repo_name
stringlengths 6
130
| hexsha
list | file_path
list | code
list | apis
list |
|---|---|---|---|---|
VoidHaruhi/OpenHGNN | [
"1371c238e7a4cab777333128619c0e4dd9bec2de"
]
| [
"openhgnn/trainerflow/base_flow.py"
]
| [
"import os\nimport torch\nfrom abc import ABC, abstractmethod\n\nfrom ..tasks import build_task\nfrom ..layers.HeteroLinear import HeteroFeature\nfrom ..utils import get_nodes_dict\n\n\nclass BaseFlow(ABC):\n candidate_optimizer = {\n 'Adam': torch.optim.Adam,\n 'SGD': torch.optim.SGD\n }\n\n def __init__(self, args):\n super(BaseFlow, self).__init__()\n self.evaluator = None\n self.evaluate_interval = 1\n self.load_from_checkpoint = True\n if hasattr(args, '_checkpoint'):\n self._checkpoint = os.path.join(args._checkpoint,\n f\"{args.model}_{args.dataset}.pt\")\n else:\n if self.load_from_checkpoint:\n self._checkpoint = os.path.join(\"./openhgnn/output/{}\".format(args.model),\n f\"{args.model}_{args.dataset}.pt\")\n else:\n self._checkpoint = None\n\n if not hasattr(args, 'HGB_results_path') and args.dataset[:3] == 'HGB':\n args.HGB_results_path = os.path.join(\"./openhgnn/output/{}/{}_{}.txt\".format(args.model, args.dataset[5:], args.seed))\n\n self.args = args\n self.model_name = args.model\n self.device = args.device\n self.task = build_task(args)\n self.hg = self.task.get_graph().to(self.device)\n self.args.meta_paths = self.task.dataset.meta_paths\n self.args.meta_paths_dict = self.task.dataset.meta_paths_dict\n self.patience = args.patience\n self.max_epoch = args.max_epoch\n self.optimizer = None\n self.loss_fn = self.task.get_loss_fn()\n\n def preprocess_feature(self):\n r\"\"\"\n Every trainerflow should run the preprocess_feature if you want to get a feature preprocessing.\n The Parameters in input_feature will be added into optimizer and input_feature will be added into the model.\n\n Attributes\n -----------\n input_feature : HeteroFeature\n It will return the processed feature if call it.\n\n \"\"\"\n if hasattr(self.args, 'activation'):\n act = self.args.activation\n else:\n act = None\n # useful type selection\n if self.args.dataset[:3] == 'HGB':\n if self.args.feat == 0:\n print(\"feat0, pass!\")\n pass\n elif self.args.feat == 1:\n h_dict = self.hg.ndata.pop('h')\n if h_dict.get(self.category, False):\n self.hg.ndata['h'] = {self.category: h_dict[self.category]}\n print('feat1, preserve target nodes!')\n elif self.args.feat == 2:\n self.hg.ndata.pop('h')\n print('feat2, drop features!')\n\n if isinstance(self.hg.ndata['h'], dict):\n self.input_feature = HeteroFeature(self.hg.ndata['h'], get_nodes_dict(self.hg), self.args.hidden_dim, act=act).to(self.device)\n elif isinstance(self.hg.ndata['h'], torch.Tensor):\n self.input_feature = HeteroFeature({self.hg.ntypes[0]: self.hg.ndata['h']}, get_nodes_dict(self.hg), self.args.hidden_dim, act=act).to(self.device)\n # else:\n # self.input_feature = HeteroFeature({}, get_nodes_dict(self.hg), self.args.hidden_dim,\n # act=act).to(self.device)\n self.optimizer.add_param_group({'params': self.input_feature.parameters()})\n self.model.add_module('feature', self.input_feature)\n\n @abstractmethod\n def train(self):\n pass\n\n def _full_train_step(self):\n r\"\"\"\n Train with a full_batch graph\n \"\"\"\n raise NotImplementedError\n\n def _mini_train_step(self):\n r\"\"\"\n Train with a mini_batch seed nodes graph\n \"\"\"\n raise NotImplementedError\n\n def _full_test_step(self):\n r\"\"\"\n Test with a full_batch graph\n \"\"\"\n raise NotImplementedError\n\n def _mini_test_step(self):\n r\"\"\"\n Test with a mini_batch seed nodes graph\n \"\"\"\n raise NotImplementedError\n\n def load_from_pretrained(self):\n if self.load_from_checkpoint:\n try:\n ck_pt = torch.load(self._checkpoint)\n self.model.load_state_dict(ck_pt)\n except FileNotFoundError:\n print(f\"'{self._checkpoint}' doesn't exists\")\n return self.model\n\n def save_checkpoint(self):\n if self._checkpoint and hasattr(self.model, \"_parameters()\"):\n torch.save(self.model.state_dict(), self._checkpoint)\n"
]
| [
[
"torch.load"
]
]
|
jiyangchen/benchmarks | [
"cfc870bd2871072accf8cef14b03edcb963bf62b"
]
| [
"scripts/tf_cnn_benchmarks/variable_mgr.py"
]
| [
"# Copyright 2017 The TensorFlow Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n# ==============================================================================\n\"\"\"Defines VariableMgr and subclasses used to manage variables.\n\n\"\"\"\n\nfrom __future__ import print_function\n\nimport tensorflow as tf\n\nimport allreduce\nimport variable_mgr_util\n\n\nclass VariableMgr(object):\n \"\"\"Abstract superclass for class used by BenchmarkCNN to control variables.\n\n Functions on this class are used to control how variables are created and\n managed, and how gradients are computed and applied.\n \"\"\"\n\n def __init__(self, benchmark_cnn):\n self.benchmark_cnn = benchmark_cnn\n self.staging_delta_ops = []\n\n # A variable for automatic loss scaling.\n self.grad_has_inf_nan = None\n\n def each_tower_has_variables(self):\n \"\"\"Returns True if each GPU tower of the model has separate variables.\"\"\"\n assert False, 'Must be implemented in subclass'\n\n def supports_staged_vars(self):\n \"\"\"Whether staged variable management is supported.\"\"\"\n return False\n\n def create_outer_variable_scope(self, device_num):\n \"\"\"Create the tf.variable_scope around all model graph operations.\"\"\"\n del device_num # unused by this implementation\n assert False, 'Must be implemented in subclass'\n\n def preprocess_device_grads(self, device_grads):\n \"\"\"Preprocess the device gradients prior to applying them.\n\n Args:\n device_grads: List of lists of (gradient, variable) tuples.\n device_grads[t][g] = (gradient, variable), where t is the index of the\n tower and g is the index of the gradient-variable pair.\n\n Returns: a tuple of (apply_gradients_devices, gradient_state).\n gradient_state is an opaque structure that should be passed to\n get_gradients_to_apply() and append_apply_gradients_ops() (in that order).\n apply_gradients_devices is a list of devices where the gradients will be\n applied with get_gradients_to_apply() and append_apply_gradients_ops().\n \"\"\"\n del device_grads # unused by this implementation\n assert False, 'Must be implemented in subclass'\n\n def get_gradients_to_apply(self, device_num, gradient_state):\n \"\"\"Returns the [(gradient, variable)] list to apply for device_num.\n\n Args:\n device_num: indexes into apply_gradients_devices, which was returned by an\n earlier call to preprocess_device_grads.\n gradient_state: from previous call to apply_gradients_devices.\n \"\"\"\n del device_num, gradient_state # unused by this implementation\n assert False, 'Must be implemented in subclass'\n\n def append_apply_gradients_ops(self, gradient_state, opt, grads, training_ops,\n loss_scale_params):\n \"\"\"Adds training ops for grads to 'training_ops'.\n\n\n\n Args:\n gradient_state: from previous call to apply_gradients_devices.\n opt: the underlying optimizer\n grads: [(grad, var)] to apply\n training_ops: list to which to add ops\n loss_scale_params: parameters for loss scaling.\n \"\"\"\n del gradient_state # unused by this implementation\n\n def get_apply_gradients_ops_func():\n \"\"\"Returns the apply_gradients op.\"\"\"\n return [opt.apply_gradients(grads)]\n\n variable_mgr_util.append_gradients_with_loss_scale(\n training_ops, get_apply_gradients_ops_func, loss_scale_params,\n self.grad_has_inf_nan)\n\n def get_post_init_ops(self):\n \"\"\"Returns ops that should run post-initialization.\"\"\"\n return []\n\n def get_devices(self):\n \"\"\"Returns devices to use for computation; includes replica selection.\"\"\"\n assert False, 'Must be implemented in subclass'\n\n def savable_variables(self):\n \"\"\"Returns a list/dict of savable variables to pass to tf.train.Saver.\"\"\"\n return tf.global_variables()\n\n def trainable_variables_on_device(self,\n rel_device_num,\n abs_device_num,\n writable=False):\n \"\"\"Return the set of trainable variables on device.\n\n Args:\n rel_device_num: local worker device index.\n abs_device_num: global graph device index.\n writable: whether to get a reference to the underlying variable.\n\n Returns:\n The set of trainable variables on the specified device.\n \"\"\"\n del rel_device_num, writable\n if self.each_tower_has_variables():\n params = [\n v for v in tf.trainable_variables()\n if v.name.startswith('v%s/' % abs_device_num)\n ]\n else:\n params = tf.trainable_variables()\n return params\n\n\nclass VariableMgrIndependent(VariableMgr):\n \"\"\"VariableMgr that implements the --independent mode for local jobs.\n\n Each GPU has its own copy of the variables, and gradients are\n not shared between towers. This can be used to check\n performance when no data is moved between GPUs.\n \"\"\"\n\n def each_tower_has_variables(self):\n return True\n\n def create_outer_variable_scope(self, device_num):\n return tf.variable_scope('v%s' % device_num)\n\n def preprocess_device_grads(self, device_grads):\n return (self.benchmark_cnn.devices, device_grads)\n\n def get_gradients_to_apply(self, device_num, gradient_state):\n device_grads = gradient_state\n return device_grads[device_num]\n\n def get_devices(self):\n return self.benchmark_cnn.raw_devices\n\n\nclass VariableMgrLocalFetchFromPS(VariableMgr):\n \"\"\"VariableMgr that implements the --parameter_server mode for local jobs.\n\n Variables are stored on a parameter server. For each step, each tower gets\n a copy of the variables from the parameter server, and sends its gradients\n to the param server.\n \"\"\"\n\n def each_tower_has_variables(self):\n return False\n\n def create_outer_variable_scope(self, device_num):\n return tf.variable_scope('v', reuse=bool(device_num))\n\n def preprocess_device_grads(self, device_grads):\n return ([self.benchmark_cnn.param_server_device], device_grads)\n\n def get_gradients_to_apply(self, device_num, gradient_state):\n assert device_num == 0\n device_grads = gradient_state\n agg_grads, self.grad_has_inf_nan = (\n variable_mgr_util.\n aggregate_gradients_using_copy_with_variable_colocation(\n device_grads,\n use_mean=True,\n check_inf_nan=self.benchmark_cnn.enable_auto_loss_scale))\n return agg_grads\n\n def get_devices(self):\n raw_devices = self.benchmark_cnn.raw_devices\n if self.benchmark_cnn.local_parameter_device_flag == 'gpu':\n return [\n variable_mgr_util.ParamServerDeviceSetter(d, raw_devices)\n for d in raw_devices\n ]\n else:\n return [\n tf.train.replica_device_setter(\n worker_device=d,\n ps_device=self.benchmark_cnn.param_server_device,\n ps_tasks=1) for d in raw_devices\n ]\n\n\nclass VariableMgrLocalFetchFromStagedPS(VariableMgrLocalFetchFromPS):\n \"\"\"Implements fetching a local variable through staging buffers.\n \"\"\"\n\n def __init__(self, benchmark_cnn):\n super(VariableMgrLocalFetchFromStagedPS, self).__init__(benchmark_cnn)\n # A data structure to track where the variables are used on each device.\n # Indexed by device_num and var_name, each entry stores the \"put\" and \"get\"\n # ops used for that variable on that device:\n # staging_vars_on_devices[device_num][var_name] == (put_op, get_op)\n self.staging_vars_on_devices = [\n dict() for _ in self.benchmark_cnn.raw_devices\n ]\n\n def supports_staged_vars(self):\n return True\n\n def create_outer_variable_scope(self, device_num):\n self._custom_getter = variable_mgr_util.StagedVariableGetter(\n device_num, self.benchmark_cnn.raw_devices, None, self)\n return tf.variable_scope(\n 'v', reuse=bool(device_num), custom_getter=self._custom_getter)\n\n def trainable_variables_on_device(self,\n rel_device_num,\n abs_device_num,\n writable=False):\n return self._custom_getter.trainable_variables_on_device(\n rel_device_num, abs_device_num, writable=writable)\n\n\nclass VariableMgrLocalReplicated(VariableMgr):\n \"\"\"VariableMgr that implements the --replicated mode for local jobs.\n\n Each GPU has its own copy of the variables. To apply gradients,\n either a local all-reduce algorithm is applied or a regular\n cross-device aggregation is used to replicate the combined\n gradients to all towers.\n \"\"\"\n\n def __init__(self, benchmark_cnn, all_reduce_spec, agg_small_grads_max_bytes,\n agg_small_grads_max_group):\n super(VariableMgrLocalReplicated, self).__init__(benchmark_cnn)\n if all_reduce_spec:\n spec = allreduce.parse_all_reduce_spec(all_reduce_spec)\n if len(spec) != 1:\n raise ValueError(\n 'replicated mode does not support hybrid all-reduce strategies')\n self._all_reduce_spec = spec[0]\n else:\n self._all_reduce_spec = None\n self._agg_small_grads_max_bytes = agg_small_grads_max_bytes\n self._agg_small_grads_max_group = agg_small_grads_max_group\n\n def each_tower_has_variables(self):\n return True\n\n def create_outer_variable_scope(self, device_num):\n return tf.variable_scope('v%s' % device_num)\n\n def preprocess_device_grads(self, device_grads):\n if self._all_reduce_spec:\n aggregated_device_grads = allreduce.sum_gradients_all_reduce(\n ['/job:localhost'],\n device_grads,\n 1,\n self._all_reduce_spec.alg,\n self._all_reduce_spec.shards,\n self.benchmark_cnn.gpu_indices,\n agg_small_grads_max_bytes=self._agg_small_grads_max_bytes,\n agg_small_grads_max_group=self._agg_small_grads_max_group)\n else:\n agg_grads, self.grad_has_inf_nan = (\n variable_mgr_util.\n aggregate_gradients_using_copy_with_device_selection(\n self.benchmark_cnn,\n device_grads,\n use_mean=False,\n check_inf_nan=self.benchmark_cnn.enable_auto_loss_scale))\n aggregated_device_grads = []\n for arr in device_grads:\n aggregated_device_grads.append(\n [(g, v) for (_, v), (g, _) in zip(arr, agg_grads)])\n return self.benchmark_cnn.devices, aggregated_device_grads\n\n def get_gradients_to_apply(self, device_num, gradient_state):\n device_grads = gradient_state\n return device_grads[device_num]\n\n def get_post_init_ops(self):\n # Copy initialized values for variables on GPU 0 to other GPUs.\n global_vars = tf.global_variables()\n var_by_name = dict([(v.name, v) for v in global_vars])\n post_init_ops = []\n for v in global_vars:\n split_name = v.name.split('/')\n # TODO(b/62630508): use more specific prefix than v or v0.\n if split_name[0] == 'v0' or not v.name.startswith('v'):\n continue\n split_name[0] = 'v0'\n copy_from = var_by_name['/'.join(split_name)]\n post_init_ops.append(v.assign(copy_from.read_value()))\n return post_init_ops\n\n def savable_variables(self):\n \"\"\"Return the set of variables used for saving/loading the model.\"\"\"\n params = []\n for v in tf.global_variables():\n split_name = v.name.split('/')\n if split_name[0] == 'v0' or not v.name.startswith('v'):\n params.append(v)\n return params\n\n def get_devices(self):\n return self.benchmark_cnn.raw_devices\n\n\nclass VariableMgrDistributedAllReduce(VariableMgr):\n \"\"\"VariableMgr that implements the --distributed_all_reduce mode.\n\n Each GPU has its own copy of the variables. To apply gradients,\n the specified all-reduce algorithm is used to reduce the gradients\n and replicate the final value to all GPUs.\n \"\"\"\n\n def __init__(self, benchmark_cnn, all_reduce_spec, job_name, num_workers,\n agg_small_grads_max_bytes, agg_small_grads_max_group):\n super(VariableMgrDistributedAllReduce, self).__init__(benchmark_cnn)\n if not all_reduce_spec:\n raise ValueError(\n 'distributed_all_reduce requires a non-empty all_reduce_spec')\n self._all_reduce_spec = allreduce.parse_all_reduce_spec(all_reduce_spec)\n self._all_reduce_device_prefixes = (\n allreduce.build_all_reduce_device_prefixes(job_name, num_workers))\n self._num_workers = num_workers\n self._agg_small_grads_max_bytes = agg_small_grads_max_bytes\n self._agg_small_grads_max_group = agg_small_grads_max_group\n if not self._all_reduce_spec:\n raise ValueError('all_reduce_spec must be specified')\n\n def each_tower_has_variables(self):\n return True\n\n def create_outer_variable_scope(self, device_num):\n \"\"\"Create a scope for the named device.\n\n Args:\n device_num: index of device for variable scope. (Note that\n device_num spans all processes in cluster since a single global\n graph is used.)\n\n Returns:\n the requested variable_scope\n \"\"\"\n return tf.variable_scope('v%s' % device_num)\n\n def preprocess_device_grads(self, device_grads):\n remaining_grads = device_grads\n aggregated_grads = []\n for spec_tuple in self._all_reduce_spec:\n if spec_tuple.limit < 0:\n this_grads = remaining_grads\n remaining_grads = []\n else:\n (this_grads, remaining_grads) = allreduce.split_grads_by_size(\n spec_tuple.limit, remaining_grads)\n if this_grads:\n range_agg_grads = allreduce.sum_gradients_all_reduce(\n self._all_reduce_device_prefixes,\n this_grads,\n self._num_workers,\n spec_tuple.alg,\n spec_tuple.shards,\n self.benchmark_cnn.gpu_indices,\n agg_small_grads_max_bytes=self._agg_small_grads_max_bytes,\n agg_small_grads_max_group=self._agg_small_grads_max_group)\n if not aggregated_grads:\n aggregated_grads = range_agg_grads\n else:\n assert len(aggregated_grads) == len(range_agg_grads)\n for i in range(len(aggregated_grads)):\n aggregated_grads[i] += range_agg_grads[i]\n assert not remaining_grads\n full_device_set = []\n for grads in device_grads:\n g, v = grads[0]\n del v\n full_device_set.append(g.device)\n return (full_device_set, aggregated_grads)\n\n def get_gradients_to_apply(self, device_num, gradient_state):\n device_grads = gradient_state\n if device_num >= len(device_grads):\n raise ValueError('device_num %d exceeds length of device_grads (%d)' %\n (device_num, len(device_grads)))\n return device_grads[device_num]\n\n def get_post_init_ops(self):\n \"\"\"Copy initialized values for variables to other devices.\"\"\"\n global_vars = tf.global_variables()\n var_by_name = dict([(v.name, v) for v in global_vars])\n post_init_ops = []\n for v in global_vars:\n split_name = v.name.split('/')\n # TODO(b/62630508): use more specific prefix than v or v0.\n if split_name[0] == 'v0' or not v.name.startswith('v'):\n continue\n split_name[0] = 'v0'\n copy_from = var_by_name['/'.join(split_name)]\n post_init_ops.append(v.assign(copy_from.read_value()))\n return post_init_ops\n\n def savable_variables(self):\n \"\"\"Return the set of variables used for saving/loading the model.\"\"\"\n params = []\n for v in tf.global_variables():\n split_name = v.name.split('/')\n if split_name[0] == 'v0' or not v.name.startswith('v'):\n params.append(v)\n return params\n\n def get_devices(self):\n return self.benchmark_cnn.raw_devices\n\n\nclass VariableMgrDistributedFetchFromPS(VariableMgr):\n \"\"\"Implements --variable_update=parameter_server mode for distributed jobs.\n\n Variables are stored on a parameter server. For each step, each tower gets\n a copy of the variables from the parameter server, and sends its gradients\n to the param server.\n \"\"\"\n\n def each_tower_has_variables(self):\n return False\n\n def create_outer_variable_scope(self, device_num):\n if self.benchmark_cnn.local_parameter_device_flag == 'gpu':\n caching_devices = self.benchmark_cnn.raw_devices\n else:\n caching_devices = [self.benchmark_cnn.cpu_device]\n custom_getter = variable_mgr_util.OverrideCachingDevice(\n caching_devices, self.benchmark_cnn.cpu_device, 1024 * 64)\n return tf.variable_scope(\n 'v', reuse=bool(device_num), custom_getter=custom_getter)\n\n def preprocess_device_grads(self, device_grads):\n # Returns (gradient_devices, gradient_state)\n return ([self.benchmark_cnn.param_server_device], device_grads)\n\n def get_gradients_to_apply(self, device_num, gradient_state):\n assert device_num == 0\n agg_grads, self.grad_has_inf_nan = (\n variable_mgr_util.aggregate_gradients_using_copy(\n gradient_state,\n use_mean=True,\n check_inf_nan=self.benchmark_cnn.enable_auto_loss_scale))\n return agg_grads\n\n def get_devices(self):\n ps_strategy = tf.contrib.training.GreedyLoadBalancingStrategy(\n self.benchmark_cnn.num_ps, tf.contrib.training.byte_size_load_fn)\n return [\n tf.train.replica_device_setter(\n worker_device=d,\n cluster=self.benchmark_cnn.cluster_manager.get_cluster_spec(),\n ps_strategy=ps_strategy) for d in self.benchmark_cnn.raw_devices\n ]\n\n\nclass VariableMgrDistributedFetchFromStagedPS(\n VariableMgrDistributedFetchFromPS):\n \"\"\"Extends VariableMgrDistributedFetchFromPS for --staged_vars.\"\"\"\n\n def __init__(self, benchmark_cnn):\n super(VariableMgrDistributedFetchFromStagedPS, self).__init__(benchmark_cnn)\n self.staging_vars_on_devices = [\n dict() for _ in self.benchmark_cnn.raw_devices\n ]\n self.staged_vars_on_cpu = {}\n\n def create_outer_variable_scope(self, device_num):\n self._custom_getter = variable_mgr_util.StagedVariableGetter(\n device_num, self.benchmark_cnn.raw_devices,\n self.benchmark_cnn.cpu_device, self)\n return tf.variable_scope(\n 'v', reuse=bool(device_num), custom_getter=self._custom_getter)\n\n def supports_staged_vars(self):\n return True\n\n def trainable_variables_on_device(self,\n rel_device_num,\n abs_device_num,\n writable=False):\n return self._custom_getter.trainable_variables_on_device(\n rel_device_num, abs_device_num, writable=writable)\n\n\nclass VariableMgrDistributedReplicated(VariableMgr):\n \"\"\"VariableMgr that implements the --distributed_replicated mode.\n\n Each GPU has a copy of the variables, and updates its copy after the\n parameter servers are all updated with the gradients from all servers. Only\n works with cross_replica_sync=true. Unlike 'replicated', does not use nccl\n all-reduce for replicating within a server.\n \"\"\"\n\n def each_tower_has_variables(self):\n return True\n\n def create_outer_variable_scope(self, device_num):\n return tf.variable_scope(\n 'v%s' % device_num,\n custom_getter=variable_mgr_util.OverrideToLocalVariableIfNotPsVar())\n\n def preprocess_device_grads(self, device_grads):\n return ([self.benchmark_cnn.param_server_device], device_grads)\n\n def get_gradients_to_apply(self, device_num, gradient_state):\n device_grads = gradient_state # From 2nd result of preprocess_device_grads.\n\n avg_grads, self.grad_has_inf_nan = (\n variable_mgr_util.aggregate_gradients_using_copy_with_device_selection(\n self.benchmark_cnn,\n device_grads,\n use_mean=True,\n check_inf_nan=self.benchmark_cnn.enable_auto_loss_scale))\n\n # Make shadow variable on a parameter server for each original trainable\n # variable.\n for i, (g, v) in enumerate(avg_grads):\n my_name = variable_mgr_util.PS_SHADOW_VAR_PREFIX + '/' + v.name\n if my_name.endswith(':0'):\n my_name = my_name[:-2]\n new_v = tf.get_variable(\n my_name,\n dtype=v.dtype.base_dtype,\n initializer=v.initial_value,\n trainable=True)\n avg_grads[i] = (g, new_v)\n return avg_grads\n\n def append_apply_gradients_ops(self, gradient_state, opt, grads, training_ops,\n loss_scale_params):\n device_grads = gradient_state # From 2nd result of preprocess_device_grads.\n\n def get_apply_gradients_ops_func():\n \"\"\"Returns a list of ops for updating gradients.\"\"\"\n apply_gradients_ops = []\n # For each variable, apply the combined gradients for this server on\n # the parameter server, and then wait for all other servers to do this.\n for i, (g, v) in enumerate(grads):\n apply_gradient_op = opt.apply_gradients([(g, v)])\n barrier = self.benchmark_cnn.add_sync_queues_and_barrier(\n 'replicate_variable_%s' % i, [apply_gradient_op])\n with tf.control_dependencies([barrier]):\n with tf.device(self.benchmark_cnn.cpu_device):\n updated_value = v.read_value()\n for my_d in range(len(self.benchmark_cnn.devices)):\n apply_gradients_ops.append(\n device_grads[my_d][i][1].assign(updated_value))\n return apply_gradients_ops\n\n variable_mgr_util.append_gradients_with_loss_scale(\n training_ops, get_apply_gradients_ops_func, loss_scale_params,\n self.grad_has_inf_nan)\n\n def _strip_port(self, s):\n if s.endswith(':0'):\n return s[:-2]\n return s\n\n def get_post_init_ops(self):\n # Copy initialized variables for variables on the parameter server\n # to the local copy of the variable.\n\n local_vars = tf.local_variables()\n local_var_by_name = dict(\n [(self._strip_port(v.name), v) for v in local_vars])\n post_init_ops = []\n for v in tf.global_variables():\n if v.name.startswith(variable_mgr_util.PS_SHADOW_VAR_PREFIX + '/v0/'):\n prefix = self._strip_port(\n v.name[len(variable_mgr_util.PS_SHADOW_VAR_PREFIX + '/v0'):])\n for i in range(self.benchmark_cnn.num_gpus):\n name = 'v%s%s' % (i, prefix)\n if name in local_var_by_name:\n copy_to = local_var_by_name[name]\n post_init_ops.append(copy_to.assign(v.read_value()))\n return post_init_ops\n\n def _remove_shadow_var_prefix_if_present(self, var_name):\n if var_name.startswith(variable_mgr_util.PS_SHADOW_VAR_PREFIX + '/'):\n return var_name[len(variable_mgr_util.PS_SHADOW_VAR_PREFIX + '/'):]\n else:\n return var_name\n\n def var_dict_name(self, v):\n return self._strip_port(self._remove_shadow_var_prefix_if_present(v.name))\n\n def savable_variables(self):\n \"\"\"Returns a list/dict of savable variables to pass to tf.train.Saver.\"\"\"\n params = {}\n for v in tf.global_variables():\n assert (v.name.startswith(variable_mgr_util.PS_SHADOW_VAR_PREFIX + '/v0/')\n or v.name in ('global_step:0', 'loss_scale:0',\n 'loss_scale_normal_steps:0')), (\n 'Invalid global variable: %s' % v)\n # We store variables in the checkpoint with the shadow variable prefix\n # removed so we can evaluate checkpoints in non-distributed replicated\n # mode. The checkpoints can also be loaded for training in\n # distributed_replicated mode.\n name = self._strip_port(self._remove_shadow_var_prefix_if_present(v.name))\n params[name] = v\n for v in tf.local_variables():\n # Non-trainable variables, such as batch norm moving averages, do not have\n # corresponding global shadow variables, so we add them here. Trainable\n # local variables have corresponding global shadow variables, which were\n # added in the global variable loop above.\n if v.name.startswith('v0/') and v not in tf.trainable_variables():\n params[self._strip_port(v.name)] = v\n return params\n\n def get_devices(self):\n return self.benchmark_cnn.raw_devices\n"
]
| [
[
"tensorflow.get_variable",
"tensorflow.device",
"tensorflow.control_dependencies",
"tensorflow.global_variables",
"tensorflow.local_variables",
"tensorflow.contrib.training.GreedyLoadBalancingStrategy",
"tensorflow.trainable_variables",
"tensorflow.train.replica_device_setter",
"tensorflow.variable_scope"
]
]
|
ult-processor/tensortrade | [
"c2848f3bc33295085c31b8ad774c6e12f23210ea"
]
| [
"tensortrade/features/scalers/standard_normalizer.py"
]
| [
"# Copyright 2019 The TensorTrade Authors.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License\n\nimport pandas as pd\nimport numpy as np\n\nfrom typing import Union, List, Tuple\nfrom sklearn.preprocessing import StandardScaler\n\nfrom tensortrade.features.transformer import Transformer, TransformableList\n\n\nclass StandardNormalizer(Transformer):\n \"\"\"A transformer for normalizing values within a feature pipeline by removing the mean and scaling to unit variance.\"\"\"\n\n def __init__(self, columns: Union[List[str], str] = None):\n \"\"\"\n Arguments:\n columns (optional): A list of column names to normalize.\n \"\"\"\n self._columns = columns\n self._scaler = StandardScaler()\n\n def transform(self, X: TransformableList, y: TransformableList = None):\n if self._columns is None:\n return self._scaler.fit_transform(X, y)\n\n return self._scaler.fit_transform(X[self._columns], y)\n"
]
| [
[
"sklearn.preprocessing.StandardScaler"
]
]
|
laurencer/functorch | [
"1bc4093b09f7a69606ff3fd2e6c76ffd55d4ac13"
]
| [
"test/test_pythonkey.py"
]
| [
"# Copyright (c) Facebook, Inc. and its affiliates.\n# All rights reserved.\n#\n# This source code is licensed under the BSD-style license found in the\n# LICENSE file in the root directory of this source tree.\n\nfrom torch.testing._internal.common_utils import TestCase, run_tests\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport unittest\nimport functools\nimport itertools\nimport warnings\nimport math\nfrom typing import Callable, Type\nfrom torch.testing._internal.common_device_type import instantiate_device_type_tests, \\\n skipCUDAIfNoMagma, onlyOnCPUAndCUDA, onlyCPU\nimport types\nfrom functools import partial\n\nimport functorch\nfrom functorch import (\n grad, vjp, vmap, jacrev, grad_and_value,\n make_functional_deprecated_v1, make_functional_with_buffers_deprecated_v1, make_fx, nnc_jit\n)\n\n# NB: numpy is a testing dependency!\nimport numpy as np\n\nclass TestPythonKey(TestCase):\n def test_make_fx(self, device):\n def f(x):\n return torch.sin(x)\n inp = torch.randn(3)\n fx_f = make_fx(f)(inp)\n\n new_inp = torch.randn(3)\n self.assertEqual(fx_f(new_inp), f(new_inp))\n\n def test_make_fx_grad(self, device):\n def f(x):\n return torch.sin(x).sum()\n inp = torch.randn(3)\n f = grad(f)\n fx_f = make_fx(f)(inp)\n\n new_inp = torch.randn(3)\n self.assertEqual(fx_f(new_inp), f(new_inp))\n\n def test_make_fx_vmap(self, device):\n def f(x):\n return torch.sin(x)\n inp = torch.randn(5, 3)\n f = vmap(f)\n fx_f = make_fx(f)(inp)\n new_inp = torch.randn(5, 3)\n self.assertEqual(fx_f(new_inp), f(new_inp))\n\n def test_make_fx_jacrev(self, device):\n def f(x):\n return x.sin().sum()\n inp = torch.randn(3)\n f = jacrev(jacrev(f))\n fx_f = make_fx(f)(inp)\n new_inp = torch.randn(3)\n self.assertEqual(fx_f(new_inp), f(new_inp))\n\n def test_make_fx_jvp(self, device):\n def f(x):\n return torch.sin(x).sum()\n\n primals = torch.randn(3)\n _, vjp_fn = vjp(f, primals)\n cotangent = torch.randn(())\n fx_f = make_fx(vjp_fn)(cotangent, True, True)\n new_cotangent = torch.randn(())\n self.assertEqual(fx_f(new_cotangent, True, True), vjp_fn(new_cotangent))\n\n def test_nnc_jit(self, device):\n def f(x):\n return torch.sin(x)\n\n jit_f = nnc_jit(f)\n\n inp = torch.randn(3)\n self.assertEqual(jit_f(inp), f(inp))\n\n def test_nnc_scalar(self, device):\n def f(x):\n return torch.sin(x)\n\n jit_f = nnc_jit(f)\n\n inp = torch.randn(())\n self.assertEqual(jit_f(inp), f(inp))\n\n def test_nnc_pytrees(self, device):\n def f(x):\n return [torch.sin(x[0])]\n\n jit_f = nnc_jit(f)\n\n inp = [torch.randn(3)]\n self.assertEqual(jit_f(inp), f(inp))\n\n def test_external_calls(self, device):\n def f(a, b):\n return torch.mv(a, b)\n jit_f = nnc_jit(f)\n inp = [torch.randn(3, 3), torch.randn(3)]\n self.assertEqual(jit_f(*inp), f(*inp))\n\n def test_nnc_passthrough(self, device):\n def f(x, y):\n return x + y, y\n inp = (torch.randn(3), torch.randn(3))\n jit_f = nnc_jit(f)\n self.assertEqual(jit_f(*inp), f(*inp))\n\n def f(x):\n x['a'] = x['a'] * 2\n return x\n inp = ({'a': torch.randn(3), 'b': torch.randn(3)},)\n jit_f = nnc_jit(f)\n self.assertEqual(jit_f(*inp), f(*inp))\n\n\n\n\nonly_for = (\"cpu\")\ninstantiate_device_type_tests(\n TestPythonKey,\n globals(),\n only_for=only_for,\n)\n\n\nif __name__ == '__main__':\n run_tests()\n"
]
| [
[
"torch.randn",
"torch.mv",
"torch.testing._internal.common_utils.run_tests",
"torch.sin"
]
]
|
NASLab/GroundROS | [
"6673db009ffcff59500eb1e3d5873111282e7749"
]
| [
"src/experimental_results/navigation_test/path_planning_analysis.py"
]
| [
"# python experimental tests for Husky\n\nimport numpy as np\nfrom numpy import sin, cos, pi\nimport matplotlib.pyplot as plt\n\nyaw_bound = 2 * pi / 180\nyaw_calibrate = pi / 180 * (0)\nx_offset_calibrate = .23\ny_offset_calibrate = -.08\n\n\ndata = np.load('pos.npy')[1:]\n# print len(data)\nerror_long = data[:, 0]\nerror_lat = data[:, 1]\nref_x = [value for value in data[:, 2]]\n# print ref_x[:30]\nref_y = [value for value in data[:, 3]]\npos_x = [value for value in data[:, 4]][0::1]\npos_y = [value for value in data[:, 5]][0::1]\npos_theta = data[:, 6]\n# print data\ntime = data[:, 7] - data[0, 7]\nvel = data[:, 8]\n# plt.plot(ref_x, ref_y, 'ro')\n# plt.gca().set_aspect('equal', adjustable='box')\nf0 = plt.figure(1, figsize=(9, 9))\nax0 = f0.add_subplot(111)\n# ax0.plot(ref_x, ref_y, '--', lw=3, label='Reference Trajectory')\nax0.plot(pos_x[0], pos_y[0], 'ms', markersize=10, label='Start Point')\n# ax0.plot(pos_x, pos_y, 'go', label='Robot Trajectory')\nenv_data = np.load('env.npy')[1:]\nx = [[]] * len(env_data)\ny = [[]] * len(env_data)\n# print len(env_data)\n\nm=5\nfor i in range(m, len(env_data) - m):\n if len(env_data[i]) > 0:\n x[i] = env_data[i][0]\n y[i] = env_data[i][1]\n yaw = env_data[i][2]\n\n # filter some of the readings; comment to see the effect\n if len(env_data[i + m]) == 0 or abs(yaw - env_data[i - m][2]) > yaw_bound or abs(yaw - env_data[i + m][2]) > yaw_bound:\n continue\n\n readings = env_data[i][3]\n readings_x = [[]] * len(readings)\n readings_y = [[]] * len(readings)\n k = 0\n for j in range(len(readings)):\n # lidar readings in lidar frame\n x_temp = readings[j][0] * cos(-readings[j][1])\n y_temp = readings[j][0] * sin(-readings[j][1])\n\n # lidar readings in robot frame\n x_temp2 = x_temp * \\\n cos(yaw_calibrate) - y_temp * \\\n sin(yaw_calibrate) + x_offset_calibrate\n y_temp2 = y_temp * \\\n cos(yaw_calibrate) + x_temp * \\\n sin(yaw_calibrate) + y_offset_calibrate\n\n # lidar readings in global frame\n readings_x[k] = x_temp2 * cos(yaw) - y_temp2 * sin(yaw) + x[i]\n readings_y[k] = y_temp2 * cos(yaw) + x_temp2 * sin(yaw) + y[i]\n k += 1\n\n ax0.plot(readings_x, readings_y, 'r.')\n\n# for i in range(len(env_data)):\n# if len(env_data[i])>0:\n# x[i] = env_data[i][0]\n# y[i] = env_data[i][1]\n# yaw = env_data[i][2]\n# print yaw\n# readings = env_data[i][3]\n# readings_x = [[]]*len(readings)\n# readings_y = [[]]*len(readings)\n# print len(readings),len(readings_x)\n# k=0\n# for j in range(len(readings)):\n# if i<200:\n# print k,j,len(readings_x)\n# readings_x[k] = x[i] + readings[j][0]*sin(pi/2-yaw+readings[j][1])\n# readings_y[k] = y[i] + readings[j][0]*cos(pi/2-yaw+readings[j][1])\n# k+=1\n# ax0.plot(readings_x, readings_y,'r.')\n\nax0.plot([], [], 'r.', label='Lidar Reading')\n# print x\nax0.plot([value for value in x if value],\n [value for value in y if value], 'g', lw=3,label='Robot\\'s Trajectory')\n\n\n# env_y = np.load('env.npy')[1]\n# env_x = [value for value in env_x if value]\n# env_y = [value for value in env_y if value]\n# ax0.plot(env_x, env_y, 'r.', )\nax0.plot(-.5, 2.7, 'cs', markersize=10, label='Destination')\nax0.legend()\nax0.axis('equal')\nax0.set_xlim(-3.5, 3.5)\nax0.set_ylim(-3, 4)\nax0.set_xlabel('X (m)')\nax0.set_ylabel('Y (m)')\n# ax0.axis('equal')\n\nplt.tight_layout()\nplt.draw()\nplt.pause(.1) # <-------\nraw_input(\"<Hit Enter To Close>\")\nplt.close(f0)\n"
]
| [
[
"matplotlib.pyplot.tight_layout",
"numpy.cos",
"matplotlib.pyplot.draw",
"numpy.sin",
"matplotlib.pyplot.close",
"numpy.load",
"matplotlib.pyplot.pause",
"matplotlib.pyplot.figure"
]
]
|
Salonijain27/cuml | [
"887575445ee0b162f4ac02feae86c0d7de14f793"
]
| [
"python/cuml/test/test_mbsgd_classifier.py"
]
| [
"# Copyright (c) 2019, NVIDIA CORPORATION.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n\nimport numpy as np\nimport pytest\n\nfrom cuml.linear_model import MBSGDClassifier as cumlMBSGClassifier\nfrom cuml.test.utils import unit_param, quality_param, stress_param\n\nfrom sklearn.linear_model import SGDClassifier\nfrom sklearn.datasets.samples_generator import make_classification\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.model_selection import train_test_split\n\n\[email protected](scope=\"module\", params=[\n unit_param([500, 20, 10, np.float32]),\n unit_param([500, 20, 10, np.float64]),\n quality_param([5000, 100, 50, np.float32]),\n quality_param([5000, 100, 50, np.float64]),\n stress_param([500000, 1000, 500, np.float32]),\n stress_param([500000, 1000, 500, np.float64]),\n], ids=['500-20-10-f32', '500-20-10-f64',\n '5000-100-50-f32', '5000-100-50-f64',\n '500000-1000-500-f32', '500000-1000-500-f64'])\ndef make_dataset(request):\n nrows, ncols, n_info, datatype = request.param\n X, y = make_classification(n_samples=nrows, n_informative=n_info,\n n_features=ncols, random_state=0)\n X = X.astype(datatype)\n y = y.astype(datatype)\n X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.8,\n random_state=10)\n\n y_train = y_train.astype(datatype)\n y_test = y_test.astype(datatype)\n\n return nrows, X_train, X_test, y_train, y_test\n\n\[email protected](\n # Grouped those tests to reduce the total number of individual tests\n # while still keeping good coverage of the different features of MBSGD\n ('lrate', 'penalty', 'loss'), [\n ('constant', 'none', 'log'),\n ('invscaling', 'l2', 'hinge'),\n ('adaptive', 'l1', 'squared_loss'),\n ('constant', 'elasticnet', 'hinge'),\n ]\n)\ndef test_mbsgd_classifier(lrate, penalty, loss, make_dataset):\n nrows, X_train, X_test, y_train, y_test = make_dataset\n\n cu_mbsgd_classifier = cumlMBSGClassifier(learning_rate=lrate, eta0=0.005,\n epochs=100, fit_intercept=True,\n batch_size=2, tol=0.0,\n penalty=penalty)\n\n cu_mbsgd_classifier.fit(X_train, y_train)\n cu_pred = cu_mbsgd_classifier.predict(X_test).to_array()\n cu_acc = accuracy_score(cu_pred, y_test)\n\n if nrows < 500000:\n skl_sgd_classifier = SGDClassifier(learning_rate=lrate, eta0=0.005,\n max_iter=100, fit_intercept=True,\n tol=0.0, penalty=penalty,\n random_state=0)\n\n skl_sgd_classifier.fit(X_train, y_train)\n skl_pred = skl_sgd_classifier.predict(X_test)\n skl_acc = accuracy_score(skl_pred, y_test)\n assert cu_acc >= skl_acc - 0.06\n\n\ndef test_mbsgd_classifier_default(make_dataset):\n nrows, X_train, X_test, y_train, y_test = make_dataset\n\n cu_mbsgd_classifier = cumlMBSGClassifier()\n\n cu_mbsgd_classifier.fit(X_train, y_train)\n cu_pred = cu_mbsgd_classifier.predict(X_test).to_array()\n cu_acc = accuracy_score(cu_pred, y_test)\n\n if nrows < 500000:\n skl_sgd_classifier = SGDClassifier()\n\n skl_sgd_classifier.fit(X_train, y_train)\n skl_pred = skl_sgd_classifier.predict(X_test)\n skl_acc = accuracy_score(skl_pred, y_test)\n assert cu_acc >= skl_acc - 0.05\n"
]
| [
[
"sklearn.datasets.samples_generator.make_classification",
"sklearn.model_selection.train_test_split",
"sklearn.linear_model.SGDClassifier",
"sklearn.metrics.accuracy_score"
]
]
|
Howdy-Personally/detrold | [
"5b3685db5fc114dd7770fd9a13ebe86f01756fc0"
]
| [
"util/misc.py"
]
| [
"# Copyright (c) Facebook, Inc. and its affiliates. All Rights Reserved\r\n\"\"\"\r\nMisc functions, including distributed helpers.\r\n\r\nMostly copy-paste from torchvision references.\r\n\"\"\"\r\nimport os\r\nimport subprocess\r\nimport time\r\nfrom collections import defaultdict, deque\r\nimport datetime\r\nimport pickle\r\nfrom typing import Optional, List\r\n\r\nimport torch\r\nimport torch.distributed as dist\r\nfrom torch import Tensor\r\n\r\n# needed due to empty tensor bug in pytorch and torchvision 0.5\r\nimport torchvision\r\nif float(torchvision.__version__[:3]) < 0.7:\r\n from torchvision.ops import _new_empty_tensor\r\n from torchvision.ops.misc import _output_size\r\n\r\n\r\nclass SmoothedValue(object):\r\n \"\"\"Track a series of values and provide access to smoothed values over a\r\n window or the global series average.\r\n \"\"\"\r\n\r\n def __init__(self, window_size=20, fmt=None):\r\n if fmt is None:\r\n fmt = \"{median:.4f} ({global_avg:.4f})\"\r\n self.deque = deque(maxlen=window_size)\r\n self.total = 0.0\r\n self.count = 0\r\n self.fmt = fmt\r\n\r\n def update(self, value, n=1):\r\n self.deque.append(value)\r\n self.count += n\r\n self.total += value * n\r\n\r\n def synchronize_between_processes(self):\r\n \"\"\"\r\n Warning: does not synchronize the deque!\r\n \"\"\"\r\n if not is_dist_avail_and_initialized():\r\n return\r\n t = torch.tensor([self.count, self.total], dtype=torch.float64, device='cuda')\r\n dist.barrier()\r\n dist.all_reduce(t)\r\n t = t.tolist()\r\n self.count = int(t[0])\r\n self.total = t[1]\r\n\r\n @property\r\n def median(self):\r\n d = torch.tensor(list(self.deque))\r\n return d.median().item()\r\n\r\n @property\r\n def avg(self):\r\n d = torch.tensor(list(self.deque), dtype=torch.float32)\r\n return d.mean().item()\r\n\r\n @property\r\n def global_avg(self):\r\n return self.total / self.count\r\n\r\n @property\r\n def max(self):\r\n return max(self.deque)\r\n\r\n @property\r\n def value(self):\r\n return self.deque[-1]\r\n\r\n def __str__(self):\r\n return self.fmt.format(\r\n median=self.median,\r\n avg=self.avg,\r\n global_avg=self.global_avg,\r\n max=self.max,\r\n value=self.value)\r\n\r\n\r\ndef all_gather(data):\r\n \"\"\"\r\n Run all_gather on arbitrary picklable data (not necessarily tensors)\r\n Args:\r\n data: any picklable object\r\n Returns:\r\n list[data]: list of data gathered from each rank\r\n \"\"\"\r\n world_size = get_world_size()\r\n if world_size == 1:\r\n return [data]\r\n\r\n # serialized to a Tensor\r\n buffer = pickle.dumps(data)\r\n storage = torch.ByteStorage.from_buffer(buffer)\r\n tensor = torch.ByteTensor(storage).to(\"cuda\")\r\n\r\n # obtain Tensor size of each rank\r\n local_size = torch.tensor([tensor.numel()], device=\"cuda\")\r\n size_list = [torch.tensor([0], device=\"cuda\") for _ in range(world_size)]\r\n dist.all_gather(size_list, local_size)\r\n size_list = [int(size.item()) for size in size_list]\r\n max_size = max(size_list)\r\n\r\n # receiving Tensor from all ranks\r\n # we pad the tensor because torch all_gather does not support\r\n # gathering tensors of different shapes\r\n tensor_list = []\r\n for _ in size_list:\r\n tensor_list.append(torch.empty((max_size,), dtype=torch.uint8, device=\"cuda\"))\r\n if local_size != max_size:\r\n padding = torch.empty(size=(max_size - local_size,), dtype=torch.uint8, device=\"cuda\")\r\n tensor = torch.cat((tensor, padding), dim=0)\r\n dist.all_gather(tensor_list, tensor)\r\n\r\n data_list = []\r\n for size, tensor in zip(size_list, tensor_list):\r\n buffer = tensor.cpu().numpy().tobytes()[:size]\r\n data_list.append(pickle.loads(buffer))\r\n\r\n return data_list\r\n\r\n\r\ndef reduce_dict(input_dict, average=True):\r\n \"\"\"\r\n Args:\r\n input_dict (dict): all the values will be reduced\r\n average (bool): whether to do average or sum\r\n Reduce the values in the dictionary from all processes so that all processes\r\n have the averaged results. Returns a dict with the same fields as\r\n input_dict, after reduction.\r\n \"\"\"\r\n world_size = get_world_size()\r\n if world_size < 2:\r\n return input_dict\r\n with torch.no_grad():\r\n names = []\r\n values = []\r\n # sort the keys so that they are consistent across processes\r\n for k in sorted(input_dict.keys()):\r\n names.append(k)\r\n values.append(input_dict[k])\r\n values = torch.stack(values, dim=0)\r\n dist.all_reduce(values)\r\n if average:\r\n values /= world_size\r\n reduced_dict = {k: v for k, v in zip(names, values)}\r\n return reduced_dict\r\n\r\n\r\nclass MetricLogger(object):\r\n def __init__(self, delimiter=\"\\t\"):\r\n self.meters = defaultdict(SmoothedValue)\r\n self.delimiter = delimiter\r\n\r\n def update(self, **kwargs):\r\n for k, v in kwargs.items():\r\n if isinstance(v, torch.Tensor):\r\n v = v.item()\r\n assert isinstance(v, (float, int))\r\n self.meters[k].update(v)\r\n\r\n def __getattr__(self, attr):\r\n if attr in self.meters:\r\n return self.meters[attr]\r\n if attr in self.__dict__:\r\n return self.__dict__[attr]\r\n raise AttributeError(\"'{}' object has no attribute '{}'\".format(\r\n type(self).__name__, attr))\r\n\r\n def __str__(self):\r\n loss_str = []\r\n for name, meter in self.meters.items():\r\n loss_str.append(\r\n \"{}: {}\".format(name, str(meter))\r\n )\r\n return self.delimiter.join(loss_str)\r\n\r\n def synchronize_between_processes(self):\r\n for meter in self.meters.values():\r\n meter.synchronize_between_processes()\r\n\r\n def add_meter(self, name, meter):\r\n self.meters[name] = meter\r\n\r\n def log_every(self, iterable, print_freq, header=None):\r\n i = 0\r\n if not header:\r\n header = ''\r\n start_time = time.time()\r\n end = time.time()\r\n iter_time = SmoothedValue(fmt='{avg:.4f}')\r\n data_time = SmoothedValue(fmt='{avg:.4f}')\r\n space_fmt = ':' + str(len(str(len(iterable)))) + 'd'\r\n if torch.cuda.is_available():\r\n log_msg = self.delimiter.join([\r\n header,\r\n '[{0' + space_fmt + '}/{1}]',\r\n 'eta: {eta}',\r\n '{meters}',\r\n 'time: {time}',\r\n 'data: {data}',\r\n 'max mem: {memory:.0f}'\r\n ])\r\n else:\r\n log_msg = self.delimiter.join([\r\n header,\r\n '[{0' + space_fmt + '}/{1}]',\r\n 'eta: {eta}',\r\n '{meters}',\r\n 'time: {time}',\r\n 'data: {data}'\r\n ])\r\n MB = 1024.0 * 1024.0\r\n for obj in iterable:\r\n data_time.update(time.time() - end)\r\n yield obj\r\n iter_time.update(time.time() - end)\r\n if i % print_freq == 0 or i == len(iterable) - 1:\r\n eta_seconds = iter_time.global_avg * (len(iterable) - i)\r\n eta_string = str(datetime.timedelta(seconds=int(eta_seconds)))\r\n if torch.cuda.is_available():\r\n print(log_msg.format(\r\n i, len(iterable), eta=eta_string,\r\n meters=str(self),\r\n time=str(iter_time), data=str(data_time),\r\n memory=torch.cuda.max_memory_allocated() / MB))\r\n else:\r\n print(log_msg.format(\r\n i, len(iterable), eta=eta_string,\r\n meters=str(self),\r\n time=str(iter_time), data=str(data_time)))\r\n i += 1\r\n end = time.time()\r\n total_time = time.time() - start_time\r\n total_time_str = str(datetime.timedelta(seconds=int(total_time)))\r\n print('{} Total time: {} ({:.4f} s / it)'.format(\r\n header, total_time_str, total_time / len(iterable)))\r\n\r\n\r\ndef get_sha():\r\n cwd = os.path.dirname(os.path.abspath(__file__))\r\n\r\n def _run(command):\r\n return subprocess.check_output(command, cwd=cwd).decode('ascii').strip()\r\n sha = 'N/A'\r\n diff = \"clean\"\r\n branch = 'N/A'\r\n try:\r\n sha = _run(['git', 'rev-parse', 'HEAD'])\r\n subprocess.check_output(['git', 'diff'], cwd=cwd)\r\n diff = _run(['git', 'diff-index', 'HEAD'])\r\n diff = \"has uncommited changes\" if diff else \"clean\"\r\n branch = _run(['git', 'rev-parse', '--abbrev-ref', 'HEAD'])\r\n except Exception:\r\n pass\r\n message = f\"sha: {sha}, status: {diff}, branch: {branch}\"\r\n return message\r\n\r\n\r\ndef collate_fn(batch):\r\n batch = list(zip(*batch))\r\n batch[0] = nested_tensor_from_tensor_list(batch[0])\r\n return tuple(batch)\r\n\r\n\r\ndef _max_by_axis(the_list):\r\n # type: (List[List[int]]) -> List[int]\r\n maxes = the_list[0]\r\n for sublist in the_list[1:]:\r\n for index, item in enumerate(sublist):\r\n maxes[index] = max(maxes[index], item)\r\n return maxes\r\n\r\n\r\nclass NestedTensor(object):\r\n def __init__(self, tensors, mask: Optional[Tensor]):\r\n self.tensors = tensors\r\n self.mask = mask\r\n\r\n def to(self, device):\r\n # type: (Device) -> NestedTensor # noqa\r\n cast_tensor = self.tensors.to(device)\r\n mask = self.mask\r\n if mask is not None:\r\n assert mask is not None\r\n cast_mask = mask.to(device)\r\n else:\r\n cast_mask = None\r\n return NestedTensor(cast_tensor, cast_mask)\r\n\r\n def decompose(self):\r\n return self.tensors, self.mask\r\n\r\n def __repr__(self):\r\n return str(self.tensors)\r\n\r\n\r\ndef nested_tensor_from_tensor_list(tensor_list: List[Tensor]):\r\n # TODO make this more general\r\n if tensor_list[0].ndim == 3:\r\n if torchvision._is_tracing():\r\n # nested_tensor_from_tensor_list() does not export well to ONNX\r\n # call _onnx_nested_tensor_from_tensor_list() instead\r\n return _onnx_nested_tensor_from_tensor_list(tensor_list)\r\n\r\n # TODO make it support different-sized images\r\n max_size = _max_by_axis([list(img.shape) for img in tensor_list])\r\n # min_size = tuple(min(s) for s in zip(*[img.shape for img in tensor_list]))\r\n batch_shape = [len(tensor_list)] + max_size\r\n b, c, h, w = batch_shape\r\n dtype = tensor_list[0].dtype\r\n device = tensor_list[0].device\r\n tensor = torch.zeros(batch_shape, dtype=dtype, device=device)\r\n mask = torch.ones((b, h, w), dtype=torch.bool, device=device)\r\n for img, pad_img, m in zip(tensor_list, tensor, mask):\r\n pad_img[: img.shape[0], : img.shape[1], : img.shape[2]].copy_(img)\r\n m[: img.shape[1], :img.shape[2]] = False\r\n else:\r\n raise ValueError('not supported')\r\n return NestedTensor(tensor, mask)\r\n\r\n\r\n# _onnx_nested_tensor_from_tensor_list() is an implementation of\r\n# nested_tensor_from_tensor_list() that is supported by ONNX tracing.\r\[email protected]\r\ndef _onnx_nested_tensor_from_tensor_list(tensor_list: List[Tensor]) -> NestedTensor:\r\n max_size = []\r\n for i in range(tensor_list[0].dim()):\r\n max_size_i = torch.max(torch.stack([img.shape[i] for img in tensor_list]).to(torch.float32)).to(torch.int64)\r\n max_size.append(max_size_i)\r\n max_size = tuple(max_size)\r\n\r\n # work around for\r\n # pad_img[: img.shape[0], : img.shape[1], : img.shape[2]].copy_(img)\r\n # m[: img.shape[1], :img.shape[2]] = False\r\n # which is not yet supported in onnx\r\n padded_imgs = []\r\n padded_masks = []\r\n for img in tensor_list:\r\n padding = [(s1 - s2) for s1, s2 in zip(max_size, tuple(img.shape))]\r\n padded_img = torch.nn.functional.pad(img, (0, padding[2], 0, padding[1], 0, padding[0]))\r\n padded_imgs.append(padded_img)\r\n\r\n m = torch.zeros_like(img[0], dtype=torch.int, device=img.device)\r\n padded_mask = torch.nn.functional.pad(m, (0, padding[2], 0, padding[1]), \"constant\", 1)\r\n padded_masks.append(padded_mask.to(torch.bool))\r\n\r\n tensor = torch.stack(padded_imgs)\r\n mask = torch.stack(padded_masks)\r\n\r\n return NestedTensor(tensor, mask=mask)\r\n\r\n\r\ndef setup_for_distributed(is_master):\r\n \"\"\"\r\n This function disables printing when not in master process\r\n \"\"\"\r\n import builtins as __builtin__\r\n builtin_print = __builtin__.print\r\n\r\n def print(*args, **kwargs):\r\n force = kwargs.pop('force', False)\r\n if is_master or force:\r\n builtin_print(*args, **kwargs)\r\n\r\n __builtin__.print = print\r\n\r\n\r\ndef is_dist_avail_and_initialized():\r\n if not dist.is_available():\r\n return False\r\n if not dist.is_initialized():\r\n return False\r\n return True\r\n\r\n\r\ndef get_world_size():\r\n if not is_dist_avail_and_initialized():\r\n return 1\r\n return dist.get_world_size()\r\n\r\n\r\ndef get_rank():\r\n if not is_dist_avail_and_initialized():\r\n return 0\r\n return dist.get_rank()\r\n\r\n\r\ndef is_main_process():\r\n return get_rank() == 0\r\n\r\n\r\ndef save_on_master(*args, **kwargs):\r\n if is_main_process():\r\n torch.save(*args, **kwargs)\r\n\r\n\r\ndef init_distributed_mode(args):\r\n if 'RANK' in os.environ and 'WORLD_SIZE' in os.environ:\r\n args.rank = int(os.environ[\"RANK\"])\r\n args.world_size = int(os.environ['WORLD_SIZE'])\r\n args.gpu = int(os.environ['LOCAL_RANK'])\r\n elif 'SLURM_PROCID' in os.environ:\r\n args.rank = int(os.environ['SLURM_PROCID'])\r\n args.gpu = args.rank % torch.cuda.device_count()\r\n else:\r\n print('Not using distributed mode')\r\n args.distributed = False\r\n return\r\n\r\n args.distributed = True\r\n\r\n torch.cuda.set_device(args.gpu)\r\n args.dist_backend = 'nccl'\r\n print('| distributed init (rank {}): {}'.format(\r\n args.rank, args.dist_url), flush=True)\r\n torch.distributed.init_process_group(backend=args.dist_backend, init_method=args.dist_url,\r\n world_size=args.world_size, rank=args.rank)\r\n torch.distributed.barrier()\r\n setup_for_distributed(args.rank == 0)\r\n\r\n\r\[email protected]_grad()\r\ndef accuracy(output, target, topk=(1,)):\r\n \"\"\"Computes the precision@k for the specified values of k\"\"\"\r\n if target.numel() == 0:\r\n return [torch.zeros([], device=output.device)]\r\n maxk = max(topk)\r\n batch_size = target.size(0)\r\n\r\n _, pred = output.topk(maxk, 1, True, True)\r\n pred = pred.t()\r\n correct = pred.eq(target.view(1, -1).expand_as(pred))\r\n\r\n res = []\r\n for k in topk:\r\n correct_k = correct[:k].view(-1).float().sum(0)\r\n res.append(correct_k.mul_(100.0 / batch_size))\r\n return res\r\n\r\n\r\ndef interpolate(input, size=None, scale_factor=None, mode=\"nearest\", align_corners=None):\r\n # type: (Tensor, Optional[List[int]], Optional[float], str, Optional[bool]) -> Tensor\r\n \"\"\"\r\n Equivalent to nn.functional.interpolate, but with support for empty batch sizes.\r\n This will eventually be supported natively by PyTorch, and this\r\n class can go away.\r\n \"\"\"\r\n if float(torchvision.__version__[:3]) < 0.7:\r\n if input.numel() > 0:\r\n return torch.nn.functional.interpolate(\r\n input, size, scale_factor, mode, align_corners\r\n )\r\n\r\n output_shape = _output_size(2, input, size, scale_factor)\r\n output_shape = list(input.shape[:-2]) + list(output_shape)\r\n return _new_empty_tensor(input, output_shape)\r\n else:\r\n return torchvision.ops.misc.interpolate(input, size, scale_factor, mode, align_corners)\r\n"
]
| [
[
"torch.cat",
"torch.zeros",
"torch.no_grad",
"torch.cuda.is_available",
"torch.nn.functional.interpolate",
"torch.distributed.get_rank",
"torch.ByteStorage.from_buffer",
"torch.save",
"torch.ones",
"torch.distributed.init_process_group",
"torch.distributed.barrier",
"torch.tensor",
"torch.nn.functional.pad",
"torch.empty",
"torch.zeros_like",
"torch.distributed.is_initialized",
"torch.distributed.is_available",
"torch.stack",
"torch.distributed.get_world_size",
"torch.cuda.device_count",
"torch.ByteTensor",
"torch.cuda.set_device",
"torch.distributed.all_gather",
"torch.cuda.max_memory_allocated",
"torch.distributed.all_reduce"
]
]
|
MSZhang19/captum | [
"a189a1607f37bcf8a27decec516d5afb1709a161"
]
| [
"captum/attr/_utils/attribution.py"
]
| [
"#!/usr/bin/env python3\nfrom typing import Callable\n\nimport torch\nimport torch.nn.functional as F\n\nfrom .common import (\n _run_forward,\n _format_input_baseline,\n _format_tensor_into_tuples,\n _format_additional_forward_args,\n _validate_input,\n _validate_target,\n _tensorize_baseline,\n)\nfrom .gradient import compute_gradients\n\n\nclass Attribution:\n r\"\"\"\n All attribution algorithms extend this class. It enforces its child classes\n to extend and override core `attribute` method.\n \"\"\"\n\n def __init__(self, forward_func):\n r\"\"\"\n Args:\n forward_func (callable or torch.nn.Module): This can either be an instance\n of pytorch model or any modification of model's forward\n function.\n \"\"\"\n self.forward_func = forward_func\n\n attribute: Callable\n r\"\"\"\n This method computes and returns the attribution values for each input tensor.\n Deriving classes are responsible for implementing its logic accordingly.\n\n Specific attribution algorithms that extend this class take relevant\n arguments.\n\n Args:\n\n inputs (tensor or tuple of tensors): Input for which attribution\n is computed. It can be provided as a single tensor or\n a tuple of multiple tensors. If multiple input tensors\n are provided, the batch sizes must be aligned accross all\n tensors.\n\n\n Returns:\n\n *tensor* or tuple of *tensors* of **attributions**:\n - **attributions** (*tensor* or tuple of *tensors*):\n Attribution values for each\n input tensor. The `attributions` have the same shape and\n dimensionality as the inputs.\n If a single tensor is provided as inputs, a single tensor\n is returned. If a tuple is provided for inputs, a tuple of\n corresponding sized tensors is returned.\n\n \"\"\"\n\n def has_convergence_delta(self):\n r\"\"\"\n This method informs the user whether the attribution algorithm provides\n a convergence delta (aka an approximation error) or not. Convergence\n delta may serve as a proxy of correctness of attribution algorithm's\n approximation. If deriving attribution class provides a\n `compute_convergence_delta` method, it should\n override both `compute_convergence_delta` and `has_convergence_delta` methods.\n\n Returns:\n bool:\n Returns whether the attribution algorithm\n provides a convergence delta (aka approximation error) or not.\n\n \"\"\"\n return False\n\n def compute_convergence_delta(self, attributions, *args):\n r\"\"\"\n The attribution algorithms which derive `Attribution` class and provide\n convergence delta (aka approximation error) should implement this method.\n Convergence delta can be computed based on certain properties of the\n attribution alogrithms.\n\n Args:\n\n attributions (tensor or tuple of tensors): Attribution scores that\n are precomputed by an attribution algorithm.\n Attributions can be provided in form of a single tensor\n or a tuple of those. It is assumed that attribution\n tensor's dimension 0 corresponds to the number of\n examples, and if multiple input tensors are provided,\n the examples must be aligned appropriately.\n *args (optional): Additonal arguments that are used by the\n sub-classes depending on the specific implementation\n of `compute_convergence_delta`.\n\n Returns:\n\n *tensor* of **deltas**:\n - **deltas** (*tensor*):\n Depending on specific implementaion of\n sub-classes, convergence delta can be returned per\n sample in form of a tensor or it can be aggregated\n across multuple samples and returned in form of a\n single floating point tensor.\n \"\"\"\n raise NotImplementedError(\n \"Deriving sub-class should implement\" \" compute_convergence_delta method\"\n )\n\n\nclass GradientAttribution(Attribution):\n r\"\"\"\n All gradient based attribution algorithms extend this class. It requires a\n forward function, which most commonly is the forward function of the model\n that we want to interpret or the model itself.\n \"\"\"\n\n def __init__(self, forward_func):\n r\"\"\"\n Args:\n\n forward_func (callable or torch.nn.Module): This can either be an instance\n of pytorch model or any modification of model's forward\n function.\n \"\"\"\n Attribution.__init__(self, forward_func)\n self.gradient_func = compute_gradients\n\n def compute_convergence_delta(\n self,\n attributions,\n start_point,\n end_point,\n target=None,\n additional_forward_args=None,\n ):\n r\"\"\"\n Here we provide a specific implementation for `compute_convergence_delta`\n which is based on a common property among gradient-based attribution algorithms.\n In the literature sometimes it is also called completeness axiom. Completeness\n axiom states that the sum of the attribution must be equal to the differences of\n NN Models's function at its end and start points. In other words:\n sum(attributions) - (F(end_point) - F(start_point)) is close to zero.\n Returned delta of this method is defined as above stated difference.\n\n This implementation assumes that both the `start_point` and `end_point` have\n the same shape and dimensionality. It also assumes that the target must have\n the same number of examples as the `start_point` and the `end_point` in case\n it is provided in form of a list or a non-singleton tensor.\n\n Args:\n\n attributions (tensor or tuple of tensors): Precomputed attribution\n scores. The user can compute those using any attribution\n algorithm. It is assumed the the shape and the\n dimensionality of attributions must match the shape and\n the dimensionality of `start_point` and `end_point`.\n It also assumes that the attribution tensor's\n dimension 0 corresponds to the number of\n examples, and if multiple input tensors are provided,\n the examples must be aligned appropriately.\n start_point (tensor or tuple of tensors, optional): `start_point`\n is passed as an input to model's forward function. It\n is the starting point of attributions' approximation.\n It is assumed that both `start_point` and `end_point`\n have the same shape and dimensionality.\n end_point (tensor or tuple of tensors): `end_point`\n is passed as an input to model's forward function. It\n is the end point of attributions' approximation.\n It is assumed that both `start_point` and `end_point`\n have the same shape and dimensionality.\n target (int, tuple, tensor or list, optional): Output indices for\n which gradients are computed (for classification cases,\n this is usually the target class).\n If the network returns a scalar value per example,\n no target index is necessary.\n For general 2D outputs, targets can be either:\n\n - a single integer or a tensor containing a single\n integer, which is applied to all input examples\n\n - a list of integers or a 1D tensor, with length matching\n the number of examples in inputs (dim 0). Each integer\n is applied as the target for the corresponding example.\n\n For outputs with > 2 dimensions, targets can be either:\n\n - A single tuple, which contains #output_dims - 1\n elements. This target index is applied to all examples.\n\n - A list of tuples with length equal to the number of\n examples in inputs (dim 0), and each tuple containing\n #output_dims - 1 elements. Each tuple is applied as the\n target for the corresponding example.\n\n Default: None\n additional_forward_args (any, optional): If the forward function\n requires additional arguments other than the inputs for\n which attributions should not be computed, this argument\n can be provided. It must be either a single additional\n argument of a Tensor or arbitrary (non-tuple) type or a\n tuple containing multiple additional arguments including\n tensors or any arbitrary python types. These arguments\n are provided to forward_func in order following the\n arguments in inputs.\n For a tensor, the first dimension of the tensor must\n correspond to the number of examples.\n `additional_forward_args` is used both for `start_point`\n and `end_point` when computing the forward pass.\n Default: None\n\n Returns:\n\n *tensor* of **deltas**:\n - **deltas** (*tensor*):\n This implementation returns convergence delta per\n sample. Deriving sub-classes may do any type of aggregation\n of those values, if necessary.\n \"\"\"\n end_point, start_point = _format_input_baseline(end_point, start_point)\n additional_forward_args = _format_additional_forward_args(\n additional_forward_args\n )\n # tensorizing start_point in case it is a scalar or one example baseline\n # If the batch size is large we could potentially also tensorize only one\n # sample and expand the output to the rest of the elements in the batch\n start_point = _tensorize_baseline(end_point, start_point)\n\n attributions = _format_tensor_into_tuples(attributions)\n\n # verify that the attributions and end_point match on 1st dimension\n for attribution, end_point_tnsr in zip(attributions, end_point):\n assert end_point_tnsr.shape[0] == attribution.shape[0], (\n \"Attributions tensor and the end_point must match on the first\"\n \" dimension but found attribution: {} and end_point: {}\".format(\n attribution.shape[0], end_point_tnsr.shape[0]\n )\n )\n\n num_samples = end_point[0].shape[0]\n _validate_input(end_point, start_point)\n _validate_target(num_samples, target)\n\n def _sum_rows(input):\n return input.view(input.shape[0], -1).sum(1)\n\n with torch.no_grad():\n start_point = _sum_rows(\n _run_forward(\n self.forward_func, start_point, target, additional_forward_args\n )\n )\n\n end_point = _sum_rows(\n _run_forward(\n self.forward_func, end_point, target, additional_forward_args\n )\n )\n row_sums = [_sum_rows(attribution) for attribution in attributions]\n attr_sum = torch.stack([sum(row_sum) for row_sum in zip(*row_sums)])\n return attr_sum - (end_point - start_point)\n\n\nclass PerturbationAttribution(Attribution):\n r\"\"\"\n All perturbation based attribution algorithms extend this class. It requires a\n forward function, which most commonly is the forward function of the model\n that we want to interpret or the model itself.\n \"\"\"\n\n def __init__(self, forward_func):\n r\"\"\"\n Args:\n\n forward_func (callable or torch.nn.Module): This can either be an instance\n of pytorch model or any modification of model's forward\n function.\n \"\"\"\n Attribution.__init__(self, forward_func)\n\n\nclass InternalAttribution(Attribution):\n r\"\"\"\n Shared base class for LayerAttrubution and NeuronAttribution,\n attribution types that require a model and a particular layer.\n \"\"\"\n\n def __init__(self, forward_func, layer, device_ids=None):\n r\"\"\"\n Args:\n\n forward_func (callable or torch.nn.Module): This can either be an instance\n of pytorch model or any modification of model's forward\n function.\n layer (torch.nn.Module): Layer for which output attributions are computed.\n Output size of attribute matches that of layer output.\n device_ids (list(int)): Device ID list, necessary only if forward_func\n applies a DataParallel model, which allows reconstruction of\n intermediate outputs from batched results across devices.\n If forward_func is given as the DataParallel model itself,\n then it is not necessary to provide this argument.\n \"\"\"\n Attribution.__init__(self, forward_func)\n self.layer = layer\n self.device_ids = device_ids\n\n\nclass LayerAttribution(InternalAttribution):\n r\"\"\"\n Layer attribution provides attribution values for the given layer, quanitfying\n the importance of each neuron within the given layer's output. The output\n attribution of calling attribute on a LayerAttribution object always matches\n the size of the layer output.\n \"\"\"\n\n def __init__(self, forward_func, layer, device_ids=None):\n r\"\"\"\n Args:\n\n forward_func (callable or torch.nn.Module): This can either be an instance\n of pytorch model or any modification of model's forward\n function.\n layer (torch.nn.Module): Layer for which output attributions are computed.\n Output size of attribute matches that of layer output.\n device_ids (list(int)): Device ID list, necessary only if forward_func\n applies a DataParallel model, which allows reconstruction of\n intermediate outputs from batched results across devices.\n If forward_func is given as the DataParallel model itself,\n then it is not necessary to provide this argument.\n \"\"\"\n InternalAttribution.__init__(self, forward_func, layer, device_ids)\n\n def interpolate(layer_attribution, interpolate_dims, interpolate_mode=\"nearest\"):\n r\"\"\"\n Interpolates given 3D, 4D or 5D layer attribution to given dimensions.\n This is often utilized to upsample the attribution of a convolutional layer\n to the size of an input, which allows visualizing in the input space.\n\n Args:\n\n layer_attribution (torch.Tensor): Tensor of given layer attributions.\n interpolate_dims (int or tuple): Upsampled dimensions. The\n number of elements must be the number of dimensions\n of layer_attribution - 2, since the first dimension\n corresponds to number of examples and the second is\n assumed to correspond to the number of channels.\n interpolate_mode (str): Method for interpolation, which\n must be a valid input interpolation mode for\n torch.nn.functional. These methods are\n \"nearest\", \"area\", \"linear\" (3D-only), \"bilinear\"\n (4D-only), \"bicubic\" (4D-only), \"trilinear\" (5D-only)\n based on the number of dimensions of the given layer\n attribution.\n\n Returns:\n *tensor* of upsampled **attributions**:\n - **attributions** (*tensor*):\n Upsampled layer attributions with first 2 dimensions matching\n slayer_attribution and remaining dimensions given by\n interpolate_dims.\n \"\"\"\n return F.interpolate(layer_attribution, interpolate_dims, mode=interpolate_mode)\n\n\nclass NeuronAttribution(InternalAttribution):\n r\"\"\"\n Neuron attribution provides input attribution for a given neuron, quanitfying\n the importance of each input feature in the activation of a particular neuron.\n Calling attribute on a NeuronAttribution object requires also providing\n the index of the neuron in the output of the given layer for which attributions\n are required.\n The output attribution of calling attribute on a NeuronAttribution object\n always matches the size of the input.\n \"\"\"\n\n def __init__(self, forward_func, layer, device_ids=None):\n r\"\"\"\n Args:\n\n forward_func (callable or torch.nn.Module): This can either be an instance\n of pytorch model or any modification of model's forward\n function.\n layer (torch.nn.Module): Layer for which output attributions are computed.\n Output size of attribute matches that of layer output.\n device_ids (list(int)): Device ID list, necessary only if forward_func\n applies a DataParallel model, which allows reconstruction of\n intermediate outputs from batched results across devices.\n If forward_func is given as the DataParallel model itself,\n then it is not necessary to provide this argument.\n \"\"\"\n InternalAttribution.__init__(self, forward_func, layer, device_ids)\n\n attribute: Callable\n r\"\"\"\n This method computes and returns the neuron attribution values for each\n input tensor. Deriving classes are responsible for implementing\n its logic accordingly.\n\n Specific attribution algorithms that extend this class take relevant\n arguments.\n\n Args:\n\n inputs: A single high dimensional input tensor or a tuple of them.\n neuron_index (int or tuple): Tuple providing index of neuron in output\n of given layer for which attribution is desired. Length of\n this tuple must be one less than the number of\n dimensions in the output of the given layer (since\n dimension 0 corresponds to number of examples).\n\n Returns:\n\n *tensor* or tuple of *tensors* of **attributions**:\n - **attributions** (*tensor* or tuple of *tensors*):\n Attribution values for\n each input vector. The `attributions` have the\n dimensionality of inputs.\n \"\"\"\n"
]
| [
[
"torch.no_grad",
"torch.nn.functional.interpolate"
]
]
|
markpp/object_detectors | [
"8a6cac32ec2d8b578c0d301feceef19390343e85"
]
| [
"pytorch/MaskRCNN/MaskRCNN.py"
]
| [
"import torch\n\nimport torchvision\nfrom torchvision.models.detection.faster_rcnn import FastRCNNPredictor\nfrom torchvision.models.detection.mask_rcnn import MaskRCNNPredictor\n\nimport numpy as np\nimport cv2\n\ndef create_model(num_classes,pretrained=True):\n # load an instance segmentation model pre-trained on COCO\n model = torchvision.models.detection.maskrcnn_resnet50_fpn(pretrained=pretrained)\n\n # get the number of input features for the classifier\n in_features = model.roi_heads.box_predictor.cls_score.in_features\n # replace the pre-trained head with a new one\n model.roi_heads.box_predictor = FastRCNNPredictor(in_features, num_classes)\n\n # now get the number of input features for the mask classifier\n in_features_mask = model.roi_heads.mask_predictor.conv5_mask.in_channels\n hidden_layer = 256\n # and replace the mask predictor with a new one\n model.roi_heads.mask_predictor = MaskRCNNPredictor(in_features_mask, hidden_layer, num_classes)\n\n return model\n\ndef _evaluate_iou(target, pred):\n \"\"\"\n Evaluate intersection over union (IOU) for target from dataset and output prediction\n from model\n \"\"\"\n if pred[\"boxes\"].shape[0] == 0:\n # no box detected\n return torch.tensor(0.0, device=pred[\"boxes\"].device)\n return torchvision.ops.box_iou(target[\"boxes\"], pred[\"boxes\"]).diag().mean()\n\ndef _plot_boxes(imgs, targets, preds):\n \"\"\"\n Plot the target and prediction boxes\n \"\"\"\n dets = []\n for img, tar, pred in zip(imgs, targets, preds):\n out = img.cpu()\n out[0] = out[0] * 0.229 + 0.485\n out[1] = out[1] * 0.224 + 0.456\n out[2] = out[2] * 0.225 + 0.406\n out = out.mul(255).permute(1, 2, 0).byte().numpy()\n for b,l in zip(tar[\"boxes\"],tar[\"labels\"]):\n x1, y1, x2, y2 = [int(x) for x in b.tolist()]\n cv2.rectangle(out,(x1, y1),(x2, y2),(255,0,0),3)\n for b,l,s in zip(pred[\"boxes\"],pred[\"labels\"],pred[\"scores\"]):\n score = s.item()\n if score > 0.25:\n x1, y1, x2, y2 = [int(x) for x in b.tolist()]\n cv2.rectangle(out,(x1, y1),(x2, y2),(0,0,255),2)\n cv2.putText(out,\"{:.2f}\".format(score), (x1, y1), cv2.FONT_HERSHEY_SIMPLEX, 0.5, (0,0,255), 2)\n\n if len(dets):\n dets = np.concatenate((dets, out), axis=1)\n else:\n dets = out\n return dets\n"
]
| [
[
"numpy.concatenate",
"torch.tensor"
]
]
|
ViugiNick/sentiment-discovery | [
"7f5ab28918a6fc29318a30f557b9454f0f5cc26a"
]
| [
"multiproc.py"
]
| [
"import torch\nimport sys\nimport os\nimport subprocess\n\nargslist = list(sys.argv)[1:]\n\nLOGDIR = 'distributed_logs'\nif '--save' in argslist:\n savepath = os.path.splitext(os.path.basename(argslist[argslist.index('--save')+1]))[0]\nelse:\n savepath = 'model'\nLOGDIR = os.path.join(LOGDIR, savepath)\nif not os.path.exists(LOGDIR):\n os.makedirs(LOGDIR)\n\nif '--world_size' in argslist:\n world_size = int(argslist[argslist.index('--world_size')+1])\nelse:\n world_size = torch.cuda.device_count()\n argslist.append('--world_size')\n argslist.append(str(world_size))\n\nfor i in range(world_size):\n if '--rank' in argslist:\n argslist[argslist.index('--rank')+1] = str(i)\n else:\n argslist.append('--rank')\n argslist.append(str(i))\n #stdout = open(os.path.join(LOGDIR, str(i)+\".log\"), \"w\")\n stdout = None if i == 0 else open(os.path.join(LOGDIR, str(i)+\".log\"), \"w\")\n call = subprocess.Popen\n if i == world_size-1:\n call = subprocess.call\n call([str(sys.executable)]+argslist, stdout=stdout)\n\n\n"
]
| [
[
"torch.cuda.device_count"
]
]
|
andre91998/Image_Processing | [
"e507b4b95a64d76bbabb63f8148317879dbc80d3"
]
| [
"src/log.py"
]
| [
"\n# coding: utf-8\n\n# # Function log\n# \n# Laplacian of Gaussian image.\n# \n# ## Synopse\n# 1D, 2D Laplacian of Gaussian image.\n# \n# - **g = ialog(s, mu, sigma)**\n# - Output:\n# - **g**: Image.\n# - Input:\n# - **s**: Image. [rows cols], output image dimensions.\n# - **mu**: Image. [row0 col0], center of the function.\n# - **sigma**: Double. standart deviation.\n\n# ## Description\n# \n# Creates a Laplacian of Gaussian image with dimensions given by s, origin given by $\\mu$ and standart deviation given by $\\sigma$ . This function is used in the Marr-Hildreth filter.\n# \n\n# ## Function code\n\n# In[3]:\n\nimport numpy as np\n\ndef log(s, mu, sigma):\n mu = np.array(mu)\n s = np.array(s)\n if np.product(np.shape(s)) == 1:\n x = np.arange(s)\n coord_center = (x-mu)**2\n else:\n (rr, cc) = np.indices( s)\n coord_center = (rr-mu[0])**2 + (cc-mu[1])**2\n\n gauss_factor = coord_center/(2.*sigma**2)\n gauss_factor_r = np.ravel(gauss_factor)\n \n exp_factor = np.exp(-gauss_factor_r)\n exp_factor = exp_factor.reshape( np.shape(coord_center))\n \n g = -(((1 - gauss_factor )/ (sigma**4 * np.pi)) * exp_factor)\n return g\n\n\n# ## Examples\n\n# In[1]:\n\ntesting = (__name__ == \"__main__\")\n\nif testing:\n get_ipython().system(' jupyter nbconvert --to python log.ipynb')\n import sys\n import os\n ea979path = os.path.abspath('../../')\n if ea979path not in sys.path:\n sys.path.append(ea979path)\n import ea979.src as ia\n \n import matplotlib.pyplot as plt\n get_ipython().magic('matplotlib inline')\n\n\n# ### Example 1D\n# \n# #### Numerical example:\n\n# In[2]:\n\nif testing:\n s, mu, sigma = 5, 3, 0.8\n F = ia.log(s, mu, sigma)\n\n print('image dimensions = ', s)\n print('center of function = ', mu)\n print('spread factor =', sigma)\n print('Laplacian of Gaussian image : \\n', F.round(2))\n\n\n# #### Image exemple:\n\n# In[3]:\n\nif testing:\n \n \n s, mu, sigma = 256, 128, 8\n F = ia.log(s, mu, sigma)\n\n print('image dimensions = ', s)\n print('center of function = ', mu)\n print('spread factor =', sigma)\n\n plt.plot(F)\n plt.title('Laplacian of Gaussian')\n\n\n# ### Example 2D\n# \n# #### Numerical exemple.\n\n# In[4]:\n\nif testing:\n s, mu, sigma = [5, 7], [3, 4], .5\n F = ia.log(s, mu, sigma)\n print('image dimensions = ', s)\n print('center of function = ', mu)\n print('spread factor =', sigma)\n print('Laplacian of Gaussian image : \\n', F.round(2))\n\n\n# #### Generating a image 2D 128x128, centered at 64x64 and sigma 4:\n\n# In[5]:\n\nif testing:\n s, mu, sigma = [128, 128], [64, 64], 4\n F = ia.log(s, mu, sigma)\n print('image dimensions = ', s)\n print('center of function = ', mu)\n print('spread factor =', sigma)\n ia.adshow(ia.normalize(F), 'Laplacian of Gaussian')\n\n\n# #### Generating a image 2D 256x256, centered at 128x128 and sigma 20\n\n# In[6]:\n\nif testing:\n s, mu, sigma = [256, 256], [128, 128], 20\n F = ia.log(s, mu, sigma)\n print('image dimensions = ', s)\n print('center of function = ', mu)\n print('spread factor =', sigma)\n ia.adshow(ia.normalize(F), 'Laplacian of Gaussian')\n\n\n# ## Measuring time:\n\n# In[7]:\n\nif testing:\n s, mu, sigma = [256, 256], [128, 128], 20\n print('Computational time is:')\n get_ipython().magic('timeit ia.log(s, mu, sigma)')\n\n\n# ## Equation\n# \n# \n# $$\n# \\begin{matrix}\n# LoG = \\frac{1}{\\pi\\sigma^4}\n# \\left[\n# 1 - \\frac{(x-\\mu_x)^2}{2\\sigma^2}\n# \\right]\n# e \\frac{-(x-\\mu_x)^2}{2\\sigma^2}\n# \\end{matrix}\n# $$\n# \n# $$\n# \\begin{matrix}\n# LoG = \\frac{1}{\\pi\\sigma^4}\n# \\left[\n# 1 - \\frac{(r-\\mu_r)^2 + (c-\\mu_c)^2}{2\\sigma^2}\n# \\right]\n# e \\frac{(r-\\mu_r)^2 + (c-\\mu_c)^2}{2\\sigma^2}\n# \\end{matrix}\n# $$\n\n# ## See Also\n# \n# - [logfilter]() - Laplacian of Gaussian filter.\n# - [Blob detection wikipedia](http://en.wikipedia.org/wiki/Blob_detection)\n# - [Laplacian of Gaussian](http://homepages.inf.ed.ac.uk/rbf/HIPR2/log.htm)\n\n# In[ ]:\n\n\n\n"
]
| [
[
"matplotlib.pyplot.title",
"numpy.arange",
"numpy.indices",
"matplotlib.pyplot.plot",
"numpy.shape",
"numpy.ravel",
"numpy.array",
"numpy.exp"
]
]
|
GireeshS22/Kaggle-submissions | [
"d772d3fc8fafb1719972a1e58b5106876c9dfba3"
]
| [
"AV/Ship prediction/train.py"
]
| [
"# -*- coding: utf-8 -*-\r\n\"\"\"\r\nCreated on Fri Apr 12 10:08:12 2019\r\n\r\n@author: tornado\r\n\"\"\"\r\n\r\nimport pandas as pd\r\n\r\nfrom keras.layers import Input, TimeDistributed, Bidirectional, Conv2D, BatchNormalization, MaxPooling2D, Flatten, LSTM, Dense, Lambda, GRU, Activation\r\nfrom keras.optimizers import SGD, Adam\r\nfrom keras.models import Model\r\nfrom keras.callbacks import ModelCheckpoint, CSVLogger, TensorBoard\r\nfrom keras.layers.merge import add, concatenate\r\n\r\nimport keras.backend as K\r\n#from time_distributed_read_images import generate_arrays_from_file, generate_val_arrays_from_file\r\nfrom read_images import generate_arrays_from_file, generate_val_arrays_from_file\r\n\r\nfrom config import height, width, train_file, mini_batch_size\r\n\r\nfrom sklearn.model_selection import train_test_split\r\n\r\n#%%\r\ninput_data = Input(shape= (height, width, 3), name= \"the_input\")\r\n\r\nconv = Conv2D(filters=64, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv1\")(input_data)\r\nbn = BatchNormalization(name = \"batch_norm1\")(conv)\r\nconv = Conv2D(filters=64, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv2\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm2\")(conv)\r\npooling = MaxPooling2D(pool_size=(2,2), name = \"max_pool1\")(bn)\r\n\r\nconv = Conv2D(filters=128, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv3\")(pooling)\r\nbn = BatchNormalization(name = \"batch_norm3\")(conv)\r\nconv = Conv2D(filters=128, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv4\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm4\")(conv)\r\npooling = MaxPooling2D(pool_size=(2,2), name = \"max_pool2\")(bn)\r\n\r\nconv = Conv2D(filters=256, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv5\")(pooling)\r\nbn = BatchNormalization(name = \"batch_norm5\")(conv)\r\nconv = Conv2D(filters=256, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv6\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm6\")(conv)\r\nconv = Conv2D(filters=256, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv7\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm7\")(conv)\r\npooling = MaxPooling2D(pool_size=(2,1), name = \"max_pool3\")(bn)\r\n\r\nconv = Conv2D(filters=512, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv8\")(pooling)\r\nbn = BatchNormalization(name = \"batch_norm8\")(conv)\r\nconv = Conv2D(filters=512, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv9\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm9\")(conv)\r\nconv = Conv2D(filters=512, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv10\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm10\")(conv)\r\npooling = MaxPooling2D(pool_size=(2,1), name = \"max_pool4\")(bn)\r\n\r\nconv = Conv2D(filters=512, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv11\")(pooling)\r\nbn = BatchNormalization(name = \"batch_norm11\")(conv)\r\nconv = Conv2D(filters=512, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv12\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm12\")(conv)\r\nconv = Conv2D(filters=512, kernel_size=(1, 1), activation='relu', kernel_initializer='he_normal', name = \"conv13\")(bn)\r\nbn = BatchNormalization(name = \"batch_norm13\")(conv)\r\npooling = MaxPooling2D(pool_size=(2,1), name = \"max_pool5\")(bn)\r\n\r\nflatten = Flatten(name = \"flatten\")(pooling)\r\n\r\ndense = Dense(5, name = \"dense\")(flatten)\r\ny_pred = Activation('softmax', name='label')(dense)\r\n\r\nModel(inputs = input_data, outputs = y_pred).summary()\r\nmodel = Model(inputs = input_data, outputs = y_pred)\r\n\r\n# clipnorm seems to speeds up convergence\r\n#opt = SGD(lr=0.02, decay=1e-6, momentum=0.9, nesterov=True, clipnorm=5)\r\nopt = Adam(lr = 0.0005)\r\n\r\n# the loss calc occurs elsewhere, so use a dummy lambda func for the loss\r\nmodel.compile(loss=\"categorical_crossentropy\", optimizer=opt, metrics=['accuracy'])\r\n\r\nmodel.load_weights(\"gdrive/My Drive/Colab/Ship/Checkpoints/td.weights.last.hdf5\")\r\n\r\n#%%\r\n#from keras.utils import plot_model\r\n#plot_model(model, to_file='model.png', show_shapes=True)\r\n\r\n#%%\r\ndata = pd.read_csv(train_file)\r\ntraining, validation = train_test_split(data, test_size = 0.10)\r\n\r\ntraining = training.reset_index().drop(columns = [\"index\"])\r\nvalidation = validation.reset_index().drop(columns = [\"index\"])\r\n\r\nroundofffortraining = (len(training) // mini_batch_size) * mini_batch_size\r\nroundoffforvalidation = (len(training) // mini_batch_size) * mini_batch_size\r\n\r\ntraining = training[:roundofffortraining]\r\nvalidation = validation[:roundoffforvalidation]\r\n\r\nprint(\"Training on \", str(len(training)), \" samples\")\r\nprint(\"Validating on \", str(len(validation)), \" samples\")\r\n\r\nprint(training.head())\r\n\r\n#%%\r\nfilepath=\"gdrive/My Drive/Colab/Ship/Checkpoints/weights.{acc:.3f}a-{loss:.3f}l.hdf5\"\r\ncheckpoint = ModelCheckpoint(filepath, monitor='acc', verbose=1, save_best_only=True, mode='max')\r\n\r\ncheckpoint_all = ModelCheckpoint(\"gdrive/My Drive/Colab/Ship/Checkpoints/td.weights.last.hdf5\", monitor='loss', verbose=1, save_best_only=False)\r\n\r\ntraininglog = CSVLogger(\"gdrive/My Drive/Colab/Ship/Checkpoints/logs.csv\", separator=',', append=True)\r\n\r\ntensorboard = TensorBoard(log_dir=\"gdrive/My Drive/Colab/Ship/Checkpoints\")\r\n\r\ncallbacks_list = [checkpoint, checkpoint_all, traininglog, tensorboard]\r\n\r\n#%%\r\nmodel.fit_generator(generator = generate_arrays_from_file(training), \r\n steps_per_epoch=len(training) // mini_batch_size, \r\n epochs=3000, \r\n callbacks=callbacks_list,\r\n validation_data = generate_val_arrays_from_file(validation),\r\n validation_steps=100,\r\n initial_epoch=0\r\n )"
]
| [
[
"pandas.read_csv",
"sklearn.model_selection.train_test_split"
]
]
|
dericke/pandas | [
"efa85af76db3d76e175abbb4a98c0dddb88f57a0"
]
| [
"pandas/core/indexes/base.py"
]
| [
"from copy import copy as copy_func\nfrom datetime import datetime\nimport operator\nfrom textwrap import dedent\nfrom typing import TYPE_CHECKING, Any, FrozenSet, Hashable, Union\nimport warnings\n\nimport numpy as np\n\nfrom pandas._libs import algos as libalgos, index as libindex, lib\nimport pandas._libs.join as libjoin\nfrom pandas._libs.lib import is_datetime_array, no_default\nfrom pandas._libs.tslibs import OutOfBoundsDatetime, Timestamp\nfrom pandas._libs.tslibs.period import IncompatibleFrequency\nfrom pandas._libs.tslibs.timezones import tz_compare\nfrom pandas._typing import Label\nfrom pandas.compat import set_function_name\nfrom pandas.compat.numpy import function as nv\nfrom pandas.util._decorators import Appender, Substitution, cache_readonly, doc\n\nfrom pandas.core.dtypes import concat as _concat\nfrom pandas.core.dtypes.cast import (\n maybe_cast_to_integer_array,\n validate_numeric_casting,\n)\nfrom pandas.core.dtypes.common import (\n ensure_categorical,\n ensure_int64,\n ensure_object,\n ensure_platform_int,\n is_bool,\n is_bool_dtype,\n is_categorical_dtype,\n is_datetime64_any_dtype,\n is_dtype_equal,\n is_extension_array_dtype,\n is_float,\n is_float_dtype,\n is_hashable,\n is_integer,\n is_integer_dtype,\n is_interval_dtype,\n is_iterator,\n is_list_like,\n is_object_dtype,\n is_period_dtype,\n is_scalar,\n is_signed_integer_dtype,\n is_timedelta64_dtype,\n is_unsigned_integer_dtype,\n)\nfrom pandas.core.dtypes.concat import concat_compat\nfrom pandas.core.dtypes.generic import (\n ABCCategorical,\n ABCDataFrame,\n ABCDatetimeIndex,\n ABCIntervalIndex,\n ABCMultiIndex,\n ABCPandasArray,\n ABCPeriodIndex,\n ABCRangeIndex,\n ABCSeries,\n ABCTimedeltaIndex,\n)\nfrom pandas.core.dtypes.missing import array_equivalent, isna\n\nfrom pandas.core import ops\nfrom pandas.core.accessor import CachedAccessor\nimport pandas.core.algorithms as algos\nfrom pandas.core.arrays import ExtensionArray\nfrom pandas.core.base import IndexOpsMixin, PandasObject\nimport pandas.core.common as com\nfrom pandas.core.indexers import deprecate_ndim_indexing\nfrom pandas.core.indexes.frozen import FrozenList\nimport pandas.core.missing as missing\nfrom pandas.core.ops import get_op_result_name\nfrom pandas.core.ops.invalid import make_invalid_op\nfrom pandas.core.strings import StringMethods\n\nfrom pandas.io.formats.printing import (\n PrettyDict,\n default_pprint,\n format_object_attrs,\n format_object_summary,\n pprint_thing,\n)\n\nif TYPE_CHECKING:\n from pandas import Series\n\n\n__all__ = [\"Index\"]\n\n_unsortable_types = frozenset((\"mixed\", \"mixed-integer\"))\n\n_index_doc_kwargs = dict(\n klass=\"Index\",\n inplace=\"\",\n target_klass=\"Index\",\n raises_section=\"\",\n unique=\"Index\",\n duplicated=\"np.ndarray\",\n)\n_index_shared_docs = dict()\nstr_t = str\n\n\ndef _make_comparison_op(op, cls):\n def cmp_method(self, other):\n if isinstance(other, (np.ndarray, Index, ABCSeries, ExtensionArray)):\n if other.ndim > 0 and len(self) != len(other):\n raise ValueError(\"Lengths must match to compare\")\n\n if is_object_dtype(self) and isinstance(other, ABCCategorical):\n left = type(other)(self._values, dtype=other.dtype)\n return op(left, other)\n elif is_object_dtype(self) and isinstance(other, ExtensionArray):\n # e.g. PeriodArray\n with np.errstate(all=\"ignore\"):\n result = op(self.values, other)\n\n elif is_object_dtype(self) and not isinstance(self, ABCMultiIndex):\n # don't pass MultiIndex\n with np.errstate(all=\"ignore\"):\n result = ops.comp_method_OBJECT_ARRAY(op, self.values, other)\n\n else:\n with np.errstate(all=\"ignore\"):\n result = op(self.values, np.asarray(other))\n\n if is_bool_dtype(result):\n return result\n return ops.invalid_comparison(self, other, op)\n\n name = f\"__{op.__name__}__\"\n return set_function_name(cmp_method, name, cls)\n\n\ndef _make_arithmetic_op(op, cls):\n def index_arithmetic_method(self, other):\n if isinstance(other, (ABCSeries, ABCDataFrame, ABCTimedeltaIndex)):\n return NotImplemented\n\n from pandas import Series\n\n result = op(Series(self), other)\n if isinstance(result, tuple):\n return (Index(result[0]), Index(result[1]))\n return Index(result)\n\n name = f\"__{op.__name__}__\"\n # TODO: docstring?\n return set_function_name(index_arithmetic_method, name, cls)\n\n\nclass InvalidIndexError(Exception):\n pass\n\n\n_o_dtype = np.dtype(object)\n_Identity = object\n\n\ndef _new_Index(cls, d):\n \"\"\"\n This is called upon unpickling, rather than the default which doesn't\n have arguments and breaks __new__.\n \"\"\"\n # required for backward compat, because PI can't be instantiated with\n # ordinals through __new__ GH #13277\n if issubclass(cls, ABCPeriodIndex):\n from pandas.core.indexes.period import _new_PeriodIndex\n\n return _new_PeriodIndex(cls, **d)\n\n if issubclass(cls, ABCMultiIndex):\n if \"labels\" in d and \"codes\" not in d:\n # GH#23752 \"labels\" kwarg has been replaced with \"codes\"\n d[\"codes\"] = d.pop(\"labels\")\n\n return cls.__new__(cls, **d)\n\n\nclass Index(IndexOpsMixin, PandasObject):\n \"\"\"\n Immutable ndarray implementing an ordered, sliceable set. The basic object\n storing axis labels for all pandas objects.\n\n Parameters\n ----------\n data : array-like (1-dimensional)\n dtype : NumPy dtype (default: object)\n If dtype is None, we find the dtype that best fits the data.\n If an actual dtype is provided, we coerce to that dtype if it's safe.\n Otherwise, an error will be raised.\n copy : bool\n Make a copy of input ndarray.\n name : object\n Name to be stored in the index.\n tupleize_cols : bool (default: True)\n When True, attempt to create a MultiIndex if possible.\n\n See Also\n --------\n RangeIndex : Index implementing a monotonic integer range.\n CategoricalIndex : Index of :class:`Categorical` s.\n MultiIndex : A multi-level, or hierarchical Index.\n IntervalIndex : An Index of :class:`Interval` s.\n DatetimeIndex : Index of datetime64 data.\n TimedeltaIndex : Index of timedelta64 data.\n PeriodIndex : Index of Period data.\n Int64Index : A special case of :class:`Index` with purely integer labels.\n UInt64Index : A special case of :class:`Index` with purely unsigned integer labels.\n Float64Index : A special case of :class:`Index` with purely float labels.\n\n Notes\n -----\n An Index instance can **only** contain hashable objects\n\n Examples\n --------\n >>> pd.Index([1, 2, 3])\n Int64Index([1, 2, 3], dtype='int64')\n\n >>> pd.Index(list('abc'))\n Index(['a', 'b', 'c'], dtype='object')\n \"\"\"\n\n # tolist is not actually deprecated, just suppressed in the __dir__\n _deprecations: FrozenSet[str] = (\n PandasObject._deprecations\n | IndexOpsMixin._deprecations\n | frozenset([\"contains\", \"set_value\"])\n )\n\n # To hand over control to subclasses\n _join_precedence = 1\n\n # Cython methods; see github.com/cython/cython/issues/2647\n # for why we need to wrap these instead of making them class attributes\n # Moreover, cython will choose the appropriate-dtyped sub-function\n # given the dtypes of the passed arguments\n def _left_indexer_unique(self, left, right):\n return libjoin.left_join_indexer_unique(left, right)\n\n def _left_indexer(self, left, right):\n return libjoin.left_join_indexer(left, right)\n\n def _inner_indexer(self, left, right):\n return libjoin.inner_join_indexer(left, right)\n\n def _outer_indexer(self, left, right):\n return libjoin.outer_join_indexer(left, right)\n\n _typ = \"index\"\n _data: Union[ExtensionArray, np.ndarray]\n _id = None\n _name: Label = None\n # MultiIndex.levels previously allowed setting the index name. We\n # don't allow this anymore, and raise if it happens rather than\n # failing silently.\n _no_setting_name: bool = False\n _comparables = [\"name\"]\n _attributes = [\"name\"]\n _is_numeric_dtype = False\n _can_hold_na = True\n\n # would we like our indexing holder to defer to us\n _defer_to_indexing = False\n\n # prioritize current class for _shallow_copy_with_infer,\n # used to infer integers as datetime-likes\n _infer_as_myclass = False\n\n _engine_type = libindex.ObjectEngine\n # whether we support partial string indexing. Overridden\n # in DatetimeIndex and PeriodIndex\n _supports_partial_string_indexing = False\n\n _accessors = {\"str\"}\n\n str = CachedAccessor(\"str\", StringMethods)\n\n # --------------------------------------------------------------------\n # Constructors\n\n def __new__(\n cls, data=None, dtype=None, copy=False, name=None, tupleize_cols=True, **kwargs,\n ) -> \"Index\":\n\n from pandas.core.indexes.range import RangeIndex\n\n name = maybe_extract_name(name, data, cls)\n\n if isinstance(data, ABCPandasArray):\n # ensure users don't accidentally put a PandasArray in an index.\n data = data.to_numpy()\n\n # range\n if isinstance(data, RangeIndex):\n return RangeIndex(start=data, copy=copy, dtype=dtype, name=name)\n elif isinstance(data, range):\n return RangeIndex.from_range(data, dtype=dtype, name=name)\n\n # categorical\n elif is_categorical_dtype(data) or is_categorical_dtype(dtype):\n # Delay import for perf. https://github.com/pandas-dev/pandas/pull/31423\n from pandas.core.indexes.category import CategoricalIndex\n\n return _maybe_asobject(dtype, CategoricalIndex, data, copy, name, **kwargs)\n\n # interval\n elif is_interval_dtype(data) or is_interval_dtype(dtype):\n # Delay import for perf. https://github.com/pandas-dev/pandas/pull/31423\n from pandas.core.indexes.interval import IntervalIndex\n\n return _maybe_asobject(dtype, IntervalIndex, data, copy, name, **kwargs)\n\n elif (\n is_datetime64_any_dtype(data)\n or is_datetime64_any_dtype(dtype)\n or \"tz\" in kwargs\n ):\n # Delay import for perf. https://github.com/pandas-dev/pandas/pull/31423\n from pandas import DatetimeIndex\n\n return _maybe_asobject(dtype, DatetimeIndex, data, copy, name, **kwargs)\n\n elif is_timedelta64_dtype(data) or is_timedelta64_dtype(dtype):\n # Delay import for perf. https://github.com/pandas-dev/pandas/pull/31423\n from pandas import TimedeltaIndex\n\n return _maybe_asobject(dtype, TimedeltaIndex, data, copy, name, **kwargs)\n\n elif is_period_dtype(data) or is_period_dtype(dtype):\n # Delay import for perf. https://github.com/pandas-dev/pandas/pull/31423\n from pandas import PeriodIndex\n\n return _maybe_asobject(dtype, PeriodIndex, data, copy, name, **kwargs)\n\n # extension dtype\n elif is_extension_array_dtype(data) or is_extension_array_dtype(dtype):\n if not (dtype is None or is_object_dtype(dtype)):\n # coerce to the provided dtype\n ea_cls = dtype.construct_array_type()\n data = ea_cls._from_sequence(data, dtype=dtype, copy=False)\n else:\n data = np.asarray(data, dtype=object)\n\n # coerce to the object dtype\n data = data.astype(object)\n return Index(data, dtype=object, copy=copy, name=name, **kwargs)\n\n # index-like\n elif isinstance(data, (np.ndarray, Index, ABCSeries)):\n # Delay import for perf. https://github.com/pandas-dev/pandas/pull/31423\n from pandas.core.indexes.numeric import (\n Float64Index,\n Int64Index,\n UInt64Index,\n )\n\n if dtype is not None:\n # we need to avoid having numpy coerce\n # things that look like ints/floats to ints unless\n # they are actually ints, e.g. '0' and 0.0\n # should not be coerced\n # GH 11836\n data = _maybe_cast_with_dtype(data, dtype, copy)\n dtype = data.dtype # TODO: maybe not for object?\n\n # maybe coerce to a sub-class\n if is_signed_integer_dtype(data.dtype):\n return Int64Index(data, copy=copy, dtype=dtype, name=name)\n elif is_unsigned_integer_dtype(data.dtype):\n return UInt64Index(data, copy=copy, dtype=dtype, name=name)\n elif is_float_dtype(data.dtype):\n return Float64Index(data, copy=copy, dtype=dtype, name=name)\n elif issubclass(data.dtype.type, np.bool) or is_bool_dtype(data):\n subarr = data.astype(\"object\")\n else:\n subarr = com.asarray_tuplesafe(data, dtype=object)\n\n # asarray_tuplesafe does not always copy underlying data,\n # so need to make sure that this happens\n if copy:\n subarr = subarr.copy()\n\n if dtype is None:\n new_data, new_dtype = _maybe_cast_data_without_dtype(subarr)\n if new_dtype is not None:\n return cls(\n new_data, dtype=new_dtype, copy=False, name=name, **kwargs\n )\n\n if kwargs:\n raise TypeError(f\"Unexpected keyword arguments {repr(set(kwargs))}\")\n if subarr.ndim > 1:\n # GH#13601, GH#20285, GH#27125\n raise ValueError(\"Index data must be 1-dimensional\")\n return cls._simple_new(subarr, name)\n\n elif data is None or is_scalar(data):\n raise cls._scalar_data_error(data)\n elif hasattr(data, \"__array__\"):\n return Index(np.asarray(data), dtype=dtype, copy=copy, name=name, **kwargs)\n else:\n if tupleize_cols and is_list_like(data):\n # GH21470: convert iterable to list before determining if empty\n if is_iterator(data):\n data = list(data)\n\n if data and all(isinstance(e, tuple) for e in data):\n # we must be all tuples, otherwise don't construct\n # 10697\n from pandas.core.indexes.multi import MultiIndex\n\n return MultiIndex.from_tuples(\n data, names=name or kwargs.get(\"names\")\n )\n # other iterable of some kind\n subarr = com.asarray_tuplesafe(data, dtype=object)\n return Index(subarr, dtype=dtype, copy=copy, name=name, **kwargs)\n\n \"\"\"\n NOTE for new Index creation:\n\n - _simple_new: It returns new Index with the same type as the caller.\n All metadata (such as name) must be provided by caller's responsibility.\n Using _shallow_copy is recommended because it fills these metadata\n otherwise specified.\n\n - _shallow_copy: It returns new Index with the same type (using\n _simple_new), but fills caller's metadata otherwise specified. Passed\n kwargs will overwrite corresponding metadata.\n\n - _shallow_copy_with_infer: It returns new Index inferring its type\n from passed values. It fills caller's metadata otherwise specified as the\n same as _shallow_copy.\n\n See each method's docstring.\n \"\"\"\n\n @property\n def asi8(self):\n \"\"\"\n Integer representation of the values.\n\n Returns\n -------\n ndarray\n An ndarray with int64 dtype.\n \"\"\"\n return None\n\n @classmethod\n def _simple_new(cls, values, name: Label = None):\n \"\"\"\n We require that we have a dtype compat for the values. If we are passed\n a non-dtype compat, then coerce using the constructor.\n\n Must be careful not to recurse.\n \"\"\"\n assert isinstance(values, np.ndarray), type(values)\n\n result = object.__new__(cls)\n result._data = values\n # _index_data is a (temporary?) fix to ensure that the direct data\n # manipulation we do in `_libs/reduction.pyx` continues to work.\n # We need access to the actual ndarray, since we're messing with\n # data buffers and strides.\n result._index_data = values\n result._name = name\n result._cache = {}\n\n return result._reset_identity()\n\n @cache_readonly\n def _constructor(self):\n return type(self)\n\n # --------------------------------------------------------------------\n # Index Internals Methods\n\n def _get_attributes_dict(self):\n \"\"\"\n Return an attributes dict for my class.\n \"\"\"\n return {k: getattr(self, k, None) for k in self._attributes}\n\n def _shallow_copy(self, values=None, name: Label = no_default):\n \"\"\"\n Create a new Index with the same class as the caller, don't copy the\n data, use the same object attributes with passed in attributes taking\n precedence.\n\n *this is an internal non-public method*\n\n Parameters\n ----------\n values : the values to create the new Index, optional\n name : Label, defaults to self.name\n \"\"\"\n name = self.name if name is no_default else name\n cache = self._cache.copy() if values is None else {}\n if values is None:\n values = self.values\n\n result = self._simple_new(values, name=name)\n result._cache = cache\n return result\n\n def _shallow_copy_with_infer(self, values, **kwargs):\n \"\"\"\n Create a new Index inferring the class with passed value, don't copy\n the data, use the same object attributes with passed in attributes\n taking precedence.\n\n *this is an internal non-public method*\n\n Parameters\n ----------\n values : the values to create the new Index, optional\n kwargs : updates the default attributes for this Index\n \"\"\"\n attributes = self._get_attributes_dict()\n attributes.update(kwargs)\n attributes[\"copy\"] = False\n if not len(values) and \"dtype\" not in kwargs:\n # TODO: what if hasattr(values, \"dtype\")?\n attributes[\"dtype\"] = self.dtype\n if self._infer_as_myclass:\n try:\n return self._constructor(values, **attributes)\n except (TypeError, ValueError):\n pass\n\n # Remove tz so Index will try non-DatetimeIndex inference\n attributes.pop(\"tz\", None)\n return Index(values, **attributes)\n\n def is_(self, other) -> bool:\n \"\"\"\n More flexible, faster check like ``is`` but that works through views.\n\n Note: this is *not* the same as ``Index.identical()``, which checks\n that metadata is also the same.\n\n Parameters\n ----------\n other : object\n other object to compare against.\n\n Returns\n -------\n True if both have same underlying data, False otherwise : bool\n \"\"\"\n # use something other than None to be clearer\n return self._id is getattr(other, \"_id\", Ellipsis) and self._id is not None\n\n def _reset_identity(self):\n \"\"\"\n Initializes or resets ``_id`` attribute with new object.\n \"\"\"\n self._id = _Identity()\n return self\n\n def _cleanup(self):\n self._engine.clear_mapping()\n\n @cache_readonly\n def _engine(self):\n # property, for now, slow to look up\n\n # to avoid a reference cycle, bind `target_values` to a local variable, so\n # `self` is not passed into the lambda.\n target_values = self._get_engine_target()\n return self._engine_type(lambda: target_values, len(self))\n\n # --------------------------------------------------------------------\n # Array-Like Methods\n\n # ndarray compat\n def __len__(self) -> int:\n \"\"\"\n Return the length of the Index.\n \"\"\"\n return len(self._data)\n\n def __array__(self, dtype=None) -> np.ndarray:\n \"\"\"\n The array interface, return my values.\n \"\"\"\n return np.asarray(self._data, dtype=dtype)\n\n def __array_wrap__(self, result, context=None):\n \"\"\"\n Gets called after a ufunc.\n \"\"\"\n result = lib.item_from_zerodim(result)\n if is_bool_dtype(result) or lib.is_scalar(result) or np.ndim(result) > 1:\n return result\n\n attrs = self._get_attributes_dict()\n return Index(result, **attrs)\n\n @cache_readonly\n def dtype(self):\n \"\"\"\n Return the dtype object of the underlying data.\n \"\"\"\n return self._data.dtype\n\n def ravel(self, order=\"C\"):\n \"\"\"\n Return an ndarray of the flattened values of the underlying data.\n\n Returns\n -------\n numpy.ndarray\n Flattened array.\n\n See Also\n --------\n numpy.ndarray.ravel\n \"\"\"\n values = self._get_engine_target()\n return values.ravel(order=order)\n\n def view(self, cls=None):\n\n # we need to see if we are subclassing an\n # index type here\n if cls is not None and not hasattr(cls, \"_typ\"):\n result = self._data.view(cls)\n else:\n result = self._shallow_copy()\n if isinstance(result, Index):\n result._id = self._id\n return result\n\n def astype(self, dtype, copy=True):\n \"\"\"\n Create an Index with values cast to dtypes. The class of a new Index\n is determined by dtype. When conversion is impossible, a ValueError\n exception is raised.\n\n Parameters\n ----------\n dtype : numpy dtype or pandas type\n Note that any signed integer `dtype` is treated as ``'int64'``,\n and any unsigned integer `dtype` is treated as ``'uint64'``,\n regardless of the size.\n copy : bool, default True\n By default, astype always returns a newly allocated object.\n If copy is set to False and internal requirements on dtype are\n satisfied, the original data is used to create a new Index\n or the original Index is returned.\n\n Returns\n -------\n Index\n Index with values cast to specified dtype.\n \"\"\"\n if is_dtype_equal(self.dtype, dtype):\n return self.copy() if copy else self\n\n elif is_categorical_dtype(dtype):\n from pandas.core.indexes.category import CategoricalIndex\n\n return CategoricalIndex(self.values, name=self.name, dtype=dtype, copy=copy)\n\n elif is_extension_array_dtype(dtype):\n return Index(np.asarray(self), name=self.name, dtype=dtype, copy=copy)\n\n try:\n casted = self.values.astype(dtype, copy=copy)\n except (TypeError, ValueError) as err:\n raise TypeError(\n f\"Cannot cast {type(self).__name__} to dtype {dtype}\"\n ) from err\n return Index(casted, name=self.name, dtype=dtype)\n\n _index_shared_docs[\n \"take\"\n ] = \"\"\"\n Return a new %(klass)s of the values selected by the indices.\n\n For internal compatibility with numpy arrays.\n\n Parameters\n ----------\n indices : list\n Indices to be taken.\n axis : int, optional\n The axis over which to select values, always 0.\n allow_fill : bool, default True\n fill_value : bool, default None\n If allow_fill=True and fill_value is not None, indices specified by\n -1 is regarded as NA. If Index doesn't hold NA, raise ValueError.\n\n Returns\n -------\n numpy.ndarray\n Elements of given indices.\n\n See Also\n --------\n numpy.ndarray.take\n \"\"\"\n\n @Appender(_index_shared_docs[\"take\"] % _index_doc_kwargs)\n def take(self, indices, axis=0, allow_fill=True, fill_value=None, **kwargs):\n if kwargs:\n nv.validate_take(tuple(), kwargs)\n indices = ensure_platform_int(indices)\n if self._can_hold_na:\n taken = self._assert_take_fillable(\n self.values,\n indices,\n allow_fill=allow_fill,\n fill_value=fill_value,\n na_value=self._na_value,\n )\n else:\n if allow_fill and fill_value is not None:\n cls_name = type(self).__name__\n raise ValueError(\n f\"Unable to fill values because {cls_name} cannot contain NA\"\n )\n taken = self.values.take(indices)\n return self._shallow_copy(taken)\n\n def _assert_take_fillable(\n self, values, indices, allow_fill=True, fill_value=None, na_value=np.nan\n ):\n \"\"\"\n Internal method to handle NA filling of take.\n \"\"\"\n indices = ensure_platform_int(indices)\n\n # only fill if we are passing a non-None fill_value\n if allow_fill and fill_value is not None:\n if (indices < -1).any():\n raise ValueError(\n \"When allow_fill=True and fill_value is not None, \"\n \"all indices must be >= -1\"\n )\n taken = algos.take(\n values, indices, allow_fill=allow_fill, fill_value=na_value\n )\n else:\n taken = values.take(indices)\n return taken\n\n _index_shared_docs[\n \"repeat\"\n ] = \"\"\"\n Repeat elements of a %(klass)s.\n\n Returns a new %(klass)s where each element of the current %(klass)s\n is repeated consecutively a given number of times.\n\n Parameters\n ----------\n repeats : int or array of ints\n The number of repetitions for each element. This should be a\n non-negative integer. Repeating 0 times will return an empty\n %(klass)s.\n axis : None\n Must be ``None``. Has no effect but is accepted for compatibility\n with numpy.\n\n Returns\n -------\n repeated_index : %(klass)s\n Newly created %(klass)s with repeated elements.\n\n See Also\n --------\n Series.repeat : Equivalent function for Series.\n numpy.repeat : Similar method for :class:`numpy.ndarray`.\n\n Examples\n --------\n >>> idx = pd.Index(['a', 'b', 'c'])\n >>> idx\n Index(['a', 'b', 'c'], dtype='object')\n >>> idx.repeat(2)\n Index(['a', 'a', 'b', 'b', 'c', 'c'], dtype='object')\n >>> idx.repeat([1, 2, 3])\n Index(['a', 'b', 'b', 'c', 'c', 'c'], dtype='object')\n \"\"\"\n\n @Appender(_index_shared_docs[\"repeat\"] % _index_doc_kwargs)\n def repeat(self, repeats, axis=None):\n repeats = ensure_platform_int(repeats)\n nv.validate_repeat(tuple(), dict(axis=axis))\n return self._shallow_copy(self._values.repeat(repeats))\n\n # --------------------------------------------------------------------\n # Copying Methods\n\n def copy(self, name=None, deep=False, dtype=None, names=None):\n \"\"\"\n Make a copy of this object.\n\n Name and dtype sets those attributes on the new object.\n\n Parameters\n ----------\n name : Label, optional\n Set name for new object.\n deep : bool, default False\n dtype : numpy dtype or pandas type, optional\n Set dtype for new object.\n names : list-like, optional\n Kept for compatibility with MultiIndex. Should not be used.\n\n Returns\n -------\n Index\n Index refer to new object which is a copy of this object.\n\n Notes\n -----\n In most cases, there should be no functional difference from using\n ``deep``, but if ``deep`` is passed it will attempt to deepcopy.\n \"\"\"\n if deep:\n new_index = self._shallow_copy(self._data.copy())\n else:\n new_index = self._shallow_copy()\n\n names = self._validate_names(name=name, names=names, deep=deep)\n new_index = new_index.set_names(names)\n\n if dtype:\n new_index = new_index.astype(dtype)\n return new_index\n\n def __copy__(self, **kwargs):\n return self.copy(**kwargs)\n\n def __deepcopy__(self, memo=None):\n \"\"\"\n Parameters\n ----------\n memo, default None\n Standard signature. Unused\n \"\"\"\n return self.copy(deep=True)\n\n # --------------------------------------------------------------------\n # Rendering Methods\n\n def __repr__(self) -> str_t:\n \"\"\"\n Return a string representation for this object.\n \"\"\"\n klass_name = type(self).__name__\n data = self._format_data()\n attrs = self._format_attrs()\n space = self._format_space()\n attrs_str = [f\"{k}={v}\" for k, v in attrs]\n prepr = f\",{space}\".join(attrs_str)\n\n # no data provided, just attributes\n if data is None:\n data = \"\"\n\n res = f\"{klass_name}({data}{prepr})\"\n\n return res\n\n def _format_space(self) -> str_t:\n\n # using space here controls if the attributes\n # are line separated or not (the default)\n\n # max_seq_items = get_option('display.max_seq_items')\n # if len(self) > max_seq_items:\n # space = \"\\n%s\" % (' ' * (len(klass) + 1))\n return \" \"\n\n @property\n def _formatter_func(self):\n \"\"\"\n Return the formatter function.\n \"\"\"\n return default_pprint\n\n def _format_data(self, name=None) -> str_t:\n \"\"\"\n Return the formatted data as a unicode string.\n \"\"\"\n # do we want to justify (only do so for non-objects)\n is_justify = True\n\n if self.inferred_type == \"string\":\n is_justify = False\n elif self.inferred_type == \"categorical\":\n if is_object_dtype(self.categories): # type: ignore\n is_justify = False\n\n return format_object_summary(\n self, self._formatter_func, is_justify=is_justify, name=name\n )\n\n def _format_attrs(self):\n \"\"\"\n Return a list of tuples of the (attr,formatted_value).\n \"\"\"\n return format_object_attrs(self)\n\n def _mpl_repr(self):\n # how to represent ourselves to matplotlib\n return self.values\n\n def format(self, name: bool = False, formatter=None, **kwargs):\n \"\"\"\n Render a string representation of the Index.\n \"\"\"\n header = []\n if name:\n header.append(\n pprint_thing(self.name, escape_chars=(\"\\t\", \"\\r\", \"\\n\"))\n if self.name is not None\n else \"\"\n )\n\n if formatter is not None:\n return header + list(self.map(formatter))\n\n return self._format_with_header(header, **kwargs)\n\n def _format_with_header(self, header, na_rep=\"NaN\", **kwargs):\n values = self.values\n\n from pandas.io.formats.format import format_array\n\n if is_categorical_dtype(values.dtype):\n values = np.array(values)\n\n elif is_object_dtype(values.dtype):\n values = lib.maybe_convert_objects(values, safe=1)\n\n if is_object_dtype(values.dtype):\n result = [pprint_thing(x, escape_chars=(\"\\t\", \"\\r\", \"\\n\")) for x in values]\n\n # could have nans\n mask = isna(values)\n if mask.any():\n result = np.array(result)\n result[mask] = na_rep\n result = result.tolist()\n\n else:\n result = _trim_front(format_array(values, None, justify=\"left\"))\n return header + result\n\n def to_native_types(self, slicer=None, **kwargs):\n \"\"\"\n Format specified values of `self` and return them.\n\n Parameters\n ----------\n slicer : int, array-like\n An indexer into `self` that specifies which values\n are used in the formatting process.\n kwargs : dict\n Options for specifying how the values should be formatted.\n These options include the following:\n\n 1) na_rep : str\n The value that serves as a placeholder for NULL values\n 2) quoting : bool or None\n Whether or not there are quoted values in `self`\n 3) date_format : str\n The format used to represent date-like values.\n\n Returns\n -------\n numpy.ndarray\n Formatted values.\n \"\"\"\n values = self\n if slicer is not None:\n values = values[slicer]\n return values._format_native_types(**kwargs)\n\n def _format_native_types(self, na_rep=\"\", quoting=None, **kwargs):\n \"\"\"\n Actually format specific types of the index.\n \"\"\"\n mask = isna(self)\n if not self.is_object() and not quoting:\n values = np.asarray(self).astype(str)\n else:\n values = np.array(self, dtype=object, copy=True)\n\n values[mask] = na_rep\n return values\n\n def _summary(self, name=None) -> str_t:\n \"\"\"\n Return a summarized representation.\n\n Parameters\n ----------\n name : str\n name to use in the summary representation\n\n Returns\n -------\n String with a summarized representation of the index\n \"\"\"\n if len(self) > 0:\n head = self[0]\n if hasattr(head, \"format\") and not isinstance(head, str):\n head = head.format()\n tail = self[-1]\n if hasattr(tail, \"format\") and not isinstance(tail, str):\n tail = tail.format()\n index_summary = f\", {head} to {tail}\"\n else:\n index_summary = \"\"\n\n if name is None:\n name = type(self).__name__\n return f\"{name}: {len(self)} entries{index_summary}\"\n\n # --------------------------------------------------------------------\n # Conversion Methods\n\n def to_flat_index(self):\n \"\"\"\n Identity method.\n\n .. versionadded:: 0.24.0\n\n This is implemented for compatibility with subclass implementations\n when chaining.\n\n Returns\n -------\n pd.Index\n Caller.\n\n See Also\n --------\n MultiIndex.to_flat_index : Subclass implementation.\n \"\"\"\n return self\n\n def to_series(self, index=None, name=None):\n \"\"\"\n Create a Series with both index and values equal to the index keys.\n\n Useful with map for returning an indexer based on an index.\n\n Parameters\n ----------\n index : Index, optional\n Index of resulting Series. If None, defaults to original index.\n name : str, optional\n Dame of resulting Series. If None, defaults to name of original\n index.\n\n Returns\n -------\n Series\n The dtype will be based on the type of the Index values.\n \"\"\"\n from pandas import Series\n\n if index is None:\n index = self._shallow_copy()\n if name is None:\n name = self.name\n\n return Series(self.values.copy(), index=index, name=name)\n\n def to_frame(self, index: bool = True, name=None):\n \"\"\"\n Create a DataFrame with a column containing the Index.\n\n .. versionadded:: 0.24.0\n\n Parameters\n ----------\n index : bool, default True\n Set the index of the returned DataFrame as the original Index.\n\n name : object, default None\n The passed name should substitute for the index name (if it has\n one).\n\n Returns\n -------\n DataFrame\n DataFrame containing the original Index data.\n\n See Also\n --------\n Index.to_series : Convert an Index to a Series.\n Series.to_frame : Convert Series to DataFrame.\n\n Examples\n --------\n >>> idx = pd.Index(['Ant', 'Bear', 'Cow'], name='animal')\n >>> idx.to_frame()\n animal\n animal\n Ant Ant\n Bear Bear\n Cow Cow\n\n By default, the original Index is reused. To enforce a new Index:\n\n >>> idx.to_frame(index=False)\n animal\n 0 Ant\n 1 Bear\n 2 Cow\n\n To override the name of the resulting column, specify `name`:\n\n >>> idx.to_frame(index=False, name='zoo')\n zoo\n 0 Ant\n 1 Bear\n 2 Cow\n \"\"\"\n from pandas import DataFrame\n\n if name is None:\n name = self.name or 0\n result = DataFrame({name: self._values.copy()})\n\n if index:\n result.index = self\n return result\n\n # --------------------------------------------------------------------\n # Name-Centric Methods\n\n @property\n def name(self):\n \"\"\"\n Return Index or MultiIndex name.\n \"\"\"\n return self._name\n\n @name.setter\n def name(self, value):\n if self._no_setting_name:\n # Used in MultiIndex.levels to avoid silently ignoring name updates.\n raise RuntimeError(\n \"Cannot set name on a level of a MultiIndex. Use \"\n \"'MultiIndex.set_names' instead.\"\n )\n maybe_extract_name(value, None, type(self))\n self._name = value\n\n def _validate_names(self, name=None, names=None, deep: bool = False):\n \"\"\"\n Handles the quirks of having a singular 'name' parameter for general\n Index and plural 'names' parameter for MultiIndex.\n \"\"\"\n from copy import deepcopy\n\n if names is not None and name is not None:\n raise TypeError(\"Can only provide one of `names` and `name`\")\n elif names is None and name is None:\n return deepcopy(self.names) if deep else self.names\n elif names is not None:\n if not is_list_like(names):\n raise TypeError(\"Must pass list-like as `names`.\")\n return names\n else:\n if not is_list_like(name):\n return [name]\n return name\n\n def _get_names(self):\n return FrozenList((self.name,))\n\n def _set_names(self, values, level=None):\n \"\"\"\n Set new names on index. Each name has to be a hashable type.\n\n Parameters\n ----------\n values : str or sequence\n name(s) to set\n level : int, level name, or sequence of int/level names (default None)\n If the index is a MultiIndex (hierarchical), level(s) to set (None\n for all levels). Otherwise level must be None\n\n Raises\n ------\n TypeError if each name is not hashable.\n \"\"\"\n if not is_list_like(values):\n raise ValueError(\"Names must be a list-like\")\n if len(values) != 1:\n raise ValueError(f\"Length of new names must be 1, got {len(values)}\")\n\n # GH 20527\n # All items in 'name' need to be hashable:\n for name in values:\n if not is_hashable(name):\n raise TypeError(f\"{type(self).__name__}.name must be a hashable type\")\n self._name = values[0]\n\n names = property(fset=_set_names, fget=_get_names)\n\n def set_names(self, names, level=None, inplace: bool = False):\n \"\"\"\n Set Index or MultiIndex name.\n\n Able to set new names partially and by level.\n\n Parameters\n ----------\n names : label or list of label\n Name(s) to set.\n level : int, label or list of int or label, optional\n If the index is a MultiIndex, level(s) to set (None for all\n levels). Otherwise level must be None.\n inplace : bool, default False\n Modifies the object directly, instead of creating a new Index or\n MultiIndex.\n\n Returns\n -------\n Index\n The same type as the caller or None if inplace is True.\n\n See Also\n --------\n Index.rename : Able to set new names without level.\n\n Examples\n --------\n >>> idx = pd.Index([1, 2, 3, 4])\n >>> idx\n Int64Index([1, 2, 3, 4], dtype='int64')\n >>> idx.set_names('quarter')\n Int64Index([1, 2, 3, 4], dtype='int64', name='quarter')\n\n >>> idx = pd.MultiIndex.from_product([['python', 'cobra'],\n ... [2018, 2019]])\n >>> idx\n MultiIndex([('python', 2018),\n ('python', 2019),\n ( 'cobra', 2018),\n ( 'cobra', 2019)],\n )\n >>> idx.set_names(['kind', 'year'], inplace=True)\n >>> idx\n MultiIndex([('python', 2018),\n ('python', 2019),\n ( 'cobra', 2018),\n ( 'cobra', 2019)],\n names=['kind', 'year'])\n >>> idx.set_names('species', level=0)\n MultiIndex([('python', 2018),\n ('python', 2019),\n ( 'cobra', 2018),\n ( 'cobra', 2019)],\n names=['species', 'year'])\n \"\"\"\n if level is not None and not isinstance(self, ABCMultiIndex):\n raise ValueError(\"Level must be None for non-MultiIndex\")\n\n if level is not None and not is_list_like(level) and is_list_like(names):\n raise TypeError(\"Names must be a string when a single level is provided.\")\n\n if not is_list_like(names) and level is None and self.nlevels > 1:\n raise TypeError(\"Must pass list-like as `names`.\")\n\n if not is_list_like(names):\n names = [names]\n if level is not None and not is_list_like(level):\n level = [level]\n\n if inplace:\n idx = self\n else:\n idx = self._shallow_copy()\n idx._set_names(names, level=level)\n if not inplace:\n return idx\n\n def rename(self, name, inplace=False):\n \"\"\"\n Alter Index or MultiIndex name.\n\n Able to set new names without level. Defaults to returning new index.\n Length of names must match number of levels in MultiIndex.\n\n Parameters\n ----------\n name : label or list of labels\n Name(s) to set.\n inplace : bool, default False\n Modifies the object directly, instead of creating a new Index or\n MultiIndex.\n\n Returns\n -------\n Index\n The same type as the caller or None if inplace is True.\n\n See Also\n --------\n Index.set_names : Able to set new names partially and by level.\n\n Examples\n --------\n >>> idx = pd.Index(['A', 'C', 'A', 'B'], name='score')\n >>> idx.rename('grade')\n Index(['A', 'C', 'A', 'B'], dtype='object', name='grade')\n\n >>> idx = pd.MultiIndex.from_product([['python', 'cobra'],\n ... [2018, 2019]],\n ... names=['kind', 'year'])\n >>> idx\n MultiIndex([('python', 2018),\n ('python', 2019),\n ( 'cobra', 2018),\n ( 'cobra', 2019)],\n names=['kind', 'year'])\n >>> idx.rename(['species', 'year'])\n MultiIndex([('python', 2018),\n ('python', 2019),\n ( 'cobra', 2018),\n ( 'cobra', 2019)],\n names=['species', 'year'])\n >>> idx.rename('species')\n Traceback (most recent call last):\n TypeError: Must pass list-like as `names`.\n \"\"\"\n return self.set_names([name], inplace=inplace)\n\n # --------------------------------------------------------------------\n # Level-Centric Methods\n\n @property\n def nlevels(self) -> int:\n \"\"\"\n Number of levels.\n \"\"\"\n return 1\n\n def _sort_levels_monotonic(self):\n \"\"\"\n Compat with MultiIndex.\n \"\"\"\n return self\n\n def _validate_index_level(self, level):\n \"\"\"\n Validate index level.\n\n For single-level Index getting level number is a no-op, but some\n verification must be done like in MultiIndex.\n\n \"\"\"\n if isinstance(level, int):\n if level < 0 and level != -1:\n raise IndexError(\n \"Too many levels: Index has only 1 level, \"\n f\"{level} is not a valid level number\"\n )\n elif level > 0:\n raise IndexError(\n f\"Too many levels: Index has only 1 level, not {level + 1}\"\n )\n elif level != self.name:\n raise KeyError(\n f\"Requested level ({level}) does not match index name ({self.name})\"\n )\n\n def _get_level_number(self, level) -> int:\n self._validate_index_level(level)\n return 0\n\n def sortlevel(self, level=None, ascending=True, sort_remaining=None):\n \"\"\"\n For internal compatibility with with the Index API.\n\n Sort the Index. This is for compat with MultiIndex\n\n Parameters\n ----------\n ascending : bool, default True\n False to sort in descending order\n\n level, sort_remaining are compat parameters\n\n Returns\n -------\n Index\n \"\"\"\n return self.sort_values(return_indexer=True, ascending=ascending)\n\n def _get_level_values(self, level):\n \"\"\"\n Return an Index of values for requested level.\n\n This is primarily useful to get an individual level of values from a\n MultiIndex, but is provided on Index as well for compatibility.\n\n Parameters\n ----------\n level : int or str\n It is either the integer position or the name of the level.\n\n Returns\n -------\n Index\n Calling object, as there is only one level in the Index.\n\n See Also\n --------\n MultiIndex.get_level_values : Get values for a level of a MultiIndex.\n\n Notes\n -----\n For Index, level should be 0, since there are no multiple levels.\n\n Examples\n --------\n >>> idx = pd.Index(list('abc'))\n >>> idx\n Index(['a', 'b', 'c'], dtype='object')\n\n Get level values by supplying `level` as integer:\n\n >>> idx.get_level_values(0)\n Index(['a', 'b', 'c'], dtype='object')\n \"\"\"\n self._validate_index_level(level)\n return self\n\n get_level_values = _get_level_values\n\n def droplevel(self, level=0):\n \"\"\"\n Return index with requested level(s) removed.\n\n If resulting index has only 1 level left, the result will be\n of Index type, not MultiIndex.\n\n .. versionadded:: 0.23.1 (support for non-MultiIndex)\n\n Parameters\n ----------\n level : int, str, or list-like, default 0\n If a string is given, must be the name of a level\n If list-like, elements must be names or indexes of levels.\n\n Returns\n -------\n Index or MultiIndex\n \"\"\"\n if not isinstance(level, (tuple, list)):\n level = [level]\n\n levnums = sorted(self._get_level_number(lev) for lev in level)[::-1]\n\n if len(level) == 0:\n return self\n if len(level) >= self.nlevels:\n raise ValueError(\n f\"Cannot remove {len(level)} levels from an index with {self.nlevels} \"\n \"levels: at least one level must be left.\"\n )\n # The two checks above guarantee that here self is a MultiIndex\n\n new_levels = list(self.levels)\n new_codes = list(self.codes)\n new_names = list(self.names)\n\n for i in levnums:\n new_levels.pop(i)\n new_codes.pop(i)\n new_names.pop(i)\n\n if len(new_levels) == 1:\n\n # set nan if needed\n mask = new_codes[0] == -1\n result = new_levels[0].take(new_codes[0])\n if mask.any():\n result = result.putmask(mask, np.nan)\n\n result._name = new_names[0]\n return result\n else:\n from pandas.core.indexes.multi import MultiIndex\n\n return MultiIndex(\n levels=new_levels,\n codes=new_codes,\n names=new_names,\n verify_integrity=False,\n )\n\n def _get_grouper_for_level(self, mapper, level=None):\n \"\"\"\n Get index grouper corresponding to an index level\n\n Parameters\n ----------\n mapper: Group mapping function or None\n Function mapping index values to groups\n level : int or None\n Index level\n\n Returns\n -------\n grouper : Index\n Index of values to group on.\n labels : ndarray of int or None\n Array of locations in level_index.\n uniques : Index or None\n Index of unique values for level.\n \"\"\"\n assert level is None or level == 0\n if mapper is None:\n grouper = self\n else:\n grouper = self.map(mapper)\n\n return grouper, None, None\n\n # --------------------------------------------------------------------\n # Introspection Methods\n\n @property\n def is_monotonic(self) -> bool:\n \"\"\"\n Alias for is_monotonic_increasing.\n \"\"\"\n return self.is_monotonic_increasing\n\n @property\n def is_monotonic_increasing(self) -> bool:\n \"\"\"\n Return if the index is monotonic increasing (only equal or\n increasing) values.\n\n Examples\n --------\n >>> Index([1, 2, 3]).is_monotonic_increasing\n True\n >>> Index([1, 2, 2]).is_monotonic_increasing\n True\n >>> Index([1, 3, 2]).is_monotonic_increasing\n False\n \"\"\"\n return self._engine.is_monotonic_increasing\n\n @property\n def is_monotonic_decreasing(self) -> bool:\n \"\"\"\n Return if the index is monotonic decreasing (only equal or\n decreasing) values.\n\n Examples\n --------\n >>> Index([3, 2, 1]).is_monotonic_decreasing\n True\n >>> Index([3, 2, 2]).is_monotonic_decreasing\n True\n >>> Index([3, 1, 2]).is_monotonic_decreasing\n False\n \"\"\"\n return self._engine.is_monotonic_decreasing\n\n @property\n def _is_strictly_monotonic_increasing(self) -> bool:\n \"\"\"\n Return if the index is strictly monotonic increasing\n (only increasing) values.\n\n Examples\n --------\n >>> Index([1, 2, 3])._is_strictly_monotonic_increasing\n True\n >>> Index([1, 2, 2])._is_strictly_monotonic_increasing\n False\n >>> Index([1, 3, 2])._is_strictly_monotonic_increasing\n False\n \"\"\"\n return self.is_unique and self.is_monotonic_increasing\n\n @property\n def _is_strictly_monotonic_decreasing(self) -> bool:\n \"\"\"\n Return if the index is strictly monotonic decreasing\n (only decreasing) values.\n\n Examples\n --------\n >>> Index([3, 2, 1])._is_strictly_monotonic_decreasing\n True\n >>> Index([3, 2, 2])._is_strictly_monotonic_decreasing\n False\n >>> Index([3, 1, 2])._is_strictly_monotonic_decreasing\n False\n \"\"\"\n return self.is_unique and self.is_monotonic_decreasing\n\n @cache_readonly\n def is_unique(self) -> bool:\n \"\"\"\n Return if the index has unique values.\n \"\"\"\n return self._engine.is_unique\n\n @property\n def has_duplicates(self) -> bool:\n \"\"\"\n Check if the Index has duplicate values.\n\n Returns\n -------\n bool\n Whether or not the Index has duplicate values.\n\n Examples\n --------\n >>> idx = pd.Index([1, 5, 7, 7])\n >>> idx.has_duplicates\n True\n\n >>> idx = pd.Index([1, 5, 7])\n >>> idx.has_duplicates\n False\n\n >>> idx = pd.Index([\"Watermelon\", \"Orange\", \"Apple\",\n ... \"Watermelon\"]).astype(\"category\")\n >>> idx.has_duplicates\n True\n\n >>> idx = pd.Index([\"Orange\", \"Apple\",\n ... \"Watermelon\"]).astype(\"category\")\n >>> idx.has_duplicates\n False\n \"\"\"\n return not self.is_unique\n\n def is_boolean(self) -> bool:\n \"\"\"\n Check if the Index only consists of booleans.\n\n Returns\n -------\n bool\n Whether or not the Index only consists of booleans.\n\n See Also\n --------\n is_integer : Check if the Index only consists of integers.\n is_floating : Check if the Index is a floating type.\n is_numeric : Check if the Index only consists of numeric data.\n is_object : Check if the Index is of the object dtype.\n is_categorical : Check if the Index holds categorical data.\n is_interval : Check if the Index holds Interval objects.\n is_mixed : Check if the Index holds data with mixed data types.\n\n Examples\n --------\n >>> idx = pd.Index([True, False, True])\n >>> idx.is_boolean()\n True\n\n >>> idx = pd.Index([\"True\", \"False\", \"True\"])\n >>> idx.is_boolean()\n False\n\n >>> idx = pd.Index([True, False, \"True\"])\n >>> idx.is_boolean()\n False\n \"\"\"\n return self.inferred_type in [\"boolean\"]\n\n def is_integer(self) -> bool:\n \"\"\"\n Check if the Index only consists of integers.\n\n Returns\n -------\n bool\n Whether or not the Index only consists of integers.\n\n See Also\n --------\n is_boolean : Check if the Index only consists of booleans.\n is_floating : Check if the Index is a floating type.\n is_numeric : Check if the Index only consists of numeric data.\n is_object : Check if the Index is of the object dtype.\n is_categorical : Check if the Index holds categorical data.\n is_interval : Check if the Index holds Interval objects.\n is_mixed : Check if the Index holds data with mixed data types.\n\n Examples\n --------\n >>> idx = pd.Index([1, 2, 3, 4])\n >>> idx.is_integer()\n True\n\n >>> idx = pd.Index([1.0, 2.0, 3.0, 4.0])\n >>> idx.is_integer()\n False\n\n >>> idx = pd.Index([\"Apple\", \"Mango\", \"Watermelon\"])\n >>> idx.is_integer()\n False\n \"\"\"\n return self.inferred_type in [\"integer\"]\n\n def is_floating(self) -> bool:\n \"\"\"\n Check if the Index is a floating type.\n\n The Index may consist of only floats, NaNs, or a mix of floats,\n integers, or NaNs.\n\n Returns\n -------\n bool\n Whether or not the Index only consists of only consists of floats, NaNs, or\n a mix of floats, integers, or NaNs.\n\n See Also\n --------\n is_boolean : Check if the Index only consists of booleans.\n is_integer : Check if the Index only consists of integers.\n is_numeric : Check if the Index only consists of numeric data.\n is_object : Check if the Index is of the object dtype.\n is_categorical : Check if the Index holds categorical data.\n is_interval : Check if the Index holds Interval objects.\n is_mixed : Check if the Index holds data with mixed data types.\n\n Examples\n --------\n >>> idx = pd.Index([1.0, 2.0, 3.0, 4.0])\n >>> idx.is_floating()\n True\n\n >>> idx = pd.Index([1.0, 2.0, np.nan, 4.0])\n >>> idx.is_floating()\n True\n\n >>> idx = pd.Index([1, 2, 3, 4, np.nan])\n >>> idx.is_floating()\n True\n\n >>> idx = pd.Index([1, 2, 3, 4])\n >>> idx.is_floating()\n False\n \"\"\"\n return self.inferred_type in [\"floating\", \"mixed-integer-float\", \"integer-na\"]\n\n def is_numeric(self) -> bool:\n \"\"\"\n Check if the Index only consists of numeric data.\n\n Returns\n -------\n bool\n Whether or not the Index only consists of numeric data.\n\n See Also\n --------\n is_boolean : Check if the Index only consists of booleans.\n is_integer : Check if the Index only consists of integers.\n is_floating : Check if the Index is a floating type.\n is_object : Check if the Index is of the object dtype.\n is_categorical : Check if the Index holds categorical data.\n is_interval : Check if the Index holds Interval objects.\n is_mixed : Check if the Index holds data with mixed data types.\n\n Examples\n --------\n >>> idx = pd.Index([1.0, 2.0, 3.0, 4.0])\n >>> idx.is_numeric()\n True\n\n >>> idx = pd.Index([1, 2, 3, 4.0])\n >>> idx.is_numeric()\n True\n\n >>> idx = pd.Index([1, 2, 3, 4])\n >>> idx.is_numeric()\n True\n\n >>> idx = pd.Index([1, 2, 3, 4.0, np.nan])\n >>> idx.is_numeric()\n True\n\n >>> idx = pd.Index([1, 2, 3, 4.0, np.nan, \"Apple\"])\n >>> idx.is_numeric()\n False\n \"\"\"\n return self.inferred_type in [\"integer\", \"floating\"]\n\n def is_object(self) -> bool:\n \"\"\"\n Check if the Index is of the object dtype.\n\n Returns\n -------\n bool\n Whether or not the Index is of the object dtype.\n\n See Also\n --------\n is_boolean : Check if the Index only consists of booleans.\n is_integer : Check if the Index only consists of integers.\n is_floating : Check if the Index is a floating type.\n is_numeric : Check if the Index only consists of numeric data.\n is_categorical : Check if the Index holds categorical data.\n is_interval : Check if the Index holds Interval objects.\n is_mixed : Check if the Index holds data with mixed data types.\n\n Examples\n --------\n >>> idx = pd.Index([\"Apple\", \"Mango\", \"Watermelon\"])\n >>> idx.is_object()\n True\n\n >>> idx = pd.Index([\"Apple\", \"Mango\", 2.0])\n >>> idx.is_object()\n True\n\n >>> idx = pd.Index([\"Watermelon\", \"Orange\", \"Apple\",\n ... \"Watermelon\"]).astype(\"category\")\n >>> idx.is_object()\n False\n\n >>> idx = pd.Index([1.0, 2.0, 3.0, 4.0])\n >>> idx.is_object()\n False\n \"\"\"\n return is_object_dtype(self.dtype)\n\n def is_categorical(self) -> bool:\n \"\"\"\n Check if the Index holds categorical data.\n\n Returns\n -------\n bool\n True if the Index is categorical.\n\n See Also\n --------\n CategoricalIndex : Index for categorical data.\n is_boolean : Check if the Index only consists of booleans.\n is_integer : Check if the Index only consists of integers.\n is_floating : Check if the Index is a floating type.\n is_numeric : Check if the Index only consists of numeric data.\n is_object : Check if the Index is of the object dtype.\n is_interval : Check if the Index holds Interval objects.\n is_mixed : Check if the Index holds data with mixed data types.\n\n Examples\n --------\n >>> idx = pd.Index([\"Watermelon\", \"Orange\", \"Apple\",\n ... \"Watermelon\"]).astype(\"category\")\n >>> idx.is_categorical()\n True\n\n >>> idx = pd.Index([1, 3, 5, 7])\n >>> idx.is_categorical()\n False\n\n >>> s = pd.Series([\"Peter\", \"Victor\", \"Elisabeth\", \"Mar\"])\n >>> s\n 0 Peter\n 1 Victor\n 2 Elisabeth\n 3 Mar\n dtype: object\n >>> s.index.is_categorical()\n False\n \"\"\"\n return self.inferred_type in [\"categorical\"]\n\n def is_interval(self) -> bool:\n \"\"\"\n Check if the Index holds Interval objects.\n\n Returns\n -------\n bool\n Whether or not the Index holds Interval objects.\n\n See Also\n --------\n IntervalIndex : Index for Interval objects.\n is_boolean : Check if the Index only consists of booleans.\n is_integer : Check if the Index only consists of integers.\n is_floating : Check if the Index is a floating type.\n is_numeric : Check if the Index only consists of numeric data.\n is_object : Check if the Index is of the object dtype.\n is_categorical : Check if the Index holds categorical data.\n is_mixed : Check if the Index holds data with mixed data types.\n\n Examples\n --------\n >>> idx = pd.Index([pd.Interval(left=0, right=5),\n ... pd.Interval(left=5, right=10)])\n >>> idx.is_interval()\n True\n\n >>> idx = pd.Index([1, 3, 5, 7])\n >>> idx.is_interval()\n False\n \"\"\"\n return self.inferred_type in [\"interval\"]\n\n def is_mixed(self) -> bool:\n \"\"\"\n Check if the Index holds data with mixed data types.\n\n Returns\n -------\n bool\n Whether or not the Index holds data with mixed data types.\n\n See Also\n --------\n is_boolean : Check if the Index only consists of booleans.\n is_integer : Check if the Index only consists of integers.\n is_floating : Check if the Index is a floating type.\n is_numeric : Check if the Index only consists of numeric data.\n is_object : Check if the Index is of the object dtype.\n is_categorical : Check if the Index holds categorical data.\n is_interval : Check if the Index holds Interval objects.\n\n Examples\n --------\n >>> idx = pd.Index(['a', np.nan, 'b'])\n >>> idx.is_mixed()\n True\n\n >>> idx = pd.Index([1.0, 2.0, 3.0, 5.0])\n >>> idx.is_mixed()\n False\n \"\"\"\n warnings.warn(\n \"Index.is_mixed is deprecated and will be removed in a future version. \"\n \"Check index.inferred_type directly instead.\",\n FutureWarning,\n stacklevel=2,\n )\n return self.inferred_type in [\"mixed\"]\n\n def holds_integer(self) -> bool:\n \"\"\"\n Whether the type is an integer type.\n \"\"\"\n return self.inferred_type in [\"integer\", \"mixed-integer\"]\n\n @cache_readonly\n def inferred_type(self) -> str_t:\n \"\"\"\n Return a string of the type inferred from the values.\n \"\"\"\n return lib.infer_dtype(self, skipna=False)\n\n @cache_readonly\n def is_all_dates(self) -> bool:\n \"\"\"\n Whether or not the index values only consist of dates.