha256=e57a10f985ecada50b63b780c8c94993a9c88220d22e86cf95d9edd6849fb0c1\n Stored in directory: /root/.cache/pip/wheels/6e/f8/83/534c52482d6da64622ddbf72cd93c35d2ef2881b78fd08ff0c\n Building wheel for thriftpy2 (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for thriftpy2: filename=thriftpy2-0.4.14-cp37-cp37m-linux_x86_64.whl size=944229 sha256=a726ea63447440ad4e81f049aff2d238172bd719e370b86b314126b8b9b56b0b\n Stored in directory: /root/.cache/pip/wheels/2a/f5/49/9c0d851aa64b58db72883cf9393cc824d536bdf13f5c83cff4\n Building wheel for gpustat (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for gpustat: filename=gpustat-1.0.0b1-py3-none-any.whl size=15979 sha256=a125e11ecb6ea252334188ef2356e3cef9a94fef6d1c3b2ae7028a76777ff3b9\n Stored in directory: /root/.cache/pip/wheels/1a/16/e2/3e2437fba4c4b6a97a97bd96fce5d14e66cff5c4966fb1cc8c\nSuccessfully built finrl elegantrl pyfolio empyrical exchange-calendars gputil thriftpy2 gpustat\nInstalling collected packages: requests, multidict, frozenlist, yarl, lxml, deprecated, asynctest, async-timeout, aiosignal, redis, pyyaml, pycares, ply, platformdirs, opencensus-context, msgpack, hiredis, distlib, blessed, aiohttp, websockets, websocket-client, virtualenv, thriftpy2, tensorboardX, stable-baselines3, ray, pymysql, pyluach, pybullet, py-spy, psycopg2-binary, opencensus, nodeenv, mock, identify, gpustat, empyrical, cryptography, colorful, cfgv, box2d-py, aioredis, aiohttp-cors, aiodns, yfinance, wrds, stockstats, pyfolio, pre-commit, lz4, jqdatasdk, gputil, exchange-calendars, elegantrl, ccxt, alpaca-trade-api, finrl\n Attempting uninstall: requests\n Found existing installation: requests 2.23.0\n Uninstalling requests-2.23.0:\n Successfully uninstalled requests-2.23.0\n Attempting uninstall: lxml\n Found existing installation: lxml 4.2.6\n Uninstalling lxml-4.2.6:\n Successfully uninstalled lxml-4.2.6\n Attempting uninstall: pyyaml\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\n Attempting uninstall: msgpack\n Found existing installation: msgpack 1.0.3\n Uninstalling msgpack-1.0.3:\n Successfully uninstalled msgpack-1.0.3\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ngoogle-colab 1.0.0 requires requests~=2.23.0, but you have requests 2.27.1 which is incompatible.\ndatascience 0.10.6 requires folium==0.2.1, but you have folium 0.8.3 which is incompatible.\u001b[0m\nSuccessfully installed aiodns-3.0.0 aiohttp-3.8.1 aiohttp-cors-0.7.0 aioredis-1.3.1 aiosignal-1.2.0 alpaca-trade-api-1.2.3 async-timeout-4.0.2 asynctest-0.13.0 blessed-1.19.0 box2d-py-2.3.8 ccxt-1.67.31 cfgv-3.3.1 colorful-0.5.4 cryptography-36.0.1 deprecated-1.2.13 distlib-0.3.4 elegantrl-0.3.3 empyrical-0.5.5 exchange-calendars-3.5 finrl-0.3.4 frozenlist-1.2.0 gpustat-1.0.0b1 gputil-1.4.0 hiredis-2.0.0 identify-2.4.3 jqdatasdk-1.8.10 lxml-4.7.1 lz4-3.1.10 mock-4.0.3 msgpack-1.0.2 multidict-5.2.0 nodeenv-1.6.0 opencensus-0.8.0 opencensus-context-0.1.2 platformdirs-2.4.1 ply-3.11 pre-commit-2.16.0 psycopg2-binary-2.9.3 py-spy-0.3.11 pybullet-3.2.1 pycares-4.1.2 pyfolio-0.9.2+75.g4b901f6 pyluach-1.3.0 pymysql-1.0.2 pyyaml-6.0 ray-1.9.2 redis-4.1.0 requests-2.27.1 stable-baselines3-1.3.0 stockstats-0.4.1 tensorboardX-2.4.1 thriftpy2-0.4.14 virtualenv-20.13.0 websocket-client-1.2.3 websockets-9.1 wrds-3.1.1 yarl-1.7.2 yfinance-0.1.69\n"
]
],
[
[
"\n<a id='1.2'></a>\n## 2.2. Check if the additional packages needed are present, if not install them. \n* Yahoo Finance API\n* pandas\n* numpy\n* matplotlib\n* stockstats\n* OpenAI gym\n* stable-baselines\n* tensorflow\n* pyfolio",
"_____no_output_____"
],
[
"<a id='1.3'></a>\n## 2.3. Import Packages",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt\n# matplotlib.use('Agg')\nimport datetime\n\n%matplotlib inline\nfrom finrl import config\nfrom finrl import config_tickers\nfrom finrl.finrl_meta.preprocessor.yahoodownloader import YahooDownloader\nfrom finrl.finrl_meta.preprocessor.preprocessors import FeatureEngineer, data_split\nfrom finrl.finrl_meta.env_stock_trading.env_stocktrading import StockTradingEnv\nfrom finrl.agents.stablebaselines3.models import DRLAgent,DRLEnsembleAgent\nfrom finrl.plot import backtest_stats, backtest_plot, get_daily_return, get_baseline\n\nfrom pprint import pprint\n\nimport sys\nsys.path.append(\"../FinRL-Library\")\n\nimport itertools",
"_____no_output_____"
]
],
[
[
"<a id='1.4'></a>\n## 2.4. Create Folders",
"_____no_output_____"
]
],
[
[
"import os\nif not os.path.exists(\"./\" + config.DATA_SAVE_DIR):\n os.makedirs(\"./\" + config.DATA_SAVE_DIR)\nif not os.path.exists(\"./\" + config.TRAINED_MODEL_DIR):\n os.makedirs(\"./\" + config.TRAINED_MODEL_DIR)\nif not os.path.exists(\"./\" + config.TENSORBOARD_LOG_DIR):\n os.makedirs(\"./\" + config.TENSORBOARD_LOG_DIR)\nif not os.path.exists(\"./\" + config.RESULTS_DIR):\n os.makedirs(\"./\" + config.RESULTS_DIR)",
"_____no_output_____"
]
],
[
[
"<a id='2'></a>\n# Part 3. Download Data\nYahoo Finance is a website that provides stock data, financial news, financial reports, etc. All the data provided by Yahoo Finance is free.\n* FinRL uses a class **YahooDownloader** to fetch data from Yahoo Finance API\n* Call Limit: Using the Public API (without authentication), you are limited to 2,000 requests per hour per IP (or up to a total of 48,000 requests a day).\n",
"_____no_output_____"
],
[
"\n\n-----\nclass YahooDownloader:\n Provides methods for retrieving daily stock data from\n Yahoo Finance API\n\n Attributes\n ----------\n start_date : str\n start date of the data (modified from config.py)\n end_date : str\n end date of the data (modified from config.py)\n ticker_list : list\n a list of stock tickers (modified from config.py)\n\n Methods\n -------\n fetch_data()\n Fetches data from yahoo API\n",
"_____no_output_____"
]
],
[
[
"# from config.py start_date is a string\nconfig.START_DATE",
"_____no_output_____"
],
[
"print(config_tickers.DOW_30_TICKER)",
"['AXP', 'AMGN', 'AAPL', 'BA', 'CAT', 'CSCO', 'CVX', 'GS', 'HD', 'HON', 'IBM', 'INTC', 'JNJ', 'KO', 'JPM', 'MCD', 'MMM', 'MRK', 'MSFT', 'NKE', 'PG', 'TRV', 'UNH', 'CRM', 'VZ', 'V', 'WBA', 'WMT', 'DIS', 'DOW']\n"
],
[
"df = YahooDownloader(start_date = '2009-01-01',\n end_date = '2021-07-06',\n ticker_list = config_tickers.DOW_30_TICKER).fetch_data()",
"[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\n[*********************100%***********************] 1 of 1 completed\nShape of DataFrame: (91841, 8)\n"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.tail()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.sort_values(['date','tic']).head()",
"_____no_output_____"
],
[
"len(df.tic.unique())",
"_____no_output_____"
],
[
"df.tic.value_counts()",
"_____no_output_____"
]
],
[
[
"# Part 4: Preprocess Data\nData preprocessing is a crucial step for training a high quality machine learning model. We need to check for missing data and do feature engineering in order to convert the data into a model-ready state.\n* Add technical indicators. In practical trading, various information needs to be taken into account, for example the historical stock prices, current holding shares, technical indicators, etc. In this article, we demonstrate two trend-following technical indicators: MACD and RSI.\n* Add turbulence index. Risk-aversion reflects whether an investor will choose to preserve the capital. It also influences one's trading strategy when facing different market volatility level. To control the risk in a worst-case scenario, such as financial crisis of 2007–2008, FinRL employs the financial turbulence index that measures extreme asset price fluctuation.",
"_____no_output_____"
]
],
[
[
"tech_indicators = ['macd',\n 'rsi_30',\n 'cci_30',\n 'dx_30']",
"_____no_output_____"
],
[
"fe = FeatureEngineer(\n use_technical_indicator=True,\n tech_indicator_list = tech_indicators,\n use_turbulence=True,\n user_defined_feature = False)\n\nprocessed = fe.preprocess_data(df)\nprocessed = processed.copy()\nprocessed = processed.fillna(0)\nprocessed = processed.replace(np.inf,0)",
"Successfully added technical indicators\nSuccessfully added turbulence index\n"
],
[
"processed.sample(5)",
"_____no_output_____"
]
],
[
[
"<a id='4'></a>\n# Part 5. Design Environment\nConsidering the stochastic and interactive nature of the automated stock trading tasks, a financial task is modeled as a **Markov Decision Process (MDP)** problem. The training process involves observing stock price change, taking an action and reward's calculation to have the agent adjusting its strategy accordingly. By interacting with the environment, the trading agent will derive a trading strategy with the maximized rewards as time proceeds.\n\nOur trading environments, based on OpenAI Gym framework, simulate live stock markets with real market data according to the principle of time-driven simulation.\n\nThe action space describes the allowed actions that the agent interacts with the environment. Normally, action a includes three actions: {-1, 0, 1}, where -1, 0, 1 represent selling, holding, and buying one share. Also, an action can be carried upon multiple shares. We use an action space {-k,…,-1, 0, 1, …, k}, where k denotes the number of shares to buy and -k denotes the number of shares to sell. For example, \"Buy 10 shares of AAPL\" or \"Sell 10 shares of AAPL\" are 10 or -10, respectively. The continuous action space needs to be normalized to [-1, 1], since the policy is defined on a Gaussian distribution, which needs to be normalized and symmetric.",
"_____no_output_____"
]
],
[
[
"stock_dimension = len(processed.tic.unique())\nstate_space = 1 + 2*stock_dimension + len(tech_indicators)*stock_dimension\nprint(f\"Stock Dimension: {stock_dimension}, State Space: {state_space}\")\n",
"Stock Dimension: 29, State Space: 175\n"
],
[
"env_kwargs = {\n \"hmax\": 100, \n \"initial_amount\": 1000000, \n \"buy_cost_pct\": 0.001, \n \"sell_cost_pct\": 0.001, \n \"state_space\": state_space, \n \"stock_dim\": stock_dimension, \n \"tech_indicator_list\": tech_indicators,\n \"action_space\": stock_dimension, \n \"reward_scaling\": 1e-4,\n \"print_verbosity\":5\n \n}",
"_____no_output_____"
]
],
[
[
"<a id='5'></a>\n# Part 6: Implement DRL Algorithms\n* The implementation of the DRL algorithms are based on **OpenAI Baselines** and **Stable Baselines**. Stable Baselines is a fork of OpenAI Baselines, with a major structural refactoring, and code cleanups.\n* FinRL library includes fine-tuned standard DRL algorithms, such as DQN, DDPG,\nMulti-Agent DDPG, PPO, SAC, A2C and TD3. We also allow users to\ndesign their own DRL algorithms by adapting these DRL algorithms.\n\n* In this notebook, we are training and validating 3 agents (A2C, PPO, DDPG) using Rolling-window Ensemble Method ([reference code](https://github.com/AI4Finance-LLC/Deep-Reinforcement-Learning-for-Automated-Stock-Trading-Ensemble-Strategy-ICAIF-2020/blob/80415db8fa7b2179df6bd7e81ce4fe8dbf913806/model/models.py#L92))",
"_____no_output_____"
]
],
[
[
"rebalance_window = 63 # rebalance_window is the number of days to retrain the model\nvalidation_window = 63 # validation_window is the number of days to do validation and trading (e.g. if validation_window=63, then both validation and trading period will be 63 days)\ntrain_start = '2009-01-01'\ntrain_end = '2020-04-01'\nval_test_start = '2020-04-01'\nval_test_end = '2021-07-20'\n\nensemble_agent = DRLEnsembleAgent(df=processed,\n train_period=(train_start,train_end),\n val_test_period=(val_test_start,val_test_end),\n rebalance_window=rebalance_window, \n validation_window=validation_window, \n **env_kwargs)",
"_____no_output_____"
],
[
"A2C_model_kwargs = {\n 'n_steps': 5,\n 'ent_coef': 0.01,\n 'learning_rate': 0.0005\n }\n\nPPO_model_kwargs = {\n \"ent_coef\":0.01,\n \"n_steps\": 2048,\n \"learning_rate\": 0.00025,\n \"batch_size\": 64\n }\n\nDDPG_model_kwargs = {\n #\"action_noise\":\"ornstein_uhlenbeck\",\n \"buffer_size\": 100_000,\n \"learning_rate\": 0.000005,\n \"batch_size\": 64\n }\n\ntimesteps_dict = {'a2c' : 30_000, \n 'ppo' : 100_000, \n 'ddpg' : 10_000\n }\n\n\ntimesteps_dict = {'a2c' : 10_000, \n 'ppo' : 10_000, \n 'ddpg' : 10_000\n }",
"_____no_output_____"
],
[
"df_summary = ensemble_agent.run_ensemble_strategy(A2C_model_kwargs,\n PPO_model_kwargs,\n DDPG_model_kwargs,\n timesteps_dict)",
"============Start Ensemble Strategy============\n============================================\nturbulence_threshold: 200.92993079372604\n======Model training from: 2009-01-01 to 2020-04-02\n======A2C Training========\n{'n_steps': 5, 'ent_coef': 0.01, 'learning_rate': 0.0005}\nUsing cpu device\nLogging to tensorboard_log/a2c/a2c_126_1\n----------------------------------------\n| time/ | |\n| fps | 59 |\n| iterations | 100 |\n| time_elapsed | 8 |\n| total_timesteps | 500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0328 |\n| learning_rate | 0.0005 |\n| n_updates | 99 |\n| policy_loss | -7.49 |\n| reward | -0.051769912 |\n| std | 1 |\n| value_loss | 0.344 |\n----------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 70 |\n| iterations | 200 |\n| time_elapsed | 14 |\n| total_timesteps | 1000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0206 |\n| learning_rate | 0.0005 |\n| n_updates | 199 |\n| policy_loss | -122 |\n| reward | -1.4591484 |\n| std | 1 |\n| value_loss | 9.47 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 75 |\n| iterations | 300 |\n| time_elapsed | 19 |\n| total_timesteps | 1500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.000771 |\n| learning_rate | 0.0005 |\n| n_updates | 299 |\n| policy_loss | -404 |\n| reward | 6.2669177 |\n| std | 1 |\n| value_loss | 86.4 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 78 |\n| iterations | 400 |\n| time_elapsed | 25 |\n| total_timesteps | 2000 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 399 |\n| policy_loss | -41.2 |\n| reward | 2.9870415 |\n| std | 1 |\n| value_loss | 8.41 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 80 |\n| iterations | 500 |\n| time_elapsed | 31 |\n| total_timesteps | 2500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | 0.00274 |\n| learning_rate | 0.0005 |\n| n_updates | 499 |\n| policy_loss | 470 |\n| reward | -8.643984 |\n| std | 1 |\n| value_loss | 133 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 81 |\n| iterations | 600 |\n| time_elapsed | 36 |\n| total_timesteps | 3000 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -0.808 |\n| learning_rate | 0.0005 |\n| n_updates | 599 |\n| policy_loss | 34.4 |\n| reward | 0.82639563 |\n| std | 1 |\n| value_loss | 2.14 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 700 |\n| time_elapsed | 42 |\n| total_timesteps | 3500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | 0.0166 |\n| learning_rate | 0.0005 |\n| n_updates | 699 |\n| policy_loss | 39.5 |\n| reward | 1.6315455 |\n| std | 1 |\n| value_loss | 1.53 |\n-------------------------------------\n----------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 800 |\n| time_elapsed | 47 |\n| total_timesteps | 4000 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -1.19e-07 |\n| learning_rate | 0.0005 |\n| n_updates | 799 |\n| policy_loss | -107 |\n| reward | -0.075020224 |\n| std | 1 |\n| value_loss | 10.4 |\n----------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 84 |\n| iterations | 900 |\n| time_elapsed | 53 |\n| total_timesteps | 4500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -0.16 |\n| learning_rate | 0.0005 |\n| n_updates | 899 |\n| policy_loss | 12.6 |\n| reward | -2.6403275 |\n| std | 1 |\n| value_loss | 0.708 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1000 |\n| time_elapsed | 58 |\n| total_timesteps | 5000 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -0.0825 |\n| learning_rate | 0.0005 |\n| n_updates | 999 |\n| policy_loss | 9.93 |\n| reward | 0.6558525 |\n| std | 1 |\n| value_loss | 1.33 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1100 |\n| time_elapsed | 64 |\n| total_timesteps | 5500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.132 |\n| learning_rate | 0.0005 |\n| n_updates | 1099 |\n| policy_loss | -567 |\n| reward | -7.434152 |\n| std | 1 |\n| value_loss | 206 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1200 |\n| time_elapsed | 69 |\n| total_timesteps | 6000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0284 |\n| learning_rate | 0.0005 |\n| n_updates | 1199 |\n| policy_loss | -52.7 |\n| reward | -2.3101468 |\n| std | 1.01 |\n| value_loss | 2.58 |\n--------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1300 |\n| time_elapsed | 75 |\n| total_timesteps | 6500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 1299 |\n| policy_loss | -115 |\n| reward | 0.21878022 |\n| std | 1.01 |\n| value_loss | 9.42 |\n--------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1400 |\n| time_elapsed | 80 |\n| total_timesteps | 7000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 1399 |\n| policy_loss | 53.8 |\n| reward | 0.87620676 |\n| std | 1.01 |\n| value_loss | 3.67 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 87 |\n| iterations | 1500 |\n| time_elapsed | 86 |\n| total_timesteps | 7500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.126 |\n| learning_rate | 0.0005 |\n| n_updates | 1499 |\n| policy_loss | 128 |\n| reward | -1.743883 |\n| std | 1.01 |\n| value_loss | 13 |\n-------------------------------------\n------------------------------------\n| time/ | |\n| fps | 87 |\n| iterations | 1600 |\n| time_elapsed | 91 |\n| total_timesteps | 8000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.181 |\n| learning_rate | 0.0005 |\n| n_updates | 1599 |\n| policy_loss | 31.5 |\n| reward | 2.94982 |\n| std | 1.01 |\n| value_loss | 21 |\n------------------------------------\n------------------------------------------\n| time/ | |\n| fps | 87 |\n| iterations | 1700 |\n| time_elapsed | 97 |\n| total_timesteps | 8500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.0969 |\n| learning_rate | 0.0005 |\n| n_updates | 1699 |\n| policy_loss | -1.06e+03 |\n| reward | -0.00018015769 |\n| std | 1.01 |\n| value_loss | 1.26e+03 |\n------------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 87 |\n| iterations | 1800 |\n| time_elapsed | 102 |\n| total_timesteps | 9000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0.32 |\n| learning_rate | 0.0005 |\n| n_updates | 1799 |\n| policy_loss | -3.74 |\n| reward | -0.731615 |\n| std | 1.01 |\n| value_loss | 0.0184 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 87 |\n| iterations | 1900 |\n| time_elapsed | 108 |\n| total_timesteps | 9500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 1899 |\n| policy_loss | -94 |\n| reward | -0.577778 |\n| std | 1.01 |\n| value_loss | 6.25 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 88 |\n| iterations | 2000 |\n| time_elapsed | 113 |\n| total_timesteps | 10000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0.0354 |\n| learning_rate | 0.0005 |\n| n_updates | 1999 |\n| policy_loss | 268 |\n| reward | -0.7198939 |\n| std | 1.01 |\n| value_loss | 52.7 |\n--------------------------------------\n======A2C Validation from: 2020-04-02 to 2020-07-02\nA2C Sharpe Ratio: 0.25033460143894093\n======PPO Training========\n{'ent_coef': 0.01, 'n_steps': 2048, 'learning_rate': 0.00025, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ppo/ppo_126_1\n------------------------------------\n| time/ | |\n| fps | 97 |\n| iterations | 1 |\n| time_elapsed | 21 |\n| total_timesteps | 2048 |\n| train/ | |\n| reward | -0.04487341 |\n------------------------------------\nday: 2830, episode: 5\nbegin_total_asset: 1000000.00\nend_total_asset: 2196151.30\ntotal_reward: 1196151.30\ntotal_cost: 335848.27\ntotal_trades: 78804\nSharpe: 0.497\n=================================\n-----------------------------------------\n| time/ | |\n| fps | 94 |\n| iterations | 2 |\n| time_elapsed | 43 |\n| total_timesteps | 4096 |\n| train/ | |\n| approx_kl | 0.018230084 |\n| clip_fraction | 0.245 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | -0.00354 |\n| learning_rate | 0.00025 |\n| loss | 7.6 |\n| n_updates | 10 |\n| policy_gradient_loss | -0.0305 |\n| reward | -1.5173061 |\n| std | 1 |\n| value_loss | 14.3 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 93 |\n| iterations | 3 |\n| time_elapsed | 65 |\n| total_timesteps | 6144 |\n| train/ | |\n| approx_kl | 0.019515125 |\n| clip_fraction | 0.233 |\n| clip_range | 0.2 |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0247 |\n| learning_rate | 0.00025 |\n| loss | 55.8 |\n| n_updates | 20 |\n| policy_gradient_loss | -0.0225 |\n| reward | 2.814237 |\n| std | 1.01 |\n| value_loss | 54.5 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 92 |\n| iterations | 4 |\n| time_elapsed | 88 |\n| total_timesteps | 8192 |\n| train/ | |\n| approx_kl | 0.024547426 |\n| clip_fraction | 0.261 |\n| clip_range | 0.2 |\n| entropy_loss | -41.3 |\n| explained_variance | 0.00199 |\n| learning_rate | 0.00025 |\n| loss | 22.5 |\n| n_updates | 30 |\n| policy_gradient_loss | -0.0196 |\n| reward | 1.2317431 |\n| std | 1.01 |\n| value_loss | 49.9 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 92 |\n| iterations | 5 |\n| time_elapsed | 110 |\n| total_timesteps | 10240 |\n| train/ | |\n| approx_kl | 0.025651447 |\n| clip_fraction | 0.245 |\n| clip_range | 0.2 |\n| entropy_loss | -41.4 |\n| explained_variance | -0.0224 |\n| learning_rate | 0.00025 |\n| loss | 11.2 |\n| n_updates | 40 |\n| policy_gradient_loss | -0.0189 |\n| reward | 1.1878457 |\n| std | 1.01 |\n| value_loss | 28.9 |\n-----------------------------------------\n======PPO Validation from: 2020-04-02 to 2020-07-02\nPPO Sharpe Ratio: 0.19630374643213191\n======DDPG Training========\n{'buffer_size': 100000, 'learning_rate': 5e-06, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ddpg/ddpg_126_1\nday: 2830, episode: 10\nbegin_total_asset: 1000000.00\nend_total_asset: 2902215.69\ntotal_reward: 1902215.69\ntotal_cost: 1846.50\ntotal_trades: 39965\nSharpe: 0.629\n=================================\n-----------------------------------\n| time/ | |\n| episodes | 4 |\n| fps | 44 |\n| time_elapsed | 255 |\n| total_timesteps | 11324 |\n| train/ | |\n| actor_loss | 45.4 |\n| critic_loss | 391 |\n| learning_rate | 5e-06 |\n| n_updates | 8493 |\n| reward | -13.366796 |\n-----------------------------------\n======DDPG Validation from: 2020-04-02 to 2020-07-02\n======Best Model Retraining from: 2009-01-01 to 2020-07-02\n======Trading from: 2020-07-02 to 2020-10-01\n============================================\nturbulence_threshold: 200.92993079372604\n======Model training from: 2009-01-01 to 2020-07-02\n======A2C Training========\n{'n_steps': 5, 'ent_coef': 0.01, 'learning_rate': 0.0005}\nUsing cpu device\nLogging to tensorboard_log/a2c/a2c_189_1\n--------------------------------------\n| time/ | |\n| fps | 87 |\n| iterations | 100 |\n| time_elapsed | 5 |\n| total_timesteps | 500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -0.0404 |\n| learning_rate | 0.0005 |\n| n_updates | 99 |\n| policy_loss | -42.7 |\n| reward | 0.27108225 |\n| std | 1 |\n| value_loss | 1.71 |\n--------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 200 |\n| time_elapsed | 11 |\n| total_timesteps | 1000 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | 0.036 |\n| learning_rate | 0.0005 |\n| n_updates | 199 |\n| policy_loss | 4.02 |\n| reward | -1.6978885 |\n| std | 1 |\n| value_loss | 1.61 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 300 |\n| time_elapsed | 17 |\n| total_timesteps | 1500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -0.797 |\n| learning_rate | 0.0005 |\n| n_updates | 299 |\n| policy_loss | -336 |\n| reward | 7.4013658 |\n| std | 1 |\n| value_loss | 72.2 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 400 |\n| time_elapsed | 23 |\n| total_timesteps | 2000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0249 |\n| learning_rate | 0.0005 |\n| n_updates | 399 |\n| policy_loss | 144 |\n| reward | 0.18983567 |\n| std | 1 |\n| value_loss | 14.7 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 500 |\n| time_elapsed | 29 |\n| total_timesteps | 2500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | 0.00651 |\n| learning_rate | 0.0005 |\n| n_updates | 499 |\n| policy_loss | 399 |\n| reward | -5.953611 |\n| std | 1 |\n| value_loss | 113 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 600 |\n| time_elapsed | 34 |\n| total_timesteps | 3000 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -1.17 |\n| learning_rate | 0.0005 |\n| n_updates | 599 |\n| policy_loss | 81.1 |\n| reward | 0.2980301 |\n| std | 1 |\n| value_loss | 6.01 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 700 |\n| time_elapsed | 40 |\n| total_timesteps | 3500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | 0.546 |\n| learning_rate | 0.0005 |\n| n_updates | 699 |\n| policy_loss | -59.3 |\n| reward | 1.7133454 |\n| std | 1 |\n| value_loss | 2.1 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 800 |\n| time_elapsed | 46 |\n| total_timesteps | 4000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0664 |\n| learning_rate | 0.0005 |\n| n_updates | 799 |\n| policy_loss | 36.4 |\n| reward | 1.9195006 |\n| std | 1 |\n| value_loss | 2.08 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 900 |\n| time_elapsed | 52 |\n| total_timesteps | 4500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.037 |\n| learning_rate | 0.0005 |\n| n_updates | 899 |\n| policy_loss | 124 |\n| reward | 0.44107372 |\n| std | 1.01 |\n| value_loss | 10.7 |\n--------------------------------------\n---------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1000 |\n| time_elapsed | 57 |\n| total_timesteps | 5000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 999 |\n| policy_loss | -46.1 |\n| reward | -0.85359025 |\n| std | 1.01 |\n| value_loss | 3.56 |\n---------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1100 |\n| time_elapsed | 63 |\n| total_timesteps | 5500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0407 |\n| learning_rate | 0.0005 |\n| n_updates | 1099 |\n| policy_loss | -96.3 |\n| reward | -11.516248 |\n| std | 1.01 |\n| value_loss | 14 |\n--------------------------------------\n---------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1200 |\n| time_elapsed | 69 |\n| total_timesteps | 6000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0572 |\n| learning_rate | 0.0005 |\n| n_updates | 1199 |\n| policy_loss | -82.4 |\n| reward | 0.005680397 |\n| std | 1.01 |\n| value_loss | 6 |\n---------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1300 |\n| time_elapsed | 75 |\n| total_timesteps | 6500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.378 |\n| learning_rate | 0.0005 |\n| n_updates | 1299 |\n| policy_loss | 100 |\n| reward | 0.16837326 |\n| std | 1.01 |\n| value_loss | 9.2 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1400 |\n| time_elapsed | 81 |\n| total_timesteps | 7000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.108 |\n| learning_rate | 0.0005 |\n| n_updates | 1399 |\n| policy_loss | 56.7 |\n| reward | 0.2755015 |\n| std | 1.01 |\n| value_loss | 2.95 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1500 |\n| time_elapsed | 87 |\n| total_timesteps | 7500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.00349 |\n| learning_rate | 0.0005 |\n| n_updates | 1499 |\n| policy_loss | -270 |\n| reward | -0.9525704 |\n| std | 1.01 |\n| value_loss | 42.1 |\n--------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1600 |\n| time_elapsed | 92 |\n| total_timesteps | 8000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.39 |\n| learning_rate | 0.0005 |\n| n_updates | 1599 |\n| policy_loss | 56.1 |\n| reward | -0.5067834 |\n| std | 1.01 |\n| value_loss | 2.25 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1700 |\n| time_elapsed | 98 |\n| total_timesteps | 8500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 5.96e-08 |\n| learning_rate | 0.0005 |\n| n_updates | 1699 |\n| policy_loss | 42.3 |\n| reward | 2.8328996 |\n| std | 1.01 |\n| value_loss | 26.3 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1800 |\n| time_elapsed | 104 |\n| total_timesteps | 9000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.148 |\n| learning_rate | 0.0005 |\n| n_updates | 1799 |\n| policy_loss | 2.71 |\n| reward | 1.2313448 |\n| std | 1.01 |\n| value_loss | 0.228 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 1900 |\n| time_elapsed | 110 |\n| total_timesteps | 9500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.842 |\n| learning_rate | 0.0005 |\n| n_updates | 1899 |\n| policy_loss | 108 |\n| reward | 1.0191236 |\n| std | 1.01 |\n| value_loss | 7.19 |\n-------------------------------------\n------------------------------------\n| time/ | |\n| fps | 86 |\n| iterations | 2000 |\n| time_elapsed | 115 |\n| total_timesteps | 10000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.166 |\n| learning_rate | 0.0005 |\n| n_updates | 1999 |\n| policy_loss | -79.9 |\n| reward | 3.596779 |\n| std | 1.01 |\n| value_loss | 5.47 |\n------------------------------------\n======A2C Validation from: 2020-07-02 to 2020-10-01\nA2C Sharpe Ratio: 0.3324629886764393\n======PPO Training========\n{'ent_coef': 0.01, 'n_steps': 2048, 'learning_rate': 0.00025, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ppo/ppo_189_1\n-------------------------------------\n| time/ | |\n| fps | 93 |\n| iterations | 1 |\n| time_elapsed | 22 |\n| total_timesteps | 2048 |\n| train/ | |\n| reward | -0.085719466 |\n-------------------------------------\nday: 2893, episode: 5\nbegin_total_asset: 1000000.00\nend_total_asset: 3087465.25\ntotal_reward: 2087465.25\ntotal_cost: 354063.72\ntotal_trades: 80864\nSharpe: 0.626\n=================================\n-----------------------------------------\n| time/ | |\n| fps | 90 |\n| iterations | 2 |\n| time_elapsed | 45 |\n| total_timesteps | 4096 |\n| train/ | |\n| approx_kl | 0.019241996 |\n| clip_fraction | 0.246 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | -0.021 |\n| learning_rate | 0.00025 |\n| loss | 5.07 |\n| n_updates | 10 |\n| policy_gradient_loss | -0.0243 |\n| reward | 1.500187 |\n| std | 1 |\n| value_loss | 14.7 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 90 |\n| iterations | 3 |\n| time_elapsed | 68 |\n| total_timesteps | 6144 |\n| train/ | |\n| approx_kl | 0.014936658 |\n| clip_fraction | 0.147 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | 0.0165 |\n| learning_rate | 0.00025 |\n| loss | 63.9 |\n| n_updates | 20 |\n| policy_gradient_loss | -0.0182 |\n| reward | 0.18779926 |\n| std | 1 |\n| value_loss | 95.5 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 89 |\n| iterations | 4 |\n| time_elapsed | 91 |\n| total_timesteps | 8192 |\n| train/ | |\n| approx_kl | 0.019090343 |\n| clip_fraction | 0.208 |\n| clip_range | 0.2 |\n| entropy_loss | -41.3 |\n| explained_variance | 0.00491 |\n| learning_rate | 0.00025 |\n| loss | 19.6 |\n| n_updates | 30 |\n| policy_gradient_loss | -0.0183 |\n| reward | -1.1587315 |\n| std | 1.01 |\n| value_loss | 85.3 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 89 |\n| iterations | 5 |\n| time_elapsed | 114 |\n| total_timesteps | 10240 |\n| train/ | |\n| approx_kl | 0.021126334 |\n| clip_fraction | 0.254 |\n| clip_range | 0.2 |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0162 |\n| learning_rate | 0.00025 |\n| loss | 11.6 |\n| n_updates | 40 |\n| policy_gradient_loss | -0.0232 |\n| reward | -2.061006 |\n| std | 1.01 |\n| value_loss | 20.6 |\n-----------------------------------------\n======PPO Validation from: 2020-07-02 to 2020-10-01\nPPO Sharpe Ratio: 0.12120565668544925\n======DDPG Training========\n{'buffer_size': 100000, 'learning_rate': 5e-06, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ddpg/ddpg_189_1\nday: 2893, episode: 10\nbegin_total_asset: 1000000.00\nend_total_asset: 2726077.37\ntotal_reward: 1726077.37\ntotal_cost: 1765.83\ntotal_trades: 41996\nSharpe: 0.558\n=================================\n----------------------------------\n| time/ | |\n| episodes | 4 |\n| fps | 43 |\n| time_elapsed | 264 |\n| total_timesteps | 11576 |\n| train/ | |\n| actor_loss | 88.8 |\n| critic_loss | 640 |\n| learning_rate | 5e-06 |\n| n_updates | 8682 |\n| reward | 2.7025018 |\n----------------------------------\n======DDPG Validation from: 2020-07-02 to 2020-10-01\n======Best Model Retraining from: 2009-01-01 to 2020-10-01\n======Trading from: 2020-10-01 to 2020-12-31\n============================================\nturbulence_threshold: 200.92993079372604\n======Model training from: 2009-01-01 to 2020-10-01\n======A2C Training========\n{'n_steps': 5, 'ent_coef': 0.01, 'learning_rate': 0.0005}\nUsing cpu device\nLogging to tensorboard_log/a2c/a2c_252_1\n---------------------------------------\n| time/ | |\n| fps | 79 |\n| iterations | 100 |\n| time_elapsed | 6 |\n| total_timesteps | 500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.125 |\n| learning_rate | 0.0005 |\n| n_updates | 99 |\n| policy_loss | -35.5 |\n| reward | -0.09041673 |\n| std | 1.01 |\n| value_loss | 2.62 |\n---------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 75 |\n| iterations | 200 |\n| time_elapsed | 13 |\n| total_timesteps | 1000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.077 |\n| learning_rate | 0.0005 |\n| n_updates | 199 |\n| policy_loss | -14.5 |\n| reward | 0.3587879 |\n| std | 1.01 |\n| value_loss | 0.584 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 78 |\n| iterations | 300 |\n| time_elapsed | 19 |\n| total_timesteps | 1500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0709 |\n| learning_rate | 0.0005 |\n| n_updates | 299 |\n| policy_loss | -253 |\n| reward | 4.5632515 |\n| std | 1.01 |\n| value_loss | 46.9 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 80 |\n| iterations | 400 |\n| time_elapsed | 24 |\n| total_timesteps | 2000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 399 |\n| policy_loss | -21.1 |\n| reward | 1.0681273 |\n| std | 1.01 |\n| value_loss | 4.81 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 81 |\n| iterations | 500 |\n| time_elapsed | 30 |\n| total_timesteps | 2500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.139 |\n| learning_rate | 0.0005 |\n| n_updates | 499 |\n| policy_loss | 423 |\n| reward | -6.0229154 |\n| std | 1.01 |\n| value_loss | 131 |\n--------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 82 |\n| iterations | 600 |\n| time_elapsed | 36 |\n| total_timesteps | 3000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.000959 |\n| learning_rate | 0.0005 |\n| n_updates | 599 |\n| policy_loss | -6.28 |\n| reward | -1.3853685 |\n| std | 1.01 |\n| value_loss | 1.27 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 82 |\n| iterations | 700 |\n| time_elapsed | 42 |\n| total_timesteps | 3500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.522 |\n| learning_rate | 0.0005 |\n| n_updates | 699 |\n| policy_loss | -56 |\n| reward | -1.983197 |\n| std | 1.01 |\n| value_loss | 2.56 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 82 |\n| iterations | 800 |\n| time_elapsed | 48 |\n| total_timesteps | 4000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -1.46 |\n| learning_rate | 0.0005 |\n| n_updates | 799 |\n| policy_loss | -36.1 |\n| reward | 1.8168931 |\n| std | 1.01 |\n| value_loss | 1.48 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 82 |\n| iterations | 900 |\n| time_elapsed | 54 |\n| total_timesteps | 4500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 899 |\n| policy_loss | -30.7 |\n| reward | 2.7901127 |\n| std | 1.01 |\n| value_loss | 4.68 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1000 |\n| time_elapsed | 60 |\n| total_timesteps | 5000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.0115 |\n| learning_rate | 0.0005 |\n| n_updates | 999 |\n| policy_loss | 96.8 |\n| reward | 2.1106558 |\n| std | 1.01 |\n| value_loss | 16.5 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1100 |\n| time_elapsed | 65 |\n| total_timesteps | 5500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0.188 |\n| learning_rate | 0.0005 |\n| n_updates | 1099 |\n| policy_loss | 605 |\n| reward | 0.8484186 |\n| std | 1.01 |\n| value_loss | 210 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1200 |\n| time_elapsed | 71 |\n| total_timesteps | 6000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.202 |\n| learning_rate | 0.0005 |\n| n_updates | 1199 |\n| policy_loss | 81.4 |\n| reward | -2.480206 |\n| std | 1.01 |\n| value_loss | 5.93 |\n-------------------------------------\n---------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1300 |\n| time_elapsed | 77 |\n| total_timesteps | 6500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -1.19e-07 |\n| learning_rate | 0.0005 |\n| n_updates | 1299 |\n| policy_loss | 43.9 |\n| reward | -0.26437733 |\n| std | 1.01 |\n| value_loss | 3.52 |\n---------------------------------------\n---------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1400 |\n| time_elapsed | 83 |\n| total_timesteps | 7000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0.00658 |\n| learning_rate | 0.0005 |\n| n_updates | 1399 |\n| policy_loss | 181 |\n| reward | -0.63620466 |\n| std | 1.01 |\n| value_loss | 26 |\n---------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1500 |\n| time_elapsed | 89 |\n| total_timesteps | 7500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.0235 |\n| learning_rate | 0.0005 |\n| n_updates | 1499 |\n| policy_loss | 270 |\n| reward | -0.5455963 |\n| std | 1.01 |\n| value_loss | 40.8 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1600 |\n| time_elapsed | 95 |\n| total_timesteps | 8000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.112 |\n| learning_rate | 0.0005 |\n| n_updates | 1599 |\n| policy_loss | -52.3 |\n| reward | -2.071636 |\n| std | 1.01 |\n| value_loss | 4.2 |\n-------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1700 |\n| time_elapsed | 101 |\n| total_timesteps | 8500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0406 |\n| learning_rate | 0.0005 |\n| n_updates | 1699 |\n| policy_loss | 322 |\n| reward | 0.5182562 |\n| std | 1.01 |\n| value_loss | 70.6 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1800 |\n| time_elapsed | 107 |\n| total_timesteps | 9000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.0319 |\n| learning_rate | 0.0005 |\n| n_updates | 1799 |\n| policy_loss | -0.937 |\n| reward | 0.67256796 |\n| std | 1.01 |\n| value_loss | 0.705 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 1900 |\n| time_elapsed | 113 |\n| total_timesteps | 9500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 1899 |\n| policy_loss | -153 |\n| reward | 1.9791864 |\n| std | 1.01 |\n| value_loss | 14.5 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 83 |\n| iterations | 2000 |\n| time_elapsed | 119 |\n| total_timesteps | 10000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | 0.0338 |\n| learning_rate | 0.0005 |\n| n_updates | 1999 |\n| policy_loss | -123 |\n| reward | -1.0705515 |\n| std | 1.01 |\n| value_loss | 17.1 |\n--------------------------------------\n======A2C Validation from: 2020-10-01 to 2020-12-31\nA2C Sharpe Ratio: 0.157398297864907\n======PPO Training========\n{'ent_coef': 0.01, 'n_steps': 2048, 'learning_rate': 0.00025, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ppo/ppo_252_1\n-----------------------------------\n| time/ | |\n| fps | 91 |\n| iterations | 1 |\n| time_elapsed | 22 |\n| total_timesteps | 2048 |\n| train/ | |\n| reward | 0.30793157 |\n-----------------------------------\nday: 2956, episode: 5\nbegin_total_asset: 1000000.00\nend_total_asset: 3942705.65\ntotal_reward: 2942705.65\ntotal_cost: 374440.05\ntotal_trades: 82889\nSharpe: 0.797\n=================================\n-----------------------------------------\n| time/ | |\n| fps | 89 |\n| iterations | 2 |\n| time_elapsed | 46 |\n| total_timesteps | 4096 |\n| train/ | |\n| approx_kl | 0.017636795 |\n| clip_fraction | 0.207 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | -0.00652 |\n| learning_rate | 0.00025 |\n| loss | 4.89 |\n| n_updates | 10 |\n| policy_gradient_loss | -0.0222 |\n| reward | 0.14401561 |\n| std | 1 |\n| value_loss | 11.8 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 88 |\n| iterations | 3 |\n| time_elapsed | 69 |\n| total_timesteps | 6144 |\n| train/ | |\n| approx_kl | 0.019762876 |\n| clip_fraction | 0.223 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | -0.00246 |\n| learning_rate | 0.00025 |\n| loss | 17.1 |\n| n_updates | 20 |\n| policy_gradient_loss | -0.0192 |\n| reward | 1.1803926 |\n| std | 1 |\n| value_loss | 50.7 |\n-----------------------------------------\n----------------------------------------\n| time/ | |\n| fps | 88 |\n| iterations | 4 |\n| time_elapsed | 92 |\n| total_timesteps | 8192 |\n| train/ | |\n| approx_kl | 0.01684109 |\n| clip_fraction | 0.199 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | -0.0116 |\n| learning_rate | 0.00025 |\n| loss | 77.2 |\n| n_updates | 30 |\n| policy_gradient_loss | -0.0217 |\n| reward | 1.0463831 |\n| std | 1 |\n| value_loss | 68.2 |\n----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 88 |\n| iterations | 5 |\n| time_elapsed | 115 |\n| total_timesteps | 10240 |\n| train/ | |\n| approx_kl | 0.024190305 |\n| clip_fraction | 0.278 |\n| clip_range | 0.2 |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0346 |\n| learning_rate | 0.00025 |\n| loss | 4.4 |\n| n_updates | 40 |\n| policy_gradient_loss | -0.0212 |\n| reward | -1.2749629 |\n| std | 1.01 |\n| value_loss | 19.7 |\n-----------------------------------------\n======PPO Validation from: 2020-10-01 to 2020-12-31\nPPO Sharpe Ratio: 0.20250342417658126\n======DDPG Training========\n{'buffer_size': 100000, 'learning_rate': 5e-06, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ddpg/ddpg_252_1\nday: 2956, episode: 10\nbegin_total_asset: 1000000.00\nend_total_asset: 3957618.19\ntotal_reward: 2957618.19\ntotal_cost: 1601.16\ntotal_trades: 56009\nSharpe: 0.762\n=================================\n---------------------------------\n| time/ | |\n| episodes | 4 |\n| fps | 44 |\n| time_elapsed | 268 |\n| total_timesteps | 11828 |\n| train/ | |\n| actor_loss | -5.37 |\n| critic_loss | 178 |\n| learning_rate | 5e-06 |\n| n_updates | 8871 |\n| reward | 4.856528 |\n---------------------------------\n======DDPG Validation from: 2020-10-01 to 2020-12-31\n======Best Model Retraining from: 2009-01-01 to 2020-12-31\n======Trading from: 2020-12-31 to 2021-04-05\n============================================\nturbulence_threshold: 200.92993079372604\n======Model training from: 2009-01-01 to 2020-12-31\n======A2C Training========\n{'n_steps': 5, 'ent_coef': 0.01, 'learning_rate': 0.0005}\nUsing cpu device\nLogging to tensorboard_log/a2c/a2c_315_1\n-------------------------------------\n| time/ | |\n| fps | 84 |\n| iterations | 100 |\n| time_elapsed | 5 |\n| total_timesteps | 500 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | -0.828 |\n| learning_rate | 0.0005 |\n| n_updates | 99 |\n| policy_loss | -18 |\n| reward | 0.2629864 |\n| std | 1 |\n| value_loss | 0.645 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 200 |\n| time_elapsed | 11 |\n| total_timesteps | 1000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.00115 |\n| learning_rate | 0.0005 |\n| n_updates | 199 |\n| policy_loss | -57 |\n| reward | -1.3294735 |\n| std | 1 |\n| value_loss | 2.91 |\n--------------------------------------\n------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 300 |\n| time_elapsed | 17 |\n| total_timesteps | 1500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0673 |\n| learning_rate | 0.0005 |\n| n_updates | 299 |\n| policy_loss | -330 |\n| reward | 5.483449 |\n| std | 1.01 |\n| value_loss | 70.3 |\n------------------------------------\n---------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 400 |\n| time_elapsed | 23 |\n| total_timesteps | 2000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0331 |\n| learning_rate | 0.0005 |\n| n_updates | 399 |\n| policy_loss | -86.5 |\n| reward | -0.29608092 |\n| std | 1 |\n| value_loss | 19.8 |\n---------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 500 |\n| time_elapsed | 29 |\n| total_timesteps | 2500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 1.76e-05 |\n| learning_rate | 0.0005 |\n| n_updates | 499 |\n| policy_loss | 746 |\n| reward | -11.729683 |\n| std | 1 |\n| value_loss | 387 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 600 |\n| time_elapsed | 35 |\n| total_timesteps | 3000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.338 |\n| learning_rate | 0.0005 |\n| n_updates | 599 |\n| policy_loss | 170 |\n| reward | -16.74081 |\n| std | 1 |\n| value_loss | 26 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 700 |\n| time_elapsed | 41 |\n| total_timesteps | 3500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 699 |\n| policy_loss | 71.1 |\n| reward | -0.4083204 |\n| std | 1 |\n| value_loss | 4.58 |\n--------------------------------------\n------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 800 |\n| time_elapsed | 46 |\n| total_timesteps | 4000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0553 |\n| learning_rate | 0.0005 |\n| n_updates | 799 |\n| policy_loss | -99.9 |\n| reward | 2.392244 |\n| std | 1.01 |\n| value_loss | 6.52 |\n------------------------------------\n------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 900 |\n| time_elapsed | 52 |\n| total_timesteps | 4500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.378 |\n| learning_rate | 0.0005 |\n| n_updates | 899 |\n| policy_loss | 83.9 |\n| reward | 2.506633 |\n| std | 1.01 |\n| value_loss | 7.72 |\n------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1000 |\n| time_elapsed | 58 |\n| total_timesteps | 5000 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.595 |\n| learning_rate | 0.0005 |\n| n_updates | 999 |\n| policy_loss | -67.9 |\n| reward | 0.69298357 |\n| std | 1.01 |\n| value_loss | 4.27 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1100 |\n| time_elapsed | 64 |\n| total_timesteps | 5500 |\n| train/ | |\n| entropy_loss | -41.4 |\n| explained_variance | -0.563 |\n| learning_rate | 0.0005 |\n| n_updates | 1099 |\n| policy_loss | 120 |\n| reward | 10.568737 |\n| std | 1.01 |\n| value_loss | 13.2 |\n-------------------------------------\n------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1200 |\n| time_elapsed | 70 |\n| total_timesteps | 6000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0285 |\n| learning_rate | 0.0005 |\n| n_updates | 1199 |\n| policy_loss | -607 |\n| reward | 6.724224 |\n| std | 1.01 |\n| value_loss | 200 |\n------------------------------------\n---------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1300 |\n| time_elapsed | 75 |\n| total_timesteps | 6500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0284 |\n| learning_rate | 0.0005 |\n| n_updates | 1299 |\n| policy_loss | 22.8 |\n| reward | -0.28616953 |\n| std | 1 |\n| value_loss | 0.979 |\n---------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1400 |\n| time_elapsed | 81 |\n| total_timesteps | 7000 |\n| train/ | |\n| entropy_loss | -41.2 |\n| explained_variance | 0.0225 |\n| learning_rate | 0.0005 |\n| n_updates | 1399 |\n| policy_loss | 35.3 |\n| reward | -4.7152042 |\n| std | 1 |\n| value_loss | 1.87 |\n--------------------------------------\n------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1500 |\n| time_elapsed | 87 |\n| total_timesteps | 7500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.0126 |\n| learning_rate | 0.0005 |\n| n_updates | 1499 |\n| policy_loss | -225 |\n| reward | 4.668683 |\n| std | 1 |\n| value_loss | 47.6 |\n------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1600 |\n| time_elapsed | 93 |\n| total_timesteps | 8000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0.0403 |\n| learning_rate | 0.0005 |\n| n_updates | 1599 |\n| policy_loss | -93.7 |\n| reward | 0.66578263 |\n| std | 1 |\n| value_loss | 5.76 |\n--------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1700 |\n| time_elapsed | 99 |\n| total_timesteps | 8500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.16 |\n| learning_rate | 0.0005 |\n| n_updates | 1699 |\n| policy_loss | 220 |\n| reward | -11.292281 |\n| std | 1.01 |\n| value_loss | 35.6 |\n--------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1800 |\n| time_elapsed | 105 |\n| total_timesteps | 9000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 1799 |\n| policy_loss | 54.6 |\n| reward | -2.0646114 |\n| std | 1.01 |\n| value_loss | 15.3 |\n--------------------------------------\n-------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 1900 |\n| time_elapsed | 110 |\n| total_timesteps | 9500 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | -0.483 |\n| learning_rate | 0.0005 |\n| n_updates | 1899 |\n| policy_loss | 36.2 |\n| reward | 1.1640425 |\n| std | 1.01 |\n| value_loss | 2.17 |\n-------------------------------------\n--------------------------------------\n| time/ | |\n| fps | 85 |\n| iterations | 2000 |\n| time_elapsed | 116 |\n| total_timesteps | 10000 |\n| train/ | |\n| entropy_loss | -41.3 |\n| explained_variance | 0 |\n| learning_rate | 0.0005 |\n| n_updates | 1999 |\n| policy_loss | 114 |\n| reward | -1.5702732 |\n| std | 1.01 |\n| value_loss | 11.2 |\n--------------------------------------\n======A2C Validation from: 2020-12-31 to 2021-04-05\nA2C Sharpe Ratio: 0.1821600849726842\n======PPO Training========\n{'ent_coef': 0.01, 'n_steps': 2048, 'learning_rate': 0.00025, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ppo/ppo_315_1\n-----------------------------------\n| time/ | |\n| fps | 91 |\n| iterations | 1 |\n| time_elapsed | 22 |\n| total_timesteps | 2048 |\n| train/ | |\n| reward | 0.12615822 |\n-----------------------------------\nday: 3019, episode: 5\nbegin_total_asset: 1000000.00\nend_total_asset: 4990034.90\ntotal_reward: 3990034.90\ntotal_cost: 391126.43\ntotal_trades: 84743\nSharpe: 0.902\n=================================\n----------------------------------------\n| time/ | |\n| fps | 89 |\n| iterations | 2 |\n| time_elapsed | 45 |\n| total_timesteps | 4096 |\n| train/ | |\n| approx_kl | 0.01619377 |\n| clip_fraction | 0.236 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | 0.0004 |\n| learning_rate | 0.00025 |\n| loss | 6.52 |\n| n_updates | 10 |\n| policy_gradient_loss | -0.0221 |\n| reward | -3.2302363 |\n| std | 1 |\n| value_loss | 15.6 |\n----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 88 |\n| iterations | 3 |\n| time_elapsed | 69 |\n| total_timesteps | 6144 |\n| train/ | |\n| approx_kl | 0.016321607 |\n| clip_fraction | 0.186 |\n| clip_range | 0.2 |\n| entropy_loss | -41.2 |\n| explained_variance | 0.00344 |\n| learning_rate | 0.00025 |\n| loss | 53.1 |\n| n_updates | 20 |\n| policy_gradient_loss | -0.0126 |\n| reward | -0.37026227 |\n| std | 1 |\n| value_loss | 68.3 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 88 |\n| iterations | 4 |\n| time_elapsed | 92 |\n| total_timesteps | 8192 |\n| train/ | |\n| approx_kl | 0.018890683 |\n| clip_fraction | 0.221 |\n| clip_range | 0.2 |\n| entropy_loss | -41.3 |\n| explained_variance | 0.00564 |\n| learning_rate | 0.00025 |\n| loss | 20.5 |\n| n_updates | 30 |\n| policy_gradient_loss | -0.0192 |\n| reward | -0.12967756 |\n| std | 1.01 |\n| value_loss | 66.8 |\n-----------------------------------------\n-----------------------------------------\n| time/ | |\n| fps | 88 |\n| iterations | 5 |\n| time_elapsed | 116 |\n| total_timesteps | 10240 |\n| train/ | |\n| approx_kl | 0.025792193 |\n| clip_fraction | 0.284 |\n| clip_range | 0.2 |\n| entropy_loss | -41.4 |\n| explained_variance | -0.0129 |\n| learning_rate | 0.00025 |\n| loss | 3.81 |\n| n_updates | 40 |\n| policy_gradient_loss | -0.0253 |\n| reward | 0.29261595 |\n| std | 1.01 |\n| value_loss | 10.1 |\n-----------------------------------------\n======PPO Validation from: 2020-12-31 to 2021-04-05\nPPO Sharpe Ratio: 0.29777940379649687\n======DDPG Training========\n{'buffer_size': 100000, 'learning_rate': 5e-06, 'batch_size': 64}\nUsing cpu device\nLogging to tensorboard_log/ddpg/ddpg_315_1\nday: 3019, episode: 10\nbegin_total_asset: 1000000.00\nend_total_asset: 6083808.47\ntotal_reward: 5083808.47\ntotal_cost: 1441.78\ntotal_trades: 41663\nSharpe: 0.886\n=================================\n---------------------------------\n| time/ | |\n| episodes | 4 |\n| fps | 44 |\n| time_elapsed | 274 |\n| total_timesteps | 12080 |\n| train/ | |\n| actor_loss | 36.3 |\n| critic_loss | 46.9 |\n| learning_rate | 5e-06 |\n| n_updates | 9060 |\n| reward | 1.169599 |\n---------------------------------\n======DDPG Validation from: 2020-12-31 to 2021-04-05\n======Best Model Retraining from: 2009-01-01 to 2021-04-05\n======Trading from: 2021-04-05 to 2021-07-02\nEnsemble Strategy took: 36.559605312347415 minutes\n"
],
[
"df_summary",
"_____no_output_____"
]
],
[
[
"<a id='6'></a>\n# Part 7: Backtest Our Strategy\nBacktesting plays a key role in evaluating the performance of a trading strategy. Automated backtesting tool is preferred because it reduces the human error. We usually use the Quantopian pyfolio package to backtest our trading strategies. It is easy to use and consists of various individual plots that provide a comprehensive image of the performance of a trading strategy.",
"_____no_output_____"
]
],
[
[
"unique_trade_date = processed[(processed.date > val_test_start)&(processed.date <= val_test_end)].date.unique()",
"_____no_output_____"
],
[
"df_trade_date = pd.DataFrame({'datadate':unique_trade_date})\n\ndf_account_value=pd.DataFrame()\nfor i in range(rebalance_window+validation_window, len(unique_trade_date)+1,rebalance_window):\n temp = pd.read_csv('results/account_value_trade_{}_{}.csv'.format('ensemble',i))\n df_account_value = df_account_value.append(temp,ignore_index=True)\nsharpe=(252**0.5)*df_account_value.account_value.pct_change(1).mean()/df_account_value.account_value.pct_change(1).std()\nprint('Sharpe Ratio: ',sharpe)\ndf_account_value=df_account_value.join(df_trade_date[validation_window:].reset_index(drop=True))",
"Sharpe Ratio: 1.5454149316420212\n"
],
[
"df_account_value.head()",
"_____no_output_____"
],
[
"%matplotlib inline\ndf_account_value.account_value.plot()",
"_____no_output_____"
]
],
[
[
"<a id='6.1'></a>\n## 7.1 BackTestStats\npass in df_account_value, this information is stored in env class\n",
"_____no_output_____"
]
],
[
[
"print(\"==============Get Backtest Results===========\")\nnow = datetime.datetime.now().strftime('%Y%m%d-%Hh%M')\n\nperf_stats_all = backtest_stats(account_value=df_account_value)\nperf_stats_all = pd.DataFrame(perf_stats_all)",
"==============Get Backtest Results===========\nAnnual return 0.228941\nCumulative returns 0.228941\nAnnual volatility 0.140345\nSharpe ratio 1.545415\nCalmar ratio 2.432010\nStability 0.898921\nMax drawdown -0.094137\nOmega ratio 1.298442\nSortino ratio 2.237300\nSkew NaN\nKurtosis NaN\nTail ratio 1.065696\nDaily value at risk -0.016821\ndtype: float64\n"
],
[
"#baseline stats\nprint(\"==============Get Baseline Stats===========\")\nbaseline_df = get_baseline(\n ticker=\"^DJI\", \n start = df_account_value.loc[0,'date'],\n end = df_account_value.loc[len(df_account_value)-1,'date'])\n\nstats = backtest_stats(baseline_df, value_col_name = 'close')",
"==============Get Baseline Stats===========\n\r[*********************100%***********************] 1 of 1 completed\nShape of DataFrame: (251, 8)\nAnnual return 0.337432\nCumulative returns 0.335890\nAnnual volatility 0.146098\nSharpe ratio 2.072078\nCalmar ratio 3.778308\nStability 0.944970\nMax drawdown -0.089308\nOmega ratio 1.411593\nSortino ratio 3.102218\nSkew NaN\nKurtosis NaN\nTail ratio 1.078766\nDaily value at risk -0.017205\ndtype: float64\n"
]
],
[
[
"<a id='6.2'></a>\n## 7.2 BackTestPlot",
"_____no_output_____"
]
],
[
[
"print(\"==============Compare to DJIA===========\")\n%matplotlib inline\n# S&P 500: ^GSPC\n# Dow Jones Index: ^DJI\n# NASDAQ 100: ^NDX\nbacktest_plot(df_account_value, \n baseline_ticker = '^DJI', \n baseline_start = df_account_value.loc[0,'date'],\n baseline_end = df_account_value.loc[len(df_account_value)-1,'date'])",
"==============Compare to DJIA===========\n[*********************100%***********************] 1 of 1 completed\nShape of DataFrame: (251, 8)\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbab61f15dcae403a01035619eb44697dfc161bc
| 119,104 |
ipynb
|
Jupyter Notebook
|
Workshop/GRU_212e.ipynb
|
ShepherdCode/ShepherdML
|
fd8d71c63f7bd788ea0052294d93e43246254a12
|
[
"MIT"
] | null | null | null |
Workshop/GRU_212e.ipynb
|
ShepherdCode/ShepherdML
|
fd8d71c63f7bd788ea0052294d93e43246254a12
|
[
"MIT"
] | 4 |
2020-03-24T18:05:09.000Z
|
2020-12-22T17:42:54.000Z
|
Workshop/GRU_212e.ipynb
|
ShepherdCode/ShepherdML
|
fd8d71c63f7bd788ea0052294d93e43246254a12
|
[
"MIT"
] | null | null | null | 120.672746 | 58,862 | 0.714174 |
[
[
[
"# GRU 212\n* Operate on 16000 GenCode 34 seqs.\n* 5-way cross validation. Save best model per CV.\n* Report mean accuracy from final re-validation with best 5.\n* Use Adam with a learn rate decay schdule.",
"_____no_output_____"
]
],
[
[
"NC_FILENAME='ncRNA.gc34.processed.fasta'\nPC_FILENAME='pcRNA.gc34.processed.fasta'\nDATAPATH=\"\"\ntry:\n from google.colab import drive\n IN_COLAB = True\n PATH='/content/drive/'\n drive.mount(PATH)\n DATAPATH=PATH+'My Drive/data/' # must end in \"/\"\n NC_FILENAME = DATAPATH+NC_FILENAME\n PC_FILENAME = DATAPATH+PC_FILENAME\nexcept:\n IN_COLAB = False\n DATAPATH=\"\" \n\nEPOCHS=200\nSPLITS=1\nK=3\nVOCABULARY_SIZE=4**K+1 # e.g. K=3 => 64 DNA K-mers + 'NNN'\nEMBED_DIMEN=16\nFILENAME='GRU212'\nNEURONS=64\nDROP=0.0\nACT=\"tanh\"",
"Mounted at /content/drive/\n"
],
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom sklearn.model_selection import ShuffleSplit\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.model_selection import RepeatedKFold\nfrom sklearn.model_selection import StratifiedKFold\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom keras.wrappers.scikit_learn import KerasRegressor\nfrom keras.models import Sequential\nfrom keras.layers import Bidirectional\nfrom keras.layers import GRU\nfrom keras.layers import Dense\nfrom keras.layers import LayerNormalization\nimport time\ndt='float32'\ntf.keras.backend.set_floatx(dt)",
"_____no_output_____"
]
],
[
[
"## Build model",
"_____no_output_____"
]
],
[
[
"def compile_model(model):\n adam_default_learn_rate = 0.001\n schedule = tf.keras.optimizers.schedules.ExponentialDecay(\n initial_learning_rate = adam_default_learn_rate*10,\n #decay_steps=100000, decay_rate=0.96, staircase=True)\n decay_steps=10000, decay_rate=0.99, staircase=True)\n # learn rate = initial_learning_rate * decay_rate ^ (step / decay_steps)\n alrd = tf.keras.optimizers.Adam(learning_rate=schedule)\n bc=tf.keras.losses.BinaryCrossentropy(from_logits=False)\n print(\"COMPILE...\")\n #model.compile(loss=bc, optimizer=alrd, metrics=[\"accuracy\"])\n model.compile(loss=bc, optimizer=\"adam\", metrics=[\"accuracy\"])\n print(\"...COMPILED\")\n return model\n\ndef build_model():\n embed_layer = keras.layers.Embedding(\n #VOCABULARY_SIZE, EMBED_DIMEN, input_length=1000, input_length=1000, mask_zero=True)\n #input_dim=[None,VOCABULARY_SIZE], output_dim=EMBED_DIMEN, mask_zero=True)\n input_dim=VOCABULARY_SIZE, output_dim=EMBED_DIMEN, mask_zero=True)\n #rnn1_layer = keras.layers.Bidirectional(\n rnn1_layer = keras.layers.GRU(NEURONS, return_sequences=True, \n input_shape=[1000,EMBED_DIMEN], activation=ACT, dropout=DROP) #)#bi\n #rnn2_layer = keras.layers.Bidirectional(\n rnn2_layer = keras.layers.GRU(NEURONS, return_sequences=False, \n activation=ACT, dropout=DROP) #)#bi\n dense1_layer = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt)\n #drop1_layer = keras.layers.Dropout(DROP)\n dense2_layer = keras.layers.Dense(NEURONS, activation=ACT,dtype=dt)\n #drop2_layer = keras.layers.Dropout(DROP)\n output_layer = keras.layers.Dense(1, activation=\"sigmoid\", dtype=dt)\n mlp = keras.models.Sequential()\n mlp.add(embed_layer)\n mlp.add(rnn1_layer)\n mlp.add(rnn2_layer)\n mlp.add(dense1_layer)\n #mlp.add(drop1_layer)\n mlp.add(dense2_layer)\n #mlp.add(drop2_layer)\n mlp.add(output_layer)\n mlpc = compile_model(mlp)\n return mlpc",
"_____no_output_____"
]
],
[
[
"## Load and partition sequences",
"_____no_output_____"
]
],
[
[
"# Assume file was preprocessed to contain one line per seq.\n# Prefer Pandas dataframe but df does not support append.\n# For conversion to tensor, must avoid python lists.\ndef load_fasta(filename,label):\n DEFLINE='>'\n labels=[]\n seqs=[]\n lens=[]\n nums=[]\n num=0\n with open (filename,'r') as infile:\n for line in infile:\n if line[0]!=DEFLINE:\n seq=line.rstrip()\n num += 1 # first seqnum is 1\n seqlen=len(seq)\n nums.append(num)\n labels.append(label)\n seqs.append(seq)\n lens.append(seqlen)\n df1=pd.DataFrame(nums,columns=['seqnum'])\n df2=pd.DataFrame(labels,columns=['class'])\n df3=pd.DataFrame(seqs,columns=['sequence'])\n df4=pd.DataFrame(lens,columns=['seqlen'])\n df=pd.concat((df1,df2,df3,df4),axis=1)\n return df\n\ndef separate_X_and_y(data):\n y= data[['class']].copy()\n X= data.drop(columns=['class','seqnum','seqlen'])\n return (X,y)\n\n",
"_____no_output_____"
]
],
[
[
"## Make K-mers",
"_____no_output_____"
]
],
[
[
"def make_kmer_table(K):\n npad='N'*K\n shorter_kmers=['']\n for i in range(K):\n longer_kmers=[]\n for mer in shorter_kmers:\n longer_kmers.append(mer+'A')\n longer_kmers.append(mer+'C')\n longer_kmers.append(mer+'G')\n longer_kmers.append(mer+'T')\n shorter_kmers = longer_kmers\n all_kmers = shorter_kmers\n kmer_dict = {}\n kmer_dict[npad]=0\n value=1\n for mer in all_kmers:\n kmer_dict[mer]=value\n value += 1\n return kmer_dict\n\nKMER_TABLE=make_kmer_table(K)\n\ndef strings_to_vectors(data,uniform_len):\n all_seqs=[]\n for seq in data['sequence']:\n i=0\n seqlen=len(seq)\n kmers=[]\n while i < seqlen-K+1 -1: # stop at minus one for spaced seed\n #kmer=seq[i:i+2]+seq[i+3:i+5] # SPACED SEED 2/1/2 for K=4\n kmer=seq[i:i+K] \n i += 1\n value=KMER_TABLE[kmer]\n kmers.append(value)\n pad_val=0\n while i < uniform_len:\n kmers.append(pad_val)\n i += 1\n all_seqs.append(kmers)\n pd2d=pd.DataFrame(all_seqs)\n return pd2d # return 2D dataframe, uniform dimensions",
"_____no_output_____"
],
[
"def make_kmers(MAXLEN,train_set):\n (X_train_all,y_train_all)=separate_X_and_y(train_set)\n X_train_kmers=strings_to_vectors(X_train_all,MAXLEN)\n # From pandas dataframe to numpy to list to numpy\n num_seqs=len(X_train_kmers)\n tmp_seqs=[]\n for i in range(num_seqs):\n kmer_sequence=X_train_kmers.iloc[i]\n tmp_seqs.append(kmer_sequence)\n X_train_kmers=np.array(tmp_seqs)\n tmp_seqs=None\n labels=y_train_all.to_numpy()\n return (X_train_kmers,labels)",
"_____no_output_____"
],
[
"def make_frequencies(Xin):\n Xout=[]\n VOCABULARY_SIZE= 4**K + 1 # plus one for 'NNN'\n for seq in Xin:\n freqs =[0] * VOCABULARY_SIZE\n total = 0\n for kmerval in seq:\n freqs[kmerval] += 1\n total += 1\n for c in range(VOCABULARY_SIZE):\n freqs[c] = freqs[c]/total\n Xout.append(freqs)\n Xnum = np.asarray(Xout)\n return (Xnum)\ndef make_slice(data_set,min_len,max_len):\n slice = data_set.query('seqlen <= '+str(max_len)+' & seqlen>= '+str(min_len))\n return slice",
"_____no_output_____"
]
],
[
[
"## Cross validation",
"_____no_output_____"
]
],
[
[
"def do_cross_validation(X,y,given_model):\n cv_scores = []\n fold=0\n splitter = ShuffleSplit(n_splits=SPLITS, test_size=0.1, random_state=37863)\n for train_index,valid_index in splitter.split(X):\n fold += 1\n X_train=X[train_index] # use iloc[] for dataframe\n y_train=y[train_index]\n X_valid=X[valid_index]\n y_valid=y[valid_index] \n # Avoid continually improving the same model.\n model = compile_model(keras.models.clone_model(given_model))\n bestname=DATAPATH+FILENAME+\".cv.\"+str(fold)+\".best\"\n mycallbacks = [keras.callbacks.ModelCheckpoint(\n filepath=bestname, save_best_only=True, \n monitor='val_accuracy', mode='max')] \n print(\"FIT\")\n start_time=time.time()\n history=model.fit(X_train, y_train, # batch_size=10, default=32 works nicely\n epochs=EPOCHS, verbose=1, # verbose=1 for ascii art, verbose=0 for none\n callbacks=mycallbacks,\n validation_data=(X_valid,y_valid) )\n end_time=time.time()\n elapsed_time=(end_time-start_time) \n print(\"Fold %d, %d epochs, %d sec\"%(fold,EPOCHS,elapsed_time))\n pd.DataFrame(history.history).plot(figsize=(8,5))\n plt.grid(True)\n plt.gca().set_ylim(0,1)\n plt.show()\n best_model=keras.models.load_model(bestname)\n scores = best_model.evaluate(X_valid, y_valid, verbose=0)\n print(\"%s: %.2f%%\" % (best_model.metrics_names[1], scores[1]*100))\n cv_scores.append(scores[1] * 100) \n print()\n print(\"%d-way Cross Validation mean %.2f%% (+/- %.2f%%)\" % (fold, np.mean(cv_scores), np.std(cv_scores)))",
"_____no_output_____"
]
],
[
[
"## Train on RNA lengths 200-1Kb",
"_____no_output_____"
]
],
[
[
"MINLEN=200\nMAXLEN=1000\nprint(\"Load data from files.\")\nnc_seq=load_fasta(NC_FILENAME,0)\npc_seq=load_fasta(PC_FILENAME,1)\ntrain_set=pd.concat((nc_seq,pc_seq),axis=0)\nnc_seq=None\npc_seq=None\nprint(\"Ready: train_set\")\n#train_set\nsubset=make_slice(train_set,MINLEN,MAXLEN)# One array to two: X and y\nprint (\"Data reshape\")\n(X_train,y_train)=make_kmers(MAXLEN,subset)\n#print (\"Data prep\")\n#X_train=make_frequencies(X_train)",
"Load data from files.\nReady: train_set\nData reshape\n"
],
[
"print (\"Compile the model\")\nmodel=build_model()\nprint (\"Summarize the model\")\nprint(model.summary()) # Print this only once\nmodel.save(DATAPATH+FILENAME+'.model')\n",
"Compile the model\nCOMPILE...\n...COMPILED\nSummarize the model\nModel: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (None, None, 16) 1040 \n_________________________________________________________________\ngru (GRU) (None, None, 64) 15744 \n_________________________________________________________________\ngru_1 (GRU) (None, 64) 24960 \n_________________________________________________________________\ndense (Dense) (None, 64) 4160 \n_________________________________________________________________\ndense_1 (Dense) (None, 64) 4160 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 65 \n=================================================================\nTotal params: 50,129\nTrainable params: 50,129\nNon-trainable params: 0\n_________________________________________________________________\nNone\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/training/tracking/tracking.py:111: Model.state_updates (from tensorflow.python.keras.engine.training) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis property should not be used in TensorFlow 2.0, as updates are applied automatically.\nWARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/training/tracking/tracking.py:111: Layer.updates (from tensorflow.python.keras.engine.base_layer) is deprecated and will be removed in a future version.\nInstructions for updating:\nThis property should not be used in TensorFlow 2.0, as updates are applied automatically.\nINFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.model/assets\n"
],
[
"print (\"Cross valiation\")\ndo_cross_validation(X_train,y_train,model) \nprint (\"Done\")",
"Cross valiation\nCOMPILE...\n...COMPILED\nFIT\nEpoch 1/200\n453/453 [==============================] - ETA: 0s - loss: 0.6305 - accuracy: 0.6491INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 53s 116ms/step - loss: 0.6305 - accuracy: 0.6491 - val_loss: 0.6030 - val_accuracy: 0.6710\nEpoch 2/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6304 - accuracy: 0.6566 - val_loss: 0.6514 - val_accuracy: 0.6530\nEpoch 3/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.6557 - accuracy: 0.6397 - val_loss: 0.6451 - val_accuracy: 0.6530\nEpoch 4/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.6547 - accuracy: 0.6397 - val_loss: 0.6449 - val_accuracy: 0.6530\nEpoch 5/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.6540 - accuracy: 0.6397 - val_loss: 0.6452 - val_accuracy: 0.6530\nEpoch 6/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.6542 - accuracy: 0.6397 - val_loss: 0.6470 - val_accuracy: 0.6530\nEpoch 7/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6536 - accuracy: 0.6397 - val_loss: 0.6447 - val_accuracy: 0.6530\nEpoch 8/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.6541 - accuracy: 0.6397 - val_loss: 0.6464 - val_accuracy: 0.6530\nEpoch 9/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6543 - accuracy: 0.6397 - val_loss: 0.6470 - val_accuracy: 0.6530\nEpoch 10/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6538 - accuracy: 0.6397 - val_loss: 0.6448 - val_accuracy: 0.6530\nEpoch 11/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6541 - accuracy: 0.6397 - val_loss: 0.6446 - val_accuracy: 0.6530\nEpoch 12/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6541 - accuracy: 0.6397 - val_loss: 0.6512 - val_accuracy: 0.6530\nEpoch 13/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6535 - accuracy: 0.6397 - val_loss: 0.6458 - val_accuracy: 0.6530\nEpoch 14/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.6535 - accuracy: 0.6397 - val_loss: 0.6461 - val_accuracy: 0.6530\nEpoch 15/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.6535 - accuracy: 0.6397 - val_loss: 0.6453 - val_accuracy: 0.6530\nEpoch 16/200\n453/453 [==============================] - 34s 74ms/step - loss: 0.6534 - accuracy: 0.6397 - val_loss: 0.6445 - val_accuracy: 0.6530\nEpoch 17/200\n453/453 [==============================] - 33s 74ms/step - loss: 0.6536 - accuracy: 0.6384 - val_loss: 0.6424 - val_accuracy: 0.6530\nEpoch 18/200\n453/453 [==============================] - ETA: 0s - loss: 0.6275 - accuracy: 0.6565INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.6275 - accuracy: 0.6565 - val_loss: 0.5141 - val_accuracy: 0.7523\nEpoch 19/200\n453/453 [==============================] - ETA: 0s - loss: 0.4795 - accuracy: 0.7749INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.4795 - accuracy: 0.7749 - val_loss: 0.4184 - val_accuracy: 0.8088\nEpoch 20/200\n453/453 [==============================] - 34s 74ms/step - loss: 0.4271 - accuracy: 0.8062 - val_loss: 0.4316 - val_accuracy: 0.8001\nEpoch 21/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.4119 - accuracy: 0.8127 - val_loss: 0.4248 - val_accuracy: 0.8063\nEpoch 22/200\n453/453 [==============================] - ETA: 0s - loss: 0.4114 - accuracy: 0.8132INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.4114 - accuracy: 0.8132 - val_loss: 0.3721 - val_accuracy: 0.8287\nEpoch 23/200\n453/453 [==============================] - ETA: 0s - loss: 0.3924 - accuracy: 0.8270INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.3924 - accuracy: 0.8270 - val_loss: 0.3867 - val_accuracy: 0.8305\nEpoch 24/200\n453/453 [==============================] - ETA: 0s - loss: 0.3887 - accuracy: 0.8266INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 107ms/step - loss: 0.3887 - accuracy: 0.8266 - val_loss: 0.3563 - val_accuracy: 0.8405\nEpoch 25/200\n453/453 [==============================] - ETA: 0s - loss: 0.3774 - accuracy: 0.8343INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.3774 - accuracy: 0.8343 - val_loss: 0.3595 - val_accuracy: 0.8411\nEpoch 26/200\n453/453 [==============================] - ETA: 0s - loss: 0.3718 - accuracy: 0.8362INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 107ms/step - loss: 0.3718 - accuracy: 0.8362 - val_loss: 0.3457 - val_accuracy: 0.8436\nEpoch 27/200\n453/453 [==============================] - ETA: 0s - loss: 0.3660 - accuracy: 0.8410INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.3660 - accuracy: 0.8410 - val_loss: 0.3410 - val_accuracy: 0.8529\nEpoch 28/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.3611 - accuracy: 0.8404 - val_loss: 0.3596 - val_accuracy: 0.8380\nEpoch 29/200\n453/453 [==============================] - ETA: 0s - loss: 0.3505 - accuracy: 0.8467INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.3505 - accuracy: 0.8467 - val_loss: 0.3121 - val_accuracy: 0.8672\nEpoch 30/200\n453/453 [==============================] - ETA: 0s - loss: 0.3368 - accuracy: 0.8553INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 109ms/step - loss: 0.3368 - accuracy: 0.8553 - val_loss: 0.3052 - val_accuracy: 0.8734\nEpoch 31/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.3421 - accuracy: 0.8494 - val_loss: 0.3112 - val_accuracy: 0.8647\nEpoch 32/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.3339 - accuracy: 0.8542 - val_loss: 0.3210 - val_accuracy: 0.8529\nEpoch 33/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3401 - accuracy: 0.8492 - val_loss: 0.3341 - val_accuracy: 0.8560\nEpoch 34/200\n453/453 [==============================] - ETA: 0s - loss: 0.3219 - accuracy: 0.8641INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 49s 108ms/step - loss: 0.3219 - accuracy: 0.8641 - val_loss: 0.3067 - val_accuracy: 0.8746\nEpoch 35/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.3449 - accuracy: 0.8519 - val_loss: 0.3140 - val_accuracy: 0.8634\nEpoch 36/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3309 - accuracy: 0.8594 - val_loss: 0.3475 - val_accuracy: 0.8461\nEpoch 37/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3172 - accuracy: 0.8615 - val_loss: 0.3681 - val_accuracy: 0.8386\nEpoch 38/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3476 - accuracy: 0.8499 - val_loss: 0.3101 - val_accuracy: 0.8634\nEpoch 39/200\n453/453 [==============================] - ETA: 0s - loss: 0.3095 - accuracy: 0.8737INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 50s 110ms/step - loss: 0.3095 - accuracy: 0.8737 - val_loss: 0.2870 - val_accuracy: 0.8839\nEpoch 40/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3713 - accuracy: 0.8374 - val_loss: 0.3475 - val_accuracy: 0.8386\nEpoch 41/200\n453/453 [==============================] - 35s 76ms/step - loss: 0.3605 - accuracy: 0.8436 - val_loss: 0.3717 - val_accuracy: 0.8218\nEpoch 42/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.3540 - accuracy: 0.8450 - val_loss: 0.3486 - val_accuracy: 0.8430\nEpoch 43/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.3446 - accuracy: 0.8503 - val_loss: 0.3320 - val_accuracy: 0.8485\nEpoch 44/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3424 - accuracy: 0.8507 - val_loss: 0.3320 - val_accuracy: 0.8492\nEpoch 45/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3459 - accuracy: 0.8512 - val_loss: 0.3092 - val_accuracy: 0.8647\nEpoch 46/200\n453/453 [==============================] - 35s 76ms/step - loss: 0.3276 - accuracy: 0.8586 - val_loss: 0.3103 - val_accuracy: 0.8603\nEpoch 47/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.3166 - accuracy: 0.8651 - val_loss: 0.2928 - val_accuracy: 0.8715\nEpoch 48/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3025 - accuracy: 0.8718 - val_loss: 0.2806 - val_accuracy: 0.8839\nEpoch 49/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.2887 - accuracy: 0.8815 - val_loss: 0.4335 - val_accuracy: 0.8169\nEpoch 50/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3924 - accuracy: 0.8261 - val_loss: 0.3667 - val_accuracy: 0.8392\nEpoch 51/200\n453/453 [==============================] - 34s 75ms/step - loss: 0.3546 - accuracy: 0.8445 - val_loss: 0.3396 - val_accuracy: 0.8516\nEpoch 52/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3439 - accuracy: 0.8499 - val_loss: 0.3307 - val_accuracy: 0.8504\nEpoch 53/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3394 - accuracy: 0.8508 - val_loss: 0.3624 - val_accuracy: 0.8417\nEpoch 54/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3337 - accuracy: 0.8575 - val_loss: 0.3265 - val_accuracy: 0.8572\nEpoch 55/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3231 - accuracy: 0.8619 - val_loss: 0.3096 - val_accuracy: 0.8659\nEpoch 56/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3105 - accuracy: 0.8689 - val_loss: 0.3047 - val_accuracy: 0.8579\nEpoch 57/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.3025 - accuracy: 0.8732 - val_loss: 0.3227 - val_accuracy: 0.8579\nEpoch 58/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2857 - accuracy: 0.8819 - val_loss: 0.3030 - val_accuracy: 0.8721\nEpoch 59/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2783 - accuracy: 0.8870 - val_loss: 0.2879 - val_accuracy: 0.8771\nEpoch 60/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3084 - accuracy: 0.8690 - val_loss: 0.3569 - val_accuracy: 0.8361\nEpoch 61/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3175 - accuracy: 0.8633 - val_loss: 0.3282 - val_accuracy: 0.8436\nEpoch 62/200\n453/453 [==============================] - 35s 76ms/step - loss: 0.2931 - accuracy: 0.8770 - val_loss: 0.2875 - val_accuracy: 0.8802\nEpoch 63/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2902 - accuracy: 0.8766 - val_loss: 0.2899 - val_accuracy: 0.8783\nEpoch 64/200\n453/453 [==============================] - 35s 76ms/step - loss: 0.3087 - accuracy: 0.8695 - val_loss: 0.3627 - val_accuracy: 0.8436\nEpoch 65/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3440 - accuracy: 0.8500 - val_loss: 0.3475 - val_accuracy: 0.8399\nEpoch 66/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3396 - accuracy: 0.8539 - val_loss: 0.3981 - val_accuracy: 0.8231\nEpoch 67/200\n453/453 [==============================] - 35s 76ms/step - loss: 0.3372 - accuracy: 0.8532 - val_loss: 0.3304 - val_accuracy: 0.8523\nEpoch 68/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.3248 - accuracy: 0.8582 - val_loss: 0.3560 - val_accuracy: 0.8355\nEpoch 69/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3168 - accuracy: 0.8610 - val_loss: 0.3528 - val_accuracy: 0.8436\nEpoch 70/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.3150 - accuracy: 0.8675 - val_loss: 0.3184 - val_accuracy: 0.8616\nEpoch 71/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.3072 - accuracy: 0.8677 - val_loss: 0.3153 - val_accuracy: 0.8678\nEpoch 72/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2988 - accuracy: 0.8706 - val_loss: 0.3103 - val_accuracy: 0.8572\nEpoch 73/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2896 - accuracy: 0.8780 - val_loss: 0.3042 - val_accuracy: 0.8703\nEpoch 74/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2802 - accuracy: 0.8859 - val_loss: 0.2927 - val_accuracy: 0.8833\nEpoch 75/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.2743 - accuracy: 0.8864 - val_loss: 0.3052 - val_accuracy: 0.8628\nEpoch 76/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.2680 - accuracy: 0.8912 - val_loss: 0.3050 - val_accuracy: 0.8752\nEpoch 77/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.2835 - accuracy: 0.8822 - val_loss: 0.2949 - val_accuracy: 0.8796\nEpoch 78/200\n453/453 [==============================] - ETA: 0s - loss: 0.2718 - accuracy: 0.8874INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 52s 115ms/step - loss: 0.2718 - accuracy: 0.8874 - val_loss: 0.2880 - val_accuracy: 0.8883\nEpoch 79/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2535 - accuracy: 0.8969 - val_loss: 0.2920 - val_accuracy: 0.8827\nEpoch 80/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.2525 - accuracy: 0.8976 - val_loss: 0.2817 - val_accuracy: 0.8858\nEpoch 81/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.2512 - accuracy: 0.8977 - val_loss: 0.3178 - val_accuracy: 0.8703\nEpoch 82/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2686 - accuracy: 0.8871 - val_loss: 0.2972 - val_accuracy: 0.8821\nEpoch 83/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2434 - accuracy: 0.9011 - val_loss: 0.3296 - val_accuracy: 0.8616\nEpoch 84/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2540 - accuracy: 0.8939 - val_loss: 0.3162 - val_accuracy: 0.8734\nEpoch 85/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2414 - accuracy: 0.9013 - val_loss: 0.3167 - val_accuracy: 0.8759\nEpoch 86/200\n453/453 [==============================] - ETA: 0s - loss: 0.2234 - accuracy: 0.9106INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 50s 111ms/step - loss: 0.2234 - accuracy: 0.9106 - val_loss: 0.2865 - val_accuracy: 0.8914\nEpoch 87/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.2158 - accuracy: 0.9151 - val_loss: 0.3179 - val_accuracy: 0.8790\nEpoch 88/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2099 - accuracy: 0.9189 - val_loss: 0.3026 - val_accuracy: 0.8734\nEpoch 89/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1972 - accuracy: 0.9232 - val_loss: 0.3006 - val_accuracy: 0.8889\nEpoch 90/200\n453/453 [==============================] - ETA: 0s - loss: 0.2301 - accuracy: 0.9071INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 51s 113ms/step - loss: 0.2301 - accuracy: 0.9071 - val_loss: 0.2787 - val_accuracy: 0.8970\nEpoch 91/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2134 - accuracy: 0.9159 - val_loss: 0.2908 - val_accuracy: 0.8870\nEpoch 92/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2234 - accuracy: 0.9120 - val_loss: 0.2900 - val_accuracy: 0.8852\nEpoch 93/200\n453/453 [==============================] - ETA: 0s - loss: 0.2158 - accuracy: 0.9150INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 51s 113ms/step - loss: 0.2158 - accuracy: 0.9150 - val_loss: 0.2766 - val_accuracy: 0.8982\nEpoch 94/200\n453/453 [==============================] - ETA: 0s - loss: 0.2190 - accuracy: 0.9152INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 50s 111ms/step - loss: 0.2190 - accuracy: 0.9152 - val_loss: 0.2702 - val_accuracy: 0.9032\nEpoch 95/200\n453/453 [==============================] - ETA: 0s - loss: 0.1976 - accuracy: 0.9240INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 51s 112ms/step - loss: 0.1976 - accuracy: 0.9240 - val_loss: 0.2666 - val_accuracy: 0.9081\nEpoch 96/200\n453/453 [==============================] - ETA: 0s - loss: 0.1906 - accuracy: 0.9247INFO:tensorflow:Assets written to: /content/drive/My Drive/data/GRU212.cv.1.best/assets\n453/453 [==============================] - 51s 113ms/step - loss: 0.1906 - accuracy: 0.9247 - val_loss: 0.2488 - val_accuracy: 0.9131\nEpoch 97/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1839 - accuracy: 0.9316 - val_loss: 0.2976 - val_accuracy: 0.8895\nEpoch 98/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.2136 - accuracy: 0.9178 - val_loss: 0.2602 - val_accuracy: 0.9069\nEpoch 99/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1820 - accuracy: 0.9310 - val_loss: 0.2842 - val_accuracy: 0.8876\nEpoch 100/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.1960 - accuracy: 0.9235 - val_loss: 0.3218 - val_accuracy: 0.8678\nEpoch 101/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.2146 - accuracy: 0.9147 - val_loss: 0.2916 - val_accuracy: 0.8945\nEpoch 102/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.2237 - accuracy: 0.9078 - val_loss: 0.3195 - val_accuracy: 0.8771\nEpoch 103/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1974 - accuracy: 0.9213 - val_loss: 0.3016 - val_accuracy: 0.8901\nEpoch 104/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1770 - accuracy: 0.9325 - val_loss: 0.3146 - val_accuracy: 0.8932\nEpoch 105/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1754 - accuracy: 0.9334 - val_loss: 0.2833 - val_accuracy: 0.9038\nEpoch 106/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1679 - accuracy: 0.9342 - val_loss: 0.2737 - val_accuracy: 0.9112\nEpoch 107/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1566 - accuracy: 0.9399 - val_loss: 0.2631 - val_accuracy: 0.9131\nEpoch 108/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1474 - accuracy: 0.9441 - val_loss: 0.2628 - val_accuracy: 0.9094\nEpoch 109/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1965 - accuracy: 0.9195 - val_loss: 0.4364 - val_accuracy: 0.8287\nEpoch 110/200\n453/453 [==============================] - 36s 81ms/step - loss: 0.2141 - accuracy: 0.9149 - val_loss: 0.2839 - val_accuracy: 0.9007\nEpoch 111/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.1750 - accuracy: 0.9334 - val_loss: 0.2764 - val_accuracy: 0.9063\nEpoch 112/200\n453/453 [==============================] - 38s 83ms/step - loss: 0.1762 - accuracy: 0.9314 - val_loss: 0.3089 - val_accuracy: 0.8821\nEpoch 113/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.1637 - accuracy: 0.9353 - val_loss: 0.2825 - val_accuracy: 0.8982\nEpoch 114/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.1476 - accuracy: 0.9421 - val_loss: 0.2990 - val_accuracy: 0.9081\nEpoch 115/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.2025 - accuracy: 0.9165 - val_loss: 0.3348 - val_accuracy: 0.8672\nEpoch 116/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1929 - accuracy: 0.9227 - val_loss: 0.3170 - val_accuracy: 0.8790\nEpoch 117/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.1680 - accuracy: 0.9345 - val_loss: 0.2959 - val_accuracy: 0.8920\nEpoch 118/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.1579 - accuracy: 0.9387 - val_loss: 0.2751 - val_accuracy: 0.9081\nEpoch 119/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.1576 - accuracy: 0.9409 - val_loss: 0.3208 - val_accuracy: 0.8908\nEpoch 120/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1950 - accuracy: 0.9229 - val_loss: 0.3422 - val_accuracy: 0.8653\nEpoch 121/200\n453/453 [==============================] - 37s 83ms/step - loss: 0.2065 - accuracy: 0.9162 - val_loss: 0.3388 - val_accuracy: 0.8672\nEpoch 122/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.1932 - accuracy: 0.9217 - val_loss: 0.3131 - val_accuracy: 0.8839\nEpoch 123/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1916 - accuracy: 0.9235 - val_loss: 0.3457 - val_accuracy: 0.8796\nEpoch 124/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.2029 - accuracy: 0.9197 - val_loss: 0.3701 - val_accuracy: 0.8566\nEpoch 125/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1811 - accuracy: 0.9280 - val_loss: 0.3261 - val_accuracy: 0.8777\nEpoch 126/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.1712 - accuracy: 0.9324 - val_loss: 0.3124 - val_accuracy: 0.8895\nEpoch 127/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.2705 - accuracy: 0.8873 - val_loss: 0.3962 - val_accuracy: 0.8411\nEpoch 128/200\n453/453 [==============================] - 37s 82ms/step - loss: 0.2500 - accuracy: 0.8953 - val_loss: 0.3608 - val_accuracy: 0.8541\nEpoch 129/200\n453/453 [==============================] - 37s 81ms/step - loss: 0.2201 - accuracy: 0.9084 - val_loss: 0.3745 - val_accuracy: 0.8622\nEpoch 130/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.1980 - accuracy: 0.9194 - val_loss: 0.3504 - val_accuracy: 0.8653\nEpoch 131/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1870 - accuracy: 0.9263 - val_loss: 0.3177 - val_accuracy: 0.8827\nEpoch 132/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1774 - accuracy: 0.9315 - val_loss: 0.3724 - val_accuracy: 0.8641\nEpoch 133/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1773 - accuracy: 0.9278 - val_loss: 0.3447 - val_accuracy: 0.8783\nEpoch 134/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.2132 - accuracy: 0.9118 - val_loss: 0.3498 - val_accuracy: 0.8665\nEpoch 135/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1837 - accuracy: 0.9264 - val_loss: 0.3311 - val_accuracy: 0.8883\nEpoch 136/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1648 - accuracy: 0.9333 - val_loss: 0.3615 - val_accuracy: 0.8821\nEpoch 137/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1433 - accuracy: 0.9433 - val_loss: 0.3443 - val_accuracy: 0.8889\nEpoch 138/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1339 - accuracy: 0.9480 - val_loss: 0.3577 - val_accuracy: 0.8914\nEpoch 139/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1544 - accuracy: 0.9405 - val_loss: 0.3535 - val_accuracy: 0.8901\nEpoch 140/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.2321 - accuracy: 0.9037 - val_loss: 0.3755 - val_accuracy: 0.8485\nEpoch 141/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2213 - accuracy: 0.9077 - val_loss: 0.3730 - val_accuracy: 0.8572\nEpoch 142/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1932 - accuracy: 0.9202 - val_loss: 0.3686 - val_accuracy: 0.8665\nEpoch 143/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.1769 - accuracy: 0.9282 - val_loss: 0.3813 - val_accuracy: 0.8603\nEpoch 144/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1593 - accuracy: 0.9356 - val_loss: 0.3510 - val_accuracy: 0.8796\nEpoch 145/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1533 - accuracy: 0.9387 - val_loss: 0.4014 - val_accuracy: 0.8628\nEpoch 146/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.1472 - accuracy: 0.9428 - val_loss: 0.3949 - val_accuracy: 0.8641\nEpoch 147/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1364 - accuracy: 0.9456 - val_loss: 0.3981 - val_accuracy: 0.8808\nEpoch 148/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1389 - accuracy: 0.9435 - val_loss: 0.4436 - val_accuracy: 0.8616\nEpoch 149/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1167 - accuracy: 0.9547 - val_loss: 0.4435 - val_accuracy: 0.8678\nEpoch 150/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1413 - accuracy: 0.9436 - val_loss: 0.3964 - val_accuracy: 0.8541\nEpoch 151/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1747 - accuracy: 0.9287 - val_loss: 0.4349 - val_accuracy: 0.8454\nEpoch 152/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1432 - accuracy: 0.9431 - val_loss: 0.4872 - val_accuracy: 0.8498\nEpoch 153/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1292 - accuracy: 0.9485 - val_loss: 0.5019 - val_accuracy: 0.8547\nEpoch 154/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1134 - accuracy: 0.9543 - val_loss: 0.5625 - val_accuracy: 0.8461\nEpoch 155/200\n453/453 [==============================] - 36s 80ms/step - loss: 0.1017 - accuracy: 0.9607 - val_loss: 0.5504 - val_accuracy: 0.8448\nEpoch 156/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1036 - accuracy: 0.9610 - val_loss: 0.5165 - val_accuracy: 0.8585\nEpoch 157/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0951 - accuracy: 0.9643 - val_loss: 0.5832 - val_accuracy: 0.8529\nEpoch 158/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.0944 - accuracy: 0.9641 - val_loss: 0.5291 - val_accuracy: 0.8616\nEpoch 159/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.0957 - accuracy: 0.9651 - val_loss: 0.4683 - val_accuracy: 0.8703\nEpoch 160/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1219 - accuracy: 0.9537 - val_loss: 0.4675 - val_accuracy: 0.8659\nEpoch 161/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0962 - accuracy: 0.9636 - val_loss: 0.4884 - val_accuracy: 0.8752\nEpoch 162/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0837 - accuracy: 0.9682 - val_loss: 0.4629 - val_accuracy: 0.8771\nEpoch 163/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0901 - accuracy: 0.9642 - val_loss: 0.4772 - val_accuracy: 0.8771\nEpoch 164/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.0837 - accuracy: 0.9671 - val_loss: 0.4851 - val_accuracy: 0.8727\nEpoch 165/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.0920 - accuracy: 0.9649 - val_loss: 0.5216 - val_accuracy: 0.8672\nEpoch 166/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0977 - accuracy: 0.9629 - val_loss: 0.3943 - val_accuracy: 0.8839\nEpoch 167/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1709 - accuracy: 0.9314 - val_loss: 0.4033 - val_accuracy: 0.8696\nEpoch 168/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1353 - accuracy: 0.9466 - val_loss: 0.4367 - val_accuracy: 0.8783\nEpoch 169/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1083 - accuracy: 0.9572 - val_loss: 0.4595 - val_accuracy: 0.8690\nEpoch 170/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0985 - accuracy: 0.9614 - val_loss: 0.4684 - val_accuracy: 0.8759\nEpoch 171/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0886 - accuracy: 0.9648 - val_loss: 0.5465 - val_accuracy: 0.8659\nEpoch 172/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1051 - accuracy: 0.9607 - val_loss: 0.3906 - val_accuracy: 0.8852\nEpoch 173/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.0995 - accuracy: 0.9614 - val_loss: 0.4058 - val_accuracy: 0.8883\nEpoch 174/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1028 - accuracy: 0.9606 - val_loss: 0.4578 - val_accuracy: 0.8727\nEpoch 175/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1571 - accuracy: 0.9374 - val_loss: 0.4033 - val_accuracy: 0.8715\nEpoch 176/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1502 - accuracy: 0.9404 - val_loss: 0.3933 - val_accuracy: 0.8783\nEpoch 177/200\n453/453 [==============================] - 34s 76ms/step - loss: 0.1322 - accuracy: 0.9488 - val_loss: 0.3913 - val_accuracy: 0.8746\nEpoch 178/200\n453/453 [==============================] - 35s 76ms/step - loss: 0.1337 - accuracy: 0.9476 - val_loss: 0.3742 - val_accuracy: 0.8696\nEpoch 179/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1476 - accuracy: 0.9425 - val_loss: 0.3835 - val_accuracy: 0.8703\nEpoch 180/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1339 - accuracy: 0.9469 - val_loss: 0.4159 - val_accuracy: 0.8591\nEpoch 181/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1274 - accuracy: 0.9474 - val_loss: 0.4365 - val_accuracy: 0.8790\nEpoch 182/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1022 - accuracy: 0.9605 - val_loss: 0.4043 - val_accuracy: 0.8814\nEpoch 183/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0857 - accuracy: 0.9667 - val_loss: 0.4831 - val_accuracy: 0.8740\nEpoch 184/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0812 - accuracy: 0.9689 - val_loss: 0.4784 - val_accuracy: 0.8678\nEpoch 185/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0804 - accuracy: 0.9705 - val_loss: 0.4843 - val_accuracy: 0.8727\nEpoch 186/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0965 - accuracy: 0.9624 - val_loss: 0.5097 - val_accuracy: 0.8616\nEpoch 187/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.0793 - accuracy: 0.9694 - val_loss: 0.5210 - val_accuracy: 0.8703\nEpoch 188/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.1031 - accuracy: 0.9600 - val_loss: 0.5172 - val_accuracy: 0.8659\nEpoch 189/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0866 - accuracy: 0.9678 - val_loss: 0.4990 - val_accuracy: 0.8591\nEpoch 190/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0883 - accuracy: 0.9677 - val_loss: 0.5327 - val_accuracy: 0.8690\nEpoch 191/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0631 - accuracy: 0.9774 - val_loss: 0.5244 - val_accuracy: 0.8690\nEpoch 192/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0523 - accuracy: 0.9810 - val_loss: 0.5622 - val_accuracy: 0.8684\nEpoch 193/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0562 - accuracy: 0.9803 - val_loss: 0.5898 - val_accuracy: 0.8634\nEpoch 194/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.0712 - accuracy: 0.9731 - val_loss: 0.5036 - val_accuracy: 0.8715\nEpoch 195/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.0880 - accuracy: 0.9673 - val_loss: 0.4881 - val_accuracy: 0.8808\nEpoch 196/200\n453/453 [==============================] - 35s 77ms/step - loss: 0.0794 - accuracy: 0.9698 - val_loss: 0.5320 - val_accuracy: 0.8765\nEpoch 197/200\n453/453 [==============================] - 36s 78ms/step - loss: 0.1350 - accuracy: 0.9488 - val_loss: 0.3910 - val_accuracy: 0.8603\nEpoch 198/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.2026 - accuracy: 0.9180 - val_loss: 0.3561 - val_accuracy: 0.8721\nEpoch 199/200\n453/453 [==============================] - 36s 79ms/step - loss: 0.1372 - accuracy: 0.9476 - val_loss: 0.3678 - val_accuracy: 0.8790\nEpoch 200/200\n453/453 [==============================] - 35s 78ms/step - loss: 0.1141 - accuracy: 0.9552 - val_loss: 0.4328 - val_accuracy: 0.8678\nFold 1, 200 epochs, 7380 sec\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbab761242b146571ad93d5a2dcd8278821cd819
| 95,455 |
ipynb
|
Jupyter Notebook
|
module1-statistics-probability-and-inference/LS_DS_141_Statistics_Probability_and_Inference.ipynb
|
PWalis/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments
|
5522f9bba4b8273359ce7e6437898e000f22827b
|
[
"MIT"
] | null | null | null |
module1-statistics-probability-and-inference/LS_DS_141_Statistics_Probability_and_Inference.ipynb
|
PWalis/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments
|
5522f9bba4b8273359ce7e6437898e000f22827b
|
[
"MIT"
] | null | null | null |
module1-statistics-probability-and-inference/LS_DS_141_Statistics_Probability_and_Inference.ipynb
|
PWalis/DS-Unit-1-Sprint-4-Statistical-Tests-and-Experiments
|
5522f9bba4b8273359ce7e6437898e000f22827b
|
[
"MIT"
] | null | null | null | 41.233261 | 9,648 | 0.374522 |
[
[
[
"# Lambda School Data Science Module 141\n## Statistics, Probability, and Inference",
"_____no_output_____"
],
[
"## Prepare - examine what's available in SciPy\n\nAs we delve into statistics, we'll be using more libraries - in particular the [stats package from SciPy](https://docs.scipy.org/doc/scipy/reference/tutorial/stats.html).",
"_____no_output_____"
]
],
[
[
"from scipy import stats\ndir(stats)",
"_____no_output_____"
],
[
"# As usual, lots of stuff here! There's our friend, the normal distribution\nnorm = stats.norm()\nprint(norm.mean())\nprint(norm.std())\nprint(norm.var())",
"0.0\n1.0\n1.0\n"
],
[
"# And a new friend - t\nt1 = stats.t(5) # 5 is df \"shape\" parameter\nprint(t1.mean())\nprint(t1.std())\nprint(t1.var())",
"0.0\n1.2909944487358056\n1.6666666666666667\n"
]
],
[
[
"\n\n*(Picture from [Wikipedia](https://en.wikipedia.org/wiki/Student's_t-distribution#/media/File:Student_t_pdf.svg))*\n\nThe t-distribution is \"normal-ish\" - the larger the parameter (which reflects its degrees of freedom - more input data/features will increase it), the closer to true normal.",
"_____no_output_____"
]
],
[
[
"t2 = stats.t(30) # Will be closer to normal\nprint(t2.mean())\nprint(t2.std())\nprint(t2.var())",
"0.0\n1.0350983390135313\n1.0714285714285714\n"
]
],
[
[
"Why is it different from normal? To better reflect the tendencies of small data and situations with unknown population standard deviation. In other words, the normal distribution is still the nice pure ideal in the limit (thanks to the central limit theorem), but the t-distribution is much more useful in many real-world situations.\n\nHistory sidenote - this is \"Student\":\n\n\n\n*(Picture from [Wikipedia](https://en.wikipedia.org/wiki/File:William_Sealy_Gosset.jpg))*\n\nHis real name is William Sealy Gosset, and he published under the pen name \"Student\" because he was not an academic. He was a brewer, working at Guinness and using trial and error to determine the best ways to yield barley. He's also proof that, even 100 years ago, you don't need official credentials to do real data science!",
"_____no_output_____"
],
[
"## Live Lecture - let's perform and interpret a t-test\n\nWe'll generate our own data, so we can know and alter the \"ground truth\" that the t-test should find. We will learn about p-values and how to interpret \"statistical significance\" based on the output of a hypothesis test. We will also dig a bit deeper into how the test statistic is calculated based on the sample error, and visually what it looks like to have 1 or 2 \"tailed\" t-tests.",
"_____no_output_____"
]
],
[
[
"# Just making som data to play around with\nlambda_heights = [72,72,77,72,73,67,64,58,63,78]\n\nimport pandas as pd\ndf = pd.DataFrame({'heights': lambda_heights})\ndf.head(10)",
"_____no_output_____"
],
[
"df.heights.mean()",
"_____no_output_____"
],
[
"# Making random data 0's and 1's\nimport random \nrandom.seed(10)\n\npopulation = []\n\nfor _ in range(1000):\n population.append(random.randint(0,1))\n \nprint(population)\nprint(len(population))",
"[0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 1, 0, 0, 0, 0, 0, 1, 1, 0, 1, 1, 1, 1, 0, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 1, 1, 0, 0, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 0, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 0, 0, 0, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 1, 1, 0, 1, 1, 0, 1, 0, 0, 1, 0, 0, 1, 0, 1, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 1, 1, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 1, 1, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 1, 1, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 0, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 0, 1, 1, 1, 0, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 0, 1, 0, 0, 1, 0]\n1000\n"
],
[
"# Take a sample from the data randomly\nsample = random.sample(population, 100)\nprint(sample)\nprint(len(sample))",
"[0, 0, 0, 0, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0]\n100\n"
],
[
"df = pd.DataFrame({'likes_coke': sample})\ndf.head()\n\ndf.likes_coke.mean()",
"_____no_output_____"
],
[
"df.plot.hist()",
"_____no_output_____"
],
[
"df.likes_coke.describe()",
"_____no_output_____"
],
[
"import numpy as np\n\ndef mean(list):\n average = np.sum(list)/len(list)\n return average \n\nprint('Population Mean:', mean(population))\nprint('Sample Mean:', mean(sample))",
"Population Mean: 0.507\nSample Mean: 0.46\n"
],
[
"def variance(list):\n n = len(list)\n return np.sum((list - mean(list))**2)/(n-1)\n\nvariance(df.likes_coke)",
"_____no_output_____"
],
[
"def stddev(list):\n var = variance(list)\n return var**(1/2)\n\nstddev(df.likes_coke)",
"_____no_output_____"
],
[
"n = len(df.likes_coke)\nt_stat = (mean(df.likes_coke) - .5)/(stddev(df.likes_coke)/n**(1/2))\nprint(t_stat)",
"-0.7985494095046901\n"
],
[
"import scipy\nscipy.stats.ttest_1samp(df['likes_coke'], .5)",
"_____no_output_____"
]
],
[
[
"#P-Value\nP-value is a threshold that we set for ourselves to denote \"statistical significance\" Statistical Significance means - the odds of me getting unlucky that I'm willing to deal with. The probability that I would have to see that says that these two differences are not just due to chance.\n\n5% - Will only accept this result as reliable or significant if I calculate that this outcome has a 5% chance or less of happening just due to chance.\n\nThe probability that the pattern in our data that we're seeing could be produced by random data.",
"_____no_output_____"
],
[
"## Assignment - apply the t-test to real data\n\nYour assignment is to determine which issues have \"statistically significant\" differences between political parties in this [1980s congressional voting data](https://archive.ics.uci.edu/ml/datasets/Congressional+Voting+Records). The data consists of 435 instances (one for each congressperson), a class (democrat or republican), and 16 binary attributes (yes or no for voting for or against certain issues). Be aware - there are missing values!\n\nYour goals:\n\n1. Load and clean the data (or determine the best method to drop observations when running tests)\n2. Using hypothesis testing, find an issue that democrats support more than republicans with p < 0.01\n3. Using hypothesis testing, find an issue that republicans support more than democrats with p < 0.01\n4. Using hypothesis testing, find an issue where the difference between republicans and democrats has p > 0.1 (i.e. there may not be much of a difference)\n\nNote that this data will involve *2 sample* t-tests, because you're comparing averages across two groups (republicans and democrats) rather than a single group against a null hypothesis.\n\nStretch goals:\n\n1. Refactor your code into functions so it's easy to rerun with arbitrary variables\n2. Apply hypothesis testing to your personal project data (for the purposes of this notebook you can type a summary of the hypothesis you formed and tested)",
"_____no_output_____"
]
],
[
[
"column_names = ['Class Name', 'handicapped-infants', 'water-project-cost-sharing','adoption-of-the-budget-resolution', 'physician-fee-freeze', 'el-salvador-aid',\n 'religious-groups-in-schools', 'anti-satellite-test-ban', 'aid-to-nicaraguan-contras', 'mx-missile', 'immigration', 'synfuels-corporation-cutback', \n 'education-spending', 'superfund-right-to-sue', 'crime', 'duty-free-exports', 'export-administration-act-south-africa']\ndf = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/voting-records/house-votes-84.data', header=None, names=column_names, na_values='?')\ndf.head()",
"_____no_output_____"
],
[
"df = df.fillna(-1)",
"_____no_output_____"
],
[
"for column in df.columns:\n df[column] = df[column].replace({'n':0, 'y':1})\n\n ",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"rep = df[df['Class Name']=='republican']\ndem= df[df['Class Name']=='democrat']",
"_____no_output_____"
],
[
"rep.head()",
"_____no_output_____"
],
[
"dem.head()",
"_____no_output_____"
],
[
"for column in rep.columns:\n print(rep[column].value_counts(normalize=True),'\\n#####################################################\\n')",
"republican 1.0\nName: Class Name, dtype: float64 \n#####################################################\n\n 0 0.797619\n 1 0.184524\n-1 0.017857\nName: handicapped-infants, dtype: float64 \n#####################################################\n\n 1 0.446429\n 0 0.434524\n-1 0.119048\nName: water-project-cost-sharing, dtype: float64 \n#####################################################\n\n 0 0.845238\n 1 0.130952\n-1 0.023810\nName: adoption-of-the-budget-resolution, dtype: float64 \n#####################################################\n\n 1 0.970238\n-1 0.017857\n 0 0.011905\nName: physician-fee-freeze, dtype: float64 \n#####################################################\n\n 1 0.934524\n 0 0.047619\n-1 0.017857\nName: el-salvador-aid, dtype: float64 \n#####################################################\n\n 1 0.886905\n 0 0.101190\n-1 0.011905\nName: religious-groups-in-schools, dtype: float64 \n#####################################################\n\n 0 0.732143\n 1 0.232143\n-1 0.035714\nName: anti-satellite-test-ban, dtype: float64 \n#####################################################\n\n 0 0.791667\n 1 0.142857\n-1 0.065476\nName: aid-to-nicaraguan-contras, dtype: float64 \n#####################################################\n\n 0 0.869048\n 1 0.113095\n-1 0.017857\nName: mx-missile, dtype: float64 \n#####################################################\n\n 1 0.547619\n 0 0.434524\n-1 0.017857\nName: immigration, dtype: float64 \n#####################################################\n\n 0 0.821429\n 1 0.125000\n-1 0.053571\nName: synfuels-corporation-cutback, dtype: float64 \n#####################################################\n\n 1 0.803571\n 0 0.119048\n-1 0.077381\nName: education-spending, dtype: float64 \n#####################################################\n\n 1 0.809524\n 0 0.130952\n-1 0.059524\nName: superfund-right-to-sue, dtype: float64 \n#####################################################\n\n 1 0.940476\n-1 0.041667\n 0 0.017857\nName: crime, dtype: float64 \n#####################################################\n\n 0 0.845238\n 1 0.083333\n-1 0.071429\nName: duty-free-exports, dtype: float64 \n#####################################################\n\n 1 0.571429\n 0 0.297619\n-1 0.130952\nName: export-administration-act-south-africa, dtype: float64 \n#####################################################\n\n"
],
[
"for column in dem.columns:\n print(dem[column].value_counts(normalize=True),'\\n#####################################################\\n')",
"democrat 1.0\nName: Class Name, dtype: float64 \n#####################################################\n\n 1 0.584270\n 0 0.382022\n-1 0.033708\nName: handicapped-infants, dtype: float64 \n#####################################################\n\n 1 0.449438\n 0 0.445693\n-1 0.104869\nName: water-project-cost-sharing, dtype: float64 \n#####################################################\n\n 1 0.865169\n 0 0.108614\n-1 0.026217\nName: adoption-of-the-budget-resolution, dtype: float64 \n#####################################################\n\n 0 0.917603\n 1 0.052434\n-1 0.029963\nName: physician-fee-freeze, dtype: float64 \n#####################################################\n\n 0 0.749064\n 1 0.205993\n-1 0.044944\nName: el-salvador-aid, dtype: float64 \n#####################################################\n\n 0 0.505618\n 1 0.460674\n-1 0.033708\nName: religious-groups-in-schools, dtype: float64 \n#####################################################\n\n 1 0.749064\n 0 0.220974\n-1 0.029963\nName: anti-satellite-test-ban, dtype: float64 \n#####################################################\n\n 1 0.816479\n 0 0.168539\n-1 0.014981\nName: aid-to-nicaraguan-contras, dtype: float64 \n#####################################################\n\n 1 0.704120\n 0 0.224719\n-1 0.071161\nName: mx-missile, dtype: float64 \n#####################################################\n\n 0 0.520599\n 1 0.464419\n-1 0.014981\nName: immigration, dtype: float64 \n#####################################################\n\n 1 0.483146\n 0 0.471910\n-1 0.044944\nName: synfuels-corporation-cutback, dtype: float64 \n#####################################################\n\n 0 0.797753\n 1 0.134831\n-1 0.067416\nName: education-spending, dtype: float64 \n#####################################################\n\n 0 0.670412\n 1 0.273408\n-1 0.056180\nName: superfund-right-to-sue, dtype: float64 \n#####################################################\n\n 0 0.625468\n 1 0.337079\n-1 0.037453\nName: crime, dtype: float64 \n#####################################################\n\n 1 0.599251\n 0 0.340824\n-1 0.059925\nName: duty-free-exports, dtype: float64 \n#####################################################\n\n 1 0.647940\n-1 0.307116\n 0 0.044944\nName: export-administration-act-south-africa, dtype: float64 \n#####################################################\n\n"
],
[
"print('All Mean:', mean(dem['handicapped-infants']))\nprint('Sample Mean:', mean(dem['handicapped-infants']))\nscipy.stats.ttest_1samp(dem['handicapped-infants'], .5)",
"All Mean: 0.550561797752809\nSample Mean: 0.550561797752809\n"
],
[
"print('All Mean:', mean(rep['handicapped-infants']))\nprint('Sample Mean:', mean(rep['handicapped-infants']))\nscipy.stats.ttest_1samp(rep['handicapped-infants'], .5)",
"All Mean: 0.16666666666666666\nSample Mean: 0.16666666666666666\n"
],
[
"rep_nonan = rep[rep['handicapped-infants']!=-1]\ndem_nonan = dem[dem['handicapped-infants']!=-1]",
"_____no_output_____"
],
[
"scipy.stats.ttest_1samp(dem_nonan['handicapped-infants'], .5)",
"_____no_output_____"
],
[
"scipy.stats.ttest_1samp(rep_nonan['handicapped-infants'], .5)",
"_____no_output_____"
]
],
[
[
"Democrats support Handicapped Infants more than Republicans",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"# Resources\n\n- https://homepage.divms.uiowa.edu/~mbognar/applets/t.html\n- https://rpsychologist.com/d3/tdist/\n- https://gallery.shinyapps.io/tdist/\n- https://en.wikipedia.org/wiki/Standard_deviation#Sample_standard_deviation_of_metabolic_rate_of_northern_fulmars",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbab79b786113e559411126c7fa3c8568c13bbc1
| 17,342 |
ipynb
|
Jupyter Notebook
|
Chapter 8/Ch8_book.ipynb
|
gnperdue/DeepReinforcementLearningInAction
|
3d32324fac0c6dff1b88d35004d9efc07185fb3f
|
[
"MIT"
] | 2 |
2019-12-12T21:10:48.000Z
|
2021-08-23T16:41:36.000Z
|
Chapter 8/Ch8_book.ipynb
|
Samper-Escudero/DeepReinforcementLearningInAction
|
3d32324fac0c6dff1b88d35004d9efc07185fb3f
|
[
"MIT"
] | null | null | null |
Chapter 8/Ch8_book.ipynb
|
Samper-Escudero/DeepReinforcementLearningInAction
|
3d32324fac0c6dff1b88d35004d9efc07185fb3f
|
[
"MIT"
] | null | null | null | 31.588342 | 111 | 0.520009 |
[
[
[
"# Chaper 8 - Intrinsic Curiosity Module\n#### Deep Reinforcement Learning *in Action*",
"_____no_output_____"
],
[
"##### Listing 8.1",
"_____no_output_____"
]
],
[
[
"import gym\nfrom nes_py.wrappers import BinarySpaceToDiscreteSpaceEnv #A\nimport gym_super_mario_bros\nfrom gym_super_mario_bros.actions import SIMPLE_MOVEMENT, COMPLEX_MOVEMENT #B\nenv = gym_super_mario_bros.make('SuperMarioBros-v0')\nenv = BinarySpaceToDiscreteSpaceEnv(env, COMPLEX_MOVEMENT) #C",
"_____no_output_____"
],
[
"done = True\nfor step in range(2500): #D\n if done:\n state = env.reset()\n state, reward, done, info = env.step(env.action_space.sample())\n env.render()\nenv.close()",
"_____no_output_____"
]
],
[
[
"##### Listing 8.2",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom skimage.transform import resize #A\nimport numpy as np\n\ndef downscale_obs(obs, new_size=(42,42), to_gray=True):\n if to_gray:\n return resize(obs, new_size, anti_aliasing=True).max(axis=2) #B\n else:\n return resize(obs, new_size, anti_aliasing=True)",
"_____no_output_____"
],
[
"plt.imshow(env.render(\"rgb_array\"))\nplt.imshow(downscale_obs(env.render(\"rgb_array\")))",
"_____no_output_____"
]
],
[
[
"##### Listing 8.4",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch import nn\nfrom torch import optim\nimport torch.nn.functional as F\nfrom collections import deque\n\ndef prepare_state(state): #A\n return torch.from_numpy(downscale_obs(state, to_gray=True)).float().unsqueeze(dim=0)\n\n\ndef prepare_multi_state(state1, state2): #B\n state1 = state1.clone()\n tmp = torch.from_numpy(downscale_obs(state2, to_gray=True)).float()\n state1[0][0] = state1[0][1]\n state1[0][1] = state1[0][2]\n state1[0][2] = tmp\n return state1\n\n\ndef prepare_initial_state(state,N=3): #C\n state_ = torch.from_numpy(downscale_obs(state, to_gray=True)).float()\n tmp = state_.repeat((N,1,1))\n return tmp.unsqueeze(dim=0)",
"_____no_output_____"
]
],
[
[
"##### Listing 8.5",
"_____no_output_____"
]
],
[
[
"def policy(qvalues, eps=None): #A\n if eps is not None:\n if torch.rand(1) < eps:\n return torch.randint(low=0,high=7,size=(1,))\n else:\n return torch.argmax(qvalues)\n else:\n return torch.multinomial(F.softmax(F.normalize(qvalues)), num_samples=1) #B",
"_____no_output_____"
]
],
[
[
"##### Listing 8.6",
"_____no_output_____"
]
],
[
[
"from random import shuffle\nimport torch\nfrom torch import nn\nfrom torch import optim\nimport torch.nn.functional as F\n\nclass ExperienceReplay:\n def __init__(self, N=500, batch_size=100):\n self.N = N #A\n self.batch_size = batch_size #B\n self.memory = [] \n self.counter = 0\n \n def add_memory(self, state1, action, reward, state2):\n self.counter +=1 \n if self.counter % 500 == 0: #C\n self.shuffle_memory()\n \n if len(self.memory) < self.N: #D\n self.memory.append( (state1, action, reward, state2) )\n else:\n rand_index = np.random.randint(0,self.N-1)\n self.memory[rand_index] = (state1, action, reward, state2)\n \n def shuffle_memory(self): #E\n shuffle(self.memory)\n \n def get_batch(self): #F\n if len(self.memory) < self.batch_size:\n batch_size = len(self.memory)\n else:\n batch_size = self.batch_size\n if len(self.memory) < 1:\n print(\"Error: No data in memory.\")\n return None\n #G\n ind = np.random.choice(np.arange(len(self.memory)),batch_size,replace=False)\n batch = [self.memory[i] for i in ind] #batch is a list of tuples\n state1_batch = torch.stack([x[0].squeeze(dim=0) for x in batch],dim=0)\n action_batch = torch.Tensor([x[1] for x in batch]).long()\n reward_batch = torch.Tensor([x[2] for x in batch])\n state2_batch = torch.stack([x[3].squeeze(dim=0) for x in batch],dim=0)\n return state1_batch, action_batch, reward_batch, state2_batch",
"_____no_output_____"
]
],
[
[
"##### Listing 8.7",
"_____no_output_____"
]
],
[
[
"class Phi(nn.Module): #A\n def __init__(self):\n super(Phi, self).__init__()\n self.conv1 = nn.Conv2d(3, 32, kernel_size=(3,3), stride=2, padding=1)\n self.conv2 = nn.Conv2d(32, 32, kernel_size=(3,3), stride=2, padding=1)\n self.conv3 = nn.Conv2d(32, 32, kernel_size=(3,3), stride=2, padding=1)\n self.conv4 = nn.Conv2d(32, 32, kernel_size=(3,3), stride=2, padding=1)\n\n def forward(self,x):\n x = F.normalize(x)\n y = F.elu(self.conv1(x))\n y = F.elu(self.conv2(y))\n y = F.elu(self.conv3(y))\n y = F.elu(self.conv4(y)) #size [1, 32, 3, 3] batch, channels, 3 x 3\n y = y.flatten(start_dim=1) #size N, 288\n return y\n\nclass Gnet(nn.Module): #B\n def __init__(self):\n super(Gnet, self).__init__()\n self.linear1 = nn.Linear(576,256)\n self.linear2 = nn.Linear(256,12)\n\n def forward(self, state1,state2):\n x = torch.cat( (state1, state2) ,dim=1)\n y = F.relu(self.linear1(x))\n y = self.linear2(y)\n y = F.softmax(y,dim=1)\n return y\n\nclass Fnet(nn.Module): #C\n def __init__(self):\n super(Fnet, self).__init__()\n self.linear1 = nn.Linear(300,256)\n self.linear2 = nn.Linear(256,288)\n\n def forward(self,state,action):\n action_ = torch.zeros(action.shape[0],12) #D\n indices = torch.stack( (torch.arange(action.shape[0]), action.squeeze()), dim=0)\n indices = indices.tolist()\n action_[indices] = 1.\n x = torch.cat( (state,action_) ,dim=1)\n y = F.relu(self.linear1(x))\n y = self.linear2(y)\n return y",
"_____no_output_____"
]
],
[
[
"##### Listing 8.8",
"_____no_output_____"
]
],
[
[
"class Qnetwork(nn.Module):\n def __init__(self):\n super(Qnetwork, self).__init__()\n #in_channels, out_channels, kernel_size, stride=1, padding=0\n self.conv1 = nn.Conv2d(in_channels=3, out_channels=32, kernel_size=(3,3), stride=2, padding=1)\n self.conv2 = nn.Conv2d(32, 32, kernel_size=(3,3), stride=2, padding=1)\n self.conv3 = nn.Conv2d(32, 32, kernel_size=(3,3), stride=2, padding=1)\n self.conv4 = nn.Conv2d(32, 32, kernel_size=(3,3), stride=2, padding=1)\n self.linear1 = nn.Linear(288,100)\n self.linear2 = nn.Linear(100,12)\n \n def forward(self,x):\n x = F.normalize(x)\n y = F.elu(self.conv1(x))\n y = F.elu(self.conv2(y))\n y = F.elu(self.conv3(y))\n y = F.elu(self.conv4(y))\n y = y.flatten(start_dim=2)\n y = y.view(y.shape[0], -1, 32)\n y = y.flatten(start_dim=1)\n y = F.elu(self.linear1(y))\n y = self.linear2(y) #size N, 12\n return y",
"_____no_output_____"
]
],
[
[
"##### Listing 8.9",
"_____no_output_____"
]
],
[
[
"params = {\n 'batch_size':150,\n 'beta':0.2,\n 'lambda':0.1,\n 'eta': 1.0,\n 'gamma':0.2,\n 'max_episode_len':100,\n 'min_progress':15,\n 'action_repeats':6,\n 'frames_per_state':3\n}\n\nreplay = ExperienceReplay(N=1000, batch_size=params['batch_size'])\nQmodel = Qnetwork()\nencoder = Phi()\nforward_model = Fnet()\ninverse_model = Gnet()\nforward_loss = nn.MSELoss(reduction='none')\ninverse_loss = nn.CrossEntropyLoss(reduction='none')\nqloss = nn.MSELoss()\nall_model_params = list(Qmodel.parameters()) + list(encoder.parameters()) #A\nall_model_params += list(forward_model.parameters()) + list(inverse_model.parameters())\nopt = optim.Adam(lr=0.001, params=all_model_params)",
"_____no_output_____"
]
],
[
[
"##### Listing 8.10",
"_____no_output_____"
]
],
[
[
"def loss_fn(q_loss, inverse_loss, forward_loss):\n loss_ = (1 - params['beta']) * inverse_loss\n loss_ += params['beta'] * forward_loss\n loss_ = loss_.sum() / loss_.flatten().shape[0]\n loss = loss_ + params['lambda'] * q_loss\n return loss\n\ndef reset_env():\n \"\"\"\n Reset the environment and return a new initial state\n \"\"\"\n env.reset()\n state1 = prepare_initial_state(env.render('rgb_array'))\n return state1",
"_____no_output_____"
]
],
[
[
"##### Listing 8.11",
"_____no_output_____"
]
],
[
[
"def ICM(state1, action, state2, forward_scale=1., inverse_scale=1e4):\n state1_hat = encoder(state1) #A\n state2_hat = encoder(state2)\n state2_hat_pred = forward_model(state1_hat.detach(), action.detach()) #B\n forward_pred_err = forward_scale * forward_loss(state2_hat_pred, \\\n state2_hat.detach()).sum(dim=1).unsqueeze(dim=1)\n pred_action = inverse_model(state1_hat, state2_hat) #C\n inverse_pred_err = inverse_scale * inverse_loss(pred_action, \\\n action.detach().flatten()).unsqueeze(dim=1)\n return forward_pred_err, inverse_pred_err",
"_____no_output_____"
]
],
[
[
"##### Listing 8.12",
"_____no_output_____"
]
],
[
[
"def minibatch_train(use_extrinsic=True):\n state1_batch, action_batch, reward_batch, state2_batch = replay.get_batch() \n action_batch = action_batch.view(action_batch.shape[0],1) #A\n reward_batch = reward_batch.view(reward_batch.shape[0],1)\n \n forward_pred_err, inverse_pred_err = ICM(state1_batch, action_batch, state2_batch) #B\n i_reward = (1. / params['eta']) * forward_pred_err #C\n reward = i_reward.detach() #D\n if use_explicit: #E\n reward += reward_batch \n qvals = Qmodel(state2_batch) #F\n reward += params['gamma'] * torch.max(qvals)\n reward_pred = Qmodel(state1_batch)\n reward_target = reward_pred.clone()\n indices = torch.stack( (torch.arange(action_batch.shape[0]), \\\n action_batch.squeeze()), dim=0)\n indices = indices.tolist()\n reward_target[indices] = reward.squeeze()\n q_loss = 1e5 * qloss(F.normalize(reward_pred), F.normalize(reward_target.detach()))\n return forward_pred_err, inverse_pred_err, q_loss",
"_____no_output_____"
]
],
[
[
"##### Listing 8.13",
"_____no_output_____"
]
],
[
[
"epochs = 5000\nenv.reset()\nstate1 = prepare_initial_state(env.render('rgb_array'))\neps=0.15\nlosses = []\nepisode_length = 0\nswitch_to_eps_greedy = 1000\nstate_deque = deque(maxlen=params['frames_per_state'])\ne_reward = 0.\nlast_x_pos = env.env.env._x_position #A\nep_lengths = []\nuse_explicit = False\nfor i in range(epochs):\n opt.zero_grad()\n episode_length += 1\n q_val_pred = Qmodel(state1) #B\n if i > switch_to_eps_greedy: #C\n action = int(policy(q_val_pred,eps))\n else:\n action = int(policy(q_val_pred))\n for j in range(params['action_repeats']): #D\n state2, e_reward_, done, info = env.step(action)\n last_x_pos = info['x_pos']\n if done:\n state1 = reset_env()\n break\n e_reward += e_reward_\n state_deque.append(prepare_state(state2))\n state2 = torch.stack(list(state_deque),dim=1) #E\n replay.add_memory(state1, action, e_reward, state2) #F\n e_reward = 0\n if episode_length > params['max_episode_len']: #G\n if (info['x_pos'] - last_x_pos) < params['min_progress']:\n done = True\n else:\n last_x_pos = info['x_pos']\n if done:\n ep_lengths.append(info['x_pos'])\n state1 = reset_env()\n last_x_pos = env.env.env._x_position\n episode_length = 0\n else:\n state1 = state2\n if len(replay.memory) < params['batch_size']:\n continue\n forward_pred_err, inverse_pred_err, q_loss = minibatch_train(use_extrinsic=False) #H\n loss = loss_fn(q_loss, forward_pred_err, inverse_pred_err) #I\n loss_list = (q_loss.mean(), forward_pred_err.flatten().mean(),\\\n inverse_pred_err.flatten().mean())\n losses.append(loss_list)\n loss.backward()\n opt.step()",
"_____no_output_____"
]
],
[
[
"##### Test Trained Agent",
"_____no_output_____"
]
],
[
[
"done = True\nstate_deque = deque(maxlen=params['frames_per_state'])\nfor step in range(5000):\n if done:\n env.reset()\n state1 = prepare_initial_state(env.render('rgb_array'))\n q_val_pred = Qmodel(state1)\n action = int(policy(q_val_pred,eps))\n state2, reward, done, info = env.step(action)\n state2 = prepare_multi_state(state1,state2)\n state1=state2\n env.render()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbab893c1371af0d5496b208a0d3e1d5a6cf23b6
| 18,646 |
ipynb
|
Jupyter Notebook
|
GAN/.ipynb_checkpoints/GAN_MNIST-checkpoint.ipynb
|
Las-Desire/Paper-Desire
|
146102f5b3238de350cc7732bb2a8640fedda871
|
[
"MIT"
] | null | null | null |
GAN/.ipynb_checkpoints/GAN_MNIST-checkpoint.ipynb
|
Las-Desire/Paper-Desire
|
146102f5b3238de350cc7732bb2a8640fedda871
|
[
"MIT"
] | null | null | null |
GAN/.ipynb_checkpoints/GAN_MNIST-checkpoint.ipynb
|
Las-Desire/Paper-Desire
|
146102f5b3238de350cc7732bb2a8640fedda871
|
[
"MIT"
] | null | null | null | 104.752809 | 1,840 | 0.679181 |
[
[
[
"import tensorflow as tf\nimport numpy as np\nimport matplotlib.pyplot as plt",
"D:\\LLAS\\lib\\site-packages\\h5py\\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
],
[
"from tensorflow.examples.tutorials.mnist import input_data\nmnist = input_data.read_data_sets(\"./mnist/data/\",one_hot=True)",
"Extracting ./mnist/data/train-images-idx3-ubyte.gz\nExtracting ./mnist/data/train-labels-idx1-ubyte.gz\nExtracting ./mnist/data/t10k-images-idx3-ubyte.gz\nExtracting ./mnist/data/t10k-labels-idx1-ubyte.gz\n"
],
[
"total_epoch = 200\nbatch_size = 100\nlearning_rate = 0.01\nn_hidden = 256\nn_input = 28*28\nn_noise = 128\nn_class = 10",
"_____no_output_____"
],
[
"#Real data\nX = tf.placeholder(tf.float32,[None,n_input])\n#Noise data\nZ = tf.placeholder(tf.float32,[None,n_noise])\n#Y Hint Data\nY = tf.placeholder(tf.float32,[None,n_class])",
"_____no_output_____"
],
[
"def generater(noise, labels):\n with tf.variable_scope('Generater'):\n inputs = tf.concat([noise, labels],1)\n hidden = tf.layers.dense(inputs,n_hidden,tf.nn.relu)\n output = tf.layers.dense(hidden,n_input,activation=tf.nn.sigmoid)\n \n return output",
"_____no_output_____"
],
[
"def discriminaster(inputs, labels, reuse=None):\n with tf.variable_scope('Discriminaster') as scope:\n if(reuse):\n scope.reuse_variables()\n \n inputs = tf.concat([inputs, labels], 1)\n hidden = tf.layers.Dense(inputs, n_hidden, activation=tf.nn.relu)\n output = tf.layers.Dense(hidden , 1, activation=None)\n \n return output",
"_____no_output_____"
],
[
"def get_noise(batch_size, n_noise):\n return np.random.uniform(-1., 1., size = [batch_size, n_noise])",
"_____no_output_____"
],
[
"G = generater(Z, Y)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbab900b9170b5a5be11ec0480a4b5548436ab22
| 508,198 |
ipynb
|
Jupyter Notebook
|
julia.ipynb
|
renatoac/jupyternotebooks
|
f78e624907712780e78ead6de40892fde8a93a23
|
[
"MIT"
] | 403 |
2019-05-07T23:40:45.000Z
|
2022-03-31T11:14:07.000Z
|
julia.ipynb
|
renatoac/jupyternotebooks
|
f78e624907712780e78ead6de40892fde8a93a23
|
[
"MIT"
] | 514 |
2019-05-07T17:00:14.000Z
|
2022-03-31T20:09:16.000Z
|
julia.ipynb
|
renatoac/jupyternotebooks
|
f78e624907712780e78ead6de40892fde8a93a23
|
[
"MIT"
] | 108 |
2019-05-07T23:40:47.000Z
|
2022-03-30T00:15:19.000Z
| 6,049.97619 | 506,586 | 0.966234 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbab92d5c2290e3201bc147d76f39577bc9fca78
| 68,313 |
ipynb
|
Jupyter Notebook
|
matrix_tree/day4.ipynb
|
666bury/dw_matrix-
|
429d676c9502bf1f12d6ebed9189128a2a7bd250
|
[
"MIT"
] | null | null | null |
matrix_tree/day4.ipynb
|
666bury/dw_matrix-
|
429d676c9502bf1f12d6ebed9189128a2a7bd250
|
[
"MIT"
] | null | null | null |
matrix_tree/day4.ipynb
|
666bury/dw_matrix-
|
429d676c9502bf1f12d6ebed9189128a2a7bd250
|
[
"MIT"
] | null | null | null | 68,313 | 68,313 | 0.825085 |
[
[
[
"import pandas as pd\nimport numpy as np\n\nimport tensorflow as tf\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.layers import Conv2D, MaxPool2D, Dense, Flatten, Dropout\nfrom tensorflow.keras.utils import to_categorical\n\nimport os\nimport datetime\n\n%load_ext tensorboard\n\nimport matplotlib.pyplot as plt\nfrom skimage import color, exposure\n\nfrom sklearn.metrics import accuracy_score",
"_____no_output_____"
],
[
"cd '/content/drive/My Drive/Colab Notebooks/Matrix/dw_matrix-/matrix_tree'",
"/content/drive/My Drive/Colab Notebooks/Matrix/dw_matrix-/matrix_tree\n"
],
[
"train = pd.read_pickle('data/train.p')\ntest = pd.read_pickle('data/test.p')\n\nX_train, y_train = train['features'], train['labels']\nX_test, y_test = test['features'], test['labels']",
"_____no_output_____"
],
[
"y_train",
"_____no_output_____"
],
[
"len(np.unique(y_train))",
"_____no_output_____"
],
[
"to_categorical(y_train)[0]",
"_____no_output_____"
],
[
"np.unique(y_train)",
"_____no_output_____"
],
[
"if y_train.ndim == 1: y_train = to_categorical(y_train)\nif y_test.ndim == 1: y_test = to_categorical(y_test)",
"_____no_output_____"
],
[
"input_shape = X_train.shape[1:]\nnum_classes = y_train.shape[1]",
"_____no_output_____"
],
[
"def get_cnn_v1(input_shape, num_classes):\n return Sequential([\n Conv2D(filters=64, kernel_size=(3,3), activation='relu', input_shape=input_shape),\n Flatten(),\n Dense(num_classes, activation='softmax') \n ])\n\ndef train_model(model, X_train, y_train, params_fit={}):\n model.compile(loss='categorical_crossentropy', optimizer='Adam', metrics=['accuracy'])\n\n logdir = os.path.join('logs', datetime.datetime.now().strftime(\"%Y%m%d-%H%M%S\"))\n tensorboard_callback = tf.keras.callbacks.TensorBoard(logdir, histogram_freq=1)\n\n model.fit(X_train, \n y_train,\n batch_size=params_fit.get('batch_size', 128),\n epochs=params_fit.get('epochs', 5),\n verbose=params_fit.get('verbose', 1),\n validation_data=params_fit.get('validation_data', (X_train, y_train)),\n callbacks=[tensorboard_callback]\n )\n \n return model",
"_____no_output_____"
],
[
"model = get_cnn_v1(input_shape, num_classes)\nmodel_trained = train_model(model, X_train, y_train)",
"Epoch 1/5\n272/272 [==============================] - 7s 24ms/step - loss: 34.8265 - accuracy: 0.7571 - val_loss: 0.2518 - val_accuracy: 0.9411\nEpoch 2/5\n272/272 [==============================] - 6s 24ms/step - loss: 0.2217 - accuracy: 0.9501 - val_loss: 0.1226 - val_accuracy: 0.9724\nEpoch 3/5\n272/272 [==============================] - 6s 23ms/step - loss: 0.1421 - accuracy: 0.9695 - val_loss: 0.1157 - val_accuracy: 0.9747\nEpoch 4/5\n272/272 [==============================] - 6s 23ms/step - loss: 0.1510 - accuracy: 0.9684 - val_loss: 0.0760 - val_accuracy: 0.9842\nEpoch 5/5\n272/272 [==============================] - 6s 23ms/step - loss: 0.1149 - accuracy: 0.9747 - val_loss: 0.0717 - val_accuracy: 0.9830\n"
],
[
"y_pred_prob = model_trained.predict(X_test)\ny_pred_prob",
"_____no_output_____"
],
[
"y_pred_prob[400]",
"_____no_output_____"
],
[
"np.sum([1.8309039e-12, 8.3091247e-01, 3.9765782e-12, 1.9530295e-18,\n 1.6908756e-01, 1.4008565e-10, 8.5738119e-21, 2.2408531e-24,\n 5.4893188e-22, 1.8297317e-11, 8.1105787e-23, 6.6644010e-25,\n 3.4779175e-28, 1.8838944e-17, 2.9940773e-32, 3.4073123e-32,\n 3.2674885e-20, 1.8934992e-31, 2.8295588e-18, 3.7037542e-33,\n 1.0314749e-23, 8.9143100e-26, 1.9489775e-29, 6.2612767e-23,\n 4.4554421e-27, 3.4258839e-25, 3.1946413e-24, 5.3853781e-32,\n 5.1257782e-19, 1.5215109e-30, 9.1346340e-31, 2.7345416e-26,\n 7.5727543e-33, 8.0035479e-26, 0.0000000e+00, 1.3156333e-30,\n 0.0000000e+00, 6.8313645e-28, 3.5761218e-32, 1.6024535e-34,\n 1.2879560e-27, 2.0423062e-31, 0.0000000e+00])",
"_____no_output_____"
],
[
"df = pd.read_csv('data/signnames.csv')\nlabels_dict = df.to_dict()['b']",
"_____no_output_____"
],
[
"def predict(model_trained, X_test, y_test, scoring=accuracy_score):\n\n y_test_norm = np.argmax(y_test, axis=1)\n\n y_pred_prob = model_trained.predict(X_test)\n y_pred = np.argmax(y_pred_prob, axis=1)\n\n return scoring(y_test_norm, y_pred)",
"_____no_output_____"
],
[
"predict(model_trained, X_test, y_test)",
"_____no_output_____"
],
[
"def train_and_predict(model):\n model_trained = train_model(model, X_train, y_train)\n return predict(model_trained, X_test, y_test)",
"_____no_output_____"
],
[
"def get_cnn_v2(input_shape, num_classes):\n return Sequential([\n Conv2D(filters=32, kernel_size=(3,3), activation='relu', input_shape=input_shape),\n MaxPool2D(),\n Dropout(0.3),\n\n Conv2D(filters=64, kernel_size=(3,3), activation='relu'),\n MaxPool2D(),\n\n Flatten(),\n \n Dense(1024, activation='relu'),\n Dropout(0.3),\n\n Dense(num_classes, activation='softmax') \n ])\n\ntrain_and_predict(get_cnn_v2(input_shape, num_classes))",
"Epoch 1/5\n272/272 [==============================] - 6s 23ms/step - loss: 5.1678 - accuracy: 0.3870 - val_loss: 0.9546 - val_accuracy: 0.7200\nEpoch 2/5\n272/272 [==============================] - 6s 22ms/step - loss: 0.7689 - accuracy: 0.7739 - val_loss: 0.3025 - val_accuracy: 0.9345\nEpoch 3/5\n272/272 [==============================] - 6s 23ms/step - loss: 0.3962 - accuracy: 0.8862 - val_loss: 0.1311 - val_accuracy: 0.9713\nEpoch 4/5\n272/272 [==============================] - 6s 23ms/step - loss: 0.2593 - accuracy: 0.9238 - val_loss: 0.0882 - val_accuracy: 0.9783\nEpoch 5/5\n272/272 [==============================] - 6s 22ms/step - loss: 0.1873 - accuracy: 0.9467 - val_loss: 0.0576 - val_accuracy: 0.9863\n"
],
[
"def get_cnn_v3(input_shape, num_classes):\n return Sequential([\n Conv2D(filters=32, kernel_size=(3,3), activation='relu', input_shape=input_shape),\n Conv2D(filters=32, kernel_size=(3,3), activation='relu', input_shape=input_shape),\n MaxPool2D(),\n Dropout(0.3),\n\n Conv2D(filters=64, kernel_size=(3,3), activation='relu'),\n Conv2D(filters=64, kernel_size=(3,3), activation='relu'),\n MaxPool2D(),\n Dropout(0.3),\n\n Flatten(),\n \n Dense(1024, activation='relu'),\n Dropout(0.3),\n\n Dense(num_classes, activation='softmax') \n ])\n\ntrain_and_predict(get_cnn_v3(input_shape, num_classes))",
"Epoch 1/5\n272/272 [==============================] - 10s 37ms/step - loss: 2.0662 - accuracy: 0.5823 - val_loss: 0.2549 - val_accuracy: 0.9540\nEpoch 2/5\n272/272 [==============================] - 9s 35ms/step - loss: 0.4017 - accuracy: 0.8853 - val_loss: 0.0812 - val_accuracy: 0.9832\nEpoch 3/5\n272/272 [==============================] - 10s 35ms/step - loss: 0.2128 - accuracy: 0.9396 - val_loss: 0.0312 - val_accuracy: 0.9933\nEpoch 4/5\n272/272 [==============================] - 9s 35ms/step - loss: 0.1412 - accuracy: 0.9593 - val_loss: 0.0186 - val_accuracy: 0.9958\nEpoch 5/5\n272/272 [==============================] - 10s 35ms/step - loss: 0.1130 - accuracy: 0.9665 - val_loss: 0.0181 - val_accuracy: 0.9951\n"
],
[
"def get_cnn_v4(input_shape, num_classes):\n return Sequential([\n Conv2D(filters=32, kernel_size=(3,3), activation='relu', input_shape=input_shape),\n Conv2D(filters=32, kernel_size=(3,3), activation='relu', padding='same'),\n MaxPool2D(),\n Dropout(0.3),\n\n Conv2D(filters=64, kernel_size=(3,3), activation='relu', padding='same'),\n Conv2D(filters=64, kernel_size=(3,3), activation='relu'),\n MaxPool2D(),\n Dropout(0.3),\n\n Conv2D(filters=64, kernel_size=(3,3), activation='relu', padding='same'),\n Conv2D(filters=64, kernel_size=(3,3), activation='relu'),\n MaxPool2D(),\n Dropout(0.3),\n\n Flatten(),\n \n Dense(1024, activation='relu'),\n Dropout(0.3),\n\n Dense(num_classes, activation='softmax') \n ])\n\n# get_cnn_v4(input_shape, num_classes).summary()\ntrain_and_predict(get_cnn_v4(input_shape, num_classes))",
"Epoch 1/5\n272/272 [==============================] - 12s 44ms/step - loss: 2.5766 - accuracy: 0.3089 - val_loss: 0.7375 - val_accuracy: 0.8151\nEpoch 2/5\n272/272 [==============================] - 11s 42ms/step - loss: 0.6774 - accuracy: 0.7965 - val_loss: 0.1631 - val_accuracy: 0.9684\nEpoch 3/5\n272/272 [==============================] - 11s 42ms/step - loss: 0.2896 - accuracy: 0.9140 - val_loss: 0.0701 - val_accuracy: 0.9826\nEpoch 4/5\n272/272 [==============================] - 11s 42ms/step - loss: 0.1908 - accuracy: 0.9443 - val_loss: 0.0460 - val_accuracy: 0.9874\nEpoch 5/5\n272/272 [==============================] - 11s 42ms/step - loss: 0.1553 - accuracy: 0.9550 - val_loss: 0.0304 - val_accuracy: 0.9912\n"
],
[
"def get_cnn_v5(input_shape, num_classes):\n return Sequential([\n Conv2D(filters=32, kernel_size=(3,3), activation='relu', input_shape=input_shape),\n Conv2D(filters=32, kernel_size=(3,3), activation='relu', padding='same'),\n MaxPool2D(),\n Dropout(0.3),\n\n Conv2D(filters=64, kernel_size=(3,3), activation='relu', padding='same'),\n Conv2D(filters=64, kernel_size=(3,3), activation='relu'),\n MaxPool2D(),\n Dropout(0.3),\n\n Conv2D(filters=64, kernel_size=(3,3), activation='relu', padding='same'),\n Conv2D(filters=64, kernel_size=(3,3), activation='relu'),\n MaxPool2D(),\n Dropout(0.3),\n\n Flatten(),\n \n Dense(1024, activation='relu'),\n Dropout(0.3),\n\n Dense(1024, activation='relu'),\n Dropout(0.3),\n\n Dense(num_classes, activation='softmax') \n ])\n\n# get_cnn_v4(input_shape, num_classes).summary()\ntrain_and_predict(get_cnn_v5(input_shape, num_classes))",
"Epoch 1/5\n272/272 [==============================] - 13s 46ms/step - loss: 2.4799 - accuracy: 0.3028 - val_loss: 0.9428 - val_accuracy: 0.6848\nEpoch 2/5\n272/272 [==============================] - 12s 45ms/step - loss: 0.8827 - accuracy: 0.7146 - val_loss: 0.2386 - val_accuracy: 0.9324\nEpoch 3/5\n272/272 [==============================] - 12s 44ms/step - loss: 0.4470 - accuracy: 0.8597 - val_loss: 0.0997 - val_accuracy: 0.9709\nEpoch 4/5\n272/272 [==============================] - 12s 44ms/step - loss: 0.2733 - accuracy: 0.9160 - val_loss: 0.0658 - val_accuracy: 0.9807\nEpoch 5/5\n272/272 [==============================] - 12s 44ms/step - loss: 0.2195 - accuracy: 0.9335 - val_loss: 0.0572 - val_accuracy: 0.9833\n"
],
[
"X_train_gray = color.rgb2gray(X_train).reshape(-1, 32, 32, 1)\nX_test_gray = color.rgb2gray(X_test).reshape(-1, 32, 32, 1)",
"_____no_output_____"
],
[
"model = get_cnn_v5((32,32,1), num_classes)\nmodel_trained = train_model(model, X_train_gray, y_train, params_fit={})\npredict(model_trained, X_test_gray, y_test, scoring=accuracy_score)\n# model.compile(loss='category_crosse')",
"Epoch 1/5\n272/272 [==============================] - 12s 46ms/step - loss: 2.6317 - accuracy: 0.2575 - val_loss: 0.7301 - val_accuracy: 0.7656\nEpoch 2/5\n272/272 [==============================] - 12s 44ms/step - loss: 0.5906 - accuracy: 0.8090 - val_loss: 0.1707 - val_accuracy: 0.9514\nEpoch 3/5\n272/272 [==============================] - 12s 44ms/step - loss: 0.2622 - accuracy: 0.9170 - val_loss: 0.0601 - val_accuracy: 0.9815\nEpoch 4/5\n272/272 [==============================] - 12s 44ms/step - loss: 0.1671 - accuracy: 0.9467 - val_loss: 0.0443 - val_accuracy: 0.9871\nEpoch 5/5\n272/272 [==============================] - 12s 45ms/step - loss: 0.1307 - accuracy: 0.9588 - val_loss: 0.0305 - val_accuracy: 0.9917\n"
],
[
"plt.imshow(color.rgb2gray(X_train[0]), cmap=plt.get_cmap('gray'))",
"_____no_output_____"
],
[
"X_train[0].shape",
"_____no_output_____"
],
[
"color.rgb2gray(X_train[0]).shape",
"_____no_output_____"
],
[
"def preproc_img(img):\n hsv = color.rgb2hsv(img)\n hsv[:, :, 2] = exposure.equalize_adapthist(hsv[:, :, 2])\n img = color.hsv2rgb(hsv)\n\n return img",
"_____no_output_____"
],
[
"plt.imshow(preproc_img(X_train[400]))",
"_____no_output_____"
],
[
"plt.imshow(X_train[400])",
"_____no_output_____"
],
[
"labels_dict[ np.argmax(y_pred_prob[400]) ]",
"_____no_output_____"
],
[
"plt.imshow(X_test[400])",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbaba1f260081b36c8ec043e7ee44d91528e8ebd
| 337,848 |
ipynb
|
Jupyter Notebook
|
docs/notebooks/Grammars.ipynb
|
leonbett/fuzzingbook
|
fbb46c825306b4b79c67b9375393b42a1283c680
|
[
"MIT"
] | 1 |
2022-02-09T22:01:26.000Z
|
2022-02-09T22:01:26.000Z
|
docs/notebooks/Grammars.ipynb
|
leonbett/fuzzingbook
|
fbb46c825306b4b79c67b9375393b42a1283c680
|
[
"MIT"
] | null | null | null |
docs/notebooks/Grammars.ipynb
|
leonbett/fuzzingbook
|
fbb46c825306b4b79c67b9375393b42a1283c680
|
[
"MIT"
] | null | null | null | 38.110321 | 757 | 0.508519 |
[
[
[
"# Fuzzing with Grammars\n\nIn the chapter on [\"Mutation-Based Fuzzing\"](MutationFuzzer.ipynb), we have seen how to use extra hints – such as sample input files – to speed up test generation. In this chapter, we take this idea one step further, by providing a _specification_ of the legal inputs to a program. Specifying inputs via a _grammar_ allows for very systematic and efficient test generation, in particular for complex input formats. Grammars also serve as the base for configuration fuzzing, API fuzzing, GUI fuzzing, and many more.",
"_____no_output_____"
]
],
[
[
"from bookutils import YouTubeVideo\nYouTubeVideo('mswyS3Wok1c')",
"_____no_output_____"
]
],
[
[
"**Prerequisites**\n\n* You should know how basic fuzzing works, e.g. from the [Chapter introducing fuzzing](Fuzzer.ipynb).\n* Knowledge on [mutation-based fuzzing](MutationFuzzer.ipynb) and [coverage](Coverage.ipynb) is _not_ required yet, but still recommended.",
"_____no_output_____"
]
],
[
[
"import bookutils",
"_____no_output_____"
],
[
"from typing import List, Dict, Union, Any, Tuple, Optional",
"_____no_output_____"
],
[
"import Fuzzer",
"_____no_output_____"
]
],
[
[
"## Synopsis\n<!-- Automatically generated. Do not edit. -->\n\nTo [use the code provided in this chapter](Importing.ipynb), write\n\n```python\n>>> from fuzzingbook.Grammars import <identifier>\n```\n\nand then make use of the following features.\n\n\nThis chapter introduces _grammars_ as a simple means to specify input languages, and to use them for testing programs with syntactically valid inputs. A grammar is defined as a mapping of nonterminal symbols to lists of alternative expansions, as in the following example:\n\n```python\n>>> US_PHONE_GRAMMAR: Grammar = {\n>>> \"<start>\": [\"<phone-number>\"],\n>>> \"<phone-number>\": [\"(<area>)<exchange>-<line>\"],\n>>> \"<area>\": [\"<lead-digit><digit><digit>\"],\n>>> \"<exchange>\": [\"<lead-digit><digit><digit>\"],\n>>> \"<line>\": [\"<digit><digit><digit><digit>\"],\n>>> \"<lead-digit>\": [\"2\", \"3\", \"4\", \"5\", \"6\", \"7\", \"8\", \"9\"],\n>>> \"<digit>\": [\"0\", \"1\", \"2\", \"3\", \"4\", \"5\", \"6\", \"7\", \"8\", \"9\"]\n>>> }\n>>> \n>>> assert is_valid_grammar(US_PHONE_GRAMMAR)\n```\nNonterminal symbols are enclosed in angle brackets (say, `<digit>`). To generate an input string from a grammar, a _producer_ starts with the start symbol (`<start>`) and randomly chooses a random expansion for this symbol. It continues the process until all nonterminal symbols are expanded. The function `simple_grammar_fuzzer()` does just that:\n\n```python\n>>> [simple_grammar_fuzzer(US_PHONE_GRAMMAR) for i in range(5)]\n['(692)449-5179',\n '(519)230-7422',\n '(613)761-0853',\n '(979)881-3858',\n '(810)914-5475']\n```\nIn practice, though, instead of `simple_grammar_fuzzer()`, you should use [the `GrammarFuzzer` class](GrammarFuzzer.ipynb) or one of its [coverage-based](GrammarCoverageFuzzer.ipynb), [probabilistic-based](ProbabilisticGrammarFuzzer.ipynb), or [generator-based](GeneratorGrammarFuzzer.ipynb) derivatives; these are more efficient, protect against infinite growth, and provide several additional features.\n\nThis chapter also introduces a [grammar toolbox](#A-Grammar-Toolbox) with several helper functions that ease the writing of grammars, such as using shortcut notations for character classes and repetitions, or extending grammars \n\n",
"_____no_output_____"
],
[
"## Input Languages\n\nAll possible behaviors of a program can be triggered by its input. \"Input\" here can be a wide range of possible sources: We are talking about data that is read from files, from the environment, or over the network, data input by the user, or data acquired from interaction with other resources. The set of all these inputs determines how the program will behave – including its failures. When testing, it is thus very helpful to think about possible input sources, how to get them under control, and _how to systematically test them_.",
"_____no_output_____"
],
[
"For the sake of simplicity, we will assume for now that the program has only one source of inputs; this is the same assumption we have been using in the previous chapters, too. The set of valid inputs to a program is called a _language_. Languages range from the simple to the complex: the CSV language denotes the set of valid comma-separated inputs, whereas the Python language denotes the set of valid Python programs. We commonly separate data languages and programming languages, although any program can also be treated as input data (say, to a compiler). The [Wikipedia page on file formats](https://en.wikipedia.org/wiki/List_of_file_formats) lists more than 1,000 different file formats, each of which is its own language.",
"_____no_output_____"
],
[
"To formally describe languages, the field of *formal languages* has devised a number of *language specifications* that describe a language. *Regular expressions* represent the simplest class of these languages to denote sets of strings: The regular expression `[a-z]*`, for instance, denotes a (possibly empty) sequence of lowercase letters. *Automata theory* connects these languages to automata that accept these inputs; *finite state machines*, for instance, can be used to specify the language of regular expressions.",
"_____no_output_____"
],
[
"Regular expressions are great for not-too-complex input formats, and the associated finite state machines have many properties that make them great for reasoning. To specify more complex inputs, though, they quickly encounter limitations. At the other end of the language spectrum, we have *universal grammars* that denote the language accepted by *Turing machines*. A Turing machine can compute anything that can be computed; and with Python being Turing-complete, this means that we can also use a Python program $p$ to specify or even enumerate legal inputs. But then, computer science theory also tells us that each such testing program has to be written specifically for the program to be tested, which is not the level of automation we want.",
"_____no_output_____"
],
[
"## Grammars\n\nThe middle ground between regular expressions and Turing machines is covered by *grammars*. Grammars are among the most popular (and best understood) formalisms to formally specify input languages. Using a grammar, one can express a wide range of the properties of an input language. Grammars are particularly great for expressing the *syntactical structure* of an input, and are the formalism of choice to express nested or recursive inputs. The grammars we use are so-called *context-free grammars*, one of the easiest and most popular grammar formalisms.",
"_____no_output_____"
],
[
"### Rules and Expansions\n\nA grammar consists of a *start symbol* and a set of *expansion rules* (or simply *rules*) which indicate how the start symbol (and other symbols) can be expanded. As an example, consider the following grammar, denoting a sequence of two digits:\n\n```\n<start> ::= <digit><digit>\n<digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9\n```\n\nTo read such a grammar, start with the start symbol (`<start>`). An expansion rule `<A> ::= <B>` means that the symbol on the left side (`<A>`) can be replaced by the string on the right side (`<B>`). In the above grammar, `<start>` would be replaced by `<digit><digit>`.\n\nIn this string again, `<digit>` would be replaced by the string on the right side of the `<digit>` rule. The special operator `|` denotes *expansion alternatives* (or simply *alternatives*), meaning that any of the digits can be chosen for an expansion. Each `<digit>` thus would be expanded into one of the given digits, eventually yielding a string between `00` and `99`. There are no further expansions for `0` to `9`, so we are all set.",
"_____no_output_____"
],
[
"The interesting thing about grammars is that they can be *recursive*. That is, expansions can make use of symbols expanded earlier – which would then be expanded again. As an example, consider a grammar that describes integers:\n\n```\n<start> ::= <integer>\n<integer> ::= <digit> | <digit><integer>\n<digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9\n```\n\nHere, a `<integer>` is either a single digit, or a digit followed by another integer. The number `1234` thus would be represented as a single digit `1`, followed by the integer `234`, which in turn is a digit `2`, followed by the integer `34`.",
"_____no_output_____"
],
[
"If we wanted to express that an integer can be preceded by a sign (`+` or `-`), we would write the grammar as\n\n```\n<start> ::= <number>\n<number> ::= <integer> | +<integer> | -<integer>\n<integer> ::= <digit> | <digit><integer>\n<digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9\n```\n\nThese rules formally define the language: Anything that can be derived from the start symbol is part of the language; anything that cannot is not.",
"_____no_output_____"
]
],
[
[
"from bookutils import quiz",
"_____no_output_____"
],
[
"quiz(\"Which of these strings cannot be produced \"\n \"from the above `<start>` symbol?\",\n [\n \"`007`\",\n \"`-42`\",\n \"`++1`\",\n \"`3.14`\"\n ], \"[27 ** (1/3), 256 ** (1/4)]\")",
"_____no_output_____"
]
],
[
[
"### Arithmetic Expressions\n\nLet us expand our grammar to cover full *arithmetic expressions* – a poster child example for a grammar. We see that an expression (`<expr>`) is either a sum, or a difference, or a term; a term is either a product or a division, or a factor; and a factor is either a number or a parenthesized expression. Almost all rules can have recursion, and thus allow arbitrary complex expressions such as `(1 + 2) * (3.4 / 5.6 - 789)`.\n\n```\n<start> ::= <expr>\n<expr> ::= <term> + <expr> | <term> - <expr> | <term>\n<term> ::= <term> * <factor> | <term> / <factor> | <factor>\n<factor> ::= +<factor> | -<factor> | (<expr>) | <integer> | <integer>.<integer>\n<integer> ::= <digit><integer> | <digit>\n<digit> ::= 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9\n```\n\nIn such a grammar, if we start with `<start>` and then expand one symbol after another, randomly choosing alternatives, we can quickly produce one valid arithmetic expression after another. Such *grammar fuzzing* is highly effective as it comes to produce complex inputs, and this is what we will implement in this chapter.",
"_____no_output_____"
]
],
[
[
"quiz(\"Which of these strings cannot be produced \"\n \"from the above `<start>` symbol?\",\n [\n \"`1 + 1`\",\n \"`1+1`\",\n \"`+1`\",\n \"`+(1)`\",\n ], \"4 ** 0.5\")",
"_____no_output_____"
]
],
[
[
"## Representing Grammars in Python\n\nOur first step in building a grammar fuzzer is to find an appropriate format for grammars. To make the writing of grammars as simple as possible, we use a format that is based on strings and lists. Our grammars in Python take the format of a _mapping_ between symbol names and expansions, where expansions are _lists_ of alternatives. A one-rule grammar for digits thus takes the form",
"_____no_output_____"
]
],
[
[
"DIGIT_GRAMMAR = {\n \"<start>\":\n [\"0\", \"1\", \"2\", \"3\", \"4\", \"5\", \"6\", \"7\", \"8\", \"9\"]\n}",
"_____no_output_____"
]
],
[
[
"### Excursion: A `Grammar` Type",
"_____no_output_____"
],
[
"Let us define a type for grammars, such that we can check grammar types statically. ",
"_____no_output_____"
],
[
"A first attempt at a grammar type would be that each symbol (a string) is mapped to a list of expansions (strings):",
"_____no_output_____"
]
],
[
[
"SimpleGrammar = Dict[str, List[str]]",
"_____no_output_____"
]
],
[
[
"However, our `opts()` feature for adding optional attributes, which we will introduce later in this chapter, also allows expansions to be _pairs_ that consist of strings and options, where options are mappings of strings to values:",
"_____no_output_____"
]
],
[
[
"Option = Dict[str, Any]",
"_____no_output_____"
]
],
[
[
"Hence, an expansion is either a string – or a pair of a string and an option.",
"_____no_output_____"
]
],
[
[
"Expansion = Union[str, Tuple[str, Option]]",
"_____no_output_____"
]
],
[
[
"With this, we can now define a `Grammar` as a mapping of strings to `Expansion` lists.",
"_____no_output_____"
],
[
"### End of Excursion",
"_____no_output_____"
],
[
"We can capture the grammar structure in a _`Grammar`_ type, in which each symbol (a string) is mapped to a list of expansions (strings):",
"_____no_output_____"
]
],
[
[
"Grammar = Dict[str, List[Expansion]]",
"_____no_output_____"
]
],
[
[
"With this `Grammar` type, the full grammar for arithmetic expressions looks like this:",
"_____no_output_____"
]
],
[
[
"EXPR_GRAMMAR: Grammar = {\n \"<start>\":\n [\"<expr>\"],\n\n \"<expr>\":\n [\"<term> + <expr>\", \"<term> - <expr>\", \"<term>\"],\n\n \"<term>\":\n [\"<factor> * <term>\", \"<factor> / <term>\", \"<factor>\"],\n\n \"<factor>\":\n [\"+<factor>\",\n \"-<factor>\",\n \"(<expr>)\",\n \"<integer>.<integer>\",\n \"<integer>\"],\n\n \"<integer>\":\n [\"<digit><integer>\", \"<digit>\"],\n\n \"<digit>\":\n [\"0\", \"1\", \"2\", \"3\", \"4\", \"5\", \"6\", \"7\", \"8\", \"9\"]\n}",
"_____no_output_____"
]
],
[
[
"In the grammar, every symbol can be defined exactly once. We can access any rule by its symbol...",
"_____no_output_____"
]
],
[
[
"EXPR_GRAMMAR[\"<digit>\"]",
"_____no_output_____"
]
],
[
[
"....and we can check whether a symbol is in the grammar:",
"_____no_output_____"
]
],
[
[
"\"<identifier>\" in EXPR_GRAMMAR",
"_____no_output_____"
]
],
[
[
"Note that we assume that on the left hand side of a rule (i.e., the key in the mapping) is always a single symbol. This is the property that gives our grammars the characterization of _context-free_.",
"_____no_output_____"
],
[
"## Some Definitions",
"_____no_output_____"
],
[
"We assume that the canonical start symbol is `<start>`:",
"_____no_output_____"
]
],
[
[
"START_SYMBOL = \"<start>\"",
"_____no_output_____"
]
],
[
[
"The handy `nonterminals()` function extracts the list of nonterminal symbols (i.e., anything between `<` and `>`, except spaces) from an expansion.",
"_____no_output_____"
]
],
[
[
"import re",
"_____no_output_____"
],
[
"RE_NONTERMINAL = re.compile(r'(<[^<> ]*>)')",
"_____no_output_____"
],
[
"def nonterminals(expansion):\n # In later chapters, we allow expansions to be tuples,\n # with the expansion being the first element\n if isinstance(expansion, tuple):\n expansion = expansion[0]\n\n return RE_NONTERMINAL.findall(expansion)",
"_____no_output_____"
],
[
"assert nonterminals(\"<term> * <factor>\") == [\"<term>\", \"<factor>\"]\nassert nonterminals(\"<digit><integer>\") == [\"<digit>\", \"<integer>\"]\nassert nonterminals(\"1 < 3 > 2\") == []\nassert nonterminals(\"1 <3> 2\") == [\"<3>\"]\nassert nonterminals(\"1 + 2\") == []\nassert nonterminals((\"<1>\", {'option': 'value'})) == [\"<1>\"]",
"_____no_output_____"
]
],
[
[
"Likewise, `is_nonterminal()` checks whether some symbol is a nonterminal:",
"_____no_output_____"
]
],
[
[
"def is_nonterminal(s):\n return RE_NONTERMINAL.match(s)",
"_____no_output_____"
],
[
"assert is_nonterminal(\"<abc>\")\nassert is_nonterminal(\"<symbol-1>\")\nassert not is_nonterminal(\"+\")",
"_____no_output_____"
]
],
[
[
"## A Simple Grammar Fuzzer\n\nLet us now put the above grammars to use. We will build a very simple grammar fuzzer that starts with a start symbol (`<start>`) and then keeps on expanding it. To avoid expansion to infinite inputs, we place a limit (`max_nonterminals`) on the number of nonterminals. Furthermore, to avoid being stuck in a situation where we cannot reduce the number of symbols any further, we also limit the total number of expansion steps.",
"_____no_output_____"
]
],
[
[
"import random",
"_____no_output_____"
],
[
"class ExpansionError(Exception):\n pass",
"_____no_output_____"
],
[
"def simple_grammar_fuzzer(grammar: Grammar, \n start_symbol: str = START_SYMBOL,\n max_nonterminals: int = 10,\n max_expansion_trials: int = 100,\n log: bool = False) -> str:\n \"\"\"Produce a string from `grammar`.\n `start_symbol`: use a start symbol other than `<start>` (default).\n `max_nonterminals`: the maximum number of nonterminals \n still left for expansion\n `max_expansion_trials`: maximum # of attempts to produce a string\n `log`: print expansion progress if True\"\"\"\n\n term = start_symbol\n expansion_trials = 0\n\n while len(nonterminals(term)) > 0:\n symbol_to_expand = random.choice(nonterminals(term))\n expansions = grammar[symbol_to_expand]\n expansion = random.choice(expansions)\n # In later chapters, we allow expansions to be tuples,\n # with the expansion being the first element\n if isinstance(expansion, tuple):\n expansion = expansion[0]\n\n new_term = term.replace(symbol_to_expand, expansion, 1)\n\n if len(nonterminals(new_term)) < max_nonterminals:\n term = new_term\n if log:\n print(\"%-40s\" % (symbol_to_expand + \" -> \" + expansion), term)\n expansion_trials = 0\n else:\n expansion_trials += 1\n if expansion_trials >= max_expansion_trials:\n raise ExpansionError(\"Cannot expand \" + repr(term))\n\n return term",
"_____no_output_____"
]
],
[
[
"Let us see how this simple grammar fuzzer obtains an arithmetic expression from the start symbol:",
"_____no_output_____"
]
],
[
[
"simple_grammar_fuzzer(grammar=EXPR_GRAMMAR, max_nonterminals=3, log=True)",
"<start> -> <expr> <expr>\n<expr> -> <term> + <expr> <term> + <expr>\n<term> -> <factor> <factor> + <expr>\n<factor> -> <integer> <integer> + <expr>\n<integer> -> <digit> <digit> + <expr>\n<digit> -> 6 6 + <expr>\n<expr> -> <term> - <expr> 6 + <term> - <expr>\n<expr> -> <term> 6 + <term> - <term>\n<term> -> <factor> 6 + <factor> - <term>\n<factor> -> -<factor> 6 + -<factor> - <term>\n<term> -> <factor> 6 + -<factor> - <factor>\n<factor> -> (<expr>) 6 + -(<expr>) - <factor>\n<factor> -> (<expr>) 6 + -(<expr>) - (<expr>)\n<expr> -> <term> 6 + -(<term>) - (<expr>)\n<expr> -> <term> 6 + -(<term>) - (<term>)\n<term> -> <factor> 6 + -(<factor>) - (<term>)\n<factor> -> +<factor> 6 + -(+<factor>) - (<term>)\n<factor> -> +<factor> 6 + -(++<factor>) - (<term>)\n<term> -> <factor> 6 + -(++<factor>) - (<factor>)\n<factor> -> (<expr>) 6 + -(++(<expr>)) - (<factor>)\n<factor> -> <integer> 6 + -(++(<expr>)) - (<integer>)\n<expr> -> <term> 6 + -(++(<term>)) - (<integer>)\n<integer> -> <digit> 6 + -(++(<term>)) - (<digit>)\n<digit> -> 9 6 + -(++(<term>)) - (9)\n<term> -> <factor> * <term> 6 + -(++(<factor> * <term>)) - (9)\n<term> -> <factor> 6 + -(++(<factor> * <factor>)) - (9)\n<factor> -> <integer> 6 + -(++(<integer> * <factor>)) - (9)\n<integer> -> <digit> 6 + -(++(<digit> * <factor>)) - (9)\n<digit> -> 2 6 + -(++(2 * <factor>)) - (9)\n<factor> -> +<factor> 6 + -(++(2 * +<factor>)) - (9)\n<factor> -> -<factor> 6 + -(++(2 * +-<factor>)) - (9)\n<factor> -> -<factor> 6 + -(++(2 * +--<factor>)) - (9)\n<factor> -> -<factor> 6 + -(++(2 * +---<factor>)) - (9)\n<factor> -> -<factor> 6 + -(++(2 * +----<factor>)) - (9)\n<factor> -> <integer>.<integer> 6 + -(++(2 * +----<integer>.<integer>)) - (9)\n<integer> -> <digit> 6 + -(++(2 * +----<digit>.<integer>)) - (9)\n<integer> -> <digit> 6 + -(++(2 * +----<digit>.<digit>)) - (9)\n<digit> -> 1 6 + -(++(2 * +----1.<digit>)) - (9)\n<digit> -> 7 6 + -(++(2 * +----1.7)) - (9)\n"
]
],
[
[
"By increasing the limit of nonterminals, we can quickly get much longer productions:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(simple_grammar_fuzzer(grammar=EXPR_GRAMMAR, max_nonterminals=5))",
"7 / +48.5\n-5.9 / 9 - 4 * +-(-+++((1 + (+7 - (-1 * (++-+7.7 - -+-4.0))))) * +--4 - -(6) + 64)\n8.2 - 27 - -9 / +((+9 * --2 + --+-+-((-1 * +(8 - 5 - 6)) * (-((-+(((+(4))))) - ++4) / +(-+---((5.6 - --(3 * -1.8 * +(6 * +-(((-(-6) * ---+6)) / +--(+-+-7 * (-0 * (+(((((2)) + 8 - 3 - ++9.0 + ---(--+7 / (1 / +++6.37) + (1) / 482) / +++-+0)))) * -+5 + 7.513)))) - (+1 / ++((-84)))))))) * ++5 / +-(--2 - -++-9.0)))) / 5 * --++090\n1 - -3 * 7 - 28 / 9\n(+9) * +-5 * ++-926.2 - (+9.03 / -+(-(-6) / 2 * +(-+--(8) / -(+1.0) - 5 + 4)) * 3.5)\n8 + -(9.6 - 3 - -+-4 * +77)\n-(((((++((((+((++++-((+-37))))))))))))) / ++(-(+++(+6)) * -++-(+(++(---6 * (((7)) * (1) / (-7.6 * 535338) + +256) * 0) * 0))) - 4 + +1\n5.43\n(9 / -405 / -23 - +-((+-(2 * (13))))) + +6 - +8 - 934\n-++2 - (--+715769550) / 8 / (1)\n"
]
],
[
[
"Note that while fuzzer does the job in most cases, it has a number of drawbacks.",
"_____no_output_____"
]
],
[
[
"quiz(\"What drawbacks does `simple_grammar_fuzzer()` have?\",\n [\n \"It has a large number of string search and replace operations\",\n \"It may fail to produce a string (`ExpansionError`)\",\n \"It often picks some symbol to expand \"\n \"that does not even occur in the string\",\n \"All of the above\"\n ], \"1 << 2\")",
"_____no_output_____"
]
],
[
[
"Indeed, `simple_grammar_fuzzer()` is rather inefficient due to the large number of search and replace operations, and it may even fail to produce a string. On the other hand, the implementation is straightforward and does the job in most cases. For this chapter, we'll stick to it; in the [next chapter](GrammarFuzzer.ipynb), we'll show how to build a more efficient one.",
"_____no_output_____"
],
[
"## Visualizing Grammars as Railroad Diagrams",
"_____no_output_____"
],
[
"With grammars, we can easily specify the format for several of the examples we discussed earlier. The above arithmetic expressions, for instance, can be directly sent into `bc` (or any other program that takes arithmetic expressions). Before we introduce a few additional grammars, let us give a means to _visualize_ them, giving an alternate view to aid their understanding.",
"_____no_output_____"
],
[
"_Railroad diagrams_, also called _syntax diagrams_, are a graphical representation of context-free grammars. They are read left to right, following possible \"rail\" tracks; the sequence of symbols encountered on the track defines the language. To produce railroad diagrams, we implement a function `syntax_diagram()`.",
"_____no_output_____"
],
[
"### Excursion: Implementing `syntax_diagram()`",
"_____no_output_____"
],
[
"We use [RailroadDiagrams](RailroadDiagrams.ipynb), an external library for visualization.",
"_____no_output_____"
]
],
[
[
"from RailroadDiagrams import NonTerminal, Terminal, Choice, HorizontalChoice, Sequence\nfrom RailroadDiagrams import show_diagram",
"_____no_output_____"
],
[
"from IPython.display import SVG",
"_____no_output_____"
]
],
[
[
"We first define the method `syntax_diagram_symbol()` to visualize a given symbol. Terminal symbols are denoted as ovals, whereas nonterminal symbols (such as `<term>`) are denoted as rectangles.",
"_____no_output_____"
]
],
[
[
"def syntax_diagram_symbol(symbol: str) -> Any:\n if is_nonterminal(symbol):\n return NonTerminal(symbol[1:-1])\n else:\n return Terminal(symbol)",
"_____no_output_____"
],
[
"SVG(show_diagram(syntax_diagram_symbol('<term>')))",
"_____no_output_____"
]
],
[
[
"We define `syntax_diagram_expr()` to visualize expansion alternatives.",
"_____no_output_____"
]
],
[
[
"def syntax_diagram_expr(expansion: Expansion) -> Any:\n # In later chapters, we allow expansions to be tuples,\n # with the expansion being the first element\n if isinstance(expansion, tuple):\n expansion = expansion[0]\n\n symbols = [sym for sym in re.split(RE_NONTERMINAL, expansion) if sym != \"\"]\n if len(symbols) == 0:\n symbols = [\"\"] # special case: empty expansion\n\n return Sequence(*[syntax_diagram_symbol(sym) for sym in symbols])",
"_____no_output_____"
],
[
"SVG(show_diagram(syntax_diagram_expr(EXPR_GRAMMAR['<term>'][0])))",
"_____no_output_____"
]
],
[
[
"This is the first alternative of `<term>` – a `<factor>` followed by `*` and a `<term>`.",
"_____no_output_____"
],
[
"Next, we define `syntax_diagram_alt()` for displaying alternate expressions.",
"_____no_output_____"
]
],
[
[
"from itertools import zip_longest",
"_____no_output_____"
],
[
"def syntax_diagram_alt(alt: List[Expansion]) -> Any:\n max_len = 5\n alt_len = len(alt)\n if alt_len > max_len:\n iter_len = alt_len // max_len\n alts = list(zip_longest(*[alt[i::iter_len] for i in range(iter_len)]))\n exprs = [[syntax_diagram_expr(expr) for expr in alt\n if expr is not None] for alt in alts]\n choices = [Choice(len(expr) // 2, *expr) for expr in exprs]\n return HorizontalChoice(*choices)\n else:\n return Choice(alt_len // 2, *[syntax_diagram_expr(expr) for expr in alt])",
"_____no_output_____"
],
[
"SVG(show_diagram(syntax_diagram_alt(EXPR_GRAMMAR['<digit>'])))",
"_____no_output_____"
]
],
[
[
"We see that a `<digit>` can be any single digit from `0` to `9`.",
"_____no_output_____"
],
[
"Finally, we define `syntax_diagram()` which given a grammar, displays the syntax diagram of its rules.",
"_____no_output_____"
]
],
[
[
"def syntax_diagram(grammar: Grammar) -> None:\n from IPython.display import SVG, display\n\n for key in grammar:\n print(\"%s\" % key[1:-1])\n display(SVG(show_diagram(syntax_diagram_alt(grammar[key]))))",
"_____no_output_____"
]
],
[
[
"### End of Excursion",
"_____no_output_____"
],
[
"Let us use `syntax_diagram()` to produce a railroad diagram of our expression grammar:",
"_____no_output_____"
]
],
[
[
"syntax_diagram(EXPR_GRAMMAR)",
"start\n"
]
],
[
[
"This railroad representation will come in handy as it comes to visualizing the structure of grammars – especially for more complex grammars.",
"_____no_output_____"
],
[
"## Some Grammars\n\nLet us create (and visualize) some more grammars and use them for fuzzing.",
"_____no_output_____"
],
[
"### A CGI Grammar\n\nHere's a grammar for `cgi_decode()` introduced in the [chapter on coverage](Coverage.ipynb).",
"_____no_output_____"
]
],
[
[
"CGI_GRAMMAR: Grammar = {\n \"<start>\":\n [\"<string>\"],\n\n \"<string>\":\n [\"<letter>\", \"<letter><string>\"],\n\n \"<letter>\":\n [\"<plus>\", \"<percent>\", \"<other>\"],\n\n \"<plus>\":\n [\"+\"],\n\n \"<percent>\":\n [\"%<hexdigit><hexdigit>\"],\n\n \"<hexdigit>\":\n [\"0\", \"1\", \"2\", \"3\", \"4\", \"5\", \"6\", \"7\",\n \"8\", \"9\", \"a\", \"b\", \"c\", \"d\", \"e\", \"f\"],\n\n \"<other>\": # Actually, could be _all_ letters\n [\"0\", \"1\", \"2\", \"3\", \"4\", \"5\", \"a\", \"b\", \"c\", \"d\", \"e\", \"-\", \"_\"],\n}",
"_____no_output_____"
],
[
"syntax_diagram(CGI_GRAMMAR)",
"start\n"
]
],
[
[
"In contrast to [basic fuzzing](Fuzzer.ipynb) or [mutation-based fuzzing](MutationFuzzer.ipynb), the grammar quickly produces all sorts of combinations:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(simple_grammar_fuzzer(grammar=CGI_GRAMMAR, max_nonterminals=10))",
"+%9a\n+++%ce+\n+_\n+%c6c\n++\n+%cd+5\n1%ee\n%b9%d5\n%96\n%57d%42\n"
]
],
[
[
"### A URL Grammar\n\nThe same properties we have seen for CGI input also hold for more complex inputs. Let us use a grammar to produce a large number of valid URLs:",
"_____no_output_____"
]
],
[
[
"URL_GRAMMAR: Grammar = {\n \"<start>\":\n [\"<url>\"],\n \"<url>\":\n [\"<scheme>://<authority><path><query>\"],\n \"<scheme>\":\n [\"http\", \"https\", \"ftp\", \"ftps\"],\n \"<authority>\":\n [\"<host>\", \"<host>:<port>\", \"<userinfo>@<host>\", \"<userinfo>@<host>:<port>\"],\n \"<host>\": # Just a few\n [\"cispa.saarland\", \"www.google.com\", \"fuzzingbook.com\"],\n \"<port>\":\n [\"80\", \"8080\", \"<nat>\"],\n \"<nat>\":\n [\"<digit>\", \"<digit><digit>\"],\n \"<digit>\":\n [\"0\", \"1\", \"2\", \"3\", \"4\", \"5\", \"6\", \"7\", \"8\", \"9\"],\n \"<userinfo>\": # Just one\n [\"user:password\"],\n \"<path>\": # Just a few\n [\"\", \"/\", \"/<id>\"],\n \"<id>\": # Just a few\n [\"abc\", \"def\", \"x<digit><digit>\"],\n \"<query>\":\n [\"\", \"?<params>\"],\n \"<params>\":\n [\"<param>\", \"<param>&<params>\"],\n \"<param>\": # Just a few\n [\"<id>=<id>\", \"<id>=<nat>\"],\n}",
"_____no_output_____"
],
[
"syntax_diagram(URL_GRAMMAR)",
"start\n"
]
],
[
[
"Again, within milliseconds, we can produce plenty of valid inputs.",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(simple_grammar_fuzzer(grammar=URL_GRAMMAR, max_nonterminals=10))",
"https://user:[email protected]:80/\nhttp://fuzzingbook.com?def=56&x89=3&x46=48&def=def\nftp://cispa.saarland/?x71=5&x35=90&def=abc\nhttps://cispa.saarland:80/def?def=7&x23=abc\nhttps://fuzzingbook.com:80/\nhttps://fuzzingbook.com:80/abc?def=abc&abc=x14&def=abc&abc=2&def=38\nftps://fuzzingbook.com/x87\nhttps://user:[email protected]:6?def=54&x44=abc\nhttp://fuzzingbook.com:80?x33=25&def=8\nhttp://fuzzingbook.com:8080/def\n"
]
],
[
[
"### A Natural Language Grammar\n\nFinally, grammars are not limited to *formal languages* such as computer inputs, but can also be used to produce *natural language*. This is the grammar we used to pick a title for this book:",
"_____no_output_____"
]
],
[
[
"TITLE_GRAMMAR: Grammar = {\n \"<start>\": [\"<title>\"],\n \"<title>\": [\"<topic>: <subtopic>\"],\n \"<topic>\": [\"Generating Software Tests\", \"<fuzzing-prefix>Fuzzing\", \"The Fuzzing Book\"],\n \"<fuzzing-prefix>\": [\"\", \"The Art of \", \"The Joy of \"],\n \"<subtopic>\": [\"<subtopic-main>\",\n \"<subtopic-prefix><subtopic-main>\",\n \"<subtopic-main><subtopic-suffix>\"],\n \"<subtopic-main>\": [\"Breaking Software\",\n \"Generating Software Tests\",\n \"Principles, Techniques and Tools\"],\n \"<subtopic-prefix>\": [\"\", \"Tools and Techniques for \"],\n \"<subtopic-suffix>\": [\" for <reader-property> and <reader-property>\",\n \" for <software-property> and <software-property>\"],\n \"<reader-property>\": [\"Fun\", \"Profit\"],\n \"<software-property>\": [\"Robustness\", \"Reliability\", \"Security\"],\n}",
"_____no_output_____"
],
[
"syntax_diagram(TITLE_GRAMMAR)",
"start\n"
],
[
"from typing import Set",
"_____no_output_____"
],
[
"titles: Set[str] = set()\nwhile len(titles) < 10:\n titles.add(simple_grammar_fuzzer(\n grammar=TITLE_GRAMMAR, max_nonterminals=10))\ntitles",
"_____no_output_____"
]
],
[
[
"(If you find that there is redundancy (\"Robustness and Robustness\") in here: In [our chapter on coverage-based fuzzing](GrammarCoverageFuzzer.ipynb), we will show how to cover each expansion only once. And if you like some alternatives more than others, [probabilistic grammar fuzzing](ProbabilisticGrammarFuzzer.ipynb) will be there for you.)",
"_____no_output_____"
],
[
"## Grammars as Mutation Seeds",
"_____no_output_____"
],
[
"One very useful property of grammars is that they produce mostly valid inputs. From a syntactical standpoint, the inputs are actually _always_ valid, as they satisfy the constraints of the given grammar. (Of course, one needs a valid grammar in the first place.) However, there are also _semantical_ properties that cannot be easily expressed in a grammar. If, say, for a URL, the port range is supposed to be between 1024 and 2048, this is hard to write in a grammar. If one has to satisfy more complex constraints, one quickly reaches the limits of what a grammar can express.",
"_____no_output_____"
],
[
"One way around this is to attach constraints to grammars, as we will discuss [later in this book](ConstraintFuzzer.ipynb). Another possibility is to put together the strengths of grammar-based fuzzing and [mutation-based fuzzing](MutationFuzzer.ipynb). The idea is to use the grammar-generated inputs as *seeds* for further mutation-based fuzzing. This way, we can explore not only _valid_ inputs, but also check out the _boundaries_ between valid and invalid inputs. This is particularly interesting as slightly invalid inputs allow to find parser errors (which are often abundant). As with fuzzing in general, it is the unexpected which reveals errors in programs.",
"_____no_output_____"
],
[
"To use our generated inputs as seeds, we can feed them directly into the mutation fuzzers introduced earlier:",
"_____no_output_____"
]
],
[
[
"from MutationFuzzer import MutationFuzzer # minor dependency",
"_____no_output_____"
],
[
"number_of_seeds = 10\nseeds = [\n simple_grammar_fuzzer(\n grammar=URL_GRAMMAR,\n max_nonterminals=10) for i in range(number_of_seeds)]\nseeds",
"_____no_output_____"
],
[
"m = MutationFuzzer(seeds)",
"_____no_output_____"
],
[
"[m.fuzz() for i in range(20)]",
"_____no_output_____"
]
],
[
[
"While the first 10 `fuzz()` calls return the seeded inputs (as designed), the later ones again create arbitrary mutations. Using `MutationCoverageFuzzer` instead of `MutationFuzzer`, we could again have our search guided by coverage – and thus bring together the best of multiple worlds.",
"_____no_output_____"
],
[
"## A Grammar Toolbox\n\nLet us now introduce a few techniques that help us writing grammars.",
"_____no_output_____"
],
[
"### Escapes\n\nWith `<` and `>` delimiting nonterminals in our grammars, how can we actually express that some input should contain `<` and `>`? The answer is simple: Just introduce a symbol for them.",
"_____no_output_____"
]
],
[
[
"simple_nonterminal_grammar: Grammar = {\n \"<start>\": [\"<nonterminal>\"],\n \"<nonterminal>\": [\"<left-angle><identifier><right-angle>\"],\n \"<left-angle>\": [\"<\"],\n \"<right-angle>\": [\">\"],\n \"<identifier>\": [\"id\"] # for now\n}",
"_____no_output_____"
]
],
[
[
"In `simple_nonterminal_grammar`, neither the expansion for `<left-angle>` nor the expansion for `<right-angle>` can be mistaken as a nonterminal. Hence, we can produce as many as we want.",
"_____no_output_____"
],
[
"### Extending Grammars\n\nIn the course of this book, we frequently run into the issue of creating a grammar by _extending_ an existing grammar with new features. Such an extension is very much like subclassing in object-oriented programming.",
"_____no_output_____"
],
[
"To create a new grammar $g'$ from an existing grammar $g$, we first copy $g$ into $g'$, and then go and extend existing rules with new alternatives and/or add new symbols. Here's an example, extending the above `nonterminal` grammar with a better rule for identifiers:",
"_____no_output_____"
]
],
[
[
"import copy",
"_____no_output_____"
],
[
"nonterminal_grammar = copy.deepcopy(simple_nonterminal_grammar)\nnonterminal_grammar[\"<identifier>\"] = [\"<idchar>\", \"<identifier><idchar>\"]\nnonterminal_grammar[\"<idchar>\"] = ['a', 'b', 'c', 'd'] # for now",
"_____no_output_____"
],
[
"nonterminal_grammar",
"_____no_output_____"
]
],
[
[
"Since such an extension of grammars is a common operation, we introduce a custom function `extend_grammar()` which first copies the given grammar and then updates it from a dictionary, using the Python dictionary `update()` method:",
"_____no_output_____"
]
],
[
[
"def extend_grammar(grammar: Grammar, extension: Grammar = {}) -> Grammar:\n new_grammar = copy.deepcopy(grammar)\n new_grammar.update(extension)\n return new_grammar",
"_____no_output_____"
]
],
[
[
"This call to `extend_grammar()` extends `simple_nonterminal_grammar` to `nonterminal_grammar` just like the \"manual\" example above:",
"_____no_output_____"
]
],
[
[
"nonterminal_grammar = extend_grammar(simple_nonterminal_grammar,\n {\n \"<identifier>\": [\"<idchar>\", \"<identifier><idchar>\"],\n # for now\n \"<idchar>\": ['a', 'b', 'c', 'd']\n }\n )",
"_____no_output_____"
]
],
[
[
"### Character Classes",
"_____no_output_____"
],
[
"In the above `nonterminal_grammar`, we have enumerated only the first few letters; indeed, enumerating all letters or digits in a grammar manually, as in `<idchar> ::= 'a' | 'b' | 'c' ...` is a bit painful.",
"_____no_output_____"
],
[
"However, remember that grammars are part of a program, and can thus also be constructed programmatically. We introduce a function `srange()` which constructs a list of characters in a string:",
"_____no_output_____"
]
],
[
[
"import string",
"_____no_output_____"
],
[
"def srange(characters: str) -> List[Expansion]:\n \"\"\"Construct a list with all characters in the string\"\"\"\n return [c for c in characters]",
"_____no_output_____"
]
],
[
[
"If we pass it the constant `string.ascii_letters`, which holds all ASCII letters, `srange()` returns a list of all ASCII letters:",
"_____no_output_____"
]
],
[
[
"string.ascii_letters",
"_____no_output_____"
],
[
"srange(string.ascii_letters)[:10]",
"_____no_output_____"
]
],
[
[
"We can use such constants in our grammar to quickly define identifiers:",
"_____no_output_____"
]
],
[
[
"nonterminal_grammar = extend_grammar(nonterminal_grammar,\n {\n \"<idchar>\": (srange(string.ascii_letters) + \n srange(string.digits) + \n srange(\"-_\"))\n }\n )",
"_____no_output_____"
],
[
"[simple_grammar_fuzzer(nonterminal_grammar, \"<identifier>\") for i in range(10)]",
"_____no_output_____"
]
],
[
[
"The shortcut `crange(start, end)` returns a list of all characters in the ASCII range of `start` to (including) `end`:",
"_____no_output_____"
]
],
[
[
"def crange(character_start: str, character_end: str) -> List[Expansion]:\n return [chr(i)\n for i in range(ord(character_start), ord(character_end) + 1)]",
"_____no_output_____"
]
],
[
[
"We can use this to express ranges of characters:",
"_____no_output_____"
]
],
[
[
"crange('0', '9')",
"_____no_output_____"
],
[
"assert crange('a', 'z') == srange(string.ascii_lowercase)",
"_____no_output_____"
]
],
[
[
"### Grammar Shortcuts",
"_____no_output_____"
],
[
"In the above `nonterminal_grammar`, as in other grammars, we have to express repetitions of characters using _recursion_, that is, by referring to the original definition:",
"_____no_output_____"
]
],
[
[
"nonterminal_grammar[\"<identifier>\"]",
"_____no_output_____"
]
],
[
[
"It could be a bit easier if we simply could state that a nonterminal should be a non-empty sequence of letters – for instance, as in\n\n```\n<identifier> = <idchar>+\n```\n\nwhere `+` denotes a non-empty repetition of the symbol it follows.",
"_____no_output_____"
],
[
"Operators such as `+` are frequently introduced as handy _shortcuts_ in grammars. Formally, our grammars come in the so-called [Backus-Naur form](https://en.wikipedia.org/wiki/Backus-Naur_form), or *BNF* for short. Operators _extend_ BNF to so-called _extended BNF*, or *EBNF* for short:\n\n* The form `<symbol>?` indicates that `<symbol>` is optional – that is, it can occur 0 or 1 times.\n* The form `<symbol>+` indicates that `<symbol>` can occur 1 or more times repeatedly.\n* The form `<symbol>*` indicates that `<symbol>` can occur 0 or more times. (In other words, it is an optional repetition.)\n\nTo make matters even more interesting, we would like to use _parentheses_ with the above shortcuts. Thus, `(<foo><bar>)?` indicates that the sequence of `<foo>` and `<bar>` is optional.",
"_____no_output_____"
],
[
"Using such operators, we can define the identifier rule in a simpler way. To this end, let us create a copy of the original grammar and modify the `<identifier>` rule:",
"_____no_output_____"
]
],
[
[
"nonterminal_ebnf_grammar = extend_grammar(nonterminal_grammar,\n {\n \"<identifier>\": [\"<idchar>+\"]\n }\n )",
"_____no_output_____"
]
],
[
[
"Likewise, we can simplify the expression grammar. Consider how signs are optional, and how integers can be expressed as sequences of digits.",
"_____no_output_____"
]
],
[
[
"EXPR_EBNF_GRAMMAR: Grammar = {\n \"<start>\":\n [\"<expr>\"],\n\n \"<expr>\":\n [\"<term> + <expr>\", \"<term> - <expr>\", \"<term>\"],\n\n \"<term>\":\n [\"<factor> * <term>\", \"<factor> / <term>\", \"<factor>\"],\n\n \"<factor>\":\n [\"<sign>?<factor>\", \"(<expr>)\", \"<integer>(.<integer>)?\"],\n\n \"<sign>\":\n [\"+\", \"-\"],\n\n \"<integer>\":\n [\"<digit>+\"],\n\n \"<digit>\":\n srange(string.digits)\n}",
"_____no_output_____"
]
],
[
[
"Let us implement a function `convert_ebnf_grammar()` that takes such an EBNF grammar and automatically translates it into a BNF grammar.",
"_____no_output_____"
],
[
"#### Excursion: Implementing `convert_ebnf_grammar()`",
"_____no_output_____"
],
[
"Our aim is to convert EBNF grammars such as the ones above into a regular BNF grammar. This is done by four rules:\n\n1. An expression `(content)op`, where `op` is one of `?`, `+`, `*`, becomes `<new-symbol>op`, with a new rule `<new-symbol> ::= content`.\n2. An expression `<symbol>?` becomes `<new-symbol>`, where `<new-symbol> ::= <empty> | <symbol>`.\n3. An expression `<symbol>+` becomes `<new-symbol>`, where `<new-symbol> ::= <symbol> | <symbol><new-symbol>`.\n4. An expression `<symbol>*` becomes `<new-symbol>`, where `<new-symbol> ::= <empty> | <symbol><new-symbol>`.\n\nHere, `<empty>` expands to the empty string, as in `<empty> ::= `. (This is also called an *epsilon expansion*.)",
"_____no_output_____"
],
[
"If these operators remind you of _regular expressions_, this is not by accident: Actually, any basic regular expression can be converted into a grammar using the above rules (and character classes with `crange()`, as defined above).",
"_____no_output_____"
],
[
"Applying these rules on the examples above yields the following results:\n\n* `<idchar>+` becomes `<idchar><new-symbol>` with `<new-symbol> ::= <idchar> | <idchar><new-symbol>`. \n* `<integer>(.<integer>)?` becomes `<integer><new-symbol>` with `<new-symbol> ::= <empty> | .<integer>`.",
"_____no_output_____"
],
[
"Let us implement these rules in three steps.",
"_____no_output_____"
],
[