\n \"\"\"\n return is_datetime_array(ensure_object(self.values))\n\n # --------------------------------------------------------------------\n # Pickle Methods\n\n def __reduce__(self):\n d = dict(data=self._data)\n d.update(self._get_attributes_dict())\n return _new_Index, (type(self), d), None\n\n # --------------------------------------------------------------------\n # Null Handling Methods\n\n _na_value = np.nan\n \"\"\"The expected NA value to use with this index.\"\"\"\n\n @cache_readonly\n def _isnan(self):\n \"\"\"\n Return if each value is NaN.\n \"\"\"\n if self._can_hold_na:\n return isna(self)\n else:\n # shouldn't reach to this condition by checking hasnans beforehand\n values = np.empty(len(self), dtype=np.bool_)\n values.fill(False)\n return values\n\n @cache_readonly\n def _nan_idxs(self):\n if self._can_hold_na:\n return self._isnan.nonzero()[0]\n else:\n return np.array([], dtype=np.int64)\n\n @cache_readonly\n def hasnans(self) -> bool:\n \"\"\"\n Return if I have any nans; enables various perf speedups.\n \"\"\"\n if self._can_hold_na:\n return bool(self._isnan.any())\n else:\n return False\n\n def isna(self):\n \"\"\"\n Detect missing values.\n\n Return a boolean same-sized object indicating if the values are NA.\n NA values, such as ``None``, :attr:`numpy.NaN` or :attr:`pd.NaT`, get\n mapped to ``True`` values.\n Everything else get mapped to ``False`` values. Characters such as\n empty strings `''` or :attr:`numpy.inf` are not considered NA values\n (unless you set ``pandas.options.mode.use_inf_as_na = True``).\n\n Returns\n -------\n numpy.ndarray\n A boolean array of whether my values are NA.\n\n See Also\n --------\n Index.notna : Boolean inverse of isna.\n Index.dropna : Omit entries with missing values.\n isna : Top-level isna.\n Series.isna : Detect missing values in Series object.\n\n Examples\n --------\n Show which entries in a pandas.Index are NA. The result is an\n array.\n\n >>> idx = pd.Index([5.2, 6.0, np.NaN])\n >>> idx\n Float64Index([5.2, 6.0, nan], dtype='float64')\n >>> idx.isna()\n array([False, False, True])\n\n Empty strings are not considered NA values. None is considered an NA\n value.\n\n >>> idx = pd.Index(['black', '', 'red', None])\n >>> idx\n Index(['black', '', 'red', None], dtype='object')\n >>> idx.isna()\n array([False, False, False, True])\n\n For datetimes, `NaT` (Not a Time) is considered as an NA value.\n\n >>> idx = pd.DatetimeIndex([pd.Timestamp('1940-04-25'),\n ... pd.Timestamp(''), None, pd.NaT])\n >>> idx\n DatetimeIndex(['1940-04-25', 'NaT', 'NaT', 'NaT'],\n dtype='datetime64[ns]', freq=None)\n >>> idx.isna()\n array([False, True, True, True])\n \"\"\"\n return self._isnan\n\n isnull = isna\n\n def notna(self):\n \"\"\"\n Detect existing (non-missing) values.\n\n Return a boolean same-sized object indicating if the values are not NA.\n Non-missing values get mapped to ``True``. Characters such as empty\n strings ``''`` or :attr:`numpy.inf` are not considered NA values\n (unless you set ``pandas.options.mode.use_inf_as_na = True``).\n NA values, such as None or :attr:`numpy.NaN`, get mapped to ``False``\n values.\n\n Returns\n -------\n numpy.ndarray\n Boolean array to indicate which entries are not NA.\n\n See Also\n --------\n Index.notnull : Alias of notna.\n Index.isna: Inverse of notna.\n notna : Top-level notna.\n\n Examples\n --------\n Show which entries in an Index are not NA. The result is an\n array.\n\n >>> idx = pd.Index([5.2, 6.0, np.NaN])\n >>> idx\n Float64Index([5.2, 6.0, nan], dtype='float64')\n >>> idx.notna()\n array([ True, True, False])\n\n Empty strings are not considered NA values. None is considered a NA\n value.\n\n >>> idx = pd.Index(['black', '', 'red', None])\n >>> idx\n Index(['black', '', 'red', None], dtype='object')\n >>> idx.notna()\n array([ True, True, True, False])\n \"\"\"\n return ~self.isna()\n\n notnull = notna\n\n def fillna(self, value=None, downcast=None):\n \"\"\"\n Fill NA/NaN values with the specified value.\n\n Parameters\n ----------\n value : scalar\n Scalar value to use to fill holes (e.g. 0).\n This value cannot be a list-likes.\n downcast : dict, default is None\n A dict of item->dtype of what to downcast if possible,\n or the string 'infer' which will try to downcast to an appropriate\n equal type (e.g. float64 to int64 if possible).\n\n Returns\n -------\n Index\n\n See Also\n --------\n DataFrame.fillna : Fill NaN values of a DataFrame.\n Series.fillna : Fill NaN Values of a Series.\n \"\"\"\n self._assert_can_do_op(value)\n if self.hasnans:\n result = self.putmask(self._isnan, value)\n if downcast is None:\n # no need to care metadata other than name\n # because it can't have freq if\n return Index(result, name=self.name)\n return self._shallow_copy()\n\n def dropna(self, how=\"any\"):\n \"\"\"\n Return Index without NA/NaN values.\n\n Parameters\n ----------\n how : {'any', 'all'}, default 'any'\n If the Index is a MultiIndex, drop the value when any or all levels\n are NaN.\n\n Returns\n -------\n Index\n \"\"\"\n if how not in (\"any\", \"all\"):\n raise ValueError(f\"invalid how option: {how}\")\n\n if self.hasnans:\n return self._shallow_copy(self._values[~self._isnan])\n return self._shallow_copy()\n\n # --------------------------------------------------------------------\n # Uniqueness Methods\n\n def unique(self, level=None):\n \"\"\"\n Return unique values in the index. Uniques are returned in order\n of appearance, this does NOT sort.\n\n Parameters\n ----------\n level : int or str, optional, default None\n Only return values from specified level (for MultiIndex).\n\n .. versionadded:: 0.23.0\n\n Returns\n -------\n Index without duplicates\n\n See Also\n --------\n unique\n Series.unique\n \"\"\"\n if level is not None:\n self._validate_index_level(level)\n result = super().unique()\n return self._shallow_copy(result)\n\n def drop_duplicates(self, keep=\"first\"):\n \"\"\"\n Return Index with duplicate values removed.\n\n Parameters\n ----------\n keep : {'first', 'last', ``False``}, default 'first'\n - 'first' : Drop duplicates except for the first occurrence.\n - 'last' : Drop duplicates except for the last occurrence.\n - ``False`` : Drop all duplicates.\n\n Returns\n -------\n deduplicated : Index\n\n See Also\n --------\n Series.drop_duplicates : Equivalent method on Series.\n DataFrame.drop_duplicates : Equivalent method on DataFrame.\n Index.duplicated : Related method on Index, indicating duplicate\n Index values.\n\n Examples\n --------\n Generate an pandas.Index with duplicate values.\n\n >>> idx = pd.Index(['lama', 'cow', 'lama', 'beetle', 'lama', 'hippo'])\n\n The `keep` parameter controls which duplicate values are removed.\n The value 'first' keeps the first occurrence for each\n set of duplicated entries. The default value of keep is 'first'.\n\n >>> idx.drop_duplicates(keep='first')\n Index(['lama', 'cow', 'beetle', 'hippo'], dtype='object')\n\n The value 'last' keeps the last occurrence for each set of duplicated\n entries.\n\n >>> idx.drop_duplicates(keep='last')\n Index(['cow', 'beetle', 'lama', 'hippo'], dtype='object')\n\n The value ``False`` discards all sets of duplicated entries.\n\n >>> idx.drop_duplicates(keep=False)\n Index(['cow', 'beetle', 'hippo'], dtype='object')\n \"\"\"\n return super().drop_duplicates(keep=keep)\n\n def duplicated(self, keep=\"first\"):\n \"\"\"\n Indicate duplicate index values.\n\n Duplicated values are indicated as ``True`` values in the resulting\n array. Either all duplicates, all except the first, or all except the\n last occurrence of duplicates can be indicated.\n\n Parameters\n ----------\n keep : {'first', 'last', False}, default 'first'\n The value or values in a set of duplicates to mark as missing.\n\n - 'first' : Mark duplicates as ``True`` except for the first\n occurrence.\n - 'last' : Mark duplicates as ``True`` except for the last\n occurrence.\n - ``False`` : Mark all duplicates as ``True``.\n\n Returns\n -------\n numpy.ndarray\n\n See Also\n --------\n Series.duplicated : Equivalent method on pandas.Series.\n DataFrame.duplicated : Equivalent method on pandas.DataFrame.\n Index.drop_duplicates : Remove duplicate values from Index.\n\n Examples\n --------\n By default, for each set of duplicated values, the first occurrence is\n set to False and all others to True:\n\n >>> idx = pd.Index(['lama', 'cow', 'lama', 'beetle', 'lama'])\n >>> idx.duplicated()\n array([False, False, True, False, True])\n\n which is equivalent to\n\n >>> idx.duplicated(keep='first')\n array([False, False, True, False, True])\n\n By using 'last', the last occurrence of each set of duplicated values\n is set on False and all others on True:\n\n >>> idx.duplicated(keep='last')\n array([ True, False, True, False, False])\n\n By setting keep on ``False``, all duplicates are True:\n\n >>> idx.duplicated(keep=False)\n array([ True, False, True, False, True])\n \"\"\"\n return super().duplicated(keep=keep)\n\n def _get_unique_index(self, dropna: bool = False):\n \"\"\"\n Returns an index containing unique values.\n\n Parameters\n ----------\n dropna : bool, default False\n If True, NaN values are dropped.\n\n Returns\n -------\n uniques : index\n \"\"\"\n if self.is_unique and not dropna:\n return self\n\n values = self.values\n\n if not self.is_unique:\n values = self.unique()\n if not isinstance(self, ABCMultiIndex):\n # extract an array to pass to _shallow_copy\n values = values._data\n\n if dropna:\n try:\n if self.hasnans:\n values = values[~isna(values)]\n except NotImplementedError:\n pass\n\n return self._shallow_copy(values)\n\n # --------------------------------------------------------------------\n # Arithmetic & Logical Methods\n\n def __add__(self, other):\n if isinstance(other, (ABCSeries, ABCDataFrame)):\n return NotImplemented\n from pandas import Series\n\n return Index(Series(self) + other)\n\n def __radd__(self, other):\n from pandas import Series\n\n return Index(other + Series(self))\n\n def __iadd__(self, other):\n # alias for __add__\n return self + other\n\n def __sub__(self, other):\n return Index(np.array(self) - other)\n\n def __rsub__(self, other):\n # wrap Series to ensure we pin name correctly\n from pandas import Series\n\n return Index(other - Series(self))\n\n def __and__(self, other):\n return self.intersection(other)\n\n def __or__(self, other):\n return self.union(other)\n\n def __xor__(self, other):\n return self.symmetric_difference(other)\n\n def __nonzero__(self):\n raise ValueError(\n f\"The truth value of a {type(self).__name__} is ambiguous. \"\n \"Use a.empty, a.bool(), a.item(), a.any() or a.all().\"\n )\n\n __bool__ = __nonzero__\n\n # --------------------------------------------------------------------\n # Set Operation Methods\n\n def _get_reconciled_name_object(self, other):\n \"\"\"\n If the result of a set operation will be self,\n return self, unless the name changes, in which\n case make a shallow copy of self.\n \"\"\"\n name = get_op_result_name(self, other)\n if self.name != name:\n return self._shallow_copy(name=name)\n return self\n\n def _union_incompatible_dtypes(self, other, sort):\n \"\"\"\n Casts this and other index to object dtype to allow the formation\n of a union between incompatible types.\n\n Parameters\n ----------\n other : Index or array-like\n sort : False or None, default False\n Whether to sort the resulting index.\n\n * False : do not sort the result.\n * None : sort the result, except when `self` and `other` are equal\n or when the values cannot be compared.\n\n Returns\n -------\n Index\n \"\"\"\n this = self.astype(object, copy=False)\n # cast to Index for when `other` is list-like\n other = Index(other).astype(object, copy=False)\n return Index.union(this, other, sort=sort).astype(object, copy=False)\n\n def _is_compatible_with_other(self, other) -> bool:\n \"\"\"\n Check whether this and the other dtype are compatible with each other.\n Meaning a union can be formed between them without needing to be cast\n to dtype object.\n\n Parameters\n ----------\n other : Index or array-like\n\n Returns\n -------\n bool\n \"\"\"\n return type(self) is type(other) and is_dtype_equal(self.dtype, other.dtype)\n\n def _validate_sort_keyword(self, sort):\n if sort not in [None, False]:\n raise ValueError(\n \"The 'sort' keyword only takes the values of \"\n f\"None or False; {sort} was passed.\"\n )\n\n def union(self, other, sort=None):\n \"\"\"\n Form the union of two Index objects.\n\n If the Index objects are incompatible, both Index objects will be\n cast to dtype('object') first.\n\n .. versionchanged:: 0.25.0\n\n Parameters\n ----------\n other : Index or array-like\n sort : bool or None, default None\n Whether to sort the resulting Index.\n\n * None : Sort the result, except when\n\n 1. `self` and `other` are equal.\n 2. `self` or `other` has length 0.\n 3. Some values in `self` or `other` cannot be compared.\n A RuntimeWarning is issued in this case.\n\n * False : do not sort the result.\n\n .. versionadded:: 0.24.0\n\n .. versionchanged:: 0.24.1\n\n Changed the default value from ``True`` to ``None``\n (without change in behaviour).\n\n Returns\n -------\n union : Index\n\n Examples\n --------\n Union matching dtypes\n\n >>> idx1 = pd.Index([1, 2, 3, 4])\n >>> idx2 = pd.Index([3, 4, 5, 6])\n >>> idx1.union(idx2)\n Int64Index([1, 2, 3, 4, 5, 6], dtype='int64')\n\n Union mismatched dtypes\n\n >>> idx1 = pd.Index(['a', 'b', 'c', 'd'])\n >>> idx2 = pd.Index([1, 2, 3, 4])\n >>> idx1.union(idx2)\n Index(['a', 'b', 'c', 'd', 1, 2, 3, 4], dtype='object')\n \"\"\"\n self._validate_sort_keyword(sort)\n self._assert_can_do_setop(other)\n\n if not self._is_compatible_with_other(other):\n return self._union_incompatible_dtypes(other, sort=sort)\n\n return self._union(other, sort=sort)\n\n def _union(self, other, sort):\n \"\"\"\n Specific union logic should go here. In subclasses, union behavior\n should be overwritten here rather than in `self.union`.\n\n Parameters\n ----------\n other : Index or array-like\n sort : False or None, default False\n Whether to sort the resulting index.\n\n * False : do not sort the result.\n * None : sort the result, except when `self` and `other` are equal\n or when the values cannot be compared.\n\n Returns\n -------\n Index\n \"\"\"\n if not len(other) or self.equals(other):\n return self._get_reconciled_name_object(other)\n\n if not len(self):\n return other._get_reconciled_name_object(self)\n\n # TODO(EA): setops-refactor, clean all this up\n lvals = self._values\n rvals = other._values\n\n if sort is None and self.is_monotonic and other.is_monotonic:\n try:\n result = self._outer_indexer(lvals, rvals)[0]\n except TypeError:\n # incomparable objects\n result = list(lvals)\n\n # worth making this faster? a very unusual case\n value_set = set(lvals)\n result.extend([x for x in rvals if x not in value_set])\n result = Index(result)._values # do type inference here\n else:\n # find indexes of things in \"other\" that are not in \"self\"\n if self.is_unique:\n indexer = self.get_indexer(other)\n indexer = (indexer == -1).nonzero()[0]\n else:\n indexer = algos.unique1d(self.get_indexer_non_unique(other)[1])\n\n if len(indexer) > 0:\n other_diff = algos.take_nd(rvals, indexer, allow_fill=False)\n result = concat_compat((lvals, other_diff))\n\n else:\n result = lvals\n\n if sort is None:\n try:\n result = algos.safe_sort(result)\n except TypeError as err:\n warnings.warn(\n f\"{err}, sort order is undefined for incomparable objects\",\n RuntimeWarning,\n stacklevel=3,\n )\n\n # for subclasses\n return self._wrap_setop_result(other, result)\n\n def _wrap_setop_result(self, other, result):\n name = get_op_result_name(self, other)\n return self._shallow_copy(result, name=name)\n\n # TODO: standardize return type of non-union setops type(self vs other)\n def intersection(self, other, sort=False):\n \"\"\"\n Form the intersection of two Index objects.\n\n This returns a new Index with elements common to the index and `other`.\n\n Parameters\n ----------\n other : Index or array-like\n sort : False or None, default False\n Whether to sort the resulting index.\n\n * False : do not sort the result.\n * None : sort the result, except when `self` and `other` are equal\n or when the values cannot be compared.\n\n .. versionadded:: 0.24.0\n\n .. versionchanged:: 0.24.1\n\n Changed the default from ``True`` to ``False``, to match\n the behaviour of 0.23.4 and earlier.\n\n Returns\n -------\n intersection : Index\n\n Examples\n --------\n >>> idx1 = pd.Index([1, 2, 3, 4])\n >>> idx2 = pd.Index([3, 4, 5, 6])\n >>> idx1.intersection(idx2)\n Int64Index([3, 4], dtype='int64')\n \"\"\"\n self._validate_sort_keyword(sort)\n self._assert_can_do_setop(other)\n other = ensure_index(other)\n\n if self.equals(other):\n return self._get_reconciled_name_object(other)\n\n if not is_dtype_equal(self.dtype, other.dtype):\n this = self.astype(\"O\")\n other = other.astype(\"O\")\n return this.intersection(other, sort=sort)\n\n # TODO(EA): setops-refactor, clean all this up\n lvals = self._values\n rvals = other._values\n\n if self.is_monotonic and other.is_monotonic:\n try:\n result = self._inner_indexer(lvals, rvals)[0]\n except TypeError:\n pass\n else:\n return self._wrap_setop_result(other, result)\n\n try:\n indexer = Index(rvals).get_indexer(lvals)\n indexer = indexer.take((indexer != -1).nonzero()[0])\n except (InvalidIndexError, IncompatibleFrequency):\n # InvalidIndexError raised by get_indexer if non-unique\n # IncompatibleFrequency raised by PeriodIndex.get_indexer\n indexer = algos.unique1d(Index(rvals).get_indexer_non_unique(lvals)[0])\n indexer = indexer[indexer != -1]\n\n taken = other.take(indexer)\n res_name = get_op_result_name(self, other)\n\n if sort is None:\n taken = algos.safe_sort(taken.values)\n return self._shallow_copy(taken, name=res_name)\n\n taken.name = res_name\n return taken\n\n def difference(self, other, sort=None):\n \"\"\"\n Return a new Index with elements from the index that are not in\n `other`.\n\n This is the set difference of two Index objects.\n\n Parameters\n ----------\n other : Index or array-like\n sort : False or None, default None\n Whether to sort the resulting index. By default, the\n values are attempted to be sorted, but any TypeError from\n incomparable elements is caught by pandas.\n\n * None : Attempt to sort the result, but catch any TypeErrors\n from comparing incomparable elements.\n * False : Do not sort the result.\n\n .. versionadded:: 0.24.0\n\n .. versionchanged:: 0.24.1\n\n Changed the default value from ``True`` to ``None``\n (without change in behaviour).\n\n Returns\n -------\n difference : Index\n\n Examples\n --------\n >>> idx1 = pd.Index([2, 1, 3, 4])\n >>> idx2 = pd.Index([3, 4, 5, 6])\n >>> idx1.difference(idx2)\n Int64Index([1, 2], dtype='int64')\n >>> idx1.difference(idx2, sort=False)\n Int64Index([2, 1], dtype='int64')\n \"\"\"\n self._validate_sort_keyword(sort)\n self._assert_can_do_setop(other)\n\n if self.equals(other):\n # pass an empty np.ndarray with the appropriate dtype\n return self._shallow_copy(self._data[:0])\n\n other, result_name = self._convert_can_do_setop(other)\n\n this = self._get_unique_index()\n\n indexer = this.get_indexer(other)\n indexer = indexer.take((indexer != -1).nonzero()[0])\n\n label_diff = np.setdiff1d(np.arange(this.size), indexer, assume_unique=True)\n the_diff = this.values.take(label_diff)\n if sort is None:\n try:\n the_diff = algos.safe_sort(the_diff)\n except TypeError:\n pass\n\n return this._shallow_copy(the_diff, name=result_name)\n\n def symmetric_difference(self, other, result_name=None, sort=None):\n \"\"\"\n Compute the symmetric difference of two Index objects.\n\n Parameters\n ----------\n other : Index or array-like\n result_name : str\n sort : False or None, default None\n Whether to sort the resulting index. By default, the\n values are attempted to be sorted, but any TypeError from\n incomparable elements is caught by pandas.\n\n * None : Attempt to sort the result, but catch any TypeErrors\n from comparing incomparable elements.\n * False : Do not sort the result.\n\n .. versionadded:: 0.24.0\n\n .. versionchanged:: 0.24.1\n\n Changed the default value from ``True`` to ``None``\n (without change in behaviour).\n\n Returns\n -------\n symmetric_difference : Index\n\n Notes\n -----\n ``symmetric_difference`` contains elements that appear in either\n ``idx1`` or ``idx2`` but not both. Equivalent to the Index created by\n ``idx1.difference(idx2) | idx2.difference(idx1)`` with duplicates\n dropped.\n\n Examples\n --------\n >>> idx1 = pd.Index([1, 2, 3, 4])\n >>> idx2 = pd.Index([2, 3, 4, 5])\n >>> idx1.symmetric_difference(idx2)\n Int64Index([1, 5], dtype='int64')\n\n You can also use the ``^`` operator:\n\n >>> idx1 ^ idx2\n Int64Index([1, 5], dtype='int64')\n \"\"\"\n self._validate_sort_keyword(sort)\n self._assert_can_do_setop(other)\n other, result_name_update = self._convert_can_do_setop(other)\n if result_name is None:\n result_name = result_name_update\n\n this = self._get_unique_index()\n other = other._get_unique_index()\n indexer = this.get_indexer(other)\n\n # {this} minus {other}\n common_indexer = indexer.take((indexer != -1).nonzero()[0])\n left_indexer = np.setdiff1d(\n np.arange(this.size), common_indexer, assume_unique=True\n )\n left_diff = this._values.take(left_indexer)\n\n # {other} minus {this}\n right_indexer = (indexer == -1).nonzero()[0]\n right_diff = other._values.take(right_indexer)\n\n the_diff = concat_compat([left_diff, right_diff])\n if sort is None:\n try:\n the_diff = algos.safe_sort(the_diff)\n except TypeError:\n pass\n\n attribs = self._get_attributes_dict()\n attribs[\"name\"] = result_name\n if \"freq\" in attribs:\n attribs[\"freq\"] = None\n return self._shallow_copy_with_infer(the_diff, **attribs)\n\n def _assert_can_do_setop(self, other):\n if not is_list_like(other):\n raise TypeError(\"Input must be Index or array-like\")\n return True\n\n def _convert_can_do_setop(self, other):\n if not isinstance(other, Index):\n other = Index(other, name=self.name)\n result_name = self.name\n else:\n result_name = get_op_result_name(self, other)\n return other, result_name\n\n # --------------------------------------------------------------------\n # Indexing Methods\n\n def get_loc(self, key, method=None, tolerance=None):\n \"\"\"\n Get integer location, slice or boolean mask for requested label.\n\n Parameters\n ----------\n key : label\n method : {None, 'pad'/'ffill', 'backfill'/'bfill', 'nearest'}, optional\n * default: exact matches only.\n * pad / ffill: find the PREVIOUS index value if no exact match.\n * backfill / bfill: use NEXT index value if no exact match\n * nearest: use the NEAREST index value if no exact match. Tied\n distances are broken by preferring the larger index value.\n tolerance : int or float, optional\n Maximum distance from index value for inexact matches. The value of\n the index at the matching location most satisfy the equation\n ``abs(index[loc] - key) <= tolerance``.\n\n Returns\n -------\n loc : int if unique index, slice if monotonic index, else mask\n\n Examples\n --------\n >>> unique_index = pd.Index(list('abc'))\n >>> unique_index.get_loc('b')\n 1\n\n >>> monotonic_index = pd.Index(list('abbc'))\n >>> monotonic_index.get_loc('b')\n slice(1, 3, None)\n\n >>> non_monotonic_index = pd.Index(list('abcb'))\n >>> non_monotonic_index.get_loc('b')\n array([False, True, False, True])\n \"\"\"\n if method is None:\n if tolerance is not None:\n raise ValueError(\n \"tolerance argument only valid if using pad, \"\n \"backfill or nearest lookups\"\n )\n casted_key = self._maybe_cast_indexer(key)\n try:\n return self._engine.get_loc(casted_key)\n except KeyError as err:\n raise KeyError(key) from err\n\n if tolerance is not None:\n tolerance = self._convert_tolerance(tolerance, np.asarray(key))\n\n indexer = self.get_indexer([key], method=method, tolerance=tolerance)\n if indexer.ndim > 1 or indexer.size > 1:\n raise TypeError(\"get_loc requires scalar valued input\")\n loc = indexer.item()\n if loc == -1:\n raise KeyError(key)\n return loc\n\n _index_shared_docs[\n \"get_indexer\"\n ] = \"\"\"\n Compute indexer and mask for new index given the current index. The\n indexer should be then used as an input to ndarray.take to align the\n current data to the new index.\n\n Parameters\n ----------\n target : %(target_klass)s\n method : {None, 'pad'/'ffill', 'backfill'/'bfill', 'nearest'}, optional\n * default: exact matches only.\n * pad / ffill: find the PREVIOUS index value if no exact match.\n * backfill / bfill: use NEXT index value if no exact match\n * nearest: use the NEAREST index value if no exact match. Tied\n distances are broken by preferring the larger index value.\n limit : int, optional\n Maximum number of consecutive labels in ``target`` to match for\n inexact matches.\n tolerance : optional\n Maximum distance between original and new labels for inexact\n matches. The values of the index at the matching locations most\n satisfy the equation ``abs(index[indexer] - target) <= tolerance``.\n\n Tolerance may be a scalar value, which applies the same tolerance\n to all values, or list-like, which applies variable tolerance per\n element. List-like includes list, tuple, array, Series, and must be\n the same size as the index and its dtype must exactly match the\n index's type.\n\n Returns\n -------\n indexer : ndarray of int\n Integers from 0 to n - 1 indicating that the index at these\n positions matches the corresponding target values. Missing values\n in the target are marked by -1.\n %(raises_section)s\n Examples\n --------\n >>> index = pd.Index(['c', 'a', 'b'])\n >>> index.get_indexer(['a', 'b', 'x'])\n array([ 1, 2, -1])\n\n Notice that the return value is an array of locations in ``index``\n and ``x`` is marked by -1, as it is not in ``index``.\n \"\"\"\n\n @Appender(_index_shared_docs[\"get_indexer\"] % _index_doc_kwargs)\n def get_indexer(\n self, target, method=None, limit=None, tolerance=None\n ) -> np.ndarray:\n method = missing.clean_reindex_fill_method(method)\n target = ensure_index(target)\n if tolerance is not None:\n tolerance = self._convert_tolerance(tolerance, target)\n\n # Treat boolean labels passed to a numeric index as not found. Without\n # this fix False and True would be treated as 0 and 1 respectively.\n # (GH #16877)\n if target.is_boolean() and self.is_numeric():\n return ensure_platform_int(np.repeat(-1, target.size))\n\n pself, ptarget = self._maybe_promote(target)\n if pself is not self or ptarget is not target:\n return pself.get_indexer(\n ptarget, method=method, limit=limit, tolerance=tolerance\n )\n\n if not is_dtype_equal(self.dtype, target.dtype):\n this = self.astype(object)\n target = target.astype(object)\n return this.get_indexer(\n target, method=method, limit=limit, tolerance=tolerance\n )\n\n if not self.is_unique:\n raise InvalidIndexError(\n \"Reindexing only valid with uniquely valued Index objects\"\n )\n\n if method == \"pad\" or method == \"backfill\":\n indexer = self._get_fill_indexer(target, method, limit, tolerance)\n elif method == \"nearest\":\n indexer = self._get_nearest_indexer(target, limit, tolerance)\n else:\n if tolerance is not None:\n raise ValueError(\n \"tolerance argument only valid if doing pad, \"\n \"backfill or nearest reindexing\"\n )\n if limit is not None:\n raise ValueError(\n \"limit argument only valid if doing pad, \"\n \"backfill or nearest reindexing\"\n )\n\n indexer = self._engine.get_indexer(target._get_engine_target())\n\n return ensure_platform_int(indexer)\n\n def _convert_tolerance(self, tolerance, target):\n # override this method on subclasses\n tolerance = np.asarray(tolerance)\n if target.size != tolerance.size and tolerance.size > 1:\n raise ValueError(\"list-like tolerance size must match target index size\")\n return tolerance\n\n def _get_fill_indexer(\n self, target: \"Index\", method: str_t, limit=None, tolerance=None\n ) -> np.ndarray:\n\n target_values = target._get_engine_target()\n\n if self.is_monotonic_increasing and target.is_monotonic_increasing:\n engine_method = (\n self._engine.get_pad_indexer\n if method == \"pad\"\n else self._engine.get_backfill_indexer\n )\n indexer = engine_method(target_values, limit)\n else:\n indexer = self._get_fill_indexer_searchsorted(target, method, limit)\n if tolerance is not None:\n indexer = self._filter_indexer_tolerance(target_values, indexer, tolerance)\n return indexer\n\n def _get_fill_indexer_searchsorted(\n self, target: \"Index\", method: str_t, limit=None\n ) -> np.ndarray:\n \"\"\"\n Fallback pad/backfill get_indexer that works for monotonic decreasing\n indexes and non-monotonic targets.\n \"\"\"\n if limit is not None:\n raise ValueError(\n f\"limit argument for {repr(method)} method only well-defined \"\n \"if index and target are monotonic\"\n )\n\n side = \"left\" if method == \"pad\" else \"right\"\n\n # find exact matches first (this simplifies the algorithm)\n indexer = self.get_indexer(target)\n nonexact = indexer == -1\n indexer[nonexact] = self._searchsorted_monotonic(target[nonexact], side)\n if side == \"left\":\n # searchsorted returns \"indices into a sorted array such that,\n # if the corresponding elements in v were inserted before the\n # indices, the order of a would be preserved\".\n # Thus, we need to subtract 1 to find values to the left.\n indexer[nonexact] -= 1\n # This also mapped not found values (values of 0 from\n # np.searchsorted) to -1, which conveniently is also our\n # sentinel for missing values\n else:\n # Mark indices to the right of the largest value as not found\n indexer[indexer == len(self)] = -1\n return indexer\n\n def _get_nearest_indexer(self, target: \"Index\", limit, tolerance) -> np.ndarray:\n \"\"\"\n Get the indexer for the nearest index labels; requires an index with\n values that can be subtracted from each other (e.g., not strings or\n tuples).\n \"\"\"\n left_indexer = self.get_indexer(target, \"pad\", limit=limit)\n right_indexer = self.get_indexer(target, \"backfill\", limit=limit)\n\n target_values = target._values\n left_distances = np.abs(self._values[left_indexer] - target_values)\n right_distances = np.abs(self._values[right_indexer] - target_values)\n\n op = operator.lt if self.is_monotonic_increasing else operator.le\n indexer = np.where(\n op(left_distances, right_distances) | (right_indexer == -1),\n left_indexer,\n right_indexer,\n )\n if tolerance is not None:\n indexer = self._filter_indexer_tolerance(target_values, indexer, tolerance)\n return indexer\n\n def _filter_indexer_tolerance(\n self,\n target: Union[\"Index\", np.ndarray, ExtensionArray],\n indexer: np.ndarray,\n tolerance,\n ) -> np.ndarray:\n distance = abs(self._values[indexer] - target)\n indexer = np.where(distance <= tolerance, indexer, -1)\n return indexer\n\n # --------------------------------------------------------------------\n # Indexer Conversion Methods\n\n def _get_partial_string_timestamp_match_key(self, key):\n \"\"\"\n Translate any partial string timestamp matches in key, returning the\n new key.\n\n Only relevant for MultiIndex.\n \"\"\"\n # GH#10331\n return key\n\n def _validate_positional_slice(self, key: slice):\n \"\"\"\n For positional indexing, a slice must have either int or None\n for each of start, stop, and step.\n \"\"\"\n self._validate_indexer(\"positional\", key.start, \"iloc\")\n self._validate_indexer(\"positional\", key.stop, \"iloc\")\n self._validate_indexer(\"positional\", key.step, \"iloc\")\n\n def _convert_slice_indexer(self, key: slice, kind: str_t):\n \"\"\"\n Convert a slice indexer.\n\n By definition, these are labels unless 'iloc' is passed in.\n Floats are not allowed as the start, step, or stop of the slice.\n\n Parameters\n ----------\n key : label of the slice bound\n kind : {'loc', 'getitem'}\n \"\"\"\n assert kind in [\"loc\", \"getitem\"], kind\n\n # potentially cast the bounds to integers\n start, stop, step = key.start, key.stop, key.step\n\n # figure out if this is a positional indexer\n def is_int(v):\n return v is None or is_integer(v)\n\n is_index_slice = is_int(start) and is_int(stop) and is_int(step)\n is_positional = is_index_slice and not (\n self.is_integer() or self.is_categorical()\n )\n\n if kind == \"getitem\":\n \"\"\"\n called from the getitem slicers, validate that we are in fact\n integers\n \"\"\"\n if self.is_integer() or is_index_slice:\n self._validate_indexer(\"slice\", key.start, \"getitem\")\n self._validate_indexer(\"slice\", key.stop, \"getitem\")\n self._validate_indexer(\"slice\", key.step, \"getitem\")\n return key\n\n # convert the slice to an indexer here\n\n # if we are mixed and have integers\n if is_positional:\n try:\n # Validate start & stop\n if start is not None:\n self.get_loc(start)\n if stop is not None:\n self.get_loc(stop)\n is_positional = False\n except KeyError:\n pass\n\n if com.is_null_slice(key):\n # It doesn't matter if we are positional or label based\n indexer = key\n elif is_positional:\n if kind == \"loc\":\n # GH#16121, GH#24612, GH#31810\n warnings.warn(\n \"Slicing a positional slice with .loc is not supported, \"\n \"and will raise TypeError in a future version. \"\n \"Use .loc with labels or .iloc with positions instead.\",\n FutureWarning,\n stacklevel=6,\n )\n indexer = key\n else:\n indexer = self.slice_indexer(start, stop, step, kind=kind)\n\n return indexer\n\n def _convert_listlike_indexer(self, keyarr):\n \"\"\"\n Parameters\n ----------\n keyarr : list-like\n Indexer to convert.\n\n Returns\n -------\n indexer : numpy.ndarray or None\n Return an ndarray or None if cannot convert.\n keyarr : numpy.ndarray\n Return tuple-safe keys.\n \"\"\"\n if isinstance(keyarr, Index):\n keyarr = self._convert_index_indexer(keyarr)\n else:\n keyarr = self._convert_arr_indexer(keyarr)\n\n indexer = self._convert_list_indexer(keyarr)\n return indexer, keyarr\n\n def _convert_arr_indexer(self, keyarr):\n \"\"\"\n Convert an array-like indexer to the appropriate dtype.\n\n Parameters\n ----------\n keyarr : array-like\n Indexer to convert.\n\n Returns\n -------\n converted_keyarr : array-like\n \"\"\"\n keyarr = com.asarray_tuplesafe(keyarr)\n return keyarr\n\n def _convert_index_indexer(self, keyarr):\n \"\"\"\n Convert an Index indexer to the appropriate dtype.\n\n Parameters\n ----------\n keyarr : Index (or sub-class)\n Indexer to convert.\n\n Returns\n -------\n converted_keyarr : Index (or sub-class)\n \"\"\"\n return keyarr\n\n def _convert_list_indexer(self, keyarr):\n \"\"\"\n Convert a list-like indexer to the appropriate dtype.\n\n Parameters\n ----------\n keyarr : Index (or sub-class)\n Indexer to convert.\n kind : iloc, loc, optional\n\n Returns\n -------\n positional indexer or None\n \"\"\"\n return None\n\n def _invalid_indexer(self, form: str_t, key):\n \"\"\"\n Consistent invalid indexer message.\n \"\"\"\n raise TypeError(\n f\"cannot do {form} indexing on {type(self).__name__} with these \"\n f\"indexers [{key}] of type {type(key).__name__}\"\n )\n\n # --------------------------------------------------------------------\n # Reindex Methods\n\n def _can_reindex(self, indexer):\n \"\"\"\n Check if we are allowing reindexing with this particular indexer.\n\n Parameters\n ----------\n indexer : an integer indexer\n\n Raises\n ------\n ValueError if its a duplicate axis\n \"\"\"\n # trying to reindex on an axis with duplicates\n if not self.is_unique and len(indexer):\n raise ValueError(\"cannot reindex from a duplicate axis\")\n\n def reindex(self, target, method=None, level=None, limit=None, tolerance=None):\n \"\"\"\n Create index with target's values (move/add/delete values\n as necessary).\n\n Parameters\n ----------\n target : an iterable\n\n Returns\n -------\n new_index : pd.Index\n Resulting index.\n indexer : np.ndarray or None\n Indices of output values in original index.\n \"\"\"\n # GH6552: preserve names when reindexing to non-named target\n # (i.e. neither Index nor Series).\n preserve_names = not hasattr(target, \"name\")\n\n # GH7774: preserve dtype/tz if target is empty and not an Index.\n target = ensure_has_len(target) # target may be an iterator\n\n if not isinstance(target, Index) and len(target) == 0:\n if isinstance(self, ABCRangeIndex):\n values = range(0)\n else:\n values = self._data[:0] # appropriately-dtyped empty array\n target = self._simple_new(values, name=self.name)\n else:\n target = ensure_index(target)\n\n if level is not None:\n if method is not None:\n raise TypeError(\"Fill method not supported if level passed\")\n _, indexer, _ = self._join_level(\n target, level, how=\"right\", return_indexers=True\n )\n else:\n if self.equals(target):\n indexer = None\n else:\n # check is_overlapping for IntervalIndex compat\n if self.is_unique and not getattr(self, \"is_overlapping\", False):\n indexer = self.get_indexer(\n target, method=method, limit=limit, tolerance=tolerance\n )\n else:\n if method is not None or limit is not None:\n raise ValueError(\n \"cannot reindex a non-unique index \"\n \"with a method or limit\"\n )\n indexer, missing = self.get_indexer_non_unique(target)\n\n if preserve_names and target.nlevels == 1 and target.name != self.name:\n target = target.copy()\n target.name = self.name\n\n return target, indexer\n\n def _reindex_non_unique(self, target):\n \"\"\"\n Create a new index with target's values (move/add/delete values as\n necessary) use with non-unique Index and a possibly non-unique target.\n\n Parameters\n ----------\n target : an iterable\n\n Returns\n -------\n new_index : pd.Index\n Resulting index.\n indexer : np.ndarray or None\n Indices of output values in original index.\n\n \"\"\"\n target = ensure_index(target)\n indexer, missing = self.get_indexer_non_unique(target)\n check = indexer != -1\n new_labels = self.take(indexer[check])\n new_indexer = None\n\n if len(missing):\n length = np.arange(len(indexer))\n\n missing = ensure_platform_int(missing)\n missing_labels = target.take(missing)\n missing_indexer = ensure_int64(length[~check])\n cur_labels = self.take(indexer[check]).values\n cur_indexer = ensure_int64(length[check])\n\n new_labels = np.empty(tuple([len(indexer)]), dtype=object)\n new_labels[cur_indexer] = cur_labels\n new_labels[missing_indexer] = missing_labels\n\n # a unique indexer\n if target.is_unique:\n\n # see GH5553, make sure we use the right indexer\n new_indexer = np.arange(len(indexer))\n new_indexer[cur_indexer] = np.arange(len(cur_labels))\n new_indexer[missing_indexer] = -1\n\n # we have a non_unique selector, need to use the original\n # indexer here\n else:\n\n # need to retake to have the same size as the indexer\n indexer[~check] = -1\n\n # reset the new indexer to account for the new size\n new_indexer = np.arange(len(self.take(indexer)))\n new_indexer[~check] = -1\n\n new_index = self._shallow_copy_with_infer(new_labels)\n return new_index, indexer, new_indexer\n\n # --------------------------------------------------------------------\n # Join Methods\n\n def join(self, other, how=\"left\", level=None, return_indexers=False, sort=False):\n \"\"\"\n Compute join_index and indexers to conform data\n structures to the new index.\n\n Parameters\n ----------\n other : Index\n how : {'left', 'right', 'inner', 'outer'}\n level : int or level name, default None\n return_indexers : bool, default False\n sort : bool, default False\n Sort the join keys lexicographically in the result Index. If False,\n the order of the join keys depends on the join type (how keyword).\n\n Returns\n -------\n join_index, (left_indexer, right_indexer)\n \"\"\"\n other = ensure_index(other)\n self_is_mi = isinstance(self, ABCMultiIndex)\n other_is_mi = isinstance(other, ABCMultiIndex)\n\n # try to figure out the join level\n # GH3662\n if level is None and (self_is_mi or other_is_mi):\n\n # have the same levels/names so a simple join\n if self.names == other.names:\n pass\n else:\n return self._join_multi(other, how=how, return_indexers=return_indexers)\n\n # join on the level\n if level is not None and (self_is_mi or other_is_mi):\n return self._join_level(\n other, level, how=how, return_indexers=return_indexers\n )\n\n if len(other) == 0 and how in (\"left\", \"outer\"):\n join_index = self._shallow_copy()\n if return_indexers:\n rindexer = np.repeat(-1, len(join_index))\n return join_index, None, rindexer\n else:\n return join_index\n\n if len(self) == 0 and how in (\"right\", \"outer\"):\n join_index = other._shallow_copy()\n if return_indexers:\n lindexer = np.repeat(-1, len(join_index))\n return join_index, lindexer, None\n else:\n return join_index\n\n if self._join_precedence < other._join_precedence:\n how = {\"right\": \"left\", \"left\": \"right\"}.get(how, how)\n result = other.join(\n self, how=how, level=level, return_indexers=return_indexers\n )\n if return_indexers:\n x, y, z = result\n result = x, z, y\n return result\n\n if not is_dtype_equal(self.dtype, other.dtype):\n this = self.astype(\"O\")\n other = other.astype(\"O\")\n return this.join(other, how=how, return_indexers=return_indexers)\n\n _validate_join_method(how)\n\n if not self.is_unique and not other.is_unique:\n return self._join_non_unique(\n other, how=how, return_indexers=return_indexers\n )\n elif not self.is_unique or not other.is_unique:\n if self.is_monotonic and other.is_monotonic:\n return self._join_monotonic(\n other, how=how, return_indexers=return_indexers\n )\n else:\n return self._join_non_unique(\n other, how=how, return_indexers=return_indexers\n )\n elif self.is_monotonic and other.is_monotonic:\n try:\n return self._join_monotonic(\n other, how=how, return_indexers=return_indexers\n )\n except TypeError:\n pass\n\n if how == \"left\":\n join_index = self\n elif how == \"right\":\n join_index = other\n elif how == \"inner\":\n # TODO: sort=False here for backwards compat. It may\n # be better to use the sort parameter passed into join\n join_index = self.intersection(other, sort=False)\n elif how == \"outer\":\n # TODO: sort=True here for backwards compat. It may\n # be better to use the sort parameter passed into join\n join_index = self.union(other)\n\n if sort:\n join_index = join_index.sort_values()\n\n if return_indexers:\n if join_index is self:\n lindexer = None\n else:\n lindexer = self.get_indexer(join_index)\n if join_index is other:\n rindexer = None\n else:\n rindexer = other.get_indexer(join_index)\n return join_index, lindexer, rindexer\n else:\n return join_index\n\n def _join_multi(self, other, how, return_indexers=True):\n from pandas.core.indexes.multi import MultiIndex\n from pandas.core.reshape.merge import _restore_dropped_levels_multijoin\n\n # figure out join names\n self_names = set(com.not_none(*self.names))\n other_names = set(com.not_none(*other.names))\n overlap = self_names & other_names\n\n # need at least 1 in common\n if not overlap:\n raise ValueError(\"cannot join with no overlapping index names\")\n\n self_is_mi = isinstance(self, ABCMultiIndex)\n other_is_mi = isinstance(other, ABCMultiIndex)\n\n if self_is_mi and other_is_mi:\n\n # Drop the non-matching levels from left and right respectively\n ldrop_names = list(self_names - overlap)\n rdrop_names = list(other_names - overlap)\n\n # if only the order differs\n if not len(ldrop_names + rdrop_names):\n self_jnlevels = self\n other_jnlevels = other.reorder_levels(self.names)\n else:\n self_jnlevels = self.droplevel(ldrop_names)\n other_jnlevels = other.droplevel(rdrop_names)\n\n # Join left and right\n # Join on same leveled multi-index frames is supported\n join_idx, lidx, ridx = self_jnlevels.join(\n other_jnlevels, how, return_indexers=True\n )\n\n # Restore the dropped levels\n # Returned index level order is\n # common levels, ldrop_names, rdrop_names\n dropped_names = ldrop_names + rdrop_names\n\n levels, codes, names = _restore_dropped_levels_multijoin(\n self, other, dropped_names, join_idx, lidx, ridx\n )\n\n # Re-create the multi-index\n multi_join_idx = MultiIndex(\n levels=levels, codes=codes, names=names, verify_integrity=False\n )\n\n multi_join_idx = multi_join_idx.remove_unused_levels()\n\n return multi_join_idx, lidx, ridx\n\n jl = list(overlap)[0]\n\n # Case where only one index is multi\n # make the indices into mi's that match\n flip_order = False\n if self_is_mi:\n self, other = other, self\n flip_order = True\n # flip if join method is right or left\n how = {\"right\": \"left\", \"left\": \"right\"}.get(how, how)\n\n level = other.names.index(jl)\n result = self._join_level(\n other, level, how=how, return_indexers=return_indexers\n )\n\n if flip_order:\n if isinstance(result, tuple):\n return result[0], result[2], result[1]\n return result\n\n def _join_non_unique(self, other, how=\"left\", return_indexers=False):\n from pandas.core.reshape.merge import _get_join_indexers\n\n # We only get here if dtypes match\n assert self.dtype == other.dtype\n\n if is_extension_array_dtype(self.dtype):\n lvalues = self._data._values_for_argsort()\n rvalues = other._data._values_for_argsort()\n else:\n lvalues = self._values\n rvalues = other._values\n\n left_idx, right_idx = _get_join_indexers(\n [lvalues], [rvalues], how=how, sort=True\n )\n\n left_idx = ensure_platform_int(left_idx)\n right_idx = ensure_platform_int(right_idx)\n\n join_index = np.asarray(lvalues.take(left_idx))\n mask = left_idx == -1\n np.putmask(join_index, mask, rvalues.take(right_idx))\n\n join_index = self._wrap_joined_index(join_index, other)\n\n if return_indexers:\n return join_index, left_idx, right_idx\n else:\n return join_index\n\n def _join_level(\n self, other, level, how=\"left\", return_indexers=False, keep_order=True\n ):\n \"\"\"\n The join method *only* affects the level of the resulting\n MultiIndex. Otherwise it just exactly aligns the Index data to the\n labels of the level in the MultiIndex.\n\n If ```keep_order == True```, the order of the data indexed by the\n MultiIndex will not be changed; otherwise, it will tie out\n with `other`.\n \"\"\"\n from pandas.core.indexes.multi import MultiIndex\n\n def _get_leaf_sorter(labels):\n \"\"\"\n Returns sorter for the inner most level while preserving the\n order of higher levels.\n \"\"\"\n if labels[0].size == 0:\n return np.empty(0, dtype=\"int64\")\n\n if len(labels) == 1:\n lab = ensure_int64(labels[0])\n sorter, _ = libalgos.groupsort_indexer(lab, 1 + lab.max())\n return sorter\n\n # find indexers of beginning of each set of\n # same-key labels w.r.t all but last level\n tic = labels[0][:-1] != labels[0][1:]\n for lab in labels[1:-1]:\n tic |= lab[:-1] != lab[1:]\n\n starts = np.hstack(([True], tic, [True])).nonzero()[0]\n lab = ensure_int64(labels[-1])\n return lib.get_level_sorter(lab, ensure_int64(starts))\n\n if isinstance(self, MultiIndex) and isinstance(other, MultiIndex):\n raise TypeError(\"Join on level between two MultiIndex objects is ambiguous\")\n\n left, right = self, other\n\n flip_order = not isinstance(self, MultiIndex)\n if flip_order:\n left, right = right, left\n how = {\"right\": \"left\", \"left\": \"right\"}.get(how, how)\n\n level = left._get_level_number(level)\n old_level = left.levels[level]\n\n if not right.is_unique:\n raise NotImplementedError(\n \"Index._join_level on non-unique index is not implemented\"\n )\n\n new_level, left_lev_indexer, right_lev_indexer = old_level.join(\n right, how=how, return_indexers=True\n )\n\n if left_lev_indexer is None:\n if keep_order or len(left) == 0:\n left_indexer = None\n join_index = left\n else: # sort the leaves\n left_indexer = _get_leaf_sorter(left.codes[: level + 1])\n join_index = left[left_indexer]\n\n else:\n left_lev_indexer = ensure_int64(left_lev_indexer)\n rev_indexer = lib.get_reverse_indexer(left_lev_indexer, len(old_level))\n\n new_lev_codes = algos.take_nd(\n rev_indexer, left.codes[level], allow_fill=False\n )\n\n new_codes = list(left.codes)\n new_codes[level] = new_lev_codes\n\n new_levels = list(left.levels)\n new_levels[level] = new_level\n\n if keep_order: # just drop missing values. o.w. keep order\n left_indexer = np.arange(len(left), dtype=np.intp)\n mask = new_lev_codes != -1\n if not mask.all():\n new_codes = [lab[mask] for lab in new_codes]\n left_indexer = left_indexer[mask]\n\n else: # tie out the order with other\n if level == 0: # outer most level, take the fast route\n ngroups = 1 + new_lev_codes.max()\n left_indexer, counts = libalgos.groupsort_indexer(\n new_lev_codes, ngroups\n )\n\n # missing values are placed first; drop them!\n left_indexer = left_indexer[counts[0] :]\n new_codes = [lab[left_indexer] for lab in new_codes]\n\n else: # sort the leaves\n mask = new_lev_codes != -1\n mask_all = mask.all()\n if not mask_all:\n new_codes = [lab[mask] for lab in new_codes]\n\n left_indexer = _get_leaf_sorter(new_codes[: level + 1])\n new_codes = [lab[left_indexer] for lab in new_codes]\n\n # left_indexers are w.r.t masked frame.\n # reverse to original frame!\n if not mask_all:\n left_indexer = mask.nonzero()[0][left_indexer]\n\n join_index = MultiIndex(\n levels=new_levels,\n codes=new_codes,\n names=left.names,\n verify_integrity=False,\n )\n\n if right_lev_indexer is not None:\n right_indexer = algos.take_nd(\n right_lev_indexer, join_index.codes[level], allow_fill=False\n )\n else:\n right_indexer = join_index.codes[level]\n\n if flip_order:\n left_indexer, right_indexer = right_indexer, left_indexer\n\n if return_indexers:\n left_indexer = (\n None if left_indexer is None else ensure_platform_int(left_indexer)\n )\n right_indexer = (\n None if right_indexer is None else ensure_platform_int(right_indexer)\n )\n return join_index, left_indexer, right_indexer\n else:\n return join_index\n\n def _join_monotonic(self, other, how=\"left\", return_indexers=False):\n # We only get here with matching dtypes\n assert other.dtype == self.dtype\n\n if self.equals(other):\n ret_index = other if how == \"right\" else self\n if return_indexers:\n return ret_index, None, None\n else:\n return ret_index\n\n if is_extension_array_dtype(self.dtype):\n sv = self._data._values_for_argsort()\n ov = other._data._values_for_argsort()\n else:\n sv = self._values\n ov = other._values\n\n if self.is_unique and other.is_unique:\n # We can perform much better than the general case\n if how == \"left\":\n join_index = self\n lidx = None\n ridx = self._left_indexer_unique(sv, ov)\n elif how == \"right\":\n join_index = other\n lidx = self._left_indexer_unique(ov, sv)\n ridx = None\n elif how == \"inner\":\n join_index, lidx, ridx = self._inner_indexer(sv, ov)\n join_index = self._wrap_joined_index(join_index, other)\n elif how == \"outer\":\n join_index, lidx, ridx = self._outer_indexer(sv, ov)\n join_index = self._wrap_joined_index(join_index, other)\n else:\n if how == \"left\":\n join_index, lidx, ridx = self._left_indexer(sv, ov)\n elif how == \"right\":\n join_index, ridx, lidx = self._left_indexer(ov, sv)\n elif how == \"inner\":\n join_index, lidx, ridx = self._inner_indexer(sv, ov)\n elif how == \"outer\":\n join_index, lidx, ridx = self._outer_indexer(sv, ov)\n join_index = self._wrap_joined_index(join_index, other)\n\n if return_indexers:\n lidx = None if lidx is None else ensure_platform_int(lidx)\n ridx = None if ridx is None else ensure_platform_int(ridx)\n return join_index, lidx, ridx\n else:\n return join_index\n\n def _wrap_joined_index(self, joined, other):\n name = get_op_result_name(self, other)\n return Index(joined, name=name)\n\n # --------------------------------------------------------------------\n # Uncategorized Methods\n\n @property\n def values(self) -> np.ndarray:\n \"\"\"\n Return an array representing the data in the Index.\n\n .. warning::\n\n We recommend using :attr:`Index.array` or\n :meth:`Index.to_numpy`, depending on whether you need\n a reference to the underlying data or a NumPy array.\n\n Returns\n -------\n array: numpy.ndarray or ExtensionArray\n\n See Also\n --------\n Index.array : Reference to the underlying data.\n Index.to_numpy : A NumPy array representing the underlying data.\n \"\"\"\n return self._data.view(np.ndarray)\n\n @cache_readonly\n @doc(IndexOpsMixin.array)\n def array(self) -> ExtensionArray:\n array = self._data\n if isinstance(array, np.ndarray):\n from pandas.core.arrays.numpy_ import PandasArray\n\n array = PandasArray(array)\n return array\n\n @property\n def _values(self) -> Union[ExtensionArray, np.ndarray]:\n \"\"\"\n The best array representation.\n\n This is an ndarray or ExtensionArray.\n\n ``_values`` are consistent between``Series`` and ``Index``.\n\n It may differ from the public '.values' method.\n\n index | values | _values |\n ----------------- | --------------- | ------------- |\n Index | ndarray | ndarray |\n CategoricalIndex | Categorical | Categorical |\n DatetimeIndex | ndarray[M8ns] | DatetimeArray |\n DatetimeIndex[tz] | ndarray[M8ns] | DatetimeArray |\n PeriodIndex | ndarray[object] | PeriodArray |\n IntervalIndex | IntervalArray | IntervalArray |\n\n See Also\n --------\n values\n \"\"\"\n return self._data\n\n def _get_engine_target(self) -> np.ndarray:\n \"\"\"\n Get the ndarray that we can pass to the IndexEngine constructor.\n \"\"\"\n return self._values\n\n @doc(IndexOpsMixin.memory_usage)\n def memory_usage(self, deep: bool = False) -> int:\n result = super().memory_usage(deep=deep)\n\n # include our engine hashtable\n result += self._engine.sizeof(deep=deep)\n return result\n\n def where(self, cond, other=None):\n \"\"\"\n Replace values where the condition is False.\n\n The replacement is taken from other.\n\n Parameters\n ----------\n cond : bool array-like with the same length as self\n Condition to select the values on.\n other : scalar, or array-like, default None\n Replacement if the condition is False.\n\n Returns\n -------\n pandas.Index\n A copy of self with values replaced from other\n where the condition is False.\n\n See Also\n --------\n Series.where : Same method for Series.\n DataFrame.where : Same method for DataFrame.\n\n Examples\n --------\n >>> idx = pd.Index(['car', 'bike', 'train', 'tractor'])\n >>> idx\n Index(['car', 'bike', 'train', 'tractor'], dtype='object')\n >>> idx.where(idx.isin(['car', 'train']), 'other')\n Index(['car', 'other', 'train', 'other'], dtype='object')\n \"\"\"\n if other is None:\n other = self._na_value\n\n dtype = self.dtype\n values = self.values\n\n if is_bool(other) or is_bool_dtype(other):\n\n # bools force casting\n values = values.astype(object)\n dtype = None\n\n values = np.where(cond, values, other)\n\n if self._is_numeric_dtype and np.any(isna(values)):\n # We can't coerce to the numeric dtype of \"self\" (unless\n # it's float) if there are NaN values in our output.\n dtype = None\n\n return self._shallow_copy_with_infer(values, dtype=dtype)\n\n # construction helpers\n @classmethod\n def _scalar_data_error(cls, data):\n # We return the TypeError so that we can raise it from the constructor\n # in order to keep mypy happy\n return TypeError(\n f\"{cls.__name__}(...) must be called with a collection of some \"\n f\"kind, {repr(data)} was passed\"\n )\n\n @classmethod\n def _string_data_error(cls, data):\n raise TypeError(\n \"String dtype not supported, you may need \"\n \"to explicitly cast to a numeric type\"\n )\n\n def _coerce_scalar_to_index(self, item):\n \"\"\"\n We need to coerce a scalar to a compat for our index type.\n\n Parameters\n ----------\n item : scalar item to coerce\n \"\"\"\n dtype = self.dtype\n\n if self._is_numeric_dtype and isna(item):\n # We can't coerce to the numeric dtype of \"self\" (unless\n # it's float) if there are NaN values in our output.\n dtype = None\n\n return Index([item], dtype=dtype, **self._get_attributes_dict())\n\n def _to_safe_for_reshape(self):\n \"\"\"\n Convert to object if we are a categorical.\n \"\"\"\n return self\n\n def _convert_for_op(self, value):\n \"\"\"\n Convert value to be insertable to ndarray.\n \"\"\"\n return value\n\n def _assert_can_do_op(self, value):\n \"\"\"\n Check value is valid for scalar op.\n \"\"\"\n if not is_scalar(value):\n raise TypeError(f\"'value' must be a scalar, passed: {type(value).__name__}\")\n\n @property\n def _has_complex_internals(self) -> bool:\n \"\"\"\n Indicates if an index is not directly backed by a numpy array\n \"\"\"\n # used to avoid libreduction code paths, which raise or require conversion\n return False\n\n def _is_memory_usage_qualified(self) -> bool:\n \"\"\"\n Return a boolean if we need a qualified .info display.\n \"\"\"\n return self.is_object()\n\n def is_type_compatible(self, kind) -> bool:\n \"\"\"\n Whether the index type is compatible with the provided type.\n \"\"\"\n return kind == self.inferred_type\n\n def __contains__(self, key: Any) -> bool:\n \"\"\"\n Return a boolean indicating whether the provided key is in the index.\n\n Parameters\n ----------\n key : label\n The key to check if it is present in the index.\n\n Returns\n -------\n bool\n Whether the key search is in the index.\n\n Raises\n ------\n TypeError\n If the key is not hashable.\n\n See Also\n --------\n Index.isin : Returns an ndarray of boolean dtype indicating whether the\n list-like key is in the index.\n\n Examples\n --------\n >>> idx = pd.Index([1, 2, 3, 4])\n >>> idx\n Int64Index([1, 2, 3, 4], dtype='int64')\n\n >>> 2 in idx\n True\n >>> 6 in idx\n False\n \"\"\"\n hash(key)\n try:\n return key in self._engine\n except (OverflowError, TypeError, ValueError):\n return False\n\n def __hash__(self):\n raise TypeError(f\"unhashable type: {repr(type(self).__name__)}\")\n\n def __setitem__(self, key, value):\n raise TypeError(\"Index does not support mutable operations\")\n\n def __getitem__(self, key):\n \"\"\"\n Override numpy.ndarray's __getitem__ method to work as desired.\n\n This function adds lists and Series as valid boolean indexers\n (ndarrays only supports ndarray with dtype=bool).\n\n If resulting ndim != 1, plain ndarray is returned instead of\n corresponding `Index` subclass.\n\n \"\"\"\n # There's no custom logic to be implemented in __getslice__, so it's\n # not overloaded intentionally.\n getitem = self._data.__getitem__\n promote = self._shallow_copy\n\n if is_scalar(key):\n key = com.cast_scalar_indexer(key)\n return getitem(key)\n\n if isinstance(key, slice):\n # This case is separated from the conditional above to avoid\n # pessimization of basic indexing.\n return promote(getitem(key))\n\n if com.is_bool_indexer(key):\n key = np.asarray(key, dtype=bool)\n\n result = getitem(key)\n if not is_scalar(result):\n if np.ndim(result) > 1:\n deprecate_ndim_indexing(result)\n return result\n return promote(result)\n else:\n return result\n\n def _can_hold_identifiers_and_holds_name(self, name) -> bool:\n \"\"\"\n Faster check for ``name in self`` when we know `name` is a Python\n identifier (e.g. in NDFrame.__getattr__, which hits this to support\n . key lookup). For indexes that can't hold identifiers (everything\n but object & categorical) we just return False.\n\n https://github.com/pandas-dev/pandas/issues/19764\n \"\"\"\n if self.is_object() or self.is_categorical():\n return name in self\n return False\n\n def append(self, other):\n \"\"\"\n Append a collection of Index options together.\n\n Parameters\n ----------\n other : Index or list/tuple of indices\n\n Returns\n -------\n appended : Index\n \"\"\"\n to_concat = [self]\n\n if isinstance(other, (list, tuple)):\n to_concat = to_concat + list(other)\n else:\n to_concat.append(other)\n\n for obj in to_concat:\n if not isinstance(obj, Index):\n raise TypeError(\"all inputs must be Index\")\n\n names = {obj.name for obj in to_concat}\n name = None if len(names) > 1 else self.name\n\n return self._concat(to_concat, name)\n\n def _concat(self, to_concat, name):\n\n typs = _concat.get_dtype_kinds(to_concat)\n\n if len(typs) == 1:\n return self._concat_same_dtype(to_concat, name=name)\n return Index._concat_same_dtype(self, to_concat, name=name)\n\n def _concat_same_dtype(self, to_concat, name):\n \"\"\"\n Concatenate to_concat which has the same class.\n \"\"\"\n # must be overridden in specific classes\n klasses = (\n ABCDatetimeIndex,\n ABCTimedeltaIndex,\n ABCPeriodIndex,\n ExtensionArray,\n ABCIntervalIndex,\n )\n to_concat = [\n x.astype(object) if isinstance(x, klasses) else x for x in to_concat\n ]\n\n self = to_concat[0]\n attribs = self._get_attributes_dict()\n attribs[\"name\"] = name\n\n to_concat = [x._values if isinstance(x, Index) else x for x in to_concat]\n\n return self._shallow_copy_with_infer(np.concatenate(to_concat), **attribs)\n\n def putmask(self, mask, value):\n \"\"\"\n Return a new Index of the values set with the mask.\n\n Returns\n -------\n Index\n\n See Also\n --------\n numpy.ndarray.putmask\n \"\"\"\n values = self.values.copy()\n try:\n np.putmask(values, mask, self._convert_for_op(value))\n if is_period_dtype(self.dtype):\n # .values cast to object, so we need to cast back\n values = type(self)(values)._data\n return self._shallow_copy(values)\n except (ValueError, TypeError) as err:\n if is_object_dtype(self):\n raise err\n\n # coerces to object\n return self.astype(object).putmask(mask, value)\n\n def equals(self, other: Any) -> bool:\n \"\"\"\n Determine if two Index object are equal.\n\n The things that are being compared are:\n\n * The elements inside the Index object.\n * The order of the elements inside the Index object.\n\n Parameters\n ----------\n other : Any\n The other object to compare against.\n\n Returns\n -------\n bool\n True if \"other\" is an Index and it has the same elements and order\n as the calling index; False otherwise.\n\n Examples\n --------\n >>> idx1 = pd.Index([1, 2, 3])\n >>> idx1\n Int64Index([1, 2, 3], dtype='int64')\n >>> idx1.equals(pd.Index([1, 2, 3]))\n True\n\n The elements inside are compared\n\n >>> idx2 = pd.Index([\"1\", \"2\", \"3\"])\n >>> idx2\n Index(['1', '2', '3'], dtype='object')\n\n >>> idx1.equals(idx2)\n False\n\n The order is compared\n\n >>> ascending_idx = pd.Index([1, 2, 3])\n >>> ascending_idx\n Int64Index([1, 2, 3], dtype='int64')\n >>> descending_idx = pd.Index([3, 2, 1])\n >>> descending_idx\n Int64Index([3, 2, 1], dtype='int64')\n >>> ascending_idx.equals(descending_idx)\n False\n\n The dtype is *not* compared\n\n >>> int64_idx = pd.Int64Index([1, 2, 3])\n >>> int64_idx\n Int64Index([1, 2, 3], dtype='int64')\n >>> uint64_idx = pd.UInt64Index([1, 2, 3])\n >>> uint64_idx\n UInt64Index([1, 2, 3], dtype='uint64')\n >>> int64_idx.equals(uint64_idx)\n True\n \"\"\"\n if self.is_(other):\n return True\n\n if not isinstance(other, Index):\n return False\n\n if is_object_dtype(self.dtype) and not is_object_dtype(other.dtype):\n # if other is not object, use other's logic for coercion\n return other.equals(self)\n\n if isinstance(other, ABCMultiIndex):\n # d-level MultiIndex can equal d-tuple Index\n return other.equals(self)\n\n if is_extension_array_dtype(other.dtype):\n # All EA-backed Index subclasses override equals\n return other.equals(self)\n\n return array_equivalent(self._values, other._values)\n\n def identical(self, other) -> bool:\n \"\"\"\n Similar to equals, but check that other comparable attributes are\n also equal.\n\n Returns\n -------\n bool\n If two Index objects have equal elements and same type True,\n otherwise False.\n \"\"\"\n return (\n self.equals(other)\n and all(\n (\n getattr(self, c, None) == getattr(other, c, None)\n for c in self._comparables\n )\n )\n and type(self) == type(other)\n )\n\n def asof(self, label):\n \"\"\"\n Return the label from the index, or, if not present, the previous one.\n\n Assuming that the index is sorted, return the passed index label if it\n is in the index, or return the previous index label if the passed one\n is not in the index.\n\n Parameters\n ----------\n label : object\n The label up to which the method returns the latest index label.\n\n Returns\n -------\n object\n The passed label if it is in the index. The previous label if the\n passed label is not in the sorted index or `NaN` if there is no\n such label.\n\n See Also\n --------\n Series.asof : Return the latest value in a Series up to the\n passed index.\n merge_asof : Perform an asof merge (similar to left join but it\n matches on nearest key rather than equal key).\n Index.get_loc : An `asof` is a thin wrapper around `get_loc`\n with method='pad'.\n\n Examples\n --------\n `Index.asof` returns the latest index label up to the passed label.\n\n >>> idx = pd.Index(['2013-12-31', '2014-01-02', '2014-01-03'])\n >>> idx.asof('2014-01-01')\n '2013-12-31'\n\n If the label is in the index, the method returns the passed label.\n\n >>> idx.asof('2014-01-02')\n '2014-01-02'\n\n If all of the labels in the index are later than the passed label,\n NaN is returned.\n\n >>> idx.asof('1999-01-02')\n nan\n\n If the index is not sorted, an error is raised.\n\n >>> idx_not_sorted = pd.Index(['2013-12-31', '2015-01-02',\n ... '2014-01-03'])\n >>> idx_not_sorted.asof('2013-12-31')\n Traceback (most recent call last):\n ValueError: index must be monotonic increasing or decreasing\n \"\"\"\n try:\n loc = self.get_loc(label, method=\"pad\")\n except KeyError:\n return self._na_value\n else:\n if isinstance(loc, slice):\n loc = loc.indices(len(self))[-1]\n return self[loc]\n\n def asof_locs(self, where, mask):\n \"\"\"\n Find the locations (indices) of the labels from the index for\n every entry in the `where` argument.\n\n As in the `asof` function, if the label (a particular entry in\n `where`) is not in the index, the latest index label up to the\n passed label is chosen and its index returned.\n\n If all of the labels in the index are later than a label in `where`,\n -1 is returned.\n\n `mask` is used to ignore NA values in the index during calculation.\n\n Parameters\n ----------\n where : Index\n An Index consisting of an array of timestamps.\n mask : array-like\n Array of booleans denoting where values in the original\n data are not NA.\n\n Returns\n -------\n numpy.ndarray\n An array of locations (indices) of the labels from the Index\n which correspond to the return values of the `asof` function\n for every element in `where`.\n \"\"\"\n locs = self.values[mask].searchsorted(where.values, side=\"right\")\n locs = np.where(locs > 0, locs - 1, 0)\n\n result = np.arange(len(self))[mask].take(locs)\n\n first = mask.argmax()\n result[(locs == 0) & (where.values < self.values[first])] = -1\n\n return result\n\n def sort_values(self, return_indexer: bool = False, ascending: bool = True):\n \"\"\"\n Return a sorted copy of the index.\n\n Return a sorted copy of the index, and optionally return the indices\n that sorted the index itself.\n\n Parameters\n ----------\n return_indexer : bool, default False\n Should the indices that would sort the index be returned.\n ascending : bool, default True\n Should the index values be sorted in an ascending order.\n\n Returns\n -------\n sorted_index : pandas.Index\n Sorted copy of the index.\n indexer : numpy.ndarray, optional\n The indices that the index itself was sorted by.\n\n See Also\n --------\n Series.sort_values : Sort values of a Series.\n DataFrame.sort_values : Sort values in a DataFrame.\n\n Examples\n --------\n >>> idx = pd.Index([10, 100, 1, 1000])\n >>> idx\n Int64Index([10, 100, 1, 1000], dtype='int64')\n\n Sort values in ascending order (default behavior).\n\n >>> idx.sort_values()\n Int64Index([1, 10, 100, 1000], dtype='int64')\n\n Sort values in descending order, and also get the indices `idx` was\n sorted by.\n\n >>> idx.sort_values(ascending=False, return_indexer=True)\n (Int64Index([1000, 100, 10, 1], dtype='int64'), array([3, 1, 0, 2]))\n \"\"\"\n _as = self.argsort()\n if not ascending:\n _as = _as[::-1]\n\n sorted_index = self.take(_as)\n\n if return_indexer:\n return sorted_index, _as\n else:\n return sorted_index\n\n def sort(self, *args, **kwargs):\n \"\"\"\n Use sort_values instead.\n \"\"\"\n raise TypeError(\"cannot sort an Index object in-place, use sort_values instead\")\n\n def shift(self, periods=1, freq=None):\n \"\"\"\n Shift index by desired number of time frequency increments.\n\n This method is for shifting the values of datetime-like indexes\n by a specified time increment a given number of times.\n\n Parameters\n ----------\n periods : int, default 1\n Number of periods (or increments) to shift by,\n can be positive or negative.\n freq : pandas.DateOffset, pandas.Timedelta or str, optional\n Frequency increment to shift by.\n If None, the index is shifted by its own `freq` attribute.\n Offset aliases are valid strings, e.g., 'D', 'W', 'M' etc.\n\n Returns\n -------\n pandas.Index\n Shifted index.\n\n See Also\n --------\n Series.shift : Shift values of Series.\n\n Notes\n -----\n This method is only implemented for datetime-like index classes,\n i.e., DatetimeIndex, PeriodIndex and TimedeltaIndex.\n\n Examples\n --------\n Put the first 5 month starts of 2011 into an index.\n\n >>> month_starts = pd.date_range('1/1/2011', periods=5, freq='MS')\n >>> month_starts\n DatetimeIndex(['2011-01-01', '2011-02-01', '2011-03-01', '2011-04-01',\n '2011-05-01'],\n dtype='datetime64[ns]', freq='MS')\n\n Shift the index by 10 days.\n\n >>> month_starts.shift(10, freq='D')\n DatetimeIndex(['2011-01-11', '2011-02-11', '2011-03-11', '2011-04-11',\n '2011-05-11'],\n dtype='datetime64[ns]', freq=None)\n\n The default value of `freq` is the `freq` attribute of the index,\n which is 'MS' (month start) in this example.\n\n >>> month_starts.shift(10)\n DatetimeIndex(['2011-11-01', '2011-12-01', '2012-01-01', '2012-02-01',\n '2012-03-01'],\n dtype='datetime64[ns]', freq='MS')\n \"\"\"\n raise NotImplementedError(f\"Not supported for type {type(self).__name__}\")\n\n def argsort(self, *args, **kwargs) -> np.ndarray:\n \"\"\"\n Return the integer indices that would sort the index.\n\n Parameters\n ----------\n *args\n Passed to `numpy.ndarray.argsort`.\n **kwargs\n Passed to `numpy.ndarray.argsort`.\n\n Returns\n -------\n numpy.ndarray\n Integer indices that would sort the index if used as\n an indexer.\n\n See Also\n --------\n numpy.argsort : Similar method for NumPy arrays.\n Index.sort_values : Return sorted copy of Index.\n\n Examples\n --------\n >>> idx = pd.Index(['b', 'a', 'd', 'c'])\n >>> idx\n Index(['b', 'a', 'd', 'c'], dtype='object')\n\n >>> order = idx.argsort()\n >>> order\n array([1, 0, 3, 2])\n\n >>> idx[order]\n Index(['a', 'b', 'c', 'd'], dtype='object')\n \"\"\"\n result = self.asi8\n if result is None:\n result = np.array(self)\n return result.argsort(*args, **kwargs)\n\n def get_value(self, series: \"Series\", key):\n \"\"\"\n Fast lookup of value from 1-dimensional ndarray. Only use this if you\n know what you're doing.\n\n Returns\n -------\n scalar or Series\n \"\"\"\n self._check_indexing_error(key)\n\n try:\n # GH 20882, 21257\n # First try to convert the key to a location\n # If that fails, raise a KeyError if an integer\n # index, otherwise, see if key is an integer, and\n # try that\n loc = self.get_loc(key)\n except KeyError:\n if not self._should_fallback_to_positional():\n raise\n elif is_integer(key):\n # If the Index cannot hold integer, then this is unambiguously\n # a locational lookup.\n loc = key\n else:\n raise\n\n return self._get_values_for_loc(series, loc, key)\n\n def _check_indexing_error(self, key):\n if not is_scalar(key):\n # if key is not a scalar, directly raise an error (the code below\n # would convert to numpy arrays and raise later any way) - GH29926\n raise InvalidIndexError(key)\n\n def _should_fallback_to_positional(self) -> bool:\n \"\"\"\n If an integer key is not found, should we fall back to positional indexing?\n \"\"\"\n if len(self) > 0 and (self.holds_integer() or self.is_boolean()):\n return False\n return True\n\n def _get_values_for_loc(self, series: \"Series\", loc, key):\n \"\"\"\n Do a positional lookup on the given Series, returning either a scalar\n or a Series.\n\n Assumes that `series.index is self`\n\n key is included for MultiIndex compat.\n \"\"\"\n if is_integer(loc):\n return series._values[loc]\n\n return series.iloc[loc]\n\n def set_value(self, arr, key, value):\n \"\"\"\n Fast lookup of value from 1-dimensional ndarray.\n\n .. deprecated:: 1.0\n\n Notes\n -----\n Only use this if you know what you're doing.\n \"\"\"\n warnings.warn(\n (\n \"The 'set_value' method is deprecated, and \"\n \"will be removed in a future version.\"\n ),\n FutureWarning,\n stacklevel=2,\n )\n loc = self._engine.get_loc(key)\n validate_numeric_casting(arr.dtype, value)\n arr[loc] = value\n\n _index_shared_docs[\n \"get_indexer_non_unique\"\n ] = \"\"\"\n Compute indexer and mask for new index given the current index. The\n indexer should be then used as an input to ndarray.take to align the\n current data to the new index.\n\n Parameters\n ----------\n target : %(target_klass)s\n\n Returns\n -------\n indexer : ndarray of int\n Integers from 0 to n - 1 indicating that the index at these\n positions matches the corresponding target values. Missing values\n in the target are marked by -1.\n missing : ndarray of int\n An indexer into the target of the values not found.\n These correspond to the -1 in the indexer array.\n \"\"\"\n\n @Appender(_index_shared_docs[\"get_indexer_non_unique\"] % _index_doc_kwargs)\n def get_indexer_non_unique(self, target):\n target = ensure_index(target)\n pself, ptarget = self._maybe_promote(target)\n if pself is not self or ptarget is not target:\n return pself.get_indexer_non_unique(ptarget)\n\n if is_categorical_dtype(target.dtype):\n tgt_values = np.asarray(target)\n else:\n tgt_values = target._get_engine_target()\n\n indexer, missing = self._engine.get_indexer_non_unique(tgt_values)\n return ensure_platform_int(indexer), missing\n\n def get_indexer_for(self, target, **kwargs):\n \"\"\"\n Guaranteed return of an indexer even when non-unique.\n\n This dispatches to get_indexer or get_indexer_non_unique\n as appropriate.\n\n Returns\n -------\n numpy.ndarray\n List of indices.\n \"\"\"\n if self.is_unique:\n return self.get_indexer(target, **kwargs)\n indexer, _ = self.get_indexer_non_unique(target, **kwargs)\n return indexer\n\n def _maybe_promote(self, other):\n # A hack, but it works\n\n if self.inferred_type == \"date\" and isinstance(other, ABCDatetimeIndex):\n return type(other)(self), other\n elif self.inferred_type == \"boolean\":\n if not is_object_dtype(self.dtype):\n return self.astype(\"object\"), other.astype(\"object\")\n return self, other\n\n def groupby(self, values) -> PrettyDict[Hashable, np.ndarray]:\n \"\"\"\n Group the index labels by a given array of values.\n\n Parameters\n ----------\n values : array\n Values used to determine the groups.\n\n Returns\n -------\n dict\n {group name -> group labels}\n \"\"\"\n # TODO: if we are a MultiIndex, we can do better\n # that converting to tuples\n if isinstance(values, ABCMultiIndex):\n values = values.values\n values = ensure_categorical(values)\n result = values._reverse_indexer()\n\n # map to the label\n result = {k: self.take(v) for k, v in result.items()}\n\n return PrettyDict(result)\n\n def map(self, mapper, na_action=None):\n \"\"\"\n Map values using input correspondence (a dict, Series, or function).\n\n Parameters\n ----------\n mapper : function, dict, or Series\n Mapping correspondence.\n na_action : {None, 'ignore'}\n If 'ignore', propagate NA values, without passing them to the\n mapping correspondence.\n\n Returns\n -------\n applied : Union[Index, MultiIndex], inferred\n The output of the mapping function applied to the index.\n If the function returns a tuple with more than one element\n a MultiIndex will be returned.\n \"\"\"\n from pandas.core.indexes.multi import MultiIndex\n\n new_values = super()._map_values(mapper, na_action=na_action)\n\n attributes = self._get_attributes_dict()\n\n # we can return a MultiIndex\n if new_values.size and isinstance(new_values[0], tuple):\n if isinstance(self, MultiIndex):\n names = self.names\n elif attributes.get(\"name\"):\n names = [attributes.get(\"name\")] * len(new_values[0])\n else:\n names = None\n return MultiIndex.from_tuples(new_values, names=names)\n\n attributes[\"copy\"] = False\n if not new_values.size:\n # empty\n attributes[\"dtype\"] = self.dtype\n\n return Index(new_values, **attributes)\n\n # TODO: De-duplicate with map, xref GH#32349\n def _transform_index(self, func, level=None) -> \"Index\":\n \"\"\"\n Apply function to all values found in index.\n\n This includes transforming multiindex entries separately.\n Only apply function to one level of the MultiIndex if level is specified.\n \"\"\"\n if isinstance(self, ABCMultiIndex):\n if level is not None:\n items = [\n tuple(func(y) if i == level else y for i, y in enumerate(x))\n for x in self\n ]\n else:\n items = [tuple(func(y) for y in x) for x in self]\n return type(self).from_tuples(items, names=self.names)\n else:\n items = [func(x) for x in self]\n return Index(items, name=self.name, tupleize_cols=False)\n\n def isin(self, values, level=None):\n \"\"\"\n Return a boolean array where the index values are in `values`.\n\n Compute boolean array of whether each index value is found in the\n passed set of values. The length of the returned boolean array matches\n the length of the index.\n\n Parameters\n ----------\n values : set or list-like\n Sought values.\n level : str or int, optional\n Name or position of the index level to use (if the index is a\n `MultiIndex`).\n\n Returns\n -------\n is_contained : ndarray\n NumPy array of boolean values.\n\n See Also\n --------\n Series.isin : Same for Series.\n DataFrame.isin : Same method for DataFrames.\n\n Notes\n -----\n In the case of `MultiIndex` you must either specify `values` as a\n list-like object containing tuples that are the same length as the\n number of levels, or specify `level`. Otherwise it will raise a\n ``ValueError``.\n\n If `level` is specified:\n\n - if it is the name of one *and only one* index level, use that level;\n - otherwise it should be a number indicating level position.\n\n Examples\n --------\n >>> idx = pd.Index([1,2,3])\n >>> idx\n Int64Index([1, 2, 3], dtype='int64')\n\n Check whether each index value in a list of values.\n\n >>> idx.isin([1, 4])\n array([ True, False, False])\n\n >>> midx = pd.MultiIndex.from_arrays([[1,2,3],\n ... ['red', 'blue', 'green']],\n ... names=('number', 'color'))\n >>> midx\n MultiIndex([(1, 'red'),\n (2, 'blue'),\n (3, 'green')],\n names=['number', 'color'])\n\n Check whether the strings in the 'color' level of the MultiIndex\n are in a list of colors.\n\n >>> midx.isin(['red', 'orange', 'yellow'], level='color')\n array([ True, False, False])\n\n To check across the levels of a MultiIndex, pass a list of tuples:\n\n >>> midx.isin([(1, 'red'), (3, 'red')])\n array([ True, False, False])\n\n For a DatetimeIndex, string values in `values` are converted to\n Timestamps.\n\n >>> dates = ['2000-03-11', '2000-03-12', '2000-03-13']\n >>> dti = pd.to_datetime(dates)\n >>> dti\n DatetimeIndex(['2000-03-11', '2000-03-12', '2000-03-13'],\n dtype='datetime64[ns]', freq=None)\n\n >>> dti.isin(['2000-03-11'])\n array([ True, False, False])\n \"\"\"\n if level is not None:\n self._validate_index_level(level)\n return algos.isin(self, values)\n\n def _get_string_slice(self, key: str_t, use_lhs: bool = True, use_rhs: bool = True):\n # this is for partial string indexing,\n # overridden in DatetimeIndex, TimedeltaIndex and PeriodIndex\n raise NotImplementedError\n\n def slice_indexer(self, start=None, end=None, step=None, kind=None):\n \"\"\"\n For an ordered or unique index, compute the slice indexer for input\n labels and step.\n\n Parameters\n ----------\n start : label, default None\n If None, defaults to the beginning.\n end : label, default None\n If None, defaults to the end.\n step : int, default None\n kind : str, default None\n\n Returns\n -------\n indexer : slice\n\n Raises\n ------\n KeyError : If key does not exist, or key is not unique and index is\n not ordered.\n\n Notes\n -----\n This function assumes that the data is sorted, so use at your own peril\n\n Examples\n --------\n This is a method on all index types. For example you can do:\n\n >>> idx = pd.Index(list('abcd'))\n >>> idx.slice_indexer(start='b', end='c')\n slice(1, 3, None)\n\n >>> idx = pd.MultiIndex.from_arrays([list('abcd'), list('efgh')])\n >>> idx.slice_indexer(start='b', end=('c', 'g'))\n slice(1, 3, None)\n \"\"\"\n start_slice, end_slice = self.slice_locs(start, end, step=step, kind=kind)\n\n # return a slice\n if not is_scalar(start_slice):\n raise AssertionError(\"Start slice bound is non-scalar\")\n if not is_scalar(end_slice):\n raise AssertionError(\"End slice bound is non-scalar\")\n\n return slice(start_slice, end_slice, step)\n\n def _maybe_cast_indexer(self, key):\n \"\"\"\n If we have a float key and are not a floating index, then try to cast\n to an int if equivalent.\n \"\"\"\n if not self.is_floating():\n return com.cast_scalar_indexer(key)\n return key\n\n def _validate_indexer(self, form: str_t, key, kind: str_t):\n \"\"\"\n If we are positional indexer, validate that we have appropriate\n typed bounds must be an integer.\n \"\"\"\n assert kind in [\"getitem\", \"iloc\"]\n\n if key is None:\n pass\n elif is_integer(key):\n pass\n else:\n self._invalid_indexer(form, key)\n\n def _maybe_cast_slice_bound(self, label, side: str_t, kind):\n \"\"\"\n This function should be overloaded in subclasses that allow non-trivial\n casting on label-slice bounds, e.g. datetime-like indices allowing\n strings containing formatted datetimes.\n\n Parameters\n ----------\n label : object\n side : {'left', 'right'}\n kind : {'loc', 'getitem'} or None\n\n Returns\n -------\n label : object\n\n Notes\n -----\n Value of `side` parameter should be validated in caller.\n \"\"\"\n assert kind in [\"loc\", \"getitem\", None]\n\n # We are a plain index here (sub-class override this method if they\n # wish to have special treatment for floats/ints, e.g. Float64Index and\n # datetimelike Indexes\n # reject them\n if is_float(label):\n self._invalid_indexer(\"slice\", label)\n\n # we are trying to find integer bounds on a non-integer based index\n # this is rejected (generally .loc gets you here)\n elif is_integer(label):\n self._invalid_indexer(\"slice\", label)\n\n return label\n\n def _searchsorted_monotonic(self, label, side=\"left\"):\n if self.is_monotonic_increasing:\n return self.searchsorted(label, side=side)\n elif self.is_monotonic_decreasing:\n # np.searchsorted expects ascending sort order, have to reverse\n # everything for it to work (element ordering, search side and\n # resulting value).\n pos = self[::-1].searchsorted(\n label, side=\"right\" if side == \"left\" else \"left\"\n )\n return len(self) - pos\n\n raise ValueError(\"index must be monotonic increasing or decreasing\")\n\n def get_slice_bound(self, label, side: str_t, kind) -> int:\n \"\"\"\n Calculate slice bound that corresponds to given label.\n\n Returns leftmost (one-past-the-rightmost if ``side=='right'``) position\n of given label.\n\n Parameters\n ----------\n label : object\n side : {'left', 'right'}\n kind : {'loc', 'getitem'} or None\n\n Returns\n -------\n int\n Index of label.\n \"\"\"\n assert kind in [\"loc\", \"getitem\", None]\n\n if side not in (\"left\", \"right\"):\n raise ValueError(\n \"Invalid value for side kwarg, must be either \"\n f\"'left' or 'right': {side}\"\n )\n\n original_label = label\n\n # For datetime indices label may be a string that has to be converted\n # to datetime boundary according to its resolution.\n label = self._maybe_cast_slice_bound(label, side, kind)\n\n # we need to look up the label\n try:\n slc = self.get_loc(label)\n except KeyError as err:\n try:\n return self._searchsorted_monotonic(label, side)\n except ValueError:\n # raise the original KeyError\n raise err\n\n if isinstance(slc, np.ndarray):\n # get_loc may return a boolean array or an array of indices, which\n # is OK as long as they are representable by a slice.\n if is_bool_dtype(slc):\n slc = lib.maybe_booleans_to_slice(slc.view(\"u1\"))\n else:\n slc = lib.maybe_indices_to_slice(slc.astype(\"i8\"), len(self))\n if isinstance(slc, np.ndarray):\n raise KeyError(\n f\"Cannot get {side} slice bound for non-unique \"\n f\"label: {repr(original_label)}\"\n )\n\n if isinstance(slc, slice):\n if side == \"left\":\n return slc.start\n else:\n return slc.stop\n else:\n if side == \"right\":\n return slc + 1\n else:\n return slc\n\n def slice_locs(self, start=None, end=None, step=None, kind=None):\n \"\"\"\n Compute slice locations for input labels.\n\n Parameters\n ----------\n start : label, default None\n If None, defaults to the beginning.\n end : label, default None\n If None, defaults to the end.\n step : int, defaults None\n If None, defaults to 1.\n kind : {'loc', 'getitem'} or None\n\n Returns\n -------\n start, end : int\n\n See Also\n --------\n Index.get_loc : Get location for a single label.\n\n Notes\n -----\n This method only works if the index is monotonic or unique.\n\n Examples\n --------\n >>> idx = pd.Index(list('abcd'))\n >>> idx.slice_locs(start='b', end='c')\n (1, 3)\n \"\"\"\n inc = step is None or step >= 0\n\n if not inc:\n # If it's a reverse slice, temporarily swap bounds.\n start, end = end, start\n\n # GH 16785: If start and end happen to be date strings with UTC offsets\n # attempt to parse and check that the offsets are the same\n if isinstance(start, (str, datetime)) and isinstance(end, (str, datetime)):\n try:\n ts_start = Timestamp(start)\n ts_end = Timestamp(end)\n except (ValueError, TypeError):\n pass\n else:\n if not tz_compare(ts_start.tzinfo, ts_end.tzinfo):\n raise ValueError(\"Both dates must have the same UTC offset\")\n\n start_slice = None\n if start is not None:\n start_slice = self.get_slice_bound(start, \"left\", kind)\n if start_slice is None:\n start_slice = 0\n\n end_slice = None\n if end is not None:\n end_slice = self.get_slice_bound(end, \"right\", kind)\n if end_slice is None:\n end_slice = len(self)\n\n if not inc:\n # Bounds at this moment are swapped, swap them back and shift by 1.\n #\n # slice_locs('B', 'A', step=-1): s='B', e='A'\n #\n # s='A' e='B'\n # AFTER SWAP: | |\n # v ------------------> V\n # -----------------------------------\n # | | |A|A|A|A| | | | | |B|B| | | | |\n # -----------------------------------\n # ^ <------------------ ^\n # SHOULD BE: | |\n # end=s-1 start=e-1\n #\n end_slice, start_slice = start_slice - 1, end_slice - 1\n\n # i == -1 triggers ``len(self) + i`` selection that points to the\n # last element, not before-the-first one, subtracting len(self)\n # compensates that.\n if end_slice == -1:\n end_slice -= len(self)\n if start_slice == -1:\n start_slice -= len(self)\n\n return start_slice, end_slice\n\n def delete(self, loc):\n \"\"\"\n Make new Index with passed location(-s) deleted.\n\n Parameters\n ----------\n loc : int or list of int\n Location of item(-s) which will be deleted.\n Use a list of locations to delete more than one value at the same time.\n\n Returns\n -------\n Index\n New Index with passed location(-s) deleted.\n\n See Also\n --------\n numpy.delete : Delete any rows and column from NumPy array (ndarray).\n\n Examples\n --------\n >>> idx = pd.Index(['a', 'b', 'c'])\n >>> idx.delete(1)\n Index(['a', 'c'], dtype='object')\n\n >>> idx = pd.Index(['a', 'b', 'c'])\n >>> idx.delete([0, 2])\n Index(['b'], dtype='object')\n \"\"\"\n return self._shallow_copy(np.delete(self._data, loc))\n\n def insert(self, loc: int, item):\n \"\"\"\n Make new Index inserting new item at location.\n\n Follows Python list.append semantics for negative values.\n\n Parameters\n ----------\n loc : int\n item : object\n\n Returns\n -------\n new_index : Index\n \"\"\"\n # Note: this method is overridden by all ExtensionIndex subclasses,\n # so self is never backed by an EA.\n arr = np.asarray(self)\n item = self._coerce_scalar_to_index(item)._values\n idx = np.concatenate((arr[:loc], item, arr[loc:]))\n return self._shallow_copy_with_infer(idx)\n\n def drop(self, labels, errors: str_t = \"raise\"):\n \"\"\"\n Make new Index with passed list of labels deleted.\n\n Parameters\n ----------\n labels : array-like\n errors : {'ignore', 'raise'}, default 'raise'\n If 'ignore', suppress error and existing labels are dropped.\n\n Returns\n -------\n dropped : Index\n\n Raises\n ------\n KeyError\n If not all of the labels are found in the selected axis\n \"\"\"\n arr_dtype = \"object\" if self.dtype == \"object\" else None\n labels = com.index_labels_to_array(labels, dtype=arr_dtype)\n indexer = self.get_indexer(labels)\n mask = indexer == -1\n if mask.any():\n if errors != \"ignore\":\n raise KeyError(f\"{labels[mask]} not found in axis\")\n indexer = indexer[~mask]\n return self.delete(indexer)\n\n # --------------------------------------------------------------------\n # Generated Arithmetic, Comparison, and Unary Methods\n\n @classmethod\n def _add_comparison_methods(cls):\n \"\"\"\n Add in comparison methods.\n \"\"\"\n cls.__eq__ = _make_comparison_op(operator.eq, cls)\n cls.__ne__ = _make_comparison_op(operator.ne, cls)\n cls.__lt__ = _make_comparison_op(operator.lt, cls)\n cls.__gt__ = _make_comparison_op(operator.gt, cls)\n cls.__le__ = _make_comparison_op(operator.le, cls)\n cls.__ge__ = _make_comparison_op(operator.ge, cls)\n\n @classmethod\n def _add_numeric_methods_add_sub_disabled(cls):\n \"\"\"\n Add in the numeric add/sub methods to disable.\n \"\"\"\n cls.__add__ = make_invalid_op(\"__add__\")\n cls.__radd__ = make_invalid_op(\"__radd__\")\n cls.__iadd__ = make_invalid_op(\"__iadd__\")\n cls.__sub__ = make_invalid_op(\"__sub__\")\n cls.__rsub__ = make_invalid_op(\"__rsub__\")\n cls.__isub__ = make_invalid_op(\"__isub__\")\n\n @classmethod\n def _add_numeric_methods_disabled(cls):\n \"\"\"\n Add in numeric methods to disable other than add/sub.\n \"\"\"\n cls.__pow__ = make_invalid_op(\"__pow__\")\n cls.__rpow__ = make_invalid_op(\"__rpow__\")\n cls.__mul__ = make_invalid_op(\"__mul__\")\n cls.__rmul__ = make_invalid_op(\"__rmul__\")\n cls.__floordiv__ = make_invalid_op(\"__floordiv__\")\n cls.__rfloordiv__ = make_invalid_op(\"__rfloordiv__\")\n cls.__truediv__ = make_invalid_op(\"__truediv__\")\n cls.__rtruediv__ = make_invalid_op(\"__rtruediv__\")\n cls.__mod__ = make_invalid_op(\"__mod__\")\n cls.__divmod__ = make_invalid_op(\"__divmod__\")\n cls.__neg__ = make_invalid_op(\"__neg__\")\n cls.__pos__ = make_invalid_op(\"__pos__\")\n cls.__abs__ = make_invalid_op(\"__abs__\")\n cls.__inv__ = make_invalid_op(\"__inv__\")\n\n @classmethod\n def _add_numeric_methods_binary(cls):\n \"\"\"\n Add in numeric methods.\n \"\"\"\n cls.__add__ = _make_arithmetic_op(operator.add, cls)\n cls.__radd__ = _make_arithmetic_op(ops.radd, cls)\n cls.__sub__ = _make_arithmetic_op(operator.sub, cls)\n cls.__rsub__ = _make_arithmetic_op(ops.rsub, cls)\n cls.__rpow__ = _make_arithmetic_op(ops.rpow, cls)\n cls.__pow__ = _make_arithmetic_op(operator.pow, cls)\n\n cls.__truediv__ = _make_arithmetic_op(operator.truediv, cls)\n cls.__rtruediv__ = _make_arithmetic_op(ops.rtruediv, cls)\n\n # TODO: rmod? rdivmod?\n cls.__mod__ = _make_arithmetic_op(operator.mod, cls)\n cls.__floordiv__ = _make_arithmetic_op(operator.floordiv, cls)\n cls.__rfloordiv__ = _make_arithmetic_op(ops.rfloordiv, cls)\n cls.__divmod__ = _make_arithmetic_op(divmod, cls)\n cls.__mul__ = _make_arithmetic_op(operator.mul, cls)\n cls.__rmul__ = _make_arithmetic_op(ops.rmul, cls)\n\n @classmethod\n def _add_numeric_methods_unary(cls):\n \"\"\"\n Add in numeric unary methods.\n \"\"\"\n\n def _make_evaluate_unary(op, opstr: str_t):\n def _evaluate_numeric_unary(self):\n\n attrs = self._get_attributes_dict()\n return Index(op(self.values), **attrs)\n\n _evaluate_numeric_unary.__name__ = opstr\n return _evaluate_numeric_unary\n\n cls.__neg__ = _make_evaluate_unary(operator.neg, \"__neg__\")\n cls.__pos__ = _make_evaluate_unary(operator.pos, \"__pos__\")\n cls.__abs__ = _make_evaluate_unary(np.abs, \"__abs__\")\n cls.__inv__ = _make_evaluate_unary(lambda x: -x, \"__inv__\")\n\n @classmethod\n def _add_numeric_methods(cls):\n cls._add_numeric_methods_unary()\n cls._add_numeric_methods_binary()\n\n @classmethod\n def _add_logical_methods(cls):\n \"\"\"\n Add in logical methods.\n \"\"\"\n _doc = \"\"\"\n %(desc)s\n\n Parameters\n ----------\n *args\n These parameters will be passed to numpy.%(outname)s.\n **kwargs\n These parameters will be passed to numpy.%(outname)s.\n\n Returns\n -------\n %(outname)s : bool or array_like (if axis is specified)\n A single element array_like may be converted to bool.\"\"\"\n\n _index_shared_docs[\"index_all\"] = dedent(\n \"\"\"\n\n See Also\n --------\n Index.any : Return whether any element in an Index is True.\n Series.any : Return whether any element in a Series is True.\n Series.all : Return whether all elements in a Series are True.\n\n Notes\n -----\n Not a Number (NaN), positive infinity and negative infinity\n evaluate to True because these are not equal to zero.\n\n Examples\n --------\n **all**\n\n True, because nonzero integers are considered True.\n\n >>> pd.Index([1, 2, 3]).all()\n True\n\n False, because ``0`` is considered False.\n\n >>> pd.Index([0, 1, 2]).all()\n False\n\n **any**\n\n True, because ``1`` is considered True.\n\n >>> pd.Index([0, 0, 1]).any()\n True\n\n False, because ``0`` is considered False.\n\n >>> pd.Index([0, 0, 0]).any()\n False\n \"\"\"\n )\n\n _index_shared_docs[\"index_any\"] = dedent(\n \"\"\"\n\n See Also\n --------\n Index.all : Return whether all elements are True.\n Series.all : Return whether all elements are True.\n\n Notes\n -----\n Not a Number (NaN), positive infinity and negative infinity\n evaluate to True because these are not equal to zero.\n\n Examples\n --------\n >>> index = pd.Index([0, 1, 2])\n >>> index.any()\n True\n\n >>> index = pd.Index([0, 0, 0])\n >>> index.any()\n False\n \"\"\"\n )\n\n def _make_logical_function(name: str_t, desc: str_t, f):\n @Substitution(outname=name, desc=desc)\n @Appender(_index_shared_docs[\"index_\" + name])\n @Appender(_doc)\n def logical_func(self, *args, **kwargs):\n result = f(self.values)\n if (\n isinstance(result, (np.ndarray, ABCSeries, Index))\n and result.ndim == 0\n ):\n # return NumPy type\n return result.dtype.type(result.item())\n else: # pragma: no cover\n return result\n\n logical_func.__name__ = name\n return logical_func\n\n cls.all = _make_logical_function(\n \"all\", \"Return whether all elements are True.\", np.all\n )\n cls.any = _make_logical_function(\n \"any\", \"Return whether any element is True.\", np.any\n )\n\n @classmethod\n def _add_logical_methods_disabled(cls):\n \"\"\"\n Add in logical methods to disable.\n \"\"\"\n cls.all = make_invalid_op(\"all\")\n cls.any = make_invalid_op(\"any\")\n\n @property\n def shape(self):\n \"\"\"\n Return a tuple of the shape of the underlying data.\n \"\"\"\n # not using \"(len(self), )\" to return \"correct\" shape if the values\n # consists of a >1 D array (see GH-27775)\n # overridden in MultiIndex.shape to avoid materializing the values\n return self._values.shape\n\n\nIndex._add_numeric_methods_disabled()\nIndex._add_logical_methods()\nIndex._add_comparison_methods()\n\n\ndef ensure_index_from_sequences(sequences, names=None):\n \"\"\"\n Construct an index from sequences of data.\n\n A single sequence returns an Index. Many sequences returns a\n MultiIndex.\n\n Parameters\n ----------\n sequences : sequence of sequences\n names : sequence of str\n\n Returns\n -------\n index : Index or MultiIndex\n\n Examples\n --------\n >>> ensure_index_from_sequences([[1, 2, 3]], names=[\"name\"])\n Int64Index([1, 2, 3], dtype='int64', name='name')\n\n >>> ensure_index_from_sequences([[\"a\", \"a\"], [\"a\", \"b\"]], names=[\"L1\", \"L2\"])\n MultiIndex([('a', 'a'),\n ('a', 'b')],\n names=['L1', 'L2'])\n\n See Also\n --------\n ensure_index\n \"\"\"\n from pandas.core.indexes.multi import MultiIndex\n\n if len(sequences) == 1:\n if names is not None:\n names = names[0]\n return Index(sequences[0], name=names)\n else:\n return MultiIndex.from_arrays(sequences, names=names)\n\n\ndef ensure_index(index_like, copy: bool = False):\n \"\"\"\n Ensure that we have an index from some index-like object.\n\n Parameters\n ----------\n index_like : sequence\n An Index or other sequence\n copy : bool, default False\n\n Returns\n -------\n index : Index or MultiIndex\n\n See Also\n --------\n ensure_index_from_sequences\n\n Examples\n --------\n >>> ensure_index(['a', 'b'])\n Index(['a', 'b'], dtype='object')\n\n >>> ensure_index([('a', 'a'), ('b', 'c')])\n Index([('a', 'a'), ('b', 'c')], dtype='object')\n\n >>> ensure_index([['a', 'a'], ['b', 'c']])\n MultiIndex([('a', 'b'),\n ('a', 'c')],\n )\n \"\"\"\n if isinstance(index_like, Index):\n if copy:\n index_like = index_like.copy()\n return index_like\n if hasattr(index_like, \"name\"):\n return Index(index_like, name=index_like.name, copy=copy)\n\n if is_iterator(index_like):\n index_like = list(index_like)\n\n # must check for exactly list here because of strict type\n # check in clean_index_list\n if isinstance(index_like, list):\n if type(index_like) != list:\n index_like = list(index_like)\n\n converted, all_arrays = lib.clean_index_list(index_like)\n\n if len(converted) > 0 and all_arrays:\n from pandas.core.indexes.multi import MultiIndex\n\n return MultiIndex.from_arrays(converted)\n else:\n index_like = converted\n else:\n # clean_index_list does the equivalent of copying\n # so only need to do this if not list instance\n if copy:\n index_like = copy_func(index_like)\n\n return Index(index_like)\n\n\ndef ensure_has_len(seq):\n \"\"\"\n If seq is an iterator, put its values into a list.\n \"\"\"\n try:\n len(seq)\n except TypeError:\n return list(seq)\n else:\n return seq\n\n\ndef _trim_front(strings):\n \"\"\"\n Trims zeros and decimal points.\n \"\"\"\n trimmed = strings\n while len(strings) > 0 and all(x[0] == \" \" for x in trimmed):\n trimmed = [x[1:] for x in trimmed]\n return trimmed\n\n\ndef _validate_join_method(method: str):\n if method not in [\"left\", \"right\", \"inner\", \"outer\"]:\n raise ValueError(f\"do not recognize join method {method}\")\n\n\ndef default_index(n):\n from pandas.core.indexes.range import RangeIndex\n\n return RangeIndex(0, n, name=None)\n\n\ndef maybe_extract_name(name, obj, cls) -> Label:\n \"\"\"\n If no name is passed, then extract it from data, validating hashability.\n \"\"\"\n if name is None and isinstance(obj, (Index, ABCSeries)):\n # Note we don't just check for \"name\" attribute since that would\n # pick up e.g. dtype.name\n name = obj.name\n\n # GH#29069\n if not is_hashable(name):\n raise TypeError(f\"{cls.__name__}.name must be a hashable type\")\n\n return name\n\n\ndef _maybe_cast_with_dtype(data: np.ndarray, dtype: np.dtype, copy: bool) -> np.ndarray:\n \"\"\"\n If a dtype is passed, cast to the closest matching dtype that is supported\n by Index.\n\n Parameters\n ----------\n data : np.ndarray\n dtype : np.dtype\n copy : bool\n\n Returns\n -------\n np.ndarray\n \"\"\"\n # we need to avoid having numpy coerce\n # things that look like ints/floats to ints unless\n # they are actually ints, e.g. '0' and 0.0\n # should not be coerced\n # GH 11836\n if is_integer_dtype(dtype):\n inferred = lib.infer_dtype(data, skipna=False)\n if inferred == \"integer\":\n data = maybe_cast_to_integer_array(data, dtype, copy=copy)\n elif inferred in [\"floating\", \"mixed-integer-float\"]:\n if isna(data).any():\n raise ValueError(\"cannot convert float NaN to integer\")\n\n if inferred == \"mixed-integer-float\":\n data = maybe_cast_to_integer_array(data, dtype)\n\n # If we are actually all equal to integers,\n # then coerce to integer.\n try:\n data = _try_convert_to_int_array(data, copy, dtype)\n except ValueError:\n data = np.array(data, dtype=np.float64, copy=copy)\n\n elif inferred == \"string\":\n pass\n else:\n data = data.astype(dtype)\n elif is_float_dtype(dtype):\n inferred = lib.infer_dtype(data, skipna=False)\n if inferred == \"string\":\n pass\n else:\n data = data.astype(dtype)\n else:\n data = np.array(data, dtype=dtype, copy=copy)\n\n return data\n\n\ndef _maybe_cast_data_without_dtype(subarr):\n \"\"\"\n If we have an arraylike input but no passed dtype, try to infer\n a supported dtype.\n\n Parameters\n ----------\n subarr : np.ndarray, Index, or Series\n\n Returns\n -------\n converted : np.ndarray or ExtensionArray\n dtype : np.dtype or ExtensionDtype\n \"\"\"\n # Runtime import needed bc IntervalArray imports Index\n from pandas.core.arrays import (\n IntervalArray,\n PeriodArray,\n DatetimeArray,\n TimedeltaArray,\n )\n\n inferred = lib.infer_dtype(subarr, skipna=False)\n\n if inferred == \"integer\":\n try:\n data = _try_convert_to_int_array(subarr, False, None)\n return data, data.dtype\n except ValueError:\n pass\n\n return subarr, object\n\n elif inferred in [\"floating\", \"mixed-integer-float\", \"integer-na\"]:\n # TODO: Returns IntegerArray for integer-na case in the future\n return subarr, np.float64\n\n elif inferred == \"interval\":\n try:\n data = IntervalArray._from_sequence(subarr, copy=False)\n return data, data.dtype\n except ValueError:\n # GH27172: mixed closed Intervals --> object dtype\n pass\n elif inferred == \"boolean\":\n # don't support boolean explicitly ATM\n pass\n elif inferred != \"string\":\n if inferred.startswith(\"datetime\"):\n try:\n data = DatetimeArray._from_sequence(subarr, copy=False)\n return data, data.dtype\n except (ValueError, OutOfBoundsDatetime):\n # GH 27011\n # If we have mixed timezones, just send it\n # down the base constructor\n pass\n\n elif inferred.startswith(\"timedelta\"):\n data = TimedeltaArray._from_sequence(subarr, copy=False)\n return data, data.dtype\n elif inferred == \"period\":\n try:\n data = PeriodArray._from_sequence(subarr)\n return data, data.dtype\n except IncompatibleFrequency:\n pass\n\n return subarr, subarr.dtype\n\n\ndef _try_convert_to_int_array(\n data: np.ndarray, copy: bool, dtype: np.dtype\n) -> np.ndarray:\n \"\"\"\n Attempt to convert an array of data into an integer array.\n\n Parameters\n ----------\n data : The data to convert.\n copy : bool\n Whether to copy the data or not.\n dtype : np.dtype\n\n Returns\n -------\n int_array : data converted to either an ndarray[int64] or ndarray[uint64]\n\n Raises\n ------\n ValueError if the conversion was not successful.\n \"\"\"\n if not is_unsigned_integer_dtype(dtype):\n # skip int64 conversion attempt if uint-like dtype is passed, as\n # this could return Int64Index when UInt64Index is what's desired\n try:\n res = data.astype(\"i8\", copy=False)\n if (res == data).all():\n return res # TODO: might still need to copy\n except (OverflowError, TypeError, ValueError):\n pass\n\n # Conversion to int64 failed (possibly due to overflow) or was skipped,\n # so let's try now with uint64.\n try:\n res = data.astype(\"u8\", copy=False)\n if (res == data).all():\n return res # TODO: might still need to copy\n except (OverflowError, TypeError, ValueError):\n pass\n\n raise ValueError\n\n\ndef _maybe_asobject(dtype, klass, data, copy: bool, name: Label, **kwargs):\n \"\"\"\n If an object dtype was specified, create the non-object Index\n and then convert it to object.\n\n Parameters\n ----------\n dtype : np.dtype, ExtensionDtype, str\n klass : Index subclass\n data : list-like\n copy : bool\n name : hashable\n **kwargs\n\n Returns\n -------\n Index\n\n Notes\n -----\n We assume that calling .astype(object) on this klass will make a copy.\n \"\"\"\n\n # GH#23524 passing `dtype=object` to DatetimeIndex is invalid,\n # will raise in the where `data` is already tz-aware. So\n # we leave it out of this step and cast to object-dtype after\n # the DatetimeIndex construction.\n\n if is_dtype_equal(_o_dtype, dtype):\n # Note we can pass copy=False because the .astype below\n # will always make a copy\n index = klass(data, copy=False, name=name, **kwargs)\n return index.astype(object)\n\n return klass(data, dtype=dtype, copy=copy, name=name, **kwargs)\n"
]
| [
[
"pandas.core.indexes.multi.MultiIndex",
"pandas.core.indexes.range.RangeIndex",
"pandas.core.dtypes.common.ensure_object",
"numpy.where",
"pandas.core.dtypes.common.is_interval_dtype",
"pandas.core.common.cast_scalar_indexer",
"pandas.core.arrays.PeriodArray._from_sequence",
"pandas.core.dtypes.common.is_iterator",
"pandas.core.dtypes.common.is_float_dtype",
"pandas.core.dtypes.common.is_categorical_dtype",
"pandas._libs.join.outer_join_indexer",
"pandas.core.dtypes.common.is_list_like",
"pandas.core.indexes.numeric.UInt64Index",
"numpy.delete",
"numpy.array",
"pandas.core.algorithms.take",
"pandas.core.dtypes.common.is_bool_dtype",
"pandas.core.arrays.DatetimeArray._from_sequence",
"pandas.core.common.is_null_slice",
"pandas.core.dtypes.missing.isna",
"pandas.io.formats.printing.pprint_thing",
"pandas.Series",
"pandas._libs.tslibs.Timestamp",
"numpy.asarray",
"pandas._libs.join.inner_join_indexer",
"numpy.concatenate",
"pandas._libs.lib.clean_index_list",
"pandas.core.indexes.numeric.Int64Index",
"pandas.core.dtypes.common.is_unsigned_integer_dtype",
"pandas.core.common.asarray_tuplesafe",
"pandas.io.formats.printing.PrettyDict",
"pandas.io.formats.format.format_array",
"pandas.core.indexes.multi.MultiIndex.from_tuples",
"pandas.core.algorithms.take_nd",
"pandas.core.algorithms.safe_sort",
"numpy.ndim",
"numpy.errstate",
"pandas.core.indexers.deprecate_ndim_indexing",
"pandas._libs.algos.groupsort_indexer",
"pandas.core.arrays.TimedeltaArray._from_sequence",
"pandas.core.dtypes.common.is_integer",
"pandas._libs.lib.infer_dtype",
"pandas.util._decorators.doc",
"pandas._libs.lib.item_from_zerodim",
"numpy.empty",
"pandas.core.dtypes.cast.maybe_cast_to_integer_array",
"pandas.core.dtypes.common.is_extension_array_dtype",
"pandas.core.dtypes.common.ensure_categorical",
"pandas.core.dtypes.common.is_dtype_equal",
"pandas._libs.lib.is_scalar",
"pandas.core.indexes.frozen.FrozenList",
"pandas.core.indexes.numeric.Float64Index",
"pandas.core.indexes.range.RangeIndex.from_range",
"pandas.core.reshape.merge._get_join_indexers",
"numpy.hstack",
"pandas.util._decorators.Substitution",
"pandas.core.common.not_none",
"pandas.core.dtypes.common.ensure_int64",
"pandas.core.dtypes.concat.concat_compat",
"numpy.repeat",
"pandas.core.arrays.numpy_.PandasArray",
"pandas._libs.tslibs.timezones.tz_compare",
"pandas.core.ops.invalid.make_invalid_op",
"pandas.core.dtypes.common.is_integer_dtype",
"pandas._libs.join.left_join_indexer_unique",
"pandas.util._decorators.Appender",
"pandas.core.dtypes.cast.validate_numeric_casting",
"pandas.core.dtypes.common.is_timedelta64_dtype",
"pandas.core.dtypes.common.is_hashable",
"pandas.core.dtypes.common.is_period_dtype",
"pandas.core.indexes.period._new_PeriodIndex",
"pandas.core.indexes.category.CategoricalIndex",
"pandas.core.reshape.merge._restore_dropped_levels_multijoin",
"pandas.core.arrays.IntervalArray._from_sequence",
"pandas.core.ops.invalid_comparison",
"pandas.core.dtypes.common.is_bool",
"pandas.core.ops.comp_method_OBJECT_ARRAY",
"pandas.core.algorithms.isin",
"pandas.core.common.is_bool_indexer",
"pandas.core.dtypes.common.is_scalar",
"pandas.core.dtypes.missing.array_equivalent",
"pandas.core.accessor.CachedAccessor",
"pandas.io.formats.printing.format_object_summary",
"numpy.dtype",
"pandas.core.dtypes.common.is_signed_integer_dtype",
"numpy.arange",
"pandas.core.ops.get_op_result_name",
"pandas.compat.set_function_name",
"pandas.core.dtypes.common.is_float",
"pandas.core.dtypes.common.is_datetime64_any_dtype",
"pandas.core.dtypes.common.ensure_platform_int",
"pandas.io.formats.printing.format_object_attrs",
"numpy.abs",
"pandas.core.dtypes.concat.get_dtype_kinds",
"pandas._libs.join.left_join_indexer",
"pandas.core.dtypes.common.is_object_dtype",
"pandas.core.indexes.multi.MultiIndex.from_arrays",
"pandas._libs.lib.maybe_convert_objects",
"pandas.core.common.index_labels_to_array"
]
]
|
melvyniandrag/quadpy | [
"ae28fc17351be8e76909033f03d71776c7ef8280"
]
| [
"tools/xiao_gimbutas/import_xiao_gimbutas_tet.py"
]
| [
"# -*- coding: utf-8 -*-\n#\n\"\"\"\nParse Fortran code to extract points and weight of the Xiao-Gimbutas schemes.\n\"\"\"\nimport numpy\n\n\n# TODO the first two functions could go into a helper and be shared with tri\ndef _parsed_strings_to_array(strings):\n return numpy.array(\n \"\".join(strings).replace(\"&\", \"\").replace(\"/\", \"\").replace(\"D\", \"e\").split(\",\"),\n dtype=float,\n )\n\n\ndef _parse():\n # The Fortran file contains multiple sections like\n # ```\n # data xs / &\n # -.1685037180276000D+00,0.2783799427534418D-01, &\n # [...]\n # data ys / &\n # 0.1910914916271708D+00,-.2304932838839657D-01, &\n # [...]\n # data zs / &\n # -.3896267314585163D+00,0.5481350663241830D+00, &\n # [...]\n # data ws / &\n # 0.1287213727402025D+00,0.2179034339695993D+00, &\n # [...]\n # ```\n # (Sometimes single columns.)\n # Find those and extract the data.\n data = []\n\n with open(\"tet.txt\", \"r\") as f:\n while True:\n line = f.readline()\n if not line:\n # EOF\n break\n\n line = line.strip()\n\n # skip if not at the start of a data block\n if line[:7] != \"data xs\":\n continue\n\n # start of a data block\n xstr = []\n while line[-1] == \"&\":\n line = f.readline().strip()\n xstr.append(line)\n\n line = f.readline().strip()\n assert line[:7] == \"data ys\"\n ystr = []\n while line[-1] == \"&\":\n line = f.readline().strip()\n ystr.append(line)\n\n line = f.readline().strip()\n assert line[:7] == \"data zs\"\n zstr = []\n while line[-1] == \"&\":\n line = f.readline().strip()\n zstr.append(line)\n\n line = f.readline().strip()\n assert line[:7] == \"data ws\"\n wstr = []\n while line[-1] == \"&\":\n line = f.readline().strip()\n wstr.append(line)\n\n points = numpy.column_stack(\n [\n _parsed_strings_to_array(xstr),\n _parsed_strings_to_array(ystr),\n _parsed_strings_to_array(zstr),\n ]\n )\n weights = _parsed_strings_to_array(wstr)\n data.append((points, weights))\n\n return data\n\n\ndef _extract_bary_data(data):\n # The points are given in terms of coordinates of a reference tetrahedron. Convert\n # to barycentric coordinates, and check their symmetry there.\n t0 = [-1, -1 / numpy.sqrt(3), -1 / numpy.sqrt(6)]\n t1 = [+0, +2 / numpy.sqrt(3), -1 / numpy.sqrt(6)]\n t2 = [+1, -1 / numpy.sqrt(3), -1 / numpy.sqrt(6)]\n t3 = [+0, +0, 3 / numpy.sqrt(6)]\n\n T = numpy.array([[t1[k] - t0[k], t2[k] - t0[k], t3[k] - t0[k]] for k in range(3)])\n\n all_dicts = []\n\n ref_weight = 0.9709835434146467\n\n for k, item in enumerate(data):\n d = {\"degree\": k + 1}\n points, weights = item\n\n b = (points - t0).T\n sol = numpy.linalg.solve(T, b)\n bary = numpy.column_stack(\n [sol[0], sol[1], sol[2], 1.0 - sol[0] - sol[1] - sol[2]]\n )\n\n idx = numpy.argsort(weights)\n d[\"weights\"] = (weights[idx] / ref_weight).tolist()\n d[\"bary\"] = bary[idx].tolist()\n all_dicts.append(d)\n\n return all_dicts\n\n\ndef _main():\n data = _parse()\n all_dicts = _extract_bary_data(data)\n\n # Write the json files.\n\n # Getting floats in scientific notation in python.json is almost impossible, so do\n # some work here. Compare with <https://stackoverflow.com/a/1733105/353337>.\n class PrettyFloat(float):\n def __repr__(self):\n return \"{:.16e}\".format(self)\n\n def pretty_floats(obj):\n if isinstance(obj, float):\n return PrettyFloat(obj)\n elif isinstance(obj, dict):\n return dict((k, pretty_floats(v)) for k, v in obj.items())\n elif isinstance(obj, (list, tuple, numpy.ndarray)):\n return list(map(pretty_floats, obj))\n return obj\n\n for d in all_dicts:\n degree = d[\"degree\"]\n print(d)\n with open(\"xg{:02d}.json\".format(degree), \"w\") as f:\n string = (\n pretty_floats(d)\n .__repr__()\n .replace(\"'\", '\"')\n .replace(\"{\", \"{\\n \")\n .replace(\"[[\", \"[\\n [\")\n .replace(\"], [\", \"],\\n [\")\n .replace(']], \"', ']\\n ],\\n \"')\n .replace(\"}\", \"\\n}\")\n )\n f.write(string)\n return\n\n\nif __name__ == \"__main__\":\n _main()\n"
]
| [
[
"numpy.argsort",
"numpy.linalg.solve",
"numpy.sqrt",
"numpy.column_stack"
]
]
|
kimager/CRRLpy | [
"7209f18f7b2d25c85ea1938e5ae8474511823d6b"
]
| [
"crrlpy/frec_calc.py"
]
| [
"#!/usr/bin/env python\n\n__docformat__ = 'reStructuredText'\n\nimport argparse\n\nimport numpy as np\n\nfrom scipy.constants import c, m_e, physical_constants\nfrom astropy import units as u\n\ndef line_freq(Z, R_X, n, dn):\n \"\"\"\n Uses the Rydberg formula to get the frequency\n of a transition to quantum number n for a given atom.\n \n :param Z: Charge of the atom.\n :type Z: int\n :param R_X:\n :type R_X: float\n :param n: Principal quantum number of the transition. :math:`n+\\\\Delta n\\\\rightarrow n`.\n :type n: int\n :param dn: Difference between the principal quantum number of the initial state \\\n and the final state. :math:`\\\\Delta n=n_{f}-n_{i}`.\n :type dn: int\n :returns: The frequency of the transition in MHz.\n :rtype: float\n \"\"\"\n \n return (Z**2)*R_X*c*((1./(n**2))-(1./((n + dn)**2)))\n\ndef set_specie(specie):\n \"\"\"\n Sets atomic constants based on the atomic specie.\n \n :param specie: Atomic specie.\n :type specie: string\n :returns: Array with the atomic mass in a.m.u., ionization potential, abundance relative to HI, :math:`V_{X}-V_{H}` and the electric charge.\n \n :Example:\n \n >>> set_specie('CI')\n [12.0, 11.4, 0.0003, 149.5, 1.0]\n\n \"\"\"\n \n # data for species (table 1 RG92)\n # [atomic.mass, ion.potential, abundance, V_X-V_H, Z]\n if 'HI' in specie:\n X = [1.0078,13.6,1.0,0.0,1.0]\n name = 'HI'\n if 'HeI' in specie:\n X = [4.0026,24.6,0.1,122.1,1.0]\n name = 'HeI'\n if 'CI' in specie:\n #X = [12.0000,11.4,3.e-4,149.5,6.0]\n X = [12.0000,11.4,3.e-4,149.5,1.0]\n name = 'CI'\n if 'NI' in specie:\n X = [14.0067,1,1,1,1.0]\n name = 'NI'\n if 'SI' in specie:\n #X = [37.9721,10.3,2.e-5,158.0,16.0]\n X = [37.9721,10.3,2.e-5,158.0,1.0]\n name = 'SI'\n # isotopes\n if 'CI13' in specie:\n X = [13.00335,-1.0,-1.0,-1.0,1.0]\n name = 'CI13'\n if 'CI14' in specie:\n X = [14.003241,-1.0,-1.0,-1.0,1.0]\n name = 'CI14'\n \n return X\n\ndef set_trans(dn):\n \"\"\"\n Sets a name depending on the difference between atomic levels.\n \n :param dn: Separation between :math:`n_{i}` and :math:`n_{f}`, :math:`\\\\Delta n=n_{i}-n_{f}`.\n :type dn: int\n :returns: alpha, beta, gamma, delta or epsilon depending on :math:`\\\\Delta n`.\n :rtype: string\n \n :Example:\n \n >>> set_trans(5)\n 'epsilon'\n \"\"\"\n \n if dn == 1:\n name = 'alpha'\n if dn == 2:\n name = 'beta'\n if dn == 3:\n name = 'gamma'\n if dn == 4:\n name = 'delta'\n if dn == 5:\n name = 'epsilon'\n if dn == 6:\n name = 'zeta'\n if dn == 7:\n name = 'eta'\n \n return name\n\ndef set_dn(name):\n \"\"\"\n Sets the value of Delta n depending on the transition name.\n \n :param name: Name of the transition.\n :type name: string\n :returns: :math:`\\\\Delta n` for the given transition.\n :rtype: int\n \n :Example:\n \n >>> set_dn('CIalpha')\n 1\n >>> set_dn('CIdelta')\n 4\n \"\"\"\n \n if 'alpha' in name:\n dn = 1\n elif 'beta' in name:\n dn = 2\n elif 'gamma' in name:\n dn = 3\n elif 'delta' in name:\n dn = 4\n elif 'epsilon' in name:\n dn = 5\n elif 'zeta' in name:\n dn = 6\n elif 'eta' in name:\n dn = 7\n \n return dn\n\ndef make_line_list(line, n_min=1, n_max=1500, unitless=True):\n \"\"\"\n Creates a list of frequencies for the corresponding line. The frequencies are in MHz.\n \n :param line: Line to compute the frequencies for.\n :type line: string\n :param n_min: Minimum n number to include in the list.\n :type n_min: int\n :param n_max: Maximum n number to include in the list.\n :type n_max: int\n :param unitless: If True the list will have no units. If not the list will be of astropy.units.Quantity_ objects.\n :type unitless: bool\n :returns: 3 lists with the line name, principal quantum number and frequency of the transitions.\n :rtype: list\n \n .. _astropy.units.Quantity: http://docs.astropy.org/en/stable/api/astropy.units.Quantity.html#astropy.units.Quantity\n \"\"\"\n \n n = np.arange(n_min, n_max)\n \n # Define the electron mass in atomic mass units\n m_e_amu = m_e/physical_constants['atomic mass constant'][0]\n \n # set the specie\n X = set_specie(line)\n dn = set_dn(line)\n trans = set_trans(dn)\n \n M_X = X[0]\n R_X = 10.97373/(1.0 + (m_e_amu/M_X))\n Z = X[4]\n \n freq = line_freq(Z, R_X, n, dn)\n \n if not unitless:\n freq = freq*u.MHz\n \n return line, n, freq, trans\n\ndef main():\n \"\"\"\n Main body of the program. Useful for calling as a script.\n \"\"\"\n \n parser = argparse.ArgumentParser()\n parser.add_argument('-i', '--n_min', type=int,\n dest='n_min', default=1, help=\"Minimum n number\")\n parser.add_argument('-n', '--n_max', type=int,\n dest='n_max', default=10000, help=\"Maximum n number\")\n parser.add_argument('-l', '--line', dest='line', default='CI', type=str,\n help=\"Line name. E.g., CIalpha, HeIbeta, HIalpha, CI13alpha, CI14gamma or SIepsilon\")\n args = parser.parse_args()\n \n n_min = args.n_min\n n_max = args.n_max\n line = args.line\n \n line, n, freq, trans = make_line_list(line, n_min, n_max)\n \n specie = line[:line.index(trans)]\n \n # Write the line list to a file\n out = 'RRL_{0}{1}.txt'.format(specie, trans)\n with open(out, 'w') as outf:\n outf.write('#SPECIES-NAME, TRANSITION-TYPE, N-LEVEL, FREQUENCY-[MHZ]\\n')\n for i, ni in enumerate(n):\n outf.write('{0} {1} {2} {3}\\n'.format(specie, trans, ni, freq[i]))\n\nif __name__ == '__main__':\n main()\n"
]
| [
[
"numpy.arange"
]
]
|
abakhru/stock_prediction | [
"bfb4483ac888bc67e2a8928fdf037d23acbf48f9"
]
| [
"stock_predictions/search_stock.py"
]
| [
"#!/usr/bin/env python\nimport json\nfrom datetime import datetime\n\nimport click\nimport requests\nimport yfinance\nfrom pandas import DataFrame\n\nfrom stock_predictions import ROOT\nfrom stock_predictions.logger import LOGGER\nfrom stock_predictions.utils import pretty_print_df\n\nLOGGER.setLevel('INFO')\nfinal_df = DataFrame(columns=['Symbol', 'Name', 'URL', 'High', 'Low', 'Open', 'Close', 'Volume'])\n\n\nclass SearchStockSymbol:\n def __init__(\n self,\n company_name,\n exchange='NASDAQ',\n start_date=\"2020-01-01\",\n end_date=datetime.now().strftime(\"%Y-%m-%d\"),\n ):\n self.company_name = company_name\n self.stock_symbol = None\n self.exchange_name = exchange\n self.start_date = start_date\n self.end_date = end_date\n self.search_stock_symbol()\n\n def search_stock_symbol(self, retry=2):\n url = (\n f'http://d.yimg.com/autoc.finance.yahoo.com/autoc?query='\n f'{self.company_name}&callback=YAHOO.Finance.SymbolSuggest.ssCallback&'\n f'lang=en'\n )\n if self.company_name in final_df['Name'].values:\n LOGGER.warning(f'{self.company_name} already processed')\n return\n response = requests.get(url)\n assert response.status_code == 200\n data = json.loads(\n response.text.replace('YAHOO.Finance.SymbolSuggest.ssCallback(', '').replace(\");\", \"\")\n )\n if retry <= 0:\n LOGGER.critical(\n f'Stock symbol for \"{self.company_name.capitalize()}\" '\n f'not found in {self.exchange_name} exchange'\n )\n return\n try:\n nasdaq_result = [\n i for i in data['ResultSet']['Result'] if i['exchDisp'] == self.exchange_name\n ][0]\n LOGGER.debug(f'{self.exchange_name} only:\\n{json.dumps(nasdaq_result, indent=4)}')\n self.company_name = nasdaq_result['name']\n self.stock_symbol = nasdaq_result['symbol']\n LOGGER.info(f'[{self.stock_symbol}] ==> {self.company_name}')\n except IndexError as _:\n LOGGER.debug(f'All Exchanges results:\\n{json.dumps(data, sort_keys=True, indent=True)}')\n self.company_name = self.company_name.split()[0]\n LOGGER.error(f'Retrying with only the first part of company: {self.company_name}')\n self.search_stock_symbol(retry=retry - 1)\n # sys.exit()\n\n def ticker_details(self):\n if self.stock_symbol is None:\n return\n ticker = yfinance.Ticker(self.stock_symbol)\n data = ticker.history(interval=\"1d\", start=self.start_date, end=self.end_date)\n # data = DataReader(self.stock_symbol, data_source='yahoo', start=self.start_date,\n # end=self.end_date)\n LOGGER.debug(f'[{self.stock_symbol}] details:\\n{data.tail()}')\n LOGGER.info(f'[{self.end_date}] Price: {data[\"Close\"][-1]}')\n ab = data.drop(['Dividends', 'Stock Splits'], axis=1)\n cd = ab.tail(1)\n cd.insert(0, 'Symbol', self.stock_symbol)\n cd.insert(1, 'Name', self.company_name)\n cd.insert(2, 'URL', f'https://finance.yahoo.com/quote/{self.stock_symbol}')\n global final_df\n final_df = final_df.append(cd)\n return data\n\n\[email protected]()\[email protected]('-s', '--stock', default='FB', help='Stock name for prediction')\[email protected]('-e', '--exchange', default='NASDAQ', help='Exchange name to search in')\ndef main(stock, exchange):\n p = SearchStockSymbol(stock, exchange)\n p.ticker_details()\n\n\nif __name__ == '__main__':\n m = ROOT.joinpath('data', 't.txt').read_text().splitlines()\n s = [i.split(' ', 1)[1:] for i in m]\n b = [i[0].strip() for i in s]\n c = [i.split('EQ')[0].replace('-', '') for i in b]\n for i in c:\n p = SearchStockSymbol(company_name=i, exchange='NSE')\n p.ticker_details()\n LOGGER.info(pretty_print_df(final_df.drop_duplicates()))\n # main()\n"
]
| [
[
"pandas.DataFrame"
]
]
|
VijaySingh-GSLab/venue_recommender | [
"3c3bddc19c2f3be71833b85c5a1de2522771f0b3"
]
| [
"utils.py"
]
| [
"import numpy as np\nimport pandas as pd\nfrom scipy import stats\nfrom scipy.spatial import distance\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom matplotlib.ticker import MaxNLocator\nfrom pathlib import Path\nimport plotly.graph_objects as go\n\nMAPBOX_TOKEN = 'pk.eyJ1IjoidmlqYXlzaW5naGdzbGFiIiwiYSI6ImNrbDE2dzB2bjBzZm4ydWxibWIyeG5kYXcifQ.zXZCjXTH-S2UjRmrblp76g'\n\nSEED = 100\nDO_PRINT = True\nMAX_MATCH = 50\nMIN_VENUES_REQUIRED = 3\n\nlist_color_0 = ['#e6194b', '#3cb44b', '#ffe119', '#4363d8', '#f58231', '#911eb4', '#46f0f0', '#f032e6', '#bcf60c',\n '#fabebe', '#008080', '#e6beff', '#9a6324', '#fffac8', '#800000', '#aaffc3', '#808000', '#ffd8b1',\n '#000075', '#808080', '#ffffff', '#fff008']\nlist_palette_0 = ['Blues_r', 'OrRd_r', 'BuGn_r', 'PuRd_r', 'BuPu_r', 'Wistia_r', 'binary_r', 'Blues_r', 'OrRd_r', 'BuGn_r', 'PuRd_r', 'BuPu_r', 'Wistia_r', 'binary_r' ]\n\nPROJECT_PATH = Path.cwd()\nDATA_PATH = PROJECT_PATH.joinpath('dataset')\nDATA_PATH_RAW = DATA_PATH.joinpath('raw_data')\nDATA_PATH_ARTIFACTS = PROJECT_PATH.joinpath('artifacts')\nDATA_PATH_ARTIFACTS_APPDATA = DATA_PATH_ARTIFACTS.joinpath('app_data')\n\ncol_grain = 'Neighborhood'\ncol_feature = 'Venue Category' # Venue\ncol_feature_name = 'Venue'\ncol_latitude = 'Neighborhood_Latitude'\ncol_longitude = 'Neighborhood_Longitude'\n#col_latitude = 'location_lat'\n#col_longitude = 'location_lng'\n\ncolList_coordinates = [col_latitude, col_longitude]\ncolList_rawData = [col_grain, col_feature, col_feature_name] + colList_coordinates\n#colList_rawData = [col_grain, col_feature]\ncolList_meta = [col_grain]\n\n\n#LIST_CITY_DATA_FILE_NAME = ['toronto_venues.csv', 'new_york_venues.csv']\nLIST_CITY_DATA_FILE_NAME = ['Delhi_venues.csv', 'Bangalore_venues.csv', 'Pune_venues.csv', 'Hyderabad_venues.csv',\n 'Mumbai_venues.csv',\n 'Toronto_venues.csv', 'London_venues.csv', 'Austin_venues.csv', 'New York_venues.csv']\n\n\nLIST_CITY = [i.split('_venues.csv')[0] for i in LIST_CITY_DATA_FILE_NAME]\n\n\nDICT_CITY_NAME_MAPPER = {\n 'toronto': 'Toronto',\n 'delhi': 'Delhi',\n 'bangalore': 'Banglore',\n 'pune': 'Pune',\n 'hyderabad': 'Hyderabad',\n 'mumbai': 'Mumbai'\n}\n\n\ndef get_data_path(data_type):\n path = None\n if data_type == 'raw':\n path = DATA_PATH_RAW\n elif data_type == 'artifact_app':\n path = DATA_PATH_ARTIFACTS_APPDATA\n else:\n raise Exception('invalid data_type : {}'.format(data_type))\n return path\n\n\ndef read_data_file(file_name=None, data_type='raw'):\n if data_type == 'artifact_app':\n file_name = '{}_{}'.format('app', file_name)\n\n path = get_data_path(data_type)\n read_path = path.joinpath(file_name)\n #read_path = str(read_path)\n #print(read_path)\n if data_type == 'raw':\n X = pd.read_csv(read_path, usecols=colList_rawData, engine='python')\n X = X[colList_rawData]\n else:\n print('='*50)\n print(read_path)\n X = pd.read_csv(read_path, engine='python')\n print('=' * 50)\n return X\n\n\ndef save_data_file(X=None, file_name=None, data_type='raw'):\n if data_type == 'artifact_app':\n file_name = '{}_{}'.format('app', file_name)\n path = get_data_path(data_type)\n X.to_csv(path_or_buf=path.joinpath(file_name), index=False)\n return X\n\n\ndef neighborhood_haiving_min_n_venues(X_raw=None, min_venues=4):\n X = X_raw.copy()\n X['count'] = 1\n X = X.groupby(by=[col_grain, col_feature], as_index=False).count()\n\n arr = X[col_grain].value_counts()\n return arr[arr >= min_venues].index.values\n\n\ndef pre_process_raw_data(X=None):\n X = X[[col_grain, col_feature]]\n\n ls_neighborhood = neighborhood_haiving_min_n_venues(X_raw=X, min_venues=MIN_VENUES_REQUIRED)\n X = X[X[col_grain].isin(ls_neighborhood)]\n\n X['count'] = 1\n X = pd.pivot_table(X, values='count', index=col_grain, columns=col_feature, aggfunc=np.sum, fill_value=0)\n X = X.drop(columns=[col_grain], errors='ignore')\n X = X.reset_index()\n\n return X\n\n\ndef prepare_app_data_from_raw_data(file_name, return_app_data=False):\n X_raw = read_data_file(file_name, data_type='raw')\n X = pre_process_raw_data(X_raw)\n\n save_data_file(X=X, file_name=file_name, data_type='artifact_app')\n\n if return_app_data:\n return X\n\n\ndef get_common_feature_list(X_source=None, X_dest=None):\n list_features = set(X_source.columns.values).intersection(set(X_dest.columns.values))\n list_features = list(list_features-set(colList_meta))\n return list_features\n\n\ndef calculate_distance(v1, v2):\n return distance.euclidean(v1, v2)\n\n\ndef verbose_print(msg):\n if DO_PRINT:\n print(msg)\n\n\ndef vector_similarity_score(v1, arr_v2, precise_match=False):\n arr_d = []\n if precise_match:\n ls = np.nonzero(v1)\n for v2 in arr_v2:\n d = calculate_distance(np.take(v1, ls), np.take(v2, ls))\n arr_d.append(d)\n else:\n for v2 in arr_v2:\n d = calculate_distance(v1, v2)\n arr_d.append(d)\n return arr_d\n\n\ndef mask_array(v):\n return (v > 0).astype(np.int)\n\n\ndef find_top_match(d_arr=None, arr_vec=None, n=1):\n ls = np.array(d_arr).argsort()[:n]\n return np.array(d_arr)[ls], arr_vec[ls]\n\n\ndef sort_array(vec_1, arr_vec):\n ls = vec_1.argsort()[::-1]\n # return vec_1[ls], arr_vec[ls]\n return vec_1[ls], arr_vec.T[ls].T\n\n\ndef find_best_match(vec_1, arr_vec_match):\n vec_1_st, arr_vec_st = sort_array(vec_1, arr_vec_match.copy()) # col wise\n arr_vec_diff = arr_vec_st - vec_1_st\n\n #print(vec_1_st)\n # print(vec_1_st, arr_vec_st, arr_vec_diff)\n\n arr_sl = []\n for v in arr_vec_diff:\n sl, *vals = stats.linregress(np.arange(0, len(v)), v)\n arr_sl.append(sl)\n\n ls = np.array(arr_sl).argsort()\n return np.array(arr_sl)[ls], arr_vec_diff[ls], arr_vec_st[ls], arr_vec_match[ls]\n # return np.array(arr_sl), arr_vec_diff, arr_vec_st, arr_vec_match\n\n\ndef matching_vector_index(arr_vec_match, arr_vec):\n arr_i = []\n for v in arr_vec_match:\n arr = np.argwhere((v == arr_vec).all(1))[0]\n arr_i.extend(list(arr))\n\n return arr_i\n\n\ndef perform_match(vec_1=None, arr_vec=None, precise_match=False, num_match=1):\n #print(vec_1)\n vec_1_mk = mask_array(vec_1)\n arr_vec_mk = mask_array(arr_vec)\n\n d_arr = vector_similarity_score(vec_1_mk, arr_vec_mk, precise_match=precise_match)\n d_arr_filt, arr_vec_match = find_top_match(d_arr, arr_vec, num_match)\n\n ls_sl, arr_vec_diff, arr_vec_st, arr_vec_match = find_best_match(vec_1, arr_vec_match)\n\n list_ind_match = matching_vector_index(arr_vec_match, arr_vec)\n\n # for d, sl, vec_diff, vec_st, vec_match in zip(d_arr_filt, ls_sl, arr_vec_diff, arr_vec_st, arr_vec_match):\n # print('{} --> {}, {} : {}, {}'.format(vec_match, vec_st, vec_diff, np.round(sl,2), np.round(1.2,2)))\n\n return list_ind_match\n\n\ndef prepare_sorted_match_df(X_source=None, X_match=None, nv=1, colList_features=None):\n X = X_source.copy()\n X['index'] = -1\n X = X.set_index('index')\n\n df = pd.concat(objs=[X[colList_features], X_match[colList_features]])\n\n # X_source is already a selected row\n vec_source = X_source.values[0]\n ls_sort = vec_source.argsort()[::-1][:nv]\n df = df.iloc[:, ls_sort].copy()\n return df\n\n\ndef preapre_venue_plot_data(X_match_sorted=None, X_meta_mapper=None, colList_features=None):\n X = X_match_sorted.copy()\n ls_df_order = X.index.values # use later for sorting\n\n # order of X is imp\n X = X.reset_index()\n X = pd.merge(left=X_meta_mapper, right=X, on='index', how='right')\n\n #X = X.drop_duplicates(subset=[col_grain], keep='first') # new change in app\n\n ls_features = [i for i in colList_features if i in X.columns.values]\n plot_df = pd.melt(frame=X, id_vars=col_grain, value_vars=ls_features)\n return X, plot_df\n\n\ndef get_source_vector(X=None, source_name=None, return_df=False, colList_features=None):\n if return_df:\n return X.loc[X['Neighborhood'] == source_name][colList_features].copy()\n else:\n return X.loc[X['Neighborhood'] == source_name][colList_features].values[0]\n\n\ndef get_match_df(X=None, list_ind=None, colList_features=None):\n df = X[colList_features].copy()\n df = df.loc[list_ind]\n\n return df\n\n\ndef get_sorted_list_of_features(X_source_selected=None, colList_features=None):\n vec_source = X_source_selected.values[0]\n vec_source = vec_source.argsort()[::-1]\n\n arr = X_source_selected[colList_features].columns.values\n arr = arr[vec_source]\n return arr\n\n\ndef prepare_meta_mapper(X_dest=None, colList_meta=None, source_name=None):\n X = X_dest.copy()\n X = X[colList_meta].reset_index()\n X.loc[X.index.max()+1] = [-1, source_name]\n return X\n\n\ndef perform_match_wrapper(X_source=None, X_dest=None, source_name=None, num_match=None, precise_match=True,\n colList_features=None, colList_meta=None):\n if num_match is None:\n if len(X_dest) <= 50:\n num_match = len(X_dest)-2\n else:\n num_match = MAX_MATCH\n\n # input data\n X_meta_mapper = prepare_meta_mapper(X_dest=X_dest, colList_meta=colList_meta, source_name=source_name)\n vec_1 = get_source_vector(X=X_source, source_name=source_name, colList_features=colList_features, return_df=False)\n #arr_vec = X_dest.drop(columns=[col_grain]).values # ph2 change\n arr_vec = X_dest[colList_features].values\n\n # matching\n list_ind_match = perform_match(vec_1=vec_1, arr_vec=arr_vec, precise_match=precise_match, num_match=num_match)\n X_match = get_match_df(X=X_dest, list_ind=list_ind_match, colList_features=colList_features)\n\n return X_match, X_meta_mapper\n\n\ndef visualize_venue_match_results_wrapper(X_source=None, X_match=None, X_meta_mapper=None,\n source_name=None, colList_features=None, num_match=1, num_venues=1,\n show_plot=False):\n X_match = X_match.head(num_match)\n # prepare plot data\n X_source_selected = get_source_vector(X=X_source, source_name=source_name, return_df=True,\n colList_features=colList_features)\n X_match_sorted = prepare_sorted_match_df(X_source=X_source_selected, X_match=X_match,\n colList_features=colList_features, nv=num_venues)\n X_match_sorted_named, plot_df = preapre_venue_plot_data(X_match_sorted=X_match_sorted, X_meta_mapper=X_meta_mapper,\n colList_features=colList_features)\n\n # plot\n ls_feature_sorted = get_sorted_list_of_features(X_source_selected=X_source_selected,\n colList_features=colList_features)\n plot = plot_venue_match_data(plot_df=X_match_sorted_named, num_match=num_match, num_venues=num_venues,\n colList_features=colList_features, show_plot=show_plot)\n\n return X_match_sorted_named, plot\n\n\ndef plot_venue_match_data(plot_df=None, num_match=-1, num_venues=-1, colList_features=None, show_plot=False):\n\n # drop dup entry of nbhd\n #mac_vc = plot_df[col_grain].value_counts().values[0]\n #if mac_vc > 1:\n # plot_df = plot_df.drop_duplicates(subset=[col_grain])\n # num_match = num_match-1\n\n sns.set_style('darkgrid')\n fig, axis = plt.subplots(num_match, 1, figsize=(1.5 * num_venues, num_match * 4))\n fig.subplots_adjust(hspace=0.5, wspace=0.5)\n\n list_features = [i for i in plot_df.columns if i in colList_features]\n list_match = plot_df[col_grain].values\n ylim = plot_df[list_features].max().max()\n\n for i_main in range(num_match):\n data_temp = plot_df.loc[i_main][list_features]\n ls_venues = data_temp.index.values\n ls_count = data_temp.values\n ax = axis[i_main]\n sns.barplot(x=ls_venues, y=ls_count, ax=ax, palette=sns.color_palette(list_palette_0[i_main], num_venues, 0.9))\n\n ax.set_ylabel(ylabel='', fontsize=12, color='red')\n ax.yaxis.set_major_locator(MaxNLocator(integer=True))\n\n ylim = max(ls_count)\n ax.set_ylim(0, ylim + 1)\n\n ax.set_xticklabels(ls_venues, rotation=15, horizontalalignment='right', size=12)\n # ax.text((len(ls_venues)-1)//2.5, ylim, list_match[i_main], fontsize=15)\n # ax.text(int(len(ls_venues)*0.4), ylim-1, list_match[i_main], fontsize=15)\n\n if i_main == 0:\n # ax.text(int(len(ls_venues)*0.4), ylim-1, 'Source City\\n{}'.format(list_match[i_main]), fontsize=12)\n ax.set_title('{}\\n(Source City)'.format(list_match[i_main]), loc='center', pad=-10,\n fontdict={'verticalalignment': 'center_baseline'})\n else:\n # ax.text(int(len(ls_venues)*0.4), ylim-1, list_match[i_main], fontsize=12)\n ax.set_title('{}'.format(list_match[i_main]), loc='center', pad=-10,\n fontdict={'verticalalignment': 'center_baseline'})\n\n main_label = 'Comparison of suggested Neighborhood with the base location\\nnum neighborhood : {}\\nnum top venues : {}'.format(\n num_match, num_venues)\n fig.text(0.5, 1.02, main_label, ha='center', va='center', rotation='horizontal', size=18, color='blue')\n\n plt.tight_layout()\n\n if show_plot:\n plt.show()\n\n return fig\n\n\ndef generate_ui_df(X_match_sorted_named=None):\n df = X_match_sorted_named.drop(columns=['index']).T.copy()\n df.columns = df.iloc[0]\n df = df.iloc[1:]\n return df\n\n\ndef plot_nbhd_on_map(plot_df=None, marker_size=20, map_zoom=11, show_plot=False, col_text=col_grain):\n ls_symbol = ['triangle' for i in range(len(plot_df))]\n\n fig = go.Figure(go.Scattermapbox(\n mode=\"markers+text\",\n lon=plot_df[col_longitude], lat=plot_df[col_latitude],\n marker={'size': marker_size, 'symbol': ls_symbol},\n hovertext=plot_df[col_text],\n text=plot_df[col_text], textposition=\"bottom right\"))\n\n fig.update_layout(\n mapbox={\n 'accesstoken': MAPBOX_TOKEN,\n 'center': go.layout.mapbox.Center(\n lat=plot_df[col_latitude].mean(),\n lon=plot_df[col_longitude].mean()\n ),\n 'style': \"outdoors\", 'zoom': map_zoom},\n showlegend=False)\n\n # fig.update_layout(mapbox_style=\"open-street-map\")\n fig.update_layout(margin={\"r\": 0, \"t\": 0, \"l\": 0, \"b\": 0})\n\n if show_plot:\n fig.show()\n return fig\n"
]
| [
[
"pandas.concat",
"pandas.merge",
"matplotlib.pyplot.tight_layout",
"pandas.read_csv",
"numpy.nonzero",
"numpy.take",
"matplotlib.pyplot.subplots",
"scipy.spatial.distance.euclidean",
"matplotlib.ticker.MaxNLocator",
"numpy.array",
"matplotlib.pyplot.show",
"pandas.melt",
"pandas.pivot_table"
]
]
|
hbendekgey/FairSurrogates | [
"d81747817c866a67a6a324f8aea2f02cc3ece5c9"
]
| [
"CelebA/sim_dp.py"
]
| [