"##### Creating New Symbols\n\nFirst, we need a mechanism to create new symbols. This is fairly straightforward.",
"_____no_output_____"
]
],
[
[
"def new_symbol(grammar: Grammar, symbol_name: str = \"<symbol>\") -> str:\n \"\"\"Return a new symbol for `grammar` based on `symbol_name`\"\"\"\n if symbol_name not in grammar:\n return symbol_name\n\n count = 1\n while True:\n tentative_symbol_name = symbol_name[:-1] + \"-\" + repr(count) + \">\"\n if tentative_symbol_name not in grammar:\n return tentative_symbol_name\n count += 1",
"_____no_output_____"
],
[
"assert new_symbol(EXPR_EBNF_GRAMMAR, '<expr>') == '<expr-1>'",
"_____no_output_____"
]
],
[
[
"##### Expanding Parenthesized Expressions",
"_____no_output_____"
],
[
"Next, we need a means to extract parenthesized expressions from our expansions and expand them according to the rules above. Let's start with extracting expressions:",
"_____no_output_____"
]
],
[
[
"RE_PARENTHESIZED_EXPR = re.compile(r'\\([^()]*\\)[?+*]')",
"_____no_output_____"
],
[
"def parenthesized_expressions(expansion: Expansion) -> List[str]:\n # In later chapters, we allow expansions to be tuples,\n # with the expansion being the first element\n if isinstance(expansion, tuple):\n expansion = expansion[0]\n\n return re.findall(RE_PARENTHESIZED_EXPR, expansion)",
"_____no_output_____"
],
[
"assert parenthesized_expressions(\"(<foo>)* (<foo><bar>)+ (+<foo>)? <integer>(.<integer>)?\") == [\n '(<foo>)*', '(<foo><bar>)+', '(+<foo>)?', '(.<integer>)?']",
"_____no_output_____"
]
],
[
[
"We can now use these to apply rule number 1, above, introducing new symbols for expressions in parentheses.",
"_____no_output_____"
]
],
[
[
"def convert_ebnf_parentheses(ebnf_grammar: Grammar) -> Grammar:\n \"\"\"Convert a grammar in extended BNF to BNF\"\"\"\n grammar = extend_grammar(ebnf_grammar)\n for nonterminal in ebnf_grammar:\n expansions = ebnf_grammar[nonterminal]\n\n for i in range(len(expansions)):\n expansion = expansions[i]\n if not isinstance(expansion, str):\n expansion = expansion[0]\n\n while True:\n parenthesized_exprs = parenthesized_expressions(expansion)\n if len(parenthesized_exprs) == 0:\n break\n\n for expr in parenthesized_exprs:\n operator = expr[-1:]\n contents = expr[1:-2]\n\n new_sym = new_symbol(grammar)\n\n exp = grammar[nonterminal][i]\n opts = None\n if isinstance(exp, tuple):\n (exp, opts) = exp\n assert isinstance(exp, str)\n\n expansion = exp.replace(expr, new_sym + operator, 1)\n if opts:\n grammar[nonterminal][i] = (expansion, opts)\n else:\n grammar[nonterminal][i] = expansion\n\n grammar[new_sym] = [contents]\n\n return grammar",
"_____no_output_____"
]
],
[
[
"This does the conversion as sketched above:",
"_____no_output_____"
]
],
[
[
"convert_ebnf_parentheses({\"<number>\": [\"<integer>(.<integer>)?\"]})",
"_____no_output_____"
]
],
[
[
"It even works for nested parenthesized expressions:",
"_____no_output_____"
]
],
[
[
"convert_ebnf_parentheses({\"<foo>\": [\"((<foo>)?)+\"]})",
"_____no_output_____"
]
],
[
[
"##### Expanding Operators\n\nAfter expanding parenthesized expressions, we now need to take care of symbols followed by operators (`?`, `*`, `+`). As with `convert_ebnf_parentheses()`, above, we first extract all symbols followed by an operator.",
"_____no_output_____"
]
],
[
[
"RE_EXTENDED_NONTERMINAL = re.compile(r'(<[^<> ]*>[?+*])')",
"_____no_output_____"
],
[
"def extended_nonterminals(expansion: Expansion) -> List[str]:\n # In later chapters, we allow expansions to be tuples,\n # with the expansion being the first element\n if isinstance(expansion, tuple):\n expansion = expansion[0]\n\n return re.findall(RE_EXTENDED_NONTERMINAL, expansion)",
"_____no_output_____"
],
[
"assert extended_nonterminals(\n \"<foo>* <bar>+ <elem>? <none>\") == ['<foo>*', '<bar>+', '<elem>?']",
"_____no_output_____"
]
],
[
[
"Our converter extracts the symbol and the operator, and adds new symbols according to the rules laid out above.",
"_____no_output_____"
]
],
[
[
"def convert_ebnf_operators(ebnf_grammar: Grammar) -> Grammar:\n \"\"\"Convert a grammar in extended BNF to BNF\"\"\"\n grammar = extend_grammar(ebnf_grammar)\n for nonterminal in ebnf_grammar:\n expansions = ebnf_grammar[nonterminal]\n\n for i in range(len(expansions)):\n expansion = expansions[i]\n extended_symbols = extended_nonterminals(expansion)\n\n for extended_symbol in extended_symbols:\n operator = extended_symbol[-1:]\n original_symbol = extended_symbol[:-1]\n assert original_symbol in ebnf_grammar, \\\n f\"{original_symbol} is not defined in grammar\"\n\n new_sym = new_symbol(grammar, original_symbol)\n\n exp = grammar[nonterminal][i]\n opts = None\n if isinstance(exp, tuple):\n (exp, opts) = exp\n assert isinstance(exp, str)\n \n new_exp = exp.replace(extended_symbol, new_sym, 1)\n if opts:\n grammar[nonterminal][i] = (new_exp, opts)\n else:\n grammar[nonterminal][i] = new_exp\n\n if operator == '?':\n grammar[new_sym] = [\"\", original_symbol]\n elif operator == '*':\n grammar[new_sym] = [\"\", original_symbol + new_sym]\n elif operator == '+':\n grammar[new_sym] = [\n original_symbol, original_symbol + new_sym]\n\n return grammar",
"_____no_output_____"
],
[
"convert_ebnf_operators({\"<integer>\": [\"<digit>+\"], \"<digit>\": [\"0\"]})",
"_____no_output_____"
]
],
[
[
"##### All Together\n\nWe can combine the two, first extending parentheses and then operators:",
"_____no_output_____"
]
],
[
[
"def convert_ebnf_grammar(ebnf_grammar: Grammar) -> Grammar:\n return convert_ebnf_operators(convert_ebnf_parentheses(ebnf_grammar))",
"_____no_output_____"
]
],
[
[
"#### End of Excursion",
"_____no_output_____"
],
[
"Here's an example of using `convert_ebnf_grammar()`:",
"_____no_output_____"
]
],
[
[
"convert_ebnf_grammar({\"<authority>\": [\"(<userinfo>@)?<host>(:<port>)?\"]})",
"_____no_output_____"
],
[
"expr_grammar = convert_ebnf_grammar(EXPR_EBNF_GRAMMAR)\nexpr_grammar",
"_____no_output_____"
]
],
[
[
"Success! We have nicely converted the EBNF grammar into BNF.",
"_____no_output_____"
],
[
"With character classes and EBNF grammar conversion, we have two powerful tools that make the writing of grammars easier. We will use these again and again as it comes to working with grammars.",
"_____no_output_____"
],
[
"### Grammar Extensions",
"_____no_output_____"
],
[
"During the course of this book, we frequently want to specify _additional information_ for grammars, such as [_probabilities_](ProbabilisticGrammarFuzzer.ipynb) or [_constraints_](GeneratorGrammarFuzzer.ipynb). To support these extensions, as well as possibly others, we define an _annotation_ mechanism.",
"_____no_output_____"
],
[
"Our concept for annotating grammars is to add _annotations_ to individual expansions. To this end, we allow that an expansion cannot only be a string, but also a _pair_ of a string and a set of attributes, as in\n\n```python\n \"<expr>\":\n [(\"<term> + <expr>\", opts(min_depth=10)),\n (\"<term> - <expr>\", opts(max_depth=2)),\n \"<term>\"]\n```\n\nHere, the `opts()` function would allow us to express annotations that apply to the individual expansions; in this case, the addition would be annotated with a `min_depth` value of 10, and the subtraction with a `max_depth` value of 2. The meaning of these annotations is left to the individual algorithms dealing with the grammars; the general idea, though, is that they can be ignored.",
"_____no_output_____"
],
[
"#### Excursion: Implementing `opts()`",
"_____no_output_____"
],
[
"Our `opts()` helper function returns a mapping of its arguments to values:",
"_____no_output_____"
]
],
[
[
"def opts(**kwargs: Any) -> Dict[str, Any]:\n return kwargs",
"_____no_output_____"
],
[
"opts(min_depth=10)",
"_____no_output_____"
]
],
[
[
"To deal with both expansion strings and pairs of expansions and annotations, we access the expansion string and the associated annotations via designated helper functions, `exp_string()` and `exp_opts()`:",
"_____no_output_____"
]
],
[
[
"def exp_string(expansion: Expansion) -> str:\n \"\"\"Return the string to be expanded\"\"\"\n if isinstance(expansion, str):\n return expansion\n return expansion[0]",
"_____no_output_____"
],
[
"exp_string((\"<term> + <expr>\", opts(min_depth=10)))",
"_____no_output_____"
],
[
"def exp_opts(expansion: Expansion) -> Dict[str, Any]:\n \"\"\"Return the options of an expansion. If options are not defined, return {}\"\"\"\n if isinstance(expansion, str):\n return {}\n return expansion[1]",
"_____no_output_____"
],
[
"def exp_opt(expansion: Expansion, attribute: str) -> Any:\n \"\"\"Return the given attribution of an expansion.\n If attribute is not defined, return None\"\"\"\n return exp_opts(expansion).get(attribute, None)",
"_____no_output_____"
],
[
"exp_opts((\"<term> + <expr>\", opts(min_depth=10)))",
"_____no_output_____"
],
[
"exp_opt((\"<term> - <expr>\", opts(max_depth=2)), 'max_depth')",
"_____no_output_____"
]
],
[
[
"Finally, we define a helper function that sets a particular option:",
"_____no_output_____"
]
],
[
[
"def set_opts(grammar: Grammar, symbol: str, expansion: Expansion, \n opts: Option = {}) -> None:\n \"\"\"Set the options of the given expansion of grammar[symbol] to opts\"\"\"\n expansions = grammar[symbol]\n for i, exp in enumerate(expansions):\n if exp_string(exp) != exp_string(expansion):\n continue\n\n new_opts = exp_opts(exp)\n if opts == {} or new_opts == {}:\n new_opts = opts\n else:\n for key in opts:\n new_opts[key] = opts[key]\n\n if new_opts == {}:\n grammar[symbol][i] = exp_string(exp)\n else:\n grammar[symbol][i] = (exp_string(exp), new_opts)\n\n return\n\n raise KeyError(\n \"no expansion \" +\n repr(symbol) +\n \" -> \" +\n repr(\n exp_string(expansion)))",
"_____no_output_____"
]
],
[
[
"#### End of Excursion",
"_____no_output_____"
],
[
"## Checking Grammars\n\nSince grammars are represented as strings, it is fairly easy to introduce errors. So let us introduce a helper function that checks a grammar for consistency.",
"_____no_output_____"
],
[
"The helper function `is_valid_grammar()` iterates over a grammar to check whether all used symbols are defined, and vice versa, which is very useful for debugging; it also checks whether all symbols are reachable from the start symbol. You don't have to delve into details here, but as always, it is important to get the input data straight before we make use of it.",
"_____no_output_____"
],
[
"### Excursion: Implementing `is_valid_grammar()`",
"_____no_output_____"
]
],
[
[
"import sys",
"_____no_output_____"
],
[
"def def_used_nonterminals(grammar: Grammar, start_symbol: \n str = START_SYMBOL) -> Tuple[Optional[Set[str]], \n Optional[Set[str]]]:\n \"\"\"Return a pair (`defined_nonterminals`, `used_nonterminals`) in `grammar`.\n In case of error, return (`None`, `None`).\"\"\"\n\n defined_nonterminals = set()\n used_nonterminals = {start_symbol}\n\n for defined_nonterminal in grammar:\n defined_nonterminals.add(defined_nonterminal)\n expansions = grammar[defined_nonterminal]\n if not isinstance(expansions, list):\n print(repr(defined_nonterminal) + \": expansion is not a list\",\n file=sys.stderr)\n return None, None\n\n if len(expansions) == 0:\n print(repr(defined_nonterminal) + \": expansion list empty\",\n file=sys.stderr)\n return None, None\n\n for expansion in expansions:\n if isinstance(expansion, tuple):\n expansion = expansion[0]\n if not isinstance(expansion, str):\n print(repr(defined_nonterminal) + \": \"\n + repr(expansion) + \": not a string\",\n file=sys.stderr)\n return None, None\n\n for used_nonterminal in nonterminals(expansion):\n used_nonterminals.add(used_nonterminal)\n\n return defined_nonterminals, used_nonterminals",
"_____no_output_____"
],
[
"def reachable_nonterminals(grammar: Grammar,\n start_symbol: str = START_SYMBOL) -> Set[str]:\n reachable = set()\n\n def _find_reachable_nonterminals(grammar, symbol):\n nonlocal reachable\n reachable.add(symbol)\n for expansion in grammar.get(symbol, []):\n for nonterminal in nonterminals(expansion):\n if nonterminal not in reachable:\n _find_reachable_nonterminals(grammar, nonterminal)\n\n _find_reachable_nonterminals(grammar, start_symbol)\n return reachable",
"_____no_output_____"
],
[
"def unreachable_nonterminals(grammar: Grammar,\n start_symbol=START_SYMBOL) -> Set[str]:\n return grammar.keys() - reachable_nonterminals(grammar, start_symbol)",
"_____no_output_____"
],
[
"def opts_used(grammar: Grammar) -> Set[str]:\n used_opts = set()\n for symbol in grammar:\n for expansion in grammar[symbol]:\n used_opts |= set(exp_opts(expansion).keys())\n return used_opts",
"_____no_output_____"
],
[
"def is_valid_grammar(grammar: Grammar,\n start_symbol: str = START_SYMBOL, \n supported_opts: Set[str] = set()) -> bool:\n \"\"\"Check if the given `grammar` is valid.\n `start_symbol`: optional start symbol (default: `<start>`)\n `supported_opts`: options supported (default: none)\"\"\"\n\n defined_nonterminals, used_nonterminals = \\\n def_used_nonterminals(grammar, start_symbol)\n if defined_nonterminals is None or used_nonterminals is None:\n return False\n\n # Do not complain about '<start>' being not used,\n # even if start_symbol is different\n if START_SYMBOL in grammar:\n used_nonterminals.add(START_SYMBOL)\n\n for unused_nonterminal in defined_nonterminals - used_nonterminals:\n print(repr(unused_nonterminal) + \": defined, but not used\",\n file=sys.stderr)\n for undefined_nonterminal in used_nonterminals - defined_nonterminals:\n print(repr(undefined_nonterminal) + \": used, but not defined\",\n file=sys.stderr)\n\n # Symbols must be reachable either from <start> or given start symbol\n unreachable = unreachable_nonterminals(grammar, start_symbol)\n msg_start_symbol = start_symbol\n\n if START_SYMBOL in grammar:\n unreachable = unreachable - \\\n reachable_nonterminals(grammar, START_SYMBOL)\n if start_symbol != START_SYMBOL:\n msg_start_symbol += \" or \" + START_SYMBOL\n\n for unreachable_nonterminal in unreachable:\n print(repr(unreachable_nonterminal) + \": unreachable from \" + msg_start_symbol,\n file=sys.stderr)\n\n used_but_not_supported_opts = set()\n if len(supported_opts) > 0:\n used_but_not_supported_opts = opts_used(\n grammar).difference(supported_opts)\n for opt in used_but_not_supported_opts:\n print(\n \"warning: option \" +\n repr(opt) +\n \" is not supported\",\n file=sys.stderr)\n\n return used_nonterminals == defined_nonterminals and len(unreachable) == 0",
"_____no_output_____"
]
],
[
[
"### End of Excursion",
"_____no_output_____"
],
[
"Let us make use of `is_valid_grammar()`. Our grammars defined above pass the test:",
"_____no_output_____"
]
],
[
[
"assert is_valid_grammar(EXPR_GRAMMAR)\nassert is_valid_grammar(CGI_GRAMMAR)\nassert is_valid_grammar(URL_GRAMMAR)",
"_____no_output_____"
]
],
[
[
"The check can also be applied to EBNF grammars:",
"_____no_output_____"
]
],
[
[
"assert is_valid_grammar(EXPR_EBNF_GRAMMAR)",
"_____no_output_____"
]
],
[
[
"These ones do not pass the test, though:",
"_____no_output_____"
]
],
[
[
"assert not is_valid_grammar({\"<start>\": [\"<x>\"], \"<y>\": [\"1\"]}) # type: ignore",
"'<y>': defined, but not used\n'<x>': used, but not defined\n'<y>': unreachable from <start>\n"
],
[
"assert not is_valid_grammar({\"<start>\": \"123\"}) # type: ignore",
"'<start>': expansion is not a list\n"
],
[
"assert not is_valid_grammar({\"<start>\": []}) # type: ignore",
"'<start>': expansion list empty\n"
],
[
"assert not is_valid_grammar({\"<start>\": [1, 2, 3]}) # type: ignore",
"'<start>': 1: not a string\n"
]
],
[
[
"(The `#type: ignore` annotations avoid static checkers flagging the above as errors).",
"_____no_output_____"
],
[
"From here on, we will always use `is_valid_grammar()` when defining a grammar.",
"_____no_output_____"
],
[
"## Synopsis\n\nThis chapter introduces _grammars_ as a simple means to specify input languages, and to use them for testing programs with syntactically valid inputs. A grammar is defined as a mapping of nonterminal symbols to lists of alternative expansions, as in the following example:",
"_____no_output_____"
]
],
[
[
"US_PHONE_GRAMMAR: Grammar = {\n \"<start>\": [\"<phone-number>\"],\n \"<phone-number>\": [\"(<area>)<exchange>-<line>\"],\n \"<area>\": [\"<lead-digit><digit><digit>\"],\n \"<exchange>\": [\"<lead-digit><digit><digit>\"],\n \"<line>\": [\"<digit><digit><digit><digit>\"],\n \"<lead-digit>\": [\"2\", \"3\", \"4\", \"5\", \"6\", \"7\", \"8\", \"9\"],\n \"<digit>\": [\"0\", \"1\", \"2\", \"3\", \"4\", \"5\", \"6\", \"7\", \"8\", \"9\"]\n}\n\nassert is_valid_grammar(US_PHONE_GRAMMAR)",
"_____no_output_____"
]
],
[
[
"Nonterminal symbols are enclosed in angle brackets (say, `<digit>`). To generate an input string from a grammar, a _producer_ starts with the start symbol (`<start>`) and randomly chooses a random expansion for this symbol. It continues the process until all nonterminal symbols are expanded. The function `simple_grammar_fuzzer()` does just that:",
"_____no_output_____"
]
],
[
[
"[simple_grammar_fuzzer(US_PHONE_GRAMMAR) for i in range(5)]",
"_____no_output_____"
]
],
[
[
"In practice, though, instead of `simple_grammar_fuzzer()`, you should use [the `GrammarFuzzer` class](GrammarFuzzer.ipynb) or one of its [coverage-based](GrammarCoverageFuzzer.ipynb), [probabilistic-based](ProbabilisticGrammarFuzzer.ipynb), or [generator-based](GeneratorGrammarFuzzer.ipynb) derivatives; these are more efficient, protect against infinite growth, and provide several additional features.",
"_____no_output_____"
],
[
"This chapter also introduces a [grammar toolbox](#A-Grammar-Toolbox) with several helper functions that ease the writing of grammars, such as using shortcut notations for character classes and repetitions, or extending grammars ",
"_____no_output_____"
],
[
"## Lessons Learned\n\n* Grammars are powerful tools to express and produce syntactically valid inputs.\n* Inputs produced from grammars can be used as is, or used as seeds for mutation-based fuzzing.\n* Grammars can be extended with character classes and operators to make writing easier.",
"_____no_output_____"
],
[
"## Next Steps\n\nAs they make a great foundation for generating software tests, we use grammars again and again in this book. As a sneak preview, we can use grammars to [fuzz configurations](ConfigurationFuzzer.ipynb):\n\n```\n<options> ::= <option>*\n<option> ::= -h | --version | -v | -d | -i | --global-config <filename>\n```",
"_____no_output_____"
],
[
"We can use grammars for [fuzzing functions and APIs](APIFuzzer.ipynb) and [fuzzing graphical user interfaces](WebFuzzer.ipynb):\n\n```\n<call-sequence> ::= <call>*\n<call> ::= urlparse(<url>) | urlsplit(<url>)\n```",
"_____no_output_____"
],
[
"We can assign [probabilities](ProbabilisticGrammarFuzzer.ipynb) and [constraints](GeneratorGrammarFuzzer.ipynb) to individual expansions:\n\n```\n<term>: 50% <factor> * <term> | 30% <factor> / <term> | 20% <factor>\n<integer>: <digit>+ { <integer> >= 100 }\n```",
"_____no_output_____"
],
[
"All these extras become especially valuable as we can\n\n1. _infer grammars automatically_, dropping the need to specify them manually, and\n2. _guide them towards specific goals_ such as coverage or critical functions;\n\nwhich we also discuss for all techniques in this book.",
"_____no_output_____"
],
[
"To get there, however, we still have bit of homework to do. In particular, we first have to learn how to \n\n* [create an efficient grammar fuzzer](GrammarFuzzer.ipynb)",
"_____no_output_____"
],
[
"## Background\n\nAs one of the foundations of human language, grammars have been around as long as human language existed. The first _formalization_ of generative grammars was by Dakṣiputra Pāṇini in 350 BC \\cite{Panini350bce}. As a general means to express formal languages for both data and programs, their role in computer science cannot be overstated. The seminal work by Chomsky \\cite{Chomsky1956} introduced the central models of regular languages, context-free grammars, context-sensitive grammars, and universal grammars as they are used (and taught) in computer science as a means to specify input and programming languages ever since.",
"_____no_output_____"
],
[
"The use of grammars for _producing_ test inputs goes back to Burkhardt \\cite{Burkhardt1967}, to be later rediscovered and applied by Hanford \\cite{Hanford1970} and Purdom \\cite{Purdom1972}. The most important use of grammar testing since then has been *compiler testing*. Actually, grammar-based testing is one important reason why compilers and Web browsers work as they should:\n\n* The [CSmith](https://embed.cs.utah.edu/csmith/) tool \\cite{Yang2011} specifically targets C programs, starting with a C grammar and then applying additional steps, such as referring to variables and functions defined earlier or ensuring integer and type safety. Their authors have used it \"to find and report more than 400 previously unknown compiler bugs.\"\n\n* The [LangFuzz](http://issta2016.cispa.saarland/interview-with-christian-holler/) work \\cite{Holler2012}, which shares two authors with this book, uses a generic grammar to produce outputs, and is used day and night to generate JavaScript programs and test their interpreters; as of today, it has found more than 2,600 bugs in browsers such as Mozilla Firefox, Google Chrome, and Microsoft Edge.\n\n* The [EMI Project](http://web.cs.ucdavis.edu/~su/emi-project/) \\cite{Le2014} uses grammars to stress-test C compilers, transforming known tests into alternative programs that should be semantically equivalent over all inputs. Again, this has led to more than 100 bugs in C compilers being fixed.\n\n* [Grammarinator](https://github.com/renatahodovan/grammarinator) \\cite{Hodovan2018} is an open-source grammar fuzzer (written in Python!), using the popular ANTLR format as grammar specification. Like LangFuzz, it uses the grammar for both parsing and producing, and has found more than 100 issues in the *JerryScript* lightweight JavaScript engine and an associated platform.\n\n* [Domato](https://github.com/googleprojectzero/domato) is a generic grammar generation engine that is specifically used for fuzzing DOM input. It has revealed a number of security issues in popular Web browsers.",
"_____no_output_____"
],
[
"Compilers and Web browsers, of course, are not only domains where grammars are needed for testing, but also domains where grammars are well-known. Our claim in this book is that grammars can be used to generate almost _any_ input, and our aim is to empower you to do precisely that.",
"_____no_output_____"
],
[
"## Exercises",
"_____no_output_____"
],
[
"### Exercise 1: A JSON Grammar\n\nTake a look at the [JSON specification](http://www.json.org) and derive a grammar from it:\n\n* Use _character classes_ to express valid characters\n* Use EBNF to express repetitions and optional parts\n* Assume that\n - a string is a sequence of digits, ASCII letters, punctuation and space characters without quotes or escapes\n - whitespace is just a single space.\n* Use `is_valid_grammar()` to ensure the grammar is valid.\n\nFeed the grammar into `simple_grammar_fuzzer()`. Do you encounter any errors, and why?",
"_____no_output_____"
],
[
"**Solution.** This is a fairly straightforward translation:",
"_____no_output_____"
]
],
[
[
"CHARACTERS_WITHOUT_QUOTE = (string.digits\n + string.ascii_letters\n + string.punctuation.replace('\"', '').replace('\\\\', '')\n + ' ')",
"_____no_output_____"
],
[
"JSON_EBNF_GRAMMAR: Grammar = {\n \"<start>\": [\"<json>\"],\n\n \"<json>\": [\"<element>\"],\n\n \"<element>\": [\"<ws><value><ws>\"],\n\n \"<value>\": [\"<object>\", \"<array>\", \"<string>\", \"<number>\",\n \"true\", \"false\", \"null\", \"'; DROP TABLE STUDENTS\"],\n\n \"<object>\": [\"{<ws>}\", \"{<members>}\"],\n\n \"<members>\": [\"<member>(,<members>)*\"],\n\n \"<member>\": [\"<ws><string><ws>:<element>\"],\n\n \"<array>\": [\"[<ws>]\", \"[<elements>]\"],\n\n \"<elements>\": [\"<element>(,<elements>)*\"],\n\n \"<element>\": [\"<ws><value><ws>\"],\n\n \"<string>\": ['\"' + \"<characters>\" + '\"'],\n \n \"<characters>\": [\"<character>*\"],\n\n \"<character>\": srange(CHARACTERS_WITHOUT_QUOTE),\n\n \"<number>\": [\"<int><frac><exp>\"],\n\n \"<int>\": [\"<digit>\", \"<onenine><digits>\", \"-<digits>\", \"-<onenine><digits>\"],\n\n \"<digits>\": [\"<digit>+\"],\n\n \"<digit>\": ['0', \"<onenine>\"],\n\n \"<onenine>\": crange('1', '9'),\n\n \"<frac>\": [\"\", \".<digits>\"],\n\n \"<exp>\": [\"\", \"E<sign><digits>\", \"e<sign><digits>\"],\n\n \"<sign>\": [\"\", '+', '-'],\n\n # \"<ws>\": srange(string.whitespace)\n\n \"<ws>\": [\" \"]\n}\n\nassert is_valid_grammar(JSON_EBNF_GRAMMAR)",
"_____no_output_____"
],
[
"JSON_GRAMMAR = convert_ebnf_grammar(JSON_EBNF_GRAMMAR)",
"_____no_output_____"
],
[
"from ExpectError import ExpectError",
"_____no_output_____"
],
[
"for i in range(50):\n with ExpectError():\n print(simple_grammar_fuzzer(JSON_GRAMMAR, '<object>'))",
"{ \"\" : '; DROP TABLE STUDENTS , \"/h?O \" : [ ] , \"\" : \"\" , \"x\" : false , \"\" : null }\n{ }\n{ }\n{ }\n{ }\n{ }\n{ }\n{ }\n{ }\n{ \"\" : \".qF\" , \"\" : '; DROP TABLE STUDENTS , \"\" : 47 }\n{ }\n{ \"7\" : { \"y\" : \"\" } , \"\" : false , \"X\" : \"N7|:\" , \"\" : [ true ] , \"\" : [ ] , \"\" : { } }\n{ \"Hm\" : false }\n{ }\n{ \"\" : [ ] }\n{ \"\" : [ ] , \"9z6}l\" : null }\n{ }\n{ \"#\" : false , \"D\" : { \"\" : true } , \"t\" : 90 , \"g\" : [ '; DROP TABLE STUDENTS ] , \"\" : [ false ] , \"=R5\" : [ ] , \" \" : '; DROP TABLE STUDENTS , \"`l\" : { \"\" : \"?'L\" , \"E\" : null , \"\" : [ 70.3076998940e6 ] , \"Ju\" : true } }\n{ }\n{ \"\" : true , \"\" : \"%7y\" , \"!\" : false , \"\" : true , \"\" : { \"\" : [ ] , \"\" : -096860E+0 , \"U\" : 0E-5 } }\n{ \"'ia\" : [ true , '; DROP TABLE STUDENTS , null , [ false , { } ] , true ] }\n{ \"\" : null }\n{ \"@meB1T]\" : 0.0 , \"\" : null , \"\" : true , \"7\" : 208.00E4 , \"\" : true , \"\" : 70e+10 , \"\" : \"\" , \"5zJ\" : [ false , false ] }\n{ }\n{ \"\" : \"H\" , \"d;\" : '; DROP TABLE STUDENTS }\n{ }\n{ }\n{ }\n{ }\n{ \"Y!Z\" : \".i\" , \"h\" : '; DROP TABLE STUDENTS }\n{ }\n{ }\n{ \"W5#\" : [ ] }\n{ \"\" : -64.0e-06 , \"\" : [ { \"p[f\" : false , \"\" : '; DROP TABLE STUDENTS , \"m\" : [ ] , \"\" : true , \"8D\" : -0 , \"@R\" : true } ] }\n{ }\n{ \"\" : '; DROP TABLE STUDENTS }\n{ }\n{ }\n{ \"r\" : '; DROP TABLE STUDENTS , \"zJzjT\" : 6.59 }\n{ }\n{ \"oh\" : false }\n{ }\n{ }\n{ \"Ue5\" : \"\" }\n{ \"c\" : [ false , 304e+008520 , null , false , '; DROP TABLE STUDENTS , \"m[MD\" , [ false ] ] }\n{ }\n{ \"N\" : '; DROP TABLE STUDENTS , \"\" : '; DROP TABLE STUDENTS }\n"
]
],
[
[
"We get these errors because `simple_grammar_fuzzer()` first expands to a maximum number of elements, and then is limited because every further expansion would _increase_ the number of nonterminals, even though these may eventually reduce the string length. This issue is addressed in the [next chapter](GrammarFuzzer.ipynb), introducing a more solid algorithm for producing strings from grammars.",
"_____no_output_____"
],
[
"### Exercise 2: Finding Bugs\n\nThe name `simple_grammar_fuzzer()` does not come by accident: The way it expands grammars is limited in several ways. What happens if you apply `simple_grammar_fuzzer()` on `nonterminal_grammar` and `expr_grammar`, as defined above, and why?",
"_____no_output_____"
],
[
"**Solution**. `nonterminal_grammar` does not work because `simple_grammar_fuzzer()` eventually tries to expand the just generated nonterminal:",
"_____no_output_____"
]
],
[
[
"from ExpectError import ExpectError, ExpectTimeout",
"_____no_output_____"
],
[
"with ExpectError():\n simple_grammar_fuzzer(nonterminal_grammar, log=True)",
"<start> -> <nonterminal> <nonterminal>\n<nonterminal> -> <left-angle><identifier><right-angle> <left-angle><identifier><right-angle>\n<left-angle> -> < <<identifier><right-angle>\n<identifier> -> <identifier><idchar> <<identifier><idchar><right-angle>\n<right-angle> -> > <<identifier><idchar>>\n<identifier> -> <identifier><idchar> <<identifier><idchar><idchar>>\n<idchar> -> y <<identifier>y<idchar>>\n<identifier> -> <identifier><idchar> <<identifier><idchar>y<idchar>>\n<idchar> -> n <<identifier>ny<idchar>>\n<identifier> -> <idchar> <<idchar>ny<idchar>>\n<idchar> -> S <Sny<idchar>>\n<idchar> -> Z <SnyZ>\n"
]
],
[
[
"For `expr_grammar`, things are even worse, as `simple_grammar_fuzzer()` can start a series of infinite expansions:",
"_____no_output_____"
]
],
[
[
"with ExpectTimeout(1):\n for i in range(10):\n print(simple_grammar_fuzzer(expr_grammar))",
"Traceback (most recent call last):\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_13175/557333580.py\", line 3, in <module>\n print(simple_grammar_fuzzer(expr_grammar))\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_13175/2839760968.py\", line 17, in simple_grammar_fuzzer\n symbol_to_expand = random.choice(nonterminals(term))\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_13175/2431549160.py\", line 7, in nonterminals\n return RE_NONTERMINAL.findall(expansion)\n File \"/Users/zeller/Projects/fuzzingbook/notebooks/Timeout.ipynb\", line 43, in timeout_handler\n raise TimeoutError()\nTimeoutError (expected)\n"
]
],
[
[
"Both issues are addressed and discussed in the [next chapter](GrammarFuzzer.ipynb), introducing a more solid algorithm for producing strings from grammars.",
"_____no_output_____"
],
[
"### Exercise 3: Grammars with Regular Expressions\n\nIn a _grammar extended with regular expressions_, we can use the special form\n```\n/regex/\n```\nto include regular expressions in expansions. For instance, we can have a rule\n```\n<integer> ::= /[+-]?[0-9]+/\n```\nto quickly express that an integer is an optional sign, followed by a sequence of digits.",
"_____no_output_____"
],
[
"#### Part 1: Convert regular expressions\n\nWrite a converter `convert_regex(r)` that takes a regular expression `r` and creates an equivalent grammar. Support the following regular expression constructs:\n\n* `*`, `+`, `?`, `()` should work just in EBNFs, above.\n* `a|b` should translate into a list of alternatives `[a, b]`.\n* `.` should match any character except newline.\n* `[abc]` should translate into `srange(\"abc\")`\n* `[^abc]` should translate into the set of ASCII characters _except_ `srange(\"abc\")`.\n* `[a-b]` should translate into `crange(a, b)`\n* `[^a-b]` should translate into the set of ASCII characters _except_ `crange(a, b)`.\n\nExample: `convert_regex(r\"[0-9]+\")` should yield a grammar such as\n```python\n{\n \"<start>\": [\"<s1>\"],\n \"<s1>\": [ \"<s2>\", \"<s1><s2>\" ],\n \"<s2>\": crange('0', '9')\n}\n```",
"_____no_output_____"
],
[
"**Solution.** Left as exercise to the reader.",
"_____no_output_____"
],
[
"#### Part 2: Identify and expand regular expressions\n\nWrite a converter `convert_regex_grammar(g)` that takes a EBNF grammar `g` containing regular expressions in the form `/.../` and creates an equivalent BNF grammar. Support the regular expression constructs as above.\n\nExample: `convert_regex_grammar({ \"<integer>\" : \"/[+-]?[0-9]+/\" })` should yield a grammar such as\n```python\n{\n \"<integer>\": [\"<s1><s3>\"],\n \"<s1>\": [ \"\", \"<s2>\" ],\n \"<s2>\": srange(\"+-\"),\n \"<s3>\": [ \"<s4>\", \"<s4><s3>\" ],\n \"<s4>\": crange('0', '9')\n}\n```",
"_____no_output_____"
],
[
"Optional: Support _escapes_ in regular expressions: `\\c` translates to the literal character `c`; `\\/` translates to `/` (and thus does not end the regular expression); `\\\\` translates to `\\`.",
"_____no_output_____"
],
[
"**Solution.** Left as exercise to the reader.",
"_____no_output_____"
],
[
"### Exercise 4: Defining Grammars as Functions (Advanced)\n\nTo obtain a nicer syntax for specifying grammars, one can make use of Python constructs which then will be _parsed_ by an additional function. For instance, we can imagine a grammar definition which uses `|` as a means to separate alternatives:",
"_____no_output_____"
]
],
[
[
"def expression_grammar_fn():\n start = \"<expr>\"\n expr = \"<term> + <expr>\" | \"<term> - <expr>\"\n term = \"<factor> * <term>\" | \"<factor> / <term>\" | \"<factor>\"\n factor = \"+<factor>\" | \"-<factor>\" | \"(<expr>)\" | \"<integer>.<integer>\" | \"<integer>\"\n integer = \"<digit><integer>\" | \"<digit>\"\n digit = '0' | '1' | '2' | '3' | '4' | '5' | '6' | '7' | '8' | '9'",
"_____no_output_____"
]
],
[
[
"If we execute `expression_grammar_fn()`, this will yield an error. Yet, the purpose of `expression_grammar_fn()` is not to be executed, but to be used as _data_ from which the grammar will be constructed.",
"_____no_output_____"
]
],
[
[
"with ExpectError():\n expression_grammar_fn()",
"Traceback (most recent call last):\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_13175/1271268731.py\", line 2, in <module>\n expression_grammar_fn()\n File \"/var/folders/n2/xd9445p97rb3xh7m1dfx8_4h0006ts/T/ipykernel_13175/3029408019.py\", line 3, in expression_grammar_fn\n expr = \"<term> + <expr>\" | \"<term> - <expr>\"\nTypeError: unsupported operand type(s) for |: 'str' and 'str' (expected)\n"
]
],
[
[
"To this end, we make use of the `ast` (abstract syntax tree) and `inspect` (code inspection) modules.",
"_____no_output_____"
]
],
[
[
"import ast\nimport inspect",
"_____no_output_____"
]
],
[
[
"First, we obtain the source code of `expression_grammar_fn()`...",
"_____no_output_____"
]
],
[
[
"source = inspect.getsource(expression_grammar_fn)\nsource",
"_____no_output_____"
]
],
[
[
"... which we then parse into an abstract syntax tree:",
"_____no_output_____"
]
],
[
[
"tree = ast.parse(source)",
"_____no_output_____"
]
],
[
[
"We can now parse the tree to find operators and alternatives. `get_alternatives()` iterates over all nodes `op` of the tree; If the node looks like a binary _or_ (`|` ) operation, we drill deeper and recurse. If not, we have reached a single production, and we try to get the expression from the production. We define the `to_expr` parameter depending on how we want to represent the production. In this case, we represent a single production by a single string.",
"_____no_output_____"
]
],
[
[
"def get_alternatives(op, to_expr=lambda o: o.s):\n if isinstance(op, ast.BinOp) and isinstance(op.op, ast.BitOr):\n return get_alternatives(op.left, to_expr) + [to_expr(op.right)]\n return [to_expr(op)]",
"_____no_output_____"
]
],
[
[
"`funct_parser()` takes the abstract syntax tree of a function (say, `expression_grammar_fn()`) and iterates over all assignments:",
"_____no_output_____"
]
],
[
[
"def funct_parser(tree, to_expr=lambda o: o.s):\n return {assign.targets[0].id: get_alternatives(assign.value, to_expr)\n for assign in tree.body[0].body}",
"_____no_output_____"
]
],
[
[
"The result is a grammar in our regular format:",
"_____no_output_____"
]
],
[
[
"grammar = funct_parser(tree)\nfor symbol in grammar:\n print(symbol, \"::=\", grammar[symbol])",
"start ::= ['<expr>']\nexpr ::= ['<term> + <expr>', '<term> - <expr>']\nterm ::= ['<factor> * <term>', '<factor> / <term>', '<factor>']\nfactor ::= ['+<factor>', '-<factor>', '(<expr>)', '<integer>.<integer>', '<integer>']\ninteger ::= ['<digit><integer>', '<digit>']\ndigit ::= ['0', '1', '2', '3', '4', '5', '6', '7', '8', '9']\n"
]
],
[
[
"#### Part 1 (a): One Single Function\n\nWrite a single function `define_grammar(fn)` that takes a grammar defined as function (such as `expression_grammar_fn()`) and returns a regular grammar.",
"_____no_output_____"
],
[
"**Solution**. This is straightforward:",
"_____no_output_____"
]
],
[
[
"def define_grammar(fn, to_expr=lambda o: o.s):\n source = inspect.getsource(fn)\n tree = ast.parse(source)\n grammar = funct_parser(tree, to_expr)\n return grammar",
"_____no_output_____"
],
[
"define_grammar(expression_grammar_fn)",
"_____no_output_____"
]
],
[
[
"**Note.** Python allows us to directly bind the generated grammar to the name `expression_grammar_fn` using function decorators. This can be used to ensure that we do not have a faulty function lying around:\n\n```python\n@define_grammar\ndef expression_grammar():\n start = \"<expr>\"\n expr = \"<term> + <expr>\" | \"<term> - <expr>\"\n #...\n```",
"_____no_output_____"
],
[
"#### Part 1 (b): Alternative representations",
"_____no_output_____"
],
[
"We note that the grammar representation we designed previously does not allow simple generation of alternatives such as `srange()` and `crange()`. Further, one may find the string representation of expressions limiting. It turns out that it is simple to extend our grammar definition to support grammars such as below:",
"_____no_output_____"
]
],
[
[
"def define_name(o):\n return o.id if isinstance(o, ast.Name) else o.s",
"_____no_output_____"
],
[
"def define_expr(op):\n if isinstance(op, ast.BinOp) and isinstance(op.op, ast.Add):\n return (*define_expr(op.left), define_name(op.right))\n return (define_name(op),)",
"_____no_output_____"
],
[
"def define_ex_grammar(fn):\n return define_grammar(fn, define_expr)",
"_____no_output_____"
]
],
[
[
"The grammar:\n\n```python\n@define_ex_grammar\ndef expression_grammar():\n start = expr\n expr = (term + '+' + expr\n | term + '-' + expr)\n term = (factor + '*' + term\n | factor + '/' + term\n | factor)\n factor = ('+' + factor\n | '-' + factor\n | '(' + expr + ')'\n | integer + '.' + integer\n | integer)\n integer = (digit + integer\n | digit)\n digit = '0' | '1' | '2' | '3' | '4' | '5' | '6' | '7' | '8' | '9'\n \nfor symbol in expression_grammar:\n print(symbol, \"::=\", expression_grammar[symbol])\n```",
"_____no_output_____"
],
[
"**Note.** The grammar data structure thus obtained is a little more detailed than the standard data structure. It represents each production as a tuple.",
"_____no_output_____"
],
[
"We note that we have not enabled `srange()` or `crange()` in the above grammar. How would you go about adding these? (*Hint:* wrap `define_expr()` to look for `ast.Call`)",
"_____no_output_____"
],
[
"#### Part 2: Extended Grammars\n\nIntroduce an operator `*` that takes a pair `(min, max)` where `min` and `max` are the minimum and maximum number of repetitions, respectively. A missing value `min` stands for zero; a missing value `max` for infinity.",
"_____no_output_____"
]
],
[
[
"def identifier_grammar_fn():\n identifier = idchar * (1,)",
"_____no_output_____"
]
],
[
[
"With the `*` operator, we can generalize the EBNF operators – `?` becomes (0,1), `*` becomes (0,), and `+` becomes (1,). Write a converter that takes an extended grammar defined using `*`, parse it, and convert it into BNF.",
"_____no_output_____"
],
[
"**Solution.** No solution yet :-)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbaba285075229e93159d96af58e1b297e40d931
| 7,890 |
ipynb
|
Jupyter Notebook
|
code/ARAX/Examples/ICEES+ queries plus ARAXi example1.ipynb
|
RTXteam/RTX
|
eabc4f4445d69c71d428acc8e9fa9865a85fb593
|
[
"MIT"
] | 31 |
2018-03-05T20:01:10.000Z
|
2022-02-01T03:31:22.000Z
|
code/ARAX/Examples/ICEES+ queries plus ARAXi example1.ipynb
|
RTXteam/RTX
|
eabc4f4445d69c71d428acc8e9fa9865a85fb593
|
[
"MIT"
] | 1,774 |
2018-03-06T01:55:03.000Z
|
2022-03-31T03:09:04.000Z
|
code/ARAX/Examples/ICEES+ queries plus ARAXi example1.ipynb
|
RTXteam/RTX
|
eabc4f4445d69c71d428acc8e9fa9865a85fb593
|
[
"MIT"
] | 19 |
2018-05-10T00:43:19.000Z
|
2022-03-08T19:26:16.000Z
| 33.012552 | 136 | 0.565146 |
[
[
[
"import requests\nimport json\nimport re",
"_____no_output_____"
],
[
"# Setting the base URL for the ARAX reasoner and its endpoint\nendpoint_url = 'https://arax.rtx.ai/api/rtx/v1/query'\n# Given we have some chemical substances which are linked to asthma exacerbations for a certain cohort of patients, \n# we want to find what diseases are associated with them\n# This DSL command extracts the pathways to view which diseases are associated with those chemicals. \n# We do this by creating a dict of the request, specifying a start previous Message and the list of DSL commands\nquery = {\"previous_message_processing_plan\": {\"processing_actions\": [\n \"add_qnode(curie=CHEMBL.COMPOUND:CHEMBL896, type= chemical_substance, id=n0)\",\n \"add_qnode(type=protein, id=n1)\",\n \"add_qnode(type=disease, id=n2)\",\n \"add_qedge(source_id=n0, target_id=n1, id=e0)\",\n \"add_qedge(source_id=n1, target_id=n2, id=e1)\",\n \"expand()\",\n #\"expand(kp=RTX-KG2)\".\n \"resultify()\",\n \"filter_results(action=limit_number_of_results, max_results=20)\",\n \"return(message=true, store=true)\",\n ]}}",
"_____no_output_____"
],
[
"# Sending the request to RTX and check the status\nprint(f\"Executing query at {endpoint_url}\\nPlease wait...\")\nresponse_content = requests.post(endpoint_url, json=query, headers={'accept': 'application/json'})\nstatus_code = response_content.status_code\nif status_code != 200:\n print(\"ERROR returned with status \"+str(status_code))\n print(response_content.json())\nelse:\n print(f\"Response returned with status {status_code}\")",
"Executing query at https://arax.rtx.ai/api/rtx/v1/query\nPlease wait...\nResponse returned with status 200\n"
],
[
"# Unpack respsonse from JSON and display the information log\nresponse_dict = response_content.json()\nfor message in response_dict['log']:\n if message['level'] >= 20:\n print(message['prefix']+message['message'])",
"_____no_output_____"
],
[
"# These URLs provide direct access to resulting data and GUI\nif 'id' in response_dict and response_dict['id'] is not None:\n print(f\"Data: {response_dict['id']}\")\n match = re.search(r'(\\d+)$', response_dict['id'])\n if match:\n print(f\"GUI: https://arax.rtx.ai/?m={match.group(1)}\")\nelse:\n print(\"No id was returned in response\")",
"Data: https://arax.rtx.ai/api/rtx/v1/message/2199\nGUI: https://arax.rtx.ai/?m=2199\n"
],
[
"# Or you can view the entire Translator API response Message\nprint(json.dumps(response_dict, indent=2, sort_keys=True))",
"_____no_output_____"
],
[
"# Setting the base URL for the ARAX reasoner and its endpoint\nendpoint_url = 'https://arax.rtx.ai/api/rtx/v1/query'\n# Given we have some chemical substances which are linked to asthma exacerbations for a certain cohort of patients, we want to \n# find what diseases are associated with them\n# This DSL command extracts the pathways to view which phenotypes are associated with those chemicals. \n# We do this by creating a dict of the request, specifying a start previous Message and the list of DSL commands\nquery = {\"previous_message_processing_plan\": {\"processing_actions\": [\n \"add_qnode(curie=CHEMBL.COMPOUND:CHEMBL896, type= chemical_substance, id=n0)\",\n \"add_qnode(type=protein, id=n1)\",\n \"add_qnode(type=phenotypic_feature, id=n2)\",\n \"add_qedge(source_id=n0, target_id=n1, id=e0)\",\n \"add_qedge(source_id=n1, target_id=n2, id=e1)\",\n \"expand()\",\n #\"expand(kp=RTX-KG2)\".\n \"resultify()\",\n \"filter_results(action=limit_number_of_results, max_results=20)\",\n \"return(message=true, store=true)\",\n ]}}",
"_____no_output_____"
],
[
"# Sending the request to RTX and check the status\nprint(f\"Executing query at {endpoint_url}\\nPlease wait...\")\nresponse_content = requests.post(endpoint_url, json=query, headers={'accept': 'application/json'})\nstatus_code = response_content.status_code\nif status_code != 200:\n print(\"ERROR returned with status \"+str(status_code))\n print(response_content.json())\nelse:\n print(f\"Response returned with status {status_code}\")",
"Executing query at https://arax.rtx.ai/api/rtx/v1/query\nPlease wait...\nResponse returned with status 200\n"
],
[
"# Unpack respsonse from JSON and display the information log\nresponse_dict = response_content.json()\nfor message in response_dict['log']:\n if message['level'] >= 20:\n print(message['prefix']+message['message'])",
"_____no_output_____"
],
[
"# These URLs provide direct access to resulting data and GUI\nif 'id' in response_dict and response_dict['id'] is not None:\n print(f\"Data: {response_dict['id']}\")\n match = re.search(r'(\\d+)$', response_dict['id'])\n if match:\n print(f\"GUI: https://arax.rtx.ai/?m={match.group(1)}\")\nelse:\n print(\"No id was returned in response\")",
"Data: https://arax.rtx.ai/api/rtx/v1/message/2200\nGUI: https://arax.rtx.ai/?m=2200\n"
],
[
"# Or you can view the entire Translator API response Message\nprint(json.dumps(response_dict, indent=2, sort_keys=True))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbaba86fb1112be8f7cf8b6570b80f2da2586ddc
| 32,795 |
ipynb
|
Jupyter Notebook
|
workshops/docs/modules/notebooks/intro.ipynb
|
rksin8/python-workshop-base
|
ed235a9ef212463ddce3e7b1c1cc8f01ef29ca15
|
[
"CC-BY-4.0"
] | null | null | null |
workshops/docs/modules/notebooks/intro.ipynb
|
rksin8/python-workshop-base
|
ed235a9ef212463ddce3e7b1c1cc8f01ef29ca15
|
[
"CC-BY-4.0"
] | null | null | null |
workshops/docs/modules/notebooks/intro.ipynb
|
rksin8/python-workshop-base
|
ed235a9ef212463ddce3e7b1c1cc8f01ef29ca15
|
[
"CC-BY-4.0"
] | null | null | null | 20.80901 | 323 | 0.465345 |
[
[
[
"# Python: the basics",
"_____no_output_____"
],
[
"Python is a general purpose programming language that supports rapid development\nof scripts and applications.\n\nPython's main advantages:\n\n* Open Source software, supported by Python Software Foundation\n* Available on all major platforms (ie. Windows, Linux and MacOS) \n* It is a general-purpose programming language, designed for readability\n* Supports multiple programming paradigms ('functional', 'object oriented')\n* Very large community with a rich ecosystem of third-party packages",
"_____no_output_____"
],
[
"## Interpreter\n\nPython is an interpreted language[*](https://softwareengineering.stackexchange.com/a/24560) which can be used in two ways:\n\n* \"Interactive\" Mode: It functions like an \"advanced calculator\", executing\n one command at a time:\n \n```bash\nuser:host:~$ python\nPython 3.5.1 (default, Oct 23 2015, 18:05:06)\n[GCC 4.8.3] on linux2\nType \"help\", \"copyright\", \"credits\" or \"license\" for more information.\n>>> 2 + 2\n4\n>>> print(\"Hello World\")\nHello World\n```",
"_____no_output_____"
],
[
"* \"Scripting\" Mode: Executing a series of \"commands\" saved in text file,\n usually with a `.py` extension after the name of your file:\n\n```bash\nuser:host:~$ python my_script.py\nHello World\n```",
"_____no_output_____"
],
[
"## Using interactive Python in Jupyter-style notebooks\n\nA convenient and powerful way to use interactive-mode Python is via a Jupyter Notebook, or similar browser-based interface.\n\nThis particularly lends itself to data analysis since the notebook records a history of commands and shows output and graphs immediately in the browser.\n\nThere are several ways you can run a Jupyter(-style) notebook - locally installed on your computer or hosted as a service on the web. Today we will use a Jupyter notebook service provided by Google: https://colab.research.google.com (Colaboratory).\n\n### Jupyter-style notebooks: a quick tour\n\nGo to https://colab.research.google.com and login with your Google account.\n\nSelect ***NEW NOTEBOOK → NEW PYTHON 3 NOTEBOOK*** - a new notebook will be created.\n\n---\n\nType some Python code in the top cell, eg:\n\n```python\nprint(\"Hello Jupyter !\")\n```\n\n***Shift-Enter*** to run the contents of the cell\n\n---\n\nYou can add new cells.\n\n***Insert → Insert Code Cell***\n\n---\n\nNOTE: When the text on the left hand of the cell is: `In [*]` (with an asterisk rather than a number), the cell is still running. It's usually best to wait until one cell has finished running before running the next.\n\nLet's begin writing some code in our notebook.",
"_____no_output_____"
]
],
[
[
"print(\"Hello Jupyter !\")",
"Hello Jupyter !\n"
]
],
[
[
"In Jupyter/Collaboratory, just typing the name of a variable in the cell prints its representation:",
"_____no_output_____"
]
],
[
[
"message = \"Hello again !\"\nmessage",
"_____no_output_____"
],
[
"# A 'hash' symbol denotes a comment\n# This is a comment. Anything after the 'hash' symbol on the line is ignored by the Python interpreter\n\nprint(\"No comment\") # comment",
"No comment\n"
]
],
[
[
"## Variables and data types\n### Integers, floats, strings",
"_____no_output_____"
]
],
[
[
"a = 5",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"type(a)",
"_____no_output_____"
]
],
[
[
"Adding a decimal point creates a `float`",
"_____no_output_____"
]
],
[
[
"b = 5.0",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
],
[
"type(b)",
"_____no_output_____"
]
],
[
[
"`int` and `float` are collectively called 'numeric' types\n\n(There are also other numeric types like `hex` for hexidemical and `complex` for complex numbers)",
"_____no_output_____"
],
[
"## Challenge - Types\n\nWhat is the **type** of the variable `letters` defined below ?\n\n`letters = \"ABACBS\"`\n\n* A) `int`\n* B) `str`\n* C) `float`\n* D) `text`\n\nWrite some code the outputs the type - paste your answer into the Etherpad.",
"_____no_output_____"
],
[
"## Solution\n\nOption B - `str`.",
"_____no_output_____"
]
],
[
[
"letters = \"ABACBS\"\ntype(letters)",
"_____no_output_____"
]
],
[
[
"### Strings",
"_____no_output_____"
]
],
[
[
"some_words = \"Python3 strings are Unicode (UTF-8) ❤❤❤ 😸 蛇\"",
"_____no_output_____"
],
[
"some_words",
"_____no_output_____"
],
[
"type(some_words)",
"_____no_output_____"
]
],
[
[
"The variable `some_words` is of type `str`, short for \"string\". Strings hold\nsequences of characters, which can be letters, numbers, punctuation\nor more exotic forms of text (even emoji!).",
"_____no_output_____"
],
[
"## Operators\n\nWe can perform mathematical calculations in Python using the basic operators:\n\n`+` `-` `*` `/` `%` `//` `**`",
"_____no_output_____"
]
],
[
[
"2 + 2 # Addition",
"_____no_output_____"
],
[
"6 * 7 # Multiplication",
"_____no_output_____"
],
[
"5/2 # Division",
"_____no_output_____"
],
[
"13 % 5 # Modulo",
"_____no_output_____"
],
[
"13 // 5 # Floor Division",
"_____no_output_____"
],
[
"2 ** 16 # Power",
"_____no_output_____"
],
[
"# int + int = int\na = 5\na + 1",
"_____no_output_____"
],
[
"# float + int = float\nb = 5.0\nb + 1",
"_____no_output_____"
],
[
"a + b",
"_____no_output_____"
]
],
[
[
"```python\nsome_words = \"I'm a string\"\na = 6\na + some_words\n```\n\n",
"_____no_output_____"
],
[
"Outputs:\n\n```\n---------------------------------------------------------------------------\nTypeError Traceback (most recent call last)\n<ipython-input-1-781eba7cf148> in <module>()\n 1 some_words = \"I'm a string\"\n 2 a = 6\n----> 3 a + some_words\n\nTypeError: unsupported operand type(s) for +: 'int' and 'str'\n```",
"_____no_output_____"
]
],
[
[
"str(a) + \" \" + some_words",
"_____no_output_____"
],
[
"# Shorthand: operators with assignment\na += 1\na\n\n# Equivalent to:\n# a = a + 1",
"_____no_output_____"
]
],
[
[
"### Boolean operations\n\nWe can also use comparison and logic operators:\n`<, >, ==, !=, <=, >=` and statements of identity such as\n`and, or, not`. The data type returned by this is\ncalled a _boolean_.\n",
"_____no_output_____"
]
],
[
[
"3 > 4",
"_____no_output_____"
],
[
"True and True",
"_____no_output_____"
],
[
"True or False",
"_____no_output_____"
]
],
[
[
"## Lists and sequence types",
"_____no_output_____"
],
[
"### Lists",
"_____no_output_____"
]
],
[
[
"numbers = [2, 4, 6, 8, 10]\nnumbers",
"_____no_output_____"
],
[
"# `len` get the length of a list\nlen(numbers)",
"_____no_output_____"
],
[
"# Lists can contain multiple data types, including other lists\nmixed_list = [\"asdf\", 2, 3.142, numbers, ['a','b','c']]\nmixed_list",
"_____no_output_____"
]
],
[
[
"You can retrieve items from a list by their *index*. In Python, the first item has an index of 0 (zero).",
"_____no_output_____"
]
],
[
[
"numbers[0]",
"_____no_output_____"
],
[
"numbers[3]",
"_____no_output_____"
]
],
[
[
"You can also assign a new value to any position in the list.",
"_____no_output_____"
]
],
[
[
"numbers[3] = numbers[3] * 100\nnumbers",
"_____no_output_____"
]
],
[
[
"You can append items to the end of the list.",
"_____no_output_____"
]
],
[
[
"numbers.append(12)\nnumbers",
"_____no_output_____"
]
],
[
[
"You can add multiple items to the end of a list with `extend`.",
"_____no_output_____"
]
],
[
[
"numbers.extend([14, 16, 18])\nnumbers",
"_____no_output_____"
]
],
[
[
"### Loops\n\nA for loop can be used to access the elements in a list or other Python data structure one at a time. We will learn about loops in other lesson.",
"_____no_output_____"
]
],
[
[
"for num in numbers:\n print(num)",
"2\n4\n6\n800\n10\n12\n14\n16\n18\n"
]
],
[
[
"**Indentation** is very important in Python. Note that the second line in the\nexample above is indented, indicating the code that is the body of the loop.",
"_____no_output_____"
],
[
"To find out what methods are available for an object, we can use the built-in `help` command:",
"_____no_output_____"
]
],
[
[
"help(numbers)",
"Help on list object:\n\nclass list(object)\n | list() -> new empty list\n | list(iterable) -> new list initialized from iterable's items\n | \n | Methods defined here:\n | \n | __add__(self, value, /)\n | Return self+value.\n | \n | __contains__(self, key, /)\n | Return key in self.\n | \n | __delitem__(self, key, /)\n | Delete self[key].\n | \n | __eq__(self, value, /)\n | Return self==value.\n | \n | __ge__(self, value, /)\n | Return self>=value.\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __getitem__(...)\n | x.__getitem__(y) <==> x[y]\n | \n | __gt__(self, value, /)\n | Return self>value.\n | \n | __iadd__(self, value, /)\n | Implement self+=value.\n | \n | __imul__(self, value, /)\n | Implement self*=value.\n | \n | __init__(self, /, *args, **kwargs)\n | Initialize self. See help(type(self)) for accurate signature.\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __le__(self, value, /)\n | Return self<=value.\n | \n | __len__(self, /)\n | Return len(self).\n | \n | __lt__(self, value, /)\n | Return self<value.\n | \n | __mul__(self, value, /)\n | Return self*value.\n | \n | __ne__(self, value, /)\n | Return self!=value.\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n | \n | __repr__(self, /)\n | Return repr(self).\n | \n | __reversed__(...)\n | L.__reversed__() -- return a reverse iterator over the list\n | \n | __rmul__(self, value, /)\n | Return value*self.\n | \n | __setitem__(self, key, value, /)\n | Set self[key] to value.\n | \n | __sizeof__(...)\n | L.__sizeof__() -- size of L in memory, in bytes\n | \n | append(...)\n | L.append(object) -> None -- append object to end\n | \n | clear(...)\n | L.clear() -> None -- remove all items from L\n | \n | copy(...)\n | L.copy() -> list -- a shallow copy of L\n | \n | count(...)\n | L.count(value) -> integer -- return number of occurrences of value\n | \n | extend(...)\n | L.extend(iterable) -> None -- extend list by appending elements from the iterable\n | \n | index(...)\n | L.index(value, [start, [stop]]) -> integer -- return first index of value.\n | Raises ValueError if the value is not present.\n | \n | insert(...)\n | L.insert(index, object) -- insert object before index\n | \n | pop(...)\n | L.pop([index]) -> item -- remove and return item at index (default last).\n | Raises IndexError if list is empty or index is out of range.\n | \n | remove(...)\n | L.remove(value) -> None -- remove first occurrence of value.\n | Raises ValueError if the value is not present.\n | \n | reverse(...)\n | L.reverse() -- reverse *IN PLACE*\n | \n | sort(...)\n | L.sort(key=None, reverse=False) -> None -- stable sort *IN PLACE*\n | \n | ----------------------------------------------------------------------\n | Data and other attributes defined here:\n | \n | __hash__ = None\n\n"
]
],
[
[
"### Tuples\n\nA tuple is similar to a list in that it's an ordered sequence of elements.\nHowever, tuples can not be changed once created (they are \"immutable\"). Tuples\nare created by placing comma-separated values inside parentheses `()`.",
"_____no_output_____"
]
],
[
[
"tuples_are_immutable = (\"bar\", 100, 200, \"foo\")\ntuples_are_immutable",
"_____no_output_____"
],
[
"tuples_are_immutable[1]",
"_____no_output_____"
]
],
[
[
"```python\ntuples_are_immutable[1] = 666\n```",
"_____no_output_____"
],
[
"Outputs:\n\n```\n---------------------------------------------------------------------------\nTypeError Traceback (most recent call last)\n<ipython-input-39-c91965b0815a> in <module>()\n----> 1 tuples_are_immutable[1] = 666\n\nTypeError: 'tuple' object does not support item assignment\n```",
"_____no_output_____"
],
[
"### Dictionaries\n\nDictionaries are a container that store key-value pairs. They are unordered. \n\nOther programming languages might call this a 'hash', 'hashtable' or 'hashmap'.",
"_____no_output_____"
]
],
[
[
"pairs = {'Apple': 1, 'Orange': 2, 'Pear': 4}\npairs",
"_____no_output_____"
],
[
"pairs['Orange']",
"_____no_output_____"
],
[
"pairs['Orange'] = 16\npairs",
"_____no_output_____"
]
],
[
[
"The `items` method returns a sequence of the key-value pairs as tuples.\n\n`values` returns a sequence of just the values.\n\n`keys` returns a sequence of just the keys.\n\n---\nIn Python 3, the `.items()`, `.values()` and `.keys()` methods return a ['dictionary view' object](https://docs.python.org/3/library/stdtypes.html#dictionary-view-objects) that behaves like a list or tuple in for loops but doesn't support indexing. 'Dictionary views' stay in sync even when the dictionary changes.\n\nYou can turn them into a normal list or tuple with the `list()` or `tuple()` functions.",
"_____no_output_____"
]
],
[
[
"pairs.items()\n# list(pairs.items())",
"_____no_output_____"
],
[
"pairs.values()\n# list(pairs.values())",
"_____no_output_____"
],
[
"pairs.keys()\n# list(pairs.keys())",
"_____no_output_____"
],
[
"len(pairs)",
"_____no_output_____"
],
[
"dict_of_dicts = {'first': {1:2, 2: 4, 4: 8, 8: 16}, 'second': {'a': 2.2, 'b': 4.4}}\ndict_of_dicts",
"_____no_output_____"
]
],
[
[
"## Challenge - Dictionaries\n\nGiven the dictionary:\n\n```python\njam_ratings = {'Plum': 6, 'Apricot': 2, 'Strawberry': 8}\n```\n\nHow would you change the value associated with the key `Apricot` to `9`.\n\nA) `jam_ratings = {'apricot': 9}`\n\nB) `jam_ratings[9] = 'Apricot'`\n\nC) `jam_ratings['Apricot'] = 9`\n\nD) `jam_ratings[2] = 'Apricot'`",
"_____no_output_____"
],
[
"## Solution - Dictionaries\n\nThe correct answer is **C**.\n\n**A** assigns the name `jam_ratings` to a new dictionary with only the key `apricot` - not only are the other jam ratings now missing, but strings used as dictionary keys are *case sensitive* - `apricot` is not the same key as `Apricot`.\n\n**B** mixes up the value and the key. Assigning to a dictionary uses the form: `dictionary[key] = value`.\n\n**C** is correct. Bonus - another way to do this would be `jam_ratings.update({'Apricot': 9})` or even `jam_ratings.update(Apricot=9)`.\n\n**D** mixes up the value and the key (and doesn't actually include the new value to be assigned, `9`, anywhere). `2` is the original *value*, `Apricot` is the key. Assigning to a dictionary uses the form: `dictionary[key] = value`.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbabbc66a9d0781091ab41025c9056f8784343ca
| 5,846 |
ipynb
|
Jupyter Notebook
|
Python Scope.ipynb
|
Vinaykumargond/Python-Tutorial
|
2bc90f0472c96d783250c715471fc35a703eeaf2
|
[
"Apache-2.0"
] | null | null | null |
Python Scope.ipynb
|
Vinaykumargond/Python-Tutorial
|
2bc90f0472c96d783250c715471fc35a703eeaf2
|
[
"Apache-2.0"
] | null | null | null |
Python Scope.ipynb
|
Vinaykumargond/Python-Tutorial
|
2bc90f0472c96d783250c715471fc35a703eeaf2
|
[
"Apache-2.0"
] | null | null | null | 18.5 | 249 | 0.497605 |
[
[
[
"# Python Scope",
"_____no_output_____"
]
],
[
[
"A variable is only available from inside the region it is created. This is called scope.",
"_____no_output_____"
]
],
[
[
"# Local Scope",
"_____no_output_____"
]
],
[
[
"A variable created inside a function belongs to the local scope of that function, and can only be used inside that function.",
"_____no_output_____"
]
],
[
[
"A variable created inside a function is available inside that function:",
"_____no_output_____"
]
],
[
[
"def myfunc():\n x = 300\n print(x)\n\nmyfunc()",
"300\n"
]
],
[
[
"# Function Inside Function",
"_____no_output_____"
]
],
[
[
"As explained in the example above, the variable x is not available outside the function, but it is available for any function inside the function:",
"_____no_output_____"
]
],
[
[
"The local variable can be accessed from a function within the function:",
"_____no_output_____"
]
],
[
[
"def myfunc():\n x = 300\n def myinnerfunc():\n print(x)\n myinnerfunc()\n\nmyfunc()",
"300\n"
]
],
[
[
"# Global Scope",
"_____no_output_____"
]
],
[
[
"A variable created in the main body of the Python code is a global variable and belongs to the global scope.",
"_____no_output_____"
],
[
"Global variables are available from within any scope, global and local.",
"_____no_output_____"
]
],
[
[
"A variable created outside of a function is global and can be used by anyone:",
"_____no_output_____"
]
],
[
[
"x = 300\n\ndef myfunc():\n print(x)\n\nmyfunc()\n\nprint(x)",
"300\n300\n"
]
],
[
[
"# Naming Variables",
"_____no_output_____"
]
],
[
[
"If you operate with the same variable name inside and outside of a function, Python will treat them as two separate variables, one available in the global scope (outside the function) and one available in the local scope (inside the function):",
"_____no_output_____"
]
],
[
[
"The function will print the local x, and then the code will print the global x:",
"_____no_output_____"
]
],
[
[
"x = 300\n\ndef myfunc():\n x = 200\n print(x)\n\nmyfunc()\n\nprint(x)",
"200\n300\n"
]
],
[
[
"# Global Keyword",
"_____no_output_____"
]
],
[
[
"If you need to create a global variable, but are stuck in the local scope, you can use the global keyword.",
"_____no_output_____"
],
[
"The global keyword makes the variable global.",
"_____no_output_____"
]
],
[
[
"If you use the global keyword, the variable belongs to the global scope:",
"_____no_output_____"
]
],
[
[
"def myfunc():\n global x\n x = 300\n\nmyfunc()\n\nprint(x)",
"300\n"
]
],
[
[
"Also, use the global keyword if you want to make a change to a global variable inside a function.",
"_____no_output_____"
],
[
"To change the value of a global variable inside a function, refer to the variable by using the global keyword:",
"_____no_output_____"
]
],
[
[
"x = 300\n\ndef myfunc():\n global x\n x = 200\n\nmyfunc()\n\nprint(x)",
"200\n"
]
]
] |
[
"markdown",
"raw",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"markdown",
"raw",
"markdown",
"code",
"raw",
"code"
] |
[
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw",
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"raw",
"raw"
],
[
"markdown"
],
[
"code"
],
[
"raw",
"raw"
],
[
"code"
]
] |
cbabbffad1da9f90ffc6a623821bb887c7623b46
| 3,715 |
ipynb
|
Jupyter Notebook
|
chatbot.ipynb
|
nlnadialigia/chatbot-inteligente
|
5a5795eaf95681f0e830ae6df13a83e271c8b334
|
[
"MIT"
] | null | null | null |
chatbot.ipynb
|
nlnadialigia/chatbot-inteligente
|
5a5795eaf95681f0e830ae6df13a83e271c8b334
|
[
"MIT"
] | null | null | null |
chatbot.ipynb
|
nlnadialigia/chatbot-inteligente
|
5a5795eaf95681f0e830ae6df13a83e271c8b334
|
[
"MIT"
] | null | null | null | 20.190217 | 77 | 0.49852 |
[
[
[
"## Criado um chatbot Inteligente",
"_____no_output_____"
]
],
[
[
"from chatterbot import ChatBot\nfrom chatterbot.trainers import ListTrainer",
"_____no_output_____"
],
[
"chatbot = ChatBot('BotNew')",
"[nltk_data] Downloading package averaged_perceptron_tagger to\n[nltk_data] /home/nlnadialigia/nltk_data...\n[nltk_data] Package averaged_perceptron_tagger is already up-to-\n[nltk_data] date!\n[nltk_data] Downloading package stopwords to\n[nltk_data] /home/nlnadialigia/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n"
],
[
"conversa = [\n 'Coe',\n 'E aí, tranquilo?',\n 'Tranquilo',\n 'Qual a boa de hoje?',\n 'A Hashtag tṕa ensinando Python e até chatbot',\n 'Caraca',\n 'Maneiro',\n 'Irado'\n]",
"_____no_output_____"
],
[
"trainer = ListTrainer(chatbot)",
"_____no_output_____"
],
[
"trainer.train(conversa)",
"List Trainer: [####################] 100%\n"
],
[
"chatbot.get_response('Qual a boa?')",
"_____no_output_____"
],
[
"chatbot.get_response('Hashtag')",
"_____no_output_____"
],
[
"chatbot.storage.drop()",
"_____no_output_____"
],
[
"while True:\n mensagem = input('Mande uma mensagem para o chatbot: ')\n if mensagem == 'parar':\n break\n resposta = chatbot.get_response(mensagem)\n print(resposta)\n ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbabc688563243a1aad900f10736a440f02da340
| 15,516 |
ipynb
|
Jupyter Notebook
|
Example/Psi4Numpy/01-Psi4Numpy-Basics/1f_tensor-manipulation.ipynb
|
yychuang/109-2-compchem-lite
|
cbf17e542f9447e89fb48de1b28759419ffff956
|
[
"BSD-3-Clause"
] | 214 |
2017-03-01T08:04:48.000Z
|
2022-03-23T08:52:04.000Z
|
Example/Psi4Numpy/01-Psi4Numpy-Basics/1f_tensor-manipulation.ipynb
|
yychuang/109-2-compchem-lite
|
cbf17e542f9447e89fb48de1b28759419ffff956
|
[
"BSD-3-Clause"
] | 100 |
2017-03-03T13:20:20.000Z
|
2022-03-05T18:20:27.000Z
|
Example/Psi4Numpy/01-Psi4Numpy-Basics/1f_tensor-manipulation.ipynb
|
yychuang/109-2-compchem-lite
|
cbf17e542f9447e89fb48de1b28759419ffff956
|
[
"BSD-3-Clause"
] | 150 |
2017-02-17T19:44:47.000Z
|
2022-03-22T05:52:43.000Z
| 37.387952 | 362 | 0.590358 |
[
[
[
"# Tensor Manipulation: Psi4 and NumPy manipulation routines\nContracting tensors together forms the core of the Psi4NumPy project. First let us consider the popluar [Einstein Summation Notation](https://en.wikipedia.org/wiki/Einstein_notation) which allows for very succinct descriptions of a given tensor contraction.\n\nFor example, let us consider a [inner (dot) product](https://en.wikipedia.org/wiki/Dot_product):\n$$c = \\sum_{ij} A_{ij} * B_{ij}$$\n\nWith the Einstein convention, all indices that are repeated are considered summed over, and the explicit summation symbol is dropped:\n$$c = A_{ij} * B_{ij}$$\n\nThis can be extended to [matrix multiplication](https://en.wikipedia.org/wiki/Matrix_multiplication):\n\\begin{align}\n\\rm{Conventional}\\;\\;\\; C_{ik} &= \\sum_{j} A_{ij} * B_{jk} \\\\\n\\rm{Einstein}\\;\\;\\; C &= A_{ij} * B_{jk} \\\\\n\\end{align}\n\nWhere the $C$ matrix has *implied* indices of $C_{ik}$ as the only repeated index is $j$.\n\nHowever, there are many cases where this notation fails. Thus we often use the generalized Einstein convention. To demonstrate let us examine a [Hadamard product](https://en.wikipedia.org/wiki/Hadamard_product_(matrices)):\n$$C_{ij} = \\sum_{ij} A_{ij} * B_{ij}$$\n\n\nThis operation is nearly identical to the dot product above, and is not able to be written in pure Einstein convention. The generalized convention allows for the use of indices on the left hand side of the equation:\n$$C_{ij} = A_{ij} * B_{ij}$$\n\nUsually it should be apparent within the context the exact meaning of a given expression.\n\nFinally we also make use of Matrix notation:\n\\begin{align}\n{\\rm Matrix}\\;\\;\\; \\bf{D} &= \\bf{A B C} \\\\\n{\\rm Einstein}\\;\\;\\; D_{il} &= A_{ij} B_{jk} C_{kl}\n\\end{align}\n\nNote that this notation is signified by the use of bold characters to denote matrices and consecutive matrices next to each other imply a chain of matrix multiplications! ",
"_____no_output_____"
],
[
"## Einsum\n\nTo perform most operations we turn to [NumPy's einsum function](https://docs.scipy.org/doc/numpy/reference/generated/numpy.einsum.html) which allows the Einsten convention as an input. In addition to being much easier to read, manipulate, and change, it is also much more efficient that a pure Python implementation.\n\nTo begin let us consider the construction of the following tensor (which you may recognize):\n$$G_{pq} = 2.0 * I_{pqrs} D_{rs} - 1.0 * I_{prqs} D_{rs}$$ \n\nFirst let us import our normal suite of modules:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport psi4\nimport time",
"_____no_output_____"
]
],
[
[
"We can then use conventional Python loops and einsum to perform the same task. Keep size relatively small as these 4-index tensors grow very quickly in size.",
"_____no_output_____"
]
],
[
[
"size = 20\n\nif size > 30:\n raise Exception(\"Size must be smaller than 30.\")\nD = np.random.rand(size, size)\nI = np.random.rand(size, size, size, size)\n\n# Build the fock matrix using loops, while keeping track of time\ntstart_loop = time.time()\nGloop = np.zeros((size, size))\nfor p in range(size):\n for q in range(size):\n for r in range(size):\n for s in range(size):\n Gloop[p, q] += 2 * I[p, q, r, s] * D[r, s]\n Gloop[p, q] -= I[p, r, q, s] * D[r, s]\n\ng_loop_time = time.time() - tstart_loop\n\n# Build the fock matrix using einsum, while keeping track of time\ntstart_einsum = time.time()\nJ = np.einsum('pqrs,rs', I, D, optimize=True)\nK = np.einsum('prqs,rs', I, D, optimize=True)\nG = 2 * J - K\n\neinsum_time = time.time() - tstart_einsum\n\n# Make sure the correct answer is obtained\nprint('The loop and einsum fock builds match: %s\\n' % np.allclose(G, Gloop))\n# Print out relative times for explicit loop vs einsum Fock builds\nprint('Time for loop G build: %14.4f seconds' % g_loop_time)\nprint('Time for einsum G build: %14.4f seconds' % einsum_time)\nprint('G builds with einsum are {:3.4f} times faster than Python loops!'.format(g_loop_time / einsum_time))",
"The loop and einsum fock builds match: True\n\nTime for loop G build: 0.4252 seconds\nTime for einsum G build: 0.0277 seconds\nG builds with einsum are 15.3570 times faster than Python loops!\n"
]
],
[
[
"As you can see, the einsum function is considerably faster than the pure Python loops and, in this author's opinion, much cleaner and easier to use.",
"_____no_output_____"
],
[
"## Dot\n\nNow let us turn our attention to a more canonical matrix multiplication example such as:\n$$D_{il} = A_{ij} B_{jk} C_{kl}$$\n\nWe could perform this operation using einsum; however, matrix multiplication is an extremely common operation in all branches of linear algebra. Thus, these functions have been optimized to be more efficient than the `einsum` function. The matrix product will explicitly compute the following operation:\n$$C_{ij} = A_{ij} * B_{ij}$$\n\nThis can be called with [NumPy's dot function](https://docs.scipy.org/doc/numpy/reference/generated/numpy.dot.html#numpy.dot).",
"_____no_output_____"
]
],
[
[
"size = 200\nA = np.random.rand(size, size)\nB = np.random.rand(size, size)\nC = np.random.rand(size, size)\n\n# First compute the pair product\ntmp_dot = np.dot(A, B)\ntmp_einsum = np.einsum('ij,jk->ik', A, B, optimize=True)\nprint(\"Pair product allclose: %s\" % np.allclose(tmp_dot, tmp_einsum))",
"Pair product allclose: True\n"
]
],
[
[
"Now that we have proved exactly what the dot product does, let us consider the full chain and do a timing comparison:",
"_____no_output_____"
]
],
[
[
"D_dot = np.dot(A, B).dot(C)\nD_einsum = np.einsum('ij,jk,kl->il', A, B, C, optimize=True)\nprint(\"Chain multiplication allclose: %s\" % np.allclose(D_dot, D_einsum))\n\nprint(\"\\nnp.dot time:\")\n%timeit np.dot(A, B).dot(C)\n\nprint(\"\\nnp.einsum time\")\n# no optimization here for illustrative purposes!\n%timeit np.einsum('ij,jk,kl->il', A, B, C)",
"Chain multiplication allclose: True\n\nnp.dot time:\n1.25 ms ± 255 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)\n\nnp.einsum time\n1.89 s ± 56.4 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
]
],
[
[
"On most machines the `np.dot` times are roughly ~2,000 times faster. The reason is twofold:\n - The `np.dot` routines typically call [Basic Linear Algebra Subprograms (BLAS)](https://en.wikipedia.org/wiki/Basic_Linear_Algebra_Subprograms). The BLAS routines are highly optimized and threaded versions of the code.\n - The `np.einsum` code will not factorize the operation by default; Thus, the overall cost is ${\\cal O}(N^4)$ (as there are four indices) rather than the factored $(\\bf{A B}) \\bf{C}$ which runs ${\\cal O}(N^3)$.\n \nThe first issue is difficult to overcome; however, the second issue can be resolved by the following:",
"_____no_output_____"
]
],
[
[
"print(\"np.einsum factorized time:\")\n# no optimization here for illustrative purposes!\n%timeit np.einsum('ik,kl->il', np.einsum('ij,jk->ik', A, B), C)",
"np.einsum factorized time:\n6.93 ms ± 294 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)\n"
]
],
[
[
"On most machines the factorized `einsum` expression is only ~10 times slower than `np.dot`. While a massive improvement, this is a clear demonstration the BLAS usage is usually recommended. It is a tradeoff between speed and readability. The Psi4NumPy project tends to lean toward `einsum` usage except in case where the benefit is too large to pass up.\n\nStarting in NumPy 1.12, the [einsum function](https://docs.scipy.org/doc/numpy/reference/generated/numpy.einsum.html) has a `optimize` flag which will automatically factorize the einsum code for you using a greedy algorithm, leading to considerable speedups at almost no cost:",
"_____no_output_____"
]
],
[
[
"print(\"\\nnp.einsum optimized time\")\n%timeit np.einsum('ij,jk,kl->il', A, B, C, optimize=True)",
"\nnp.einsum optimized time\n1.57 ms ± 210 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)\n"
]
],
[
[
"In this example, using `optimize=True` for automatic factorization is only 25% slower than `np.dot`. Furthermore, it is ~5 times faster than factorizing the expression by hand, which represents a very good trade-off between speed and readability. When unsure, `optimize=True` is strongly recommended.",
"_____no_output_____"
],
[
"## Complex tensor manipulations\nLet us consider a popular index transformation example:\n$$M_{pqrs} = C_{pi} C_{qj} I_{ijkl} C_{rk} C_{sl}$$\n\nHere, a naive `einsum` call would scale like $\\mathcal{O}(N^8)$ which translates to an extremely costly computation for all but the smallest $N$.",
"_____no_output_____"
]
],
[
[
"# Grab orbitals\nsize = 15\nif size > 15:\n raise Exception(\"Size must be smaller than 15.\")\n \nC = np.random.rand(size, size)\nI = np.random.rand(size, size, size, size)\n\n# Numpy einsum N^8 transformation.\nprint(\"\\nStarting Numpy's N^8 transformation...\")\nn8_tstart = time.time()\n# no optimization here for illustrative purposes!\nMO_n8 = np.einsum('pI,qJ,pqrs,rK,sL->IJKL', C, C, I, C, C)\nn8_time = time.time() - n8_tstart\nprint(\"...transformation complete in %.3f seconds.\" % (n8_time))\n\n# Numpy einsum N^5 transformation.\nprint(\"\\n\\nStarting Numpy's N^5 transformation with einsum...\")\nn5_tstart = time.time()\n# no optimization here for illustrative purposes!\nMO_n5 = np.einsum('pA,pqrs->Aqrs', C, I)\nMO_n5 = np.einsum('qB,Aqrs->ABrs', C, MO_n5)\nMO_n5 = np.einsum('rC,ABrs->ABCs', C, MO_n5)\nMO_n5 = np.einsum('sD,ABCs->ABCD', C, MO_n5)\nn5_time = time.time() - n5_tstart\nprint(\"...transformation complete in %.3f seconds.\" % n5_time)\nprint(\"\\nN^5 %4.2f faster than N^8 algorithm!\" % (n8_time / n5_time))\nprint(\"Allclose: %s\" % np.allclose(MO_n8, MO_n5))\n\n# Numpy einsum optimized transformation.\nprint(\"\\nNow Numpy's optimized transformation...\")\nn8_tstart = time.time()\nMO_n8 = np.einsum('pI,qJ,pqrs,rK,sL->IJKL', C, C, I, C, C, optimize=True)\nn8_time_opt = time.time() - n8_tstart\nprint(\"...optimized transformation complete in %.3f seconds.\" % (n8_time_opt))\n\n# Numpy GEMM N^5 transformation.\n# Try to figure this one out!\nprint(\"\\n\\nStarting Numpy's N^5 transformation with dot...\")\ndgemm_tstart = time.time()\nMO = np.dot(C.T, I.reshape(size, -1))\nMO = np.dot(MO.reshape(-1, size), C)\nMO = MO.reshape(size, size, size, size).transpose(1, 0, 3, 2)\n\nMO = np.dot(C.T, MO.reshape(size, -1))\nMO = np.dot(MO.reshape(-1, size), C)\nMO = MO.reshape(size, size, size, size).transpose(1, 0, 3, 2)\ndgemm_time = time.time() - dgemm_tstart\nprint(\"...transformation complete in %.3f seconds.\" % dgemm_time)\nprint(\"\\nAllclose: %s\" % np.allclose(MO_n8, MO))\nprint(\"N^5 %4.2f faster than N^8 algorithm!\" % (n8_time / dgemm_time))",
"\nStarting Numpy's N^8 transformation...\n...transformation complete in 26.988 seconds.\n\n\nStarting Numpy's N^5 transformation with einsum...\n...transformation complete in 0.004 seconds.\n\nN^5 6144.96 faster than N^8 algorithm!\nAllclose: True\n\nNow Numpy's optimized transformation...\n...optimized transformation complete in 0.002 seconds.\n\n\nStarting Numpy's N^5 transformation with dot...\n...transformation complete in 0.001 seconds.\n\nAllclose: True\nN^5 18744.20 faster than N^8 algorithm!\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbabdab066950b35c3532b5141139f6117c2c80a
| 113,253 |
ipynb
|
Jupyter Notebook
|
solutions/10_joint.ipynb
|
jonathonfletcher/BiteSizeBayes
|
6ef5c268deccdff3b3fa5fa6da6fca7945f3c38d
|
[
"MIT"
] | 116 |
2020-01-20T15:04:49.000Z
|
2022-03-28T07:42:33.000Z
|
solutions/10_joint.ipynb
|
jonathonfletcher/BiteSizeBayes
|
6ef5c268deccdff3b3fa5fa6da6fca7945f3c38d
|
[
"MIT"
] | 5 |
2020-02-02T14:12:50.000Z
|
2020-10-26T12:01:21.000Z
|
solutions/10_joint.ipynb
|
jonathonfletcher/BiteSizeBayes
|
6ef5c268deccdff3b3fa5fa6da6fca7945f3c38d
|
[
"MIT"
] | 28 |
2020-01-25T07:45:47.000Z
|
2022-02-16T13:29:43.000Z
| 84.517164 | 13,980 | 0.83943 |
[
[
[
"# Joint Probability",
"_____no_output_____"
],
[
"This notebook is part of [Bite Size Bayes](https://allendowney.github.io/BiteSizeBayes/), an introduction to probability and Bayesian statistics using Python.\n\nCopyright 2020 Allen B. Downey\n\nLicense: [Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0)](https://creativecommons.org/licenses/by-nc-sa/4.0/)",
"_____no_output_____"
],
[
"The following cell downloads `utils.py`, which contains some utility function we'll need.",
"_____no_output_____"
]
],
[
[
"from os.path import basename, exists\n\ndef download(url):\n filename = basename(url)\n if not exists(filename):\n from urllib.request import urlretrieve\n local, _ = urlretrieve(url, filename)\n print('Downloaded ' + local)\n\ndownload('https://github.com/AllenDowney/BiteSizeBayes/raw/master/utils.py')",
"_____no_output_____"
]
],
[
[
"If everything we need is installed, the following cell should run with no error messages.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Review\n\nSo far we have been working with distributions of only one variable. In this notebook we'll take a step toward multivariate distributions, starting with two variables.\n\nWe'll use cross-tabulation to compute a **joint distribution**, then use the joint distribution to compute **conditional distributions** and **marginal distributions**.\n\nWe will re-use `pmf_from_seq`, which I introduced in a previous notebook.",
"_____no_output_____"
]
],
[
[
"def pmf_from_seq(seq):\n \"\"\"Make a PMF from a sequence of values.\n \n seq: sequence\n \n returns: Series representing a PMF\n \"\"\"\n pmf = pd.Series(seq).value_counts(sort=False).sort_index()\n pmf /= pmf.sum()\n return pmf",
"_____no_output_____"
]
],
[
[
"## Cross tabulation\n\nTo understand joint distributions, I'll start with cross tabulation. And to demonstrate cross tabulation, I'll generate a dataset of colors and fruits.\n\nHere are the possible values.",
"_____no_output_____"
]
],
[
[
"colors = ['red', 'yellow', 'green']\nfruits = ['apple', 'banana', 'grape']",
"_____no_output_____"
]
],
[
[
"And here's a random sample of 100 fruits.",
"_____no_output_____"
]
],
[
[
"np.random.seed(2)\nfruit_sample = np.random.choice(fruits, 100, replace=True)",
"_____no_output_____"
]
],
[
[
"We can use `pmf_from_seq` to compute the distribution of fruits.",
"_____no_output_____"
]
],
[
[
"pmf_fruit = pmf_from_seq(fruit_sample)\npmf_fruit",
"_____no_output_____"
]
],
[
[
"And here's what it looks like.",
"_____no_output_____"
]
],
[
[
"pmf_fruit.plot.bar(color='C0')\n\nplt.ylabel('Probability')\nplt.title('Distribution of fruit');",
"_____no_output_____"
]
],
[
[
"Similarly, here's a random sample of colors.",
"_____no_output_____"
]
],
[
[
"color_sample = np.random.choice(colors, 100, replace=True)",
"_____no_output_____"
]
],
[
[
"Here's the distribution of colors.",
"_____no_output_____"
]
],
[
[
"pmf_color = pmf_from_seq(color_sample)\npmf_color",
"_____no_output_____"
]
],
[
[
"And here's what it looks like.",
"_____no_output_____"
]
],
[
[
"pmf_color.plot.bar(color='C1')\n\nplt.ylabel('Probability')\nplt.title('Distribution of colors');",
"_____no_output_____"
]
],
[
[
"Looking at these distributions, we know the proportion of each fruit, ignoring color, and we know the proportion of each color, ignoring fruit type.\n\nBut if we only have the distributions and not the original data, we don't know how many apples are green, for example, or how many yellow fruits are bananas.\n\nWe can compute that information using `crosstab`, which computes the number of cases for each combination of fruit type and color.",
"_____no_output_____"
]
],
[
[
"xtab = pd.crosstab(color_sample, fruit_sample, \n rownames=['color'], colnames=['fruit'])\nxtab",
"_____no_output_____"
]
],
[
[
"The result is a DataFrame with colors along the rows and fruits along the columns.",
"_____no_output_____"
],
[
"## Heatmap\n\nThe following function plots a cross tabulation using a pseudo-color plot, also known as a heatmap.\n\nIt represents each element of the cross tabulation with a colored square, where the color corresponds to the magnitude of the element.\n\nThe following function generates a heatmap using the Matplotlib function `pcolormesh`:",
"_____no_output_____"
]
],
[
[
"def plot_heatmap(xtab):\n \"\"\"Make a heatmap to represent a cross tabulation.\n \n xtab: DataFrame containing a cross tabulation\n \"\"\"\n\n plt.pcolormesh(xtab)\n\n # label the y axis\n ys = xtab.index\n plt.ylabel(ys.name)\n locs = np.arange(len(ys)) + 0.5\n plt.yticks(locs, ys)\n\n # label the x axis\n xs = xtab.columns\n plt.xlabel(xs.name)\n locs = np.arange(len(xs)) + 0.5\n plt.xticks(locs, xs)\n \n plt.colorbar()\n plt.gca().invert_yaxis()",
"_____no_output_____"
],
[
"plot_heatmap(xtab)",
"_____no_output_____"
]
],
[
[
"## Joint Distribution\n\nA cross tabulation represents the \"joint distribution\" of two variables, which is a complete description of two distributions, including all of the conditional distributions.\n\nIf we normalize `xtab` so the sum of the elements is 1, the result is a joint PMF:",
"_____no_output_____"
]
],
[
[
"joint = xtab / xtab.to_numpy().sum()\njoint",
"_____no_output_____"
]
],
[
[
"Each column in the joint PMF represents the conditional distribution of color for a given fruit.\n\nFor example, we can select a column like this:",
"_____no_output_____"
]
],
[
[
"col = joint['apple']\ncol",
"_____no_output_____"
]
],
[
[
"If we normalize it, we get the conditional distribution of color for a given fruit.",
"_____no_output_____"
]
],
[
[
"col / col.sum()",
"_____no_output_____"
]
],
[
[
"Each row of the cross tabulation represents the conditional distribution of fruit for each color.\n\nIf we select a row and normalize it, like this:",
"_____no_output_____"
]
],
[
[
"row = xtab.loc['red']\nrow / row.sum()",
"_____no_output_____"
]
],
[
[
"The result is the conditional distribution of fruit type for a given color.",
"_____no_output_____"
],
[
"## Conditional distributions\n\nThe following function takes a joint PMF and computes conditional distributions:",
"_____no_output_____"
]
],
[
[
"def conditional(joint, name, value):\n \"\"\"Compute a conditional distribution.\n \n joint: DataFrame representing a joint PMF\n name: string name of an axis\n value: value to condition on\n \n returns: Series representing a conditional PMF\n \"\"\"\n if joint.columns.name == name:\n cond = joint[value]\n elif joint.index.name == name:\n cond = joint.loc[value]\n return cond / cond.sum()",
"_____no_output_____"
]
],
[
[
"The second argument is a string that identifies which axis we want to select; in this example, `'fruit'` means we are selecting a column, like this:",
"_____no_output_____"
]
],
[
[
"conditional(joint, 'fruit', 'apple')",
"_____no_output_____"
]
],
[
[
"And `'color'` means we are selecting a row, like this:",
"_____no_output_____"
]
],
[
[
"conditional(joint, 'color', 'red')",
"_____no_output_____"
]
],
[
[
"**Exercise:** Compute the conditional distribution of color for bananas. What is the probability that a banana is yellow?",
"_____no_output_____"
]
],
[
[
"# Solution\n\ncond = conditional(joint, 'fruit', 'banana')\ncond",
"_____no_output_____"
],
[
"# Solution\n\ncond['yellow']",
"_____no_output_____"
]
],
[
[
"## Marginal distributions\n\nGiven a joint distribution, we can compute the unconditioned distribution of either variable.\n\nIf we sum along the rows, which is axis 0, we get the distribution of fruit type, regardless of color.",
"_____no_output_____"
]
],
[
[
"joint.sum(axis=0)",
"_____no_output_____"
]
],
[
[
"If we sum along the columns, which is axis 1, we get the distribution of color, regardless of fruit type.",
"_____no_output_____"
]
],
[
[
"joint.sum(axis=1)",
"_____no_output_____"
]
],
[
[
"These distributions are called \"[marginal](https://en.wikipedia.org/wiki/Marginal_distribution#Multivariate_distributions)\" because of the way they are often displayed. We'll see an example later.\n\nAs we did with conditional distributions, we can write a function that takes a joint distribution and computes the marginal distribution of a given variable:",
"_____no_output_____"
]
],
[
[
"def marginal(joint, name):\n \"\"\"Compute a marginal distribution.\n \n joint: DataFrame representing a joint PMF\n name: string name of an axis\n \n returns: Series representing a marginal PMF\n \"\"\"\n if joint.columns.name == name:\n return joint.sum(axis=0)\n elif joint.index.name == name:\n return joint.sum(axis=1)",
"_____no_output_____"
]
],
[
[
"Here's the marginal distribution of fruit.",
"_____no_output_____"
]
],
[
[
"pmf_fruit = marginal(joint, 'fruit')\npmf_fruit",
"_____no_output_____"
]
],
[
[
"And the marginal distribution of color:",
"_____no_output_____"
]
],
[
[
"pmf_color = marginal(joint, 'color')\npmf_color",
"_____no_output_____"
]
],
[
[
"The sum of the marginal PMF is the same as the sum of the joint PMF, so if the joint PMF was normalized, the marginal PMF should be, too.",
"_____no_output_____"
]
],
[
[
"joint.to_numpy().sum()",
"_____no_output_____"
],
[
"pmf_color.sum()",
"_____no_output_____"
]
],
[
[
"However, due to floating point error, the total might not be exactly 1.",
"_____no_output_____"
]
],
[
[
"pmf_fruit.sum()",
"_____no_output_____"
]
],
[
[
"**Exercise:** The following cells load the data from the General Social Survey that we used in Notebooks 1 and 2.",
"_____no_output_____"
]
],
[
[
"# Load the data file\nimport os\n\nif not os.path.exists('gss_bayes.csv'):\n !wget https://github.com/AllenDowney/BiteSizeBayes/raw/master/gss_bayes.csv",
"_____no_output_____"
],
[
"gss = pd.read_csv('gss_bayes.csv', index_col=0)",
"_____no_output_____"
]
],
[
[
"As an exercise, you can use this data to explore the joint distribution of two variables:\n\n* `partyid` encodes each respondent's political affiliation, that is, the party the belong to. [Here's the description](https://gssdataexplorer.norc.org/variables/141/vshow).\n\n* `polviews` encodes their political alignment on a spectrum from liberal to conservative. [Here's the description](https://gssdataexplorer.norc.org/variables/178/vshow).",
"_____no_output_____"
],
[
"The values for `partyid` are\n\n```\n0\tStrong democrat\n1\tNot str democrat\n2\tInd,near dem\n3\tIndependent\n4\tInd,near rep\n5\tNot str republican\n6\tStrong republican\n7\tOther party\n```",
"_____no_output_____"
],
[
"The values for `polviews` are:\n\n```\n1\tExtremely liberal\n2\tLiberal\n3\tSlightly liberal\n4\tModerate\n5\tSlightly conservative\n6\tConservative\n7\tExtremely conservative\n```",
"_____no_output_____"
],
[
"1. Make a cross tabulation of `gss['partyid']` and `gss['polviews']` and normalize it to make a joint PMF.\n\n2. Use `plot_heatmap` to display a heatmap of the joint distribution. What patterns do you notice?\n\n3. Use `marginal` to compute the marginal distributions of `partyid` and `polviews`, and plot the results.\n\n4. Use `conditional` to compute the conditional distribution of `partyid` for people who identify themselves as \"Extremely conservative\" (`polviews==7`). How many of them are \"strong Republicans\" (`partyid==6`)?\n\n5. Use `conditional` to compute the conditional distribution of `polviews` for people who identify themselves as \"Strong Democrat\" (`partyid==0`). How many of them are \"Extremely liberal\" (`polviews==1`)?",
"_____no_output_____"
]
],
[
[
"# Solution\n\nxtab2 = pd.crosstab(gss['partyid'], gss['polviews'])\njoint2 = xtab2 / xtab2.to_numpy().sum()",
"_____no_output_____"
],
[
"# Solution\n\nplot_heatmap(joint2)\n\nplt.xlabel('polviews')\nplt.title('Joint distribution of polviews and partyid');",
"_____no_output_____"
],
[
"# Solution\n\nmarginal(joint2, 'polviews').plot.bar(color='C2')\n\nplt.ylabel('Probability')\nplt.title('Distribution of polviews');",
"_____no_output_____"
],
[
"# Solution\n\nmarginal(joint2, 'polviews').plot.bar(color='C3')\n\nplt.ylabel('Probability')\nplt.title('Distribution of polviews');",
"_____no_output_____"
],
[
"# Solution\n\ncond1 = conditional(joint2, 'polviews', 7)\ncond1.plot.bar(label='Extremely conservative', color='C4')\n\nplt.ylabel('Probability')\nplt.title('Distribution of partyid')\n\ncond1[6]",
"_____no_output_____"
],
[
"# Solution\n\ncond2 = conditional(joint2, 'partyid', 0)\ncond2.plot.bar(label='Strong democrat', color='C6')\n\nplt.ylabel('Probability')\nplt.title('Distribution of polviews')\n\ncond2[1]",
"_____no_output_____"
]
],
[
[
"## Review\n\nIn this notebook we started with cross tabulation, which we normalized to create a joint distribution, which describes the distribution of two (or more) variables and all of their conditional distributions.\n\nWe used heatmaps to visualize cross tabulations and joint distributions.\n\nThen we defined `conditional` and `marginal` functions that take a joint distribution and compute conditional and marginal distributions for each variables.\n\nAs an exercise, you had a chance to apply the same methods to explore the relationship between political alignment and party affiliation using data from the General Social Survey.\n\nYou might have noticed that we did not use Bayes's Theorem in this notebook. [In the next notebook](https://colab.research.google.com/github/AllenDowney/BiteSizeBayes/blob/master/11_faceoff.ipynb) we'll take the ideas from this notebook and apply them Bayesian inference.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbabdecafb2fcaa0abd1f95e8d1f8d6a17965ccb
| 148,867 |
ipynb
|
Jupyter Notebook
|
notebooks/knn.ipynb
|
srensi/tmprss2
|
639ccafedd0000b364ee07ffa41fc036e1996337
|
[
"MIT"
] | 8 |
2020-05-12T23:11:11.000Z
|
2022-03-20T23:29:46.000Z
|
notebooks/knn.ipynb
|
srensi/tmprss2
|
639ccafedd0000b364ee07ffa41fc036e1996337
|
[
"MIT"
] | 25 |
2020-05-19T20:09:19.000Z
|
2020-09-04T17:57:48.000Z
|
notebooks/knn.ipynb
|
srensi/tmprss2
|
639ccafedd0000b364ee07ffa41fc036e1996337
|
[
"MIT"
] | 7 |
2020-05-05T23:07:49.000Z
|
2021-09-01T02:42:51.000Z
| 169.745724 | 25,600 | 0.886402 |
[
[
[
"# KNN\nHere we use K Nearest Neighbors algorithm to perform classification and regression",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport sys\nimport pandas as pd\n\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn.model_selection import train_test_split\nfrom rdkit import Chem, DataStructs\nfrom sklearn.neighbors import KNeighborsRegressor, KNeighborsClassifier\nfrom tqdm.notebook import tqdm",
"_____no_output_____"
]
],
[
[
"## Load Data",
"_____no_output_____"
]
],
[
[
"# training data:\nassays = pd.read_pickle('../processed_data/combined_dataset.pkl')\nassays = assays[assays.activity_target.isin(['Active', 'Inactive'])] # get rid of any 'Inconclusive'\nassays = assays.dropna(subset=['acvalue_scaled_to_tmprss2']) # only use data that could be scaled\n\ndcm = pd.read_pickle('../processed_data/DarkChemicalMatter_processed.pkl.gz')\n\n# testing data:\nscreening_data = pd.read_pickle('../processed_data/screening_data_processed.pkl')",
"_____no_output_____"
]
],
[
[
"# Classification",
"_____no_output_____"
],
[
"## Load training data",
"_____no_output_____"
]
],
[
[
"# set up features (X) and labels (y) for knn\nX_assays = np.stack(assays.morgan_fingerprint)\ny_assays = assays.activity_target.values\nassays_hist = plt.hist(y_assays)",
"_____no_output_____"
],
[
"X_dcm = np.stack(dcm.sample(frac=.1).morgan_fingerprint)\ny_dcm = ['Inactive'] * len(X_dcm)\ndcm_hist = plt.hist(y_dcm)",
"_____no_output_____"
]
],
[
[
"### Validation",
"_____no_output_____"
]
],
[
[
"# make a validation set out of some of the assays and some of the dcm\npercent_test_assays = .3 # Make the val set a bit less skewed than the train set\npercent_test_dcm = .1 #\nrandom_state = 3 # for reproducibility of train/val split\n\ntrain_X_assays, val_X_assays, train_y_assays, test_y_assays = train_test_split(X_assays, y_assays, test_size=percent_test_assays, random_state=random_state)\ntrain_X_dcm, val_X_dcm, train_y_dcm, test_y_dcm = train_test_split(X_dcm, y_dcm, test_size=percent_test_dcm, random_state=random_state)\n\nplt.figure()\nplt.bar(['assays', 'dcm'], [len(train_X_assays), len(train_X_dcm)])\nplt.title('training data')\n\nplt.figure()\nplt.bar(['assays', 'dcm'], [len(val_X_assays), len(val_X_dcm)])\nplt.title('val data')\n\ntrain_X = np.concatenate([train_X_assays, train_X_dcm], axis=0)\nval_X = np.concatenate([val_X_assays, val_X_dcm], axis=0)\ntrain_y = np.concatenate([train_y_assays, train_y_dcm], axis=0)\ntest_y = np.concatenate([test_y_assays, test_y_dcm], axis=0)",
"_____no_output_____"
]
],
[
[
"## Optimize KNN Classifier using Validation Data",
"_____no_output_____"
]
],
[
[
"# optimize knn, test a couple ks\n\nks = np.arange(1, 14, 2)\n\naccuracies = []\nactive_accuracies = []\ninactive_accuracies = []\nfor k in tqdm(ks):\n nbrs = KNeighborsClassifier(n_neighbors=k, metric='jaccard', algorithm='ball_tree', n_jobs=32)\n nbrs.fit(train_X, train_y)\n pred = nbrs.predict(val_X)\n accuracies.append(np.count_nonzero(pred == test_y) / len(test_y))\n if np.count_nonzero(test_y == 'Inactive') == 0:\n inactive_accuracies.append(1) # all inactive classified correctly: vacuously true\n else:\n inactive_accuracies.append(np.count_nonzero((pred == test_y) & (pred == 'Inactive')) / np.count_nonzero(test_y == 'Inactive'))\n if np.count_nonzero(test_y == 'Active') == 0:\n active_accuracies.append(1)\n else:\n active_accuracies.append(np.count_nonzero((pred == test_y) & (test_y == 'Active')) / np.count_nonzero(test_y == 'Active'))\n\nplt.figure()\nplt.plot(ks, accuracies, label='overall')\nplt.plot(ks, active_accuracies, label='active')\nplt.plot(ks, inactive_accuracies, label='inactive')\nplt.xlabel(\"k\")\nplt.ylabel(\"accuracy\")\nplt.title('Classification Accuracy')\nplt.legend()",
"_____no_output_____"
]
],
[
[
"From the above experiment, we can see that k=5 does the best on active compounds; we'll choose this.",
"_____no_output_____"
],
[
"## Test on the Screening Data",
"_____no_output_____"
]
],
[
[
"# set up train and test\nX_train = np.concatenate([X_assays, X_dcm])\ny_train = np.concatenate([y_assays, y_dcm])\n\nX_test = np.stack(screening_data.morgan_fingerprint)\n\nprint(\"Training set size:\", len(X_train))\nprint(\"Test set size:\", len(X_test))",
"Training set size: 14359\nTest set size: 21011\n"
],
[
"nbrs = KNeighborsClassifier(n_neighbors=3, metric='jaccard', algorithm='ball_tree', weights='distance', n_jobs=32) # turns out it gets much faster with many jobs (even 8x more jobs than my laptop's 4 physical cores). 64 is slower than 32 though, overhead catches up I guess.\nnbrs.fit(train_X, train_y)\n\n# chunk the test set in order to get some sense of progress\npred_activity = []\nfor test_chunk in tqdm(np.array_split(X_test, 100)):\n pred_activity.append(nbrs.predict(test_chunk))\npred_activity = np.concatenate(pred_activity)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(1, 2, figsize=(8, 3))\nax[0].hist(y_train)\nax[0].set_title('training labels')\nax[1].hist(pred_activity)\nax[1].set_title('predicted labels')\nt = plt.suptitle('Label Distributions')",
"_____no_output_____"
]
],
[
[
"We can see the screening data mostly comes back as inactive. The distribution is similar to the training distribution, which could mean the model is biased by the training distribution, but this isn't necessarily true. Could use a test with different training data distribution to see.",
"_____no_output_____"
],
[
"# Regression\nNow that we have identified active compounds out of the screening data, we can regress the activity of these compounds using our assay data.",
"_____no_output_____"
],
[
"## Validation\n### Load Train Data\nFeatures are still morgan fingerprints, labels are log activity values. Where available, the activity values are scaled to tmprss2 based on correlation between target activities. Where correlation was unavailable, activity values are unscaled.\n",
"_____no_output_____"
]
],
[
[
"X_assays = np.stack(assays.morgan_fingerprint)\ny_assays = np.log10(assays.acvalue_scaled_to_tmprss2)\nassert y_assays.isna().sum() == 0",
"_____no_output_____"
]
],
[
[
"### Regression Cross-Validation",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import cross_val_score\nks = np.arange(1, 23, 2)\nRMSE = []\nfor k in tqdm(ks):\n knn_cv = KNeighborsRegressor(n_neighbors=k, metric='jaccard', weights='distance')\n RMSE.append(-cross_val_score(knn_cv, X_assays, y_assays, cv=10, scoring='neg_root_mean_squared_error'))\nplt.plot(ks, RMSE, '.')\nplt.plot(ks, np.median(RMSE, axis=1), label='median')\nplt.plot(ks, np.mean(RMSE, axis=1), label='mean')\nplt.xticks(ks)\nplt.legend()\nplt.ylabel('RMSE')\nplt.xlabel('k')\nplt.title('10-fold Cross Validation')",
"_____no_output_____"
]
],
[
[
"From the cross-validation, it seems k=7 is a reasonable choice.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import mean_squared_error\nbest_k_regr = 7\nbest_RMSE = np.median(RMSE, axis=1)[3]\n\nX_train, X_test, y_train, y_test = train_test_split(X_assays, y_assays, test_size=.25, random_state=1)\nnbrs = KNeighborsRegressor(n_neighbors=best_k_regr, metric='jaccard', weights='distance')\nnbrs.fit(X_train, y_train)\ny_pred = nbrs.predict(X_test)\n\nplt.plot(y_test, y_pred, '.')\nplt.xlabel('True Activity Values (log)')\nplt.ylabel('Predicted Activity Values (log)')\nbnds = [np.min([y_test, y_pred])*1.1, np.max([y_test, y_pred])*1.2]\nplt.axis('square')\nplt.xlim(bnds)\nplt.ylim(bnds)\nplt.plot(np.linspace(*bnds), np.linspace(*bnds), 'k--', label='y=x')\nplt.legend()\nplt.title('Sample Validation (3/4 train, 1/4 test)')\nprint(f'RMSE={mean_squared_error(y_test, y_pred, squared=False)}')",
"RMSE=0.7764739168696334\n"
]
],
[
[
"Here you can see that the distribution as a whole looks good, but the accuracy in the low-end is poor. Since we care about the low end, this is concerning.",
"_____no_output_____"
],
[
"### Load Test Data\nThe test data consists of all the screening molecules which were marked 'active' in classification above.",
"_____no_output_____"
]
],
[
[
"active_screening_data = screening_data[pred_activity=='Active'].copy()\nX_test_active = np.stack(active_screening_data.morgan_fingerprint)",
"_____no_output_____"
],
[
"nbrs = KNeighborsRegressor(n_neighbors=best_k_regr, metric='jaccard', weights='distance')\nnbrs.fit(X_assays, y_assays)\npred_acvalue = nbrs.predict(X_test_active)\n\nactive_screening_data.insert(loc=2, column='predicted_acvalue(log10)', value=pred_acvalue)",
"_____no_output_____"
],
[
"# a look at the predicted activity distributions by dataset source:\nfrom seaborn import violinplot\nviolinplot(x='source', y='predicted_acvalue(log10)', data=active_screening_data)",
"_____no_output_____"
],
[
"# and the top hits!\nactive_screening_data.sort_values(by='predicted_acvalue(log10)', inplace=True)\nactive_screening_data['name'] = active_screening_data.name.str.upper()\nactive_screening_data.drop(columns=['morgan_fingerprint'], inplace=True)\nactive_screening_data.drop_duplicates(subset=['name'], inplace=True)\nactive_screening_data.head(20)",
"_____no_output_____"
]
],
[
[
"Nafamostat comes in on top, which is reassuring. The rest of the results... unclear. Not a ton of trust in the KNN, the large errors in the low-end on the regression test is concerning.",
"_____no_output_____"
]
],
[
[
"# store the results!\nactive_screening_data['RMSE'] = best_RMSE\nactive_screening_data.to_csv('../results/knn_results.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbabe681990b9580c3132a30061d8dc88d2af233
| 36,971 |
ipynb
|
Jupyter Notebook
|
docs/tutorials/dispersion_micropolar.ipynb
|
nicoguaro/continuum_mechanics
|
f8149b69b8461784f6ed721294cd1a49ffdfa3d7
|
[
"MIT"
] | 21 |
2018-12-09T15:02:51.000Z
|
2022-02-16T09:28:38.000Z
|
docs/tutorials/dispersion_micropolar.ipynb
|
nicoguaro/continuum_mechanics
|
f8149b69b8461784f6ed721294cd1a49ffdfa3d7
|
[
"MIT"
] | 223 |
2019-05-06T16:31:50.000Z
|
2022-03-31T21:21:03.000Z
|
docs/tutorials/dispersion_micropolar.ipynb
|
nicoguaro/continuum_mechanics
|
f8149b69b8461784f6ed721294cd1a49ffdfa3d7
|
[
"MIT"
] | 7 |
2020-01-29T10:03:52.000Z
|
2022-02-25T19:34:37.000Z
| 81.97561 | 7,312 | 0.726245 |
[
[
[
"# Dispersion relations in a micropolar medium",
"_____no_output_____"
],
[
"We are interested in computing the dispersion relations in a\nhomogeneous micropolar solid.",
"_____no_output_____"
],
[
"## Wave propagation in micropolar solids",
"_____no_output_____"
],
[
"The equations of motion for a micropolar solid are given by [[1, 2]](#References)\n\n\\begin{align}\n&c_1^2\n\\nabla\\nabla\\cdot\\mathbf{u}- c_2^2\\nabla\\times\\nabla\\times\\mathbf{u} + K^2\\nabla\\times\\boldsymbol{\\theta} = -\\omega^2 \\mathbf{u} \\, ,\\\\\n&c_3^2 \\nabla\\nabla\\cdot\\boldsymbol{\\theta} - c_4^2\\nabla\\times\\nabla\\times\\boldsymbol{\\theta} + Q^2\\nabla\\times\\mathbf{u} - 2Q^2\\boldsymbol{\\theta} = -\\omega^2 \\boldsymbol{\\theta} \\, \n\\end{align}\n\n\nwhere $\\mathbf{u}$ is the displacement vector and $\\boldsymbol{\\theta}$ is the microrrotations vector,\nand where: $c_1$ represents the phase/group speed for the longitudinal wave \n($P$) that is non-dispersive as in the classical case, $c_2$ represents the \nhigh-frequency limit phase/group speed for a transverse wave ($S$) that is dispersive unlike the classical counterpart, $c_3$ represents the high-frequency limit phase/group speed \nfor a longitudinal-rotational wave ($LR$) with a corkscrew-like motion that is \ndispersive and does not have a classical counterpart, $c_4$ represents \nthe high-frequency limit phase/group speed for a transverse-rotational wave ($TR$) that is dispersive and does not have a classical counterpart, $Q$ represents the cut-off frequency for rotational \nwaves appearance, and $K$ quantifies the difference between the low-frequency \nand high-frequency phase/group speed for the S-wave. These parameters are defined by:\n\n\\begin{align}\nc_1^2 = \\frac{\\lambda +2\\mu}{\\rho},\\quad &c_3^2 =\\frac{\\beta + 2\\eta}{J},\\\\\nc_2^2 = \\frac{\\mu +\\alpha}{\\rho},\\quad &c_4^2 =\\frac{\\eta + \\varepsilon}{J},\\\\\nQ^2= \\frac{2\\alpha}{J},\\quad &K^2 =\\frac{2\\alpha}{\\rho} \\, ,\n\\end{align}\n\n\n",
"_____no_output_____"
],
[
"## Dispersion relations",
"_____no_output_____"
],
[
"To identify types of propagating waves that can arise in the micropolar medium \nit is convenient to expand the displacement and rotation vectors in terms of \nscalar and vector potentials\n\n\\begin{align}\n\\mathbf{u} &= \\nabla \\phi + \\nabla\\times\\boldsymbol{\\Gamma}\\, ,\\\\\n\\boldsymbol{\\theta} &= \\nabla \\tau + \\nabla\\times\\mathbf{E}\\, ,\n\\end{align}\n\nsubject to the conditions:\n\n\\begin{align}\n&\\nabla\\cdot\\boldsymbol{\\Gamma} = 0\\\\\n&\\nabla\\cdot\\mathbf{E} = 0\\, .\n\\end{align}\n\nUsing the above in the displacements equations of motion yields the following\nequations, after some manipulations\n\n\\begin{align}\nc_1^2 \\nabla^2 \\phi &= \\frac{\\partial^2 \\phi}{\\partial t^2}\\, ,\\\\\nc_3^2 \\nabla^2 \\tau - 2Q^2\\tau &= \\frac{\\partial^2 \\tau}{\\partial t^2}\\, ,\\\\\n\\begin{bmatrix}\nc_2^2 \\nabla^2 &K^2\\nabla\\times\\, ,\\\\\nQ^2\\nabla\\times &c_4^2\\nabla^2 - 2Q^2\n\\end{bmatrix}\n\\begin{Bmatrix} \\boldsymbol{\\Gamma}\\\\ \\mathbf{E}\\end{Bmatrix} &=\n\\frac{\\partial^2}{\\partial t^2} \\begin{Bmatrix} \\boldsymbol{\\Gamma}\\\\ \\mathbf{E}\\end{Bmatrix} \\, ,\n\\end{align}\n\n\nwhere we can see that the equations for the scalar potentials are uncoupled,\nwhile the ones for the vector potentials are coupled.\n\nWriting the vector potentials as plane waves of amplitude $ \\mathbf{A}$ and $ \n\\mathbf{B}$, wave number $\\kappa$ and circular frequency $\\omega$ that propagate \nalong the \\(x\\) axis, \n\n\\begin{align}\n\\boldsymbol{\\Gamma} &= \\mathbf{A}\\exp(i\\kappa x - i\\omega t)\\\\\n\\mathbf{E} &= \\mathbf{B}\\exp(i\\kappa x - i\\omega t)\\, .\n\\end{align}\n\nWe can do these calculations using some the functions available functions in the package.",
"_____no_output_____"
]
],
[
[
"from sympy import Matrix, diff, symbols, exp, I, sqrt\nfrom sympy import simplify, expand, solve, limit\nfrom sympy import init_printing, pprint, factor\nfrom continuum_mechanics.vector import lap_vec, curl, div",
"_____no_output_____"
],
[
"init_printing()",
"_____no_output_____"
],
[
"A1, A2, A3, B1, B2, B3 = symbols(\"A1 A2 A3 B1 B2 B3\")\nkappa, omega, t, x = symbols(\"kappa omega t x\")\nc1, c2, c3, c4, K, Q = symbols(\"c1 c2 c3 c4 K Q\", positive=True)",
"_____no_output_____"
]
],
[
[
"We define the vector potentials $\\boldsymbol{\\Gamma}$ and $\\mathbf{E}$.",
"_____no_output_____"
]
],
[
[
"Gamma = Matrix([A1, A2, A3]) * exp(I*kappa*x - I*omega*t)\nE = Matrix([B1, B2, B3]) * exp(I*kappa*x - I*omega*t)",
"_____no_output_____"
]
],
[
[
"And compute the equations using the vector operators. Namely,\nthe Laplace ([`vector.lap_vec()`](https://continuum-mechanics.readthedocs.io/en/latest/modules.html#vector.lap_vec) and the curl\n([`vector.curl()`](https://continuum-mechanics.readthedocs.io/en/latest/modules.html#vector.curl))\noperators.",
"_____no_output_____"
]
],
[
[
"eq1 = c2**2 * lap_vec(Gamma) + K**2*curl(E) - Gamma.diff(t, 2)\neq2 = Q**2 * curl(Gamma) + c4**2*lap_vec(E) - 2*Q**2*E - E.diff(t, 2)\neq1 = simplify(eq1/exp(I*kappa*x - I*omega*t))\neq2 = simplify(eq2/exp(I*kappa*x - I*omega*t))\neq = eq1.col_join(eq2)",
"_____no_output_____"
]
],
[
[
"We can compute the matrix for the system using [`.jacobian()`](https://docs.sympy.org/1.5.1/modules/matrices/matrices.html#sympy.matrices.matrices.MatrixCalculus.jacobian) ",
"_____no_output_____"
]
],
[
[
"M = eq.jacobian([A1, A2, A3, B1, B2, B3])\nM",
"_____no_output_____"
]
],
[
[
"And, we are interested in the determinant of the matrix $M$.",
"_____no_output_____"
]
],
[
[
"factor(M.det())",
"_____no_output_____"
]
],
[
[
"The roots for this polynomial (in $\\omega^2$) represent the dispersion\nrelations.",
"_____no_output_____"
]
],
[
[
"disps = solve(M.det(), omega**2)\nfor disp in disps:\n display(disp)",
"_____no_output_____"
]
],
[
[
"## References",
"_____no_output_____"
],
[
"1. Nowacki, W. (1986). Theory of asymmetric elasticity. Pergamon Press, Headington Hill Hall, Oxford OX 3 0 BW, UK, 1986.\n\n2. Guarín-Zapata, N., Gomez, J., Valencia, C., Dargush, G. F., & Hadjesfandiari, A. R. (2020). Finite element modeling of micropolar-based phononic crystals. Wave Motion, 92, 102406.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbabf0f1cb354841365de90650b479ed13547594
| 83,421 |
ipynb
|
Jupyter Notebook
|
Magno-Valeriano-Price-Pattern-Detection-Prediction-Copy1.ipynb
|
dabideee13/Price-Pattern-Prediction
|
632d961fc08777adab8eeb7ecbf16ac7cc71a3a7
|
[
"MIT"
] | null | null | null |
Magno-Valeriano-Price-Pattern-Detection-Prediction-Copy1.ipynb
|
dabideee13/Price-Pattern-Prediction
|
632d961fc08777adab8eeb7ecbf16ac7cc71a3a7
|
[
"MIT"
] | null | null | null |
Magno-Valeriano-Price-Pattern-Detection-Prediction-Copy1.ipynb
|
dabideee13/Price-Pattern-Prediction
|
632d961fc08777adab8eeb7ecbf16ac7cc71a3a7
|
[
"MIT"
] | null | null | null | 60.93572 | 5,194 | 0.589372 |
[
[
[
"\n#### A multiclass classification problem\n\nby Aries P. Valeriano and Dave Emmanuel Q. Magno",
"_____no_output_____"
],
[
"## Executive Summary\n\nThe goal of this project is to a build a prediction model that make use of stock chart pattern, in particular double top to predict the next movement of stock price if it will decrease further, increase, or stay still within the next 10 days. If successful, traders can use this prediction model to make a data driven decision on their next trade.\n\nTo achieve this goal, we will create our own dataset first. This can be done by determining the minima and maxima of the time series dataset for every stock from various industry. Then, we will refer to it to detect the double top stock chart pattern. There are 5 points/prices that consists of maxima and minima that forms the pattern, these will become the descriptive features that we will use to predict the target feature which is the movement of stock price within the next 10 days. We also include the indexes of the start and end of the pattern, as well as the industry of the stocks where the pattern occurs.\n\nNow that we have the dataset. We will perform data exploration to visualize the double top pattern and verify the multicollinearity of the descriptive features because obviously they are correlated to each other since each are part of double top pattern. Moreover, we performed also feature engineer to get additional features that could help increase the predictive accuracy of the model.\n\nAfter data exploration, we proceed to predictive modelling. Note that the target feature consists of 3 levels. Thus, we will fit multiclass models to our dataset such as multiclass KNN, multinomial logistic regression, multiclass SVM etc.. Fortunately, this can be done simultaneously by using pycaret machine learning library, moreover it automatically split the dataset which we set to 80/20, and further perform data exploration such as normalization, in which we set it as minmax scaler, one hat encoding for nominal feature (industry). And as a result, the model the produces the highest F1 score is the gradient boosting classifier, a sequential ensemble approach with 0.5217 score. This imply that with 52.17% accuracy, we can predict the movement of stocks prices within 10 days after the double top stock chart pattern happen.",
"_____no_output_____"
],
[
"## Jupyter Display Settings",
"_____no_output_____"
]
],
[
[
"%%javascript\nIPython.OutputArea.prototype._should_scroll = function(lines) {\n return false;\n}",
"_____no_output_____"
],
[
"from IPython.core.display import HTML\nHTML(\"\"\"\n<style>\n.output_png {\n display: table-cell;\n text-align: center;\n vertical-align: middle;\n}\n</style>\n\"\"\")",
"_____no_output_____"
]
],
[
[
"## Prerequisites",
"_____no_output_____"
]
],
[
[
"from collections import Counter\nfrom typing import Union\nfrom itertools import combinations\n\nfrom pycaret.classification import *\nfrom sklearn.datasets import dump_svmlight_file\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.model_selection import RepeatedKFold, train_test_split\nfrom sklearn.metrics import precision_score\nfrom imblearn.under_sampling import NearMiss\nimport xgboost as xgb\nfrom xgboost import XGBClassifier\n\nimport pandas as pd\nfrom dfply import *\n\nimport plotly.express as px\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nsns.set()\n%matplotlib inline\n\nfrom price_detection_tools import import_",
"_____no_output_____"
]
],
[
[
"## Tools",
"_____no_output_____"
]
],
[
[
"@dfpipe\ndef extract_date(df_: pd.DataFrame) -> pd.DataFrame:\n df = df_.copy(deep=True)\n for i, col_name in zip(range(2), ['dateF', 'dateE']):\n df[col_name] = df.date.apply(lambda x: x[i])\n return df\n\n@dfpipe\ndef drop_(df_: pd.DataFrame) -> pd.DataFrame:\n to_drop = ['fw_ret_1', 'fw_ret_2', 'fw_ret_3']\n return df_.drop([to_drop])\n\n@dfpipe\ndef get_average(df_: pd.DataFrame) -> pd.DataFrame:\n df = df_.copy(deep=True)\n f1_idx = df.columns.get_loc('f1')\n f5_idx = df.columns.get_loc('f5')\n averages = df.iloc[:, f1_idx:f5_idx + 1].mean(axis=1)\n averages.rename('averages', inplace=True)\n return pd.concat([df_, averages], axis=1)\n\ndef lump_categories(data: Union[pd.DataFrame,\n pd.Series],\n percentage: float = 0.010):\n \n def set_threshold(df: pd.DataFrame,\n percentage: float = 0.010) -> float:\n \"\"\"Sets threshold to be a percentage of the shape\n of dataframe.\"\"\"\n return df.shape[0] * percentage\n\n if isinstance(data, pd.DataFrame): pass\n if isinstance(data, pd.Series):\n data = data.to_frame()\n return data.apply(lambda x: x.mask(\n x.map(x.value_counts()) < set_threshold(df_copy),\n 'Others'))\n\ndef encode_label(series: pd.Series):\n label_encoder = LabelEncoder()\n label_encoder.fit(series)\n return label_encoder.transform(series)\n\ndef get_height(fA_: pd.Series,\n fB_: pd.Series) -> pd.DataFrame:\n return np.abs(fA_ - fB_)\n\n@dfpipe\ndef add_height_features(df_: pd.DataFrame) -> pd.DataFrame:\n df = df_.copy(deep=True)\n heights = ['h{}'.format(i + 1) for i in range(10)]\n feats = ['f{}'.format(i + 1) for i in range(5)]\n \n for height, comb in zip(heights,\n combinations(feats, 2)):\n df[height] = get_height(df[comb[0]],\n df[comb[1]])\n return df",
"_____no_output_____"
]
],
[
[
"## Data Description\n\nIt contains a target feature (label) that have 3 levels, decrease \"1\", neutral \"2\", increase \"3\" and 8 descriptive features in which 5 of it (f1, f2, f3, f4, f5) consist of minima and maxima that forms a double top stock chart pattern, 2 of it (dateF, dateE) are the indexes of the start and end of the pattern lastly, industry of the stock where the pattern occur.\n\nHowever, before arriving at this dataset, web scraping of stocks historical dataset for various industries at https://finance.yahoo.com/ are performed, see above table, then closing price from it was utilize. Moreover, minima and maxima of time series dataset for every stock were determined. Next, detection of double top stock chart pattern by setting threshold for the minima and maxima that forms the pattern, and lastly, determine its corresponding target feature by comparing the highest maxima or lowest minima within the pattern and the maximum or minimum prices within the next 10 days after the pattern is observed. Target is labeled increase \"2\" if the maximum price within the next 10 days is greater than the highest maxima within the pattern, decrease \"0\" if the minimum price within the next 10 days is lesser than the lowest minima within the pattern, neutral \"0\" if neither or both happens. ",
"_____no_output_____"
],
[
"## Data Exploration and Preprocessing\n\nIn here, we have done feature engineer. Created 3 additional descriptive features, which are the the absolute difference between f1 and f3 , also between f3 and f5, then we took the sum of the values from f1 to f5. These features were named d1, d2, and sum respectively. These additional features will help increase the predictive accuracy of the model built later on.",
"_____no_output_____"
]
],
[
[
"df = import_('trial.csv').drop(['increment', 'ema', 'window'], axis=1)",
"_____no_output_____"
],
[
"df_copy = df.copy(deep=True)",
"_____no_output_____"
]
],
[
[
"#### Label encoding of 'industry'",
"_____no_output_____"
],
[
"#### Lump categories",
"_____no_output_____"
]
],
[
[
"df_copy['industry_lumped'] = lump_categories(df_copy['industry'])",
"_____no_output_____"
],
[
"df_copy = df_copy.drop('industry', axis=1)",
"_____no_output_____"
],
[
"df_copy['industry_coded'] = encode_label(df_copy['industry_lumped'])",
"_____no_output_____"
],
[
"df_copy = df_copy.drop('industry_lumped', axis=1)",
"_____no_output_____"
],
[
"data = (df_copy >>\n extract_date >>\n drop(['date']) >>\n drop(['fw_ret_1', \n 'fw_ret_2',\n 'fw_ret_3']) >>\n add_height_features)",
"_____no_output_____"
]
],
[
[
"#### Get the size or width of the whole pattern",
"_____no_output_____"
]
],
[
[
"data['pattern_width'] = np.abs(data.dateF - data.dateE)",
"_____no_output_____"
],
[
"data['w1'] = get_height(data.idx1, data.idx3)\ndata['w2'] = get_height(data.idx3, data.idx5)\ndata['w3'] = get_height(data.idx2, data.idx4)",
"_____no_output_____"
],
[
"indices = ['idx1', 'idx2', 'idx3', 'idx4', 'idx5']",
"_____no_output_____"
],
[
"data = data.drop(indices, axis=1)",
"_____no_output_____"
]
],
[
[
"#### Data validation",
"_____no_output_____"
],
[
"Drop values with 0 widths, invalid pattern.",
"_____no_output_____"
]
],
[
[
"data = data[data.w1 != 0]\ndata = data[data.w2 != 0]\ndata = data[data.w3 != 0]",
"_____no_output_____"
]
],
[
[
"#### Drop 'dateF' and 'dateE' after getting pattern width",
"_____no_output_____"
]
],
[
[
"data = data.drop(['dateF', 'dateE'], axis=1)",
"_____no_output_____"
],
[
"Counter(data.label)",
"_____no_output_____"
],
[
"undersample = NearMiss(version=3)",
"_____no_output_____"
],
[
"def undersample_(X: pd.DataFrame, \n y: pd.Series,\n version: int = 1):\n \n undersample = NearMiss(version=version)\n return undersample.fit_resample(X, y)",
"_____no_output_____"
],
[
"X, y = undersample_(data.drop('label', axis=1),\n data['label'])",
"_____no_output_____"
],
[
"Counter(y)",
"_____no_output_____"
]
],
[
[
"#### Change the coding of the label",
"_____no_output_____"
]
],
[
[
"X, y = data.drop('label', axis=1), data['label']",
"_____no_output_____"
],
[
"mapping = {1: 0, 2: 1, 3: 2}",
"_____no_output_____"
],
[
"y = y.replace(mapping)",
"_____no_output_____"
],
[
"to_filter = ['f1', 'f2', 'f3', 'f4', 'f5', \n 'industry_coded', 'pattern_width',\n 'h1', 'h2', 'h4', 'h6', 'h9',\n 'w1', 'w2', 'w3']",
"_____no_output_____"
],
[
"X = X[to_filter]",
"_____no_output_____"
],
[
"to_corr = ['w1', 'w2', 'w3', 'pattern_width', 'f1',\n 'h1', 'h2', 'h4', 'h6', 'h9']",
"_____no_output_____"
],
[
"fig=plt.figure(figsize=(12,10), dpi= 100)\nsns.heatmap(X[to_corr].corr(method='spearman'), annot=True)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Model selection\n\nThe dataset we have consists of quantitative features and a single categorical feature which is the target feature. The target feature contains multiple levels. Therefore, we will fit several models that are multiclass to our dataset, in particular multiclass KNN, multinomial logistic regression, multiclass SVM etc. to find the best predictive model of this project. Fortunately, we can fit these models to our dataset simultaneously using pycaret machine learning library. \nMoreover, pycaret also automatically normalized then splits dataset if specified, do one hat encoding for nominal features, perform cross validation, tuned hyperparameter for every model etc.. In our case, we specify the normalization method as minmax scaler then split it to 80% training set and 20% testing set, then let it perform 5 fold cross validation with 10 repetition.",
"_____no_output_____"
]
],
[
[
"data = pd.concat([X, y], axis=1)",
"_____no_output_____"
],
[
"cv = RepeatedKFold(n_splits=5, n_repeats=10, random_state=13)",
"_____no_output_____"
],
[
"exp_name = setup(data=data,\n target='label',\n normalize=True,\n normalize_method='minmax',\n train_size=0.8,\n data_split_stratify=True,\n fold_strategy=cv,\n remove_outliers=True,\n use_gpu=True,\n session_id=13,\n pca=True,\n pca_method='linear')",
"_____no_output_____"
],
[
"best_model = compare_models()",
"_____no_output_____"
],
[
"get_config('X')",
"_____no_output_____"
]
],
[
[
"Since this project aims to predict stock prices given double top stock chart pattern for trading purposes. Both false negative and false positive are crucial. That is, in the case of false negative, if we predict a decrease in price after the pattern then decided to sale the stocks because we won't get any more profit however, it actually increases, then we just lose the opportunity to earn more. \n\nOn the other hand, in the case of false positive, if we predict an increase in price after the pattern then decided to buy stocks so we can sale it during the increase however, it actually decreases, then we just lose some money. Also, the target feature has imbalanced classes. Therefore, we will emphasize the F1 score over accuracy and any other performance metrics for this project to measure the predictive power of the model built.\n\nThe above output shows the list of predictive performance for several models after we fit it to our data. Notice that the model that produces the highest F1 score is the Gradient descent classifier, a sequential ensemble approach, with 0.5217 score. This imply that the model we built can predict the target feature given f1, f2, f3, f4, f5, dateF, dateE, and industry with 52.17% accuracy.\n\nMoreover, we will further interpret other performance metrics but this is just for better understanding of the predictive model performance. After all, we already interpreted F1 score, the appropriate performance metric for this project.\n\nNow, notice that the model that produces the highest accuracy is still Gradient boosting classifier with value 0.5616, followed by Ada boost classifier with value 0.5579, both are sequential ensemble approach. These implies that the models we built can predict the target feature given f1, f2, f3, f4, f5, dateF, dateE, and industry with 56.16% and 55.79% accuracy respectively. \n\nAda Boost Classifier also produces the highest Precision with value 0.5210. This suggest that for the number of predictions that the model made, 52.19% of it are correct. Whereas, Gradient boosting classifier produces the highest AUC and Recall (Sensitivity) with values 0.6750 and 0.4883 respectively. AUC value suggest that the model have 67.50% accuracy to predict the target feature that will or will not occur. And recall value imply that, for the target feature that should occur, we predicted 48.83% of it.",
"_____no_output_____"
],
[
"## Tuned hyperparameters",
"_____no_output_____"
]
],
[
[
"best_model",
"_____no_output_____"
],
[
"plot_model(best_model, plot='confusion_matrix')",
"_____no_output_____"
],
[
"plot_model(best_model)",
"_____no_output_____"
],
[
"plot_model(best_model, plot='class_report')",
"_____no_output_____"
],
[
"plot_model(best_model, plot='pr')",
"_____no_output_____"
],
[
"plot_model(best_model, plot='boundary')",
"_____no_output_____"
],
[
"plot_model(best_model, plot='learning')",
"_____no_output_____"
]
],
[
[
"## Evaluate predictive accuracy of the model built",
"_____no_output_____"
]
],
[
[
"test_scores = predict_model(best_model)",
"_____no_output_____"
],
[
"test_scores",
"_____no_output_____"
],
[
"test_scores.Score.mean()",
"_____no_output_____"
]
],
[
[
"The predictive model built when use for test set, gives us a predictive accuracy of 62.40%. This suggest that we predicted the movement of stock prices given new quiries of double top stock chart pattern (test set) with 62.40% accuracy.",
"_____no_output_____"
],
[
"## Conclusion and Recommendation",
"_____no_output_____"
],
[
"The dataset created consists of target feature with 3 levels (decrease/1, neutral/2, increase/3) and 7 descriptive features, in which 5 of them (f1, f2, f3, f4, f5) forms a double stock chart pattern and the other two are indexes of the start and end of the pattern. After we tried to fit several multiclass models on this dataset, we found out that its accuracy is less than 50%. Thus, we performed feature engineer by taking the absolute difference between f1 and f3, f3 and f5, and get the average from f1 to f5. These 3 additional descriptive features actually helped the predictive model we built to increase its accuracy from below 50% to a little bit higher than 50%. The predictive model that produces this result is the gradient boosting classifier, a sequential ensemble approach that gives an F1 score of 0.5217. This only imply that the predictive model built can predict the movement of the stock prices given double top stock chart pattern with 52.17% accuracy. Furthermore, when model performance was evaluated with the test set, it produces a 62.40% predictive accuracy.",
"_____no_output_____"
],
[
"## References",
"_____no_output_____"
],
[
"* https://medium.com/analytics-vidhya/accuracy-vs-f1-score-6258237beca2\n* https://pycaret.org/classification/\n* https://finance.yahoo.com/",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbac07e698b0dbb757924f94c6a612bce8cc0166
| 34,522 |
ipynb
|
Jupyter Notebook
|
intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb
|
avijit2527/UdacityPytorch
|
0b4abf4b1e1d1e3f9c158d22357d20d0c45b398f
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb
|
avijit2527/UdacityPytorch
|
0b4abf4b1e1d1e3f9c158d22357d20d0c45b398f
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 4 - Fashion-MNIST (Exercises).ipynb
|
avijit2527/UdacityPytorch
|
0b4abf4b1e1d1e3f9c158d22357d20d0c45b398f
|
[
"MIT"
] | null | null | null | 144.443515 | 23,872 | 0.880424 |
[
[
[
"# Classifying Fashion-MNIST\n\nNow it's your turn to build and train a neural network. You'll be using the [Fashion-MNIST dataset](https://github.com/zalandoresearch/fashion-mnist), a drop-in replacement for the MNIST dataset. MNIST is actually quite trivial with neural networks where you can easily achieve better than 97% accuracy. Fashion-MNIST is a set of 28x28 greyscale images of clothes. It's more complex than MNIST, so it's a better representation of the actual performance of your network, and a better representation of datasets you'll use in the real world.\n\n<img src='assets/fashion-mnist-sprite.png' width=500px>\n\nIn this notebook, you'll build your own neural network. For the most part, you could just copy and paste the code from Part 3, but you wouldn't be learning. It's important for you to write the code yourself and get it to work. Feel free to consult the previous notebooks though as you work through this.\n\nFirst off, let's load the dataset through torchvision.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision import datasets, transforms\nimport helper\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])\n# Download and load the training data\ntrainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)\n\n# Download and load the test data\ntestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"Here we can see one of the images.",
"_____no_output_____"
]
],
[
[
"image, label = next(iter(trainloader))\nhelper.imshow(image[0,:]);",
"_____no_output_____"
]
],
[
[
"## Building the network\n\nHere you should define your network. As with MNIST, each image is 28x28 which is a total of 784 pixels, and there are 10 classes. You should include at least one hidden layer. We suggest you use ReLU activations for the layers and to return the logits or log-softmax from the forward pass. It's up to you how many layers you add and the size of those layers.",
"_____no_output_____"
]
],
[
[
"# TODO: Define your network architecture here\nfrom torch import nn\nfrom torch import optim\nmodel = nn.Sequential(nn.Linear(784,500),\n nn.ReLU(),\n nn.Linear(500,350),\n nn.ReLU(),\n nn.Linear(350,200),\n nn.ReLU(),\n nn.Linear(200,100),\n nn.ReLU(),\n nn.Linear(100,10),\n nn.LogSoftmax(dim = 1))\n\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.parameters(), lr = 0.003)\n",
"_____no_output_____"
]
],
[
[
"# Train the network\n\nNow you should create your network and train it. First you'll want to define [the criterion](http://pytorch.org/docs/master/nn.html#loss-functions) ( something like `nn.CrossEntropyLoss`) and [the optimizer](http://pytorch.org/docs/master/optim.html) (typically `optim.SGD` or `optim.Adam`).\n\nThen write the training code. Remember the training pass is a fairly straightforward process:\n\n* Make a forward pass through the network to get the logits \n* Use the logits to calculate the loss\n* Perform a backward pass through the network with `loss.backward()` to calculate the gradients\n* Take a step with the optimizer to update the weights\n\nBy adjusting the hyperparameters (hidden units, learning rate, etc), you should be able to get the training loss below 0.4.",
"_____no_output_____"
]
],
[
[
"# TODO: Create the network, define the criterion and optimizer\n",
"_____no_output_____"
],
[
"# TODO: Train the network here\nepoch = 5\nfor x in range(epoch):\n cumulative_loss = 0\n for images, labels in trainloader:\n optimizer.zero_grad()\n images = images.view(images.shape[0],-1)\n output = model(images)\n loss = criterion(output, labels)\n cumulative_loss = cumulative_loss + loss\n loss.backward()\n optimizer.step()\n else:\n print(f\"Training loss: {cumulative_loss/len(trainloader)}\")",
"Training loss: 0.5541394352912903\nTraining loss: 0.41243255138397217\nTraining loss: 0.373406320810318\nTraining loss: 0.34406355023384094\nTraining loss: 0.3256939947605133\n"
],
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport helper\n\n# Test out your network!\n\ndataiter = iter(testloader)\nimages, labels = dataiter.next()\nimg = images[0]\n# Convert 2D image to 1D vector\nimg = img.resize_(1, 784)\n\n# TODO: Calculate the class probabilities (softmax) for img\nwith torch.no_grad():\n logps = model(img)\nps = torch.exp(logps)\n\n# Plot the image and probabilities\nhelper.view_classify(img.resize_(1, 28, 28), ps, version='Fashion')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbac0eb26b7f29bd4ae9313b48274b3535c35f66
| 95,479 |
ipynb
|
Jupyter Notebook
|
Models/BenchMarkRecreation_SimpleLinearModel.ipynb
|
jpnevrones/PenguinRandomWalk
|
ddaa1b425cc8bb156ea4ebc1863b2428980c4807
|
[
"MIT"
] | null | null | null |
Models/BenchMarkRecreation_SimpleLinearModel.ipynb
|
jpnevrones/PenguinRandomWalk
|
ddaa1b425cc8bb156ea4ebc1863b2428980c4807
|
[
"MIT"
] | null | null | null |
Models/BenchMarkRecreation_SimpleLinearModel.ipynb
|
jpnevrones/PenguinRandomWalk
|
ddaa1b425cc8bb156ea4ebc1863b2428980c4807
|
[
"MIT"
] | null | null | null | 54.034522 | 20,376 | 0.607118 |
[
[
[
"%matplotlib inline\n\nfrom __future__ import print_function\nfrom __future__ import division\n\nimport os\n\nimport pandas as pd\nimport numpy as np\nfrom tqdm import tqdm_notebook\n\nfrom matplotlib import pyplot as plt\nfrom matplotlib.colors import rgb2hex\nimport seaborn as sns\n\nimport statsmodels.api as sm\n\n# let's not pollute this blog post with warnings\nfrom warnings import filterwarnings\nfilterwarnings('ignore')",
"_____no_output_____"
],
[
"observations = pd.read_csv(os.path.join('data', 'training_set_observations.csv'), index_col=0)\nobservations.head()",
"_____no_output_____"
],
[
"observations.columns",
"_____no_output_____"
],
[
"observations.describe()",
"_____no_output_____"
],
[
"\ncorr = observations.select_dtypes(include = ['float64', 'int64']).iloc[:, 1:].corr()\nplt.figure(figsize=(12, 12))\nsns.heatmap(corr, vmax=1, square=True)",
"_____no_output_____"
],
[
"\nprint(\n \"We have {} penguin observations from {} to {} at {} unique sites in the Antarctic!\" \\\n .format(observations.shape[0],\n observations.season_starting.min(),\n observations.season_starting.max(),\n observations.site_id.nunique())\n)\n",
"We have 2952 penguin observations from 1895 to 2013 at 619 unique sites in the Antarctic!\n"
],
[
"\n# How many observations do we have for each species?\nobservations.common_name.value_counts()",
"_____no_output_____"
],
[
"\n# How many differnet sites do we see each species at?\n(observations.groupby(\"common_name\")\n .site_id\n .nunique())",
"_____no_output_____"
],
[
"\n# How many count types do we have for each species?\n(observations.groupby(\"common_name\")\n .count_type\n .value_counts())",
"_____no_output_____"
],
[
"\nnest_counts = pd.read_csv(\n os.path.join('data', 'training_set_nest_counts.csv'),\n index_col=[0,1]\n )\n\n# Let's look at the first 10 rows, and the last 10 columns\nnest_counts.iloc[:10, -10:]",
"_____no_output_____"
],
[
"\n# get a sort order for the sites with the most observations\nsorted_idx = (pd.notnull(nest_counts)\n .sum(axis=1)\n .sort_values(ascending=False)\n .index)\n\n# get the top 25 most common sites and divide by the per-series mean\nto_plot = nest_counts.loc[sorted_idx].head(25)\nto_plot = to_plot.divide(to_plot.mean(axis=1), axis=0)\n\n# plot the data\nplt.gca().matshow(to_plot,\n cmap='viridis')\nplt.show()\n",
"_____no_output_____"
],
[
"def preprocess_timeseries(timeseries, first_year, fillna_value=0):\n \"\"\" Takes one of the timeseries dataframes, removes\n columns before `first_year`, and fills NaN values\n with the preceeding value. Then backfills any\n remaining NaNs.\n \n As a courtesy, also turns year column name into\n integers for easy comparisons.\n \"\"\"\n # column type\n timeseries.columns = timeseries.columns.astype(int)\n \n # subset to just data after first_year\n timeseries = timeseries.loc[:, timeseries.columns >= first_year]\n \n # Forward fill count values. This is a strong assumption.\n timeseries.fillna(method=\"ffill\", axis=1, inplace=True)\n timeseries.fillna(method=\"bfill\", axis=1, inplace=True)\n \n # For sites with no observations, fill with fill_na_value\n timeseries.fillna(fillna_value, inplace=True)\n \n return timeseries\n\nnest_counts = preprocess_timeseries(nest_counts,\n 1980,\n fillna_value=0.0)\nnest_counts.head()",
"_____no_output_____"
],
[
"# get the top 25 most common sites and divide by the per-series mean\nto_plot = nest_counts.loc[sorted_idx].head(25)\nto_plot = to_plot.divide(to_plot.mean(axis=1), axis=0)\n\nplt.gca().matshow(to_plot,\n cmap='viridis')\nplt.show()",
"_____no_output_____"
],
[
"e_n_values = pd.read_csv(\n os.path.join('data', 'training_set_e_n.csv'),\n index_col=[0,1]\n )\n\n# Process error data to match our nest_counts data\ne_n_values = preprocess_timeseries(e_n_values, 1980, fillna_value=0.05)\ne_n_values.head()",
"_____no_output_____"
],
[
"def amape(y_true, y_pred, accuracies):\n \"\"\" Adjusted MAPE\n \"\"\"\n not_nan_mask = ~np.isnan(y_true)\n \n # calculate absolute error\n abs_error = (np.abs(y_true[not_nan_mask] - y_pred[not_nan_mask]))\n \n # calculate the percent error (replacing 0 with 1\n # in order to avoid divide-by-zero errors).\n pct_error = abs_error / np.maximum(1, y_true[not_nan_mask])\n \n # adjust error by count accuracies\n adj_error = pct_error / accuracies[not_nan_mask]\n \n # return the mean as a percentage\n return np.mean(adj_error)",
"_____no_output_____"
],
[
"# Let's confirm the best possible score is 0!\namape(nest_counts.values,\n nest_counts.values,\n e_n_values.values)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.isotonic import IsotonicRegression\nfrom sklearn.tree import DecisionTreeRegressor\n\ndef train_model_per_row(ts, acc, split_year=2010):\n # Split into train/test to tune our parameter\n train = ts.iloc[ts.index < split_year]\n \n test = ts.iloc[ts.index >= split_year]\n test_acc = acc.iloc[acc.index >= split_year]\n \n # Store best lag parameter\n best_mape = np.inf \n best_lag = None\n \n # Test linear regression models with the most recent\n # 2 points through using all of the points\n for lag in range(2, train.shape[0]):\n # fit the model\n temp_model = LinearRegression()\n #temp_model = DecisionTreeRegressor(max_depth=4)\n\n temp_model.fit(\n train.index[-lag:].values.reshape(-1, 1),\n train[-lag:]\n )\n \n # make our predictions on the test set\n preds = temp_model.predict(\n test.index.values.reshape(-1, 1)\n )\n\n # calculate the score using the custom metric\n mape = amape(test.values,\n preds,\n test_acc.values)\n\n # if it's the best score yet, hold on to the parameter\n if mape < best_mape:\n best_mape = mape\n best_lag = lag\n \n\n # return model re-trained on entire dataset\n final_model = LinearRegression()\n #final_model = DecisionTreeRegressor(max_depth=4)\n\n final_model.fit(\n ts.index[-best_lag:].values.reshape(-1, 1),\n ts[-best_lag:]\n )\n\n return final_model, best_mape ,best_lag",
"_____no_output_____"
],
[
"models = {}\navg_Best_Mape = 0.0\navg_best_lag = 0.0\niteration = 0\nfor i, row in tqdm_notebook(nest_counts.iterrows(),\n total=nest_counts.shape[0]):\n acc = e_n_values.loc[i]\n models[i], best_mape, best_lag = train_model_per_row(row, acc)\n avg_Best_Mape = avg_Best_Mape + best_mape\n avg_best_lag = avg_best_lag + best_lag\n iteration = iteration + 1\navg_Best_Mape = avg_Best_Mape / iteration\navg_best_lag = avg_best_lag / iteration \nprint(\"Avg Best Mape : {0}\".format(avg_Best_Mape))\nprint(\"Avg Best Lag : {0}\".format(avg_best_lag))",
"\nAvg Best Mape : 57.66601321778737\nAvg Best Lag : 3.779320987654321\n"
],
[
"submission_format = pd.read_csv(\n os.path.join('data','submission_format.csv'),\n index_col=[0, 1]\n)\n\nprint(submission_format.shape)\nsubmission_format.head()",
"(648, 4)\n"
],
[
"preds = []\n\n# For every row in the submission file\nfor i, row in tqdm_notebook(submission_format.iterrows(),\n total=submission_format.shape[0]):\n \n # get the model for this site + common_name\n model = models[i]\n \n # make predictions using the model\n row_predictions = model.predict(\n submission_format.columns.values.reshape(-1, 1)\n )\n \n # keep our predictions, rounded to nearest whole number\n preds.append(np.round(row_predictions))\n\n# Create a dataframe that we can write out to a CSV\nprediction_df = pd.DataFrame(preds,\n index=submission_format.index,\n columns=submission_format.columns)\n\nprediction_df.head()",
"\n"
],
[
"prediction_df.to_csv('predictions.csv')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbac19d2abd2f9b1ca78efbad5a28e2739b8b87c
| 106,234 |
ipynb
|
Jupyter Notebook
|
doc/_build/Notebooks/Workflow.ipynb
|
esowc/UNSEEN-open
|
f7f9921a78d7357116b3dd12ea46f5a323be145b
|
[
"MIT"
] | 7 |
2020-05-14T05:48:16.000Z
|
2021-07-29T03:18:10.000Z
|
doc/_build/_sources/Notebooks/Workflow.ipynb.txt
|
esowc/UNSEEN-open
|
f7f9921a78d7357116b3dd12ea46f5a323be145b
|
[
"MIT"
] | null | null | null |
doc/_build/_sources/Notebooks/Workflow.ipynb.txt
|
esowc/UNSEEN-open
|
f7f9921a78d7357116b3dd12ea46f5a323be145b
|
[
"MIT"
] | 4 |
2020-12-09T13:38:11.000Z
|
2022-03-01T09:41:15.000Z
| 57.579404 | 8,715 | 0.54203 |
[
[
[
"# UNSEEN-open\n\nIn this project, the aim is to build an open, reproducible, and transferable workflow for UNSEEN.\n<!-- -- an increasingly popular method that exploits seasonal prediction systems to assess and anticipate climate extremes beyond the observed record. The approach uses pooled forecasts as plausible alternate realities. Instead of the 'single realization' of reality, pooled forecasts can be exploited to better assess the likelihood of infrequent events. -->\nThe workflow consists of four steps, as illustrated below:",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"In this project, UNSEEN-open is applied to assess two extreme events in 2020: February 2020 UK precipitation and the 2020 Siberian heatwave. \n\nFebruary average precipitation was the highest on record in the UK: with what frequency of occurrence can February extreme precipitation events such as the 2020 event be expected?\n\nThe Siberian heatwave has broken the records as well. Could such an event be anticipation with UNSEEN? And to what extend can we expect changes in the frequency of occurrence and magnitude of these kind of events?",
"_____no_output_____"
],
[
"## Overview\n\nHere we provide an overview of the steps taken to apply UNSEEN-open.",
"_____no_output_____"
],
[
"### Download",
"_____no_output_____"
],
[
"We want to download February precipitation over the UK and March-May average temperature over Siberia. We retrieve all SEAS5 seasonal forecasts that are forecasting the target months (i.e. February and MAM) and we retrieve ERA5 reanalysis for the same regions and variables for evaluation. ",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nsys.path.insert(0, os.path.abspath('../../'))\nos.chdir(os.path.abspath('../../'))",
"_____no_output_____"
],
[
"import src.cdsretrieve as retrieve\nimport src.preprocess as preprocess",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"retrieve.retrieve_SEAS5(variables = ['2m_temperature','2m_dewpoint_temperature'], target_months = [3,4,5], \n area = [70, -11, 30, 120], years=np.arange(1981, 2021), folder = '../Siberia_example/SEAS5/')",
"2020-09-16 12:42:52,756 INFO Welcome to the CDS\n2020-09-16 12:42:52,758 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:42:55,461 INFO Request is queued\n2020-09-16 12:43:44,892 INFO Request is completed\n2020-09-16 12:43:44,895 INFO Downloading http://136.156.133.41/cache-compute-0013/cache/data1/adaptor.mars.external-1600256610.5590868-10484-1-43f9f2fc-9209-4306-ba84-5fc2f56c8cbb.nc to ../Siberia_example/SEAS5/201702.nc (3.2M)\n2020-09-16 12:43:45,155 INFO Download rate 12.3M/s \n2020-09-16 12:43:46,261 INFO Welcome to the CDS\n2020-09-16 12:43:46,263 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:43:47,913 INFO Request is queued\n2020-09-16 12:43:52,712 INFO Request is running\n2020-09-16 12:44:01,189 INFO Request is completed\n2020-09-16 12:44:01,191 INFO Downloading http://136.156.132.198/cache-compute-0003/cache/data3/adaptor.mars.external-1600256631.4983242-12958-16-44a30635-2276-47c7-9a9c-312b21ba1879.nc to ../Siberia_example/SEAS5/201701.nc (3.2M)\n2020-09-16 12:44:01,333 INFO Download rate 22.3M/s \n2020-09-16 12:44:03,029 INFO Welcome to the CDS\n2020-09-16 12:44:03,031 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:44:04,786 INFO Request is queued\n2020-09-16 12:44:18,071 INFO Request is running\n2020-09-16 12:44:25,689 INFO Request is completed\n2020-09-16 12:44:25,691 INFO Downloading http://136.156.132.210/cache-compute-0005/cache/data4/adaptor.mars.external-1600256656.8702655-11521-21-84570425-df1d-45cf-ae6a-2f220e1f1c62.nc to ../Siberia_example/SEAS5/201612.nc (1.6M)\n2020-09-16 12:44:25,780 INFO Download rate 17.7M/s\n2020-09-16 12:44:27,551 INFO Welcome to the CDS\n2020-09-16 12:44:27,552 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:44:29,188 INFO Request is queued\n2020-09-16 12:45:18,630 INFO Request is completed\n2020-09-16 12:45:18,631 INFO Downloading http://136.156.133.46/cache-compute-0015/cache/data1/adaptor.mars.external-1600256707.239661-17582-21-c50e6007-95da-4019-9324-d423e1862177.nc to ../Siberia_example/SEAS5/201802.nc (3.2M)\n2020-09-16 12:45:18,775 INFO Download rate 22.4M/s \n2020-09-16 12:45:19,725 INFO Welcome to the CDS\n2020-09-16 12:45:19,726 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:45:21,293 INFO Request is queued\n2020-09-16 12:45:42,197 INFO Request is running\n2020-09-16 12:45:53,616 INFO Request is completed\n2020-09-16 12:45:53,618 INFO Downloading http://136.156.132.110/cache-compute-0001/cache/data2/adaptor.mars.external-1600256736.302646-2916-7-906ebfa9-4208-4d26-9642-9090ea44be6e.nc to ../Siberia_example/SEAS5/201801.nc (3.2M)\n2020-09-16 12:45:53,784 INFO Download rate 19.1M/s \n2020-09-16 12:45:56,421 INFO Welcome to the CDS\n2020-09-16 12:45:56,423 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:45:58,590 INFO Request is queued\n2020-09-16 12:46:30,897 INFO Request is completed\n2020-09-16 12:46:30,898 INFO Downloading http://136.156.133.36/cache-compute-0010/cache/data1/adaptor.mars.external-1600256780.4346173-24644-14-b7e1baa7-a597-484a-ad07-4704018026fc.nc to ../Siberia_example/SEAS5/201712.nc (3.2M)\n2020-09-16 12:46:31,063 INFO Download rate 19.3M/s \n2020-09-16 12:46:32,348 INFO Welcome to the CDS\n2020-09-16 12:46:32,349 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:46:33,083 INFO Request is queued\n2020-09-16 12:47:22,511 INFO Request is running\n2020-09-16 12:47:48,182 INFO Request is completed\n2020-09-16 12:47:48,184 INFO Downloading http://136.156.133.39/cache-compute-0012/cache/data9/adaptor.mars.external-1600256837.9028022-8098-12-c831d16e-b859-42cf-b5be-ef68bfe6d486.nc to ../Siberia_example/SEAS5/201902.nc (3.2M)\n2020-09-16 12:47:48,334 INFO Download rate 21.3M/s \n2020-09-16 12:47:49,241 INFO Welcome to the CDS\n2020-09-16 12:47:49,243 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:47:52,161 INFO Request is queued\n2020-09-16 12:48:41,595 INFO Request is running\n2020-09-16 12:49:07,268 INFO Request is completed\n2020-09-16 12:49:07,269 INFO Downloading http://136.156.133.42/cache-compute-0014/cache/data5/adaptor.mars.external-1600256919.2350588-1054-28-68c7f028-c6c0-4e48-acf8-123db9b7fa2e.nc to ../Siberia_example/SEAS5/201901.nc (3.2M)\n2020-09-16 12:49:07,419 INFO Download rate 21.4M/s \n2020-09-16 12:49:08,744 INFO Welcome to the CDS\n2020-09-16 12:49:08,747 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:49:10,318 INFO Request is queued\n2020-09-16 12:49:23,593 INFO Request is running\n2020-09-16 12:49:31,209 INFO Request is completed\n2020-09-16 12:49:31,210 INFO Downloading http://136.156.132.110/cache-compute-0001/cache/data7/adaptor.mars.external-1600256959.9121842-3051-15-ae0c3334-05f6-4503-a76a-9e94b580d344.nc to ../Siberia_example/SEAS5/201812.nc (3.2M)\n2020-09-16 12:49:31,323 INFO Download rate 27.9M/s\n2020-09-16 12:49:32,568 INFO Welcome to the CDS\n2020-09-16 12:49:32,569 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:49:33,136 INFO Request is queued\n2020-09-16 12:49:54,013 INFO Request is running\n2020-09-16 12:50:05,432 INFO Request is completed\n2020-09-16 12:50:05,434 INFO Downloading http://136.156.133.25/cache-compute-0008/cache/data5/adaptor.mars.external-1600256989.3033717-31909-16-4100686f-0c78-4337-bb28-9ec75058c74b.nc to ../Siberia_example/SEAS5/202002.nc (3.2M)\n2020-09-16 12:50:05,610 INFO Download rate 18.1M/s \n2020-09-16 12:50:05,797 INFO Welcome to the CDS\n2020-09-16 12:50:05,798 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:50:06,347 INFO Request is queued\n2020-09-16 12:50:11,146 INFO Request is running\n2020-09-16 12:50:14,540 INFO Request is completed\n2020-09-16 12:50:14,542 INFO Downloading http://136.156.132.105/cache-compute-0000/cache/data9/adaptor.mars.external-1600257009.4601834-28351-14-4b0c98c4-0453-42a5-b214-f96628f6588f.nc to ../Siberia_example/SEAS5/202001.nc (3.2M)\n2020-09-16 12:50:14,741 INFO Download rate 16M/s \n2020-09-16 12:50:15,321 INFO Welcome to the CDS\n2020-09-16 12:50:15,323 INFO Sending request to https://cds.climate.copernicus.eu/api/v2/resources/seasonal-monthly-single-levels\n2020-09-16 12:50:15,528 INFO Request is queued\n2020-09-16 12:50:16,546 INFO Request is running\n2020-09-16 12:50:23,771 INFO Request is completed\n2020-09-16 12:50:23,773 INFO Downloading http://136.156.132.105/cache-compute-0000/cache/data5/adaptor.mars.external-1600257016.6044283-28180-19-953e8ab1-ad1d-459a-b87e-881b25166155.nc to ../Siberia_example/SEAS5/201912.nc (3.2M)\n2020-09-16 12:50:23,920 INFO Download rate 21.7M/s \n"
],
[
"retrieve.retrieve_SEAS5(variables = 'total_precipitation', target_months = [2], \n area = [60, -11, 50, 2], folder = '../UK_example/SEAS5/')",
"_____no_output_____"
],
[
"retrieve.retrieve_ERA5(variables = ['2m_temperature','2m_dewpoint_temperature'], target_months = [3,4,5], \n area = [70, -11, 30, 120], folder = '../Siberia_example/ERA5/')",
"_____no_output_____"
],
[
"retrieve.retrieve_ERA5(variables = 'total_precipitation', target_months = [2], \n area = [60, -11, 50, 2], folder = '../UK_example/ERA5/')",
"_____no_output_____"
]
],
[
[
"### Preprocess",
"_____no_output_____"
],
[
"In the preprocessing step, we first merge all downloaded files into one netcdf file.\nThen the rest of the preprocessing depends on the definition of the extreme event. For example, for the UK case study, we want to extract the UK average precipitation while for the Siberian heatwave we will just used the defined area to spatially average over. For the MAM season, we still need to take the seasonal average, while for the UK we already have the average February precipitation. ",
"_____no_output_____"
]
],
[
[
"SEAS5_Siberia = preprocess.merge_SEAS5(folder = '../Siberia_example/SEAS5/', target_months = [3,4,5])\nSEAS5_Siberia",
"Lead time: 02\n1\n12\n"
],
[
"SEAS5_Siberia.sel(latitude = 60, longitude = -10, time = '2000-03', number = 24, leadtime = 3).load()",
"_____no_output_____"
],
[
"SEAS5_UK = preprocess.merge_SEAS5(folder = '../UK_example/SEAS5/', target_months = [2])\nSEAS5_UK",
"Lead time: 1\n12\n11\n10\n9\n"
]
],
[
[
"### Read more\nJump into the respective sections for more detail:\n\n* **Download**\n * [1. Retrieve](1.Download/1.Retrieve.ipynb) \n* **Pre-process**\n * [2.1 Merge](2.Preprocess/2.1Merge.ipynb) \n * [2.2 Mask](2.Preprocess/2.2Mask.ipynb)\n * [2.3 Upscale](2.Preprocess/2.3Upscale.ipynb)\n* **Evaluate**\n * [3. Evaluate](3.Evaluate/3.Evaluate.ipynb)\n* **Illustrate**\n ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbac1ea4a33565c854e5b0ce976bdff9478eb833
| 17,931 |
ipynb
|
Jupyter Notebook
|
notebooks/04_Working_with_Tabular_Data.ipynb
|
luzpaz/pyviz
|
15d3e8bf8ea99b650f2f2911616b4c5e66e7841a
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/04_Working_with_Tabular_Data.ipynb
|
luzpaz/pyviz
|
15d3e8bf8ea99b650f2f2911616b4c5e66e7841a
|
[
"BSD-3-Clause"
] | null | null | null |
notebooks/04_Working_with_Tabular_Data.ipynb
|
luzpaz/pyviz
|
15d3e8bf8ea99b650f2f2911616b4c5e66e7841a
|
[
"BSD-3-Clause"
] | null | null | null | 41.411085 | 639 | 0.645251 |
[
[
[
"<style>div.container { width: 100% }</style>\n<img style=\"float:left; vertical-align:text-bottom;\" height=\"65\" width=\"172\" src=\"assets/PyViz_logo_wm_line.png\" />\n<div style=\"float:right; vertical-align:text-bottom;\"><h2>Tutorial 04. Working with tabular data</h2></div>",
"_____no_output_____"
],
[
"As we have already discovered, HoloViews elements are simple wrappers around your data that provide a semantically meaningful representation. The real power of HoloViews becomes most evident when working with larger, multi-dimensional datasets, whether they are tabular like in a database or CSV file, or gridded like large datasets of images.\n\nTabular data (also called columnar data) is one of the most common, general, and versatile data formats, corresponding to how data is laid out in a spreadsheet. There are many different ways to put data into a tabular format, but for interactive analysis having [**tidy data**](http://www.jeannicholashould.com/tidy-data-in-python.html) provides flexibility and simplicity. Here we will show how to make your data tidy as a first step, but see [hvPlot](http://hvplot.pyviz.org) for convenient ways to work with non-tidy data directly.\n\nIn this tutorial all the information you have learned in the previous sections will finally really pay off. We will discover how to facet data and use different element types to explore and visualize the data contained in a real dataset, using many of the same libraries introduced earlier along with some statistics methods from SciPy:\n\n<div style=\"margin: 10px\">\n<a href=\"http://holoviews.org\"><img style=\"margin:8px; display:inline; object-fit:scale-down; max-height:150px\" src=\"./assets/holoviews.png\"/></a>\n<a href=\"http://bokeh.pydata.org\"><img style=\"margin:8px; display:inline; object-fit:scale-down; max-height:150px\" src=\"./assets/bokeh.png\"/></a>\n<a href=\"http://pandas.pydata.org\"><img style=\"margin:8px; display:inline; object-fit:scale-down; max-height:140px\" src=\"./assets/pandas.png\"/></a>\n<a href=\"http://numpy.org\"><img style=\"margin:8px; display:inline; object-fit:scale-down; max-height:150px\" src=\"./assets/numpy.png\"/></a>\n<a href=\"https://docs.scipy.org/doc/scipy/reference\"><img style=\"margin:8px; display:inline; object-fit:scale-down; max-height:150px\" src=\"./assets/scipy.png\"/></a>\n</div>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport scipy.stats as ss\nimport pandas as pd\nimport holoviews as hv\nhv.extension('bokeh')\n%opts Curve Scatter Bars [tools=['hover']]",
"_____no_output_____"
]
],
[
[
"## What is tabular, tidy data?",
"_____no_output_____"
]
],
[
[
"macro_df = pd.read_csv('../data/macro.csv')\nmacro_df.head()",
"_____no_output_____"
]
],
[
[
"For tidy data, the **columns** of the table represent **variables** or **dimensions** and the **rows** represent **observations**. \n\nThe opposite of tidy data is so-called **wide** data. To see what wide data looks like, we can use the pandas ``pivot_table`` method:",
"_____no_output_____"
]
],
[
[
"wide = macro_df.pivot_table('unem', 'year', 'country')\nwide.head(5)",
"_____no_output_____"
]
],
[
[
"In this wide format we can see that each column represents the unemployment figures for one country indexed, and each row a particular year. A wide table can represent data very concisely in some cases, but it is difficult to work with in practice because it does not make dimensions easily accessible for plotting or analysis. To go from wide to tidy data you can use the ``pd.melt`` function: ",
"_____no_output_____"
]
],
[
[
"melted = pd.melt(wide.reset_index(), id_vars='year', value_name='unemployment')\nmelted.head()",
"_____no_output_____"
]
],
[
[
"## Declaring Dimensions on a Dataset\n\nA HoloViews `Dataset` is similar to a HoloViews Element, without having any specific metadata that lets it visualize itself. A Dataset is useful for specifying a set of `Dimension`s that apply to the data, which will later be inherited by any visualizable elements that you create from the Dataset.\n\nA HoloViews Dimension is the same concept as a **dependent** or **independent** variable in mathematics. In HoloViews such variables are called value dimensions and key dimensions (respectively). In `macro_df`, the ``'country'`` and ``'year'`` are the independent variables, so when we create a Dataset we will declare those as key dimensions. The remaining columns will automatically be inferred to be value dimensions:",
"_____no_output_____"
]
],
[
[
"macro = hv.Dataset(macro_df, ['country', 'year'])\nmacro",
"_____no_output_____"
]
],
[
[
"One of the first things we'll want to do with our Dimensions is to give them more sensible labels using ``redim.label``:",
"_____no_output_____"
]
],
[
[
"macro = macro.redim.label(growth='GDP Growth', unem='Unemployment', year='Year', country='Country')",
"_____no_output_____"
]
],
[
[
"Notice how HoloViews differs from using a plotting library directly in this case -- here we can annotate the data *once* to capture our knowledge about what those columns represent, and those annotations will then feed directly into any plots later derived from this data. In a plotting program, you would normally need to supply such metadata every single time you make a new plot, which is tedious, error prone, and discourages easy exploration. In the rest of this tutorial we'll see how we can explore and reveal this annotated data.\n\n\n## Groupby\n\nThe great thing about a tidy tabular Dataset is that we can easily group the data by a particular variable, allowing us to plot or analyze each subset separately. Let's say for instance that we want to break the macro-economic data down by 'year'. Using the groupby method we can easily split the Dataset into subsets by year:",
"_____no_output_____"
]
],
[
[
"print(macro.groupby('Year'))",
"_____no_output_____"
]
],
[
[
"The resulting object has a top-level data structure called a [``HoloMap``](http://holoviews.org/reference/containers/bokeh/HoloMap.html), indexed by year. A HoloMap (like its dynamically generated equivalent [``DynamicMap``](http://holoviews.org/reference/containers/bokeh/DynamicMap.html)) is a potentially many-dimensional indexable container for Elements and Datasets, allowing them to be explored easily. \n\nHowever, we cannot visualize this particular HoloMap, because a Dataset has no visual representation. We haven't yet told HoloViews whether the various dependent variables here are continuous, discrete, binned, or any of the other properties the data could have that would allow a specific Element to be chosen.",
"_____no_output_____"
],
[
"## Mapping dimensions to elements\n\nLuckily, as soon as you choose what sort of Element you want a given column to be, you can make this data visualizable using the convenient ``.to`` method, which allows us to group the dataset and map dimensions to elements in a single step.\n\nThe ``.to`` method of a Dataset takes up to four main arguments:\n\n1. The element you want to convert to\n2. The key dimensions (i.e., independent variables) to display\n3. The dependent variables to display, if any\n4. The dimensions to group by, if any\n\nAs a first simple example let's go through such a declaration:\n\n1. We will use a ``Curve``, to declare that the variables are continuous\n2. Our independent variable will be the 'year'\n3. Our dependent variable will be 'unem'\n4. We will ``groupby`` the 'country'.",
"_____no_output_____"
]
],
[
[
"curves = macro.to(hv.Curve, 'year', 'unem', groupby='country')\nprint(curves)\ncurves",
"_____no_output_____"
]
],
[
[
"If you look at the printed output you will see that instead of a simple ``Curve`` we got a ``HoloMap`` of ``Curve`` Elements for each country. Each Curve is now visualizable, but we haven't told HoloViews what to do with the ``Country``, and so to make the entire structure visualizable, HoloViews creates a widget where you can select which country you want to view. Additional value dimensions would result in additional widgets here.\n\nAlternatively we could also group by the year and view the unemployment rate by country as Bars instead. If we simply want to groupby all remaining key dimensions (in this case just the year) we can leave out the groupby argument:",
"_____no_output_____"
]
],
[
[
"%%opts Bars [width=600 xrotation=45]\nbars = macro.sort('country').to(hv.Bars, 'country', 'unem')\nbars",
"_____no_output_____"
],
[
"\n# Exercise: Create a hv.HeatMap using ``macro.to``, declaring kdims 'year' and 'country', and vdims 'growth'\n# You'll need to declare ``width`` and/or ``xrotation`` plot options for HeatMap to make the plot readable\n# You can also add ``tools=['hover']`` to get more info on each cell\n",
"_____no_output_____"
]
],
[
[
"## Displaying distributions\n\nOften we want to summarize the distribution of values, e.g. to reveal the distribution of unemployment rates for each OECD country across all measurements. This means we want to ignore the 'year' dimension in our dataset, letting it be summarized instead. To stop HoloViews from grouping by the extra variable, we pass an empty list to the groupby argument. In this case we can easily declare the ``BoxWhisker`` directly, but omitting a key dimension from the ``groupby`` can be useful in cases when there are more dimensions:",
"_____no_output_____"
]
],
[
[
"%%opts BoxWhisker [width=800 xrotation=30] (box_fill_color=Palette('Category20'))\nmacro.to(hv.BoxWhisker, 'country', 'growth', groupby=[])\n# Is equivalent to:\nhv.BoxWhisker(macro, kdims=['country'], vdims=['growth'])",
"_____no_output_____"
],
[
"\n# Exercise: Display the distribution of GDP growth by year using the BoxWhisker element\n",
"_____no_output_____"
]
],
[
[
"## Faceting dimensions\n\nOnce the data has been grouped into a ``HoloMap`` as we did above, we can further use the grouping capabilities by using the ``.grid``, ``.layout`` and ``.overlay`` methods to lay the groups out on the page rather than flipping through them with a set of widgets.",
"_____no_output_____"
],
[
"#### NdOverlay",
"_____no_output_____"
]
],
[
[
"%%opts Scatter [width=800 height=400 size_index='growth'] (color=Palette('Category20') size=5)\n%%opts NdOverlay [legend_position='left']\nndoverlay = macro.to(hv.Scatter, 'year', ['unem', 'growth']).overlay()\nprint(ndoverlay)\nndoverlay.relabel('OECD Unemployment 1960 - 1990')",
"_____no_output_____"
]
],
[
[
"#### GridSpace",
"_____no_output_____"
]
],
[
[
"%%opts GridSpace [shared_yaxis=True]\nsubset = macro.select(country=['Austria', 'Belgium', 'Netherlands', 'West Germany'])\ngrid = subset.to(hv.Bars, 'year', 'unem').grid()\nprint(grid)\ngrid",
"_____no_output_____"
]
],
[
[
"To understand what is actually going on here, let's rewrite this example in a slightly different way. Instead of using the convenient ``.to`` or ``.groupby`` methods, we can express the same thing by explicitly iterating over the countries we want to look at, selecting the subset of the data for that country using the ``.select`` and then passing these plots to the container we want.\n\nIn the example above that means we ``select`` by 'country' on the macro ``Dataset``, pass the selection to ``Bars`` elements, and declare the key and value dimension to display. We then pass the dictionary of ``Bars`` elements to the ``GridSpace`` container and declare the kdim of the container as 'Country':",
"_____no_output_____"
]
],
[
[
"countries = ['Austria', 'Belgium', 'Netherlands', 'West Germany']\nhv.GridSpace({country: hv.Bars(macro.select(country=country), 'year', 'unem') for country in countries},\n kdims=['Country'])",
"_____no_output_____"
]
],
[
[
"As you can see, ``.to`` is much simpler and less error-prone in practice.\n\n#### NdLayout",
"_____no_output_____"
]
],
[
[
"%%opts Curve [width=200 height=200]\nndlayout = subset.to(hv.Curve, 'year', 'unem').layout()\nprint(ndlayout)\nndlayout",
"_____no_output_____"
],
[
"## Exercise: Recreate the plot above using hv.NdLayout and using macro.select just as we did for the GridSpace above\n",
"_____no_output_____"
]
],
[
[
"## Aggregating\n\nAnother common operation is computing aggregates (summary transformations that collapse some dimensions of the data down to scalar values like a mean or a variance). We can compute and visualize these easily using the ``aggregate`` method. The aggregate method lets you declare the dimension(s) to aggregate by and a function to aggregate with (with an optional secondary function to compute the spread if desired). Once we have computed the aggregate we can simply pass it to the [``Curve``](http://holoviews.org/reference/elements/bokeh/Curve.html) and [``ErrorBars``](http://holoviews.org/reference/elements/bokeh/ErrorBars.html):",
"_____no_output_____"
]
],
[
[
"%%opts Curve [width=600]\nagg = macro.reindex(vdims=['growth']).aggregate('year', function=np.mean, spreadfn=np.std)\nhv.Curve(agg) * hv.ErrorBars(agg)",
"_____no_output_____"
],
[
"\n# Exercise: Display aggregate GDP growth by country, building it up in a series of steps\n# Step 1. First, aggregate the data by country rather than by year, using\n# np.mean and ss.sem as the function and spreadfn, respectively, then\n# make a `Bars` element from the resulting ``agg``\n\n",
"_____no_output_____"
],
[
"\n# Step 2: You should now have a bars plot, but with no error bars. Now add ErrorBars as above. \n# Hint: You'll want to make the plot wider and use an xrotation to see the labels clearly\n",
"_____no_output_____"
]
],
[
[
"<!--\nagg = macro.reindex(vdims=['growth']).aggregate('year', function=np.mean, spreadfn=np.std)\nhv.Curve(agg) * hv.ErrorBars(agg)\n-->\n\n## Onward\n\n* Go through the Tabular Data [getting started](http://holoviews.org/getting_started/Tabular_Datasets.html) and [user guide](http://holoviews.org/user_guide/Tabular_Datasets.html).\n* Learn about slicing, indexing and sampling in the [Indexing and Selecting Data](http://holoviews.org/user_guide/Indexing_and_Selecting_Data.html) user guide.\n\nThe [next section](./05_Working_with_Gridded_Data.ipynb) shows a similar approach, but for working with gridded data, in multidimensional array formats.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbac231b8b9531f3bb81af144871fef1f8504317
| 49,020 |
ipynb
|
Jupyter Notebook
|
latexdemo.ipynb
|
mhy12345/waveform-analysis
|
b71cd456f024eda3ecc04f5f6a69910ce229e86e
|
[
"MIT"
] | null | null | null |
latexdemo.ipynb
|
mhy12345/waveform-analysis
|
b71cd456f024eda3ecc04f5f6a69910ce229e86e
|
[
"MIT"
] | null | null | null |
latexdemo.ipynb
|
mhy12345/waveform-analysis
|
b71cd456f024eda3ecc04f5f6a69910ce229e86e
|
[
"MIT"
] | 1 |
2021-12-06T05:47:02.000Z
|
2021-12-06T05:47:02.000Z
| 41.297388 | 437 | 0.548103 |
[
[
[
"import os\nimport math\n\nimport matplotlib.pyplot as plt\nfrom mpl_toolkits.axisartist.axislines import AxesZero\nimport matplotlib.gridspec as gridspec\nfrom matplotlib import cm, transforms\nimport matplotlib.ticker as mtick\nfrom mpl_axes_aligner import align\nimport numpy as np\nnp.seterr(divide='ignore')\nfrom scipy.stats import poisson, norm, lognorm\nfrom scipy import optimize as opti\nimport pandas as pd\nfrom tqdm import tqdm\nfrom scipy import special\nfrom scipy.stats import norm\nfrom scipy.stats import multivariate_normal\nfrom scipy.signal import savgol_filter\nimport h5py\nimport torch\nfrom torchviz import make_dot\n\nimport wf_func as wff\n\nnp.random.seed(0)",
"_____no_output_____"
],
[
"Mu = 4\nTau = 20\nSigma = 5\nfile = '4.0-20-5'",
"_____no_output_____"
],
[
"with h5py.File('waveform/' + file + '.h5', 'r', libver='latest', swmr=True) as ipt:\n ent = ipt['Readout/Waveform'][:]\n tru = ipt['SimTriggerInfo/PEList'][:]\n gmu = ipt['SimTriggerInfo/PEList'].attrs['gmu']\n gsigma = ipt['SimTriggerInfo/PEList'].attrs['gsigma']\n t0truth = ipt['SimTruth/T'][:]",
"_____no_output_____"
],
[
"def normcombine(x, m, s, a):\n return a[0] * norm.pdf((x - m[0]) / s[0]) + a[1] * norm.pdf((x - m[1]) / s[1])\n\ndef normcombine2d(x, m, s, a, rho):\n return a[0, 0] * multivariate_normal.pdf(x, mean=[m[0, 0], m[1, 0]], cov=matrix(s[0, 0], s[1, 0], rho[0, 0])) + a[0, 1] * multivariate_normal.pdf(x, mean=[m[0, 0], m[1, 1]], cov=matrix(s[0, 0], s[1, 1], rho[0, 1])) + a[1, 0] * multivariate_normal.pdf(x, mean=[m[0, 1], m[1, 0]], cov=matrix(s[0, 1], s[1, 0], rho[1, 0])) + a[1, 1] * multivariate_normal.pdf(x, mean=[m[0, 1], m[1, 1]], cov=matrix(s[0, 1], s[1, 1], rho[1, 1]))\n\ndef matrix(sx, sy, rho):\n return np.array([[sx ** 2, rho * sx * sy], [rho * sx * sy, sy ** 2]])\n\ndef chargehist(t):\n c = norm.pdf(t, loc=gmu, scale=gsigma)\n# q1 = 150.8\n# sigma = 37.59\n# w = 2.433e-5\n# alpha = 0.01335\n# mu = 2.851e-5\n# c = np.exp(-mu)*(w*alpha*np.exp(-alpha*t))\n# c = c + mu*np.exp(-mu)*(\n# (1-w)/(sigma*np.sqrt(2*np.pi))*np.exp(-(t-q1)**2/(2*sigma**2))+\n# w*(alpha/2*np.exp(-alpha*(t-q1-alpha/2*sigma**2))*(1+special.erf(t-q1-alpha*sigma**2)/(np.sqrt(2)*sigma))))\n return c",
"_____no_output_____"
],
[
"Thres = wff.Thres\nstd = 1.\nspe_pre = wff.read_model('spe.h5', 1)\np = spe_pre[0]['parameters']\nwindow = wff.window\nt_auto = np.arange(window).reshape(window, 1) - np.arange(window).reshape(1, window)\nmnecpu = wff.spe((t_auto + np.abs(t_auto)) / 2, p[0], p[1], p[2])",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(8, 6))\nt = np.arange(-4 * 5, 5 * 20, 0.1)\n# gs = gridspec.GridSpec(1, 1, figure=fig, left=0.15, right=0.95, top=0.95, bottom=0.15, wspace=0.4, hspace=0.5)\n# ax = fig.add_subplot(gs[0, 0])\nax = fig.add_axes((.125, .12, .775, .77))\nax.plot(t, wff.convolve_exp_norm(t, 20, 0), label=r'$(20,0)$', color='g')\nax.plot(t, wff.convolve_exp_norm(t, 0, 5), label=r'$(0,5)$', color='r')\nax.plot(t, wff.convolve_exp_norm(t, 20, 5), label=r'$(20,5)$', color='b')\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.grid()\nax.set_xlim(xmin=-4 * int(5))\nax.set_ylabel(r'$\\mathrm{PDF}$')\nax.legend(title=r'$(\\tau_l, \\sigma_l)/\\si{ns}$', loc='upper right')\nax.set_ylim(0, ax.get_ylim()[1] * 1.05)\n# ax.annotate(r'$t_{0}$', xy=(0, 0), xytext=(5, 0.01), arrowprops=dict(facecolor='k', shrink=0.1, width=0.1, headwidth=2))\nfig.savefig('Note/figures/profile.pgf')\nfig.savefig('Note/figures/profile.pdf')\nplt.close()\nax.get_position()",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(8, 4))\n# fig.tight_layout()\ngs = gridspec.GridSpec(1, 1, figure=fig, left=0.05, right=0.97, top=0.97, bottom=0.1, wspace=0.3, hspace=0.3)\nax = fig.add_subplot(gs[0, 0])\n\nax.spines['left'].set_position(('data', 0))\nax.spines['bottom'].set_position(('data', 0))\nax.plot(1, 0, '>k', transform=ax.get_yaxis_transform(), clip_on=False)\nax.plot(0, 1, '^k', transform=ax.get_xaxis_transform(), clip_on=False)\n\nt = np.linspace(0, 6, 201)\nax.plot(t, lognorm.pdf(t, loc=0, s=0.3), color='darkorange')\nax.plot(t, lognorm.pdf(t, loc=3, s=0.3), color='darkblue')\nax.fill_between(t, 0, lognorm.pdf(t, loc=0, s=0.3), color='darkorange', alpha=0.5)\nax.fill_between(t, 0, lognorm.pdf(t, loc=3, s=0.3), color='darkblue', alpha=0.5)\nax.set_xlim(0, 6)\nax.set_ylim(bottom=1e-3)\nax.set_xticks([])\nax.set_yticks([])\nax.set_xlabel(r'$\\mathrm{Time}$')\nax.set_ylabel(r'$\\mathrm{Voltage}$')\nax.spines['top'].set_visible(False)\nax.spines['right'].set_visible(False)\nax.annotate(s='', xy=(1.5, 1), xytext=(3.5, 1), arrowprops=dict(arrowstyle='<->'))\nax.text(x=2.2, y=1.1, s=r'$\\sim D_w$')\nax.text(x=0.7, y=0.3, s=r'$\\sim \\mathrm{RSS}$')\nax.text(x=3.7, y=0.3, s=r'$\\sim \\mathrm{RSS}$')\nfig.savefig('Note/figures/tab.pgf')\nfig.savefig('Note/figures/tab.pdf')\nfig.clf()\nplt.close(fig)",
"_____no_output_____"
],
[
"i = 2\ncid = ent[i]['ChannelID']\neid = ent[i]['TriggerNo']\ntruth = np.sort(tru[(tru['TriggerNo'] == eid) & (tru['PMTId'] == cid)], kind='stable', order=['TriggerNo', 'PMTId', 'HitPosInWindow'])\nwave = ent[i]['Waveform'].astype(np.float) * spe_pre[cid]['epulse']\ndf = pd.DataFrame(truth)\ndf = df.rename(columns={'HitPosInWindow':'HitTime'})\ncharge = df['Charge'].copy()\nhittime = df['HitTime'].copy()\ndf = df.astype({'Charge': 'float32'})\ndf = df.astype({'TriggerNo' : 'str', 'PMTId' : 'str', 'HitTime' : 'str', 'Charge': 'str'})\ndf['HitTime'] = ['{:.02f}'.format(s) for s in hittime]\ndf['Charge'] = ['{:.02f}'.format(s) for s in charge]\ndf",
"_____no_output_____"
],
[
"ind = np.argwhere(wave > spe_pre[cid]['std'] * 5).flatten()\nxmin = ((ind.min() - spe_pre[cid]['mar_l']) // 20 - 1) * 20\nxmax = max(((ind.max() + spe_pre[cid]['mar_r']) // 20 + 1) * 20, xmin + 200)",
"_____no_output_____"
],
[
"TRIALS = 8000\nn = 1\nb_t0 = [0., 600.]\n\n# initialization\nA, y, tlist, t0_t, t0_delta, cha, left_wave, right_wave = wff.initial_params(wave[::wff.nshannon], spe_pre[cid], Tau, Sigma, gmu, Thres['lucyddm'], p, is_t0=True, is_delta=False, n=n)\n# assert len(np.unique(np.diff(tlist))) == 1\ns_cha = np.cumsum(cha)\n# moving average filter of size 2*n+1\ncha = np.pad(s_cha[2*n+1:], (n+1, n), 'edge') - np.pad(s_cha[:-(2*n+1)], (n+1, n), 'edge')\ncha += 1e-8 # for completeness of the random walk.\np_cha = cha / np.sum(cha)\nmu_t = abs(y.sum() / gmu)\n\n# Eq. (9) where the columns of A are taken to be unit-norm.\nmus = np.sqrt(np.diag(np.matmul(A.T, A)))\nassert np.std(mus) < 1e-4, 'mus must be equal'\nmus = mus[0]\nA = A / mus\np1 = mu_t * wff.convolve_exp_norm(tlist - t0_t, Tau, Sigma) / n + 1e-8\nsig2w = spe_pre[cid]['std'] ** 2\nsig2s = (gsigma * mus / gmu) ** 2\n\nnu_star, T_star, c_star, es_history, NPE_evo = wff.metropolis_fbmp(y, A, sig2w, sig2s, mus, p1, p_cha, mu_t)\n\nilp_cha = np.log(cha.sum()) - np.log(cha)\nguess = ilp_cha[es_history['loc'].astype(int)]\nes_history['loc'] = np.interp(es_history['loc'], xp=np.arange(0.5, len(tlist)), fp=tlist)\nans = opti.fmin_l_bfgs_b(lambda x: -np.sum(wff.log_convolve_exp_norm(es_history['loc'] - x, Tau, Sigma)), x0=[t0_t], approx_grad=True, bounds=[b_t0], maxfun=500000)\nt00 = ans[0].item() if ans[-1]['warnflag'] == 0 else t0_t\ndef fit():\n mu = mu_t\n b_mu = [max(1e-8, mu - 5 * np.sqrt(mu)), mu + 5 * np.sqrt(mu)]\n def agg_NPE(t0):\n log_f = wff.log_convolve_exp_norm(es_history['loc'] - t0, Tau, Sigma) + guess\n return wff.jit_agg_NPE(es_history['step'], log_f, TRIALS)\n\n def t_t0(t0):\n nonlocal mu\n NPE, f_agg = agg_NPE(t0)\n ans = opti.fmin_l_bfgs_b(lambda μ: μ - special.logsumexp(NPE * np.log(μ / mu) + f_agg), x0=[mu], approx_grad=True, bounds=[b_mu], maxfun=500000)\n mu = ans[0].item()\n return ans[1]\n\n ans = opti.fmin_l_bfgs_b(t_t0, x0=[t00], approx_grad=True, bounds=[b_t0], maxfun=500000)\n t0 = ans[0].item()\n return mu, t0\nmu, t0 = fit()\n\nj = 0\nxmmse_most = np.zeros(len(tlist))\nwhile np.all(xmmse_most <= 0):\n maxindex = nu_star.argsort()[::-1][j]\n zx = y - np.dot(A, mus * c_star[maxindex])\n Phi_s = wff.Phi(y, A, c_star[maxindex], mus, sig2s, sig2w)\n invPhi = np.linalg.inv(Phi_s)\n xmmse_most = mus * c_star[maxindex] + np.matmul(np.diagflat(sig2s * c_star[maxindex]), np.matmul(A.T, np.matmul(invPhi, zx)))\n j += 0\npet = np.repeat(tlist[xmmse_most > 0], c_star[maxindex][xmmse_most > 0])\ncha = np.repeat(xmmse_most[xmmse_most > 0] / mus / c_star[maxindex][xmmse_most > 0], c_star[maxindex][xmmse_most > 0])\n\npet, pwe = wff.clip(pet, cha, 0.0)\npwe = pwe\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=0.5)\nax.plot(wave, label='Waveform')\nax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nwave_ylim = ax.get_ylim()\nfig.savefig('Note/figures/fbmp.pgf')\nfig.savefig('Note/figures/fbmp.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p)\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [198. 212. 222. 228. 247.], Weight = [0.61287108 0.89353614 1.06551214 0.53047119 0.5244661 ]\nwdist = 0.6358741149424056, cdiff = 0.026090974619535037\nRSS = 17.77534343983739\n-3.9656086721994654\n"
],
[
"# fig = plt.figure(figsize=(8, 6))\n# # fig.tight_layout()\n# ax = fig.add_axes((.125, .12, .775, .77))\n# ax.plot(wave, label='Waveform')\n# ax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\n# ax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\n# ax.set_xlim(0, len(wave))\n# wave_ylim = ax.get_ylim()\n# ax.set_ylim(wave_ylim[0] * 1.05, wave_ylim[1] * 1.05)\n# ax.legend()\n# fig.savefig('Note/figures/wave.pgf')\n# fig.savefig('Note/figures/wave.pdf')\n# fig.savefig('Note/figures/wave.png')\n# fig.clf()\n# plt.close(fig)\n\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(truth['HitPosInWindow'], 0, truth['Charge'] / gmu, color='r', label='Charge', linewidth=1.0)\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nax2.set_ylim(wave_ylim[0] / 30, wave_ylim[1] / 30)\nax.plot(wave, label='Waveform')\n# ax.set_xlim(xmin, xmax)\nax.set_xlim(0, len(wave) / 2)\nax.set_ylim(wave_ylim[0] * 0.7, wave_ylim[1] * 0.7)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nalign.yaxes(ax, 0, ax2, 0)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nfig.savefig('Note/figures/wave.pgf')\nfig.savefig('Note/figures/wave.pdf')\nfig.savefig('Note/figures/wave.png')\nfig.clf()\nplt.close(fig)\n\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\n# gs = gridspec.GridSpec(1, 1, figure=fig, left=0.15, right=0.85, top=0.95, bottom=0.15, wspace=0.4, hspace=0.5)\n# ax = fig.add_subplot(gs[0, 0])\nax = fig.add_axes((.125, .12, .775, .77))\nt = np.arange(0, 100, 0.1)\nax.plot(t, wff.spe(t, p[0], p[1], p[2]), color='b', label='Single PE response')\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.grid()\nax.set_xlim(0, 80)\nax.set_ylim(wave_ylim[0] * 0.7, wave_ylim[1] * 0.7)\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax.legend()\nfig.savefig('Note/figures/spe.pgf')\nfig.savefig('Note/figures/spe.pdf')\nfig.savefig('Note/figures/spe.png')\nplt.close()\n\n# fig = plt.figure(figsize=(8, 6))\n# # fig.tight_layout()\n# ax = fig.add_axes((.125, .12, .775, .77))\n# ax.vlines(truth['HitPosInWindow'], 0, truth['Charge'] / gmu, color='r', label='Charge')\n# ax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\n# ax.set_ylabel(r'$\\mathrm{Charge}$')\n# ax.set_xlim(0, len(wave))\n# ax.set_ylim(wave_ylim[0] / 20, wave_ylim[1] / 20)\n# ax.axhline(y=0, color='k', linestyle='dashed', alpha=0.5)\n# ax.legend()\n# fig.savefig('Note/figures/charge.pgf')\n# fig.savefig('Note/figures/charge.pdf')\n# fig.savefig('Note/figures/charge.png')\n# fig.clf()\n# plt.close(fig)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\n# gs = gridspec.GridSpec(1, 1, figure=fig, left=0.15, right=0.85, top=0.95, bottom=0.15, wspace=0.4, hspace=0.5)\n# ax = fig.add_subplot(gs[0, 0])\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(truth['HitPosInWindow'], 0, truth['Charge'] / gmu, color='r', label='Charge')\nax.plot(wave, label='Waveform')\nax.hlines(2, 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nax.set_ylim(bottom=-5)\nax2.set_ylim(bottom=-5 / gmu)\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/goal.pgf')\nfig.savefig('Note/figures/goal.pdf')\nfig.clf()\nplt.close(fig)",
"_____no_output_____"
],
[
"print(wave.sum())\nprint(truth['Charge'][truth['Charge'] > 0].sum()) # made by noise",
"600.612084468333\n580.2596844449434\n"
],
[
"t = np.load('result/takara/char/Channel00/cnn_testing_record_2021-07-30_17:15:10.npz')['arr_0']\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax.plot(np.arange(1, len(t)+1), t, label=r'$D_w$', color='C1')\nax.set_xlabel(r'$\\mathrm{epoch}$')\nax.set_ylabel(r'$\\mathrm{Wasserstein\\ Distance}/\\si{ns}$')\nax.legend()\nax.grid()\nfig.savefig('Note/figures/epoch.pgf')\nfig.savefig('Note/figures/epoch.pdf')\nfig.clf()\nplt.close(fig)",
"_____no_output_____"
],
[
"pet, pwe = wff.threshold(wave, spe_pre[cid])\npet, pwe = wff.clip(pet, pwe, Thres['threshold'])\noutput = np.zeros(window)\noutput[pet] = pwe\nalpha = opti.fmin_l_bfgs_b(lambda alpha: wff.rss_alpha(alpha, output, wave, mnecpu), x0=[0.01], approx_grad=True, bounds=[[1e-20, np.inf]], maxfun=50000)[0]\npwe = pwe * alpha\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=0.5)\nax.plot(wave, label='Waveform')\nax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nax2.annotate('', xy=(pet.mean(), pwe.max()*1.1), xytext=(pet.mean()+pet.ptp(), pwe.max()*1.1), arrowprops=dict(facecolor='k', shrink=0.01, width=2, headwidth=4))\nax2.set_ylim(top=pwe.max()*1.2)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/threshold.pgf')\nfig.savefig('Note/figures/threshold.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p, fold='/tmp')\nt0 = wff.likelihoodt0(pet, char=pwe * gmu, gmu=gmu, Tau=Tau, Sigma=Sigma, mode='charge')[0]\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [195 196 197 198 199 200 201 202 203 205 208 209 210 211 212 213 214 215\n 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233\n 234 235 236 244 245 246 247 248 249 250 251], Weight = [0.03584503 0.04490778 0.05110216 0.05317444 0.05945964 0.06187086\n 0.05573373 0.04979399 0.04034561 0.03863693 0.04677279 0.04984948\n 0.08103905 0.09004558 0.09130871 0.10570219 0.07561306 0.0813948\n 0.08146041 0.06363937 0.09169068 0.12561056 0.12280308 0.12547056\n 0.12715693 0.12155163 0.13276326 0.13767504 0.12497751 0.12037777\n 0.11556223 0.10702699 0.09377507 0.08716678 0.06529191 0.04768089\n 0.04531403 0.05371381 0.03920505 0.0472688 0.05136636 0.04668365\n 0.0619234 0.05624993 0.04690911 0.04493587 0.04083298]\nwdist = 3.1087242325850317, cdiff = -14.081985436358435\nRSS = 369.5632725931966\n-0.6859650837487834\n"
],
[
"pet, pwe = wff.findpeak(wave, spe_pre[cid])\npet, pwe = wff.clip(pet, pwe, Thres['findpeak'])\noutput = np.zeros(window)\noutput[pet] = pwe\nalpha = opti.fmin_l_bfgs_b(lambda alpha: wff.rss_alpha(alpha, output, wave, mnecpu), x0=[0.01], approx_grad=True, bounds=[[1e-20, np.inf]], maxfun=50000)[0]\npwe = pwe * alpha\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=1.5)\nax.plot(wave, label='Waveform')\nax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nloc = pet + spe_pre[cid]['peak_c']\nloc = loc[loc < window]\namp = wave[loc]\nfor j in range(len(loc)):\n ax.annotate('', xy=(loc[j], amp[j]+5), xytext=(loc[j], amp[j]+15), arrowprops=dict(facecolor='k', shrink=0.01, width=0.5, headwidth=2))\nax2.annotate('', xy=(pet.mean(), pwe.max()*1.1), xytext=(pet.mean()+pet.ptp(), pwe.max()*1.1), arrowprops=dict(facecolor='k', shrink=0.01, width=2, headwidth=4))\nax2.set_ylim(top=pwe.max()*1.2)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/findpeak.pgf')\nfig.savefig('Note/figures/findpeak.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p, fold='/tmp')\nt0 = wff.likelihoodt0(pet, char=pwe * gmu, gmu=gmu, Tau=Tau, Sigma=Sigma, mode='charge')[0]\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [200 213 223 248], Weight = [0.61116178 1.04412873 1.20068987 0.55563809]\nwdist = 2.3999066086436542, cdiff = -34.41134854495826\nRSS = 266.9323131608509\n-1.9290449992176946\n"