"import sys\nimport torch\nimport torchvision\nimport torch.optim as optim\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom tqdm import tqdm, trange\nfrom time import sleep\nfrom PIL import Image\nfrom torchvision import transforms\n\nif len(sys.argv) != 4:\n print(\"Usage: python simulate.py lambda_fair formulation GPU_index\")\n exit(1)\nlam_fair = float(sys.argv[1])\nform = sys.argv[2]\ngpui = int(sys.argv[3])\n\npreprocess = transforms.Compose([\n transforms.Resize(256),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),\n])\n\ntrainset = torchvision.datasets.CelebA(root='./data', split='train', target_type='attr', transform=preprocess)\nvalidset = torchvision.datasets.CelebA(root='./data', split='valid', target_type='attr', transform=preprocess)\ntestset = torchvision.datasets.CelebA(root='./data', split='test', target_type='attr', transform=preprocess)\n\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=32, shuffle=True, num_workers=2)\nvalidloader = torch.utils.data.DataLoader(validset, batch_size=32, shuffle=True, num_workers=2)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=32, shuffle=False, num_workers=2)\n\nattrs = '5_o_Clock_Shadow Arched_Eyebrows Attractive Bags_Under_Eyes Bald Bangs Big_Lips Big_Nose Black_Hair Blond_Hair Blurry Brown_Hair Bushy_Eyebrows Chubby Double_Chin Eyeglasses Goatee Gray_Hair Heavy_Makeup High_Cheekbones Male Mouth_Slightly_Open Mustache Narrow_Eyes No_Beard Oval_Face Pale_Skin Pointy_Nose Receding_Hairline Rosy_Cheeks Sideburns Smiling Straight_Hair Wavy_Hair Wearing_Earrings Wearing_Hat Wearing_Lipstick Wearing_Necklace Wearing_Necktie Young '.split()\n\nmodel = torch.hub.load('pytorch/vision:v0.6.0', 'wide_resnet50_2', pretrained=False) #pretrained = True if you want\nmodel.fc = nn.Linear(2048, 1, bias=True)\nmodel.load_state_dict(torch.load(\"baseline\"))\n\nti = attrs.index(\"Smiling\")\nsi = attrs.index(\"Male\")\n\n(Pmale, Pfem) = (trainset.attr[:,si].float().mean(), 1 - trainset.attr[:,si].float().mean())\n\nploss = nn.BCEWithLogitsLoss()\nif form == \"logistic\":\n def floss(outputs, sens_attr):\n return -lam_fair/outputs.shape[0] * (F.logsigmoid(outputs[sens_attr]).sum()/Pmale + F.logsigmoid(-outputs[~sens_attr]).sum()/Pfem)\nelif form == \"hinge\":\n baseline = torch.tensor(0.).to(device)\n def floss(outputs, sens_attr):\n return lam_fair/outputs.shape[0] * (torch.max(baseline,1-outputs[sens_attr]).sum()/Pmale + torch.max(baseline,1+outputs[~sens_attr]).sum()/Pfem)\nelse:\n def floss(outputs, sens_attr):\n return lam_fair/outputs.shape[0] * (-outputs[sens_attr].sum()/Pmale + outputs[~sens_attr].sum()/Pfem)\n\noptimizer = optim.Adam(model.parameters(), lr=0.01)\nscheduler = optim.lr_scheduler.ReduceLROnPlateau(optimizer, 'min', verbose=True)\n\ndevice = torch.device(\"cuda:\" + str(gpui) if torch.cuda.is_available() else \"cpu\")\nmodel.to(device)\ntorch.cuda.empty_cache()\n\ndef calc_loss(data):\n inputs, labels = data\n inputs, labels, sens_attr = inputs.to(device), labels[:,ti].float().to(device), labels[:,si].bool().to(device)\n optimizer.zero_grad()\n outputs = model(inputs).reshape(-1)\n loss = ploss(outputs, labels) + floss(outputs, sens_attr)\n loss.backward()\n preds = (outputs >= 0).float()\n unfairness = torch.tensor([preds[ sens_attr].sum(), preds[ sens_attr].shape[0],\n preds[~sens_attr].sum(), preds[~sens_attr].shape[0]]) #msmiling, m, fsmiling, f\n return ((labels == preds).float().mean(), loss, unfairness)\n\n\nprint_every = 200\nvalid_batches = 32\nfor epoch in range(3): # loop over the dataset multiple times\n\n running_loss = 0.0\n running_acc = 0.0\n running_unfair = 0.0\n for i, data in tqdm(enumerate(trainloader, 0)):\n # get the inputs; data is a list of [inputs, labels]\n (acc, loss, unfair) = calc_loss(data)\n optimizer.step()\n\n # print statistics\n running_loss += loss.item()\n running_acc += acc.item()\n running_unfair += unfair\n\n if i % print_every == (print_every - 1): # print every 200 mini-batches\n print('[%d, %5d]' % (epoch + 1, i + 1))\n valid_loss = 0.0\n valid_acc = 0.0\n valid_iter = iter(validloader)\n for vi in range(valid_batches):\n (new_valid_acc, new_valid_loss, _) = calc_loss(next(valid_iter))\n valid_loss += new_valid_loss\n valid_acc += new_valid_acc.item()\n scheduler.step(valid_loss)\n print('Training Accuracy: %.3f, Validation Accuracy: %.3f, Unfairness: %.3f' % (running_acc / print_every,\n valid_acc/valid_batches,\n running_unfair[2]/running_unfair[3] -\n running_unfair[0]/running_unfair[1]))\n print('Training Loss: %.3f, Validation Loss: %.3f' % (running_loss / print_every, valid_loss/valid_batches))\n print('')\n running_loss = 0.0\n running_acc = 0.0\n running_unfair = 0.0\n sleep(1)\n\nprint('Finished Training')\n\ntorch.save(model.state_dict(), form + str(lam_fair))\n"
]
| [
[
"torch.optim.lr_scheduler.ReduceLROnPlateau",
"torch.max",
"torch.load",
"torch.utils.data.DataLoader",
"torch.cuda.empty_cache",
"torch.nn.functional.logsigmoid",
"torch.tensor",
"torch.nn.Linear",
"torch.nn.BCEWithLogitsLoss",
"torch.cuda.is_available",
"torch.hub.load"
]
]
|
Chris2L/gnss-ins-sim | [
"2d2ba0fac193afd10b4d7758cc85d14457f17001"
]
| [
"demo_free_integration_long_time.py"
]
| [
"# -*- coding: utf-8 -*-\n# Filename: demo_free_integration_long_time.py\n\n\"\"\"\nA simple free integration (strapdown inertial navigation) demo of Sim.\nCreated on 2018-01-23\n@author: dongxiaoguang\n\"\"\"\n\nimport os\nimport math\nimport numpy as np\nfrom gnss_ins_sim.sim import imu_model\nfrom gnss_ins_sim.sim import ins_sim\n\n# globals\nD2R = math.pi/180\n\nmotion_def_path = os.path.abspath('.//demo_motion_def_files//')\nfs = 200.0 # IMU sample frequency\n\ndef test_free_integration():\n '''\n test Sim\n '''\n #### IMU model, typical for IMU381\n imu_err = {'gyro_b': np.array([0.0, 0.0, 0.0]),\n 'gyro_arw': np.array([0.25, 0.25, 0.25]) * 0.0,\n 'gyro_b_stability': np.array([3.5, 3.5, 3.5]) * 0.0,\n 'gyro_b_corr': np.array([100.0, 100.0, 100.0]),\n 'accel_b': np.array([0.0e-3, 0.0e-3, 0.0e-3]),\n 'accel_vrw': np.array([0.03119, 0.03009, 0.04779]) * 0.0,\n 'accel_b_stability': np.array([4.29e-5, 5.72e-5, 8.02e-5]) * 0.0,\n 'accel_b_corr': np.array([200.0, 200.0, 200.0]),\n 'mag_std': np.array([0.2, 0.2, 0.2]) * 0.0\n }\n # do not generate GPS and magnetometer data\n imu = imu_model.IMU(accuracy=imu_err, axis=6, gps=False)\n\n #### Algorithm\n # Free integration in a virtual inertial frame\n from demo_algorithms import free_integration\n '''\n Free integration requires initial states (position, velocity and attitude).\n You should provide theses values when you create the algorithm object.\n '''\n ini_pos_vel_att = np.genfromtxt(motion_def_path+\"//motion_def-long_drive.csv\",\\\n delimiter=',', skip_header=1, max_rows=1)\n ini_pos_vel_att[0] = ini_pos_vel_att[0] * D2R\n ini_pos_vel_att[1] = ini_pos_vel_att[1] * D2R\n ini_pos_vel_att[6:9] = ini_pos_vel_att[6:9] * D2R\n # add initial states error if needed\n ini_vel_err = np.array([0.0, 0.0, 0.0]) # initial velocity error in the body frame, m/s\n ini_att_err = np.array([0.0, 0.0, 0.0]) # initial Euler angles error, deg\n ini_pos_vel_att[3:6] += ini_vel_err\n ini_pos_vel_att[6:9] += ini_att_err * D2R\n # create the algorith object\n algo = free_integration.FreeIntegration(ini_pos_vel_att)\n\n #### start simulation\n sim = ins_sim.Sim([fs, 0.0, 0.0],\n motion_def_path+\"//motion_def-long_drive.csv\",\n ref_frame=0,\n imu=imu,\n mode=None,\n env=None,\n algorithm=algo)\n # run the simulation once\n sim.run(1)\n # generate simulation results, summary\n # do not save data, generate .kml file\n sim.results('', err_stats_start=-1, gen_kml=True)\n\nif __name__ == '__main__':\n test_free_integration()\n"
]
| [
[
"numpy.array",
"numpy.genfromtxt"
]
]
|
dadi-vardhan/Assurance-cases-for-LEC | [
"cbd6e8b296c624fdd734ac4ee0fbf4308a114026"
]
| [
"robo_cup_experiments/test.py"
]
| [
"from cv2 import sepFilter2D\nimport torch\nimport os\nimport numpy as np\nimport pytorch_lightning as pl\nfrom datamodule import MNISTDataModule\nfrom model import MnistModel\nfrom torchvision import datasets\nfrom torchvision.transforms import ToTensor\nfrom utils import get_device\nfrom torchmetrics.functional.classification.accuracy import accuracy\nfrom torch.utils.data import DataLoader\nfrom eval_metrics import eval_metrics\n\n\ndm = MNISTDataModule(os.getcwd())\n\nmodel = MnistModel.load_from_checkpoint(\n checkpoint_path=\"/home/dadi_vardhan/RandD/Assurance-cases-for-LEC/MNIST-lightning/Untitled/AC-71/checkpoints/epoch=18-step=1025.ckpt\").eval()\n\ndevice =get_device()\n# init trainer with whatever options\ntrainer = pl.Trainer(checkpoint_callback=True,\n gpus=1)\n\n# test (pass in the model)\n# trainer.fit(model)\n# trainer.test(model, dm)\n\n\n# train_data = datasets.MNIST(root = 'data',train = True,transform = ToTensor(),download = True)\n# test_data = datasets.MNIST(\n# root = 'data', \n# train = False, \n# download=True,)\n# preds = []\n# targets =[]\n# nums = np.random.randint(0,500,10)\n# for i in nums: \n# img = test_data.data[i].reshape(1,28,28)\n# img = img.unsqueeze_(0)\n# print(img)\n# target = test_data.targets[i]\n# targets.append(target)\n# logits = model(img)\n# pred = torch.argmax(logits)\n# preds.append(pred)\n# print(f\"pred : {pred} and label : {target}\")\n\n# acc = accuracy(preds, targets)\n# print(\"acc: \",acc)\n\n#testing\nimport numpy as np\nclasses = ('Zero', 'One', 'Two', 'Three', 'Four',\n 'Five', 'Six', 'Seven', 'Eight', 'Nine')\n\nmodel.freeze()\ntest_loader = DataLoader(datasets.MNIST(os.getcwd(), train=False, download=True, transform=ToTensor()), batch_size=1028, shuffle=True)\n\ny_true, y_pred = [],[]\nfor i, (x, y) in enumerate(test_loader):\n y_hat = model.forward(x).argmax(axis=1).cpu().detach().numpy()\n y = y.cpu().detach().numpy()\n\n y_true.append(y)\n y_pred.append(y_hat)\n\n if i == len(test_loader):\n break\ny_true = np.hstack(y_true)\ny_pred = np.hstack(y_pred)\n\nem_mon = eval_metrics(y_true, y_pred,classes=classes)\nprint(f\"accracy-mon:{em_mon.accuracy()}\")\nprint(em_mon.classify_report())\n\n\n\n"
]
| [
[
"numpy.hstack"
]
]
|
zenseact/development_kit | [
"c52a8c8a625b0acd60c158bb2330cf35ee7dde11"
]
| [
"plot_gps_on_image.py"
]
| [
"\"\"\"Module to perform OxTS extraction and visualize GPS track projection on image plane.\"\"\"\nfrom datetime import datetime\n\nimport cv2\nimport numpy as np\nimport pandas as pd\nimport pyproj\nfrom pytz import utc\n\nfrom constants import (\n DEFAULT_COL_VALUES,\n DISTORTION,\n ECEF_XYZ,\n EXTRINSICS,\n INTRINSICS,\n OXTS_COLS,\n OXTS_OPTIONAL_COLS,\n)\n\n\n# pylint: disable=C0103\nECEF = pyproj.Proj(proj=\"geocent\", ellps=\"WGS84\", datum=\"WGS84\")\nLLA = pyproj.Proj(proj=\"latlong\", ellps=\"WGS84\", datum=\"WGS84\")\nGPS_EPOCH = datetime(1980, 1, 6, tzinfo=utc)\nPATH_POINTS = np.arange(5, 201, 5)\n\n\ndef move(T, P, *, vec_dim=None):\n \"\"\"Apply transform to points.\n\n Notes\n -----\n If the dimensionality of `P` is known, and you have a very large batch\n of `T` or `P`, consider passing the `vec_dim` argument to the function.\n This will switch to a codepath that depending on the situation can be\n up to 1000x faster to compute.\n\n \"\"\"\n R, t = R_t(T)\n return matvec(R, P, nrow=vec_dim) + t\n\n\ndef T_from_R_t(R, t):\n \"\"\"Combine rotation matrix and translation vec into a transform.\"\"\"\n t = t[..., None]\n o = np.zeros_like(R[..., -1:, :])\n i = np.ones_like(t[..., -1:, :])\n return np.concatenate(\n [np.concatenate([R, t], axis=-1), np.concatenate([o, i], axis=-1)], axis=-2\n )\n\n\ndef T_inv(T, *, vec_dim=None):\n \"\"\"Compute inverse transform.\n\n Notes\n -----\n This computes an inverse of a transform matrix of form\n\n ```\n T T\n inv |R t| = |R -R t|\n |0 1| |0 1 |\n ```\n\n If the dimension of the problem (for the matrix `T` of shape NxN the\n the dimension is N-1) is known, and you have a very large batch of `T`,\n consider passing the `vec_dim` argument to the function. This will\n switch to a codepath that depending on the situation can be up to 1000x\n faster to compute.\n\n \"\"\"\n R, t = R_t(T)\n return T_from_R_t(mT(R), -matvec(mT(R), t, nrow=vec_dim))\n\n\ndef mT(M):\n \"\"\"Compute matrix transpose in the last two dimensions.\"\"\"\n return np.einsum(\"...ij->...ji\", M)\n\n\ndef Ry(r):\n \"\"\"Construct a rotation matrix around Y-axis.\"\"\"\n return _Ry(np.cos(r), np.sin(r))\n\n\ndef Rz(r):\n \"\"\"Construct a rotation matrix around Z-axis.\"\"\"\n return _Rz(np.cos(r), np.sin(r))\n\n\ndef Rx(r):\n \"\"\"Construct a rotation matrix around X-axis.\"\"\"\n return _Rx(np.cos(r), np.sin(r))\n\n\ndef R_t(T):\n \"\"\"Decompose a transform into rotation matrix and translation vec.\"\"\"\n return T[..., :-1, :-1], T[..., :-1, -1]\n\n\ndef _Rx(c, s):\n \"\"\"Construct a rotation matrix around X-axis given cos and sin.\n\n The `c` and `s` MUST satisfy c^2 + s^2 = 1 and have the same shape.\n\n See https://en.wikipedia.org/wiki/Rotation_matrix#Basic_rotations.\n\n \"\"\"\n o = np.zeros_like(c)\n i = np.ones_like(o)\n return _tailstack2([[i, o, o], [o, c, -s], [o, s, c]])\n\n\ndef _Ry(c, s):\n o = np.zeros_like(c)\n i = np.ones_like(o)\n return _tailstack2([[c, o, s], [o, i, o], [-s, o, c]])\n\n\ndef _Rz(c, s):\n o = np.zeros_like(c)\n i = np.ones_like(o)\n return _tailstack2([[c, -s, o], [s, c, o], [o, o, i]])\n\n\ndef rotation_matrix(roll, pitch, yaw):\n \"\"\"Compute extrinsic x-y-z (intrinsic z-y'-x'') rotation matrix.\n\n This works e.g. for converting ENU/NED to ISO/OXTS vehicle frame\n given roll, pitch and yaw/heading angles.\n\n See https://support.oxts.com/hc/en-us/articles/\n 115002859149-OxTS-Reference-Frames-and-ISO8855-Reference-Frames\n for more context.\n\n WARNING: If you are not sure this function works for your use case,\n consider composing rotations yourself using functions `Rx`, `Ry` and\n `Rz`.\n\n \"\"\"\n return Rz(yaw) @ Ry(pitch) @ Rx(roll)\n\n\ndef _tailstack2(nxm):\n \"\"\"Stack a list of lists of tensors in the tail dimensions.\n\n Take list of length N of lists of length M of arrays of shape (...),\n and return an array of shape (..., N, M). This does not broadcast,\n so the shape (...) MUST be the same for all arrays.\n\n \"\"\"\n # pylint: disable=unnecessary-comprehension\n return np.stack([np.stack([m for m in n], axis=-1) for n in nxm], axis=-2)\n\n\ndef to_unit_norm(a):\n \"\"\"Normalize (batch of) vectors to unit norm in the last dimension.\"\"\"\n return a / np.linalg.norm(a, axis=-1, keepdims=True)\n\n\ndef to_homogenous(a):\n \"\"\"Append ones to the last dimension of `a`.\"\"\"\n return np.concatenate([a, np.ones_like(a[..., 0:1])], axis=-1)\n\n\ndef matvec(A, b, *, nrow=None):\n \"\"\"Multiply vector by matrix.\n\n Notes\n -----\n If you have very large batches of small-dimensional\n matrices / vectors, you should consider passing the\n number of rows in the matrix with the `nrow` argument,\n as this will switch to a codepath that depending on\n the situation can be up to 1000x faster to compute.\n\n \"\"\"\n if nrow is not None:\n return np.stack([inner(A[..., i, :], b) for i in range(nrow)], axis=-1)\n return (A @ b[..., None])[..., 0]\n\n\ndef inner(a, b):\n \"\"\"Compute broadcastable inner product in the last dimension.\"\"\"\n return np.sum(a * b, axis=-1)\n\n\ndef kannala_project(P, K, dist):\n \"\"\"Project 3D -> pixel coordinates under the Kannala camera model.\n\n Parameters\n ----------\n P: tensor of shape `(..., 3)`\n 3D coordinates\n K: tensor of shape `(..., 3, 3)`\n Camera matrix\n dist: tensor of shape `(..., 4)`\n Distortion coefficients\n\n Returns\n -------\n p: tensor of shape `(..., 2)`\n Projected pixel coordinates\n\n \"\"\"\n xy = P[..., :2]\n radius = np.linalg.norm(xy, axis=-1, keepdims=True)\n theta = np.arctan2(radius, P[..., 2:3])\n dist_angle = theta * (\n 1\n + dist[..., 0:1] * theta ** 2\n + dist[..., 1:2] * theta ** 4\n + dist[..., 2:3] * theta ** 6\n + dist[..., 3:4] * theta ** 8\n )\n\n uv_distorted = dist_angle * np.where(radius != 0, xy / radius, np.zeros_like(xy))\n uv_hom = to_homogenous(uv_distorted)\n\n return matvec(K[..., :2, :], uv_hom, nrow=2)\n\n\ndef convert_h5_to_pandas(oxts_h5):\n \"\"\"Convert HDF5 OXTS output into a Pandas DataFrame.\"\"\"\n\n def column_from_samples(oxts_samples, col):\n return [\n sample[col]\n if col not in OXTS_OPTIONAL_COLS\n else (sample[col] if col in sample.dtype.names else DEFAULT_COL_VALUES[col])\n for sample in oxts_samples\n ]\n\n oxts_dict = {col: column_from_samples(oxts_h5, col) for col in OXTS_COLS + OXTS_OPTIONAL_COLS}\n oxts_dataframe = pd.DataFrame.from_dict(oxts_dict).set_index(\"timestamp\")\n return oxts_dataframe\n\n\ndef convert_lla_to_ecef(lat_deg, lon_deg, alt):\n \"\"\"Convert LLA to ECEF and return separately X, Y, Z coords.\"\"\"\n ecef_x, ecef_y, ecef_z = pyproj.transform(LLA, ECEF, lon_deg, lat_deg, alt, radians=False)\n return ecef_x, ecef_y, ecef_z\n\n\ndef preprocess_oxts(oxts):\n \"\"\"Convert the raw oxts frame into convenient format.\n\n Make altitude ellipsoidal.\n Pre-compute ECEF coordinates for the oxts frame.\n Pre-compute traveled distance.\n Fix pitch.\n Compute UTC timestamps from GPS time.\n\n \"\"\"\n # Undulation is the difference between the WGS84 elipsoid model\n # of the earth and the altitude on the earth surface. At the sea\n # level, undulation values are negligible.\n # This value is generated by the oxts reciever using the\n # altitude table EGM96\n alt_ellips = oxts.posAlt.values + oxts.undulation.values\n ecef_x, ecef_y, ecef_z = convert_lla_to_ecef(oxts.posLat.values, oxts.posLon.values, alt_ellips)\n ecef = np.stack([ecef_x, ecef_y, ecef_z], axis=-1)\n traveled = np.cumsum(np.linalg.norm(ecef[1:] - ecef[:-1], axis=-1))\n traveled = np.concatenate([[0], traveled])\n\n def misalignment_to_float(field):\n value, _, _, units = field\n if units == b\"Radians\":\n return np.degrees(value)\n return np.array(value)\n\n # pylint: disable=invalid-name\n def apply_misalignment_to_column(o, column):\n try:\n return o[column] + o[f\"{column}Missalignment\"].map(misalignment_to_float)\n except KeyError: # This happens for newly preprocessed drives\n return o[column]\n\n return (\n oxts.reset_index()\n .rename(columns={\"timestamp\": \"time_gps\"})\n .assign(\n posAlt=alt_ellips,\n ecef_x=ecef_x,\n ecef_y=ecef_y,\n ecef_z=ecef_z,\n traveled=traveled,\n pitch=lambda o: apply_misalignment_to_column(o, \"pitch\"),\n heading=lambda o: apply_misalignment_to_column(o, \"heading\"),\n time_utc=lambda o: GPS_EPOCH + pd.to_timedelta(o.time_gps + o.leapSeconds, unit=\"s\"),\n )\n .drop(columns=[\"undulation\"])\n .drop(columns=[\"pitchMissalignment\", \"headingMissalignment\"], errors=\"ignore\")\n .set_index(\"time_utc\")\n )\n\n\ndef get_initial_position(oxts, time_utc):\n \"\"\"Interpolate between OXTS points nearest to a UTC time.\"\"\"\n prev_idx = oxts.index.get_loc(time_utc, method=\"ffill\")\n prev_row = oxts.iloc[prev_idx]\n prev_time = oxts.index[prev_idx].timestamp()\n next_row = oxts.iloc[prev_idx + 1]\n next_time = oxts.index[prev_idx + 1].timestamp()\n this_time = time_utc.timestamp()\n alpha = (this_time - next_time) / (prev_time - next_time)\n return (alpha * prev_row + (1 - alpha) * next_row).rename(time_utc)\n\n\ndef _idx_of(iterable, condition=bool):\n return next((i for i, e in enumerate(iterable) if condition(e)), None)\n\n\ndef interpolate_minus_plus_oxts(oxts_minus, oxts_plus, start_traveled):\n \"\"\"Interpolate between closer and further points to get path point estimates.\"\"\"\n traveled_minus = oxts_minus.traveled.values - start_traveled\n traveled_plus = oxts_plus.traveled.values - start_traveled\n alpha = (PATH_POINTS - traveled_plus) / (traveled_minus - traveled_plus)\n new_idx = oxts_minus.index + (1 - alpha) * (oxts_plus.index - oxts_minus.index)\n return alpha[..., None] * oxts_minus.set_index(new_idx) + (\n 1 - alpha[..., None]\n ) * oxts_plus.set_index(new_idx)\n\n\ndef _find_oxts_path_points(oxts, frame_time_utc):\n # Find start of the relevant oxts portion\n oxts_0 = get_initial_position(oxts, frame_time_utc)\n start_idx = oxts.index.get_loc(frame_time_utc, method=\"ffill\")\n oxts_onward = oxts.iloc[start_idx:]\n\n # Find the end of the relevant oxts portion\n start_traveled = oxts_0.traveled\n # This will throw a ValueError if the travel distance of\n # PATH_POINTS[-1] from the oxts_0 is not reached until the\n # end of the `oxts` frame\n end_idx = _idx_of(oxts_onward.traveled.values, lambda x: x > start_traveled + PATH_POINTS[-1])\n if end_idx is None:\n raise ValueError(\n f\"The OXTS segment ends less than {PATH_POINTS[-1]} m \"\n f\"away from the starting position at {frame_time_utc}.\"\n )\n oxts_segment = oxts_onward.iloc[: end_idx + 1]\n\n # Find path point candidates in the selected oxts portion\n path_points_idxs = np.argmax(\n oxts_segment.traveled.values[..., None] > start_traveled + PATH_POINTS, axis=0\n )\n\n if len(np.unique(path_points_idxs)) < len(PATH_POINTS):\n raise ValueError(\n \"There are some path points missing in the OXTS segment. \"\n \"There is possibly a jump in GPS coordinates.\"\n )\n\n # Combine with previous points of candidates and interpolate\n oxts_path_points = interpolate_minus_plus_oxts(\n oxts_segment.iloc[path_points_idxs - 1], oxts_segment.iloc[path_points_idxs], start_traveled\n )\n\n return oxts_0, oxts_path_points\n\n\ndef ecef_to_enu_rotation(lat_deg, lon_deg):\n \"\"\"Compute rotation matrix from ECEF to ENU at given coordinates.\"\"\"\n sl = np.sin(np.radians(lat_deg))\n cl = np.cos(np.radians(lat_deg))\n sp = np.sin(np.radians(lon_deg))\n cp = np.cos(np.radians(lon_deg))\n\n o = np.zeros_like(sl)\n\n return np.stack(\n [\n np.stack([-sp, cp, o], axis=-1),\n np.stack([-cp * sl, -sp * sl, cl], axis=-1),\n np.stack([cp * cl, sp * cl, sl], axis=-1),\n ],\n axis=-2,\n )\n\n\ndef nwu_to_ref_rotation_from_points(points_x_nwu):\n \"\"\"Compute matrix with heading and pitch by aligning path points.\"\"\"\n\n def c_s_from_a_b(a, b):\n c = inner(a, b)\n s = np.cross(a, b)\n return c, s\n\n heading_nwu = to_unit_norm(points_x_nwu[..., [0, 1]])\n R_head = _Rz(*c_s_from_a_b(heading_nwu, np.array([1, 0])))\n\n points_x_fl = matvec(R_head, points_x_nwu)\n pitch_fl = to_unit_norm(points_x_fl[..., [0, 2]])\n\n _cp, _sp = c_s_from_a_b(pitch_fl, np.array([1, 0]))\n R_pitch = _Ry(_cp, -_sp)\n\n return R_pitch @ R_head\n\n\ndef _find_pitch_at_point(point, oxts):\n def median_direction(a):\n return to_unit_norm(np.median(to_unit_norm(a).reshape(-1, a.shape[-1]), axis=0))\n\n enu_R_ecef = ecef_to_enu_rotation(point.posLat, point.posLon)\n nwu_R_enu = rotation_matrix(0, 0, np.radians(-90))\n nwu_R_ecef = nwu_R_enu @ enu_R_ecef\n\n relevant_oxts = oxts[(oxts.traveled > point.traveled) & (oxts.traveled < point.traveled + 2)]\n nwu = matvec(nwu_R_ecef, relevant_oxts[ECEF_XYZ].values - point[ECEF_XYZ].values)\n ref_R_nwu = nwu_to_ref_rotation_from_points(median_direction(nwu))\n sy = ref_R_nwu[..., 0, 2]\n cy = ref_R_nwu[..., 2, 2]\n return np.degrees(np.arctan2(sy, cy))\n\n\ndef enu_to_ref_frame_rotation(heading_deg, pitch_deg, roll_deg):\n \"\"\"Compute rotation matrix from ISO 8855 ENU to vehicle reference frame.\n\n Refer to https://support.oxts.com/hc/en-us/articles/\n 115002859149-OxTS-Reference-Frames-and-ISO8855-Reference-Frames\n\n \"\"\"\n o = np.zeros_like(heading_deg)\n i = np.ones_like(o)\n A = rotation_matrix(o, o, np.radians(90 * i))\n B = np.eye(3)\n C = rotation_matrix(np.radians(180 * i), o, o)\n HPR = rotation_matrix(np.radians(roll_deg), np.radians(pitch_deg), np.radians(heading_deg))\n\n return mT(C) @ mT(HPR) @ A @ B @ C\n\n\n# pylint: disable=too-many-arguments\ndef ecef_to_ref_frame_transform(lat_deg, lon_deg, heading_deg, pitch_deg, roll_deg, ecef_xyz):\n \"\"\"Compute 4x4 transformation from ECEF to OXTS reference frame.\"\"\"\n enu_R_ecef = ecef_to_enu_rotation(lat_deg, lon_deg)\n ref_R_enu = enu_to_ref_frame_rotation(heading_deg, pitch_deg, roll_deg)\n ref_R_ecef = ref_R_enu @ enu_R_ecef\n ecef_t_ref = ecef_xyz\n ref_t_ecef = -matvec(ref_R_ecef, ecef_t_ref)\n return T_from_R_t(ref_R_ecef, ref_t_ecef)\n\n\n# pylint: disable=invalid-name\ndef odometry_from_oxts(oxts, oxts_0=None):\n \"\"\"Compute 4x4 odometry transform matrices relative to reference position.\n\n If `oxts_0` is a pandas.Series like a row in `oxts` dataframe, then it\n will be used as the reference position. If it's `None`, the first row\n `oxts.iloc[0]` will be used for that.\n\n \"\"\"\n if oxts_0 is None:\n oxts_0 = oxts.iloc[0]\n\n ref_T_ecef = ecef_to_ref_frame_transform(\n oxts.posLat.values,\n oxts.posLon.values,\n oxts.heading.values,\n oxts.pitch.values,\n oxts.roll.values,\n oxts[ECEF_XYZ].values,\n )\n ref0_T_ecef = ecef_to_ref_frame_transform(\n oxts_0.posLat,\n oxts_0.posLon,\n oxts_0.heading,\n oxts_0.pitch,\n oxts_0.roll,\n oxts_0[ECEF_XYZ].values,\n )\n ref0_T_ref = ref0_T_ecef @ T_inv(ref_T_ecef)\n return ref0_T_ref\n\n\ndef generate_odometry(oxts, frame_time_utc, pitch_from_points=False):\n \"\"\"Given oxts log generate car odometry relative to frame.\n\n Args:\n oxts(pd.DataFrame):\n OXTS dataframe (e.g. coming from\n query_oxts -> convert_h5_to_pandas -> preprocess_oxts)\n frame_time_utc(datetime):\n Time for the initial path point. Must have UTC timezone.\n pitch_from_points(bool):\n Indicates whether to compute pitch from neighboring points,\n or to take the values from the oxts frame.\n\n Returns:\n array of shape (len(PATH_POINTS), 4, 4) with odometry relative to\n the frame at PATH_POINTS travel distances from the frame.\n\n \"\"\"\n oxts_0, oxts_path_points = _find_oxts_path_points(oxts, frame_time_utc)\n\n if pitch_from_points:\n oxts_0 = oxts_0.copy() # in case someone needs the original\n oxts_0.pitch = _find_pitch_at_point(oxts_0, oxts)\n oxts_path_points = oxts_path_points.assign(\n pitch=[_find_pitch_at_point(point, oxts) for _, point in oxts_path_points.iterrows()]\n )\n\n # Compute odometry relative to oxts_0\n odometry = odometry_from_oxts(oxts_path_points, oxts_0)\n return odometry\n\n\ndef get_path_from_oxts(oxts_h5: np.ndarray, frame_time_utc: datetime):\n \"\"\"Demonstrate how to get a HP GT path from only a frame_id.\n\n Args:\n oxts_h5: OxTS data for sequence\n frame_time_utc:\n\n \"\"\"\n oxts_dataframe = convert_h5_to_pandas(oxts_h5)\n preprocessed_oxts = preprocess_oxts(oxts_dataframe)\n odometry = generate_odometry(preprocessed_oxts, frame_time_utc)\n return R_t(odometry)[1]\n\n\ndef draw_line(image, line, color):\n \"\"\"Draw a line in image.\"\"\"\n return cv2.polylines(\n image.copy(), [np.round(line).astype(np.int32)], isClosed=False, color=color, thickness=10\n )\n\n\ndef _get_path_in_cam(path: np.ndarray, calib: dict):\n return kannala_project(\n move(T_inv(calib[EXTRINSICS]), path - [0, 0, 0.3]),\n calib[INTRINSICS],\n calib[DISTORTION],\n )\n\n\ndef visualize_gps_on_image(\n oxts_data: np.ndarray, frame_time: datetime, calib: dict, image: np.ndarray\n):\n \"\"\"Visualize GPS track on image.\"\"\"\n path_3d = get_path_from_oxts(oxts_data, frame_time)\n path_on_image = _get_path_in_cam(path_3d, calib)\n image = draw_line(image, path_on_image, (50, 100, 200))\n return image\n"
]
| [
[
"numpy.radians",
"numpy.einsum",
"numpy.arctan2",
"numpy.concatenate",
"numpy.round",
"numpy.zeros_like",
"numpy.cross",
"numpy.ones_like",
"numpy.unique",
"numpy.arange",
"numpy.eye",
"numpy.stack",
"numpy.sin",
"numpy.argmax",
"pandas.DataFrame.from_dict",
"numpy.array",
"numpy.sum",
"numpy.degrees",
"numpy.linalg.norm",
"numpy.cos",
"pandas.to_timedelta"
]
]
|
iimuz/til | [
"b100438e8ce2f369331b3be215a4b9cdce9ffda5",
"b100438e8ce2f369331b3be215a4b9cdce9ffda5"
]
| [
"machine_learning/tf_autoencoder/src/models/dense_ae.py",
"machine_learning/tf_eager_mnist/model.py"
]
| [
"# third party\nimport tensorflow as tf\nfrom tensorflow.keras import layers\n\n# my packages\nfrom src.data import history\n\n\nclass Autoencoder(tf.keras.Model):\n \"\"\"単純な全結合Autoencoder\n \"\"\"\n\n def __init__(self, input_dim: int) -> None:\n \"\"\"Initialize\n\n Args:\n input_dim (int): 入力次元数\n \"\"\"\n super(Autoencoder, self).__init__()\n dims = 32\n self.encoder = _make_encoder(input_dim, dims)\n self.decoder = _make_decoder(dims, input_dim)\n\n def call(self, inputs: tf.Tensor) -> tf.Tensor:\n code = self.encoder(inputs)\n reconstruct = self.decoder(code)\n return reconstruct\n\n @tf.function\n def train_step(\n self,\n inputs: tf.Tensor,\n loss_obj: tf.keras.losses.Loss,\n optimizer: tf.keras.optimizers.Optimizer,\n batch_history: history.Batch = None,\n ) -> None:\n \"\"\"1バッチに対する学習\n\n Args:\n inputs (tf.Tensor): バッチ\n loss_obj (tf.keras.losses.Loss): 損失計算オブジェクト\n optimizer (tf.keras.optimizers.Optimizer): 最適化関数\n batch_history (history.Batch, optional): 計算履歴出力用. Defaults to None.\n \"\"\"\n with tf.GradientTape() as tape:\n reconstruct = self(inputs)\n loss = loss_obj(inputs, reconstruct)\n gradients = tape.gradient(loss, self.trainable_variables)\n optimizer.apply_gradients(zip(gradients, self.trainable_variables))\n\n if batch_history is not None:\n batch_history.loss(loss)\n\n\ndef reconstruct(model: Autoencoder, inputs: tf.Tensor) -> tf.Tensor:\n \"\"\"入力データをモデルを利用して再構成する\n\n Args:\n model (Autoencoder): 利用するモデル\n inputs (tf.Tensor): 入力データ\n\n Returns:\n tf.Tensor: 再構成結果\n \"\"\"\n return model(inputs)\n\n\ndef _make_decoder(input_dim: int, output_dim: int) -> tf.keras.Model:\n \"\"\"decoderを生成する\n\n Args:\n input_dim (int): 入力次元\n output_dim (int): 出力次元\n\n Returns:\n tf.keras.Model: decoderモデル\n \"\"\"\n model = tf.keras.Sequential([layers.Dense(output_dim, activation=\"relu\")])\n\n return model\n\n\ndef _make_encoder(input_dim: int, output_dim: int) -> tf.keras.Model:\n \"\"\"encoderを生成する\n\n Args:\n input_dim (int): 入力次元\n output_dim (int): 出力次元\n\n Returns:\n tf.keras.Model: decoderモデル\n \"\"\"\n model = tf.keras.Sequential([layers.Dense(output_dim, activation=\"relu\")])\n\n return model\n",
"import tensorflow as tf\n\nfrom logging import getLogger\n\nlogger = getLogger(__name__)\n\n\nclass MNISTModel(tf.keras.Model):\n def __init__(self):\n super(MNISTModel, self).__init__()\n self.flatten = tf.keras.layers.Flatten()\n self.d1 = tf.keras.layers.Dense(128, activation=\"relu\")\n self.d2 = tf.keras.layers.Dense(10, activation=\"softmax\")\n\n def call(self, x):\n x = self.flatten(x)\n x = self.d1(x)\n x = self.d2(x)\n\n return x\n\n\nclass Trainer:\n def __init__(self):\n self.loss = tf.keras.losses.SparseCategoricalCrossentropy()\n self.optimizer = tf.keras.optimizers.Adam()\n self.train_loss = tf.keras.metrics.Mean(name=\"train_loss\")\n self.train_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(\n name=\"train_accuracy\"\n )\n\n @tf.function\n def train_step(self, model, images, labels):\n with tf.GradientTape() as tape:\n predictions = model(images)\n loss_val = self.loss(labels, predictions)\n gradients = tape.gradient(loss_val, model.trainable_variables)\n self.optimizer.apply_gradients(zip(gradients, model.trainable_variables))\n self.train_loss(loss_val)\n self.train_accuracy(labels, predictions)\n\n\nclass Predictor:\n def __init__(self):\n self.loss = tf.keras.losses.SparseCategoricalCrossentropy()\n self.predict_loss = tf.keras.metrics.Mean(name=\"predict_loss\")\n self.predict_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(\n name=\"predict_accuracy\"\n )\n\n @tf.function\n def predict_step(self, model, images, labels):\n predictions = model(images)\n loss_val = self.loss(labels, predictions)\n self.predict_loss(loss_val)\n self.predict_accuracy(labels, predictions)\n\n\nclass Checkpoint:\n def __init__(self, network, optimizer=None):\n args = {\"net\": network}\n if optimizer is not None:\n args[\"optimizer\"] = optimizer\n self.ckpt = tf.train.Checkpoint(**args)\n self.manager = tf.train.CheckpointManager(\n self.ckpt, \"_data/ckpts\", max_to_keep=3\n )\n self.ckpt.restore(self.manager.latest_checkpoint)\n if self.manager.latest_checkpoint:\n logger.info(f\"Restored from {self.manager.latest_checkpoint}\")\n else:\n logger.info(\"Initializing from scratch.\")\n\n def save_counter(self):\n return self.ckpt.save_counter.numpy()\n\n def save(self):\n return self.manager.save()\n"
]
| [
[
"tensorflow.keras.layers.Dense",
"tensorflow.GradientTape"
],
[
"tensorflow.train.CheckpointManager",
"tensorflow.keras.losses.SparseCategoricalCrossentropy",
"tensorflow.keras.layers.Dense",
"tensorflow.train.Checkpoint",
"tensorflow.keras.optimizers.Adam",
"tensorflow.keras.metrics.SparseCategoricalAccuracy",
"tensorflow.keras.layers.Flatten",
"tensorflow.keras.metrics.Mean",
"tensorflow.GradientTape"
]
]
|
dmpelt/pywt | [
"5fc0f769dfdd6d21c2d0b2fa4fa8cdb7e887bb9f"
]
| [
"pywt/_dwt.py"
]
| [
"import numpy as np\n\nfrom ._extensions._pywt import Wavelet, Modes, _check_dtype\nfrom ._extensions._dwt import (dwt_single, dwt_axis, idwt_single, idwt_axis,\n upcoef as _upcoef, downcoef as _downcoef,\n dwt_max_level as _dwt_max_level,\n dwt_coeff_len as _dwt_coeff_len)\n\n__all__ = [\"dwt\", \"idwt\", \"downcoef\", \"upcoef\", \"dwt_max_level\", \"dwt_coeff_len\"]\n\n\ndef dwt_max_level(data_len, filter_len):\n \"\"\"\n dwt_max_level(data_len, filter_len)\n\n Compute the maximum useful level of decomposition.\n\n Parameters\n ----------\n data_len : int\n Input data length.\n filter_len : int\n Wavelet filter length.\n\n Returns\n -------\n max_level : int\n Maximum level.\n\n Examples\n --------\n >>> import pywt\n >>> w = pywt.Wavelet('sym5')\n >>> pywt.dwt_max_level(data_len=1000, filter_len=w.dec_len)\n 6\n >>> pywt.dwt_max_level(1000, w)\n 6\n \"\"\"\n if isinstance(filter_len, Wavelet):\n filter_len = filter_len.dec_len\n\n return _dwt_max_level(data_len, filter_len)\n\n\ndef dwt_coeff_len(data_len, filter_len, mode):\n \"\"\"\n dwt_coeff_len(data_len, filter_len, mode='symmetric')\n\n Returns length of dwt output for given data length, filter length and mode\n\n Parameters\n ----------\n data_len : int\n Data length.\n filter_len : int\n Filter length.\n mode : str, optional (default: 'symmetric')\n Signal extension mode, see Modes\n\n Returns\n -------\n len : int\n Length of dwt output.\n\n Notes\n -----\n For all modes except periodization::\n\n len(cA) == len(cD) == floor((len(data) + wavelet.dec_len - 1) / 2)\n\n for periodization mode (\"per\")::\n\n len(cA) == len(cD) == ceil(len(data) / 2)\n\n \"\"\"\n if isinstance(filter_len, Wavelet):\n filter_len = filter_len.dec_len\n\n return _dwt_coeff_len(data_len, filter_len, Modes.from_object(mode))\n\n\ndef dwt(data, wavelet, mode='symmetric', axis=-1):\n \"\"\"\n dwt(data, wavelet, mode='symmetric', axis=-1)\n\n Single level Discrete Wavelet Transform.\n\n Parameters\n ----------\n data : array_like\n Input signal\n wavelet : Wavelet object or name\n Wavelet to use\n mode : str, optional\n Signal extension mode, see Modes\n axis: int, optional\n Axis over which to compute the DWT. If not given, the\n last axis is used.\n\n\n Returns\n -------\n (cA, cD) : tuple\n Approximation and detail coefficients.\n\n Notes\n -----\n Length of coefficients arrays depends on the selected mode.\n For all modes except periodization:\n\n ``len(cA) == len(cD) == floor((len(data) + wavelet.dec_len - 1) / 2)``\n\n For periodization mode (\"per\"):\n\n ``len(cA) == len(cD) == ceil(len(data) / 2)``\n\n Examples\n --------\n >>> import pywt\n >>> (cA, cD) = pywt.dwt([1, 2, 3, 4, 5, 6], 'db1')\n >>> cA\n array([ 2.12132034, 4.94974747, 7.77817459])\n >>> cD\n array([-0.70710678, -0.70710678, -0.70710678])\n\n \"\"\"\n if np.iscomplexobj(data):\n data = np.asarray(data)\n cA_r, cD_r = dwt(data.real, wavelet, mode)\n cA_i, cD_i = dwt(data.imag, wavelet, mode)\n return (cA_r + 1j*cA_i, cD_r + 1j*cD_i)\n\n # accept array_like input; make a copy to ensure a contiguous array\n dt = _check_dtype(data)\n data = np.array(data, dtype=dt)\n mode = Modes.from_object(mode)\n if not isinstance(wavelet, Wavelet):\n wavelet = Wavelet(wavelet)\n\n if axis < 0:\n axis = axis + data.ndim\n if not 0 <= axis < data.ndim:\n raise ValueError(\"Axis greater than data dimensions\")\n\n if data.ndim == 1:\n cA, cD = dwt_single(data, wavelet, mode)\n # TODO: Check whether this makes a copy\n cA, cD = np.asarray(cA, dt), np.asarray(cD, dt)\n else:\n cA, cD = dwt_axis(data, wavelet, mode, axis=axis)\n\n return (cA, cD)\n\n\ndef idwt(cA, cD, wavelet, mode='symmetric', axis=-1):\n \"\"\"\n idwt(cA, cD, wavelet, mode='symmetric', axis=-1)\n\n Single level Inverse Discrete Wavelet Transform.\n\n Parameters\n ----------\n cA : array_like or None\n Approximation coefficients. If None, will be set to array of zeros\n with same shape as `cD`.\n cD : array_like or None\n Detail coefficients. If None, will be set to array of zeros\n with same shape as `cA`.\n wavelet : Wavelet object or name\n Wavelet to use\n mode : str, optional (default: 'symmetric')\n Signal extension mode, see Modes\n axis: int, optional\n Axis over which to compute the inverse DWT. If not given, the\n last axis is used.\n\n\n Returns\n -------\n rec: array_like\n Single level reconstruction of signal from given coefficients.\n\n \"\"\"\n # TODO: Lots of possible allocations to eliminate (zeros_like, asarray(rec))\n # accept array_like input; make a copy to ensure a contiguous array\n\n if cA is None and cD is None:\n raise ValueError(\"At least one coefficient parameter must be \"\n \"specified.\")\n\n # for complex inputs: compute real and imaginary separately then combine\n if np.iscomplexobj(cA) or np.iscomplexobj(cD):\n if cA is None:\n cD = np.asarray(cD)\n cA = np.zeros_like(cD)\n elif cD is None:\n cA = np.asarray(cA)\n cD = np.zeros_like(cA)\n return (idwt(cA.real, cD.real, wavelet, mode) +\n 1j*idwt(cA.imag, cD.imag, wavelet, mode))\n\n if cA is not None:\n dt = _check_dtype(cA)\n cA = np.array(cA, dtype=dt)\n if cD is not None:\n dt = _check_dtype(cD)\n cD = np.array(cD, dtype=dt)\n\n if cA is not None and cD is not None:\n if cA.dtype != cD.dtype:\n # need to upcast to common type\n cA = cA.astype(np.float64)\n cD = cD.astype(np.float64)\n elif cA is None:\n cA = np.zeros_like(cD)\n elif cD is None:\n cD = np.zeros_like(cA)\n\n # cA and cD should be same dimension by here\n ndim = cA.ndim\n\n mode = Modes.from_object(mode)\n if not isinstance(wavelet, Wavelet):\n wavelet = Wavelet(wavelet)\n\n if axis < 0:\n axis = axis + ndim\n if not 0 <= axis < ndim:\n raise ValueError(\"Axis greater than coefficient dimensions\")\n\n if ndim == 1:\n rec = idwt_single(cA, cD, wavelet, mode)\n else:\n rec = idwt_axis(cA, cD, wavelet, mode, axis=axis)\n\n return rec\n\n\ndef downcoef(part, data, wavelet, mode='symmetric', level=1):\n \"\"\"\n downcoef(part, data, wavelet, mode='symmetric', level=1)\n\n Partial Discrete Wavelet Transform data decomposition.\n\n Similar to `pywt.dwt`, but computes only one set of coefficients.\n Useful when you need only approximation or only details at the given level.\n\n Parameters\n ----------\n part : str\n Coefficients type:\n\n * 'a' - approximations reconstruction is performed\n * 'd' - details reconstruction is performed\n\n data : array_like\n Input signal.\n wavelet : Wavelet object or name\n Wavelet to use\n mode : str, optional\n Signal extension mode, see `Modes`. Default is 'symmetric'.\n level : int, optional\n Decomposition level. Default is 1.\n\n Returns\n -------\n coeffs : ndarray\n 1-D array of coefficients.\n\n See Also\n --------\n upcoef\n\n \"\"\"\n if np.iscomplexobj(data):\n return (downcoef(part, data.real, wavelet, mode, level) +\n 1j*downcoef(part, data.imag, wavelet, mode, level))\n # accept array_like input; make a copy to ensure a contiguous array\n dt = _check_dtype(data)\n data = np.array(data, dtype=dt)\n if part not in 'ad':\n raise ValueError(\"Argument 1 must be 'a' or 'd', not '%s'.\" % part)\n mode = Modes.from_object(mode)\n if not isinstance(wavelet, Wavelet):\n wavelet = Wavelet(wavelet)\n return np.asarray(_downcoef(part == 'a', data, wavelet, mode, level))\n\n\ndef upcoef(part, coeffs, wavelet, level=1, take=0):\n \"\"\"\n upcoef(part, coeffs, wavelet, level=1, take=0)\n\n Direct reconstruction from coefficients.\n\n Parameters\n ----------\n part : str\n Coefficients type:\n * 'a' - approximations reconstruction is performed\n * 'd' - details reconstruction is performed\n coeffs : array_like\n Coefficients array to recontruct\n wavelet : Wavelet object or name\n Wavelet to use\n level : int, optional\n Multilevel reconstruction level. Default is 1.\n take : int, optional\n Take central part of length equal to 'take' from the result.\n Default is 0.\n\n Returns\n -------\n rec : ndarray\n 1-D array with reconstructed data from coefficients.\n\n See Also\n --------\n downcoef\n\n Examples\n --------\n >>> import pywt\n >>> data = [1,2,3,4,5,6]\n >>> (cA, cD) = pywt.dwt(data, 'db2', 'smooth')\n >>> pywt.upcoef('a', cA, 'db2') + pywt.upcoef('d', cD, 'db2')\n array([-0.25 , -0.4330127 , 1. , 2. , 3. ,\n 4. , 5. , 6. , 1.78589838, -1.03108891])\n >>> n = len(data)\n >>> pywt.upcoef('a', cA, 'db2', take=n) + pywt.upcoef('d', cD, 'db2', take=n)\n array([ 1., 2., 3., 4., 5., 6.])\n\n \"\"\"\n if np.iscomplexobj(coeffs):\n return (upcoef(part, coeffs.real, wavelet, level, take) +\n 1j*upcoef(part, coeffs.imag, wavelet, level, take))\n # accept array_like input; make a copy to ensure a contiguous array\n dt = _check_dtype(coeffs)\n coeffs = np.array(coeffs, dtype=dt)\n if not isinstance(wavelet, Wavelet):\n wavelet = Wavelet(wavelet)\n if part not in 'ad':\n raise ValueError(\"Argument 1 must be 'a' or 'd', not '%s'.\" % part)\n return np.asarray(_upcoef(part == 'a', coeffs, wavelet, level, take))\n"
]
| [
[
"numpy.asarray",
"numpy.array",
"numpy.zeros_like",
"numpy.iscomplexobj"
]
]
|
geometrikal/coral_usb_ros | [
"46341a30ec91d887f631353f1e7b26680d75e8d1"
]
| [
"node_scripts/edgetpu_human_pose_estimator.py"
]
| [
"#!/usr/bin/env python\n\n\nimport matplotlib\nmatplotlib.use(\"Agg\") # NOQA\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport os\nimport sys\n\n# OpenCV import for python3.5\nsys.path.remove('/opt/ros/{}/lib/python2.7/dist-packages'.format(os.getenv('ROS_DISTRO'))) # NOQA\nimport cv2 # NOQA\nsys.path.append('/opt/ros/{}/lib/python2.7/dist-packages'.format(os.getenv('ROS_DISTRO'))) # NOQA\n\nfrom chainercv.visualizations import vis_point\nfrom cv_bridge import CvBridge\nimport rospkg\nimport rospy\n\nfrom dynamic_reconfigure.server import Server\nfrom geometry_msgs.msg import Point\nfrom geometry_msgs.msg import Pose\nfrom jsk_recognition_msgs.msg import PeoplePose\nfrom jsk_recognition_msgs.msg import PeoplePoseArray\nfrom jsk_topic_tools import ConnectionBasedTransport\nfrom sensor_msgs.msg import Image\n\nfrom coral_usb.cfg import EdgeTPUHumanPoseEstimatorConfig\nfrom coral_usb import PoseEngine\n\n\nclass EdgeTPUHumanPoseEstimator(ConnectionBasedTransport):\n\n def __init__(self):\n super(EdgeTPUHumanPoseEstimator, self).__init__()\n rospack = rospkg.RosPack()\n pkg_path = rospack.get_path('coral_usb')\n self.bridge = CvBridge()\n self.classifier_name = rospy.get_param(\n '~classifier_name', rospy.get_name())\n model_file = os.path.join(\n pkg_path,\n './python/coral_usb/posenet/models/'\n 'posenet_mobilenet_v1_075_481_641_quant_decoder_edgetpu.tflite')\n model_file = rospy.get_param('~model_file', model_file)\n\n self.engine = PoseEngine(model_file, mirror=False)\n self.resized_H = self.engine.image_height\n self.resized_W = self.engine.image_width\n\n # dynamic reconfigure\n self.srv = Server(\n EdgeTPUHumanPoseEstimatorConfig, self.config_callback)\n\n self.pub_pose = self.advertise(\n '~output/poses', PeoplePoseArray, queue_size=1)\n self.pub_image = self.advertise(\n '~output/image', Image, queue_size=1)\n\n def subscribe(self):\n self.sub_image = rospy.Subscriber(\n '~input', Image, self.image_cb, queue_size=1, buff_size=2**26)\n\n def unsubscribe(self):\n self.sub_image.unregister()\n\n @property\n def visualize(self):\n return self.pub_image.get_num_connections() > 0\n\n def config_callback(self, config, level):\n self.score_thresh = config.score_thresh\n self.joint_score_thresh = config.joint_score_thresh\n return config\n\n def image_cb(self, msg):\n img = self.bridge.imgmsg_to_cv2(msg, desired_encoding='rgb8')\n resized_img = cv2.resize(img, (self.resized_W, self.resized_H))\n H, W, _ = img.shape\n y_scale = self.resized_H / H\n x_scale = self.resized_W / W\n\n poses, _ = self.engine.DetectPosesInImage(resized_img.astype(np.uint8))\n\n poses_msg = PeoplePoseArray()\n poses_msg.header = msg.header\n points = []\n visibles = []\n for pose in poses:\n if pose.score < self.score_thresh:\n continue\n pose_msg = PeoplePose()\n point = []\n visible = []\n for lbl, keypoint in pose.keypoints.items():\n resized_key_y, resized_key_x = keypoint.yx\n key_y = resized_key_y / y_scale\n key_x = resized_key_x / x_scale\n point.append((key_y, key_x))\n if keypoint.score < self.joint_score_thresh:\n visible.append(False)\n continue\n pose_msg.limb_names.append(lbl)\n pose_msg.scores.append(keypoint.score)\n pose_msg.poses.append(\n Pose(position=Point(x=key_x, y=key_y)))\n visible.append(True)\n poses_msg.poses.append(pose_msg)\n points.append(point)\n visibles.append(visible)\n self.pub_pose.publish(poses_msg)\n\n points = np.array(points, dtype=np.int32)\n visibles = np.array(visibles, dtype=np.bool)\n\n if self.visualize:\n fig = plt.figure(\n tight_layout={'pad': 0})\n ax = plt.Axes(fig, [0., 0., 1., 1.])\n ax.axis('off')\n fig.add_axes(ax)\n vis_point(img.transpose((2, 0, 1)), points, visibles, ax=ax)\n fig.canvas.draw()\n w, h = fig.canvas.get_width_height()\n vis_img = np.fromstring(\n fig.canvas.tostring_rgb(), dtype=np.uint8)\n vis_img.shape = (h, w, 3)\n fig.clf()\n plt.close()\n vis_msg = self.bridge.cv2_to_imgmsg(vis_img, 'rgb8')\n # BUG: https://answers.ros.org/question/316362/sensor_msgsimage-generates-float-instead-of-int-with-python3/ # NOQA\n vis_msg.step = int(vis_msg.step)\n vis_msg.header = msg.header\n self.pub_image.publish(vis_msg)\n\n\nif __name__ == '__main__':\n rospy.init_node('edgetpu_human_pose_estimator')\n detector = EdgeTPUHumanPoseEstimator()\n rospy.spin()\n"
]
| [
[
"matplotlib.use",
"matplotlib.pyplot.Axes",
"matplotlib.pyplot.close",
"numpy.array",
"matplotlib.pyplot.figure"
]
]
|
maker-tj/WA-SUPER-BPD | [
"afe8977cb3fb3ba3db2d7f3361e286dd4221fa7d"
]
| [
"vis_flux.py"
]
| [
"import sys\nimport scipy.io as sio\nimport math\nimport numpy as np\nimport cv2\nimport matplotlib\nmatplotlib.use('agg')\nimport pylab as plt\nfrom matplotlib import cm\nimport torch\n\n\ndef label2color(label): # 定义颜色标签\n\n label = label.astype(np.uint16) # astype生成的矩阵是无符号整型\n \n height, width = label.shape\n color3u = np.zeros((height, width, 3), dtype=np.uint8)\n unique_labels = np.unique(label) # 除去重复标签并进行排序之后输出的标签\n\n if unique_labels[-1] >= 2**24: # 标签数组的最后一位大于2**24为溢出\n raise RuntimeError('Error: label overflow!') # 标签溢出\n\n for i in range(len(unique_labels)): # 遍历每一个标签\n \n binary = '{:024b}'.format(unique_labels[i]) # 格式限定符{:b}表示二进制\n # r g b 3*8=24 2**8*3=256*3\n # 切片提取每一个标签里面的rgb,并将二进制转化为十进制\n r = int(binary[::3][::-1], 2) # 每三个值为一个切片(三个值分别为rgb),从每个切片的第1个值开始取,从后往前排列\n g = int(binary[1::3][::-1], 2) # 每三个值为一个切片,从每个切片的第2个值开始取,从后往前排列\n b = int(binary[2::3][::-1], 2) # 每三个值为一个切片,从每个切片的第3个值开始取,从后往前排列\n\n color3u[label == unique_labels[i]] = np.array([r, g, b])\n\n return color3u\n\n\ndef vis_flux(vis_image, pred_flux, gt_flux, gt_mask, image_name, save_dir):\n # .data.cpu().numpy():将tensor转换成numpy的格式\n vis_image = vis_image.data.cpu().numpy()[0, ...] # 可视化图像\n pred_flux = pred_flux.data.cpu().numpy()[0, ...] # 通过model预测的bpd\n\n # 对真实BPD进行归一化保证norm中间图相似\n gt_flux = 0.999999 * gt_flux / (gt_flux.norm(p=2, dim=1) + 1e-9)\n # np.set_printoptions(suppress=True)\n\n gt_flux = gt_flux.data.cpu().numpy()[0, ...] # 真实的bpd\n gt_mask = gt_mask.data.cpu().numpy()[0, ...] # 掩膜\n \n image_name = image_name[0]\n # print(image_name)\n\n norm_pred = np.sqrt(pred_flux[1,:,:]**2 + pred_flux[0,:,:]**2) # 预测L2范数距离\n angle_pred = 180/math.pi*np.arctan2(pred_flux[1,:,:], pred_flux[0,:,:]) # 两个像素点的预测BPD的夹角\n\n # print(torch.from_numpy(np.sqrt(gt_flux[1,:,:]**2 + gt_flux[0,:,:]**2)).norm(p=2, dim=1).shape)\n # print(np.sqrt(gt_flux[1,:,:]**2 + gt_flux[0,:,:]**2).shape)\n norm_gt = np.sqrt(gt_flux[1, :, :] ** 2 + gt_flux[0, :, :] ** 2)\n angle_gt = 180/math.pi*np.arctan2(gt_flux[1,:,:], gt_flux[0,:,:]) # 真实角度\n\n fig = plt.figure(figsize=(10,6))\n\n ax0 = fig.add_subplot(231)\n ax0.imshow(vis_image[:,:,::-1])\n\n ax2 = fig.add_subplot(233)\n ax2.set_title('Angle_gt')\n ax2.set_autoscale_on(True)\n im2 = ax2.imshow(angle_gt, cmap=cm.jet)\n plt.colorbar(im2, shrink=0.5)\n\n ax3 = fig.add_subplot(234)\n color_mask = label2color(gt_mask)\n ax3.imshow(color_mask)\n\n ax4 = fig.add_subplot(235)\n ax4.set_title('Norm_pred')\n ax4.set_autoscale_on(True)\n im4 = ax4.imshow(norm_pred, cmap=cm.jet)\n plt.colorbar(im4,shrink=0.5)\n\n ax1 = fig.add_subplot(232)\n ax1.set_title('Norm_gt')\n ax1.set_autoscale_on(True) # 在绘图命令上应用自动缩放\n im1 = ax1.imshow(norm_gt, cmap=cm.jet)\n # print(norm_gt)\n plt.colorbar(im4, shrink=0.5) # 添加颜色渐变条\n\n ax5 = fig.add_subplot(236)\n ax5.set_title('Angle_pred')\n ax5.set_autoscale_on(True)\n im5 = ax5.imshow(angle_pred, cmap=cm.jet)\n plt.colorbar(im5, shrink=0.5)\n\n # plt.savefig(save_dir + image_name + '.png') # 将图片存入固定路径\n plt.savefig('D:\\\\A-work\\\\our_SuperBPD\\\\images\\\\my_images'+ image_name + '.png')\n plt.close(fig) # 关闭图像窗口\n\n# label2color(np.array([[255,255,255],[255,255,255]]))\n"
]
| [
[
"numpy.sqrt",
"numpy.unique",
"matplotlib.use",
"numpy.arctan2",
"numpy.array",
"numpy.zeros"
]
]
|
Eurus-Holmes/CHABCNet | [
"8d3985c7680981e58751d043880b5b5a818cc1d3"
]
| [
"adet/data/augmentation.py"
]
| [
"import numpy as np\nfrom fvcore.transforms import transform as T\n\nfrom detectron2.data.transforms import RandomCrop, StandardAugInput\nfrom detectron2.structures import BoxMode\n\n\nclass InstanceAugInput(StandardAugInput):\n \"\"\"\n Keep the old behavior of instance-aware augmentation\n \"\"\"\n\n def __init__(self, *args, **kwargs):\n instances = kwargs.pop(\"instances\", None)\n super().__init__(*args, **kwargs)\n if instances is not None:\n self.instances = instances\n\n\ndef gen_crop_transform_with_instance(crop_size, image_size, instances, crop_box=True):\n \"\"\"\n Generate a CropTransform so that the cropping region contains\n the center of the given instance.\n\n Args:\n crop_size (tuple): h, w in pixels\n image_size (tuple): h, w\n instance (dict): an annotation dict of one instance, in Detectron2's\n dataset format.\n \"\"\"\n instance = (np.random.choice(instances),)\n instance = instance[0]\n crop_size = np.asarray(crop_size, dtype=np.int32)\n bbox = BoxMode.convert(instance[\"bbox\"], instance[\"bbox_mode\"], BoxMode.XYXY_ABS)\n center_yx = (bbox[1] + bbox[3]) * 0.5, (bbox[0] + bbox[2]) * 0.5\n # print(image_size[0], center_yx[0], image_size[1], center_yx[1])\n # if not (image_size[0] >= center_yx[0] and image_size[1] >= center_yx[1]):\n # center_yx = (bbox[1] + bbox[3]) * 0.1, (bbox[0] + bbox[2]) * 0.1\n assert (\n image_size[0] >= center_yx[0] and image_size[1] >= center_yx[1]\n ), \"The annotation bounding box is outside of the image!\"\n assert (\n image_size[0] >= crop_size[0] and image_size[1] >= crop_size[1]\n ), \"Crop size is larger than image size!\"\n\n min_yx = np.maximum(np.floor(center_yx).astype(np.int32) - crop_size, 0)\n max_yx = np.maximum(np.asarray(image_size, dtype=np.int32) - crop_size, 0)\n max_yx = np.minimum(max_yx, np.ceil(center_yx).astype(np.int32))\n\n y0 = np.random.randint(min_yx[0], max_yx[0] + 1)\n x0 = np.random.randint(min_yx[1], max_yx[1] + 1)\n\n # if some instance is cropped extend the box\n if not crop_box:\n num_modifications = 0\n modified = True\n\n # convert crop_size to float\n crop_size = crop_size.astype(np.float32)\n while modified:\n modified, x0, y0, crop_size = adjust_crop(x0, y0, crop_size, instances)\n num_modifications += 1\n if num_modifications > 100:\n raise ValueError(\n \"Cannot finished cropping adjustment within 100 tries (#instances {}).\".format(\n len(instances)\n )\n )\n return T.CropTransform(0, 0, image_size[1], image_size[0])\n\n return T.CropTransform(*map(int, (x0, y0, crop_size[1], crop_size[0])))\n\n\ndef adjust_crop(x0, y0, crop_size, instances, eps=1e-3):\n modified = False\n\n x1 = x0 + crop_size[1]\n y1 = y0 + crop_size[0]\n\n for instance in instances:\n bbox = BoxMode.convert(\n instance[\"bbox\"], instance[\"bbox_mode\"], BoxMode.XYXY_ABS\n )\n\n if bbox[0] < x0 - eps and bbox[2] > x0 + eps:\n crop_size[1] += x0 - bbox[0]\n x0 = bbox[0]\n modified = True\n\n if bbox[0] < x1 - eps and bbox[2] > x1 + eps:\n crop_size[1] += bbox[2] - x1\n x1 = bbox[2]\n modified = True\n\n if bbox[1] < y0 - eps and bbox[3] > y0 + eps:\n crop_size[0] += y0 - bbox[1]\n y0 = bbox[1]\n modified = True\n\n if bbox[1] < y1 - eps and bbox[3] > y1 + eps:\n crop_size[0] += bbox[3] - y1\n y1 = bbox[3]\n modified = True\n\n return modified, x0, y0, crop_size\n\n\nclass RandomCropWithInstance(RandomCrop):\n \"\"\" Instance-aware cropping.\n \"\"\"\n\n def __init__(self, crop_type, crop_size, crop_instance=True):\n \"\"\"\n Args:\n crop_instance (bool): if False, extend cropping boxes to avoid cropping instances\n \"\"\"\n super().__init__(crop_type, crop_size)\n self.crop_instance = crop_instance\n self.input_args = (\"image\", \"instances\")\n\n def get_transform(self, img, instances):\n image_size = img.shape[:2]\n crop_size = self.get_crop_size(image_size)\n return gen_crop_transform_with_instance(\n crop_size, image_size, instances, crop_box=self.crop_instance\n )\n"
]
| [
[
"numpy.random.choice",
"numpy.asarray",
"numpy.ceil",
"numpy.floor",
"numpy.random.randint"
]
]
|
anandnet/Bone-Fracture-Detection- | [
"562ce7b12a0a92a937863b82b3769332fa0e5499"
]
| [
"manual/main.py"
]
| [
"import cv2\nimport numpy as np\n\nfrom pre_process import _reshape_img, get_model\n\nimg_name=\"new\"\n\"\"\"\nCurrently, `img_name` will be used to get resized image from `images/resized` folder\nand original image from `images/Fractured Bone` so it expects the same named image file\nto be available in both the folders.\n\nAll names starting with F{n} are available in both the folders. 1<= n <=100\n\"\"\"\n\nmodel_name= \"ridge_model\"\n\n\n\nimg_file= 'images/resized/{}'.format(img_name)\norig_img= 'images/Fractured Bone/{}'.format(img_name)\n\n#for image read\ntry:\n\timg_t=cv2.imread(img_file+\".jpg\",cv2.IMREAD_COLOR)\n\timg=cv2.imread(orig_img+\".jpg\",cv2.IMREAD_COLOR)\n\tshape= img.shape\nexcept (AttributeError,FileNotFoundError):\n\ttry:\n\t\timg_t=cv2.imread(img_file+\".JPG\",cv2.IMREAD_COLOR)\n\t\timg=cv2.imread(orig_img+\".JPG\",cv2.IMREAD_COLOR)\n\t\tshape=img.shape\n\texcept (AttributeError,FileNotFoundError):\n\t\timg_t=cv2.imread(img_file+\".png\",cv2.IMREAD_COLOR)\n\t\timg=cv2.imread(orig_img+\".png\",cv2.IMREAD_COLOR)\n\t\tshape=img.shape\n\n\t#else: raise FileNotFoundError(\"No image file {img_file}.jpg or {img_file}.JPG\".format(img_file=img_file))\n#else:\n#\traise FileNotFoundError(\"No image file {img_file}.jpg or {img_file}.JPG\".format(img_file=img_file))\n\n\n#details of Imge\nprint(\"\\nShape: \",shape)\nprint(\"\\nSize: \",img.size)\nprint(\"\\nDType: \",img.dtype)\n\n#==============Manual edge ditect=====================\ndef segment_img(_img,limit):\n\tfor i in range(0,_img.shape[0]-1):\n\t\tfor j in range(0,_img.shape[1]-1): \n\t\t\tif int(_img[i,j+1])-int(_img[i,j])>=limit:\n\t\t\t\t_img[i,j]=0\n\t\t\telif(int(_img[i,j-1])-int(_img[i,j])>=limit):\n\t\t\t\t_img[i,j]=0\n\t\n\treturn _img\n#======================================================\n\ngray=cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)\n\n#for i in range(0,gray.shape[0]):\n#\tfor j in range(0,gray.shape[1]): \n#\t\tif (int(gray[i,j]))<=100:\n#\t\t\tgray[i,j]=100\n\n#gray=segment_img(gray,15)\ncv2.imshow(\"GrayEdited\",gray)\nmedian = cv2.medianBlur(gray,5)\n\nmodel= get_model(model_name)\npred_thresh= model.predict([_reshape_img(img_t)])\nbool,threshold_img=cv2.threshold(median,pred_thresh,255,cv2.THRESH_BINARY)\n#blur=cv2.GaussianBlur(threshold_img,(7,7),0)\ncv2.imshow(\"threshold\",threshold_img)\n\n\ninitial=[]\nfinal=[]\nline=[]\n#count=[]\n#for i in range(0,256):\n#\tcount.append(0)\n\nfor i in range(0,gray.shape[0]):\n\ttmp_initial=[]\n\ttmp_final=[]\n\tfor j in range(0,gray.shape[1]-1):\n\t\t#count[gray[i,j]]+=1\n\t\tif threshold_img[i,j]==0 and (threshold_img[i,j+1])==255:\n\t\t\ttmp_initial.append((i,j))\n\t\t\t#img[i,j]=[255,0,0]\n\t\tif threshold_img[i,j]==255 and (threshold_img[i,j+1])==0:\n\t\t\ttmp_final.append((i,j))\n\t\t\t#img[i,j]=[255,0,0]\n\t\n\tx= [each for each in zip(tmp_initial,tmp_final)]\n\tx.sort(key= lambda each: each[1][1]-each[0][1])\n\ttry:\n\t\tline.append(x[len(x)-1])\n\texcept IndexError: pass\n\n#print(count)\n\n\nerr= 15\ndanger_points=[]\n\n#store distances\ndist_list=[]\n\nfor i in range(1,len(line)-1):\n\tdist_list.append(line[i][1][1]-line[i][0][1])\n\ttry:\n\t\tprev_= line[i-3]\n\t\tnext_= line[i+3]\n\n\t\tdist_prev= prev_[1][1]-prev_[0][1]\n\t\tdist_next= next_[1][1]-next_[0][1]\n\t\tdiff= abs(dist_next-dist_prev)\n\t\tif diff>err:\n\t\t\t#print(\"Dist: {}\".format(abs(dist_next-dist_prev)))\n\t\t\t#print(line[i])\n\t\t\tdata=(diff, line[i])\n\t\t\t#print(data)\n\t\t\tif len(danger_points):\n\t\t\t\tprev_data=danger_points[len(danger_points)-1]\n\t\t\t\t#print(prev_data)\n\t\t\t\t#print(\"here1....\")\n\t\t\t\tif abs(prev_data[0]-data[0])>2 or data[1][0]-prev_data[1][0]!=1:\n\t\t\t\t\t#print(\"here2....\")\n\t\t\t\t\tprint(data)\n\t\t\t\t\tdanger_points.append(data)\n\t\t\telse:\n\t\t\t\tprint(data)\n\t\t\t\tdanger_points.append(data)\n\texcept Exception as e:\n\t\tprint(e)\n\t\tpass\n\n\t#print(each)\n\tstart,end= line[i]\n\t#raise ZeroDivisionError\n\tmid=int((start[0]+end[0])/2),int((start[1]+end[1])/2)\n\t#img[mid[0],mid[1]]=[0,0,255]\n\nfor i in range(0,len(danger_points)-1,2):\n\ttry:\n\t\tstart_rect=danger_points[i][1][0][::-1]\n\t\tstart_rect=(start_rect[0]-40, start_rect[1]-40)\n \n\t\tend_rect= danger_points[i+1][1][1][::-1]\n\t\tend_rect= (end_rect[0]+40, end_rect[1]+40)\n \n\t\tcv2.rectangle(img,start_rect,end_rect,(0,255,0),2)\n\texcept:\n\t\tprint(\"Pair not found\")\n\n#blur= cv2.GaussianBlur(img,(5,5),0)\n\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfig, (ax1, ax2)= plt.subplots(2,1)\n\nfig2, ax3= plt.subplots(1,1)\n\nx= np.arange(1,gray.shape[0]-1)\ny= dist_list\n\n#print(len(x),len(y))\n\ncv2.calcHist(gray,[0],None,[256],[0,256])\n\ntry:\n\tax1.plot(x,y)\nexcept:\n\tprint(\"Could not plot\")\nimg= np.rot90(img)\nax2.imshow(img)\n\n#count= range(256)\n#ax3.hist(count, 255, weights=count, range=[0,256])\nax3.hist(gray.ravel(),256,[0,256])\n\nplt.show()\n\n\n\n#wait for key pressing\ncv2.waitKey(0)\n\n#Distroy all the cv windows\ncv2.destroyAllWindows()\n"
]
| [
[
"numpy.rot90",
"numpy.arange",
"matplotlib.pyplot.show",
"matplotlib.pyplot.subplots"
]
]
|
JiajShi/VSR-colorization | [
"800f5a3f3cbbaf12936b272b430b5e3f8d07b794"
]
| [
"VSR+colorization.py"
]
| [
"#!/usr/bin/env python\n# coding: utf-8\n\n# # 1.**安装PaddleGAN**\n# \n# PaddleGAN的安装目前支持Clone GitHub和Gitee两种方式.\n\n# In[ ]:\n\n\n# 当前目录在: /home/aistudio/, 该目录即:左边文件和文件夹所在的目录\n# 克隆最新的PaddleGAN仓库到当前目录\n# !git clone https://github.com/PaddlePaddle/PaddleGAN.git\n# github下载慢,从gitee clone:\nget_ipython().system('git clone https://gitee.com/paddlepaddle/PaddleGAN.git')\nget_ipython().run_line_magic('cd', 'PaddleGAN/')\nget_ipython().system('pip install -v -e .')\n\n\n# # 2.**PaddleGAN 中使用的模型介绍**\n# ## 2.1补帧模型 DAIN\n# DAIN的全称是Depth-Aware Video Frame Interpolation,即深度感知视频帧插值,DAIN模型通过探索深度的信息来显式检测遮挡。\n# \n# 在这篇研究中,研究人员提出了一种通过探索深度信息来检测遮挡的方法。\n# 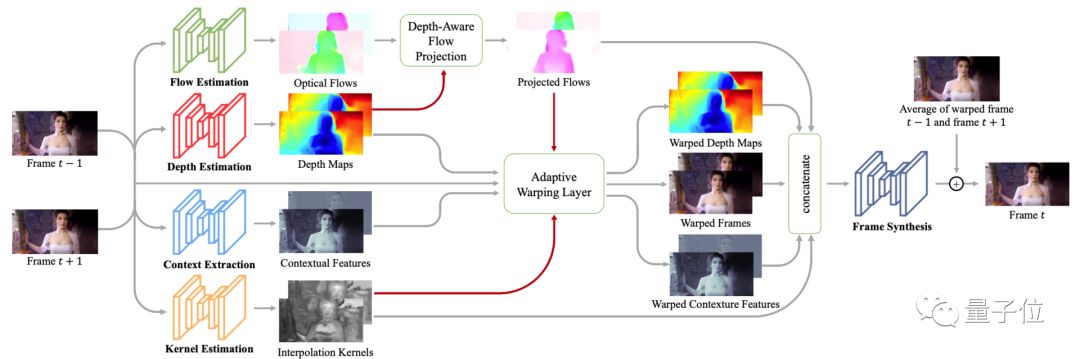\n# 上图是DAIN的体系架构:给定两个时刻的输入帧,先估计光流和深度图,然后使用建议的深度感知流投影层生成中间流。\n# \n# 之后,模型基于光流和局部插值内核对输入帧、深度图和上下文特征进行扭曲,合成输出帧。\n# \n# 这种模型紧凑、高效且完全可微分。定量和定性的结果表明,DAIN在各种数据集上均优于最新的帧插值方法。\n# \n# 简单来说,作者开发了一个深度感知光流投影层来合成中间流,中间流对较远的对象进行采样。此外,学习分层功能以从相邻像素收集上下文信息。\n# \n# 【1】论文地址:[https://arxiv.org/pdf/1904.00830.pdf](http://)\n# \n# *\"Depth-Aware Video Frame Interpolation\"*\n# \n# 【2】项目地址:[https://github.com/baowenbo/DAIN*](http://)\n# \n# \n# \n# ```\n# ppgan.apps.DAINPredictor(\n# output_path='output',\n# weight_path=None,\n# time_step=None,\n# use_gpu=True,\n# remove_duplicates=False)\n# ```\n# #### 参数\n# \n# - `output_path (str,可选的)`: 输出的文件夹路径,默认值:`output`.\n# - `weight_path (None,可选的)`: 载入的权重路径,如果没有设置,则从云端下载默认的权重到本地。默认值:`None`。\n# - `time_step (int)`: 补帧的时间系数,如果设置为0.5,则原先为每秒30帧的视频,补帧后变为每秒60帧。\n# - `remove_duplicates (bool,可选的)`: 是否删除重复帧,默认值:`False`.\n# \n# ## 2.2上色模型 DeOldifyPredictor\n# DeOldify采用自注意力机制的生成对抗网络,生成器是一个U-NET结构的网络。在图像的上色方面有着较好的效果。\n# \n# DeOldify使用了一种名为NoGAN的新型GAN训练方法,用来解决在使用由一个鉴别器和一个生成器组成的正常对抗性网络架构进行训练时出现的主要问题。典型地,GAN训练同时训练鉴别器和生成器,生成器一开始是完全随机的,随着时间的推移,它会欺骗鉴别器,鉴别器试图辨别出图像是生成的还是真实的。NoGan提供了与通常的GAN训练相同的好处,同时花费更少的时间来训练GAN架构(通常计算时间相当长)。相反,它对生成器进行了预先训练,使其利用常规损失函数,变得更强大、更快、更可靠;大部分的训练时间是用更直接、快速和可靠的传统方法分别预训练生成器和鉴别器。**这里的一个关键观点是,那些更 \"传统 \"的方法通常可以得到你所需要的大部分结果,而GAN可以用来缩小现实性方面的差距。**\n# \n# 其步骤如下:\n# \n# *Step1.以传统的方式只用特征损失(feature loss)训练生成器。*\n# \n# *Step2.接下来,从中生成图像,并作为一个基本的二进制分类器训练鉴别器区分这些输出和真实图像。*\n# \n# *Step3.最后,在GAN设置中一起训练生成器和鉴别器。*\n# \n# 【1】暂无论文\n# \n# 【2】项目地址:[https://github.com/jantic/DeOldify](http://)\n# \n# \n# \n# ```\n# ppgan.apps.DeOldifyPredictor(output='output', weight_path=None, render_factor=32)\n# ```\n# #### 参数\n# \n# - `output_path (str,可选的)`: 输出的文件夹路径,默认值:`output`.\n# - `weight_path (None,可选的)`: 载入的权重路径,如果没有设置,则从云端下载默认的权重到本地。默认值:`None`。\n# - `render_factor (int)`: 会将该参数乘以16后作为输入帧的resize的值,如果该值设置为32,\n# 则输入帧会resize到(32 * 16, 32 * 16)的尺寸再输入到网络中。\n# \n# ## 2.3上色模型 DeepRemasterPredictor\n# DeepRemaster 模型基于时空卷积神经网络和自注意力机制。并且能够根据输入的任意数量的参考帧对图片进行上色。\n# \n# \n# ```\n# ppgan.apps.DeepRemasterPredictor(\n# output='output',\n# weight_path=None,\n# colorization=False,\n# reference_dir=None,\n# mindim=360):\n# ```\n# #### 参数\n# \n# - `output_path (str,可选的)`: 输出的文件夹路径,默认值:`output`.\n# - `weight_path (None,可选的)`: 载入的权重路径,如果没有设置,则从云端下载默认的权重到本地。默认值:`None`。\n# - `colorization (bool)`: 是否对输入视频上色,如果选项设置为 `True` ,则参考帧的文件夹路径也必须要设置。默认值:`False`。\n# - `reference_dir (bool)`: 参考帧的文件夹路径。默认值:`None`。\n# - `mindim (bool)`: 输入帧重新resize后的短边的大小。默认值:360。\n# \n# ## 2.4超分辨率模型 RealSRPredictor\n# RealSR模型通过估计各种模糊内核以及实际噪声分布,为现实世界的图像设计一种新颖的真实图片降采样框架。基于该降采样框架,可以获取与真实世界图像共享同一域的低分辨率图像。并且提出了一个旨在提高感知度的真实世界超分辨率模型。对合成噪声数据和真实世界图像进行的大量实验表明,该模型能够有效降低了噪声并提高了视觉质量。\n# \n# > 在CVPR-NTIRE-2020真实图像超分比赛中以明显优势获得双赛道冠军。\n# \n# **算法创新设计**,与已有的超分辨率方法相比,RealSR的创新主要体现在三个方面:\n# \n# 1. RealSR采用了自主设计的新型图片退化方法,通过分析真实图片中的模糊和噪声,模拟真实图片的退化过程。\n# \n# 2. 不需要成对的训练数据,利用无标记的数据即可进行训练。\n# \n# 3. 可以处理低分辨率图像中的模糊噪声问题,得到更加清晰干净的高分辨结果。\n# \n# 【1】论文地址:[https://arxiv.org/pdf/1904.00523.pdf](http://) \n# \n# *\"Toward Real-World Single Image Super-Resolution: A New Benchmark and A New Model\"*\n# \n# 【2】项目地址:[https://github.com/Tencent/Real-SR](http://)\n# \n# \n# \n# ```\n# ppgan.apps.RealSRPredictor(output='output', weight_path=None)\n# ```\n# #### 参数\n# \n# - `output_path (str,可选的)`: 输出的文件夹路径,默认值:`output`.\n# - `weight_path (None,可选的)`: 载入的权重路径,如果没有设置,则从云端下载默认的权重到本地。默认值:`None`。\n# \n# ## 2.5超分辨率模型 EDVRPredictor\n# EDVR模型提出了一个新颖的视频具有增强可变形卷积的还原框架:第一,为了处理大动作而设计的一个金字塔,级联和可变形(PCD)对齐模块,使用可变形卷积以从粗到精的方式在特征级别完成对齐;第二,提出时空注意力机制(TSA)融合模块,在时间和空间上都融合了注意机制,用以增强复原的功能。\n# \n# > 在CVPR 2019 Workshop NTIRE 2019 视频恢复比赛中,来自商汤科技、港中文、南洋理工、深圳先进技术研究院的联合研究团队使用EDVR获得了全部四个赛道的所有冠军!\n# \n# **算法创新设计**:\n# \n# 1. 图像对齐(Alignment)。\n# \n# 视频相邻帧存在一定的抖动,必须先对齐才能进一步处理融合。以往这可以使用光流算法处理,但本文中作者发明了一种新的网络模块PCD对齐模块,使用Deformable卷积进行视频的对齐,整个过程可以端到端训练。\n# \n# 2. 时空信息融合(Fusion)。\n# \n# 挖掘时域(视频前后帧)和空域(同一帧内部)的信息融合。本文中作者发明了一种时空注意力模型进行信息融合。\n# \n# EDVR算法架构:\n# 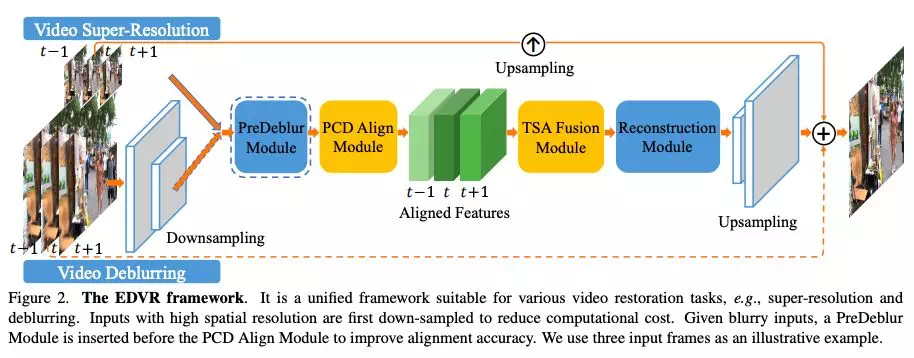\n# \n# 其中PCD 对齐模块,使用金字塔结构级联的Deformable卷积构建,如图: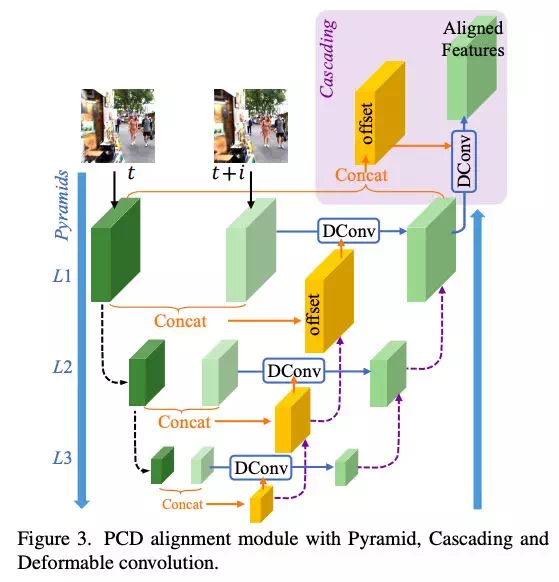\n# \n# 时空注意力融合模型TSA如图: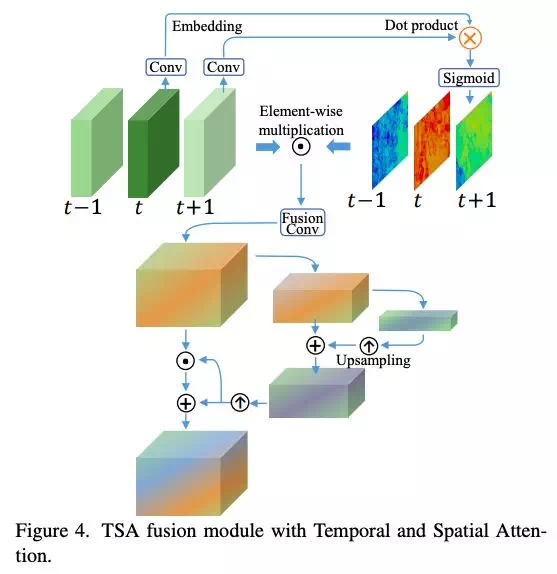\n# \n# \n# 【1】论文地址:[https://arxiv.org/pdf/1905.02716.pdf](http://)\n# \n# *\"EDVR: Video Restoration with Enhanced Deformable Convolutional Networks\"*\n# \n# 【2】项目地址:[https://github.com/xinntao/EDVR](http://)\n# \n# \n# \n# ```\n# ppgan.apps.EDVRPredictor(output='output', weight_path=None)\n# ```\n# #### 参数\n# \n# - `output_path (str,可选的)`: 输出的文件夹路径,默认值:`output`.\n# - `weight_path (None,可选的)`: 载入的权重路径,如果没有设置,则从云端下载默认的权重到本地。默认值:`None`。\n\n# # **3.使用 PaddleGAN 进行视频修复**\n\n# ## 3.1import-导入可视化需要的包\n\n# In[ ]:\n\n\nimport cv2\nimport imageio\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport matplotlib.animation as animation\nfrom IPython.display import HTML\nimport warnings\nwarnings.filterwarnings(\"ignore\")\nimport paddle\nprint(\"本项目Paddle版本号:\"+ paddle.__version__)\n\n\n# ## 3.2定义函数用于展示视频\n\n# In[ ]:\n\n\n# 定义函数用于展示视频\ndef display(driving, fps, size=(8, 6)):\n fig = plt.figure(figsize=size)\n\n ims = []\n for i in range(len(driving)):\n cols = []\n cols.append(driving[i])\n\n im = plt.imshow(np.concatenate(cols, axis=1), animated=True)\n plt.axis('off')\n ims.append([im])\n\n video = animation.ArtistAnimation(fig, ims, interval=1000.0/fps, repeat_delay=1000)\n\n plt.close()\n return video\n\n\n# ## 3.3用于处理的原始视频展示\n\n# In[ ]:\n\n\nvideo_path = '/home/aistudio/Peking_5s.mp4' # 需要处理的视频的路径\nvideo_frames = imageio.mimread(video_path, memtest=False)\ncap = cv2.VideoCapture(video_path) # 打开视频文件\nfps = cap.get(cv2.CAP_PROP_FPS) # 获得视频的原分辨率\nHTML(display(video_frames, fps).to_html5_video()) # Html5 video展示需要处理的原始黑白视频\n\n\n# ## 3.4调用模型,视频处理过程\n\n# In[23]:\n\n\n# 使用插帧(DAIN), 上色(DeOldify), 超分(EDVR, RealSR)模型对该视频进行修复\n\"\"\"\n input参数表示输入的视频路径\n proccess_order 表示使用的模型和顺序(目前支持)\n output表示处理后的视频的存放文件夹\n\"\"\"\nget_ipython().run_line_magic('cd', '/home/aistudio/PaddleGAN/applications/')\nget_ipython().system('python tools/video-enhance.py --input /home/aistudio/Peking_5s.mp4 --process_order DAIN DeOldify EDVR --output output_dir')\n\n\n# ## 3.5处理后的视频展示\n\n# In[24]:\n\n\n# 处理好的视频路径如下, 注:如果视频太大耗时久又可能会报错,最好下载到本地来看。\noutput_video_path = '/home/aistudio/PaddleGAN/applications/output_dir/EDVR/Peking_5s_deoldify_out_edvr_out.mp4'\n# 加载过长视频会造成内存溢出,可以在网页上展示处理后的19秒的视频\n# output_video_path = '/home/aistudio/moderntimes_output19.mp4'\nvideo_frames = imageio.mimread(output_video_path, memtest=False)\ncap = cv2.VideoCapture(output_video_path) # 打开处理后的视频文件 \nfps = cap.get(cv2.CAP_PROP_FPS) # 获得视频的原分辨率\nHTML(display(video_frames, fps).to_html5_video()) # 展示处理后的视频\n\n\n# ## 3.6音频处理\n\n# In[ ]:\n\n\n# 完整版Peking_5s.mp4,添加了音频,需要下载到本地播放\n# 以上过程没有考虑视频的音频,这部分代码用于音频的添加\nvideo_frames = imageio.mimread(output_video_path2, memtest=False)\ncap = cv2.VideoCapture(output_video_path2) \nfps = cap.get(cv2.CAP_PROP_FPS) # 获得视频的原分辨率\nHTML(display(video_frames, fps).to_html5_video())\n\n"
]
| [
[
"matplotlib.animation.ArtistAnimation",
"numpy.concatenate",
"matplotlib.pyplot.close",
"matplotlib.pyplot.axis",
"matplotlib.pyplot.figure"
]
]
|
EmanuelCastanho/PMLDataPipeline | [
"3f613a46a42898f78a90d956c0c6af364778c529"
]
| [
"download_features.py"
]
| [
"'''This script downloads and processes modeling traning data \n in to a format that is ready for the modeling script.\n To run use the following command\n\n python download_features -feature_list=data/features.csv -o output_dir\n \n This will generate EOPatches with each training data point at the center and will\n create a folder for each that contains the EOPatch and some plot's to help contextualize \n the datapoint.\n\n The expected format for the input file is a csv with one column per feature\n\n Lat, Lon, date, label\n\n Where label is on of \"Debris\", \"Water\", \"Spume\", \"Timber\", \"Pumice\", \"Seaweed\"\n'''\n\nimport pandas as pd \nimport geopanads as gp\nimport argparse\nfrom plasticfinder.tasks.combined_masks import CombineMask\nfrom plasticfinder.tasks.cloud_classifier import cloud_classifier_task\nfrom plasticfinder.tasks.water_detection import WaterDetector\nfrom plasticfinder.tasks.ndwi import ndwi_task\nfrom plasticfinder.tasks.ndvi import ndvi_task\nfrom plasticfinder.tasks.fdi import CalcFDI\nfrom plasticfinder.tasks.input_tasks import input_task,true_color,add_l2a\nfrom plasticfinder.tasks.local_Norm import LocalNormalization\nfrom eolearn.core import SaveTask, LinearWorkflow, LoadTask\nfrom eolearn.core.constants import OverwritePermission\n\n\ndef load_fetures_from_file(file, buffer_x=500, buffer_y=500):\n '''A function to load in the list of feature targets and generates a buffer around each of the specified extent\n \n Parameters:\n file (str): Location of the file specifying the training targets\n buffer_x (float): Amount in meters to buffer the point by. Default is 500m.\n buffer_y (float): Amount in meters to buffer the point by. Default is 500m.\n\n Returns:\n features (GeoDataFrame): A GeoPandas GeoDataFrame \n ''' \n\n features = pd.read_csv('data/features.csv')\n features = gp.GeoDataFrame(features,geometry = features.apply(lambda x: Point(x.Lon,x.Lat),axis=1), crs='EPSG:4326')\n bounds = gp.GeoSeries(features.to_crs(\"EPSG:3857\").apply(lambda x : box(x.geometry.x - buffer_x, x.geometry.y - buffer_y, x.geometry.x + buffer_x, x.geometry.y + buffer_y),axis=1), crs='EPSG:3857').to_crs('EPSG:4326')\n features = features.assign(date_start= pd.to_datetime(features.date.str[0:10], format='%Y_%m_%d') - timedelta(days=1), date_end = pd.to_datetime(features.date.str[0:10],format='%Y_%m_%d') +timedelta(days=1))\n\n # features[features['file']=='S2B_MSI_2019_04_24_07_36_19_T36JUN_L2R'].date_start = datetime.datetime(year = 2019, month=5, day=23)\n # features[features['file']=='S2B_MSI_2019_04_24_07_36_19_T36JUN_L2R'].date_end = datetime.datetime(year = 2019, month=5, day=25)\n return features\n\n\ndef process_feature(feature, feature_index): \n '''A function to download a given target pixel and it's surroundings as an EOPatch\n \n Parameters:\n feature (GeoSeries): A row from the GeoDataFrame produced by load_fetures_from_file\n feature_index (int): The integer used in saving the EOPatch to disk.\n \n\n Returns:\n Nothing \n ''' \n\n save = SaveTask(path=f'{base_dir}/feature_{feature_index}/', overwrite_permission=OverwritePermission.OVERWRITE_PATCH)\n train_test_workflow = LinearWorkflow(input_task,true_color,add_l2a,ndvi,ndwi,add_fdi,cloud_detection,water_detection,combine_mask,save )\n\n feature_result = train_test_workflow.execute({\n input_task: {\n 'bbox':BBox(bounds.iloc[feature_index],bbox_list[0].crs),\n 'time_interval': [feature.date_start, feature.date_end]\n },\n combine_mask:{\n 'use_water': False #(target.reduced_label != 'Timber')\n },\n add_fdi:{\n 'band_layer': USE_BANDS,\n 'band_names': band_names\n }\n })\n patch = feature_result.eopatch()\n return patch \n\n\nif __name__=='__main__':\n\n parser = argparse.ArgumentParser(description='Script to download target features and their surroundings as EOPatches')\n parser.add_argument('--features', type=str, help='The location of a csv that holds the data for each feature')\n \n\n args = parser.parse_args()\n\n feature_file = args.features\n output_dir = 'data/Training'\n \n features = load_fetures_from_file(feature_file)\n \n for feature_index in range(0, features.shape[0],1):\n target = features.iloc[feature_index]\n try:\n print(\"running \", feature_index, ' of ', features.shape[0])\n patch = process_feature(target,feature_index)\n fig,axs = plot_masks_and_vals(patch, features.iloc[feature_index:feature_index+1])\n fig.savefig(f'{base_dir}/feature_{feature_index}/mask_vals.png')\n plt.close(fig)\n except:\n print(\"Could not download feature \", feature_index)\n\n for feature_index in range(0,features.shape[0],1):\n target = features.iloc[feature_index]\n try:\n patch = apply_local_norm(target,feature_index, method='median', window_size=40)\n except Exception as e :\n print(\"Could not process feature \", feature_index)\n"
]
| [
[
"pandas.read_csv",
"pandas.to_datetime"
]
]
|
czyzq/mstrio-py | [
"b25fd19936b659d503a7eaaa96c8d0b4e118cb7c",
"b25fd19936b659d503a7eaaa96c8d0b4e118cb7c"
]
| [
"mstrio/users_and_groups/user.py",
"mstrio/utils/acl.py"
]
| [
"from datetime import datetime\nfrom typing import List, Optional, TYPE_CHECKING, Union\n\nfrom pandas import DataFrame, read_csv\nfrom requests.exceptions import HTTPError\n\nfrom mstrio import config\nfrom mstrio.access_and_security.privilege_mode import PrivilegeMode\nfrom mstrio.access_and_security.security_role import SecurityRole\nfrom mstrio.api import users\nfrom mstrio.connection import Connection\nfrom mstrio.users_and_groups.user_connections import UserConnections\nfrom mstrio.utils import helper, time_helper\nfrom mstrio.utils.acl import TrusteeACLMixin\nfrom mstrio.utils.entity import DeleteMixin, Entity, ObjectTypes\n\nif TYPE_CHECKING:\n from mstrio.access_and_security.privilege import Privilege\n from mstrio.access_and_security.security_filter import SecurityFilter\n from mstrio.server.project import Project\n from mstrio.users_and_groups.user_group import UserGroup\n\n\ndef create_users_from_csv(connection: Connection, csv_file: str) -> List[\"User\"]:\n \"\"\"Create new user objects from csv file. Possible header values for the\n users are the same as in the `User.create()` method.\n\n Args:\n connection: MicroStrategy connection object returned by\n `connection.Connection()`\n csv_file: path to file containing at minimum 'username' and 'full_name'\n headers'.\n \"\"\"\n return User._create_users_from_csv(connection=connection, csv_file=csv_file)\n\n\ndef list_users(connection: Connection, name_begins: Optional[str] = None,\n abbreviation_begins: Optional[str] = None, to_dictionary: bool = False,\n limit: Optional[int] = None, **filters) -> Union[List[\"User\"], List[dict]]:\n \"\"\"Get list of user objects or user dicts. Optionally filter the users by\n specifying 'name_begins', 'abbreviation_begins' or other filters.\n\n Wildcards available for name_begins and abbreviation_begins:\n ? - any character\n * - 0 or more of any characters\n e.g name_begins = ?onny wil return Sonny and Tonny\n\n Args:\n connection: MicroStrategy connection object returned by\n `connection.Connection()`\n name_begins: characters that the user name must begin with.\n abbreviation_begins: characters that the abbreviation must begin with.\n to_dictionary: If True returns dict, by default (False) returns\n User objects.\n limit: limit the number of elements returned. If `None` (default), all\n objects are returned.\n **filters: Available filter parameters: ['id', 'name', 'abbreviation',\n 'description', 'type', 'subtype', 'date_created', 'date_modified',\n 'version', 'acg', 'icon_path', 'owner', 'initials']\n\n Examples:\n >>> list_users(connection, name_begins='user', initials='UR')\n \"\"\"\n return User._get_users(\n connection=connection,\n name_begins=name_begins,\n abbreviation_begins=abbreviation_begins,\n to_dictionary=to_dictionary,\n limit=limit,\n **filters,\n )\n\n\nclass User(Entity, DeleteMixin, TrusteeACLMixin):\n \"\"\"Object representation of MicroStrategy User object.\n\n Attributes:\n connection: A MicroStrategy connection object\n id: User ID\n name: User name\n username: User username\n full_name: full name of the User\n intitials: User initials, derived from user's last name or username\n abbreviation: User login name\n description: User description\n memberships: IDs and names of direct parent groups for user\n security_roles: security roles that the user is a member of\n addresses: addresses for the user\n privileges: user privileges per project\n trust_id: Unique user ID provided by trusted authentication provider\n enabled: Specifies if user is allowed to log in\n owner: owner ID and name\n ancestors: List of ancestor folders\n password_modifiable: If user password can be modified\n require_new_password: If user is required to change new password\n standard_auth: If standard authentication is allowed for user\n date_created: Creation time, DateTime object\n date_modified: Last modification time, DateTime object\n type: Object type\n subtype: Object subtype\n ext_type: Object extended type\n version: Version ID\n acg: Access rights (See EnumDSSXMLAccessRightFlags for possible values)\n acl: Object access control list\n \"\"\"\n\n _PATCH_PATH_TYPES = {\n \"name\": str,\n \"username\": str,\n \"full_name\": str,\n \"description\": str,\n \"password\": str,\n \"enabled\": bool,\n \"password_modifiable\": bool,\n \"password_expiration_date\": datetime,\n \"standard_auth\": bool,\n \"require_new_password\": bool,\n \"ldapdn\": str,\n \"trust_id\": str,\n \"abbreviation\": str,\n }\n _OBJECT_TYPE = ObjectTypes.USER\n _API_GETTERS = {\n ('id', 'name', 'type', 'subtype', 'ext_type', 'abbreviation', 'date_created',\n 'date_modified', 'version', 'owner', 'ancestors', 'username', 'full_name', 'enabled',\n 'password_modifiable', 'require_new_password', 'standard_auth', 'memberships', 'acg',\n 'acl', 'trust_id', 'initials'): users.get_user_info,\n 'addresses': users.get_addresses,\n 'security_roles': users.get_user_security_roles,\n 'privileges': users.get_user_privileges,\n }\n _API_PATCH: dict = {\n ('name', 'abbreviation', 'username', 'full_name', 'enabled', 'password', 'description',\n 'password_modifiable', 'require_new_password', 'password_expiration_date',\n 'standard_auth', 'ldapdn', 'trust_id', 'initials', 'privileges', 'memberships',\n 'addresses', 'security_roles'): (users.update_user_info, 'patch')\n }\n _FROM_DICT_MAP = {\n **Entity._FROM_DICT_MAP,\n 'password_expiration_date': time_helper.DatetimeFormats.FULLDATETIME,\n }\n\n def __init__(self, connection: Connection, username: Optional[str] = None,\n name: Optional[str] = None, id: Optional[str] = None) -> None:\n \"\"\"Initialize User object by passing username, name, or id.\n When `id` is provided (not `None`), `username` and `name` are omitted.\n When `id` is not provided (`None`) and `username` is provided (not\n `None`), `name` is omitted.\n\n Args:\n connection: MicroStrategy connection object returned by\n `connection.Connection()`\n id: ID of User\n username: username of User\n name: name of User\n \"\"\"\n if id is None and name is None and username is None:\n helper.exception_handler(\n \"Please specify either 'id', 'username' or 'name' parameter in the constructor.\")\n\n if id is None:\n users = User._get_user_ids(\n connection=connection,\n abbreviation_begins=username,\n abbreviation=username,\n ) if username is not None else User._get_user_ids(\n connection=connection,\n name_begins=name,\n name=name,\n )\n\n if users:\n [id] = users\n else:\n temp_name = name if name else username\n helper.exception_handler(\"There is no user: '{}'\".format(temp_name),\n exception_type=ValueError)\n\n super().__init__(connection=connection, object_id=id, username=username, name=name)\n\n def _init_variables(self, **kwargs) -> None:\n super()._init_variables(**kwargs)\n self.username = kwargs.get(\"username\")\n self.full_name = kwargs.get(\"full_name\")\n self.enabled = kwargs.get(\"enabled\")\n self.password_modifiable = kwargs.get(\"password_modifiable\")\n self.standard_auth = kwargs.get(\"standard_auth\")\n self.require_new_password = kwargs.get(\"require_new_password\")\n self.ldapdn = kwargs.get(\"ldapdn\")\n self.trust_id = kwargs.get(\"trust_id\")\n\n self._initials = kwargs.get(\"initials\")\n self._addresses = kwargs.get(\"addresses\")\n self._memberships = kwargs.get(\"memberships\")\n self._security_roles = kwargs.get(\"security_roles\")\n self._privileges = kwargs.get(\"privileges\")\n self._security_filters = kwargs.get(\"security_filters\")\n\n @classmethod\n def create(cls, connection: Connection, username: str, full_name: str,\n password: Optional[str] = None, description: Optional[str] = None,\n enabled: bool = True, password_modifiable: bool = True,\n password_expiration_date: Optional[Union[str, datetime]] = None,\n require_new_password: bool = True, standard_auth: bool = True,\n ldapdn: Optional[str] = None, trust_id: Optional[str] = None,\n database_auth_login: Optional[str] = None,\n memberships: Optional[list] = None) -> \"User\":\n \"\"\"Create a new user on the I-Server. Returns User object.\n\n Args:\n connection: MicroStrategy connection object returned by\n `connection.Connection()`\n username: username of user\n full_name: user full name\n password: user password\n description: user description\n enabled: specifies if user is allowed to log in\n password_modifiable: Specifies if user password can be modified\n password_expiration_date: Expiration date of user password either\n as a datetime or string: \"yyyy-MM-dd HH:mm:ss\" in UTC\n require_new_password: Specifies if user is required to provide a new\n password.\n standard_auth: Specifies whether standard authentication is allowed\n for user.\n ldapdn: User's LDAP distinguished name\n trust_id: Unique user ID provided by trusted authentication provider\n database_auth_login: Database Authentication Login\n memberships: specify User Group IDs which User will be member off.\n \"\"\"\n password_expiration_date = time_helper.map_datetime_to_str(\n name='password_expiration_date', date=password_expiration_date,\n string_to_date_map=cls._FROM_DICT_MAP)\n body = {\n \"username\": username,\n \"fullName\": full_name,\n \"description\": description,\n \"password\": password,\n \"enabled\": enabled,\n \"passwordModifiable\": password_modifiable,\n \"passwordExpirationDate\": password_expiration_date,\n \"requireNewPassword\": require_new_password,\n \"standardAuth\": standard_auth,\n \"ldapdn\": ldapdn,\n \"trustId\": trust_id,\n \"databaseAuthLogin\": database_auth_login,\n \"memberships\": memberships\n }\n body = helper.delete_none_values(body)\n response = users.create_user(connection, body, username).json()\n if config.verbose:\n print(\"Successfully created user named: '{}' with ID: '{}'\".format(\n response.get('username'), response.get('id')))\n return cls.from_dict(source=response, connection=connection)\n\n @classmethod\n def _create_users_from_csv(cls, connection: Connection, csv_file: str) -> List[\"User\"]:\n func = cls.create\n args = func.__code__.co_varnames[:func.__code__.co_argcount]\n df = read_csv(csv_file, na_filter=False, usecols=lambda x: x in args)\n user_list = []\n all_param_value_dict = df.to_dict('records')\n\n for param_value_dict in all_param_value_dict:\n param_value_dict['connection'] = connection\n try:\n temp_user = func(**param_value_dict)\n except HTTPError:\n pass\n else:\n user_list.append(temp_user)\n\n return user_list\n\n @classmethod\n def _get_users(cls, connection: Connection, name_begins: Optional[str] = None,\n abbreviation_begins: Optional[str] = None, to_dictionary: bool = False,\n limit: Optional[int] = None, **filters) -> Union[List[\"User\"], List[dict]]:\n msg = \"Error getting information for a set of users.\"\n objects = helper.fetch_objects_async(connection, users.get_users_info,\n users.get_users_info_async, limit=limit,\n chunk_size=1000, error_msg=msg,\n name_begins=name_begins,\n abbreviation_begins=abbreviation_begins,\n filters=filters)\n if to_dictionary:\n return objects\n else:\n return [cls.from_dict(source=obj, connection=connection) for obj in objects]\n\n @classmethod\n def _get_user_ids(cls, connection: Connection, name_begins: Optional[str] = None,\n abbreviation_begins: Optional[str] = None, limit: Optional[int] = None,\n **filters) -> List[str]:\n user_dicts = User._get_users(\n connection=connection,\n name_begins=name_begins,\n abbreviation_begins=abbreviation_begins,\n to_dictionary=True,\n limit=limit,\n **dict(filters),\n )\n return [user['id'] for user in user_dicts]\n\n def alter(self, username: Optional[str] = None, full_name: Optional[str] = None,\n description: Optional[str] = None, password: Optional[str] = None,\n enabled: Optional[bool] = None, password_modifiable: Optional[bool] = None,\n password_expiration_date: Optional[str] = None, standard_auth: Optional[bool] = None,\n require_new_password: Optional[bool] = None, ldapdn: Optional[str] = None,\n trust_id: Optional[str] = None, database_auth_login: Optional[str] = None) -> None:\n \"\"\"Alter user properties.\n\n Args:\n username: username of user\n full_name: user full name\n description: user description\n password: user password\n enabled: specifies if user is allowed to log in\n password_modifiable: Specifies if user password can be modified\n password_expiration_date: Expiration date of user password,\n \"yyyy-MM-dd HH:mm:ss\" in UTC\n standard_auth: Specifies whether standard authentication is allowed\n for user.\n require_new_password: Specifies if user is required to provide a new\n password.\n ldapdn: User's LDAP distinguished name\n trust_id: Unique user ID provided by trusted authentication provider\n database_auth_login: Database Authentication Login\n \"\"\"\n func = self.alter\n args = func.__code__.co_varnames[:func.__code__.co_argcount]\n defaults = func.__defaults__ # type: ignore\n default_dict = dict(zip(args[-len(defaults):], defaults)) if defaults else {}\n local = locals()\n properties = {}\n for property_key in default_dict.keys():\n if local[property_key] is not None:\n properties[property_key] = local[property_key]\n\n self._alter_properties(**properties)\n\n def add_address(self, name: Optional[str] = None, address: Optional[str] = None,\n default: bool = True) -> None:\n \"\"\"Add new address to the user object.\n\n Args:\n name: User-specified name for the address\n address: The actual value of the address i.e. email address\n associated with this address name/id\n default: Specifies whether this address is the default address\n (change isDefault parameter).\n \"\"\"\n helper.validate_param_value('address', address, str, regex=r\"[^@]+@[^@]+\\.[^@]+\",\n valid_example=\"[email protected]\")\n helper.validate_param_value('name', name, str)\n helper.validate_param_value('default', default, bool)\n body = {\n \"name\": name,\n \"deliveryMode\": \"EMAIL\",\n \"device\": \"GENERIC_EMAIL\",\n \"value\": address,\n \"default\": default\n }\n response = users.create_address(self.connection, self.id, body)\n if response.ok:\n if config.verbose:\n print(\"Added address '{}' for user '{}'\".format(address, self.name))\n setattr(self, \"_addresses\", response.json().get('addresses'))\n\n def update_address(self, id: str, name: Optional[str] = None, address: Optional[str] = None,\n default: Optional[bool] = None) -> None:\n \"\"\"Update existing address. The address ID has to be specified\n as the name is not unique.\n\n Args:\n id: ID of the address\n name: New user-specified name for the address\n address: New address value\n default: Whether the address should be (un)marked as default\n \"\"\"\n if id is None:\n helper.exception_handler(\"Please specify 'id' parameter in the method.\")\n body = {}\n if name is not None:\n helper.validate_param_value('name', name, str)\n body[\"name\"] = name\n if address is not None:\n helper.validate_param_value('address', address, str, regex=r\"[^@]+@[^@]+\\.[^@]+\",\n valid_example=\"[email protected]\")\n body[\"value\"] = address\n if default is not None:\n helper.validate_param_value('default', default, bool)\n response = users.update_address(self.connection, self.id, id, body)\n if response.ok:\n if config.verbose:\n print(f\"Updated address with ID '{id}' for user '{self.name}'\")\n self.fetch(\"addresses\")\n\n def remove_address(self, name: Optional[str] = None, address: Optional[str] = None,\n id: Optional[str] = None) -> None:\n \"\"\"Remove existing address from the user object. Specify either address\n ID or name. Warning, address names are not unique and can potentially\n remove multiple addresses.\n\n Args:\n name: User-specified name for the address\n address: The actual value of the address i.e. email address\n associated with this address name/id\n id: ID of the address.\n \"\"\"\n initial_addresses = self.addresses\n if name is None and id is None and address is None or (name and id and address):\n helper.exception_handler(\n \"Please specify either 'name' or 'id' parameter in the method.\")\n if id is not None:\n addresses = helper.filter_list_of_dicts(initial_addresses, id=id)\n new_addresses = helper.filter_list_of_dicts(initial_addresses, id='!={}'.format(id))\n elif address is not None:\n addresses = helper.filter_list_of_dicts(initial_addresses, value=address)\n new_addresses = helper.filter_list_of_dicts(initial_addresses,\n value='!={}'.format(address))\n elif name is not None:\n addresses = helper.filter_list_of_dicts(initial_addresses, name=name)\n new_addresses = helper.filter_list_of_dicts(initial_addresses,\n name='!={}'.format(name))\n\n for addr in addresses:\n response = users.delete_address(self.connection, id=self.id, address_id=addr['id'])\n if response.ok:\n if config.verbose:\n print(\"Removed address '{}' with id {} from user '{}'\".format(\n addr['name'], addr['id'], self.name))\n setattr(self, \"_addresses\", new_addresses)\n\n def add_to_user_groups(\n self, user_groups: Union[str, \"UserGroup\", List[Union[str, \"UserGroup\"]]]) -> None:\n \"\"\"Adds this User to user groups specified in user_groups.\n\n Args:\n user_groups: list of `UserGroup` objects or IDs\n \"\"\"\n succeeded, failed = self._update_nested_properties(user_groups, \"memberships\", \"add\")\n if succeeded and config.verbose:\n print(\"Added user '{}' to group(s): {}\".format(self.name, succeeded))\n elif failed and config.verbose:\n print(\"User {} is already a member of {}\".format(self.name, failed))\n\n def remove_from_user_groups(\n self, user_groups: Union[str, \"UserGroup\", List[Union[str, \"UserGroup\"]]]) -> None:\n \"\"\"Removes this User from user groups specified in user_groups.\n\n Args:\n user_groups: list of `UserGroup` objects or IDs\n \"\"\"\n succeeded, failed = self._update_nested_properties(user_groups, \"memberships\", \"remove\")\n if succeeded and config.verbose:\n print(\"Removed user '{}' from group(s): {}\".format(self.name, succeeded))\n elif failed and config.verbose:\n print(\"User {} is not in {} group(s)\".format(self.name, failed))\n\n def remove_from_all_user_groups(self) -> None:\n \"\"\"Removes this User from all user groups.\"\"\"\n memberships = getattr(self, 'memberships')\n existing_ids = [obj.get('id') for obj in memberships]\n self.remove_from_user_groups(user_groups=existing_ids)\n\n def assign_security_role(self, security_role: Union[SecurityRole, str],\n project: Union[\"Project\", str] = None) -> None: # NOSONAR\n \"\"\"Assigns a Security Role to the user for given project.\n\n Args:\n security_role: Security Role ID or object\n project: Project name or object\n \"\"\"\n\n security_role = security_role if isinstance(security_role, SecurityRole) else SecurityRole(\n self.connection, id=str(security_role))\n\n security_role.grant_to([self.id], project)\n if config.verbose:\n print(\"Assigned Security Role '{}' to user: '{}'\".format(security_role.name,\n self.name))\n\n def revoke_security_role(self, security_role: Union[SecurityRole, str],\n project: Union[\"Project\", str] = None) -> None: # NOSONAR\n \"\"\"Removes a Security Role from the user for given project.\n\n Args:\n security_role: Security Role ID or object\n project: Project name or object\n \"\"\"\n\n security_role = security_role if isinstance(security_role, SecurityRole) else SecurityRole(\n self.connection, id=str(security_role))\n\n security_role.revoke_from([self.id], project)\n if config.verbose:\n print(\"Revoked Security Role '{}' from user: '{}'\".format(\n security_role.name, self.name))\n\n def list_security_filters(self, projects: Optional[Union[str, List[str]]] = None,\n to_dictionary: bool = False) -> dict:\n \"\"\"Get the list of security filters for user. They can be filtered by\n the projects' ids.\n\n Args:\n projects (str or list of str, optional): collection of projects' ids\n which is used for filtering data\n to_dictionary: If True returns security filters as dicts, by default\n (False) returns them as objects.\n\n Returns:\n Dictionary with project names as keys and list with security\n filters as values. In case of no securtiy filter for the given user\n in the particular project, then this project is not placed in\n the dictionary.\n \"\"\"\n from mstrio.access_and_security.security_filter import SecurityFilter\n objects_ = users.get_security_filters(self.connection, self.id, projects).json()\n projects_ = objects_.get(\"projects\")\n\n objects_ = {\n project_.get(\"name\"): project_.get(\"securityFilters\")\n for project_ in projects_\n if project_.get(\"securityFilters\")\n }\n\n self._security_filters = {\n name:\n [SecurityFilter.from_dict(sec_filter, self.connection) for sec_filter in sec_filters]\n for (name, sec_filters) in objects_.items()\n }\n if to_dictionary:\n return objects_\n return self._security_filters\n\n def apply_security_filter(self, security_filter: Union[\"SecurityFilter\", str]) -> bool:\n \"\"\"Apply a security filter to the user.\n\n Args:\n security_filter (string or object): identifier of security filter or\n `SecurityFilter` object which will be applied to the user.\n Returns:\n True when applying was successful. False otherwise.\n \"\"\"\n if isinstance(security_filter, str):\n from mstrio.access_and_security.security_filter import SecurityFilter\n security_filter = SecurityFilter.from_dict({\"id\": security_filter}, self.connection)\n return security_filter.apply(self.id)\n\n def revoke_security_filter(self, security_filter: Union[\"SecurityFilter\", str]) -> bool:\n \"\"\"Revoke a security filter from the user.\n\n Args:\n security_filter (string or object): identifier of security filter or\n `SecurityFilter` object which will be revoked from the user.\n\n Returns:\n True when revoking was successful. False otherwise.\n \"\"\"\n if isinstance(security_filter, str):\n from mstrio.access_and_security.security_filter import SecurityFilter\n security_filter = SecurityFilter.from_dict({\"id\": security_filter}, self.connection)\n return security_filter.revoke(self.id)\n\n def grant_privilege(self, privilege: Union[str, List[str], \"Privilege\",\n List[\"Privilege\"]]) -> None:\n \"\"\"Grant privileges directly to the user.\n\n Args:\n privilege: list of privilege objects, ids or names\n \"\"\"\n from mstrio.access_and_security.privilege import Privilege\n privileges = [\n priv['id'] for priv in Privilege._validate_privileges(self.connection, privilege)\n ]\n existing_ids = [\n privilege['privilege']['id'] for privilege in self.list_privileges(mode='GRANTED')\n ]\n succeeded, failed = self._update_nested_properties(privileges, \"privileges\", \"add\",\n existing_ids)\n\n if succeeded:\n self.fetch('privileges') # fetch the object properties and set object attributes\n if config.verbose:\n print(\"Granted privilege(s) {} to '{}'\".format(succeeded, self.name))\n if failed and config.verbose:\n print(\"User '{}' already has privilege(s) {}\".format(self.name, failed))\n\n def revoke_privilege(self, privilege: Union[str, List[str], \"Privilege\",\n List[\"Privilege\"]]) -> None:\n \"\"\"Revoke directly granted user privileges.\n\n Args:\n privilege: list of privilege objects, ids or names\n \"\"\"\n from mstrio.access_and_security.privilege import Privilege\n privileges = set(\n [priv['id'] for priv in Privilege._validate_privileges(self.connection, privilege)])\n existing_ids = [\n privilege['privilege']['id'] for privilege in self.list_privileges(mode='ALL')\n ]\n directly_granted = set(\n [privilege['privilege']['id'] for privilege in self.list_privileges(mode='GRANTED')])\n to_revoke = list(privileges.intersection(directly_granted))\n not_directly_granted = list(\n (set(existing_ids) - directly_granted).intersection(privileges))\n\n if not_directly_granted:\n msg = (f\"Privileges {sorted(not_directly_granted)} are inherited and will be ommited. \"\n \"Only directly granted privileges can be revoked by this method.\")\n helper.exception_handler(msg, exception_type=Warning)\n\n succeeded, failed = self._update_nested_properties(to_revoke, \"privileges\", \"remove\",\n existing_ids)\n if succeeded:\n self.fetch('privileges') # fetch the object properties and set object attributes\n if config.verbose:\n print(\"Revoked privilege(s) {} from '{}'\".format(succeeded, self.name))\n if failed and config.verbose:\n print(\"User '{}' does not have privilege(s) {}\".format(self.name, failed))\n\n def revoke_all_privileges(self, force: bool = False) -> None:\n \"\"\"Revoke directly granted user privileges.\n\n Args:\n force: If True, no additional prompt will be shown before revoking\n all privileges from User.\n \"\"\"\n user_input = 'N'\n if not force:\n user_input = input(\n \"Are you sure you want to revoke all privileges from user '{}'? [Y/N]: \".format(\n self.name))\n if force or user_input == 'Y':\n to_revoke = [\n privilege['privilege']['id'] for privilege in self.list_privileges(mode='GRANTED')\n ]\n if to_revoke:\n self.revoke_privilege(privilege=to_revoke)\n else:\n print(\"User '{}' does not have any directly granted privileges\".format(self.name))\n\n def list_privileges(self, mode: Union[PrivilegeMode, str] = PrivilegeMode.ALL,\n to_dataframe: bool = False) -> list:\n \"\"\"List privileges for user.\n\n Args:\n mode: Filter by source of privileges. One of: `ALL`, `INHERITED`,\n or `GRANTED`. See: `privilege.PrivilegeMode` enum.\n to_dataframe: If True, return a `DataFrame` object containing\n privileges.\n \"\"\"\n self.fetch('privileges')\n\n def to_df(priv_list):\n priv_dict = {}\n for priv in priv_list:\n priv_dict[priv['privilege']['id']] = priv['privilege']['name']\n df = DataFrame.from_dict(priv_dict, orient='index', columns=['Name'])\n df.index.name = 'ID'\n return df\n\n if not isinstance(mode, PrivilegeMode):\n try:\n mode = PrivilegeMode(mode)\n except ValueError:\n msg = (\"Wrong privilege mode has been passed, allowed modes are \"\n \"['ALL'/'INHERITED'/'GRANTED']. See: `privilege.PrivilegeMode` enum.\")\n helper.exception_handler(msg, ValueError)\n\n privileges = list()\n if mode == PrivilegeMode.ALL:\n privileges = self.privileges\n elif mode == PrivilegeMode.INHERITED:\n for privilege in self.privileges:\n is_inherited = False\n for source in privilege['sources']:\n is_inherited = not source['direct'] or is_inherited\n if is_inherited:\n privileges.append(privilege)\n else: # GRANTED\n for privilege in self.privileges:\n is_granted = False\n for source in privilege['sources']:\n is_granted = source['direct'] or is_granted\n if is_granted:\n privileges.append(privilege)\n\n return to_df(privileges) if to_dataframe else privileges\n\n def disconnect(self, nodes: Union[str, List[str]] = None) -> None:\n \"\"\"Disconnect all active user connection sessions for the specified\n node.\n\n Args:\n nodes: list of node names\n \"\"\"\n temp_connections = UserConnections(self.connection)\n temp_connections.disconnect_users(users=self, nodes=nodes)\n\n def delete(self, force: bool = False) -> bool:\n \"\"\"Deletes the user.\n Deleting the user will not remove the user's shared files.\n\n Args:\n force: If True, no additional prompt will be shown before deleting\n User.\n Returns:\n True for success. False otherwise.\n \"\"\"\n return super().delete(force=force)\n\n @property\n def memberships(self):\n return self._memberships\n\n @property\n def initials(self):\n return self._initials\n\n @property\n def addresses(self):\n return self._addresses\n\n @property\n def security_roles(self):\n return self._security_roles\n\n @property\n def privileges(self):\n return self._privileges\n\n @property\n def security_filters(self):\n if not self._security_filters:\n self.list_security_filters()\n return self._security_filters\n",
"from enum import Enum, IntFlag\nfrom typing import Any, Dict, List, Optional, TYPE_CHECKING, TypeVar, Union\n\nimport pandas as pd\nfrom requests import HTTPError\n\nfrom mstrio.api import objects\nfrom mstrio.connection import Connection\nfrom mstrio.utils.helper import Dictable, exception_handler, filter_obj_list\nfrom mstrio.types import ObjectTypes\n\nif TYPE_CHECKING:\n from mstrio.users_and_groups import UserOrGroup\n from mstrio.utils.entity import Entity\n from mstrio.server import Project\n\n\nclass Rights(IntFlag):\n \"\"\"\"Enumeration constants used to specify the access granted attribute of\n the DSS objects. \"\"\"\n EXECUTE = 0b10000000\n USE = 0b01000000\n CONTROL = 0b00100000\n DELETE = 0b00010000\n WRITE = 0b00001000\n READ = 0b00000100\n USE_EXECUTE = 0b00000010 # This constant is deprecated\n BROWSE = 0b00000001\n INHERITABLE = 0b100000000000000000000000000000\n\n\nclass Permissions(Enum):\n \"\"\"Enumeration constants used to specify combination of Rights values\n similar to workstation Security Access.\n\n TODO: This has to be string-based to discern between 'Denied All'\n and 'Full Control', which have the same mask.\n \"\"\"\n DENIED_ALL = 'Denied All'\n DEFAULT_ALL = 'Default All'\n CONSUME = 'Consume'\n VIEW = 'View'\n MODIFY = 'Modify'\n FULL_CONTROL = 'Full Control'\n\n\nclass AggregatedRights(IntFlag):\n \"\"\"Enumeration constants used to specify combination of Rights values.\"\"\"\n NONE = 0b00000000\n CONSUME = 0b01000101\n VIEW = 0b11000101\n MODIFY = 0b11011101\n ALL = 0b11111111\n\n\nAGGREGATED_RIGHTS_MAP = {\n Permissions.VIEW: AggregatedRights.VIEW,\n Permissions.MODIFY: AggregatedRights.MODIFY,\n Permissions.FULL_CONTROL: AggregatedRights.ALL,\n Permissions.DENIED_ALL: AggregatedRights.ALL,\n Permissions.DEFAULT_ALL: AggregatedRights.NONE,\n Permissions.CONSUME: AggregatedRights.CONSUME,\n}\n\nT = TypeVar(\"T\")\n\n\nclass ACE(Dictable):\n\n _FROM_DICT_MAP = {\n 'rights': Rights,\n }\n\n def __init__(self, deny: bool, entry_type: int, rights: Union[Rights, int], trustee_id: str,\n trustee_name: str, trustee_type: int, trustee_subtype: int, inheritable: bool):\n \"\"\"Set ACL object.\n\n Args:\n deny: Specifies whether access is denied\n entry_type: Access control entry type (1 for object access).\n Possible values can be found in EnumDSSXMLAccessEntryType\n rights: Rights assigned to the designated trustee\n trustee_id: User ID of the designated trustee\n trustee_name: User name of the designated trustee\n trustee_type: Type of the designated trustee\n trustee_subtype: Sub-type of the designated trustee\n inheritable: Specifies whether access control is inherited\n \"\"\"\n\n self.deny = deny\n self.entry_type = entry_type\n self.rights = rights if isinstance(rights, Rights) else Rights(rights)\n self.trustee_id = trustee_id\n self.trustee_name = trustee_name\n self.trustee_type = trustee_type\n self.trustee_subtype = trustee_subtype\n self.inheritable = inheritable\n\n def __eq__(self, other: object) -> bool:\n if len(self.__dict__) != len(other.__dict__):\n return False\n for attr in self.__dict__:\n if getattr(self, attr) != getattr(other, attr):\n return False\n return True\n\n @classmethod\n def from_dict(cls, source: Dict[str, Any], connection: Connection):\n\n def translate_names(name: str):\n if name == \"type\":\n return \"entry_type\"\n return name\n\n modified_source = {translate_names(key): val for key, val in source.items()}\n return super().from_dict(modified_source, connection)\n\n def to_dict(self, camel_case=True):\n\n def translate_names(name: str):\n if name == \"entry_type\" or name == \"entryType\":\n return \"type\"\n return name\n\n result_dict = super().to_dict(camel_case=camel_case)\n return {translate_names(key): val for key, val in result_dict.items()}\n\n\nclass ACLMixin:\n \"\"\"ACLMixin class adds Access Control List (ACL) management for supporting\n objects.\n\n An ACL is a set of permissions on objects so that users or user groups have\n control over individual objects in the system. Those permissions decide\n whether or not a user can perform a particular class of operations on a\n particular object. For example, a user may have permissions to view and\n execute a report , but cannot modify the report definition or delete the\n report.\n\n NOTE: Must be mixedin with Entity or its subclasses.\n \"\"\"\n\n def list_acl(self, to_dataframe: bool = False, **filters) -> Union[pd.DataFrame, list]:\n \"\"\"Get Access Control List (ACL) for this object. Optionally filter\n ACLs by specifying filters.\n\n Args:\n to_dataframe(bool, optional): if True, return datasets as\n pandas DataFrame\n **filters: Available filter parameters: [deny, type, rights,\n trustee_id, trustee_name, trustee_type, trustee_subtype,\n inheritable]\n\n Examples:\n >>> list_acl(deny=True, trustee_name=\"John\")\n \"\"\"\n acl = filter_obj_list(self.acl, **filters)\n if to_dataframe:\n return pd.DataFrame(acl)\n else:\n return acl\n\n def acl_add(self: \"Entity\", rights: Union[int, Rights, AggregatedRights],\n trustees: Union[\"UserOrGroup\", List[\"UserOrGroup\"]], denied: bool = False,\n inheritable: bool = False, propagate_to_children: Optional[bool] = None) -> None:\n \"\"\"Add Access Control Element (ACE) to the object ACL.\n\n Note:\n Argument `propagate_to_children` is used only for objects with\n type `ObjectTypes.FOLDER`.\n\n Args:\n rights: The degree to which the user or group is granted or denied\n access to the object. The available permissions are defined in\n `Rights` and `AggregatedRights` Enums\n trustees: list of trustees (`User` or `UserGroup` objects or ids) to\n update the ACE for\n denied: flag to indicate granted or denied access to the object\n inheritable: Applies only to folders. If set, any objects placed in\n the folder inherit the folder's entry in the ACL.\n propagate_to_children: used for folder objects only, default value\n is None, if set to True/False adds `propagateACLToChildren`\n keyword to the request body and sets its value accordingly\n\n Examples:\n >>> obj.acl_add(rights=Rights.BROWSE | Rights.EXECUTE,\n >>> trustees=user_obj, denied=True)\n \"\"\"\n self._update_acl(op=\"ADD\", rights=rights, trustees=trustees, denied=denied,\n inheritable=inheritable, propagate_to_children=propagate_to_children)\n\n def acl_remove(self: \"Entity\", rights: Union[int, Rights, AggregatedRights],\n trustees: Union[\"UserOrGroup\", List[\"UserOrGroup\"]], denied: bool = False,\n inheritable: bool = False,\n propagate_to_children: Optional[bool] = None) -> None:\n \"\"\"Remove Access Control Element (ACE) from the object ACL.\n\n Note:\n Argument `propagate_to_children` is used only for objects with\n type `ObjectTypes.FOLDER`.\n\n Args:\n rights: The degree to which the user or group is granted or denied\n access to the object. The available permissions are defined in\n `Rights` and `AggregatedRights` Enums\n trustees: list of trustees (`User` or `UserGroup` objects or ids) to\n update the ACE for\n denied: flag to indicate granted or denied access to the object\n inheritable: Applies only to folders. If set, any objects placed in\n the folder inherit the folder's entry in the ACL.\n propagate_to_children: used for folder objects only, default value\n is None, if set to True/False adds `propagateACLToChildren`\n keyword to the request body and sets its value accordingly\n\n Examples:\n >>> obj.acl_remove(rights=Rights.BROWSE | Rights.EXECUTE,\n >>> trustees=user_obj, denied=True)\n \"\"\"\n self._update_acl(op=\"REMOVE\", rights=rights, trustees=trustees, denied=denied,\n inheritable=inheritable, propagate_to_children=propagate_to_children)\n\n def acl_alter(self: \"Entity\", rights: Union[int, Rights, AggregatedRights],\n trustees: Union[\"UserOrGroup\", List[\"UserOrGroup\"]], denied: bool = False,\n inheritable: bool = False, propagate_to_children: Optional[bool] = None) -> None:\n \"\"\"Alter an existing Access Control Element (ACE) of the object ACL.\n\n Note:\n Argument `propagate_to_children` is used only for objects with\n type `ObjectTypes.FOLDER`.\n\n Args:\n rights: The degree to which the user or group is granted or denied\n access to the object. The available permissions are defined in\n `Rights` and `AggregatedRights` Enums\n trustees: list of trustees (`User` or `UserGroup` objects or ids) to\n update the ACE for\n denied: flag to indicate granted or denied access to the object\n inheritable: Applies only to folders. If set, any objects placed in\n the folder inherit the folder's entry in the ACL.\n propagate_to_children: used for folder objects only, default value\n is None, if set to True/False adds `propagateACLToChildren`\n keyword to the request body and sets its value accordingly\n\n Examples:\n >>> obj.acl_alter(rights=Rights.BROWSE | Rights.EXECUTE,\n >>> trustees=user_obj, denied=True)\n \"\"\"\n self._update_acl(op=\"REPLACE\", rights=rights, trustees=trustees, denied=denied,\n inheritable=inheritable, propagate_to_children=propagate_to_children)\n\n def _update_acl(self: \"Entity\", op: str, rights: Union[int, Rights, AggregatedRights],\n trustees: Union[\"UserOrGroup\", List[\"UserOrGroup\"]],\n propagate_to_children: Optional[bool] = None, denied: bool = False,\n inheritable: bool = False) -> None:\n \"\"\"Updates the ACL for this object, performs operation defined by the\n `op` parameter on all objects from `trustees` list.\n\n Note:\n Argument `propagate_to_children` is used only for objects with\n type `ObjectTypes.FOLDER`.\n\n Args:\n op: ACL update operator, available values are \"ADD\", \"REMOVE\" and\n \"REPLACE\"\n rights: value of rights to use by the operator\n trustees: list of trustees to update the ACE for\n propagate_to_children: used for folder objects only, default value\n is None, if set to True/False adds `propagateACLToChildren`\n keyword to the request body and sets its value accordingly\n denied: flag to indicate granted or denied access to the object\n inheritable: Applies only to folders. If set, any objects placed in\n the folder inherit the folder's entry in the ACL.\n \"\"\"\n # TODO merge duplicated code with _modify_rights function\n # TODO move (op, rights, ids, propagate_to_children=None,\n # denied=None, inheritable=None, types=None) to\n # separate AccesControlEntry class\n if op not in [\"ADD\", \"REMOVE\", \"REPLACE\"]:\n raise ValueError(\"Wrong ACL operator passed. Please use ADD, REMOVE or REPLACE\")\n\n # convert Enums\n rights = rights.value if isinstance(rights, Enum) else rights\n if rights not in range(256) and rights not in range(536_870_912, 536_871_167):\n msg = (\"Wrong `rights` value, please provide value in range 0-255 or combination of \"\n \"Rights enums\")\n raise ValueError(msg)\n if not isinstance(trustees, list):\n trustees = [trustees]\n trustee_ids = [trustee if isinstance(trustee, str) else trustee.id for trustee in trustees]\n body = {\n \"acl\": [\n {\n \"op\": op,\n \"trustee\": id,\n \"rights\": rights,\n \"type\": 1, # EnumDSSXMLAccessEntryType\n \"denied\": denied,\n \"inheritable\": inheritable\n } for id in trustee_ids\n ]\n }\n\n if isinstance(propagate_to_children, bool) and self._OBJECT_TYPE is ObjectTypes.FOLDER:\n body[\"propagateACLToChildren\"] = propagate_to_children\n\n response = objects.update_object(connection=self.connection, id=self.id, body=body,\n object_type=self._OBJECT_TYPE.value)\n if response.ok:\n response = response.json()\n self._set_object(**response)\n\n\nclass TrusteeACLMixin:\n \"\"\"TrusteeACLMixin class adds ACL management for Trustee classes.\n\n Objects currently supporting this Mixin are: (`User` and `UserGroup`).\n \"\"\"\n\n def set_permission(self, permission: Union[Permissions, str],\n to_objects: Union[str, List[str]], object_type: \"ObjectTypes\",\n project: Optional[Union[str, \"Project\"]] = None,\n propagate_to_children: Optional[bool] = None) -> None:\n \"\"\"Set permission to perform actions on given object(s).\n\n Function is used to set permission of the trustee to perform given\n actions on the provided objects. Within one execution of the function\n permission will be set in the same manner for each of the provided\n objects. The only available values of permission are: 'View', 'Modify',\n 'Full Control', 'Denied All', 'Default All'. Permission is the\n predefined set of rights. All objects to which the rights will be given\n have to be of the same type which is also provided.\n\n Args:\n permission: The Permission which defines set of rights. See:\n `Permissions` enum\n to_objects: List of object ids on access list for which the\n permissions will be set\n object_type: Type of objects on access list. See: `ObjectTypes` enum\n project: Object or id of Project where the object is\n located. If not passed, Project (project_id) selected in\n Connection object is used\n propagate_to_children: Flag used in the request to determine if\n those rights will be propagated to children of the trustee\n Returns:\n None\n \"\"\"\n\n if not isinstance(permission, Permissions):\n try:\n permission = Permissions(permission)\n except ValueError:\n msg = (\"Invalid `permission` value. Available values are: 'View', \"\n \"'Modify', 'Full Control', 'Denied All', 'Default All'. \"\n \"See: Permissions enum.\")\n exception_handler(msg)\n right_value = AGGREGATED_RIGHTS_MAP[permission].value\n denied = permission is Permissions.DENIED_ALL\n\n # those 2 tries are for clearing current rights (set to default values)\n try:\n _modify_rights(connection=self.connection, trustee_id=self.id, op='REMOVE',\n rights=AggregatedRights.ALL.value, ids=to_objects,\n object_type=object_type, project=project, denied=(not denied),\n propagate_to_children=propagate_to_children, verbose=False)\n except HTTPError:\n pass\n try:\n _modify_rights(connection=self.connection, trustee_id=self.id, op='REMOVE',\n rights=AggregatedRights.ALL.value, ids=to_objects,\n object_type=object_type, project=project, denied=denied,\n propagate_to_children=propagate_to_children, verbose=False)\n except HTTPError:\n pass\n\n if not permission == Permissions.DEFAULT_ALL:\n _modify_rights(connection=self.connection, trustee_id=self.id, op='ADD',\n rights=right_value, ids=to_objects, object_type=object_type,\n project=project, denied=denied,\n propagate_to_children=propagate_to_children)\n\n def set_custom_permissions(self, to_objects: Union[str, List[str]],\n object_type: Union[\"ObjectTypes\", int],\n project: Optional[Union[str, \"Project\"]] = None,\n execute: Optional[str] = None, use: Optional[str] = None,\n control: Optional[str] = None, delete: Optional[str] = None,\n write: Optional[str] = None, read: Optional[str] = None,\n browse: Optional[str] = None) -> None:\n \"\"\"Set custom permissions to perform actions on given object(s).\n\n Function is used to set rights of the trustee to perform given actions\n on the provided objects. Within one execution of the function rights\n will be set in the same manner for each of the provided objects.\n None of the rights is necessary, but if provided then only possible\n values are 'grant' (to grant right), 'deny' (to deny right), 'default'\n (to reset right) or None which is default value and means that nothing\n will be changed for this right. All objects to which the rights will be\n given have to be of the same type which is also provided.\n\n Args:\n to_objects: (str, list(str)): List of object ids on access list to\n which the permissions will be set\n object_type (int): Type of objects on access list\n project (str, Project): Object or id of Project in which\n the object is located. If not passed, Project\n (project_id) selected in Connection object is used.\n execute (str): value for right \"Execute\". Available are 'grant',\n 'deny', 'default' or None\n use (str): value for right \"Use\". Available are 'grant',\n 'deny', 'default' or None\n control (str): value for right \"Control\". Available are 'grant',\n 'deny', 'default' or None\n delete (str): value for right \"Delete\". Available are 'grant',\n 'deny', 'default' or None\n write (str): value for right \"Write\". Available are 'grant',\n 'deny', 'default' or None\n read (str): value for right \"Read\". Available are 'grant',\n 'deny', 'default' or None\n browse (str): value for right \"Browse. Available are 'grant',\n 'deny', 'default' or None\n Returns:\n None\n \"\"\"\n\n def modify_custom_rights(connection, trustee_id: str, right: Union[Rights, List[Rights]],\n to_objects: List[str], object_type: \"ObjectTypes\", denied: bool,\n project: Optional[Union[str,\n \"Project\"]] = None, default: bool = False,\n propagate_to_children: Optional[bool] = None) -> None:\n\n right_value = _get_custom_right_value(right)\n try:\n _modify_rights(connection=connection, trustee_id=trustee_id, op='REMOVE',\n rights=right_value, ids=to_objects, object_type=object_type,\n project=project, denied=(not denied),\n propagate_to_children=propagate_to_children, verbose=False)\n except HTTPError:\n pass\n\n op = 'REMOVE' if default else 'ADD'\n verbose = not default\n try:\n _modify_rights(connection=connection, trustee_id=trustee_id, op=op,\n rights=right_value, ids=to_objects, object_type=object_type,\n project=project, denied=denied,\n propagate_to_children=propagate_to_children, verbose=verbose)\n except HTTPError:\n pass\n\n rights_dict = {\n Rights.EXECUTE: execute,\n Rights.USE: use,\n Rights.CONTROL: control,\n Rights.DELETE: delete,\n Rights.WRITE: write,\n Rights.READ: read,\n Rights.BROWSE: browse\n }\n if not set(rights_dict.values()).issubset({'grant', 'deny', 'default', None}):\n msg = (\"Invalid value of the right. Available values are 'grant', 'deny', \"\n \"'default' or None.\")\n raise ValueError(msg)\n\n grant_list = [right for right, value in rights_dict.items() if value == 'grant']\n deny_list = [right for right, value in rights_dict.items() if value == 'deny']\n default_list = [right for right, value in rights_dict.items() if value == 'default']\n\n modify_custom_rights(connection=self.connection, trustee_id=self.id, right=grant_list,\n to_objects=to_objects, object_type=object_type, denied=False,\n project=project)\n modify_custom_rights(connection=self.connection, trustee_id=self.id, right=deny_list,\n to_objects=to_objects, object_type=object_type, denied=True,\n project=project)\n modify_custom_rights(connection=self.connection, trustee_id=self.id, right=default_list,\n to_objects=to_objects, object_type=object_type, denied=True,\n project=project, default=True)\n\n\ndef _modify_rights(connection, trustee_id: str, op: str, rights: int, object_type: \"ObjectTypes\",\n ids: List[str], project: Optional[Union[str, \"Project\"]] = None,\n propagate_to_children: Optional[bool] = None, denied: Optional[bool] = None,\n inheritable: Optional[bool] = None, verbose: bool = True) -> None:\n if op not in [\"ADD\", \"REMOVE\", \"REPLACE\"]:\n raise ValueError(\"Wrong ACL operator passed. Please use ADD, REMOVE or REPLACE\")\n\n # convert Enums\n rights = rights.value if isinstance(rights, Enum) else rights\n if rights not in range(256) and rights not in range(536_870_912, 536_871_168):\n msg = (\"Wrong `rights` value, please provide value in range 0-255 or combination of \"\n \"Rights enums\")\n exception_handler(msg)\n\n if project:\n project = project if isinstance(project, str) else project.id\n\n if isinstance(ids, str):\n ids = [ids]\n # TODO decide what to do about this code\n for id in ids:\n response = objects.get_object_info(connection=connection, id=id,\n object_type=object_type.value, project_id=project)\n if inheritable is None:\n tmp = [\n ace['inheritable']\n for ace in response.json().get('acl', [])\n if ace['trusteeId'] == trustee_id and ace['deny'] == denied\n ]\n inheritable = False if not tmp else tmp[0]\n\n body = {\n \"acl\": [{\n \"op\": op,\n \"trustee\": trustee_id,\n \"rights\": rights,\n \"type\": 1, # EnumDSSXMLAccessEntryType\n \"denied\": denied,\n \"inheritable\": inheritable\n }]\n }\n\n if isinstance(propagate_to_children, bool):\n body[\"propagateACLToChildren\"] = propagate_to_children\n\n _ = objects.update_object(connection=connection, id=id, body=body,\n object_type=object_type.value, project_id=project,\n verbose=verbose)\n\n\ndef _parse_acl_rights_bin_to_dict(rights_bin: int) -> Dict[Rights, bool]:\n # TODO move this to ENUM?\n return {right: rights_bin & right.value != 0 for right in Rights}\n\n\ndef _parse_acl_rights_dict_to_bin(rights_dict: Dict[Rights, bool]) -> int:\n output = 0\n for right, given in rights_dict.items():\n if given:\n output |= right.value\n return output\n\n\ndef _get_custom_right_value(right: Union[Rights, List[Rights]]) -> int:\n # TODO move this to ENUM?\n right_value = 0\n if not isinstance(right, list):\n right = [right]\n for r in right:\n if not isinstance(r, Rights):\n try:\n r = Rights[r.upper()]\n except ValueError:\n msg = (f\"Invalid custom `right` value: {r}. Available values are: EXECUTE, USE, \"\n \"CONTROL, DELETE, WRITE, READ, BROWSE. See: the Rights enum.\")\n raise ValueError(msg)\n right_value |= r.value\n return right_value\n"
]
| [
[
"pandas.read_csv",
"pandas.DataFrame.from_dict"
],
[
"pandas.DataFrame"
]
]
|
Eng-RSMY/OpenPNM | [
"a0a057d0f6346c515792459b1da97f05bab383c1"
]
| [
"OpenPNM/Utilities/__topology__.py"
]
| [
"# -*- coding: utf-8 -*-\n\"\"\"\n===============================================================================\nNetwork.tools.topology: Assorted topological manipulation methods\n===============================================================================\n\n\"\"\"\nimport scipy as _sp\nimport numpy as _np\nimport scipy.sparse as _sprs\nimport scipy.spatial as _sptl\nfrom OpenPNM.Base import logging as _logging\nfrom OpenPNM.Base import Controller as _controller\nlogger = _logging.getLogger(__name__)\n_ctrl = _controller()\n\n\nclass topology(object):\n\n def extend(self, network, pore_coords=[], throat_conns=[], labels=[]):\n r'''\n Add individual pores and/or throats to the network from a list of coords\n or conns. This is an in-place operation, meaning the received Network\n object will be altered directly.\n\n Parameters\n ----------\n network : OpenPNM Network Object\n The Network to which pores or throats should be added\n pore_coords : array_like\n The coordinates of the pores to add\n throat_conns : array_like\n The throat connections to add\n labels : string, or list of strings, optional\n A list of labels to apply to the new pores and throats\n\n Notes\n -----\n This needs to be enhanced so that it increases the size of all pore\n and throat props and labels on ALL associated Phase objects. At the\n moment it throws an error is there are any associated Phases.\n\n '''\n if (network._phases != []):\n raise Exception('Network has active Phases, cannot proceed')\n\n logger.info('Extending network')\n Np_old = network.num_pores()\n Nt_old = network.num_throats()\n Np = Np_old + int(_sp.size(pore_coords)/3)\n Nt = Nt_old + int(_sp.size(throat_conns)/2)\n # Adjust 'all' labels\n del network['pore.all'], network['throat.all']\n network['pore.all'] = _sp.ones((Np,), dtype=bool)\n network['throat.all'] = _sp.ones((Nt,), dtype=bool)\n # Add coords and conns\n if _sp.size(pore_coords) > 0:\n coords = _sp.vstack((network['pore.coords'], pore_coords))\n network['pore.coords'] = coords\n if _sp.size(throat_conns) > 0:\n conns = _sp.vstack((network['throat.conns'], throat_conns))\n network['throat.conns'] = conns\n # Increase size of any prop or label arrays on Network\n for item in list(network.keys()):\n if item.split('.')[1] not in ['coords', 'conns', 'all']:\n if item.split('.')[0] == 'pore':\n N = Np\n else:\n N = Nt\n if network[item].dtype == bool:\n temp = _sp.where(network[item])[0]\n network[item] = _sp.zeros((N,), dtype=bool)\n network[item][temp] = True\n elif network[item].dtype == object:\n temp = network[item]\n network[item] = _sp.ndarray((N,), dtype=object)\n network[item][_sp.arange(0, _sp.shape(temp)[0])] = temp\n else:\n temp = network[item]\n try:\n network[item] = _sp.ones((N, _sp.shape(temp)[1]),\n dtype=float)*_sp.nan\n except:\n network[item] = _sp.ones((N,), dtype=float)*_sp.nan\n network[item][_sp.arange(0, _sp.shape(temp)[0])] = temp\n # Apply labels, if supplied\n if labels != []:\n # Convert labels to list if necessary\n if type(labels) is str:\n labels = [labels]\n for label in labels:\n # Remove pore or throat from label, if present\n label = label.split('.')[-1]\n if _sp.size(pore_coords) > 0:\n Ps = _sp.r_[Np_old:Np]\n if 'pore.'+label not in network.labels():\n network['pore.'+label] = False\n network['pore.'+label][Ps] = True\n if _sp.size(throat_conns) > 0:\n Ts = _sp.r_[Nt_old:Nt]\n if 'throat.'+label not in network.labels():\n network['throat.'+label] = False\n network['throat.'+label][Ts] = True\n # Regnerate the adjacency matrices\n network._update_network()\n\n def trim(self, network, pores=[], throats=[]):\n '''\n Remove pores or throats from the network. This is an in-place operation,\n meaning the received Network object will be altered directly.\n\n Parameters\n ----------\n network : OpenPNM Network Object\n The Network from which pores or throats should be removed\n pores (or throats) : array_like\n A boolean mask of length Np (or Nt) or a list of indices of the\n pores (or throats) to be removed.\n\n Notes\n -----\n Trimming only adjusts Phase, Geometry, and Physics objects. Trimming a\n Network that has already been used to run simulations will break those\n simulation objects.\n\n Examples\n --------\n >>> import OpenPNM\n >>> pn = OpenPNM.Network.TestNet()\n >>> pn.Np\n 125\n >>> pn.Nt\n 300\n >>> pn.trim(pores=[1])\n >>> pn.Np\n 124\n >>> pn.Nt\n 296\n\n '''\n ctrl = network.controller\n for net in ctrl.networks():\n if net._parent is network:\n raise Exception('This Network has been cloned, cannot trim')\n if (_sp.size(pores) > 0) and (_sp.size(throats) > 0):\n raise Exception('Cannot delete pores and throats simultaneously')\n elif _sp.size(pores) > 0:\n pores = _sp.array(pores, ndmin=1)\n Pkeep = _sp.ones((network.num_pores(),), dtype=bool)\n Pkeep[pores] = False\n Tkeep = _sp.ones((network.num_throats(),), dtype=bool)\n Ts = network.find_neighbor_throats(pores)\n if len(Ts) > 0:\n Tkeep[Ts] = False\n elif _sp.size(throats) > 0:\n throats = _sp.array(throats, ndmin=1)\n Tkeep = _sp.ones((network.num_throats(),), dtype=bool)\n Tkeep[throats] = False\n Pkeep = network['pore.all'].copy()\n else:\n logger.warning('No pores or throats recieved')\n return\n\n # Trim all associated objects\n for item in network._geometries+network._physics+network._phases:\n Pnet = network['pore.'+item.name]*Pkeep\n Tnet = network['throat.'+item.name]*Tkeep\n temp = network.map_pores(pores=_sp.where(Pnet)[0],\n target=item,\n return_mapping=True)\n Ps = temp['target']\n temp = network.map_throats(throats=_sp.where(Tnet)[0],\n target=item,\n return_mapping=True)\n Ts = temp['target']\n # Then resize 'all\n item.update({'pore.all': _sp.ones((_sp.sum(Pnet),), dtype=bool)})\n item.update({'throat.all': _sp.ones((_sp.sum(Tnet),), dtype=bool)})\n # Overwrite remaining data and info\n for key in list(item.keys()):\n if key.split('.')[1] not in ['all']:\n temp = item.pop(key)\n if key.split('.')[0] == 'throat':\n logger.debug('Trimming {a} from {b}'.format(a=key,\n b=item.name))\n item[key] = temp[Ts]\n if key.split('.')[0] == 'pore':\n logger.debug('Trimming {a} from {b}'.format(a=key,\n b=item.name))\n item[key] = temp[Ps]\n\n # Remap throat connections\n Pmap = _sp.ones((network.Np,), dtype=int)*-1\n Pmap[Pkeep] = _sp.arange(0, _sp.sum(Pkeep))\n tpore1 = network['throat.conns'][:, 0]\n tpore2 = network['throat.conns'][:, 1]\n Tnew1 = Pmap[tpore1[Tkeep]]\n Tnew2 = Pmap[tpore2[Tkeep]]\n # Write 'all' label specifically\n network.update({'throat.all': _sp.ones((_sp.sum(Tkeep),), dtype=bool)})\n network.update({'pore.all': _sp.ones((_sp.sum(Pkeep),), dtype=bool)})\n # Write throat connections specifically\n network.update({'throat.conns': _sp.vstack((Tnew1, Tnew2)).T})\n # Overwrite remaining data and info\n for item in list(network.keys()):\n if item.split('.')[-1] not in ['conns', 'all']:\n temp = network.pop(item)\n if item.split('.')[0] == 'throat':\n logger.debug('Trimming {a} from {b}'.format(a=item,\n b=network.name))\n network[item] = temp[Tkeep]\n if item.split('.')[0] == 'pore':\n logger.debug('Trimming {a} from {b}'.format(a=item,\n b=network.name))\n network[item] = temp[Pkeep]\n\n # Reset network graphs\n network._update_network(mode='regenerate')\n\n # Check Network health\n health = network.check_network_health()\n if health['trim_pores'] != []:\n logger.warning('Isolated pores exist! Run check_network_health to ID \\\n which pores to remove.')\n pass\n\n def clone_pores(self, network, pores, apply_label=['clone'], mode='parents'):\n r'''\n Clones the specified pores and adds them to the network\n\n Parameters\n ----------\n network : OpenPNM Network Object\n The Network object to which the new pores are to be added\n pores : array_like\n List of pores to clone\n apply_labels : string, or list of strings\n The labels to apply to the clones, default is 'clone'\n mode : string\n Controls the connections between parents and clones. Options are:\n\n - 'parents': (Default) Each clone is connected only to its parent\n - 'siblings': Clones are only connected to each other in the same manner\n as parents were connected\n - 'isolated': No connections between parents or siblings\n '''\n if (network._geometries != []):\n logger.warning('Network has active Geometries, new pores must be \\\n assigned a Geometry')\n if (network._phases != []):\n raise Exception('Network has active Phases, cannot proceed')\n\n logger.debug('Cloning pores')\n apply_label = list(apply_label)\n # Clone pores\n Np = network.num_pores()\n Nt = network.num_throats()\n parents = _sp.array(pores, ndmin=1)\n pcurrent = network['pore.coords']\n pclone = pcurrent[pores, :]\n pnew = _sp.concatenate((pcurrent, pclone), axis=0)\n Npnew = _sp.shape(pnew)[0]\n clones = _sp.arange(Np, Npnew)\n # Add clone labels to network\n for item in apply_label:\n if 'pore.' + item not in network.keys():\n network['pore.'+item] = False\n if 'throat.' + item not in network.keys():\n network['throat.'+item] = False\n # Add connections between parents and clones\n if mode == 'parents':\n tclone = _sp.vstack((parents, clones)).T\n self.extend(network=network, pore_coords=pclone, throat_conns=tclone)\n if mode == 'siblings':\n ts = network.find_neighbor_throats(pores=pores, mode='intersection')\n tclone = network['throat.conns'][ts] + network.num_pores()\n self.extend(network=network, pore_coords=pclone, throat_conns=tclone)\n if mode == 'isolated':\n self.extend(network=network, pore_coords=pclone)\n # Apply provided labels to cloned pores\n for item in apply_label:\n network['pore.'+item][network.pores('all') >= Np] = True\n network['throat.'+item][network.throats('all') >= Nt] = True\n\n # Any existing adjacency and incidence matrices will be invalid\n network._update_network()\n\n def stitch(self, network, donor, P_network, P_donor, method='nearest',\n len_max=_sp.inf, label_suffix=''):\n r'''\n Stitches a second a network to the current network.\n\n Parameters\n ----------\n networK : OpenPNM Network Object\n The Network that will to which to donor Network will be attached\n\n donor : OpenPNM Network Object\n The Network to stitch on to the current Network\n\n P_network : array_like\n The pores on the current Network\n\n P_donor : array_like\n The pores on the donor Network\n\n label_suffix : string or None\n Some text to append to each label in the donor Network before\n inserting them into the recipient. The default is to append no\n text, but a common option would be to append the donor Network's\n name. To insert none of the donor labels, use None.\n\n len_max : float\n Set a length limit on length of new throats\n\n method : string (default = 'delaunay')\n The method to use when making pore to pore connections. Options are:\n\n - 'delaunay' : Use a Delaunay tessellation\n - 'nearest' : Connects each pore on the receptor network to its nearest\n pore on the donor network\n\n Notes\n -----\n Before stitching it is necessary to translate the pore coordinates of\n one of the Networks so that it is positioned correctly relative to the\n other.\n\n Examples\n --------\n >>> import OpenPNM\n >>> pn = OpenPNM.Network.TestNet()\n >>> pn2 = OpenPNM.Network.TestNet()\n >>> [pn.Np, pn.Nt]\n [125, 300]\n >>> [pn2.Np, pn2.Nt]\n [125, 300]\n >>> pn2['pore.coords'][:, 2] += 5.0\n >>> pn.stitch(donor=pn2, P_network=pn.pores('top'),\n ... P_donor=pn2.pores('bottom'), method='nearest', len_max=1.0)\n >>> [pn.Np, pn.Nt]\n [250, 625]\n\n '''\n # Ensure Networks have no associated objects yet\n if (len(network._simulation()) > 1) or (len(donor._simulation()) > 1):\n raise Exception('Cannot stitch a Network with active sibling objects')\n # Get the initial number of pores and throats\n N_init = {}\n N_init['pore'] = network.Np\n N_init['throat'] = network.Nt\n if method == 'nearest':\n P1 = P_network\n P2 = P_donor + N_init['pore'] # Increment pores on donor\n C1 = network['pore.coords'][P_network]\n C2 = donor['pore.coords'][P_donor]\n D = _sp.spatial.distance.cdist(C1, C2)\n [P1_ind, P2_ind] = _sp.where(D <= len_max)\n conns = _sp.vstack((P1[P1_ind], P2[P2_ind])).T\n else:\n raise RuntimeError('<{}> method not supported'.format(method))\n\n # Enter donor's pores into the Network\n self.extend(network=network, pore_coords=donor['pore.coords'])\n\n # Enter donor's throats into the Network\n self.extend(network=network, throat_conns=donor['throat.conns'] +\n N_init['pore'])\n\n # Trim throats that are longer then given len_max\n C1 = network['pore.coords'][conns[:, 0]]\n C2 = network['pore.coords'][conns[:, 1]]\n L = _sp.sum((C1 - C2)**2, axis=1)**0.5\n conns = conns[L <= len_max]\n\n # Add donor labels to recipient network\n if label_suffix is not None:\n if label_suffix != '':\n label_suffix = '_'+label_suffix\n for label in donor.labels():\n element = label.split('.')[0]\n locations = _sp.where(network._get_indices(element) >=\n N_init[element])[0]\n try:\n network[label + label_suffix]\n except:\n network[label + label_suffix] = False\n network[label+label_suffix][locations] = donor[label]\n\n # Add the new stitch throats to the Network\n self.extend(network=network, throat_conns=conns, labels='stitched')\n\n # Remove donor from Controller, if present\n # This check allows for the reuse of a donor Network multiple times\n if donor in _ctrl.values():\n _ctrl.purge_object(donor)\n\n def connect_pores(self, network, pores1, pores2, labels=[]):\n r'''\n Returns the possible connections between two group of pores.\n\n Parameters\n ----------\n networK : OpenPNM Network Object\n\n pores1 : array_like\n The first group of pores on the network\n\n pores2 : array_like\n The second group of pores on the network\n\n Notes\n -----\n It creates the connections in a format which is acceptable by\n the default OpenPNM connection key ('throat.conns') and adds them to\n the network.\n\n Examples\n --------\n >>> import OpenPNM\n >>> pn = OpenPNM.Network.TestNet()\n >>> pn.Nt\n 300\n >>> pn.connect_pores(pores1=[22, 32], pores2=[16, 80, 68])\n >>> pn.Nt\n 306\n >>> pn['throat.conns'][300:306]\n array([[16, 22],\n [22, 80],\n [22, 68],\n [16, 32],\n [32, 80],\n [32, 68]])\n\n '''\n size1 = _sp.size(pores1)\n size2 = _sp.size(pores2)\n array1 = _sp.repeat(pores1, size2)\n array2 = _sp.tile(pores2, size1)\n conns = _sp.vstack([array1, array2]).T\n self.extend(network=network, throat_conns=conns, labels=labels)\n\n def find_centroid(coords=None):\n r'''\n It finds the coordinates of the centroid of the sent pores.\n '''\n l = _np.float64(len(coords))\n x, y, z = coords.T\n sx = _np.sum(x)\n sy = _np.sum(y)\n sz = _np.sum(z)\n c = _np.array([sx/l, sy/l, sz/l], ndmin=1)\n return c\n\n def find_pores_distance(network, pores1=None, pores2=None):\n r'''\n It finds the distance between two group of pores.\n '''\n from scipy.spatial.distance import cdist\n p1 = _sp.array(pores1, ndmin=1)\n p2 = _sp.array(pores2, ndmin=1)\n coords = network['pore.coords']\n return cdist(coords[p1], coords[p2])\n\n def subdivide(self, network, pores, shape, labels=[]):\n r'''\n It trim the pores and replace them by cubic networks with the sent shape.\n\n Parameters\n ----------\n network : OpenPNM Network Object\n\n pores : array_like\n The first group of pores to be replaced\n\n shape : array_like\n The shape of cubic networks in the target locations\n\n Notes\n -----\n - It works only for cubic networks.\n\n Examples\n --------\n >>> import OpenPNM\n >>> pn = OpenPNM.Network.Cubic(shape=[5,6,5], spacing=0.001)\n >>> pn.Np\n 150\n >>> nano_pores = [2,13,14,15]\n >>> pn.subdivide(pores=nano_pores, shape=[4,7,3], labels='nano')\n >>> pn.Np\n 482\n >>> assert pn.Np == (150+4*(4*7*3)-4)\n\n '''\n mro = [item.__name__ for item in network.__class__.__mro__]\n if 'Cubic' not in mro:\n raise Exception('Subdivide is only supported for Cubic Networks')\n from OpenPNM.Network import Cubic\n pores = _sp.array(pores, ndmin=1)\n\n # Checks to find boundary pores in the selected pores\n try:\n b = network.pores('boundary')\n if (_sp.in1d(pores, b)).any():\n raise Exception('boundary pores cannot be subdivided!')\n except KeyError:\n pass\n\n # Assigning right shape and division\n if _sp.size(shape) != 2 and _sp.size(shape) != 3:\n raise Exception('Subdivide not implemented for Networks other than 2D \\\n and 3D')\n elif _sp.size(shape) == 3 and 1 not in shape:\n div = _sp.array(shape, ndmin=1)\n single_dim = None\n else:\n single_dim = _sp.where(_sp.array(network._shape) == 1)[0]\n if _sp.size(single_dim) == 0:\n single_dim = None\n if _sp.size(shape) == 3:\n div = _sp.array(shape, ndmin=1)\n else:\n div = _sp.zeros(3, dtype=_sp.int32)\n if single_dim is None:\n dim = 2\n else:\n dim = single_dim\n div[dim] = 1\n div[-_sp.array(div, ndmin=1, dtype=bool)] = _sp.array(shape, ndmin=1)\n\n # Creating small network and handling labels\n network_spacing = network._spacing\n new_net_spacing = network_spacing/div\n new_net = Cubic(shape=div, spacing=new_net_spacing)\n main_labels = ['left', 'right', 'front', 'back', 'top', 'bottom']\n if single_dim is not None:\n label_groups = _sp.array([['front', 'back'],\n ['left', 'right'],\n ['top', 'bottom']])\n non_single_labels = label_groups[_sp.array([0, 1, 2]) != single_dim]\n for l in main_labels:\n new_net['pore.surface_' + l] = False\n network['pore.surface_' + l] = False\n if single_dim is None:\n new_net['pore.surface_' + l][new_net.pores(labels=l)] = True\n else:\n for ind in [0, 1]:\n loc = (non_single_labels[ind] == l)\n temp_pores = new_net.pores(non_single_labels[ind][loc])\n new_net['pore.surface_' + l][temp_pores] = True\n\n old_coords = _sp.copy(new_net['pore.coords'])\n if labels == []:\n labels = ['pore.subdivided_' + new_net.name]\n for P in pores:\n # Shifting the new network to the right location and attaching it to the\n # main network\n shift = network['pore.coords'][P] - network_spacing/2\n new_net['pore.coords'] += shift\n Pn = network.find_neighbor_pores(pores=P)\n try:\n Pn_new_net = network.pores(labels)\n except:\n Pn_new_net = []\n Pn_old_net = Pn[~_sp.in1d(Pn, Pn_new_net)]\n Np1 = network.Np\n self.extend(pore_coords=new_net['pore.coords'],\n throat_conns=new_net['throat.conns'] + Np1,\n labels=labels, network=network)\n\n # Moving the temporary labels to the big network\n for l in main_labels:\n network['pore.surface_'+l][Np1:] = new_net['pore.surface_'+l]\n\n # Stitching the old pores of the main network to the new extended pores\n surf_pores = network.pores('surface_*')\n surf_coord = network['pore.coords'][surf_pores]\n for neighbor in Pn:\n neighbor_coord = network['pore.coords'][neighbor]\n dist = [round(_sp.inner(neighbor_coord-x, neighbor_coord-x),\n 20) for x in surf_coord]\n nearest_neighbor = surf_pores[dist == _sp.amin(dist)]\n if neighbor in Pn_old_net:\n coplanar_labels = network.labels(pores=nearest_neighbor)\n new_neighbors = network.pores(coplanar_labels,\n mode='intersection')\n # This might happen to the edge of the small network\n if _sp.size(new_neighbors) == 0:\n labels = network.labels(pores=nearest_neighbor,\n mode='intersection')\n common_label = [l for l in labels if 'surface_' in l]\n new_neighbors = network.pores(common_label)\n elif neighbor in Pn_new_net:\n new_neighbors = nearest_neighbor\n self.connect_pores(network=network, pores1=neighbor,\n pores2=new_neighbors, labels=labels)\n\n # Removing temporary labels\n for l in main_labels:\n network['pore.surface_' + l] = False\n new_net['pore.coords'] = _sp.copy(old_coords)\n\n network._label_surfaces()\n for l in main_labels:\n del network['pore.surface_'+l]\n self.trim(network=network, pores=pores)\n\n def trim_occluded_throats(self, network, mask='all'):\n r\"\"\"\n Remove throats with zero area from the network and also remove\n pores that are isolated (as a result or otherwise)\n\n Parameters\n ----------\n network : OpenPNM Network Object\n\n mask : string\n Applies routine only to pores and throats with this label\n \"\"\"\n occluded_ts = network['throat.area'] == 0\n if _sp.sum(occluded_ts) > 0:\n # Apply mask\n occluded_ts *= network[\"throat.\"+mask]\n self.trim(network=network, throats=occluded_ts)\n # Also get rid of isolated pores\n isolated_ps = network.check_network_health()['isolated_pores']\n if _sp.size(isolated_ps) > 0:\n # Convert to Bool array and apply mask\n temp_array = _sp.zeros(network.num_pores()).astype(bool)\n temp_array[isolated_ps] = True\n isolated_ps = temp_array * network[\"pore.\"+mask]\n self.trim(network=network, pores=isolated_ps)\n\n def merge_pores(self, network, pores, labels=['merged']):\n r\"\"\"\n Combines a selection of pores into a new single pore located at the\n centroid of the selected pores and connected to all of their neighbors.\n\n Parameters\n ----------\n network : OpenPNM Network Object\n\n pores : array_like\n The list of pores which are to be combined into a new single pore\n\n labels : string or list of strings\n The labels to apply to the new pore and new throat connections\n\n Notes\n -----\n The selection of pores should be chosen carefully, preferrable so that\n they all form a continuous cluster. For instance, it is recommended\n to use the ``find_nearby_pores`` method to find all pores within a\n certain distance of a given pore, and these can then be merged without\n causing any abnormal connections.\n\n Examples\n --------\n >>> import OpenPNM as op\n >>> pn = op.Network.Cubic(shape=[20,20,1])\n >>> topo = op.Utilities.topology()\n >>> P = pn.find_nearby_pores(pores=111, distance=5, flatten=True)\n >>> topo.merge_pores(network=pn, pores=P, labels=['merged'])\n >>> print(pn.Np)\n 321\n >>> pn.pores('merged')\n array([320])\n >>> pn.num_throats('merged')\n 32\n\n \"\"\"\n Pn = network.find_neighbor_pores(pores=pores,\n mode='union',\n flatten=True,\n excl_self=True)\n xyz = _sp.mean(network['pore.coords'][pores], axis=0)\n self.extend(network, pore_coords=xyz, labels=labels)\n Pnew = network.Ps[-1]\n self.connect_pores(network, pores1=Pnew, pores2=Pn, labels=labels)\n self.trim(network=network, pores=pores)\n"