],
[
"pet, pwe = wff.waveformfft(wave, spe_pre[cid])\npet, pwe = wff.clip(pet, pwe, Thres['fftrans'])\noutput = np.zeros(window)\noutput[pet] = pwe\nalpha = opti.fmin_l_bfgs_b(lambda alpha: wff.rss_alpha(alpha, output, wave, mnecpu), x0=[0.01], approx_grad=True, bounds=[[1e-20, np.inf]], maxfun=50000)[0]\npwe = pwe * alpha\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=0.5)\nax.plot(wave, label='Waveform')\nax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/fftrans.pgf')\nfig.savefig('Note/figures/fftrans.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p, fold='/tmp')\nt0 = wff.likelihoodt0(pet, char=pwe * gmu, gmu=gmu, Tau=Tau, Sigma=Sigma, mode='charge')[0]\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [198 199 200 210 211 212 213 214 220 221 222 223 224 225 226 227 246 247\n 248], Weight = [0.14995905 0.16136076 0.14971286 0.15427833 0.19532405 0.20962768\n 0.19607729 0.16101529 0.1266714 0.1781401 0.22298341 0.24987798\n 0.25239387 0.23044018 0.18988035 0.14052016 0.14142437 0.14293999\n 0.12627225]\nwdist = 2.029237712534919, cdiff = -39.646304087048016\nRSS = 124.67317512606327\n-1.1599673109834612\n"
],
[
"pet, pwe = wff.lucyddm(wave, spe_pre[cid]['spe'])\npet, pwe = wff.clip(pet, pwe, Thres['lucyddm'])\noutput = np.zeros(window)\noutput[pet] = pwe\nalpha = opti.fmin_l_bfgs_b(lambda alpha: wff.rss_alpha(alpha, output, wave, mnecpu), x0=[0.01], approx_grad=True, bounds=[[1e-20, np.inf]], maxfun=50000)[0]\npwe = pwe * alpha\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=0.5)\nax.plot(wave, label='Waveform')\nax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/lucyddm.pgf')\nfig.savefig('Note/figures/lucyddm.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p, fold='/tmp')\nt0 = wff.likelihoodt0(pet, char=pwe * gmu, gmu=gmu, Tau=Tau, Sigma=Sigma, mode='charge')[0]\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [199 212 213 222 228 247], Weight = [0.45509249 0.48911629 0.54882022 1.04444101 0.43353076 0.50499002]\nwdist = 1.0994500816814046, cdiff = -24.111976439534768\nRSS = 70.25593260038926\n-0.7472914331635252\n"
],
[
"with h5py.File('result/takara/char/' + file + '.h5', 'r', libver='latest', swmr=True) as ipt:\n photoelec = ipt['photoelectron'][:]\ns = photoelec[(photoelec['TriggerNo'] == eid) & (photoelec['ChannelID'] == cid)]\npet = s['HitPosInWindow']\npwe = s['Charge']\npwe = pwe / gmu\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=0.5)\nax.plot(wave, label='Waveform')\n# ax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/takara.pgf')\nfig.savefig('Note/figures/takara.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p, fold='/tmp')\nt0 = wff.likelihoodt0(pet, char=pwe * gmu, gmu=gmu, Tau=Tau, Sigma=Sigma, mode='charge')[0]\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [198 199 212 213 222 228 247], Weight = [0.57807553 0.07023228 0.37157281 0.51505843 1.00229091 0.57173777\n 0.51790512]\nwdist = 0.6408859192063601, cdiff = 0.028682484579321738\nRSS = 10.958349058233402\n-3.449534874550608\n"
],
[
"with h5py.File('result/mcmc/char/' + file + '.h5', 'r', libver='latest', swmr=True) as ipt:\n photoelec = ipt['photoelectron'][:]\ns = photoelec[(photoelec['TriggerNo'] == eid) & (photoelec['ChannelID'] == cid)]\npet = s['HitPosInWindow']\npwe = s['Charge']\npwe = pwe / gmu\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=0.5)\nax.plot(wave, label='Waveform')\nax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/mcmc.pgf')\nfig.savefig('Note/figures/mcmc.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p, fold='/tmp')\nt0 = wff.likelihoodt0(pet, char=pwe * gmu, gmu=gmu, Tau=Tau, Sigma=Sigma, mode='charge')[0]\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [195. 196. 196.5 197. 198. 198.5 199. 199.5 200. 201.5 202. 202.5\n 203. 209.5 210. 212.5 213. 214. 217.5 219. 221.5 222. 224.5 227.5\n 228. 229.5 230. 230.5 246. 246.5 247.5 251. ], Weight = [0.00006055 0.0000123 0.00002088 0.00001721 0.00001772 0.62653152\n 0.00004476 0.00002066 0.00000857 0.00000206 0.00002402 0.00000339\n 0.00001742 0.00003347 0.00000176 0.88393095 0.0000119 0.00002502\n 0.00000106 0.00000154 0.49642209 0.49695779 0.00000266 0.5833009\n 0.00001748 0.00001247 0.00001806 0.00000023 0.00003033 0.52634848\n 0.00002249 0.00000373]\nwdist = 0.695623374533694, cdiff = -2.043179450830088\nRSS = 8.949425328589244\n-2.9015390278386803\n"
],
[
"# pet, pwe = wff.xiaopeip(wave, spe_pre[cid], eta=0)\npet, pwe = wff.xiaopeip(wave, spe_pre[cid], Tau, Sigma, Thres['lucyddm'], p, eta=0)\npet, pwe = wff.clip(pet, pwe, Thres['xiaopeip'])\nfig = plt.figure(figsize=(8, 6))\n# fig.tight_layout()\nax = fig.add_axes((.125, .12, .775, .77))\nax2 = ax.twinx()\nax2.vlines(pet, 0, pwe, color='r', label='Charge', linewidth=0.5)\nax.plot(wave, label='Waveform')\n# ax.hlines(5 * spe_pre[cid]['std'], 0, window, color='g', label='Threshold')\nax.set_xlim(xmin, xmax)\nlines, labels = ax.get_legend_handles_labels()\nlines2, labels2 = ax2.get_legend_handles_labels()\nax2.yaxis.get_major_formatter().set_powerlimits((0, 1))\nax2.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax2.legend(lines + lines2, labels + labels2)\nax.set_xlabel(r'$\\mathrm{t}/\\si{ns}$')\nax.set_ylabel(r'$\\mathrm{Voltage}/\\si{mV}$')\nax2.set_ylabel(r'$\\mathrm{Charge}$')\nalign.yaxes(ax, 0, ax2, 0)\nfig.savefig('Note/figures/xiaopeip.pgf')\nfig.savefig('Note/figures/xiaopeip.pdf')\nfig.clf()\nplt.close(fig)\nwff.demo(pet, pwe, truth, spe_pre[cid], window, wave, cid, p, fold='/tmp')\nt0 = wff.likelihoodt0(pet, char=pwe * gmu, gmu=gmu, Tau=Tau, Sigma=Sigma, mode='charge')[0]\nprint((t0 - t0truth[i]['T0']).item())",
"PEnum is 5\ntruth HitPosInWindow = [198.30013112 212.60177919 222.16899484 227.93643283 246.79015495], Weight = [0.61947338 0.95673715 1.01986389 0.52680905 0.50381012]\ntruth RSS = 1003.2304605600609\nHitPosInWindow = [195. 196. 197. 198. 199. 200. 201. 202. 212. 213. 221. 222. 223. 227.\n 228. 246. 247.], Weight = [0.0659943 0.01389369 0.00915128 0.21940894 0.29132406 0.00109253\n 0.0212546 0.02775144 0.4397345 0.4458349 0.20819241 0.76179933\n 0.04677314 0.11259772 0.44952796 0.06116492 0.46108861]\nwdist = 0.7822434792350879, cdiff = 1.5824878343872444\nRSS = 8.85458283626419\n-3.2778238764112757\n"
]
],
[
[
"with h5py.File('result/mcmc/solu/' + file + '.h5', 'r', libver='latest', swmr=True) as soluf, h5py.File('waveform/' + file + '.h5', 'r', libver='latest', swmr=True) as wavef:\n start = wavef['SimTruth/T'][:]\n time = soluf['starttime'][:]\ndata = time['tswave'] - start['T0']\nvali = np.abs(data - np.mean(data)) < 5 * np.std(data, ddof=-1)\ndata = data[vali]\nfig = plt.figure(figsize=(8, 6))\nax = fig.add_axes((.1, .45, .85, .45))\nax.hist(data, bins=100, density=1)\nax.set_ylabel('arb. unit')\nax.set_xticks([])\nax.set_yticks([])\nax.set_xlim(data.min() - 0.05, data.max() + 0.05)\nax.set_ylim(0, ax.get_ylim()[1] * 1.05)\naxb = fig.add_axes((.1, .15, .85, .3))\naxb.boxplot(data, vert=False, sym='', patch_artist=True)\naxb.set_xlabel(r'$\\Delta t_{0}/\\si{ns}$')\naxb.set_yticks([])\naxb.set_xlim(ax.get_xlim())\nfig.savefig('Note/figures/mcmct0hist.pgf')\nfig.savefig('Note/figures/mcmct0hist.pdf')\nplt.close(fig)",
"_____no_output_____"
]
],
[
[
"methods = ['lucyddm', 'xiaopeip', 'takara', 'fbmp', 'mcmc']\n\nfor m in methods:\n with h5py.File('result/' + m + '/dist/' + file + '.h5', 'r', libver='latest', swmr=True) as distfile:\n dt = distfile['Record'][:]\n N = np.percentile(dt['wdist'], 95)\n M = 500\n\n penum = np.unique(dt['NPE'])\n l = min(50, penum.max())\n wdist_stats = np.full((l, 6), np.nan)\n edist_stats = np.full((l, 6), np.nan)\n for i in range(l):\n vali = dt['NPE'] == i+1\n if np.sum(vali) == 0:\n continue\n dtwpi = dt['wdist'][vali]\n dtepi = dt['RSS'][vali]\n wdist_stats[i, 0] = np.median(dtwpi)\n wdist_stats[i, 1] = np.median(np.abs(dtwpi - np.median(dtwpi)))\n wdist_stats[i, 2] = np.mean(dtwpi)\n wdist_stats[i, 3] = np.std(dtwpi)\n wdist_stats[i, 4] = np.percentile(dtwpi, 5)\n wdist_stats[i, 5] = np.percentile(dtwpi, 95)\n edist_stats[i, 0] = np.median(dtepi)\n edist_stats[i, 1] = np.median(np.abs(dtepi - np.median(dtepi)))\n edist_stats[i, 2] = np.mean(dtepi)\n edist_stats[i, 3] = np.std(dtepi)\n edist_stats[i, 4] = np.percentile(dtepi, 5)\n edist_stats[i, 5] = np.percentile(dtepi, 95)\n\n L = len(dt)\n data = dt['wdist']\n fig = plt.figure(figsize=(8, 6))\n ax1 = fig.add_axes((.125, .12, .6, .77))\n boxdict = ax1.boxplot(np.array([dt['wdist'][dt['NPE'] == i+1] for i in range(l)], dtype=np.object), sym='', patch_artist=True)\n ax1.set_xticks(np.arange(1, 16, 2))\n ax1.set_xticklabels(np.arange(1, 16, 2).astype(str))\n ax1.plot(np.arange(1, l + 1), wdist_stats[:, 0], label=r'$D_w$')\n ax1.set_xlim(0, l + 1)\n ax1.set_ylim(0, max([boxdict['whiskers'][2 * i + 1].get_xydata()[1, 1] for i in range(l)]) * 1.05)\n ax1.set_xlabel(r'$N_{\\mathrm{PE}}$')\n ax1.set_ylabel(r'$\\mathrm{Wasserstein\\ Distance}/\\si{ns}$')\n ax1.legend()\n ax2 = fig.add_axes((.725, .12, .175, .77))\n ax2.hist(data, bins=np.arange(0, data.max(), np.percentile(data, 98) / 40), density=1, orientation='horizontal')\n ax2.set_xlabel(r'$\\mathrm{arb.\\ unit}$')\n ax2.set_xlim(0, ax2.get_xlim()[1] * 1.05)\n ax2.set_xticks([])\n ax2.set_yticks([])\n ax2.set_ylim(ax1.get_ylim())\n fig.savefig('Note/figures/' + m + 'chargestats.pgf')\n fig.savefig('Note/figures/' + m + 'chargestats.pdf')\n plt.close(fig)",
"_____no_output_____"
],
[
"t = np.arange(0, 1000, 0.1) / gmu\n\npdf = np.zeros_like(t)\ntlist = np.arange(-50, 200)\n\nfor mu in tqdm(Mu * wff.convolve_exp_norm(tlist, Tau, Sigma)):\n for i in range(1, 15):\n pdf += mu * poisson.pmf(i, mu) * norm.pdf(t, loc=1, scale=gsigma / gmu / np.sqrt(i))",
"100%|██████████| 250/250 [00:01<00:00, 125.09it/s]\n"
],
[
"methods = ['lucyddm', 'xiaopeip', 'takara', 'fbmp']\ncolors = {'truth':'k', 'lucyddm':'y', 'xiaopeip':'c', 'takara':'C0', 'fbmp':'r'}\nfig = plt.figure(figsize=(10, 6))\nfig.tight_layout()\nax = fig.add_axes((.1, .12, .85, .80))\nt = np.arange(0, 1000, 0.1) / gmu\n# ax.plot(t, norm.pdf(t, loc=1, scale=gsigma / gmu) / (1 - norm.cdf(0, loc=1, scale=gsigma / gmu)), color=colors['truth'], alpha=0.2)\nax.plot(t, pdf / pdf.sum() / np.diff(t)[0], label='$\\mathrm{ChargePDF}$', color=colors['truth'])\n# th = 160 * 5 * 1e-4\nth = 10 / gmu\nlabels = {'truth':'\\mathrm{Truth}', 'lucyddm':'\\mathrm{LucyDDM}', 'xiaopeip':'\\mathrm{Fitting}', 'takara':'\\mathrm{CNN}', 'fbmp':'\\mathrm{FSMP}', 'fbmpwave':'\\mathrm{FSMP}'}\nfor m in methods:\n ch = h5py.File('result/' + m + '/char/' + file + '.h5', 'r', libver='latest', swmr=True)\n cha = ch['photoelectron']['Charge'] / gmu\n ax.hist(cha[cha > th], bins=np.linspace(th, 400 / gmu, 101), label='$'+labels[m]+'$', histtype='step', density=True, color=colors[m], linewidth=2.)\nax.set_xlim(10 / gmu, 310 / gmu)\nax.set_yticks([])\n# ax.yaxis.get_major_formatter().set_powerlimits((0, 1))\n# ax.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax.legend()\n# ax.set_xlabel('$\\mathrm{Charge}/\\si{mV\\cdot ns}$')\nax.set_xlabel('$\\mathrm{Charge}$')\nax.set_ylabel(r'$\\mathrm{Normalized\\ Count}$')\nplt.savefig('Note/figures/recchargehist.png')\nplt.savefig('Note/figures/recchargehist.pdf')\nplt.savefig('Note/figures/recchargehist.pgf')\nplt.show()",
"_____no_output_____"
],
[
"t = np.arange(0, 1000, 0.1) / gmu\n\npdf = np.zeros_like(t)\nb = 0.5\ntlist = np.arange(-50, 200, b)\n\nfor mu in tqdm(25 * wff.convolve_exp_norm(tlist, Tau, Sigma) * b):\n for i in range(1, 15):\n pdf += mu * poisson.pmf(i, mu) * norm.pdf(t, loc=1, scale=gsigma / gmu / np.sqrt(i))",
"100%|██████████| 500/500 [00:03<00:00, 126.79it/s]\n"
],
[
"methods = ['lucyddm', 'xiaopeip', 'takara', 'fbmp']\ncolors = {'truth':'k', 'lucyddm':'y', 'xiaopeip':'c', 'takara':'C0', 'fbmp':'r'}\nfig = plt.figure(figsize=(10, 6))\nfig.tight_layout()\nax = fig.add_axes((.1, .12, .85, .80))\nax.plot(t, norm.pdf(t, loc=1, scale=gsigma / gmu) / (1 - norm.cdf(0, loc=1, scale=gsigma / gmu)), color=colors['truth'], alpha=0.2)\nax.plot(t, pdf / pdf.sum() / np.diff(t)[0], label='$\\mathrm{ChargePDF}$', color=colors['truth'])\n# th = 160 * 5 * 1e-4\nth = 10 / gmu\nlabels = {'truth':'\\mathrm{Truth}', 'lucyddm':'\\mathrm{LucyDDM}', 'xiaopeip':'\\mathrm{Fitting}', 'takara':'\\mathrm{CNN}', 'fbmp':'\\mathrm{FSMP}', 'fbmpwave':'\\mathrm{FSMP}'}\nfor m in methods:\n ch = h5py.File('result/' + m + '/char/15.0-20-5' + '.h5', 'r', libver='latest', swmr=True)\n cha = ch['photoelectron']['Charge'] / gmu\n ax.hist(cha[cha > th], bins=np.linspace(th, 400 / gmu, 101), label='$'+labels[m]+'$', histtype='step', density=True, color=colors[m], linewidth=2.)\nax.set_xlim(10 / gmu, 310 / gmu)\nax.set_yticks([])\n# ax.yaxis.get_major_formatter().set_powerlimits((0, 1))\n# ax.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))\nax.legend()\n# ax.set_xlabel('$\\mathrm{Charge}/\\si{mV\\cdot ns}$')\nax.set_xlabel('$\\mathrm{Charge}$')\nax.set_ylabel(r'$\\mathrm{Normalized\\ Count}$')\nplt.savefig('Note/figures/recchargehist25.png')\n# plt.savefig('Note/figures/recchargehist.pdf')\n# plt.savefig('Note/figures/recchargehist.pgf')\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"raw",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"raw"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbac3ea7545a0a8175fd353fa472ca43cc4aa57d
| 4,067 |
ipynb
|
Jupyter Notebook
|
04_utils.ipynb
|
netdata/netdata-airflow-utils
|
e7eb7b06fe427d9a5610eb93bfc409f1cc35675f
|
[
"Apache-2.0"
] | null | null | null |
04_utils.ipynb
|
netdata/netdata-airflow-utils
|
e7eb7b06fe427d9a5610eb93bfc409f1cc35675f
|
[
"Apache-2.0"
] | null | null | null |
04_utils.ipynb
|
netdata/netdata-airflow-utils
|
e7eb7b06fe427d9a5610eb93bfc409f1cc35675f
|
[
"Apache-2.0"
] | null | null | null | 29.686131 | 137 | 0.544136 |
[
[
[
"# default_exp utils",
"_____no_output_____"
]
],
[
[
"# Utils\n\n> General purpose utils functions",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
],
[
"#export\n\nimport os\nfrom typing import Dict, Any\n",
"_____no_output_____"
],
[
"#export\n\n\ndef dest(destination_dataset_table, prefix_dataset='tmp', return_dataset_only=False, return_table_only=False) -> Dict[str, Any]:\n \"\"\"If `AIRFLOW_ENV != PROD` then write results to `prefix_dataset` instead.\n\n :param destination_dataset_table: destination to write results to.\n :return: destination_dataset_table: destination to write results to with prefix added if needed.\n \"\"\"\n\n AIRFLOW_ENV = os.environ.get(\"AIRFLOW_ENV\", \"UNK\")\n\n if AIRFLOW_ENV != 'PROD':\n destination_dataset_table_list = destination_dataset_table.replace(':', '.').split('.')\n destination_project = destination_dataset_table_list[0]\n destination_dataset = prefix_dataset\n destination_table = f'{destination_dataset_table_list[1]}_{destination_dataset_table_list[2]}'\n destination_dataset_table = f'{destination_project}.{destination_dataset}.{destination_table}'\n\n destination_parts = destination_dataset_table.split('.')\n\n if return_dataset_only == True:\n return destination_parts[1]\n elif return_table_only == True:\n return destination_parts[2]\n else:\n return destination_dataset_table\n\n\ndef dest_dict(destination_dataset_table, prefix_dataset='tmp') -> Dict[str, str]:\n \"\"\"Wrapper for `dest()` but to return as dict.\n \"\"\"\n destination_dataset_table = dest(destination_dataset_table, prefix_dataset)\n destination_parts = destination_dataset_table.split('.')\n return {\n \"projectId\": destination_parts[0],\n \"datasetId\": destination_parts[1],\n \"tableId\": destination_parts[2]\n }\n\n\ndef sched(schedule: Any) -> Any:\n \"\"\"If AIRFLOW_ENV != PROD then schedule should be `@once`.\n\n :param schedule: schedule for prod.\n :return: schedule: `schedule` if prod else `@once`.\n \"\"\"\n\n AIRFLOW_ENV = os.environ.get(\"AIRFLOW_ENV\", \"UNK\")\n\n if AIRFLOW_ENV == 'PROD':\n return schedule\n else:\n return '@once'",
"_____no_output_____"
],
[
"#tests\n#hide\nos.environ.pop('AIRFLOW_ENV', None)\nassert sched('foo') == '@once'\nos.environ['AIRFLOW_ENV'] = 'PROD'\nassert sched('foo') == 'foo'\n\nassert dest_dict('p.d.t', prefix_dataset='') == {'projectId': 'p', 'datasetId': 'd', 'tableId': 't'}\n\nos.environ.pop('AIRFLOW_ENV', None)\nassert dest('p.d.t') == 'p.tmp.d_t'\nassert dest('p.d.t', return_dataset_only=True) == 'tmp'\nassert dest('p.d.t', return_table_only=True) == 'd_t'",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbac3f3574e8f713690d79cb3be772b6781aad01
| 719,907 |
ipynb
|
Jupyter Notebook
|
projects/customer_segments/customer_segments.ipynb
|
anandsaha/ml-nanodegree
|
b16c98eb7f8580f64ead501de5eb5d57e07b0275
|
[
"MIT"
] | null | null | null |
projects/customer_segments/customer_segments.ipynb
|
anandsaha/ml-nanodegree
|
b16c98eb7f8580f64ead501de5eb5d57e07b0275
|
[
"MIT"
] | null | null | null |
projects/customer_segments/customer_segments.ipynb
|
anandsaha/ml-nanodegree
|
b16c98eb7f8580f64ead501de5eb5d57e07b0275
|
[
"MIT"
] | null | null | null | 290.168077 | 265,194 | 0.897699 |
[
[
[
"# Machine Learning Engineer Nanodegree\n## Unsupervised Learning\n## Project: Creating Customer Segments",
"_____no_output_____"
],
[
"Welcome to the third project of the Machine Learning Engineer Nanodegree! In this notebook, some template code has already been provided for you, and it will be your job to implement the additional functionality necessary to successfully complete this project. Sections that begin with **'Implementation'** in the header indicate that the following block of code will require additional functionality which you must provide. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `'TODO'` statement. Please be sure to read the instructions carefully!\n\nIn addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide. \n\n>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.",
"_____no_output_____"
],
[
"## Getting Started\n\nIn this project, you will analyze a dataset containing data on various customers' annual spending amounts (reported in *monetary units*) of diverse product categories for internal structure. One goal of this project is to best describe the variation in the different types of customers that a wholesale distributor interacts with. Doing so would equip the distributor with insight into how to best structure their delivery service to meet the needs of each customer.\n\nThe dataset for this project can be found on the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Wholesale+customers). For the purposes of this project, the features `'Channel'` and `'Region'` will be excluded in the analysis — with focus instead on the six product categories recorded for customers.\n\nRun the code block below to load the wholesale customers dataset, along with a few of the necessary Python libraries required for this project. You will know the dataset loaded successfully if the size of the dataset is reported.",
"_____no_output_____"
]
],
[
[
"# Import libraries necessary for this project\nimport numpy as np\nimport pandas as pd\nfrom IPython.display import display # Allows the use of display() for DataFrames\n\n# Import supplementary visualizations code visuals.py\nimport visuals as vs\n\n# Pretty display for notebooks\n%matplotlib inline\n\n# Load the wholesale customers dataset\ntry:\n data = pd.read_csv(\"customers.csv\")\n data.drop(['Region', 'Channel'], axis = 1, inplace = True)\n print \"Wholesale customers dataset has {} samples with {} features each.\".format(*data.shape)\nexcept:\n print \"Dataset could not be loaded. Is the dataset missing?\"",
"Wholesale customers dataset has 440 samples with 6 features each.\n"
]
],
[
[
"## Data Exploration\nIn this section, you will begin exploring the data through visualizations and code to understand how each feature is related to the others. You will observe a statistical description of the dataset, consider the relevance of each feature, and select a few sample data points from the dataset which you will track through the course of this project.\n\nRun the code block below to observe a statistical description of the dataset. Note that the dataset is composed of six important product categories: **'Fresh'**, **'Milk'**, **'Grocery'**, **'Frozen'**, **'Detergents_Paper'**, and **'Delicatessen'**. Consider what each category represents in terms of products you could purchase.",
"_____no_output_____"
]
],
[
[
"# Display a description of the dataset\ndisplay(data.describe())",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 440 entries, 0 to 439\nData columns (total 6 columns):\nFresh 440 non-null int64\nMilk 440 non-null int64\nGrocery 440 non-null int64\nFrozen 440 non-null int64\nDetergents_Paper 440 non-null int64\nDelicatessen 440 non-null int64\ndtypes: int64(6)\nmemory usage: 20.7 KB\n"
]
],
[
[
"### Implementation: Selecting Samples\nTo get a better understanding of the customers and how their data will transform through the analysis, it would be best to select a few sample data points and explore them in more detail. In the code block below, add **three** indices of your choice to the `indices` list which will represent the customers to track. It is suggested to try different sets of samples until you obtain customers that vary significantly from one another.",
"_____no_output_____"
]
],
[
[
"# TODO: Select three indices of your choice you wish to sample from the dataset\nindices = [181, 93, 85]\n\n# Create a DataFrame of the chosen samples\nsamples = pd.DataFrame(data.loc[indices], columns = data.keys()).reset_index(drop = True)\nprint \"Chosen samples of wholesale customers dataset:\"\ndisplay(samples)",
"Chosen samples of wholesale customers dataset:\n"
],
[
"data.rank(pct=True).iloc[indices]",
"_____no_output_____"
],
[
"import seaborn as sns\nimport matplotlib.pyplot as plt\nfig, ax = plt.subplots(figsize=(10,5)) \nsns.heatmap((100 * data.rank(pct=True)).iloc[indices], vmin=1, vmax = 100, annot=True, ax=ax)",
"_____no_output_____"
]
],
[
[
"### Question 1\nConsider the total purchase cost of each product category and the statistical description of the dataset above for your sample customers. \n*What kind of establishment (customer) could each of the three samples you've chosen represent?* \n**Hint:** Examples of establishments include places like markets, cafes, and retailers, among many others. Avoid using names for establishments, such as saying *\"McDonalds\"* when describing a sample customer as a restaurant.",
"_____no_output_____"
],
[
"**Answer:**\n\nFrom the heat map, it can be seen that:\n\n* Customer #1 has maximum spendings on Fresh in it's category. But can also be seen that this customer has high spending across all the categories (80% +). This is most probably a super market.\n\n* Customer #2 has high spendings on Frozen which is 11 times the mean of Frozen category and maximum in its category. Seems to be primarily an ice cream parlor. (Has moderate spendings on Deli, seems keeps perishable items)\n\n* Customer #3 has highest spendings on Grocery (11 times the mean), Detergents_Paper (14 times the mean). Seems to be primarily a grocery store, also stocking misc items like Milk, detergents and paper.",
"_____no_output_____"
],
[
"### Implementation: Feature Relevance\nOne interesting thought to consider is if one (or more) of the six product categories is actually relevant for understanding customer purchasing. That is to say, is it possible to determine whether customers purchasing some amount of one category of products will necessarily purchase some proportional amount of another category of products? We can make this determination quite easily by training a supervised regression learner on a subset of the data with one feature removed, and then score how well that model can predict the removed feature.\n\nIn the code block below, you will need to implement the following:\n - Assign `new_data` a copy of the data by removing a feature of your choice using the `DataFrame.drop` function.\n - Use `sklearn.cross_validation.train_test_split` to split the dataset into training and testing sets.\n - Use the removed feature as your target label. Set a `test_size` of `0.25` and set a `random_state`.\n - Import a decision tree regressor, set a `random_state`, and fit the learner to the training data.\n - Report the prediction score of the testing set using the regressor's `score` function.",
"_____no_output_____"
]
],
[
[
"# TODO: Make a copy of the DataFrame, using the 'drop' function to drop the given feature\n\nfrom sklearn.cross_validation import train_test_split\nfrom sklearn import tree\nimport operator\n\ndef dt_predict(spending_head):\n new_data = data.copy()\n target_label = new_data.ix[:, [spending_head]].copy() \n new_data.drop([spending_head], axis = 1, inplace = True)\n\n new_data = new_data.values\n target_label = target_label.values\n\n # TODO: Split the data into training and testing sets using the given feature as the target\n X_train, X_test, y_train, y_test = train_test_split(new_data, target_label, test_size=0.25, random_state=1)\n\n # TODO: Create a decision tree regressor and fit it to the training set\n regressor = tree.DecisionTreeRegressor()\n regressor.fit(X_train, y_train)\n\n # TODO: Report the score of the prediction using the testing set\n score = regressor.score(X_test, y_test)\n #print(\"Score for predicting '{0}' is {1}\".format(spending_head, score))\n return score\n\n\n\n \nprediction_ranking = {}\n\nprediction_ranking['Fresh'] = dt_predict('Fresh')\nprediction_ranking['Milk'] = dt_predict('Milk')\nprediction_ranking['Grocery'] = dt_predict('Grocery')\nprediction_ranking['Frozen'] = dt_predict('Frozen')\nprediction_ranking['Detergents_Paper'] = dt_predict('Detergents_Paper')\nprediction_ranking['Delicatessen'] = dt_predict('Delicatessen')\n\nprint(\"\\nRanking (asc)\")\nprint(\"===============================\")\nsorted_prediction_ranking = sorted(prediction_ranking.items(), key=operator.itemgetter(1))\nfor item, score in sorted_prediction_ranking:\n print(score,item)\n\n",
"\nRanking (asc)\n===============================\n(-10.712047529258234, 'Delicatessen')\n(-0.8763369473667586, 'Frozen')\n(-0.56890403760690522, 'Fresh')\n(0.32123561964289604, 'Milk')\n(0.82752742421106262, 'Detergents_Paper')\n(0.83712289844409138, 'Grocery')\n"
]
],
[
[
"### Question 2\n*Which feature did you attempt to predict? What was the reported prediction score? Is this feature necessary for identifying customers' spending habits?* \n**Hint:** The coefficient of determination, `R^2`, is scored between 0 and 1, with 1 being a perfect fit. A negative `R^2` implies the model fails to fit the data.",
"_____no_output_____"
],
[
"**Answer:**\n\n*Which feature did you attempt to predict? What was the reported prediction score?*\n\nI tried all. The 'Grocery' feature could be predicted with 84% accuracy.\n\n*Is this feature necessary for identifying customers' spending habits?*\n\nI would argue that __yes it is necessary__. Though 83% is the highest score amongst all, it is not high enough to ignore this feature. The Grocery spending is not fully explained by the other features. \n\nAlso it is not deterministic. \n\na. I experimented with the 'random_state' parameter. When set to 0, I got accuracy of 73%. When set to 42, gives accuracy of 68%. And when set to 1, both Grocery and Detergents_Paper gets an accuracy of 82%! So, the accuracy depends on the train/test split.\n\nb. Everytime the cell is refreshed, I get different values of the scores. 'Detergents_Paper' comes quite close to Grocery.\n\nWe don't have enough data to conclude that Grocery is predictable.\n\n__Score is not consistent and is highly sensitive to the train/test split.__",
"_____no_output_____"
],
[
"### Visualize Feature Distributions\nTo get a better understanding of the dataset, we can construct a scatter matrix of each of the six product features present in the data. If you found that the feature you attempted to predict above is relevant for identifying a specific customer, then the scatter matrix below may not show any correlation between that feature and the others. Conversely, if you believe that feature is not relevant for identifying a specific customer, the scatter matrix might show a correlation between that feature and another feature in the data. Run the code block below to produce a scatter matrix.",
"_____no_output_____"
]
],
[
[
"# Produce a scatter matrix for each pair of features in the data\npd.scatter_matrix(data, alpha = 0.3, figsize = (14,8), diagonal = 'kde');",
"_____no_output_____"
]
],
[
[
"### Question 3\n*Are there any pairs of features which exhibit some degree of correlation? Does this confirm or deny your suspicions about the relevance of the feature you attempted to predict? How is the data for those features distributed?* \n**Hint:** Is the data normally distributed? Where do most of the data points lie? ",
"_____no_output_____"
],
[
"**Answer:**\n\n*Are there any pairs of features which exhibit some degree of correlation? *\n\nIndeed, Grocery, Milk and Detergents_Paper show high degree of correlation between them. As can be seen from the plots between Grocery and Detergents_Paper, the plot is quite cohesive along the diagonal, which expnains their high score. Milk comes distant 3rd, with the dots scatterd more.\n\n*Does this confirm or deny your suspicions about the relevance of the feature you attempted to predict?*\n\nYes. These 3 has been the top 3 in the ranking of scores in the previous exercise. But ofcourse, Grocery and Detergent_Paper are close together, with Milk a distant 3rd. \n\n*How is the data for those features distributed?*\n\nThe distribution is not normal. The distributions suggest that there are many customers spending less on these items, and lesser customers spending more. Among all these items, 'Fresh' has the highest spread (variance).\n",
"_____no_output_____"
],
[
"## Data Preprocessing\nIn this section, you will preprocess the data to create a better representation of customers by performing a scaling on the data and detecting (and optionally removing) outliers. Preprocessing data is often times a critical step in assuring that results you obtain from your analysis are significant and meaningful.",
"_____no_output_____"
],
[
"### Implementation: Feature Scaling\nIf data is not normally distributed, especially if the mean and median vary significantly (indicating a large skew), it is most [often appropriate](http://econbrowser.com/archives/2014/02/use-of-logarithms-in-economics) to apply a non-linear scaling — particularly for financial data. One way to achieve this scaling is by using a [Box-Cox test](http://scipy.github.io/devdocs/generated/scipy.stats.boxcox.html), which calculates the best power transformation of the data that reduces skewness. A simpler approach which can work in most cases would be applying the natural logarithm.\n\nIn the code block below, you will need to implement the following:\n - Assign a copy of the data to `log_data` after applying logarithmic scaling. Use the `np.log` function for this.\n - Assign a copy of the sample data to `log_samples` after applying logarithmic scaling. Again, use `np.log`.",
"_____no_output_____"
]
],
[
[
"# TODO: Scale the data using the natural logarithm\nlog_data = np.log(data)\n\n# TODO: Scale the sample data using the natural logarithm\nlog_samples = pd.DataFrame(log_data.loc[indices], columns = log_data.keys()).reset_index(drop = True)\n\n# Produce a scatter matrix for each pair of newly-transformed features\npd.scatter_matrix(log_data, alpha = 0.3, figsize = (14,8), diagonal = 'kde');",
"_____no_output_____"
],
[
"# Exploration\nX = data.ix[:, ['Detergents_Paper']].values\nimport matplotlib.pyplot as plt \n\nX_log = np.log(X)\nplt.hist(X_log, bins=20)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Observation\nAfter applying a natural logarithm scaling to the data, the distribution of each feature should appear much more normal. For any pairs of features you may have identified earlier as being correlated, observe here whether that correlation is still present (and whether it is now stronger or weaker than before).\n\nRun the code below to see how the sample data has changed after having the natural logarithm applied to it.",
"_____no_output_____"
]
],
[
[
"# Display the log-transformed sample data\ndisplay(log_samples)",
"_____no_output_____"
]
],
[
[
"### Implementation: Outlier Detection\nDetecting outliers in the data is extremely important in the data preprocessing step of any analysis. The presence of outliers can often skew results which take into consideration these data points. There are many \"rules of thumb\" for what constitutes an outlier in a dataset. Here, we will use [Tukey's Method for identfying outliers](http://datapigtechnologies.com/blog/index.php/highlighting-outliers-in-your-data-with-the-tukey-method/): An *outlier step* is calculated as 1.5 times the interquartile range (IQR). A data point with a feature that is beyond an outlier step outside of the IQR for that feature is considered abnormal.\n\nIn the code block below, you will need to implement the following:\n - Assign the value of the 25th percentile for the given feature to `Q1`. Use `np.percentile` for this.\n - Assign the value of the 75th percentile for the given feature to `Q3`. Again, use `np.percentile`.\n - Assign the calculation of an outlier step for the given feature to `step`.\n - Optionally remove data points from the dataset by adding indices to the `outliers` list.\n\n**NOTE:** If you choose to remove any outliers, ensure that the sample data does not contain any of these points! \nOnce you have performed this implementation, the dataset will be stored in the variable `good_data`.",
"_____no_output_____"
]
],
[
[
"index_counts = {}\n\n# For each feature find the data points with extreme high or low values\nfor feature in log_data.keys():\n \n # TODO: Calculate Q1 (25th percentile of the data) for the given feature\n Q1 = np.percentile(log_data[feature], 25)\n \n # TODO: Calculate Q3 (75th percentile of the data) for the given feature\n Q3 = np.percentile(log_data[feature], 75)\n \n # TODO: Use the interquartile range to calculate an outlier step (1.5 times the interquartile range)\n step = 1.5 * (Q3 - Q1)\n \n # Display the outliers\n print \"Data points considered outliers for the feature '{}':\".format(feature)\n outliers = log_data[~((log_data[feature] >= Q1 - step) & (log_data[feature] <= Q3 + step))]\n \n for item in outliers.index.values:\n if item in index_counts.keys():\n index_counts[item] += 1\n else:\n index_counts[item] = 1\n \n display(outliers)\n \nthe_outliers = []\nprint('Data points considered outliers for more than one feature:')\nfor item in index_counts.keys():\n if index_counts[item] > 1:\n print(item)\n the_outliers.append(item)\n\n# OPTIONAL: Select the indices for data points you wish to remove\noutliers = the_outliers\n\n# Remove the outliers, if any were specified\ngood_data = log_data.drop(log_data.index[outliers]).reset_index(drop = True)\n",
"Data points considered outliers for the feature 'Fresh':\n"
]
],
[
[
"### Question 4\n*Are there any data points considered outliers for more than one feature based on the definition above? Should these data points be removed from the dataset? If any data points were added to the `outliers` list to be removed, explain why.* ",
"_____no_output_____"
],
[
"**Answer:**\n\n*Are there any data points considered outliers for more than one feature based on the definition above? *\n\nYes, there were 5 such datapoints: 128, 154, 65, 66, 75\n\n*Should these data points be removed from the dataset? *\n\nYes, they should be removed, else they will misguide the clustering model. For e.g., K-means algorithm heavily depends on the distance from datapoints to the centroid. Any outliers will shift the center of gravity towards itself. This will lead to misrepresentation of the cluster.\n\n*If any data points were added to the outliers list to be removed, explain why.*\n\nI added the outliers identified above to the outlier's list. This is because down below I am using K Means clustering and don't want the outliers to affect the clustering.\n",
"_____no_output_____"
],
[
"## Feature Transformation\nIn this section you will use principal component analysis (PCA) to draw conclusions about the underlying structure of the wholesale customer data. Since using PCA on a dataset calculates the dimensions which best maximize variance, we will find which compound combinations of features best describe customers.",
"_____no_output_____"
],
[
"### Implementation: PCA\n\nNow that the data has been scaled to a more normal distribution and has had any necessary outliers removed, we can now apply PCA to the `good_data` to discover which dimensions about the data best maximize the variance of features involved. In addition to finding these dimensions, PCA will also report the *explained variance ratio* of each dimension — how much variance within the data is explained by that dimension alone. Note that a component (dimension) from PCA can be considered a new \"feature\" of the space, however it is a composition of the original features present in the data.\n\nIn the code block below, you will need to implement the following:\n - Import `sklearn.decomposition.PCA` and assign the results of fitting PCA in six dimensions with `good_data` to `pca`.\n - Apply a PCA transformation of `log_samples` using `pca.transform`, and assign the results to `pca_samples`.",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition.pca import PCA\n\n\n\n# TODO: Apply PCA by fitting the good data with the same number of dimensions as features\npca = PCA(n_components=6)\n\npca.fit(good_data)\n\n# TODO: Transform log_samples using the PCA fit above\npca_samples = pca.transform(log_samples)\n\n# Generate PCA results plot\npca_results = vs.pca_results(good_data, pca)",
"_____no_output_____"
]
],
[
[
"### Question 5\n*How much variance in the data is explained* ***in total*** *by the first and second principal component? What about the first four principal components? Using the visualization provided above, discuss what the first four dimensions best represent in terms of customer spending.* \n**Hint:** A positive increase in a specific dimension corresponds with an *increase* of the *positive-weighted* features and a *decrease* of the *negative-weighted* features. The rate of increase or decrease is based on the individual feature weights.",
"_____no_output_____"
],
[
"**Answer:**\n\nThe total variation explained by first and second component is: 0.4424 + 0.2766 = 0.719\n\nThe total variation explained by first four components is: 0.4424 + 0.2766 + 0.1162 + 0.0962 = 0.9314\n\nThus, the first four components could explain more than 90% of variation.\n\nBreaking down individually,\n\n* Component 1: The dominant feature explained by this component is Detergents_Paper (about 0.75 weightage). Milk and Grocery come close second (around 0.45). This agrees with our earlier observation that there is a coorelation between DetergentPaper, Milk and Grocery.\n* Component 2: The dominant feature explained by this component is Fresh (0.7), followed by Deli and Frozen (0.5). This component suggests that these group of items are often sold together.\n* Component 3: The dominant feature explained by this component is Fresh (0.7) and Deli (-0.7), and they are inversely weighted. This component differentiates between exclusive fresh outlets and exclusive deli outlets. \n* Component 4: The dominant feature explained by this component is Frozen (0.8), with Deli (-0.5) coming second. They are inversely weighted. This component differentiates between exclusive Ice cream parlours/Frozen food outlets and exclusive Delis.",
"_____no_output_____"
],
[
"### Observation\nRun the code below to see how the log-transformed sample data has changed after having a PCA transformation applied to it in six dimensions. Observe the numerical value for the first four dimensions of the sample points. Consider if this is consistent with your initial interpretation of the sample points.",
"_____no_output_____"
]
],
[
[
"# Display sample log-data after having a PCA transformation applied\ndisplay(pd.DataFrame(np.round(pca_samples, 4), columns = pca_results.index.values))",
"_____no_output_____"
]
],
[
[
"### Implementation: Dimensionality Reduction\nWhen using principal component analysis, one of the main goals is to reduce the dimensionality of the data — in effect, reducing the complexity of the problem. Dimensionality reduction comes at a cost: Fewer dimensions used implies less of the total variance in the data is being explained. Because of this, the *cumulative explained variance ratio* is extremely important for knowing how many dimensions are necessary for the problem. Additionally, if a signifiant amount of variance is explained by only two or three dimensions, the reduced data can be visualized afterwards.\n\nIn the code block below, you will need to implement the following:\n - Assign the results of fitting PCA in two dimensions with `good_data` to `pca`.\n - Apply a PCA transformation of `good_data` using `pca.transform`, and assign the results to `reduced_data`.\n - Apply a PCA transformation of `log_samples` using `pca.transform`, and assign the results to `pca_samples`.",
"_____no_output_____"
]
],
[
[
"# TODO: Apply PCA by fitting the good data with only two dimensions\npca = PCA(n_components=2)\npca.fit(good_data)\n\n# TODO: Transform the good data using the PCA fit above\nreduced_data = pca.transform(good_data)\n\n# TODO: Transform log_samples using the PCA fit above\npca_samples = pca.transform(log_samples)\n\n# Create a DataFrame for the reduced data\nreduced_data = pd.DataFrame(reduced_data, columns = ['Dimension 1', 'Dimension 2'])",
"_____no_output_____"
]
],
[
[
"### Observation\nRun the code below to see how the log-transformed sample data has changed after having a PCA transformation applied to it using only two dimensions. Observe how the values for the first two dimensions remains unchanged when compared to a PCA transformation in six dimensions.",
"_____no_output_____"
]
],
[
[
"# Display sample log-data after applying PCA transformation in two dimensions\ndisplay(pd.DataFrame(np.round(pca_samples, 4), columns = ['Dimension 1', 'Dimension 2']))",
"_____no_output_____"
]
],
[
[
"## Visualizing a Biplot\nA biplot is a scatterplot where each data point is represented by its scores along the principal components. The axes are the principal components (in this case `Dimension 1` and `Dimension 2`). In addition, the biplot shows the projection of the original features along the components. A biplot can help us interpret the reduced dimensions of the data, and discover relationships between the principal components and original features.\n\nRun the code cell below to produce a biplot of the reduced-dimension data.",
"_____no_output_____"
]
],
[
[
"# Create a biplot\nvs.biplot(good_data, reduced_data, pca)",
"_____no_output_____"
]
],
[
[
"### Observation\n\nOnce we have the original feature projections (in red), it is easier to interpret the relative position of each data point in the scatterplot. For instance, a point the lower right corner of the figure will likely correspond to a customer that spends a lot on `'Milk'`, `'Grocery'` and `'Detergents_Paper'`, but not so much on the other product categories. \n\nFrom the biplot, which of the original features are most strongly correlated with the first component? What about those that are associated with the second component? Do these observations agree with the pca_results plot you obtained earlier?",
"_____no_output_____"
],
[
"## Clustering\n\nIn this section, you will choose to use either a K-Means clustering algorithm or a Gaussian Mixture Model clustering algorithm to identify the various customer segments hidden in the data. You will then recover specific data points from the clusters to understand their significance by transforming them back into their original dimension and scale. ",
"_____no_output_____"
],
[
"### Question 6\n*What are the advantages to using a K-Means clustering algorithm? What are the advantages to using a Gaussian Mixture Model clustering algorithm? Given your observations about the wholesale customer data so far, which of the two algorithms will you use and why?*",
"_____no_output_____"
],
[
"**Answer:**\n\n* K Means uses hard assignment to assign a data point to a cluster. It starts with the random cluster centers (the number of clusters is determined by the user). For each iteration, we do two things: a) The datapoints in the set are assigned to the nearest center of cluster. b) The center of clusters are recalculated based on center of mass of the datapoints assigned to them. KMeans is faster to train than GMM, better for higher dimentional data and easy to interpret. However, it's approach of hard assignment may lead to wrong groupings and may not work well with certain shapes of clusters.\n\n* GMM works with probability. The datapoints are not assigned directly. Rather the probability of each datapoint to be part of a cluster is calculated. This is a soft assignment. It works well with non linear geometric distribution. However it is difficult to initialize for high dimentional data, is slower (many parameters to be fitted to the data) and is difficult to interpret.\n\n\nFor our dataset, I will go with K Means, since there seems to be a pattern of spending involved. Hence there can be a clear demarcations between clusters.\n\n\nReference:\n[1] https://www.quora.com/What-is-the-difference-between-K-means-and-the-mixture-model-of-Gaussian",
"_____no_output_____"
],
[
"### Implementation: Creating Clusters\nDepending on the problem, the number of clusters that you expect to be in the data may already be known. When the number of clusters is not known *a priori*, there is no guarantee that a given number of clusters best segments the data, since it is unclear what structure exists in the data — if any. However, we can quantify the \"goodness\" of a clustering by calculating each data point's *silhouette coefficient*. The [silhouette coefficient](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.silhouette_score.html) for a data point measures how similar it is to its assigned cluster from -1 (dissimilar) to 1 (similar). Calculating the *mean* silhouette coefficient provides for a simple scoring method of a given clustering.\n\nIn the code block below, you will need to implement the following:\n - Fit a clustering algorithm to the `reduced_data` and assign it to `clusterer`.\n - Predict the cluster for each data point in `reduced_data` using `clusterer.predict` and assign them to `preds`.\n - Find the cluster centers using the algorithm's respective attribute and assign them to `centers`.\n - Predict the cluster for each sample data point in `pca_samples` and assign them `sample_preds`.\n - Import `sklearn.metrics.silhouette_score` and calculate the silhouette score of `reduced_data` against `preds`.\n - Assign the silhouette score to `score` and print the result.",
"_____no_output_____"
]
],
[
[
"# TODO: Apply your clustering algorithm of choice to the reduced data \nfrom sklearn.metrics import silhouette_score\nfrom sklearn.cluster import KMeans\n\ndef calculate_clusters(num_clusters):\n clusterer = KMeans(n_clusters=num_clusters, random_state=42)\n clusterer.fit(reduced_data)\n\n # TODO: Predict the cluster for each data point\n preds = clusterer.predict(reduced_data)\n\n # TODO: Find the cluster centers\n centers = clusterer.cluster_centers_\n\n # TODO: Predict the cluster for each transformed sample data point\n sample_preds = clusterer.predict(pca_samples)\n\n # TODO: Calculate the mean silhouette coefficient for the number of clusters chosen\n score = silhouette_score(reduced_data, clusterer.labels_, metric='euclidean')\n\n print(num_clusters, score)\n \n return preds, centers, sample_preds\n \nfor i in range(9):\n calculate_clusters(i + 2)\n \n\n# Final cluster size of 2\npreds, centers, sample_preds = calculate_clusters(2)",
"(2, 0.4262810154691084)\n(3, 0.39689092644980506)\n(4, 0.33184127600936941)\n(5, 0.34999779752629762)\n(6, 0.36588522063299811)\n(7, 0.36480120797880022)\n(8, 0.36764075649149885)\n(9, 0.35987901460536154)\n(10, 0.36415528051550899)\n(2, 0.4262810154691084)\n"
]
],
[
[
"### Question 7\n*Report the silhouette score for several cluster numbers you tried. Of these, which number of clusters has the best silhouette score?* ",
"_____no_output_____"
],
[
"**Answer:**\n\nSilhouette scores for cluster size from 2 to 10 were tried, with the best score obtained for size of 2 (0.41).",
"_____no_output_____"
],
[
"### Cluster Visualization\nOnce you've chosen the optimal number of clusters for your clustering algorithm using the scoring metric above, you can now visualize the results by executing the code block below. Note that, for experimentation purposes, you are welcome to adjust the number of clusters for your clustering algorithm to see various visualizations. The final visualization provided should, however, correspond with the optimal number of clusters. ",
"_____no_output_____"
]
],
[
[
"# Display the results of the clustering from implementation\nvs.cluster_results(reduced_data, preds, centers, pca_samples)",
"_____no_output_____"
]
],
[
[
"### Implementation: Data Recovery\nEach cluster present in the visualization above has a central point. These centers (or means) are not specifically data points from the data, but rather the *averages* of all the data points predicted in the respective clusters. For the problem of creating customer segments, a cluster's center point corresponds to *the average customer of that segment*. Since the data is currently reduced in dimension and scaled by a logarithm, we can recover the representative customer spending from these data points by applying the inverse transformations.\n\nIn the code block below, you will need to implement the following:\n - Apply the inverse transform to `centers` using `pca.inverse_transform` and assign the new centers to `log_centers`.\n - Apply the inverse function of `np.log` to `log_centers` using `np.exp` and assign the true centers to `true_centers`.\n",
"_____no_output_____"
]
],
[
[
"# TODO: Inverse transform the centers\nlog_centers = pca.inverse_transform(centers)\n\n# TODO: Exponentiate the centers\ntrue_centers = np.exp(log_centers)\n\n# Display the true centers\nsegments = ['Segment {}'.format(i) for i in range(0,len(centers))]\ntrue_centers = pd.DataFrame(np.round(true_centers), columns = data.keys())\ntrue_centers.index = segments\ndisplay(true_centers)",
"_____no_output_____"
],
[
"import seaborn as sns\nsns.heatmap((true_centers-data.mean())/data.std(ddof=1), annot=True, cbar=False, square=True)\n",
"_____no_output_____"
]
],
[
[
"### Question 8\nConsider the total purchase cost of each product category for the representative data points above, and reference the statistical description of the dataset at the beginning of this project. *What set of establishments could each of the customer segments represent?* \n**Hint:** A customer who is assigned to `'Cluster X'` should best identify with the establishments represented by the feature set of `'Segment X'`.",
"_____no_output_____"
],
[
"**Answer:**\n\nThe normalized cluster expenditures is given by the heat map. Interpreting these numbers:\n\n* Segment 0: The spendings in this segment is dominated by Fresh & Frozen, followed by almost equal but less spending on Milk, Grocery and Detergent_Paper. This seems to be profile of an outlet selling perishable items. \n\n* Segment 1: The spendings in this segment is dominated by Grocery, Milk and Detergents_Paper. Definitely characterizes a grocer who sells retail stuff.\n\n",
"_____no_output_____"
],
[
"### Question 9\n*For each sample point, which customer segment from* ***Question 8*** *best represents it? Are the predictions for each sample point consistent with this?*\n\nRun the code block below to find which cluster each sample point is predicted to be.",
"_____no_output_____"
]
],
[
[
"# Display the predictions\nfor i, pred in enumerate(sample_preds):\n print \"Sample point\", i, \"predicted to be in Cluster\", pred",
"Sample point 0 predicted to be in Cluster 1\nSample point 1 predicted to be in Cluster 0\nSample point 2 predicted to be in Cluster 1\n"
]
],
[
[
"**Answer:**\n\nThis was my earlier prediction (copy pasted from above)\n\n* Customer #1 has maximum spendings on Fresh in it's category. But can also be seen that this customer has high spending across all the categories (80% +). This is most probably a super market.\n\n* Customer #2 has high spendings on Frozen which is 11 times the mean of Frozen category and maximum in its category. Seems to be primarily an ice cream parlor. (Has moderate spendings on Deli, seems keeps perishable items)\n\n* Customer #3 has highest spendings on Grocery (11 times the mean), Detergents_Paper (14 times the mean). Seems to be primarily a grocery store, also stocking misc items like Milk, detergents and paper\n\n\nPost clustering, this is what the prediction was:\n* Customer #1: Grocer.\n* Customer #2: Sells perishable items.\n* Customer #3: Grocer.\n\nCustomer #2 and #3 are consistent with what my earlier guess was. Customer #1 is off the chart.",
"_____no_output_____"
],
[
"## Conclusion",
"_____no_output_____"
],
[
"In this final section, you will investigate ways that you can make use of the clustered data. First, you will consider how the different groups of customers, the ***customer segments***, may be affected differently by a specific delivery scheme. Next, you will consider how giving a label to each customer (which *segment* that customer belongs to) can provide for additional features about the customer data. Finally, you will compare the ***customer segments*** to a hidden variable present in the data, to see whether the clustering identified certain relationships.",
"_____no_output_____"
],
[
"### Question 10\nCompanies will often run [A/B tests](https://en.wikipedia.org/wiki/A/B_testing) when making small changes to their products or services to determine whether making that change will affect its customers positively or negatively. The wholesale distributor is considering changing its delivery service from currently 5 days a week to 3 days a week. However, the distributor will only make this change in delivery service for customers that react positively. *How can the wholesale distributor use the customer segments to determine which customers, if any, would react positively to the change in delivery service?* \n**Hint:** Can we assume the change affects all customers equally? How can we determine which group of customers it affects the most?",
"_____no_output_____"
],
[
"**Answer:**\n\nFrom cluster analysis, we found that segment 1 deals with perishable items. Hence one can argue that changing the delivery service from 5 to 3 days will be difficult for this segment. These items cannot be stored for more than a day or two without degradation. \n\nIt might be doable without any decrease in customer satisfcation, for segment 0. This is where we have grocery and milk. Milk can be refrigerated for 2-3 days.\n\nHence, going by our analysis, our hunch is that segment 0 customers will react positively to the service, but not segment 1 customers.\n\nTo test out, we can do a couple of things:\n\n* We initially carry out a survey to gauge customer's willingness to the change\n* We carry out A/B testing with a smaller representative sample of customers ",
"_____no_output_____"
],
[
"### Question 11\nAdditional structure is derived from originally unlabeled data when using clustering techniques. Since each customer has a ***customer segment*** it best identifies with (depending on the clustering algorithm applied), we can consider *'customer segment'* as an **engineered feature** for the data. Assume the wholesale distributor recently acquired ten new customers and each provided estimates for anticipated annual spending of each product category. Knowing these estimates, the wholesale distributor wants to classify each new customer to a ***customer segment*** to determine the most appropriate delivery service. \n*How can the wholesale distributor label the new customers using only their estimated product spending and the* ***customer segment*** *data?* \n**Hint:** A supervised learner could be used to train on the original customers. What would be the target variable?",
"_____no_output_____"
],
[
"**Answer:**\n\nWe can treat this as a classification problem where we have to classify a new customer into segment 0 or 1.\n\nWe can use many of the classification algorithms like SVM, KNN or Decision Tree/Random forest.\n\nThe parameters are the estimates and the output is the segment. \n\nThe model can be trained on the existing customer data.",
"_____no_output_____"
],
[
"### Visualizing Underlying Distributions\n\nAt the beginning of this project, it was discussed that the `'Channel'` and `'Region'` features would be excluded from the dataset so that the customer product categories were emphasized in the analysis. By reintroducing the `'Channel'` feature to the dataset, an interesting structure emerges when considering the same PCA dimensionality reduction applied earlier to the original dataset.\n\nRun the code block below to see how each data point is labeled either `'HoReCa'` (Hotel/Restaurant/Cafe) or `'Retail'` the reduced space. In addition, you will find the sample points are circled in the plot, which will identify their labeling.",
"_____no_output_____"
]
],
[
[
"# Display the clustering results based on 'Channel' data\nvs.channel_results(reduced_data, outliers, pca_samples)",
"_____no_output_____"
]
],
[
[
"### Question 12\n*How well does the clustering algorithm and number of clusters you've chosen compare to this underlying distribution of Hotel/Restaurant/Cafe customers to Retailer customers? Are there customer segments that would be classified as purely 'Retailers' or 'Hotels/Restaurants/Cafes' by this distribution? Would you consider these classifications as consistent with your previous definition of the customer segments?*",
"_____no_output_____"
],
[
"**Answer:**\n\nData points 1 and 2 are consistant with this new classification between HoReCa and Retailer. Datapoint 1 indeed represent segment dealing with perishable items, which is the HoReCa case. And Datapoint 2 was our grocer, which here is the retailer.\n\nDatapoint 0 has been misclassified here as HoReCa, though earlier we had classified it as a Grocer.",
"_____no_output_____"
],
[
"> **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to \n**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbac588d569b2167e2a429eb7e17a1292894a427
| 744,926 |
ipynb
|
Jupyter Notebook
|
SumanthLazarus_MovieReviewSentimentAnalysis-TheLionKing(2019)_EDA_Visualization.ipynb
|
SumanthLazarus/Movie-Review-Sentiment-Analysis-NLP
|
511946309f22b3b247abdd7cc2b028f1af4bab14
|
[
"MIT"
] | null | null | null |
SumanthLazarus_MovieReviewSentimentAnalysis-TheLionKing(2019)_EDA_Visualization.ipynb
|
SumanthLazarus/Movie-Review-Sentiment-Analysis-NLP
|
511946309f22b3b247abdd7cc2b028f1af4bab14
|
[
"MIT"
] | null | null | null |
SumanthLazarus_MovieReviewSentimentAnalysis-TheLionKing(2019)_EDA_Visualization.ipynb
|
SumanthLazarus/Movie-Review-Sentiment-Analysis-NLP
|
511946309f22b3b247abdd7cc2b028f1af4bab14
|
[
"MIT"
] | null | null | null | 152.181001 | 183,860 | 0.840446 |
[
[
[
"## Movie Review Sentiment Analysis - The Lion King (2019) EDA and Visualization\n\n## Agenda\n - Data Extraction (Web-scrapping)\n - Visualization\n - Regular Expression for special character removal\n - Removal of accented characters and expanding contractions\n - Tokenisation\n - Stop Word Removal\n - Stemming and Lemmatization\n - TF-IDF Matrix\n - Clustering\n - SVD using scikitlearn",
"_____no_output_____"
],
[
"#### Import Libraries",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nimport requests\nimport time\n\nimport re\n\nimport random\nrandom.seed(123)\n\n#ignore warnings\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"Data Extraction/ Web scrapping\n------\n#### NOTE: Time to execute below chunk = 25 min\n#### Load from local file 'data_0.csv'",
"_____no_output_____"
]
],
[
[
"data=pd.read_csv('data_0.csv')\ndata.head()",
"_____no_output_____"
],
[
"# headers = {\n# 'Referer': 'https://www.rottentomatoes.com/m/the_lion_king_2019/reviews?type=user',\n# 'User-Agent': 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/74.0.3729.108 Safari/537.36',\n# 'X-Requested-With': 'XMLHttpRequest',\n# }\n\n# url = 'https://www.rottentomatoes.com/napi/movie/9057c2cf-7cab-317f-876f-e50b245ca76e/reviews/user'\n# s = requests.Session() # A Requests session. Provides cookie persistence, connection-pooling, and configuration.",
"_____no_output_____"
],
[
"# start_time = time.time()\n# data = pd.DataFrame() #initializing empty dataframe, payload parameter values\n# end = ''\n# start = ''\n\n# for i in list(range(1,301)):\n# payload = {\n# 'direction': 'next',\n# 'endCursor': end,\n# 'startCursor': start,\n# }\n# r = s.get(url, headers=headers, params=payload) # GET Call. Sends a GET request. Returns :class:`Response` object.\n# data= data.append(r.json()['reviews'], ignore_index=True) #append Review info to dataframe\n# end=r.json()['pageInfo']['endCursor'] # update 'start' key for new page\n# time.sleep(5)\n# print('Web scrap completed in %s s' % (str(time.time()-start_time))) \n\n# Store to Data local to local file\n# data.to_csv('data.csv', index=False)",
"_____no_output_____"
]
],
[
[
"Data Pre-processing and EDA\n---------",
"_____no_output_____"
]
],
[
[
"data.shape",
"_____no_output_____"
],
[
"data.dtypes",
"_____no_output_____"
],
[
"data.describe()",
"_____no_output_____"
],
[
"data.describe(exclude='float64')",
"_____no_output_____"
],
[
"import ast\ndata['userId'] = data['user'].apply(lambda x: ast.literal_eval(x)['userId']) # string literal evaluation --> Data structure (dictionary)",
"_____no_output_____"
],
[
"data.describe(exclude='float64')",
"_____no_output_____"
],
[
"# Setting 'userId' as Index\ndata.set_index('userId', inplace=True)\ndata.head()",
"_____no_output_____"
],
[
"data.drop(['displayName','displayImageUrl','hasProfanity','hasSpoilers','rating','isSuperReviewer',\n 'isVerified','timeFromCreation','updateDate','user',],\n axis=1,inplace=True)\n\ndata.columns",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
]
],
[
[
"### Feature Engineering\n\n1. Convert *score* --> **sentiment**:\\\n i. if score > 3, **sentiment** = 0 [POSITIVE Review]\\\n ii. if score =< 3, **sentiment** = 1 [NEGATIVE Review]\\\n\n2. Organize *createDate* --> **date**, **time**\n\n**NOTE**: our Target Level for prediction is Negative Review.",
"_____no_output_____"
]
],
[
[
"# Predic\ndata['sentiment'] = data['score'].apply(lambda x: 0 if x>3 else 1)\ndata.describe()",
"_____no_output_____"
],
[
"print('Count of Review Sentiments:\\n')\nprint(data['sentiment'].value_counts(),'\\n')\n\nprint('Frequency % of Review Sentiments:\\n')\nprint(data['sentiment'].value_counts(normalize=True)*100) # <30% of All reviews are Negative",
"Count of Review Sentiments:\n\n0 2170\n1 830\nName: sentiment, dtype: int64 \n\nFrequency % of Review Sentiments:\n\n0 72.333333\n1 27.666667\nName: sentiment, dtype: float64\n"
],
[
"data.drop('score', axis=1,inplace=True)\ndata.head()",
"_____no_output_____"
],
[
"from datetime import datetime as dt\n\ndata['createDate'] = pd.to_datetime(data['createDate'], infer_datetime_format=True)\ndata['date'] = data['createDate'].dt.date # Date of review\ndata['time'] = data['createDate'].dt.time # Time of review\ndata['weekday'] = data['createDate'].dt.weekday #weekday number : Monday=0, Sunday=6",
"_____no_output_____"
],
[
"data['weekday'].value_counts()",
"_____no_output_____"
],
[
"data.dtypes",
"_____no_output_____"
],
[
"data.describe(exclude='int64')",
"_____no_output_____"
],
[
"data.drop('createDate', axis=1,inplace=True) #Drop original Datetime variable\ndata.dtypes",
"_____no_output_____"
],
[
"# Reviews begin on 1st August 2018, collected till 18th August 2018\nx = data['date'].value_counts().sort_index().index.values\ny = data['date'].value_counts().sort_index().values\nprint(pd.DataFrame({'Date':x,'Review Count': y}).head(),'\\n')\nprint(pd.DataFrame({'Date':x,'Review Count': y}).tail(),)",
" Date Review Count\n0 2019-08-01 152\n1 2019-08-02 252\n2 2019-08-03 258\n3 2019-08-04 294\n4 2019-08-05 318 \n\n Date Review Count\n13 2019-08-14 107\n14 2019-08-15 93\n15 2019-08-16 67\n16 2019-08-17 88\n17 2019-08-18 46\n"
],
[
"with sns.axes_style('white'):\n sns.set(rc={'figure.figsize':(10,5)})\n plt.plot(list(range(1,19)),y, color='blue') \n plt.xlabel('Day since Release', fontsize=15)\n plt.ylabel('Review Count', fontsize=15)\n xtick_location = list(range(1,19))\n xtick_labels = list(range(1,19))\n plt.xticks(ticks=xtick_location, labels=xtick_labels, rotation=0, fontsize=12, horizontalalignment='center', alpha=.9)\n plt.yticks(fontsize=12,)\n plt.title(\"Reviews Added Daily\", fontsize=18)\n plt.grid(axis='both', alpha=.3)\n plt.show()\n \n# Plot shows a Rise in Reviews during initial weeks, followed by a decline post Day 7",
"_____no_output_____"
],
[
"with sns.axes_style('white'):\n sns.set(rc={'figure.figsize':(10,5)})\n\n g = sns.countplot(x='weekday', data=data, palette='Set1', saturation=0.7)\n v_list = [str(round(i,1))+' %' for i in (data['weekday'].value_counts(normalize=True)*100).sort_index()]\n\n for v, p in zip(v_list, g.patches): # Annotate the point 'xy' with Frequency%\n g.annotate(v, (p.get_x() + p.get_width() / 2., p.get_height()), ha='center', va='center',\\\n xytext = (0, 10), textcoords = 'offset points')\n xtick_location = list(range(0,7))\n xtick_labels = ['Mon','Tue','Wed','Thu','Fri','Sat','Sun'] \n plt.xticks(ticks=xtick_location, labels=xtick_labels, rotation=0, fontsize=12, horizontalalignment='center', alpha=.9)\n plt.xlabel('Weekday(s)',fontsize=15)\n plt.ylabel('Movie Review(s)',fontsize=15)\n plt.title('Frequency % of Reviews by Weekday', fontsize=18)\n plt.show()\n\n# Shows a dip in reviews(viewership) on Tuesday and Wednesdays",
"_____no_output_____"
],
[
"# Trend of Positive and Negative Reviews grouped by Weekday(s)",
"_____no_output_____"
],
[
"with sns.axes_style('white'):\n sns.set(rc={'figure.figsize':(10,5)})\n g = sns.countplot(\"weekday\", data=data, hue='sentiment',hue_order=[1,0], palette='Set1') \n v_list = [str(int(i))+' %' for i in data.groupby('weekday')['sentiment'].value_counts(normalize=True)*100]\n v_1 = [y for x,y in enumerate(v_list) if x%2==0] # Frequency% of Positive Review(s)\n v_2 = [y for x,y in enumerate(v_list) if x%2!=0] # Frequency% of Negative Review(s)\n v_list = v_2+v_1\n for v, p in zip(v_list, g.patches): # Annotate the point 'xy' with Frequency%\n g.annotate(v, (p.get_x() + p.get_width() / 2., \n p.get_height()), ha='center', va='center', xytext = (0, 10), textcoords = 'offset points')\n xtick_location = list(range(0,7))\n xtick_labels = ['Mon','Tue','Wed','Thu','Fri','Sat','Sun']\n plt.xticks(ticks=xtick_location, labels=xtick_labels, rotation=0, fontsize=12, horizontalalignment='center', alpha=.9)\n plt.xlabel('Weekday(s)',fontsize=15)\n plt.ylabel('Movie Review(s)',fontsize=15)\n plt.title('Frequency % of Reviews by Weekday, Sentiment', fontsize=18)\n g.legend(['1-Negative', '0-Positive'], loc=0)",
"_____no_output_____"
]
],
[
[
"#### Calendar Heat Map of Count of Negative Review(s) over 18 days of Available data",
"_____no_output_____"
]
],
[
[
"# Preparing Dateframe for Calendar Heat Map\nneg_review=data[data['sentiment']>0].groupby('date')['sentiment'].value_counts() # Count of Negative Reviews\ndata_cmap = pd.DataFrame(data={'neg_review':neg_review})\ndata_cmap.index = data_cmap.index.droplevel(1) # Drop Second Index level\nindex=list(data_cmap.index.values)\nindex = [np.datetime64(i) for i in index] # converting timestamp to np.datetime64()\ndata_cmap.index = index\ndata_cmap.head() # Date indexed Dataframe",
"_____no_output_____"
],
[
"# !pip install calmap\nimport calmap\n\n# Plot\nplt.figure(figsize=(16,10), dpi= 80)\ncalmap.calendarplot(data=data_cmap['2019']['neg_review'] ,fig_kws={'figsize': (16,10)},\\\n yearlabel_kws={'color':'black', 'fontsize':18})\nplt.title('Calendar Heat Map of Negative Reviews (last 18 days)', fontsize=18)\nplt.xlabel('Month', fontsize=15)\nplt.show()",
"_____no_output_____"
]
],
[
[
"**NOTE:** The First 7 days had the largest contribution of Negative Reviews ()",
"_____no_output_____"
]
],
[
[
"data.dtypes",
"_____no_output_____"
],
[
"# Dropping 'date', 'time' info\ndata.drop(['date','time','weekday'], axis=1, inplace=True)\ndata.head()",
"_____no_output_____"
]
],
[
[
"Text Preprocessing\n--------",
"_____no_output_____"
]
],
[
[
"data.dtypes",
"_____no_output_____"
],
[
"# !pip install -U spacy\n# !python -m spacy download en_core_web_sm\nimport spacy\nnlp = spacy.load(\"en_core_web_sm\")\n\nimport re\nimport random\nrandom.seed(123)",
"_____no_output_____"
],
[
"#Duplicating the original text extracted before proceeeding with preprocessing steps\n\noriginal_data = data.copy()\nprint(data.keys())\nprint(original_data.keys())",
"Index(['review', 'sentiment'], dtype='object')\nIndex(['review', 'sentiment'], dtype='object')\n"
]
],
[
[
"### LowerCase all text",
"_____no_output_____"
]
],
[
[
"data['review'] = [text.strip().lower() for text in data['review']] # remove Trailing/Leading whitespaces\ndata['review'][:10]",
"_____no_output_____"
],
[
"# eg of 'review' to preprocess\ndata['review'][100]",
"_____no_output_____"
]
],
[
[
"### Removal/Replacement of: Contractions, Accented Characters, Symbols/Markdown Characters\n**Contraction-Expansion Map:**",
"_____no_output_____"
]
],
[
[
"CONTRACTION_MAP = {\n\"ain't\": \"is not\", \"aren't\": \"are not\", \"can't\": \"cannot\", \"can't've\": \"cannot have\", \"'cause\": \"because\", \"could've\": \"could have\",\n\"couldn't\": \"could not\", \"couldn't've\": \"could not have\", \"didn't\": \"did not\", \"doesn't\": \"does not\", \"don't\": \"do not\", \"hadn't\": \"had not\",\n\"hadn't've\": \"had not have\", \"hasn't\": \"has not\", \"haven't\": \"have not\", \"he'd\": \"he would\", \"he'd've\": \"he would have\",\n\"he'll\": \"he will\", \"he'll've\": \"he will have\", \"he's\": \"he is\",\"how'd\": \"how did\",\"how'd'y\": \"how do you\",\"how'll\": \"how will\",\"how's\": \"how is\",\n\"I'd\": \"I would\",\"I'd've\": \"I would have\",\"I'll\": \"I will\",\"I'll've\": \"I will have\",\"I'm\": \"I am\",\"I've\": \"I have\",\"i'd\": \"i would\",\n\"i'd've\": \"i would have\",\"i'll\": \"i will\",\"i'll've\": \"i will have\",\"i'm\": \"i am\",\"i've\": \"i have\",\"isn't\": \"is not\",\"it'd\": \"it would\",\n\"it'd've\": \"it would have\",\"it'll\": \"it will\",\"it'll've\": \"it will have\",\"it's\": \"it is\",\"let's\": \"let us\",\"ma'am\": \"madam\",\"mayn't\": \"may not\",\n\"might've\": \"might have\",\"mightn't\": \"might not\",\"mightn't've\": \"might not have\",\"must've\": \"must have\",\"mustn't\": \"must not\",\n\"mustn't've\": \"must not have\",\"needn't\": \"need not\",\"needn't've\": \"need not have\",\"o'clock\": \"of the clock\",\"oughtn't\": \"ought not\",\n\"oughtn't've\": \"ought not have\",\"shan't\": \"shall not\",\"sha'n't\": \"shall not\",\"shan't've\": \"shall not have\",\"she'd\": \"she would\",\n\"she'd've\": \"she would have\",\"she'll\": \"she will\",\"she'll've\": \"she will have\",\"she's\": \"she is\",\"should've\": \"should have\",\"shouldn't\": \"should not\",\n\"shouldn't've\": \"should not have\",\"so've\": \"so have\",\"so's\": \"so as\",\"that'd\": \"that would\",\"that'd've\": \"that would have\",\"that's\": \"that is\",\n\"there'd\": \"there would\",\"there'd've\": \"there would have\",\"there's\": \"there is\",\"they'd\": \"they would\",\"they'd've\": \"they would have\",\n\"they'll\": \"they will\",\"they'll've\": \"they will have\",\"they're\": \"they are\",\"they've\": \"they have\",\"to've\": \"to have\",\"wasn't\": \"was not\",\n\"we'd\": \"we would\",\"we'd've\": \"we would have\",\"we'll\": \"we will\",\"we'll've\": \"we will have\",\"we're\": \"we are\",\"we've\": \"we have\",\"weren't\": \"were not\",\n\"what'll\": \"what will\",\"what'll've\": \"what will have\",\"what're\": \"what are\",\"what's\": \"what is\",\"what've\": \"what have\",\"when's\": \"when is\",\n\"when've\": \"when have\",\"where'd\": \"where did\",\"where's\": \"where is\",\"where've\": \"where have\",\"who'll\": \"who will\",\"who'll've\": \"who will have\",\n\"who's\": \"who is\",\"who've\": \"who have\",\"why's\": \"why is\",\"why've\": \"why have\",\"will've\": \"will have\",\"won't\": \"will not\",\"won't've\": \"will not have\",\n\"would've\": \"would have\",\"wouldn't\": \"would not\",\"wouldn't've\": \"would not have\",\"y'all\": \"you all\",\"y'all'd\": \"you all would\",\n\"y'all'd've\": \"you all would have\",\"y'all're\": \"you all are\",\"y'all've\": \"you all have\",\"you'd\": \"you would\",\"you'd've\": \"you would have\",\n\"you'll\": \"you will\",\"you'll've\": \"you will have\",\"you're\": \"you are\",\"you've\": \"you have\"\n}",
"_____no_output_____"
],
[
"# from contractions import CONTRACTION_MAP\nimport unicodedata\n\ndef expand_contractions(text, contraction_mapping=CONTRACTION_MAP):\n \n # Create 're' object by re.compile(pattern, repl, string) \n contractions_pattern = re.compile('({})'.format('|'.join(contraction_mapping.keys())), \n flags=re.IGNORECASE|re.DOTALL)\n # re.IGNORECASE : Make search case-insensiitive\n # re.DOTALL: Make the '.' special character match any character at all, including a newline\n \n # To Expand the Contracted Words\n def expand_match(contraction):\n match = contraction.group(0) \n expanded_contraction = contraction_mapping.get(match)\\\n if contraction_mapping.get(match)\\\n else contraction_mapping.get(match.lower()) \n return expanded_contraction # match, 'replaced by -->',expanded_contraction\n \n # string substitution: regex.sub(replacement, subject) \n expanded_text = contractions_pattern.sub(expand_match, text)\n expanded_text = re.sub(pattern=\"'\", repl=\"\", string=expanded_text) # Remove apostrophe\n return expanded_text # Returns expanded text",
"_____no_output_____"
],
[
"# Removes accented characters and emojis too\ndef remove_accented_chars(text):\n text = unicodedata.normalize('NFKD', text).encode('ascii', 'ignore').decode('utf-8', 'ignore')\n return text",
"_____no_output_____"
],
[
"def scrub_words(text):\n #Replace \\xao characters in text\n text = re.sub('\\xa0', ' ', text)\n \n #Replace non ascii, non-Words and Digits/Numerals\n text = re.sub(\"(\\\\W|\\\\d)\",' ',text) # \\W: non-alphanumeric character, \\d: decimal digit\n \n #Replace new line characters and following text untill space\n text = re.sub('\\n(\\w*?)[\\s]', '', text) # \\n: newline char, \\w: any alphanumeric character, \n # *: matches zero or more occurrences, ?: matches Zero or One occurrence of the pattern left to it.\n # (a|b|c)xz: group sub-patterns to match, [abc]: set of characters to match, \\s: whitespace \n # |: is used for alternation a|b\n \n #Remove html markup\n text = re.sub(\"<.*?>\", ' ', text)\n return text",
"_____no_output_____"
],
[
"# Test: expand_contractions()\ntxt = \"They aren't sick, you shouldn't worry!\"\nprint(expand_contractions(txt),'\\n')\n\n# Test: remove_accented_chars()\ntxt = 'Demain, dès l’aube, à l’heure où blanchit la campagne, Je partirai. J’irai par la forêt, j’irai par la montagne.'\nprint('Non-Accented Text:',remove_accented_chars(txt),'\\n')\n\n# Test: scrub_words()\ntxt = \"Love, Love, \\n\\n\\t, Love this movie!!😍😍😍❤️❤️❤️,&*(@)$&Lion King is the best#(@#$)\"\nprint('Scrubbed Text:',scrub_words(txt))",
"They are not sick, you should not worry! \n\nNon-Accented Text: Demain, des laube, a lheure ou blanchit la campagne, Je partirai. Jirai par la foret, jirai par la montagne. \n\nScrubbed Text: Love Love Love this movie Lion King is the best \n"
],
[
"print('Average Review length:', np.mean([len(i) for i in data['review']]))",
"Average Review length: 146.767\n"
]
],
[
[
"#### Invoking the above defined functions",
"_____no_output_____"
]
],
[
[
"# data['review']= [expand_contractions(re.sub('’', \"'\", text)) for text in data['review']]\ndata['review'] = data['review'].apply(lambda x: expand_contractions(re.sub('’', \"'\", x)))",
"_____no_output_____"
],
[
"# Apply remove_accented_chars()\ndata['review'] = data['review'].apply(lambda x: remove_accented_chars(re.sub('’', \"'\", x)))",
"_____no_output_____"
],
[
"# Apply scrub_words()\ndata['review'] = data['review'].apply(lambda x: scrub_words(re.sub('’', \"'\", x)))",
"_____no_output_____"
]
],
[
[
"#### Checking the integrity of the data after initial preprocessing steps",
"_____no_output_____"
]
],
[
[
"print(len(data['review']))\nprint(len(original_data['review']),'\\n')\n\nprint('Original Text:', original_data['review'][2784])\nprint(\"-\"*20)\nprint('Processed Text:',data['review'][2784])",
"3000\n3000 \n\nOriginal Text: an absolutely must see! we loved it! so sweet and funny. the cinematography is amazing!\n--------------------\nProcessed Text: an absolutely must see we loved it so sweet and funny the cinematography is amazing \n"
]
],
[
[
"### Adding new column \"word_count\" which specifies the number of tokens in each document",
"_____no_output_____"
]
],
[
[
"data['word_count'] = data['review'].apply(lambda x: len(x.split(' '))) # tokenize words separated by single space\ndata[['review','word_count']].iloc[1000:1005,:]",
"_____no_output_____"
],
[
"print('Mean Review Length:',data['word_count'].mean())\nprint('Minimum Review Length:',data['word_count'].min())\nprint('Max Review Length:',data['word_count'].max())",
"Mean Review Length: 31.346\nMinimum Review Length: 2\nMax Review Length: 891\n"
]
],
[
[
"### Lemmatization, Stemming, Tokenization and Stopwords.",
"_____no_output_____"
]
],
[
[
"## load spacy's English stopwords as variable called 'stopwords'\n\nstopwords = spacy.lang.en.stop_words.STOP_WORDS\nprint('Number of stop words: %d' % len(stopwords))\nprint('First ten stop words: %s' % list(stopwords)[:10])\n# stopwords.remove('no')\n# stopwords.remove('not')",
"Number of stop words: 326\nFirst ten stop words: ['be', 'rather', 'her', 'moreover', 'seeming', 'something', \"'s\", 'fifteen', 'must', 'everywhere']\n"
],
[
"len(stopwords) # stopwords is a set()",
"_____no_output_____"
],
[
"## Adding Custom stopwords to the spacy stopword list\nfor w in stopwords:\n nlp.vocab[w].is_stop = True",
"_____no_output_____"
],
[
"## Use NLTK for stemming.\n## load nltk's SnowballStemmer as variable 'stemmer'\nfrom nltk.stem.snowball import SnowballStemmer\nstemmer = SnowballStemmer(\"english\")",
"_____no_output_____"
],
[
"# Here I define a tokenizer and stemmer which returns the set of stems (excluding stop words) in the text that it is passed\n\ndef tokenize_and_stem(doc, remove_stopwords = True):\n # first tokenize by sentence, then by word to ensure that punctuation is caught as it's own token\n if remove_stopwords:\n tokens = [word.text for word in doc if not word.is_stop]\n else:\n tokens = [word.text for word in doc]\n \n #print(tokens[:5])\n filtered_tokens = []\n # filter out any tokens not containing letters (e.g., numeric tokens, raw punctuation)\n for token in tokens:\n if re.search('[a-zA-Z]', token):\n filtered_tokens.append(token)\n \n #print(\"ended re.search\")\n stems = [stemmer.stem(t) for t in filtered_tokens]\n #print(\"returning stems\")\n return stems\n\ndef tokenize_and_lemmatize(doc, remove_stopwords = True):\n \n # spaCy will convert word to lower case and changing past tense, \n # gerund form (other tenses as well) to present tense. Also, “they” normalize to “-PRON-” which is pronoun.\n\n if remove_stopwords:\n tokens = [word for word in doc if not word.is_stop]\n else:\n tokens = [word for word in doc]\n #print(\"Completed tokenization\")\n filtered_tokens = []\n # filter out any tokens not containing letters (e.g., numeric tokens, raw punctuation)\n for token in tokens:\n if re.search('[a-zA-Z]', token.text):\n filtered_tokens.append(token)\n \n #print(\"ended re.search\")\n lemma = [t.lemma_ for t in filtered_tokens]\n #print(\"returning lemms\")\n return lemma\n\n\ndef tokenize_only(doc, remove_stopwords = True):\n # first tokenize by sentence, then by word to ensure that punctuation is caught as it's own token\n if remove_stopwords:\n tokens = [word.text for word in doc if not word.is_stop]\n else:\n tokens = [word.text for word in doc]\n filtered_tokens = []\n # filter out any tokens not containing letters (e.g., numeric tokens, raw punctuation)\n for token in tokens:\n if re.search('[a-zA-Z]', token):\n filtered_tokens.append(token)\n return filtered_tokens",
"_____no_output_____"
]
],
[
[
"We are trying to create four seperate lists:\n1. Clean Review Lemmatized (w/o stopwords)\n2. Clean Review Stemmed (w/o stop words)\n3. Review Lemmatized (w stopwords)\n4. Review Stemmed (w stopwords)",
"_____no_output_____"
],
[
"# NOTE: Time to execute below chunk = 503.5 s (9 min)\n## Load from local file 'data_txt_preprocessed.csv'",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv('data_txt_preprocessed.csv', index_col='userId')\n# data.head()\nimport ast\n# string literal evaluation --> data structure\ndata['clean_review_stemmed'] = data['clean_review_stemmed'].apply(lambda x: ast.literal_eval(x))\ndata['clean_review_lemmatized'] = data['clean_review_lemmatized'].apply(lambda x: ast.literal_eval(x))\ndata['clean_review_tokenized'] = data['clean_review_tokenized'].apply(lambda x: ast.literal_eval(x))\ndata['review_stemmed'] = data['review_stemmed'].apply(lambda x: ast.literal_eval(x))\ndata['review_lemmatized'] = data['review_lemmatized'].apply(lambda x: ast.literal_eval(x))\ndata['review_tokenized'] = data['review_tokenized'].apply(lambda x: ast.literal_eval(x))",
"_____no_output_____"
],
[
"data.iloc[1000:1005,:]",
"_____no_output_____"
]
],
[
[
"## Naming Conventions followed ####\n## 'clean' describes Review that does not contain stopwords\n## 'all' describes Review that contains stopwords.\n# Stemmed - Lemmatized - Tokenized\n\nst = time.time() # start time\n\n# stemmed, lemmatized, tokenized reviews\n\n# w.o Stopwords \ndata['clean_review_lemmatized'] = data['review'].apply(lambda x: tokenize_and_lemmatize(nlp(x)))\ndata['clean_review_stemmed'] = data['review'].apply(lambda x: tokenize_and_stem(nlp(x)))\ndata['clean_review_tokenized'] = data['review'].apply(lambda x: tokenize_only(nlp(x)))\n\n# w. Stopwords\ndata['review_lemmatized'] = data['review'].apply(lambda x: tokenize_and_lemmatize(nlp(x), False))\ndata['review_stemmed'] = data['review'].apply(lambda x: tokenize_and_stem(nlp(x), False))\ndata['review_tokenized'] = data['review'].apply(lambda x: tokenize_only(nlp(x), False))\n\nprint('Execution Time:', time.time()-st)",
"_____no_output_____"
]
],
[
[
"# Vocab List w.o stopwords\nclean_vocab_lemmatized = []\nclean_vocab_stemmed = []\nclean_vocab_tokenized = []\n\n# Vocab List w stopwords\nall_vocab_lemmatized = []\nall_vocab_tokenized = []\n\nfor i,j,k in zip(data['clean_review_lemmatized'],data['clean_review_stemmed'], data['clean_review_tokenized']):\n clean_vocab_lemmatized.extend(i)\n clean_vocab_stemmed.extend(j)\n clean_vocab_tokenized.extend(k)\n \nfor i,j in zip(data['review_lemmatized'], data['review_tokenized']):\n all_vocab_lemmatized.extend(i) \n all_vocab_tokenized.extend(j)\n",
"_____no_output_____"
],
[
"print(len(clean_vocab_lemmatized))\nprint(len(clean_vocab_stemmed))\nprint(len(clean_vocab_tokenized),'\\n')\nprint(len(all_vocab_lemmatized))\nprint(len(all_vocab_tokenized))",
"35727\n35727\n35727 \n\n80818\n80818\n"
],
[
"print(data['review'][1000],'\\n')\nprint(data['clean_review_lemmatized'][1000],'\\n')\nprint(data['clean_review_stemmed'][1000],'\\n')\nprint(data['review_stemmed'][1000],'\\n')\nprint(data['review_lemmatized'][1000],'\\n')",
"i really liked it i really thought some of the scenes were done really well and the music i thought was just as good as the cartoon like i really liked the hakuna matata scene and the can you feel the love tonight i though those both sounded really well overall i really liked it for a disney remake the scenes were more intense than the cartoon especially at the end when simba was fighting scar \n\n['like', 'think', 'scene', 'music', 'think', 'good', 'cartoon', 'like', 'like', 'hakuna', 'matata', 'scene', 'feel', 'love', 'tonight', 'sound', 'overall', 'like', 'disney', 'remake', 'scene', 'intense', 'cartoon', 'especially', 'end', 'simba', 'fight', 'scar'] \n\n['like', 'thought', 'scene', 'music', 'thought', 'good', 'cartoon', 'like', 'like', 'hakuna', 'matata', 'scene', 'feel', 'love', 'tonight', 'sound', 'overal', 'like', 'disney', 'remak', 'scene', 'intens', 'cartoon', 'especi', 'end', 'simba', 'fight', 'scar'] \n\n['i', 'realli', 'like', 'it', 'i', 'realli', 'thought', 'some', 'of', 'the', 'scene', 'were', 'done', 'realli', 'well', 'and', 'the', 'music', 'i', 'thought', 'was', 'just', 'as', 'good', 'as', 'the', 'cartoon', 'like', 'i', 'realli', 'like', 'the', 'hakuna', 'matata', 'scene', 'and', 'the', 'can', 'you', 'feel', 'the', 'love', 'tonight', 'i', 'though', 'those', 'both', 'sound', 'realli', 'well', 'overal', 'i', 'realli', 'like', 'it', 'for', 'a', 'disney', 'remak', 'the', 'scene', 'were', 'more', 'intens', 'than', 'the', 'cartoon', 'especi', 'at', 'the', 'end', 'when', 'simba', 'was', 'fight', 'scar'] \n\n['i', 'really', 'like', '-PRON-', 'i', 'really', 'think', 'some', 'of', 'the', 'scene', 'be', 'do', 'really', 'well', 'and', 'the', 'music', 'i', 'think', 'be', 'just', 'as', 'good', 'as', 'the', 'cartoon', 'like', 'i', 'really', 'like', 'the', 'hakuna', 'matata', 'scene', 'and', 'the', 'can', '-PRON-', 'feel', 'the', 'love', 'tonight', 'i', 'though', 'those', 'both', 'sound', 'really', 'well', 'overall', 'i', 'really', 'like', '-PRON-', 'for', 'a', 'disney', 'remake', 'the', 'scene', 'be', 'more', 'intense', 'than', 'the', 'cartoon', 'especially', 'at', 'the', 'end', 'when', 'simba', 'be', 'fight', 'scar'] \n\n"
]
],
[
[
"Text Data Visualization\n----------",
"_____no_output_____"
]
],
[
[
"# Creating Dataframe for tokens in Review's Vocabulary\nall_vocab_frame = pd.DataFrame({'words': all_vocab_tokenized}, index = all_vocab_lemmatized)\nprint ('There are ' + str(all_vocab_frame.shape[0]) + ' words in all_vocab_frame')\n\nclean_vocab_frame = pd.DataFrame({'words': clean_vocab_tokenized}, index = clean_vocab_lemmatized)\nprint ('There are ' + str(clean_vocab_frame.shape[0]) + ' words in clean_vocab_frame')",
"There are 80818 words in all_vocab_frame\nThere are 35727 words in clean_vocab_frame\n"
]
],
[
[
"### Plotting Most frequent words before and after stopword removal",
"_____no_output_____"
]
],
[
[
"values, counts = np.unique(clean_vocab_frame, return_counts=True)\nall_values, all_counts = np.unique(all_vocab_frame, return_counts=True)",
"_____no_output_____"
],
[
"sorted_indices = np.argsort(-counts)\nprint(sorted_indices)\nall_sorted_indices = np.argsort(-all_counts)\nprint(all_sorted_indices)",
"[2521 2726 1729 ... 1941 1926 4435]\n[4158 2187 4530 ... 2076 2117 4692]\n"
],
[
"values = values[sorted_indices]\ncounts = counts[sorted_indices]\n\nall_values = all_values[all_sorted_indices]\nall_counts = all_counts[all_sorted_indices]",
"_____no_output_____"
],
[
"font = {'family' : 'DejaVu Sans',\n 'weight' : 'bold'}\nplt.rc('font', **font)\nplt.figure(figsize=(15,10))\n\n# Frequency plot of words w/o stopwords\nplt.subplot(1,2,1)\nplt.barh(values[:15], counts[:15], color='blue')\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=15)\nplt.title('Word Frequency: w/o Stopwords', fontsize=20)\n\n\n# Frequency plot of words with stopwords\nplt.subplot(1,2,2)\nplt.barh(all_values[:15], all_counts[:15], color='mediumspringgreen')\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=15)\nplt.title('Word Frequency: w. Stopwords', fontsize=20)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Observations from the Frequency Plots\n\n1) The most occuring words present in both the graphs are quite different.\\\n2) Words in graph 1 (without stopwords) better describes the themes within the Reviews written",
"_____no_output_____"
],
[
"### Wordcloud of Review words (Lemmatized)",
"_____no_output_____"
]
],
[
[
"# Word Cloud string\nclean_review_wordcloud=[]\nfor i in data['clean_review_lemmatized']:\n clean_review_wordcloud+=i\nclean_string = \" \".join(clean_review_wordcloud)",
"_____no_output_____"
],
[
"# !pip install wordcloud\nfrom wordcloud import WordCloud\nwordcloud = WordCloud(max_font_size=100, width = 600,height=300,max_words=50, background_color=\"white\").generate(clean_string)\nimport matplotlib.pyplot as plt\n%matplotlib inline\nplt.figure(figsize=(30,50))\nplt.imshow(wordcloud)\nplt.axis(False)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Word Frequency by Sentimentiment Groups",
"_____no_output_____"
]
],
[
[
"data.head(2)",
"_____no_output_____"
],
[
"# grouby sentiment\ngrouped_text = data.groupby('sentiment')['clean_review_tokenized']\n\n# Fetch entire tokenized text for specific group\nfrom itertools import chain\nfrequent_words_sentiment_df = pd.DataFrame(columns={\"values\", \"counts\", \"sentiment\"})\n\nfor num in range(2): # 2 Sentiment levels\n values, counts = np.unique(list(chain.from_iterable(grouped_text.get_group(num))), return_counts=True)\n # Create single List of Tokenized Reviews; lazily evaluates by taking a single iterable argument at a time\n \n sorted_indices = np.argsort(-counts) # returns indices of sorted 'counts' in reversed order \n frequent_words_sentiment_df = frequent_words_sentiment_df.append({\"values\":values[sorted_indices], \"counts\":counts[sorted_indices], \"sentiment\": num}, ignore_index=True)\n # Append word values in decreasing count order grouped by sentiment\n\nfrequent_words_sentiment_df.head() # words sorted by counts order",
"_____no_output_____"
],
[
"font = {'family' : ' DejaVu Sans', 'weight' : 'bold', 'size': 15}\n\nplt.rc('font', **font)\nplt.figure(figsize=(18,10))\n\nplt.subplot(1,2,1)\nplt.barh(frequent_words_sentiment_df.loc[1,'values'][:15], frequent_words_sentiment_df.loc[1,'counts'][:15], color='royalblue')\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=15)\nplt.title('Words Frequency: Sentiment 1', fontsize='20')\n\nplt.subplot(1,2,2)\nplt.barh(frequent_words_sentiment_df.loc[0,'values'][:15], frequent_words_sentiment_df.loc[0,'counts'][:15], color='blue')\nplt.gca().invert_yaxis()\nplt.title('Words Frequency: Sentiment 0', fontsize='20')\nplt.yticks(fontsize=15)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Observations:\n1. Generic words common to both Sentiment reviews cloud the differences between the classes (top words common to both: 'movie','original', 'like' etc.\n\n\n2. Difference in count of occurences of key words: ",
"_____no_output_____"
]
],
[
[
"print('Count of \"good\" in Sentiment-1:',frequent_words_sentiment_df.loc[1,'counts'][np.where(frequent_words_sentiment_df.loc[1,'values']=='good')][0])\nprint('Count of \"good\" in Sentiment-0:',frequent_words_sentiment_df.loc[0,'counts'][np.where(frequent_words_sentiment_df.loc[0,'values']=='good')][0],'\\n')\nprint('Count of \"no\" in Sentiment-1:',frequent_words_sentiment_df.loc[1,'counts'][np.where(frequent_words_sentiment_df.loc[1,'values']=='no')][0])\nprint('Count of \"no\" in Sentiment-0:',frequent_words_sentiment_df.loc[0,'counts'][np.where(frequent_words_sentiment_df.loc[0,'values']=='no')][0],'\\n')\nprint('Count of \"bad\" in Sentiment-1:',frequent_words_sentiment_df.loc[1,'counts'][np.where(frequent_words_sentiment_df.loc[1,'values']=='bad')][0])\nprint('Count of \"bad\" in Sentiment-0:',frequent_words_sentiment_df.loc[0,'counts'][np.where(frequent_words_sentiment_df.loc[0,'values']=='bad')][0],'\\n')\nprint('Count of \"boring\" in Sentiment-1:',frequent_words_sentiment_df.loc[1,'counts'][np.where(frequent_words_sentiment_df.loc[1,'values']=='boring')][0])\nprint('Count of \"boring\" in Sentiment-0:',frequent_words_sentiment_df.loc[0,'counts'][np.where(frequent_words_sentiment_df.loc[0,'values']=='boring')][0],'\\n')\nprint('Count of \"lacked\" in Sentiment-1:',frequent_words_sentiment_df.loc[1,'counts'][np.where(frequent_words_sentiment_df.loc[1,'values']=='lacked')][0])\nprint('Count of \"lacked\" in Sentiment-0:',frequent_words_sentiment_df.loc[0,'counts'][np.where(frequent_words_sentiment_df.loc[0,'values']=='lacked')][0],'\\n')",
"Count of \"good\" in Sentiment-1: 161\nCount of \"good\" in Sentiment-0: 345 \n\nCount of \"no\" in Sentiment-1: 123\nCount of \"no\" in Sentiment-0: 51 \n\nCount of \"bad\" in Sentiment-1: 55\nCount of \"bad\" in Sentiment-0: 24 \n\nCount of \"boring\" in Sentiment-1: 49\nCount of \"boring\" in Sentiment-0: 7 \n\nCount of \"lacked\" in Sentiment-1: 48\nCount of \"lacked\" in Sentiment-0: 15 \n\n"
]
],
[
[
"### Word Frequency of Pure Negative, Pure Positive tokens of the Sentiment groups\n- To better understand the divergence of the sentiments established between the two groups, we must remove the intersecting tokens present in both classes.",
"_____no_output_____"
]
],
[
[
"# Review Words in sentiment=1 not in sentiment=0\nneg_tokens = list(set(frequent_words_sentiment_df.loc[1,'values'])-set(frequent_words_sentiment_df.loc[0,'values']))\n# 1136 Pure Negative Words found\n\nneg_index = np.array([list(frequent_words_sentiment_df.loc[1,'values']).index(i) for i in neg_tokens]) # index location\nneg_counts = frequent_words_sentiment_df.loc[1,'counts'][neg_index] # counts of words\nneg_tokens = np.array(neg_tokens)\n\n# Sort Tokens by Descending Count order\nindex = np.argsort(-neg_counts)\nneg_counts = neg_counts[index]\nneg_tokens = neg_tokens[index]\n\n# Review Words in sentiment=0 not in sentiment=1\npos_tokens = list(set(frequent_words_sentiment_df.loc[0,'values'])-set(frequent_words_sentiment_df.loc[1,'values']))\n# 1136 Pure positive Words found\n\npos_index = np.array([list(frequent_words_sentiment_df.loc[0,'values']).index(i) for i in pos_tokens]) # index location\npos_counts = frequent_words_sentiment_df.loc[0,'counts'][pos_index] # counts of words\npos_tokens = np.array(pos_tokens)\n\n# Sort Tokens by Descending Count order\nindex = np.argsort(-pos_counts)\npos_counts = pos_counts[index]\npos_tokens = pos_tokens[index]",
"_____no_output_____"
],
[
"font = {'family' : ' DejaVu Sans', 'weight' : 'bold'}\n\nplt.rc('font', **font)\nplt.figure(figsize=(30,18))\n\nplt.subplot(1,2,1)\nplt.barh(neg_tokens[:15], neg_counts[:15], color='tomato')\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=25)\nplt.xticks(fontsize=20)\nplt.title('Pure Negative Word Frequency: Sentiment 1', fontsize='30')\n\nplt.subplot(1,2,2)\nplt.barh(pos_tokens[:15], pos_counts[:15], color='royalblue')\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=25)\nplt.xticks(fontsize=20)\nplt.title('Pure Positive Word Frequency: Sentiment 0', fontsize='30')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Observations:\n1. Note the Top Words, Nouns and adjacectives in the classes <u>correlate</U> to the **Negative and Positive sentiments** expressed by the reviewers.",
"_____no_output_____"
],
[
"# TF-IDF",
"_____no_output_____"
],
[
"# TF-IDF Explaination:\n\n\n\n- TF: Term Frequency, which measures how frequently a term occurs in a document. The term frequency is often divided by the document length (aka. the total number of terms in the document) as a way of <u>normalization</u>:\n\n**TF(t) = (Number of times term t appears in a document) / (Total number of terms in the document)**",
"_____no_output_____"
],
[
"- IDF: Inverse Document Frequency, which measures how important a term is.<u>Frequent Terms are weighed down, while Rare Terms are scaled up</u>, by computing the following:\n\n**IDF(t) = log_e(Total number of documents / Number of documents with term t in it)**",
"_____no_output_____"
]
],
[
[
"## tfidf vectorizer needs sentence and not token. Hence we need to combine all the tokens back to form a string\ndata['clean_review_stemmed'] = [' '.join(text) for text in data['clean_review_stemmed']]\ndata['clean_review_lemmatized'] = [' '.join(text) for text in data['clean_review_lemmatized']]",
"_____no_output_____"
],
[
"data['clean_review_lemmatized'][0]",
"_____no_output_____"
]
],
[
[
"### Creating the `tfidf_matrix`",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import TfidfVectorizer\n\n#define vectorizer parameters\n\n# max_df : cutoff to exclude highly populated words in all doc eg: stopwords\n# min_df : cutoff to exclude highly rare words in all doc eg: rarewords, no semantic value across corpus\n# ngram_range : type of ngrams to include (min_ngram, max_ngram) (default=(1, 1))\n# max_features : features dimension cutoff\ntfidf_vectorizer = TfidfVectorizer(max_df=0.9, max_features=1500, #(0.05, 0.001)\n min_df=0.2,\n use_idf=True, ngram_range=(1,1))\n\ntfidf_matrix = tfidf_vectorizer.fit_transform(data['clean_review_lemmatized'])\n\nprint(tfidf_matrix.shape)",
"(3000, 3)\n"
],
[
"# Terms: Main latent themes of the Text\n# vocabulary_: Main latent Features of the Text\n# tfidf_vectorizer.vocabulary_\n\nterms = tfidf_vectorizer.get_feature_names()\nterms",
"_____no_output_____"
],
[
"tfidf_matrix.todense() # todense() : Return a dense matrix representation of matrix.",
"_____no_output_____"
]
],
[
[
"Unsupervised Learning\n-------\n### 1. K-mean Clustering",
"_____no_output_____"
],
[
"### Fitting the elbow curve to identify right number of clusters/topics",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\nfrom sklearn.cluster import KMeans\nfrom scipy.spatial.distance import cdist #cluster distance\nimport joblib\nSum_of_squared_distances = []\nK = range(1,6)\nfor k in K:\n kmeanModel = KMeans(n_clusters=k, random_state=123)\n kmeanModel.fit(tfidf_matrix)\n Sum_of_squared_distances.append(kmeanModel.inertia_)\n ",
"_____no_output_____"
],
[
"Sum_of_squared_distances",
"_____no_output_____"
],
[
"# Plot the elbow\n\n# Distortion, on the y-axis, corresponds to our cost function: \n# the sum of squared difference between each data point and the centroid, i.e., the cluster centre.\n\n# As K increases the corresponding distortion value will tend to zero, \n# because you end up having just one data point per cluster. With only one data point in per cluster, \n# the centroid is the data point itself, so the distortion will be equal to zero.\n\nfont = {'family' : 'normal',\n 'weight' : 'bold',\n 'size' : 10}\n\nplt.rc('font', **font)\nplt.plot(K, Sum_of_squared_distances, 'bx-')\nplt.xlabel('k')\nplt.ylabel('Sum_of_squared_distances')\nplt.title('Elbow Method For Optimal k')\nplt.show()\n",
"findfont: Font family ['normal'] not found. Falling back to DejaVu Sans.\nfindfont: Font family ['normal'] not found. Falling back to DejaVu Sans.\n"
],
[
"# Based on Elbow cure, we choose 4 clusters\nnum_clusters = 4\n\nkm = KMeans(n_clusters=num_clusters)\n\nkm.fit(tfidf_matrix)\n#km.labels_\nclusters = km.labels_.tolist()\n#km.cluster_centers\ncenters = km.cluster_centers_\nprint(f\"the cluster centers are {centers}\")",
"the cluster centers are [[0.03579355 0.97705253 0.04971069]\n [0. 0. 0. ]\n [0.01508697 0.22601083 0.91680239]\n [0.88066786 0.21829916 0.13143605]]\n"
]
],
[
[
"### Getting the top words from each cluster",
"_____no_output_____"
]
],
[
[
"print(km.cluster_centers_)\nprint(km.cluster_centers_.shape)",
"[[0.03579355 0.97705253 0.04971069]\n [0. 0. 0. ]\n [0.01508697 0.22601083 0.91680239]\n [0.88066786 0.21829916 0.13143605]]\n(4, 3)\n"
],
[
"# Sort Index of original list\nkm.cluster_centers_.argsort()",
"_____no_output_____"
],
[
"## Reversing the list so that index of max element is in 0th index\nkm.cluster_centers_.argsort()[:,::-1]",
"_____no_output_____"
],
[
"print(\"Top terms per cluster:\")\n\n#sort cluster centers by proximity to centroid and picking the top 6 words per cluster\norder_centroids = km.cluster_centers_.argsort()[:, ::-1] # Reverse the ndarray column order, returns same 'n' col array\nfor i in range(num_clusters):\n print()\n print(\"Top words in Cluster-%d :\" % i, end='')\n print()\n for ind in order_centroids[i, :3]: #replace 6 with n words per cluster\n print('%s' % terms[ind].split(' '), end=',')",
"Top terms per cluster:\n\nTop words in Cluster-0 :\n['movie'],['original'],['love'],\nTop words in Cluster-1 :\n['original'],['movie'],['love'],\nTop words in Cluster-2 :\n['original'],['movie'],['love'],\nTop words in Cluster-3 :\n['love'],['movie'],['original'],"
],
[
"data['cluster_group'] = clusters\n# data.pop('clean_text', None)\ndata.head()",
"_____no_output_____"
],
[
"data.keys()",
"_____no_output_____"
],
[
"cluster_df = pd.DataFrame(data)",
"_____no_output_____"
],
[
"cluster_df['cluster_group'].value_counts()",
"_____no_output_____"
]
],
[
[
"#### Fetching the most frequent words among each cluster\n\nStep 1) Tokenize the entire text <br>\nStep 2) Group the tokenized text by cluster id (output is list of lists: [[],[],[]])<br>\nStep 3) Unlist the array of lists for each cluster group using chain function from itertools",
"_____no_output_____"
]
],
[
[
"cluster_df.groupby('sentiment')['cluster_group'].value_counts()",
"_____no_output_____"
],
[
"##Step 1\ncluster_df['clean_review_tokenized'] = [text.split(' ') for text in cluster_df['clean_review_lemmatized']]",
"_____no_output_____"
],
[
"##Step 2: Create pandas SeriesGroupBy object\n## Fetch entire tokenized text for specific group\ngrouped_text = cluster_df.groupby('cluster_group')['clean_review_tokenized']",
"_____no_output_____"
],
[
"from itertools import chain",
"_____no_output_____"
],
[
"frequent_words_df = pd.DataFrame(columns={\"values\", \"counts\", \"cluster_id\"})",
"_____no_output_____"
],
[
"for num in range(num_clusters):\n values, counts = np.unique(list(chain.from_iterable(grouped_text.get_group(num))), return_counts=True)\n # eg: returns an 1D array of unique words from tokenized reviews\n # chain() constructor taking a single iterable argument that evaluates lazily;\n \n sorted_indices = np.argsort(-counts) # returns indices of sorted list in reversed order\n # Create Cluster df of values(word list) sorted by counts \n frequent_words_df = frequent_words_df.append({\"values\":values[sorted_indices], \"counts\":counts[sorted_indices], \"cluster_id\": num}, ignore_index=True)",
"_____no_output_____"
],
[
"frequent_words_df.head() # words sorted by counts order",
"_____no_output_____"
]
],
[
[
"### Plotting Top Words in Clusters 0, 1, 2, 3",
"_____no_output_____"
]
],
[
[
"font = {'family' : 'DejaVu Sans',\n 'weight' : 'bold',\n 'size' : 35}\n\nplt.rc('font', **font)\n\nfig = plt.figure(figsize=(15,20))\nplt.subplot(2,2,1)\nplt.barh(frequent_words_df.loc[0,'values'][:8], frequent_words_df.loc[0,'counts'][:8])\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=15)\nplt.title('Words Frequency: Cluster 0', fontsize=20)\n\nplt.subplot(2,2,2)\nplt.barh(frequent_words_df.loc[1,'values'][:8], frequent_words_df.loc[1,'counts'][:8])\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=15)\nplt.title('Words Frequency: Cluster 1', fontsize=20)\n\nplt.subplot(2,2,3)\nplt.barh(frequent_words_df.loc[2,'values'][:8], frequent_words_df.loc[2,'counts'][:8])\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=15)\nplt.title('Words Frequency: Cluster 2', fontsize=20)\n\nplt.subplot(2,2,4)\nplt.barh(frequent_words_df.loc[3,'values'][:8], frequent_words_df.loc[3,'counts'][:8])\nplt.gca().invert_yaxis()\nplt.yticks(fontsize=15)\nplt.title('Words Frequency: Cluster 3', fontsize=20)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### Observations:\nWords/Themes populated in the Clusters' Reviews describe:\n\nCluster 0: describes 'love' for the movie, again consisting of potentially shorter reviews\n\nCluster 1: positive adjectives like 'great', 'like' etc\n\nCluster 2: 'movie', also consist of possible shorter reviews that describe the movie in 1 sentence\n\nCluster 3: talks about originality of the movie/remake of the 2019 Lion King version",
"_____no_output_____"
],
[
"### 2. Truncated SVD (Latent Semantic Analysis - LSA) using Scikitlearn\nTopic Modelling by Matrix Decomposition",
"_____no_output_____"
],
[
"Upon Truncated SVD processing, we obtain 2 Matrices\n\n1. U ∈ ℝ^(m ⨉ t) emerges as our Document-specific Topic allocation matrix : m-document vector, t-topic\n2. V ∈ ℝ^(n ⨉ t) becomes our Topic-specific Term allocation matrix : n-term vector, t-topic\n\n<u>In both U and V, the columns correspond to one of our t topics. </u>",
"_____no_output_____"
],
[
"#### Import Libraries",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import TfidfVectorizer\n\ntfidf_vectorizer = TfidfVectorizer(max_df=0.9, max_features=1500, #(0.05, 0.001)\n min_df=0.2,\n use_idf=True, ngram_range=(1,1))\n\ntfidf_matrix = tfidf_vectorizer.fit_transform(data['clean_review_lemmatized'])\n\nprint(tfidf_matrix.shape)",
"(3000, 3)\n"
],
[
"from sklearn.decomposition import TruncatedSVD",
"_____no_output_____"
],
[
"# Importing tfidf vectorized documents\nprint(tfidf_matrix.shape)\ntfidf_matrix.todense()",
"(3000, 3)\n"
]
],
[
[
"### Creating the `svd_matrix` from the `tfidf_matrix`",
"_____no_output_____"
]
],
[
[
"# Select No. of Latent Themes to extract from text\nn_components = 2\nsvd_model = TruncatedSVD(n_components=n_components, algorithm='randomized',n_iter=20,random_state=143)\n\nsvd_matrix = svd_model.fit(tfidf_matrix)\n\nsvd_matrix",
"_____no_output_____"
],
[
"# explained_variance_ratio_\nprint(f\"Explained Variance Ratio : {svd_matrix.explained_variance_ratio_}\") \nprint(f\"Total Explained Variance : {round(svd_matrix.explained_variance_ratio_.sum() * 100, 2)} %\")\n\n# singular_values_ : explains the Top 2 Latent Topics found in Text\nprint(f\"The singular values are {svd_matrix.singular_values_}\")",
"Explained Variance Ratio : [0.29173502 0.3830367 ]\nTotal Explained Variance : 67.48 %\nThe singular values are [31.05889539 23.21106784]\n"
]
],
[
[
"i.e\nC-1 explains 30% of variation\\\nC-2 explains 38% of variation",
"_____no_output_____"
],
[
"### Picking the few most important words in each topic\n\nThe components of svd_model are our topics and we can access them using svdmodel.components.<br>\nlet's print a few most important words in each of the 4 topics and see how our model has done.",
"_____no_output_____"
]
],
[
[
"# Components describe the Theme of Text (represented by Singular Values, Singular Vectors)\n# Theme = 2, \nsvd_model.components_",
"_____no_output_____"
],
[
"# Term vs Topic Strength\nfor i, comp in enumerate(svd_model.components_):\n print(f\"The component is {comp} and shape is {comp.shape}\") # Expl\n terms_comp = zip(terms, comp)\n sorted_terms = sorted(terms_comp, key= lambda x:x[1], reverse=True)[:6]\n print(\"Topic \"+str(i)+\": \")\n for t in sorted_terms:\n print(f\"{t[0]} -- {t[1]}\")\n print(\" \")",
"The component is [0.29933712 0.84659996 0.44007477] and shape is (3,)\nTopic 0: \nmovie -- 0.8465999579544627\noriginal -- 0.44007476736593937\nlove -- 0.29933711817834313\n \nThe component is [ 0.04572337 -0.47342075 0.87964889] and shape is (3,)\nTopic 1: \noriginal -- 0.8796488906374453\nlove -- 0.04572337221063354\nmovie -- -0.47342074567344294\n \n"
]
],
[
[
"### Tagging each document with a topic\n### Creating the `doc_topic_matrix`\n`doc_topic_matrix` is the resultant SVD Output",
"_____no_output_____"
]
],
[
[
"# 2 Singular Values, 2 Components (Eigenvalues, Eigenvectors - Strength of Variation)\n# Documents - 3000, Topic - 2\ndoc_topic_matrix = svd_matrix.transform(tfidf_matrix)\nprint(doc_topic_matrix,'\\n')\n\nsvd_categories = np.argmax(doc_topic_matrix, axis=1) # Returns the indices of the maximum values along an axis.\nprint(doc_topic_matrix.shape,'\\n')\nprint(svd_categories)",
"[[ 0.93584811 -0.20875454]\n [ 0.44007477 0.87964889]\n [ 0.88547331 0.35877625]\n ...\n [ 0. 0. ]\n [ 0.52977825 0.79339121]\n [ 0.29933712 0.04572337]] \n\n(3000, 2) \n\n[0 1 0 ... 0 1 0]\n"
],
[
"data['SVD_group'] = svd_categories",
"_____no_output_____"
],
[
"pd.DataFrame(data).head(6)",
"_____no_output_____"
],
[
"print(data.groupby('sentiment')['SVD_group'].value_counts())",
"sentiment SVD_group\n0 0 1865\n 1 305\n1 0 645\n 1 185\nName: SVD_group, dtype: int64\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"raw",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"raw"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbac5e2155ed69728d12731e163f740f23c0fa2a
| 499,577 |
ipynb
|
Jupyter Notebook
|
02-Camera-Calibration/.ipynb_checkpoints/DistortionCorrection-checkpoint.ipynb
|
vyasparthm/AutonomousDriving
|
4a767442a7e661dd71f4e160f3c071dc02614ab6
|
[
"MIT"
] | null | null | null |
02-Camera-Calibration/.ipynb_checkpoints/DistortionCorrection-checkpoint.ipynb
|
vyasparthm/AutonomousDriving
|
4a767442a7e661dd71f4e160f3c071dc02614ab6
|
[
"MIT"
] | null | null | null |
02-Camera-Calibration/.ipynb_checkpoints/DistortionCorrection-checkpoint.ipynb
|
vyasparthm/AutonomousDriving
|
4a767442a7e661dd71f4e160f3c071dc02614ab6
|
[
"MIT"
] | null | null | null | 2,601.963542 | 390,124 | 0.961902 |
[
[
[
"## Distortion Correction\n\nThis notebook will explain about correcting Distortion and generate an undistorted image.\n\nThere are two main steps to this process: use chessboard images to obtain image points and object points, and then use the OpenCV functions `cv2.calibrateCamera()` and `cv2.undistort()` to compute the calibration and undistortion.\n",
"_____no_output_____"
],
[
"### Generating Imagepoints and ObjectPoints\n\nThis below cell will generate image points and object points as we did in earlier notebook.",
"_____no_output_____"
]
],
[
[
"\n\n#Import required libraries\nimport numpy as np\nimport matplotlib.image as mpimg\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom matplotlib import patches\nimport glob\nimport cv2\nimport os\n\n\n\n\n# These arrays will be used to store object points and image points from all input images.\nobjpts = [] # 3d points in real world space\nimgpts = [] # 2d points in image plane\n\n# prepare object points, like (0,0,0), (1,0,0), (2,0,0) ....,(6,5,0)\nobjp = np.zeros((6*9,3), np.float32)\nobjp[:,:2] = np.mgrid[0:9,0:6].T.reshape(-1,2)\n\n# Making a list of calibration images from input images\nfiles = os.listdir('camera_cal')\n\n# looking through the list and searching for chessboard corners\nfor fname in files:\n if fname.startswith('calibration'):\n img = mpimg.imread('camera_cal/'+fname)\n gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n\n # Finding the chessboard corners using inbuilt opencv feature findChessboardCorners\n ret, corners = cv2.findChessboardCorners(gray, (9,6), None)\n\n # If found,we add object points and image points to arrays imagepts and objpts\n if ret == True:\n imgpts.append(corners)\n objpts.append(objp)\n\n # Draw and display the corners using opencv\n img = cv2.drawChessboardCorners(img, (9,6), corners, ret) \n \n # Save the image in a premade folder\n plt.imsave('./output_images/ChessboardCorners/'+fname, img)\n\n\n# Display the last image with Chessboard corners drawn\nplt.imshow(img)\n\n",
"_____no_output_____"
],
[
"import pickle\n# Lets define a simple function to generate undistorted images\n\ndef cal_undistort(img, objpoints,imgpoints):\n gray = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)\n ret, mtx, dist, rvecs, tvecs = cv2.calibrateCamera(objpoints,imgpoints,gray.shape[::-1],None, None)\n dist_pickle = {}\n dist_pickle[\"mtx\"] = mtx\n dist_pickle[\"dist\"] = dist\n pickle.dump(dist_pickle, open(\"camera_cal/dist_pickle.p\",\"wb\"))\n undist = cv2.undistort(img,mtx,dist, None, mtx)\n return undist\n\n\n# lets read in the image\nimg = cv2.imread('camera_cal/calibration1.jpg')\n\n#We will use objpoints and imgpoints from above\n\nundistorted_img = cal_undistort(img,objpts,imgpts)\n\nfig,ax = plt.subplots(1,2, figsize = (15,30))\nfig.tight_layout()\nax[0].imshow(img)\nax[0].set_title('Original', fontsize= 20, color = 'g')\nax[1].imshow(undistorted_img)\nax[1].set_title('Undistorted', fontsize= 20, color = 'r') ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbac60101c8500d1906c1b4864fbe6c6553eb8a4
| 10,605 |
ipynb
|
Jupyter Notebook
|
utils.ipynb
|
limiteinductive/DataBox
|
357bee0eb71e14c1674b3a58ed7114f8e8472c5c
|
[
"Apache-2.0"
] | 3 |
2021-11-30T13:03:22.000Z
|
2021-12-21T16:08:55.000Z
|
utils.ipynb
|
limiteinductive/DataBox
|
357bee0eb71e14c1674b3a58ed7114f8e8472c5c
|
[
"Apache-2.0"
] | null | null | null |
utils.ipynb
|
limiteinductive/DataBox
|
357bee0eb71e14c1674b3a58ed7114f8e8472c5c
|
[
"Apache-2.0"
] | null | null | null | 21.911157 | 206 | 0.455257 |
[
[
[
"# default_exp utils",
"_____no_output_____"
]
],
[
[
"# Utils\n\n> Collection of useful functions.",
"_____no_output_____"
]
],
[
[
"#hide\nfrom nbdev.showdoc import *",
"_____no_output_____"
],
[
"#export\nimport os\nimport numpy as np\n\nfrom typing import Iterable, TypeVar, Generator\nfrom plum import dispatch\nfrom pathlib import Path\nfrom functools import reduce\n\nfunction = type(lambda: ())\nT = TypeVar('T')",
"_____no_output_____"
]
],
[
[
"## Basics",
"_____no_output_____"
]
],
[
[
"#export\ndef identity(x: T) -> T:\n \"\"\"Indentity function.\"\"\"\n\n return x",
"_____no_output_____"
],
[
"#export\ndef simplify(x): \n \"\"\"Return an object of an iterable if it is lonely.\"\"\"\n \n\n @dispatch\n def _simplify(x): \n if callable(x):\n try:\n return x()\n except TypeError:\n pass\n return x\n\n @dispatch\n def _simplify(i: Iterable): return next(i.__iter__()) if len(i) == 1 else i\n\n return _simplify(x)",
"_____no_output_____"
]
],
[
[
"The simplify function is used to de-nest an iterable with a single element in it, as for instance [1], while leaving everything else constant. It can also exchange a function for its default argument.",
"_____no_output_____"
]
],
[
[
"simplify({1})",
"_____no_output_____"
],
[
"simplify(simplify)(lambda x='lul': 2*x)",
"_____no_output_____"
],
[
"#export\ndef listify(x, *args):\n \"\"\"Convert `x` to a `list`.\"\"\"\n if args:\n x = (x,) + args\n\n if x is None:\n result = []\n elif isinstance(x, list): result = x\n elif isinstance(x, str) or hasattr(x, \"__array__\") or hasattr(x, \"iloc\"):\n result = [x]\n elif isinstance(x, (Iterable, Generator)):\n result = list(x)\n else:\n result = [x]\n \n return result",
"_____no_output_____"
]
],
[
[
"What's very convenient is that it leaves lists invariant (it doen't nest them into a new list).",
"_____no_output_____"
]
],
[
[
"listify([1, 2])",
"_____no_output_____"
],
[
"listify(1, 2, 3)",
"_____no_output_____"
],
[
"#export\ndef setify(x, *args):\n \"\"\"Convert `x` to a `set`.\"\"\"\n\n return set(listify(x, *args))",
"_____no_output_____"
],
[
"setify(1, 2, 3)",
"_____no_output_____"
],
[
"#export\ndef tuplify(x, *args):\n \"\"\"Convert `x` to a `tuple`.\"\"\"\n\n return tuple(listify(x, *args))",
"_____no_output_____"
],
[
"tuplify(1)",
"_____no_output_____"
],
[
"#export\ndef merge_tfms(*tfms):\n \"\"\"Merge two dictionnaries by stacking common key into list.\"\"\"\n\n def _merge_tfms(tf1, tf2):\n return {\n k: simplify(listify(setify(listify(tf1.get(k)) + listify(tf2.get(k)))))\n for k in {**tf1, **tf2}\n }\n \n return reduce(_merge_tfms, tfms, dict())",
"_____no_output_____"
],
[
"\nmerge_tfms(\n {'animals': ['cats', 'dog'], 'colors': 'blue'}, \n {'animals': 'cats', 'colors': 'red', 'OS': 'i use arch btw'}\n)",
"_____no_output_____"
],
[
"#export\ndef compose(*functions):\n \"\"\"Compose an arbitrary number of functions.\"\"\"\n\n def _compose(fn1, fn2):\n return lambda x: fn1(fn2(x))\n\n return reduce(_compose, functions, identity)",
"_____no_output_____"
],
[
"#export\ndef pipe(*functions):\n \"\"\"Pipe an arbitrary number of functions.\"\"\"\n\n return compose(*functions[::-1])",
"_____no_output_____"
],
[
"#export\ndef flow(data, *functions):\n \"\"\"Flow `data` through a list of functions.\"\"\"\n\n return pipe(*functions)(data)",
"_____no_output_____"
]
],
[
[
"## File manipulation helper",
"_____no_output_____"
]
],
[
[
"#export\ndef get_files(path, extensions=None, recurse=False, folders=None, followlinks=True):\n \"\"\"Get all those file names.\"\"\"\n path = Path(path)\n folders = listify(folders)\n extensions = setify(extensions)\n extensions = {e.lower() for e in extensions}\n\n def simple_getter(p, fs, extensions=None):\n p = Path(p)\n res = [\n p / f\n for f in fs\n if not f.startswith(\".\")\n and ((not extensions) or f'.{f.split(\".\")[-1].lower()}' in extensions)\n ]\n return res\n\n if recurse:\n result = []\n for i, (p, d, f) in enumerate(os.walk(path, followlinks=followlinks)):\n if len(folders) != 0 and i == 0:\n d[:] = [o for o in d if o in folders]\n else:\n d[:] = [o for o in d if not o.startswith(\".\")]\n if len(folders) != 0 and i == 0 and \".\" not in folders:\n continue\n result += simple_getter(p, f, extensions)\n else:\n f = [o.name for o in os.scandir(path) if o.is_file()]\n result = simple_getter(path, f, extensions)\n return list(map(str, result))",
"_____no_output_____"
],
[
"# export\nfrom fastcore.all import *\n\n@patch\ndef decompress(self: Path, dest='.'): \n pass",
"_____no_output_____"
],
[
"#export\n@patch\ndef compress(self: Path, dest='.', keep_copy=True):\n pass",
"_____no_output_____"
],
[
"#export\ndef save_array(array, fname, suffix):\n \"\"\"Save an array with the given name and suffix.\"\"\"\n if not suffix.startswith(\".\"):\n suffix = \".\" + suffix\n\n fname = Path(fname)\n\n return np.save(array, fname.with_suffix(suffix))",
"_____no_output_____"
],
[
"def save_dataset(data):\n return 'NotImplementedError'",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbac72a2f165f9f4d0281f0098ad88fd78a81bad
| 6,502 |
ipynb
|
Jupyter Notebook
|
misc/.ipynb_checkpoints/fun with numpy arrays-checkpoint.ipynb
|
EmilMachine/misc_notebooks
|
af02d7f4feaf9159d8dd18027e0e12c31798a608
|
[
"MIT"
] | null | null | null |
misc/.ipynb_checkpoints/fun with numpy arrays-checkpoint.ipynb
|
EmilMachine/misc_notebooks
|
af02d7f4feaf9159d8dd18027e0e12c31798a608
|
[
"MIT"
] | null | null | null |
misc/.ipynb_checkpoints/fun with numpy arrays-checkpoint.ipynb
|
EmilMachine/misc_notebooks
|
af02d7f4feaf9159d8dd18027e0e12c31798a608
|
[
"MIT"
] | null | null | null | 25.007692 | 239 | 0.436481 |
[
[
[
"import numpy as np",
"_____no_output_____"
],
[
"l = list(range(0,100))",
"_____no_output_____"
]
],
[
[
"# numpy arrays",
"_____no_output_____"
]
],
[
[
"l\na = np.array(l)\na",
"_____no_output_____"
],
[
"a[(a > 20) & (a<80)]",
"_____no_output_____"
],
[
"b = np.random.randint(0,100,100)",
"_____no_output_____"
],
[
"b",
"_____no_output_____"
],
[
"b[(a > 20) & (a<80)]",
"_____no_output_____"
],
[
"(a > 20) & (a<80)",
"_____no_output_____"
]
],
[
[
"# Lists",
"_____no_output_____"
]
],
[
[
"l2 = list(b)\n\ncut = [j for (i,j) in zip(l,l2) if i>20 and i<80 ]\nprint(cut)",
"[34, 10, 93, 91, 18, 9, 74, 36, 0, 8, 82, 92, 64, 27, 66, 53, 79, 53, 69, 35, 14, 36, 12, 71, 12, 95, 0, 47, 16, 19, 54, 14, 65, 46, 13, 53, 64, 8, 16, 25, 29, 69, 22, 36, 68, 4, 10, 57, 64, 60, 61, 11, 36, 91, 20, 8, 93, 58, 43]\n"
]
],
[
[
"what is it doing?",
"_____no_output_____"
]
],
[
[
"# zip is pull them together\nlist(zip([\"a\",\"b\",\"c\"],[1,2,3]))",
"_____no_output_____"
],
[
"# we then grab all the js (stuff from list 2) while doing conditions (if statemnet) on list 1",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbac72a8d1bb9d95a5a54387a391bfcbf4f02615
| 172,384 |
ipynb
|
Jupyter Notebook
|
Introduction to Matplotlib.ipynb
|
DonAvery/my-ds-ml-education-journey
|
1813101d7eb89a79c7a6ddd900627ea9653980b3
|
[
"MIT"
] | null | null | null |
Introduction to Matplotlib.ipynb
|
DonAvery/my-ds-ml-education-journey
|
1813101d7eb89a79c7a6ddd900627ea9653980b3
|
[
"MIT"
] | null | null | null |
Introduction to Matplotlib.ipynb
|
DonAvery/my-ds-ml-education-journey
|
1813101d7eb89a79c7a6ddd900627ea9653980b3
|
[
"MIT"
] | null | null | null | 290.698145 | 25,004 | 0.935475 |
[
[
[
"# Introduction to Matplotlib",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"plt.plot()",
"_____no_output_____"
],
[
"# The semicolon prevents the [] from printing also\nplt.plot();",
"_____no_output_____"
],
[
"plt.plot()\nplt.show()",
"_____no_output_____"
],
[
"plt.plot([1,2,3,4]);",
"_____no_output_____"
],
[
"x = [1, 2, 3, 4]\ny = [11, 22, 33, 44]\nplt.plot(x, y);",
"_____no_output_____"
],
[
"# 1st method\nfig = plt.figure() # creates a figure\nax = fig.add_subplot() # adds some axes\nplt.show()",
"_____no_output_____"
],
[
"# 2nd method\nfig = plt.figure() # creates a figure\nax = fig.add_axes([1, 1, 1, 1])\nax.plot(x, y) # add some data\nplt.show()",
"_____no_output_____"
],
[
"# 3rd method (recommended)\nfig, ax = plt.subplots()\nax.plot(x, y); # add some data\ntype(fig), type(ax)",
"_____no_output_____"
]
],
[
[
"## Matplotlib example workflow",
"_____no_output_____"
]
],
[
[
"# 0. import matplotlib and get it ready for plotting in Jupyter\n%matplotlib inline\nimport matplotlib.pyplot as plt\n\n# 1. Prepare data\nx = [1, 2, 3, 4]\ny = [11, 22, 33, 44]\n\n# 2. Setup plot\nfig, ax = plt.subplots(figsize=(10,10)) # (width, height)\n\n# 3. Plot the data\nax.plot(x, y)\n\n# 4. Customize plot\nax.set(title=\"Simple Plot\",\n xlabel=\"x-axis\",\n ylabel=\"y-axis\")\n\n# 5. Save & show (you save the whole figure)\nfig.savefig(\"sample-plot.png\")",
"_____no_output_____"
]
],
[
[
"## Making figures with Numpy arrays\n\nWe want:\n* Line plot\n* Scatter plot\n* Bar plot\n* Histogram\n* Subplots",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"# Create some data\nx = np.linspace(0, 10, 100)\nx[:10]",
"_____no_output_____"
],
[
"# Plot the data and create a line plot\nfig, ax = plt.subplots()\nax.plot(x, x**2)",
"_____no_output_____"
],
[
"# Use the same data to make a scatter\nfig, ax = plt.subplots()\nax.scatter(x, np.exp(x));",
"_____no_output_____"
],
[
"# Another scatter plot\nfig, ax = plt.subplots()\nax.scatter(x, np.sin(x));",
"_____no_output_____"
],
[
"# Make a plot from dictionary\nnut_butter_prices = {\"Almond butter\": 10,\n \"Peanut butter\": 8,\n \"Cashew butter\": 12}\nfig, ax = plt.subplots()\nax.bar(nut_butter_prices.keys(), nut_butter_prices.values())\nax.set(title=\"Dan's Nut Butter Store\", \n ylabel=\"Price ($)\");",
"_____no_output_____"
],
[
"fig, ax=plt.subplots()\nax.barh(list(nut_butter_prices.keys()), list(nut_butter_prices.values()));",
"_____no_output_____"
],
[
"# Make some data for histograms and plot it\nx = np.random.randn(1000)\nfig, ax=plt.subplots()\nax.hist(x);",
"_____no_output_____"
]
],
[
[
"## Two options for subplots\n",
"_____no_output_____"
]
],
[
[
"# Subplot option 1\nfig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(nrows=2,\n ncols=2,\n figsize=(10, 10))\n# Plot to each different axis\nax1.plot(x, x/2);\nax2.scatter(np.random.random(10), np.random.random(10));\nax3.bar(nut_butter_prices.keys(), nut_butter_prices.values());\nax4.hist(np.random.randn(1000));",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbac7bbaa656c4dcb00711c4b6e393fa0926cfe2
| 860,475 |
ipynb
|
Jupyter Notebook
|
springboard_modules/the art of storytelling (Lending Club).ipynb
|
tanaysd/Data-Science-Springboard
|
951e20109e14a784c2c8746f352f712e653ba250
|
[
"MIT"
] | null | null | null |
springboard_modules/the art of storytelling (Lending Club).ipynb
|
tanaysd/Data-Science-Springboard
|
951e20109e14a784c2c8746f352f712e653ba250
|
[
"MIT"
] | null | null | null |
springboard_modules/the art of storytelling (Lending Club).ipynb
|
tanaysd/Data-Science-Springboard
|
951e20109e14a784c2c8746f352f712e653ba250
|
[
"MIT"
] | null | null | null | 507.355542 | 548,970 | 0.910492 |
[
[
[
"# Module 7.2 | Apply Data Storytelling to Lending Club loan data",
"_____no_output_____"
],
[
"Guiding Principles for EDA/ Visualizations\n1. Graphical Integrity\n2. Keep it simple\n3. Use the right display\n4. Use color strategically\n5. Tell a story with Data\n\nGuiding Principles for Effective Storytelling\n1. Audience (Know Your Audience)\n2. Engaging & Memorable\n3. Answer concise questions\n4. Carefully designed story (Beginning, Middle, End)\n5. Moves audience (call to action)\n\nGuiding Principles for Effective Presentations\n1. Clarity of Message (1 big idea)\n2. Clarity of Slides\n3. Clarity of Delivery\n\nIMAC\n(Inferential Goal,\nModel,\nAlgorithms, \nConclusion & Checking)\n\nAll models are wrong, some are useful.",
"_____no_output_____"
],
[
"Data Source -- https://www.kaggle.com/wendykan/lending-club-loan-data",
"_____no_output_____"
]
],
[
[
"%autosave 60",
"_____no_output_____"
],
[
"#import necessary modules\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\npd.set_option('max_columns', None)\n\nimport nltk\nimport collections as co\nfrom wordcloud import WordCloud, STOPWORDS\n\n%matplotlib inline",
"_____no_output_____"
],
[
"#read loans.csv as a dataframe\nloans_df = pd.read_csv('loan.csv',low_memory=False, engine='c')",
"_____no_output_____"
],
[
"loans_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 887379 entries, 0 to 887378\nData columns (total 75 columns):\nid 887379 non-null int64\nmember_id 887379 non-null int64\nloan_amnt 887379 non-null float64\nfunded_amnt 887379 non-null float64\nfunded_amnt_inv 887379 non-null float64\nterm 887379 non-null object\nint_rate 887379 non-null float64\ninstallment 887379 non-null float64\ngrade 887379 non-null object\nsub_grade 887379 non-null object\nemp_title 835922 non-null object\nemp_length 887379 non-null object\nhome_ownership 887379 non-null object\nannual_inc 887375 non-null float64\nverification_status 887379 non-null object\nissue_d 887379 non-null object\nloan_status 887379 non-null object\npymnt_plan 887379 non-null object\nurl 887379 non-null object\ndesc 126029 non-null object\npurpose 887379 non-null object\ntitle 887228 non-null object\nzip_code 887379 non-null object\naddr_state 887379 non-null object\ndti 887379 non-null float64\ndelinq_2yrs 887350 non-null float64\nearliest_cr_line 887350 non-null object\ninq_last_6mths 887350 non-null float64\nmths_since_last_delinq 433067 non-null float64\nmths_since_last_record 137053 non-null float64\nopen_acc 887350 non-null float64\npub_rec 887350 non-null float64\nrevol_bal 887379 non-null float64\nrevol_util 886877 non-null float64\ntotal_acc 887350 non-null float64\ninitial_list_status 887379 non-null object\nout_prncp 887379 non-null float64\nout_prncp_inv 887379 non-null float64\ntotal_pymnt 887379 non-null float64\ntotal_pymnt_inv 887379 non-null float64\ntotal_rec_prncp 887379 non-null float64\ntotal_rec_int 887379 non-null float64\ntotal_rec_late_fee 887379 non-null float64\nrecoveries 887379 non-null float64\ncollection_recovery_fee 887379 non-null float64\nlast_pymnt_d 869720 non-null object\nlast_pymnt_amnt 887379 non-null float64\nnext_pymnt_d 634408 non-null object\nlast_credit_pull_d 887326 non-null object\ncollections_12_mths_ex_med 887234 non-null float64\nmths_since_last_major_derog 221703 non-null float64\npolicy_code 887379 non-null float64\napplication_type 887379 non-null object\nannual_inc_joint 511 non-null float64\ndti_joint 509 non-null float64\nverification_status_joint 511 non-null object\nacc_now_delinq 887350 non-null float64\ntot_coll_amt 817103 non-null float64\ntot_cur_bal 817103 non-null float64\nopen_acc_6m 21372 non-null float64\nopen_il_6m 21372 non-null float64\nopen_il_12m 21372 non-null float64\nopen_il_24m 21372 non-null float64\nmths_since_rcnt_il 20810 non-null float64\ntotal_bal_il 21372 non-null float64\nil_util 18617 non-null float64\nopen_rv_12m 21372 non-null float64\nopen_rv_24m 21372 non-null float64\nmax_bal_bc 21372 non-null float64\nall_util 21372 non-null float64\ntotal_rev_hi_lim 817103 non-null float64\ninq_fi 21372 non-null float64\ntotal_cu_tl 21372 non-null float64\ninq_last_12m 21372 non-null float64\nloan_status_bin 887379 non-null object\ndtypes: float64(49), int64(2), object(24)\nmemory usage: 507.8+ MB\n"
],
[
"loans_df.head()",
"_____no_output_____"
],
[
"loans_df.describe()",
"_____no_output_____"
]
],
[
[
"## Q1. What is the the distribution of loans by loan amount?",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"ticks\")\n\nfig, axs = plt.subplots(2,1,figsize=(20,20))\n\nsns.distplot(loans_df.loan_amnt, ax=axs[0], hist=True, kde=True, bins=40)\naxs[0].set(xlabel='Loan Amount', \n ylabel='% Distribution',title='Density Plot of Loan Amount')\n\nsns.violinplot(loans_df.loan_amnt, ax=axs[1], color='0.6')\naxs[1].set(xlabel='Loan Amount', \n ylabel='Distribution',title='Violin Plot of Loan Amount')\n\nsns.despine()\n\nplt.show()",
"_____no_output_____"
],
[
"loans_df['loan_status'].unique()",
"_____no_output_____"
],
[
"#define a function to classify loan status into one of the following bins ('Fully Paid', 'Default', 'Current')\ndef loan_status_bin(text):\n if text in ('Fully Paid', 'Does not meet the credit policy. Status:Fully Paid'):\n return 'Fully Paid'\n elif text in ('Current', 'Issued'):\n return 'Current'\n elif text in ('Charged Off', 'Default', 'Does not meet the credit policy. Status:Charged Off'):\n return 'Default'\n elif text in ('Late (16-30 days)', 'Late (31-120 days)', 'In Grace Period'):\n return 'Late'\n else:\n 'UNKNOWN STATUS'",
"_____no_output_____"
],
[
"#create a new attribute 'loan_status_bin' in the dataframe\nloans_df['loan_status_bin']=loans_df['loan_status'].apply(loan_status_bin)\nloans_df['loan_status_bin'].unique()",
"_____no_output_____"
]
],
[
[
"## Q2. What is the distribution of loans by loan status represented as a pie plot, and a violin plot?",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"ticks\")\nfig, axs = plt.subplots(1,2,figsize=(18,8))\n\nloans_df.groupby('loan_status_bin').size().plot(kind='pie', ax=axs[0]);\naxs[0].set(title='Pie Plot of Loan Status bin')\n\nsns.violinplot(x=loans_df['term'], y=loans_df['loan_amnt'], hue=loans_df['loan_status_bin'], ax=axs[1])\naxs[1].set(xlabel='Loan Status bin', \n ylabel='Loan Amount',title='Violin Plot of Loan Term, Loan Status and Loan Amount')\naxs[1].legend(loc=4)\n\n\nsns.despine()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Q3. Why are people borrowing money?",
"_____no_output_____"
]
],
[
[
"plt.rcParams['figure.figsize'] = (16,16)\nlist_wc = list()\nloans_df['title'].apply(lambda x: list_wc.append(x))\nstring_wc=str(list_wc)\n\nwordcloud = WordCloud(stopwords=STOPWORDS, background_color='white', max_words=200, width=800, height=400).generate(string_wc)\n\nplt.imshow(wordcloud, interpolation='bilinear')\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Q4. Who is borrowing money?",
"_____no_output_____"
]
],
[
[
"int_rate, emp_length, home_ownership, addr_state, dti, term",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbac8dbe33b786900e9519fdc1b762a1fc7047e5
| 11,651 |
ipynb
|
Jupyter Notebook
|
CoreBx_island_v6.ipynb
|
csherwood-usgs/CoreBx
|
481a1252c95a3ee3c9f83c6969c154cb12946626
|
[
"CC0-1.0"
] | null | null | null |
CoreBx_island_v6.ipynb
|
csherwood-usgs/CoreBx
|
481a1252c95a3ee3c9f83c6969c154cb12946626
|
[
"CC0-1.0"
] | null | null | null |
CoreBx_island_v6.ipynb
|
csherwood-usgs/CoreBx
|
481a1252c95a3ee3c9f83c6969c154cb12946626
|
[
"CC0-1.0"
] | null | null | null | 36.071207 | 263 | 0.538409 |
[
[
[
"### CoreBx_island_v6 - Try to process entire N. Core Banks\n\nInterpolate the North Core Banks DEMs onto rotated 1-m grid and save each as a .nc file.\n\nVersioning jumped from v2 to v5, trying to be consistent with versions in processing notebooks.\n\nNew invV5\n* Files are switched to the \"merged DEMs\" that Jin-Si made, so the rapid iteration can occur.\n* Box is re-adjusted to accomodate the whole island. The resulting array is huge, but manageble.\n\nNew in v2\n* Now 4D maps, two made made during visit to Santa Cruz and two ftp'd from Andy\n* Apr. 9 - changed to _v3 for Sep map\n* Now does the interpolation without the loop\n* Apr. 21 - moved origin to SE to accomodate curvature in NE end of island. Add 400 m to size of array.\n* Watch file names, esp. underline (or not) after \"1m_DEM\"\n\nNew in v6\n* Added maps through Sep 28 2020\n\nTODO: The alongshore/cross-shore names are switched.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport xarray as xr\n# from dask.distributed import LocalCluster\nfrom scipy import interpolate, signal\n%matplotlib inline",
"_____no_output_____"
],
[
"# define all of the functions by runnng this python file\n%run -i CoreBx_funcs",
"_____no_output_____"
],
[
"def make_grid(name=None,e0=None,n0=None,xlen=None,ylen=None,dxdy=None,theta=None):\n nx = int((1./dxdy)*xlen)\n ny = int((1./dxdy)*ylen)\n\n xcoords = np.linspace(0.5*dxdy,xlen-0.5*dxdy,nx)\n ycoords = np.linspace(0.5*dxdy,ylen-0.5*dxdy,ny)\n\n # these will be the coordinates in rotated space\n xrot, yrot = np.meshgrid(xcoords, ycoords ,sparse=False, indexing='xy')\n\n print('Shape of xrot, yrot: ',np.shape(xrot),np.shape(yrot))\n shp = np.shape(xrot)\n xu, yu = box2UTMh(xrot.flatten(), yrot.flatten(), e0, n0, theta)\n xu=np.reshape(xu,shp)\n yu=np.reshape(yu,shp)\n # write the UTM coords of the corners to an ASCII file\n corners = np.asarray( [[xu[0][0],yu[0][0]],\\\n [xu[0][-1],yu[0][-1]],\\\n [xu[-1][-1],yu[-1][-1]],\\\n [xu[-1][0],yu[-1][0]],\\\n [xu[0][0],yu[0][0]]])\n\n print('corners x, corners y]n',corners[0,:],corners[1,:])\n print(corners)\n fn = name+'.csv'\n np.savetxt(fn, corners, delimiter=\",\")\n return xu, yu, xrot, yrot, xcoords, ycoords",
"_____no_output_____"
],
[
"# April 9, 2020: Replaced \"2019-09-12-13_1m_DEM_4D_crop.tif\",\\\n# with _v3 and re-ran on my desktop\n\n# May 4 - Changed to use Jin-Si's merged dems\n\nfdir = \"C:/crs/proj/2019_DorianOBX/Santa_Cruz_Products/merged_dems/\"\n#fdir = \"D:/crs/proj/2019_DorianOBX/Santa_Cruz_Products/clipped_dems/\"\n\n\nfnames = (\\\n \"C:/crs/proj/2019_DorianOBX/Santa_Cruz_Products/merged_dems/2019-08-30_1m_DEM_4D_crop2_m.tif\",\\\n \"C:/crs/proj/2019_DorianOBX/Santa_Cruz_Products/merged_dems/2019-09-12-13_1m_DEM_4D_v3_m.tif\",\\\n \"C:/crs/proj/2019_DorianOBX/SfM_OBX_results/dems/NCB_20191011_DEM_1m_lidarMerge_NAD83_2011_UTM18N_NAVD88_crs.tif\",\\\n \"C:/crs/proj/2019_DorianOBX/Santa_Cruz_Products/merged_dems/2019-11-26_1m_DEM_4D_crop_m.tif\",\\\n \"C:/crs/proj/2019_DorianOBX/SfM_OBX_results/dems/NCB_20200208-09_DEM_1m_4D_NAD83_2011_UTM18N_NAVD88_crs.tif\",\\\n# \"C:/crs/proj/2019_DorianOBX/SfM_OBX_results/dems/NCB_20200508-09_DEM_1m_4D_NAD83_2011_UTM18N_NAVD88_crs.tif\",\\\n# \"C:/crs/proj/2019_DorianOBX/SfM_OBX_results/dems/NCB_20200802_DEM_1m_4D_NAD83_2011_UTM18N_NAVD88_cog.tif\",\\\n# \"C:/crs/proj/2019_DorianOBX/SfM_OBX_results/dems/NCB_20200805-09_DEM_1m_4D_NAD83_2011_UTM18N_NAVD88_cog.tif\",\\\n# \"C:/crs/proj/2019_DorianOBX/SfM_OBX_results/dems/NCB_20200928_DEM_1m_4D_NAD83_2011_UTM18N_NAVD88_cog.tif\")\n\ntitles = ([\\\n \"Aug 30 2019 pre-Dorian\",\\\n \"Sep 12-13 2019 post-Dorian\",\\\n \"Oct 11 2019 lidar merge\",\\\n \"Nov 26 2019 post-Nor'easter\"])\n# \"Feb 8-9 2020\",\\\n# \"May 8-9 2020\",\\\n# \"Aug 2 2020 pre-Isaias\",\\\n# \"Aug 5-9 2020 post-Isaias\",\\\n# \"Sep 28 2020 post-Teddy\"])\nnf = len(fnames)\n\nfill_fnames = ('EBK_201909_YesLidar_Comb_Extent_m.tif')\nfill_titles = ('Sep_fill')\n\n# optional median-filter smoothing of original maps\nsmooth = False\n# kernal size...this should be an odd number >= dxy/0.1\nksize = 3",
"_____no_output_____"
],
[
"# Make an array of dicts, where analysis region is defined by:\n# name\n# e0 - UTM Easting of origin [m]\n# n0 - UTM Northing of origin [m]\n# xlen - Length of alongshore axis [m]\n# ylen - Length of cross-shore axis [m]\n# dxdy - grid size (must be isotropic right now) [m]\n# theta - rotation CCW from x-axis [deg]\n\nr = {'name':\"ncorebx\",\"e0\": 378500.,\"n0\": 3856350.,\"xlen\": 36000.,\"ylen\": 1100.,\"dxdy\": 1.,\"theta\": 42.}\n\n# move the origin 400 m SE\nxo,yo = xycoord(400.,42.+90)\nprint(xo,yo)\nr['e0']=r['e0']+xo\nr['n0']=r['n0']+yo\n\n# add 400 m to ylen\nr['ylen']=r['ylen']+400.\n\n# that was called ncorebx_v4\n# move that origin 460 m sw\nxo,yo = xycoord(460., 42.+180.)\nr['e0']=r['e0']+xo\nr['n0']=r['n0']+yo\n# add 650 m to ylen\nr['xlen']=r['xlen']+650.\nr['name']='ncorebx_v6'\n\nprint(r)\n\nprint(r['name'])\nxu,yu,xrot,yrot,xcoords,ycoords = make_grid(**r)\nny,nx = np.shape(xu)\nprint(ny,nx)",
"_____no_output_____"
],
[
"%%time\ndslist=[]\nfor i, fn in enumerate(fnames):\n iswarned = False\n\n print(i, fn)\n\n # open the tif with XArray as a DataArray\n da = xr.open_rasterio(fn)\n\n print( np.shape(np.flipud(da['y'].values)), np.shape(da['x'].values), np.shape( np.flipud(da.values)) )\n x = da['x'].values\n y = np.flipud(da['y'].values)\n\n # Not sure how da.values got a singleton dimension, but squeeze gets rid of it.\n # However, make sure to squeeze before flipping\n z = np.flipud(np.squeeze(da.values))\n print(np.shape(x),np.shape(y),np.shape(z))\n\n if(smooth):\n # smooth with 2D running median\n zs = signal.medfilt2d(z, kernel_size=ksize)\n else:\n zs = z\n\n f = interpolate.RegularGridInterpolator( (y, x), zs, method='linear') \n\n # Array for interpolated elevations\n zi=np.NaN*np.ones((ny,nx))\n \n # this is the fast iteration, which only works when all of the source points fall inside the target box\n try:\n zi=f((yu,xu))\n\n # this is a slow iteration through all of the points, but allows us to skip ones that are outside\n except:\n if(not iswarned):\n print(\"Warning: using slow iteration.\")\n iswarned = True\n for ij in np.ndindex(zi.shape):\n try:\n zi[ij]=f((yu[ij],xu[ij]))\n except:\n zi[ij]=np.NaN\n\n da = xr.DataArray(zi,dims=['Alongshore','Cross-shore'],coords={'Alongshore': ycoords, 'Cross-shore':xcoords })\n da = da.chunk()\n dslist.append(da)\n\ndsa = xr.concat(dslist, dim='map')\nfn = r['name']+'.nc'\ndsa.to_netcdf(fn)",
"_____no_output_____"
],
[
"%%time\n# Read in the fill map and make netcdf files\nfn = fdir+fill_fnames\nprint(fn)\n\n# open the tif with XArray as a DataArray\ndaf = xr.open_rasterio(fn)\n\nprint( np.shape(np.flipud(daf['y'].values)), np.shape(daf['x'].values), np.shape( np.flipud(daf.values)) )\nx = daf['x'].values\ny = np.flipud(daf['y'].values)\n\n# Not sure how da.values got a singleton dimension, but squeeze gets rid of it.\n# However, make sure to squeeze before flipping\nz = np.flipud(np.squeeze(daf.values))\nprint(np.shape(x),np.shape(y),np.shape(z))\n\nf = interpolate.RegularGridInterpolator( (y, x), z, method='linear') \n\n# Array for interpolated elevations\nzi=np.NaN*np.ones((ny,nx))\n\n# this is a slow iteration through all of the points, but allows us to skip ones that are outside\n# for ij in np.ndindex(zi.shape):\n# try:\n# zi[ij]=f((yu[ij],xu[ij]))\n# except:\n# zi[ij]=np.NaN\n\n# this is the fast technique.\nzi=f((yu,xu))\n\nda = xr.DataArray(zi,dims=['Alongshore','Cross-shore'],coords={'Alongshore': ycoords, 'Cross-shore':xcoords })\nda = da.chunk()\n\nfno = r['name']+'_Sep_fill.nc'\nda.to_netcdf(fno)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbaca79a4500a5b474164cf13f47f362a5df05c5
| 13,143 |
ipynb
|
Jupyter Notebook
|
9. AMLD2019 - Document Digitization/code/processFeatures.ipynb
|
lordzsolt/Data-Science
|
02241b541108f3b8e6031f260ddde0dd733988d0
|
[
"MIT"
] | null | null | null |
9. AMLD2019 - Document Digitization/code/processFeatures.ipynb
|
lordzsolt/Data-Science
|
02241b541108f3b8e6031f260ddde0dd733988d0
|
[
"MIT"
] | null | null | null |
9. AMLD2019 - Document Digitization/code/processFeatures.ipynb
|
lordzsolt/Data-Science
|
02241b541108f3b8e6031f260ddde0dd733988d0
|
[
"MIT"
] | null | null | null | 32.134474 | 140 | 0.53169 |
[
[
[
"# Notebook setup",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk import sent_tokenize, word_tokenize\nimport os\nimport string\nimport re\nfrom nltk.sentiment.vader import SentimentIntensityAnalyzer\n\nnltk.download('punkt')\nnltk.download('vader_lexicon');",
"[nltk_data] Downloading package punkt to /Users/zkovacs/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n[nltk_data] Downloading package vader_lexicon to\n[nltk_data] /Users/zkovacs/nltk_data...\n[nltk_data] Package vader_lexicon is already up-to-date!\n"
],
[
"# Disable output to reduce execution time.\noutput = False",
"_____no_output_____"
],
[
"outPath = \"../training_set/ocr_output/\"\n\nfor (dirpath, dirnames, filenames) in os.walk(outPath):\n break\n\nif '.DS_Store' in filenames :\n filenames.remove('.DS_Store')",
"_____no_output_____"
]
],
[
[
"# Features",
"_____no_output_____"
],
[
"## Useful functions",
"_____no_output_____"
]
],
[
[
"def readFile(filename):\n f = open(outPath+filename, 'r', encoding=\"cp1252\") #for MAC ?\n rawText = f.read()\n text = rawText.replace(\"\\n\\n\", \"%EOL%\").replace(\"\\n\",\" \").replace(\"%EOL%\",\"\\n\")\n return text\n\n\ndef removePunctuation(text):\n return text.translate(str.maketrans('', '', string.punctuation))\n\n\ndef findWithKeywords(text, anyKeywords=[], allKeywords=[], excludedKeywords=[]):\n text = text.replace(\"\\n\\n\", \"%EOL%\").replace(\"\\n\",\" \").replace(\"%EOL%\",\"\\n\")\n sentences = sent_tokenize(text)\n matched = []\n for sentence in sentences:\n if len(anyKeywords) > 0 and not any(keyword in sentence.lower() for keyword in anyKeywords):\n continue\n if len(allKeywords) and not all(keyword in sentence.lower() for keyword in allKeywords):\n continue\n if not any(keyword in sentence.lower() for keyword in excludedKeywords):\n matched.append(sentence)\n\n return \"\\n\\n\".join(matched)\n\n\ndef findWithKeywordsSentenceWindow(text, anyKeywords=[], allKeywords=[], excludedKeywords=[], windowSize=1):\n text = text.replace(\"\\n\\n\", \"%EOL%\").replace(\"\\n\",\" \").replace(\"%EOL%\",\"\\n\")\n sentences = sent_tokenize(text)\n matched = []\n \n for index in range(0, len(sentences) - windowSize):\n sentence = sentences[index] + '\\n\\n' + sentences[index + 1]\n if len(anyKeywords) > 0 and not any(keyword in sentence.lower() for keyword in anyKeywords):\n continue\n if len(allKeywords) and not all(keyword in sentence.lower() for keyword in allKeywords):\n continue\n if not any(keyword in sentence.lower() for keyword in excludedKeywords):\n matched.append(sentence)\n\n return \"\\n\\n\".join(matched)\n\n\ndef findSentencesWithAnyKeywords(text, keywords, excludedKeywords=[]):\n return findWithKeywords(text, anyKeywords=keywords, excludedKeywords=excludedKeywords)\n\n\ndef findSentencesWithAllKeywords(text, keywords, excludedKeywords=[]):\n return findWithKeywords(text, allKeywords=keywords, excludedKeywords=excludedKeywords)\n\ndef findDirectorNumberText(text):\n return findSentencesWithAllKeywords(text,[\"number of directors\"], [\"chair\", \"vacancy\", \"vacancies\", \"quorum\"])\n\ndef findFirstNumberAfterWord(text, paramWord=\"\"):\n numWords = [\n \"zero\", \"one\", \"two\", \"three\", \"four\", \"five\", \"six\", \"seven\", \"eight\",\n \"nine\", \"ten\", \"eleven\", \"twelve\", \"thirteen\", \"fourteen\", \"fifteen\",\n \"sixteen\", \"seventeen\", \"eighteen\", \"nineteen\", \"twenty\"]\n listWords = word_tokenize(text)\n for word in listWords[listWords.index(paramWord):]:\n word = removePunctuation(word)\n if word in numWords:\n return numWords.index(word)\n if word.isdigit():\n return word\n return \"\"",
"_____no_output_____"
]
],
[
[
"## Is the company empowered to borrow?",
"_____no_output_____"
]
],
[
[
"def findCanBorrowText(text):\n return (\n findSentencesWithAnyKeywords(text, [\"any business\", \"issue debt\", \"indebtedness\"])\n + \" \"\n + findWithKeywords(text, anyKeywords=[\"borrow\", \"raise\"], allKeywords=[\"money\"])\n )\n\n\ndef canBorrow(text):\n canBorrowText = findCanBorrowText(text)\n if canBorrowText.strip() == \"\":\n return \"no\"\n return getSentiment(canBorrowText)\n\n\ndef getSentiment(text):\n if text.strip() == \"\":\n return \"\"\n sentimentAnalyzer = SentimentIntensityAnalyzer()\n scores = sentimentAnalyzer.polarity_scores(text)\n aggregated_score = scores[\"compound\"]\n return \"yes\" if aggregated_score > 0 else \"no\"\n\n\nfor filename in filenames:\n text = readFile(filename)\n if output:\n print(filename)\n print(canBorrow(text))\n print(\"\\n\")",
"_____no_output_____"
]
],
[
[
"## What is the size of the board of directors? Minimum and maximum.",
"_____no_output_____"
]
],
[
[
"def findMinDirectors(fullText):\n directorText = findDirectorNumberText(fullText)\n if \"no minimum\" in directorText:\n return \"noMin\" \n if \"minimum\" in directorText:\n return findFirstNumberAfterWord(directorText, \"minimum\")\n if \"less\" in directorText: # for cases of \"not less than\" and \"shall not be less than\"\n return findFirstNumberAfterWord(directorText, \"less\")\n return \"1\"\n\ndef findMaxDirectors(fullText):\n directorText = findDirectorNumberText(fullText)\n if \"no maximum\" in directorText:\n return \"noMax\" \n if \"maximum\" in directorText:\n return findFirstNumberAfterWord(directorText, \"maximum\")\n if \"more\" in directorText: # for cases of \"not more than\" and \"shall not be more than\"\n return findFirstNumberAfterWord(directorText, \"more\")\n return \"noMax\" # TODO: Use noMax if ran out of ideas\n\nfor filename in filenames:\n text = readFile(filename)\n if output:\n print(filename)\n print(findDirectorNumberText(text))\n print(findMinDirectors(text))\n print(findMaxDirectors(text))\n print(\"\\n\")",
"_____no_output_____"
]
],
[
[
"## Are the directors empowered to borrow?",
"_____no_output_____"
]
],
[
[
"def findDirectorsCanBorrowText(text):\n return (\n findWithKeywords(text, anyKeywords=[\"borrow\", \"debt\", \"incur\", \"indebtedness\"], allKeywords=[\"directors may\"])\n + \" \" \n + findWithKeywords(text, anyKeywords=[\"borrow\", \"debt\", \"incur\", \"indebtedness\"], allKeywords=[\"directors can\"])\n )\n\ndef findBoardCanBorrowText(text):\n return findWithKeywords(text, anyKeywords=[\"borrow\", \"debt\", \"incur\", \"indebtedness\"], allKeywords=[\"the board may\"])\n\ndef canDirectorsBorrow(text):\n directorsText = findDirectorsCanBorrowText(text)\n if directorsText.strip() != \"\":\n return getSentiment(directorsText)\n boardText = findBoardCanBorrowText(text)\n if boardText.strip() != \"\":\n return \"no\"\n return \"yes\"",
"_____no_output_____"
]
],
[
[
"## Is a resolution of directors required to borrow?",
"_____no_output_____"
]
],
[
[
"def resolutionNeeded(text):\n directorsText = findDirectorsCanBorrowText(text);\n if canDirectorsBorrow(directorsText):\n if \"resolution\" in directorsText.lower(): \n return \"yes\"\n else:\n return \"no\"\n else:\n return \"no\"\n\nfor filename in filenames:\n text = readFile(filename)\n if output:\n print(filename)\n print(findDirectorsCanBorrowText(text))\n print(canDirectorsBorrow(text))\n print(resolutionNeeded(text))\n print(\"\\n\")",
"_____no_output_____"
]
],
[
[
"## What is the quorum for such a resolution?",
"_____no_output_____"
]
],
[
[
"def findQuorumText(text, keywords=[\"quorum\", \"number\"]):\n return findWithKeywordsSentenceWindow(text, allKeywords=keywords, anyKeywords=[\"directors\", \"shareholders\"], windowSize=2)\n\ndef findQuorum(fullText):\n quorumText = findQuorumText(fullText)\n if quorumText.strip() == \"\":\n quorumText = findQuorumText(text, keywords=[\"quorum\", \"meeting\"])\n match = re.search(r'not less than (.*?) of the', quorumText)\n if match:\n matched = match.group(1)\n return matched.translate(str.maketrans('-—',' '))\n else:\n return \"2\"\n \nfor filename in filenames:\n text = readFile(filename)\n if output:\n print(filename)\n print(findQuorumText(text))\n print(\"quorum : \" + findQuorum(text))\n print(\"\\n\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbacb178bf8253a433e0c9253e1ab0e5c38e87bd
| 103,643 |
ipynb
|
Jupyter Notebook
|
demos/Generate_ROI_labeled_arrays.ipynb
|
Nikea/Demos
|
a528c19509e34de6dae507dc404e715dd4c7a052
|
[
"BSD-3-Clause"
] | 13 |
2016-03-04T09:33:10.000Z
|
2020-12-15T17:14:26.000Z
|
demos/Generate_ROI_labeled_arrays.ipynb
|
scikit-beam/scikit-beam-examples
|
a528c19509e34de6dae507dc404e715dd4c7a052
|
[
"BSD-3-Clause"
] | 32 |
2015-12-11T19:55:41.000Z
|
2019-06-25T13:20:39.000Z
|
demos/Generate_ROI_labeled_arrays.ipynb
|
Nikea/Demos
|
a528c19509e34de6dae507dc404e715dd4c7a052
|
[
"BSD-3-Clause"
] | 13 |
2015-12-17T20:11:02.000Z
|
2021-07-13T16:18:40.000Z
| 137.640106 | 18,226 | 0.88088 |
[
[
[
"# Generate Region of Interests (ROI) labeled arrays for simple shapes\nThis example notebook explain the use of analysis module \"skbeam/core/roi\" https://github.com/scikit-beam/scikit-beam/blob/master/skbeam/core/roi.py ",
"_____no_output_____"
]
],
[
[
"import skbeam.core.roi as roi\nimport skbeam.core.correlation as corr\n\nimport numpy as np\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom matplotlib.ticker import MaxNLocator\nfrom matplotlib.colors import LogNorm\n\nimport xray_vision.mpl_plotting as mpl_plot",
"_____no_output_____"
]
],
[
[
"### Easily switch between interactive and static matplotlib plots",
"_____no_output_____"
]
],
[
[
"interactive_mode = False\n\nimport matplotlib as mpl\nif interactive_mode:\n %matplotlib notebook\nelse:\n %matplotlib inline\n\nbackend = mpl.get_backend()\ncmap='viridis'",
"_____no_output_____"
]
],
[
[
"## Draw annular (ring-shaped) regions of interest",
"_____no_output_____"
]
],
[
[
"center = (100., 100.) # center of the rings\n\n# Image shape which is used to determine the maximum extent of output pixel coordinates\nimg_shape = (200, 205) \n\nfirst_q = 10.0 # inner radius of the inner-most ring\ndelta_q = 5.0 #ring thickness\n\nnum_rings = 7 # number of Q rings\n\n# step or spacing, spacing between rings \none_step_q = 5.0 # one spacing between rings\n\nstep_q = [2.5, 3.0, 5.8] # differnt spacing between rings",
"_____no_output_____"
]
],
[
[
"### Test when there is same spacing between rings",
"_____no_output_____"
]
],
[
[
"# inner and outer radius for each ring\nedges = roi.ring_edges(first_q, width=delta_q, spacing=one_step_q,\n num_rings=num_rings)\nedges",
"_____no_output_____"
],
[
"#Elements not inside any ROI are zero; elements inside each\n#ROI are 1, 2, 3, corresponding to the order they are specified in edges.\nlabel_array = roi.rings(edges, center, img_shape)\n\n# plot the figure\nfig, axes = plt.subplots(figsize=(6, 5))\naxes.set_title(\"Same spacing between rings\")\nim = mpl_plot.show_label_array(axes, label_array, cmap)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Test when there is different spacing between rings",
"_____no_output_____"
]
],
[
[
"# inner and outer radius for each ring\n\nedges = roi.ring_edges(first_q, width=delta_q, spacing=step_q,\n num_rings=4)\nprint(\"edges when there is different spacing between rings\", edges)",
"edges when there is different spacing between rings [[ 10. 15. ]\n [ 17.5 22.5]\n [ 25.5 30.5]\n [ 36.3 41.3]]\n"
],
[
"#Elements not inside any ROI are zero; elements inside each\n#ROI are 1, 2, 3, corresponding to the order they are specified in edges.\nlabel_array = roi.rings(edges, center, img_shape)\n\n# plot the figure\nfig, axes = plt.subplots(figsize=(6, 5))\naxes.set_title(\"Different spacing between rings\")\naxes.set_xlim(50, 150)\naxes.set_ylim(50, 150)\nim = mpl_plot.show_label_array(axes, label_array, cmap)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"### Test when there is no spacing between rings",
"_____no_output_____"
]
],
[
[
"# inner and outer radius for each ring\nedges = roi.ring_edges(first_q, width=delta_q, num_rings=num_rings)\nedges",
"_____no_output_____"
],
[
"#Elements not inside any ROI are zero; elements inside each\n#ROI are 1, 2, 3, corresponding to the order they are specified in edges.\nlabel_array = roi.rings(edges, center, img_shape)\n\n# plot the figure\nfig, axes = plt.subplots(figsize=(6, 5))\naxes.set_title(\"There is no spacing between rings\")\naxes.set_xlim(50, 150)\naxes.set_ylim(50, 150)\nim = mpl_plot.show_label_array(axes, label_array, cmap)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Generate a ROI of Segmented Rings¶",
"_____no_output_____"
]
],
[
[
"center = (75, 75) # center of the rings\n\n#Image shape which is used to determine the maximum extent of output pixel coordinates\nimg_shape = (150, 140) \n\nfirst_q = 5.0 # inner radius of the inner-most ring\ndelta_q = 5.0 #ring thickness\nnum_rings = 4 # number of rings\n\nslicing = 4 # number of pie slices or list of angles in radians\nspacing = 4 # margin between rings, 0 by default",
"_____no_output_____"
]
],
[
[
"\n\n#### find the inner and outer radius of each ring\n",
"_____no_output_____"
]
],
[
[
"# inner and outer radius for each ring\nedges = roi.ring_edges(first_q, width=delta_q, spacing=spacing,\n num_rings=num_rings)\n\nedges",
"_____no_output_____"
],
[
"#Elements not inside any ROI are zero; elements inside each\n#ROI are 1, 2, 3, corresponding to the order they are specified in edges.\nlabel_array = roi.segmented_rings(edges, slicing, center,\n img_shape, offset_angle=0)\n\n# plot the figure\nfig, axes = plt.subplots(figsize=(6, 5))\naxes.set_title(\"Segmented Rings\")\naxes.set_xlim(38, 120)\naxes.set_ylim(38, 120)\nim = mpl_plot.show_label_array(axes, label_array, cmap)\nplt.show()\n",
"_____no_output_____"
]
],
[
[
"## Segmented rings using list of angles in radians",
"_____no_output_____"
]
],
[
[
"slicing = np.radians([0, 60, 120, 240, 300])\nslicing",
"_____no_output_____"
],
[
"#Elements not inside any ROI are zero; elements inside each\n#ROI are 1, 2, 3, corresponding to the order they are specified in edges.\nlabel_array = roi.segmented_rings(edges, slicing, center,\n img_shape, offset_angle=0)\n\n# plot the figure\nfig, axes = plt.subplots(figsize=(6, 5))\naxes.set_title(\"Segmented Rings\")\naxes.set_xlim(38, 120)\naxes.set_ylim(38, 120)\nim = mpl_plot.show_label_array(axes, label_array, cmap=\"gray\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Generate a ROI of Pies",
"_____no_output_____"
]
],
[
[
"first_q = 0 \n\n# inner and outer radius for each ring\nedges = roi.ring_edges(first_q, width=50, num_rings=1)\n\nedges",
"_____no_output_____"
],
[
"slicing = 10 # number of pie slices or list of angles in radians\n\n#Elements not inside any ROI are zero; elements inside each\n#ROI are 1, 2, 3, corresponding to the order they are specified in edges.\nlabel_array = roi.segmented_rings(edges, slicing, center,\n img_shape, offset_angle=0)\n\n# plot the figure\nfig, axes = plt.subplots(figsize=(6, 5))\naxes.set_title(\"Pies\")\naxes.set_xlim(20, 140)\naxes.set_ylim(20, 140)\nim = mpl_plot.show_label_array(axes, label_array, cmap)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Rectangle region of interests.",
"_____no_output_____"
]
],
[
[
"# Image shape which is used to determine the maximum extent of output pixel coordinates\nshape = (15, 26)\n\n# coordinates of the upper-left corner and width and height of each rectangle\nroi_data = np.array(([2, 2, 6, 3], [6, 7, 8, 5], [8, 18, 5, 10]),\n dtype=np.int64)\n\n#Elements not inside any ROI are zero; elements inside each ROI are 1, 2, 3, corresponding\n# to the order they are specified in coords.\nlabel_array = roi.rectangles(roi_data, shape)\nroi_inds, pixel_list = roi.extract_label_indices(label_array)",
"_____no_output_____"
]
],
[
[
"## Generate Bar ROI's",
"_____no_output_____"
]
],
[
[
"edges = [[3, 4], [5, 7], [12, 15]]\nedges",
"_____no_output_____"
]
],
[
[
"## Create Horizontal bars and Vertical bars",
"_____no_output_____"
]
],
[
[
"h_label_array = roi.bar(edges, (20, 25)) # Horizontal Bars",
"_____no_output_____"
],
[
"v_label_array = roi.bar(edges, (20, 25), horizontal=False) # Vertical Bars",
"_____no_output_____"
]
],
[
[
"## Create Box ROI's",
"_____no_output_____"
]
],
[
[
"b_label_array = roi.box((20, 25), edges)",
"_____no_output_____"
]
],
[
[
"## Plot bar rois, box rois and rectangle rois",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(2, 2, figsize=(12, 10))\naxes[1, 0].set_title(\"Horizontal Bars\")\nim = mpl_plot.show_label_array(axes[1, 0], h_label_array, cmap)\naxes[0, 1].set_title(\"Vertical Bars\")\nim = mpl_plot.show_label_array(axes[0, 1], v_label_array, cmap)\naxes[1, 1].set_title(\"Box Rois\")\nim = mpl_plot.show_label_array(axes[1, 1], b_label_array, cmap)\naxes[0, 0].set_title(\"Rectangle Rois\")\nim = mpl_plot.show_label_array(axes[0, 0], label_array, cmap)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Create line ROI's",
"_____no_output_____"
]
],
[
[
"label_lines= roi.lines(([0, 45, 50, 256], [56, 60, 80, 150]), (150, 250))\n# plot the figure\nfig, axes = plt.subplots(figsize=(6, 5))\naxes.set_title(\"Lines\")\nim = mpl_plot.show_label_array(axes, label_lines, cmap)\nplt.show()",
"_____no_output_____"
],
[
"import skbeam\nprint(skbeam.__version__)",
"0.0.8+42.g679637f\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbacb62368ec8c0ebf28ac8eed0be3bf29279c99
| 28,032 |
ipynb
|
Jupyter Notebook
|
notebooks/data_collection.ipynb
|
arijitmondal-94/app-review-sentiment-anslysis-using-bert
|
78edceb6c2077348d6b3f2904477d6cad00c1671
|
[
"MIT"
] | null | null | null |
notebooks/data_collection.ipynb
|
arijitmondal-94/app-review-sentiment-anslysis-using-bert
|
78edceb6c2077348d6b3f2904477d6cad00c1671
|
[
"MIT"
] | null | null | null |
notebooks/data_collection.ipynb
|
arijitmondal-94/app-review-sentiment-anslysis-using-bert
|
78edceb6c2077348d6b3f2904477d6cad00c1671
|
[
"MIT"
] | null | null | null | 106.992366 | 17,057 | 0.657855 |
[
[
[
"import json\nimport pandas as pd\n\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n\nfrom pygments import highlight\nfrom pygments.lexers import JsonLexer\nfrom pygments.formatters import TerminalFormatter \nfrom tqdm import tqdm\n\nfrom google_play_scraper import Sort, reviews, app\n\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\nsns.set(style='whitegrid', palette='muted', font_scale=1.2)",
"_____no_output_____"
]
],
[
[
"## Collecting app data",
"_____no_output_____"
]
],
[
[
"app_packages = [\n 'com.anydo',\n 'com.todoist',\n 'com.ticktick.task',\n 'com.habitrpg.android.habitica',\n 'cc.forestapp',\n 'com.oristats.habitbull',\n 'com.levor.liferpgtasks',\n 'com.habitnow',\n 'com.microsoft.todos',\n 'prox.lab.calclock',\n 'com.artfulagenda.app',\n 'com.tasks.android',\n 'com.appgenix.bizcal',\n 'com.appxy.planner',\n 'com.android.chrome'\n]",
"_____no_output_____"
],
[
"app_infos = []\n\nfor ap in tqdm(app_packages):\n info = app(ap, lang='en', country='us')\n del info['comments']\n app_infos.append(info)",
"100%|██████████| 14/14 [00:04<00:00, 3.14it/s]\n"
],
[
"def print_json(json_object):\n json_str = json.dumps(\n json_object,\n indent=2,\n sort_keys=True,\n default=str\n )\n print(highlight(json_str, JsonLexer(), TerminalFormatter()))\nprint_json(app_infos[0])",
"{\n \u001b[94m\"adSupported\"\u001b[39;49;00m: \u001b[34mnull\u001b[39;49;00m,\n \u001b[94m\"androidVersion\"\u001b[39;49;00m: \u001b[33m\"Varies\"\u001b[39;49;00m,\n \u001b[94m\"androidVersionText\"\u001b[39;49;00m: \u001b[33m\"Varies with device\"\u001b[39;49;00m,\n \u001b[94m\"appId\"\u001b[39;49;00m: \u001b[33m\"com.anydo\"\u001b[39;49;00m,\n \u001b[94m\"containsAds\"\u001b[39;49;00m: \u001b[34mfalse\u001b[39;49;00m,\n \u001b[94m\"contentRating\"\u001b[39;49;00m: \u001b[33m\"Everyone\"\u001b[39;49;00m,\n \u001b[94m\"contentRatingDescription\"\u001b[39;49;00m: \u001b[34mnull\u001b[39;49;00m,\n \u001b[94m\"currency\"\u001b[39;49;00m: \u001b[33m\"USD\"\u001b[39;49;00m,\n \u001b[94m\"description\"\u001b[39;49;00m: \u001b[33m\"\\ud83e\\udd47 <b>\\\"#1 to do list app out there\\u201d</b> - WSJ\\r\\n\\ud83c\\udfc6 <b>Editor's Choice</b> by Google\\r\\n\\r\\nOver 30M people rely on Any.do to stay organized and get more done.\\r\\nIt's a simple to do list app with reminders, planner & calendar - all in one.\\r\\n\\r\\n<b>\\ud83e\\udd47 \\\"A MUST HAVE APP\\\" (Lifehacker, NYTimes, USA TODAY).</b>\\r\\n\\r\\nAny.do is a free to-do list, planner & calendar app for managing and organizing your daily tasks, to-do lists, notes, reminders, checklists, calendar events, grocery lists and more.\\r\\n\\r\\n\\ud83d\\udcc5 Organize your tasks & to-do list\\r\\n\\r\\n\\u2022 ADVANCED CALENDAR & DAILY PLANNER - Keep your to-do list and calendar events always at hand with our calendar widget. Any.do to-do list & planner support daily calendar view, 3-day Calendar view, Weekly calendar view & agenda view, with built-in reminders. Review and organize your calendar events and to do list side by side.\\r\\n\\r\\n\\u2022 SYNCS SEAMLESSLY - Keeps all your to do list, tasks, reminders, notes, calendar & agenda always in sync so you\\u2019ll never forget a thing. Sync your phone\\u2019s calendar, google calendar, Facebook events, outlook calendar or any other calendar so you don\\u2019t forget an important event.\\r\\n\\r\\n\\u2022 SET REMINDERS - One time reminders, recurring reminders, Location reminders & voice reminders. NEW! Easily create tasks and get reminders in WhatsApp.\\r\\n\\r\\n\\u2022 WORK TOGETHER - Share your to do list and assign tasks with your friends, family & colleagues from your task list to collaborate and get more done. \\r\\n\\r\\n---\\r\\n\\r\\nALL-IN-ONE PLANNER & CALENDAR APP FOR GETTING THINGS DONE\\r\\nCreate and set reminders with voice to your to do list. \\r\\nFor better task management flow we added a calendar integration to keep your agenda always up to date. \\r\\nFor better productivity, we added recurring reminders, location reminders, one-time reminder, sub-tasks, notes & file attachments. \\r\\nTo keep your to do list up to date, we\\u2019ve added a daily planner and focus mode.\\r\\n\\r\\nINTEGRATIONS\\r\\nAny.do To do list, Calendar, planner & Reminders Integrates with Google Calendar, Outlook, WhatsApp, Slack, Gmail, Google Tasks, Evernote, Trello, Wunderlist, Todoist, Zapier, Asana, Microsoft to-do, Salesforce, OneNote, Google Assistant, Amazon Alexa, Office 365, Exchange, Jira & More.\\r\\n\\r\\nTO DO LIST, CALENDAR, PLANNER & REMINDERS MADE SIMPLE\\r\\nDesigned to keep you on top of your to do list, tasks and calendar events with no hassle. With intuitive drag and drop of tasks, swiping to mark to-do's as complete, and shaking your device to remove completed from your to do list - you can stay organized and enjoy every minute of it.\\r\\n\\r\\nPOWERFUL TO DO LIST TASK MANAGEMENT\\r\\nAdd a to do list item straight from your email / Gmail / Outlook inbox by forwarding [email protected]. Attach files from your computer, Dropbox, or Google Drive to your to- tasks.\\r\\n\\r\\nDAILY PLANNER & LIFE ORGANIZER\\r\\nAny.do is a to do list, a calendar, an inbox, a notepad, a checklist, task list, a board for post its or sticky notes, a task & project management tool, a reminder app, a daily planner, a family organizer, an agenda, a bill planner and overall the simplest productivity tool you will ever have. \\r\\n\\r\\nSHARE LISTS, ASSIGN & ORGANIZE TASKS\\r\\nTo plan & organize projects has never been easier. Now you can share lists between family members, assign tasks to each other, chat and much more. Any.do will help you and the people around you stay in-sync and get reminders so that you can focus on what matters, knowing you had a productive day and crossed off your to do list.\\r\\n\\r\\nGROCERY LIST & SHOPPING LIST\\r\\nAny.do task list, calendar, agenda, reminders & planner is also great for shopping lists at the grocery store. Simply create a list on Any.do, share it with your loved ones and see them adding their shopping items in real-time.\"\u001b[39;49;00m,\n \u001b[94m\"descriptionHTML\"\u001b[39;49;00m: \u001b[33m\"\\ud83e\\udd47 <b>"#1 to do list app out there\\u201d</b> - WSJ<br>\\ud83c\\udfc6 <b>Editor's Choice</b> by Google<br><br>Over 30M people rely on Any.do to stay organized and get more done.<br>It's a simple to do list app with reminders, planner & calendar - all in one.<br><br><b>\\ud83e\\udd47 "A MUST HAVE APP" (Lifehacker, NYTimes, USA TODAY).</b><br><br>Any.do is a free to-do list, planner & calendar app for managing and organizing your daily tasks, to-do lists, notes, reminders, checklists, calendar events, grocery lists and more.<br><br>\\ud83d\\udcc5 Organize your tasks & to-do list<br><br>\\u2022 ADVANCED CALENDAR & DAILY PLANNER - Keep your to-do list and calendar events always at hand with our calendar widget. Any.do to-do list & planner support daily calendar view, 3-day Calendar view, Weekly calendar view & agenda view, with built-in reminders. Review and organize your calendar events and to do list side by side.<br><br>\\u2022 SYNCS SEAMLESSLY - Keeps all your to do list, tasks, reminders, notes, calendar & agenda always in sync so you\\u2019ll never forget a thing. Sync your phone\\u2019s calendar, google calendar, Facebook events, outlook calendar or any other calendar so you don\\u2019t forget an important event.<br><br>\\u2022 SET REMINDERS - One time reminders, recurring reminders, Location reminders & voice reminders. NEW! Easily create tasks and get reminders in WhatsApp.<br><br>\\u2022 WORK TOGETHER - Share your to do list and assign tasks with your friends, family & colleagues from your task list to collaborate and get more done. <br><br>---<br><br>ALL-IN-ONE PLANNER & CALENDAR APP FOR GETTING THINGS DONE<br>Create and set reminders with voice to your to do list. <br>For better task management flow we added a calendar integration to keep your agenda always up to date. <br>For better productivity, we added recurring reminders, location reminders, one-time reminder, sub-tasks, notes & file attachments. <br>To keep your to do list up to date, we\\u2019ve added a daily planner and focus mode.<br><br>INTEGRATIONS<br>Any.do To do list, Calendar, planner & Reminders Integrates with Google Calendar, Outlook, WhatsApp, Slack, Gmail, Google Tasks, Evernote, Trello, Wunderlist, Todoist, Zapier, Asana, Microsoft to-do, Salesforce, OneNote, Google Assistant, Amazon Alexa, Office 365, Exchange, Jira & More.<br><br>TO DO LIST, CALENDAR, PLANNER & REMINDERS MADE SIMPLE<br>Designed to keep you on top of your to do list, tasks and calendar events with no hassle. With intuitive drag and drop of tasks, swiping to mark to-do's as complete, and shaking your device to remove completed from your to do list - you can stay organized and enjoy every minute of it.<br><br>POWERFUL TO DO LIST TASK MANAGEMENT<br>Add a to do list item straight from your email / Gmail / Outlook inbox by forwarding [email protected]. Attach files from your computer, Dropbox, or Google Drive to your to- tasks.<br><br>DAILY PLANNER & LIFE ORGANIZER<br>Any.do is a to do list, a calendar, an inbox, a notepad, a checklist, task list, a board for post its or sticky notes, a task & project management tool, a reminder app, a daily planner, a family organizer, an agenda, a bill planner and overall the simplest productivity tool you will ever have. <br><br>SHARE LISTS, ASSIGN & ORGANIZE TASKS<br>To plan & organize projects has never been easier. Now you can share lists between family members, assign tasks to each other, chat and much more. Any.do will help you and the people around you stay in-sync and get reminders so that you can focus on what matters, knowing you had a productive day and crossed off your to do list.<br><br>GROCERY LIST & SHOPPING LIST<br>Any.do task list, calendar, agenda, reminders & planner is also great for shopping lists at the grocery store. Simply create a list on Any.do, share it with your loved ones and see them adding their shopping items in real-time.\"\u001b[39;49;00m,\n \u001b[94m\"developer\"\u001b[39;49;00m: \u001b[33m\"Any.do To-do list & Calendar\"\u001b[39;49;00m,\n \u001b[94m\"developerAddress\"\u001b[39;49;00m: \u001b[33m\"Any.do Inc.\\n\\n6 Agripas Street, Tel Aviv\\n6249106 ISRAEL\"\u001b[39;49;00m,\n \u001b[94m\"developerEmail\"\u001b[39;49;00m: \u001b[33m\"[email protected]\"\u001b[39;49;00m,\n \u001b[94m\"developerId\"\u001b[39;49;00m: \u001b[33m\"5304780265295461149\"\u001b[39;49;00m,\n \u001b[94m\"developerInternalID\"\u001b[39;49;00m: \u001b[33m\"5304780265295461149\"\u001b[39;49;00m,\n \u001b[94m\"developerWebsite\"\u001b[39;49;00m: \u001b[33m\"https://www.any.do\"\u001b[39;49;00m,\n \u001b[94m\"editorsChoice\"\u001b[39;49;00m: \u001b[34mfalse\u001b[39;49;00m,\n \u001b[94m\"free\"\u001b[39;49;00m: \u001b[34mtrue\u001b[39;49;00m,\n \u001b[94m\"genre\"\u001b[39;49;00m: \u001b[33m\"Productivity\"\u001b[39;49;00m,\n \u001b[94m\"genreId\"\u001b[39;49;00m: \u001b[33m\"PRODUCTIVITY\"\u001b[39;49;00m,\n \u001b[94m\"headerImage\"\u001b[39;49;00m: \u001b[33m\"https://play-lh.googleusercontent.com/umhDP6phYKbuh-WzrSu6_bgFmAWP4qf9WsktFVBWDCMmkIwArJmyQKka7A1VcjFoEQc\"\u001b[39;49;00m,\n \u001b[94m\"histogram\"\u001b[39;49;00m: [\n \u001b[34m33055\u001b[39;49;00m,\n \u001b[34m11068\u001b[39;49;00m,\n \u001b[34m18270\u001b[39;49;00m,\n \u001b[34m34643\u001b[39;49;00m,\n \u001b[34m267437\u001b[39;49;00m\n ],\n \u001b[94m\"icon\"\u001b[39;49;00m: \u001b[33m\"https://play-lh.googleusercontent.com/zgOLUXCHkF91H8xuMTMLT17smwgLPwSBjUlKVWF-cZRFjlv-Uvtman7DiHEii54fbEE\"\u001b[39;49;00m,\n \u001b[94m\"inAppProductPrice\"\u001b[39;49;00m: \u001b[33m\"$0.99 - $83.88 per item\"\u001b[39;49;00m,\n \u001b[94m\"installs\"\u001b[39;49;00m: \u001b[33m\"10,000,000+\"\u001b[39;49;00m,\n \u001b[94m\"minInstalls\"\u001b[39;49;00m: \u001b[34m10000000\u001b[39;49;00m,\n \u001b[94m\"offersIAP\"\u001b[39;49;00m: \u001b[34mtrue\u001b[39;49;00m,\n \u001b[94m\"originalPrice\"\u001b[39;49;00m: \u001b[34mnull\u001b[39;49;00m,\n \u001b[94m\"price\"\u001b[39;49;00m: \u001b[34m0\u001b[39;49;00m,\n \u001b[94m\"privacyPolicy\"\u001b[39;49;00m: \u001b[33m\"https://www.any.do/privacy\"\u001b[39;49;00m,\n \u001b[94m\"ratings\"\u001b[39;49;00m: \u001b[34m364473\u001b[39;49;00m,\n \u001b[94m\"recentChanges\"\u001b[39;49;00m: \u001b[33m\"- Dark mode is finally here, hooray! \\r\\n- Recurring reminders issue was fixed\\r\\n- We added a save button to the task details screen\\r\\n- Filter your tasks for today, the next 7 days, and all tasks\\r\\n- Universal search with tasks, events, sub-tasks, notes, lists & tags\\r\\n- Keep track of how many tasks you\\u2019ve completed with Any.do\\r\\n- Upgraded sorting by time (the 2nd most requested feature!)\\r\\n- New and stunning reminders & snoozing redesign\\r\\n- Import tasks into Any.do from your clipboard\"\u001b[39;49;00m,\n \u001b[94m\"recentChangesHTML\"\u001b[39;49;00m: \u001b[33m\"- Dark mode is finally here, hooray! <br>- Recurring reminders issue was fixed<br>- We added a save button to the task details screen<br>- Filter your tasks for today, the next 7 days, and all tasks<br>- Universal search with tasks, events, sub-tasks, notes, lists & tags<br>- Keep track of how many tasks you\\u2019ve completed with Any.do<br>- Upgraded sorting by time (the 2nd most requested feature!)<br>- New and stunning reminders & snoozing redesign<br>- Import tasks into Any.do from your clipboard\"\u001b[39;49;00m,\n \u001b[94m\"released\"\u001b[39;49;00m: \u001b[33m\"Nov 10, 2011\"\u001b[39;49;00m,\n \u001b[94m\"reviews\"\u001b[39;49;00m: \u001b[34m129051\u001b[39;49;00m,\n \u001b[94m\"sale\"\u001b[39;49;00m: \u001b[34mfalse\u001b[39;49;00m,\n \u001b[94m\"saleText\"\u001b[39;49;00m: \u001b[34mnull\u001b[39;49;00m,\n \u001b[94m\"saleTime\"\u001b[39;49;00m: \u001b[34mnull\u001b[39;49;00m,\n \u001b[94m\"score\"\u001b[39;49;00m: \u001b[34m4.3508196\u001b[39;49;00m,\n \u001b[94m\"screenshots\"\u001b[39;49;00m: [\n \u001b[33m\"https://play-lh.googleusercontent.com/cGoZDZRRpHMKadBQeLaRwGQmKvTv3_nNkWKZkwT6VOBBEU-OvAfBX6XY3WD3wwy2lY0\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/49E2H7BYGrJkbH1dx-eQBgZGpgme8Wcfh8crvMyLSbCNZHbY8AI3YpR1fm1trpTtXoF5\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/rwql50U-W5Vn34MKy0rrzVlZwSF8Xj-utk7ymJAa4vCKgxUqyV6YT1YrduGkv01bug\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/zN8pIcOhwIuZ2sqeaWQkC6-iwVMSMWnY4QZAtKEgu9rFV76BASXxX6_XZcF3em3yBSc3\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/x7WDgU_nqe_82ehBrRYyP6OdCFgL5lCHVVaEXAvP3J7mhEaJBdo3NHTkhvoZZ9P5l2OH\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/cZBjFbEnGmqom0VDI5GBwe-KWZetnBxS4L-lumWNnFnq5tIVAAak6NqdvJaQp5_8CGg\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/OCmp0hmEVO8NUFhSRWe7Zv8WcxdMhKR6Lu5U6fkr8SCL2Dibakr-dQog07R_Li4MRMMq\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/4b_0LOovdrOVBowvua4loQzycriBSMubOQN3HZCPGZUUglo2VzSGb2n0368RMWRJN0A\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/ndr_2Hm3vaWzgY4-mUYqFwsw49KfovhkN5nbFpPG4HUg9fZjXQjfa3TYf-OYNB32SUDB\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/UZULO6XshlbT5o6pjksSqrhJ_g-pR-wJBYXq2h1Yoo6pDFhKFjyivnjCavGfGKIWCw\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/zPnaXYbEo_LEMhgfzW_O1nv-IccJa1xy26vWIbpSrChGaL-KzINBLk3GBs48v7j-fg\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/P_bSjsp8OAcqKF903GtrpamILuJhAJB-9OO5QkYnEUHoxfB7lWtbBK2ZVvnXJ8Y0SFI\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/9Jv2TQJ8nmEVuluW3JQ5ELDWeX_IuHIZX3NO6rFUTviCCKtkSVT8tCN208Zzq0Qgce-y\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/nnY_hLmuLBwiOBfIokKVg1v2vXe3WngVaKAejPe0Na1yXbtEGNhCf_a6sstmWZkbFyQ\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/5LoWLPyekVCtbxaPH_PUe9Q0Eblt7EIXXff6pnnhohkPhzG-FBdTqK7QBhCDA9jJx_w\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/Hv5TfdnmXbJ7_mQZdiXgGlZ6XbDj5kN7lwnj_Cg10ym81PW46PRKT4nDfJjnpNIvwVA\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/gaLzc8DFPM_JlIyRv96gnB9TrcAkd6H80EKe2-UOoR6m7WBGvCBZ9tv5CEOKu8yAX2IJ\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/fNde6nXNd_AmYM9UZRROu1WBkoKOiSGJAloM_jL2NCk08-ovo6OX34cZu2WeiQ7LPiM\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/x1s1I6gwi4oxMpoejQfccfLFKshCxj1-6ebTpg8JamQxzQtKDi7e0N_Hc_Hw3i01QUs\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/b7FN0ez09iJO0TMlIwbNO8AWnjN7bTLhctZWe1DvSu30kqoZdkgo_9R_QjAf0_YcDnU\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/2hLQGgdn14CfEngt9MCgxAqJqz32IFAtaTzooiI7cG_VUVjVJzIasBEpadUYrg-1ZJ8\"\u001b[39;49;00m,\n \u001b[33m\"https://play-lh.googleusercontent.com/OVfp00Cb60TbgoIFoDo2aZhfCWO4zzvTyl07IH2wHNuf9mgXA1EV1y7EwhMJhPtmFzs\"\u001b[39;49;00m\n ],\n \u001b[94m\"size\"\u001b[39;49;00m: \u001b[33m\"Varies with device\"\u001b[39;49;00m,\n \u001b[94m\"summary\"\u001b[39;49;00m: \u001b[33m\"To-do list & Tasks \\u2705 Calendar \\ud83d\\udcc5\\u05bf Reminder \\ud83d\\udd14 Checklist \\ud83d\\udcdd Agenda - All-in-one\"\u001b[39;49;00m,\n \u001b[94m\"summaryHTML\"\u001b[39;49;00m: \u001b[33m\"To-do list & Tasks \\u2705 Calendar \\ud83d\\udcc5\\u05bf Reminder \\ud83d\\udd14 Checklist \\ud83d\\udcdd Agenda - All-in-one\"\u001b[39;49;00m,\n \u001b[94m\"title\"\u001b[39;49;00m: \u001b[33m\"Any.do: To do list, Task, Reminders & Planner\"\u001b[39;49;00m,\n \u001b[94m\"updated\"\u001b[39;49;00m: \u001b[34m1614496999\u001b[39;49;00m,\n \u001b[94m\"url\"\u001b[39;49;00m: \u001b[33m\"https://play.google.com/store/apps/details?id=com.anydo&hl=en&gl=us\"\u001b[39;49;00m,\n \u001b[94m\"version\"\u001b[39;49;00m: \u001b[33m\"Varies with device\"\u001b[39;49;00m,\n \u001b[94m\"video\"\u001b[39;49;00m: \u001b[34mnull\u001b[39;49;00m,\n \u001b[94m\"videoImage\"\u001b[39;49;00m: \u001b[34mnull\u001b[39;49;00m\n}\n\n"
],
[
"df_app_infos = pd.DataFrame(app_infos)\ndf_app_infos.to_csv('./data/app_data.csv', index=None, header=True)",
"_____no_output_____"
]
],
[
[
"## Scraping app data",
"_____no_output_____"
]
],
[
[
"app_reviews = []\n\nfor app in tqdm(app_packages):\n for score in range(1, 6):\n for sort_order in [Sort.MOST_RELEVANT, Sort.NEWEST]:\n rvs = reviews(\n app, \n lang='en',\n country='us',\n sort=sort_order,\n count=200 if score == 3 else 100,\n filter_score_with=score\n )[0]\n\n for r in rvs:\n r['sortOrder'] = 'most_relevant' if sort_order == Sort.MOST_RELEVANT else 'newest'\n r['appId'] = app\n app_reviews.extend(rvs)",
"100%|██████████| 14/14 [01:30<00:00, 6.47s/it]\n"
],
[
"df_app_reviews = pd.DataFrame(app_reviews)\ndf_app_reviews.head()",
"_____no_output_____"
],
[
"df_app_reviews.to_csv('./data/app_review.csv', index=None, header=True)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbace5502aea0bb47bd374b981b422fab5ce3ddd
| 55,215 |
ipynb
|
Jupyter Notebook
|
notebooks/cerf_spatial.ipynb
|
KristianNelson/nelson_etal_2021_scidata
|
1000b27532c42bcb5994b664b4fd7869a766d0fe
|
[
"BSD-2-Clause"
] | null | null | null |
notebooks/cerf_spatial.ipynb
|
KristianNelson/nelson_etal_2021_scidata
|
1000b27532c42bcb5994b664b4fd7869a766d0fe
|
[
"BSD-2-Clause"
] | null | null | null |
notebooks/cerf_spatial.ipynb
|
KristianNelson/nelson_etal_2021_scidata
|
1000b27532c42bcb5994b664b4fd7869a766d0fe
|
[
"BSD-2-Clause"
] | 1 |
2021-06-24T15:50:31.000Z
|
2021-06-24T15:50:31.000Z
| 146.071429 | 34,144 | 0.890102 |
[
[
[
"%matplotlib inline\n\nimport os\n\nimport numpy as np\nimport pandas as pd\nimport geopandas as gpd\nimport fiona\n\nfrom matplotlib import pyplot\nfrom rasterio.plot import show\nfrom rasterio import features\n\n\nimport rasterio",
"_____no_output_____"
],
[
"# our polygon exclusion shapefile\nf = '/Users/d3y010/projects/mcmanamay/global_energy_potential/gis/species_ranges/botw_whooping_crane_mollweide.shp'\n\n# read it into a geopandas data frame\ngdf = gpd.read_file(f)",
"_____no_output_____"
]
],
[
[
"#### Select our field, we want it to be 1 for full excluded",
"_____no_output_____"
]
],
[
[
"gdf['value'] = 1",
"_____no_output_____"
]
],
[
[
"#### Only keep fields that we need; create fid field",
"_____no_output_____"
]
],
[
[
"gdf['fid'] = gdf.index\ngdf = gdf[['fid', 'value', 'geometry']]",
"_____no_output_____"
]
],
[
[
"#### See the coordinate reference system",
"_____no_output_____"
]
],
[
[
"gdf.crs",
"_____no_output_____"
]
],
[
[
"#### Reproject to Albers",
"_____no_output_____"
]
],
[
[
"pdf = gdf.to_crs('+proj=aea +lat_1=29.5 +lat_2=45.5 +lat_0=37.5 +lon_0=-96 +x_0=0 +y_0=0 +ellps=GRS80 +datum=NAD83 +units=m +no_defs')",
"_____no_output_____"
],
[
"pdf.plot()",
"_____no_output_____"
],
[
"pdf.crs",
"_____no_output_____"
]
],
[
[
"#### Check out our template raster",
"_____no_output_____"
]
],
[
[
"template_raster = '/Users/d3y010/projects/cerf/suitability/_common/PNNL_Land_Mask_CONUS.img'\n",
"_____no_output_____"
],
[
"rast = rasterio.open(template_raster)",
"_____no_output_____"
],
[
"rast.meta",
"_____no_output_____"
]
],
[
[
"### TODO: Get proj4s for shapefile reprojection from raster template CRS",
"_____no_output_____"
]
],
[
[
"rast.crs",
"_____no_output_____"
],
[
"rast.close()",
"_____no_output_____"
]
],
[
[
"#### Convert the geodataframe to a raster using our template",
"_____no_output_____"
]
],
[
[
"def rasterize(geodataframe, template_raster, outras, field):\n \n with rasterio.open(template_raster) as src:\n kwargs = src.meta.copy()\n kwargs.update({\n 'driver': 'GTiff',\n 'compress': 'lzw'\n })\n \n #windows = src.block_windows(1) \n \n with rasterio.open(outras, 'w', **kwargs) as dst:\n \n #for idx, window in windows:\n \n out_arr = src.read(1) #, window=window)\n \n # this is where we create a generator of geom, value pairs to use in rasterizing\n shapes = ((geom,value) for geom, value in zip(geodataframe.geometry, geodataframe[field]))\n\n burned = features.rasterize(shapes=shapes, fill=0, out=out_arr, transform=src.transform, all_touched=True)\n\n dst.write_band(1, burned) #, window=window) ",
"_____no_output_____"
],
[
"out_raster = '/Users/d3y010/Desktop/species.tif'\n\nrasterize(pdf, template_raster, out_raster, 'value')",
"_____no_output_____"
]
],
[
[
"#### Examine new raster",