]
| [
[
"scipy.amin",
"scipy.sum",
"scipy.zeros",
"scipy.copy",
"scipy.ones",
"scipy.ndarray",
"scipy.shape",
"scipy.mean",
"scipy.array",
"scipy.vstack",
"scipy.where",
"scipy.inner",
"scipy.spatial.distance.cdist",
"scipy.size",
"scipy.arange",
"numpy.array",
"scipy.tile",
"numpy.sum",
"scipy.repeat",
"scipy.in1d",
"scipy.concatenate"
]
]
|
AaronWChen/RHCR | [
"e5d5ac06c46d7cc4052a688338d9371518d1f445"
]
| [
"model_training_v2/mnist/mnist_saved_model_tf2.py"
]
| [
"\"\"\"\nThis script is a refactor of the original mnist_saved_model.py originally included in the repo. \nThe original was written in 2016 by Google and the original tutorial does not seem to be available \nanymore. \n\nAlso, TensorFlow 2 debuted in September 2019, so refactoring to TF 2 seemed like a good idea.\n\nIt is based on the tutorial \"Multi-class classification with MNIST\" colab notebook:\nhttps://colab.research.google.com/github/google/eng-edu/blob/master/ml/cc/exercises/multi-class_classification_with_MNIST.ipynb?utm_source=mlcc&utm_campaign=colab-external&utm_medium=referral&utm_content=multiclass_tf2-colab&hl=en\n\nRequires Python 3, TensorFlow 2\nEasiest way to work is to pip install the requirements.txt file\n\nTrain and export a simple Softmax Regression TensorFlow model.\nThe model is from the TensorFlow \"MNIST For ML Beginner\" tutorial. This program\nsimply follows all its training instructions, and uses TensorFlow SavedModel to\nexport the trained model with proper signatures that can be loaded by standard\ntensorflow_model_server.\n\nUsage: mnist_saved_model.py [--training_iteration=x] [--model_version=y] export_dir\n\"\"\"\n\nfrom __future__ import absolute_import, division, print_function, unicode_literals\nimport os\nimport sys\nimport tensorflow as tf\nfrom tensorflow.keras import Layers\nimport mnist_input_data\nimport numpy as np\nimport pandas as pd\n\ntf.compat.v1.app.flags.DEFINE_integer('training_iteration', 1000,\n 'number of training iterations.')\ntf.compat.v1.app.flags.DEFINE_integer('model_version', 1, 'version number of the model.')\ntf.compat.v1.app.flags.DEFINE_string('work_dir', '/tmp', 'Working directory.')\nFLAGS = tf.compat.v1.app.flags.FLAGS\n\n"
]
| [
[
"tensorflow.compat.v1.app.flags.DEFINE_integer",
"tensorflow.compat.v1.app.flags.DEFINE_string"
]
]
|
hrlblab/CircleNet | [
"219aa47fa4dc4f362b28448c0dcd41b29c4f1166"
]
| [
"src/lib/datasets/dataset/kidpath_new.py"
]
| [
"from __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\n\nimport pycocotools.coco as coco\nimport datasets.eval_protocals.kidpath_circle as kidpath_circle\nfrom datasets.eval_protocals.circle_eval import CIRCLEeval\nfrom pycocotools.cocoeval import COCOeval\n\nimport numpy as np\nimport json\nimport os\n\nimport torch.utils.data as data\n\n\nclass KidPath(data.Dataset):\n num_classes = 1\n default_resolution = [512, 512]\n mean = np.array([0.40789654, 0.44719302, 0.47026115],\n dtype=np.float32).reshape(1, 1, 3)\n std = np.array([0.28863828, 0.27408164, 0.27809835],\n dtype=np.float32).reshape(1, 1, 3)\n\n def __init__(self, opt, split):\n super(KidPath, self).__init__()\n self.data_dir = os.path.join(opt.data_dir, 'kidpath')\n self.img_dir = os.path.join(self.data_dir, '{}'.format(split))\n if split == 'test':\n self.annot_path = os.path.join(\n self.data_dir, 'kidneypath_test2019.json').format(split)\n else:\n if opt.task == 'exdet':\n self.annot_path = os.path.join(\n self.data_dir, 'kidneypath_extreme_{}2017.json').format(split)\n else:\n self.annot_path = os.path.join(\n self.data_dir, 'kidneypath_{}2019.json').format(split)\n self.max_objs = 128\n self.class_name = [\n '__background__', 'glomerulus']\n self._valid_ids = [1]\n self.cat_ids = {v: i for i, v in enumerate(self._valid_ids)}\n self.voc_color = [(v // 32 * 64 + 64, (v // 8) % 4 * 64, v % 8 * 32) \\\n for v in range(1, self.num_classes + 1)]\n self._data_rng = np.random.RandomState(123)\n self._eig_val = np.array([0.2141788, 0.01817699, 0.00341571],\n dtype=np.float32)\n self._eig_vec = np.array([\n [-0.58752847, -0.69563484, 0.41340352],\n [-0.5832747, 0.00994535, -0.81221408],\n [-0.56089297, 0.71832671, 0.41158938]\n ], dtype=np.float32)\n # self.mean = np.array([0.485, 0.456, 0.406], np.float32).reshape(1, 1, 3)\n # self.std = np.array([0.229, 0.224, 0.225], np.float32).reshape(1, 1, 3)\n\n self.split = split\n self.opt = opt\n\n print('==> initializing kidpath 2019 {} data.'.format(split))\n self.coco = coco.COCO(self.annot_path)\n self.images = self.coco.getImgIds()\n self.num_samples = len(self.images)\n\n self.circle = kidpath_circle.CIRCLE(self.annot_path)\n self.images_circle = self.circle.getImgIds()\n self.num_samples_circle = len(self.images_circle)\n\n print('Loaded {} {} samples'.format(split, self.num_samples))\n\n def _to_float(self, x):\n return float(\"{:.2f}\".format(x))\n\n def convert_eval_format(self, all_bboxes):\n # import pdb; pdb.set_trace()\n detections = []\n for image_id in all_bboxes:\n for cls_ind in all_bboxes[image_id]:\n category_id = self._valid_ids[cls_ind - 1]\n for bbox in all_bboxes[image_id][cls_ind]:\n bbox[2] -= bbox[0]\n bbox[3] -= bbox[1]\n score = bbox[4]\n bbox_out = list(map(self._to_float, bbox[0:4]))\n\n detection = {\n \"image_id\": int(image_id),\n \"category_id\": int(category_id),\n \"bbox\": bbox_out,\n \"score\": float(\"{:.2f}\".format(score))\n }\n if len(bbox) > 5:\n extreme_points = list(map(self._to_float, bbox[5:13]))\n detection[\"extreme_points\"] = extreme_points\n detections.append(detection)\n return detections\n\n def convert_eval_circle_format(self, all_circles):\n # import pdb; pdb.set_trace()\n detections = []\n for image_id in all_circles:\n for cls_ind in all_circles[image_id]:\n try:\n category_id = self._valid_ids[cls_ind - 1]\n except:\n aaa =1\n for circle in all_circles[image_id][cls_ind]:\n score = circle[3]\n circle_out = list(map(self._to_float, circle[0:3]))\n\n detection = {\n \"image_id\": int(image_id),\n \"category_id\": int(category_id),\n \"score\": float(\"{:.2f}\".format(score)),\n 'circle_center': [circle_out[0], circle_out[1]],\n 'circle_radius': circle_out[2]\n }\n if len(circle) > 5:\n extreme_points = list(map(self._to_float, circle[5:13]))\n detection[\"extreme_points\"] = extreme_points\n\n # output_h = 512 # hard coded\n # output_w = 512 # hard coded\n # cp = [0, 0]\n # cp[0] = circle_out[0]\n # cp[1] = circle_out[1]\n # cr = circle_out[2]\n # if cp[0] - cr < 0 or cp[0] + cr > output_w:\n # continue\n # if cp[1] - cr < 0 or cp[1] + cr > output_h:\n # continue\n\n detections.append(detection)\n return detections\n\n def __len__(self):\n return self.num_samples\n\n def save_results(self, results, save_dir):\n json.dump(self.convert_eval_format(results),\n open('{}/results.json'.format(save_dir), 'w'))\n\n def run_eval(self, results, save_dir):\n # result_json = os.path.join(save_dir, \"results.json\")\n # detections = self.convert_eval_format(results)\n # json.dump(detections, open(result_json, \"w\"))\n self.save_results(results, save_dir)\n coco_dets = self.coco.loadRes('{}/results.json'.format(save_dir))\n coco_eval = COCOeval(self.coco, coco_dets, \"bbox\")\n coco_eval.evaluate()\n coco_eval.accumulate()\n coco_eval.summarize()\n\n\n def save_circle_results(self, results, save_dir):\n json.dump(self.convert_eval_circle_format(results),\n open('{}/results.json'.format(save_dir), 'w'))\n\n def run_circle_eval(self, results, save_dir):\n # result_json = os.path.join(save_dir, \"results.json\")\n # detections = self.convert_eval_format(results)\n # json.dump(detections, open(result_json, \"w\"))\n self.save_circle_results(results, save_dir)\n circle_dets = self.circle.loadRes('{}/results.json'.format(save_dir))\n # circle_eval = CIRCLEeval(self.circle, circle_dets, \"circle\")\n circle_eval = CIRCLEeval(self.circle, circle_dets, \"circle_box\")\n circle_eval.evaluate()\n circle_eval.accumulate()\n circle_eval.summarize()\n"
]
| [
[
"numpy.array",
"numpy.random.RandomState"
]
]
|
sukruc/parking-model | [
"82b9e5cb33095a82f86ac6a94da8865890ff6d01"
]
| [
"taxiv2.py"
]
| [
"from gym.envs.toy_text import TaxiEnv\nimport gym.envs.toy_text.discrete as discrete\nimport numpy as np\nimport json\n\nMAP = [\n \"+---------+\",\n \"|R: | : :G|\",\n \"| : | : : |\",\n \"| : : : : |\",\n \"| | : | : |\",\n \"|Y| : |B: |\",\n \"+---------+\",\n]\n\nclass TaxiEnvTr(TaxiEnv):\n def decode(self, i):\n out = []\n out.append(i % 4)\n i = i // 4\n out.append(i % 5)\n i = i // 5\n out.append(i % 5)\n i = i // 5\n out.append(i)\n # assert 0 <= i < 5\n return reversed(out)\n\n def __init__(self, rowreps=1, colreps=1):\n self.desc = np.asarray(MAP, dtype='c')\n\n self.desc = np.concatenate([self.desc[:,:3], np.repeat(self.desc[:, 3:5], colreps, axis=1), self.desc[:,5:]], axis=1)\n self.desc = np.concatenate([self.desc[:2,:], np.repeat(self.desc[2:3, :], rowreps, axis=0), self.desc[3:,:]], axis=0)\n\n self.rowreps = rowreps\n num_rows = self.desc.shape[0] - 2\n num_columns = self.desc.shape[1] - 6 - (colreps - 1) * 2\n\n self.num_rows = num_rows\n self.num_cols = num_columns\n\n num_states = num_rows * num_columns * 5 * 4\n max_row = num_rows - 1\n self.locs = locs = [(0, 0), (0, 4), (max_row, 0), (max_row, 3)]\n max_col = num_columns - 1\n initial_state_distrib = np.zeros(num_states)\n num_actions = 6\n P = {state: {action: []\n for action in range(num_actions)} for state in range(num_states)}\n for row in range(num_rows):\n for col in range(num_columns):\n for pass_idx in range(len(locs) + 1): # +1 for being inside taxi\n for dest_idx in range(len(locs)):\n state = self.encode(row, col, pass_idx, dest_idx)\n if pass_idx < 4 and pass_idx != dest_idx:\n initial_state_distrib[state] += 1\n for action in range(num_actions):\n # defaults\n new_row, new_col, new_pass_idx = row, col, pass_idx\n reward = -1 # default reward when there is no pickup/dropoff\n done = False\n taxi_loc = (row, col)\n\n if action == 0:\n new_row = min(row + 1, max_row)\n elif action == 1:\n new_row = max(row - 1, 0)\n try:\n self.desc[1 + row, 2 * col + 2]\n except IndexError as e:\n print(2 * col + 2)\n raise e\n if action == 2 and self.desc[1 + row, 2 * col + 2] == b\":\":\n new_col = min(col + 1, max_col)\n elif action == 3 and self.desc[1 + row, 2 * col] == b\":\":\n new_col = max(col - 1, 0)\n elif action == 4: # pickup\n if (pass_idx < 4 and taxi_loc == locs[pass_idx]):\n new_pass_idx = 4\n else: # passenger not at location\n reward = -10\n elif action == 5: # dropoff\n if (taxi_loc == locs[dest_idx]) and pass_idx == 4:\n new_pass_idx = dest_idx\n done = True\n reward = 20\n elif (taxi_loc in locs) and pass_idx == 4:\n new_pass_idx = locs.index(taxi_loc)\n else: # dropoff at wrong location\n reward = -10\n new_state = self.encode(\n new_row, new_col, new_pass_idx, dest_idx)\n P[state][action].append(\n (1.0, new_state, reward, done))\n initial_state_distrib /= initial_state_distrib.sum()\n discrete.DiscreteEnv.__init__(\n self, num_states, num_actions, P, initial_state_distrib)\n\n @property\n def transition(self):\n initial_state = self.s\n tr = np.zeros((self.nA, self.nS, self.nS))\n\n done_states = []\n\n for a in range(self.nA):\n for s in range(self.nS):\n self.s = s\n sp, reward, done, proba = self.step(a)\n if done:\n done_states.append(sp)\n tr[a, s, sp] = proba['prob']\n\n for s in done_states:\n for a in range(self.nA):\n tr[a, s] = np.zeros(self.nS)\n tr[a, s, s] = 1.0\n\n self.s = initial_state\n return tr\n\n @property\n def rewards(self):\n rw = np.zeros((self.nA, self.nS, self.nS))\n\n for a in range(6):\n for s in range(self.nS):\n self.s = s\n sp, reward, done, proba = self.step(a)\n rw[a, s, sp] = reward\n self.reset()\n return rw\n\n def export_config(self, filename):\n with open(filename, 'w') as f:\n json.dump({'rowreps': self.rowreps}, f)\n\n @classmethod\n def from_config(cls, filename):\n with open(filename) as f:\n params = json.load(f)\n return cls(**params)\n"
]
| [
[
"numpy.asarray",
"numpy.repeat",
"numpy.zeros"
]
]
|
lsauthie/nlp | [
"8d562443ffea056a66db6ec87823d9198255884d"
]
| [
"main.py"
]
| [
"import cosine\nimport fileprocessing\nimport re\nimport sys\nimport colorama\nfrom colorama import Fore, Style\ncolorama.init()\n\ncfg_data = fileprocessing.read_json() #get config data\nlist_q = []\nlist_db = []\n\ncosine_ob = '' #initiate the object\n\n#h_p_x functions should help printing information in the console\n#this is a simple helper function to print textual information - improve standardization\ndef h_p_console(level, txt=''):\n #level 1 - information\n if level == 1:\n print('')\n print(\"{:-^40s}\".format(txt), end='\\n\\n')\n #level 2 - error\n elif level == 2:\n print(\"ERROR >> \" + txt, end='\\n\\n')\n elif level == 3:\n print(\"INFO >> \" + txt, end='\\n\\n')\n \n\n#print colored keywords [] out of s string\ndef h_p_color(string, keywords): \n \n #note: because the word is replaced with the input from user, we might lost upper case if the word is at beginning of the sentence\n for k in keywords:\n insensitive_k = re.compile(re.escape(k), re.IGNORECASE)\n string = insensitive_k.sub(Fore.YELLOW+k+Style.RESET_ALL, string)\n \n return string\n \n# --- end h_p_x functions ---\n \n \n#using keywords to identify the best suitable answer - if inexistant return ''\n#origin defined from where the function has been called (standalone or extended)\ndef sys_exit():\n print('')\n h_p_console(1,\"Goodbye !!!\")\n fileprocessing.write_json(cfg_data)\n sys.exit()\n \ndef search_keyWords(origin='extended'):\n \n input_list = [x[1] for x in list_db]\n \n list_keywords = []\n \n keyword = 'start'\n while keyword != '':\n if origin != 'standalone':\n h_p_console(1, \"2.2.0 Cumulative Keywords Search\")\n h_p_console(3, \"Nbre of answers found in DB: \" + str(len(input_list)))\n keyword = input(\"Please enter \\n[Keyword(s)] you want to search - use ',' as separator \\n[b] Back to main menu \\n[s] New search \\n[Inx] If for a prefered option:\\n>> \")\n \n if keyword == 'b':\n h_p_console(1)\n return ''\n else:\n h_p_console(1, \"Cumulative Keywords Search\")\n h_p_console(3, \"Nbre of answers found in DB: \" + str(len(input_list)))\n keyword = input(\"Please enter \\n[Keyword(s)] you want to search - use ',' as separator \\n[s] New search \\n>> \")\n \n\n if keyword == 's':\n list_keywords = []\n input_list = [x[1] for x in list_db]\n elif len(keyword) < 3 or not keyword:\n try:\n if not keyword:\n h_p_console(1)\n return ''\n else:\n h_p_console(1)\n return input_list[int(keyword)]\n except:\n h_p_console(2,\"Please make sure to enter a digit which is smaller than 3 char or a keyword bigger than 3 char\")\n pass\n else:\n k = keyword.split(',')\n list_keywords += k\n for i in k:\n input_list = [x for x in input_list if i.lower() in x.lower()] #look for the keyword in the response\n \n input_list = list(set(input_list))\n \n h_p_console(1)\n print(\"Number of instances found: \" + str(len(input_list)))\n \n if len(input_list) > 0:\n \n if len(input_list) > int(cfg_data['default']['nb_responses_displayed']): #if less than 5 - is automatically displayed\n output = input(\"How many instances do you want to print out: \")\n if not output:\n output = '1'\n print('')\n else:\n output = '5'\n \n try:\n output = int(output)\n except:\n h_p_console(2, \"No digit - apply 1 per default\")\n output = '1'\n \n h_p_console(1)\n for ix, i in enumerate(input_list[:int(output)]):\n print(Fore.CYAN+\"{:4s} {:<2d}\".format('Inx', ix)+Style.RESET_ALL)\n print(\"{:<s}\".format( h_p_color(i, list_keywords)))\n \n else:\n input_list = [x[1] for x in list_db]\n\n\ndef initial_run(model='jaccard'):\n \n global cosine_ob\n \n cosine_ob = cosine.Cosine(list_db, model) \n output_list = [] #[[q_id, q, ratio, db.q, db.a]] - focus only on the best ratio\n \n try:\n list_q = fileprocessing.read_csv(cfg_data['default']['questions_name'])\n except:\n h_p_console(2, \"The file with questions does not exist - please refer to documentation\")\n sys_exit()\n \n #for q in list_q[:2]: #[:2] used to limit the number of iteration while testing\n for q_id, q in enumerate(list_q):\n question = q[0]\n #best_fit = compare.run_check(question,list_db) #[[r,q,a],[r,q,a]]\n best_fit = cosine_ob.main(question) #[[r,q,a],[r,q,a]]\n output_list.append([q_id]+[question]+best_fit[0]) #only the best ratio - this part should be adapted if we want all result\n q_id += 1 #to define the id of the question\n \n fileprocessing.write_csv(cfg_data['default']['output_name'],output_list)\n\n#used to pass a question\ndef manual_run(question):\n \n global cosine_ob\n \n if cosine_ob == '': #object not initialized\n cosine_ob = cosine.Cosine(list_db) \n \n best_fit = cosine_ob.main(question, 5)\n #best_fit = compare.run_check(question,list_db, 5) #[[r,q,a],[r,q,a]]\n \n h_p_console(1, \"5 Alternatives\")\n \n for ix, i in enumerate(best_fit): #index is used to make choice possible\n print(Fore.CYAN+\"{:4s} {:<2d} - {:4s} {:.2f}\".format('Inx', ix, 'Ratio', i[0])+Style.RESET_ALL)\n print('Q_db: ' + i[1])\n print('R_db: ' + i[2])\n \n return best_fit\n \ndef work_on_result():\n \n load_output = fileprocessing.read_csv(cfg_data['default']['output_name']) #[[q_id, q, ratio, db.q, db.a]]\n\n #compute ratio distribution\n list_r = []\n [list_r.append(float(x[2])) for x in load_output]\n \n import numpy as np\n h_p_console(1, \"Percentiles\")\n print(\"Percentiles: 10, 25, 50, 75, 90 >> \", end='')\n print([round(x,2) for x in np.percentile(list_r, [10, 25, 50, 75, 90])])\n\n ratio_min = input(\"All questions with a ratio < [x.yy] will be manually reviewed: \")\n h_p_console(1)\n if not ratio_min:\n ratio_min = 1\n\n #work on questions with a ratio smaller than the one which is defined\n while True:\n \n nb_questions = 0\n for inx, x in enumerate(load_output): #Note: we nmust loop on all elements because we are modifying the list\n if float(x[2]) <= float(ratio_min):\n nb_questions += 1\n print(\"{:4s} {:<4d} - {:4s} {:.2f}\".format('Q_id', inx, 'Ratio', float(x[2])))\n print('Q_in: ' + x[1])\n print('Q_db: ' + x[3], end='\\n\\n')\n \n if nb_questions == 0:\n break\n\n inx_digit = False\n question_context = ''\n while not inx_digit:\n h_p_console(1)\n inx = input(\"Enter the [Q_id] for a manual search: \")\n question_context = str(inx)\n try:\n inx = int(inx)\n break\n except:\n print(\"Please enter a digit\")\n\n while True:\n h_p_console(1, '2.0.0 Manual Search')\n print(\"QUESTION: \" + load_output[inx][1], end='\\n\\n')\n choice = input('[0] - Look for 5 different alternatives \\n[1] - Enter a modified question manually \\n[2] - Key words search in response:\\n[3] - Back to list of questions\\n>> ')\n\n if choice == '0':\n #look for 5 different alternatives\n #inx = 0\n h_p_console(1,\"2.1.0 Existing Alternatives\")\n new_search = manual_run(load_output[inx][1]) #[[r,q,a],[r,q,a]]\n\n input_res = input(\"Choose the best option [inx] or [n] for a new search: \")\n #input_res = 1\n if input_res == 'n' or not input_res:\n pass\n else:\n input_res = int(input_res)\n oa = load_output[inx] #[q_id, q, ratio, db.q, db.a] - extract from load_output\n oa[2] = '1'\n oa[3] = new_search[input_res][1]\n oa[4] = new_search[input_res][2]\n\n fileprocessing.write_csv(cfg_data['default']['output_name'],load_output) #save a copy\n break #exit the while\n\n elif choice == '1':\n #add the question manually and complete same process - replace the q_db question with this one in the file\n h_p_console(1,\"2.2.0 Manual Question\")\n question_manual = input(\"Please enter the question manually, 5 alternatives will be looked up: \")\n new_search = manual_run(question_manual) #[[r,q,a],[r,q,a]]\n\n input_res = input(\"Choose the best option [inx] or [n] for a new search: \")\n #input_res = 1\n if input_res == 'n' or not input_res:\n pass\n else:\n input_res = int(input_res)\n oa = load_output[inx] #[q_id, q, ratio, db.q, db.a] - extract from load_output\n oa[2] = '1'\n oa[3] = new_search[input_res][1]\n oa[4] = new_search[input_res][2]\n\n fileprocessing.write_csv(cfg_data['default']['output_name'],load_output) #save a copy\n break #exit the while\n\n elif choice == '2':\n #look for an answer using key words\n response = search_keyWords() #db.q, db.a\n if response == '':\n pass\n else:\n oa = load_output[inx] #[q_id, q, ratio, db.q, db.a] - extract from load_output\n oa[2] = '1'\n oa[3] = oa[1] #in this case we don't consider the db.q as we are looking directly for the response, furthermore the db as more than one question for a same answer\n oa[4] = response\n\n fileprocessing.write_csv(cfg_data['default']['output_name'],load_output) #save a copy\n break\n else:\n break\n\ntry:\n list_db = fileprocessing.read_csv(cfg_data['default']['db_name'])\nexcept Exception as e:\n h_p_console(2, str(e))\n sys_exit()\n\n#Can be commented depending on what we want to do\n\nh_p_console(1,\"Welcome\")\n\nwith_model = input(\"Do you want to:\\n[1] - Use Jaccard on keywords \\n[2] - Use true similarity on keywords \\n>> \")\nif with_model == '1':\n model = 'jaccard'\nelse:\n model = 'cosine'\n\nh_p_console(3, 'The script will run using: ' + model)\n \ntry:\n \n to_do = input(\"Do you want to:\\n[1] - Run the script to find the best options \\n[2] - Refine the result \\n[3] - Both sequentially \\n[4] - Search responses with keywords\\n>> \")\n if to_do == '1':\n print(\"... Please be patient - it might take a while\", end='\\n\\n')\n initial_run(model)\n print(\"... Completed the script is closing\")\n elif to_do == '2':\n work_on_result()\n elif to_do == '4':\n search_keyWords('standalone')\n else:\n print(\"... Please be patient - it might take a while\", end='\\n\\n')\n initial_run(model)\n work_on_result()\n\nexcept KeyboardInterrupt:\n sys_exit()\n\nsys_exit()"
]
| [
[
"numpy.percentile"
]
]
|
thocoo/gamma-desk | [
"9cb63a65fe23e30e155b3beca862f369b7fa1b7e"
]
| [
"gdesk/panels/imgview/imgdata.py"
]
| [
"import pathlib\nimport collections\nimport queue\nimport threading\nimport math\n\nfrom qtpy import QtGui, QtCore\nfrom qtpy.QtGui import QImage\n\nimport numpy as np\n\nfrom ... import gui, config\n\nfrom ...utils.shared import SharedArray\nfrom ...utils import imconvert\n\nfrom .dimensions import DimRanges\nfrom . import fasthist\n\nhere = pathlib.Path(__file__).absolute().parent\n\ntry:\n from .numba_func import map_values_mono, map_values_rgbswap, map_values_rgb\n has_numba = True\n \nexcept:\n has_numba = False\n\n#https://doc.qt.io/qtforpython-5.12/PySide2/QtGui/QPainter.html#composition-modes\nCOMPMODE = dict() \nCOMPMODE['sourceover'] = QtGui.QPainter.CompositionMode_SourceOver \nCOMPMODE['plus'] = QtGui.QPainter.CompositionMode_Plus \nCOMPMODE['multiply'] = QtGui.QPainter.CompositionMode_Multiply \nCOMPMODE['screen'] = QtGui.QPainter.CompositionMode_Screen \nCOMPMODE['overlay'] = QtGui.QPainter.CompositionMode_Overlay \nCOMPMODE['darken'] = QtGui.QPainter.CompositionMode_Darken \nCOMPMODE['lighten'] = QtGui.QPainter.CompositionMode_Lighten \n \nclass SelectRoi(DimRanges):\n\n \"\"\"\n Selection widget range data.\n \"\"\"\n\n def __init__(self, height, width, update_statistics_func=None):\n DimRanges.__init__(self, (height, width))\n self.update_statistics_func = update_statistics_func\n \n @property\n def yr(self):\n return self.rngs[0]\n \n @property \n def xr(self):\n return self.rngs[1]\n\n def ensure_rising(self):\n for rng in self.rngs:\n if rng.start > rng.stop:\n rng.start, rng.stop = rng.stop, rng.start\n\n def copy(self):\n s = SelectRoi(self.rngs[0].maxstop, self.rngs[1].maxstop)\n s.inherite(self)\n return s\n \n def update_statistics(self):\n if not self.update_statistics_func is None:\n self.update_statistics_func()\n \nclass ImageStatistics(object):\n\n def __init__(self):\n self._cache = dict()\n self.arr2d = None \n \n def attach_arr2d(self, arr2d):\n self.arr2d = arr2d\n self.clear()\n \n @property\n def dtype(self):\n return self.arr2d.dtype \n \n def clear(self):\n self._cache.clear()\n \n def step_for_bins(self, bins):\n if self.dtype in ['float16', 'float32', 'float64']:\n return math.ceil(65536 / bins) \n \n if len(self._cache.keys()) == 0:\n self.calc_histogram()\n \n hist1 = self._cache['hist'] \n \n return math.ceil(len(hist1) / bins)\n \n def histogram(self, step=1):\n if len(self._cache.keys()) == 0:\n self.calc_histogram()\n \n hist1 = self._cache['hist'] \n \n if step > 1:\n bins = len(hist1) // step\n left = len(hist1) % step\n tmp = hist1[:step*bins] \n if left > 0:\n hist = np.r_[tmp.reshape(bins, step).sum(1), hist1[step*bins:].sum()] \n else:\n hist = tmp.reshape(bins, step).sum(1)\n return hist\n else:\n return hist1 \n \n def starts(self, step=1):\n if len(self._cache.keys()) == 0:\n self.calc_histogram()\n \n starts1 = self._cache['starts'] \n\n if step > 1:\n return starts1[::step]\n else: \n return starts1 \n \n def calc_histogram(self, bins=None, step=None): \n if self.dtype in ['int8', 'uint8', 'int16', 'uint16']:\n hist, starts, stepsize = fasthist.hist16bit(self.arr2d, bins=None, step=1, use_numba=True)\n \n elif self.dtype in ['int32', 'uint32', 'float16', 'float32', 'float64']:\n hist, starts, stepsize = fasthist.histfloat(self.arr2d, bins=65536, step=None, pow2snap=False, use_numba=True)\n \n self._cache['hist'] = hist\n self._cache['starts'] = starts\n self._cache['stepsize'] = stepsize \n \n @property \n def bins(self):\n return len(self.starts())\n \n def stepsize(self, step):\n return self._cache['stepsize'] * step\n \n def n(self):\n return self.arr2d.shape[0] * self.arr2d.shape[1]\n \n def sum(self):\n return (self.histogram() * self.starts()).sum()\n \n def mean(self):\n return self.sum() / self.n()\n\n def sumsq(self):\n return (self.histogram() * self.starts()**2).sum()\n \n def min(self):\n non_zeros_indices = np.argwhere(self.histogram() > 0)\n min_index = non_zeros_indices[0][0]\n max_index = non_zeros_indices[-1][0]\n return self.starts()[min_index]\n \n def max(self):\n non_zeros_indices = np.argwhere(self.histogram() > 0)\n max_index = non_zeros_indices[-1][0]\n return self.starts()[max_index] \n \n def std(self):\n n = self.n()\n result = ((self.sumsq() - ((self.sum() * 1.0) ** 2) / n) / (n - 1)) ** 0.5\n return result\n \n\nclass ImageData(object): \n def __init__(self):\n self.qimg = None\n self.map8 = None\n self.sharray = None\n self.imghist = ArrayHistory(config['image'].get(\"history_size\", 500e6))\n \n arr = np.ones((1,1),'uint8') * 128\n self.selroi = SelectRoi(1, 1, self.update_roi_statistics)\n self.chanstats = dict()\n \n self.show_array(arr)\n self.layers = collections.OrderedDict()\n \n def load_by_qt(self, path):\n self.qimg = QImage(str(path))\n \n def show_array(self, array=None, black=0, white=256, colormap=None, gamma=1, log=True):\n with gui.qapp.waitCursor():\n threadcount = config['image']['threads'] \n use_numba = config['image']['numba'] and has_numba \n \n \n if array is None:\n #offset and gain adjust of current viewer\n array = self.sharray\n shape = array.shape\n dtype = array.dtype \n # for stat in self.chanstats.values():\n # stat.clear()\n \n else:\n if log and not self.sharray is None:\n self.imghist.push(self.sharray)\n \n if isinstance(array, SharedArray):\n self.sharray = array \n shape = array.shape\n dtype = array.dtype \n \n else:\n shape = array.shape\n dtype = array.dtype\n self.sharray = SharedArray(shape, dtype)\n self.sharray[:] = array \n\n self.chanstats.clear()\n \n if len(shape) == 2:\n self.chanstats['K'] = ImageStatistics()\n self.chanstats['K'].attach_arr2d(self.sharray.ndarray)\n self.chanstats['RK'] = ImageStatistics()\n self.update_roi_statistics()\n else:\n self.chanstats['R'] = ImageStatistics()\n self.chanstats['G'] = ImageStatistics()\n self.chanstats['B'] = ImageStatistics()\n self.chanstats['R'].attach_arr2d(self.sharray.ndarray[:,:,0])\n self.chanstats['G'].attach_arr2d(self.sharray.ndarray[:,:,1])\n self.chanstats['B'].attach_arr2d(self.sharray.ndarray[:,:,2]) \n self.chanstats['RR'] = ImageStatistics()\n self.chanstats['RG'] = ImageStatistics()\n self.chanstats['RB'] = ImageStatistics()\n self.update_roi_statistics()\n \n height, width, *ignore = shape\n self.height, self.width = height, width\n \n if self.selroi.isfullrange():\n self.selroi.xr.maxstop = width\n self.selroi.yr.maxstop = height\n self.selroi.reset()\n else:\n self.selroi.xr.maxstop = width\n self.selroi.yr.maxstop = height\n self.selroi.clip()\n \n natrange = imconvert.natural_range(self.sharray.dtype) \n gain = natrange / (white - black)\n self.array8bit, self.qimg = imconvert.process_ndarray_to_qimage_8bit(\n self.sharray.ndarray, black, gain, colormap, refer=True, shared=config[\"image\"].get(\"qimg_shared_mem\", False),\n gamma=gamma)\n \n def get_natural_range(self):\n return imconvert.natural_range(self.sharray.dtype)\n \n def set_mask(self, array=None, composition='sourceover'):\n self.set_layer('mask', array, composition) \n \n def set_layer(self, name, array=None, composition='sourceover'):\n if array is None:\n if name in self.layers.keys():\n self.layers.pop(name)\n return\n \n assert array.ndim == 2\n assert array.dtype in ['uint8', 'bool']\n \n height, width = array.shape\n \n compmode = COMPMODE[composition.lower()]\n \n qimage = QImage(memoryview(array), width, height, width, QImage.Format_Indexed8)\n qimage.setColorTable(imconvert.make_color_table('mask'))\n self.layers[name] = {'array': array, 'qimage': qimage, 'composition': compmode} \n \n \n def update_roi_statistics(self):\n slices = self.selroi.getslices()\n \n clr_slices = {'RK': slices,\n 'RR': (slices[0], slices[1], 0),\n 'RG': (slices[0], slices[1], 1),\n 'RB': (slices[0], slices[1], 2)}\n \n for clr, chanstat in self.chanstats.items(): \n if not clr in clr_slices.keys(): continue\n chanstat.attach_arr2d(self.sharray[clr_slices[clr]])\n\n def update_array8bit_by_slices(self, slices):\n def takemap(source_slice, target_slice):\n self.array8bit[target_slice] = np.take(self.map8, self.sharray.ndarray[source_slice])\n \n threads = []\n for (source_slice, target_slice) in slices:\n threads.append(threading.Thread(target=takemap, args=(source_slice, target_slice)))\n \n for thread in threads:\n thread.start()\n \n for thread in threads:\n thread.join() \n \n @property\n def statarr(self):\n return self.sharray.ndarray \n \n def get_number_of_bytes(self): \n nbytes = 0\n nbytes += self.sharray.ndarray.nbytes\n nbytes += self.array8bit.nbytes\n return nbytes\n \nclass ArrayHistory(object):\n\n def __init__(self, max_size=4):\n self.max_size = max_size \n self.prior_arrays = []\n self.next_arrays = []\n \n def push(self, array):\n overflow = self.reduce_size_to_max_byte_size(array.size) \n if overflow < 0: self.prior_arrays.append(array)\n self.next_arrays.clear() \n \n def reduce_size_to_max_byte_size(self, add_size=0):\n current_size = add_size\n for arr in self.prior_arrays:\n current_size += arr.size\n overflow = current_size - self.max_size\n while overflow > 0 and len(self.prior_arrays) > 0:\n array = self.prior_arrays.pop(0) \n overflow -= array.size \n return overflow\n \n def prior(self, current_array):\n array = self.prior_arrays.pop(-1)\n self.next_arrays.append(current_array)\n return array\n \n def next(self, current_array):\n array = self.next_arrays.pop(-1)\n self.prior_arrays.append(current_array) \n return array\n \n def __len__(self):\n return len(self.prior_arrays) + len(self.next_arrays)\n \n def prior_length(self):\n return len(self.prior_arrays)\n \n def next_length(self):\n return len(self.next_arrays) \n \n def clear(self):\n self.stack.clear()"
]
| [
[
"numpy.take",
"numpy.ones"
]
]
|
NHPatterson/napari | [
"d8d2d301b71af79a6a7f8f4bafa1f55a18317dd8"
]
| [
"napari/_vispy/experimental/tiled_image_visual.py"
]
| [
"\"\"\"TiledImageVisual class\n\nA visual that draws tiles using a texture atlas.\n\nUltimately TiledImageVisual cannot depend on OctreeChunk. And Octree\ncode should not depend on TiledImageVisual! So there really can be\nno class or named tuple that gets passed between them.\n\nInstead, we'll probably just have a function signature that takes things\nlike the pos, size and depth of each tile as separate arguments. But\nfor now both do depend on OctreeChunk.\n\"\"\"\nfrom typing import List, Set\n\nimport numpy as np\n\nfrom napari.layers.image.experimental.octree_chunk import OctreeChunkKey\n\nfrom ...layers.image.experimental import OctreeChunk\nfrom ..vendored import ImageVisual\nfrom ..vendored.image import _build_color_transform\nfrom .texture_atlas import TextureAtlas2D\nfrom .tile_set import TileSet\n\n# Shape of she whole texture in tiles. Hardcode for now. TODO_OCTREE: We\n# need to calculate this based on the tile size. Long term we want to\n# support huge tiles. So big images/layers will get small tiles, but if the\n# layer can fit within the max texture size, as big as (16384, 16384) then\n# it might be more efficient to put that in a single tiles.\n#\n# This might require that we can support multiple TexturAtlas2D objects\n# per TiledImageVisual.\nSHAPE_IN_TILES = (16, 16)\n\n\nclass TiledImageVisual(ImageVisual):\n \"\"\"An image that is drawn using one or more chunks or tiles.\n\n A regular ImageVisual is a single image drawn as a single rectangle\n with a single texture. A tiled TiledImageVisual also has a single\n texture, but that texture is a TextureAtlas2D.\n\n A texture atlas is basically a single texture that contains smaller\n textures within it, like a quilt. In our cases the smaller textures are\n all the same size, for example (256, 256). For example a 4k x 4k\n texture can hold 256 different (256, 256) tiles.\n\n When the TiledImageVisual draws, it draws a single list of quads. Each\n quad's texture coordinates potentially refers to a different texture in\n the atlas.\n\n The quads can be located anywhere, even in 3D. TiledImageVisual does\n not know if it's drawing an octree or a grid, or just a scatter of tiles.\n A key point is while the texture tiles are all the same size, the quads\n can all be different sizes.\n\n For example, one quad might have a (256, 256) texture, but it's\n physically tiny on the screen. While the next quad is also showing a\n (256, 256) texture, it has to be, but that quad is really big on that\n same screen. This ability to have different size quads comes in handy\n for octree rendering, where we often draw chunks from multiple levels\n of the octree at the same time.\n\n Adding or removing tiles from a TiledImageVisual is efficient. Only the\n bytes in the the tile(s) being updated are sent to the card. The Vispy\n method BaseTexture.set_data() has an \"offset\" argument. When setting\n texture data with an offset under the hood Vispy calls\n glTexSubImage2D(). It will only update the rectangular region within\n the texture that's being updated. This is critical to making\n TiledImageVisual efficient.\n\n In addition, uploading new tiles does not cause the shader to be\n rebuilt. This is another reason TiledImageVisual is faster than\n creating a stand-alone ImageVisuals, where each new ImageVisual results\n in a shader build today. If that were fixed TiledImageVisual would\n still be faster, but the speed gap would be smaller.\n\n Parameters\n ----------\n tile_shape : np.ndarray\n The shape of one tile like (256, 256, 3).\n \"\"\"\n\n def __init__(self, tile_shape: np.ndarray, *args, **kwargs):\n self.tile_shape = tile_shape\n\n self._tiles: TileSet = TileSet() # The tiles we are drawing.\n\n self._clim = np.array([0, 1]) # TOOD_OCTREE: need to support clim\n\n # Initialize our parent ImageVisual.\n super().__init__(*args, **kwargs)\n\n # We must create the texture atlas *after* calling super().__init__\n # because super().__init__ creates self._interpolation which we\n # our _create_texture_atlas references.\n #\n # The unfreeze/freeze stuff is just a vispy thing to guard against\n # adding attributes after construction, which often leads to bugs,\n # so we have to toggle it off here. Not a big deal.\n self.unfreeze()\n self._texture_atlas = self._create_texture_atlas(tile_shape)\n self.freeze()\n\n def _create_texture_atlas(self, tile_shape: np.ndarray) -> TextureAtlas2D:\n \"\"\"Create texture atlas up front or if we change texture shape.\n\n Attributes\n ----------\n tile_shape : np.ndarray\n The shape of our tiles such as (256, 256, 4).\n\n Return\n ------\n TextureAtlas2D\n The newly created texture atlas.\n \"\"\"\n interp = 'linear' if self._interpolation == 'bilinear' else 'nearest'\n return TextureAtlas2D(tile_shape, SHAPE_IN_TILES, interpolation=interp)\n\n def set_data(self, image) -> None:\n \"\"\"Set data of the ImageVisual.\n\n VispyImageLayer._on_display_change calls this with an empty image, but\n we can just ignore it. When created we are \"empty\" by virtue of not\n drawing any tiles yet.\n \"\"\"\n\n def set_tile_shape(self, tile_shape: np.ndarray) -> None:\n \"\"\"Set the shape of our tiles.\n\n All tiles are the same shape in terms of texels. However they might\n be drawn different physical sizes. For example drawing a single\n view into a quadtree might end up drawing some tiles 2X or 4X\n bigger than others. Typically you want to draw the \"best available\"\n data which might be on a different level.\n\n Parameters\n ----------\n tile_shape : np.ndarray\n Our tiles shape like (256, 256, 4)\n \"\"\"\n\n # Clear all our previous tile information and set the new shape.\n self._tiles.clear()\n self.tile_shape = tile_shape\n\n # Create the new atlas and tell the shader about it.\n self._texture_atlas = self._create_texture_atlas(tile_shape)\n self._data_lookup_fn['texture'] = self._texture_atlas\n\n @property\n def size(self):\n # TODO_OCTREE: Who checks this? Need to compute the size...\n #\n # ImageVisual.size() does\n # return self._data.shape[:2][::-1]\n #\n # We don't have a self._data so what do we put here? Maybe need\n # a bounds for all the currently drawable tiles?\n # return self._texture_atlas.texture_shape[:2]\n #\n return (1024, 1024)\n\n @property\n def num_tiles(self) -> int:\n \"\"\"Return the number tiles currently being drawn.\n\n Return\n ------\n int\n The number of tiles currently being drawn.\n \"\"\"\n return self._texture_atlas.num_slots_used\n\n @property\n def octree_chunks(self) -> List[OctreeChunk]:\n \"\"\"Return data for the chunks we are drawing.\n\n List[OctreeChunk]\n The data for the chunks we are drawing.\n \"\"\"\n return self._tiles.chunks\n\n def add_chunks(self, chunks: List[OctreeChunk]) -> int:\n \"\"\"Any one or more chunks that we are not already drawing.\n\n Parameters\n ----------\n chunks : List[OctreeChunk]\n Chunks that we may or may not already be drawing.\n\n Return\n ------\n int\n The number of chunks that still need to be added.\n \"\"\"\n # Get only the new chunks, the ones we are not currently drawing.\n new_chunks = [\n octree_chunk\n for octree_chunk in chunks\n if not self._tiles.contains_octree_chunk(octree_chunk)\n ]\n\n # Add one or more of the new chunks.\n while new_chunks:\n self.add_one_chunk(new_chunks.pop(0)) # Add the first one.\n\n # In the future we might add several chunks here. We want\n # to add as many as we can without tanking the framerate\n # too much. But for now we just add one, because we\n # were seeing adding taking 40ms for one (256, 256) tile!\n #\n # But if that improves, we might want to multiple tiles here,\n # up to some budget limit. Although not the cost of adding\n # most happens later when glFlush() is called.\n break\n\n # Return how many chunks we did NOT add. The system should continue\n # to poll and draw until we return 0.\n return len(new_chunks)\n\n def add_one_chunk(self, octree_chunk: OctreeChunk) -> None:\n \"\"\"Add one chunk to the tiled image.\n\n Parameters\n ----------\n octree_chunk : OctreeChunk\n The chunk we are adding.\n\n Return\n ------\n int\n The newly added chunk's index.\n \"\"\"\n # Add to the texture atlas.\n atlas_tile = self._texture_atlas.add_tile(octree_chunk)\n\n if atlas_tile is None:\n # TODO_OCTREE: No slot was available in the atlas. That's bad,\n # but not sure what we should do in this case.\n return\n\n # Add our mapping between chunks and atlas tiles.\n self._tiles.add(octree_chunk, atlas_tile)\n\n # Call self._build_vertex_data() the next time we are drawn, so\n # can update things to draw this new chunk.\n self._need_vertex_update = True\n\n @property\n def chunk_set(self) -> Set[OctreeChunkKey]:\n \"\"\"Return the set of chunks we are drawing.\n\n Return\n ------\n Set[OctreeChunkKey]\n The set of chunks we are drawing.\n \"\"\"\n return self._tiles.chunk_set\n\n def prune_tiles(self, drawable_set: Set[OctreeChunk]) -> None:\n \"\"\"Remove tiles that are not part of the drawable set.\n\n drawable_set : Set[OctreeChunk]\n The set of currently drawable chunks.\n \"\"\"\n for tile_data in list(self._tiles.tile_data):\n if tile_data.octree_chunk not in drawable_set:\n # print(f\"REMOVE: {tile_data.octree_chunk}\")\n tile_index = tile_data.atlas_tile.index\n self._remove_tile(tile_index)\n\n def _remove_tile(self, tile_index: int) -> None:\n \"\"\"Remove one tile from the tiled image.\n\n Parameters\n ----------\n tile_index : int\n The tile to remove.\n \"\"\"\n try:\n self._tiles.remove(tile_index)\n self._texture_atlas.remove_tile(tile_index)\n\n # Must rebuild to remove this from what we are drawing.\n self._need_vertex_update = True\n except IndexError as exc:\n # Fatal error right now, but maybe in weird situation we should\n # ignore this error? Let's see when it happens.\n raise RuntimeError(f\"Tile index {tile_index} not found.\") from exc\n\n def _build_vertex_data(self) -> None:\n \"\"\"Build vertex and texture coordinate buffers.\n\n This overrides ImageVisual._build_vertex_data(), it is called from\n our _prepare_draw().\n\n This is the heart of tiled rendering. Instead of drawing one quad\n with one texture, we draw one quad per tile. And for each quad we\n set its texture coordinates so that it will pull from the right\n slot in the atlas.\n\n As the card draws the tiles, the locations it samples from the\n texture will hop around in the atlas texture.\n\n Today we only have one atlas texture, but in the future we might\n have multiple atlas textures. If so, we'll want to sort the quads\n to minimize the number of texture swaps. Sample from different\n tiles in one atlas texture is fast, but switching texture is\n slower.\n \"\"\"\n if len(self._tiles) == 0:\n return # Nothing to draw.\n\n verts = np.zeros((0, 2), dtype=np.float32)\n tex_coords = np.zeros((0, 2), dtype=np.float32)\n\n # TODO_OCTREE: We can probably avoid vstack here? Maybe create one\n # vertex buffer sized according to the max number of tiles we\n # expect? But grow it if we exceed our guess?\n for tile_data in self._tiles.tile_data_sorted:\n atlas_tile = tile_data.atlas_tile\n verts = np.vstack((verts, atlas_tile.verts))\n tex_coords = np.vstack((tex_coords, atlas_tile.tex_coords))\n\n # Set the base ImageVisual's _subdiv_ buffers. ImageVisual has two\n # modes: imposter and subdivision. So far TiledImageVisual\n # implicitly is always in subdivision mode. Not sure if we'd ever\n # support imposter, or if that even makes sense with tiles?\n self._subdiv_position.set_data(verts)\n self._subdiv_texcoord.set_data(tex_coords)\n self._need_vertex_update = False\n\n def _build_texture(self) -> None:\n \"\"\"Override of ImageVisual._build_texture().\n\n TODO_OCTREE: This needs work. Need to do the clim stuff in in the\n base ImageVisual._build_texture but do it for each tile?\n \"\"\"\n self._clim = np.array([0, 1])\n\n self._texture_limits = np.array([0, 1]) # hardcode\n self._need_colortransform_update = True\n\n self._need_texture_upload = False\n\n def _prepare_draw(self, view) -> None:\n \"\"\"Override of ImageVisual._prepare_draw()\n\n TODO_OCTREE: See how much this changes from base class, if we can\n avoid too much duplication. Or factor out some common methods.\n \"\"\"\n if self._need_interpolation_update:\n # Call the base ImageVisual._build_interpolation()\n self._build_interpolation()\n\n # But override to use our texture atlas.\n self._data_lookup_fn['texture'] = self._texture_atlas\n\n # We call our own _build_texture\n if self._need_texture_upload:\n self._build_texture()\n\n # TODO_OCTREE: how does colortransform change for tiled?\n if self._need_colortransform_update:\n prg = view.view_program\n grayscale = len(self.tile_shape) == 2 or self.tile_shape[2] == 1\n self.shared_program.frag[\n 'color_transform'\n ] = _build_color_transform(\n grayscale, self.clim_normalized, self.gamma, self.cmap\n )\n self._need_colortransform_update = False\n prg['texture2D_LUT'] = (\n self.cmap.texture_lut()\n if (hasattr(self.cmap, 'texture_lut'))\n else None\n )\n\n # We call our own _build_vertex_data()\n if self._need_vertex_update:\n self._build_vertex_data()\n\n # Call the normal ImageVisual._update_method() unchanged.\n if view._need_method_update:\n self._update_method(view)\n"
]
| [
[
"numpy.array",
"numpy.zeros",
"numpy.vstack"
]
]
|
OSUKED/Outage-Watch | [
"d2795e3ef971c906e633cf669c2d70511349a1aa"
]
| [
"outage/retrieval.py"
]
| [
"# AUTOGENERATED! DO NOT EDIT! File to edit: nbs/01-retrieval.ipynb (unless otherwise specified).\n\n__all__ = ['extract_ukpn_single_incident_ids', 'extract_ukpn_multiple_incident_urls',\n 'extract_ukpn_multiple_incident_ids', 'get_ukpn_incident_detail_url', 'is_ukpn_incident_active',\n 'extract_ukpn_relevant_info', 'get_ukpn_incidents_info', 'save_json_data', 'clean_ssen_incident_info',\n 'get_ssen_incidents_info', 'get_wpd_incident_feed', 'wpd_incident_id_to_url', 'clean_wpd_incident_info',\n 'get_wpd_incidents_info', 'get_raw_sp_incidents_info', 'clean_sp_incident_info', 'get_sp_incidents_info',\n 'get_np_auth', 'get_np_r', 'get_np_raw_incidents_info', 'clean_np_incident_info', 'get_np_incidents_info',\n 'get_enw_incidents_page', 'check_num_results', 'get_enw_raw_incidents', 'clean_enw_incident_info',\n 'get_enw_incidents_info']\n\n# Cell\nimport json\nimport pandas as pd\n\nimport html\nimport requests\nfrom bs4 import BeautifulSoup as bs\nfrom warnings import warn\n\nfrom ipypb import track\n\n# Cell\nextract_ukpn_single_incident_ids = lambda r_json: [\n incident['PanelContentUrl'].split('incidentId=')[1]\n for incident\n in r_json['Incidents']\n if incident['PowerCutType'] != 'Multiple'\n]\n\n# Cell\nextract_ukpn_multiple_incident_urls = lambda r_json: [\n f\"https://www.ukpowernetworks.co.uk{incident['PanelContentUrl']}\"\n for incident\n in r_json['Incidents']\n if incident['PowerCutType'] == 'Multiple'\n]\n\n# Cell\ndef extract_ukpn_multiple_incident_ids(multiple_incident_urls):\n incident_ids = []\n\n for multiple_incident_url in track(multiple_incident_urls, label='Multiple Ids'):\n r = requests.get(multiple_incident_url)\n soup = bs(r.text, features='lxml')\n\n incident_ids += [\n link['data-url'].split('incidentId=')[1]\n for link\n in soup.find('div', {'class': 'multiple-incidents--wrapper mb-4'}).findAll('a')\n ]\n\n return incident_ids\n\n# Cell\nget_ukpn_incident_detail_url = lambda incident_id: f'https://www.ukpowernetworks.co.uk/Incidents/getincidentdetails?incidentid={incident_id}'\n\n# Cell\ndef is_ukpn_incident_active(r_json):\n if r_json['PowerCutType'] == 'Restored':\n return False\n else:\n return True\n\ndef extract_ukpn_relevant_info(r_json):\n incident_info = {\n 'incident_active': is_ukpn_incident_active(r_json),\n 'restored_time': r_json['UKPNIncident']['RestoredDateTime'],\n 'estimated_restored_time': r_json['UKPNIncident']['EstimatedRestorationDate'],\n 'planned_time': r_json['UKPNIncident']['PlannedDate'],\n 'received_time': r_json['UKPNIncident']['ReceivedDate'],\n 'postcodes_impacted': r_json['FullPostcodeData'],\n 'description': r_json['IncidentCategoryCustomerFriendlyDescription'],\n 'incident_url': f\"https://www.ukpowernetworks.co.uk/power-cut/map?incidentid={r_json['IncidentReference']}\"\n }\n\n return incident_info\n\n# Cell\ndef get_ukpn_incidents_info(incidents_url='https://www.ukpowernetworks.co.uk/Incidents/GetIncidents'):\n r_json = requests.get(incidents_url).json()\n\n incident_ids = (\n extract_ukpn_single_incident_ids(r_json) +\n extract_ukpn_multiple_incident_ids(extract_ukpn_multiple_incident_urls(r_json))\n )\n\n raw_incidents_info = dict()\n cleaned_incidents_info = dict()\n\n for incident_id in track(incident_ids, label='Details'):\n try:\n incident_detail_url = get_ukpn_incident_detail_url(incident_id)\n r_json = requests.get(incident_detail_url).json()\n\n raw_incidents_info[incident_id] = r_json\n cleaned_incidents_info[incident_id] = extract_ukpn_relevant_info(r_json)\n except:\n warn(f'Failed to retrieve incident details for: {incident_id}')\n\n return raw_incidents_info, cleaned_incidents_info\n\n# Cell\ndef save_json_data(data, filename, data_dir='data/raw'):\n with open(f'{data_dir}/{filename}.json', 'w') as fp:\n json.dump(data, fp)\n\n# Cell\ndef clean_ssen_incident_info(incident):\n ssen_relevant_dates_name_mapping = {\n 'LoggedAtUtc': 'received_time',\n 'EstimatedArrivalOnSiteUtc': 'estimated_arrival_time',\n 'EstimatedRestorationTimeUtc': 'estimated_restored_time'\n }\n\n cleaned_incident_info = dict()\n\n for old_dt_name, new_dt_name in ssen_relevant_dates_name_mapping.items():\n dt_str = incident[old_dt_name].split('(')[1].split(')')[0]\n\n if dt_str != '-62135596800000':\n try:\n dt = pd.to_datetime(int(dt_str)*1e6).strftime('%Y-%m-%d %H:%M:%S')\n except:\n warn(f'{dt_str} could not be parsed to a date')\n dt = None\n else:\n dt = None\n\n cleaned_incident_info[new_dt_name] = dt\n\n cleaned_incident_info['postcodes_impacted'] = incident['AffectedAreas']\n cleaned_incident_info['description'] = incident['Message']\n cleaned_incident_info['incident_active'] = True\n cleaned_incident_info['incident_url'] = 'https://www.ssen.co.uk/Powertrack/'\n\n return cleaned_incident_info\n\n# Cell\ndef get_ssen_incidents_info(incidents_url='https://www.ssen.co.uk/Sse_Components/Views/Controls/FormControls/PowerTrackHandler.ashx'):\n raw_incidents_info = requests.get(incidents_url, verify=False).json()\n cleaned_incidents_info = dict()\n\n for incident in track(raw_incidents_info['Faults']):\n incident_ref = incident['Reference']\n cleaned_incidents_info[incident_ref] = clean_ssen_incident_info(incident)\n\n return raw_incidents_info, cleaned_incidents_info\n\n# Cell\ndef get_wpd_incident_feed(url='https://powercuts.westernpower.co.uk'):\n r = requests.get(url)\n\n raw_incidents_info = json.loads(html.unescape(r.text.split('data-ng-init=\"init(')[1].split(')\"></div><div id=\"powercuts\"')[0]))\n feed = json.loads(raw_incidents_info['feed'])\n\n return feed\n\n# Cell\nwpd_incident_id_to_url = lambda incident_id: f'https://powercuts.westernpower.co.uk/__powercuts/getIncidentById?incidentId={incident_id}'\n\n# Cell\ndef clean_wpd_incident_info(incident):\n wpd_relevant_dates_name_mapping = {\n 'startTime': 'received_time',\n 'etr': 'estimated_restored_time'\n }\n\n cleaned_incident_info = dict()\n\n for old_dt_name, new_dt_name in wpd_relevant_dates_name_mapping.items():\n if incident[old_dt_name] is not None:\n cleaned_incident_info[new_dt_name] = pd.to_datetime(incident[old_dt_name]*1e6).strftime('%Y-%m-%d %H:%M:%S')\n else:\n cleaned_incident_info[new_dt_name] = None\n\n cleaned_incident_info['postcodes_impacted'] = incident['postcodes']\n cleaned_incident_info['description'] = incident['status']\n cleaned_incident_info['incident_active'] = bool(1 - incident['restored'])\n cleaned_incident_info['incident_url'] = f\"https://powercuts.westernpower.co.uk/incident/{incident['id']}\"\n\n return cleaned_incident_info\n\n# Cell\ndef get_wpd_incidents_info(incidents_url='https://powercuts.westernpower.co.uk'):\n cleaned_incidents_info = dict()\n\n raw_incidents_info = get_wpd_incident_feed(incidents_url)\n incident_ids = [incident['id'] for incident in raw_incidents_info['incidents']]\n\n for incident_id in incident_ids:\n incident_url = wpd_incident_id_to_url(incident_id)\n r_json = requests.get(incident_url).json()\n cleaned_incidents_info[incident_id] = clean_wpd_incident_info(r_json)\n\n return raw_incidents_info, cleaned_incidents_info\n\n# Cell\nget_raw_sp_incidents_info = lambda sp_map_url='https://www.spenergynetworks.co.uk/pages/power_cuts_map.aspx': json.loads(requests.get(sp_map_url).text.split('arrPowercutsPostcodes:')[1].split('strPagePathListView')[0].replace(',\\r\\n', '').strip())\n\n# Cell\ndef clean_sp_incident_info(incident_info):\n def clean_dt(dt):\n if dt is None:\n return ''\n else:\n return pd.to_datetime(dt, utc=True).strftime('%Y-%m-%d %H:%M')\n\n cleaned_incident_info = dict()\n\n cleaned_incident_info['received_time'] = clean_dt(incident_info['CREATION_DATE'])\n cleaned_incident_info['estimated_restored_time'] = clean_dt(incident_info['EST_REST_DATE'])\n cleaned_incident_info['postcodes_impacted'] = incident_info['POSTCODES']\n cleaned_incident_info['description'] = incident_info['MAIN_MESSAGE']\n cleaned_incident_info['incident_active'] = incident_info['HISTORIC_FLAG'] is None\n cleaned_incident_info['incident_url'] = f\"https://www.spenergynetworks.co.uk/pages/power_cuts_map.aspx?incRef={incident_info['INCIDENT_REF']}\"\n\n return cleaned_incident_info\n\n# Cell\ndef get_sp_incidents_info(sp_map_url='https://www.spenergynetworks.co.uk/pages/power_cuts_map.aspx'):\n cleaned_incidents_info = dict()\n\n raw_incidents_info = get_raw_sp_incidents_info(sp_map_url)\n\n for incident_info in raw_incidents_info:\n incident_id = incident_info['INCIDENT_REF']\n cleaned_incidents_info[incident_id] = clean_sp_incident_info(incident_info)\n\n return raw_incidents_info, cleaned_incidents_info\n\n# Cell\ndef get_np_auth(np_main_url='https://www.northernpowergrid.com/power-cuts'):\n s = requests.Session()\n\n r = s.get(np_main_url)\n soup = bs(r.text, features='lxml')\n\n authenticity_token = soup.find('input', attrs={'name': 'authenticityToken'})['value']\n\n return s, authenticity_token\n\n# Cell\ndef get_np_r(\n np_main_url='https://www.northernpowergrid.com/power-cuts',\n np_incidents_url='https://www.northernpowergrid.com/powercutsgetallbyincno'\n):\n s, authenticity_token = get_np_auth(np_main_url)\n\n data = {\n 'method': 'incno',\n 'categoryFilters': 'Service Cutout Change,Asset repairs by Troublecall,Metering,Emergency Disconnection,Emergency Disconnection (Charge),Cat A,Cat B,Cat C',\n 'authenticityToken': authenticity_token\n }\n\n r = s.post(np_incidents_url, data=data)\n\n return r\n\n# Cell\nget_np_raw_incidents_info = lambda r: json.loads(r.json()['data'])\n\n# Cell\ndef clean_np_incident_info(incident_info):\n def clean_dt(dt):\n if dt is None:\n return ''\n else:\n return pd.to_datetime(dt, unit='ms').strftime('%Y-%m-%d %H:%M')\n\n cleaned_incident_info = dict()\n\n cleaned_incident_info['received_time'] = clean_dt(incident_info['logged'])\n cleaned_incident_info['estimated_restored_time'] = clean_dt(incident_info['estimatedTimeTillResolution'])\n cleaned_incident_info['postcodes_impacted'] = incident_info['postcodes']\n cleaned_incident_info['description'] = f\"{incident_info['category']} {incident_info['status']}\"\n cleaned_incident_info['incident_active'] = (incident_info['totalConfirmedOff'] + incident_info['totalPredictedOff']) > 0\n cleaned_incident_info['incident_url'] = 'https://www.northernpowergrid.com/power-cuts'\n\n return cleaned_incident_info\n\n# Cell\ndef get_np_incidents_info(\n np_main_url='https://www.northernpowergrid.com/power-cuts',\n np_incidents_url='https://www.northernpowergrid.com/powercutsgetallbyincno'\n):\n cleaned_incidents_info = dict()\n\n r = get_np_r(np_main_url, np_incidents_url)\n raw_incidents_info = get_np_raw_incidents_info(r)\n\n for incident_id, incident_info in raw_incidents_info['powercuts'].items():\n cleaned_incidents_info[incident_id] = clean_np_incident_info(incident_info)\n\n return raw_incidents_info, cleaned_incidents_info\n\n# Cell\nget_enw_incidents_page = lambda page=1, page_size=10000: f'https://www.enwl.co.uk/power-outages/search?pageSize={page_size}&postcodeOrReferenceNumber=&pageNumber={page}&includeCurrent=true&includeResolved=true&includeTodaysPlanned=true&includeFuturePlanned=true&includeCancelledPlanned=true'\n\n# Cell\ndef check_num_results(r_json):\n num_total_results = r_json['TotalResults']\n num_results_returned = len(r_json['Items'])\n\n if num_total_results != num_results_returned:\n warn(f'Only {num_results_returned} items were returned for Electricity North West when there are {num_total_results} in total')\n\n return\n\n# Cell\ndef get_enw_raw_incidents(page=1, page_size=10000):\n url = get_enw_incidents_page(page=page, page_size=page_size)\n raw_incidents = requests.get(url).json()\n check_num_results(raw_incidents)\n\n return raw_incidents\n\n# Cell\ndef clean_enw_incident_info(incident_info):\n def clean_dt(dt):\n if dt is None:\n return ''\n else:\n return pd.to_datetime(dt).strftime('%Y-%m-%d %H:%M')\n\n cleaned_incident_info = dict()\n cleaned_incident_info['received_time'] = clean_dt(incident_info['date'])\n cleaned_incident_info['estimated_restored_time'] = clean_dt(incident_info['estimatedTimeOfRestorationMajority'])\n cleaned_incident_info['postcodes_impacted'] = incident_info['AffectedPostcodes'].strip().split(', ')\n cleaned_incident_info['description'] = incident_info['AdditionalFaultInfo']\n cleaned_incident_info['incident_active'] = incident_info['FaultLabel'] in ['CurrentFault', 'Live power cut']\n cleaned_incident_info['incident_url'] = 'https://www.enwl.co.uk/power-cuts/power-cuts-power-cuts-live-power-cut-information-fault-list/fault-list'\n\n return cleaned_incident_info\n\n# Cell\ndef get_enw_incidents_info(page=1, page_size=10000):\n cleaned_incidents_info = dict()\n\n raw_incidents_info = get_enw_raw_incidents(page=page, page_size=page_size)\n\n for incident_info in raw_incidents_info['Items']:\n incident_id = incident_info['faultNumber']\n cleaned_incidents_info[incident_id] = clean_enw_incident_info(incident_info)\n\n return raw_incidents_info, cleaned_incidents_info"
]
| [
[
"pandas.to_datetime"
]
]
|
StanfordMSL/mslquad | [
"c319ecf4ba1063075221b67f12f4e017992f28fc"
]
| [
"mslquad/scripts/trajectory_path.py"
]
| [
"#!/usr/bin/env python\n\n#Sergio Esteban SURF 2019\n#takes waypoints from .CSV files and commands the drone\nimport rospy\nimport std_msgs.msg\nfrom geometry_msgs.msg import Pose, PoseStamped\nimport numpy as np\n\nclass Conductor:\n\n\tdef __init__(self):\n\t\trospy.init_node('poseConductor', anonymous=True)\n\t\tself.sub = rospy.Subscriber('mavros/local_position/pose',PoseStamped,self.poseCallBack)\n\t\tself.pub = rospy.Publisher('command/pose', Pose, queue_size = 10)\n\n\t\tself.file_path = rospy.get_param('~file_path')\n\t\tself.distTol = rospy.get_param('~tolerance')\n\t\tself.waypoints = np.loadtxt(open(self.file_path),delimiter=',',skiprows=1)\n\t\t\n\t\trospy.sleep(2)\n\n\tdef poseCallBack(self,message):\n\t\tself.pose = message.pose\n\n\tdef get_distance(self):\n\t\tcurrent = np.array((self.pose.position.x,\n\t\t\t\t \t\t self.pose.position.y,\n\t\t\t\t self.pose.position.z))\n\t\tdestination = np.array((self.goalPose.position.x,\n\t\t\t\t\t self.goalPose.position.y,\n\t\t\t\t\t self.goalPose.position.z))\n\t\treturn np.linalg.norm(destination - current)\n\n\tdef runPath(self):\n\t\twhile not rospy.is_shutdown():\n\t\t\tfor point in self.waypoints:\n\t\t\t\tself.goalPose = Pose()\n\t\t\t\tself.goalPose.position.x = point[0]\n\t\t\t\tself.goalPose.position.y = point[1]\n\t\t\t\tself.goalPose.position.z = point[2]\n\t\t\t\tself.next_target(self.goalPose)\n\n\tdef next_target(self,pose):\n\t\tself.pub.publish(pose)\n\n\t\tdistance = self.get_distance()\n\t\twhile distance > self.distTol:\n\t\t\tdistance = self.get_distance()\n\t\t\tprint(\"Pose: %.3f, %.3f, %.3f | Distance: %.3f\" % (self.pose.position.x,self.pose.position.y,self.pose.position.z,distance))\n\t\t\trospy.sleep(0.1)\n\n\t\tprint(\"\\t\\n*** Here at %.3f, %.3f, %.3f ***\\n\" % (pose.position.x,pose.position.y,pose.position.z))\n\t\trospy.sleep(1)\n\n\nif __name__ == '__main__':\n\n\tuser = \" \"\n\twhile user != \"x\":\n\t\tuser = raw_input(\"Enter 'x' to begin: \")\n\n\tconductor = Conductor()\n\tconductor.runPath()\n\n\n\n\n\n"
]
| [
[
"numpy.array",
"numpy.linalg.norm"
]
]
|
burkovae/evidently | [
"015f7d24811b956a624f379ebf2d58c98cc3b652"
]
| [
"src/evidently/analyzers/classification_performance_analyzer.py"
]
| [
"#!/usr/bin/env python\n# coding: utf-8\nfrom typing import List\nfrom typing import Optional\nfrom typing import Dict\n\nimport pandas as pd\nimport numpy as np\nfrom dataclasses import dataclass\nfrom sklearn import metrics\n\nfrom evidently import ColumnMapping\nfrom evidently.analyzers.base_analyzer import Analyzer\nfrom evidently.analyzers.base_analyzer import BaseAnalyzerResult\nfrom evidently.analyzers.utils import process_columns\nfrom evidently.analyzers.utils import calculate_confusion_by_classes\n\n\n@dataclass\nclass ConfusionMatrix:\n labels: List[str]\n values: list\n\n\n@dataclass\nclass PerformanceMetrics:\n \"\"\"Class for performance metrics values\"\"\"\n accuracy: float\n precision: float\n recall: float\n f1: float\n metrics_matrix: Dict[str, Dict]\n confusion_matrix: ConfusionMatrix\n confusion_by_classes: Dict[str, Dict[str, int]]\n\n\n@dataclass\nclass ClassificationPerformanceAnalyzerResults(BaseAnalyzerResult):\n reference_metrics: Optional[PerformanceMetrics] = None\n current_metrics: Optional[PerformanceMetrics] = None\n\n\ndef _calculate_performance_metrics(\n *, data: pd.DataFrame, target_column: str, prediction_column: str, target_names: List[str]\n) -> PerformanceMetrics:\n data.replace([np.inf, -np.inf], np.nan, inplace=True)\n data.dropna(axis=0, how='any', inplace=True)\n\n # calculate quality metrics\n accuracy_score = metrics.accuracy_score(data[target_column], data[prediction_column])\n avg_precision = metrics.precision_score(data[target_column], data[prediction_column], average='macro')\n avg_recall = metrics.recall_score(data[target_column], data[prediction_column], average='macro')\n avg_f1 = metrics.f1_score(data[target_column], data[prediction_column], average='macro')\n\n # calculate class support and metrics matrix\n metrics_matrix = metrics.classification_report(\n data[target_column],\n data[prediction_column],\n output_dict=True)\n\n # calculate confusion matrix\n confusion_matrix = metrics.confusion_matrix(data[target_column], data[prediction_column])\n labels = target_names if target_names else sorted(set(data[target_column]))\n confusion_by_classes = calculate_confusion_by_classes(confusion_matrix, labels)\n\n return PerformanceMetrics(\n accuracy=accuracy_score,\n precision=avg_precision,\n recall=avg_recall,\n f1=avg_f1,\n metrics_matrix=metrics_matrix,\n confusion_matrix=ConfusionMatrix(labels=labels, values=confusion_matrix.tolist()),\n confusion_by_classes=confusion_by_classes\n )\n\n\nclass ClassificationPerformanceAnalyzer(Analyzer):\n @staticmethod\n def get_results(analyzer_results) -> ClassificationPerformanceAnalyzerResults:\n return analyzer_results[ClassificationPerformanceAnalyzer]\n\n def calculate(self,\n reference_data: pd.DataFrame,\n current_data: Optional[pd.DataFrame],\n column_mapping: ColumnMapping) -> ClassificationPerformanceAnalyzerResults:\n if reference_data is None:\n raise ValueError('reference_data should be present')\n\n columns = process_columns(reference_data, column_mapping)\n result = ClassificationPerformanceAnalyzerResults(columns=columns)\n target_column = columns.utility_columns.target\n prediction_column = columns.utility_columns.prediction\n target_names = columns.target_names\n\n if target_column is not None and prediction_column is not None:\n result.reference_metrics = _calculate_performance_metrics(\n data=reference_data,\n target_column=target_column,\n prediction_column=prediction_column,\n target_names=target_names\n )\n\n if current_data is not None:\n result.current_metrics = _calculate_performance_metrics(\n data=current_data,\n target_column=target_column,\n prediction_column=prediction_column,\n target_names=target_names\n )\n\n return result\n"
]
| [
[
"sklearn.metrics.precision_score",
"sklearn.metrics.classification_report",
"sklearn.metrics.confusion_matrix",
"sklearn.metrics.f1_score",
"sklearn.metrics.recall_score",
"sklearn.metrics.accuracy_score"
]
]
|
RyanWangZf/pytorch-deeplab-xception | [
"d01239585e582e485c143694728ffb5c2c2f6eba"
]
| [
"data/construction/split_train_val.py"
]
| [
"import numpy as np\nimport os, pdb\nimport shutil\nnp.random.seed(2020)\n\ndef check_exist_path(path):\n if not os.path.exists(path):\n os.makedirs(path)\n\n\n# create save dirs\nsave_tr_dir = \"train\"\nsave_tr_img_dir = os.path.join(save_tr_dir, \"image_npy\")\ncheck_exist_path(save_tr_img_dir)\nsave_tr_label_dir = os.path.join(save_tr_dir, \"label_npy\")\ncheck_exist_path(save_tr_label_dir)\n\nsave_va_dir = \"val\"\nsave_va_img_dir = os.path.join(save_va_dir, \"image_npy\")\ncheck_exist_path(save_va_img_dir)\nsave_va_label_dir = os.path.join(save_va_dir, \"label_npy\")\ncheck_exist_path(save_va_label_dir)\n\n# split\nread_img_dir = \"image_npy\"\nread_label_dir = \"label_npy\"\nimg_files = os.listdir(read_img_dir)\n\nall_index = np.arange(len(img_files))\nnp.random.shuffle(all_index)\n\n# set test sample number\nnum_te = 200\ntr_index = all_index[num_te:]\nva_index = all_index[:num_te]\n\n#tr_ratio = 0.7\n#tr_index = all_index[:int(len(all_index)*tr_ratio)]\n#va_index = all_index[int(len(all_index)*tr_ratio):]\n\nf_tr = open(\"train.txt\",\"w\")\nf_va = open(\"val.txt\",\"w\")\n\nfor tri in tr_index:\n # cp images\n img_filename = img_files[tri]\n ori_path = os.path.join(read_img_dir, img_filename)\n target_path = os.path.join(save_tr_img_dir, img_filename)\n shutil.copyfile(ori_path, target_path)\n # cp labels\n ori_path_l = os.path.join(read_label_dir, img_filename)\n target_path_l = os.path.join(save_tr_label_dir, img_filename)\n shutil.copyfile(ori_path_l, target_path_l)\n f_tr.write(img_filename + \"\\n\")\n \n\nfor vai in va_index:\n # cp images\n img_filename = img_files[vai]\n ori_path = os.path.join(read_img_dir, img_filename)\n target_path = os.path.join(save_va_img_dir, img_filename)\n shutil.copyfile(ori_path, target_path)\n # cp labels\n ori_path_l = os.path.join(read_label_dir, img_filename)\n target_path_l = os.path.join(save_va_label_dir, img_filename)\n shutil.copyfile(ori_path_l, target_path_l)\n f_va.write(img_filename + \"\\n\")\n\nf_tr.close()\nf_va.close()\nprint(\"done\")\n\n\n \n"
]
| [
[
"numpy.random.shuffle",
"numpy.random.seed"
]
]
|
Brollof/Advent-of-Code | [
"c9cdf52660acc997aa7947509ca9933adcf11ca8"
]
| [
"2020/Day 20/20_1.py"
]
| [
"import re\nimport numpy as np\nfrom copy import deepcopy as dc, copy\nfrom math import sqrt\n\n\nLEFT = (1 << 0)\nRIGHT = (1 << 1)\nUP = (1 << 2)\nDOWN = (1 << 3)\n\n\nclass Tile:\n def __init__(self, raw_data=None):\n self.raw_data = raw_data\n tile_str = raw_data.splitlines()\n self.id = int(tile_str[0].split(' ')[1][:-1])\n self.grid = np.array([list(row) for row in tile_str[1:]])\n\n def __eq__(self, o):\n return self.id == o.id\n\n def __repr__(self):\n return f\"{self.id}\"\n\n\n def print_all(self):\n print(\"Original:\")\n self.print_grid()\n\n print(\"\\nLeft 1\")\n self.rotateLeft()\n self.print_grid()\n\n print(\"\\nLeft 2\")\n self.rotateLeft()\n self.print_grid()\n\n print(\"\\nLeft 3\")\n self.rotateLeft()\n self.print_grid()\n\n # back to original\n self.rotateLeft()\n\n print('\\nFlipped X')\n self.flipx()\n self.print_grid()\n\n print('\\nFlipped X, left 1')\n self.rotateLeft()\n self.print_grid()\n\n print('\\nFlipped X, left 2')\n self.rotateLeft()\n self.print_grid()\n\n print('\\nFlipped X, left 3')\n self.rotateLeft()\n self.print_grid()\n\n # back to original\n self.rotateLeft()\n self.flipx()\n\n print('\\nFlipped Y')\n self.flipy()\n self.print_grid()\n\n print('\\nFlipped Y, left 1')\n self.rotateLeft()\n self.print_grid()\n\n print('\\nFlipped Y, left 2')\n self.rotateLeft()\n self.print_grid()\n\n print('\\nFlipped Y, left 3')\n self.rotateLeft()\n self.print_grid()\n\n def print_grid(self):\n print('\\n'.join(''.join(row) for row in self.grid))\n\n def rotateLeft(self, k=1):\n self.grid = np.rot90(self.grid, k=k)\n\n def flipx(self):\n self.grid = np.fliplr(self.grid)\n\n def flipy(self):\n self.grid = np.flipud(self.grid)\n\n def check(self, o, d):\n if d == 'top': # check my top edge against other bottom\n return list(self.grid[0]) == list(o.grid[-1])\n if d == 'bot':\n return list(self.grid[-1]) == list(o.grid[0])\n if d == 'left':\n return [row[0] for row in self.grid] == [row[-1] for row in o.grid]\n if d == 'right':\n return [row[-1] for row in self.grid] == [row[0] for row in o.grid]\n\n\ntiles = []\n\nwith open('input.txt') as file:\n data = file.read()\n\n\nfor tile_str in data.split('\\n\\n'):\n tiles.append(Tile(tile_str))\n\nN = int(sqrt(len(tiles)))\n\nall_tiles = []\n# generate all possibilities\nfor tile in tiles:\n # ORIGINAL\n t = dc(tile)\n all_tiles.append(t)\n\n # ROTATE 1\n t = dc(tile)\n t.rotateLeft()\n all_tiles.append(t)\n\n # ROTATE 2\n t = dc(tile)\n t.rotateLeft()\n t.rotateLeft()\n all_tiles.append(t)\n\n # ROTATE 3\n t = dc(tile)\n t.rotateLeft()\n t.rotateLeft()\n t.rotateLeft()\n all_tiles.append(t)\n\n # FLIP X\n t = dc(tile)\n t.flipx()\n all_tiles.append(t)\n\n # FLIP X, ROTATE 1\n t = dc(tile)\n t.flipx()\n t.rotateLeft()\n all_tiles.append(t)\n\n # FLIP X, ROTATE 2\n t = dc(tile)\n t.flipx()\n t.rotateLeft()\n t.rotateLeft()\n all_tiles.append(t)\n\n # FLIP X, ROTATE 3\n t = dc(tile)\n t.flipx()\n t.rotateLeft()\n t.rotateLeft()\n t.rotateLeft()\n all_tiles.append(t)\n\ndef can_use(tile, grid):\n for row in grid:\n for el in row:\n if el and tile == el:\n return False\n return True\n\ndef find(x, y, grid):\n new = []\n for tile in all_tiles:\n if not can_use(tile, grid):\n continue\n if x - 1 >= 0 and grid[y][x - 1] is not None:\n if not tile.check(grid[y][x - 1], 'left'):\n continue\n if x + 1 < N and grid[y][x + 1] is not None:\n if not tile.check(grid[y][x + 1], 'right'):\n continue\n if y - 1 >= 0 and grid[y - 1][x] is not None:\n if not tile.check(grid[y - 1][x], 'top'):\n continue\n if y + 1 < N and grid[y + 1][x] is not None:\n if not tile.check(grid[y + 1][x], 'bot'):\n continue\n new.append(tile)\n return new\n\n\ndef is_full(grid):\n for row in grid:\n for el in row:\n if el is None:\n return False\n return True\n\n\ndef find_nejbs(x, y, start=None, grid=None):\n if not grid:\n grid = [[None] * N for _ in range(N)]\n\n if is_full(grid):\n return grid\n\n if start:\n grid[y][x] = start\n dx = [0, 0, 1, -1]\n dy = [1, -1, 0, 0]\n to_find = []\n for i in range(len(dx)):\n nx = x + dx[i]\n ny = y + dy[i]\n if 0 <= nx < N and 0 <= ny < N and grid[ny][nx] is None:\n to_find.append((nx, ny))\n\n for nx, ny in to_find:\n fits = find(nx, ny, grid)\n # assert len(fits) == 1, f\"{fits}\"\n if fits:\n grid[ny][nx] = fits[0]\n return find_nejbs(nx, ny, grid=grid)\n\n\nfor i in range(len(all_tiles)):\n print(i)\n g = find_nejbs(0, 0, start=all_tiles[i])\n if g:\n # print(g)\n part1 = g[0][0].id * g[0][N-1].id * g[N-1][0].id * g[N-1][N-1].id\n print(part1)\n # break\n"
]
| [
[
"numpy.fliplr",
"numpy.rot90",
"numpy.flipud"
]
]
|
wiVlad/DPMMSubClusters.jl | [
"64fa5a5a37bef85b715bbd056c66d47ffe6922a5"
]
| [
"paper_utils.py"
]
| [
"# Change directory to VSCode workspace root so that relative path loads work correctly. Turn this addition off with the DataScience.changeDirOnImportExport setting\n# ms-toolsai.jupyter added\nimport os\n\n\nimport pickle\nfrom datetime import datetime as date\nfrom pathlib import Path\nfrom timeit import default_timer as timer\n\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport torch.backends.cudnn as cudnn\nimport torch.nn as nn\n\n# import warnings\nfrom sklearn.cluster import KMeans\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.metrics.cluster import adjusted_rand_score as ari_score\nfrom sklearn.metrics.cluster import normalized_mutual_info_score\nfrom sklearn.mixture import BayesianGaussianMixture, GaussianMixture\n\n\ntry:\n os.chdir(os.path.join(os.getcwd(), \"/home/vlad/projects/bnpy\"))\n print(os.getcwd())\nexcept:\n pass\n\nimport bnpy\n\nfrom dpmmpython.dpmmwrapper import DPMMPython\nfrom dpmmpython.priors import niw\n\n\ndef save_obj(obj, path):\n with open(path + \".pkl\", \"wb\") as f:\n pickle.dump(obj, f, pickle.HIGHEST_PROTOCOL)\n\n\ndef load_obj(path):\n with open(path + \".pkl\", \"rb\") as f:\n return pickle.load(f)\n\n\ndef add_results(res_dict, name, labels, prediction, time):\n\n res_dict[\"method\"].append(name)\n res_dict[\"k_mae\"].append(\n np.abs(len(np.unique(labels)) - len(np.unique(prediction)))\n )\n res_dict[\"NMI\"].append(\n normalized_mutual_info_score(\n prediction.astype(int), np.array(labels), average_method=\"arithmetic\"\n )\n )\n res_dict[\"ARI\"].append(ari_score(labels[::2], prediction[::2]))\n res_dict[\"Time\"].append(time)\n return res_dict\n\n\ndef run_synthetic_data_comparisons(\n D: int,\n K: int,\n N: int,\n var_scale: int,\n alpha: int,\n iters: int,\n burnout: int,\n repeats: int,\n):\n\n results = {\n \"method\": [],\n \"k_mae\": [],\n \"NMI\": [],\n \"ARI\": [],\n \"Time\": [],\n }\n\n i = 0\n while i < repeats:\n\n # generate dataset\n data, labels = DPMMPython.generate_gaussian_data(N, D, K, var_scale)\n\n prior = niw(1, np.zeros(D), 100, np.eye(D) * 0.5)\n # run DPGMM\n\n if D == 2:\n start = timer()\n dpmm_splitnet_results = DPMMPython.fit(\n data,\n alpha,\n iterations=iters,\n burnout=burnout,\n verbose=False,\n init_type=\"splitnet_2d\",\n )[0]\n dpmm_net_time = timer() - start\n\n elif D <= 10:\n start = timer()\n dpmm_splitnet_results = DPMMPython.fit(\n data,\n alpha,\n iterations=ITERS,\n burnout=BURNOUT,\n verbose=False,\n init_type=\"splitnet_10d\",\n )[0]\n dpmm_net_time = timer() - start\n\n else:\n start = timer()\n dpmm_splitnet_results = DPMMPython.fit(\n data,\n alpha,\n iterations=ITERS,\n burnout=BURNOUT,\n verbose=False,\n init_type=\"splitnet_128d\",\n )[0]\n dpmm_net_time = timer() - start\n\n if len(np.unique(dpmm_splitnet_results)) < K // 2:\n print(\"failed.\")\n else:\n start = timer()\n dpmm_rand_results = DPMMPython.fit(\n data,\n alpha,\n iterations=iters,\n burnout=burnout,\n verbose=False,\n init_type=\"none\",\n )[0]\n dpmm_rand_time = timer() - start\n\n start = timer()\n dpmm_kmeans_results = DPMMPython.fit(\n data,\n alpha,\n iterations=iters,\n burnout=burnout,\n verbose=False,\n init_type=\"kmeans\",\n )[0]\n dpmm_kmeans_time = timer() - start\n\n # run kmeans\n start = timer()\n kmeans = KMeans(n_clusters=K).fit(data.T)\n kmeans_time = timer() - start\n kmeans_labels = kmeans.labels_\n\n # run GMM\n start = timer()\n gmm = GaussianMixture(n_components=K, covariance_type=\"full\").fit(data.T)\n gmm_labels = gmm.predict(data.T)\n gmm_time = timer() - start\n\n # sklearn DPGMM\n start = timer()\n dpgmm = BayesianGaussianMixture(\n n_components=2 * K,\n covariance_type=\"full\",\n weight_concentration_prior=alpha,\n weight_concentration_prior_type=\"dirichlet_process\",\n mean_precision_prior=1e2,\n covariance_prior=1e0 * np.eye(D),\n init_params=\"kmeans\",\n max_iter=iters,\n verbose=0,\n ).fit(data.T)\n dpgmm_labels = dpgmm.predict(data.T)\n dpgmmsk_time = timer() - start\n\n # moVB\n\n # pass data NxD\n data_bnpy = bnpy.data.XData(data.T)\n\n start = timer()\n model, run_info = bnpy.run(\n data_bnpy,\n \"DPMixtureModel\",\n \"Gauss\",\n \"memoVB\",\n nTask=1,\n nBatch=1,\n K=1,\n nLap=iters,\n moves=\"birth,merge,shuffle\",\n gt=labels,\n gamma0=alpha,\n )\n\n moVB_time = timer() - start\n LP = model.calc_local_params(data_bnpy)\n moVB_labels = LP[\"resp\"].argmax(axis=1)\n\n # calc metrics and aggregate\n results = add_results(\n results, \"k-means\", labels, kmeans_labels, kmeans_time\n )\n results = add_results(results, \"EM-GMM\", labels, gmm_labels, gmm_time)\n results = add_results(\n results, \"DPGMM (SKlearn's)\", labels, dpgmm_labels, dpgmmsk_time\n )\n results = add_results(\n results, \"DPGMM-Random\", labels, dpmm_rand_results, dpmm_rand_time\n )\n results = add_results(\n results, \"DPGMM-k-means\", labels, dpmm_kmeans_results, dpmm_kmeans_time\n )\n results = add_results(\n results, \"DPGMM-SplitNet\", labels, dpmm_splitnet_results, dpmm_net_time\n )\n results = add_results(results, \"moVB\", labels, moVB_labels, moVB_time)\n\n i += 1\n print(f\"Finished iteration {i}\")\n\n return results\n\n\ndef run_datasets_comparisons(\n data, labels, alpha: int, iters: int, burnout: int, repeats: int\n):\n\n results = {\n \"method\": [],\n \"k_mae\": [],\n \"NMI\": [],\n \"ARI\": [],\n # \"Acc\": [],\n }\n\n N, D = data.shape\n K = len(np.unique(labels))\n\n for i in range(repeats):\n\n # run kmeans\n kmeans = KMeans(n_clusters=K).fit(data)\n kmeans_labels = kmeans.labels_\n\n # run GMM\n gmm = GaussianMixture(n_components=K, covariance_type=\"full\").fit(data)\n gmm_labels = gmm.predict(data)\n\n # sklearn DPGMM\n dpgmm = BayesianGaussianMixture(\n n_components=2 * K,\n covariance_type=\"full\",\n weight_concentration_prior=1e2,\n weight_concentration_prior_type=\"dirichlet_process\",\n mean_precision_prior=1e2,\n covariance_prior=1e0 * np.eye(D),\n init_params=\"kmeans\",\n max_iter=ITERS,\n verbose=0,\n ).fit(data)\n dpgmm_labels = dpgmm.predict(data)\n\n prior = niw(1, np.zeros(D), D + 2, np.eye(D) * 0.5)\n # run DPGMM\n # dpmm_rand_results = DPMMPython.fit(data.T ,alpha, prior = prior, iterations=iters, burnout=burnout, verbose=False, init_type=\"none\")[0]\n # dpmm_kmeans_results = DPMMPython.fit(data.T ,alpha, prior = prior, iterations=iters, burnout=burnout, verbose=False, init_type=\"kmeans\")[0]/\n dpmm_rand_results = DPMMPython.fit(\n data.T,\n alpha,\n iterations=iters,\n burnout=burnout,\n verbose=False,\n init_type=\"none\",\n )[0]\n dpmm_kmeans_results = DPMMPython.fit(\n data.T,\n alpha,\n iterations=iters,\n burnout=burnout,\n verbose=False,\n init_type=\"kmeans\",\n )[0]\n if D == 2:\n dpmm_splitnet_results = DPMMPython.fit(\n data.T,\n alpha,\n iterations=iters,\n burnout=burnout,\n verbose=False,\n init_type=\"splitnet_2d\",\n )[0]\n elif D <= 10:\n dpmm_splitnet_results = DPMMPython.fit(\n data.T,\n alpha,\n iterations=ITERS,\n burnout=BURNOUT,\n verbose=False,\n init_type=\"splitnet_10d\",\n )[0]\n else:\n dpmm_splitnet_results = DPMMPython.fit(\n data.T,\n alpha,\n iterations=ITERS,\n burnout=BURNOUT,\n verbose=False,\n init_type=\"splitnet_128d\",\n )[0]\n\n # calc metrics and aggregate\n\n results = add_results(results, \"k-means\", labels, kmeans_labels)\n results = add_results(results, \"EM-GMM\", labels, gmm_labels)\n results = add_results(results, \"DPGMM (SKlearn's)\", labels, dpgmm_labels)\n results = add_results(results, \"DPGMM-Random\", labels, dpmm_rand_results)\n results = add_results(results, \"DPGMM-k-means\", labels, dpmm_kmeans_results)\n results = add_results(results, \"DPGMM-SplitNet\", labels, dpmm_splitnet_results)\n\n print(f\"Finished iteration {i}\")\n\n return results\n"
]
| [
[
"sklearn.cluster.KMeans",
"numpy.unique",
"numpy.eye",
"sklearn.mixture.GaussianMixture",
"numpy.array",
"numpy.zeros",
"sklearn.metrics.cluster.adjusted_rand_score"
]
]
|
prstolpe/rrc_simulation | [
"b430fe4e575641cdd64945cf57d0dd67a0eea17a"
]
| [
"tests/test_reset_joints.py"
]
| [
"#!/usr/bin/env python3\nimport unittest\nimport numpy as np\n\nfrom rrc_simulation.sim_finger import SimFinger\nfrom rrc_simulation import sample\n\n\nclass TestResetJoints(unittest.TestCase):\n \"\"\"\n This test verifies that the state of the finger(s) gets reset correctly.\n\n So, all the 1DOF joints of the finger(s) should be at the *exact* positions\n and have the *exact* same velocities to which we want the joints to get\n reset to.\n \"\"\"\n\n def test_reproduce_reset_state(self):\n \"\"\"\n Send hundred states (positions + velocities) to all the 1DOF joints\n of the fingers and assert they exactly reach these states.\n \"\"\"\n finger = SimFinger(finger_type=\"fingerone\")\n\n for _ in range(100):\n state_positions = sample.random_joint_positions(\n finger.number_of_fingers\n )\n state_velocities = [pos * 10 for pos in state_positions]\n\n reset_state = finger.reset_finger_positions_and_velocities(\n state_positions, state_velocities\n )\n\n reset_positions = reset_state.position\n reset_velocities = reset_state.velocity\n\n np.testing.assert_array_equal(\n reset_positions, state_positions, verbose=True\n )\n np.testing.assert_array_equal(\n reset_velocities, state_velocities, verbose=True\n )\n\n\nif __name__ == \"__main__\":\n unittest.main()\n"
]
| [
[
"numpy.testing.assert_array_equal"
]
]
|
maxim1um/PathFinding | [
"272391a0434c3fe1c17ef7f8e507a240183788f3"
]
| [
"A-Star/animation_demo.py"
]
| [
"\"\"\"\n================\npyplot animation\n================\n\nGenerating an animation by calling `~.pyplot.pause` between plotting commands.\n\nThe method shown here is only suitable for simple, low-performance use. For\nmore demanding applications, look at the :mod:`animation` module and the\nexamples that use it.\n\nNote that calling `time.sleep` instead of `~.pyplot.pause` would *not* work.\n\"\"\"\n\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nnp.random.seed(19680801)\ndata = np.random.random((50, 50, 50))\n\nfig, ax = plt.subplots()\n\ndataSize = len(data)\n\nfor i in range(len(data)):\n test = data[i]\n ax.cla()\n ax.imshow(data[i])\n ax.set_title(\"frame {}\".format(i))\n # Note that using time.sleep does *not* work here!\n plt.pause(0.1)\n"
]
| [
[
"numpy.random.random",
"matplotlib.pyplot.subplots",
"numpy.random.seed",
"matplotlib.pyplot.pause"
]
]
|
10088/MockingBird | [
"6a793cea8488ad40fcad6ab30f9d82bc920ac114"
]
| [
"vocoder/fregan/dwt.py"
]
| [
"# Copyright (c) 2019, Adobe Inc. All rights reserved.\n#\n# This work is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike\n# 4.0 International Public License. To view a copy of this license, visit\n# https://creativecommons.org/licenses/by-nc-sa/4.0/legalcode.\n\n# DWT code borrow from https://github.com/LiQiufu/WaveSNet/blob/12cb9d24208c3d26917bf953618c30f0c6b0f03d/DWT_IDWT/DWT_IDWT_layer.py\n\n\nimport pywt\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\n\n__all__ = ['DWT_1D']\nPad_Mode = ['constant', 'reflect', 'replicate', 'circular']\n\n\nclass DWT_1D(nn.Module):\n def __init__(self, pad_type='reflect', wavename='haar',\n stride=2, in_channels=1, out_channels=None, groups=None,\n kernel_size=None, trainable=False):\n\n super(DWT_1D, self).__init__()\n self.trainable = trainable\n self.kernel_size = kernel_size\n if not self.trainable:\n assert self.kernel_size == None\n self.in_channels = in_channels\n self.out_channels = self.in_channels if out_channels == None else out_channels\n self.groups = self.in_channels if groups == None else groups\n assert isinstance(self.groups, int) and self.in_channels % self.groups == 0\n self.stride = stride\n assert self.stride == 2\n self.wavename = wavename\n self.pad_type = pad_type\n assert self.pad_type in Pad_Mode\n self.get_filters()\n self.initialization()\n\n def get_filters(self):\n wavelet = pywt.Wavelet(self.wavename)\n band_low = torch.tensor(wavelet.rec_lo)\n band_high = torch.tensor(wavelet.rec_hi)\n length_band = band_low.size()[0]\n self.kernel_size = length_band if self.kernel_size == None else self.kernel_size\n assert self.kernel_size >= length_band\n a = (self.kernel_size - length_band) // 2\n b = - (self.kernel_size - length_band - a)\n b = None if b == 0 else b\n self.filt_low = torch.zeros(self.kernel_size)\n self.filt_high = torch.zeros(self.kernel_size)\n self.filt_low[a:b] = band_low\n self.filt_high[a:b] = band_high\n\n def initialization(self):\n self.filter_low = self.filt_low[None, None, :].repeat((self.out_channels, self.in_channels // self.groups, 1))\n self.filter_high = self.filt_high[None, None, :].repeat((self.out_channels, self.in_channels // self.groups, 1))\n if torch.cuda.is_available():\n self.filter_low = self.filter_low.cuda()\n self.filter_high = self.filter_high.cuda()\n if self.trainable:\n self.filter_low = nn.Parameter(self.filter_low)\n self.filter_high = nn.Parameter(self.filter_high)\n if self.kernel_size % 2 == 0:\n self.pad_sizes = [self.kernel_size // 2 - 1, self.kernel_size // 2 - 1]\n else:\n self.pad_sizes = [self.kernel_size // 2, self.kernel_size // 2]\n\n def forward(self, input):\n assert isinstance(input, torch.Tensor)\n assert len(input.size()) == 3\n assert input.size()[1] == self.in_channels\n input = F.pad(input, pad=self.pad_sizes, mode=self.pad_type)\n return F.conv1d(input, self.filter_low.to(input.device), stride=self.stride, groups=self.groups), \\\n F.conv1d(input, self.filter_high.to(input.device), stride=self.stride, groups=self.groups)\n"
]
| [
[
"torch.nn.Parameter",
"torch.zeros",
"torch.tensor",
"torch.cuda.is_available",
"torch.nn.functional.pad"
]
]
|
yydv98/FinNetIndicators | [
"5559b2039e173175b8efed89916d4e8601f31414"
]
| [
"network_entropy.py"
]
| [
"# Code calculates network entropy for degree and remaining degree distribution\n# Reference for Network Entropy: R.V. Sole and S. Valverde, Information theory of complex networks: on evolution and architectural constraints. Complex networks. Springer Berlin Heidelberg, 2004. 189-207.\n# Packages required: igraph, numpy\n# python network_entropy.py inputfile_folder_path 1/0 outputfile\n# 1 -- weighted\n# 0 -- unweighted\n\"\"\"\n=================================================================================================\nIf you are using this code, kindly cite the following articles:\n(1) S. Venkatesan, R.P. Vivek-Ananth, R.P. Sreejith, P. Mangalapandi, A.A. Hassanali & A. Samal, Network approach towards understanding the crazing in glassy amorphous polymers, Journal of Statistical Mechanics: Theory and Experiment 043305 (2018).\n(2) A. Samal, H.K. Pharasi, S. J. Ramaia, H. Kannan, E. Saucan, J. Jost & A. Chakraborti, Network geometry and market instability, R. Soc. Open Sci. 8: 201734 (2021).\n(3) A. Samal, S. Kumar, Y. Yadav & A. Chakraborti, Network-centric indicators for fragility in global financial indices, Front. Phys. 8: 624373 (2021).\n=================================================================================================\n\"\"\"\n\nimport igraph as ig\nimport numpy as np\nimport sys\nimport glob as gb\nimport re\n\n#edge files folder path\ninfile=sys.argv[1]\nweight=bool(sys.argv[2]) #input 1 if weighted; 0 if unweighted\noutfile=sys.argv[3]\n\n#function input: dictionary of degree/remaining degree distribution\ndef entropy_dg(P):\n\tent=0\n\tfor key in P:\n\t\tprob=float(P[key])/num_nodes\n\t\tent+=prob*np.log2(prob)\n\treturn -1*ent\ndef entropy_rdg(P):\n\tent=0\n\tfor key in P:\n\t\tent+=P[key]*np.log2(P[key])\n\treturn -1*ent\n\n#calculate remaining degree distribution from degree distribution\ndef rem_deg_dist(P):\n\trem_deg=[]\n\tmean_P=0\n\tfor key in P:\n\t\tprob=float(P[key])/num_nodes\n\t\tmean_P+=float(key)*prob\n\tprint ('\\n<E> ',mean_P,'\\n')\n\tfor key in P:\n \t\tprob=float(P[key])/num_nodes\n \t\tq=float(key)*prob/float(mean_P)\n \t\trem_deg.append((key-1,q))\n\treturn dict(rem_deg)\n\nedgefiles=infile\n\nf=open(outfile,'w')\n\nfor fil in gb.glob(edgefiles):\n\n\t#Read the edge list; Change parameter weights and directed for network of study\n\tp=ig.Graph.Read_Ncol(fil,weights=weight,directed=False,names=True)\n\t\n\t#number of nodes\n\tnum_nodes=p.vcount()\n\n\tig.summary(p)\n\n\tdg_dis=p.degree()\n\tdg_dis_count=[]\n\tfor i in set(dg_dis):\n\t\tdg_dis_count.append((i,dg_dis.count(i)))\n\n\tedg=entropy_dg(dict(dg_dis_count))\n\tprint ('\\nEntropy of degree distribution for the given network is ', edg)\n\ta=rem_deg_dist(dict(dg_dis_count))\n\terdg=entropy_rdg(a)\n\tprint ('\\nEntropy of remaining degree distribution for the given network is ',erdg,'\\n')\n\n\tf.write(fil+'\\t'+str(erdg)+'\\n')\n\nf.close()\n"
]
| [
[
"numpy.log2"
]
]
|
mli0603/Bi3D | [
"dfcccdae3c3e0a6fc847e5cec6129dc5b497b906"
]
| [
"src/models/RefineNet3D.py"
]
| [
"# Copyright (c) 2020, NVIDIA CORPORATION. All rights reserved.\n#\n# NVIDIA CORPORATION and its licensors retain all intellectual property\n# and proprietary rights in and to this software, related documentation\n# and any modifications thereto. Any use, reproduction, disclosure or\n# distribution of this software and related documentation without an express\n# license agreement from NVIDIA CORPORATION is strictly prohibited.\n\nimport torch\nimport torch.nn as nn\nimport numpy as np\n\n__all__ = [\"segregnet3d\"]\n\nfrom models.GCNet import conv3d_relu\nfrom models.GCNet import deconv3d_relu\nfrom models.GCNet import feature3d\n\n\ndef net_init(net):\n\n for m in net.modules():\n if isinstance(m, nn.Linear):\n m.weight.data = fanin_init(m.weight.data.size())\n elif isinstance(m, nn.Conv3d):\n n = m.kernel_size[0] * m.kernel_size[1] * m.kernel_size[2] * m.out_channels\n m.weight.data.normal_(0, np.sqrt(2.0 / n))\n elif isinstance(m, nn.Conv2d):\n n = m.kernel_size[0] * m.kernel_size[1] * m.out_channels\n m.weight.data.normal_(0, np.sqrt(2.0 / n))\n elif isinstance(m, nn.Conv1d):\n n = m.kernel_size[0] * m.out_channels\n m.weight.data.normal_(0, np.sqrt(2.0 / n))\n elif isinstance(m, nn.BatchNorm3d):\n m.weight.data.fill_(1)\n m.bias.data.zero_()\n elif isinstance(m, nn.BatchNorm2d):\n m.weight.data.fill_(1)\n m.bias.data.zero_()\n elif isinstance(m, nn.BatchNorm1d):\n m.weight.data.fill_(1)\n m.bias.data.zero_()\n\n\nclass SegRegNet3D(nn.Module):\n def __init__(self, F=16):\n\n super(SegRegNet3D, self).__init__()\n\n self.conf_preprocess = conv3d_relu(1, F, kernel_size=3, stride=1)\n self.layer3d = feature3d(F)\n\n net_init(self)\n\n def forward(self, fL, conf_volume):\n\n fL_stack = fL[:, :, None, :, :].repeat(1, 1, int(conf_volume.shape[2]), 1, 1)\n conf_vol_preprocess = self.conf_preprocess(conf_volume)\n input_volume = torch.cat((fL_stack, conf_vol_preprocess), dim=1)\n oL = self.layer3d(input_volume)\n\n return oL\n\n\ndef segregnet3d(options, data=None):\n\n print(\"==> USING SegRegNet3D\")\n for key in options:\n if \"regnet\" in key:\n print(\"{} : {}\".format(key, options[key]))\n\n model = SegRegNet3D(F=options[\"regnet_out_planes\"])\n if data is not None:\n model.load_state_dict(data[\"state_dict\"])\n\n return model\n"
]
| [
[
"numpy.sqrt",
"torch.cat"
]
]
|
davicn/tuh_seizures | [
"0b1e6983baba3ffba0aa9942c3d22ab1e14c1e4b"
]
| [
"scripts/getFeatures.py"
]
| [
"import numpy as np\nimport pandas as pd\nimport mne\nimport os\nfrom functions import gpuFeatures as gf\n\nPATH = os.getcwd()\nHOME = os.path.expanduser(\"~\")\n\nw = pd.read_csv(PATH+'/docs/class_w.csv', header=None)\nn = pd.read_csv(PATH+'/docs/class_n.csv', header=None)\n\nuse = ['EEG FP1-REF', 'EEG FP2-REF', 'EEG F3-REF', 'EEG F4-REF', 'EEG C3-REF',\n 'EEG C4-REF', 'EEG P3-REF', 'EEG P4-REF', 'EEG O1-REF', 'EEG O2-REF',\n 'EEG F7-REF', 'EEG F8-REF', 'EEG T3-REF', 'EEG T4-REF', 'EEG T5-REF',\n 'EEG T6-REF', 'EEG T1-REF', 'EEG T2-REF', 'EEG FZ-REF', 'EEG CZ-REF',\n 'EEG PZ-REF']\n\nt = .25\nfs = 256\n\nw = np.unique(w.iloc[:,0].to_numpy())\nn = np.unique(n.iloc[:,0].to_numpy())\n\n# eegs = []\nw_eegs_var = []\nw_eegs_eng = []\nw_ecgs_var = []\nw_ecgs_eng = []\nw_emgs_var = []\nw_emgs_eng = []\n\nn_eegs_eng = []\nn_eegs_var = []\nn_ecgs_eng = []\nn_ecgs_var = []\nn_emgs_var = []\nn_emgs_eng = []\n\nfor i in range(len(w)):\n raw = mne.io.read_raw_edf(\n HOME + '/Documentos/tuh_edf/' + w[i].replace('tse', 'edf'), preload=False, verbose=False)\n\n d = raw.to_data_frame().T\n eeg = d.loc[use, :].to_numpy()\n# ecg = d.loc['EEG EKG1-REF', :].to_numpy()\n# emg = d.loc['EMG-REF', :].to_numpy()\n\n eeg_var = gf.variancia(eeg, t, fs, True)\n eeg_eng = gf.energia(eeg, t, fs, True)\n\n# emg_var = gf.variancia(emg, t, fs, False)\n# emg_eng = gf.energia(emg, t, fs, False)\n# ecg_var = gf.variancia(ecg, t, fs, False)\n# ecg_eng = gf.energia(ecg, t, fs, False)\n\n print(eeg_var.shape)\n print(eeg_eng.shape)\n\n w_eegs_var.append(eeg_var)\n w_eegs_eng.append(eeg_eng)\n\n\n# w_emgs_var.append(emg_var.cpu().numpy())\n# w_emgs_eng.append(emg_eng.cpu().numpy())\n# w_ecgs_var.append(ecg_var.cpu().numpy())\n# w_ecgs_eng.append(ecg_eng.cpu().numpy())\n\n# print(w[i])\nnp.save(\"w_eeg_var.npy\",w_eegs_var)\nnp.save(\"w_eeg_eng.npy\",w_eegs_eng)\n# np.save(\"w_emg_var.npy\",w_emgs_var)\n# np.save(\"w_emg_eng.npy\",w_emgs_eng)\n# np.save(\"w_ecg_var.npy\",w_ecgs_var)\n# np.save(\"w_ecg_eng.npy\",w_ecgs_eng)\n\n\nfor i in range(len(n)):\n raw = mne.io.read_raw_edf(\n HOME + '/Documentos/tuh_edf/' + n[i].replace('tse', 'edf'), preload=False, verbose=False)\n\n d = raw.to_data_frame().T\n eeg = d.loc[use, :].to_numpy()\n# ecg = d.loc['EEG EKG1-REF', :].to_numpy()\n# emg = d.loc['EMG-REF', :].to_numpy()\n eeg_var = gf.variancia(eeg, t, fs, True)\n eeg_eng = gf.energia(eeg, t, fs, True)\n# # eeg_var = gf.variancia(eeg, t, fs, True)\n# emg_eng = gf.energia(emg, t, fs, False)\n# emg_var = gf.variancia(emg, t, fs, False)\n# ecg_var = gf.variancia(ecg, t, fs, False)\n# ecg_eng = gf.energia(ecg, t, fs, False)\n print(eeg_var.shape)\n print(eeg_eng.shape)\n\n n_eegs_var.append(eeg_var)\n n_eegs_eng.append(eeg_eng)\n\n# # print(eeg_var)\n# n_emgs_var.append(emg_var.cpu().numpy())\n# n_emgs_eng.append(emg_eng.cpu().numpy())\n# n_ecgs_var.append(ecg_var.cpu().numpy())\n# n_ecgs_eng.append(ecg_eng.cpu().numpy())\n\n# print(n[i])\nnp.save(\"n_eeg_var.npy\",n_eegs_var)\nnp.save(\"n_eeg_eng.npy\",n_eegs_eng)\n# np.save(\"n_ecg_var.npy\",n_ecgs_var)\n# np.save(\"n_emg_var.npy\",n_emgs_var)\n# np.save(\"n_ecg_eng.npy\",n_ecgs_eng)\n# np.save(\"n_emg_eng.npy\",n_emgs_eng)\n\n\n# np.save('tamanhos2.npy',np.array(lens))\n\n# w = np.load(PATH + '/data/w_train.npy', allow_pickle=True)\n# w = pd.DataFrame(w)\n# w.columns = ['path', 'start', 'end', 'type', 'fs', 'duration', 'montage']\n\n# n = np.load(PATH + '/data/n_train.npy', allow_pickle=True)\n# n = pd.DataFrame(n)\n# n.columns = ['path', 'fs', 'duration', 'montage']\n"
]
| [
[
"pandas.read_csv",
"numpy.save"
]
]
|
EchoWho/AnytimeNeuralNetwork_public | [
"f2f04a836b984250aca6180afedfcab29be9ead9"
]
| [
"tensorpack/dataflow/format.py"
]
| [
"# -*- coding: utf-8 -*-\n# File: format.py\n\n\nimport numpy as np\nimport six\nfrom six.moves import range\nimport os\nimport struct\n\nfrom ..utils import logger\nfrom ..utils.utils import get_tqdm\nfrom ..utils.timer import timed_operation\nfrom ..utils.loadcaffe import get_caffe_pb\nfrom ..utils.serialize import loads\nfrom ..utils.argtools import log_once\nfrom .base import RNGDataFlow, DataFlow, DataFlowReentrantGuard\nfrom .common import MapData\n\n__all__ = ['HDF5Data', 'LMDBData', 'LMDBDataDecoder', 'LMDBDataPoint', 'LMDBDataPointIndexed',\n 'CaffeLMDB', 'SVMLightData', 'TFRecordData', 'BinaryData', 'NPZData']\n\n\"\"\"\nAdapters for different data format.\n\"\"\"\n\n\nclass HDF5Data(RNGDataFlow):\n \"\"\"\n Zip data from different paths in an HDF5 file.\n\n Warning:\n The current implementation will load all data into memory. (TODO)\n \"\"\"\n# TODO\n\n def __init__(self, filename, data_paths, shuffle=True):\n \"\"\"\n Args:\n filename (str): h5 data file.\n data_paths (list): list of h5 paths to zipped.\n For example `['images', 'labels']`.\n shuffle (bool): shuffle all data.\n \"\"\"\n self.f = h5py.File(filename, 'r')\n logger.info(\"Loading {} to memory...\".format(filename))\n self.dps = [self.f[k].value for k in data_paths]\n lens = [len(k) for k in self.dps]\n assert all([k == lens[0] for k in lens])\n self._size = lens[0]\n self.shuffle = shuffle\n\n def size(self):\n return self._size\n\n def get_data(self):\n idxs = list(range(self._size))\n if self.shuffle:\n self.rng.shuffle(idxs)\n for k in idxs:\n yield [dp[k] for dp in self.dps]\n\n\nclass LMDBData(RNGDataFlow):\n \"\"\"\n Read a LMDB database and produce (k,v) raw bytes pairs.\n The raw bytes are usually not what you're interested in.\n You might want to use\n :class:`LMDBDataDecoder`, :class:`LMDBDataPoint`, or apply a\n mapper function after :class:`LMDBData`.\n \"\"\"\n def __init__(self, lmdb_path, shuffle=True, keys=None):\n \"\"\"\n Args:\n lmdb_path (str): a directory or a file.\n shuffle (bool): shuffle the keys or not.\n keys (list[str] or str): list of str as the keys, used only when shuffle is True.\n It can also be a format string e.g. ``{:0>8d}`` which will be\n formatted with the indices from 0 to *total_size - 1*.\n\n If not provided, it will then look in the database for ``__keys__`` which\n :func:`dump_dataflow_to_lmdb` used to store the list of keys.\n If still not found, it will iterate over the database to find\n all the keys.\n \"\"\"\n self._lmdb_path = lmdb_path\n self._shuffle = shuffle\n\n self._open_lmdb()\n self._size = self._txn.stat()['entries']\n self._set_keys(keys)\n logger.info(\"Found {} entries in {}\".format(self._size, self._lmdb_path))\n self._guard = DataFlowReentrantGuard()\n\n def _set_keys(self, keys=None):\n def find_keys(txn, size):\n logger.warn(\"Traversing the database to find keys is slow. Your should specify the keys.\")\n keys = []\n with timed_operation(\"Loading LMDB keys ...\", log_start=True), \\\n get_tqdm(total=size) as pbar:\n for k in self._txn.cursor():\n assert k[0] != b'__keys__'\n keys.append(k[0])\n pbar.update()\n return keys\n\n self.keys = self._txn.get(b'__keys__')\n if self.keys is not None:\n self.keys = loads(self.keys)\n self._size -= 1 # delete this item\n\n if self._shuffle: # keys are necessary when shuffle is True\n if keys is None:\n if self.keys is None:\n self.keys = find_keys(self._txn, self._size)\n else:\n # check if key-format like '{:0>8d}' was given\n if isinstance(keys, six.string_types):\n self.keys = map(lambda x: keys.format(x), list(np.arange(self._size)))\n else:\n self.keys = keys\n\n def _open_lmdb(self):\n self._lmdb = lmdb.open(self._lmdb_path,\n subdir=os.path.isdir(self._lmdb_path),\n readonly=True, lock=False, readahead=True,\n map_size=1099511627776 * 2, max_readers=100)\n self._txn = self._lmdb.begin()\n\n def reset_state(self):\n self._lmdb.close()\n super(LMDBData, self).reset_state()\n self._open_lmdb()\n\n def size(self):\n return self._size\n\n def get_data(self):\n with self._guard:\n if not self._shuffle:\n c = self._txn.cursor()\n while c.next():\n k, v = c.item()\n if k != b'__keys__':\n yield [k, v]\n else:\n self.rng.shuffle(self.keys)\n for k in self.keys:\n v = self._txn.get(k)\n yield [k, v]\n\n\nclass LMDBDataDecoder(MapData):\n \"\"\" Read a LMDB database and produce a decoded output.\"\"\"\n def __init__(self, lmdb_data, decoder):\n \"\"\"\n Args:\n lmdb_data: a :class:`LMDBData` instance.\n decoder (k,v -> dp | None): a function taking k, v and returning a datapoint,\n or return None to discard.\n \"\"\"\n def f(dp):\n return decoder(dp[0], dp[1])\n super(LMDBDataDecoder, self).__init__(lmdb_data, f)\n\n\nclass LMDBDataPoint(MapData):\n \"\"\"\n Read a LMDB file and produce deserialized datapoints.\n It **only** accepts the database produced by\n :func:`tensorpack.dataflow.dftools.dump_dataflow_to_lmdb`,\n which uses :func:`tensorpack.utils.serialize.dumps` for serialization.\n\n Example:\n .. code-block:: python\n\n ds = LMDBDataPoint(\"/data/ImageNet.lmdb\", shuffle=False) # read and decode\n\n # The above is equivalent to:\n ds = LMDBData(\"/data/ImageNet.lmdb\", shuffle=False) # read\n ds = LMDBDataPoint(ds) # decode\n # Sometimes it makes sense to separate reading and decoding\n # to be able to make decoding parallel.\n \"\"\"\n\n def __init__(self, *args, **kwargs):\n \"\"\"\n Args:\n args, kwargs: Same as in :class:`LMDBData`.\n\n In addition, args[0] can be a :class:`LMDBData` instance.\n In this case args[0] has to be the only argument.\n \"\"\"\n if isinstance(args[0], DataFlow):\n ds = args[0]\n assert len(args) == 1 and len(kwargs) == 0, \\\n \"No more arguments are allowed if LMDBDataPoint is called with a LMDBData instance!\"\n else:\n ds = LMDBData(*args, **kwargs)\n\n def f(dp):\n return loads(dp[1])\n super(LMDBDataPoint, self).__init__(ds, f)\n\n\nclass LMDBDataPointIndexed(MapData):\n \"\"\" Read a LMDB file and produce deserialized values.\n This can work with :func:`tensorpack.dataflow.dftools.dump_dataflow_to_lmdb`. \n \n The input DS can be processed already (with join, shuffle, etc), so the index\n of key and val are specified. \n\n Further more, there could be multiple LMDB data fused together before local shuffle.\n \"\"\"\n def __init__(self, ds, index=1):\n def f(dp):\n # replace key and serialized val with val\n dp[index-1:index+1] = loads(dp[index])\n return dp\n super(LMDBDataPointIndexed, self).__init__(ds, f)\n\n\ndef CaffeLMDB(lmdb_path, shuffle=True, keys=None):\n \"\"\"\n Read a Caffe LMDB file where each value contains a ``caffe.Datum`` protobuf.\n Produces datapoints of the format: [HWC image, label].\n\n Note that Caffe LMDB format is not efficient: it stores serialized raw\n arrays rather than JPEG images.\n\n Args:\n lmdb_path, shuffle, keys: same as :class:`LMDBData`.\n\n Returns:\n a :class:`LMDBDataDecoder` instance.\n\n Example:\n .. code-block:: python\n\n ds = CaffeLMDB(\"/tmp/validation\", keys='{:0>8d}')\n \"\"\"\n\n cpb = get_caffe_pb()\n lmdb_data = LMDBData(lmdb_path, shuffle, keys)\n\n def decoder(k, v):\n try:\n datum = cpb.Datum()\n datum.ParseFromString(v)\n img = np.fromstring(datum.data, dtype=np.uint8)\n img = img.reshape(datum.channels, datum.height, datum.width)\n except Exception:\n log_once(\"Cannot read key {}\".format(k), 'warn')\n return None\n return [img.transpose(1, 2, 0), datum.label]\n logger.warn(\"Caffe LMDB format doesn't store jpeg-compressed images, \\\n it's not recommended due to its inferior performance.\")\n return LMDBDataDecoder(lmdb_data, decoder)\n\n\nclass SVMLightData(RNGDataFlow):\n \"\"\" Read X,y from a svmlight file, and produce [X_i, y_i] pairs. \"\"\"\n\n def __init__(self, filename, shuffle=True):\n \"\"\"\n Args:\n filename (str): input file\n shuffle (bool): shuffle the data\n \"\"\"\n import sklearn.datasets # noqa\n self.X, self.y = sklearn.datasets.load_svmlight_file(filename)\n self.X = np.asarray(self.X.todense())\n self.shuffle = shuffle\n\n def size(self):\n return len(self.y)\n\n def get_data(self):\n idxs = np.arange(self.size())\n if self.shuffle:\n self.rng.shuffle(idxs)\n for id in idxs:\n yield [self.X[id, :], self.y[id]]\n\n\nclass TFRecordData(DataFlow):\n \"\"\"\n Produce datapoints from a TFRecord file, assuming each record is\n serialized by :func:`serialize.dumps`.\n This class works with :func:`dftools.dump_dataflow_to_tfrecord`.\n \"\"\"\n def __init__(self, path, size=None):\n \"\"\"\n Args:\n path (str): path to the tfrecord file\n size (int): total number of records, because this metadata is not\n stored in the tfrecord file.\n \"\"\"\n self._path = path\n self._size = int(size)\n\n def size(self):\n if self._size:\n return self._size\n return super(TFRecordData, self).size()\n\n def get_data(self):\n gen = tf.python_io.tf_record_iterator(self._path)\n for dp in gen:\n yield loads(dp)\n\nclass BinaryData(DataFlow):\n \"\"\"\n Produce data points from a binary file. A format for each row is given \n to the init so that we can decode the binary data one row at a time.\n \"\"\"\n def __init__(self, path, n_val_per_row=None):\n self.dp_format = 'f' * n_val_per_row\n self.row_n_bytes = 4 * n_val_per_row\n self.path = path\n self.statinfo = os.stat(self.path)\n self._size = self.statinfo.st_size / 4 / n_val_per_row\n\n def size(self):\n return self._size \n\n def get_data(self):\n with open(self.path, 'rb') as fin:\n for _ in range(self._size):\n yield [np.asarray(struct.unpack(self.dp_format, \n fin.read(self.row_n_bytes)),dtype=np.float32)]\n\nclass NPZData(DataFlow):\n\n def __init__(self, path, keys, select_range=None):\n self.path = path\n self.keys = keys\n self.data = np.load(path)\n\n if select_range is not None:\n start = select_range[0]\n end = select_range[1]\n self.data_temp={}\n for key in keys:\n self.data_temp[key] = self.data[key][start:end]\n self.data=self.data_temp\n\n def size(self):\n return self.data[self.keys[0]].shape[0] \n\n def get_data(self):\n for i in range(self.size()):\n yield [self.data[key][i] for key in self.keys]\n\n\nfrom ..utils.develop import create_dummy_class # noqa\ntry:\n import h5py\nexcept ImportError:\n HDF5Data = create_dummy_class('HDF5Data', 'h5py') # noqa\n\ntry:\n import lmdb\nexcept ImportError:\n for klass in ['LMDBData', 'LMDBDataDecoder', 'LMDBDataPoint', 'CaffeLMDB']:\n globals()[klass] = create_dummy_class(klass, 'lmdb')\n\ntry:\n import tensorflow as tf\nexcept ImportError:\n TFRecordData = create_dummy_class('TFRecordData', 'tensorflow') # noqa\n"
]
| [
[
"tensorflow.python_io.tf_record_iterator",
"numpy.load",
"numpy.fromstring",
"numpy.arange"
]
]
|
jusjusjus/autograd-hacks | [
"c12556d03e40cccaa0e70e14b0120b723002ed9e"
]
| [
"autograd_hacks/test_autograd_hacks.py"
]
| [
"\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport pytest\n\nfrom . import autograd_hacks\n\n\nclass StriddenNet(nn.Module):\n def __init__(self):\n super().__init__()\n self.conv1 = nn.Conv2d(1, 20, 5, stride=2, padding=2)\n self.conv2 = nn.Conv2d(20, 30, 5, stride=2, padding=2)\n self.fc1_input_size = 7 * 7 * 30\n self.fc1 = nn.Linear(self.fc1_input_size, 500)\n self.fc2 = nn.Linear(500, 10)\n\n def forward(self, x):\n batch_size = x.shape[0]\n x = F.relu(self.conv1(x))\n x = F.relu(self.conv2(x))\n x = x.view(batch_size, self.fc1_input_size)\n x = F.relu(self.fc1(x))\n x = self.fc2(x)\n return x\n\n\nclass SimpleNet(nn.Module):\n \"\"\"Lenet-5 from https://github.com/pytorch/examples/blob/master/mnist/main.py\"\"\"\n def __init__(self):\n super().__init__()\n self.linear = nn.Linear(28 * 28, 10)\n\n def forward(self, x):\n x = torch.flatten(x, 1)\n return self.linear(x)\n\n\nclass Net(nn.Module):\n \"\"\"Lenet-5 from https://github.com/pytorch/examples/blob/master/mnist/main.py\"\"\"\n def __init__(self):\n super().__init__()\n self.conv1 = nn.Conv2d(1, 20, 5)\n self.conv2 = nn.Conv2d(20, 50, 5)\n self.fc1 = nn.Linear(4 * 4 * 50, 500)\n self.fc2 = nn.Linear(500, 10)\n\n def forward(self, x):\n x = F.relu(self.conv1(x))\n x = F.max_pool2d(x, 2, 2)\n x = F.relu(self.conv2(x))\n x = F.max_pool2d(x, 2, 2)\n x = x.view(-1, 4 * 4 * 50)\n x = F.relu(self.fc1(x))\n x = self.fc2(x)\n return x\n\n\nclass TinyNet(nn.Module):\n \"\"\"Tiny LeNet-5 for Hessian testing\"\"\"\n\n def __init__(self):\n super().__init__()\n self.conv1 = nn.Conv2d(1, 2, 2, 1)\n self.conv2 = nn.Conv2d(2, 2, 2, 1)\n self.fc1 = nn.Linear(2, 2)\n self.fc2 = nn.Linear(2, 10)\n\n def forward(self, x): # 28x28\n x = F.max_pool2d(x, 4, 4) # 7x7\n x = F.relu(self.conv1(x)) # 6x6\n x = F.max_pool2d(x, 2, 2) # 3x3\n x = F.relu(self.conv2(x)) # 2x2\n x = F.max_pool2d(x, 2, 2) # 1x1\n x = x.view(-1, 2 * 1 * 1) # C * W * H\n x = F.relu(self.fc1(x))\n x = self.fc2(x)\n return x\n\n\n# Autograd helpers, from https://gist.github.com/apaszke/226abdf867c4e9d6698bd198f3b45fb7\ndef jacobian(y: torch.Tensor, x: torch.Tensor, create_graph=False):\n jac = []\n flat_y = y.reshape(-1)\n grad_y = torch.zeros_like(flat_y)\n for i in range(len(flat_y)):\n grad_y[i] = 1.\n grad_x, = torch.autograd.grad(flat_y, x, grad_y, retain_graph=True, create_graph=create_graph)\n jac.append(grad_x.reshape(x.shape))\n grad_y[i] = 0.\n return torch.stack(jac).reshape(y.shape + x.shape)\n\n\ndef hessian(y: torch.Tensor, x: torch.Tensor):\n return jacobian(jacobian(y, x, create_graph=True), x)\n\n\[email protected](\"Net\", [Net, TinyNet, SimpleNet, StriddenNet])\ndef test_grad1(Net):\n torch.manual_seed(1)\n model = Net()\n loss_fn = nn.CrossEntropyLoss()\n\n n = 4\n data = torch.rand(n, 1, 28, 28)\n targets = torch.LongTensor(n).random_(0, 10)\n\n autograd_hacks.add_hooks(model)\n output = model(data)\n loss_fn(output, targets).backward(retain_graph=True)\n autograd_hacks.compute_grad1(model)\n autograd_hacks.disable_hooks()\n\n # Compare values against autograd\n losses = torch.stack([loss_fn(output[i:i+1], targets[i:i+1])\n for i in range(len(data))])\n\n for layer in model.modules():\n if not autograd_hacks.is_supported(layer):\n continue\n\n for param in layer.parameters():\n assert torch.allclose(param.grad, param.grad1[0].mean(dim=0))\n assert torch.allclose(jacobian(losses, param), param.grad1[0])\n\n\ndef test_applying_backwards_twice_fails():\n torch.manual_seed(42)\n model = Net()\n loss_fn = nn.CrossEntropyLoss()\n\n data = torch.rand(5, 1, 28, 28)\n targets = torch.LongTensor(5).random_(0, 10)\n\n autograd_hacks.add_hooks(model)\n output = model(data)\n loss_fn(output, targets).backward()\n output = model(data)\n with pytest.raises(AssertionError):\n loss_fn(output, targets).backward()\n\n\ndef test_grad1_for_multiple_connected_passes():\n torch.manual_seed(42)\n model = SimpleNet()\n loss_fn = nn.CrossEntropyLoss(reduction='sum')\n\n def get_data(batch_size):\n return (torch.rand(batch_size, 1, 28, 28),\n torch.LongTensor(batch_size).random_(0, 10))\n\n n = 5\n autograd_hacks.add_hooks(model)\n\n data, targets = get_data(n)\n output = model(data)\n loss1 = loss_fn(output, targets)\n data, targets = get_data(n)\n output = model(data)\n loss2 = loss_fn(output, targets)\n loss = loss1 - loss2\n loss.backward()\n\n autograd_hacks.compute_grad1(model)\n autograd_hacks.disable_hooks()\n\n for n, p in model.named_parameters():\n grad1 = p.grad1[0] + p.grad1[1]\n assert p.grad.shape == grad1.shape[1:]\n assert torch.allclose(p.grad, grad1.mean(dim=0), atol=1e-7)\n\n\[email protected](\"hess_type\", ['CrossEntropy', 'LeastSquares'])\ndef test_hess(hess_type):\n torch.manual_seed(1)\n model = TinyNet()\n\n def least_squares_loss(data_, targets_):\n assert len(data_) == len(targets_)\n err = data_ - targets_\n return torch.sum(err * err) / 2 / len(data_)\n\n n = 3\n data = torch.rand(n, 1, 28, 28)\n\n autograd_hacks.add_hooks(model)\n output = model(data)\n\n if hess_type == 'LeastSquares':\n targets = torch.rand(output.shape)\n loss_fn = least_squares_loss\n elif hess_type == 'CrossEntropy':\n targets = torch.LongTensor(n).random_(0, 10)\n loss_fn = nn.CrossEntropyLoss()\n else:\n raise ValueError(f\"Unknown hessian type\")\n\n autograd_hacks.backprop_hess(output, hess_type)\n autograd_hacks.clear_backprops(model)\n autograd_hacks.backprop_hess(output, hess_type)\n\n autograd_hacks.compute_hess(model)\n autograd_hacks.disable_hooks()\n\n for layer in model.modules():\n if not autograd_hacks.is_supported(layer):\n continue\n\n for param in layer.parameters():\n loss = loss_fn(output, targets)\n hess_autograd = hessian(loss, param)\n hess = param.hess\n assert torch.allclose(hess, hess_autograd.reshape(hess.shape))\n"
]
| [
[
"torch.nn.CrossEntropyLoss",
"torch.LongTensor",
"torch.manual_seed",
"torch.nn.Conv2d",
"torch.zeros_like",
"torch.sum",
"torch.nn.Linear",
"torch.rand",
"torch.flatten",
"torch.stack",
"torch.nn.functional.max_pool2d",
"torch.autograd.grad"
]
]
|
mkzirncz1/ResNet-PyTorch | [
"2a19be30cc65eb5cb8cc41341b22bd040c022546"
]
| [
"examples/simple/test.py"
]
| [
"# Copyright 2020 Lorna Authors. All Rights Reserved.\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n# ==============================================================================\n\n\"\"\"Example\nIn this simple example, we load an image, pre-process it, and classify it with a pretrained ResNet.\n\"\"\"\nimport json\n\nimport torch\nimport torchvision.transforms as transforms\nfrom PIL import Image\n\nfrom resnet_pytorch import ResNet\n\n# Open image\ninput_image = Image.open(\"img.jpg\")\n\n# Preprocess image\npreprocess = transforms.Compose([\n transforms.Resize(256),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),\n])\ninput_tensor = preprocess(input_image)\ninput_batch = input_tensor.unsqueeze(0) # create a mini-batch as expected by the model\n\n# Load class names\nlabels_map = json.load(open(\"labels_map.txt\"))\nlabels_map = [labels_map[str(i)] for i in range(1000)]\n\n# Classify with ResNet18\nmodel = ResNet.from_pretrained(\"resnet18\")\nmodel.eval()\n\n# move the input and model to GPU for speed if available\nif torch.cuda.is_available():\n input_batch = input_batch.to(\"cuda\")\n model.to(\"cuda\")\n\nwith torch.no_grad():\n logits = model(input_batch)\npreds = torch.topk(logits, k=5).indices.squeeze(0).tolist()\n\n#print\nprint(\"-----\")\nfor idx in preds:\n label = labels_map[idx]\n prob = torch.softmax(logits, dim=1)[0, idx].item()\n print(f\"{label:<75} ({prob * 100:.2f}%)\")\n"
]
| [
[
"torch.topk",
"torch.softmax",
"torch.no_grad",
"torch.cuda.is_available"
]
]
|
hrabeale/arrow | [
"4009b62086dfa43a4fd8bfa714772716e6531c6f"
]
| [
"python/pyarrow/tests/test_convert_builtin.py"
]
| [
"# -*- coding: utf-8 -*-\n# Licensed to the Apache Software Foundation (ASF) under one\n# or more contributor license agreements. See the NOTICE file\n# distributed with this work for additional information\n# regarding copyright ownership. The ASF licenses this file\n# to you under the Apache License, Version 2.0 (the\n# \"License\"); you may not use this file except in compliance\n# with the License. You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing,\n# software distributed under the License is distributed on an\n# \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY\n# KIND, either express or implied. See the License for the\n# specific language governing permissions and limitations\n# under the License.\n\nimport pytest\n\nfrom pyarrow.compat import unittest, u # noqa\nimport pyarrow as pa\n\nimport datetime\nimport decimal\nimport itertools\nimport numpy as np\nimport six\nimport pytz\n\n\nint_type_pairs = [\n (np.int8, pa.int8()),\n (np.int16, pa.int64()),\n (np.int32, pa.int32()),\n (np.int64, pa.int64()),\n (np.uint8, pa.uint8()),\n (np.uint16, pa.uint64()),\n (np.uint32, pa.uint32()),\n (np.uint64, pa.uint64())]\n\n\nnp_int_types, _ = zip(*int_type_pairs)\n\n\nclass StrangeIterable:\n def __init__(self, lst):\n self.lst = lst\n\n def __iter__(self):\n return self.lst.__iter__()\n\n\ndef test_iterable_types():\n arr1 = pa.array(StrangeIterable([0, 1, 2, 3]))\n arr2 = pa.array((0, 1, 2, 3))\n\n assert arr1.equals(arr2)\n\n\ndef test_empty_iterable():\n arr = pa.array(StrangeIterable([]))\n assert len(arr) == 0\n assert arr.null_count == 0\n assert arr.type == pa.null()\n assert arr.to_pylist() == []\n\n\ndef test_limited_iterator_types():\n arr1 = pa.array(iter(range(3)), type=pa.int64(), size=3)\n arr2 = pa.array((0, 1, 2))\n assert arr1.equals(arr2)\n\n\ndef test_limited_iterator_size_overflow():\n arr1 = pa.array(iter(range(3)), type=pa.int64(), size=2)\n arr2 = pa.array((0, 1))\n assert arr1.equals(arr2)\n\n\ndef test_limited_iterator_size_underflow():\n arr1 = pa.array(iter(range(3)), type=pa.int64(), size=10)\n arr2 = pa.array((0, 1, 2))\n assert arr1.equals(arr2)\n\n\ndef test_iterator_without_size():\n expected = pa.array((0, 1, 2))\n arr1 = pa.array(iter(range(3)))\n assert arr1.equals(expected)\n # Same with explicit type\n arr1 = pa.array(iter(range(3)), type=pa.int64())\n assert arr1.equals(expected)\n\n\ndef test_infinite_iterator():\n expected = pa.array((0, 1, 2))\n arr1 = pa.array(itertools.count(0), size=3)\n assert arr1.equals(expected)\n # Same with explicit type\n arr1 = pa.array(itertools.count(0), type=pa.int64(), size=3)\n assert arr1.equals(expected)\n\n\ndef _as_list(xs):\n return xs\n\n\ndef _as_tuple(xs):\n return tuple(xs)\n\n\ndef _as_dict_values(xs):\n dct = {k: v for k, v in enumerate(xs)}\n return six.viewvalues(dct)\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\ndef test_sequence_types(seq):\n arr1 = pa.array(seq([1, 2, 3]))\n arr2 = pa.array([1, 2, 3])\n\n assert arr1.equals(arr2)\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\ndef test_sequence_boolean(seq):\n expected = [True, None, False, None]\n arr = pa.array(seq(expected))\n assert len(arr) == 4\n assert arr.null_count == 2\n assert arr.type == pa.bool_()\n assert arr.to_pylist() == expected\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\ndef test_sequence_numpy_boolean(seq):\n expected = [np.bool(True), None, np.bool(False), None]\n arr = pa.array(seq(expected))\n assert len(arr) == 4\n assert arr.null_count == 2\n assert arr.type == pa.bool_()\n assert arr.to_pylist() == expected\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\ndef test_empty_list(seq):\n arr = pa.array(seq([]))\n assert len(arr) == 0\n assert arr.null_count == 0\n assert arr.type == pa.null()\n assert arr.to_pylist() == []\n\n\ndef test_sequence_all_none():\n arr = pa.array([None, None])\n assert len(arr) == 2\n assert arr.null_count == 2\n assert arr.type == pa.null()\n assert arr.to_pylist() == [None, None]\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\[email protected](\"np_scalar_pa_type\", int_type_pairs)\ndef test_sequence_integer(seq, np_scalar_pa_type):\n np_scalar, pa_type = np_scalar_pa_type\n expected = [1, None, 3, None,\n np.iinfo(np_scalar).min, np.iinfo(np_scalar).max]\n arr = pa.array(seq(expected), type=pa_type)\n assert len(arr) == 6\n assert arr.null_count == 2\n assert arr.type == pa_type\n assert arr.to_pylist() == expected\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\ndef test_sequence_integer_inferred(seq):\n expected = [1, None, 3, None]\n arr = pa.array(seq(expected))\n assert len(arr) == 4\n assert arr.null_count == 2\n assert arr.type == pa.int64()\n assert arr.to_pylist() == expected\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\[email protected](\"np_scalar_pa_type\", int_type_pairs)\ndef test_sequence_numpy_integer(seq, np_scalar_pa_type):\n np_scalar, pa_type = np_scalar_pa_type\n expected = [np_scalar(1), None, np_scalar(3), None,\n np_scalar(np.iinfo(np_scalar).min),\n np_scalar(np.iinfo(np_scalar).max)]\n arr = pa.array(seq(expected), type=pa_type)\n assert len(arr) == 6\n assert arr.null_count == 2\n assert arr.type == pa_type\n assert arr.to_pylist() == expected\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\[email protected](\"np_scalar_pa_type\", int_type_pairs)\ndef test_sequence_numpy_integer_inferred(seq, np_scalar_pa_type):\n np_scalar, pa_type = np_scalar_pa_type\n expected = [np_scalar(1), None, np_scalar(3), None]\n if np_scalar != np.uint64:\n expected += [np_scalar(np.iinfo(np_scalar).min),\n np_scalar(np.iinfo(np_scalar).max)]\n else:\n # max(uint64) is too large for the inferred int64 type\n expected += [0, np.iinfo(np.int64).max]\n arr = pa.array(seq(expected))\n assert len(arr) == 6\n assert arr.null_count == 2\n assert arr.type == pa.int64()\n assert arr.to_pylist() == expected\n\n\[email protected](\"bits\", [8, 16, 32, 64])\ndef test_signed_integer_overflow(bits):\n ty = getattr(pa, \"int%d\" % bits)()\n # XXX ideally would raise OverflowError\n with pytest.raises((ValueError, pa.ArrowException)):\n pa.array([2 ** (bits - 1)], ty)\n with pytest.raises((ValueError, pa.ArrowException)):\n pa.array([-2 ** (bits - 1) - 1], ty)\n\n\[email protected](\"bits\", [8, 16, 32, 64])\ndef test_unsigned_integer_overflow(bits):\n ty = getattr(pa, \"uint%d\" % bits)()\n # XXX ideally would raise OverflowError\n with pytest.raises((ValueError, pa.ArrowException)):\n pa.array([2 ** bits], ty)\n with pytest.raises((ValueError, pa.ArrowException)):\n pa.array([-1], ty)\n\n\ndef test_garbage_collection():\n import gc\n\n # Force the cyclic garbage collector to run\n gc.collect()\n\n bytes_before = pa.total_allocated_bytes()\n pa.array([1, None, 3, None])\n gc.collect()\n assert pa.total_allocated_bytes() == bytes_before\n\n\ndef test_sequence_double():\n data = [1.5, 1, None, 2.5, None, None]\n arr = pa.array(data)\n assert len(arr) == 6\n assert arr.null_count == 3\n assert arr.type == pa.float64()\n assert arr.to_pylist() == data\n\n\[email protected](\"seq\", [_as_list, _as_tuple, _as_dict_values])\[email protected](\"np_scalar\", [np.float16, np.float32, np.float64])\ndef test_sequence_numpy_double(seq, np_scalar):\n data = [np_scalar(1.5), np_scalar(1), None, np_scalar(2.5), None, None]\n arr = pa.array(seq(data))\n assert len(arr) == 6\n assert arr.null_count == 3\n assert arr.type == pa.float64()\n assert arr.to_pylist() == data\n\n\ndef test_sequence_unicode():\n data = [u'foo', u'bar', None, u'mañana']\n arr = pa.array(data)\n assert len(arr) == 4\n assert arr.null_count == 1\n assert arr.type == pa.string()\n assert arr.to_pylist() == data\n\n\ndef test_sequence_bytes():\n u1 = b'ma\\xc3\\xb1ana'\n data = [b'foo',\n u1.decode('utf-8'), # unicode gets encoded,\n bytearray(b'bar'),\n None]\n for ty in [None, pa.binary()]:\n arr = pa.array(data, type=ty)\n assert len(arr) == 4\n assert arr.null_count == 1\n assert arr.type == pa.binary()\n assert arr.to_pylist() == [b'foo', u1, b'bar', None]\n\n\ndef test_sequence_utf8_to_unicode():\n # ARROW-1225\n data = [b'foo', None, b'bar']\n arr = pa.array(data, type=pa.string())\n assert arr[0].as_py() == u'foo'\n\n # test a non-utf8 unicode string\n val = (u'mañana').encode('utf-16-le')\n with pytest.raises(pa.ArrowException):\n pa.array([val], type=pa.string())\n\n\ndef test_sequence_fixed_size_bytes():\n data = [b'foof', None, bytearray(b'barb'), b'2346']\n arr = pa.array(data, type=pa.binary(4))\n assert len(arr) == 4\n assert arr.null_count == 1\n assert arr.type == pa.binary(4)\n assert arr.to_pylist() == [b'foof', None, b'barb', b'2346']\n\n\ndef test_fixed_size_bytes_does_not_accept_varying_lengths():\n data = [b'foo', None, b'barb', b'2346']\n with pytest.raises(pa.ArrowInvalid):\n pa.array(data, type=pa.binary(4))\n\n\ndef test_sequence_date():\n data = [datetime.date(2000, 1, 1), None, datetime.date(1970, 1, 1),\n datetime.date(2040, 2, 26)]\n arr = pa.array(data)\n assert len(arr) == 4\n assert arr.type == pa.date64()\n assert arr.null_count == 1\n assert arr[0].as_py() == datetime.date(2000, 1, 1)\n assert arr[1].as_py() is None\n assert arr[2].as_py() == datetime.date(1970, 1, 1)\n assert arr[3].as_py() == datetime.date(2040, 2, 26)\n\n\ndef test_sequence_date32():\n data = [datetime.date(2000, 1, 1), None]\n arr = pa.array(data, type=pa.date32())\n\n data2 = [10957, None]\n arr2 = pa.array(data2, type=pa.date32())\n\n for x in [arr, arr2]:\n assert len(x) == 2\n assert x.type == pa.date32()\n assert x.null_count == 1\n assert x[0].as_py() == datetime.date(2000, 1, 1)\n assert x[1] is pa.NA\n\n # Overflow\n data3 = [2**32, None]\n with pytest.raises(pa.ArrowException):\n pa.array(data3, type=pa.date32())\n\n\ndef test_sequence_timestamp():\n data = [\n datetime.datetime(2007, 7, 13, 1, 23, 34, 123456),\n None,\n datetime.datetime(2006, 1, 13, 12, 34, 56, 432539),\n datetime.datetime(2010, 8, 13, 5, 46, 57, 437699)\n ]\n arr = pa.array(data)\n assert len(arr) == 4\n assert arr.type == pa.timestamp('us')\n assert arr.null_count == 1\n assert arr[0].as_py() == datetime.datetime(2007, 7, 13, 1,\n 23, 34, 123456)\n assert arr[1].as_py() is None\n assert arr[2].as_py() == datetime.datetime(2006, 1, 13, 12,\n 34, 56, 432539)\n assert arr[3].as_py() == datetime.datetime(2010, 8, 13, 5,\n 46, 57, 437699)\n\n\ndef test_sequence_numpy_timestamp():\n data = [\n np.datetime64(datetime.datetime(2007, 7, 13, 1, 23, 34, 123456)),\n None,\n np.datetime64(datetime.datetime(2006, 1, 13, 12, 34, 56, 432539)),\n np.datetime64(datetime.datetime(2010, 8, 13, 5, 46, 57, 437699))\n ]\n arr = pa.array(data)\n assert len(arr) == 4\n assert arr.type == pa.timestamp('us')\n assert arr.null_count == 1\n assert arr[0].as_py() == datetime.datetime(2007, 7, 13, 1,\n 23, 34, 123456)\n assert arr[1].as_py() is None\n assert arr[2].as_py() == datetime.datetime(2006, 1, 13, 12,\n 34, 56, 432539)\n assert arr[3].as_py() == datetime.datetime(2010, 8, 13, 5,\n 46, 57, 437699)\n\n\ndef test_sequence_timestamp_with_unit():\n data = [\n datetime.datetime(2007, 7, 13, 1, 23, 34, 123456),\n ]\n\n s = pa.timestamp('s')\n ms = pa.timestamp('ms')\n us = pa.timestamp('us')\n ns = pa.timestamp('ns')\n\n arr_s = pa.array(data, type=s)\n assert len(arr_s) == 1\n assert arr_s.type == s\n assert arr_s[0].as_py() == datetime.datetime(2007, 7, 13, 1,\n 23, 34, 0)\n\n arr_ms = pa.array(data, type=ms)\n assert len(arr_ms) == 1\n assert arr_ms.type == ms\n assert arr_ms[0].as_py() == datetime.datetime(2007, 7, 13, 1,\n 23, 34, 123000)\n\n arr_us = pa.array(data, type=us)\n assert len(arr_us) == 1\n assert arr_us.type == us\n assert arr_us[0].as_py() == datetime.datetime(2007, 7, 13, 1,\n 23, 34, 123456)\n\n arr_ns = pa.array(data, type=ns)\n assert len(arr_ns) == 1\n assert arr_ns.type == ns\n assert arr_ns[0].as_py() == datetime.datetime(2007, 7, 13, 1,\n 23, 34, 123456)\n\n\ndef test_sequence_timestamp_from_int_with_unit():\n data = [1]\n\n s = pa.timestamp('s')\n ms = pa.timestamp('ms')\n us = pa.timestamp('us')\n ns = pa.timestamp('ns')\n\n arr_s = pa.array(data, type=s)\n assert len(arr_s) == 1\n assert arr_s.type == s\n assert str(arr_s[0]) == \"Timestamp('1970-01-01 00:00:01')\"\n\n arr_ms = pa.array(data, type=ms)\n assert len(arr_ms) == 1\n assert arr_ms.type == ms\n assert str(arr_ms[0]) == \"Timestamp('1970-01-01 00:00:00.001000')\"\n\n arr_us = pa.array(data, type=us)\n assert len(arr_us) == 1\n assert arr_us.type == us\n assert str(arr_us[0]) == \"Timestamp('1970-01-01 00:00:00.000001')\"\n\n arr_ns = pa.array(data, type=ns)\n assert len(arr_ns) == 1\n assert arr_ns.type == ns\n assert str(arr_ns[0]) == \"Timestamp('1970-01-01 00:00:00.000000001')\"\n\n with pytest.raises(pa.ArrowException):\n class CustomClass():\n pass\n pa.array([1, CustomClass()], type=ns)\n pa.array([1, CustomClass()], type=pa.date32())\n pa.array([1, CustomClass()], type=pa.date64())\n\n\ndef test_sequence_mixed_nesting_levels():\n pa.array([1, 2, None])\n pa.array([[1], [2], None])\n pa.array([[1], [2], [None]])\n\n with pytest.raises(pa.ArrowInvalid):\n pa.array([1, 2, [1]])\n\n with pytest.raises(pa.ArrowInvalid):\n pa.array([1, 2, []])\n\n with pytest.raises(pa.ArrowInvalid):\n pa.array([[1], [2], [None, [1]]])\n\n\ndef test_sequence_list_of_int():\n data = [[1, 2, 3], [], None, [1, 2]]\n arr = pa.array(data)\n assert len(arr) == 4\n assert arr.null_count == 1\n assert arr.type == pa.list_(pa.int64())\n assert arr.to_pylist() == data\n\n\ndef test_sequence_mixed_types_fails():\n data = ['a', 1, 2.0]\n with pytest.raises(pa.ArrowException):\n pa.array(data)\n\n\ndef test_sequence_mixed_types_with_specified_type_fails():\n data = ['-10', '-5', {'a': 1}, '0', '5', '10']\n\n type = pa.string()\n with pytest.raises(TypeError):\n pa.array(data, type=type)\n\n\ndef test_sequence_decimal():\n data = [decimal.Decimal('1234.183'), decimal.Decimal('8094.234')]\n type = pa.decimal128(precision=7, scale=3)\n arr = pa.array(data, type=type)\n assert arr.to_pylist() == data\n\n\ndef test_sequence_decimal_different_precisions():\n data = [\n decimal.Decimal('1234234983.183'), decimal.Decimal('80943244.234')\n ]\n type = pa.decimal128(precision=13, scale=3)\n arr = pa.array(data, type=type)\n assert arr.to_pylist() == data\n\n\ndef test_sequence_decimal_no_scale():\n data = [decimal.Decimal('1234234983'), decimal.Decimal('8094324')]\n type = pa.decimal128(precision=10)\n arr = pa.array(data, type=type)\n assert arr.to_pylist() == data\n\n\ndef test_sequence_decimal_negative():\n data = [decimal.Decimal('-1234.234983'), decimal.Decimal('-8.094324')]\n type = pa.decimal128(precision=10, scale=6)\n arr = pa.array(data, type=type)\n assert arr.to_pylist() == data\n\n\ndef test_sequence_decimal_no_whole_part():\n data = [decimal.Decimal('-.4234983'), decimal.Decimal('.0103943')]\n type = pa.decimal128(precision=7, scale=7)\n arr = pa.array(data, type=type)\n assert arr.to_pylist() == data\n\n\ndef test_sequence_decimal_large_integer():\n data = [decimal.Decimal('-394029506937548693.42983'),\n decimal.Decimal('32358695912932.01033')]\n type = pa.decimal128(precision=23, scale=5)\n arr = pa.array(data, type=type)\n assert arr.to_pylist() == data\n\n\ndef test_range_types():\n arr1 = pa.array(range(3))\n arr2 = pa.array((0, 1, 2))\n assert arr1.equals(arr2)\n\n\ndef test_empty_range():\n arr = pa.array(range(0))\n assert len(arr) == 0\n assert arr.null_count == 0\n assert arr.type == pa.null()\n assert arr.to_pylist() == []\n\n\ndef test_structarray():\n ints = pa.array([None, 2, 3], type=pa.int64())\n strs = pa.array([u'a', None, u'c'], type=pa.string())\n bools = pa.array([True, False, None], type=pa.bool_())\n arr = pa.StructArray.from_arrays(\n [ints, strs, bools],\n ['ints', 'strs', 'bools'])\n\n expected = [\n {'ints': None, 'strs': u'a', 'bools': True},\n {'ints': 2, 'strs': None, 'bools': False},\n {'ints': 3, 'strs': u'c', 'bools': None},\n ]\n\n pylist = arr.to_pylist()\n assert pylist == expected, (pylist, expected)\n\n\ndef test_struct_from_dicts():\n ty = pa.struct([pa.field('a', pa.int32()),\n pa.field('b', pa.string()),\n pa.field('c', pa.bool_())])\n arr = pa.array([], type=ty)\n assert arr.to_pylist() == []\n\n data = [{'a': 5, 'b': 'foo', 'c': True},\n {'a': 6, 'b': 'bar', 'c': False}]\n arr = pa.array(data, type=ty)\n assert arr.to_pylist() == data\n\n # With omitted values\n data = [{'a': 5, 'c': True},\n None,\n {},\n {'a': None, 'b': 'bar'}]\n arr = pa.array(data, type=ty)\n expected = [{'a': 5, 'b': None, 'c': True},\n None,\n {'a': None, 'b': None, 'c': None},\n {'a': None, 'b': 'bar', 'c': None}]\n assert arr.to_pylist() == expected\n\n\ndef test_struct_from_tuples():\n ty = pa.struct([pa.field('a', pa.int32()),\n pa.field('b', pa.string()),\n pa.field('c', pa.bool_())])\n\n data = [(5, 'foo', True),\n (6, 'bar', False)]\n expected = [{'a': 5, 'b': 'foo', 'c': True},\n {'a': 6, 'b': 'bar', 'c': False}]\n arr = pa.array(data, type=ty)\n assert arr.to_pylist() == expected\n\n # With omitted values\n data = [(5, 'foo', None),\n None,\n (6, None, False)]\n expected = [{'a': 5, 'b': 'foo', 'c': None},\n None,\n {'a': 6, 'b': None, 'c': False}]\n arr = pa.array(data, type=ty)\n assert arr.to_pylist() == expected\n\n # Invalid tuple size\n for tup in [(5, 'foo'), (), ('5', 'foo', True, None)]:\n with pytest.raises(ValueError, match=\"(?i)tuple size\"):\n pa.array([tup], type=ty)\n\n\ndef test_struct_from_mixed_sequence():\n # It is forbidden to mix dicts and tuples when initializing a struct array\n ty = pa.struct([pa.field('a', pa.int32()),\n pa.field('b', pa.string()),\n pa.field('c', pa.bool_())])\n data = [(5, 'foo', True),\n {'a': 6, 'b': 'bar', 'c': False}]\n with pytest.raises(TypeError):\n pa.array(data, type=ty)\n\n\ndef test_structarray_from_arrays_coerce():\n # ARROW-1706\n ints = [None, 2, 3]\n strs = [u'a', None, u'c']\n bools = [True, False, None]\n ints_nonnull = [1, 2, 3]\n\n arrays = [ints, strs, bools, ints_nonnull]\n result = pa.StructArray.from_arrays(arrays,\n ['ints', 'strs', 'bools',\n 'int_nonnull'])\n expected = pa.StructArray.from_arrays(\n [pa.array(ints, type='int64'),\n pa.array(strs, type='utf8'),\n pa.array(bools),\n pa.array(ints_nonnull, type='int64')],\n ['ints', 'strs', 'bools', 'int_nonnull'])\n\n with pytest.raises(ValueError):\n pa.StructArray.from_arrays(arrays)\n\n assert result.equals(expected)\n\n\ndef test_decimal_array_with_none_and_nan():\n values = [decimal.Decimal('1.234'), None, np.nan, decimal.Decimal('nan')]\n array = pa.array(values)\n assert array.type == pa.decimal128(4, 3)\n assert array.to_pylist() == values[:2] + [None, None]\n\n array = pa.array(values, type=pa.decimal128(10, 4))\n assert array.to_pylist() == [decimal.Decimal('1.2340'), None, None, None]\n\n\[email protected]('tz,name', [\n (pytz.FixedOffset(90), '+01:30'),\n (pytz.FixedOffset(-90), '-01:30'),\n (pytz.utc, 'UTC'),\n (pytz.timezone('America/New_York'), 'America/New_York')\n])\ndef test_timezone_string(tz, name):\n assert pa.lib.tzinfo_to_string(tz) == name\n assert pa.lib.string_to_tzinfo(name) == tz\n"