"_____no_output_____"
]
],
[
[
"with rasterio.open(out_raster) as rast:\n show(rast)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbacecc5c7691c3019e148c4063ba53f30519199
| 112,343 |
ipynb
|
Jupyter Notebook
|
python_tools/model_3_Neural_network_Keras.ipynb
|
PHoogestraat/Where-is-my-BIke
|
f2528e87f0d8e00028565a08da25b66d74b22659
|
[
"MIT"
] | null | null | null |
python_tools/model_3_Neural_network_Keras.ipynb
|
PHoogestraat/Where-is-my-BIke
|
f2528e87f0d8e00028565a08da25b66d74b22659
|
[
"MIT"
] | null | null | null |
python_tools/model_3_Neural_network_Keras.ipynb
|
PHoogestraat/Where-is-my-BIke
|
f2528e87f0d8e00028565a08da25b66d74b22659
|
[
"MIT"
] | 1 |
2021-04-04T17:50:36.000Z
|
2021-04-04T17:50:36.000Z
| 39.62716 | 1,636 | 0.506947 |
[
[
[
"# Neural Networks with Keras\n\n513/513 - 0s - loss: 1.7734 - acc: 0.2710\n\nLoss: 1.7734287705337792, Accuracy: 0.2709551751613617\n\nuses minmaxscaler\n\nref: 21-Machine-Learning/3/Activities/02-Evr_First_Neural_Network/Solved/First_Neural_Network.ipynb#Model-Summary",
"_____no_output_____"
]
],
[
[
"# Dependencies\nimport pandas as pd\nfrom sklearn.preprocessing import LabelEncoder\nimport numpy as np\nfrom sklearn import tree\nimport os\nfrom sklearn.datasets import make_classification",
"_____no_output_____"
]
],
[
[
"Import csv that has been optimized for 10 freatures",
"_____no_output_____"
]
],
[
[
"# import processed data\n\npath = \"data/\"\nfile = \"Best_disposition_data.csv\"\npath_file = path + file\n\ndf = pd.read_csv(path_file)\ndf = df.drop(\"Unnamed: 0\", axis=1)\ndf",
"_____no_output_____"
]
],
[
[
"# Data Preprocessing\nIt is really important to scale our data before using multilayer perceptron models.\n\nWithout scaling, it is often difficult for the training cycle to converge",
"_____no_output_____"
]
],
[
[
"X = df.drop(\"disposition\", axis=1)\ny = df[\"disposition\"]\nprint(X.shape, y.shape)",
"(2052, 11) (2052,)\n"
],
[
"from sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import LabelEncoder, MinMaxScaler\nfrom tensorflow.keras.utils import to_categorical",
"C:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:516: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:517: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:518: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:519: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:520: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\framework\\dtypes.py:525: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:541: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:542: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:543: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:544: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:545: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\nC:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorboard\\compat\\tensorflow_stub\\dtypes.py:550: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
],
[
"X_train, X_test, y_train, y_test = train_test_split(\n X, y, random_state=1)",
"_____no_output_____"
],
[
"# MinMaxScaler is used\n\nX_scaler = MinMaxScaler().fit(X_train)\nX_train_scaled = X_scaler.transform(X_train)\nX_test_scaled = X_scaler.transform(X_test)\n",
"_____no_output_____"
],
[
"# Step 1: Label-encode data set\nlabel_encoder = LabelEncoder()\nlabel_encoder.fit(y_train)\nencoded_y_train = label_encoder.transform(y_train)\nencoded_y_test = label_encoder.transform(y_test)",
"_____no_output_____"
],
[
"#n= 0\n#for label, original_class in zip(encoded_y, y):\n# print('Original Class: ' + str(original_class))\n# print('Encoded Label: ' + str(label)) \n# n=n+1\n# print(n)\n# print('-' * 12)",
"_____no_output_____"
],
[
"# Step 2: Convert encoded labels to one-hot-encoding\ny_train_categorical = to_categorical(encoded_y_train)\ny_test_categorical = to_categorical(encoded_y_test)",
"_____no_output_____"
]
],
[
[
"## Creating our Model\n\nDecide what kind of model to apply to our data. \n\n For numerical data, we use a regressor model. \n\n For categorical data, we use a classifier model. \n\nIn this example, we will use a classifier to build the following network:",
"_____no_output_____"
],
[
"## Defining our Model Architecture (the layers)\n\nCreate a sequential model",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.models import Sequential\n\nmodel = Sequential()",
"_____no_output_____"
],
[
"from tensorflow.keras.layers import Dense\nfrom tensorflow.python.ops.init_ops import VarianceScaling\n\n\nnumber_inputs = 11\nnumber_hidden_nodes = 12\nmodel.add(Dense(units=number_hidden_nodes,\n activation='relu', input_dim=number_inputs))",
"WARNING:tensorflow:From C:\\Users\\phoog\\anaconda3\\envs\\PythonData\\lib\\site-packages\\tensorflow\\python\\ops\\init_ops.py:1251: calling VarianceScaling.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.\nInstructions for updating:\nCall initializer instance with the dtype argument instead of passing it to the constructor\n"
],
[
"number_classes = 6\nmodel.add(Dense(units=number_classes, activation='softmax'))",
"_____no_output_____"
]
],
[
[
"## Model Summary",
"_____no_output_____"
]
],
[
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 12) 144 \n_________________________________________________________________\ndense_1 (Dense) (None, 6) 78 \n=================================================================\nTotal params: 222\nTrainable params: 222\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"## Compile the Model\n\nNow that we have our model architecture defined, we must compile the model using a loss function and optimizer. We can also specify additional training metrics such as accuracy.",
"_____no_output_____"
]
],
[
[
"# Use categorical crossentropy for categorical data and mean squared error for regression\n# Hint: your output layer in this example is using software for logistic regression (categorical)\n# If your output layer activation was `linear` then you may want to use `mse` for loss\nmodel.compile(optimizer='adam',\n loss='categorical_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## Training the Model\nFinally, we train our model using our training data\nTraining consists of updating our weights using our optimizer and loss function. In this example, we choose 1000 iterations (loops) of training that are called epochs.\n\nWe also choose to shuffle our training data and increase the detail printed out during each training cycle.",
"_____no_output_____"
]
],
[
[
"# Fit (train) the model\nmodel.fit(\n X_train_scaled,\n y_train_categorical,\n epochs=1000,\n shuffle=True,\n verbose=2\n)",
"Epoch 1/1000\n1539/1539 - 0s - loss: 1.7774 - acc: 0.2359\nEpoch 2/1000\n1539/1539 - 0s - loss: 1.7682 - acc: 0.2424\nEpoch 3/1000\n1539/1539 - 0s - loss: 1.7632 - acc: 0.2482\nEpoch 4/1000\n1539/1539 - 0s - loss: 1.7604 - acc: 0.2521\nEpoch 5/1000\n1539/1539 - 0s - loss: 1.7576 - acc: 0.2521\nEpoch 6/1000\n1539/1539 - 0s - loss: 1.7550 - acc: 0.2495\nEpoch 7/1000\n1539/1539 - 0s - loss: 1.7530 - acc: 0.2508\nEpoch 8/1000\n1539/1539 - 0s - loss: 1.7514 - acc: 0.2528\nEpoch 9/1000\n1539/1539 - 0s - loss: 1.7503 - acc: 0.2554\nEpoch 10/1000\n1539/1539 - 0s - loss: 1.7473 - acc: 0.2586\nEpoch 11/1000\n1539/1539 - 0s - loss: 1.7460 - acc: 0.2586\nEpoch 12/1000\n1539/1539 - 0s - loss: 1.7442 - acc: 0.2625\nEpoch 13/1000\n1539/1539 - 0s - loss: 1.7434 - acc: 0.2736\nEpoch 14/1000\n1539/1539 - 0s - loss: 1.7417 - acc: 0.2762\nEpoch 15/1000\n1539/1539 - 0s - loss: 1.7403 - acc: 0.2755\nEpoch 16/1000\n1539/1539 - 0s - loss: 1.7391 - acc: 0.2801\nEpoch 17/1000\n1539/1539 - 0s - loss: 1.7377 - acc: 0.2833\nEpoch 18/1000\n1539/1539 - 0s - loss: 1.7368 - acc: 0.2820\nEpoch 19/1000\n1539/1539 - 0s - loss: 1.7355 - acc: 0.2840\nEpoch 20/1000\n1539/1539 - 0s - loss: 1.7347 - acc: 0.2853\nEpoch 21/1000\n1539/1539 - 0s - loss: 1.7332 - acc: 0.2827\nEpoch 22/1000\n1539/1539 - 0s - loss: 1.7326 - acc: 0.2891\nEpoch 23/1000\n1539/1539 - 0s - loss: 1.7322 - acc: 0.2930\nEpoch 24/1000\n1539/1539 - 0s - loss: 1.7305 - acc: 0.2878\nEpoch 25/1000\n1539/1539 - 0s - loss: 1.7302 - acc: 0.2911\nEpoch 26/1000\n1539/1539 - 0s - loss: 1.7290 - acc: 0.2950\nEpoch 27/1000\n1539/1539 - 0s - loss: 1.7292 - acc: 0.2930\nEpoch 28/1000\n1539/1539 - 0s - loss: 1.7290 - acc: 0.2930\nEpoch 29/1000\n1539/1539 - 0s - loss: 1.7267 - acc: 0.2924\nEpoch 30/1000\n1539/1539 - 0s - loss: 1.7268 - acc: 0.2878\nEpoch 31/1000\n1539/1539 - 0s - loss: 1.7258 - acc: 0.2930\nEpoch 32/1000\n1539/1539 - 0s - loss: 1.7253 - acc: 0.2995\nEpoch 33/1000\n1539/1539 - 0s - loss: 1.7244 - acc: 0.2911\nEpoch 34/1000\n1539/1539 - 0s - loss: 1.7244 - acc: 0.2924\nEpoch 35/1000\n1539/1539 - 0s - loss: 1.7244 - acc: 0.2930\nEpoch 36/1000\n1539/1539 - 0s - loss: 1.7234 - acc: 0.2924\nEpoch 37/1000\n1539/1539 - 0s - loss: 1.7232 - acc: 0.2930\nEpoch 38/1000\n1539/1539 - 0s - loss: 1.7224 - acc: 0.2937\nEpoch 39/1000\n1539/1539 - 0s - loss: 1.7221 - acc: 0.2898\nEpoch 40/1000\n1539/1539 - 0s - loss: 1.7213 - acc: 0.2878\nEpoch 41/1000\n1539/1539 - 0s - loss: 1.7209 - acc: 0.2885\nEpoch 42/1000\n1539/1539 - 0s - loss: 1.7208 - acc: 0.2898\nEpoch 43/1000\n1539/1539 - 0s - loss: 1.7200 - acc: 0.2904\nEpoch 44/1000\n1539/1539 - 0s - loss: 1.7198 - acc: 0.2878\nEpoch 45/1000\n1539/1539 - 0s - loss: 1.7199 - acc: 0.2917\nEpoch 46/1000\n1539/1539 - 0s - loss: 1.7196 - acc: 0.2924\nEpoch 47/1000\n1539/1539 - 0s - loss: 1.7190 - acc: 0.2911\nEpoch 48/1000\n1539/1539 - 0s - loss: 1.7185 - acc: 0.2917\nEpoch 49/1000\n1539/1539 - 0s - loss: 1.7186 - acc: 0.2878\nEpoch 50/1000\n1539/1539 - 0s - loss: 1.7184 - acc: 0.2917\nEpoch 51/1000\n1539/1539 - 0s - loss: 1.7182 - acc: 0.2937\nEpoch 52/1000\n1539/1539 - 0s - loss: 1.7174 - acc: 0.2904\nEpoch 53/1000\n1539/1539 - 0s - loss: 1.7173 - acc: 0.2911\nEpoch 54/1000\n1539/1539 - 0s - loss: 1.7172 - acc: 0.2898\nEpoch 55/1000\n1539/1539 - 0s - loss: 1.7173 - acc: 0.2891\nEpoch 56/1000\n1539/1539 - 0s - loss: 1.7170 - acc: 0.2891\nEpoch 57/1000\n1539/1539 - 0s - loss: 1.7178 - acc: 0.2878\nEpoch 58/1000\n1539/1539 - 0s - loss: 1.7163 - acc: 0.2904\nEpoch 59/1000\n1539/1539 - 0s - loss: 1.7155 - acc: 0.2924\nEpoch 60/1000\n1539/1539 - 0s - loss: 1.7157 - acc: 0.2917\nEpoch 61/1000\n1539/1539 - 0s - loss: 1.7151 - acc: 0.2924\nEpoch 62/1000\n1539/1539 - 0s - loss: 1.7154 - acc: 0.2898\nEpoch 63/1000\n1539/1539 - 0s - loss: 1.7150 - acc: 0.2937\nEpoch 64/1000\n1539/1539 - 0s - loss: 1.7146 - acc: 0.2891\nEpoch 65/1000\n1539/1539 - 0s - loss: 1.7152 - acc: 0.2950\nEpoch 66/1000\n1539/1539 - 0s - loss: 1.7142 - acc: 0.2898\nEpoch 67/1000\n1539/1539 - 0s - loss: 1.7145 - acc: 0.2943\nEpoch 68/1000\n1539/1539 - 0s - loss: 1.7139 - acc: 0.2878\nEpoch 69/1000\n1539/1539 - 0s - loss: 1.7134 - acc: 0.2943\nEpoch 70/1000\n1539/1539 - 0s - loss: 1.7142 - acc: 0.2924\nEpoch 71/1000\n1539/1539 - 0s - loss: 1.7134 - acc: 0.2937\nEpoch 72/1000\n1539/1539 - 0s - loss: 1.7129 - acc: 0.2917\nEpoch 73/1000\n1539/1539 - 0s - loss: 1.7133 - acc: 0.2865\nEpoch 74/1000\n1539/1539 - 0s - loss: 1.7136 - acc: 0.2891\nEpoch 75/1000\n1539/1539 - 0s - loss: 1.7128 - acc: 0.2891\nEpoch 76/1000\n1539/1539 - 0s - loss: 1.7126 - acc: 0.2911\nEpoch 77/1000\n1539/1539 - 0s - loss: 1.7123 - acc: 0.2930\nEpoch 78/1000\n1539/1539 - 0s - loss: 1.7125 - acc: 0.2911\nEpoch 79/1000\n1539/1539 - 0s - loss: 1.7125 - acc: 0.2878\nEpoch 80/1000\n1539/1539 - 0s - loss: 1.7115 - acc: 0.2917\nEpoch 81/1000\n1539/1539 - 0s - loss: 1.7114 - acc: 0.2917\nEpoch 82/1000\n1539/1539 - 0s - loss: 1.7114 - acc: 0.2950\nEpoch 83/1000\n1539/1539 - 0s - loss: 1.7112 - acc: 0.2904\nEpoch 84/1000\n1539/1539 - 0s - loss: 1.7118 - acc: 0.2872\nEpoch 85/1000\n1539/1539 - 0s - loss: 1.7109 - acc: 0.2898\nEpoch 86/1000\n1539/1539 - 0s - loss: 1.7108 - acc: 0.2911\nEpoch 87/1000\n1539/1539 - 0s - loss: 1.7105 - acc: 0.2911\nEpoch 88/1000\n1539/1539 - 0s - loss: 1.7121 - acc: 0.2943\nEpoch 89/1000\n1539/1539 - 0s - loss: 1.7107 - acc: 0.2989\nEpoch 90/1000\n1539/1539 - 0s - loss: 1.7107 - acc: 0.2930\nEpoch 91/1000\n1539/1539 - 0s - loss: 1.7098 - acc: 0.2943\nEpoch 92/1000\n1539/1539 - 0s - loss: 1.7100 - acc: 0.2924\nEpoch 93/1000\n1539/1539 - 0s - loss: 1.7097 - acc: 0.2956\nEpoch 94/1000\n1539/1539 - 0s - loss: 1.7101 - acc: 0.2898\nEpoch 95/1000\n1539/1539 - 0s - loss: 1.7093 - acc: 0.2943\nEpoch 96/1000\n1539/1539 - 0s - loss: 1.7095 - acc: 0.2891\nEpoch 97/1000\n1539/1539 - 0s - loss: 1.7095 - acc: 0.2950\nEpoch 98/1000\n1539/1539 - 0s - loss: 1.7086 - acc: 0.2898\nEpoch 99/1000\n1539/1539 - 0s - loss: 1.7088 - acc: 0.2917\nEpoch 100/1000\n1539/1539 - 0s - loss: 1.7087 - acc: 0.2956\nEpoch 101/1000\n1539/1539 - 0s - loss: 1.7084 - acc: 0.2937\nEpoch 102/1000\n1539/1539 - 0s - loss: 1.7086 - acc: 0.2956\nEpoch 103/1000\n1539/1539 - 0s - loss: 1.7090 - acc: 0.2924\nEpoch 104/1000\n1539/1539 - 0s - loss: 1.7077 - acc: 0.2982\nEpoch 105/1000\n1539/1539 - 0s - loss: 1.7078 - acc: 0.2950\nEpoch 106/1000\n1539/1539 - 0s - loss: 1.7086 - acc: 0.2982\nEpoch 107/1000\n1539/1539 - 0s - loss: 1.7086 - acc: 0.2937\nEpoch 108/1000\n1539/1539 - 0s - loss: 1.7068 - acc: 0.2956\nEpoch 109/1000\n1539/1539 - 0s - loss: 1.7075 - acc: 0.2950\nEpoch 110/1000\n1539/1539 - 0s - loss: 1.7077 - acc: 0.2963\nEpoch 111/1000\n1539/1539 - 0s - loss: 1.7070 - acc: 0.2963\nEpoch 112/1000\n1539/1539 - 0s - loss: 1.7067 - acc: 0.2924\nEpoch 113/1000\n1539/1539 - 0s - loss: 1.7071 - acc: 0.2937\nEpoch 114/1000\n1539/1539 - 0s - loss: 1.7068 - acc: 0.2917\nEpoch 115/1000\n1539/1539 - 0s - loss: 1.7066 - acc: 0.2969\nEpoch 116/1000\n1539/1539 - 0s - loss: 1.7064 - acc: 0.2969\nEpoch 117/1000\n1539/1539 - 0s - loss: 1.7060 - acc: 0.2930\nEpoch 118/1000\n1539/1539 - 0s - loss: 1.7066 - acc: 0.2943\nEpoch 119/1000\n1539/1539 - 0s - loss: 1.7065 - acc: 0.2982\nEpoch 120/1000\n1539/1539 - 0s - loss: 1.7069 - acc: 0.2956\nEpoch 121/1000\n1539/1539 - 0s - loss: 1.7060 - acc: 0.2976\nEpoch 122/1000\n1539/1539 - 0s - loss: 1.7060 - acc: 0.2950\nEpoch 123/1000\n1539/1539 - 0s - loss: 1.7049 - acc: 0.2969\nEpoch 124/1000\n1539/1539 - 0s - loss: 1.7053 - acc: 0.2963\nEpoch 125/1000\n1539/1539 - 0s - loss: 1.7049 - acc: 0.2995\nEpoch 126/1000\n1539/1539 - 0s - loss: 1.7051 - acc: 0.2995\nEpoch 127/1000\n1539/1539 - 0s - loss: 1.7052 - acc: 0.2950\nEpoch 128/1000\n1539/1539 - 0s - loss: 1.7051 - acc: 0.2950\nEpoch 129/1000\n1539/1539 - 0s - loss: 1.7051 - acc: 0.2969\nEpoch 130/1000\n1539/1539 - 0s - loss: 1.7048 - acc: 0.2963\nEpoch 131/1000\n1539/1539 - 0s - loss: 1.7044 - acc: 0.2930\nEpoch 132/1000\n1539/1539 - 0s - loss: 1.7037 - acc: 0.2995\nEpoch 133/1000\n1539/1539 - 0s - loss: 1.7038 - acc: 0.3008\nEpoch 134/1000\n1539/1539 - 0s - loss: 1.7050 - acc: 0.3008\nEpoch 135/1000\n1539/1539 - 0s - loss: 1.7034 - acc: 0.2982\nEpoch 136/1000\n1539/1539 - 0s - loss: 1.7040 - acc: 0.2956\nEpoch 137/1000\n1539/1539 - 0s - loss: 1.7037 - acc: 0.3021\nEpoch 138/1000\n1539/1539 - 0s - loss: 1.7034 - acc: 0.3002\nEpoch 139/1000\n1539/1539 - 0s - loss: 1.7030 - acc: 0.2982\nEpoch 140/1000\n1539/1539 - 0s - loss: 1.7030 - acc: 0.3002\nEpoch 141/1000\n1539/1539 - 0s - loss: 1.7038 - acc: 0.3002\n"
]
],
[
[
"# Save the Trained Model",
"_____no_output_____"
]
],
[
[
"#Save the mod#el\nmodel.save(\"z3_Nueral_network_model.h5\")",
"_____no_output_____"
]
],
[
[
"# Quantifying the Model\nTesting data to validate our model. Determine the validity of model (i.e. the ability to predict new and previously unseen data points)",
"_____no_output_____"
]
],
[
[
"# Evaluate the model using the testing data\nmodel_loss, model_accuracy = model.evaluate(\n X_test_scaled, y_test_categorical, verbose=2)\nprint(f\"Loss: {model_loss}, Accuracy: {model_accuracy}\")",
"513/513 - 0s - loss: 1.8079 - acc: 0.2339\nLoss: 1.8079279228957772, Accuracy: 0.23391813039779663\n"
]
],
[
[
"## Making Predictions with new data\n\nUse trained model to make predictions using `model.predict`\n\n## Making Predictions with new data\n<b>Label asignments</b>\n\n\n<b>Test Data was taken from the first row:</b>\n\n\nnew_data2\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nnew_data = np.array([[2012,12, 49, 7, 23, 0, 281.34, 70, 1016, 2]])\nnew_data_2 = np.array([[2017,7, 28, 6, 13, 27, 292.06, 88, 1017, 1]]) ",
"_____no_output_____"
],
[
"# Test 1\nprint(f\"Predicted class: {model.predict_classes(new_data)}\")",
"_____no_output_____"
],
[
"# Test 2\nprint(f\"Predicted class: {model.predict_classes(new_data)}\")",
"_____no_output_____"
]
],
[
[
"# Evaluate the Model",
"_____no_output_____"
]
],
[
[
"# Load the model\n#from tensorflow.keras.models import load_model\n#from tensorflow.python.ops.init_ops import Zeros\n\n#model = load_model(\"z_Nueral_network_model.h5\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbacf1fad5eb872c3c3d8f2741335d7c053f094a
| 41,736 |
ipynb
|
Jupyter Notebook
|
tests/old_version_comparison.ipynb
|
nostosgenomics/MMSplice_MTSplice
|
da429002484effed03183032507d0eb829272cac
|
[
"MIT"
] | null | null | null |
tests/old_version_comparison.ipynb
|
nostosgenomics/MMSplice_MTSplice
|
da429002484effed03183032507d0eb829272cac
|
[
"MIT"
] | null | null | null |
tests/old_version_comparison.ipynb
|
nostosgenomics/MMSplice_MTSplice
|
da429002484effed03183032507d0eb829272cac
|
[
"MIT"
] | null | null | null | 97.971831 | 9,560 | 0.778872 |
[
[
[
"# Example code to apply the modular splicing model",
"_____no_output_____"
],
[
"Example test variants come from ClinVar BRCA1 variants",
"_____no_output_____"
],
[
"## Splicing delta PSI prediction",
"_____no_output_____"
]
],
[
[
"from mmsplice.vcf_dataloader import SplicingVCFDataloader\nfrom mmsplice import MMSplice, predict_all_table\n\nfrom mmsplice.utils import max_varEff",
"Using TensorFlow backend.\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/concise/utils/plot.py:115: FutureWarning: arrays to stack must be passed as a \"sequence\" type such as list or tuple. Support for non-sequence iterables such as generators is deprecated as of NumPy 1.16 and will raise an error in the future.\n min_coords = np.vstack(data.min(0) for data in polygons_data).min(0)\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/concise/utils/plot.py:116: FutureWarning: arrays to stack must be passed as a \"sequence\" type such as list or tuple. Support for non-sequence iterables such as generators is deprecated as of NumPy 1.16 and will raise an error in the future.\n max_coords = np.vstack(data.max(0) for data in polygons_data).max(0)\nWARNING: Logging before flag parsing goes to stderr.\nW0812 20:34:13.562207 140080037934848 deprecation_wrapper.py:119] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/optimizers.py:790: The name tf.train.Optimizer is deprecated. Please use tf.compat.v1.train.Optimizer instead.\n\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/sklearn/externals/joblib/__init__.py:15: DeprecationWarning: sklearn.externals.joblib is deprecated in 0.21 and will be removed in 0.23. Please import this functionality directly from joblib, which can be installed with: pip install joblib. If this warning is raised when loading pickled models, you may need to re-serialize those models with scikit-learn 0.21+.\n warnings.warn(msg, category=DeprecationWarning)\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/sklearn/base.py:306: UserWarning: Trying to unpickle estimator HuberRegressor from version 0.19.2 when using version 0.21.2. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/sklearn/base.py:306: UserWarning: Trying to unpickle estimator StandardScaler from version 0.19.2 when using version 0.21.2. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/sklearn/base.py:306: UserWarning: Trying to unpickle estimator LogisticRegression from version 0.19.2 when using version 0.21.2. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/sklearn/base.py:306: UserWarning: Trying to unpickle estimator Pipeline from version 0.19.2 when using version 0.21.2. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n"
],
[
"import pkg_resources\nassert pkg_resources.get_distribution(\"mmsplice\").version == '0.2.7'\npkg_resources.get_distribution(\"mmsplice\").version",
"_____no_output_____"
],
[
"gtf = '../tests/data/test.gtf'\nvcf = '../tests/data/test.vcf.gz'\nfasta = '../tests/data/hg19.nochr.chr17.fa'",
"_____no_output_____"
],
[
"dl = SplicingVCFDataloader(gtf, fasta, vcf,\n split_seq=False,encode=False, overhang=(100,100))",
"_____no_output_____"
],
[
"model = MMSplice()",
"W0812 20:34:17.349804 140080037934848 deprecation_wrapper.py:119] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:95: The name tf.reset_default_graph is deprecated. Please use tf.compat.v1.reset_default_graph instead.\n\nW0812 20:34:17.353101 140080037934848 deprecation_wrapper.py:119] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:98: The name tf.placeholder_with_default is deprecated. Please use tf.compat.v1.placeholder_with_default instead.\n\nW0812 20:34:17.363179 140080037934848 deprecation_wrapper.py:119] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:102: The name tf.get_default_graph is deprecated. Please use tf.compat.v1.get_default_graph instead.\n\nW0812 20:34:17.369028 140080037934848 deprecation_wrapper.py:119] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:517: The name tf.placeholder is deprecated. Please use tf.compat.v1.placeholder instead.\n\nW0812 20:34:17.373578 140080037934848 deprecation_wrapper.py:119] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:4138: The name tf.random_uniform is deprecated. Please use tf.random.uniform instead.\n\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/engine/saving.py:292: UserWarning: No training configuration found in save file: the model was *not* compiled. Compile it manually.\n warnings.warn('No training configuration found in save file: '\nW0812 20:34:17.809584 140080037934848 deprecation.py:506] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:3445: calling dropout (from tensorflow.python.ops.nn_ops) with keep_prob is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.\nW0812 20:34:17.983376 140080037934848 deprecation.py:323] From /home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/tensorflow/python/ops/nn_impl.py:180: add_dispatch_support.<locals>.wrapper (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.\nInstructions for updating:\nUse tf.where in 2.0, which has the same broadcast rule as np.where\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/engine/saving.py:292: UserWarning: No training configuration found in save file: the model was *not* compiled. Compile it manually.\n warnings.warn('No training configuration found in save file: '\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/keras/engine/saving.py:292: UserWarning: No training configuration found in save file: the model was *not* compiled. Compile it manually.\n warnings.warn('No training configuration found in save file: '\n"
],
[
"predictions = predict_all_table(model, dl, assembly=True, pathogenicity=True,\n splicing_efficiency=True, split_seq=False)",
"0it [00:00, ?it/s]/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/mmsplice/mmsplice.py:162: UserWarning: None GT donor\n warnings.warn(\"None GT donor\", UserWarning)\n/home/muhammedhasan/Projects/MMSplice/old-env/lib/python3.7/site-packages/mmsplice/mmsplice.py:164: UserWarning: None AG donor\n warnings.warn(\"None AG donor\", UserWarning)\n5it [00:16, 2.95s/it]\n"
],
[
"def exon_annotation_mapping(exon):\n loc = exon.split('_')\n start = int(loc[1]) - 1\n loc[1] = str(start)\n return '%s:%s-%s' % tuple(loc)",
"_____no_output_____"
],
[
"predictions['exons'] = predictions['exons'].map(exon_annotation_mapping)\npredictions = predictions.set_index(['ID', 'exons'])",
"_____no_output_____"
],
[
"import pandas as pd\n# run notebooks example notebook file to obtain this file\ndf = pd.read_csv('../notebooks/pred.csv')\ndf = df.set_index(['ID', 'exons'])",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df_join = predictions.join(df, how='inner')[['mmsplice_dlogitPsi', 'delta_logit_psi']]",
"_____no_output_____"
],
[
"df_join.head()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nplt.scatter(df_join['mmsplice_dlogitPsi'], df_join['delta_logit_psi'])",
"_____no_output_____"
],
[
"(df_join['delta_logit_psi'] - df_join['mmsplice_dlogitPsi']).hist(bins=50)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbad004f09b59bcf5d4bb3d5df2af86b53100499
| 3,614 |
ipynb
|
Jupyter Notebook
|
examples/auto-sklearn.ipynb
|
Ennosigaeon/xautoml
|
6e49ee8b2ffb6d19dcfd9cbe8b3397416c9b5ded
|
[
"BSD-3-Clause"
] | 4 |
2022-02-27T08:54:08.000Z
|
2022-03-30T21:19:29.000Z
|
examples/auto-sklearn.ipynb
|
Ennosigaeon/xautoml
|
6e49ee8b2ffb6d19dcfd9cbe8b3397416c9b5ded
|
[
"BSD-3-Clause"
] | 1 |
2022-02-28T09:41:00.000Z
|
2022-03-02T07:44:17.000Z
|
examples/auto-sklearn.ipynb
|
Ennosigaeon/xautoml
|
6e49ee8b2ffb6d19dcfd9cbe8b3397416c9b5ded
|
[
"BSD-3-Clause"
] | 2 |
2022-03-01T00:38:09.000Z
|
2022-03-21T09:38:49.000Z
| 23.316129 | 328 | 0.550083 |
[
[
[
"# Credit Prediction\n\n## Load Data\nThis dataset classifies people described by a set of attributes as good or bad credit risks.",
"_____no_output_____"
]
],
[
[
"from xautoml.util.datasets import openml_task\n\nX_train, y_train = openml_task(31, 0, train=True)\nX_train",
"_____no_output_____"
]
],
[
[
"## Install auto-sklearn\nIf you haven't installed auto-sklearn yet, you can install it via\n\n```\npip install auto-sklearn\n```",
"_____no_output_____"
],
[
"## Start the Model Building\n\nUse `auto-sklearn` to train a classifier on the training data.\n\nBy default, `auto-sklearn` only keeps the models used in the ensemble in memory and deletes all other models after completing the optimization. If you want to analyse all constructed models, you will have to provide `delete_tmp_folder_after_terminate`, `max_models_on_disc` and `tmp_folder` has done in the following cell.",
"_____no_output_____"
]
],
[
[
"import shutil\nimport os\nimport autosklearn.classification\nfrom autosklearn.metrics import accuracy\n\nworkdir = './_auto-sklearn_/'\nif os.path.exists(workdir):\n shutil.rmtree(workdir)\n\nautoml = autosklearn.classification.AutoSklearnClassifier(\n time_left_for_this_task=60,\n per_run_time_limit=20,\n metric=accuracy,\n # Optional: Set the following three parameters to analyse all models generate by auto-sklearn. Otherwise, you can only inspect the top 50 models.\n delete_tmp_folder_after_terminate=False,\n max_models_on_disc=None,\n tmp_folder=workdir\n\n)\nautoml.fit(X_train, y_train, dataset_name='credit-g')",
"_____no_output_____"
]
],
[
[
"## Visualize the Optimization Run in XAutoML",
"_____no_output_____"
]
],
[
[
"from xautoml.main import XAutoML\nfrom xautoml.adapter import import_auto_sklearn\n\nX_test, y_test = openml_task(31, 0, test=True)\n\nrh = import_auto_sklearn(automl)\nmain = XAutoML(rh, X_test, y_test)\nmain",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbad0b9b9194e07b141564fe28a0e07fd6958ed5
| 119,754 |
ipynb
|
Jupyter Notebook
|
SBERT_CORD19_QA_CrossEncoders.ipynb
|
Nikoletos-K/QA-with-SBERT-for-CORD19
|
21675d38201b277b86268109f7a9a9dd40057d7c
|
[
"MIT"
] | 3 |
2021-03-31T06:18:14.000Z
|
2021-09-13T13:11:28.000Z
|
SBERT_CORD19_QA_CrossEncoders.ipynb
|
Nikoletos-K/QA-with-SBERT-for-CORD19
|
21675d38201b277b86268109f7a9a9dd40057d7c
|
[
"MIT"
] | null | null | null |
SBERT_CORD19_QA_CrossEncoders.ipynb
|
Nikoletos-K/QA-with-SBERT-for-CORD19
|
21675d38201b277b86268109f7a9a9dd40057d7c
|
[
"MIT"
] | null | null | null | 40.132038 | 334 | 0.538095 |
[
[
[
"<p align=\"center\">\n <img src=\"http://www.di.uoa.gr/themes/corporate_lite/logo_el.png\" title=\"Department of Informatics and Telecommunications - University of Athens\"/> </p>\n\n---\n<h1 align=\"center\"> \n Artificial Intelligence\n</h1>\n<h1 align=\"center\" > \n Deep Learning for Natural Language Processing\n</h1>\n\n---\n<h2 align=\"center\"> \n <b>Konstantinos Nikoletos</b>\n</h2>\n\n<h3 align=\"center\"> \n <b>Winter 2020-2021</b>\n</h3>\n\n\n---\n---",
"_____no_output_____"
],
[
"\n### __Task__ \nThis exercise is about developing a document retrieval system to return titles of scientific\npapers containing the answer to a given user question. You will use the first version of\nthe COVID-19 Open Research Dataset (CORD-19) in your work (articles in the folder\ncomm use subset).\n\n\nFor example, for the question “What are the coronaviruses?”, your system can return the\npaper title “Distinct Roles for Sialoside and Protein Receptors in Coronavirus Infection”\nsince this paper contains the answer to the asked question.\n\n\nTo achieve the goal of this exercise, you will need first to read the paper Sentence-BERT:\nSentence Embeddings using Siamese BERT-Networks, in order to understand how you\ncan create sentence embeddings. In the related work of this paper, you will also find other\napproaches for developing your model. For example, you can using Glove embeddings,\netc. In this link, you can find the extended versions of this dataset to test your model, if\nyou want. You are required to:\n\n\n<ol type=\"a\">\n <li>Preprocess the provided dataset. You will decide which data of each paper is useful\nto your model in order to create the appropriate embeddings. You need to explain\nyour decisions.</li>\n <li>Implement at least 2 different sentence embedding approaches (see the related work\nof the Sentence-BERT paper), in order for your model to retrieve the titles of the\npapers related to a given question.</li>\n <li>Compare your 2 models based on at least 2 different criteria of your choice. Explain\nwhy you selected these criteria, your implementation choices, and the results. Some\nquestions you can pose are included here. You will need to provide the extra questions\nyou posed to your model and the results of all the questions as well.</li>\n</ol>\n\n### __Notebook__ \n\n\nSame implementation as Sentence Bert notebook but with adding CrossEncoders that I read that they perform even better \n\n\n---\n---",
"_____no_output_____"
],
[
"__Import__ of essential libraries\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd \nimport sys # only needed to determine Python version number\nimport matplotlib # only needed to determine Matplotlib version \nimport nltk\nfrom nltk.stem import WordNetLemmatizer\nimport pprint\nimport torch\nimport torch.nn as nn\nimport torch.optim as optim\nfrom torchtext import data\nimport logging\nnltk.download('punkt')\nnltk.download('wordnet')\nnltk.download('stopwords')\nnltk.download('averaged_perceptron_tagger')",
"[nltk_data] Downloading package punkt to /root/nltk_data...\n[nltk_data] Package punkt is already up-to-date!\n[nltk_data] Downloading package wordnet to /root/nltk_data...\n[nltk_data] Package wordnet is already up-to-date!\n[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Package stopwords is already up-to-date!\n[nltk_data] Downloading package averaged_perceptron_tagger to\n[nltk_data] /root/nltk_data...\n[nltk_data] Package averaged_perceptron_tagger is already up-to-\n[nltk_data] date!\n"
]
],
[
[
"Selecting device (GPU - CUDA if available)",
"_____no_output_____"
]
],
[
[
"# First checking if GPU is available\ntrain_on_gpu=torch.cuda.is_available()\n\nif(train_on_gpu):\n print('Training on GPU.')\nelse:\n print('No GPU available, training on CPU.')",
"Training on GPU.\n"
]
],
[
[
"# Loading data\n---",
"_____no_output_____"
]
],
[
[
"# Opening data file\nimport io\nfrom google.colab import drive\nfrom os import listdir\nfrom os.path import isfile, join\nimport json\n\ndrive.mount('/content/drive',force_remount=True)",
"Mounted at /content/drive\n"
]
],
[
[
"Loading the dictionary if it has been created",
"_____no_output_____"
]
],
[
[
"#@title Select number of papers that will be feeded in the model { vertical-output: true, display-mode: \"both\" }\nnumber_of_papers = \"9000\" #@param [\"1000\",\"3000\", \"6000\",\"9000\"]\nimport pickle\n\nCORD19_Dataframe = r\"/content/drive/My Drive/AI_4/CORD19_SentenceMap_\"+number_of_papers+\".pkl\"\nwith open(CORD19_Dataframe, 'rb') as drivef:\n CORD19Dictionary = pickle.load(drivef)",
"_____no_output_____"
]
],
[
[
"OR the summary of the papers",
"_____no_output_____"
]
],
[
[
"#@title Select number of summarized papers that will be feeded in the model { vertical-output: true, display-mode: \"both\" }\nnumber_of_papers = \"9000\" #@param [\"1000\", \"3000\", \"6000\", \"9000\"]\nimport pickle\n\nCORD19_Dataframe = r\"/content/drive/My Drive/AI_4/CORD19_SentenceMap_Summarized_\"+number_of_papers+\".pkl\"\nwith open(CORD19_Dataframe, 'rb') as drivef:\n CORD19Dictionary = pickle.load(drivef)",
"_____no_output_____"
]
],
[
[
"## Queries\n---",
"_____no_output_____"
]
],
[
[
"query_list = [\n 'What are the coronoviruses?',\n 'What was discovered in Wuhuan in December 2019?',\n 'What is Coronovirus Disease 2019?',\n 'What is COVID-19?',\n 'What is caused by SARS-COV2?', 'How is COVID-19 spread?',\n 'Where was COVID-19 discovered?','How does coronavirus spread?'\n]\n\nproposed_answers = [\n 'Coronaviruses (CoVs) are common human and animal pathogens that can transmit zoonotically and cause severe respiratory disease syndromes. ',\n 'In December 2019, a novel coronavirus, called COVID-19, was discovered in Wuhan, China, and has spread to different cities in China as well as to 24 other countries.',\n 'Coronavirus Disease 2019 (COVID-19) is an emerging disease with a rapid increase in cases and deaths since its first identification in Wuhan, China, in December 2019.',\n 'COVID-19 is a viral respiratory illness caused by a new coronavirus called SARS-CoV-2.',\n 'Coronavirus disease (COVID-19) is caused by SARS-COV2 and represents the causative agent of a potentially fatal disease that is of great global public health concern.', \n 'First, although COVID-19 is spread by the airborne route, air disinfection of cities and communities is not known to be effective for disease control and needs to be stopped.',\n 'In December 2019, a novel coronavirus, called COVID-19, was discovered in Wuhan, China, and has spread to different cities in China as well as to 24 other countries.',\n 'The new coronavirus was reported to spread via droplets, contact and natural aerosols from human-to-human.'\n]\n\nmyquery_list = [\n \"How long can the coronavirus survive on surfaces?\",\n \"What means COVID-19?\",\n \"Is COVID19 worse than flue?\",\n \"When the vaccine will be ready?\",\n \"Whats the proteins that consist COVID-19?\",\n \"Whats the symptoms of COVID-19?\",\n \"How can I prevent COVID-19?\",\n \"What treatments are available for COVID-19?\",\n \"Is hand sanitizer effective against COVID-19?\",\n \"Am I at risk for serious complications from COVID-19 if I smoke cigarettes?\",\n \"Are there any FDA-approved drugs (medicines) for COVID-19?\",\n \"How are people tested?\",\n \"Why is the disease being called coronavirus disease 2019, COVID-19?\",\n \"Am I at risk for COVID-19 from mail, packages, or products?\",\n \"What is community spread?\",\n \"How can I protect myself?\",\n \"What is a novel coronavirus?\",\n \"Was Harry Potter a good magician?\"\n]",
"_____no_output_____"
]
],
[
[
"# Results dataframes",
"_____no_output_____"
]
],
[
[
"resultsDf = pd.DataFrame(columns=['Number of papers','Embeddings creation time'])\n\nqueriesDf = pd.DataFrame(columns=['Query','Proposed_answer','Model_answer','Cosine_similarity'])\nqueriesDf['Query'] = query_list\nqueriesDf['Proposed_answer'] = proposed_answers\n\nmyQueriesDf = pd.DataFrame(columns=['Query','Model_answer','Cosine_similarity'])\nmyQueriesDf['Query'] = myquery_list\n\nqueriesDf",
"_____no_output_____"
]
],
[
[
"# SBERT\n---",
"_____no_output_____"
]
],
[
[
"!pip install -U sentence-transformers",
"Requirement already up-to-date: sentence-transformers in /usr/local/lib/python3.6/dist-packages (0.4.1.2)\nRequirement already satisfied, skipping upgrade: torch>=1.6.0 in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (1.7.0+cu101)\nRequirement already satisfied, skipping upgrade: numpy in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (1.19.5)\nRequirement already satisfied, skipping upgrade: tqdm in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (4.41.1)\nRequirement already satisfied, skipping upgrade: sentencepiece in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (0.1.95)\nRequirement already satisfied, skipping upgrade: transformers<5.0.0,>=3.1.0 in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (4.3.2)\nRequirement already satisfied, skipping upgrade: scikit-learn in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (0.22.2.post1)\nRequirement already satisfied, skipping upgrade: nltk in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (3.2.5)\nRequirement already satisfied, skipping upgrade: scipy in /usr/local/lib/python3.6/dist-packages (from sentence-transformers) (1.4.1)\nRequirement already satisfied, skipping upgrade: future in /usr/local/lib/python3.6/dist-packages (from torch>=1.6.0->sentence-transformers) (0.16.0)\nRequirement already satisfied, skipping upgrade: dataclasses in /usr/local/lib/python3.6/dist-packages (from torch>=1.6.0->sentence-transformers) (0.8)\nRequirement already satisfied, skipping upgrade: typing-extensions in /usr/local/lib/python3.6/dist-packages (from torch>=1.6.0->sentence-transformers) (3.7.4.3)\nRequirement already satisfied, skipping upgrade: sacremoses in /usr/local/lib/python3.6/dist-packages (from transformers<5.0.0,>=3.1.0->sentence-transformers) (0.0.43)\nRequirement already satisfied, skipping upgrade: regex!=2019.12.17 in /usr/local/lib/python3.6/dist-packages (from transformers<5.0.0,>=3.1.0->sentence-transformers) (2019.12.20)\nRequirement already satisfied, skipping upgrade: requests in /usr/local/lib/python3.6/dist-packages (from transformers<5.0.0,>=3.1.0->sentence-transformers) (2.23.0)\nRequirement already satisfied, skipping upgrade: tokenizers<0.11,>=0.10.1 in /usr/local/lib/python3.6/dist-packages (from transformers<5.0.0,>=3.1.0->sentence-transformers) (0.10.1)\nRequirement already satisfied, skipping upgrade: packaging in /usr/local/lib/python3.6/dist-packages (from transformers<5.0.0,>=3.1.0->sentence-transformers) (20.9)\nRequirement already satisfied, skipping upgrade: importlib-metadata; python_version < \"3.8\" in /usr/local/lib/python3.6/dist-packages (from transformers<5.0.0,>=3.1.0->sentence-transformers) (3.4.0)\nRequirement already satisfied, skipping upgrade: filelock in /usr/local/lib/python3.6/dist-packages (from transformers<5.0.0,>=3.1.0->sentence-transformers) (3.0.12)\nRequirement already satisfied, skipping upgrade: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn->sentence-transformers) (1.0.0)\nRequirement already satisfied, skipping upgrade: six in /usr/local/lib/python3.6/dist-packages (from nltk->sentence-transformers) (1.15.0)\nRequirement already satisfied, skipping upgrade: click in /usr/local/lib/python3.6/dist-packages (from sacremoses->transformers<5.0.0,>=3.1.0->sentence-transformers) (7.1.2)\nRequirement already satisfied, skipping upgrade: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->transformers<5.0.0,>=3.1.0->sentence-transformers) (3.0.4)\nRequirement already satisfied, skipping upgrade: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->transformers<5.0.0,>=3.1.0->sentence-transformers) (1.24.3)\nRequirement already satisfied, skipping upgrade: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->transformers<5.0.0,>=3.1.0->sentence-transformers) (2020.12.5)\nRequirement already satisfied, skipping upgrade: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->transformers<5.0.0,>=3.1.0->sentence-transformers) (2.10)\nRequirement already satisfied, skipping upgrade: pyparsing>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from packaging->transformers<5.0.0,>=3.1.0->sentence-transformers) (2.4.7)\nRequirement already satisfied, skipping upgrade: zipp>=0.5 in /usr/local/lib/python3.6/dist-packages (from importlib-metadata; python_version < \"3.8\"->transformers<5.0.0,>=3.1.0->sentence-transformers) (3.4.0)\n"
]
],
[
[
"# Selecting transformer and Cross Encoder",
"_____no_output_____"
]
],
[
[
"from sentence_transformers import SentenceTransformer, util, CrossEncoder\nimport torch\nimport time\n\nencoder = SentenceTransformer('msmarco-distilbert-base-v2')\ncross_encoder = CrossEncoder('cross-encoder/ms-marco-TinyBERT-L-6')",
"_____no_output_____"
]
],
[
[
"# Initializing corpus",
"_____no_output_____"
]
],
[
[
"corpus = list(CORD19Dictionary.keys())",
"_____no_output_____"
]
],
[
[
"# Creating the embeddings",
"_____no_output_____"
],
[
"Encoding the papers",
"_____no_output_____"
]
],
[
[
"%%time\ncorpus_embeddings = encoder.encode(corpus, convert_to_tensor=True, show_progress_bar=True,device='cuda')",
"_____no_output_____"
]
],
[
[
"# Saving corpus as tensors to drive",
"_____no_output_____"
]
],
[
[
"corpus_embeddings_path = r\"/content/drive/My Drive/AI_4/corpus_embeddings_6000_CrossEncoder.pt\"\ntorch.save(corpus_embeddings,corpus_embeddings_path)",
"_____no_output_____"
]
],
[
[
"# Loading embeddings if have been created and saved\n\n\n\n---",
"_____no_output_____"
]
],
[
[
"corpus_embeddings_path = r\"/content/drive/My Drive/AI_4/corpus_embeddings_6000_CrossEncoder.pt\"\nwith open(corpus_embeddings_path, 'rb') as f:\n corpus_embeddings = torch.load(f)",
"_____no_output_____"
]
],
[
[
"# Evaluation\n---\n",
"_____no_output_____"
]
],
[
[
"import re\nfrom nltk import tokenize\nfrom termcolor import colored\n\n\ndef paperTitle(answer,SentenceMap):\n record = SentenceMap[answer]\n print(\"Paper title:\",record[1])\n print(\"Paper id: \",record[0]) \n \ndef evaluation(query_list,top_k,resultsDf):\n query_answers = []\n scores = []\n\n for query in query_list:\n #Encode the query using the bi-encoder and find potentially relevant corpus\n start_time = time.time()\n question_embedding = encoder.encode(query, convert_to_tensor=True,device='cuda')\n hits = util.semantic_search(question_embedding, corpus_embeddings, top_k=top_k)\n hits = hits[0] # Get the hits for the first query\n\n #Now, score all retrieved corpus with the cross_encoder\n cross_inp = [[query, corpus[hit['corpus_id']]] for hit in hits]\n cross_scores = cross_encoder.predict(cross_inp)\n \n #Sort results by the cross-encoder scores\n for idx in range(len(cross_scores)):\n hits[idx]['cross-score'] = cross_scores[idx]\n\n hits = sorted(hits, key=lambda x: x['cross-score'], reverse=True)\n end_time = time.time()\n\n #Output of top-5 hits\n print(\"\\n\\n======================\\n\\n\")\n print(\"Query:\",colored(query,'green') )\n \n print(\"Results (after {:.3f} seconds):\".format(end_time - start_time))\n iter=0\n for hit in hits[0:top_k]:\n print(\"\\n-> \",iter+1)\n answer = ' '.join([re.sub(r\"^\\[.*\\]\", \"\", x) for x in corpus[hit['corpus_id']].split()])\n if len(tokenize.word_tokenize(answer)) > 1:\n print(\"Score: {:.4f}\".format(hit['cross-score']))\n \n paperTitle(corpus[hit['corpus_id']],CORD19Dictionary)\n print(\"Anser size: \",len(tokenize.word_tokenize(answer)))\n print(\"Anser: \")\n if iter==0:\n query_answers.append(answer)\n scores.append(hit['cross-score'].item())\n iter+=1\n print(colored(answer,'yellow'))\n \n \n resultsDf['Model_answer'] = query_answers\n resultsDf['Cosine_similarity'] = scores\n",
"_____no_output_____"
],
[
"top_k = 3\nevaluation(query_list,top_k,queriesDf)",
"\n\n======================\n\n\nQuery: \u001b[32mWhat are the coronoviruses?\u001b[0m\nResults (after 0.839 seconds):\n\n-> 1\nScore: 0.0639\nPaper title: Citation: Interactions Between Enteroviruses and the Inflammasome: New Insights Into Viral Pathogenesis\nPaper id: 423e1f15afb86012057acacc26d0766aa4bc582a\nAnser size: 7\nAnser: \n\u001b[33mEnteroviruses are the members of Picornaviridae.\u001b[0m\n\n-> 2\nScore: 0.0185\nPaper title: Full Genome Virus Detection in Fecal Samples Using Sensitive Nucleic Acid Preparation, Deep Sequencing, and a Novel Iterative Sequence Classification Algorithm\nPaper id: ab98d1b125aa0704e63adef426b27abd32e935f0\nAnser size: 14\nAnser: \n\u001b[33mCosavirus is a new genus in the Picornaviridae family first described in 2008 .\u001b[0m\n\n-> 3\nScore: 0.0073\nPaper title: Identification of diverse viruses in upper respiratory samples in dromedary camels from United Arab Emirates\nPaper id: 04b5f15cca91a7b810216682780f8ea6e1ab3046\nAnser size: 2\nAnser: \n\u001b[33mOrthonairoviruses.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat was discovered in Wuhuan in December 2019?\u001b[0m\nResults (after 0.525 seconds):\n\n-> 1\nScore: 0.7336\nPaper title: Transmission routes of 2019-nCoV and controls in dental practice\nPaper id: 9756bb3c608ed790d2306fc8db815a694eeca45f\nAnser size: 16\nAnser: \n\u001b[33mAn emergent pneumonia outbreak originated in Wuhan City, in the late December 2019 1 .\u001b[0m\n\n-> 2\nScore: 0.0006\nPaper title: Molecular Sciences Effects of AntagomiRs on Different Lung Diseases in Human, Cellular, and Animal Models\nPaper id: 3aed588044335032787a5eb91ee61afadcd4a006\nAnser size: 6\nAnser: \n\u001b[33m2019 or Liu et al.,\u001b[0m\n\n-> 3\nScore: 0.0002\nPaper title: Estimated effectiveness of symptom and risk screening to prevent the spread of COVID-19\nPaper id: a70e7c4d8ee484ce956e91c8700d0c9310bbdbbc\nAnser size: 6\nAnser: \n\u001b[33m2020; Liu et al.,\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat is Coronovirus Disease 2019?\u001b[0m\nResults (after 0.523 seconds):\n\n-> 1\nScore: 0.6516\nPaper title: \nPaper id: af000c5a8e181550fd16291e5d4f0f70ca9161a1\nAnser size: 12\nAnser: \n\u001b[33mCOVID-19: coronavirus disease 2019; PPE: personal protective equipment.\u001b[0m\n\n-> 2\nScore: 0.2219\nPaper title: \nPaper id: 19ff77e874c0706f794908e9b6878314671d385a\nAnser size: 9\nAnser: \n\u001b[33mNaming 2019-nCoV as SARS-CoV-2 is therefore truly misleading.\u001b[0m\n\n-> 3\nScore: 0.1593\nPaper title: \nPaper id: 82210c1cb5ac59acd1468cedcaf6fb8d951f4903\nAnser size: 14\nAnser: \n\u001b[33mThe infective pathogen was later identified as a novel coronavirus, called 2019-nCoV .\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat is COVID-19?\u001b[0m\nResults (after 0.551 seconds):\n\n-> 1\nScore: 0.9631\nPaper title: \nPaper id: af000c5a8e181550fd16291e5d4f0f70ca9161a1\nAnser size: 12\nAnser: \n\u001b[33mCOVID-19: coronavirus disease 2019; PPE: personal protective equipment.\u001b[0m\n\n-> 2\nScore: 0.6902\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n-> 3\nScore: 0.1879\nPaper title: Clinical Medicine Incubation Period and Other Epidemiological Characteristics of 2019 Novel Coronavirus Infections with Right Truncation: A Statistical Analysis of Publicly Available Case Data\nPaper id: 210a892deb1c61577f6fba58505fd65356ce6636\nAnser size: 16\nAnser: \n\u001b[33mIt remains to be seen if this will be the case for COVID-19 as well.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat is caused by SARS-COV2?\u001b[0m\nResults (after 0.551 seconds):\n\n-> 1\nScore: 0.3216\nPaper title: \nPaper id: 0eb44c0cc59184754a0a2cd8ee3c8b2302a8927c\nAnser size: 13\nAnser: \n\u001b[33mWe thus assumed that a SARS-related CoV is involved in the outbreak.\u001b[0m\n\n-> 2\nScore: 0.0962\nPaper title: Middle East respiratory syndrome coronavirus infection: virus-host cell interactions and implications on pathogenesis\nPaper id: 12f712c348c26e092759d804778defe2d2d4af6f\nAnser size: 7\nAnser: \n\u001b[33mThe SARS-CoV can infect human macrophages.\u001b[0m\n\n-> 3\nScore: 0.0673\nPaper title: Potential Factors Influencing Repeated SARS Outbreaks in China\nPaper id: 655537fc8cc52bccf43cf7189ab060d3097caa7a\nAnser size: 12\nAnser: \n\u001b[33mThe risk of SARS-CoV-2 infection will remain for a long time.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mHow is COVID-19 spread?\u001b[0m\nResults (after 0.547 seconds):\n\n-> 1\nScore: 0.9799\nPaper title: The novel coronavirus outbreak in Wuhan, China\nPaper id: 5ba8056230c17ec133169d79aacf61ed7d4b458b\nAnser size: 14\nAnser: \n\u001b[33mThe COVID-19 has then rapidly spread to all over China and the world.\u001b[0m\n\n-> 2\nScore: 0.9631\nPaper title: The novel coronavirus outbreak in Wuhan, China\nPaper id: 5ba8056230c17ec133169d79aacf61ed7d4b458b\nAnser size: 18\nAnser: \n\u001b[33mIt is found that the COVID-19 can be transmitted through droplets, contact, aerosol, etc.\u001b[0m\n\n-> 3\nScore: 0.2594\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhere was COVID-19 discovered?\u001b[0m\nResults (after 0.530 seconds):\n\n-> 1\nScore: 0.9490\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 17\nAnser: \n\u001b[33mThe epidemic of COVID-19 is caused by a novel virus first detected in Wuhan, China.\u001b[0m\n\n-> 2\nScore: 0.4963\nPaper title: The novel coronavirus outbreak in Wuhan, China\nPaper id: 5ba8056230c17ec133169d79aacf61ed7d4b458b\nAnser size: 14\nAnser: \n\u001b[33mThe COVID-19 has then rapidly spread to all over China and the world.\u001b[0m\n\n-> 3\nScore: 0.0428\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mHow does coronavirus spread?\u001b[0m\nResults (after 0.548 seconds):\n\n-> 1\nScore: 0.1335\nPaper title: Analysis of the codon usage pattern in Middle East Respiratory Syndrome Coronavirus\nPaper id: 627ada5c21fb8d0e43b37999fa66bf41ca36c353\nAnser size: 13\nAnser: \n\u001b[33mThis may hint that coronavirus does not spread so widely in humans.\u001b[0m\n\n-> 2\nScore: 0.0041\nPaper title: Tropical Medicine and Infectious Disease Potential Intermediate Hosts for Coronavirus Transmission: No Evidence of Clade 2c Coronaviruses in Domestic Livestock from Ghana\nPaper id: 95cc317541d97e3dbaa1662894fdbed842098910\nAnser size: 2\nAnser: \n\u001b[33mcoronavirus.\u001b[0m\n\n-> 3\nScore: 0.0041\nPaper title: Population genetics, community of parasites, and resistance to rodenticides in an urban brown rat (Rattus norvegicus) population\nPaper id: c8d60caf44017989b3b9633350fc1d2efda570a5\nAnser size: 2\nAnser: \n\u001b[33mCoronavirus.\u001b[0m\n"
],
[
"top_k = 3\nevaluation(myquery_list,top_k,myQueriesDf)",
"\n\n======================\n\n\nQuery: \u001b[32mHow long can the coronavirus survive on surfaces?\u001b[0m\nResults (after 0.537 seconds):\n\n-> 1\nScore: 0.9850\nPaper title: Outbreak of Novel Coronavirus (SARS-Cov-2): First Evidences From International Scientific Literature and Pending Questions\nPaper id: 7b7c71218f8d7ea1a1f8f702e4262b839bf7cc8a\nAnser size: 15\nAnser: \n\u001b[33mOn inanimate surfaces, human coronaviruses can remain infectious for up to 9 days.\u001b[0m\n\n-> 2\nScore: 0.7655\nPaper title: Characterisation of the canine faecal virome in healthy dogs and dogs with acute diarrhoea using shotgun metagenomics\nPaper id: fcb1ba715b2516823fee057cbb0f8276c76d19d7\nAnser size: 21\nAnser: \n\u001b[33mCanine coronavirus can be shed in faeces in high numbers for up to 156 days [44, 45] .\u001b[0m\n\n-> 3\nScore: 0.1069\nPaper title: Human Coronaviruses: Insights into Environmental Resistance and Its Influence on the Development of New Antiseptic Strategies\nPaper id: d171f82b892a2afafc2bc8a5458219dc04c8fd8d\nAnser size: 21\nAnser: \n\u001b[33mHuman coronavirus infections occur mainly in winter, with a short incubation time [19, 23, 24] .\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat means COVID-19?\u001b[0m\nResults (after 0.534 seconds):\n\n-> 1\nScore: 0.9272\nPaper title: \nPaper id: af000c5a8e181550fd16291e5d4f0f70ca9161a1\nAnser size: 12\nAnser: \n\u001b[33mCOVID-19: coronavirus disease 2019; PPE: personal protective equipment.\u001b[0m\n\n-> 2\nScore: 0.7040\nPaper title: \nPaper id: 19ff77e874c0706f794908e9b6878314671d385a\nAnser size: 13\nAnser: \n\u001b[33mThe new name is also not consistent with the disease name COVID-19.\u001b[0m\n\n-> 3\nScore: 0.5282\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mIs COVID19 worse than flue?\u001b[0m\nResults (after 0.521 seconds):\n\n-> 1\nScore: 0.0644\nPaper title: Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV\nPaper id: f294f0df7468a8ac9e27776cc15fa20297a9f040\nAnser size: 11\nAnser: \n\u001b[33mIn comparison, COVID-19 showed similar trends with SARS patients .\u001b[0m\n\n-> 2\nScore: 0.0359\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n-> 3\nScore: 0.0022\nPaper title: The effect of corticosteroids on mortality of patients with influenza pneumonia: a systematic review and meta-analysis\nPaper id: ff220214e91fabc8d302d1605cd9bac44fac507f\nAnser size: 10\nAnser: \n\u001b[33mCorticosteroids could increase mortality in patients with influenza pneumonia.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhen the vaccine will be ready?\u001b[0m\nResults (after 0.536 seconds):\n\n-> 1\nScore: 0.2100\nPaper title: Mast cells and influenza A virus: association with allergic responses and beyond\nPaper id: 29621887690af716dac0c244eeb95bce74fa8755\nAnser size: 10\nAnser: \n\u001b[33mCurrent vaccine strategies take approximately 6 months for production.\u001b[0m\n\n-> 2\nScore: 0.1330\nPaper title: Rapid and simple colorimetric detection of multiple influenza viruses infecting humans using a reverse transcriptional loop- mediated isothermal amplification (RT-LAMP) diagnostic platform\nPaper id: dd12c39ca963dca8336d7f30c8842d892ec8236c\nAnser size: 15\nAnser: \n\u001b[33mHowever, vaccine production usually takes 6-12 months to prepare for newly emerging viruses.\u001b[0m\n\n-> 3\nScore: 0.1195\nPaper title: Vaccination to Conserved Influenza Antigens in Mice Using a Novel Simian Adenovirus Vector, PanAd3, Derived from the Bonobo Pan paniscus\nPaper id: 532e417a66dbe4822a3f8f9b496c105ccc7dd412\nAnser size: 15\nAnser: \n\u001b[33mNew vaccines are often required, and take about 6 months to become available .\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhats the proteins that consist COVID-19?\u001b[0m\nResults (after 0.532 seconds):\n\n-> 1\nScore: 0.0004\nPaper title: Integrin b3 Is Required in Infection and Proliferation of Classical Swine Fever Virus\nPaper id: e50473adb66bac4a176d80051d63f415d2dbd5a8\nAnser size: 14\nAnser: \n\u001b[33mCSFV contains 4 structural proteins: C, Erns, E1 and E2.\u001b[0m\n\n-> 2\nScore: 0.0004\nPaper title: Proteome and phosphoproteome analysis of honeybee (Apis mellifera) venom collected from electrical stimulation and manual extraction of the venom gland Proteome and phosphoproteome analysis of honeybee (Apis mellifera) venom collected from electrical stimulation and manual extraction of the venom gland\nPaper id: 1c8a0fb2f60c243f71d16d5fefb5b51d7978869e\nAnser size: 15\nAnser: \n\u001b[33mIn GV, 27 proteins were specifically expressed: 4 toxins and 23 non-toxins.\u001b[0m\n\n-> 3\nScore: 0.0002\nPaper title: The Interplay between Dengue Virus and the Human Innate Immune System: A Game of Hide and Seek\nPaper id: ac8b5e9b4a49a1062eddf4fc48a19778e66e9a78\nAnser size: 3\nAnser: \n\u001b[33mThis proteins.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhats the symptoms of COVID-19?\u001b[0m\nResults (after 0.525 seconds):\n\n-> 1\nScore: 0.1345\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n-> 2\nScore: 0.0006\nPaper title: Characterization of Host and Bacterial Contributions to Lung Barrier Dysfunction Following Co-infection with 2009 Pandemic Influenza and Methicillin Resistant Staphylococcus aureus\nPaper id: edee1fd45587a0a71d88a5db58cc81342840e2f6\nAnser size: 5\nAnser: \n\u001b[33mInitial signs and symptoms include\u001b[0m\n\n-> 3\nScore: 0.0002\nPaper title: Effect of Pullet Vaccination on Development and Longevity of Immunity\nPaper id: e192e65a6546583fe49086c4d3ac29a0620d5bd5\nAnser size: 2\nAnser: \n\u001b[33mClinical Signs\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mHow can I prevent COVID-19?\u001b[0m\nResults (after 0.531 seconds):\n\n-> 1\nScore: 0.2469\nPaper title: Clinical Medicine Incubation Period and Other Epidemiological Characteristics of 2019 Novel Coronavirus Infections with Right Truncation: A Statistical Analysis of Publicly Available Case Data\nPaper id: 210a892deb1c61577f6fba58505fd65356ce6636\nAnser size: 16\nAnser: \n\u001b[33mIt remains to be seen if this will be the case for COVID-19 as well.\u001b[0m\n\n-> 2\nScore: 0.1603\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n-> 3\nScore: 0.0029\nPaper title: Identify-Isolate-Inform: A Modified Tool for Initial Detection and Management of Middle East Respiratory Syndrome Patients in the Emergency Department\nPaper id: e8ae9d6178f8322e2f9b2453ef13bb312427bd15\nAnser size: 8\nAnser: \n\u001b[33mPrevention of MERS-CoV transmission involves avoiding exposure.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat treatments are available for COVID-19?\u001b[0m\nResults (after 0.531 seconds):\n\n-> 1\nScore: 0.9569\nPaper title: Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV\nPaper id: f294f0df7468a8ac9e27776cc15fa20297a9f040\nAnser size: 17\nAnser: \n\u001b[33mAs effective drugs for SARS, hormones and interferons can also be used to treat COVID-19 .\u001b[0m\n\n-> 2\nScore: 0.0802\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n-> 3\nScore: 0.0433\nPaper title: Clinical Medicine Incubation Period and Other Epidemiological Characteristics of 2019 Novel Coronavirus Infections with Right Truncation: A Statistical Analysis of Publicly Available Case Data\nPaper id: 210a892deb1c61577f6fba58505fd65356ce6636\nAnser size: 16\nAnser: \n\u001b[33mIt remains to be seen if this will be the case for COVID-19 as well.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mIs hand sanitizer effective against COVID-19?\u001b[0m\nResults (after 0.534 seconds):\n\n-> 1\nScore: 0.2058\nPaper title: Respiratory viral infections in children with asthma: do they matter and can we prevent them?\nPaper id: bbc2824ce7dff3d23d060b7abbe96cba28095fb8\nAnser size: 15\nAnser: \n\u001b[33mThe use of alcohol-based hand sanitizers is also effective [54, 55] .\u001b[0m\n\n-> 2\nScore: 0.0120\nPaper title: Cell Discovery Phase-adjusted estimation of the number of Coronavirus Disease 2019 cases in Wuhan, China\nPaper id: 6abb30ae61aa5e41f16a28b9437940d5d76d745b\nAnser size: 19\nAnser: \n\u001b[33mIn response to the outbreak of COVID-19, a series of prompt public health measures have been taken.\u001b[0m\n\n-> 3\nScore: 0.0036\nPaper title: \nPaper id: af000c5a8e181550fd16291e5d4f0f70ca9161a1\nAnser size: 12\nAnser: \n\u001b[33mCOVID-19: coronavirus disease 2019; PPE: personal protective equipment.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mAm I at risk for serious complications from COVID-19 if I smoke cigarettes?\u001b[0m\nResults (after 0.523 seconds):\n\n-> 1\nScore: 0.0151\nPaper title: Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV\nPaper id: f294f0df7468a8ac9e27776cc15fa20297a9f040\nAnser size: 16\nAnser: \n\u001b[33mreported that people who have not been exposed to SARS-CoV-2 are all susceptible to COVID-19 .\u001b[0m\n\n-> 2\nScore: 0.0042\nPaper title: Clinical Medicine Incubation Period and Other Epidemiological Characteristics of 2019 Novel Coronavirus Infections with Right Truncation: A Statistical Analysis of Publicly Available Case Data\nPaper id: 210a892deb1c61577f6fba58505fd65356ce6636\nAnser size: 16\nAnser: \n\u001b[33mIt remains to be seen if this will be the case for COVID-19 as well.\u001b[0m\n\n-> 3\nScore: 0.0014\nPaper title: \nPaper id: af000c5a8e181550fd16291e5d4f0f70ca9161a1\nAnser size: 12\nAnser: \n\u001b[33mCOVID-19: coronavirus disease 2019; PPE: personal protective equipment.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mAre there any FDA-approved drugs (medicines) for COVID-19?\u001b[0m\nResults (after 0.519 seconds):\n\n-> 1\nScore: 0.0156\nPaper title: \nPaper id: 19ff77e874c0706f794908e9b6878314671d385a\nAnser size: 13\nAnser: \n\u001b[33mThe new name is also not consistent with the disease name COVID-19.\u001b[0m\n\n-> 2\nScore: 0.0143\nPaper title: Clinical Medicine Incubation Period and Other Epidemiological Characteristics of 2019 Novel Coronavirus Infections with Right Truncation: A Statistical Analysis of Publicly Available Case Data\nPaper id: 210a892deb1c61577f6fba58505fd65356ce6636\nAnser size: 16\nAnser: \n\u001b[33mIt remains to be seen if this will be the case for COVID-19 as well.\u001b[0m\n\n-> 3\nScore: 0.0028\nPaper title: Human Ebola virus infection in West Africa: a review of available therapeutic agents that target different steps of the life cycle of Ebola virus-mutable host cell therapeutic targets for Ebola virus, Cocktail therapeutic intervention for RNA virus Multilingual abstract\nPaper id: 06190bfcbc53a5d5d17e0a60a3a0f6488d8ae1db\nAnser size: 11\nAnser: \n\u001b[33mThese medications are FDA-approved for the treatment of other diseases.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mHow are people tested?\u001b[0m\nResults (after 0.520 seconds):\n\n-> 1\nScore: 0.0092\nPaper title: A Human DPP4-Knockin Mouse's Susceptibility to Infection by Authentic and Pseudotyped MERS-CoV\nPaper id: 874e540a730ee1060365af8d2caa03f537508e33\nAnser size: 11\nAnser: \n\u001b[33mStudent's t-tests were used to assess differences between groups.\u001b[0m\n\n-> 2\nScore: 0.0075\nPaper title: Virology Journal A focus reduction neutralization assay for hepatitis C virus neutralizing antibodies\nPaper id: ee8dca216514deeed4c9415bc2ad8a78dc3d9670\nAnser size: 11\nAnser: \n\u001b[33mStudent's t-test was used to compare data between groups.\u001b[0m\n\n-> 3\nScore: 0.0006\nPaper title: Ecohealth research in Southeast Asia: past, present and the way forward\nPaper id: 1495c1fa93db3b9a5d12b5ae15ff0c8639b83452\nAnser size: 8\nAnser: \n\u001b[33mHow can these be tested in practice?\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhy is the disease being called coronavirus disease 2019, COVID-19?\u001b[0m\nResults (after 0.528 seconds):\n\n-> 1\nScore: 0.9373\nPaper title: \nPaper id: af000c5a8e181550fd16291e5d4f0f70ca9161a1\nAnser size: 12\nAnser: \n\u001b[33mCOVID-19: coronavirus disease 2019; PPE: personal protective equipment.\u001b[0m\n\n-> 2\nScore: 0.9238\nPaper title: Cell Discovery Phase-adjusted estimation of the number of Coronavirus Disease 2019 cases in Wuhan, China\nPaper id: 6abb30ae61aa5e41f16a28b9437940d5d76d745b\nAnser size: 20\nAnser: \n\u001b[33mWorld Health Organization (WHO) now has named the disease Coronavirus Disease 2019 (COVID- 19) 3 .\u001b[0m\n\n-> 3\nScore: 0.8515\nPaper title: \nPaper id: 82210c1cb5ac59acd1468cedcaf6fb8d951f4903\nAnser size: 14\nAnser: \n\u001b[33mThe infective pathogen was later identified as a novel coronavirus, called 2019-nCoV .\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mAm I at risk for COVID-19 from mail, packages, or products?\u001b[0m\nResults (after 0.520 seconds):\n\n-> 1\nScore: 0.1850\nPaper title: First two months of the 2019 Coronavirus Disease (COVID-19) epidemic in China: real- time surveillance and evaluation with a second derivative model\nPaper id: 469ed0f00c09e2637351c9735c306f27acf3aace\nAnser size: 8\nAnser: \n\u001b[33mThis is particularly true for the COVID-19.\u001b[0m\n\n-> 2\nScore: 0.0283\nPaper title: Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV\nPaper id: f294f0df7468a8ac9e27776cc15fa20297a9f040\nAnser size: 16\nAnser: \n\u001b[33mreported that people who have not been exposed to SARS-CoV-2 are all susceptible to COVID-19 .\u001b[0m\n\n-> 3\nScore: 0.0033\nPaper title: \nPaper id: af000c5a8e181550fd16291e5d4f0f70ca9161a1\nAnser size: 12\nAnser: \n\u001b[33mCOVID-19: coronavirus disease 2019; PPE: personal protective equipment.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat is community spread?\u001b[0m\nResults (after 0.526 seconds):\n\n-> 1\nScore: 0.2350\nPaper title: An Opportunistic Pathogen Afforded Ample Opportunities: Middle East Respiratory Syndrome Coronavirus\nPaper id: 32da24606ad160166f08cf05349eaadd580ccff0\nAnser size: 9\nAnser: \n\u001b[33mCommunity spread and subclinical transmission need more attention.\u001b[0m\n\n-> 2\nScore: 0.0135\nPaper title: People at Risk of Influenza Pandemics: The Evolution of Perception and Behavior\nPaper id: 51f8792fd26cd2c094c1b2d0e5539902fb6221da\nAnser size: 15\nAnser: \n\u001b[33mEfforts were then put into preventing spread of the disease at the community level.\u001b[0m\n\n-> 3\nScore: 0.0002\nPaper title: Comparative Analysis of the Effectiveness of Three Immunization Strategies in Controlling Disease Outbreaks in Realistic Social Networks\nPaper id: b16b23aad25d88c3af9ccd50b754cd4d9e8762fe\nAnser size: 3\nAnser: \n\u001b[33mCommunity-Bridge Immunization.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mHow can I protect myself?\u001b[0m\nResults (after 0.529 seconds):\n\n-> 1\nScore: 0.0502\nPaper title: BMC Public Health Healthcare workers' attitudes to working during pandemic influenza: a qualitative study\nPaper id: c337fa83ebb25e4600c0f9333ee0cb0fa938e947\nAnser size: 8\nAnser: \n\u001b[33mGet as much protection as you can.\u001b[0m\n\n-> 2\nScore: 0.0354\nPaper title: Need of surveillance response systems to combat Ebola outbreaks and other emerging infectious diseases in African countries\nPaper id: 70f3c90a651224f9292378da905af4ec635d5f43\nAnser size: 18\nAnser: \n\u001b[33mMoreover, people who don't have the knowledge should be educated on how to protect themselves.\u001b[0m\n\n-> 3\nScore: 0.0019\nPaper title: a Stakeholder Survey on live Bird market closures policy for controlling Highly pathogenic avian influenza in Vietnam\nPaper id: 17fe16cf66ebbe693a2e75dda11d14513fec7519\nAnser size: 13\nAnser: \n\u001b[33mTo mitigate these risks, the following safeguards were put in place.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWhat is a novel coronavirus?\u001b[0m\nResults (after 0.520 seconds):\n\n-> 1\nScore: 0.1118\nPaper title: Population genetics, community of parasites, and resistance to rodenticides in an urban brown rat (Rattus norvegicus) population\nPaper id: c8d60caf44017989b3b9633350fc1d2efda570a5\nAnser size: 2\nAnser: \n\u001b[33mCoronavirus.\u001b[0m\n\n-> 2\nScore: 0.1118\nPaper title: Tropical Medicine and Infectious Disease Potential Intermediate Hosts for Coronavirus Transmission: No Evidence of Clade 2c Coronaviruses in Domestic Livestock from Ghana\nPaper id: 95cc317541d97e3dbaa1662894fdbed842098910\nAnser size: 2\nAnser: \n\u001b[33mcoronavirus.\u001b[0m\n\n-> 3\nScore: 0.0265\nPaper title: Retargeting of Viruses to Generate Oncolytic Agents\nPaper id: bd44d72a9c41b1c382bd180da10a1f7ef38d2d56\nAnser size: 2\nAnser: \n\u001b[33mCoronaviruses.\u001b[0m\n\n\n======================\n\n\nQuery: \u001b[32mWas Harry Potter a good magician?\u001b[0m\nResults (after 0.515 seconds):\n\n-> 1\nScore: 0.0002\nPaper title: Immunoproteomic analysis of bacterial proteins of Actinobacillus pleuropneumoniae serotype 1\nPaper id: 47fb645312a069e65bb7557c58204244c3c92953\nAnser size: 12\nAnser: \n\u001b[33mThe vaccines elicited humoral immune responses and protective efficacy in mice .\u001b[0m\n\n-> 2\nScore: 0.0002\nPaper title: \nPaper id: fa137f1562d599f03605b83bc68f91e5105110d9\nAnser size: 5\nAnser: \n\u001b[33mEhrlich's magic bullet.\u001b[0m\n\n-> 3\nScore: 0.0002\nPaper title: Knowledge and attitudes of university students toward pandemic influenza: a cross-sectional study from Turkey\nPaper id: 545def8771357b4cb2875f5795a0760e97534cc9\nAnser size: 12\nAnser: \n\u001b[33mSurprisingly, a higher proportion believed that herbal remedies were effective.\u001b[0m\n"
]
],
[
[
"# Overall results",
"_____no_output_____"
],
[
"## 6000 papers with no summarization \n---\n\n### Time needed for creating the embeddings: \n- CPU times: \n - user 13min 10s\n - sys: 5min 40s\n - total: 18min 51s\n- Wall time: 18min 26s\n\n### Remarks\nBest results among the notebooks so far, almost 5/7 questions are answered and from mine 7/17. I expected better results since Cross Encoders enhance much the performance of Sentence Bert.\n\n__Top-k__ \n\nTop-2 and 3 have lots of answers, as I noticed that are better that the first one. Also good results and with some tunning would be nearly to the wanted.\n\n\n",
"_____no_output_____"
],
[
"### Results",
"_____no_output_____"
]
],
[
[
"with pd.option_context('display.max_colwidth', None):\n display(queriesDf)",
"_____no_output_____"
],
[
"with pd.option_context('display.max_colwidth', None):\n display(myQueriesDf)",
"_____no_output_____"
]
],
[
[
"## 9000 papers with no summarization \n---\n\nSession crashed due to RAM\n",
"_____no_output_____"
],
[
"## 6000 papers with paraphrase-distilroberta-base-v1 model and summarization \n---\n\n### Time needed for creating the embeddings: \n- CPU times: \n - user: 1min 18s\n - sys: 22.8 s\n - total: 1min 37s\n- Wall time: 1min 37s\n\n### Remarks\nNot good results. From these results I think that the BERT summarizer parameters were not the appropriate and I should experiment with them. I shouldn't have so strict summarization and I may over summarized the papers.\n\n\n__Top-k__ \n\nNot good.\n\n",
"_____no_output_____"
],
[
"### Results",
"_____no_output_____"
]
],
[
[
"with pd.option_context('display.max_colwidth', None):\n display(queriesDf)",
"_____no_output_____"
],
[
"with pd.option_context('display.max_colwidth', None):\n display(myQueriesDf)",
"_____no_output_____"
]
],
[
[
"## 9000 papers with summarization \n---\n\n### Time needed for creating the embeddings: \n- CPU times: \n - user: 1min 48s\n - sys: 32.6 s\n - total: 2min 20s\n- Wall time: 2min 16s\n\n### Remarks\nAgain not good results and this is due my summarization tunning.\n\n** Again I didn't have the time to re run and process again.\n",
"_____no_output_____"
],
[
"### Results",
"_____no_output_____"
]
],
[
[
"with pd.option_context('display.max_colwidth', None):\n display(queriesDf)",
"_____no_output_____"
],
[
"with pd.option_context('display.max_colwidth', None):\n display(myQueriesDf)",
"_____no_output_____"
]
],
[
[
"# References\n\n[1] https://colab.research.google.com/drive/1l6stpYdRMmeDBK_vw0L5NitdiAuhdsAr?usp=sharing#scrollTo=D_hDi8KzNgMM\n\n[2] https://www.sbert.net/docs/package_reference/cross_encoder.html",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbad0e43d59902adee21ba0c21447b08d1b541f9
| 8,290 |
ipynb
|
Jupyter Notebook
|
content/getting_started/setup.ipynb
|
nealcaren/textminingwithpython
|
dbfbd5cd9c4fe0002f1260656575e203e38e422b
|
[
"MIT"
] | 1 |
2020-05-15T17:03:31.000Z
|
2020-05-15T17:03:31.000Z
|
content/getting_started/setup.ipynb
|
nealcaren/textminingwithpython
|
dbfbd5cd9c4fe0002f1260656575e203e38e422b
|
[
"MIT"
] | null | null | null |
content/getting_started/setup.ipynb
|
nealcaren/textminingwithpython
|
dbfbd5cd9c4fe0002f1260656575e203e38e422b
|
[
"MIT"
] | null | null | null | 45.549451 | 729 | 0.672497 |
[
[
[
"# Setup\n\nBefore attending the workshp you should set up a scientific Python computing environment using the [Anaconda python distribution by Continuum Analytics](https://www.continuum.io/downloads). This page describes how. If this doesn't work, let [me](mailto:[email protected]) know and I will set you up with a virtual environment you can use on my server.\n\n",
"_____no_output_____"
],
[
"\n## Why Python?\nAs is true in human language, there are hundreds of computer programming languages. While each has its own merit, the major languages for scientific computing are C, C++, R, MATLAB, Python, Java, and Fortran. MATLAB and Python are similar in syntax and typically read as if they were written in plain english. This makes both languages a useful tool for teaching but they are also very powerful languages and are very actively used in real-life research. MATLAB is proprietary while Python is open source. A benefit of being open source is that anyone can write and release Python packages. For science, there are many wonderful community-driven packages such as NumPy, SciPy, scikit-image, and Pandas just to name a few.\n",
"_____no_output_____"
],
[
"## Installing Python 3.7 with Anaconda\n\nThere are several scientific Python distributions available for MacOS, Windows, and Linux. The most popular, [Anaconda](https://www.continuum.io/why-anaconda), is specifically designed for scientific computing and data science work. For this course, we will use the Anaconda Python 3.7 distribution. To install the correct version, follow the instructions below.\n1. Navigate to the [Anaconda download page](https://www.anaconda.com/distribution/) and download the Python 3.7 graphical installer.\n2. Launch the installer and follow the onscreen instructions.\n3. Congratulations! You now have the beginnings of a scientific Python distribution.",
"_____no_output_____"
],
[
"## What is a Jupyter notebook?\n\n[Jupyter](http://jupyter.org/) is a browser-based system to write code, math, and text in the same document so you can clearly explain the concepts and practices used in your program. Jupyter is not only for Python, but can be used with R, Juila, MATLAB, and about 35 other languages as of this writing. All files are saved as a [JSON](http://www.json.org/) formatted text file with the extension `.ipynb`.\n\n",
"_____no_output_____"
],
[
"## How to launch the notebook\n\nA Jupyter Notebook server can either be launched from the command line or from a GUI program installed along with anaconda called Navigator.\n\n",
"_____no_output_____"
],
[
"### Launching from the Anaconda Navigator\n\nInstalling Python 3 from Anaconda should also install a GUI application called [Anaconda Navigator](https://docs.continuum.io/anaconda/navigator). From here, you can launch several applications such as a QTconsole, the Spyder IDE, and a data visualization software called GlueViz. We are interested in the Jupyter Notebook application tab, which is shown boxed in red below:\n\n\n\nBy clicking on 'Launch', you will instantiate a Jupyter notebook server which should open in a new window.\n\n\n\n\n",
"_____no_output_____"
],
[
"### Launching from the terminal\n\nTo launch a notebook server from the command line, simply open a terminal emulator (Terminal.app on OSX or gitbash on windows) and navigate to the directory you would like to set up a server by typing `cd path/to/folder`\nOnce you are in the correct folder, you can launch a notebook server by typing:\n\n```\njupyter notebook\n```\n\nThis will open a screen in your default internet browser with a server containing your notebooks. Its address will be [`http://localhost:8888`](http://localhost:8888/) and is only available on your computer. **Note that once you start a server, you must keep the terminal window open.** This is where the 'guts' of the python kernel is.\n\n",
"_____no_output_____"
],
[
"## Interacting with the notebook\nIf everything launched correctly, you should be able to see a screen which looks something like this:\n\n\n\nTo start a new python window, click on the right-hand side of the application window and select `New`. This will give you a bunch of options for new notebook kernels. In the above screen shot, there are two available Python kernels and one Matlab kernel. When starting a notebook, you should choose `Python 3` if it is available. If you have just a tab that says \"Python\", choose that one.\n\nOnce you start a new notebook, you will be brought to the following screen.\n\n\n\nWelcome to the Jupyter notebook! There are many available buttons for you to click. However, the three most important components of the notebook are highlighted in colored boxes. In blue is the name of the notebook. By clicking this, you can rename the notebook. In red is the cell formatting assignment. By default, it is registered as code, but it can also be set to markdown as described later.\n\nFinally, in purple, is the code cell. In this cell, you can type an execute Python code as well as text that will be formatted in a nicely readable format.",
"_____no_output_____"
],
[
"## Writing code\n\nAll code you write in the notebook will be in the code cell. You can write single lines, to entire loops, to complete functions. As an example, we can write and evaluate a print statement in a code cell, as is shown below. To exectue the code, we can simply hit `shift + enter` while our cursor is in the code cell.\n",
"_____no_output_____"
]
],
[
[
"# This is a comment and is not read by Python\nprint('Hello! This is the print function. Python will print this line below')",
"_____no_output_____"
]
],
[
[
"The box with the gray background contains the python code while the output is in the box with the white background.\n ",
"_____no_output_____"
],
[
"## Next Steps\n\nNow that you have a Python environment up and running, proceed to the [Python] notebook to learn the basics of the language. ",
"_____no_output_____"
],
[
"*Note: This is a modified version of Griffin Chure's [Setting Up Python For Scientific Computing for Bi 1 - Principles of Biology](http://bi1.caltech.edu/code/t0a_setting_up_python.html). This work is licensed under a [Creative Commons Attribution License CC-BY 4.0](https://creativecommons.org/licenses/by/4.0/).*",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbad25ccd2ac9608824424866e51406d8139f20f
| 1,759 |
ipynb
|
Jupyter Notebook
|
tests/notebooks/exception.ipynb
|
lsst-sqre/sciencemonkey
|
02638bb883093c4c225251d32d0b01fe79778c2a
|
[
"MIT"
] | null | null | null |
tests/notebooks/exception.ipynb
|
lsst-sqre/sciencemonkey
|
02638bb883093c4c225251d32d0b01fe79778c2a
|
[
"MIT"
] | 26 |
2021-02-18T03:44:39.000Z
|
2022-03-21T19:34:52.000Z
|
tests/notebooks/exception.ipynb
|
lsst-sqre/mobu
|
3bf84601f287c8c34eb54f842d9ef25a88178e6a
|
[
"MIT"
] | null | null | null | 30.859649 | 548 | 0.550313 |
[
[
[
"This is a notebook that will throw an exception to test error handling and reporting.",
"_____no_output_____"
]
],
[
[
"foo = {\"bar\": \"baz\"}\nfoo[\"nothing\"]",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
cbad3b6a1d2cf46b5bc69056260c1abf0e94e7d7
| 306,858 |
ipynb
|
Jupyter Notebook
|
mcmc_retrievals_distributed.ipynb
|
simonpf/predictive_uncertainty
|
01a0b27f2902e57c49131d5c27faf5494318a064
|
[
"MIT"
] | 4 |
2018-07-30T01:59:48.000Z
|
2019-03-10T14:10:03.000Z
|
mcmc_retrievals_distributed.ipynb
|
simonpf/predictive_uncertainty
|
01a0b27f2902e57c49131d5c27faf5494318a064
|
[
"MIT"
] | null | null | null |
mcmc_retrievals_distributed.ipynb
|
simonpf/predictive_uncertainty
|
01a0b27f2902e57c49131d5c27faf5494318a064
|
[
"MIT"
] | 3 |
2018-07-30T02:00:52.000Z
|
2020-12-05T08:22:35.000Z
| 41.310985 | 1,405 | 0.613466 |
[
[
[
"### Distributed MCMC Retrieval\n\nThis notebook runs the MCMC retrievals on a local cluster using `ipyparallel`.\n",
"_____no_output_____"
]
],
[
[
"import ipyparallel as ipp\nc = ipp.Client(profile='gold')\nlview = c.load_balanced_view()",
"_____no_output_____"
]
],
[
[
"## Retrieval Setup",
"_____no_output_____"
]
],
[
[
"%%px\n%env ARTS_BUILD_PATH=/home/simonpf/build/arts\n%env ARTS_INCLUDE_PATH=/home/simonpf/src/atms_simulations/:/home/simonpf/src/arts/controlfiles\n%env ARTS_DATA_PATH=/home/simonpf/src/arts_xml/\n%env OMP_NUM_THREADS=1\n\nimport sys\nsys.path.insert(1,\"/home/simonpf/src/atms_simulations/\")\nsys.path.insert(1, \"/home/simonpf/src/typhon/\")\n\nimport os\nos.chdir(\"/home/simonpf/src/atms_simulations\")\n\n# This is important otherwise engines just crash.\nimport matplotlib; matplotlib.use(\"agg\")",
"_____no_output_____"
],
[
"from typhon.arts.workspace import Workspace\nimport atms\nimport numpy as np\n\nws = Workspace()\nchannels = [0,15,16,17,19]\natms.setup_atmosphere(ws)\natms.setup_sensor(ws, channels)\natms.checks(ws)\nws.yCalc()",
"Loading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[43647152]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[43647152]: {\nARTS[43647152]: - verbosityInit\nARTS[43647152]: - scat_speciesSet\nARTS[43647152]: - MatrixSet\nARTS[43647152]: - Tensor4SetConstant\nARTS[43647152]: - ArrayOfStringSet\nARTS[43647152]: - Touch\nARTS[43647152]: - FlagOff\nARTS[43647152]: - MatrixSet\nARTS[43647152]: - NumericSet\nARTS[43647152]: - ArrayOfStringSet\nARTS[43647152]: - Tensor3SetConstant\nARTS[43647152]: - Tensor3SetConstant\nARTS[43647152]: - Tensor3SetConstant\nARTS[43647152]: - Tensor3SetConstant\nARTS[43647152]: - Tensor3SetConstant\nARTS[43647152]: - Tensor3SetConstant\nARTS[43647152]: - IndexSet\nARTS[43647152]: - IndexSet\nARTS[43647152]: - IndexSet\nARTS[43647152]: - IndexSet\nARTS[43647152]: - FlagOff\nARTS[43647152]: - output_file_formatSetAscii\nARTS[43647152]: - StringSet\nARTS[43647152]: - IndexSet\nARTS[43647152]: - abs_lineshapeDefine\nARTS[43647152]: - NumericSet\nARTS[43647152]: - NumericSet\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - IndexSet\nARTS[43647152]: - IndexSet\nARTS[43647152]: - NumericSet\nARTS[43647152]: - NumericSet\nARTS[43647152]: - nlteOff\nARTS[43647152]: - partition_functionsInitFromBuiltin\nARTS[43647152]: - IndexSet\nARTS[43647152]: }\n\nARTS[43647152]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[43647152]: {\nARTS[43647152]: - abs_cont_descriptionInit\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: - abs_cont_descriptionAppend\nARTS[43647152]: }\n\nARTS[43647152]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[43647152]: {\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - FlagOff\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - FlagOff\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - AgendaCreate\nARTS[43647152]: - AgendaSet\nARTS[43647152]: }\n\nARTS[43647152]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[43647152]: {\nARTS[43647152]: - isotopologue_ratiosInitFromBuiltin\nARTS[43647152]: - refellipsoidEarth\nARTS[43647152]: - NumericSet\nARTS[43647152]: - AgendaSet\nARTS[43647152]: - NumericSet\nARTS[43647152]: }\n\nARTS[43647152]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[43647152]: {\nARTS[43647152]: - ArrayOfArrayOfIndexCreate\nARTS[43647152]: - ArrayOfIndexCreate\nARTS[43647152]: - VectorCreate\nARTS[43647152]: - ArrayOfIndexCreate\nARTS[43647152]: - NumericCreate\nARTS[43647152]: - VectorCreate\nARTS[43647152]: - IndexCreate\nARTS[43647152]: }\n\nARTS[43647152]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[43647152]: {\nARTS[43647152]: - MatrixSet\nARTS[43647152]: - MatrixSet\nARTS[43647152]: - ArrayOfStringSet\nARTS[43647152]: - VectorSet\nARTS[43647152]: - ArrayOfIndexSet\nARTS[43647152]: - VectorSet\nARTS[43647152]: - Extract\nARTS[43647152]: - nrowsGet\nARTS[43647152]: - VectorSetConstant\nARTS[43647152]: - Delete\nARTS[43647152]: }\n\nARTS[43647152]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[43647152]: {\nARTS[43647152]: - Select\nARTS[43647152]: - Select\nARTS[43647152]: - Select\nARTS[43647152]: - Select\nARTS[43647152]: - Select\nARTS[43647152]: - f_gridMetMM\nARTS[43647152]: - sensor_responseMetMM\nARTS[43647152]: }\n\n"
],
[
"%%px\nfrom typhon.arts.workspace import Workspace\nimport atms\nimport numpy as np\n\nws = Workspace()\nchannels = [0,15,16,17,19]\natms.setup_atmosphere(ws)\natms.setup_sensor(ws, channels)\natms.checks(ws)\nws.yCalc()",
"[stdout:0] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[47768304]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[47768304]: {\nARTS[47768304]: - verbosityInit\nARTS[47768304]: - scat_speciesSet\nARTS[47768304]: - MatrixSet\nARTS[47768304]: - Tensor4SetConstant\nARTS[47768304]: - ArrayOfStringSet\nARTS[47768304]: - Touch\nARTS[47768304]: - FlagOff\nARTS[47768304]: - MatrixSet\nARTS[47768304]: - NumericSet\nARTS[47768304]: - ArrayOfStringSet\nARTS[47768304]: - Tensor3SetConstant\nARTS[47768304]: - Tensor3SetConstant\nARTS[47768304]: - Tensor3SetConstant\nARTS[47768304]: - Tensor3SetConstant\nARTS[47768304]: - Tensor3SetConstant\nARTS[47768304]: - Tensor3SetConstant\nARTS[47768304]: - IndexSet\nARTS[47768304]: - IndexSet\nARTS[47768304]: - IndexSet\nARTS[47768304]: - IndexSet\nARTS[47768304]: - FlagOff\nARTS[47768304]: - output_file_formatSetAscii\nARTS[47768304]: - StringSet\nARTS[47768304]: - IndexSet\nARTS[47768304]: - abs_lineshapeDefine\nARTS[47768304]: - NumericSet\nARTS[47768304]: - NumericSet\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - IndexSet\nARTS[47768304]: - IndexSet\nARTS[47768304]: - NumericSet\nARTS[47768304]: - NumericSet\nARTS[47768304]: - nlteOff\nARTS[47768304]: - partition_functionsInitFromBuiltin\nARTS[47768304]: - IndexSet\nARTS[47768304]: }\n\nARTS[47768304]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[47768304]: {\nARTS[47768304]: - abs_cont_descriptionInit\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: - abs_cont_descriptionAppend\nARTS[47768304]: }\n\nARTS[47768304]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[47768304]: {\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - FlagOff\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - FlagOff\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - AgendaCreate\nARTS[47768304]: - AgendaSet\nARTS[47768304]: }\n\nARTS[47768304]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[47768304]: {\nARTS[47768304]: - isotopologue_ratiosInitFromBuiltin\nARTS[47768304]: - refellipsoidEarth\nARTS[47768304]: - NumericSet\nARTS[47768304]: - AgendaSet\nARTS[47768304]: - NumericSet\nARTS[47768304]: }\n\nARTS[47768304]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[47768304]: {\nARTS[47768304]: - ArrayOfArrayOfIndexCreate\nARTS[47768304]: - ArrayOfIndexCreate\nARTS[47768304]: - VectorCreate\nARTS[47768304]: - ArrayOfIndexCreate\nARTS[47768304]: - NumericCreate\nARTS[47768304]: - VectorCreate\nARTS[47768304]: - IndexCreate\nARTS[47768304]: }\n\nARTS[47768304]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[47768304]: {\nARTS[47768304]: - MatrixSet\nARTS[47768304]: - MatrixSet\nARTS[47768304]: - ArrayOfStringSet\nARTS[47768304]: - VectorSet\nARTS[47768304]: - ArrayOfIndexSet\nARTS[47768304]: - VectorSet\nARTS[47768304]: - Extract\nARTS[47768304]: - nrowsGet\nARTS[47768304]: - VectorSetConstant\nARTS[47768304]: - Delete\nARTS[47768304]: }\n\nARTS[47768304]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[47768304]: {\nARTS[47768304]: - Select\nARTS[47768304]: - Select\nARTS[47768304]: - Select\nARTS[47768304]: - Select\nARTS[47768304]: - Select\nARTS[47768304]: - f_gridMetMM\nARTS[47768304]: - sensor_responseMetMM\nARTS[47768304]: }\n\n[stdout:1] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[67700528]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[67700528]: {\nARTS[67700528]: - verbosityInit\nARTS[67700528]: - scat_speciesSet\nARTS[67700528]: - MatrixSet\nARTS[67700528]: - Tensor4SetConstant\nARTS[67700528]: - ArrayOfStringSet\nARTS[67700528]: - Touch\nARTS[67700528]: - FlagOff\nARTS[67700528]: - MatrixSet\nARTS[67700528]: - NumericSet\nARTS[67700528]: - ArrayOfStringSet\nARTS[67700528]: - Tensor3SetConstant\nARTS[67700528]: - Tensor3SetConstant\nARTS[67700528]: - Tensor3SetConstant\nARTS[67700528]: - Tensor3SetConstant\nARTS[67700528]: - Tensor3SetConstant\nARTS[67700528]: - Tensor3SetConstant\nARTS[67700528]: - IndexSet\nARTS[67700528]: - IndexSet\nARTS[67700528]: - IndexSet\nARTS[67700528]: - IndexSet\nARTS[67700528]: - FlagOff\nARTS[67700528]: - output_file_formatSetAscii\nARTS[67700528]: - StringSet\nARTS[67700528]: - IndexSet\nARTS[67700528]: - abs_lineshapeDefine\nARTS[67700528]: - NumericSet\nARTS[67700528]: - NumericSet\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - IndexSet\nARTS[67700528]: - IndexSet\nARTS[67700528]: - NumericSet\nARTS[67700528]: - NumericSet\nARTS[67700528]: - nlteOff\nARTS[67700528]: - partition_functionsInitFromBuiltin\nARTS[67700528]: - IndexSet\nARTS[67700528]: }\n\nARTS[67700528]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[67700528]: {\nARTS[67700528]: - abs_cont_descriptionInit\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: - abs_cont_descriptionAppend\nARTS[67700528]: }\n\nARTS[67700528]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[67700528]: {\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - FlagOff\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - FlagOff\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - AgendaCreate\nARTS[67700528]: - AgendaSet\nARTS[67700528]: }\n\nARTS[67700528]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[67700528]: {\nARTS[67700528]: - isotopologue_ratiosInitFromBuiltin\nARTS[67700528]: - refellipsoidEarth\nARTS[67700528]: - NumericSet\nARTS[67700528]: - AgendaSet\nARTS[67700528]: - NumericSet\nARTS[67700528]: }\n\nARTS[67700528]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[67700528]: {\nARTS[67700528]: - ArrayOfArrayOfIndexCreate\nARTS[67700528]: - ArrayOfIndexCreate\nARTS[67700528]: - VectorCreate\nARTS[67700528]: - ArrayOfIndexCreate\nARTS[67700528]: - NumericCreate\nARTS[67700528]: - VectorCreate\nARTS[67700528]: - IndexCreate\nARTS[67700528]: }\n\nARTS[67700528]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[67700528]: {\nARTS[67700528]: - MatrixSet\nARTS[67700528]: - MatrixSet\nARTS[67700528]: - ArrayOfStringSet\nARTS[67700528]: - VectorSet\nARTS[67700528]: - ArrayOfIndexSet\nARTS[67700528]: - VectorSet\nARTS[67700528]: - Extract\nARTS[67700528]: - nrowsGet\nARTS[67700528]: - VectorSetConstant\nARTS[67700528]: - Delete\nARTS[67700528]: }\n\nARTS[67700528]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[67700528]: {\nARTS[67700528]: - Select\nARTS[67700528]: - Select\nARTS[67700528]: - Select\nARTS[67700528]: - Select\nARTS[67700528]: - Select\nARTS[67700528]: - f_gridMetMM\nARTS[67700528]: - sensor_responseMetMM\nARTS[67700528]: }\n\n[stdout:2] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[68319552]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[68319552]: {\nARTS[68319552]: - verbosityInit\nARTS[68319552]: - scat_speciesSet\nARTS[68319552]: - MatrixSet\nARTS[68319552]: - Tensor4SetConstant\nARTS[68319552]: - ArrayOfStringSet\nARTS[68319552]: - Touch\nARTS[68319552]: - FlagOff\nARTS[68319552]: - MatrixSet\nARTS[68319552]: - NumericSet\nARTS[68319552]: - ArrayOfStringSet\nARTS[68319552]: - Tensor3SetConstant\nARTS[68319552]: - Tensor3SetConstant\nARTS[68319552]: - Tensor3SetConstant\nARTS[68319552]: - Tensor3SetConstant\nARTS[68319552]: - Tensor3SetConstant\nARTS[68319552]: - Tensor3SetConstant\nARTS[68319552]: - IndexSet\nARTS[68319552]: - IndexSet\nARTS[68319552]: - IndexSet\nARTS[68319552]: - IndexSet\nARTS[68319552]: - FlagOff\nARTS[68319552]: - output_file_formatSetAscii\nARTS[68319552]: - StringSet\nARTS[68319552]: - IndexSet\nARTS[68319552]: - abs_lineshapeDefine\nARTS[68319552]: - NumericSet\nARTS[68319552]: - NumericSet\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - IndexSet\nARTS[68319552]: - IndexSet\nARTS[68319552]: - NumericSet\nARTS[68319552]: - NumericSet\nARTS[68319552]: - nlteOff\nARTS[68319552]: - partition_functionsInitFromBuiltin\nARTS[68319552]: - IndexSet\nARTS[68319552]: }\n\nARTS[68319552]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[68319552]: {\nARTS[68319552]: - abs_cont_descriptionInit\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: - abs_cont_descriptionAppend\nARTS[68319552]: }\n\nARTS[68319552]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[68319552]: {\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - FlagOff\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - FlagOff\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - AgendaCreate\nARTS[68319552]: - AgendaSet\nARTS[68319552]: }\n\nARTS[68319552]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[68319552]: {\nARTS[68319552]: - isotopologue_ratiosInitFromBuiltin\nARTS[68319552]: - refellipsoidEarth\nARTS[68319552]: - NumericSet\nARTS[68319552]: - AgendaSet\nARTS[68319552]: - NumericSet\nARTS[68319552]: }\n\nARTS[68319552]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[68319552]: {\nARTS[68319552]: - ArrayOfArrayOfIndexCreate\nARTS[68319552]: - ArrayOfIndexCreate\nARTS[68319552]: - VectorCreate\nARTS[68319552]: - ArrayOfIndexCreate\nARTS[68319552]: - NumericCreate\nARTS[68319552]: - VectorCreate\nARTS[68319552]: - IndexCreate\nARTS[68319552]: }\n\nARTS[68319552]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[68319552]: {\nARTS[68319552]: - MatrixSet\nARTS[68319552]: - MatrixSet\nARTS[68319552]: - ArrayOfStringSet\nARTS[68319552]: - VectorSet\nARTS[68319552]: - ArrayOfIndexSet\nARTS[68319552]: - VectorSet\nARTS[68319552]: - Extract\nARTS[68319552]: - nrowsGet\nARTS[68319552]: - VectorSetConstant\nARTS[68319552]: - Delete\nARTS[68319552]: }\n\nARTS[68319552]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[68319552]: {\nARTS[68319552]: - Select\nARTS[68319552]: - Select\nARTS[68319552]: - Select\nARTS[68319552]: - Select\nARTS[68319552]: - Select\nARTS[68319552]: - f_gridMetMM\nARTS[68319552]: - sensor_responseMetMM\nARTS[68319552]: }\n\n[stdout:3] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[61976304]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[61976304]: {\nARTS[61976304]: - verbosityInit\nARTS[61976304]: - scat_speciesSet\nARTS[61976304]: - MatrixSet\nARTS[61976304]: - Tensor4SetConstant\nARTS[61976304]: - ArrayOfStringSet\nARTS[61976304]: - Touch\nARTS[61976304]: - FlagOff\nARTS[61976304]: - MatrixSet\nARTS[61976304]: - NumericSet\nARTS[61976304]: - ArrayOfStringSet\nARTS[61976304]: - Tensor3SetConstant\nARTS[61976304]: - Tensor3SetConstant\nARTS[61976304]: - Tensor3SetConstant\nARTS[61976304]: - Tensor3SetConstant\nARTS[61976304]: - Tensor3SetConstant\nARTS[61976304]: - Tensor3SetConstant\nARTS[61976304]: - IndexSet\nARTS[61976304]: - IndexSet\nARTS[61976304]: - IndexSet\nARTS[61976304]: - IndexSet\nARTS[61976304]: - FlagOff\nARTS[61976304]: - output_file_formatSetAscii\nARTS[61976304]: - StringSet\nARTS[61976304]: - IndexSet\nARTS[61976304]: - abs_lineshapeDefine\nARTS[61976304]: - NumericSet\nARTS[61976304]: - NumericSet\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - IndexSet\nARTS[61976304]: - IndexSet\nARTS[61976304]: - NumericSet\nARTS[61976304]: - NumericSet\nARTS[61976304]: - nlteOff\nARTS[61976304]: - partition_functionsInitFromBuiltin\nARTS[61976304]: - IndexSet\nARTS[61976304]: }\n\nARTS[61976304]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[61976304]: {\nARTS[61976304]: - abs_cont_descriptionInit\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: - abs_cont_descriptionAppend\nARTS[61976304]: }\n\nARTS[61976304]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[61976304]: {\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - FlagOff\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - FlagOff\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - AgendaCreate\nARTS[61976304]: - AgendaSet\nARTS[61976304]: }\n\nARTS[61976304]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[61976304]: {\nARTS[61976304]: - isotopologue_ratiosInitFromBuiltin\nARTS[61976304]: - refellipsoidEarth\nARTS[61976304]: - NumericSet\nARTS[61976304]: - AgendaSet\nARTS[61976304]: - NumericSet\nARTS[61976304]: }\n\nARTS[61976304]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[61976304]: {\nARTS[61976304]: - ArrayOfArrayOfIndexCreate\nARTS[61976304]: - ArrayOfIndexCreate\nARTS[61976304]: - VectorCreate\nARTS[61976304]: - ArrayOfIndexCreate\nARTS[61976304]: - NumericCreate\nARTS[61976304]: - VectorCreate\nARTS[61976304]: - IndexCreate\nARTS[61976304]: }\n\nARTS[61976304]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[61976304]: {\nARTS[61976304]: - MatrixSet\nARTS[61976304]: - MatrixSet\nARTS[61976304]: - ArrayOfStringSet\nARTS[61976304]: - VectorSet\nARTS[61976304]: - ArrayOfIndexSet\nARTS[61976304]: - VectorSet\nARTS[61976304]: - Extract\nARTS[61976304]: - nrowsGet\nARTS[61976304]: - VectorSetConstant\nARTS[61976304]: - Delete\nARTS[61976304]: }\n\nARTS[61976304]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[61976304]: {\nARTS[61976304]: - Select\nARTS[61976304]: - Select\nARTS[61976304]: - Select\nARTS[61976304]: - Select\nARTS[61976304]: - Select\nARTS[61976304]: - f_gridMetMM\nARTS[61976304]: - sensor_responseMetMM\nARTS[61976304]: }\n\n[stdout:4] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[64811280]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[64811280]: {\nARTS[64811280]: - verbosityInit\nARTS[64811280]: - scat_speciesSet\nARTS[64811280]: - MatrixSet\nARTS[64811280]: - Tensor4SetConstant\nARTS[64811280]: - ArrayOfStringSet\nARTS[64811280]: - Touch\nARTS[64811280]: - FlagOff\nARTS[64811280]: - MatrixSet\nARTS[64811280]: - NumericSet\nARTS[64811280]: - ArrayOfStringSet\nARTS[64811280]: - Tensor3SetConstant\nARTS[64811280]: - Tensor3SetConstant\nARTS[64811280]: - Tensor3SetConstant\nARTS[64811280]: - Tensor3SetConstant\nARTS[64811280]: - Tensor3SetConstant\nARTS[64811280]: - Tensor3SetConstant\nARTS[64811280]: - IndexSet\nARTS[64811280]: - IndexSet\nARTS[64811280]: - IndexSet\nARTS[64811280]: - IndexSet\nARTS[64811280]: - FlagOff\nARTS[64811280]: - output_file_formatSetAscii\nARTS[64811280]: - StringSet\nARTS[64811280]: - IndexSet\nARTS[64811280]: - abs_lineshapeDefine\nARTS[64811280]: - NumericSet\nARTS[64811280]: - NumericSet\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - IndexSet\nARTS[64811280]: - IndexSet\nARTS[64811280]: - NumericSet\nARTS[64811280]: - NumericSet\nARTS[64811280]: - nlteOff\nARTS[64811280]: - partition_functionsInitFromBuiltin\nARTS[64811280]: - IndexSet\nARTS[64811280]: }\n\nARTS[64811280]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[64811280]: {\nARTS[64811280]: - abs_cont_descriptionInit\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: - abs_cont_descriptionAppend\nARTS[64811280]: }\n\nARTS[64811280]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[64811280]: {\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - FlagOff\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - FlagOff\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - AgendaCreate\nARTS[64811280]: - AgendaSet\nARTS[64811280]: }\n\nARTS[64811280]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[64811280]: {\nARTS[64811280]: - isotopologue_ratiosInitFromBuiltin\nARTS[64811280]: - refellipsoidEarth\nARTS[64811280]: - NumericSet\nARTS[64811280]: - AgendaSet\nARTS[64811280]: - NumericSet\nARTS[64811280]: }\n\nARTS[64811280]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[64811280]: {\nARTS[64811280]: - ArrayOfArrayOfIndexCreate\nARTS[64811280]: - ArrayOfIndexCreate\nARTS[64811280]: - VectorCreate\nARTS[64811280]: - ArrayOfIndexCreate\nARTS[64811280]: - NumericCreate\nARTS[64811280]: - VectorCreate\nARTS[64811280]: - IndexCreate\nARTS[64811280]: }\n\nARTS[64811280]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[64811280]: {\nARTS[64811280]: - MatrixSet\nARTS[64811280]: - MatrixSet\nARTS[64811280]: - ArrayOfStringSet\nARTS[64811280]: - VectorSet\nARTS[64811280]: - ArrayOfIndexSet\nARTS[64811280]: - VectorSet\nARTS[64811280]: - Extract\nARTS[64811280]: - nrowsGet\nARTS[64811280]: - VectorSetConstant\nARTS[64811280]: - Delete\nARTS[64811280]: }\n\nARTS[64811280]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[64811280]: {\nARTS[64811280]: - Select\nARTS[64811280]: - Select\nARTS[64811280]: - Select\nARTS[64811280]: - Select\nARTS[64811280]: - Select\nARTS[64811280]: - f_gridMetMM\nARTS[64811280]: - sensor_responseMetMM\nARTS[64811280]: }\n\n[stdout:5] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[54546288]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[54546288]: {\nARTS[54546288]: - verbosityInit\nARTS[54546288]: - scat_speciesSet\nARTS[54546288]: - MatrixSet\nARTS[54546288]: - Tensor4SetConstant\nARTS[54546288]: - ArrayOfStringSet\nARTS[54546288]: - Touch\nARTS[54546288]: - FlagOff\nARTS[54546288]: - MatrixSet\nARTS[54546288]: - NumericSet\nARTS[54546288]: - ArrayOfStringSet\nARTS[54546288]: - Tensor3SetConstant\nARTS[54546288]: - Tensor3SetConstant\nARTS[54546288]: - Tensor3SetConstant\nARTS[54546288]: - Tensor3SetConstant\nARTS[54546288]: - Tensor3SetConstant\nARTS[54546288]: - Tensor3SetConstant\nARTS[54546288]: - IndexSet\nARTS[54546288]: - IndexSet\nARTS[54546288]: - IndexSet\nARTS[54546288]: - IndexSet\nARTS[54546288]: - FlagOff\nARTS[54546288]: - output_file_formatSetAscii\nARTS[54546288]: - StringSet\nARTS[54546288]: - IndexSet\nARTS[54546288]: - abs_lineshapeDefine\nARTS[54546288]: - NumericSet\nARTS[54546288]: - NumericSet\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - IndexSet\nARTS[54546288]: - IndexSet\nARTS[54546288]: - NumericSet\nARTS[54546288]: - NumericSet\nARTS[54546288]: - nlteOff\nARTS[54546288]: - partition_functionsInitFromBuiltin\nARTS[54546288]: - IndexSet\nARTS[54546288]: }\n\nARTS[54546288]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[54546288]: {\nARTS[54546288]: - abs_cont_descriptionInit\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: - abs_cont_descriptionAppend\nARTS[54546288]: }\n\nARTS[54546288]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[54546288]: {\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - FlagOff\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - FlagOff\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - AgendaCreate\nARTS[54546288]: - AgendaSet\nARTS[54546288]: }\n\nARTS[54546288]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[54546288]: {\nARTS[54546288]: - isotopologue_ratiosInitFromBuiltin\nARTS[54546288]: - refellipsoidEarth\nARTS[54546288]: - NumericSet\nARTS[54546288]: - AgendaSet\nARTS[54546288]: - NumericSet\nARTS[54546288]: }\n\nARTS[54546288]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[54546288]: {\nARTS[54546288]: - ArrayOfArrayOfIndexCreate\nARTS[54546288]: - ArrayOfIndexCreate\nARTS[54546288]: - VectorCreate\nARTS[54546288]: - ArrayOfIndexCreate\nARTS[54546288]: - NumericCreate\nARTS[54546288]: - VectorCreate\nARTS[54546288]: - IndexCreate\nARTS[54546288]: }\n\nARTS[54546288]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[54546288]: {\nARTS[54546288]: - MatrixSet\nARTS[54546288]: - MatrixSet\nARTS[54546288]: - ArrayOfStringSet\nARTS[54546288]: - VectorSet\nARTS[54546288]: - ArrayOfIndexSet\nARTS[54546288]: - VectorSet\nARTS[54546288]: - Extract\nARTS[54546288]: - nrowsGet\nARTS[54546288]: - VectorSetConstant\nARTS[54546288]: - Delete\nARTS[54546288]: }\n\nARTS[54546288]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[54546288]: {\nARTS[54546288]: - Select\nARTS[54546288]: - Select\nARTS[54546288]: - Select\nARTS[54546288]: - Select\nARTS[54546288]: - Select\nARTS[54546288]: - f_gridMetMM\nARTS[54546288]: - sensor_responseMetMM\nARTS[54546288]: }\n\n[stdout:6] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[39689808]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[39689808]: {\nARTS[39689808]: - verbosityInit\nARTS[39689808]: - scat_speciesSet\nARTS[39689808]: - MatrixSet\nARTS[39689808]: - Tensor4SetConstant\nARTS[39689808]: - ArrayOfStringSet\nARTS[39689808]: - Touch\nARTS[39689808]: - FlagOff\nARTS[39689808]: - MatrixSet\nARTS[39689808]: - NumericSet\nARTS[39689808]: - ArrayOfStringSet\nARTS[39689808]: - Tensor3SetConstant\nARTS[39689808]: - Tensor3SetConstant\nARTS[39689808]: - Tensor3SetConstant\nARTS[39689808]: - Tensor3SetConstant\nARTS[39689808]: - Tensor3SetConstant\nARTS[39689808]: - Tensor3SetConstant\nARTS[39689808]: - IndexSet\nARTS[39689808]: - IndexSet\nARTS[39689808]: - IndexSet\nARTS[39689808]: - IndexSet\nARTS[39689808]: - FlagOff\nARTS[39689808]: - output_file_formatSetAscii\nARTS[39689808]: - StringSet\nARTS[39689808]: - IndexSet\nARTS[39689808]: - abs_lineshapeDefine\nARTS[39689808]: - NumericSet\nARTS[39689808]: - NumericSet\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - IndexSet\nARTS[39689808]: - IndexSet\nARTS[39689808]: - NumericSet\nARTS[39689808]: - NumericSet\nARTS[39689808]: - nlteOff\nARTS[39689808]: - partition_functionsInitFromBuiltin\nARTS[39689808]: - IndexSet\nARTS[39689808]: }\n\nARTS[39689808]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[39689808]: {\nARTS[39689808]: - abs_cont_descriptionInit\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: - abs_cont_descriptionAppend\nARTS[39689808]: }\n\nARTS[39689808]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[39689808]: {\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - FlagOff\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - FlagOff\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - AgendaCreate\nARTS[39689808]: - AgendaSet\nARTS[39689808]: }\n\nARTS[39689808]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[39689808]: {\nARTS[39689808]: - isotopologue_ratiosInitFromBuiltin\nARTS[39689808]: - refellipsoidEarth\nARTS[39689808]: - NumericSet\nARTS[39689808]: - AgendaSet\nARTS[39689808]: - NumericSet\nARTS[39689808]: }\n\nARTS[39689808]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[39689808]: {\nARTS[39689808]: - ArrayOfArrayOfIndexCreate\nARTS[39689808]: - ArrayOfIndexCreate\nARTS[39689808]: - VectorCreate\nARTS[39689808]: - ArrayOfIndexCreate\nARTS[39689808]: - NumericCreate\nARTS[39689808]: - VectorCreate\nARTS[39689808]: - IndexCreate\nARTS[39689808]: }\n\nARTS[39689808]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[39689808]: {\nARTS[39689808]: - MatrixSet\nARTS[39689808]: - MatrixSet\nARTS[39689808]: - ArrayOfStringSet\nARTS[39689808]: - VectorSet\nARTS[39689808]: - ArrayOfIndexSet\nARTS[39689808]: - VectorSet\nARTS[39689808]: - Extract\nARTS[39689808]: - nrowsGet\nARTS[39689808]: - VectorSetConstant\nARTS[39689808]: - Delete\nARTS[39689808]: }\n\nARTS[39689808]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[39689808]: {\nARTS[39689808]: - Select\nARTS[39689808]: - Select\nARTS[39689808]: - Select\nARTS[39689808]: - Select\nARTS[39689808]: - Select\nARTS[39689808]: - f_gridMetMM\nARTS[39689808]: - sensor_responseMetMM\nARTS[39689808]: }\n\n[stdout:7] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[40289504]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[40289504]: {\nARTS[40289504]: - verbosityInit\nARTS[40289504]: - scat_speciesSet\nARTS[40289504]: - MatrixSet\nARTS[40289504]: - Tensor4SetConstant\nARTS[40289504]: - ArrayOfStringSet\nARTS[40289504]: - Touch\nARTS[40289504]: - FlagOff\nARTS[40289504]: - MatrixSet\nARTS[40289504]: - NumericSet\nARTS[40289504]: - ArrayOfStringSet\nARTS[40289504]: - Tensor3SetConstant\nARTS[40289504]: - Tensor3SetConstant\nARTS[40289504]: - Tensor3SetConstant\nARTS[40289504]: - Tensor3SetConstant\nARTS[40289504]: - Tensor3SetConstant\nARTS[40289504]: - Tensor3SetConstant\nARTS[40289504]: - IndexSet\nARTS[40289504]: - IndexSet\nARTS[40289504]: - IndexSet\nARTS[40289504]: - IndexSet\nARTS[40289504]: - FlagOff\nARTS[40289504]: - output_file_formatSetAscii\nARTS[40289504]: - StringSet\nARTS[40289504]: - IndexSet\nARTS[40289504]: - abs_lineshapeDefine\nARTS[40289504]: - NumericSet\nARTS[40289504]: - NumericSet\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - IndexSet\nARTS[40289504]: - IndexSet\nARTS[40289504]: - NumericSet\nARTS[40289504]: - NumericSet\nARTS[40289504]: - nlteOff\nARTS[40289504]: - partition_functionsInitFromBuiltin\nARTS[40289504]: - IndexSet\nARTS[40289504]: }\n\nARTS[40289504]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[40289504]: {\nARTS[40289504]: - abs_cont_descriptionInit\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: - abs_cont_descriptionAppend\nARTS[40289504]: }\n\nARTS[40289504]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[40289504]: {\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - FlagOff\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - FlagOff\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - AgendaCreate\nARTS[40289504]: - AgendaSet\nARTS[40289504]: }\n\nARTS[40289504]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[40289504]: {\nARTS[40289504]: - isotopologue_ratiosInitFromBuiltin\nARTS[40289504]: - refellipsoidEarth\nARTS[40289504]: - NumericSet\nARTS[40289504]: - AgendaSet\nARTS[40289504]: - NumericSet\nARTS[40289504]: }\n\nARTS[40289504]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[40289504]: {\nARTS[40289504]: - ArrayOfArrayOfIndexCreate\nARTS[40289504]: - ArrayOfIndexCreate\nARTS[40289504]: - VectorCreate\nARTS[40289504]: - ArrayOfIndexCreate\nARTS[40289504]: - NumericCreate\nARTS[40289504]: - VectorCreate\nARTS[40289504]: - IndexCreate\nARTS[40289504]: }\n\nARTS[40289504]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[40289504]: {\nARTS[40289504]: - MatrixSet\nARTS[40289504]: - MatrixSet\nARTS[40289504]: - ArrayOfStringSet\nARTS[40289504]: - VectorSet\nARTS[40289504]: - ArrayOfIndexSet\nARTS[40289504]: - VectorSet\nARTS[40289504]: - Extract\nARTS[40289504]: - nrowsGet\nARTS[40289504]: - VectorSetConstant\nARTS[40289504]: - Delete\nARTS[40289504]: }\n\nARTS[40289504]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[40289504]: {\nARTS[40289504]: - Select\nARTS[40289504]: - Select\nARTS[40289504]: - Select\nARTS[40289504]: - Select\nARTS[40289504]: - Select\nARTS[40289504]: - f_gridMetMM\nARTS[40289504]: - sensor_responseMetMM\nARTS[40289504]: }\n\n[stdout:8] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[61390224]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[61390224]: {\nARTS[61390224]: - verbosityInit\nARTS[61390224]: - scat_speciesSet\nARTS[61390224]: - MatrixSet\nARTS[61390224]: - Tensor4SetConstant\nARTS[61390224]: - ArrayOfStringSet\nARTS[61390224]: - Touch\nARTS[61390224]: - FlagOff\nARTS[61390224]: - MatrixSet\nARTS[61390224]: - NumericSet\nARTS[61390224]: - ArrayOfStringSet\nARTS[61390224]: - Tensor3SetConstant\nARTS[61390224]: - Tensor3SetConstant\nARTS[61390224]: - Tensor3SetConstant\nARTS[61390224]: - Tensor3SetConstant\nARTS[61390224]: - Tensor3SetConstant\nARTS[61390224]: - Tensor3SetConstant\nARTS[61390224]: - IndexSet\nARTS[61390224]: - IndexSet\nARTS[61390224]: - IndexSet\nARTS[61390224]: - IndexSet\nARTS[61390224]: - FlagOff\nARTS[61390224]: - output_file_formatSetAscii\nARTS[61390224]: - StringSet\nARTS[61390224]: - IndexSet\nARTS[61390224]: - abs_lineshapeDefine\nARTS[61390224]: - NumericSet\nARTS[61390224]: - NumericSet\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - IndexSet\nARTS[61390224]: - IndexSet\nARTS[61390224]: - NumericSet\nARTS[61390224]: - NumericSet\nARTS[61390224]: - nlteOff\nARTS[61390224]: - partition_functionsInitFromBuiltin\nARTS[61390224]: - IndexSet\nARTS[61390224]: }\n\nARTS[61390224]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[61390224]: {\nARTS[61390224]: - abs_cont_descriptionInit\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: - abs_cont_descriptionAppend\nARTS[61390224]: }\n\nARTS[61390224]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[61390224]: {\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - FlagOff\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - FlagOff\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - AgendaCreate\nARTS[61390224]: - AgendaSet\nARTS[61390224]: }\n\nARTS[61390224]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[61390224]: {\nARTS[61390224]: - isotopologue_ratiosInitFromBuiltin\nARTS[61390224]: - refellipsoidEarth\nARTS[61390224]: - NumericSet\nARTS[61390224]: - AgendaSet\nARTS[61390224]: - NumericSet\nARTS[61390224]: }\n\nARTS[61390224]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[61390224]: {\nARTS[61390224]: - ArrayOfArrayOfIndexCreate\nARTS[61390224]: - ArrayOfIndexCreate\nARTS[61390224]: - VectorCreate\nARTS[61390224]: - ArrayOfIndexCreate\nARTS[61390224]: - NumericCreate\nARTS[61390224]: - VectorCreate\nARTS[61390224]: - IndexCreate\nARTS[61390224]: }\n\nARTS[61390224]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[61390224]: {\nARTS[61390224]: - MatrixSet\nARTS[61390224]: - MatrixSet\nARTS[61390224]: - ArrayOfStringSet\nARTS[61390224]: - VectorSet\nARTS[61390224]: - ArrayOfIndexSet\nARTS[61390224]: - VectorSet\nARTS[61390224]: - Extract\nARTS[61390224]: - nrowsGet\nARTS[61390224]: - VectorSetConstant\nARTS[61390224]: - Delete\nARTS[61390224]: }\n\nARTS[61390224]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[61390224]: {\nARTS[61390224]: - Select\nARTS[61390224]: - Select\nARTS[61390224]: - Select\nARTS[61390224]: - Select\nARTS[61390224]: - Select\nARTS[61390224]: - f_gridMetMM\nARTS[61390224]: - sensor_responseMetMM\nARTS[61390224]: }\n\n[stdout:9] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[41346640]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[41346640]: {\nARTS[41346640]: - verbosityInit\nARTS[41346640]: - scat_speciesSet\nARTS[41346640]: - MatrixSet\nARTS[41346640]: - Tensor4SetConstant\nARTS[41346640]: - ArrayOfStringSet\nARTS[41346640]: - Touch\nARTS[41346640]: - FlagOff\nARTS[41346640]: - MatrixSet\nARTS[41346640]: - NumericSet\nARTS[41346640]: - ArrayOfStringSet\nARTS[41346640]: - Tensor3SetConstant\nARTS[41346640]: - Tensor3SetConstant\nARTS[41346640]: - Tensor3SetConstant\nARTS[41346640]: - Tensor3SetConstant\nARTS[41346640]: - Tensor3SetConstant\nARTS[41346640]: - Tensor3SetConstant\nARTS[41346640]: - IndexSet\nARTS[41346640]: - IndexSet\nARTS[41346640]: - IndexSet\nARTS[41346640]: - IndexSet\nARTS[41346640]: - FlagOff\nARTS[41346640]: - output_file_formatSetAscii\nARTS[41346640]: - StringSet\nARTS[41346640]: - IndexSet\nARTS[41346640]: - abs_lineshapeDefine\nARTS[41346640]: - NumericSet\nARTS[41346640]: - NumericSet\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - IndexSet\nARTS[41346640]: - IndexSet\nARTS[41346640]: - NumericSet\nARTS[41346640]: - NumericSet\nARTS[41346640]: - nlteOff\nARTS[41346640]: - partition_functionsInitFromBuiltin\nARTS[41346640]: - IndexSet\nARTS[41346640]: }\n\nARTS[41346640]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[41346640]: {\nARTS[41346640]: - abs_cont_descriptionInit\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: - abs_cont_descriptionAppend\nARTS[41346640]: }\n\nARTS[41346640]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[41346640]: {\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - FlagOff\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - FlagOff\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - AgendaCreate\nARTS[41346640]: - AgendaSet\nARTS[41346640]: }\n\nARTS[41346640]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[41346640]: {\nARTS[41346640]: - isotopologue_ratiosInitFromBuiltin\nARTS[41346640]: - refellipsoidEarth\nARTS[41346640]: - NumericSet\nARTS[41346640]: - AgendaSet\nARTS[41346640]: - NumericSet\nARTS[41346640]: }\n\nARTS[41346640]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[41346640]: {\nARTS[41346640]: - ArrayOfArrayOfIndexCreate\nARTS[41346640]: - ArrayOfIndexCreate\nARTS[41346640]: - VectorCreate\nARTS[41346640]: - ArrayOfIndexCreate\nARTS[41346640]: - NumericCreate\nARTS[41346640]: - VectorCreate\nARTS[41346640]: - IndexCreate\nARTS[41346640]: }\n\nARTS[41346640]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[41346640]: {\nARTS[41346640]: - MatrixSet\nARTS[41346640]: - MatrixSet\nARTS[41346640]: - ArrayOfStringSet\nARTS[41346640]: - VectorSet\nARTS[41346640]: - ArrayOfIndexSet\nARTS[41346640]: - VectorSet\nARTS[41346640]: - Extract\nARTS[41346640]: - nrowsGet\nARTS[41346640]: - VectorSetConstant\nARTS[41346640]: - Delete\nARTS[41346640]: }\n\nARTS[41346640]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[41346640]: {\nARTS[41346640]: - Select\nARTS[41346640]: - Select\nARTS[41346640]: - Select\nARTS[41346640]: - Select\nARTS[41346640]: - Select\nARTS[41346640]: - f_gridMetMM\nARTS[41346640]: - sensor_responseMetMM\nARTS[41346640]: }\n\n[stdout:10] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[41141504]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[41141504]: {\nARTS[41141504]: - verbosityInit\nARTS[41141504]: - scat_speciesSet\nARTS[41141504]: - MatrixSet\nARTS[41141504]: - Tensor4SetConstant\nARTS[41141504]: - ArrayOfStringSet\nARTS[41141504]: - Touch\nARTS[41141504]: - FlagOff\nARTS[41141504]: - MatrixSet\nARTS[41141504]: - NumericSet\nARTS[41141504]: - ArrayOfStringSet\nARTS[41141504]: - Tensor3SetConstant\nARTS[41141504]: - Tensor3SetConstant\nARTS[41141504]: - Tensor3SetConstant\nARTS[41141504]: - Tensor3SetConstant\nARTS[41141504]: - Tensor3SetConstant\nARTS[41141504]: - Tensor3SetConstant\nARTS[41141504]: - IndexSet\nARTS[41141504]: - IndexSet\nARTS[41141504]: - IndexSet\nARTS[41141504]: - IndexSet\nARTS[41141504]: - FlagOff\nARTS[41141504]: - output_file_formatSetAscii\nARTS[41141504]: - StringSet\nARTS[41141504]: - IndexSet\nARTS[41141504]: - abs_lineshapeDefine\nARTS[41141504]: - NumericSet\nARTS[41141504]: - NumericSet\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - IndexSet\nARTS[41141504]: - IndexSet\nARTS[41141504]: - NumericSet\nARTS[41141504]: - NumericSet\nARTS[41141504]: - nlteOff\nARTS[41141504]: - partition_functionsInitFromBuiltin\nARTS[41141504]: - IndexSet\nARTS[41141504]: }\n\nARTS[41141504]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[41141504]: {\nARTS[41141504]: - abs_cont_descriptionInit\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: - abs_cont_descriptionAppend\nARTS[41141504]: }\n\nARTS[41141504]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[41141504]: {\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - FlagOff\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - FlagOff\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - AgendaCreate\nARTS[41141504]: - AgendaSet\nARTS[41141504]: }\n\nARTS[41141504]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[41141504]: {\nARTS[41141504]: - isotopologue_ratiosInitFromBuiltin\nARTS[41141504]: - refellipsoidEarth\nARTS[41141504]: - NumericSet\nARTS[41141504]: - AgendaSet\nARTS[41141504]: - NumericSet\nARTS[41141504]: }\n\nARTS[41141504]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[41141504]: {\nARTS[41141504]: - ArrayOfArrayOfIndexCreate\nARTS[41141504]: - ArrayOfIndexCreate\nARTS[41141504]: - VectorCreate\nARTS[41141504]: - ArrayOfIndexCreate\nARTS[41141504]: - NumericCreate\nARTS[41141504]: - VectorCreate\nARTS[41141504]: - IndexCreate\nARTS[41141504]: }\n\nARTS[41141504]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[41141504]: {\nARTS[41141504]: - MatrixSet\nARTS[41141504]: - MatrixSet\nARTS[41141504]: - ArrayOfStringSet\nARTS[41141504]: - VectorSet\nARTS[41141504]: - ArrayOfIndexSet\nARTS[41141504]: - VectorSet\nARTS[41141504]: - Extract\nARTS[41141504]: - nrowsGet\nARTS[41141504]: - VectorSetConstant\nARTS[41141504]: - Delete\nARTS[41141504]: }\n\nARTS[41141504]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[41141504]: {\nARTS[41141504]: - Select\nARTS[41141504]: - Select\nARTS[41141504]: - Select\nARTS[41141504]: - Select\nARTS[41141504]: - Select\nARTS[41141504]: - f_gridMetMM\nARTS[41141504]: - sensor_responseMetMM\nARTS[41141504]: }\n\n[stdout:11] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[36735968]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[36735968]: {\nARTS[36735968]: - verbosityInit\nARTS[36735968]: - scat_speciesSet\nARTS[36735968]: - MatrixSet\nARTS[36735968]: - Tensor4SetConstant\nARTS[36735968]: - ArrayOfStringSet\nARTS[36735968]: - Touch\nARTS[36735968]: - FlagOff\nARTS[36735968]: - MatrixSet\nARTS[36735968]: - NumericSet\nARTS[36735968]: - ArrayOfStringSet\nARTS[36735968]: - Tensor3SetConstant\nARTS[36735968]: - Tensor3SetConstant\nARTS[36735968]: - Tensor3SetConstant\nARTS[36735968]: - Tensor3SetConstant\nARTS[36735968]: - Tensor3SetConstant\nARTS[36735968]: - Tensor3SetConstant\nARTS[36735968]: - IndexSet\nARTS[36735968]: - IndexSet\nARTS[36735968]: - IndexSet\nARTS[36735968]: - IndexSet\nARTS[36735968]: - FlagOff\nARTS[36735968]: - output_file_formatSetAscii\nARTS[36735968]: - StringSet\nARTS[36735968]: - IndexSet\nARTS[36735968]: - abs_lineshapeDefine\nARTS[36735968]: - NumericSet\nARTS[36735968]: - NumericSet\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - IndexSet\nARTS[36735968]: - IndexSet\nARTS[36735968]: - NumericSet\nARTS[36735968]: - NumericSet\nARTS[36735968]: - nlteOff\nARTS[36735968]: - partition_functionsInitFromBuiltin\nARTS[36735968]: - IndexSet\nARTS[36735968]: }\n\nARTS[36735968]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[36735968]: {\nARTS[36735968]: - abs_cont_descriptionInit\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: - abs_cont_descriptionAppend\nARTS[36735968]: }\n\nARTS[36735968]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[36735968]: {\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - FlagOff\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - FlagOff\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - AgendaCreate\nARTS[36735968]: - AgendaSet\nARTS[36735968]: }\n\nARTS[36735968]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[36735968]: {\nARTS[36735968]: - isotopologue_ratiosInitFromBuiltin\nARTS[36735968]: - refellipsoidEarth\nARTS[36735968]: - NumericSet\nARTS[36735968]: - AgendaSet\nARTS[36735968]: - NumericSet\nARTS[36735968]: }\n\nARTS[36735968]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[36735968]: {\nARTS[36735968]: - ArrayOfArrayOfIndexCreate\nARTS[36735968]: - ArrayOfIndexCreate\nARTS[36735968]: - VectorCreate\nARTS[36735968]: - ArrayOfIndexCreate\nARTS[36735968]: - NumericCreate\nARTS[36735968]: - VectorCreate\nARTS[36735968]: - IndexCreate\nARTS[36735968]: }\n\nARTS[36735968]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[36735968]: {\nARTS[36735968]: - MatrixSet\nARTS[36735968]: - MatrixSet\nARTS[36735968]: - ArrayOfStringSet\nARTS[36735968]: - VectorSet\nARTS[36735968]: - ArrayOfIndexSet\nARTS[36735968]: - VectorSet\nARTS[36735968]: - Extract\nARTS[36735968]: - nrowsGet\nARTS[36735968]: - VectorSetConstant\nARTS[36735968]: - Delete\nARTS[36735968]: }\n\nARTS[36735968]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[36735968]: {\nARTS[36735968]: - Select\nARTS[36735968]: - Select\nARTS[36735968]: - Select\nARTS[36735968]: - Select\nARTS[36735968]: - Select\nARTS[36735968]: - f_gridMetMM\nARTS[36735968]: - sensor_responseMetMM\nARTS[36735968]: }\n\n[stdout:12] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[46610688]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[46610688]: {\nARTS[46610688]: - verbosityInit\nARTS[46610688]: - scat_speciesSet\nARTS[46610688]: - MatrixSet\nARTS[46610688]: - Tensor4SetConstant\nARTS[46610688]: - ArrayOfStringSet\nARTS[46610688]: - Touch\nARTS[46610688]: - FlagOff\nARTS[46610688]: - MatrixSet\nARTS[46610688]: - NumericSet\nARTS[46610688]: - ArrayOfStringSet\nARTS[46610688]: - Tensor3SetConstant\nARTS[46610688]: - Tensor3SetConstant\nARTS[46610688]: - Tensor3SetConstant\nARTS[46610688]: - Tensor3SetConstant\nARTS[46610688]: - Tensor3SetConstant\nARTS[46610688]: - Tensor3SetConstant\nARTS[46610688]: - IndexSet\nARTS[46610688]: - IndexSet\nARTS[46610688]: - IndexSet\nARTS[46610688]: - IndexSet\nARTS[46610688]: - FlagOff\nARTS[46610688]: - output_file_formatSetAscii\nARTS[46610688]: - StringSet\nARTS[46610688]: - IndexSet\nARTS[46610688]: - abs_lineshapeDefine\nARTS[46610688]: - NumericSet\nARTS[46610688]: - NumericSet\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - IndexSet\nARTS[46610688]: - IndexSet\nARTS[46610688]: - NumericSet\nARTS[46610688]: - NumericSet\nARTS[46610688]: - nlteOff\nARTS[46610688]: - partition_functionsInitFromBuiltin\nARTS[46610688]: - IndexSet\nARTS[46610688]: }\n\nARTS[46610688]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[46610688]: {\nARTS[46610688]: - abs_cont_descriptionInit\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: - abs_cont_descriptionAppend\nARTS[46610688]: }\n\nARTS[46610688]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[46610688]: {\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - FlagOff\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - FlagOff\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - AgendaCreate\nARTS[46610688]: - AgendaSet\nARTS[46610688]: }\n\nARTS[46610688]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[46610688]: {\nARTS[46610688]: - isotopologue_ratiosInitFromBuiltin\nARTS[46610688]: - refellipsoidEarth\nARTS[46610688]: - NumericSet\nARTS[46610688]: - AgendaSet\nARTS[46610688]: - NumericSet\nARTS[46610688]: }\n\nARTS[46610688]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[46610688]: {\nARTS[46610688]: - ArrayOfArrayOfIndexCreate\nARTS[46610688]: - ArrayOfIndexCreate\nARTS[46610688]: - VectorCreate\nARTS[46610688]: - ArrayOfIndexCreate\nARTS[46610688]: - NumericCreate\nARTS[46610688]: - VectorCreate\nARTS[46610688]: - IndexCreate\nARTS[46610688]: }\n\nARTS[46610688]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[46610688]: {\nARTS[46610688]: - MatrixSet\nARTS[46610688]: - MatrixSet\nARTS[46610688]: - ArrayOfStringSet\nARTS[46610688]: - VectorSet\nARTS[46610688]: - ArrayOfIndexSet\nARTS[46610688]: - VectorSet\nARTS[46610688]: - Extract\nARTS[46610688]: - nrowsGet\nARTS[46610688]: - VectorSetConstant\nARTS[46610688]: - Delete\nARTS[46610688]: }\n\nARTS[46610688]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[46610688]: {\nARTS[46610688]: - Select\nARTS[46610688]: - Select\nARTS[46610688]: - Select\nARTS[46610688]: - Select\nARTS[46610688]: - Select\nARTS[46610688]: - f_gridMetMM\nARTS[46610688]: - sensor_responseMetMM\nARTS[46610688]: }\n\n[stdout:13] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[38633296]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[38633296]: {\nARTS[38633296]: - verbosityInit\nARTS[38633296]: - scat_speciesSet\nARTS[38633296]: - MatrixSet\nARTS[38633296]: - Tensor4SetConstant\nARTS[38633296]: - ArrayOfStringSet\nARTS[38633296]: - Touch\nARTS[38633296]: - FlagOff\nARTS[38633296]: - MatrixSet\nARTS[38633296]: - NumericSet\nARTS[38633296]: - ArrayOfStringSet\nARTS[38633296]: - Tensor3SetConstant\nARTS[38633296]: - Tensor3SetConstant\nARTS[38633296]: - Tensor3SetConstant\nARTS[38633296]: - Tensor3SetConstant\nARTS[38633296]: - Tensor3SetConstant\nARTS[38633296]: - Tensor3SetConstant\nARTS[38633296]: - IndexSet\nARTS[38633296]: - IndexSet\nARTS[38633296]: - IndexSet\nARTS[38633296]: - IndexSet\nARTS[38633296]: - FlagOff\nARTS[38633296]: - output_file_formatSetAscii\nARTS[38633296]: - StringSet\nARTS[38633296]: - IndexSet\nARTS[38633296]: - abs_lineshapeDefine\nARTS[38633296]: - NumericSet\nARTS[38633296]: - NumericSet\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - IndexSet\nARTS[38633296]: - IndexSet\nARTS[38633296]: - NumericSet\nARTS[38633296]: - NumericSet\nARTS[38633296]: - nlteOff\nARTS[38633296]: - partition_functionsInitFromBuiltin\nARTS[38633296]: - IndexSet\nARTS[38633296]: }\n\nARTS[38633296]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[38633296]: {\nARTS[38633296]: - abs_cont_descriptionInit\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: - abs_cont_descriptionAppend\nARTS[38633296]: }\n\nARTS[38633296]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[38633296]: {\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - FlagOff\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - FlagOff\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - AgendaCreate\nARTS[38633296]: - AgendaSet\nARTS[38633296]: }\n\nARTS[38633296]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[38633296]: {\nARTS[38633296]: - isotopologue_ratiosInitFromBuiltin\nARTS[38633296]: - refellipsoidEarth\nARTS[38633296]: - NumericSet\nARTS[38633296]: - AgendaSet\nARTS[38633296]: - NumericSet\nARTS[38633296]: }\n\nARTS[38633296]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[38633296]: {\nARTS[38633296]: - ArrayOfArrayOfIndexCreate\nARTS[38633296]: - ArrayOfIndexCreate\nARTS[38633296]: - VectorCreate\nARTS[38633296]: - ArrayOfIndexCreate\nARTS[38633296]: - NumericCreate\nARTS[38633296]: - VectorCreate\nARTS[38633296]: - IndexCreate\nARTS[38633296]: }\n\nARTS[38633296]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[38633296]: {\nARTS[38633296]: - MatrixSet\nARTS[38633296]: - MatrixSet\nARTS[38633296]: - ArrayOfStringSet\nARTS[38633296]: - VectorSet\nARTS[38633296]: - ArrayOfIndexSet\nARTS[38633296]: - VectorSet\nARTS[38633296]: - Extract\nARTS[38633296]: - nrowsGet\nARTS[38633296]: - VectorSetConstant\nARTS[38633296]: - Delete\nARTS[38633296]: }\n\nARTS[38633296]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[38633296]: {\nARTS[38633296]: - Select\nARTS[38633296]: - Select\nARTS[38633296]: - Select\nARTS[38633296]: - Select\nARTS[38633296]: - Select\nARTS[38633296]: - f_gridMetMM\nARTS[38633296]: - sensor_responseMetMM\nARTS[38633296]: }\n\n[stdout:14] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[53923056]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[53923056]: {\nARTS[53923056]: - verbosityInit\nARTS[53923056]: - scat_speciesSet\nARTS[53923056]: - MatrixSet\nARTS[53923056]: - Tensor4SetConstant\nARTS[53923056]: - ArrayOfStringSet\nARTS[53923056]: - Touch\nARTS[53923056]: - FlagOff\nARTS[53923056]: - MatrixSet\nARTS[53923056]: - NumericSet\nARTS[53923056]: - ArrayOfStringSet\nARTS[53923056]: - Tensor3SetConstant\nARTS[53923056]: - Tensor3SetConstant\nARTS[53923056]: - Tensor3SetConstant\nARTS[53923056]: - Tensor3SetConstant\nARTS[53923056]: - Tensor3SetConstant\nARTS[53923056]: - Tensor3SetConstant\nARTS[53923056]: - IndexSet\nARTS[53923056]: - IndexSet\nARTS[53923056]: - IndexSet\nARTS[53923056]: - IndexSet\nARTS[53923056]: - FlagOff\nARTS[53923056]: - output_file_formatSetAscii\nARTS[53923056]: - StringSet\nARTS[53923056]: - IndexSet\nARTS[53923056]: - abs_lineshapeDefine\nARTS[53923056]: - NumericSet\nARTS[53923056]: - NumericSet\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - IndexSet\nARTS[53923056]: - IndexSet\nARTS[53923056]: - NumericSet\nARTS[53923056]: - NumericSet\nARTS[53923056]: - nlteOff\nARTS[53923056]: - partition_functionsInitFromBuiltin\nARTS[53923056]: - IndexSet\nARTS[53923056]: }\n\nARTS[53923056]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[53923056]: {\nARTS[53923056]: - abs_cont_descriptionInit\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: - abs_cont_descriptionAppend\nARTS[53923056]: }\n\nARTS[53923056]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[53923056]: {\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - FlagOff\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - FlagOff\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - AgendaCreate\nARTS[53923056]: - AgendaSet\nARTS[53923056]: }\n\nARTS[53923056]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[53923056]: {\nARTS[53923056]: - isotopologue_ratiosInitFromBuiltin\nARTS[53923056]: - refellipsoidEarth\nARTS[53923056]: - NumericSet\nARTS[53923056]: - AgendaSet\nARTS[53923056]: - NumericSet\nARTS[53923056]: }\n\nARTS[53923056]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[53923056]: {\nARTS[53923056]: - ArrayOfArrayOfIndexCreate\nARTS[53923056]: - ArrayOfIndexCreate\nARTS[53923056]: - VectorCreate\nARTS[53923056]: - ArrayOfIndexCreate\nARTS[53923056]: - NumericCreate\nARTS[53923056]: - VectorCreate\nARTS[53923056]: - IndexCreate\nARTS[53923056]: }\n\nARTS[53923056]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[53923056]: {\nARTS[53923056]: - MatrixSet\nARTS[53923056]: - MatrixSet\nARTS[53923056]: - ArrayOfStringSet\nARTS[53923056]: - VectorSet\nARTS[53923056]: - ArrayOfIndexSet\nARTS[53923056]: - VectorSet\nARTS[53923056]: - Extract\nARTS[53923056]: - nrowsGet\nARTS[53923056]: - VectorSetConstant\nARTS[53923056]: - Delete\nARTS[53923056]: }\n\nARTS[53923056]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[53923056]: {\nARTS[53923056]: - Select\nARTS[53923056]: - Select\nARTS[53923056]: - Select\nARTS[53923056]: - Select\nARTS[53923056]: - Select\nARTS[53923056]: - f_gridMetMM\nARTS[53923056]: - sensor_responseMetMM\nARTS[53923056]: }\n\n[stdout:15] \nLoading ARTS API from: /home/simonpf/build/arts/src/libarts_api.so\nARTS[63539808]: Executing /home/simonpf/src/arts/controlfiles/general/general.arts\nARTS[63539808]: {\nARTS[63539808]: - verbosityInit\nARTS[63539808]: - scat_speciesSet\nARTS[63539808]: - MatrixSet\nARTS[63539808]: - Tensor4SetConstant\nARTS[63539808]: - ArrayOfStringSet\nARTS[63539808]: - Touch\nARTS[63539808]: - FlagOff\nARTS[63539808]: - MatrixSet\nARTS[63539808]: - NumericSet\nARTS[63539808]: - ArrayOfStringSet\nARTS[63539808]: - Tensor3SetConstant\nARTS[63539808]: - Tensor3SetConstant\nARTS[63539808]: - Tensor3SetConstant\nARTS[63539808]: - Tensor3SetConstant\nARTS[63539808]: - Tensor3SetConstant\nARTS[63539808]: - Tensor3SetConstant\nARTS[63539808]: - IndexSet\nARTS[63539808]: - IndexSet\nARTS[63539808]: - IndexSet\nARTS[63539808]: - IndexSet\nARTS[63539808]: - FlagOff\nARTS[63539808]: - output_file_formatSetAscii\nARTS[63539808]: - StringSet\nARTS[63539808]: - IndexSet\nARTS[63539808]: - abs_lineshapeDefine\nARTS[63539808]: - NumericSet\nARTS[63539808]: - NumericSet\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - IndexSet\nARTS[63539808]: - IndexSet\nARTS[63539808]: - NumericSet\nARTS[63539808]: - NumericSet\nARTS[63539808]: - nlteOff\nARTS[63539808]: - partition_functionsInitFromBuiltin\nARTS[63539808]: - IndexSet\nARTS[63539808]: }\n\nARTS[63539808]: Executing /home/simonpf/src/arts/controlfiles/general/continua.arts\nARTS[63539808]: {\nARTS[63539808]: - abs_cont_descriptionInit\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: - abs_cont_descriptionAppend\nARTS[63539808]: }\n\nARTS[63539808]: Executing /home/simonpf/src/arts/controlfiles/general/agendas.arts\nARTS[63539808]: {\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - FlagOff\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - FlagOff\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - AgendaCreate\nARTS[63539808]: - AgendaSet\nARTS[63539808]: }\n\nARTS[63539808]: Executing /home/simonpf/src/arts/controlfiles/general/planet_earth.arts\nARTS[63539808]: {\nARTS[63539808]: - isotopologue_ratiosInitFromBuiltin\nARTS[63539808]: - refellipsoidEarth\nARTS[63539808]: - NumericSet\nARTS[63539808]: - AgendaSet\nARTS[63539808]: - NumericSet\nARTS[63539808]: }\n\nARTS[63539808]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/prepare_metmm.arts\nARTS[63539808]: {\nARTS[63539808]: - ArrayOfArrayOfIndexCreate\nARTS[63539808]: - ArrayOfIndexCreate\nARTS[63539808]: - VectorCreate\nARTS[63539808]: - ArrayOfIndexCreate\nARTS[63539808]: - NumericCreate\nARTS[63539808]: - VectorCreate\nARTS[63539808]: - IndexCreate\nARTS[63539808]: }\n\nARTS[63539808]: Executing /home/simonpf/src/atms_simulations/sensor_atms.arts\nARTS[63539808]: {\nARTS[63539808]: - MatrixSet\nARTS[63539808]: - MatrixSet\nARTS[63539808]: - ArrayOfStringSet\nARTS[63539808]: - VectorSet\nARTS[63539808]: - ArrayOfIndexSet\nARTS[63539808]: - VectorSet\nARTS[63539808]: - Extract\nARTS[63539808]: - nrowsGet\nARTS[63539808]: - VectorSetConstant\nARTS[63539808]: - Delete\nARTS[63539808]: }\n\nARTS[63539808]: Executing /home/simonpf/src/arts/controlfiles/instruments/metmm/sensor_descriptions/apply_metmm.arts\nARTS[63539808]: {\nARTS[63539808]: - Select\nARTS[63539808]: - Select\nARTS[63539808]: - Select\nARTS[63539808]: - Select\nARTS[63539808]: - Select\nARTS[63539808]: - f_gridMetMM\nARTS[63539808]: - sensor_responseMetMM\nARTS[63539808]: }\n\n"
]
],
[
[
"## A Priori State\n\nThe simulations are based on the a priori assumptions, that the profiles of specific humidity, temperature and ozone vary independently and that the relative variations can be described by Log-Gaussian distributions.",
"_____no_output_____"
]
],
[
[
"%%px\nqt_mean = np.load(\"data/qt_mean.npy\").ravel()\nqt_cov = np.load(\"data/qt_cov.npy\")\nqt_cov_inv = np.linalg.inv(qt_cov)",
"_____no_output_____"
]
],
[
[
"## Jumping Functions\n\nThe jumping functions are used inside the MCMC iteration and propose new atmospheric states for specific humidity, temperature and ozone, respectively. The proposed states are generated from random walks that use scaled versions of the a priori covariances.",
"_____no_output_____"
]
],
[
[
"%%px\nimport numpy as np\nfrom typhon.retrieval.mcmc import RandomWalk\nc = (1.0 / np.sqrt(qt_mean.size)) ** 2\nrw_qt = RandomWalk(c * qt_cov)\n\ndef j_qt(ws, x, revert = False):\n if revert:\n x_new = x\n else:\n x_new = rw_qt.step(x)\n q_new = (np.exp(x_new[14::-1]).reshape((15,)))\n q_new = atms.mmr2vmr(ws, q_new, \"h2o\")\n ws.vmr_field.value[0, :, 0, 0] = q_new\n ws.t_field.value[:, 0, 0] = x_new[:14:-1]\n ws.sst = np.maximum(ws.t_field.value[0, 0, 0], 270.0)\n return x_new",
"_____no_output_____"
]
],
[
[
"## A Priori Distributions\n\nThese functions return the likelihood (up to an additive constant) of a given state for each of the variables. Note that the states of specific humidity, temperature and ozone are given by the logs of the relative variations. ",
"_____no_output_____"
]
],
[
[
"%%px\ndef p_a_qt(x):\n dx = x - qt_mean\n l = - 0.5 * np.dot(dx, np.dot(qt_cov_inv, dx))\n return l",
"_____no_output_____"
]
],
[
[
"## Measurement Uncertainty\n\nWe assume that uncertainty of the measured brightness temperatures can be described by independent Gaussian error with a standard deviation of $1 K$.",
"_____no_output_____"
]
],
[
[
"%%px\ncovmat_y = np.diag(np.ones(len(channels)))\ncovmat_y_inv = np.linalg.inv(covmat_y)\n\ndef p_y(y, yf):\n dy = y - yf\n l = - 0.5 * np.dot(dy, np.dot(covmat_y_inv, dy))\n return l",
"_____no_output_____"
]
],
[
[
"# Running MCMC\n",
"_____no_output_____"
],
[
"### The Simulated Measurement\n\nFor the simulated measurement, we sample a state from the a priori distribution of atmsopheric states and simulate the measured brightness temperatures.",
"_____no_output_____"
],
[
"A simple heuristic is applied to ensure that reasonable acceptance rates are obtained during the MCMC simulations. After the initial burn-in phase, 1000 simulation steps are performed. If the acceptance rates during this simulation are too low/high that covariance matrices of the corresponding random walks are scaled by a factor 0.1 / 9.0, respectively.",
"_____no_output_____"
]
],
[
[
"%%px\ndef adapt_covariances(a):\n if (np.sum(a[:, 0]) / a.shape[0]) < 0.2:\n rw_qt.covmat *= 0.7\n if (np.sum(a[:, 0]) / a.shape[0]) > 0.4:\n rw_qt.covmat *= 1.5",
"_____no_output_____"
],
[
"%%px\nfrom typhon.retrieval.mcmc import MCMC\nfrom atms import vmr2cd\n\ndist = atms.StateDistribution()\nn_burn_in = 500\nn_prod = 5000\ndrop = 10",
"_____no_output_____"
],
[
"from typhon.retrieval.mcmc import MCMC\nfrom atms import vmr2cd\n\ndef run_retrieval(i):\n \n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n\n # Generate True State\n dist.sample(ws)\n ws.yCalc()\n y_true = np.copy(ws.y)\n q_true = np.copy(ws.vmr_field.value[0, :, 0, 0].ravel())\n t_true = np.copy(ws.t_field.value[:, 0, 0].ravel())\n cwv_true = atms.vmr2cd(ws)\n \n dist.a_priori(ws)\n qt = np.zeros(qt_mean.size)\n\n # Add Noise\n y_true += np.random.randn(*y_true.shape)\n \n #try:\n mcmc = MCMC([[qt, p_a_qt, j_qt]], y_true, p_y, [vmr2cd])\n\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_1, s_1, _, _ = mcmc.run(ws, n_prod)\n\n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_2, s_2, _, _ = mcmc.run(ws, n_prod)\n\n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_3, s_3, _, _ = mcmc.run(ws, n_prod)\n\n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_4, s_4, _, _ = mcmc.run(ws, n_prod)\n\n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_5, s_5, _, _ = mcmc.run(ws, n_prod)\n\n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_6, s_6, _, _ = mcmc.run(ws, n_prod)\n\n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_7, s_7, _, _ = mcmc.run(ws, n_prod)\n\n # Reset covariance matrices.\n rw_qt.covmat = np.copy(c * qt_cov)\n qt_0 = dist.sample_factors()\n _, _, _, a = mcmc.warm_up(ws, [qt_0], n_burn_in)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, 200)\n adapt_covariances(a)\n _, _, _, a = mcmc.run(ws, n_burn_in)\n hist_8, s_8, _, _ = mcmc.run(ws, n_prod)\n\n profiles_q = np.stack([hist_1[0][::drop, :15],\n hist_2[0][::drop, :15],\n hist_3[0][::drop, :15],\n hist_4[0][::drop, :15],\n hist_5[0][::drop, :15],\n hist_6[0][::drop, :15],\n hist_7[0][::drop, :15],\n hist_8[0][::drop, :15]])\n profiles_t = np.stack([hist_1[0][::drop, 15:],\n hist_2[0][::drop, 15:],\n hist_3[0][::drop, 15:],\n hist_4[0][::drop, 15:],\n hist_5[0][::drop, 15:],\n hist_6[0][::drop, 15:],\n hist_7[0][::drop, 15:],\n hist_8[0][::drop, 15:]])\n cwv = np.stack([s_1[::drop], s_2[::drop], s_3[::drop], s_4[::drop],\n s_5[::drop],s_6[::drop],s_7[::drop],s_8[::drop]], axis=0)\n return y_true, q_true, cwv_true, profiles_q, profiles_t, cwv ",
"_____no_output_____"
]
],
[
[
"## Running the Retrievals",
"_____no_output_____"
]
],
[
[
"import numpy as np\nids = np.arange(3500)\nrs = lview.map_async(run_retrieval, ids)",
"_____no_output_____"
],
[
"from atms import create_output_file\n\nroot_group, v_y_true, v_cwv_true, v_cwv ,v_h2o = create_output_file(\"data/mcmc_retrievals_5.nc\", 5, 15)\nfor y_true, h2o_true, cwv_true, profiles_q, profiles_t, cwv in rs:\n if not y_true is None:\n t = v_cwv_true.shape[0]\n print(\"saving simulation: \" + str(t))\n steps=cwv.size\n v_y_true[t,:] = y_true\n ws.vmr_field.value[0,:,:,:] = h2o_true.reshape(-1,1,1)\n v_cwv_true[t] = cwv_true\n v_cwv[t, :steps] = cwv[:]\n v_h2o[t, :steps,:] = profiles_q.ravel().reshape(-1, 15)\n else:\n print(\"failure in simulation: \" + str(t))\n print(h2o_true)\n print(cwv_true)\n print(profiles)",
"loading existing file: data/mcmc_retrievals_5.nc\nsaving simulation: 9046\nsaving simulation: 9047\nsaving simulation: 9048\nsaving simulation: 9049\nsaving simulation: 9050\nsaving simulation: 9051\nsaving simulation: 9052\nsaving simulation: 9053\nsaving simulation: 9054\nsaving simulation: 9055\nsaving simulation: 9056\nsaving simulation: 9057\nsaving simulation: 9058\nsaving simulation: 9059\nsaving simulation: 9060\nsaving simulation: 9061\nsaving simulation: 9062\nsaving simulation: 9063\nsaving simulation: 9064\nsaving simulation: 9065\nsaving simulation: 9066\nsaving simulation: 9067\nsaving simulation: 9068\nsaving simulation: 9069\nsaving simulation: 9070\nsaving simulation: 9071\nsaving simulation: 9072\nsaving simulation: 9073\nsaving simulation: 9074\nsaving simulation: 9075\nsaving simulation: 9076\nsaving simulation: 9077\nsaving simulation: 9078\nsaving simulation: 9079\nsaving simulation: 9080\nsaving simulation: 9081\nsaving simulation: 9082\nsaving simulation: 9083\nsaving simulation: 9084\nsaving simulation: 9085\nsaving simulation: 9086\nsaving simulation: 9087\nsaving simulation: 9088\nsaving simulation: 9089\nsaving simulation: 9090\nsaving simulation: 9091\nsaving simulation: 9092\nsaving simulation: 9093\nsaving simulation: 9094\nsaving simulation: 9095\nsaving simulation: 9096\nsaving simulation: 9097\nsaving simulation: 9098\nsaving simulation: 9099\nsaving simulation: 9100\nsaving simulation: 9101\nsaving simulation: 9102\nsaving simulation: 9103\nsaving simulation: 9104\nsaving simulation: 9105\nsaving simulation: 9106\nsaving simulation: 9107\nsaving simulation: 9108\nsaving simulation: 9109\nsaving simulation: 9110\nsaving simulation: 9111\nsaving simulation: 9112\nsaving simulation: 9113\nsaving simulation: 9114\nsaving simulation: 9115\nsaving simulation: 9116\nsaving simulation: 9117\nsaving simulation: 9118\nsaving simulation: 9119\nsaving simulation: 9120\nsaving simulation: 9121\nsaving simulation: 9122\nsaving simulation: 9123\nsaving simulation: 9124\nsaving simulation: 9125\nsaving simulation: 9126\nsaving simulation: 9127\nsaving simulation: 9128\nsaving simulation: 9129\nsaving simulation: 9130\nsaving simulation: 9131\nsaving simulation: 9132\nsaving simulation: 9133\nsaving simulation: 9134\nsaving simulation: 9135\nsaving simulation: 9136\nsaving simulation: 9137\nsaving simulation: 9138\nsaving simulation: 9139\nsaving simulation: 9140\nsaving simulation: 9141\nsaving simulation: 9142\nsaving simulation: 9143\nsaving simulation: 9144\nsaving simulation: 9145\nsaving simulation: 9146\nsaving simulation: 9147\nsaving simulation: 9148\nsaving simulation: 9149\nsaving simulation: 9150\nsaving simulation: 9151\nsaving simulation: 9152\nsaving simulation: 9153\nsaving simulation: 9154\nsaving simulation: 9155\nsaving simulation: 9156\nsaving simulation: 9157\nsaving simulation: 9158\nsaving simulation: 9159\nsaving simulation: 9160\nsaving simulation: 9161\nsaving simulation: 9162\nsaving simulation: 9163\nsaving simulation: 9164\nsaving simulation: 9165\nsaving simulation: 9166\nsaving simulation: 9167\nsaving simulation: 9168\nsaving simulation: 9169\nsaving simulation: 9170\nsaving simulation: 9171\nsaving simulation: 9172\nsaving simulation: 9173\nsaving simulation: 9174\nsaving simulation: 9175\nsaving simulation: 9176\nsaving simulation: 9177\nsaving simulation: 9178\nsaving simulation: 9179\nsaving simulation: 9180\nsaving simulation: 9181\nsaving simulation: 9182\nsaving simulation: 9183\nsaving simulation: 9184\nsaving simulation: 9185\nsaving simulation: 9186\nsaving simulation: 9187\nsaving simulation: 9188\nsaving simulation: 9189\nsaving simulation: 9190\nsaving simulation: 9191\nsaving simulation: 9192\nsaving simulation: 9193\nsaving simulation: 9194\nsaving simulation: 9195\nsaving simulation: 9196\nsaving simulation: 9197\nsaving simulation: 9198\nsaving simulation: 9199\nsaving simulation: 9200\nsaving simulation: 9201\nsaving simulation: 9202\nsaving simulation: 9203\nsaving simulation: 9204\nsaving simulation: 9205\nsaving simulation: 9206\nsaving simulation: 9207\nsaving simulation: 9208\nsaving simulation: 9209\nsaving simulation: 9210\nsaving simulation: 9211\nsaving simulation: 9212\nsaving simulation: 9213\nsaving simulation: 9214\nsaving simulation: 9215\nsaving simulation: 9216\nsaving simulation: 9217\nsaving simulation: 9218\nsaving simulation: 9219\nsaving simulation: 9220\nsaving simulation: 9221\nsaving simulation: 9222\nsaving simulation: 9223\nsaving simulation: 9224\nsaving simulation: 9225\nsaving simulation: 9226\nsaving simulation: 9227\nsaving simulation: 9228\nsaving simulation: 9229\nsaving simulation: 9230\nsaving simulation: 9231\nsaving simulation: 9232\nsaving simulation: 9233\nsaving simulation: 9234\nsaving simulation: 9235\nsaving simulation: 9236\nsaving simulation: 9237\nsaving simulation: 9238\nsaving simulation: 9239\nsaving simulation: 9240\nsaving simulation: 9241\nsaving simulation: 9242\nsaving simulation: 9243\nsaving simulation: 9244\nsaving simulation: 9245\nsaving simulation: 9246\nsaving simulation: 9247\nsaving simulation: 9248\nsaving simulation: 9249\nsaving simulation: 9250\nsaving simulation: 9251\nsaving simulation: 9252\nsaving simulation: 9253\nsaving simulation: 9254\nsaving simulation: 9255\nsaving simulation: 9256\nsaving simulation: 9257\nsaving simulation: 9258\nsaving simulation: 9259\nsaving simulation: 9260\nsaving simulation: 9261\nsaving simulation: 9262\nsaving simulation: 9263\nsaving simulation: 9264\nsaving simulation: 9265\nsaving simulation: 9266\nsaving simulation: 9267\nsaving simulation: 9268\nsaving simulation: 9269\nsaving simulation: 9270\nsaving simulation: 9271\nsaving simulation: 9272\nsaving simulation: 9273\nsaving simulation: 9274\nsaving simulation: 9275\nsaving simulation: 9276\nsaving simulation: 9277\nsaving simulation: 9278\nsaving simulation: 9279\nsaving simulation: 9280\nsaving simulation: 9281\nsaving simulation: 9282\nsaving simulation: 9283\nsaving simulation: 9284\nsaving simulation: 9285\nsaving simulation: 9286\nsaving simulation: 9287\nsaving simulation: 9288\nsaving simulation: 9289\nsaving simulation: 9290\nsaving simulation: 9291\nsaving simulation: 9292\nsaving simulation: 9293\nsaving simulation: 9294\nsaving simulation: 9295\nsaving simulation: 9296\nsaving simulation: 9297\nsaving simulation: 9298\nsaving simulation: 9299\nsaving simulation: 9300\nsaving simulation: 9301\nsaving simulation: 9302\nsaving simulation: 9303\nsaving simulation: 9304\nsaving simulation: 9305\nsaving simulation: 9306\nsaving simulation: 9307\nsaving simulation: 9308\nsaving simulation: 9309\nsaving simulation: 9310\nsaving simulation: 9311\nsaving simulation: 9312\nsaving simulation: 9313\nsaving simulation: 9314\nsaving simulation: 9315\nsaving simulation: 9316\nsaving simulation: 9317\nsaving simulation: 9318\nsaving simulation: 9319\nsaving simulation: 9320\nsaving simulation: 9321\nsaving simulation: 9322\nsaving simulation: 9323\nsaving simulation: 9324\nsaving simulation: 9325\nsaving simulation: 9326\nsaving simulation: 9327\nsaving simulation: 9328\nsaving simulation: 9329\nsaving simulation: 9330\nsaving simulation: 9331\nsaving simulation: 9332\nsaving simulation: 9333\nsaving simulation: 9334\nsaving simulation: 9335\nsaving simulation: 9336\nsaving simulation: 9337\nsaving simulation: 9338\nsaving simulation: 9339\nsaving simulation: 9340\nsaving simulation: 9341\nsaving simulation: 9342\nsaving simulation: 9343\nsaving simulation: 9344\nsaving simulation: 9345\nsaving simulation: 9346\nsaving simulation: 9347\nsaving simulation: 9348\nsaving simulation: 9349\nsaving simulation: 9350\nsaving simulation: 9351\nsaving simulation: 9352\nsaving simulation: 9353\nsaving simulation: 9354\nsaving simulation: 9355\nsaving simulation: 9356\nsaving simulation: 9357\nsaving simulation: 9358\nsaving simulation: 9359\nsaving simulation: 9360\nsaving simulation: 9361\nsaving simulation: 9362\nsaving simulation: 9363\nsaving simulation: 9364\nsaving simulation: 9365\nsaving simulation: 9366\nsaving simulation: 9367\nsaving simulation: 9368\nsaving simulation: 9369\nsaving simulation: 9370\nsaving simulation: 9371\nsaving simulation: 9372\nsaving simulation: 9373\nsaving simulation: 9374\nsaving simulation: 9375\nsaving simulation: 9376\nsaving simulation: 9377\nsaving simulation: 9378\nsaving simulation: 9379\nsaving simulation: 9380\nsaving simulation: 9381\nsaving simulation: 9382\nsaving simulation: 9383\nsaving simulation: 9384\nsaving simulation: 9385\nsaving simulation: 9386\nsaving simulation: 9387\n"
],
[
"import matplotlib_settings\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"root_group.close()",
"_____no_output_____"
],
[
"\nroot_group, v_y_true, v_cwv_true, v_cwv ,v_h2o = create_output_file(\"data/mcmc_retrievals_5.nc\", 5, 27)",
"_____no_output_____"
],
[
"for i in range(1000, 1100):\n plt.plot(v_cwv[i, :])\n plt.gca().axhline(v_cwv_true[i], c = 'k', ls = '--')",
"_____no_output_____"
],
[
"v_h2o[118, 250:500, :].shape",
"_____no_output_____"
],
[
"plt.plot(np.mean(profs_t[2, 0:200], axis = 0), p)\nplt.plot(np.mean(profs_t[2, 200:400], axis = 0), p)\nplt.title(\"Temperature Profiles\")\nplt.xlabel(\"T [K]\")\nplt.ylabel(\"P [hPa]\")\nplt.gca().invert_yaxis()",
"_____no_output_____"
],
[
"p = np.load(\"data/p_grid.npy\")\nprofiles_t[1, :, :].shape",
"_____no_output_____"
],
[
"plt.plot(np.mean(np.exp(profs_q[1, 0:200]) * 18.0 / 28.9, axis = 0), p)\nplt.plot(np.mean(np.exp(profs_q[1, 200:400]) * 18.0/ 28.9, axis = 0), p)\nplt.gca().invert_yaxis()\nplt.title(\"Water Vapor Profiles\")\nplt.xlabel(\"$H_2O$ [mol / mol]\")\nplt.ylabel(\"P [hPa]\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbad3d87600f48ec3f1734c7c42f9f0b6c12bd00
| 881 |
ipynb
|
Jupyter Notebook
|
notebooks/07.00-Interfacing_with_Laboratory_Equipment.ipynb
|
jckantor/cbe61622
|
bdc08e6c4f0674c5e991617945cafd1b121d6b4b
|
[
"MIT"
] | 2 |
2021-11-22T20:36:35.000Z
|
2021-12-07T07:52:10.000Z
|
notebooks/07.00-Interfacing_with_Laboratory_Equipment.ipynb
|
jckantor/cbe-virtual-laboratory
|
bdc08e6c4f0674c5e991617945cafd1b121d6b4b
|
[
"MIT"
] | null | null | null |
notebooks/07.00-Interfacing_with_Laboratory_Equipment.ipynb
|
jckantor/cbe-virtual-laboratory
|
bdc08e6c4f0674c5e991617945cafd1b121d6b4b
|
[
"MIT"
] | 1 |
2021-12-11T20:39:32.000Z
|
2021-12-11T20:39:32.000Z
| 17.979592 | 63 | 0.5437 |
[
[
[
"# Interfacing with Laboratory Equipment\n",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
cbad3f3b48ec6f320fa4b38133c142b2875cc1c7
| 649,440 |
ipynb
|
Jupyter Notebook
|
analysis/validated-boundaries-vs-government-unit-density.ipynb
|
tbuffington7/data
|
6ef19c9ad5b3c9cea6fcdf13d04e7edf51aa7eb9
|
[
"MIT"
] | null | null | null |
analysis/validated-boundaries-vs-government-unit-density.ipynb
|
tbuffington7/data
|
6ef19c9ad5b3c9cea6fcdf13d04e7edf51aa7eb9
|
[
"MIT"
] | null | null | null |
analysis/validated-boundaries-vs-government-unit-density.ipynb
|
tbuffington7/data
|
6ef19c9ad5b3c9cea6fcdf13d04e7edf51aa7eb9
|
[
"MIT"
] | null | null | null | 41.638777 | 5,708 | 0.338873 |
[
[
[
"## Validated boundaries to government unit incident density comparison\n\nThe backing theory for this notebook is proving that we will be able to use the highest-density (fire count vs government unit area) government unit to determine a department's boundary for departments that do not have boundaries.",
"_____no_output_____"
]
],
[
[
"import psycopg2\nfrom psycopg2.extras import RealDictCursor\nimport pandas as pd\n# import geopandas as gpd\n# from shapely import wkb\n# from shapely.geometry import mapping as to_geojson\n# import folium\n\npd.options.display.max_columns = None\npd.options.display.max_rows = None\n#pd.set_option('display.float_format', lambda x: '%.3f' % x)\n\n%matplotlib inline",
"/Users/joe/.pyenv/versions/2.7.14/envs/firecares-data/lib/python2.7/site-packages/psycopg2/__init__.py:144: UserWarning: The psycopg2 wheel package will be renamed from release 2.8; in order to keep installing from binary please use \"pip install psycopg2-binary\" instead. For details see: <http://initd.org/psycopg/docs/install.html#binary-install-from-pypi>.\n \"\"\")\n"
],
[
"conn = psycopg2.connect('service=firecares')\nnfirs = psycopg2.connect('service=nfirs')",
"_____no_output_____"
]
],
[
[
"### DB migration/setup",
"_____no_output_____"
]
],
[
[
"# Create materialized view of all usgs govt units in FireCARES\n\nq = \"\"\"\ncreate materialized view if not exists usgs_governmentunits as\n(\n select id, county_name as name, 'countyorequivalent' as source, geom from usgs_countyorequivalent where geom is not null\n union\n select id, place_name as name, 'incorporatedplace' as source, geom from usgs_incorporatedplace where geom is not null\n union\n select id, minorcivildivision_name as name, 'minorcivildivision' as source, geom from usgs_minorcivildivision where geom is not null\n union\n select id, name, 'nativeamericanarea' as source, geom from usgs_nativeamericanarea where geom is not null\n union\n select id, name, 'reserve' as source, geom from usgs_reserve where geom is not null\n union\n select id, state_name as name, 'stateorterritoryhigh' as source, geom from usgs_stateorterritoryhigh where geom is not null\n union\n select id, place_name as name, 'unincorporatedplace' as source, geom from usgs_unincorporatedplace where geom is not null\n);\n\ncreate unique index on usgs_governmentunits (id, source);\ncreate index on usgs_governmentunits using gist (geom);\n\"\"\"\n \nwith conn.cursor() as c:\n c.execute(q)\n \nconn.commit()",
"_____no_output_____"
],
[
"# Link remote firecares usgs_governmentunits view to nfirs-local usgs_government units\n\nq = \"\"\"\ncreate foreign table usgs_governmentunits (id integer, name character varying(120), source text, geom geometry)\nserver firecares\noptions (table_name 'usgs_governmentunits');\n\"\"\"\n\nwith nfirs.cursor() as c:\n c.execute(q)\n \nnfirs.commit()",
"_____no_output_____"
],
[
"# Old nfirs.firestation_firedepartment foreign table columns needed to be synced\n\nq = \"\"\"\nalter foreign TABLE firestation_firedepartment add column archived boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column domain_name character varying(255);\nalter foreign TABLE firestation_firedepartment add column owned_tracts_geom public.geometry(MultiPolygon,4326);\nalter foreign TABLE firestation_firedepartment add column display_metrics boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column boundary_verified boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column cfai_accredited boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column ems_transport boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column staffing_verified boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column stations_verified boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column census_override boolean NOT NULL;\nalter foreign TABLE firestation_firedepartment add column additional_fdids character varying(255);\n\"\"\"\n\nwith nfirs.cursor() as c:\n c.execute(q)\nnfirs.commit()",
"_____no_output_____"
],
[
"q = \"\"\"\ncreate foreign table if not exists firecares_core_address (id integer NOT NULL,\n address_line1 character varying(100) NOT NULL,\n address_line2 character varying(100),\n city character varying(50) NOT NULL,\n state_province character varying(40) NOT NULL,\n postal_code character varying(10) NOT NULL,\n geom public.geometry(Point,4326),\n geocode_results text,\n country_id character varying(2) NOT NULL)\nserver firecares\noptions (table_name 'firecares_core_address');\n\"\"\"\n\nwith nfirs.cursor() as c:\n c.execute(q)\nnfirs.commit()",
"_____no_output_____"
]
],
[
[
"### Processing",
"_____no_output_____"
]
],
[
[
"q = \"\"\"\nselect id, fdid, state, name\nfrom firestation_firedepartment\nwhere boundary_verified = true;\n\"\"\"\n\nwith nfirs.cursor(cursor_factory=RealDictCursor) as c:\n c.execute(q)\n fds = c.fetchall()",
"_____no_output_____"
],
[
"q = \"\"\"\nwith fires as (select * from joint_buildingfires\n inner join joint_incidentaddress\n using (fdid, inc_no, inc_date, state, exp_no)\n where state = %(state)s and fdid = %(fdid)s\n),\ngovt_units as (\n select gu.name, gu.source, gu.id, gu.geom, fd.id as fc_id, fd.geom as fd_geom, ST_Distance(addr.geom, ST_Centroid(gu.geom)) as distance_to_headquarters\n from firestation_firedepartment fd\n inner join firecares_core_address addr\n on addr.id = fd.headquarters_address_id\n join usgs_governmentunits gu\n on ST_Intersects(ST_Buffer(addr.geom, 0.05), gu.geom)\n where \n fd.fdid = %(fdid)s and fd.state = %(state)s and source != 'stateorterritoryhigh'\n )\nselect gu.fc_id, count(fires) / ST_Area(gu.geom) as density, count(fires), ST_Area(ST_SymDifference(gu.fd_geom, gu.geom)) / ST_Area(gu.fd_geom) as percent_difference_to_verified_boundary, ST_Area(gu.geom), gu.distance_to_headquarters, gu.name, gu.id, gu.source from fires\ninner join govt_units gu\non ST_Intersects(fires.geom, gu.geom)\ngroup by gu.name, gu.id, gu.geom, gu.source, gu.distance_to_headquarters, gu.fd_geom, gu.fc_id\norder by ST_Area(gu.geom) / count(fires) asc;\n\"\"\"\n\nfor fd in fds:\n with nfirs.cursor(cursor_factory=RealDictCursor) as c:\n print 'Analyzing: {} (id: {} fdid: {} {})'.format(fd['name'], fd['id'], fd['fdid'], fd['state'])\n c.execute(q, dict(fdid=fd['fdid'], state=fd['state']))\n items = c.fetchall()\n df = pd.DataFrame(items)\n df.to_csv('./boundary-analysis-{}.csv'.format(fd['id']))",
"Analyzing: Southington Fire Department (id: 95681 fdid: 02230 CT)\n"
]
],
[
[
"### Results",
"_____no_output_____"
]
],
[
[
"from glob import glob",
"_____no_output_____"
],
[
"df = None\nfor f in glob(\"boundary-analysis*.csv\"):\n if df is not None:\n df = df.append(pd.read_csv(f))\n else:\n df = pd.read_csv(f)",
"_____no_output_____"
],
[
"df.rename(columns={'Unnamed: 0': 'rank'}, inplace=True)\nselected_government_units = df[df['rank'] == 0].set_index('fc_id')\ntotal_validated_department_count = len(selected_government_units)\nperfect_fits = len(selected_government_units[selected_government_units['percent_difference_to_verified_boundary'] == 0])",
"_____no_output_____"
],
[
"print 'Perfect fits: {}/{} ({:.2%})'.format(perfect_fits, total_validated_department_count, float(perfect_fits) / total_validated_department_count)\nprint 'Machine-selected government unit area difference mean: {:.2%}'.format(df[df['rank'] == 0].percent_difference_to_verified_boundary.mean())",
"Perfect fits: 25/71 (35.21%)\nMachine-selected government unit area difference mean: 40.29%\n"
],
[
"selected_government_units['percent_difference_to_verified_boundary'].hist(bins=50)",
"_____no_output_____"
],
[
"selected_government_units",
"_____no_output_____"
],
[
"df.set_index('fc_id')",
"_____no_output_____"
],
[
"df.to_csv('./validated-boundary-vs-government-unit-density.csv')",
"_____no_output_____"
],
[
"pd.read_csv('./validated-boundary-vs-government-unit-density.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbad62203b5c931bf091bb9beb829a046e3de779
| 9,431 |
ipynb
|
Jupyter Notebook
|
RF_notebooks/Parameters_search.ipynb
|
haddocking/MD-scoring
|
f744cf4abf4eed8c6b742332042d473d99107941
|
[
"Apache-2.0"
] | null | null | null |
RF_notebooks/Parameters_search.ipynb
|
haddocking/MD-scoring
|
f744cf4abf4eed8c6b742332042d473d99107941
|
[
"Apache-2.0"
] | null | null | null |
RF_notebooks/Parameters_search.ipynb
|
haddocking/MD-scoring
|
f744cf4abf4eed8c6b742332042d473d99107941
|
[
"Apache-2.0"
] | 1 |
2021-11-23T10:24:36.000Z
|
2021-11-23T10:24:36.000Z
| 31.754209 | 172 | 0.591242 |
[
[
[
"# Search for best parameters for Random Forest classifier",
"_____no_output_____"
],
[
"## Read data",
"_____no_output_____"
]
],
[
[
"# Pandas is used for data manipulation\nimport pandas as pd\ntime='80_100'\n\n# Read in data as a dataframe\nfeatures = pd.read_csv('../features/features_training1/features_{}.csv'.format(time))\n\nimport numpy as np\nimport seaborn as sns\nimport matplotlib.pyplot as plt\n# One Hot Encoding\nfeatures_num=features.to_numpy()\nfeatures[:] = np.nan_to_num(features_num)\nnp.where(pd.isnull(features_num))\nfeatures.describe(include='all')\n\n# Extract features and labels and print feature names\nlabels = features['quality']\nfeatures = features.drop('quality', axis = 1)\n\nlabels[1:6]\nnames=features.columns\nprint(names)\n\ny = labels.map({'native':1,\"non-native\":0})\nx = features.values\n# Convert to numpy arrays\nfeatures = np.array(x)\nlabels = np.array(y)",
"_____no_output_____"
]
],
[
[
"## Specify training and test sets",
"_____no_output_____"
]
],
[
[
"# Training and Testing Sets\nfrom sklearn.model_selection import train_test_split\n\ntrain_features, test_features, train_labels, test_labels = train_test_split(features, labels, \n test_size = 0.25, random_state = 42)\n\n",
"_____no_output_____"
]
],
[
[
"## Set a base model with RF classifier ",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\n\nbase_model = RandomForestClassifier(n_estimators = 10,random_state = 42)\n\nfrom pprint import pprint\n\n# Look at parameters used by our current forest\nprint('Parameters currently in use:\\n')\npprint(base_model.get_params())\n\nfrom sklearn import metrics\nbase_model.fit(train_features,train_labels);\npred_labels=base_model.predict(test_features)\nbase_accuracy=metrics.accuracy_score(test_labels, pred_labels)\nprint(\"Base model Accuracy:\",metrics.accuracy_score(test_labels, pred_labels))\n",
"_____no_output_____"
]
],
[
[
"## Random Search with Cross Validation",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV\n\n# Number of trees in random forest\nn_estimators = [int(x) for x in np.linspace(start = 200, stop = 2000, num = 10)]\n# Number of features to consider at every split\nmax_features = ['auto', 'sqrt']\n# Maximum number of levels in tree\nmax_depth = [int(x) for x in np.linspace(10, 110, num = 11)]\nmax_depth.append(None)\n# Minimum number of samples required to split a node\nmin_samples_split = [2, 5, 10]\n# Minimum number of samples required at each leaf node\nmin_samples_leaf = [1, 2, 4]\n# Method of selecting samples for training each tree\nbootstrap = [True, False]\n\n# Create the random grid\nrandom_grid = {'n_estimators': n_estimators,\n 'max_features': max_features,\n 'max_depth': max_depth,\n 'min_samples_split': min_samples_split,\n 'min_samples_leaf': min_samples_leaf,\n 'bootstrap': bootstrap}\n\npprint(random_grid)\n\n# Use the random grid to search for best hyperparameters\n# First create the base model to tune\nrf = RandomForestClassifier(random_state = 42)\n# Random search of parameters, using 3 fold cross validation, \n# search across 100 different combinations, and use all available cores\nrf_random = RandomizedSearchCV(estimator=rf, param_distributions=random_grid,\n n_iter = 100, scoring='neg_mean_absolute_error', \n cv = 3, verbose=2, random_state=42, n_jobs=-1,\n return_train_score=True)\n\n# Fit the random search model\nrf_random.fit(train_features, train_labels);\n\nrf_random.best_params_",
"_____no_output_____"
]
],
[
[
"### Evaluate the Best Random Search Model",
"_____no_output_____"
]
],
[
[
"best_random = rf_random.best_estimator_\nbest_random.fit(train_features,train_labels);\npred_labels=best_random.predict(test_features)\nrandom_accuracy=metrics.accuracy_score(test_labels, pred_labels)\nprint(\"Best random model Accuracy:\",metrics.accuracy_score(test_labels, pred_labels))\nprint('Improvement of {:0.2f}%.'.format( 100 * (random_accuracy - base_accuracy) / base_accuracy))",
"_____no_output_____"
]
],
[
[
"## Grid Search \n\nWe can now perform grid search building on the result from the random search. \nWe will test a range of hyperparameters around the best values returned by random search. ",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\n# Create the parameter grid based on the results of random search \nparam_grid = {\n 'bootstrap': [True],\n 'max_depth': [5, 10, 50, 110],\n 'min_samples_leaf': [1, 3, 5],\n 'min_samples_split': [2, 8, 12],\n 'n_estimators': [100, 300, 1000, 1500],\n 'max_features' : ['auto', 'sqrt'],\n 'oob_score' : [ True],\n 'warm_start' : [False, True]\n}\n\n# Create a base model\nrf = RandomForestClassifier(random_state = 42)\n\n# Instantiate the grid search model\ngrid_search = GridSearchCV(estimator = rf, param_grid = param_grid, \n cv = 5, n_jobs = -1, verbose = 2, return_train_score=True)\n\n# Fit the grid search to the data\ngid_search.fit(train_features, train_labels);\ngrid_search.best_params_\n\n",
"_____no_output_____"
]
],
[
[
"### Test RF classifier with the best parameters",
"_____no_output_____"
]
],
[
[
"rf_param = RandomForestClassifier(bootstrap= True, max_depth=50, max_features='auto', min_samples_leaf=1, min_samples_split=2, n_estimators = 1000,oob_score= True,\n random_state = 42)\n\nrf_param.fit(train_features, train_labels);\n\npred_labels_best=rf.param.predict(test_features)\nbest_accuracy=metrics.accuracy_score(test_labels, pred_labels_best)\ngrid_accuracy=metrics.accuracy_score(test_labels, pred_labels)\nprint(\"Best Grid model Accuracy:\",metrics.accuracy_score(test_labels, pred_labels))",
"_____no_output_____"
]
],
[
[
"#### Evaluate the Best Model from Grid Search",
"_____no_output_____"
]
],
[
[
"pred_labels_best=rf_param.predict(test_features)\nbest_accuracy=metrics.accuracy_score(test_labels, pred_labels_best)\nprint(\"Base model Accuracy:\",metrics.accuracy_score(test_labels, pred_labels_best))\nprint(rf_param.oob_score_)\nd = grid_search.best_estimator_\ngrid_accuracy =metrics.accuracy_score(test_labels, pred_labels)\nprint(\"Best Grid model Accuracy:\",metrics.accuracy_score(test_labels, pred_labels))\nprint(best_grid.oob_score_)\nbest_grid = grid_search.best_estimator_\ngrid_accuracy =metrics.accuracy_score(test_labels, pred_labels)\nprint(\"Best Grid model Accuracy:\",metrics.accuracy_score(test_labels, pred_labels))\nprint(best_grid.oob_score_)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbad70122b098db6d59ad0633a65535bfd97413f
| 3,620 |
ipynb
|
Jupyter Notebook
|
Operators in Python/Variables & Input.ipynb
|
damanpreetkour/Data-Analysis-Course-ETG
|
7af89d4b1d07bc27ab26f666e3e016165ca13a70
|
[
"Apache-2.0"
] | 305 |
2021-08-23T14:11:49.000Z
|
2022-03-24T17:47:32.000Z
|
Operators in Python/Variables & Input.ipynb
|
damanpreetkour/Data-Analysis-Course-ETG
|
7af89d4b1d07bc27ab26f666e3e016165ca13a70
|
[
"Apache-2.0"
] | 1 |
2021-09-04T14:28:51.000Z
|
2021-09-04T14:28:51.000Z
|
Operators in Python/Variables & Input.ipynb
|
damanpreetkour/Data-Analysis-Course-ETG
|
7af89d4b1d07bc27ab26f666e3e016165ca13a70
|
[
"Apache-2.0"
] | 462 |
2021-08-23T14:15:46.000Z
|
2022-03-25T06:54:21.000Z
| 15.670996 | 56 | 0.446961 |
[
[
[
"# Variables with Python",
"_____no_output_____"
]
],
[
[
"first_name = 'Ashish'\nlast_name = 'Jangra'\n\nage = 22",
"_____no_output_____"
],
[
"first_name",
"_____no_output_____"
],
[
"type(last_name)",
"_____no_output_____"
],
[
"type(age)",
"_____no_output_____"
],
[
"first_name",
"_____no_output_____"
],
[
"first_name = 'Rahul'",
"_____no_output_____"
],
[
"last_name = 'Singh'",
"_____no_output_____"
],
[
"last_name",
"_____no_output_____"
]
],
[
[
"# Input",
"_____no_output_____"
]
],
[
[
"first_name = raw_input(\"Enter the first name: \")",
"Enter the first name: Ashish\n"
],
[
"type(int(first_name))",
"_____no_output_____"
],
[
"print first_name",
"Ashish\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbad7c5870036ef70304e974fbdac38215216a65
| 142,973 |
ipynb
|
Jupyter Notebook
|
demo_of_random_forest.ipynb
|
yshimizu12/EnsembleDecisionTree
|
85d1841ccc383804b97b4b0a75d8bb37767ebffb
|
[
"MIT"
] | null | null | null |
demo_of_random_forest.ipynb
|
yshimizu12/EnsembleDecisionTree
|
85d1841ccc383804b97b4b0a75d8bb37767ebffb
|
[
"MIT"
] | null | null | null |
demo_of_random_forest.ipynb
|
yshimizu12/EnsembleDecisionTree
|
85d1841ccc383804b97b4b0a75d8bb37767ebffb
|
[
"MIT"
] | null | null | null | 266.243948 | 63,824 | 0.919649 |
[
[
[
"# ランダムフォレストのデモプログラム",
"_____no_output_____"
],
[
"ランダムフォレストのデモプログラムです。 \nランダムフォレストの中身に関してはこちら↓で解説しています。 \nhttps://yuyumoyuyu.com/2021/02/21/ensembledecisiontree/",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom matplotlib.colors import ListedColormap\nimport matplotlib.figure as figure\nimport numpy as np\n\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn.model_selection import train_test_split\n\nfrom sklearn.metrics import confusion_matrix\n\nimport time\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# クラス分類",
"_____no_output_____"
],
[
"### 決定木との比較",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier\nfrom sklearn.ensemble import RandomForestClassifier as RFC\n\nfrom sklearn.datasets import make_moons",
"_____no_output_____"
],
[
"# sklearnのデータセットを用いる\n\nX, y = make_moons(n_samples=500, noise=0.3, random_state=6)\n\nplt.figure()\n\ncmap = ListedColormap(('red', 'blue'))\nplt.scatter(X[:, 0], X[:, 1], marker='o', c=y, cmap=cmap, s=8)\n\nplt.xlabel(\"x1\")\nplt.ylabel(\"x2\")\n\nplt.show()",
"_____no_output_____"
],
[
"# 入力データは2次元の座標データ\nprint(\"X =\\n\", X[:10])",
"X =\n [[ 1.05769609e-01 1.42222019e-02]\n [-6.69715288e-01 6.91297509e-01]\n [-1.20374501e+00 3.70471637e-01]\n [ 2.40648591e+00 5.59849147e-01]\n [ 1.07922887e+00 3.46245244e-01]\n [ 1.95305802e+00 2.78931158e-01]\n [ 1.15826758e+00 -4.38060183e-01]\n [ 6.66709932e-01 -2.39615329e-03]\n [-6.55169079e-01 2.55762239e-01]\n [ 1.42811074e+00 -3.32526975e-01]]\n"
],
[
"# 学習データをtrain/test分割\nX_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y,\n random_state=42)",
"_____no_output_____"
],
[
"# 分類木の学習\ntree = DecisionTreeClassifier(max_depth=5, random_state=0)\ntree.fit(X_train, y_train)\n\nprint(\"Accuracy on training set: {:.3f}\".format(tree.score(X_train, y_train)))\nprint(\"Accuracy on test set: {:.3f}\".format(tree.score(X_test, y_test)))",
"Accuracy on training set: 0.933\nAccuracy on test set: 0.904\n"
],
[
"# ランダムフォレスト分類器の学習\n\nrfc = RFC(max_depth=5, n_estimators=10, random_state=0)\nrfc.fit(X_train, y_train)\n\nprint(\"Accuracy on training set: {:.3f}\".format(rfc.score(X_train, y_train)))\nprint(\"Accuracy on test set: {:.3f}\".format(rfc.score(X_test, y_test)))",
"Accuracy on training set: 0.933\nAccuracy on test set: 0.928\n"
],
[
"# 学習結果を可視化\nx1 = np.linspace(-2.0, 3.0, 100)\nx2 = np.linspace(-1.5, 2.0, 100)\nx1_mesh, x2_mesh = np.meshgrid(x1, x2) \n\nz1 = tree.predict(np.array([x1_mesh.ravel(), x2_mesh.ravel()]).T)\nz1 = z1.reshape(x1_mesh.shape)\nz2 = rfc.predict(np.array([x1_mesh.ravel(), x2_mesh.ravel()]).T)\nz2 = z2.reshape(x1_mesh.shape)\n\nz_list = [z1, z2]\ntitles = ['Decision Tree', 'Random Forest']\n\nfig, axes = plt.subplots(1, 2, figsize=(8,4))\n\nfor ax, z, title in zip(axes, z_list, titles):\n ax.contourf(x1_mesh, x2_mesh, z, cmap=cmap, alpha=0.1, linestyles=None)\n ax.scatter(X[:, 0], X[:, 1], marker='o', c=y, cmap=cmap, s=5)\n\n ax.set_xlabel(\"x1\")\n ax.set_ylabel(\"x2\")\n\n ax.set_title(title)\n\nfig.tight_layout()",
"_____no_output_____"
]
],
[
[
"アンサンブル学習により,高精度な分類が可能に",
"_____no_output_____"
],
[
"### ハイパーパラメータ最適化",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import load_iris",
"_____no_output_____"
],
[
"# Irisデータセットを用いる\niris = load_iris()\n\n# 学習データをtrain/test分割\nX_train_i, X_test_i, y_train_i, y_test_i = train_test_split(iris.data,\n iris.target,\n stratify=iris.target,\n random_state=0)",
"_____no_output_____"
],
[
"# グリッドサーチによるハイパーパラメータ最適化\nt1 = time.time()\n\nprint(\"RFC\\n\")\nparams = {\n 'max_depth': [2,3,5,10], # treeの深さの最大値\n 'max_features': [1,3,'auto'], # treeの構築に使用する特徴量の数\n 'min_samples_split': [2,3,5], # ノード分割に必要な最小サンプルサイズ\n 'min_samples_leaf': [1,3,5], # 葉を構成するのに必要な最低サンプル数\n 'n_estimators': [10,30,50,100,300] # treeの数\n }\n\nprint(\"parameters: \\n{}\\n\".format(params))\n\ngrid_search = GridSearchCV(RFC(), params, cv=5, return_train_score=True)\ngrid_search.fit(X_train_i, y_train_i)\n\nprint(\"best parameters: {}\".format(grid_search.best_params_))\nprint(\"best cross-validation score: {:.3f}\".format(grid_search.best_score_))\n\nprint(\"\\nelapsed time: {:.3f} sec\".format(time.time()-t1))",
"RFC\n\nparameters: \n{'max_depth': [2, 3, 5, 10], 'max_features': [1, 3, 'auto'], 'min_samples_split': [2, 3, 5], 'min_samples_leaf': [1, 3, 5], 'n_estimators': [10, 30, 50, 100, 300]}\n\nbest parameters: {'max_depth': 3, 'max_features': 1, 'min_samples_leaf': 3, 'min_samples_split': 5, 'n_estimators': 30}\nbest cross-validation score: 0.964\n\nelapsed time: 287.895 sec\n"
],
[
"rfc = RFC(**grid_search.best_params_).fit(X_train_i, y_train_i)\n\nprint(\"==Training set==\")\nprint(\"Score: {:.3f}\".format(rfc.score(X_train_i, y_train_i)))\nprint(\"Confusion matrix:\\n\", confusion_matrix(y_train_i,rfc.predict(X_train_i),labels=sorted(set(y_train_i))))\n\nprint(\"\\n==Test set==\")\nprint(\"Score: {:.3f}\".format(rfc.score(X_test_i, y_test_i)))\nprint(\"Confusion matrix:\\n\", confusion_matrix(y_test_i,rfc.predict(X_test_i),labels=sorted(set(y_test_i))))",
"==Training set==\nScore: 0.955\nConfusion matrix:\n [[37 0 0]\n [ 0 35 2]\n [ 0 3 35]]\n\n==Test set==\nScore: 0.974\nConfusion matrix:\n [[13 0 0]\n [ 0 13 0]\n [ 0 1 11]]\n"
]
],
[
[
"# 回帰木",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestRegressor as RFR\n\nfrom sklearn.datasets import load_boston",
"_____no_output_____"
],
[
"# Bostonデータセットを用いる\nboston = load_boston()\n\n# 学習データをtrain/test分割\nX_train_b, X_test_b, y_train_b, y_test_b = train_test_split(boston.data,\n boston.target,\n random_state=0)",
"_____no_output_____"
],
[
"# グリッドサーチによるハイパーパラメータ最適化\nt1 = time.time()\n\nprint(\"RFR\\n\")\nparams = {\n 'max_depth': [10,20,30], # treeの深さの最大値\n 'max_features': [3,5,10,'auto'], # treeの構築に使用する特徴量の数\n 'min_samples_split': [2,3,5], # ノード分割に必要な最小サンプルサイズ\n 'min_samples_leaf': [1,3,5], # 葉を構成するのに必要な最低サンプル数\n 'n_estimators': [10,30,50,100] # treeの数\n }\n\nprint(\"parameters: \\n{}\\n\".format(params))\n\ngrid_search = GridSearchCV(RFR(), params, cv=5, return_train_score=True)\ngrid_search.fit(X_train_b, y_train_b)\n\nprint(\"best parameters: {}\".format(grid_search.best_params_))\nprint(\"best cross-validation score: {:.3f}\".format(grid_search.best_score_))\n\nprint(\"\\nelapsed time: {:.3f} sec\".format(time.time()-t1))",
"RFR\n\nparameters: \n{'max_depth': [10, 20, 30], 'max_features': [3, 5, 10, 'auto'], 'min_samples_split': [2, 3, 5], 'min_samples_leaf': [1, 3, 5], 'n_estimators': [10, 30, 50, 100]}\n\nbest parameters: {'max_depth': 20, 'max_features': 5, 'min_samples_leaf': 1, 'min_samples_split': 2, 'n_estimators': 50}\nbest cross-validation score: 0.880\n\nelapsed time: 145.429 sec\n"
],
[
"# 可視化\nrfr = RFR(**grid_search.best_params_).fit(X_train_b, y_train_b)\n\n# trainデータ\ny_train_est = rfr.predict(X_train_b)\n\nplt.figure(figsize=figure.figaspect(1))\nplt.scatter(y_train_b, y_train_est)\ny_max = max( y_train_b.max(), y_train_est.max() )\ny_min = min( y_train_b.min(), y_train_est.min() )\nplt.plot([y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min)],\n [y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min)], 'k-')\nplt.ylim(y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min))\nplt.xlim(y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min))\nplt.xlabel('Actual y')\nplt.ylabel('Estimated y')\nplt.show()\nprint(\" Training set score: {:.3f}\".format(rfr.score(X_train_b, y_train_b))) \n\n# testデータ\ny_test_pred = rfr.predict(X_test_b)\n\nplt.figure(figsize=figure.figaspect(1))\nplt.scatter(y_test_b, y_test_pred)\ny_max = max( y_test_b.max(), y_test_pred.max() )\ny_min = min( y_test_b.min(), y_test_pred.min() )\nplt.plot([y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min)],\n [y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min)], 'k-')\nplt.ylim(y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min))\nplt.xlim(y_min - 0.05 * (y_max - y_min), y_max + 0.05 * (y_max - y_min))\nplt.xlabel('Actual y')\nplt.ylabel('Predicted y')\nplt.show()\nprint(\" Test set score: {:.3f}\".format(rfr.score(X_test_b, y_test_b)))\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbad7d4d2c0538f766c8ec436467512989dbc7a5
| 45,088 |
ipynb
|
Jupyter Notebook
|
notebooks/trade_demo/wip/04_08_2021/Setup Italy Node with Data.ipynb
|
Noob-can-Compile/PySyft
|
156cf93489b16dd0205b0058d4d23d56b3a91ab8
|
[
"Apache-2.0"
] | 8,428 |
2017-08-10T09:17:49.000Z
|
2022-03-31T08:20:14.000Z
|
notebooks/trade_demo/wip/04_08_2021/Setup Italy Node with Data.ipynb
|
Noob-can-Compile/PySyft
|
156cf93489b16dd0205b0058d4d23d56b3a91ab8
|
[
"Apache-2.0"
] | 4,779 |
2017-08-09T23:19:00.000Z
|
2022-03-29T11:49:36.000Z
|
notebooks/trade_demo/wip/04_08_2021/Setup Italy Node with Data.ipynb
|
Noob-can-Compile/PySyft
|
156cf93489b16dd0205b0058d4d23d56b3a91ab8
|
[
"Apache-2.0"
] | 2,307 |
2017-08-10T08:52:12.000Z
|
2022-03-30T05:36:07.000Z
| 35.755749 | 1,363 | 0.434084 |
[
[
[
"from hagrid import cli, grammar",
"_____no_output_____"
],
[
"args = [\"on\", \"docker\"]\n# args = []\nverb = cli.get_launch_verb()\nargs = grammar.launch_shorthand_support(args=args)\nargs",
"_____no_output_____"
],
[
"args = [\"domain\",\"on\", \"docker\"]\n# args = []\nverb = cli.get_launch_verb()\nargs = grammar.launch_shorthand_support(args=args)\nargs",
"_____no_output_____"
],
[
"verb = cli.get_launch_verb()\nargs = cli.launch_shorthand_support(args=args, verb=verb)\ngrammar = cli.parse_grammar(args=tuple(args), verb=verb)\nverb.load_grammar(grammar=grammar)\ncmd = cli.create_launch_cmd(verb=verb, kwargs={}, ignore_docker_version_check=True)",
"\n `\n `.+yys/.`\n ``/NMMMNNs`\n `./shNMMMMMMNs`` `..`\n `-smNMMNNMMMMMMN/.``......`\n `.yNMMMMNmmmmNNMMm/.`....`\n `:sdNMMMMMMNNNNddddds-`.`` `--. `\n `.+dNNNNMMMMMMMMMNNNNmddohmh//hddy/.```..`\n `-hNMMMMMMMMMMMMNNdmNNMNNdNNd:sdyoo+/++:..`\n ../mMMMMMMMMMMMMMMNNmmmmNMNmNNmdmd/hNNNd+:`\n `:mMNNMMMMMMMMMMMMNMNNmmmNNNNNdNNd/NMMMMm::\n `:mMNNNMMMMMMMMMMMMMMMNNNNdNMNNmmNd:smMMmh//\n ``/mMMMMMMMMMMMMMMMMMMMMMMNmdmNNMMNNNy/osoo/-`\n `-sNMMMMMMMMMMMMMMMMMMMMMMMMNNmmMMMMNh-....`\n `/dNMMMMMMMMMMMMMMMMMMMMMMMMMMMNNMMMNy.`\n ``.omNNMMMMMMMMMMMMNMMMMMMMNmmmmNNMMMMN+`\n `:hmNNMMMMMMMMMMMNo/ohNNNNho+os+-+hNys/`\n -mNNNNNNMMMMMMMMm+``-yNdd+/mMMMms.-:`\n .+dmNNNNMMMMMMNd:``:dNN+y`oMMMMMm-.`\n `+dmmmNNNmmmmy+. `-+m/s/+MMMMm/--\n `+mmmhNy/-...``` ``-.-sosyys+/-`\n ``.smmmsoo`` .oh+-:/:.\n `.:odmdh/```` `.+d+``````\n ```/sydNdhy+.` ``-sds.\n `:hdmhs::-```` `oNs.`\n```.sdmh/`` `-ym+`\n ``ssy+` `-yms.`\n `` `:hNy-``\n ` `-yMN/```\n `-yNhy-\n `/yNd/`\n `:dNMs``\n `.+mNy/.`\n `.+hNMMs``\n `:dMMMMh.`\nLaunching a domain PyGrid node on port 8081!\n\n - TYPE: domain\n - NAME: on_docker\n - TAG: 93435e816d0c4c12222f840e10f8cdd0\n - PORT: 8081\n - DOCKER: n/a\n - TAIL: True\n\n\n"
],
[
"cmd",
"_____no_output_____"
],
[
"import syft as sy\nimport pandas as pd",
"_____no_output_____"
],
[
"obj = sy.deserialize(sy.serialize({\"asdf\":3,\"fdsa\":4}))",
"_____no_output_____"
],
[
"obj",
"_____no_output_____"
],
[
"node = sy.login(email=\"[email protected]\", password=\"changethis\", port=8081)",
"Connecting to http://localhost:8081..."