]
| [
[
"numpy.bool",
"numpy.iinfo"
]
]
|
SuzukiDaishi/AutoVC.pytorch | [
"0ce6f2dd5b6e34f812c56fc1466bb3444ef837bd"
]
| [
"train.py"
]
| [
"import os\nimport json\nimport argparse\nimport torch\nimport random\nimport torch.optim as optim\nfrom model_vc import StyleEncoder\nfrom model_vc import Generator\nfrom dataset import AudiobookDataset\nfrom dataset import train_collate\nfrom dataset import test_collate\nfrom utils.dsp import save_wav\nimport numpy as np\nfrom hparams import hparams as hp\n\n \ndef save_checkpoint(device, model, optimizer, checkpoint_dir, epoch):\n checkpoint_path = os.path.join(\n checkpoint_dir, \"checkpoint_step{:06d}.pth\".format(epoch))\n optimizer_state = optimizer.state_dict()\n torch.save({\n \"model\": model.state_dict(),\n \"optimizer\": optimizer_state,\n \"epoch\": epoch\n }, checkpoint_path)\n print(\"Saved checkpoint:\", checkpoint_path)\n\ndef load_checkpoint(path, model, device, optimizer, reset_optimizer=False):\n print(\"Load checkpoint from: {}\".format(path))\n checkpoint = torch.load(path, map_location=device)\n model.load_state_dict(checkpoint[\"model\"])\n if not reset_optimizer:\n optimizer_state = checkpoint[\"optimizer\"]\n if optimizer_state is not None:\n print(\"Load optimizer state from {}\".format(path))\n optimizer.load_state_dict(checkpoint[\"optimizer\"])\n epoch = checkpoint['epoch'] \n return epoch\n\ndef train(args, model, device, train_loader, optimizer, epoch, sigma=1.0):\n model.train()\n train_loss = 0\n\n for batch_idx, (m, e) in enumerate(train_loader):\n m = m.to(device)\n e = e.to(device)\n \n model.zero_grad()\n\n mel_outputs, mel_outputs_postnet, codes = model(m, e, e)\n\n m_rec = mel_outputs_postnet\n codes_rec = model(m_rec, e, None)\n\n L_recon = ((mel_outputs_postnet - m) ** 2).sum(dim=(1,2)).mean()\n L_recon0 = ((mel_outputs - m) ** 2).sum(dim=(1,2)).mean()\n L_content = torch.abs(codes - codes_rec).sum(dim=1).mean()\n\n loss = L_recon + L_recon0 + L_content\n\n loss.backward()\n optimizer.step()\n\n train_loss += loss.item() * len(m)\n\n if batch_idx % 10 == 0:\n print('Train Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n epoch, batch_idx * len(m), len(train_loader.dataset),\n 100. * batch_idx / len(train_loader), loss.item()))\n \n train_loss /= len(train_loader.dataset)\n print('\\nTrain set: Average loss: {:.4f}\\n'.format(train_loss))\n\ndef test(model, device, test_loader, checkpoint_dir, epoch, sigma=1.0):\n print(\"Using averaged model for evaluation\")\n model.eval()\n \n test_loss = 0\n\n with torch.no_grad():\n for batch_idx, (m, e) in enumerate(test_loader):\n m = m.to(device)\n e = e.to(device)\n \n mel_outputs, mel_outputs_postnet, codes = model(m, e, e)\n\n m_rec = mel_outputs_postnet\n codes_rec = model(m_rec, e, None)\n\n L_recon = ((mel_outputs_postnet - m) ** 2).sum(dim=(1,2)).mean()\n L_recon0 = ((mel_outputs - m) ** 2).sum(dim=(1,2)).mean()\n L_content = torch.abs(codes - codes_rec).sum(dim=1).mean()\n\n loss = L_recon + L_recon0 + L_content\n\n if batch_idx % 100 == 0:\n print('Val Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n epoch, batch_idx * len(m), len(test_loader.dataset),\n 100. * batch_idx / len(test_loader), loss.item()))\n test_loss += loss.item()\n\n test_loss /= len(test_loader.dataset)\n print('\\nTest set: Average loss: {:.4f}\\n'.format(test_loss))\n\nif __name__ == '__main__':\n parser = argparse.ArgumentParser(description='Train or run some neural net')\n parser.add_argument('-d', '--data', type=str, default='./data', help='dataset directory')\n parser.add_argument('--checkpoint', type=str, default=None,\n help='The path to checkpoint')\n parser.add_argument('--epochs', type=int, default=600,\n help='number of epochs to train (default: 14)')\n parser.add_argument('--batch-size', type=int, default=8, metavar='N',\n help='input batch size for training (default: 64)')\n parser.add_argument('--lr', type=float, default=1e-4, metavar='LR',\n help='learning rate (default: 1.0)')\n parser.add_argument('--no-cuda', action='store_true', default=False,\n help='disables CUDA training')\n args = parser.parse_args()\n\n torch.manual_seed(0)\n np.random.seed(0)\n random.seed(0)\n\n data_path = args.data\n\n use_cuda = not args.no_cuda and torch.cuda.is_available()\n device = torch.device(\"cuda\" if use_cuda else \"cpu\")\n \n kwargs = {'num_workers': 8, 'pin_memory': True} if use_cuda else {}\n\n torch.autograd.set_detect_anomaly(True)\n \n with open(os.path.join(data_path, 'train_data.json'), 'r') as f:\n train_data = json.load(f)\n\n with open(os.path.join(data_path, 'test_data.json'), 'r') as f:\n test_data = json.load(f)\n\n train_loader = torch.utils.data.DataLoader(\n AudiobookDataset(train_data),\n collate_fn=train_collate,\n batch_size=args.batch_size, shuffle=True, **kwargs)\n\n test_loader = torch.utils.data.DataLoader(\n AudiobookDataset(test_data),\n collate_fn=test_collate,\n batch_size=1, shuffle=False, **kwargs)\n\n model = Generator(hp.dim_neck, hp.dim_emb, hp.dim_pre, hp.freq).to(device)\n optimizer = optim.Adam(model.parameters(), lr=args.lr)\n\n current_epoch = 0\n if args.checkpoint:\n current_epoch = load_checkpoint(args.checkpoint, model, device, optimizer)\n \n checkpoint_dir = 'checkpoints'\n os.makedirs(checkpoint_dir, exist_ok=True)\n\n for epoch in range(current_epoch + 1, args.epochs + 1):\n print(f'epoch {epoch}')\n train(args, model, device, train_loader, optimizer, epoch)\n\n if epoch % 10 == 0:\n test(model, device, test_loader, checkpoint_dir, epoch)\n save_checkpoint(device, model, optimizer, checkpoint_dir, epoch)\n"
]
| [
[
"torch.abs",
"torch.autograd.set_detect_anomaly",
"numpy.random.seed",
"torch.load",
"torch.manual_seed",
"torch.no_grad",
"torch.cuda.is_available",
"torch.device"
]
]
|
kyawakyawa/image-filters-python | [
"0bcc8ef583834deba4697f17d0e627ce695bfd07"
]
| [
"main.py"
]
| [
"\"\"\"\nMIT License\n\nCopyright (c) 2021 kyawakyawa\n\nPermission is hereby granted, free of charge, to any person obtaining a copy\nof this software and associated documentation files (the \"Software\"), to deal\nin the Software without restriction, including without limitation the rights\nto use, copy, modify, merge, publish, distribute, sublicense, and/or sell\ncopies of the Software, and to permit persons to whom the Software is\nfurnished to do so, subject to the following conditions:\n\nThe above copyright notice and this permission notice shall be included in all\ncopies or substantial portions of the Software.\n\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\nIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\nFITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\nAUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\nLIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\nOUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\nSOFTWARE.\n\"\"\"\n\n\nimport argparse\nimport math\nimport torch\nimport torchvision\nfrom utils import try_gpu, to_gray_scale\nfrom box_filter import box_filter\nfrom shift_filter import shift_filter\nfrom first_order_derivative_operation import (\n x_derivative_operator,\n y_derivative_operator,\n x_prewitt_filter,\n y_prewitt_filter,\n x_sobel_filter,\n y_sobel_filter,\n steerable_filter,\n)\nfrom second_order_derivative_operation import (\n x_2nd_derivative_operator,\n y_2nd_derivative_operator,\n laplacian_filter,\n)\nfrom gaussian_filter import gaussian_filter\nfrom laplacian_of_gaussian import laplacian_of_gaussian\nfrom garbor_filter import garbor_filter\n\nfrom PIL import Image\n\n\nif __name__ == \"__main__\":\n parser = argparse.ArgumentParser()\n\n parser.add_argument(\n \"-i\",\n \"--image\",\n action=\"store\",\n nargs=1,\n const=None,\n default=None,\n type=str,\n choices=None,\n help=\"filepath to image\",\n metavar=None,\n )\n\n args = parser.parse_args()\n\n im = Image.open(args.image[0])\n\n torch.set_grad_enabled(False)\n\n im = torchvision.transforms.functional.to_tensor(im)\n\n H, W = im.shape[1:3]\n\n im = try_gpu(im)\n\n im = im.unsqueeze(0)\n\n # box filter\n im_box_filterd = box_filter(im, 5, 2)\n\n im_box_filterd = im_box_filterd.squeeze()\n\n im_box_filterd = torchvision.transforms.functional.to_pil_image(\n im_box_filterd\n )\n\n im_box_filterd.save(\"output-box-filtered.jpg\")\n\n # shift filter\n im_shift_filterd = shift_filter(im, 5, 2)\n\n im_shift_filterd = im_shift_filterd.squeeze()\n\n im_shift_filterd = torchvision.transforms.functional.to_pil_image(\n im_shift_filterd\n )\n\n im_shift_filterd.save(\"output-shift-filtered.jpg\")\n\n # x derivative operator\n im_x_derivative_operator = to_gray_scale(im)\n im_x_derivative_operator = x_derivative_operator(im_x_derivative_operator)\n im_x_derivative_operator = torch.abs(im_x_derivative_operator)\n mx = torch.max(im_x_derivative_operator)\n im_x_derivative_operator = im_x_derivative_operator / mx\n im_x_derivative_operator = im_x_derivative_operator.squeeze()\n im_x_derivative_operator = torchvision.transforms.functional.to_pil_image(\n im_x_derivative_operator\n )\n\n im_x_derivative_operator.save(\"output-x-derivative-operator.jpg\")\n\n # y derivative operator\n im_y_derivative_operator = to_gray_scale(im)\n im_y_derivative_operator = y_derivative_operator(im_y_derivative_operator)\n im_y_derivative_operator = torch.abs(im_y_derivative_operator)\n mx = torch.max(im_y_derivative_operator)\n im_y_derivative_operator = im_y_derivative_operator / mx\n im_y_derivative_operator = im_y_derivative_operator.squeeze()\n im_y_derivative_operator = torchvision.transforms.functional.to_pil_image(\n im_y_derivative_operator\n )\n\n im_y_derivative_operator.save(\"output-y-derivative-operator.jpg\")\n\n # x prewitt filter\n im_x_prewitt_filter = to_gray_scale(im)\n im_x_prewitt_filter = x_prewitt_filter(im_x_prewitt_filter)\n im_x_prewitt_filter = torch.abs(im_x_prewitt_filter)\n mx = torch.max(im_x_prewitt_filter)\n im_x_prewitt_filter = im_x_prewitt_filter / mx\n im_x_prewitt_filter = im_x_prewitt_filter.squeeze()\n im_x_prewitt_filter = torchvision.transforms.functional.to_pil_image(\n im_x_prewitt_filter\n )\n\n im_x_prewitt_filter.save(\"output-x-prewitt-filter.jpg\")\n\n # y prewitt filter\n im_y_prewitt_filter = to_gray_scale(im)\n im_y_prewitt_filter = y_prewitt_filter(im_y_prewitt_filter)\n im_y_prewitt_filter = torch.abs(im_y_prewitt_filter)\n mx = torch.max(im_y_prewitt_filter)\n im_y_prewitt_filter = im_y_prewitt_filter / mx\n im_y_prewitt_filter = im_y_prewitt_filter.squeeze()\n im_y_prewitt_filter = torchvision.transforms.functional.to_pil_image(\n im_y_prewitt_filter\n )\n\n im_y_prewitt_filter.save(\"output-y-prewitt-filter.jpg\")\n\n # x sobel filter\n im_x_sobel_filter = to_gray_scale(im)\n im_x_sobel_filter = x_sobel_filter(im_x_sobel_filter)\n im_x_sobel_filter = torch.abs(im_x_sobel_filter)\n mx = torch.max(im_x_sobel_filter)\n im_x_sobel_filter = im_x_sobel_filter / mx\n im_x_sobel_filter = im_x_sobel_filter.squeeze()\n im_x_sobel_filter = torchvision.transforms.functional.to_pil_image(\n im_x_sobel_filter\n )\n\n im_x_sobel_filter.save(\"output-x-sobel-filter.jpg\")\n\n # y sobel filter\n im_y_sobel_filter = to_gray_scale(im)\n im_y_sobel_filter = y_sobel_filter(im_y_sobel_filter)\n im_y_sobel_filter = torch.abs(im_y_sobel_filter)\n mx = torch.max(im_y_sobel_filter)\n im_y_sobel_filter = im_y_sobel_filter / mx\n im_y_sobel_filter = im_y_sobel_filter.squeeze()\n im_y_sobel_filter = torchvision.transforms.functional.to_pil_image(\n im_y_sobel_filter\n )\n\n im_y_sobel_filter.save(\"output-y-sobel-filter.jpg\")\n\n # steerable filter\n im_steerable_filter = to_gray_scale(im)\n im_steerable_filter = steerable_filter(\n im_steerable_filter, math.radians(45)\n )\n im_steerable_filter = torch.abs(im_steerable_filter)\n mx = torch.max(im_steerable_filter)\n im_steerable_filter = im_steerable_filter / mx\n im_steerable_filter = im_steerable_filter.squeeze()\n im_steerable_filter = torchvision.transforms.functional.to_pil_image(\n im_steerable_filter\n )\n\n im_steerable_filter.save(\"output-steerable_filter.jpg\")\n\n # x 2nd derivative operator\n im_x_2nd_derivative_operator = to_gray_scale(im)\n im_x_2nd_derivative_operator = x_2nd_derivative_operator(\n im_x_2nd_derivative_operator\n )\n im_x_2nd_derivative_operator = torch.abs(im_x_2nd_derivative_operator)\n mx = torch.max(im_x_2nd_derivative_operator)\n im_x_2nd_derivative_operator = im_x_2nd_derivative_operator / mx\n im_x_2nd_derivative_operator = im_x_2nd_derivative_operator.squeeze()\n im_x_2nd_derivative_operator = (\n torchvision.transforms.functional.to_pil_image(\n im_x_2nd_derivative_operator\n )\n )\n\n im_x_2nd_derivative_operator.save(\"output-x-2nd-derivative-operator.jpg\")\n\n # y 2nd derivative operator\n im_y_2nd_derivative_operator = to_gray_scale(im)\n im_y_2nd_derivative_operator = y_2nd_derivative_operator(\n im_y_2nd_derivative_operator\n )\n im_y_2nd_derivative_operator = torch.abs(im_y_2nd_derivative_operator)\n mx = torch.max(im_y_2nd_derivative_operator)\n im_y_2nd_derivative_operator = im_y_2nd_derivative_operator / mx\n im_y_2nd_derivative_operator = im_y_2nd_derivative_operator.squeeze()\n im_y_2nd_derivative_operator = (\n torchvision.transforms.functional.to_pil_image(\n im_y_2nd_derivative_operator\n )\n )\n\n im_y_2nd_derivative_operator.save(\"output-y-2nd-derivative-operator.jpg\")\n\n # laplacian filter\n im_laplacian_filter = to_gray_scale(im)\n im_laplacian_filter = laplacian_filter(im_laplacian_filter)\n im_laplacian_filter = torch.abs(im_laplacian_filter)\n mx = torch.max(im_laplacian_filter)\n im_laplacian_filter = im_laplacian_filter / mx\n im_laplacian_filter = im_laplacian_filter.squeeze()\n im_laplacian_filter = torchvision.transforms.functional.to_pil_image(\n im_laplacian_filter\n )\n\n im_laplacian_filter.save(\"output-laplacian-filter.jpg\")\n\n # gaussian filter\n im_gaussian_filter = gaussian_filter(im, 9, 4, 3)\n im_gaussian_filter = torch.abs(im_gaussian_filter)\n mx = torch.max(im_gaussian_filter)\n im_gaussian_filter = im_gaussian_filter / mx\n im_gaussian_filter = im_gaussian_filter.squeeze()\n im_gaussian_filter = torchvision.transforms.functional.to_pil_image(\n im_gaussian_filter\n )\n\n im_gaussian_filter.save(\"output-gaussian-filter.jpg\")\n\n # laplacian of gaussian filter\n im_laplacian_of_gaussian = to_gray_scale(im)\n im_laplacian_of_gaussian = laplacian_of_gaussian(\n im_laplacian_of_gaussian, 9, 4, 1\n )\n im_laplacian_of_gaussian = torch.abs(im_laplacian_of_gaussian)\n mx = torch.max(im_laplacian_of_gaussian)\n im_laplacian_of_gaussian = im_laplacian_of_gaussian / mx\n im_laplacian_of_gaussian = im_laplacian_of_gaussian.squeeze()\n im_laplacian_of_gaussian = torchvision.transforms.functional.to_pil_image(\n im_laplacian_of_gaussian\n )\n\n im_laplacian_of_gaussian.save(\"output-laplacian-of-gaussian.jpg\")\n\n # garbor filter\n im_garbor_filter = to_gray_scale(im)\n im_garbor_filter = garbor_filter(\n im, 21, 10, math.pi / 4, 2.0, 2.0, 2 * math.pi, 0.0\n )\n im_garbor_filter = torch.abs(im_garbor_filter)\n mx = torch.max(im_garbor_filter)\n im_garbor_filter = im_garbor_filter / mx\n im_garbor_filter = im_garbor_filter.squeeze()\n im_garbor_filter = torchvision.transforms.functional.to_pil_image(\n im_garbor_filter\n )\n\n im_garbor_filter.save(\"output-garbor-filter.jpg\")\n"
]
| [
[
"torch.abs",
"torch.set_grad_enabled",
"torch.max"
]
]
|
Windxy/YOLO-F | [
"b334471613816238f7d968e538aca1b3f30b9482"
]
| [
"utils/utils.py"
]
| [
"from __future__ import division\nimport os\nimport math\nimport time\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.autograd import Variable\nimport numpy as np\nfrom PIL import Image, ImageDraw, ImageFont\nimport matplotlib.pyplot as plt\n\nclass DecodeBox(nn.Module):\n def __init__(self, anchors, num_classes, img_size):\n super(DecodeBox, self).__init__()\n self.anchors = anchors\n self.num_anchors = len(anchors)\n self.num_classes = num_classes\n self.bbox_attrs = 5 + num_classes\n self.img_size = img_size\n\n def forward(self, input):\n # input为bs,3*(1+4+num_classes),13,13\n\n # 一共多少张图片\n batch_size = input.size(0)\n # 13,13\n input_height = input.size(2)\n input_width = input.size(3)\n\n # 计算步长\n # 每一个特征点对应原来的图片上多少个像素点\n # 如果特征层为13x13的话,一个特征点就对应原来的图片上的32个像素点\n # 416/13 = 32\n stride_h = self.img_size[1] / input_height\n stride_w = self.img_size[0] / input_width\n\n # 把先验框的尺寸调整成特征层大小的形式\n # 计算出先验框在特征层上对应的宽高\n scaled_anchors = [(anchor_width / stride_w, anchor_height / stride_h) for anchor_width, anchor_height in self.anchors]\n\n # bs,3*(5+num_classes),13,13 -> bs,3,13,13,(5+num_classes)\n prediction = input.view(batch_size, self.num_anchors,\n self.bbox_attrs, input_height, input_width).permute(0, 1, 3, 4, 2).contiguous()\n\n # 先验框的中心位置的调整参数\n x = torch.sigmoid(prediction[..., 0]) \n y = torch.sigmoid(prediction[..., 1])\n # 先验框的宽高调整参数\n w = prediction[..., 2] # Width\n h = prediction[..., 3] # Height\n\n # 获得置信度,是否有物体\n conf = torch.sigmoid(prediction[..., 4])\n # 种类置信度\n pred_cls = torch.sigmoid(prediction[..., 5:]) # Cls pred.\n\n FloatTensor = torch.cuda.FloatTensor if x.is_cuda else torch.FloatTensor\n LongTensor = torch.cuda.LongTensor if x.is_cuda else torch.LongTensor\n\n # 生成网格,先验框中心,网格左上角 batch_size,3,13,13\n grid_x = torch.linspace(0, input_width - 1, input_width).repeat(input_width, 1).repeat(\n batch_size * self.num_anchors, 1, 1).view(x.shape).type(FloatTensor)\n grid_y = torch.linspace(0, input_height - 1, input_height).repeat(input_height, 1).t().repeat(\n batch_size * self.num_anchors, 1, 1).view(y.shape).type(FloatTensor)\n\n # 生成先验框的宽高\n anchor_w = FloatTensor(scaled_anchors).index_select(1, LongTensor([0]))\n anchor_h = FloatTensor(scaled_anchors).index_select(1, LongTensor([1]))\n anchor_w = anchor_w.repeat(batch_size, 1).repeat(1, 1, input_height * input_width).view(w.shape)\n anchor_h = anchor_h.repeat(batch_size, 1).repeat(1, 1, input_height * input_width).view(h.shape)\n \n # 计算调整后的先验框中心与宽高\n pred_boxes = FloatTensor(prediction[..., :4].shape)\n pred_boxes[..., 0] = x.data + grid_x\n pred_boxes[..., 1] = y.data + grid_y\n pred_boxes[..., 2] = torch.exp(w.data) * anchor_w\n pred_boxes[..., 3] = torch.exp(h.data) * anchor_h\n\n # fig = plt.figure()\n # ax = fig.add_subplot(121)\n # if input_height==13:\n # plt.ylim(0,13)\n # plt.xlim(0,13)\n # elif input_height==26:\n # plt.ylim(0,26)\n # plt.xlim(0,26)\n # elif input_height==52:\n # plt.ylim(0,52)\n # plt.xlim(0,52)\n # plt.scatter(grid_x.cpu(),grid_y.cpu())\n\n # anchor_left = grid_x - anchor_w/2 \n # anchor_top = grid_y - anchor_h/2 \n\n # rect1 = plt.Rectangle([anchor_left[0,0,5,5],anchor_top[0,0,5,5]],anchor_w[0,0,5,5],anchor_h[0,0,5,5],color=\"r\",fill=False)\n # rect2 = plt.Rectangle([anchor_left[0,1,5,5],anchor_top[0,1,5,5]],anchor_w[0,1,5,5],anchor_h[0,1,5,5],color=\"r\",fill=False)\n # rect3 = plt.Rectangle([anchor_left[0,2,5,5],anchor_top[0,2,5,5]],anchor_w[0,2,5,5],anchor_h[0,2,5,5],color=\"r\",fill=False)\n\n # ax.add_patch(rect1)\n # ax.add_patch(rect2)\n # ax.add_patch(rect3)\n\n # ax = fig.add_subplot(122)\n # if input_height==13:\n # plt.ylim(0,13)\n # plt.xlim(0,13)\n # elif input_height==26:\n # plt.ylim(0,26)\n # plt.xlim(0,26)\n # elif input_height==52:\n # plt.ylim(0,52)\n # plt.xlim(0,52)\n # plt.scatter(grid_x.cpu(),grid_y.cpu())\n # plt.scatter(pred_boxes[0,:,5,5,0].cpu(),pred_boxes[0,:,5,5,1].cpu(),c='r')\n\n # pre_left = pred_boxes[...,0] - pred_boxes[...,2]/2 \n # pre_top = pred_boxes[...,1] - pred_boxes[...,3]/2 \n\n # rect1 = plt.Rectangle([pre_left[0,0,5,5],pre_top[0,0,5,5]],pred_boxes[0,0,5,5,2],pred_boxes[0,0,5,5,3],color=\"r\",fill=False)\n # rect2 = plt.Rectangle([pre_left[0,1,5,5],pre_top[0,1,5,5]],pred_boxes[0,1,5,5,2],pred_boxes[0,1,5,5,3],color=\"r\",fill=False)\n # rect3 = plt.Rectangle([pre_left[0,2,5,5],pre_top[0,2,5,5]],pred_boxes[0,2,5,5,2],pred_boxes[0,2,5,5,3],color=\"r\",fill=False)\n\n # ax.add_patch(rect1)\n # ax.add_patch(rect2)\n # ax.add_patch(rect3)\n\n # plt.show()\n # 用于将输出调整为相对于416x416的大小\n _scale = torch.Tensor([stride_w, stride_h] * 2).type(FloatTensor)\n output = torch.cat((pred_boxes.view(batch_size, -1, 4) * _scale,\n conf.view(batch_size, -1, 1), pred_cls.view(batch_size, -1, self.num_classes)), -1)\n return output.data\n \ndef letterbox_image(image, size):\n iw, ih = image.size\n w, h = size\n scale = min(w/iw, h/ih)\n nw = int(iw*scale)\n nh = int(ih*scale)\n\n image = image.resize((nw,nh), Image.BICUBIC)\n new_image = Image.new('RGB', size, (128,128,128))\n new_image.paste(image, ((w-nw)//2, (h-nh)//2))\n return new_image\n\ndef yolo_correct_boxes(top, left, bottom, right, input_shape, image_shape):\n new_shape = image_shape*np.min(input_shape/image_shape)\n\n offset = (input_shape-new_shape)/2./input_shape\n scale = input_shape/new_shape\n\n box_yx = np.concatenate(((top+bottom)/2,(left+right)/2),axis=-1)/input_shape\n box_hw = np.concatenate((bottom-top,right-left),axis=-1)/input_shape\n\n box_yx = (box_yx - offset) * scale\n box_hw *= scale\n\n box_mins = box_yx - (box_hw / 2.)\n box_maxes = box_yx + (box_hw / 2.)\n boxes = np.concatenate([\n box_mins[:, 0:1],\n box_mins[:, 1:2],\n box_maxes[:, 0:1],\n box_maxes[:, 1:2]\n ],axis=-1)\n # print(np.shape(boxes))\n boxes *= np.concatenate([image_shape, image_shape],axis=-1)\n return boxes\n\ndef bbox_iou(box1, box2, x1y1x2y2=True):\n \"\"\"\n 计算IOU\n \"\"\"\n if not x1y1x2y2:\n b1_x1, b1_x2 = box1[:, 0] - box1[:, 2] / 2, box1[:, 0] + box1[:, 2] / 2\n b1_y1, b1_y2 = box1[:, 1] - box1[:, 3] / 2, box1[:, 1] + box1[:, 3] / 2\n b2_x1, b2_x2 = box2[:, 0] - box2[:, 2] / 2, box2[:, 0] + box2[:, 2] / 2\n b2_y1, b2_y2 = box2[:, 1] - box2[:, 3] / 2, box2[:, 1] + box2[:, 3] / 2\n else:\n b1_x1, b1_y1, b1_x2, b1_y2 = box1[:, 0], box1[:, 1], box1[:, 2], box1[:, 3]\n b2_x1, b2_y1, b2_x2, b2_y2 = box2[:, 0], box2[:, 1], box2[:, 2], box2[:, 3]\n\n inter_rect_x1 = torch.max(b1_x1, b2_x1)\n inter_rect_y1 = torch.max(b1_y1, b2_y1)\n inter_rect_x2 = torch.min(b1_x2, b2_x2)\n inter_rect_y2 = torch.min(b1_y2, b2_y2)\n\n inter_area = torch.clamp(inter_rect_x2 - inter_rect_x1 + 1, min=0) * \\\n torch.clamp(inter_rect_y2 - inter_rect_y1 + 1, min=0)\n \n b1_area = (b1_x2 - b1_x1 + 1) * (b1_y2 - b1_y1 + 1)\n b2_area = (b2_x2 - b2_x1 + 1) * (b2_y2 - b2_y1 + 1)\n\n iou = inter_area / (b1_area + b2_area - inter_area + 1e-16)\n\n return iou\n\n\ndef non_max_suppression(prediction, num_classes, conf_thres=0.5, nms_thres=0.4):\n # 求左上角和右下角\n box_corner = prediction.new(prediction.shape)\n box_corner[:, :, 0] = prediction[:, :, 0] - prediction[:, :, 2] / 2\n box_corner[:, :, 1] = prediction[:, :, 1] - prediction[:, :, 3] / 2\n box_corner[:, :, 2] = prediction[:, :, 0] + prediction[:, :, 2] / 2\n box_corner[:, :, 3] = prediction[:, :, 1] + prediction[:, :, 3] / 2\n prediction[:, :, :4] = box_corner[:, :, :4]\n\n output = [None for _ in range(len(prediction))]\n for image_i, image_pred in enumerate(prediction):\n # 利用置信度进行第一轮筛选\n conf_mask = (image_pred[:, 4] >= conf_thres).squeeze()\n image_pred = image_pred[conf_mask]\n\n if not image_pred.size(0):\n continue\n\n # 获得种类及其置信度\n class_conf, class_pred = torch.max(image_pred[:, 5:5 + num_classes], 1, keepdim=True)\n\n # 获得的内容为(x1, y1, x2, y2, obj_conf, class_conf, class_pred)\n detections = torch.cat((image_pred[:, :5], class_conf.float(), class_pred.float()), 1)\n\n # 获得种类\n unique_labels = detections[:, -1].cpu().unique()\n\n if prediction.is_cuda:\n unique_labels = unique_labels.cuda()\n\n for c in unique_labels:\n # 获得某一类初步筛选后全部的预测结果\n detections_class = detections[detections[:, -1] == c]\n # 按照存在物体的置信度排序\n _, conf_sort_index = torch.sort(detections_class[:, 4], descending=True)\n detections_class = detections_class[conf_sort_index]\n # 进行非极大抑制\n max_detections = []\n while detections_class.size(0):\n # 取出这一类置信度最高的,一步一步往下判断,判断重合程度是否大于nms_thres,如果是则去除掉\n max_detections.append(detections_class[0].unsqueeze(0))\n if len(detections_class) == 1:\n break\n ious = bbox_iou(max_detections[-1], detections_class[1:])\n detections_class = detections_class[1:][ious < nms_thres]\n # 堆叠\n max_detections = torch.cat(max_detections).data\n # Add max detections to outputs\n output[image_i] = max_detections if output[image_i] is None else torch.cat(\n (output[image_i], max_detections))\n\n return output\n\ndef merge_bboxes(bboxes, cutx, cuty):\n merge_bbox = []\n for i in range(len(bboxes)):\n for box in bboxes[i]:\n tmp_box = []\n x1,y1,x2,y2 = box[0], box[1], box[2], box[3]\n\n if i == 0:\n if y1 > cuty or x1 > cutx:\n continue\n if y2 >= cuty and y1 <= cuty:\n y2 = cuty\n if y2-y1 < 5:\n continue\n if x2 >= cutx and x1 <= cutx:\n x2 = cutx\n if x2-x1 < 5:\n continue\n \n if i == 1:\n if y2 < cuty or x1 > cutx:\n continue\n\n if y2 >= cuty and y1 <= cuty:\n y1 = cuty\n if y2-y1 < 5:\n continue\n \n if x2 >= cutx and x1 <= cutx:\n x2 = cutx\n if x2-x1 < 5:\n continue\n\n if i == 2:\n if y2 < cuty or x2 < cutx:\n continue\n\n if y2 >= cuty and y1 <= cuty:\n y1 = cuty\n if y2-y1 < 5:\n continue\n\n if x2 >= cutx and x1 <= cutx:\n x1 = cutx\n if x2-x1 < 5:\n continue\n\n if i == 3:\n if y1 > cuty or x2 < cutx:\n continue\n\n if y2 >= cuty and y1 <= cuty:\n y2 = cuty\n if y2-y1 < 5:\n continue\n\n if x2 >= cutx and x1 <= cutx:\n x1 = cutx\n if x2-x1 < 5:\n continue\n\n tmp_box.append(x1)\n tmp_box.append(y1)\n tmp_box.append(x2)\n tmp_box.append(y2)\n tmp_box.append(box[-1])\n merge_bbox.append(tmp_box)\n return merge_bbox"
]
| [
[
"torch.sigmoid",
"torch.linspace",
"torch.max",
"torch.Tensor",
"numpy.min",
"torch.cat",
"torch.min",
"numpy.concatenate",
"torch.exp",
"torch.sort",
"torch.clamp"
]
]
|
daBawse167/torch-toolbox | [
"a1c0440127bca00201dfa0d0e0dcbafb65628632"
]
| [
"torchtoolbox/tools/summary.py"
]
| [
"# -*- coding: utf-8 -*-\n# @Author : DevinYang([email protected])\n__all__ = ['summary']\n\nfrom collections import OrderedDict\nimport torch\nimport torch.nn as nn\nimport numpy as np\n\n\ndef _flops_str(flops):\n preset = [(1e12, 'T'), (1e9, 'G'), (1e6, 'M'), (1e3, 'K')]\n\n for p in preset:\n if flops // p[0] > 0:\n N = flops / p[0]\n ret = \"%.1f%s\" % (N, p[1])\n return ret\n ret = \"%.1f\" % flops\n return ret\n\n\ndef _cac_grad_params(p, w):\n t, n = 0, 0\n if w.requires_grad:\n t += p\n else:\n n += p\n return t, n\n\n\ndef _cac_conv(layer, input, output):\n # bs, ic, ih, iw = input[0].shape\n oh, ow = output.shape[-2:]\n kh, kw = layer.kernel_size\n ic, oc = layer.in_channels, layer.out_channels\n g = layer.groups\n\n tb_params = 0\n ntb__params = 0\n flops = 0\n if hasattr(layer, 'weight') and hasattr(layer.weight, 'shape'):\n params = np.prod(layer.weight.shape)\n t, n = _cac_grad_params(params, layer.weight)\n tb_params += t\n ntb__params += n\n flops += (2 * ic * kh * kw - 1) * oh * ow * (oc // g)\n if hasattr(layer, 'bias') and hasattr(layer.bias, 'shape'):\n params = np.prod(layer.bias.shape)\n t, n = _cac_grad_params(params, layer.bias)\n tb_params += t\n ntb__params += n\n flops += oh * ow * (oc // g)\n return tb_params, ntb__params, flops\n\n\ndef _cac_xx_norm(layer, input, output):\n tb_params = 0\n ntb__params = 0\n if hasattr(layer, 'weight') and hasattr(layer.weight, 'shape'):\n params = np.prod(layer.weight.shape)\n t, n = _cac_grad_params(params, layer.weight)\n tb_params += t\n ntb__params += n\n if hasattr(layer, 'bias') and hasattr(layer.bias, 'shape'):\n params = np.prod(layer.bias.shape)\n t, n = _cac_grad_params(params, layer.bias)\n tb_params += t\n ntb__params += n\n if hasattr(layer, 'running_mean') and hasattr(layer.running_mean, 'shape'):\n params = np.prod(layer.running_mean.shape)\n ntb__params += params\n if hasattr(layer, 'running_var') and hasattr(layer.running_var, 'shape'):\n params = np.prod(layer.running_var.shape)\n ntb__params += params\n in_shape = input[0]\n flops = np.prod(in_shape.shape)\n if layer.affine:\n flops *= 2\n return tb_params, ntb__params, flops\n\n\ndef _cac_linear(layer, input, output):\n ic, oc = layer.in_features, layer.out_features\n\n tb_params = 0\n ntb__params = 0\n flops = 0\n if hasattr(layer, 'weight') and hasattr(layer.weight, 'shape'):\n params = np.prod(layer.weight.shape)\n t, n = _cac_grad_params(params, layer.weight)\n tb_params += t\n ntb__params += n\n flops += (2 * ic - 1) * oc\n if hasattr(layer, 'bias') and hasattr(layer.bias, 'shape'):\n params = np.prod(layer.bias.shape)\n t, n = _cac_grad_params(params, layer.bias)\n tb_params += t\n ntb__params += n\n flops += oc\n return tb_params, ntb__params, flops\n\n\[email protected]_grad()\ndef summary(model, x, return_results=False):\n \"\"\"\n\n Args:\n model (nn.Module): model to summary\n x (torch.Tensor): input data\n return_results (bool): return results\n\n Returns:\n\n \"\"\"\n # change bn work way\n model.eval()\n\n def register_hook(layer):\n\n def hook(layer, input, output):\n model_name = str(layer.__class__.__name__)\n module_idx = len(model_summary)\n s_key = '{}-{}'.format(model_name, module_idx + 1)\n model_summary[s_key] = OrderedDict()\n model_summary[s_key]['input_shape'] = list(input[0].shape)\n if isinstance(output, (tuple, list)):\n model_summary[s_key]['output_shape'] = [\n list(o.shape) for o in output\n ]\n else:\n model_summary[s_key]['output_shape'] = list(output.shape)\n tb_params = 0\n ntb__params = 0\n flops = 0\n\n if isinstance(layer, nn.Conv2d):\n tb_params, ntb__params, flops = _cac_conv(layer, input, output)\n elif isinstance(layer, (nn.BatchNorm2d, nn.GroupNorm)):\n tb_params, ntb__params, flops = _cac_xx_norm(\n layer, input, output)\n elif isinstance(layer, nn.Linear):\n tb_params, ntb__params, flops = _cac_linear(\n layer, input, output)\n\n model_summary[s_key]['trainable_params'] = tb_params\n model_summary[s_key]['non_trainable_params'] = ntb__params\n model_summary[s_key]['params'] = tb_params + ntb__params\n model_summary[s_key]['flops'] = flops\n\n if not isinstance(layer, (nn.Sequential, nn.ModuleList,\n nn.Identity, nn.ModuleDict)):\n hooks.append(layer.register_forward_hook(hook))\n\n model_summary = OrderedDict()\n hooks = []\n model.apply(register_hook)\n model(x)\n for h in hooks:\n h.remove()\n\n print('-' * 80)\n line_new = \"{:>20} {:>25} {:>15} {:>15}\".format(\n \"Layer (type)\", \"Output Shape\", \"Params\", \"FLOPs(M+A) #\")\n print(line_new)\n print('=' * 80)\n total_params = 0\n trainable_params = 0\n total_flops = 0\n for layer in model_summary:\n line_new = \"{:>20} {:>25} {:>15} {:>15}\".format(\n layer,\n str(model_summary[layer]['output_shape']),\n model_summary[layer]['params'],\n model_summary[layer]['flops'],\n )\n print(line_new)\n total_params += model_summary[layer]['params']\n trainable_params += model_summary[layer]['trainable_params']\n total_flops += model_summary[layer]['flops']\n\n param_str = _flops_str(total_params)\n flop_str = _flops_str(total_flops)\n flop_str_m = _flops_str(total_flops // 2)\n param_size = total_params * 4 / (1024 ** 2)\n print('=' * 80)\n print(' Total parameters: {:,} {}'.format(total_params, param_str))\n print(' Trainable parameters: {:,}'.format(trainable_params))\n print(\n 'Non-trainable parameters: {:,}'.format(total_params - trainable_params))\n print('Total flops(M) : {:,} {}'.format(total_flops // 2, flop_str_m))\n print('Total flops(M+A): {:,} {}'.format(total_flops, flop_str))\n print('-' * 80)\n print('Parameters size (MB): {:.2f}'.format(param_size))\n if return_results:\n return total_params, total_flops\n"
]
| [
[
"torch.no_grad",
"numpy.prod"
]
]
|
mjasieczko/new_offer_success_predictor | [
"d6bf146f25729c96f611ffdbe57ecf9b9afa82b6"
]
| [
"src/application/app_utils.py"
]
| [
"import pickle\nimport warnings\nfrom pathlib import Path\n\nimport pandas as pd\n\nfrom data.data_manager import DataManager\nfrom data.data_processor import DataProcessor, TestDataProcessor\nfrom ml_preprocessing.categorical_encoders import LeaveOneOutEncoder\n\nwarnings.filterwarnings('ignore')\n\n\ndef run(arg_unseen_data_path: Path, arg_output_path: Path):\n \"\"\"\n main script of new_offer_success_predictor repo\n\n predicts offer acceptance probabilities (probabilities of success i.e. customer will\n accept our offer) using the best overall model (via roc_auc, recall, accuracy and precision\n metrics)\n\n saves results to excel file (xlsx) in form:\n customer_name | success_probability\n 1 x\n 2 y\n etc.\n \"\"\"\n \"\"\"\n read train data to help encode test set\n \"\"\"\n DM = DataManager()\n train_df = DM.load_data()\n\n \"\"\"\n firefighting\n \"\"\"\n arg_unseen_data_path = Path(Path(str(arg_unseen_data_path).split('.')[0]))\n\n \"\"\"\n read unseen data to predict their class\n \"\"\"\n DM_unseen = DataManager(local_path=Path(arg_unseen_data_path),\n project_path=Path(''),\n filename='',\n suffix='.parquet',\n csv_suffix='.csv')\n unseen_df = DM_unseen.load_data()\n\n \"\"\"\n only for testing reasons\n \"\"\"\n if 'accepted' in unseen_df.columns:\n unseen_df = unseen_df.drop(columns=['accepted'])\n\n \"\"\"\n process both train and test data\n \"\"\"\n customer_names = unseen_df.reset_index()[['name']].rename(columns={'name': 'customer_name'})\n DP = DataProcessor(train_df=train_df)\n processed_train_df = DP.perform_initial_features_engineering()\n TDP = TestDataProcessor(not_processed_train_df=train_df,\n processed_train_df=processed_train_df,\n test_df=unseen_df,\n sneaky_peaky=True)\n processed_unseen_df = TDP.perform_initial_features_engineering()\n\n columns_to_encode = ['offer_class',\n 'gender',\n 'customer_type',\n 'center',\n 'phone_calls',\n 'cc_len',\n 'cc_startswith']\n \"\"\"\n encoding test set\n \"\"\"\n enc = LeaveOneOutEncoder(train_df=processed_train_df,\n test_df=processed_unseen_df,\n columns_to_encode=columns_to_encode,\n target_column='target',\n random_state=42,\n mean=1,\n std=0.05)\n _, test_df_encoded = enc.fit()\n test_df_encoded_ohemails = test_df_encoded.copy(deep=True)\n\n \"\"\"\n dictionary for email ohe mapping\n \"\"\"\n email_ohe_names = {0: '0_emails',\n 1: '1_email',\n 2: '2_emails',\n 3: '3_emails',\n 4: '4_emails',\n 5: '5_emails'}\n\n test_df_encoded_ohemails = (\n pd.concat([test_df_encoded_ohemails, pd.get_dummies(test_df_encoded_ohemails['emails'])],\n axis=1).rename(columns=email_ohe_names)).drop(columns=['emails'])\n\n \"\"\"\n features used to predict on test set\n \"\"\"\n test_columns = ['log_salary', 'log_estimated_expenses_knn', 'log_offer_value_knn',\n 'nan_age', 'not_nan_age', '0_emails', '1_email', '2_emails',\n '3_emails', '4_emails', '5_emails', 'encoded_offer_class',\n 'encoded_gender', 'encoded_customer_type', 'encoded_center',\n 'encoded_phone_calls', 'encoded_cc_len', 'encoded_cc_startswith']\n unseen_data = test_df_encoded_ohemails[test_columns]\n\n \"\"\"\n load model\n \"\"\"\n models_path = (Path('/Users/mjasiecz/PycharmProjects/new_offer_success_predictor/models/final_model.pickle'))\n if not models_path.exists():\n print('be sure to change model path my friend :)')\n final_model = pickle.load(open(models_path, 'rb'))\n\n \"\"\"\n predict probability\n \"\"\"\n probabilities = pd.DataFrame(\n {'success_probability': final_model.predict_proba(unseen_data)[:, 1]}\n )\n result = pd.merge(customer_names,\n probabilities,\n how='inner',\n on=customer_names.index).drop(columns='key_0')\n\n \"\"\"\n generate results\n \"\"\"\n result.to_excel(arg_output_path,\n sheet_name='cust_prob_list',\n engine='xlsxwriter')\n\n print('results were generated to: '+str(arg_output_path))\n"
]
| [
[
"pandas.merge",
"pandas.get_dummies"
]
]
|
Nyquixt/multiview-human-pose-estimation-pytorch | [
"d1469ee19f1761281aa738287c6fca0fb1fd9866"
]
| [
"lib/utils/pose_utils.py"
]
| [
"# ------------------------------------------------------------------------------\n# Copyright (c) Microsoft Corporation. All rights reserved.\n# Licensed under the MIT License.\n# Written by Chunyu Wang ([email protected])\n# ------------------------------------------------------------------------------\n\nimport numpy as np\nfrom numpy.linalg import inv\nfrom sklearn.preprocessing import normalize\n\n\nclass PoseUtils:\n\n def estimate_camera(self, pose_2d, pose_3d, indices=None):\n \"\"\"Estimate camera parameters given 2d-3d pose pair.\n Args:\n pose_2d: [n_joint, 2] 2d pose array\n pose_3d: Correspondent [n_joint, 3] 3d pose array\n indices: Indices used to do least square. At least 4 independent points should provided. \n All of the points will be used if not specified.\n Returns:\n A [2, 3] projection matrix and a [2] translation matrix.\n \"\"\"\n if indices is None:\n indices = np.arange(pose_2d.shape[0])\n pose_2d = pose_2d.reshape([-1, 2])\n pose_3d = pose_3d.reshape([-1, 3])\n pose_2d_mean = np.mean(pose_2d, axis=0)\n pose_3d_mean = np.mean(pose_3d, axis=0)\n pose_2d = pose_2d - pose_2d_mean\n pose_3d = pose_3d - pose_3d_mean\n\n M = np.matmul(pose_2d[indices].T, np.linalg.pinv(pose_3d.T))\n U, s, Vt = np.linalg.svd(M)\n R = np.matmul(np.matmul(U, np.array([[1, 0, 0], [0, 1, 0]])), Vt)\n M = np.matmul(np.diag(s), R)\n t = pose_2d_mean - np.matmul(M, pose_3d_mean)\n\n R3 = np.cross(R[0, :], R[1, :])\n R3 = np.reshape(R3, (1, 3))\n R3 = normalize(R3)\n\n camera = {'R': np.concatenate((R, R3), axis=0), 's': s, 't': t}\n return camera\n\n def align_3d_to_2d(self, pose_2d, pose_3d, camera, rootIdx):\n \"\"\" Given the 2d and 3d poses, we align 3D pose to the 2D image frame, z of root is zero\n Args:\n pose_2d: [n_joint, 2] 2d pose array\n pose_3d: Correspondent [n_joint, 3] 3d pose array\n Returns:\n aligned3d: Correspondent [n_joint, 3] 3d pose array \n \"\"\"\n R = camera['R']\n s = np.mean(camera['s'])\n t = np.reshape(camera['t'], (2, 1))\n translation = np.dot(inv(R), np.vstack((t / s, s)))\n aligned3d = s * np.dot(R, (pose_3d + translation.T).T).T\n return aligned3d - np.array([0, 0, aligned3d[rootIdx, 2]])\n\n def procrustes(self, A, B, scaling=True, reflection='best'):\n \"\"\" A port of MATLAB's `procrustes` function to Numpy.\n $$ \\min_{R, T, S} \\sum_i^N || A_i - R B_i + T ||^2. $$\n Use notation from [course note]\n (https://fling.seas.upenn.edu/~cis390/dynamic/slides/CIS390_Lecture11.pdf).\n Args:\n A: Matrices of target coordinates.\n B: Matrices of input coordinates. Must have equal numbers of points\n (rows), but B may have fewer dimensions (columns) than A.\n scaling: if False, the scaling component of the transformation is forced\n to 1\n reflection:\n if 'best' (default), the transformation solution may or may not\n include a reflection component, depending on which fits the data\n best. setting reflection to True or False forces a solution with\n reflection or no reflection respectively.\n Returns:\n d: The residual sum of squared errors, normalized according to a measure\n of the scale of A, ((A - A.mean(0))**2).sum().\n Z: The matrix of transformed B-values.\n tform: A dict specifying the rotation, translation and scaling that\n maps A --> B.\n \"\"\"\n assert A.shape[0] == B.shape[0]\n n, dim_x = A.shape\n _, dim_y = B.shape\n\n # remove translation\n A_bar = A.mean(0)\n B_bar = B.mean(0)\n A0 = A - A_bar\n B0 = B - B_bar\n\n # remove scale\n ssX = (A0**2).sum()\n ssY = (B0**2).sum()\n A_norm = np.sqrt(ssX)\n B_norm = np.sqrt(ssY)\n A0 /= A_norm\n B0 /= B_norm\n\n if dim_y < dim_x:\n B0 = np.concatenate((B0, np.zeros(n, dim_x - dim_y)), 0)\n\n # optimum rotation matrix of B\n A = np.dot(A0.T, B0)\n U, s, Vt = np.linalg.svd(A)\n V = Vt.T\n R = np.dot(V, U.T)\n\n if reflection is not 'best':\n # does the current solution use a reflection?\n have_reflection = np.linalg.det(R) < 0\n\n # if that's not what was specified, force another reflection\n if reflection != have_reflection:\n V[:, -1] *= -1\n s[-1] *= -1\n R = np.dot(V, U.T)\n\n S_trace = s.sum()\n if scaling:\n # optimum scaling of B\n scale = S_trace * A_norm / B_norm\n\n # standarised distance between A and scale*B*R + c\n d = 1 - S_trace**2\n\n # transformed coords\n Z = A_norm * S_trace * np.dot(B0, R) + A_bar\n else:\n scale = 1\n d = 1 + ssY / ssX - 2 * S_trace * B_norm / A_norm\n Z = B_norm * np.dot(B0, R) + A_bar\n\n # transformation matrix\n if dim_y < dim_x:\n R = R[:dim_y, :]\n translation = A_bar - scale * np.dot(B_bar, R)\n\n # transformation values\n tform = {'rotation': R, 'scale': scale, 'translation': translation}\n return d, Z, tform\n\n\nif __name__ == '__main__':\n pose2d = np.array(\n [[115.42669678, 102.42271423], [99.8081665, 100.83456421], [\n 97.40727234, 154.66975403\n ], [93.27631378, 198.52540588], [130.71669006, 103.99852753], [\n 127.00919342, 156.07492065\n ], [116.97068024, 199.52674866], [116.74355316, 72.27806854],\n [117.79602051, 41.93487549], [119.92079926, 31.99210548], [\n 119.03995514, 17.96786118\n ], [135.2973175, 41.59934235], [165.03352356, 48.2557373],\n [193.47923279, 49.47089005], [98.16778564, 41.83195496],\n [66.04647827, 52.59766006], [39.56548309, 56.02058792]])\n\n pose3d = np.array([[-81.77583313, -552.1887207, 4291.38916016], [\n -211.30630493, -559.60925293, 4243.83203125\n ], [-236.88369751, -112.57989502,\n 4357.40820312], [-287.40042114, 277.21643066, 4596.80908203], [\n 47.75523758, -544.76800537, 4338.94726562\n ], [16.53305054, -103.14489746,\n 4470.79199219], [-74.98716736, 291.76489258, 4688.68212891], [\n -69.65709686, -795.61425781, 4218.54589844\n ], [-60.29067993, -1038.8338623,\n 4161.70996094], [-41.86167908, -1097.89172363, 4069.44775391], [\n -48.99368286, -1212.49609375, 4063.14331055\n ], [85.64931488, -1060.91186523,\n 4238.390625], [344.31176758, -1031.31384277, 4349.76513672],\n [592.30114746, -1025.36315918, 4360.91748047], [\n -219.86462402, -1028.68896484, 4115.89404297\n ], [-473.94354248, -924.9197998, 4046.16748047],\n [-662.23358154, -865.71044922, 3895.49780273]])\n\n box = np.array([234, 125, 736, 627])\n utils = PoseUtils()\n camera = utils.estimate_camera(pose2d, pose3d)\n print(camera['R'])\n aligned3d = utils.align_3d_to_2d(pose2d, pose3d, camera, 0)\n print(aligned3d)\n\n\n import cv2\n image = cv2.imread('./s_11_act_16_subact_02_ca_04_000001.jpg')\n print(image.shape)\n image = image[125:627, 234:736, :]\n npoints = aligned3d.shape[0]\n for i in range(npoints):\n cv2.circle(image, (int(aligned3d[i, 0]*502.0/224.0), int(aligned3d[i, 1]*502.0/224.0)), radius=14, color=(0,255,0))\n\n cv2.imshow('image', image)\n cv2.waitKey(0)\n\n\n"
]
| [
[
"numpy.diag",
"numpy.dot",
"numpy.linalg.svd",
"numpy.sqrt",
"numpy.reshape",
"numpy.arange",
"numpy.linalg.inv",
"numpy.matmul",
"numpy.concatenate",
"numpy.linalg.pinv",
"sklearn.preprocessing.normalize",
"numpy.linalg.det",
"numpy.mean",
"numpy.cross",
"numpy.array",
"numpy.zeros",
"numpy.vstack"
]
]
|
joaopfonseca/research | [
"ac4ad6fa05b5985050c63dc9e4e18cd00965e09b"
]
| [
"research/utils/_data.py"
]
| [
"\"\"\"\nData I/O utils. Later on I might add other data handling utilities.\n\"\"\"\nfrom os import listdir\nfrom os.path import isdir, join\nimport pandas as pd\nfrom sqlite3 import connect\n\n\ndef load_datasets(data_dir, suffix=\"\", target_exists=True, **read_csv_kwargs):\n \"\"\"Load datasets from sqlite database and/or csv files.\"\"\"\n assert isdir(data_dir), \"`data_dir` must be a directory.\"\n\n # Filter data by suffix\n dat_names = [dat for dat in listdir(data_dir) if dat.endswith(suffix)]\n\n # Read data\n datasets = []\n for dat_name in dat_names:\n data_path = join(data_dir, dat_name)\n\n # Handle csv data\n if dat_name.endswith(\".csv\"):\n ds = pd.read_csv(data_path, **read_csv_kwargs)\n name = dat_name.replace(\".csv\", \"\").replace(\"_\", \" \").upper()\n if target_exists:\n ds = (ds.iloc[:, :-1], ds.iloc[:, -1])\n datasets.append((name, ds))\n\n # Handle sqlite database\n elif dat_name.endswith(\".db\"):\n with connect(data_path) as connection:\n datasets_names = [\n name[0]\n for name in connection.execute(\n \"SELECT name FROM sqlite_master WHERE type='table';\"\n )\n ]\n for dataset_name in datasets_names:\n ds = pd.read_sql(f'select * from \"{dataset_name}\"', connection)\n if target_exists:\n ds = (ds.iloc[:, :-1], ds.iloc[:, -1])\n datasets.append((dataset_name.replace(\"_\", \" \").upper(), ds))\n return datasets\n"
]
| [
[
"pandas.read_csv",
"pandas.read_sql"
]
]
|
Zacchaeus14/lang-seg | [
"ad1196a4d33830f3219dbe2260a69364a745f094"
]
| [
"modules/lsegmentation_module.py"
]
| [
"import types\nimport time\nimport random\nimport clip\nimport torch\nimport torch.nn as nn\nimport torchvision.transforms as transforms\n\nfrom argparse import ArgumentParser\n\nimport pytorch_lightning as pl\n\nfrom data import get_dataset, get_available_datasets\n\nfrom encoding.models import get_segmentation_model\nfrom encoding.nn import SegmentationLosses\n\nfrom encoding.utils import batch_pix_accuracy, batch_intersection_union\n\n# add mixed precision\nimport torch.cuda.amp as amp\nimport numpy as np\n\nfrom encoding.utils import SegmentationMetric\n\nclass LSegmentationModule(pl.LightningModule):\n def __init__(self, data_path, dataset, batch_size, base_lr, max_epochs, **kwargs):\n super().__init__()\n\n self.data_path = data_path\n self.batch_size = batch_size\n self.base_lr = base_lr / 16 * batch_size\n self.lr = self.base_lr\n\n self.epochs = max_epochs\n self.other_kwargs = kwargs\n self.enabled = False #True mixed precision will make things complicated and leading to NAN error\n self.scaler = amp.GradScaler(enabled=self.enabled)\n\n def forward(self, x, labelset=''):\n return self.net(x, labelset=labelset)\n\n def evaluate(self, x, target=None):\n pred = self.net.forward(x)\n # print('pred shape', pred.shape)\n # print('target shape', target.shape)\n if isinstance(pred, (tuple, list)):\n pred = pred[0]\n if target is None:\n return pred\n correct, labeled = batch_pix_accuracy(pred.data, target.data)\n inter, union = batch_intersection_union(pred.data, target.data, self.nclass)\n\n return correct, labeled, inter, union\n\n def evaluate_random(self, x, labelset, target=None):\n pred = self.net.forward(x, labelset)\n # print('pred shape', pred.shape)\n if isinstance(pred, (tuple, list)):\n pred = pred[0]\n if target is None:\n return pred\n correct, labeled = batch_pix_accuracy(pred.data, target.data)\n inter, union = batch_intersection_union(pred.data, target.data, self.nclass)\n\n return correct, labeled, inter, union\n \n\n def training_step(self, batch, batch_nb):\n if self.dataset == 'vizwiz':\n img, target, question = batch\n question = question[0]\n # print('forwarding viwiz')\n # print('question', question)\n # print('target', target)\n else:\n img, target = batch\n with amp.autocast(enabled=self.enabled):\n if self.dataset == 'vizwiz':\n out = self(img, labelset=['other', question])\n else:\n out = self(img)\n # print('out shape', out.shape, '; target shape', target.shape)\n multi_loss = isinstance(out, tuple)\n if multi_loss:\n loss = self.criterion(*out, target)\n else:\n loss = self.criterion(out, target)\n loss = self.scaler.scale(loss)\n final_output = out[0] if multi_loss else out\n train_pred, train_gt = self._filter_invalid(final_output, target)\n if train_gt.nelement() != 0:\n self.train_accuracy(train_pred, train_gt)\n self.log(\"train_loss\", loss)\n return loss\n\n def training_epoch_end(self, outs):\n self.log(\"train_acc_epoch\", self.train_accuracy.compute())\n\n def validation_step(self, batch, batch_nb):\n if self.dataset == 'vizwiz':\n img, target, question = batch\n question = question[0]\n # print('forwarding viwiz')\n # print('question', question)\n # print('target', target)\n else:\n img, target = batch\n if self.dataset == 'vizwiz':\n out = self(img, labelset=['other', question])\n else:\n out = self(img)\n # print('out shape', out.shape, '; target shape', target.shape)\n multi_loss = isinstance(out, tuple)\n if multi_loss:\n val_loss = self.criterion(*out, target)\n else:\n val_loss = self.criterion(out, target)\n final_output = out[0] if multi_loss else out\n valid_pred, valid_gt = self._filter_invalid(final_output, target)\n self.val_iou.update(target, final_output)\n pixAcc, iou = self.val_iou.get()\n self.log(\"val_loss_step\", val_loss)\n self.log(\"pix_acc_step\", pixAcc)\n self.log(\n \"val_acc_step\",\n self.val_accuracy(valid_pred, valid_gt),\n )\n self.log(\"val_iou\", iou)\n\n def validation_epoch_end(self, outs):\n pixAcc, iou = self.val_iou.get()\n self.log(\"val_acc_epoch\", self.val_accuracy.compute())\n self.log(\"val_iou_epoch\", iou)\n self.log(\"pix_acc_epoch\", pixAcc)\n\n self.val_iou.reset()\n\n def _filter_invalid(self, pred, target):\n valid = target != self.other_kwargs[\"ignore_index\"]\n _, mx = torch.max(pred, dim=1)\n return mx[valid], target[valid]\n\n def configure_optimizers(self):\n params_list = [\n {\"params\": self.net.pretrained.parameters(), \"lr\": self.base_lr},\n ]\n if self.unfreeze_text:\n print(\"Unfreeze text encoder\")\n params_list.append(\n {\"params\": self.net.clip_pretrained.transformer.parameters(), \"lr\": self.base_lr}\n )\n if hasattr(self.net, \"scratch\"):\n print(\"Found output scratch\")\n params_list.append(\n {\"params\": self.net.scratch.parameters(), \"lr\": self.base_lr * 10}\n )\n if hasattr(self.net, \"auxlayer\"):\n print(\"Found auxlayer\")\n params_list.append(\n {\"params\": self.net.auxlayer.parameters(), \"lr\": self.base_lr * 10}\n )\n if hasattr(self.net, \"scale_inv_conv\"):\n print(self.net.scale_inv_conv)\n print(\"Found scaleinv layers\")\n params_list.append(\n {\n \"params\": self.net.scale_inv_conv.parameters(),\n \"lr\": self.base_lr * 10,\n }\n )\n params_list.append(\n {\"params\": self.net.scale2_conv.parameters(), \"lr\": self.base_lr * 10}\n )\n params_list.append(\n {\"params\": self.net.scale3_conv.parameters(), \"lr\": self.base_lr * 10}\n )\n params_list.append(\n {\"params\": self.net.scale4_conv.parameters(), \"lr\": self.base_lr * 10}\n )\n\n if self.other_kwargs[\"midasproto\"]:\n print(\"Using midas optimization protocol\")\n \n opt = torch.optim.Adam(\n params_list,\n lr=self.base_lr,\n betas=(0.9, 0.999),\n weight_decay=self.other_kwargs[\"weight_decay\"],\n )\n sch = torch.optim.lr_scheduler.LambdaLR(\n opt, lambda x: pow(1.0 - x / self.epochs, 0.9)\n )\n\n else:\n opt = torch.optim.SGD(\n params_list,\n lr=self.base_lr,\n momentum=0.9,\n weight_decay=self.other_kwargs[\"weight_decay\"],\n )\n sch = torch.optim.lr_scheduler.LambdaLR(\n opt, lambda x: pow(1.0 - x / self.epochs, 0.9)\n )\n return [opt], [sch]\n\n def train_dataloader(self):\n return torch.utils.data.DataLoader(\n self.trainset,\n batch_size=self.batch_size,\n shuffle=True,\n num_workers=16,\n worker_init_fn=lambda x: random.seed(time.time() + x),\n )\n\n def val_dataloader(self):\n return torch.utils.data.DataLoader(\n self.valset,\n batch_size=self.batch_size,\n shuffle=False,\n num_workers=16,\n )\n\n def get_trainset(self, dset, augment=False, **kwargs):\n print(kwargs)\n if augment == True:\n mode = \"train_x\"\n else:\n mode = \"train\"\n\n print('trainset mode', mode)\n dset = get_dataset(\n dset,\n root=self.data_path,\n split=\"train\",\n mode=mode,\n transform=self.train_transform,\n **kwargs\n )\n\n self.num_classes = dset.num_class\n self.train_accuracy = pl.metrics.Accuracy()\n\n return dset\n\n def get_valset(self, dset, augment=False, **kwargs):\n self.val_accuracy = pl.metrics.Accuracy()\n self.val_iou = SegmentationMetric(self.num_classes)\n\n if augment == True:\n mode = \"val_x\"\n else:\n mode = \"val\"\n\n print('valset mode', mode)\n return get_dataset(\n dset,\n root=self.data_path,\n split=\"val\",\n mode=mode,\n transform=self.val_transform,\n **kwargs\n )\n\n\n def get_criterion(self, **kwargs):\n return SegmentationLosses(\n se_loss=kwargs[\"se_loss\"], \n aux=kwargs[\"aux\"], \n nclass=self.num_classes, \n se_weight=kwargs[\"se_weight\"], \n aux_weight=kwargs[\"aux_weight\"], \n ignore_index=kwargs[\"ignore_index\"], \n )\n\n @staticmethod\n def add_model_specific_args(parent_parser):\n parser = ArgumentParser(parents=[parent_parser], add_help=False)\n parser.add_argument(\n \"--data_path\", type=str, help=\"path where dataset is stored\"\n )\n parser.add_argument(\n \"--dataset\",\n choices=get_available_datasets(),\n default=\"ade20k\",\n help=\"dataset to train on\",\n )\n parser.add_argument(\n \"--batch_size\", type=int, default=16, help=\"size of the batches\"\n )\n parser.add_argument(\n \"--base_lr\", type=float, default=0.004, help=\"learning rate\"\n )\n parser.add_argument(\"--momentum\", type=float, default=0.9, help=\"SGD momentum\")\n parser.add_argument(\n \"--weight_decay\", type=float, default=1e-4, help=\"weight_decay\"\n )\n parser.add_argument(\n \"--aux\", action=\"store_true\", default=False, help=\"Auxilary Loss\"\n )\n parser.add_argument(\n \"--aux-weight\",\n type=float,\n default=0.2,\n help=\"Auxilary loss weight (default: 0.2)\",\n )\n parser.add_argument(\n \"--se-loss\",\n action=\"store_true\",\n default=False,\n help=\"Semantic Encoding Loss SE-loss\",\n )\n parser.add_argument(\n \"--se-weight\", type=float, default=0.2, help=\"SE-loss weight (default: 0.2)\"\n )\n\n parser.add_argument(\n \"--midasproto\", action=\"store_true\", default=False, help=\"midasprotocol\"\n )\n\n parser.add_argument(\n \"--ignore_index\",\n type=int,\n default=-1,\n help=\"numeric value of ignore label in gt\",\n )\n parser.add_argument(\n \"--augment\",\n action=\"store_true\",\n default=False,\n help=\"Use extended augmentations\",\n )\n\n parser.add_argument(\n \"--unfreeze_text\",\n default=False,\n action=\"store_true\",\n help=\"unfreeze text encoder\",\n )\n\n return parser\n"
]
| [
[
"torch.optim.Adam",
"torch.max",
"torch.utils.data.DataLoader",
"torch.cuda.amp.autocast",
"torch.cuda.amp.GradScaler",
"torch.optim.SGD"
]
]
|
FangHao1993/LineBot-X-PhotoHub- | [
"77164642f610e56dbb720a1d7debc70efdb42382"
]
| [
"object_detection/evaluate2.py"
]
| [
"#! /usr/bin/env python\n# coding=utf-8\n#================================================================\n# Copyright (C) 2019 * Ltd. All rights reserved.\n#\n# Editor : VIM\n# File name : evaluate.py\n# Author : YunYang1994\n# Created date: 2019-02-21 15:30:26\n# Description :\n#\n#================================================================\n\nimport cv2\nimport os\nimport shutil\nimport numpy as np\nimport tensorflow as tf\nimport core.utils as utils\nfrom core.config import cfg\nfrom core.yolov3 import YOLOV3\nfrom collections import Counter\nclass YoloTest(object):\n def __init__(self):\n self.input_size = cfg.TEST.INPUT_SIZE\n self.anchor_per_scale = cfg.YOLO.ANCHOR_PER_SCALE\n self.classes = utils.read_class_names(cfg.YOLO.CLASSES)\n self.num_classes = len(self.classes)\n self.anchors = np.array(utils.get_anchors(cfg.YOLO.ANCHORS))\n self.score_threshold = cfg.TEST.SCORE_THRESHOLD\n self.iou_threshold = cfg.TEST.IOU_THRESHOLD\n self.moving_ave_decay = cfg.YOLO.MOVING_AVE_DECAY\n self.annotation_path = cfg.TEST.ANNOT_PATH\n self.weight_file = cfg.TEST.WEIGHT_FILE\n self.write_image = cfg.TEST.WRITE_IMAGE\n self.write_image_path = cfg.TEST.WRITE_IMAGE_PATH\n self.show_label = cfg.TEST.SHOW_LABEL\n\n with tf.name_scope('input'):\n self.input_data = tf.placeholder(dtype=tf.float32, name='input_data')\n self.trainable = tf.placeholder(dtype=tf.bool, name='trainable')\n\n model = YOLOV3(self.input_data, self.trainable)\n self.pred_sbbox, self.pred_mbbox, self.pred_lbbox = model.pred_sbbox, model.pred_mbbox, model.pred_lbbox\n\n with tf.name_scope('ema'):\n ema_obj = tf.train.ExponentialMovingAverage(self.moving_ave_decay)\n\n self.sess = tf.Session(config=tf.ConfigProto(allow_soft_placement=True))\n self.saver = tf.train.Saver(ema_obj.variables_to_restore())\n self.saver.restore(self.sess, self.weight_file)\n\n def predict(self, image):\n\n org_image = np.copy(image)\n org_h, org_w, _ = org_image.shape\n\n image_data = utils.image_preporcess(image, [self.input_size, self.input_size])\n image_data = image_data[np.newaxis, ...]\n\n pred_sbbox, pred_mbbox, pred_lbbox = self.sess.run(\n [self.pred_sbbox, self.pred_mbbox, self.pred_lbbox],\n feed_dict={\n self.input_data: image_data,\n self.trainable: False\n }\n )\n\n pred_bbox = np.concatenate([np.reshape(pred_sbbox, (-1, 5 + self.num_classes)),\n np.reshape(pred_mbbox, (-1, 5 + self.num_classes)),\n np.reshape(pred_lbbox, (-1, 5 + self.num_classes))], axis=0)\n bboxes = utils.postprocess_boxes(pred_bbox, (org_h, org_w), self.input_size, self.score_threshold)\n bboxes = utils.nms(bboxes, self.iou_threshold)\n\n return bboxes\n\n def evaluate(self):\n predicted_dir_path = './mAP/predicted'\n ground_truth_dir_path = './mAP/ground-truth'\n if os.path.exists(predicted_dir_path): shutil.rmtree(predicted_dir_path)\n if os.path.exists(ground_truth_dir_path): shutil.rmtree(ground_truth_dir_path)\n if os.path.exists(self.write_image_path): shutil.rmtree(self.write_image_path)\n os.mkdir(predicted_dir_path)\n os.mkdir(ground_truth_dir_path)\n os.mkdir(self.write_image_path)\n\n with open(self.annotation_path, 'r') as annotation_file:\n for num, line in enumerate(annotation_file):\n annotation = line.strip().split()\n image_path = annotation[0]\n image_name = image_path.split('/')[-1]\n image = cv2.imread(image_path)\n bbox_data_gt = np.array([list(map(int, box.split(','))) for box in annotation[1:]])\n\n if len(bbox_data_gt) == 0:\n bboxes_gt=[]\n classes_gt=[]\n else:\n bboxes_gt, classes_gt = bbox_data_gt[:, :4], bbox_data_gt[:, 4]\n ground_truth_path = os.path.join(ground_truth_dir_path, str(num) + '.txt')\n\n print('=> ground truth of %s:' % image_name)\n num_bbox_gt = len(bboxes_gt)\n with open(ground_truth_path, 'w') as f:\n for i in range(num_bbox_gt):\n class_name = self.classes[classes_gt[i]]\n xmin, ymin, xmax, ymax = list(map(str, bboxes_gt[i]))\n bbox_mess = ' '.join([class_name, xmin, ymin, xmax, ymax]) + '\\n'\n f.write(bbox_mess)\n print('\\t' + str(bbox_mess).strip())\n print('=> predict result of %s:' % image_name)\n predict_result_path = os.path.join(predicted_dir_path, str(num) + '.txt')\n bboxes_pr = self.predict(image)\n\n if self.write_image:\n image = utils.draw_bbox(image, bboxes_pr, show_label=self.show_label)\n cv2.imwrite(self.write_image_path+image_name, image)\n \n with open(\"./mAP/predicted/123.txt\", 'a') as f:\n class_names = []\n for bbox in bboxes_pr: \n coor = np.array(bbox[:4], dtype=np.int32)\n score = bbox[4]\n class_ind = int(bbox[5]) \n class_name = self.classes[class_ind]\n score = '%.4f' % score\n xmin, ymin, xmax, ymax = list(map(str, coor))\n# bbox_mess = ' '.join([class_name, image_path]) + '\\n'\n class_names.append(class_name)\n# bbox_mess = ' '.join([class_name, score, xmin, ymin, xmax, ymax]) + '\\n'\n bbox_mess = ' '.join\n# f.write(bbox_mess)\n print('\\t' + str(bbox_mess).strip())\n id_name = f'{dict(Counter(class_names))}@{image_path}\\n' \n f.write(id_name)\n if num == 9:\n break\n def voc_2012_test(self, voc2012_test_path):\n\n img_inds_file = os.path.join(voc2012_test_path, 'ImageSets', 'Main', 'test.txt')\n with open(img_inds_file, 'r') as f:\n txt = f.readlines()\n image_inds = [line.strip() for line in txt]\n\n results_path = 'results/VOC2012/Main'\n if os.path.exists(results_path):\n shutil.rmtree(results_path)\n os.makedirs(results_path)\n\n for image_ind in image_inds:\n image_path = os.path.join(voc2012_test_path, 'JPEGImages', image_ind + '.jpg')\n image = cv2.imread(image_path)\n\n print('predict result of %s:' % image_ind)\n bboxes_pr = self.predict(image)\n\n for bbox in bboxes_pr: \n coor = np.array(bbox[:4], dtype=np.int32)\n score = bbox[4]\n class_ind = int(bbox[5])\n class_name = self.classes[class_ind]\n score = '%.4f' % score\n xmin, ymin, xmax, ymax = list(map(str, coor))\n bbox_mess = ' '.join([image_ind, score, xmin, ymin, xmax, ymax]) + '\\n'\n with open(os.path.join(results_path, 'comp4_det_test_' + class_name + '.txt'), 'a') as f:\n f.write(bbox_mess)\n print('\\t' + str(bbox_mess).strip())\n\n\nif __name__ == '__main__': YoloTest().evaluate()\n\n\n\n"
]
| [
[
"numpy.reshape",
"tensorflow.placeholder",
"tensorflow.ConfigProto",
"numpy.copy",
"tensorflow.train.ExponentialMovingAverage",
"tensorflow.name_scope",
"numpy.array"
]
]
|
eloqute/WaveRNN | [
"036674b2e3745e22f15f6f945661f5f9d8a63003"
]
| [
"models/tacotron.py"
]
| [
"import os\nimport numpy as np\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom pathlib import Path\nfrom typing import Union\n\n\nclass HighwayNetwork(nn.Module):\n def __init__(self, size):\n super().__init__()\n self.W1 = nn.Linear(size, size)\n self.W2 = nn.Linear(size, size)\n self.W1.bias.data.fill_(0.)\n\n def forward(self, x):\n x1 = self.W1(x)\n x2 = self.W2(x)\n g = torch.sigmoid(x2)\n y = g * F.relu(x1) + (1. - g) * x\n return y\n\n\nclass Encoder(nn.Module):\n def __init__(self, embed_dims, num_chars, cbhg_channels, K, num_highways, dropout):\n super().__init__()\n self.embedding = nn.Embedding(num_chars, embed_dims)\n self.pre_net = PreNet(embed_dims)\n self.cbhg = CBHG(K=K, in_channels=cbhg_channels, channels=cbhg_channels,\n proj_channels=[cbhg_channels, cbhg_channels],\n num_highways=num_highways)\n\n def forward(self, x):\n x = self.embedding(x)\n x = self.pre_net(x)\n x.transpose_(1, 2)\n x = self.cbhg(x)\n return x\n\n\nclass BatchNormConv(nn.Module):\n def __init__(self, in_channels, out_channels, kernel, relu=True):\n super().__init__()\n self.conv = nn.Conv1d(in_channels, out_channels, kernel, stride=1, padding=kernel // 2, bias=False)\n self.bnorm = nn.BatchNorm1d(out_channels)\n self.relu = relu\n\n def forward(self, x):\n x = self.conv(x)\n x = F.relu(x) if self.relu is True else x\n return self.bnorm(x)\n\n\nclass CBHG(nn.Module):\n def __init__(self, K, in_channels, channels, proj_channels, num_highways):\n super().__init__()\n\n # List of all rnns to call `flatten_parameters()` on\n self._to_flatten = []\n\n self.bank_kernels = [i for i in range(1, K + 1)]\n self.conv1d_bank = nn.ModuleList()\n for k in self.bank_kernels:\n conv = BatchNormConv(in_channels, channels, k)\n self.conv1d_bank.append(conv)\n\n self.maxpool = nn.MaxPool1d(kernel_size=2, stride=1, padding=1)\n\n self.conv_project1 = BatchNormConv(len(self.bank_kernels) * channels, proj_channels[0], 3)\n self.conv_project2 = BatchNormConv(proj_channels[0], proj_channels[1], 3, relu=False)\n\n # Fix the highway input if necessary\n if proj_channels[-1] != channels:\n self.highway_mismatch = True\n self.pre_highway = nn.Linear(proj_channels[-1], channels, bias=False)\n else:\n self.highway_mismatch = False\n\n self.highways = nn.ModuleList()\n for i in range(num_highways):\n hn = HighwayNetwork(channels)\n self.highways.append(hn)\n\n self.rnn = nn.GRU(channels, channels, batch_first=True, bidirectional=True)\n self._to_flatten.append(self.rnn)\n\n # Avoid fragmentation of RNN parameters and associated warning\n self._flatten_parameters()\n\n def forward(self, x):\n # Although we `_flatten_parameters()` on init, when using DataParallel\n # the model gets replicated, making it no longer guaranteed that the\n # weights are contiguous in GPU memory. Hence, we must call it again\n self._flatten_parameters()\n\n # Save these for later\n residual = x\n seq_len = x.size(-1)\n conv_bank = []\n\n # Convolution Bank\n for conv in self.conv1d_bank:\n c = conv(x) # Convolution\n conv_bank.append(c[:, :, :seq_len])\n\n # Stack along the channel axis\n conv_bank = torch.cat(conv_bank, dim=1)\n\n # dump the last padding to fit residual\n x = self.maxpool(conv_bank)[:, :, :seq_len]\n\n # Conv1d projections\n x = self.conv_project1(x)\n x = self.conv_project2(x)\n\n # Residual Connect\n x = x + residual\n\n # Through the highways\n x = x.transpose(1, 2)\n if self.highway_mismatch is True:\n x = self.pre_highway(x)\n for h in self.highways: x = h(x)\n\n # And then the RNN\n x, _ = self.rnn(x)\n return x\n\n def _flatten_parameters(self):\n \"\"\"Calls `flatten_parameters` on all the rnns used by the WaveRNN. Used\n to improve efficiency and avoid PyTorch yelling at us.\"\"\"\n [m.flatten_parameters() for m in self._to_flatten]\n\nclass PreNet(nn.Module):\n def __init__(self, in_dims, fc1_dims=256, fc2_dims=128, dropout=0.5):\n super().__init__()\n self.fc1 = nn.Linear(in_dims, fc1_dims)\n self.fc2 = nn.Linear(fc1_dims, fc2_dims)\n self.p = dropout\n\n def forward(self, x):\n x = self.fc1(x)\n x = F.relu(x)\n x = F.dropout(x, self.p, training=self.training)\n x = self.fc2(x)\n x = F.relu(x)\n x = F.dropout(x, self.p, training=self.training)\n return x\n\n\nclass Attention(nn.Module):\n def __init__(self, attn_dims):\n super().__init__()\n self.W = nn.Linear(attn_dims, attn_dims, bias=False)\n self.v = nn.Linear(attn_dims, 1, bias=False)\n\n def forward(self, encoder_seq_proj, query, t):\n\n # print(encoder_seq_proj.shape)\n # Transform the query vector\n query_proj = self.W(query).unsqueeze(1)\n\n # Compute the scores\n u = self.v(torch.tanh(encoder_seq_proj + query_proj))\n scores = F.softmax(u, dim=1)\n\n return scores.transpose(1, 2)\n\n\nclass LSA(nn.Module):\n def __init__(self, attn_dim, kernel_size=31, filters=32):\n super().__init__()\n self.conv = nn.Conv1d(2, filters, padding=(kernel_size - 1) // 2, kernel_size=kernel_size, bias=False)\n self.L = nn.Linear(filters, attn_dim, bias=True)\n self.W = nn.Linear(attn_dim, attn_dim, bias=True)\n self.v = nn.Linear(attn_dim, 1, bias=False)\n self.cumulative = None\n self.attention = None\n\n def init_attention(self, encoder_seq_proj):\n device = next(self.parameters()).device # use same device as parameters\n b, t, c = encoder_seq_proj.size()\n self.cumulative = torch.zeros(b, t, device=device)\n self.attention = torch.zeros(b, t, device=device)\n\n def forward(self, encoder_seq_proj, query, t):\n\n if t == 0: self.init_attention(encoder_seq_proj)\n\n processed_query = self.W(query).unsqueeze(1)\n\n location = torch.cat([self.cumulative.unsqueeze(1), self.attention.unsqueeze(1)], dim=1)\n processed_loc = self.L(self.conv(location).transpose(1, 2))\n\n u = self.v(torch.tanh(processed_query + encoder_seq_proj + processed_loc))\n u = u.squeeze(-1)\n\n # Smooth Attention\n scores = torch.sigmoid(u) / torch.sigmoid(u).sum(dim=1, keepdim=True)\n # scores = F.softmax(u, dim=1)\n self.attention = scores\n self.cumulative += self.attention\n\n return scores.unsqueeze(-1).transpose(1, 2)\n\n\nclass Decoder(nn.Module):\n # Class variable because its value doesn't change between classes\n # yet ought to be scoped by class because its a property of a Decoder\n max_r = 20\n def __init__(self, n_mels, decoder_dims, lstm_dims):\n super().__init__()\n self.register_buffer('r', torch.tensor(1, dtype=torch.int))\n self.n_mels = n_mels\n self.prenet = PreNet(n_mels)\n self.attn_net = LSA(decoder_dims)\n self.attn_rnn = nn.GRUCell(decoder_dims + decoder_dims // 2, decoder_dims)\n self.rnn_input = nn.Linear(2 * decoder_dims, lstm_dims)\n self.res_rnn1 = nn.LSTMCell(lstm_dims, lstm_dims)\n self.res_rnn2 = nn.LSTMCell(lstm_dims, lstm_dims)\n self.mel_proj = nn.Linear(lstm_dims, n_mels * self.max_r, bias=False)\n\n def zoneout(self, prev, current, p=0.1):\n device = next(self.parameters()).device # Use same device as parameters\n mask = torch.zeros(prev.size(), device=device).bernoulli_(p)\n return prev * mask + current * (1 - mask)\n\n def forward(self, encoder_seq, encoder_seq_proj, prenet_in,\n hidden_states, cell_states, context_vec, t):\n\n # Need this for reshaping mels\n batch_size = encoder_seq.size(0)\n\n # Unpack the hidden and cell states\n attn_hidden, rnn1_hidden, rnn2_hidden = hidden_states\n rnn1_cell, rnn2_cell = cell_states\n\n # PreNet for the Attention RNN\n prenet_out = self.prenet(prenet_in)\n\n # Compute the Attention RNN hidden state\n attn_rnn_in = torch.cat([context_vec, prenet_out], dim=-1)\n attn_hidden = self.attn_rnn(attn_rnn_in.squeeze(1), attn_hidden)\n\n # Compute the attention scores\n scores = self.attn_net(encoder_seq_proj, attn_hidden, t)\n\n # Dot product to create the context vector\n context_vec = scores @ encoder_seq\n context_vec = context_vec.squeeze(1)\n\n # Concat Attention RNN output w. Context Vector & project\n x = torch.cat([context_vec, attn_hidden], dim=1)\n x = self.rnn_input(x)\n\n # Compute first Residual RNN\n rnn1_hidden_next, rnn1_cell = self.res_rnn1(x, (rnn1_hidden, rnn1_cell))\n if self.training:\n rnn1_hidden = self.zoneout(rnn1_hidden, rnn1_hidden_next)\n else:\n rnn1_hidden = rnn1_hidden_next\n x = x + rnn1_hidden\n\n # Compute second Residual RNN\n rnn2_hidden_next, rnn2_cell = self.res_rnn2(x, (rnn2_hidden, rnn2_cell))\n if self.training:\n rnn2_hidden = self.zoneout(rnn2_hidden, rnn2_hidden_next)\n else:\n rnn2_hidden = rnn2_hidden_next\n x = x + rnn2_hidden\n\n # Project Mels\n mels = self.mel_proj(x)\n mels = mels.view(batch_size, self.n_mels, self.max_r)[:, :, :self.r]\n hidden_states = (attn_hidden, rnn1_hidden, rnn2_hidden)\n cell_states = (rnn1_cell, rnn2_cell)\n\n return mels, scores, hidden_states, cell_states, context_vec\n\n\nclass Tacotron(nn.Module):\n def __init__(self, embed_dims, num_chars, encoder_dims, decoder_dims, n_mels, fft_bins, postnet_dims,\n encoder_K, lstm_dims, postnet_K, num_highways, dropout, stop_threshold):\n super().__init__()\n self.n_mels = n_mels\n self.lstm_dims = lstm_dims\n self.decoder_dims = decoder_dims\n self.encoder = Encoder(embed_dims, num_chars, encoder_dims,\n encoder_K, num_highways, dropout)\n self.encoder_proj = nn.Linear(decoder_dims, decoder_dims, bias=False)\n self.decoder = Decoder(n_mels, decoder_dims, lstm_dims)\n self.postnet = CBHG(postnet_K, n_mels, postnet_dims, [256, 80], num_highways)\n self.post_proj = nn.Linear(postnet_dims * 2, fft_bins, bias=False)\n\n self.init_model()\n self.num_params()\n\n self.register_buffer('step', torch.zeros(1, dtype=torch.long))\n self.register_buffer('stop_threshold', torch.tensor(stop_threshold, dtype=torch.float32))\n\n @property\n def r(self):\n return self.decoder.r.item()\n\n @r.setter\n def r(self, value):\n self.decoder.r = self.decoder.r.new_tensor(value, requires_grad=False)\n\n def forward(self, x, m, generate_gta=False):\n device = next(self.parameters()).device # use same device as parameters\n\n self.step += 1\n\n if generate_gta:\n self.eval()\n else:\n self.train()\n\n batch_size, _, steps = m.size()\n\n # Initialise all hidden states and pack into tuple\n attn_hidden = torch.zeros(batch_size, self.decoder_dims, device=device)\n rnn1_hidden = torch.zeros(batch_size, self.lstm_dims, device=device)\n rnn2_hidden = torch.zeros(batch_size, self.lstm_dims, device=device)\n hidden_states = (attn_hidden, rnn1_hidden, rnn2_hidden)\n\n # Initialise all lstm cell states and pack into tuple\n rnn1_cell = torch.zeros(batch_size, self.lstm_dims, device=device)\n rnn2_cell = torch.zeros(batch_size, self.lstm_dims, device=device)\n cell_states = (rnn1_cell, rnn2_cell)\n\n # <GO> Frame for start of decoder loop\n go_frame = torch.zeros(batch_size, self.n_mels, device=device)\n\n # Need an initial context vector\n context_vec = torch.zeros(batch_size, self.decoder_dims, device=device)\n\n # Project the encoder outputs to avoid\n # unnecessary matmuls in the decoder loop\n encoder_seq = self.encoder(x)\n encoder_seq_proj = self.encoder_proj(encoder_seq)\n\n # Need a couple of lists for outputs\n mel_outputs, attn_scores = [], []\n\n # Run the decoder loop\n for t in range(0, steps, self.r):\n prenet_in = m[:, :, t - 1] if t > 0 else go_frame\n mel_frames, scores, hidden_states, cell_states, context_vec = \\\n self.decoder(encoder_seq, encoder_seq_proj, prenet_in,\n hidden_states, cell_states, context_vec, t)\n mel_outputs.append(mel_frames)\n attn_scores.append(scores)\n\n # Concat the mel outputs into sequence\n mel_outputs = torch.cat(mel_outputs, dim=2)\n\n # Post-Process for Linear Spectrograms\n postnet_out = self.postnet(mel_outputs)\n linear = self.post_proj(postnet_out)\n linear = linear.transpose(1, 2)\n\n # For easy visualisation\n attn_scores = torch.cat(attn_scores, 1)\n # attn_scores = attn_scores.cpu().data.numpy()\n\n return mel_outputs, linear, attn_scores\n\n def generate(self, x, steps=2000):\n self.eval()\n device = next(self.parameters()).device # use same device as parameters\n\n batch_size = 1\n x = torch.as_tensor(x, dtype=torch.long, device=device).unsqueeze(0)\n\n # Need to initialise all hidden states and pack into tuple for tidyness\n attn_hidden = torch.zeros(batch_size, self.decoder_dims, device=device)\n rnn1_hidden = torch.zeros(batch_size, self.lstm_dims, device=device)\n rnn2_hidden = torch.zeros(batch_size, self.lstm_dims, device=device)\n hidden_states = (attn_hidden, rnn1_hidden, rnn2_hidden)\n\n # Need to initialise all lstm cell states and pack into tuple for tidyness\n rnn1_cell = torch.zeros(batch_size, self.lstm_dims, device=device)\n rnn2_cell = torch.zeros(batch_size, self.lstm_dims, device=device)\n cell_states = (rnn1_cell, rnn2_cell)\n\n # Need a <GO> Frame for start of decoder loop\n go_frame = torch.zeros(batch_size, self.n_mels, device=device)\n\n # Need an initial context vector\n context_vec = torch.zeros(batch_size, self.decoder_dims, device=device)\n\n # Project the encoder outputs to avoid\n # unnecessary matmuls in the decoder loop\n encoder_seq = self.encoder(x)\n encoder_seq_proj = self.encoder_proj(encoder_seq)\n\n # Need a couple of lists for outputs\n mel_outputs, attn_scores = [], []\n\n # Run the decoder loop\n for t in range(0, steps, self.r):\n prenet_in = mel_outputs[-1][:, :, -1] if t > 0 else go_frame\n mel_frames, scores, hidden_states, cell_states, context_vec = \\\n self.decoder(encoder_seq, encoder_seq_proj, prenet_in,\n hidden_states, cell_states, context_vec, t)\n mel_outputs.append(mel_frames)\n attn_scores.append(scores)\n # Stop the loop if silent frames present\n if (mel_frames < self.stop_threshold).all() and t > 10: break\n\n # Concat the mel outputs into sequence\n mel_outputs = torch.cat(mel_outputs, dim=2)\n\n # Post-Process for Linear Spectrograms\n postnet_out = self.postnet(mel_outputs)\n linear = self.post_proj(postnet_out)\n\n\n linear = linear.transpose(1, 2)[0].cpu().data.numpy()\n mel_outputs = mel_outputs[0].cpu().data.numpy()\n\n # For easy visualisation\n attn_scores = torch.cat(attn_scores, 1)\n attn_scores = attn_scores.cpu().data.numpy()[0]\n\n self.train()\n\n return mel_outputs, linear, attn_scores\n\n def init_model(self):\n for p in self.parameters():\n if p.dim() > 1: nn.init.xavier_uniform_(p)\n\n def get_step(self):\n return self.step.data.item()\n\n def reset_step(self):\n # assignment to parameters or buffers is overloaded, updates internal dict entry\n self.step = self.step.data.new_tensor(1)\n\n def log(self, path, msg):\n with open(path, 'a') as f:\n print(msg, file=f)\n\n def load(self, path: Union[str, Path]):\n # Use device of model params as location for loaded state\n device = next(self.parameters()).device\n state_dict = torch.load(path, map_location=device)\n\n # Backwards compatibility with old saved models\n if 'r' in state_dict and not 'decoder.r' in state_dict:\n self.r = state_dict['r']\n\n self.load_state_dict(state_dict, strict=False)\n\n def save(self, path: Union[str, Path]):\n # No optimizer argument because saving a model should not include data\n # only relevant in the training process - it should only be properties\n # of the model itself. Let caller take care of saving optimzier state.\n torch.save(self.state_dict(), path)\n\n def num_params(self, print_out=True):\n parameters = filter(lambda p: p.requires_grad, self.parameters())\n parameters = sum([np.prod(p.size()) for p in parameters]) / 1_000_000\n if print_out:\n print('Trainable Parameters: %.3fM' % parameters)\n return parameters\n"
]
| [
[
"torch.nn.functional.softmax",
"torch.cat",
"torch.nn.functional.dropout",
"torch.zeros",
"torch.load",
"torch.nn.GRU",
"torch.nn.Embedding",
"torch.tanh",
"torch.tensor",
"torch.nn.LSTMCell",
"torch.nn.MaxPool1d",
"torch.nn.functional.relu",
"torch.nn.GRUCell",
"torch.nn.BatchNorm1d",
"torch.sigmoid",
"torch.nn.ModuleList",
"torch.nn.Linear",
"torch.nn.Conv1d",
"torch.as_tensor",
"torch.nn.init.xavier_uniform_"
]
]
|
gaecom/Paddle | [
"d39d8bee4ee34d44f7012a0e5715f00f3ea31a90"
]
| [
"python/paddle/fluid/tests/unittests/test_dygraph_to_static_basic.py"
]
| [
"# Copyright (c) 2019 PaddlePaddle Authors. All Rights Reserved.\n#\n# Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# http://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.\n\nfrom __future__ import print_function\n\nimport numpy as np\nimport paddle.fluid as fluid\nimport unittest\n\nfrom paddle.fluid.dygraph.jit import dygraph_to_static_output\n\nnp.random.seed(1)\n\n\ndef dyfunc_with_if_else(x_v):\n if fluid.layers.mean(x_v).numpy()[0] > 5:\n x_v = x_v - 1\n else:\n x_v = x_v + 1\n return x_v\n\n\ndef dyfunc_with_if_else2(x):\n i, j = 0, 0\n if fluid.layers.reduce_mean(x).numpy()[0] > x.numpy()[i][j]:\n y = fluid.layers.relu(x)\n else:\n x_pow = fluid.layers.pow(x, 2)\n y = fluid.layers.tanh(x_pow)\n return y\n\n\ndef nested_if_else(x_v):\n batch_size = x_v.shape[0]\n feat_size = x_v.shape[-1]\n bias = fluid.layers.fill_constant([feat_size], dtype='float32', value=1)\n if fluid.layers.mean(x_v).numpy()[0] < 0:\n y = x_v + bias\n w = fluid.layers.fill_constant([feat_size], dtype='float32', value=10)\n if y.numpy()[0] < 10:\n tmp = y * w\n y = fluid.layers.relu(tmp)\n if fluid.layers.mean(y).numpy()[0] < batch_size:\n y = fluid.layers.abs(y)\n else:\n tmp = fluid.layers.fill_constant(\n [feat_size], dtype='float32', value=-1)\n y = y - tmp\n else:\n y = x_v - bias\n return y\n\n\nclass TestDygraphIfElse(unittest.TestCase):\n \"\"\"\n TestCase for the transformation from control flow `if/else`\n dependent on tensor in Dygraph into Static `fluid.layers.cond`.\n \"\"\"\n\n def setUp(self):\n self.x = np.random.random([10, 16]).astype('float32')\n self.dyfunc = dyfunc_with_if_else\n\n def _run_static(self):\n main_program = fluid.Program()\n with fluid.program_guard(main_program):\n x_v = fluid.layers.assign(self.x)\n # Transform into static graph\n out = dygraph_to_static_output(self.dyfunc)(x_v)\n exe = fluid.Executor(fluid.CPUPlace())\n ret = exe.run(main_program, fetch_list=out)\n return ret\n\n def _run_dygraph(self):\n with fluid.dygraph.guard():\n x_v = fluid.dygraph.to_variable(self.x)\n ret = self.dyfunc(x_v)\n return ret.numpy()\n\n def test_ast_to_func(self):\n self.assertTrue((self._run_dygraph() == self._run_static()).all())\n\n\nclass TestDygraphIfElse2(TestDygraphIfElse):\n def setUp(self):\n self.x = np.random.random([10, 16]).astype('float32')\n self.dyfunc = dyfunc_with_if_else2\n\n\nclass TestDygraphIfElse3(TestDygraphIfElse):\n def setUp(self):\n self.x = np.random.random([10, 16]).astype('float32')\n self.dyfunc = nested_if_else\n\n\nif __name__ == '__main__':\n unittest.main()\n"
]
| [
[
"numpy.random.random",
"numpy.random.seed"
]
]
|
KasiaOtko/Master_thesis | [
"41630cb2d4b7e82ceacc146437645acacbe41428"
]
| [
"src/models/gnn_classifiers/GraphSAGE_model.py"
]
| [
"import torch\nimport torch.nn.functional as F\nfrom torch import batch_norm, nn\nfrom torch_geometric.nn import BatchNorm, Linear, SAGEConv # type: ignore\n\n\nclass SAGE(nn.Module):\n def __init__(\n self, in_channels: int, hidden_channels: int, out_channels: int, dropout: float, num_layers: int, aggr: str, linear_l: bool\n ) -> None:\n super().__init__()\n self.num_layers = num_layers\n self.linear_l - linear_l\n self.convs = torch.nn.ModuleList()\n self.skips = torch.nn.ModuleList()\n self.batch_norms = torch.nn.ModuleList()\n\n self.lin1 = Linear(in_channels, hidden_channels)\n if self.linear_l:\n self.convs.append(SAGEConv(hidden_channels, hidden_channels, aggr = aggr, root_weight = False))\n self.skips.append(Linear(hidden_channels, hidden_channels))\n else:\n self.convs.append(SAGEConv(in_channels, hidden_channels, aggr = aggr, root_weight = False))\n self.skips.append(Linear(in_channels, hidden_channels))\n self.batch_norms.append(BatchNorm(hidden_channels))\n\n for _ in range(num_layers - 2):\n self.convs.append(SAGEConv(hidden_channels, hidden_channels, aggr = aggr, root_weight = False))\n self.skips.append(Linear(hidden_channels, hidden_channels))\n self.batch_norms.append(BatchNorm(hidden_channels))\n\n if self.linear_l:\n self.convs.append(SAGEConv(hidden_channels, hidden_channels, aggr = aggr, root_weight = False))\n self.skips.append(Linear(hidden_channels, hidden_channels))\n self.batch_norms.append(BatchNorm(hidden_channels))\n self.lin2 = Linear(hidden_channels, out_channels)\n else:\n self.convs.append(SAGEConv(hidden_channels, out_channels, aggr = aggr, root_weight = False))\n self.skips.append(Linear(hidden_channels, out_channels))\n self.dropout = nn.Dropout(dropout)\n\n\n def forward(self, x: torch.Tensor, edge_index: torch.Tensor) -> torch.Tensor:\n if x.ndim != 2:\n raise ValueError(\"Expected input is not a 2D tensor,\"\n f\"instead it is a {x.ndim}D tensor.\")\n\n if self.linear_l:\n x = self.lin1(x).relu()\n for i, (conv, skip, batch_norm) in enumerate(zip(self.convs, self.skips, self.batch_norms)):\n x = conv(x, edge_index) + skip(x)\n if i == self.num_layers - 1:\n h = x.clone()\n x = batch_norm(x)\n x = F.relu(x)\n x = self.dropout(x)\n x = self.lin2(x)\n\n else:\n\n for i, (conv, skip, batch_norm) in enumerate(zip(self.convs[:-1], self.skips[:-1], self.batch_norms)):\n\n x = conv(x, edge_index) + skip(x)\n if i == self.num_layers - 2:\n h = x.clone()\n x = batch_norm(x)\n x = F.relu(x)\n x = self.dropout(x)\n x = self.convs[-1](x, edge_index) + self.skips[-1](x)\n\n return h, x"
]
| [
[
"torch.nn.Dropout",
"torch.nn.ModuleList",
"torch.nn.functional.relu",
"torch.batch_norm"
]
]
|
RebeccaLeh/todolists | [
"234e5e873374aca9d1af1f40f95832da680decea"
]
| [
"export_pitcount.pyt"
]
| [
"import xlsxwriter , os, arcpy, sys, string, math, traceback, numpy\r\nimport pandas as df\r\n\r\nclass Toolbox(object):\r\n def __init__(self):\r\n \"\"\"Define the toolbox (the name of the toolbox is the name of the\r\n .pyt file).\"\"\"\r\n self.label = \"Toolbox\"\r\n self.alias = \"\"\r\n\r\n # List of tool classes associated with this toolbox\r\n self.tools = [ExcelOutput]\r\n\r\n###############################################################################################################################\r\n############################################################### RISK LAYER #########################################################\r\n#################################################################################################################################\r\n\r\nclass ExcelOutput(object):\r\n def __init__(self):\r\n \"\"\"Define the tool (tool name is the name of the class).\"\"\"\r\n self.label = \"Create Excel Output\"\r\n self.description = \"This tool creates a data output for HUD reporting on PIT count\"\r\n self.canRunInBackground = False\r\n\r\n def getParameterInfo(self):\r\n inputLayer = arcpy.Parameter(\r\n displayName = \"Input Features\",\r\n name = \"in_features\",\r\n datatype = \"GPFeatureLayer\",\r\n parameterType = \"Required\",\r\n direction = \"Input\")\r\n\r\n outputLocation = arcpy.Parameter(\r\n displayName=\"Output Location\",\r\n name=\"out_xlsx\",\r\n datatype=\"DEFile\",\r\n parameterType=\"Required\",\r\n direction=\"Output\")\r\n\r\n params = [inputLayer, outputLocation] \r\n\r\n return params\r\n\r\n def isLicensed(self):\r\n \"\"\"Set whether tool is licensed to execute.\"\"\"\r\n return True\r\n\r\n def updateParameters(self, parameters):\r\n \"\"\"Modify the values and properties of parameters before internal\r\n validation is performed. This method is called whenever a parameter\r\n has been changed.\"\"\"\r\n\r\n return\r\n\r\n def updateMessages(self, parameters):\r\n \"\"\"Modify the messages created by internal validation for each tool\r\n parameter. This method is called after internal validation.\"\"\"\r\n return\r\n\r\n def execute(self, parameters, messages):\r\n \"\"\"The source code of the tool.\"\"\"\r\n\r\n arcpy.AddMessage(\"preparing Excel file...\")\r\n \r\n inputLayer, outputLocation = parameters \r\n pitCount = inputLayer.valueAsText\r\n xLocate = outputLocation.valueAsText\r\n\r\n ###SHELTER TYPE Function\r\n def headingvET(familyType, familyFullName, startRow):\r\n header = [familyFullName, \" \", 'Sheltered ES', 'Sheltered TH', 'Sheltered SH', 'Unsheltered', 'Totals']\r\n #write worksheet header\r\n worksheet_data.write_row(('A'+(str(3+ startRow))), header, cell_format_title)\r\n \r\n if sleeplo1 in familyType:\r\n #Emergency Shelter\r\n dfES = familyType[familyType[sleeplo1]=='EmergencyShelter']\r\n dfESV = len(dfES[dfES[vet1]==\"Yes\"].index)\r\n dfESHH = dfES[parid1].nunique()\r\n dfESunq = len(dfES.index)\r\n #Transitional Housing\r\n dfTH = familyType[familyType[sleeplo1] == 'TransitionalHousing']\r\n dfTHV = len(dfTH[dfTH[vet1]==\"Yes\"].index)\r\n dfTHHH = dfTH[parid1].nunique()\r\n dfTHunq = len(dfTH.index)\r\n #Safe Haven\r\n dfSH = familyType[familyType[sleeplo1] == 'SafeHavens']\r\n dfSHHH = dfSH[parid1].nunique()\r\n dfSHV = len(dfSH[dfSH[vet1]==\"Yes\"].index)\r\n dfSHunq = len(dfSH.index)\r\n #Unsheltered\r\n dfUN = familyType[familyType[shelt1]==\"No\"]\r\n dfUNHH = dfUN[parid1].nunique()\r\n dfUNV = len(dfUN[dfUN[vet1]==\"Yes\"].index)\r\n dfUNunq = len(dfUN.index)\r\n\r\n #HH and uniue\r\n household = ['Total Number of Households', \" \",dfESHH, dfTHHH, dfSHHH, dfUNHH]\r\n unique = ['Total Number of Persons', \" \",dfESunq, dfTHunq, dfSHunq, dfUNunq]\r\n vets = ['Total number of Veterans', \"\", dfESV, dfTHV, dfSHV, dfUNV]\r\n #writetools\r\n worksheet_data.write_row(('A'+(str(4 + startRow))), household)\r\n worksheet_data.write_row(('A'+(str(5 + startRow))), unique)\r\n worksheet_data.write_row(('A'+(str(6+ startRow))), vets)\r\n\r\n def heading(familyType, familyFullName, startRow):\r\n header = [familyFullName, \" \", 'Sheltered ES', 'Sheltered TH', 'Sheltered SH', 'Unsheltered', 'Totals']\r\n #write worksheet header\r\n worksheet_data.write_row(('A'+(str(3+ startRow))), header, cell_format_title)\r\n \r\n if sleeplo1 in familyType:\r\n #Emergency Shelter\r\n dfES = familyType[familyType[sleeplo1]=='EmergencyShelter']\r\n dfESHH = dfES[parid1].nunique()\r\n dfESunq = len(dfES.index)\r\n #Transitional Housing\r\n dfTH = familyType[familyType[sleeplo1] == 'TransitionalHousing']\r\n dfTHHH = dfTH[parid1].nunique()\r\n dfTHunq = len(dfTH.index)\r\n #Safe Haven\r\n dfSH = familyType[familyType[sleeplo1] == 'SafeHavens']\r\n dfSHHH = dfSH[parid1].nunique()\r\n dfSHunq = len(dfSH.index)\r\n #Unsheltered\r\n dfUN = familyType[familyType[shelt1]==\"No\"]\r\n dfUNHH = dfUN[parid1].nunique()\r\n dfUNunq = len(dfUN.index)\r\n\r\n #HH and uniue\r\n household = ['Total Number of Households', \" \",dfESHH, dfTHHH, dfSHHH, dfUNHH]\r\n unique = ['Total Number of Persons', \" \",dfESunq, dfTHunq, dfSHunq, dfUNunq]\r\n #writetools\r\n worksheet_data.write_row(('A'+(str(4 + startRow))), household)\r\n worksheet_data.write_row(('A'+(str(5 + startRow))), unique)\r\n #age\r\n dfESu18 = len(dfES[dfES[age1]<18].index)\r\n dfESu24 = len(dfES[dfES[age1].between(17,25, inclusive=False)].index)\r\n dfESo24 = len(dfES[dfES[age1]>24].index)\r\n dfTHu18 = len(dfTH[dfTH[age1]<18].index)\r\n dfTHu24 = len(dfTH[dfTH[age1].between(17,25, inclusive=False)].index)\r\n dfTHo24 = len(dfTH[dfTH[age1]>24].index)\r\n dfSHu18 = len(dfSH[dfSH[age1]<18].index)\r\n dfSHu24 = len(dfSH[dfSH[age1].between(17,25, inclusive=False)].index)\r\n dfSHo24 = len(dfSH[dfSH[age1]>24].index)\r\n dfUNu18 = len(dfUN[dfUN[age1]<18].index)\r\n dfUNu24 = len(dfUN[dfUN[age1].between(17,25, inclusive=False)].index)\r\n dfUNo24 = len(dfUN[dfUN[age1]>24].index)\r\n\r\n ################ age\r\n u18 = [\"\", \"Under Age 18\", dfESu18, dfTHu18, dfSHu18, dfUNu18]\r\n u24 = [\"\", \"Age 18-24\", dfESu24, dfTHu24, dfSHu24, dfUNu24]\r\n o24 = [\"\", \"Over Age 24\", dfESo24, dfESo24, dfSHo24, dfUNo24]\r\n #write age to sheet\r\n worksheet_data.write_row(('A'+(str(6 + startRow))), u18)\r\n worksheet_data.write_row(('A'+(str(7 + startRow))), u24)\r\n worksheet_data.write_row(('A'+(str(8 + startRow))), o24)\r\n\r\n def demograph(familyType, extraWords, startRow):\r\n bold = workbook.add_format({'bold':True})\r\n if sleeplo1 in familyType:\r\n #chronic\r\n chro2 = df.DataFrame()\r\n disab = familyType[(familyType[mental1]==\"Yes\") | (familyType[substan1]==\"Yes\") | (familyType[hiv1]==\"Yes\")]\r\n chro = disab[(disab[timhom1]==\"4 or more times\") & (disab[totdayhom1]>364)]\r\n chro1 = disab[disab[numday1]>364]\r\n chro2 = chro2.append([chro1, chro]) \r\n chronic = familyType[(familyType[31]==\"nonsense\")]\r\n if len(chro2)>0:\r\n uniqChro = chro2[parid1].unique()\r\n #create an empty dataframe to use\r\n for ID in uniqChro:\r\n HHSep = familyType[familyType[parid1]==ID]\r\n chronic = chronic.append(HHSep) \r\n\r\n #Emergency Shelter\r\n dfES = familyType[familyType[sleeplo1]=='EmergencyShelter']\r\n dfESChro = len(chronic[chronic[sleeplo1]==\"EmergencyShelter\"].index)\r\n dfESC = chronic[chronic[sleeplo1]==\"EmergencyShelter\"]\r\n if parid1 in dfESC:\r\n dfESChH = dfESC[parid1].nunique()\r\n else:\r\n dfesChH = len(chronic).index\r\n dfESHH = dfES[parid1].nunique()\r\n dfESunq = len(dfES.index)\r\n #Transitional Housing\r\n dfTH = familyType[familyType[sleeplo1] == 'TransitionalHousing']\r\n dfTHHH = dfTH[parid1].nunique()\r\n dfTHunq = len(dfTH.index)\r\n #Safe Haven\r\n dfSH = familyType[familyType[sleeplo1] == 'SafeHavens']\r\n dfSHHH = dfSH[parid1].nunique()\r\n dfSHChro = len(chronic[chronic[sleeplo1]==\"SafeHavens\"].index)\r\n dfSHC = chronic[chronic[sleeplo1]==\"SafeHavens\"]\r\n if parid1 in dfSHC:\r\n dfSHChH = dfSHC[parid1].nunique()\r\n else:\r\n dfSHChH = len(chronic).index\r\n dfSHunq = len(dfSH.index)\r\n #Unsheltered\r\n dfUN = familyType[familyType[shelt1]==\"No\"]\r\n dfUNHH = dfUN[parid1].nunique()\r\n dfUNChro = len(chronic[chronic[shelt1]==\"No\"].index)\r\n dfUNC = chronic[chronic[shelt1]==\"No\"]\r\n if parid1 in dfUNC:\r\n dfUNChH = dfUNC[parid1].nunique()\r\n else:\r\n dfUNChH = len(chronic).index \r\n dfUNunq = len(dfUN.index)\r\n\r\n #gender\r\n dfESF = len(dfES[dfES[gend1]=='Female'].index)\r\n dfESM = len(dfES[dfES[gend1]=='Male'].index)\r\n dfEST = len(dfES[dfES[gend1]== \"Transgender\"].index)\r\n dfESN = len(dfES[dfES[gend1]== \"DontIdentify\"].index)\r\n dfTHF = len(dfTH[dfTH[gend1]=='Female'].index)\r\n dfTHM = len(dfTH[dfTH[gend1]=='Male'].index)\r\n dfTHT = len(dfTH[dfTH[gend1]== \"Transgender\"].index)\r\n dfTHN = len(dfTH[dfTH[gend1]== \"DontIdentify\"].index)\r\n dfSHF = len(dfSH[dfSH[gend1]=='Female'].index)\r\n dfSHM = len(dfSH[dfSH[gend1]=='Male'].index)\r\n dfSHT = len(dfSH[dfSH[gend1]== \"Transgender\"].index)\r\n dfSHN = len(dfSH[dfSH[gend1]== \"DontIdentify\"].index)\r\n dfUNF = len(dfUN[dfUN[gend1]=='Female'].index)\r\n dfUNM = len(dfUN[dfUN[gend1]=='Male'].index)\r\n dfUNT = len(dfUN[dfUN[gend1]== \"Transgender\"].index)\r\n dfUNN = len(dfUN[dfUN[gend1]== \"DontIdentify\"].index)\r\n ############### gender\r\n gender = [\"Gender\", extraWords]\r\n female = [\" \", \"Female\",dfESF, dfTHF, dfSHF, dfUNF]\r\n male = [\" \", \"Male\",dfESM, dfTHM, dfSHM, dfUNM]\r\n trans = [\" \", \"Transgender\", dfEST, dfTHT, dfSHT, dfUNT]\r\n noncon = [\"\", \"Gender Non-Conforming\", dfESN, dfTHN, dfSHN, dfUNN]\r\n totgen = [\" \", \"Total number of persons for which gender is known\"] \r\n #write gender\r\n worksheet_data.write_row(('A'+(str(9 + startRow))), gender, bold)\r\n worksheet_data.write_row(('A'+(str(10 + startRow))), female)\r\n worksheet_data.write_row(('A'+(str(11 + startRow))), male)\r\n worksheet_data.write_row(('A'+(str(12 + startRow))), trans)\r\n worksheet_data.write_row(('A'+(str(13 + startRow))), noncon)\r\n worksheet_data.write_row(('A'+(str(14 + startRow))), totgen)\r\n \r\n #ethnicity\r\n dfESHis = len(dfES[dfES[ethnicity]== \"Yes\"].index)\r\n dfESNohi = len(dfES[dfES[ethnicity]== \"No\"].index)\r\n dfTHHis = len(dfTH[dfTH[ethnicity]== \"Yes\"].index)\r\n dfTHNohi = len(dfTH[dfTH[ethnicity]== \"No\"].index)\r\n dfSHHis = len(dfSH[dfSH[ethnicity]== \"Yes\"].index)\r\n dfSHNohi = len(dfSH[dfSH[ethnicity]== \"No\"].index)\r\n dfUNHis = len(dfUN[dfUN[ethnicity]== \"Yes\"].index)\r\n dfUNNohi = len(dfUN[dfUN[ethnicity]== \"No\"].index)\r\n #compile\r\n ethn = [\"Ethnicity\", extraWords]\r\n NOspan = [\"\", \"Non-Hispanic/Non-Latino\", dfESNohi, dfTHNohi, dfSHNohi, dfUNNohi]\r\n spans = [ \"\", \"Hispanic/Latino\", dfESHis, dfTHHis, dfSHHis, dfUNHis]\r\n totspan = [\"\", \"Total number of persons for which ethnicity is known\"] \r\n #write\r\n worksheet_data.write_row(('A'+(str(15 +startRow))), ethn, bold) \r\n worksheet_data.write_row(('A'+(str(16 + startRow))), NOspan) \r\n worksheet_data.write_row(('A'+(str(17 + startRow))), spans)\r\n worksheet_data.write_row(('A'+(str(18 + startRow))), totspan)\r\n\r\n #Ethnicity and race\r\n dfESWh = len(dfES[dfES[race1]== \"White\"].index)\r\n dfESBl = len(dfES[dfES[race1]== \"Black\"].index)\r\n dfESAs = len(dfES[dfES[race1]== \"Asian\"].index)\r\n dfESAi = len(dfES[dfES[race1]== \"AmercanIndian\"].index)\r\n dfESHi = len(dfES[dfES[race1]==\"NativeHawaiian\"].index)\r\n dfESMul = len(dfES[dfES[race1].str.contains(',', na=False)].index)\r\n dfTHWh = len(dfTH[dfTH[race1]== \"White\"].index)\r\n dfTHBl = len(dfTH[dfTH[race1]== \"Black\"].index)\r\n dfTHAs = len(dfTH[dfTH[race1]== \"Asian\"].index)\r\n dfTHAi = len(dfTH[dfTH[race1]== \"AmercanIndian\"].index)\r\n dfTHHi = len(dfTH[dfTH[race1]==\"NativeHawaiian\"].index)\r\n dfTHMul = len(dfTH[dfTH[race1].str.contains(',', na=False)].index)\r\n dfSHWh = len(dfSH[dfSH[race1]== \"White\"].index)\r\n dfSHBl = len(dfSH[dfSH[race1]== \"Black\"].index)\r\n dfSHAs = len(dfSH[dfSH[race1]== \"Asian\"].index)\r\n dfSHAi = len(dfSH[dfSH[race1]== \"AmercanIndian\"].index)\r\n dfSHHi = len(dfSH[dfSH[race1]==\"NativeHawaiian\"].index)\r\n dfSHMul = len(dfSH[dfSH[race1].str.contains(',', na=False)].index)\r\n dfUNWh = len(dfUN[dfUN[race1]== \"White\"].index)\r\n dfUNBl = len(dfUN[dfUN[race1]== \"Black\"].index)\r\n dfUNAs = len(dfUN[dfUN[race1]== \"Asian\"].index)\r\n dfUNAi = len(dfUN[dfUN[race1]== \"AmericanIndian\"].index)\r\n dfUNHi = len(dfUN[dfUN[race1]==\"NativeHawaiian\"].index)\r\n dfUNMul = len(dfUN[dfUN[race1].str.contains(',', na=False)].index)\r\n \r\n #summup\r\n racehead = [\"Race\", extraWords]\r\n white = [\"\", \"White\", dfESWh, dfTHWh, dfSHWh, dfUNWh]\r\n black = [\"\", \"Black of African-American\", dfESBl, dfTHBl, dfSHBl, dfUNBl]\r\n asian = [\"\", \"Asian\", dfESAs, dfTHAs, dfSHAs, dfUNAs]\r\n amerInd = [\"\", \"American Indian or Alaskan Native\", dfESAi, dfTHAi, dfSHAi, dfUNAi]\r\n paci = [\"\", \"Native Hawaiian or Other Pacific Islander\", dfESHi, dfTHHi, dfSHHi, dfUNHi]\r\n mult = [\"\", \"Multiple Races\", dfESMul, dfTHMul, dfSHMul, dfUNMul]\r\n totrac= [\"\", \"Total Number of persons for which race is known\"] \r\n #WRITE WORKSHEET\r\n worksheet_data.write_row(('A'+(str(19 + startRow))), racehead, bold)\r\n worksheet_data.write_row(('A'+(str(20 + startRow))), white)\r\n worksheet_data.write_row(('A'+(str(21 + startRow))), black)\r\n worksheet_data.write_row(('A'+(str(22 + startRow))), asian)\r\n worksheet_data.write_row(('A'+(str(23 + startRow))), amerInd)\r\n worksheet_data.write_row(('A'+(str(24 + startRow))), paci)\r\n worksheet_data.write_row(('A'+(str(25 + startRow))), mult)\r\n worksheet_data.write_row(('A'+(str(26 + startRow))), totrac)\r\n\r\n #habitual\r\n chronicHom = [\"\", \"Total Number of Persons\", 0,\"NA\",0,0]\r\n chronicHH = [\"\", \"Total Number of Households\", 0,\"NA\",0,0]\r\n if len(chronic)>0:\r\n chronicHH = [\"\", \"Total Number of Households\", dfESChH, \"NA\", dfSHChH, dfUNChH]\r\n chronicHom = [\"\", \"Total Number of Persons\",dfESChro, \"NA\", dfSHChro, dfUNChro]\r\n chronicHead = [\"Chronically Homeless\", \"(all)\"]\r\n \r\n worksheet_data.write_row(('A'+(str(27 + startRow))), chronicHead, bold)\r\n worksheet_data.write_row(('A'+(str(28 + startRow))), chronicHH)\r\n worksheet_data.write_row(('A'+(str(29 + startRow))), chronicHom)\r\n\r\n #get field names to use in query \r\n fields = arcpy.ListFields(pitCount)\r\n fieldNames = []\r\n\r\n for field in fields:\r\n fieldNames.append(field.name)\r\n arcpy.AddMessage(fieldNames)\r\n relate1 = fieldNames.index(\"relationship\")\r\n sleeplo1 = fieldNames.index(\"sleep_location_individual\")\r\n age1 = fieldNames.index(\"age\")\r\n gend1 = fieldNames.index(\"gender\")\r\n ethnicity = fieldNames.index(\"hispanic\")\r\n race1 = fieldNames.index(\"race\")\r\n parid1 = fieldNames.index(\"parentglobalid\")\r\n timhom1 = fieldNames.index(\"times_homeless\")\r\n numday1 = fieldNames.index(\"days_homeless\")\r\n totdayhom1 = fieldNames.index(\"days_total_homeless\")\r\n vet1 = fieldNames.index(\"veteran_full\")\r\n shelt1 = fieldNames.index(\"sheltered_individuals\")\r\n hiv1 = fieldNames.index(\"has_hiv\")\r\n mental1 = fieldNames.index(\"psychiatric_condition\")\r\n substan1 = fieldNames.index(\"substance_use\")\r\n domvi1 = fieldNames.index(\"been_abused\")\r\n surv = fieldNames.index(\"household_survey_type\")\r\n\r\n #do one search cursor to get all data into a list \r\n dataList = []\r\n with arcpy.da.SearchCursor(pitCount, \"*\") as cursor:\r\n for row in cursor:\r\n dataList.append(row) \r\n \r\n ##################using Pandas get each type of household into a seprate list \r\n listo = df.DataFrame(dataList)\r\n panda = listo[listo[surv]==\"Interview\"]\r\n #Unique Households and seperate them into families and single \r\n family = df.DataFrame()\r\n single = df.DataFrame()\r\n HH = panda[parid1].unique()\r\n for ID in HH:\r\n HHSep = panda.loc[panda[parid1] == ID]\r\n if len(HHSep)>1:\r\n family = family.append(HHSep)\r\n else:\r\n single = single.append(HHSep)\r\n \r\n #Get Family Type\r\n #########seprate sub groups (not family units)\r\n if age1 in family:\r\n U18Fam = family.loc[family.loc[:,age1]<18]\r\n YTHFam = family.loc[family[age1]<25]\r\n ADYFam = family.loc[family[age1]>17]\r\n ADFam = family.loc[family[age1]>24]\r\n\r\n ########do the same for singles (not family units)\r\n #make variables that are empty \r\n if age1 in single:\r\n ADSing = single[single[age1]>17]\r\n CHSing = single[single[age1]<18]\r\n YTSing = single[single[age1]<24]\r\n else:\r\n ADSing = panda[panda[1]==\"Yellow\"]\r\n CHSing = panda[panda[1]==\"Yellow\"]\r\n YTSing = panda[panda[1]==\"Yellow\"]\r\n\r\n \r\n workbook = xlsxwriter.Workbook(xLocate)\r\n #####Format options\r\n # Create a format to use in the merged range.\r\n merge_format = workbook.add_format({\r\n 'bold': 1,\r\n 'border': 1,\r\n 'align': 'center',\r\n 'valign': 'vcenter',\r\n 'fg_color': 'gray',\r\n 'font_size':14,\r\n 'font_color':\"white\"})\r\n #title header\r\n bold = workbook.add_format({'bold':True})\r\n worksheet_data = workbook.add_worksheet(\"Interview Data\")\r\n worksheet_data.merge_range('A1:G1', 'PRE-EXTRAPOLATED DATA', merge_format)\r\n worksheet_data.merge_range('A2:G2', 'ALL HOUSEHOLDS', merge_format)\r\n worksheet_data.merge_range('A84:G84', \"VETERAN HOUSEHOLDS\", merge_format)\r\n worksheet_data.merge_range('A135:G135', \"YOUTH HOUSEHOLDS\", merge_format)\r\n worksheet_data.merge_range('A193:G193', \"ADDITIONAL HOMELESS POPULATIONS\", merge_format)\r\n #column names\r\n worksheet_data.set_column(2,7, 15)\r\n worksheet_data.set_column(1,1,30)\r\n worksheet_data.set_column(0,0,40, bold )\r\n #usable formating for title of sub headings\r\n cell_format_title = workbook.add_format({'bold':True, 'bg_color':'#C0C0C0'})\r\n cell_format_total = workbook.add_format({'bold':True})\r\n #set totals to bold\r\n worksheet_data.set_column(6, 6, None, cell_format_total)\r\n\r\n ############################################get households with at least one adult and one child \r\n ##through this we also get HH with prenting youth\r\n ADCH = df.DataFrame()\r\n CHFAM = df.DataFrame()\r\n ADNOCH = df.DataFrame()\r\n for ID in HH:\r\n ch = U18Fam.loc[U18Fam[parid1] == ID]\r\n ad = ADYFam.loc[ADYFam[parid1] == ID]\r\n if len(ad.index)>0 and len(ch.index)>0:\r\n ADCH = ADCH.append([ad,ch])\r\n elif len(ad.index)==0 and len(ch.index)>0:\r\n CHFAM = CHFAM.append(ch)\r\n elif len(ad.index)>0 and len(ch.index)==0:\r\n ADNOCH = ADNOCH.append(ad)\r\n\r\n ##shelter type\r\n heading(ADCH, 'Households with At Least One Adult and One Child', 0)\r\n demograph(ADCH, \"(adults & children)\", 0)\r\n #####chronic homelessness status\r\n \r\n ###########################################get households with no children \r\n noChildren = ADNOCH.append(ADSing)\r\n ##shelter type\r\n heading(noChildren, 'Households without Children', 27)\r\n demograph(noChildren, \"\", 27)\r\n ##########################################Get households with only children \r\n #families with children\r\n childOnly = CHFAM.append(CHSing)\r\n \r\n ##shelter type\r\n heading(childOnly, 'Households with only Chidlren (under18)', 54)\r\n demograph(childOnly, \"\", 54)\r\n ############################################Veterans\r\n vets = df.DataFrame()\r\n allVets = panda.loc[panda[vet1]==\"Yes\"]\r\n if parid1 in allVets:\r\n uniqVets = allVets[parid1].unique()\r\n for ID in uniqVets:\r\n HHSep = panda.loc[panda[parid1] == ID]\r\n vets = vets.append(HHSep)\r\n vetNOCH = df.DataFrame()\r\n vetCH = df.DataFrame()\r\n if age1 in vets:\r\n U18Vet = vets[vets[age1]<18]\r\n ADVet = vets[vets[age1]>17]\r\n\r\n for ID in uniqVets:\r\n ch = U18Vet[U18Vet[parid1] == ID]\r\n ad = ADVet[ADVet[parid1] == ID]\r\n if len(ad.index)>0 and len(ch.index)>0:\r\n vetCH = vetCH.append([ad,ch])\r\n elif len(ad.index)>0 and len(ch.index)==0:\r\n vetNOCH = vetNOCH.append(ad)\r\n if vet1 in vetCH: \r\n vetsofvetCH = vetCH[vetCH[vet1]==\"Yes\"]\r\n else:\r\n vetsofvetCH = vetCH\r\n if vet1 in vetNOCH:\r\n vetsofvetNOCH = vetNOCH[vetNOCH[vet1]==\"Yes\"]\r\n else:\r\n vetsofvetNOCH = vetCH\r\n\r\n #households with children\r\n headingvET(vetCH, \"Veteran HH with At Least One Adult and One Child\", 82)\r\n demograph(vetsofvetCH, \"(Veterans Only)\", 80)\r\n ############################################veterans households without childre\r\n ##shelter type\r\n headingvET(vetNOCH, \"Veteran HH without Children\", 107)\r\n demograph(vetsofvetNOCH, \"(Veterans Only)\", 105)\r\n #####chronic homelessness status\r\n\r\n ############################################youth unaccompanied \r\n UNYTH = df.DataFrame()\r\n for ID in HH:\r\n yt = YTHFam.loc[YTHFam[parid1] == ID]\r\n ad = ADFam.loc[ADFam[parid1] == ID]\r\n if len(yt.index)>0 and len(ad.index)==0:\r\n UNYTH = UNYTH.append(yt)\r\n\r\n ###Get parenting youth seprate from more than one yth living together \r\n PARYth = df.DataFrame()\r\n NOparyth = df.DataFrame()\r\n childrenof = df.DataFrame()\r\n if parid1 in UNYTH: \r\n ythNum = UNYTH[parid1].unique()\r\n for ID in ythNum:\r\n HHSep = UNYTH.loc[UNYTH[parid1] ==ID]\r\n lenfam = HHSep[HHSep[relate1]==\"Child\"]\r\n if len(lenfam.index)>0:\r\n PARYth = PARYth.append(HHSep)\r\n childrenof = childrenof.append(lenfam)\r\n else:\r\n NOparyth = NOparyth.append(HHSep)\r\n ythUnaccSi = UNYTH.append(YTSing)\r\n ythUnacc = ythUnaccSi.append(NOparyth)\r\n\r\n def parYthWrite(familyType, title, startRow):\r\n unique = [\"\", title, 0, 0 , 0 , 0]\r\n if len(familyType)>0:\r\n #Emergency Shelter\r\n dfES = familyType[familyType[sleeplo1]=='EmergencyShelter']\r\n dfESunq = len(dfES.index)\r\n #Transitional Housing\r\n dfTH = familyType[familyType[sleeplo1] == 'TransitionalHousing']\r\n dfTHunq = len(dfTH.index)\r\n #Safe Haven\r\n dfSH = familyType[familyType[sleeplo1] == 'SafeHavens']\r\n dfSHunq = len(dfSH.index)\r\n #Unsheltered\r\n dfUN = familyType[familyType[shelt1]==\"No\"]\r\n dfUNunq = len(dfUN.index)\r\n\r\n #HH and uniue\r\n unique = [\"\",title,dfESunq, dfTHunq, dfSHunq, dfUNunq]\r\n #writetools\r\n worksheet_data.write_row(('A'+(str(startRow))), unique)\r\n \r\n heading(ythUnacc, \"Unaccompanied Youth Households\", 133)\r\n ###unaccompanied youth write out \r\n demograph(ythUnacc, \"(unaccompanied youth)\", 133)\r\n \r\n #####chronic homelessness status\r\n\r\n ############################################Parenting youth 14\r\n ##shelter type\r\n headerPar = [\"Parenting Youth Households\", \" \", 'Sheltered ES', 'Sheltered TH', 'Sheltered SH', 'Unsheltered', 'Totals']\r\n #write worksheet header\r\n worksheet_data.write_row(('A163'), headerPar, cell_format_title)\r\n if parid1 in PARYth:\r\n parentsOnly = PARYth[PARYth[relate1].isnull()]\r\n parentsover18 = parentsOnly[parentsOnly[age1]>17]\r\n parentsunder18 = parentsOnly[parentsOnly[age1]<18]\r\n \r\n parentsoveruniq= parentsover18[parid1].unique()\r\n otheruniq = childrenof[parid1].unique()\r\n parentsunderuniq= parentsunder18[parid1].unique()\r\n childofYth = df.DataFrame()\r\n childofChild = df.DataFrame()\r\n for ID in parentsoveruniq:\r\n youthFam = childrenof.loc[childrenof[parid1]==ID]\r\n childofYth = childofYth.append(youthFam)\r\n for ID in parentsunderuniq:\r\n childFam = childrenof.loc[childrenof[parid1]==ID]\r\n childofChild = childofChild.append(childFam)\r\n\r\n parYthWrite(parentsunder18, \"Number of parenting youth under Age 18\",168)\r\n parYthWrite(parentsover18, \"Number of parenting youth Age 18-24\",170)\r\n parYthWrite(childofYth, \" Children in HH with parenting youth Age 18-24\", 171)\r\n parYthWrite(childofChild, \" Children in HH with parenting youth under Age 18\",169)\r\n demograph(parentsOnly, \"(youth parents only)\", 163)\r\n #####chronic homelessness status\r\n \r\n def parYthTop(familyType, title, startRow):\r\n unique = [title, \"\", 0, 0 , 0 , 0]\r\n if len(familyType)>0:\r\n #Emergency Shelter\r\n dfES = familyType[familyType[sleeplo1]=='EmergencyShelter']\r\n dfESunq = len(dfES.index)\r\n #Transitional Housing\r\n dfTH = familyType[familyType[sleeplo1] == 'TransitionalHousing']\r\n dfTHunq = len(dfTH.index)\r\n #Safe Haven\r\n dfSH = familyType[familyType[sleeplo1] == 'SafeHavens']\r\n dfSHunq = len(dfSH.index)\r\n #Unsheltered\r\n dfUN = familyType[familyType[shelt1]==\"No\"]\r\n dfUNunq = len(dfUN.index)\r\n #HH and uniue\r\n unique = [title,\"\", dfESunq, dfTHunq, dfSHunq, dfUNunq]\r\n\r\n #writetools\r\n worksheet_data.write_row(('A'+(str(startRow))), unique)\r\n\r\n if parid1 in PARYth:\r\n parYthTop(parentsOnly, \"Total Number of Parenting Youth Households\", 164)\r\n parYthTop(PARYth, \"Total Number of Persons in Parenting Youth Households\", 165)\r\n parYthTop(parentsOnly, \"Total Parenting Youth\", 166)\r\n parYthTop(childrenof, \"Total Children in Parenting Youth Household\", 167)\r\n ##############################################Adult aids, mental illness etc.\r\n ####Final Formatting\r\n #gender\r\n def sumppl(column, i):\r\n formula = \"{=SUM(\"+column+(str(i-4)+\":\"+column+(str(i-1))+\")}\")\r\n worksheet_data.write_array_formula(column+(str(i))+column+(str(i)), formula)\r\n #ethnicity\r\n def sumeth(column, i):\r\n formula = \"{=SUM(\"+column+(str(i-2)+\":\"+column+(str(i-1))+\")}\")\r\n worksheet_data.write_array_formula(column+(str(i))+column+(str(i)), formula)\r\n #race\r\n def sumrace(column, i):\r\n formula = \"{=SUM(\"+column+(str(i-6)+\":\"+column+(str(i-1))+\")}\")\r\n worksheet_data.write_array_formula(column+(str(i))+column+(str(i)), formula)\r\n #groups\r\n totgend = {(\"C\",14), (\"D\",14),(\"E\",14), (\"F\",14),(\"C\",41), (\"D\",41),(\"E\",41), (\"F\",41),(\"C\",68), (\"D\",68),(\"E\",68), (\"F\",68),(\"C\",94), (\"D\",94), (\"E\",94), (\"F\",94),(\"C\",119), (\"D\",119),(\"E\",119), (\"F\",119),(\"C\",147), (\"D\",147),(\"E\",147), (\"F\",147),(\"C\",177), (\"D\",177),(\"E\",177), (\"F\",177)}\r\n toteth = {(\"C\",18), (\"D\",18),(\"E\",18), (\"F\",18),(\"C\",45), (\"D\",45),(\"E\",45), (\"F\",45),(\"C\",72), (\"D\",72),(\"E\",72), (\"F\",72),(\"C\",98), (\"D\",98), (\"E\",98), (\"F\",98),(\"C\",123), (\"D\",123),(\"E\",123), (\"F\",123),(\"C\",151), (\"D\",151),(\"E\",151), (\"F\",151),(\"C\",181), (\"D\",181),(\"E\",181), (\"F\",181)}\r\n totrace = {(\"C\",26), (\"D\",26),(\"E\",26), (\"F\",26),(\"C\",53), (\"D\",53),(\"E\",53), (\"F\",53),(\"C\",80), (\"D\",80),(\"E\",80), (\"F\",80),(\"C\",106), (\"D\",106), (\"E\",106), (\"F\",106),(\"C\",131), (\"D\",131),(\"E\",131), (\"F\",131),(\"C\",159), (\"D\",159),(\"E\",159), (\"F\",159),(\"C\",189), (\"D\",189),(\"E\",189), (\"F\",189)}\r\n for (a,b) in totgend:\r\n sumppl(a,b)\r\n for (a,b) in toteth:\r\n sumeth(a,b)\r\n for (a,b) in totrace:\r\n sumrace(a,b)\r\n ##final summation\r\n rows = list(range(4,30))\r\n rows2 = list(range(31,57))\r\n rows3 = list(range(58,84))\r\n rows4 = list(range(86,110))\r\n rows5 = list(range(111,135))\r\n rows6 = list(range(137,163))\r\n rows7 = list(range(164,193))\r\n rows8 = list(range(193,199))\r\n def rowsum(rows):\r\n for i in rows:\r\n formula = \"{=SUM(C\"+(str(i)+\":F\"+(str(i))+\")}\")\r\n worksheet_data.write_array_formula('G'+(str(i))+'G'+(str(i)), formula)\r\n rowsum(rows)\r\n rowsum(rows2)\r\n rowsum(rows3)\r\n rowsum(rows4)\r\n rowsum(rows5)\r\n rowsum(rows6)\r\n rowsum(rows7)\r\n rowsum(rows8)\r\n\r\n HIV = panda[panda[hiv1]==\"Yes\"]\r\n DoVi = panda[panda[domvi1] ==\"Yes\"]\r\n ment = panda[panda[mental1]==\"Yes\"]\r\n sub = panda[panda[substan1]==\"Yes\"]\r\n \r\n def adultD(thing, name, row):\r\n dfES = len(thing[thing[sleeplo1]=='EmergencyShelter'].index)\r\n dfSH = len(thing[thing[sleeplo1] == 'SafeHavens'].index)\r\n dfTH = len(thing[thing[sleeplo1] == 'TransitionalHousing'].index)\r\n dfUN = len(thing[thing[shelt1]==\"No\"].index)\r\n all = [name, \"\", dfES, dfTH, dfSH, dfUN] \r\n worksheet_data.write_row(('A'+(str(row))), all)\r\n\r\n header = [\"\", \" \", 'Sheltered ES', 'Sheltered TH', 'Sheltered SH', 'Unsheltered', 'Totals']\r\n #write worksheet header\r\n worksheet_data.write_row('A194', header, cell_format_title)\r\n \r\n adultD(ment, \"Adults with Serious Mental Illness\", 195)\r\n adultD(sub, \"Adults with a Substance Abuse Disorder\", 196)\r\n adultD(HIV, \"Adults with HIV/AIDS\", 197)\r\n adultD(DoVi, \"Adult Survivors of Domestic Violence\", 198)\r\n\r\n #write\r\n workbook.close()\r\n \r\n return\r\n\r\n ##############################scratch"
]
| [
[
"pandas.DataFrame"
]
]
|
hitesh-ag1/expense-tracker | [
"bf5238ef4dbed82aff151f53b2e0784eca7213d3"
]
| [
"todayFuncs.py"
]
| [
"import numpy as np\nimport pandas as pd\nfrom datetime import datetime\n\ndef todayFilter(data):\n data = data[(data.index.year == datetime.today().year) & (data.index.month == datetime.today().month) &(data.index.day == datetime.today().day)]\n return data\n\ndef transToday(data):\n trans = (data)[['amt', 'payee', 'type']]\n trans.loc[trans.type == 'Payment', 'amt'] = np.negative(trans.loc[trans.type == 'Payment', 'amt']).values\n trans = trans.groupby('payee',).amt.sum()\n trans = trans.to_dict()\n return trans\n\ndef transTodaySummarybyType(data):\n da = (data)[['amt', 'type']]\n da = ((da.groupby([pd.Grouper(freq='D'), 'type']).amt.sum()).droplevel('date')).to_dict()\n return da\n\ndef transTodaySummarybyCat(data):\n da = (data)[['amt', 'category', 'type']]\n da = ((da.groupby([pd.Grouper(freq='D'), 'category', 'type']).amt.sum()).droplevel('date'))\n da.loc[da.index.get_level_values('type') == 'Payment'] = np.negative(da.loc[da.index.get_level_values('type') == 'Payment'])\n da = da.droplevel('type')\n da = da.to_dict()\n return da\n # summary = data.groupby([pd.Grouper(freq='D'), 'category', 'type']).amt.sum()\n # day_summary = todayMultiIndexFilter(summary)\n # day_summary = day_summary.droplevel('datetime')\n # day_summary.loc[day_summary.index.get_level_values('type') == 'Payment'] = np.negative(day_summary.loc[day_summary.index.get_level_values('type') == 'Payment'])\n # day_summary = day_summary.droplevel('type')\n # day_summary = day_summary.to_dict()\n"
]
| [
[
"numpy.negative",
"pandas.Grouper"
]
]
|
fellajimed/SparseScatNet | [
"9725e066b4ddb899c8c1ae225ca4a4292fc04e64",
"9725e066b4ddb899c8c1ae225ca4a4292fc04e64"
]
| [
"models/Classifier.py",
"models/LinearProj.py"
]
| [
"import torch.nn as nn\n\n\nclass Classifier(nn.Module):\n def __init__(self, n_space, nb_channels_in, classifier_type='mlp', nb_classes=1000,\n nb_hidden_units=2048, nb_l_mlp=2, dropout_p_mlp=0.3, avg_ker_size=1):\n super(Classifier, self).__init__()\n\n self.nb_classes = nb_classes\n\n assert (classifier_type in ['fc', 'mlp'])\n\n self.bn = nn.BatchNorm2d(nb_channels_in)\n\n self.avg_ker_size = avg_ker_size\n if self.avg_ker_size > 1:\n n = n_space - avg_ker_size + 1\n else:\n n = n_space\n\n in_planes = nb_channels_in * (n ** 2)\n\n if classifier_type == 'mlp':\n classif_modules = [nn.Linear(in_planes, nb_hidden_units)]\n\n for i in range(nb_l_mlp-1):\n classif_modules.append(nn.ReLU(inplace=True))\n classif_modules.append(nn.Dropout(p=dropout_p_mlp))\n classif_modules.append(nn.Linear(nb_hidden_units, nb_hidden_units))\n\n classif_modules.append(nn.ReLU(inplace=True))\n classif_modules.append(nn.Dropout(p=dropout_p_mlp))\n classif_modules.append(nn.Linear(nb_hidden_units, nb_classes))\n\n self.classifier = nn.Sequential(*classif_modules)\n\n elif classifier_type == 'fc':\n self.classifier = nn.Linear(in_planes, nb_classes)\n\n def forward(self, x):\n x = self.bn(x)\n if self.avg_ker_size > 1:\n ## if the size of the images is too small you might need to add : padding=1\n x = nn.functional.avg_pool2d(x, self.avg_ker_size, stride=1)\n x = x.view(x.size(0), -1)\n output = self.classifier(x)\n return output\n",
"import torch.nn as nn\nimport torch\n\n\nclass LinearProj(nn.Module):\n def __init__(self, standardization, proj, L_kernel_size=3):\n super(LinearProj, self).__init__()\n self.standardization = standardization\n self.proj = proj\n self.L_kernel_size = L_kernel_size\n\n def forward(self, x):\n output = self.standardization(x)\n if self.L_kernel_size > 1:\n ## using reflect as a mode for the padding gives errors related to the dimensions of the tensor\n output = nn.functional.pad(output, ((self.L_kernel_size-1)//2,)*4)#, mode='reflect')\n output = self.proj(output)\n\n output = torch.div(output, output.norm(p=2, dim=1, keepdim=True))\n return output"
]
| [
[
"torch.nn.Sequential",
"torch.nn.Dropout",
"torch.nn.functional.avg_pool2d",
"torch.nn.Linear",
"torch.nn.BatchNorm2d",
"torch.nn.ReLU"
],
[
"torch.nn.functional.pad"
]
]
|
braniii/numba | [
"565a778cbcd0f363bf82e31292ef1b1a3e77a390"
]
| [
"numba/cuda/compiler.py"
]
| [
"import collections\nimport ctypes\nimport functools\nimport inspect\nimport os\nimport subprocess\nimport sys\nimport tempfile\n\nimport numpy as np\n\nfrom numba import _dispatcher\nfrom numba.core.typing.templates import AbstractTemplate, ConcreteTemplate\nfrom numba.core import (types, typing, utils, funcdesc, serialize, config,\n compiler, sigutils)\nfrom numba.core.typeconv.rules import default_type_manager\nfrom numba.core.compiler import (CompilerBase, DefaultPassBuilder,\n compile_result)\nfrom numba.core.compiler_lock import global_compiler_lock\nfrom numba.core.compiler_machinery import (LoweringPass, PassManager,\n register_pass)\nfrom numba.core.dispatcher import OmittedArg\nfrom numba.core.errors import NumbaDeprecationWarning\nfrom numba.core.typed_passes import IRLegalization, NativeLowering\nfrom numba.core.typing.typeof import Purpose, typeof\nfrom warnings import warn\nimport numba\nfrom .cudadrv.devices import get_context\nfrom .cudadrv.libs import get_cudalib\nfrom .cudadrv import nvvm, driver\nfrom .errors import missing_launch_config_msg, normalize_kernel_dimensions\nfrom .api import get_current_device\nfrom .args import wrap_arg\n\n\n@register_pass(mutates_CFG=True, analysis_only=False)\nclass CUDABackend(LoweringPass):\n\n _name = \"cuda_backend\"\n\n def __init__(self):\n LoweringPass.__init__(self)\n\n def run_pass(self, state):\n \"\"\"\n Back-end: Packages lowering output in a compile result\n \"\"\"\n lowered = state['cr']\n signature = typing.signature(state.return_type, *state.args)\n\n state.cr = compile_result(\n typing_context=state.typingctx,\n target_context=state.targetctx,\n typing_error=state.status.fail_reason,\n type_annotation=state.type_annotation,\n library=state.library,\n call_helper=lowered.call_helper,\n signature=signature,\n fndesc=lowered.fndesc,\n )\n return True\n\n\nclass CUDACompiler(CompilerBase):\n def define_pipelines(self):\n dpb = DefaultPassBuilder\n pm = PassManager('cuda')\n\n untyped_passes = dpb.define_untyped_pipeline(self.state)\n pm.passes.extend(untyped_passes.passes)\n\n typed_passes = dpb.define_typed_pipeline(self.state)\n pm.passes.extend(typed_passes.passes)\n\n lowering_passes = self.define_cuda_lowering_pipeline(self.state)\n pm.passes.extend(lowering_passes.passes)\n\n pm.finalize()\n return [pm]\n\n def define_cuda_lowering_pipeline(self, state):\n pm = PassManager('cuda_lowering')\n # legalise\n pm.add_pass(IRLegalization,\n \"ensure IR is legal prior to lowering\")\n\n # lower\n pm.add_pass(NativeLowering, \"native lowering\")\n pm.add_pass(CUDABackend, \"cuda backend\")\n\n pm.finalize()\n return pm\n\n\n@global_compiler_lock\ndef compile_cuda(pyfunc, return_type, args, debug=False, inline=False,\n fastmath=False):\n from .descriptor import cuda_target\n typingctx = cuda_target.typingctx\n targetctx = cuda_target.targetctx\n\n flags = compiler.Flags()\n # Do not compile (generate native code), just lower (to LLVM)\n flags.set('no_compile')\n flags.set('no_cpython_wrapper')\n flags.set('no_cfunc_wrapper')\n if debug:\n flags.set('debuginfo')\n if inline:\n flags.set('forceinline')\n if fastmath:\n flags.set('fastmath')\n # Run compilation pipeline\n cres = compiler.compile_extra(typingctx=typingctx,\n targetctx=targetctx,\n func=pyfunc,\n args=args,\n return_type=return_type,\n flags=flags,\n locals={},\n pipeline_class=CUDACompiler)\n\n library = cres.library\n library.finalize()\n\n return cres\n\n\ndef compile_kernel(pyfunc, args, link, debug=False, inline=False,\n fastmath=False, extensions=[], max_registers=None, opt=True):\n return _Kernel(pyfunc, args, link, debug=debug, inline=inline,\n fastmath=fastmath, extensions=extensions,\n max_registers=max_registers, opt=opt)\n\n\n@global_compiler_lock\ndef compile_ptx(pyfunc, args, debug=False, device=False, fastmath=False,\n cc=None, opt=True):\n \"\"\"Compile a Python function to PTX for a given set of argument types.\n\n :param pyfunc: The Python function to compile.\n :param args: A tuple of argument types to compile for.\n :param debug: Whether to include debug info in the generated PTX.\n :type debug: bool\n :param device: Whether to compile a device function. Defaults to ``False``,\n to compile global kernel functions.\n :type device: bool\n :param fastmath: Whether to enable fast math flags (ftz=1, prec_sqrt=0,\n prec_div=, and fma=1)\n :type fastmath: bool\n :param cc: Compute capability to compile for, as a tuple ``(MAJOR, MINOR)``.\n Defaults to ``(5, 2)``.\n :type cc: tuple\n :param opt: Enable optimizations. Defaults to ``True``.\n :type opt: bool\n :return: (ptx, resty): The PTX code and inferred return type\n :rtype: tuple\n \"\"\"\n cres = compile_cuda(pyfunc, None, args, debug=debug)\n resty = cres.signature.return_type\n if device:\n llvm_module = cres.library._final_module\n nvvm.fix_data_layout(llvm_module)\n else:\n fname = cres.fndesc.llvm_func_name\n tgt = cres.target_context\n lib, kernel = tgt.prepare_cuda_kernel(cres.library, fname,\n cres.signature.args, debug=debug)\n llvm_module = lib._final_module\n\n options = {\n 'debug': debug,\n 'fastmath': fastmath,\n }\n\n cc = cc or config.CUDA_DEFAULT_PTX_CC\n opt = 3 if opt else 0\n arch = nvvm.get_arch_option(*cc)\n llvmir = str(llvm_module)\n ptx = nvvm.llvm_to_ptx(llvmir, opt=opt, arch=arch, **options)\n return ptx.decode('utf-8'), resty\n\n\ndef compile_ptx_for_current_device(pyfunc, args, debug=False, device=False,\n fastmath=False, opt=True):\n \"\"\"Compile a Python function to PTX for a given set of argument types for\n the current device's compute capabilility. This calls :func:`compile_ptx`\n with an appropriate ``cc`` value for the current device.\"\"\"\n cc = get_current_device().compute_capability\n return compile_ptx(pyfunc, args, debug=-debug, device=device,\n fastmath=fastmath, cc=cc, opt=True)\n\n\ndef disassemble_cubin(cubin):\n # nvdisasm only accepts input from a file, so we need to write out to a\n # temp file and clean up afterwards.\n fd = None\n fname = None\n try:\n fd, fname = tempfile.mkstemp()\n with open(fname, 'wb') as f:\n f.write(cubin)\n\n try:\n cp = subprocess.run(['nvdisasm', fname], check=True,\n stdout=subprocess.PIPE,\n stderr=subprocess.PIPE)\n except FileNotFoundError as e:\n if e.filename == 'nvdisasm':\n msg = (\"nvdisasm is required for SASS inspection, and has not \"\n \"been found.\\n\\nYou may need to install the CUDA \"\n \"toolkit and ensure that it is available on your \"\n \"PATH.\\n\")\n raise RuntimeError(msg)\n return cp.stdout.decode('utf-8')\n finally:\n if fd is not None:\n os.close(fd)\n if fname is not None:\n os.unlink(fname)\n\n\nclass DeviceFunctionTemplate(serialize.ReduceMixin):\n \"\"\"Unmaterialized device function\n \"\"\"\n def __init__(self, pyfunc, debug, inline, opt):\n self.py_func = pyfunc\n self.debug = debug\n self.inline = inline\n self.opt = opt\n self._compileinfos = {}\n name = getattr(pyfunc, '__name__', 'unknown')\n self.__name__ = f\"{name} <CUDA device function>\".format(name)\n\n def _reduce_states(self):\n return dict(py_func=self.py_func, debug=self.debug, inline=self.inline)\n\n @classmethod\n def _rebuild(cls, py_func, debug, inline):\n return compile_device_template(py_func, debug=debug, inline=inline)\n\n def compile(self, args):\n \"\"\"Compile the function for the given argument types.\n\n Each signature is compiled once by caching the compiled function inside\n this object.\n\n Returns the `CompileResult`.\n \"\"\"\n if args not in self._compileinfos:\n cres = compile_cuda(self.py_func, None, args, debug=self.debug,\n inline=self.inline)\n first_definition = not self._compileinfos\n self._compileinfos[args] = cres\n libs = [cres.library]\n\n if first_definition:\n # First definition\n cres.target_context.insert_user_function(self, cres.fndesc,\n libs)\n else:\n cres.target_context.add_user_function(self, cres.fndesc, libs)\n\n else:\n cres = self._compileinfos[args]\n\n return cres\n\n def inspect_llvm(self, args):\n \"\"\"Returns the LLVM-IR text compiled for *args*.\n\n Parameters\n ----------\n args: tuple[Type]\n Argument types.\n\n Returns\n -------\n llvmir : str\n \"\"\"\n # Force a compilation to occur if none has yet - this can be needed if\n # the user attempts to inspect LLVM IR or PTX before the function has\n # been called for the given arguments from a jitted kernel.\n self.compile(args)\n cres = self._compileinfos[args]\n mod = cres.library._final_module\n return str(mod)\n\n def inspect_ptx(self, args, nvvm_options={}):\n \"\"\"Returns the PTX compiled for *args* for the currently active GPU\n\n Parameters\n ----------\n args: tuple[Type]\n Argument types.\n nvvm_options : dict; optional\n See `CompilationUnit.compile` in `numba/cuda/cudadrv/nvvm.py`.\n\n Returns\n -------\n ptx : bytes\n \"\"\"\n llvmir = self.inspect_llvm(args)\n # Make PTX\n cuctx = get_context()\n device = cuctx.device\n cc = device.compute_capability\n arch = nvvm.get_arch_option(*cc)\n opt = 3 if self.opt else 0\n ptx = nvvm.llvm_to_ptx(llvmir, opt=opt, arch=arch, **nvvm_options)\n return ptx\n\n\ndef compile_device_template(pyfunc, debug=False, inline=False, opt=True):\n \"\"\"Create a DeviceFunctionTemplate object and register the object to\n the CUDA typing context.\n \"\"\"\n from .descriptor import cuda_target\n\n dft = DeviceFunctionTemplate(pyfunc, debug=debug, inline=inline, opt=opt)\n\n class device_function_template(AbstractTemplate):\n key = dft\n\n def generic(self, args, kws):\n assert not kws\n return dft.compile(args).signature\n\n def get_template_info(cls):\n basepath = os.path.dirname(os.path.dirname(numba.__file__))\n code, firstlineno = inspect.getsourcelines(pyfunc)\n path = inspect.getsourcefile(pyfunc)\n sig = str(utils.pysignature(pyfunc))\n info = {\n 'kind': \"overload\",\n 'name': getattr(cls.key, '__name__', \"unknown\"),\n 'sig': sig,\n 'filename': utils.safe_relpath(path, start=basepath),\n 'lines': (firstlineno, firstlineno + len(code) - 1),\n 'docstring': pyfunc.__doc__\n }\n return info\n\n typingctx = cuda_target.typingctx\n typingctx.insert_user_function(dft, device_function_template)\n return dft\n\n\ndef compile_device(pyfunc, return_type, args, inline=True, debug=False):\n return DeviceFunction(pyfunc, return_type, args, inline=True, debug=False)\n\n\ndef declare_device_function(name, restype, argtypes):\n from .descriptor import cuda_target\n typingctx = cuda_target.typingctx\n targetctx = cuda_target.targetctx\n sig = typing.signature(restype, *argtypes)\n extfn = ExternFunction(name, sig)\n\n class device_function_template(ConcreteTemplate):\n key = extfn\n cases = [sig]\n\n fndesc = funcdesc.ExternalFunctionDescriptor(\n name=name, restype=restype, argtypes=argtypes)\n typingctx.insert_user_function(extfn, device_function_template)\n targetctx.insert_user_function(extfn, fndesc)\n return extfn\n\n\nclass DeviceFunction(serialize.ReduceMixin):\n\n def __init__(self, pyfunc, return_type, args, inline, debug):\n self.py_func = pyfunc\n self.return_type = return_type\n self.args = args\n self.inline = True\n self.debug = False\n cres = compile_cuda(self.py_func, self.return_type, self.args,\n debug=self.debug, inline=self.inline)\n self.cres = cres\n\n class device_function_template(ConcreteTemplate):\n key = self\n cases = [cres.signature]\n\n cres.typing_context.insert_user_function(\n self, device_function_template)\n cres.target_context.insert_user_function(self, cres.fndesc,\n [cres.library])\n\n def _reduce_states(self):\n return dict(py_func=self.py_func, return_type=self.return_type,\n args=self.args, inline=self.inline, debug=self.debug)\n\n @classmethod\n def _rebuild(cls, py_func, return_type, args, inline, debug):\n return cls(py_func, return_type, args, inline, debug)\n\n def __repr__(self):\n fmt = \"<DeviceFunction py_func={0} signature={1}>\"\n return fmt.format(self.py_func, self.cres.signature)\n\n\nclass ExternFunction(object):\n def __init__(self, name, sig):\n self.name = name\n self.sig = sig\n\n\nclass ForAll(object):\n def __init__(self, kernel, ntasks, tpb, stream, sharedmem):\n if ntasks < 0:\n raise ValueError(\"Can't create ForAll with negative task count: %s\"\n % ntasks)\n self.kernel = kernel\n self.ntasks = ntasks\n self.thread_per_block = tpb\n self.stream = stream\n self.sharedmem = sharedmem\n\n def __call__(self, *args):\n if self.ntasks == 0:\n return\n\n if self.kernel.specialized:\n kernel = self.kernel\n else:\n kernel = self.kernel.specialize(*args)\n blockdim = self._compute_thread_per_block(kernel)\n griddim = (self.ntasks + blockdim - 1) // blockdim\n\n return kernel[griddim, blockdim, self.stream, self.sharedmem](*args)\n\n def _compute_thread_per_block(self, kernel):\n tpb = self.thread_per_block\n # Prefer user-specified config\n if tpb != 0:\n return tpb\n # Else, ask the driver to give a good config\n else:\n ctx = get_context()\n kwargs = dict(\n func=kernel._func.get(),\n b2d_func=0, # dynamic-shared memory is constant to blksz\n memsize=self.sharedmem,\n blocksizelimit=1024,\n )\n _, tpb = ctx.get_max_potential_block_size(**kwargs)\n return tpb\n\n\nclass CachedPTX(object):\n \"\"\"A PTX cache that uses compute capability as a cache key\n \"\"\"\n def __init__(self, name, llvmir, options):\n self.name = name\n self.llvmir = llvmir\n self.cache = {}\n self._extra_options = options.copy()\n\n def get(self, cc=None):\n \"\"\"\n Get PTX for the current active context.\n \"\"\"\n if not cc:\n cuctx = get_context()\n device = cuctx.device\n cc = device.compute_capability\n\n ptx = self.cache.get(cc)\n if ptx is None:\n arch = nvvm.get_arch_option(*cc)\n ptx = nvvm.llvm_to_ptx(self.llvmir, arch=arch,\n **self._extra_options)\n self.cache[cc] = ptx\n if config.DUMP_ASSEMBLY:\n print((\"ASSEMBLY %s\" % self.name).center(80, '-'))\n print(ptx.decode('utf-8'))\n print('=' * 80)\n return ptx\n\n\nclass CachedCUFunction(serialize.ReduceMixin):\n \"\"\"\n Get or compile CUDA function for the current active context\n\n Uses device ID as key for cache.\n \"\"\"\n\n def __init__(self, entry_name, ptx, linking, max_registers):\n self.entry_name = entry_name\n self.ptx = ptx\n self.linking = linking\n self.cache = {}\n self.ccinfos = {}\n self.cubins = {}\n self.max_registers = max_registers\n\n def get(self):\n cuctx = get_context()\n device = cuctx.device\n cufunc = self.cache.get(device.id)\n if cufunc is None:\n ptx = self.ptx.get()\n\n # Link\n linker = driver.Linker(max_registers=self.max_registers)\n linker.add_ptx(ptx)\n for path in self.linking:\n linker.add_file_guess_ext(path)\n cubin, size = linker.complete()\n compile_info = linker.info_log\n module = cuctx.create_module_image(cubin)\n\n # Load\n cufunc = module.get_function(self.entry_name)\n\n # Populate caches\n self.cache[device.id] = cufunc\n self.ccinfos[device.id] = compile_info\n # We take a copy of the cubin because it's owned by the linker\n cubin_ptr = ctypes.cast(cubin, ctypes.POINTER(ctypes.c_char))\n cubin_data = np.ctypeslib.as_array(cubin_ptr, shape=(size,)).copy()\n self.cubins[device.id] = cubin_data\n return cufunc\n\n def get_sass(self):\n self.get() # trigger compilation\n device = get_context().device\n return disassemble_cubin(self.cubins[device.id])\n\n def get_info(self):\n self.get() # trigger compilation\n cuctx = get_context()\n device = cuctx.device\n ci = self.ccinfos[device.id]\n return ci\n\n def _reduce_states(self):\n \"\"\"\n Reduce the instance for serialization.\n Pre-compiled PTX code string is serialized inside the `ptx` (CachedPTX).\n Loaded CUfunctions are discarded. They are recreated when unserialized.\n \"\"\"\n if self.linking:\n msg = ('cannot pickle CUDA kernel function with additional '\n 'libraries to link against')\n raise RuntimeError(msg)\n return dict(entry_name=self.entry_name, ptx=self.ptx,\n linking=self.linking, max_registers=self.max_registers)\n\n @classmethod\n def _rebuild(cls, entry_name, ptx, linking, max_registers):\n \"\"\"\n Rebuild an instance.\n \"\"\"\n return cls(entry_name, ptx, linking, max_registers)\n\n\nclass _Kernel(serialize.ReduceMixin):\n '''\n CUDA Kernel specialized for a given set of argument types. When called, this\n object launches the kernel on the device.\n '''\n\n @global_compiler_lock\n def __init__(self, py_func, argtypes, link=None, debug=False, inline=False,\n fastmath=False, extensions=None, max_registers=None, opt=True):\n super().__init__()\n\n self.py_func = py_func\n self.argtypes = argtypes\n self.debug = debug\n self.extensions = extensions or []\n\n cres = compile_cuda(self.py_func, types.void, self.argtypes,\n debug=self.debug,\n inline=inline,\n fastmath=fastmath)\n fname = cres.fndesc.llvm_func_name\n args = cres.signature.args\n lib, kernel = cres.target_context.prepare_cuda_kernel(cres.library,\n fname,\n args,\n debug=self.debug)\n\n options = {\n 'debug': self.debug,\n 'fastmath': fastmath,\n 'opt': 3 if opt else 0\n }\n\n llvm_ir = str(lib._final_module)\n pretty_name = cres.fndesc.qualname\n ptx = CachedPTX(pretty_name, llvm_ir, options=options)\n\n if not link:\n link = []\n\n # A kernel needs cooperative launch if grid_sync is being used.\n self.cooperative = 'cudaCGGetIntrinsicHandle' in ptx.llvmir\n # We need to link against cudadevrt if grid sync is being used.\n if self.cooperative:\n link.append(get_cudalib('cudadevrt', static=True))\n\n cufunc = CachedCUFunction(kernel.name, ptx, link, max_registers)\n\n # populate members\n self.entry_name = kernel.name\n self.signature = cres.signature\n self._type_annotation = cres.type_annotation\n self._func = cufunc\n self.call_helper = cres.call_helper\n self.link = link\n\n @property\n def argument_types(self):\n return tuple(self.signature.args)\n\n @classmethod\n def _rebuild(cls, cooperative, name, argtypes, cufunc, link, debug,\n call_helper, extensions):\n \"\"\"\n Rebuild an instance.\n \"\"\"\n instance = cls.__new__(cls)\n # invoke parent constructor\n super(cls, instance).__init__()\n # populate members\n instance.cooperative = cooperative\n instance.entry_name = name\n instance.argument_types = tuple(argtypes)\n instance.link = tuple(link)\n instance._type_annotation = None\n instance._func = cufunc\n instance.debug = debug\n instance.call_helper = call_helper\n instance.extensions = extensions\n return instance\n\n def _reduce_states(self):\n \"\"\"\n Reduce the instance for serialization.\n Compiled definitions are serialized in PTX form.\n Type annotation are discarded.\n Thread, block and shared memory configuration are serialized.\n Stream information is discarded.\n \"\"\"\n return dict(cooperative=self.cooperative, name=self.entry_name,\n argtypes=self.argtypes, cufunc=self._func, link=self.link,\n debug=self.debug, call_helper=self.call_helper,\n extensions=self.extensions)\n\n def bind(self):\n \"\"\"\n Force binding to current CUDA context\n \"\"\"\n self._func.get()\n\n @property\n def ptx(self):\n '''\n PTX code for this kernel.\n '''\n return self._func.ptx.get().decode('utf8')\n\n @property\n def device(self):\n \"\"\"\n Get current active context\n \"\"\"\n return get_current_device()\n\n @property\n def regs_per_thread(self):\n '''\n The number of registers used by each thread for this kernel.\n '''\n return self._func.get().attrs.regs\n\n def inspect_llvm(self):\n '''\n Returns the LLVM IR for this kernel.\n '''\n return str(self._func.ptx.llvmir)\n\n def inspect_asm(self, cc):\n '''\n Returns the PTX code for this kernel.\n '''\n return self._func.ptx.get(cc).decode('ascii')\n\n def inspect_sass(self):\n '''\n Returns the SASS code for this kernel.\n\n Requires nvdisasm to be available on the PATH.\n '''\n return self._func.get_sass()\n\n def inspect_types(self, file=None):\n '''\n Produce a dump of the Python source of this function annotated with the\n corresponding Numba IR and type information. The dump is written to\n *file*, or *sys.stdout* if *file* is *None*.\n '''\n if self._type_annotation is None:\n raise ValueError(\"Type annotation is not available\")\n\n if file is None:\n file = sys.stdout\n\n print(\"%s %s\" % (self.entry_name, self.argument_types), file=file)\n print('-' * 80, file=file)\n print(self._type_annotation, file=file)\n print('=' * 80, file=file)\n\n def max_cooperative_grid_blocks(self, blockdim, dynsmemsize=0):\n '''\n Calculates the maximum number of blocks that can be launched for this\n kernel in a cooperative grid in the current context, for the given block\n and dynamic shared memory sizes.\n\n :param blockdim: Block dimensions, either as a scalar for a 1D block, or\n a tuple for 2D or 3D blocks.\n :param dynsmemsize: Dynamic shared memory size in bytes.\n :return: The maximum number of blocks in the grid.\n '''\n ctx = get_context()\n cufunc = self._func.get()\n\n if isinstance(blockdim, tuple):\n blockdim = functools.reduce(lambda x, y: x * y, blockdim)\n active_per_sm = ctx.get_active_blocks_per_multiprocessor(cufunc,\n blockdim,\n dynsmemsize)\n sm_count = ctx.device.MULTIPROCESSOR_COUNT\n return active_per_sm * sm_count\n\n def launch(self, args, griddim, blockdim, stream=0, sharedmem=0):\n # Prepare kernel\n cufunc = self._func.get()\n\n if self.debug:\n excname = cufunc.name + \"__errcode__\"\n excmem, excsz = cufunc.module.get_global_symbol(excname)\n assert excsz == ctypes.sizeof(ctypes.c_int)\n excval = ctypes.c_int()\n excmem.memset(0, stream=stream)\n\n # Prepare arguments\n retr = [] # hold functors for writeback\n\n kernelargs = []\n for t, v in zip(self.argument_types, args):\n self._prepare_args(t, v, stream, retr, kernelargs)\n\n stream_handle = stream and stream.handle or None\n\n # Invoke kernel\n driver.launch_kernel(cufunc.handle,\n *griddim,\n *blockdim,\n sharedmem,\n stream_handle,\n kernelargs,\n cooperative=self.cooperative)\n\n if self.debug:\n driver.device_to_host(ctypes.addressof(excval), excmem, excsz)\n if excval.value != 0:\n # An error occurred\n def load_symbol(name):\n mem, sz = cufunc.module.get_global_symbol(\"%s__%s__\" %\n (cufunc.name,\n name))\n val = ctypes.c_int()\n driver.device_to_host(ctypes.addressof(val), mem, sz)\n return val.value\n\n tid = [load_symbol(\"tid\" + i) for i in 'zyx']\n ctaid = [load_symbol(\"ctaid\" + i) for i in 'zyx']\n code = excval.value\n exccls, exc_args, loc = self.call_helper.get_exception(code)\n # Prefix the exception message with the source location\n if loc is None:\n locinfo = ''\n else:\n sym, filepath, lineno = loc\n filepath = os.path.abspath(filepath)\n locinfo = 'In function %r, file %s, line %s, ' % (sym,\n filepath,\n lineno,)\n # Prefix the exception message with the thread position\n prefix = \"%stid=%s ctaid=%s\" % (locinfo, tid, ctaid)\n if exc_args:\n exc_args = (\"%s: %s\" % (prefix, exc_args[0]),) + \\\n exc_args[1:]\n else:\n exc_args = prefix,\n raise exccls(*exc_args)\n\n # retrieve auto converted arrays\n for wb in retr:\n wb()\n\n def _prepare_args(self, ty, val, stream, retr, kernelargs):\n \"\"\"\n Convert arguments to ctypes and append to kernelargs\n \"\"\"\n\n # map the arguments using any extension you've registered\n for extension in reversed(self.extensions):\n ty, val = extension.prepare_args(\n ty,\n val,\n stream=stream,\n retr=retr)\n\n if isinstance(ty, types.Array):\n devary = wrap_arg(val).to_device(retr, stream)\n\n c_intp = ctypes.c_ssize_t\n\n meminfo = ctypes.c_void_p(0)\n parent = ctypes.c_void_p(0)\n nitems = c_intp(devary.size)\n itemsize = c_intp(devary.dtype.itemsize)\n data = ctypes.c_void_p(driver.device_pointer(devary))\n kernelargs.append(meminfo)\n kernelargs.append(parent)\n kernelargs.append(nitems)\n kernelargs.append(itemsize)\n kernelargs.append(data)\n for ax in range(devary.ndim):\n kernelargs.append(c_intp(devary.shape[ax]))\n for ax in range(devary.ndim):\n kernelargs.append(c_intp(devary.strides[ax]))\n\n elif isinstance(ty, types.Integer):\n cval = getattr(ctypes, \"c_%s\" % ty)(val)\n kernelargs.append(cval)\n\n elif ty == types.float64:\n cval = ctypes.c_double(val)\n kernelargs.append(cval)\n\n elif ty == types.float32:\n cval = ctypes.c_float(val)\n kernelargs.append(cval)\n\n elif ty == types.boolean:\n cval = ctypes.c_uint8(int(val))\n kernelargs.append(cval)\n\n elif ty == types.complex64:\n kernelargs.append(ctypes.c_float(val.real))\n kernelargs.append(ctypes.c_float(val.imag))\n\n elif ty == types.complex128:\n kernelargs.append(ctypes.c_double(val.real))\n kernelargs.append(ctypes.c_double(val.imag))\n\n elif isinstance(ty, (types.NPDatetime, types.NPTimedelta)):\n kernelargs.append(ctypes.c_int64(val.view(np.int64)))\n\n elif isinstance(ty, types.Record):\n devrec = wrap_arg(val).to_device(retr, stream)\n kernelargs.append(devrec)\n\n elif isinstance(ty, types.BaseTuple):\n assert len(ty) == len(val)\n for t, v in zip(ty, val):\n self._prepare_args(t, v, stream, retr, kernelargs)\n\n else:\n raise NotImplementedError(ty, val)\n\n\nclass _KernelConfiguration:\n def __init__(self, dispatcher, griddim, blockdim, stream, sharedmem):\n self.dispatcher = dispatcher\n self.griddim = griddim\n self.blockdim = blockdim\n self.stream = stream\n self.sharedmem = sharedmem\n\n def __call__(self, *args):\n return self.dispatcher.call(args, self.griddim, self.blockdim,\n self.stream, self.sharedmem)\n\n\nclass StopUsingCCDict(dict):\n def __getitem__(self, key):\n if len(key) > 1 and isinstance(key[0], tuple):\n msg = \"dicts returned by inspect functions should be keyed on \" \\\n \"argument types only\"\n warn(msg, category=NumbaDeprecationWarning)\n return super().__getitem__(key[1])\n return super().__getitem__(key)\n\n\nclass Dispatcher(_dispatcher.Dispatcher, serialize.ReduceMixin):\n '''\n CUDA Dispatcher object. When configured and called, the dispatcher will\n specialize itself for the given arguments (if no suitable specialized\n version already exists) & compute capability, and launch on the device\n associated with the current context.\n\n Dispatcher objects are not to be constructed by the user, but instead are\n created using the :func:`numba.cuda.jit` decorator.\n '''\n\n # Whether to fold named arguments and default values. Default values are\n # presently unsupported on CUDA, so we can leave this as False in all\n # cases.\n _fold_args = False\n\n def __init__(self, py_func, sigs, targetoptions):\n self.py_func = py_func\n self.sigs = []\n self.link = targetoptions.pop('link', (),)\n self._can_compile = True\n\n # Specializations for given sets of argument types\n self.specializations = {}\n\n # A mapping of signatures to compile results\n self.overloads = collections.OrderedDict()\n\n self.targetoptions = targetoptions\n\n # defensive copy\n self.targetoptions['extensions'] = \\\n list(self.targetoptions.get('extensions', []))\n\n from .descriptor import cuda_target\n\n self.typingctx = cuda_target.typingctx\n\n self._tm = default_type_manager\n\n pysig = utils.pysignature(py_func)\n arg_count = len(pysig.parameters)\n argnames = tuple(pysig.parameters)\n default_values = self.py_func.__defaults__ or ()\n defargs = tuple(OmittedArg(val) for val in default_values)\n can_fallback = False # CUDA cannot fallback to object mode\n\n try:\n lastarg = list(pysig.parameters.values())[-1]\n except IndexError:\n has_stararg = False\n else:\n has_stararg = lastarg.kind == lastarg.VAR_POSITIONAL\n\n exact_match_required = False\n\n _dispatcher.Dispatcher.__init__(self, self._tm.get_pointer(),\n arg_count, self._fold_args, argnames,\n defargs, can_fallback, has_stararg,\n exact_match_required)\n\n if sigs:\n if len(sigs) > 1:\n raise TypeError(\"Only one signature supported at present\")\n self.compile(sigs[0])\n self._can_compile = False\n\n def configure(self, griddim, blockdim, stream=0, sharedmem=0):\n griddim, blockdim = normalize_kernel_dimensions(griddim, blockdim)\n return _KernelConfiguration(self, griddim, blockdim, stream, sharedmem)\n\n def __getitem__(self, args):\n if len(args) not in [2, 3, 4]:\n raise ValueError('must specify at least the griddim and blockdim')\n return self.configure(*args)\n\n def forall(self, ntasks, tpb=0, stream=0, sharedmem=0):\n \"\"\"Returns a 1D-configured kernel for a given number of tasks.\n\n This assumes that:\n\n - the kernel maps the Global Thread ID ``cuda.grid(1)`` to tasks on a\n 1-1 basis.\n - the kernel checks that the Global Thread ID is upper-bounded by\n ``ntasks``, and does nothing if it is not.\n\n :param ntasks: The number of tasks.\n :param tpb: The size of a block. An appropriate value is chosen if this\n parameter is not supplied.\n :param stream: The stream on which the configured kernel will be\n launched.\n :param sharedmem: The number of bytes of dynamic shared memory required\n by the kernel.\n :return: A configured kernel, ready to launch on a set of arguments.\"\"\"\n\n return ForAll(self, ntasks, tpb=tpb, stream=stream, sharedmem=sharedmem)\n\n @property\n def extensions(self):\n '''\n A list of objects that must have a `prepare_args` function. When a\n specialized kernel is called, each argument will be passed through\n to the `prepare_args` (from the last object in this list to the\n first). The arguments to `prepare_args` are:\n\n - `ty` the numba type of the argument\n - `val` the argument value itself\n - `stream` the CUDA stream used for the current call to the kernel\n - `retr` a list of zero-arg functions that you may want to append\n post-call cleanup work to.\n\n The `prepare_args` function must return a tuple `(ty, val)`, which\n will be passed in turn to the next right-most `extension`. After all\n the extensions have been called, the resulting `(ty, val)` will be\n passed into Numba's default argument marshalling logic.\n '''\n return self.targetoptions['extensions']\n\n def __call__(self, *args, **kwargs):\n # An attempt to launch an unconfigured kernel\n raise ValueError(missing_launch_config_msg)\n\n def call(self, args, griddim, blockdim, stream, sharedmem):\n '''\n Compile if necessary and invoke this kernel with *args*.\n '''\n if self.specialized:\n kernel = next(iter(self.overloads.values()))\n else:\n kernel = _dispatcher.Dispatcher._cuda_call(self, *args)\n\n kernel.launch(args, griddim, blockdim, stream, sharedmem)\n\n def _compile_for_args(self, *args, **kws):\n # Based on _DispatcherBase._compile_for_args.\n assert not kws\n argtypes = [self.typeof_pyval(a) for a in args]\n return self.compile(tuple(argtypes))\n\n def _search_new_conversions(self, *args, **kws):\n # Based on _DispatcherBase._search_new_conversions\n assert not kws\n args = [self.typeof_pyval(a) for a in args]\n found = False\n for sig in self.nopython_signatures:\n conv = self.typingctx.install_possible_conversions(args, sig.args)\n if conv:\n found = True\n return found\n\n def typeof_pyval(self, val):\n # Based on _DispatcherBase.typeof_pyval, but differs from it to support\n # the CUDA Array Interface.\n try:\n return typeof(val, Purpose.argument)\n except ValueError:\n if numba.cuda.is_cuda_array(val):\n # When typing, we don't need to synchronize on the array's\n # stream - this is done when the kernel is launched.\n return typeof(numba.cuda.as_cuda_array(val, sync=False),\n Purpose.argument)\n else:\n raise\n\n @property\n def nopython_signatures(self):\n # Based on _DispatcherBase.nopython_signatures\n return [kernel.signature for kernel in self.overloads.values()]\n\n def specialize(self, *args):\n '''\n Create a new instance of this dispatcher specialized for the given\n *args*.\n '''\n cc = get_current_device().compute_capability\n argtypes = tuple(\n [self.typingctx.resolve_argument_type(a) for a in args])\n if self.specialized:\n raise RuntimeError('Dispatcher already specialized')\n\n specialization = self.specializations.get((cc, argtypes))\n if specialization:\n return specialization\n\n targetoptions = self.targetoptions\n targetoptions['link'] = self.link\n specialization = Dispatcher(self.py_func, [types.void(*argtypes)],\n targetoptions)\n self.specializations[cc, argtypes] = specialization\n return specialization\n\n def disable_compile(self, val=True):\n self._can_compile = not val\n\n @property\n def specialized(self):\n \"\"\"\n True if the Dispatcher has been specialized.\n \"\"\"\n return len(self.sigs) == 1 and not self._can_compile\n\n @property\n def definition(self):\n warn('Use overloads instead of definition',\n category=NumbaDeprecationWarning)\n # There is a single definition only when the dispatcher has been\n # specialized.\n if not self.specialized:\n raise ValueError(\"Dispatcher needs to be specialized to get the \"\n \"single definition\")\n return next(iter(self.overloads.values()))\n\n @property\n def definitions(self):\n warn('Use overloads instead of definitions',\n category=NumbaDeprecationWarning)\n return self.overloads\n\n @property\n def _func(self):\n if self.specialized:\n return next(iter(self.overloads.values()))._func\n else:\n return {sig: defn._func for sig, defn in self.overloads.items()}\n\n def get_regs_per_thread(self, signature=None):\n '''\n Returns the number of registers used by each thread in this kernel for\n the device in the current context.\n\n :param signature: The signature of the compiled kernel to get register\n usage for. This may be omitted for a specialized\n kernel.\n :return: The number of registers used by the compiled variant of the\n kernel for the given signature and current device.\n '''\n if signature is not None:\n return self.definitions[signature.args].regs_per_thread\n if self.specialized:\n return self.definition.regs_per_thread\n else:\n return {sig: defn.regs_per_thread\n for sig, defn in self.definitions.items()}\n\n def compile(self, sig):\n '''\n Compile and bind to the current context a version of this kernel\n specialized for the given signature.\n '''\n argtypes, return_type = sigutils.normalize_signature(sig)\n assert return_type is None or return_type == types.none\n if self.specialized:\n return next(iter(self.overloads.values()))\n else:\n kernel = self.overloads.get(argtypes)\n if kernel is None:\n if not self._can_compile:\n raise RuntimeError(\"Compilation disabled\")\n kernel = _Kernel(self.py_func, argtypes, link=self.link,\n **self.targetoptions)\n # Inspired by _DispatcherBase.add_overload, but differs slightly\n # because we're inserting a _Kernel object instead of a compiled\n # function.\n c_sig = [a._code for a in argtypes]\n self._insert(c_sig, kernel, cuda=True)\n self.overloads[argtypes] = kernel\n\n kernel.bind()\n self.sigs.append(sig)\n return kernel\n\n def inspect_llvm(self, signature=None, compute_capability=None):\n '''\n Return the LLVM IR for this kernel.\n\n :param signature: A tuple of argument types.\n :param compute_capability: Deprecated: accepted but ignored, provided\n only for backwards compatibility.\n :return: The LLVM IR for the given signature, or a dict of LLVM IR\n for all previously-encountered signatures. If the dispatcher\n is specialized, the IR for the single specialization is\n returned even if no signature was provided.\n\n '''\n if compute_capability is not None:\n warn('passing compute_capability has no effect on the LLVM IR',\n category=NumbaDeprecationWarning)\n if signature is not None:\n return self.overloads[signature].inspect_llvm()\n elif self.specialized:\n warn('inspect_llvm will always return a dict in future',\n category=NumbaDeprecationWarning)\n return next(iter(self.overloads.values())).inspect_llvm()\n else:\n return StopUsingCCDict((sig, defn.inspect_llvm())\n for sig, defn in self.overloads.items())\n\n def inspect_asm(self, signature=None, compute_capability=None):\n '''\n Return this kernel's PTX assembly code for for the device in the\n current context.\n\n :param signature: A tuple of argument types.\n :param compute_capability: Deprecated: accepted but ignored, provided\n only for backwards compatibility.\n :return: The PTX code for the given signature, or a dict of PTX codes\n for all previously-encountered signatures. If the dispatcher\n is specialized, the PTX code for the single specialization is\n returned even if no signature was provided.\n '''\n if compute_capability is not None:\n msg = 'The compute_capability kwarg is deprecated'\n warn(msg, category=NumbaDeprecationWarning)\n\n cc = compute_capability or get_current_device().compute_capability\n if signature is not None:\n return self.overloads[signature].inspect_asm(cc)\n elif self.specialized:\n warn('inspect_asm will always return a dict in future',\n category=NumbaDeprecationWarning)\n return next(iter(self.overloads.values())).inspect_asm(cc)\n else:\n return StopUsingCCDict((sig, defn.inspect_asm(cc))\n for sig, defn in self.overloads.items())\n\n def inspect_sass(self, signature=None, compute_capability=None):\n '''\n Return this kernel's SASS assembly code for for the device in the\n current context.\n\n :param signature: A tuple of argument types.\n :param compute_capability: Deprecated: accepted but ignored, provided\n only for backwards compatibility.\n :return: The SASS code for the given signature, or a dict of SASS codes\n for all previously-encountered signatures. If the dispatcher\n is specialized, the SASS code for the single specialization is\n returned even if no signature was provided.\n\n SASS for the device in the current context is returned.\n\n Requires nvdisasm to be available on the PATH.\n '''\n if compute_capability is not None:\n warn('passing compute_capability has no effect on the SASS code',\n category=NumbaDeprecationWarning)\n if signature is not None:\n return self.overloads[signature].inspect_sass()\n elif self.specialized:\n warn('inspect_sass will always return a dict in future',\n category=NumbaDeprecationWarning)\n return next(iter(self.overloads.values())).inspect_sass()\n else:\n return StopUsingCCDict((sig, defn.inspect_sass())\n for sig, defn in self.overloads.items())\n\n def inspect_types(self, file=None):\n '''\n Produce a dump of the Python source of this function annotated with the\n corresponding Numba IR and type information. The dump is written to\n *file*, or *sys.stdout* if *file* is *None*.\n '''\n if file is None:\n file = sys.stdout\n\n for _, defn in self.overloads.items():\n defn.inspect_types(file=file)\n\n @property\n def ptx(self):\n if self.specialized:\n warn('ptx will always return a dict in future',\n category=NumbaDeprecationWarning)\n return next(iter(self.overloads.values())).ptx\n else:\n return StopUsingCCDict((sig, defn.ptx)\n for sig, defn in self.overloads.items())\n\n def bind(self):\n for defn in self.overloads.values():\n defn.bind()\n\n @classmethod\n def _rebuild(cls, py_func, sigs, targetoptions):\n \"\"\"\n Rebuild an instance.\n \"\"\"\n instance = cls(py_func, sigs, targetoptions)\n return instance\n\n def _reduce_states(self):\n \"\"\"\n Reduce the instance for serialization.\n Compiled definitions are discarded.\n \"\"\"\n return dict(py_func=self.py_func, sigs=self.sigs,\n targetoptions=self.targetoptions)\n"
]
| [
[
"numpy.ctypeslib.as_array"
]
]
|
rogerlew/statsmodels | [
"3c460db864d2039126593d6b61849c8796296adf"
]
| [
"statsmodels/tsa/tests/test_ar.py"
]
| [
"\"\"\"\nTest AR Model\n\"\"\"\nimport statsmodels.api as sm\nfrom statsmodels.tsa.ar_model import AR\nfrom numpy.testing import (assert_almost_equal, assert_equal, #assert_allclose,\n assert_)\nfrom results import results_ar\nimport numpy as np\nimport numpy.testing as npt\nfrom pandas import Series, Index\n\nDECIMAL_6 = 6\nDECIMAL_5 = 5\nDECIMAL_4 = 4\n\nclass CheckAR(object):\n def test_params(self):\n assert_almost_equal(self.res1.params, self.res2.params, DECIMAL_6)\n\n def test_bse(self):\n bse = np.sqrt(np.diag(self.res1.cov_params())) # no dof correction\n # for compatability with Stata\n assert_almost_equal(bse, self.res2.bse_stata, DECIMAL_6)\n assert_almost_equal(self.res1.bse, self.res2.bse_gretl, DECIMAL_5)\n\n def test_llf(self):\n assert_almost_equal(self.res1.llf, self.res2.llf, DECIMAL_6)\n\n def test_fpe(self):\n assert_almost_equal(self.res1.fpe, self.res2.fpe, DECIMAL_6)\n\n def test_pickle(self):\n from statsmodels.compatnp.py3k import BytesIO\n fh = BytesIO()\n #test wrapped results load save pickle\n self.res1.save(fh)\n fh.seek(0,0)\n res_unpickled = self.res1.__class__.load(fh)\n assert_(type(res_unpickled) is type(self.res1))\n\nclass TestAROLSConstant(CheckAR):\n \"\"\"\n Test AR fit by OLS with a constant.\n \"\"\"\n @classmethod\n def setupClass(cls):\n data = sm.datasets.sunspots.load()\n cls.res1 = AR(data.endog).fit(maxlag=9, method='cmle')\n cls.res2 = results_ar.ARResultsOLS(constant=True)\n\n def test_predict(self):\n model = self.res1.model\n params = self.res1.params\n assert_almost_equal(model.predict(params),self.res2.FVOLSnneg1start0,\n DECIMAL_4)\n assert_almost_equal(model.predict(params),self.res2.FVOLSnneg1start9,\n DECIMAL_4)\n assert_almost_equal(model.predict(params, start=100),\n self.res2.FVOLSnneg1start100, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=9, end=200),\n self.res2.FVOLSn200start0, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=200, end=400),\n self.res2.FVOLSn200start200, DECIMAL_4)\n #assert_almost_equal(model.predict(params, n=200,start=-109),\n # self.res2.FVOLSn200startneg109, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=308, end=424),\n self.res2.FVOLSn100start325, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=9, end=310),\n self.res2.FVOLSn301start9, DECIMAL_4)\n assert_almost_equal(model.predict(params),\n self.res2.FVOLSdefault, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=308, end=316),\n self.res2.FVOLSn4start312, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=308, end=327),\n self.res2.FVOLSn15start312, DECIMAL_4)\n\n\nclass TestAROLSNoConstant(CheckAR):\n \"\"\"f\n Test AR fit by OLS without a constant.\n \"\"\"\n @classmethod\n def setupClass(cls):\n data = sm.datasets.sunspots.load()\n cls.res1 = AR(data.endog).fit(maxlag=9,method='cmle',trend='nc')\n cls.res2 = results_ar.ARResultsOLS(constant=False)\n\n def test_predict(self):\n model = self.res1.model\n params = self.res1.params\n assert_almost_equal(model.predict(params),self.res2.FVOLSnneg1start0,\n DECIMAL_4)\n assert_almost_equal(model.predict(params),self.res2.FVOLSnneg1start9,\n DECIMAL_4)\n assert_almost_equal(model.predict(params, start=100),\n self.res2.FVOLSnneg1start100, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=9, end=200),\n self.res2.FVOLSn200start0, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=200, end=400),\n self.res2.FVOLSn200start200, DECIMAL_4)\n #assert_almost_equal(model.predict(params, n=200,start=-109),\n # self.res2.FVOLSn200startneg109, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=308,end=424),\n self.res2.FVOLSn100start325, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=9, end=310),\n self.res2.FVOLSn301start9, DECIMAL_4)\n assert_almost_equal(model.predict(params),\n self.res2.FVOLSdefault, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=308, end=316),\n self.res2.FVOLSn4start312, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=308, end=327),\n self.res2.FVOLSn15start312, DECIMAL_4)\n\n #class TestARMLEConstant(CheckAR):\nclass TestARMLEConstant(object):\n @classmethod\n def setupClass(cls):\n data = sm.datasets.sunspots.load()\n cls.res1 = AR(data.endog).fit(maxlag=9,method=\"mle\", disp=-1)\n cls.res2 = results_ar.ARResultsMLE(constant=True)\n\n def test_predict(self):\n model = self.res1.model\n params = self.res1.params\n assert_almost_equal(model.predict(params), self.res2.FVMLEdefault,\n DECIMAL_4)\n assert_almost_equal(model.predict(params, start=9, end=308),\n self.res2.FVMLEstart9end308, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=100, end=308),\n self.res2.FVMLEstart100end308, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=0, end=200),\n self.res2.FVMLEstart0end200, DECIMAL_4)\n\n # Note: factor 0.5 in below two tests needed to meet precision on OS X.\n assert_almost_equal(0.5 * model.predict(params, start=200, end=333),\n 0.5 * self.res2.FVMLEstart200end334, DECIMAL_4)\n assert_almost_equal(0.5 * model.predict(params, start=308, end=333),\n 0.5 * self.res2.FVMLEstart308end334, DECIMAL_4)\n\n assert_almost_equal(model.predict(params, start=9,end=309),\n self.res2.FVMLEstart9end309, DECIMAL_4)\n assert_almost_equal(model.predict(params, end=301),\n self.res2.FVMLEstart0end301, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=4, end=312),\n self.res2.FVMLEstart4end312, DECIMAL_4)\n assert_almost_equal(model.predict(params, start=2, end=7),\n self.res2.FVMLEstart2end7, DECIMAL_4)\n\n def test_dynamic_predict(self):\n res1 = self.res1\n res2 = self.res2\n\n # assert_raises pre-sample\n\n # 9, 51\n start, end = 9, 51\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn[start:end+1], DECIMAL_4)\n\n # 9, 308\n start, end = 9, 308\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn[start:end+1], DECIMAL_4)\n\n # 9, 333\n start, end = 9, 333\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn[start:end+1], DECIMAL_4)\n\n # 100, 151\n start, end = 100, 151\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn2[start:end+1], DECIMAL_4)\n\n # 100, 308\n start, end = 100, 308\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn2[start:end+1], DECIMAL_4)\n\n # 100, 333\n start, end = 100, 333\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn2[start:end+1], DECIMAL_4)\n\n # 308, 308\n start, end = 308, 308\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn3[start:end+1], DECIMAL_4)\n\n # 308, 333\n start, end = 308, 333\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn3[start:end+1], DECIMAL_4)\n\n # 309, 333\n start, end = 309, 333\n fv = res1.predict(start, end, dynamic=True)\n assert_almost_equal(fv, res2.fcdyn4[start:end+1], DECIMAL_4)\n\n # None, None\n start, end = None, None\n fv = res1.predict(dynamic=True)\n assert_almost_equal(fv, res2.fcdyn[9:309], DECIMAL_4)\n\n\nclass TestAutolagAR(object):\n @classmethod\n def setupClass(cls):\n data = sm.datasets.sunspots.load()\n endog = data.endog\n results = []\n for lag in range(1,16+1):\n endog_tmp = endog[16-lag:]\n r = AR(endog_tmp).fit(maxlag=lag)\n # See issue #324 for why we're doing these corrections vs. R\n # results\n k_ar = r.k_ar\n k_trend = r.k_trend\n log_sigma2 = np.log(r.sigma2)\n #import ipdb; ipdb.set_trace()\n aic = r.aic\n aic = (aic - log_sigma2) * (1 + k_ar)/(1 + k_ar + k_trend)\n aic += log_sigma2\n\n hqic = r.hqic\n hqic = (hqic - log_sigma2) * (1 + k_ar)/(1 + k_ar + k_trend)\n hqic += log_sigma2\n\n bic = r.bic\n bic = (bic - log_sigma2) * (1 + k_ar)/(1 + k_ar + k_trend)\n bic += log_sigma2\n\n\n results.append([aic, hqic, bic, r.fpe])\n res1 = np.asarray(results).T.reshape(4,-1, order='C')\n # aic correction to match R\n cls.res1 = res1\n cls.res2 = results_ar.ARLagResults(\"const\").ic\n\n def test_ic(self):\n\n npt.assert_almost_equal(self.res1, self.res2, DECIMAL_6)\n\ndef test_ar_dates():\n # just make sure they work\n data = sm.datasets.sunspots.load()\n dates = sm.tsa.datetools.dates_from_range('1700', length=len(data.endog))\n endog = Series(data.endog, index=dates)\n ar_model = sm.tsa.AR(endog, freq='A').fit(maxlag=9, method='mle', disp=-1)\n pred = ar_model.predict(start='2005', end='2015')\n predict_dates = sm.tsa.datetools.dates_from_range('2005', '2015')\n try:\n from pandas import DatetimeIndex\n predict_dates = DatetimeIndex(predict_dates, freq='infer')\n except:\n pass\n assert_equal(ar_model.data.predict_dates, predict_dates)\n assert_equal(pred.index, predict_dates)\n\ndef test_ar_named_series():\n dates = sm.tsa.datetools.dates_from_range(\"2011m1\", length=72)\n y = Series(np.random.randn(72), name=\"foobar\", index=dates)\n results = sm.tsa.AR(y).fit(2)\n assert_(results.params.index.equals(Index([\"const\", \"L1.foobar\",\n \"L2.foobar\"])))\n\ndef test_ar_start_params():\n # fix 236\n # smoke test\n data = sm.datasets.sunspots.load()\n res = AR(data.endog).fit(maxlag=9, start_params=0.1*np.ones(10.),\n method=\"mle\", disp=-1)\n\n\n#TODO: likelihood for ARX model?\n#class TestAutolagARX(object):\n# def setup(self):\n# data = sm.datasets.macrodata.load()\n# endog = data.data.realgdp\n# exog = data.data.realint\n# results = []\n# for lag in range(1, 26):\n# endog_tmp = endog[26-lag:]\n# exog_tmp = exog[26-lag:]\n# r = AR(endog_tmp, exog_tmp).fit(maxlag=lag, trend='ct')\n# results.append([r.aic, r.hqic, r.bic, r.fpe])\n# self.res1 = np.asarray(results).T.reshape(4,-1, order='C')\n\n\n\n"
]
| [
[
"numpy.testing.assert_equal",
"numpy.log",
"pandas.Series",
"numpy.asarray",
"pandas.Index",
"pandas.DatetimeIndex",
"numpy.ones",
"numpy.testing.assert_almost_equal",
"numpy.random.randn"
]
]
|
ereidelbach/cfbAnalysis | [
"55366a4305b3f99adab491b38426258b0a28e62f"
]
| [
"src/data/scrape_Salaries.py"
]
| [
"#!/usr/bin/env python3\n# -*- coding: utf-8 -*-\n\"\"\"\nCreated on Thu Sep 21 13:56:10 2017\n\n@author: ejreidelbach\n\n:DESCRIPTION:\n - Scrapes Coaching Salary Data from USA Today's Website\n\n:REQUIRES:\n - Refer to `Package Import` section below for required packages\n \n:TODO:\n - NONE\n \n\"\"\"\n#==============================================================================\n# Package Import\n#==============================================================================\nfrom bs4 import BeautifulSoup\nimport os\nimport pandas as pd\nimport pathlib\nimport requests\n\n#==============================================================================\n# Reference Variable Declaration\n#==============================================================================\n# Specify the USA Today Coaching Websites\nurl_usatoday = ['http://sports.usatoday.com/ncaa/salaries/',\n 'http://sports.usatoday.com/ncaa/salaries/mens-basketball/coach',\n 'http://sports.usatoday.com/ncaa/salaries/football/assistant',\n 'http://sports.usatoday.com/ncaa/salaries/football/strength']\n\nfilenames_hist = ['salary_fb_head_team_hist.csv',\n 'salary_fb_head_team_coach.csv',\n 'salary_bb_head_hist.csv',\n 'salary_fb_asst_hist.csv',\n 'salary_fb_str_hist.csv'] \n\n# 11 teams don't have team names (`list_orig`) that match the official \n# NCAA stats page (`list_new`)\nlist_orig = [\"Alabama at Birmingham\", \"Central Florida\", \"Miami (Ohio)\",\n \"Miami (Fla.)\", \"Mississippi\", \"Nevada-Las Vegas\",\n \"Southern California\", \"Southern Mississippi\", \"Texas Christian\",\n \"Texas-El Paso\", \"Texas-San Antonio\", \"Texas AM\"]\nlist_new = [\"UAB\",\"UCF\",\"Miami (OH)\",\"Miami (FL)\",\"Ole Miss\",\"UNLV\",\"USC\",\n \"Southern Miss\",\"TCU\",\"UTEP\",\"UTSA\",\"Texas A&M\"]\n\n#==============================================================================\n# Function Definitions\n#==============================================================================\ndef directoryCheck():\n '''\n Purpose: Run a check of the /data/raw/Salary/ folder to see if such a \n folder exists. If it doesn't, create it.\n \n Input:\n - NONE\n \n Outpu:\n - NONE\n '''\n # Check for the team folder\n pathlib.Path('data/raw/Salary/').mkdir(parents=True, exist_ok=True)\n\ndef team_info(url):\n '''\n Scrape team information from all universities on file\n \n Input: \n - url: string containing the specific URL to be scraped\n \n Ouptput:\n - team_info.csv: file containing all university-related info\n - Nothing is returned by this function\n ''' \n # Request the site and soupify the data\n r = requests.get(url)\n soup = BeautifulSoup(r.content,'xml')\n \n # Extract the salary data for all recorded years\n table = soup.find('table', \n class_='datatable datatable-salaries fixed-column')\n rows = table.findAll('tr')\n \n # Create storage variables\n school_dict = {}\n df_team_info = pd.DataFrame()\n\n # First row will be the column headings, the rest will be coach/school info\n for j in range(len(rows)):\n # For all body rows, extract the school ID ('passid') and the coach ID \n # ('data-coach')\n if j != 0:\n cols = rows[j].findAll('td')\n if (len(cols) > 1): # Ignore any rows which have no column info\n temp_dict = {}\n temp_dict['schoolID'] = cols[1]['data-passid']\n school_dict[cols[1].text] = temp_dict\n \n # Using the school and coach IDs, extract the historical salary data for \n # every school\n key_count = 0\n for key in school_dict:\n key_count += 1\n \n # Request information for the specific school\n print(\"Downloading data for #\" + str(key_count) + \": \" + str(key))\n s = requests.get(\n 'http://sports.usatoday.com/ajaxservice/ncaa/salaries__school__' \n + str(school_dict[key]['schoolID']))\n\n # Create temporary storage variables\n temp_json = s.json()\n \n # Populate a separate table with important info concerning each school\n # Ex: Team Abbrevation, ID number, Conference, and Logo URL\n team_dict = {}\n team_dict['school'] = key\n team_dict['teamID'] = temp_json['position']\n team_dict['conference'] = temp_json['profile']['conference']\n try:\n team_dict['teamABBR'] = temp_json['profile']['team_abbr']\n team_dict['icon_url'] = (('http://www.gannett-cdn.com/media/SMG/' + \n 'sports_logos/ncaa-whitebg/110/') +\n str(temp_json['profile']['team_abbr']) + '.png')\n except:\n pass\n temp_df_team = pd.DataFrame([team_dict])\n \n # Append the temporarily created dataframe for the team to the master\n # dataframe containing info for all teams\n df_team_info = df_team_info.append(temp_df_team)\n \n # Wait 6 seconds before querying the next team so we don't overload the \n # the USA Today server and risk getting locked out\n #time.sleep(6)\n \n # Convert names of select schools to match their format on the NCAA website\n df_team_info[['school']] = df_team_info[['school']].replace(list_orig,list_new)\n \n print(\"Writing data to: team_info.csv\")\n df_team_info.to_csv(pathlib.Path('data/raw/Info','team_info.csv'), \n index=False) \n\ndef current_year(url, fname):\n '''\n Scrape salary info for all TEAMS for the most recent year (i.e. current)\n \n Input: \n - url: string containing the specific URL to be scraped\n - fname: string containing the filename (CSV) to which the data \n should be written\n \n Ouptput:\n - fname.csv: file containing the year's salary information\n - Nothing is returned by this function\n '''\n # Request the site, soupify the data, and extract the main table (table 0)\n r = requests.get(url)\n soup = BeautifulSoup(r.content,'xml')\n table = soup.find_all('table')[0]\n\n #---- Extract the data for the current year \n # Extract the nested dataframe we want\n df_salary = pd.read_html(str(table))[0]\n # Drop Rank column as we won't need it\n df_salary.drop('Rk', axis=1, inplace=True) \n # Drop footer that made it into the table \n df_salary = df_salary[df_salary.School != 'School']\n \n # Convert the salary column to a numeric value\n df_salary[df_salary.columns[3:]] = df_salary[df_salary.columns[3:]].replace(\n '[\\$,]','', regex=True).apply(pd.to_numeric, errors='coerce')\n \n # Convert school names to values that match the NCAA website\n df_salary[['School']] = df_salary[['School']].replace(list_orig,list_new)\n \n # Write the current year's salaries to csv files\n df_salary.to_csv(pathlib.Path('data/raw/Salary', fname), index=False) \n\ndef all_years_team(url, fname):\n '''\n Scrape historical salary information for all TEAMS throughout all \n available years\n \n Input: \n - url: string containing the specific URL to be scraped\n - fname: string containing the filename (CSV) to which the data \n should be written\n \n Ouptput:\n - fname.csv: file containing all historical salary information\n - Nothing is returned by this function\n ''' \n # Request the site and soupify the data\n r = requests.get(url)\n soup = BeautifulSoup(r.content,'xml')\n \n # Extract the salary data for all recorded years\n table = soup.find('table', class_='datatable datatable-salaries fixed-column')\n rows = table.findAll('tr')\n \n # Create storage variables\n salary_dict = {}\n df_salary_hist = pd.DataFrame()\n \n # First row will be the column headings, the rest will be coach/school info\n for j in range(len(rows)):\n # For all body rows, extract the school ID ('passid') ]\n # and the coach ID ('data-coach')\n if j != 0:\n cols = rows[j].findAll('td')\n if (len(cols) > 1): # Ignore any rows which have no column info\n temp_dict = {}\n temp_dict['schoolID'] = cols[1]['data-passid']\n temp_dict['coachID'] = cols[1]['data-coach']\n salary_dict[cols[1].text] = temp_dict\n \n # Using the school and coach IDs, extract the historical salary data \n # for every school\n key_count = 0\n for key in salary_dict:\n key_count += 1\n \n # Request information for the specific school\n print(\"Downloading \" + str(fname.split('_')[2]) + \" data for #\" \n + str(key_count) + \": \" + str(key))\n s = requests.get(\n str('http://sports.usatoday.com/ajaxservice/ncaa/salaries__school__' \n + str(salary_dict[key]['schoolID'])))\n\n # Create temporary storage variables\n temp_json = s.json()\n temp_df = pd.DataFrame() \n \n # For every row of data the school has, extract it and add in the team name\n for k in range(len(temp_json['rows'])):\n temp_df = temp_df.append(pd.DataFrame.from_dict(\n temp_json['rows'][k]), ignore_index = True)\n temp_df['school'] = key\n temp_df['conference'] = temp_json['profile']['conference']\n \n # Append the temporarily created dataframe to the master salary dataframe\n df_salary_hist = df_salary_hist.append(temp_df)\n \n # Wait 6 seconds before querying the next team so we don't overload the \n # the USA Today server and risk getting locked out\n #time.sleep(6)\n\n # Convert the salary columns from strings to numeric values\n df_salary_hist[df_salary_hist.columns[2:7]] = df_salary_hist[\n df_salary_hist.columns[2:7]].replace('[\\$,]','',regex=True).apply(\n pd.to_numeric, errors='coerce')\n \n # Convert missing values to NaN\n df_salary_hist = df_salary_hist.fillna(0)\n \n # Convert the names of select schools to match their format on the NCAA website\n df_salary_hist[['school']] = df_salary_hist[['school']].replace(\n list_orig,list_new)\n \n # Write the historical salaries to csv files\n print(\"Writing data to: \" + str(fname))\n df_salary_hist.to_csv(pathlib.Path('data/raw/Salary', fname), index=False)\n\ndef all_years_coach(url, fname):\n '''\n Scrape historical salary information for all COACHES throughout all\n available years\n \n Input: \n - url: string containing the specific URL to be scraped\n - fname: string containing the filename (CSV) to which the data \n should be written\n \n Ouptput:\n - fname.csv: file containing all historical salary information\n - Nothing is returned by this function\n '''\n # Request the site and soupify the data\n r = requests.get(url)\n soup = BeautifulSoup(r.content,'xml')\n \n # Extract the salary data for all recorded years\n table = soup.find('table', \n class_='datatable datatable-salaries fixed-column')\n rows = table.findAll('tr')\n \n # Create storage variables\n salary_dict = {}\n df_coach_info = pd.DataFrame()\n df_salary_hist = pd.DataFrame()\n \n # First row will be the column headings, the rest will be coach/school info\n for j in range(len(rows)):\n # For all body rows, extract the school ID ('passid') and the \n # coach ID ('data-coach')\n if j != 0:\n cols = rows[j].findAll('td')\n if (len(cols) > 1): # Ignore any rows which have no column info\n salary_dict[cols[3].text] = cols[1]['data-coach']\n \n # Using the school and coach IDs, extract the historical salary data for \n # every school\n key_count = 0\n for key in salary_dict:\n key_count += 1\n \n # Request information for the specific school\n print(\"Downloading \" + str(fname.split('_')[2]) + \" data for #\" \n + str(key_count) + \": \" + str(key))\n s = requests.get(\n 'http://sports.usatoday.com/ajaxservice/ncaa/salaries__coach__'\n + str(salary_dict[key]))\n\n # Create temporary storage variables\n temp_json = s.json()\n temp_df = pd.DataFrame() \n \n # For every row of data the school has, add in the team name\n for year in temp_json['rows']:\n nested_dict = {'coach':key}\n for cat in year:\n nested_dict[cat] = year[cat]['value']\n temp_df = temp_df.append(pd.DataFrame([nested_dict]))\n \n # Populate a separate table with important info concerning each school\n # Ex: Team Abbrevation, ID number, Conference, and Logo URL\n coach_dict = {}\n coach_dict['coachID'] = temp_json['position']\n coach_dict.update(temp_json['profile'])\n try:\n coach_dict.pop('team_rgb')\n except:\n pass\n try:\n coach_dict['head_shot'] = ('http://www.gannett-cdn.com/media/' +\n str(coach_dict['head_shot']))\n except:\n pass\n temp_df_coach = pd.DataFrame([coach_dict])\n \n # Append the temporarily created dataframe for the team to the master\n # dataframe containing info for all teams\n df_salary_hist = df_salary_hist.append(temp_df)\n df_coach_info = df_coach_info.append(temp_df_coach)\n \n # Wait 6 seconds before querying the next team so we don't overload the \n # the USA Today server and risk getting locked out\n #time.sleep(6)\n\n # Convert the salary columns from strings to numeric values\n if 'strength' not in url:\n col_list = ['last_year_bonus', 'max_bonus', \n 'other_pay', 'school_pay', 'total_pay'] \n df_salary_hist[col_list] = df_salary_hist[col_list].replace(\n '[\\$,]','',regex=True).apply(pd.to_numeric, errors='coerce') \n else:\n col_list = ['max_bonus', 'other_pay', 'school_pay', 'total_pay'] \n df_salary_hist[col_list] = df_salary_hist[col_list].replace(\n '[\\$,]','',regex=True).apply(pd.to_numeric, errors='coerce')\n \n # Convert missing values to NaN\n df_salary_hist = df_salary_hist.fillna(0)\n \n # Convert the names of select schools to match their format on the NCAA website\n df_salary_hist[['school_name']] = df_salary_hist[['school_name']].replace(\n list_orig,list_new)\n df_coach_info[['school_name']] = df_coach_info[['school_name']].replace(\n list_orig,list_new)\n \n # Write the historical salaries to a csv file\n print(\"Writing data to: \" + str(fname))\n df_salary_hist.to_csv(pathlib.Path('data/raw/Salary', fname), index=False)\n \n # Write the information obtained for coaches to a csv file\n print(\"Writing data to: team_info.csv\")\n output_name = \"coach_info_\" + fname.split('_')[1] + \"_\" + fname.split(\n '_')[2] + \".csv\"\n df_coach_info.to_csv(\n pathlib.Path('data/raw/Salary', output_name), index=False) \n\n#==============================================================================\n# Working Code\n#==============================================================================\n \n# Set the project working directory\npath_project = pathlib.Path(__file__).resolve().parents[2]\n#path_project = pathlib.Path('/home/ejreidelbach/Projects/cfbAnalysis')\nos.chdir(path_project)\n\n# Ensure the data/raw/Salary folder exists\ndirectoryCheck()\n\n# Scrape information for all Universities\nteam_info(url_usatoday[0])\n\n#------------------------------------------------------------------------------\n# Football Head Coach Salaries\n#------------------------------------------------------------------------------\n# Head Football Coach Salaries by University \nall_years_team(url_usatoday[0], filenames_hist[0])\n\n# Head Coach Salaries by Coach\nall_years_coach(url_usatoday[0], filenames_hist[1])\n\n# Current Head Coach Salaries\ncurrent_year(url_usatoday[0], 'current_head.csv')\n\n#------------------------------------------------------------------------------\n# Football Assistant Coach Salaries\n#------------------------------------------------------------------------------\n# Assistant Coach Salaries by Coach\nall_years_coach(url_usatoday[2], filenames_hist[3])\n\n# Current Assistant Coach Salaries\ncurrent_year(url_usatoday[2], 'current_asst.csv')\n\n#------------------------------------------------------------------------------\n# Football Strength Coach Salaries\n#------------------------------------------------------------------------------\n# Strength Coach Salaries by University (only data for 2016)\nall_years_coach(url_usatoday[3], filenames_hist[4])\n\n# Current Strength Coach Salaries\ncurrent_year(url_usatoday[3], 'current_strength.csv')"
]
| [
[
"pandas.DataFrame",
"pandas.DataFrame.from_dict"
]
]
|
Chellison/rcp2 | [
"779f2eb787cce342200f0c43e0988f1008f091ac"
]
| [
"src/data/DataLoaders.py"
]
| [
"from pathlib import Path\nimport numpy as np\nimport pandas as pd\nfrom src import utils\n\n\nclass ARCPData():\n # american red cross preparedness data \n def __init__(self, ACS, file_name = 'ARC Preparedness Data.csv' ):\n self.data = None\n self.file_name = utils.DATA['master'] / file_name \n self.Load()\n self.standardizeColumnNames(ACS)\n\n def Load(self):\n\n self.data = pd.read_csv(self.file_name)\n\n def standardizeColumnNames(self, ACS):\n \"\"\"\n Standardizes column names\n \"\"\"\n\n df = self.data\n df.columns = map(str.lower, df.columns)\n df.columns = df.columns.str.replace(', ', '_')\n df.columns = df.columns.str.replace('-', '_')\n df.columns = df.columns.str.replace('/', '_')\n df.columns = df.columns.str.replace('(', '_')\n df.columns = df.columns.str.replace(')', '_')\n df.columns = df.columns.str.replace(' ', '_')\n df.dropna(inplace = True)\n # trim geoid leading saftey marks \n df['geoid'] = df['geoid'].str[2:]\n\n df = df[df['geoid'].isin(ACS.tot_pop.index)]\n\n self.data = df \n \n\nclass ACSData():\n # TODO: typechecking\n \n def __init__(self,year = 2016,level = 'block_group', pop_thresh = 0):\n\n self.file_name = utils.DATA['acs'] / \"acs_{}_data.csv\".format(year)\n self.level = level\n self.data = None\n self.tot_pop = None\n self.pop_thresh = pop_thresh\n self.Load()\n self.Clean(self.data)\n self.Munge(self.data,self.tot_pop, self.pop_thresh, self.level)\n\n \n def Load(self):\n self.data = pd.read_csv(self.file_name, dtype = {'GEOID':'object'}, index_col = 1)\n\n def Clean(self,ACS):\n \n ## Cleans ACS data \n # 'ACS' - ACS variable from LoadACS\n # 'self.level' - geography level to munge the data to\n # levels can be found in utils.GEOID\n # #Note: this can function can only aggregate data \n\n # Ensures GEOID variable is in the correct format and sets it as the dataframe index\n ACS.reset_index(inplace = True)\n ACS['GEOID'] = ACS['geoid'].str[2:]\n \n ACS.set_index(['GEOID'],inplace = True)\n\n ACS.drop('geoid','columns',inplace =True)\n\n \n # Removes extraneous features (i.e. non-numeric) in the dataframe\n if 'Unnamed: 0' in ACS.columns:\n ACS.drop('Unnamed: 0','columns',inplace= True)\n \n if 'NAME' in ACS.columns:\n ACS.drop('NAME','columns',inplace= True)\n \n #if 'inc_pcincome' in ACS.columns:\n # ACS.drop('inc_pcincome','columns',inplace= True)\n \n\n \n self.tot_pop = ACS[['tot_population']].groupby('GEOID').sum()\n # Drop all total count columns in ACS and keeps all percentage columns\n #cols = ACS.columns.to_list()\n #print(cols)\n #for col in cols:\n # if col.find('tot') != -1 : \n # print(col)\n # ACS.drop(col,'columns', inplace = True)\n \n \n\n \n # Remove missing values from dataframe\n ACS.replace([np.inf, -np.inf], np.nan,inplace = True)\n #ACS.dropna(inplace = True)\n\n \n self.data = ACS\n\n \n def Munge(self,ACS,tot_pop, pop_thresh,level='block_group'):\n\n ## ACS Munging\n \n #ACS.drop(ACS.loc[:, 'state':'in_poverty'], inplace = True, axis = 1)\n #print(ACS.columns)\n #education adjustment \n ACS['educ_less_12th'] = ACS.loc[:,'educ_nursery_4th':'educ_12th_no_diploma'].sum(axis =1 )\n ACS['educ_high_school'] = ACS.loc[:,'educ_high_school_grad':'educ_some_col_no_grad'].sum(axis =1 )\n ACS.drop(ACS.loc[:, 'educ_nursery_4th':'educ_some_col_no_grad'], inplace = True, axis = 1)\n\n # house age adjustment \n ACS['house_yr_pct_before_1960'] =ACS.loc[:,'house_yr_pct_1950_1959':'house_yr_pct_earlier_1939'].sum(axis =1 )\n ACS['house_yr_pct_after_2000'] = ACS.loc[:, 'house_yr_pct_2014_plus':'house_yr_pct_2000_2009'].sum(axis = 1 )\n ACS['house_yr_pct_1960_2000'] = ACS.loc[:, 'house_yr_pct_1990_1999':'house_yr_pct_1960_1969'].sum(axis = 1 )\n ACS.drop(ACS.loc[:, 'house_yr_pct_2014_plus':'house_yr_pct_earlier_1939'], inplace = True, axis = 1)\n \n # housing Price adjustment\n ACS['house_val_less_50K']=ACS.loc[:,'house_val_less_10K':'house_val_40K_50K'].sum(axis =1 )\n ACS['house_val_50_100K']=ACS.loc[:,'house_val_50K_60K':'house_val_90K_100K'].sum(axis =1 )\n ACS['house_val_100K_300K']=ACS.loc[:,'house_val_100K_125K':'house_val_250K_300K'].sum(axis =1 )\n ACS['house_val_300K_500K']=ACS.loc[:,'house_val_300K_400K':'house_val_400K_500K'].sum(axis =1 )\n ACS['house_val_more_500K'] = ACS.loc[:,'house_val_500K_750K':'house_val_more_2M'].sum(axis = 1)\n ACS.drop(ACS.loc[:, 'house_val_less_10K':'house_val_more_2M'], inplace = True, axis = 1)\n \n ACS['race_pct_black_or_amind'] = ACS.loc[:,'race_pct_black'] \\\n + ACS.loc[:,'race_pct_amind']\n\n ACS['pct_alt_heat'] = ACS.loc[:,'heat_pct_fueloil_kerosene'] \\\n + ACS.loc[:,'heat_pct_coal'] \\\n + ACS.loc[:,'heat_pct_wood'] \\\n + ACS.loc[:,'heat_pct_bottled_tank_lpgas']\n\n \n \n #print(ACS.columns)\n self.data = ACS\n\n \n # munge to appropriate level \n\n if self.level =='block_group':\n #ACS data already at block_group level\n self.tot_pop = tot_pop\n else:\n Data = self.data \n Data = Data.multiply(tot_pop['tot_population'],axis= 'index')\n \n Data.index , tot_pop.index = Data.index.str[0:utils.GEOID[level]], \\\n tot_pop.index.str[0:utils.GEOID[level]]\n\n Data, tot_pop = Data.groupby(Data.index).sum(), \\\n tot_pop.groupby(tot_pop.index).sum()\n\n self.data = Data.divide(tot_pop['tot_population'],axis = 'index')\n self.tot_pop = tot_pop\n #only get geoids with population greater than user defined value\n self.tot_pop = self.tot_pop[self.tot_pop['tot_population']>=self.pop_thresh]\n self.data = self.data[self.data.index.isin(self.tot_pop.index)]\n\nclass SVIData():\n # TODO: typechecking\n # level and year are fixe\n def __init__(self,ACS):\n\n self.file_name = utils.DATA['svi'] / \"SVI Tract Data.csv\"\n self.data = None\n self.Load()\n self.Clean(ACS)\n \n def Load(self):\n self.data = pd.read_csv(self.file_name, encoding='ISO-8859-1')\n self.data['Tract'] = self.data['GEOID'].str[2:]\n \n def Clean(self, ACS):\n ACS['Tract'] = ACS.index.str[:-1]\n ACS['geos'] = ACS.index\n merged = ACS.merge(self.data, how = 'left', left_on = 'Tract' , right_on ='Tract')\n \n merged.set_index('geos', inplace=True)\n cols = ['inc_pct_poverty','RPL_THEME1', 'RPL_THEME2', 'RPL_THEME3','RPL_THEME4']\n self.data = merged[cols]\n \n \nclass NFIRSData():\n \n def __init__(self,level,tot_pop,pop_thresh = 0, sev=False, min_loss = 10000):\n self.file_name = utils.DATA['master'] /'NFIRS Fire Incident Data.csv'\n self.tot_pop = tot_pop\n self.level = level\n self.severeFiresOnly = sev\n self.pop_thresh = pop_thresh\n self.data = None\n self.fires = None\n self.top10 = None\n self.severeFire = None\n self.min_loss = min_loss\n self.Load()\n # self.Clean(self.data)\n # munge to appropriate level \n self.Munge(self.data, self.tot_pop,self.level, self.min_loss, self.pop_thresh)\n\n def set_sev_loss(self, min_loss):\n self.min_loss = min_loss\n nfirs = self.data\n nfirs['severe_fire'] = 'not_sev_fire'\n sev_fire_mask = (nfirs['oth_death'] > 0) | (nfirs['oth_inj'] > 0) | (nfirs['tot_loss'] >= self.min_loss) | (nfirs['tot_units_affected'] > 1)\n nfirs.loc[sev_fire_mask,'severe_fire'] = 'sev_fire'\n nfirs['min_loss'] = np.where(nfirs['tot_loss']>=self.min_loss,'had_min_loss','no_min_loss')\n self.data = nfirs\n\n return\n\n\n\n def Load(self):\n cols_to_use = ['state','fdid','inc_date','oth_inj','oth_death','prop_loss',\n 'cont_loss','tot_loss','tot_units_affected','geoid']\n\n # Specify particular data type for geoid column\n col_dtypes = {'geoid':str}\n\n # utils.DATA['master'] / self.file_name\n\n #Read in NFIRS dataframe\n Data_path = self.file_name\n \n Data = pd.read_csv(Data_path,\n dtype = col_dtypes,\n usecols = cols_to_use,\n encoding='latin-1')\n self.data = Data\n \n \n\n def Munge(self, nfirs, tot_pop, level, min_loss, pop_thresh):\n #NFIRS Munging\n\n #Convert inc_date column values to python datetime type\n nfirs['inc_date'] = pd.to_datetime(nfirs['inc_date'], infer_datetime_format=True)\n\n\n\n # Ensure correct calculation of tot_loss column \n nfirs['tot_loss'] = nfirs['prop_loss'] + nfirs['cont_loss']\n\n # # Create mask for new severe fire variable\n # sev_fire_mask = (nfirs['oth_death'] > 0) | (nfirs['oth_inj'] > 0) | (nfirs['tot_loss'] >= 10000)\n\n # # By default assigns values of severe fire column as not severe\n # nfirs['severe_fire'] = 'not_sev_fire'\n\n # # Applies filter to severe fire column to label the severe fire instances correctly\n # nfirs.loc[sev_fire_mask,'severe_fire'] = 'sev_fire'\n\n self.set_sev_loss(min_loss)\n\n # Create new NFIRS variables based on specified thresholds of existing variables in dataframe\n nfirs['had_inj'] = np.where(nfirs['oth_inj']>0,'had_inj','no_inj')\n nfirs['had_death'] = np.where(nfirs['oth_death']>0,'had_death','no_death')\n \n\n # Extract just the numeric portion of the geoid\n nfirs['geoid'] = nfirs['geoid'].str.strip('#_')\n\n # Add a year column to be used to groupby in addition to geoid\n nfirs['year'] = nfirs['inc_date'].dt.year.astype('str')\n nfirs.set_index('geoid',inplace = True)\n\n\n # package \n self.data = nfirs\n#-------------------------\n nfirs = self.data\n L = utils.GEOID[level]\n # shorten geoid to desired geography\n nfirs.index = nfirs.index.str[0:L]\n\n # subset to severe fires if requested \n if self.severeFiresOnly:\n nfirs_geos = nfirs.index.unique()\n nfirs_sev = nfirs[nfirs['severe_fire'] == 'sev_fire' ]\n\n fires = pd.crosstab(nfirs_sev.index, nfirs_sev['year'])\n # ensure no geographies were lost in restriction\n missing_geos = nfirs_geos.difference(fires.index)\n fires = fires.reindex(fires.index.append(missing_geos ) )\n\n else:\n # create a list of number of fires per year for each geography\n fires = pd.crosstab(nfirs.index, nfirs['year'])\n \n # Grab total population values pulled from ACS dataframe and assign to each census block in NFIRS dataframe\n #fires = fires.merge(tot_pop, how = 'left', left_index = True, right_index = True)\n #change order to keep ACS geoids\n fires = tot_pop.merge(fires, how = 'left', left_index = True, right_index = True)\n #fires = tot_pop.merge(fires, how = 'right', left_index = True, right_index = True)\n fires.index = fires.index.rename('geoid')\n\n\n # Remove resulting infinity values and zeros following merge \n # note: We keep resulting NA's as NA's to show gaps in data collection \n # use NA tolerant algo or change or add line to drop all rows with NAs\n fires.replace([np.inf, -np.inf,0], np.nan,inplace = True)\n\n # drop rows with low population count\n fires = fires[fires['tot_population'] >= pop_thresh ] \n\n # population adjustment to fires per_n_people \n per_n_people = 1000\n min_year,max_year = nfirs['year'].min(), nfirs['year'].max()\n fires_noAdjustment = fires.copy()\n fires.loc[:,min_year:max_year] = fires.loc[:,min_year:max_year].div(fires['tot_population'], axis = 'index') * per_n_people\n \n # remove population\n fires.drop('tot_population',axis = 1, inplace = True)\n\n # find top decile in terms of number of adjusted fires each year\n top10 = fires > fires.quantile(.9)\n\n self.fires = fires\n self.fires_noAdjustment = fires_noAdjustment\n self.top10 = top10\n"
]
| [
[
"pandas.crosstab",
"pandas.to_datetime",
"pandas.read_csv",
"numpy.where"
]
]
|
yhcc/fastNLP | [
"5ec58e3b868f07d9b8ac36568168165304d499d4",
"5ec58e3b868f07d9b8ac36568168165304d499d4"
]
| [
"fastNLP/core/field.py",
"fastNLP/modules/encoder/char_embedding.py"
]
| [
"import torch\n\n\nclass Field(object):\n \"\"\"A field defines a data type.\n\n \"\"\"\n\n def __init__(self, is_target: bool):\n self.is_target = is_target\n\n def index(self, vocab):\n raise NotImplementedError\n\n def get_length(self):\n raise NotImplementedError\n\n def to_tensor(self, padding_length):\n raise NotImplementedError\n\n def contents(self):\n raise NotImplementedError\n\nclass TextField(Field):\n def __init__(self, text, is_target):\n \"\"\"\n :param text: list of strings\n :param is_target: bool\n \"\"\"\n super(TextField, self).__init__(is_target)\n self.text = text\n self._index = None\n\n def index(self, vocab):\n if self._index is None:\n self._index = [vocab[c] for c in self.text]\n else:\n raise RuntimeError(\"Replicate indexing of this field.\")\n return self._index\n\n def get_length(self):\n \"\"\"Fetch the length of the text field.\n\n :return length: int, the length of the text.\n\n \"\"\"\n return len(self.text)\n\n def to_tensor(self, padding_length: int):\n \"\"\"Convert text field to tensor.\n\n :param padding_length: int\n :return tensor: torch.LongTensor, of shape [padding_length, ]\n \"\"\"\n pads = []\n if self._index is None:\n raise RuntimeError(\"Indexing not done before to_tensor in TextField.\")\n if padding_length > self.get_length():\n pads = [0] * (padding_length - self.get_length())\n return torch.LongTensor(self._index + pads)\n\n def contents(self):\n return self.text.copy()\n\nclass LabelField(Field):\n \"\"\"The Field representing a single label. Can be a string or integer.\n\n \"\"\"\n def __init__(self, label, is_target=True):\n super(LabelField, self).__init__(is_target)\n self.label = label\n self._index = None\n\n def get_length(self):\n \"\"\"Fetch the length of the label field.\n\n :return length: int, the length of the label, always 1.\n \"\"\"\n return 1\n\n def index(self, vocab):\n if self._index is None:\n if isinstance(self.label, str):\n self._index = vocab[self.label]\n return self._index\n\n def to_tensor(self, padding_length):\n if self._index is None:\n if isinstance(self.label, int):\n return torch.tensor(self.label)\n elif isinstance(self.label, str):\n raise RuntimeError(\"Field {} not indexed. Call index method.\".format(self.label))\n else:\n raise RuntimeError(\n \"Not support type for LabelField. Expect str or int, got {}.\".format(type(self.label)))\n else:\n return torch.LongTensor([self._index])\n\n def contents(self):\n return [self.label]\n\nclass SeqLabelField(Field):\n def __init__(self, label_seq, is_target=True):\n super(SeqLabelField, self).__init__(is_target)\n self.label_seq = label_seq\n self._index = None\n\n def get_length(self):\n return len(self.label_seq)\n\n def index(self, vocab):\n if self._index is None:\n self._index = [vocab[c] for c in self.label_seq]\n return self._index\n\n def to_tensor(self, padding_length):\n pads = [0] * (padding_length - self.get_length())\n if self._index is None:\n if self.get_length() == 0:\n return torch.LongTensor(pads)\n elif isinstance(self.label_seq[0], int):\n return torch.LongTensor(self.label_seq + pads)\n elif isinstance(self.label_seq[0], str):\n raise RuntimeError(\"Field {} not indexed. Call index method.\".format(self.label))\n else:\n raise RuntimeError(\n \"Not support type for SeqLabelField. Expect str or int, got {}.\".format(type(self.label)))\n else:\n return torch.LongTensor(self._index + pads)\n\n def contents(self):\n return self.label_seq.copy()\n\n\nclass CharTextField(Field):\n def __init__(self, text, max_word_len, is_target=False):\n super(CharTextField, self).__init__(is_target)\n self.text = text\n self.max_word_len = max_word_len\n self._index = []\n\n def get_length(self):\n return len(self.text)\n\n def contents(self):\n return self.text.copy()\n\n def index(self, char_vocab):\n if len(self._index) == 0:\n for word in self.text:\n char_index = [char_vocab[ch] for ch in word]\n if self.max_word_len >= len(char_index):\n char_index += [0] * (self.max_word_len - len(char_index))\n else:\n self._index.clear()\n raise RuntimeError(\"Word {} has more than {} characters. \".format(word, self.max_word_len))\n self._index.append(char_index)\n return self._index\n\n def to_tensor(self, padding_length):\n \"\"\"\n\n :param padding_length: int, the padding length of the word sequence.\n :return : tensor of shape (padding_length, max_word_len)\n \"\"\"\n pads = [[0] * self.max_word_len] * (padding_length - self.get_length())\n return torch.LongTensor(self._index + pads)\n",
"import torch\nimport torch.nn.functional as F\nfrom torch import nn\n\nfrom fastNLP.modules.utils import initial_parameter\n\n\n# from torch.nn.init import xavier_uniform\nclass ConvCharEmbedding(nn.Module):\n\n def __init__(self, char_emb_size=50, feature_maps=(40, 30, 30), kernels=(3, 4, 5), initial_method=None):\n \"\"\"\n Character Level Word Embedding\n :param char_emb_size: the size of character level embedding. Default: 50\n say 26 characters, each embedded to 50 dim vector, then the input_size is 50.\n :param feature_maps: tuple of int. The length of the tuple is the number of convolution operations\n over characters. The i-th integer is the number of filters (dim of out channels) for the i-th\n convolution.\n :param kernels: tuple of int. The width of each kernel.\n \"\"\"\n super(ConvCharEmbedding, self).__init__()\n self.convs = nn.ModuleList([\n nn.Conv2d(1, feature_maps[i], kernel_size=(char_emb_size, kernels[i]), bias=True, padding=(0, 4))\n for i in range(len(kernels))])\n\n initial_parameter(self, initial_method)\n\n def forward(self, x):\n \"\"\"\n :param x: [batch_size * sent_length, word_length, char_emb_size]\n :return: [batch_size * sent_length, sum(feature_maps), 1]\n \"\"\"\n x = x.contiguous().view(x.size(0), 1, x.size(1), x.size(2))\n # [batch_size*sent_length, channel, width, height]\n x = x.transpose(2, 3)\n # [batch_size*sent_length, channel, height, width]\n return self.convolute(x).unsqueeze(2)\n\n def convolute(self, x):\n feats = []\n for conv in self.convs:\n y = conv(x)\n # [batch_size*sent_length, feature_maps[i], 1, width - kernels[i] + 1]\n y = torch.squeeze(y, 2)\n # [batch_size*sent_length, feature_maps[i], width - kernels[i] + 1]\n y = F.tanh(y)\n y, __ = torch.max(y, 2)\n # [batch_size*sent_length, feature_maps[i]]\n feats.append(y)\n return torch.cat(feats, 1) # [batch_size*sent_length, sum(feature_maps)]\n\n\nclass LSTMCharEmbedding(nn.Module):\n \"\"\"\n Character Level Word Embedding with LSTM with a single layer.\n :param char_emb_size: int, the size of character level embedding. Default: 50\n say 26 characters, each embedded to 50 dim vector, then the input_size is 50.\n :param hidden_size: int, the number of hidden units. Default: equal to char_emb_size.\n \"\"\"\n\n def __init__(self, char_emb_size=50, hidden_size=None, initial_method=None):\n super(LSTMCharEmbedding, self).__init__()\n self.hidden_size = char_emb_size if hidden_size is None else hidden_size\n\n self.lstm = nn.LSTM(input_size=char_emb_size,\n hidden_size=self.hidden_size,\n num_layers=1,\n bias=True,\n batch_first=True)\n initial_parameter(self, initial_method)\n\n def forward(self, x):\n \"\"\"\n :param x:[ n_batch*n_word, word_length, char_emb_size]\n :return: [ n_batch*n_word, char_emb_size]\n \"\"\"\n batch_size = x.shape[0]\n h0 = torch.empty(1, batch_size, self.hidden_size)\n h0 = nn.init.orthogonal_(h0)\n c0 = torch.empty(1, batch_size, self.hidden_size)\n c0 = nn.init.orthogonal_(c0)\n\n _, hidden = self.lstm(x, (h0, c0))\n return hidden[0].squeeze().unsqueeze(2)\n"
]
| [
[
"torch.LongTensor",
"torch.tensor"
],
[
"torch.empty",
"torch.max",
"torch.nn.LSTM",
"torch.cat",
"torch.nn.Conv2d",
"torch.nn.init.orthogonal_",
"torch.nn.functional.tanh",
"torch.squeeze"
]
]
|
dacb/eisy | [
"bb948590b03124ddb90d112c07ecf60908735daf"
]
| [
"eisy/data/simulation/circuits.py"
]
| [
"import numpy as np\n\n\ndef freq_gen(high_freq, low_freq, decades=7):\n '''\n Function that generates the frequency range used to investigate the\n impedance response of an electrical circuit Frequency Generator with\n logspaced freqencies\n\n Parameters\n ----------\n high_freq : single value (int or float)\n initial frequency value (high frequency domain) [Hz]\n high_freq : single value (int or float)\n final frequency value (low frequency domain) [Hz]\n decades : integer\n number of frequency decades to be used as range. Default value\n is set to be 7 [-]\n\n Returns\n ----------\n [0] = frequency range [Hz]\n [1] = Angular frequency range [1/s]\n '''\n f_decades = np.log10(high_freq) - np.log10(low_freq)\n f_range = np.logspace(np.log10(high_freq), np.log10(low_freq),\n np.around(decades*f_decades), endpoint=True)\n w_range = 2 * np.pi * f_range\n return f_range, w_range\n\n\ndef cir_RC_parallel(angular_freq, resistance='none', capacitance='none',\n peak_frequency='none'):\n '''\n Function that simulates the impedance response of a resistor and a\n capacitor in a parallel configuration.\n String representation for this circuit: -(RC)-\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n resistance : single value (int or float)\n Solution resistance [ohm]\n capacitance : single value (int or float)\n Electrode capacitance [F]\n peak_frequency : single value (int or float)\n Peak frequency of RC circuit [Hz]\n\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [ohm]\n '''\n circuit = '-(RC)-'\n if resistance == 'none':\n resistance = (1/(capacitance*(2*np.pi*peak_frequency)))\n elif capacitance == 'none':\n capacitance = (1/(resistance*(2*np.pi*peak_frequency)))\n # compute the impedance response as a complex array\n Z_complex = (resistance/(1+resistance*capacitance*(angular_freq*1j)))\n return Z_complex\n\n\ndef cir_RC_series(angular_freq, resistance='none', capacitance='none',\n peak_frequency='none'):\n '''\n Function that simulates the impedance response of a resistor and a\n capacitor in a series configuration.\n This circuit configuration is used to simulate the response of an ideally\n polarizable electrode, also known as a blocking electrode.\n String representation for this circuit: -R-C-\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n resistance : single value (int or float)\n Solution resistance [ohm]\n capacitance : single value (int or float)\n Capacitance of an electrode surface [F]\n peak_frequency : single value (int or float)\n Peak frequency of RC circuit [Hz]\n\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [ohm]\n '''\n circuit = '-R-C-'\n if (resistance, capacitance, peak_frequency) == 'none':\n raise AssertionError('No circuit element value was provided. Cannot\\\n compute the impedance response')\n elif (resistance, capacitance) == 'none':\n raise AssertionError('Not enough circuit element values were provided.\\\n Cannot compute the impedance response')\n elif resistance == 'none':\n resistance = (1/(capacitance*(2*np.pi*peak_frequency)))\n elif capacitance == 'none':\n capacitance = (1/(resistance*(2*np.pi*peak_frequency)))\n # compute the impedance response as a complex array\n Z_complex = resistance + 1/(capacitance*(angular_freq*1j))\n return Z_complex\n\n\ndef cir_RQ_parallel(angular_freq, resistance='none',\n constant_phase_element='none', alpha='none',\n peak_frequency='none'):\n '''\n Function that simulates the impedance response of a resistor and a\n constant phase element in a parallel configuration.\n String representation for this circuit: -(RQ)-\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n resistance : single value (int or float)\n Solution resistance [Ohm]\n constant_phase_element : single value (int or float)\n Constant phase angle [s^(alpha-1)/ohm]\n alpha : single value -float\n Exponent of the constant phase element. Should be a value between\n 0 and 1 [-]\n peak_frequency : single value (int or float)\n Peak frequency of RC circuit [Hz]\n\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [Ohm]\n '''\n circuit = '-(RQ)-'\n if (resistance, constant_phase_element, alpha, peak_frequency) == 'none':\n raise AssertionError('No circuit element value was provided. Cannot\\\n compute the impedance response')\n elif (resistance, constant_phase_element, alpha) == 'none':\n raise AssertionError('Not enough circuit element values were provided.\\\n Cannot compute the impedance response')\n elif resistance == 'none':\n resistance = (1/(constant_phase_element*(2*np.pi*peak_frequency\n ) ** alpha))\n elif constant_phase_element == 'none':\n constant_phase_element = (1/(resistance*(2*np.pi*peak_frequency\n ) ** alpha))\n elif alpha == 'none':\n alpha = np.log(constant_phase_element *\n resistance)/np.log(1/(2*np.pi * peak_frequency))\n Z_complex = (resistance/(1+resistance*constant_phase_element*(\n angular_freq*1j)**alpha))\n return Z_complex\n\n\ndef cir_RQ_series(angular_freq, resistance='none',\n constant_phase_element='none', alpha='none',\n peak_frequency='none'):\n '''\n Function that simulates the impedance response of a resistor and a\n constant phase element in a series configuration.\n This circuit configuration is used to simulate the response of a\n blocking electrode with distribution of reactivity.\n String representation for this circuit: -R-Q-\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n resistance : single value (int or float)\n Solution resistance [ohm]\n constant_phase_element : single value (int or float)\n Constant phase angle [s^(alpha-1)/ohm]\n alpha : single value -float\n Exponent of the constant phase element. Should be a value between\n 0 and 1 [-]\n peak_frequency : single value (int or float)\n Peak frequency of RC circuit [Hz]\n\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [Oom]\n '''\n circuit = '-R-Q-'\n if (resistance, constant_phase_element, alpha, peak_frequency) == 'none':\n raise AssertionError('No circuit element value was provided. Cannot\\\n compute the impedance response')\n elif (resistance, constant_phase_element, alpha) == 'none':\n raise AssertionError('Not enough circuit element values were provided.\\\n Cannot compute the impedance response')\n elif resistance == 'none':\n resistance = (1/(constant_phase_element*(2*np.pi*peak_frequency) **\n alpha))\n elif constant_phase_element == 'none':\n constant_phase_element = (1/(resistance*(2*np.pi*peak_frequency) **\n alpha))\n elif alpha == 'none':\n alpha = np.log(constant_phase_element *\n resistance)/np.log(1/(2*np.pi * peak_frequency))\n # compute the impedance response as a complex array\n Z_complex = resistance + 1/(constant_phase_element*(\n angular_freq*1j)**alpha)\n return Z_complex\n\n\ndef cir_RsRC(angular_freq, solution_resistance,\n parallel_resistance='none', capacitance='none',\n peak_frequency='none'):\n ''''\n Function that simulates the impedance response of a solution resistor in\n series with a resistor in parallel with a capacitor.\n This circuit configuration is used to simulate the response of an ideally\n polarizable electrode, also known as a blocking electrode.\n String representation for this circuit: -Rs-(RC)-\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n solution_resistance : single value (int or float)\n Solution resistance [ohm]\n parallel_resistance : single value (int or float)\n resistance of the element in parallel with\n the capacitor [ohm]\n capacitance : single value (int or float)\n Capacitance of an electrode surface [F]\n peak_frequency : single value (int or float)\n Peak frequency of the parallel RC circuit [Hz]\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [Ohm]\n '''\n circuit = '-Rs-(RC)-'\n # compute the impedance response as a complex array\n if (parallel_resistance, capacitance, peak_frequency) == 'none':\n raise AssertionError('No circuit element value was provided. Cannot\\\n compute the impedance response')\n elif (parallel_resistance, capacitance) == 'none':\n raise AssertionError('Not enough circuit element values were provided.\\\n Cannot compute the impedance response')\n elif parallel_resistance == 'none':\n parallel_resistance = (1/(capacitance*(2*np.pi*peak_frequency)))\n elif capacitance == 'none':\n capacitance = (1/(parallel_resistance*(2*np.pi*peak_frequency)))\n Z_parallel = (parallel_resistance/(1 + parallel_resistance *\n capacitance * (angular_freq*1j)))\n Z_complex = solution_resistance + Z_parallel\n return Z_complex\n\n\ndef cir_RsRQRQ(angular_freq, solution_resistance='none',\n parallel_resistance_1='none', constant_phase_element_1='none',\n alpha_1='none', parallel_resistance_2='none',\n constant_phase_element_2='none', alpha_2='none',\n peak_frequency_1='none', peak_frequency_2='none'):\n '''\n Function that simulates the impedance response of a solution resistor in\n series with two sets of a resistor in parallel with a constant phase\n elements.\n String representation for this circuit: -Rs-(RQ)-(RQ)-\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n solution_resistance : single value (int or float)\n Solution resistance [ohm]\n parallel_resistance_1 : single value (int or float)\n first combination of resistor in parallel with\n constant phase element [ohm]\n constant_phase_element_1 : single value (int or float)\n First constant phas angle [s^(alpha-1)/ohm]\n alpha_1 : single value -float\n Exponent of the first constant phase element.\n Should be a value between 0 and 1 [-]\n parallel_resistance_2 : single value (int or float)\n Second combination of resistor in parallel with\n constant phase element [ohm]\n constant_phase_element_2 : single value (int or float)\n Second Constant phas angle [s^(alpha-1)/ohm]\n alpha_2 : single value -float\n Exponent of the second constant phase element.\n Should be a value between 0 and 1 [-]\n peak_frequency_1 : single value (int or float)\n Peak frequency of the first parallel RQ circuit [Hz]\n peak_frequency_2 : single value (int or float)\n Peak frequency of the second parallel RQ circuit [Hz]\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [Ohm]\n '''\n circuit = '-Rs-(RQ)-(RQ)-'\n\n if (parallel_resistance_1, constant_phase_element_1, peak_frequency_1,\n parallel_resistance_2, constant_phase_element_2,\n peak_frequency_2) == 'none':\n raise AssertionError('No circuit element value was provided. Cannot\\\n compute the impedance response')\n elif (parallel_resistance_1, constant_phase_element_1,\n parallel_resistance_2, constant_phase_element_2) == 'none':\n raise AssertionError('Not enough circuit element values were provided.\\\n Cannot compute the impedance response')\n\n if parallel_resistance_1 == 'none':\n parallel_resistance_1 = (1/(constant_phase_element_1 *\n (2*np.pi*peak_frequency_1)**alpha_1))\n elif constant_phase_element_1 == 'none':\n constant_phase_element_1 = (1/(parallel_resistance_1 *\n (2*np.pi*peak_frequency_1)**alpha_1))\n if parallel_resistance_2 == 'none':\n parallel_resistance_2 = (1/(constant_phase_element_2 *\n (2*np.pi*peak_frequency_2)**alpha_2))\n elif constant_phase_element_2 == 'none':\n constant_phase_element_2 = (1/(parallel_resistance_2 *\n (2*np.pi*peak_frequency_2)**alpha_2))\n\n Z_parallel_1 = (parallel_resistance_1 /\n (1+parallel_resistance_1*constant_phase_element_1\n * (angular_freq*1j)**alpha_1))\n Z_parallel_2 = (parallel_resistance_2 /\n (1+parallel_resistance_2*constant_phase_element_2\n * (angular_freq*1j)**alpha_2))\n Z_complex = solution_resistance + Z_parallel_1 + Z_parallel_2\n\n return Z_complex\n\n\ndef cir_RsRCRC(angular_freq, solution_resistance,\n parallel_resistance_1='none', capacitance_1='none',\n parallel_resistance_2='none', capacitance_2='none',\n peak_frequency_1='none', peak_frequency_2='none'):\n '''\n Function that simulates the impedance response of a solution resistor in\n series with two sets of a resistor in parallel with a capacitor.\n String representation for this circuit: -Rs-(RC)-(RC)-\n\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n solution_resistance : single value (int or float)\n Solution resistance [ohm]\n parallel_resistance_1 : single value (int or float)\n first combination of resistor in parallel with\n capacitor [ohm]\n capacitance_1 : single value (int or float)\n Capacitance of an electrode surface whichi is part of the\n first combination of RC in parallel [F]\n parallel_resistance_2 : single value (int or float)\n second combination of resistor in parallel with\n capacitor [ohm]\n capacitance_2 : single value (int or float)\n Capacitance of an electrode surface whichi is part of the\n second combination of RC in parallel [F]\n peak_frequency_1 : single value (int or float)\n Peak frequency of the first parallel RC circuit [Hz]\n peak_frequency_2 : single value (int or float)\n Peak frequency of the second parallel RC circuit [Hz]\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [Ohm]\n '''\n circuit = '-Rs-(RC)-(RC)-'\n\n if (parallel_resistance_1, capacitance_1, peak_frequency_1,\n parallel_resistance_2, capacitance_2, peak_frequency_2) == 'none':\n raise AssertionError('No circuit element value was provided. Cannot\\\n compute the impedance response')\n elif (parallel_resistance_1, capacitance_1,\n parallel_resistance_2, capacitance_2) == 'none':\n raise AssertionError('Not enough circuit element values were provided.\\\nCannot compute the impedance response')\n\n if parallel_resistance_1 == 'none':\n parallel_resistance_1 = (1/(capacitance_1*(2*np.pi *\n peak_frequency_1)))\n elif capacitance_1 == 'none':\n capacitance_1 = (1/(parallel_resistance_1*(2*np.pi *\n peak_frequency_1)))\n if parallel_resistance_2 == 'none':\n parallel_resistance_2 = (1/(capacitance_2*(2*np.pi *\n peak_frequency_2)))\n elif capacitance_2 == 'none':\n capacitance_2 = (1/(parallel_resistance_2*(2*np.pi *\n peak_frequency_2)))\n\n Z_parallel_1 = (parallel_resistance_1/(1 + parallel_resistance_1 *\n capacitance_1*(angular_freq*1j)))\n Z_parallel_2 = (parallel_resistance_2/(1 + parallel_resistance_2 *\n capacitance_2*(angular_freq*1j)))\n Z_complex = solution_resistance + Z_parallel_1 + Z_parallel_2\n return Z_complex\n\n\ndef cir_Randles_simplified(angular_freq, solution_resistance,\n parallel_resistance, alpha='none', sigma='none',\n Q='none', fs='none'):\n '''\n Return the impedance of a Randles circuit with a simplified Warburg element\n This form of the Randles circuit is only meant for to simulate\n semi-infinate linear diffusion\n String representation for this circuit: -Rs-(Q-(RW)-)-\n\n Parameters\n ----------\n angular_freq : array-like\n Angular frequency [1/s]\n solution_resistance : single value (int or float)\n Solution resistance [ohm]\n parallel_resistance : single value (int or float)\n resistance of the element in parallel with\n the capacitor [ohm]\n capacitance : single value (int or float)\n Capacitance of an electrode surface [F]\n [[Need to add new parameters!!!!]]\n Returns\n ---------\n Z_complex : array-like\n impedance response of the circuit under investigation [Ohm]\n '''\n circuit = '-Rs-(Q-(RW)-)-'\n if parallel_resistance == 'none':\n parallel_resistance = (1/(Q*(2*np.pi*fs)**alpha))\n elif sigma == 'none':\n sigma = (1/(parallel_resistance*(2*np.pi*fs)**alpha))\n elif alpha == 'none':\n alpha = np.log(Q*parallel_resistance)/np.log(1/(2*np.pi*fs))\n\n Z_Q = 1/(Q*(angular_freq*1j)**alpha)\n Z_R = parallel_resistance\n Z_w = sigma*(angular_freq**(-0.5))-1j*sigma*(angular_freq**(-0.5))\n\n return solution_resistance + 1/(1/Z_Q + 1/(Z_R+Z_w))\n"
]
| [
[
"numpy.around",
"numpy.log",
"numpy.log10"
]
]
|
KTH-dESA/GEOSeMOSYS_Kenya | [
"bb2a525538359845f2e995ca5513d98ec031ce7d"
]
| [
"src/Download_files.py"
]
| [
"\"\"\"\nModule: Download_files\n=============================\n\nA module that downloads data that is required for the GEOSeMOSYS analysis and unzips them and places them in a new folder\n----------------------------------------------------------------------------------------------------------------------------------------------------------------------------\n\nModule author: Nandi Moksnes <[email protected]>\n\n\"\"\"\n\nfrom urllib.request import Request, urlopen\nimport shutil\nimport zipfile\nfrom gzip import open as gzopen\nimport tarfile\nimport pandas as pd\nimport requests\nimport json\nimport os\nfrom tkinter import *\n\n# Retrieve access token\ndef download_viirs(url_viirs, temp):\n \"\"\"\n This function downloads the night time light data from EOG and places it in param temp. It requires a login to that webpage which is free.\n :param url_viirs:\n :param temp:\n :return:\n \"\"\"\n def get_input():\n pwd= entry1.get()\n uname = entry2.get()\n\n params = {\n 'client_id': 'eogdata_oidc',\n 'client_secret': '2677ad81-521b-4869-8480-6d05b9e57d48',\n 'username': uname,\n 'password': pwd,\n 'grant_type': 'password'\n }\n token_url = 'https://eogauth.mines.edu/auth/realms/master/protocol/openid-connect/token'\n response = requests.post(token_url, data=params)\n master.destroy()\n access_token_dict = json.loads(response.text)\n access_token = access_token_dict.get('access_token')\n # Submit request with token bearer\n ## Change data_url variable to the file you want to download\n data_url = url_viirs\n auth = 'Bearer ' + access_token\n headers = {'Authorization': auth}\n response = requests.get(data_url, headers=headers)\n # Write response to output file\n output_file = \"../%s/%s\" % (temp, 'VNL_v2_npp_2020_global_vcmslcfg_c202102150000.average_masked.tif.gz')\n with open(output_file, 'wb') as f:\n f.write(response.content)\n\n master = Tk()\n\n label1 = Label(master, text=\"Username Eart Observation group\")\n label1.pack()\n label1.config(justify=CENTER)\n label2 = Label(master, text=\"Password Eart Observation group\", width=30)\n label2.pack()\n label2.config(justify=CENTER)\n\n entry1 = Entry(master)\n entry2 = Entry(master)\n\n button = Button(master, text=\"Submit\")\n button.config()\n\n entry2 = Entry(master, width=30)\n entry2.pack()\n entry1 = Entry(master, width=30)\n entry1.pack()\n\n buttone1 = Button(master, text=\"Submit\")\n buttone1.pack()\n buttone1.config(command = get_input)\n\n master.mainloop()\n\n\n\ndef download_url_data(url,temp):\n \"\"\" This function downloads the data from URL in url (comma separated file) and place them in temp folder.\n\n :param url:\n :param temp:\n :return:\n \"\"\"\n\n def create_dir(dir):\n if not os.path.exists(dir):\n os.makedirs(dir)\n create_dir(('../' + temp))\n url_adress = pd.read_csv(url, header=None, sep=',')\n for i, row in url_adress.iterrows():\n try:\n req = Request(row[0], headers={'User-Agent': 'Chrome'})\n with urlopen(req) as response, open(\"../%s/%s\" % (temp, row[1]), 'wb') as out_file:\n shutil.copyfileobj(response, out_file)\n except Exception as e:\n print(e)\n return()\n\n\ndef unzip_all(url, fromfolder, tofolder):\n\n \"\"\" This function unzips the data from URL (url) in \"fromfolder\" and place them in \"tofolder\".\n\n :param url:\n :param fromfolder:\n :param tofolder:\n :return:\n \"\"\"\n\n def create_dir(dir):\n if not os.path.exists(dir):\n os.makedirs(dir)\n\n create_dir((tofolder))\n\n url_adress = pd.read_csv(url, header=None, sep=',')\n def unzip(infolder, outfolder):\n with zipfile.ZipFile(infolder, 'r') as zip_ref:\n zip_ref.extractall(outfolder)\n return ()\n\n def extract(tar_url, out_path):\n tar = tarfile.open(tar_url, 'r')\n for item in tar:\n tar.extract(item, out_path)\n if item.name.find(\".tgz\") != -1 or item.name.find(\".tar\") != -1:\n\n extract(item.name, \"./\" + item.name[:item.name.rfind('/')])\n def gzip_e(fin, fou):\n with gzopen(fin, 'rb') as s_file, \\\n open(fou, 'wb') as d_file:\n shutil.copyfileobj(s_file, d_file)\n\n\n for i, row in url_adress.iterrows():\n _, filename = os.path.split(row[1])\n name, ending = os.path.splitext(filename)\n if ending == '.zip':\n unzip(os.path.join(fromfolder, row[1]), os.path.join(tofolder, name))\n elif ending == '.gz':\n gzip_e(os.path.join(fromfolder, row[1]), os.path.join(tofolder, name))\n else:\n shutil.copy(os.path.join(fromfolder, row[1]), os.path.join(tofolder, row[1]))\n\n\n"
]
| [
[
"pandas.read_csv"
]
]
|
tonybutzer/etm_v3_tony | [
"ec32d94a8730247af90072c49bdb9f81d036af9b"
]
| [
"Validate/aws_functions.py"
]
| [
"import os\n\nimport pandas as pd\nimport requests\nimport json\nimport sys\nimport boto3\nfrom botocore.exceptions import ClientError\n\n\n# aws stuff here\n\n##-----------------------------------------------------------------------------------------------------------\n# S3 AWS UTILITY FUNCIONS\n\n# def s3_bucket_analyze(bucket, prefix):\n# def s3_bucket_prefixes(bucket_p, performer, prefix_with_slash):\n\n##-----------------------------------------------------------------------------------------------------------\n\ndef _get_bucket(s3_full_path):\n bucket = s3_full_path.split('/')[2]\n pre_path = '/'.join(s3_full_path.split('/')[3:])\n return (bucket, pre_path)\n\ndef s3_upload_chs(local_file_name, full_s3_url, delete_local=True):\n\n _save_chs_global_keys\n REGION_NAME='us-west-2'\n\n print (local_file_name, full_s3_url, delete_local)\n\n (bucket, bucket_filepath) = _get_bucket(full_s3_url)\n \n s3 = boto3.client('s3', \n aws_access_key_id=chs_AccessKeyId, \n aws_secret_access_key=chs_SecretAccessKey, \n aws_session_token=chs_Token,\n region_name=REGION_NAME\n )\n \n with open(local_file_name, \"rb\") as f:\n s3.upload_fileobj(f, bucket, bucket_filepath)\n if delete_local:\n os.remove(local_file_name)\n\ndef _s3_list_pseudo_subdirs(bucket, prefix_with_slash):\n\n subfolder_list = []\n #Make sure you provide / in the end\n\n a = prefix_with_slash.split('/')\n prefix_with_slash='/'.join(a[1:])\n prefix = prefix_with_slash\n\n print(f'prefix_with_slash, {prefix_with_slash}')\n\n #client = boto3.client('s3')\n session = boto3.Session(profile_name='smart')\n client = session.client('s3')\n\n try:\n result = client.list_objects(Bucket=bucket, Prefix=prefix, Delimiter='/')\n try:\n for o in result.get('CommonPrefixes'):\n #print ('sub folder : ', o.get('Prefix'))\n subfolder_list.append(o.get('Prefix'))\n return subfolder_list\n except:\n return ['emptyDir', 'Maybe']\n except ClientError as e:\n if e.response['Error']['Code'] == 'NoSuchBucket':\n print('no such bucket', bucket)\n return ['NoSuchBucket', 'Really_No_such_bucket']\n else:\n print(\"Unexpected error: %s\" % e)\n return ['Unexpected', 'Really_unexpected']\n\ndef s3_bucket_prefixes(bucket_p, performer, prefix_with_slash):\n\n bucket = f'{bucket_p}{performer}'\n folders = _s3_list_pseudo_subdirs(bucket, prefix_with_slash)\n return folders\n\n# \n\n\ndef s3_bucket_analyze(bucket, prefix):\n\n objs = []\n\n print(\"bucket\", bucket)\n print(\"prefix\", prefix)\n\n bucket_name = bucket\n prefix = prefix\n\n s3 = boto3.resource('s3')\n bucket = s3.Bucket(bucket_name)\n cnt=0\n storage_class_h = {'STANDARD' : 0,\n 'GLACIER' : 0,\n 'INTELLIGENT_TIERING' : 0,\n }\n sum_class_h = {'STANDARD' : 0,\n 'GLACIER' : 0,\n 'INTELLIGENT_TIERING' : 0,\n }\n sum = 0\n for obj in bucket.objects.filter(Prefix=prefix):\n storage_class_h[obj.storage_class] = storage_class_h[obj.storage_class] + 1\n cnt = cnt + 1\n if not cnt%1000:\n print(bucket, \"bucket object count = \", cnt, flush=True)\n sum = sum + obj.size\n sum_class_h[obj.storage_class] = sum_class_h[obj.storage_class] + obj.size\n my_obj = {\n 'bucket_name':obj.bucket_name,\n 'key':obj.key,\n 'size':obj.size,\n 'class':obj.storage_class\n }\n objs.append(my_obj)\n\n print (\"COUNT=\", cnt)\n for ky in storage_class_h.keys():\n print(ky, storage_class_h[ky])\n sum = sum_class_h[ky]\n print(ky, sum_class_h[ky])\n gig = sum/(1024*1024*1024)\n print (ky, \"GBYTES=\", gig)\n if ky == 'GLACIER': \n cost=.007\n else: \n cost=.023\n print (ky, \"Cost/Month=\", gig * cost)\n print (\"----\" * 25)\n print(\"END LOOP\")\n my_key = 'STANDARD'\n ret_gbytes = sum_class_h[my_key]/(1024*1024*1024)\n if (ret_gbytes < 1):\n ret_gbytes = 1\n ret_costs = .023 * ret_gbytes\n print('G:', ret_gbytes, ret_costs)\n return objs\n \n \n\n##-------------------------------------------------------------------------------------------------------------------\n# Authentication Functions\n#\n# def auth_init(smart=True, chs=True):\n# def auth_user_scope_chs():\n# def auth_user_scope_smart():\n#\n##-------------------------------------------------------------------------------------------------------------------\n\ndef _return_chs_session_keys():\n url_loop_local = 'http://169.254.169.254/latest/meta-data/iam/security-credentials/lsds-developer-ec2'\n r = requests.get(url_loop_local)\n chs_auth = json.loads(r.text)\n return chs_auth\n\n\ndef _get_aws_credentials_carefully_from_file(cred_file_path):\n ''' return an env dict for docker use\n and for setting the env in notebooks\n not necessary pretty - but gets us authenticated to access the necessary buckets\n '''\n\n\n # read text file into pandas DataFrame\n df = pd.read_table(cred_file_path, delimiter=\"=\").T\n secret_be_careful = df['aws_secret_access_key '].values[0].replace(' ', '')\n secret_key = df['aws_access_key_id '].values[0].replace(' ', '')\n\n access_key = 'aws_access_key_id'.upper()\n access_secret = 'aws_secret_access_key'.upper()\n\n\n docker_env_dict = {\n access_key:secret_key,\n access_secret:secret_be_careful\n }\n return docker_env_dict\n\n\ndef _set_aws_environment(docker_env_dict):\n for i in docker_env_dict:\n #print(i, docker_env_dict[i])\n os.environ[i]=docker_env_dict[i]\n\n\ndef _smart_environment_set(): \n '''check if the aws environ var set - if NOT then set them!\n - always return True\n '''\n key=\"AWS_ACCESS_KEY_ID\"\n\n if key in os.environ:\n print('SMART Global Environment set correctly YES!', os.environ['AWS_ACCESS_KEY_ID'][0])\n return True\n else:\n print(\"Key does not exist - setting them from credentials\")\n aws_credentials_file='/home/ec2-user/.aws/credentials'\n aws_env_dict = _get_aws_credentials_carefully_from_file(aws_credentials_file)\n #print(aws_env_dict)\n _set_aws_environment(aws_env_dict)\n return True\n \n\ndef _save_smart_global_keys():\n global smart_AccessKeyId \n global smart_SecretAccessKey\n global smart_Token\n\n if _smart_environment_set():\n smart_AccessKeyId = os.environ['AWS_ACCESS_KEY_ID']\n smart_SecretAccessKey = os.environ['AWS_SECRET_ACCESS_KEY']\n smart_Token = ''\n else:\n print('something wrong in key chain no smart key pair')\n \n\ndef _save_chs_global_keys():\n c_auth = _return_chs_session_keys()\n global chs_AccessKeyId \n chs_AccessKeyId = c_auth['AccessKeyId']\n global chs_SecretAccessKey\n chs_SecretAccessKey = c_auth['SecretAccessKey']\n global chs_Token\n chs_Token = c_auth['Token']\n\n\ndef auth_init(smart=True, chs=True):\n if smart:\n _save_smart_global_keys()\n \n if chs:\n _save_chs_global_keys()\n \n\ndef auth_user_scope_chs():\n os.environ['AWS_ACCESS_KEY_ID'] = chs_AccessKeyId\n os.environ['AWS_SECRET_ACCESS_KEY'] = chs_SecretAccessKey\n os.environ['AWS_SESSION_TOKEN'] = chs_Token\n\n os.environ['AWS_DEFAULT_REGION'] = 'us-west-2'\n os.environ['AWS_REQUEST_PAYER'] = 'requester'\n print('CHS', os.environ['AWS_ACCESS_KEY_ID'])\n\n\n\ndef auth_user_scope_smart():\n os.environ['AWS_ACCESS_KEY_ID'] = smart_AccessKeyId\n os.environ['AWS_SECRET_ACCESS_KEY'] = smart_SecretAccessKey\n os.environ['AWS_SESSION_TOKEN'] = smart_Token\n\n os.environ['AWS_DEFAULT_REGION'] = 'us-west-2'\n os.environ['AWS_REQUEST_PAYER'] = 'requester'\n print('Smart', os.environ['AWS_ACCESS_KEY_ID'])\n\n\n\n"
]
| [
[
"pandas.read_table"
]
]
|
H-C-Orsted-Gym/SO2-opgave | [
"e594ada8f6b9490c0e72f52f08199b7ef8a6f956"
]
| [
"main.py"
]
| [
"# ALLE KRAV HER:\n\n# - pH skal ligge på 7 - 7,4\n# - Frit klor skal ligge på 0.5-3mg/l\n# - z \n\n# ALLE MÅDER MAN LAVER JUSTERINGER PÅ:\n\n\n# I vores \n\n# HOCL koncentration = frit Klor koncentration / volumen \n# 4.5 pH == HOCL\n\n# HOCl/OCl-\n# Disse chlorforbindelser benævnes ofte samlet\n# som frit chlor. \n\n# pis = ammonium\n# ammonium og frit klor = bundet klor\n# Stop børnene i at pisse i vandet\n# monoklorit = poolstank\n\nfrom numpy import random\nimport time\nimport datetime\nnuværendepH = 7\nnuværendeFritKlor = 1\nnuværendeBundetKlor = 0\n\ndef CheckWater(førpH, førFritKlor, førBundetKlor):\n\tglobal nuværendepH\n\tnuværendepH = round(random.uniform(førpH - 0.1, førpH + 0.1), 2)\n\tprint(\"Nuværende pH værdi er: \" + str(nuværendepH))\n\n\tglobal nuværendeFritKlor\n\tnuværendeFritKlor = round(random.uniform(førFritKlor - 0.1, førFritKlor + 0.1), 2)\n\tif nuværendeFritKlor <= 0:\n\t\tnuværendeFritKlor = 0\n\tprint(\"Nuværende frit klor mængde er: \" + str(nuværendeFritKlor) + \" mg/L\")\n\n\t\n\tglobal nuværendeBundetKlor\n\tnuværendeBundetKlor = round(random.uniform(førBundetKlor - 0.1, førBundetKlor + 0.1), 2)\n\tif nuværendeBundetKlor <= 0:\n\t\tnuværendeBundetKlor = 0\n\tprint(\"Nuværende bundet klor mængde er: \" + str(nuværendeBundetKlor) + \" mg/L\")\n\n\t\n\t\n\t\ndef CheckData(pH, fritKlor, bundetKlor):\n\tif pH >=7.4:\n\t\twhile pH >= 7.4:\n\t\t\tpH -= 0.2\n\t\tglobal nuværendepH\n\t\tnuværendepH = round(pH, 2)\n\t\tprint(\"Vi recirkulerer vandet, for at fjerne noget af kloret. \\npH'en er nu: \" + str(nuværendepH))\n\tif pH < 7:\n\t\twhile pH < 7:\n\t\t\tpH += 0.2\n\t\tnuværendepH = round(pH, 2)\n\t\tprint(\"Vi tilføjer klor for at hæve pH værdien. \\npH'en er nu: \" + str(nuværendepH))\n\t\t\n\tif fritKlor >=3:\n\t\twhile fritKlor >= 3:\n\t\t\tfritKlor -= 0.2\n\t\tglobal nuværendeFritKlor\n\t\tnuværendeFritKlor = fritKlor\n\t\tprint(\"\\nVi recirkulerer vandet, for at fjerne noget af kloret. \\nfritKlor koncentrationen er nu: \" + str(fritKlor) + \" mg/L\")\n\tif fritKlor < 0.5:\n\t\twhile fritKlor < 0.5:\n\t\t\tfritKlor += 0.2\n\t\tnuværendeFritKlor = fritKlor\n\t\tprint(\"\\nVi tilsætter lidt klor. \\nfritKlor koncentrationen er nu: \" + str(fritKlor) + \" mg/L\")\n\n\tif bundetKlor >=1:\n\t\twhile bundetKlor >= 1:\n\t\t\tbundetKlor -= 0.2\n\t\tglobal nuværendeBundetKlor\n\t\tnuværendeBundetKlor = bundetKlor\n\t\tprint(\"Vi filtrere vand ud.\\n bundetKlor koncentrationen er nu er nu: \" + str(bundetKlor) + \" mg/L\")\n\ndef getTime():\n\tx = datetime.datetime.now()\n\trealTime = x.strftime(\"%d/%m/%Y - %H:%M:%S\")\n\n\treturn realTime\n\nwhile True: \n\ttime.sleep(5)\n\tprint(\"-----------------\" + getTime() + \"-----------------\")\n\tCheckWater(nuværendepH,nuværendeFritKlor,nuværendeBundetKlor)\n\tCheckData(nuværendepH,nuværendeFritKlor,nuværendeBundetKlor)\n\tprint(\"-------------------------------------------------------\")\n\tprint(\"\\n\\n\")\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\n\t "
]
| [
[
"numpy.random.uniform"
]
]
|
supriyascode/MODIS-Aggregation | [
"cc6ce992b81c358e6b7cb2635a5b8ff72d6e99f9"
]
| [
"benchmarking/spark/monthly-aggregation-pixel-level-spark-sampling.py"
]
| [
"from netCDF4 import Dataset\nimport numpy as np\nimport glob\nimport matplotlib.pyplot as plt\nimport time\nimport h5py\nimport sys\nimport xarray as xr\nfrom pyspark.sql import SparkSession\n\ndef aggregateOneFileData(M06_file, M03_file, sampling_rate):\n \"\"\"Aggregate one file from MYD06_L2 and its corresponding file from MYD03. Read 'Cloud_Mask_1km' variable from the MYD06_L2 file, read 'Latitude' and 'Longitude' variables from the MYD03 file. Group Cloud_Mask_1km values based on their (lat, lon) grid.\n Args:\n M06_file (string): File path for M06_file.\n M03_file (string): File path for corresponding M03_file.\n sampling_rate (int): sampling rate where 1 means no sampling. \n\n Returns:\n (cloud_pix, total_pix) (tuple): cloud_pix is an 2D(180*360) numpy array for cloud pixel count of each grid, total_pix is an 2D(180*360) numpy array for total pixel count of each grid.\n \"\"\"\n #print(M06_file)\n #print(M03_file)\n \n #output as a dictionary\n output_dictionaray = {} \n output_list = []\n total_pix = np.zeros((180, 360))\n cloud_pix = np.zeros((180, 360))\n #read 'Cloud_Mask_1km' variable from the MYD06_L2 file, whose shape is (2030, 1354)\n ncfile = Dataset(M06_file,'r')\n d06 = ncfile.variables['Cloud_Mask_1km'][:,:,0]\n ncfile.close()\n # sampling data with 1/3 ratio (pick 1st, 4th, 7th, ...) in both latitude and longitude direction. d06CM's shape is (677, 452)\n d06CM = d06[::sampling_rate,::sampling_rate]\n ds06_decoded = (np.array(d06CM, dtype = \"byte\") & 0b00000110) >> 1\n ds06_1d = ds06_decoded.ravel()\n \n \n #[print(\"one element:\" + str(a)) for a in ds06_1d]\n # shape of d03_lat and d03_lon: (2030, 1354)\n ncfile = Dataset(M03_file,'r')\n d03_lat = ncfile.variables['Latitude'][:,:]\n d03_lon = ncfile.variables['Longitude'][:,:]\n ncfile.close()\n \n # sampling data with 1/3 ratio, shape of lat and lon: (677, 452), then convert data from 2D to 1D, then add offset to change value range from (-90, 90) to (0, 180) for lat.\n lat = (d03_lat[::sampling_rate,::sampling_rate].ravel() + 89.5).astype(int)\n lon = (d03_lon[::sampling_rate,::sampling_rate].ravel() + 179.5).astype(int)\n lat = np.where(lat > -1, lat, 0)\n lon = np.where(lon > -1, lon, 0)\n \n #create one element <(lat, lon), (cloud_pixel_number, total_pixel_number)> for each pixel and add it to output list\n for i in range(0, ds06_1d.size):\n #print(\"one element:\" + str(lat[i]) + \",\" + str(lon[i]) + \", \" + str(ds06_1d[i]))\n output_list.append(((lat[i], lon[i]), (1 if ds06_1d[i] == 0 else 0, 1)))\n \n #print(\"output for \" + str(M06_file) + \":\" + str(output_list)) \n return output_list\n\ndef save_output(cf, node_num, sampling_rate):\n cf1 = xr.DataArray(cf)\n output_file_name = \"monthlyCloudFraction-pixel-level-\" + node_num + \"-nodes-\" + sampling_rate + \"-sampling\"\n cf1.to_netcdf(output_file_name + \".nc\")\n plt.figure(figsize=(14, 7))\n plt.contourf(range(-180, 180), range(-90, 90), cf, 100, cmap=\"jet\")\n plt.xlabel(\"Longitude\", fontsize=14)\n plt.ylabel(\"Latitude\", fontsize=14)\n plt.title(\"Level 3 Cloud Fraction Aggregation for January 2008\", fontsize=16)\n plt.colorbar()\n plt.savefig(output_file_name + \".png\")\n\n\nif __name__ =='__main__':\n\n #node_num = int(sys.argv[1])\n if len(sys.argv) == 3:\n sampling_rate = int(sys.argv[2])\n else:\n sampling_rate = 3 # Default sampling rate is 3\n print (\"running on \" + sys.argv[1] + \"nodes with \" + str(sampling_rate) + \" sampling.\")\n\n M06_dir = \"/umbc/xfs1/cybertrn/common/Data/Satellite_Observations/MODIS/MYD06_L2/\"\n M03_dir = \"/umbc/xfs1/cybertrn/common/Data/Satellite_Observations/MODIS/MYD03/\"\n M06_files = sorted(glob.glob(M06_dir + \"MYD06_L2.A2008*\"))\n file_num = len(M06_files)\n M03_files = sorted(glob.glob(M03_dir + \"MYD03.A2008*\"))\n file_pairs = zip(M06_files, M03_files)\n #print(list(file_pairs))\n #print(len(list(file_pairs))) \n\n t0 = time.time()\n \n # Initiate and process the parallel by Spark\n spark = SparkSession\\\n .builder\\\n .appName(\"MODIS_agg\")\\\n .getOrCreate()\n sc = spark.sparkContext\n result = sc.parallelize(list(file_pairs), file_num)\\\n .flatMap(lambda x: aggregateOneFileData(x[0], x[1], sampling_rate))\\\n .reduceByKey(lambda x, y: (x[0] + y[0], x[1] + y[1]))\\\n .collect()\n spark.stop() # Stop Spark\n \n #print(result)\n \n global_cloud_pix = np.zeros((180, 360))\n global_total_pix = np.zeros((180, 360))\n for element in result:\n #print(\"element:\" + str(element))\n global_cloud_pix[element[0][0], element[0][1]] = element[1][0]\n global_total_pix[element[0][0], element[0][1]] = element[1][1]\n \n total_cloud_fraction = (global_cloud_pix/global_total_pix)\n print(\"total_cloud_fraction:\" + str(total_cloud_fraction))\n print(\"total_cloud_fraction.shape:\" + str(total_cloud_fraction.shape))\n \n #total_cloud_fraction = (global_cloud_pix/global_total_pix).reshape([lat_bnd,lon_bnd])\n save_output(total_cloud_fraction, sys.argv[1], sys.argv[2])\n\n #calculate execution time\n t1 = time.time()\n total = t1-t0\n print(\"total execution time (Seconds):\" + str(total))\n"
]
| [
[
"matplotlib.pyplot.title",
"matplotlib.pyplot.figure",
"matplotlib.pyplot.savefig",
"matplotlib.pyplot.colorbar",
"matplotlib.pyplot.xlabel",
"numpy.array",
"numpy.zeros",
"numpy.where",
"matplotlib.pyplot.ylabel"
]
]
|
Sapphirine/oscar-winner-prediction | [
"7b323bac082937911ff09f8d87a0ecfd66642a02"
]
| [
"src/predict.py"
]
| [
"import cPickle\nimport json\nimport logging\nimport os\n\nimport numpy as np\nimport pandas as pd\nfrom sklearn.metrics import classification_report\nfrom sklearn.preprocessing import scale\n\nfrom models import Adaboost, LogisticReg, SVM\n\nlogging.basicConfig(level=logging.INFO)\nlogger = logging.getLogger(__name__ if __name__ != '__main__' else [])\n\nCOLUMNS = ('winner', 'Metascore', 'imdbRating', 'imdbVotes',\n 'pv-release-1m', 'pv-oscar-1m', 'review_sentiment')\nDATA_CACHE = '../cache/dataset.p'\nOUTPUT_FOLDER = '../out/imdb/unweighted/'\n\n\nCONFIG = [{'name': 'logit',\n 'clf': LogisticReg(),\n 'params': {'C': [0.001, 0.01, 1, 100, 1000, 10000],\n 'class_weight': ['balanced'],\n 'n_jobs': 1,\n 'verbose': True}},\n {'name': 'svm-rbf',\n 'clf': SVM(),\n 'params': {'C': [0.001, 0.01, 1, 100, 1000, 10000],\n 'kernel': 'rbf',\n 'gamma': 'auto',\n 'class_weight': 'balanced',\n 'degree': [2, 3],\n 'verbose': True,\n 'max_iter': -1}},\n {'name': 'svm-linear',\n 'clf': SVM(),\n 'params': {'C': [0.001, 0.01, 1, 100, 1000, 10000],\n 'kernel': 'linear',\n 'class_weight': 'balanced',\n 'verbose': True,\n 'max_iter': -1}},\n {'name': 'svm-poly',\n 'clf': SVM(),\n 'params': {'C': [0.001, 0.01, 1, 100, 1000, 10000],\n 'kernel': 'poly',\n 'gamma': 'auto',\n 'class_weight': 'balanced',\n 'degree': [2, 3, 4],\n 'verbose': True,\n 'max_iter': -1}},\n {'name': 'adaboost',\n 'clf': Adaboost(),\n 'params': {'max_depth': [1, 3],\n 'n_estimators': [1, 2, 4, 8, 16, 32],\n 'learning_rate': 1.0}}]\n\n\ndef run_predictions(clf, X, y, show_plot=False, **params):\n best_params = clf.tune(X, y, **params)\n names = ['lose', 'win']\n report = classification_report(y, clf.predict(X), target_names=names)\n logger.info('Training set classification report\\n\\n%s\\n' % report)\n roc_result = clf.mean_roc(X, y, show_plot, **best_params)\n return roc_result\n\n\ndef save_roc(roc_result, path):\n folder = os.path.split(path)[0]\n if not os.path.exists(folder):\n os.makedirs(folder)\n with open(path, 'wb') as fo:\n output = roc_result._asdict()\n for k, v in output.items():\n if isinstance(v, np.ndarray):\n output[k] = v.tolist()\n json.dump(output, fo)\n fo.write('\\n')\n logger.info('Saved: %s' % path)\n\n\ndef driver(name, clf, params, X, y):\n roc_result = run_predictions(clf, X, y, **params)\n logger.info('%s roc auc: %f'\n % (name, roc_result.mean_auc))\n path = OUTPUT_FOLDER + name + '.json'\n save_roc(roc_result, path)\n\n\ndef main():\n if not os.path.exists(DATA_CACHE):\n logger.error('No data cached: %s' % DATA_CACHE)\n\n with open(DATA_CACHE, 'rb') as fi:\n data = cPickle.load(fi)\n assert isinstance(data, pd.DataFrame)\n\n data = data.ix[:, COLUMNS]\n y = data.ix[:, 'winner']\n X = data.drop('winner', 1)\n X = scale(X)\n X = pd.DataFrame(X)\n\n for setting in CONFIG:\n name = setting['name']\n clf = setting['clf']\n params = setting['params']\n driver(name, clf, params, X, y)\n\n\nif __name__ == '__main__':\n main()\n"
]
| [
[
"sklearn.preprocessing.scale",
"pandas.DataFrame"
]
]
|
IrohXu/MAE-ViT-pytorch | [
"725ded18ff688af4d933c2b8b501864d0ef1bfc2"
]
| [
"dataloader.py"
]
| [
"import os\nimport torch\nfrom torch.utils.data import Dataset, DataLoader\nfrom PIL import Image\nimport cv2\nimport numpy as np\n\nfrom torchvision.transforms import ToTensor\nfrom torchvision import datasets, transforms\n\nimport random\n\nclass RandomMaskingGenerator:\n def __init__(self, input_size, mask_ratio):\n if not isinstance(input_size, tuple):\n input_size = (input_size,) * 2\n\n self.height, self.width = input_size\n\n self.num_patches = self.height * self.width\n self.num_mask = int(mask_ratio * self.num_patches)\n\n def __repr__(self):\n repr_str = \"Maks: total patches {}, mask patches {}\".format(\n self.num_patches, self.num_mask\n )\n return repr_str\n\n def __call__(self):\n mask = np.hstack([\n np.zeros(self.num_patches - self.num_mask),\n np.ones(self.num_mask),\n ])\n np.random.shuffle(mask)\n return mask\n\n\nclass TusimpleMAE(Dataset):\n def __init__(self, dataset, transform=None):\n self._gt_img_list = []\n\n self.transform = transform\n \n self.generate_mask = RandomMaskingGenerator(24, 0.75)\n\n with open(dataset, 'r') as file:\n for _info in file:\n _info = _info.strip('\\n')\n _info = _info.split(' ')[0]\n info_tmp = os.path.join(dataset, _info)\n self._gt_img_list.append(info_tmp)\n\n # self._shuffle()\n\n def _shuffle(self):\n # randomly shuffle all list identically\n c = list(zip(self._gt_img_list))\n random.shuffle(c)\n self._gt_img_list = zip(*c)\n\n def __len__(self):\n return len(self._gt_img_list)\n\n def __getitem__(self, idx):\n # load all\n\n img = Image.open(self._gt_img_list[idx])\n # optional transformations\n if self.transform:\n img = self.transform(img)\n\n mask = self.generate_mask()\n\n # we could split the instance label here, each instance in one channel (basically a binary mask for each)\n return img, mask"
]
| [
[
"numpy.zeros",
"numpy.random.shuffle",
"numpy.ones"
]
]
|
HawkTom/EAPY | [
"c9dabbfb233f4a135bf9560bebcf6c01ec9baf94"
]
| [
"pso/wPSO.py"
]
| [
"# !/usr/bin/env python\n\"\"\"\nwPSO.py\n\nDescription: the implemention of modified particle swarm optimization\n\nRefrence paper:\nShi, Yuhui, and Russell Eberhart. \"A modified particle swarm optimizer.\" \nEvolutionary Computation Proceedings, 1998. IEEE World Congress on Computational Intelligence.\n\nMember variables:\n\n Name: wPSO\n FERuntime: time for fitness evaluation\n FENum: number of fitness evaluation\n runtime: time for whole algorithm\n optimalX: optimal solution for problem\n optimalY: optimal value for problem\n convergeCurve: the procedure of convergence\n convergeCurveInterval: inverval between two saved points\n w(weight): default w_max = 0.9, w_min = 0.4 \n learningRate: default = 2.0\n\nMember function:\n \n setParameters(weight, learningRate): setting parameters\n\n optimize(cfp, ap, printLog): the main process of optimization\n\n cfp: config for continue function parameters\n ap: config for algorithm parameters\n printLog: determine whether to print Log after opitmization\n (true default)\n\nExample: \n\n agent = wPSO()\n agent.optimize(cfp, ap, printLog=True) # cfp ap need to config at first\n\n\"\"\"\n\nfrom function import continueFunction as cF\nimport numpy as np\nimport time\nimport sys\nimport copy\n\n__all__ = ['wPSO']\n\n\nclass wPSO:\n\n def __init__(self):\n self.Name = \"wPSO\"\n self.FERuntime = 0\n self.FENum = 0\n self.setParameters()\n\n # setting weight, learning rate of PSO\n def setParameters(self, weight=[0.9, 0.4], learningRate=2.0):\n self.w = weight\n self.learningRate = learningRate\n\n def optimize(self, cfp, ap, printLog=True):\n runtimeStart = time.clock()\n self.__mainLoop(cfp, ap, printLog)\n self.runtime = time.clock() - runtimeStart\n\n def __mainLoop(self, cfp, ap, printLog):\n np.random.seed(ap.initialSeed)\n popSize = ap.populationSize\n Dim = cfp.funcDim\n function = getattr(cF, cfp.funcName)\n lowerBoundX = np.kron(np.ones((popSize, 1)), cfp.funcLowerBound)\n upperBoundX = np.kron(np.ones((popSize, 1)), cfp.funcUpperBound)\n\n lowerInitBoundX = np.kron(\n np.ones((popSize, 1)), cfp.funcInitLowerBound)\n upperInitBoundX = np.kron(\n np.ones((popSize, 1)), cfp.funcInitUpperBound)\n\n upperBoundV = 0.2 * (upperBoundX - lowerBoundX)\n lowerBoundV = -1 * upperBoundV\n # initial X position and velocity\n X = (upperInitBoundX - lowerInitBoundX) * \\\n np.random.random_sample((popSize, Dim)) + lowerInitBoundX\n V = (upperBoundV - lowerBoundV) * \\\n np.random.random_sample((popSize, Dim)) + lowerBoundV\n start = time.clock()\n y = function(X)\n self.FERuntime += (time.clock()-start)\n self.FENum += popSize\n # initialize personal best X and y\n personBestX, personBestY = copy.deepcopy(X), copy.deepcopy(y)\n # initialize global best X and y\n gBestX, gBestY = X[np.argmin(y), :], np.min(y)\n self.convergeCurve = [y[0], gBestY]\n maxGen, gen = ap.iterationMax, 0\n while self.FENum < ap.FEMax:\n wk = self.w[0] - (self.w[0] - self.w[1]) * gen / maxGen\n for pi in range(popSize):\n # update and limit V\n V[pi, :] = wk * V[pi, :] + self.learningRate * np.random.random_sample((1, Dim)) * (\n personBestX[pi, :] - X[pi, :]) + self.learningRate * np.random.random_sample((1, Dim)) * (gBestX - X[pi, :])\n V[pi, :][V[pi, :] < lowerBoundV[pi, :]\n ] = lowerBoundV[pi, :][V[pi, :] < lowerBoundV[pi, :]]\n V[pi, :][V[pi, :] > upperBoundV[pi, :]\n ] = upperBoundV[pi, :][V[pi, :] > upperBoundV[pi, :]]\n # update X\n X[pi, :] = X[pi, :] + V[pi, :]\n X[pi, :][X[pi, :] < lowerBoundX[pi, :]\n ] = lowerBoundX[pi, :][X[pi, :] < lowerBoundX[pi, :]]\n X[pi, :][X[pi, :] > upperBoundX[pi, :]\n ] = upperBoundX[pi, :][X[pi, :] > upperBoundX[pi, :]]\n # update personal and global best X and y\n start = time.clock()\n y[pi] = function(X[pi, :][np.newaxis, :])\n self.FERuntime += (time.clock() - start)\n self.FENum += 1\n if y[pi] < personBestY[pi]:\n personBestX[pi, :] = X[pi, :]\n personBestY[pi] = y[pi]\n if personBestY[pi] < gBestY:\n gBestX = personBestX[pi, :]\n gBestY = personBestY[pi]\n if self.FENum % popSize == 0:\n self.convergeCurve.append(gBestY)\n gen = gen + 1\n # print('Gen:{0} BestV: {1} \\n'.format(self.FENum, gBestY))\n self.optimalX = gBestX\n self.optimalY = gBestY\n self.convergeCurveIntrval = popSize\n if printLog:\n # summary\n print('$--------Result--------$\\n')\n print('*Function: {0}\\tDimension: {1}\\t FEMax: {2}\\n'.format(\n cfp.funcName, cfp.funcDim, self.FENum))\n print('Optimal Y : {0} \\n'.format(self.optimalY))\n"
]
| [
[
"numpy.random.seed",
"numpy.min",
"numpy.random.random_sample",
"numpy.ones",
"numpy.argmin"
]
]
|
kevintli/mural | [
"e18e7d1a72b561fab1b5da026806e3417a9c63db"
]
| [
"examples/sawyer_pick/variants.py"
]
| [
"from copy import deepcopy\nfrom ray import tune\nimport numpy as np\nimport tensorflow as tf\n\nfrom softlearning.misc.utils import get_git_rev, deep_update\nfrom softlearning.misc.generate_goal_examples import (\n DOOR_TASKS, PUSH_TASKS, PICK_TASKS)\n# from softlearning.misc.get_multigoal_example_pools import (\n# get_example_pools_from_variant)\n# import dsuite\nimport os\n\nDEFAULT_KEY = '__DEFAULT_KEY__'\n\nM = 256\nN = 2\n\nREPARAMETERIZE = True\n\nNUM_COUPLING_LAYERS = 2\n\n\"\"\"\nPolicy params\n\"\"\"\n\nGAUSSIAN_POLICY_PARAMS_BASE = {\n 'type': 'GaussianPolicy',\n 'kwargs': {\n 'hidden_layer_sizes': (M, ) * N,\n 'squash': True,\n 'observation_keys': None,\n 'observation_preprocessors_params': {}\n }\n}\n\nGAUSSIAN_POLICY_PARAMS_FOR_DOMAIN = {\n 'Ant': {\n 'kwargs': {\n 'observation_keys': ('state_observation', 'xy_observation')\n }\n }\n}\n\nPOLICY_PARAMS_BASE = {\n 'GaussianPolicy': GAUSSIAN_POLICY_PARAMS_BASE,\n}\n\nPOLICY_PARAMS_BASE.update({\n 'gaussian': POLICY_PARAMS_BASE['GaussianPolicy'],\n})\n\nPOLICY_PARAMS_FOR_DOMAIN = {\n 'GaussianPolicy': GAUSSIAN_POLICY_PARAMS_FOR_DOMAIN,\n}\n\nPOLICY_PARAMS_FOR_DOMAIN.update({\n 'gaussian': POLICY_PARAMS_FOR_DOMAIN['GaussianPolicy'],\n})\n\nMAX_PATH_LENGTH_PER_DOMAIN = {\n DEFAULT_KEY: 100,\n 'Point2D': 100,\n 'DClaw': 100,\n 'Ant': 400,\n}\n\n\"\"\"\nAlgorithm params\n\"\"\"\n\nALGORITHM_PARAMS_BASE = {\n 'type': 'SAC',\n\n 'kwargs': {\n 'epoch_length': 1000,\n 'train_every_n_steps': 1,\n 'n_train_repeat': 1,\n 'eval_render_kwargs': {},\n 'eval_n_episodes': 3,\n 'eval_deterministic': True,\n 'save_training_video_frequency': 5,\n 'discount': 0.99,\n 'tau': 5e-3,\n 'reward_scale': 1.0,\n 'ext_reward_coeff': 1,\n # 'normalize_ext_reward_gamma': 0.99,\n 'rnd_int_rew_coeff': 0,\n },\n 'rnd_params': {\n 'convnet_params': {\n 'conv_filters': (16, 32, 64),\n 'conv_kernel_sizes': (3, 3, 3),\n 'conv_strides': (2, 2, 2),\n 'normalization_type': None,\n },\n 'fc_params': {\n 'hidden_layer_sizes': (256, 256),\n 'output_size': 512,\n },\n }\n}\n\nALGORITHM_PARAMS_ADDITIONAL = {\n 'SAC': {\n 'type': 'SAC',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'n_initial_exploration_steps': int(1e3),\n 'n_epochs': 200,\n\n # 'rnd_int_rew_coeff': tune.sample_from([1, 5, 10]),\n # 'normalize_ext_reward_gamma': tune.grid_search([0.99]),\n },\n # 'rnd_params': {\n # 'convnet_params': {\n # 'conv_filters': (16, 32, 64),\n # 'conv_kernel_sizes': (3, 3, 3),\n # 'conv_strides': (2, 2, 2),\n # 'normalization_type': None,\n # },\n # 'fc_params': {\n # 'hidden_layer_sizes': (256, 256),\n # 'output_size': 512,\n # },\n # }\n },\n 'SACClassifier': {\n 'type': 'SACClassifier',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'classifier_lr': 1e-4,\n 'classifier_batch_size': 128,\n 'n_initial_exploration_steps': int(1e3),\n 'n_classifier_train_steps': 10000,\n 'classifier_optim_name': 'adam',\n 'reward_type': 'logits',\n 'n_epochs': 200,\n 'mixup_alpha': 1.0,\n }\n },\n 'SQIL': {\n 'type': 'SQIL',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'n_initial_exploration_steps': int(1e3),\n 'n_epochs': 200,\n 'goal_negative_ratio': 1.0,\n 'lambda_samp': 1.0,\n }\n },\n 'RAQ': {\n 'type': 'RAQ',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'classifier_lr': 1e-4,\n 'classifier_batch_size': 128,\n 'n_initial_exploration_steps': int(1e3),\n 'n_classifier_train_steps': 10,\n 'classifier_optim_name': 'adam',\n 'reward_type': 'logits',\n 'active_query_frequency': 1,\n 'n_epochs': 200,\n 'mixup_alpha': 1.0,\n }\n },\n 'VICE': {\n 'type': 'VICE',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'classifier_lr': 1e-4,\n 'classifier_batch_size': 128,\n 'n_initial_exploration_steps': int(1e3),\n # 'n_initial_classifier_train_steps': 200,\n 'n_classifier_train_steps': tune.grid_search([2]),\n 'classifier_optim_name': 'adam',\n 'n_epochs': 200,\n 'mixup_alpha': tune.grid_search([0]),\n 'save_training_video_frequency': 0,\n\n #############################################\n # Meta-NML\n #############################################\n 'discount': 0.99,\n 'use_laplace_smoothing_rewards': False,\n\n # Standard meta-NML hyperparameters\n 'use_meta_nml': True,\n 'meta_nml_reward_type': tune.grid_search(['probs']),\n 'meta_nml_train_on_positives': tune.grid_search([False]),\n 'meta_nml_uniform_train_data': False,\n 'meta_nml_layers': tune.grid_search([(512, 512)]),\n 'meta_nml_num_finetuning_layers': tune.grid_search([None]),\n 'dist_weight_thresh': tune.grid_search([0.6]),\n 'query_point_weight': tune.grid_search([1]), \n 'nml_grad_steps': tune.grid_search([1]),\n 'meta_train_sample_size': 128, \n 'meta_test_sample_size': 2048,\n 'meta_task_batch_size': 1, \n 'accumulation_steps': 16,\n 'meta_test_batch_size': 2048,\n 'equal_pos_neg_test': True, \n 'test_strategy': 'sample',\n 'points_per_meta_task': 64,\n\n # Use a custom key from the observation dict as embeddings for meta-NML distance weighting.\n # 'meta_nml_custom_embedding_key': 'state_observation',\n\n #############################################\n\n # Tune over the reward scaling between count based bonus and VICE reward\n # 'ext_reward_coeff': tune.grid_search([0.25]), # Needed for VICE + count-based\n # 'normalize_ext_reward_gamma': tune.grid_search([1]),\n 'use_env_intrinsic_reward': tune.grid_search([False]),\n # 'rnd_int_rew_coeff': tune.sample_from([1]),\n\n #'gradient_penalty_weight': tune.grid_search([0, 0.5, 10]),\n\n #'positive_on_first_occurence': tune.grid_search([True]),\n 'positive_on_first_occurence': tune.grid_search([False]),\n },\n # === Using RND ===\n # 'rnd_params': {\n # 'convnet_params': {\n # 'conv_filters': (16, 32, 64),\n # 'conv_kernel_sizes': (3, 3, 3),\n # 'conv_strides': (2, 2, 2),\n # 'normalization_type': None,\n # },\n # 'fc_params': {\n # 'hidden_layer_sizes': (256, 256),\n # 'output_size': 512,\n # },\n # }\n },\n 'DynamicsAwareEmbeddingVICE': {\n 'type': 'DynamicsAwareEmbeddingVICE',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'classifier_lr': 1e-4,\n 'classifier_batch_size': 128,\n 'n_initial_exploration_steps': int(1e3),\n 'n_classifier_train_steps': tune.grid_search([2]),\n 'classifier_optim_name': 'adam',\n 'n_epochs': 200,\n 'mixup_alpha': tune.grid_search([1.]),\n 'save_training_video_frequency': 0,\n\n # === EMBEDDING TRAINING PARAMS ===\n 'use_ground_truth_distances': tune.grid_search([True]),\n 'train_distance_fn_every_n_steps': tune.grid_search([16]),\n 'ddl_batch_size': 256,\n 'ddl_clip_length': tune.grid_search([None]),\n\n 'normalize_distance_targets': tune.grid_search([False]),\n # 'use_l2_distance_targets': tune.grid_search([True, False]),\n\n # Tune over the reward scaling between count based bonus and VICE reward\n 'ext_reward_coeff': tune.grid_search([0.25]),\n 'normalize_ext_reward_gamma': tune.grid_search([1]),\n 'use_env_intrinsic_reward': tune.grid_search([False]),\n # 'rnd_int_rew_coeff': tune.sample_from([1]),\n\n 'positive_on_first_occurence': tune.grid_search([False]),\n },\n # === Using RND ===\n # 'rnd_params': {\n # 'convnet_params': {\n # 'conv_filters': (16, 32, 64),\n # 'conv_kernel_sizes': (3, 3, 3),\n # 'conv_strides': (2, 2, 2),\n # 'normalization_type': None,\n # },\n # 'fc_params': {\n # 'hidden_layer_sizes': (256, 256),\n # 'output_size': 512,\n # },\n # }\n },\n 'VICEDynamicsAware': {\n 'type': 'VICEDynamicsAware',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'classifier_lr': 1e-4,\n 'classifier_batch_size': 128,\n 'n_initial_exploration_steps': int(1e3),\n 'n_classifier_train_steps': 2,\n 'classifier_optim_name': 'adam',\n 'mixup_alpha': tune.grid_search([1.]),\n\n 'train_dynamics_model_every_n_steps': tune.grid_search([16, 64]),\n 'dynamics_model_lr': 3e-4,\n 'dynamics_model_batch_size': 256,\n\n # 'normalize_ext_reward_gamma': tune.grid_search([0.99, 1]),\n # 'use_env_intrinsic_reward': tune.grid_search([True, False]),\n }\n },\n 'MultiVICEGAN': {\n 'type': 'MultiVICEGAN',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'her_iters': tune.grid_search([0]),\n # === BELOW FOR RND RESET CONTROLLER ===\n 'rnd_int_rew_coeffs': tune.sample_from([[1, 1]]),\n 'ext_reward_coeffs': [1, 0], # 0 corresponds to reset policy\n # === BELOW FOR 2 GOALS ===\n # 'rnd_int_rew_coeffs': tune.sample_from([[0, 0]]),\n # 'ext_reward_coeffs': [1, 1],\n 'n_initial_exploration_steps': int(1e4),\n 'normalize_ext_reward_gamma': 0.99,\n 'share_pool': False,\n 'n_classifier_train_steps': 5,\n 'classifier_optim_name': 'adam',\n 'n_epochs': 200,\n 'mixup_alpha': 1.0,\n 'eval_n_episodes': 15, # 15 for free screw\n # 'eval_n_episodes': 8, # 8 for beads, fixed screw\n },\n 'rnd_params': {\n 'convnet_params': {\n 'conv_filters': (16, 32, 64),\n 'conv_kernel_sizes': (3, 3, 3),\n 'conv_strides': (2, 2, 2),\n 'normalization_type': None,\n },\n 'fc_params': {\n 'hidden_layer_sizes': (256, 256),\n 'output_size': 512,\n },\n },\n },\n 'VICEGAN': {\n 'type': 'VICEGAN',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'rnd_int_rew_coeff': tune.sample_from([5]),\n # Only train with RND reset controller\n 'ext_reward_coeff': 0, # 0 corresponds to reset policy\n # 'normalize_ext_reward_gamma': 0.99,\n 'n_initial_exploration_steps': int(1e4),\n 'n_classifier_train_steps': 0,\n 'classifier_lr': 1e-4,\n 'classifier_batch_size': 128,\n 'classifier_optim_name': 'adam',\n 'n_epochs': 1500,\n 'mixup_alpha': 1.0,\n 'normalize_ext_reward_gamma': 0.99,\n 'rnd_int_rew_coeff': 0,\n 'eval_n_episodes': 8,\n },\n },\n 'VICERAQ': {\n 'type': 'VICERAQ',\n 'kwargs': {\n 'reparameterize': REPARAMETERIZE,\n 'lr': 3e-4,\n 'target_update_interval': 1,\n 'tau': 5e-3,\n 'target_entropy': 'auto',\n 'action_prior': 'uniform',\n 'classifier_lr': 1e-4,\n 'classifier_batch_size': 256,\n 'n_initial_exploration_steps': int(1e3),\n 'n_classifier_train_steps': 10,\n 'classifier_optim_name': 'adam',\n 'active_query_frequency': 10,\n 'n_epochs': 500,\n 'eval_n_episodes': 3,\n 'mixup_alpha': tune.grid_search([1.0]),\n }\n },\n}\n\nDEFAULT_NUM_EPOCHS = 200\nNUM_CHECKPOINTS = 10\n\nCLASSIFIER_PARAMS_BASE = {\n 'type': 'feedforward_classifier',\n 'kwargs': {\n 'hidden_layer_sizes': (M,) * N,\n 'observation_keys': None,\n 'kernel_regularizer_lambda': tune.grid_search([5e-3]),\n },\n}\n\nCLASSIFIER_SAMPLER_PARAMS_PER_UNIVERSE_DOMAIN_TASK_VISION = {\n 'gym': {\n 'Point2D': {\n **{\n env: {'observation_keys': ('pixels', 'state_observation'), }\n for env in ('Maze-v0', )\n },\n },\n 'StateSawyer': {\n **{\n env: {'observation_keys': ('pixels', 'state'), }\n for env in ('PickAndPlace3DEnv-v0', )\n },\n }\n }\n}\n\nCLASSIFIER_SAMPLER_PARAMS_PER_UNIVERSE_DOMAIN_TASK = {}\n\n# Can optionally specify different classifier params when `from_vision = True`.\n# Otherwise, will default to whatever is in CLASSIFIER_PARAMS_PER_UNIVERSE_DOMAIN_TASK\nCLASSIFIER_PARAMS_PER_UNIVERSE_DOMAIN_TASK_VISION = {\n 'gym': {\n 'Point2D': {\n **{\n env: {'observation_keys': ('pixels', ), }\n for env in ('Maze-v0', )\n },\n },\n }\n}\n\nCLASSIFIER_PARAMS_PER_UNIVERSE_DOMAIN_TASK = {\n 'gym': {\n 'Ant': {\n **{\n env: {\n 'observation_keys': ('state_observation', ),\n }\n for env in ('MazeOneWallEnv-v0', 'MazeSEnv-v0', 'v0')\n },\n },\n 'Point2D': {\n **{\n env: {'observation_keys': ('state_observation', ), }\n for env in ('Maze-v0', )\n },\n # === Initialize an online embedding preprocessor ===\n **{\n env: {\n 'observation_keys': ('state_observation', ),\n 'observation_preprocessors_params': {\n 'state_observation': {\n 'type': 'EmbeddingPreprocessor',\n # (TODO) Figure out the best way to provide params\n 'kwargs': {\n 'hidden_layer_sizes': (M, ) * N,\n 'observation_keys': None,\n\n # # Output dimension if using an embedding\n 'embedding_dim': 16,\n\n # Use weight decay for distance fn\n 'kernel_regularizer': tune.grid_search([None]),\n }\n },\n }\n }\n for env in (\n # 'Maze-v0', \n 'Fixed-v0'\n )\n },\n },\n 'Pusher2D': {\n **{\n key: {\n 'observation_keys': tune.grid_search([\n ('object_pos', ),\n ('gripper_qpos', 'object_pos'),\n ]),\n }\n for key in (\n 'Simple-v0',\n )\n },\n },\n 'DClaw': {\n **{\n key: {'observation_keys': ('object_xy_position', )}\n for key in (\n 'TranslatePuckFixed-v0',\n )\n },\n # **{\n # key: {'observation_keys': ('object_position', 'object_quaternion')}\n # for key in (\n # 'LiftDDFixed-v0',\n # )\n # },\n **{\n key: {'observation_keys': ('pixels', )}\n for key in (\n 'TurnResetFree-v0',\n # 'TurnFreeValve3ResetFree-v0',\n 'SlideBeadsResetFree-v0',\n 'TurnFreeValve3Hardware-v0',\n )\n },\n **{\n key: {'observation_keys': ('pixels', 'goal_index')}\n for key in (\n 'TurnMultiGoalResetFree-v0',\n 'TurnFreeValve3ResetFreeSwapGoal-v0',\n )\n },\n **{\n key: {\n 'observation_keys': (\n 'object_xy_position',\n 'object_z_orientation_cos',\n 'object_z_orientation_sin')\n }\n for key in (\n 'TurnFreeValve3ResetFree-v0',\n )\n },\n **{\n env: {'observation_keys': ('object_angle_cos',\n 'object_angle_sin'), }\n for env in ('TurnFixed-v0', 'TurnFixedHardware-v0',)\n },\n },\n 'SawyerDhandInHand': {\n **{\n env: {\n 'observation_keys': (\n \"object_xyz\", # 3\n ),\n }\n for env in (\n 'Valve3RepositionFixed-v0',\n 'Valve3PickupFixed-v0'\n )\n },\n }\n }\n}\n\nDYNAMICS_MODEL_PARAMS_BASE = {\n # 'type': 'feedforward_classifier',\n 'kwargs': {\n 'action_input': True,\n 'encoder_kwargs': {\n 'hidden_layer_sizes': (64, 64),\n },\n 'decoder_kwargs': {\n 'hidden_layer_sizes': (64, 64),\n },\n 'dynamics_latent_dim': 16,\n }\n}\n\n\"\"\"\nDistance Estimator params\n\"\"\"\n\nDISTANCE_FN_PARAMS_BASE = {\n 'type': 'feedforward_distance_fn',\n 'kwargs': {\n 'hidden_layer_sizes': (M, ) * N,\n 'observation_keys': None,\n\n # Output dimension if using an embedding\n 'embedding_dim': 2,\n\n # Use weight decay for distance fn\n # 'kernel_regularizer': tune.grid_search([None, tf.keras.regularizers.l2(5e-4)]),\n }\n}\n\nDISTANCE_FN_KWARGS_UNIVERSE_DOMAIN_TASK = {\n 'gym': {\n 'Point2D': {\n **{\n key: {'observation_keys': ('state_observation', )}\n for key in (\n 'Fixed-v0',\n 'SingleWall-v0',\n 'Maze-v0',\n 'BoxWall-v1',\n )\n },\n **{\n key: {'observation_keys': ('onehot_observation', )}\n for key in (\n # 'Fixed-v0',\n # 'SingleWall-v0',\n # 'Maze-v0',\n # 'BoxWall-v1',\n )\n },\n },\n 'Pusher2D': {\n **{\n key: {'observation_keys': ('object_pos', )}\n for key in (\n 'Simple-v0',\n )\n },\n },\n }\n}\n\n\n\nENVIRONMENT_PARAMS_PER_UNIVERSE_DOMAIN_TASK_STATE = {\n 'gym': {\n 'Ant': {\n 'MazeOneWallEnv-v0': {\n # === Dense Manhattan distance reward ===\n # 'reward_type': 'xy_manhattan',\n\n # === Sparse reward + count bonus ===\n 'reward_type': 'xy_sparse',\n 'target_radius': 2.0,\n 'n_bins': 36,\n 'count_bonus_coeff': 0.5,\n 'vel_in_state': False,\n\n 'observation_keys': ('state_observation', 'xy_observation'),\n 'init_qpos': [-5.5, 4.5, 0.565, 1., 0., 0., 0., 0., 1., 0., -1., 0., -1., 0., 1.],\n # 'terminate_when_unhealthy': True,\n # 'done_penalty': -1000,\n },\n 'MazeSEnv-v0': {\n # === Dense Manhattan distance reward ===\n # 'reward_type': 'xy_manhattan',\n\n # === Sparse reward + count bonus ===\n 'reward_type': 'xy_sparse',\n 'target_radius': 1.0,\n 'n_bins': 36,\n 'count_bonus_coeff': tune.grid_search([1]),\n\n # Low gear ratio ant env with S-shaped walls\n 'observation_keys': ('state_observation', 'xy_observation', ),\n 'init_qpos': [-5.5, 4.5, 0.565, 1., 0., 0., 0., 0., 1., 0., -1., 0., -1., 0., 1.],\n # 'diagnostics_goal': np.array([5.5, -4.5]),\n # 'terminate_when_unhealthy': True,\n # 'done_penalty': -1000,\n },\n 'v0': {\n # Standard normal gear ratio ant env with no walls\n\n # === Dense L2 reward ===\n 'reward_type': 'xy_dense',\n\n # === Sparse reward + count bonus ===\n # 'reward_type': 'xy_sparse',\n # 'target_radius': 0.5,\n # 'n_bins': 16,\n # 'count_bonus_coeff': 0.5,\n\n 'observation_keys': ('state_observation', 'xy_observation',),\n 'use_low_gear_ratio': True,\n # 'terminate_when_unhealthy': True,\n # 'done_penalty': -1000,\n }\n },\n 'Point2D': {\n # === Point Mass ===\n 'Fixed-v0': {\n 'action_scale': tune.grid_search([0.5]),\n 'images_are_rgb': True,\n # 'init_pos_range': ((-2, -3), (-3, -3)), # Fixed reset\n 'init_pos_range': None, # Random reset\n # 'target_pos_range': ((3, 3), (3, 3)), # Set the goal to (x, y) = (2, 2)\n 'target_pos_range': ((0, 0), (0, 0)), # Set the goal to (x, y) = (0, 0)\n 'render_onscreen': False,\n 'observation_keys': ('state_observation', ),\n # 'n_bins': 50,\n # 'observation_keys': ('onehot_observation', ),\n },\n 'SingleWall-v0': {\n # 'boundary_distance': tune.grid_search([4, 8]),\n 'action_scale': tune.grid_search([0.5]),\n 'images_are_rgb': True,\n 'init_pos_range': None, # Random reset\n 'target_pos_range': ((0, 3), (0, 3)), # Set the goal to (x, y) = (2, 2)\n 'render_onscreen': False,\n 'observation_keys': ('state_observation', ),\n },\n 'BoxWall-v1': {\n 'action_scale': tune.grid_search([0.5]),\n 'images_are_rgb': True,\n 'init_pos_range': None, # Random reset\n # 'target_pos_range': ((3.5, 3.5), (3.5, 3.5)),\n 'target_pos_range': ((0, 3), (0, 3)),\n 'render_onscreen': False,\n 'observation_keys': ('onehot_observation', ),\n # 'observation_keys': ('state_observation', ),\n },\n 'Maze-v0': {\n 'action_scale': 0.5,\n 'images_are_rgb': True,\n\n # === Use environment's count-based reward ===\n 'reward_type': 'none',\n 'use_count_reward': False,\n 'n_bins': tune.grid_search([50]), # Number of bins to discretize the space with\n\n # === EASY ===\n # 'wall_shape': 'easy-maze',\n # 'init_pos_range': ((-2.5, -2.5), (-2.5, -2.5)),\n # 'target_pos_range': ((2.5, -2.5), (2.5, -2.5)),\n # === MEDIUM ===\n # 'wall_shape': 'medium-maze',\n # 'init_pos_range': ((-3, -3), (-3, -3)),\n # 'target_pos_range': ((3, 3), (3, 3)),\n # 'shuffle_states': True,\n # === HARD ===\n # 'wall_shape': 'hard-maze',\n # 'init_pos_range': ((-3, -3), (-3, -3)),\n # 'target_pos_range': ((-0.5, 1.25), (-0.5, 1.25)),\n # === HORIZONTAL (3 walls) ===\n # 'wall_shape': 'horizontal-maze',\n # 'init_pos_range': ((-3, -3), (-3, -3)),\n # 'target_pos_range': ((-3, 3), (-3, 3)),\n # === MULTI-GOAL ===\n 'wall_shape': 'double-medium-maze',\n 'init_pos_range': ((0, 0), (0, 0)),\n 'target_pos_range': ((-3.4, 2.8), (-3.0, 3.2)),\n 'sparse_goals': (np.array([[2.75, -0.5], [3.75, -0.5], [2.75, 0.5], [3.75, 0.5], [3.25, 0]]), 0.3),\n\n 'render_onscreen': False,\n 'observation_keys': ('state_observation', ),\n },\n },\n 'Pusher2D': {\n 'Simple-v0': {\n 'init_qpos_range': ((0, 0, 0), (0, 0, 0)),\n 'init_object_pos_range': ((1, 0), (1, 0)),\n 'target_pos_range': ((2, 2), (2, 2)),\n 'reset_gripper': True,\n 'reset_object': True,\n 'observation_keys': (\n 'gripper_qpos',\n 'gripper_qvel',\n 'object_pos',\n # 'target_pos'\n ),\n },\n },\n 'SawyerDhandInHand': {\n 'Valve3RepositionFixed-v0': {\n 'reset_every_n_episodes': 1,\n 'init_xyz_range_params': {\n \"type\": \"DiscreteRange\",\n \"values\": [np.array([0.72 + 0.15, 0.15 + 0.15, 0.75])],\n # \"values\": [np.array([0.72, 0.15, 0.75])],\n },\n 'target_xyz_range_params': {\n \"type\": \"DiscreteRange\",\n \"values\": [np.array([0.72 - 0.15, 0.15 - 0.15, 0.75])],\n # \"values\": [np.array([0.72, 0.15, 0.75])],\n },\n # \"init_euler_range_params\": {\n # \"type\": \"UniformRange\",\n # \"values\": [\n # np.array([np.pi / 2, -np.pi, 0]),\n # np.array([np.pi / 2, np.pi, 0])\n # ],\n # },\n 'readjust_to_object_in_reset': tune.grid_search([True]),\n 'readjust_hand_xyz': True,\n \"readjust_hand_euler\": False,\n 'reward_keys_and_weights': {\n # 'object_to_target_xy_sparse_reward': 1.0,\n # 'object_to_hand_xyz_sparse_reward': 1.0,\n 'span_dist': 0.0,\n },\n \"observation_keys\": (\n \"object_xyz\",\n #####################\n \"dhand_qpos\",\n \"sawyer_arm_qpos\",\n # \"dhand_qvel\",\n # \"sawyer_arm_qvel\",\n \"mocap_pos\",\n # \"object_z_orientation_cos\",\n ),\n },\n },\n 'DClaw': {\n # === FIXED SCREW RANDOM RESET EVAL TASK BELOW ===\n 'TurnFixed-v0': {\n # 'reward_keys_and_weights': { # <- this reward doesn't actually get used for VICE\n # 'object_to_target_angle_distance_reward': 1,\n # 'sparse_reward': 1,\n # },\n 'init_pos_range': (0, 0),\n 'target_pos_range': (np.pi, np.pi),\n 'reward_keys_and_weights': {\n 'object_to_target_angle_distance_reward': 1\n },\n # 'observation_keys': (\n # 'object_angle_cos',\n # 'object_angle_sin',\n # 'claw_qpos',\n # 'last_action'\n # ),\n },\n 'TurnResetFree-v0': {\n 'init_object_pos_range': (0., 0.),\n 'target_pos_range': (-np.pi, np.pi),\n 'reward_keys': ('object_to_target_angle_dist_cost', )\n },\n 'TurnFreeValve3ResetFree-v0': {\n 'init_qpos_range': (\n (0, 0, 0, 0, 0, 0),\n (0, 0, 0, 0, 0, 0),\n ),\n 'target_qpos_range': [\n (0, 0, 0, 0, 0, np.pi),\n (0, 0, 0, 0, 0, 0),\n ],\n 'reset_fingers': True,\n 'swap_goal_upon_completion': False,\n 'observation_keys': (\n 'object_xy_position',\n 'object_z_orientation_cos',\n 'object_z_orientation_sin',\n 'claw_qpos',\n 'last_action'\n ),\n 'reward_keys_and_weights': {\n 'object_to_target_position_distance_reward': 2,\n 'object_to_target_orientation_distance_reward': 1,\n },\n },\n 'TurnFreeValve3Fixed-v0': {\n 'observation_keys': (\n 'claw_qpos',\n 'last_action',\n 'object_xy_position',\n 'object_z_orientation_cos',\n 'object_z_orientation_sin',\n ),\n 'init_qpos_range': ((0, 0, 0, 0, 0, 0), ) * 2,\n 'target_qpos_range': ((0, 0, 0, 0, 0, np.pi), ) * 2,\n },\n 'TurnFreeValve3MultiGoalResetFree-v0': {\n 'goals': ((0, 0, 0, 0, 0, np.pi), (0, 0, 0, 0, 0, 0)),\n # 'goals': (\n # (0.01, 0.01, 0, 0, 0, 0),\n # (0.01, -0.01, 0, 0, 0, np.pi / 2),\n # (-0.01, -0.01, 0, 0, 0, np.pi),\n # (-0.01, 0.01, 0, 0, 0, -np.pi / 2),\n # ),\n 'goal_completion_position_threshold': 0.04,\n 'goal_completion_orientation_threshold': 0.15,\n 'swap_goals_upon_completion': False,\n },\n 'TurnFreeValve3MultiGoal-v0': {\n 'goals': ((0, 0, 0, 0, 0, np.pi), (0, 0, 0, 0, 0, 0)),\n # 'goals': (\n # (0.01, 0.01, 0, 0, 0, 0),\n # (0.01, -0.01, 0, 0, 0, np.pi / 2),\n # (-0.01, -0.01, 0, 0, 0, np.pi),\n # (-0.01, 0.01, 0, 0, 0, -np.pi / 2),\n # ),\n 'swap_goals_upon_completion': False,\n 'random_goal_sampling': True,\n },\n 'TurnFreeValve3ResetFreeSwapGoal': {\n 'init_angle_range': (0., 0.),\n 'reward_keys_and_weights': {\n 'object_to_target_position_distance_cost': 2,\n 'object_to_target_orientation_distance_cost': 1,\n },\n },\n 'TurnFreeValve3ResetFreeSwapGoalEval': {\n 'reward_keys_and_weights': {\n 'object_to_target_position_distance_cost': 2,\n 'object_to_target_orientation_distance_cost': 1,\n },\n },\n # === LIFTING === \n 'LiftDDFixed-v0': {\n 'init_qpos_range': tune.grid_search([\n (\n (0, 0, 0.041, 1.017, 0, 0),\n (0, 0, 0.041, 1.017, 0, 0),\n ),\n # (\n # (0, 0, 0.041, -np.pi, -np.pi, -np.pi),\n # (0, 0, 0.041, np.pi, np.pi, np.pi),\n # )\n ]),\n 'target_qpos_range': [\n (0, 0, 0.045, 0, 0, 0)\n ],\n 'reward_keys_and_weights': {\n # Dense reward (want z reward to be 10x in magnitude)\n 'object_to_target_z_position_distance_reward': 10,\n 'object_to_target_xy_position_distance_reward': 0.1,\n 'object_to_target_orientation_distance_reward': 0,\n\n # 'sparse_position_reward': 1\n },\n 'observation_keys': (\n 'object_position',\n 'object_quaternion',\n 'claw_qpos',\n 'last_action'\n ),\n # Camera settings for video\n 'camera_settings': {\n 'distance': 0.35,\n 'elevation': -15,\n 'lookat': (0, 0, 0.05),\n },\n },\n\n # === Single Object Translation Tasks ===\n 'TranslatePuckFixed-v0': {\n 'init_qpos_range': (\n (-0.08, -0.08, 0, 0, 0, 0),\n (0.08, 0.08, 0, 0, 0, 0),\n ),\n 'target_qpos_range': (\n (0, 0, 0, 0, 0, 0),\n (0, 0, 0, 0, 0, 0),\n ),\n 'n_bins': 100,\n 'observation_keys': (\n 'claw_qpos',\n 'last_action',\n 'object_xy_position',\n ),\n 'reward_keys_and_weights': {\n 'object_to_target_position_distance_reward': 1,\n },\n # Camera settings for video\n 'camera_settings': {\n 'distance': 0.35,\n 'elevation': -15,\n 'lookat': (0, 0, 0.05),\n },\n },\n # === Multi-Object Translation Tasks ===\n 'TranslateMultiPuckFixed-v0': {\n 'init_qpos_ranges': (\n ((0.05, 0.05, 0, 0, 0, 0), (0.05, 0.05, 0, 0, 0, 0)),\n ((-0.05, -0.05, 0, 0, 0, 0), (-0.05, -0.05, 0, 0, 0, 0)),\n ),\n 'target_qpos_ranges': (\n ((0.05, -0.05, 0, 0, 0, 0), (0.05, -0.05, 0, 0, 0, 0)),\n ((-0.05, 0.05, 0, 0, 0, 0), (-0.05, 0.05, 0, 0, 0, 0)),\n ),\n 'observation_keys': (\n 'claw_qpos',\n 'last_action',\n 'object1_xy_position',\n 'object2_xy_position',\n ),\n 'reward_keys_and_weights': {\n 'object1_to_target_position_distance_log_reward': 1,\n 'object2_to_target_position_distance_log_reward': 1,\n }\n },\n }\n },\n}\n\n\nFREE_SCREW_VISION_KWARGS = {\n 'pixel_wrapper_kwargs': {\n 'pixels_only': False,\n 'normalize': False,\n 'render_kwargs': {\n 'width': 32,\n 'height': 32,\n 'camera_id': -1,\n },\n },\n 'camera_settings': {\n 'azimuth': 180,\n 'distance': 0.38,\n 'elevation': -36,\n 'lookat': (0.04, 0.008, 0.026),\n },\n}\nFIXED_SCREW_VISION_KWARGS = {\n 'pixel_wrapper_kwargs': {\n 'pixels_only': False,\n 'normalize': False,\n 'render_kwargs': {\n 'width': 32,\n 'height': 32,\n 'camera_id': -1,\n }\n },\n 'camera_settings': {\n 'azimuth': 180,\n 'distance': 0.3,\n 'elevation': -50,\n 'lookat': np.array([0.02, 0.004, 0.09]),\n },\n}\nSLIDE_BEADS_VISION_KWARGS = {\n 'pixel_wrapper_kwargs': {\n 'pixels_only': False,\n 'normalize': False,\n 'render_kwargs': {\n 'width': 32,\n 'height': 32,\n 'camera_id': -1,\n },\n },\n 'camera_settings': {\n 'azimuth': 90,\n 'distance': 0.37,\n 'elevation': -45,\n 'lookat': (0, 0.046, -0.016),\n },\n}\n\nENVIRONMENT_PARAMS_PER_UNIVERSE_DOMAIN_TASK_VISION = {\n 'gym': {\n 'Point2D': {\n 'Maze-v0': {\n 'action_scale': 0.5,\n 'images_are_rgb': True,\n\n # === Use environment's count-based reward ===\n 'reward_type': 'none',\n 'use_count_reward': False,\n 'n_bins': tune.grid_search([50]), # Number of bins to discretize the space with\n\n # === EASY ===\n # 'wall_shape': 'easy-maze',\n # 'init_pos_range': ((-2.5, -2.5), (-2.5, -2.5)),\n # 'target_pos_range': ((2.5, -2.5), (2.5, -2.5)),\n # === MEDIUM ===\n 'wall_shape': 'medium-maze',\n 'init_pos_range': ((-3, -3), (-3, -3)),\n 'target_pos_range': ((3, 3), (3, 3)),\n # === HARD ===\n # 'wall_shape': 'hard-maze',\n # 'init_pos_range': ((-3, -3), (-3, -3)),\n # 'target_pos_range': ((-0.5, 1.25), (-0.5, 1.25)),\n # === HORIZONTAL (3 walls) ===\n # 'wall_shape': 'horizontal-maze',\n # 'init_pos_range': ((-3, -3), (-3, -3)),\n # 'target_pos_range': ((-3, 3), (-3, 3)),\n\n 'render_onscreen': False,\n 'observation_keys': ('pixels', 'state_observation'),\n 'convert_obs_to_image': True,\n 'show_goal': False,\n 'ball_pixel_radius': 1,\n 'pixel_wrapper_kwargs': {\n 'render_kwargs': {\n 'mode': 'rgb_array',\n 'width': 48,\n 'height': 48,\n 'invert_colors': True,\n },\n 'pixels_only': False,\n 'normalize': False,\n },\n },\n },\n 'StateSawyer': {\n 'PickAndPlace3DEnv-v0': {\n 'observation_keys': ('pixels', 'state', ),\n 'pixel_wrapper_kwargs': {\n 'render_kwargs': {\n 'mode': 'rgb_array',\n 'width': 28,\n 'height': 28,\n },\n 'pixels_only': False,\n 'normalize': False,\n },\n },\n },\n 'Image48Sawyer': {\n 'PickAndPlace3DEnv-v0': {\n 'observation_keys': ('pixels',),\n 'pixel_wrapper_kwargs': {\n 'render_kwargs': {\n 'mode': 'rgb_array',\n 'width': 48,\n 'height': 48,\n },\n 'pixels_only': False,\n 'normalize': False,\n },\n },\n },\n 'DClaw': {\n # === FIXED SCREW RANDOM RESET EVAL TASK BELOW ===\n 'TurnFixed-v0': {\n **FIXED_SCREW_VISION_KWARGS,\n # 'init_pos_range': (-np.pi, np.pi), # Random reset between -pi, pi\n # Reset to every 45 degrees between -pi and pi\n 'init_pos_range': list(np.arange(-np.pi, np.pi, np.pi / 4)),\n\n # === GOAL = -90 DEGREES ===\n # Single goal + RND reset controller\n 'target_pos_range': [-np.pi / 2, -np.pi / 2],\n # 2 goal + no RND reset controller\n # 'target_pos_range': [-np.pi / 2, np.pi / 2],\n # 1 goal + no RND reset controller\n # 'target_pos_range': [-np.pi / 2],\n 'observation_keys': (\n 'pixels',\n 'claw_qpos',\n 'last_action',\n # == BELOW JUST FOR LOGGING ==\n 'object_angle_cos',\n 'object_angle_sin',\n ),\n },\n # === FIXED SCREW RESET FREE TASK BELOW ===\n 'TurnResetFree-v0': {\n **FIXED_SCREW_VISION_KWARGS,\n 'reset_fingers': True,\n 'init_pos_range': (0, 0),\n # Single goal + RND reset controller\n 'target_pos_range': [-np.pi / 2, -np.pi / 2],\n # 2 goal + no RND reset controller\n # 'target_pos_range': [-np.pi / 2, np.pi / 2],\n # 1 goal + no RND reset controller\n # 'target_pos_range': [-np.pi / 2],\n 'observation_keys': (\n 'claw_qpos',\n 'pixels',\n 'last_action',\n # === BELOW JUST FOR LOGGING ===\n 'object_angle_cos',\n 'object_angle_sin',\n ),\n },\n 'TurnFreeValve3Fixed-v0': {\n 'observation_keys': (\n 'claw_qpos',\n 'last_action',\n 'object_xy_position',\n 'object_z_orientation_cos',\n 'object_z_orientation_sin',\n ),\n 'init_qpos_range': ((0, 0, 0, 0, 0, 0), ) * 2,\n 'target_qpos_range': ((0, 0, 0, 0, 0, np.pi), ) * 2,\n },\n # Random evaluation environment for free screw\n # 'TurnFreeValve3Fixed-v0': {\n # **FREE_SCREW_VISION_KWARGS,\n # # Random init evaluations\n # # 'init_qpos_range': (\n # # (-0.08, -0.08, 0, 0, 0, -np.pi),\n # # (0.08, 0.08, 0, 0, 0, np.pi)\n # # ),\n # # Evaluations from fixed set of inits\n # 'init_qpos_range': [\n # (0, 0, 0, 0, 0, 0),\n # (0, 0, 0, 0, 0, -np.pi),\n # (0, 0, 0, 0, 0, -np.pi / 2),\n # (0, 0, 0, 0, 0, np.pi / 2),\n # (-0.05, 0.075, 0, 0, 0, -np.pi),\n # (-0.075, 0.05, 0, 0, 0, -np.pi / 2),\n # (-0.05, 0.05, 0, 0, 0, -3 * np.pi / 4),\n # (-0.07, 0.07, 0, 0, 0, np.pi / 4),\n # (0, 0.075, 0, 0, 0, -np.pi),\n # (0.05, 0.075, 0, 0, 0, -np.pi),\n # (0.075, 0.05, 0, 0, 0, np.pi / 2),\n # (0.05, 0.05, 0, 0, 0, 3 * np.pi / 4),\n # (0.07, 0.07, 0, 0, 0, -np.pi / 4),\n # (-0.05, -0.075, 0, 0, 0, 0),\n # (-0.075, -0.05, 0, 0, 0, -np.pi / 2),\n # (-0.05, -0.05, 0, 0, 0, -np.pi / 4),\n # (-0.07, -0.07, 0, 0, 0, 3 * np.pi / 4),\n # (0, -0.075, 0, 0, 0, 0),\n # (0.05, -0.075, 0, 0, 0, 0),\n # (0.075, -0.05, 0, 0, 0, np.pi / 2),\n # (0.05, -0.05, 0, 0, 0, np.pi / 4),\n # (0.07, -0.07, 0, 0, 0, -3 * np.pi / 4),\n # (-0.075, 0, 0, 0, 0, -np.pi / 2),\n # (0.075, 0, 0, 0, 0, np.pi / 2),\n # ],\n # 'cycle_inits': True,\n\n # # 1 goal for RND reset controller\n # # 'target_qpos_range': [\n # # (0, 0, 0, 0, 0, -np.pi / 2),\n # # (0, 0, 0, 0, 0, -np.pi / 2),\n # # ],\n # # 2 goal, no RND reset controller\n # # 'target_qpos_range': [\n # # (0, 0, 0, 0, 0, -np.pi / 2),\n # # (0, 0, 0, 0, 0, np.pi / 2),\n # # ],\n # # 2 goals\n # 'target_qpos_range': [\n # # (top left, center)\n # # (-0.05, -0.05, 0, 0, 0, -np.pi / 2),\n # # (0, 0, 0, 0, 0, np.pi / 2),\n # # bottom right, top right\n # (0.075, 0.075, 0, 0, 0, -np.pi),\n # (-0.075, 0.075, 0, 0, 0, -np.pi)\n # ],\n # 'observation_keys': (\n # 'pixels',\n # 'claw_qpos',\n # 'last_action',\n # # === BELOW IS JUST FOR LOGGING ===\n # 'object_xy_position',\n # 'object_z_orientation_cos',\n # 'object_z_orientation_sin',\n # ),\n # },\n 'TurnFreeValve3ResetFree-v0': {\n **FREE_SCREW_VISION_KWARGS,\n 'init_qpos_range': [(0, 0, 0, 0, 0, 0)],\n # Below needs to be 2 for a MultiVICEGAN run, since the goals switch\n # Single goal + RND reset controller\n # 'target_qpos_range': [\n # (0, 0, 0, 0, 0, -np.pi / 2),\n # (0, 0, 0, 0, 0, -np.pi / 2), # Second goal is arbitrary\n # ],\n # 2 goal, no RND reset controller\n # 'target_qpos_range': [\n # (0, 0, 0, 0, 0, -np.pi / 2),\n # (0, 0, 0, 0, 0, np.pi / 2),\n # ],\n # 2 goals\n 'target_qpos_range': [\n # (top left, center)\n # (-0.05, -0.05, 0, 0, 0, -np.pi / 2),\n # (0, 0, 0, 0, 0, np.pi / 2),\n # bottom right, top right\n (0.075, 0.075, 0, 0, 0, -np.pi),\n (-0.075, 0.075, 0, 0, 0, -np.pi)\n ],\n 'swap_goal_upon_completion': False,\n 'observation_keys': (\n 'pixels',\n 'claw_qpos',\n 'last_action',\n # === BELOW IS JUST FOR LOGGING ===\n 'object_xy_position',\n 'object_z_orientation_cos',\n 'object_z_orientation_sin',\n ),\n },\n # === FREE SCREW HARDWARE ===\n 'TurnFreeValve3Hardware-v0': {\n 'pixel_wrapper_kwargs': {\n 'pixels_only': False,\n 'normalize': False,\n 'render_kwargs': {\n 'width': 32,\n 'height': 32,\n 'camera_id': -1,\n 'box_warp': True,\n }\n },\n 'observation_keys': (\n 'claw_qpos',\n 'pixels',\n 'last_action',\n ),\n 'device_path': '/dev/ttyUSB0',\n 'camera_config': {\n 'topic': '/kinect2_001161563647/qhd/image_color',\n 'image_shape': (256, 256, 3),\n }\n },\n 'TurnFreeValve3ResetFreeSwapGoal-v0': {\n **FREE_SCREW_VISION_KWARGS,\n 'reset_fingers': True,\n 'reset_frequency': 0,\n 'goals': [\n (0, 0, 0, 0, 0, np.pi / 2),\n (0, 0, 0, 0, 0, -np.pi / 2),\n ],\n 'observation_keys': (\n 'claw_qpos',\n 'last_action',\n 'target_xy_position',\n 'target_z_orientation_cos',\n 'target_z_orientation_sin',\n 'goal_index',\n 'pixels',\n # === BELOW IS JUST FOR LOGGING ===\n 'object_xy_position',\n 'object_orientation_cos',\n 'object_orientation_sin',\n ),\n },\n 'TurnFreeValve3ResetFreeSwapGoalEval-v0': {\n **FREE_SCREW_VISION_KWARGS,\n 'goals': [\n (0, 0, 0, 0, 0, np.pi / 2),\n (0, 0, 0, 0, 0, -np.pi / 2),\n ],\n 'observation_keys': (\n 'claw_qpos',\n 'last_action',\n 'target_xy_position',\n 'target_z_orientation_cos',\n 'target_z_orientation_sin',\n 'goal_index',\n 'pixels',\n # === BELOW IS JUST FOR LOGGING ===\n 'object_xy_position',\n 'object_orientation_cos',\n 'object_orientation_sin',\n ),\n },\n 'LiftDDFixed-v0': {\n 'reward_keys_and_weights': {\n 'object_to_target_z_position_distance_reward': 1,\n 'object_to_target_xy_position_distance_reward': 0,\n 'object_to_target_orientation_distance_reward': 0, #tune.sample_from([1, 5]), #5,\n },\n 'target_qpos_range': [(0, 0, 0.05, 0, 0, 0)],\n 'pixel_wrapper_kwargs': {\n 'observation_key': 'pixels',\n 'pixels_only': False,\n 'render_kwargs': {\n 'width': 32,\n 'height': 32,\n },\n },\n 'observation_keys': (\n 'claw_qpos',\n 'object_position',\n 'object_quaternion',\n 'last_action',\n 'target_position',\n 'target_quaternion',\n 'pixels',\n ),\n 'camera_settings': {\n 'azimuth': 180,\n 'distance': 0.26,\n 'elevation': -40,\n 'lookat': (0, 0, 0.06),\n }\n },\n 'LiftDDResetFree-v0': {\n 'reward_keys_and_weights': {\n 'object_to_target_z_position_distance_reward': 1,\n 'object_to_target_xy_position_distance_reward': 0,\n 'object_to_target_orientation_distance_reward': 0, #tune.sample_from([1, 5]), #5,\n },\n # 'target_qpos_range': (\n # (-0.1, -0.1, 0.0, 0, 0, 0),\n # (0.1, 0.1, 0.0, 0, 0, 0), # bgreen side up\n # ),\n 'target_qpos_range': [(0, 0, 0.05, 0, 0, 0)],\n 'pixel_wrapper_kwargs': {\n 'observation_key': 'pixels',\n 'pixels_only': False,\n 'render_kwargs': {\n 'width': 32,\n 'height': 32,\n },\n },\n 'observation_keys': (\n 'claw_qpos',\n 'object_position',\n 'object_quaternion',\n 'last_action',\n 'target_position',\n 'target_quaternion',\n 'pixels',\n ),\n 'camera_settings': {\n 'azimuth': 180,\n 'distance': 0.26,\n 'elevation': -40,\n 'lookat': (0, 0, 0.06),\n }\n },\n # Sliding Tasks\n 'SlideBeadsFixed-v0': {\n **SLIDE_BEADS_VISION_KWARGS,\n 'num_objects': 4,\n # Random init\n # 'init_qpos_range': (\n # (-0.0475, -0.0475, -0.0475, -0.0475),\n # (0.0475, 0.0475, 0.0475, 0.0475),\n # ),\n 'init_qpos_range': [\n (-0.0475, -0.0475, -0.0475, -0.0475), # 4 left\n (0.0475, 0.0475, 0.0475, 0.0475), # 4 right\n (0, 0, 0, 0), # 4 middle\n (-0.0475, -0.0475, -0.0475, 0.0475), # 3 left, 1 right\n (-0.0475, 0.0475, 0.0475, 0.0475), # 1 left, 3 right\n (-0.0475, -0.0475, 0.0475, 0.0475), # 2 left, 2 right\n (-0.0475, -0.02375, 0.02375, 0.0475), # even spaced\n (-0.0475, 0, 0, 0.0475), # slides, and 2 in the middle\n ],\n 'cycle_inits': True,\n # Goal we want to evaluate:\n 'target_qpos_range': [\n # 4 left\n # (-0.0475, -0.0475, -0.0475, -0.0475),\n # 2 left, 2 right\n (-0.0475, -0.0475, 0.0475, 0.0475),\n (-0.0475, -0.0475, 0.0475, 0.0475),\n # Remove below for 1 goal reset free\n # (0, 0, 0, 0)\n ],\n 'observation_keys': (\n 'claw_qpos',\n 'last_action',\n 'pixels',\n # === BELOW JUST FOR LOGGING ===\n 'objects_positions',\n 'objects_target_positions',\n ),\n },\n 'SlideBeadsResetFree-v0': {\n **SLIDE_BEADS_VISION_KWARGS,\n 'init_qpos_range': [(0, 0, 0, 0)],\n 'num_objects': 4,\n 'target_qpos_range': [\n # 4 left\n # (-0.0475, -0.0475, -0.0475, -0.0475),\n # 2 left, 2 right\n (-0.0475, -0.0475, 0.0475, 0.0475),\n (-0.0475, -0.0475, 0.0475, 0.0475),\n # This second one is arbitrary for training env\n # (0, 0, 0, 0),\n ],\n 'reset_fingers': False,\n 'observation_keys': (\n 'pixels',\n 'claw_qpos',\n 'last_action',\n # === BELOW JUST FOR LOGGING ===\n 'objects_target_positions',\n 'objects_positions',\n ),\n },\n 'SlideBeadsResetFreeEval-v0': {\n 'reward_keys_and_weights': {\n 'objects_to_targets_mean_distance_reward': 1,\n },\n 'init_qpos_range': [(0, 0)],\n 'num_objects': 4,\n 'target_qpos_range': [\n (0, 0, 0, 0),\n (-0.0475, -0.0475, 0.0475, 0.0475),\n ],\n # 'target_qpos_range': [\n # (0, 0),\n # (-0.0825, 0.0825),\n # (0.0825, 0.0825),\n # (-0.04, 0.04),\n # (-0.0825, -0.0825),\n # ],\n 'cycle_goals': True,\n 'pixel_wrapper_kwargs': {\n 'observation_key': 'pixels',\n 'pixels_only': False,\n 'render_kwargs': {\n 'width': 32,\n 'height': 32,\n },\n },\n 'observation_keys': (\n 'claw_qpos',\n 'objects_positions',\n 'last_action',\n 'objects_target_positions',\n 'pixels',\n ),\n 'camera_settings': {\n 'azimuth': 90,\n 'lookat': (0, 0.04581637, -0.01614516),\n 'elevation': -45,\n 'distance': 0.37,\n },\n },\n\n\n }\n },\n}\n\n\"\"\"\nHelper methods for retrieving universe/domain/task specific params.