],
[
"istat_data = pd.read_csv('../datasets/it - feb 2021.csv')[0:50000]",
"_____no_output_____"
],
[
"istat_data = node.load_dataset(assets={\"dec2020\":istat_data}, \n name=\"iStat Trade Data - first 50K rows\", \n description=\"\"\"A collection of reports from Italy's statistics \n bureau about how much it thinks it imports and exports from other countries.\"\"\")",
"_____no_output_____"
],
[
"sy.serialize(['a','b'])",
"_____no_output_____"
],
[
"node.datasets.all()",
"_____no_output_____"
],
[
"ca.users.create(name=\"Andrew Trask\",\n email=\"[email protected]\",\n password=\"abc123\")\n",
"Ignoring: user with email:[email protected] already exists\n"
],
[
"ca.users",
"_____no_output_____"
],
[
"ca.store",
"_____no_output_____"
],
[
"ca.requests.pandas",
"_____no_output_____"
],
[
"ca.requests[0].accept()",
"_____no_output_____"
],
[
"ca.users",
"_____no_output_____"
],
[
"d = ca.datasets[-1]",
"_____no_output_____"
],
[
"d.sample",
"_____no_output_____"
],
[
"ca.datasets[0]\n",
"_____no_output_____"
],
[
"# un = sy.login(email=\"[email protected]\", password=\"changethis\", port=8082)",
"_____no_output_____"
],
[
"# ca.users.update(user_id=1, name=\"Bob\")\n# un.users.update(user_id=1, name=\"Alice\")",
"_____no_output_____"
],
[
"un.users.all(pandas=True)",
"_____no_output_____"
],
[
"# us = sy.login(email=\"[email protected]\", password=\"changethis\", port=8082)\n# it = sy.login(email=\"[email protected]\", password=\"changethis\", port=8083)\n# ne = sy.login(email=\"[email protected]\", password=\"changethis\", port=8084)\n\n",
"_____no_output_____"
],
[
"y = p + p",
"_____no_output_____"
],
[
"y.get()",
"[2021-06-30T16:06:51.320382+0000][CRITICAL][logger]][207] UnknownPrivateException has been triggered.\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbad7fad5309586e14f547e6783508d0c9e208fe
| 7,160 |
ipynb
|
Jupyter Notebook
|
human2anime.ipynb
|
AliennCheng/stylegan2
|
796449b30bfb01b68fe8c173075860cef3795355
|
[
"BSD-Source-Code"
] | null | null | null |
human2anime.ipynb
|
AliennCheng/stylegan2
|
796449b30bfb01b68fe8c173075860cef3795355
|
[
"BSD-Source-Code"
] | null | null | null |
human2anime.ipynb
|
AliennCheng/stylegan2
|
796449b30bfb01b68fe8c173075860cef3795355
|
[
"BSD-Source-Code"
] | null | null | null | 32.995392 | 246 | 0.634497 |
[
[
[
"# Face Transformation from Human to Anime\nThis notebook demonstrates how to transform a human face to an anime face using StyleGAN2.\n\nContact: [email protected]",
"_____no_output_____"
],
[
"## Data Preparation\nFirst of all, we need a dataset to train the model.\nI downloaded the [danbooru2019-portrait](https://www.gwern.net/Crops#danbooru2019-portraits) dataset and manually pick 1000 images from it.\n\nThen we run the dataset tool provided by StyleGAN2 to create TensorFlow records.\n\n#### Note\n1. As [Doron Adler's work](https://twitter.com/Buntworthy/status/1297976798236598274) shows, 317 images should be enough.\n2. Beware of mode issues. The model never learn something not (or seldomly) appearing in the dataset. Due to the extreme gender ratio of the anime portraits, the model would eventually learn how to generate anime faces excluding male faces.",
"_____no_output_____"
]
],
[
[
"!python dataset_tool.py create_from_images datasets/custom_512 datasets_img/",
"_____no_output_____"
]
],
[
[
"## Train a StyleGAN2 network to generate anime portrait\nWe need a network to generate anime portrait.\n\nI suggest transfer learning from a well-trained network so that we can not only save the time but also somehow keep the latent space of the source network.\n\nHere I fine-tuned the model [stylegan2-ffhq-512-config-f](https://twitter.com/AydaoAI/status/1269689136019116032?s=20) with the dataset above.",
"_____no_output_____"
]
],
[
[
"!python run_training.py --result-dir=D://Anime/results/ --data-dir=datasets/ --dataset=custom_512 --config=config-f --total-kimg=200",
"_____no_output_____"
]
],
[
[
"## Align a human face image\nAn aligned face image is more understandable for model. Furthermore, we need the image cropped in a suitable manner.",
"_____no_output_____"
]
],
[
[
"!python align_images.py images/raw images/aligned",
"_____no_output_____"
]
],
[
[
"So far we have the essential materials prepared, a human face image, a ffhq model and a anime face model, both of which share the latent space (or similar at least).\n\nThen what we need to do next is to tranform the human face to an anime face.\n\nHere we have some choices to get our works done:\n1. Extract the latent code of the given human face image, then simply insert it to the anime model.\n2. Blend the human model and the anime model to get a mixture of them. That would be closer to the original image but the models might conflict and generate a hybrid.\n3. With both models generating paired images, learn a pix2pix model.",
"_____no_output_____"
],
[
"## Extract the latent code of the face image\nWe need to attain the latent code corresponding to the given human image so that we can make use of it, and [rolux have done this](https://github.com/rolux/stylegan2encoder)!\n\n> Note that we should replace *training/dataset.py* by *training/dataset-toonify.py*, and replace *dataset_tool.py* by *dataset_tool-toonify.py* here. The **-s2.py* files are backups.",
"_____no_output_____"
]
],
[
[
"!python project_images.py --num-steps 500 images/aligned images/generated --network-pkl=pretrained_models/stylegan2-ffhq-512-config-f.pkl",
"_____no_output_____"
]
],
[
[
"## Generate Anime face with the latent code\nWe get the latent code of the given image now.\n\nThe simplest way to generate the anime face image is by inserting the code directly into the anime model.",
"_____no_output_____"
],
[
"> In this way, we assume the latent space between the human face model and the anime face model was the same.\n>\n> However, in fact, we just fine-tuned it so we could not guarantee such assumption would hold.\n>\n> So the output image would be a little different from the original image.",
"_____no_output_____"
]
],
[
[
"!python generate_from_latent.py",
"_____no_output_____"
]
],
[
[
"## Blend the models\nAs [Justin Pinkney's work](https://colab.research.google.com/drive/1tputbmA9EaXs9HL9iO21g7xN7jz_Xrko?usp=sharing) shows, StyleGAN2 models can be blended easily.\n\nWe can get a blended model to generate an image between a human face and an anime face.\n\n> Similarly we can transform a human face to a blended face.\n>\n> If you want to do so, you need to revise the file *generate_from_latent.py*.\n>\n> Replace *pretrained_models/ffhq-to-anime-512-config-f.pkl* by *pretrained_models/blended.pkl*.",
"_____no_output_____"
]
],
[
[
"!python blend_models.py --low_res_pkl=pretrained_models/stylegan2-ffhq-512-config-f.pkl --high_res_pkl=pretrained_models/ffhq-to-anime-512-config-f.pkl --resolution=32 --output_pkl=pretrained_models/blended.pkl",
"_____no_output_____"
]
],
[
[
"## Train a pix2pix model\nWIP",
"_____no_output_____"
],
[
"## Reference\n[Analyzing and Improving the Image Quality of StyleGAN](https://arxiv.org/abs/1912.04958)\n\n[Toonify yourself by Justin Pinkney](https://www.justinpinkney.com/toonify-yourself/)\n\n[stylegan2encoder by rolux](https://github.com/rolux/stylegan2encoder)\n\n[Making Anime Faces With StyleGAN](https://www.gwern.net/Faces)\n\n[malnyun_faces by bryandlee](https://github.com/bryandlee/malnyun_faces)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbad96c26e29e8908c97357c2fc65ed5c2d3d2c6
| 1,983 |
ipynb
|
Jupyter Notebook
|
Untitled18.ipynb
|
colinpeterjaison/Python
|
81fcf5015785099b47d0a128a6ee289a5403f6cf
|
[
"MIT"
] | null | null | null |
Untitled18.ipynb
|
colinpeterjaison/Python
|
81fcf5015785099b47d0a128a6ee289a5403f6cf
|
[
"MIT"
] | null | null | null |
Untitled18.ipynb
|
colinpeterjaison/Python
|
81fcf5015785099b47d0a128a6ee289a5403f6cf
|
[
"MIT"
] | null | null | null | 20.234694 | 98 | 0.496722 |
[
[
[
"import numpy as np",
"_____no_output_____"
],
[
"x=np.array([1,2,3,4,5,6,7,8,9,10,11,12,13])\ny=np.array([1859.4,2110.2,2018,1966,2166,2118,2054.3,2273,2501.95,2536.25,2433,2365,2451.2])",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"y",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
cbad9be38f21b3935c7e6795b62876467c071fbe
| 1,959 |
ipynb
|
Jupyter Notebook
|
Untitled.ipynb
|
muhammadrazaali-RAZA/Evaluation-of-Subjective-Answers-Using-Semantic-Similarity
|
7d2d055cfa237c3618587dc374cf9ed4c43fa3a4
|
[
"MIT"
] | null | null | null |
Untitled.ipynb
|
muhammadrazaali-RAZA/Evaluation-of-Subjective-Answers-Using-Semantic-Similarity
|
7d2d055cfa237c3618587dc374cf9ed4c43fa3a4
|
[
"MIT"
] | null | null | null |
Untitled.ipynb
|
muhammadrazaali-RAZA/Evaluation-of-Subjective-Answers-Using-Semantic-Similarity
|
7d2d055cfa237c3618587dc374cf9ed4c43fa3a4
|
[
"MIT"
] | null | null | null | 22.77907 | 75 | 0.481368 |
[
[
[
"import tkinter\nfrom tkinter import *\n\n\nmaster = tkinter.Tk()\nmaster.geometry(\"750x500\")\n\nlistbox = Listbox(master)\nlistbox.place(x=3,y=0)\n\n\nfor item in [\"one\", \"two\", \"three\", \"four\"]:\n listbox.insert(END, item)\n\nenable = {'button 1','button 2', 'button 3'}\n\ndef onselect(evt):\n # Note here that Tkinter passes an event object to onselect()\n w = evt.widget\n \n x=0\n index = int(w.curselection()[0])\n value = w.get(index)\n print ('You selected item %d: \"%s\"' % (index, value))\n for item in enable:\n checkboxes = Checkbutton(master, text=item, variable=item)\n checkboxes.place(x=300,y=0+x)\n x+=50\n\nlistbox.bind('<<ListboxSelect>>', onselect)\n\nprint(enable)\n\nmainloop()",
"{'button 1', 'button 2', 'button 3'}\nYou selected item 2: \"three\"\nYou selected item 1: \"two\"\nYou selected item 0: \"one\"\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbad9cd65a07f6fbae9ccbd34774ebdc17304e14
| 145,367 |
ipynb
|
Jupyter Notebook
|
DEMO_Counter_speech.ipynb
|
binny-mathew/Countering_Hate_Speech_ICWSM2019
|
1a598369b3e9f4a000c2d3b04a51cb4965a2bf38
|
[
"MIT"
] | 16 |
2019-04-09T13:20:46.000Z
|
2020-12-15T03:03:31.000Z
|
DEMO_Counter_speech.ipynb
|
binny-mathew/Countering_Hate_Speech
|
1a598369b3e9f4a000c2d3b04a51cb4965a2bf38
|
[
"MIT"
] | 4 |
2019-12-16T21:43:17.000Z
|
2020-11-13T18:10:28.000Z
|
DEMO_Counter_speech.ipynb
|
binny-mathew/Countering_Hate_Speech
|
1a598369b3e9f4a000c2d3b04a51cb4965a2bf38
|
[
"MIT"
] | 3 |
2021-08-08T08:12:02.000Z
|
2022-03-23T04:00:32.000Z
| 126.07719 | 27,480 | 0.817207 |
[
[
[
"# Introduction \n\n\n\n\nThis notebook provides a demo to use the methods used in the paper with new data. If new to collaboratory ,please check the following [link](https://medium.com/lean-in-women-in-tech-india/google-colab-the-beginners-guide-5ad3b417dfa) to know how to run the code.",
"_____no_output_____"
],
[
"### Import the required libraries:",
"_____no_output_____"
]
],
[
[
"#import \n\nfrom gensim.test.utils import datapath, get_tmpfile\nfrom gensim.models import KeyedVectors\nfrom gensim.scripts.glove2word2vec import glove2word2vec\nimport os\nimport joblib\nimport json\nimport pandas as pd\nimport numpy as np\n\n###ipywigets\nfrom __future__ import print_function\nfrom ipywidgets import interact, interactive, fixed, interact_manual\nimport ipywidgets as widgets\n\n\nfrom sklearn import *\nfrom sklearn.model_selection import *\nfrom sklearn.metrics import *\n\nimport nltk\nnltk.download('stopwords')\n ",
"[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Unzipping corpora/stopwords.zip.\n"
],
[
"#copy the git clone address here \n \n!git clone https://github.com/binny-mathew/Countering_Hate_Speech.git \n#Best binary classifier was XGBclassifier \n#Best multilabel classifier was XGBclassifier\n\nbest_binary_classifier = joblib.load('Countering_Hate_Speech/Best_model/XGB_classifier_task_1.joblib.pkl')\nbest_multiclass_classifier = joblib.load('Countering_Hate_Speech/Best_model/XGB_classifier_task_3.joblib.pkl')\nbest_black_classifier = joblib.load('Countering_Hate_Speech/Best_model/black_XGB_classifier_task_2.joblib.pkl')\nbest_jew_classifier = joblib.load('Countering_Hate_Speech/Best_model/jew_XGB_classifier_task_2.joblib.pkl')\nbest_lgbt_classifier = joblib.load('Countering_Hate_Speech/Best_model/lgbt_XGB_classifier_task_2.joblib.pkl')\n\n",
"/usr/local/lib/python3.6/dist-packages/sklearn/base.py:253: UserWarning: Trying to unpickle estimator LogisticRegression from version 0.20.2 when using version 0.20.3. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/base.py:253: UserWarning: Trying to unpickle estimator LabelBinarizer from version 0.20.2 when using version 0.20.3. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/base.py:253: UserWarning: Trying to unpickle estimator _ConstantPredictor from version 0.20.2 when using version 0.20.3. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n/usr/local/lib/python3.6/dist-packages/sklearn/base.py:253: UserWarning: Trying to unpickle estimator OneVsRestClassifier from version 0.20.2 when using version 0.20.3. This might lead to breaking code or invalid results. Use at your own risk.\n UserWarning)\n"
]
],
[
[
"###Word Embeddings Loaded Here",
"_____no_output_____"
]
],
[
[
"####downloading the word embeddings\n!wget http://nlp.stanford.edu/data/glove.840B.300d.zip\n!unzip glove.840B.300d.zip\n####extracting the glove model file \n#import zipfile\n#archive = zipfile.ZipFile('glove.840B.300d.zip', 'r')\nGLOVE_MODEL_FILE ='glove.840B.300d.txt'\n\n\nimport numpy as np\n\n## change the embedding dimension according to the model\nEMBEDDING_DIM = 300\n###change the method type\n\n\n### method two\ndef loadGloveModel2(glove_file):\n tmp_file = get_tmpfile(\"test_crawl_200.txt\")\n\n # call glove2word2vec script\n # default way (through CLI): python -m gensim.scripts.glove2word2vec --input <glove_file> --output <w2v_file>\n glove2word2vec(glove_file, tmp_file)\n model=KeyedVectors.load_word2vec_format(tmp_file)\n return model\n\n\n",
"--2019-03-28 11:17:48-- http://nlp.stanford.edu/data/glove.840B.300d.zip\nResolving nlp.stanford.edu (nlp.stanford.edu)... 171.64.67.140\nConnecting to nlp.stanford.edu (nlp.stanford.edu)|171.64.67.140|:80... connected.\nHTTP request sent, awaiting response... 302 Found\nLocation: https://nlp.stanford.edu/data/glove.840B.300d.zip [following]\n--2019-03-28 11:17:48-- https://nlp.stanford.edu/data/glove.840B.300d.zip\nConnecting to nlp.stanford.edu (nlp.stanford.edu)|171.64.67.140|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 2176768927 (2.0G) [application/zip]\nSaving to: ‘glove.840B.300d.zip’\n\nglove.840B.300d.zip 100%[===================>] 2.03G 6.61MB/s in 5m 19s \n\n2019-03-28 11:23:07 (6.52 MB/s) - ‘glove.840B.300d.zip’ saved [2176768927/2176768927]\n\nArchive: glove.840B.300d.zip\n inflating: glove.840B.300d.txt \n"
],
[
"word2vec_model = loadGloveModel2(GLOVE_MODEL_FILE)",
"_____no_output_____"
]
],
[
[
"## Dataset is loaded here",
"_____no_output_____"
]
],
[
[
"#@title Select the type of file used \n\ntype_of_file = 'X.json' #@param ['X.json','X.csv']",
"_____no_output_____"
]
],
[
[
"### File type information\n\nIf the file type is **.json** then each element should contain the following fields:-\n1. Community\n2. CounterSpeech\n3. Category\n4. commentText\n5. id\n\nIf the file type is **.csv** then it must have the following columns:-\n1. Community\n2. CounterSpeech\n3. Category\n4. commentText\n5. id\n\n\nNote:- If you don't have the Category or Community add an dummy element or column ",
"_____no_output_____"
]
],
[
[
"####CHANGE THE PATH OF THE FILE\npath_of_file='Countering_Hate_Speech/Data/Counterspeech_Dataset.json'\n\ndef convert_class_label(input_text):\n if input_text:\n return 'counter'\n else:\n return 'noncounter'\n\n\n\nif(type_of_file=='X.json'):\n with open(path_of_file) as fp:\n train_data = json.load(fp)\n pd_train = pd.DataFrame(columns=['id','class','community','category','text'])\n\n for count, each in enumerate(train_data):\n try:\n pd_train.loc[count] = [each['id'], convert_class_label(each['CounterSpeech']), each['Community'],each['Category'],each['commentText']]\n except:\n pass\n print('Training Data Loading Completed...')\nelif(type_of_file=='X.csv'):\n pd_train=pd.read_csv(path_of_the_file)\n \npd_train.head() ",
"Training Data Loading Completed...\n"
],
[
"#@title How your dataframe should look like after extraction {display-mode: \"form\"}\n\n# This code will be hidden when the notebook is loaded.\n\npath_of_data_file='Countering_Hate_Speech/Data/Counterspeech_Dataset.json'\n\ndef convert_class_label(input_text):\n if input_text:\n return 'counter'\n else:\n return 'noncounter'\nwith open(path_of_data_file) as fp:\n train_data = json.load(fp)\npd_train_sample = pd.DataFrame(columns=['id','class','community','category','text'])\n\nfor count, each in enumerate(train_data):\n try:\n pd_train_sample.loc[count] = [each['id'], convert_class_label(each['CounterSpeech']), each['Community'],each['Category'],each['commentText']]\n except:\n pass\nprint('Training Data Loading Completed...')\npd_train_sample.head()",
"Training Data Loading Completed...\n"
],
[
"pd_train['text'].replace('', np.nan, inplace=True)\npd_train.dropna(subset=['text'], inplace=True)",
"_____no_output_____"
],
[
"import sys\n\n####features module has the necessary function for feature generation \nfrom Countering_Hate_Speech.utils import features\nfrom Countering_Hate_Speech.utils import multi_features\n###tokenize module has the tokenization funciton\nfrom Countering_Hate_Speech.utils.tokenize import *\n###helper prints confusion matrix and stores results\nfrom Countering_Hate_Speech.utils.helper import *\n###common preprocessing imports\nfrom Countering_Hate_Speech.utils.commen_preprocess import *\n",
"....start....cleaning\n"
]
],
[
[
"#### Next few sections cover three different classifiers namely - \n\n* Binary classification \n* Multlabel classification\n* Cross community \n\nYou can run the cells corresponding to the result you want to analyse.\n\n",
"_____no_output_____"
],
[
"### **Binary Classification**",
"_____no_output_____"
]
],
[
[
"X,y= features.combine_tf_rem_google_rem_embed(pd_train,word2vec_model)\n\nlabel_map = {\n 'counter': 0,\n 'noncounter': 1\n }\ntemp=[]\nfor data in y:\n temp.append(label_map[data])\n\ny=np.array(temp)\n\n\ny_pred=best_binary_classifier.predict(X)\nreport = classification_report(y, y_pred)\ncm=confusion_matrix(y, y_pred)\nplt=plot_confusion_matrix(cm,normalize= True,target_names = ['counter','non_counter'],title = \"Confusion Matrix\")\nplt.savefig('Confusion_matrix.png')\ndf_result=pandas_classification_report(y,y_pred)\ndf_result.to_csv('Classification_Report.csv', sep=',')\nprint(\"You can download the files from the file directory now \")",
"INFO:tensorflow:Saver not created because there are no variables in the graph to restore\n"
]
],
[
[
"### **Multilabel Classification**",
"_____no_output_____"
]
],
[
[
"import scipy\n\npd_train_multilabel =pd_train.copy()\npd_train_multilabel =pd_train_multilabel[pd_train_multilabel['category']!='Default']\nlist1=[[],[],[],[],[],[],[],[],[],[]]\nfor ele in pd_train_multilabel['category']:\n temp=[]\n if type(ele) is int:\n ele =str(ele)\n for i in range(0,len(ele),2):\n temp.append(ord(ele[i])-ord('0'))\n #print(temp)\n if(len(temp)==0):\n print(temp)\n for i in range(0,10):\n if i+1 in temp:\n list1[i].append(1)\n else:\n list1[i].append(0)\ny_train=np.array([np.array(xi) for xi in list1])\n\n### final dataframe for the task created \npd_train_multilabel = pd.DataFrame({'text':list(pd_train_multilabel['text']),'cat0':list1[0],'cat1':list1[1],'cat2':list1[2],'cat3':list1[3],'cat4':list1[4],'cat5':list1[5],'cat6':list1[6],'cat7':list1[7],'cat8':list1[8],'cat9':list1[9]})\n### drop the entries having blank entries\npd_train_multilabel['text'].replace('', np.nan, inplace=True)\npd_train_multilabel.dropna(subset=['text'], inplace=True)\n\n\n\n\n\nX,y= multi_features.combine_tf_rem_google_rem_embed(pd_train_multilabel,word2vec_model)\npath='multilabel_res'\nos.makedirs(path, exist_ok=True)\nX = np.array(X)\ny = np.array(y)\ny_pred = best_multiclass_classifier.predict(X)\nif(scipy.sparse.issparse(y_pred)):\n ham,acc,pre,rec,f1=calculate_score(y,y_pred.toarray())\n accuracy_test=accuracy_score(y,y_pred.toarray())\n\nelse:\n ham,acc,pre,rec,f1=calculate_score(y,y_pred)\n accuracy_test=my_accuracy_score(y,y_pred)\n\n \nfor i in range(10):\n df_result=pandas_classification_report(y[:,i],y_pred[:,i])\n df_result.to_csv(path+'/report'+str(i)+'.csv')\n\n\nf = open(path+'/final_report.txt', \"w\")\nf.write(\"best_model\") \nf.write(\"The hard metric score is :- \" + str(accuracy_test))\nf.write(\"The accuracy is :- \" + str(acc))\nf.write(\"The precision is :- \" + str(pre))\nf.write(\"The recall is :- \" + str(rec))\nf.write(\"The f1_score is :- \" + str(f1))\nf.write(\"The hamming loss is :-\" + str(ham))\nf.close()\n \n \n!zip -r mulitlabel_results.zip multilabel_res\n",
"INFO:tensorflow:Saver not created because there are no variables in the graph to restore\n"
]
],
[
[
"### **Cross CommunityClassification**",
"_____no_output_____"
]
],
[
[
"pd_cross=pd_train.copy()\npart_j=pd_cross.loc[pd_train['community']=='jews']\npart_b=pd_cross.loc[pd_train['community']=='black']\npart_l=pd_cross.loc[pd_train['community']=='lgbt']\nX_black,y_black= features.combine_tf_rem_google_rem_embed(part_b,word2vec_model)\nX_jew,y_jew= features.combine_tf_rem_google_rem_embed(part_j,word2vec_model)\nX_lgbt,y_lgbt= features.combine_tf_rem_google_rem_embed(part_l,word2vec_model)\n",
"INFO:tensorflow:Saver not created because there are no variables in the graph to restore\n"
],
[
"label_map = {\n 'counter': 0,\n 'noncounter': 1\n }\ntemp=[]\nfor data in y_black:\n temp.append(label_map[data])\n\ny_black=np.array(temp)\n\n\ny_pred_black=best_black_classifier.predict(X_black)\nreport = classification_report(y_black, y_pred_black)\ncm=confusion_matrix(y_black, y_pred_black)\nplt=plot_confusion_matrix(cm,normalize= True,target_names = ['counter','non_counter'],title = \"Confusion Matrix\")\nplt.savefig('black_Confusion_matrix.png')\ndf_result=pandas_classification_report(y_black,y_pred_black)\ndf_result.to_csv('black_Classification_Report.csv', sep=',')\nprint(\"You can download the files from the file directory now \")",
"You can download the files from the file directory now \n"
],
[
"label_map = {\n 'counter': 0,\n 'noncounter': 1\n }\ntemp=[]\nfor data in y_jew:\n temp.append(label_map[data])\n\ny_jew=np.array(temp)\n\n\ny_pred_jew=best_jew_classifier.predict(X_jew)\nreport = classification_report(y_jew, y_pred_jew)\ncm=confusion_matrix(y_jew, y_pred_jew)\nplt=plot_confusion_matrix(cm,normalize= True,target_names = ['counter','non_counter'],title = \"Confusion Matrix\")\nplt.savefig('jew_Confusion_matrix.png')\ndf_result=pandas_classification_report(y_jew,y_pred_jew)\ndf_result.to_csv('jew_Classification_Report.csv', sep=',')\nprint(\"You can download the files from the file directory now \")",
"_____no_output_____"
],
[
"label_map = {\n 'counter': 0,\n 'noncounter': 1\n }\ntemp=[]\nfor data in y_lgbt:\n temp.append(label_map[data])\n\ny_lgbt=np.array(temp)\n\n\ny_pred_lgbt=best_lgbt_classifier.predict(X_lgbt)\nreport = classification_report(y_lgbt, y_pred_lgbt)\ncm=confusion_matrix(y_lgbt, y_pred_lgbt)\nplt=plot_confusion_matrix(cm,normalize= True,target_names = ['counter','non_counter'],title = \"Confusion Matrix\")\nplt.savefig('lgbt_Confusion_matrix.png')\ndf_result=pandas_classification_report(y_lgbt,y_pred_lgbt)\ndf_result.to_csv('lgbt_Classification_Report.csv', sep=',')\nprint(\"You can download the files from the file directory now \")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbad9f63bb7f00d048b5060cc67c482edbfe5239
| 3,300 |
ipynb
|
Jupyter Notebook
|
examples/example.ipynb
|
arnabuchiha/CarND-Advanced-Lane-Lines
|
6c5a861325e29c8662ff45b48cca7298cb078d3b
|
[
"MIT"
] | 2 |
2019-01-13T18:00:31.000Z
|
2020-09-29T19:30:57.000Z
|
examples/example.ipynb
|
arnabuchiha/CarND-Advanced-Lane-Lines
|
6c5a861325e29c8662ff45b48cca7298cb078d3b
|
[
"MIT"
] | null | null | null |
examples/example.ipynb
|
arnabuchiha/CarND-Advanced-Lane-Lines
|
6c5a861325e29c8662ff45b48cca7298cb078d3b
|
[
"MIT"
] | null | null | null | 29.20354 | 120 | 0.567879 |
[
[
[
"## Advanced Lane Finding Project\n\nThe goals / steps of this project are the following:\n\n* Compute the camera calibration matrix and distortion coefficients given a set of chessboard images.\n* Apply a distortion correction to raw images.\n* Use color transforms, gradients, etc., to create a thresholded binary image.\n* Apply a perspective transform to rectify binary image (\"birds-eye view\").\n* Detect lane pixels and fit to find the lane boundary.\n* Determine the curvature of the lane and vehicle position with respect to center.\n* Warp the detected lane boundaries back onto the original image.\n* Output visual display of the lane boundaries and numerical estimation of lane curvature and vehicle position.\n\n---\n## First, I'll compute the camera calibration using chessboard images",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport cv2\nimport glob\nimport matplotlib.pyplot as plt\n%matplotlib qt\n\n# prepare object points, like (0,0,0), (1,0,0), (2,0,0) ....,(6,5,0)\nobjp = np.zeros((6*9,3), np.float32)\nobjp[:,:2] = np.mgrid[0:9,0:6].T.reshape(-1,2)\n\n# Arrays to store object points and image points from all the images.\nobjpoints = [] # 3d points in real world space\nimgpoints = [] # 2d points in image plane.\n\n# Make a list of calibration images\nimages = glob.glob('../camera_cal/calibration*.jpg')\n\n# Step through the list and search for chessboard corners\nfor fname in images:\n img = cv2.imread(fname)\n gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)\n\n # Find the chessboard corners\n ret, corners = cv2.findChessboardCorners(gray, (9,6),None)\n\n # If found, add object points, image points\n if ret == True:\n objpoints.append(objp)\n imgpoints.append(corners)\n\n # Draw and display the corners\n img = cv2.drawChessboardCorners(img, (9,6), corners, ret)\n cv2.imshow('img',img)\n cv2.waitKey(500)\n\ncv2.destroyAllWindows()",
"_____no_output_____"
]
],
[
[
"## And so on and so forth...",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbadb39e278b9393315ac7c036f4e6b59a7cf382
| 2,115 |
ipynb
|
Jupyter Notebook
|
Euler 231 - The prime factorisation of binomial coefficients.ipynb
|
Radcliffe/project-euler
|
5eb0c56e2bd523f3dc5329adb2fbbaf657e7fa38
|
[
"MIT"
] | 6 |
2016-05-11T18:55:35.000Z
|
2019-12-27T21:38:43.000Z
|
Euler 231 - The prime factorisation of binomial coefficients.ipynb
|
Radcliffe/project-euler
|
5eb0c56e2bd523f3dc5329adb2fbbaf657e7fa38
|
[
"MIT"
] | null | null | null |
Euler 231 - The prime factorisation of binomial coefficients.ipynb
|
Radcliffe/project-euler
|
5eb0c56e2bd523f3dc5329adb2fbbaf657e7fa38
|
[
"MIT"
] | null | null | null | 21.15 | 84 | 0.48747 |
[
[
[
"# Euler Problem 231",
"_____no_output_____"
],
[
"The binomial coefficient 10C3 = 120.\n120 = 23 × 3 × 5 = 2 × 2 × 2 × 3 × 5, and 2 + 2 + 2 + 3 + 5 = 14.\nSo the sum of the terms in the prime factorisation of 10C3 is 14.\n\nFind the sum of the terms in the prime factorisation of 20000000C15000000. ",
"_____no_output_____"
]
],
[
[
"def digitsum(n, p):\n \"\"\"Sum of digits of n in base p\"\"\"\n s = 0\n while n:\n s += n % p\n n //= p\n return s\n\n\ndef binom(n, k, p):\n \"\"\"Exponent of p in the prime factorization of (n choose k).\"\"\"\n return (digitsum(k, p) + digitsum(n - k, p) - digitsum(n, p)) // (p - 1)\n",
"_____no_output_____"
],
[
"from primesieve import primes\n\ndef sum_of_prime_factors_binom(n, k):\n return sum(p * binom(n, k, p) for p in primes(n))\n",
"_____no_output_____"
],
[
"print(sum_of_prime_factors_binom(20000000, 15000000))",
"7526965179680\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbadc82bf452dcd1138b361383ba6100d25744b3
| 235,189 |
ipynb
|
Jupyter Notebook
|
knn.ipynb
|
acauligi/ee364b-project
|
66547286c3f40f5fe8dc48e1dc5d99308420deac
|
[
"MIT"
] | null | null | null |
knn.ipynb
|
acauligi/ee364b-project
|
66547286c3f40f5fe8dc48e1dc5d99308420deac
|
[
"MIT"
] | 5 |
2021-06-08T21:45:26.000Z
|
2022-03-12T00:34:24.000Z
|
knn.ipynb
|
acauligi/ee364b-project
|
66547286c3f40f5fe8dc48e1dc5d99308420deac
|
[
"MIT"
] | null | null | null | 42.94905 | 2,072 | 0.613753 |
[
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"import numpy as np\nimport cvxpy as cp\nfrom scipy import stats\n\nimport torch\nfrom torch import nn\nimport torch.nn.functional as F\nfrom torch.autograd import Variable\nfrom torch.utils.data import DataLoader\nimport torch.optim as optim\nimport os\nimport random\n\nimport sys\nsys.path.insert(0, './mlopt-micp')\nsys.path.insert(0, './mlopt-micp/cartpole')\n\nimport optimizer\nfrom problem import Cartpole\nfrom src.ae import Encoder, get_cartpole_encoder",
"_____no_output_____"
],
[
"def euclidean_dist(x,y):\n # x: NxD\n # y: MxD\n n = x.size(0)\n m = y.size(0)\n d = x.size(1)\n assert d == y.size(1)\n \n x = x.unsqueeze(1).expand(n, m, d)\n y = y.unsqueeze(0).expand(n, m, d)\n return torch.pow(x-y, 2).sum(2)",
"_____no_output_____"
],
[
"pp = Cartpole()",
"_____no_output_____"
],
[
"print('Total number of classes: {}'.format(pp.n_strategies))\nprint('Length of feature vector: {}'.format(pp.n_features))",
"Total number of classes: 581\nLength of feature vector: 13\n"
],
[
"dim_in, dim_z = pp.n_features, 4#pp.n_strategies\n\nenc = get_cartpole_encoder(dim_in, dim_z).cuda()\nenc(torch.from_numpy(pp.features[:2]).float().cuda())\n\n# training parameters\nTRAINING_ITERATIONS = int(5000)\nBATCH_SIZE = int(10)\nCHECKPOINT_AFTER = int(1250)\nSAVEPOINT_AFTER = int(2500)\n\nrand_idx = list(np.arange(0, pp.n_strategies-1))\n\nindices = [rand_idx[ii * BATCH_SIZE:(ii + 1) * BATCH_SIZE] for ii in range((len(rand_idx) + BATCH_SIZE - 1) // BATCH_SIZE)]\nrandom.shuffle(indices)\n\nenc_dict = {}\nstr_dict = {}\nfor ii in range(len(pp.features)):\n str_idx = int(pp.labels[ii,0])\n str_dict[ii] = str_idx\n if str_idx in enc_dict.keys():\n enc_dict[str_idx] += [ii]\n else:\n enc_dict[str_idx] = [ii]\n \nfeats = torch.from_numpy(pp.features).float().cuda()\n\npp.training_batch_percentage = 1.\npp.construct_strategies()\nstrat_lookup = {}\nfor k, v in pp.strategy_dict.items():\n strat_lookup[v[0]] = v[1:]\npp.training_batch_percentage = 0.9\npp.n_evals = 5\npp.training_batch_percentage=0.9",
"_____no_output_____"
],
[
"#nearest neighbors\npp.training_batch_percentage=0.9\ntrain_set_length = int(pp.training_batch_percentage*pp.n_probs)\nY = feats[:train_set_length,:]\n\n#classifier\ndef nn_classifier(x,Y,k=1):\n dist_inds = torch.argsort(torch.cdist(Y,x[None,:]),dim=0).cpu().numpy()\n strats_sorted = pp.labels[dist_inds,0].astype(int)\n return int(stats.mode(strats_sorted[:k])[0])\n #_, unique_inds = np.unique(strats_sorted,return_index=True)\n #return np.concatenate([strats_sorted[index] for index in sorted(unique_inds)])\n\nnn_classifier(feats[train_set_length+8,:],Y,k=5)",
"_____no_output_____"
],
[
"#test script\nn_train_strategies = pp.n_strategies #store how many strats in train set\nc_k = torch.zeros((n_train_strategies,4)) \nembeddings = enc(feats) #embed training points\nfor ii in range(n_train_strategies): #compute train centroids\n inds = enc_dict[ii]\n c_k[ii,:] = torch.mean(embeddings[inds,:],axis=0).cuda()\n\n#compute strategy dictionary for all problems\npp.training_batch_percentage = 1.\npp.construct_strategies()\nstrat_lookup = {}\nfor k, v in pp.strategy_dict.items():\n strat_lookup[v[0]] = v[1:]\n\n#setup for test\ntest_feats = torch.from_numpy(pp.features[int(0.9*pp.n_probs):,:]).float().cuda()\ntest_enc = enc(test_feats).cuda()\ntest_dists = torch.cdist(test_enc,c_k.cuda()).detach().cpu().numpy()\ntest_start = int(0.9*pp.n_probs)\nn_test = int(0.1*pp.n_probs)\nind_max = np.argsort(test_dists)[:,:pp.n_evals]\nfeasible = np.zeros(n_test)\ncosts = np.zeros(n_test)",
"_____no_output_____"
],
[
"prob_success = False\npp.n_evals = 1\nk=5\n\nfor ii in range(n_test):\n #strats_sorted = nn_classifier(feats[test_start+ii,:],Y);\n #for jj in range(pp.n_evals):\n y_guess = strat_lookup[nn_classifier(feats[test_start+ii,:],Y,k=k)]\n #y_guess = strat_lookup[int(pp.labels[ii,0])]\n try:\n prob_success, cost, solve_time = pp.solve_mlopt_prob_with_idx(ii+test_start, y_guess)\n if prob_success:\n feasible[ii] = 1.\n costs[ii] = cost\n print('Succeded at {} with {} tries'.format(ii,1))\n except (KeyboardInterrupt, SystemExit):\n raise\n except:\n print('mosek failed at '.format(ii))",
"Succeded at 1 with 1 tries\nSucceded at 2 with 1 tries\nSucceded at 3 with 1 tries\nSucceded at 6 with 1 tries\nSucceded at 7 with 1 tries\nSucceded at 8 with 1 tries\nSucceded at 11 with 1 tries\nSucceded at 13 with 1 tries\nSucceded at 16 with 1 tries\nSucceded at 17 with 1 tries\nSucceded at 18 with 1 tries\nSucceded at 19 with 1 tries\nSucceded at 20 with 1 tries\nSucceded at 23 with 1 tries\nSucceded at 25 with 1 tries\nSucceded at 31 with 1 tries\nSucceded at 32 with 1 tries\nSucceded at 33 with 1 tries\nSucceded at 34 with 1 tries\nSucceded at 35 with 1 tries\nSucceded at 37 with 1 tries\nSucceded at 39 with 1 tries\nSucceded at 40 with 1 tries\nSucceded at 41 with 1 tries\nSucceded at 42 with 1 tries\nSucceded at 45 with 1 tries\nSucceded at 46 with 1 tries\nSucceded at 48 with 1 tries\nSucceded at 51 with 1 tries\nSucceded at 52 with 1 tries\nSucceded at 54 with 1 tries\nSucceded at 56 with 1 tries\nSucceded at 60 with 1 tries\nSucceded at 61 with 1 tries\nSucceded at 63 with 1 tries\nSucceded at 64 with 1 tries\nSucceded at 65 with 1 tries\nSucceded at 66 with 1 tries\nSucceded at 67 with 1 tries\nSucceded at 68 with 1 tries\nSucceded at 70 with 1 tries\nSucceded at 74 with 1 tries\nSucceded at 76 with 1 tries\nSucceded at 78 with 1 tries\nSucceded at 79 with 1 tries\nSucceded at 84 with 1 tries\nSucceded at 85 with 1 tries\nSucceded at 86 with 1 tries\nSucceded at 87 with 1 tries\nSucceded at 88 with 1 tries\nSucceded at 89 with 1 tries\nSucceded at 90 with 1 tries\nSucceded at 91 with 1 tries\nSucceded at 93 with 1 tries\nSucceded at 94 with 1 tries\nSucceded at 95 with 1 tries\nSucceded at 96 with 1 tries\nSucceded at 97 with 1 tries\nSucceded at 98 with 1 tries\nSucceded at 101 with 1 tries\nSucceded at 102 with 1 tries\nSucceded at 103 with 1 tries\nSucceded at 104 with 1 tries\nSucceded at 105 with 1 tries\nSucceded at 106 with 1 tries\nSucceded at 107 with 1 tries\nSucceded at 108 with 1 tries\nSucceded at 109 with 1 tries\nSucceded at 110 with 1 tries\nSucceded at 111 with 1 tries\nSucceded at 112 with 1 tries\nSucceded at 115 with 1 tries\nSucceded at 116 with 1 tries\nSucceded at 119 with 1 tries\nSucceded at 120 with 1 tries\nSucceded at 121 with 1 tries\nSucceded at 122 with 1 tries\nSucceded at 124 with 1 tries\nSucceded at 126 with 1 tries\nSucceded at 127 with 1 tries\nSucceded at 129 with 1 tries\nSucceded at 133 with 1 tries\nSucceded at 134 with 1 tries\nSucceded at 138 with 1 tries\nSucceded at 140 with 1 tries\nSucceded at 141 with 1 tries\nSucceded at 142 with 1 tries\nSucceded at 143 with 1 tries\nSucceded at 145 with 1 tries\nSucceded at 146 with 1 tries\nmosek failed at \nSucceded at 150 with 1 tries\nSucceded at 151 with 1 tries\nSucceded at 152 with 1 tries\nSucceded at 153 with 1 tries\nSucceded at 154 with 1 tries\nSucceded at 155 with 1 tries\nSucceded at 156 with 1 tries\nSucceded at 157 with 1 tries\nSucceded at 158 with 1 tries\nSucceded at 159 with 1 tries\nSucceded at 161 with 1 tries\nSucceded at 166 with 1 tries\nSucceded at 167 with 1 tries\nSucceded at 169 with 1 tries\nSucceded at 170 with 1 tries\nSucceded at 171 with 1 tries\nSucceded at 172 with 1 tries\nSucceded at 174 with 1 tries\nSucceded at 175 with 1 tries\nSucceded at 176 with 1 tries\nSucceded at 177 with 1 tries\nSucceded at 179 with 1 tries\nSucceded at 182 with 1 tries\nSucceded at 183 with 1 tries\nSucceded at 187 with 1 tries\nSucceded at 189 with 1 tries\nSucceded at 191 with 1 tries\nSucceded at 192 with 1 tries\nSucceded at 193 with 1 tries\nSucceded at 194 with 1 tries\nSucceded at 195 with 1 tries\nSucceded at 197 with 1 tries\nSucceded at 198 with 1 tries\nSucceded at 199 with 1 tries\nSucceded at 200 with 1 tries\nSucceded at 201 with 1 tries\nSucceded at 202 with 1 tries\nSucceded at 204 with 1 tries\nSucceded at 207 with 1 tries\nSucceded at 208 with 1 tries\nSucceded at 209 with 1 tries\nSucceded at 210 with 1 tries\nSucceded at 211 with 1 tries\nSucceded at 213 with 1 tries\nSucceded at 216 with 1 tries\nSucceded at 217 with 1 tries\nSucceded at 219 with 1 tries\nSucceded at 220 with 1 tries\nSucceded at 221 with 1 tries\nSucceded at 222 with 1 tries\nSucceded at 223 with 1 tries\nSucceded at 224 with 1 tries\nSucceded at 226 with 1 tries\nSucceded at 227 with 1 tries\nSucceded at 228 with 1 tries\nSucceded at 231 with 1 tries\nSucceded at 232 with 1 tries\nSucceded at 233 with 1 tries\nSucceded at 234 with 1 tries\nSucceded at 236 with 1 tries\nSucceded at 237 with 1 tries\nSucceded at 238 with 1 tries\nSucceded at 244 with 1 tries\nSucceded at 247 with 1 tries\nSucceded at 249 with 1 tries\nSucceded at 251 with 1 tries\nSucceded at 253 with 1 tries\nSucceded at 254 with 1 tries\nSucceded at 255 with 1 tries\nSucceded at 256 with 1 tries\nSucceded at 257 with 1 tries\nSucceded at 259 with 1 tries\nSucceded at 263 with 1 tries\nSucceded at 264 with 1 tries\nSucceded at 265 with 1 tries\nSucceded at 266 with 1 tries\nSucceded at 267 with 1 tries\nSucceded at 269 with 1 tries\nSucceded at 271 with 1 tries\nmosek failed at \nSucceded at 275 with 1 tries\nSucceded at 277 with 1 tries\nSucceded at 278 with 1 tries\nSucceded at 279 with 1 tries\nSucceded at 280 with 1 tries\nSucceded at 283 with 1 tries\nSucceded at 285 with 1 tries\nSucceded at 286 with 1 tries\nSucceded at 288 with 1 tries\nSucceded at 292 with 1 tries\nSucceded at 293 with 1 tries\nSucceded at 294 with 1 tries\nSucceded at 295 with 1 tries\nSucceded at 297 with 1 tries\nSucceded at 298 with 1 tries\nSucceded at 299 with 1 tries\nSucceded at 300 with 1 tries\nSucceded at 301 with 1 tries\nSucceded at 302 with 1 tries\nSucceded at 303 with 1 tries\nSucceded at 308 with 1 tries\nSucceded at 309 with 1 tries\nSucceded at 311 with 1 tries\nSucceded at 312 with 1 tries\nSucceded at 313 with 1 tries\nSucceded at 315 with 1 tries\nSucceded at 318 with 1 tries\nSucceded at 321 with 1 tries\nSucceded at 322 with 1 tries\nSucceded at 323 with 1 tries\nSucceded at 327 with 1 tries\nSucceded at 328 with 1 tries\nSucceded at 329 with 1 tries\nSucceded at 330 with 1 tries\nSucceded at 331 with 1 tries\nSucceded at 332 with 1 tries\nSucceded at 333 with 1 tries\nSucceded at 334 with 1 tries\nSucceded at 340 with 1 tries\nSucceded at 343 with 1 tries\nSucceded at 345 with 1 tries\nSucceded at 346 with 1 tries\nSucceded at 347 with 1 tries\nSucceded at 348 with 1 tries\nSucceded at 350 with 1 tries\nSucceded at 352 with 1 tries\nSucceded at 353 with 1 tries\nSucceded at 354 with 1 tries\nSucceded at 355 with 1 tries\nSucceded at 357 with 1 tries\nSucceded at 359 with 1 tries\nSucceded at 360 with 1 tries\nSucceded at 361 with 1 tries\nSucceded at 363 with 1 tries\nSucceded at 364 with 1 tries\nSucceded at 365 with 1 tries\nSucceded at 366 with 1 tries\nSucceded at 367 with 1 tries\nSucceded at 368 with 1 tries\nSucceded at 370 with 1 tries\nSucceded at 374 with 1 tries\nSucceded at 375 with 1 tries\nSucceded at 376 with 1 tries\nSucceded at 377 with 1 tries\nSucceded at 378 with 1 tries\nSucceded at 379 with 1 tries\nSucceded at 380 with 1 tries\nSucceded at 382 with 1 tries\nSucceded at 386 with 1 tries\nSucceded at 387 with 1 tries\nSucceded at 388 with 1 tries\nSucceded at 390 with 1 tries\nSucceded at 391 with 1 tries\nSucceded at 392 with 1 tries\nSucceded at 396 with 1 tries\nSucceded at 397 with 1 tries\nSucceded at 398 with 1 tries\nSucceded at 399 with 1 tries\nSucceded at 402 with 1 tries\nSucceded at 403 with 1 tries\nSucceded at 407 with 1 tries\nSucceded at 408 with 1 tries\nSucceded at 409 with 1 tries\nSucceded at 411 with 1 tries\nSucceded at 412 with 1 tries\nSucceded at 414 with 1 tries\nSucceded at 416 with 1 tries\nSucceded at 417 with 1 tries\nSucceded at 418 with 1 tries\nSucceded at 419 with 1 tries\nSucceded at 420 with 1 tries\nSucceded at 421 with 1 tries\nSucceded at 423 with 1 tries\nSucceded at 425 with 1 tries\nSucceded at 428 with 1 tries\nSucceded at 429 with 1 tries\nSucceded at 430 with 1 tries\nSucceded at 431 with 1 tries\nSucceded at 432 with 1 tries\nSucceded at 433 with 1 tries\nSucceded at 434 with 1 tries\nSucceded at 435 with 1 tries\nSucceded at 437 with 1 tries\nSucceded at 438 with 1 tries\nSucceded at 440 with 1 tries\nSucceded at 443 with 1 tries\nSucceded at 445 with 1 tries\nSucceded at 447 with 1 tries\nSucceded at 449 with 1 tries\nSucceded at 452 with 1 tries\nSucceded at 453 with 1 tries\nSucceded at 454 with 1 tries\nSucceded at 455 with 1 tries\nSucceded at 456 with 1 tries\nSucceded at 457 with 1 tries\nSucceded at 458 with 1 tries\nSucceded at 459 with 1 tries\nSucceded at 461 with 1 tries\nSucceded at 462 with 1 tries\nSucceded at 463 with 1 tries\nSucceded at 464 with 1 tries\nSucceded at 466 with 1 tries\nSucceded at 468 with 1 tries\nSucceded at 469 with 1 tries\nSucceded at 472 with 1 tries\nSucceded at 473 with 1 tries\nSucceded at 474 with 1 tries\nSucceded at 476 with 1 tries\nSucceded at 478 with 1 tries\nSucceded at 479 with 1 tries\nSucceded at 480 with 1 tries\nSucceded at 481 with 1 tries\nSucceded at 486 with 1 tries\nSucceded at 488 with 1 tries\nSucceded at 489 with 1 tries\nSucceded at 490 with 1 tries\nSucceded at 492 with 1 tries\nSucceded at 493 with 1 tries\nSucceded at 496 with 1 tries\nSucceded at 497 with 1 tries\nSucceded at 500 with 1 tries\nSucceded at 501 with 1 tries\nSucceded at 502 with 1 tries\nSucceded at 503 with 1 tries\nSucceded at 504 with 1 tries\nSucceded at 505 with 1 tries\nSucceded at 506 with 1 tries\nSucceded at 508 with 1 tries\nSucceded at 509 with 1 tries\nSucceded at 510 with 1 tries\nSucceded at 512 with 1 tries\nSucceded at 513 with 1 tries\nSucceded at 514 with 1 tries\nSucceded at 515 with 1 tries\nSucceded at 518 with 1 tries\nSucceded at 520 with 1 tries\nSucceded at 521 with 1 tries\nSucceded at 522 with 1 tries\nSucceded at 526 with 1 tries\nSucceded at 529 with 1 tries\nSucceded at 530 with 1 tries\nSucceded at 531 with 1 tries\nSucceded at 534 with 1 tries\nSucceded at 538 with 1 tries\nSucceded at 539 with 1 tries\nSucceded at 540 with 1 tries\nSucceded at 542 with 1 tries\nSucceded at 543 with 1 tries\nSucceded at 545 with 1 tries\nSucceded at 546 with 1 tries\nSucceded at 547 with 1 tries\nSucceded at 548 with 1 tries\nSucceded at 549 with 1 tries\nSucceded at 552 with 1 tries\nSucceded at 554 with 1 tries\nSucceded at 555 with 1 tries\nSucceded at 558 with 1 tries\nSucceded at 559 with 1 tries\nSucceded at 561 with 1 tries\nSucceded at 562 with 1 tries\nSucceded at 563 with 1 tries\nSucceded at 565 with 1 tries\nSucceded at 566 with 1 tries\nSucceded at 567 with 1 tries\nSucceded at 568 with 1 tries\nSucceded at 571 with 1 tries\nSucceded at 572 with 1 tries\nSucceded at 573 with 1 tries\nSucceded at 574 with 1 tries\nSucceded at 575 with 1 tries\nSucceded at 576 with 1 tries\nSucceded at 577 with 1 tries\nSucceded at 578 with 1 tries\nSucceded at 579 with 1 tries\nSucceded at 580 with 1 tries\nSucceded at 581 with 1 tries\nSucceded at 582 with 1 tries\nSucceded at 583 with 1 tries\nSucceded at 585 with 1 tries\nSucceded at 586 with 1 tries\nSucceded at 587 with 1 tries\nSucceded at 588 with 1 tries\nSucceded at 589 with 1 tries\nSucceded at 590 with 1 tries\nSucceded at 591 with 1 tries\nSucceded at 592 with 1 tries\nSucceded at 593 with 1 tries\nSucceded at 594 with 1 tries\nSucceded at 595 with 1 tries\nSucceded at 596 with 1 tries\nSucceded at 597 with 1 tries\nSucceded at 600 with 1 tries\nSucceded at 601 with 1 tries\nSucceded at 602 with 1 tries\nSucceded at 603 with 1 tries\nSucceded at 605 with 1 tries\nSucceded at 606 with 1 tries\nSucceded at 607 with 1 tries\nSucceded at 608 with 1 tries\nSucceded at 609 with 1 tries\nSucceded at 610 with 1 tries\nSucceded at 611 with 1 tries\nSucceded at 612 with 1 tries\nSucceded at 617 with 1 tries\nSucceded at 618 with 1 tries\nSucceded at 619 with 1 tries\nSucceded at 620 with 1 tries\nSucceded at 621 with 1 tries\nSucceded at 622 with 1 tries\nSucceded at 627 with 1 tries\nSucceded at 628 with 1 tries\nSucceded at 630 with 1 tries\nSucceded at 632 with 1 tries\nSucceded at 633 with 1 tries\nSucceded at 636 with 1 tries\nSucceded at 637 with 1 tries\nSucceded at 639 with 1 tries\nSucceded at 640 with 1 tries\nSucceded at 641 with 1 tries\nSucceded at 642 with 1 tries\nSucceded at 644 with 1 tries\nSucceded at 646 with 1 tries\nSucceded at 647 with 1 tries\nSucceded at 648 with 1 tries\nSucceded at 650 with 1 tries\nSucceded at 651 with 1 tries\nSucceded at 652 with 1 tries\nSucceded at 657 with 1 tries\nSucceded at 658 with 1 tries\nSucceded at 660 with 1 tries\nSucceded at 661 with 1 tries\nSucceded at 662 with 1 tries\nSucceded at 663 with 1 tries\nSucceded at 664 with 1 tries\nSucceded at 665 with 1 tries\nSucceded at 667 with 1 tries\nSucceded at 668 with 1 tries\nSucceded at 669 with 1 tries\nSucceded at 670 with 1 tries\nSucceded at 671 with 1 tries\nSucceded at 672 with 1 tries\nSucceded at 673 with 1 tries\nSucceded at 674 with 1 tries\nSucceded at 675 with 1 tries\nSucceded at 678 with 1 tries\nSucceded at 679 with 1 tries\nSucceded at 680 with 1 tries\nSucceded at 681 with 1 tries\nSucceded at 685 with 1 tries\nSucceded at 686 with 1 tries\nSucceded at 687 with 1 tries\nSucceded at 690 with 1 tries\nSucceded at 691 with 1 tries\nSucceded at 692 with 1 tries\nSucceded at 693 with 1 tries\nSucceded at 694 with 1 tries\nSucceded at 695 with 1 tries\nSucceded at 697 with 1 tries\nSucceded at 699 with 1 tries\nSucceded at 700 with 1 tries\nSucceded at 705 with 1 tries\nSucceded at 706 with 1 tries\nSucceded at 707 with 1 tries\nSucceded at 708 with 1 tries\nSucceded at 710 with 1 tries\nSucceded at 711 with 1 tries\nSucceded at 713 with 1 tries\nSucceded at 714 with 1 tries\nSucceded at 715 with 1 tries\nSucceded at 716 with 1 tries\nSucceded at 717 with 1 tries\nSucceded at 718 with 1 tries\nSucceded at 719 with 1 tries\nSucceded at 720 with 1 tries\nSucceded at 721 with 1 tries\nSucceded at 722 with 1 tries\nSucceded at 723 with 1 tries\nSucceded at 724 with 1 tries\nSucceded at 726 with 1 tries\nSucceded at 727 with 1 tries\nSucceded at 728 with 1 tries\nSucceded at 729 with 1 tries\nSucceded at 730 with 1 tries\nSucceded at 731 with 1 tries\nSucceded at 732 with 1 tries\nSucceded at 733 with 1 tries\nSucceded at 734 with 1 tries\nSucceded at 735 with 1 tries\nSucceded at 736 with 1 tries\nSucceded at 737 with 1 tries\nSucceded at 738 with 1 tries\nSucceded at 739 with 1 tries\nSucceded at 740 with 1 tries\nSucceded at 742 with 1 tries\nSucceded at 743 with 1 tries\nSucceded at 744 with 1 tries\nSucceded at 745 with 1 tries\nSucceded at 746 with 1 tries\nSucceded at 747 with 1 tries\nSucceded at 748 with 1 tries\nSucceded at 749 with 1 tries\nSucceded at 750 with 1 tries\nSucceded at 751 with 1 tries\nSucceded at 753 with 1 tries\nSucceded at 755 with 1 tries\nmosek failed at \nSucceded at 757 with 1 tries\nSucceded at 758 with 1 tries\nSucceded at 759 with 1 tries\nSucceded at 761 with 1 tries\nSucceded at 763 with 1 tries\nSucceded at 764 with 1 tries\nSucceded at 765 with 1 tries\nSucceded at 769 with 1 tries\nSucceded at 771 with 1 tries\nSucceded at 772 with 1 tries\nSucceded at 773 with 1 tries\nSucceded at 778 with 1 tries\nSucceded at 779 with 1 tries\nSucceded at 780 with 1 tries\nSucceded at 782 with 1 tries\nSucceded at 783 with 1 tries\nSucceded at 784 with 1 tries\nSucceded at 785 with 1 tries\nSucceded at 786 with 1 tries\nSucceded at 788 with 1 tries\nSucceded at 791 with 1 tries\nSucceded at 792 with 1 tries\nSucceded at 794 with 1 tries\nSucceded at 795 with 1 tries\nSucceded at 796 with 1 tries\nSucceded at 798 with 1 tries\nSucceded at 799 with 1 tries\nSucceded at 800 with 1 tries\nSucceded at 802 with 1 tries\nSucceded at 804 with 1 tries\nSucceded at 805 with 1 tries\nSucceded at 807 with 1 tries\nSucceded at 810 with 1 tries\nSucceded at 811 with 1 tries\nSucceded at 812 with 1 tries\nSucceded at 813 with 1 tries\nSucceded at 814 with 1 tries\nSucceded at 815 with 1 tries\nSucceded at 816 with 1 tries\nSucceded at 817 with 1 tries\nSucceded at 818 with 1 tries\nSucceded at 819 with 1 tries\nSucceded at 820 with 1 tries\nSucceded at 822 with 1 tries\nSucceded at 823 with 1 tries\nSucceded at 824 with 1 tries\nSucceded at 825 with 1 tries\nSucceded at 826 with 1 tries\nSucceded at 827 with 1 tries\nSucceded at 829 with 1 tries\nSucceded at 831 with 1 tries\nSucceded at 832 with 1 tries\nSucceded at 833 with 1 tries\nSucceded at 834 with 1 tries\nSucceded at 836 with 1 tries\nSucceded at 838 with 1 tries\nSucceded at 841 with 1 tries\nSucceded at 842 with 1 tries\nSucceded at 843 with 1 tries\nSucceded at 845 with 1 tries\nSucceded at 846 with 1 tries\nSucceded at 847 with 1 tries\nSucceded at 849 with 1 tries\nSucceded at 851 with 1 tries\nSucceded at 853 with 1 tries\nSucceded at 854 with 1 tries\nSucceded at 855 with 1 tries\nSucceded at 856 with 1 tries\nSucceded at 857 with 1 tries\nSucceded at 859 with 1 tries\nSucceded at 860 with 1 tries\nSucceded at 861 with 1 tries\nSucceded at 864 with 1 tries\nSucceded at 865 with 1 tries\nSucceded at 866 with 1 tries\nSucceded at 867 with 1 tries\nSucceded at 868 with 1 tries\nSucceded at 869 with 1 tries\nSucceded at 870 with 1 tries\nSucceded at 871 with 1 tries\nSucceded at 872 with 1 tries\nSucceded at 873 with 1 tries\nSucceded at 874 with 1 tries\nSucceded at 876 with 1 tries\nSucceded at 877 with 1 tries\nSucceded at 878 with 1 tries\nSucceded at 879 with 1 tries\nSucceded at 880 with 1 tries\nSucceded at 881 with 1 tries\nSucceded at 884 with 1 tries\nSucceded at 885 with 1 tries\nSucceded at 886 with 1 tries\nSucceded at 887 with 1 tries\nSucceded at 888 with 1 tries\nSucceded at 889 with 1 tries\nSucceded at 891 with 1 tries\nSucceded at 892 with 1 tries\nSucceded at 893 with 1 tries\nSucceded at 894 with 1 tries\nSucceded at 896 with 1 tries\nSucceded at 897 with 1 tries\nSucceded at 898 with 1 tries\nSucceded at 899 with 1 tries\nSucceded at 900 with 1 tries\nSucceded at 901 with 1 tries\nSucceded at 904 with 1 tries\nSucceded at 905 with 1 tries\nSucceded at 906 with 1 tries\nSucceded at 907 with 1 tries\nSucceded at 909 with 1 tries\nSucceded at 910 with 1 tries\nSucceded at 911 with 1 tries\nSucceded at 914 with 1 tries\nSucceded at 915 with 1 tries\nSucceded at 917 with 1 tries\nSucceded at 918 with 1 tries\nSucceded at 920 with 1 tries\nSucceded at 921 with 1 tries\nSucceded at 924 with 1 tries\nSucceded at 925 with 1 tries\nSucceded at 926 with 1 tries\nSucceded at 927 with 1 tries\nSucceded at 931 with 1 tries\nSucceded at 935 with 1 tries\nSucceded at 936 with 1 tries\nSucceded at 937 with 1 tries\nSucceded at 938 with 1 tries\nSucceded at 939 with 1 tries\nSucceded at 942 with 1 tries\nSucceded at 944 with 1 tries\nSucceded at 945 with 1 tries\nSucceded at 946 with 1 tries\nSucceded at 947 with 1 tries\nSucceded at 950 with 1 tries\nSucceded at 951 with 1 tries\nSucceded at 952 with 1 tries\nSucceded at 953 with 1 tries\nSucceded at 954 with 1 tries\nSucceded at 955 with 1 tries\nSucceded at 956 with 1 tries\nSucceded at 958 with 1 tries\nSucceded at 959 with 1 tries\nSucceded at 961 with 1 tries\nSucceded at 963 with 1 tries\nSucceded at 964 with 1 tries\nSucceded at 965 with 1 tries\nSucceded at 966 with 1 tries\nSucceded at 970 with 1 tries\nSucceded at 971 with 1 tries\nSucceded at 972 with 1 tries\nSucceded at 973 with 1 tries\nSucceded at 976 with 1 tries\nSucceded at 977 with 1 tries\nSucceded at 978 with 1 tries\nSucceded at 979 with 1 tries\nSucceded at 982 with 1 tries\nSucceded at 984 with 1 tries\nSucceded at 986 with 1 tries\nSucceded at 987 with 1 tries\nSucceded at 988 with 1 tries\nSucceded at 989 with 1 tries\nSucceded at 990 with 1 tries\nSucceded at 991 with 1 tries\nSucceded at 993 with 1 tries\nSucceded at 996 with 1 tries\nSucceded at 998 with 1 tries\nSucceded at 999 with 1 tries\nSucceded at 1000 with 1 tries\nSucceded at 1002 with 1 tries\nSucceded at 1003 with 1 tries\nSucceded at 1007 with 1 tries\nSucceded at 1011 with 1 tries\nSucceded at 1014 with 1 tries\nSucceded at 1015 with 1 tries\nSucceded at 1017 with 1 tries\nSucceded at 1018 with 1 tries\nSucceded at 1020 with 1 tries\nSucceded at 1022 with 1 tries\nSucceded at 1023 with 1 tries\nSucceded at 1024 with 1 tries\nSucceded at 1025 with 1 tries\nSucceded at 1026 with 1 tries\nSucceded at 1028 with 1 tries\nSucceded at 1030 with 1 tries\nSucceded at 1031 with 1 tries\nSucceded at 1032 with 1 tries\nSucceded at 1033 with 1 tries\nSucceded at 1035 with 1 tries\nSucceded at 1037 with 1 tries\nSucceded at 1039 with 1 tries\nSucceded at 1040 with 1 tries\nSucceded at 1046 with 1 tries\nSucceded at 1047 with 1 tries\nSucceded at 1050 with 1 tries\nSucceded at 1051 with 1 tries\nSucceded at 1052 with 1 tries\nSucceded at 1053 with 1 tries\nSucceded at 1056 with 1 tries\nSucceded at 1058 with 1 tries\nSucceded at 1059 with 1 tries\nSucceded at 1060 with 1 tries\nSucceded at 1061 with 1 tries\nSucceded at 1062 with 1 tries\nSucceded at 1063 with 1 tries\nSucceded at 1065 with 1 tries\nSucceded at 1066 with 1 tries\nSucceded at 1067 with 1 tries\nSucceded at 1068 with 1 tries\nSucceded at 1069 with 1 tries\nSucceded at 1071 with 1 tries\nSucceded at 1072 with 1 tries\nSucceded at 1073 with 1 tries\nSucceded at 1074 with 1 tries\nSucceded at 1075 with 1 tries\nSucceded at 1076 with 1 tries\nSucceded at 1077 with 1 tries\nSucceded at 1078 with 1 tries\nSucceded at 1079 with 1 tries\nSucceded at 1080 with 1 tries\nSucceded at 1081 with 1 tries\nSucceded at 1082 with 1 tries\nSucceded at 1084 with 1 tries\nSucceded at 1085 with 1 tries\nSucceded at 1086 with 1 tries\nSucceded at 1087 with 1 tries\nSucceded at 1088 with 1 tries\nSucceded at 1090 with 1 tries\nSucceded at 1091 with 1 tries\nSucceded at 1092 with 1 tries\nSucceded at 1094 with 1 tries\nSucceded at 1095 with 1 tries\nSucceded at 1096 with 1 tries\nSucceded at 1097 with 1 tries\nSucceded at 1099 with 1 tries\nSucceded at 1100 with 1 tries\nSucceded at 1102 with 1 tries\nSucceded at 1103 with 1 tries\nSucceded at 1104 with 1 tries\nSucceded at 1106 with 1 tries\nSucceded at 1107 with 1 tries\nSucceded at 1108 with 1 tries\nSucceded at 1110 with 1 tries\nSucceded at 1111 with 1 tries\nSucceded at 1113 with 1 tries\nSucceded at 1117 with 1 tries\nSucceded at 1118 with 1 tries\nSucceded at 1121 with 1 tries\nSucceded at 1122 with 1 tries\nSucceded at 1123 with 1 tries\nSucceded at 1124 with 1 tries\nSucceded at 1125 with 1 tries\nSucceded at 1126 with 1 tries\nSucceded at 1128 with 1 tries\nSucceded at 1129 with 1 tries\nSucceded at 1130 with 1 tries\nSucceded at 1131 with 1 tries\nSucceded at 1134 with 1 tries\nSucceded at 1135 with 1 tries\nSucceded at 1136 with 1 tries\nSucceded at 1137 with 1 tries\nSucceded at 1138 with 1 tries\nSucceded at 1141 with 1 tries\nSucceded at 1142 with 1 tries\nSucceded at 1143 with 1 tries\nSucceded at 1144 with 1 tries\nSucceded at 1145 with 1 tries\nSucceded at 1146 with 1 tries\nSucceded at 1147 with 1 tries\nSucceded at 1148 with 1 tries\nSucceded at 1149 with 1 tries\nSucceded at 1150 with 1 tries\nSucceded at 1152 with 1 tries\nSucceded at 1153 with 1 tries\nSucceded at 1154 with 1 tries\nSucceded at 1155 with 1 tries\nSucceded at 1157 with 1 tries\nSucceded at 1160 with 1 tries\nSucceded at 1161 with 1 tries\nSucceded at 1162 with 1 tries\nSucceded at 1163 with 1 tries\nSucceded at 1164 with 1 tries\nSucceded at 1165 with 1 tries\nSucceded at 1166 with 1 tries\nSucceded at 1168 with 1 tries\nSucceded at 1170 with 1 tries\nSucceded at 1171 with 1 tries\nSucceded at 1172 with 1 tries\nSucceded at 1173 with 1 tries\nSucceded at 1174 with 1 tries\nSucceded at 1175 with 1 tries\nSucceded at 1177 with 1 tries\nSucceded at 1178 with 1 tries\nSucceded at 1180 with 1 tries\nSucceded at 1181 with 1 tries\nSucceded at 1182 with 1 tries\nSucceded at 1183 with 1 tries\nSucceded at 1184 with 1 tries\nSucceded at 1186 with 1 tries\nSucceded at 1188 with 1 tries\nSucceded at 1189 with 1 tries\nSucceded at 1191 with 1 tries\nSucceded at 1192 with 1 tries\nSucceded at 1193 with 1 tries\nSucceded at 1194 with 1 tries\nSucceded at 1195 with 1 tries\nSucceded at 1199 with 1 tries\nSucceded at 1200 with 1 tries\nSucceded at 1201 with 1 tries\nSucceded at 1203 with 1 tries\nSucceded at 1204 with 1 tries\nSucceded at 1205 with 1 tries\nSucceded at 1206 with 1 tries\nSucceded at 1208 with 1 tries\nSucceded at 1209 with 1 tries\nSucceded at 1210 with 1 tries\nSucceded at 1211 with 1 tries\nSucceded at 1213 with 1 tries\nSucceded at 1214 with 1 tries\nSucceded at 1215 with 1 tries\nSucceded at 1216 with 1 tries\nSucceded at 1217 with 1 tries\nSucceded at 1218 with 1 tries\nSucceded at 1222 with 1 tries\nSucceded at 1223 with 1 tries\nSucceded at 1224 with 1 tries\nSucceded at 1226 with 1 tries\nSucceded at 1227 with 1 tries\nSucceded at 1229 with 1 tries\nSucceded at 1233 with 1 tries\nSucceded at 1235 with 1 tries\nSucceded at 1236 with 1 tries\nSucceded at 1237 with 1 tries\nSucceded at 1238 with 1 tries\nSucceded at 1239 with 1 tries\nSucceded at 1240 with 1 tries\nSucceded at 1241 with 1 tries\nSucceded at 1242 with 1 tries\nSucceded at 1243 with 1 tries\nSucceded at 1244 with 1 tries\nSucceded at 1247 with 1 tries\nSucceded at 1248 with 1 tries\nSucceded at 1249 with 1 tries\nSucceded at 1250 with 1 tries\nSucceded at 1251 with 1 tries\nSucceded at 1253 with 1 tries\nSucceded at 1254 with 1 tries\nSucceded at 1255 with 1 tries\nSucceded at 1256 with 1 tries\nSucceded at 1258 with 1 tries\nSucceded at 1259 with 1 tries\nSucceded at 1260 with 1 tries\nSucceded at 1261 with 1 tries\nSucceded at 1262 with 1 tries\nSucceded at 1264 with 1 tries\nSucceded at 1265 with 1 tries\nSucceded at 1267 with 1 tries\nSucceded at 1268 with 1 tries\nSucceded at 1269 with 1 tries\nSucceded at 1270 with 1 tries\nSucceded at 1272 with 1 tries\nSucceded at 1274 with 1 tries\nSucceded at 1278 with 1 tries\nSucceded at 1279 with 1 tries\nSucceded at 1280 with 1 tries\nSucceded at 1282 with 1 tries\nSucceded at 1283 with 1 tries\nSucceded at 1284 with 1 tries\nSucceded at 1285 with 1 tries\nSucceded at 1286 with 1 tries\nSucceded at 1288 with 1 tries\nSucceded at 1289 with 1 tries\nSucceded at 1290 with 1 tries\nSucceded at 1291 with 1 tries\nSucceded at 1292 with 1 tries\nSucceded at 1293 with 1 tries\nSucceded at 1295 with 1 tries\nSucceded at 1299 with 1 tries\nSucceded at 1300 with 1 tries\nSucceded at 1301 with 1 tries\nSucceded at 1302 with 1 tries\nSucceded at 1303 with 1 tries\nSucceded at 1304 with 1 tries\nSucceded at 1305 with 1 tries\nSucceded at 1306 with 1 tries\nSucceded at 1307 with 1 tries\nSucceded at 1308 with 1 tries\nSucceded at 1309 with 1 tries\nSucceded at 1310 with 1 tries\nSucceded at 1311 with 1 tries\nSucceded at 1312 with 1 tries\nSucceded at 1314 with 1 tries\nSucceded at 1315 with 1 tries\nSucceded at 1316 with 1 tries\nSucceded at 1317 with 1 tries\nSucceded at 1320 with 1 tries\nSucceded at 1321 with 1 tries\nSucceded at 1322 with 1 tries\nSucceded at 1323 with 1 tries\nSucceded at 1324 with 1 tries\nSucceded at 1326 with 1 tries\nSucceded at 1327 with 1 tries\nSucceded at 1328 with 1 tries\nSucceded at 1330 with 1 tries\nSucceded at 1331 with 1 tries\nSucceded at 1333 with 1 tries\nSucceded at 1335 with 1 tries\nSucceded at 1336 with 1 tries\nSucceded at 1337 with 1 tries\nSucceded at 1339 with 1 tries\nSucceded at 1340 with 1 tries\nSucceded at 1341 with 1 tries\nSucceded at 1342 with 1 tries\nSucceded at 1343 with 1 tries\nSucceded at 1344 with 1 tries\nSucceded at 1345 with 1 tries\nSucceded at 1346 with 1 tries\nSucceded at 1348 with 1 tries\nSucceded at 1349 with 1 tries\nSucceded at 1350 with 1 tries\nSucceded at 1352 with 1 tries\nSucceded at 1353 with 1 tries\nSucceded at 1354 with 1 tries\nSucceded at 1355 with 1 tries\nSucceded at 1357 with 1 tries\nSucceded at 1361 with 1 tries\nSucceded at 1362 with 1 tries\nSucceded at 1366 with 1 tries\nSucceded at 1368 with 1 tries\nSucceded at 1369 with 1 tries\nSucceded at 1370 with 1 tries\nSucceded at 1372 with 1 tries\nSucceded at 1373 with 1 tries\nSucceded at 1374 with 1 tries\nSucceded at 1375 with 1 tries\nSucceded at 1376 with 1 tries\nSucceded at 1377 with 1 tries\nSucceded at 1380 with 1 tries\nSucceded at 1381 with 1 tries\nSucceded at 1383 with 1 tries\nSucceded at 1384 with 1 tries\nSucceded at 1386 with 1 tries\nSucceded at 1387 with 1 tries\nSucceded at 1388 with 1 tries\nSucceded at 1389 with 1 tries\nSucceded at 1393 with 1 tries\nSucceded at 1394 with 1 tries\nSucceded at 1395 with 1 tries\nSucceded at 1396 with 1 tries\nSucceded at 1397 with 1 tries\nSucceded at 1399 with 1 tries\nSucceded at 1400 with 1 tries\nSucceded at 1401 with 1 tries\nSucceded at 1402 with 1 tries\nSucceded at 1403 with 1 tries\nSucceded at 1405 with 1 tries\nSucceded at 1408 with 1 tries\nSucceded at 1409 with 1 tries\nSucceded at 1410 with 1 tries\nSucceded at 1411 with 1 tries\nSucceded at 1413 with 1 tries\nSucceded at 1415 with 1 tries\nSucceded at 1416 with 1 tries\nSucceded at 1417 with 1 tries\nSucceded at 1418 with 1 tries\nSucceded at 1419 with 1 tries\nSucceded at 1420 with 1 tries\nSucceded at 1421 with 1 tries\nSucceded at 1423 with 1 tries\nSucceded at 1426 with 1 tries\nSucceded at 1427 with 1 tries\nSucceded at 1428 with 1 tries\nSucceded at 1430 with 1 tries\nSucceded at 1431 with 1 tries\nSucceded at 1432 with 1 tries\nSucceded at 1433 with 1 tries\nSucceded at 1434 with 1 tries\nSucceded at 1436 with 1 tries\nSucceded at 1437 with 1 tries\nSucceded at 1438 with 1 tries\nSucceded at 1439 with 1 tries\nSucceded at 1440 with 1 tries\nSucceded at 1441 with 1 tries\nSucceded at 1442 with 1 tries\nSucceded at 1443 with 1 tries\nSucceded at 1444 with 1 tries\nSucceded at 1445 with 1 tries\nSucceded at 1446 with 1 tries\nSucceded at 1447 with 1 tries\nSucceded at 1449 with 1 tries\nSucceded at 1450 with 1 tries\nSucceded at 1451 with 1 tries\nSucceded at 1452 with 1 tries\nSucceded at 1453 with 1 tries\nSucceded at 1454 with 1 tries\nSucceded at 1455 with 1 tries\nSucceded at 1456 with 1 tries\nSucceded at 1457 with 1 tries\nSucceded at 1459 with 1 tries\nSucceded at 1460 with 1 tries\nSucceded at 1463 with 1 tries\nSucceded at 1464 with 1 tries\nSucceded at 1467 with 1 tries\nSucceded at 1468 with 1 tries\nSucceded at 1469 with 1 tries\nSucceded at 1470 with 1 tries\nSucceded at 1471 with 1 tries\nSucceded at 1473 with 1 tries\nSucceded at 1474 with 1 tries\nSucceded at 1475 with 1 tries\nSucceded at 1476 with 1 tries\nSucceded at 1477 with 1 tries\nSucceded at 1478 with 1 tries\nSucceded at 1479 with 1 tries\nSucceded at 1480 with 1 tries\nSucceded at 1481 with 1 tries\nSucceded at 1482 with 1 tries\nSucceded at 1483 with 1 tries\nSucceded at 1484 with 1 tries\nSucceded at 1486 with 1 tries\nSucceded at 1487 with 1 tries\nSucceded at 1488 with 1 tries\nSucceded at 1489 with 1 tries\nSucceded at 1490 with 1 tries\nSucceded at 1491 with 1 tries\nSucceded at 1492 with 1 tries\nSucceded at 1493 with 1 tries\nSucceded at 1494 with 1 tries\nSucceded at 1496 with 1 tries\nSucceded at 1499 with 1 tries\nSucceded at 1500 with 1 tries\nSucceded at 1501 with 1 tries\nSucceded at 1502 with 1 tries\nSucceded at 1503 with 1 tries\nSucceded at 1505 with 1 tries\nSucceded at 1507 with 1 tries\nSucceded at 1508 with 1 tries\nSucceded at 1510 with 1 tries\nSucceded at 1512 with 1 tries\nSucceded at 1513 with 1 tries\nSucceded at 1514 with 1 tries\nSucceded at 1515 with 1 tries\nSucceded at 1518 with 1 tries\nSucceded at 1519 with 1 tries\nSucceded at 1521 with 1 tries\nSucceded at 1522 with 1 tries\nSucceded at 1523 with 1 tries\nSucceded at 1524 with 1 tries\nSucceded at 1526 with 1 tries\nSucceded at 1527 with 1 tries\nSucceded at 1528 with 1 tries\nSucceded at 1529 with 1 tries\nSucceded at 1533 with 1 tries\nSucceded at 1535 with 1 tries\nSucceded at 1536 with 1 tries\nSucceded at 1537 with 1 tries\nSucceded at 1538 with 1 tries\nSucceded at 1539 with 1 tries\nSucceded at 1541 with 1 tries\nSucceded at 1543 with 1 tries\nSucceded at 1544 with 1 tries\nSucceded at 1545 with 1 tries\nSucceded at 1547 with 1 tries\nSucceded at 1548 with 1 tries\nSucceded at 1550 with 1 tries\nmosek failed at \nSucceded at 1554 with 1 tries\nSucceded at 1555 with 1 tries\nSucceded at 1557 with 1 tries\nSucceded at 1558 with 1 tries\nSucceded at 1559 with 1 tries\nSucceded at 1560 with 1 tries\nSucceded at 1561 with 1 tries\nSucceded at 1562 with 1 tries\nSucceded at 1563 with 1 tries\nSucceded at 1565 with 1 tries\nSucceded at 1566 with 1 tries\nSucceded at 1567 with 1 tries\nSucceded at 1568 with 1 tries\nSucceded at 1570 with 1 tries\nSucceded at 1571 with 1 tries\nSucceded at 1572 with 1 tries\nSucceded at 1573 with 1 tries\nSucceded at 1574 with 1 tries\nSucceded at 1575 with 1 tries\nSucceded at 1576 with 1 tries\nSucceded at 1579 with 1 tries\nSucceded at 1581 with 1 tries\nSucceded at 1582 with 1 tries\nSucceded at 1585 with 1 tries\nSucceded at 1587 with 1 tries\nSucceded at 1589 with 1 tries\nSucceded at 1590 with 1 tries\nSucceded at 1591 with 1 tries\nSucceded at 1592 with 1 tries\nSucceded at 1593 with 1 tries\nSucceded at 1594 with 1 tries\nSucceded at 1595 with 1 tries\nSucceded at 1596 with 1 tries\nSucceded at 1597 with 1 tries\nSucceded at 1603 with 1 tries\nSucceded at 1604 with 1 tries\nSucceded at 1610 with 1 tries\nSucceded at 1611 with 1 tries\nSucceded at 1612 with 1 tries\nSucceded at 1613 with 1 tries\nSucceded at 1614 with 1 tries\nSucceded at 1615 with 1 tries\nSucceded at 1616 with 1 tries\nSucceded at 1617 with 1 tries\nSucceded at 1618 with 1 tries\nSucceded at 1619 with 1 tries\nSucceded at 1620 with 1 tries\nSucceded at 1621 with 1 tries\nSucceded at 1622 with 1 tries\nSucceded at 1623 with 1 tries\nSucceded at 1624 with 1 tries\nSucceded at 1625 with 1 tries\nSucceded at 1626 with 1 tries\nSucceded at 1627 with 1 tries\nSucceded at 1628 with 1 tries\nSucceded at 1630 with 1 tries\nSucceded at 1631 with 1 tries\nSucceded at 1633 with 1 tries\nSucceded at 1634 with 1 tries\nSucceded at 1635 with 1 tries\nSucceded at 1636 with 1 tries\nSucceded at 1637 with 1 tries\nSucceded at 1638 with 1 tries\nSucceded at 1639 with 1 tries\nSucceded at 1640 with 1 tries\nSucceded at 1642 with 1 tries\nSucceded at 1644 with 1 tries\nSucceded at 1646 with 1 tries\nSucceded at 1647 with 1 tries\nSucceded at 1648 with 1 tries\nSucceded at 1649 with 1 tries\nSucceded at 1652 with 1 tries\nSucceded at 1653 with 1 tries\nSucceded at 1654 with 1 tries\nSucceded at 1655 with 1 tries\nSucceded at 1656 with 1 tries\nSucceded at 1657 with 1 tries\nSucceded at 1658 with 1 tries\nSucceded at 1661 with 1 tries\nSucceded at 1664 with 1 tries\nSucceded at 1665 with 1 tries\nSucceded at 1666 with 1 tries\nSucceded at 1668 with 1 tries\nSucceded at 1669 with 1 tries\nSucceded at 1670 with 1 tries\nSucceded at 1671 with 1 tries\nSucceded at 1675 with 1 tries\nSucceded at 1676 with 1 tries\nSucceded at 1678 with 1 tries\nSucceded at 1681 with 1 tries\nSucceded at 1684 with 1 tries\nSucceded at 1685 with 1 tries\nSucceded at 1686 with 1 tries\nSucceded at 1687 with 1 tries\nSucceded at 1688 with 1 tries\nSucceded at 1689 with 1 tries\nSucceded at 1690 with 1 tries\nSucceded at 1694 with 1 tries\nSucceded at 1695 with 1 tries\nSucceded at 1696 with 1 tries\nSucceded at 1697 with 1 tries\nSucceded at 1700 with 1 tries\nSucceded at 1702 with 1 tries\nSucceded at 1705 with 1 tries\nSucceded at 1706 with 1 tries\nSucceded at 1708 with 1 tries\nSucceded at 1709 with 1 tries\nSucceded at 1711 with 1 tries\nSucceded at 1712 with 1 tries\nSucceded at 1713 with 1 tries\nSucceded at 1714 with 1 tries\nSucceded at 1715 with 1 tries\nSucceded at 1717 with 1 tries\nSucceded at 1718 with 1 tries\nSucceded at 1719 with 1 tries\nSucceded at 1720 with 1 tries\nSucceded at 1721 with 1 tries\nSucceded at 1722 with 1 tries\nSucceded at 1723 with 1 tries\nSucceded at 1724 with 1 tries\nSucceded at 1725 with 1 tries\nSucceded at 1726 with 1 tries\nSucceded at 1727 with 1 tries\nSucceded at 1728 with 1 tries\nSucceded at 1729 with 1 tries\nSucceded at 1730 with 1 tries\nSucceded at 1731 with 1 tries\nSucceded at 1732 with 1 tries\nSucceded at 1733 with 1 tries\nSucceded at 1734 with 1 tries\nSucceded at 1736 with 1 tries\nSucceded at 1737 with 1 tries\nSucceded at 1738 with 1 tries\nSucceded at 1739 with 1 tries\nSucceded at 1740 with 1 tries\nSucceded at 1742 with 1 tries\nSucceded at 1743 with 1 tries\nSucceded at 1744 with 1 tries\nSucceded at 1745 with 1 tries\nSucceded at 1747 with 1 tries\nSucceded at 1749 with 1 tries\nSucceded at 1750 with 1 tries\nSucceded at 1752 with 1 tries\nSucceded at 1753 with 1 tries\nSucceded at 1754 with 1 tries\nSucceded at 1755 with 1 tries\nSucceded at 1760 with 1 tries\nSucceded at 1761 with 1 tries\nSucceded at 1763 with 1 tries\nSucceded at 1764 with 1 tries\nSucceded at 1766 with 1 tries\nSucceded at 1769 with 1 tries\nSucceded at 1770 with 1 tries\nSucceded at 1772 with 1 tries\nSucceded at 1774 with 1 tries\nSucceded at 1776 with 1 tries\nSucceded at 1779 with 1 tries\nSucceded at 1781 with 1 tries\nSucceded at 1782 with 1 tries\nSucceded at 1783 with 1 tries\nSucceded at 1784 with 1 tries\nSucceded at 1785 with 1 tries\nSucceded at 1786 with 1 tries\nSucceded at 1787 with 1 tries\nSucceded at 1789 with 1 tries\nSucceded at 1790 with 1 tries\nSucceded at 1791 with 1 tries\nSucceded at 1794 with 1 tries\nSucceded at 1796 with 1 tries\nSucceded at 1798 with 1 tries\nSucceded at 1799 with 1 tries\nSucceded at 1800 with 1 tries\nSucceded at 1801 with 1 tries\nSucceded at 1802 with 1 tries\nSucceded at 1803 with 1 tries\nSucceded at 1804 with 1 tries\nSucceded at 1805 with 1 tries\nSucceded at 1806 with 1 tries\nSucceded at 1808 with 1 tries\nSucceded at 1809 with 1 tries\nSucceded at 1810 with 1 tries\nSucceded at 1811 with 1 tries\nSucceded at 1813 with 1 tries\nSucceded at 1814 with 1 tries\nSucceded at 1816 with 1 tries\nSucceded at 1817 with 1 tries\nSucceded at 1818 with 1 tries\nSucceded at 1819 with 1 tries\nSucceded at 1823 with 1 tries\nSucceded at 1824 with 1 tries\nSucceded at 1825 with 1 tries\nSucceded at 1827 with 1 tries\nSucceded at 1828 with 1 tries\nSucceded at 1831 with 1 tries\nSucceded at 1833 with 1 tries\nSucceded at 1834 with 1 tries\nSucceded at 1836 with 1 tries\nSucceded at 1837 with 1 tries\nSucceded at 1840 with 1 tries\nSucceded at 1841 with 1 tries\nSucceded at 1842 with 1 tries\nSucceded at 1844 with 1 tries\nSucceded at 1846 with 1 tries\nSucceded at 1847 with 1 tries\nSucceded at 1848 with 1 tries\nSucceded at 1849 with 1 tries\nSucceded at 1850 with 1 tries\nSucceded at 1851 with 1 tries\nSucceded at 1852 with 1 tries\nSucceded at 1853 with 1 tries\nSucceded at 1854 with 1 tries\nSucceded at 1855 with 1 tries\nSucceded at 1856 with 1 tries\nSucceded at 1857 with 1 tries\nSucceded at 1859 with 1 tries\nSucceded at 1861 with 1 tries\nSucceded at 1862 with 1 tries\nSucceded at 1864 with 1 tries\nSucceded at 1865 with 1 tries\nSucceded at 1866 with 1 tries\nSucceded at 1868 with 1 tries\nSucceded at 1870 with 1 tries\nSucceded at 1871 with 1 tries\nSucceded at 1872 with 1 tries\nSucceded at 1873 with 1 tries\nSucceded at 1874 with 1 tries\nSucceded at 1875 with 1 tries\nSucceded at 1876 with 1 tries\nSucceded at 1877 with 1 tries\nSucceded at 1878 with 1 tries\nSucceded at 1879 with 1 tries\nSucceded at 1881 with 1 tries\nSucceded at 1882 with 1 tries\nSucceded at 1883 with 1 tries\nSucceded at 1885 with 1 tries\nSucceded at 1887 with 1 tries\nSucceded at 1888 with 1 tries\nSucceded at 1889 with 1 tries\nSucceded at 1890 with 1 tries\nSucceded at 1892 with 1 tries\nSucceded at 1893 with 1 tries\nSucceded at 1895 with 1 tries\nSucceded at 1896 with 1 tries\nmosek failed at \nSucceded at 1902 with 1 tries\nSucceded at 1904 with 1 tries\nSucceded at 1905 with 1 tries\nSucceded at 1907 with 1 tries\nSucceded at 1908 with 1 tries\nSucceded at 1910 with 1 tries\nSucceded at 1911 with 1 tries\nSucceded at 1912 with 1 tries\nSucceded at 1913 with 1 tries\nSucceded at 1914 with 1 tries\nSucceded at 1915 with 1 tries\nSucceded at 1916 with 1 tries\nSucceded at 1918 with 1 tries\nSucceded at 1919 with 1 tries\nSucceded at 1921 with 1 tries\nSucceded at 1922 with 1 tries\nSucceded at 1925 with 1 tries\nSucceded at 1926 with 1 tries\nSucceded at 1927 with 1 tries\nSucceded at 1928 with 1 tries\nSucceded at 1929 with 1 tries\nSucceded at 1930 with 1 tries\nSucceded at 1931 with 1 tries\nSucceded at 1933 with 1 tries\nSucceded at 1934 with 1 tries\nSucceded at 1935 with 1 tries\nSucceded at 1937 with 1 tries\nSucceded at 1939 with 1 tries\nSucceded at 1942 with 1 tries\nSucceded at 1943 with 1 tries\nSucceded at 1944 with 1 tries\nSucceded at 1949 with 1 tries\nSucceded at 1950 with 1 tries\nSucceded at 1952 with 1 tries\nSucceded at 1953 with 1 tries\nSucceded at 1955 with 1 tries\nSucceded at 1956 with 1 tries\nSucceded at 1958 with 1 tries\nSucceded at 1959 with 1 tries\nSucceded at 1960 with 1 tries\nSucceded at 1962 with 1 tries\nSucceded at 1966 with 1 tries\nSucceded at 1968 with 1 tries\nSucceded at 1969 with 1 tries\nSucceded at 1971 with 1 tries\nSucceded at 1972 with 1 tries\nSucceded at 1973 with 1 tries\nSucceded at 1974 with 1 tries\nSucceded at 1975 with 1 tries\nSucceded at 1978 with 1 tries\nSucceded at 1979 with 1 tries\nSucceded at 1980 with 1 tries\nSucceded at 1981 with 1 tries\nSucceded at 1982 with 1 tries\nSucceded at 1984 with 1 tries\nSucceded at 1985 with 1 tries\nSucceded at 1986 with 1 tries\nSucceded at 1990 with 1 tries\nSucceded at 1992 with 1 tries\nSucceded at 1993 with 1 tries\nSucceded at 1995 with 1 tries\nSucceded at 1997 with 1 tries\nSucceded at 1999 with 1 tries\nSucceded at 2000 with 1 tries\nSucceded at 2001 with 1 tries\nSucceded at 2002 with 1 tries\nSucceded at 2003 with 1 tries\nSucceded at 2004 with 1 tries\nSucceded at 2005 with 1 tries\nSucceded at 2006 with 1 tries\nSucceded at 2007 with 1 tries\nSucceded at 2008 with 1 tries\nSucceded at 2010 with 1 tries\nSucceded at 2013 with 1 tries\nSucceded at 2014 with 1 tries\nSucceded at 2016 with 1 tries\nSucceded at 2017 with 1 tries\nSucceded at 2018 with 1 tries\nSucceded at 2020 with 1 tries\nSucceded at 2022 with 1 tries\nSucceded at 2023 with 1 tries\nSucceded at 2025 with 1 tries\nSucceded at 2027 with 1 tries\nSucceded at 2028 with 1 tries\nSucceded at 2030 with 1 tries\nSucceded at 2031 with 1 tries\nSucceded at 2032 with 1 tries\nSucceded at 2033 with 1 tries\nSucceded at 2034 with 1 tries\nSucceded at 2035 with 1 tries\nSucceded at 2037 with 1 tries\nSucceded at 2038 with 1 tries\nSucceded at 2039 with 1 tries\nSucceded at 2042 with 1 tries\nSucceded at 2046 with 1 tries\nSucceded at 2047 with 1 tries\nSucceded at 2048 with 1 tries\nSucceded at 2050 with 1 tries\nSucceded at 2051 with 1 tries\nSucceded at 2056 with 1 tries\nSucceded at 2057 with 1 tries\nSucceded at 2059 with 1 tries\nSucceded at 2060 with 1 tries\nSucceded at 2061 with 1 tries\nSucceded at 2062 with 1 tries\nSucceded at 2063 with 1 tries\nSucceded at 2065 with 1 tries\nSucceded at 2066 with 1 tries\nSucceded at 2067 with 1 tries\nSucceded at 2070 with 1 tries\nSucceded at 2072 with 1 tries\nSucceded at 2074 with 1 tries\nSucceded at 2075 with 1 tries\nSucceded at 2076 with 1 tries\nSucceded at 2077 with 1 tries\nSucceded at 2079 with 1 tries\nSucceded at 2080 with 1 tries\nSucceded at 2081 with 1 tries\nSucceded at 2083 with 1 tries\nSucceded at 2084 with 1 tries\nSucceded at 2085 with 1 tries\nSucceded at 2087 with 1 tries\nSucceded at 2088 with 1 tries\nSucceded at 2089 with 1 tries\nSucceded at 2091 with 1 tries\nSucceded at 2092 with 1 tries\nSucceded at 2093 with 1 tries\nSucceded at 2095 with 1 tries\nSucceded at 2096 with 1 tries\nSucceded at 2097 with 1 tries\nSucceded at 2098 with 1 tries\nSucceded at 2099 with 1 tries\nSucceded at 2100 with 1 tries\nSucceded at 2101 with 1 tries\nSucceded at 2102 with 1 tries\nSucceded at 2103 with 1 tries\nSucceded at 2104 with 1 tries\nSucceded at 2105 with 1 tries\nSucceded at 2106 with 1 tries\nSucceded at 2108 with 1 tries\nSucceded at 2109 with 1 tries\nSucceded at 2110 with 1 tries\nSucceded at 2114 with 1 tries\nSucceded at 2115 with 1 tries\nSucceded at 2117 with 1 tries\nSucceded at 2118 with 1 tries\nSucceded at 2120 with 1 tries\nSucceded at 2121 with 1 tries\nSucceded at 2122 with 1 tries\nSucceded at 2123 with 1 tries\nSucceded at 2124 with 1 tries\nSucceded at 2125 with 1 tries\nSucceded at 2126 with 1 tries\nSucceded at 2129 with 1 tries\nSucceded at 2130 with 1 tries\nSucceded at 2131 with 1 tries\nSucceded at 2132 with 1 tries\nSucceded at 2133 with 1 tries\nSucceded at 2135 with 1 tries\nSucceded at 2136 with 1 tries\nSucceded at 2137 with 1 tries\nSucceded at 2138 with 1 tries\nSucceded at 2139 with 1 tries\nSucceded at 2140 with 1 tries\nSucceded at 2141 with 1 tries\nSucceded at 2142 with 1 tries\nSucceded at 2144 with 1 tries\nSucceded at 2146 with 1 tries\nSucceded at 2147 with 1 tries\nSucceded at 2148 with 1 tries\nSucceded at 2149 with 1 tries\nSucceded at 2153 with 1 tries\nSucceded at 2154 with 1 tries\nSucceded at 2156 with 1 tries\nSucceded at 2157 with 1 tries\nSucceded at 2158 with 1 tries\nSucceded at 2159 with 1 tries\nSucceded at 2160 with 1 tries\nSucceded at 2161 with 1 tries\nSucceded at 2163 with 1 tries\nSucceded at 2164 with 1 tries\nSucceded at 2165 with 1 tries\nSucceded at 2166 with 1 tries\nSucceded at 2167 with 1 tries\nSucceded at 2168 with 1 tries\nSucceded at 2172 with 1 tries\nSucceded at 2173 with 1 tries\nSucceded at 2174 with 1 tries\nSucceded at 2175 with 1 tries\nSucceded at 2176 with 1 tries\nmosek failed at \nSucceded at 2181 with 1 tries\nSucceded at 2182 with 1 tries\nSucceded at 2183 with 1 tries\nSucceded at 2184 with 1 tries\nSucceded at 2185 with 1 tries\nSucceded at 2188 with 1 tries\nSucceded at 2189 with 1 tries\nSucceded at 2191 with 1 tries\nSucceded at 2193 with 1 tries\nSucceded at 2195 with 1 tries\nSucceded at 2196 with 1 tries\nSucceded at 2197 with 1 tries\nSucceded at 2199 with 1 tries\nSucceded at 2200 with 1 tries\nSucceded at 2201 with 1 tries\nSucceded at 2202 with 1 tries\nSucceded at 2203 with 1 tries\nSucceded at 2204 with 1 tries\nSucceded at 2207 with 1 tries\nSucceded at 2208 with 1 tries\nSucceded at 2209 with 1 tries\nSucceded at 2211 with 1 tries\nSucceded at 2212 with 1 tries\nSucceded at 2213 with 1 tries\nSucceded at 2217 with 1 tries\nSucceded at 2219 with 1 tries\nSucceded at 2220 with 1 tries\nSucceded at 2221 with 1 tries\nSucceded at 2222 with 1 tries\nSucceded at 2223 with 1 tries\nSucceded at 2224 with 1 tries\nSucceded at 2225 with 1 tries\nSucceded at 2227 with 1 tries\nSucceded at 2230 with 1 tries\nSucceded at 2232 with 1 tries\nSucceded at 2233 with 1 tries\nSucceded at 2234 with 1 tries\nSucceded at 2235 with 1 tries\nSucceded at 2237 with 1 tries\nSucceded at 2238 with 1 tries\nSucceded at 2239 with 1 tries\nSucceded at 2241 with 1 tries\nSucceded at 2242 with 1 tries\nSucceded at 2243 with 1 tries\nSucceded at 2244 with 1 tries\nSucceded at 2246 with 1 tries\nSucceded at 2248 with 1 tries\nSucceded at 2249 with 1 tries\nSucceded at 2250 with 1 tries\nSucceded at 2251 with 1 tries\nSucceded at 2252 with 1 tries\nSucceded at 2253 with 1 tries\nSucceded at 2254 with 1 tries\nSucceded at 2255 with 1 tries\nSucceded at 2256 with 1 tries\nSucceded at 2258 with 1 tries\nSucceded at 2259 with 1 tries\nSucceded at 2261 with 1 tries\nSucceded at 2263 with 1 tries\nSucceded at 2264 with 1 tries\nSucceded at 2265 with 1 tries\nSucceded at 2267 with 1 tries\nSucceded at 2268 with 1 tries\nSucceded at 2272 with 1 tries\nSucceded at 2273 with 1 tries\nSucceded at 2274 with 1 tries\nSucceded at 2276 with 1 tries\nSucceded at 2277 with 1 tries\nSucceded at 2280 with 1 tries\nSucceded at 2281 with 1 tries\nSucceded at 2282 with 1 tries\nSucceded at 2284 with 1 tries\nSucceded at 2285 with 1 tries\nSucceded at 2286 with 1 tries\nSucceded at 2287 with 1 tries\nSucceded at 2290 with 1 tries\nSucceded at 2291 with 1 tries\nSucceded at 2293 with 1 tries\nSucceded at 2297 with 1 tries\nSucceded at 2298 with 1 tries\nSucceded at 2299 with 1 tries\nSucceded at 2300 with 1 tries\nSucceded at 2304 with 1 tries\nSucceded at 2305 with 1 tries\nSucceded at 2306 with 1 tries\nSucceded at 2307 with 1 tries\nSucceded at 2308 with 1 tries\nSucceded at 2309 with 1 tries\nSucceded at 2310 with 1 tries\nSucceded at 2311 with 1 tries\nSucceded at 2312 with 1 tries\nSucceded at 2315 with 1 tries\nSucceded at 2316 with 1 tries\nSucceded at 2317 with 1 tries\nSucceded at 2319 with 1 tries\nSucceded at 2321 with 1 tries\nSucceded at 2322 with 1 tries\nSucceded at 2323 with 1 tries\nSucceded at 2325 with 1 tries\nSucceded at 2326 with 1 tries\nSucceded at 2327 with 1 tries\nSucceded at 2328 with 1 tries\nSucceded at 2329 with 1 tries\nSucceded at 2330 with 1 tries\nSucceded at 2331 with 1 tries\nSucceded at 2332 with 1 tries\nSucceded at 2336 with 1 tries\nSucceded at 2337 with 1 tries\nSucceded at 2338 with 1 tries\nSucceded at 2339 with 1 tries\nSucceded at 2340 with 1 tries\nSucceded at 2341 with 1 tries\nSucceded at 2342 with 1 tries\nSucceded at 2345 with 1 tries\nSucceded at 2347 with 1 tries\nSucceded at 2350 with 1 tries\nSucceded at 2352 with 1 tries\nSucceded at 2356 with 1 tries\nSucceded at 2358 with 1 tries\nSucceded at 2361 with 1 tries\nSucceded at 2362 with 1 tries\nSucceded at 2363 with 1 tries\nSucceded at 2364 with 1 tries\nSucceded at 2366 with 1 tries\nSucceded at 2368 with 1 tries\nSucceded at 2369 with 1 tries\nSucceded at 2372 with 1 tries\nSucceded at 2373 with 1 tries\nSucceded at 2376 with 1 tries\nSucceded at 2377 with 1 tries\nSucceded at 2378 with 1 tries\nSucceded at 2379 with 1 tries\nSucceded at 2380 with 1 tries\nSucceded at 2381 with 1 tries\nSucceded at 2382 with 1 tries\nSucceded at 2383 with 1 tries\nSucceded at 2384 with 1 tries\nSucceded at 2386 with 1 tries\nSucceded at 2387 with 1 tries\nSucceded at 2388 with 1 tries\nSucceded at 2389 with 1 tries\nSucceded at 2390 with 1 tries\nSucceded at 2391 with 1 tries\nSucceded at 2392 with 1 tries\nSucceded at 2394 with 1 tries\nSucceded at 2395 with 1 tries\nSucceded at 2396 with 1 tries\nSucceded at 2397 with 1 tries\nSucceded at 2398 with 1 tries\nSucceded at 2399 with 1 tries\nSucceded at 2400 with 1 tries\nSucceded at 2403 with 1 tries\nSucceded at 2404 with 1 tries\nSucceded at 2405 with 1 tries\nSucceded at 2406 with 1 tries\nSucceded at 2407 with 1 tries\nSucceded at 2408 with 1 tries\nSucceded at 2409 with 1 tries\nmosek failed at \nSucceded at 2412 with 1 tries\nSucceded at 2413 with 1 tries\nSucceded at 2414 with 1 tries\nSucceded at 2415 with 1 tries\nSucceded at 2416 with 1 tries\nSucceded at 2417 with 1 tries\nSucceded at 2419 with 1 tries\nSucceded at 2420 with 1 tries\nSucceded at 2421 with 1 tries\nSucceded at 2422 with 1 tries\nSucceded at 2423 with 1 tries\nSucceded at 2424 with 1 tries\nSucceded at 2426 with 1 tries\nSucceded at 2427 with 1 tries\nSucceded at 2428 with 1 tries\nSucceded at 2431 with 1 tries\nSucceded at 2432 with 1 tries\nSucceded at 2433 with 1 tries\nSucceded at 2436 with 1 tries\nSucceded at 2438 with 1 tries\nSucceded at 2441 with 1 tries\nSucceded at 2442 with 1 tries\nSucceded at 2444 with 1 tries\nSucceded at 2446 with 1 tries\nSucceded at 2448 with 1 tries\nSucceded at 2451 with 1 tries\nSucceded at 2452 with 1 tries\nSucceded at 2454 with 1 tries\nSucceded at 2455 with 1 tries\nSucceded at 2456 with 1 tries\nSucceded at 2458 with 1 tries\nSucceded at 2459 with 1 tries\nSucceded at 2462 with 1 tries\nSucceded at 2463 with 1 tries\nSucceded at 2465 with 1 tries\nSucceded at 2466 with 1 tries\nSucceded at 2468 with 1 tries\nSucceded at 2469 with 1 tries\nSucceded at 2470 with 1 tries\nSucceded at 2471 with 1 tries\nSucceded at 2473 with 1 tries\nSucceded at 2474 with 1 tries\nSucceded at 2475 with 1 tries\nSucceded at 2476 with 1 tries\nSucceded at 2477 with 1 tries\nSucceded at 2478 with 1 tries\nSucceded at 2479 with 1 tries\nSucceded at 2480 with 1 tries\nSucceded at 2481 with 1 tries\nSucceded at 2483 with 1 tries\nSucceded at 2485 with 1 tries\nSucceded at 2486 with 1 tries\nSucceded at 2487 with 1 tries\nSucceded at 2488 with 1 tries\nSucceded at 2489 with 1 tries\nSucceded at 2490 with 1 tries\nSucceded at 2491 with 1 tries\nSucceded at 2493 with 1 tries\nSucceded at 2494 with 1 tries\nSucceded at 2496 with 1 tries\nSucceded at 2498 with 1 tries\nSucceded at 2499 with 1 tries\nSucceded at 2501 with 1 tries\nSucceded at 2502 with 1 tries\nSucceded at 2503 with 1 tries\nSucceded at 2505 with 1 tries\nSucceded at 2506 with 1 tries\nSucceded at 2507 with 1 tries\nSucceded at 2508 with 1 tries\nSucceded at 2510 with 1 tries\nSucceded at 2511 with 1 tries\nSucceded at 2513 with 1 tries\nSucceded at 2516 with 1 tries\nSucceded at 2517 with 1 tries\nSucceded at 2521 with 1 tries\nSucceded at 2522 with 1 tries\nSucceded at 2523 with 1 tries\nSucceded at 2524 with 1 tries\nSucceded at 2525 with 1 tries\nSucceded at 2526 with 1 tries\nSucceded at 2527 with 1 tries\nSucceded at 2528 with 1 tries\nSucceded at 2529 with 1 tries\nSucceded at 2530 with 1 tries\nSucceded at 2533 with 1 tries\nSucceded at 2535 with 1 tries\nSucceded at 2536 with 1 tries\nSucceded at 2538 with 1 tries\nSucceded at 2539 with 1 tries\nSucceded at 2540 with 1 tries\nSucceded at 2541 with 1 tries\nSucceded at 2542 with 1 tries\nSucceded at 2544 with 1 tries\nSucceded at 2547 with 1 tries\nSucceded at 2548 with 1 tries\nSucceded at 2550 with 1 tries\nSucceded at 2553 with 1 tries\nSucceded at 2554 with 1 tries\nSucceded at 2556 with 1 tries\nSucceded at 2557 with 1 tries\nSucceded at 2560 with 1 tries\nSucceded at 2561 with 1 tries\nSucceded at 2562 with 1 tries\nSucceded at 2566 with 1 tries\nSucceded at 2568 with 1 tries\nSucceded at 2569 with 1 tries\nSucceded at 2571 with 1 tries\nSucceded at 2572 with 1 tries\nSucceded at 2574 with 1 tries\nSucceded at 2575 with 1 tries\nSucceded at 2577 with 1 tries\nSucceded at 2580 with 1 tries\nSucceded at 2581 with 1 tries\nSucceded at 2584 with 1 tries\nSucceded at 2585 with 1 tries\nSucceded at 2587 with 1 tries\nSucceded at 2588 with 1 tries\nSucceded at 2589 with 1 tries\nSucceded at 2590 with 1 tries\nSucceded at 2591 with 1 tries\nSucceded at 2592 with 1 tries\nSucceded at 2594 with 1 tries\nSucceded at 2595 with 1 tries\nSucceded at 2596 with 1 tries\nSucceded at 2597 with 1 tries\nSucceded at 2598 with 1 tries\nSucceded at 2599 with 1 tries\nSucceded at 2600 with 1 tries\nSucceded at 2601 with 1 tries\nSucceded at 2603 with 1 tries\nSucceded at 2604 with 1 tries\nSucceded at 2605 with 1 tries\nSucceded at 2608 with 1 tries\nSucceded at 2609 with 1 tries\nSucceded at 2610 with 1 tries\nSucceded at 2611 with 1 tries\nSucceded at 2612 with 1 tries\nSucceded at 2613 with 1 tries\nSucceded at 2614 with 1 tries\nSucceded at 2615 with 1 tries\nSucceded at 2617 with 1 tries\nSucceded at 2618 with 1 tries\nSucceded at 2619 with 1 tries\nSucceded at 2620 with 1 tries\nSucceded at 2621 with 1 tries\nSucceded at 2622 with 1 tries\nSucceded at 2623 with 1 tries\nSucceded at 2624 with 1 tries\nSucceded at 2625 with 1 tries\nSucceded at 2630 with 1 tries\nSucceded at 2632 with 1 tries\nSucceded at 2633 with 1 tries\nSucceded at 2634 with 1 tries\nSucceded at 2635 with 1 tries\nSucceded at 2637 with 1 tries\nSucceded at 2638 with 1 tries\nSucceded at 2640 with 1 tries\nSucceded at 2641 with 1 tries\nSucceded at 2642 with 1 tries\nSucceded at 2643 with 1 tries\nSucceded at 2644 with 1 tries\nSucceded at 2645 with 1 tries\nSucceded at 2646 with 1 tries\nSucceded at 2647 with 1 tries\nSucceded at 2648 with 1 tries\nSucceded at 2650 with 1 tries\nSucceded at 2651 with 1 tries\nSucceded at 2653 with 1 tries\nSucceded at 2654 with 1 tries\nSucceded at 2655 with 1 tries\nSucceded at 2656 with 1 tries\nSucceded at 2657 with 1 tries\nSucceded at 2658 with 1 tries\nSucceded at 2659 with 1 tries\nSucceded at 2661 with 1 tries\nSucceded at 2663 with 1 tries\nSucceded at 2664 with 1 tries\nSucceded at 2668 with 1 tries\nSucceded at 2671 with 1 tries\nSucceded at 2674 with 1 tries\nSucceded at 2675 with 1 tries\nSucceded at 2676 with 1 tries\nSucceded at 2678 with 1 tries\nSucceded at 2679 with 1 tries\nSucceded at 2680 with 1 tries\nSucceded at 2681 with 1 tries\nSucceded at 2682 with 1 tries\nSucceded at 2684 with 1 tries\nSucceded at 2686 with 1 tries\nSucceded at 2688 with 1 tries\nSucceded at 2689 with 1 tries\nSucceded at 2692 with 1 tries\nSucceded at 2694 with 1 tries\nSucceded at 2695 with 1 tries\nSucceded at 2697 with 1 tries\nSucceded at 2698 with 1 tries\nSucceded at 2699 with 1 tries\nSucceded at 2700 with 1 tries\nSucceded at 2701 with 1 tries\nSucceded at 2704 with 1 tries\nSucceded at 2707 with 1 tries\nSucceded at 2711 with 1 tries\nSucceded at 2713 with 1 tries\nSucceded at 2714 with 1 tries\nSucceded at 2716 with 1 tries\nSucceded at 2718 with 1 tries\nSucceded at 2719 with 1 tries\nSucceded at 2720 with 1 tries\nSucceded at 2721 with 1 tries\nSucceded at 2722 with 1 tries\nSucceded at 2725 with 1 tries\nSucceded at 2726 with 1 tries\nSucceded at 2727 with 1 tries\nSucceded at 2728 with 1 tries\nSucceded at 2729 with 1 tries\nSucceded at 2731 with 1 tries\nSucceded at 2732 with 1 tries\nSucceded at 2736 with 1 tries\nSucceded at 2737 with 1 tries\nSucceded at 2738 with 1 tries\nSucceded at 2739 with 1 tries\nSucceded at 2740 with 1 tries\nSucceded at 2741 with 1 tries\nSucceded at 2744 with 1 tries\nSucceded at 2745 with 1 tries\nSucceded at 2746 with 1 tries\nSucceded at 2747 with 1 tries\nSucceded at 2748 with 1 tries\nSucceded at 2753 with 1 tries\nSucceded at 2755 with 1 tries\nSucceded at 2756 with 1 tries\nSucceded at 2757 with 1 tries\nSucceded at 2759 with 1 tries\nSucceded at 2761 with 1 tries\nSucceded at 2762 with 1 tries\nSucceded at 2763 with 1 tries\nSucceded at 2764 with 1 tries\nSucceded at 2767 with 1 tries\nSucceded at 2768 with 1 tries\nSucceded at 2770 with 1 tries\nSucceded at 2771 with 1 tries\nSucceded at 2774 with 1 tries\nSucceded at 2776 with 1 tries\nSucceded at 2777 with 1 tries\nSucceded at 2778 with 1 tries\nSucceded at 2779 with 1 tries\nSucceded at 2780 with 1 tries\nSucceded at 2781 with 1 tries\nSucceded at 2782 with 1 tries\nSucceded at 2785 with 1 tries\nSucceded at 2787 with 1 tries\nSucceded at 2788 with 1 tries\nSucceded at 2790 with 1 tries\nSucceded at 2792 with 1 tries\nSucceded at 2793 with 1 tries\nSucceded at 2796 with 1 tries\nSucceded at 2798 with 1 tries\nSucceded at 2799 with 1 tries\nSucceded at 2802 with 1 tries\nSucceded at 2803 with 1 tries\nSucceded at 2805 with 1 tries\nSucceded at 2806 with 1 tries\nSucceded at 2808 with 1 tries\nSucceded at 2809 with 1 tries\nSucceded at 2812 with 1 tries\nSucceded at 2813 with 1 tries\nSucceded at 2815 with 1 tries\nSucceded at 2816 with 1 tries\nSucceded at 2817 with 1 tries\nSucceded at 2818 with 1 tries\nSucceded at 2820 with 1 tries\nSucceded at 2821 with 1 tries\nSucceded at 2822 with 1 tries\nSucceded at 2823 with 1 tries\nSucceded at 2824 with 1 tries\nSucceded at 2826 with 1 tries\nSucceded at 2828 with 1 tries\nSucceded at 2829 with 1 tries\nSucceded at 2830 with 1 tries\nSucceded at 2831 with 1 tries\nSucceded at 2832 with 1 tries\nSucceded at 2833 with 1 tries\nSucceded at 2834 with 1 tries\nSucceded at 2835 with 1 tries\nSucceded at 2840 with 1 tries\nSucceded at 2843 with 1 tries\nSucceded at 2844 with 1 tries\nSucceded at 2845 with 1 tries\nSucceded at 2849 with 1 tries\nSucceded at 2850 with 1 tries\nSucceded at 2856 with 1 tries\nSucceded at 2857 with 1 tries\nSucceded at 2858 with 1 tries\nSucceded at 2860 with 1 tries\nSucceded at 2861 with 1 tries\nSucceded at 2862 with 1 tries\nSucceded at 2864 with 1 tries\nSucceded at 2865 with 1 tries\nSucceded at 2868 with 1 tries\nSucceded at 2869 with 1 tries\nSucceded at 2870 with 1 tries\nSucceded at 2871 with 1 tries\nSucceded at 2872 with 1 tries\nSucceded at 2873 with 1 tries\nSucceded at 2874 with 1 tries\nSucceded at 2877 with 1 tries\nSucceded at 2878 with 1 tries\nSucceded at 2879 with 1 tries\nSucceded at 2880 with 1 tries\nSucceded at 2881 with 1 tries\nSucceded at 2882 with 1 tries\nSucceded at 2883 with 1 tries\nSucceded at 2884 with 1 tries\nSucceded at 2886 with 1 tries\nSucceded at 2887 with 1 tries\nSucceded at 2889 with 1 tries\nSucceded at 2890 with 1 tries\nSucceded at 2891 with 1 tries\nSucceded at 2892 with 1 tries\nSucceded at 2893 with 1 tries\nSucceded at 2895 with 1 tries\nSucceded at 2897 with 1 tries\nSucceded at 2898 with 1 tries\nSucceded at 2899 with 1 tries\nSucceded at 2900 with 1 tries\nSucceded at 2901 with 1 tries\nSucceded at 2903 with 1 tries\nSucceded at 2908 with 1 tries\nSucceded at 2909 with 1 tries\nSucceded at 2911 with 1 tries\nSucceded at 2913 with 1 tries\nSucceded at 2915 with 1 tries\nSucceded at 2916 with 1 tries\nSucceded at 2917 with 1 tries\nSucceded at 2918 with 1 tries\nSucceded at 2919 with 1 tries\nSucceded at 2920 with 1 tries\nSucceded at 2921 with 1 tries\nSucceded at 2922 with 1 tries\nSucceded at 2924 with 1 tries\nSucceded at 2927 with 1 tries\nSucceded at 2928 with 1 tries\nSucceded at 2929 with 1 tries\nSucceded at 2930 with 1 tries\nSucceded at 2932 with 1 tries\nSucceded at 2933 with 1 tries\nSucceded at 2934 with 1 tries\nSucceded at 2936 with 1 tries\nSucceded at 2937 with 1 tries\nSucceded at 2938 with 1 tries\nSucceded at 2939 with 1 tries\nSucceded at 2941 with 1 tries\nSucceded at 2942 with 1 tries\nSucceded at 2943 with 1 tries\nSucceded at 2944 with 1 tries\nSucceded at 2945 with 1 tries\nSucceded at 2946 with 1 tries\nSucceded at 2947 with 1 tries\nSucceded at 2949 with 1 tries\nSucceded at 2950 with 1 tries\nSucceded at 2951 with 1 tries\nSucceded at 2952 with 1 tries\nSucceded at 2954 with 1 tries\nSucceded at 2955 with 1 tries\nSucceded at 2956 with 1 tries\nSucceded at 2958 with 1 tries\nSucceded at 2959 with 1 tries\nSucceded at 2961 with 1 tries\nSucceded at 2963 with 1 tries\nSucceded at 2965 with 1 tries\nSucceded at 2966 with 1 tries\nSucceded at 2967 with 1 tries\nSucceded at 2968 with 1 tries\nSucceded at 2969 with 1 tries\nSucceded at 2970 with 1 tries\nSucceded at 2971 with 1 tries\nSucceded at 2972 with 1 tries\nSucceded at 2973 with 1 tries\nSucceded at 2974 with 1 tries\nSucceded at 2975 with 1 tries\nSucceded at 2976 with 1 tries\nSucceded at 2977 with 1 tries\nSucceded at 2978 with 1 tries\nSucceded at 2980 with 1 tries\nSucceded at 2981 with 1 tries\nSucceded at 2982 with 1 tries\nSucceded at 2984 with 1 tries\nSucceded at 2986 with 1 tries\nSucceded at 2987 with 1 tries\nSucceded at 2988 with 1 tries\nSucceded at 2989 with 1 tries\nSucceded at 2990 with 1 tries\nSucceded at 2991 with 1 tries\nSucceded at 2992 with 1 tries\nSucceded at 2993 with 1 tries\nSucceded at 2994 with 1 tries\nSucceded at 2995 with 1 tries\nSucceded at 2996 with 1 tries\nSucceded at 2999 with 1 tries\nSucceded at 3000 with 1 tries\nSucceded at 3001 with 1 tries\nSucceded at 3002 with 1 tries\nSucceded at 3003 with 1 tries\nSucceded at 3004 with 1 tries\nSucceded at 3005 with 1 tries\nSucceded at 3007 with 1 tries\nSucceded at 3009 with 1 tries\nSucceded at 3010 with 1 tries\nSucceded at 3011 with 1 tries\nSucceded at 3014 with 1 tries\nSucceded at 3015 with 1 tries\nSucceded at 3016 with 1 tries\nSucceded at 3017 with 1 tries\nSucceded at 3018 with 1 tries\nSucceded at 3021 with 1 tries\nSucceded at 3022 with 1 tries\nSucceded at 3023 with 1 tries\nSucceded at 3024 with 1 tries\nSucceded at 3025 with 1 tries\nSucceded at 3026 with 1 tries\nSucceded at 3030 with 1 tries\nSucceded at 3031 with 1 tries\nSucceded at 3033 with 1 tries\nSucceded at 3035 with 1 tries\nSucceded at 3036 with 1 tries\nSucceded at 3037 with 1 tries\nSucceded at 3038 with 1 tries\nSucceded at 3040 with 1 tries\nSucceded at 3042 with 1 tries\nSucceded at 3043 with 1 tries\nSucceded at 3045 with 1 tries\nSucceded at 3047 with 1 tries\nSucceded at 3048 with 1 tries\nSucceded at 3049 with 1 tries\nSucceded at 3051 with 1 tries\nSucceded at 3052 with 1 tries\nSucceded at 3053 with 1 tries\nSucceded at 3054 with 1 tries\nSucceded at 3055 with 1 tries\nSucceded at 3056 with 1 tries\nSucceded at 3057 with 1 tries\nSucceded at 3059 with 1 tries\nSucceded at 3060 with 1 tries\nSucceded at 3062 with 1 tries\nSucceded at 3063 with 1 tries\nSucceded at 3064 with 1 tries\nSucceded at 3066 with 1 tries\nSucceded at 3067 with 1 tries\nSucceded at 3069 with 1 tries\nSucceded at 3070 with 1 tries\nSucceded at 3071 with 1 tries\nSucceded at 3072 with 1 tries\nSucceded at 3073 with 1 tries\nSucceded at 3076 with 1 tries\nSucceded at 3077 with 1 tries\nSucceded at 3078 with 1 tries\nSucceded at 3079 with 1 tries\nSucceded at 3080 with 1 tries\nSucceded at 3081 with 1 tries\nSucceded at 3082 with 1 tries\nSucceded at 3084 with 1 tries\nSucceded at 3087 with 1 tries\nSucceded at 3088 with 1 tries\nSucceded at 3089 with 1 tries\nSucceded at 3090 with 1 tries\nSucceded at 3091 with 1 tries\nSucceded at 3092 with 1 tries\nSucceded at 3093 with 1 tries\nSucceded at 3094 with 1 tries\nSucceded at 3096 with 1 tries\nSucceded at 3097 with 1 tries\nSucceded at 3099 with 1 tries\nSucceded at 3100 with 1 tries\nSucceded at 3102 with 1 tries\nSucceded at 3103 with 1 tries\nSucceded at 3104 with 1 tries\nSucceded at 3105 with 1 tries\nSucceded at 3107 with 1 tries\nSucceded at 3108 with 1 tries\nSucceded at 3109 with 1 tries\nSucceded at 3110 with 1 tries\nSucceded at 3111 with 1 tries\nSucceded at 3112 with 1 tries\nSucceded at 3116 with 1 tries\nSucceded at 3118 with 1 tries\nSucceded at 3119 with 1 tries\nSucceded at 3120 with 1 tries\nSucceded at 3123 with 1 tries\nSucceded at 3124 with 1 tries\nSucceded at 3127 with 1 tries\nSucceded at 3128 with 1 tries\nSucceded at 3129 with 1 tries\nSucceded at 3130 with 1 tries\nSucceded at 3131 with 1 tries\nSucceded at 3132 with 1 tries\nSucceded at 3133 with 1 tries\nSucceded at 3135 with 1 tries\nSucceded at 3136 with 1 tries\nSucceded at 3137 with 1 tries\nSucceded at 3140 with 1 tries\nSucceded at 3141 with 1 tries\nSucceded at 3143 with 1 tries\nSucceded at 3144 with 1 tries\nSucceded at 3145 with 1 tries\nSucceded at 3146 with 1 tries\nSucceded at 3147 with 1 tries\nSucceded at 3148 with 1 tries\nSucceded at 3149 with 1 tries\nSucceded at 3150 with 1 tries\nSucceded at 3151 with 1 tries\nSucceded at 3152 with 1 tries\nSucceded at 3153 with 1 tries\nSucceded at 3154 with 1 tries\nSucceded at 3156 with 1 tries\nSucceded at 3157 with 1 tries\nSucceded at 3158 with 1 tries\nSucceded at 3160 with 1 tries\nSucceded at 3161 with 1 tries\nSucceded at 3162 with 1 tries\nSucceded at 3163 with 1 tries\nSucceded at 3165 with 1 tries\nSucceded at 3166 with 1 tries\nSucceded at 3167 with 1 tries\nSucceded at 3168 with 1 tries\nSucceded at 3170 with 1 tries\nSucceded at 3171 with 1 tries\nSucceded at 3172 with 1 tries\nSucceded at 3173 with 1 tries\nSucceded at 3176 with 1 tries\nSucceded at 3178 with 1 tries\nSucceded at 3180 with 1 tries\nSucceded at 3181 with 1 tries\nSucceded at 3182 with 1 tries\nSucceded at 3183 with 1 tries\nSucceded at 3184 with 1 tries\nSucceded at 3186 with 1 tries\nSucceded at 3187 with 1 tries\nSucceded at 3188 with 1 tries\nSucceded at 3190 with 1 tries\nSucceded at 3191 with 1 tries\nSucceded at 3193 with 1 tries\nSucceded at 3194 with 1 tries\nSucceded at 3195 with 1 tries\nSucceded at 3196 with 1 tries\nSucceded at 3197 with 1 tries\nSucceded at 3198 with 1 tries\nSucceded at 3199 with 1 tries\nSucceded at 3200 with 1 tries\nSucceded at 3201 with 1 tries\nSucceded at 3202 with 1 tries\nSucceded at 3203 with 1 tries\nSucceded at 3204 with 1 tries\nSucceded at 3205 with 1 tries\nSucceded at 3207 with 1 tries\nSucceded at 3209 with 1 tries\nSucceded at 3211 with 1 tries\nSucceded at 3212 with 1 tries\nSucceded at 3213 with 1 tries\nSucceded at 3214 with 1 tries\nSucceded at 3215 with 1 tries\nSucceded at 3217 with 1 tries\nSucceded at 3219 with 1 tries\nSucceded at 3222 with 1 tries\nSucceded at 3225 with 1 tries\nSucceded at 3229 with 1 tries\nSucceded at 3230 with 1 tries\nSucceded at 3231 with 1 tries\nSucceded at 3232 with 1 tries\nSucceded at 3233 with 1 tries\nSucceded at 3235 with 1 tries\nSucceded at 3236 with 1 tries\nSucceded at 3237 with 1 tries\nSucceded at 3238 with 1 tries\nSucceded at 3240 with 1 tries\nSucceded at 3241 with 1 tries\nSucceded at 3242 with 1 tries\nSucceded at 3243 with 1 tries\nSucceded at 3245 with 1 tries\nSucceded at 3246 with 1 tries\nSucceded at 3250 with 1 tries\nSucceded at 3251 with 1 tries\nSucceded at 3252 with 1 tries\nSucceded at 3253 with 1 tries\nSucceded at 3255 with 1 tries\nSucceded at 3257 with 1 tries\nSucceded at 3258 with 1 tries\nSucceded at 3259 with 1 tries\nSucceded at 3261 with 1 tries\nSucceded at 3262 with 1 tries\nSucceded at 3264 with 1 tries\nSucceded at 3265 with 1 tries\nSucceded at 3267 with 1 tries\nSucceded at 3270 with 1 tries\nSucceded at 3271 with 1 tries\nSucceded at 3273 with 1 tries\nSucceded at 3274 with 1 tries\nSucceded at 3275 with 1 tries\nSucceded at 3276 with 1 tries\nSucceded at 3278 with 1 tries\nSucceded at 3280 with 1 tries\nSucceded at 3281 with 1 tries\nSucceded at 3282 with 1 tries\nSucceded at 3283 with 1 tries\nSucceded at 3284 with 1 tries\nSucceded at 3285 with 1 tries\nSucceded at 3287 with 1 tries\nSucceded at 3290 with 1 tries\nSucceded at 3291 with 1 tries\nSucceded at 3293 with 1 tries\nSucceded at 3296 with 1 tries\nSucceded at 3298 with 1 tries\nSucceded at 3299 with 1 tries\nSucceded at 3304 with 1 tries\nSucceded at 3305 with 1 tries\nSucceded at 3307 with 1 tries\nSucceded at 3309 with 1 tries\nSucceded at 3310 with 1 tries\nSucceded at 3311 with 1 tries\nSucceded at 3313 with 1 tries\nSucceded at 3315 with 1 tries\nSucceded at 3316 with 1 tries\nSucceded at 3317 with 1 tries\nSucceded at 3318 with 1 tries\nSucceded at 3319 with 1 tries\nSucceded at 3320 with 1 tries\nSucceded at 3325 with 1 tries\nSucceded at 3328 with 1 tries\nSucceded at 3331 with 1 tries\nSucceded at 3332 with 1 tries\nSucceded at 3333 with 1 tries\nSucceded at 3334 with 1 tries\nSucceded at 3335 with 1 tries\nSucceded at 3336 with 1 tries\nSucceded at 3337 with 1 tries\nSucceded at 3338 with 1 tries\nSucceded at 3339 with 1 tries\nSucceded at 3340 with 1 tries\nSucceded at 3343 with 1 tries\nSucceded at 3344 with 1 tries\nSucceded at 3345 with 1 tries\nSucceded at 3346 with 1 tries\nSucceded at 3347 with 1 tries\nSucceded at 3349 with 1 tries\nSucceded at 3350 with 1 tries\nSucceded at 3351 with 1 tries\nSucceded at 3353 with 1 tries\nSucceded at 3354 with 1 tries\nSucceded at 3356 with 1 tries\nSucceded at 3357 with 1 tries\nSucceded at 3358 with 1 tries\nSucceded at 3363 with 1 tries\nSucceded at 3364 with 1 tries\nSucceded at 3365 with 1 tries\nSucceded at 3366 with 1 tries\nSucceded at 3368 with 1 tries\nSucceded at 3370 with 1 tries\nSucceded at 3371 with 1 tries\nSucceded at 3372 with 1 tries\nSucceded at 3373 with 1 tries\nSucceded at 3376 with 1 tries\nSucceded at 3377 with 1 tries\nSucceded at 3378 with 1 tries\nSucceded at 3379 with 1 tries\nSucceded at 3380 with 1 tries\nSucceded at 3381 with 1 tries\nSucceded at 3382 with 1 tries\nSucceded at 3383 with 1 tries\nSucceded at 3384 with 1 tries\nSucceded at 3385 with 1 tries\nSucceded at 3388 with 1 tries\nSucceded at 3389 with 1 tries\nSucceded at 3390 with 1 tries\nSucceded at 3391 with 1 tries\nSucceded at 3394 with 1 tries\nSucceded at 3395 with 1 tries\nSucceded at 3397 with 1 tries\nSucceded at 3398 with 1 tries\nSucceded at 3399 with 1 tries\nSucceded at 3401 with 1 tries\nSucceded at 3402 with 1 tries\nSucceded at 3403 with 1 tries\nSucceded at 3404 with 1 tries\nSucceded at 3405 with 1 tries\nSucceded at 3407 with 1 tries\nSucceded at 3408 with 1 tries\nSucceded at 3409 with 1 tries\nSucceded at 3410 with 1 tries\nSucceded at 3411 with 1 tries\nSucceded at 3412 with 1 tries\nSucceded at 3413 with 1 tries\nSucceded at 3414 with 1 tries\nSucceded at 3415 with 1 tries\nSucceded at 3416 with 1 tries\nSucceded at 3417 with 1 tries\nSucceded at 3418 with 1 tries\nSucceded at 3420 with 1 tries\nSucceded at 3421 with 1 tries\nSucceded at 3422 with 1 tries\nSucceded at 3423 with 1 tries\nSucceded at 3424 with 1 tries\nSucceded at 3425 with 1 tries\nSucceded at 3426 with 1 tries\nSucceded at 3428 with 1 tries\nSucceded at 3429 with 1 tries\nSucceded at 3431 with 1 tries\nSucceded at 3434 with 1 tries\nSucceded at 3435 with 1 tries\nSucceded at 3437 with 1 tries\nSucceded at 3440 with 1 tries\nSucceded at 3441 with 1 tries\nSucceded at 3443 with 1 tries\nSucceded at 3444 with 1 tries\nSucceded at 3445 with 1 tries\nSucceded at 3446 with 1 tries\nSucceded at 3447 with 1 tries\nSucceded at 3448 with 1 tries\nSucceded at 3449 with 1 tries\nSucceded at 3450 with 1 tries\nSucceded at 3453 with 1 tries\nSucceded at 3454 with 1 tries\nSucceded at 3455 with 1 tries\nSucceded at 3456 with 1 tries\nSucceded at 3458 with 1 tries\nSucceded at 3460 with 1 tries\nSucceded at 3464 with 1 tries\nSucceded at 3465 with 1 tries\nSucceded at 3466 with 1 tries\nSucceded at 3468 with 1 tries\nSucceded at 3470 with 1 tries\nSucceded at 3472 with 1 tries\nSucceded at 3473 with 1 tries\nSucceded at 3476 with 1 tries\nSucceded at 3477 with 1 tries\nSucceded at 3478 with 1 tries\nSucceded at 3479 with 1 tries\nSucceded at 3481 with 1 tries\nSucceded at 3482 with 1 tries\nSucceded at 3483 with 1 tries\nSucceded at 3484 with 1 tries\nSucceded at 3488 with 1 tries\nSucceded at 3489 with 1 tries\nSucceded at 3490 with 1 tries\nSucceded at 3491 with 1 tries\nSucceded at 3494 with 1 tries\nSucceded at 3495 with 1 tries\nSucceded at 3496 with 1 tries\nSucceded at 3497 with 1 tries\nSucceded at 3498 with 1 tries\nSucceded at 3501 with 1 tries\nSucceded at 3502 with 1 tries\nSucceded at 3503 with 1 tries\nSucceded at 3504 with 1 tries\nSucceded at 3505 with 1 tries\nSucceded at 3506 with 1 tries\nSucceded at 3507 with 1 tries\nSucceded at 3508 with 1 tries\nSucceded at 3509 with 1 tries\nSucceded at 3511 with 1 tries\nSucceded at 3512 with 1 tries\nSucceded at 3513 with 1 tries\nSucceded at 3516 with 1 tries\nSucceded at 3517 with 1 tries\nSucceded at 3518 with 1 tries\nSucceded at 3519 with 1 tries\nSucceded at 3520 with 1 tries\nSucceded at 3521 with 1 tries\nSucceded at 3522 with 1 tries\nSucceded at 3523 with 1 tries\nSucceded at 3524 with 1 tries\nSucceded at 3526 with 1 tries\nSucceded at 3528 with 1 tries\nSucceded at 3529 with 1 tries\nSucceded at 3530 with 1 tries\nSucceded at 3531 with 1 tries\nSucceded at 3533 with 1 tries\nSucceded at 3536 with 1 tries\nSucceded at 3537 with 1 tries\nSucceded at 3539 with 1 tries\nSucceded at 3542 with 1 tries\nSucceded at 3543 with 1 tries\nSucceded at 3545 with 1 tries\nSucceded at 3546 with 1 tries\nSucceded at 3547 with 1 tries\nSucceded at 3548 with 1 tries\nSucceded at 3550 with 1 tries\nSucceded at 3551 with 1 tries\nSucceded at 3553 with 1 tries\nSucceded at 3554 with 1 tries\nSucceded at 3555 with 1 tries\nSucceded at 3556 with 1 tries\nSucceded at 3557 with 1 tries\nSucceded at 3558 with 1 tries\nSucceded at 3560 with 1 tries\nSucceded at 3562 with 1 tries\nSucceded at 3563 with 1 tries\nSucceded at 3564 with 1 tries\nSucceded at 3566 with 1 tries\nSucceded at 3568 with 1 tries\nSucceded at 3569 with 1 tries\nSucceded at 3570 with 1 tries\nSucceded at 3572 with 1 tries\nSucceded at 3573 with 1 tries\nSucceded at 3574 with 1 tries\nSucceded at 3575 with 1 tries\nSucceded at 3576 with 1 tries\nSucceded at 3577 with 1 tries\nSucceded at 3578 with 1 tries\nSucceded at 3579 with 1 tries\nSucceded at 3581 with 1 tries\nSucceded at 3582 with 1 tries\nSucceded at 3585 with 1 tries\nSucceded at 3586 with 1 tries\nSucceded at 3590 with 1 tries\nSucceded at 3591 with 1 tries\nSucceded at 3592 with 1 tries\nSucceded at 3594 with 1 tries\nSucceded at 3595 with 1 tries\nSucceded at 3596 with 1 tries\nSucceded at 3597 with 1 tries\nSucceded at 3598 with 1 tries\nSucceded at 3599 with 1 tries\nSucceded at 3601 with 1 tries\nSucceded at 3603 with 1 tries\nSucceded at 3605 with 1 tries\nSucceded at 3607 with 1 tries\nSucceded at 3609 with 1 tries\nSucceded at 3610 with 1 tries\nSucceded at 3613 with 1 tries\nSucceded at 3617 with 1 tries\nSucceded at 3619 with 1 tries\nSucceded at 3620 with 1 tries\nSucceded at 3621 with 1 tries\nSucceded at 3624 with 1 tries\nSucceded at 3625 with 1 tries\nSucceded at 3626 with 1 tries\nSucceded at 3627 with 1 tries\nSucceded at 3630 with 1 tries\nSucceded at 3631 with 1 tries\nSucceded at 3634 with 1 tries\nSucceded at 3636 with 1 tries\nSucceded at 3637 with 1 tries\nSucceded at 3639 with 1 tries\nSucceded at 3640 with 1 tries\nSucceded at 3641 with 1 tries\nSucceded at 3642 with 1 tries\nSucceded at 3643 with 1 tries\nSucceded at 3646 with 1 tries\nSucceded at 3647 with 1 tries\nSucceded at 3648 with 1 tries\nSucceded at 3649 with 1 tries\nSucceded at 3651 with 1 tries\nSucceded at 3652 with 1 tries\nSucceded at 3654 with 1 tries\nSucceded at 3656 with 1 tries\nSucceded at 3657 with 1 tries\nSucceded at 3659 with 1 tries\nSucceded at 3660 with 1 tries\nSucceded at 3661 with 1 tries\nSucceded at 3664 with 1 tries\nSucceded at 3665 with 1 tries\nSucceded at 3667 with 1 tries\nSucceded at 3668 with 1 tries\nSucceded at 3669 with 1 tries\nSucceded at 3670 with 1 tries\nmosek failed at \nSucceded at 3672 with 1 tries\nSucceded at 3673 with 1 tries\nSucceded at 3675 with 1 tries\nSucceded at 3676 with 1 tries\nSucceded at 3678 with 1 tries\nSucceded at 3679 with 1 tries\nSucceded at 3680 with 1 tries\nSucceded at 3681 with 1 tries\nSucceded at 3682 with 1 tries\nSucceded at 3683 with 1 tries\nSucceded at 3684 with 1 tries\nSucceded at 3685 with 1 tries\nSucceded at 3687 with 1 tries\nSucceded at 3688 with 1 tries\nSucceded at 3689 with 1 tries\nSucceded at 3690 with 1 tries\nSucceded at 3691 with 1 tries\nSucceded at 3693 with 1 tries\nSucceded at 3694 with 1 tries\nSucceded at 3696 with 1 tries\nSucceded at 3698 with 1 tries\nSucceded at 3699 with 1 tries\nSucceded at 3700 with 1 tries\nSucceded at 3701 with 1 tries\nSucceded at 3702 with 1 tries\nSucceded at 3704 with 1 tries\nSucceded at 3706 with 1 tries\nSucceded at 3708 with 1 tries\nSucceded at 3709 with 1 tries\nSucceded at 3710 with 1 tries\nSucceded at 3711 with 1 tries\nSucceded at 3714 with 1 tries\nSucceded at 3715 with 1 tries\nSucceded at 3717 with 1 tries\nSucceded at 3718 with 1 tries\nSucceded at 3720 with 1 tries\nSucceded at 3722 with 1 tries\nSucceded at 3723 with 1 tries\nSucceded at 3724 with 1 tries\nSucceded at 3725 with 1 tries\nSucceded at 3726 with 1 tries\nSucceded at 3727 with 1 tries\nSucceded at 3728 with 1 tries\nSucceded at 3729 with 1 tries\nSucceded at 3730 with 1 tries\nSucceded at 3731 with 1 tries\nSucceded at 3732 with 1 tries\nSucceded at 3734 with 1 tries\nSucceded at 3735 with 1 tries\nSucceded at 3736 with 1 tries\nSucceded at 3737 with 1 tries\nSucceded at 3738 with 1 tries\nSucceded at 3739 with 1 tries\nSucceded at 3740 with 1 tries\nSucceded at 3741 with 1 tries\nSucceded at 3742 with 1 tries\nSucceded at 3743 with 1 tries\nSucceded at 3744 with 1 tries\nSucceded at 3745 with 1 tries\nSucceded at 3747 with 1 tries\nSucceded at 3748 with 1 tries\nSucceded at 3749 with 1 tries\nSucceded at 3751 with 1 tries\nSucceded at 3752 with 1 tries\nSucceded at 3753 with 1 tries\nSucceded at 3754 with 1 tries\nSucceded at 3755 with 1 tries\nSucceded at 3756 with 1 tries\nSucceded at 3757 with 1 tries\nSucceded at 3758 with 1 tries\nSucceded at 3759 with 1 tries\nSucceded at 3760 with 1 tries\nSucceded at 3761 with 1 tries\nSucceded at 3762 with 1 tries\nSucceded at 3766 with 1 tries\nSucceded at 3767 with 1 tries\nSucceded at 3769 with 1 tries\nSucceded at 3770 with 1 tries\nSucceded at 3772 with 1 tries\nSucceded at 3776 with 1 tries\nSucceded at 3777 with 1 tries\nSucceded at 3779 with 1 tries\nSucceded at 3780 with 1 tries\nSucceded at 3782 with 1 tries\nSucceded at 3783 with 1 tries\nSucceded at 3784 with 1 tries\nSucceded at 3786 with 1 tries\nSucceded at 3787 with 1 tries\nSucceded at 3788 with 1 tries\nSucceded at 3789 with 1 tries\nSucceded at 3790 with 1 tries\nSucceded at 3792 with 1 tries\nSucceded at 3793 with 1 tries\nSucceded at 3794 with 1 tries\nSucceded at 3796 with 1 tries\nSucceded at 3797 with 1 tries\nSucceded at 3798 with 1 tries\nSucceded at 3800 with 1 tries\nSucceded at 3801 with 1 tries\nSucceded at 3802 with 1 tries\nSucceded at 3803 with 1 tries\nSucceded at 3804 with 1 tries\nSucceded at 3805 with 1 tries\nSucceded at 3806 with 1 tries\nSucceded at 3807 with 1 tries\nSucceded at 3808 with 1 tries\nSucceded at 3810 with 1 tries\nSucceded at 3811 with 1 tries\nSucceded at 3814 with 1 tries\nSucceded at 3815 with 1 tries\nSucceded at 3816 with 1 tries\nSucceded at 3818 with 1 tries\nSucceded at 3821 with 1 tries\nSucceded at 3824 with 1 tries\nSucceded at 3825 with 1 tries\nSucceded at 3827 with 1 tries\nSucceded at 3828 with 1 tries\nSucceded at 3830 with 1 tries\nSucceded at 3831 with 1 tries\nSucceded at 3832 with 1 tries\nSucceded at 3833 with 1 tries\nSucceded at 3836 with 1 tries\nSucceded at 3837 with 1 tries\nSucceded at 3838 with 1 tries\nSucceded at 3839 with 1 tries\nSucceded at 3840 with 1 tries\nSucceded at 3841 with 1 tries\nSucceded at 3843 with 1 tries\nSucceded at 3845 with 1 tries\nSucceded at 3846 with 1 tries\nSucceded at 3849 with 1 tries\nSucceded at 3851 with 1 tries\nSucceded at 3853 with 1 tries\nSucceded at 3854 with 1 tries\nSucceded at 3855 with 1 tries\nSucceded at 3856 with 1 tries\nSucceded at 3858 with 1 tries\nSucceded at 3863 with 1 tries\nSucceded at 3864 with 1 tries\nSucceded at 3865 with 1 tries\nSucceded at 3867 with 1 tries\nSucceded at 3869 with 1 tries\nSucceded at 3870 with 1 tries\nSucceded at 3871 with 1 tries\nSucceded at 3872 with 1 tries\nSucceded at 3874 with 1 tries\nSucceded at 3875 with 1 tries\nSucceded at 3877 with 1 tries\nSucceded at 3878 with 1 tries\nSucceded at 3881 with 1 tries\nSucceded at 3882 with 1 tries\nSucceded at 3884 with 1 tries\nSucceded at 3885 with 1 tries\nSucceded at 3886 with 1 tries\nSucceded at 3887 with 1 tries\nSucceded at 3888 with 1 tries\nSucceded at 3889 with 1 tries\nSucceded at 3891 with 1 tries\nSucceded at 3892 with 1 tries\nSucceded at 3893 with 1 tries\nSucceded at 3894 with 1 tries\nSucceded at 3895 with 1 tries\nSucceded at 3897 with 1 tries\nSucceded at 3899 with 1 tries\nSucceded at 3900 with 1 tries\nSucceded at 3902 with 1 tries\nSucceded at 3903 with 1 tries\nSucceded at 3904 with 1 tries\nSucceded at 3905 with 1 tries\nSucceded at 3908 with 1 tries\nSucceded at 3909 with 1 tries\nSucceded at 3912 with 1 tries\nSucceded at 3913 with 1 tries\nSucceded at 3914 with 1 tries\nSucceded at 3915 with 1 tries\nSucceded at 3917 with 1 tries\nSucceded at 3919 with 1 tries\nSucceded at 3920 with 1 tries\nSucceded at 3921 with 1 tries\nSucceded at 3922 with 1 tries\nSucceded at 3924 with 1 tries\nSucceded at 3927 with 1 tries\nSucceded at 3929 with 1 tries\nSucceded at 3930 with 1 tries\nSucceded at 3933 with 1 tries\nSucceded at 3937 with 1 tries\nSucceded at 3938 with 1 tries\nSucceded at 3939 with 1 tries\nSucceded at 3940 with 1 tries\nSucceded at 3942 with 1 tries\nSucceded at 3943 with 1 tries\nSucceded at 3944 with 1 tries\nSucceded at 3945 with 1 tries\nSucceded at 3946 with 1 tries\nSucceded at 3948 with 1 tries\nSucceded at 3950 with 1 tries\nSucceded at 3951 with 1 tries\nSucceded at 3953 with 1 tries\nSucceded at 3954 with 1 tries\nSucceded at 3955 with 1 tries\nSucceded at 3956 with 1 tries\nSucceded at 3957 with 1 tries\nSucceded at 3959 with 1 tries\nSucceded at 3960 with 1 tries\nSucceded at 3961 with 1 tries\nSucceded at 3962 with 1 tries\nSucceded at 3963 with 1 tries\nSucceded at 3965 with 1 tries\nSucceded at 3966 with 1 tries\nSucceded at 3968 with 1 tries\nSucceded at 3969 with 1 tries\nSucceded at 3970 with 1 tries\nSucceded at 3971 with 1 tries\nSucceded at 3972 with 1 tries\nSucceded at 3973 with 1 tries\nSucceded at 3975 with 1 tries\nSucceded at 3976 with 1 tries\nSucceded at 3977 with 1 tries\nSucceded at 3978 with 1 tries\nSucceded at 3980 with 1 tries\nSucceded at 3983 with 1 tries\nSucceded at 3984 with 1 tries\nSucceded at 3985 with 1 tries\nSucceded at 3986 with 1 tries\nSucceded at 3987 with 1 tries\nSucceded at 3988 with 1 tries\nSucceded at 3989 with 1 tries\nSucceded at 3990 with 1 tries\nSucceded at 3994 with 1 tries\nSucceded at 3995 with 1 tries\nSucceded at 3996 with 1 tries\nSucceded at 3998 with 1 tries\nSucceded at 3999 with 1 tries\nSucceded at 4000 with 1 tries\nSucceded at 4001 with 1 tries\nSucceded at 4002 with 1 tries\nSucceded at 4003 with 1 tries\nSucceded at 4004 with 1 tries\nSucceded at 4005 with 1 tries\nSucceded at 4007 with 1 tries\nSucceded at 4008 with 1 tries\nSucceded at 4009 with 1 tries\nSucceded at 4010 with 1 tries\nSucceded at 4011 with 1 tries\nSucceded at 4012 with 1 tries\nSucceded at 4014 with 1 tries\nSucceded at 4015 with 1 tries\nSucceded at 4016 with 1 tries\nSucceded at 4017 with 1 tries\nSucceded at 4018 with 1 tries\nSucceded at 4019 with 1 tries\nSucceded at 4020 with 1 tries\nSucceded at 4021 with 1 tries\nSucceded at 4022 with 1 tries\nSucceded at 4023 with 1 tries\nSucceded at 4024 with 1 tries\nSucceded at 4025 with 1 tries\nSucceded at 4028 with 1 tries\nSucceded at 4029 with 1 tries\nSucceded at 4030 with 1 tries\nSucceded at 4032 with 1 tries\nSucceded at 4033 with 1 tries\nSucceded at 4034 with 1 tries\nSucceded at 4035 with 1 tries\nSucceded at 4037 with 1 tries\nSucceded at 4038 with 1 tries\nSucceded at 4039 with 1 tries\nSucceded at 4042 with 1 tries\nSucceded at 4043 with 1 tries\nSucceded at 4045 with 1 tries\nSucceded at 4046 with 1 tries\nSucceded at 4048 with 1 tries\nSucceded at 4049 with 1 tries\nSucceded at 4051 with 1 tries\nSucceded at 4052 with 1 tries\nSucceded at 4053 with 1 tries\nSucceded at 4054 with 1 tries\nSucceded at 4055 with 1 tries\nSucceded at 4056 with 1 tries\nSucceded at 4058 with 1 tries\nSucceded at 4059 with 1 tries\nSucceded at 4060 with 1 tries\nSucceded at 4061 with 1 tries\nSucceded at 4064 with 1 tries\nSucceded at 4068 with 1 tries\nSucceded at 4069 with 1 tries\nSucceded at 4071 with 1 tries\nSucceded at 4074 with 1 tries\nSucceded at 4075 with 1 tries\nSucceded at 4077 with 1 tries\nSucceded at 4080 with 1 tries\nSucceded at 4082 with 1 tries\nSucceded at 4084 with 1 tries\nSucceded at 4086 with 1 tries\nSucceded at 4087 with 1 tries\nSucceded at 4088 with 1 tries\nSucceded at 4090 with 1 tries\nSucceded at 4091 with 1 tries\nSucceded at 4092 with 1 tries\nSucceded at 4093 with 1 tries\nSucceded at 4095 with 1 tries\nSucceded at 4096 with 1 tries\nSucceded at 4097 with 1 tries\nSucceded at 4099 with 1 tries\nSucceded at 4100 with 1 tries\nSucceded at 4101 with 1 tries\nSucceded at 4102 with 1 tries\nSucceded at 4104 with 1 tries\nSucceded at 4105 with 1 tries\nSucceded at 4107 with 1 tries\nSucceded at 4109 with 1 tries\nSucceded at 4110 with 1 tries\nSucceded at 4111 with 1 tries\nSucceded at 4113 with 1 tries\nSucceded at 4116 with 1 tries\nSucceded at 4117 with 1 tries\nSucceded at 4118 with 1 tries\nSucceded at 4119 with 1 tries\nSucceded at 4120 with 1 tries\nSucceded at 4121 with 1 tries\nSucceded at 4123 with 1 tries\nSucceded at 4124 with 1 tries\nSucceded at 4128 with 1 tries\nSucceded at 4129 with 1 tries\nSucceded at 4130 with 1 tries\nSucceded at 4131 with 1 tries\nSucceded at 4132 with 1 tries\nSucceded at 4133 with 1 tries\nSucceded at 4134 with 1 tries\nSucceded at 4136 with 1 tries\nSucceded at 4138 with 1 tries\nSucceded at 4139 with 1 tries\nSucceded at 4140 with 1 tries\nSucceded at 4143 with 1 tries\nSucceded at 4144 with 1 tries\nSucceded at 4145 with 1 tries\nSucceded at 4148 with 1 tries\nSucceded at 4149 with 1 tries\nSucceded at 4153 with 1 tries\nSucceded at 4155 with 1 tries\nSucceded at 4156 with 1 tries\nSucceded at 4157 with 1 tries\nSucceded at 4158 with 1 tries\nSucceded at 4160 with 1 tries\nSucceded at 4161 with 1 tries\nSucceded at 4162 with 1 tries\nSucceded at 4163 with 1 tries\nSucceded at 4164 with 1 tries\nSucceded at 4165 with 1 tries\nSucceded at 4166 with 1 tries\nSucceded at 4167 with 1 tries\nSucceded at 4168 with 1 tries\nSucceded at 4171 with 1 tries\nSucceded at 4172 with 1 tries\nSucceded at 4175 with 1 tries\nSucceded at 4177 with 1 tries\nSucceded at 4178 with 1 tries\nSucceded at 4179 with 1 tries\nSucceded at 4180 with 1 tries\nSucceded at 4181 with 1 tries\nSucceded at 4184 with 1 tries\nSucceded at 4185 with 1 tries\nSucceded at 4187 with 1 tries\nSucceded at 4189 with 1 tries\nSucceded at 4190 with 1 tries\nSucceded at 4191 with 1 tries\nSucceded at 4192 with 1 tries\nSucceded at 4193 with 1 tries\nSucceded at 4195 with 1 tries\nSucceded at 4197 with 1 tries\nSucceded at 4199 with 1 tries\nSucceded at 4200 with 1 tries\nSucceded at 4201 with 1 tries\nSucceded at 4202 with 1 tries\nSucceded at 4205 with 1 tries\nSucceded at 4207 with 1 tries\nSucceded at 4208 with 1 tries\nSucceded at 4209 with 1 tries\nSucceded at 4210 with 1 tries\nSucceded at 4211 with 1 tries\nSucceded at 4213 with 1 tries\nSucceded at 4214 with 1 tries\nSucceded at 4216 with 1 tries\nSucceded at 4217 with 1 tries\nSucceded at 4219 with 1 tries\nSucceded at 4223 with 1 tries\nSucceded at 4224 with 1 tries\nSucceded at 4226 with 1 tries\nSucceded at 4230 with 1 tries\nSucceded at 4231 with 1 tries\nSucceded at 4233 with 1 tries\nSucceded at 4234 with 1 tries\nSucceded at 4235 with 1 tries\nSucceded at 4237 with 1 tries\nSucceded at 4238 with 1 tries\nSucceded at 4241 with 1 tries\nSucceded at 4242 with 1 tries\nSucceded at 4243 with 1 tries\nSucceded at 4244 with 1 tries\nSucceded at 4247 with 1 tries\nSucceded at 4248 with 1 tries\nSucceded at 4249 with 1 tries\nSucceded at 4251 with 1 tries\nSucceded at 4253 with 1 tries\nSucceded at 4254 with 1 tries\nSucceded at 4257 with 1 tries\nSucceded at 4258 with 1 tries\nSucceded at 4259 with 1 tries\nSucceded at 4260 with 1 tries\nSucceded at 4262 with 1 tries\nSucceded at 4263 with 1 tries\nSucceded at 4264 with 1 tries\nSucceded at 4265 with 1 tries\nSucceded at 4266 with 1 tries\nSucceded at 4267 with 1 tries\nSucceded at 4268 with 1 tries\nSucceded at 4269 with 1 tries\nSucceded at 4270 with 1 tries\nSucceded at 4271 with 1 tries\nSucceded at 4272 with 1 tries\nSucceded at 4274 with 1 tries\nSucceded at 4276 with 1 tries\nSucceded at 4277 with 1 tries\nSucceded at 4278 with 1 tries\nSucceded at 4279 with 1 tries\nSucceded at 4283 with 1 tries\nSucceded at 4284 with 1 tries\nSucceded at 4285 with 1 tries\nSucceded at 4286 with 1 tries\nSucceded at 4287 with 1 tries\nSucceded at 4290 with 1 tries\nSucceded at 4292 with 1 tries\nSucceded at 4293 with 1 tries\nSucceded at 4295 with 1 tries\nSucceded at 4296 with 1 tries\nSucceded at 4301 with 1 tries\nSucceded at 4302 with 1 tries\nSucceded at 4303 with 1 tries\nSucceded at 4305 with 1 tries\nSucceded at 4306 with 1 tries\nSucceded at 4307 with 1 tries\nSucceded at 4309 with 1 tries\nSucceded at 4310 with 1 tries\nSucceded at 4312 with 1 tries\nSucceded at 4316 with 1 tries\nSucceded at 4317 with 1 tries\nSucceded at 4318 with 1 tries\nSucceded at 4319 with 1 tries\nSucceded at 4320 with 1 tries\nSucceded at 4324 with 1 tries\nSucceded at 4325 with 1 tries\nSucceded at 4326 with 1 tries\nSucceded at 4327 with 1 tries\nSucceded at 4328 with 1 tries\nSucceded at 4330 with 1 tries\nSucceded at 4333 with 1 tries\nSucceded at 4334 with 1 tries\nSucceded at 4338 with 1 tries\nSucceded at 4339 with 1 tries\nSucceded at 4341 with 1 tries\nSucceded at 4342 with 1 tries\nSucceded at 4343 with 1 tries\nSucceded at 4345 with 1 tries\nSucceded at 4347 with 1 tries\nSucceded at 4348 with 1 tries\nSucceded at 4349 with 1 tries\nSucceded at 4351 with 1 tries\nSucceded at 4352 with 1 tries\nSucceded at 4353 with 1 tries\nSucceded at 4354 with 1 tries\nSucceded at 4355 with 1 tries\nSucceded at 4357 with 1 tries\nSucceded at 4358 with 1 tries\nSucceded at 4359 with 1 tries\nSucceded at 4360 with 1 tries\nSucceded at 4361 with 1 tries\nSucceded at 4362 with 1 tries\nSucceded at 4363 with 1 tries\nSucceded at 4364 with 1 tries\nSucceded at 4365 with 1 tries\nSucceded at 4366 with 1 tries\nSucceded at 4367 with 1 tries\nSucceded at 4368 with 1 tries\nSucceded at 4371 with 1 tries\nSucceded at 4372 with 1 tries\nSucceded at 4373 with 1 tries\nSucceded at 4374 with 1 tries\nSucceded at 4376 with 1 tries\nSucceded at 4377 with 1 tries\nSucceded at 4379 with 1 tries\nSucceded at 4381 with 1 tries\nSucceded at 4382 with 1 tries\nSucceded at 4384 with 1 tries\nSucceded at 4385 with 1 tries\nSucceded at 4386 with 1 tries\nSucceded at 4387 with 1 tries\nSucceded at 4389 with 1 tries\nSucceded at 4390 with 1 tries\nSucceded at 4392 with 1 tries\nSucceded at 4393 with 1 tries\nSucceded at 4394 with 1 tries\nSucceded at 4395 with 1 tries\nSucceded at 4398 with 1 tries\nSucceded at 4399 with 1 tries\nSucceded at 4400 with 1 tries\nSucceded at 4401 with 1 tries\nSucceded at 4402 with 1 tries\nSucceded at 4404 with 1 tries\nSucceded at 4405 with 1 tries\nSucceded at 4406 with 1 tries\nSucceded at 4407 with 1 tries\nSucceded at 4408 with 1 tries\nSucceded at 4409 with 1 tries\nSucceded at 4410 with 1 tries\nSucceded at 4412 with 1 tries\nSucceded at 4413 with 1 tries\nSucceded at 4415 with 1 tries\nSucceded at 4416 with 1 tries\nSucceded at 4417 with 1 tries\nSucceded at 4419 with 1 tries\nSucceded at 4421 with 1 tries\nSucceded at 4422 with 1 tries\nSucceded at 4423 with 1 tries\nSucceded at 4427 with 1 tries\nSucceded at 4429 with 1 tries\nSucceded at 4430 with 1 tries\nSucceded at 4432 with 1 tries\nSucceded at 4433 with 1 tries\nSucceded at 4434 with 1 tries\nSucceded at 4438 with 1 tries\nSucceded at 4439 with 1 tries\nSucceded at 4440 with 1 tries\nSucceded at 4441 with 1 tries\nSucceded at 4443 with 1 tries\nSucceded at 4444 with 1 tries\nSucceded at 4445 with 1 tries\nSucceded at 4446 with 1 tries\nSucceded at 4447 with 1 tries\nSucceded at 4448 with 1 tries\nSucceded at 4449 with 1 tries\nSucceded at 4450 with 1 tries\nSucceded at 4451 with 1 tries\nSucceded at 4452 with 1 tries\nSucceded at 4454 with 1 tries\nSucceded at 4455 with 1 tries\nSucceded at 4456 with 1 tries\nSucceded at 4458 with 1 tries\nSucceded at 4461 with 1 tries\nSucceded at 4462 with 1 tries\nSucceded at 4466 with 1 tries\nSucceded at 4467 with 1 tries\nSucceded at 4468 with 1 tries\nSucceded at 4470 with 1 tries\nSucceded at 4473 with 1 tries\nSucceded at 4475 with 1 tries\nSucceded at 4476 with 1 tries\nSucceded at 4477 with 1 tries\nSucceded at 4479 with 1 tries\nSucceded at 4480 with 1 tries\nSucceded at 4482 with 1 tries\nSucceded at 4485 with 1 tries\nSucceded at 4486 with 1 tries\nSucceded at 4488 with 1 tries\nSucceded at 4489 with 1 tries\nSucceded at 4490 with 1 tries\nSucceded at 4491 with 1 tries\nSucceded at 4494 with 1 tries\nSucceded at 4495 with 1 tries\nSucceded at 4496 with 1 tries\nSucceded at 4499 with 1 tries\nSucceded at 4500 with 1 tries\nSucceded at 4501 with 1 tries\nSucceded at 4502 with 1 tries\nSucceded at 4503 with 1 tries\nSucceded at 4506 with 1 tries\nSucceded at 4507 with 1 tries\nSucceded at 4508 with 1 tries\nSucceded at 4509 with 1 tries\nSucceded at 4511 with 1 tries\nSucceded at 4512 with 1 tries\nSucceded at 4513 with 1 tries\nSucceded at 4515 with 1 tries\nSucceded at 4516 with 1 tries\nSucceded at 4517 with 1 tries\nSucceded at 4518 with 1 tries\nSucceded at 4519 with 1 tries\nSucceded at 4520 with 1 tries\nSucceded at 4521 with 1 tries\nSucceded at 4522 with 1 tries\nSucceded at 4523 with 1 tries\nSucceded at 4524 with 1 tries\nSucceded at 4525 with 1 tries\nSucceded at 4526 with 1 tries\nSucceded at 4528 with 1 tries\nSucceded at 4530 with 1 tries\nSucceded at 4533 with 1 tries\nSucceded at 4535 with 1 tries\nSucceded at 4536 with 1 tries\nSucceded at 4537 with 1 tries\nSucceded at 4538 with 1 tries\nSucceded at 4539 with 1 tries\nSucceded at 4540 with 1 tries\nSucceded at 4543 with 1 tries\nSucceded at 4544 with 1 tries\nSucceded at 4546 with 1 tries\nSucceded at 4547 with 1 tries\nSucceded at 4549 with 1 tries\nSucceded at 4550 with 1 tries\nSucceded at 4551 with 1 tries\nSucceded at 4552 with 1 tries\nSucceded at 4554 with 1 tries\nSucceded at 4555 with 1 tries\nSucceded at 4556 with 1 tries\nSucceded at 4557 with 1 tries\nmosek failed at \nSucceded at 4560 with 1 tries\nSucceded at 4562 with 1 tries\nSucceded at 4563 with 1 tries\nSucceded at 4564 with 1 tries\nSucceded at 4565 with 1 tries\nSucceded at 4566 with 1 tries\nSucceded at 4568 with 1 tries\nSucceded at 4572 with 1 tries\nSucceded at 4574 with 1 tries\nSucceded at 4577 with 1 tries\nSucceded at 4579 with 1 tries\nSucceded at 4582 with 1 tries\nSucceded at 4583 with 1 tries\nSucceded at 4588 with 1 tries\nSucceded at 4589 with 1 tries\nSucceded at 4590 with 1 tries\nSucceded at 4594 with 1 tries\nSucceded at 4596 with 1 tries\nSucceded at 4597 with 1 tries\nSucceded at 4600 with 1 tries\nSucceded at 4601 with 1 tries\nSucceded at 4603 with 1 tries\nSucceded at 4604 with 1 tries\nSucceded at 4606 with 1 tries\nSucceded at 4607 with 1 tries\nSucceded at 4608 with 1 tries\nSucceded at 4609 with 1 tries\nSucceded at 4611 with 1 tries\nSucceded at 4612 with 1 tries\nSucceded at 4613 with 1 tries\nSucceded at 4615 with 1 tries\nSucceded at 4616 with 1 tries\nSucceded at 4617 with 1 tries\nSucceded at 4618 with 1 tries\nSucceded at 4619 with 1 tries\nSucceded at 4623 with 1 tries\nSucceded at 4624 with 1 tries\nSucceded at 4625 with 1 tries\nSucceded at 4627 with 1 tries\nSucceded at 4630 with 1 tries\nSucceded at 4633 with 1 tries\nSucceded at 4634 with 1 tries\nSucceded at 4635 with 1 tries\nSucceded at 4636 with 1 tries\nSucceded at 4637 with 1 tries\nSucceded at 4638 with 1 tries\nSucceded at 4643 with 1 tries\nSucceded at 4644 with 1 tries\nSucceded at 4645 with 1 tries\nSucceded at 4646 with 1 tries\nSucceded at 4647 with 1 tries\nSucceded at 4648 with 1 tries\nSucceded at 4652 with 1 tries\nSucceded at 4653 with 1 tries\nSucceded at 4654 with 1 tries\nSucceded at 4655 with 1 tries\nSucceded at 4656 with 1 tries\nSucceded at 4657 with 1 tries\nSucceded at 4658 with 1 tries\nSucceded at 4659 with 1 tries\nSucceded at 4660 with 1 tries\nSucceded at 4663 with 1 tries\nSucceded at 4664 with 1 tries\nSucceded at 4668 with 1 tries\nSucceded at 4669 with 1 tries\nSucceded at 4670 with 1 tries\nSucceded at 4672 with 1 tries\nSucceded at 4673 with 1 tries\nSucceded at 4674 with 1 tries\nSucceded at 4675 with 1 tries\nSucceded at 4676 with 1 tries\nSucceded at 4678 with 1 tries\nSucceded at 4682 with 1 tries\nSucceded at 4683 with 1 tries\nSucceded at 4684 with 1 tries\nSucceded at 4686 with 1 tries\nSucceded at 4688 with 1 tries\nSucceded at 4690 with 1 tries\nSucceded at 4692 with 1 tries\nSucceded at 4693 with 1 tries\nSucceded at 4694 with 1 tries\nSucceded at 4695 with 1 tries\nSucceded at 4697 with 1 tries\nSucceded at 4698 with 1 tries\nSucceded at 4700 with 1 tries\nSucceded at 4701 with 1 tries\nSucceded at 4702 with 1 tries\nSucceded at 4703 with 1 tries\nSucceded at 4704 with 1 tries\nSucceded at 4705 with 1 tries\nSucceded at 4706 with 1 tries\nSucceded at 4707 with 1 tries\nSucceded at 4709 with 1 tries\nSucceded at 4711 with 1 tries\nSucceded at 4713 with 1 tries\nSucceded at 4714 with 1 tries\nSucceded at 4717 with 1 tries\nSucceded at 4718 with 1 tries\nSucceded at 4719 with 1 tries\nSucceded at 4720 with 1 tries\nSucceded at 4723 with 1 tries\nSucceded at 4724 with 1 tries\nSucceded at 4725 with 1 tries\nSucceded at 4726 with 1 tries\nSucceded at 4730 with 1 tries\nSucceded at 4731 with 1 tries\nSucceded at 4732 with 1 tries\nSucceded at 4733 with 1 tries\nSucceded at 4734 with 1 tries\nSucceded at 4738 with 1 tries\nSucceded at 4739 with 1 tries\nSucceded at 4740 with 1 tries\nSucceded at 4741 with 1 tries\nSucceded at 4743 with 1 tries\nSucceded at 4744 with 1 tries\nSucceded at 4745 with 1 tries\nSucceded at 4747 with 1 tries\nSucceded at 4748 with 1 tries\nSucceded at 4749 with 1 tries\nSucceded at 4751 with 1 tries\nSucceded at 4753 with 1 tries\nSucceded at 4754 with 1 tries\nSucceded at 4757 with 1 tries\nSucceded at 4759 with 1 tries\nSucceded at 4767 with 1 tries\nSucceded at 4768 with 1 tries\nSucceded at 4772 with 1 tries\nSucceded at 4774 with 1 tries\nSucceded at 4776 with 1 tries\nSucceded at 4779 with 1 tries\nSucceded at 4780 with 1 tries\nSucceded at 4781 with 1 tries\nSucceded at 4784 with 1 tries\nSucceded at 4787 with 1 tries\nSucceded at 4789 with 1 tries\nSucceded at 4790 with 1 tries\nSucceded at 4794 with 1 tries\nSucceded at 4797 with 1 tries\nSucceded at 4798 with 1 tries\nSucceded at 4799 with 1 tries\nSucceded at 4800 with 1 tries\nSucceded at 4801 with 1 tries\nSucceded at 4804 with 1 tries\nSucceded at 4805 with 1 tries\nSucceded at 4806 with 1 tries\nSucceded at 4809 with 1 tries\nSucceded at 4810 with 1 tries\nSucceded at 4811 with 1 tries\nSucceded at 4812 with 1 tries\nSucceded at 4814 with 1 tries\nSucceded at 4815 with 1 tries\nSucceded at 4816 with 1 tries\nSucceded at 4817 with 1 tries\nSucceded at 4818 with 1 tries\nSucceded at 4819 with 1 tries\nSucceded at 4820 with 1 tries\nSucceded at 4821 with 1 tries\nSucceded at 4823 with 1 tries\nSucceded at 4824 with 1 tries\nSucceded at 4826 with 1 tries\nSucceded at 4828 with 1 tries\nSucceded at 4829 with 1 tries\nSucceded at 4830 with 1 tries\nSucceded at 4832 with 1 tries\nSucceded at 4833 with 1 tries\nSucceded at 4834 with 1 tries\nSucceded at 4835 with 1 tries\nSucceded at 4838 with 1 tries\nSucceded at 4839 with 1 tries\nSucceded at 4840 with 1 tries\nSucceded at 4841 with 1 tries\nSucceded at 4845 with 1 tries\nSucceded at 4847 with 1 tries\nSucceded at 4848 with 1 tries\nSucceded at 4849 with 1 tries\nSucceded at 4850 with 1 tries\nSucceded at 4851 with 1 tries\nSucceded at 4855 with 1 tries\nSucceded at 4856 with 1 tries\nSucceded at 4857 with 1 tries\nSucceded at 4859 with 1 tries\nSucceded at 4862 with 1 tries\nSucceded at 4863 with 1 tries\nSucceded at 4864 with 1 tries\nSucceded at 4865 with 1 tries\nSucceded at 4869 with 1 tries\nSucceded at 4870 with 1 tries\nSucceded at 4873 with 1 tries\nSucceded at 4874 with 1 tries\nSucceded at 4876 with 1 tries\nmosek failed at \nSucceded at 4879 with 1 tries\nSucceded at 4881 with 1 tries\nSucceded at 4882 with 1 tries\nSucceded at 4883 with 1 tries\nSucceded at 4885 with 1 tries\nSucceded at 4886 with 1 tries\nSucceded at 4888 with 1 tries\nSucceded at 4890 with 1 tries\nSucceded at 4891 with 1 tries\nSucceded at 4892 with 1 tries\nSucceded at 4893 with 1 tries\nSucceded at 4894 with 1 tries\nSucceded at 4898 with 1 tries\nSucceded at 4899 with 1 tries\nSucceded at 4901 with 1 tries\nSucceded at 4903 with 1 tries\nSucceded at 4905 with 1 tries\nSucceded at 4906 with 1 tries\nSucceded at 4908 with 1 tries\nSucceded at 4909 with 1 tries\nSucceded at 4911 with 1 tries\nSucceded at 4912 with 1 tries\nSucceded at 4913 with 1 tries\nSucceded at 4914 with 1 tries\nSucceded at 4916 with 1 tries\nSucceded at 4917 with 1 tries\nSucceded at 4919 with 1 tries\nSucceded at 4920 with 1 tries\nSucceded at 4922 with 1 tries\nSucceded at 4923 with 1 tries\nSucceded at 4924 with 1 tries\nSucceded at 4925 with 1 tries\nSucceded at 4926 with 1 tries\nSucceded at 4927 with 1 tries\nSucceded at 4928 with 1 tries\nSucceded at 4929 with 1 tries\nSucceded at 4930 with 1 tries\nSucceded at 4931 with 1 tries\nSucceded at 4932 with 1 tries\nSucceded at 4933 with 1 tries\nSucceded at 4934 with 1 tries\nSucceded at 4935 with 1 tries\nSucceded at 4938 with 1 tries\nSucceded at 4941 with 1 tries\nSucceded at 4942 with 1 tries\nSucceded at 4943 with 1 tries\nSucceded at 4946 with 1 tries\nSucceded at 4947 with 1 tries\nSucceded at 4948 with 1 tries\nSucceded at 4949 with 1 tries\nSucceded at 4950 with 1 tries\nSucceded at 4951 with 1 tries\nSucceded at 4952 with 1 tries\nSucceded at 4954 with 1 tries\nSucceded at 4956 with 1 tries\nSucceded at 4957 with 1 tries\nSucceded at 4959 with 1 tries\nSucceded at 4960 with 1 tries\nSucceded at 4962 with 1 tries\nSucceded at 4963 with 1 tries\nSucceded at 4964 with 1 tries\nSucceded at 4966 with 1 tries\nSucceded at 4967 with 1 tries\nSucceded at 4968 with 1 tries\nSucceded at 4969 with 1 tries\nSucceded at 4970 with 1 tries\nSucceded at 4971 with 1 tries\nSucceded at 4975 with 1 tries\nSucceded at 4976 with 1 tries\nSucceded at 4977 with 1 tries\nSucceded at 4978 with 1 tries\nSucceded at 4979 with 1 tries\nSucceded at 4980 with 1 tries\nSucceded at 4981 with 1 tries\nSucceded at 4982 with 1 tries\nSucceded at 4983 with 1 tries\nSucceded at 4985 with 1 tries\nSucceded at 4986 with 1 tries\nSucceded at 4987 with 1 tries\nSucceded at 4988 with 1 tries\nSucceded at 4991 with 1 tries\nSucceded at 4992 with 1 tries\nSucceded at 4993 with 1 tries\nSucceded at 4994 with 1 tries\nSucceded at 4995 with 1 tries\nSucceded at 4996 with 1 tries\nSucceded at 4997 with 1 tries\nSucceded at 4999 with 1 tries\nSucceded at 5000 with 1 tries\nSucceded at 5002 with 1 tries\nSucceded at 5004 with 1 tries\nSucceded at 5005 with 1 tries\nSucceded at 5006 with 1 tries\nSucceded at 5007 with 1 tries\nSucceded at 5008 with 1 tries\nSucceded at 5009 with 1 tries\nSucceded at 5010 with 1 tries\nSucceded at 5011 with 1 tries\nSucceded at 5012 with 1 tries\nSucceded at 5013 with 1 tries\nSucceded at 5015 with 1 tries\nSucceded at 5016 with 1 tries\nSucceded at 5017 with 1 tries\nSucceded at 5019 with 1 tries\nSucceded at 5020 with 1 tries\nSucceded at 5022 with 1 tries\nSucceded at 5028 with 1 tries\nSucceded at 5029 with 1 tries\nSucceded at 5030 with 1 tries\nSucceded at 5031 with 1 tries\nSucceded at 5032 with 1 tries\nSucceded at 5033 with 1 tries\nSucceded at 5034 with 1 tries\nSucceded at 5036 with 1 tries\nSucceded at 5038 with 1 tries\nSucceded at 5040 with 1 tries\nSucceded at 5041 with 1 tries\nSucceded at 5042 with 1 tries\nSucceded at 5043 with 1 tries\nSucceded at 5044 with 1 tries\nSucceded at 5047 with 1 tries\nSucceded at 5048 with 1 tries\nSucceded at 5050 with 1 tries\nSucceded at 5051 with 1 tries\nSucceded at 5052 with 1 tries\nSucceded at 5053 with 1 tries\nSucceded at 5054 with 1 tries\nSucceded at 5055 with 1 tries\nSucceded at 5056 with 1 tries\nSucceded at 5057 with 1 tries\nSucceded at 5058 with 1 tries\nSucceded at 5059 with 1 tries\nSucceded at 5060 with 1 tries\nSucceded at 5062 with 1 tries\nSucceded at 5063 with 1 tries\nSucceded at 5067 with 1 tries\nSucceded at 5069 with 1 tries\nSucceded at 5070 with 1 tries\nSucceded at 5071 with 1 tries\nSucceded at 5073 with 1 tries\nSucceded at 5074 with 1 tries\nSucceded at 5075 with 1 tries\nSucceded at 5076 with 1 tries\nSucceded at 5081 with 1 tries\nSucceded at 5082 with 1 tries\nSucceded at 5083 with 1 tries\nSucceded at 5084 with 1 tries\nSucceded at 5085 with 1 tries\nSucceded at 5086 with 1 tries\nSucceded at 5089 with 1 tries\nSucceded at 5091 with 1 tries\nSucceded at 5092 with 1 tries\nSucceded at 5093 with 1 tries\nSucceded at 5094 with 1 tries\nSucceded at 5095 with 1 tries\nSucceded at 5096 with 1 tries\nSucceded at 5097 with 1 tries\nSucceded at 5099 with 1 tries\nSucceded at 5101 with 1 tries\nSucceded at 5102 with 1 tries\nSucceded at 5103 with 1 tries\nSucceded at 5105 with 1 tries\nSucceded at 5106 with 1 tries\nSucceded at 5107 with 1 tries\nSucceded at 5108 with 1 tries\nSucceded at 5109 with 1 tries\nSucceded at 5110 with 1 tries\nSucceded at 5112 with 1 tries\nSucceded at 5114 with 1 tries\nSucceded at 5115 with 1 tries\nSucceded at 5116 with 1 tries\nSucceded at 5117 with 1 tries\nSucceded at 5118 with 1 tries\nSucceded at 5119 with 1 tries\nSucceded at 5121 with 1 tries\nSucceded at 5122 with 1 tries\nSucceded at 5123 with 1 tries\nSucceded at 5125 with 1 tries\nSucceded at 5127 with 1 tries\nSucceded at 5129 with 1 tries\nSucceded at 5130 with 1 tries\nSucceded at 5131 with 1 tries\nSucceded at 5133 with 1 tries\nSucceded at 5134 with 1 tries\nSucceded at 5135 with 1 tries\nSucceded at 5136 with 1 tries\nSucceded at 5137 with 1 tries\nSucceded at 5138 with 1 tries\nSucceded at 5139 with 1 tries\nSucceded at 5140 with 1 tries\nSucceded at 5142 with 1 tries\nSucceded at 5144 with 1 tries\nSucceded at 5145 with 1 tries\nSucceded at 5146 with 1 tries\nSucceded at 5148 with 1 tries\nSucceded at 5149 with 1 tries\nSucceded at 5152 with 1 tries\nSucceded at 5155 with 1 tries\nSucceded at 5156 with 1 tries\nSucceded at 5158 with 1 tries\nSucceded at 5159 with 1 tries\nSucceded at 5161 with 1 tries\nSucceded at 5163 with 1 tries\nSucceded at 5164 with 1 tries\nSucceded at 5165 with 1 tries\nSucceded at 5166 with 1 tries\nSucceded at 5167 with 1 tries\nSucceded at 5169 with 1 tries\nSucceded at 5170 with 1 tries\nSucceded at 5172 with 1 tries\nSucceded at 5174 with 1 tries\nSucceded at 5175 with 1 tries\nSucceded at 5178 with 1 tries\nSucceded at 5179 with 1 tries\nSucceded at 5180 with 1 tries\nSucceded at 5182 with 1 tries\nSucceded at 5184 with 1 tries\nSucceded at 5185 with 1 tries\nSucceded at 5187 with 1 tries\nSucceded at 5188 with 1 tries\nSucceded at 5189 with 1 tries\nSucceded at 5192 with 1 tries\nSucceded at 5196 with 1 tries\nSucceded at 5200 with 1 tries\nSucceded at 5201 with 1 tries\nSucceded at 5203 with 1 tries\nSucceded at 5204 with 1 tries\nSucceded at 5205 with 1 tries\nSucceded at 5206 with 1 tries\nSucceded at 5207 with 1 tries\nSucceded at 5208 with 1 tries\nSucceded at 5209 with 1 tries\nSucceded at 5211 with 1 tries\nSucceded at 5212 with 1 tries\nSucceded at 5213 with 1 tries\nSucceded at 5215 with 1 tries\nSucceded at 5216 with 1 tries\nSucceded at 5217 with 1 tries\nSucceded at 5218 with 1 tries\nSucceded at 5219 with 1 tries\nSucceded at 5221 with 1 tries\nSucceded at 5222 with 1 tries\nSucceded at 5223 with 1 tries\nSucceded at 5224 with 1 tries\nSucceded at 5225 with 1 tries\nSucceded at 5226 with 1 tries\nSucceded at 5228 with 1 tries\nSucceded at 5229 with 1 tries\nSucceded at 5230 with 1 tries\nSucceded at 5231 with 1 tries\nSucceded at 5233 with 1 tries\nSucceded at 5234 with 1 tries\nSucceded at 5236 with 1 tries\nSucceded at 5238 with 1 tries\nSucceded at 5239 with 1 tries\nSucceded at 5240 with 1 tries\nSucceded at 5241 with 1 tries\nSucceded at 5242 with 1 tries\nSucceded at 5243 with 1 tries\nSucceded at 5246 with 1 tries\nSucceded at 5247 with 1 tries\nSucceded at 5251 with 1 tries\nSucceded at 5252 with 1 tries\nSucceded at 5254 with 1 tries\nSucceded at 5255 with 1 tries\nSucceded at 5256 with 1 tries\nSucceded at 5258 with 1 tries\nSucceded at 5259 with 1 tries\nSucceded at 5260 with 1 tries\nSucceded at 5261 with 1 tries\nSucceded at 5262 with 1 tries\nSucceded at 5263 with 1 tries\nSucceded at 5266 with 1 tries\nSucceded at 5267 with 1 tries\nSucceded at 5269 with 1 tries\nSucceded at 5270 with 1 tries\nSucceded at 5271 with 1 tries\nSucceded at 5272 with 1 tries\nSucceded at 5275 with 1 tries\nSucceded at 5278 with 1 tries\nSucceded at 5279 with 1 tries\nSucceded at 5280 with 1 tries\nSucceded at 5281 with 1 tries\nSucceded at 5283 with 1 tries\nSucceded at 5286 with 1 tries\nSucceded at 5287 with 1 tries\nSucceded at 5288 with 1 tries\nSucceded at 5289 with 1 tries\nSucceded at 5291 with 1 tries\nSucceded at 5292 with 1 tries\nSucceded at 5293 with 1 tries\nSucceded at 5294 with 1 tries\nSucceded at 5295 with 1 tries\nSucceded at 5296 with 1 tries\nSucceded at 5297 with 1 tries\nSucceded at 5299 with 1 tries\nSucceded at 5301 with 1 tries\nSucceded at 5302 with 1 tries\nSucceded at 5304 with 1 tries\nSucceded at 5306 with 1 tries\nSucceded at 5307 with 1 tries\nSucceded at 5308 with 1 tries\nSucceded at 5311 with 1 tries\nSucceded at 5312 with 1 tries\nSucceded at 5313 with 1 tries\nSucceded at 5314 with 1 tries\nSucceded at 5315 with 1 tries\nSucceded at 5317 with 1 tries\nSucceded at 5318 with 1 tries\nSucceded at 5319 with 1 tries\nSucceded at 5320 with 1 tries\nSucceded at 5322 with 1 tries\nSucceded at 5324 with 1 tries\nSucceded at 5325 with 1 tries\nSucceded at 5328 with 1 tries\nSucceded at 5329 with 1 tries\nSucceded at 5330 with 1 tries\nSucceded at 5331 with 1 tries\nSucceded at 5332 with 1 tries\nSucceded at 5333 with 1 tries\nSucceded at 5334 with 1 tries\nSucceded at 5337 with 1 tries\nSucceded at 5338 with 1 tries\nSucceded at 5339 with 1 tries\nSucceded at 5340 with 1 tries\nSucceded at 5341 with 1 tries\nSucceded at 5342 with 1 tries\nSucceded at 5345 with 1 tries\nSucceded at 5347 with 1 tries\nSucceded at 5348 with 1 tries\nSucceded at 5349 with 1 tries\nSucceded at 5350 with 1 tries\nSucceded at 5352 with 1 tries\nSucceded at 5353 with 1 tries\nSucceded at 5354 with 1 tries\nSucceded at 5355 with 1 tries\nSucceded at 5356 with 1 tries\nSucceded at 5357 with 1 tries\nSucceded at 5359 with 1 tries\nSucceded at 5362 with 1 tries\nSucceded at 5364 with 1 tries\nSucceded at 5366 with 1 tries\nSucceded at 5367 with 1 tries\nSucceded at 5368 with 1 tries\nSucceded at 5369 with 1 tries\nSucceded at 5370 with 1 tries\nSucceded at 5371 with 1 tries\nSucceded at 5372 with 1 tries\nSucceded at 5373 with 1 tries\nSucceded at 5374 with 1 tries\nSucceded at 5376 with 1 tries\nSucceded at 5377 with 1 tries\nSucceded at 5378 with 1 tries\nSucceded at 5380 with 1 tries\nSucceded at 5381 with 1 tries\nSucceded at 5383 with 1 tries\nSucceded at 5384 with 1 tries\nSucceded at 5385 with 1 tries\nSucceded at 5386 with 1 tries\nSucceded at 5387 with 1 tries\nSucceded at 5388 with 1 tries\nSucceded at 5389 with 1 tries\nSucceded at 5390 with 1 tries\nSucceded at 5391 with 1 tries\nSucceded at 5392 with 1 tries\nSucceded at 5394 with 1 tries\nSucceded at 5395 with 1 tries\nSucceded at 5396 with 1 tries\nSucceded at 5397 with 1 tries\nSucceded at 5399 with 1 tries\nSucceded at 5400 with 1 tries\nSucceded at 5401 with 1 tries\nSucceded at 5402 with 1 tries\nSucceded at 5403 with 1 tries\nSucceded at 5404 with 1 tries\nSucceded at 5406 with 1 tries\nSucceded at 5407 with 1 tries\nSucceded at 5408 with 1 tries\nSucceded at 5409 with 1 tries\nSucceded at 5410 with 1 tries\nSucceded at 5413 with 1 tries\nSucceded at 5414 with 1 tries\nSucceded at 5416 with 1 tries\nSucceded at 5419 with 1 tries\nSucceded at 5420 with 1 tries\nSucceded at 5421 with 1 tries\nSucceded at 5424 with 1 tries\nSucceded at 5425 with 1 tries\nSucceded at 5426 with 1 tries\nSucceded at 5427 with 1 tries\nSucceded at 5428 with 1 tries\nSucceded at 5432 with 1 tries\nSucceded at 5433 with 1 tries\nSucceded at 5434 with 1 tries\nSucceded at 5437 with 1 tries\nSucceded at 5438 with 1 tries\nSucceded at 5439 with 1 tries\nSucceded at 5440 with 1 tries\nSucceded at 5441 with 1 tries\nSucceded at 5443 with 1 tries\nSucceded at 5444 with 1 tries\nSucceded at 5445 with 1 tries\nSucceded at 5447 with 1 tries\nSucceded at 5451 with 1 tries\nSucceded at 5452 with 1 tries\nSucceded at 5455 with 1 tries\nSucceded at 5457 with 1 tries\nSucceded at 5459 with 1 tries\nSucceded at 5460 with 1 tries\nSucceded at 5461 with 1 tries\nSucceded at 5462 with 1 tries\nSucceded at 5463 with 1 tries\nSucceded at 5465 with 1 tries\nSucceded at 5467 with 1 tries\nSucceded at 5468 with 1 tries\nSucceded at 5469 with 1 tries\nSucceded at 5470 with 1 tries\nSucceded at 5471 with 1 tries\nSucceded at 5472 with 1 tries\nSucceded at 5473 with 1 tries\nSucceded at 5477 with 1 tries\nSucceded at 5478 with 1 tries\nSucceded at 5479 with 1 tries\nSucceded at 5482 with 1 tries\nSucceded at 5483 with 1 tries\nSucceded at 5486 with 1 tries\nSucceded at 5487 with 1 tries\nSucceded at 5490 with 1 tries\nSucceded at 5491 with 1 tries\nSucceded at 5493 with 1 tries\nSucceded at 5494 with 1 tries\nSucceded at 5495 with 1 tries\nSucceded at 5496 with 1 tries\nSucceded at 5497 with 1 tries\nSucceded at 5498 with 1 tries\nSucceded at 5499 with 1 tries\nSucceded at 5501 with 1 tries\nSucceded at 5502 with 1 tries\nSucceded at 5503 with 1 tries\nSucceded at 5505 with 1 tries\nSucceded at 5506 with 1 tries\nSucceded at 5507 with 1 tries\nSucceded at 5508 with 1 tries\nSucceded at 5509 with 1 tries\nSucceded at 5510 with 1 tries\nSucceded at 5513 with 1 tries\nSucceded at 5514 with 1 tries\nSucceded at 5515 with 1 tries\nSucceded at 5516 with 1 tries\nSucceded at 5517 with 1 tries\nSucceded at 5518 with 1 tries\nSucceded at 5519 with 1 tries\nSucceded at 5521 with 1 tries\nSucceded at 5522 with 1 tries\nSucceded at 5524 with 1 tries\nSucceded at 5525 with 1 tries\nSucceded at 5527 with 1 tries\nSucceded at 5529 with 1 tries\nSucceded at 5531 with 1 tries\nSucceded at 5532 with 1 tries\nSucceded at 5534 with 1 tries\nSucceded at 5536 with 1 tries\nSucceded at 5537 with 1 tries\nSucceded at 5538 with 1 tries\nSucceded at 5540 with 1 tries\nSucceded at 5541 with 1 tries\nSucceded at 5544 with 1 tries\nSucceded at 5545 with 1 tries\nSucceded at 5546 with 1 tries\nSucceded at 5547 with 1 tries\nSucceded at 5548 with 1 tries\nSucceded at 5549 with 1 tries\nSucceded at 5551 with 1 tries\nSucceded at 5553 with 1 tries\nSucceded at 5556 with 1 tries\nSucceded at 5558 with 1 tries\nSucceded at 5559 with 1 tries\nSucceded at 5560 with 1 tries\nSucceded at 5561 with 1 tries\nSucceded at 5563 with 1 tries\nSucceded at 5564 with 1 tries\nSucceded at 5566 with 1 tries\nSucceded at 5567 with 1 tries\nSucceded at 5568 with 1 tries\nSucceded at 5570 with 1 tries\nSucceded at 5571 with 1 tries\nSucceded at 5572 with 1 tries\nSucceded at 5573 with 1 tries\nSucceded at 5576 with 1 tries\nSucceded at 5578 with 1 tries\nSucceded at 5580 with 1 tries\nSucceded at 5581 with 1 tries\nSucceded at 5582 with 1 tries\nSucceded at 5583 with 1 tries\nSucceded at 5584 with 1 tries\nSucceded at 5585 with 1 tries\nSucceded at 5588 with 1 tries\nSucceded at 5589 with 1 tries\nSucceded at 5590 with 1 tries\nSucceded at 5591 with 1 tries\nSucceded at 5592 with 1 tries\nSucceded at 5593 with 1 tries\nSucceded at 5594 with 1 tries\nSucceded at 5595 with 1 tries\nSucceded at 5596 with 1 tries\nSucceded at 5597 with 1 tries\nSucceded at 5601 with 1 tries\nSucceded at 5602 with 1 tries\nSucceded at 5605 with 1 tries\nSucceded at 5606 with 1 tries\nSucceded at 5607 with 1 tries\nSucceded at 5608 with 1 tries\nSucceded at 5609 with 1 tries\nSucceded at 5612 with 1 tries\nSucceded at 5613 with 1 tries\nSucceded at 5614 with 1 tries\nSucceded at 5615 with 1 tries\nSucceded at 5616 with 1 tries\nSucceded at 5617 with 1 tries\nSucceded at 5618 with 1 tries\nSucceded at 5619 with 1 tries\nSucceded at 5620 with 1 tries\nSucceded at 5621 with 1 tries\nSucceded at 5622 with 1 tries\nSucceded at 5623 with 1 tries\nSucceded at 5624 with 1 tries\nSucceded at 5625 with 1 tries\nSucceded at 5627 with 1 tries\nSucceded at 5628 with 1 tries\nSucceded at 5630 with 1 tries\nSucceded at 5631 with 1 tries\nSucceded at 5632 with 1 tries\nSucceded at 5633 with 1 tries\nSucceded at 5634 with 1 tries\nSucceded at 5635 with 1 tries\nSucceded at 5636 with 1 tries\nSucceded at 5637 with 1 tries\nSucceded at 5639 with 1 tries\nSucceded at 5642 with 1 tries\nSucceded at 5643 with 1 tries\nSucceded at 5644 with 1 tries\nSucceded at 5645 with 1 tries\nSucceded at 5646 with 1 tries\nSucceded at 5647 with 1 tries\nSucceded at 5648 with 1 tries\nSucceded at 5649 with 1 tries\nSucceded at 5650 with 1 tries\nSucceded at 5652 with 1 tries\nSucceded at 5654 with 1 tries\nSucceded at 5655 with 1 tries\nSucceded at 5657 with 1 tries\nSucceded at 5658 with 1 tries\nSucceded at 5659 with 1 tries\nSucceded at 5662 with 1 tries\nSucceded at 5663 with 1 tries\nSucceded at 5666 with 1 tries\nSucceded at 5668 with 1 tries\nSucceded at 5670 with 1 tries\nSucceded at 5671 with 1 tries\nSucceded at 5672 with 1 tries\nSucceded at 5673 with 1 tries\nSucceded at 5674 with 1 tries\nSucceded at 5675 with 1 tries\nSucceded at 5681 with 1 tries\nSucceded at 5682 with 1 tries\nSucceded at 5684 with 1 tries\nSucceded at 5685 with 1 tries\nSucceded at 5686 with 1 tries\nSucceded at 5687 with 1 tries\nSucceded at 5688 with 1 tries\nSucceded at 5689 with 1 tries\nSucceded at 5690 with 1 tries\nSucceded at 5692 with 1 tries\nSucceded at 5693 with 1 tries\nSucceded at 5694 with 1 tries\nSucceded at 5695 with 1 tries\nSucceded at 5696 with 1 tries\nSucceded at 5697 with 1 tries\nSucceded at 5698 with 1 tries\nSucceded at 5700 with 1 tries\nSucceded at 5702 with 1 tries\nSucceded at 5705 with 1 tries\nSucceded at 5706 with 1 tries\nSucceded at 5707 with 1 tries\nSucceded at 5708 with 1 tries\nSucceded at 5711 with 1 tries\nSucceded at 5712 with 1 tries\nSucceded at 5715 with 1 tries\nSucceded at 5717 with 1 tries\nSucceded at 5720 with 1 tries\nSucceded at 5722 with 1 tries\nSucceded at 5723 with 1 tries\nSucceded at 5724 with 1 tries\nSucceded at 5726 with 1 tries\nSucceded at 5727 with 1 tries\nSucceded at 5731 with 1 tries\nSucceded at 5733 with 1 tries\nSucceded at 5736 with 1 tries\nSucceded at 5738 with 1 tries\nSucceded at 5739 with 1 tries\nSucceded at 5740 with 1 tries\nSucceded at 5741 with 1 tries\nSucceded at 5742 with 1 tries\nSucceded at 5743 with 1 tries\nSucceded at 5745 with 1 tries\nSucceded at 5747 with 1 tries\nSucceded at 5748 with 1 tries\nSucceded at 5749 with 1 tries\nSucceded at 5752 with 1 tries\nSucceded at 5753 with 1 tries\nSucceded at 5755 with 1 tries\nSucceded at 5756 with 1 tries\nSucceded at 5758 with 1 tries\nSucceded at 5759 with 1 tries\nSucceded at 5760 with 1 tries\nSucceded at 5762 with 1 tries\nSucceded at 5763 with 1 tries\nSucceded at 5764 with 1 tries\nSucceded at 5765 with 1 tries\nSucceded at 5766 with 1 tries\nSucceded at 5769 with 1 tries\nSucceded at 5770 with 1 tries\nSucceded at 5771 with 1 tries\nSucceded at 5772 with 1 tries\nSucceded at 5773 with 1 tries\nSucceded at 5775 with 1 tries\nSucceded at 5777 with 1 tries\nSucceded at 5779 with 1 tries\nSucceded at 5781 with 1 tries\nSucceded at 5782 with 1 tries\nSucceded at 5783 with 1 tries\nSucceded at 5784 with 1 tries\nSucceded at 5785 with 1 tries\nSucceded at 5786 with 1 tries\nSucceded at 5787 with 1 tries\nSucceded at 5789 with 1 tries\nSucceded at 5790 with 1 tries\nSucceded at 5791 with 1 tries\nSucceded at 5793 with 1 tries\nSucceded at 5795 with 1 tries\nSucceded at 5799 with 1 tries\nSucceded at 5800 with 1 tries\nSucceded at 5804 with 1 tries\nSucceded at 5805 with 1 tries\nSucceded at 5806 with 1 tries\nSucceded at 5809 with 1 tries\nSucceded at 5810 with 1 tries\nSucceded at 5811 with 1 tries\nSucceded at 5812 with 1 tries\nSucceded at 5814 with 1 tries\nSucceded at 5815 with 1 tries\nSucceded at 5816 with 1 tries\nSucceded at 5817 with 1 tries\nSucceded at 5818 with 1 tries\nSucceded at 5819 with 1 tries\nSucceded at 5821 with 1 tries\nSucceded at 5822 with 1 tries\nSucceded at 5823 with 1 tries\nSucceded at 5826 with 1 tries\nSucceded at 5828 with 1 tries\nSucceded at 5829 with 1 tries\nSucceded at 5831 with 1 tries\nSucceded at 5832 with 1 tries\nSucceded at 5834 with 1 tries\nSucceded at 5836 with 1 tries\nSucceded at 5837 with 1 tries\nSucceded at 5839 with 1 tries\nSucceded at 5840 with 1 tries\nSucceded at 5842 with 1 tries\nSucceded at 5843 with 1 tries\nSucceded at 5844 with 1 tries\nSucceded at 5846 with 1 tries\nSucceded at 5847 with 1 tries\nSucceded at 5848 with 1 tries\nSucceded at 5849 with 1 tries\nSucceded at 5850 with 1 tries\nSucceded at 5851 with 1 tries\nSucceded at 5854 with 1 tries\nSucceded at 5855 with 1 tries\nSucceded at 5857 with 1 tries\nSucceded at 5858 with 1 tries\nSucceded at 5859 with 1 tries\nSucceded at 5860 with 1 tries\nSucceded at 5863 with 1 tries\nSucceded at 5865 with 1 tries\nSucceded at 5867 with 1 tries\nSucceded at 5868 with 1 tries\nSucceded at 5869 with 1 tries\nSucceded at 5870 with 1 tries\nSucceded at 5872 with 1 tries\nSucceded at 5875 with 1 tries\nSucceded at 5876 with 1 tries\nSucceded at 5877 with 1 tries\nSucceded at 5879 with 1 tries\nSucceded at 5880 with 1 tries\nSucceded at 5881 with 1 tries\nSucceded at 5882 with 1 tries\nSucceded at 5883 with 1 tries\nSucceded at 5884 with 1 tries\nSucceded at 5885 with 1 tries\nSucceded at 5886 with 1 tries\nSucceded at 5887 with 1 tries\nSucceded at 5888 with 1 tries\nSucceded at 5890 with 1 tries\nSucceded at 5892 with 1 tries\nSucceded at 5893 with 1 tries\nSucceded at 5894 with 1 tries\nSucceded at 5896 with 1 tries\nSucceded at 5897 with 1 tries\nSucceded at 5898 with 1 tries\nSucceded at 5899 with 1 tries\nSucceded at 5901 with 1 tries\nSucceded at 5903 with 1 tries\nSucceded at 5904 with 1 tries\nSucceded at 5905 with 1 tries\nSucceded at 5906 with 1 tries\nSucceded at 5907 with 1 tries\nSucceded at 5908 with 1 tries\nSucceded at 5909 with 1 tries\nSucceded at 5911 with 1 tries\nSucceded at 5912 with 1 tries\nSucceded at 5913 with 1 tries\nSucceded at 5915 with 1 tries\nSucceded at 5916 with 1 tries\nSucceded at 5917 with 1 tries\nSucceded at 5918 with 1 tries\nSucceded at 5919 with 1 tries\nSucceded at 5920 with 1 tries\nSucceded at 5921 with 1 tries\nSucceded at 5922 with 1 tries\nSucceded at 5923 with 1 tries\nSucceded at 5926 with 1 tries\nSucceded at 5927 with 1 tries\nSucceded at 5928 with 1 tries\nSucceded at 5929 with 1 tries\nSucceded at 5930 with 1 tries\nSucceded at 5931 with 1 tries\nSucceded at 5936 with 1 tries\nSucceded at 5937 with 1 tries\nSucceded at 5938 with 1 tries\nSucceded at 5939 with 1 tries\nSucceded at 5940 with 1 tries\nSucceded at 5941 with 1 tries\nSucceded at 5942 with 1 tries\nSucceded at 5943 with 1 tries\nSucceded at 5945 with 1 tries\nSucceded at 5946 with 1 tries\nSucceded at 5947 with 1 tries\nSucceded at 5948 with 1 tries\nSucceded at 5949 with 1 tries\nSucceded at 5951 with 1 tries\nSucceded at 5953 with 1 tries\nSucceded at 5954 with 1 tries\nSucceded at 5956 with 1 tries\nSucceded at 5958 with 1 tries\nSucceded at 5959 with 1 tries\nSucceded at 5960 with 1 tries\nSucceded at 5961 with 1 tries\nSucceded at 5962 with 1 tries\nSucceded at 5963 with 1 tries\nSucceded at 5964 with 1 tries\nSucceded at 5965 with 1 tries\nSucceded at 5966 with 1 tries\nSucceded at 5967 with 1 tries\nSucceded at 5968 with 1 tries\nSucceded at 5969 with 1 tries\nSucceded at 5970 with 1 tries\nSucceded at 5971 with 1 tries\nSucceded at 5972 with 1 tries\nSucceded at 5973 with 1 tries\nSucceded at 5974 with 1 tries\nSucceded at 5980 with 1 tries\nSucceded at 5981 with 1 tries\nSucceded at 5982 with 1 tries\nSucceded at 5984 with 1 tries\nSucceded at 5985 with 1 tries\nSucceded at 5988 with 1 tries\nSucceded at 5989 with 1 tries\nSucceded at 5990 with 1 tries\nSucceded at 5992 with 1 tries\nSucceded at 5993 with 1 tries\nSucceded at 5994 with 1 tries\nSucceded at 5995 with 1 tries\nSucceded at 5997 with 1 tries\nSucceded at 5998 with 1 tries\nSucceded at 5999 with 1 tries\nSucceded at 6000 with 1 tries\nSucceded at 6001 with 1 tries\nSucceded at 6003 with 1 tries\nSucceded at 6005 with 1 tries\nSucceded at 6006 with 1 tries\nSucceded at 6007 with 1 tries\nSucceded at 6009 with 1 tries\nSucceded at 6010 with 1 tries\nSucceded at 6011 with 1 tries\nSucceded at 6012 with 1 tries\nSucceded at 6013 with 1 tries\nSucceded at 6014 with 1 tries\nSucceded at 6015 with 1 tries\nSucceded at 6016 with 1 tries\nSucceded at 6017 with 1 tries\nSucceded at 6018 with 1 tries\nSucceded at 6023 with 1 tries\nSucceded at 6025 with 1 tries\nSucceded at 6027 with 1 tries\nSucceded at 6028 with 1 tries\nSucceded at 6029 with 1 tries\nSucceded at 6030 with 1 tries\nSucceded at 6031 with 1 tries\nSucceded at 6033 with 1 tries\nSucceded at 6035 with 1 tries\nSucceded at 6036 with 1 tries\nSucceded at 6037 with 1 tries\nSucceded at 6038 with 1 tries\nSucceded at 6041 with 1 tries\nSucceded at 6042 with 1 tries\nSucceded at 6045 with 1 tries\nSucceded at 6046 with 1 tries\nSucceded at 6047 with 1 tries\nSucceded at 6048 with 1 tries\nSucceded at 6050 with 1 tries\nSucceded at 6051 with 1 tries\nSucceded at 6053 with 1 tries\nSucceded at 6054 with 1 tries\nSucceded at 6055 with 1 tries\nSucceded at 6056 with 1 tries\nSucceded at 6057 with 1 tries\nSucceded at 6058 with 1 tries\nSucceded at 6059 with 1 tries\nSucceded at 6060 with 1 tries\nSucceded at 6061 with 1 tries\nSucceded at 6062 with 1 tries\nSucceded at 6063 with 1 tries\nSucceded at 6064 with 1 tries\nSucceded at 6065 with 1 tries\nSucceded at 6066 with 1 tries\nSucceded at 6068 with 1 tries\nSucceded at 6070 with 1 tries\nSucceded at 6072 with 1 tries\nSucceded at 6074 with 1 tries\nSucceded at 6076 with 1 tries\nSucceded at 6077 with 1 tries\nSucceded at 6079 with 1 tries\nSucceded at 6082 with 1 tries\nSucceded at 6084 with 1 tries\nSucceded at 6087 with 1 tries\nSucceded at 6088 with 1 tries\nSucceded at 6089 with 1 tries\nSucceded at 6090 with 1 tries\nSucceded at 6091 with 1 tries\nSucceded at 6092 with 1 tries\nSucceded at 6093 with 1 tries\nSucceded at 6094 with 1 tries\nSucceded at 6096 with 1 tries\nSucceded at 6100 with 1 tries\nSucceded at 6101 with 1 tries\nSucceded at 6103 with 1 tries\nSucceded at 6104 with 1 tries\nSucceded at 6105 with 1 tries\nSucceded at 6107 with 1 tries\nSucceded at 6108 with 1 tries\nSucceded at 6110 with 1 tries\nSucceded at 6111 with 1 tries\nSucceded at 6112 with 1 tries\nSucceded at 6115 with 1 tries\nSucceded at 6116 with 1 tries\nSucceded at 6117 with 1 tries\nSucceded at 6118 with 1 tries\nSucceded at 6121 with 1 tries\nSucceded at 6122 with 1 tries\nSucceded at 6123 with 1 tries\nSucceded at 6125 with 1 tries\nSucceded at 6126 with 1 tries\nSucceded at 6127 with 1 tries\nSucceded at 6129 with 1 tries\nSucceded at 6130 with 1 tries\nSucceded at 6133 with 1 tries\nSucceded at 6134 with 1 tries\nSucceded at 6137 with 1 tries\nSucceded at 6138 with 1 tries\nSucceded at 6139 with 1 tries\nSucceded at 6140 with 1 tries\nSucceded at 6141 with 1 tries\nSucceded at 6143 with 1 tries\nSucceded at 6146 with 1 tries\nSucceded at 6148 with 1 tries\nSucceded at 6149 with 1 tries\nSucceded at 6150 with 1 tries\nSucceded at 6151 with 1 tries\nSucceded at 6152 with 1 tries\nSucceded at 6153 with 1 tries\nSucceded at 6156 with 1 tries\nSucceded at 6157 with 1 tries\nSucceded at 6159 with 1 tries\nSucceded at 6161 with 1 tries\nSucceded at 6162 with 1 tries\nSucceded at 6163 with 1 tries\nSucceded at 6164 with 1 tries\nSucceded at 6165 with 1 tries\nSucceded at 6167 with 1 tries\nSucceded at 6168 with 1 tries\nSucceded at 6169 with 1 tries\nSucceded at 6170 with 1 tries\nSucceded at 6172 with 1 tries\nSucceded at 6173 with 1 tries\nSucceded at 6174 with 1 tries\nSucceded at 6177 with 1 tries\nSucceded at 6178 with 1 tries\nSucceded at 6179 with 1 tries\nSucceded at 6181 with 1 tries\nSucceded at 6183 with 1 tries\nSucceded at 6184 with 1 tries\nSucceded at 6185 with 1 tries\nSucceded at 6188 with 1 tries\nSucceded at 6190 with 1 tries\nSucceded at 6192 with 1 tries\nSucceded at 6195 with 1 tries\nSucceded at 6196 with 1 tries\nSucceded at 6197 with 1 tries\nSucceded at 6198 with 1 tries\nSucceded at 6199 with 1 tries\nSucceded at 6200 with 1 tries\nSucceded at 6201 with 1 tries\nSucceded at 6202 with 1 tries\nSucceded at 6203 with 1 tries\nSucceded at 6207 with 1 tries\nSucceded at 6208 with 1 tries\nSucceded at 6209 with 1 tries\nSucceded at 6210 with 1 tries\nSucceded at 6211 with 1 tries\nSucceded at 6213 with 1 tries\nSucceded at 6214 with 1 tries\nSucceded at 6215 with 1 tries\nSucceded at 6218 with 1 tries\nSucceded at 6219 with 1 tries\nSucceded at 6220 with 1 tries\nSucceded at 6221 with 1 tries\nSucceded at 6223 with 1 tries\nSucceded at 6224 with 1 tries\nSucceded at 6225 with 1 tries\nSucceded at 6226 with 1 tries\nSucceded at 6228 with 1 tries\nSucceded at 6230 with 1 tries\nSucceded at 6232 with 1 tries\nSucceded at 6233 with 1 tries\nSucceded at 6234 with 1 tries\nSucceded at 6235 with 1 tries\nSucceded at 6236 with 1 tries\nSucceded at 6238 with 1 tries\nSucceded at 6240 with 1 tries\nSucceded at 6242 with 1 tries\nSucceded at 6243 with 1 tries\nSucceded at 6244 with 1 tries\nSucceded at 6246 with 1 tries\nSucceded at 6247 with 1 tries\nSucceded at 6249 with 1 tries\nSucceded at 6251 with 1 tries\nSucceded at 6253 with 1 tries\nSucceded at 6254 with 1 tries\nSucceded at 6255 with 1 tries\nSucceded at 6257 with 1 tries\nSucceded at 6259 with 1 tries\nSucceded at 6261 with 1 tries\nSucceded at 6262 with 1 tries\nSucceded at 6263 with 1 tries\nSucceded at 6264 with 1 tries\nSucceded at 6265 with 1 tries\nSucceded at 6266 with 1 tries\nSucceded at 6267 with 1 tries\nSucceded at 6270 with 1 tries\nSucceded at 6272 with 1 tries\nSucceded at 6273 with 1 tries\nSucceded at 6274 with 1 tries\nSucceded at 6277 with 1 tries\nSucceded at 6279 with 1 tries\nSucceded at 6282 with 1 tries\nSucceded at 6283 with 1 tries\nSucceded at 6284 with 1 tries\nSucceded at 6285 with 1 tries\nSucceded at 6287 with 1 tries\nSucceded at 6292 with 1 tries\nSucceded at 6293 with 1 tries\nSucceded at 6294 with 1 tries\nSucceded at 6296 with 1 tries\nSucceded at 6297 with 1 tries\nSucceded at 6299 with 1 tries\nSucceded at 6301 with 1 tries\nSucceded at 6302 with 1 tries\nSucceded at 6303 with 1 tries\nSucceded at 6304 with 1 tries\nSucceded at 6305 with 1 tries\nSucceded at 6306 with 1 tries\nSucceded at 6307 with 1 tries\nSucceded at 6309 with 1 tries\nSucceded at 6310 with 1 tries\nSucceded at 6312 with 1 tries\nSucceded at 6314 with 1 tries\nSucceded at 6315 with 1 tries\nSucceded at 6316 with 1 tries\nSucceded at 6317 with 1 tries\nSucceded at 6318 with 1 tries\nSucceded at 6319 with 1 tries\nSucceded at 6320 with 1 tries\nSucceded at 6321 with 1 tries\nSucceded at 6323 with 1 tries\nSucceded at 6324 with 1 tries\nSucceded at 6325 with 1 tries\nSucceded at 6326 with 1 tries\nSucceded at 6327 with 1 tries\nSucceded at 6328 with 1 tries\nSucceded at 6329 with 1 tries\nSucceded at 6330 with 1 tries\nSucceded at 6331 with 1 tries\nSucceded at 6332 with 1 tries\nSucceded at 6333 with 1 tries\nSucceded at 6334 with 1 tries\nSucceded at 6335 with 1 tries\nSucceded at 6337 with 1 tries\nSucceded at 6338 with 1 tries\nSucceded at 6340 with 1 tries\nSucceded at 6341 with 1 tries\nSucceded at 6342 with 1 tries\nSucceded at 6343 with 1 tries\nSucceded at 6344 with 1 tries\nSucceded at 6345 with 1 tries\nSucceded at 6347 with 1 tries\nSucceded at 6349 with 1 tries\nSucceded at 6350 with 1 tries\nSucceded at 6351 with 1 tries\nSucceded at 6352 with 1 tries\nSucceded at 6354 with 1 tries\nSucceded at 6355 with 1 tries\nSucceded at 6356 with 1 tries\nSucceded at 6357 with 1 tries\nSucceded at 6359 with 1 tries\nSucceded at 6360 with 1 tries\nSucceded at 6362 with 1 tries\nSucceded at 6363 with 1 tries\nSucceded at 6365 with 1 tries\nSucceded at 6366 with 1 tries\nSucceded at 6367 with 1 tries\nSucceded at 6368 with 1 tries\nSucceded at 6369 with 1 tries\nSucceded at 6370 with 1 tries\nSucceded at 6371 with 1 tries\nSucceded at 6372 with 1 tries\nSucceded at 6373 with 1 tries\nSucceded at 6374 with 1 tries\nSucceded at 6376 with 1 tries\nSucceded at 6377 with 1 tries\nSucceded at 6378 with 1 tries\nSucceded at 6380 with 1 tries\nSucceded at 6381 with 1 tries\nSucceded at 6383 with 1 tries\nSucceded at 6384 with 1 tries\nSucceded at 6386 with 1 tries\nSucceded at 6388 with 1 tries\nSucceded at 6391 with 1 tries\nSucceded at 6392 with 1 tries\nSucceded at 6393 with 1 tries\nSucceded at 6395 with 1 tries\nSucceded at 6397 with 1 tries\nSucceded at 6399 with 1 tries\nSucceded at 6401 with 1 tries\nSucceded at 6403 with 1 tries\nSucceded at 6405 with 1 tries\nSucceded at 6406 with 1 tries\nSucceded at 6407 with 1 tries\nSucceded at 6408 with 1 tries\nSucceded at 6409 with 1 tries\nSucceded at 6410 with 1 tries\nSucceded at 6412 with 1 tries\nSucceded at 6416 with 1 tries\nSucceded at 6417 with 1 tries\nSucceded at 6418 with 1 tries\nSucceded at 6420 with 1 tries\nSucceded at 6421 with 1 tries\nSucceded at 6422 with 1 tries\nSucceded at 6423 with 1 tries\nSucceded at 6424 with 1 tries\nSucceded at 6425 with 1 tries\nSucceded at 6427 with 1 tries\nSucceded at 6428 with 1 tries\nSucceded at 6429 with 1 tries\nSucceded at 6430 with 1 tries\nSucceded at 6431 with 1 tries\nSucceded at 6432 with 1 tries\nSucceded at 6433 with 1 tries\nSucceded at 6434 with 1 tries\nSucceded at 6436 with 1 tries\nSucceded at 6437 with 1 tries\nSucceded at 6440 with 1 tries\nSucceded at 6441 with 1 tries\nSucceded at 6444 with 1 tries\nSucceded at 6445 with 1 tries\nSucceded at 6446 with 1 tries\nSucceded at 6447 with 1 tries\nSucceded at 6449 with 1 tries\nSucceded at 6450 with 1 tries\nSucceded at 6451 with 1 tries\nSucceded at 6452 with 1 tries\nSucceded at 6453 with 1 tries\nSucceded at 6458 with 1 tries\nSucceded at 6463 with 1 tries\nSucceded at 6464 with 1 tries\nSucceded at 6466 with 1 tries\nSucceded at 6467 with 1 tries\nSucceded at 6468 with 1 tries\nSucceded at 6469 with 1 tries\nSucceded at 6470 with 1 tries\nSucceded at 6472 with 1 tries\nSucceded at 6474 with 1 tries\nSucceded at 6476 with 1 tries\nSucceded at 6477 with 1 tries\nSucceded at 6478 with 1 tries\nSucceded at 6479 with 1 tries\nSucceded at 6480 with 1 tries\nSucceded at 6482 with 1 tries\nSucceded at 6485 with 1 tries\nSucceded at 6486 with 1 tries\nSucceded at 6487 with 1 tries\nSucceded at 6488 with 1 tries\nSucceded at 6490 with 1 tries\nSucceded at 6491 with 1 tries\nmosek failed at \nSucceded at 6493 with 1 tries\nSucceded at 6494 with 1 tries\nSucceded at 6495 with 1 tries\nSucceded at 6497 with 1 tries\nSucceded at 6498 with 1 tries\nSucceded at 6500 with 1 tries\nSucceded at 6502 with 1 tries\nSucceded at 6504 with 1 tries\nSucceded at 6505 with 1 tries\nSucceded at 6508 with 1 tries\nSucceded at 6509 with 1 tries\nSucceded at 6510 with 1 tries\nSucceded at 6512 with 1 tries\nSucceded at 6513 with 1 tries\nSucceded at 6514 with 1 tries\nSucceded at 6516 with 1 tries\nSucceded at 6517 with 1 tries\nSucceded at 6520 with 1 tries\nSucceded at 6523 with 1 tries\nSucceded at 6524 with 1 tries\nSucceded at 6525 with 1 tries\nSucceded at 6526 with 1 tries\nSucceded at 6527 with 1 tries\nSucceded at 6528 with 1 tries\nSucceded at 6529 with 1 tries\nSucceded at 6531 with 1 tries\nSucceded at 6534 with 1 tries\nSucceded at 6535 with 1 tries\nSucceded at 6536 with 1 tries\nSucceded at 6538 with 1 tries\nSucceded at 6539 with 1 tries\nSucceded at 6541 with 1 tries\nSucceded at 6542 with 1 tries\nSucceded at 6543 with 1 tries\nSucceded at 6544 with 1 tries\nSucceded at 6545 with 1 tries\nSucceded at 6546 with 1 tries\nSucceded at 6547 with 1 tries\nSucceded at 6549 with 1 tries\nSucceded at 6550 with 1 tries\nSucceded at 6551 with 1 tries\nSucceded at 6553 with 1 tries\nSucceded at 6554 with 1 tries\nSucceded at 6556 with 1 tries\nSucceded at 6557 with 1 tries\nSucceded at 6560 with 1 tries\nSucceded at 6564 with 1 tries\nSucceded at 6565 with 1 tries\nSucceded at 6568 with 1 tries\nSucceded at 6569 with 1 tries\nSucceded at 6570 with 1 tries\nSucceded at 6571 with 1 tries\nSucceded at 6572 with 1 tries\nSucceded at 6573 with 1 tries\nSucceded at 6575 with 1 tries\nSucceded at 6576 with 1 tries\nSucceded at 6577 with 1 tries\nSucceded at 6579 with 1 tries\nSucceded at 6581 with 1 tries\nSucceded at 6582 with 1 tries\nSucceded at 6583 with 1 tries\nSucceded at 6584 with 1 tries\nSucceded at 6586 with 1 tries\nSucceded at 6587 with 1 tries\nSucceded at 6588 with 1 tries\nSucceded at 6589 with 1 tries\nSucceded at 6590 with 1 tries\nSucceded at 6592 with 1 tries\nSucceded at 6596 with 1 tries\nSucceded at 6597 with 1 tries\nSucceded at 6598 with 1 tries\nSucceded at 6599 with 1 tries\nSucceded at 6600 with 1 tries\nSucceded at 6602 with 1 tries\nSucceded at 6605 with 1 tries\nSucceded at 6606 with 1 tries\nSucceded at 6610 with 1 tries\nSucceded at 6612 with 1 tries\nSucceded at 6613 with 1 tries\nSucceded at 6614 with 1 tries\nSucceded at 6615 with 1 tries\nSucceded at 6616 with 1 tries\nSucceded at 6618 with 1 tries\nSucceded at 6619 with 1 tries\nSucceded at 6620 with 1 tries\nSucceded at 6621 with 1 tries\nSucceded at 6622 with 1 tries\nSucceded at 6623 with 1 tries\nSucceded at 6624 with 1 tries\nSucceded at 6626 with 1 tries\nSucceded at 6627 with 1 tries\nSucceded at 6629 with 1 tries\nSucceded at 6630 with 1 tries\nSucceded at 6631 with 1 tries\nSucceded at 6632 with 1 tries\nSucceded at 6633 with 1 tries\nSucceded at 6635 with 1 tries\nSucceded at 6636 with 1 tries\nSucceded at 6639 with 1 tries\nSucceded at 6640 with 1 tries\nSucceded at 6643 with 1 tries\nSucceded at 6644 with 1 tries\nSucceded at 6646 with 1 tries\nSucceded at 6648 with 1 tries\nSucceded at 6649 with 1 tries\nSucceded at 6650 with 1 tries\nSucceded at 6652 with 1 tries\nSucceded at 6654 with 1 tries\nSucceded at 6655 with 1 tries\nSucceded at 6657 with 1 tries\nmosek failed at \nSucceded at 6661 with 1 tries\nSucceded at 6662 with 1 tries\nSucceded at 6663 with 1 tries\nSucceded at 6664 with 1 tries\nSucceded at 6665 with 1 tries\nSucceded at 6666 with 1 tries\nSucceded at 6667 with 1 tries\nSucceded at 6668 with 1 tries\nSucceded at 6669 with 1 tries\nSucceded at 6671 with 1 tries\nSucceded at 6672 with 1 tries\nSucceded at 6673 with 1 tries\nSucceded at 6676 with 1 tries\nSucceded at 6677 with 1 tries\nSucceded at 6678 with 1 tries\nSucceded at 6680 with 1 tries\nSucceded at 6681 with 1 tries\nSucceded at 6682 with 1 tries\nSucceded at 6684 with 1 tries\nSucceded at 6685 with 1 tries\nSucceded at 6686 with 1 tries\nSucceded at 6687 with 1 tries\nSucceded at 6688 with 1 tries\nSucceded at 6689 with 1 tries\nSucceded at 6691 with 1 tries\nSucceded at 6693 with 1 tries\nSucceded at 6694 with 1 tries\nSucceded at 6695 with 1 tries\nSucceded at 6697 with 1 tries\nSucceded at 6698 with 1 tries\nSucceded at 6699 with 1 tries\nSucceded at 6700 with 1 tries\nSucceded at 6703 with 1 tries\nSucceded at 6704 with 1 tries\nSucceded at 6705 with 1 tries\nSucceded at 6706 with 1 tries\nSucceded at 6707 with 1 tries\nSucceded at 6708 with 1 tries\nSucceded at 6709 with 1 tries\nSucceded at 6710 with 1 tries\nSucceded at 6711 with 1 tries\nSucceded at 6713 with 1 tries\nSucceded at 6714 with 1 tries\nSucceded at 6715 with 1 tries\nSucceded at 6716 with 1 tries\nSucceded at 6720 with 1 tries\nSucceded at 6722 with 1 tries\nSucceded at 6724 with 1 tries\nSucceded at 6725 with 1 tries\nSucceded at 6726 with 1 tries\nSucceded at 6727 with 1 tries\nSucceded at 6729 with 1 tries\nSucceded at 6730 with 1 tries\nSucceded at 6731 with 1 tries\nSucceded at 6732 with 1 tries\nSucceded at 6735 with 1 tries\nSucceded at 6736 with 1 tries\nSucceded at 6737 with 1 tries\nSucceded at 6738 with 1 tries\nSucceded at 6739 with 1 tries\nSucceded at 6742 with 1 tries\nSucceded at 6744 with 1 tries\nSucceded at 6745 with 1 tries\nSucceded at 6747 with 1 tries\nSucceded at 6748 with 1 tries\nSucceded at 6749 with 1 tries\nSucceded at 6751 with 1 tries\nSucceded at 6752 with 1 tries\nSucceded at 6754 with 1 tries\nSucceded at 6756 with 1 tries\nSucceded at 6757 with 1 tries\nSucceded at 6758 with 1 tries\nSucceded at 6759 with 1 tries\nSucceded at 6760 with 1 tries\nSucceded at 6761 with 1 tries\nSucceded at 6763 with 1 tries\nSucceded at 6767 with 1 tries\nSucceded at 6768 with 1 tries\nSucceded at 6771 with 1 tries\nSucceded at 6773 with 1 tries\nSucceded at 6775 with 1 tries\nSucceded at 6776 with 1 tries\nSucceded at 6778 with 1 tries\nSucceded at 6779 with 1 tries\nSucceded at 6783 with 1 tries\nSucceded at 6784 with 1 tries\nSucceded at 6785 with 1 tries\nSucceded at 6787 with 1 tries\nSucceded at 6789 with 1 tries\nSucceded at 6792 with 1 tries\nSucceded at 6793 with 1 tries\nSucceded at 6794 with 1 tries\nSucceded at 6795 with 1 tries\nSucceded at 6797 with 1 tries\nSucceded at 6799 with 1 tries\nSucceded at 6800 with 1 tries\nSucceded at 6801 with 1 tries\nSucceded at 6802 with 1 tries\nSucceded at 6804 with 1 tries\nSucceded at 6806 with 1 tries\nSucceded at 6807 with 1 tries\nSucceded at 6808 with 1 tries\nSucceded at 6809 with 1 tries\nSucceded at 6810 with 1 tries\nSucceded at 6811 with 1 tries\nSucceded at 6812 with 1 tries\nSucceded at 6815 with 1 tries\nSucceded at 6817 with 1 tries\nSucceded at 6821 with 1 tries\nSucceded at 6822 with 1 tries\nSucceded at 6824 with 1 tries\nSucceded at 6826 with 1 tries\nSucceded at 6827 with 1 tries\nSucceded at 6830 with 1 tries\nSucceded at 6831 with 1 tries\nSucceded at 6832 with 1 tries\nSucceded at 6835 with 1 tries\nSucceded at 6836 with 1 tries\nSucceded at 6837 with 1 tries\nSucceded at 6838 with 1 tries\nSucceded at 6839 with 1 tries\nSucceded at 6840 with 1 tries\nSucceded at 6845 with 1 tries\nSucceded at 6846 with 1 tries\nSucceded at 6848 with 1 tries\nSucceded at 6849 with 1 tries\nSucceded at 6850 with 1 tries\nSucceded at 6851 with 1 tries\nSucceded at 6852 with 1 tries\nSucceded at 6853 with 1 tries\nSucceded at 6854 with 1 tries\nSucceded at 6855 with 1 tries\nSucceded at 6856 with 1 tries\nSucceded at 6857 with 1 tries\nSucceded at 6858 with 1 tries\nSucceded at 6859 with 1 tries\nSucceded at 6860 with 1 tries\nSucceded at 6861 with 1 tries\nSucceded at 6864 with 1 tries\nSucceded at 6866 with 1 tries\nSucceded at 6867 with 1 tries\nSucceded at 6868 with 1 tries\nSucceded at 6870 with 1 tries\nSucceded at 6871 with 1 tries\nSucceded at 6873 with 1 tries\nSucceded at 6874 with 1 tries\nSucceded at 6875 with 1 tries\nSucceded at 6876 with 1 tries\nSucceded at 6877 with 1 tries\nSucceded at 6878 with 1 tries\nSucceded at 6879 with 1 tries\nSucceded at 6880 with 1 tries\nSucceded at 6882 with 1 tries\nSucceded at 6883 with 1 tries\nSucceded at 6884 with 1 tries\nSucceded at 6885 with 1 tries\nSucceded at 6886 with 1 tries\nSucceded at 6887 with 1 tries\nSucceded at 6888 with 1 tries\nSucceded at 6890 with 1 tries\nSucceded at 6891 with 1 tries\nSucceded at 6893 with 1 tries\nSucceded at 6896 with 1 tries\nSucceded at 6897 with 1 tries\nSucceded at 6899 with 1 tries\nSucceded at 6900 with 1 tries\nSucceded at 6902 with 1 tries\nSucceded at 6904 with 1 tries\nSucceded at 6905 with 1 tries\nSucceded at 6907 with 1 tries\nSucceded at 6911 with 1 tries\nSucceded at 6912 with 1 tries\nSucceded at 6913 with 1 tries\nSucceded at 6914 with 1 tries\nSucceded at 6915 with 1 tries\nSucceded at 6916 with 1 tries\nSucceded at 6921 with 1 tries\nSucceded at 6924 with 1 tries\nSucceded at 6926 with 1 tries\nSucceded at 6927 with 1 tries\nSucceded at 6929 with 1 tries\nSucceded at 6932 with 1 tries\nSucceded at 6933 with 1 tries\nSucceded at 6934 with 1 tries\nSucceded at 6935 with 1 tries\nSucceded at 6936 with 1 tries\nSucceded at 6937 with 1 tries\nSucceded at 6940 with 1 tries\nSucceded at 6941 with 1 tries\nSucceded at 6942 with 1 tries\nSucceded at 6943 with 1 tries\nSucceded at 6944 with 1 tries\nSucceded at 6945 with 1 tries\nSucceded at 6946 with 1 tries\nSucceded at 6949 with 1 tries\nSucceded at 6950 with 1 tries\nSucceded at 6951 with 1 tries\nSucceded at 6952 with 1 tries\nSucceded at 6953 with 1 tries\nSucceded at 6954 with 1 tries\nSucceded at 6956 with 1 tries\nSucceded at 6957 with 1 tries\nSucceded at 6958 with 1 tries\nSucceded at 6960 with 1 tries\nSucceded at 6962 with 1 tries\nSucceded at 6963 with 1 tries\nSucceded at 6964 with 1 tries\nSucceded at 6965 with 1 tries\nSucceded at 6966 with 1 tries\nSucceded at 6968 with 1 tries\nSucceded at 6970 with 1 tries\nSucceded at 6971 with 1 tries\nSucceded at 6972 with 1 tries\nSucceded at 6974 with 1 tries\nSucceded at 6976 with 1 tries\nSucceded at 6977 with 1 tries\nSucceded at 6978 with 1 tries\nSucceded at 6979 with 1 tries\nSucceded at 6980 with 1 tries\nSucceded at 6982 with 1 tries\nSucceded at 6983 with 1 tries\nSucceded at 6984 with 1 tries\nSucceded at 6985 with 1 tries\nSucceded at 6986 with 1 tries\nSucceded at 6988 with 1 tries\nSucceded at 6990 with 1 tries\nSucceded at 6991 with 1 tries\nSucceded at 6992 with 1 tries\nSucceded at 6993 with 1 tries\nSucceded at 6995 with 1 tries\nSucceded at 6996 with 1 tries\nSucceded at 6998 with 1 tries\nSucceded at 6999 with 1 tries\nSucceded at 7000 with 1 tries\nmosek failed at \nSucceded at 7003 with 1 tries\nSucceded at 7004 with 1 tries\nSucceded at 7006 with 1 tries\nSucceded at 7008 with 1 tries\nSucceded at 7009 with 1 tries\nSucceded at 7011 with 1 tries\nSucceded at 7012 with 1 tries\nSucceded at 7013 with 1 tries\nSucceded at 7014 with 1 tries\nSucceded at 7017 with 1 tries\nSucceded at 7018 with 1 tries\nSucceded at 7019 with 1 tries\nSucceded at 7020 with 1 tries\nSucceded at 7021 with 1 tries\nSucceded at 7022 with 1 tries\nSucceded at 7024 with 1 tries\nSucceded at 7025 with 1 tries\nSucceded at 7026 with 1 tries\nSucceded at 7028 with 1 tries\nSucceded at 7030 with 1 tries\nSucceded at 7031 with 1 tries\nSucceded at 7034 with 1 tries\nSucceded at 7035 with 1 tries\nSucceded at 7037 with 1 tries\nSucceded at 7038 with 1 tries\nSucceded at 7039 with 1 tries\nSucceded at 7041 with 1 tries\nSucceded at 7042 with 1 tries\nSucceded at 7043 with 1 tries\nSucceded at 7046 with 1 tries\nSucceded at 7047 with 1 tries\nSucceded at 7049 with 1 tries\nSucceded at 7050 with 1 tries\nSucceded at 7052 with 1 tries\nSucceded at 7053 with 1 tries\nSucceded at 7056 with 1 tries\nSucceded at 7057 with 1 tries\nSucceded at 7058 with 1 tries\nSucceded at 7059 with 1 tries\nSucceded at 7060 with 1 tries\nSucceded at 7061 with 1 tries\nSucceded at 7063 with 1 tries\nSucceded at 7065 with 1 tries\nSucceded at 7068 with 1 tries\nSucceded at 7069 with 1 tries\nSucceded at 7071 with 1 tries\nSucceded at 7075 with 1 tries\nSucceded at 7076 with 1 tries\nSucceded at 7077 with 1 tries\nSucceded at 7078 with 1 tries\nSucceded at 7080 with 1 tries\nSucceded at 7081 with 1 tries\nSucceded at 7082 with 1 tries\nSucceded at 7083 with 1 tries\nSucceded at 7084 with 1 tries\nSucceded at 7086 with 1 tries\nSucceded at 7087 with 1 tries\nSucceded at 7091 with 1 tries\nSucceded at 7094 with 1 tries\nSucceded at 7095 with 1 tries\nSucceded at 7096 with 1 tries\nSucceded at 7097 with 1 tries\nSucceded at 7099 with 1 tries\nSucceded at 7100 with 1 tries\nSucceded at 7101 with 1 tries\nSucceded at 7102 with 1 tries\nSucceded at 7103 with 1 tries\nSucceded at 7104 with 1 tries\nSucceded at 7105 with 1 tries\nSucceded at 7106 with 1 tries\nSucceded at 7107 with 1 tries\nSucceded at 7109 with 1 tries\nSucceded at 7112 with 1 tries\nSucceded at 7113 with 1 tries\nSucceded at 7114 with 1 tries\nSucceded at 7116 with 1 tries\nSucceded at 7117 with 1 tries\nSucceded at 7120 with 1 tries\nSucceded at 7121 with 1 tries\nSucceded at 7122 with 1 tries\nSucceded at 7123 with 1 tries\nSucceded at 7124 with 1 tries\nSucceded at 7126 with 1 tries\nSucceded at 7128 with 1 tries\nSucceded at 7129 with 1 tries\nSucceded at 7132 with 1 tries\nSucceded at 7133 with 1 tries\nSucceded at 7134 with 1 tries\nSucceded at 7135 with 1 tries\nSucceded at 7136 with 1 tries\nSucceded at 7137 with 1 tries\nSucceded at 7138 with 1 tries\nSucceded at 7141 with 1 tries\nSucceded at 7143 with 1 tries\nSucceded at 7144 with 1 tries\nSucceded at 7145 with 1 tries\nSucceded at 7146 with 1 tries\nSucceded at 7147 with 1 tries\nSucceded at 7152 with 1 tries\nSucceded at 7153 with 1 tries\nSucceded at 7156 with 1 tries\nSucceded at 7157 with 1 tries\nSucceded at 7158 with 1 tries\nSucceded at 7160 with 1 tries\nSucceded at 7161 with 1 tries\nSucceded at 7163 with 1 tries\nSucceded at 7164 with 1 tries\nSucceded at 7165 with 1 tries\nSucceded at 7166 with 1 tries\nSucceded at 7168 with 1 tries\nSucceded at 7170 with 1 tries\nSucceded at 7172 with 1 tries\nSucceded at 7173 with 1 tries\nSucceded at 7176 with 1 tries\nSucceded at 7177 with 1 tries\nSucceded at 7179 with 1 tries\nSucceded at 7180 with 1 tries\nSucceded at 7181 with 1 tries\nSucceded at 7183 with 1 tries\nSucceded at 7186 with 1 tries\nSucceded at 7187 with 1 tries\nSucceded at 7190 with 1 tries\nSucceded at 7192 with 1 tries\nSucceded at 7194 with 1 tries\nSucceded at 7195 with 1 tries\nSucceded at 7197 with 1 tries\nSucceded at 7198 with 1 tries\nSucceded at 7199 with 1 tries\nSucceded at 7200 with 1 tries\nSucceded at 7201 with 1 tries\nSucceded at 7202 with 1 tries\nSucceded at 7208 with 1 tries\nSucceded at 7209 with 1 tries\nSucceded at 7213 with 1 tries\nSucceded at 7214 with 1 tries\nSucceded at 7215 with 1 tries\nSucceded at 7216 with 1 tries\nSucceded at 7217 with 1 tries\nSucceded at 7219 with 1 tries\nSucceded at 7221 with 1 tries\nSucceded at 7222 with 1 tries\nSucceded at 7223 with 1 tries\nSucceded at 7225 with 1 tries\nSucceded at 7226 with 1 tries\nSucceded at 7227 with 1 tries\nSucceded at 7229 with 1 tries\nSucceded at 7230 with 1 tries\nSucceded at 7231 with 1 tries\nSucceded at 7232 with 1 tries\nSucceded at 7233 with 1 tries\nSucceded at 7237 with 1 tries\nSucceded at 7238 with 1 tries\nSucceded at 7239 with 1 tries\nSucceded at 7240 with 1 tries\nSucceded at 7241 with 1 tries\nSucceded at 7245 with 1 tries\nSucceded at 7247 with 1 tries\nSucceded at 7248 with 1 tries\nSucceded at 7249 with 1 tries\nSucceded at 7251 with 1 tries\nSucceded at 7252 with 1 tries\nSucceded at 7253 with 1 tries\nSucceded at 7254 with 1 tries\nSucceded at 7255 with 1 tries\nSucceded at 7256 with 1 tries\nSucceded at 7257 with 1 tries\nSucceded at 7258 with 1 tries\nSucceded at 7259 with 1 tries\nSucceded at 7260 with 1 tries\nSucceded at 7261 with 1 tries\nSucceded at 7262 with 1 tries\nSucceded at 7263 with 1 tries\nSucceded at 7264 with 1 tries\nSucceded at 7265 with 1 tries\nSucceded at 7267 with 1 tries\nSucceded at 7268 with 1 tries\nSucceded at 7270 with 1 tries\nSucceded at 7272 with 1 tries\nSucceded at 7273 with 1 tries\nSucceded at 7274 with 1 tries\nSucceded at 7275 with 1 tries\nSucceded at 7277 with 1 tries\nSucceded at 7278 with 1 tries\nSucceded at 7279 with 1 tries\nSucceded at 7280 with 1 tries\nSucceded at 7281 with 1 tries\nSucceded at 7282 with 1 tries\nSucceded at 7283 with 1 tries\nSucceded at 7285 with 1 tries\nSucceded at 7288 with 1 tries\nSucceded at 7289 with 1 tries\nSucceded at 7290 with 1 tries\nSucceded at 7291 with 1 tries\nSucceded at 7293 with 1 tries\nSucceded at 7294 with 1 tries\nSucceded at 7295 with 1 tries\nSucceded at 7296 with 1 tries\nSucceded at 7297 with 1 tries\nSucceded at 7299 with 1 tries\nSucceded at 7300 with 1 tries\nSucceded at 7302 with 1 tries\nSucceded at 7303 with 1 tries\nSucceded at 7304 with 1 tries\nSucceded at 7306 with 1 tries\nSucceded at 7307 with 1 tries\nSucceded at 7309 with 1 tries\nSucceded at 7310 with 1 tries\nSucceded at 7311 with 1 tries\nSucceded at 7312 with 1 tries\nSucceded at 7313 with 1 tries\nSucceded at 7316 with 1 tries\nSucceded at 7317 with 1 tries\nSucceded at 7318 with 1 tries\nSucceded at 7319 with 1 tries\nSucceded at 7320 with 1 tries\nSucceded at 7322 with 1 tries\nSucceded at 7323 with 1 tries\nSucceded at 7325 with 1 tries\nSucceded at 7326 with 1 tries\nSucceded at 7327 with 1 tries\nSucceded at 7329 with 1 tries\nSucceded at 7330 with 1 tries\nSucceded at 7331 with 1 tries\nSucceded at 7332 with 1 tries\nSucceded at 7333 with 1 tries\nSucceded at 7334 with 1 tries\nSucceded at 7335 with 1 tries\nSucceded at 7336 with 1 tries\nSucceded at 7338 with 1 tries\nSucceded at 7339 with 1 tries\nSucceded at 7340 with 1 tries\nSucceded at 7342 with 1 tries\nSucceded at 7343 with 1 tries\nSucceded at 7344 with 1 tries\nSucceded at 7346 with 1 tries\nSucceded at 7347 with 1 tries\nSucceded at 7348 with 1 tries\nSucceded at 7350 with 1 tries\nSucceded at 7351 with 1 tries\nSucceded at 7353 with 1 tries\nSucceded at 7354 with 1 tries\nSucceded at 7355 with 1 tries\nSucceded at 7357 with 1 tries\nSucceded at 7358 with 1 tries\nSucceded at 7359 with 1 tries\nSucceded at 7360 with 1 tries\nSucceded at 7361 with 1 tries\nSucceded at 7363 with 1 tries\nSucceded at 7364 with 1 tries\nSucceded at 7365 with 1 tries\nSucceded at 7367 with 1 tries\nSucceded at 7368 with 1 tries\nSucceded at 7369 with 1 tries\nSucceded at 7372 with 1 tries\nSucceded at 7373 with 1 tries\nSucceded at 7374 with 1 tries\nSucceded at 7375 with 1 tries\nSucceded at 7376 with 1 tries\nSucceded at 7377 with 1 tries\nSucceded at 7378 with 1 tries\nSucceded at 7379 with 1 tries\nSucceded at 7380 with 1 tries\nSucceded at 7381 with 1 tries\nSucceded at 7382 with 1 tries\nSucceded at 7383 with 1 tries\nSucceded at 7384 with 1 tries\nSucceded at 7385 with 1 tries\nSucceded at 7386 with 1 tries\nSucceded at 7389 with 1 tries\nSucceded at 7390 with 1 tries\nSucceded at 7391 with 1 tries\nSucceded at 7392 with 1 tries\nSucceded at 7393 with 1 tries\nSucceded at 7396 with 1 tries\nSucceded at 7398 with 1 tries\nSucceded at 7399 with 1 tries\nSucceded at 7400 with 1 tries\nSucceded at 7405 with 1 tries\nSucceded at 7407 with 1 tries\nSucceded at 7408 with 1 tries\nSucceded at 7409 with 1 tries\nSucceded at 7411 with 1 tries\nSucceded at 7412 with 1 tries\nSucceded at 7415 with 1 tries\nSucceded at 7416 with 1 tries\nSucceded at 7417 with 1 tries\nSucceded at 7418 with 1 tries\nSucceded at 7419 with 1 tries\nSucceded at 7421 with 1 tries\nSucceded at 7422 with 1 tries\nSucceded at 7423 with 1 tries\nSucceded at 7424 with 1 tries\nSucceded at 7425 with 1 tries\nSucceded at 7427 with 1 tries\nSucceded at 7428 with 1 tries\nSucceded at 7429 with 1 tries\nSucceded at 7431 with 1 tries\nSucceded at 7432 with 1 tries\nSucceded at 7433 with 1 tries\nSucceded at 7434 with 1 tries\nSucceded at 7435 with 1 tries\nSucceded at 7436 with 1 tries\nSucceded at 7437 with 1 tries\nSucceded at 7442 with 1 tries\nSucceded at 7443 with 1 tries\nSucceded at 7445 with 1 tries\nSucceded at 7447 with 1 tries\nSucceded at 7449 with 1 tries\nSucceded at 7450 with 1 tries\nSucceded at 7452 with 1 tries\nSucceded at 7453 with 1 tries\nSucceded at 7455 with 1 tries\nSucceded at 7456 with 1 tries\nSucceded at 7458 with 1 tries\nSucceded at 7459 with 1 tries\nSucceded at 7460 with 1 tries\nSucceded at 7463 with 1 tries\nSucceded at 7465 with 1 tries\nSucceded at 7466 with 1 tries\nSucceded at 7468 with 1 tries\nSucceded at 7469 with 1 tries\nSucceded at 7471 with 1 tries\nSucceded at 7474 with 1 tries\nSucceded at 7475 with 1 tries\nSucceded at 7476 with 1 tries\nSucceded at 7477 with 1 tries\nSucceded at 7478 with 1 tries\nSucceded at 7479 with 1 tries\nSucceded at 7480 with 1 tries\nSucceded at 7481 with 1 tries\nSucceded at 7482 with 1 tries\nSucceded at 7483 with 1 tries\nSucceded at 7484 with 1 tries\nSucceded at 7485 with 1 tries\nSucceded at 7488 with 1 tries\nSucceded at 7489 with 1 tries\nSucceded at 7490 with 1 tries\nSucceded at 7492 with 1 tries\nSucceded at 7493 with 1 tries\nSucceded at 7494 with 1 tries\nSucceded at 7496 with 1 tries\nSucceded at 7498 with 1 tries\nSucceded at 7499 with 1 tries\nSucceded at 7502 with 1 tries\nSucceded at 7504 with 1 tries\nSucceded at 7505 with 1 tries\nSucceded at 7506 with 1 tries\nSucceded at 7509 with 1 tries\nSucceded at 7510 with 1 tries\nSucceded at 7511 with 1 tries\nSucceded at 7512 with 1 tries\nSucceded at 7513 with 1 tries\nSucceded at 7514 with 1 tries\nSucceded at 7515 with 1 tries\nSucceded at 7516 with 1 tries\nSucceded at 7517 with 1 tries\nSucceded at 7519 with 1 tries\nSucceded at 7520 with 1 tries\nSucceded at 7521 with 1 tries\nSucceded at 7522 with 1 tries\nSucceded at 7525 with 1 tries\n"
],
[
"global_acc = sum(sum(np.equal(ind_max,pp.labels[test_start:,0][:,None])))/(0.1*pp.n_probs)\nglobal_acc",
"_____no_output_____"
],
[
"sum(feasible)/ii",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbade343d0d2932203917e227c70caf276544373
| 2,498 |
ipynb
|
Jupyter Notebook
|
data/read_xml.ipynb
|
janhaa/fchd
|
11f20f945c2d7aa94552c190277d2a607ed0cec5
|
[
"MIT"
] | 648 |
2018-09-25T08:59:30.000Z
|
2022-03-31T10:25:07.000Z
|
data/read_xml.ipynb
|
shaozhuqing/FCHD-Fully-Convolutional-Head-Detector
|
1823c077dc7f162628ad63d1cb2404939bd54446
|
[
"MIT"
] | 64 |
2018-09-26T09:00:17.000Z
|
2021-12-02T09:13:41.000Z
|
data/read_xml.ipynb
|
shaozhuqing/FCHD-Fully-Convolutional-Head-Detector
|
1823c077dc7f162628ad63d1cb2404939bd54446
|
[
"MIT"
] | 205 |
2018-09-26T05:41:54.000Z
|
2022-01-09T09:08:24.000Z
| 18.781955 | 109 | 0.486789 |
[
[
[
"import numpy as np \nimport os\nimport xml.etree.ElementTree as ET",
"_____no_output_____"
],
[
"print len(os.listdir('./JPEGImages/'))",
"224740\n"
],
[
"print len(os.listdir('./Annotations/'))",
"224740\n"
],
[
"anno = ET.parse(os.path.join('./Annotations/mov_017_073500.xml'))\nbbox = list()\nlabel = list()\nfor obj in anno.findall('object'):\n bndbox_anno = obj.find('bndbox')\n # subtract 1 to make pixel indexes 0-based\n bbox.append([float(bndbox_anno.find(tag).text) - 1 for tag in ('ymin', 'xmin', 'ymax', 'xmax')])\n ",
"_____no_output_____"
],
[
"bbox",
"_____no_output_____"
],
[
"import shutil ",
"_____no_output_____"
],
[
"shutil.move('./Annotations/mov_007_121337.xml', '../')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbadea0bccd072094aed727d9b91c6bb753f87d7
| 7,801 |
ipynb
|
Jupyter Notebook
|
7. Homogeneity Metrics.ipynb
|
csoehnel/DSR-FundamentalsOfML
|
6f7dd08ff9549ef2c0da5f8b453c2f6fa42cce98
|
[
"MIT"
] | null | null | null |
7. Homogeneity Metrics.ipynb
|
csoehnel/DSR-FundamentalsOfML
|
6f7dd08ff9549ef2c0da5f8b453c2f6fa42cce98
|
[
"MIT"
] | null | null | null |
7. Homogeneity Metrics.ipynb
|
csoehnel/DSR-FundamentalsOfML
|
6f7dd08ff9549ef2c0da5f8b453c2f6fa42cce98
|
[
"MIT"
] | null | null | null | 28.162455 | 205 | 0.507499 |
[
[
[
"from IPython.display import HTML\ncss_file = './custom.css'\nHTML(open(css_file, \"r\").read())",
"_____no_output_____"
]
],
[
[
"# Homogeneity Metrics\n\n© 2018 Daniel Voigt Godoy",
"_____no_output_____"
],
[
"## 1. Definitions\n\n### Entropy\n\n***Entropy*** is a measure of ***uncertainty*** associated with a given ***distribution q(y)***.\n\nFrom Wikipedia:\n\n ...is the average rate at which information is produced by a stochastic source of data. \n \n ...when a low-probability event occurs, the event carries more \"information\" (\"surprisal\")... \n\n$$\nH(q) = -\\sum_{c=1}^{C}{q(y_c) \\cdot log(q(y_c))}\n$$\n\nwhere:\n - ***q*** is the ***distribution*** (as in the distribution of red and green balls)\n - ***y*** are the ***labels*** (the respective colors of each ball)\n - ***C*** is the number of ***classes*** (as in ***red*** and ***green*** - 2 classes)\n - ***q(yc) represents the proportion of balls having the same color c***\n\n### Gini Impurity\n\n***Gini Impurity*** is a measure of ***heterogeneity*** associated with a given ***distribution q(y)***.\n\n$$\nG(q) = \\sum_{c=1}^{C}{q(y_c) \\cdot (1 - q(y_c))}\n$$\n\nFrom Wikipedia:\n\n ...is a measure of how often a randomly chosen element from the set would be incorrectly labeled if it was randomly labeled according to the distribution of labels in the subset.",
"_____no_output_____"
]
],
[
[
"from intuitiveml.Information import *",
"_____no_output_____"
],
[
"X, y = data(10)\nmyinfo = plotInfo(X, y)\nvb = VBox(build_figure(myinfo), layout={'align_items': 'center'})",
"_____no_output_____"
]
],
[
[
"## 2. Experiment\n\nThere are 10 balls (data points) of two possible colors (***classes***). Each ball has its own color (***label***), red or green.\n\nThe slider control at the bottom allows you to change the number of red balls and, consequently, the number of green balls (the total stays the same) - so, you are changing the ***distribution***.\n\nThis change will have an impact on both ***entropy*** and ***gini impurity*** measures.\n\nUse the slider to play with different configurations and answer the ***questions*** below.",
"_____no_output_____"
]
],
[
[
"vb",
"_____no_output_____"
]
],
[
[
"#### Questions:\n\n1. How to maximize (minimize) Entropy?\n2. How to maximize (minimize) Gini Impurity?\n3. What's the entropy when all balls have the same color?\n4. What kind of distribution yields the maximum Entropy?\n5. Using the formula, compute the ***entropy*** if you had 3 red balls\n6. Using the formula, compute the ***gini impurity*** if you had 7 red balls",
"_____no_output_____"
],
[
"3.) Zero\n4.) What kind of distribution yields the maximum Entropy? Uniform",
"_____no_output_____"
],
[
"#### This material is copyright Daniel Voigt Godoy and made available under the Creative Commons Attribution (CC-BY) license ([link](https://creativecommons.org/licenses/by/4.0/)). \n\n#### Code is also made available under the MIT License ([link](https://opensource.org/licenses/MIT)).",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\nHTML('''<script>\n function code_toggle() {\n if (code_shown){\n $('div.input').hide('500');\n $('#toggleButton').val('Show Code')\n } else {\n $('div.input').show('500');\n $('#toggleButton').val('Hide Code')\n }\n code_shown = !code_shown\n }\n\n $( document ).ready(function(){\n code_shown=false;\n $('div.input').hide()\n });\n</script>\n<form action=\"javascript:code_toggle()\"><input type=\"submit\" id=\"toggleButton\" value=\"Show Code\"></form>''')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
] |
cbadf020586cddbc3a69f84c1c952b19bc0f4076
| 7,431 |
ipynb
|
Jupyter Notebook
|
Assignment3.ipynb
|
RCDayrit/LinearAlgebra2021
|
7cdf4171ff613270ec87073c9a96f8e9fae447c2
|
[
"Apache-2.0"
] | null | null | null |
Assignment3.ipynb
|
RCDayrit/LinearAlgebra2021
|
7cdf4171ff613270ec87073c9a96f8e9fae447c2
|
[
"Apache-2.0"
] | null | null | null |
Assignment3.ipynb
|
RCDayrit/LinearAlgebra2021
|
7cdf4171ff613270ec87073c9a96f8e9fae447c2
|
[
"Apache-2.0"
] | null | null | null | 23.221875 | 234 | 0.369129 |
[
[
[
"<a href=\"https://colab.research.google.com/github/RCDayrit/LinearAlgebra2021/blob/main/Assignment3.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#Linear Algebra for ECE\n\n## Matrices",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport scipy.linalg as la\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"$$\nA = \\left\\{\n \\begin{array}\\\n x + y \\\\ \n 4x - 10y\n \\end{array}\n\\right. \\\\\nB = \\left\\{\n \\begin{array}\\\n x+y+z \\\\ \n 3x -2y -z \\\\\n -x + 4y +2z\n \\end{array}\n\\right. $$",
"_____no_output_____"
],
[
"$$\nA=\\begin{bmatrix} 1 & 1 \\\\ 4 & {-10}\\end{bmatrix} \\\\\nB=\\begin{bmatrix} 1 & 1 & 1 \\\\ 3 & -2 & -1 \\\\ -1 & 4 & 2\\end{bmatrix}\n$$",
"_____no_output_____"
],
[
"$$A=\\begin{bmatrix}\na_{(0,0)}&a_{(0,1)}&\\dots&a_{(0,j-1)}\\\\\na_{(1,0)}&a_{(1,1)}&\\dots&a_{(1,j-1)}\\\\\n\\vdots&\\vdots&\\ddots&\\vdots&\\\\\na_{(i-1,0)}&a_{(i-1,1)}&\\dots&a_{(i-1,j-1)}\n\\end{bmatrix}\n$$",
"_____no_output_____"
]
],
[
[
"def describe_mat(matrix):\n print(f'Matrix:\\n{matrix}\\n\\nShape:\\t{matrix.shape}\\nRank:\\t{matrix.ndim}\\n')",
"_____no_output_____"
],
[
"## Declaring a 2 x 2 matrix\nA = np.array([\n [1, 2],\n [3, 1]\n])\ndescribe_mat(A)",
"Matrix:\n[[1 2]\n [3 1]]\n\nShape:\t(2, 2)\nRank:\t2\n\n"
],
[
"## Declaring a 3 x 2 matrix\nB = np.array([\n [8, 2],\n [5, 4],\n [1, 1]\n])\ndescribe_mat(B)",
"Matrix:\n[[8 2]\n [5 4]\n [1 1]]\n\nShape:\t(3, 2)\nRank:\t2\n\n"
],
[
"G = np.array([])",
"_____no_output_____"
],
[
"H = np.array([1,2,3,4,5])\ndescribe_mat(H)",
"Matrix:\n[1 2 3 4 5]\n\nShape:\t(5,)\nRank:\t1\n\n"
],
[
"",
"_____no_output_____"
],
[
"## Declaring a Row matrix\n\nrow_mat_1D = np.array([\n 1, 3 , 2\n]) ## this is a 1-D Matrix with a Shape of (3.). it's not really considered as a row matrix.\n\nrow_mat_2D = np.array([\n 1, 2, 3 \n\n])\ndescribe_mat(row_mat_1D)\ndescribe_mat(row_mat_2D)",
"Matrix:\n[1 3 2]\n\nShape:\t(3,)\nRank:\t1\n\nMatrix:\n[1 2 3]\n\nShape:\t(3,)\nRank:\t1\n\n"
],
[
"zero_mat_row = np.zeros((1,2))\n\nprint(f'Zero Row Matrix: \\n{zero_mat_row}')",
"Zero Row Matrix: \n[[0. 0.]]\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbadfe0ecfac67d0def3a56e5c90f952a69d9cd3
| 8,599 |
ipynb
|
Jupyter Notebook
|
topic_entropy.ipynb
|
anselmrothe/dtm
|
a76cbfa94edbd230234d5a75d85262676694675b
|
[
"MIT"
] | 11 |
2017-10-25T19:12:49.000Z
|
2021-08-30T08:14:38.000Z
|
topic_entropy.ipynb
|
anselmrothe/dtm
|
a76cbfa94edbd230234d5a75d85262676694675b
|
[
"MIT"
] | null | null | null |
topic_entropy.ipynb
|
anselmrothe/dtm
|
a76cbfa94edbd230234d5a75d85262676694675b
|
[
"MIT"
] | 2 |
2018-06-23T03:04:11.000Z
|
2021-08-30T08:14:44.000Z
| 29.248299 | 146 | 0.559716 |
[
[
[
"import logging\nfrom gensim.models import ldaseqmodel\nfrom gensim.corpora import Dictionary, bleicorpus, textcorpus\nimport numpy as np\nfrom gensim.matutils import hellinger\nimport time\nimport pickle\nimport pyLDAvis\nimport matplotlib.pyplot as plt\nfrom scipy.stats import entropy\nimport pandas as pd\nfrom numpy.linalg import norm\n",
"_____no_output_____"
],
[
"alldata_new = pickle.load(open('output/dtm_processed_output.p', 'rb'))\n# load data\ndoc_year=alldata_new['docs_per_year']\ndoc_ids =[0]+list(np.cumsum(doc_year))\n\nterm_topic = alldata_new['term_topic']# term_topic is n_years*n_topics*n_terms\nterms = alldata_new['terms']\n\ndoc_topicyrs = alldata_new['doc_topic']\n\ndoc_topic = []\nfor year in range(len(term_topic)): \n doc_topic.append(alldata_new['doc_topic'][doc_ids[year]:doc_ids[year+1]])# doc_topic is nyear*n_docs given year*n_topics\n# rename topics by the hand-picked names\ntopic_labels = pickle.load(open('topicnames.p','rb'))\n ",
"_____no_output_____"
],
[
"def totvar(p,q):\n maxdist=np.max(abs(p-q))\n maxid=np.argmax(abs(p-q))\n return [maxdist,maxid]\ndef JSD(P, Q):\n _P = P / norm(P, ord=1)\n _Q = Q / norm(Q, ord=1)\n _M = 0.5 * (_P + _Q)\n dist=0.5 * (entropy(_P, _M) + entropy(_Q, _M)) \n return dist",
"_____no_output_____"
],
[
"# entropy change within a topic -- which topic's content has changed most in the past years\nepsilon = 1e-15\nntopics = 20\ntopicdelta=np.empty((ntopics,len(term_topic))) # distance from previous year: jenson-shannon distance\ntopicshift=np.empty(ntopics) # distance from 2000 to 2017\ntopicdelta_tv=np.empty((ntopics,len(term_topic))) # distance from previous year: total variance\ntopicshift_tv=np.empty(ntopics) # distance from 2000 to 2017:total variance\n\n\ndeltaterm=[]\nshiftterm=[]\nfor k in range(ntopics):\n sftterms=[]\n for iyear in range(len(term_topic)): \n topic = term_topic[iyear][k]\n # why not using KL: 1) avoid asymetry 2) avoid inf\n topic = topic/sum(topic)\n topicdelta[k,iyear] = JSD(topic,term_topic[max(iyear-1,0)][k]) # jensen-shannon distance\n [topicdelta_tv[k,iyear],maxterm]=totvar(topic,term_topic[max(iyear-1,0)][k]) # maxterm: term of biggest change from previous year\n sftterms.append(terms[maxterm])\n topicshift[k] = JSD(term_topic[-1][k],term_topic[0][k])\n [topicshift_tv[k],maxterm]=totvar(term_topic[-1][k],term_topic[0][k])\n shiftterm.append(terms[maxterm]) # biggest shift from 2017 to 2000\n deltaterm.append(sftterms) # biggest delta from prev year: max term for every year",
"_____no_output_____"
],
[
"deltaterm[4]",
"_____no_output_____"
],
[
"shiftidx=np.argsort(-topicshift)\nfor idx in shiftidx:\n print(topic_labels[idx]+': %.3f'%topicshift[idx])\n\nprint('total variance:')\nshiftidx=np.argsort(-topicshift_tv)\nfor idx in shiftidx:\n print(topic_labels[idx]+': %.3f'%topicshift_tv[idx]+' max shift word:'+shiftterm[idx])",
"_____no_output_____"
],
[
"#TODO: get the raise and fall terms for each topic...just copy the other code; set the jsd as titles",
"_____no_output_____"
],
[
"# calculate the topic distribution for each year (should correspond to the topic evolution trend...can't find that code right now)\nntopics = len(topic_labels)\nptop_years = []\nentrop_years = []\nfor iyear in range(len(term_topic)): \n ptopics = np.zeros(ntopics)\n for doc in doc_topic[iyear]:\n ptopics+=doc\n ptopics = ptopics/sum(ptopics)\n ptop_years.append(ptopics)\n entrop_years.append(entropy(ptopics))\nprint(entrop_years)\n\n# plot the entropy change across years\nyears = np.arange(len(term_topic))+2000\nplt.plot(years,entrop_years,'-o')\nplt.xlabel('year')\nplt.title('entropy of topic distribution')\nplt.show()\n\n# could be done: find the paper with highest / lowest entropy; find the topic with highest/lowest entropy",
"_____no_output_____"
],
[
"# KL-divergence across years\nkl_years = []\n\ngap=1\nfor iyear in range(len(term_topic)-gap): \n# kl_years.append(entropy(ptop_years[iyear],ptop_years[iyear+gap]))\n kl_years.append(entropy(ptop_years[iyear+gap],ptop_years[iyear]))# sanity check: reverse the direction of KL. not differen\nplt.plot(years[gap:],kl_years,'-o')\nplt.xlabel('year')\nplt.title('KL div with the previous year')\nplt.show()\n\n# TODO: eye-balling the distribution overlayed",
"_____no_output_____"
]
],
[
[
"**tentative conclusion**\n- the diversity of topics seem to increase over years\n- 2002 has a relatively less diverse topic distribution while 2013 was pretty diverse.\n\n- the year-to-year difference has been decreasing across years...it's like the field is changing more slowly? doesn't make sense to me...",
"_____no_output_____"
]
],
[
[
"# entropy of topics\nfor iyear in range(len(term_topic)):\n print('\\n Year='+str(years[iyear]))\n entrop_terms=[]\n for k in range(ntopics):\n topic = term_topic[iyear][k] # already normalized\n entrop_terms.append(entropy(topic))\n sorted_H = np.sort(entrop_terms)\n idx = np.argsort(entrop_terms)\n [print(topic_labels[idx[j]]+':'+str(sorted_H[j])) for j in range(len(idx))]\n# turns out the ranking of entropy is pretty stable over the years.",
"_____no_output_____"
],
[
"sum(term_topic[iyear][3])",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbae05bcd1939ef58275e6d1e5b5a7b5d0f7352d
| 49,247 |
ipynb
|
Jupyter Notebook
|
workbooks/analysis1.ipynb
|
webclinic017/Algo-Owls
|
889ed0ca694399e072fcb5b294db4dcbaba3ea04
|
[
"MIT"
] | 1 |
2021-10-01T21:29:46.000Z
|
2021-10-01T21:29:46.000Z
|
workbooks/analysis1.ipynb
|
markmurdock11/Algo-Owls
|
889ed0ca694399e072fcb5b294db4dcbaba3ea04
|
[
"MIT"
] | null | null | null |
workbooks/analysis1.ipynb
|
markmurdock11/Algo-Owls
|
889ed0ca694399e072fcb5b294db4dcbaba3ea04
|
[
"MIT"
] | 2 |
2021-03-05T18:28:23.000Z
|
2021-10-01T21:29:43.000Z
| 96.942913 | 2,892 | 0.471338 |
[
[
[
"\"\"\"\nThis file was created with the purpose of developing\na random forest classifier to identify market squeeze\n\nThis squeeze classification depends of the comparison of 2 indicators:\n2 std of a 20 period bollinger bands and 2 atr of a 20 period keltner channel\n\nour definition of squeeze: \nwhen the upper bollinger band (bbup) is less or equal to upper keltner band (kcup)\nAND lower bollinger band (bblo) is above or equal to lower keltner channel (kclo)\n\"\"\"\n\n",
"_____no_output_____"
],
[
"\"\"\"\nTo develop the random forest model, a csv file was prepared extracting prices, bollinger bands and squeeze \nclassification from tradestation.\n\nA custom squeeze_id indicator was developed in easylanguage to obtain a column with values ranging 0 \nor 1 depending upon the market being on a squeeze or not (based on the requirements specified above)\n\n\"\"\"\n",
"_____no_output_____"
]
],
[
[
"# Import libraries and dependencies\nimport pandas as pd\nimport numpy as np\nfrom pathlib import Path\n%matplotlib inline\n\nimport warnings\nwarnings.filterwarnings('ignore')\n",
"_____no_output_____"
],
[
"csv_path = Path('../Resources/ts_squeeze_jpm.csv')\ncsv_path",
"_____no_output_____"
],
[
"ts_file_df = pd.read_csv(csv_path, parse_dates=[['Date', 'Time']])\nts_file_df.tail()",
"_____no_output_____"
],
[
"# set index as Date_Time and drop MidLine.1 column (it is a duplicate of MidLine)\nts_file_df.set_index(pd.to_datetime(ts_file_df['Date_Time'], infer_datetime_format=True), inplace=True)\nts_file_df.drop(columns=['Date_Time', 'MidLine.1'], inplace=True)\nts_file_df.head()\n",
"_____no_output_____"
],
[
"# Set a variable list of features to feed to our model\n\nx_var_list = ['Open', 'High', 'Low', 'Close', 'Up', 'Down', 'kcup', 'kclo', 'MidLine', 'bbup', 'bblo', 'FastEMA', 'SlowEMA']\nts_file_df[x_var_list].head()",
"_____no_output_____"
],
[
"# Shift DataFrame values by 1\nts_file_df[x_var_list] = ts_file_df[x_var_list].shift(1)\nts_file_df[x_var_list].head()",
"_____no_output_____"
],
[
"ts_file_df.head()",
"_____no_output_____"
],
[
"ts_file_df.dropna(inplace=True)\nts_file_df.head()",
"_____no_output_____"
],
[
"# Construct training start and training end dates\n\ntraining_start = ts_file_df.index.min().strftime(format='%Y-%m-%d')\ntraining_end = '2019-01-11'\n\n# Construct test start and test end dates\n\ntesting_start = '2019-01-12'\ntesting_end = '2019-06-12'\n\n# Construct validating start and validating end dates\n\nvali_start = '2019-06-13'\nvali_end = '2020-01-12'\n\n# Confirming training, testing and validating dates\nprint(f\"Training Start: {training_start}\")\nprint(f\"Training End: {training_end}\")\nprint(f\"Testing Start: {testing_start}\")\nprint(f\"Testing End: {testing_end}\")\nprint(f\"validating Start: {vali_start}\")\nprint(f\"validating end: {vali_end}\")",
"Training Start: 2018-01-12\nTraining End: 2019-01-11\nTesting Start: 2019-01-12\nTesting End: 2019-06-12\nvalidating Start: 2019-06-13\nvalidating end: 2020-01-12\n"
],
[
"# Construct the X_train and y_train datasets\nX_train = ts_file_df[x_var_list][training_start:training_end]\ny_train = ts_file_df['squeeze'][training_start:training_end]\n\nX_train.head()",
"_____no_output_____"
],
[
"y_train.tail()",
"_____no_output_____"
],
[
"# Construct the X test and y test datasets\n\nX_test = ts_file_df[x_var_list][testing_start:testing_end]\ny_test = ts_file_df['squeeze'][testing_start:testing_end]\n\nX_test.head()",
"_____no_output_____"
],
[
"y_test.head()",
"_____no_output_____"
],
[
"# Construct the X valid and y validation datasets\n\nX_vali = ts_file_df[x_var_list][vali_start:vali_end]\ny_vali = ts_file_df['squeeze'][vali_start:vali_end]\n\nX_vali.head()",
"_____no_output_____"
],
[
"y_vali.tail()",
"_____no_output_____"
],
[
"# Import SKLearn library and Classes\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.datasets import make_classification",
"_____no_output_____"
],
[
"# Fit SKLearn regression with training datasets:\nmodel = RandomForestClassifier(n_estimators=1000, max_depth=5, random_state=1)\nmodel.fit(X_train, y_train)\n\n# Make predictions of \"y\" values from the X_test dataset\npredictions = model.predict(X_test)\n\n# Assemble actual y_test with predicted values\n\ncompare_predict_df = y_test.to_frame()\ncompare_predict_df[\"predict_squeeze\"] = predictions\n\ncompare_predict_df",
"_____no_output_____"
],
[
"# Save the pre-trained model\nfrom joblib import dump, load\ndump(model, 'random_forest_model_squeeze.joblib')",
"_____no_output_____"
],
[
"\"\"\"\nBelow the exporting code to csv files\n\"\"\"",
"_____no_output_____"
],
[
"X_testoutput_path = Path('../Resources/X_test.csv')\nX_test.to_csv(X_testoutput_path)",
"_____no_output_____"
],
[
"model_results_path = Path('../Resources/results.csv')\ncompare_predict_df.to_csv(model_results_path)",
"_____no_output_____"
],
[
"# different datasets to csv files for reconfigurations\n\nX_valioutput_path = Path(\"../Resources/X_vali.csv\")\nX_vali.to_csv(X_valioutput_path)\n\ny_valioutput_path = Path(\"../Resources/y_vali.csv\")\ny_vali.to_csv(y_valioutput_path)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbae0d349e13647c9f8f2b1b50d8f62eb423b01d
| 261,402 |
ipynb
|
Jupyter Notebook
|
coursera/ml_yandex/course6/identification/check/week3_visual_analysis_and_fe2.ipynb
|
VadimKirilchuk/education
|
ebddb2fb971ff1f3991e71fcb17ce83b95c4a397
|
[
"Apache-2.0"
] | null | null | null |
coursera/ml_yandex/course6/identification/check/week3_visual_analysis_and_fe2.ipynb
|
VadimKirilchuk/education
|
ebddb2fb971ff1f3991e71fcb17ce83b95c4a397
|
[
"Apache-2.0"
] | null | null | null |
coursera/ml_yandex/course6/identification/check/week3_visual_analysis_and_fe2.ipynb
|
VadimKirilchuk/education
|
ebddb2fb971ff1f3991e71fcb17ce83b95c4a397
|
[
"Apache-2.0"
] | null | null | null | 151.977907 | 44,392 | 0.861765 |
[
[
[
"<center>\n<img src=\"https://habrastorage.org/web/677/8e1/337/6778e1337c3d4b159d7e99df94227cb2.jpg\"/>\n## Специализация \"Машинное обучение и анализ данных\"\n<center>Автор материала: программист-исследователь Mail.Ru Group, старший преподаватель Факультета Компьютерных Наук ВШЭ [Юрий Кашницкий](https://yorko.github.io/)",
"_____no_output_____"
],
[
"# <center> Capstone проект №1 <br> Идентификация пользователей по посещенным веб-страницам\n<img src='http://i.istockimg.com/file_thumbview_approve/21546327/5/stock-illustration-21546327-identification-de-l-utilisateur.jpg'>\n\n# <center>Неделя 3. Визуальный анализ данных и построение признаков\n\nНа 3 неделе мы займемся визуальным анализом данных и построением признаков. Сначала мы вместе построим и проанализируем несколько признаков, потом Вы сможете сами придумать и описать различные признаки. \n\n**План 3 недели:**\n - Часть 1. Построение признаков\n - Часть 2. Визуальный анализ данных\n - Часть 3. Дальнейшее построение признаков\n - Часть 4. Проверка построенных признаков\n\n**В этой части проекта Вам могут быть полезны видеозаписи следующих лекций курса \"Поиск структуры в данных\":**\n - [Задача визуализации](https://www.coursera.org/learn/unsupervised-learning/lecture/hlvlT/zadacha-vizualizatsii)\n - [Визуализация данных в sklearn](https://www.coursera.org/learn/unsupervised-learning/lecture/ityMo/vizualizatsiia-dannykh-v-sklearn)\n \n**Также в задании будет использоваться библиотека Seaborn (ее можно дополнительно установить командой *pip install seaborn*), будет полезно обращаться к документациям [Matplotlib](http://matplotlib.org/users/) и [Seaborn](http://seaborn.pydata.org/), а также к примерам визуализации, описанным на StackOverflow.**\n\n",
"_____no_output_____"
],
[
"### Задание\n1. Заполните код в этой тетрадке \n2. Если вы проходите специализацию Яндеса и МФТИ, пошлите тетрадку в соответствующем Peer Review. <br> Если вы проходите курс ODS, выберите ответы в [веб-форме](https://docs.google.com/forms/d/1EbjK7-hF-Gepi6RH-K5I2XeiYGRoY0LNDx03QmLu9Xo). ",
"_____no_output_____"
],
[
"## Часть 1. Построение признаков",
"_____no_output_____"
]
],
[
[
"# отключим всякие предупреждения Anaconda\nimport warnings\nwarnings.filterwarnings('ignore')\nfrom glob import glob\nimport os\nfrom tqdm import tqdm\nimport numpy as np\nimport pandas as pd\nimport re\nimport datetime\nfrom itertools import chain\npd.set_option('display.max.columns', 25)\nimport pickle\n#pip install seaborn\nimport seaborn as sns\n%matplotlib inline\nfrom matplotlib import pyplot as plt",
"_____no_output_____"
],
[
"# Поменяйте на свой путь к данным\nPATH_TO_DATA = 'capstone_user_identification'",
"_____no_output_____"
]
],
[
[
"**Создайте на основе функций *prepare_train_set* и *prepare_sparse_train_set_window* новую – *prepare_train_set_with_fe*, (от \"feature engineering\"), создайте следующие признаки:**\n- `session_timespan` – продолжительность сессии (разница между максимальным и минимальным временем посещения сайтов в сессии, в секундах)\n- `#unique_sites` – число уникальных сайтов в сессии \n- `start_hour` – час начала сессии (то есть час в записи минимального timestamp среди десяти)\n- `day_of_week` – день недели (то есть день недели в записи минимального timestamp среди десяти)\n\nФункция должна возвращать новый DataFrame (как возвращала функция *prepare_train_set*), только признаков должно быть на 4 больше. Порядок, в котором добавляются признаки: *site1*, ... *site10*, *session_timespan*, *#unique_sites*, *start_hour*, *day_of_week* и *user_id* (это видно и чуть ниже по тому, как функция вызывается).",
"_____no_output_____"
]
],
[
[
"def boundaries(N, session_length, window_size):\n ran = lambda x: x+session_length\n slice_list=[(i, ran(i) if ran(i) < N else N) for i in range(0, N, window_size)]\n return slice_list\n\ndef prepare_train_set_with_fe(path_to_csv_files, site_freq_path, feature_names,\n session_length=10, window_size=10):\n user_re = re.compile(\"user([\\d]+)[.]\")\n list_times_incomplete = []\n list_sites = []\n list_users = []\n list_timediffs = []\n \n with open(site_freq_path,\"rb\") as f:\n site_freq = pickle.load(f)\n\n for file in tqdm(glob(path_to_csv_files+'/*')):\n sites_raw = pd.read_csv(file)['site'].apply(lambda x: site_freq[x][0])\n timestamps_raw = pd.read_csv(file)['timestamp']\n\n indices = boundaries(len(sites_raw),session_length,window_size)\n list_users += [int(re.search(user_re, file).group(1))] * len(indices)\n list_times_incomplete += [timestamps_raw.values[ind[0]:ind[1]].reshape(-1) for ind in indices]\n list_sites += [sites_raw.values[ind[0]:ind[1]].reshape(-1) for ind in indices]\n \n\n list_times = [list(map(np.datetime64, i)) for i in list_times_incomplete]\n total_time = [(i[-1]-i[0]).astype(int) for i in list_times]\n unique = [len(np.unique(i)) for i in list_sites]\n \n for session in list_times:\n localdiff = [(session[i]-session[i-1]).astype(int) for i in range(1, len(session))]\n list_timediffs.append(localdiff)\n \n df_tstamps = pd.DataFrame(list_times, columns=[f'time{i}' for i in range(session_length)])\n df_sites = pd.DataFrame(list_sites)\n df_timediffs = pd.DataFrame(list_timediffs) \n df = pd.concat([df_sites, df_timediffs], axis=1)\n df = df.fillna(0).astype('int')\n \n df['total'] = total_time\n df['unique'] = unique\n df['hours'] = df_tstamps['time0'].dt.hour\n df['days'] = df_tstamps['time0'].dt.dayofweek\n df['user_id'] = list_users\n df.columns = feature_names\n \n return df",
"_____no_output_____"
]
],
[
[
"**Проверим функцию на игрушечном примере.**",
"_____no_output_____"
]
],
[
[
"feature_names = ['site' + str(i) for i in range(1,11)] + \\\n ['time_diff' + str(j) for j in range(1,10)] + \\\n ['session_timespan', '#unique_sites', 'start_hour', \n 'day_of_week', 'target']\ntrain_data_toy = prepare_train_set_with_fe(os.path.join(PATH_TO_DATA, \n '3users'), \n site_freq_path=os.path.join(PATH_TO_DATA, \n 'site_freq_3users.pkl'),\n feature_names=feature_names, session_length=10)",
"100%|████████████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 8.31it/s]\n"
],
[
"train_data_toy",
"_____no_output_____"
]
],
[
[
"**Примените функцию *prepare_train_set_with_fe* к данным по 10 пользователям, укажите *session_length*=10.**",
"_____no_output_____"
]
],
[
[
"%%time\ntrain_data_10users = prepare_train_set_with_fe(os.path.join(PATH_TO_DATA, \n '10users'), \n site_freq_path=os.path.join(PATH_TO_DATA, \n 'site_freq_10users.pkl'),\n feature_names=feature_names, session_length=10)",
"100%|██████████████████████████████████████████████████████████████████████████████████| 10/10 [00:01<00:00, 9.93it/s]\n"
],
[
"train_data_10users.head()",
"_____no_output_____"
]
],
[
[
"**Примените функцию *prepare_train_set_with_fe* к данным по 150 пользователям, укажите *session_length*=10.**",
"_____no_output_____"
]
],
[
[
"%%time\ntrain_data_150users = prepare_train_set_with_fe(os.path.join(PATH_TO_DATA, \n '150users'), \n site_freq_path=os.path.join(PATH_TO_DATA, \n 'site_freq_150users.pkl'),\n feature_names=feature_names, session_length=10)",
"100%|████████████████████████████████████████████████████████████████████████████████| 150/150 [00:09<00:00, 16.19it/s]\n"
]
],
[
[
"**Сохраните в pickle-файлы признаки *session_timespan*, *#unique_sites*, *start_hour* и *day_of_week* для 10 и 150 пользователей.**",
"_____no_output_____"
]
],
[
[
"new_features_10users = train_data_10users.loc[:,['session_timespan', '#unique_sites', 'start_hour', 'day_of_week']]\nnew_features_150users = train_data_150users.loc[:,['session_timespan', '#unique_sites', 'start_hour', 'day_of_week']]",
"_____no_output_____"
],
[
"with open(os.path.join(PATH_TO_DATA, \n 'new_features_10users.pkl'), 'wb') as new_features_10users_pkl:\n pickle.dump(new_features_10users, new_features_10users_pkl)\nwith open(os.path.join(PATH_TO_DATA, \n 'new_features_150users.pkl'), 'wb') as new_features_150users_pkl:\n pickle.dump(new_features_150users, new_features_150users_pkl)",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Вопрос 1. </font> Выведите медианную продолжительность сессии (*session_timespan*) для сессий 10 пользователей.**",
"_____no_output_____"
]
],
[
[
"np.median(train_data_10users.session_timespan)",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Вопрос 2. </font> Выведите медианный день недели, в который началась сессия, для сессий 10 пользователей.**",
"_____no_output_____"
]
],
[
[
"np.median(train_data_10users.day_of_week)",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Вопрос 3. </font>Выведите медианный час начала сессии для сессий 150 пользователей.**",
"_____no_output_____"
]
],
[
[
"np.median(train_data_150users.start_hour)",
"_____no_output_____"
]
],
[
[
"**<font color='red'>Вопрос 4. </font>Выведите медианное значение числа уникальных сайтов в сессиях 150 пользователей.**",
"_____no_output_____"
]
],
[
[
"np.median(train_data_150users['#unique_sites'])",
"_____no_output_____"
]
],
[
[
"## Часть 2. Визуальный анализ данных",
"_____no_output_____"
],
[
"**Забавы ради, потехи для дадим пользователям имена и ассоциируем с ними цвета.**",
"_____no_output_____"
]
],
[
[
"id_name_dict = {128: 'Mary-Kate', 39: 'Ashley', 207: 'Lindsey', 127: 'Naomi', 237: 'Avril',\n 33: 'Bob', 50: 'Bill', 31: 'John', 100: 'Dick', 241: 'Ed'}\ntrain_data_10users['target'] = train_data_10users['target'].map(id_name_dict)",
"_____no_output_____"
],
[
"color_dic = {'Mary-Kate': 'pink', 'Ashley': 'darkviolet', 'Lindsey':'blueviolet', \n 'Naomi': 'hotpink', 'Avril': 'orchid', \n 'Bob': 'firebrick', 'Bill': 'gold', 'John': 'forestgreen', \n 'Dick': 'slategrey', 'Ed':'brown'}",
"_____no_output_____"
]
],
[
[
"**1. Постройте гистограмму распределения длины сессии в секундах (*session_timespan*). Ограничьте по *x* значением 200 (иначе слишком тяжелый хвост). Сделайте гистограмму цвета *darkviolet*, подпишите оси по-русски.**",
"_____no_output_____"
]
],
[
[
"train_data_10users['session_timespan'].hist(color='darkviolet', range=(0, 200))\nplt.xlabel('Длина сессии')\nplt.ylabel('Частота')",
"_____no_output_____"
]
],
[
[
"**2. Постройте гистограмму распределения числа уникальных сайтов в сессии (*#unique_sites*). Сделайте гистограмму цвета *aqua*, подпишите оси по-русски.**",
"_____no_output_____"
]
],
[
[
"train_data_10users['#unique_sites'].hist(color='aqua')\nplt.xlabel('Число уникальных сайтов')\nplt.ylabel('Частота')",
"_____no_output_____"
]
],
[
[
"**3. Постройте гистограммы распределения числа уникальных сайтов в сессии (*#unique_sites*) для каждого из 10 пользователей по отдельности. Используйте *subplots*, чтоб разместить все 10 картинок на одной большой. Пометьте легендой каждую картинку, на легенде должно быть написано имя пользователя. Для каждого пользователя раскрасьте гистограмму его/ее цветом (*color_dic*). Подпишите оси по-русски в каждой из 10 гистограмм.**",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=3, ncols=4, figsize=(16, 10))\n\n# как вариант, можно и по-другому\n \nindices = [(i, j) for i in range(3) for j in range(4)]\nfor idx, (user, sub_df) in enumerate(train_data_10users.groupby('target')): \n sub_df['#unique_sites'].hist(ax=axes[indices[idx]], color=color_dic[user])\n axes[indices[idx]].legend([user])",
"_____no_output_____"
]
],
[
[
"**4. Постройте гистограмму распределения часа начала сессии (*start_hour*). Сделайте гистограмму цвета *darkgreen*, подпишите оси по-русски.**",
"_____no_output_____"
]
],
[
[
"train_data_10users['start_hour'].hist(color='darkgreen')\nplt.xlabel('Час начала сессии')\nplt.ylabel('Частота')",
"_____no_output_____"
]
],
[
[
"**5. Постройте гистограммы распределения часа начала сессии (*start_hour*) для каждого из 10 пользователей по отдельности. Используйте *subplots*, чтоб разместить все 10 картинок на одной большой. Пометьте легендой каждую картинку, на легенде должно быть написано имя пользователя. Для каждого пользователя раскрасьте гистограмму его/ее цветом (*color_dic*). Подпишите оси по-русски в каждой из 10 гистограмм.**",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=3, ncols=4, figsize=(16, 10))\n\n# как вариант, можно и по-другому\n\nindices = [(i, j) for i in range(3) for j in range(4)]\nfor idx, (user, sub_df) in enumerate(train_data_10users.groupby('target')): \n sub_df['start_hour'].hist(ax=axes[indices[idx]], color=color_dic[user])\n axes[indices[idx]].legend([user])",
"_____no_output_____"
]
],
[
[
"**6. Постройте гистограмму распределения дня недели, в который началась сессия (*day_of_week*). Сделайте гистограмму цвета *sienna*, подпишите оси по-русски.**",
"_____no_output_____"
]
],
[
[
"train_data_10users['day_of_week'].hist(color='sienna')\nplt.xlabel('День недели начала сессии')\nplt.ylabel('Частота')",
"_____no_output_____"
]
],
[
[
"**7. Постройте гистограммы распределения дня недели, в который началась сессия (*day_of_week*) для каждого из 10 пользователей по отдельности. Используйте *subplots*, чтоб разместить все 10 картинок на одной большой. Измените метки по оси *X* на ['Пн', 'Вт', 'Ср', 'Чт', 'Пт', 'Сб', 'Вс'] – метод *set_xticklabels*. Пометьте легендой каждую картинку, на легенде должно быть написано имя пользователя. Для каждого пользователя раскрасьте гистограмму его/ее цветом (*color_dic*). Подпишите по-русски название каждой из 10 гистограмм.**",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=3, ncols=4, figsize=(16, 10))\n\n# как вариант, можно и по-другому\nweekdays = ['Пн', 'Вт', 'Ср', 'Чт', 'Пт', 'Сб', 'Вс']\n\nindices = [(i, j) for i in range(3) for j in range(4)]\nfor idx, (user, sub_df) in enumerate(train_data_10users.groupby('target')): \n sub_df['day_of_week'].hist(ax=axes[indices[idx]], color=color_dic[user])\n axes[indices[idx]].legend([user])\n axes[indices[idx]].set_xticklabels(weekdays)",
"_____no_output_____"
]
],
[
[
"**8. Сделайте выводы про каждого пользователя по построенным графикам.**",
"_____no_output_____"
],
[
"Bill и Ashley часто заходят на один сайт. Dick и Mary-Kate - на два. У остальных распределение по уникальным сайтам примерно одинаковое. Ashley чаще всего заходит куда-либо днем. У Avril пик приходится на обед и вечер. У Bill - утро и обед. Bob, по-видимому, не сидит в интернете позднее 17 часов. У Dick и John пик приходится на утро и обед. John и Lindsey редко заходит в интернет вечером. У Mary-Kate пик приходится на вечер. Naomi почти не заходит в интернет утром, пик приходится на обед. Ashley чаще всего заходит в интернет по четвергам. У Avril распределение по дням недели примерно равномерно. У Bill и Lindsey пик приходится на начало недели, к концу происходит постепенный спад. У John наблюдается обратная тенденция. У Dick два пика приходятся на среду и выходные. Во вторник и четверг он почти не сидит в интернете. У Mary-Kate и Naomi распределение в целом равномерно, но пик приходится на вторую половину недели.",
"_____no_output_____"
],
[
"**Загрузите сохраненный ранее в pickle-файл частотный словарь сайтов для 10 пользователей. **",
"_____no_output_____"
],
[
"**Определите топ-10 самых посещаемых сайтов (*top10_sites*) и соответствующие кол-ва посещений (*top10_freqs*).**",
"_____no_output_____"
]
],
[
[
"with open(f'{PATH_TO_DATA}\\\\site_freq_10users.pkl', 'rb') as f:\n vocab = pickle.load(f)\n\nvocab_sort = list(vocab.items())\nvocab_sort.sort(key=lambda x: x[1][1], reverse=True)\n\ntop10_freqs = [i[1][1] for i in vocab_sort[:10]]\ntop10_sites = [i[0] for i in vocab_sort[:10]]",
"_____no_output_____"
]
],
[
[
"**9. Нарисуйте *seaborn barplot*, показывающий частоты посещений топ-10 сайтов. Сделайте подписи сайтов вертикальными, иначе они сливаются (*xticks*).**",
"_____no_output_____"
]
],
[
[
"sns.barplot(x=top10_sites, y=top10_freqs)\nplt.xticks(rotation='vertical')",
"_____no_output_____"
]
],
[
[
"## Часть 3. Дальнейшее построение признаков",
"_____no_output_____"
],
[
"Это задание творческое, тут надо придумать, как еще учесть время посещения веб-страниц и прочие признаки. \n\nНа следующей неделе мы будем использовать \"мешок\" сайтов для классификации сессий по принадлежности разным пользователям, а эти новые признаки, которые Вы сейчас создадите, потом добавим и посмотрим, улучшается ли модель. Поэтому можно их создать в виде отдельных матриц и сохранить их также отдельно. \n\nВ этой части задания Вы можете построить и визуально исследовать самые разные признаки (ничто фантазию не ограничивает):\n- год, месяц и день начала сессии\n- час начала сессии (с учетом года, месяца и дня)\n- время суток\n- среднее время пребывания на сайте, посчитать можно, скажем, для топ-30 популярных сайтов\n- индикаторы посещения популярных сайтов (скажем, тоже для топ-30 популярных сайтов)\n- частота посещения Facebook\n- ...",
"_____no_output_____"
],
[
"**Напишите функцию для создания новых признаков и примените ее к исходным данным – каталогам с 10 и 150 файлами. Сделайте это только для набора данных, полученного с параметрами *session_length=10* и *window_size=10*. Сериализуйте полученные матрицы с помощью pickle. Функция может возвращать как только новые признаки, так и старые с новыми. При этом сигнатура функции может быть другой – тут уже свобода выбора.**",
"_____no_output_____"
]
],
[
[
"def feature_engineering(path_to_csv_files, site_freq_path, features, session_length=10):\n user_re = re.compile(\"user([\\d]+)[.]\")\n list_times_incomplete = []\n list_sites = []\n list_users = []\n list_timediffs = []\n \n with open(site_freq_path,\"rb\") as f:\n site_freq = pickle.load(f)\n\n for file in tqdm(glob(path_to_csv_files+'/*')):\n sites_raw = pd.read_csv(file)['site'].apply(lambda x: site_freq[x][0])\n timestamps_raw = pd.read_csv(file)['timestamp']\n\n indices = boundaries(len(sites_raw),session_length, 10)\n list_users += [int(re.search(user_re, file).group(1))] * len(indices)\n list_times_incomplete += [timestamps_raw.values[ind[0]:ind[1]].reshape(-1) for ind in indices]\n list_sites += [sites_raw.values[ind[0]:ind[1]].reshape(-1) for ind in indices]\n \n\n list_times = [list(map(np.datetime64, i)) for i in list_times_incomplete]\n total_time = [(i[-1]-i[0]).astype(int) for i in list_times]\n unique = [len(np.unique(i)) for i in list_sites]\n facebook_id = site_freq.get('www.facebook.com', (-1, -1))[0]\n google_id = site_freq.get('www.google.com', (-1, -1))[0]\n facebook_count = [list(i).count(facebook_id) for i in list_sites]\n google_count = [list(i).count(google_id) for i in list_sites]\n total_time = [(i[-1]-i[0]).astype(int) for i in list_times] \n \n \n for session in list_times:\n localdiff = [(session[i]-session[i-1]).astype(int) for i in range(1, len(session))]\n list_timediffs.append(localdiff)\n \n df_tstamps = pd.DataFrame(list_times, columns=[f'time{i}' for i in range(session_length)])\n df_sites = pd.DataFrame(list_sites, columns=[f'site{i}' for i in range(1, session_length+1)])\n df_timediffs = pd.DataFrame(list_timediffs, columns=[f'time{i}' for i in range(1, session_length)]) \n df = pd.concat([df_sites, df_timediffs], axis=1)\n df = df.fillna(0).astype('int')\n \n df['session_timespan'] = total_time\n df['#unique_sites'] = unique\n df['start_hour'] = df_tstamps['time0'].dt.hour\n df['day_of_week'] = df_tstamps['time0'].dt.dayofweek\n df['target'] = list_users\n df['facebook_visits'] = facebook_count\n df['google_visits'] = google_count\n df = df.loc[:, features]\n \n return df, df.values",
"_____no_output_____"
],
[
"features = ['facebook_visits', 'google_visits']",
"_____no_output_____"
],
[
"new_features_10users, _ = feature_engineering(os.path.join(PATH_TO_DATA, '10users'), site_freq_path=os.path.join(PATH_TO_DATA, \n 'site_freq_10users.pkl'), features=features)",
"100%|██████████████████████████████████████████████████████████████████████████████████| 10/10 [00:00<00:00, 9.17it/s]\n"
],
[
"new_features_150users, _ = feature_engineering(os.path.join(PATH_TO_DATA, '150users'), site_freq_path=os.path.join(PATH_TO_DATA, \n 'site_freq_150users.pkl'), features=features)",
"100%|████████████████████████████████████████████████████████████████████████████████| 150/150 [00:07<00:00, 30.73it/s]\n"
]
],
[
[
"**10. Постройте картинки для новых признаков, поисследуйте их, прокомментируйте результаты.**",
"_____no_output_____"
]
],
[
[
"fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(12, 4))\nnew_features_10users['facebook_visits'].hist(ax=ax1, color='red')\nnew_features_10users['google_visits'].hist(ax=ax2, color='blue')",
"_____no_output_____"
],
[
"new_features_10users['facebook_visits'].value_counts()",
"_____no_output_____"
],
[
"new_features_10users['google_visits'].value_counts()",
"_____no_output_____"
]
],
[
[
"Мы видим, что доля пользователей, посещающих гугл, больше, чем доля пользователей, не посещающих его. Доля пользователей, посетивших гугл 10 раз за сессию, также больше.",
"_____no_output_____"
]
],
[
[
"fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(12, 4))\nnew_features_150users['facebook_visits'].hist(ax=ax1, color='red')\nnew_features_150users['google_visits'].hist(ax=ax2, color='blue')",
"_____no_output_____"
],
[
"new_features_150users['facebook_visits'].value_counts()",
"_____no_output_____"
],
[
"new_features_150users['google_visits'].value_counts()",
"_____no_output_____"
]
],
[
[
"Мы видим, что тенденция сохраняется и для 150 пользователей",
"_____no_output_____"
],
[
"**В конце сохраните в pickle-файлы только те признаки, которые, как Вы предполагаете, помогут идентифицировать пользователя более точно. Это касается и признаков, которые мы вместе создали в начале (*session_timespan, #unique_sites, start_hour, day_of_week*), и Ваших собственных. Можно создать все эти признаки не только для сессий из 10 сайтов, но и для других сочетаний параметров *session_length* и *window_size*.**",
"_____no_output_____"
]
],
[
[
"features = ['session_timespan', '#unique_sites', 'start_hour', 'day_of_week']",
"_____no_output_____"
],
[
"selected_features_10users, _ = feature_engineering(os.path.join(PATH_TO_DATA, '10users'), site_freq_path=os.path.join(PATH_TO_DATA, \n 'site_freq_10users.pkl'), features=features)\nselected_features_150users, _ = feature_engineering(os.path.join(PATH_TO_DATA, '150users'), site_freq_path=os.path.join(PATH_TO_DATA, \n 'site_freq_150users.pkl'), features=features) ",
"100%|██████████████████████████████████████████████████████████████████████████████████| 10/10 [00:00<00:00, 19.80it/s]\n100%|████████████████████████████████████████████████████████████████████████████████| 150/150 [00:07<00:00, 20.27it/s]\n"
],
[
"with open(os.path.join(PATH_TO_DATA, \n 'selected_features_10users.pkl'), 'wb') as selected_features_10users_pkl:\n pickle.dump(selected_features_10users, selected_features_10users_pkl, \n protocol=2)\nwith open(os.path.join(PATH_TO_DATA, \n 'selected_features_150users.pkl'), 'wb') as selected_features_150users_pkl:\n pickle.dump(selected_features_150users, selected_features_150users_pkl, \n protocol=2)",
"_____no_output_____"
]
],
[
[
"### Критерии оценки работы (только для Peer Review в специализации):\n- Верно ли отображена гистограмма session_timespan из п. 1? (max. 3 балла)\n- Верно ли отображена гистограмма #unique_sites из п. 2? (max. 3 балла)\n- Верно ли отображены гистограммы #unique_sites по каждому пользователю из п. 3? (max. 6 баллов)\n- Верно ли отображена гистограмма start_hour из п. 4? (max. 3 балла)\n- Верно ли отображены гистограммы start_hour по каждому пользователю из п. 5? (max. 6 баллов)\n- Верно ли отображена гистограмма day_of_week из п. 6? (max. 3 балла)\n- Верно ли отображены гистограммы day_of_week по каждому пользователю из п. 7? (max. 6 баллов)\n- Насколько сделанные выводы в п. 8 соответствуют построенным картинкам? (max. 6 баллов)\n- Верно ли отображен barplot для 10 популярных сайтов из п. 9? (max. 6 баллов)\n- Правильно ли посчитана медианная продолжительность сессий в п. 10? (max. 3 балла)\n- Правильно ли посчитан медианный день недели начала сессии в п. 11? (max. 3 балла)\n- Правильно ли посчитан медианный час начала сессии в п. 12? (max. 3 балла)\n- Правильно ли посчитано медианное значение числа уникальных сайтов в сессиях 150 пользователей п. 13? (max. 3 балла)\n- Есть ли оригинальные построенные признаки и картинки к ним? Оцените также и качество картинок. (max. 8 баллов)",
"_____no_output_____"
],
[
"## Пути улучшения\nЧто еще можно добавить по 3 части проекта:\n- IPython-widgets, интерактив и анимация (стоящие статьи по этому ремеслу – [раз](https://habrahabr.ru/post/308162/) и [два](https://habrahabr.ru/company/ods/blog/323210/))\n- можно попробовать изобразить исходные данные в некотором пространстве, например, Word2Vec, потом выделить главные компоненты или t-SNE (только пользуйтесь эффективными реализациями типа [Multicore-TSNE](https://github.com/DmitryUlyanov/Multicore-TSNE), не Sklearn) и раскрасить по целевому классу. Но нет гарантий, что получится что-то значимо отличающееся от каши\n\nНа следующей неделе мы наконец приступим к обучению моделей классификации. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbae0ecee554753dae771e5918e04b9ec737d01b
| 636,858 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/lab1-Python-checkpoint.ipynb
|
ubco-mds-2020-labs/dashboard-project-cryptocurrency_db
|
7ebac16fc0012408f098bd3bed7d427f6ad052a8
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/lab1-Python-checkpoint.ipynb
|
ubco-mds-2020-labs/dashboard-project-cryptocurrency_db
|
7ebac16fc0012408f098bd3bed7d427f6ad052a8
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/lab1-Python-checkpoint.ipynb
|
ubco-mds-2020-labs/dashboard-project-cryptocurrency_db
|
7ebac16fc0012408f098bd3bed7d427f6ad052a8
|
[
"MIT"
] | 1 |
2021-03-04T19:16:54.000Z
|
2021-03-04T19:16:54.000Z
| 664.778706 | 169,906 | 0.945785 |
[
[
[
"# 531 - Lab 1 - Visualizing world health data\n\nThere are two versions of this lab, one in Python and one in R.\nThe R lab will use `ggplot` and the Python lab will use `Altair`.\n\nThis is the Python version.\n\nPlease choose a version to complete, though keep in mind that you are required to alternate between completing the R labs and the Python labs to get experience using both languages.",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n## Submission instructions\nrubric={mechanics:2}\n\n<p>You receive marks for submitting your lab correctly, please follow these instructions:</p>\n\n<ul>\n <li><a href=\"https://ubc-mds.github.io/resources_pages/general_lab_instructions/\">\n Follow the general lab instructions.</a></li>\n <li><a href=\"https://github.com/UBC-MDS/public/tree/master/rubric\">\n Click here to view a description of the rubrics used to grade the questions</a></li>\n <li>Push your <code>.ipynb</code> file to your GitHub repository for this lab.</li>\n <li>Upload a <code>.html</code> version of your assignment to Canvas.\n <ul>\n <li> Either manually or using the last cell of this notebook.</li>\n </ul>\n </li>\n <li>Include a clickable link to your GitHub repo for the lab just below this cell\n <ul>\n <li>It should look something like this https://github.ubc.ca/MDS-2020-21/DSCI_531_labX_yourcwl.</li>\n </ul>\n </li>\n</ul>\n</div>",
"_____no_output_____"
],
[
"https://github.com/ubco-mds-2020-labs/data-550-lab-1-group-11",
"_____no_output_____"
]
],
[
[
"# Run this cell to ensure that altair plots show up in the exported HTML\n# and that the R cell magic works\nimport altair as alt\n\n# Save a vega-lite spec and a PNG blob for each plot in the notebook\nalt.renderers.enable('mimetype')\n# Handle large data sets without embedding them in the notebook\nalt.data_transformers.enable('data_server')\n",
"_____no_output_____"
]
],
[
[
"# 1. Get motivated!\n\nYou have already worked with the Gapminder world health data set in the previous block\nand we will revisit an updated version of it in this lab.\nThe Gapminder foundation strives to educate people about the public health status\nin countries all around the world\nand fight devastating misconceptions that hinder world development.\nThis information is important both for our capacity to make considerate choices as individuals,\nand from an industry perspective in understanding where markets are emerging.\nIn their research,\nGapminder has discovered that most people don't really know what the world looks like today.\nDo you?\n[Take this 7-8 min quiz to find out](https://forms.gapminder.org/s3/test-2018).\n\nThis quiz is not easy,\nso don't worry if you get a low score.\nI took this quiz for the first time a few years back and I didn't do too well myself =)\nIt is primarily meant to spark your curiosity to learn more about this lab's data set!\nWhen you are done,\n[please submit your score in this Google form](https://docs.google.com/forms/d/e/1FAIpQLSc2B0wlF-QWqAeJnHbu534WT-Twhpetk_4uUMM3LZvV0wv0mg/viewform?usp=sf_link).\nThis is anonymous,\nI just want to explore if we can use the the distribution of scores\nfor something interesting in class or future labs.",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n### Question 1.1\nrubric={reasoning:1,writing:1}\n\n<p>To answer the first lab question\n<a href=https://www.youtube.com/watch?v=usdJgEwMinM>watch this 20 min video of Hans Rosling</a>\na public health professor at Karolinska Institute\nwho founded Gapminder together with his son and his son's wife.\nAlthough the video is almost 15 years old,\nit is a formidable demonstration on how to present data in a way that engages your audience\nwhile conveying a strong, important message.\n(The original clip has over 3 million views,\nbut I linked you one of better video quality).</p>\n\n<p>Briefly describe (<=90 words)\nwhat you think is the most important message conveyed in the video\nand which data visualization you think was the most effective\nin getting this message across to the viewers.</p>\n \n</div>",
"_____no_output_____"
],
[
"# YOUR ANSWER GOES HERE\n\nAverage people (60% of total population) takes 24% of the total income. \nThe most effective message in my opinion was how Asia (who were the poorest in 1970) and caused the bigger hump in the continuous histogram moved towards average by 2003 by moving out of poverty and the hump in the graph reduced.\n\nAlso, within Asia, the progress of South Korea (speed and direction of development), and the country move much faster if they are healthy first than wealthy first",
"_____no_output_____"
],
[
"# 2. The Gapminder bubble chart\n\nThe \"bubble chart\" have become quite famous from their appearance in the Gapminder talks,\nand are widely used in other areas as well.\nLet's start by recreating a simple version of this chart ourselves!\n\nThere will be some data wrangling involved in this lab,\nand since 531 is primarily about visualization and this is the first lab,\nI will give you some hints for most data wrangling parts of this lab.\nOften I will link documentation or StackOverflow,\nso that you get practice finding information on these sources,\nand sometimes you will need to search them yourself if I haven't included a link.\n\nTo make this more interesting,\nI have compiled a more recent version of the Gapminder dataset,\nwhich contains values up until 2018 for most of the features.\nWe will not use all the columns in the data set,\nbut here is a description of what they contain\nthat you can refer back to throughout the lab.\n\n| Column | Description |\n|-----------------------|----------------------------------------------------------------------------------------------|\n| country | Country name |\n| year | Year of observation |\n| population | Population in the country at each year |\n| region | Continent the country belongs to |\n| sub_region | Sub-region the country belongs to |\n| income_group | Income group [as specified by the world bank in 2018] |\n| life_expectancy | The mean number of years a newborn would <br>live if mortality patterns remained constant |\n| income | GDP per capita (in USD) <em>adjusted <br>for differences in purchasing power</em> |\n| children_per_woman | Average number of children born per woman |\n| child_mortality | Deaths of children under 5 years <break>of age per 1000 live births |\n| pop_density | Average number of people per km<sup>2</sup> |\n| co2_per_capita | CO2 emissions from fossil fuels (tonnes per capita) |\n| years_in_school_men | Mean number of years in primary, secondary,<br>and tertiary school for 25-36 years old men |\n| years_in_school_women | Mean number of years in primary, secondary,<br>and tertiary school for 25-36 years old women |\n\n[as specified by the world bank in 2018]: https://datahelpdesk.worldbank.org/knowledgebase/articles/378833-how-are-the-income-group-thresholds-determined",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n### Question 2\nrubric={accuracy:1,quality:1,viz:2}\n\n<h4>Python</h4>\n<ol type=\"1\">\n<li>I have uploaded the <a href=https://raw.githubusercontent.com/UofTCoders/workshops-dc-py/master/data/processed/world-data-gapminder.csv> 2018 Gapminder data at this URL.</a> Use <code>read_csv</code> from <code>pandas</code> to load the data directly from the URL and assign it a suitable variable name. Set the <code>parse_dates</code> parameter to <code>['year']</code> to ensure that Altair recognizes this columns as time data.</li>\n<li>Now let’s create a similar bubble chart to what you saw in the video:\n<ul>\n<li>Filter the dataframe to only keep observations from a single year, 1962. You can create a new data frame variable or perform the filtering directly as you pass the data to Altair. Dates can be matched as strings when filtering.</li>\n<li>Use a circle mark to recreate the appearance of the plot in the video.</li>\n<li>Encode the proper variables so that children per woman is on the x-axis, life expectancy on the y-axis, and so that the circles’ color corresponds to their region, and the size reflects the population.</li>\n</ul></li>\n</ol>\n<p> Don't worry about getting axis labels and sizes to be exactly like in the video,\nwe will return to this code later in the lab to customize it.</p>\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE\nimport pandas as pd\n\ndf = pd.read_csv(\"https://raw.githubusercontent.com/UofTCoders/workshops-dc-py/master/data/processed/world-data-gapminder.csv\", parse_dates=['year'])",
"_____no_output_____"
],
[
"import datetime as dt \ndf_1962 = df[df['year'].dt.year == 1962]",
"_____no_output_____"
],
[
"import altair as alt\n\nalt.Chart(df_1962).mark_point().encode(\n alt.X('children_per_woman'),\n alt.Y('life_expectancy'),\n size='region',\n color='region'\n)\n",
"_____no_output_____"
]
],
[
[
"# 3. Education balance\n\nA common misconception is that women around the world go to school many years less than men. Let’s find out what the data actually says about this.",
"_____no_output_____"
]
],
[
[
"df['ratio'] = df['years_in_school_women']/ df['years_in_school_men']\ndf1 = df.loc[df[\"year\"].between('1970-01-01', '2015-12-31')]\ndf2 = df1.groupby(['income_group','year'], as_index=False).agg({\"ratio\": \"mean\"})\n",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n### Question 3\nrubric={accuracy:2,quality:1,viz:2}\n\n<h4>\nPython\n</h4>\n<ol type=\"1\">\n<li>Compute a new column in your dataframe that represents the ratio between the number of years in school for women and men (calculate it so that the value 1 means as many years for both, and 0.5 means half as many for women compared to men).</li>\n<li>Filter the dataframe to only contain value from 1970 - 2015, since those are the years where the education data has been recorded. Again you can either create a new variable or perform the filtering as you pass the data to the plotting function.</li>\n<li>Create a line plot showing how the ratio of women’s of men’s years in school has changed over time. Group the data by income group and plot the mean for each group.</li>\n<li>Use layering to add a square mark for every data point in your line plot (so one per yearly mean in each group).</li>\n</ol>\n\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE\n\nalt.Chart(df2).mark_line().encode(\n x='year',\n y='ratio',\n color='income_group',\n shape=alt.Shape('income_group', scale=alt.Scale(range=['square', 'square', 'square', 'square']), legend=None)\n)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-warning\" style=\"color:black\">\n\n### Question 3.1 (Optional)\nrubric={accuracy:1}\n\n<h4>\nPython\n</h4>\nAdd <a href=https://altair-viz.github.io/gallery/line_with_ci.html> a confidence interval band</a>\nto your line + square plot by assigning the plot in the previous question to a variable name\nand then using layering to add the band.\nThe default in the link above is a 95% bootstrapped confidence interval.\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE",
"_____no_output_____"
]
],
[
[
"# 4. Family planning\n\nAnother common misconception is that saving the lives of children in low income countries\nwill lead to overpopulation.\nRather,\nlower child mortality is actually correlated with smaller family sizes.\nAs more children survive,\nparents feel more secure with a smaller family size.\nLet's have a look in the data to see how this relationship has evolved over time.\n\nIn the plots we are going to make,\nit is important to note that it is not possible to tell causation,\njust correlation.\nHowever,\nin the [Gapminder](https://www.gapminder.org/videos/) video library\nthere are a few videos on this topic\n(including [this](https://www.gapminder.org/answers/will-saving-poor-children-lead-to-overpopulation/)\nand [this](https://www.gapminder.org/videos/population-growth-explained-with-ikea-boxes/) one),\ndiscussing how reducing poverty can help slow down population growth\nthrough decreased family sizes.\nCurrent estimates suggest that the word population\nwill stabilize around 11 billion people\nand the average number of children per woman\nwill be close to two worldwide in year 2100.",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n### Question 4\nrubric={accuracy:1,viz:2,reasoning:1}\n\n<h4>\nPython\n</h4>\n<ol type=\"1\">\n<li>Filter the data to include only the years 1918, 1938, 1958, 1978, 1998, and 2018. To do this, you need to write out the full date strings, <code>'1918-01-01'</code> etc, or use <code>pd.to_datetime</code> with <code>format=%Y</code> on a list of the year integers only, up to you which one.</li>\n<li>Use filled circles to make a scatter plot with children per women on the x-axis, child mortality on the y-axis, and the circles colored by the income group.</li>\n<li>Facet your data into six subplots, one for each year laid out in 3 columns and 2 rows. To avoid taking too much space, set the width and height of the plots to suitable numbers.</li>\n<li>Briefly describe your interpretation of the data. Does it support what was written in the introduction to this section of the lab? Why / why not?</li>\n</ol>\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE\ndf_1918 = df[df['year'].dt.year == 1918]\ndf_1938 = df[df['year'].dt.year == 1938]\ndf_1958 = df[df['year'].dt.year == 1958]\ndf_1978 = df[df['year'].dt.year == 1978]\ndf_1998 = df[df['year'].dt.year == 1998]\ndf_2018 = df[df['year'].dt.year == 2018]\n\ndf4 = pd.concat([df_1918, df_1938, df_1958, df_1978, df_1998, df_2018])\ndf4\n\nalt.Chart(df4).mark_point().encode(\n x='children_per_woman',\n y='child_mortality',\n color='income_group',\n facet=alt.Facet('year', columns=3)\n).properties(\n width=180,\n height=180\n)\n",
"_____no_output_____"
]
],
[
[
"YOUR ANSWER TO 4 GOES HERE\nGradually over the year, regions are moving towards less children per woman and we can see child_mortality is decreasing",
"_____no_output_____"
],
[
"# 5. Carbon dioxide emissions\n\nCO2 emissions are often talked about in it's relation to climate change.\nLet's explore the data to see which countries emits the most CO2 per capita\nand which regions has emitted the most in total over time.",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n### Question 5\nrubric={accuracy:1,quality:1,viz:2}\n\n<h4>\nPython\n</h4>\n<ol type=\"1\">\n<li>Filter the data to include only the most recent year when <code>'co2_per_capita'</code> was measured (it is up to you how you find out which year this is).</li>\n<li>Use the data frame <code>nlargest</code> method to select the top 40 countries in CO2 production per capita for that year.</li>\n<li>Since we have only one value per country per year, let’s create a bar chart to visualize it. Encode the CO2 per capita as on the x-axis, the country on the y-axis, and the region as the color.</li>\n<li>Sort your bar chart so that the highest CO2 per capita is the closest to the x-axis (the bottom of the chart). <a href=\"https://altair-viz.github.io/gallery/bar_chart_sorted.html\">Here is an example of how to sort in Altair</a>.</li>\n</ol>\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE\ndf_2014 = df[df['year'].dt.year == 2014]\ndf_2014_large = df_2014.nlargest(40,\"co2_per_capita\")\nalt.Chart(df_2014_large).mark_bar().encode(\n x=alt.X('co2_per_capita'),\n y=alt.Y('country', sort='x'),\n color='region'\n).properties(\n width=400,\n height=400\n)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n### Question 5.1\nrubric={accuracy:1,quality:1,viz:2}\n\n<h4>\nPython\n</h4>\n<ol type=\"1\">\n<li>in addition to the co2 per capita, the total population also matter for a country’s overall co2 emissions. compute a new column in your data set called <code>'co2_total'</code> which contains the total co2 emissions per observation.</li>\n<li>plot this new column over time in an area chart, but instead of plotting one area for each country, plot one for each region which represents the sum of all countries co2 emissions in that region.</li>\n</ol>\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE\ndf['co2_total'] = df['co2_per_capita']*df['population']\n\nalt.Chart(df).mark_bar().encode(\n x=alt.X('year'),\n y='co2_total',\n color='region'\n).properties(\n width=600,\n height=600\n)",
"_____no_output_____"
]
],
[
[
"# 6. Income distribution\n\nIn his talk back in 2003, Rosling showed a projection of how the world income distribution would look like in 2015. Let’s eyeball if the suggested trend was accurate.",
"_____no_output_____"
],
[
"<div class=\"alert alert-warning\" style=\"color:black\">\n\n### Question 6 (Optional)\nrubric={accuracy:1,viz:1}\n\n<h4>Python</h4>\n<ol type=\"1\">\n<li>Wrangle your data to include the years 1979, 1991, 2003 and 2015.</li>\n<li>Create a histogram (binned bar chart) of the income distribution with an appropriate number of bins.</li>\n<li>Facet by year and make the plots smaller so that they fit in a single row.</li>\n<li>It is a little hard to tell if the data is exactly the same as the prediction since we are not using a log scale and a histogram instead of a density plot (we’ll learn about these things later). But in general, briefly explain whether you think the trend is the same or not?</li>\n</ol>\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE",
"_____no_output_____"
]
],
[
[
"# 7. Chart beautification\n\n\nLet's make our charts from question 2 look more like the Gapminder bubble chart! Beautifying charts can take a long time, but it is also satisfying when you end up with a really nice looking chart in the end. We will learn more about how to create charts for communication later, but these parameters are usually enough to create basic communication charts and to help you in your data exploration.",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\" style=\"color:black\">\n\n### Question 7\nrubric={accuracy:2,quality:1,viz:1}\n\n<h4>\nPython\n</h4>\n<ol type=\"1\">\n<li>Copy in your code from question 2.1 and confirm that your scatter plot is generated properly so that you didn't miss to copy anything.</li>\n<li>Add a title of your choice to the chart.</li>\n<li>Set the x-axis and y-axis scale so that they don’t include zero and are zoomed in to the extent of the data instead.</li>\n<li>Set proper titles for the axis and the legends, which include spaces instead of underscores and are capitalized.</li>\n<li>Some of the dots are really hard to see because they are so small and it is a bit difficult to distinguish the changes in size as well. Let’s make everything bigger and emphasize the size difference by using the <a href=\"https://altair-viz.github.io/gallery/airport_connections.html\">range argument to <code>alt.Scale</code></a> (there is a lot of other things going on in this example, so just focus on how they specify <code>size</code>).</li>\n<li>Enlarge the axis title font by finding and setting the <a href=\"https://altair-viz.github.io/user_guide/configuration.html?highlight=titlefont#axis-configuration\">right parameter of <code>.configure_axis</code></a></li>\n</ol>\n\n</div>",
"_____no_output_____"
]
],
[
[
"# YOUR PYTHON ANSWER GOES HERE\n\nalt.Chart(df_1962, title = 'Data of 1962').mark_point().encode(\n alt.X('children_per_woman', scale=alt.Scale(zero=False), title='Children Per Woman'),\n alt.Y('life_expectancy', scale=alt.Scale(zero=False), title='Life Expectancy'),\n color='region',\n size=alt.Size('region', scale=alt.Scale(range=[0, 200]))\n).configure_axis(labelFontSize=10, titleFontSize=20\n).configure_title(fontSize=30)",
"_____no_output_____"
]
],
[
[
"---\n\n# Submission to Canvas\n\nWhen you are ready to submit your assignment do the following:\n\n1. Run all cells in your notebook to make sure there are no errors by doing `Kernel -> Restart Kernel and Run All Cells...`\n2. Convert your notebook to .html format using the `convert_notebook()` function below or by `File -> Export Notebook As... -> Export Notebook to HTML`\n3. Submit your exported .html file to Canvas.\n4. Don't forget to also push all your work (including the .ipynb file) to GitHub.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbae1c84ef6dbc20a904b703f39a42cf271d7075
| 772,484 |
ipynb
|
Jupyter Notebook
|
examples/example-3-meg-functional-connectivity.ipynb
|
vagechirkov/neurolib
|
593092f16d53b310f16744dd778135164ac50aa4
|
[
"MIT"
] | null | null | null |
examples/example-3-meg-functional-connectivity.ipynb
|
vagechirkov/neurolib
|
593092f16d53b310f16744dd778135164ac50aa4
|
[
"MIT"
] | null | null | null |
examples/example-3-meg-functional-connectivity.ipynb
|
vagechirkov/neurolib
|
593092f16d53b310f16744dd778135164ac50aa4
|
[
"MIT"
] | null | null | null | 588.33511 | 200,576 | 0.937561 |
[
[
[
"# Modeling resting-state MEG-Data",
"_____no_output_____"
],
[
"In this example we will learn how to use `neurolib` to simulate resting state functional connectivity of MEG recordings. \n\nIn the first part of the notebook, we will compute the frequency specific functional connectivity matrix of an examplary resting state MEG recording from the [YouR-Study](https://doi.org/10.1186/s12888-017-1206-5) *Uhlhaas, P.J., Gajwani, R., Gross, J. et al. The Youth Mental Health Risk and Resilience Study (YouR-Study). BMC Psychiatry 17, 43 (2017)*.\n\n\nTo this end we will: \n\n* Band-Pass filter the signal \n* Apply the `hilbert`-transformation to extract the signal envelope\n* Orthogonalize the signal envelopes of two examplary regions\n* Low-Pass filter the signal envelopes\n* and compute the pairwise envelope correlations which yields the `functional connectivity` matrix.\n\nWe follow the approach presented in *[Hipp, J., Hawellek, D., Corbetta, M. et al.](https://doi.org/10.1038/nn.3101), Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat Neurosci 15, 884–890 (2012)*\n\nIn the second part of this notebook, we will use a whole-brain model to simulate brain activity and compute functional connectivity matrix of the simulated signal envelope, as was done for the empirical MEG data. The parameters of this model have been previously optimized with `neurolib`'s evolutionary algorithms (not shown here).\n\nFinally, we will compute the fit (Pearson correlation) of the simulated functional connectivity to the empirical MEG data, which was used as a fitting objective in a previous optimization procedure.",
"_____no_output_____"
]
],
[
[
"# change to the root directory of the project\nimport os\nif os.getcwd().split(\"/\")[-1] == \"examples\":\n os.chdir('..')\n \n# This will reload all imports as soon as the code changes\n%load_ext autoreload\n%autoreload 2 ",
"_____no_output_____"
],
[
"import os\nimport numpy as np\nimport xarray as xr\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nimport ipywidgets as widgets\nfrom IPython.utils import io\nimport warnings\nwarnings.simplefilter(action='ignore', category=FutureWarning)\nimport time\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"## Empirical Functional Connectivity",
"_____no_output_____"
],
[
"### Load MEG-Data\n\nFirst off, let's load the MEG data using the `Signal` class from `neurolib`. Our example data has already been preprocessed and projected into source space using the [AAL2](https://www.gin.cnrs.fr/en/tools/aal/) atlas.",
"_____no_output_____"
]
],
[
[
"from neurolib.utils.signal import Signal \n\nsignal = Signal.from_file(os.path.join('examples', 'data','rs-meg.nc'))\nregion_labels = signal.data.regions.values\nnr_regions = len(region_labels)\ndisplay(signal.data)",
"_____no_output_____"
]
],
[
[
"### Band-Pass filter and Hilbert transform",
"_____no_output_____"
],
[
"We will now filter the signal into the desidered frequency band and apply the [hilbert transform](https://en.wikipedia.org/wiki/Hilbert_transform) on the band-passed filtered signal. This will provide us with the analytic representation of the signal, which we can then use to extract the signal's envelope and its phase.\n\nIn the following, we plot each processing step for an example target region that you can chose using the widgets below *(default: left Precentral Gyrus)*. Furthermore, we can also choose the frequency range that we'd like to filter the signal in *(default: alpha (8-12Hz))*.",
"_____no_output_____"
]
],
[
[
"print('Select a region from the AAL2 atlas and a frequency range')\n# Select a Region \ntarget = widgets.Select(options=region_labels, value='PreCG.L', description='Regions', \n tooltips=['Description of slow', 'Description of regular', 'Description of fast'], \n layout=widgets.Layout(width='50%', height='150px'))\ndisplay(target)\n\n# Select Frequency Range\nfreq = widgets.IntRangeSlider(min=1, max=46, description='Frequency (Hz)', value=[8, 12], layout=widgets.Layout(width='80%'), \n style={'description_width': 'initial'})\ndisplay(freq)",
"Select a region from the AAL2 atlas and a frequency range\n"
],
[
"# Define how many timepoints you'd like to plot\nplot_timepoints = 1000\n\n# Plot unfiltered Signal\nfig, ax = plt.subplots(2,1,figsize=(12,8), sharex=True)\nsns.lineplot(x=signal.data.time[:plot_timepoints], y=signal.data.sel(regions=target.value)[:plot_timepoints], \n ax=ax[0], color='k', alpha=0.6)\nax[0].set_title(f'Unfiltered Signal ({target.value})');\n\n# Band Pass Filter the Signal\nsignal.filter(freq.value[0], freq.value[1], inplace=True);\n\n# Apply hilbert-transform to extract the signal envelope\ncomplex_signal = signal.hilbert_transform('complex', inplace=False)\nsignal_env = np.abs(complex_signal.data)\n\n# Plot filtered Signal and Signal Envelope\nsns.lineplot(x=signal.data.time[:plot_timepoints], y=signal.data.sel(regions=target.value)[:plot_timepoints], \n ax=ax[1], label='Bandpass-Filtered Signal')\nsns.lineplot(x=signal_env.time[:plot_timepoints], y=signal_env.sel(regions=target.value)[:plot_timepoints], \n ax=ax[1], label='Signal Envelope')\nax[1].set_title(f'Filtered Signal ({target.value})');\nax[1].legend(bbox_to_anchor=(1.2, 1),borderaxespad=0)\nsns.despine(trim=True)",
"Setting up band-pass filter from 8 - 12 Hz\n\nFIR filter parameters\n---------------------\nDesigning a one-pass, zero-phase, non-causal bandpass filter:\n- Windowed time-domain design (firwin) method\n- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation\n- Lower passband edge: 8.00\n- Lower transition bandwidth: 2.00 Hz (-6 dB cutoff frequency: 7.00 Hz)\n- Upper passband edge: 12.00 Hz\n- Upper transition bandwidth: 3.00 Hz (-6 dB cutoff frequency: 13.50 Hz)\n- Filter length: 165 samples (1.650 sec)\n\n"
]
],
[
[
"### Orthogonalized signal envelope\n\nNow we are going to address the main methodological issue of MEG when it comes to the analysis of the cortical\nfunctional connectivity structure, i.e. its low spatial resolution. The electric field\ngenerated by any given neural source spreads widely over the cortex so that the signal captured at the MEG sensors is a complex mixture of signals from multiple underlying neural sources.\n\nTo account for the effect of electric field spread on our MEG connectivity measures, we adapted the orthogonalization approach by *Hipp, J., Hawellek, D., Corbetta, M. et al. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat Neurosci 15, 884–890 (2012) __[link](https://doi.org/10.1038/nn.3101)__*.\n\nThe basic idea here is that a signal generated by one neural source and measured at two separate sensors must have exactly the same phase at both sensors. In contrast, signals from different neural sources have different phases. And thus it is possible to eliminate the effect of a reference signal on the target signal by removing the signal component that has the same phase as a reference region.\n\nFormally, this can be expressed as: $Y_{\\perp X}(t,f) = imag\\big(\\ Y(t,f)\\ \\frac{X(t,f)^\\star}{|X(t,f)|}\\ \\big)\\ \\label{eq:orth}$. Here, $Y$ represents the analytic signal from our target regions that is being orthogonalized with respect to the signal from region $X$.\n\nUsing the widgets below, you can choose the reference region $X$ *(default: right Precentral Gyrus)*",
"_____no_output_____"
]
],
[
[
"print('Select a reference region for the orthogonalization')\n# Select a Region \nreferenz = widgets.Select(options=region_labels, value='PreCG.R', description='Regions',\n tooltips=['Description of slow', 'Description of regular', 'Description of fast'],\n layout=widgets.Layout(width='50%', height='150px'))\ndisplay(referenz)",
"Select a reference region for the orthogonalization\n"
],
[
"# Perform Orthogonalization\nsignal_conj = complex_signal.data.conj()\nconj_div_env = signal_conj/signal_env\north_signal = (complex_signal.data.sel(regions=target.value) * conj_div_env.sel(regions=referenz.value)).imag\north_env = np.abs(orth_signal)\n\n# Plot \nfig, ax = plt.subplots(2,1,figsize=(12,8), sharex=True)\nsns.lineplot(x=signal.data.time[:plot_timepoints], y=signal.data.sel(regions=referenz.value)[:plot_timepoints], ax=ax[0])\nsns.lineplot(x=signal_env.time[:plot_timepoints], y=signal_env.sel(regions=referenz.value)[:plot_timepoints], ax=ax[0])\nax[0].set_title(f'Referenz Region X ({referenz.value})');\nsns.lineplot(x=signal.data.time[:plot_timepoints], y=signal.data.sel(regions=target.value)[:plot_timepoints], \n ax=ax[1], label='Bandpass-Filtered Signal')\nsns.lineplot(x=signal_env.time[:plot_timepoints], y=signal_env.sel(regions=target.value)[:plot_timepoints], \n ax=ax[1], label='Signal Envelope')\nsns.lineplot(x = orth_env.time[:plot_timepoints], y=orth_env[:plot_timepoints], ax=ax[1], label='Orthogonalized Envelope')\nax[1].legend(bbox_to_anchor=(1.2, 1),borderaxespad=0)\nax[1].set_title(f'Target Region Y ({target.value})');\nsns.despine(trim=True)",
"_____no_output_____"
]
],
[
[
"### Low-Pass filtering of the envelopes \n\nAs a last step, before calculating the envelope correlations, we need to low-pass filter the signal envelopes since the connectivity measures of (ultra)-low frequency components of the MEG-signal correspond best to the functional connectivity as measured using fMRI.\n\nBelow, you can choose the low-pass frequency *(default: 0.2 Hz)*.",
"_____no_output_____"
]
],
[
[
"low_pass = widgets.FloatSlider(value=0.2, min=0, max=2.0, step=0.1, description='Low-Pass Frequency (Hz)', \n disabled=False, readout=True, readout_format='.1f', layout=widgets.Layout(width='80%'), \n style={'description_width': 'initial'})\ndisplay(low_pass)",
"_____no_output_____"
],
[
"with io.capture_output() as captured:\n low_orth_env = Signal(orth_env).filter(low_freq=None, high_freq=low_pass.value, inplace=False)\n low_signal_env = Signal(signal_env.sel(regions=referenz.value)).filter(low_freq=None, high_freq=low_pass.value, inplace=False)\n\n# Plot\nfig, ax = plt.subplots(1,2,figsize=(15,4), sharey=True)\nsns.lineplot(x=signal_env.time[:plot_timepoints], y=signal_env.sel(regions=referenz.value)[:plot_timepoints], ax=ax[0])\nsns.lineplot(x=low_signal_env.data.time[:plot_timepoints], y=low_signal_env.data[:plot_timepoints], ax=ax[0])\nax[0].set_title(f'Referenz Region X ({referenz.value})');\nsns.lineplot(x = orth_env.time[:plot_timepoints], y=orth_env[:plot_timepoints], ax=ax[1], label='Orthogonalized Envelope')\nsns.lineplot(x = low_orth_env.data.time[:plot_timepoints], y=low_orth_env.data[:plot_timepoints], ax=ax[1], label='Low-Passed Orthogonalized Envelope')\nax[1].legend(bbox_to_anchor=(1, -0.18),borderaxespad=0)\nax[1].set_title(f'Target Region Y ({target.value})');\nsns.despine(trim=True)\nprint(f'Orthogonalized envelope correlation between {referenz.value} and {target.value}: ', np.round(np.corrcoef(low_orth_env.data,low_signal_env.data)[0,1],2))",
"Orthogonalized envelope correlation between PreCG.R and PreCG.L: 0.13\n"
]
],
[
[
"### Computing the functional connectivity matrix\n\nWe will now define a function that iterates over each pair of brain regions and performs the previously presented processing steps, i.e. that extracts the envelopes, performs the orthogonalization, applies the low-pass filter, and returns the functional connectivity matrix that contains the pairwise envelope correlations. \n\nThis step may take a minute.",
"_____no_output_____"
]
],
[
[
"def orth_fc(signal, low_pass): \n nr_regions = signal.data.shape[0]\n progress = widgets.IntProgress(min=0, max=nr_regions, description=('Calculating FC Matrix'),\n layout=widgets.Layout(width='80%'), style={'description_width': 'initial'})\n display(progress)\n complex_signal = signal.hilbert_transform('complex', inplace=False)\n signal_env = signal.hilbert_transform('amplitude', inplace=False);\n conj_div_env = complex_signal.data.conj()/signal_env.data \n \n # Low-pass filter Signal envelope\n with io.capture_output() as captured:\n signal_env.filter(low_freq=None, high_freq=low_pass)\n \n corr = []\n for complex_region in complex_signal.data: \n orth_signal = (complex_region * conj_div_env).imag\n orth_env = np.abs(orth_signal).T\n orth_env = Signal(orth_env)\n with io.capture_output() as captured:\n orth_env.filter(low_freq=None, high_freq=low_pass)\n corr_mat = np.corrcoef(orth_env.data, signal_env.data)\n corr.append(np.diag(corr_mat, k=nr_regions))\n progress.value += 1\n\n fc = np.array(corr)\n # Since the orthogonalization process is not symmetric we take the mean of both directions.\n fc = (fc.T + fc) / 2.\n np.fill_diagonal(fc,0)\n return fc\n\n# Execute Function\nfc = orth_fc(signal, low_pass.value)",
"_____no_output_____"
]
],
[
[
"Let's now plot the functional connectivity matrix. We label only every second row/column since right and left regions alternate in the AAL2 atlas.",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(10,8))\nsns.heatmap(fc, square=True, ax=ax, cmap='YlGnBu', linewidth=0.005, cbar_kws={\"shrink\": .8})\nticks = [tick[:-2] for tick in region_labels[::2]]\nax.set_xticks(np.arange(0,94,2)); ax.set_yticks(np.arange(0,94,2)) \nax.set_xticklabels(ticks, rotation=90, fontsize=8); ax.set_yticklabels(ticks, rotation=0, fontsize=8);",
"_____no_output_____"
]
],
[
[
"#### Exclude subcortical regions\nFor the following whole-brain simulation we are only interested in the cortical regions. So we'll now exclude all subcortical regions: \n* Hippocampus: 41 - 44\n* Amygdala: 45-46\n* Basal Ganglia: 75-80\n* Thalamus: 81-82\n\n> Attention: AAL indices start with 1",
"_____no_output_____"
]
],
[
[
"exclude = list(range(40, 46)) + list(range(74, 82))\ntmp = np.delete(fc, exclude, axis=0)\nemp_fc = np.delete(tmp, exclude, axis=1)\n# Exclude regions from the list of region labels\nemp_labels = np.delete(region_labels, exclude)",
"_____no_output_____"
]
],
[
[
"## Whole-brain model\n\nIn this part of the notebook, we will use `neurolib` to simulate the functional connectivity. We will therefore:\n\n* Load structural connectivity matrices from the *Human Connectome Project* and initiate the whole-brain model using the Wilson-Cowan model to simulate each brain region\n* Set the *global coupling strength*, *exc. background input*, and the *noise strength* parameters of the model\n* Run the simulation\n* Compute the functional connectivity using the signal envelopes\n\nPlease refer to the `wc-minimal` example for an introduction to the Wilson-Cowan model.",
"_____no_output_____"
],
[
"#### Initiate whole-brain model",
"_____no_output_____"
]
],
[
[
"# Let's import the neurolib\nfrom neurolib.models.wc import WCModel\nfrom neurolib.utils.loadData import Dataset\n\n# First we load the structural data set from the Human Connectome Project \nds = Dataset(\"hcp\")\n\n# We initiate the Wilson-Cowan model\nwc = WCModel(Cmat = ds.Cmat, Dmat = ds.Dmat, seed=0)",
"_____no_output_____"
]
],
[
[
"#### Parameter settings\n\nYou may now choose parameters settings for the *global coupling*, the *excitatory background input*, and the *noise strength*, which will be used when we run the model. The final fit between simulated and empirical connectivity matrices will depend on the parameters choosen here.",
"_____no_output_____"
]
],
[
[
"global_coupling = widgets.FloatSlider(value=6.55, min=0., max=20.0, step=0.01, description='Global Coupling', \n disabled=False, readout=True, readout_format='.2f', layout=widgets.Layout(width='80%'), \n style={'description_width': 'initial'})\nexc_drive = widgets.FloatSlider(value=1.58, min=0.0, max=4.0, step=0.01, description='Exc. Background Drive', \n disabled=False, readout=True, readout_format='.2f', layout=widgets.Layout(width='80%'), \n style={'description_width': 'initial'})\ninh_drive = widgets.FloatSlider(value=2.83, min=0.0, max=4.0, step=0.01, description='Inh. Background Drive', \n disabled=False, readout=True, readout_format='.2f', layout=widgets.Layout(width='80%'), \n style={'description_width': 'initial'})\nnoise_level = widgets.FloatSlider(value=0.02, min=0.001, max=0.05, step=0.001, description='Noise Level', \n disabled=False, readout=True, readout_format='.3f', layout=widgets.Layout(width='80%'), \n style={'description_width': 'initial'})\ndisplay(global_coupling)\ndisplay(exc_drive)\ndisplay(inh_drive)\ndisplay(noise_level)",
"_____no_output_____"
]
],
[
[
"#### Run the simulation\n\nLet's now run the whole-brain model using the defined parameter settings. This may take some time since we're simulating a complete minute here. ",
"_____no_output_____"
]
],
[
[
"# Let's set the previously defined parameters\n# note: the duraiton here is short for testing:\nwc.params['duration'] = 10*1000 \n\n# use longer simulation for real run:\n#wc.params['duration'] = 1*60*1000 \n\nwc.params['K_gl'] = global_coupling.value\nwc.params['exc_ext'] = exc_drive.value\nwc.params['inh_ext'] = inh_drive.value\nwc.params['sigma_ou'] = noise_level.value\n# Run the model\nwc.run()",
"_____no_output_____"
]
],
[
[
"### Simulated functional connectivity\n\nWe'll now compute the functional connectivity matrix containing the pairwise envelope correlations between all cortical regions of the AAL2 atlas. We'll thus follow the processing steps as before, i.e. band-pass filter the signal, extract the signal envelopes using the hilbert transformation, low-pass filter the envelopes and compute the pairwise pearson correlations. Note that we don't apply the orthogonalization scheme here, since this was only done to account to the electric field spread in the empirical data. ",
"_____no_output_____"
]
],
[
[
"# Create xr DataArray from the simulated excitatory timeseries (keeping the region labels)\nsim_signal = xr.DataArray(wc.exc[:, int(1000/wc.params.dt):], dims=(\"regions\", \"time\"), coords={\"regions\": emp_labels, \"time\": wc.t[int(1000/wc.params.dt):]/1000}, \n attrs={'atlas':'AAL2'})\n\n# Initialize Figure\nfig, ax = plt.subplots(figsize=(12,4))\n\n# Filter signal\nsim_signal = Signal(sim_signal)\nsim_signal.resample(to_frequency=100)\nwith io.capture_output() as captured:\n sim_signal.filter(freq.value[0], freq.value[1], inplace=True);\nsns.lineplot(x=sim_signal.data.time[:plot_timepoints], y=sim_signal.data.sel(regions=target.value)[:plot_timepoints], ax=ax, label='Filtered Signal')\n\n# Extract signal envelope \nsim_signal.hilbert_transform('amplitude', inplace=True)\nsns.lineplot(x=sim_signal.data.time[:plot_timepoints], y=sim_signal.data.sel(regions=target.value)[:plot_timepoints], ax=ax, label='Signal Envelope')\n\n# Low-Pass Filter\nwith io.capture_output() as captured:\n sim_signal.filter(low_freq=None, high_freq=low_pass.value, inplace=True)\nsns.lineplot(x=sim_signal.data.time[:plot_timepoints], y=sim_signal.data.sel(regions=target.value)[:plot_timepoints], ax=ax, label='Low-Pass Signal Envelope')\nax.legend(bbox_to_anchor=(1.2, 1),borderaxespad=0)\nax.set_title(f'Simulated Signal of Target Region Y ({target.value})');\nsns.despine(trim=True)",
"_____no_output_____"
]
],
[
[
"To compute the simulated functional connectivity matrix we use the `fc` functions from neurolib. ",
"_____no_output_____"
]
],
[
[
"import neurolib.utils.functions as func\n\n# Compute the functional connectivity matrix\nsim_fc = func.fc(sim_signal.data)\n\n# Set diagonal to zero\nnp.fill_diagonal(sim_fc, 0)\n\n# Plot Empirical and simulated connectivity matrix\nfig, ax = plt.subplots(1,2, figsize=(16,10))\nsns.heatmap(emp_fc, square=True, ax=ax[0], cmap='YlGnBu', linewidth=0.005, cbar_kws={\"shrink\": .5})\nax[0].set_title('Empirical FC',pad=10);\nsns.heatmap(sim_fc, square=True, ax=ax[1], cmap='YlGnBu', linewidth=0.005, cbar_kws={\"shrink\": .5})\nax[1].set_title('Simulated FC',pad=10);\nticks = [tick[:-2] for tick in emp_labels[::2]]\nfor ax in ax:\n ax.set_xticks(np.arange(0,80,2)); ax.set_yticks(np.arange(0,80,2)) \n ax.set_xticklabels(ticks, rotation=90, fontsize=8); ax.set_yticklabels(ticks, rotation=0, fontsize=8);",
"_____no_output_____"
]
],
[
[
"## Model fit\n\nLastly, we may evaluate the model fit by computing the pearson correlation between our simulated functional connectivity matrix and the empirical one. Additionally we'll also plot the correlation between structural and functional connectivity matrices to have a reference. ",
"_____no_output_____"
]
],
[
[
"# Compare structural and simulated connectivity to the empirical functional connectivity\nstruct_emp = np.corrcoef(emp_fc.flatten(), ds.Cmat.flatten())[0,1]\nsim_emp = np.corrcoef(emp_fc.flatten(), sim_fc.flatten())[0,1]\n\n# Plot\nfig, ax = plt.subplots(figsize=(6,6))\nsplot = sns.barplot(x=['Structural Connectivity', 'Simulated Connectivity'], y=[struct_emp, sim_emp], ax=ax)\nax.set_title('Correlation to Empiral Functional Connectivity', pad=10)\nfor p in splot.patches:\n splot.annotate(format(p.get_height(), '.2f'), \n (p.get_x() + p.get_width() / 2., p.get_height()), \n ha = 'center', va = 'center', \n size=20, color='white',\n xytext = (0, -12), \n textcoords = 'offset points')\nsns.despine()\nprint(f\"Parameters: \\tGlobal Coupling: {wc.params['K_gl']}\\n\\t\\tExc. Background Drive: {wc.params['exc_ext']}\")\nprint(f\"\\t\\tNoise Level: {wc.params['sigma_ou']}\")",
"Parameters: \tGlobal Coupling: 6.55\n\t\tExc. Background Drive: 1.58\n\t\tNoise Level: 0.02\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbae1cc59be7efd6fa9168c92887277fdd9dae36
| 156,981 |
ipynb
|
Jupyter Notebook
|
pytorch/0422_FFNetworksWithPyTorch.ipynb
|
bhanu0925/DeepLearning
|
747445df8622e74eff8fae6aabb8cb156cbc16da
|
[
"MIT"
] | 20 |
2020-01-04T16:35:48.000Z
|
2022-03-29T20:47:43.000Z
|
pytorch/0422_FFNetworksWithPyTorch.ipynb
|
bhanu0925/DeepLearning
|
747445df8622e74eff8fae6aabb8cb156cbc16da
|
[
"MIT"
] | null | null | null |
pytorch/0422_FFNetworksWithPyTorch.ipynb
|
bhanu0925/DeepLearning
|
747445df8622e74eff8fae6aabb8cb156cbc16da
|
[
"MIT"
] | 25 |
2020-01-17T14:48:39.000Z
|
2022-01-26T08:29:08.000Z
| 151.088547 | 61,168 | 0.859703 |
[
[
[
"## Outline\n\n* Recap of data\n* Feedforward network with Pytorch tensors and autograd\n* Using Pytorch's NN -> Functional, Linear, Sequential & Pytorch's Optim\n* Moving things to CUDA",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport math\nimport matplotlib.pyplot as plt\nimport matplotlib.colors\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score, mean_squared_error, log_loss\nfrom tqdm import tqdm_notebook \nimport seaborn as sns\nimport time\nfrom IPython.display import HTML\nimport warnings\nwarnings.filterwarnings('ignore')\n\nfrom sklearn.preprocessing import OneHotEncoder\nfrom sklearn.datasets import make_blobs\n\nimport torch",
"_____no_output_____"
],
[
"torch.manual_seed(0)",
"_____no_output_____"
],
[
"my_cmap = matplotlib.colors.LinearSegmentedColormap.from_list(\"\", [\"red\",\"yellow\",\"green\"])",
"_____no_output_____"
]
],
[
[
"## Generate Dataset",
"_____no_output_____"
]
],
[
[
"data, labels = make_blobs(n_samples=1000, centers=4, n_features=2, random_state=0)\nprint(data.shape, labels.shape)",
"(1000, 2) (1000,)\n"
],
[
"plt.scatter(data[:,0], data[:,1], c=labels, cmap=my_cmap)\nplt.show()",
"_____no_output_____"
],
[
"X_train, X_val, Y_train, Y_val = train_test_split(data, labels, stratify=labels, random_state=0)\nprint(X_train.shape, X_val.shape, labels.shape)",
"(750, 2) (250, 2) (1000,)\n"
]
],
[
[
"## Using torch tensors and autograd",
"_____no_output_____"
]
],
[
[
"X_train, Y_train, X_val, Y_val = map(torch.tensor, (X_train, Y_train, X_val, Y_val))",
"_____no_output_____"
],
[
"print(X_train.shape, Y_train.shape)",
"torch.Size([750, 2]) torch.Size([750])\n"
],
[
"def model(x):\n a1 = torch.matmul(x, weights1) + bias1 # (N, 2) x (2, 2) -> (N, 2)\n h1 = a1.sigmoid() # (N, 2)\n a2 = torch.matmul(h1, weights2) + bias2 # (N, 2) x (2, 4) -> (N, 4)\n h2 = a2.exp()/a2.exp().sum(-1).unsqueeze(-1) # (N, 4)\n return h2",
"_____no_output_____"
],
[
"y_hat = torch.tensor([[0.1, 0.2, 0.3, 0.4], [0.8, 0.1, 0.05, 0.05]])\ny = torch.tensor([2, 0])\n\n(-y_hat[range(y_hat.shape[0]), y].log()).mean().item()\n\n(torch.argmax(y_hat, dim=1) == y).float().mean().item()",
"_____no_output_____"
],
[
"def loss_fn(y_hat, y):\n return -(y_hat[range(y.shape[0]), y].log()).mean()",
"_____no_output_____"
],
[
"def accuracy(y_hat, y):\n pred = torch.argmax(y_hat, dim=1)\n return (pred == y).float().mean()",
"_____no_output_____"
],
[
"torch.manual_seed(0)\nweights1 = torch.randn(2, 2) / math.sqrt(2)\nweights1.requires_grad_()\nbias1 = torch.zeros(2, requires_grad=True)\n\nweights2 = torch.randn(2, 4) / math.sqrt(2)\nweights2.requires_grad_()\nbias2 = torch.zeros(4, requires_grad=True)\n\nlearning_rate = 0.2\nepochs = 10000\n\nX_train = X_train.float()\nY_train = Y_train.long()\n\nloss_arr = []\nacc_arr = []\n\nfor epoch in range(epochs):\n y_hat = model(X_train)\n loss = loss_fn(y_hat, Y_train)\n loss.backward()\n loss_arr.append(loss.item())\n acc_arr.append(accuracy(y_hat, Y_train))\n\n with torch.no_grad():\n weights1 -= weights1.grad * learning_rate\n bias1 -= bias1.grad * learning_rate\n weights2 -= weights2.grad * learning_rate\n bias2 -= bias2.grad * learning_rate\n weights1.grad.zero_()\n bias1.grad.zero_()\n weights2.grad.zero_()\n bias2.grad.zero_()\n\nplt.plot(loss_arr, 'r-')\nplt.plot(acc_arr, 'b-')\nplt.show()\nprint('Loss before training', loss_arr[0])\nprint('Loss after training', loss_arr[-1])",
"_____no_output_____"
]
],
[
[
"## Using NN.Functional",
"_____no_output_____"
]
],
[
[
"import torch.nn.functional as F",
"_____no_output_____"
],
[
"torch.manual_seed(0)\nweights1 = torch.randn(2, 2) / math.sqrt(2)\nweights1.requires_grad_()\nbias1 = torch.zeros(2, requires_grad=True)\n\nweights2 = torch.randn(2, 4) / math.sqrt(2)\nweights2.requires_grad_()\nbias2 = torch.zeros(4, requires_grad=True)\n\nlearning_rate = 0.2\nepochs = 10000\n\nloss_arr = []\nacc_arr = []\n\nfor epoch in range(epochs):\n y_hat = model(X_train)\n loss = F.cross_entropy(y_hat, Y_train)\n loss.backward()\n loss_arr.append(loss.item())\n acc_arr.append(accuracy(y_hat, Y_train))\n\n with torch.no_grad():\n weights1 -= weights1.grad * learning_rate\n bias1 -= bias1.grad * learning_rate\n weights2 -= weights2.grad * learning_rate\n bias2 -= bias2.grad * learning_rate\n weights1.grad.zero_()\n bias1.grad.zero_()\n weights2.grad.zero_()\n bias2.grad.zero_()\n\nplt.plot(loss_arr, 'r-')\nplt.plot(acc_arr, 'b-')\nplt.show()\nprint('Loss before training', loss_arr[0])\nprint('Loss after training', loss_arr[-1])",
"_____no_output_____"
]
],
[
[
"## Using NN.Parameter",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn",
"_____no_output_____"
],
[
"class FirstNetwork(nn.Module):\n \n def __init__(self):\n super().__init__()\n torch.manual_seed(0)\n self.weights1 = nn.Parameter(torch.randn(2, 2) / math.sqrt(2))\n self.bias1 = nn.Parameter(torch.zeros(2))\n self.weights2 = nn.Parameter(torch.randn(2, 4) / math.sqrt(2))\n self.bias2 = nn.Parameter(torch.zeros(4))\n \n def forward(self, X):\n a1 = torch.matmul(X, self.weights1) + self.bias1\n h1 = a1.sigmoid()\n a2 = torch.matmul(h1, self.weights2) + self.bias2\n h2 = a2.exp()/a2.exp().sum(-1).unsqueeze(-1)\n return h2",
"_____no_output_____"
],
[
"def fit(epochs = 1000, learning_rate = 1):\n loss_arr = []\n acc_arr = []\n for epoch in range(epochs):\n y_hat = fn(X_train)\n loss = F.cross_entropy(y_hat, Y_train)\n loss_arr.append(loss.item())\n acc_arr.append(accuracy(y_hat, Y_train))\n\n loss.backward()\n with torch.no_grad():\n for param in fn.parameters():\n param -= learning_rate * param.grad\n fn.zero_grad()\n \n plt.plot(loss_arr, 'r-')\n plt.plot(acc_arr, 'b-')\n plt.show() \n print('Loss before training', loss_arr[0])\n print('Loss after training', loss_arr[-1])",
"_____no_output_____"
],
[
"fn = FirstNetwork()\nfit()",
"_____no_output_____"
]
],
[
[
"## Using NN.Linear and Optim",
"_____no_output_____"
]
],
[
[
"class FirstNetwork_v1(nn.Module):\n \n def __init__(self):\n super().__init__()\n torch.manual_seed(0)\n self.lin1 = nn.Linear(2, 2)\n self.lin2 = nn.Linear(2, 4)\n \n def forward(self, X):\n a1 = self.lin1(X)\n h1 = a1.sigmoid()\n a2 = self.lin2(h1)\n h2 = a2.exp()/a2.exp().sum(-1).unsqueeze(-1)\n return h2",
"_____no_output_____"
],
[
"fn = FirstNetwork_v1()\nfit()",
"_____no_output_____"
],
[
"from torch import optim",
"_____no_output_____"
],
[
"def fit_v1(epochs = 1000, learning_rate = 1):\n loss_arr = []\n acc_arr = []\n opt = optim.SGD(fn.parameters(), lr=learning_rate)\n \n for epoch in range(epochs):\n y_hat = fn(X_train)\n loss = F.cross_entropy(y_hat, Y_train)\n loss_arr.append(loss.item())\n acc_arr.append(accuracy(y_hat, Y_train))\n\n loss.backward()\n opt.step()\n opt.zero_grad()\n \n plt.plot(loss_arr, 'r-')\n plt.plot(acc_arr, 'b-')\n plt.show() \n print('Loss before training', loss_arr[0])\n print('Loss after training', loss_arr[-1])",
"_____no_output_____"
],
[
"fn = FirstNetwork_v1()\nfit_v1()",
"_____no_output_____"
]
],
[
[
"## Using NN.Sequential",
"_____no_output_____"
]
],
[
[
"class FirstNetwork_v2(nn.Module):\n \n def __init__(self):\n super().__init__()\n torch.manual_seed(0)\n self.net = nn.Sequential(\n nn.Linear(2, 2), \n nn.Sigmoid(), \n nn.Linear(2, 4), \n nn.Softmax()\n )\n\n def forward(self, X):\n return self.net(X)",
"_____no_output_____"
],
[
"fn = FirstNetwork_v2()\nfit_v1()",
"_____no_output_____"
],
[
"def fit_v2(x, y, model, opt, loss_fn, epochs = 1000):\n \n for epoch in range(epochs):\n loss = loss_fn(model(x), y)\n\n loss.backward()\n opt.step()\n opt.zero_grad()\n \n return loss.item()",
"_____no_output_____"
],
[
"fn = FirstNetwork_v2()\nloss_fn = F.cross_entropy\nopt = optim.SGD(fn.parameters(), lr=1)\nfit_v2(X_train, Y_train, fn, opt, loss_fn)",
"_____no_output_____"
]
],
[
[
"## Running it on GPUs",
"_____no_output_____"
]
],
[
[
"device = torch.device(\"cuda\")\n\nX_train=X_train.to(device)\nY_train=Y_train.to(device)\nfn = FirstNetwork_v2()\nfn.to(device)\ntic = time.time()\nprint('Final loss', fit_v2(X_train, Y_train, fn, opt, loss_fn))\ntoc = time.time()\nprint('Time taken', toc - tic)",
"Final loss 1.395159363746643\nTime taken 0.7891602516174316\n"
],
[
"class FirstNetwork_v3(nn.Module):\n \n def __init__(self):\n super().__init__()\n torch.manual_seed(0)\n self.net = nn.Sequential(\n nn.Linear(2, 1024*4), \n nn.Sigmoid(), \n nn.Linear(1024*4, 4), \n nn.Softmax()\n )\n\n def forward(self, X):\n return self.net(X)",
"_____no_output_____"
],
[
"device = torch.device(\"cpu\")\n\nX_train=X_train.to(device)\nY_train=Y_train.to(device)\nfn = FirstNetwork_v3()\nfn.to(device)\ntic = time.time()\nprint('Final loss', fit_v2(X_train, Y_train, fn, opt, loss_fn))\ntoc = time.time()\nprint('Time taken', toc - tic)",
"Final loss 1.3890225887298584\nTime taken 29.352728128433228\n"
]
],
[
[
"## Exercises\n\n1. Try out a deeper neural network, eg. 2 hidden layers\n2. Try out different parameters in the optimizer (eg. try momentum, nestrov) -> check `optim.SGD` docs\n3. Try out other optimization methods (eg. RMSProp and Adam) which are supported in `optim`\n4. Try out different initialisation methods which are supported in `nn.init` ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbae1ff9cbc1d5dd30115eeb8b65aafb0b517ee7
| 321,471 |
ipynb
|
Jupyter Notebook
|
notebooks/FIG_partition.ipynb
|
kLabUM/hydraulic-controller-placement
|
b2cfbee19bb41d69702f4c218c9dba80bd6e4fae
|
[
"MIT"
] | null | null | null |
notebooks/FIG_partition.ipynb
|
kLabUM/hydraulic-controller-placement
|
b2cfbee19bb41d69702f4c218c9dba80bd6e4fae
|
[
"MIT"
] | null | null | null |
notebooks/FIG_partition.ipynb
|
kLabUM/hydraulic-controller-placement
|
b2cfbee19bb41d69702f4c218c9dba80bd6e4fae
|
[
"MIT"
] | 3 |
2019-01-18T21:04:28.000Z
|
2020-09-04T14:45:02.000Z
| 543.025338 | 97,684 | 0.944253 |
[
[
[
"# Import modules",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom matplotlib import gridspec\nfrom matplotlib import cm\nfrom pysheds.grid import Grid\nfrom matplotlib import colors\nimport seaborn as sns\nimport warnings\nfrom partition import differentiated_linear_weights, controller_placement_algorithm\n\nwarnings.filterwarnings('ignore')\nsns.set()\nsns.set_palette('husl', 8)\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Generate graph",
"_____no_output_____"
]
],
[
[
"grid = Grid.from_raster('../data/n30w100_dir', data_name='dir')",
"_____no_output_____"
],
[
"dirmap = (64, 128, 1, 2, 4, 8, 16, 32)\n\n# Specify pour point\nx, y = -97.294167, 32.73750\n\n# Delineate the catchment\ngrid.catchment(data='dir', x=x, y=y, dirmap=dirmap, out_name='catch',\n recursionlimit=15000, xytype='label')\n\n# Clip the bounding box to the catchment\ngrid.clip_to('catch', pad=(1,1,1,1))\n\n# Compute flow distance\ngrid.accumulation(data='catch', out_name='acc', dirmap=dirmap)\ngrid.flow_distance(data='catch', x=x, y=y, dirmap=dirmap, out_name='dist', xytype='label')\ndist = grid.view('dist', nodata=0, dtype=np.float64)",
"_____no_output_____"
],
[
"dist_weights = (np.where(grid.view('acc') >= 100, 0.1, 0) \n + np.where((0 < grid.view('acc')) & (grid.view('acc') <= 100), 1, 0)).ravel()",
"_____no_output_____"
],
[
"dists = grid.flow_distance(data='catch', x=x, y=y, weights=dist_weights,\n dirmap=dirmap, out_name='dist', xytype='label', inplace=False)",
"_____no_output_____"
],
[
"weights = differentiated_linear_weights(dists)",
"_____no_output_____"
]
],
[
[
"# Determine weighted accumulation",
"_____no_output_____"
]
],
[
[
"acc = grid.accumulation(data='catch', dirmap=dirmap, inplace=False)\nwacc = grid.accumulation(data='catch', weights=weights, dirmap=dirmap, inplace=False)",
"_____no_output_____"
],
[
"ratio = np.where(grid.mask & acc.astype(bool), wacc / acc, np.nan).ravel()",
"_____no_output_____"
],
[
"mask = (dists != 0)\nhist, bin_edges = np.histogram(dists[mask].ravel(), range=(0,dists.max()+1e-5), bins=40)",
"_____no_output_____"
]
],
[
[
"# Ratio of accumulation within critical range to total accumulation",
"_____no_output_____"
]
],
[
[
"k = 7\nc = 2000\nfdir = grid.view('catch')",
"_____no_output_____"
],
[
"subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, grid=grid,\n compute_weights=differentiated_linear_weights,\n dist_weights=dist_weights)\nixy, ixx = np.unravel_index(ixes, wacc.shape)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(12, 4))\n\nfig.patch.set_alpha(0)\ngs = gridspec.GridSpec(1, 2, width_ratios=[2, 3])\nax0 = plt.subplot(gs[0])\nax1 = plt.subplot(gs[1])\ncmap = cm.get_cmap('plasma', len(subs))\n\n\nim = np.zeros_like(wacc)\n\nfor i, sub in enumerate(subs):\n im += (1 + i)*(sub != 0).astype(int)\n\nim[im == 0] = np.nan\n\nim0 = ax0.imshow(im, cmap=cmap, zorder=2)\nax0.scatter(ixx, ixy, zorder=4, c='k', s=15, marker='x')\nax0.grid(zorder=-1)\nax0.xaxis.set_ticklabels([])\nax0.yaxis.set_ticklabels([])\nax0.set_title('Ordered partitions (k = 7)', size=14)\nplt.colorbar(im0, ax=ax0)\n\nplotlist = [np.bincount(np.digitize(dists.flat[np.where(sub.ravel())[0]], bin_edges[1:]),\n minlength=len(bin_edges) - 1).astype(int)\n for sub in subs]\n\nax1.stackplot(bin_edges[1:], *plotlist, linewidth=0.7,\n colors=sns.color_palette('plasma', k), edgecolor='0.4')\nax1.set_xlim(0, int(dists.max()))\nax1.set_title('Stacked width function of partitions', size=14)\nax1.set_xlabel('Normalized travel time [-]', size=13)\nax1.set_ylabel('Frequency', size=13)\nax1.yaxis.tick_right()\nax1.yaxis.set_label_position('right')\nplt.tight_layout()\nplt.savefig('../img/partitions_k7_phi10.png', bbox_inches='tight', dpi=200)",
"_____no_output_____"
],
[
"k = 15\nc = 900\nfdir = grid.view('catch')",
"_____no_output_____"
],
[
"subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, grid=grid,\n compute_weights=differentiated_linear_weights,\n dist_weights=dist_weights)\nixy, ixx = np.unravel_index(ixes, wacc.shape)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(12, 4))\n\nfig.patch.set_alpha(0)\ngs = gridspec.GridSpec(1, 2, width_ratios=[2, 3])\nax0 = plt.subplot(gs[0])\nax1 = plt.subplot(gs[1])\ncmap = cm.get_cmap('plasma', len(subs))\n\n\nim = np.zeros_like(wacc)\n\nfor i, sub in enumerate(subs):\n im += (1 + i)*(sub != 0).astype(int)\n\nim[im == 0] = np.nan\n\nim0 = ax0.imshow(im, cmap=cmap, zorder=2)\nax0.scatter(ixx, ixy, zorder=4, c='k', s=15, marker='x')\nax0.grid(zorder=-1)\nax0.xaxis.set_ticklabels([])\nax0.yaxis.set_ticklabels([])\nax0.set_title('Ordered partitions (k = 15)', size=14)\nplt.colorbar(im0, ax=ax0)\n\nplotlist = [np.bincount(np.digitize(dists.flat[np.where(sub.ravel())[0]], bin_edges[1:]), minlength=40).astype(int)\n for sub in subs]\n\nax1.stackplot(bin_edges[1:], *plotlist, linewidth=0.4,\n colors=sns.color_palette('plasma', k), edgecolor='0.6')\nax1.set_xlim(0, int(dists.max()))\nax1.set_title('Stacked width function of partitions', size=14)\nax1.set_xlabel('Normalized travel time [-]', size=13)\nax1.set_ylabel('Frequency', size=13)\nax1.yaxis.tick_right()\nax1.yaxis.set_label_position('right')\nplt.tight_layout()\nplt.savefig('../img/partitions_k15_phi10.png', bbox_inches='tight', dpi=200)",
"_____no_output_____"
],
[
"k = 10\nc = 1350\nfdir = grid.view('catch')",
"_____no_output_____"
],
[
"subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, grid=grid,\n compute_weights=differentiated_linear_weights,\n dist_weights=dist_weights)\nixy, ixx = np.unravel_index(ixes, wacc.shape)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(12, 4))\n\nfig.patch.set_alpha(0)\ngs = gridspec.GridSpec(1, 2, width_ratios=[2, 3])\nax0 = plt.subplot(gs[0])\nax1 = plt.subplot(gs[1])\ncmap = cm.get_cmap('plasma', len(subs))\n\n\nim = np.zeros_like(wacc)\n\nfor i, sub in enumerate(subs):\n im += (1 + i)*(sub != 0).astype(int)\n\nim[im == 0] = np.nan\n\nim0 = ax0.imshow(im, cmap=cmap, zorder=2)\nax0.scatter(ixx, ixy, zorder=4, c='k', s=15, marker='x')\nax0.grid(zorder=-1)\nax0.xaxis.set_ticklabels([])\nax0.yaxis.set_ticklabels([])\nax0.set_title('Ordered partitions (k = 10)', size=14)\nplt.colorbar(im0, ax=ax0)\n\nplotlist = [np.bincount(np.digitize(dists.flat[np.where(sub.ravel())[0]], bin_edges[1:]), minlength=40).astype(int)\n for sub in subs]\n\nax1.stackplot(bin_edges[1:], *plotlist, linewidth=0.4,\n colors=sns.color_palette('plasma', k), edgecolor='0.4')\nax1.set_xlim(0, int(dists.max()))\nax1.set_title('Stacked width function of partitions', size=14)\nax1.set_xlabel('Normalized travel time [-]', size=13)\nax1.set_ylabel('Frequency', size=13)\nax1.yaxis.tick_right()\nax1.yaxis.set_label_position('right')\nplt.tight_layout()\nplt.savefig('../img/partitions_k10_phi10.png', bbox_inches='tight', dpi=200)",
"_____no_output_____"
],
[
"k = 25\nc = 530\nfdir = grid.view('catch')",
"_____no_output_____"
],
[
"subs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, grid=grid,\n compute_weights=differentiated_linear_weights,\n dist_weights=dist_weights)\nixy, ixx = np.unravel_index(ixes, wacc.shape)",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(12, 4))\n\nfig.patch.set_alpha(0)\ngs = gridspec.GridSpec(1, 2, width_ratios=[2, 3])\nax0 = plt.subplot(gs[0])\nax1 = plt.subplot(gs[1])\ncmap = cm.get_cmap('plasma', len(subs))\n\n\nim = np.zeros_like(wacc)\n\nfor i, sub in enumerate(subs):\n im += (1 + i)*(sub != 0).astype(int)\n\nim[im == 0] = np.nan\n\nim0 = ax0.imshow(im, cmap=cmap, zorder=2)\nax0.scatter(ixx, ixy, zorder=4, c='k', s=15, marker='x')\nax0.grid(zorder=-1)\nax0.xaxis.set_ticklabels([])\nax0.yaxis.set_ticklabels([])\nax0.set_title('Ordered partitions (k = 25)', size=14)\nplt.colorbar(im0, ax=ax0)\n\nplotlist = [np.bincount(np.digitize(dists.flat[np.where(sub.ravel())[0]], bin_edges[1:]), minlength=40).astype(int)\n for sub in subs]\n\nax1.stackplot(bin_edges[1:], *plotlist, linewidth=0.1,\n colors=sns.color_palette('plasma', k), edgecolor='0.2')\nax1.set_xlim(0, int(dists.max()))\nax1.set_title('Stacked width function of partitions', size=14)\nax1.set_xlabel('Normalized travel time [-]', size=13)\nax1.set_ylabel('Frequency', size=13)\nax1.yaxis.tick_right()\nax1.yaxis.set_label_position('right')\nplt.tight_layout()\nplt.savefig('../img/partitions_k25_phi10.png', bbox_inches='tight', dpi=200)",
"_____no_output_____"
],
[
"1350 / np.count_nonzero(grid.mask)",
"_____no_output_____"
],
[
"900 / np.count_nonzero(grid.mask)",
"_____no_output_____"
],
[
"# Time run",
"_____no_output_____"
],
[
"k = 15\nc = 900\nfdir = grid.view('catch')",
"_____no_output_____"
],
[
"%%timeit\nsubs, ixes = controller_placement_algorithm(fdir, c, k, weights=weights, grid=grid,\n compute_weights=differentiated_linear_weights,\n dist_weights=dist_weights)",
"2.89 s ± 14.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbae2e4930ab05e4dd4041fe041d28ce445f4943
| 85,892 |
ipynb
|
Jupyter Notebook
|
notebooks/evaluation/malware_detection.ipynb
|
dmacko232/AndroidMalwareLabeling
|
f0d89a157a34d788d6849b9d4aea8464a1de6d14
|
[
"MIT"
] | null | null | null |
notebooks/evaluation/malware_detection.ipynb
|
dmacko232/AndroidMalwareLabeling
|
f0d89a157a34d788d6849b9d4aea8464a1de6d14
|
[
"MIT"
] | null | null | null |
notebooks/evaluation/malware_detection.ipynb
|
dmacko232/AndroidMalwareLabeling
|
f0d89a157a34d788d6849b9d4aea8464a1de6d14
|
[
"MIT"
] | null | null | null | 140.117455 | 13,884 | 0.866181 |
[
[
[
"# Evaluation - Malware Detection",
"_____no_output_____"
]
],
[
[
"PREDICTIONS_DIRPATH = \"../../results/evaluation/predictions/malware_detection\"\nTEST_Y_PATH = \"../../data/prepared/malware_detection/test_y.csv\"\nTRAIN_Y_PATH = \"../../data/prepared/malware_detection/train_y.csv\"\nOUTPUT_MATRIX_PATH = \"../../results/evaluation/plots/confusion_matrix_malware_detection.svg\"",
"_____no_output_____"
],
[
"!pip install -q pandas",
"_____no_output_____"
],
[
"import sys\n\nimport pandas as pd\n\n# add directory to path in order to import own module\nsys.path.insert(0, \"../..\")\nfrom android_malware_labeling.evaluation.evaluation import (\n evaluate_binary_predictions,\n evaluate_imbalanced_multiclass_prediction,\n evaluate_binary_prediction,\n plot_conf_matrix\n)\nfrom android_malware_labeling.evaluation.utils import load_predictions",
"_____no_output_____"
],
[
"predictions = load_predictions(PREDICTIONS_DIRPATH, TRAIN_Y_PATH)\ntest_y = pd.read_csv(TEST_Y_PATH, index_col=0, squeeze=True)",
"_____no_output_____"
],
[
"scores = evaluate_binary_predictions(test_y, predictions, positive_class=True)\nscores.sort_values(by=[\"accuracy\"])",
"_____no_output_____"
],
[
"plot_conf_matrix(test_y, predictions[\"random_forest\"], label_mapping={True: \"malicious\", False: \"benign\"}, output_path=OUTPUT_MATRIX_PATH)",
"_____no_output_____"
],
[
"evaluate_imbalanced_multiclass_prediction(test_y, predictions[\"random_forest\"]\n ).drop(labels=[\"TOTAL\"]).drop(labels=[\"cohen kappa\"], axis=1)",
"_____no_output_____"
],
[
"plot_conf_matrix(test_y, predictions[\"mlp\"], label_mapping={True: \"malicious\", False: \"benign\"})",
"_____no_output_____"
],
[
"plot_conf_matrix(test_y, predictions[\"naive_bayes\"], label_mapping={True: \"malicious\", False: \"benign\"})",
"_____no_output_____"
],
[
"plot_conf_matrix(test_y, predictions[\"logistic_regression\"], label_mapping={True: \"malicious\", False: \"benign\"})",
"_____no_output_____"
],
[
"plot_conf_matrix(test_y, predictions[\"lgbm\"], label_mapping={True: \"malicious\", False: \"benign\"})",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbae416aa9ea9fe608bb77b540a3c0acee3260c2
| 18,161 |
ipynb
|
Jupyter Notebook
|
NSF_COA_affiliation_tool.ipynb
|
tanaes/NSF_COA_GooSchol_parser
|
2c8e14ad26d5a13cd6071376fae363730c8a65ae
|
[
"MIT"
] | null | null | null |
NSF_COA_affiliation_tool.ipynb
|
tanaes/NSF_COA_GooSchol_parser
|
2c8e14ad26d5a13cd6071376fae363730c8a65ae
|
[
"MIT"
] | null | null | null |
NSF_COA_affiliation_tool.ipynb
|
tanaes/NSF_COA_GooSchol_parser
|
2c8e14ad26d5a13cd6071376fae363730c8a65ae
|
[
"MIT"
] | null | null | null | 35.540117 | 438 | 0.504873 |
[
[
[
"# NSF COA author/affiliation tool\n\nInspired by [this awesome tool](https://github.com/ejfertig/NSFBiosketch) from Dr. Elana Fertig, but unable to get it to run in time due to a java install problem with the xlsx package in my perpetually infuriating R environment, I whipped up something similar for the Pythonistas. \n\nThis tool will take a list of PMIDs and return the list of authors and affiliations, along with most recent authorship date. ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom pymed import PubMed\nfrom time import sleep",
"_____no_output_____"
]
],
[
[
"## Import papers\nImport a list of your publication PMIDs, one per line in a plaintext file",
"_____no_output_____"
]
],
[
[
"pmids = []\nwith open('PMID-export.txt', 'r') as f:\n for line in f:\n pmids.append(line.strip())",
"_____no_output_____"
],
[
"pmids",
"_____no_output_____"
]
],
[
[
"We'll sort them in chronological order, to ensure we get the most recent conflict dates per author",
"_____no_output_____"
]
],
[
[
"pmids.sort()",
"_____no_output_____"
],
[
"# Create a PubMed object that GraphQL can use to query\n# Note that the parameters are not required but kindly requested by PubMed Central\n# https://www.ncbi.nlm.nih.gov/pmc/tools/developers/\n\npubmed = PubMed(tool=\"BioSketchify\", email=\"[email protected]\")\n",
"_____no_output_____"
]
],
[
[
"## Retrieve and parse PubMed entries\n\nQuery PubMed one publication at a time, and parse the author and affiliation list.\n\nDue to API limits, we have to limit the rate at which we query.",
"_____no_output_____"
]
],
[
[
"authors = {}\n\nfor pmid in pmids:\n results = pubmed.query(pmid, max_results=1)\n for article in results:\n for author in article.authors:\n name = '%s, %s' % (author['lastname'], author['firstname'])\n year = article.publication_date.year\n affiliation = author['affiliation']\n authors[name] = (year, affiliation)\n print(article.title)\n sleep(1)\n ",
"Cephaloticoccus gen. nov., a new genus of 'Verrucomicrobia' containing two novel species isolated from Cephalotes ant guts.\nDissecting host-associated communities with DNA barcodes.\nGut microbiota of dung beetles correspond to dietary specializations of adults and larvae.\nBy their own devices: invasive Argentine ants have shifted diet without clear aid from symbiotic microbes.\nUnraveling the processes shaping mammalian gut microbiomes over evolutionary time.\nCorrigendum: Cephaloticoccus gen. nov., a new genus of 'Verrucomicrobia' containing two novel species isolated from Cephalotes ant guts.\nThe structured diversity of specialized gut symbionts of the New World army ants.\nAnt-plant mutualism: a dietary by-product of a tropical ant's macronutrient requirements.\nDramatic Differences in Gut Bacterial Densities Correlate with Diet and Habitat in Rainforest Ants.\nA communal catalogue reveals Earth's multiscale microbial diversity.\nThe human microbiome in evolution.\nImproving saliva shotgun metagenomics by chemical host DNA depletion.\nHerbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome.\nBest practices for analysing microbiomes.\nAuthor Correction: Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome.\nGenome Evolution of Bartonellaceae Symbionts of Ants at the Opposite Ends of the Trophic Scale.\nEvolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes.\nAre microbiome studies ready for hypothesis-driven research?\nSocial behaviour in bees influences the abundance of \nQiita: rapid, web-enabled microbiome meta-analysis.\nThe genetic basis for adaptation of model-designed syntrophic co-cultures.\nNot all animals need a microbiome.\nIs there convergence of gut microbes in blood-feeding vertebrates?\nQuantifying and Understanding Well-to-Well Contamination in Microbiome Research.\nAdapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext).\nMetaMiner: A Scalable Peptidogenomics Approach for Discovery of Ribosomal Peptide Natural Products with Blind Modifications from Microbial Communities.\nOptimizing sequencing protocols for leaderboard metagenomics by combining long and short reads.\nPhylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea.\nComparative Analyses of Vertebrate Gut Microbiomes Reveal Convergence between Birds and Bats.\n"
]
],
[
[
"Make an author dataframe, with blank columns for \"Organization\" and \"Department\"",
"_____no_output_____"
]
],
[
[
"author_df = pd.DataFrame.from_dict(authors, orient='index', columns=['year','affiliation'])\nauthor_df['Organization'] = ''\nauthor_df['Department'] = ''\n\nauthor_df.head()",
"_____no_output_____"
]
],
[
[
"## Split affiliation into department and organization\n\nThis might be optional, but PubMed stores affiliation in a single column, and NSF requests 'Organization' be in its own column. This function will loop over the author dataframe, and present each comma-separated element of the 'affiliation' value to you and prompt for input. Press 1 to store that chunk to the 'Department' column, 2 to store that chunk to the 'Organization' column, and any other key to move to the next author.\n\nIt will only parse authors that have no entry for the required 'Organization' column, so if you miss that and re-run this cell it will pick up where you left off.",
"_____no_output_____"
]
],
[
[
"print(\"Enter 1 for Department, 2 for Organization, or nothing to skip rest\")\n\nfor i, author in author_df.iterrows():\n if author['Organization'] != '':\n continue\n try:\n for bit in author['affiliation'].split(','):\n\n print(bit)\n choice = input(\"Input:\")\n if choice == '1':\n author_df.loc[i, 'Department'] = author_df.loc[i, 'Department'] + bit\n elif choice == '2':\n author_df.loc[i, 'Organization'] = author_df.loc[i, 'Organization'] + bit\n else:\n break\n except:\n continue\n ",
"Enter 1 for Department, 2 for Organization, or nothing to skip rest\nLajuma Research Centre\nInput:2\n Louis Trichardt (Makhado)\nInput:\nEstacion Biologica Corrientes (MACN-BR) - CONICET\nInput:2\n Corrientes\nInput:\n"
],
[
"author_df.head()",
"_____no_output_____"
]
],
[
[
"## Export author dataframe to CSV file\n\nYou can now open this in your favorite spreadsheet column to clean it up and add to the NSF workbook.",
"_____no_output_____"
]
],
[
[
"author_df.to_csv('authors_with_affiliations.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbae4cffa93c59151ee2502e6adc3d968a229c85
| 73,743 |
ipynb
|
Jupyter Notebook
|
dev/12_optimizer.ipynb
|
cs224/fastai_dev
|
c1c5a3d5198c7047522384652a8793c13c6c831b
|
[
"Apache-2.0"
] | null | null | null |
dev/12_optimizer.ipynb
|
cs224/fastai_dev
|
c1c5a3d5198c7047522384652a8793c13c6c831b
|
[
"Apache-2.0"
] | null | null | null |
dev/12_optimizer.ipynb
|
cs224/fastai_dev
|
c1c5a3d5198c7047522384652a8793c13c6c831b
|
[
"Apache-2.0"
] | null | null | null | 39.245875 | 10,868 | 0.616709 |
[
[
[
"# default_exp optimizer",
"_____no_output_____"
],
[
"#export\nfrom local.torch_basics import *\nfrom local.test import *",
"_____no_output_____"
],
[
"from local.notebook.showdoc import *",
"_____no_output_____"
]
],
[
[
"# Optimizer\n\n> Define the general fastai optimizer and the variants",
"_____no_output_____"
],
[
"## Optimizer -",
"_____no_output_____"
]
],
[
[
"#export\nclass _BaseOptimizer():\n \"Common functionality between `Optimizer` and `OptimWrapper`\"\n def all_params(self, n=slice(None), with_grad=False):\n res = L((p,pg,self.state[p],hyper) for pg,hyper in zip(self.param_groups[n],self.hypers[n]) for p in pg)\n return L(o for o in res if o[0].grad is not None) if with_grad else res\n\n def _set_require_grad(self, rg, p,pg,state,h): p.requires_grad_(rg or state.get('force_train', False))\n def freeze_to(self, n):\n self.frozen_idx = n if n >= 0 else len(self.param_groups) + n\n if self.frozen_idx >= len(self.param_groups):\n warn(f\"Freezing {self.frozen_idx} groups; model has {len(self.param_groups)}; whole model is frozen.\")\n for o in self.all_params(slice(n, None)): self._set_require_grad(True, *o)\n for o in self.all_params(slice(None, n)): self._set_require_grad(False, *o)\n\n def freeze(self):\n assert(len(self.param_groups)>1)\n self.freeze_to(-1)\n\n def unfreeze(self): self.freeze_to(0)\n def set_hypers(self, **kwargs): L(kwargs.items()).starmap(self.set_hyper)\n def _set_hyper(self, k, v):\n for v_,h in zip(v, self.hypers): h[k] = v_\n\n def set_hyper(self, k, v):\n if isinstance(v, slice):\n if v.start: v = even_mults(v.start, v.stop, len(self.param_groups))\n else: v = [v.stop/10]*(len(self.param_groups)-1) + [v.stop]\n v = L(v, use_list=None)\n if len(v)==1: v = v*len(self.param_groups)\n assert len(v) == len(self.hypers), f\"Trying to set {len(v)} values for {k} but there are {len(self.param_groups)} parameter groups.\"\n self._set_hyper(k, v)",
"_____no_output_____"
],
[
"add_docs(_BaseOptimizer, \n all_params=\"List of param_groups, parameters, and hypers\",\n freeze_to=\"Freeze parameter groups up to `n`\",\n freeze=\"Freeze up to last parameter group\",\n unfreeze=\"Unfreeze the entire model\",\n set_hypers=\"`set_hyper` for all `kwargs`\",\n set_hyper=\"Set the value(s) in `v` for hyper-parameter `k`\")",
"_____no_output_____"
],
[
"# export\nclass Optimizer(_BaseOptimizer):\n \"Base optimizer class for the fastai library, updating `params` with `steppers`\"\n _keep_on_clear = ['force_train', 'do_wd']\n def __init__(self, params, steppers, stats=None, train_bn=True, **defaults):\n params = L(params)\n self.steppers,self.stats,self.state,self.train_bn = L(steppers),L(stats),defaultdict(dict),train_bn\n defaults = merge(*self.stats.attrgot('defaults'), *self.steppers.attrgot('defaults'), defaults)\n self.param_groups = L(L(p) for p in params) if isinstance(params[0], (L,list)) else L([params])\n #self.step_func = compose(*steppers)\n self.hypers = L({} for _ in range_of(self.param_groups))\n self.set_hypers(**defaults)\n self.frozen_idx = 0\n\n def zero_grad(self):\n for p,*_ in self.all_params(with_grad=True):\n p.grad.detach_()\n p.grad.zero_()\n\n def step(self):\n for p,pg,state,hyper in self.all_params(with_grad=True):\n for stat in self.stats: state = stat(state, p, **hyper)\n for step in self.steppers: step(p, **{**state, **hyper})\n self.state[p] = state\n\n def clear_state(self):\n for p,pg,state,hyper in self.all_params():\n self.state[p] = {k: state[k] for k in self._keep_on_clear if k in state}\n\n def state_dict(self):\n state = [self.state[p] for p,*_ in self.all_params()]\n return {'state': state, 'hypers': self.hypers}\n\n def load_state_dict(self, sd):\n assert len(sd[\"hypers\"]) == len(self.param_groups)\n assert len(sd[\"state\"]) == sum([len(pg) for pg in self.param_groups])\n self.hypers = sd['hypers']\n self.state = {p: s for p,s in zip(self.all_params().itemgot(0), sd['state'])}",
"_____no_output_____"
],
[
"add_docs(Optimizer, \n zero_grad=\"Standard PyTorch API: Zero all the grad attributes of the parameters\",\n step=\"Standard PyTorch API: Update the stats and execute the steppers in on all parameters that have a grad\",\n state_dict=\"Return the state of the optimizer in a dictionary\",\n load_state_dict=\"Load the content of `sd`\",\n clear_state=\"Reset the state of the optimizer\")",
"_____no_output_____"
]
],
[
[
"### Initializing an Optimizer",
"_____no_output_____"
],
[
"`params` will be used to create the `param_groups` of the optimizer. If it's a collection (or a generator) of parameters, it will be a `L` containing one `L` with all the parameters. To define multiple parameter groups `params` should be passed as a collection (or a generator) of `L`s.\n\n> Note: In PyTorch, `model.parameters()` returns a generator with all the parameters, that you can directly pass to `Optimizer`.",
"_____no_output_____"
]
],
[
[
"opt = Optimizer([1,2,3], noop)\ntest_eq(opt.param_groups, [[1,2,3]])\nopt = Optimizer(range(3), noop)\ntest_eq(opt.param_groups, [[0,1,2]])\nopt = Optimizer([[1,2],[3]], noop)\ntest_eq(opt.param_groups, [[1,2],[3]])\nopt = Optimizer(([o,o+1] for o in range(0,4,2)), noop)\ntest_eq(opt.param_groups, [[0,1],[2,3]])",
"_____no_output_____"
]
],
[
[
"`steppers` is a list of functions that will be composed when applying the step. For instance, you can compose a function making the SGD step, with another one applying weight decay. Additionally, each `stepper` can have a `defaults` attribute that contains hyper-parameters and their default value. Those are all gathered at initialization, and new values can be passed to override those defaults with the `defaults` kwargs. The steppers will be called by `Optimizer.step` (which is the standard PyTorch name), and gradients can be cleared with `Optimizer.zero_grad` (also a standard PyTorch name).\n\nOnce the defaults have all been pulled off, they are copied as many times as there are `param_groups` and stored in `hypers`. To apply different hyper-parameters to different groups (differential learning rates, or no weight decay for certain layers for instance), you will need to adjsut those values after the init. ",
"_____no_output_____"
]
],
[
[
"def tst_arg(p, lr=0, **kwargs): return p\ntst_arg.defaults = dict(lr=1e-2)\n\ndef tst_arg2(p, lr2=0, **kwargs): return p\ntst_arg2.defaults = dict(lr2=1e-3)\n\ndef tst_arg3(p, mom=0, **kwargs): return p\ntst_arg3.defaults = dict(mom=0.9)\n\ndef tst_arg4(p, **kwargs): return p\n\nopt = Optimizer([1,2,3], [tst_arg,tst_arg2], tst_arg3)\ntest_eq(opt.hypers, [{'lr2': 1e-3, 'mom': 0.9, 'lr': 1e-2}])\nopt = Optimizer([1,2,3], tst_arg, lr=0.1)\ntest_eq(opt.hypers, [{'lr': 0.1}])\nopt = Optimizer([[1,2],[3]], tst_arg)\ntest_eq(opt.hypers, [{'lr': 1e-2}, {'lr': 1e-2}])\nopt = Optimizer([[1,2],[3]], tst_arg, lr=0.1)\ntest_eq(opt.hypers, [{'lr': 0.1}, {'lr': 0.1}])",
"_____no_output_____"
]
],
[
[
"For each hyper-parameter, you can pass a slice or a collection to set them, if there are multiple parameter groups. A slice will be converted to a log-uniform collection from its beginning to its end, or if it only has an end `e`, to a collection of as many values as there are parameter groups that are `...,e/10,e/10,e`.\n\nSetting an yper-paramter with a collection that has a different number of elements than the optimizer has paramter groups will raise an error.",
"_____no_output_____"
]
],
[
[
"opt = Optimizer([[1,2],[3]], tst_arg, lr=[0.1,0.2])\ntest_eq(opt.hypers, [{'lr': 0.1}, {'lr': 0.2}])\nopt = Optimizer([[1,2],[3],[4]], tst_arg, lr=slice(1e-2))\ntest_eq(opt.hypers, [{'lr': 1e-3}, {'lr': 1e-3}, {'lr': 1e-2}])\nopt = Optimizer([[1,2],[3],[4]], tst_arg, lr=slice(1e-4,1e-2))\ntest_eq(opt.hypers, [{'lr': 1e-4}, {'lr': 1e-3}, {'lr': 1e-2}])\ntest_fail(lambda: Optimizer([[1,2],[3],[4]], tst_arg, lr=np.array([0.1,0.2])))",
"_____no_output_____"
]
],
[
[
"### Basic steppers",
"_____no_output_____"
],
[
"To be able to give examples of optimizer steps, we will need some steppers, like the following:",
"_____no_output_____"
]
],
[
[
"#export\ndef sgd_step(p, lr, **kwargs):\n p.data.add_(-lr, p.grad.data)\n return p",
"_____no_output_____"
],
[
"def tst_param(val, grad=None):\n \"Create a tensor with `val` and a gradient of `grad` for testing\"\n res = tensor([val]).float()\n res.grad = tensor([val/10 if grad is None else grad]).float()\n return res",
"_____no_output_____"
],
[
"p = tst_param(1., 0.1)\np = sgd_step(p, 1.)\ntest_eq(p, tensor([0.9]))\ntest_eq(p.grad, tensor([0.1]))",
"_____no_output_____"
],
[
"#export\ndef weight_decay(p, lr, wd, do_wd=True, **kwargs):\n \"Weight decay as decaying `p` with `lr*wd`\"\n if do_wd and wd!=0: p.data.mul_(1 - lr*wd)\n return p\nweight_decay.defaults = dict(wd=0.)",
"_____no_output_____"
],
[
"p = tst_param(1., 0.1)\np = weight_decay(p, 1., 0.1)\ntest_eq(p, tensor([0.9]))\ntest_eq(p.grad, tensor([0.1]))",
"_____no_output_____"
],
[
"#export\ndef l2_reg(p, lr, wd, do_wd=True, **kwargs):\n \"L2 regularization as adding `wd*p` to `p.grad`\"\n if do_wd and wd!=0: p.grad.data.add_(wd, p.data)\n return p\nl2_reg.defaults = dict(wd=0.)",
"_____no_output_____"
],
[
"p = tst_param(1., 0.1)\np = l2_reg(p, 1., 0.1)\ntest_eq(p, tensor([1.]))\ntest_eq(p.grad, tensor([0.2]))",
"_____no_output_____"
]
],
[
[
"> Warning: Weight decay and L2 regularization is the same thing for basic SGD, but for more complex optimizers, they are very different. See [Decoupled Weight Decay Regularization](https://arxiv.org/abs/1711.05101) for more information.",
"_____no_output_____"
],
[
"### Making the step",
"_____no_output_____"
]
],
[
[
"show_doc(Optimizer.step)",
"_____no_output_____"
]
],
[
[
"This method will loop over all param groups, then all parameters for which `grad` is not None and call each function in `stepper`, passing it the parameter `p` with the hyper-parameters in the corresponding dict in `hypers`.",
"_____no_output_____"
]
],
[
[
"#test basic step\nr = L.range(4)\ndef tst_params(): return r.map(tst_param)\n\nparams = tst_params()\nopt = Optimizer(params, sgd_step, lr=0.1)\nopt.step()\ntest_close([p.item() for p in params], r.map(mul(0.99)))",
"_____no_output_____"
],
[
"#test two steps\nparams = tst_params()\nopt = Optimizer(params, [weight_decay, sgd_step], lr=0.1, wd=0.1)\nopt.step()\ntest_close([p.item() for p in params], r.map(mul(0.98)))",
"_____no_output_____"
],
[
"#test None gradients are ignored\nparams = tst_params()\nopt = Optimizer(params, sgd_step, lr=0.1)\nparams[-1].grad = None\nopt.step()\ntest_close([p.item() for p in params], [0., 0.99, 1.98, 3.])",
"_____no_output_____"
],
[
"#test discriminative lrs\nparams = tst_params()\nopt = Optimizer([params[:2], params[2:]], sgd_step, lr=0.1)\nopt.hypers[0]['lr'] = 0.01\nopt.step()\ntest_close([p.item() for p in params], [0., 0.999, 1.98, 2.97])",
"_____no_output_____"
],
[
"show_doc(Optimizer.zero_grad)",
"_____no_output_____"
],
[
"params = tst_params()\nopt = Optimizer(params, [weight_decay, sgd_step], lr=0.1, wd=0.1)\nopt.zero_grad()\n[test_eq(p.grad, tensor([0.])) for p in params];",
"_____no_output_____"
]
],
[
[
"`Optimizer` has `stats` which are functions taking the state associated with a parameter. `stats` use that parameter, plus the optimizer hyper-parameters, to update the state. \nThat state can then be used by any stepper. The best example is a momentum calculation. \n`stats` are initialized to an empty dictionary the first time we try to access it, and after that the `stat` function will have to be properly initialized.",
"_____no_output_____"
]
],
[
[
"def tst_stat(state, p, **kwargs): \n state['sum'] = state.get('sum', torch.zeros_like(p)) + p.data\n return state\ntst_stat.defaults = {'mom': 0.9}\n\n#Test Optimizer init\nopt = Optimizer([1,2,3], noop, stats=tst_stat)\ntest_eq(opt.hypers, [{'mom': 0.9}])\nopt = Optimizer([1,2,3], noop, stats=tst_stat, mom=0.99)\ntest_eq(opt.hypers, [{'mom': 0.99}])\n\n#Test stat\nx = torch.randn(4,5)\nstate = tst_stat({}, x)\nassert 'sum' in state\ntest_eq(state['sum'], x)\nstate = tst_stat(state, x)\ntest_eq(state['sum'], 2*x)",
"_____no_output_____"
]
],
[
[
"## Statistics",
"_____no_output_____"
]
],
[
[
"# export\ndef average_grad(state, p, mom, dampening=False, **kwargs):\n \"Keeps track of the avg grads of `p` in `state` with `mom`.\"\n if 'grad_avg' not in state: state['grad_avg'] = torch.zeros_like(p.grad.data)\n damp = 1-mom if dampening else 1.\n state['grad_avg'].mul_(mom).add_(damp, p.grad.data)\n return state\n\naverage_grad.defaults = dict(mom=0.9)",
"_____no_output_____"
]
],
[
[
"`dampening=False` gives the classical formula for momentum in SGD: \n```\nnew_val = old_val * mom + grad\n```\nwhereas `dampening=True` makes it an exponential moving average:\n```\nnew_val = old_val * mom + grad * (1-mom)\n```",
"_____no_output_____"
]
],
[
[
"p = tst_param([1,2,3], [4,5,6])\nstate = {}\nstate = average_grad(state, p, mom=0.9)\ntest_eq(state['grad_avg'], p.grad)\nstate = average_grad(state, p, mom=0.9)\ntest_eq(state['grad_avg'], p.grad * 1.9)\n#Test dampening\nstate = {}\nstate = average_grad(state, p, mom=0.9, dampening=True)\ntest_eq(state['grad_avg'], 0.1*p.grad)\nstate = average_grad(state, p, mom=0.9, dampening=True)\ntest_eq(state['grad_avg'], (0.1*0.9+0.1)*p.grad)",
"_____no_output_____"
],
[
"# export\ndef average_sqr_grad(state, p, sqr_mom, dampening=True, **kwargs):\n if 'sqr_avg' not in state: state['sqr_avg'] = torch.zeros_like(p.grad.data)\n damp = 1-sqr_mom if dampening else 1.\n state['sqr_avg'].mul_(sqr_mom).addcmul_(damp, p.grad.data, p.grad.data)\n return state\n\naverage_sqr_grad.defaults = dict(sqr_mom=0.99)",
"_____no_output_____"
]
],
[
[
"`dampening=False` gives the classical formula for momentum in SGD: \n```\nnew_val = old_val * mom + grad**2\n```\nwhereas `dampening=True` makes it an exponential moving average:\n```\nnew_val = old_val * mom + (grad**2) * (1-mom)\n```",
"_____no_output_____"
]
],
[
[
"p = tst_param([1,2,3], [4,5,6])\nstate = {}\nstate = average_sqr_grad(state, p, sqr_mom=0.99, dampening=False)\ntest_eq(state['sqr_avg'], p.grad.pow(2))\nstate = average_sqr_grad(state, p, sqr_mom=0.99, dampening=False)\ntest_eq(state['sqr_avg'], p.grad.pow(2) * 1.99)\n#Test dampening\nstate = {}\nstate = average_sqr_grad(state, p, sqr_mom=0.99)\ntest_close(state['sqr_avg'], 0.01*p.grad.pow(2))\nstate = average_sqr_grad(state, p, sqr_mom=0.99)\ntest_close(state['sqr_avg'], (0.01*0.99+0.01)*p.grad.pow(2))",
"_____no_output_____"
]
],
[
[
"### Freezing part of the model",
"_____no_output_____"
]
],
[
[
"show_doc(Optimizer.freeze)",
"_____no_output_____"
],
[
"show_doc(Optimizer.freeze_to)",
"_____no_output_____"
],
[
"show_doc(Optimizer.unfreeze)",
"_____no_output_____"
],
[
"#Freezing the first layer\nparams = [tst_params(), tst_params(), tst_params()]\nopt = Optimizer(params, sgd_step, lr=0.1)\nopt.freeze_to(1)\nreq_grad = Self.requires_grad()\ntest_eq(L(params[0]).map(req_grad), [False]*4)\nfor i in {1,2}: test_eq(L(params[i]).map(req_grad), [True]*4)\n \n#Unfreezing\nopt.unfreeze()\nfor i in range(2): test_eq(L(params[i]).map(req_grad), [True]*4)\n\n#TODO: test warning\n# opt.freeze_to(3)",
"_____no_output_____"
]
],
[
[
"Parameters such as batchnorm weights/bias can be marked to always be in training mode, just put `force_train=true` in their state.",
"_____no_output_____"
]
],
[
[
"params = [tst_params(), tst_params(), tst_params()]\nopt = Optimizer(params, sgd_step, lr=0.1)\nfor p in L(params[1])[[1,3]]: opt.state[p] = {'force_train': True}\nopt.freeze()\ntest_eq(L(params[0]).map(req_grad), [False]*4)\ntest_eq(L(params[1]).map(req_grad), [False, True, False, True])\ntest_eq(L(params[2]).map(req_grad), [True]*4)",
"_____no_output_____"
]
],
[
[
"### Serializing",
"_____no_output_____"
]
],
[
[
"show_doc(Optimizer.state_dict)",
"_____no_output_____"
],
[
"show_doc(Optimizer.load_state_dict)",
"_____no_output_____"
],
[
"p = tst_param([1,2,3], [4,5,6])\nopt = Optimizer(p, noop, stats=average_grad)\nopt.step()\ntest_eq(opt.state[p]['grad_avg'], tensor([[4., 5., 6.]]))\n\nsd = opt.state_dict()\np1 = tst_param([10,20,30], [40,50,60])\nopt = Optimizer(p1, noop, stats=average_grad, mom=0.99)\ntest_eq(opt.hypers[0]['mom'], 0.99)\ntest_eq(opt.state, {})\n\nopt.load_state_dict(sd)\ntest_eq(opt.hypers[0]['mom'], 0.9)\ntest_eq(opt.state[p1]['grad_avg'], tensor([[4., 5., 6.]]))",
"_____no_output_____"
],
[
"show_doc(Optimizer.clear_state)",
"_____no_output_____"
],
[
"p = tst_param([1,2,3], [4,5,6])\nopt = Optimizer(p, noop, stats=average_grad)\nopt.state[p] = {'force_train': True}\nopt.step()\ntest_eq(opt.state[p]['grad_avg'], tensor([[4., 5., 6.]]))\n\nopt.clear_state()\ntest_eq(opt.state[p], {'force_train': True})",
"_____no_output_____"
]
],
[
[
"## Optimizers",
"_____no_output_____"
],
[
"### SGD with momentum",
"_____no_output_____"
]
],
[
[
"#export\ndef momentum_step(p, lr, grad_avg, **kwargs):\n \"Step for SGD with momentum with `lr`\"\n p.data.add_(-lr, grad_avg)\n return p",
"_____no_output_____"
],
[
"#export\ndef SGD(params, lr, mom=0., wd=0., decouple_wd=True):\n \"A `Optimizer` for SGD with `lr` and `mom` and `params`\"\n steppers = [weight_decay] if decouple_wd else [l2_reg]\n steppers.append(sgd_step if mom==0 else momentum_step)\n if mom == 0.: return Optimizer(params, steppers, lr=lr, wd=wd)\n else: return Optimizer(params, steppers, stats=average_grad, lr=lr, mom=mom, wd=wd)",
"_____no_output_____"
]
],
[
[
"Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).",
"_____no_output_____"
]
],
[
[
"#Vanilla SGD\nparams = tst_params()\nopt = SGD(params, lr=0.1)\nopt.step()\ntest_close([p.item() for p in params], [i*0.99 for i in range(4)])\nopt.step()\n[p.item() for p in params]\ntest_close([p.item() for p in params], [i*0.98 for i in range(4)])",
"_____no_output_____"
],
[
"#SGD with momentum\nparams = tst_params()\nopt = SGD(params, lr=0.1, mom=0.9)\nassert isinstance(opt, Optimizer)\nopt.step()\ntest_close([p.item() for p in params], [i*0.99 for i in range(4)])\nopt.step()\n[p.item() for p in params]\ntest_close([p.item() for p in params], [i*(1 - 0.1 * (0.1 + 0.1*1.9)) for i in range(4)])\nfor i,p in enumerate(params): test_close(opt.state[p]['grad_avg'].item(), i*0.19)",
"_____no_output_____"
]
],
[
[
"Test weight decay, notice how we can see that L2 regularization is different from weight decay even for simple SGD with momentum.",
"_____no_output_____"
]
],
[
[
"params = tst_params()\n#Weight decay\nopt = SGD(params, lr=0.1, mom=0.9, wd=0.1)\nopt.step()\ntest_close([p.item() for p in params], [i*0.98 for i in range(4)])\n#L2 reg\nopt = SGD(params, lr=0.1, mom=0.9, wd=0.1, decouple_wd=False)\nopt.step()\ntest_close([p.item() for p in params], [i*0.97 for i in range(4)])",
"_____no_output_____"
]
],
[
[
"### RMSProp",
"_____no_output_____"
]
],
[
[
"#export\ndef rms_prop_step(p, lr, sqr_avg, eps, grad_avg=None, **kwargs):\n \"Step for SGD with momentum with `lr`\"\n denom = sqr_avg.sqrt().add_(eps)\n p.data.addcdiv_(-lr, (grad_avg if grad_avg is not None else p.grad), denom)\n return p\n\nrms_prop_step.defaults = dict(eps=1e-8)",
"_____no_output_____"
],
[
"#export\ndef RMSProp(params, lr, sqr_mom=0.99, mom=0., wd=0., decouple_wd=True):\n \"A `Optimizer` for RMSProp with `lr`, `sqr_mom`, `mom` and `params`\"\n steppers = [weight_decay] if decouple_wd else [l2_reg]\n steppers.append(rms_prop_step)\n stats = [average_sqr_grad] if mom==0. else [average_grad, average_sqr_grad]\n return Optimizer(params, steppers, stats=stats, lr=lr, mom=mom, sqr_mom=sqr_mom, wd=wd)",
"_____no_output_____"
]
],
[
[
"RMSProp was introduced by Geoffrey Hinton in his [course](http://www.cs.toronto.edu/~tijmen/csc321/slides/lecture_slides_lec6.pdf). What is named `sqr_mom` here is the `alpha` in the course. Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).",
"_____no_output_____"
]
],
[
[
"#Without momentum\nimport math\nparams = tst_param([1,2,3], [0.1,0.2,0.3])\nopt = RMSProp(params, lr=0.1)\nopt.step()\ntest_close(params[0], tensor([0.,1.,2.]))\nopt.step()\nstep = - 0.1 * 0.1 / (math.sqrt((0.01*0.99+0.01) * 0.1**2) + 1e-8)\ntest_close(params[0], tensor([step, 1+step, 2+step]))",
"_____no_output_____"
],
[
"#With momentum\nparams = tst_param([1,2,3], [0.1,0.2,0.3])\nopt = RMSProp(params, lr=0.1, mom=0.9)\nopt.step()\ntest_close(params[0], tensor([0.,1.,2.]))\nopt.step()\nstep = - 0.1 * (0.1 + 0.9*0.1) / (math.sqrt((0.01*0.99+0.01) * 0.1**2) + 1e-8)\ntest_close(params[0], tensor([step, 1+step, 2+step]))",
"_____no_output_____"
]
],
[
[
"### Adam",
"_____no_output_____"
]
],
[
[
"#export\ndef step_stat(state, p, **kwargs):\n \"Register the number of steps done in `state` for `p`\"\n if 'step' not in state: state['step'] = 0\n state['step'] += 1\n return state",
"_____no_output_____"
],
[
"p = tst_param(1,0.1)\nstate = {}\nstate = step_stat(state, p)\ntest_eq(state['step'], 1)\nfor _ in range(5): state = step_stat(state, p)\ntest_eq(state['step'], 6)",
"_____no_output_____"
],
[
"#export\ndef debias(mom, damp, step): return damp * (1 - mom**step) / (1-mom)",
"_____no_output_____"
],
[
"#export\ndef adam_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, **kwargs):\n \"Step for Adam with `lr` on `p`\"\n debias1 = debias(mom, 1-mom, step)\n debias2 = debias(sqr_mom, 1-sqr_mom, step)\n p.data.addcdiv_(-lr / debias1, grad_avg, (sqr_avg/debias2).sqrt() + eps)\n return p\n\nadam_step._defaults = dict(eps=1e-5)",
"_____no_output_____"
],
[
"#export\ndef Adam(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0., decouple_wd=True):\n \"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`\"\n steppers = [weight_decay] if decouple_wd else [l2_reg]\n steppers.append(adam_step)\n stats = [partial(average_grad, dampening=True), average_sqr_grad, step_stat]\n return Optimizer(params, steppers, stats=stats, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)",
"_____no_output_____"
]
],
[
[
"Adam was introduced by Diederik P. Kingma and Jimmy Ba in [Adam: A Method for Stochastic Optimization](https://arxiv.org/abs/1412.6980). For consistency accross optimizers, we renamed `beta1` and `beta2` in the paper to `mom` and `sqr_mom`. Note that our defaults also differ from the paper (0.99 for `sqr_mom` or `beta2`, 1e-5 for `eps`). Those values seem to be better from our experiments in a wide range of situations.\n\nOptional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).\n\n> Note: Don't forget that `eps` is an hyper-parameter you can change. Some models won't train without a very high `eps` like 0.1 (intuitively, the higher `eps` is, the closer we are to normal SGD). The usual default of 1e-8 is often too extreme in the sense we don't manage to get as good results as with SGD. ",
"_____no_output_____"
]
],
[
[
"params = tst_param([1,2,3], [0.1,0.2,0.3])\nopt = Adam(params, lr=0.1)\nopt.step()\nstep = -0.1 * 0.1 / (math.sqrt(0.1**2) + 1e-8)\ntest_close(params[0], tensor([1+step, 2+step, 3+step]))\nopt.step()\ntest_close(params[0], tensor([1+2*step, 2+2*step, 3+2*step]), eps=1e-3)",
"_____no_output_____"
]
],
[
[
"### RAdam",
"_____no_output_____"
],
[
"RAdam (for rectified Adam) was introduced by Zhang et al. in [On the Variance of the Adaptive Learning Rate and Beyond](https://arxiv.org/abs/1907.08610) to slightly modify the Adam optimizer to be more stable at the beginning of training (and thus not require a long warmup). They use an estimate of the variance of the moving average of the squared gradients (the term in the denominator of traditional Adam) and rescale this moving average by this term before performing the update.",
"_____no_output_____"
]
],
[
[
"#export\ndef radam_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, **kwargs):\n \"Step for RAdam with `lr` on `p`\"\n debias1 = debias(mom, 1-mom, step)\n debias2 = debias(sqr_mom, 1-sqr_mom, step)\n r_inf = 2/(1-sqr_mom) - 1\n r = r_inf - 2*step*sqr_mom**step/(1-sqr_mom**step)\n if r > 4:\n v = math.sqrt(((r-4) * (r-2) * r_inf)/((r_inf-4)*(r_inf-2)*r))\n p.data.addcdiv_(-lr*v / debias1, grad_avg, (sqr_avg/debias2).sqrt() + eps)\n else: p.data.add_(-lr / debias1, grad_avg)\n return p\n\nradam_step._defaults = dict(eps=1e-5)",
"_____no_output_____"
],
[
"#export\ndef RAdam(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0., decouple_wd=True):\n \"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`\"\n steppers = [weight_decay] if decouple_wd else [l2_reg]\n steppers.append(radam_step)\n stats = [partial(average_grad, dampening=True), average_sqr_grad, step_stat]\n return Optimizer(params, steppers, stats=stats, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)",
"_____no_output_____"
]
],
[
[
"This is the effective correction apported to the adam step for 500 iterations in RAdam. We can see how it goes from 0 to 1, mimicking the effect of a warm-up.",
"_____no_output_____"
]
],
[
[
"beta = 0.99\nr_inf = 2/(1-beta) - 1\nrs = np.array([r_inf - 2*s*beta**s/(1-beta**s) for s in range(5,500)])\nv = np.sqrt(((rs-4) * (rs-2) * r_inf)/((r_inf-4)*(r_inf-2)*rs))\nplt.plot(v)",
"_____no_output_____"
],
[
"params = tst_param([1,2,3], [0.1,0.2,0.3])\nopt = RAdam(params, lr=0.1)\n#The r factor is lower than 5 during the first 5 steps so updates use the aveage of gradients (all the same)\nr_inf = 2/(1-0.99) - 1\nfor i in range(4): \n r = r_inf - 2*(i+1)*0.99**(i+1)/(1-0.99**(i+1))\n assert r <= 4\n opt.step()\np = tensor([0.96, 1.92, 2.88])\ntest_close(params[0], p)\n\n#The r factor is greater than 4 for the sixth step so we update with RAdam\nr = r_inf - 2*5*0.99**5/(1-0.99**5)\nassert r > 4\nopt.step()\nv = math.sqrt(((r-4) * (r-2) * r_inf)/((r_inf-4)*(r_inf-2)*r))\nstep = -0.1*0.1*v/(math.sqrt(0.1**2) + 1e-8)\ntest_close(params[0], p+step)",
"_____no_output_____"
]
],
[
[
"### LARS/LARC",
"_____no_output_____"
]
],
[
[
"#export\ndef larc_layer_lr(state, p, lr, trust_coeff, wd, eps, clip=True, **kwargs):\n \"Computes the local lr before weight decay is applied\"\n p_norm,g_norm = torch.norm(p.data),torch.norm(p.grad.data)\n local_lr = lr*trust_coeff * (p_norm) / (g_norm + p_norm * wd + eps)\n state['local_lr'] = min(lr, local_lr) if clip else local_lr\n return state\nlarc_layer_lr.defaults = dict(trust_coeff=0.02, wd=0., eps=1e-8)",
"_____no_output_____"
],
[
"#export\ndef larc_step(p, local_lr, grad_avg=None, **kwargs):\n p.data.add_(-local_lr, p.grad.data if grad_avg is None else grad_avg)\n \"Step for LARC `local_lr` on `p`\"\n return p",
"_____no_output_____"
],
[
"#export\ndef Larc(params, lr, mom=0.9, clip=True, trust_coeff=0.02, eps=1e-8, wd=0., decouple_wd=True):\n \"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`\"\n steppers = [weight_decay] if decouple_wd else [l2_reg]\n steppers.append(larc_step)\n stats = [] if mom==0. else [average_grad]\n stats.append(partial(larc_layer_lr, clip=clip))\n return Optimizer(params, steppers, stats=stats, lr=lr, mom=mom, trust_coeff=trust_coeff, eps=eps, wd=wd)",
"_____no_output_____"
]
],
[
[
"The LARS optimizer was first introduced in [Large Batch Training of Convolutional Networks](https://arxiv.org/abs/1708.03888) then refined in its LARC variant (original LARS is with `clip=False`). A learning rate is computed for each individual layer with a certain `trust_coefficient`, then clipped to be always less than `lr`.\n\nOptional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).",
"_____no_output_____"
]
],
[
[
"params = [tst_param([1,2,3], [0.1,0.2,0.3]), tst_param([1,2,3], [0.01,0.02,0.03])]\nopt = Larc(params, lr=0.1)\nopt.step()\n#First param local lr is 0.02 < lr so it's not clipped\ntest_close(opt.state[params[0]]['local_lr'], 0.02)\n#Second param local lr is 0.2 > lr so it's clipped\ntest_eq(opt.state[params[1]]['local_lr'], 0.1)\ntest_close(params[0], tensor([0.998,1.996,2.994]))\ntest_close(params[1], tensor([0.999,1.998,2.997]))",
"_____no_output_____"
],
[
"params = [tst_param([1,2,3], [0.1,0.2,0.3]), tst_param([1,2,3], [0.01,0.02,0.03])]\nopt = Larc(params, lr=0.1, clip=False)\nopt.step()\n#No clipping\ntest_close(opt.state[params[0]]['local_lr'], 0.02)\ntest_close(opt.state[params[1]]['local_lr'], 0.2)\ntest_close(params[0], tensor([0.998,1.996,2.994]))\ntest_close(params[1], tensor([0.998,1.996,2.994]))",
"_____no_output_____"
]
],
[
[
"### LAMB",
"_____no_output_____"
]
],
[
[
"#export\ndef lamb_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, **kwargs):\n \"Step for LAMB with `lr` on `p`\"\n debias1 = debias(mom, 1-mom, step)\n debias2 = debias(sqr_mom, 1-sqr_mom, step)\n r1 = p.data.pow(2).mean().sqrt()\n step = (grad_avg/debias1) / ((sqr_avg/debias2).sqrt()+eps)\n r2 = step.pow(2).mean().sqrt()\n q = 1 if r1 == 0 or r2 == 0 else min(r1/r2,10)\n p.data.add_(-lr * q, step)\n return p\nlamb_step._defaults = dict(eps=1e-6, wd=0.)",
"_____no_output_____"
],
[
"#export\ndef Lamb(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0., decouple_wd=True):\n \"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`\"\n steppers = [weight_decay] if decouple_wd else [l2_reg]\n steppers.append(lamb_step)\n stats = [partial(average_grad, dampening=True), average_sqr_grad, step_stat]\n return Optimizer(params, steppers, stats=stats, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)",
"_____no_output_____"
]
],
[
[
"LAMB was introduced in [Large Batch Optimization for Deep Learning: Training BERT in 76 minutes](https://arxiv.org/abs/1904.00962). Intuitively, it's LARC applied to Adam. As in `Adam`, we renamed `beta1` and `beta2` in the paper to `mom` and `sqr_mom`. Note that our defaults also differ from the paper (0.99 for `sqr_mom` or `beta2`, 1e-5 for `eps`). Those values seem to be better from our experiments in a wide range of situations.\n\nOptional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).",
"_____no_output_____"
]
],
[
[
"params = tst_param([1,2,3], [0.1,0.2,0.3])\nopt = Lamb(params, lr=0.1)\nopt.step()\ntest_close(params[0], tensor([0.7840,1.7840,2.7840]), eps=1e-3)",
"_____no_output_____"
]
],
[
[
"## Lookahead -",
"_____no_output_____"
],
[
"Lookahead was introduced by Zhang et al. in [Lookahead Optimizer: k steps forward, 1 step back](https://arxiv.org/abs/1907.08610). It can be run on top of any optimizer and consists in having the final weights of the model be a moving average. In practice, we update our model using the internal optimizer but keep a copy of old weights that and every `k` steps, we change the weghts by a moving average of the *fast weights* (the ones updated by the inner optimizer) with the *slow weights* (the copy of old weights). Those *slow weights* act like a stability mechanism.",
"_____no_output_____"
]
],
[
[
"#export\nclass Lookahead(Optimizer, GetAttr):\n \"Wrap `opt` in a lookahead optimizer\"\n _default='opt'\n def __init__(self, opt, k=6, alpha=0.5): \n store_attr(self, 'opt,k,alpha')\n self._init_state()\n \n def step(self):\n if self.slow_weights is None: self._copy_weights()\n self.opt.step()\n self.count += 1\n if self.count%self.k != 0: return\n for slow_pg,fast_pg in zip(self.slow_weights,self.param_groups):\n for slow_p,fast_p in zip(slow_pg,fast_pg):\n slow_p.data.add_(self.alpha, fast_p.data-slow_p.data)\n fast_p.data.copy_(slow_p.data)\n \n def clear_state(self):\n self.opt.clear_state()\n self._init_state()\n \n def state_dict(self):\n state = self.opt.state_dict()\n state.update({'count': self.count, 'slow_weights': self.slow_weights})\n return state\n \n def load_state_dict(self, sd):\n self.count = sd.pop('count')\n self.slow_weights = sd.pop('slow_weights')\n self.opt.load_state_dict(sd)\n \n def _init_state(self): self.count,self.slow_weights = 0,None\n def _copy_weights(self): self.slow_weights = L(L(p.clone().detach() for p in pg) for pg in self.param_groups)\n \n @property\n def param_groups(self): return self.opt.param_groups\n @param_groups.setter\n def param_groups(self, v): self.opt.param_groups = v",
"_____no_output_____"
],
[
"params = tst_param([1,2,3], [0.1,0.2,0.3])\np,g = params[0].data.clone(),tensor([0.1,0.2,0.3])\nopt = LookAhead(SGD(params, lr=0.1))\nfor k in range(5): opt.step()\n#first 5 steps are normal SGD steps\ntest_close(params[0], p - 0.5*g)\n#Since k=6, sixth step is a moving average of the 6 SGD steps with the intial weight\nopt.step()\ntest_close(params[0], p * 0.5 + (p-0.6*g) * 0.5)",
"_____no_output_____"
]
],
[
[
"## OptimWrapper -",
"_____no_output_____"
]
],
[
[
"#export\ndef detuplify_pg(d):\n res = {}\n for k,v in d.items():\n if k == 'params': continue\n if is_listy(v): res.update(**{f'{k}__{i}': v_ for i,v_ in enumerate(v)})\n else: res[k] = v\n return res",
"_____no_output_____"
],
[
"tst = {'lr': 1e-2, 'mom': 0.9, 'params':[0,1,2]}\ntest_eq(detuplify_pg(tst), {'lr': 1e-2, 'mom': 0.9})\ntst = {'lr': 1e-2, 'betas': (0.9,0.999), 'params':[0,1,2]}\ntest_eq(detuplify_pg(tst), {'lr': 1e-2, 'betas__0': 0.9, 'betas__1': 0.999})",
"_____no_output_____"
],
[
"#export\ndef set_item_pg(pg, k, v):\n if '__' not in k: pg[k] = v\n else:\n name,idx = k.split('__')\n pg[name] = tuple(v if i==int(idx) else pg[name][i] for i in range_of(pg[name]))\n return pg",
"_____no_output_____"
],
[
"tst = {'lr': 1e-2, 'mom': 0.9, 'params':[0,1,2]}\ntest_eq(set_item_pg(tst, 'lr', 1e-3), {'lr': 1e-3, 'mom': 0.9, 'params':[0,1,2]})\ntst = {'lr': 1e-2, 'betas': (0.9,0.999), 'params':[0,1,2]}\ntest_eq(set_item_pg(tst, 'betas__0', 0.95), {'lr': 1e-2, 'betas': (0.95,0.999), 'params':[0,1,2]})",
"_____no_output_____"
],
[
"#export\npytorch_hp_map = {'momentum': 'mom', 'weight_decay': 'wd', 'alpha': 'sqr_mom', 'betas__0': 'mom', 'betas__1': 'sqr_mom'}",
"_____no_output_____"
],
[
"#export\nclass OptimWrapper(_BaseOptimizer, GetAttr):\n _xtra=['zero_grad', 'step', 'state_dict', 'load_state_dict']\n _default='opt'\n def __init__(self, opt, hp_map=None):\n self.opt = opt\n if hp_map is None: hp_map = pytorch_hp_map\n self.fwd_map = {k: hp_map[k] if k in hp_map else k for k in detuplify_pg(opt.param_groups[0]).keys()}\n self.bwd_map = {v:k for k,v in self.fwd_map.items()}\n self.state = defaultdict(dict, {})\n self.frozen_idx = 0\n\n @property\n def param_groups(self): return [pg['params'] for pg in self.opt.param_groups]\n @param_groups.setter\n def param_groups(self, v):\n for pg,v_ in zip(self.opt.param_groups,v): pg['params'] = v_\n\n @property\n def hypers(self):\n return [{self.fwd_map[k]:v for k,v in detuplify_pg(pg).items() if k != 'params'} for pg in self.opt.param_groups]\n\n def _set_hyper(self, k, v):\n for pg,v_ in zip(self.opt.param_groups,v): pg = set_item_pg(pg, self.bwd_map[k], v_)\n\n def clear_state(self): self.opt.state = defaultdict(dict, {})",
"_____no_output_____"
],
[
"sgd = SGD([tensor([1,2,3])], lr=1e-3, mom=0.9, wd=1e-2)\ntst_sgd = OptimWrapper(torch.optim.SGD([tensor([1,2,3])], lr=1e-3, momentum=0.9, weight_decay=1e-2))\n#Access to param_groups\ntest_eq(tst_sgd.param_groups, sgd.param_groups)\n#Set param_groups\ntst_sgd.param_groups = [[tensor([4,5,6])]]\ntest_eq(tst_sgd.opt.param_groups[0]['params'], [tensor(4,5,6)])\n#Access to hypers\ntest_eq(tst_sgd.hypers, [{**sgd.hypers[0], 'dampening': 0., 'nesterov': False}])\n#Set hypers\ntst_sgd.set_hyper('mom', 0.95)\ntest_eq(tst_sgd.opt.param_groups[0]['momentum'], 0.95)",
"_____no_output_____"
],
[
"tst_sgd = OptimWrapper(torch.optim.SGD([{'params': [tensor([1,2,3])], 'lr': 1e-3}, \n {'params': [tensor([4,5,6])], 'lr': 1e-2}], momentum=0.9, weight_decay=1e-2))\nsgd = SGD([[tensor([1,2,3])], [tensor([4,5,6])]], lr=[1e-3, 1e-2], mom=0.9, wd=1e-2)\n#Access to param_groups\ntest_eq(tst_sgd.param_groups, sgd.param_groups)\n#Set param_groups\ntst_sgd.param_groups = [[tensor([4,5,6])], [tensor([1,2,3])]]\ntest_eq(tst_sgd.opt.param_groups[0]['params'], [tensor(4,5,6)])\ntest_eq(tst_sgd.opt.param_groups[1]['params'], [tensor(1,2,3)])\n#Access to hypers\ntest_eq(tst_sgd.hypers, [{**sgd.hypers[i], 'dampening': 0., 'nesterov': False} for i in range(2)])\n#Set hypers\ntst_sgd.set_hyper('mom', 0.95)\ntest_eq([pg['momentum'] for pg in tst_sgd.opt.param_groups], [0.95,0.95])\ntst_sgd.set_hyper('lr', [1e-4,1e-3])\ntest_eq([pg['lr'] for pg in tst_sgd.opt.param_groups], [1e-4,1e-3])",
"_____no_output_____"
],
[
"#hide\n#check it works with tuply hp names like in Adam\ntst_adam = OptimWrapper(torch.optim.Adam([tensor([1,2,3])], lr=1e-2, betas=(0.9, 0.99)))\ntest_eq(tst_adam.hypers, [{'lr': 0.01, 'mom': 0.9, 'sqr_mom': 0.99, 'eps': 1e-08, 'wd': 0, 'amsgrad': False}])\ntst_adam.set_hyper('mom', 0.95)\ntest_eq(tst_adam.opt.param_groups[0]['betas'], (0.95, 0.99))\ntst_adam.set_hyper('sqr_mom', 0.9)\ntest_eq(tst_adam.opt.param_groups[0]['betas'], (0.95, 0.9))",
"_____no_output_____"
],
[
"def _mock_train(m, x, y, opt):\n m.train()\n for i in range(0, 100, 25):\n z = m(x[i:i+25])\n loss = F.mse_loss(z, y[i:i+25])\n loss.backward()\n opt.step()\n opt.zero_grad()",
"_____no_output_____"
],
[
"m = nn.Linear(4,5)\nx = torch.randn(100, 3, 4)\ny = torch.randn(100, 3, 5)\ntry:\n torch.save(m.state_dict(), 'tmp.pth')\n wgt,bias = m.weight.data.clone(),m.bias.data.clone()\n\n m.load_state_dict(torch.load('tmp.pth'))\n opt1 = OptimWrapper(torch.optim.AdamW(m.parameters(), betas=(0.9, 0.99), eps=1e-5, weight_decay=1e-2))\n _mock_train(m, x.clone(), y.clone(), opt1)\n wgt1,bias1 = m.weight.data.clone(),m.bias.data.clone()\n\n m.load_state_dict(torch.load('tmp.pth'))\n opt2 = Adam(m.parameters(), 1e-3, wd=1e-2)\n _mock_train(m, x.clone(), y.clone(), opt2)\n wgt2,bias2 = m.weight.data.clone(),m.bias.data.clone()\n \n test_close(wgt1,wgt2,eps=1e-3)\n test_close(bias1,bias2,eps=1e-3)\nfinally: os.remove('tmp.pth')",
"_____no_output_____"
],
[
"m = nn.Linear(4,5)\nx = torch.randn(100, 3, 4)\ny = torch.randn(100, 3, 5)\ntry:\n torch.save(m.state_dict(), 'tmp.pth')\n wgt,bias = m.weight.data.clone(),m.bias.data.clone()\n\n m.load_state_dict(torch.load('tmp.pth'))\n opt1 = OptimWrapper(torch.optim.Adam(m.parameters(), betas=(0.9, 0.99), eps=1e-5, weight_decay=1e-2))\n _mock_train(m, x.clone(), y.clone(), opt1)\n wgt1,bias1 = m.weight.data.clone(),m.bias.data.clone()\n\n m.load_state_dict(torch.load('tmp.pth'))\n opt2 = Adam(m.parameters(), 1e-3, wd=1e-2, decouple_wd=False)\n _mock_train(m, x.clone(), y.clone(), opt2)\n wgt2,bias2 = m.weight.data.clone(),m.bias.data.clone()\n \n test_close(wgt1,wgt2,eps=1e-3)\n test_close(bias1,bias2,eps=1e-3)\nfinally: os.remove('tmp.pth')",
"_____no_output_____"
]
],
[
[
"## Export -",
"_____no_output_____"
]
],
[
[
"#hide\nfrom local.notebook.export import *\nnotebook2script(all_fs=True)",
"Converted 00_test.ipynb.\nConverted 01_core_foundation.ipynb.\nConverted 01a_core_utils.ipynb.\nConverted 01b_core_dispatch.ipynb.\nConverted 01c_core_transform.ipynb.\nConverted 02_core_script.ipynb.\nConverted 03_torchcore.ipynb.\nConverted 03a_layers.ipynb.\nConverted 04_data_load.ipynb.\nConverted 05_data_core.ipynb.\nConverted 06_data_transforms.ipynb.\nConverted 07_data_block.ipynb.\nConverted 08_vision_core.ipynb.\nConverted 09_vision_augment.ipynb.\nConverted 09a_vision_data.ipynb.\nConverted 10_pets_tutorial.ipynb.\nConverted 11_vision_models_xresnet.ipynb.\nConverted 12_optimizer.ipynb.\nConverted 13_learner.ipynb.\nConverted 13a_metrics.ipynb.\nConverted 14_callback_schedule.ipynb.\nConverted 14a_callback_data.ipynb.\nConverted 15_callback_hook.ipynb.\nConverted 15a_vision_models_unet.ipynb.\nConverted 16_callback_progress.ipynb.\nConverted 17_callback_tracker.ipynb.\nConverted 18_callback_fp16.ipynb.\nConverted 19_callback_mixup.ipynb.\nConverted 20_interpret.ipynb.\nConverted 20a_distributed.ipynb.\nConverted 21_vision_learner.ipynb.\nConverted 22_tutorial_imagenette.ipynb.\nConverted 23_tutorial_transfer_learning.ipynb.\nConverted 30_text_core.ipynb.\nConverted 31_text_data.ipynb.\nConverted 32_text_models_awdlstm.ipynb.\nConverted 33_text_models_core.ipynb.\nConverted 34_callback_rnn.ipynb.\nConverted 35_tutorial_wikitext.ipynb.\nConverted 36_text_models_qrnn.ipynb.\nConverted 37_text_learner.ipynb.\nConverted 38_tutorial_ulmfit.ipynb.\nConverted 40_tabular_core.ipynb.\nConverted 41_tabular_model.ipynb.\nConverted 42_tabular_rapids.ipynb.\nConverted 50_data_block_examples.ipynb.\nConverted 60_medical_imaging.ipynb.\nConverted 65_medical_text.ipynb.\nConverted 70_callback_wandb.ipynb.\nConverted 71_callback_tensorboard.ipynb.\nConverted 90_notebook_core.ipynb.\nConverted 91_notebook_export.ipynb.\nConverted 92_notebook_showdoc.ipynb.\nConverted 93_notebook_export2html.ipynb.\nConverted 94_notebook_test.ipynb.\nConverted 95_index.ipynb.\nConverted 96_data_external.ipynb.\nConverted 97_utils_test.ipynb.\nConverted notebook2jekyll.ipynb.\nConverted xse_resnext.ipynb.\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbae5820231f11c8e91cddf15ae9fbd804272654
| 599,082 |
ipynb
|
Jupyter Notebook
|
examples/hospital.ipynb
|
ruszukorhaz/ThinkBayes2
|
fdddef305a4727d70359123b27a22727a878e9c1
|
[
"MIT"
] | 1 |
2021-07-04T13:30:36.000Z
|
2021-07-04T13:30:36.000Z
|
examples/hospital.ipynb
|
ruszukorhaz/ThinkBayes2
|
fdddef305a4727d70359123b27a22727a878e9c1
|
[
"MIT"
] | null | null | null |
examples/hospital.ipynb
|
ruszukorhaz/ThinkBayes2
|
fdddef305a4727d70359123b27a22727a878e9c1
|
[
"MIT"
] | null | null | null | 209.396015 | 23,968 | 0.904676 |
[
[
[
"# Grid algorithm for the beta-binomial hierarchical model\n\n[Bayesian Inference with PyMC](https://allendowney.github.io/BayesianInferencePyMC)\n\nCopyright 2021 Allen B. Downey\n\nLicense: [Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0)](https://creativecommons.org/licenses/by-nc-sa/4.0/)",
"_____no_output_____"
]
],
[
[
"# If we're running on Colab, install PyMC and ArviZ\nimport sys\nIN_COLAB = 'google.colab' in sys.modules\n\nif IN_COLAB:\n !pip install pymc3\n !pip install arviz",
"_____no_output_____"
],
[
"# PyMC generates a FutureWarning we don't need to deal with yet\n\nimport warnings\nwarnings.filterwarnings(\"ignore\", category=FutureWarning)",
"_____no_output_____"
],
[
"import seaborn as sns\n\ndef plot_hist(sample, **options):\n \"\"\"Plot a histogram of goals.\n \n sample: sequence of values\n \"\"\"\n sns.histplot(sample, stat='probability', discrete=True,\n alpha=0.5, **options)",
"_____no_output_____"
],
[
"def plot_kde(sample, **options):\n \"\"\"Plot a distribution using KDE.\n \n sample: sequence of values\n \"\"\"\n sns.kdeplot(sample, cut=0, **options)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\ndef legend(**options):\n \"\"\"Make a legend only if there are labels.\"\"\"\n handles, labels = plt.gca().get_legend_handles_labels()\n if len(labels):\n plt.legend(**options)",
"_____no_output_____"
],
[
"def decorate(**options):\n plt.gca().set(**options)\n legend()\n plt.tight_layout()",
"_____no_output_____"
],
[
"def decorate_heads(ylabel='Probability'):\n \"\"\"Decorate the axes.\"\"\"\n plt.xlabel('Number of heads (k)')\n plt.ylabel(ylabel)\n plt.title('Distribution of heads')\n legend()",
"_____no_output_____"
],
[
"def decorate_proportion(ylabel='Likelihood'):\n \"\"\"Decorate the axes.\"\"\"\n plt.xlabel('Proportion of heads (x)')\n plt.ylabel(ylabel)\n plt.title('Distribution of proportion')\n legend()",
"_____no_output_____"
],
[
"from empiricaldist import Cdf\n\ndef compare_cdf(pmf, sample):\n pmf.make_cdf().plot(label='grid')\n Cdf.from_seq(sample).plot(label='mcmc')\n print(pmf.mean(), sample.mean())\n decorate()",
"_____no_output_____"
]
],
[
[
"## The Grid Algorithm",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom scipy.stats import gamma\n\nalpha = 4\nbeta = 0.5\n\nqs = np.linspace(0.1, 25, 100)\nps = gamma(alpha, scale=1/beta).pdf(qs)",
"_____no_output_____"
],
[
"from empiricaldist import Pmf\n\nprior_alpha = Pmf(ps, qs)\nprior_alpha.normalize()\n\nprior_alpha.index.name = 'alpha'\nprior_alpha.shape",
"_____no_output_____"
],
[
"prior_alpha.plot()\nprior_alpha.mean()",
"_____no_output_____"
],
[
"qs = np.linspace(0.1, 25, 90)\nps = gamma(alpha, scale=1/beta).pdf(qs)\nprior_beta = Pmf(ps, qs)\nprior_beta.normalize()\n\nprior_beta.index.name = 'beta'\nprior_beta.shape",
"_____no_output_____"
],
[
"prior_beta.plot()\nprior_beta.mean()",
"_____no_output_____"
],
[
"def make_hyper(prior_alpha, prior_beta):\n PA, PB = np.meshgrid(prior_alpha.ps, prior_beta.ps, indexing='ij')\n hyper = PA * PB\n return hyper\n\nhyper = make_hyper(prior_alpha, prior_beta)\nhyper.shape",
"_____no_output_____"
],
[
"import pandas as pd\nfrom utils import plot_contour\n\nplot_contour(pd.DataFrame(hyper))",
"_____no_output_____"
]
],
[
[
"## Make Prior",
"_____no_output_____"
]
],
[
[
"from scipy.stats import beta as betadist\n\nxs = np.linspace(0.01, 0.99, 80)\nprior_x = Pmf(betadist.pdf(xs, 2, 2), xs)\nprior_x.plot()",
"_____no_output_____"
],
[
"from scipy.stats import beta as betadist\n\ndef make_prior(hyper, prior_alpha, prior_beta, xs):\n \n A, B, X = np.meshgrid(prior_alpha.qs, prior_beta.qs, xs, indexing='ij')\n ps = betadist.pdf(X, A, B)\n\n totals = ps.sum(axis=2)\n\n nc = hyper / totals\n\n shape = nc.shape + (1,)\n prior = ps * nc.reshape(shape)\n\n return prior",
"_____no_output_____"
],
[
"xs = np.linspace(0.01, 0.99, 80)\nprior = make_prior(hyper, prior_alpha, prior_beta, xs)\nprior.sum()",
"_____no_output_____"
],
[
"def marginal(joint, axis):\n axes = [i for i in range(3) if i != axis]\n return joint.sum(axis=tuple(axes))",
"_____no_output_____"
],
[
"prior_a = Pmf(marginal(prior, 0), prior_alpha.qs)\nprior_alpha.plot()\nprior_a.plot()\nprior_a.mean()",
"_____no_output_____"
],
[
"prior_b = Pmf(marginal(prior, 1), prior_beta.qs)\nprior_beta.plot()\nprior_b.plot()",
"_____no_output_____"
],
[
"prior_x = Pmf(marginal(prior, 2), xs)\nprior_x.plot()",
"_____no_output_____"
]
],
[
[
"## The Update",
"_____no_output_____"
]
],
[
[
"from scipy.stats import binom\n\nn = 250\nks = 140\nX, K = np.meshgrid(xs, ks)\nlike_x = binom.pmf(K, n, X).prod(axis=0)\nlike_x.shape",
"_____no_output_____"
],
[
"plt.plot(xs, like_x)",
"_____no_output_____"
],
[
"def update(prior, data):\n n, ks = data\n X, K = np.meshgrid(xs, ks)\n like_x = binom.pmf(K, n, X).prod(axis=0)\n\n posterior = prior * like_x\n posterior /= posterior.sum()\n return posterior",
"_____no_output_____"
],
[
"data = 250, 140\nposterior = update(prior, data)",
"_____no_output_____"
],
[
"marginal_x = Pmf(marginal(posterior, 2), xs)\nmarginal_x.plot()\nmarginal_x.mean()",
"_____no_output_____"
],
[
"marginal_alpha = Pmf(marginal(posterior, 0), prior_alpha.qs)\nmarginal_alpha.plot()\nmarginal_alpha.mean()",
"_____no_output_____"
],
[
"marginal_beta = Pmf(marginal(posterior, 1), prior_beta.qs)\nmarginal_beta.plot()\nmarginal_beta.mean()",
"_____no_output_____"
]
],
[
[
"## One coin with PyMC",
"_____no_output_____"
]
],
[
[
"import pymc3 as pm\n\nn = 250\nwith pm.Model() as model1:\n alpha = pm.Gamma('alpha', alpha=4, beta=0.5)\n beta = pm.Gamma('beta', alpha=4, beta=0.5)\n x1 = pm.Beta('x1', alpha, beta)\n k1 = pm.Binomial('k1', n=n, p=x1, observed=140)\n pred = pm.sample_prior_predictive(1000)",
"_____no_output_____"
]
],
[
[
"Here's the graphical representation of the model.",
"_____no_output_____"
]
],
[
[
"pm.model_to_graphviz(model1)",
"_____no_output_____"
],
[
"from utils import kde_from_sample\n\nkde_from_sample(pred['alpha'], prior_alpha.qs).plot()\nprior_alpha.plot()",
"_____no_output_____"
],
[
"kde_from_sample(pred['beta'], prior_beta.qs).plot()\nprior_beta.plot()",
"_____no_output_____"
],
[
"kde_from_sample(pred['x1'], prior_x.qs).plot()\nprior_x.plot()",
"_____no_output_____"
]
],
[
[
"Now let's run the sampler.",
"_____no_output_____"
]
],
[
[
"with model1:\n trace1 = pm.sample(500)",
"Auto-assigning NUTS sampler...\nInitializing NUTS using jitter+adapt_diag...\nMultiprocess sampling (4 chains in 4 jobs)\nNUTS: [x1, beta, alpha]\n"
]
],
[
[
"Here are the posterior distributions for the two coins.",
"_____no_output_____"
]
],
[
[
"compare_cdf(marginal_alpha, trace1['alpha'])",
"8.805918336878161 8.877890425680253\n"
],
[
"compare_cdf(marginal_beta, trace1['beta'])",
"7.540121805297106 7.693727396265147\n"
],
[
"compare_cdf(marginal_x, trace1['x1'])",
"0.5586974454638711 0.5601591970725471\n"
]
],
[
[
"## Two coins",
"_____no_output_____"
]
],
[
[
"def get_hyper(joint):\n return joint.sum(axis=2)",
"_____no_output_____"
],
[
"posterior_hyper = get_hyper(posterior)\nposterior_hyper.shape",
"_____no_output_____"
],
[
"prior2 = make_prior(posterior_hyper, prior_alpha, prior_beta, xs)",
"_____no_output_____"
],
[
"data = 250, 110\nposterior2 = update(prior2, data)",
"_____no_output_____"
],
[
"marginal_alpha2 = Pmf(marginal(posterior2, 0), prior_alpha.qs)\nmarginal_alpha2.plot()\nmarginal_alpha2.mean()",
"_____no_output_____"
],
[
"marginal_beta2 = Pmf(marginal(posterior2, 1), prior_beta.qs)\nmarginal_beta2.plot()\nmarginal_beta2.mean()",
"_____no_output_____"
],
[
"marginal_x2 = Pmf(marginal(posterior2, 2), xs)\nmarginal_x2.plot()\nmarginal_x2.mean()",
"_____no_output_____"
]
],
[
[
"## Two coins with PyMC",
"_____no_output_____"
]
],
[
[
"with pm.Model() as model2:\n alpha = pm.Gamma('alpha', alpha=4, beta=0.5)\n beta = pm.Gamma('beta', alpha=4, beta=0.5)\n x1 = pm.Beta('x1', alpha, beta)\n x2 = pm.Beta('x2', alpha, beta)\n k1 = pm.Binomial('k1', n=n, p=x1, observed=140)\n k2 = pm.Binomial('k2', n=n, p=x2, observed=110)",
"_____no_output_____"
]
],
[
[
"Here's the graph for this model.",
"_____no_output_____"
]
],
[
[
"pm.model_to_graphviz(model2)",
"_____no_output_____"
]
],
[
[
"\n\nLet's run the sampler.",
"_____no_output_____"
]
],
[
[
"with model2:\n trace2 = pm.sample(500)",
"Auto-assigning NUTS sampler...\nInitializing NUTS using jitter+adapt_diag...\nMultiprocess sampling (4 chains in 4 jobs)\nNUTS: [x2, x1, beta, alpha]\n"
]
],
[
[
"And here are the results.",
"_____no_output_____"
]
],
[
[
"kde_from_sample(trace2['alpha'], marginal_alpha.qs).plot()\nmarginal_alpha2.plot()\ntrace2['alpha'].mean(), marginal_alpha2.mean()",
"_____no_output_____"
],
[
"kde_from_sample(trace2['beta'], marginal_beta.qs).plot()\nmarginal_beta2.plot()\ntrace2['beta'].mean(), marginal_beta2.mean()",
"_____no_output_____"
],
[
"kde_from_sample(trace2['x2'], marginal_x.qs).plot()\nmarginal_x2.plot()",
"_____no_output_____"
]
],
[
[
"## Heart Attack Data\n\nThis example is based on [Chapter 10 of *Probability and Bayesian Modeling*](https://bayesball.github.io/BOOK/bayesian-hierarchical-modeling.html#example-deaths-after-heart-attack); it uses data on death rates due to heart attack for patients treated at various hospitals in New York City.\n\nWe can use Pandas to read the data into a `DataFrame`.",
"_____no_output_____"
]
],
[
[
"import os\n\nfilename = 'DeathHeartAttackManhattan.csv'\nif not os.path.exists(filename):\n !wget https://github.com/AllenDowney/BayesianInferencePyMC/raw/main/DeathHeartAttackManhattan.csv",
"_____no_output_____"
],
[
"import pandas as pd\n\ndf = pd.read_csv(filename)\ndf",
"_____no_output_____"
]
],
[
[
"The columns we need are `Cases`, which is the number of patients treated at each hospital, and `Deaths`, which is the number of those patients who died.",
"_____no_output_____"
]
],
[
[
"# shuffled = df.sample(frac=1)",
"_____no_output_____"
],
[
"data_ns = df['Cases'].values\ndata_ks = df['Deaths'].values",
"_____no_output_____"
]
],
[
[
"Here's a hierarchical model that estimates the death rate for each hospital, and simultaneously estimates the distribution of rates across hospitals.",
"_____no_output_____"
],
[
"## Hospital Data with grid",
"_____no_output_____"
]
],
[
[
"alpha = 4\nbeta = 0.5\n\nqs = np.linspace(0.1, 25, 100)\nps = gamma(alpha, scale=1/beta).pdf(qs)\n\nprior_alpha = Pmf(ps, qs)\nprior_alpha.normalize()\n\nprior_alpha.index.name = 'alpha'",
"_____no_output_____"
],
[
"qs = np.linspace(0.1, 50, 90)\nps = gamma(alpha, scale=1/beta).pdf(qs)\nprior_beta = Pmf(ps, qs)\nprior_beta.normalize()\n\nprior_beta.index.name = 'beta'\nprior_beta.shape",
"_____no_output_____"
],
[
"prior_alpha.plot()\nprior_beta.plot()\nprior_alpha.mean()",
"_____no_output_____"
],
[
"hyper = make_hyper(prior_alpha, prior_beta)\nhyper.shape",
"_____no_output_____"
],
[
"xs = np.linspace(0.01, 0.99, 80)\nprior = make_prior(hyper, prior_alpha, prior_beta, xs)\nprior.shape",
"_____no_output_____"
],
[
"for data in zip(data_ns, data_ks):\n print(data)\n posterior = update(prior, data)\n hyper = get_hyper(posterior)\n prior = make_prior(hyper, prior_alpha, prior_beta, xs)",
"(129, 4)\n(35, 1)\n(228, 18)\n(84, 7)\n(291, 24)\n(270, 16)\n(46, 6)\n(293, 19)\n(241, 15)\n(105, 13)\n(353, 25)\n(250, 11)\n(41, 4)\n"
],
[
"marginal_alpha = Pmf(marginal(posterior, 0), prior_alpha.qs)\nmarginal_alpha.plot()\nmarginal_alpha.mean()",
"_____no_output_____"
],
[
"marginal_beta = Pmf(marginal(posterior, 1), prior_beta.qs)\nmarginal_beta.plot()\nmarginal_beta.mean()",
"_____no_output_____"
],
[
"marginal_x = Pmf(marginal(posterior, 2), prior_x.qs)\nmarginal_x.plot()\nmarginal_x.mean()",
"_____no_output_____"
]
],
[
[
"## Hospital Data with PyMC",
"_____no_output_____"
]
],
[
[
"with pm.Model() as model4:\n alpha = pm.Gamma('alpha', alpha=4, beta=0.5)\n beta = pm.Gamma('beta', alpha=4, beta=0.5)\n xs = pm.Beta('xs', alpha, beta, shape=len(data_ns))\n ks = pm.Binomial('ks', n=data_ns, p=xs, observed=data_ks)\n trace4 = pm.sample(500)",
"Auto-assigning NUTS sampler...\nInitializing NUTS using jitter+adapt_diag...\nMultiprocess sampling (4 chains in 4 jobs)\nNUTS: [xs, beta, alpha]\n"
]
],
[
[
"Here's the graph representation of the model, showing that the observable is an array of 13 values.",
"_____no_output_____"
]
],
[
[
"pm.model_to_graphviz(model4)",
"_____no_output_____"
]
],
[
[
"Here's the trace.",
"_____no_output_____"
]
],
[
[
"kde_from_sample(trace4['alpha'], marginal_alpha.qs).plot()\nmarginal_alpha.plot()\ntrace4['alpha'].mean(), marginal_alpha.mean()",
"_____no_output_____"
],
[
"kde_from_sample(trace4['beta'], marginal_beta.qs).plot()\nmarginal_beta.plot()\ntrace4['beta'].mean(), marginal_beta.mean()",
"_____no_output_____"
],
[
"trace_xs = trace4['xs'].transpose()\ntrace_xs.shape",
"_____no_output_____"
],
[
"kde_from_sample(trace_xs[-1], marginal_x.qs).plot()\nmarginal_x.plot()\ntrace_xs[-1].mean(), marginal_x.mean()",
"_____no_output_____"
],
[
"xs = np.linspace(0.01, 0.99, 80)\nhyper = get_hyper(posterior)\npost_all = make_prior(hyper, prior_alpha, prior_beta, xs)",
"_____no_output_____"
],
[
"def forget(posterior, data):\n n, ks = data\n X, K = np.meshgrid(xs, ks)\n like_x = binom.pmf(K, n, X).prod(axis=0)\n\n prior = posterior / like_x\n prior /= prior.sum()\n return prior",
"_____no_output_____"
],
[
"def get_marginal_x(post_all, data):\n prior = forget(post_all, data)\n hyper = get_hyper(prior)\n prior = make_prior(hyper, prior_alpha, prior_beta, xs)\n posterior = update(prior, data)\n marginal_x = Pmf(marginal(posterior, 2), prior_x.qs)\n return marginal_x",
"_____no_output_____"
],
[
"data = 270, 16\nmarginal_x = get_marginal_x(post_all, data)",
"/tmp/ipykernel_256314/2625109009.py:6: RuntimeWarning: divide by zero encountered in true_divide\n prior = posterior / like_x\n/tmp/ipykernel_256314/2625109009.py:7: RuntimeWarning: invalid value encountered in true_divide\n prior /= prior.sum()\n"
],
[
"kde_from_sample(trace_xs[0], marginal_x.qs).plot()\nmarginal_x.plot()\ntrace_xs[0].mean(), marginal_x.mean()",
"_____no_output_____"
]
],
[
[
"## One at a time",
"_____no_output_____"
]
],
[
[
"prior.shape, prior.sum()",
"_____no_output_____"
],
[
"likelihood = np.empty((len(df), len(xs)))\n\nfor i, row in df.iterrows():\n n = row['Cases']\n k = row['Deaths']\n likelihood[i] = binom.pmf(k, n, xs)",
"_____no_output_____"
],
[
"prod = likelihood.prod(axis=0)\nprod.shape",
"_____no_output_____"
],
[
"i = 3\nall_but_one = prod / likelihood[i]",
"_____no_output_____"
],
[
"prior",
"_____no_output_____"
],
[
"hyper_i = get_hyper(prior * all_but_one)\nhyper_i.sum()",
"_____no_output_____"
],
[
"prior_i = make_prior(hyper_i, prior_alpha, prior_beta, xs)",
"_____no_output_____"
],
[
"data = df.loc[i, 'Cases'], df.loc[i, 'Deaths']\ndata",
"_____no_output_____"
],
[
"posterior_i = update(prior_i, data)",
"_____no_output_____"
],
[
"marginal_alpha = Pmf(marginal(posterior_i, 0), prior_alpha.qs)\nmarginal_beta = Pmf(marginal(posterior_i, 1), prior_beta.qs)\nmarginal_x = Pmf(marginal(posterior_i, 2), prior_x.qs)",
"_____no_output_____"
],
[
"compare_cdf(marginal_alpha, trace4['alpha'])",
"2.2118989111174576 2.152194951973725\n"
],
[
"compare_cdf(marginal_beta, trace4['beta'])",
"21.347689380761715 20.44533534199944\n"
],
[
"compare_cdf(marginal_x, trace_xs[i])",
"0.08568040914334583 0.08557547064806013\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbae586911303eb163e68b535f70090772bf85ca
| 256,057 |
ipynb
|
Jupyter Notebook
|
Damian-Project2/Data_Visualizations.ipynb
|
9161AD/cosc3570-introdatascience-fa18
|
7cc87ed71946ce34d61f420453fb932a5221c140
|
[
"MIT"
] | 13 |
2018-08-30T04:57:09.000Z
|
2019-12-02T08:48:26.000Z
|
Damian-Project2/Data_Visualizations.ipynb
|
9161AD/cosc3570-introdatascience-fa18
|
7cc87ed71946ce34d61f420453fb932a5221c140
|
[
"MIT"
] | 3 |
2018-10-18T22:54:08.000Z
|
2018-12-17T20:00:59.000Z
|
Damian-Project2/Data_Visualizations.ipynb
|
9161AD/cosc3570-introdatascience-fa18
|
7cc87ed71946ce34d61f420453fb932a5221c140
|
[
"MIT"
] | 29 |
2018-10-17T15:58:20.000Z
|
2020-06-16T04:28:24.000Z
| 86.564233 | 61,068 | 0.696423 |
[
[
[
"from datascience import *\nimport numpy as np\n%matplotlib inline\n\nimport pandas as pd\nimport matplotlib.pyplot as plt\nplt.style.use('seaborn')\n\nfrom scipy import stats\nfrom scipy.stats import norm",
"_____no_output_____"
],
[
"import matplotlib\nmatplotlib.__version__",
"_____no_output_____"
],
[
"\nimport seaborn as sns\nsns.set(color_codes = True)\n",
"_____no_output_____"
],
[
"#Data or Fe-based, Cuprates, Hydrides\n#There were no high T hydrides in the original data set \nfeatures8 = pd.read_csv(\"https://raw.githubusercontent.com/9161AD/superconduct-/master/features_H_Cu_Fe2.csv\")\nfeatures8",
"_____no_output_____"
],
[
"len(features8)",
"_____no_output_____"
],
[
"# We Remove the one outlier that contains Hg but no Cu to isolate the Hydrides\n#Already determined All Fe based SCs contain Cu \nfeatures_Hydrides1 = features8[~features8.material_name.str.contains(\"Cu\")]\nfeatures_Hydrides2 = features_Hydrides1[~features_Hydrides1.material_name.str.contains(\"Hg\")]\nfeatures_Hydrides3 = features_Hydrides2[~features_Hydrides2.material_name.str.contains(\"Hf\")]\nfeatures_Hydrides4 = features_Hydrides3[~features_Hydrides3.material_name.str.contains(\"Hs\")]\nfeatures_Hydrides5 = features_Hydrides4[~features_Hydrides4.material_name.str.contains(\"Ho\")]\nfeatures_Hydrides6 = features_Hydrides5[~features_Hydrides5.material_name.str.contains(\"Fe\")]\n\nfeatures_Hydrides6.head()\n",
"_____no_output_____"
],
[
"#Hydrides Groups\nHydrides = features_Hydrides6.assign(Group='Hydride')[['Group'] + features_Hydrides6.columns.tolist()]\nHydrides = Hydrides.drop(Hydrides.columns[1], axis=1)\nHydrides.head()\n",
"_____no_output_____"
],
[
"len(Hydrides)\n",
"_____no_output_____"
],
[
"(len(Hydrides)/len(features8)) * 100 \n#9% Hydrides",
"_____no_output_____"
],
[
"#Cuprate Groups --> Isolating Fe then picking out Cu\nfeatures_Cuprates1 = features8[~features8.material_name.str.contains(\"Fe\")]\nfeatures_Cuprates2 = features_Cuprates1[features_Cuprates1.material_name.str.contains(\"Cu\")]\n\n",
"_____no_output_____"
],
[
"#Cuprates Groups\nCuprates = features_Cuprates2.assign(Group='Cuprate')[['Group'] + features_Cuprates2.columns.tolist()]\nCuprates = Cuprates.drop(Cuprates.columns[1], axis=1)\nCuprates.head()",
"_____no_output_____"
],
[
"len(Cuprates)\n",
"_____no_output_____"
],
[
"len(Cuprates)\n(len(Cuprates)/len(features8)) * 100 \n#60 % Cuprates",
"_____no_output_____"
],
[
"features_Fe = features8[features8.material_name.str.contains(\"Fe\")]\n",
"_____no_output_____"
],
[
"#Iron Groups\nIron_Based = features_Fe.assign(Group='Iron-Based')[['Group'] + features_Fe.columns.tolist()]\nIron_Based = Iron_Based.drop(Iron_Based.columns[1], axis=1)\nIron_Based.head()\n",
"_____no_output_____"
],
[
"len(Iron_Based)",
"_____no_output_____"
],
[
"(len(Iron_Based)/len(features8)) * 100 \n# 7% Iron Based ",
"_____no_output_____"
],
[
"#Isolated 3 desired Classes",
"_____no_output_____"
],
[
"Classes = Hydrides.append(Cuprates).append(Iron_Based)",
"_____no_output_____"
],
[
"len(Classes)",
"_____no_output_____"
],
[
"(len(Classes)) / 21263 * 100 \n#Now down to 5.66 % of dataset",
"_____no_output_____"
],
[
"Box1 = sns.violinplot(x='Group', y='critical_temp', data=Classes)\nplt\nplt.title(\"Classes Critical Temperature Distributions\", loc = \"left\")\nplt.xlabel(\"Class\")\nplt.ylabel(\"Critical Temperature (K)\")",
"_____no_output_____"
],
[
"#Superposition of Jitter with Boxplot\nBox2 =sns.boxplot(x='Group', y='critical_temp', data = Classes)\nBox2 = sns.stripplot(x='Group', y = 'critical_temp', data= Classes, color = \"orange\", jitter = 0.2, size = 2.5) \nplt.title(\"Classes Critical Temperature Distributions\", loc = \"left\")\nplt.xlabel(\"Class\")\nplt.ylabel(\"Critical Temperature (K)\")\n",
"_____no_output_____"
],
[
"g = sns.pairplot(Classes, vars=[\"critical_temp\", \"number_of_elements\"], hue = \"Group\")",
"_____no_output_____"
],
[
"import seaborn as sns; sns.set(style=\"ticks\", color_codes=True, hue = \"Group\")\ng = sns.pairplot(Classes)\ng",
"_____no_output_____"
],
[
"#Normalized for all classes\n#features8.hist('critical_temp', bins = 16, range = (10,160), color = 'r', density=1)\n#plots.title('Critical Temperature for Iron-Based,Cuprates,Hydrides-- High T Superconductors')\n#plots.xlabel(\"Temperature (K)\")\n#plots.ylabel(\"Count\")",
"_____no_output_____"
],
[
"import statsmodels.formula.api as smf\n#Begins groundwork for setting a linear regression \nmodel = 'critical_temp ~ %s'%(\" + \".join(Classes.columns.values[2:]))\n",
"_____no_output_____"
],
[
"#Multiple Regression Analysis on 3 combined classes \nlinear_regression = smf.ols(model, data = Classes).fit()\nlinear_regression.summary()",
"_____no_output_____"
],
[
"import statsmodels.formula.api as smf\n#Begins groundwork for setting a linear regression \nmodel = 'critical_temp ~ %s'%(\" + \".join(Hydrides.columns.values[2:]))\n",
"_____no_output_____"
],
[
"#Train Test on Combined Classes\n#X contains predictors \nX1 = Classes.drop(['Group','material_name','critical_temp'],1)\nX1.head()",
"_____no_output_____"
],
[
"#Make Y a true column vector containing the mpg for each superconductor\nY1 = Classes[['critical_temp']]\n\n#Removed Material_names because they are not statistical predictors\n#, rather just labels \n\nZ1 = Classes[['Group', 'material_name']]\nfrom sklearn.model_selection import train_test_split\n# Split X and y into X_\n\n#test size = 0.66 to match previous literature\nX1_train, X1_test, Y1_train, Y1_test = train_test_split(X1, Y1, test_size=0.66, random_state=1)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.model_selection import train_test_split\nlineReg = LinearRegression()\nlineReg.fit(X1_train, Y1_train)\nlineReg.score(X1_test, Y1_test)\n\n#Recent Literature had 74% for full data set. I matched this as well\n#priro to splitting up by class\n#See how reducing to single classes affects correlation",
"_____no_output_____"
],
[
"#Train Test on HYDRIDES\n",
"_____no_output_____"
],
[
"#X2 contains predictors \nX2 = Hydrides.drop(['Group','material_name','critical_temp'],1)\nlen(X2)",
"_____no_output_____"
],
[
"#Make Y2 a true column vector containing the mpg for each superconductor\nY2 = Hydrides[['critical_temp']]\n\n#Removed Material_names because they are not statistical predictors\n#, rather just labels \n\nZ2 = Hydrides[['Group', 'material_name']]\nfrom sklearn.model_selection import train_test_split\n# Split X and y into X_\n\n#test size = 0.66 to match previous literature\nX2_train, X2_test, Y2_train, Y2_test = train_test_split(X2, Y2, test_size=0.66, random_state=1)\n",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.model_selection import train_test_split\nlineReg = LinearRegression()\nlineReg.fit(X2_train, Y2_train)\nlineReg.score(X2_test, Y2_test)\n",
"_____no_output_____"
],
[
"#Test Cuprates Variable-3",
"_____no_output_____"
],
[
"#X2 contains predictors \nX3 = Cuprates.drop(['Group','material_name','critical_temp'],1)\nY3 = Cuprates[['critical_temp']]\nZ3 = Cuprates[['Group', 'material_name']]\nfrom sklearn.model_selection import train_test_split\nX3_train, X3_test, Y3_train, Y3_test = train_test_split(X3, Y3, test_size=0.66, random_state=1)\n",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.model_selection import train_test_split\nlineReg = LinearRegression()\nlineReg.fit(X3_train, Y3_train)\nabs(lineReg.score(X3_test, Y3_test))\n",
"_____no_output_____"
],
[
"#Test Fe-Based Variable - 4",
"_____no_output_____"
],
[
"#X4 contains predictors \nX4 = Iron_Based.drop(['Group','material_name','critical_temp'],1)\nY4 = Iron_Based[['critical_temp']]\nZ4 = Iron_Based[['Group', 'material_name']]\nfrom sklearn.model_selection import train_test_split\nX4_train, X4_test, Y4_train, Y4_test = train_test_split(X4, Y4, test_size=0.66, random_state=1)\n",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.model_selection import train_test_split\nlineReg = LinearRegression()\nlineReg.fit(X4_train, Y4_train)\nabs(lineReg.score(X4_test, Y4_test))\n",
"_____no_output_____"
],
[
"Groups = ['Hydrides', 'Cuprates', 'Iron-Based']\nNumber_Entries =[len(Hydrides),len(Cuprates),len(Iron_Based)]\nMR_Scores = [-0.78, 0.31, 0.27]\nSummary = pd.DataFrame({'Class': Groups,\n 'Number of Materials': Number_Entries,\n 'Coeffieicent of Multiple Determination': MR_Scores,\n })\nSummary",
"_____no_output_____"
],
[
"sns.lmplot(x='Number of Materials', y='MR_Scores', data=Summary)\nplt.ylim(-1,1)\nplt.xlim(0,1000)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbae5fb7f90db44d3c830ea22abf02cebea09743
| 5,434 |
ipynb
|
Jupyter Notebook
|
generate_reference.ipynb
|
andrewfullard/carsus-refdata
|
f11840c2615481eabfd4f5b9d05c377d77bf4647
|
[
"BSD-3-Clause"
] | null | null | null |
generate_reference.ipynb
|
andrewfullard/carsus-refdata
|
f11840c2615481eabfd4f5b9d05c377d77bf4647
|
[
"BSD-3-Clause"
] | 3 |
2021-05-03T16:00:37.000Z
|
2022-03-09T07:30:15.000Z
|
generate_reference.ipynb
|
andrewfullard/carsus-refdata
|
f11840c2615481eabfd4f5b9d05c377d77bf4647
|
[
"BSD-3-Clause"
] | 3 |
2019-07-02T19:56:45.000Z
|
2022-02-17T16:28:26.000Z
| 31.964706 | 109 | 0.621273 |
[
[
[
"# Generate reference data\n\nMake reference data for `carsus/io/output/base.py` module.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom pandas.testing import assert_frame_equal, assert_series_equal",
"_____no_output_____"
],
[
"from carsus.io.nist import NISTWeightsComp, NISTIonizationEnergies\nfrom carsus.io.kurucz import GFALLReader\nfrom carsus.io.zeta import KnoxLongZeta\nfrom carsus.io.chianti_ import ChiantiReader\nfrom carsus.io.cmfgen import CMFGENEnergyLevelsParser, CMFGENOscillatorStrengthsParser, CMFGENReader\nfrom carsus.io.output import TARDISAtomData",
"_____no_output_____"
],
[
"GFALL_IONS = \"H-Si\"\nCHIANTI_IONS = \"H-He\"\nCMFGEN_IONS = \"Si_I-II\"",
"_____no_output_____"
],
[
"fname = f\"test_data_ku_{GFALL_IONS}_ch_{CHIANTI_IONS}_cm_{CMFGEN_IONS}.h5\"\nrefdata = pd.HDFStore(fname)",
"_____no_output_____"
],
[
"atomic_weights = NISTWeightsComp()\nionization_energies = NISTIonizationEnergies(GFALL_IONS)\ngfall_reader = GFALLReader(ions=GFALL_IONS)\nchianti_reader = ChiantiReader(ions=CHIANTI_IONS, collisions=True, priority=20)\nzeta_data = KnoxLongZeta()",
"_____no_output_____"
],
[
"si_0_lvl = CMFGENEnergyLevelsParser('./cmfgen/energy_levels/SiI_OSC')\nsi_0_osc = CMFGENOscillatorStrengthsParser('./cmfgen/energy_levels/SiI_OSC')\nsi_1_lvl = CMFGENEnergyLevelsParser('./cmfgen/energy_levels/si2_osc_kurucz')\nsi_1_osc = CMFGENOscillatorStrengthsParser('./cmfgen/energy_levels/si2_osc_kurucz')\n\ncmfgen_data = {'Si 0': {'levels': si_0_lvl, 'lines': si_0_osc},\n 'Si 1': {'levels': si_1_lvl, 'lines': si_1_osc},}\n\ncmfgen_reader = CMFGENReader(cmfgen_data, priority=20)",
"_____no_output_____"
],
[
"atom_data = TARDISAtomData(atomic_weights,\n ionization_energies,\n gfall_reader,\n zeta_data,\n chianti_reader,\n cmfgen_reader)",
"_____no_output_____"
],
[
"atomic_weights = atom_data.atomic_weights.base.loc[1:14] # H-Si to do: make more consistent\nionization_energies = atom_data.ionization_energies.base # to do: make more consistent\nlevels_all = atom_data._get_all_levels_data().drop(columns=[\"ds_id\"])\nlevels = atom_data.levels.drop(columns=[\"ds_id\"])\nlevels_prepared = atom_data.levels_prepared\nlines_all = atom_data._get_all_lines_data().drop(columns=[\"ds_id\"])\nlines = atom_data.lines.drop(columns=[\"ds_id\"])\nlines_prepared = atom_data.lines_prepared\nmacro_atom = atom_data.macro_atom\nmacro_atom_prepared = atom_data.macro_atom_prepared\nmacro_atom_references = atom_data.macro_atom_references\nmacro_atom_references_prepared = atom_data.macro_atom_references_prepared\ncollisions = atom_data.collisions.drop(columns=[\"btemp\", \"bscups\"])\ncollisions_prepared = atom_data.collisions_prepared\nzeta_data = atom_data.zeta_data.base # to do: make more consistent",
"_____no_output_____"
],
[
"refdata.put('atomic_weights', atomic_weights)\nrefdata.put('ionization_energies', ionization_energies)\nrefdata.put('levels_all', levels_all)\nrefdata.put('levels', levels)\nrefdata.put('levels_prepared', levels_prepared)\nrefdata.put('lines_all', lines_all)\nrefdata.put('lines', lines)\nrefdata.put('lines_prepared', lines_prepared)\nrefdata.put('macro_atom', macro_atom)\nrefdata.put('macro_atom_prepared', macro_atom_prepared)\nrefdata.put('macro_atom_references', macro_atom_references)\nrefdata.put('macro_atom_references_prepared', macro_atom_references_prepared)\nrefdata.put('collisions', collisions)\nrefdata.put('collisions_prepared', collisions_prepared)\nrefdata.put('zeta_data', zeta_data)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbae77976288c35877c3f0ad5a7ba73c2e559062
| 55,478 |
ipynb
|
Jupyter Notebook
|
04 scrape for missing addresses.ipynb
|
rajachak/DSI-Project5
|
454fe50c42d3aadda3511b3c64daf722d6c43469
|
[
"MIT"
] | null | null | null |
04 scrape for missing addresses.ipynb
|
rajachak/DSI-Project5
|
454fe50c42d3aadda3511b3c64daf722d6c43469
|
[
"MIT"
] | null | null | null |
04 scrape for missing addresses.ipynb
|
rajachak/DSI-Project5
|
454fe50c42d3aadda3511b3c64daf722d6c43469
|
[
"MIT"
] | 2 |
2018-03-21T21:03:08.000Z
|
2018-04-11T15:39:42.000Z
| 39.854885 | 162 | 0.347291 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport requests\nfrom bs4 import BeautifulSoup\nfrom selenium import webdriver\nimport sys\nimport re\nimport webbrowser\nfrom time import sleep",
"_____no_output_____"
],
[
"# https://stackoverflow.com/questions/4028904/how-to-get-the-home-directory-in-python\nfrom os.path import expanduser\nfrom os import listdir\nexecutable_path = expanduser(\"~\") + \"/chromedriver\"\n\nif 'chromedriver.exe' not in listdir(expanduser(\"~\")):\n print('chomedriver.exe not found in the home directory! Refer to Selenium docs.')\n sys.exit()\n\ndriver = webdriver.Chrome(executable_path=executable_path)",
"chomedriver.exe not found in the home directory! Refer to Selenium docs.\n"
],
[
"from selenium import webdriver\nfrom bs4 import BeautifulSoup\n\nimport pandas as pd\n\n\n# https://stackoverflow.com/questions/4028904/how-to-get-the-home-directory-in-python\nfrom os.path import expanduser\nfrom os import listdir\nexecutable_path = expanduser(\"~\") + \"/chromedriver\"\n\nif 'chromedriver.exe' not in listdir(expanduser(\"~\")):\n print('chomedriver.exe not found in the home directory! Refer to Selenium docs.')\n sys.exit()\n\ndriver = webdriver.Chrome(executable_path=executable_path)",
"chomedriver.exe not found in the home directory! Refer to Selenium docs.\n"
],
[
"#opportunities = pd.read_csv('opportunities.csv')\n#use this if restarting scraper\nopportunities = pd.read_csv('opportunities_scraped.csv')",
"_____no_output_____"
],
[
"opportunities.head()",
"_____no_output_____"
],
[
"#opportunities['scraped_address']=np.nan",
"_____no_output_____"
],
[
"opportunities[opportunities.scraped_address!='not found'].head()",
"_____no_output_____"
],
[
"opportunities.head(33)",
"_____no_output_____"
],
[
"driver = webdriver.Chrome(executable_path=\"/Users/rajchakrabarty/Downloads/chromedriver\")",
"_____no_output_____"
],
[
"#export to csv\nopportunities.to_csv('opportunities_scraped.csv',index_label=False)",
"_____no_output_____"
],
[
"\ndef scrape():\n driver = webdriver.Chrome(executable_path=\"/Users/rajchakrabarty/Downloads/chromedriver\")\n counter = 0\n for i, row in opportunities[(opportunities['scraped_address'].isnull()==True)].iterrows():\n search_term = ''\n url = ''\n address = ''\n search_term = row['Opportunity Name'].replace(' ','+')+'+address'\n url = 'https://www.google.com/search?\\&q='+search_term\n driver.get(url)\n search_bar = driver.find_element_by_id('lst-ib')\n search_bar.clear()\n search_bar.send_keys(search_term)\n response = driver.page_source\n html = BeautifulSoup(response, 'lxml')\n span = html.find('span',{'class':'LrzXr'})\n #print (i, search_term)\n try:\n address = span.text\n except:\n address = 'not found'\n #print(address)\n opportunities.loc[opportunities['Opportunity ID'] == row['Opportunity ID']\n ,'scraped_address'] = address\n counter += 1\n if counter%20 == 0:\n print(i, search_term)\n print(address)\n #sleep(10)\n\nprint('start')\nkeep_going = True\nwhile keep_going:\n try:\n scrape()\n keep_going = False\n except:\n opportunities.to_csv('opportunities_scraped.csv',index_label=False)\n \nopportunities.to_csv('opportunities_scraped.csv',index_label=False)\n",
"start\n2344 Palantir+-+Multi+Office+Expansion+address\n1025 Thomas Jefferson St NW #600, Washington, DC 20007\n2364 Clear+Channel+UK+address\nnot found\n2402 Mercy+Housing-MSA+address\nnot found\n2422 Macerich+Tysons+Corner+address\n1961 Chain Bridge Rd, Tysons, VA 22102\n2460 Concert+Properties-+2+buildings+-+TORONTO+address\nnot found\n2480 Persimmon+Capital-+Huntington+Gardens+address\nnot found\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbae77de05055220b34d2cd6df65847e2cd82e73
| 27,863 |
ipynb
|
Jupyter Notebook
|
docs/source/tutorials/local_neuroglancer.ipynb
|
dokato/pyroglancer
|
b6263c08c4da836d652a1fa7d7d50eb7b49be10b
|
[
"BSD-3-Clause"
] | null | null | null |
docs/source/tutorials/local_neuroglancer.ipynb
|
dokato/pyroglancer
|
b6263c08c4da836d652a1fa7d7d50eb7b49be10b
|
[
"BSD-3-Clause"
] | 12 |
2021-01-21T16:27:48.000Z
|
2022-01-24T12:13:53.000Z
|
docs/source/tutorials/local_neuroglancer.ipynb
|
dokato/pyroglancer
|
b6263c08c4da836d652a1fa7d7d50eb7b49be10b
|
[
"BSD-3-Clause"
] | 2 |
2021-03-17T09:30:50.000Z
|
2021-07-27T13:56:57.000Z
| 35.359137 | 152 | 0.592793 |
[
[
[
"# Starting a local neuroglancer session with FAFB dataset",
"_____no_output_____"
],
[
"### This example shows how to start a local neuroglancer session and further add neurons, synapses, neuropil meshes from a public catmaid instance",
"_____no_output_____"
],
[
"### Import neccesary library modules now",
"_____no_output_____"
]
],
[
[
"import navis\nimport fafbseg\nimport pymaid",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport os\nfrom copy import deepcopy",
"_____no_output_____"
],
[
"import io\nfrom PIL import Image",
"_____no_output_____"
],
[
"from pyroglancer.layers import create_nglayer, setlayerproperty\nfrom pyroglancer.localserver import startdataserver, closedataserver\nfrom pyroglancer.ngviewer import openviewer, closeviewer,setviewerstate, get_ngscreenshot\nfrom pyroglancer.ngspaces import create_ngspace\nfrom pyroglancer.createconfig import createconfig",
"_____no_output_____"
]
],
[
[
"### Set configurations to fetch from data from CATMAID",
"_____no_output_____"
]
],
[
[
"publicurl = 'https://fafb.catmaid.virtualflybrain.org/'",
"_____no_output_____"
],
[
"working_rm = pymaid.CatmaidInstance(publicurl, api_token=None, project_id = 1)",
"INFO : Global CATMAID instance set. Caching is ON. (pymaid)\nINFO - 2021-05-19 22:00:43,902 - client - Global CATMAID instance set. Caching is ON.\n"
]
],
[
[
"### Get sample skids and neuropil meshes from CATMAID",
"_____no_output_____"
]
],
[
[
"sample_skids = ['40637','27295','57311','2863104','57323']",
"_____no_output_____"
],
[
"catmiad_neuronlist=pymaid.get_neurons(sample_skids,remote_instance = working_rm)",
"Make nrn: 0%| | 0/5 [00:00<?, ?it/s] INFO - 2021-05-19 22:00:44,720 - utils - NumExpr defaulting to 8 threads.\n \r"
],
[
"vols = pymaid.get_volume(['AL_L', 'AL_R'], color=(255, 0, 0, .2))",
" \r"
],
[
"vols['AL_R'].id = 200\nvols['AL_L'].id = 300\nvols",
"_____no_output_____"
]
],
[
[
"### Start the dataserver to host precomputed data..",
"_____no_output_____"
]
],
[
[
"startdataserver()",
"Serving data from: "
]
],
[
[
"### Start a basic neuroglancer local session with all FAFB configurations..",
"_____no_output_____"
]
],
[
[
"configdata = [dict(\n ngspace='FAFB',\n dimension=dict(x=1, y=1,z=1,units='um'),\n voxelsize=dict(x=4,y=4,z=40,units='nm'),\n layers=dict(\n fafb_v14_clahe=dict(\n type='image',\n source='precomputed://gs://neuroglancer-fafb-data/fafb_v14/fafb_v14_clahe'),\n fafb_surf=dict(\n type='surfacemesh',\n source='vtk://https://storage.googleapis.com/neuroglancer-fafb-data/elmr-data/FAFB.surf.vtk.gz'\n ))\n )]",
"/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7\nServing directory at http://127.0.0.1:8000\n"
],
[
"configfileloc = '/Users/sri/.pyroglancer/config_temp.yml'",
"_____no_output_____"
],
[
"createconfig(configdata, configfileloc)",
"setting default config file loc\n"
],
[
"layer_kws = {'ngspace': 'FAFB'}",
"_____no_output_____"
],
[
"create_ngspace(layer_kws)",
"config file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nNeuroglancer viewer created at: http://127.0.0.1:58817/v/a51569d4574735f355dd0a0f034e16ffafb1eae9/\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: image\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nUsing layout : xy-3d\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: surfacemesh\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nUsing layout : xy-3d\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: synapsepred\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nUsing layout : xy-3d\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: segmentation\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nUsing layout : xy-3d\n"
]
],
[
[
"### Add skids to neuroglancer layers..",
"_____no_output_____"
]
],
[
[
"tmpviewer = create_nglayer(layer_kws = {'type': 'skeletons',\n 'source': catmiad_neuronlist,\n 'name':'catmaid_skels',\n 'color': 'green',\n 'alpha': 0.5})",
"config file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: skeletons\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/skeletons/40637\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/skeletons/27295\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/skeletons/57311\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/skeletons/2863104\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/skeletons/57323\ncreating: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/skeletons/seg_props\nUsing layout : xy-3d\n"
]
],
[
[
"### Add synapses to neuroglancer layers..",
"_____no_output_____"
]
],
[
[
"tmpviewer = create_nglayer(layer_kws = {'type': 'synapses',\n 'linked_layername': 'catmaid_skels',\n 'source': catmiad_neuronlist})",
"config file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: synapses\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nflushing stuff..\npresynapse stuff at: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7 /precomputed/catmaid_skels/presynapses\npostsynapse stuff at: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7 /precomputed/catmaid_skels/postsynapses\nsynapses info path: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels\ncreating: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/presynapses\ncreating: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/postsynapses\nAdding neuron: 40637\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/presynapses/presynapses_cell/40637\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/postsynapses/postsynapses_cell/40637\nAdding neuron: 27295\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/presynapses/presynapses_cell/27295\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/postsynapses/postsynapses_cell/27295\nAdding neuron: 57311\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/presynapses/presynapses_cell/57311\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/postsynapses/postsynapses_cell/57311\nAdding neuron: 2863104\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/presynapses/presynapses_cell/2863104\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/postsynapses/postsynapses_cell/2863104\nAdding neuron: 57323\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/presynapses/presynapses_cell/57323\nmaking: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/catmaid_skels/postsynapses/postsynapses_cell/57323\nUsing layout : xy-3d\n"
]
],
[
[
"### Add neuropil meshes to neuroglancer layers..",
"_____no_output_____"
]
],
[
[
"tmpviewer = create_nglayer(layer_kws = {'type': 'volumes','source': [vols['AL_R'],vols['AL_L']],\n 'name': 'neuropils','color': ['magenta', 'blue'], 'alpha': 0.3})",
"config file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: volumes\nmesh/200\nSeg id is: 200\nFull filepath: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/neuropils/mesh/200\nmesh/300\nSeg id is: 300\nFull filepath: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/neuropils/mesh/300\ncreating: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/neuropils/mesh/segment_properties\ncreating: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/neuropils/mesh/segment_names\nUsing layout : xy-3d\n"
]
],
[
[
"### Add annotations meshes to neuroglancer layers..",
"_____no_output_____"
]
],
[
[
"temp_pts = pd.DataFrame([[123072, 47001, 3375]],columns=['x','y','z'])\ntemp_pts = pd.DataFrame([[123072, 47001, 3375], [120000, 17001, 3000]], columns=['x', 'y', 'z'])\ntemp_pts['description'] = ['center_pt','above_pt']",
"_____no_output_____"
],
[
"#plot landmarks..\ntmpviewer = create_nglayer(layer_kws = {'type': 'points','name': 'landmarks',\n \"annotationstatetype\": 'precomputed',\n 'source': temp_pts,'color': 'orange'})",
"config file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nLayer created: points\nconfig file loc is at: None\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nDimensions are in : FAFB\nusing default location at: /Users/sri/.pyroglancer/config_temp.yml\nusing voxel space with scale: [4, 4, 40]\ncreating: /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/landmarks\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/landmarks/spatial0/0_0_0\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/landmarks/by_id/0\n/private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7/precomputed/landmarks/by_id/1\nUsing layout : xy-3d\n"
]
],
[
[
"### Set settings of the viewer/segments",
"_____no_output_____"
]
],
[
[
"tmpviewer = setlayerproperty(tmpviewer, property_kws = {'name': 'synapses_buhmann2019','visibility': False})",
"_____no_output_____"
],
[
"tmpviewer = setlayerproperty(tmpviewer, property_kws = {'name': 'catmaid_skels','segments': sample_skids})",
"127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/skeletons/2863104 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/skeletons/57323 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/skeletons/40637 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/skeletons/57311 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/skeletons/27295 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/2863104 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/40637 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/27295 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/57323 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/57311 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/2863104 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/57311 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/40637 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/27295 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:05] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/57323 HTTP/1.1\" 200 -\n"
],
[
"tmpviewer = setlayerproperty(tmpviewer, property_kws = {'name': 'neuropils','segments': [vols['AL_R'].id, vols['AL_L'].id]})",
"127.0.0.1 - - [19/May/2021 22:01:10] \"GET /precomputed/neuropils/mesh/300:0 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:10] \"GET /precomputed/neuropils/mesh/200:0 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:10] \"GET /precomputed/neuropils/mesh/200 HTTP/1.1\" 200 -\n127.0.0.1 - - [19/May/2021 22:01:10] \"GET /precomputed/neuropils/mesh/300 HTTP/1.1\" 200 -\n"
],
[
"tmpviewer = setviewerstate(axis_lines = False, bounding_box = False)",
"_____no_output_____"
],
[
"#adjust the zoom factor a bit according your settings, screen, viewer state before etc.\ntmpviewer = setviewerstate(tmpviewer, axis_lines=False, bounding_box=False, layout='3d', zoom_factor = 208000)",
"127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/skeletons/info HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/presynapses/info HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/postsynapses/info HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/neuropils/mesh/info HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/landmarks/info HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/skeletons/seg_props/info HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/neuropils/mesh/segment_properties/info HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/57311 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/40637 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/57311 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/57323 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/57323 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/27295 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/27295 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/2863104 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/presynapses/presynapses_cell/2863104 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/postsynapses/postsynapses_cell/40637 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/landmarks/spatial0/0_0_0 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/neuropils/mesh/200:0 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/neuropils/mesh/300:0 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/skeletons/57311 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/skeletons/2863104 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/skeletons/57323 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/skeletons/40637 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/catmaid_skels/skeletons/27295 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/neuropils/mesh/200 HTTP/1.1\" 304 -\n127.0.0.1 - - [19/May/2021 22:01:59] \"GET /precomputed/neuropils/mesh/300 HTTP/1.1\" 304 -\n"
]
],
[
[
"### Screenshot of the neuroglancer instance",
"_____no_output_____"
]
],
[
[
"screenshot = get_ngscreenshot(tmpviewer, viewer_size=[1000, 1000])",
"_____no_output_____"
],
[
"imageStream = io.BytesIO(screenshot.image)\nimageFile = Image.open(imageStream)",
"_____no_output_____"
],
[
"current_folder = globals()['_dh'][0]",
"_____no_output_____"
],
[
"imagefilepath = os.path.join(current_folder, 'pics/local_neuroglancersession.png')\nimagefilepath",
"_____no_output_____"
],
[
"imageFile.save(imagefilepath)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"### Close the viewer and dataserver",
"_____no_output_____"
]
],
[
[
"closeviewer()",
"closing already existing ng viewer\n"
],
[
"closedataserver()",
"Closing server at http://127.0.0.1:8000\nCleaning directory at /private/var/folders/_l/lrfvj_8j3ps0c37ncbr3c8dh0000gn/T/tmp37r6z4p7\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbaea6e1c96f206fe3c1004a800de899bbbe7448
| 10,368 |
ipynb
|
Jupyter Notebook
|
notebooks/BB84.ipynb
|
gitkarma/quantum-computing-course
|
4b78a3bbc6942fb134d726e7c6230fb95d0748fb
|
[
"MIT"
] | 25 |
2019-05-20T06:53:58.000Z
|
2021-03-06T04:36:25.000Z
|
notebooks/BB84.ipynb
|
gitkarma/quantum-computing-course
|
4b78a3bbc6942fb134d726e7c6230fb95d0748fb
|
[
"MIT"
] | null | null | null |
notebooks/BB84.ipynb
|
gitkarma/quantum-computing-course
|
4b78a3bbc6942fb134d726e7c6230fb95d0748fb
|
[
"MIT"
] | 34 |
2020-09-01T13:09:01.000Z
|
2021-03-01T06:29:41.000Z
| 39.272727 | 219 | 0.447724 |
[
[
[
"# BB84 Quantum Key Distribution (QKD) Protocol using Qiskit\n\nThis notebook is a _demonstration_ of the BB84 Protocol for QKD using Qiskit. \nBB84 is a quantum key distribution scheme developed by Charles Bennett and Gilles Brassard in 1984 ([paper]).\nThe first three sections of the paper are readable and should give you all the necessary information required. \n\n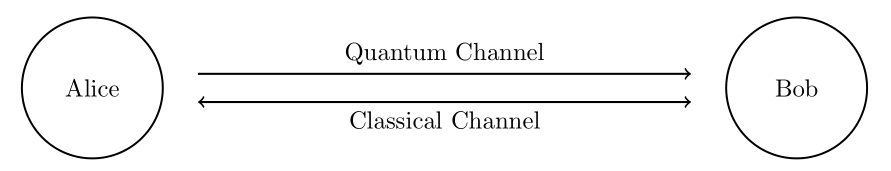\n\n[paper]: http://researcher.watson.ibm.com/researcher/files/us-bennetc/BB84highest.pdf \n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt\n# Importing standard Qiskit libraries\nfrom qiskit import QuantumCircuit, execute\nfrom qiskit.providers.aer import QasmSimulator\nfrom qiskit.visualization import *",
"_____no_output_____"
]
],
[
[
"## Choosing bases and encoding states\n\nAlice generates two binary strings. One encodes the basis for each qubit:\n\n$0 \\rightarrow$ Computational basis\n\n$1 \\rightarrow$ Hadamard basis\n\nThe other encodes the state:\n\n$0 \\rightarrow|0\\rangle$ or $|+\\rangle $ \n\n$1 \\rightarrow|1\\rangle$ or $|-\\rangle $ \n\nBob also generates a binary string and uses the same convention to choose a basis for measurement\n",
"_____no_output_____"
]
],
[
[
"num_qubits = 32\n\nalice_basis = np.random.randint(2, size=num_qubits)\nalice_state = np.random.randint(2, size=num_qubits)\nbob_basis = np.random.randint(2, size=num_qubits)\n\n\nprint(f\"Alice's State:\\t {np.array2string(alice_state, separator='')}\")\nprint(f\"Alice's Bases:\\t {np.array2string(alice_basis, separator='')}\")\nprint(f\"Bob's Bases:\\t {np.array2string(bob_basis, separator='')}\")",
"Alice's State:\t [01111000001010100100110011000110]\nAlice's Bases:\t [00011010011000001100111111110101]\nBob's Bases:\t [11010000001111010111110000101010]\n"
]
],
[
[
"## Creating the circuit\n\nBased on the following results:\n\n$X|0\\rangle = |1\\rangle$\n\n$H|0\\rangle = |+\\rangle$\n\n$ HX|0\\rangle = |-\\rangle$\n\nOur algorithm to construct the circuit is as follows:\n\n1. Whenever Alice wants to encode 1 in a qubit, she applies an $X$ gate to the qubit. To encode 0, no action is needed.\n2. Wherever she wants to encode it in the Hadamard basis, she applies an $H$ gate. No action is necessary to encode a qubit in the computational basis.\n\n3. She then _sends_ the qubits to Bob (symbolically represented in this circuit using wires)\n\n4. Bob measures the qubits according to his binary string. To measure a qubit in the Hadamard basis, he applies an $H$ gate to the corresponding qubit and then performs a mesurement on the computational basis. \n\n",
"_____no_output_____"
]
],
[
[
"def make_bb84_circ(enc_state, enc_basis, meas_basis):\n '''\n enc_state: array of 0s and 1s denoting the state to be encoded\n enc_basis: array of 0s and 1s denoting the basis to be used for encoding\n 0 -> Computational Basis\n 1 -> Hadamard Basis\n meas_basis: array of 0s and 1s denoting the basis to be used for measurement\n 0 -> Computational Basis\n 1 -> Hadamard Basis\n '''\n num_qubits = len(enc_state)\n \n bb84_circ = QuantumCircuit(num_qubits)\n\n # Sender prepares qubits\n for index in range(len(enc_basis)):\n if enc_state[index] == 1:\n bb84_circ.x(index)\n if enc_basis[index] == 1:\n bb84_circ.h(index)\n bb84_circ.barrier() \n\n # Receiver measures the received qubits\n for index in range(len(meas_basis)):\n if meas_basis[index] == 1:\n bb84_circ.h(index)\n\n bb84_circ.barrier() \n bb84_circ.measure_all()\n \n return bb84_circ\n",
"_____no_output_____"
]
],
[
[