\n\"\"\"\n\n\ndef get_policy_params(universe, domain, task):\n policy_params = GAUSSIAN_POLICY_PARAMS_BASE.copy()\n return policy_params\n\n\ndef get_max_path_length(universe, domain, task):\n max_path_length = MAX_PATH_LENGTH_PER_DOMAIN.get(domain) or \\\n MAX_PATH_LENGTH_PER_DOMAIN[DEFAULT_KEY]\n return max_path_length\n\n\ndef get_environment_params(universe, domain, task, from_vision):\n if from_vision:\n params = ENVIRONMENT_PARAMS_PER_UNIVERSE_DOMAIN_TASK_VISION\n else:\n params = ENVIRONMENT_PARAMS_PER_UNIVERSE_DOMAIN_TASK_STATE\n\n environment_params = (\n params.get(universe, {}).get(domain, {}).get(task, {}))\n return environment_params\n\n\ndef get_classifier_params(universe, domain, task, from_vision):\n classifier_params = CLASSIFIER_PARAMS_BASE.copy()\n if from_vision:\n params = CLASSIFIER_PARAMS_PER_UNIVERSE_DOMAIN_TASK_VISION.get(\n universe, {}).get(domain, {}).get(task, {})\n if not classifier_params or not from_vision:\n params = CLASSIFIER_PARAMS_PER_UNIVERSE_DOMAIN_TASK.get(\n universe, {}).get(domain, {}).get(task, {})\n\n classifier_params['kwargs'].update(params)\n return classifier_params\n\n\ndef get_dynamics_model_params(universe, domain, task):\n params = DYNAMICS_MODEL_PARAMS_BASE.copy()\n # classifier_params['kwargs'].update(\n # CLASSIFIER_PARAMS_PER_UNIVERSE_DOMAIN_TASK.get(\n # universe, {}).get(domain, {}).get(task, {}))\n return params\n\n\ndef get_distance_fn_params(universe, domain, task):\n distance_fn_params = DISTANCE_FN_PARAMS_BASE.copy()\n distance_fn_params['kwargs'].update(\n DISTANCE_FN_KWARGS_UNIVERSE_DOMAIN_TASK.get(\n universe, {}).get(domain, {}).get(task, {}))\n return distance_fn_params\n\n\ndef get_checkpoint_frequency(spec):\n config = spec.get('config', spec)\n checkpoint_frequency = (\n config\n ['algorithm_params']\n ['kwargs']\n ['n_epochs']\n ) // NUM_CHECKPOINTS\n\n return checkpoint_frequency\n\n\ndef is_image_env(universe, domain, task, variant_spec):\n return ('image' in task.lower()\n or 'image' in domain.lower()\n or 'pixel_wrapper_kwargs' in (\n variant_spec['environment_params']['training']['kwargs']))\n\n\n\"\"\"\nPreprocessor params\n\"\"\"\nSTATE_PREPROCESSOR_PARAMS = {\n 'ReplicationPreprocessor': {\n 'type': 'ReplicationPreprocessor',\n 'kwargs': {\n 'n': 0,\n 'scale_factor': 1,\n }\n },\n 'RandomNNPreprocessor': {\n 'type': 'RandomNNPreprocessor',\n 'kwargs': {\n 'hidden_layer_sizes': (32, 32),\n 'activation': 'linear',\n 'output_activation': 'linear',\n }\n },\n 'RandomMatrixPreprocessor': {\n 'type': 'RandomMatrixPreprocessor',\n 'kwargs': {\n 'output_size_scale_factor': 1,\n 'coefficient_range': (-1., 1.),\n }\n },\n 'None': None,\n}\n\n\nfrom softlearning.misc.utils import PROJECT_PATH, NFS_PATH\nPIXELS_PREPROCESSOR_PARAMS = {\n 'StateEstimatorPreprocessor': {\n 'type': 'StateEstimatorPreprocessor',\n 'kwargs': {\n 'input_shape': (32, 32, 3),\n 'num_hidden_units': 512,\n 'num_hidden_layers': 2,\n 'state_estimator_path': os.path.join(PROJECT_PATH,\n 'softlearning',\n 'models',\n 'state_estimators',\n 'state_estimator_from_vae_latents.h5'),\n # === INCLUDE A PRETRAINED VAE ===\n 'preprocessor_params': {\n 'type': 'VAEPreprocessor',\n 'kwargs': {\n 'encoder_path': os.path.join(PROJECT_PATH,\n 'softlearning',\n 'models',\n 'vae_16_dim_beta_3_invisible_claw_l2_reg',\n 'encoder_16_dim_3.0_beta.h5'),\n 'decoder_path': os.path.join(PROJECT_PATH,\n 'softlearning',\n 'models',\n 'vae_16_dim_beta_3_invisible_claw_l2_reg',\n 'decoder_16_dim_3.0_beta.h5'),\n 'trainable': False,\n 'image_shape': (32, 32, 3),\n 'latent_dim': 16,\n 'include_decoder': False,\n }\n }\n }\n },\n 'VAEPreprocessor': {\n 'type': 'VAEPreprocessor',\n 'kwargs': {\n 'trainable': False,\n # === Bead manipulation ===\n # 'image_shape': (32, 32, 3),\n # 'latent_dim': 16,\n # 'encoder_path': os.path.join(PROJECT_PATH,\n # 'softlearning',\n # 'models',\n # 'slide_beads_vae_16_230iters',\n # 'encoder_16_dim_1_beta.h5'),\n # === Free Screw ===\n 'image_shape': (32, 32, 3),\n 'latent_dim': 32, # 8,\n 'encoder_path': os.path.join(PROJECT_PATH,\n 'softlearning',\n 'models',\n 'hardware_free_screw_vae_black_box',\n 'encoder_32_dim_0.5_beta_final.h5'),\n # 'encoder_path': os.path.join(PROJECT_PATH,\n # 'softlearning',\n # 'models',\n # 'hardware_free_screw_vae_rnd_filtered_warped_include_claw',\n # 'encoder_8_dim_0.5_beta_final.h5'),\n # === Fixed Screw ===\n # 'image_shape': (32, 32, 3),\n # 'latent_dim': 16,\n # 'encoder_path': os.path.join(PROJECT_PATH,\n # 'softlearning',\n # 'models',\n # 'fixed_screw_16_dim_beta_half',\n # 'encoder_16_dim_0.5_beta_210.h5')\n },\n },\n 'RAEPreprocessor': {\n 'type': 'RAEPreprocessor',\n 'kwargs': {\n 'trainable': True,\n 'image_shape': (32, 32, 3),\n 'latent_dim': 32,\n },\n 'shared': True,\n },\n 'ConvnetPreprocessor': tune.grid_search([\n {\n 'type': 'ConvnetPreprocessor',\n 'kwargs': {\n 'conv_filters': (8, ) * 2,\n 'conv_kernel_sizes': (5, ) * 2,\n 'conv_strides': (2, ) * 2,\n 'normalization_type': normalization_type,\n 'downsampling_type': 'conv',\n 'output_kwargs': {\n 'type': 'flatten',\n }\n },\n # Specify a `weights_path` here if you want to load in a pretrained convnet\n }\n for normalization_type in (None, )\n ]),\n}\n\n\n\"\"\"\nConfiguring variant specs\n\"\"\"\n\n\ndef get_variant_spec_base(universe, domain, task, task_eval,\n policy, algorithm, from_vision):\n algorithm_params = ALGORITHM_PARAMS_BASE\n algorithm_params = deep_update(\n algorithm_params,\n ALGORITHM_PARAMS_ADDITIONAL.get(algorithm, {})\n )\n\n import tensorflow as tf\n num_goals = 2\n variant_spec = {\n 'git_sha': get_git_rev(),\n 'num_goals': num_goals, # TODO: Separate classifier_rl with multigoal\n 'environment_params': {\n 'training': {\n 'domain': domain,\n 'task': task,\n 'universe': universe,\n 'kwargs': get_environment_params(universe, domain, task, from_vision),\n },\n 'evaluation': {\n 'domain': domain,\n 'task': task_eval,\n 'universe': universe,\n 'kwargs': (\n tune.sample_from(lambda spec: (\n spec.get('config', spec)\n ['environment_params']\n ['training']\n .get('kwargs')\n ))\n if task == task_eval\n else get_environment_params(universe, domain, task_eval, from_vision)),\n },\n },\n 'policy_params': deep_update(\n POLICY_PARAMS_BASE[policy],\n POLICY_PARAMS_FOR_DOMAIN[policy].get(domain, {})\n ),\n 'exploration_policy_params': {\n 'type': 'UniformPolicy',\n 'kwargs': {\n 'observation_keys': tune.sample_from(lambda spec: (\n spec.get('config', spec)\n ['policy_params']\n ['kwargs']\n .get('observation_keys')\n ))\n },\n },\n 'Q_params': {\n 'type': 'double_feedforward_Q_function',\n 'kwargs': {\n 'hidden_layer_sizes': (M, M),\n 'observation_keys': tune.sample_from(lambda spec: (\n spec.get('config', spec)\n ['policy_params']\n ['kwargs']\n .get('observation_keys')\n )), # None means everything, pass in all keys but the goal_index\n 'observation_preprocessors_params': {},\n 'kernel_regularizer': tune.grid_search([\n None,\n # tf.keras.regularizers.l2(5e-4)\n ]),\n }\n },\n 'distance_fn_params': get_distance_fn_params(universe, domain, task),\n 'algorithm_params': algorithm_params,\n 'replay_pool_params': {\n 'type': 'SimpleReplayPool',\n 'kwargs': {\n 'max_size': int(15e5),\n }\n },\n 'sampler_params': {\n 'type': 'SimpleSampler',\n 'kwargs': {\n # 'max_path_length': get_max_path_length(universe, domain, task),\n # 'min_pool_size': get_max_path_length(universe, domain, task),\n 'max_path_length': get_max_path_length(universe, domain, task),\n 'min_pool_size': 50,\n 'batch_size': 256, # tune.grid_search([128, 256]),\n 'store_last_n_paths': 20,\n }\n },\n 'run_params': {\n 'seed': tune.sample_from(\n lambda spec: np.random.randint(0, 10000)),\n 'checkpoint_at_end': True,\n 'checkpoint_frequency': tune.sample_from(get_checkpoint_frequency),\n 'checkpoint_replay_pool': False,\n },\n }\n\n # Filter out parts of the state relating to the object when training from pixels\n env_kwargs = variant_spec['environment_params']['training']['kwargs']\n if from_vision and \"device_path\" not in env_kwargs.keys():\n env_obs_keys = env_kwargs['observation_keys']\n\n non_image_obs_keys = tuple(key for key in env_obs_keys if key != 'pixels')\n variant_spec['replay_pool_params']['kwargs']['obs_save_keys'] = non_image_obs_keys\n\n non_object_obs_keys = tuple(key for key in env_obs_keys if 'object' not in key)\n variant_spec['policy_params']['kwargs']['observation_keys'] = variant_spec[\n 'exploration_policy_params']['kwargs']['observation_keys'] = variant_spec[\n 'Q_params']['kwargs']['observation_keys'] = non_object_obs_keys\n # variant_spec['exploration_policy_params']['kwargs']['observation_keys'] += ('state_observation',)\n\n if 'Hardware' in task:\n env_kwargs['num_goals'] = num_goals\n return variant_spec\n\n\ndef get_variant_spec_classifier(universe,\n domain,\n task,\n task_eval,\n policy,\n algorithm,\n n_goal_examples,\n from_vision,\n *args,\n **kwargs):\n variant_spec = get_variant_spec_base(\n universe, domain, task, task_eval, policy, algorithm, from_vision, *args, **kwargs)\n\n variant_spec['reward_classifier_params'] = get_classifier_params(universe, domain, task, from_vision)\n variant_spec['dynamics_model_params'] = get_dynamics_model_params(universe, domain, task)\n variant_spec['data_params'] = {\n 'n_goal_examples': n_goal_examples,\n 'n_goal_examples_validation_max': 100,\n }\n\n ## For meta-NML, assign rewards once when states are encountered,\n ## instead of recomputing them each time during training\n if (variant_spec['algorithm_params']['kwargs'].get('use_meta_nml', False) or \n variant_spec['algorithm_params']['kwargs'].get('use_laplace_smoothing_rewards', False)):\n print(\"[Meta-NML] Using ClassifierSampler rewards\")\n variant_spec['sampler_params']['type'] = 'ClassifierSampler'\n if from_vision:\n params = ENVIRONMENT_PARAMS_PER_UNIVERSE_DOMAIN_TASK_VISION\n else:\n params = ENVIRONMENT_PARAMS_PER_UNIVERSE_DOMAIN_TASK_STATE\n variant_spec['sampler_params']['kwargs'] = {\n **variant_spec['sampler_params'].get('kwargs', {}),\n **(CLASSIFIER_SAMPLER_PARAMS_PER_UNIVERSE_DOMAIN_TASK_VISION if from_vision \\\n else CLASSIFIER_SAMPLER_PARAMS_PER_UNIVERSE_DOMAIN_TASK)\n .get(universe, {}).get(domain, {}).get(task, {}),\n }\n # Add classifier rewards to the replay pool\n from softlearning.replay_pools.flexible_replay_pool import Field\n variant_spec['replay_pool_params']['kwargs']['extra_fields'] = {\n 'learned_rewards': Field(\n name='learned_rewards',\n dtype='float32',\n shape=(1, )\n )\n }\n\n if algorithm in ['RAQ', 'VICERAQ']:\n if task in DOOR_TASKS:\n is_goal_key = 'angle_success'\n elif task in PUSH_TASKS:\n is_goal_key = 'puck_success'\n elif task in PICK_TASKS:\n is_goal_key = 'obj_success'\n else:\n raise NotImplementedError('Success metric not defined for task')\n\n variant_spec.update({\n 'sampler_params': {\n 'type': 'ActiveSampler',\n 'kwargs': {\n 'is_goal_key': is_goal_key,\n 'max_path_length': get_max_path_length(universe, domain, task),\n 'min_pool_size': get_max_path_length(universe, domain, task),\n 'batch_size': 256,\n }\n },\n 'replay_pool_params': {\n 'type': 'ActiveReplayPool',\n 'kwargs': {\n 'max_size': 1e6,\n }\n },\n })\n return variant_spec\n\n\nCLASSIFIER_ALGS = (\n 'SACClassifier',\n 'RAQ',\n 'VICE',\n 'VICEDynamicsAware',\n 'DynamicsAwareEmbeddingVICE',\n 'VICEGAN',\n 'VICERAQ',\n 'VICEGANTwoGoal',\n 'VICEGANMultiGoal',\n 'MultiVICEGAN'\n)\n\n\ndef get_variant_spec(args):\n universe, domain = args.universe, args.domain\n task, task_eval, algorithm, n_epochs = (\n args.task, args.task_evaluation, args.algorithm, args.n_epochs)\n\n from_vision = args.vision\n\n if algorithm in CLASSIFIER_ALGS:\n variant_spec = get_variant_spec_classifier(\n universe, domain, task, task_eval, args.policy, algorithm,\n args.n_goal_examples, from_vision)\n else:\n variant_spec = get_variant_spec_base(\n universe, domain, task, task_eval, args.policy, algorithm, from_vision)\n\n if args.algorithm in ('RAQ', 'VICERAQ'):\n active_query_frequency = args.active_query_frequency\n variant_spec['algorithm_params']['kwargs'][\n 'active_query_frequency'] = active_query_frequency\n\n variant_spec['algorithm_params']['kwargs']['n_epochs'] = n_epochs\n\n preprocessor_type = args.preprocessor_type\n\n if is_image_env(universe, domain, task, variant_spec):\n assert preprocessor_type in PIXELS_PREPROCESSOR_PARAMS\n preprocessor_params = PIXELS_PREPROCESSOR_PARAMS[preprocessor_type]\n\n variant_spec['policy_params']['kwargs']['hidden_layer_sizes'] = (M, M)\n variant_spec['policy_params']['kwargs'][\n 'observation_preprocessors_params'] = {\n 'pixels': deepcopy(preprocessor_params)\n }\n\n variant_spec['Q_params']['kwargs']['hidden_layer_sizes'] = (\n tune.sample_from(lambda spec: (deepcopy(\n spec.get('config', spec)\n ['policy_params']\n ['kwargs']\n ['hidden_layer_sizes']\n )))\n )\n variant_spec['Q_params']['kwargs'][\n 'observation_preprocessors_params'] = (\n tune.sample_from(lambda spec: (deepcopy(\n spec.get('config', spec)\n ['policy_params']\n ['kwargs']\n ['observation_preprocessors_params']\n )))\n )\n if args.algorithm in CLASSIFIER_ALGS:\n reward_classifier_preprocessor_params = {\n 'type': 'ConvnetPreprocessor',\n 'kwargs': {\n 'conv_filters': (64, 64, 64),\n 'conv_kernel_sizes': (3, ) * 3,\n 'conv_strides': (2, 2, 2),\n 'normalization_type': None,\n 'downsampling_type': 'conv',\n 'output_kwargs': {\n 'type': 'flatten',\n }\n },\n }\n (variant_spec\n ['reward_classifier_params']\n ['kwargs']\n ['observation_preprocessors_params']) = {\n 'pixels': reward_classifier_preprocessor_params\n }\n\n if args.checkpoint_replay_pool is not None:\n variant_spec['run_params']['checkpoint_replay_pool'] = (\n args.checkpoint_replay_pool)\n\n return variant_spec\n"
]
| [
[
"numpy.arange",
"numpy.array",
"numpy.random.randint"
]
]
|
amirziai/cs224n-project | [
"64f62d8894d2cea10c96c4c83815aa404e1e1056",
"64f62d8894d2cea10c96c4c83815aa404e1e1056"
]
| [
"experiments.py",
"archive/pytorch/run.py"
]
| [
"import os\nfrom itertools import product\nfrom typing import Dict, Set, List, Any, Tuple\nfrom uuid import uuid4\n\nimport pandas as pd\nfrom joblib import Parallel, delayed\n\nimport params\nimport run\nfrom run import ExperimentResults\nfrom utils import log, merge_dicts, uuid_to_str, pickle_object, unpickle\n\nParamSet = Dict[str, Any]\nParamGrid = List[ParamSet]\nRunnerUUID = str\n\n\nclass ExperimentRunner:\n def __init__(self, experiment_parameters: Dict[str, Set[Any]], n_jobs: int):\n self.experiment_parameters = experiment_parameters\n self.n_jobs = n_jobs\n self.uuid: str = uuid_to_str(uuid4())\n\n @staticmethod\n def _get_param_grid(parameters: Dict[str, Set[Any]]) -> ParamGrid:\n return [dict(zip(parameters.keys(), t)) for t in product(*parameters.values())]\n\n @staticmethod\n def _file_path_experiment_results(runner_uuid: RunnerUUID) -> str:\n return f'results/{runner_uuid}_experiment_results.pkl'\n\n def _experiment_result_exists(self, runner_uuid: RunnerUUID) -> bool:\n return os.path.isfile(self._file_path_experiment_results(runner_uuid))\n\n def _param_run(self, param_set: ParamSet) -> Tuple[ExperimentResults, RunnerUUID]:\n log(f'Running param set: {param_set}')\n runner = run.Runner(**param_set)\n if self._experiment_result_exists(runner.uuid):\n log('Loading experiment results from cache')\n experiment_results = unpickle(self._file_path_experiment_results(runner.uuid))\n else:\n experiment_results = runner.run()\n pickle_object(experiment_results, self._file_path_experiment_results(runner.uuid))\n\n return experiment_results, runner.uuid\n\n def run(self):\n param_grid = self._get_param_grid(self.experiment_parameters)\n if self.n_jobs > 1:\n run_output = Parallel(n_jobs=self.n_jobs)(delayed(self._param_run)(param) for param in param_grid)\n else:\n run_output = [self._param_run(param) for param in param_grid]\n results_enriched = [\n merge_dicts(result._asdict(), param_set, {'runner_uuid': runner_uuid}, {'experiment_uuid': self.uuid})\n for (result, runner_uuid), param_set in zip(run_output, param_grid)\n ]\n pd.DataFrame(results_enriched).to_csv(f'results/results_{self.uuid}.csv', index=False)\n\n\nif __name__ == '__main__':\n experiment1 = ExperimentRunner(params.experiment1, n_jobs=params.n_jobs)\n experiment1.run()\n",
"#!/usr/bin/env python3\n# -*- coding: utf-8 -*-\n\n\"\"\"\nSemantic parsing\nrun.py: Run Script for semantic parsing given a domain\nAmir Ziai <[email protected]>\nAdapted from CS224N NMT assignment code\n\nUsage:\n run.py train --file-path-train=<file> --file-path-dev=<file> [options]\n run.py decode --file-path-train=<file> --file-path-dev=<file> [options]\n run.py train decode --file-path-train=<file> --file-path-dev=<file> [options]\n\nOptions:\n -h --help show this screen.\n --domain-name=<str> dataset domain name [default: geoquery]\n --file-path-train=<file> train file\n --file-path-dev=<file> dev file\n --vocab-size=<int> vocab size [default: 10000]\n --seed=<int> seed [default: 0]\n --freq-cutoff=<int> tokens less freuqent than this are cut out from vocab [default: 1]\n --embed-size=<int> embedding size [default: 256]\n --hidden-size=<int> hidden size [default: 256]\n --dropout=<float> dropout [default: 0.3]\n --uniform-init=<float> uniformly initialize all parameters [default: 0.1]\n --lr=<float> learning rate [default: 0.001]\n --lr-decay=<float> learning rate decay [default: 0.5]\n --cuda use GPU\n --batch-size-train=<int> batch size for training [default: 32]\n --batch-size-dev=<int> batch size for dev [default: 128]\n --clip-grad=<float> gradient clipping [default: 5.0]\n --log-every=<int> log every [default: 10]\n --valid-niter=<int> perform validation after how many iterations [default: 100]\n --max-epoch=<int> max epoch [default: 30]\n --patience=<int> wait for how many iterations to decay learning rate [default: 5]\n --file-path-model=<file> model save path [default: model.bin]\n --max-num-trial=<int> terminate training after how many trials [default: 5]\n --beam-size=<int> beam size [default: 5]\n --max-decoding-time-step=<int> maximum number of decoding time steps [default: 70]\n\"\"\"\nimport math\nimport sys\nimport time\nfrom typing import List\n\nimport numpy as np\nimport torch\nfrom docopt import docopt\nfrom tqdm import tqdm\n\nimport domains\nfrom domains import DomainData\nfrom nmt_model import NMT, Hypothesis\nfrom utils import log, batch_iter, unzip\nfrom vocab import Vocab\n\n\nclass Runner:\n def __init__(self,\n domain_name: str,\n file_path_train: str,\n file_path_dev: str,\n vocab_size: int,\n seed: int,\n freq_cutoff: int,\n embed_size: int,\n hidden_size: int,\n dropout: float,\n uniform_init: float,\n lr: float,\n lr_decay: float,\n cuda: bool,\n batch_size_train: int,\n batch_size_dev: int,\n clip_grad: float,\n log_every: int,\n valid_niter: int,\n max_epoch: int,\n patience: int,\n file_path_model: str,\n max_num_trial: int,\n beam_size: int,\n max_decoding_time_step: int):\n # params\n self.domain_name = domain_name\n self.file_path_train = file_path_train\n self.file_path_dev = file_path_dev\n self.vocab_size = vocab_size\n self.freq_cutoff = freq_cutoff\n self.embed_size = embed_size\n self.hidden_size = hidden_size\n self.dropout = dropout\n self.uniform_init = uniform_init\n self.lr = lr\n self.lr_decay = lr_decay\n self.cuda = cuda\n self.batch_size_train = batch_size_train\n self.batch_size_dev = batch_size_dev\n self.clip_grad = clip_grad\n self.log_every = log_every\n self.valid_niter = valid_niter\n self.max_epoch = max_epoch\n self.patience = patience\n self.file_path_model = file_path_model\n self.max_num_trial = max_num_trial\n self.beam_size = beam_size\n self.max_decoding_time_step = max_decoding_time_step\n self.seed = seed\n\n # seed the random number generators\n torch.manual_seed(self.seed)\n if self.cuda:\n torch.cuda.manual_seed(self.seed)\n np.random.seed(self.seed)\n\n # important things to populate later\n self.domain = None\n self.data_train = None\n self.data_dev = None\n self.vocab = None\n self.model = None\n self.device = None\n self.optimizer = None\n\n @staticmethod\n def _load_data(file_path: str, domain: domains.Domain) -> DomainData:\n dataset = []\n with open(file_path) as f:\n for line in f:\n x_str, y_str = line.rstrip('\\n').split('\\t')\n if domain:\n y_str = domain.preprocess_lf(y_str)\n\n x = x_str.split()\n # TODO: </s> seems to already exist somewhere\n # y = ['<s>'] + y_str.split() + ['</s>']\n y = ['<s>'] + y_str.split() # + ['</s>']\n dataset.append((x, y))\n\n return dataset\n\n def _init_params(self):\n if np.abs(self.uniform_init) > 0.:\n log('uniformly initialize parameters [-%f, +%f]' % (self.uniform_init, self.uniform_init))\n for p in self.model.parameters():\n p.data.uniform_(-self.uniform_init, self.uniform_init)\n\n def _vocab_mask(self) -> None:\n vocab_mask = torch.ones(len(self.vocab.tgt))\n vocab_mask[self.vocab.tgt['<pad>']] = 0\n\n def _init_device(self) -> None:\n self.device = torch.device(\"cuda:0\" if self.cuda else \"cpu\")\n log('use device: %s' % self.device)\n self.model = self.model.to(self.device)\n\n def _init_optimizer(self) -> None:\n self.optimizer = torch.optim.Adam(self.model.parameters(), lr=self.lr)\n\n def _evaluate_perplexity(self) -> float:\n was_training = self.model.training\n self.model.eval()\n\n cum_loss = 0.\n cum_tgt_words = 0.\n\n # no_grad() signals backend to throw away all gradients\n with torch.no_grad():\n for src_sents, tgt_sents in batch_iter(self.data_dev, self.batch_size_dev):\n loss = -self.model(src_sents, tgt_sents).sum()\n\n cum_loss += loss.item()\n tgt_word_num_to_predict = sum(len(s[1:]) for s in tgt_sents) # omitting leading `<s>`\n cum_tgt_words += tgt_word_num_to_predict\n\n ppl = np.exp(cum_loss / cum_tgt_words)\n\n if was_training:\n self.model.train()\n\n return ppl\n\n def _train_loop(self):\n num_trial = 0\n train_iter = patience = cum_loss = report_loss = cum_tgt_words = report_tgt_words = 0\n cum_examples = report_examples = epoch = valid_num = 0\n hist_valid_scores = []\n train_time = begin_time = time.time()\n log('begin Maximum Likelihood training')\n\n log(f'Train dataset has {len(self.data_train)} items')\n early_stop = False\n\n while epoch < self.max_epoch and not early_stop:\n epoch += 1\n\n # TODO: add augmenter here to extend data\n # TODO: add concatenated examples\n\n for src_sents, tgt_sents in batch_iter(self.data_train, batch_size=self.batch_size_train, shuffle=True):\n train_iter += 1\n\n self.optimizer.zero_grad()\n\n batch_size = len(src_sents)\n\n example_losses = -self.model(src_sents, tgt_sents) # (batch_size,)\n batch_loss = example_losses.sum()\n loss = batch_loss / batch_size\n\n loss.backward()\n\n # clip gradient\n grad_norm = torch.nn.utils.clip_grad_norm_(self.model.parameters(), self.clip_grad)\n\n self.optimizer.step()\n\n batch_losses_val = batch_loss.item()\n report_loss += batch_losses_val\n cum_loss += batch_losses_val\n\n # TODO: why remove <s>?\n tgt_words_num_to_predict = sum(len(s[1:]) for s in tgt_sents) # omitting leading `<s>`\n report_tgt_words += tgt_words_num_to_predict\n cum_tgt_words += tgt_words_num_to_predict\n report_examples += batch_size\n cum_examples += batch_size\n\n if train_iter % self.log_every == 0:\n log('epoch %d, iter %d, avg. loss %.2f, avg. ppl %.2f ' \\\n 'cum. examples %d, speed %.2f words/sec, time elapsed %.2f sec' % (epoch, train_iter,\n report_loss /\n report_examples,\n math.exp(\n report_loss /\n report_tgt_words),\n cum_examples,\n report_tgt_words / (\n time.time() -\n train_time),\n time.time() - begin_time))\n train_time = time.time()\n report_loss = report_tgt_words = report_examples = 0.\n\n # perform validation\n if train_iter % self.valid_niter == 0:\n log('epoch %d, iter %d, cum. loss %.2f, cum. ppl %.2f cum. examples %d' % (epoch,\n train_iter,\n cum_loss / cum_examples,\n np.exp(cum_loss /\n cum_tgt_words),\n cum_examples))\n\n cum_loss = cum_examples = cum_tgt_words = 0.\n valid_num += 1\n\n print('begin validation ...', file=sys.stderr)\n\n # dev batch size can be a bit larger\n dev_ppl = self._evaluate_perplexity()\n valid_metric = -dev_ppl\n\n print('validation: iter %d, dev. ppl %f' % (train_iter, dev_ppl), file=sys.stderr)\n\n is_better = len(hist_valid_scores) == 0 or valid_metric > max(hist_valid_scores)\n hist_valid_scores.append(valid_metric)\n\n if is_better:\n patience = 0\n log('save currently the best model to [%s]' % self.file_path_model)\n self.model.save(self.file_path_model)\n\n # also save the optimizers' state\n torch.save(self.optimizer.state_dict(), self.file_path_model + '.optim')\n elif patience < self.patience:\n patience += 1\n print('hit patience %d' % patience, file=sys.stderr)\n\n if patience == self.patience:\n num_trial += 1\n print('hit #%d trial' % num_trial, file=sys.stderr)\n if num_trial == self.max_num_trial:\n log('early stop!')\n early_stop = True\n break\n\n # decay lr, and restore from previously best checkpoint\n lr = self.optimizer.param_groups[0]['lr'] * float(self.lr_decay)\n log('load previously best model and decay learning rate to %f' % lr)\n\n # load model\n params = torch.load(self.file_path_model, map_location=lambda storage, loc: storage)\n self.model.load_state_dict(params['state_dict'])\n self.model = self.model.to(self.device)\n\n log('restore parameters of the optimizers')\n self.optimizer.load_state_dict(torch.load(self.file_path_model + '.optim'))\n\n # set new lr\n for param_group in self.optimizer.param_groups:\n param_group['lr'] = lr\n\n # reset patience\n patience = 0\n\n if not early_stop and epoch >= self.max_epoch:\n log(f'reached epoch {epoch} which is >= {self.max_epoch} maximum number of epochs!')\n\n def train(self) -> None:\n log('training')\n\n # get data by domain\n self.domain = domains.new(self.domain_name)\n self.data_train = self._load_data(self.file_path_train, self.domain)\n self.data_dev = self._load_data(self.file_path_dev, self.domain)\n train_src, train_tgt = unzip(self.data_train)\n\n # build the vocabulary from the training data\n self.vocab = Vocab.build(train_src, train_tgt, vocab_size=self.vocab_size, freq_cutoff=self.freq_cutoff)\n\n # build the model with the provided params\n self.model = NMT(embed_size=self.embed_size,\n hidden_size=self.hidden_size, dropout_rate=self.dropout, vocab=self.vocab)\n\n # model architecture and parameters\n log(str(self.model.train()))\n\n # init\n self._init_params()\n self._vocab_mask()\n self._init_optimizer()\n\n # train\n self._train_loop()\n\n log('training done!')\n\n def _load_model(self) -> None:\n if not self.model:\n log(\"load model from {}\".format(self.file_path_model))\n self.model = NMT.load(self.file_path_model)\n self._init_device()\n\n @staticmethod\n def _log_accuracy_metrics(is_correct_list: List[int], tokens_correct_list: List[int], y_len_list: List[int],\n denotation_correct_list: List) -> None:\n # Overall metrics\n num_examples = len(is_correct_list)\n num_correct = sum(is_correct_list)\n num_tokens_correct = sum(tokens_correct_list)\n num_tokens = sum(y_len_list)\n seq_accuracy = float(num_correct) / num_examples\n token_accuracy = float(num_tokens_correct) / num_tokens\n\n # sequence-level accuracy\n log('Sequence-level accuracy: %d/%d = %g' % (num_correct, num_examples, seq_accuracy))\n log('Token-level accuracy: %d/%d = %g' % (num_tokens_correct, num_tokens, token_accuracy))\n\n # denotation-level accuracy\n if denotation_correct_list:\n denotation_correct = sum(denotation_correct_list)\n denotation_accuracy = float(denotation_correct) / num_examples\n log('Denotation-level accuracy: %d/%d = %g' % (denotation_correct, num_examples, denotation_accuracy))\n\n def _evaluate(self, data: DomainData, hypotheses: List[List[Hypothesis]]) -> None:\n xs, ys = unzip(data)\n true_answers = [' '.join(y) for y in ys]\n\n derivs, denotation_correct_list = self.domain.compare_answers(true_answers, hypotheses)\n\n is_correct_list = []\n tokens_correct_list = []\n y_len_list = []\n\n for x, y, y_str, deriv in zip(xs, ys, true_answers, derivs):\n y_pred_toks = deriv.value\n y_pred_str = ' '.join(y_pred_toks)\n\n # Compute accuracy metrics\n is_correct = (y_pred_str == y_str)\n tokens_correct = sum(a == b for a, b in zip(y_pred_toks, y))\n is_correct_list.append(is_correct)\n tokens_correct_list.append(tokens_correct)\n y_len_list.append(len(y))\n\n self._log_accuracy_metrics(is_correct_list, tokens_correct_list, y_len_list, denotation_correct_list)\n\n def decode(self) -> None:\n # data\n log(\"load test source sentences from [{}]\".format(self.file_path_dev))\n if not self.data_dev:\n self.data_dev = self._load_data(self.file_path_dev, self.domain)\n if not self.domain:\n self.domain = domains.new(self.domain_name)\n\n dev_src, dev_tgt = unzip(self.data_dev)\n\n # model\n self._load_model()\n\n hypotheses = self.beam_search(dev_src)\n self._evaluate(self.data_dev, hypotheses)\n\n def beam_search(self, data_src: List[List[str]]) -> List[List[Hypothesis]]:\n \"\"\"\n Run beam search to construct hypotheses for a list of natural language utterances.\n @param data_src: List of list of tokens.\n @returns hypotheses: List of Hypothesis logical forms.\n \"\"\"\n was_training = self.model.training\n self.model.eval()\n\n hypotheses = []\n with torch.no_grad():\n for src_sent in tqdm(data_src, desc='Decoding', file=sys.stdout):\n example_hyps = self.model.beam_search(src_sent, beam_size=self.beam_size,\n max_decoding_time_step=self.max_decoding_time_step)\n\n hypotheses.append(example_hyps)\n\n if was_training:\n self.model.train(was_training)\n\n return hypotheses\n\n\ndef main() -> None:\n \"\"\"\n Parse args and run.\n \"\"\"\n args = docopt(__doc__)\n\n # Check PyTorch version\n assert torch.__version__ == \"1.0.0\", f\"You have PyTorch=={torch.__version__} and you should have version 1.0.0\"\n\n runner = Runner(\n domain_name=args['--domain-name'],\n file_path_train=args['--file-path-train'],\n file_path_dev=args['--file-path-dev'],\n vocab_size=int(args['--vocab-size']),\n seed=int(args['--seed']),\n freq_cutoff=int(args['--freq-cutoff']),\n embed_size=int(args['--embed-size']),\n hidden_size=int(args['--hidden-size']),\n dropout=float(args['--dropout']),\n uniform_init=float(args['--uniform-init']),\n lr=float(args['--lr']),\n lr_decay=float(args['--lr-decay']),\n cuda=args['--cuda'],\n batch_size_train=int(args['--batch-size-train']),\n batch_size_dev=int(args['--batch-size-dev']),\n clip_grad=float(args['--clip-grad']),\n log_every=int(args['--log-every']),\n valid_niter=int(args['--valid-niter']),\n max_epoch=int(args['--max-epoch']),\n patience=int(args['--patience']),\n file_path_model=args['--file-path-model'],\n max_num_trial=int(args['--max-num-trial']),\n beam_size=int(args['--beam-size']),\n max_decoding_time_step=int(args['--max-decoding-time-step'])\n )\n\n if args['train']:\n runner.train()\n\n if args['decode']:\n runner.decode()\n\n\nif __name__ == '__main__':\n main()\n"
]
| [
[
"pandas.DataFrame"
],
[
"numpy.abs",
"torch.cuda.manual_seed",
"numpy.random.seed",
"torch.load",
"torch.manual_seed",
"torch.no_grad",
"torch.device",
"numpy.exp"
]
]
|
gabririgo/hummingbot | [
"7ae2c7cc04f0bfdb2838371450b838872db314c9"
]
| [
"test/test_cross_exchange_market_making.py"
]
| [
"#!/usr/bin/env python\n\nfrom os.path import join, realpath\nimport sys;\n\n\nsys.path.insert(0, realpath(join(__file__, \"../../\")))\n\nfrom nose.plugins.attrib import attr\n\nfrom hummingbot.strategy.market_symbol_pair import MarketSymbolPair\nfrom decimal import Decimal\nimport logging; logging.basicConfig(level=logging.ERROR)\nimport pandas as pd\nfrom typing import List\nimport unittest\n\nfrom hummingbot.core.utils.exchange_rate_conversion import ExchangeRateConversion\nfrom hummingsim.backtest.backtest_market import BacktestMarket\nfrom hummingsim.backtest.market import (\n AssetType,\n Market,\n MarketConfig,\n QuantizationParams\n)\nfrom hummingsim.backtest.mock_order_book_loader import MockOrderBookLoader\nfrom hummingbot.core.clock import (\n Clock,\n ClockMode\n)\nfrom hummingbot.core.event.event_logger import EventLogger\nfrom hummingbot.core.event.events import (\n MarketEvent,\n OrderBookTradeEvent,\n TradeType,\n OrderType,\n OrderFilledEvent,\n BuyOrderCompletedEvent,\n SellOrderCompletedEvent,\n TradeFee,\n)\nfrom hummingbot.core.data_type.order_book import OrderBook\nfrom hummingbot.core.data_type.order_book_row import OrderBookRow\nfrom hummingbot.core.data_type.limit_order import LimitOrder\nfrom hummingbot.strategy.cross_exchange_market_making import CrossExchangeMarketMakingStrategy\nfrom hummingbot.strategy.cross_exchange_market_making.cross_exchange_market_pair import CrossExchangeMarketPair\n\n\n@attr('stable')\nclass HedgedMarketMakingUnitTest(unittest.TestCase):\n start: pd.Timestamp = pd.Timestamp(\"2019-01-01\", tz=\"UTC\")\n end: pd.Timestamp = pd.Timestamp(\"2019-01-01 01:00:00\", tz=\"UTC\")\n start_timestamp: float = start.timestamp()\n end_timestamp: float = end.timestamp()\n maker_symbols: List[str] = [\"COINALPHA-WETH\", \"COINALPHA\", \"WETH\"]\n taker_symbols: List[str] = [\"coinalpha/eth\", \"COINALPHA\", \"ETH\"]\n\n @classmethod\n def setUpClass(cls):\n ExchangeRateConversion.set_global_exchange_rate_config({\n \"conversion_required\": {\n \"WETH\": {\"default\": 1.0, \"source\": \"None\"},\n \"QETH\": {\"default\": 0.95, \"source\": \"None\"}\n }\n })\n\n def setUp(self):\n self.clock: Clock = Clock(ClockMode.BACKTEST, 1.0, self.start_timestamp, self.end_timestamp)\n self.maker_market: BacktestMarket = BacktestMarket()\n self.taker_market: BacktestMarket = BacktestMarket()\n self.maker_data: MockOrderBookLoader = MockOrderBookLoader(*self.maker_symbols)\n self.taker_data: MockOrderBookLoader = MockOrderBookLoader(*self.taker_symbols)\n self.maker_data.set_balanced_order_book(1.0, 0.5, 1.5, 0.01, 10)\n self.taker_data.set_balanced_order_book(1.0, 0.5, 1.5, 0.001, 4)\n self.maker_market.add_data(self.maker_data)\n self.taker_market.add_data(self.taker_data)\n self.maker_market.set_balance(\"COINALPHA\", 5)\n self.maker_market.set_balance(\"WETH\", 5)\n self.maker_market.set_balance(\"QETH\", 5)\n self.taker_market.set_balance(\"COINALPHA\", 5)\n self.taker_market.set_balance(\"ETH\", 5)\n self.maker_market.set_quantization_param(\n QuantizationParams(\n self.maker_symbols[0], 5, 5, 5, 5\n )\n )\n self.taker_market.set_quantization_param(\n QuantizationParams(\n self.taker_symbols[0], 5, 5, 5, 5\n )\n )\n\n self.market_pair: CrossExchangeMarketPair = CrossExchangeMarketPair(\n MarketSymbolPair(self.maker_market, *self.maker_symbols),\n MarketSymbolPair(self.taker_market, *self.taker_symbols),\n 2\n )\n\n logging_options: int = (CrossExchangeMarketMakingStrategy.OPTION_LOG_ALL &\n (~CrossExchangeMarketMakingStrategy.OPTION_LOG_NULL_ORDER_SIZE))\n self.strategy: CrossExchangeMarketMakingStrategy = CrossExchangeMarketMakingStrategy(\n [self.market_pair],\n 0.0004,\n order_size_portfolio_ratio_limit=0.3,\n logging_options=logging_options\n )\n self.logging_options = logging_options\n self.clock.add_iterator(self.maker_market)\n self.clock.add_iterator(self.taker_market)\n self.clock.add_iterator(self.strategy)\n\n self.maker_order_fill_logger: EventLogger = EventLogger()\n self.taker_order_fill_logger: EventLogger = EventLogger()\n self.cancel_order_logger: EventLogger = EventLogger()\n self.maker_market.add_listener(MarketEvent.OrderFilled, self.maker_order_fill_logger)\n self.taker_market.add_listener(MarketEvent.OrderFilled, self.taker_order_fill_logger)\n self.maker_market.add_listener(MarketEvent.OrderCancelled, self.cancel_order_logger)\n\n def simulate_maker_market_trade(self, is_buy: bool, quantity: float):\n maker_symbol: str = self.maker_symbols[0]\n order_book: OrderBook = self.maker_market.get_order_book(maker_symbol)\n trade_price: float = order_book.get_price(True) if is_buy else order_book.get_price(False)\n trade_event: OrderBookTradeEvent = OrderBookTradeEvent(\n maker_symbol,\n self.clock.current_timestamp,\n TradeType.BUY if is_buy else TradeType.SELL,\n trade_price,\n quantity\n )\n order_book.apply_trade(trade_event)\n\n @staticmethod\n def simulate_order_book_widening(order_book: OrderBook, top_bid: float, top_ask: float):\n bid_diffs: List[OrderBookRow] = []\n ask_diffs: List[OrderBookRow] = []\n update_id: int = order_book.last_diff_uid + 1\n for row in order_book.bid_entries():\n if row.price > top_bid:\n bid_diffs.append(OrderBookRow(row.price, 0, update_id))\n else:\n break\n for row in order_book.ask_entries():\n if row.price < top_ask:\n ask_diffs.append(OrderBookRow(row.price, 0, update_id))\n else:\n break\n order_book.apply_diffs(bid_diffs, ask_diffs, update_id)\n\n @staticmethod\n def simulate_limit_order_fill(market: Market, limit_order: LimitOrder):\n quote_currency_traded: float = float(limit_order.price * limit_order.quantity)\n base_currency_traded: float = float(limit_order.quantity)\n quote_currency: str = limit_order.quote_currency\n base_currency: str = limit_order.base_currency\n config: MarketConfig = market.config\n\n if limit_order.is_buy:\n market.set_balance(quote_currency, market.get_balance(quote_currency) - quote_currency_traded)\n market.set_balance(base_currency, market.get_balance(base_currency) + base_currency_traded)\n market.trigger_event(MarketEvent.OrderFilled, OrderFilledEvent(\n market.current_timestamp,\n limit_order.client_order_id,\n limit_order.symbol,\n TradeType.BUY,\n OrderType.LIMIT,\n float(limit_order.price),\n float(limit_order.quantity),\n TradeFee(0.0)\n ))\n market.trigger_event(MarketEvent.BuyOrderCompleted, BuyOrderCompletedEvent(\n market.current_timestamp,\n limit_order.client_order_id,\n base_currency,\n quote_currency,\n base_currency if config.buy_fees_asset is AssetType.BASE_CURRENCY else quote_currency,\n base_currency_traded,\n quote_currency_traded,\n 0.0,\n OrderType.LIMIT\n ))\n else:\n market.set_balance(quote_currency, market.get_balance(quote_currency) + quote_currency_traded)\n market.set_balance(base_currency, market.get_balance(base_currency) - base_currency_traded)\n market.trigger_event(MarketEvent.OrderFilled, OrderFilledEvent(\n market.current_timestamp,\n limit_order.client_order_id,\n limit_order.symbol,\n TradeType.SELL,\n OrderType.LIMIT,\n float(limit_order.price),\n float(limit_order.quantity),\n TradeFee(0.0)\n ))\n market.trigger_event(MarketEvent.SellOrderCompleted, SellOrderCompletedEvent(\n market.current_timestamp,\n limit_order.client_order_id,\n base_currency,\n quote_currency,\n base_currency if config.sell_fees_asset is AssetType.BASE_CURRENCY else quote_currency,\n base_currency_traded,\n quote_currency_traded,\n 0.0,\n OrderType.LIMIT\n ))\n\n def test_both_sides_profitable(self):\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(1, len(self.strategy.active_asks))\n\n bid_order: LimitOrder = self.strategy.active_bids[0][1]\n ask_order: LimitOrder = self.strategy.active_asks[0][1]\n self.assertEqual(Decimal(\"0.99501\"), bid_order.price)\n self.assertEqual(Decimal(\"1.0049\"), ask_order.price)\n self.assertEqual(Decimal(\"3.0\"), bid_order.quantity)\n self.assertEqual(Decimal(\"3.0\"), ask_order.quantity)\n\n self.simulate_maker_market_trade(True, 1.0)\n\n self.clock.backtest_til(self.start_timestamp + 10)\n self.assertEqual(1, len(self.maker_order_fill_logger.event_log))\n self.assertEqual(1, len(self.taker_order_fill_logger.event_log))\n\n maker_fill: OrderFilledEvent = self.maker_order_fill_logger.event_log[0]\n taker_fill: OrderFilledEvent = self.taker_order_fill_logger.event_log[0]\n self.assertEqual(TradeType.SELL, maker_fill.trade_type)\n self.assertEqual(TradeType.BUY, taker_fill.trade_type)\n self.assertAlmostEqual(1.0049, maker_fill.price)\n self.assertAlmostEqual(1.0005, taker_fill.price)\n self.assertAlmostEqual(3.0, maker_fill.amount)\n self.assertAlmostEqual(3.0, taker_fill.amount)\n\n def test_bid_side_profitable(self):\n self.maker_data.order_book.apply_diffs([], [OrderBookRow(1.0, 5, 2)], 2)\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(0, len(self.strategy.active_asks))\n\n def test_ask_side_profitable(self):\n self.maker_data.order_book.apply_diffs([OrderBookRow(1.0, 5, 2)], [], 2)\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(0, len(self.strategy.active_bids))\n self.assertEqual(1, len(self.strategy.active_asks))\n\n def test_neither_side_profitable(self):\n self.simulate_order_book_widening(self.taker_data.order_book, 0.95, 1.05)\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(0, len(self.strategy.active_bids))\n self.assertEqual(0, len(self.strategy.active_asks))\n\n def test_market_changed_to_unprofitable(self):\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(1, len(self.strategy.active_asks))\n self.assertEqual(0, len(self.cancel_order_logger.event_log))\n\n self.maker_data.order_book.apply_diffs([], [OrderBookRow(1.0, 15, 2)], 2)\n self.clock.backtest_til(self.start_timestamp + 10)\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(0, len(self.strategy.active_asks))\n self.assertEqual(1, len(self.cancel_order_logger.event_log))\n\n def test_market_became_wider(self):\n self.clock.backtest_til(self.start_timestamp + 5)\n\n bid_order: LimitOrder = self.strategy.active_bids[0][1]\n ask_order: LimitOrder = self.strategy.active_asks[0][1]\n self.assertEqual(Decimal(\"0.99501\"), bid_order.price)\n self.assertEqual(Decimal(\"1.0049\"), ask_order.price)\n self.assertEqual(Decimal(\"3.0\"), bid_order.quantity)\n self.assertEqual(Decimal(\"3.0\"), ask_order.quantity)\n\n self.simulate_order_book_widening(self.maker_data.order_book, 0.99, 1.01)\n\n self.clock.backtest_til(self.start_timestamp + 90)\n self.assertEqual(2, len(self.cancel_order_logger.event_log))\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(1, len(self.strategy.active_asks))\n\n bid_order = self.strategy.active_bids[0][1]\n ask_order = self.strategy.active_asks[0][1]\n self.assertEqual(Decimal(\"0.98501\"), bid_order.price)\n self.assertEqual(Decimal(\"1.0149\"), ask_order.price)\n\n def test_market_became_narrower(self):\n self.clock.backtest_til(self.start_timestamp + 5)\n bid_order: LimitOrder = self.strategy.active_bids[0][1]\n ask_order: LimitOrder = self.strategy.active_asks[0][1]\n self.assertEqual(Decimal(\"0.99501\"), bid_order.price)\n self.assertEqual(Decimal(\"1.0049\"), ask_order.price)\n self.assertEqual(Decimal(\"3.0\"), bid_order.quantity)\n self.assertEqual(Decimal(\"3.0\"), ask_order.quantity)\n\n self.maker_data.order_book.apply_diffs([OrderBookRow(0.996, 30, 2)], [OrderBookRow(1.004, 30, 2)], 2)\n\n self.clock.backtest_til(self.start_timestamp + 10)\n self.assertEqual(2, len(self.cancel_order_logger.event_log))\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(1, len(self.strategy.active_asks))\n\n bid_order = self.strategy.active_bids[0][1]\n ask_order = self.strategy.active_asks[0][1]\n self.assertEqual(Decimal(\"0.99601\"), bid_order.price)\n self.assertEqual(Decimal(\"1.0039\"), ask_order.price)\n\n def test_top_depth_tolerance(self):\n self.maker_data.order_book.apply_diffs([OrderBookRow(0.999, 0.1, 2), OrderBookRow(0.998, 0.1, 2)],\n [OrderBookRow(1.001, 0.1, 2), OrderBookRow(1.002, 0.1, 2)],\n 2)\n self.clock.backtest_til(self.start_timestamp + 5)\n bid_order: LimitOrder = self.strategy.active_bids[0][1]\n ask_order: LimitOrder = self.strategy.active_asks[0][1]\n self.assertEqual(Decimal(\"0.99501\"), bid_order.price)\n self.assertEqual(Decimal(\"1.0049\"), ask_order.price)\n self.assertEqual(Decimal(\"3.0\"), bid_order.quantity)\n self.assertEqual(Decimal(\"3.0\"), ask_order.quantity)\n\n self.maker_data.order_book.apply_diffs([OrderBookRow(0.996, 30, 3)], [], 3)\n\n self.clock.backtest_til(self.start_timestamp + 10)\n self.assertEqual(1, len(self.cancel_order_logger.event_log))\n bid_order: LimitOrder = self.strategy.active_bids[0][1]\n self.assertEqual(Decimal(\"0.99601\"), bid_order.price)\n\n def test_order_fills_after_cancellation(self):\n self.clock.backtest_til(self.start_timestamp + 5)\n bid_order: LimitOrder = self.strategy.active_bids[0][1]\n ask_order: LimitOrder = self.strategy.active_asks[0][1]\n self.assertEqual(Decimal(\"0.99501\"), bid_order.price)\n self.assertEqual(Decimal(\"1.0049\"), ask_order.price)\n self.assertEqual(Decimal(\"3.0\"), bid_order.quantity)\n self.assertEqual(Decimal(\"3.0\"), ask_order.quantity)\n\n bid_order: LimitOrder = self.strategy.active_bids[0][1]\n ask_order: LimitOrder = self.strategy.active_asks[0][1]\n\n self.maker_data.order_book.apply_diffs([OrderBookRow(0.996, 30, 2)], [OrderBookRow(1.004, 30, 2)], 2)\n\n self.clock.backtest_til(self.start_timestamp + 10)\n self.assertEqual(2, len(self.cancel_order_logger.event_log))\n self.assertEqual(Decimal(\"0.99601\"), self.strategy.active_bids[0][1].price)\n self.assertEqual(Decimal(\"1.0039\"), self.strategy.active_asks[0][1].price)\n self.assertEqual(0, len(self.taker_order_fill_logger.event_log))\n\n self.clock.backtest_til(self.start_timestamp + 20)\n self.simulate_limit_order_fill(self.maker_market, bid_order)\n self.simulate_limit_order_fill(self.maker_market, ask_order)\n\n self.clock.backtest_til(self.start_timestamp + 25)\n fill_events: List[OrderFilledEvent] = self.taker_order_fill_logger.event_log\n bid_hedges: List[OrderFilledEvent] = [evt for evt in fill_events if evt.trade_type is TradeType.SELL]\n ask_hedges: List[OrderFilledEvent] = [evt for evt in fill_events if evt.trade_type is TradeType.BUY]\n self.assertEqual(1, len(bid_hedges))\n self.assertEqual(1, len(ask_hedges))\n self.assertGreater(\n self.maker_market.get_balance(self.maker_symbols[2]) + self.taker_market.get_balance(self.taker_symbols[2]),\n 10\n )\n\n def test_profitability_without_conversion(self):\n self.clock.remove_iterator(self.strategy)\n self.strategy: CrossExchangeMarketMakingStrategy = CrossExchangeMarketMakingStrategy(\n [self.market_pair],\n 0.01,\n order_size_portfolio_ratio_limit=0.3,\n logging_options=self.logging_options\n )\n self.clock.add_iterator(self.strategy)\n\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(0, len(self.strategy.active_bids))\n self.assertEqual(0, len(self.strategy.active_asks))\n self.assertEqual((False, False), self.strategy.has_market_making_profit_potential(\n self.market_pair\n ))\n\n self.clock.remove_iterator(self.strategy)\n self.strategy: CrossExchangeMarketMakingStrategy = CrossExchangeMarketMakingStrategy(\n [self.market_pair],\n 0.004,\n order_size_portfolio_ratio_limit=0.3,\n logging_options=self.logging_options\n )\n self.clock.add_iterator(self.strategy)\n self.clock.backtest_til(self.start_timestamp + 10)\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(1, len(self.strategy.active_asks))\n self.assertEqual((True, True), self.strategy.has_market_making_profit_potential(\n self.market_pair\n ))\n\n def test_profitability_with_conversion(self):\n self.clock.remove_iterator(self.strategy)\n self.market_pair: CrossExchangeMarketPair = CrossExchangeMarketPair(\n MarketSymbolPair(self.maker_market, *[\"COINALPHA-QETH\", \"COINALPHA\", \"QETH\"]),\n MarketSymbolPair(self.taker_market, *self.taker_symbols),\n 2\n )\n self.maker_data: MockOrderBookLoader = MockOrderBookLoader(\"COINALPHA-QETH\", \"COINALPHA\", \"QETH\")\n self.maker_data.set_balanced_order_book(1.05263, 0.55, 1.55, 0.01, 10)\n self.maker_market.add_data(self.maker_data)\n self.strategy: CrossExchangeMarketMakingStrategy = CrossExchangeMarketMakingStrategy(\n [self.market_pair],\n 0.01,\n order_size_portfolio_ratio_limit=0.3,\n logging_options=self.logging_options\n )\n self.clock.add_iterator(self.strategy)\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(0, len(self.strategy.active_bids))\n self.assertEqual(0, len(self.strategy.active_asks))\n self.assertEqual((False, False), self.strategy.has_market_making_profit_potential(\n self.market_pair\n ))\n\n self.clock.remove_iterator(self.strategy)\n self.strategy: CrossExchangeMarketMakingStrategy = CrossExchangeMarketMakingStrategy(\n [self.market_pair],\n 0.003,\n order_size_portfolio_ratio_limit=0.3,\n logging_options=self.logging_options\n )\n self.clock.add_iterator(self.strategy)\n self.clock.backtest_til(self.start_timestamp + 10)\n self.assertEqual(1, len(self.strategy.active_bids))\n self.assertEqual(1, len(self.strategy.active_asks))\n self.assertEqual((True, True), self.strategy.has_market_making_profit_potential(\n self.market_pair\n ))\n\n def test_price_and_size_limit_calculation(self):\n # Test the case where the profitable hedging depth is less than order size limit based on balance.\n self.taker_data.set_balanced_order_book(1.0, 0.5, 1.5, 0.001, 3)\n bid_price, bid_size_limit = self.strategy.get_market_making_price_and_size_limit(\n self.market_pair,\n True\n )\n ask_price, ask_size_limit = self.strategy.get_market_making_price_and_size_limit(\n self.market_pair,\n False\n )\n self.assertEqual((Decimal(\"0.99501\"), Decimal(\"4.9749\")), (bid_price, bid_size_limit))\n self.assertEqual((Decimal(\"1.0049\"), Decimal(\"4.9507\")), (ask_price, ask_size_limit))\n\n # Test the case where the profitable hedging depth is equal to order size limit based on balance.\n self.taker_data.set_balanced_order_book(1.0, 0.5, 1.5, 0.001, 4)\n bid_price, bid_size_limit = self.strategy.get_market_making_price_and_size_limit(\n self.market_pair,\n True\n )\n ask_price, ask_size_limit = self.strategy.get_market_making_price_and_size_limit(\n self.market_pair,\n False\n )\n self.assertEqual((Decimal(\"0.99501\"), Decimal(\"4.9749\")), (bid_price, bid_size_limit))\n self.assertEqual((Decimal(\"1.0049\"), Decimal(\"4.9507\")), (ask_price, ask_size_limit))\n\n # Test the case where the hedging trade is numerically profitable but below the min profit setting.\n self.simulate_order_book_widening(self.taker_data.order_book, 0.995, 1.005)\n bid_price, bid_size_limit = self.strategy.get_market_making_price_and_size_limit(\n self.market_pair,\n True\n )\n ask_price, ask_size_limit = self.strategy.get_market_making_price_and_size_limit(\n self.market_pair,\n False\n )\n self.assertEqual((Decimal(\"0.99501\"), Decimal(\"0\")), (bid_price, bid_size_limit))\n self.assertEqual((Decimal(\"1.0049\"), Decimal(\"0\")), (ask_price, ask_size_limit))\n\n # Make sure the strategy doesn't emit any orders in this case\n self.clock.backtest_til(self.start_timestamp + 5)\n self.assertEqual(0, len(self.strategy.active_bids))\n self.assertEqual(0, len(self.strategy.active_asks))\n self.assertEqual(0, len(self.cancel_order_logger.event_log))\n"
]
| [
[
"pandas.Timestamp"
]
]
|
Photic/mlAllTheCars | [
"b54c9efaf8626bfdbef11508664f096dee06d299"
]
| [
"src/R_functions.py"
]
| [
"# Some of the functions in this file has been copyed from https://towardsdatascience.com/how-to-write-your-favorite-r-functions-in-python-11e1e9c29089\n\ndef mean(array):\n \"\"\"\n Calculates the mean of an array/vector\n \"\"\"\n import numpy as np\n array=np.array(array)\n result= np.mean(array)\n return result\n\ndef sd(array):\n \"\"\"\n Calculates the standard deviation of an array/vector\n \"\"\"\n import statistics\n return statistics.stdev(array)\n\ndef median(array):\n \"\"\"\n Calculates the median of an array/vector\n \"\"\"\n import numpy as np\n array=np.array(array)\n result= np.median(array)\n return result\n\ndef var(array):\n \"\"\"\n Calculates the variance of an array/vector\n \"\"\"\n import statistics\n return statistics.variance(array)\n\ndef cov(x,y=None):\n \"\"\"\n Calculates the covariance between two arrays/vectors or of a single matrix\n \"\"\"\n import numpy as np\n array1=np.array(x)\n if y!=None:\n array2=np.array(y)\n if array1.shape!=array2.shape:\n print(\"Error: incompatible dimensions\")\n return None\n covmat=np.cov(array1,array2)\n result=covmat[0][1]\n elif len(array1.shape)==1:\n result=float(np.cov(array1))\n else:\n result=np.cov(array1)\n return result\n\ndef cor(x, y):\n \"\"\"\n Calculate Sample Correlation between two arrays\n \"\"\"\n return (cov(x,y) / (sd(x) * sd(y)))\n\ndef dif(x, y, afrund = None):\n \"\"\"\n Calculate the difference between two arrays\n \"\"\"\n result = []\n\n if afrund == None:\n afrund = 2\n\n for index, element in enumerate(x):\n result.append(round((element - y[index]), afrund))\n\n return result\n\ndef summary(array):\n \"\"\"\n Calculates the Tuckey Five-number (min/median/max/1st quartile/3rd quartile) of an array/vector\n \"\"\"\n import numpy as np\n return {'min' : np.min(array), 'Q1' : np.percentile(array,25), 'median' : np.median(array), 'mean' : np.mean(array), 'Q3' : np.percentile(array,75), 'max' : np.max(array)}\n\ndef IQR(array):\n \"\"\"\n Calculates the inter-quartile range of an array/vector\n \"\"\"\n import numpy as np\n array=np.array(array)\n result = np.percentile(array,75)-np.percentile(array,25)\n \n return result\n\n\"\"\"\nProbability distributions\n\"\"\"\n\n#=====================\n# Uniform distribution\n#=====================\n\ndef dunif(x, minimum=0,maximum=1):\n \"\"\"\n Calculates the point estimate of the uniform distribution\n \"\"\"\n from scipy.stats import uniform\n result=uniform.pdf(x=x,loc=minimum,scale=maximum-minimum)\n return result\n\ndef punif(q, minimum=0,maximum=1):\n \"\"\"\n Calculates the cumulative of the uniform distribution\n \"\"\"\n from scipy.stats import uniform\n result=uniform.cdf(x=q,loc=minimum,scale=maximum-minimum)\n return result\n\ndef quantile(array):\n \"\"\"\n Calculate and show R Quantile 0%, 25%, 50%, 75%, 100%\n \"\"\"\n import numpy as np\n return {\n 'Q0' : np.percentile(array, 0), \n 'Q1' : np.percentile(array, 25), \n 'Q2' : np.percentile(array, 50), \n 'Q3' : np.percentile(array, 75), \n 'Q4' : np.percentile(array, 100)\n }\n\ndef qunif(p, minimum=0,maximum=1):\n \"\"\"\n Calculates the quantile function of the uniform distribution\n \"\"\"\n from scipy.stats import uniform\n result=uniform.ppf(q=p,loc=minimum,scale=maximum-minimum)\n return result\n\ndef runif(n, minimum=0,maximum=1):\n \"\"\"\n Generates random variables from the uniform distribution\n \"\"\"\n from scipy.stats import uniform\n result=uniform.rvs(size=n,loc=minimum,scale=maximum-minimum)\n return result\n\n#======================\n# Binomial distribution\n#======================\n\ndef dbinom(x,size,prob=0.5):\n \"\"\"\n Calculates the point estimate of the binomial distribution\n \"\"\"\n from scipy.stats import binom\n result=binom.pmf(k=x,n=size,p=prob,loc=0)\n return result\n\ndef pbinom(q,size,prob=0.5):\n \"\"\"\n Calculates the cumulative of the binomial distribution\n \"\"\"\n from scipy.stats import binom\n result=binom.cdf(k=q,n=size,p=prob,loc=0)\n return result\n\ndef qbinom(p, size, prob=0.5):\n \"\"\"\n Calculates the quantile function from the binomial distribution\n \"\"\"\n from scipy.stats import binom\n result=binom.ppf(q=p,n=size,p=prob,loc=0)\n return result\n\ndef rbinom(n,size,prob=0.5):\n \"\"\"\n Generates random variables from the binomial distribution\n \"\"\"\n from scipy.stats import binom\n result=binom.rvs(n=size,p=prob,size=n)\n return result\n\n#=====================\n# Normal distribution\n#=====================\n\ndef dnorm(x,mean=0,sd =1):\n \"\"\"\n Calculates the density of the Normal distribution\n \"\"\"\n from scipy.stats import norm\n result=norm.pdf(x,loc=mean,scale=sd)\n return result\n\ndef pnorm(q,mean=0,sd=1):\n \"\"\"\n Calculates the cumulative of the normal distribution\n \"\"\"\n from scipy.stats import norm\n result=norm.cdf(x=q,loc=mean,scale=sd)\n return result\n\ndef qnorm(p,mean=0,sd=1):\n \"\"\"\n Calculates the quantile function of the normal distribution\n \"\"\"\n from scipy.stats import norm\n result=norm.ppf(q=p,loc=mean,scale=sd)\n return result\n\ndef rnorm(n,mean=0,sd=1):\n \"\"\"\n Generates random variables from the normal distribution\n \"\"\"\n from scipy.stats import norm\n result=norm.rvs(size=n,loc=mean,scale=sd)\n return result\n\n#=====================\n# Poisson distribution\n#=====================\n\ndef dpois(x,mu):\n \"\"\"\n Calculates the density/point estimate of the Poisson distribution\n \"\"\"\n from scipy.stats import poisson\n result=poisson.pmf(k=x,mu=mu)\n return result\n\ndef ppois(q,mu):\n \"\"\"\n Calculates the cumulative of the Poisson distribution\n \"\"\"\n from scipy.stats import poisson\n result=poisson.cdf(k=q,mu=mu)\n return result\n\ndef qpois(p,mu):\n \"\"\"\n Calculates the quantile function of the Poisson distribution\n \"\"\"\n from scipy.stats import poisson\n result=poisson.ppf(q=p,mu=mu)\n return result\n\ndef rpois(n,mu):\n \"\"\"\n Generates random variables from the Poisson distribution\n \"\"\"\n from scipy.stats import poisson\n result=poisson.rvs(size=n,mu=mu)\n return result\n\n#=====================\n# chi^2-distribution\n#=====================\n\ndef dchisq(x,df,ncp=0):\n \"\"\"\n Calculates the density/point estimate of the chi-square distribution\n \"\"\"\n from scipy.stats import chi2,ncx2\n if ncp==0:\n result=chi2.pdf(x=x,df=df,loc=0,scale=1)\n else:\n result=ncx2.pdf(x=x,df=df,nc=ncp,loc=0,scale=1)\n return result\n\ndef pchisq(q,df,ncp=0):\n \"\"\"\n Calculates the cumulative of the chi-square distribution\n \"\"\"\n from scipy.stats import chi2,ncx2\n if ncp==0:\n result=chi2.cdf(x=q,df=df,loc=0,scale=1)\n else:\n result=ncx2.cdf(x=q,df=df,nc=ncp,loc=0,scale=1)\n return result\n\ndef qchisq(p,df,ncp=0):\n \"\"\"\n Calculates the quantile function of the chi-square distribution\n \"\"\"\n from scipy.stats import chi2,ncx2\n if ncp==0:\n result=chi2.ppf(q=p,df=df,loc=0,scale=1)\n else:\n result=ncx2.ppf(q=p,df=df,nc=ncp,loc=0,scale=1)\n return result\n\ndef rchisq(n,df,ncp=0):\n \"\"\"\n Generates random variables from the chi-square distribution\n \"\"\"\n from scipy.stats import chi2,ncx2\n if ncp==0:\n result=chi2.rvs(size=n,df=df,loc=0,scale=1)\n else:\n result=ncx2.rvs(size=n,df=df,nc=ncp,loc=0,scale=1)\n return result\n\n#==============================\n# ### Student's t-distribution\n#==============================\n\ndef dt(x,df,ncp=0):\n \"\"\"\n Calculates the density/point estimate of the t-distribution\n \"\"\"\n from scipy.stats import t,nct\n if ncp==0:\n result=t.pdf(x=x,df=df,loc=0,scale=1)\n else:\n result=nct.pdf(x=x,df=df,nc=ncp,loc=0,scale=1)\n return result\n\ndef pt(q,df,ncp=0):\n \"\"\"\n Calculates the cumulative of the t-distribution\n \"\"\"\n from scipy.stats import t,nct\n if ncp==0:\n result=t.cdf(x=q,df=df,loc=0,scale=1)\n else:\n result=nct.cdf(x=q,df=df,nc=ncp,loc=0,scale=1)\n return result\n\ndef qt(p,df,ncp=0):\n \"\"\"\n Calculates the quantile function of the t-distribution\n \"\"\"\n from scipy.stats import t,nct\n if ncp==0:\n result=t.ppf(q=p,df=df,loc=0,scale=1)\n else:\n result=nct.ppf(q=p,df=df,nc=ncp,loc=0,scale=1)\n return result\n\ndef rt(n,df,ncp=0):\n \"\"\"\n Generates random variables from the t-distribution\n \"\"\"\n from scipy.stats import t,nct\n if ncp==0:\n result=t.rvs(size=n,df=df,loc=0,scale=1)\n else:\n result=nct.rvs(size=n,df=df,nc=ncp,loc=0,scale=1)\n return result\n\n#================\n# F-distribution\n#================\n\ndef df(x,df1,df2,ncp=0):\n \"\"\"\n Calculates the density/point estimate of the F-distribution\n \"\"\"\n from scipy.stats import f,ncf\n if ncp==0:\n result=f.pdf(x=x,dfn=df1,dfd=df2,loc=0,scale=1)\n else:\n result=ncf.pdf(x=x,dfn=df1,dfd=df2,nc=ncp,loc=0,scale=1)\n return result\n\ndef pf(q,df1,df2,ncp=0):\n \"\"\"\n Calculates the cumulative of the F-distribution\n \"\"\"\n from scipy.stats import f,ncf\n if ncp==0:\n result=f.cdf(x=q,dfn=df1,dfd=df2,loc=0,scale=1)\n else:\n result=ncf.cdf(x=q,dfn=df1,dfd=df2,nc=ncp,loc=0,scale=1)\n return result\n\ndef qf(p,df1,df2,ncp=0):\n \"\"\"\n Calculates the quantile function of the F-distribution\n \"\"\"\n from scipy.stats import f,ncf\n if ncp==0:\n result=f.ppf(q=p,dfn=df1,dfd=df2,loc=0,scale=1)\n else:\n result=ncf.ppf(q=p,dfn=df1,dfd=df2,nc=ncp,loc=0,scale=1)\n return result\n\ndef rf(n,df1,df2,ncp=0):\n \"\"\"\n Calculates the quantile function of the F-distribution\n \"\"\"\n from scipy.stats import f,ncf\n if ncp==0:\n result=f.rvs(size=n,dfn=df1,dfd=df2,loc=0,scale=1)\n else:\n result=ncf.rvs(size=n,dfn=df1,dfd=df2,nc=ncp,loc=0,scale=1)\n return result\n\n#===================\n# Beta distribution\n#===================\n\ndef dbeta(x,shape1,shape2):\n \"\"\"\n Calculates the density/point estimate of the Beta-distribution\n \"\"\"\n from scipy.stats import beta\n result=beta.pdf(x=x,a=shape1,b=shape2,loc=0,scale=1)\n return result\n\ndef pbeta(q,shape1,shape2):\n \"\"\"\n Calculates the cumulative of the Beta-distribution\n \"\"\"\n from scipy.stats import beta\n result=beta.cdf(x=q,a=shape1,b=shape2,loc=0,scale=1)\n return result\n\ndef qbeta(p,shape1,shape2):\n \"\"\"\n Calculates the cumulative of the Beta-distribution\n \"\"\"\n from scipy.stats import beta\n result=beta.ppf(q=p,a=shape1,b=shape2,loc=0,scale=1)\n return result\n\ndef rbeta(n,shape1,shape2):\n \"\"\"\n Calculates the cumulative of the Beta-distribution\n \"\"\"\n from scipy.stats import beta\n result=beta.rvs(size=n,a=shape1,b=shape2,loc=0,scale=1)\n return result\n\n#========================\n# ### Gamma distribution\n#========================\n\ndef dgamma(x,shape,rate=1):\n \"\"\"\n Calculates the density/point estimate of the Gamma-distribution\n \"\"\"\n from scipy.stats import gamma\n result=rate*gamma.pdf(x=rate*x,a=shape,loc=0,scale=1)\n return result\n\ndef pgamma(q,shape,rate=1):\n \"\"\"\n Calculates the cumulative of the Gamma-distribution\n \"\"\"\n from scipy.stats import gamma\n result=gamma.cdf(x=rate*q,a=shape,loc=0,scale=1)\n return result\n\ndef qgamma(p,shape,rate=1):\n \"\"\"\n Calculates the cumulative of the Gamma-distribution\n \"\"\"\n from scipy.stats import gamma\n result=(1/rate)*gamma.ppf(q=p,a=shape,loc=0,scale=1)\n return result\n\ndef rgamma(n,shape,rate=1):\n \"\"\"\n Calculates the cumulative of the Gamma-distribution\n \"\"\"\n from scipy.stats import gamma\n result=gamma.rvs(size=n,a=shape,loc=0,scale=1)\n return result"
]
| [
[
"scipy.stats.norm.ppf",
"scipy.stats.ncf.ppf",
"scipy.stats.ncf.pdf",
"scipy.stats.norm.cdf",
"scipy.stats.ncx2.pdf",
"numpy.max",
"numpy.mean",
"scipy.stats.uniform.rvs",
"scipy.stats.binom.ppf",
"scipy.stats.binom.rvs",
"scipy.stats.gamma.cdf",
"scipy.stats.f.rvs",
"scipy.stats.t.pdf",
"scipy.stats.ncf.cdf",
"scipy.stats.poisson.cdf",
"scipy.stats.chi2.rvs",
"scipy.stats.f.ppf",
"scipy.stats.beta.ppf",
"scipy.stats.beta.cdf",
"scipy.stats.t.cdf",
"scipy.stats.t.rvs",
"scipy.stats.chi2.ppf",
"scipy.stats.uniform.cdf",
"scipy.stats.nct.rvs",
"numpy.min",
"scipy.stats.uniform.ppf",
"numpy.median",
"scipy.stats.binom.pmf",
"scipy.stats.ncx2.ppf",
"scipy.stats.nct.pdf",
"numpy.cov",
"scipy.stats.norm.rvs",
"scipy.stats.poisson.ppf",
"scipy.stats.binom.cdf",
"scipy.stats.poisson.pmf",
"numpy.array",
"scipy.stats.gamma.ppf",
"scipy.stats.ncx2.cdf",
"scipy.stats.chi2.pdf",
"scipy.stats.ncx2.rvs",
"scipy.stats.norm.pdf",
"scipy.stats.poisson.rvs",
"scipy.stats.beta.pdf",
"scipy.stats.beta.rvs",
"scipy.stats.uniform.pdf",
"scipy.stats.gamma.pdf",
"scipy.stats.nct.cdf",
"numpy.percentile",
"scipy.stats.f.cdf",
"scipy.stats.t.ppf",
"scipy.stats.gamma.rvs",
"scipy.stats.nct.ppf",
"scipy.stats.ncf.rvs",
"scipy.stats.chi2.cdf",
"scipy.stats.f.pdf"
]
]
|
xbe/qcc | [
"90a35057c7fea37187a0cf253eb65613702c51e3"
]
| [
"src/solovay_kitaev.py"
]
| [
"# python3\n\"\"\"Example: Solovay-Kitaev Algorithm for gate approximation.\"\"\"\n\nimport math\nimport random\n\nfrom absl import app\nimport numpy as np\n\nfrom src.lib import helper\nfrom src.lib import ops\nfrom src.lib import state\n\n\ndef to_su2(U):\n \"\"\"Convert a 2x2 unitary to a unitary with determinant 1.0.\"\"\"\n\n return np.sqrt(1 / np.linalg.det(U)) * U\n\n\ndef trace_dist(U, V):\n \"\"\"Compute trace distance between two 2x2 matrices.\"\"\"\n\n return np.real(0.5 * np.trace(np.sqrt((U - V).adjoint() @ (U - V))))\n\n\ndef create_unitaries(base, limit):\n \"\"\"Create all combinations of all base gates, up to length 'limit'.\"\"\"\n\n # Create bitstrings up to bitstring length limit-1:\n # 0, 1, 00, 01, 10, 11, 000, 001, 010, ...\n #\n # Multiply together the 2 base operators, according to their index.\n # Note: This can be optimized, by remembering the last 2^x results\n # and multiplying them with base gets 0, 1.\n #\n gate_list = []\n for width in range(limit):\n for bits in helper.bitprod(width):\n U = ops.Identity()\n for bit in bits:\n U = U @ base[bit]\n gate_list.append(U)\n return gate_list\n\n\ndef find_closest_u(gate_list, u):\n \"\"\"Find the one gate in the list closest to u.\"\"\"\n\n # Linear search over list of gates - is _very_ slow.\n # This can be optimized by using kd-trees.\n #\n min_dist = 10\n min_u = ops.Identity()\n for gate in gate_list:\n tr_dist = trace_dist(gate, u)\n if tr_dist < min_dist:\n min_dist = tr_dist\n min_u = gate\n return min_u\n\n\ndef u_to_bloch(U):\n \"\"\"Compute angle and axis for a unitary.\"\"\"\n\n angle = np.real(np.arccos((U[0, 0] + U[1, 1])/2))\n sin = np.sin(angle)\n if sin < 1e-10:\n axis = [0, 0, 1]\n else:\n nx = (U[0, 1] - U[1, 1]) / (2j * sin)\n ny = (U[0, 1] - U[1, 0]) / (2j * sin)\n nz = (U[1, 1] - U[0, 0]) / (2j * sin)\n axis = [nx, ny, nz]\n return axis, angle\n\n\ndef gc_decomp(U):\n \"\"\"Group Commutator Decomposition.\"\"\"\n\n def diagonalize(U):\n _, V = np.linalg.eig(U)\n return ops.Operator(V)\n\n # Because of moderate numerical instability, it can happen\n # that the trace is just a tad over 2.000000. If this happens,\n # we tolerate it and set the trace to exactly 2.000000.\n tr = np.trace(U)\n if tr > 2.0:\n tr = 2.0\n\n # We know how to compute theta from u_to_bloch().\n theta = 2.0 * np.arccos(np.real(tr / 2))\n # The angle phi comes from eq 10 in 'The Solovay-Kitaev Algorithm' by\n # Dawson, Nielsen.\n phi = 2.0 * np.arcsin(np.sqrt(np.sqrt((0.5 - 0.5 * np.cos(theta / 2)))))\n\n axis, _ = u_to_bloch(U)\n V = ops.RotationX(phi)\n if axis[2] < 0:\n W = ops.RotationY(2 * np.pi - phi)\n else:\n W = ops.RotationY(phi)\n\n V1 = diagonalize(U)\n V2 = diagonalize(V @ W @ V.adjoint() @ W.adjoint())\n S = V1 @ V2.adjoint()\n V_tilde = S @ V @ S.adjoint()\n W_tilde = S @ W @ S.adjoint()\n return V_tilde, W_tilde\n\n\ndef sk_algo(U, gates, n):\n \"\"\"Solovay-Kitaev Algorithm.\"\"\"\n\n if n == 0:\n return find_closest_u(gates, U)\n else:\n U_next = sk_algo(U, gates, n-1)\n V, W = gc_decomp(U @ U_next.adjoint())\n V_next = sk_algo(V, gates, n-1)\n W_next = sk_algo(W, gates, n-1)\n return V_next @ W_next @ V_next.adjoint() @ W_next.adjoint() @ U_next\n\n\ndef main(argv):\n if len(argv) > 1:\n raise app.UsageError('Too many command-line arguments.')\n\n num_experiments = 10\n depth = 8\n recursion = 4\n print('SK algorithm - depth: {}, recursion: {}, experiments: {}'.\n format(depth, recursion, num_experiments))\n\n base = [to_su2(ops.Hadamard()), to_su2(ops.Tgate())]\n gates = create_unitaries(base, depth)\n sum_dist = 0.0\n for i in range(num_experiments):\n U = (ops.RotationX(2.0 * np.pi * random.random()) @\n ops.RotationY(2.0 * np.pi * random.random()) @\n ops.RotationZ(2.0 * np.pi * random.random()))\n\n U_approx = sk_algo(U, gates, recursion)\n\n dist = trace_dist(U, U_approx)\n sum_dist += dist\n\n phi1 = U(state.zero)\n phi2 = U_approx(state.zero)\n print('[{:2d}]: Trace Dist: {:.4f} State: {:6.4f}%'.\n format(i, dist,\n 100.0 * (1.0 - np.real(np.dot(phi1, phi2.conj())))))\n\n print('Gates: {}, Mean Trace Dist:: {:.4f}'.\n format(len(gates), sum_dist / num_experiments))\n\n\nif __name__ == '__main__':\n app.run(main)\n"
]
| [
[
"numpy.linalg.eig",
"numpy.arccos",
"numpy.cos",
"numpy.sin",
"numpy.linalg.det",
"numpy.real",
"numpy.trace"
]
]
|
BenDavisonPetch/ahfhalotools | [
"d150ac66294e29feb3f43baa0cea135ac12fcb9b"
]
| [
"examples/clusterComp/HaloDatavsHaloDataScatterPlot.py"
]
| [
"import numpy as np\nimport ahfhalotools.filetools as ft\nfrom ahfhalotools.objects import Cluster\nimport ahfhalotools.analysis as analysis\nfrom astropy.cosmology import FlatLambdaCDM\nfrom astropy.cosmology import WMAP9\nfrom astropy import units as au\nimport matplotlib.pyplot as plt\nimport matplotlib.colors as mcolors\nimport matplotlib.cm as cm\nimport scicm\n\n#define base file name for full, untruncated files\nfileNameBaseGX = \"{simName}-NewMDCLUSTER_{clusterNum:0=4d}.snap_{snap:0=3d}.z{z:.3f}\"\nfileNameBaseGiz = fileNameBaseGX\nfileNameBaseMus = \"{simName}-NewMDCLUSTER_{clusterNum:0=4d}.z{z:.3f}\"\n#define directory to read from\ndirectory = \"TruncData/{simName}/\"\n\n\n#cluster numbers\n#clusterNums = np.arange(1,325)\nclusterNums = np.arange(1,325)\n\n#up to 128\nmusClusters1 = ft.loadClusters(np.arange(1,22), [128], \"GadgetMUSIC\",\n directory = directory,\n fileBaseFmt = fileNameBaseMus,\n printProgress = True, skipmtree = True)\n\n#up to 17\nmusClusters2 = ft.loadClusters(np.arange(22,325), [17], \"GadgetMUSIC\",\n directory = directory,\n fileBaseFmt = fileNameBaseMus,\n printProgress = True, skipmtree = True)\n\ngizClusters = ft.loadClusters(clusterNums, [128], \"GIZMO\",\n directory = directory, printProgress = True,\n skipmtree = True)\n\ngxClusters = ft.loadClusters(clusterNums, [128], \"GadgetX\",\n directory = directory, printProgress = True,\n skipmtree = True)\n\n## done loading data\n# histogram of N(rho) vs rho at z = 0\n\n# get critical density at z=0 using parameters from simulations\nflcdm = FlatLambdaCDM(H0=WMAP9.H(0), Om0=0.307, Ob0=0.048)\nrhoCrit = flcdm.critical_density(0)\n# units of density calculated from profile data is Msol kpc^-3 h^2\n# so must convert units to Msol kpc^-3\nrhoCrit = rhoCrit.to(au.solMass / (au.kpc)**3)\n\n## ------------ PLOT ------------- ##\n## plots a scatter plot of halodata vs halodata for each set of clusters\n## parameters for plotting\nhaloID = 128000000000001\nxQuantity = \"Qvir\"\nyQuantity = \"mbp_offset\"\nxlabel = \"Virial Ratio, $Q_{vir}$\"\nylabel = \"MBP offset / $kpc \\\\; h^{-1}$\"\nfigtitle = \"MBP offset vs Virial Ratio for central clusters at z=0\"\nlogxScale = True\nlogyScale = True\n\n# whether or not to try and plot the GadgetMUSIC clusters numbers 22 and up\n# don't set to True if trying to plot at not z = 0\nincludeExtraMusClusters = True\n\n# sets of clusters\nclustersSet = [gxClusters, gizClusters, musClusters1]\nclustersNames = [\"GadgetX\", \"GIZMO\", \"GadgetMUSIC\"]\n\nfig, ax = plt.subplots(1,3,figsize = (12,6), sharey = \"all\", sharex = \"all\")\n\nfor i, clusters in enumerate(clustersSet):\n #default color for points\n #get values of whatever quantity specified\n xvalues = [cluster.getHaloData(haloID, xQuantity) for cluster in clusters]\n yvalues = [cluster.getHaloData(haloID, yQuantity) for cluster in clusters]\n\n #plot scatterplot\n ax[i].scatter(xvalues,yvalues,s=15,label=\"128 Snapshot Clusters\")\n\n #plot extra music data\n if i == 2 and includeExtraMusClusters:\n xvalues = []\n yvalues = []\n # add values from the second set of gadgetMUSIC clusters\n # (has to be done separately as *most* (not all) of the clusters after\n # cluster 22 only have 17 snapshots)\n for cluster in musClusters2:\n xQ = cluster.getHaloData(17000000000001, xQuantity)\n yQ = cluster.getHaloData(17000000000001, yQuantity)\n #check if they are -1 (ie not loaded into memory)\n if xQ == -1:\n xQ = cluster.getHaloData(128000000000001, xQuantity)\n yQ = cluster.getHaloData(128000000000001, yQuantity)\n\n xvalues.append(xQ)\n yvalues.append(yQ)\n\n ax[i].scatter(xvalues,yvalues,s=15,label=\"17 Snapshot Clusters\")\n ax[i].legend()\n\n #set axis title and labels\n ax[i].set_title(clustersNames[i])\n ax[i].set_xlabel(xlabel)\n ax[i].set_ylabel(ylabel)\n ax[i].grid(b=True)\n if logxScale: ax[i].set_xscale(\"log\")\n if logyScale: ax[i].set_yscale(\"log\")\n\n#set figure title\nfig.suptitle(figtitle)\n\nplt.tight_layout()\nplt.show()\n"
]
| [
[
"numpy.arange",
"matplotlib.pyplot.tight_layout",
"matplotlib.pyplot.subplots",
"matplotlib.pyplot.show"
]
]
|
gbisschoff/Z-model | [
"98e34799ccedfc52a6f5f6958f5e0ebeca1aa5f3"
]
| [
"src/z_model/exposure_at_default.py"
]
| [
"from numpy import array, repeat, arange, cumprod, cumsum, maximum, minimum, ceil\nfrom pandas import Series\nfrom .account import Account\nfrom .effective_interest_rate import EffectiveInterestRate\nfrom .assumptions import EADAssumptions\nfrom .scenarios import Scenario\n\nclass ConstantExposureAtDefault:\n def __init__(self, exposure_at_default: float):\n self.exposure_at_default = exposure_at_default\n\n def __getitem__(self, account: Account):\n return Series(self.exposure_at_default, index=account.remaining_life_index)\n\n\nclass AmortisingExposureAtDefault:\n def __init__(self, effective_interest_rate: EffectiveInterestRate, fixed_fees: float = .0, fees_pct: float = .0, prepayment_pct: float = .0, **kwargs):\n self.effective_interest_rate = effective_interest_rate\n self.fixed_fees = fixed_fees\n self.fees_pct = fees_pct\n self.prepayment_pct = prepayment_pct\n\n def __getitem__(self, account: Account):\n balance = account.outstanding_balance\n t = arange(account.remaining_life) + 1\n eir = self.effective_interest_rate[account]\n df_t0 = 1 / cumprod(1 + eir)\n\n is_pmt_period = ((account.remaining_life - t) % (12 / account.contractual_freq) == 0)\n n_pmts = cumsum(is_pmt_period)\n\n pmt = account.contractual_payment * is_pmt_period\n cf = pmt * (1 + self.prepayment_pct) - self.fixed_fees\n cf_t0 = cf * df_t0\n cum_cf_t = cumsum(cf_t0) / df_t0\n\n balance_t = balance / df_t0 - cum_cf_t\n # Fees are charged before payment is deducted\n fees_pct_amt = cumsum((balance_t + pmt) * self.fees_pct * df_t0)/df_t0\n balance_t_pfees = maximum(balance_t + fees_pct_amt,0)\n\n arrears_allowance = account.contractual_payment * 3\n remaining_allowance = max(arrears_allowance - account.current_arrears, 0)\n remaining_allowance_t = ceil(remaining_allowance / account.contractual_payment)\n\n arrears_t0 = account.contractual_payment * (n_pmts <= remaining_allowance_t) * is_pmt_period * df_t0\n arrears_t = minimum(cumsum(arrears_t0) / df_t0, remaining_allowance)\n\n ead = maximum((balance_t_pfees + arrears_t) / account.outstanding_balance, 0)\n\n return Series(ead, index=account.remaining_life_index)\n\n\nclass CCFExposureAtDefault:\n def __init__(self, ccf_method: str, ccf: float, **kwargs):\n self.ccf_method = ccf_method.upper()\n self.ccf = ccf\n\n def __getitem__(self, account: Account):\n if self.ccf_method.upper() == 'METHOD 1':\n return Series(\n self.ccf, account.remaining_life,\n index=account.remaining_life_index\n )\n elif self.ccf_method.upper() == 'METHOD 2':\n return Series(\n account.limit * self.ccf / account.outstanding_balance,\n index=account.remaining_life_index\n )\n elif self.ccf_method.upper() == 'METHOD 3':\n return Series(\n (account.outstanding_balance + (account.limit - account.outstanding_balance) * self.ccf) /\n account.outstanding_balance,\n index=account.remaining_life_index\n )\n else:\n raise ValueError(f'CCF Method ({self.ccf_method}) not supported.')\n\n\nclass ExposureAtDefault:\n @classmethod\n def from_assumptions(cls, assumptions: EADAssumptions, scenario: Scenario, eir: EffectiveInterestRate):\n if assumptions.type.upper() =='CONSTANT':\n return ConstantExposureAtDefault(\n exposure_at_default=assumptions.exposure_at_default\n )\n elif assumptions.type.upper() =='AMORTISING':\n return AmortisingExposureAtDefault(\n effective_interest_rate=eir,\n fixed_fees=assumptions.fees_fixed,\n fees_pct=assumptions.fees_pct,\n prepayment_pct=assumptions.prepayment_pct\n )\n elif assumptions.type.upper() =='CCF':\n return CCFExposureAtDefault(\n ccf_method=assumptions.ccf_method,\n ccf=assumptions.ccf\n )\n else:\n raise ValueError(f'Invalid EAD method: {assumptions.type}')\n"
]
| [
[
"numpy.maximum",
"pandas.Series",
"numpy.arange",
"numpy.cumsum",
"numpy.ceil",
"numpy.cumprod"
]
]
|
batikim09/translate | [
"ae275549c414a68af783a87cb2eacc303a0553b0",
"ae275549c414a68af783a87cb2eacc303a0553b0"
]
| [
"pytorch_translate/research/adversarial/adversarial_utils.py",
"pytorch_translate/test/test_beam_decode.py"
]
| [
"#!/usr/bin/env python3\n\nimport re\n\nimport torch\nimport torch.nn.functional as F\n\n\n# Delimiter for the word to word file\nblank_delim = re.compile(r\"[ \\t]+\")\n\n\ndef pairwise_dot_product(src_embeds, vocab_embeds, cosine=False):\n \"\"\"Compute the cosine similarity between each word in the vocab and each\n word in the source\n\n If `cosine=True` this returns the pairwise cosine similarity\"\"\"\n # Normlize vectors for the cosine similarity\n if cosine:\n src_embeds = F.normalize(src_embeds, dim=-1, p=2)\n vocab_embeds = F.normalize(vocab_embeds, dim=-1, p=2)\n # Take the dot product\n dot_product = torch.einsum(\"bij,kj->bik\", (src_embeds, vocab_embeds))\n return dot_product\n\n\ndef pairwise_distance(src_embeds, vocab_embeds, squared=False):\n \"\"\"Compute the euclidean distance between each word in the vocab and each\n word in the source\"\"\"\n # We will compute the squared norm first to avoid having to compute all\n # the directions (which would have space complexity B x T x |V| x d)\n # First compute the squared norm of each word vector\n vocab_sq_norm = vocab_embeds.norm(p=2, dim=-1) ** 2\n src_sq_norm = src_embeds.norm(p=2, dim=-1) ** 2\n # Take the dot product\n dot_product = pairwise_dot_product(src_embeds, vocab_embeds)\n # Reshape for broadcasting\n # 1 x 1 x |V|\n vocab_sq_norm = vocab_sq_norm.unsqueeze(0).unsqueeze(0)\n # B x T x 1\n src_sq_norm = src_sq_norm.unsqueeze(2)\n # Compute squared difference\n sq_norm = vocab_sq_norm + src_sq_norm - 2 * dot_product\n # Either return the squared norm or return the sqrt\n if squared:\n return sq_norm\n else:\n # Relu + epsilon for numerical stability\n sq_norm = F.relu(sq_norm) + 1e-20\n # Take the square root\n return sq_norm.sqrt()\n\n\ndef sample_gumbel_trick(logits, temperature=1.0, num_samples=None, dim=-1):\n \"\"\"Use the gumbel trick to sample from a distribution parametrized by logits\n\n For references on the Gumbel trick see eg.:\n - Original paper:\n stat.ucla.edu/~gpapan/pubs/confr/PapandreouYuille_PerturbAndMap_ieee-c-iccv11.pdf\n - Nice blog post:\n http://irenechen.net/blog/2017/08/17/gumbel-trick.html\n \"\"\"\n # Sample from the Gumbel distribution\n uniform_noise = torch.rand_like(logits)\n gumbel_noise = -torch.log(-torch.log(uniform_noise + 1e-20) + 1e-20)\n # For stable behaviour at both high and low temperature we either rescale\n # the logprob or the gumbel noise so that we never end up with huge noise\n # or logits. In any case, all the following expressions are equal up to\n # a multiplicative constant (which doesn't matter because we're only\n # interested in the softmax)\n if temperature > 1:\n # High temperature: rescale the logits\n noisy_logits = logits / temperature + gumbel_noise\n elif temperature == 1:\n # Temperature = 1: no rescaling needed\n noisy_logits = logits + gumbel_noise\n else:\n # Low temperatures: rescale the noise\n noisy_logits = logits + gumbel_noise * temperature\n # Use the Gumbel trick to sample\n if num_samples is None:\n # The behavior is different for num_samples=None and num_samples=1\n # if num_samples=None we reduce the singleton dim\n _, samples = noisy_logits.max(dim=dim)\n else:\n # I am not 100% sure that this is valid for more num_samples>1 (ie.\n # that it corresponds to sampling multiple times without replacement)\n _, samples = noisy_logits.topk(num_samples, dim=dim)\n\n return samples\n\n\ndef load_one_to_many_dict(filename):\n \"\"\"Load a mapping from words to lists of words\n\n The expected format is `[word] [alternative_1] [alternative_2]...`\n The separating character can be either tab or space.\"\"\"\n dic = {}\n with open(filename, \"r\") as dict_file:\n for line in dict_file:\n try:\n fields = blank_delim.split(line.strip())\n if len(fields) <= 1:\n continue\n word = fields[0]\n alternatives = fields[1:]\n if word not in dic:\n dic[word] = set()\n for alt in alternatives:\n dic[word].add(alt)\n except ValueError:\n continue\n\n return dic\n\n\ndef tile(tensor, dim, repeat):\n \"\"\"Repeat each element `repeat` times along dimension `dim`\"\"\"\n # We will insert a new dim in the tensor and torch.repeat it\n # First we get the repeating counts\n repeat_dims = [1] * len(tensor.size())\n repeat_dims.insert(dim + 1, repeat)\n # And the final dims\n new_dims = list(tensor.size())\n new_dims[dim] = 2 * tensor.size(dim)\n # Now unsqueeze, repeat and reshape\n return tensor.unsqueeze(dim + 1).repeat(*repeat_dims).view(*new_dims)\n\n\ndef detach_sample(sample):\n \"\"\"Detach sample to save memory\"\"\"\n\n if len(sample) == 0:\n return {}\n\n def _detach(maybe_tensor):\n if torch.is_tensor(maybe_tensor):\n return maybe_tensor.detach()\n elif isinstance(maybe_tensor, dict):\n return {key: _detach(val) for key, val in maybe_tensor.items()}\n elif isinstance(maybe_tensor, list):\n return [_detach(val) for val in maybe_tensor]\n else:\n return maybe_tensor\n\n return _detach(sample)\n\n\ndef clone_sample(sample):\n \"\"\"Clone sample to save memory\"\"\"\n\n if len(sample) == 0:\n return {}\n\n def _clone(maybe_tensor):\n if torch.is_tensor(maybe_tensor):\n return maybe_tensor.clone()\n elif isinstance(maybe_tensor, dict):\n return {key: _clone(val) for key, val in maybe_tensor.items()}\n elif isinstance(maybe_tensor, list):\n return [_clone(val) for val in maybe_tensor]\n else:\n return maybe_tensor\n\n return _clone(sample)\n",
"#!/usr/bin/env python3\n\nimport unittest\nfrom typing import Any, List\n\nimport numpy as np\nimport torch\nfrom pytorch_translate import char_source_model # noqa (must be after rnn)\nfrom pytorch_translate import rnn # noqa\nfrom pytorch_translate import beam_decode, tasks\nfrom pytorch_translate.test import utils as test_utils\n\n\nclass TestBeamDecode(unittest.TestCase):\n @unittest.skipIf(torch.cuda.device_count() < 1, \"No GPU available for test.\")\n def test_basic_generate(self):\n test_args = test_utils.ModelParamsDict()\n _, src_dict, tgt_dict = test_utils.prepare_inputs(test_args)\n task = tasks.DictionaryHolderTask(src_dict, tgt_dict)\n model = task.build_model(test_args)\n translator = beam_decode.SequenceGenerator([model], task.target_dictionary)\n src_tokens = torch.LongTensor([[0, 0, 0], [0, 0, 0]])\n src_lengths = torch.LongTensor([3, 3])\n encoder_input = (src_tokens, src_lengths)\n translator.generate(encoder_input, maxlen=7)\n\n @unittest.skipIf(torch.cuda.device_count() < 1, \"No GPU available for test.\")\n def test_char_rnn_generate(self):\n test_args = test_utils.ModelParamsDict(sequence_lstm=True)\n test_args.arch = \"char_source\"\n test_args.char_source_dict_size = 126\n test_args.char_embed_dim = 8\n test_args.char_rnn_units = 12\n test_args.char_rnn_layers = 2\n\n _, src_dict, tgt_dict = test_utils.prepare_inputs(test_args)\n task = tasks.DictionaryHolderTask(src_dict, tgt_dict)\n model = task.build_model(test_args)\n translator = beam_decode.SequenceGenerator([model], task.target_dictionary)\n src_tokens = torch.LongTensor([[0, 0, 0], [0, 0, 0]])\n src_lengths = torch.LongTensor([3, 3])\n char_inds = torch.LongTensor(np.zeros((2, 3, 5)))\n word_lengths = torch.LongTensor([[5, 5, 5], [5, 5, 5]])\n encoder_input = (src_tokens, src_lengths, char_inds, word_lengths)\n translator.generate(encoder_input, maxlen=7)\n\n @unittest.skipIf(torch.cuda.device_count() < 1, \"No GPU available for test.\")\n def test_gather_probs_with_vr(self):\n \"\"\" Tests gather_probs when there is vocab reduction \"\"\"\n all_translation_tokens: List[Any] = [\n torch.LongTensor([3, 7, 8, 9]),\n torch.LongTensor([0, 3, 5]),\n ]\n all_probs: List[Any] = [\n torch.FloatTensor(\n [[0.25, 0.25, 0.25, 0.25], [0.25, 0.25, 0.25, 0.25]]\n ).cuda(),\n torch.FloatTensor([[0.4, 0.5, 0.1], [0.4, 0.5, 0.1]]).cuda(),\n ]\n avg_probs, possible_translation_tokens = beam_decode.SequenceGenerator.gather_probs(\n all_translation_tokens=all_translation_tokens, all_probs=all_probs\n )\n avg_probs = avg_probs.detach().cpu().numpy()\n possible_translation_tokens = possible_translation_tokens.detach().cpu().numpy()\n\n avg_probs_ref = sorted([0.4, 0.75, 0.1, 0.25, 0.25, 0.25])\n possible_translation_tokens_ref = sorted([0, 3, 5, 7, 8, 9])\n\n np.testing.assert_allclose(\n actual=np.sort(avg_probs[0]), desired=np.array(avg_probs_ref), atol=1e-5\n )\n np.testing.assert_allclose(\n actual=np.sort(possible_translation_tokens),\n desired=np.array(possible_translation_tokens_ref),\n )\n np.testing.assert_allclose(\n actual=np.sort(possible_translation_tokens),\n desired=np.array(possible_translation_tokens_ref),\n )\n\n @unittest.skipIf(torch.cuda.device_count() < 1, \"No GPU available for test.\")\n def test_gather_probs_without_vr(self):\n \"\"\" Tests gather_probs when there is no vocab reduction \"\"\"\n all_probs: List[Any] = [\n torch.FloatTensor([[0.25, 0.25, 0.25, 0.25], [0.25, 0.25, 0.25, 0.25]]),\n torch.FloatTensor([[0.4, 0.2, 0.1, 0.3], [0.4, 0.2, 0.1, 0.3]]),\n ]\n all_translation_tokens: List[Any] = [None, None]\n avg_probs, possible_translation_tokens = beam_decode.SequenceGenerator.gather_probs(\n all_translation_tokens=all_translation_tokens, all_probs=all_probs\n )\n\n assert possible_translation_tokens is None\n avg_probs_ref = [0.65, 0.45, 0.35, 0.55]\n np.testing.assert_allclose(\n actual=avg_probs[0], desired=np.array(avg_probs_ref), atol=1e-5\n )\n np.testing.assert_allclose(\n actual=avg_probs[1], desired=np.array(avg_probs_ref), atol=1e-5\n )\n"
]
| [
[
"torch.nn.functional.normalize",
"torch.rand_like",
"torch.einsum",
"torch.is_tensor",
"torch.nn.functional.relu",
"torch.log"
],
[
"torch.LongTensor",
"numpy.sort",
"torch.FloatTensor",
"torch.cuda.device_count",
"numpy.array",
"numpy.zeros"
]
]
|
Jaimedlrm/entropytriangle | [
"46076aa6e9e06777df4dcf885cd951afdf1de168"
]
| [
"entropytriangle/ternary/helpers.py"
]
| [
"\"\"\"\nHelper functions and utilities for projecting to the simplex and various tasks.\n\"\"\"\n\nimport numpy\n\n\n### Constants ###\n\nSQRT3 = numpy.sqrt(3)\nSQRT3OVER2 = SQRT3 / 2.\n\n### Auxilliary Functions ###\n\n\ndef unzip(l):\n \"\"\"[(a1, b1), ..., (an, bn)] ----> ([a1, ..., an], [b1, ..., bn])\"\"\"\n return list(zip(*l))\n\n\ndef normalize(l):\n \"\"\"\n Normalizes input list.\n\n Parameters\n ----------\n l: list\n The list to be normalized\n\n Returns\n -------\n The normalized list or numpy array\n\n Raises\n ------\n ValueError, if the list sums to zero\n \"\"\"\n\n s = float(sum(l))\n if s == 0:\n raise ValueError(\"Cannot normalize list with sum 0\")\n return [x / s for x in l]\n\n\ndef simplex_iterator(scale, boundary=True):\n \"\"\"\n Systematically iterates through a lattice of points on the 2-simplex.\n\n Parameters\n ----------\n scale: Int\n The normalized scale of the simplex, i.e. N such that points (x,y,z)\n satisify x + y + z == N\n\n boundary: bool, True\n Include the boundary points (tuples where at least one\n coordinate is zero)\n\n Yields\n ------\n 3-tuples, There are binom(n+2, 2) points (the triangular\n number for scale + 1, less 3*(scale+1) if boundary=False\n \"\"\"\n\n start = 0\n if not boundary:\n start = 1\n for i in range(start, scale + (1 - start)):\n for j in range(start, scale + (1 - start) - i):\n k = scale - i - j\n yield (i, j, k)\n\n\n## Ternary Projections ##\n\n## Ternary Projections ##\n\ndef rotate_point(p, angle=None):\n \n \n \"\"\"\n Rotate the point p \"angle\" degrees, multiplying the point by the rotation matrix\n\n p = [x,y]\n\n rotation matrix\n | cos(angle) , -sin(angle) |\n | sin(angle) , cos(angle) |\n\n \"\"\"\n if(isinstance(angle, type(None))):\n return p\n\n rotation_matrix = [[numpy.cos(numpy.deg2rad(angle)) , -numpy.sin(numpy.deg2rad(angle))] , [numpy.sin(numpy.deg2rad(angle)) , numpy.cos(numpy.deg2rad(angle))]]\n rotated_point = numpy.dot(rotation_matrix,p)\n return rotated_point\n\ndef permute_point(p, permutation=None):\n \"\"\"\n Permutes the point according to the permutation keyword argument. The\n default permutation is \"012\" which does not change the order of the\n coordinate. To rotate counterclockwise, use \"120\" and to rotate clockwise\n use \"201\".\"\"\"\n if not permutation:\n return p\n return [p[int(permutation[i])] for i in range(len(p))]\n\n\ndef project_point(p, permutation=None, angle = None):\n \"\"\"\n Maps (x,y,z) coordinates to planar simplex.\n\n Parameters\n ----------\n p: 3-tuple\n The point to be projected p = (x, y, z)\n permutation: string, None, equivalent to \"012\"\n The order of the coordinates, counterclockwise from the origin\n \"\"\"\n permuted = permute_point(p, permutation=permutation)\n a = permuted[0]\n b = permuted[1]\n x = a + b/2.\n y = SQRT3OVER2 * b\n \n point = numpy.array([x,y])\n rotated_point = rotate_point(point, angle = angle)\n\n return rotated_point\n\n\n\ndef project_sequence(s, permutation=None, angle = None):\n \"\"\"\n Projects a point or sequence of points using `project_point` to lists xs, ys\n for plotting with Matplotlib.\n\n Parameters\n ----------\n s, Sequence-like\n The sequence of points (3-tuples) to be projected.\n\n Returns\n -------\n xs, ys: The sequence of projected points in coordinates as two lists \n \"\"\"\n\n xs, ys = unzip([project_point(p, permutation=permutation, angle = angle) for p in s])\n return xs, ys\n\n\n# Convert coordinates for custom plots with limits\n\ndef convert_coordinates(q, conversion, axisorder):\n \"\"\"\n Convert a 3-tuple in data coordinates into to simplex data\n coordinates for plotting.\n\n Parameters\n ----------\n q: 3-tuple\n the point to be plotted in data coordinates\n\n conversion: dict\n keys = ['b','l','r']\n values = lambda function giving the conversion\n axisorder: String giving the order of the axes for the coordinate tuple\n e.g. 'blr' for bottom, left, right coordinates.\n\n Returns\n -------\n p: 3-tuple\n The point converted to simplex coordinates.\n \"\"\"\n p = []\n for k in range(3):\n p.append(conversion[axisorder[k]](q[k]))\n\n return tuple(p)\n\n\ndef get_conversion(scale, limits):\n \"\"\"\n Get the conversion equations for each axis.\n\n limits: dict of min and max values for the axes in the order blr.\n \"\"\"\n fb = float(scale) / float(limits['b'][1] - limits['b'][0])\n fl = float(scale) / float(limits['l'][1] - limits['l'][0])\n fr = float(scale) / float(limits['r'][1] - limits['r'][0])\n\n conversion = {\"b\": lambda x: (x - limits['b'][0]) * fb,\n \"l\": lambda x: (x - limits['l'][0]) * fl,\n \"r\": lambda x: (x - limits['r'][0]) * fr}\n\n return conversion\n \n\ndef convert_coordinates_sequence(qs, scale, limits, axisorder):\n \"\"\"\n Take a sequence of 3-tuples in data coordinates and convert them\n to simplex coordinates for plotting. This is needed for custom\n plots where the scale of the simplex axes is set within limits rather\n than being defined by the scale parameter.\n\n Parameters\n ----------\n qs, sequence of 3-tuples\n The points to be plotted in data coordinates.\n\n scale: int\n The scale parameter for the plot.\n limits: dict\n keys = ['b','l','r']\n values = min,max data values for this axis.\n axisorder: String giving the order of the axes for the coordinate tuple\n e.g. 'blr' for bottom, left, right coordinates.\n\n Returns\n -------\n s, list of 3-tuples\n the points converted to simplex coordinates\n \"\"\"\n conversion = get_conversion(scale, limits)\n \n return [convert_coordinates(q, conversion, axisorder) for q in qs]\n"
]
| [
[
"numpy.dot",
"numpy.deg2rad",
"numpy.array",
"numpy.sqrt"
]
]
|
mjam03/epl | [
"ded3a931059f967366b1cc791ab466266b9a5332"
]
| [
"epl/features_parse.py"
]
| [
"import datetime as dt\nfrom functools import reduce\nimport numpy as np\nimport pandas as pd\n\nfrom epl.query import create_and_query, create_conn, get_table_columns, query_creator, query_db, table_exists\n\nFEATURE_KEY_COLS = ['Date', 'Team']\nFEATURE_ID_COLS = ['Country', 'Div', 'Season']\n\n\ndef create_features_key_col(df):\n '''\n Returns orig df with a new key column\n Key is concat of str() date with team name\n '''\n if 'Date' in df.columns and 'Team' in df.columns:\n df['Key'] = df['Date'].apply(\n lambda x: x.strftime('%Y-%m-%d')+'_') + df['Team']\n return df\n else:\n print('Date and Team must be present in the df columns')\n return None\n\n\ndef get_current_feature_keys(table_name, uat=False):\n '''\n Returns list of keys currently in the features table\n Used to calc what new matches need features computed\n '''\n if table_exists(table_name, uat=uat):\n try:\n df = create_and_query(table_name, uat=uat, cols=FEATURE_KEY_COLS)\n df = create_features_key_col(df)\n curr_keys = list(df.Key.values)\n return curr_keys\n except:\n print('Failed trying to query {} table'.format(table_name))\n return []\n else:\n print(\"{} table doesn't exist\".format(table_name))\n return []\n\n\ndef get_new_matches(fixtures=False, uat=False):\n '''\n Returns df of [Date, Team] for matches with no matching feature data\n Gets current feature and match keys, diffs and returns matches\n '''\n # first query feature table to get the key cols\n if fixtures:\n curr_feat_keys = get_current_feature_keys('fixtures_features', uat=uat)\n else:\n curr_feat_keys = get_current_feature_keys('features', uat=uat)\n\n # next query matches table for the key cols for comparison\n desired_cols = ['Date', 'HomeTeam', 'AwayTeam'] + FEATURE_ID_COLS\n if fixtures:\n curr_match_keys = create_and_query(\n 'fixtures', uat=uat, cols=desired_cols)\n curr_match_keys = curr_match_keys.drop_duplicates()\n else:\n curr_match_keys = create_and_query(\n 'matches', uat=uat, cols=desired_cols)\n\n # transform HomeTeam and AwayTeam into singular Team column\n curr_match_keys = convert_home_away_df(curr_match_keys)\n curr_match_keys = create_features_key_col(curr_match_keys)\n\n new_matches = curr_match_keys[~curr_match_keys.Key.isin(curr_feat_keys)]\n new_matches = new_matches.drop(columns=['Key'])\n return new_matches\n\n\ndef get_feat_col_names(feats, streak_length, avg_type):\n '''\n Returns a dict of {[feat col name]: [feat_col_base]}\n e.g. for 'GF' feat, sl = 3, avg_type='Avg'\n output would be {AvgGF_3: GF}\n '''\n feat_cols = [avg_type+x+'_'+str(streak_length) for x in feats]\n return dict(zip(feat_cols, feats))\n\n\ndef get_feat_required_cols(feats):\n '''\n Returns dict of {feat_base: req_cols}\n '''\n return {k: list(v.values()) for k, v in feats.items()}\n\n\ndef create_col_map(feats, streak_length, avg_type):\n '''\n Returns dict of feat_col_name to component data\n e.g. {'AvgGF_3': {'GF': {'Home': 'FTHG', 'Away': 'FTAG'}}}\n '''\n col_map = {k: {v: feats[v]} for k, v in get_feat_col_names(\n feats.keys(), streak_length, avg_type).items()}\n return col_map\n\n\ndef convert_home_away_df(df):\n '''\n Returns df with HomeTeam / AwayTeam cols melted into {Team, Location}\n Required as feature data stored as {Date,Team}\n for ease of creation and time series analysis\n '''\n col_map = {'HomeTeam': 'Home', 'AwayTeam': 'Away'}\n other_cols = [x for x in df.columns if x not in col_map.keys()]\n\n df = pd.melt(df, id_vars=other_cols, var_name='Location',\n value_vars=col_map.keys(), value_name='Team')\n df['Location'] = df['Location'].map(col_map)\n\n new_key = ['Date', 'Team', 'Location']\n new_cols = new_key + [x for x in df.columns if x not in new_key]\n df = df.sort_values(new_key)\n df = df[new_cols]\n return df\n\n\ndef create_base_feat_cols(df, fts, loc):\n '''\n Returns df with base cols required added on\n df: df of raw match data with required raw cols e.g. FTHG, FTAG\n fts: dict of feature col_map\n loc: str enum of ['All', 'Home', 'Away']\n '''\n df_new = df.copy()\n for k, v in fts.items():\n # get building blocks\n ft = list(v.values())[0]\n col_name = list(v.keys())[0]\n\n finish_a_or_h = (('A' == col_name[-1:]) or ('H' == col_name[-1:]))\n if finish_a_or_h and (col_name[:-1] in df_new.columns):\n df_new[col_name] = df_new[col_name[:-1]]\n else:\n if 'PPG' in col_name:\n # need to map to points\n h_map = {'H': 3, 'D': 1, 'A': 0}\n a_map = {'H': 0, 'D': 1, 'A': 3}\n if loc == 'All':\n df_new[col_name] = np.where(df.Location == 'Home', df_new[ft['Home']].map(\n h_map), df_new[ft['Away']].map(a_map))\n elif loc == 'Home':\n df_new[col_name] = df_new[ft['Home']].map(h_map)\n elif loc == 'Away':\n df_new[col_name] = df_new[ft['Away']].map(a_map)\n else:\n if loc == 'All':\n df_new[col_name] = np.where(\n df_new.Location == 'Home', df_new[ft['Home']], df_new[ft['Away']])\n elif loc == 'Home':\n df_new[col_name] = df_new[ft['Home']]\n elif loc == 'Away':\n df_new[col_name] = df_new[ft['Away']]\n\n return df_new\n\n\ndef split_home_away_feats(feats):\n '''\n Splits feature col map into 3 consti dicts\n Required to then create relevant feats separately\n '''\n # need to split out\n all_feats = {}\n home_feats = {}\n away_feats = {}\n\n for k, v in feats.items():\n\n const_cols = list(v.values())[0]\n home_away = const_cols.keys()\n if 'Home' in home_away and 'Away' in home_away:\n all_feats[k] = v\n elif 'Away' not in home_away:\n home_feats[k] = v\n elif 'Home'not in home_away:\n away_feats[k] = v\n\n return all_feats, home_feats, away_feats\n\n\ndef get_feats_raw_data(feats, uat=False):\n '''\n Returns raw matches df used for feature calculation\n Gets the required key cols as well as extra ID cols\n '''\n\n # get the required raw data columns from the required feats dict\n req_cols_dict = get_feat_required_cols(feats)\n req_cols = list(set([j for i in req_cols_dict.values() for j in i]))\n\n # define query\n key_cols = ['Date', 'HomeTeam', 'AwayTeam']\n all_cols = key_cols + req_cols\n if table_exists('matches', uat=uat):\n df = create_and_query('matches', uat=uat, cols=all_cols)\n df = convert_home_away_df(df)\n df = df.sort_values(FEATURE_KEY_COLS[::-1])\n return df\n else:\n print(\"matches table doesn't exist - need to create it first\")\n return None\n\n\ndef calc_rolling_avg(df, feats, streak_length):\n '''\n Accept df of raw data, col_map dict of feats and avg length\n Computes rolling avg data for the specified features and renames cols\n '''\n\n # get the cols to avg then rename\n col_to_avg = [list(x.keys())[0] for x in feats.values()]\n col_rename_dict = dict(zip(col_to_avg, feats.keys()))\n\n # get the key cols and create new df with only data we need\n key_cols = FEATURE_KEY_COLS + ['Location']\n # df_ft forms our basis to which we will lj on the avg features\n df_ft = df[key_cols + col_to_avg]\n\n # create mean, shift and remove incorrectly shifted values\n df_feats = df_ft[col_to_avg].groupby(\n df_ft['Team']).rolling(streak_length).mean()\n\n # rename cols and reset index to only orig index (not Team included)\n df_feats = df_feats.rename(columns=col_rename_dict)\n df_feats = df_feats.reset_index()\n df_feats = df_feats.set_index('level_1')\n\n # lj onto the original data, sort and return\n feat_cols = col_rename_dict.values()\n df_out = pd.merge(\n left=df_ft, right=df_feats[feat_cols], how='left', left_index=True, right_index=True)\n df_out = df_out.sort_values(FEATURE_KEY_COLS[::-1])\n return df_out\n\n\ndef merge_home_away(ft_dfs, all_feats, home_feats, away_feats, shift=True):\n\n # join them and handle conditional ffill\n key_cols = FEATURE_KEY_COLS + ['Location']\n # order list of dfs so 'All' is first\n ft_d = [ft_dfs['All'], ft_dfs['Home'], ft_dfs['Away']]\n df_merged = reduce(lambda left, right: pd.merge(\n left, right, on=key_cols, how='left'), ft_d)\n\n # now we want to ffill all home and away cols\n # NaNs created when joining home / away on to all match data\n # as e.g. AvgGFA_3 doesn't exist on rows where Location == 'Home'\n # we thus need to ffill this PER TEAM from the previous Away result\n\n # create list of home and away specific feature cols\n home_away_cols = list(home_feats.keys()) + list(away_feats.keys())\n # ffill these cols once grouped by team\n df_merged[home_away_cols] = df_merged[['Team'] +\n home_away_cols].groupby(['Team']).ffill()\n\n if shift:\n # now we need to shift games down 1\n # this is to prevent taking into account the score in todays game\n # i.e. all avg features should be backward looking (and not include obs today)\n # e.g. 0-0 draw in last 3 games but today match 6-0\n # AvgGF_3 should be 0, not (6+0+0)/3 = 2\n # however we only do this for home and away feats for those games\n # the above fwd fill procedure handles away feats for home matches and vice versa\n df_merged.update(df_merged[all_feats.keys()].groupby(\n df_merged['Team']).shift(1))\n df_merged.update(df_merged[df_merged.Location == 'Home'][home_feats.keys()].groupby(\n df_merged['Team']).shift(1))\n df_merged.update(df_merged[df_merged.Location == 'Away'][away_feats.keys()].groupby(\n df_merged['Team']).shift(1))\n\n return df_merged\n\n\ndef handle_feats(feat_list, fixtures=False):\n\n # for each list of features, applies correct function to args\n # concats them all together columnwise at the end\n feat_dfs = []\n\n # get the new matches we need to compute for\n df_new_matches = get_new_matches(fixtures=fixtures)\n if len(df_new_matches) == 0:\n print('No new matches to process features for')\n return None\n\n # now iterate over each feature set, get raw data, compute and return\n for feat_desc in feat_list:\n if feat_desc['feat_type'] == 'avg':\n # unpack args\n feats = feat_desc['feat_dict']\n streak_length = feat_desc['streak']\n avg_type = feat_desc['avg_type']\n\n # get the raw data required to calc the features\n df_raw = get_feats_raw_data(feats)\n # restrict to only teams in the new matches df\n df_raw = df_raw[df_raw.Team.isin(df_new_matches.Team.unique())]\n\n # create map from feat_name to base_name to construction\n # split out by All / Home / Away for sequential calc\n col_map = create_col_map(feats, streak_length, avg_type)\n all_feats, home_feats, away_feats = split_home_away_feats(col_map)\n\n # create base cols e.g. GF / GA for use to calc e.g. AvgGF_3\n c_dict = {'All': all_feats, 'Home': home_feats, 'Away': away_feats}\n for k, v in c_dict.items():\n if len(v) > 0:\n df_raw = create_base_feat_cols(df_raw, v, k)\n elif k == 'All':\n # if all feats blank then issue so report\n print('All features is blank - probably an error: {}'.format(v))\n\n # compute feats\n ft_dfs = {}\n for k, v in c_dict.items():\n # if not all, then restrict data to only home/away games\n if k != 'All':\n df_r = df_raw[df_raw.Location == k]\n df_f = calc_rolling_avg(df_r, v, streak_length)\n else:\n df_f = calc_rolling_avg(df_raw, v, streak_length)\n # add to dict\n ft_dfs[k] = df_f\n\n df_feats = merge_home_away(\n ft_dfs, all_feats, home_feats, away_feats, shift=not(fixtures))\n\n # now we have our correctly offset feats\n # we need to select just the cols we need\n id_cols = FEATURE_KEY_COLS + ['Location']\n df_feats = df_feats[id_cols + list(col_map.keys())]\n\n feat_dfs.append(df_feats)\n else:\n print('Not supported yet')\n\n # now we concat them altogether along common key\n key_cols = FEATURE_KEY_COLS + ['Location']\n df_merged = reduce(lambda left, right: pd.merge(\n left, right, on=key_cols, how='outer'), feat_dfs)\n\n # now we only return the matches we needed\n if fixtures:\n # drop location for fixtures as we want most recent game regardless\n df_merged = df_merged.drop(columns=['Location'])\n # sort by date, then team for asof join\n df_merged = df_merged.sort_values(FEATURE_KEY_COLS)\n # backwards as of join the data on\n df_final = pd.merge_asof(df_new_matches, df_merged, on='Date', by=[\n 'Team'], direction='backward', allow_exact_matches=False)\n else:\n df_final = pd.merge(left=df_new_matches,\n right=df_merged, how='left', on=key_cols)\n return df_final\n\n\ndef get_requested_feat_cols(feat_list):\n\n feats = []\n for f in feat_list:\n fts = f['feat_dict']\n sl = f['streak']\n avg_type = f['avg_type']\n ft_col_map = create_col_map(fts, sl, avg_type)\n for k in ft_col_map.keys():\n feats.append(k)\n\n return feats\n\n\ndef process_feature_data(feat_list, fixtures=False, uat=False):\n '''\n Handles process of:\n - Identifying new matches that require features\n - Checking if the features requested line up to the columns in the feature table already\n - Compute new features data\n - Set/append down into sqlite\n '''\n if fixtures:\n table_name = 'fixtures_features'\n else:\n table_name = 'features'\n # first check if the columns requested are equal to those in the table\n # will handle this in future but for now just throw an error\n req_feats = get_requested_feat_cols(feat_list)\n new_feat_cols = FEATURE_KEY_COLS + \\\n ['Location'] + FEATURE_ID_COLS + req_feats\n\n cols_match = False\n if table_exists(table_name, uat=uat):\n curr_cols = get_table_columns(table_name, uat=uat)\n # use sets as all cols should be unique names and this gens order\n cols_match = (set(curr_cols) == set(new_feat_cols))\n else:\n print(\"Table doesn't exist yet so fine to set new col schema\")\n cols_match = True\n\n # if cols match then can go ahead and process\n if cols_match:\n df = handle_feats(feat_list, fixtures=fixtures)\n else:\n # for now given sqlite limitations, check if new feats > existing cols\n # if so then we delete the current features table and set new one\n if set(curr_cols).issubset(set(new_feat_cols)):\n print(\n 'Old cols subset of new requested {} - deleting and recreating'.format(table_name))\n conn = create_conn(uat=uat)\n cur = conn.cursor()\n cur.execute('DROP TABLE {}'.format(table_name))\n conn.commit()\n conn.execute('VACUUM')\n conn.close()\n df = handle_feats(feat_list, fixtures=fixtures)\n else:\n print(\n 'New requested cols do not match the existing {} columns'.format(table_name))\n return None\n\n # now we have a df for the new matches\n if df is None:\n # then no new matches and exit\n print('Exiting feature processing for table {}'.format(table_name))\n return None\n else:\n # we need to set the table down into sql\n try:\n conn = create_conn(uat=uat)\n df.to_sql(table_name, conn, if_exists='append', index=False)\n except:\n print('Failed to set down / append to {} table post calc'.format(table_name))\n\n return df\n\n\nif __name__ == '__main__':\n None\n"
]
| [
[
"pandas.merge",
"numpy.where",
"pandas.merge_asof"
]
]
|
davidinouye/failing-loudly | [
"6eee78530d48b83cef41ee5a4faa82d6fd1d892b"
]
| [
"shift_tester.py"
]
| [
"# -------------------------------------------------\n# IMPORTS\n# -------------------------------------------------\n\nimport numpy as np\nimport torch\nfrom torch import *\nfrom torch_two_sample import *\nfrom scipy.stats import ks_2samp, binom_test, chisquare, chi2_contingency, anderson_ksamp\nfrom scipy.spatial import distance\n\nfrom shared_utils import *\n\n# -------------------------------------------------\n# SHIFT TESTER\n# -------------------------------------------------\n\nclass ShiftTester:\n\n def __init__(self, dim=TestDimensionality.One, sign_level=0.05, ot=None, mt=None):\n self.dim = dim\n self.sign_level = sign_level\n self.ot = ot\n self.mt = mt\n\n def test_shift(self, X_tr, X_te):\n if self.ot is not None:\n return self.one_dimensional_test(X_tr, X_te)\n elif self.mt is not None:\n return self.multi_dimensional_test(X_tr, X_te)\n\n def test_chi2_shift(self, X_tr, X_te, nb_classes):\n\n # Calculate observed and expected counts\n freq_exp = np.zeros(nb_classes)\n freq_obs = np.zeros(nb_classes)\n\n unique_tr, counts_tr = np.unique(X_tr, return_counts=True)\n total_counts_tr = np.sum(counts_tr)\n unique_te, counts_te = np.unique(X_te, return_counts=True)\n total_counts_te = np.sum(counts_te)\n\n for i in range(len(unique_tr)):\n val = counts_tr[i]\n freq_exp[unique_tr[i]] = val\n \n for i in range(len(unique_te)):\n freq_obs[unique_te[i]] = counts_te[i]\n\n if np.amin(freq_exp) == 0 or np.amin(freq_obs) == 0:\n # The chi-squared test using contingency tables is not well defined if zero-element classes exist, which\n # might happen in the low-sample regime. In this case, we calculate the standard chi-squared test.\n for i in range(len(unique_tr)):\n val = counts_tr[i] / total_counts_tr * total_counts_te\n freq_exp[unique_tr[i]] = val\n _, p_val = chisquare(freq_obs, f_exp=freq_exp)\n else:\n # In almost all cases, we resort to obtaining a p-value from the chi-squared test's contingency table.\n freq_conc = np.array([freq_exp, freq_obs])\n _, p_val, _, _ = chi2_contingency(freq_conc)\n \n return p_val\n\n def test_shift_bin(self, k, n, test_rate):\n p_val = binom_test(k, n, test_rate)\n return p_val\n\n def one_dimensional_test(self, X_tr, X_te):\n p_vals = []\n\n # For each dimension we conduct a separate KS test\n for i in range(X_tr.shape[1]):\n feature_tr = X_tr[:, i]\n feature_te = X_te[:, i]\n\n t_val, p_val = None, None\n\n if self.ot == OnedimensionalTest.KS:\n\n # Compute KS statistic and p-value\n t_val, p_val = ks_2samp(feature_tr, feature_te)\n elif self.ot == OnedimensionalTest.AD:\n t_val, _, p_val = anderson_ksamp([feature_tr.tolist(), feature_te.tolist()])\n\n p_vals.append(p_val)\n\n # Apply the Bonferroni correction to bound the family-wise error rate. This can be done by picking the minimum\n # p-value from all individual tests.\n p_vals = np.array(p_vals)\n p_val = min(np.min(p_vals), 1.0)\n\n return p_val, p_vals\n\n def multi_dimensional_test(self, X_tr, X_te):\n\n # torch_two_sample somehow wants the inputs to be explicitly casted to float 32.\n X_tr = X_tr.astype(np.float32)\n X_te = X_te.astype(np.float32)\n\n p_val = None\n\n # We provide a couple of different tests, although we only report results for MMD in the paper.\n if self.mt == MultidimensionalTest.MMD:\n mmd_test = MMDStatistic(len(X_tr), len(X_te))\n\n # As per the original MMD paper, the median distance between all points in the aggregate sample from both\n # distributions is a good heuristic for the kernel bandwidth, which is why compute this distance here.\n if len(X_tr.shape) == 1:\n X_tr = X_tr.reshape((len(X_tr),1))\n X_te = X_te.reshape((len(X_te),1))\n all_dist = distance.cdist(X_tr, X_te, 'euclidean')\n else:\n all_dist = distance.cdist(X_tr, X_te, 'euclidean')\n median_dist = np.median(all_dist)\n\n # Calculate MMD.\n t_val, matrix = mmd_test(torch.autograd.Variable(torch.tensor(X_tr)),\n torch.autograd.Variable(torch.tensor(X_te)),\n alphas=[1/median_dist], ret_matrix=True)\n p_val = mmd_test.pval(matrix)\n elif self.mt == MultidimensionalTest.Energy:\n energy_test = EnergyStatistic(len(X_tr), len(X_te))\n t_val, matrix = energy_test(torch.autograd.Variable(torch.tensor(X_tr)),\n torch.autograd.Variable(torch.tensor(X_te)),\n ret_matrix=True)\n p_val = energy_test.pval(matrix)\n elif self.mt == MultidimensionalTest.FR:\n fr_test = FRStatistic(len(X_tr), len(X_te))\n t_val, matrix = fr_test(torch.autograd.Variable(torch.tensor(X_tr)),\n torch.autograd.Variable(torch.tensor(X_te)),\n norm=2, ret_matrix=True)\n p_val = fr_test.pval(matrix)\n elif self.mt == MultidimensionalTest.KNN:\n knn_test = KNNStatistic(len(X_tr), len(X_te), 20)\n t_val, matrix = knn_test(torch.autograd.Variable(torch.tensor(X_tr)),\n torch.autograd.Variable(torch.tensor(X_te)),\n norm=2, ret_matrix=True)\n p_val = knn_test.pval(matrix)\n \n return p_val, np.array([])\n"
]
| [
[
"scipy.stats.ks_2samp",
"scipy.stats.chi2_contingency",
"numpy.unique",
"numpy.min",
"scipy.stats.binom_test",
"numpy.median",
"numpy.amin",
"scipy.spatial.distance.cdist",
"torch.tensor",
"scipy.stats.chisquare",
"numpy.array",
"numpy.zeros",
"numpy.sum"
]
]
|
SiddeshSambasivam/pytorch | [
"ef5ac15f592f335eeb314bfceaf5d9af953ad15b"
]
| [
"test/test_jit.py"
]
| [