"## Creating the key\n\nAlice and Bob only keep the bits where their bases match.\n\nThe following outcomes are possible for each bit sent using the BB84 protocol\n\n| Alice's bit \t| Alice's basis \t| Alice's State \t| Bob's basis \t| Bob's outcome \t| Bob's bit \t| Probability \t|\n|----------------------\t|------------------------\t|------------------------\t|----------------------\t|------------------------\t|--------------------\t|--------------------\t|\n| 0 \t| C \t| 0 \t| C \t| 0 \t| 0 \t| 1/8 \t|\n| 0 \t| C \t| 0 \t| H \t| + \t| 0 \t| 1/16 \t|\n| 0 \t| C \t| 0 \t| H \t| - \t| 1 \t| 1/16 \t|\n| 0 \t| H \t| + \t| C \t| 0 \t| 0 \t| 1/16 \t|\n| 0 \t| H \t| + \t| C \t| 1 \t| 1 \t| 1/16 \t|\n| 0 \t| H \t| + \t| H \t| + \t| 0 \t| 1/8 \t|\n| 1 \t| C \t| 1 \t| C \t| 1 \t| 1 \t| 1/8 \t|\n| 1 \t| C \t| 1 \t| H \t| + \t| 0 \t| 1/16 \t|\n| 1 \t| C \t| 1 \t| H \t| - \t| 1 \t| 1/16 \t|\n| 1 \t| H \t| - \t| C \t| 0 \t| 0 \t| 1/16 \t|\n| 1 \t| H \t| - \t| C \t| 1 \t| 1 \t| 1/16 \t|\n| 1 \t| H \t| - \t| H \t| - \t| 1 \t| 1/8 \t|\n\n\\begin{align*}\nP_{\\text{same basis}} &= P_A(C)\\times P_B(C) + P_A(H)\\times P_B(H)\\\\\n&= \\frac{1}{2} \\times \\frac{1}{2} + \\frac{1}{2} \\times \\frac{1}{2} \\\\ \n&= \\frac{1}{2}\n\\end{align*}\n\nThus, on average, only half of the total bits will be in the final key. It is also interesting to note that half of the key bits will be 0 and the other half will be 1 (again, on average)",
"_____no_output_____"
]
],
[
[
"bb84_circ = make_bb84_circ(alice_state, alice_basis, bob_basis)\ntemp_key = execute(bb84_circ.reverse_bits(),backend=QasmSimulator(),shots=1).result().get_counts().most_frequent()\nkey = ''\nfor i in range(num_qubits):\n if alice_basis[i] == bob_basis[i]: # Only choose bits where Alice and Bob chose the same basis\n key += str(temp_key[i])\nprint(f'The length of the key is {len(key)}')\nprint(f\"The key contains {(key).count('0')} zeroes and {(key).count('1')} ones\")\nprint(f\"Key: {key}\")",
"The length of the key is 11\nThe key contains 4 zeroes and 7 ones\nKey: 11000111110\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbaeab516cd3c0ee33a62209a5037dc82fbb7999
| 27,366 |
ipynb
|
Jupyter Notebook
|
summarystats/toolboxone.ipynb
|
nrslvsjs/intro_stats_with_tidyverse
|
118f21a58f8b92be2f3d26598da27b7418d68c54
|
[
"MIT"
] | 15 |
2019-02-01T21:49:00.000Z
|
2022-01-11T04:38:33.000Z
|
summarystats/toolboxone.ipynb
|
nrslvsjs/intro_stats_with_tidyverse
|
118f21a58f8b92be2f3d26598da27b7418d68c54
|
[
"MIT"
] | null | null | null |
summarystats/toolboxone.ipynb
|
nrslvsjs/intro_stats_with_tidyverse
|
118f21a58f8b92be2f3d26598da27b7418d68c54
|
[
"MIT"
] | 8 |
2019-03-05T00:34:18.000Z
|
2021-11-29T06:07:18.000Z
| 84.987578 | 15,808 | 0.784441 |
[
[
[
"**[Back to Fan's Intro Stat Table of Content](https://fanwangecon.github.io/Stat4Econ/)**\n\n# Rescaling Standard Deviation and Covariance\n\nWe have various tools at our disposal to summarize variables and the relationship between variables. Imagine that we have multiple toolboxes. This is the first one. There are two levels to this toolbox.\n\n## Three Basic Tools\n\nOur three basic tools are:\n\n1. (sample) Mean of X (or Y)\n2. (sample) Standard Deviation of X (or Y)\n3. (sample) Covariance of X and Y\n\n## Two Rescaling Tools\n\nAdditionally, we have two tools that combine the tools from the first level:\n\n1. Coefficient of Variation = (Standard Deviation)/(Mean)\n2. Correlation = (Covariance of X and Y)/((Standard Deviation of X)*(Standard Deviation of Y))\n\nThe tools on the second level rescale the standard deviation and covariance statistics. \n",
"_____no_output_____"
],
[
"# Data Examples\n\n**The dataset, *EPIStateEduWage2017.csv*, can be downloaded [here](../data/EPIStateEduWage2017.csv).**\n\n## College Education Share and Hourly Wage\n\nTwo variables:\n\n1. Fraction of individual with college degree in a state\n + this is in Fraction units, the minimum is 0.00, the maximum is 100 percent, which is 1.00\n2. Average hourly salary in the state\n + this is in Dollar units",
"_____no_output_____"
]
],
[
[
"# Load in Data Tools\n# For Reading/Loading Data\nlibrary(readr)\nlibrary(tibble)\nlibrary(dplyr)\nlibrary(ggplot2)\n# Load in Data\ndf_wgedu <- read_csv('../data/EPIStateEduWage2017.csv')",
"Parsed with column specification:\ncols(\n State = col_character(),\n Share.College.Edu = col_double(),\n Hourly.Salary = col_double()\n)\n"
]
],
[
[
"## A Scatter Plot\n\nWe can Visualize the Data with a Scatter Plot. There seems to be a positive relationship between the share of individuals in a state with a college education, and the average hourly salary in that state.\n\nWhile most states are along the trend line, we have some states, like WY, that are outliers. WY has a high hourly salary but low share with college education.",
"_____no_output_____"
]
],
[
[
"# Control Graph Size\noptions(repr.plot.width = 5, repr.plot.height = 5)\n# Draw Scatter Plot\n# 1. specify x and y\n# 2. label each state\n# 3. add in trend line\nscatter <- ggplot(df_wgedu, aes(x=Share.College.Edu, y=Hourly.Salary)) +\n geom_point(size=1) +\n geom_text(aes(label=State), size=3, hjust=-.2, vjust=-.2) +\n geom_smooth(method=lm) +\n labs(title = 'Hourly Wage and College Share by States',\n x = 'Fraction with College Education',\n y = 'Hourly Wage',\n caption = 'Economic Policy Institute\\n www.epi.org/data/') +\n theme_bw()\nprint(scatter)",
"_____no_output_____"
]
],
[
[
"## Standard Deviations and Coefficient of Variation\n\nThe two variables above are in different units. We first calculate the mean, standard deviation, and covariance. With just these, it is hard to compare the standard deviation of the two variables, which are on different scales.\n\nThe sample standard deviations for the two variables are: $0.051$ and $1.51$, in fraction and dollar units. Can we say the hourly salary has a larger standard deviation? But it is just a different scale. $1.51$ is a large number, but that does not mean that variable has greater variation than the fraction with college education variable. \n\nConverting the Statistics to Coefficient of Variations, now we have: $0.16$ and $0.09$. Because of the division, these are both in fraction units--standard deviations as a fraction of the mean. Now these are more comparable.",
"_____no_output_____"
]
],
[
[
"# We can compute the three basic statistics\nstats.msdv <- list(\n # Mean, SD and Var for the College Share variable\n Shr.Coll.Mean = mean(df_wgedu$Share.College.Edu), \n Shr.Coll.Std = sd(df_wgedu$Share.College.Edu),\n Shr.Coll.Var = var(df_wgedu$Share.College.Edu),\n \n # Mean, SD and Var for the Hourly Wage Variable\n Hr.Wage.Mean = mean(df_wgedu$Hourly.Salary), \n Hr.Wage.Std = sd(df_wgedu$Hourly.Salary),\n Hr.Wage.Var = var(df_wgedu$Hourly.Salary)\n )\n\n# We can compute the three basic statistics\nstats.coefvari <- list( \n # Coefficient of Variation\n Shr.Coll.Coef.Variation = (stats.msdv$Shr.Coll.Std)/(stats.msdv$Shr.Coll.Mean),\n Hr.Wage.Coef.Variation = (stats.msdv$Hr.Wage.Std)/(stats.msdv$Hr.Wage.Mean)\n )\n\n# Let's Print the Statistics we Computed\nas_tibble(stats.msdv)\nas_tibble(stats.coefvari)",
"_____no_output_____"
]
],
[
[
"## Covariance and Correlation\n\nFor covariance, hard to tell whether it is large or small. To make comparisons possible, we calculate the coefficient of variations and correlation statistics.\n\nThe covariance we get is positive: $0.06$, but is this actually large positive relationship? $0.06$ seems like a small number. \n\nRescaling covariance to correlation, the correlation between the two variables is: $0.78$. Since the correlation of two variable is below $-1$ and $+1$, we can now say actually the two variables are very positively related. A higher share of individuals with a college education is strongly positively correlated with a higher hourly salary. ",
"_____no_output_____"
]
],
[
[
"# We can compute the three basic statistics\nstates.covcor <- list( \n # Covariance between the two variables\n Shr.Wage.Cov = cov(df_wgedu$Hourly.Salary,\n df_wgedu$Share.College.Edu), \n # Correlation \n Shr.Wage.Cor = cor(df_wgedu$Hourly.Salary, df_wgedu$Share.College.Edu),\n Shr.Wage.Cor.Formula = (cov(df_wgedu$Hourly.Salary, df_wgedu$Share.College.Edu)\n /(stats.msdv$Shr.Coll.Std*stats.msdv$Hr.Wage.Std))\n )\n\n# Let's Print the Statistics we Computed\nas_tibble(states.covcor)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbaeb60dd135ff3ffd867d70b3c8cb407086cfcf
| 2,560 |
ipynb
|
Jupyter Notebook
|
Intro_to_Python.ipynb
|
dess99/Elective1-3
|
4660054cf2373b641018fad897b01721679f99f2
|
[
"Apache-2.0"
] | null | null | null |
Intro_to_Python.ipynb
|
dess99/Elective1-3
|
4660054cf2373b641018fad897b01721679f99f2
|
[
"Apache-2.0"
] | null | null | null |
Intro_to_Python.ipynb
|
dess99/Elective1-3
|
4660054cf2373b641018fad897b01721679f99f2
|
[
"Apache-2.0"
] | null | null | null | 22.654867 | 230 | 0.444922 |
[
[
[
"<a href=\"https://colab.research.google.com/github/dess99/Elective1-3/blob/main/Intro_to_Python.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"##Python Indention",
"_____no_output_____"
]
],
[
[
"if 5<2:\n print(\"Five is less than two\")\nelse:\n print(\"Five is greater than two\")",
"Five is greater than two\n"
]
],
[
[
"##Python Comments",
"_____no_output_____"
]
],
[
[
"#This is a program that displays Hello, World\n\nprint(\"Hello World\")\nprint(\"Welcome to Python Programming\")",
"Hello World\nWelcome to Python Programming\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbaec6677f86ceeb902ae812790a51f4dee08b38
| 1,685 |
ipynb
|
Jupyter Notebook
|
docs/pep8.ipynb
|
0101011/Visual-PEPs
|
afc3205b11cb75eabbb967b810bf2dc4b527a66b
|
[
"MIT"
] | null | null | null |
docs/pep8.ipynb
|
0101011/Visual-PEPs
|
afc3205b11cb75eabbb967b810bf2dc4b527a66b
|
[
"MIT"
] | null | null | null |
docs/pep8.ipynb
|
0101011/Visual-PEPs
|
afc3205b11cb75eabbb967b810bf2dc4b527a66b
|
[
"MIT"
] | null | null | null | 20.059524 | 79 | 0.526409 |
[
[
[
"# PEP 8",
"_____no_output_____"
],
[
"## Indentation",
"_____no_output_____"
],
[
"1. Use 4 spaces per indentation level\n2. Align wrapped elements vertically or using a hanging indent",
"_____no_output_____"
]
],
[
[
"# Align arguments with opening delimiter.\nfoo = long_function_name(var_one, var_two,\n var_three, var_four)",
"_____no_output_____"
],
[
"# Additional 4 spaces included to distinguish arguments from the rest.\ndef long_function_name(\n var_one, var_two, var_three,\n var_four):\n print(var_one)",
"_____no_output_____"
],
[
"# Hanging indents should add a level.\nfoo = long_function_name(\n var_one, var_two,\n var_three, var_four)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbaeef9e0d18ca3b2423f7462be29f24fea4673a
| 10,171 |
ipynb
|
Jupyter Notebook
|
examples/data/.ipynb_checkpoints/Testing-checkpoint.ipynb
|
joshlam123/dsc_project
|
b3bd8b803ed898121261d78316282b02106c5659
|
[
"MIT"
] | 1 |
2020-05-07T04:59:52.000Z
|
2020-05-07T04:59:52.000Z
|
examples/data/.ipynb_checkpoints/Testing-checkpoint.ipynb
|
joshlam123/dsc_project
|
b3bd8b803ed898121261d78316282b02106c5659
|
[
"MIT"
] | 6 |
2020-04-06T03:35:03.000Z
|
2022-02-27T01:12:06.000Z
|
examples/data/.ipynb_checkpoints/Testing-checkpoint.ipynb
|
joshlam123/dsc_project
|
b3bd8b803ed898121261d78316282b02106c5659
|
[
"MIT"
] | null | null | null | 44.414847 | 2,034 | 0.364861 |
[
[
[
"import json\nimport os\nimport numpy as np\nimport networkx as nx",
"_____no_output_____"
],
[
"class PageRank:\n def __init__(self, epsilon=10e-3):\n self.epsilon = epsilon\n \n def makeGraphProcess(self, fileName):\n self.openGraph(fileName)\n \n def openGraph(self, fileName:str):\n graphProblem = os.getcwd() + f'/prob/{fileName}.json'\n\n with open(graphProblem, 'r+') as f:\n graphProblem = json.loads(f.read())\n \n self.graph = self.processGraph(graphProblem)\n \n def processGraph(self, graph):\n G = nx.Graph()\n for k,v in graphProblem.items():\n G.add_node(k, value=v['Value'], outgoingEdges=v['OutgoingEdges'])\n for x,y in v['OutgoingEdges'].items():\n G.add_edge(k, x)\n return G\n \n def pageRankAlgorithm(self, alpha=0.85):\n err = 0 \n \n while err >= self.epsilon:\n previous = self.graph\n r = dict.fromkeys(previous.keys(), 0) \n \n danglesum = alpha * sum(xlast[n] for n in dangling_nodes) \n \n for vertex in self.graph.edges: \n R[vertex] = 0.15 + alpha * sum([])\n M[i] = R[i]/\n SendM[i] to all N_out(i)\n err=abs(r[n]−previous[n])",
"_____no_output_____"
],
[
"pr = PageRank()\npr.makeGraphProcess('randM20')\npr.graph.edges",
"_____no_output_____"
],
[
"G = nx.Graph()\ngraphProblem = os.getcwd() + '/prob/randM20.json'\n\nwith open(graphProblem, 'r+') as f:\n graphProblem = json.loads(f.read())\n\nprint(graphProblem)",
"{'1': {'Value': 5, 'OutgoingEdges': {'10': 14, '14': 2, '15': 19, '17': 1, '18': 17, '4': 16, '7': 12, '8': 5}}, '10': {'Value': 14, 'OutgoingEdges': {'1': 5, '12': 16, '13': 14, '17': 1, '19': 3, '2': 1, '3': 6, '4': 16, '6': 2}}, '11': {'Value': 13, 'OutgoingEdges': {'10': 14, '14': 2, '2': 1, '4': 16}}, '12': {'Value': 16, 'OutgoingEdges': {'1': 5, '17': 1, '18': 17, '2': 1, '4': 16, '6': 2, '7': 12, '9': 0}}, '13': {'Value': 14, 'OutgoingEdges': {'1': 5}}, '14': {'Value': 2, 'OutgoingEdges': {'13': 14, '17': 1, '2': 1, '9': 0}}, '15': {'Value': 19, 'OutgoingEdges': {'1': 5, '16': 0, '3': 6, '7': 12, '8': 5}}, '16': {'Value': 0, 'OutgoingEdges': {'1': 5}}, '17': {'Value': 1, 'OutgoingEdges': {'1': 5, '10': 14, '11': 13, '12': 16, '15': 19, '16': 0, '17': 1, '18': 17, '2': 1, '6': 2, '8': 5, '9': 0}}, '18': {'Value': 17, 'OutgoingEdges': {'11': 13, '13': 14, '15': 19, '17': 1, '18': 17, '19': 3, '2': 1, '3': 6, '5': 1, '7': 12}}, '19': {'Value': 3, 'OutgoingEdges': {'11': 13, '12': 16, '13': 14, '14': 2, '15': 19, '16': 0, '17': 1, '18': 17, '19': 3, '4': 16, '5': 1, '6': 2, '8': 5}}, '2': {'Value': 1, 'OutgoingEdges': {'11': 13, '12': 16, '13': 14, '19': 3, '7': 12, '8': 5}}, '20': {'Value': 12, 'OutgoingEdges': {'10': 14, '12': 16, '14': 2, '15': 19, '17': 1, '19': 3, '2': 1, '6': 2, '8': 5}}, '3': {'Value': 6, 'OutgoingEdges': {'1': 5, '10': 14, '11': 13, '17': 1, '5': 1}}, '4': {'Value': 16, 'OutgoingEdges': {'1': 5, '10': 14, '12': 16, '14': 2, '15': 19, '16': 0, '17': 1, '19': 3, '3': 6, '5': 1, '6': 2, '9': 0}}, '5': {'Value': 1, 'OutgoingEdges': {'11': 13, '12': 16, '16': 0, '2': 1, '5': 1}}, '6': {'Value': 2, 'OutgoingEdges': {'1': 5, '10': 14, '11': 13, '12': 16, '13': 14, '14': 2, '17': 1, '4': 16, '5': 1, '7': 12}}, '7': {'Value': 12, 'OutgoingEdges': {'11': 13, '12': 16, '15': 19, '19': 3, '2': 1, '5': 1, '8': 5}}, '8': {'Value': 5, 'OutgoingEdges': {'18': 17, '19': 3}}, '9': {'Value': 0, 'OutgoingEdges': {'10': 14, '14': 2, '2': 1, '3': 6, '4': 16, '5': 1, '6': 2, '9': 0}}}\n"
],
[
"for k,v in graphProblem.items():\n G.add_node(k, value=v['Value'], outgoingEdges=v['OutgoingEdges'])\n for x,y in v['OutgoingEdges'].items():\n G.add_edge(k, x)\n\nG.nodes.data()\nG.edges",
"_____no_output_____"
],
[
"G['1']",
"_____no_output_____"
],
[
"a = dict.fromkeys(G, 1.0 / G.number_of_nodes()) ",
"_____no_output_____"
],
[
"dict.fromkeys(a.keys(), 0) ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbaf050b62118330b62833f06a10bfbd9db7b260
| 4,090 |
ipynb
|
Jupyter Notebook
|
2-intermediate-python/4-iterations.ipynb
|
abdulazeezoj/mslearn-intro-to-python-space-exploration
|
7176e38fa248862b6b478b35f6d3aabe68b5d14e
|
[
"MIT"
] | 1 |
2022-01-11T18:07:15.000Z
|
2022-01-11T18:07:15.000Z
|
2-intermediate-python/4-iterations.ipynb
|
abdulazeezoj/mslearn-intro-to-python-space-exploration
|
7176e38fa248862b6b478b35f6d3aabe68b5d14e
|
[
"MIT"
] | null | null | null |
2-intermediate-python/4-iterations.ipynb
|
abdulazeezoj/mslearn-intro-to-python-space-exploration
|
7176e38fa248862b6b478b35f6d3aabe68b5d14e
|
[
"MIT"
] | null | null | null | 43.510638 | 464 | 0.601222 |
[
[
[
"Iterations, in programming, let coders repeat a set of instructions until a condition is met. Think about this as being stuck in a loop that will continue until something tells you to break out.\n\n## While loop\n\nThe `while` loop is one of two iteration types you'll learn about. In this loop, you must specify a condition first and then include the code that you want the loop to iterate over. The loop first checks if the condition is `True` and if it is, then it looks at the code inside the loop. When the condition becomes `False`, the code in the loop is skipped over and the program continues executing the rest of your code.\n\nIf the condition in the loop is `False` to begin with, the code within the loop never executes. During a single loop, the program then goes through the loop and runs the code. Once the code is finished, it looks back at the condition to see if it is still `True`. It's necessary to change the variables in your loop to eventually have a condition that is `False`, or else an infinite loop will occur.\n\nAs shown in the code below, to write a `while` loop, you must first type \"while\" and then the condition you'll check before every loop. End the line with a colon and be sure to indent the next line, which will be the actual loop. The code below prints out a countdown for a rocket. As you can see, the countdown variable in the condition section decreases in every loop until it reaches -1, at which point the condition is `False` and the loop ends.\n\nPredict what will happen when you run this code, then click the run button to verify you've understood.",
"_____no_output_____"
]
],
[
[
"countdown = 5\r\n\r\nwhile countdown >= 0:\r\n print(countdown)\r\n countdown = countdown - 1 \r\nprint(\"Lift Off\")",
"_____no_output_____"
]
],
[
[
"In the following example, the condition is never met and the loop continues forever (if we don't stop it). In this code, the developer forgot to decrease the timer variable, so the condition is always true.",
"_____no_output_____"
]
],
[
[
"# Trying to find life outside our planet\r\ntimer = 10\r\nwhile timer > 0:\r\n print(\"Hello, I am from Earth\")",
"_____no_output_____"
]
],
[
[
"This is an infinite loop and you must either wait for Python to terminate it or select the stop button at the top of the window. It's best to avoid infinite loops, if that wasn't already apparent.\n\n## For loop\n\n`For` loops essentially perform the same task as `while` loops: they tend to focus on iterating a set number of times. `For` loops are great when you want to go through a list and look at every single element. In the code below, we make a list and then go through all the elements and print them out.",
"_____no_output_____"
]
],
[
[
"planets = \"Mars\", \"Saturn\", \"Jupiter\"\n\nfor planet in planets:\n print(planet)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbaf071a1236278b4ae36684d4ffa98ce56effa5
| 68,195 |
ipynb
|
Jupyter Notebook
|
Correlation-Analysis-between-Naver-Daum-Google-Search-Ranking/jupyter_exe.ipynb
|
Kormap/Side-Projects
|
9e61d5b062cc6823cfebc18370f7caae622ea571
|
[
"MIT"
] | null | null | null |
Correlation-Analysis-between-Naver-Daum-Google-Search-Ranking/jupyter_exe.ipynb
|
Kormap/Side-Projects
|
9e61d5b062cc6823cfebc18370f7caae622ea571
|
[
"MIT"
] | null | null | null |
Correlation-Analysis-between-Naver-Daum-Google-Search-Ranking/jupyter_exe.ipynb
|
Kormap/Side-Projects
|
9e61d5b062cc6823cfebc18370f7caae622ea571
|
[
"MIT"
] | 1 |
2022-03-19T10:37:38.000Z
|
2022-03-19T10:37:38.000Z
| 88.911343 | 25,288 | 0.626468 |
[
[
[
"'''\n -*- coding: utf-8 -*-\n\n 2019 - 2학기 - 정보융합학부 데이터사이언스\n 빅데이터 처리 및 응용 과목 지정 프로젝트\n\n 주제 : \" 네이버 - 다음 - 구글 실시간 검색어 순위 크롤링 및 분석 \"\n\n Blog : https://blog.naver.com/sooftware\n GitHub : https://github.com/sh951011\n\n Kwangwoon University Electronic-Communication Dept. 2014707073 김수환\n\n'''\nfrom PyQt5 import QtCore, QtGui, QtWidgets, Qt\nfrom multi import MultiCrawler\nfrom matplotlink import MatplotWidget\nfrom keyword_trend import connect_btn, KeywordTrendWindow\nimport logging\nimport sys\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom matplotlib import font_manager, rc\nfrom matplotlib import style\nfrom datetime import datetime\nimport queue\n\n# MAIN WINDOW ==\nMAIN_WINDOW_WIDTH = 1280\nMAIN_WINDOW_HEIGHT = 1000\nRANK_NUM = 10\n# ==========================\n\n# TITLE ==\nTITLE_COORD_X = 160\nTITLE_COORD_Y = 25\nTITLE_WIDTH = 1000\nTITLE_HEIGHT = 50\nMIDDLE_COORD_Y = 410\nMIDDLE_WIDTH = 300\nMIDDLE_HEIGHT = TITLE_HEIGHT\nMIDDLE1_COORD_X = 85\nMIDDLE2_COORD_X = 275\nMIDDLE3_COORD_X = 650\nMIDDLE4_COORD_X = 1020\n# ==========================\n\n# RANK CONTAINERS ==\nRANK_WIDTH = 350\nRANK_HEIGHT = 30\nRANK_COORD_X = 150\nRANK_COORD_Y = 485\nRANK_GAP_X = 380\nRANK_GAP_Y = 50\nSHOW_RANK_WIDTH = 60\nSHOW_RANK_HEIGHT = RANK_HEIGHT\n# ==========================\n\n# CORR ==\nCORR_COORD_X = 180\nCORR_COORD_Y = 365\nCORR_WIDTH = 80\nCORR_HEIGHT = 30\n# =========================\n\n# TIME ==\nTIME_COORD_X = 50\nTIME_COORD_Y = CORR_COORD_Y\nTIME_WIDTH = 380\nTIME_HEIGHT = CORR_HEIGHT\n# =========================\n\n# MATPLOT ==\nPLOT_COORD_X = 50\nPLOT_COORD_Y = 90\nPLOT_GAP_X = 420\nPLOT_WIDTH = TIME_WIDTH\nPLOT_HEIGHT = 270\nPLOT_COMMENT_COORD_X = 560\nPLOT_COMMENT_GAP_X = 80\nPLOT_COMMENT_COORD_Y = TIME_COORD_Y\nPLOT_COMMENT_WIDTH = 60\nPLOT_COMMENT_HEIGHT = TIME_HEIGHT\n# ==========================\n\n# KEYWORD ==\nKEYWORD_HEIGHT = 25\nKEYWORD_WIDTH = PLOT_WIDTH\nKEYWORD_COORD_X = PLOT_COORD_X + 2 * PLOT_GAP_X\nKEYWORD_COORD_Y = 105\nKEYWORD_GAP_Y = 43\n# =========================\n\n# FONT ==\nMARGUN_FONT = \"맑은 고딕\"\nNANUM_BOLD_FONT = \"나눔스퀘어 ExtraBold\"\nNANUM_FONT = \"나눔스퀘어\"\nTITLE_FONT_SIZE = 24\nMEDIUM_FONT_SIZE = 14\nRANK_FONT_SIZE = 12\n# ==========================\n\n# Basic Setting ==\nlogger = logging.getLogger('root')\nFORMAT = \"[%(asctime)s %(filename)s:%(lineno)s - %(funcName)s()] %(message)s\"\nlogging.basicConfig(stream=sys.stdout, level=logging.DEBUG, format=FORMAT)\nlogger.setLevel(logging.INFO)\nfont_name = font_manager.FontProperties(fname=\"c:/Windows/Fonts/malgun.ttf\").get_name()\nrc('font', family=font_name)\nstyle.use('ggplot')\n# ==========================\n\nclass MainWindow(object):\n def __init__(self):\n self.main_window = None\n self.centralwidget = None\n self.queue = queue.Queue(3) # Multi Threading을 위한 큐\n self.data_list = list() # 크롤링한 데이터들을 합친 리스트\n self.n_ranks = None # Naver 검색어 저장\n self.d_ranks = None # Daum 검색어 저장\n self.g_ranks = None # Google 검색어 저장\n self.rank_containers_list = None\n\n # 정수만을 입력받기 위한 처리\n while True:\n self.update_period = input(\"Enter Update Peroid (sec) : \")\n if self.update_period.isdecimal():\n break\n\n def setup(self, main_window):\n # Main_Window Set ===\n translate = QtCore.QCoreApplication.translate\n main_window.resize(MAIN_WINDOW_WIDTH, MAIN_WINDOW_HEIGHT)\n main_window.setMinimumSize(QtCore.QSize(MAIN_WINDOW_WIDTH, MAIN_WINDOW_HEIGHT))\n self.centralwidget = QtWidgets.QWidget(main_window)\n main_window.setWindowTitle(translate(\"MainWindow\", \"Naver Daum Google Search Ranking\"))\n # ============================================\n\n # Matplot ===\n self.keywords_score = MatplotWidget(self.centralwidget)\n self.keywords_score.setGeometry(QtCore.QRect(PLOT_COORD_X + PLOT_GAP_X, PLOT_COORD_Y, PLOT_WIDTH, PLOT_HEIGHT))\n self.corr = MatplotWidget(self.centralwidget)\n self.corr.setGeometry(QtCore.QRect(PLOT_COORD_X, PLOT_COORD_Y, PLOT_WIDTH, PLOT_HEIGHT))\n denote_colors = ['#c2c2f0', '#ff9999', '#ffb3e6']\n denote_text = ['Naver','Daum','Google']\n self.color_denote = [0] * 3\n for i, label in enumerate(self.color_denote):\n label = QtWidgets.QLabel(self.centralwidget)\n label.setGeometry(QtCore.QRect(PLOT_COMMENT_COORD_X + i * PLOT_COMMENT_GAP_X, TIME_COORD_Y, PLOT_COMMENT_WIDTH, TIME_HEIGHT))\n font = QtGui.QFont(NANUM_BOLD_FONT)\n font.setPointSize(11)\n label.setFont(font)\n label.setAlignment(QtCore.Qt.AlignCenter)\n label.setStyleSheet(\"color: \" + denote_colors[i] + \";\")\n label.setText(denote_text[i])\n # ============================================\n\n # Title ===\n self.title = QtWidgets.QLabel(self.centralwidget)\n self.title.setGeometry(QtCore.QRect(TITLE_COORD_X, TITLE_COORD_Y, TITLE_WIDTH, TITLE_HEIGHT))\n font = QtGui.QFont(NANUM_BOLD_FONT)\n font.setPointSize(TITLE_FONT_SIZE)\n self.title.setFont(font)\n self.title.setAlignment(QtCore.Qt.AlignCenter)\n self.title.setText(translate(\"MainWindow\", \"Naver - Daum - Google 실시간 검색어 순위\"))\n self.title.setStyleSheet(\"color: purple;\")\n # =============================\n\n # Time ===\n now = datetime.now()\n time = str(now.year)\n format_ = [now.month, now.day, now.hour, now.minute]\n delimiters = ['-', '-', ' ', ':']\n for i, item in enumerate(format_):\n time += delimiters[i] + str(item)\n self.time_plot = QtWidgets.QLabel(self.centralwidget)\n self.time_plot.setGeometry(QtCore.QRect(TIME_COORD_X, TIME_COORD_Y, TIME_WIDTH, TIME_HEIGHT))\n font = QtGui.QFont(MARGUN_FONT)\n font.setPointSize(12)\n self.time_plot.setFont(font)\n self.time_plot.setAlignment(QtCore.Qt.AlignCenter)\n self.time_plot.setText(translate(\"MainWindow\", \"<\"+time+\"> 기준\"))\n # ============================\n\n # Middle ===\n labels = ['순위', 'Naver', 'Daum', 'Google']\n colors = ['black', 'green', 'brown', 'blue']\n geometrys = [\n [MIDDLE1_COORD_X, MIDDLE_COORD_Y, MIDDLE_WIDTH, MIDDLE_HEIGHT],\n [MIDDLE2_COORD_X, MIDDLE_COORD_Y, MIDDLE_WIDTH, MIDDLE_HEIGHT],\n [MIDDLE3_COORD_X, MIDDLE_COORD_Y, MIDDLE_WIDTH, MIDDLE_HEIGHT],\n [MIDDLE4_COORD_X, MIDDLE_COORD_Y, MIDDLE_WIDTH, MIDDLE_HEIGHT]\n ]\n fonts = [MARGUN_FONT] + [NANUM_BOLD_FONT] * 3\n font_sizes = [MEDIUM_FONT_SIZE] + [TITLE_FONT_SIZE] * 3\n\n for i in range(4):\n self.middle = QtWidgets.QLabel(self.centralwidget)\n self.middle.setGeometry(QtCore.QRect(geometrys[i][0], geometrys[i][1], geometrys[i][2], geometrys[i][3]))\n font = QtGui.QFont(fonts[i])\n font.setPointSize(font_sizes[i])\n self.middle.setFont(font)\n self.middle.setText(translate(\"MainWindow\", labels[i]))\n self.middle.setStyleSheet(\"color: \" + colors[i] + \";\")\n # ===========================\n\n # Keyword Label ===\n self.max_keyword_label = [0] * 3\n self.min_keyword_label = [0] * 3\n blue_font = QtGui.QFont(NANUM_BOLD_FONT)\n blue_font.setPointSize(13)\n black_font = QtGui.QFont(NANUM_BOLD_FONT)\n black_font.setPointSize(12)\n for i in range(3):\n self.max_keyword_label[i] = QtWidgets.QLabel(self.centralwidget)\n self.max_keyword_label[i].setGeometry(QtCore.QRect(KEYWORD_COORD_X,KEYWORD_COORD_Y + 2 * i * KEYWORD_GAP_Y,KEYWORD_WIDTH,KEYWORD_HEIGHT))\n self.max_keyword_label[i].setFont(blue_font)\n self.max_keyword_label[i].setAlignment(QtCore.Qt.AlignCenter)\n self.max_keyword_label[i].setStyleSheet(\"color: blue;\")\n self.min_keyword_label[i] = QtWidgets.QLabel(self.centralwidget)\n self.min_keyword_label[i].setGeometry(QtCore.QRect(KEYWORD_COORD_X,KEYWORD_COORD_Y + KEYWORD_GAP_Y + 2 * i * KEYWORD_GAP_Y,KEYWORD_WIDTH,KEYWORD_HEIGHT))\n self.min_keyword_label[i].setFont(black_font)\n self.min_keyword_label[i].setAlignment(QtCore.Qt.AlignCenter)\n self.min_keyword_label[i].setStyleSheet(\"color: black;\")\n self.keyword_comment = QtWidgets.QLabel(self.centralwidget)\n self.keyword_comment.setGeometry(QtCore.QRect(KEYWORD_COORD_X, CORR_COORD_Y, KEYWORD_WIDTH, KEYWORD_HEIGHT))\n font = QtGui.QFont(MARGUN_FONT)\n font.setPointSize(11)\n self.keyword_comment.setFont(font)\n self.keyword_comment.setAlignment(QtCore.Qt.AlignCenter)\n # ============================\n\n # Rank Containers ===\n def _create_rank_containers(self):\n self.rank_containers_list = list()\n for i in range(3):\n self.rank_containers_list.append(list())\n\n for i, rank_containers in enumerate(self.rank_containers_list):\n for j in range(RANK_NUM):\n rank_containers.append(QtWidgets.QPushButton(self.centralwidget))\n\n #rank_containers.append(QtWidgets.QLabel(self.centralwidget))\n for j, rank in enumerate(rank_containers):\n rank.setGeometry(QtCore.QRect(RANK_COORD_X + RANK_GAP_X * i,\n RANK_COORD_Y + RANK_GAP_Y * j,\n RANK_WIDTH, RANK_HEIGHT))\n font = QtGui.QFont(MARGUN_FONT)\n font.setPointSize(RANK_FONT_SIZE)\n rank.setFont(font)\n rank.setStyleSheet(\"border-radius: 5px;\\n\"\"color: black;\\n\")\n\n for i in range(10):\n rank_label = QtWidgets.QLabel(self.centralwidget)\n rank_label.setGeometry(QtCore.QRect(MIDDLE1_COORD_X, RANK_COORD_Y + RANK_GAP_Y * i, SHOW_RANK_WIDTH, SHOW_RANK_HEIGHT))\n font = QtGui.QFont(NANUM_FONT)\n font.setPointSize(MEDIUM_FONT_SIZE)\n rank_label.setFont(font)\n rank_label.setObjectName(\"rank_label\" + str(i))\n rank_label.setText(translate(\"MainWindow\", \" \" + str(i+1) + \"위\"))\n # ================================================\n\n # Event Connect\n def _set_connect(self):\n self.crawling() # 처음 실행시 크롤링 실행\n connect_btn(self.rank_containers_list, self.keyword_clicked) # 각 검색어 버튼들 keyword_clicked와 연결\n self.timer = QtCore.QTimer() # 타이머 설정\n self.timer.setInterval(1000 * int(self.update_period)) # ms단위라 1000과 입력받은 주기 (sec)을 곱해주면 해당 초만큼 주기적으로 실행\n self.timer.timeout.connect(self.crawling)\n self.timer.start()\n\n _create_rank_containers(self)\n _set_connect(self)\n main_window.setCentralWidget(self.centralwidget)\n\n # 키워드 클릭시 실행\n def keyword_clicked(self, keyword):\n rank_changes = KeywordTrendWindow(keyword=keyword)\n rank_changes.show()\n\n # 크롤링\n def crawling(self):\n # Naver - Daum - Google에서 수집한 검색어 순위를 포맷에 맞춰 csv 파일로 저장\n def update_data(self):\n columns = ['Time', 'Search Engine']\n columns += ['Rank' + str(i) for i in range(1, 11)]\n data_list = list()\n for i in range(3):\n data_list += self.queue.get() # Thread들이 저장해놓은 데이터 get\n\n self.data_list = data_list\n\n new_data = pd.DataFrame(np.array(data_list).reshape(3, 12), columns=columns) # 새로 수집한 데이터\n read_data = pd.read_csv('./data/data.csv', encoding='utf-8') # 기존 엑셀 파일 데이터\n merge_data = pd.concat([read_data, new_data], sort=False) # (기존 + New) 병합\n merge_data.to_csv('./data/data.csv', encoding='utf-8', sep=',', index=False) # 병합 데이터 저장\n\n # 검색어 종합 스코어 Top5를 Pie chart로 표시\n # - 3포털에 대해서 점수를 합산해서 상위 스코어 5개를 표시한다\n def update_pie(self):\n # 각 키워드별로 점수 계산\n # 각 포털 1위는 10점 2위는 9점 ... 10위는 1점 순위권 밖은 0점을 준다\n def _get_keywords_score(self):\n scores = [] # 키워드에 대한 점수를 저장하는 리스트\n keywords = [] # 키워드를 저장하는 리스트\n except_case = [0,1,12,13,24,25] # csv파일의 'Time', 'Search Engine' 컬럼에 해당하는 인덱스는 예외처리\n k = 0\n self.g_ranks = self.data_list[2:12] # google 검색어 저장\n self.n_ranks = self.data_list[14:24] # Naver 검색어 저장\n self.d_ranks = self.data_list[26:36] # Daum 검색어 저장\n\n # self.data_list (3 포털 검색어가 저장된 리스트) 에서 키워드를 하나씩 뽑는다\n for i, keyword in enumerate(self.data_list):\n if i in except_case: # 미리 선언해놓은 except_case면 건너뛴다\n continue\n score = 10 - ( k % 10 ) # score가 10 ~ 1점이 나오도록 한다 => score는 총 30개가 나오는데 10 9 ... 1이 3번 반복된다\n k += 1\n # keywords에 keyword가 없다면 새로 삽입과, score를 계산한다(\n if keyword not in keywords:\n keywords.append(keyword)\n scores.append(score)\n # keywords에 keyword가 있다면 (다른 포털 keyword와 일치하는 경우)\n # 해당 keyword의 index를 계산하여 점수를 더해준다\n else:\n index = keywords.index(keyword)\n scores[index] += (score)\n\n scores, keywords = zip(*sorted(zip(scores, keywords), reverse = True)) # sort together (scores를 기준으로 keywords도 같이 정렬)\n return keywords, scores\n\n # 테스트\n def _top5_engines_score(self, keywords):\n self.g_ranks = self.data_list[2:12] # google 검색어 저장\n self.n_ranks = self.data_list[14:24] # Naver 검색어 저장\n self.d_ranks = self.data_list[26:36] # Daum 검색어 저장\n g_scores = list()\n n_scores = list()\n d_scores = list()\n\n for keyword in keywords:\n if keyword in self.g_ranks:\n g_scores.append(10 - self.g_ranks.index(keyword))\n else:\n g_scores.append(0)\n if keyword in self.n_ranks:\n n_scores.append(10 - self.n_ranks.index(keyword))\n else:\n n_scores.append(0)\n if keyword in self.d_ranks:\n d_scores.append(10 - self.d_ranks.index(keyword))\n else:\n d_scores.append(0)\n\n return n_scores, d_scores, g_scores\n\n # get_keywords_score로 계산한 Top5를 파이차트로 draw\n def _draw(self, keywords, scores, n_scores, d_scores, g_scores):\n explode = [0.07] * 5\n outer_colors = ['#ff6666', '#ffcc99', '#99ff99', '#66b3ff', 'skyblue']\n inner_colors = ['#c2c2f0', '#ff9999', '#ffb3e6'] * 5\n site_ratio = list()\n for i in range(5):\n site_ratio.extend([n_scores[i],d_scores[i],g_scores[i]])\n\n self.keywords_score.canvas.axes.clear()\n self.keywords_score.canvas.axes.pie(scores, labels=keywords, shadow=True,\n startangle=90, colors = outer_colors, explode = explode)\n self.keywords_score.canvas.axes.pie(site_ratio, shadow=True,radius=0.75,\n startangle=90, colors = inner_colors, explode = explode * 3)\n circle = plt.Circle((0, 0), 0.5, color='white')\n self.keywords_score.canvas.axes.add_artist(circle)\n self.keywords_score.canvas.axes.set_title(\"종합 검색어 스코어 Top 5\")\n self.keywords_score.canvas.axes.grid(linewidth=0.2)\n self.keywords_score.canvas.draw()\n\n keywords, scores = _get_keywords_score(self)\n n_scores, d_scores, g_scores = _top5_engines_score(self, keywords)\n _draw(self, keywords[:5], scores[:5], n_scores, d_scores, g_scores)\n\n # 1 - 10위까지 키워드 중 순위 차이가 가장 큰 키워드와 작은 키워드 표시\n def update_keywords(self,ranks1, ranks2,engine1,engine2,loc = 0):\n # 1 - 10위까지 키워드들 중에 중복되는 키워드들의 distance를 계산\n def _get_distances(self, ranks1, ranks2):\n # 중복되는 키워드를 추출\n def _extract_keywords(self, ranks1, ranks2):\n keywords = list()\n for item in ranks1:\n if item in ranks2:\n keywords.append(item)\n return keywords\n # 중복 키워드들의 distance 계산\n def _cal_distance(self, keywords, ranks1, ranks2):\n distances = list()\n for keyword in keywords:\n distances.append(abs(ranks1.index(keyword) - ranks2.index(keyword)))\n return distances\n\n keywords = _extract_keywords(self, ranks1=ranks1, ranks2=ranks2)\n distances = _cal_distance(self, keywords=keywords,ranks1=ranks1,ranks2=ranks2)\n\n return keywords, distances\n\n # get_distances()로 계산한 distance 기준으로 키워드 Set\n def _set_keywords(self, keywords, distances, engine1, engine2, loc = 0):\n # 계산한 distance 기준으로 max_corr, min_corr을 계산\n def _get_max_n_min_corr(keywords, distances):\n # 중복되는 키워드가 없는 경우 '해당없음'으로 표시\n if len(distances) == 0:\n return '해당없음', '해당없음'\n # distance가 가장 작은 키워드가 max_corr, distance가 가장 큰 키워드가 min_corr\n return keywords[(distances.index(min(distances)))], keywords[(distances.index(max(distances)))]\n\n # max_corr, min_corr 키워드 Set\n # 추가로 현재시간 업데이트도 같이 함\n def _set_text(self, max_corr_keyword, min_corr_keyword, engine1, engine2, loc):\n translate = QtCore.QCoreApplication.translate\n self.max_keyword_label[loc].setText(translate(\"MainWindow\", engine1 + \" - \" + engine2 + \" \" + max_corr_keyword))\n self.min_keyword_label[loc].setText(translate(\"MainWindow\", engine1 + \" - \" + engine2 + \" \" + min_corr_keyword))\n self.keyword_comment.setText(translate(\"MainWindow\", \"blue : max distance black : min distance\"))\n now = datetime.now()\n time = str(now.year)\n format_ = [now.month, now.day, now.hour, now.minute]\n delimiters = ['-', '-', ' ', ':']\n for i, item in enumerate(format_):\n time += delimiters[i] + str(item)\n self.time_plot.setText(translate(\"MainWindow\", \"<\" + time + \"> 기준\"))\n\n max_corr_keyword, min_corr_keyword = _get_max_n_min_corr(keywords, distances)\n _set_text(self, max_corr_keyword, min_corr_keyword, engine1=engine1, engine2=engine2, loc=loc)\n\n keywords, distances = _get_distances(self, ranks1=ranks1,ranks2=ranks2)\n _set_keywords(self, keywords=keywords,distances=distances,engine1=engine1,engine2=engine2, loc=loc)\n\n # 상관관계\n def update_corrs(self, ranks1, ranks2, engine1, engine2):\n # ranks1과 ranks2의 상관관계 계산\n # 순위 1-10위까지만을 이용하여 비교를 하고,\n # 매치되지 않는 키워드들도 있으므로 기존 상관관계 계산법에 따르지 않고 새로 정의\n # 상관관계는 보통 -1.0 ~ 1.0 의 값을 가지지만, 검색어 순위의 상관관계에서\n # 음의 상관관계는 나올 수가 없다고 판단하고, 0.0 ~ 1.0 으로 제한을 둠\n # 계산시, N사 1등과 D사 10등이 있다고 하면, 기존 상관관계 계산법으로는 낮은 상관관계가 나오겠지만,\n # 검색어 10위라는 값이 이미 어느 정도 상관관계가 있다고 판단하여, 통상적으로 강한 상관관계라고 판단하기 시작하는 0.3의 값을 부여해줌.\n def _get_corrs(ranks1, ranks2):\n ranks1.reverse()\n ranks2.reverse()\n corrs = list()\n\n for i, keyword1 in enumerate(ranks1):\n corrs.append(0)\n for j, keyword2 in enumerate(ranks2):\n if keyword1 == keyword2:\n # 순위가 같다면 corr == 1\n if i == j:\n corrs[i] = 1.0\n # 랭킹에 있는데 순위가 다르다면\n # Naver는 1위 Daum은 10위라고 한다면, 둘이 상관관계가 어느 정도 있다고 판단하고\n # 통상적으로 어느 정도 상관관계가 있다고 판단하는 0.3을 준다\n # 그 사이의 값들은 0.3 ~ 1.0 까지 중 distance 를 고려하여 corr을 계산한다\n else:\n corrs[i] = ( 1 - (0.7 / 9) * abs(i-j) )\n break\n return corrs\n\n # get_corrs로 계산한 corr들을 plot\n def _draw(self, corrs, engine1='Naver', engine2='Daum'):\n self.corr.canvas.axes.clear()\n self.corr.canvas.axes.scatter([str(x) for x in range(10)], corrs, color='lightcoral', label = \"corr \" + str(round(np.mean(corrs),2)))\n self.corr.canvas.axes.legend(fontsize='medium', loc='upper left')\n self.corr.canvas.axes.set_xticklabels(['Rank10', '.', '.', '.', '.', '.', '.', '.', '.', 'Rank1'])\n self.corr.canvas.axes.set_title(engine1 + ' - ' + engine2 + ' corr')\n self.corr.canvas.axes.grid(linewidth=0.2)\n self.corr.canvas.draw()\n\n\n corrs = _get_corrs(ranks1, ranks2)\n _draw(self, corrs, engine1=engine1, engine2=engine2)\n\n # web_crawling Func Execute code\n multi_crawler = MultiCrawler(self.rank_containers_list, self.queue) # Multi Threading Crawling\n multi_crawler.start() # Multi Thread Run\n multi_crawler.join() # Wait Threads\n update_data(self)\n update_pie(self)\n update_corrs(self, ranks1=self.n_ranks, ranks2=self.d_ranks, engine1='Naver', engine2='Daum')\n engine_list = [['Naver', 'Daum'], ['Daum', 'Google'], ['Google', 'Naver']]\n self.g_ranks.reverse() # 왜 g_ranks가 뒤집어져 있는지 아직 확인 못함\n ranks_list = [[self.n_ranks, self.d_ranks], [self.d_ranks, self.g_ranks], [self.g_ranks, self.n_ranks]]\n for i in range(3):\n update_keywords(self, ranks1=ranks_list[i][0],\n ranks2=ranks_list[i][1], engine1=engine_list[i][0],\n engine2=engine_list[i][1], loc=i)\n\nif __name__ == '__main__':\n app = QtWidgets.QApplication(sys.argv)\n main_window = QtWidgets.QMainWindow()\n process = MainWindow()\n process.setup(main_window)\n main_window.show()\n sys.exit(app.exec_())",
"[2019-11-18 01:19:46,482 remote_connection.py:388 - _request()] POST http://127.0.0.1:56769/session {\"capabilities\": {\"firstMatch\": [{}], \"alwaysMatch\": {\"browserName\": \"chrome\", \"platformName\": \"any\", \"goog:chromeOptions\": {\"extensions\": [], \"args\": []}}}, \"desiredCapabilities\": {\"browserName\": \"chrome\", \"version\": \"\", \"platform\": \"ANY\", \"goog:chromeOptions\": {\"extensions\": [], \"args\": []}}}\n[2019-11-18 01:19:46,487 connectionpool.py:205 - _new_conn()] Starting new HTTP connection (1): 127.0.0.1:56769\n[2019-11-18 01:19:48,890 connectionpool.py:393 - _make_request()] http://127.0.0.1:56769 \"POST /session HTTP/1.1\" 200 685\n[2019-11-18 01:19:48,890 remote_connection.py:440 - _request()] Finished Request\n[2019-11-18 01:19:48,892 remote_connection.py:388 - _request()] POST http://127.0.0.1:56769/session/82c93a8e3b48f1f60d52fc63b7be2ab4/url {\"url\": \"https://trends.google.co.kr/trends/trendingsearches/daily?geo=KR\"}\n[2019-11-18 01:19:55,574 connectionpool.py:393 - _make_request()] http://127.0.0.1:56769 \"POST /session/82c93a8e3b48f1f60d52fc63b7be2ab4/url HTTP/1.1\" 200 14\n[2019-11-18 01:19:55,575 remote_connection.py:440 - _request()] Finished Request\n[2019-11-18 01:19:55,969 __init__.py:415 - wrapper()] $HOME=C:\\Users\\SooHwanKim\n[2019-11-18 01:19:55,971 __init__.py:415 - wrapper()] CONFIGDIR=C:\\Users\\SooHwanKim\\.matplotlib\n[2019-11-18 01:19:55,973 __init__.py:415 - wrapper()] matplotlib data path: C:\\ProgramData\\Anaconda3\\lib\\site-packages\\matplotlib\\mpl-data\n[2019-11-18 01:19:55,981 __init__.py:1093 - rc_params_from_file()] loaded rc file C:\\ProgramData\\Anaconda3\\lib\\site-packages\\matplotlib\\mpl-data\\matplotlibrc\n[2019-11-18 01:19:55,985 __init__.py:1819 - <module>()] matplotlib version 3.0.2\n[2019-11-18 01:19:55,986 __init__.py:1820 - <module>()] interactive is False\n[2019-11-18 01:19:55,987 __init__.py:1821 - <module>()] platform is win32\n[2019-11-18 01:19:55,988 __init__.py:1822 - <module>()] loaded modules: ['sys', 'builtins', '_frozen_importlib', '_imp', '_thread', '_warnings', '_weakref', 'zipimport', '_frozen_importlib_external', '_io', 'marshal', 'nt', 'winreg', 'encodings', 'codecs', '_codecs', 'encodings.aliases', 'encodings.utf_8', '_signal', '__main__', 'encodings.latin_1', 'io', 'abc', '_abc', '_bootlocale', '_locale', 'encodings.cp949', '_codecs_kr', '_multibytecodec', 'site', 'os', 'stat', '_stat', 'ntpath', 'genericpath', 'os.path', '_collections_abc', '_sitebuiltins', 'types', 'importlib', 'importlib._bootstrap', 'importlib._bootstrap_external', 'warnings', 'importlib.util', 'importlib.abc', 'importlib.machinery', 'contextlib', 'collections', 'operator', '_operator', 'keyword', 'heapq', '_heapq', 'itertools', 'reprlib', '_collections', 'functools', '_functools', 'mpl_toolkits', 'google', 'sphinxcontrib', 'runpy', 'pkgutil', 'weakref', '_weakrefset', 'ipykernel', 'ipykernel._version', 'ipykernel.connect', '__future__', 'json', 'json.decoder', 're', 'enum', 'sre_compile', '_sre', 'sre_parse', 'sre_constants', 'copyreg', 'json.scanner', '_json', 'json.encoder', 'subprocess', 'time', 'signal', 'errno', 'threading', 'traceback', 'linecache', 'tokenize', 'token', 'msvcrt', '_winapi', 'IPython', 'IPython.core', 'IPython.core.getipython', 'IPython.core.release', 'IPython.core.application', 'atexit', 'copy', 'glob', 'fnmatch', 'posixpath', 'logging', 'collections.abc', 'string', '_string', 'shutil', 'zlib', 'bz2', '_compression', '_bz2', 'lzma', '_lzma', 'traitlets', 'traitlets.traitlets', 'inspect', 'dis', 'opcode', '_opcode', 'six', 'struct', '_struct', 'traitlets.utils', 'traitlets.utils.getargspec', 'traitlets.utils.importstring', 'ipython_genutils', 'ipython_genutils._version', 'ipython_genutils.py3compat', 'ipython_genutils.encoding', 'locale', 'platform', 'traitlets.utils.sentinel', 'traitlets.utils.bunch', 'traitlets._version', 'traitlets.config', 'traitlets.config.application', 'decorator', 'traitlets.config.configurable', 'traitlets.config.loader', 'argparse', 'gettext', 'ast', '_ast', 'ipython_genutils.path', 'random', 'math', 'hashlib', '_hashlib', '_blake2', '_sha3', 'bisect', '_bisect', '_random', 'ipython_genutils.text', 'textwrap', 'ipython_genutils.importstring', 'IPython.core.crashhandler', 'pprint', 'IPython.core.ultratb', 'pydoc', 'urllib', 'urllib.parse', 'IPython.core.debugger', 'bdb', 'IPython.utils', 'IPython.utils.PyColorize', 'IPython.utils.coloransi', 'IPython.utils.ipstruct', 'IPython.utils.colorable', 'pygments', 'pygments.util', 'IPython.utils.py3compat', 'IPython.utils.encoding', 'IPython.core.excolors', 'IPython.testing', 'IPython.testing.skipdoctest', 'pdb', 'cmd', 'code', 'codeop', 'IPython.core.display_trap', 'IPython.utils.path', 'IPython.utils.process', 'IPython.utils._process_win32', 'ctypes', '_ctypes', 'ctypes._endian', 'ctypes.wintypes', 'IPython.utils._process_common', 'shlex', 'IPython.utils.decorators', 'IPython.utils.data', 'IPython.utils.terminal', 'IPython.utils.sysinfo', 'IPython.utils._sysinfo', 'IPython.core.profiledir', 'IPython.paths', 'tempfile', 'IPython.utils.importstring', 'IPython.terminal', 'IPython.terminal.embed', 'IPython.core.compilerop', 'IPython.core.magic_arguments', 'IPython.core.error', 'IPython.utils.text', 'pathlib', 'IPython.core.magic', 'getopt', 'IPython.core.oinspect', 'IPython.core.page', 'IPython.core.display', 'binascii', 'mimetypes', 'IPython.lib', 'IPython.lib.security', 'getpass', 'IPython.lib.pretty', 'datetime', '_datetime', 'IPython.utils.openpy', 'IPython.utils.dir2', 'IPython.utils.wildcard', 'pygments.lexers', 'pygments.lexers._mapping', 'pygments.modeline', 'pygments.plugin', 'pygments.lexers.python', 'pygments.lexer', 'pygments.filter', 'pygments.filters', 'pygments.token', 'pygments.regexopt', 'pygments.unistring', 'pygments.formatters', 'pygments.formatters._mapping', 'pygments.formatters.html', 'pygments.formatter', 'pygments.styles', 'IPython.core.inputtransformer2', 'typing', 'typing.io', 'typing.re', 'IPython.core.interactiveshell', 'asyncio', 'asyncio.base_events', 'concurrent', 'concurrent.futures', 'concurrent.futures._base', 'socket', '_socket', 'selectors', 'select', 'ssl', '_ssl', 'base64', 'asyncio.constants', 'asyncio.coroutines', 'asyncio.base_futures', 'asyncio.format_helpers', 'asyncio.log', 'asyncio.events', 'contextvars', '_contextvars', 'asyncio.base_tasks', '_asyncio', 'asyncio.futures', 'asyncio.protocols', 'asyncio.sslproto', 'asyncio.transports', 'asyncio.tasks', 'asyncio.locks', 'asyncio.runners', 'asyncio.queues', 'asyncio.streams', 'asyncio.subprocess', 'asyncio.windows_events', '_overlapped', 'asyncio.base_subprocess', 'asyncio.proactor_events', 'asyncio.selector_events', 'asyncio.windows_utils', 'pickleshare', 'pickle', '_compat_pickle', '_pickle', 'IPython.core.prefilter', 'IPython.core.autocall', 'IPython.core.macro', 'IPython.core.splitinput', 'IPython.core.alias', 'IPython.core.builtin_trap', 'IPython.core.events', 'backcall', 'backcall.backcall', 'IPython.core.displayhook', 'IPython.core.displaypub', 'IPython.core.extensions', 'IPython.core.formatters', 'IPython.utils.sentinel', 'IPython.core.history', 'sqlite3', 'sqlite3.dbapi2', '_sqlite3', 'IPython.core.logger', 'IPython.core.payload', 'IPython.core.usage', 'IPython.display', 'IPython.lib.display', 'html', 'html.entities', 'IPython.utils.io', 'IPython.utils.capture', 'IPython.utils.strdispatch', 'IPython.core.hooks', 'IPython.utils.syspathcontext', 'IPython.utils.tempdir', 'IPython.utils.contexts', 'IPython.core.async_helpers', 'IPython.terminal.interactiveshell', 'prompt_toolkit', 'prompt_toolkit.application', 'prompt_toolkit.application.application', 'prompt_toolkit.buffer', 'prompt_toolkit.application.current', 'prompt_toolkit.eventloop', 'prompt_toolkit.eventloop.base', 'prompt_toolkit.log', 'prompt_toolkit.eventloop.coroutine', 'prompt_toolkit.eventloop.defaults', 'prompt_toolkit.utils', 'six.moves', 'wcwidth', 'wcwidth.wcwidth', 'wcwidth.table_wide', 'wcwidth.table_zero', 'prompt_toolkit.cache', 'prompt_toolkit.eventloop.future', 'prompt_toolkit.eventloop.context', 'prompt_toolkit.eventloop.async_generator', 'queue', '_queue', 'six.moves.queue', 'prompt_toolkit.eventloop.event', 'prompt_toolkit.application.run_in_terminal', 'prompt_toolkit.auto_suggest', 'prompt_toolkit.filters', 'prompt_toolkit.filters.base', 'prompt_toolkit.filters.app', 'prompt_toolkit.enums', 'prompt_toolkit.filters.utils', 'prompt_toolkit.filters.cli', 'prompt_toolkit.clipboard', 'prompt_toolkit.clipboard.base', 'prompt_toolkit.selection', 'prompt_toolkit.clipboard.in_memory', 'prompt_toolkit.completion', 'prompt_toolkit.completion.base', 'prompt_toolkit.completion.filesystem', 'prompt_toolkit.completion.word_completer', 'prompt_toolkit.document', 'prompt_toolkit.history', 'prompt_toolkit.search', 'prompt_toolkit.key_binding', 'prompt_toolkit.key_binding.key_bindings', 'prompt_toolkit.keys', 'prompt_toolkit.key_binding.vi_state', 'prompt_toolkit.validation', 'prompt_toolkit.input', 'prompt_toolkit.input.base', 'prompt_toolkit.input.defaults', 'prompt_toolkit.input.typeahead', 'prompt_toolkit.key_binding.bindings', 'prompt_toolkit.key_binding.bindings.page_navigation', 'prompt_toolkit.key_binding.bindings.scroll', 'prompt_toolkit.key_binding.defaults', 'prompt_toolkit.key_binding.bindings.basic', 'prompt_toolkit.key_binding.key_processor', 'prompt_toolkit.key_binding.bindings.named_commands', 'prompt_toolkit.key_binding.bindings.completion', 'prompt_toolkit.key_binding.bindings.emacs', 'prompt_toolkit.key_binding.bindings.vi', 'prompt_toolkit.input.vt100_parser', 'prompt_toolkit.input.ansi_escape_sequences', 'prompt_toolkit.key_binding.digraphs', 'prompt_toolkit.key_binding.bindings.mouse', 'prompt_toolkit.layout', 'prompt_toolkit.layout.containers', 'prompt_toolkit.layout.controls', 'prompt_toolkit.formatted_text', 'prompt_toolkit.formatted_text.base', 'prompt_toolkit.formatted_text.html', 'xml', 'xml.dom', 'xml.dom.domreg', 'xml.dom.minidom', 'xml.dom.minicompat', 'xml.dom.xmlbuilder', 'xml.dom.NodeFilter', 'prompt_toolkit.formatted_text.ansi', 'prompt_toolkit.output', 'prompt_toolkit.output.base', 'prompt_toolkit.layout.screen', 'prompt_toolkit.output.defaults', 'prompt_toolkit.output.color_depth', 'prompt_toolkit.output.vt100', 'prompt_toolkit.styles', 'prompt_toolkit.styles.base', 'prompt_toolkit.styles.defaults', 'prompt_toolkit.styles.style', 'prompt_toolkit.styles.named_colors', 'prompt_toolkit.styles.pygments', 'prompt_toolkit.styles.style_transformation', 'colorsys', 'array', 'prompt_toolkit.formatted_text.pygments', 'prompt_toolkit.formatted_text.utils', 'prompt_toolkit.lexers', 'prompt_toolkit.lexers.base', 'prompt_toolkit.lexers.pygments', 'prompt_toolkit.mouse_events', 'prompt_toolkit.layout.processors', 'prompt_toolkit.layout.utils', 'prompt_toolkit.layout.dimension', 'prompt_toolkit.layout.margins', 'prompt_toolkit.layout.layout', 'prompt_toolkit.layout.menus', 'prompt_toolkit.renderer', 'prompt_toolkit.layout.mouse_handlers', 'prompt_toolkit.key_binding.bindings.cpr', 'prompt_toolkit.key_binding.emacs_state', 'prompt_toolkit.layout.dummy', 'prompt_toolkit.application.dummy', 'prompt_toolkit.shortcuts', 'prompt_toolkit.shortcuts.dialogs', 'prompt_toolkit.key_binding.bindings.focus', 'prompt_toolkit.widgets', 'prompt_toolkit.widgets.base', 'prompt_toolkit.widgets.toolbars', 'prompt_toolkit.widgets.dialogs', 'prompt_toolkit.widgets.menus', 'prompt_toolkit.shortcuts.prompt', 'prompt_toolkit.key_binding.bindings.auto_suggest', 'prompt_toolkit.key_binding.bindings.open_in_editor', 'prompt_toolkit.shortcuts.utils', 'prompt_toolkit.shortcuts.progress_bar', 'prompt_toolkit.shortcuts.progress_bar.base', 'prompt_toolkit.shortcuts.progress_bar.formatters', 'prompt_toolkit.patch_stdout', 'pygments.style', 'IPython.terminal.debugger', 'IPython.core.completer', 'unicodedata', 'IPython.core.latex_symbols', 'IPython.utils.generics', 'jedi', 'jedi.api', 'parso', 'parso.parser', 'parso.tree', 'parso._compatibility', 'parso.pgen2', 'parso.pgen2.generator', 'parso.pgen2.grammar_parser', 'parso.python', 'parso.python.tokenize', 'parso.python.token', 'parso.utils', 'parso.grammar', 'parso.python.diff', 'difflib', 'parso.python.parser', 'parso.python.tree', 'parso.python.prefix', 'parso.cache', 'gc', 'parso.python.errors', 'parso.normalizer', 'parso.python.pep8', 'jedi._compatibility', 'jedi.parser_utils', 'jedi.debug', 'jedi.settings', 'jedi.cache', 'jedi.api.classes', 'jedi.evaluate', 'jedi.evaluate.utils', 'jedi.evaluate.imports', 'jedi.evaluate.sys_path', 'jedi.evaluate.cache', 'jedi.evaluate.base_context', 'jedi.common', 'jedi.common.context', 'jedi.evaluate.helpers', 'jedi.common.utils', 'jedi.evaluate.compiled', 'jedi.evaluate.compiled.context', 'jedi.evaluate.filters', 'jedi.evaluate.flow_analysis', 'jedi.evaluate.recursion', 'jedi.evaluate.lazy_context', 'jedi.evaluate.compiled.access', 'jedi.evaluate.compiled.getattr_static', 'jedi.evaluate.compiled.fake', 'jedi.evaluate.analysis', 'jedi.evaluate.context', 'jedi.evaluate.context.module', 'jedi.evaluate.context.klass', 'jedi.evaluate.context.function', 'jedi.evaluate.docstrings', 'jedi.evaluate.pep0484', 'jedi.evaluate.arguments', 'jedi.evaluate.context.iterable', 'jedi.evaluate.param', 'jedi.evaluate.context.asynchronous', 'jedi.evaluate.parser_cache', 'jedi.evaluate.context.instance', 'jedi.evaluate.syntax_tree', 'jedi.evaluate.finder', 'jedi.api.keywords', 'pydoc_data', 'pydoc_data.topics', 'jedi.api.interpreter', 'jedi.evaluate.compiled.mixed', 'jedi.api.helpers', 'jedi.api.completion', 'jedi.api.environment', 'filecmp', 'jedi.evaluate.compiled.subprocess', 'jedi.evaluate.compiled.subprocess.functions', 'jedi.api.exceptions', 'jedi.api.project', 'jedi.evaluate.usages', 'IPython.terminal.ptutils', 'IPython.terminal.shortcuts', 'IPython.lib.clipboard', 'IPython.terminal.magics', 'IPython.terminal.pt_inputhooks', 'IPython.terminal.prompts', 'IPython.terminal.ipapp', 'IPython.core.magics', 'IPython.core.magics.auto', 'IPython.core.magics.basic', 'IPython.core.magics.code', 'urllib.request', 'email', 'http', 'http.client', 'email.parser', 'email.feedparser', 'email.errors', 'email._policybase', 'email.header', 'email.quoprimime', 'email.base64mime', 'email.charset', 'email.encoders', 'quopri', 'email.utils', 'email._parseaddr', 'calendar', 'email.message', 'uu', 'email._encoded_words', 'email.iterators', 'urllib.error', 'urllib.response', 'nturl2path', 'IPython.core.magics.config', 'IPython.core.magics.display', 'IPython.core.magics.execution', 'timeit', 'cProfile', '_lsprof', 'profile', 'pstats', 'IPython.utils.module_paths', 'IPython.utils.timing', 'IPython.core.magics.extension', 'IPython.core.magics.history', 'IPython.core.magics.logging', 'IPython.core.magics.namespace', 'IPython.core.magics.osm', 'IPython.core.magics.pylab', 'IPython.core.pylabtools', 'IPython.core.magics.script', 'IPython.lib.backgroundjobs', 'IPython.core.shellapp', 'IPython.extensions', 'IPython.extensions.storemagic', 'IPython.utils.frame', 'jupyter_client', 'jupyter_client._version', 'jupyter_client.connect', 'zmq', 'zmq.backend', 'zmq.backend.select', 'zmq.backend.cython', 'cython_runtime', 'zmq.backend.cython.constants', '_cython_0_28_5', 'zmq.backend.cython.error', 'zmq.backend.cython.message', 'zmq.error', 'zmq.backend.cython.context', 'zmq.backend.cython.socket', 'zmq.backend.cython.utils', 'zmq.backend.cython._poll', 'zmq.backend.cython._version', 'zmq.backend.cython._device', 'zmq.sugar', 'zmq.sugar.constants', 'zmq.utils', 'zmq.utils.constant_names', 'zmq.sugar.context', 'zmq.sugar.attrsettr', 'zmq.sugar.socket', 'zmq.sugar.poll', 'zmq.utils.jsonapi', 'zmq.utils.strtypes', 'zmq.sugar.frame', 'zmq.sugar.tracker', 'zmq.sugar.version', 'zmq.sugar.stopwatch', 'jupyter_client.localinterfaces', 'jupyter_core', 'jupyter_core.version', 'jupyter_core.paths', 'jupyter_client.launcher', 'traitlets.log', 'jupyter_client.client', 'jupyter_client.channels', 'jupyter_client.channelsabc', 'jupyter_client.clientabc', 'jupyter_client.manager', 'jupyter_client.kernelspec', 'jupyter_client.managerabc', 'jupyter_client.blocking', 'jupyter_client.blocking.client', 'jupyter_client.blocking.channels', 'jupyter_client.multikernelmanager', 'uuid', 'ipykernel.kernelapp', 'tornado', 'tornado.ioloop', 'numbers', 'tornado.concurrent', 'tornado.log', 'logging.handlers', 'tornado.escape', 'tornado.util', 'tornado.speedups', 'colorama', 'colorama.initialise', 'colorama.ansitowin32', 'colorama.ansi', 'colorama.winterm', 'colorama.win32', 'tornado.stack_context', 'tornado.platform', 'tornado.platform.auto', 'tornado.platform.common', 'tornado.platform.interface', 'tornado.platform.windows', 'concurrent.futures.thread', 'zmq.eventloop', 'zmq.eventloop.ioloop', 'tornado.platform.asyncio', 'tornado.gen', 'zmq.eventloop.zmqstream', 'ipykernel.iostream', 'imp', 'jupyter_client.session', 'hmac', 'jupyter_client.jsonutil', 'dateutil', 'dateutil._version', 'dateutil.parser', 'dateutil.parser._parser', 'decimal', '_decimal', 'dateutil.relativedelta', 'dateutil._common', 'dateutil.tz', 'dateutil.tz.tz', 'dateutil.tz._common', 'dateutil.tz._factories', 'dateutil.tz.win', 'dateutil.parser.isoparser', '_strptime', 'jupyter_client.adapter', 'ipykernel.heartbeat', 'ipykernel.ipkernel', 'IPython.utils.tokenutil', 'ipykernel.comm', 'ipykernel.comm.manager', 'ipykernel.comm.comm', 'ipykernel.kernelbase', 'tornado.queues', 'tornado.locks', 'ipykernel.jsonutil', 'ipykernel.zmqshell', 'IPython.core.payloadpage', 'ipykernel.displayhook', 'ipykernel.parentpoller', 'faulthandler', 'ipykernel.datapub', 'ipykernel.serialize', 'ipykernel.pickleutil', 'ipykernel.codeutil', 'IPython.core.completerlib', 'storemagic', 'ipywidgets', 'ipywidgets._version', 'ipywidgets.widgets', 'ipywidgets.widgets.widget', 'ipywidgets.widgets.domwidget', 'ipywidgets.widgets.trait_types', 'ipywidgets.widgets.widget_layout', 'ipywidgets.widgets.widget_style', 'ipywidgets.widgets.valuewidget', 'ipywidgets.widgets.widget_core', 'ipywidgets.widgets.widget_bool', 'ipywidgets.widgets.widget_description', 'ipywidgets.widgets.widget_button', 'ipywidgets.widgets.widget_box', 'ipywidgets.widgets.docutils', 'ipywidgets.widgets.widget_float', 'ipywidgets.widgets.widget_int', 'ipywidgets.widgets.widget_color', 'ipywidgets.widgets.widget_date', 'ipywidgets.widgets.widget_output', 'ipywidgets.widgets.widget_selection', 'ipywidgets.widgets.widget_selectioncontainer', 'ipywidgets.widgets.widget_string', 'ipywidgets.widgets.widget_controller', 'ipywidgets.widgets.interaction', 'ipywidgets.widgets.widget_link', 'ipywidgets.widgets.widget_media', 'PyQt5', 'sip', 'PyQt5.sip', 'PyQt5.QtCore', 'PyQt5.QtGui', 'PyQt5.QtWidgets', 'PyQt5.QtDBus', 'PyQt5.QtNetwork', 'PyQt5.QtNetworkAuth', 'PyQt5.QtSensors', 'PyQt5.QtSerialPort', 'PyQt5.QtMultimedia', 'PyQt5.QtQml', 'PyQt5.QtXml', 'PyQt5.QtXmlPatterns', 'PyQt5.QtDesigner', 'PyQt5.QtHelp', 'PyQt5.QtMultimediaWidgets', 'PyQt5.QtOpenGL', 'PyQt5.QtPrintSupport', 'PyQt5.QtQuick', 'PyQt5.QtSql', 'PyQt5.QtSvg', 'PyQt5.QtTest', 'PyQt5.QtBluetooth', 'PyQt5.QtPositioning', 'PyQt5.QtWinExtras', 'PyQt5.QtQuickWidgets', 'PyQt5.QtWebSockets', 'PyQt5.QtWebChannel', 'PyQt5.QtLocation', 'PyQt5.QtNfc', 'PyQt5.QtRemoteObjects', 'PyQt5.Qt', 'multi', 'bs4', 'bs4.builder', 'bs4.element', 'bs4.dammit', 'chardet', 'chardet.compat', 'chardet.universaldetector', 'chardet.charsetgroupprober', 'chardet.enums', 'chardet.charsetprober', 'chardet.escprober', 'chardet.codingstatemachine', 'chardet.escsm', 'chardet.latin1prober', 'chardet.mbcsgroupprober', 'chardet.utf8prober', 'chardet.mbcssm', 'chardet.sjisprober', 'chardet.mbcharsetprober', 'chardet.chardistribution', 'chardet.euctwfreq', 'chardet.euckrfreq', 'chardet.gb2312freq', 'chardet.big5freq', 'chardet.jisfreq', 'chardet.jpcntx', 'chardet.eucjpprober', 'chardet.gb2312prober', 'chardet.euckrprober', 'chardet.cp949prober', 'chardet.big5prober', 'chardet.euctwprober', 'chardet.sbcsgroupprober', 'chardet.sbcharsetprober', 'chardet.langcyrillicmodel', 'chardet.langgreekmodel', 'chardet.langbulgarianmodel', 'chardet.langthaimodel', 'chardet.langhebrewmodel', 'chardet.hebrewprober', 'chardet.langturkishmodel', 'chardet.version', 'bs4.builder._htmlparser', 'html.parser', '_markupbase', 'bs4.builder._html5lib', 'html5lib', 'html5lib.html5parser', 'html5lib._inputstream', 'six.moves.urllib', 'webencodings', 'webencodings.labels', 'encodings.utf_16_le', 'encodings.utf_16_be', 'html5lib.constants', 'html5lib._utils', 'xml.etree', 'xml.etree.cElementTree', 'xml.etree.ElementTree', 'xml.etree.ElementPath', 'pyexpat.errors', 'pyexpat.model', 'pyexpat', '_elementtree', 'html5lib._tokenizer', 'html5lib._trie', 'html5lib._trie.py', 'html5lib._trie._base', 'html5lib.treebuilders', 'html5lib.treebuilders.base', 'html5lib.treewalkers', 'html5lib.serializer', 'xml.sax', 'xml.sax.xmlreader', 'xml.sax.handler', 'xml.sax._exceptions', 'xml.sax.saxutils', 'bs4.builder._lxml', 'lxml', 'lxml.etree', 'lxml._elementpath', 'gzip', 'selenium', 'selenium.webdriver', 'selenium.webdriver.firefox', 'selenium.webdriver.firefox.webdriver', 'selenium.webdriver.common', 'selenium.webdriver.common.desired_capabilities', 'selenium.webdriver.remote', 'selenium.webdriver.remote.webdriver', 'selenium.webdriver.remote.command', 'selenium.webdriver.remote.webelement', 'zipfile', 'selenium.common', 'selenium.common.exceptions', 'selenium.webdriver.common.by', 'selenium.webdriver.common.utils', 'selenium.webdriver.common.keys', 'selenium.webdriver.remote.remote_connection', 'urllib3', 'urllib3.connectionpool', 'urllib3.exceptions', 'urllib3.packages', 'urllib3.packages.ssl_match_hostname', 'urllib3.packages.six', 'urllib3.packages.six.moves', 'urllib3.packages.six.moves.http_client', 'urllib3.connection', 'urllib3.util', 'urllib3.util.connection', 'urllib3.util.wait', 'urllib3.contrib', 'urllib3.contrib._appengine_environ', 'urllib3.util.request', 'urllib3.util.response', 'urllib3.util.ssl_', 'urllib3.util.timeout', 'urllib3.util.retry', 'urllib3.util.url', 'urllib3._collections', 'urllib3.request', 'urllib3.filepost', 'urllib3.fields', 'urllib3.packages.six.moves.urllib', 'urllib3.packages.six.moves.urllib.parse', 'urllib3.response', 'urllib3.util.queue', 'urllib3.poolmanager', 'selenium.webdriver.remote.errorhandler', 'selenium.webdriver.remote.utils', 'selenium.webdriver.remote.switch_to', 'selenium.webdriver.common.alert', 'selenium.webdriver.remote.mobile', 'selenium.webdriver.remote.file_detector', 'selenium.webdriver.common.html5', 'selenium.webdriver.common.html5.application_cache', 'selenium.webdriver.firefox.extension_connection', 'selenium.webdriver.firefox.firefox_binary', 'selenium.webdriver.firefox.firefox_profile', 'selenium.webdriver.common.proxy', 'selenium.webdriver.firefox.options', 'selenium.webdriver.firefox.remote_connection', 'selenium.webdriver.firefox.service', 'selenium.webdriver.common.service', 'selenium.webdriver.firefox.webelement', 'selenium.webdriver.chrome', 'selenium.webdriver.chrome.webdriver', 'selenium.webdriver.chrome.remote_connection', 'selenium.webdriver.chrome.service', 'selenium.webdriver.chrome.options', 'selenium.webdriver.ie', 'selenium.webdriver.ie.webdriver', 'selenium.webdriver.ie.service', 'selenium.webdriver.ie.options', 'selenium.webdriver.edge', 'selenium.webdriver.edge.webdriver', 'selenium.webdriver.edge.service', 'selenium.webdriver.opera', 'selenium.webdriver.opera.webdriver', 'selenium.webdriver.opera.options', 'selenium.webdriver.safari', 'selenium.webdriver.safari.webdriver', 'selenium.webdriver.safari.service', 'selenium.webdriver.safari.remote_connection', 'selenium.webdriver.blackberry', 'selenium.webdriver.blackberry.webdriver', 'selenium.webdriver.support', 'selenium.webdriver.support.ui', 'selenium.webdriver.support.select', 'selenium.webdriver.support.wait', 'selenium.webdriver.phantomjs', 'selenium.webdriver.phantomjs.webdriver', 'selenium.webdriver.phantomjs.service', 'selenium.webdriver.android', 'selenium.webdriver.android.webdriver', 'selenium.webdriver.webkitgtk', 'selenium.webdriver.webkitgtk.webdriver', 'selenium.webdriver.webkitgtk.service', 'selenium.webdriver.webkitgtk.options', 'selenium.webdriver.common.action_chains', 'selenium.webdriver.common.actions', 'selenium.webdriver.common.actions.action_builder', 'selenium.webdriver.common.actions.interaction', 'selenium.webdriver.common.actions.key_actions', 'selenium.webdriver.common.actions.key_input', 'selenium.webdriver.common.actions.input_device', 'selenium.webdriver.common.actions.pointer_actions', 'selenium.webdriver.common.actions.mouse_button', 'selenium.webdriver.common.actions.pointer_input', 'selenium.webdriver.common.touch_actions', 'encodings.idna', 'stringprep', 'matplotlink', 'matplotlib', 'distutils', 'distutils.version', 'matplotlib.cbook', 'numpy', 'numpy._globals', 'numpy.__config__', 'numpy.version', 'numpy._import_tools', 'numpy.add_newdocs', 'numpy.lib', 'numpy.lib.info', 'numpy.lib.type_check', 'numpy.core', 'numpy.core.info', 'numpy.core.multiarray', 'numpy.core.umath', 'numpy.core._internal', 'numpy.compat', 'numpy.compat._inspect', 'numpy.compat.py3k', 'numpy.core.numerictypes', 'numpy.core.numeric', 'numpy.core.fromnumeric', 'numpy.core._methods', 'numpy.core.arrayprint', 'numpy.core.defchararray', 'numpy.core.records', 'numpy.core.memmap', 'numpy.core.function_base', 'numpy.core.machar', 'numpy.core.getlimits', 'numpy.core.shape_base', 'numpy.core.einsumfunc', 'numpy.testing', 'unittest', 'unittest.result', 'unittest.util', 'unittest.case', 'unittest.suite', 'unittest.loader', 'unittest.main', 'unittest.runner', 'unittest.signals', 'numpy.testing._private', 'numpy.testing._private.utils', 'numpy.lib.utils', 'numpy.testing._private.decorators', 'numpy.testing._private.nosetester', 'numpy.testing._private.pytesttester', 'numpy.lib.ufunclike', 'numpy.lib.index_tricks', 'numpy.lib.function_base', 'numpy.lib.twodim_base', 'numpy.lib.histograms', 'numpy.matrixlib', 'numpy.matrixlib.defmatrix', 'numpy.linalg', 'numpy.linalg.info', 'numpy.linalg.linalg', 'numpy.linalg.lapack_lite', 'numpy.linalg._umath_linalg', 'numpy.lib.stride_tricks', 'numpy.lib.mixins', 'numpy.lib.nanfunctions', 'numpy.lib.shape_base', 'numpy.lib.scimath', 'numpy.lib.polynomial', 'numpy.lib.arraysetops', 'numpy.lib.npyio', 'numpy.lib.format', 'numpy.lib._datasource', 'numpy.lib._iotools', 'numpy.lib.financial', 'numpy.lib.arrayterator', 'numpy.lib.arraypad', 'numpy.lib._version', 'numpy.core._multiarray_tests', 'numpy._distributor_init', 'numpy._mklinit', 'numpy.fft', 'numpy.fft.info', 'numpy.fft.fftpack', 'numpy.fft.fftpack_lite', 'numpy.fft.helper', 'mkl_fft', 'mkl_fft._pydfti', '_cython_0_29', 'mkl_fft._version', 'mkl_fft._numpy_fft', 'numpy.polynomial', 'numpy.polynomial.polynomial', 'numpy.polynomial.polyutils', 'numpy.polynomial._polybase', 'numpy.polynomial.chebyshev', 'numpy.polynomial.legendre', 'numpy.polynomial.hermite', 'numpy.polynomial.hermite_e', 'numpy.polynomial.laguerre', 'numpy.random', 'numpy.random.info', 'numpy.random.mtrand', 'mtrand', 'numpy.ctypeslib', 'numpy.ma', 'numpy.ma.core', 'numpy.ma.extras', 'matplotlib.cbook.deprecation', 'matplotlib.rcsetup', 'matplotlib.fontconfig_pattern', 'pyparsing', 'matplotlib.colors', 'matplotlib._color_data', 'cycler', 'matplotlib._version']\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbaf0b0aafb8c1e5099bdb00232aecf74158b299
| 17,568 |
ipynb
|
Jupyter Notebook
|
snake_puzzle_solver.ipynb
|
ncw/snake-puzzle
|
cfa7c68bc27b50759ba6928a42b58e43a3beefc9
|
[
"MIT"
] | 6 |
2016-09-25T20:13:50.000Z
|
2021-04-10T14:08:48.000Z
|
snake_puzzle_solver.ipynb
|
ncw/snake-puzzle
|
cfa7c68bc27b50759ba6928a42b58e43a3beefc9
|
[
"MIT"
] | null | null | null |
snake_puzzle_solver.ipynb
|
ncw/snake-puzzle
|
cfa7c68bc27b50759ba6928a42b58e43a3beefc9
|
[
"MIT"
] | null | null | null | 26.104012 | 304 | 0.504098 |
[
[
[
"My family know I like puzzles so they gave me this one recently:\n\n\n\nWhen you take it out the box it looks like this:\n\n\n\nAnd very soon after it looked like this (which explains why I've christened the puzzle \"the snake puzzle\"):\n\n\n\nThe way it works is that there is a piece of elastic running through each block. On the majority of the blocks the elastic runs straight through, but on some of the it goes through a 90 degree bend. The puzzle is trying to make it back into a cube.\n\nAfter playing with it a while, I realised that it really is quite hard so I decided to write a program to solve it.\n\nThe first thing to do is find a representation for the puzzle. Here is the one I chose.",
"_____no_output_____"
]
],
[
[
"# definition - number of straight bits, before 90 degree bend\nsnake = [3,2,2,2,1,1,1,2,2,1,1,2,1,2,1,1,2]\nassert sum(snake) == 27",
"_____no_output_____"
]
],
[
[
"If you look at the picture of it above where it is flattened you can see where the numbers came from. Start from the right hand side.\n\nThat also gives us a way of calculating how many combinations there are. At each 90 degree joint, there are 4 possible rotations (ignoring the rotations of the 180 degree blocks) so there are",
"_____no_output_____"
]
],
[
[
"4**len(snake)",
"_____no_output_____"
]
],
[
[
"17 billion combinations. That will include some rotations and reflections, but either way it is a big number.\n\nHowever it is very easy to know when you've gone wrong with this kind of puzzle - as soon as you place a piece outside of the boundary of the 3x3x3 block you know it is wrong and should try something different.\n\nSo how to represent the solution? The way I've chosen is to represent it as a 5x5x5 cube. This is larger than it needs to be but if we fill in the edges then we don't need to do any complicated comparisons to see if a piece is out of bounds. This is a simple trick but it saves a lot of code.\n\nI've also chosen to represent the 3d structure not as a 3d array but as a 1D array (or `list` in python speak) of length 5*5*5 = 125.\n\nTo move in the `x` direction you add 1, to move in the `y` direction you add 5 and to move in the `z` direction you move 25. This simplifies the logic of the solver considerably - we don't need to deal with vectors.\n\nThe basic definitions of the cube look like this:",
"_____no_output_____"
]
],
[
[
"N = 5\nxstride=1 # number of pieces to move in the x direction\nystride=N # number of pieces to move in the y direction\nzstride=N*N # number of pieces to move in the z direction",
"_____no_output_____"
]
],
[
[
"In our `list` we will represent empty space with `0` and space which can't be used with `-1`.",
"_____no_output_____"
]
],
[
[
"empty = 0",
"_____no_output_____"
]
],
[
[
"Now define the empty cube with the boundary round the edges.",
"_____no_output_____"
]
],
[
[
"# Define cube as 5 x 5 x 5 with filled in edges but empty middle for\n# easy edge detection\ntop = [-1]*N*N\nmiddle = [-1]*5 + [-1,0,0,0,-1]*3 + [-1]*5\ncube = top + middle*3 + top",
"_____no_output_____"
]
],
[
[
"We're going to want a function to turn `x, y, z` co-ordinates into an index in the `cube` list.",
"_____no_output_____"
]
],
[
[
"def pos(x, y, z):\n \"\"\"Convert x,y,z into position in cube list\"\"\"\n return x+y*ystride+z*zstride",
"_____no_output_____"
]
],
[
[
"So let's see what that cube looks like...",
"_____no_output_____"
]
],
[
[
"def print_cube(cube, margin=1):\n \"\"\"Print the cube\"\"\"\n for z in range(margin,N-margin):\n for y in range(margin,N-margin):\n for x in range(margin,N-margin):\n v = cube[pos(x,y,z)]\n if v == 0:\n s = \" . \"\n else:\n s = \"%02d \" % v\n print(s, sep=\"\", end=\"\")\n print()\n print()\n\nprint_cube(cube, margin = 0)",
"-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n\n-1 -1 -1 -1 -1 \n-1 . . . -1 \n-1 . . . -1 \n-1 . . . -1 \n-1 -1 -1 -1 -1 \n\n-1 -1 -1 -1 -1 \n-1 . . . -1 \n-1 . . . -1 \n-1 . . . -1 \n-1 -1 -1 -1 -1 \n\n-1 -1 -1 -1 -1 \n-1 . . . -1 \n-1 . . . -1 \n-1 . . . -1 \n-1 -1 -1 -1 -1 \n\n-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n-1 -1 -1 -1 -1 \n\n"
]
],
[
[
"Normally we'll print it without the margin.\n\nNow let's work out how to place a segment.\n\nAssuming that the last piece was placed at `position` we want to place a segment of `length` in `direction`. Note the `assert` to check we aren't placing stuff on top of previous things, or out of the edges.",
"_____no_output_____"
]
],
[
[
"def place(cube, position, direction, length, piece_number):\n \"\"\"Place a segment in the cube\"\"\"\n for _ in range(length):\n position += direction\n assert cube[position] == empty\n cube[position] = piece_number\n piece_number += 1\n return position",
"_____no_output_____"
]
],
[
[
"Let's just try placing some segments and see what happens.",
"_____no_output_____"
]
],
[
[
"cube2 = cube[:] # copy the cube\nplace(cube2, pos(0,1,1), xstride, 3, 1)\nprint_cube(cube2)",
"01 02 03 \n . . . \n . . . \n\n . . . \n . . . \n . . . \n\n . . . \n . . . \n . . . \n\n"
],
[
"place(cube2, pos(3,1,1), ystride, 2, 4)\nprint_cube(cube2)",
"01 02 03 \n . . 04 \n . . 05 \n\n . . . \n . . . \n . . . \n\n . . . \n . . . \n . . . \n\n"
],
[
"place(cube2, pos(3,3,1), zstride, 2, 6)\nprint_cube(cube2)",
"01 02 03 \n . . 04 \n . . 05 \n\n . . . \n . . . \n . . 06 \n\n . . . \n . . . \n . . 07 \n\n"
]
],
[
[
"The next thing we'll need is to undo a place. You'll see why in a moment.",
"_____no_output_____"
]
],
[
[
"def unplace(cube, position, direction, length):\n \"\"\"Remove a segment from the cube\"\"\"\n for _ in range(length):\n position += direction\n cube[position] = empty",
"_____no_output_____"
],
[
"unplace(cube2, pos(3,3,1), zstride, 2)\nprint_cube(cube2)",
"01 02 03 \n . . 04 \n . . 05 \n\n . . . \n . . . \n . . . \n\n . . . \n . . . \n . . . \n\n"
]
],
[
[
"Now let's write a function which returns whether a move is valid given a current `position` and a `direction` and a `length` of the segment we are trying to place.",
"_____no_output_____"
]
],
[
[
"def is_valid(cube, position, direction, length):\n \"\"\"Returns True if a move is valid\"\"\"\n for _ in range(length):\n position += direction\n if cube[position] != empty:\n return False\n return True",
"_____no_output_____"
],
[
"is_valid(cube2, pos(3,3,1), zstride, 2)",
"_____no_output_____"
],
[
"is_valid(cube2, pos(3,3,1), zstride, 3)",
"_____no_output_____"
]
],
[
[
"Given `is_valid` it is now straight forward to work out what moves are possible at a given time, given a `cube` with a `position`, a `direction` and a `length` we are trying to place.",
"_____no_output_____"
]
],
[
[
"# directions next piece could go in\ndirections = [xstride, -xstride, ystride, -ystride, zstride, -zstride]\n\ndef moves(cube, position, direction, length):\n \"\"\"Returns the valid moves for the current position\"\"\"\n valid_moves = []\n for new_direction in directions:\n # Can't carry on in same direction, or the reverse of the same direction\n if new_direction == direction or new_direction == -direction:\n continue\n if is_valid(cube, position, new_direction, length):\n valid_moves.append(new_direction)\n return valid_moves",
"_____no_output_____"
],
[
"moves(cube2, pos(3,3,1), ystride, 2)",
"_____no_output_____"
]
],
[
[
"So that is telling us that you can insert a segment of length 2 using a direction of `-xstride` or `zstride`. If you look at previous `print_cube()` output you'll see those are the only possible moves.\n\nNow we have all the bits to build a recursive solver.",
"_____no_output_____"
]
],
[
[
"def solve(cube, position, direction, snake, piece_number):\n \"\"\"Recursive cube solver\"\"\"\n if len(snake) == 0:\n print(\"Solution\")\n print_cube(cube)\n return\n length, snake = snake[0], snake[1:]\n valid_moves = moves(cube, position, direction, length)\n for new_direction in valid_moves:\n new_position = place(cube, position, new_direction, length, piece_number)\n solve(cube, new_position, new_direction, snake, piece_number+length)\n unplace(cube, position, new_direction, length)",
"_____no_output_____"
]
],
[
[
"This works by being passed in the `snake` of moves left. If there are no moves left then it must be solved, so we print the solution. Otherwise it takes the head off the `snake` with `length, snake = snake[0], snake[1:]` and makes the list of valid moves of that `length`.\n\nThen we `place` each move, and try to `solve` that cube using a recursive call to `solve`. We `unplace` the move so we can try again.\n\nThis very quickly runs through all the possible solutions.",
"_____no_output_____"
]
],
[
[
"# Start just off the side\nposition = pos(0,1,1)\ndirection = xstride\nlength = snake[0]\n# Place the first segment along one edge - that is the only possible place it can go\nposition = place(cube, position, direction, length, 1)\n# Now solve!\nsolve(cube, position, direction, snake[1:], length+1)",
"Solution\n01 02 03 \n20 21 04 \n07 06 05 \n\n16 15 14 \n19 22 13 \n08 11 12 \n\n17 24 25 \n18 23 26 \n09 10 27 \n\nSolution\n01 02 03 \n16 15 14 \n17 24 25 \n\n20 21 04 \n19 22 13 \n18 23 26 \n\n07 06 05 \n08 11 12 \n09 10 27 \n\n"
]
],
[
[
"Wow! It came up with 2 solutions! However they are the same solution just rotated and reflected.\n\nBut how do you use the solution? Starting from the correct end of the snake, place each piece into its corresponding number. Take the first layer of the solution as being the bottom (or top - whatever is easiest), the next layer is the middle and the one after the top.\n\n\n\nAfter a bit of fiddling around you'll get...\n\n",
"_____no_output_____"
],
[
"I hope you enjoyed that introduction to puzzle solving with computer.\n\nIf you want to try one yourselves, use the same technique to solve solitaire.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbaf144c95c09963dcc41fb14b012ca9f2e4be73
| 11,783 |
ipynb
|
Jupyter Notebook
|
Optical Flow based tracking.ipynb
|
ijzepeda/MIA
|
e34a2fe829dcad0e3d3f436eb217d127a6ea1380
|
[
"MIT"
] | null | null | null |
Optical Flow based tracking.ipynb
|
ijzepeda/MIA
|
e34a2fe829dcad0e3d3f436eb217d127a6ea1380
|
[
"MIT"
] | null | null | null |
Optical Flow based tracking.ipynb
|
ijzepeda/MIA
|
e34a2fe829dcad0e3d3f436eb217d127a6ea1380
|
[
"MIT"
] | null | null | null | 42.847273 | 306 | 0.506153 |
[
[
[
"Popular technique in compter vision, it uses image features ponts to track an object. individial feature points are tracked across succesive frames in live video\nlos vectores se llaman, Motion Vectors.\n El metodo Lucas-Kanade es el mas popular, \nFirst step, extract the features points from the current frame. for each point that is extracted, a 3x3 patch is created with the feature point at the center. we are assuming that all the points in each patch have a similarmotion. the size of this window can be adjusted depending on the situacion\n\n",
"_____no_output_____"
],
[
"import cv2\nimport numpy as np\n\n#Define a function to track the object\ndef start_tracking():\n #initialize the video capture object\n cap=cv2.VideoCapture(0)\n #Define the scaling factor for the frames\n scaling_factor = 0.5\n #number of frames to track\n num_frames_to_track = 5\n #skipping factor\n num_frames_jump = 2\n #initialize variables\n tracking_paths=[]\n frame_index = 0\n #Define tracking parameters\n tracking_params = dict(winSize = (11,11), maxLevel = 2, criteria = (cv2.TERM_CRITERIA_EPS | cv2.TERM_CRITERIA_COUNT,10,0.03))\n #Iterate until the user hits esc key\n while True:\n #capture the current frame\n _, frame = cap.read()\n #resize the frame\n frame=cv2.resize(frame, None, fx=scaling_factor, fy=scaling_factor, interpolation =cv2.INTER_AREA)\n #Convert to grayscale\n frame_gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)\n #Create a copy o the frame\n output_img= frame.copy()\n if len(tracking_paths)>0:\n #Get images\n prev_img, current_img = prev_gray, frame_gray\n #organize the featyre points\n feature_points_0= np.float32([tp[-1] for tp in tracking_paths]).reshape(-1,1,2)\n #Compute optical flow\n feature_points_1, _, _ = cv2.calcOpticalFlowPyrLK(prev_img, current_img, feature_points_0, NOne, **tracking_params)\n #compute reverse optical flow\n feature_points_0_rev, _, _ = cv2.calcOpticalFlowPyrLK(curent_img, prev_img, feature_points_1, None, **tracking_params)\n #compute the difference between dorward and reverse optical flow\n diff_feature_points = abs(feature_points_0 - feature_points_0_rev).reshape(-1,2).max(-1)\n #extrack the good points\n good_points = diff_feature_points <1\n #initialize variables\n new_tracking_paths = []\n #iterate throuhj all the good feature points\n for tp, (x,y), good_points_flag in zip(tracking_paths, feature_points_1.reshape(-1,2), good_points):\n #if the flag is not true, then continue\n if not good_points_flag:\n continue\n #Append the X and Y coordinates and check if its length grater than the threshild\n tp.append((x,y))\n if len(tp) > num_frames_to_track:\n del tp[0]\n \n new_tracking_paths.append(tp)\n #draw a circle around the feature points\n cv2.circle(output_img,(x,y), 3, (0,255,0),-1)\n #update the tracking paths\n tracking_paths = new_tracking_paths\n \n #draw lines\n cv2.polylines(output_img, [np.int32(tp) for tp in tracking_paths], False, (0,150,0))\n #Go into this if condition after skipping the right number of frames right number of frames\n if not frame_index % num_frames_jump:\n #Create a mask and draw the circles\n mask - np.zeros_like(frame_gray)\n mask[:] = 255\n for x, y in [np.int32(tp[-1]) for tp in tracking_paths]:\n cv2.circle(mask, (x,y), 6, 0 ,-1)\n #compute good features to track\n feature_points = cv2.goodFeaturesToTrack(frame_gray, mask= mask, maxCorners=500, qualityLevel = 0.3, minDistance = 7, blockSize=7)\n #check if feature points exists. if so, append them to the tracking paths\n if feature_points is not None:\n for x, y in np.float32(feature_points).reshape(-1,2):\n tracking_pathsappend([(x,y)])\n #update variables\n frame_index +=1\n prev_gray = frame_gray\n #display output\n cv2.imshow(\"optical flow\", output_img)\n #check if the user hit esc key\n c = cv2.waitKey(1)\n if c ==27:\n break\n \nif __name__==\"__init__\":\n #Start the tracker\n start_tracking()\n #Close all the windows\n cv2.destroyAllWindows()\n ",
"_____no_output_____"
]
],
[
[
"import cv2\nimport numpy as np\n\n# Define a function to track the object\ndef start_tracking():\n # Initialize the video capture object\n cap = cv2.VideoCapture(0)\n\n # Define the scaling factor for the frames\n scaling_factor = 0.5\n\n # Number of frames to track\n num_frames_to_track = 5\n\n # Skipping factor\n num_frames_jump = 2\n\n # Initialize variables\n tracking_paths = []\n frame_index = 0\n\n # Define tracking parameters\n tracking_params = dict(winSize = (11, 11), maxLevel = 2,\n criteria = (cv2.TERM_CRITERIA_EPS | cv2.TERM_CRITERIA_COUNT, \n 10, 0.03))\n\n # Iterate until the user hits the 'Esc' key\n while True:\n # Capture the current frame\n _, frame = cap.read()\n\n # Resize the frame\n frame = cv2.resize(frame, None, fx=scaling_factor, \n fy=scaling_factor, interpolation=cv2.INTER_AREA)\n\n # Convert to grayscale\n frame_gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)\n\n # Create a copy of the frame\n output_img = frame.copy()\n\n if len(tracking_paths) > 0:\n # Get images\n prev_img, current_img = prev_gray, frame_gray\n\n # Organize the feature points\n feature_points_0 = np.float32([tp[-1] for tp in \\\n tracking_paths]).reshape(-1, 1, 2)\n\n # Compute optical flow\n feature_points_1, _, _ = cv2.calcOpticalFlowPyrLK(\n prev_img, current_img, feature_points_0, \n None, **tracking_params)\n\n # Compute reverse optical flow\n feature_points_0_rev, _, _ = cv2.calcOpticalFlowPyrLK(\n current_img, prev_img, feature_points_1, \n None, **tracking_params)\n\n # Compute the difference between forward and \n # reverse optical flow\n diff_feature_points = abs(feature_points_0 - \\\n feature_points_0_rev).reshape(-1, 2).max(-1)\n\n # Extract the good points\n good_points = diff_feature_points < 1\n\n # Initialize variable\n new_tracking_paths = []\n\n # Iterate through all the good feature points \n for tp, (x, y), good_points_flag in zip(tracking_paths, \n feature_points_1.reshape(-1, 2), good_points):\n # If the flag is not true, then continue\n if not good_points_flag:\n continue\n\n # Append the X and Y coordinates and check if\n # its length greater than the threshold\n tp.append((x, y))\n if len(tp) > num_frames_to_track:\n del tp[0]\n\n new_tracking_paths.append(tp)\n\n # Draw a circle around the feature points\n cv2.circle(output_img, (x, y), 3, (0, 255, 0), -1)\n\n # Update the tracking paths\n tracking_paths = new_tracking_paths\n\n # Draw lines\n cv2.polylines(output_img, [np.int32(tp) for tp in \\\n tracking_paths], False, (0, 150, 0))\n\n # Go into this 'if' condition after skipping the \n # right number of frames\n if not frame_index % num_frames_jump:\n # Create a mask and draw the circles\n mask = np.zeros_like(frame_gray)\n mask[:] = 255\n for x, y in [np.int32(tp[-1]) for tp in tracking_paths]:\n cv2.circle(mask, (x, y), 6, 0, -1)\n\n # Compute good features to track\n feature_points = cv2.goodFeaturesToTrack(frame_gray, \n mask = mask, maxCorners = 500, qualityLevel = 0.3, \n minDistance = 7, blockSize = 7) \n\n # Check if feature points exist. If so, append them\n # to the tracking paths\n if feature_points is not None:\n for x, y in np.float32(feature_points).reshape(-1, 2):\n tracking_paths.append([(x, y)])\n\n # Update variables\n frame_index += 1\n prev_gray = frame_gray\n\n # Display output\n cv2.imshow('Optical Flow', output_img)\n\n # Check if the user hit the 'Esc' key\n c = cv2.waitKey(1)\n if c == 27:\n break\n\nif __name__ == '__main__':\n\t# Start the tracker\n start_tracking()\n\n # Close all the windows\n cv2.destroyAllWindows()\n\n",
"_____no_output_____"
]
]
] |
[
"raw",
"code"
] |
[
[
"raw",
"raw"
],
[
"code"
]
] |
cbaf1c5890d5bb9bfd7203b93aea2968a0f65b00
| 1,136 |
ipynb
|
Jupyter Notebook
|
Virtualization.ipynb
|
RonSheely/jupyter-notebooks
|
8d9b37503e59675d392f9b7c3a5a71a1c73a5373
|
[
"MIT"
] | null | null | null |
Virtualization.ipynb
|
RonSheely/jupyter-notebooks
|
8d9b37503e59675d392f9b7c3a5a71a1c73a5373
|
[
"MIT"
] | null | null | null |
Virtualization.ipynb
|
RonSheely/jupyter-notebooks
|
8d9b37503e59675d392f9b7c3a5a71a1c73a5373
|
[
"MIT"
] | null | null | null | 27.047619 | 269 | 0.62588 |
[
[
[
"How can I detrermine the default VirtualBox machine folder? Start VirtualBox and select Preferences - General - Default Machine Folder, where we can view or edit the folder name.\n\nFor my MacOS, the default folder is: /Users/ronsheely/VirtualBox VMs",
"_____no_output_____"
],
[
"How do I remove a VirtualBox machine? See section 1.12 in the VirtualBox User Manual - Removing virtual machines. To remove a virtual machine which you no longer need, right-click on it in the Manager’s VM list select “Remove” from the context menu that comes up.",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown"
]
] |
cbaf25214750bceefb099840a0b205fd31bd2ddd
| 31,389 |
ipynb
|
Jupyter Notebook
|
Python-API/pal/notebooks/bostonHousingRandomForestRegressor.ipynb
|
rschop/hana-ml-samples
|
c0fa20dacf9e48affa4073c8e2c905a98ab0047a
|
[
"BSD-Source-Code"
] | null | null | null |
Python-API/pal/notebooks/bostonHousingRandomForestRegressor.ipynb
|
rschop/hana-ml-samples
|
c0fa20dacf9e48affa4073c8e2c905a98ab0047a
|
[
"BSD-Source-Code"
] | null | null | null |
Python-API/pal/notebooks/bostonHousingRandomForestRegressor.ipynb
|
rschop/hana-ml-samples
|
c0fa20dacf9e48affa4073c8e2c905a98ab0047a
|
[
"BSD-Source-Code"
] | null | null | null | 32.193846 | 392 | 0.463379 |
[
[
[
"# Random Forest Regression Example\n\n## Boston housing prices\nThe objective is to predict the median price of a home in Boston. The variables are crime rate, zoning information,\nproportion of non-retail business, etc. This dataset has median prices in Boston for 1972. Even though the data is pretty old, the methodology for analytics is valid for more recent datasets.\n\n<b>The purpose of this demonstration is to show the use of SAP HANA's Predictive Analytics Library to created Random forest model.</b>\n\nThe dataset is from Kaggle. https://www.kaggle.com/c/boston-housing. For tutorials use only.\n\n## Housing Values in Suburbs of Boston in 1972\n\nThe <font color='red'>medv</font> variable is the target variable.\n### Data description\nThe Boston data frame has 506 rows and 14 columns.\nThis data frame contains the following columns:\n1. __crim__: per capita crime rate by town.\n2. __zn__: proportion of residential land zoned for lots over 25,000 sq.ft.\n3. __indus__: proportion of non-retail business acres per town.\n4. __chas__: Charles River dummy variable (1 if tract bounds river; 0 otherwise).\n5. __nox__: nitrogen oxides concentration (parts per 10 million).\n6. __rm__: average number of rooms per dwelling.\n7. __age__: proportion of owner-occupied units built prior to 1940.\n8. __dis__: weighted mean of distances to five Boston employment centres.\n9. __rad__: index of accessibility to radial highways.\n10. __tax__: full-value property-tax rate per \\$10000\n11. __ptratio__: pupil-teacher ratio by town.\n12. __black__: 1000(Bk - 0.63)^2 where Bk is the proportion of blacks by town.\n13. __lstat__: lower status of the population (percent).\n14. __medv__: median value of owner-occupied homes in $1000s.\n</td></tr></table>\n\n### Factoids\nThe prices in Boston across years is below. If we had a historical dataset, an analysis could be done to account for the macro trends as well.\n\nThe second graph shows the intuition we have with respect to prices in relation to crime rate. It is expected that house prices will be lower in areas where crime rates are higher.\n\nThe third figure is a chart showing how inflation may affect prices. So, for deeper analysis and prediction, we may want to consider inflation.\n\nIn this notebook, these factors are not considered. They are here to demonstrate the need for deep domain analysis.\n\n<table><tr>\n<td><img src=\"images/boston_prices_by_year.png\" alt=\"Boston home prices\" title=\"Boston housing prices\" style=\"float:left;\" /></td>\n<td><img src=\"images/Crime-Rate-and-Median-House-Prices.png\" alt=\"Boston home prices\" title=\"Boston housing prices\" /></td>\n<td><img src=\"images/Inflation_Adjusted_Housing_Prices_1890_2006.jpg\" alt=\"Inflation adjusted prices\" title=\"Inflation adjusted prices\" style=\"float:left;\" />\n</td></tr></table>\n\n\nIn this notebook, we will use the dataset for Boston housing prices and predict the price based on numerous factors.",
"_____no_output_____"
]
],
[
[
"from hana_ml import dataframe\nfrom hana_ml.algorithms.pal import clustering\nfrom hana_ml.algorithms.pal import trees\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport logging",
"_____no_output_____"
]
],
[
[
"## Load data\nThe data is loaded into 4 tables, for full, training, validation, and test sets:\n<li>BOSTON_HOUSING_PRICES</li>\n<li>BOSTON_HOUSING_PRICES_TRAINING</li>\n<li>BOSTON_HOUSING_PRICES_VALIDATION</li>\n<li>BOSTON_HOUSING_PRICES_TEST</li>\n\nTo do that, a connection is created and passed to the loader.\n\nThere is a config file, config/e2edata.ini that controls the connection parameters and whether or not to reload the data from scratch. In case the data is already loaded, there would be no need to load the data. A sample section is below. If the config parameter, reload_data is true then the tables for test, training, and validation are (re-)created and data inserted into them.\n\nAlthough this ini file has other sections, please do not modify them. Only the [hana] section should be modified.\n\n#########################<br>\n[hana]<br>\nurl=host.sjc.sap.corp<br>\nuser=username<br>\npasswd=userpassword<br>\nport=3xx15<br>\n#########################<br>",
"_____no_output_____"
]
],
[
[
"from data_load_utils import DataSets, Settings\nurl, port, user, pwd = Settings.load_config(\"../../config/e2edata.ini\")\nconnection_context = dataframe.ConnectionContext(url, port, user, pwd)\nfull_tbl, training_tbl, validation_tbl, test_tbl = DataSets.load_boston_housing_data(connection_context)",
"_____no_output_____"
]
],
[
[
"# Create Data Frames\nCreate the data frames for the full, test, training, and validation sets.\n\nLet us also do some dtaa exploration.",
"_____no_output_____"
],
[
"## Define Datasets - Training, validation, and test sets\nData frames are used keep references to data so computation on large data sets in HANA can happen in HANA. Trying to bring the entire data set into the client will likely result in out of memory exceptions.\n\nThe original/full dataset is split into training, test and validation sets. In the example below, they reside in different tables.",
"_____no_output_____"
]
],
[
[
"full_set = connection_context.table(full_tbl)\ntraining_set = connection_context.table(training_tbl)\nvalidation_set = connection_context.table(validation_tbl)\ntest_set = connection_context.table(test_tbl)",
"_____no_output_____"
]
],
[
[
"## Simple Exploration\nLet us look at the number of rows in the data set",
"_____no_output_____"
]
],
[
[
"print('Number of rows in full set: {}'.format(full_set.count()))\nprint('Number of rows in training set: {}'.format(training_set.count()))\nprint('Number of rows in validation set: {}'.format(validation_set.count()))\nprint('Number of rows in test set: {}'.format(test_set.count()))",
"Number of rows in full set: 506\nNumber of rows in training set: 315\nNumber of rows in validation set: 64\nNumber of rows in test set: 127\n"
]
],
[
[
"### Let's look at the columns",
"_____no_output_____"
]
],
[
[
"print(full_set.columns)",
"['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'BLACK', 'LSTAT', 'MEDV', 'ID']\n"
]
],
[
[
"### Let's look at the data types",
"_____no_output_____"
]
],
[
[
"full_set.dtypes()",
"_____no_output_____"
]
],
[
[
"### Set up the features and labels for the model",
"_____no_output_____"
]
],
[
[
"features=['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'BLACK', 'LSTAT']\nlabel='MEDV'",
"_____no_output_____"
]
],
[
[
"# Create model using training data\nFor demonstration, we will create two models, model and model_with_id, one where we have a unique id in the training set and one where there is none.\n\nWe are using Random Forest regression and SVM routines in this example\n\nDocumentation is <a href=\"https://help.sap.com/http.svc/rc/DRAFT/3f0dbe754b194c42a6bf3405697b711f/2.0.031/en-US/html/index.html\">here</a>",
"_____no_output_____"
],
[
"## Preprocessing\nSAP HANA Predictive Analytics Library takes DOUBLE and INTEGER data types for most numeric types. Since we have DECIMALs and TINYINTs in our data set, we cast them to the types required by PAL.",
"_____no_output_____"
]
],
[
[
"# Cast to correct types so PAL can consume it.\ndfts = training_set.cast(['CRIM', \"ZN\", \"INDUS\", \"NOX\", \"RM\", \"AGE\", \"DIS\", \"PTRATIO\", \"BLACK\", \"LSTAT\", \"MEDV\"], \"DOUBLE\")\ndfts = dfts.cast([\"CHAS\", \"RAD\", \"TAX\"], \"INTEGER\")\ndfts = dfts.to_head(\"ID\")\ndfts.head(5).collect()",
"_____no_output_____"
]
],
[
[
"## Create the model\nAlthough we had seen graphically that only a few features had an impact on housing prices, let us use all the features to create a model. We will then use the model to check for importance of the features.",
"_____no_output_____"
]
],
[
[
"# We build the model without IDs. Project only the features and the label.\ndf = dfts.select(features, label)\nmodel = trees.RandomForestRegressor(connection_context)\nmodel.fit(df, features=features, label=label)",
"_____no_output_____"
]
],
[
[
"### SQL statements executed\nCalling PAL directly would require a number of SQL statements and all that is encapsulated in the Python library functions.",
"_____no_output_____"
],
[
"## Model analysis\nLet's just see what features are most important.\nNote that we are using a sort function. The property __feature_importances___ is automatically set when the fit() method is called above.",
"_____no_output_____"
]
],
[
[
"model.feature_importances_.sort(['IMPORTANCE'], desc=True).collect()",
"_____no_output_____"
]
],
[
[
"__As you can see above, LSTAT, RM, NOX, and PTRATIO seem to have the most impact on prices.__",
"_____no_output_____"
],
[
"# Predict using test set\nLet us now do some predictions and see how well the model generalizes.\n\nThe predict() method always takes a unique identifier to identify the prediction on a specific data row. This way, the caller (python programmer) can then join with the original data set to get the rest of the values for that unique row. The test_set has columns of types that PAL does not deal with and therefore the columns are cast to the types that are accepted.\n\nIn order to look at the predicted value as well as the true value, the name of the unique identifier for rows in the result table is renamed to PREDICTED_ID. This result table is joined with the test set so the predicted and true value can be compared.\n\nFor the predictions we look at the standard error. The standard error is defined as the number of standard deviations away the prediction is from the true value.",
"_____no_output_____"
]
],
[
[
"df_test = test_set.cast(['CRIM', \"ZN\", \"INDUS\", \"NOX\", \"RM\", \"AGE\", \"DIS\", \"PTRATIO\", \"BLACK\", \"LSTAT\", \"MEDV\"], \"DOUBLE\")\ndf_test = df_test.cast([\"CHAS\", \"RAD\", \"TAX\"], \"INTEGER\")\ndf_test = df_test.to_head(\"ID\")",
"_____no_output_____"
],
[
"# Note that we are renaming the column ID in the result of predict()\nresult_df = model.predict(df_test, key= 'ID', features=features).rename_columns({'ID': 'PREDICTED_ID'})\n# Note the use of join() method to join two tables.\njdf = result_df.join(test_set, '{}.\"PREDICTED_ID\"={}.\"ID\"'.format(result_df.name, test_set.name), how='inner')",
"_____no_output_____"
]
],
[
[
"### Predictions\nLet us look at the predictions. The predicted values are in 'SCORE' and the actual values are in 'MEDV'. So, we just rename the 'SCORE' column to 'PREDICTED'\n\nIn addition, the column 'CONFIDENCE' is the standard error which is the number of standard deviations away the actual values is from the predicted value. This column is renamed to 'STANDARD_ERROR'",
"_____no_output_____"
]
],
[
[
"jdf.select(['ID', 'SCORE', 'MEDV', 'CONFIDENCE']).rename_columns({\"CONFIDENCE\": \"STANDARD_ERROR\", \"SCORE\": \"PREDICTED\"}).sort(\"STANDARD_ERROR\", desc=False).head(5).collect()",
"_____no_output_____"
]
],
[
[
"### Out of bag error\nLet us look at the out of bag errors which is a method of measuring the prediction error.\n\nHere we look at the first 4 rows",
"_____no_output_____"
]
],
[
[
"model.oob_error_.head(4).collect()",
"_____no_output_____"
]
],
[
[
"## Scoring\nWe now score the results from are test data. The scoring function we use is R^2.\n\n__In the function below, PAL is not invoked but a query is directly executed against data in HANA__",
"_____no_output_____"
]
],
[
[
"r2_score = model.score(df_test, key='ID', features=features, label=label)\nprint(\"r2 score is {}\".format(r2_score))",
"r2 score is 0.6727879210329948\n"
]
],
[
[
"## Model\nThe model is available and can be saved for later predictions",
"_____no_output_____"
]
],
[
[
"# The generated model is in the database.\nmodel.model_.head(4).collect()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbaf2d4df55f6ceb936ce5694569bd7774994702
| 756 |
ipynb
|
Jupyter Notebook
|
notebooks/models/ycocg.ipynb
|
Legendin/colour-notebooks
|
357b64e60e24468c88a7d6789003a6283c809c01
|
[
"BSD-3-Clause"
] | 13 |
2016-11-23T22:13:24.000Z
|
2021-09-28T14:52:13.000Z
|
notebooks/models/ycocg.ipynb
|
Legendin/colour-notebooks
|
357b64e60e24468c88a7d6789003a6283c809c01
|
[
"BSD-3-Clause"
] | 2 |
2015-07-13T19:38:16.000Z
|
2015-12-14T06:30:04.000Z
|
notebooks/models/ycocg.ipynb
|
colour-science/colour-ipython
|
f227bb1ebc041812de4048ae20e2b702ffb3150d
|
[
"BSD-3-Clause"
] | 9 |
2016-10-06T16:18:40.000Z
|
2020-08-01T10:04:27.000Z
| 16.085106 | 34 | 0.488095 |
[
[
[
"# !!! D . R . A . F . T !!!",
"_____no_output_____"
],
[
"# YCoCg Colour Encoding",
"_____no_output_____"
],
[
"## Bibliography",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
]
] |
cbaf501e7e5fdbb60cc13fb3a68c968c5ff5b929
| 289,996 |
ipynb
|
Jupyter Notebook
|
Transforming data/New data.ipynb
|
ikicab/Trading-in-a-Black-Box
|
0a13027ca27ce27b052d5bba459ed7621204ac3c
|
[
"MIT"
] | 2 |
2021-01-25T11:50:37.000Z
|
2021-05-18T13:38:05.000Z
|
Transforming data/New data.ipynb
|
ikicab/Trading-in-a-Black-Box
|
0a13027ca27ce27b052d5bba459ed7621204ac3c
|
[
"MIT"
] | null | null | null |
Transforming data/New data.ipynb
|
ikicab/Trading-in-a-Black-Box
|
0a13027ca27ce27b052d5bba459ed7621204ac3c
|
[
"MIT"
] | null | null | null | 32.311532 | 336 | 0.349208 |
[
[
[
"%reload_ext autoreload\n%autoreload 2\n\nfrom IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = \"all\"\n\nimport os\nimport re\nimport pickle\n\nimport pandas as pd\nimport numpy as np\n\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nfrom matplotlib import rc\nrc('text', usetex=True)\n\ndef bold_text(string):\n return r'\\textbf{{{}}}'.format(string)\n\nfrom IPython.display import Markdown\ndef printmd(string):\n \"\"\"Embed the input string into Markdown.\"\"\"\n display(Markdown(string))\n \ndef list_files(startpath):\n level_colours = {0: '#339fff', 1: '#ff5b33'}\n \n for root, dirs, files in os.walk(startpath):\n if os.path.basename(root) == startpath:\n continue\n \n level = root.replace(startpath, '').count(os.sep) - 1 \n indent = ' ' * 4 * (level)\n \n printmd('<pre>{}<b><font color={}>{}</font></b></pre>'.format(indent, level_colours[level], os.path.basename(root)))\n \n if len(files) > 0:\n print('{}{}'.format(indent, files))",
"_____no_output_____"
]
],
[
[
"# Importing data",
"_____no_output_____"
],
[
"Explore the contents of the folder with all data files",
"_____no_output_____"
]
],
[
[
"data_folder = 'session_210302'",
"_____no_output_____"
],
[
"printmd('**Data contents**')\nlist_files(data_folder)",
"_____no_output_____"
]
],
[
[
"Store all data in the form ```{(market, treatment): {'deals': df_deals, 'games': df_games, 'offers': df_offers, 'players': df_players}}```",
"_____no_output_____"
]
],
[
[
"all_data = {}\ndata_types = []\n\nfor path, folders, files in os.walk(data_folder):\n for file in files:\n treatment = tuple(path.split('\\\\')[1:])\n dtype = re.match(r'^.*_(.*)\\.csv.*$', file).group(1)\n data_types.append(dtype)\n \n if treatment not in all_data.keys():\n all_data[treatment] = {}\n \n all_data[treatment][dtype] = pd.read_csv('{}\\\\{}'.format(path, file))\n \ndata_types = set(data_types)",
"_____no_output_____"
]
],
[
[
"Check whether all .csv files share the same structure and print out the names of their columns",
"_____no_output_____"
]
],
[
[
"for dtype in data_types:\n printmd('**{}**'.format(dtype))\n data = [d[dtype] for d in all_data.values()]\n \n all([(data[0].columns.intersection(df.columns) == data[0].columns).all() for df in data])\n \n data[0].columns.to_list()",
"_____no_output_____"
]
],
[
[
"Note:\\\n```var_id``` global id\\\n```var_iid``` local id",
"_____no_output_____"
],
[
"## Game information",
"_____no_output_____"
]
],
[
[
"all_data[('Externalities', 'bystanders_negative')]['games'].columns.to_list()",
"_____no_output_____"
]
],
[
[
"Find all columns with non-constant values",
"_____no_output_____"
]
],
[
[
"for treatment, data in all_data.items():\n print(treatment, list(data['games'].columns[data['games'].nunique() > 1]))",
"('Externalities', 'bystanders_negative') ['game_id', 'game_iid', 'elapsed_time']\n('Externalities', 'bystanders_positive') ['game_id', 'game_iid', 'elapsed_time']\n('Externalities', 'normal') ['game_id', 'game_iid', 'elapsed_time']\n('LimitedAsks', 'black_box') ['game_id', 'game_iid', 'elapsed_time']\n('LimitedAsks', 'open_book') ['game_id', 'game_iid', 'elapsed_time']\n"
],
[
"for treatment, data in all_data.items():\n printmd('**{}**'.format(treatment))\n data['games'][['game_iid', 'title', 'elapsed_time']]",
"_____no_output_____"
]
],
[
[
"## Player information",
"_____no_output_____"
]
],
[
[
"all_data[('Externalities', 'bystanders_negative')]['players'].columns.to_list()",
"_____no_output_____"
]
],
[
[
"Find all columns with non-constant values",
"_____no_output_____"
]
],
[
[
"for treatment, data in all_data.items():\n print(treatment, list(data['players'].columns[data['players'].nunique() > 1]))",
"('Externalities', 'bystanders_negative') ['player_id', 'player_iid', 'game_id', 'game_iid', 'rprice', 'side', 'iddle', 'total_payoff']\n('Externalities', 'bystanders_positive') ['player_id', 'player_iid', 'game_id', 'game_iid', 'rprice', 'side', 'iddle', 'total_payoff']\n('Externalities', 'normal') ['player_id', 'player_iid', 'game_id', 'game_iid', 'rprice', 'side', 'iddle', 'total_payoff']\n('LimitedAsks', 'black_box') ['player_id', 'player_iid', 'game_id', 'game_iid', 'rprice', 'side', 'iddle', 'total_payoff']\n('LimitedAsks', 'open_book') ['player_id', 'player_iid', 'game_id', 'game_iid', 'rprice', 'side', 'iddle', 'total_payoff']\n"
]
],
[
[
"## Offer information",
"_____no_output_____"
]
],
[
[
"all_data[('Externalities', 'bystanders_negative')]['offers'].columns.to_list()",
"_____no_output_____"
],
[
"for treatment, data in all_data.items():\n printmd('**{}**'.format(treatment))\n data_offers = data['offers']\n print('status: {}'.format(set(data_offers['status'])))\n print('type: {}'.format(set(data_offers['type'])))\n \n printmd('status == ```Accepted``` if and only if the bid/ask resulted in a deal')\n set(data_offers[data_offers['status'] == 'Replaced']['matched_price'].dropna())\n set(data_offers[data_offers['status'] == 'Expired']['matched_price'].dropna())\n set(data_offers[data_offers['matched_price'].notna()]['status'])\n \n printmd('type == ```Auto``` corresponds to accepting a deal')\n data_offers[(data_offers['type'] == 'Auto') & (data_offers['matched_price'].isna())]",
"_____no_output_____"
]
],
[
[
"Add treatments information and remove redundant/unnecessary columns",
"_____no_output_____"
]
],
[
[
"all_data.keys()",
"_____no_output_____"
],
[
"treatment_names = {\n ('Externalities', 'bystanders_negative'): 'FullExtNeg',\n ('Externalities', 'bystanders_positive'): 'FullExtPos',\n ('Externalities', 'normal'): 'FullExtNorm',\n ('LimitedAsks', 'black_box'): 'BBLimS',\n ('LimitedAsks', 'open_book'): 'FullLimS'\n}",
"_____no_output_____"
],
[
"for treatment, data in all_data.items():\n #data['offers'].drop(['game_id', 'round_id', 'status'], axis=1, inplace=True)\n # Keep the status column\n data['offers'].drop(['game_id', 'round_id'], axis=1, inplace=True)\n data['offers']['treatment'] = treatment_names[treatment]\n data['offers'].rename({'game_iid': 'game', 'round_iid': 'round', 'amount': 'bid',\n 'player_id': 'id', 'matched_price': 'price'}, axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"Add ```match_id``` and ```match_time```",
"_____no_output_____"
]
],
[
[
"for treatment, data in all_data.items():\n for idx, row in data['deals'].iterrows():\n game, rnd, match_time, buyer, seller, askID, bidID, bprice, sprice = row[['game_iid', 'round_iid', 'time', 'buyer_id',\n 'seller_id', 'ask_id', 'bid_id', 'bprice', 'sprice']]\n \n game_round = (data['offers']['game'] == game) & (data['offers']['round'] == rnd)\n ask_row = (data['offers']['offer_db_id'] == askID)\n bid_row = (data['offers']['offer_db_id'] == bidID)\n \n data['offers'].loc[game_round & ask_row, 'match_time'] = match_time\n data['offers'].loc[game_round & ask_row, 'match_id'] = buyer\n data['offers'].loc[game_round & ask_row, 'price_temp'] = sprice\n \n data['offers'].loc[game_round & bid_row, 'match_time'] = match_time\n data['offers'].loc[game_round & bid_row, 'match_id'] = seller\n data['offers'].loc[game_round & bid_row, 'price_temp'] = bprice",
"_____no_output_____"
],
[
"for treatment, data in all_data.items():\n data['offers']['price'].equals(data['offers']['price_temp'])",
"_____no_output_____"
],
[
"for treatment, data in all_data.items():\n data['offers'].drop(['price_temp'], axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"Add ```valuation```",
"_____no_output_____"
]
],
[
[
"for treatment, data in all_data.items():\n for (game, idx), dfi in data['offers'].groupby(['game', 'id']):\n val = data['players'][data['players']['player_id'] == idx]['rprice'].values[0]\n data['offers'].loc[dfi.index, 'valuation'] = val",
"_____no_output_____"
]
],
[
[
"Rearrange to match the order in the rest of the data",
"_____no_output_____"
]
],
[
[
"for treatment, data in all_data.items():\n data['offers'] = data['offers'][['treatment', 'game', 'round', 'time', 'id', 'side', 'valuation',\n 'bid', 'price', 'match_id', 'match_time', 'type', 'status']]",
"_____no_output_____"
]
],
[
[
"# Merging data",
"_____no_output_____"
],
[
"Store all datasets in a single dataframe",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame()\n\nfor treatment, data in all_data.items():\n df = df.append(data['offers'], ignore_index=True)",
"_____no_output_____"
]
],
[
[
"Create globally unique subject IDs",
"_____no_output_____"
]
],
[
[
"# Create globally unique subject IDs\ndf['old_id'] = df['id']\ndf['id'] = df.groupby(['treatment', 'game', 'id']).ngroup()\n\n# Update the column with match IDs accordingly\nfor (treatment, game), df_game in df.groupby(['treatment', 'game']):\n for idx, row in df_game[df_game['match_id'].notna()].iterrows():\n df.loc[idx, 'match_id'] = df_game[df_game['old_id'] == row['match_id']]['id'].iloc[0]\n\ndf.drop(columns=['old_id'], axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"Cast the valuations to ```int```",
"_____no_output_____"
]
],
[
[
"(df['valuation'] % 1 == 0).all()",
"_____no_output_____"
],
[
"df['valuation'] = df['valuation'].astype(int)",
"_____no_output_____"
]
],
[
[
"When a buyer and a seller are automatically matched under the first-price mechanism, a new entry with the bid/ask equal to the resulting price is automatically generated for the buyer/seller who submitted the bid/ask last. Remove all such entries and copy the corresopnding prices to the entries with the bids/asks submitted last.",
"_____no_output_____"
]
],
[
[
"df[['type', 'status']].drop_duplicates()\ndf.groupby(['type', 'status']).size()",
"_____no_output_____"
]
],
[
[
"The status of type ```Auto``` can only be ```Accepted```",
"_____no_output_____"
]
],
[
[
"set(df[df['type'] == 'Auto']['status'])",
"_____no_output_____"
]
],
[
[
"The status of a bid/ask is set to ```Accepted``` if and only if it results in a deal",
"_____no_output_____"
]
],
[
[
"set(df[df['price'].notna()]['status'])\ndf[df['status'] == 'Accepted']['price'].isna().any()",
"_____no_output_____"
]
],
[
[
"Each bid–ask pair striking a deal is stored as follows: the first of the two is recorded as ``Manual``, the second as ``Auto``.",
"_____no_output_____"
]
],
[
[
"df_prices = df[df['price'].notna()]",
"_____no_output_____"
],
[
"bid_ask_pairs = {'MM': 0, 'MA': 0, 'AA': 0}\n\nfor (treatment, game, rnd), dfr in df_prices.groupby(['treatment', 'game', 'round']):\n for row_id, row in dfr.iterrows():\n if row['id'] < row['match_id']:\n id1 = row['id']\n id2 = row['match_id']\n \n types = {dfr[dfr['id'] == id1]['type'].iloc[0], dfr[dfr['id'] == id2]['type'].iloc[0]}\n\n if len(types) == 2:\n bid_ask_pairs['MA'] += 1\n elif types == {'Manual'}:\n bid_ask_pairs['MM'] += 1\n else:\n bid_ask_pairs['AA'] += 1\n\nbid_ask_pairs",
"_____no_output_____"
]
],
[
[
"```Auto``` always take place after ```Manual``` (or, possibly, simultaneously)\n\nA match is made at most 1 second after a bid and an ask are compatible",
"_____no_output_____"
]
],
[
[
"times = {'same': 0, 'M then A': 0, 'A then M': 0}\nindices = {'M then A': 0, 'A then M': 0}\ndelays_to_match = []\n\nfor (treatment, game, rnd), dfr in df_prices.groupby(['treatment', 'game', 'round']):\n for row_id, row in dfr.iterrows():\n if row['id'] < row['match_id']:\n match = dfr[dfr['id'].isin([row['id'], row['match_id']])]\n \n types = set(match['type'])\n\n if len(types) == 2:\n M_time = match[match['type'] == 'Manual']['time'].iloc[0]\n A_time = match[match['type'] == 'Auto']['time'].iloc[0]\n\n M_id = match[match['type'] == 'Manual'].index\n A_id = match[match['type'] == 'Auto'].index\n\n if M_time == A_time:\n times['same'] += 1\n elif M_time < A_time:\n times['M then A'] += 1\n else:\n times['A then M'] += 1\n\n if M_id < A_id:\n indices['M then A'] += 1\n else:\n indices['A then M'] += 1\n\n if int(match['match_time'].iloc[0]) != max(match['time']):\n delays_to_match.append(int(match['match_time'].iloc[0]) - max(match['time']))\n \ntimes\nindices\ndelays_to_match",
"_____no_output_____"
]
],
[
[
"<font color=blue>The redundant rows (automatic matching enforced by the computer) correspond to ```Auto``` bids/asks following ```Replaced``` bids/asks which were high/low enough to result in a deal</font>",
"_____no_output_____"
]
],
[
[
"df_new = df.copy()",
"_____no_output_____"
],
[
"df_new['redundant'] = False\n\nstatus = {'Accepted': 0, 'Replaced': 0, 'Expired': 0}\n\nfor (treatment, game, rnd, idx), dfi in df_new.groupby(['treatment', 'game', 'round', 'id']):\n for row_id, row in dfi.iterrows():\n if row['type'] == 'Auto':\n if len(dfi) > 1:\n preceding = dfi.loc[:row.name].iloc[-2]\n\n status[preceding['status']] += 1\n\n if preceding['status'] == 'Replaced':\n if row['side'] == 'Buyer':\n if preceding['bid'] >= row['bid']:\n df_new.loc[row.name, 'redundant'] = True\n df_new.loc[preceding.name, 'price'] = row['price']\n df_new.loc[preceding.name, 'match_id'] = row['match_id']\n df_new.loc[preceding.name, 'match_time'] = row['match_time']\n else:\n if preceding['bid'] <= row['bid']:\n df_new.loc[row.name, 'redundant'] = True\n df_new.loc[preceding.name, 'price'] = row['price']\n df_new.loc[preceding.name, 'match_id'] = row['match_id']\n df_new.loc[preceding.name, 'match_time'] = row['match_time']\n\nstatus",
"_____no_output_____"
],
[
"len(df_new)\nlen(df)\n\ndf_new.drop(['redundant', 'price', 'match_id', 'match_time'], axis=1).equals(df.drop(['price', 'match_id', 'match_time'], axis=1))",
"_____no_output_____"
],
[
"df_new = df_new[~df_new['redundant']]\ndf_new.drop('redundant', axis=1, inplace=True)",
"_____no_output_____"
],
[
"len(df_new)\ndf_new.groupby('type').size()",
"_____no_output_____"
],
[
"df_prices = df_new[df_new['price'].notna()]\n\ndelays_to_match = []\n\nfor (treatment, game, rnd), dfr in df_prices.groupby(['treatment', 'game', 'round']):\n for row_id, row in dfr.iterrows():\n if row['id'] < row['match_id']:\n match = dfr[dfr['id'].isin([row['id'], row['match_id']])]\n \n if (len(match) != 2) or (match['match_time'].count() != 2) or (match['match_id'].count() != 2) or (match['price'].count() != 2):\n 'Some data is missing'\n \n if int(match['match_time'].iloc[0]) != max(match['time']):\n delays_to_match.append(int(match['match_time'].iloc[0]) - max(match['time']))\n\ndelays_to_match",
"_____no_output_____"
],
[
"for treatment, df_treatment in df.groupby(['treatment']):\n printmd(treatment)\n \n diff = pd.merge(df, df_new, how='outer', suffixes=('','_y'), indicator=True)\n diff = diff[diff['_merge'] != 'both']\n \n diff.sort_values(['treatment', 'game', 'round', 'time', 'id']).iloc[1:51]",
"_____no_output_____"
],
[
"df = df_new.copy()",
"_____no_output_____"
]
],
[
[
"# Overview of the data",
"_____no_output_____"
]
],
[
[
"index = pd.MultiIndex.from_tuples(df[['treatment', 'game']].drop_duplicates().itertuples(index=False, name=None),\n names=['Treatment', 'Game'])\noverview = pd.DataFrame(index=index, columns=['Buyers', 'Sellers', 'Bids', 'Asks'])\n\nfor (treatment, game, side), df_side in df.groupby(['treatment', 'game', 'side']):\n if side == 'Buyer':\n overview.loc[(treatment, game), 'Buyers'] = len(set(df_side['id']))\n overview.loc[(treatment, game), 'Bids'] = len(df_side)\n elif side == 'Seller':\n overview.loc[(treatment, game), 'Sellers'] = len(set(df_side['id']))\n overview.loc[(treatment, game), 'Asks'] = len(df_side)\n else:\n print('No side provided.')",
"_____no_output_____"
],
[
"overview",
"_____no_output_____"
]
],
[
[
"# Exporting data",
"_____no_output_____"
],
[
"## Externalities",
"_____no_output_____"
]
],
[
[
"df_ext = df[df['treatment'].str.contains('Ext')].copy()",
"_____no_output_____"
]
],
[
[
"Create globally unique subject IDs",
"_____no_output_____"
]
],
[
[
"# Create globally unique subject IDs\ndf_ext['old_id'] = df_ext['id']\ndf_ext['id'] = df_ext.groupby(['treatment', 'game', 'id']).ngroup()\n\n# Update the column with match IDs accordingly\nfor (treatment, game), df_game in df_ext.groupby(['treatment', 'game']):\n for idx, row in df_game[df_game['match_id'].notna()].iterrows():\n df_ext.loc[idx, 'match_id'] = df_game[df_game['old_id'] == row['match_id']]['id'].iloc[0]\n\ndf_ext.drop(columns=['old_id'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_ext",
"_____no_output_____"
],
[
"df_ext.to_csv('../Data/data_externalities.csv', index=False)",
"_____no_output_____"
]
],
[
[
"## Restricted asks",
"_____no_output_____"
]
],
[
[
"df_LimS = df[df['treatment'].str.contains('LimS')].copy()",
"_____no_output_____"
]
],
[
[
"Create globally unique subject IDs",
"_____no_output_____"
]
],
[
[
"# Create globally unique subject IDs\ndf_LimS['old_id'] = df_LimS['id']\ndf_LimS['id'] = df_LimS.groupby(['treatment', 'game', 'id']).ngroup()\n\n# Update the column with match IDs accordingly\nfor (treatment, game), df_game in df_LimS.groupby(['treatment', 'game']):\n for idx, row in df_game[df_game['match_id'].notna()].iterrows():\n df_LimS.loc[idx, 'match_id'] = df_game[df_game['old_id'] == row['match_id']]['id'].iloc[0]\n\ndf_LimS.drop(columns=['old_id'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"df_LimS",
"_____no_output_____"
],
[
"df_LimS.to_csv('../Data/data_restricted_asks.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbaf56fb4f2e5328adcd2db073b9474fa59226c8
| 9,531 |
ipynb
|
Jupyter Notebook
|
notebooks/graph_embeding.ipynb
|
GiggleLiu/tensors_intro_tutorial
|
03b74eb2e2c839f5a802c52cde948e95ae4f0027
|
[
"MIT"
] | 3 |
2019-06-07T13:12:46.000Z
|
2019-07-04T13:58:34.000Z
|
notebooks/graph_embeding.ipynb
|
GiggleLiu/tensors_tutorial_jizhi
|
03b74eb2e2c839f5a802c52cde948e95ae4f0027
|
[
"MIT"
] | null | null | null |
notebooks/graph_embeding.ipynb
|
GiggleLiu/tensors_tutorial_jizhi
|
03b74eb2e2c839f5a802c52cde948e95ae4f0027
|
[
"MIT"
] | null | null | null | 21.661364 | 122 | 0.447697 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbaf5d0e7d21da4e5376960f840f19cb1a85b35d
| 9,293 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/TextGeneratorFinal-checkpoint.ipynb
|
arushi-bhatt/RnnTextGenerator
|
8afc7f2a5e42ab19270dacfa5b75d100397d060b
|
[
"MIT"
] | 1 |
2019-08-25T10:24:28.000Z
|
2019-08-25T10:24:28.000Z
|
TextGeneratorFinal.ipynb
|
arushi-bhatt/RnnTextGenerator
|
8afc7f2a5e42ab19270dacfa5b75d100397d060b
|
[
"MIT"
] | null | null | null |
TextGeneratorFinal.ipynb
|
arushi-bhatt/RnnTextGenerator
|
8afc7f2a5e42ab19270dacfa5b75d100397d060b
|
[
"MIT"
] | null | null | null | 47.172589 | 182 | 0.510922 |
[
[
[
"import numpy\nfrom keras.models import Sequential\nfrom keras.layers import Dense\nfrom keras.layers import Dropout\nfrom keras.layers import LSTM\nfrom keras.callbacks import ModelCheckpoint\nfrom keras.utils import np_utils\n\n\n\n# load ascii text and covert to lowercase\nfilename = \"wonderlandcopy.txt\"\nraw_text = open(filename).read()\nraw_text = raw_text.lower()\n\n\n# create mapping of unique chars to integers\nchars = sorted(list(set(raw_text)))\nchar_to_int = dict((c, i) for i, c in enumerate(chars))\n\n\nn_chars = len(raw_text)\nn_vocab = len(chars)\nprint (\"Total Characters:\", n_chars)\nprint (\"Total Vocab: \",n_vocab)\n\n\n# prepare the dataset of input to output pairs encoded as integers\nseq_length = 100\ndataX = []\ndataY = []\nfor i in range(0, n_chars - seq_length, 1):\n seq_in = raw_text[i:i + seq_length]\n seq_out = raw_text[i + seq_length]\n dataX.append([char_to_int[char] for char in seq_in])\n dataY.append(char_to_int[seq_out])\nn_patterns = len(dataX)\nprint (\"Total Patterns: \", n_patterns)\n\n\n\n# reshape X to be [samples, time steps, features]\nX = numpy.reshape(dataX, (n_patterns, seq_length, 1))\n# normalize\nX = X / float(n_vocab)\n# one hot encode the output variable\ny = np_utils.to_categorical(dataY)\n\n\n\n\n\n# define the LSTM model\nmodel = Sequential()\nmodel.add(LSTM(150, input_shape=(X.shape[1], X.shape[2])))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(y.shape[1], activation='softmax'))\nmodel.compile(loss='categorical_crossentropy', optimizer='adam')\n\n\n# define the checkpoint\nfilepath=\"weights-improvement-{epoch:02d}-{loss:.4f}.hdf5\"\ncheckpoint = ModelCheckpoint(filepath, monitor='loss', verbose=1, save_best_only=True, mode='min')\ncallbacks_list = [checkpoint]\n\nmodel.fit(X, y, epochs=20, batch_size=128, callbacks=callbacks_list)\n\n",
"Using TensorFlow backend.\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbaf6290883c7f7107b08639a929cb6acbca89d8
| 46,250 |
ipynb
|
Jupyter Notebook
|
training/Training_rgb-r_r-Copy1.ipynb
|
OpenGridMap/power-grid-detection
|
221fcf0461dc869c8c64b11fa48596f83c20e1c8
|
[
"Apache-2.0"
] | null | null | null |
training/Training_rgb-r_r-Copy1.ipynb
|
OpenGridMap/power-grid-detection
|
221fcf0461dc869c8c64b11fa48596f83c20e1c8
|
[
"Apache-2.0"
] | 1 |
2018-07-22T22:43:27.000Z
|
2018-07-22T22:43:27.000Z
|
training/Training_rgb-r_r-Copy1.ipynb
|
OpenGridMap/power-grid-detection
|
221fcf0461dc869c8c64b11fa48596f83c20e1c8
|
[
"Apache-2.0"
] | null | null | null | 79.195205 | 1,362 | 0.530443 |
[
[
[
"from __future__ import print_function\n\nimport os\nimport sys\nimport numpy as np\n\nfrom keras.optimizers import SGD, Nadam, RMSprop\nfrom keras.callbacks import CSVLogger, ModelCheckpoint\nfrom keras.regularizers import l1, l2\n\nsys.path.append(os.path.join(os.getcwd(), os.pardir))\n\nimport config\n\nfrom utils.dataset.data_generator import DataGenerator\nfrom models.cnn3 import cnn, cnn_regularized",
"Using Theano backend.\nUsing gpu device 0: GeForce GTX 680 (CNMeM is disabled, cuDNN 5005)\n"
],
[
"lr = 0.1\ndecay = 0.001\nl1 = 0.00001\nl2 = 0.00001\ndropout = 0.5\nn_epochs = 500\nbatch_size = 32\ninput_shape = (140, 140, 3)\n\nname = 'cnn_140_rgb_lr_%f_decay_%f_sgd_he_normal__l1_%f_l2_%f_dropout_%f_r' % (lr, decay, l1, l2, dropout)",
"_____no_output_____"
],
[
"print('loading model...')\n# model = cnn(input_shape=input_shape, init='he_normal')\nmodel = cnn_regularized(input_shape=input_shape, init='he_normal', l1=l1, l2=l2)\nmodel.summary()\n\noptimizer = SGD(lr=lr, clipnorm=4., nesterov=True, decay=decay)\n# optimizer = Nadam(lr=lr)\n# optimizer = RMSprop(lr=lr)\n\nprint('compiling model...')\nmodel.compile(optimizer=optimizer, loss='binary_crossentropy', metrics=['accuracy'])\nprint('done.')\n\ncsv_logger = CSVLogger('%s_training.log' % name)\nbest_model_checkpointer = ModelCheckpoint(filepath=(\"./%s_training_weights_best.hdf5\" % name), verbose=1,\n save_best_only=True)\n\ncurrent_model_checkpointer = ModelCheckpoint(filepath=(\"./%s_training_weights_current.hdf5\" % name), verbose=0)",
"loading model...\n____________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n====================================================================================================\ninput_1 (InputLayer) (None, 140, 140, 3) 0 \n____________________________________________________________________________________________________\nconvolution2d_1 (Convolution2D) (None, 140, 140, 128) 18944 input_1[0][0] \n____________________________________________________________________________________________________\nactivation_1 (Activation) (None, 140, 140, 128) 0 convolution2d_1[0][0] \n____________________________________________________________________________________________________\nmaxpooling2d_1 (MaxPooling2D) (None, 70, 70, 128) 0 activation_1[0][0] \n____________________________________________________________________________________________________\nconvolution2d_2 (Convolution2D) (None, 70, 70, 64) 204864 maxpooling2d_1[0][0] \n____________________________________________________________________________________________________\nactivation_2 (Activation) (None, 70, 70, 64) 0 convolution2d_2[0][0] \n____________________________________________________________________________________________________\nmaxpooling2d_2 (MaxPooling2D) (None, 35, 35, 64) 0 activation_2[0][0] \n____________________________________________________________________________________________________\nconvolution2d_3 (Convolution2D) (None, 35, 35, 64) 36928 maxpooling2d_2[0][0] \n____________________________________________________________________________________________________\nactivation_3 (Activation) (None, 35, 35, 64) 0 convolution2d_3[0][0] \n____________________________________________________________________________________________________\nmaxpooling2d_3 (MaxPooling2D) (None, 17, 17, 64) 0 activation_3[0][0] \n____________________________________________________________________________________________________\nflatten_1 (Flatten) (None, 18496) 0 maxpooling2d_3[0][0] \n____________________________________________________________________________________________________\ndense_1 (Dense) (None, 1024) 18940928 flatten_1[0][0] \n____________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 1024) 0 dense_1[0][0] \n____________________________________________________________________________________________________\ndense_2 (Dense) (None, 1024) 1049600 dropout_1[0][0] \n____________________________________________________________________________________________________\ndropout_2 (Dropout) (None, 1024) 0 dense_2[0][0] \n____________________________________________________________________________________________________\ndense_3 (Dense) (None, 512) 524800 dropout_2[0][0] \n____________________________________________________________________________________________________\ndense_4 (Dense) (None, 2) 1026 dense_3[0][0] \n____________________________________________________________________________________________________\nactivation_4 (Activation) (None, 2) 0 dense_4[0][0] \n====================================================================================================\nTotal params: 20777090\n____________________________________________________________________________________________________\ncompiling model...\ndone.\n"
],
[
"print('Initializing data generators...')\ntrain_data_gen = DataGenerator(dataset_file=config.train_data_file, batch_size=batch_size)\nvalidation_data_gen = DataGenerator(dataset_file=config.validation_data_file, batch_size=batch_size)\ntest_data_gen = DataGenerator(dataset_file=config.test_data_file, batch_size=batch_size)\nprint('done.')",
"Initializing data generators...\ndone.\n"
],
[
"print('Fitting model...')\nhistory = model.fit_generator(train_data_gen,\n nb_epoch=n_epochs,\n samples_per_epoch=train_data_gen.n_batches * batch_size,\n validation_data=validation_data_gen,\n nb_val_samples=validation_data_gen.n_samples,\n verbose=1,\n callbacks=[csv_logger, best_model_checkpointer, current_model_checkpointer])\nprint('done.')",
"Fitting model...\nEpoch 1/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.8059 - acc: 0.6562Epoch 00000: val_loss improved from inf to 0.48095, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 355s - loss: 2.8058 - acc: 0.6562 - val_loss: 0.4809 - val_acc: 0.7449\nEpoch 2/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.6110 - acc: 0.7498Epoch 00001: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 2.6110 - acc: 0.7497 - val_loss: 0.4885 - val_acc: 0.7619\nEpoch 3/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.4028 - acc: 0.8720Epoch 00002: val_loss improved from 0.48095 to 0.30334, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 359s - loss: 2.4027 - acc: 0.8720 - val_loss: 0.3033 - val_acc: 0.8776\nEpoch 4/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.3296 - acc: 0.8904Epoch 00003: val_loss improved from 0.30334 to 0.24943, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 360s - loss: 2.3293 - acc: 0.8906 - val_loss: 0.2494 - val_acc: 0.9025\nEpoch 5/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.2733 - acc: 0.8956Epoch 00004: val_loss improved from 0.24943 to 0.24620, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 358s - loss: 2.2733 - acc: 0.8956 - val_loss: 0.2462 - val_acc: 0.9021\nEpoch 6/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.2368 - acc: 0.9035Epoch 00005: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 2.2372 - acc: 0.9034 - val_loss: 0.2856 - val_acc: 0.8847\nEpoch 7/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.2032 - acc: 0.9080Epoch 00006: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 2.2033 - acc: 0.9078 - val_loss: 0.2500 - val_acc: 0.8988\nEpoch 8/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.1767 - acc: 0.9130Epoch 00007: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 2.1766 - acc: 0.9130 - val_loss: 0.2467 - val_acc: 0.8970\nEpoch 9/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.1490 - acc: 0.9155Epoch 00008: val_loss improved from 0.24620 to 0.22698, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 359s - loss: 2.1492 - acc: 0.9154 - val_loss: 0.2270 - val_acc: 0.9162\nEpoch 10/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.1287 - acc: 0.9183Epoch 00009: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 2.1286 - acc: 0.9183 - val_loss: 0.2323 - val_acc: 0.9096\nEpoch 11/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.1064 - acc: 0.9205Epoch 00010: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 2.1063 - acc: 0.9206 - val_loss: 0.2288 - val_acc: 0.9100\nEpoch 12/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.0878 - acc: 0.9226Epoch 00011: val_loss improved from 0.22698 to 0.22173, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 359s - loss: 2.0878 - acc: 0.9224 - val_loss: 0.2217 - val_acc: 0.9164\nEpoch 13/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.0704 - acc: 0.9280Epoch 00012: val_loss improved from 0.22173 to 0.21532, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 357s - loss: 2.0702 - acc: 0.9280 - val_loss: 0.2153 - val_acc: 0.9192\nEpoch 14/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.0521 - acc: 0.9293Epoch 00013: val_loss did not improve\n10496/10496 [==============================] - 356s - loss: 2.0520 - acc: 0.9293 - val_loss: 0.2177 - val_acc: 0.9181\nEpoch 15/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.0382 - acc: 0.9322Epoch 00014: val_loss did not improve\n10496/10496 [==============================] - 356s - loss: 2.0381 - acc: 0.9322 - val_loss: 0.2229 - val_acc: 0.9162\nEpoch 16/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.0215 - acc: 0.9330Epoch 00015: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 2.0214 - acc: 0.9330 - val_loss: 0.2209 - val_acc: 0.9190\nEpoch 17/500\n10464/10496 [============================>.] - ETA: 1s - loss: 2.0082 - acc: 0.9359Epoch 00016: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 2.0080 - acc: 0.9358 - val_loss: 0.2175 - val_acc: 0.9230\nEpoch 18/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9936 - acc: 0.9397Epoch 00017: val_loss improved from 0.21532 to 0.20964, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 358s - loss: 1.9934 - acc: 0.9397 - val_loss: 0.2096 - val_acc: 0.9261\nEpoch 19/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9820 - acc: 0.9386Epoch 00018: val_loss did not improve\n10496/10496 [==============================] - 356s - loss: 1.9817 - acc: 0.9387 - val_loss: 0.2125 - val_acc: 0.9241\nEpoch 20/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9630 - acc: 0.9443Epoch 00019: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.9628 - acc: 0.9444 - val_loss: 0.2135 - val_acc: 0.9265\nEpoch 21/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9529 - acc: 0.9442Epoch 00020: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.9527 - acc: 0.9442 - val_loss: 0.2117 - val_acc: 0.9241\nEpoch 22/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9414 - acc: 0.9477Epoch 00021: val_loss improved from 0.20964 to 0.20949, saving model to ./cnn_140_rgb_lr_0.100000_decay_0.001000_sgd_he_normal__l1_0.000010_l2_0.000010_dropout_0.500000_r_training_weights_best.hdf5\n10496/10496 [==============================] - 358s - loss: 1.9411 - acc: 0.9479 - val_loss: 0.2095 - val_acc: 0.9278\nEpoch 23/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9271 - acc: 0.9515Epoch 00022: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.9269 - acc: 0.9516 - val_loss: 0.2210 - val_acc: 0.9258\nEpoch 24/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9166 - acc: 0.9543Epoch 00023: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.9163 - acc: 0.9545 - val_loss: 0.2175 - val_acc: 0.9272\nEpoch 25/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.9037 - acc: 0.9560Epoch 00024: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.9035 - acc: 0.9561 - val_loss: 0.2172 - val_acc: 0.9254\nEpoch 26/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8934 - acc: 0.9570Epoch 00025: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8934 - acc: 0.9571 - val_loss: 0.2266 - val_acc: 0.9247\nEpoch 27/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8819 - acc: 0.9620Epoch 00026: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8817 - acc: 0.9621 - val_loss: 0.2249 - val_acc: 0.9258\nEpoch 28/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8716 - acc: 0.9620Epoch 00027: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8714 - acc: 0.9621 - val_loss: 0.2291 - val_acc: 0.9243\nEpoch 29/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8607 - acc: 0.9638Epoch 00028: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8605 - acc: 0.9638 - val_loss: 0.2274 - val_acc: 0.9309\nEpoch 30/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8522 - acc: 0.9651Epoch 00029: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8520 - acc: 0.9652 - val_loss: 0.2270 - val_acc: 0.9302\nEpoch 31/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8440 - acc: 0.9665Epoch 00030: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8437 - acc: 0.9666 - val_loss: 0.2324 - val_acc: 0.9313\nEpoch 32/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8304 - acc: 0.9709Epoch 00031: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8302 - acc: 0.9710 - val_loss: 0.2351 - val_acc: 0.9276\nEpoch 33/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8241 - acc: 0.9710Epoch 00032: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8239 - acc: 0.9711 - val_loss: 0.2348 - val_acc: 0.9274\nEpoch 34/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8147 - acc: 0.9746Epoch 00033: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8145 - acc: 0.9747 - val_loss: 0.2396 - val_acc: 0.9305\nEpoch 35/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.8094 - acc: 0.9733Epoch 00034: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.8092 - acc: 0.9734 - val_loss: 0.2424 - val_acc: 0.9311\nEpoch 36/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7999 - acc: 0.9762Epoch 00035: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7997 - acc: 0.9762 - val_loss: 0.2503 - val_acc: 0.9300\nEpoch 37/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7892 - acc: 0.9796Epoch 00036: val_loss did not improve\n10496/10496 [==============================] - 359s - loss: 1.7891 - acc: 0.9797 - val_loss: 0.2416 - val_acc: 0.9311\nEpoch 38/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7839 - acc: 0.9791Epoch 00037: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7839 - acc: 0.9790 - val_loss: 0.2592 - val_acc: 0.9294\nEpoch 39/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7773 - acc: 0.9807Epoch 00038: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7771 - acc: 0.9808 - val_loss: 0.2525 - val_acc: 0.9322\nEpoch 40/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7722 - acc: 0.9802Epoch 00039: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7720 - acc: 0.9802 - val_loss: 0.2515 - val_acc: 0.9331\nEpoch 41/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7643 - acc: 0.9829Epoch 00040: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7642 - acc: 0.9829 - val_loss: 0.2687 - val_acc: 0.9313\nEpoch 42/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7612 - acc: 0.9817Epoch 00041: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7610 - acc: 0.9817 - val_loss: 0.2683 - val_acc: 0.9305\nEpoch 43/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7545 - acc: 0.9834Epoch 00042: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.7544 - acc: 0.9834 - val_loss: 0.2766 - val_acc: 0.9313\nEpoch 44/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7483 - acc: 0.9833Epoch 00043: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7482 - acc: 0.9833 - val_loss: 0.2684 - val_acc: 0.9309\nEpoch 45/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7429 - acc: 0.9837Epoch 00044: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7428 - acc: 0.9837 - val_loss: 0.2659 - val_acc: 0.9316\nEpoch 46/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7376 - acc: 0.9854Epoch 00045: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7375 - acc: 0.9855 - val_loss: 0.2776 - val_acc: 0.9318\nEpoch 47/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7315 - acc: 0.9864Epoch 00046: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7314 - acc: 0.9865 - val_loss: 0.2616 - val_acc: 0.9327\nEpoch 48/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7249 - acc: 0.9872Epoch 00047: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7248 - acc: 0.9872 - val_loss: 0.2701 - val_acc: 0.9309\nEpoch 49/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7215 - acc: 0.9883Epoch 00048: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7214 - acc: 0.9884 - val_loss: 0.2830 - val_acc: 0.9283\nEpoch 50/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7149 - acc: 0.9882Epoch 00049: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7148 - acc: 0.9883 - val_loss: 0.2903 - val_acc: 0.9300\nEpoch 51/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7073 - acc: 0.9915Epoch 00050: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7073 - acc: 0.9915 - val_loss: 0.3132 - val_acc: 0.9313\nEpoch 52/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7072 - acc: 0.9892Epoch 00051: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7071 - acc: 0.9893 - val_loss: 0.2897 - val_acc: 0.9327\nEpoch 53/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.7030 - acc: 0.9896Epoch 00052: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.7030 - acc: 0.9897 - val_loss: 0.2944 - val_acc: 0.9340\nEpoch 54/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6987 - acc: 0.9910Epoch 00053: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6986 - acc: 0.9910 - val_loss: 0.3109 - val_acc: 0.9313\nEpoch 55/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6950 - acc: 0.9915Epoch 00054: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6949 - acc: 0.9915 - val_loss: 0.2962 - val_acc: 0.9309\nEpoch 56/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6900 - acc: 0.9899Epoch 00055: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.6899 - acc: 0.9899 - val_loss: 0.3105 - val_acc: 0.9296\nEpoch 57/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6866 - acc: 0.9909Epoch 00056: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6865 - acc: 0.9909 - val_loss: 0.2949 - val_acc: 0.9331\nEpoch 58/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6818 - acc: 0.9929Epoch 00057: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.6818 - acc: 0.9929 - val_loss: 0.2985 - val_acc: 0.9357\nEpoch 59/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6776 - acc: 0.9928Epoch 00058: val_loss did not improve\n10496/10496 [==============================] - 359s - loss: 1.6776 - acc: 0.9929 - val_loss: 0.3056 - val_acc: 0.9349\nEpoch 60/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6735 - acc: 0.9926Epoch 00059: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6734 - acc: 0.9927 - val_loss: 0.3240 - val_acc: 0.9375\nEpoch 61/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6743 - acc: 0.9923Epoch 00060: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.6742 - acc: 0.9923 - val_loss: 0.3155 - val_acc: 0.9318\nEpoch 62/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6690 - acc: 0.9924Epoch 00061: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6689 - acc: 0.9924 - val_loss: 0.3316 - val_acc: 0.9305\nEpoch 63/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6640 - acc: 0.9933Epoch 00062: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6640 - acc: 0.9933 - val_loss: 0.3263 - val_acc: 0.9322\nEpoch 64/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6603 - acc: 0.9932Epoch 00063: val_loss did not improve\n10496/10496 [==============================] - 359s - loss: 1.6603 - acc: 0.9932 - val_loss: 0.3359 - val_acc: 0.9309\nEpoch 65/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6582 - acc: 0.9943Epoch 00064: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6582 - acc: 0.9943 - val_loss: 0.3313 - val_acc: 0.9318\nEpoch 66/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6532 - acc: 0.9938Epoch 00065: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6531 - acc: 0.9938 - val_loss: 0.3311 - val_acc: 0.9335\nEpoch 67/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6517 - acc: 0.9938Epoch 00066: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6516 - acc: 0.9938 - val_loss: 0.3348 - val_acc: 0.9353\nEpoch 68/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6493 - acc: 0.9941Epoch 00067: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6492 - acc: 0.9941 - val_loss: 0.3352 - val_acc: 0.9327\nEpoch 69/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6478 - acc: 0.9947Epoch 00068: val_loss did not improve\n10496/10496 [==============================] - 359s - loss: 1.6477 - acc: 0.9948 - val_loss: 0.3549 - val_acc: 0.9327\nEpoch 70/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6423 - acc: 0.9955Epoch 00069: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.6423 - acc: 0.9955 - val_loss: 0.3352 - val_acc: 0.9340\nEpoch 71/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6411 - acc: 0.9952Epoch 00070: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6410 - acc: 0.9952 - val_loss: 0.3494 - val_acc: 0.9349\nEpoch 72/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6368 - acc: 0.9955Epoch 00071: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6368 - acc: 0.9955 - val_loss: 0.3598 - val_acc: 0.9335\nEpoch 73/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6358 - acc: 0.9947Epoch 00072: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6357 - acc: 0.9948 - val_loss: 0.3638 - val_acc: 0.9331\nEpoch 74/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6323 - acc: 0.9953Epoch 00073: val_loss did not improve\n10496/10496 [==============================] - 359s - loss: 1.6323 - acc: 0.9953 - val_loss: 0.3654 - val_acc: 0.9305\nEpoch 75/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6320 - acc: 0.9953Epoch 00074: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6320 - acc: 0.9953 - val_loss: 0.3647 - val_acc: 0.9331\nEpoch 76/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6270 - acc: 0.9963Epoch 00075: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6270 - acc: 0.9963 - val_loss: 0.3634 - val_acc: 0.9322\nEpoch 77/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6255 - acc: 0.9948Epoch 00076: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6254 - acc: 0.9949 - val_loss: 0.3591 - val_acc: 0.9318\nEpoch 78/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6232 - acc: 0.9961Epoch 00077: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6232 - acc: 0.9961 - val_loss: 0.3631 - val_acc: 0.9322\nEpoch 79/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6198 - acc: 0.9965Epoch 00078: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6198 - acc: 0.9965 - val_loss: 0.3704 - val_acc: 0.9313\nEpoch 80/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6189 - acc: 0.9960Epoch 00079: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6188 - acc: 0.9960 - val_loss: 0.3664 - val_acc: 0.9305\nEpoch 81/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6148 - acc: 0.9971Epoch 00080: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.6148 - acc: 0.9971 - val_loss: 0.3917 - val_acc: 0.9296\nEpoch 82/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6140 - acc: 0.9960Epoch 00081: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6140 - acc: 0.9960 - val_loss: 0.3701 - val_acc: 0.9327\nEpoch 83/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6139 - acc: 0.9955Epoch 00082: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6140 - acc: 0.9954 - val_loss: 0.3840 - val_acc: 0.9327\nEpoch 84/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6107 - acc: 0.9957Epoch 00083: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6107 - acc: 0.9957 - val_loss: 0.3797 - val_acc: 0.9300\nEpoch 85/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6076 - acc: 0.9968Epoch 00084: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6076 - acc: 0.9969 - val_loss: 0.3894 - val_acc: 0.9313\nEpoch 86/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6060 - acc: 0.9968Epoch 00085: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6060 - acc: 0.9968 - val_loss: 0.3959 - val_acc: 0.9300\nEpoch 87/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6039 - acc: 0.9965Epoch 00086: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6038 - acc: 0.9965 - val_loss: 0.3895 - val_acc: 0.9291\nEpoch 88/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5996 - acc: 0.9977Epoch 00087: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.5995 - acc: 0.9977 - val_loss: 0.3908 - val_acc: 0.9291\nEpoch 89/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.6006 - acc: 0.9966Epoch 00088: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.6005 - acc: 0.9966 - val_loss: 0.3934 - val_acc: 0.9305\nEpoch 90/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5978 - acc: 0.9967Epoch 00089: val_loss did not improve\n10496/10496 [==============================] - 359s - loss: 1.5977 - acc: 0.9967 - val_loss: 0.3865 - val_acc: 0.9309\nEpoch 91/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5955 - acc: 0.9970Epoch 00090: val_loss did not improve\n10496/10496 [==============================] - 360s - loss: 1.5955 - acc: 0.9970 - val_loss: 0.3922 - val_acc: 0.9318\nEpoch 92/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5935 - acc: 0.9973Epoch 00091: val_loss did not improve\n10496/10496 [==============================] - 359s - loss: 1.5935 - acc: 0.9973 - val_loss: 0.3983 - val_acc: 0.9318\nEpoch 93/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5907 - acc: 0.9972Epoch 00092: val_loss did not improve\n10496/10496 [==============================] - 360s - loss: 1.5907 - acc: 0.9972 - val_loss: 0.4099 - val_acc: 0.9327\nEpoch 94/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5899 - acc: 0.9972Epoch 00093: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.5898 - acc: 0.9972 - val_loss: 0.4004 - val_acc: 0.9300\nEpoch 95/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5883 - acc: 0.9976Epoch 00094: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.5883 - acc: 0.9976 - val_loss: 0.4048 - val_acc: 0.9287\nEpoch 96/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5874 - acc: 0.9967Epoch 00095: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.5874 - acc: 0.9967 - val_loss: 0.4025 - val_acc: 0.9291\nEpoch 97/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5850 - acc: 0.9972Epoch 00096: val_loss did not improve\n10496/10496 [==============================] - 358s - loss: 1.5850 - acc: 0.9972 - val_loss: 0.3957 - val_acc: 0.9291\nEpoch 98/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5830 - acc: 0.9975Epoch 00097: val_loss did not improve\n10496/10496 [==============================] - 357s - loss: 1.5830 - acc: 0.9975 - val_loss: 0.3947 - val_acc: 0.9287\nEpoch 99/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5812 - acc: 0.9974Epoch 00098: val_loss did not improve\n10496/10496 [==============================] - 356s - loss: 1.5812 - acc: 0.9974 - val_loss: 0.3934 - val_acc: 0.9327\nEpoch 100/500\n10464/10496 [============================>.] - ETA: 1s - loss: 1.5802 - acc: 0.9975Epoch 00099: val_loss did not improve\n10496/10496 [==============================] - 356s - loss: 1.5802 - acc: 0.9975 - val_loss: 0.4062 - val_acc: 0.9300\nEpoch 101/500\n 3968/10496 [==========>...................] - ETA: 214s - loss: 1.5792 - acc: 0.9972"
],
[
"print('Evaluating model...')\nscore = model.evaluate_generator(test_data_gen, val_samples=test_data_gen.n_samples)\nprint('done.')\n\nprint('Test score:', score[0])\nprint('Test accuracy:', score[1])",
"Evaluating model...\ndone.\nTest score: 0.410274893211\nTest accuracy: 0.931338028169\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbaf6528b9837285db44f06ca5ef2027dab64c03
| 832,822 |
ipynb
|
Jupyter Notebook
|
P1.ipynb
|
melihyazgan/Udacity_SDC
|
901ffa1eb48605cfc75e3dfc0a608d6a8a8c9e78
|
[
"MIT"
] | null | null | null |
P1.ipynb
|
melihyazgan/Udacity_SDC
|
901ffa1eb48605cfc75e3dfc0a608d6a8a8c9e78
|
[
"MIT"
] | null | null | null |
P1.ipynb
|
melihyazgan/Udacity_SDC
|
901ffa1eb48605cfc75e3dfc0a608d6a8a8c9e78
|
[
"MIT"
] | null | null | null | 1,111.911883 | 128,324 | 0.954892 |
[
[
[
"# Self-Driving Car Engineer Nanodegree\n\n\n## Project: **Finding Lane Lines on the Road** \n***\nIn this project, you will use the tools you learned about in the lesson to identify lane lines on the road. You can develop your pipeline on a series of individual images, and later apply the result to a video stream (really just a series of images). Check out the video clip \"raw-lines-example.mp4\" (also contained in this repository) to see what the output should look like after using the helper functions below. \n\nOnce you have a result that looks roughly like \"raw-lines-example.mp4\", you'll need to get creative and try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\". Ultimately, you would like to draw just one line for the left side of the lane, and one for the right.\n\nIn addition to implementing code, there is a brief writeup to complete. The writeup should be completed in a separate file, which can be either a markdown file or a pdf document. There is a [write up template](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) that can be used to guide the writing process. Completing both the code in the Ipython notebook and the writeup template will cover all of the [rubric points](https://review.udacity.com/#!/rubrics/322/view) for this project.\n\n---\nLet's have a look at our first image called 'test_images/solidWhiteRight.jpg'. Run the 2 cells below (hit Shift-Enter or the \"play\" button above) to display the image.\n\n**Note: If, at any point, you encounter frozen display windows or other confounding issues, you can always start again with a clean slate by going to the \"Kernel\" menu above and selecting \"Restart & Clear Output\".**\n\n---",
"_____no_output_____"
],
[
"**The tools you have are color selection, region of interest selection, grayscaling, Gaussian smoothing, Canny Edge Detection and Hough Tranform line detection. You are also free to explore and try other techniques that were not presented in the lesson. Your goal is piece together a pipeline to detect the line segments in the image, then average/extrapolate them and draw them onto the image for display (as below). Once you have a working pipeline, try it out on the video stream below.**\n\n---\n\n<figure>\n <img src=\"examples/line-segments-example.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your output should look something like this (above) after detecting line segments using the helper functions below </p> \n </figcaption>\n</figure>\n <p></p> \n<figure>\n <img src=\"examples/laneLines_thirdPass.jpg\" width=\"380\" alt=\"Combined Image\" />\n <figcaption>\n <p></p> \n <p style=\"text-align: center;\"> Your goal is to connect/average/extrapolate line segments to get output like this</p> \n </figcaption>\n</figure>",
"_____no_output_____"
],
[
"**Run the cell below to import some packages. If you get an `import error` for a package you've already installed, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.** ",
"_____no_output_____"
],
[
"## Import Packages",
"_____no_output_____"
]
],
[
[
"#importing some useful packages\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nimport numpy as np\nimport cv2\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Read in an Image",
"_____no_output_____"
]
],
[
[
"#reading in an image\nimage = mpimg.imread('test_images/solidWhiteRight.jpg')\n\n#printing out some stats and plotting\nprint('This image is:', type(image), 'with dimensions:', image.shape)\nplt.imshow(image) # if you wanted to show a single color channel image called 'gray', for example, call as plt.imshow(gray, cmap='gray')",
"This image is: <class 'numpy.ndarray'> with dimensions: (540, 960, 3)\n"
]
],
[
[
"## Ideas for Lane Detection Pipeline",
"_____no_output_____"
],
[
"**Some OpenCV functions (beyond those introduced in the lesson) that might be useful for this project are:**\n\n`cv2.inRange()` for color selection \n`cv2.fillPoly()` for regions selection \n`cv2.line()` to draw lines on an image given endpoints \n`cv2.addWeighted()` to coadd / overlay two images\n`cv2.cvtColor()` to grayscale or change color\n`cv2.imwrite()` to output images to file \n`cv2.bitwise_and()` to apply a mask to an image\n\n**Check out the OpenCV documentation to learn about these and discover even more awesome functionality!**",
"_____no_output_____"
],
[
"## Helper Functions",
"_____no_output_____"
],
[
"Below are some helper functions to help get you started. They should look familiar from the lesson!",
"_____no_output_____"
]
],
[
[
"import math\n\ndef grayscale(img):\n \"\"\"Applies the Grayscale transform\n This will return an image with only one color channel\n but NOTE: to see the returned image as grayscale\n (assuming your grayscaled image is called 'gray')\n you should call plt.imshow(gray, cmap='gray')\"\"\"\n return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n # Or use BGR2GRAY if you read an image with cv2.imread()\n # return cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)\n \ndef canny(img, method):\n \"\"\"Applies the Canny transform\"\"\"\n \"\"\" Bi-Modal Distribution\"\"\"\n if method == \"OTSU\":\n high_threshold, th3 = cv2.threshold(img,0,255, cv2.THRESH_BINARY+cv2.THRESH_OTSU)\n ratio = 0.5\n low_threshold = high_threshold * 0.5\n elif method == \"auto\":\n sigma = 0.33\n v = np.median(img)\n low_threshold = int(max(0, (1.0 - sigma) * v))\n high_threshold = int(min(255, (1.0 + sigma) * v))\n else:\n ratio = 1/3\n high_threshold = 150\n low_threshold = high_threshold * ratio\n \n #print(\"Lowth: {}, Highth: {}\".format(low_threshold,high_threshold) )\n return cv2.Canny(img, low_threshold, high_threshold)\n\ndef gaussian_blur(img, kernel_size):\n \"\"\"Applies a Gaussian Noise kernel\"\"\"\n return cv2.GaussianBlur(img, (kernel_size, kernel_size), 0)\n\ndef region_of_interest(img, vertices):\n \"\"\"\n Applies an image mask.\n \n Only keeps the region of the image defined by the polygon\n formed from `vertices`. The rest of the image is set to black.\n \"\"\"\n #defining a blank mask to start with\n mask = np.zeros_like(img) \n \n #defining a 3 channel or 1 channel color to fill the mask with depending on the input image\n if len(img.shape) > 2:\n channel_count = img.shape[2] # i.e. 3 or 4 depending on your image\n ignore_mask_color = (255,) * channel_count\n else:\n ignore_mask_color = 255\n \n #filling pixels inside the polygon defined by \"vertices\" with the fill color \n cv2.fillPoly(mask, vertices, ignore_mask_color)\n \n #returning the image only where mask pixels are nonzero\n masked_image = cv2.bitwise_and(img, mask)\n return masked_image\n\n\ndef draw_lines(img, lines, color=[255, 0, 0], thickness=2):\n \"\"\"\n NOTE: this is the function you might want to use as a starting point once you want to \n average/extrapolate the line segments you detect to map out the full\n extent of the lane (going from the result shown in raw-lines-example.mp4\n to that shown in P1_example.mp4). \n \n Think about things like separating line segments by their \n slope ((y2-y1)/(x2-x1)) to decide which segments are part of the left\n line vs. the right line. Then, you can average the position of each of \n the lines and extrapolate to the top and bottom of the lane.\n \n This function draws `lines` with `color` and `thickness`. \n Lines are drawn on the image inplace (mutates the image).\n If you want to make the lines semi-transparent, think about combining\n this function with the weighted_img() function below\n \"\"\"\n xLeft = []\n xRight = []\n yLeft = []\n yRight = []\n for line in lines:\n for x1,y1,x2,y2 in line:\n m = (y2-y1)/(x2-x1)\n #import pdb; pdb.set_trace()\n angle = abs(math.atan(m) * 180/math.pi)\n if angle >=20 and angle <= 70 : \n if m < 0:\n xLeft.append(x1)\n xLeft.append(x2)\n yLeft.append(y1)\n yLeft.append(y2)\n ext_line(img,xLeft,yLeft)\n else:\n xRight.append(x1)\n xRight.append(x2)\n yRight.append(y1)\n yRight.append(y2)\n ext_line(img,xRight,yRight)\n\ndef ext_line(img,x,y):\n z = np.polyfit(x,y,1)\n #z_right = np.polyfit(xRight,yRight,1)\n # extrapolate the top and bottom of the lane. y_top = vertices, and y_bottom = img.shape[0]\n #boundry = vertices[0]\n top_y = 330\n top_x = int((top_y-z[1])/z[0])\n bottom_y = img.shape[0]\n bottom_x = int((bottom_y-z[1])/z[0])\n cv2.line(img, (bottom_x, bottom_y), (top_x, top_y), [255, 0, 0], 8)\n\ndef hough_lines(img, rho, theta, threshold, min_line_len, max_line_gap):\n \"\"\"\n `img` should be the output of a Canny transform.\n \n Returns an image with hough lines drawn.\n \"\"\"\n lines = cv2.HoughLinesP(img, rho, theta, threshold, np.array([]), minLineLength=min_line_len, maxLineGap=max_line_gap)\n line_img = np.zeros((img.shape[0], img.shape[1], 3), dtype=np.uint8)\n draw_lines(line_img, lines)\n return line_img\n\n# Python 3 has support for cool math symbols.\n\ndef weighted_img(img, initial_img, α=0.8, β=1., γ=0.):\n \"\"\"\n `img` is the output of the hough_lines(), An image with lines drawn on it.\n Should be a blank image (all black) with lines drawn on it.\n \n `initial_img` should be the image before any processing.\n \n The result image is computed as follows:\n \n initial_img * α + img * β + γ\n NOTE: initial_img and img must be the same shape!\n \"\"\"\n return cv2.addWeighted(initial_img, α, img, β, γ)\n\ndef save_image(img, name):\n mpimg.imsave('./output_images/{0}'.format(name),img)",
"_____no_output_____"
]
],
[
[
"## Test Images\n\nBuild your pipeline to work on the images in the directory \"test_images\" \n**You should make sure your pipeline works well on these images before you try the videos.**",
"_____no_output_____"
]
],
[
[
"import os\ntest_images = os.listdir(\"test_images/\")",
"_____no_output_____"
]
],
[
[
"## Build a Lane Finding Pipeline",
"_____no_output_____"
],
[
"Build the pipeline and run your solution on all test_images. Make copies into the `test_images_output` directory, and you can use the images in your writeup report.\n\nTry tuning the various parameters, especially the low and high Canny thresholds as well as the Hough lines parameters.",
"_____no_output_____"
]
],
[
[
"# TODO: Build your pipeline that will draw lane lines on the test_images\n# then save them to the test_images_output directory.\n# Read in and grayscale the image\ndef my_pipeline(img):\n gray_img = grayscale(img)\n #plt.imshow(gray_img, cmap= \"gray\")\n #plt.show()\n #Define a kernel size and apply Gaussian smoothing\n kernel_size = 3\n blur_gray = gaussian_blur(gray_img,kernel_size)\n \n #Define our parameters for Canny and apply\n edges = canny(blur_gray,\"manual\") \n \n # This time we are defining a four sided polygon to mask\n imshape = img.shape\n vertices = np.array([[(0,imshape[0]),(450, 325), (490, 325), (imshape[1],imshape[0])]], dtype=np.int32)\n masked_edges = region_of_interest(edges, vertices)\n \n #Define the Hough transform parameters\n \n rho = 2 # distance resolution in pixels of the Hough grid\n theta = np.pi/180 # angular resolution in radians of the Hough grid\n threshold = 40 # minimum number of votes (intersections in Hough grid cell)\n min_line_len = 90 #minimum number of pixels making up a line\n max_line_gap = 150 # maximum gap in pixels between connectable line segments\n line_img = hough_lines(masked_edges, rho, theta, threshold, min_line_len, max_line_gap)\n end_img = weighted_img(line_img, img)\n return end_img\n\n",
"_____no_output_____"
],
[
"def plot_allimages():\n for image in test_images:\n path = 'test_images/' + str(image)\n #path = os.path.join(os.getcwd(),image_name)\n img = cv2.imread(path)\n processed_img = my_pipeline(img)\n save_image(processed_img,image)\n plt.imshow(processed_img)\n plt.show()\nplot_allimages()",
"_____no_output_____"
]
],
[
[
"## Test on Videos\n\nYou know what's cooler than drawing lanes over images? Drawing lanes over video!\n\nWe can test our solution on two provided videos:\n\n`solidWhiteRight.mp4`\n\n`solidYellowLeft.mp4`\n\n**Note: if you get an import error when you run the next cell, try changing your kernel (select the Kernel menu above --> Change Kernel). Still have problems? Try relaunching Jupyter Notebook from the terminal prompt. Also, consult the forums for more troubleshooting tips.**\n\n**If you get an error that looks like this:**\n```\nNeedDownloadError: Need ffmpeg exe. \nYou can download it by calling: \nimageio.plugins.ffmpeg.download()\n```\n**Follow the instructions in the error message and check out [this forum post](https://discussions.udacity.com/t/project-error-of-test-on-videos/274082) for more troubleshooting tips across operating systems.**",
"_____no_output_____"
]
],
[
[
"# Import everything needed to edit/save/watch video clips\nfrom moviepy.editor import VideoFileClip\nfrom IPython.display import HTML",
"_____no_output_____"
],
[
"def process_image(image):\n # NOTE: The output you return should be a color image (3 channel) for processing video below\n # TODO: put your pipeline here,\n # you should return the final output (image where lines are drawn on lanes)\n result = my_pipeline(image)\n return result\n",
"_____no_output_____"
]
],
[
[
"Let's try the one with the solid white lane on the right first ...",
"_____no_output_____"
]
],
[
[
"white_output = 'test_videos_output/solidWhiteRight.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n#clip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\").subclip(0,5)\nclip1 = VideoFileClip(\"test_videos/solidWhiteRight.mp4\")\nwhite_clip = clip1.fl_image(process_image) #NOTE: this function expects color images!!\n%time white_clip.write_videofile(white_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/solidWhiteRight.mp4\n[MoviePy] Writing video test_videos_output/solidWhiteRight.mp4\n"
]
],
[
[
"Play the video inline, or if you prefer find the video in your filesystem (should be in the same directory) and play it in your video player of choice.",
"_____no_output_____"
]
],
[
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(white_output))",
"_____no_output_____"
]
],
[
[
"## Improve the draw_lines() function\n\n**At this point, if you were successful with making the pipeline and tuning parameters, you probably have the Hough line segments drawn onto the road, but what about identifying the full extent of the lane and marking it clearly as in the example video (P1_example.mp4)? Think about defining a line to run the full length of the visible lane based on the line segments you identified with the Hough Transform. As mentioned previously, try to average and/or extrapolate the line segments you've detected to map out the full extent of the lane lines. You can see an example of the result you're going for in the video \"P1_example.mp4\".**\n\n**Go back and modify your draw_lines function accordingly and try re-running your pipeline. The new output should draw a single, solid line over the left lane line and a single, solid line over the right lane line. The lines should start from the bottom of the image and extend out to the top of the region of interest.**",
"_____no_output_____"
],
[
"Now for the one with the solid yellow lane on the left. This one's more tricky!",
"_____no_output_____"
]
],
[
[
"yellow_output = 'test_videos_output/solidYellowLeft.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip2 = VideoFileClip('test_videos/solidYellowLeft.mp4').subclip(0,5)\nclip2 = VideoFileClip('test_videos/solidYellowLeft.mp4')\nyellow_clip = clip2.fl_image(process_image)\n%time yellow_clip.write_videofile(yellow_output, audio=False)",
"[MoviePy] >>>> Building video test_videos_output/solidYellowLeft.mp4\n[MoviePy] Writing video test_videos_output/solidYellowLeft.mp4\n"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(yellow_output))",
"_____no_output_____"
]
],
[
[
"## Writeup and Submission\n\nIf you're satisfied with your video outputs, it's time to make the report writeup in a pdf or markdown file. Once you have this Ipython notebook ready along with the writeup, it's time to submit for review! Here is a [link](https://github.com/udacity/CarND-LaneLines-P1/blob/master/writeup_template.md) to the writeup template file.\n",
"_____no_output_____"
],
[
"## Optional Challenge\n\nTry your lane finding pipeline on the video below. Does it still work? Can you figure out a way to make it more robust? If you're up for the challenge, modify your pipeline so it works with this video and submit it along with the rest of your project!",
"_____no_output_____"
]
],
[
[
"challenge_output = 'test_videos_output/challenge.mp4'\n## To speed up the testing process you may want to try your pipeline on a shorter subclip of the video\n## To do so add .subclip(start_second,end_second) to the end of the line below\n## Where start_second and end_second are integer values representing the start and end of the subclip\n## You may also uncomment the following line for a subclip of the first 5 seconds\n##clip3 = VideoFileClip('test_videos/challenge.mp4').subclip(0,5)\nclip3 = VideoFileClip('test_videos/challenge.mp4')\nchallenge_clip = clip3.fl_image(process_image3)\n%time challenge_clip.write_videofile(challenge_output, audio=False)",
"_____no_output_____"
],
[
"HTML(\"\"\"\n<video width=\"960\" height=\"540\" controls>\n <source src=\"{0}\">\n</video>\n\"\"\".format(challenge_output))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbaf6a5f12daae93af21242ba64f4e3a63b1610e
| 16,792 |
ipynb
|
Jupyter Notebook
|
dask.ipynb
|
RichardScottOZ/pangeo-tutorial-gallery
|
a0ee3071f95215d2a21105a52924f91105f84247
|
[
"MIT"
] | null | null | null |
dask.ipynb
|
RichardScottOZ/pangeo-tutorial-gallery
|
a0ee3071f95215d2a21105a52924f91105f84247
|
[
"MIT"
] | null | null | null |
dask.ipynb
|
RichardScottOZ/pangeo-tutorial-gallery
|
a0ee3071f95215d2a21105a52924f91105f84247
|
[
"MIT"
] | null | null | null | 28.033389 | 670 | 0.588018 |
[
[
[
"# Dask Tutorial\n\n\n<div class=\"alert-info\">\n\n### Overview\n \n* **teaching:** 20 minutes\n* **exercises:** 0\n* **questions:**\n * How does Dask parallelize computations in Python?\n</div>",
"_____no_output_____"
],
[
"### Table of contents\n1. [**Dask primer**](#Dask-primer)\n1. [**Dask clusters**](#Dask-Clusters)\n1. [**Dask dataframe**](#Dask-Dataframe)\n1. [**Dask arrays**](#Dask-Arrays)\n1. [**Dask delayed**](#Dask-Delayed)",
"_____no_output_____"
],
[
"## Dask Primer\n\n<img src=\"http://dask.readthedocs.io/en/latest/_images/dask_horizontal.svg\" \n width=\"30%\" \n align=right\n alt=\"Dask logo\">\n\n\nDask is a flexible parallel computing library for analytic computing. Dask provides dynamic parallel task scheduling and high-level big-data collections like `dask.array` and `dask.dataframe`. More on dask here: https://docs.dask.org/en/latest/\n\n_Note: Pieces of this notebook comes from the following sources:_\n\n- https://github.com/rabernat/research_computing\n- https://github.com/dask/dask-examples",
"_____no_output_____"
],
[
"## Dask Clusters\n\nDask needs a collection of computing resources in order to perform parallel computations. Dask Clusters have different names corresponding to different computing environments (for example, [LocalCluster](https://distributed.dask.org/en/latest/local-cluster.html) for your Laptop, [PBSCluster](http://jobqueue.dask.org/) for your HPC, or [Kubernetes Cluster](http://kubernetes.dask.org/) for machines on the Cloud). Each cluster has a certain number of computing resources called 'Workers', that each get allocated CPU and RAM. The dask scheduling system maps jobs to each worker on a cluster for you, so the syntax is mostly the same once you initialize a cluster!",
"_____no_output_____"
]
],
[
[
"# Let's start simple with a LocalCluster that makes use of all the cores and RAM we have on a single machine\nfrom dask.distributed import Client, LocalCluster\ncluster = LocalCluster()\n# explicitly connect to the cluster we just created\nclient = Client(cluster)\nclient",
"_____no_output_____"
]
],
[
[
"## Dask Dataframe\n\nIf you are working with a very large Pandas dataframe, you can consider parallizing computations by turning it into a Dask Dataframe. Dask Dataframes split a dataframe into partitions along an index. They support a large subset of the Pandas API. You can find additional details and examples here https://examples.dask.org/dataframe.html\n",
"_____no_output_____"
]
],
[
[
"# Although this is small csv file, we'll reuse our same example from before!\n# Load csv results from server into a Pandas DataFrame\nimport dask.dataframe as dd\nserver = 'https://webservices.volcano.si.edu/geoserver/GVP-VOTW/ows?'\nquery = 'service=WFS&version=2.0.0&request=GetFeature&typeName=GVP-VOTW:Smithsonian_VOTW_Holocene_Volcanoes&outputFormat=csv'\n\n# blocksize=None means use a single partion\ndf = dd.read_csv(server+query, blocksize=None)",
"_____no_output_____"
],
[
"# We only see the metadata, the actual data are only computed when requested.\ndf",
"_____no_output_____"
],
[
"# We can break up the table into 4 partions to map out to each core:\ndf = df.repartition(npartitions=4)\ndf",
"_____no_output_____"
],
[
"# Let's say we want to know the minimum last eruption year for all volcanoes\nlast_eruption_year_min = df.Last_Eruption_Year.min()\nlast_eruption_year_min",
"_____no_output_____"
],
[
"# Instead of getting the actual value we see dd.Scalar, which represents a recipe for actually calculating this value\nlast_eruption_year_min.visualize(format='svg')",
"_____no_output_____"
],
[
"# To get the value call the 'compute method'\n# NOTE: this was slower than using pandas directly,,, for small data you often don't need to use parallel computing!\nlast_eruption_year_min.compute()",
"_____no_output_____"
]
],
[
[
"## Dask Arrays\n\nA dask array looks and feels a lot like a numpy array.\nHowever, a dask array doesn't directly hold any data.\nInstead, it symbolically represents the computations needed to generate the data.\nNothing is actually computed until the actual numerical values are needed.\nThis mode of operation is called \"lazy\"; it allows one to build up complex, large calculations symbolically before turning them over the scheduler for execution.\n\nIf we want to create a numpy array of all ones, we do it like this:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nshape = (1000, 4000)\nones_np = np.ones(shape)\nones_np",
"_____no_output_____"
]
],
[
[
"This array contains exactly 32 MB of data:",
"_____no_output_____"
]
],
[
[
"print('%.1f MB' % (ones_np.nbytes / 1e6))",
"_____no_output_____"
]
],
[
[
"Now let's create the same array using dask's array interface.",
"_____no_output_____"
]
],
[
[
"import dask.array as da\nones = da.ones(shape)\nones",
"_____no_output_____"
]
],
[
[
"This works, but we didn't tell dask how to split up the array, so it is not optimized for distributed computation.\n\nA crucal difference with dask is that we must specify the `chunks` argument. \"Chunks\" describes how the array is split up over many sub-arrays.\n\n\n_source: [Dask Array Documentation](http://dask.pydata.org/en/latest/array-overview.html)_\n\nThere are [several ways to specify chunks](http://dask.pydata.org/en/latest/array-creation.html#chunks).\nIn this lecture, we will use a block shape.",
"_____no_output_____"
]
],
[
[
"chunk_shape = (1000, 1000)\nones = da.ones(shape, chunks=chunk_shape)\nones",
"_____no_output_____"
]
],
[
[
"Notice that we just see a symbolic represetnation of the array, including its shape, dtype, and chunksize.\nNo data has been generated yet.\nWhen we call `.compute()` on a dask array, the computation is trigger and the dask array becomes a numpy array.",
"_____no_output_____"
]
],
[
[
"ones.compute()",
"_____no_output_____"
]
],
[
[
"In order to understand what happened when we called `.compute()`, we can visualize the dask _graph_, the symbolic operations that make up the array",
"_____no_output_____"
]
],
[
[
"ones.visualize(format='svg')",
"_____no_output_____"
]
],
[
[
"Our array has four chunks. To generate it, dask calls `np.ones` four times and then concatenates this together into one array.\n\nRather than immediately loading a dask array (which puts all the data into RAM), it is more common to reduce the data somehow. For example:",
"_____no_output_____"
]
],
[
[
"sum_of_ones = ones.sum()\nsum_of_ones.visualize(format='svg')",
"_____no_output_____"
]
],
[
[
"Here we see dask's strategy for finding the sum. This simple example illustrates the beauty of dask: it automatically designs an algorithm appropriate for custom operations with big data. \n\nIf we make our operation more complex, the graph gets more complex.",
"_____no_output_____"
]
],
[
[
"fancy_calculation = (ones * ones[::-1, ::-1]).mean()\nfancy_calculation.visualize(format='svg')",
"_____no_output_____"
]
],
[
[
"### A Bigger Calculation\n\nThe examples above were toy examples; the data (32 MB) is nowhere nearly big enough to warrant the use of dask.\n\nWe can make it a lot bigger!",
"_____no_output_____"
]
],
[
[
"bigshape = (200000, 4000)\nbig_ones = da.ones(bigshape, chunks=chunk_shape)\nbig_ones",
"_____no_output_____"
],
[
"print('%.1f MB' % (big_ones.nbytes / 1e6))",
"_____no_output_____"
]
],
[
[
"This dataset is 6.4 GB, rather than 32 MB! This is probably close to or greater than the amount of available RAM than you have in your computer. Nevertheless, dask has no problem working on it.\n\n_Do not try to `.visualize()` this array!_\n\nWhen doing a big calculation, dask also has some tools to help us understand what is happening under the hood. Let's watch the dashboard again as we do a bigger computation.",
"_____no_output_____"
]
],
[
[
"big_calc = (big_ones * big_ones[::-1, ::-1]).mean()\n\nresult = big_calc.compute()\nresult",
"_____no_output_____"
]
],
[
[
"### Reduction \n\nAll the usual numpy methods work on dask arrays.\nYou can also apply numpy function directly to a dask array, and it will stay lazy.",
"_____no_output_____"
]
],
[
[
"big_ones_reduce = (np.cos(big_ones)**2).mean(axis=1)\nbig_ones_reduce",
"_____no_output_____"
]
],
[
[
"Plotting also triggers computation, since we need the actual values",
"_____no_output_____"
]
],
[
[
"from matplotlib import pyplot as plt\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (12,8)",
"_____no_output_____"
],
[
"plt.plot(big_ones_reduce)",
"_____no_output_____"
]
],
[
[
"## Dask Delayed\n\nDask.delayed is a simple and powerful way to parallelize existing code. It allows users to delay function calls into a task graph with dependencies. Dask.delayed doesn't provide any fancy parallel algorithms like Dask.dataframe, but it does give the user complete control over what they want to build.\n\nSystems like Dask.dataframe are built with Dask.delayed. If you have a problem that is paralellizable, but isn't as simple as just a big array or a big dataframe, then dask.delayed may be the right choice for you.\n\n## Create simple functions\n\nThese functions do simple operations like add two numbers together, but they sleep for a random amount of time to simulate real work.",
"_____no_output_____"
]
],
[
[
"import time\n\ndef inc(x):\n time.sleep(0.1)\n return x + 1\n\ndef dec(x):\n time.sleep(0.1)\n return x - 1\n \ndef add(x, y):\n time.sleep(0.2)\n return x + y ",
"_____no_output_____"
]
],
[
[
"We can run them like normal Python functions below",
"_____no_output_____"
]
],
[
[
"%%time\nx = inc(1)\ny = dec(2)\nz = add(x, y)\nz",
"_____no_output_____"
]
],
[
[
"These ran one after the other, in sequence. Note though that the first two lines `inc(1)` and `dec(2)` don't depend on each other, we *could* have called them in parallel had we been clever.\n\n## Annotate functions with Dask Delayed to make them lazy\n\nWe can call `dask.delayed` on our funtions to make them lazy. Rather than compute their results immediately, they record what we want to compute as a task into a graph that we'll run later on parallel hardware.",
"_____no_output_____"
]
],
[
[
"import dask\ninc = dask.delayed(inc)\ndec = dask.delayed(dec)\nadd = dask.delayed(add)",
"_____no_output_____"
]
],
[
[
"Calling these lazy functions is now almost free. We're just constructing a graph",
"_____no_output_____"
]
],
[
[
"%%time\nx = inc(1)\ny = dec(2)\nz = add(x, y)\nz",
"_____no_output_____"
]
],
[
[
"## Visualize computation",
"_____no_output_____"
]
],
[
[
"z.visualize(format='svg', rankdir='LR')",
"_____no_output_____"
]
],
[
[
"## Run in parallel\n\nCall `.compute()` when you want your result as a normal Python object\n\nIf you started `Client()` above then you may want to watch the status page during computation.",
"_____no_output_____"
]
],
[
[
"%%time\nz.compute()",
"_____no_output_____"
]
],
[
[
"## Parallelize Normal Python code\n\nNow we use Dask in normal for-loopy Python code. This generates graphs instead of doing computations directly, but still looks like the code we had before. Dask is a convenient way to add parallelism to existing workflows.",
"_____no_output_____"
]
],
[
[
"%%time\nzs = []\nfor i in range(256):\n x = inc(i)\n y = dec(x)\n z = add(x, y)\n zs.append(z)\n \nzs = dask.persist(*zs) # trigger computation in the background",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbaf6e7195700372456948d65c315342bad06a81
| 72,355 |
ipynb
|
Jupyter Notebook
|
notebooks/debug.ipynb
|
clegaspi/aflow
|
6828642f8c637012db0681dc059787029a78f3ca
|
[
"MIT"
] | 19 |
2017-11-20T19:26:49.000Z
|
2020-10-29T01:28:02.000Z
|
notebooks/debug.ipynb
|
clegaspi/aflow
|
6828642f8c637012db0681dc059787029a78f3ca
|
[
"MIT"
] | 12 |
2017-08-02T21:23:26.000Z
|
2020-02-29T03:22:40.000Z
|
notebooks/debug.ipynb
|
clegaspi/aflow
|
6828642f8c637012db0681dc059787029a78f3ca
|
[
"MIT"
] | 14 |
2017-08-30T22:28:29.000Z
|
2021-05-17T15:16:03.000Z
| 91.472819 | 55,518 | 0.604851 |
[
[
[
"%load_ext autoreload\n%autoreload 2",
"_____no_output_____"
],
[
"from aflow.entries import Entry",
"_____no_output_____"
],
[
"a = {\n \"compound\": \"Be2O2\",\n \"auid\":\"aflow:ed51b7b3938f117f\",\n \"aurl\":\"aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Be1O1_ICSD_15620\",\n \"agl_thermal_conductivity_300K\":\"53.361\",\n \"Egap\":\"7.4494\"\n}\nA = Entry(**a)",
"_____no_output_____"
],
[
"A.kpoints",
"_____no_output_____"
],
[
"from aflow.caster import _kpoints",
"_____no_output_____"
],
[
"_kpoints(\"16,16,8;17,17,9;\\Gamma-M,M-K,K-\\Gamma,\\Gamma-A,A-L,L-H,H-A,L-M,K-H;20\")",
"_____no_output_____"
],
[
"from aflow.keywords import *",
"_____no_output_____"
],
[
"from aflow.keywords import reset\nreset()",
"_____no_output_____"
],
[
"k = ((Egap > 6) | (Egap < 21)) & (PV_cell < 13)",
"_____no_output_____"
],
[
"reset()\nk1 = ((Egap > 6) | (Egap < 21)) & ((PV_cell < 13) | (PV_cell > 2))",
"_____no_output_____"
],
[
"str(k1)",
"_____no_output_____"
],
[
"reset()",
"_____no_output_____"
],
[
"k3 = ((Egap > 0) & (Egap < 2) | (Egap == 5))",
"_____no_output_____"
],
[
"str(k3)",
"_____no_output_____"
],
[
"str(PV_cell)",
"_____no_output_____"
],
[
"str(~PV_cell)",
"_____no_output_____"
],
[
"str(PV_cell)",
"_____no_output_____"
],
[
"k = (data_source == 'aflowlib') | (species % 'Si')",
"_____no_output_____"
],
[
"str(k)",
"_____no_output_____"
],
[
"reset()",
"_____no_output_____"
],
[
"k2 = (data_source < 'aflow') & (species < 'Ag')",
"_____no_output_____"
],
[
"str(k2)",
"_____no_output_____"
],
[
"%load_ext autoreload\n%autoreload 2",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"import aflow\nfrom aflow.keywords import *",
"_____no_output_____"
],
[
"Si = aflow.search(catalog=\"icsd\").filter(species == 'Si').select(positions_cartesian)",
"_____no_output_____"
],
[
"for i, entry in enumerate(Si[90:110]):\n print(i, entry.aurl)",
"(0, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCC/Al2Mg3O12Si3_ICSD_86909')\n(1, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCC/Al5O12Yb3_ICSD_170159')\n(2, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCT/Au2Cs2I6_ICSD_186066')\n(3, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCT/Ho5Ni2Sb1_ICSD_91136')\n(4, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCT/Ho5Tl3_ICSD_639788')\n(5, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCT/Cu1Nd2O4_ICSD_69479')\n(6, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Ir1Sb1_ICSD_44481')\n(7, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Cr1Ge3Nd1_ICSD_186693')\n(8, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Mn1Sb1_ICSD_53970')\n(9, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Cl1O12Pb5V3_ICSD_15750')\n(10, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Fe2W1_ICSD_634067')\n(11, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Os2Yb1_ICSD_647873')\n(12, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Ca1Mn1O3_ICSD_168906')\n(13, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/C1Si1Ti2_ICSD_183359')\n(14, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/HEX/Cu9Dy1Mg2_ICSD_245216')\n(15, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/ORCI/Al1Co2Dy2_ICSD_107402')\n(16, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/ORCI/Au3K2_ICSD_65113')\n(17, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCT/Eu1Ni2P2_ICSD_631434')\n(18, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCT/Er1Ni2Si2_ICSD_54158')\n(19, 'aflowlib.duke.edu:AFLOWDATA/ICSD_WEB/BCT/Ce1Cu2Si2_ICSD_620939')\n"
],
[
"sorted(Si.responses[2].keys())",
"_____no_output_____"
],
[
"sisl = Si[0:10]",
"_____no_output_____"
],
[
"sisl._iter",
"_____no_output_____"
],
[
"sisl._iter, sisl._max_entry",
"_____no_output_____"
],
[
"len(Si.responses)",
"_____no_output_____"
],
[
"for entry in sisl:\n print(entry.positions_cartesian)",
"[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n[[-0. -0. -0.]]\n"
],
[
"ss = slice(0, 10)",
"_____no_output_____"
],
[
"ss.",
"_____no_output_____"
],
[
"import json\nkeys = json.loads(\"\"\"{\"__schema^2__\":{\"__comment__\":[\"The zeroth element of any object or array in this document is meta.\",\"If last element is null, element parent considered optional.\",\"If last element is '.', element value can be anything.\",\"If last element is '', element value can be nothing.\",\"This document is the AAPI schema, it is self validating and order sensitive.\",\".\"],\"class\":[\"intended for document organization, defines major section. Must be one of\",\"API only\",\"chemistry\",\"crystal\",\"electronics\",\"thermodynamics\",\"magnetics\",\"scintillation\",\"mechanical\",\"optical properties\",\"other\",\"calculation\"],\"delimiter\":[\"An ordered set of single character seperators for distinguishing plural type property values\",null],\"description\":[\"intended for popup help boxes, describes the current property: freeform text\",\".\"],\"example\":[\"Actual result that may occur in API or search context, developmental: structured text\",\".\"],\"expression\":[\"intended for materials reports, developmental. Must be one of\",\"declarative\",\"directive\",\"derivative\"],\"format\":[\"intended for printf style formating of property value: corresponds to the type attribute\",\".\"],\"inclusion\":[\"intended for search filters and materials reports. Must be one of\",\"mandatory\",\"conditional\",\"optional\",\"forbidden\"],\"search\":[[\"intended for search and stat, Must be one of\",\"equals -> exact match input (select or freeform) to value\",\"contains -> substring match (select or freeform) in value\",\"range -> bounded match (select or freeform) in value\"],\"equals\",\"contains\",\"range\",null],\"status\":[\"Development stage of property. Must be one of\",\"production\",\"development\",\"deprecated\",\"reserved\"],\"subclass\":[\"intended for document organization, defines minor section\",\"label\",\"calculation parameters\",\"computational resources\",\"version\",\"provenance\",\"real space lattice\",\"bravais lattice of the crystal\",\"point group of the crystal\",\"bravais lattice of the lattice\",\"super lattice\",\"reciprocal space lattice\",\"space group\",\"parameters\",\"\"],\"syntax\":[\"Actual setting that may be used in API or search context, developmental: structured text\",\".\"],\"title\":[\"intended for labeling property in document rendering: freeform text (HTML?)\",\".\"],\"type\":[\"intended for DB and document type handling: must be one of\",\"string\",\"strings\",\"number\",\"numbers\"],\"units\":[\"units for search filter number in HTML: optional\",null],\"verification\":[\"Optional list of property references designed to certify that the result is contextually relevant.\",null]},\"Bravais_lattice_orig\":{\"__comment__\":[\"\"],\"description\":\"Returns the Bravais lattice of the original unrelaxed structure before the calculation.\",\"title\":\"original bravais lattice\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"Bravais_lattice_orig=MCLC\",\"status\":\"production\",\"syntax\":\"$aurl/?Bravais_lattice_orig\"},\"Bravais_lattice_relax\":{\"__comment__\":[\"\"],\"description\":\"Returns the Bravais lattice of the original relaxed structure after the calculation.\",\"title\":\"relaxed bravais lattice\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"stress_tensor\"],\"example\":\"Bravais_lattice_relax=MCLC\",\"status\":\"production\",\"syntax\":\"$aurl/?Bravais_lattice_relax\"},\"Egap\":{\"__comment__\":[\"\"],\"description\":\"Band gap calculated with the approximations and pseudopotentials described by other keywords.\",\"title\":\"energy gap\",\"format\":\"%s\",\"class\":\"electronics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"Egap=2.5\",\"status\":\"production\",\"syntax\":\"$aurl/?Egap\"},\"Egap_fit\":{\"__comment__\":[\"\"],\"description\":\"Simple cross-validated correction (fit) of Egap.\",\"title\":\"fitted band gap\",\"format\":\"%s\",\"class\":\"electronics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"Egap_fit=3.5\",\"status\":\"production\",\"syntax\":\"$aurl/?Egap_fit\"},\"Egap_type\":{\"__comment__\":[\"\"],\"description\":\"Given a band gap, this keyword describes if the system is a metal, a semi-metal, an insulator with direct or indirect band gap.\",\"title\":\"band gap type\",\"format\":\"%s\",\"class\":\"electronics\",\"subclass\":\"\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"Egap_type=insulator_direct\",\"status\":\"production\",\"syntax\":\"$aurl/?Egap_type\"},\"PV_atom\":{\"__comment__\":[\"\"],\"description\":\"Pressure multiplied by volume of the atom.\",\"title\":\"atomic pressure*volume\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV/atom\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"PV_atom=12.13\",\"status\":\"production\",\"syntax\":\"$aurl/?PV_atom\"},\"PV_cell\":{\"__comment__\":[\"\"],\"description\":\"Pressure multiplied by volume of the unit cell.\",\"title\":\"unit cell pressure*volume\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"PV_cell=12.13\",\"status\":\"production\",\"syntax\":\"$aurl/?PV_cell\"},\"Pearson_symbol_orig\":{\"__comment__\":[\"\"],\"description\":\"Returns the Pearson symbol of the original-unrelaxed structure before the calculation.\",\"title\":\"original pearson symbol\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"Pearson_symbol_orig=mS32\",\"status\":\"production\",\"syntax\":\"$aurl/?Pearson_symbol_orig\"},\"Pearson_symbol_relax\":{\"__comment__\":[\"\"],\"description\":\"Returns the Pearson symbol of the relaxed structure after the calculation.\",\"title\":\"relaxed pearson symbol\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"stress_tensor\"],\"example\":\"Pearson_symbol_relax=mS32\",\"status\":\"production\",\"syntax\":\"$aurl/?Pearson_symbol_relax\"},\"Pulay_stress\":{\"__comment__\":[\"\"],\"description\":\"Returns a metric of the basis set inconsistency for the calculation.\",\"title\":\"Pulay Stress\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"kbar\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"example\":\"pulay_stress=10.0\",\"status\":\"development\",\"syntax\":\"$aurl/?pulay_stress\"},\"Pullay_stress\":{\"__comment__\":[\"\"],\"description\":\"Returns a metric of the basis set inconsistency for the calculation.\",\"title\":\"Pulay Stress\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"kbar\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"example\":\"Pullay_stress=10.0\",\"status\":\"deprecated\",\"syntax\":\"$aurl/?Pullay_stress\"},\"ael_bulk_modulus_reuss\":{\"__comment__\":[\"\"],\"description\":\"Returns the bulk modulus as calculated using the Reuss method with AEL.\",\"title\":\"AEL Reuss bulk modulus\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_bulk_modulus_reuss=105.315\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_bulk_modulus_reuss\"},\"ael_bulk_modulus_voigt\":{\"__comment__\":[\"\"],\"description\":\"Returns the bulk modulus as calculated using the Voigt method with AEL.\",\"title\":\"AEL Voigt bulk modulus\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_bulk_modulus_voiht=105.315\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_bulk_modulus_voigt\"},\"ael_bulk_modulus_vrh\":{\"__comment__\":[\"\"],\"description\":\"Returns the bulk modulus as calculated using the Voigt-Reuss-Hill average with AEL.\",\"title\":\"AEL VRH bulk modulus\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_bulk_modulus_vrh=105.315\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_bulk_modulus_vrh\"},\"ael_elastic_anisotropy\":{\"__comment__\":[\"\"],\"description\":\"Returns the elastic anisotropy as calculated with AEL.\",\"title\":\"AEL elastic anisotropy\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_elastic_anisotropy=0.0008165\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_elastic_anisotropy\"},\"ael_poisson_ratio\":{\"__comment__\":[\"\"],\"description\":\"Returns the istropic Poisson ratio as calculated with AEL.\",\"title\":\"AEL Poisson ratio\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_poisson_ratio=0.216\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_poisson_ratio\"},\"ael_shear_modulus_reuss\":{\"__comment__\":[\"\"],\"description\":\"Returns the shear modulus as calculated using the Reuss method with AEL.\",\"title\":\"AEL Reuss shear modulus\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_shear_modulus_reuss=73.787\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_shear_modulus_reuss\"},\"ael_shear_modulus_voigt\":{\"__comment__\":[\"\"],\"description\":\"Returns the shear modulus as calculated using the Voigt method with AEL.\",\"title\":\"AEL Voigt shear modulus\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_shear_modulus_voigt=73.799\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_shear_modulus_voigt\"},\"ael_shear_modulus_vrh\":{\"__comment__\":[\"\"],\"description\":\"Returns the shear modulus as calculated using the Voigt-Reuss-Hill average with AEL.\",\"title\":\"AEL VRH shear modulus\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"ael_shear_modulus_vrh=73.793\",\"status\":\"production\",\"syntax\":\"$aurl/?ael_shear_modulus_vrh\"},\"aflow_version\":{\"__comment__\":[\"\"],\"description\":\"Returns the version number of AFLOW used to perform the calculation.\",\"title\":\"aflow version\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"version\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"aflow_version=aflow30641\",\"status\":\"production\",\"syntax\":\"$aurl/?aflow_version\"},\"aflowlib_date\":{\"__comment__\":[\"\"],\"description\":\"Returns the date of the AFLOW post-processor which generated the entry for the library.\",\"title\":\"material generation date\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"version\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"aflowlib_date=20140204_13:10:39_GMT-5\",\"status\":\"production\",\"syntax\":\"$aurl/?aflowlib_date\"},\"aflowlib_entries\":{\"__comment__\":[\"\"],\"description\":\"For projects and set-layer entries, aflowlib_entries lists the available sub-entries which are associated with the $aurl of the subdirectories. By parsing $aurl/?aflowlib_entries (containing $aurl/aflowlib_entries_number entries) the user finds further locations to interrogate.\",\"title\":\"aflowlib entries\",\"format\":\"%s\",\"class\":\"API only\",\"subclass\":\"\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"conditional\",\"expression\":\"directive\",\"example\":\"aflowlib_entries=AgAl,AgAs,AgAu,AgB_h,AgBa_sv,AgBe_sv,AgBi_d,AgBr,AgCa_sv,...\",\"status\":\"production\",\"syntax\":\"$aurl/?aflowlib_entries\"},\"aflowlib_entries_number\":{\"__comment__\":[\"\"],\"description\":\"For projects and set-layer entries, aflowlib_entrieslists the available sub-entries which are associated with the $aurl of the subdirectories. By parsing $aurl/?aflowlib_entries (containing $aurl/aflowlib_entries_number entries) the user finds further locations to interrogate.\",\"title\":\"aflowlib entry count\",\"format\":\"%s\",\"class\":\"API only\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"conditional\",\"expression\":\"directive\",\"example\":\"aflowlib_entries_number=654\",\"status\":\"production\",\"syntax\":\"$aurl/?aflowlib_entries_number\"},\"aflowlib_version\":{\"__comment__\":[\"\"],\"description\":\"Returns the version of the AFLOW post-processor which generated the entry for the library.\",\"title\":\"aflowlib version\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"version\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"aflowlib_version=3.1.103\",\"status\":\"production\",\"syntax\":\"$aurl/?aflowlib_version\"},\"agl_acoustic_debye\":{\"__comment__\":[\"\"],\"description\":\"Returns the acoustic Debye temperature as calculated with AGL.\",\"title\":\"AGL acoustic Debye temperature\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"K\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_acoustic_debye=492\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_acoustic_debye\"},\"agl_bulk_modulus_isothermal_300K\":{\"__comment__\":[\"\"],\"description\":\"Returns the isothermal bulk modulus at 300K as calculated with AGL.\",\"title\":\"AGL isothermal bulk modulus 300K\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_bulk_modulus_isothermal_300K=96.6\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_bulk_modulus_isothermal_300K\"},\"agl_bulk_modulus_static_300K\":{\"__comment__\":[\"\"],\"description\":\"Returns the static bulk modulus at 300K as calculated with AGL.\",\"title\":\"AGL static bulk modulus 300K\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"GPa\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_bulk_modulus_static_300K=99.6\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_bulk_modulus_static_300K\"},\"agl_debye\":{\"__comment__\":[\"\"],\"description\":\"Returns the Debye temperature as calculated with AGL.\",\"title\":\"AGL Debye temperature\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"K\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_debye=620\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_debye\"},\"agl_gruneisen\":{\"__comment__\":[\"\"],\"description\":\"Returns the Gruneisen parameter as calculated with AGL.\",\"title\":\"AGL Gruneisen parameter\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_gruneisen=2.06\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_gruneisen\"},\"agl_heat_capacity_Cp_300K\":{\"__comment__\":[\"\"],\"description\":\"Returns the heat capacity at constant pressure as calculated with AGL at 300K.\",\"title\":\"AGL heat capacity Cp\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"kB/cell\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_heat_capacity_Cp_300K=5.502\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_heat_capacity_Cp_300K\"},\"agl_heat_capacity_Cv_300K\":{\"__comment__\":[\"\"],\"description\":\"Returns the heat capacity at constant volume as calculated with AGL at 300K.\",\"title\":\"AGL heat capacity Cv\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"kB/cell\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_heat_capacity_Cv_300K=4.901\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_heat_capacity_Cv_300K\"},\"agl_thermal_conductivity_300K\":{\"__comment__\":[\"\"],\"description\":\"Returns the thermal conductivity as calculated with AGL at 300K.\",\"title\":\"AGL thermal conductivity\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"W/m*K\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_thermal_conductivity_300K=24.41\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_thermal_conductivity_300K\"},\"agl_thermal_expansion_300K\":{\"__comment__\":[\"\"],\"description\":\"Returns the thermal expansion as calculated with AGL at 300K.\",\"title\":\"AGL thermal expansion\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"1/K\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"agl_thermal_expansion_300K=4.997e-05\",\"status\":\"production\",\"syntax\":\"$aurl/?agl_thermal_expansion_300K\"},\"auid\":{\"__comment__\":[\"\"],\"description\":\"AFLOWLIB Unique Identifier for the entry, AUID, which can be used as a publishable object identifier.\",\"title\":\"AFLOWLIB Unique Identifier\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"auid=aflow:e9c6d914c4b8d9ca\",\"status\":\"production\",\"syntax\":\"$aurl/?auid\"},\"aurl\":{\"__comment__\":[\"\"],\"description\":\"AFLOWLIB Uniform Resource Locator returns the AURL of the entry.\",\"title\":\"AFLOWLIB Uniform Resource Locator\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"aurl=aflowlib.duke.edu:AFLOWDATA/LIB3_RAW/Bi_dRh_pvTi_sv/T0003.ABC:LDAU2\",\"status\":\"production\",\"syntax\":\"$aurl/?aurl\"},\"author\":{\"__comment__\":[\"\"],\"description\":\"Returns the name (not necessarily an individual) and affiliation associated with authorship of the data.\",\"title\":\"author\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"provenance\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"author=Marco_Buongiorno_Nardelli,Ohad_Levy,Jesus_Carrete\",\"status\":\"development\",\"syntax\":\"$aurl/?author\"},\"bader_atomic_volumes\":{\"__comment__\":[\"\"],\"description\":\"Returns the volume of each atom of the primitive cell as calculated by the Bader Atoms in Molecules Analysis. This volume encapsulates the electron density associated with each atom above a threshold of 0.0001 electrons.\",\"title\":\"atomic volume per atom\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"units\":\"Å<sup>3</sup>\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"bader_atomic_volumes=15.235,12.581,13.009\",\"status\":\"production\",\"syntax\":\"$aurl/?bader_atomic_volumes\"},\"bader_net_charges\":{\"__comment__\":[\"\"],\"description\":\"Returns a comma delimited set of partial charges per atom of the primitive cell as calculated by the Bader Atoms in Molecules Analysis.\",\"title\":\"partial charge per atom\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"units\":\"electrons\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"bader_net_charges=0.125,0.125,-0.25\",\"status\":\"production\",\"syntax\":\"$aurl/?bader_net_charges\"},\"calculation_cores\":{\"__comment__\":[\"\"],\"description\":\"Number of processors/cores used for the calculation.\",\"title\":\"used CPU cores\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"computational resources\",\"type\":\"number\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"calculation_cores=32\",\"status\":\"production\",\"syntax\":\"$aurl/?calculation_cores\"},\"calculation_memory\":{\"__comment__\":[\"\"],\"description\":\"The maximum memory used for the calculation.\",\"title\":\"used RAM\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"computational resources\",\"type\":\"number\",\"units\":\"Megabytes\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"calculation_memory=32\",\"status\":\"production\",\"syntax\":\"$aurl/?calculation_memory\"},\"calculation_time\":{\"__comment__\":[\"\"],\"description\":\"Total time taken for the calculation.\",\"title\":\"used time\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"computational resources\",\"type\":\"number\",\"units\":\"seconds\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"calculation_time=32\",\"status\":\"production\",\"syntax\":\"$aurl/?calculation_time\"},\"catalog\":{\"__comment__\":[\"\"],\"description\":\"Returns the context set for the calculation.\",\"title\":\"catalog\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"version\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"catalog=icsd\",\"status\":\"production\",\"syntax\":\"$aurl/?catalog\"},\"code\":{\"__comment__\":[\"\"],\"description\":\"Returns the software name and version used to perform the simulation.\",\"title\":\"ab initio code\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"version\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"code=vasp.4.6.35\",\"status\":\"production\",\"syntax\":\"$aurl/?code\"},\"composition\":{\"__comment__\":[\"\"],\"description\":\"Returns a comma delimited composition description of the structure entry in the calculated cell.\",\"title\":\"composition\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"composition=2,6,6\",\"status\":\"production\",\"syntax\":\"$aurl/?composition\"},\"compound\":{\"__comment__\":[\"\"],\"description\":\"Returns the composition description of the compound in the calculated cell.\",\"title\":\"chemical formula\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"compound=Co2Er6Si6\",\"status\":\"production\",\"syntax\":\"$aurl/?compound\"},\"corresponding\":{\"__comment__\":[\"\"],\"description\":\"Returns the name (not necessarily an individual) and affiliation associated with the data origin concerning correspondence about data.\",\"title\":\"coresponding\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"provenance\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"[email protected]\",\"status\":\"development\",\"syntax\":\"$aurl/?corresponding\"},\"data_api\":{\"__comment__\":[\"\"],\"description\":\"AFLOWLIB version of the entry, API.}\",\"title\":\"REST API version\",\"format\":\"%s\",\"class\":\"API only\",\"subclass\":\"\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"data_api=aapi1.0\",\"status\":\"production\",\"syntax\":\"$aurl/?data_api\"},\"data_language\":{\"__comment__\":[\"\"],\"description\":\"Gives the language of the data in AFLOWLIB.\",\"title\":\"data language\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"version\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"data_language=aflowlib\",\"status\":\"production\",\"syntax\":\"$aurl/?data_language\"},\"data_source\":{\"__comment__\":[\"\"],\"description\":\"Gives the source of the data in AFLOWLIB.\",\"title\":\"data source\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"version\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"data_source=aflowlib\",\"status\":\"production\",\"syntax\":\"$aurl/?data_source\"},\"delta_electronic_energy_convergence\":{\"__comment__\":[\"\"],\"description\":\"Returns the change in energy from the last step of the convergence iteration.\",\"title\":\"Electronic Energy of Convergence Step\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"example\":\"delta_electronic_energy_convergence=6.09588e-05\",\"status\":\"development\",\"syntax\":\"$aurl/?delta_electronic_energy_convergence\"},\"delta_electronic_energy_threshold\":{\"__comment__\":[\"\"],\"description\":\"Returns the maximimum change in energy required for the convergence iteration.\",\"title\":\"Electronic Energy of Convergence Threshold\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"delta_electronic_energy_threshold=0.0001\",\"status\":\"development\",\"syntax\":\"$aurl/?delta_electronic_energy_threshold\"},\"density\":{\"__comment__\":[\"\"],\"description\":\"Returns the mass density in grams/cm3.\",\"title\":\"mass density\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"real space lattice\",\"type\":\"number\",\"units\":\"grams/cm<sup>3</sup>\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"density=7.76665\",\"status\":\"production\",\"syntax\":\"$aurl/?density\"},\"dft_type\":{\"__comment__\":[\"\"],\"description\":\"Returns information about the pseudopotential type, the exchange correlation functional used (normal or hybrid) and use of GW.\",\"title\":\"DFT type\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"parameters\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"dft_type=PAW_PBE,HSE06\",\"status\":\"production\",\"syntax\":\"$aurl/?dft_type\"},\"eentropy_atom\":{\"__comment__\":[\"\"],\"description\":\"Returns the electronic entropy of the atom used to converge the ab initio calculation (smearing).\",\"title\":\"atomistic electronic entropy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV/atom\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"eentropy_atom=0.0011\",\"status\":\"production\",\"syntax\":\"$aurl/?eentropy_atom\"},\"eentropy_cell\":{\"__comment__\":[\"\"],\"description\":\"Returns the electronic entropy of the unit cell used to converge the ab initio calculation (smearing).\",\"title\":\"unit cell electronic entropy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV/atom\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"eentropy_cell=0.0011\",\"status\":\"production\",\"syntax\":\"$aurl/?eentropy_cell\"},\"energy_atom\":{\"__comment__\":[\"\"],\"description\":\"Returns the total ab initio energy per atom- the value of energy_cell/$N$).\",\"title\":\"atomic energy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV/atom\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"energy_atom=-82.1656\",\"status\":\"production\",\"syntax\":\"$aurl/?energy_atom\"},\"energy_cell\":{\"__comment__\":[\"\"],\"description\":\"Returns the total ab initio energy of the unit cell, E. At T=0K and p=0, this is the internal energy of the system (per unit cell).\",\"title\":\"unit cell energy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"energy_cell=-82.1656\",\"status\":\"production\",\"syntax\":\"$aurl/?energy_cell\"},\"energy_cutoff\":{\"__comment__\":[\"\"],\"description\":\"Set of energy cut-offs used during the various steps of the calculations.\",\"title\":\"energy cutoff\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"parameters\",\"type\":\"numbers\",\"delimiter\":\",\",\"units\":\"eV\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"energy_cutoff=384.1,384.1,384.1\",\"status\":\"production\",\"syntax\":\"$aurl/?energy_cutoff\"},\"enthalpy_atom\":{\"__comment__\":[\"\"],\"description\":\"Returns the enthalpy per atom- the value of enthalpy_cell/N).\",\"title\":\"atomic enthalpy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV/atom\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"enthalpy_atom=-82.1656\",\"status\":\"production\",\"syntax\":\"$aurl/?enthalpy_atom\"},\"enthalpy_cell\":{\"__comment__\":[\"\"],\"description\":\"Returns the enthalpy of the system of the unit cell, H = E + PV.\",\"title\":\"unit cell enthalpy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"enthalpy_cell=-82.1656\",\"status\":\"production\",\"syntax\":\"$aurl/?enthalpy_cell\"},\"enthalpy_formation_atom\":{\"__comment__\":[\"\"],\"description\":\"Returns the formation enthalpy DeltaHFatomic per atom).\",\"title\":\"atomic formation enthalpy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV/atom\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"enthalpy_formation_atom=-33.1587\",\"status\":\"production\",\"syntax\":\"$aurl/?enthalpy_formation_atom\"},\"enthalpy_formation_cell\":{\"__comment__\":[\"\"],\"description\":\"Returns the formation enthalpy DeltaHF per unit cell.\",\"title\":\"unit cell formation enthalpy\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"eV\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"enthalpy_formation_cell=-33.1587\",\"status\":\"production\",\"syntax\":\"$aurl/?enthalpy_formation_cell\"},\"entropic_temperature\":{\"__comment__\":[\"\"],\"description\":\"Returns the entropic temperature for the structure.\",\"title\":\"entropic temperature\",\"format\":\"%s\",\"class\":\"thermodynamics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"Kelvin\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"entropic_temperature=1072.1\",\"status\":\"production\",\"syntax\":\"$aurl/?entropic_temperature\"},\"files\":{\"__comment__\":[\"\"],\"description\":\"Provides access to the input and output files used in the simulation (provenance data).\",\"title\":\"I/O files\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"conditional\",\"expression\":\"directive\",\"example\":\"files=Bi_dRh_pv.33.cif,Bi_dRh_pv.33.png,CONTCAR.relax,CONTCAR.relax1,\",\"status\":\"production\",\"syntax\":\"$aurl/?files\"},\"forces\":{\"__comment__\":[\"\"],\"description\":\"Final quantum mechanical forces (Fi,Fj,Fk) in the notation of the code.\",\"title\":\"Quantum Forces\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\";,\",\"units\":\"eV/Å\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"forces=0,-0.023928,0.000197;0,0.023928,-0.000197;...\",\"status\":\"development\",\"syntax\":\"$aurl/?forces\"},\"geometry\":{\"__comment__\":[\"\"],\"description\":\"Returns geometrical data describing the unit cell in the usual a,b,c,alpha,beta,gamma notation.\",\"title\":\"unit cell basis\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"real space lattice\",\"type\":\"numbers\",\"delimiter\":\",\",\"units\":\"Å\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"geometry=18.82,18.82,18.82,32.41,32.41,32.41\",\"status\":\"production\",\"verification\":[\"energy_cutoff\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"syntax\":\"$aurl/?geometry\"},\"keywords\":{\"__comment__\":[\"\"],\"description\":\"This includes the list of keywords available in the entry, separated by commas.\",\"title\":\"Title\",\"format\":\"%s\",\"class\":\"API only\",\"subclass\":\"\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"mandatory\",\"expression\":\"directive\",\"example\":\"keywords=aurl,auid,loop,code,compound,prototype,nspecies,natoms,...\",\"status\":\"production\",\"syntax\":\"$aurl/?keywords\"},\"kpoints\":{\"__comment__\":[\"\"],\"description\":\"Set of k-point meshes uniquely identifying the various steps of the calculations, e.g. relaxation, static and electronic band structure (specifying the k-space symmetry points of the structure).\",\"title\":\"K-point mesh\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"parameters\",\"type\":\"numbers\",\"delimiter\":\":,\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"kpoints=10,10,10;16,16,16;G-X-W-K-G-L-U-W-L-K+U-X\",\"status\":\"production\",\"syntax\":\"$aurl/?kpoints\"},\"lattice_system_orig\":{\"__comment__\":[\"\"],\"description\":\"Return the lattice system and lattice variation (Brillouin zone) of the original-unrelaxed structure before the calculation.\",\"title\":\"original lattice system\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"lattice_system_orig=rhombohedral\",\"status\":\"production\",\"syntax\":\"$aurl/?lattice_system_orig\"},\"lattice_system_relax\":{\"__comment__\":[\"\"],\"description\":\"Return the lattice system and lattice variation (Brillouin zone) of the relaxed structure after the calculation.\",\"title\":\"relaxed lattice system\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"stress_tensor\"],\"example\":\"lattice_system_relax=rhombohedral\",\"status\":\"production\",\"syntax\":\"$aurl/?lattice_system_relax\"},\"lattice_variation_orig\":{\"__comment__\":[\"\"],\"description\":\"Return the lattice system and lattice variation (Brillouin zone) of the original-unrelaxed structure before the calculation.\",\"title\":\"original lattice variation\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"lattice_variation_orig=rhombohedral\",\"status\":\"production\",\"syntax\":\"$aurl/?lattice_variation_orig\"},\"lattice_variation_relax\":{\"__comment__\":[\"\"],\"description\":\"Return the lattice system and lattice variation (Brillouin zone) of the relaxed structure after the calculation.\",\"title\":\"relaxed lattice variation\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"stress_tensor\"],\"example\":\"lattice_variation_relax=rhombohedral\",\"status\":\"production\",\"syntax\":\"$aurl/?lattice_variation_relax\"},\"ldau_TLUJ\":{\"__comment__\":[\"\"],\"description\":\"This vector of numbers contains the parameters of the DFT+U calculations, based on a corrective functional inspired by the Hubbard model.\",\"title\":\"on site coulomb interaction\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"parameters\",\"type\":\"numbers\",\"delimiter\":\";,\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"ldau_TLUJ=2;2,0,0;5,0,0;0,0,0\",\"status\":\"development\",\"syntax\":\"$aurl/?ldau_TLUJ\"},\"loop\":{\"__comment__\":[\"\"],\"description\":\"Informs the user of the type of post-processing that was performed.\",\"title\":\"process category\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"parameters\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"directive\",\"example\":\"loop=thermodynamics,bands,magnetic\",\"status\":\"production\",\"syntax\":\"$aurl/?loop\"},\"natoms\":{\"__comment__\":[\"\"],\"description\":\"Returns the number of atoms in the unit cell of the structure entry. The number can be non integer if partial occupation is considered within appropriate approximations.\",\"title\":\"unit cell atom count\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"real space lattice\",\"type\":\"number\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"natoms=12\",\"status\":\"production\",\"syntax\":\"$aurl/?natoms\"},\"nbondxx\":{\"__comment__\":[\"\"],\"description\":\"Nearest neighbors bond lengths of the relaxed structure per ordered set of species Ai,Aj greater than or equal to i.\",\"title\":\"Nearest neighbors bond lengths\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"units\":\"Å\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"nbondxx=1.2599,1.0911,1.0911,1.7818,1.2599,1.7818\",\"status\":\"production\",\"syntax\":\"$aurl/?nbondxx\"},\"node_CPU_Cores\":{\"__comment__\":[\"\"],\"description\":\"Information about the number of cores in the node/cluster where the calculation was performed.\",\"title\":\"available CPU cores\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"computational resources\",\"type\":\"number\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"node_CPU_Cores=12\",\"status\":\"production\",\"syntax\":\"$aurl/?node_CPU_Cores\"},\"node_CPU_MHz\":{\"__comment__\":[\"\"],\"description\":\"Information about the CPU speed in the node/cluster where the calculation was performed.\",\"title\":\"CPU rate\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"computational resources\",\"type\":\"number\",\"units\":\"Megahertz\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"node_CPU_MHz=12\",\"status\":\"production\",\"syntax\":\"$aurl/?node_CPU_MHz\"},\"node_CPU_Model\":{\"__comment__\":[\"\"],\"description\":\"Information about the CPU model in the node/cluster where the calculation was performed.\",\"title\":\"CPU model\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"computational resources\",\"type\":\"string\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"node_CPU_Model=12\",\"status\":\"production\",\"syntax\":\"$aurl/?node_CPU_Model\"},\"node_RAM_GB\":{\"__comment__\":[\"\"],\"description\":\"Information about the RAM in the node/cluster where the calculation was performed.\",\"title\":\"available RAM\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"Gigabytes\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"node_RAM_GB=12\",\"status\":\"production\",\"syntax\":\"$aurl/?node_RAM_GB\"},\"nspecies\":{\"__comment__\":[\"\"],\"description\":\"Returns the number of species in the system (e.g., binary = 2, ternary = 3, etc.).\",\"title\":\"species count\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"nspecies=3\",\"status\":\"production\",\"syntax\":\"$aurl/?nspecies\"},\"positions_cartesian\":{\"__comment__\":[\"\"],\"description\":\"Final Cartesian positions (xi,xj,xk) in the notation of the code.\",\"title\":\"relaxed absolute positions\",\"format\":\"%s\",\"class\":\"other\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"numbers\",\"delimiter\":\";,\",\"units\":\"Å\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"positions_cartesian=0,0,0;18.18438,0,2.85027;...\",\"status\":\"development\",\"syntax\":\"$aurl/?positions_cartesian\"},\"positions_fractional\":{\"__comment__\":[\"\"],\"description\":\"Final fractional positions (xi,xj,xk) with respect to the unit cell as specified in $geometry.\",\"title\":\"relaxed relative positions\",\"format\":\"%s\",\"class\":\"other\",\"subclass\":\"\",\"type\":\"numbers\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"positions_fractional=0,0,0;0.25,0.25,0.25;...\",\"status\":\"development\",\"syntax\":\"$aurl/?positions_fractional\"},\"pressure\":{\"__comment__\":[\"\"],\"description\":\"Returns the target pressure selected for the simulation.\",\"title\":\"external pressure\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"kbar\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"pressure=10.0\",\"status\":\"production\",\"syntax\":\"$aurl/?pressure\"},\"pressure_final\":{\"__comment__\":[\"\"],\"description\":\"Returns the external pressure achieved by the simulation.\",\"title\":\"resulting pressure\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"kbar\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"example\":\"pressure_final=10.0\",\"status\":\"development\",\"syntax\":\"$aurl/?pressure_final\"},\"pressure_residual\":{\"__comment__\":[\"\"],\"description\":\"Returns the external pressure achieved by the simulation.\",\"title\":\"residual pressure\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"kbar\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"example\":\"pressure_residual=10.0\",\"status\":\"development\",\"syntax\":\"$aurl/?pressure_residual\"},\"prototype\":{\"__comment__\":[\"\"],\"description\":\"Returns the AFLOW unrelaxed prototype which was used for the calculation.\",\"title\":\"original prototype\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"label\",\"type\":\"string\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"prototype=T0001.A2BC\",\"status\":\"production\",\"syntax\":\"$aurl/?prototype\"},\"scintillation_attenuation_length\":{\"__comment__\":[\"\"],\"description\":\"Returns the scintillation attenuation length of the compound in cm.\",\"title\":\"attenuation length\",\"format\":\"%s\",\"class\":\"scintillation\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"cm\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"scintillation_attenuation_length=2.21895\",\"status\":\"production\",\"syntax\":\"$aurl/?scintillation_attenuation_length\"},\"sg\":{\"__comment__\":[\"\"],\"description\":\"Evolution of the space group of the compound. The first, second and third string represent space group name/number before the first, after the first, and after the last relaxation of the calculation.\",\"title\":\"compound space group\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"space group\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"mandatory\",\"expression\":\"directive\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"stress_tensor\"],\"example\":\"sg=Fm-3m#225,Fm-3m#225,Fm-3m#225\",\"status\":\"production\",\"syntax\":\"$aurl/?sg\"},\"sg2\":{\"__comment__\":[\"\"],\"description\":\"Evolution of the space group of the compound. The first, second and third string represent space group name/number before the first, after the first, and after the last relaxation of the calculation.\",\"title\":\"refined compound space group\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"space group\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"mandatory\",\"expression\":\"directive\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"stress_tensor\"],\"example\":\"sg2=Fm-3m#225,Fm-3m#225,Fm-3m#225\",\"status\":\"production\",\"syntax\":\"$aurl/?sg2\"},\"spacegroup_orig\":{\"__comment__\":[\"\"],\"description\":\"Returns the spacegroup number of the original-unrelaxed structure before the calculation.\",\"title\":\"original space group number\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"number\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"spacegroup_orig=225\",\"status\":\"production\",\"syntax\":\"$aurl/?spacegroup_orig\"},\"spacegroup_relax\":{\"__comment__\":[\"\"],\"description\":\"Returns the spacegroup number of the relaxed structure after the calculation.\",\"title\":\"relaxed space group number\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"bravais lattice of the crystal\",\"type\":\"number\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"stress_tensor\"],\"example\":\"spacegroup_relax=225\",\"status\":\"production\",\"syntax\":\"$aurl/?spacegroup_relax\"},\"species\":{\"__comment__\":[\"\"],\"description\":\"Species of the atoms in this material.\",\"title\":\"atomic species\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"species=Y,Zn,Zr\",\"status\":\"production\",\"syntax\":\"$aurl/?species\"},\"species_pp\":{\"__comment__\":[\"\"],\"description\":\"Pseudopotentials of the atomic species.\",\"title\":\"species pseudopotential(s)\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"species_pp=Y,Zn,Zr\",\"status\":\"production\",\"syntax\":\"$aurl/?species_pp\"},\"species_pp_ZVAL\":{\"__comment__\":[\"\"],\"description\":\"Returns the number of valence electrons of the atomic species.\",\"title\":\"valence atoms per species\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"units\":\"electrons\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"species_pp_ZVAL=3\",\"status\":\"production\",\"syntax\":\"$aurl/?species_pp_ZVAL\"},\"species_pp_version\":{\"__comment__\":[\"\"],\"description\":\"Species of the atoms, pseudopotentials species, and pseudopotential versions.\",\"title\":\"pseudopotential species/version\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"mandatory\",\"expression\":\"declarative\",\"example\":\"species_pp_version=Y,Zn,Zr\",\"status\":\"production\",\"syntax\":\"$aurl/?species_pp_version\"},\"spinD\":{\"__comment__\":[\"\"],\"description\":\"For spin polarized calculations, the spin decomposition over the atoms of the cell.\",\"title\":\"atomic spin decomposition\",\"format\":\"%s\",\"class\":\"magnetics\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"units\":\"μ<sub>B</sub>\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"spinD=0.236,0.236,-0.023,1.005\",\"status\":\"production\",\"syntax\":\"$aurl/?spinD\"},\"spinF\":{\"__comment__\":[\"\"],\"description\":\"For spin polarized calculations, the magnetization of the cell at the Fermi level.\",\"title\":\"fermi level spin decomposition\",\"format\":\"%s\",\"class\":\"magnetics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"μ<sub>B</sub>\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"spinF=0.410879\",\"status\":\"production\",\"syntax\":\"$aurl/?spinF\"},\"spin_atom\":{\"__comment__\":[\"\"],\"description\":\"For spin polarized calculations, the magnetization per atom.\",\"title\":\"atomic spin polarization\",\"format\":\"%s\",\"class\":\"magnetics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"μ<sub>B</sub>/atom\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"spin_atom=2.16419\",\"status\":\"production\",\"syntax\":\"$aurl/?spin_atom\"},\"spin_cell\":{\"__comment__\":[\"\"],\"description\":\"For spin polarized calculations, the total magnetization of the cell.\",\"title\":\"unit cell spin polarization\",\"format\":\"%s\",\"class\":\"magnetics\",\"subclass\":\"\",\"type\":\"number\",\"units\":\"μ<sub>B</sub>\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"spin_cell=2.16419\",\"status\":\"production\",\"syntax\":\"$aurl/?spin_cell\"},\"sponsor\":{\"__comment__\":[\"\"],\"description\":\"Returns information about funding agencies and other sponsors for the data.\",\"title\":\"sponsor\",\"format\":\"%s\",\"class\":\"calculation\",\"subclass\":\"provenance\",\"type\":\"strings\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"declarative\",\"example\":\"sponsor=DOD_N000141310635,NIST_70NANB12H163\",\"status\":\"development\",\"syntax\":\"$aurl/?sponsor\"},\"stoich\":{\"__comment__\":[\"\"],\"description\":\"Similar to composition, returns a comma delimited stoichiometry description of the structure entry in the calculated cell.\",\"title\":\"unit cell stoichiometry\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"inclusion\":\"optional\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"stoichiometry=0.5,0.25,0.25\",\"status\":\"deprecated\",\"syntax\":\"$aurl/?stoichiometry\"},\"stoichiometry\":{\"__comment__\":[\"\"],\"description\":\"Similar to composition, returns a comma delimited stoichiometry description of the structure entry in the calculated cell.\",\"title\":\"unit cell stoichiometry\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"numbers\",\"delimiter\":\",\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"stoichiometry=0.5,0.25,0.25\",\"status\":\"production\",\"syntax\":\"$aurl/?stoichiometry\"},\"stress_tensor\":{\"__comment__\":[\"\"],\"description\":\"Returns the stress tensor of the completed calculation.\",\"title\":\"Stress Tensor\",\"format\":\"%s\",\"class\":\"mechanical\",\"subclass\":\"\",\"type\":\"numbers\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"example\":\"stress_tensor=-0.96,-0,-0,-0,-0.96,-0,-0,-0,-0.96\",\"status\":\"development\",\"syntax\":\"$aurl/?stress_tensor\"},\"valence_cell_iupac\":{\"__comment__\":[\"\"],\"description\":\"Returns IUPAC valence, the maximum number of univalent atoms that may combine with the atoms.\",\"title\":\"unit cell IUPAC valence\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"valence_cell_iupac=22\",\"status\":\"production\",\"syntax\":\"$aurl/?valence_cell_iupac\"},\"valence_cell_std\":{\"__comment__\":[\"\"],\"description\":\"Returns standard valence, the maximum number of univalent atoms that may combine with the atoms.\",\"title\":\"unit cell standard valence\",\"format\":\"%s\",\"class\":\"chemistry\",\"subclass\":\"\",\"type\":\"number\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"kpoints\"],\"example\":\"valence_cell_std=22\",\"status\":\"production\",\"syntax\":\"$aurl/?valence_cell_std\"},\"volume_atom\":{\"__comment__\":[\"\"],\"description\":\"Returns the volume per atom in the unit cell.\",\"title\":\"atomic volume\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"real space lattice\",\"type\":\"number\",\"units\":\"Å<sup>3</sup>/atom\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"volume_atom=100.984\",\"status\":\"production\",\"syntax\":\"$aurl/?volume_atom\"},\"volume_cell\":{\"__comment__\":[\"\"],\"description\":\"Returns the volume of the unit cell.\",\"title\":\"unit cell volume\",\"format\":\"%s\",\"class\":\"crystal\",\"subclass\":\"real space lattice\",\"type\":\"number\",\"units\":\"Å<sup>3</sup>\",\"inclusion\":\"mandatory\",\"expression\":\"derivative\",\"verification\":[\"energy_cutoff\",\"forces\",\"kpoints\",\"pressure_residual\",\"stress_tensor\"],\"example\":\"volume_cell=100.984\",\"status\":\"production\",\"syntax\":\"$aurl/?volume_cell\"}}\"\"\")",
"_____no_output_____"
],
[
"keys[\"energy_cutoff\"]",
"_____no_output_____"
],
[
"from aflow.entries import Entry",
"_____no_output_____"
],
[
"hasattr(Entry, \"Egap\")",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbaf81ae7a962ab0f4af7d8859ad02490f924281
| 20,783 |
ipynb
|
Jupyter Notebook
|
temp_analysis_bonus_1_starter.ipynb
|
sharp2454/sqlalchemy-challenge
|
99c9ec880479bfa476fe29388ce94acbbbb73bba
|
[
"ADSL"
] | null | null | null |
temp_analysis_bonus_1_starter.ipynb
|
sharp2454/sqlalchemy-challenge
|
99c9ec880479bfa476fe29388ce94acbbbb73bba
|
[
"ADSL"
] | null | null | null |
temp_analysis_bonus_1_starter.ipynb
|
sharp2454/sqlalchemy-challenge
|
99c9ec880479bfa476fe29388ce94acbbbb73bba
|
[
"ADSL"
] | null | null | null | 26.644872 | 319 | 0.371842 |
[
[
[
"# Bonus: Temperature Analysis I",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nfrom datetime import datetime as dt\nfrom scipy import stats",
"_____no_output_____"
],
[
"# \"tobs\" is \"temperature observations\"\ndf = pd.read_csv('Resources/hawaii_measurements.csv')\ndf.head()",
"_____no_output_____"
],
[
"# Convert the date column format from string to datetime\ndf.date = pd.to_datetime(df.date, infer_datetime_format=True)\n\n",
"_____no_output_____"
],
[
"# Set the date column as the DataFrame index\ndf = df.set_index(df['date'])\ndf.head()",
"_____no_output_____"
],
[
"# Drop the date column\ndf=df.drop(columns='date')\ndf.head()\n",
"_____no_output_____"
]
],
[
[
"### Compare June and December data across all years ",
"_____no_output_____"
]
],
[
[
"from scipy import stats\nfrom scipy.stats import ttest_ind",
"_____no_output_____"
],
[
"# Filter data for desired months\njune_data = df[df.index.month == 6]\njune_data\n",
"_____no_output_____"
],
[
"# Create collections of temperature data\njune_temp = june_data.tobs\njune_temp",
"_____no_output_____"
],
[
"# Identify the average temperature for June\njune_temp.mean()",
"_____no_output_____"
],
[
"# Filter data for desired months\ndecember_data = df[df.index.month == 12]\ndecember_data\n",
"_____no_output_____"
],
[
"# Create collections of temperature data\ndecember_temp = december_data.tobs\ndecember_temp\n",
"_____no_output_____"
],
[
"# Identify the average temperature for December\ndecember_temp.mean()",
"_____no_output_____"
],
[
"# Run paired t-test\nttest_ind(june_temp, december_temp)\n",
"_____no_output_____"
]
],
[
[
"### Analysis",
"_____no_output_____"
],
[
"The mean temperature difference between June and December in Hawaii is only 3.9 degrees. This is a very minimal difference, yet the extremely small pvalue shows statistical significance and suggests that the sample data provides enough evidence that you can reject the null hypothesis for the entire population. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbaf8370cb3e8473f85cd6a76e767152c0d04c5b
| 26,052 |
ipynb
|
Jupyter Notebook
|
NLPFE3.ipynb
|
AsterLaoWhy/Thinkful
|
fa5d54d02b8af6a851cc7c2cec826dc8caeb777a
|
[
"MIT"
] | null | null | null |
NLPFE3.ipynb
|
AsterLaoWhy/Thinkful
|
fa5d54d02b8af6a851cc7c2cec826dc8caeb777a
|
[
"MIT"
] | null | null | null |
NLPFE3.ipynb
|
AsterLaoWhy/Thinkful
|
fa5d54d02b8af6a851cc7c2cec826dc8caeb777a
|
[
"MIT"
] | null | null | null | 41.616613 | 283 | 0.533817 |
[
[
[
"## 1. Train your own word2vec representations as we did in our first example in the checkpoint. But, you need to experiment with the hyperparameters of the vectorization step. Modify the hyperparameters and run the classification models again. Can you wrangle any improvements?",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport sklearn\nimport spacy\nimport re\nimport nltk\nfrom nltk.corpus import gutenberg\nimport gensim\nimport warnings\nwarnings.filterwarnings(\"ignore\")\n\nnltk.download('gutenberg')\n!python -m spacy download en",
"[nltk_data] Downloading package gutenberg to\n[nltk_data] C:\\Users\\jonat\\AppData\\Roaming\\nltk_data...\n[nltk_data] Package gutenberg is already up-to-date!\n"
],
[
"# utility function for standard text cleaning\ndef text_cleaner(text):\n # visual inspection identifies a form of punctuation spaCy does not\n # recognize: the double dash '--'. Better get rid of it now!\n text = re.sub(r'--',' ',text)\n text = re.sub(\"[\\[].*?[\\]]\", \"\", text)\n text = re.sub(r\"(\\b|\\s+\\-?|^\\-?)(\\d+|\\d*\\.\\d+)\\b\", \" \", text)\n text = ' '.join(text.split())\n return text",
"_____no_output_____"
],
[
"# load and clean the data\npersuasion = gutenberg.raw('austen-persuasion.txt')\nalice = gutenberg.raw('carroll-alice.txt')\n\n# the chapter indicator is idiosyncratic\npersuasion = re.sub(r'Chapter \\d+', '', persuasion)\nalice = re.sub(r'CHAPTER .*', '', alice)\n \nalice = text_cleaner(alice)\npersuasion = text_cleaner(persuasion)",
"_____no_output_____"
],
[
"# parse the cleaned novels. This can take a bit.\nnlp = spacy.load('en_core_web_sm')\nalice_doc = nlp(alice)\npersuasion_doc = nlp(persuasion)",
"_____no_output_____"
],
[
"# group into sentences\nalice_sents = [[sent, \"Carroll\"] for sent in alice_doc.sents]\npersuasion_sents = [[sent, \"Austen\"] for sent in persuasion_doc.sents]\n\n# combine the sentences from the two novels into one data frame\nsentences = pd.DataFrame(alice_sents + persuasion_sents, columns = [\"text\", \"author\"])\nsentences.head()",
"_____no_output_____"
],
[
"# get rid off stop words and punctuation\n# and lemmatize the tokens\nfor i, sentence in enumerate(sentences[\"text\"]):\n sentences.loc[i, \"text\"] = [token.lemma_ for token in sentence if not token.is_punct and not token.is_stop]",
"_____no_output_____"
]
],
[
[
"Below, we train several word2vec models. In particular, models 1 through 3 try windows sizes of 4, 6 and 8 and models 4 through 6 try vector size of 200 instead of 100:",
"_____no_output_____"
]
],
[
[
"# train word2vec on the the sentences\nmodel1 = gensim.models.Word2Vec(\n sentences[\"text\"],\n workers=4,\n min_count=1,\n window=4,\n sg=0,\n sample=1e-3,\n size=100,\n hs=1\n)\n\nmodel2 = gensim.models.Word2Vec(\n sentences[\"text\"],\n workers=4,\n min_count=1,\n window=6,\n sg=0,\n sample=1e-3,\n size=100,\n hs=1\n)\n\nmodel3 = gensim.models.Word2Vec(\n sentences[\"text\"],\n workers=4,\n min_count=1,\n window=8,\n sg=0,\n sample=1e-3,\n size=100,\n hs=1\n)\n\nmodel4 = gensim.models.Word2Vec(\n sentences[\"text\"],\n workers=4,\n min_count=1,\n window=4,\n sg=0,\n sample=1e-3,\n size=200,\n hs=1\n)\n\nmodel5 = gensim.models.Word2Vec(\n sentences[\"text\"],\n workers=4,\n min_count=1,\n window=6,\n sg=0,\n sample=1e-3,\n size=200,\n hs=1\n)\n\nmodel6 = gensim.models.Word2Vec(\n sentences[\"text\"],\n workers=4,\n min_count=1,\n window=8,\n sg=0,\n sample=1e-3,\n size=200,\n hs=1\n)",
"_____no_output_____"
],
[
"word2vec_arr1 = np.zeros((sentences.shape[0],100))\nword2vec_arr2 = np.zeros((sentences.shape[0],100))\nword2vec_arr3 = np.zeros((sentences.shape[0],100))\nword2vec_arr4 = np.zeros((sentences.shape[0],200))\nword2vec_arr5 = np.zeros((sentences.shape[0],200))\nword2vec_arr6 = np.zeros((sentences.shape[0],200))\n\nfor i, sentence in enumerate(sentences[\"text\"]):\n word2vec_arr1[i,:] = np.mean([model1[lemma] for lemma in sentence], axis=0)\n word2vec_arr2[i,:] = np.mean([model2[lemma] for lemma in sentence], axis=0)\n word2vec_arr3[i,:] = np.mean([model3[lemma] for lemma in sentence], axis=0)\n word2vec_arr4[i,:] = np.mean([model4[lemma] for lemma in sentence], axis=0)\n word2vec_arr5[i,:] = np.mean([model5[lemma] for lemma in sentence], axis=0)\n word2vec_arr6[i,:] = np.mean([model6[lemma] for lemma in sentence], axis=0)\n\nword2vec_arr1 = pd.DataFrame(word2vec_arr1)\nword2vec_arr2 = pd.DataFrame(word2vec_arr2)\nword2vec_arr3 = pd.DataFrame(word2vec_arr3)\nword2vec_arr4 = pd.DataFrame(word2vec_arr4)\nword2vec_arr5 = pd.DataFrame(word2vec_arr5)\nword2vec_arr6 = pd.DataFrame(word2vec_arr6)\n\nsentences1 = pd.concat([sentences[[\"author\", \"text\"]],word2vec_arr1], axis=1)\nsentences1.dropna(inplace=True)\n\nsentences2 = pd.concat([sentences[[\"author\", \"text\"]],word2vec_arr2], axis=1)\nsentences2.dropna(inplace=True)\n\nsentences3 = pd.concat([sentences[[\"author\", \"text\"]],word2vec_arr3], axis=1)\nsentences3.dropna(inplace=True)\n\nsentences4 = pd.concat([sentences[[\"author\", \"text\"]],word2vec_arr4], axis=1)\nsentences4.dropna(inplace=True)\n\nsentences5 = pd.concat([sentences[[\"author\", \"text\"]],word2vec_arr5], axis=1)\nsentences5.dropna(inplace=True)\n\nsentences6 = pd.concat([sentences[[\"author\", \"text\"]],word2vec_arr6], axis=1)\nsentences6.dropna(inplace=True)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier\nfrom sklearn.model_selection import train_test_split\n\nY1 = sentences1['author']\nY2 = sentences2['author']\nY3 = sentences3['author']\nY4 = sentences4['author']\nY5 = sentences5['author']\nY6 = sentences6['author']\n\nX1 = np.array(sentences1.drop(['text','author'], 1))\nX2 = np.array(sentences2.drop(['text','author'], 1))\nX3 = np.array(sentences3.drop(['text','author'], 1))\nX4 = np.array(sentences4.drop(['text','author'], 1))\nX5 = np.array(sentences5.drop(['text','author'], 1))\nX6 = np.array(sentences6.drop(['text','author'], 1))\n\n# We split the dataset into train and test sets\nX_train1, X_test1, y_train1, y_test1 = train_test_split(X1, Y1, test_size=0.4, random_state=123)\nX_train2, X_test2, y_train2, y_test2 = train_test_split(X2, Y2, test_size=0.4, random_state=123)\nX_train3, X_test3, y_train3, y_test3 = train_test_split(X3, Y3, test_size=0.4, random_state=123)\nX_train4, X_test4, y_train4, y_test4 = train_test_split(X4, Y4, test_size=0.4, random_state=123)\nX_train5, X_test5, y_train5, y_test5 = train_test_split(X5, Y5, test_size=0.4, random_state=123)\nX_train6, X_test6, y_train6, y_test6 = train_test_split(X6, Y6, test_size=0.4, random_state=123)\n\n# Models\nlr = LogisticRegression()\nrfc = RandomForestClassifier()\ngbc = GradientBoostingClassifier()\n\nprint(\"-----------------------Word2vec Model 1------------------------------\")\nlr.fit(X_train1, y_train1)\nrfc.fit(X_train1, y_train1)\ngbc.fit(X_train1, y_train1)\nprint(\"----------------------Logistic Regression Scores----------------------\")\nprint('Training set score:', lr.score(X_train1, y_train1))\nprint('\\nTest set score:', lr.score(X_test1, y_test1))\n\nprint(\"----------------------Random Forest Scores----------------------\")\nprint('Training set score:', rfc.score(X_train1, y_train1))\nprint('\\nTest set score:', rfc.score(X_test1, y_test1))\n\nprint(\"----------------------Gradient Boosting Scores----------------------\")\nprint('Training set score:', gbc.score(X_train1, y_train1))\nprint('\\nTest set score:', gbc.score(X_test1, y_test1))\n\nprint(\"-----------------------Word2vec Model 2------------------------------\")\nlr.fit(X_train2, y_train2)\nrfc.fit(X_train2, y_train2)\ngbc.fit(X_train2, y_train2)\nprint(\"----------------------Logistic Regression Scores----------------------\")\nprint('Training set score:', lr.score(X_train2, y_train2))\nprint('\\nTest set score:', lr.score(X_test2, y_test2))\n\nprint(\"----------------------Random Forest Scores----------------------\")\nprint('Training set score:', rfc.score(X_train2, y_train2))\nprint('\\nTest set score:', rfc.score(X_test2, y_test2))\n\nprint(\"----------------------Gradient Boosting Scores----------------------\")\nprint('Training set score:', gbc.score(X_train2, y_train2))\nprint('\\nTest set score:', gbc.score(X_test2, y_test2))\n\nprint(\"-----------------------Word2vec Model 3------------------------------\")\nlr.fit(X_train3, y_train3)\nrfc.fit(X_train3, y_train3)\ngbc.fit(X_train3, y_train3)\nprint(\"----------------------Logistic Regression Scores----------------------\")\nprint('Training set score:', lr.score(X_train3, y_train3))\nprint('\\nTest set score:', lr.score(X_test3, y_test3))\n\nprint(\"----------------------Random Forest Scores----------------------\")\nprint('Training set score:', rfc.score(X_train3, y_train3))\nprint('\\nTest set score:', rfc.score(X_test3, y_test3))\n\nprint(\"----------------------Gradient Boosting Scores----------------------\")\nprint('Training set score:', gbc.score(X_train3, y_train3))\nprint('\\nTest set score:', gbc.score(X_test3, y_test3))\n\nprint(\"-----------------------Word2vec Model 4------------------------------\")\nlr.fit(X_train4, y_train4)\nrfc.fit(X_train4, y_train4)\ngbc.fit(X_train4, y_train4)\nprint(\"----------------------Logistic Regression Scores----------------------\")\nprint('Training set score:', lr.score(X_train4, y_train4))\nprint('\\nTest set score:', lr.score(X_test4, y_test4))\n\nprint(\"----------------------Random Forest Scores----------------------\")\nprint('Training set score:', rfc.score(X_train4, y_train4))\nprint('\\nTest set score:', rfc.score(X_test4, y_test4))\n\nprint(\"----------------------Gradient Boosting Scores----------------------\")\nprint('Training set score:', gbc.score(X_train4, y_train4))\nprint('\\nTest set score:', gbc.score(X_test4, y_test4))\n\nprint(\"-----------------------Word2vec Model 5------------------------------\")\nlr.fit(X_train5, y_train5)\nrfc.fit(X_train5, y_train5)\ngbc.fit(X_train5, y_train5)\nprint(\"----------------------Logistic Regression Scores----------------------\")\nprint('Training set score:', lr.score(X_train5, y_train5))\nprint('\\nTest set score:', lr.score(X_test5, y_test5))\n\nprint(\"----------------------Random Forest Scores----------------------\")\nprint('Training set score:', rfc.score(X_train5, y_train5))\nprint('\\nTest set score:', rfc.score(X_test5, y_test5))\n\nprint(\"----------------------Gradient Boosting Scores----------------------\")\nprint('Training set score:', gbc.score(X_train5, y_train5))\nprint('\\nTest set score:', gbc.score(X_test5, y_test5))\n\nprint(\"-----------------------Word2vec Model 6------------------------------\")\nlr.fit(X_train6, y_train6)\nrfc.fit(X_train6, y_train6)\ngbc.fit(X_train6, y_train6)\nprint(\"----------------------Logistic Regression Scores----------------------\")\nprint('Training set score:', lr.score(X_train6, y_train6))\nprint('\\nTest set score:', lr.score(X_test6, y_test6))\n\nprint(\"----------------------Random Forest Scores----------------------\")\nprint('Training set score:', rfc.score(X_train6, y_train6))\nprint('\\nTest set score:', rfc.score(X_test6, y_test6))\n\nprint(\"----------------------Gradient Boosting Scores----------------------\")\nprint('Training set score:', gbc.score(X_train6, y_train6))\nprint('\\nTest set score:', gbc.score(X_test6, y_test6))\n",
"-----------------------Word2vec Model 1------------------------------\n----------------------Logistic Regression Scores----------------------\nTraining set score: 0.7789954337899543\n\nTest set score: 0.7835616438356164\n----------------------Random Forest Scores----------------------\nTraining set score: 0.9917808219178083\n\nTest set score: 0.821917808219178\n----------------------Gradient Boosting Scores----------------------\nTraining set score: 0.8989345509893455\n\nTest set score: 0.819634703196347\n-----------------------Word2vec Model 2------------------------------\n----------------------Logistic Regression Scores----------------------\nTraining set score: 0.7841704718417047\n\nTest set score: 0.7876712328767124\n----------------------Random Forest Scores----------------------\nTraining set score: 0.9917808219178083\n\nTest set score: 0.826027397260274\n----------------------Gradient Boosting Scores----------------------\nTraining set score: 0.8931506849315068\n\nTest set score: 0.8242009132420092\n-----------------------Word2vec Model 3------------------------------\n----------------------Logistic Regression Scores----------------------\nTraining set score: 0.7823439878234398\n\nTest set score: 0.7931506849315069\n----------------------Random Forest Scores----------------------\nTraining set score: 0.9917808219178083\n\nTest set score: 0.8301369863013699\n----------------------Gradient Boosting Scores----------------------\nTraining set score: 0.8958904109589041\n\nTest set score: 0.836986301369863\n-----------------------Word2vec Model 4------------------------------\n"
]
],
[
[
"Model 6's performance seems to be better. In particular, the best test performance is achieved using model 6 and gradient boosting. Three random forest models also achieved the highest score when trained on model 6. \n\nMoreover, model 6's performance is also superior to that of the model in the checkpoint.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbafa8e21d0956b82d4b6c64b0cd94c7270ddd57
| 10,429 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Untitled-checkpoint.ipynb
|
FurmanCenter/ACSDownloader
|
918afc0c7baa8814da98c2e3ee11352af68c027e
|
[
"Apache-2.0"
] | 1 |
2020-04-15T15:40:18.000Z
|
2020-04-15T15:40:18.000Z
|
.ipynb_checkpoints/Untitled-checkpoint.ipynb
|
FurmanCenter/ACSDownloader
|
918afc0c7baa8814da98c2e3ee11352af68c027e
|
[
"Apache-2.0"
] | null | null | null |
.ipynb_checkpoints/Untitled-checkpoint.ipynb
|
FurmanCenter/ACSDownloader
|
918afc0c7baa8814da98c2e3ee11352af68c027e
|
[
"Apache-2.0"
] | null | null | null | 50.873171 | 782 | 0.564771 |
[
[
[
"import dl_acs\n",
"_____no_output_____"
],
[
"import pip\npip.get_installed_distributions()\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbafb22903a3b28665ccb36489aa1995b955dbd0
| 116,737 |
ipynb
|
Jupyter Notebook
|
Deep_Learning/MNIST_Dataset_Recognition.ipynb
|
Ironspine/zoli
|
8e149b3458741343ea20dd9c6023dbe61d8abf14
|
[
"Apache-2.0"
] | 6 |
2020-06-21T09:08:55.000Z
|
2021-07-28T14:54:30.000Z
|
Deep_Learning/MNIST_Dataset_Recognition.ipynb
|
Ironspine/zoli
|
8e149b3458741343ea20dd9c6023dbe61d8abf14
|
[
"Apache-2.0"
] | null | null | null |
Deep_Learning/MNIST_Dataset_Recognition.ipynb
|
Ironspine/zoli
|
8e149b3458741343ea20dd9c6023dbe61d8abf14
|
[
"Apache-2.0"
] | null | null | null | 82.850958 | 30,752 | 0.738318 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport os",
"_____no_output_____"
],
[
"from keras.datasets import mnist",
"Using TensorFlow backend.\n"
],
[
"# Digit recognition when data is in 'pixel form'\n\n(X_train, y_train), (X_test, y_test) = mnist.load_data()",
"_____no_output_____"
],
[
"# Shape of the pictures\n\nX_test[4,:,:].shape",
"_____no_output_____"
],
[
"df = pd.DataFrame(X_train[0,:,:])\ndf",
"_____no_output_____"
],
[
"img = X_test[30,:,:]\nplt.imshow(img, cmap = 'gray')\nplt.title(y_train[0])\nplt.axis('off')",
"_____no_output_____"
],
[
"from keras.utils import to_categorical\n\nX_train_new = X_train[:10000,:,:]\ny_train_new = y_train[:10000]\nX_test_new = X_test[:2500,:,:]\ny_test_new = y_test[:2500]\n\ny_train_new = to_categorical(y_train_new, 10)\ny_test_new = to_categorical(y_test_new, 10)\n\nX_train_new = X_train_new.reshape(10000, 28, 28, 1)\nX_test_new = X_test_new.reshape(2500, 28, 28, 1)",
"_____no_output_____"
],
[
"y_test_new.shape",
"_____no_output_____"
],
[
"# Convolutional Neural Network for identifying the digits\n\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Conv2D, Dropout, Flatten, MaxPooling2D",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(Conv2D(32, kernel_size = (3, 3), input_shape = (28,28,1), activation = 'relu'))\nmodel.add(MaxPooling2D((2,2)))\n\nmodel.add(Conv2D(32, kernel_size = (3, 3), activation = 'relu'))\nmodel.add(MaxPooling2D((2,2)))\n\nmodel.add(Dropout(0.2))\n\nmodel.add(Flatten())\n\nmodel.add(Dense(128, activation='relu'))\nmodel.add(Dropout(0.4))\nmodel.add(Dense(10, activation = 'softmax'))\n\nmodel.compile(loss = 'categorical_crossentropy', optimizer = 'adam', metrics = ['accuracy'])",
"_____no_output_____"
],
[
"model.fit(X_train_new, y_train_new, epochs = 15, batch_size = 128, validation_data = (X_test_new, y_test_new), verbose = 1)",
"Train on 10000 samples, validate on 2500 samples\nEpoch 1/15\n10000/10000 [==============================] - 84s 8ms/step - loss: 4.7063 - accuracy: 0.5354 - val_loss: 0.5597 - val_accuracy: 0.8320\nEpoch 2/15\n10000/10000 [==============================] - 70s 7ms/step - loss: 0.6612 - accuracy: 0.7963 - val_loss: 0.3634 - val_accuracy: 0.8980\nEpoch 3/15\n10000/10000 [==============================] - 69s 7ms/step - loss: 0.4342 - accuracy: 0.8682 - val_loss: 0.2356 - val_accuracy: 0.9288\nEpoch 4/15\n10000/10000 [==============================] - 67s 7ms/step - loss: 0.3430 - accuracy: 0.8981 - val_loss: 0.2012 - val_accuracy: 0.9428\nEpoch 5/15\n10000/10000 [==============================] - 71s 7ms/step - loss: 0.2654 - accuracy: 0.9195 - val_loss: 0.1796 - val_accuracy: 0.9468\nEpoch 6/15\n10000/10000 [==============================] - 68s 7ms/step - loss: 0.2334 - accuracy: 0.9309 - val_loss: 0.1725 - val_accuracy: 0.9480\nEpoch 7/15\n10000/10000 [==============================] - 86s 9ms/step - loss: 0.1987 - accuracy: 0.9382 - val_loss: 0.1377 - val_accuracy: 0.9552\nEpoch 8/15\n10000/10000 [==============================] - 74s 7ms/step - loss: 0.1764 - accuracy: 0.9435 - val_loss: 0.1320 - val_accuracy: 0.9596\nEpoch 9/15\n10000/10000 [==============================] - 74s 7ms/step - loss: 0.1636 - accuracy: 0.9483 - val_loss: 0.1186 - val_accuracy: 0.9588\nEpoch 10/15\n10000/10000 [==============================] - 72s 7ms/step - loss: 0.1524 - accuracy: 0.9538 - val_loss: 0.1118 - val_accuracy: 0.9620\nEpoch 11/15\n10000/10000 [==============================] - 70s 7ms/step - loss: 0.1304 - accuracy: 0.9601 - val_loss: 0.1162 - val_accuracy: 0.9604\nEpoch 12/15\n10000/10000 [==============================] - 74s 7ms/step - loss: 0.1149 - accuracy: 0.9638 - val_loss: 0.1111 - val_accuracy: 0.9636\nEpoch 13/15\n10000/10000 [==============================] - 71s 7ms/step - loss: 0.1125 - accuracy: 0.9660 - val_loss: 0.1111 - val_accuracy: 0.9668\nEpoch 14/15\n10000/10000 [==============================] - 70s 7ms/step - loss: 0.1075 - accuracy: 0.9648 - val_loss: 0.1040 - val_accuracy: 0.9680\nEpoch 15/15\n10000/10000 [==============================] - 68s 7ms/step - loss: 0.0981 - accuracy: 0.9687 - val_loss: 0.1045 - val_accuracy: 0.9692\n"
],
[
"score = model.evaluate(X_test_new, y_test_new, verbose = 0)\nprint('Test loss: ', score[0])\nprint('Accuracy: ', score[1])",
"Test loss: 0.10451674363203346\nAccuracy: 0.9692000150680542\n"
],
[
"# Plotting the accuracy\n\naccuracy = model.history.history\nplt.figure(figsize = (9,7))\nplt.plot(accuracy['loss'], lw = 2)\nplt.title('Loss')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')",
"_____no_output_____"
],
[
"from sklearn.metrics import confusion_matrix, classification_report\n\nprediction = model.predict(X_test_new)\nprediction_classes = np.argmax(prediction, axis = 1)\ny_true = np.argmax(y_test_new, axis = 1)\ncm = confusion_matrix(y_true, prediction_classes)\n\nprint(classification_report(y_true, prediction_classes))",
" precision recall f1-score support\n\n 0 0.98 0.98 0.98 219\n 1 0.98 0.99 0.98 287\n 2 0.97 0.97 0.97 276\n 3 0.96 0.97 0.96 254\n 4 0.96 0.99 0.97 275\n 5 0.97 0.97 0.97 221\n 6 0.96 0.97 0.96 225\n 7 0.96 0.97 0.97 257\n 8 0.98 0.93 0.96 242\n 9 0.99 0.95 0.97 244\n\n accuracy 0.97 2500\n macro avg 0.97 0.97 0.97 2500\nweighted avg 0.97 0.97 0.97 2500\n\n"
],
[
"import seaborn as sns\n\nplt.figure(figsize = (10,8))\nsns.heatmap(cm, annot = True, cmap = 'viridis')\n#b, t = plt.ylim()\n#plt.ylim(b + 0.5, t - 0.5)\nplt.title('Confusion Matrix')",
"_____no_output_____"
],
[
"# New prediction\n\nnew_sample = X_test_new[11:12,:,:,:]\nnew_sample.shape",
"_____no_output_____"
],
[
"new_pred = model.predict(new_sample)",
"_____no_output_____"
],
[
"new_pred = new_pred.ravel()",
"_____no_output_____"
],
[
"np.argmax(new_pred, axis = 0)",
"_____no_output_____"
],
[
"# Saving model for reproduction\n\n# model.save('conv_model.h5')",
"_____no_output_____"
],
[
"from keras.models import load_model\n\nreconstructed_model = load_model('conv_model.h5')\n\n# Let's check:\n# np.testing.assert_allclose(model.predict(new_sample), reconstructed_model.predict(new_sample))\n\n# The reconstructed model is already compiled and has retained the optimizer\n# state, so training can resume:\n# reconstructed_model.fit(test_input, test_target)",
"Using TensorFlow backend.\n"
],
[
"# Creating my own digit picture using Paint \n# Let's import them with the Pillow library\n\nfrom PIL import Image\nimport matplotlib.image as mpimg\n\nimage = Image.open('numbers/number_eight.jpg')\nimage = image.resize((28, 28))\n#image.save('numbers28/28X28number_eight.jpg')\nimage = mpimg.imread('numbers28/number00.jpg')\nplt.imshow(image)",
"_____no_output_____"
],
[
"# Converting from RGB to grayscale and making prediction\n\ndef rgb2gray(rgb):\n return np.dot(rgb[...,:3], [0.2989, 0.5870, 0.1140])\ngray = rgb2gray(image)\ngray = gray.reshape(1, 28, 28, 1)\ngray_pred = reconstructed_model.predict(gray)\nprint('Predicted value:', np.argmax(gray_pred))",
"Predicted value: 8\n"
],
[
"import matplotlib.image as mpimg\n\ndef rgb2gray(rgb):\n return np.dot(rgb[...,:3], [0.2989, 0.5870, 0.1140])\n\ndef image():\n files = [f for f in os.listdir('numbers/') if f.endswith('.jpg')]\n predictions = []\n for i in range(len(files)):\n image = Image.open('numbers/' + files[i])\n image = image.resize((28, 28)) \n image.save('numbers28/number0' + str(i) + '.jpg')\n image = mpimg.imread('numbers28/number0' + str(i) + '.jpg')\n gray = rgb2gray(image)\n gray = gray.reshape(1, 28, 28, 1)\n gray_pred = reconstructed_model.predict(gray)\n predictions.append(gray_pred.argmax())\n return predictions, image\n \ndef plot_images(predictions, images):\n truth = [8, 5, 4, 9, 1, 7, 6, 3, 2, 0]\n plt.figure(figsize = (12, 6))\n for i in range(len(truth)):\n plt.subplot(2, 5, i+1)\n plt.axis('off')\n image = mpimg.imread('numbers28/number0' + str(i) + '.jpg')\n color = 'green' if truth[i] == predictions[i] else 'red'\n plt.imshow(image)\n plt.title('Predicted value:\\n' + str(predictions[i]), size = 12, color = color)\n plt.subplots_adjust(wspace = 0.2)\n return plt.show()\n\npredictions, images = image()\nplot_images(predictions, images)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbafb9b7237efa6d2c18facee982f43b6e812054
| 587,768 |
ipynb
|
Jupyter Notebook
|
introduction_to_amazon_algorithms/deepar_electricity/DeepAR-Electricity-DataClean.ipynb
|
whn09/amazon-sagemaker-examples
|
6074e6ea5416a67894a278ceb7a0342280ee23b3
|
[
"Apache-2.0"
] | null | null | null |
introduction_to_amazon_algorithms/deepar_electricity/DeepAR-Electricity-DataClean.ipynb
|
whn09/amazon-sagemaker-examples
|
6074e6ea5416a67894a278ceb7a0342280ee23b3
|
[
"Apache-2.0"
] | null | null | null |
introduction_to_amazon_algorithms/deepar_electricity/DeepAR-Electricity-DataClean.ipynb
|
whn09/amazon-sagemaker-examples
|
6074e6ea5416a67894a278ceb7a0342280ee23b3
|
[
"Apache-2.0"
] | null | null | null | 263.691341 | 387,320 | 0.865915 |
[
[
[
"# SageMaker/DeepAR demo on electricity dataset\n\nThis notebook complements the [DeepAR introduction notebook](https://github.com/awslabs/amazon-sagemaker-examples/blob/master/introduction_to_amazon_algorithms/deepar_synthetic/deepar_synthetic.ipynb). \n\nHere, we will consider a real use case and show how to use DeepAR on SageMaker for predicting energy consumption of 370 customers over time, based on a [dataset](https://archive.ics.uci.edu/ml/datasets/ElectricityLoadDiagrams20112014) that was used in the academic papers [[1](https://media.nips.cc/nipsbooks/nipspapers/paper_files/nips29/reviews/526.html)] and [[2](https://arxiv.org/abs/1704.04110)]. \n\nIn particular, we will see how to:\n* Prepare the dataset\n* Use the SageMaker Python SDK to train a DeepAR model and deploy it\n* Make requests to the deployed model to obtain forecasts interactively\n* Illustrate advanced features of DeepAR: missing values, additional time features, non-regular frequencies and category information\n\nRunning this notebook takes around 40 min on a ml.c4.2xlarge for the training, and inference is done on a ml.m4.xlarge (the usage time will depend on how long you leave your served model running).\n\nFor more information see the DeepAR [documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar.html) or [paper](https://arxiv.org/abs/1704.04110), ",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport sys\nfrom urllib.request import urlretrieve\nimport zipfile\nfrom dateutil.parser import parse\nimport json\nfrom random import shuffle\nimport random\nimport datetime\nimport os\n\nimport boto3\nimport s3fs\nimport sagemaker\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nfrom __future__ import print_function\nfrom ipywidgets import interact, interactive, fixed, interact_manual\nimport ipywidgets as widgets\nfrom ipywidgets import IntSlider, FloatSlider, Checkbox",
"_____no_output_____"
],
[
"# set random seeds for reproducibility\nnp.random.seed(42)\nrandom.seed(42)",
"_____no_output_____"
],
[
"sagemaker_session = sagemaker.Session()",
"_____no_output_____"
]
],
[
[
"Before starting, we can override the default values for the following:\n- The S3 bucket and prefix that you want to use for training and model data. This should be within the same region as the Notebook Instance, training, and hosting.\n- The IAM role arn used to give training and hosting access to your data. See the documentation for how to create these.",
"_____no_output_____"
]
],
[
[
"s3_bucket = sagemaker.Session().default_bucket() # replace with an existing bucket if needed\ns3_prefix = 'deepar-electricity-demo-notebook' # prefix used for all data stored within the bucket\n\nrole = sagemaker.get_execution_role() # IAM role to use by SageMaker",
"_____no_output_____"
],
[
"region = sagemaker_session.boto_region_name\n\ns3_data_path = \"s3://{}/{}/data\".format(s3_bucket, s3_prefix)\ns3_output_path = \"s3://{}/{}/output\".format(s3_bucket, s3_prefix)",
"_____no_output_____"
]
],
[
[
"Next, we configure the container image to be used for the region that we are running in.",
"_____no_output_____"
]
],
[
[
"image_name = sagemaker.amazon.amazon_estimator.get_image_uri(region, \"forecasting-deepar\", \"latest\")",
"_____no_output_____"
]
],
[
[
"### Import electricity dataset and upload it to S3 to make it available for Sagemaker",
"_____no_output_____"
],
[
"As a first step, we need to download the original data set of from the UCI data set repository.",
"_____no_output_____"
]
],
[
[
"DATA_HOST = \"https://archive.ics.uci.edu\"\nDATA_PATH = \"/ml/machine-learning-databases/00321/\"\nARCHIVE_NAME = \"LD2011_2014.txt.zip\"\nFILE_NAME = ARCHIVE_NAME[:-4]",
"_____no_output_____"
],
[
"def progress_report_hook(count, block_size, total_size):\n mb = int(count * block_size // 1e6)\n if count % 500 == 0:\n sys.stdout.write(\"\\r{} MB downloaded\".format(mb))\n sys.stdout.flush()\n\nif not os.path.isfile(FILE_NAME):\n print(\"downloading dataset (258MB), can take a few minutes depending on your connection\")\n urlretrieve(DATA_HOST + DATA_PATH + ARCHIVE_NAME, ARCHIVE_NAME, reporthook=progress_report_hook)\n\n print(\"\\nextracting data archive\")\n zip_ref = zipfile.ZipFile(ARCHIVE_NAME, 'r')\n zip_ref.extractall(\"./\")\n zip_ref.close()\nelse:\n print(\"File found skipping download\")",
"_____no_output_____"
]
],
[
[
"Then, we load and parse the dataset and convert it to a collection of Pandas time series, which makes common time series operations such as indexing by time periods or resampling much easier. The data is originally recorded in 15min interval, which we could use directly. Here we want to forecast longer periods (one week) and resample the data to a granularity of 2 hours.",
"_____no_output_____"
]
],
[
[
"data = pd.read_csv(FILE_NAME, sep=\";\", index_col=0, parse_dates=True, decimal=',')\nnum_timeseries = data.shape[1]\ndata_kw = data.resample('2H').sum() / 8\ntimeseries = []\nfor i in range(num_timeseries):\n timeseries.append(np.trim_zeros(data_kw.iloc[:,i], trim='f'))",
"_____no_output_____"
]
],
[
[
"Let us plot the resulting time series for the first ten customers for the time period spanning the first two weeks of 2014.",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(5, 2, figsize=(20, 20), sharex=True)\naxx = axs.ravel()\nfor i in range(0, 10):\n timeseries[i].loc[\"2014-01-01\":\"2014-01-14\"].plot(ax=axx[i])\n axx[i].set_xlabel(\"date\") \n axx[i].set_ylabel(\"kW consumption\") \n axx[i].grid(which='minor', axis='x')",
"_____no_output_____"
]
],
[
[
"### Train and Test splits\n\nOften times one is interested in evaluating the model or tuning its hyperparameters by looking at error metrics on a hold-out test set. Here we split the available data into train and test sets for evaluating the trained model. For standard machine learning tasks such as classification and regression, one typically obtains this split by randomly separating examples into train and test sets. However, in forecasting it is important to do this train/test split based on time rather than by time series.\n\nIn this example, we will reserve the last section of each of the time series for evalutation purpose and use only the first part as training data. ",
"_____no_output_____"
]
],
[
[
"# we use 2 hour frequency for the time series\nfreq = '2H'\n\n# we predict for 7 days\nprediction_length = 7 * 12\n\n# we also use 7 days as context length, this is the number of state updates accomplished before making predictions\ncontext_length = 7 * 12",
"_____no_output_____"
]
],
[
[
"We specify here the portion of the data that is used for training: the model sees data from 2014-01-01 to 2014-09-01 for training.",
"_____no_output_____"
]
],
[
[
"start_dataset = pd.Timestamp(\"2014-01-01 00:00:00\", freq=freq)\nend_training = pd.Timestamp(\"2014-09-01 00:00:00\", freq=freq)",
"_____no_output_____"
]
],
[
[
"The DeepAR JSON input format represents each time series as a JSON object. In the simplest case each time series just consists of a start time stamp (``start``) and a list of values (``target``). For more complex cases, DeepAR also supports the fields ``dynamic_feat`` for time-series features and ``cat`` for categorical features, which we will use later.",
"_____no_output_____"
]
],
[
[
"training_data = [\n {\n \"start\": str(start_dataset),\n \"target\": ts[start_dataset:end_training - 1].tolist() # We use -1, because pandas indexing includes the upper bound \n }\n for ts in timeseries\n]\nprint(len(training_data))\n\nclean_training_data = []\nfor i in range(370):\n if len(training_data[i]['target']) != 2916:\n print(i, len(training_data[i]['target']))\n else:\n clean_training_data.append(training_data[i])",
"370\n105 2760\n106 2760\n107 2760\n108 2340\n109 2760\n110 2760\n111 2412\n112 2760\n114 2760\n115 2340\n116 2760\n119 2760\n120 2760\n121 2760\n132 2056\n159 2508\n177 540\n180 2160\n336 2724\n"
]
],
[
[
"As test data, we will consider time series extending beyond the training range: these will be used for computing test scores, by using the trained model to forecast their trailing 7 days, and comparing predictions with actual values.\nTo evaluate our model performance on more than one week, we generate test data that extends to 1, 2, 3, 4 weeks beyond the training range. This way we perform *rolling evaluation* of our model.",
"_____no_output_____"
]
],
[
[
"num_test_windows = 4\n\ntest_data = [\n {\n \"start\": str(start_dataset),\n \"target\": ts[start_dataset:end_training + k * prediction_length].tolist()\n }\n for k in range(1, num_test_windows + 1) \n for ts in timeseries\n]\nprint(len(test_data))\n\nclean_test_data = []\nfor i in range(370):\n if len(test_data[i]['target']) != 3001 or len(test_data[i+370]['target']) != 3001+84 or len(test_data[i+370*2]['target']) != 3001+84*2 or len(test_data[i+370*3]['target']) != 3001+84*3:\n print(i, len(test_data[i]['target']), len(test_data[i+370]['target']), len(test_data[i+370*2]['target']), len(test_data[i+370*3]['target']))\n else:\n clean_test_data.append(test_data[i])\n clean_test_data.append(test_data[i+370])\n clean_test_data.append(test_data[i+370*2])\n clean_test_data.append(test_data[i+370*3])",
"/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/ipykernel/__main__.py:9: FutureWarning: Addition/subtraction of integers and integer-arrays to Timestamp is deprecated, will be removed in a future version. Instead of adding/subtracting `n`, use `n * self.freq`\n"
]
],
[
[
"Let's now write the dictionary to the `jsonlines` file format that DeepAR understands (it also supports gzipped jsonlines and parquet).",
"_____no_output_____"
]
],
[
[
"def write_dicts_to_file(path, data):\n with open(path, 'wb') as fp:\n for d in data:\n fp.write(json.dumps(d).encode(\"utf-8\"))\n fp.write(\"\\n\".encode('utf-8'))",
"_____no_output_____"
],
[
"%%time\nwrite_dicts_to_file(\"clean_train.json\", clean_training_data)\nwrite_dicts_to_file(\"clean_test.json\", clean_test_data)",
"CPU times: user 3.81 s, sys: 72.9 ms, total: 3.88 s\nWall time: 3.88 s\n"
]
],
[
[
"Now that we have the data files locally, let us copy them to S3 where DeepAR can access them. Depending on your connection, this may take a couple of minutes.",
"_____no_output_____"
]
],
[
[
"s3 = boto3.resource('s3')\ndef copy_to_s3(local_file, s3_path, override=False):\n assert s3_path.startswith('s3://')\n split = s3_path.split('/')\n bucket = split[2]\n path = '/'.join(split[3:])\n buk = s3.Bucket(bucket)\n \n if len(list(buk.objects.filter(Prefix=path))) > 0:\n if not override:\n print('File s3://{}/{} already exists.\\nSet override to upload anyway.\\n'.format(s3_bucket, s3_path))\n return\n else:\n print('Overwriting existing file')\n with open(local_file, 'rb') as data:\n print('Uploading file to {}'.format(s3_path))\n buk.put_object(Key=path, Body=data)",
"_____no_output_____"
],
[
"%%time\ncopy_to_s3(\"clean_train.json\", s3_data_path + \"/train/clean_train.json\")\ncopy_to_s3(\"clean_test.json\", s3_data_path + \"/test/clean_test.json\")",
"Uploading file to s3://sagemaker-ap-northeast-1-579019700964/deepar-electricity-demo-notebook/data/train/clean_train.json\nUploading file to s3://sagemaker-ap-northeast-1-579019700964/deepar-electricity-demo-notebook/data/test/clean_test.json\nCPU times: user 506 ms, sys: 69.8 ms, total: 576 ms\nWall time: 2.09 s\n"
]
],
[
[
"Let's have a look to what we just wrote to S3.",
"_____no_output_____"
]
],
[
[
"s3filesystem = s3fs.S3FileSystem()\nwith s3filesystem.open(s3_data_path + \"/train/clean_train.json\", 'rb') as fp:\n print(fp.readline().decode(\"utf-8\")[:100] + \"...\")",
"{\"start\": \"2014-01-01 00:00:00\", \"target\": [2.6967005076142154, 2.8553299492385804, 2.53807106598985...\n"
]
],
[
[
"We are all set with our dataset processing, we can now call DeepAR to train a model and generate predictions.",
"_____no_output_____"
],
[
"### Train a model\n\nHere we define the estimator that will launch the training job.",
"_____no_output_____"
]
],
[
[
"estimator = sagemaker.estimator.Estimator(\n sagemaker_session=sagemaker_session,\n image_name=image_name,\n role=role,\n train_instance_count=1,\n train_instance_type='ml.c4.2xlarge',\n base_job_name='deepar-electricity-clean-demo',\n output_path=s3_output_path\n)",
"_____no_output_____"
]
],
[
[
"Next we need to set the hyperparameters for the training job. For example frequency of the time series used, number of data points the model will look at in the past, number of predicted data points. The other hyperparameters concern the model to train (number of layers, number of cells per layer, likelihood function) and the training options (number of epochs, batch size, learning rate...). We use default parameters for every optional parameter in this case (you can always use [Sagemaker Automated Model Tuning](https://aws.amazon.com/blogs/aws/sagemaker-automatic-model-tuning/) to tune them).",
"_____no_output_____"
]
],
[
[
"hyperparameters = {\n \"time_freq\": freq,\n \"epochs\": \"400\",\n \"early_stopping_patience\": \"40\",\n \"mini_batch_size\": \"64\",\n \"learning_rate\": \"5E-4\",\n \"context_length\": str(context_length),\n \"prediction_length\": str(prediction_length)\n}",
"_____no_output_____"
],
[
"estimator.set_hyperparameters(**hyperparameters)",
"_____no_output_____"
]
],
[
[
"We are ready to launch the training job. SageMaker will start an EC2 instance, download the data from S3, start training the model and save the trained model.\n\nIf you provide the `test` data channel as we do in this example, DeepAR will also calculate accuracy metrics for the trained model on this test. This is done by predicting the last `prediction_length` points of each time-series in the test set and comparing this to the actual value of the time-series. \n\n**Note:** the next cell may take a few minutes to complete, depending on data size, model complexity, training options.",
"_____no_output_____"
]
],
[
[
"%%time\ndata_channels = {\n \"train\": \"{}/train/clean_train.json\".format(s3_data_path),\n \"test\": \"{}/test/clean_test.json\".format(s3_data_path)\n}\n\nestimator.fit(inputs=data_channels, wait=True)",
"2019-07-17 06:36:13 Starting - Starting the training job...\n2019-07-17 06:36:15 Starting - Launching requested ML instances......\n2019-07-17 06:37:22 Starting - Preparing the instances for training...\n2019-07-17 06:38:11 Downloading - Downloading input data...\n2019-07-17 06:38:41 Training - Downloading the training image...\n2019-07-17 06:38:58 Training - Training image download completed. Training in progress.\n\u001b[31mArguments: train\u001b[0m\n\u001b[31m[07/17/2019 06:39:01 INFO 139964490479424] Reading default configuration from /opt/amazon/lib/python2.7/site-packages/algorithm/resources/default-input.json: {u'num_dynamic_feat': u'auto', u'dropout_rate': u'0.10', u'mini_batch_size': u'128', u'test_quantiles': u'[0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]', u'_tuning_objective_metric': u'', u'_num_gpus': u'auto', u'num_eval_samples': u'100', u'learning_rate': u'0.001', u'num_cells': u'40', u'num_layers': u'2', u'embedding_dimension': u'10', u'_kvstore': u'auto', u'_num_kv_servers': u'auto', u'cardinality': u'auto', u'likelihood': u'student-t', u'early_stopping_patience': u''}\u001b[0m\n\u001b[31m[07/17/2019 06:39:01 INFO 139964490479424] Reading provided configuration from /opt/ml/input/config/hyperparameters.json: {u'learning_rate': u'5E-4', u'prediction_length': u'84', u'epochs': u'400', u'time_freq': u'2H', u'context_length': u'84', u'mini_batch_size': u'64', u'early_stopping_patience': u'40'}\u001b[0m\n\u001b[31m[07/17/2019 06:39:01 INFO 139964490479424] Final configuration: {u'dropout_rate': u'0.10', u'test_quantiles': u'[0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]', u'_tuning_objective_metric': u'', u'num_eval_samples': u'100', u'learning_rate': u'5E-4', u'num_layers': u'2', u'epochs': u'400', u'embedding_dimension': u'10', u'num_cells': u'40', u'_num_kv_servers': u'auto', u'mini_batch_size': u'64', u'likelihood': u'student-t', u'num_dynamic_feat': u'auto', u'cardinality': u'auto', u'_num_gpus': u'auto', u'prediction_length': u'84', u'time_freq': u'2H', u'context_length': u'84', u'_kvstore': u'auto', u'early_stopping_patience': u'40'}\u001b[0m\n\u001b[31mProcess 1 is a worker.\u001b[0m\n\u001b[31m[07/17/2019 06:39:01 INFO 139964490479424] Detected entry point for worker worker\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Using early stopping with patience 40\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] [cardinality=auto] `cat` field was NOT found in the file `/opt/ml/input/data/train/clean_train.json` and will NOT be used for training.\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] [num_dynamic_feat=auto] `dynamic_feat` field was NOT found in the file `/opt/ml/input/data/train/clean_train.json` and will NOT be used for training.\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Training set statistics:\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Real time series\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] number of time series: 351\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] number of observations: 1023516\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] mean target length: 2916\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] min/mean/max target: 0.0/637.111702965/163325.0\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] mean abs(target): 637.111702965\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] contains missing values: no\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Small number of time series. Doing 1 number of passes over dataset per epoch.\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Test set statistics:\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Real time series\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] number of time series: 1404\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] number of observations: 4390308\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] mean target length: 3127\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] min/mean/max target: 0.0/645.502472797/163325.0\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] mean abs(target): 645.502472797\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] contains missing values: no\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] nvidia-smi took: 0.02516913414 secs to identify 0 gpus\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Number of GPUs being used: 0\u001b[0m\n\u001b[31m[07/17/2019 06:39:02 INFO 139964490479424] Create Store: local\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"get_graph.time\": {\"count\": 1, \"max\": 646.4171409606934, \"sum\": 646.4171409606934, \"min\": 646.4171409606934}}, \"EndTime\": 1563345543.542158, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345542.894919}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:03 INFO 139964490479424] Number of GPUs being used: 0\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"initialize.time\": {\"count\": 1, \"max\": 1330.6598663330078, \"sum\": 1330.6598663330078, \"min\": 1330.6598663330078}}, \"EndTime\": 1563345544.225719, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345543.542235}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:04 INFO 139964490479424] Epoch[0] Batch[0] avg_epoch_loss=6.431551\u001b[0m\n\u001b[31m[07/17/2019 06:39:04 INFO 139964490479424] #quality_metric: host=algo-1, epoch=0, batch=0 train loss <loss>=6.43155145645\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] Epoch[0] Batch[5] avg_epoch_loss=6.004621\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] #quality_metric: host=algo-1, epoch=0, batch=5 train loss <loss>=6.00462055206\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] Epoch[0] Batch [5]#011Speed: 339.96 samples/sec#011loss=6.004621\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] processed a total of 343 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"epochs\": {\"count\": 1, \"max\": 400, \"sum\": 400.0, \"min\": 400}, \"update.time\": {\"count\": 1, \"max\": 1529.4289588928223, \"sum\": 1529.4289588928223, \"min\": 1529.4289588928223}}, \"EndTime\": 1563345545.75535, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345544.225826}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=224.248112127 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] #progress_metric: host=algo-1, completed 0 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] #quality_metric: host=algo-1, epoch=0, train loss <loss>=6.00462055206\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:05 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_3e5dc2ca-f2a1-4026-9d50-2129f0548d6d-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 100.91805458068848, \"sum\": 100.91805458068848, \"min\": 100.91805458068848}}, \"EndTime\": 1563345545.856888, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345545.755441}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:06 INFO 139964490479424] Epoch[1] Batch[0] avg_epoch_loss=5.504302\u001b[0m\n\u001b[31m[07/17/2019 06:39:06 INFO 139964490479424] #quality_metric: host=algo-1, epoch=1, batch=0 train loss <loss>=5.504301548\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] Epoch[1] Batch[5] avg_epoch_loss=5.600807\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] #quality_metric: host=algo-1, epoch=1, batch=5 train loss <loss>=5.60080718994\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] Epoch[1] Batch [5]#011Speed: 341.70 samples/sec#011loss=5.600807\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] processed a total of 353 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1367.285966873169, \"sum\": 1367.285966873169, \"min\": 1367.285966873169}}, \"EndTime\": 1563345547.22434, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345545.85699}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=258.153589069 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] #progress_metric: host=algo-1, completed 0 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] #quality_metric: host=algo-1, epoch=1, train loss <loss>=5.60080718994\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_8e76f6ef-cacd-4f26-8e8d-7b3157629f4c-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 61.228036880493164, \"sum\": 61.228036880493164, \"min\": 61.228036880493164}}, \"EndTime\": 1563345547.286115, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345547.224422}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] Epoch[2] Batch[0] avg_epoch_loss=5.389429\u001b[0m\n\u001b[31m[07/17/2019 06:39:07 INFO 139964490479424] #quality_metric: host=algo-1, epoch=2, batch=0 train loss <loss>=5.38942909241\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] Epoch[2] Batch[5] avg_epoch_loss=5.640230\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] #quality_metric: host=algo-1, epoch=2, batch=5 train loss <loss>=5.64022978147\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] Epoch[2] Batch [5]#011Speed: 343.97 samples/sec#011loss=5.640230\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] processed a total of 360 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1328.6170959472656, \"sum\": 1328.6170959472656, \"min\": 1328.6170959472656}}, \"EndTime\": 1563345548.614878, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345547.286196}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=270.935188339 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] #progress_metric: host=algo-1, completed 0 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] #quality_metric: host=algo-1, epoch=2, train loss <loss>=5.64022978147\u001b[0m\n\u001b[31m[07/17/2019 06:39:08 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:09 INFO 139964490479424] Epoch[3] Batch[0] avg_epoch_loss=5.136042\u001b[0m\n\u001b[31m[07/17/2019 06:39:09 INFO 139964490479424] #quality_metric: host=algo-1, epoch=3, batch=0 train loss <loss>=5.13604211807\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] Epoch[3] Batch[5] avg_epoch_loss=5.160289\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] #quality_metric: host=algo-1, epoch=3, batch=5 train loss <loss>=5.16028881073\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] Epoch[3] Batch [5]#011Speed: 259.54 samples/sec#011loss=5.160289\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] processed a total of 339 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1709.0890407562256, \"sum\": 1709.0890407562256, \"min\": 1709.0890407562256}}, \"EndTime\": 1563345550.32447, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345548.614957}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=198.338165912 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] #progress_metric: host=algo-1, completed 1 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] #quality_metric: host=algo-1, epoch=3, train loss <loss>=5.16028881073\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_73c00610-cd84-4203-8457-03f9cef77997-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 65.78898429870605, \"sum\": 65.78898429870605, \"min\": 65.78898429870605}}, \"EndTime\": 1563345550.39085, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345550.324547}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] Epoch[4] Batch[0] avg_epoch_loss=5.071866\u001b[0m\n\u001b[31m[07/17/2019 06:39:10 INFO 139964490479424] #quality_metric: host=algo-1, epoch=4, batch=0 train loss <loss>=5.07186555862\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] Epoch[4] Batch[5] avg_epoch_loss=5.042942\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] #quality_metric: host=algo-1, epoch=4, batch=5 train loss <loss>=5.0429418087\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] Epoch[4] Batch [5]#011Speed: 345.52 samples/sec#011loss=5.042942\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] processed a total of 330 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1329.5469284057617, \"sum\": 1329.5469284057617, \"min\": 1329.5469284057617}}, \"EndTime\": 1563345551.720538, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345550.390924}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=248.184113653 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] #progress_metric: host=algo-1, completed 1 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] #quality_metric: host=algo-1, epoch=4, train loss <loss>=5.0429418087\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:11 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_7fbb70c0-ac10-4f93-aaab-660ce5423021-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 59.6461296081543, \"sum\": 59.6461296081543, \"min\": 59.6461296081543}}, \"EndTime\": 1563345551.780765, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345551.720613}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:12 INFO 139964490479424] Epoch[5] Batch[0] avg_epoch_loss=4.971841\u001b[0m\n\u001b[31m[07/17/2019 06:39:12 INFO 139964490479424] #quality_metric: host=algo-1, epoch=5, batch=0 train loss <loss>=4.9718413353\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] Epoch[5] Batch[5] avg_epoch_loss=5.113073\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] #quality_metric: host=algo-1, epoch=5, batch=5 train loss <loss>=5.11307279269\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] Epoch[5] Batch [5]#011Speed: 339.23 samples/sec#011loss=5.113073\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] processed a total of 339 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1340.6801223754883, \"sum\": 1340.6801223754883, \"min\": 1340.6801223754883}}, \"EndTime\": 1563345553.121562, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345551.780826}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=252.836367975 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] #progress_metric: host=algo-1, completed 1 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] #quality_metric: host=algo-1, epoch=5, train loss <loss>=5.11307279269\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] Epoch[6] Batch[0] avg_epoch_loss=5.073530\u001b[0m\n\u001b[31m[07/17/2019 06:39:13 INFO 139964490479424] #quality_metric: host=algo-1, epoch=6, batch=0 train loss <loss>=5.07353019714\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] processed a total of 319 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1180.3858280181885, \"sum\": 1180.3858280181885, \"min\": 1180.3858280181885}}, \"EndTime\": 1563345554.302444, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345553.121637}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=270.22381926 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] #progress_metric: host=algo-1, completed 1 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] #quality_metric: host=algo-1, epoch=6, train loss <loss>=5.02971372604\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_a8605914-674a-4380-a483-aa3248149983-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 71.80905342102051, \"sum\": 71.80905342102051, \"min\": 71.80905342102051}}, \"EndTime\": 1563345554.37486, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345554.302526}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] Epoch[7] Batch[0] avg_epoch_loss=5.069306\u001b[0m\n\u001b[31m[07/17/2019 06:39:14 INFO 139964490479424] #quality_metric: host=algo-1, epoch=7, batch=0 train loss <loss>=5.06930589676\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] Epoch[7] Batch[5] avg_epoch_loss=4.725332\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] #quality_metric: host=algo-1, epoch=7, batch=5 train loss <loss>=4.72533202171\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] Epoch[7] Batch [5]#011Speed: 336.62 samples/sec#011loss=4.725332\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] processed a total of 375 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1357.6209545135498, \"sum\": 1357.6209545135498, \"min\": 1357.6209545135498}}, \"EndTime\": 1563345555.732612, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345554.374926}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=276.194861172 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] #progress_metric: host=algo-1, completed 2 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] #quality_metric: host=algo-1, epoch=7, train loss <loss>=4.72533202171\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:15 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_9263601c-b702-4cc0-846c-57beecd9dcf9-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 64.11099433898926, \"sum\": 64.11099433898926, \"min\": 64.11099433898926}}, \"EndTime\": 1563345555.797326, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345555.732692}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:16 INFO 139964490479424] Epoch[8] Batch[0] avg_epoch_loss=4.795703\u001b[0m\n\u001b[31m[07/17/2019 06:39:16 INFO 139964490479424] #quality_metric: host=algo-1, epoch=8, batch=0 train loss <loss>=4.79570293427\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] Epoch[8] Batch[5] avg_epoch_loss=4.732907\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] #quality_metric: host=algo-1, epoch=8, batch=5 train loss <loss>=4.73290721575\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] Epoch[8] Batch [5]#011Speed: 344.87 samples/sec#011loss=4.732907\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] processed a total of 369 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1331.8541049957275, \"sum\": 1331.8541049957275, \"min\": 1331.8541049957275}}, \"EndTime\": 1563345557.129308, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345555.797395}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=277.033717712 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] #progress_metric: host=algo-1, completed 2 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] #quality_metric: host=algo-1, epoch=8, train loss <loss>=4.73290721575\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] Epoch[9] Batch[0] avg_epoch_loss=4.614312\u001b[0m\n\u001b[31m[07/17/2019 06:39:17 INFO 139964490479424] #quality_metric: host=algo-1, epoch=9, batch=0 train loss <loss>=4.61431217194\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] Epoch[9] Batch[5] avg_epoch_loss=4.686977\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] #quality_metric: host=algo-1, epoch=9, batch=5 train loss <loss>=4.68697675069\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] Epoch[9] Batch [5]#011Speed: 343.74 samples/sec#011loss=4.686977\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] processed a total of 355 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1344.3231582641602, \"sum\": 1344.3231582641602, \"min\": 1344.3231582641602}}, \"EndTime\": 1563345558.474201, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345557.129386}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=264.051258979 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] #progress_metric: host=algo-1, completed 2 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] #quality_metric: host=algo-1, epoch=9, train loss <loss>=4.68697675069\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_0411f245-80ca-47d6-b0d8-0d2f6b6dff36-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 64.1179084777832, \"sum\": 64.1179084777832, \"min\": 64.1179084777832}}, \"EndTime\": 1563345558.538889, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345558.474279}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] Epoch[10] Batch[0] avg_epoch_loss=4.568539\u001b[0m\n\u001b[31m[07/17/2019 06:39:18 INFO 139964490479424] #quality_metric: host=algo-1, epoch=10, batch=0 train loss <loss>=4.56853866577\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] Epoch[10] Batch[5] avg_epoch_loss=4.630606\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] #quality_metric: host=algo-1, epoch=10, batch=5 train loss <loss>=4.63060585658\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] Epoch[10] Batch [5]#011Speed: 333.59 samples/sec#011loss=4.630606\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] processed a total of 380 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1357.252836227417, \"sum\": 1357.252836227417, \"min\": 1357.252836227417}}, \"EndTime\": 1563345559.896279, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345558.538961}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=279.955181471 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] #progress_metric: host=algo-1, completed 2 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] #quality_metric: host=algo-1, epoch=10, train loss <loss>=4.63060585658\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:19 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_8b9422d1-5391-4697-89e2-0161de2557c4-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 89.18404579162598, \"sum\": 89.18404579162598, \"min\": 89.18404579162598}}, \"EndTime\": 1563345559.986047, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345559.896351}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:20 INFO 139964490479424] Epoch[11] Batch[0] avg_epoch_loss=4.659149\u001b[0m\n\u001b[31m[07/17/2019 06:39:20 INFO 139964490479424] #quality_metric: host=algo-1, epoch=11, batch=0 train loss <loss>=4.65914916992\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] Epoch[11] Batch[5] avg_epoch_loss=4.591857\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] #quality_metric: host=algo-1, epoch=11, batch=5 train loss <loss>=4.59185663859\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] Epoch[11] Batch [5]#011Speed: 339.65 samples/sec#011loss=4.591857\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] processed a total of 367 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1333.5058689117432, \"sum\": 1333.5058689117432, \"min\": 1333.5058689117432}}, \"EndTime\": 1563345561.319685, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345559.986122}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=275.191065222 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] #progress_metric: host=algo-1, completed 3 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] #quality_metric: host=algo-1, epoch=11, train loss <loss>=4.59185663859\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_70e25e2f-14b2-4af9-8477-f32169b14c6e-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 55.69911003112793, \"sum\": 55.69911003112793, \"min\": 55.69911003112793}}, \"EndTime\": 1563345561.375979, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345561.319762}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] Epoch[12] Batch[0] avg_epoch_loss=4.415570\u001b[0m\n\u001b[31m[07/17/2019 06:39:21 INFO 139964490479424] #quality_metric: host=algo-1, epoch=12, batch=0 train loss <loss>=4.41557025909\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] Epoch[12] Batch[5] avg_epoch_loss=4.674816\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] #quality_metric: host=algo-1, epoch=12, batch=5 train loss <loss>=4.67481605212\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] Epoch[12] Batch [5]#011Speed: 336.55 samples/sec#011loss=4.674816\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] processed a total of 333 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1334.2180252075195, \"sum\": 1334.2180252075195, \"min\": 1334.2180252075195}}, \"EndTime\": 1563345562.710338, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345561.376053}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=249.564015693 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] #progress_metric: host=algo-1, completed 3 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] #quality_metric: host=algo-1, epoch=12, train loss <loss>=4.67481605212\u001b[0m\n\u001b[31m[07/17/2019 06:39:22 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:23 INFO 139964490479424] Epoch[13] Batch[0] avg_epoch_loss=4.167605\u001b[0m\n\u001b[31m[07/17/2019 06:39:23 INFO 139964490479424] #quality_metric: host=algo-1, epoch=13, batch=0 train loss <loss>=4.16760492325\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] Epoch[13] Batch[5] avg_epoch_loss=4.560476\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] #quality_metric: host=algo-1, epoch=13, batch=5 train loss <loss>=4.56047638257\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] Epoch[13] Batch [5]#011Speed: 342.14 samples/sec#011loss=4.560476\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] processed a total of 383 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1328.2301425933838, \"sum\": 1328.2301425933838, \"min\": 1328.2301425933838}}, \"EndTime\": 1563345564.039088, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345562.710412}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=288.330249219 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] #progress_metric: host=algo-1, completed 3 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] #quality_metric: host=algo-1, epoch=13, train loss <loss>=4.56047638257\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_df7ec573-3677-4475-bd8b-29b67a4912a9-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 63.51304054260254, \"sum\": 63.51304054260254, \"min\": 63.51304054260254}}, \"EndTime\": 1563345564.103134, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345564.039161}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] Epoch[14] Batch[0] avg_epoch_loss=4.394165\u001b[0m\n\u001b[31m[07/17/2019 06:39:24 INFO 139964490479424] #quality_metric: host=algo-1, epoch=14, batch=0 train loss <loss>=4.39416503906\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] Epoch[14] Batch[5] avg_epoch_loss=4.396919\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] #quality_metric: host=algo-1, epoch=14, batch=5 train loss <loss>=4.39691865444\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] Epoch[14] Batch [5]#011Speed: 337.26 samples/sec#011loss=4.396919\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] processed a total of 364 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1351.4599800109863, \"sum\": 1351.4599800109863, \"min\": 1351.4599800109863}}, \"EndTime\": 1563345565.454722, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345564.103206}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=269.317396485 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] #progress_metric: host=algo-1, completed 3 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] #quality_metric: host=algo-1, epoch=14, train loss <loss>=4.39691865444\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_e4588199-064a-4224-8cf6-a7c95b5487dd-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 55.62615394592285, \"sum\": 55.62615394592285, \"min\": 55.62615394592285}}, \"EndTime\": 1563345565.510907, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345565.454791}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] Epoch[15] Batch[0] avg_epoch_loss=4.470998\u001b[0m\n\u001b[31m[07/17/2019 06:39:25 INFO 139964490479424] #quality_metric: host=algo-1, epoch=15, batch=0 train loss <loss>=4.47099781036\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] Epoch[15] Batch[5] avg_epoch_loss=4.421463\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] #quality_metric: host=algo-1, epoch=15, batch=5 train loss <loss>=4.42146277428\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] Epoch[15] Batch [5]#011Speed: 344.70 samples/sec#011loss=4.421463\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] processed a total of 379 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1319.8580741882324, \"sum\": 1319.8580741882324, \"min\": 1319.8580741882324}}, \"EndTime\": 1563345566.830905, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345565.51098}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=287.127086928 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] #progress_metric: host=algo-1, completed 4 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] #quality_metric: host=algo-1, epoch=15, train loss <loss>=4.42146277428\u001b[0m\n\u001b[31m[07/17/2019 06:39:26 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:27 INFO 139964490479424] Epoch[16] Batch[0] avg_epoch_loss=4.100180\u001b[0m\n\u001b[31m[07/17/2019 06:39:27 INFO 139964490479424] #quality_metric: host=algo-1, epoch=16, batch=0 train loss <loss>=4.10017967224\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] Epoch[16] Batch[5] avg_epoch_loss=4.491715\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] #quality_metric: host=algo-1, epoch=16, batch=5 train loss <loss>=4.49171471596\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] Epoch[16] Batch [5]#011Speed: 337.73 samples/sec#011loss=4.491715\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] processed a total of 338 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1346.7578887939453, \"sum\": 1346.7578887939453, \"min\": 1346.7578887939453}}, \"EndTime\": 1563345568.178173, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345566.830983}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=250.95368382 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] #progress_metric: host=algo-1, completed 4 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] #quality_metric: host=algo-1, epoch=16, train loss <loss>=4.49171471596\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] Epoch[17] Batch[0] avg_epoch_loss=4.181131\u001b[0m\n\u001b[31m[07/17/2019 06:39:28 INFO 139964490479424] #quality_metric: host=algo-1, epoch=17, batch=0 train loss <loss>=4.18113088608\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] Epoch[17] Batch[5] avg_epoch_loss=4.313446\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] #quality_metric: host=algo-1, epoch=17, batch=5 train loss <loss>=4.3134458065\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] Epoch[17] Batch [5]#011Speed: 334.40 samples/sec#011loss=4.313446\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] processed a total of 337 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1340.8808708190918, \"sum\": 1340.8808708190918, \"min\": 1340.8808708190918}}, \"EndTime\": 1563345569.519613, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345568.178241}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=251.306142942 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] #progress_metric: host=algo-1, completed 4 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] #quality_metric: host=algo-1, epoch=17, train loss <loss>=4.3134458065\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_be12fa2c-cbb9-40e5-a5f0-dbc3a0a1b7bd-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 63.91000747680664, \"sum\": 63.91000747680664, \"min\": 63.91000747680664}}, \"EndTime\": 1563345569.584077, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345569.519691}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] Epoch[18] Batch[0] avg_epoch_loss=4.307560\u001b[0m\n\u001b[31m[07/17/2019 06:39:29 INFO 139964490479424] #quality_metric: host=algo-1, epoch=18, batch=0 train loss <loss>=4.30755996704\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] Epoch[18] Batch[5] avg_epoch_loss=4.231593\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] #quality_metric: host=algo-1, epoch=18, batch=5 train loss <loss>=4.23159313202\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] Epoch[18] Batch [5]#011Speed: 338.35 samples/sec#011loss=4.231593\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] processed a total of 354 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1344.9559211730957, \"sum\": 1344.9559211730957, \"min\": 1344.9559211730957}}, \"EndTime\": 1563345570.929169, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345569.584146}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=263.182744749 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] #progress_metric: host=algo-1, completed 4 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] #quality_metric: host=algo-1, epoch=18, train loss <loss>=4.23159313202\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:30 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_a96f626e-ae3b-4201-b96a-8711a2876f63-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 59.15093421936035, \"sum\": 59.15093421936035, \"min\": 59.15093421936035}}, \"EndTime\": 1563345570.988893, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345570.929247}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:31 INFO 139964490479424] Epoch[19] Batch[0] avg_epoch_loss=4.437295\u001b[0m\n\u001b[31m[07/17/2019 06:39:31 INFO 139964490479424] #quality_metric: host=algo-1, epoch=19, batch=0 train loss <loss>=4.43729496002\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] Epoch[19] Batch[5] avg_epoch_loss=4.562665\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] #quality_metric: host=algo-1, epoch=19, batch=5 train loss <loss>=4.56266498566\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] Epoch[19] Batch [5]#011Speed: 342.79 samples/sec#011loss=4.562665\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] processed a total of 359 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1318.6931610107422, \"sum\": 1318.6931610107422, \"min\": 1318.6931610107422}}, \"EndTime\": 1563345572.307749, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345570.988989}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=272.215105876 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] #progress_metric: host=algo-1, completed 5 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] #quality_metric: host=algo-1, epoch=19, train loss <loss>=4.56266498566\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] Epoch[20] Batch[0] avg_epoch_loss=4.528052\u001b[0m\n\u001b[31m[07/17/2019 06:39:32 INFO 139964490479424] #quality_metric: host=algo-1, epoch=20, batch=0 train loss <loss>=4.52805185318\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] Epoch[20] Batch[5] avg_epoch_loss=4.304532\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] #quality_metric: host=algo-1, epoch=20, batch=5 train loss <loss>=4.30453228951\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] Epoch[20] Batch [5]#011Speed: 339.14 samples/sec#011loss=4.304532\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] processed a total of 331 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1363.713026046753, \"sum\": 1363.713026046753, \"min\": 1363.713026046753}}, \"EndTime\": 1563345573.671966, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345572.307827}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=242.70118769 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] #progress_metric: host=algo-1, completed 5 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] #quality_metric: host=algo-1, epoch=20, train loss <loss>=4.30453228951\u001b[0m\n\u001b[31m[07/17/2019 06:39:33 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:34 INFO 139964490479424] Epoch[21] Batch[0] avg_epoch_loss=4.366638\u001b[0m\n\u001b[31m[07/17/2019 06:39:34 INFO 139964490479424] #quality_metric: host=algo-1, epoch=21, batch=0 train loss <loss>=4.36663818359\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] Epoch[21] Batch[5] avg_epoch_loss=4.327943\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] #quality_metric: host=algo-1, epoch=21, batch=5 train loss <loss>=4.32794276873\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] Epoch[21] Batch [5]#011Speed: 331.89 samples/sec#011loss=4.327943\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] processed a total of 336 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1351.9010543823242, \"sum\": 1351.9010543823242, \"min\": 1351.9010543823242}}, \"EndTime\": 1563345575.024383, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345573.672032}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=248.517774142 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] #progress_metric: host=algo-1, completed 5 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] #quality_metric: host=algo-1, epoch=21, train loss <loss>=4.32794276873\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] Epoch[22] Batch[0] avg_epoch_loss=4.162799\u001b[0m\n\u001b[31m[07/17/2019 06:39:35 INFO 139964490479424] #quality_metric: host=algo-1, epoch=22, batch=0 train loss <loss>=4.16279888153\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] Epoch[22] Batch[5] avg_epoch_loss=4.310891\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] #quality_metric: host=algo-1, epoch=22, batch=5 train loss <loss>=4.31089091301\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] Epoch[22] Batch [5]#011Speed: 335.52 samples/sec#011loss=4.310891\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] processed a total of 378 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1344.7041511535645, \"sum\": 1344.7041511535645, \"min\": 1344.7041511535645}}, \"EndTime\": 1563345576.369618, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345575.02446}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=281.079151738 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] #progress_metric: host=algo-1, completed 5 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] #quality_metric: host=algo-1, epoch=22, train loss <loss>=4.31089091301\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] Epoch[23] Batch[0] avg_epoch_loss=4.133379\u001b[0m\n\u001b[31m[07/17/2019 06:39:36 INFO 139964490479424] #quality_metric: host=algo-1, epoch=23, batch=0 train loss <loss>=4.13337945938\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] Epoch[23] Batch[5] avg_epoch_loss=4.319111\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] #quality_metric: host=algo-1, epoch=23, batch=5 train loss <loss>=4.31911142667\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] Epoch[23] Batch [5]#011Speed: 340.19 samples/sec#011loss=4.319111\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] processed a total of 351 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1356.73189163208, \"sum\": 1356.73189163208, \"min\": 1356.73189163208}}, \"EndTime\": 1563345577.726886, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345576.369696}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=258.688554733 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] #progress_metric: host=algo-1, completed 6 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] #quality_metric: host=algo-1, epoch=23, train loss <loss>=4.31911142667\u001b[0m\n\u001b[31m[07/17/2019 06:39:37 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:38 INFO 139964490479424] Epoch[24] Batch[0] avg_epoch_loss=4.096683\u001b[0m\n\u001b[31m[07/17/2019 06:39:38 INFO 139964490479424] #quality_metric: host=algo-1, epoch=24, batch=0 train loss <loss>=4.09668254852\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] Epoch[24] Batch[5] avg_epoch_loss=4.245122\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] #quality_metric: host=algo-1, epoch=24, batch=5 train loss <loss>=4.24512179693\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] Epoch[24] Batch [5]#011Speed: 338.28 samples/sec#011loss=4.245122\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] processed a total of 370 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1345.11399269104, \"sum\": 1345.11399269104, \"min\": 1345.11399269104}}, \"EndTime\": 1563345579.072517, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345577.726964}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=275.046710669 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] #progress_metric: host=algo-1, completed 6 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] #quality_metric: host=algo-1, epoch=24, train loss <loss>=4.24512179693\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] Epoch[25] Batch[0] avg_epoch_loss=4.374493\u001b[0m\n\u001b[31m[07/17/2019 06:39:39 INFO 139964490479424] #quality_metric: host=algo-1, epoch=25, batch=0 train loss <loss>=4.37449264526\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] Epoch[25] Batch[5] avg_epoch_loss=4.329658\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] #quality_metric: host=algo-1, epoch=25, batch=5 train loss <loss>=4.32965779305\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] Epoch[25] Batch [5]#011Speed: 334.56 samples/sec#011loss=4.329658\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] processed a total of 340 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1362.480878829956, \"sum\": 1362.480878829956, \"min\": 1362.480878829956}}, \"EndTime\": 1563345580.435589, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345579.072595}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=249.524264457 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] #progress_metric: host=algo-1, completed 6 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] #quality_metric: host=algo-1, epoch=25, train loss <loss>=4.32965779305\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] Epoch[26] Batch[0] avg_epoch_loss=4.326764\u001b[0m\n\u001b[31m[07/17/2019 06:39:40 INFO 139964490479424] #quality_metric: host=algo-1, epoch=26, batch=0 train loss <loss>=4.32676362991\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] Epoch[26] Batch[5] avg_epoch_loss=4.389358\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] #quality_metric: host=algo-1, epoch=26, batch=5 train loss <loss>=4.38935828209\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] Epoch[26] Batch [5]#011Speed: 343.94 samples/sec#011loss=4.389358\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] processed a total of 356 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1370.8579540252686, \"sum\": 1370.8579540252686, \"min\": 1370.8579540252686}}, \"EndTime\": 1563345581.80696, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345580.435666}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=259.670158506 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] #progress_metric: host=algo-1, completed 6 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] #quality_metric: host=algo-1, epoch=26, train loss <loss>=4.38935828209\u001b[0m\n\u001b[31m[07/17/2019 06:39:41 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:42 INFO 139964490479424] Epoch[27] Batch[0] avg_epoch_loss=4.395689\u001b[0m\n\u001b[31m[07/17/2019 06:39:42 INFO 139964490479424] #quality_metric: host=algo-1, epoch=27, batch=0 train loss <loss>=4.39568853378\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] Epoch[27] Batch[5] avg_epoch_loss=4.177542\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] #quality_metric: host=algo-1, epoch=27, batch=5 train loss <loss>=4.17754181226\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] Epoch[27] Batch [5]#011Speed: 340.64 samples/sec#011loss=4.177542\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] processed a total of 343 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1336.9219303131104, \"sum\": 1336.9219303131104, \"min\": 1336.9219303131104}}, \"EndTime\": 1563345583.144378, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345581.807037}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=256.537432379 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] #progress_metric: host=algo-1, completed 7 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] #quality_metric: host=algo-1, epoch=27, train loss <loss>=4.17754181226\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_4c923526-58a4-42de-b8c7-a5056595d331-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 55.10592460632324, \"sum\": 55.10592460632324, \"min\": 55.10592460632324}}, \"EndTime\": 1563345583.20008, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345583.14446}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] Epoch[28] Batch[0] avg_epoch_loss=4.253472\u001b[0m\n\u001b[31m[07/17/2019 06:39:43 INFO 139964490479424] #quality_metric: host=algo-1, epoch=28, batch=0 train loss <loss>=4.25347232819\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] Epoch[28] Batch[5] avg_epoch_loss=3.918867\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] #quality_metric: host=algo-1, epoch=28, batch=5 train loss <loss>=3.91886734962\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] Epoch[28] Batch [5]#011Speed: 339.67 samples/sec#011loss=3.918867\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] processed a total of 336 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1326.3969421386719, \"sum\": 1326.3969421386719, \"min\": 1326.3969421386719}}, \"EndTime\": 1563345584.526591, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345583.200146}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=253.298164689 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] #progress_metric: host=algo-1, completed 7 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] #quality_metric: host=algo-1, epoch=28, train loss <loss>=3.91886734962\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_726beb3b-039b-46b7-ab67-4590feb8beb1-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 65.1559829711914, \"sum\": 65.1559829711914, \"min\": 65.1559829711914}}, \"EndTime\": 1563345584.592335, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345584.526666}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] Epoch[29] Batch[0] avg_epoch_loss=4.018377\u001b[0m\n\u001b[31m[07/17/2019 06:39:44 INFO 139964490479424] #quality_metric: host=algo-1, epoch=29, batch=0 train loss <loss>=4.01837730408\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] Epoch[29] Batch[5] avg_epoch_loss=4.119975\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] #quality_metric: host=algo-1, epoch=29, batch=5 train loss <loss>=4.11997481187\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] Epoch[29] Batch [5]#011Speed: 337.64 samples/sec#011loss=4.119975\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] processed a total of 325 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1349.6339321136475, \"sum\": 1349.6339321136475, \"min\": 1349.6339321136475}}, \"EndTime\": 1563345585.942105, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345584.592404}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=240.784089707 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] #progress_metric: host=algo-1, completed 7 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] #quality_metric: host=algo-1, epoch=29, train loss <loss>=4.11997481187\u001b[0m\n\u001b[31m[07/17/2019 06:39:45 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:46 INFO 139964490479424] Epoch[30] Batch[0] avg_epoch_loss=4.360940\u001b[0m\n\u001b[31m[07/17/2019 06:39:46 INFO 139964490479424] #quality_metric: host=algo-1, epoch=30, batch=0 train loss <loss>=4.36094045639\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] Epoch[30] Batch[5] avg_epoch_loss=4.041303\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] #quality_metric: host=algo-1, epoch=30, batch=5 train loss <loss>=4.04130307833\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] Epoch[30] Batch [5]#011Speed: 343.55 samples/sec#011loss=4.041303\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] processed a total of 362 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1340.2509689331055, \"sum\": 1340.2509689331055, \"min\": 1340.2509689331055}}, \"EndTime\": 1563345587.282872, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345585.942191}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=270.076470477 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] #progress_metric: host=algo-1, completed 7 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] #quality_metric: host=algo-1, epoch=30, train loss <loss>=4.04130307833\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] Epoch[31] Batch[0] avg_epoch_loss=4.274628\u001b[0m\n\u001b[31m[07/17/2019 06:39:47 INFO 139964490479424] #quality_metric: host=algo-1, epoch=31, batch=0 train loss <loss>=4.27462768555\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] Epoch[31] Batch[5] avg_epoch_loss=4.238626\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] #quality_metric: host=algo-1, epoch=31, batch=5 train loss <loss>=4.2386256059\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] Epoch[31] Batch [5]#011Speed: 344.00 samples/sec#011loss=4.238626\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] processed a total of 351 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1330.9760093688965, \"sum\": 1330.9760093688965, \"min\": 1330.9760093688965}}, \"EndTime\": 1563345588.614338, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345587.282948}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=263.693858777 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] #progress_metric: host=algo-1, completed 8 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] #quality_metric: host=algo-1, epoch=31, train loss <loss>=4.2386256059\u001b[0m\n\u001b[31m[07/17/2019 06:39:48 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] Epoch[32] Batch[0] avg_epoch_loss=4.238575\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] #quality_metric: host=algo-1, epoch=32, batch=0 train loss <loss>=4.23857498169\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] Epoch[32] Batch[5] avg_epoch_loss=4.295443\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] #quality_metric: host=algo-1, epoch=32, batch=5 train loss <loss>=4.29544305801\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] Epoch[32] Batch [5]#011Speed: 334.12 samples/sec#011loss=4.295443\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] processed a total of 357 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1368.5579299926758, \"sum\": 1368.5579299926758, \"min\": 1368.5579299926758}}, \"EndTime\": 1563345589.983412, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345588.614417}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=260.838301548 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] #progress_metric: host=algo-1, completed 8 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] #quality_metric: host=algo-1, epoch=32, train loss <loss>=4.29544305801\u001b[0m\n\u001b[31m[07/17/2019 06:39:49 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:50 INFO 139964490479424] Epoch[33] Batch[0] avg_epoch_loss=4.151871\u001b[0m\n\u001b[31m[07/17/2019 06:39:50 INFO 139964490479424] #quality_metric: host=algo-1, epoch=33, batch=0 train loss <loss>=4.15187120438\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] Epoch[33] Batch[5] avg_epoch_loss=4.018318\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] #quality_metric: host=algo-1, epoch=33, batch=5 train loss <loss>=4.01831789811\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] Epoch[33] Batch [5]#011Speed: 340.34 samples/sec#011loss=4.018318\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] processed a total of 349 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1359.0788841247559, \"sum\": 1359.0788841247559, \"min\": 1359.0788841247559}}, \"EndTime\": 1563345591.342997, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345589.983484}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=256.770579015 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] #progress_metric: host=algo-1, completed 8 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] #quality_metric: host=algo-1, epoch=33, train loss <loss>=4.01831789811\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] Epoch[34] Batch[0] avg_epoch_loss=4.165394\u001b[0m\n\u001b[31m[07/17/2019 06:39:51 INFO 139964490479424] #quality_metric: host=algo-1, epoch=34, batch=0 train loss <loss>=4.16539382935\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] Epoch[34] Batch[5] avg_epoch_loss=4.038558\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] #quality_metric: host=algo-1, epoch=34, batch=5 train loss <loss>=4.03855776787\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] Epoch[34] Batch [5]#011Speed: 329.03 samples/sec#011loss=4.038558\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] processed a total of 347 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1374.7379779815674, \"sum\": 1374.7379779815674, \"min\": 1374.7379779815674}}, \"EndTime\": 1563345592.718241, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345591.343075}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=252.391338646 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] #progress_metric: host=algo-1, completed 8 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] #quality_metric: host=algo-1, epoch=34, train loss <loss>=4.03855776787\u001b[0m\n\u001b[31m[07/17/2019 06:39:52 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:53 INFO 139964490479424] Epoch[35] Batch[0] avg_epoch_loss=3.923457\u001b[0m\n\u001b[31m[07/17/2019 06:39:53 INFO 139964490479424] #quality_metric: host=algo-1, epoch=35, batch=0 train loss <loss>=3.92345666885\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] Epoch[35] Batch[5] avg_epoch_loss=4.210080\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] #quality_metric: host=algo-1, epoch=35, batch=5 train loss <loss>=4.21008042494\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] Epoch[35] Batch [5]#011Speed: 336.44 samples/sec#011loss=4.210080\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] processed a total of 322 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1340.0988578796387, \"sum\": 1340.0988578796387, \"min\": 1340.0988578796387}}, \"EndTime\": 1563345594.058905, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345592.718315}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=240.259622146 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] #progress_metric: host=algo-1, completed 9 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] #quality_metric: host=algo-1, epoch=35, train loss <loss>=4.21008042494\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] Epoch[36] Batch[0] avg_epoch_loss=4.293719\u001b[0m\n\u001b[31m[07/17/2019 06:39:54 INFO 139964490479424] #quality_metric: host=algo-1, epoch=36, batch=0 train loss <loss>=4.29371881485\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] Epoch[36] Batch[5] avg_epoch_loss=4.100294\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] #quality_metric: host=algo-1, epoch=36, batch=5 train loss <loss>=4.10029375553\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] Epoch[36] Batch [5]#011Speed: 329.03 samples/sec#011loss=4.100294\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] processed a total of 336 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1364.342212677002, \"sum\": 1364.342212677002, \"min\": 1364.342212677002}}, \"EndTime\": 1563345595.42379, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345594.058984}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=246.251610008 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] #progress_metric: host=algo-1, completed 9 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] #quality_metric: host=algo-1, epoch=36, train loss <loss>=4.10029375553\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] Epoch[37] Batch[0] avg_epoch_loss=4.377960\u001b[0m\n\u001b[31m[07/17/2019 06:39:55 INFO 139964490479424] #quality_metric: host=algo-1, epoch=37, batch=0 train loss <loss>=4.37796020508\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] Epoch[37] Batch[5] avg_epoch_loss=4.148266\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] #quality_metric: host=algo-1, epoch=37, batch=5 train loss <loss>=4.14826615651\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] Epoch[37] Batch [5]#011Speed: 335.26 samples/sec#011loss=4.148266\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] processed a total of 355 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1347.426176071167, \"sum\": 1347.426176071167, \"min\": 1347.426176071167}}, \"EndTime\": 1563345596.77177, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345595.423874}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=263.443593219 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] #progress_metric: host=algo-1, completed 9 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] #quality_metric: host=algo-1, epoch=37, train loss <loss>=4.14826615651\u001b[0m\n\u001b[31m[07/17/2019 06:39:56 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:57 INFO 139964490479424] Epoch[38] Batch[0] avg_epoch_loss=4.190225\u001b[0m\n\u001b[31m[07/17/2019 06:39:57 INFO 139964490479424] #quality_metric: host=algo-1, epoch=38, batch=0 train loss <loss>=4.19022512436\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] Epoch[38] Batch[5] avg_epoch_loss=4.080905\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] #quality_metric: host=algo-1, epoch=38, batch=5 train loss <loss>=4.08090488116\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] Epoch[38] Batch [5]#011Speed: 343.68 samples/sec#011loss=4.080905\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] processed a total of 324 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1326.8139362335205, \"sum\": 1326.8139362335205, \"min\": 1326.8139362335205}}, \"EndTime\": 1563345598.099089, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345596.771846}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=244.173551939 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] #progress_metric: host=algo-1, completed 9 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] #quality_metric: host=algo-1, epoch=38, train loss <loss>=4.08090488116\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] Epoch[39] Batch[0] avg_epoch_loss=4.065939\u001b[0m\n\u001b[31m[07/17/2019 06:39:58 INFO 139964490479424] #quality_metric: host=algo-1, epoch=39, batch=0 train loss <loss>=4.06593942642\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] Epoch[39] Batch[5] avg_epoch_loss=4.186403\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] #quality_metric: host=algo-1, epoch=39, batch=5 train loss <loss>=4.18640303612\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] Epoch[39] Batch [5]#011Speed: 337.98 samples/sec#011loss=4.186403\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] processed a total of 336 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1343.0650234222412, \"sum\": 1343.0650234222412, \"min\": 1343.0650234222412}}, \"EndTime\": 1563345599.442664, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345598.099167}\n\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=250.156186115 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] #progress_metric: host=algo-1, completed 10 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] #quality_metric: host=algo-1, epoch=39, train loss <loss>=4.18640303612\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] Epoch[40] Batch[0] avg_epoch_loss=4.191908\u001b[0m\n\u001b[31m[07/17/2019 06:39:59 INFO 139964490479424] #quality_metric: host=algo-1, epoch=40, batch=0 train loss <loss>=4.19190788269\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] Epoch[40] Batch[5] avg_epoch_loss=4.087716\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] #quality_metric: host=algo-1, epoch=40, batch=5 train loss <loss>=4.08771649996\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] Epoch[40] Batch [5]#011Speed: 331.17 samples/sec#011loss=4.087716\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] processed a total of 370 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1376.7621517181396, \"sum\": 1376.7621517181396, \"min\": 1376.7621517181396}}, \"EndTime\": 1563345600.819951, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345599.442728}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=268.724667655 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] #progress_metric: host=algo-1, completed 10 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] #quality_metric: host=algo-1, epoch=40, train loss <loss>=4.08771649996\u001b[0m\n\u001b[31m[07/17/2019 06:40:00 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:01 INFO 139964490479424] Epoch[41] Batch[0] avg_epoch_loss=4.169252\u001b[0m\n\u001b[31m[07/17/2019 06:40:01 INFO 139964490479424] #quality_metric: host=algo-1, epoch=41, batch=0 train loss <loss>=4.16925239563\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] Epoch[41] Batch[5] avg_epoch_loss=4.166015\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] #quality_metric: host=algo-1, epoch=41, batch=5 train loss <loss>=4.16601471106\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] Epoch[41] Batch [5]#011Speed: 328.10 samples/sec#011loss=4.166015\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] processed a total of 353 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1362.3230457305908, \"sum\": 1362.3230457305908, \"min\": 1362.3230457305908}}, \"EndTime\": 1563345602.182792, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345600.820023}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=259.092461468 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] #progress_metric: host=algo-1, completed 10 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] #quality_metric: host=algo-1, epoch=41, train loss <loss>=4.16601471106\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] Epoch[42] Batch[0] avg_epoch_loss=4.061618\u001b[0m\n\u001b[31m[07/17/2019 06:40:02 INFO 139964490479424] #quality_metric: host=algo-1, epoch=42, batch=0 train loss <loss>=4.06161832809\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] Epoch[42] Batch[5] avg_epoch_loss=4.059570\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] #quality_metric: host=algo-1, epoch=42, batch=5 train loss <loss>=4.05956979593\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] Epoch[42] Batch [5]#011Speed: 340.98 samples/sec#011loss=4.059570\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] processed a total of 375 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1331.5250873565674, \"sum\": 1331.5250873565674, \"min\": 1331.5250873565674}}, \"EndTime\": 1563345603.514859, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345602.182877}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=281.61248097 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] #progress_metric: host=algo-1, completed 10 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] #quality_metric: host=algo-1, epoch=42, train loss <loss>=4.05956979593\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] Epoch[43] Batch[0] avg_epoch_loss=3.884916\u001b[0m\n\u001b[31m[07/17/2019 06:40:03 INFO 139964490479424] #quality_metric: host=algo-1, epoch=43, batch=0 train loss <loss>=3.88491630554\u001b[0m\n\u001b[31m[07/17/2019 06:40:04 INFO 139964490479424] processed a total of 317 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1134.6409320831299, \"sum\": 1134.6409320831299, \"min\": 1134.6409320831299}}, \"EndTime\": 1563345604.65002, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345603.514922}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:04 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=279.358415887 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:04 INFO 139964490479424] #progress_metric: host=algo-1, completed 11 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:04 INFO 139964490479424] #quality_metric: host=algo-1, epoch=43, train loss <loss>=4.11265816689\u001b[0m\n\u001b[31m[07/17/2019 06:40:04 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:05 INFO 139964490479424] Epoch[44] Batch[0] avg_epoch_loss=4.116920\u001b[0m\n\u001b[31m[07/17/2019 06:40:05 INFO 139964490479424] #quality_metric: host=algo-1, epoch=44, batch=0 train loss <loss>=4.11691951752\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] Epoch[44] Batch[5] avg_epoch_loss=4.129260\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] #quality_metric: host=algo-1, epoch=44, batch=5 train loss <loss>=4.12925982475\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] Epoch[44] Batch [5]#011Speed: 335.73 samples/sec#011loss=4.129260\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] processed a total of 364 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1351.837158203125, \"sum\": 1351.837158203125, \"min\": 1351.837158203125}}, \"EndTime\": 1563345606.002408, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345604.650083}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=269.242924406 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] #progress_metric: host=algo-1, completed 11 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] #quality_metric: host=algo-1, epoch=44, train loss <loss>=4.12925982475\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] Epoch[45] Batch[0] avg_epoch_loss=4.023134\u001b[0m\n\u001b[31m[07/17/2019 06:40:06 INFO 139964490479424] #quality_metric: host=algo-1, epoch=45, batch=0 train loss <loss>=4.02313423157\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] Epoch[45] Batch[5] avg_epoch_loss=4.377657\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] #quality_metric: host=algo-1, epoch=45, batch=5 train loss <loss>=4.37765669823\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] Epoch[45] Batch [5]#011Speed: 339.19 samples/sec#011loss=4.377657\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] processed a total of 332 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1329.103946685791, \"sum\": 1329.103946685791, \"min\": 1329.103946685791}}, \"EndTime\": 1563345607.332079, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345606.002477}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=249.7705763 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] #progress_metric: host=algo-1, completed 11 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] #quality_metric: host=algo-1, epoch=45, train loss <loss>=4.37765669823\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] Epoch[46] Batch[0] avg_epoch_loss=3.794371\u001b[0m\n\u001b[31m[07/17/2019 06:40:07 INFO 139964490479424] #quality_metric: host=algo-1, epoch=46, batch=0 train loss <loss>=3.79437088966\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] Epoch[46] Batch[5] avg_epoch_loss=3.679606\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] #quality_metric: host=algo-1, epoch=46, batch=5 train loss <loss>=3.6796058019\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] Epoch[46] Batch [5]#011Speed: 339.41 samples/sec#011loss=3.679606\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] processed a total of 352 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1330.7559490203857, \"sum\": 1330.7559490203857, \"min\": 1330.7559490203857}}, \"EndTime\": 1563345608.663403, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345607.332162}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=264.488233602 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] #progress_metric: host=algo-1, completed 11 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] #quality_metric: host=algo-1, epoch=46, train loss <loss>=3.6796058019\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] best epoch loss so far\u001b[0m\n\u001b[31m[07/17/2019 06:40:08 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/state_258813cd-ab43-4214-bf17-b715f537bfee-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 58.402061462402344, \"sum\": 58.402061462402344, \"min\": 58.402061462402344}}, \"EndTime\": 1563345608.722393, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345608.663481}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:09 INFO 139964490479424] Epoch[47] Batch[0] avg_epoch_loss=3.835938\u001b[0m\n\u001b[31m[07/17/2019 06:40:09 INFO 139964490479424] #quality_metric: host=algo-1, epoch=47, batch=0 train loss <loss>=3.83593797684\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] Epoch[47] Batch[5] avg_epoch_loss=3.817087\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] #quality_metric: host=algo-1, epoch=47, batch=5 train loss <loss>=3.81708725293\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] Epoch[47] Batch [5]#011Speed: 333.37 samples/sec#011loss=3.817087\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] processed a total of 359 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1355.4449081420898, \"sum\": 1355.4449081420898, \"min\": 1355.4449081420898}}, \"EndTime\": 1563345610.077984, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345608.722472}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=264.836535516 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] #progress_metric: host=algo-1, completed 12 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] #quality_metric: host=algo-1, epoch=47, train loss <loss>=3.81708725293\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] Epoch[48] Batch[0] avg_epoch_loss=3.899132\u001b[0m\n\u001b[31m[07/17/2019 06:40:10 INFO 139964490479424] #quality_metric: host=algo-1, epoch=48, batch=0 train loss <loss>=3.89913225174\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] Epoch[48] Batch[5] avg_epoch_loss=4.173351\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] #quality_metric: host=algo-1, epoch=48, batch=5 train loss <loss>=4.17335128784\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] Epoch[48] Batch [5]#011Speed: 334.50 samples/sec#011loss=4.173351\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] processed a total of 335 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1348.6888408660889, \"sum\": 1348.6888408660889, \"min\": 1348.6888408660889}}, \"EndTime\": 1563345611.427188, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345610.078057}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=248.369633291 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] #progress_metric: host=algo-1, completed 12 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] #quality_metric: host=algo-1, epoch=48, train loss <loss>=4.17335128784\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] Epoch[49] Batch[0] avg_epoch_loss=3.844471\u001b[0m\n\u001b[31m[07/17/2019 06:40:11 INFO 139964490479424] #quality_metric: host=algo-1, epoch=49, batch=0 train loss <loss>=3.84447050095\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] Epoch[49] Batch[5] avg_epoch_loss=4.020451\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] #quality_metric: host=algo-1, epoch=49, batch=5 train loss <loss>=4.02045110861\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] Epoch[49] Batch [5]#011Speed: 340.97 samples/sec#011loss=4.020451\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] processed a total of 362 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1325.8790969848633, \"sum\": 1325.8790969848633, \"min\": 1325.8790969848633}}, \"EndTime\": 1563345612.753598, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345611.427261}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=273.003378699 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] #progress_metric: host=algo-1, completed 12 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] #quality_metric: host=algo-1, epoch=49, train loss <loss>=4.02045110861\u001b[0m\n\u001b[31m[07/17/2019 06:40:12 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:13 INFO 139964490479424] Epoch[50] Batch[0] avg_epoch_loss=4.313016\u001b[0m\n\u001b[31m[07/17/2019 06:40:13 INFO 139964490479424] #quality_metric: host=algo-1, epoch=50, batch=0 train loss <loss>=4.31301593781\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] Epoch[50] Batch[5] avg_epoch_loss=4.178299\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] #quality_metric: host=algo-1, epoch=50, batch=5 train loss <loss>=4.17829910914\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] Epoch[50] Batch [5]#011Speed: 339.96 samples/sec#011loss=4.178299\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] processed a total of 332 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1325.4599571228027, \"sum\": 1325.4599571228027, \"min\": 1325.4599571228027}}, \"EndTime\": 1563345614.079593, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345612.753676}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=250.456606535 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] #progress_metric: host=algo-1, completed 12 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] #quality_metric: host=algo-1, epoch=50, train loss <loss>=4.17829910914\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] Epoch[51] Batch[0] avg_epoch_loss=4.015382\u001b[0m\n\u001b[31m[07/17/2019 06:40:14 INFO 139964490479424] #quality_metric: host=algo-1, epoch=51, batch=0 train loss <loss>=4.01538228989\u001b[0m\n\u001b[31m[07/17/2019 06:40:15 INFO 139964490479424] processed a total of 304 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1150.8898735046387, \"sum\": 1150.8898735046387, \"min\": 1150.8898735046387}}, \"EndTime\": 1563345615.231014, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345614.079674}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:15 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=264.11837171 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:15 INFO 139964490479424] #progress_metric: host=algo-1, completed 13 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:15 INFO 139964490479424] #quality_metric: host=algo-1, epoch=51, train loss <loss>=4.02378721237\u001b[0m\n\u001b[31m[07/17/2019 06:40:15 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:15 INFO 139964490479424] Epoch[52] Batch[0] avg_epoch_loss=3.991700\u001b[0m\n\u001b[31m[07/17/2019 06:40:15 INFO 139964490479424] #quality_metric: host=algo-1, epoch=52, batch=0 train loss <loss>=3.99169993401\u001b[0m\n\u001b[31m[07/17/2019 06:40:16 INFO 139964490479424] processed a total of 306 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1136.3389492034912, \"sum\": 1136.3389492034912, \"min\": 1136.3389492034912}}, \"EndTime\": 1563345616.367855, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345615.231087}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:16 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=269.263534643 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:16 INFO 139964490479424] #progress_metric: host=algo-1, completed 13 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:16 INFO 139964490479424] #quality_metric: host=algo-1, epoch=52, train loss <loss>=3.91396250725\u001b[0m\n\u001b[31m[07/17/2019 06:40:16 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:16 INFO 139964490479424] Epoch[53] Batch[0] avg_epoch_loss=4.308815\u001b[0m\n\u001b[31m[07/17/2019 06:40:16 INFO 139964490479424] #quality_metric: host=algo-1, epoch=53, batch=0 train loss <loss>=4.30881500244\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] Epoch[53] Batch[5] avg_epoch_loss=4.172076\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] #quality_metric: host=algo-1, epoch=53, batch=5 train loss <loss>=4.17207630475\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] Epoch[53] Batch [5]#011Speed: 339.86 samples/sec#011loss=4.172076\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] processed a total of 352 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1330.4321765899658, \"sum\": 1330.4321765899658, \"min\": 1330.4321765899658}}, \"EndTime\": 1563345617.698806, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345616.367916}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=264.554015683 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] #progress_metric: host=algo-1, completed 13 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] #quality_metric: host=algo-1, epoch=53, train loss <loss>=4.17207630475\u001b[0m\n\u001b[31m[07/17/2019 06:40:17 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:18 INFO 139964490479424] Epoch[54] Batch[0] avg_epoch_loss=4.011399\u001b[0m\n\u001b[31m[07/17/2019 06:40:18 INFO 139964490479424] #quality_metric: host=algo-1, epoch=54, batch=0 train loss <loss>=4.01139879227\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] Epoch[54] Batch[5] avg_epoch_loss=3.912181\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] #quality_metric: host=algo-1, epoch=54, batch=5 train loss <loss>=3.91218090057\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] Epoch[54] Batch [5]#011Speed: 340.63 samples/sec#011loss=3.912181\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] processed a total of 347 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1327.4219036102295, \"sum\": 1327.4219036102295, \"min\": 1327.4219036102295}}, \"EndTime\": 1563345619.026785, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345617.698877}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=261.386299872 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] #progress_metric: host=algo-1, completed 13 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] #quality_metric: host=algo-1, epoch=54, train loss <loss>=3.91218090057\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] Epoch[55] Batch[0] avg_epoch_loss=4.003117\u001b[0m\n\u001b[31m[07/17/2019 06:40:19 INFO 139964490479424] #quality_metric: host=algo-1, epoch=55, batch=0 train loss <loss>=4.00311660767\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] Epoch[55] Batch[5] avg_epoch_loss=3.986671\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] #quality_metric: host=algo-1, epoch=55, batch=5 train loss <loss>=3.98667128881\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] Epoch[55] Batch [5]#011Speed: 336.61 samples/sec#011loss=3.986671\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] processed a total of 365 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1348.2990264892578, \"sum\": 1348.2990264892578, \"min\": 1348.2990264892578}}, \"EndTime\": 1563345620.375622, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345619.026865}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=270.687576638 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] #progress_metric: host=algo-1, completed 14 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] #quality_metric: host=algo-1, epoch=55, train loss <loss>=3.98667128881\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] Epoch[56] Batch[0] avg_epoch_loss=4.139123\u001b[0m\n\u001b[31m[07/17/2019 06:40:20 INFO 139964490479424] #quality_metric: host=algo-1, epoch=56, batch=0 train loss <loss>=4.13912343979\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] Epoch[56] Batch[5] avg_epoch_loss=4.072855\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] #quality_metric: host=algo-1, epoch=56, batch=5 train loss <loss>=4.07285459836\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] Epoch[56] Batch [5]#011Speed: 339.71 samples/sec#011loss=4.072855\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] processed a total of 357 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1333.7888717651367, \"sum\": 1333.7888717651367, \"min\": 1333.7888717651367}}, \"EndTime\": 1563345621.709975, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345620.375704}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=267.635494257 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] #progress_metric: host=algo-1, completed 14 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] #quality_metric: host=algo-1, epoch=56, train loss <loss>=4.07285459836\u001b[0m\n\u001b[31m[07/17/2019 06:40:21 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:22 INFO 139964490479424] Epoch[57] Batch[0] avg_epoch_loss=4.085328\u001b[0m\n\u001b[31m[07/17/2019 06:40:22 INFO 139964490479424] #quality_metric: host=algo-1, epoch=57, batch=0 train loss <loss>=4.08532762527\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] Epoch[57] Batch[5] avg_epoch_loss=3.859712\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] #quality_metric: host=algo-1, epoch=57, batch=5 train loss <loss>=3.85971152782\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] Epoch[57] Batch [5]#011Speed: 337.91 samples/sec#011loss=3.859712\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] processed a total of 364 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1335.108995437622, \"sum\": 1335.108995437622, \"min\": 1335.108995437622}}, \"EndTime\": 1563345623.04567, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345621.710053}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=272.614230725 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] #progress_metric: host=algo-1, completed 14 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] #quality_metric: host=algo-1, epoch=57, train loss <loss>=3.85971152782\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] Epoch[58] Batch[0] avg_epoch_loss=4.170880\u001b[0m\n\u001b[31m[07/17/2019 06:40:23 INFO 139964490479424] #quality_metric: host=algo-1, epoch=58, batch=0 train loss <loss>=4.17088031769\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] Epoch[58] Batch[5] avg_epoch_loss=3.920547\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] #quality_metric: host=algo-1, epoch=58, batch=5 train loss <loss>=3.92054708799\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] Epoch[58] Batch [5]#011Speed: 339.24 samples/sec#011loss=3.920547\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] processed a total of 330 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1328.3588886260986, \"sum\": 1328.3588886260986, \"min\": 1328.3588886260986}}, \"EndTime\": 1563345624.374577, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345623.045747}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=248.40490356 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] #progress_metric: host=algo-1, completed 14 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] #quality_metric: host=algo-1, epoch=58, train loss <loss>=3.92054708799\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] Epoch[59] Batch[0] avg_epoch_loss=4.140189\u001b[0m\n\u001b[31m[07/17/2019 06:40:24 INFO 139964490479424] #quality_metric: host=algo-1, epoch=59, batch=0 train loss <loss>=4.14018917084\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] Epoch[59] Batch[5] avg_epoch_loss=3.977198\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] #quality_metric: host=algo-1, epoch=59, batch=5 train loss <loss>=3.97719784578\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] Epoch[59] Batch [5]#011Speed: 332.89 samples/sec#011loss=3.977198\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] processed a total of 368 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1353.132963180542, \"sum\": 1353.132963180542, \"min\": 1353.132963180542}}, \"EndTime\": 1563345625.728276, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345624.374662}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=271.941371455 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] #progress_metric: host=algo-1, completed 15 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] #quality_metric: host=algo-1, epoch=59, train loss <loss>=3.97719784578\u001b[0m\n\u001b[31m[07/17/2019 06:40:25 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:26 INFO 139964490479424] Epoch[60] Batch[0] avg_epoch_loss=3.723538\u001b[0m\n\u001b[31m[07/17/2019 06:40:26 INFO 139964490479424] #quality_metric: host=algo-1, epoch=60, batch=0 train loss <loss>=3.72353768349\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] Epoch[60] Batch[5] avg_epoch_loss=4.249615\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] #quality_metric: host=algo-1, epoch=60, batch=5 train loss <loss>=4.24961515268\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] Epoch[60] Batch [5]#011Speed: 339.35 samples/sec#011loss=4.249615\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] processed a total of 353 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1331.1741352081299, \"sum\": 1331.1741352081299, \"min\": 1331.1741352081299}}, \"EndTime\": 1563345627.059957, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345625.728342}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=265.160339707 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] #progress_metric: host=algo-1, completed 15 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] #quality_metric: host=algo-1, epoch=60, train loss <loss>=4.24961515268\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] Epoch[61] Batch[0] avg_epoch_loss=4.081460\u001b[0m\n\u001b[31m[07/17/2019 06:40:27 INFO 139964490479424] #quality_metric: host=algo-1, epoch=61, batch=0 train loss <loss>=4.08145952225\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] Epoch[61] Batch[5] avg_epoch_loss=3.911195\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] #quality_metric: host=algo-1, epoch=61, batch=5 train loss <loss>=3.91119523843\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] Epoch[61] Batch [5]#011Speed: 330.24 samples/sec#011loss=3.911195\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] processed a total of 342 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1360.564947128296, \"sum\": 1360.564947128296, \"min\": 1360.564947128296}}, \"EndTime\": 1563345628.421045, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345627.060021}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=251.347318957 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] #progress_metric: host=algo-1, completed 15 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] #quality_metric: host=algo-1, epoch=61, train loss <loss>=3.91119523843\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] Epoch[62] Batch[0] avg_epoch_loss=4.375705\u001b[0m\n\u001b[31m[07/17/2019 06:40:28 INFO 139964490479424] #quality_metric: host=algo-1, epoch=62, batch=0 train loss <loss>=4.37570476532\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] Epoch[62] Batch[5] avg_epoch_loss=4.093192\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] #quality_metric: host=algo-1, epoch=62, batch=5 train loss <loss>=4.09319150448\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] Epoch[62] Batch [5]#011Speed: 339.02 samples/sec#011loss=4.093192\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] processed a total of 375 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1339.2810821533203, \"sum\": 1339.2810821533203, \"min\": 1339.2810821533203}}, \"EndTime\": 1563345629.760869, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345628.421115}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=279.971712046 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] #progress_metric: host=algo-1, completed 15 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] #quality_metric: host=algo-1, epoch=62, train loss <loss>=4.09319150448\u001b[0m\n\u001b[31m[07/17/2019 06:40:29 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:30 INFO 139964490479424] Epoch[63] Batch[0] avg_epoch_loss=3.812004\u001b[0m\n\u001b[31m[07/17/2019 06:40:30 INFO 139964490479424] #quality_metric: host=algo-1, epoch=63, batch=0 train loss <loss>=3.81200432777\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] Epoch[63] Batch[5] avg_epoch_loss=4.194525\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] #quality_metric: host=algo-1, epoch=63, batch=5 train loss <loss>=4.19452480475\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] Epoch[63] Batch [5]#011Speed: 343.55 samples/sec#011loss=4.194525\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] processed a total of 367 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1331.7370414733887, \"sum\": 1331.7370414733887, \"min\": 1331.7370414733887}}, \"EndTime\": 1563345631.093164, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345629.760972}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=275.556545935 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] #progress_metric: host=algo-1, completed 16 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] #quality_metric: host=algo-1, epoch=63, train loss <loss>=4.19452480475\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] Epoch[64] Batch[0] avg_epoch_loss=3.977389\u001b[0m\n\u001b[31m[07/17/2019 06:40:31 INFO 139964490479424] #quality_metric: host=algo-1, epoch=64, batch=0 train loss <loss>=3.97738909721\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] Epoch[64] Batch[5] avg_epoch_loss=4.052506\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] #quality_metric: host=algo-1, epoch=64, batch=5 train loss <loss>=4.05250632763\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] Epoch[64] Batch [5]#011Speed: 341.21 samples/sec#011loss=4.052506\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] processed a total of 351 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1349.4391441345215, \"sum\": 1349.4391441345215, \"min\": 1349.4391441345215}}, \"EndTime\": 1563345632.443105, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345631.093241}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=260.087842586 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] #progress_metric: host=algo-1, completed 16 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] #quality_metric: host=algo-1, epoch=64, train loss <loss>=4.05250632763\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] Epoch[65] Batch[0] avg_epoch_loss=4.129368\u001b[0m\n\u001b[31m[07/17/2019 06:40:32 INFO 139964490479424] #quality_metric: host=algo-1, epoch=65, batch=0 train loss <loss>=4.12936782837\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] Epoch[65] Batch[5] avg_epoch_loss=3.977775\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] #quality_metric: host=algo-1, epoch=65, batch=5 train loss <loss>=3.97777533531\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] Epoch[65] Batch [5]#011Speed: 339.99 samples/sec#011loss=3.977775\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] processed a total of 334 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1328.5579681396484, \"sum\": 1328.5579681396484, \"min\": 1328.5579681396484}}, \"EndTime\": 1563345633.77227, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345632.443175}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=251.379013611 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] #progress_metric: host=algo-1, completed 16 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] #quality_metric: host=algo-1, epoch=65, train loss <loss>=3.97777533531\u001b[0m\n\u001b[31m[07/17/2019 06:40:33 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:34 INFO 139964490479424] Epoch[66] Batch[0] avg_epoch_loss=4.010057\u001b[0m\n\u001b[31m[07/17/2019 06:40:34 INFO 139964490479424] #quality_metric: host=algo-1, epoch=66, batch=0 train loss <loss>=4.0100569725\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] Epoch[66] Batch[5] avg_epoch_loss=3.940697\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] #quality_metric: host=algo-1, epoch=66, batch=5 train loss <loss>=3.94069739183\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] Epoch[66] Batch [5]#011Speed: 336.09 samples/sec#011loss=3.940697\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] processed a total of 362 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1339.2760753631592, \"sum\": 1339.2760753631592, \"min\": 1339.2760753631592}}, \"EndTime\": 1563345635.112051, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345633.772347}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=270.274493246 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] #progress_metric: host=algo-1, completed 16 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] #quality_metric: host=algo-1, epoch=66, train loss <loss>=3.94069739183\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] Epoch[67] Batch[0] avg_epoch_loss=3.692048\u001b[0m\n\u001b[31m[07/17/2019 06:40:35 INFO 139964490479424] #quality_metric: host=algo-1, epoch=67, batch=0 train loss <loss>=3.6920478344\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] Epoch[67] Batch[5] avg_epoch_loss=3.911003\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] #quality_metric: host=algo-1, epoch=67, batch=5 train loss <loss>=3.91100307306\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] Epoch[67] Batch [5]#011Speed: 340.30 samples/sec#011loss=3.911003\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] processed a total of 359 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1329.0901184082031, \"sum\": 1329.0901184082031, \"min\": 1329.0901184082031}}, \"EndTime\": 1563345636.441677, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345635.112121}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=270.08663478 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] #progress_metric: host=algo-1, completed 17 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] #quality_metric: host=algo-1, epoch=67, train loss <loss>=3.91100307306\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] Epoch[68] Batch[0] avg_epoch_loss=3.856553\u001b[0m\n\u001b[31m[07/17/2019 06:40:36 INFO 139964490479424] #quality_metric: host=algo-1, epoch=68, batch=0 train loss <loss>=3.85655283928\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] Epoch[68] Batch[5] avg_epoch_loss=4.071871\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] #quality_metric: host=algo-1, epoch=68, batch=5 train loss <loss>=4.07187104225\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] Epoch[68] Batch [5]#011Speed: 340.67 samples/sec#011loss=4.071871\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] processed a total of 362 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1344.0649509429932, \"sum\": 1344.0649509429932, \"min\": 1344.0649509429932}}, \"EndTime\": 1563345637.786246, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345636.441754}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=269.30938728 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] #progress_metric: host=algo-1, completed 17 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] #quality_metric: host=algo-1, epoch=68, train loss <loss>=4.07187104225\u001b[0m\n\u001b[31m[07/17/2019 06:40:37 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:38 INFO 139964490479424] Epoch[69] Batch[0] avg_epoch_loss=4.065495\u001b[0m\n\u001b[31m[07/17/2019 06:40:38 INFO 139964490479424] #quality_metric: host=algo-1, epoch=69, batch=0 train loss <loss>=4.06549549103\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] Epoch[69] Batch[5] avg_epoch_loss=4.115337\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] #quality_metric: host=algo-1, epoch=69, batch=5 train loss <loss>=4.11533677578\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] Epoch[69] Batch [5]#011Speed: 339.49 samples/sec#011loss=4.115337\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] processed a total of 335 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1326.4069557189941, \"sum\": 1326.4069557189941, \"min\": 1326.4069557189941}}, \"EndTime\": 1563345639.113198, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345637.786324}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=252.541078379 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] #progress_metric: host=algo-1, completed 17 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] #quality_metric: host=algo-1, epoch=69, train loss <loss>=4.11533677578\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] Epoch[70] Batch[0] avg_epoch_loss=3.978797\u001b[0m\n\u001b[31m[07/17/2019 06:40:39 INFO 139964490479424] #quality_metric: host=algo-1, epoch=70, batch=0 train loss <loss>=3.97879743576\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] Epoch[70] Batch[5] avg_epoch_loss=4.071744\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] #quality_metric: host=algo-1, epoch=70, batch=5 train loss <loss>=4.07174396515\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] Epoch[70] Batch [5]#011Speed: 335.44 samples/sec#011loss=4.071744\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] processed a total of 347 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1340.8889770507812, \"sum\": 1340.8889770507812, \"min\": 1340.8889770507812}}, \"EndTime\": 1563345640.454648, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345639.113272}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=258.761918342 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] #progress_metric: host=algo-1, completed 17 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] #quality_metric: host=algo-1, epoch=70, train loss <loss>=4.07174396515\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] Epoch[71] Batch[0] avg_epoch_loss=3.890575\u001b[0m\n\u001b[31m[07/17/2019 06:40:40 INFO 139964490479424] #quality_metric: host=algo-1, epoch=71, batch=0 train loss <loss>=3.89057469368\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] Epoch[71] Batch[5] avg_epoch_loss=3.914051\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] #quality_metric: host=algo-1, epoch=71, batch=5 train loss <loss>=3.91405101617\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] Epoch[71] Batch [5]#011Speed: 335.87 samples/sec#011loss=3.914051\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] processed a total of 333 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1362.9679679870605, \"sum\": 1362.9679679870605, \"min\": 1362.9679679870605}}, \"EndTime\": 1563345641.818148, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345640.454727}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=244.29928526 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] #progress_metric: host=algo-1, completed 18 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] #quality_metric: host=algo-1, epoch=71, train loss <loss>=3.91405101617\u001b[0m\n\u001b[31m[07/17/2019 06:40:41 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:42 INFO 139964490479424] Epoch[72] Batch[0] avg_epoch_loss=3.606528\u001b[0m\n\u001b[31m[07/17/2019 06:40:42 INFO 139964490479424] #quality_metric: host=algo-1, epoch=72, batch=0 train loss <loss>=3.60652756691\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] Epoch[72] Batch[5] avg_epoch_loss=3.829789\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] #quality_metric: host=algo-1, epoch=72, batch=5 train loss <loss>=3.82978936036\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] Epoch[72] Batch [5]#011Speed: 335.78 samples/sec#011loss=3.829789\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] processed a total of 343 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1375.1461505889893, \"sum\": 1375.1461505889893, \"min\": 1375.1461505889893}}, \"EndTime\": 1563345643.193833, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345641.818226}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=249.40736399 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] #progress_metric: host=algo-1, completed 18 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] #quality_metric: host=algo-1, epoch=72, train loss <loss>=3.82978936036\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] Epoch[73] Batch[0] avg_epoch_loss=4.037586\u001b[0m\n\u001b[31m[07/17/2019 06:40:43 INFO 139964490479424] #quality_metric: host=algo-1, epoch=73, batch=0 train loss <loss>=4.03758621216\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] Epoch[73] Batch[5] avg_epoch_loss=3.960642\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] #quality_metric: host=algo-1, epoch=73, batch=5 train loss <loss>=3.96064217885\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] Epoch[73] Batch [5]#011Speed: 342.50 samples/sec#011loss=3.960642\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] processed a total of 352 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1337.918996810913, \"sum\": 1337.918996810913, \"min\": 1337.918996810913}}, \"EndTime\": 1563345644.53227, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345643.193911}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=263.073163952 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] #progress_metric: host=algo-1, completed 18 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] #quality_metric: host=algo-1, epoch=73, train loss <loss>=3.96064217885\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] Epoch[74] Batch[0] avg_epoch_loss=3.997142\u001b[0m\n\u001b[31m[07/17/2019 06:40:44 INFO 139964490479424] #quality_metric: host=algo-1, epoch=74, batch=0 train loss <loss>=3.99714231491\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] Epoch[74] Batch[5] avg_epoch_loss=4.013053\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] #quality_metric: host=algo-1, epoch=74, batch=5 train loss <loss>=4.01305321852\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] Epoch[74] Batch [5]#011Speed: 336.95 samples/sec#011loss=4.013053\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] processed a total of 350 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1400.0141620635986, \"sum\": 1400.0141620635986, \"min\": 1400.0141620635986}}, \"EndTime\": 1563345645.932781, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345644.532347}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=249.977675797 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] #progress_metric: host=algo-1, completed 18 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] #quality_metric: host=algo-1, epoch=74, train loss <loss>=4.01305321852\u001b[0m\n\u001b[31m[07/17/2019 06:40:45 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:46 INFO 139964490479424] Epoch[75] Batch[0] avg_epoch_loss=3.892437\u001b[0m\n\u001b[31m[07/17/2019 06:40:46 INFO 139964490479424] #quality_metric: host=algo-1, epoch=75, batch=0 train loss <loss>=3.89243721962\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] Epoch[75] Batch[5] avg_epoch_loss=3.872168\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] #quality_metric: host=algo-1, epoch=75, batch=5 train loss <loss>=3.87216842175\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] Epoch[75] Batch [5]#011Speed: 339.13 samples/sec#011loss=3.872168\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] processed a total of 325 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1383.538007736206, \"sum\": 1383.538007736206, \"min\": 1383.538007736206}}, \"EndTime\": 1563345647.316858, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345645.932858}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=234.883105426 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] #progress_metric: host=algo-1, completed 19 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] #quality_metric: host=algo-1, epoch=75, train loss <loss>=3.87216842175\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] Epoch[76] Batch[0] avg_epoch_loss=4.009735\u001b[0m\n\u001b[31m[07/17/2019 06:40:47 INFO 139964490479424] #quality_metric: host=algo-1, epoch=76, batch=0 train loss <loss>=4.00973510742\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] Epoch[76] Batch[5] avg_epoch_loss=3.934983\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] #quality_metric: host=algo-1, epoch=76, batch=5 train loss <loss>=3.93498325348\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] Epoch[76] Batch [5]#011Speed: 341.44 samples/sec#011loss=3.934983\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] processed a total of 335 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1351.9480228424072, \"sum\": 1351.9480228424072, \"min\": 1351.9480228424072}}, \"EndTime\": 1563345648.669324, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345647.316951}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=247.769705265 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] #progress_metric: host=algo-1, completed 19 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] #quality_metric: host=algo-1, epoch=76, train loss <loss>=3.93498325348\u001b[0m\n\u001b[31m[07/17/2019 06:40:48 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:49 INFO 139964490479424] Epoch[77] Batch[0] avg_epoch_loss=3.755864\u001b[0m\n\u001b[31m[07/17/2019 06:40:49 INFO 139964490479424] #quality_metric: host=algo-1, epoch=77, batch=0 train loss <loss>=3.75586414337\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] Epoch[77] Batch[5] avg_epoch_loss=3.928259\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] #quality_metric: host=algo-1, epoch=77, batch=5 train loss <loss>=3.92825897535\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] Epoch[77] Batch [5]#011Speed: 335.74 samples/sec#011loss=3.928259\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] processed a total of 354 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1370.7859516143799, \"sum\": 1370.7859516143799, \"min\": 1370.7859516143799}}, \"EndTime\": 1563345650.040631, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345648.669403}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=258.224539915 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] #progress_metric: host=algo-1, completed 19 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] #quality_metric: host=algo-1, epoch=77, train loss <loss>=3.92825897535\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] Epoch[78] Batch[0] avg_epoch_loss=3.953550\u001b[0m\n\u001b[31m[07/17/2019 06:40:50 INFO 139964490479424] #quality_metric: host=algo-1, epoch=78, batch=0 train loss <loss>=3.95354986191\u001b[0m\n\u001b[31m[07/17/2019 06:40:51 INFO 139964490479424] processed a total of 313 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1181.4789772033691, \"sum\": 1181.4789772033691, \"min\": 1181.4789772033691}}, \"EndTime\": 1563345651.222655, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345650.040709}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:51 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=264.895089961 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:51 INFO 139964490479424] #progress_metric: host=algo-1, completed 19 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:51 INFO 139964490479424] #quality_metric: host=algo-1, epoch=78, train loss <loss>=3.9522069931\u001b[0m\n\u001b[31m[07/17/2019 06:40:51 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:51 INFO 139964490479424] Epoch[79] Batch[0] avg_epoch_loss=3.672956\u001b[0m\n\u001b[31m[07/17/2019 06:40:51 INFO 139964490479424] #quality_metric: host=algo-1, epoch=79, batch=0 train loss <loss>=3.67295598984\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] Epoch[79] Batch[5] avg_epoch_loss=3.927602\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] #quality_metric: host=algo-1, epoch=79, batch=5 train loss <loss>=3.92760165532\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] Epoch[79] Batch [5]#011Speed: 343.45 samples/sec#011loss=3.927602\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] processed a total of 364 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1358.3300113677979, \"sum\": 1358.3300113677979, \"min\": 1358.3300113677979}}, \"EndTime\": 1563345652.581554, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345651.222739}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=267.953210889 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] #progress_metric: host=algo-1, completed 20 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] #quality_metric: host=algo-1, epoch=79, train loss <loss>=3.92760165532\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] Epoch[80] Batch[0] avg_epoch_loss=3.595350\u001b[0m\n\u001b[31m[07/17/2019 06:40:52 INFO 139964490479424] #quality_metric: host=algo-1, epoch=80, batch=0 train loss <loss>=3.59534955025\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] Epoch[80] Batch[5] avg_epoch_loss=3.838286\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] #quality_metric: host=algo-1, epoch=80, batch=5 train loss <loss>=3.83828639984\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] Epoch[80] Batch [5]#011Speed: 338.77 samples/sec#011loss=3.838286\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] processed a total of 349 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1346.6310501098633, \"sum\": 1346.6310501098633, \"min\": 1346.6310501098633}}, \"EndTime\": 1563345653.928663, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345652.581634}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=259.143522687 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] #progress_metric: host=algo-1, completed 20 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] #quality_metric: host=algo-1, epoch=80, train loss <loss>=3.83828639984\u001b[0m\n\u001b[31m[07/17/2019 06:40:53 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:54 INFO 139964490479424] Epoch[81] Batch[0] avg_epoch_loss=3.763526\u001b[0m\n\u001b[31m[07/17/2019 06:40:54 INFO 139964490479424] #quality_metric: host=algo-1, epoch=81, batch=0 train loss <loss>=3.76352572441\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] Epoch[81] Batch[5] avg_epoch_loss=3.975435\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] #quality_metric: host=algo-1, epoch=81, batch=5 train loss <loss>=3.97543489933\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] Epoch[81] Batch [5]#011Speed: 330.97 samples/sec#011loss=3.975435\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] processed a total of 376 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1373.694896697998, \"sum\": 1373.694896697998, \"min\": 1373.694896697998}}, \"EndTime\": 1563345655.302915, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345653.92874}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=273.692209866 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] #progress_metric: host=algo-1, completed 20 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] #quality_metric: host=algo-1, epoch=81, train loss <loss>=3.97543489933\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] Epoch[82] Batch[0] avg_epoch_loss=3.935109\u001b[0m\n\u001b[31m[07/17/2019 06:40:55 INFO 139964490479424] #quality_metric: host=algo-1, epoch=82, batch=0 train loss <loss>=3.93510890007\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] Epoch[82] Batch[5] avg_epoch_loss=4.058915\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] #quality_metric: host=algo-1, epoch=82, batch=5 train loss <loss>=4.05891493956\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] Epoch[82] Batch [5]#011Speed: 332.82 samples/sec#011loss=4.058915\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] processed a total of 362 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1393.899917602539, \"sum\": 1393.899917602539, \"min\": 1393.899917602539}}, \"EndTime\": 1563345656.697328, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345655.302992}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=259.68208649 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] #progress_metric: host=algo-1, completed 20 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] #quality_metric: host=algo-1, epoch=82, train loss <loss>=4.05891493956\u001b[0m\n\u001b[31m[07/17/2019 06:40:56 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:57 INFO 139964490479424] Epoch[83] Batch[0] avg_epoch_loss=4.026322\u001b[0m\n\u001b[31m[07/17/2019 06:40:57 INFO 139964490479424] #quality_metric: host=algo-1, epoch=83, batch=0 train loss <loss>=4.02632188797\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] Epoch[83] Batch[5] avg_epoch_loss=3.823845\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] #quality_metric: host=algo-1, epoch=83, batch=5 train loss <loss>=3.82384490967\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] Epoch[83] Batch [5]#011Speed: 343.96 samples/sec#011loss=3.823845\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] processed a total of 374 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1348.9010334014893, \"sum\": 1348.9010334014893, \"min\": 1348.9010334014893}}, \"EndTime\": 1563345658.046743, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345656.697404}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=277.239760559 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] #progress_metric: host=algo-1, completed 21 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] #quality_metric: host=algo-1, epoch=83, train loss <loss>=3.82384490967\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] Epoch[84] Batch[0] avg_epoch_loss=4.047552\u001b[0m\n\u001b[31m[07/17/2019 06:40:58 INFO 139964490479424] #quality_metric: host=algo-1, epoch=84, batch=0 train loss <loss>=4.04755210876\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] Epoch[84] Batch[5] avg_epoch_loss=3.912242\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] #quality_metric: host=algo-1, epoch=84, batch=5 train loss <loss>=3.91224193573\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] Epoch[84] Batch [5]#011Speed: 340.60 samples/sec#011loss=3.912242\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] processed a total of 368 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1377.579927444458, \"sum\": 1377.579927444458, \"min\": 1377.579927444458}}, \"EndTime\": 1563345659.424831, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345658.04682}\n\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=267.113407225 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] #progress_metric: host=algo-1, completed 21 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] #quality_metric: host=algo-1, epoch=84, train loss <loss>=3.91224193573\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] Epoch[85] Batch[0] avg_epoch_loss=4.037418\u001b[0m\n\u001b[31m[07/17/2019 06:40:59 INFO 139964490479424] #quality_metric: host=algo-1, epoch=85, batch=0 train loss <loss>=4.03741836548\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] Epoch[85] Batch[5] avg_epoch_loss=3.818764\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] #quality_metric: host=algo-1, epoch=85, batch=5 train loss <loss>=3.81876393159\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] Epoch[85] Batch [5]#011Speed: 339.54 samples/sec#011loss=3.818764\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] processed a total of 350 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1350.4638671875, \"sum\": 1350.4638671875, \"min\": 1350.4638671875}}, \"EndTime\": 1563345660.775859, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345659.424909}\n\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=259.148154778 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] #progress_metric: host=algo-1, completed 21 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] #quality_metric: host=algo-1, epoch=85, train loss <loss>=3.81876393159\u001b[0m\n\u001b[31m[07/17/2019 06:41:00 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:41:01 INFO 139964490479424] Epoch[86] Batch[0] avg_epoch_loss=4.096306\u001b[0m\n\u001b[31m[07/17/2019 06:41:01 INFO 139964490479424] #quality_metric: host=algo-1, epoch=86, batch=0 train loss <loss>=4.09630632401\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] Epoch[86] Batch[5] avg_epoch_loss=4.127224\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] #quality_metric: host=algo-1, epoch=86, batch=5 train loss <loss>=4.12722420692\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] Epoch[86] Batch [5]#011Speed: 333.85 samples/sec#011loss=4.127224\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] processed a total of 346 examples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 1379.1429996490479, \"sum\": 1379.1429996490479, \"min\": 1379.1429996490479}}, \"EndTime\": 1563345662.155515, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345660.775936}\n\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] #throughput_metric: host=algo-1, train throughput=250.860098869 records/second\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] #progress_metric: host=algo-1, completed 21 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] #quality_metric: host=algo-1, epoch=86, train loss <loss>=4.12722420692\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] loss did not improve\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] Loading parameters from best epoch (46)\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"state.deserialize.time\": {\"count\": 1, \"max\": 24.4598388671875, \"sum\": 24.4598388671875, \"min\": 24.4598388671875}}, \"EndTime\": 1563345662.180563, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345662.155592}\n\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] stopping training now\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] #progress_metric: host=algo-1, completed 100 % of epochs\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] Final loss: 3.6796058019 (occurred at epoch 46)\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] #quality_metric: host=algo-1, train final_loss <loss>=3.6796058019\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] Worker algo-1 finished training.\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 WARNING 139964490479424] wait_for_all_workers will not sync workers since the kv store is not running distributed\u001b[0m\n\u001b[31m[07/17/2019 06:41:02 INFO 139964490479424] All workers finished. Serializing model for prediction.\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"get_graph.time\": {\"count\": 1, \"max\": 803.4188747406006, \"sum\": 803.4188747406006, \"min\": 803.4188747406006}}, \"EndTime\": 1563345662.98479, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345662.180623}\n\u001b[0m\n\u001b[31m[07/17/2019 06:41:03 INFO 139964490479424] Number of GPUs being used: 0\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"finalize.time\": {\"count\": 1, \"max\": 1043.5879230499268, \"sum\": 1043.5879230499268, \"min\": 1043.5879230499268}}, \"EndTime\": 1563345663.22494, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345662.984862}\n\u001b[0m\n\u001b[31m[07/17/2019 06:41:03 INFO 139964490479424] Serializing to /opt/ml/model/model_algo-1\u001b[0m\n\u001b[31m[07/17/2019 06:41:03 INFO 139964490479424] Saved checkpoint to \"/opt/ml/model/model_algo-1-0000.params\"\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"model.serialize.time\": {\"count\": 1, \"max\": 38.70081901550293, \"sum\": 38.70081901550293, \"min\": 38.70081901550293}}, \"EndTime\": 1563345663.26373, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345663.224996}\n\u001b[0m\n\u001b[31m[07/17/2019 06:41:03 INFO 139964490479424] Successfully serialized the model for prediction.\u001b[0m\n\u001b[31m[07/17/2019 06:41:03 INFO 139964490479424] Evaluating model accuracy on testset using 100 samples\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"model.bind.time\": {\"count\": 1, \"max\": 0.027179718017578125, \"sum\": 0.027179718017578125, \"min\": 0.027179718017578125}}, \"EndTime\": 1563345663.264388, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345663.263782}\n\u001b[0m\n\u001b[31m[07/17/2019 06:41:37 INFO 139964490479424] Number of test batches scored: 10\u001b[0m\n\u001b[31m[07/17/2019 06:42:11 INFO 139964490479424] Number of test batches scored: 20\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"model.score.time\": {\"count\": 1, \"max\": 74830.17015457153, \"sum\": 74830.17015457153, \"min\": 74830.17015457153}}, \"EndTime\": 1563345738.094538, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345663.264429}\n\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, RMSE): 603.248109197\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, mean_wQuantileLoss): 0.0691546\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.1]): 0.0473733\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.2]): 0.0680008\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.3]): 0.0796946\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.4]): 0.0853203\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.5]): 0.0858047\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.6]): 0.0819466\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.7]): 0.0735508\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.8]): 0.0601982\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #test_score (algo-1, wQuantileLoss[0.9]): 0.0405022\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #quality_metric: host=algo-1, test mean_wQuantileLoss <loss>=0.0691546276212\u001b[0m\n\u001b[31m[07/17/2019 06:42:18 INFO 139964490479424] #quality_metric: host=algo-1, test RMSE <loss>=603.248109197\u001b[0m\n\u001b[31m#metrics {\"Metrics\": {\"totaltime\": {\"count\": 1, \"max\": 196219.30289268494, \"sum\": 196219.30289268494, \"min\": 196219.30289268494}, \"setuptime\": {\"count\": 1, \"max\": 9.935855865478516, \"sum\": 9.935855865478516, \"min\": 9.935855865478516}}, \"EndTime\": 1563345738.151742, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1563345738.094606}\n\u001b[0m\n\n2019-07-17 06:42:27 Uploading - Uploading generated training model\n2019-07-17 06:42:27 Completed - Training job completed\nBillable seconds: 257\nCPU times: user 882 ms, sys: 72.7 ms, total: 955 ms\nWall time: 6min 45s\n"
]
],
[
[
"Since you pass a test set in this example, accuracy metrics for the forecast are computed and logged (see bottom of the log).\nYou can find the definition of these metrics from [our documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar.html). You can use these to optimize the parameters and tune your model or use SageMaker's [Automated Model Tuning service](https://aws.amazon.com/blogs/aws/sagemaker-automatic-model-tuning/) to tune the model for you.",
"_____no_output_____"
],
[
"### Create endpoint and predictor",
"_____no_output_____"
],
[
"Now that we have a trained model, we can use it to perform predictions by deploying it to an endpoint.\n\n**Note: Remember to delete the endpoint after running this experiment. A cell at the very bottom of this notebook will do that: make sure you run it at the end.**",
"_____no_output_____"
],
[
"To query the endpoint and perform predictions, we can define the following utility class: this allows making requests using `pandas.Series` objects rather than raw JSON strings.",
"_____no_output_____"
]
],
[
[
"class DeepARPredictor(sagemaker.predictor.RealTimePredictor):\n \n def __init__(self, *args, **kwargs):\n super().__init__(*args, content_type=sagemaker.content_types.CONTENT_TYPE_JSON, **kwargs)\n \n def predict(self, ts, cat=None, dynamic_feat=None, \n num_samples=100, return_samples=False, quantiles=[\"0.1\", \"0.5\", \"0.9\"]):\n \"\"\"Requests the prediction of for the time series listed in `ts`, each with the (optional)\n corresponding category listed in `cat`.\n \n ts -- `pandas.Series` object, the time series to predict\n cat -- integer, the group associated to the time series (default: None)\n num_samples -- integer, number of samples to compute at prediction time (default: 100)\n return_samples -- boolean indicating whether to include samples in the response (default: False)\n quantiles -- list of strings specifying the quantiles to compute (default: [\"0.1\", \"0.5\", \"0.9\"])\n \n Return value: list of `pandas.DataFrame` objects, each containing the predictions\n \"\"\"\n prediction_time = ts.index[-1] + 1\n quantiles = [str(q) for q in quantiles]\n req = self.__encode_request(ts, cat, dynamic_feat, num_samples, return_samples, quantiles)\n res = super(DeepARPredictor, self).predict(req)\n return self.__decode_response(res, ts.index.freq, prediction_time, return_samples)\n \n def __encode_request(self, ts, cat, dynamic_feat, num_samples, return_samples, quantiles):\n instance = series_to_dict(ts, cat if cat is not None else None, dynamic_feat if dynamic_feat else None)\n\n configuration = {\n \"num_samples\": num_samples,\n \"output_types\": [\"quantiles\", \"samples\"] if return_samples else [\"quantiles\"],\n \"quantiles\": quantiles\n }\n \n http_request_data = {\n \"instances\": [instance],\n \"configuration\": configuration\n }\n \n return json.dumps(http_request_data).encode('utf-8')\n \n def __decode_response(self, response, freq, prediction_time, return_samples):\n # we only sent one time series so we only receive one in return\n # however, if possible one will pass multiple time series as predictions will then be faster\n predictions = json.loads(response.decode('utf-8'))['predictions'][0]\n prediction_length = len(next(iter(predictions['quantiles'].values())))\n prediction_index = pd.DatetimeIndex(start=prediction_time, freq=freq, periods=prediction_length) \n if return_samples:\n dict_of_samples = {'sample_' + str(i): s for i, s in enumerate(predictions['samples'])}\n else:\n dict_of_samples = {}\n return pd.DataFrame(data={**predictions['quantiles'], **dict_of_samples}, index=prediction_index)\n\n def set_frequency(self, freq):\n self.freq = freq\n \ndef encode_target(ts):\n return [x if np.isfinite(x) else \"NaN\" for x in ts] \n\ndef series_to_dict(ts, cat=None, dynamic_feat=None):\n \"\"\"Given a pandas.Series object, returns a dictionary encoding the time series.\n\n ts -- a pands.Series object with the target time series\n cat -- an integer indicating the time series category\n\n Return value: a dictionary\n \"\"\"\n obj = {\"start\": str(ts.index[0]), \"target\": encode_target(ts)}\n if cat is not None:\n obj[\"cat\"] = cat\n if dynamic_feat is not None:\n obj[\"dynamic_feat\"] = dynamic_feat \n return obj",
"_____no_output_____"
]
],
[
[
"Now we can deploy the model and create and endpoint that can be queried using our custom DeepARPredictor class.",
"_____no_output_____"
]
],
[
[
"predictor = estimator.deploy(\n initial_instance_count=1,\n instance_type='ml.m4.xlarge',\n predictor_cls=DeepARPredictor)",
"----------------------------------------------------------------------------------------------------------------!"
]
],
[
[
"### Make predictions and plot results",
"_____no_output_____"
],
[
"Now we can use the `predictor` object to generate predictions.",
"_____no_output_____"
]
],
[
[
"predictor.predict(ts=timeseries[120], quantiles=[0.10, 0.5, 0.90]).head()",
"/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/ipykernel/__main__.py:19: FutureWarning: Addition/subtraction of integers and integer-arrays to Timestamp is deprecated, will be removed in a future version. Instead of adding/subtracting `n`, use `n * self.freq`\n/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/ipykernel/__main__.py:46: FutureWarning: Creating a DatetimeIndex by passing range endpoints is deprecated. Use `pandas.date_range` instead.\n"
]
],
[
[
"Below we define a plotting function that queries the model and displays the forecast.",
"_____no_output_____"
]
],
[
[
"def plot(\n predictor, \n target_ts, \n cat=None, \n dynamic_feat=None, \n forecast_date=end_training, \n show_samples=False, \n plot_history=7 * 12,\n confidence=80\n):\n print(\"calling served model to generate predictions starting from {}\".format(str(forecast_date)))\n assert(confidence > 50 and confidence < 100)\n low_quantile = 0.5 - confidence * 0.005\n up_quantile = confidence * 0.005 + 0.5\n \n # we first construct the argument to call our model\n args = {\n \"ts\": target_ts[:forecast_date],\n \"return_samples\": show_samples,\n \"quantiles\": [low_quantile, 0.5, up_quantile],\n \"num_samples\": 100\n }\n\n\n if dynamic_feat is not None:\n args[\"dynamic_feat\"] = dynamic_feat\n fig = plt.figure(figsize=(20, 6))\n ax = plt.subplot(2, 1, 1)\n else:\n fig = plt.figure(figsize=(20, 3))\n ax = plt.subplot(1,1,1)\n \n if cat is not None:\n args[\"cat\"] = cat\n ax.text(0.9, 0.9, 'cat = {}'.format(cat), transform=ax.transAxes)\n\n # call the end point to get the prediction\n prediction = predictor.predict(**args)\n\n # plot the samples\n if show_samples: \n for key in prediction.keys():\n if \"sample\" in key:\n prediction[key].plot(color='lightskyblue', alpha=0.2, label='_nolegend_')\n \n \n # plot the target\n target_section = target_ts[forecast_date-plot_history:forecast_date+prediction_length]\n target_section.plot(color=\"black\", label='target')\n \n # plot the confidence interval and the median predicted\n ax.fill_between(\n prediction[str(low_quantile)].index, \n prediction[str(low_quantile)].values, \n prediction[str(up_quantile)].values, \n color=\"b\", alpha=0.3, label='{}% confidence interval'.format(confidence)\n )\n prediction[\"0.5\"].plot(color=\"b\", label='P50')\n ax.legend(loc=2) \n \n # fix the scale as the samples may change it\n ax.set_ylim(target_section.min() * 0.5, target_section.max() * 1.5)\n \n if dynamic_feat is not None:\n for i, f in enumerate(dynamic_feat, start=1):\n ax = plt.subplot(len(dynamic_feat) * 2, 1, len(dynamic_feat) + i, sharex=ax)\n feat_ts = pd.Series(\n index=pd.DatetimeIndex(start=target_ts.index[0], freq=target_ts.index.freq, periods=len(f)),\n data=f\n )\n feat_ts[forecast_date-plot_history:forecast_date+prediction_length].plot(ax=ax, color='g')",
"_____no_output_____"
]
],
[
[
"We can interact with the function previously defined, to look at the forecast of any customer at any point in (future) time. \n\nFor each request, the predictions are obtained by calling our served model on the fly.\n\nHere we forecast the consumption of an office after week-end (note the lower week-end consumption). \nYou can select any time series and any forecast date, just click on `Run Interact` to generate the predictions from our served endpoint and see the plot.",
"_____no_output_____"
]
],
[
[
"style = {'description_width': 'initial'}",
"_____no_output_____"
],
[
"@interact_manual(\n customer_id=IntSlider(min=0, max=369, value=91, style=style), \n forecast_day=IntSlider(min=0, max=100, value=51, style=style),\n confidence=IntSlider(min=60, max=95, value=80, step=5, style=style),\n history_weeks_plot=IntSlider(min=1, max=20, value=1, style=style),\n show_samples=Checkbox(value=False),\n continuous_update=False\n)\ndef plot_interact(customer_id, forecast_day, confidence, history_weeks_plot, show_samples):\n plot(\n predictor,\n target_ts=timeseries[customer_id],\n forecast_date=end_training + datetime.timedelta(days=forecast_day),\n show_samples=show_samples,\n plot_history=history_weeks_plot * 12 * 7,\n confidence=confidence\n )",
"_____no_output_____"
]
],
[
[
"### Delete endpoints",
"_____no_output_____"
]
],
[
[
"predictor.delete_endpoint()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbafc415c2f40ced4bb2317b0eedcc2dad578b3d
| 41,769 |
ipynb
|
Jupyter Notebook
|
ipynb/OCSVM.ipynb
|
SSANGMAN/Anomaly_Detection
|
0607efbbb85e9b52f34e514c73395e5c446c771e
|
[
"MIT"
] | 3 |
2020-01-08T08:20:02.000Z
|
2020-12-09T13:07:27.000Z
|
ipynb/OCSVM.ipynb
|
SSANGMAN/Anomaly_Detection
|
0607efbbb85e9b52f34e514c73395e5c446c771e
|
[
"MIT"
] | null | null | null |
ipynb/OCSVM.ipynb
|
SSANGMAN/Anomaly_Detection
|
0607efbbb85e9b52f34e514c73395e5c446c771e
|
[
"MIT"
] | 1 |
2020-12-09T13:07:29.000Z
|
2020-12-09T13:07:29.000Z
| 128.125767 | 32,744 | 0.86988 |
[
[
[
"import numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\nimport matplotlib.font_manager\n\nfrom sklearn import svm\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import confusion_matrix\nfrom pyod.utils.data import generate_data, get_outliers_inliers\n\nimport warnings\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
]
],
[
[
"## 샘플 데이터 생성\n\nOCSVM은 Unsupervised Learning Method 중 하나이며, Novelty Detection에서 사용되는 방법 중 하나이다. \n\n따라서, 모든 데이터를 정상이라고 가정하고 모델 훈련을 수행해야 한다.\n\n샘플 데이터를 생성하는 과정은 다음과 같다.\n\n- PyoD 라이브러리를 사용하여 샘플 데이터 생성. 이 때, 실제 Outlier 비율은 전체 데이터의 5%로 지정한다.\n\n\n- 전체 데이터에서 훈련 데이터와 테스트 데이터를 분할한다.",
"_____no_output_____"
]
],
[
[
"train, test = generate_data(random_state = 42, train_only = True, contamination = 0.05)\nX_train, X_test, y_train, y_test = train_test_split(train, test, test_size = 0.2, random_state = 42)",
"_____no_output_____"
]
],
[
[
"## 모델 적합\n\n앞서 말했듯이, OCSVM은 라벨 데이터를 필요로하지 않는다. 따라서, 피쳐 데이터만을 이용해 모델을 적합시킨다.",
"_____no_output_____"
]
],
[
[
"clf = svm.OneClassSVM(nu = 0.1, kernel = 'rbf', gamma = 0.1)\nclf.fit(X_train) # Unsupervised Learning Method",
"_____no_output_____"
]
],
[
[
"## 적합 모델을 이용한 라벨 분류",
"_____no_output_____"
]
],
[
[
"class OCSVM:\n def __init__(self, nu, kernel, gamma):\n self.nu = nu\n self.kernel = kernel\n self.gamma = gamma\n self.result_df = pd.DataFrame()\n \n self.clf = svm.OneClassSVM(nu = self.nu, kernel = self.kernel, gamma = self.gamma)\n \n def fit(self, X_train, ground_truth):\n self.X_train = X_train\n self.y_train = ground_truth\n \n self.clf.fit(self.X_train)\n \n return self.clf\n \n def predict(self, X_test, is_return = False):\n self.X_test = X_test\n \n self.prediction = self.clf.predict(self.X_test)\n \n if is_return:\n \n return self.prediction\n \n def visualization(self):\n self.result_df['X1'] = self.X_train[:, 0]\n self.result_df['X2'] = self.X_train[:, 1]\n self.result_df['Prediction'] = pd.Series(self.prediction).apply(lambda x: 0 if x == 1 else 1)\n self.result_df['Actual'] = self.y_train\n \n xx, yy = np.meshgrid(np.linspace(self.result_df['X1'].min() - 1, self.result_df['X1'].max() + 1, 500),\n np.linspace(self.result_df['X2'].min() - 1, self.result_df['X2'].max() + 1, 500))\n \n z = clf.decision_function(np.c_[xx.ravel(), yy.ravel()])\n z = z.reshape(xx.shape)\n \n plt.title(\"Novelty Detection\\nNu = {}, Kernel = {}, Gamma = {}\".format(self.nu, self.kernel, self.gamma))\n plt.contourf(xx, yy, levels = np.linspace(z.min(), 0, 7), cmap = plt.cm.PuBu)\n a = plt.contourf(xx, yy, z, level = [0], linewidths = 2, color = 'darkred')\n plt.contourf(xx, yy, z, levels=[0, z.max()], colors='palevioletred')\n\n s = 40\n b1 = plt.scatter(self.X_train[:, 0], self.X_train[:, 1], c = 'white', s = s, edgecolors = 'k')\n outlier = plt.scatter(self.result_df.loc[self.result_df['Prediction'] == 1]['X1'], self.result_df.loc[self.result_df['Prediction'] == 1]['X2'],\n c = 'red', edgecolor = 'k')\n actual = plt.scatter(self.result_df.loc[self.result_df['Actual'] == 1]['X1'], self.result_df.loc[self.result_df['Actual'] == 1]['X2'],\n c = 'gold', edgecolor = 'k', alpha = 0.8)\n \n plt.axis('tight')\n plt.xlim((self.result_df['X1'].min() - 1, self.result_df['X1'].max() + 1))\n plt.ylim((self.result_df['X2'].min() - 1, self.result_df['X2'].max() + 1))\n \n plt.show()",
"_____no_output_____"
],
[
"nu = 0.1\nkernel = 'rbf'\ngamma = 0.007\n\nmodel = OCSVM(nu = nu, kernel = kernel, gamma = gamma)\nmodel.fit(X_train, y_train)",
"_____no_output_____"
],
[
"model.predict(X_train)",
"_____no_output_____"
]
],
[
[
"## 시각화",
"_____no_output_____"
]
],
[
[
"model.visualization()",
"_____no_output_____"
]
],
[
[
"그래프를 통해 알 수 있다시피, OCSVM 하이퍼파라미터의 Nu는 SVM의 c와 비슷한 의미를 가진다. 다른 의미로 말하면, 오분류 비율에 대한 최대 상한 값이라고 볼 수도 있다. 예를 들어, Nu = 0.05로 설정하면 훈련 데이터의 최대 5%가 잘 못 분류된다고 말할 수 있다.",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.