"# -*- coding: utf-8 -*-\nimport torch\n\n# This is how we include tests located in test/jit/...\n# They are included here so that they are invoked when you call `test_jit.py`,\n# do not run these test files directly.\nfrom jit.test_tracer import TestTracer, TestMixTracingScripting # noqa: F401\nfrom jit.test_recursive_script import TestRecursiveScript # noqa: F401\nfrom jit.test_type_sharing import TestTypeSharing # noqa: F401\nfrom jit.test_logging import TestLogging # noqa: F401\nfrom jit.test_backends import TestBackends, TestBackendsWithCompiler # noqa: F401\nfrom jit.test_list_dict import TestList, TestDict, TestNamedTuple, TestScriptDict, TestScriptList # noqa: F401\nfrom jit.test_async import TestAsync # noqa: F401\nfrom jit.test_data_parallel import TestDataParallel # noqa: F401\nfrom jit.test_models import TestModels # noqa: F401\nfrom jit.test_modules import TestModules # noqa: F401\nfrom jit.test_autodiff_subgraph_slicing import TestAutodiffSubgraphSlicing # noqa: F401\nfrom jit.test_custom_operators import TestCustomOperators # noqa: F401\nfrom jit.test_export_modes import TestExportModes # noqa: F401\nfrom jit.test_graph_rewrite_passes import TestGraphRewritePasses # noqa: F401\nfrom jit.test_class_type import TestClassType # noqa: F401\nfrom jit.test_builtins import TestBuiltins, TestTensorBuiltins # noqa: F401\nfrom jit.test_ignore_context_manager import TestIgnoreContextManager # noqa: F401\nfrom jit.test_symbolic_shape_analysis import TestSymbolicShapeAnalysis # noqa: F401\nfrom jit.test_unsupported_ops import TestUnsupportedOps # noqa: F401\nfrom jit.test_freezing import TestFreezing, TestFrozenOptimizations, TestMKLDNNReinplacing # noqa: F401\nfrom jit.test_peephole import TestPeephole # noqa: F401\nfrom jit.test_save_load import TestSaveLoad # noqa: F401\nfrom jit.test_module_containers import TestModuleContainers # noqa: F401\nfrom jit.test_python_bindings import TestPythonBindings # noqa: F401\nfrom jit.test_python_ir import TestPythonIr # noqa: F401\nfrom jit.test_functional_blocks import TestFunctionalBlocks # noqa: F401\nfrom jit.test_remove_mutation import TestRemoveMutation # noqa: F401\nfrom jit.test_torchbind import TestTorchbind # noqa: F401\nfrom jit.test_module_interface import TestModuleInterface # noqa: F401\nfrom jit.test_onnx_export import TestONNXExport # noqa: F401\nfrom jit.test_with import TestWith # noqa: F401\nfrom jit.test_enum import TestEnum # noqa: F401\nfrom jit.test_string_formatting import TestStringFormatting # noqa: F401\nfrom jit.test_profiler import TestProfiler # noqa: F401\nfrom jit.test_slice import TestSlice # noqa: F401\nfrom jit.test_ignorable_args import TestIgnorableArgs # noqa: F401\nfrom jit.test_hooks import TestHooks # noqa: F401\nfrom jit.test_warn import TestWarn # noqa: F401\nfrom jit.test_isinstance import TestIsinstance # noqa: F401\nfrom jit.test_cuda import TestCUDA # noqa: F401\nfrom jit.test_python_builtins import TestPythonBuiltinOP # noqa: F401\nfrom jit.test_typing import TestTyping # noqa: F401\nfrom jit.test_hash import TestHash # noqa: F401\nfrom jit.test_complex import TestComplex # noqa: F401\nfrom jit.test_jit_utils import TestJitUtils # noqa: F401\nfrom jit.test_scriptmod_ann import TestScriptModuleInstanceAttributeTypeAnnotation # noqa: F401\nfrom jit.test_types import TestTypesAndAnnotation # noqa: F401\nfrom jit.test_misc import TestMisc # noqa: F401\nfrom jit.test_pdt import TestPDT # noqa: F401\nfrom jit.test_tensor_creation_ops import TestTensorCreationOps # noqa: F401\nfrom jit.test_module_apis import TestModuleAPIs # noqa: F401\nfrom jit.test_script_profile import TestScriptProfile # noqa: F401\nfrom jit.test_convert_activation import TestFunctionalToInplaceActivation, TestInplaceToFunctionalActivation # noqa: F401\nfrom jit.test_parametrization import TestParametrization # noqa: F401\n\n# Torch\nfrom torch import Tensor\nfrom torch._C import TensorType, BoolType, parse_ir, _propagate_shapes\nfrom torch._six import PY37\nfrom torch.autograd import Variable\nfrom torch.jit.annotations import BroadcastingList2, BroadcastingList3, Any # noqa: F401\nfrom torch.nn.utils.rnn import PackedSequence\nfrom torch.testing import FileCheck\nfrom torch.testing._internal.common_utils import make_tensor\nimport torch.autograd.profiler\nimport torch.cuda\nimport torch.jit\nimport torch.jit._logging\nimport torch.jit.frontend\nimport torch.nn as nn\nimport torch.nn.functional as F\n\n# Testing utils\nfrom torch.testing._internal import jit_utils\nfrom torch.testing._internal.common_jit import check_against_reference\nfrom torch.testing._internal.common_utils import run_tests, IS_WINDOWS, TEST_WITH_UBSAN, \\\n suppress_warnings, IS_SANDCASTLE, GRAPH_EXECUTOR, ProfilingMode, \\\n freeze_rng_state, slowTest, TemporaryFileName, skipIfCompiledWithoutNumpy, \\\n enable_profiling_mode_for_profiling_tests, TEST_MKL, set_default_dtype, num_profiled_runs\nfrom torch.testing._internal.jit_utils import JitTestCase, enable_cpu_fuser, disable_autodiff_subgraph_inlining, \\\n _trace, do_input_map, get_execution_plan, make_global, \\\n execWrapper, _inline_everything, _tmp_donotuse_dont_inline_everything, \\\n RUN_CUDA\nfrom torch.testing._internal.jit_metaprogramming_utils import create_script_fn, nn_functional_tests, get_script_args, \\\n EXCLUDE_SCRIPT, additional_module_tests, EXCLUDE_SCRIPT_MODULES, \\\n get_nn_module_name_from_kwargs, script_method_template\n\nfrom torch.testing._internal.common_nn import module_tests, new_module_tests, criterion_tests\nfrom torch.testing._internal.common_methods_invocations import (\n create_input, unpack_variables)\n\n# For testing truediv in python 2\nfrom torch.testing._internal.test_module.future_div import div_int_future, div_float_future\nfrom torch.testing._internal.test_module.no_future_div import div_int_nofuture, div_float_nofuture\n\n# Standard library\nfrom collections import defaultdict, namedtuple, OrderedDict\nfrom copy import deepcopy\nfrom itertools import product\nfrom textwrap import dedent\nfrom typing import List, Dict, NamedTuple, Optional, Tuple, Union\nimport copy\nimport functools\nimport inspect\nimport io\nimport itertools\nimport math\nimport numpy as np\nimport os\nimport pickle\nimport pickletools\nimport random\nimport re\nimport shutil\nimport string\nimport sys\nimport tempfile\nimport types\nimport typing\nimport unittest\nimport warnings\nimport zipfile\n\n\ndef canonical(graph):\n return torch._C._jit_pass_canonicalize(graph).str(False)\n\ndef LSTMCellF(input, hx, cx, *params):\n return LSTMCell(input, (hx, cx), *params)\n\ndef doAutodiffCheck(testname):\n # TODO: setting false on test itself is not working\n if \"test_t_\" in testname or testname == \"test_t\":\n return False\n\n if GRAPH_EXECUTOR == ProfilingMode.SIMPLE:\n return False\n\n if GRAPH_EXECUTOR == ProfilingMode.LEGACY:\n return True\n\n\n # these tests are disabled because BailOut nodes\n # inserted by ProfilingExecutor interfere with\n # subgraph slicing of Differentiable Graphs\n test_exceptions = [\n # functional\n 'test_nn_dropout',\n 'test_nn_log_softmax',\n 'test_nn_relu',\n 'test_nn_softmax',\n 'test_nn_threshold',\n 'test_nn_lp_pool2d',\n 'test_nn_lp_pool1d',\n 'test_nn_gumbel_softmax_hard',\n 'test_nn_gumbel_softmax',\n 'test_nn_multilabel_soft_margin_loss',\n 'test_nn_batch_norm',\n 'test_nn_max_pool2d_with_indices',\n # AutogradJitGenerated\n 'test___rdiv___constant',\n 'test___rdiv___scalar_constant',\n 'test_split',\n 'test_split_dim',\n 'test_split_dim_neg0',\n 'test_split_size_list',\n 'test_split_size_list_dim',\n 'test_split_size_list_dim_neg0',\n 'test_split_with_sizes',\n 'test_split_with_sizes_dim',\n 'test_split_with_sizes_dim_neg0',\n 'test_split_with_sizes_size_0',\n 'test_nn_max_pool2d_with_indices',\n ]\n\n if testname in test_exceptions:\n return False\n return True\n\n\n# TODO: enable TE in PE when all tests are fixed\ntorch._C._jit_set_texpr_fuser_enabled(GRAPH_EXECUTOR == ProfilingMode.PROFILING)\ntorch._C._jit_set_profiling_executor(GRAPH_EXECUTOR != ProfilingMode.LEGACY)\n# even though FULL_PROFILER should be our default\n# we haven't tested every single test in this file\n# but we enable FULL_PROFILER for a large subset\n# of the tests with \"with enable_profiling_mode_for_profiling_tests\"\ntorch._C._jit_set_profiling_mode(False)\n\ndef LSTMCell(input, hidden, w_ih, w_hh, b_ih=None, b_hh=None):\n hx, cx = hidden\n gates = F.linear(input, w_ih, b_ih) + F.linear(hx, w_hh, b_hh)\n\n ingate, forgetgate, cellgate, outgate = gates.chunk(4, 1)\n ingate = torch.sigmoid(ingate)\n forgetgate = torch.sigmoid(forgetgate)\n cellgate = torch.tanh(cellgate)\n outgate = torch.sigmoid(outgate)\n\n cy = (forgetgate * cx) + (ingate * cellgate)\n hy = outgate * torch.tanh(cy)\n return hy, cy\n\n\ndef LSTMCellC(*args, **kwargs):\n hy, cy = LSTMCellF(*args, **kwargs)\n return torch.cat((hy, cy))\n\n\ndef LSTMCellS(x, hx, cx, w_ih, w_hh, b_ih, b_hh):\n gates = x.mm(w_ih.t()) + hx.mm(w_hh.t()) + b_ih + b_hh\n ingate, forgetgate, cellgate, outgate = gates.chunk(4, 1)\n ingate = torch.sigmoid(ingate)\n forgetgate = torch.sigmoid(forgetgate)\n cellgate = torch.tanh(cellgate)\n outgate = torch.sigmoid(outgate)\n cy = (forgetgate * cx) + (ingate * cellgate)\n hy = outgate * torch.tanh(cy)\n return hy, cy\n\n\n# Code reference: https://github.com/pytorch/translate/blob/master/pytorch_translate/rnn_cell.py#L27:44\ndef MiLSTMCell(x, hx, cx, w_ih, w_hh, alpha, beta_i, beta_h, bias):\n Wx = x.mm(w_ih.t())\n Uz = hx.mm(w_hh.t())\n # Section 2.1 in https://arxiv.org/pdf/1606.06630.pdf\n gates = alpha * Wx * Uz + beta_i * Wx + beta_h * Uz + bias\n # Same as LSTMCell after this point\n ingate, forgetgate, cellgate, outgate = gates.chunk(4, 1)\n ingate = ingate.sigmoid()\n forgetgate = forgetgate.sigmoid()\n cellgate = cellgate.tanh()\n outgate = outgate.sigmoid()\n cy = (forgetgate * cx) + (ingate * cellgate)\n hy = outgate * cy.tanh()\n return hy, cy\n\n\n\ndef get_lstm_inputs(device, training=False, seq_length=None):\n input_shape = (3, 10) if seq_length is None else (seq_length, 3, 10)\n input = torch.randn(*input_shape, dtype=torch.float, device=device, requires_grad=training)\n hx = torch.randn(3, 20, dtype=torch.float, device=device, requires_grad=training)\n cx = torch.randn(3, 20, dtype=torch.float, device=device, requires_grad=training)\n module = nn.LSTMCell(10, 20).to(device, torch.float) # Just to allocate weights with correct sizes\n if training:\n params = tuple(module.parameters())\n else:\n params = tuple(p.requires_grad_(False) for p in module.parameters())\n return (input, hx, cx) + params\n\n\ndef get_milstm_inputs(device, training=False):\n minibatch = 3\n input_size = 10\n hidden_size = 20\n x = torch.randn(minibatch, input_size, device=device, dtype=torch.float)\n hx = torch.randn(minibatch, hidden_size, device=device, dtype=torch.float)\n cx = torch.randn(minibatch, hidden_size, device=device, dtype=torch.float)\n\n ih = torch.randn(4 * hidden_size, input_size, device=device, dtype=torch.float, requires_grad=training)\n hh = torch.randn(4 * hidden_size, hidden_size, device=device, dtype=torch.float, requires_grad=training)\n alpha = torch.randn(4 * hidden_size, dtype=torch.float, device=device, requires_grad=training)\n ibeta = torch.randn(4 * hidden_size, dtype=torch.float, device=device, requires_grad=training)\n hbeta = torch.randn(4 * hidden_size, dtype=torch.float, device=device, requires_grad=training)\n bias = torch.randn(4 * hidden_size, dtype=torch.float, device=device, requires_grad=training)\n return x, hx, cx, ih, hh, alpha, ibeta, hbeta, bias\n\n\ndef get_fn(file_name, script_path):\n import importlib.util\n spec = importlib.util.spec_from_file_location(file_name, script_path)\n module = importlib.util.module_from_spec(spec)\n spec.loader.exec_module(module)\n fn = module.fn\n return fn\n\ndef get_grad_executor(plan_state, diff_graph_idx=None, skip_check=False):\n if diff_graph_idx is None:\n nodes = list(plan_state.graph.nodes())\n\n if not skip_check:\n nodes = list(filter(lambda n : n.kind() != \"prim::BailOut\" and n.kind() != \"prim::BailoutTemplate\", nodes))\n if len(nodes) == 1 or (len(nodes) == 2 and nodes[1].kind() == \"prim::TupleConstruct\"):\n pass\n elif len(nodes) == 2 and nodes[0].kind() == \"prim::RequiresGradCheck\" and nodes[1].kind() == \"prim::If\":\n pass\n else:\n raise RuntimeError(\"Can't get a grad_executor for a non-differentiable graph\")\n grad_executors = list(plan_state.code.grad_executor_states())\n return grad_executors[diff_graph_idx or 0]\n\n\ndef all_backward_graphs(script_module, diff_graph_idx=None):\n # Note: for Python 2 the order seems to be unstable\n ge_state = script_module.get_debug_state()\n fwd_plan = get_execution_plan(ge_state)\n grad_executor_state = get_grad_executor(fwd_plan, diff_graph_idx=diff_graph_idx)\n bwd_plans = list(grad_executor_state.execution_plans.values())\n return [p.graph.copy() for p in bwd_plans]\n\n\ndef backward_graph(script_module, diff_graph_idx=None, skip_check=False):\n ge_state = script_module.get_debug_state()\n fwd_plan = get_execution_plan(ge_state)\n grad_executor_state = get_grad_executor(fwd_plan, diff_graph_idx=diff_graph_idx, skip_check=skip_check)\n bwd_plan = get_execution_plan(grad_executor_state)\n # Running JIT passes requires that we own the graph (with a shared_ptr).\n # The debug state struct does not own its graph so we make a copy of it.\n return bwd_plan.graph.copy()\n\n\n# helper function to get sum of List[Tensor]\ndef _sum_of_list(tensorlist):\n s = 0\n for t in tensorlist:\n s += t.sum()\n return s\n\n\n# has to be at top level or Pickle complains\nclass FooToPickle(torch.nn.Module):\n def __init__(self):\n super(FooToPickle, self).__init__()\n self.bar = torch.jit.ScriptModule()\n\nclass TestJit(JitTestCase):\n @unittest.skip(\"Requires a lot of RAM\")\n def test_big(self):\n m = torch.jit.ScriptModule()\n gig = int(1024 * 1024 * 1024 / 4)\n # a small tensor in the first 4GB\n m.v0 = nn.Parameter(torch.full((2,), 1, dtype=torch.float))\n # a large tensor in the first 4GB that ends outside of it\n m.v1 = nn.Parameter(torch.full((5, gig), 2, dtype=torch.float))\n # a small tensor in >4GB space\n m.v2 = nn.Parameter(torch.full((2,), 3, dtype=torch.float))\n # s large tensor in the > 4GB space\n m.v3 = nn.Parameter(torch.full((5, gig), 4, dtype=torch.float))\n\n m2 = self.getExportImportCopy(m)\n\n self.assertEqual(tuple(m.parameters()), tuple(m2.parameters()))\n\n def test_inferred_as_tensor(self):\n with self.assertRaisesRegex(RuntimeError, \"Inferred the value for argument 'dim' to be of type 'Tensor' \"\n \"because it was not annotated with an explicit type\"):\n @torch.jit.script\n def dot(points, query, dim):\n return (points * query).sum(dim)\n\n def test_constants_pkl(self):\n # This test asserts that the serialization archive includes a `constants.pkl`\n # file. This file is used by `torch.load` to determine whether a zip file\n # is a normal eager-mode serialization zip or a jit serialization zip. If\n # you are deleting `constants.pkl`, make sure to update `torch.serialization.load`\n # so it is still able to figure out which is which.\n @torch.jit.script\n def fn(x):\n return x\n\n buf = io.BytesIO()\n torch.jit.save(fn, buf)\n buf.seek(0)\n\n files = zipfile.ZipFile(buf).filelist\n self.assertTrue(any(['archive/constants.pkl' == f.filename for f in files]))\n\n def test_script_fn_pkl(self):\n\n with self.assertRaisesRegex(pickle.PickleError, \"ScriptFunction cannot be pickled\"):\n\n @torch.jit.script\n def fn(x: torch.Tensor) -> torch.Tensor:\n return x\n\n pkl_fn = pickle.dumps(fn, protocol=0)\n\n\n def test_restore_device(self):\n class M(torch.jit.ScriptModule):\n def __init__(self, cpu_device_str):\n super(M, self).__init__()\n self.p0 = nn.Parameter(torch.tensor([0.3], dtype=torch.float,\n device=cpu_device_str))\n self.b0 = torch.tensor([0.9], dtype=torch.float,\n device=cpu_device_str)\n\n # main purpose is checking map_location works\n m = M(\"cpu\")\n m2 = self.getExportImportCopy(m)\n self.assertEqual(tuple(m.parameters()), tuple(m2.parameters()))\n self.assertEqual(tuple(m.buffers()), tuple(m2.buffers()))\n self.assertFalse(m2.p0.is_cuda)\n self.assertFalse(m2.b0.is_cuda)\n\n def test_model_save_error(self):\n with TemporaryFileName() as fname:\n with self.assertRaisesRegex(pickle.PickleError, \"not supported\"):\n torch.save(FooToPickle(), fname)\n\n @unittest.skipIf(not RUN_CUDA, \"restore device requires CUDA\")\n def test_restore_device_cuda(self):\n class MyModule(torch.jit.ScriptModule):\n def __init__(self):\n super(MyModule, self).__init__()\n self.register_buffer('b0', torch.randn(1, 3))\n self.p0 = nn.Parameter(torch.randn(2, 3))\n\n @torch.jit.script_method\n def forward(self, x):\n return x + self.b0 + self.p0\n\n m = MyModule()\n m.cuda(torch.cuda.device_count() - 1)\n cuda_device_str = 'cuda:' + str(torch.cuda.device_count() - 1)\n\n self.assertTrue(m.p0.is_cuda)\n self.assertTrue(m.b0.is_cuda)\n\n # restore to the saved devices\n m2 = self.getExportImportCopy(m)\n self.assertEqual(tuple(m.parameters()), tuple(m2.parameters()))\n self.assertEqual(tuple(m.buffers()), tuple(m2.buffers()))\n self.assertEqual(str(m2.p0.device), cuda_device_str)\n self.assertEqual(str(m2.b0.device), cuda_device_str)\n\n # restore all to cpu using string\n cpu_device_str = 'cpu'\n m3 = self.getExportImportCopy(m, map_location=cpu_device_str)\n self.assertEqual(str(m3.p0.device), cpu_device_str)\n self.assertEqual(str(m3.b0.device), cpu_device_str)\n\n # restore all to first gpu using device\n m4 = self.getExportImportCopy(\n m3, map_location=torch.device('cuda:0'))\n self.assertEqual(str(m4.p0.device), 'cuda:0')\n self.assertEqual(str(m4.b0.device), 'cuda:0')\n\n # compute and compare the results\n input = torch.rand(2, 3).cuda(torch.cuda.device_count() - 1)\n origin_result = m(input)\n self.assertEqual(origin_result, m2(input))\n self.assertEqual(origin_result, m3(input.cpu()))\n self.assertEqual(origin_result, m4(input.cuda(0)))\n\n def test_trace_retains_train(self):\n class M(torch.nn.Module):\n def forward(self, x):\n return x\n m = M()\n m.eval()\n tm = torch.jit.trace(m, (torch.rand(3)))\n self.assertEqual(tm.training, m.training)\n\n @unittest.skipIf(not RUN_CUDA, \"restore device requires CUDA\")\n def test_restore_shared_storage_on_cuda(self):\n class Foo(torch.jit.ScriptModule):\n def __init__(self):\n super(Foo, self).__init__()\n whole_tensor = torch.randn(4, 5, dtype=torch.float, device='cpu')\n self.p0 = nn.Parameter(whole_tensor.narrow(0, 0, 1))\n self.register_buffer('b0', whole_tensor.narrow(0, 3, 1))\n\n m = Foo()\n m2 = self.getExportImportCopy(m, map_location=torch.device('cuda:0'))\n self.assertEqual(tuple(m.parameters()), tuple(m2.parameters()))\n self.assertEqual(tuple(m.buffers()), tuple(m2.buffers()))\n self.assertTrue(m2.p0.is_cuda)\n self.assertTrue(m2.b0.is_cuda)\n self.assertTrue(m2.p0.is_shared())\n self.assertTrue(m2.b0.is_shared())\n self.assertEqual(m2.b0.storage().data_ptr(), m2.p0.storage().data_ptr())\n\n def test_add_relu_fusion(self):\n class M(torch.nn.Module):\n def __init__(self, relu_op):\n super(M, self).__init__()\n self.relu_op = relu_op\n\n def forward(self, a, b, c):\n tmp = torch.add(a, b)\n x = self.relu_op(tmp)\n d = torch.add(a, c)\n return x + d\n a = torch.rand((7, 11))\n a = a * -10\n a = a + 5\n b = torch.rand((7, 11))\n c = torch.rand((7, 11))\n m = torch.jit.script(M(torch.relu))\n orig_res = m(a, b, c)\n torch._C._jit_pass_fuse_add_relu(m.graph)\n buffer = io.BytesIO()\n torch.jit.save(m, buffer)\n buffer.seek(0)\n m = torch.jit.load(buffer)\n new_res = m(a, b, c)\n FileCheck().check_not(\"aten::relu(\") \\\n .check(\"aten::_add_relu(\") \\\n .run(m.graph)\n torch.testing.assert_allclose(orig_res, new_res)\n\n # add, relu_\n a = torch.rand((7, 11))\n a = a * -10\n a = a + 5\n b = torch.rand((7, 11))\n c = torch.rand((7, 11))\n m = torch.jit.script(M(torch.relu_))\n orig_res = m(a, b, c)\n torch._C._jit_pass_fuse_add_relu(m.graph)\n buffer = io.BytesIO()\n torch.jit.save(m, buffer)\n buffer.seek(0)\n m = torch.jit.load(buffer)\n new_res = m(a, b, c)\n FileCheck().check_not(\"aten::relu_(\") \\\n .check(\"aten::_add_relu(\") \\\n .run(m.graph)\n torch.testing.assert_allclose(orig_res, new_res)\n\n class Madd_(torch.nn.Module):\n def __init__(self, relu_op):\n super(Madd_, self).__init__()\n self.relu_op = relu_op\n\n def forward(self, a, b):\n x = a.add_(b)\n x = self.relu_op(x)\n return x\n\n # add_, relu_\n a = torch.rand((7, 11))\n a = a * -10\n a = a + 5\n b = torch.rand((7, 11))\n # Because in place add_ will overwrite a\n a_copy = a.clone()\n m = torch.jit.script(Madd_(torch.relu_))\n orig_res = m(a, b)\n torch._C._jit_pass_fuse_add_relu(m.graph)\n buffer = io.BytesIO()\n torch.jit.save(m, buffer)\n buffer.seek(0)\n m = torch.jit.load(buffer)\n new_res = m(a_copy, b)\n FileCheck().check_not(\"aten::add_(\") \\\n .check_not(\"aten::relu_(\") \\\n .check(\"aten::_add_relu_(\") \\\n .run(m.graph)\n torch.testing.assert_allclose(orig_res, new_res)\n # Since _add_relu_ does inplace mutation ensure\n # a_copy is modified\n torch.testing.assert_allclose(orig_res, a_copy)\n\n class Madd_out(torch.nn.Module):\n def __init__(self, relu_op):\n super(Madd_out, self).__init__()\n self.relu_op = relu_op\n\n def forward(self, a, b):\n x = torch.add(a, b, out=a)\n x = self.relu_op(x)\n return x\n a = torch.rand((7, 11))\n a = a * -10\n a = a + 5\n b = torch.rand((7, 11))\n\n # add_out, relu_\n a = torch.rand((7, 11))\n a = a * -10\n a = a + 5\n b = torch.rand((7, 11))\n # Because in place add_ will overwrite a\n a_copy = a.clone()\n m = torch.jit.script(Madd_out(torch.relu_))\n orig_res = m(a, b)\n torch._C._jit_pass_fuse_add_relu(m.graph)\n buffer = io.BytesIO()\n torch.jit.save(m, buffer)\n buffer.seek(0)\n m = torch.jit.load(buffer)\n new_res = m(a_copy, b)\n FileCheck().check_not(\"aten::add(\") \\\n .check_not(\"aten::relu_(\") \\\n .check(\"aten::_add_relu(\") \\\n .run(m.graph)\n torch.testing.assert_allclose(orig_res, new_res)\n # Since _add_relu_ with out=a does inplace mutation ensure\n # a_copy is modified\n torch.testing.assert_allclose(orig_res, a_copy)\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY, \"Simple executor doesn't have shape information\")\n def test_peephole_optimize_shape_ops(self):\n def test_input(func, input, result):\n # if result == 2 we will trigger a bailout and\n # the unprofiled graph should return the correct result\n self.assertEqual(func(input, profile_and_replay=True), result)\n gre = func.graph_for(input)\n FileCheck().check_not(\"prim::If\").run(gre)\n\n def test_dim():\n @torch.jit.script\n def func(x):\n if x.dim() == 1:\n return 1\n else:\n return 2\n\n test_input(func, torch.tensor([0.5]), 1)\n test_input(func, torch.tensor([[0.5]]), 2)\n test_dim()\n\n def test_size_index():\n @torch.jit.script\n def func(x):\n if x.size(0) == 1:\n return 1\n else:\n return 2\n\n test_input(func, torch.rand([1, 2]), 1)\n test_input(func, torch.rand([1, 3]), 1)\n\n @torch.jit.script\n def neg_index(x):\n if x.size(-2) == 1:\n return 1\n else:\n return 2\n\n test_input(neg_index, torch.rand([1, 2]), 1)\n test_input(neg_index, torch.rand([1, 3]), 1)\n\n if GRAPH_EXECUTOR == ProfilingMode.PROFILING:\n test_size_index()\n\n def test_dtype():\n @torch.jit.script\n def func(x):\n if x.dtype == torch.float32:\n return 1\n else:\n return 2\n\n test_input(func, torch.tensor(0.5, dtype=torch.float32), 1)\n test_input(func, torch.tensor(0.5, dtype=torch.int64), 2)\n test_dtype()\n\n def test_is_floating_poiint():\n @torch.jit.script\n def func(x):\n if x.is_floating_point():\n return 1\n else:\n return 2\n\n test_input(func, torch.tensor(0.5, dtype=torch.float32), 1)\n test_input(func, torch.tensor(0.5, dtype=torch.int64), 2)\n test_is_floating_poiint()\n\n def test_device():\n @torch.jit.script\n def func_1(x):\n if x.device == torch.device('cuda:0'):\n a = 0\n else:\n a = 1\n return a\n\n @torch.jit.script\n def func_2(x):\n if x.is_cuda:\n a = 0\n else:\n a = 1\n return a\n\n test_input(func_1, torch.tensor(0.5), 1)\n test_input(func_2, torch.tensor(0.5), 1)\n\n if RUN_CUDA:\n test_input(func_1, torch.tensor(0.5, device=\"cuda:0\"), 0)\n test_input(func_2, torch.tensor(0.5, device=\"cuda:0\"), 0)\n\n test_device()\n\n def test_attrs(self):\n def foo(x):\n return (\n # x.dtype, TODO: dtype long -> instance conversion\n x.device,\n x.shape,\n x.is_cuda,\n x.is_mkldnn,\n x.is_quantized,\n x.requires_grad,\n # x.layout TODO: layout long -> instance conversion\n )\n\n scripted = torch.jit.script(foo)\n x = torch.rand(3, 4)\n self.assertEqual(scripted(x), foo(x))\n\n def test_layout(self):\n @torch.jit.script\n def check(x, y):\n return x.layout == y.layout\n\n x = torch.rand(3, 4)\n y = torch.rand(3, 4)\n\n self.assertTrue(check(x, y))\n\n def test_nn_conv(self):\n class Mod(nn.Module):\n def __init__(self, conv):\n super().__init__()\n self.conv = conv\n\n def forward(self, input):\n return self.conv(input)\n\n inputs = [\n # Conv\n (Mod(nn.Conv1d(16, 33, 3, stride=2)), torch.randn(20, 16, 5)),\n (Mod(nn.Conv2d(16, 33, 3, stride=2)), torch.randn(20, 16, 5, 10)),\n (Mod(nn.Conv3d(16, 33, 3, stride=2)), torch.randn(20, 16, 3, 5, 4)),\n # ConvTransposed\n (Mod(nn.ConvTranspose1d(16, 33, 3, stride=2)), torch.randn(20, 16, 5)),\n (Mod(nn.ConvTranspose2d(16, 33, 3, stride=2)), torch.randn(20, 16, 5, 10)),\n (Mod(nn.ConvTranspose3d(16, 33, 3, stride=2)), torch.randn(20, 16, 3, 5, 4)),\n ]\n\n for m, inp in inputs:\n self.checkModule(m, (inp,))\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.PROFILING, 'Not implemented for Simple or Legacy')\n def test_debug_flush_compilation_cache(self):\n def foo(x):\n return x + 2\n\n class Mod(nn.Module):\n def __init__(self):\n super(Mod, self).__init__()\n\n def forward(self, t):\n return t + 2\n\n m = torch.jit.script(Mod())\n x = torch.rand(1, 10)\n\n with enable_profiling_mode_for_profiling_tests():\n jitted = self.checkScript(foo, (x,))\n # shouldn't throw\n states = jitted.get_debug_state()\n\n # after flushing there shouldn't be\n # no opt plan\n jitted._debug_flush_compilation_cache()\n with self.assertRaisesRegex(RuntimeError, \"INTERNAL ASSERT FAILED\"):\n states = jitted.get_debug_state()\n\n NUM_RUNS = 1\n with num_profiled_runs(NUM_RUNS):\n m(x)\n m(x)\n fwd = m._c._get_method(\"forward\")\n states = m.get_debug_state()\n\n # after flushing there shouldn't be\n # no opt plan\n fwd._debug_flush_compilation_cache()\n with self.assertRaisesRegex(RuntimeError, \"INTERNAL ASSERT FAILED\"):\n states = m.get_debug_state()\n\n def test_numel(self):\n @torch.jit.script\n def get_numel_script(x):\n return x.numel()\n\n x = torch.rand(3, 4)\n numel = get_numel_script(x)\n self.assertEqual(numel, x.numel())\n\n def test_element_size(self):\n @torch.jit.script\n def get_element_size_script(x):\n return x.element_size()\n\n x = torch.rand(3, 4)\n element_size = get_element_size_script(x)\n self.assertEqual(element_size, x.element_size())\n\n def test_Sequential(self):\n class Seq(nn.Module):\n def __init__(self):\n super(Seq, self).__init__()\n self.seq = nn.Sequential(nn.Linear(10, 20), nn.Linear(20, 30))\n\n @torch.jit.script_method\n def forward(self, x):\n for l in self.seq:\n x = l(x)\n return x\n\n m = torch.jit.script(Seq())\n assert m.graph # ensure jit was able to compile\n\n def test_ModuleList(self):\n class Mod(nn.Module):\n def __init__(self):\n super(Mod, self).__init__()\n self.model = nn.ModuleList([nn.Linear(10, 10) for _ in range(10)])\n self.model += (nn.Linear(10, 20),)\n self.model.append(nn.Linear(20, 30))\n self.model.extend([nn.Linear(30, 40), nn.Linear(40, 50)])\n\n def forward(self, v):\n for m in self.model:\n v = m(v)\n return v\n\n m = torch.jit.script(Mod())\n assert m.graph # ensure jit was able to compile\n\n def test_disabled(self):\n torch.jit._state.disable()\n try:\n def f(x, y):\n return x + y\n\n self.assertIs(torch.jit.trace(f, (torch.randn(2, 2), torch.randn(2, 2))), f)\n self.assertIs(torch.jit.script(f), f)\n\n class MyModule(torch.jit.ScriptModule):\n @torch.jit.script_method\n def method(self, x):\n return x\n\n # XXX: Unfortunately ScriptModule won't simply become Module now,\n # because that requires disabling the JIT at startup time, which\n # we can't do in here.\n # We need to or those two conditions to make it work with all versions of Python\n self.assertTrue(inspect.ismethod(MyModule.method) or inspect.isfunction(MyModule.method))\n finally:\n torch.jit._state.enable()\n\n def test_train_eval(self):\n class Sub(nn.Module):\n def forward(self, input):\n if self.training:\n return input\n else:\n return -input\n\n class MyModule(torch.jit.ScriptModule):\n def __init__(self, module):\n super(MyModule, self).__init__()\n self.module = module\n\n @torch.jit.script_method\n def forward(self, input):\n return self.module(input) + 1\n\n m = MyModule(Sub())\n input = torch.rand(3, 4)\n self.assertEqual(input + 1, m(input))\n m.eval()\n self.assertEqual(-input + 1, m(input))\n\n # test batchnorm and dropout train/eval\n input = torch.randn(6, 10)\n batchnorm = nn.BatchNorm1d(10)\n dropout = nn.Dropout(p=0.2)\n\n m_batchnorm = MyModule(batchnorm)\n self.assertEqual(batchnorm(input) + 1, m_batchnorm(input))\n batchnorm.eval()\n m_batchnorm.eval()\n self.assertEqual(batchnorm(input) + 1, m_batchnorm(input))\n\n m_dropout = MyModule(dropout)\n dropout.eval()\n m_dropout.eval()\n self.assertEqual(dropout(input) + 1, m_dropout(input))\n\n def test_nn_padding(self):\n class Mod(nn.Module):\n def __init__(self, padding):\n super().__init__()\n self.padding = padding\n\n def forward(self, input):\n return self.padding(input)\n\n inputs = [\n (Mod(nn.ConstantPad1d(2, 3.5)), torch.randn(1, 2, 4)),\n (Mod(nn.ConstantPad2d(2, 3.5)), torch.randn(1, 2, 2)),\n (Mod(nn.ConstantPad3d(3, 3.5)), torch.randn(16, 3, 10, 20, 30)),\n (Mod(nn.ReflectionPad1d(2)), torch.arange(8, dtype=torch.float).reshape(1, 2, 4)),\n (Mod(nn.ReflectionPad2d(2)), torch.arange(9, dtype=torch.float).reshape(1, 1, 3, 3)),\n (Mod(nn.ReflectionPad3d(3)), torch.randn(16, 3, 8, 32, 48)),\n (Mod(nn.ReplicationPad1d(2)), torch.arange(8, dtype=torch.float).reshape(1, 2, 4)),\n (Mod(nn.ReplicationPad2d(2)), torch.arange(9, dtype=torch.float).reshape(1, 1, 3, 3)),\n (Mod(nn.ReplicationPad3d(3)), torch.randn(16, 3, 8, 32, 48)),\n (Mod(nn.ZeroPad2d(2)), torch.randn(1, 1, 3, 3))\n ]\n\n for m, inp in inputs:\n self.checkModule(m, (inp,))\n\n def test_script_autograd_grad(self):\n def test_simple_grad(x, y):\n # type: (Tensor, Tensor) -> List[Optional[Tensor]]\n z = x + 2 * y + x * y\n return torch.autograd.grad((z.sum(), ), (x, y))\n\n def test_simple_grad_with_grad_outputs(x, y):\n # type: (Tensor, Tensor) -> List[Optional[Tensor]]\n z = x + 2 * y + x * y\n grad_outputs = torch.jit.annotate(List[Optional[torch.Tensor]], [torch.ones((2, 2)), ])\n return torch.autograd.grad((z, ), (x, y), grad_outputs)\n\n def test_one_output_not_requires_grad(x, y):\n # type: (Tensor, Tensor) -> List[Optional[Tensor]]\n z = 2 * y + y\n return torch.autograd.grad((z.sum(),), (x, y), allow_unused=True)\n\n def test_retain_graph(x, y):\n # type: (Tensor, Tensor) -> None\n z = x + 2 * y + x * y\n torch.autograd.grad((z.sum(), ), (x, y), retain_graph=True)\n torch.autograd.grad((z.sum(), ), (x, y))\n\n x = torch.randn(2, 2, requires_grad=True)\n y = torch.randn(2, 2, requires_grad=True)\n self.checkScript(test_simple_grad, (x, y), inputs_requires_grad=True)\n self.checkScript(test_simple_grad_with_grad_outputs, (x, y), inputs_requires_grad=True)\n self.checkScript(test_one_output_not_requires_grad, (x, y), inputs_requires_grad=True)\n self.checkScript(test_retain_graph, (x, y), inputs_requires_grad=True)\n\n def test_script_backward(self):\n def checkBackwardScript(fn, inputs):\n scripted_fn = torch.jit.script(fn)\n FileCheck().check(\"torch.autograd.backward\").run(scripted_fn.code)\n recording_inputs = do_input_map(lambda t: t.detach().requires_grad_(), inputs)\n\n fn(*inputs)\n scripted_fn(*recording_inputs)\n\n for inp1, inp2 in zip(inputs, recording_inputs):\n self.assertEqual(inp1.grad, inp2.grad)\n\n def test_tensor_backward(input):\n # type: (Tensor) -> None\n output = torch.relu(input)\n output = output.softmax(0)\n sum_out = output.sum()\n sum_out.backward()\n\n def test_torch_autograd_backward(input):\n # type: (Tensor) -> None\n output = torch.relu(input)\n output = output.softmax(0)\n torch.autograd.backward(output.sum())\n\n def test_torch_autograd_backward_with_grad_tensors(input):\n # type: (Tensor) -> None\n output = torch.relu(input)\n output = output.softmax(0)\n grad_outputs = torch.jit.annotate(List[Optional[torch.Tensor]], [torch.ones((2, 2)), ])\n torch.autograd.backward((output,), grad_outputs)\n\n inp = torch.randn(2, 2, requires_grad=True)\n checkBackwardScript(test_tensor_backward, (inp,))\n checkBackwardScript(test_torch_autograd_backward, (inp,))\n checkBackwardScript(test_torch_autograd_backward_with_grad_tensors, (inp,))\n\n def test_script_backward_twice(self):\n def checkBackwardTwiceScript(fn, inputs, retain_graph_=False):\n torch._C._jit_set_profiling_executor(False)\n\n with torch.jit.optimized_execution(True):\n scripted_fn = torch.jit.script(fn, inputs)\n FileCheck().check(\"prim::DifferentiableGraph\").run(scripted_fn.graph_for(*inputs))\n\n result = scripted_fn(*inputs)\n result.sum().backward(retain_graph=retain_graph_)\n if not retain_graph_:\n self.assertRaisesRegex(RuntimeError, 'Specify retain_graph=True',\n lambda: result.sum().backward())\n else:\n result.sum().backward()\n\n def test_script_backward_twice_with_saved_values(input1, input2):\n # type: (Tensor, Tensor) -> Tensor\n tmp1 = torch.mul(input1, input2)\n tmp2 = torch.abs(tmp1)\n if torch.equal(input1, input2):\n tmp2 = torch.acos(tmp2)\n else:\n tmp2 = torch.atan(tmp2)\n result = torch.add(tmp2, input2)\n return result\n\n inp1 = torch.randn(2, 2, requires_grad=True)\n inp2 = torch.randn(2, 2, requires_grad=True)\n checkBackwardTwiceScript(test_script_backward_twice_with_saved_values, (inp1, inp2), False)\n checkBackwardTwiceScript(test_script_backward_twice_with_saved_values, (inp1, inp2), True)\n\n def test_diff_subgraph_clones_constants(self):\n @torch.jit.script\n def f(x, y):\n return x + x + y + x + y + x + y + x + y + x\n\n def count_constants(graph):\n return sum(node.kind() == 'prim::Constant' for node in graph.nodes())\n\n graph = f.graph.copy()\n self.run_pass('cse', graph)\n self.run_pass('create_autodiff_subgraphs', graph)\n nodes = list(graph.nodes())\n self.assertEqual(count_constants(graph), 1)\n self.assertEqual(count_constants(nodes[1].g('Subgraph')), 1)\n\n # TODO: adapt this test to check that GraphExecutor treats them differently\n @unittest.skip(\"Need to be adjusted to Graph Executor\")\n def test_arg_configurations(self):\n \"\"\"Different arg configurations should trigger different traces\"\"\"\n x = Variable(torch.FloatTensor(4, 4).uniform_())\n x_double = Variable(x.data.double())\n x_grad = Variable(x.data.clone(), requires_grad=True)\n y = Variable(torch.randn(4))\n\n configurations = [\n (x,),\n (x_double,),\n (x_grad,),\n (y,),\n ([x, x],),\n ([x, y],),\n ]\n if torch.cuda.is_available():\n x_cuda = Variable(x.data.cuda())\n configurations += [\n (x_cuda,),\n ([x, x_cuda],),\n ([x_cuda, x],),\n ([[x_cuda, x]],),\n ]\n if torch.cuda.device_count() > 1:\n x_cuda_1 = Variable(x.data.cuda(1))\n configurations += [\n (x_cuda_1,),\n ([x_cuda, x_cuda_1],),\n ]\n\n @torch.jit.compile(nderivs=0)\n def fn(*args):\n in_vars, _ = torch._C._jit_flatten(args)\n return in_vars[0] + 1\n\n for i, config in enumerate(configurations):\n self.assertFalse(fn.has_trace_for(*config))\n fn(*config)\n self.assertTrue(fn.has_trace_for(*config))\n for unk_config in configurations[i + 1:]:\n self.assertFalse(fn.has_trace_for(*unk_config))\n self.assertEqual(fn.hits, 0)\n\n def test_torch_sum(self):\n def fn(x):\n return torch.sum(x)\n\n def fn1(x, dim: int):\n return torch.sum(x, dim)\n\n x = torch.randn(3, 4)\n self.checkScript(fn, (x, ))\n self.checkScript(fn1, (x, 1, ))\n self.checkScript(fn1, (x, 0, ))\n\n def test_cse(self):\n x = torch.tensor([0.4, 0.3], requires_grad=True)\n y = torch.tensor([0.7, 0.5], requires_grad=True)\n\n def fn(x, y):\n w = (x + y) * (x + y) * (x + y)\n t = torch.tanh(w) + torch.tanh(w)\n z = (x + y) * (x + y) * (x + y) + t\n return z\n\n g, _ = torch.jit._get_trace_graph(fn, (x, y))\n self.run_pass('cse', g)\n do_exactly = True\n FileCheck().check_count(\"add\", 1).check_count(\"mul\", 2, do_exactly) \\\n .check_count(\"tanh\", 1, do_exactly).check_count(\"add\", 2, do_exactly).check_next(\"return\") \\\n .run(str(g))\n\n self.assertExportImport(g, (x, y))\n\n def test_cse_not_introduce_aliasing(self):\n @torch.jit.script\n def tensor_alias_outputs(x):\n return x + x, x + x\n\n self.run_pass('cse', tensor_alias_outputs.graph)\n FileCheck().check_count(\"aten::add\", 2).run(tensor_alias_outputs.graph)\n\n @torch.jit.script\n def ints_alias_outputs(x):\n # type: (int) -> Tuple[int, int]\n return x + x, x + x\n\n # non-aliasing types can be CSEd\n self.run_pass('cse', ints_alias_outputs.graph)\n FileCheck().check_count(\"aten::add\", 1, exactly=True).run(ints_alias_outputs.graph)\n\n def test_recursive_cse(self):\n input_str = \"\"\"\ngraph(%x : Tensor,\n %y : Tensor,\n %20 : int):\n %2 : int = prim::Constant[value=1]()\n %3 : Tensor = aten::add(%x, %y, %2)\n %4 : int = aten::add(%2, %20)\n %5 : bool = aten::Bool(%4)\n %z : int = prim::If(%5)\n # CHECK: block\n block0():\n # CHECK-NOT: aten::add\n %z.1 : int = aten::add(%2, %20)\n -> (%z.1)\n block1():\n -> (%2)\n return (%z)\n\"\"\"\n graph = parse_ir(input_str)\n self.run_pass('cse', graph)\n FileCheck().run(input_str, graph)\n\n def test_pattern_based_rewrite(self):\n # mul(mul(mul(mul(x,y),z),x),y) --> mul(mul(mulmul(x,y,z), x), y) -->\n # --> mulmul(mulmul(x,y,z), x, y)\n input_str = \"\"\"\ngraph(%x, %y, %z):\n # CHECK-NOT: aten::mul\n # CHECK: my::fused_mulmul\n %t = aten::mul(%x, %y)\n %p = aten::mul(%t, %z)\n # CHECK: my::fused_mulmul\n %u = aten::mul(%p, %x)\n %o = aten::mul(%u, %y)\n return (%o)\"\"\"\n graph = parse_ir(input_str)\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\"\"\"\ngraph(%a, %b, %c):\n %q = aten::mul(%a, %b)\n %r = aten::mul(%q, %c)\n return (%r)\"\"\", \"\"\"\ngraph(%a, %b, %c):\n %r = my::fused_mulmul(%a, %b, %c)\n return (%r)\"\"\", graph)\n FileCheck().run(input_str, graph)\n\n # Check that overlapping matches are handled correctly\n # mul(mul(mul(x,y),z),x) --> mul(mulmul(x,y,z), x)\n input_str = \"\"\"\ngraph(%x, %y, %z):\n # CHECK-NOT: aten::mul\n # CHECK: my::fused_mulmul\n %t = aten::mul(%x, %y)\n %p = aten::mul(%t, %z)\n # CHECK-NEXT: aten::mul\n %u = aten::mul(%p, %x)\n return (%u)\"\"\"\n graph = parse_ir(input_str)\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\"\"\"\ngraph(%a, %b, %c):\n %q = aten::mul(%a, %b)\n %r = aten::mul(%q, %c)\n return (%r)\"\"\", \"\"\"\ngraph(%a, %b, %c):\n %r = my::fused_mulmul(%a, %b, %c)\n return (%r)\"\"\", graph)\n FileCheck().run(input_str, graph)\n\n # Check add(mul(x,y),z) --> muladd(x,y,z) replacement\n input_str = \"\"\"\ngraph(%x, %y, %z):\n # CHECK-NOT: aten::mul\n # CHECK-NOT: aten::add\n %c = prim::Const[value=1]()\n %t = aten::mul(%x, %y)\n %p = aten::add(%t, %z, %c)\n # CHECK: my::muladd\n # CHECK-NEXT: return\n return (%p)\"\"\"\n graph = parse_ir(input_str)\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\"\"\"\ngraph(%a, %b, %c, %d):\n %q = aten::mul(%a, %b)\n %r = aten::add(%q, %c, %d)\n return (%r)\"\"\", \"\"\"\ngraph(%a, %b, %c, %d):\n %r = my::muladd(%a, %b, %c, %d)\n return (%r)\"\"\", graph)\n FileCheck().run(input_str, graph)\n\n # Check add(mul(x,y),z) --> sub(add(x,y),z) replacement\n input_str = \"\"\"\ngraph(%x, %y, %z):\n # CHECK-NOT: aten::mul\n %c = prim::Const[value=1]()\n # CHECK: aten::add\n %t = aten::mul(%x, %y)\n # CHECK-NEXT: aten::sub\n %p = aten::add(%t, %z, %c)\n # CHECK-NOT: aten::add\n # CHECK-NEXT: return\n return (%p)\"\"\"\n graph = parse_ir(input_str)\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\"\"\"\ngraph(%a, %b, %c, %d):\n %q = aten::mul(%a, %b)\n %r = aten::add(%q, %c, %d)\n return (%r)\"\"\", \"\"\"\ngraph(%a, %b, %c, %d):\n %q = aten::add(%a, %b, %d)\n %r = aten::sub(%q, %c, %d)\n return (%r)\"\"\", graph)\n FileCheck().run(input_str, graph)\n\n # Check mul(x,y) --> x replacement\n input_str = \"\"\"\ngraph(%x, %y, %z):\n %c = prim::Const[value=1]()\n # CHECK-NOT: aten::mul\n %t = aten::mul(%x, %y)\n # CHECK: aten::add(%x, %z\n %p = aten::add(%t, %z, %c)\n # CHECK-NEXT: return\n return (%p)\"\"\"\n graph = parse_ir(input_str)\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\"\"\"\ngraph(%Pa, %Pb):\n %Pq = aten::mul(%Pa, %Pb)\n return (%Pq)\"\"\", \"\"\"\ngraph(%Ra, %Rb):\n return (%Ra)\"\"\", graph)\n FileCheck().run(input_str, graph)\n\n @_tmp_donotuse_dont_inline_everything\n def test_pattern_based_module_rewrite(self):\n # Check match::module behavior\n class Test(torch.nn.Module):\n def __init__(self):\n super(Test, self).__init__()\n self.conv = torch.nn.Conv2d(1, 20, 5, 1)\n self.bn = torch.nn.BatchNorm2d(num_features=20)\n\n def forward(self, x):\n x = self.conv(x)\n x = self.bn(x)\n return x\n m = torch.jit.script(Test())\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\"\"\"\n graph(%self, %x):\n %conv = match::module[name=\"Conv2d\"](%self)\n %y = prim::CallMethod[name=\"forward\"](%conv, %x)\n %bn = match::module[name=\"BatchNorm2d\"](%self)\n %z = prim::CallMethod[name=\"forward\"](%bn, %y)\n return (%z)\"\"\", \"\"\"\n graph(%self, %x):\n %z = my::matched_conv_bn(%self, %x)\n return (%z)\"\"\", m._c._get_method(\"forward\").graph)\n\n FileCheck().check(\"my::matched_conv_bn\").run(m._c._get_method(\"forward\").graph)\n\n def test_pattern_based_rewrite_with_source_range_preserved(self):\n class TestModule1(torch.nn.Module):\n def __init__(self):\n super(TestModule1, self).__init__()\n\n def forward(self, x, y, z, w):\n x = x + y\n x = x * z\n return w - x\n\n input_pattern = \"\"\"\n graph(%x, %y, %z, %const):\n %t = aten::add(%x, %y, %const)\n %o = aten::mul(%t, %z)\n return (%o)\"\"\"\n replacement_pattern = \"\"\"\n graph(%x, %y, %z, %const):\n %o = my::add_mul(%x, %y, %z, %const)\n return (%o)\"\"\"\n scripted_model = torch.jit.script(TestModule1())\n graph = scripted_model.graph\n value_mappings = [(\"o\", \"t\")]\n for node in graph.nodes():\n if node.kind() == \"aten::add\":\n source_range_1 = node.sourceRange()\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\n input_pattern, replacement_pattern, scripted_model.graph, value_name_pairs=value_mappings)\n graph = scripted_model.graph\n for node in graph.nodes():\n if node.kind() == \"my::add_mul\":\n source_range_2 = node.sourceRange()\n self.assertTrue(source_range_1 == source_range_2)\n\n class TestModule2(torch.nn.Module):\n def __init__(self):\n super(TestModule2, self).__init__()\n\n def forward(self, x, y, z, w):\n x = x + y\n x = x + z\n x = x * z\n x = x * w\n return x - 2\n\n # Check source range preservation for two node transforms add -> my_add\n input_pattern = \"\"\"\n graph(%x, %y, %const):\n %o = aten::add(%x, %y, %const)\n return (%o)\"\"\"\n replacement_pattern = \"\"\"\n graph(%x, %y, %const):\n %o = my::add(%x, %y, %const)\n return (%o)\"\"\"\n scripted_model = copy.deepcopy(torch.jit.script(TestModule2()))\n graph_copy = scripted_model.graph.copy()\n value_mappings = [(\"o\", \"o\")]\n source_range_add_1 = None\n for node in graph_copy.nodes():\n if source_range_add_1 is None and node.kind() == \"aten::add\":\n source_range_add_1 = node.sourceRange()\n if source_range_add_1 is not None and node.kind() == \"aten::add\":\n source_range_add_2 = node.sourceRange()\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\n input_pattern, replacement_pattern, graph_copy, value_name_pairs=value_mappings)\n source_range_my_add_1 = None\n for node in graph_copy.nodes():\n if source_range_my_add_1 is None and node.kind() == \"my::add\":\n source_range_my_add_1 = node.sourceRange()\n if source_range_my_add_1 is not None and node.kind() == \"my::add\":\n source_range_my_add_2 = node.sourceRange()\n self.assertTrue(source_range_add_1 == source_range_my_add_1)\n self.assertTrue(source_range_add_2 == source_range_my_add_2)\n\n # Check source range preservation for add-add -> double_add transform\n # fuse nodes\n input_pattern = \"\"\"\n graph(%x, %y, %z, %const):\n %t = aten::add(%x, %y, %const)\n %o = aten::add(%t, %z, %const)\n return (%o)\"\"\"\n replacement_pattern = \"\"\"\n graph(%x, %y, %z, %const):\n %o = my::double_add(%x, %y, %z, %const)\n return (%o)\"\"\"\n scripted_model = torch.jit.script(TestModule2())\n graph_copy = scripted_model.graph.copy()\n value_mappings = [(\"o\", \"t\")]\n source_range_1 = None\n source_range_2 = None\n for node in graph_copy.nodes():\n if node.kind() == \"aten::add\":\n source_range_1 = node.sourceRange()\n break\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\n input_pattern, replacement_pattern, graph_copy, value_name_pairs=value_mappings)\n for node in graph_copy.nodes():\n if node.kind() == \"my::double_add\":\n source_range_2 = node.sourceRange()\n self.assertTrue(source_range_1 == source_range_2)\n\n # Check source range preservation for mul -> add + add transform\n # split node\n input_pattern = \"\"\"\n graph(%x, %y):\n %t = aten::mul(%x, %y)\n return (%t)\"\"\"\n replacement_pattern = \"\"\"\n graph(%x, %y):\n %t = my::add(%x, %y)\n %o = my::add(%t, %y)\n return (%o)\"\"\"\n scripted_model = torch.jit.script(TestModule2())\n graph_copy = scripted_model.graph.copy()\n value_mappings = [(\"t\", \"t\"), (\"o\", \"t\")]\n source_range_mul_1 = None\n for node in graph_copy.nodes():\n if source_range_mul_1 is None and node.kind() == \"aten::mul\":\n source_range_mul_1 = node.sourceRange()\n if source_range_mul_1 is not None and node.kind() == \"aten::mul\":\n source_range_mul_2 = node.sourceRange()\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(\n input_pattern, replacement_pattern, graph_copy, value_name_pairs=value_mappings)\n source_range_add_1 = None\n for node in graph_copy.nodes():\n if source_range_add_1 is None and node.kind() == \"my::add\":\n source_range_add_1 = node.sourceRange()\n if source_range_add_1 is not None and node.kind() == \"my::add\":\n source_range_add_2 = node.sourceRange()\n self.assertTrue(source_range_mul_1 == source_range_add_1)\n self.assertTrue(source_range_mul_2 == source_range_add_2)\n\n # Check lack of source range preservation for mul-mul-> double_mul transform\n input_pattern = \"\"\"\n graph(%x, %y, %z):\n %t = aten::mul(%x, %y)\n %o = aten::mul(%t, %z)\n return (%o)\"\"\"\n replacement_pattern = \"\"\"\n graph(%x, %y, %z):\n %o = my::double_mul(%x, %y, %z)\n return (%o)\"\"\"\n scripted_model = torch.jit.script(TestModule2())\n graph_copy = scripted_model.graph.copy()\n for node in graph_copy.nodes():\n if node.kind() == \"aten::mul\":\n source_range_1 = node.sourceRange()\n torch._C._jit_pass_custom_pattern_based_rewrite_graph(input_pattern, replacement_pattern, graph_copy)\n for node in graph_copy.nodes():\n if node.kind() == \"my::double_mul\":\n source_range_2 = node.sourceRange()\n self.assertFalse(source_range_1 == source_range_2)\n\n def test_expand_quantlint(self):\n pass\n\n def test_expand_fold_quant_inputs(self):\n pass\n\n def test_shape_analysis_broadcast(self):\n def broadcast(a, b):\n return a + b\n\n x = torch.randn(3, 1, 5, requires_grad=True)\n y = torch.randn(4, 1, 8, 5, requires_grad=True)\n\n graph = torch.jit.script(broadcast).graph\n torch._C._jit_pass_complete_shape_analysis(graph, (x, y), False)\n FileCheck().check(\"Double(4, 3, 8, 5, strides=[120, 40, 5, 1], device=cpu)\").run(str(graph))\n\n def test_shape_analysis_unsqueeze_in_loop(self):\n input_str = \"\"\"graph(%x.1 : Tensor):\n %4 : bool = prim::Constant[value=1]()\n %1 : int = prim::Constant[value=2]()\n %7 : int = prim::Constant[value=0]()\n # CHECK: FloatTensor(requires_grad=0, device=cpu) = prim::Loop\n %x : Tensor = prim::Loop(%1, %4, %x.1)\n # CHECK: : FloatTensor(requires_grad=0, device=cpu)):\n block0(%i : int, %x.6 : Tensor):\n # CHECK: FloatTensor(requires_grad=0, device=cpu) = aten::unsqueeze\n %x.3 : Tensor = aten::unsqueeze(%x.6, %7)\n -> (%4, %x.3)\n return (%x)\"\"\"\n graph = parse_ir(input_str)\n torch._C._jit_pass_complete_shape_analysis(graph, (torch.zeros(2, 2, dtype=torch.float32),), False)\n FileCheck().run(input_str, graph)\n\n def test_script_tensor_type(self):\n def foo(x, t: torch.dtype):\n return x.type(t)\n scr = torch.jit.script(foo)\n x = torch.rand(3, 4)\n for t in [torch.int8, torch.float64, torch.float32,\n torch.bfloat16, torch.complex64, torch.complex128, torch.bool]:\n self.assertEqual(scr(x, t), foo(x, t))\n\n def test_shape_analysis_masked_select(self):\n input_str = \"\"\"graph(%0 : Float(),\n %1 : Bool()):\n # CHECK: Float(*, requires_grad=0, device=cpu) = aten::masked_select\n %2 : Tensor = aten::masked_select(%0, %1) # test/test_jit.py:15261:0\n return (%2)\"\"\"\n graph = parse_ir(input_str)\n x = torch.ones(1, dtype=torch.float32)[0]\n mask = x.ge(0.5)\n torch._C._jit_pass_complete_shape_analysis(graph, (x, mask), False)\n FileCheck().run(input_str, graph)\n\n # TODO: update verify to work with GraphExecutors\n @unittest.skip(\"verify needs to be updated to work with GraphExecutors\")\n def test_verify(self):\n x = torch.tensor([0.4], requires_grad=True)\n y = torch.tensor([0.7], requires_grad=True)\n\n @torch.jit.compile\n def f(x, y):\n z = torch.sigmoid(x * (x + y))\n w = torch.abs(x * x * x + y) + Variable(torch.ones(1))\n return z, w\n\n torch.jit.verify(f, (x, y), loss_fn=lambda z, w: z * w, devices=[])\n\n # TODO: adapt to a GraphExecutor test\n @unittest.skip(\"Need to instrument GraphExecutors a bit more\")\n def test_flags(self):\n x, y = torch.randn(2, 2)\n y = Variable(torch.randn(2, 2))\n\n @torch.jit.compile\n def fn(x, y):\n return (x * x + y * y + x * y).sum()\n\n grads = {}\n for rx, ry in product((True, False), repeat=2):\n x.requires_grad = rx\n y.requires_grad = ry\n\n self.assertFalse(fn.has_trace_for(x, y))\n out = fn(x, y)\n\n self.assertFalse(fn.has_trace_for(x, y))\n for v, name, compute in [(x, 'x', rx), (y, 'y', ry)]:\n if not compute:\n continue\n grad_v, = torch.autograd.grad(out, v, retain_graph=True)\n expected_grad = grads.setdefault(name, grad_v)\n self.assertEqual(grad_v, expected_grad)\n self.assertEqual(fn.has_trace_for(x, y), rx or ry)\n\n def test_python_ir(self):\n x = torch.tensor([0.4], requires_grad=True)\n y = torch.tensor([0.7], requires_grad=True)\n\n def doit(x, y):\n return torch.sigmoid(torch.tanh(x * (x + y)))\n\n g, _ = torch.jit._get_trace_graph(doit, (x, y))\n self.run_pass('dce', g)\n self.run_pass('canonicalize', g)\n g2 = torch._C.Graph()\n g_to_g2 = {}\n for node in g.inputs():\n g_to_g2[node] = g2.addInput()\n for node in g.nodes():\n n_ = g2.createClone(node, lambda x: g_to_g2[x])\n g2.appendNode(n_)\n for o, no in zip(node.outputs(), n_.outputs()):\n g_to_g2[o] = no\n\n for node in g.outputs():\n g2.registerOutput(g_to_g2[node])\n\n t_node = g2.create(\"prim::TensorTest\").t_(\"a\", torch.ones([2, 2]))\n self.assertEqual(t_node.attributeNames(), [\"a\"])\n g2.appendNode(t_node)\n self.assertTrue(torch.equal(torch.ones(2, 2), t_node.t(\"a\")))\n for node in g.nodes():\n self.assertTrue(g2.findNode(node.kind()) is not None)\n\n @unittest.skipIf(IS_SANDCASTLE, \"gtest runs these in sandcastle\")\n @unittest.skipIf(RUN_CUDA, \"covered by test_cpp_cuda\")\n @unittest.skipIf(not torch._C._jit_has_cpp_tests(), \"Tests were not built, use BUILD_TEST=1\")\n def test_cpp(self):\n from cpp.jit import tests_setup\n tests_setup.setup()\n torch._C._jit_run_cpp_tests()\n tests_setup.shutdown()\n\n def test_batchnorm(self):\n x = torch.ones(2, 2, 2, 2)\n g, outputs, inputs = torch.jit._get_trace_graph(nn.BatchNorm2d(2), x,\n _force_outplace=True, return_inputs=True)\n m = self.createFunctionFromGraph(g)\n self.assertEqual(outputs, m(*inputs))\n\n def test_dropout(self):\n x = torch.ones(2, 2)\n with torch.random.fork_rng(devices=[]):\n g, outputs, inputs = torch.jit._get_trace_graph(nn.Dropout(0.6), x, return_inputs=True)\n with torch.random.fork_rng(devices=[]):\n m = self.createFunctionFromGraph(g)\n self.assertEqual(outputs, m(*inputs))\n\n @slowTest\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY, 'Testing differentiable graph')\n def test_dropout_module_requires_grad(self):\n with enable_profiling_mode_for_profiling_tests():\n class MyModule(torch.nn.Module):\n def __init__(self, M):\n super(MyModule, self).__init__()\n self.dropout = torch.nn.Dropout(0.5)\n self.linear = torch.nn.Linear(M, M)\n\n def forward(self, input):\n input = self.dropout(input)\n output = self.linear(input)\n return output\n\n def profile(func, X):\n with torch.autograd.profiler.profile() as prof:\n func(X)\n return [e.name for e in prof.function_events]\n\n M = 1000\n scripted = torch.jit.script(MyModule(M))\n # To reduce confusion about expected behaviors:\n # requires_grad controls whether dropout is symbolically differentiated.\n # training controls whether bernoulli_ is called inside symbolic differentiation of dropout.\n # * When requires_grad == training, the expected behaviors are obvious.\n # * When requires_grad=True and training=False, bernoulli_ might still show up in the graph.\n # But it's in a branch that's not called. That's why we have separate checks for autograd\n # profiler to make sure it's not run.\n # * When requires_grad=False and training=True, bernoulli_ must be run since it's the expected\n # behavior for the dropout layer in training mode. It's independent of whether graph requires\n # gradient. In fact bernoulli_ comes from autograd instead of autodiff in this case.\n for training in (True, False):\n if training:\n scripted.train()\n else:\n scripted.eval()\n for requires_grad in (True, False):\n X = torch.randn(M, M, requires_grad=requires_grad)\n if requires_grad:\n FileCheck().check(\"aten::bernoulli_\").run(scripted.graph_for(X, profile_and_replay=True))\n self.assertEqual(training, 'aten::bernoulli_' in profile(scripted, X))\n\n @unittest.skipIf(GRAPH_EXECUTOR == ProfilingMode.SIMPLE, 'Testing differentiable graph')\n def test_dropout_func_requires_grad(self):\n def dropout_training(input):\n return F.dropout(input, 0.5, training=True)\n\n def dropout_eval(input):\n return F.dropout(input, 0.5, training=False)\n\n def profile(func, X):\n with torch.autograd.profiler.profile() as prof:\n func(X)\n return [e.name for e in prof.function_events]\n\n M = 1000\n scripted_training = torch.jit.script(dropout_training)\n scripted_eval = torch.jit.script(dropout_eval)\n # See comments in test_dropout_module_requires_grad.\n with disable_autodiff_subgraph_inlining():\n for requires_grad in (True, False):\n X = torch.randn(M, M, requires_grad=requires_grad)\n if requires_grad:\n FileCheck().check(\"aten::bernoulli_\").run(scripted_training.graph_for(X, profile_and_replay=True))\n self.assertIn('aten::bernoulli_', profile(scripted_training, X))\n self.assertNotIn('aten::bernoulli_', profile(scripted_eval, X))\n\n @unittest.skipIf(not RUN_CUDA, \"test_dropout_cuda require CUDA\")\n def test_dropout_cuda(self):\n # Dropout AD is dispatched to _fused_dropout in CUDA case,\n # which is not included in TestJitGeneratedFunctional\n def _zero_rate(t):\n return torch.true_divide((t == 0).sum(), t.numel())\n\n x = torch.ones(1000, 1000).cuda().requires_grad_()\n\n with enable_profiling_mode_for_profiling_tests():\n @torch.jit.script\n def func(x):\n return torch.nn.functional.dropout(x)\n\n with freeze_rng_state():\n out_ref = torch.nn.functional.dropout(x)\n grad_ref = torch.autograd.grad(out_ref.sum(), x)\n\n with freeze_rng_state():\n out = func(x)\n grad = torch.autograd.grad(out.sum(), x)\n\n # TODO(#40882): previously we assert exact matches between eager and JIT result:\n # self.assertEqual(out, out_ref)\n # self.assertEqual(grad, grad_ref)\n # This test was disabled during legacy -> profiling executor transition.\n # Currently JIT fused results doesn't match eager result exactly due to some changes merged in between.\n # We temporarily only check statstical difference but it should be reverted once the issue is fixed.\n self.assertEqual(_zero_rate(out), _zero_rate(out_ref), rtol=1e-3, atol=1e-4)\n self.assertEqual(_zero_rate(grad[0]), _zero_rate(grad_ref[0]), rtol=1e-3, atol=1e-4)\n\n def test_torch_ops_overloaded(self):\n with self.assertRaisesRegex(RuntimeError, \"failed to many any schema\"):\n torch.ops.aten.add(\"a\", 1)\n self.assertEqual(\"ab\", torch.ops.aten.add(\"a\", \"b\"))\n a, b = torch.rand(3, 4), torch.rand(3, 4)\n self.assertEqual(a + b, torch.ops.aten.add(a, b))\n self.assertEqual(a + 1, torch.ops.aten.add(a, 1))\n\n def test_torch_complex(self):\n def fn(real, img):\n return torch.complex(real, img)\n\n def fn_out(real, img, out):\n return torch.complex(real, img, out=out)\n self.checkScript(fn, (torch.rand(3, 4), torch.rand(3, 4), ))\n self.checkScript(fn, (torch.ones(5, 1, 4), torch.ones(5, 1, 4), ))\n self.checkScript(fn, (torch.zeros(1, 6), torch.ones(6, 1), ))\n self.checkScript(fn, (torch.zeros(1, 6), torch.zeros(6, 1), ))\n self.checkScript(fn, (torch.empty(3, 4), torch.empty(3, 4), ))\n\n real = torch.tensor([1, 2], dtype=torch.float32)\n img = torch.tensor([3, 4], dtype=torch.float32)\n out = torch.empty([3, 4], dtype=torch.complex64)\n self.checkScript(fn_out, (real, img, out, ))\n\n real = torch.tensor([5, 2], dtype=torch.float64)\n img = torch.tensor([3, 4], dtype=torch.float64)\n out = torch.empty([5, 2], dtype=torch.complex128)\n self.checkScript(fn_out, (real, img, out, ))\n\n real = torch.ones([1, 2])\n img = torch.ones([1, 2])\n out = torch.empty([1, 2], dtype=torch.complex128)\n self.checkScript(fn_out, (real, img, out, ))\n\n real = torch.ones([3, 8, 7])\n img = torch.ones([3, 8, 7])\n out = torch.empty([3, 8, 7], dtype=torch.complex128)\n self.checkScript(fn_out, (real, img, out, ))\n\n real = torch.empty([3, 2, 6])\n img = torch.empty([3, 2, 6])\n out = torch.empty([3, 2, 6], dtype=torch.complex128)\n self.checkScript(fn_out, (real, img, out, ))\n\n real = torch.zeros([1, 3])\n img = torch.empty([3, 1])\n out = torch.empty([3, 3], dtype=torch.complex128)\n self.checkScript(fn_out, (real, img, out, ))\n\n real = torch.ones([2, 5])\n img = torch.empty([2, 1])\n out = torch.empty([2, 5], dtype=torch.complex128)\n self.checkScript(fn_out, (real, img, out, ))\n\n real = torch.ones([2, 5])\n img = torch.zeros([2, 1])\n out = torch.empty([2, 5], dtype=torch.complex128)\n self.checkScript(fn_out, (real, img, out, ))\n\n def test_einsum(self):\n def check(fn, jitted, *args):\n self.assertGraphContains(jitted.graph, kind='aten::einsum')\n self.assertEqual(fn(*args), jitted(*args))\n\n def equation_format(x, y):\n return torch.einsum('i,j->ij', (x, y))\n\n def equation_format_varargs(x, y):\n return torch.einsum('i,j->ij', x, y)\n\n def sublist_format(x, y):\n return torch.einsum(x, [0], y, [1], [0, 1])\n\n x = make_tensor((5,), 'cpu', torch.float32)\n y = make_tensor((10,), 'cpu', torch.float32)\n\n for fn in [equation_format, equation_format_varargs, sublist_format]:\n check(fn, torch.jit.script(fn), x, y)\n check(fn, torch.jit.trace(fn, (x, y)), x, y)\n\n def test_python_ivalue(self):\n # Test if pure python object can be hold as IValue and conversion\n # between IValue and PyObject are correct\n # test for numpy object\n py_array = np.arange(15)\n ret_py_obj = torch._C._ivalue_debug_python_object(py_array)\n self.assertEqual(py_array, ret_py_obj)\n\n # test for function object\n ret_py_obj = torch._C._ivalue_debug_python_object(F.relu)\n self.assertEqual(F.relu, ret_py_obj)\n\n # test for memory management\n # we need to ensure IValue correctly call incref/decref to avoid\n # dangling behavior and potential memory leaks during conversions\n def test_func_scope_helper(inp):\n # create a scope and do the conversion -> ivalue -> pyobject\n # this func return a new pyobject that refcount + 1\n inp_refcount = sys.getrefcount(inp)\n ivalue_holder = torch._C._ivalue_debug_python_object(inp)\n self.assertEqual(inp_refcount + 1, sys.getrefcount(ivalue_holder))\n return ivalue_holder + 1\n\n test_input = 2200\n before_count = sys.getrefcount(test_input)\n test_func_scope_helper(test_input)\n after_count = sys.getrefcount(test_input)\n\n # after the test_func_scope_helper_call, the refcount of\n # test_input should be equal to the original refcount\n # otherwise we get either dangling pointer or memory leak!\n self.assertEqual(before_count, after_count)\n\n def test_decompose_addmm(self):\n def does_decompose():\n @torch.jit.script\n def addmm(mat, mat1, mat2):\n a = mat.addmm(mat1, mat2)\n b = mat.addmm(mat1, mat2, alpha=1.0, beta=1.0)\n return a + b\n\n mat = torch.randn(2, 2)\n mat1 = torch.randn(2, 4)\n mat2 = torch.randn(4, 2)\n\n out_ref = addmm(mat, mat1, mat2)\n self.run_pass('decompose_ops', addmm.graph)\n out_test = addmm(mat, mat1, mat2)\n self.assertEqual(out_ref, out_test)\n FileCheck().check_not(\"addmm\").run(str(addmm.graph))\n\n def doesnt_decompose():\n @torch.jit.script\n def addmm(mat, mat1, mat2, alpha, beta):\n a = mat.addmm(mat1, mat2, alpha=4.20, beta=2.0)\n b = mat.addmm(mat1, mat2, alpha=int(alpha), beta=int(beta))\n\n return a + b\n\n orig = str(addmm.graph)\n self.run_pass('decompose_ops', addmm.graph)\n self.assertTrue(orig == str(addmm.graph))\n\n does_decompose()\n doesnt_decompose()\n\n @suppress_warnings\n def test_sparse_tensors(self):\n @torch.jit.ignore\n def get_sparse():\n return torch.sparse.FloatTensor(2, 3)\n\n @torch.jit.script\n def test_is_sparse(input):\n # type: (Tensor) -> bool\n return input.is_sparse\n\n script_out_is_sparse = test_is_sparse(get_sparse())\n script_out_is_dense = test_is_sparse(torch.randn(2, 3))\n self.assertEqual(script_out_is_sparse, True)\n self.assertEqual(script_out_is_dense, False)\n\n def test_basic_sparse(input):\n output = get_sparse()\n return output, input\n\n self.checkScript(test_basic_sparse, (get_sparse(),))\n self.checkScript(test_basic_sparse, (torch.tensor([1]),))\n\n def test_sparse_sum(input):\n return torch.sparse.sum(input)\n\n self.checkScript(test_sparse_sum, (get_sparse(),))\n\n def test_sparse_mm(input1, input2):\n return torch.sparse.mm(input1, input2)\n\n self.checkScript(test_sparse_mm, (get_sparse(), torch.randn(3, 4)))\n\n def test_sparse_addmm(input, input1, input2):\n return torch.sparse.addmm(input, input1, input2)\n\n def test_sparse_addmm_alpha_beta(input, input1, input2):\n return torch.sparse.addmm(input, input1, input2, 1.3, 1.5)\n\n self.checkScript(test_sparse_addmm, (torch.randn(2, 4), get_sparse(), torch.randn(3, 4)))\n self.checkScript(test_sparse_addmm_alpha_beta, (torch.randn(2, 4), get_sparse(), torch.randn(3, 4)))\n\n @suppress_warnings\n def test_sparse_csr_tensors(self):\n @torch.jit.ignore\n def get_sparse_csr():\n return torch.randn(3, 3).to_sparse_csr()\n\n @torch.jit.script\n def test_is_sparse_csr(input):\n # type: (Tensor) -> bool\n return input.is_sparse_csr\n\n script_out_is_sparse_csr = test_is_sparse_csr(get_sparse_csr())\n script_out_is_dense_csr = test_is_sparse_csr(torch.randn(3, 3))\n\n self.assertEqual(script_out_is_sparse_csr, True)\n self.assertEqual(script_out_is_dense_csr, False)\n\n @unittest.skipIf(not RUN_CUDA, \"requires CUDA\")\n def test_device_not_equal(self):\n\n def compare_device(x: torch.device):\n return x != torch.device(\"cuda:0\")\n\n def compare_two_device(x: torch.device, y: torch.device):\n return x != y\n\n self.checkScript(compare_device, (torch.device(\"cuda:0\"),))\n self.checkScript(compare_two_device, (torch.device(\"cuda:0\"), torch.device(\"cuda:1\"), ))\n\n def test_constant_prop_simple(self):\n @torch.jit.script\n def constant_prop(input_int):\n # type: (int) -> int\n a = 2 * 3\n b = a + 2\n return b - input_int\n\n out_ref = constant_prop(2)\n self.run_pass('constant_propagation', constant_prop.graph)\n out_test = constant_prop(2)\n self.assertEqual(out_ref, out_test)\n graph_str = str(constant_prop.graph)\n self.assertTrue(\"aten::add\" not in graph_str and \"aten::mul\" not in graph_str)\n const = constant_prop.graph.findNode(\"prim::Constant\").output().toIValue()\n self.assertEqual(const, 8)\n\n def test_constant_prop_nested(self):\n @torch.jit.script\n def constant_prop(a):\n b = 2 + 1\n if bool(a < 2):\n c = b + 2\n else:\n c = b - 2\n return c\n out_ref = constant_prop(torch.tensor(2))\n self.run_pass('constant_propagation', constant_prop.graph)\n out_test = constant_prop(torch.tensor(2))\n self.assertEqual(out_ref, out_test)\n if_node = constant_prop.graph.findNode(\"prim::If\")\n for block in if_node.blocks():\n for node in block.nodes():\n self.assertTrue(node.kind() == \"prim::Constant\")\n\n def test_constant_prop_print(self):\n @torch.jit.script\n def constant_prop(input_tensor):\n a = 2 * 3\n print(a)\n b = a + 2\n return b + input_tensor\n\n self.run_pass('constant_propagation', constant_prop.graph)\n graph = constant_prop.graph\n print_node = graph.findNode(\"prim::Print\")\n self.assertTrue(print_node.input().toIValue() == 6)\n\n def test_constant_prop_rand(self):\n @torch.jit.script\n def constant_prop():\n a = torch.randn([3])\n b = a + 2\n return b\n\n self.run_pass('constant_propagation', constant_prop.graph)\n self.assertTrue(\"aten::randn\" in str(constant_prop.graph))\n\n def test_constant_prop_none(self):\n @torch.jit.script\n def typed_none():\n # type: () -> Optional[int]\n return None\n\n @torch.jit.script\n def constant_prop():\n a = typed_none()\n b = typed_none()\n if (a is None and b is None):\n a = 2\n else:\n a = 1\n return a\n\n self.run_pass('constant_propagation', constant_prop.graph)\n FileCheck().check(\"prim::Constant\").run(constant_prop.graph)\n\n def test_constant_prop_if_inline(self):\n @torch.jit.script\n def constant_prop():\n cond = True\n a = 1\n if cond:\n a = 1 * 2\n else:\n a = 1 // 0\n return a\n\n # testing that 1 // 0 error is not thrownn\n self.run_pass('constant_propagation', constant_prop.graph)\n\n def test_constant_prop_exception(self):\n # checking y = a[4] does not error in constant propagation\n def bad_index(x):\n # type: (bool)\n y = 0\n if x:\n a = [1, 2, 3]\n y = a[4]\n return y\n\n self.checkScript(bad_index, (False,))\n\n def test_constant_prop_aliasing_type(self):\n @torch.jit.script\n def foo():\n return len([1]), len(torch.tensor([2]))\n\n FileCheck().check_dag(\"aten::tensor\").check_dag(\"aten::len\").run(foo.graph)\n\n @torch.jit.script\n def fn():\n if 1 == 1:\n return 1\n else:\n return 2\n\n FileCheck().check_not(\"prim::If\").run(fn.graph)\n\n def test_unchecked_cast(self):\n def test(cond):\n # type: (bool)\n a = torch.tensor([10])\n if cond:\n b = None\n else:\n b = a\n if b is not None:\n b[0] = 5\n return a.int()\n\n self.checkScript(test, (True,))\n self.checkScript(test, (False,))\n\n def test_constant_prop_if_constant(self):\n @torch.jit.script\n def constant_prop(a, b):\n c0 = 1\n c1 = 1\n c2 = 1\n if bool(a): # -> c0, c1\n if bool(b): # -> c0\n if 1 == 1: # -> c0\n c0 = c0 + 1\n if 1 == 2:\n c1 = c1 + 1\n c2 = c2 + 1\n else: # -> c0, c1\n c1 = c1 + 1\n\n if 1 == 1: # inlined\n c0 = c0 + 1 # dynamic\n c2 = c2 + 4 # set to 5\n return a + c0 + c1 + c2\n\n graph = constant_prop.graph\n self.run_pass('constant_propagation', graph)\n ifs = graph.findAllNodes(\"prim::If\", recurse=False)\n snd_if_inlined = len(ifs) == 1\n self.assertTrue(snd_if_inlined)\n first_if = ifs[0]\n self.assertTrue(first_if.outputsSize() == 2)\n second_if = first_if.findNode(\"prim::If\", recurse=False)\n self.assertTrue(second_if.outputsSize() == 1)\n self.assertTrue(second_if.findNode(\"prim::If\") is None)\n\n def test_constant_prop_loop_constant(self):\n @torch.jit.script\n def constant_prop(cond, iter):\n # type: (bool, int) -> int\n b = 0\n while True:\n print(\"stays\")\n for _ in range(2):\n print(\"stays\")\n for _ in range(iter):\n print(\"stays\")\n while cond:\n print(\"stays\")\n while False:\n print(\"removed\")\n for _i in range(0):\n print(\"removed\")\n for _i in range(-4):\n print(\"removed\")\n return b\n\n self.run_pass('constant_propagation', constant_prop.graph)\n graph = canonical(constant_prop.graph)\n self.assertTrue(graph.count(\"removed\") == 0)\n self.assertTrue(graph.count(\"stays\") == 1) # constant gets pooled\n self.assertTrue(graph.count(\"prim::Print\") == 4)\n\n def test_constant_prop_remove_output(self):\n @torch.jit.script\n def constant_prop(iter):\n # type: (int) -> None\n a = 1\n b = 1\n c = 1\n for i in range(iter):\n if 1 == 2:\n a = 10\n if i == 5:\n b = 2\n c = 3\n print(a, b, c)\n\n graph = constant_prop.graph\n self.run_pass('constant_propagation', graph)\n self.assertTrue(graph.findNode(\"prim::Loop\").outputsSize() == 2)\n\n # TODO(gmagogsfm): Refactor this test to reduce complexity.\n def test_constant_insertion(self):\n funcs_template = dedent('''\n def func():\n return {constant_constructor}\n ''')\n\n # constants: primitives: int, double, bool, str, lists of primitives,\n # and tuples\n def check_constant(constant_constructor):\n scope = {}\n funcs_str = funcs_template.format(constant_constructor=constant_constructor)\n execWrapper(funcs_str, globals(), scope)\n cu = torch.jit.CompilationUnit(funcs_str)\n f_script = cu.func\n self.run_pass('constant_propagation', f_script.graph)\n FileCheck().check_count(\"prim::Constant\", 1, exactly=True).run(f_script.graph)\n self.assertEqual(scope['func'](), f_script())\n imported = self.getExportImportCopy(f_script)\n self.assertEqual(imported(), f_script())\n\n constants = [\"None\", \"-.5\", \"0\", \"1\", \"True\", \"False\", \"''\", \"'a'\", \"'b'\", \"torch.tensor(1)\",\n \"[True, False]\", \"[0., .5]\", \"[torch.tensor(4), torch.tensor(2)]\", \"[0, 1]\", \"['0', '1']\",\n \"[True, None]\", \"[.5, None, .2]\"]\n\n for type in [\"Tensor\", \"str\", \"int\", \"float\", \"bool\"]:\n constants.append(\"torch.jit.annotate(List[ \" + type + \"], [])\")\n\n for constant in constants:\n check_constant(constant)\n\n for key_type in [\"str\", \"int\", \"float\"]:\n for value_type in [\"Tensor\", \"bool\", \"str\", \"int\", \"float\"]:\n check_constant(\"torch.jit.annotate(Dict[ \" + key_type + \", \" + value_type + \"], {})\")\n check_constant(\"torch.jit.annotate(Dict[ \" + key_type + \", Optional[\" + value_type + \"]], {})\")\n\n for i in range(len(constants)):\n for j in range(i + 1, len(constants)):\n tup_constant = constants[i] + \", \" + constants[j]\n check_constant(tup_constant)\n\n dict_constants = []\n for i in range(len(constants)):\n # check_constant constructs the second dict with another Tensor\n # which fails the comparison\n if not isinstance(eval(constants[i]), (str, int, float)):\n continue\n for j in range(len(constants)):\n dict_constant = \"{ \" + constants[i] + \": \" + constants[j] + \"}\"\n check_constant(dict_constant)\n dict_constants.append(dict_constant)\n constants = constants + dict_constants\n\n # testing node hashing\n funcs_template = dedent('''\n def func():\n print({constant_constructor})\n ''')\n single_elem_tuples = (\"(\" + x + \",)\" for x in constants)\n input_arg = \", \".join(single_elem_tuples)\n scope = {}\n funcs_str = funcs_template.format(constant_constructor=input_arg)\n execWrapper(funcs_str, globals(), scope)\n cu = torch.jit.CompilationUnit(funcs_str)\n f_script = cu.func\n self.run_pass('constant_propagation', f_script.graph)\n # prim::None return adds one constant\n self.assertEqual(len(constants) + 1, str(f_script.graph).count(\"prim::Constant\"))\n self.run_pass('cse', f_script.graph)\n # node hashing correctly working, no CSE occurs\n self.assertEqual(len(constants) + 1, str(f_script.graph).count(\"prim::Constant\"))\n\n funcs_template = dedent('''\n def func():\n a = {constant_constructor}\n print(a)\n b = {constant_constructor}\n print(b)\n ''')\n\n # generate dicts with built-in types (excluding torch.Tensor)\n xprod = itertools.product(constants, constants)\n\n # test that equal tuples and dicts correctly work with node hashing\n for tup in (\"(\" + x + \",)\" for x in constants):\n funcs_str = funcs_template.format(constant_constructor=tup)\n scope = {}\n execWrapper(funcs_str, globals(), scope)\n cu = torch.jit.CompilationUnit(funcs_str)\n f_script = cu.func\n self.run_pass('constant_propagation_immutable_types', f_script.graph)\n num_constants = str(f_script.graph).count(\"prim::Constant\")\n self.run_pass('cse', f_script.graph)\n FileCheck().check_count(\"prim::Constant\", num_constants, exactly=True).run(f_script.graph)\n\n @unittest.skipIf(not RUN_CUDA, \"requires CUDA\")\n def test_cuda_export_restore(self):\n class Sub(torch.jit.ScriptModule):\n def __init__(self):\n super(Sub, self).__init__()\n self.weight = nn.Parameter(torch.randn(3, 4))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.mod = Sub()\n\n @torch.jit.script_method\n def forward(self, v):\n return self.mod(v)\n m = M()\n m.cuda()\n m2 = self.getExportImportCopy(m)\n m2.cuda()\n input = torch.rand(3, 4).cuda()\n self.assertEqual(m(input), m2(input))\n\n @slowTest\n def test_export_batchnorm(self):\n for mode in ['eval', 'train']:\n for clazz in [\n torch.nn.BatchNorm1d(100),\n torch.nn.BatchNorm1d(100, affine=False),\n torch.nn.BatchNorm2d(100),\n torch.nn.BatchNorm2d(100, affine=False)]:\n getattr(clazz, mode)()\n input = torch.randn(20, 100) if isinstance(clazz, torch.nn.BatchNorm1d) else \\\n torch.randn(20, 100, 35, 45)\n traced = torch.jit.trace(clazz, (input,))\n imported = self.getExportImportCopy(traced)\n x = torch.randn(20, 100) if isinstance(clazz, torch.nn.BatchNorm1d) else \\\n torch.randn(20, 100, 35, 45)\n self.assertEqual(traced(x), imported(x))\n\n def test_export_rnn(self):\n for clazz in [nn.RNN(10, 20, 2), nn.GRU(10, 20, 2)]:\n class RNNTest(torch.nn.Module):\n def __init__(self):\n super(RNNTest, self).__init__()\n self.rnn = clazz\n\n def forward(self, x, lengths, h0):\n packed = torch.nn.utils.rnn.pack_padded_sequence(x, lengths)\n out, h = self.rnn(packed, h0)\n padded_outs, _ = torch.nn.utils.rnn.pad_packed_sequence(out)\n return padded_outs\n\n test = RNNTest()\n\n traced = torch.jit.trace(test, (torch.randn(5, 3, 10), torch.LongTensor([3, 2, 1]), torch.randn(2, 3, 20)))\n imported = self.getExportImportCopy(traced)\n # NB: We make sure to pass in a batch with a different max sequence\n # length to ensure that the argument stashing for pad_packed works\n # properly.\n x, lengths, h0 = torch.randn(7, 4, 10), torch.LongTensor([7, 3, 2, 1]), torch.randn(2, 4, 20)\n self.assertEqual(traced(x, lengths, h0), imported(x, lengths, h0))\n\n def test_export_lstm(self):\n class LSTMTest(torch.nn.Module):\n def __init__(self):\n super(LSTMTest, self).__init__()\n self.rnn = nn.LSTM(10, 20, 2)\n\n def forward(self, x, lengths, hiddens):\n h0, c0 = hiddens\n packed = torch.nn.utils.rnn.pack_padded_sequence(x, lengths)\n out, (h, c) = self.rnn(packed, (h0, c0))\n padded_outs, _ = torch.nn.utils.rnn.pad_packed_sequence(out)\n return padded_outs\n\n test = LSTMTest()\n\n traced = torch.jit.trace(test, (torch.randn(5, 3, 10),\n torch.LongTensor([3, 2, 1]),\n (torch.randn(2, 3, 20), torch.randn(2, 3, 20))))\n imported = self.getExportImportCopy(traced)\n x, lengths, h0, c0 = \\\n torch.randn(7, 3, 10), torch.LongTensor([7, 5, 2]), torch.randn(2, 3, 20), torch.randn(2, 3, 20)\n self.assertEqual(traced(x, lengths, (h0, c0)), imported(x, lengths, (h0, c0)))\n\n def test_unique_state_dict(self):\n class MyModule(torch.nn.Module):\n def __init__(self):\n super(MyModule, self).__init__()\n shared_param = torch.nn.Parameter(torch.ones(1))\n self.register_parameter('w1', shared_param)\n self.register_parameter('w2', shared_param)\n\n def forward(self, input):\n return input + self.w1 + self.w2\n\n model = MyModule()\n unittest.TestCase.assertEqual(\n self, len(torch.jit._unique_state_dict(model, keep_vars=False)), 1)\n unittest.TestCase.assertEqual(\n self, len(torch.jit._unique_state_dict(model, keep_vars=True)), 1)\n\n def test_export_dropout(self):\n test = torch.nn.Dropout()\n test.eval()\n\n traced = torch.jit.trace(test, (torch.rand(3, 4),), check_trace=False)\n imported = self.getExportImportCopy(traced)\n x = torch.randn(3, 4)\n self.assertEqual(traced(x), imported(x))\n\n def test_pretty_printer(self):\n @torch.jit.script\n def if_test(a, b):\n # FIXME: use 0 instead of a.\n # c = 0\n c = a\n if bool(a < b):\n c = b\n else:\n c = a\n return c\n\n @torch.jit.script\n def if_one(a, b):\n c = b\n if bool(a < b):\n c = a\n return c\n\n @torch.jit.script\n def while_test(a, i):\n while bool(i < 3):\n a *= a\n i += 1\n return a\n\n @torch.jit.script\n def while_if_test(a, b):\n c = 0\n while bool(a < 10):\n a = a + 1\n b = b + 1\n if bool(a > b):\n c = 2\n else:\n c = 3\n return a + 1 + c\n\n @torch.jit.script\n def loop_use_test(y):\n x = y + 1\n z = x + 5\n while bool(y < 8):\n y += 1\n z = x\n return x, z\n\n @torch.jit.ignore\n def python_fn(x):\n return x + 10\n\n @torch.jit.script\n def python_op_name_test(y):\n return python_fn(y)\n\n @torch.jit.script\n def empty_int_list_test(y):\n x = torch.jit.annotate(List[int], [])\n return x[0]\n\n @torch.jit.script\n def empty_float_list_test(y):\n return [1.0, 2.0, 3.0]\n\n @torch.jit.script\n def print_weird_test(y):\n print(\"hi\\016\")\n\n self.assertExpected(if_test.code, \"if_test\")\n self.assertExpected(if_one.code, \"if_one\")\n self.assertExpected(while_test.code, \"while_test\")\n self.assertExpected(while_if_test.code, \"while_if_test\")\n self.assertExpected(loop_use_test.code, \"loop_use_test\")\n self.assertExpected(python_op_name_test.code, \"python_op_name_test\")\n self.assertExpected(empty_int_list_test.code, \"empty_int_list_test\")\n self.assertExpected(empty_float_list_test.code, \"empty_float_list_test\")\n self.assertExpected(print_weird_test.code, \"print_weird_test\")\n\n def test_cu_escaped_number(self):\n cu = torch.jit.CompilationUnit('''\n def foo(a):\n print(\"hi\\016\")\n ''')\n self.assertExpected(cu.foo.code)\n\n def test_import_method(self):\n with torch._jit_internal._disable_emit_hooks():\n class Foo(torch.jit.ScriptModule):\n def __init__(self):\n super(Foo, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x, y):\n return 2 * x + y\n\n foo = Foo()\n buffer = io.BytesIO()\n torch.jit.save(foo, buffer)\n\n buffer.seek(0)\n foo_loaded = torch.jit.load(buffer)\n self.assertExpected(foo_loaded.forward.code)\n\n @unittest.skip(\"temporarily disable the test for fwd compatibility\")\n def test_non_ascii_string(self):\n class Foo(torch.jit.ScriptModule):\n def __init__(self):\n super(Foo, self).__init__()\n self.a = \"Over \\u0e55\\u0e57 57\"\n\n @torch.jit.script_method\n def forward(self, x, y):\n return self.a + \"hi\\xA1\"\n\n foo = Foo()\n buffer = io.BytesIO()\n torch.jit.save(foo, buffer)\n\n buffer.seek(0)\n foo_loaded = torch.jit.load(buffer)\n self.assertExpected(foo_loaded.forward.code)\n\n def test_function_default_values(self):\n outer_var = torch.tensor(20)\n outer_var2 = torch.tensor(30)\n a = torch.tensor(0.5)\n b = torch.tensor(10)\n\n @torch.jit.script\n def simple_fn(x, a=a, b=b, c=outer_var + outer_var2):\n return x + a + b + c\n\n self.assertEqual(\n simple_fn(torch.ones(1)),\n torch.ones(1) + 0.5 + 10 + (20 + 30))\n self.assertEqual(\n simple_fn(torch.ones(1), torch.tensor(1), torch.tensor(3), torch.tensor(4)),\n torch.ones(1) + 1 + 3 + 4)\n\n outer_c = torch.tensor(9)\n outer_flag = torch.tensor(False)\n\n @torch.jit.script\n def bool_fn(x, a=outer_c, flag=outer_flag):\n if bool(flag):\n result = x\n else:\n result = x + a\n return result\n\n self.assertEqual(bool_fn(torch.ones(1)), torch.ones(1) + 9)\n self.assertEqual(\n bool_fn(torch.ones(1), torch.tensor(1), torch.tensor(True)),\n torch.ones(1))\n\n @torch.jit.script\n def none_fn(x=None):\n # type: (Optional[int]) -> Optional[int]\n return x\n\n self.assertEqual(none_fn(), None)\n self.assertEqual(none_fn(1), 1)\n\n @torch.jit.script\n def hints(x, a=0.5, b=10):\n # type: (Tensor, float, int) -> Tensor\n return x + a + b\n\n self.assertEqual(hints(torch.ones(1)), torch.ones(1) + 0.5 + 10)\n\n with self.assertRaisesRegex(RuntimeError, \"Expected a default value\"):\n\n @torch.jit.script\n def hints_bad_types(x, a=10, b=0.5): # noqa: T484\n # type: (Tensor, float, int) -> Tensor\n return x + a + b\n with self.assertRaisesRegex(RuntimeError, \"Expected a default value\"):\n @torch.jit.script\n def bad_no_optional(x=None):\n # type: (Dict[str, int]) -> Dict[str, int]\n return x\n\n\n def test_module_default_values(self):\n four = torch.tensor(4)\n\n class Test(torch.jit.ScriptModule):\n def __init__(self):\n super(Test, self).__init__()\n\n @torch.jit.script_method\n def forward(self, input, other=four):\n return input + other\n\n t = Test()\n self.assertEqual(t(torch.ones(1)), torch.ones(1) + 4)\n\n def test_union_to_optional(self):\n def test1(u: Union[int, None]) -> int:\n if u is not None:\n return u\n else:\n return 0\n scripted = torch.jit.script(test1)\n self.assertEqual(scripted(10), test1(10))\n\n def test2(u: Union[None, int]) -> int:\n if u is not None:\n return u\n else:\n return 0\n scripted = torch.jit.script(test2)\n self.assertEqual(scripted(40), test2(40))\n\n def test3(u: Union[float, int]) -> int:\n if u is not None:\n return u\n else:\n return 0\n expected_result = \"General Union types are not currently supported\"\n with self.assertRaisesRegex(RuntimeError, expected_result):\n torch.jit.script(test3)\n\n def test_mutable_default_values(self):\n with self.assertRaisesRegex(Exception, \"Mutable default parameters\"):\n @torch.jit.script\n def foo(x=(1, [])):\n # type: (Tuple[int, List[Tensor]])\n return x\n\n class Test(torch.nn.Module):\n def forward(self, input=[]): # noqa: B006\n return input\n\n with self.assertRaisesRegex(Exception, \"Mutable default parameters\"):\n torch.jit.script(Test())\n\n def test_warnings(self):\n import warnings\n\n def fn(x):\n if bool(x < 2):\n warnings.warn(\"x is less than 2\")\n return x\n\n class M(torch.nn.Module):\n def forward(self, x):\n if bool(x < 2):\n warnings.warn(\"x is less than 2\")\n return x\n\n\n scripted_mod = torch.jit.script(M())\n scripted_fn = torch.jit.script(fn)\n\n with warnings.catch_warnings(record=True) as warns:\n fn(torch.ones(1))\n\n with warnings.catch_warnings(record=True) as script_warns:\n scripted_fn(torch.ones(1))\n\n with warnings.catch_warnings(record=True) as script_mod_warns:\n scripted_mod(torch.ones(1))\n\n self.assertEqual(str(warns[0]), str(script_warns[0]))\n self.assertEqual(len(script_mod_warns), 1)\n self.assertEqual(str(warns[0].message), str(script_mod_warns[0].message))\n\n def test_no_erroneous_warnings(self):\n import warnings\n\n def fn(x):\n if bool(x > 0):\n warnings.warn('This should NOT be printed')\n x += 1\n return x\n\n with warnings.catch_warnings(record=True) as warns:\n fn_script = torch.jit.script(fn)\n fn_script(torch.tensor(0))\n warns = [str(w.message) for w in warns]\n self.assertEqual(len(warns), 0)\n\n @unittest.skipIf(True, \"TODO: re-enable with https://github.com/pytorch/pytorch/pull/29339\")\n def test_torch_load_error(self):\n class J(torch.jit.ScriptModule):\n def __init__(self):\n super(J, self).__init__()\n\n @torch.jit.script_method\n def forward(self, input):\n return input + 100\n\n j = J()\n with TemporaryFileName() as fname:\n j.save(fname)\n with self.assertRaisesRegex(RuntimeError, \"is a zip\"):\n torch.load(fname)\n\n def test_torch_load_zipfile_check(self):\n @torch.jit.script\n def fn(x):\n return x + 10\n\n with TemporaryFileName() as fname:\n fn.save(fname)\n with io.open(fname, 'rb') as f:\n self.assertTrue(torch.serialization._is_zipfile(f))\n\n def test_python_bindings(self):\n lstm_cell = torch.jit.script(LSTMCellS)\n\n def lstm(x, hx, cx, w_ih, w_hh, b_ih, b_hh):\n for i in range(x.size(0)):\n hx, cx = lstm_cell(x[i], hx, cx, w_ih, w_hh, b_ih, b_hh)\n return hx\n\n slstm = torch.jit.script(lstm)\n\n inputs = get_lstm_inputs('cpu', training=True, seq_length=10)\n slstm(*inputs).sum().backward()\n global fw_graph\n fw_graph = slstm.graph_for(*inputs)\n nodes = list(fw_graph.nodes())\n tested_blocks = False\n for node in nodes:\n for output in node.outputs():\n self.assertTrue(hasattr(output, 'type'))\n self.assertTrue(output.type() is not None)\n for input in node.inputs():\n self.assertTrue(hasattr(input, 'type'))\n self.assertTrue(input.type() is not None)\n for block in node.blocks():\n tested_blocks = True\n self.assertTrue(hasattr(block, 'inputs'))\n self.assertTrue(hasattr(block, 'outputs'))\n for output in block.outputs():\n self.assertTrue(hasattr(output, 'type'))\n self.assertTrue(output.type() is not None)\n for input in block.inputs():\n self.assertTrue(hasattr(input, 'type'))\n self.assertTrue(input.type() is not None)\n self.assertTrue(hasattr(block, 'returnNode'))\n self.assertTrue(type(block.returnNode()) == torch._C.Node)\n self.assertTrue(hasattr(block, 'paramNode'))\n self.assertTrue(type(block.paramNode()) == torch._C.Node)\n self.assertTrue(tested_blocks)\n\n def test_export_opnames(self):\n class Foo(torch.jit.ScriptModule):\n def __init__(self):\n super(Foo, self).__init__()\n\n def one(self, x, y):\n # type: (Tensor, Tensor) -> Tensor\n return x + y\n\n def two(self, x):\n # type: (Tensor) -> Tensor\n return 2 * x\n\n @torch.jit.script_method\n def forward(self, x):\n # type: (Tensor) -> Tensor\n return self.one(self.two(x), x)\n\n class Bar(torch.jit.ScriptModule):\n def __init__(self):\n super(Bar, self).__init__()\n self.sub = Foo()\n\n @torch.jit.script_method\n def forward(self, x):\n # type: (Tensor) -> Tensor\n return self.sub.forward(x)\n\n bar = Bar()\n ops = torch.jit.export_opnames(bar)\n expected = ['aten::add.Tensor', 'aten::mul.Scalar']\n self.assertTrue(set(expected).issubset(set(ops)))\n\n def test_pytorch_jit_env_off(self):\n import subprocess\n env = os.environ.copy()\n env['PYTORCH_JIT'] = '0'\n try:\n subprocess.check_output([sys.executable, '-c', 'import torch'], env=env)\n except subprocess.CalledProcessError as e:\n raise RuntimeError(\"Could not 'import torch' with PYTORCH_JIT=0\") from e\n\n def test_print_op_module(self):\n # Issue #19351: python2 and python3 go through different paths.\n # python2 returns '<module 'torch.ops' (built-in)>'\n # python3 uses __file__ and return\n # '<module 'torch.ops' from '/scratch/ailzhang/pytorch/torch/_ops.py'>'\n s = str(torch.ops)\n self.assertRegex(s, r'ops')\n\n def test_profiler(self):\n prev_opt = torch._C._get_graph_executor_optimize()\n torch._C._set_graph_executor_optimize(False)\n\n def other_fn(x):\n return x * 2\n\n x = torch.rand(3, 4)\n traced_other_fn = torch.jit.trace(other_fn, x)\n\n def fn(x):\n y = traced_other_fn(x)\n fut = torch.jit._fork(traced_other_fn, x)\n y = torch.jit._wait(fut)\n return y\n\n traced_fn = torch.jit.trace(fn, x)\n with torch.autograd.profiler.profile() as prof:\n traced_fn(x)\n\n # expecting to see other_fn TS function call\n # with cpu time >= mul cpu time and\n # a forked other_fn\n\n mul_events = defaultdict(int)\n other_fn_events = defaultdict(int)\n for e in prof.function_events:\n if e.name == \"aten::mul\":\n self.assertTrue(e.thread not in mul_events)\n mul_events[e.thread] = e.time_range.elapsed_us()\n elif e.name == \"other_fn\":\n self.assertTrue(e.thread not in other_fn_events)\n other_fn_events[e.thread] = e.time_range.elapsed_us()\n\n self.assertTrue(len(mul_events) == 2)\n self.assertTrue(len(other_fn_events) == 2)\n\n for thread, mul_time in mul_events.items():\n self.assertTrue(thread in other_fn_events)\n self.assertTrue(other_fn_events[thread] >= mul_time)\n\n torch._C._set_graph_executor_optimize(prev_opt)\n\n def test_hide_source_ranges_context_manager(self):\n @torch.jit.script\n def foo(x):\n return torch.add(x, x)\n\n graph = foo.graph\n source_range_regex = \"# .*\\\\.py\"\n self.assertRegex(graph.__repr__(), source_range_regex)\n with torch.jit._hide_source_ranges():\n self.assertNotRegex(graph.__repr__(), source_range_regex)\n self.assertRegex(graph.str(print_source_ranges=True), source_range_regex)\n self.assertRegex(graph.__repr__(), source_range_regex)\n\n\nclass TestFrontend(JitTestCase):\n\n def test_instancing_error(self):\n @torch.jit.ignore\n class MyScriptClass(object):\n def unscriptable(self):\n return \"a\" + 200\n\n\n class TestModule(torch.nn.Module):\n def __init__(self):\n super(TestModule, self).__init__()\n\n def forward(self, x):\n return MyScriptClass()\n\n with self.assertRaises(torch.jit.frontend.FrontendError) as cm:\n torch.jit.script(TestModule())\n\n checker = FileCheck()\n checker.check(\"Cannot instantiate class\")\n checker.check(\"def forward\")\n checker.run(str(cm.exception))\n\n\nclass TestScript(JitTestCase):\n\n # Tests that calling torch.jit.script repeated on function is allowed.\n def test_repeated_script_on_function(self):\n @torch.jit.script\n @torch.jit.script\n def fn(x):\n return x\n\n torch.jit.script(torch.jit.script(fn))\n\n def test_pretty_print_function(self):\n @torch.jit.script\n def foo(x):\n return torch.nn.functional.interpolate(x)\n\n FileCheck().check(\"interpolate\").run(foo.code)\n\n def test_inlined_graph(self):\n \"\"\"\n Check that the `inlined_graph` property correctly returns an inlined\n graph, both through function calls and method calls.\n \"\"\"\n @torch.jit.script\n def foo(x):\n return torch.add(x, x)\n\n class MyNestedMod(torch.nn.Module):\n def __init__(self):\n super(MyNestedMod, self).__init__()\n\n def forward(self, x):\n return torch.sub(x, x)\n\n\n class MyMod(torch.nn.Module):\n def __init__(self):\n super(MyMod, self).__init__()\n self.nested = MyNestedMod()\n\n def forward(self, x):\n x = self.nested(x) # sub\n x = foo(x) # add\n return torch.mul(x, x)\n\n m = torch.jit.script(MyMod())\n FileCheck().check(\"aten::sub\") \\\n .check(\"aten::add\") \\\n .check(\"aten::mul\") \\\n .run(m.inlined_graph)\n\n def test_static_method_on_module(self):\n \"\"\"\n Check that the `@staticmethod` annotation on a function on a module works.\n \"\"\"\n class MyCell(torch.nn.Module):\n def __init__(self):\n super(MyCell, self).__init__()\n\n @staticmethod\n def do_it(x, h):\n new_h = torch.tanh(x + h)\n return new_h, new_h\n\n def forward(self, x, h):\n return self.do_it(x, h)\n\n my_cell = torch.jit.script(MyCell())\n x = torch.rand(3, 4)\n h = torch.rand(3, 4)\n jitted_cell = my_cell(x, h)\n non_jitted_cell = MyCell().do_it(x, h)\n\n self.assertEqual(jitted_cell, non_jitted_cell)\n\n def test_code_with_constants(self):\n \"\"\"\n Check that the `code_with_constants` property correctly returns graph CONSTANTS in the\n CONSTANTS.cN format used in the output of the `code` property.\n \"\"\"\n @torch.jit.script\n def foo(x=torch.ones(1)):\n return x\n\n class Moddy(torch.nn.Module):\n def __init__(self):\n super(Moddy, self).__init__()\n\n def forward(self, x):\n return foo()\n\n m = torch.jit.script(Moddy())\n src, CONSTANTS = m.code_with_constants\n\n self.assertEqual(CONSTANTS.c0, torch.ones(1))\n self.assertEqual(src, m.code)\n\n def test_code_with_constants_restore(self):\n \"\"\"\n Check that the `code_with_constants` property correctly works on restoration after save() + load()\n \"\"\"\n @torch.jit.script\n def foo(x=torch.ones(1)):\n return x\n\n class Moddy(torch.nn.Module):\n def __init__(self):\n super(Moddy, self).__init__()\n\n def forward(self, x):\n return foo()\n\n m = torch.jit.script(Moddy())\n src, CONSTANTS = m.code_with_constants\n eic = self.getExportImportCopy(m)\n\n src_eic, CONSTANTS_eic = eic.code_with_constants\n\n self.assertEqual(src, src_eic)\n self.assertEqual(CONSTANTS.c0, CONSTANTS_eic.c0)\n\n\n def test_oneline_func(self):\n def fn(x): return x # noqa: E704\n\n self.checkScript(fn, (torch.ones(2, 2), ))\n\n def test_request_bailout(self):\n with enable_profiling_mode_for_profiling_tests():\n\n def fct_loop(x):\n for i in range(3):\n x = torch.cat((x, x), 0)\n return x\n\n x = torch.ones(2, 3, 4, dtype=torch.float32)\n expected = fct_loop(x)\n jitted = torch.jit.script(fct_loop)\n # profile\n jitted(x)\n # optimize\n jitted(x)\n dstate = jitted.get_debug_state()\n eplan = get_execution_plan(dstate)\n num_bailouts = eplan.code.num_bailouts()\n\n for i in range(0, num_bailouts):\n eplan.code.request_bailout(i)\n self.assertEqual(jitted(x), expected)\n\n @unittest.skip(\"bailouts are being deprecated\")\n def test_dominated_bailout(self):\n with enable_profiling_mode_for_profiling_tests():\n # functional dominated guard\n @torch.jit.script\n def foo(x):\n dim = x.dim()\n if dim == 0:\n y = int(x)\n else:\n y = x.size()[dim - 1]\n return y\n\n x = torch.zeros(2)\n self.assertEqual(foo(x), 2)\n self.assertEqual(foo(x), 2)\n g = torch.jit.last_executed_optimized_graph()\n g_s = str(g)\n g_s = g_s[0:g_s.find(\"return\")]\n FileCheck().check_count(\"prim::BailOut[\", 1, exactly=True).run(g_s)\n\n # dominated guard of non-functional value\n @torch.jit.script\n def foo(x):\n dim = x.dim()\n x.add_(3)\n if dim == 0:\n return 0\n else:\n return x.size()[dim - 1]\n\n x = torch.zeros(2)\n self.assertEqual(foo(x), 2)\n self.assertEqual(foo(x), 2)\n g = torch.jit.last_executed_optimized_graph()\n FileCheck().check(\"prim::BailOut[\").check(\"aten::add_\").check_next(\"prim::BailOut[\").check(\"return\").run(g)\n\n with torch.enable_grad():\n @torch.jit.ignore\n def disable_grad():\n torch.set_grad_enabled(False)\n\n @torch.jit.ignore\n def enable_grad():\n torch.set_grad_enabled(True)\n\n @torch.jit.script\n def foo(x):\n x = x + 1\n dim = x.dim()\n disable_grad()\n if dim == 0:\n y = int(x)\n else:\n y = x.size()[dim - 1]\n enable_grad()\n return y\n\n x = torch.zeros(2, requires_grad=True)\n self.assertEqual(foo(x), 2)\n self.assertEqual(foo(x), 2)\n g = torch.jit.last_executed_optimized_graph()\n # there should still be a Bailout after disable_grad call\n FileCheck().check(\"disable_grad\").check(\"BailOut[\").check(\"BailoutTemplate\").run(g)\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.PROFILING, \"skip if profiling isn't enabled\")\n def test_profiling_merge(self):\n @torch.jit.script\n def test_not_const(x):\n if x.size(0) == 1:\n return 1\n else:\n return 2\n\n with enable_profiling_mode_for_profiling_tests():\n with num_profiled_runs(2):\n test_not_const(torch.rand([1, 2]))\n test_not_const(torch.rand([2, 2]))\n\n graph_str = torch.jit.last_executed_optimized_graph()\n FileCheck().check(\"profiled_type=Double(*, 2, strides=[2, 1], requires_grad=0, device=cpu\").run(graph_str)\n FileCheck().check_not(\"profiled_type=Double(1, 2, strides=[2, 1], requires_grad=0, device=cpu\").run(graph_str)\n\n\n def test_nested_bailouts(self):\n @torch.jit.script\n def fct_loop(x):\n for i in range(3):\n x = torch.cat((x, x), 0)\n return x\n\n x = torch.ones(2, 3, 4, dtype=torch.float32)\n out = fct_loop(x)\n jit_trace = torch.jit.trace(fct_loop, x)\n out_trace = jit_trace(x)\n\n def test_no_self_arg_ignore_function(self):\n class MyModule(nn.Module):\n @torch.jit.ignore # noqa: B902\n def call_np(): # noqa: B902\n # type: () -> int\n return np.random.choice(2, p=[.95, .05])\n\n def forward(self):\n return self.call_np()\n\n with self.assertRaisesRegex(Exception, \"does not have a self argument\"):\n torch.jit.script(MyModule())\n\n def test_loop_liveness(self):\n with enable_profiling_mode_for_profiling_tests():\n @torch.jit.script\n def f(i):\n # type: (int) -> Tensor\n l = []\n for n in [2, 1]:\n l.append(torch.zeros(n, i))\n\n return l[0]\n\n f(2)\n f(1)\n\n def test_bailout_loop_carried_deps_name_clash(self):\n with enable_profiling_mode_for_profiling_tests():\n NUM_ITERATIONS = 10\n\n @torch.jit.script\n def fct_loop(z, size):\n # type: (int, int) -> Tuple[Tensor, List[int]]\n counters = torch.jit.annotate(List[int], [])\n j = 0\n y = torch.ones(2)\n for i in range(size):\n counters.append(i + j)\n y = torch.cat((y, torch.ones(z)), 0)\n j = j + 1\n return y, counters\n\n inputs = [1, 2, 3, 4]\n expected = [x * 2 for x in range(NUM_ITERATIONS)]\n for inp in inputs:\n results = fct_loop(inp, NUM_ITERATIONS)\n self.assertEqual(results[1], expected)\n\n def test_bailout_loop_counter_transition(self):\n with enable_profiling_mode_for_profiling_tests():\n NUM_ITERATIONS = 10\n\n @torch.jit.script\n def fct_loop(z, size):\n # type: (int, int) -> Tuple[Tensor, List[int]]\n counters = torch.jit.annotate(List[int], [])\n y = torch.ones(2)\n for i in range(size):\n counters.append(i)\n y = torch.cat((y, torch.ones(z)), 0)\n return y, counters\n\n inputs = [1, 2, 3, 4]\n expected = list(range(NUM_ITERATIONS))\n for inp in inputs:\n results = fct_loop(inp, NUM_ITERATIONS)\n self.assertEqual(results[1], expected)\n\n def test_ignored_method_binding(self):\n class Bar(torch.nn.Module):\n def __init__(self):\n super(Bar, self).__init__()\n self.x : int = 0\n\n @torch.jit.export\n def setx(self, x : int):\n self.x = x\n\n @torch.jit.export\n def getx(self):\n return self.x\n\n @torch.jit.ignore\n def ignored_getx(self):\n return self.x\n\n b = Bar()\n b.setx(123)\n sb = torch.jit.script(b)\n self.assertEqual(sb.getx(), 123)\n self.assertEqual(sb.ignored_getx(), 123)\n\n sb.setx(456)\n self.assertEqual(sb.getx(), 456)\n self.assertEqual(sb.ignored_getx(), 456)\n\n def test_set_attribute_through_optional(self):\n class A(torch.nn.Module):\n __annotations__ = {\"x\": Optional[torch.Tensor]}\n\n def __init__(self):\n super(A, self).__init__()\n self.x = None\n\n @torch.jit.ignore\n def foo(self):\n if self.x is None:\n self.x = torch.tensor([3])\n return self.x\n\n def forward(self, x):\n a = self.foo()\n return x + 1\n\n m = torch.jit.script(A())\n self.assertEqual(m.x, None)\n m(torch.rand(1))\n self.assertEqual(m.x, torch.tensor([3]))\n\n def test_mutate_constant(self):\n class M(torch.jit.ScriptModule):\n __constants__ = [\"foo\"]\n\n def __init__(self, foo):\n super(M, self).__init__()\n self.foo = foo\n\n m = M(5)\n # m has a constant attribute, but we can't\n # assign to it\n with self.assertRaises(RuntimeError):\n m.foo = 6\n\n def test_class_attribute(self):\n class M(torch.jit.ScriptModule):\n FOO = 0\n\n def __init__(self):\n super(M, self).__init__()\n self.foo = self.FOO\n m = M()\n self.assertEqual(m.foo, M.FOO)\n\n def test_class_attribute_in_script(self):\n class M(torch.jit.ScriptModule):\n FOO = 0\n\n def __init__(self):\n super(M, self).__init__()\n\n @torch.jit.script_method\n def forward(self):\n return self.FOO\n with self.assertRaises(RuntimeError):\n M()\n\n def test_not_initialized_err(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n self.foo = torch.rand(2, 3)\n with self.assertRaises(RuntimeError):\n M()\n\n def test_attribute_in_init(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.foo = torch.jit.Attribute(0.1, float)\n # we should be able to use self.foo as a float here\n assert 0.0 < self.foo\n M()\n\n def test_scriptable_fn_as_attr(self):\n class M(torch.nn.Module):\n def __init__(self, fn):\n super(M, self).__init__()\n self.fn = fn\n\n def forward(self, x):\n return self.fn(x)\n\n m = M(torch.sigmoid)\n inp = torch.rand(2, 3)\n self.checkModule(m, (inp, ))\n\n def test_sequence_parsing(self):\n tests = [\n (\"return [x, x,]\", True),\n (\"return [x x]\", \"expected ]\"),\n (\"return x, x,\", True),\n (\"return bar(x, x,)\", True),\n (\"return bar()\", \"Argument x not provided\"),\n (\"for a, b, in x, x,:\\n pass\", \"List of iterables\"),\n (\"a, b, = x, x,\\n return a + b\", True)\n ]\n for exp, result in tests:\n cu = torch.jit.CompilationUnit()\n full = \"\"\"\ndef bar(x, y):\n return x + y\ndef foo(x):\n {}\n \"\"\".format(exp)\n if isinstance(result, str):\n with self.assertRaisesRegex(RuntimeError, result):\n cu.define(full)\n else:\n cu.define(full)\n\n def test_namedtuple_python(self):\n global MyTuple, MyMod # see [local resolution in python]\n MyTuple = namedtuple('MyTuple', ['a'])\n\n @torch.jit.unused\n def fn():\n # type: () -> MyTuple\n return MyTuple(1)\n\n # Only check compilation\n @torch.jit.script\n def fn2():\n # type: () -> MyTuple\n return fn()\n\n FileCheck().check(\"NamedTuple\").run(fn2.graph)\n\n class MyMod(torch.nn.Module):\n def __init__(self):\n super(MyMod, self).__init__()\n\n @torch.jit.unused\n def fn(self):\n # type: () -> MyTuple\n return MyTuple(1)\n\n def forward(self, x):\n if 1 == 1:\n return MyTuple(torch.rand(2, 3))\n else:\n return self.fn()\n\n # shouldn't throw a type error\n torch.jit.script(MyMod())\n\n def test_unused_decorator(self):\n class MyMod(torch.nn.Module):\n def __init__(self):\n super(MyMod, self).__init__()\n\n @torch.jit.unused\n @torch.no_grad()\n def fn(self, x):\n # type: (Tensor) -> int\n return next(x) # invalid, but should be ignored\n\n def forward(self, x):\n return self.fn(x)\n\n torch.jit.script(MyMod())\n\n @_inline_everything\n def test_lazy_script(self):\n def untraceable(x):\n if x.ndim > 2:\n print(\"hello\")\n else:\n print(\"goodbye\")\n return x + 2\n\n # Non-working example\n def fn(x):\n return untraceable(x)\n\n with self.capture_stdout():\n traced_bad = torch.jit.trace(fn, [torch.ones(2, 2)])\n\n FileCheck().check_not(\"goodbye\").check_not(\"hello\").run(traced_bad.graph)\n\n # Working example\n untraceable = torch.jit.script_if_tracing(untraceable)\n\n def fn2(x):\n return untraceable(x)\n\n with self.capture_stdout():\n traced = torch.jit.trace(fn, [torch.ones(2, 2)])\n\n FileCheck().check(\"goodbye\").run(traced.graph)\n\n def foo(x: int):\n return x + 1\n\n @torch.jit.script_if_tracing\n def fee(x: int = 2):\n return foo(1) + x\n\n # test directly compiling function\n fee_compiled = torch.jit.script(fee)\n self.assertEqual(fee_compiled(), fee())\n\n # test compiling it within another function\n @torch.jit.script\n def hum():\n return fee(x=3)\n\n self.assertEqual(hum(), 5)\n\n def test_big_int_literals(self):\n def ok():\n # signed 64 bit max\n a = 9223372036854775807\n return a\n\n def toobig():\n a = 9223372036854775808\n return a\n\n def waytoobig():\n a = 99999999999999999999\n return a\n\n self.checkScript(ok, [])\n\n with self.assertRaisesRegex(RuntimeError, \"out of range\"):\n torch.jit.script(toobig)\n\n with self.assertRaisesRegex(RuntimeError, \"out of range\"):\n torch.jit.script(waytoobig)\n\n def test_hex_literals(self):\n def test1():\n return 0xaaaaaa\n\n def test2():\n return 0xaaaaaa\n\n def test3():\n return -0xaaaaaa\n\n self.checkScript(test1, [])\n self.checkScript(test2, [])\n self.checkScript(test3, [])\n\n def ok():\n a = 0x7FFFFFFFFFFFFFFF\n return a\n\n def toobig():\n a = 0xFFFFFFFFFFFFFFFF\n return a\n\n def waytoobig():\n a = 0xFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF\n return a\n\n self.checkScript(ok, [])\n\n with self.assertRaisesRegex(RuntimeError, \"out of range\"):\n torch.jit.script(toobig)\n\n with self.assertRaisesRegex(RuntimeError, \"out of range\"):\n torch.jit.script(waytoobig)\n\n def test_big_float_literals(self):\n def ok():\n # Python interprets this as inf\n a = 1.2E400\n return a\n\n def check(fn):\n self.assertTrue(fn() == ok())\n\n # checkScript doesn't work since assertEqual doesn't consider\n # `inf` == `inf`\n check(torch.jit.script(ok))\n\n cu = torch.jit.CompilationUnit()\n cu.define(dedent(inspect.getsource(ok)))\n check(cu.ok)\n\n def _test_device_type(self, dest):\n def fn(x):\n # type: (Device) -> Tuple[str, Optional[int]]\n return x.type, x.index\n\n device = torch.ones(2).to(dest).device\n self.checkScript(fn, [device])\n\n def test_device_type(self):\n self._test_device_type('cpu')\n\n @unittest.skipIf(not RUN_CUDA, \"Requires CUDA\")\n def test_device_type_cuda(self):\n self._test_device_type('cuda')\n\n def test_string_device_implicit_conversion(self):\n @torch.jit.script\n def fn(x: torch.device):\n return x\n\n self.assertEqual(fn(\"cpu\"), torch.device(\"cpu\"))\n\n with self.assertRaisesRegex(RuntimeError, \"Expected one of\"):\n fn(\"invalid_device\")\n\n def test_eval_python(self):\n def _test(m):\n self.assertTrue(m(torch.ones(2, 2)))\n self.assertTrue(m.training)\n self.assertTrue(m._c.getattr('training'))\n\n m.eval()\n\n self.assertFalse(m.training)\n self.assertFalse(m._c.getattr('training'))\n self.assertFalse(m(torch.ones(2, 2)))\n\n buffer = io.BytesIO()\n torch.jit.save(m, buffer)\n buffer.seek(0)\n\n loaded = torch.jit.load(buffer)\n\n self.assertFalse(loaded.training)\n self.assertFalse(loaded._c.getattr('training'))\n\n class M(nn.Module):\n def __init__(self):\n super(M, self).__init__()\n\n def forward(self, x):\n return self.training\n\n class OldM(torch.jit.ScriptModule):\n def __init__(self):\n super(OldM, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x):\n return self.training\n\n _test(torch.jit.script(M()))\n _test(OldM())\n\n def test_inherit_method(self):\n class A(torch.jit.ScriptModule):\n def __init__(self):\n super(A, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x):\n return x + self.bar(x)\n\n class B(A):\n def __init__(self):\n super(B, self).__init__()\n\n @torch.jit.script_method\n def bar(self, x):\n return x * x\n\n with self.assertRaisesRegex(RuntimeError, 'attribute'):\n A() # cannot use because bar is not defined\n\n v = torch.rand(3, 4)\n b = B()\n self.assertEqual(b(v), v + v * v)\n\n class C(torch.jit.ScriptModule):\n def __init__(self):\n super(C, self).__init__()\n\n @torch.jit.script_method\n def bar(self, x):\n return x\n\n class D(C, B):\n def __init__(self):\n super(D, self).__init__()\n\n self.assertEqual(D()(v), v + v)\n\n def test_tensor_subclasses(self):\n def check_subclass(x, tensor):\n template = dedent(\"\"\"\n def func(input: {}) -> {}:\n return torch.zeros((input.shape[0], 1), dtype=input.dtype)\n \"\"\")\n\n self._check_code(template.format(x, x), \"func\", [tensor])\n\n check_subclass(\"torch.LongTensor\", torch.LongTensor([[1, 2], [3, 4]]))\n check_subclass(\"torch.DoubleTensor\", torch.DoubleTensor([[1.2, 2.3], [3.4, 4.5]]))\n check_subclass(\"torch.IntTensor\", torch.IntTensor([[1, 2], [3, 4]]))\n check_subclass(\"torch.BoolTensor\", torch.BoolTensor([[False, True], [True, False]]))\n\n def check_subclass_warn(input: torch.LongTensor) -> torch.LongTensor:\n return torch.zeros((input.shape[0], 1), dtype=input.dtype)\n\n with warnings.catch_warnings(record=True) as warns:\n scripted = torch.jit.script(check_subclass_warn)\n FileCheck().check(\"TorchScript will treat type annotations of Tensor\").run(str(warns[0]))\n\n def test_first_class_module(self):\n class Foo(torch.jit.ScriptModule):\n def __init__(self):\n super(Foo, self).__init__()\n self.foo = nn.Parameter(torch.rand(3, 4))\n\n @torch.jit.script_method\n def forward(self, input):\n self.foo = input\n return self.foo\n foo = Foo()\n input = torch.rand(3, 4)\n foo.forward(input)\n self.assertEqual(input, foo.foo)\n\n @_tmp_donotuse_dont_inline_everything\n def test_first_class_calls(self):\n @torch.jit.script\n class Foo(object):\n def __init__(self, x):\n self.bar = x\n\n def stuff(self, x):\n return self.bar + x\n\n @torch.jit.script\n def foo(x):\n return x * x + Foo(x).stuff(2 * x)\n\n @torch.jit.script\n def bar(x):\n return foo(x) * foo(x)\n\n x = torch.rand(3, 4)\n self.assertEqual(bar(x), (x * x + 3 * x) * (x * x + 3 * x))\n\n def test_static_methods(self):\n class M(nn.Module):\n def __init__(self):\n super(M, self).__init__()\n\n @staticmethod\n def my_method(x):\n return x + 100\n\n def forward(self, x):\n return x + M.my_method(x)\n\n class N(nn.Module):\n def __init__(self):\n super(N, self).__init__()\n\n @staticmethod\n def my_method(x):\n return x * 100\n\n def forward(self, x):\n return x - M.my_method(x) + N.my_method(x)\n\n self.checkModule(M(), (torch.ones(2, 2),))\n\n self.checkModule(N(), (torch.ones(2, 2),))\n\n def test_invalid_prefix_annotation(self):\n with self.assertRaisesRegex(RuntimeError, \"annotation prefix in line\"):\n with self.capture_stdout() as captured:\n @torch.jit.script\n def invalid_prefix_annotation1(a):\n #type: (Int) -> Int # noqa: E265\n return a + 2\n\n with self.assertRaisesRegex(RuntimeError, \"annotation prefix in line\"):\n with self.capture_stdout() as captured:\n @torch.jit.script\n def invalid_prefix_annotation2(a):\n #type : (Int) -> Int # noqa: E265\n return a + 2\n\n with self.assertRaisesRegex(RuntimeError, \"annotation prefix in line\"):\n with self.capture_stdout() as captured:\n @torch.jit.script\n def invalid_prefix_annotation3(a):\n # type: (Int) -> Int\n return a + 2\n\n def test_builtin_function_attributes(self):\n class Add(nn.Module):\n def __init__(self):\n super(Add, self).__init__()\n self.add = torch.add\n\n def forward(self, input):\n return self.add(input, input)\n\n self.checkModule(Add(), [torch.randn(2, 2)])\n\n def test_pybind_type_comparisons(self):\n @torch.jit.script\n def f():\n return None\n\n node = list(f.graph.nodes())[0]\n t = node.outputsAt(0).type()\n self.assertIsNotNone(t)\n\n @unittest.skipIf(IS_WINDOWS and sys.version_info >= (3, 8), 'TODO: need to fix the test case')\n def test_unmatched_type_annotation(self):\n message1 = re.escape(\"Number of type annotations (2) did not match the number of function parameters (1):\")\n message2 = 'def invalid2\\\\(a\\\\):\\n\\\\s*~+\\\\.*\\\\s+<--- HERE\\n\\\\s+# type: \\\\(Int, Int\\\\) -> Int\\n\\\\s+return a \\\\+ 2'\n message3 = 'def invalid4\\\\(a\\\\):\\n\\\\s*~+\\\\.*\\\\s+<--- HERE\\n\\\\s+# type: \\\\(Int, Int\\\\) -> Int\\n\\\\s+return a \\\\+ 2'\n with self.assertRaisesRegex(RuntimeError, message1):\n @torch.jit.script\n def invalid1(a):\n # type: (Int, Int) -> Int\n return a + 2\n\n with self.assertRaisesRegex(RuntimeError, message2):\n @torch.jit.script\n def invalid2(a):\n # type: (Int, Int) -> Int\n return a + 2\n\n with self.assertRaisesRegex(RuntimeError, message1):\n def invalid3(a):\n # type: (Int, Int) -> Int\n return a + 2\n torch.jit.script(invalid3)\n\n with self.assertRaisesRegex(RuntimeError, message3):\n def invalid4(a):\n # type: (Int, Int) -> Int\n return a + 2\n torch.jit.script(invalid4)\n\n def test_is_optional(self):\n ann = Union[List[int], List[float]]\n torch._jit_internal.is_optional(ann)\n\n def test_interpreter_fuzz(self):\n import builtins\n # This test generates random tree-like programs to fuzz test\n # that the interpreter does not have a bug in its stack manipulation\n # code. An assert in that code ensures individual operators are\n # not reordered.\n templates = [\n \"torch.rand(3, 4)\",\n \"({} + {})\",\n \"-{}\",\n \"({} * {})\",\n \"torch.tanh({})\",\n \"VAR {}\",\n ]\n\n def gen_code():\n src_lines = ['def f():']\n exprs = []\n n_variables = 0\n\n def get_expr(idx):\n elem = exprs[idx]\n exprs[idx] = exprs[-1]\n exprs.pop()\n return elem\n\n def select_expr_or_var():\n idx = random.randrange(0, len(exprs) + n_variables)\n if idx < len(exprs):\n return get_expr(idx)\n else:\n return 'v{}'.format(idx - len(exprs))\n\n for i in range(50):\n n = None\n while n is None or n > len(exprs) + n_variables:\n template = random.choice(templates)\n n = template.count('{}')\n\n if 'VAR' in template:\n src_lines.append(' v{} = {}'.format(n_variables, select_expr_or_var()))\n n_variables += 1\n else:\n exprs.append(template.format(*(select_expr_or_var() for _ in range(n))))\n\n src_lines.append(' return ({})\\n'.format(''.join('v{},'.format(i) for i in range(n_variables))))\n return '\\n'.join(src_lines)\n\n for i in range(100):\n g = {'torch': torch}\n code = gen_code()\n builtins.exec(code, g, None)\n cu = torch.jit.CompilationUnit(code)\n with freeze_rng_state():\n o1 = g['f']()\n with freeze_rng_state():\n o2 = cu.f()\n self.assertEqual(o1, o2)\n\n def test_cpp_module_iterator(self):\n a = nn.Module()\n a.name = 'a'\n a.p = nn.Parameter(torch.rand(3, 4))\n a.foo = nn.Module()\n a.foo.name = 'foo'\n a.foo.register_buffer('b', torch.rand(1, 1))\n a.foo.bar = nn.Module()\n a.foo.bar.name = 'bar'\n a.foo.bar.an_int = 4\n a.another = nn.Module()\n a.another.name = 'another'\n sa = torch.jit.script(a)\n result = torch._C._jit_debug_module_iterators(sa._c)\n\n def replace(e):\n if e is a.p:\n return 'P'\n elif e is a.foo.b:\n return 'B'\n elif isinstance(e, torch._C.ScriptModule):\n return e.getattr('name')\n\n return e\n for k, v in result.items():\n for i in range(len(v)):\n if isinstance(v[i], tuple):\n n, v2 = v[i]\n v[i] = (n, replace(v2))\n else:\n v[i] = replace(v[i])\n # module type creation is not deterministic, so we have to sort\n # the result\n v.sort()\n expected = {'buffers': [],\n 'buffers_r': ['B'],\n 'children': ['another', 'foo'],\n 'modules': ['a', 'another', 'bar', 'foo'],\n 'named_attributes': [('_is_full_backward_hook', None),\n ('another', 'another'),\n ('foo', 'foo'),\n ('name', 'a'),\n ('p', 'P'),\n ('training', True)],\n 'named_attributes_r': [('_is_full_backward_hook', None),\n ('another', 'another'),\n ('another._is_full_backward_hook', None),\n ('another.name', 'another'),\n ('another.training', True),\n ('foo', 'foo'),\n ('foo._is_full_backward_hook', None),\n ('foo.b', 'B'),\n ('foo.bar', 'bar'),\n ('foo.bar._is_full_backward_hook', None),\n ('foo.bar.an_int', 4),\n ('foo.bar.name', 'bar'),\n ('foo.bar.training', True),\n ('foo.name', 'foo'),\n ('foo.training', True),\n ('name', 'a'),\n ('p', 'P'),\n ('training', True)],\n 'named_buffers': [],\n 'named_buffers_r': [('foo.b', 'B')],\n 'named_children': [('another', 'another'), ('foo', 'foo')],\n 'named_modules': [('', 'a'),\n ('another', 'another'),\n ('foo', 'foo'),\n ('foo.bar', 'bar')],\n 'named_parameters': [('p', 'P')],\n 'named_parameters_r': [('p', 'P')],\n 'parameters': ['P'],\n 'parameters_r': ['P']}\n self.assertEqual(expected, result)\n\n def test_parameter_order(self):\n m = nn.Module()\n for i, name in enumerate(string.ascii_letters):\n setattr(m, name, nn.Parameter(torch.tensor([float(i)])))\n ms = torch.jit.script(m)\n print(torch.cat(list(m.parameters())))\n print(torch.cat(list(ms.parameters())))\n self.assertEqual(list(m.parameters()), list(ms.parameters()))\n\n def test_python_op_builtins(self):\n @torch.jit.unused\n def fn(x):\n # type: (List[int]) -> int\n return sum(x)\n\n @torch.jit.script\n def script_fn(x):\n # type: (List[int]) -> int\n return fn(x)\n\n def test_submodule_twice(self):\n @torch.jit.script\n def foo(x):\n return x * x\n\n class What(torch.jit.ScriptModule):\n def __init__(self, x):\n super(What, self).__init__()\n self.foo = x\n a = What(foo)\n c = What(foo)\n\n def test_training_param(self):\n class What(torch.jit.ScriptModule):\n def __init__(self):\n super(What, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x):\n # type: (int) -> int\n if self.training:\n r = x\n else:\n r = x + 4\n # check double use of training\n if self.training:\n r = r + 1\n return r\n\n w = What()\n self.assertEqual(4, w(3))\n w.train(False)\n self.assertEqual(7, w(3))\n self.assertFalse(\"training\" in w.state_dict())\n\n def test_class_as_attribute(self):\n @torch.jit.script\n class Foo321(object):\n def __init__(self):\n self.x = 3\n\n class FooBar1234(torch.nn.Module):\n def __init__(self):\n super(FooBar1234, self).__init__()\n self.f = Foo321()\n\n def forward(self, x):\n return x + self.f.x\n\n scripted = torch.jit.script(FooBar1234())\n eic = self.getExportImportCopy(scripted)\n x = torch.rand(3, 4)\n self.assertEqual(scripted(x), eic(x))\n\n def test_module_str(self):\n class Foo(torch.nn.Module):\n def forward(self, x):\n return torch.relu(x)\n\n f = torch.jit.script(Foo())\n self.assertEqual('ScriptObject', str(f._c))\n\n def test_jitter_bug(self):\n @torch.jit.script\n def fn2(input, kernel_size):\n # type: (Tensor, List[int]) -> Tensor\n if kernel_size[0] > 1:\n _stride = [2]\n else:\n _stride = kernel_size\n print(_stride, kernel_size)\n return input\n\n @torch.jit.script\n def fn(input):\n # type: (Tensor) -> Tensor\n return fn2(input, [1])\n\n def test_parser_kwargonly(self):\n cu = torch.jit.CompilationUnit('''\n def foo(x, *, y) -> Tuple[Tensor, Tensor]:\n return x, x\n def bar(x):\n return foo(x, y=x)\n ''')\n self.assertTrue('*' in str(cu.foo.schema))\n with self.assertRaisesRegex(RuntimeError, \"not provided\"):\n torch.jit.CompilationUnit('''\n def foo(x, *, y) -> Tuple[Tensor, Tensor]:\n return x, x\n def bar(x):\n return foo(x, x)\n ''')\n\n def test_annoying_doubles(self):\n mod = types.ModuleType(\"temp\")\n mod.inf = float(\"inf\")\n mod.ninf = float(\"-inf\")\n mod.nan = float(\"nan\")\n\n with torch._jit_internal._disable_emit_hooks():\n class Foo(torch.jit.ScriptModule):\n def __init__(self):\n super(Foo, self).__init__()\n\n @torch.jit.script_method\n def forward(self):\n return math.pi, 0.1, mod.inf, mod.ninf, 2.225073858507201e-308, mod.nan\n\n foo = Foo()\n buffer = io.BytesIO()\n torch.jit.save(foo, buffer)\n\n buffer.seek(0)\n foo_loaded = torch.jit.load(buffer)\n\n r = foo()\n r2 = foo_loaded()\n # use precise assert, we are checking floating point details\n self.assertTrue(r[:-1] == r2[:-1])\n self.assertTrue(math.isnan(r[-1]) and math.isnan(r2[-1]))\n\n def test_type_annotate(self):\n\n def foo(a):\n return torch.jit.annotate(torch.Tensor, a)\n\n self.checkScript(foo, (torch.rand(3),))\n\n def bar():\n a = torch.jit.annotate(List[int], [])\n for _ in range(10):\n a.append(4)\n return a\n\n self.checkScript(bar, ())\n\n def baz(a):\n return torch.jit.annotate(float, a)\n self.checkScript(baz, (torch.rand(()),))\n\n # test annotate none types\n def annotate_none():\n return torch.jit.annotate(Optional[torch.Tensor], None)\n\n self.checkScript(annotate_none, ())\n\n\n def test_robust_op_resolution(self):\n neg = torch.add # misleading name to make sure we resolve by function\n\n def stuff(x):\n return neg(x, x)\n\n a = (torch.rand(3),)\n self.checkScript(stuff, a)\n\n def test_nested_aug_assign(self):\n @torch.jit.script\n class SomeClass(object):\n def __init__(self):\n self.num = 99\n\n def __iadd__(self, x):\n # type: (int)\n self.num += x\n return self\n\n def __eq__(self, other):\n # type: (SomeClass) -> bool\n return self.num == other.num\n\n @torch.jit.script\n class SomeOutOfPlaceClass(object):\n def __init__(self):\n self.num = 99\n\n def __add__(self, x):\n # type: (int)\n self.num = x\n return self\n\n def __eq__(self, other):\n # type: (SomeClass) -> bool\n return self.num == other.num\n\n class Child(nn.Module):\n def __init__(self):\n super().__init__()\n self.x = 2\n self.o = SomeClass()\n self.oop = SomeOutOfPlaceClass()\n self.list = [1, 2, 3]\n\n class A(nn.Module):\n def __init__(self):\n super().__init__()\n self.child = Child()\n\n def forward(self):\n self.child.x += 1\n self.child.o += 5\n self.child.oop += 5\n some_list = [1, 2]\n self.child.list += some_list\n self.child.list *= 2\n return self.child.x, self.child.o, self.child.list, self.child.oop\n\n a = A()\n sa = torch.jit.script(A())\n eager_result = a()\n script_result = sa()\n self.assertEqual(eager_result, script_result)\n self.assertEqual(a.child.x, sa.child.x)\n self.assertEqual(a.child.o, sa.child.o)\n self.assertEqual(a.child.list, sa.child.list)\n\n @torch.jit.script\n class SomeNonAddableClass(object):\n def __init__(self):\n self.num = 99\n\n def __eq__(self, other):\n # type: (SomeClass) -> bool\n return self.num == other.num\n\n # with self.assertRaisesRegex(RuntimeError, \"\")\n class A(nn.Module):\n def __init__(self):\n super().__init__()\n self.x = SomeNonAddableClass()\n\n def forward(self):\n self.x += SomeNonAddableClass()\n return self.x\n\n with self.assertRaisesRegex(RuntimeError, \"Cannot emit inplace op\"):\n torch.jit.script(A())\n\n def test_var_aug_assign(self):\n @torch.jit.script\n class SomeNonAddableClass(object):\n def __init__(self):\n self.num = 99\n\n def __eq__(self, other):\n # type: (SomeNonAddableClass) -> bool\n return self.num == other.num\n\n with self.assertRaisesRegex(RuntimeError, \"Cannot emit inplace op\"):\n @torch.jit.script\n def fn():\n a = SomeNonAddableClass()\n a += SomeNonAddableClass()\n return a\n\n @torch.jit.script\n class SomeClass(object):\n def __init__(self):\n self.num = 99\n\n def __iadd__(self, x):\n # type: (int)\n self.num += x\n return self\n\n def __eq__(self, other):\n # type: (SomeClass) -> bool\n return self.num == other.num\n\n @torch.jit.script\n class SomeOutOfPlaceClass(object):\n def __init__(self):\n self.num = 99\n\n def __add__(self, x):\n # type: (int)\n self.num = x\n return self\n\n def __eq__(self, other):\n # type: (SomeClass) -> bool\n return self.num == other.num\n\n def fn2():\n a = SomeClass()\n a_copy = a\n a += 20\n assert a is a_copy\n b = SomeOutOfPlaceClass()\n b_copy = b\n b += 99\n assert b is b_copy\n c = [1, 2, 3]\n c_copy = c\n c *= 2\n assert c is c_copy\n c += [4, 5, 6]\n d = torch.ones(2, 2)\n d_copy = d\n d += torch.ones(2, 2)\n assert d is d_copy\n return a, b, c, d\n\n self.checkScript(fn2, [])\n\n def test_nested_list_construct(self):\n def foo():\n return [[4]] + [[4, 5]]\n self.checkScript(foo, ())\n\n def test_file_line_error(self):\n def foobar(xyz):\n return torch.blargh(xyz)\n\n _, lineno = inspect.getsourcelines(foobar)\n with self.assertRaisesRegex(RuntimeError, \"test_jit.py\\\", line {}\".format(lineno + 1)):\n scripted = torch.jit.script(foobar)\n\n def test_file_line_error_class_defn(self):\n class FooBar(object):\n def baz(self, xyz):\n return torch.blargh(xyz)\n\n _, lineno = inspect.getsourcelines(FooBar)\n with self.assertRaisesRegex(RuntimeError, \"test_jit.py\\\", line {}\".format(lineno + 2)):\n torch.jit.script(FooBar)\n\n def test_file_line_graph(self):\n def foobar(xyz):\n return torch.neg(xyz)\n\n scripted = torch.jit.script(foobar)\n\n _, lineno = inspect.getsourcelines(foobar)\n fc = FileCheck().check('test_jit.py:{}:19'.format(lineno + 1))\n fc.run(scripted.graph)\n fc.run(str(scripted.graph))\n\n def test_file_line_save_load(self):\n class Scripted(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, xyz):\n return torch.neg(xyz)\n\n scripted = Scripted()\n\n # NB: not using getExportImportCopy because that takes a different\n # code path that calls CompilationUnit._import rather than\n # going through the full save/load pathway\n buffer = scripted.save_to_buffer()\n bytesio = io.BytesIO(buffer)\n scripted = torch.jit.load(bytesio)\n\n _, lineno = inspect.getsourcelines(Scripted)\n fc = FileCheck().check(':{}'.format(lineno + 3))\n fc.run(scripted.graph)\n fc.run(str(scripted.graph))\n\n def test_file_line_string(self):\n scripted = torch.jit.CompilationUnit('''\ndef foo(xyz):\n return torch.neg(xyz)\n ''')\n\n fc = FileCheck().check('<string>:3:11')\n fc.run(scripted.foo.graph)\n fc.run(str(scripted.foo.graph))\n\n def test_file_line_trace(self):\n def foobar(xyz):\n return torch.neg(xyz)\n\n scripted = torch.jit.trace(foobar, (torch.rand(3, 4)))\n\n _, lineno = inspect.getsourcelines(foobar)\n fc = FileCheck().check('test_jit.py:{}:0'.format(lineno + 1))\n fc.run(scripted.graph)\n fc.run(str(scripted.graph))\n\n def test_serialized_source_ranges(self):\n\n class FooTest(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, x, w):\n return torch.mm(x, w.t())\n\n ft = FooTest()\n loaded = self.getExportImportCopy(ft)\n _, lineno = inspect.getsourcelines(FooTest)\n\n with self.assertRaisesRegex(RuntimeError, 'test_jit.py\\\", line {}'.format(lineno + 3)):\n loaded(torch.rand(3, 4), torch.rand(30, 40))\n\n def test_serialized_source_ranges_graph(self):\n\n class FooTest3(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, x, w):\n return torch.mm(x, w.t())\n\n ft = FooTest3()\n loaded = self.getExportImportCopy(ft)\n _, lineno = inspect.getsourcelines(FooTest3)\n\n fc = FileCheck().check('test_jit.py:{}'.format(lineno + 3))\n fc.run(loaded.graph)\n\n def test_serialized_source_ranges2(self):\n\n class FooTest2(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self):\n raise RuntimeError('foo')\n\n _, lineno = inspect.getsourcelines(FooTest2)\n\n with self.assertRaisesRegex(torch.jit.Error, 'test_jit.py\\\", line {}'.format(lineno + 3)):\n ft = FooTest2()\n loaded = self.getExportImportCopy(ft)\n loaded()\n\n def test_serialized_source_ranges_dont_jitter(self):\n class FooTest3(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, lim):\n first = 1\n second = 1\n i = 1\n somenum = 5\n dontmutateme = 3\n third = 0\n while bool(i < lim):\n third = first + second\n first = second\n second = third\n j = 0\n while j < 10:\n somenum = somenum * 2\n j = j + 1\n i = i + j\n i = i + dontmutateme\n\n st = second + third\n fs = first + second\n return third, st, fs\n\n ft3 = FooTest3()\n\n def debug_records_from_mod(self, mod):\n buffer = io.BytesIO()\n torch.jit.save(ft3, buffer)\n buffer.seek(0)\n archive = zipfile.ZipFile(buffer)\n files = filter(lambda x: x.startswith('archive/code/'), archive.namelist())\n debug_files = list(filter(lambda f: f.endswith('.debug_pkl'), files))\n self.assertEqual(len(debug_files), 1)\n debug_file = archive.open(debug_files[0])\n return pickle.load(debug_file), buffer\n\n records1, buffer = debug_records_from_mod(self, ft3)\n\n buffer.seek(0)\n loaded = torch.jit.load(buffer)\n records2, buffer = debug_records_from_mod(self, loaded)\n\n buffer.seek(0)\n loaded2 = torch.jit.load(buffer)\n records3, _ = debug_records_from_mod(self, loaded2)\n\n self.assertEqual(records1, records2)\n self.assertEqual(records2, records3)\n\n def test_serialized_source_ranges_no_dups(self):\n class FooTest3(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, lim):\n first = 1\n second = 1\n i = 1\n somenum = 5\n dontmutateme = 3\n third = 0\n while bool(i < lim):\n third = first + second\n first = second\n second = third\n j = 0\n while j < 10:\n somenum = somenum * 2\n j = j + 1\n i = i + j\n i = i + dontmutateme\n\n st = second + third\n fs = first + second\n return third, st, fs\n\n ft3 = FooTest3()\n\n def debug_records_from_mod(mod):\n buffer = io.BytesIO()\n torch.jit.save(ft3, buffer)\n buffer.seek(0)\n archive = zipfile.ZipFile(buffer)\n files = list(filter(lambda x: x.startswith('archive/code/'), archive.namelist()))\n debug_files = filter(lambda f: f.endswith('.debug_pkl'), files)\n debug_files = (archive.open(f) for f in debug_files)\n debug_files = (pickle.load(f) for f in debug_files)\n return list(debug_files)\n\n debug_files = debug_records_from_mod(ft3)\n for debug_file in debug_files:\n for i in range(len(debug_file) - 1):\n offset, source_range_tag, source_range = debug_file[i]\n offset2, source_range_tag2, source_range2 = debug_file[i + 1]\n self.assertNotEqual(source_range, source_range2)\n\n def test_circular_dependency(self):\n \"\"\"\n https://github.com/pytorch/pytorch/issues/25871\n \"\"\"\n class A(torch.jit.ScriptModule):\n def __init__(self):\n super(A, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x):\n return x\n\n class B(torch.jit.ScriptModule):\n def __init__(self):\n super(B, self).__init__()\n self.foo = torch.nn.ModuleList([A()])\n\n @torch.jit.script_method\n def forward(self, x):\n for f in self.foo:\n x = f(x)\n return x\n\n class C(torch.jit.ScriptModule):\n def __init__(self):\n super(C, self).__init__()\n self.foo = torch.nn.Sequential(B())\n\n @torch.jit.script_method\n def forward(self, x):\n for f in self.foo:\n x = f(x)\n return x\n self.getExportImportCopy(C())\n\n def test_serialize_long_lines(self):\n class OrderModuleLong(torch.nn.Module):\n def forward(self, long_arg_name: List[torch.Tensor]):\n return [(long_arg_name[1],), (long_arg_name[0].argmax(),)]\n src = str(torch.jit.script(OrderModuleLong()).code)\n # make long_arg_name[1] does not get reordered after the argmax\n FileCheck().check(\"long_arg_name[1]\").check(\"argmax\").run(src)\n\n def test_tensor_shape(self):\n x = torch.empty(34, 56, 78)\n\n def f(x):\n return x.shape\n\n self.checkScript(f, (x,))\n\n\n def test_block_input_grad_in_loop(self):\n\n x = torch.randn(3, 3, requires_grad=False)\n y = torch.randn(3, 3, requires_grad=True)\n\n def grad_in_loop(x, y):\n for i in range(100):\n x = y @ x\n return x\n\n scripted = torch.jit.script(grad_in_loop)\n outer = scripted.graph_for(x, y)\n loop = outer.findNode(\"prim::Loop\")\n loop_block = next(loop.blocks())\n param_node = loop_block.paramNode()\n x_value = list(param_node.outputs())[1]\n self.assertTrue(x_value.requires_grad())\n\n def test_tensor_grad(self):\n x = torch.randn(3, 4, requires_grad=True)\n y = torch.randn(3, 4, requires_grad=False)\n\n def f_requires_grad(x):\n return x.requires_grad\n\n self.checkScript(f_requires_grad, (x,))\n self.checkScript(f_requires_grad, (y,))\n\n def f_grad(x):\n return x.grad\n\n x.sum().backward()\n self.checkScript(f_grad, (x,))\n self.checkScript(f_grad, (y,))\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY, \"shape analysis is only enabled in Legacy\")\n def test_prim_grad_undefined(self):\n\n x = torch.ones(2)\n\n def f_grad(x):\n return x.grad\n\n scripted = self.checkScript(f_grad, (x,))\n g = scripted.graph_for(x)\n\n prim_grad_node = g.findNode(\"prim::grad\")\n self.assertTrue(next(prim_grad_node.outputs()).type().undefined() is None)\n\n def test_tensor_data(self):\n x = torch.randn(3, 4, requires_grad=True)\n y = torch.randn(4, 5)\n\n def f_data(x):\n return x.data\n\n scripted_f_data = torch.jit.script(f_data)\n\n scripted_x = scripted_f_data(x)\n self.assertEqual(scripted_x, f_data(x))\n self.assertEqual(scripted_x.requires_grad, False)\n\n scripted_y = scripted_f_data(y)\n self.assertEqual(scripted_y, f_data(y))\n self.assertEqual(scripted_x.requires_grad, False)\n\n def test_tensor_dtype(self):\n x_byte = torch.empty(34, 56, 78, dtype=torch.uint8)\n x_long = torch.empty(34, 56, 78, dtype=torch.long)\n x_float32 = torch.empty(34, 56, 78, dtype=torch.float32)\n\n @torch.jit.script\n def byte(x):\n return x.dtype == torch.uint8\n\n @torch.jit.script\n def long(x):\n return x.dtype == torch.long\n\n @torch.jit.script\n def float32(x):\n return x.dtype == torch.float32\n\n self.assertTrue(byte(x_byte))\n self.assertFalse(byte(x_long))\n self.assertFalse(byte(x_float32))\n self.assertFalse(long(x_byte))\n self.assertTrue(long(x_long))\n self.assertFalse(long(x_float32))\n self.assertFalse(float32(x_byte))\n self.assertFalse(float32(x_long))\n self.assertTrue(float32(x_float32))\n\n @unittest.skipIf(not RUN_CUDA, \"device tests require CUDA\")\n def test_tensor_device(self):\n cpu = torch.empty(34, 56, 78, device='cpu')\n gpu = torch.empty(34, 56, 78, device='cuda')\n\n @torch.jit.script\n def same_device(x, y):\n return x.device == y.device\n\n self.assertTrue(same_device(cpu, cpu))\n self.assertTrue(same_device(gpu, gpu))\n self.assertFalse(same_device(cpu, gpu))\n\n @unittest.skipIf(not RUN_CUDA, \"device tests require CUDA\")\n def test_tensor_to_device(self):\n def to_device(x):\n return x.to(device=\"cuda\").to(device=torch.device(\"cpu\"))\n\n self.checkScript(to_device, (torch.ones(3, 4),))\n\n def test_tensor_to_cpu(self):\n def to_cpu(x):\n return x.cpu()\n\n x = torch.ones(3, 4)\n script_fn = torch.jit.script(to_cpu)\n self.assertEqual(to_cpu(x).device, script_fn(x).device)\n self.checkScript(to_cpu, (x,))\n\n @unittest.skipIf(not RUN_CUDA, \"device tests require CUDA\")\n def test_tensor_to_cuda(self):\n def to_cuda(x):\n return x.cuda()\n\n x = torch.ones(3, 4)\n script_fn = torch.jit.script(to_cuda)\n self.assertEqual(to_cuda(x).device, script_fn(x).device)\n self.checkScript(to_cuda, (x,))\n\n def test_generic_list_errors(self):\n with self.assertRaisesRegex(RuntimeError, \"previously matched to type\"):\n @torch.jit.script\n def foo(x):\n return [[x]] + [[1]]\n\n def test_script_cu(self):\n cu = torch.jit.CompilationUnit('''\n def foo(a):\n b = a\n return b\n ''')\n a = Variable(torch.rand(1))\n self.assertEqual(a, cu.foo(a))\n\n # because the compilation unit ingests python strings\n # to use an escape sequence escape the backslash (\\\\n = \\n)\n def test_string_cu(self):\n cu = torch.jit.CompilationUnit('''\n def foo(a):\n print(a, \"\"\"a\\\\n\\tb\\\\n\"\"\", 2, \"a\\\na\")\n return a\n ''')\n FileCheck().check(\"aa\").check(\"a\\\\n\\\\tb\\\\n\").run(str(cu.foo.graph))\n\n def test_function_compilation_caching(self):\n def fun():\n return 1 + 2\n\n fun_compiled = torch.jit.script(fun)\n # python wrapper around the script function is a different pointer,\n # but the underlying script function graph is the same\n self.assertIs(fun_compiled.graph, torch.jit.script(fun).graph)\n\n def fun():\n return 3 + 4\n\n num_ref_counts = sys.getrefcount(fun)\n\n # caching doesn't get tripped up by same qualname\n fun_compiled_2 = torch.jit.script(fun)\n self.assertIsNot(fun_compiled, fun_compiled_2)\n self.assertEqual(fun_compiled_2(), 7)\n\n # caching doesnt increase refcounts to function (holds weak reference)\n self.assertTrue(sys.getrefcount(fun), num_ref_counts)\n\n def test_string_ops(self):\n def foo():\n a = \"a\" + \"b\"\n return a + a, \"ab\" == \"b\", \"ab\" != \"b\", \"ab\" == \"ab\", \"ab\" != \"ab\"\n\n self.checkScript(foo, ())\n\n def test_string_sorted(self):\n def foo(strs: List[str]):\n return sorted(strs)\n\n FileCheck() \\\n .check(\"graph\") \\\n .check_next(\"str[] = aten::sorted\") \\\n .check_next(\"return\") \\\n .run(str(torch.jit.script(foo).graph))\n\n inputs = [\"str3\", \"str2\", \"str1\"]\n self.checkScript(foo, (inputs,))\n\n def test_string_sort(self):\n def foo(strs: List[str]):\n strs.sort()\n return strs\n\n inputs = [\"str3\", \"str2\", \"str1\"]\n self.checkScript(foo, (inputs,))\n\n def test_tuple_sorted(self):\n def foo(tups: List[Tuple[int, int]]):\n return sorted(tups)\n\n inputs = [(1, 2), (0, 2), (1, 3)]\n self.checkScript(foo, (inputs,))\n\n def test_tuple_sort(self):\n def foo(tups: List[Tuple[int, int]]):\n tups.sort()\n return tups\n\n inputs = [(1, 2), (0, 2), (1, 3)]\n self.checkScript(foo, (inputs,))\n\n def test_tuple_sort_reverse(self):\n def foo(tups: List[Tuple[int, int]]):\n tups.sort(reverse=True)\n return tups\n\n inputs = [(1, 2), (0, 2), (1, 3)]\n self.checkScript(foo, (inputs,))\n\n def test_tuple_unsortable_element_type(self):\n @torch.jit.script\n def foo():\n tups = [({1: 2}, {2: 3})]\n tups.sort()\n return tups\n\n with self.assertRaisesRegexWithHighlight(RuntimeError, \"are not sortable\", \"tups.sort\"):\n foo()\n\n def test_tuple_unsortable_diff_type(self):\n @torch.jit.script\n def foo(inputs: List[Any]):\n inputs.sort()\n return inputs\n\n inputs = [(1, 2), (\"foo\", \"bar\")]\n with self.assertRaisesRegexWithHighlight(RuntimeError, \"Only values of same type can be compared\", \"inputs.sort\"):\n foo(inputs)\n\n def test_tuple_nested_sort(self):\n def foo(inputs: List[Tuple[int, Tuple[int, str]]]):\n inputs.sort()\n return inputs\n\n inputs = [(1, (2, \"foo\")), (1, (2, \"bar\")), (1, (0, \"bar\"))]\n self.checkScript(foo, (inputs,))\n\n def test_tuple_unsortable_nested_diff_type(self):\n @torch.jit.script\n def foo(inputs: List[Any]):\n inputs.sort()\n return inputs\n\n inputs = [(1, (2, 3)), (2, (\"foo\", \"bar\"))]\n with self.assertRaisesRegexWithHighlight(RuntimeError, \"Only values of same type can be compared\", \"inputs.sort\"):\n foo(inputs)\n\n def test_string_new_line(self):\n with self.assertRaisesRegex(RuntimeError, \"expected a valid token*\"):\n torch.jit.CompilationUnit('''\n def test_while(a):\n print(\"\n a\")\n return a\n ''')\n\n def test_string_single_escape(self):\n with self.assertRaisesRegex(RuntimeError, \"expected a valid token*\"):\n torch.jit.CompilationUnit('''\n def test_while(a):\n print(\"\\\\\")\n return a\n ''')\n\n def test_script_annotation(self):\n @torch.jit.script\n def foo(a):\n return a + a + a\n s = Variable(torch.rand(2))\n self.assertEqual(s + s + s, foo(s))\n\n def test_torch_pow(self):\n def func(a, b):\n return pow(a, b)\n\n def func2(a, b, c, d):\n return pow(pow(c + a, b), d)\n\n def func3(a : int, b : float):\n # type: (int, float) -> float\n return pow(a, b)\n\n def func4():\n # type: () -> float\n return pow(2, -2)\n\n def func5(x, y):\n return pow(x.item(), y.item())\n\n def func6(a : int, b : int):\n # type: (int, int) -> float\n return pow(a, b)\n\n a = torch.rand(1)\n b = torch.rand(1)\n c = torch.rand(1)\n d = torch.rand(1)\n self.checkScript(func, (a, b))\n self.checkScript(func2, (a, b, c, d))\n self.checkScript(func3, (4, -0.5))\n self.checkScript(func4, ())\n self.checkScript(func6, (2, 4))\n\n inputs = [torch.tensor(2), torch.tensor(-2), torch.tensor(.5), torch.tensor(.2)]\n for x in inputs:\n for y in inputs:\n if x < 0:\n continue\n else:\n self.checkScript(func5, (x, y))\n\n @unittest.skipIf(not RUN_CUDA, \"device tests require CUDA\")\n def test_pow_scalar_backward_cuda(self):\n # see that scalar exponent works with cuda base (#19253)\n with enable_profiling_mode_for_profiling_tests():\n for dtype in [torch.float, torch.double]:\n @torch.jit.script\n def func(a, b):\n # type: (Tensor, float) -> Tensor\n return (a * 2) ** b\n\n a = torch.rand(1, requires_grad=True, device='cuda', dtype=dtype)\n func(a, 1, profile_and_replay=True).backward()\n\n @torch.jit.script\n def func(a, b):\n # type: (float, Tensor) -> Tensor\n return a ** (b * 2 + 1)\n\n a = torch.rand(1, requires_grad=True, device='cuda', dtype=dtype)\n func(2, a, profile_and_replay=True).backward()\n\n def _check_code(self, code_str, fn_name, inputs):\n scope = {}\n exec(code_str, globals(), scope)\n cu = torch.jit.CompilationUnit(code_str)\n self.assertEqual(cu.func(*inputs), scope[fn_name](*inputs))\n\n @unittest.skipIf(not RUN_CUDA, 'no CUDA')\n def test_scriptmodule_releases_tensors_cuda(self):\n with enable_profiling_mode_for_profiling_tests():\n @torch.jit.script\n def fn(x, y):\n return x.sigmoid() * y.tanh()\n\n def test(backward=False):\n x = torch.randn(3, 3, dtype=torch.double, device='cuda', requires_grad=True)\n y = torch.randn(3, 3, dtype=torch.double, device='cuda', requires_grad=True)\n out = fn(x, y, profile_and_replay=True)\n if backward:\n out.sum().backward()\n\n with self.assertLeaksNoCudaTensors():\n test()\n test()\n test()\n\n if GRAPH_EXECUTOR != ProfilingMode.SIMPLE:\n with self.assertLeaksNoCudaTensors():\n test(backward=True)\n test(backward=True)\n test(backward=True)\n\n def test_index(self):\n def consec(size, start=0):\n numel = torch.tensor(size).prod().item()\n return torch.arange(numel).view(size)\n\n def consec_list(size):\n return list(range(size))\n\n def random_string(size):\n letters = string.ascii_lowercase\n return \"\".join(random.choice(letters) for i in range(size))\n\n def check_indexing(indexing, tensor):\n template = dedent(\"\"\"\n def func(x):\n return x{}\n \"\"\")\n\n self._check_code(template.format(indexing), \"func\", [tensor])\n\n def check_dynamic_indexing(indexing, tensor, value1, value2):\n value1 = torch.tensor(value1)\n value2 = torch.tensor(value2)\n\n template = dedent(\"\"\"\n def func(x, value1, value2):\n i = int(value1)\n j = int(value2)\n return x{}\n \"\"\")\n\n self._check_code(template.format(indexing), \"func\", [tensor, value1, value2])\n\n # Torchscript assumes type Tensor by default, so we need this explicit\n # declaration.\n def check_indexing_list_int(indexing, list):\n template = dedent(\"\"\"\n def func(x):\n # type: (List[int]) -> Any\n return x{}\n \"\"\")\n\n self._check_code(template.format(indexing), \"func\", [list])\n\n def check_indexing_str(indexing, str):\n template = dedent(\"\"\"\n def func(x):\n # type: (str) -> Any\n return x{}\n \"\"\")\n\n self._check_code(template.format(indexing), \"func\", [str])\n\n # basic slices\n check_indexing('[0]', consec((3, 3)))\n check_indexing('[1]', consec((3, 3), 10))\n check_indexing('[2]', consec((3, 3), 19))\n check_indexing('[2]', consec((3,)))\n check_indexing('[-1]', consec((3, 3), 19))\n check_indexing('[0:2]', consec((3, 3, 3)))\n check_indexing('[1:-1]', consec((3, 3, 3)))\n check_indexing('[-3:-1]', consec((6, 3)))\n check_indexing('[1:]', consec((3, 3)))\n check_indexing('[:1]', consec((3, 3)))\n check_indexing('[:]', consec((3, 2)))\n\n # multi-dim: indexes\n check_indexing('[0, 1]', consec((3, 3)))\n check_indexing('[0, 1]', consec((3, 3, 2)))\n check_indexing('[1, 0, 2]', consec((3, 3, 3)))\n check_indexing('[2, -1]', consec((3, 3)))\n\n # multi-dim: mixed slicing and indexing\n check_indexing('[0, 1:2]', consec((3, 3)))\n check_indexing('[0, :1]', consec((3, 3, 2)))\n check_indexing('[1, 2:]', consec((3, 3, 3)))\n check_indexing('[-1, 1:, 0]', consec((3, 3, 3, 3)))\n check_indexing('[1:, -1, 0]', consec((3, 3, 3, 3)))\n check_indexing('[-1, 2:, 1:2]', consec((3, 3, 3, 3)))\n check_indexing('[-1, 1:, 0]', consec((3, 3, 3, 3)))\n check_indexing('[-1, :, 0, 2]', consec((3, 3, 3, 3)))\n\n # zero-sized slices\n check_indexing('[0:0]', consec((2, 2)))\n check_indexing('[0:0, 1]', consec((3, 3)))\n\n # trivial expression usage\n check_indexing('[1+1]', consec((3, 3)))\n check_indexing('[1:(0 + 2)]', consec((3, 3, 3)))\n\n # None for new dimensions\n check_indexing('[None, 0]', consec((3, 3)))\n check_indexing('[1, None]', consec((3, 3), 10))\n check_indexing('[None, None, 2]', consec((3, 3), 19))\n check_indexing('[None, 2, None]', consec((3,)))\n check_indexing('[0:2, None]', consec((3, 3, 3)))\n check_indexing('[None, 1:-1]', consec((3, 3, 3)))\n check_indexing('[None, -3:-1, None]', consec((6, 3)))\n check_indexing('[-1, None, 2:, None, 1:2]', consec((3, 3, 3, 3)))\n check_indexing('[None, -1, None, 2:, None, 1:2, None]', consec((3, 3, 3, 3)))\n\n # dynamic expression usage\n check_dynamic_indexing(\"[i + j]\", consec((3, 3)), 0, 1)\n check_dynamic_indexing(\"[i:j, i]\", consec((3, 3, 2)), 0, 2)\n\n # positive striding\n check_indexing_list_int('[0]', consec_list(6))\n check_indexing_list_int('[1]', consec_list(7))\n check_indexing_list_int('[2]', consec_list(8))\n check_indexing_list_int('[2]', consec_list(9))\n check_indexing_list_int('[-1]', consec_list(10))\n check_indexing_list_int('[0:2]', consec_list(11))\n check_indexing_list_int('[1:-1]', consec_list(12))\n check_indexing_list_int('[-3:-1]', consec_list(13))\n check_indexing_list_int('[1:]', consec_list(15))\n check_indexing_list_int('[:1]', consec_list(16))\n check_indexing_list_int('[:]', consec_list(17))\n check_indexing_list_int('[::]', consec_list(0))\n check_indexing_list_int('[1000::]', consec_list(0))\n check_indexing_list_int('[:1000:]', consec_list(0))\n\n # negative striding\n check_indexing_list_int('[::-1]', consec_list(7))\n check_indexing_list_int('[:3:-1]', consec_list(7))\n check_indexing_list_int('[3::-1]', consec_list(7))\n check_indexing_list_int('[1000::-1]', consec_list(7))\n check_indexing_list_int('[3:0:-1]', consec_list(7))\n check_indexing_list_int('[3:-1000:-1]', consec_list(7))\n check_indexing_list_int('[0:0:-1]', consec_list(7))\n check_indexing_list_int('[0:-1000:-1]', consec_list(7))\n\n # only step is specified\n check_indexing_list_int('[::-1]', consec_list(0))\n check_indexing_list_int('[::-1]', consec_list(7))\n check_indexing_list_int('[::-2]', consec_list(7))\n check_indexing_list_int('[::2]', consec_list(7))\n check_indexing_list_int('[::42]', consec_list(7))\n check_indexing_list_int('[::-42]', consec_list(7))\n check_indexing_list_int('[::42]', consec_list(0))\n check_indexing_list_int('[::-42]', consec_list(0))\n check_indexing_list_int('[::9223372036854775807]', consec_list(42))\n check_indexing_list_int('[::-9223372036854775807]', consec_list(42))\n with self.assertRaisesRegex(RuntimeError, \"out of bounds\"):\n check_indexing_list_int('[::-9223372036854775808]', consec_list(42))\n with self.assertRaisesRegex(RuntimeError, \"should have non-zero step\"):\n check_indexing_list_int('[::0]', consec_list(42))\n\n # striding strings\n check_indexing_str('[0]', random_string(6))\n check_indexing_str('[1]', random_string(7))\n check_indexing_str('[2]', random_string(8))\n check_indexing_str('[2]', random_string(9))\n check_indexing_str('[-1]', random_string(10))\n check_indexing_str('[0:2]', random_string(11))\n check_indexing_str('[1:-1]', random_string(12))\n check_indexing_str('[-3:-1]', random_string(13))\n check_indexing_str('[1:]', random_string(15))\n check_indexing_str('[:1]', random_string(16))\n check_indexing_str('[:]', random_string(17))\n check_indexing_str('[::]', random_string(0))\n check_indexing_str('[1000::]', random_string(0))\n check_indexing_str('[:1000:]', random_string(0))\n\n check_indexing_str('[::-1]', random_string(7))\n check_indexing_str('[:3:-1]', random_string(7))\n check_indexing_str('[3::-1]', random_string(7))\n check_indexing_str('[1000::-1]', random_string(7))\n check_indexing_str('[3:0:-1]', random_string(7))\n check_indexing_str('[3:-1000:-1]', random_string(7))\n check_indexing_str('[0:0:-1]', random_string(7))\n check_indexing_str('[0:-1000:-1]', random_string(7))\n\n check_indexing_str('[::-1]', random_string(0))\n check_indexing_str('[::-1]', random_string(7))\n check_indexing_str('[::-2]', random_string(7))\n check_indexing_str('[::2]', random_string(7))\n check_indexing_str('[::42]', random_string(7))\n check_indexing_str('[::-42]', random_string(7))\n check_indexing_str('[::42]', random_string(0))\n check_indexing_str('[::-42]', random_string(0))\n check_indexing_str('[::9223372036854775807]', random_string(42))\n check_indexing_str('[::-9223372036854775807]', random_string(42))\n with self.assertRaisesRegex(RuntimeError, \"out of bounds\"):\n check_indexing_str('[::-9223372036854775808]', random_string(42))\n with self.assertRaisesRegex(RuntimeError, \"should have non-zero step\"):\n check_indexing_str('[::0]', random_string(42))\n\n def test_module_copy_with_attributes(self):\n class Vocabulary(torch.jit.ScriptModule):\n def __init__(self, vocab_list):\n super(Vocabulary, self).__init__()\n self._vocab = torch.jit.Attribute(vocab_list, List[str])\n self.some_idx = torch.jit.Attribute(2, int)\n self.idx = torch.jit.Attribute(\n {word: i for i, word in enumerate(vocab_list)}, Dict[str, int]\n )\n\n @torch.jit.script_method\n def lookup_indices_1d(self, values):\n # type: (List[str]) -> List[int]\n result = torch.jit.annotate(List[int], [])\n # Direct list iteration not supported\n for i in range(len(values)):\n value = values[i]\n result.append(self.idx.get(value, self.some_idx))\n return result\n\n @torch.jit.script_method\n def forward(self, values):\n # type: (List[List[str]]) -> List[List[int]]\n result = torch.jit.annotate(List[List[int]], [])\n # Direct list iteration not supported\n for i in range(len(values)):\n result.append(self.lookup_indices_1d(values[i]))\n return result\n\n v = Vocabulary(list('uabcdefg'))\n v.__copy__()\n\n def test_tuple_to_opt_list(self):\n @torch.jit.script\n def foo(x):\n # type: (Optional[List[int]]) -> int\n return 1\n\n @torch.jit.script\n def tuple_call():\n return foo((1, 2))\n\n def test_keyword(self):\n @torch.jit.script\n def func(x):\n return torch.sum(x, dim=0)\n\n x = torch.rand(10, dtype=torch.float, requires_grad=True)\n y = func(x)\n y2 = torch.sum(x, dim=0)\n self.assertEqual(y, y2)\n\n def test_constant_pooling_none(self):\n @torch.jit.script\n def typed_nones(a=None, b=None, c=None):\n # type: (Optional[int], Optional[bool], Optional[Tensor]) -> Tuple[Optional[int], Optional[bool], Optional[Tensor]]\n return a, b, c\n\n @torch.jit.script\n def test(a):\n # type: (bool) -> None\n if a:\n print(typed_nones())\n else:\n print(typed_nones())\n\n graph_str = str(test.graph)\n self.assertTrue(graph_str.count(\"NoneType = prim::Constant\") == 1)\n\n def test_constant_pooling_same_identity(self):\n def foo():\n a = torch.tensor([4])\n b = (a,)\n index = len(a) - 1\n c = b[index]\n d = b[index]\n return c, d\n\n foo_script = torch.jit.script(foo)\n self.run_pass('constant_propagation', foo_script.graph)\n self.run_pass('constant_pooling', foo_script.graph)\n # even though the c & d escape scope, we are still able\n # pool them into one constant because they are the same object\n FileCheck().check_count(\"prim::Constant\", 1, exactly=True).run(foo_script.graph)\n self.assertEqual(foo(), foo_script())\n\n def test_constant_pooling_introduce_aliasing(self):\n @torch.jit.script\n def foo():\n a = torch.tensor(1)\n b = torch.tensor(1)\n return a, b\n\n self.run_pass('constant_propagation', foo.graph)\n self.run_pass('constant_pooling', foo.graph)\n # dont pool constants bc it would introduce observable alias relationship changing\n a, b = foo()\n self.assertIsNot(a, b)\n\n def test_literal(self):\n def func1(a, b):\n c = a, b\n d, e = c\n return d + e\n\n def func2(a, b):\n c = a, (a, b)\n d, e = c\n f, g = e\n return d + f + g\n\n def func3(a, b):\n # type: (float, float) -> float\n c = 0., (0., 0.)\n x = True\n while x:\n x = False\n c = a, (a, b)\n d, e = c\n f, g = e\n return d + f + g\n\n a = torch.rand(1, requires_grad=True)\n b = torch.rand(1, requires_grad=True)\n self.checkScript(func1, (a, b), optimize=True)\n self.checkScript(func2, (a, b), optimize=True)\n self.checkScript(func3, (a.item(), b.item()), optimize=True)\n\n def test_expand(self):\n @torch.jit.script\n def func(x, y):\n return x + y\n\n x = torch.rand(2, 3, dtype=torch.float, requires_grad=True)\n y = torch.rand(3, dtype=torch.float, requires_grad=True)\n out = func(x, y)\n self.assertEqual(func(x, y), x + y)\n\n grad = torch.randn(2, 3, dtype=torch.float)\n out.backward(grad)\n self.assertEqual(x.grad, grad)\n self.assertEqual(y.grad, grad.sum(dim=0))\n\n def test_sum(self):\n @torch.jit.script\n def func(x):\n return x.sum(dim=[4])\n\n @torch.jit.script\n def func2(x):\n return x.sum(dim=4)\n\n # test that shape analysis is written correctly for sum with IntArrayRef[1] dim argument\n self.run_pass('constant_propagation', func.graph)\n self.run_pass('constant_propagation', func2.graph)\n g = _propagate_shapes(func.graph, (torch.zeros(1, 1, 1, 1, 4),), False)\n g2 = _propagate_shapes(func2.graph, (torch.zeros(1, 1, 1, 1, 4),), False)\n\n def test_cat(self):\n with enable_profiling_mode_for_profiling_tests():\n @torch.jit.script\n def func(x):\n return torch.cat((x, x), dim=0)\n\n x = torch.rand(10, dtype=torch.float, requires_grad=True)\n self.assertEqual(func(x, profile_and_replay=True), torch.cat((x, x), dim=0))\n\n @torch.jit.script\n def func2(x, y):\n return torch.cat((x, x), y)\n\n with disable_autodiff_subgraph_inlining():\n x = torch.rand([2, 2]).requires_grad_()\n y = torch.tensor(1)\n\n output = func2(x, y, profile_and_replay=True)\n output_ref = torch.cat((x, x), y)\n self.assertEqual(output, output_ref)\n\n if GRAPH_EXECUTOR != ProfilingMode.SIMPLE:\n self.assertAutodiffNode(func2.graph_for(x, y), True, ['aten::cat'], [])\n\n grad = torch.autograd.grad(output.sum(), x)\n grad_ref = torch.autograd.grad(output_ref.sum(), x)\n self.assertEqual(grad, grad_ref)\n\n def test_cat_lifts(self):\n @torch.jit.script\n def foo(x):\n return torch.cat([x, x], dim=1)\n\n @torch.jit.script\n def foo2(x):\n return torch.cat([], dim=1)\n\n @torch.jit.script\n def foo3(x):\n return torch.cat([x], dim=1)\n\n for g in [foo.graph, foo2.graph, foo3.graph]:\n FileCheck().check(\"int =\").check(\"ListConstruct\").check(\"aten::cat\").run(str(g))\n\n def test_stack(self):\n with enable_profiling_mode_for_profiling_tests():\n @torch.jit.script\n def func(x):\n return torch.stack((x, x), dim=1)\n x = torch.rand(10, 10)\n self.assertEqual(func(x, profile_and_replay=True), torch.stack((x, x), dim=1))\n\n @torch.jit.script\n def func2(x, y):\n return torch.stack((x, y), dim=0)\n\n with disable_autodiff_subgraph_inlining():\n x = torch.randn([2, 2]).requires_grad_()\n y = torch.randn([2, 2]).requires_grad_()\n\n output = func2(x, y, profile_and_replay=True)\n output_ref = torch.stack((x, y), 0)\n self.assertEqual(output, output_ref)\n if GRAPH_EXECUTOR != ProfilingMode.SIMPLE:\n self.assertAutodiffNode(func2.graph_for(x, y), True, ['aten::stack'], [])\n\n grads = torch.autograd.grad(output.sum(), (x, y))\n grads_ref = torch.autograd.grad(output_ref.sum(), (x, y))\n self.assertEqual(grads, grads_ref)\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY,\n \"Profiling executor will be using different heuristics for constructing differentiable graphs\")\n def test_unbind(self):\n with enable_profiling_mode_for_profiling_tests():\n @torch.jit.script\n def func(x, y):\n # type: (Tensor, int) -> List[Tensor]\n return torch.unbind(x, y)\n\n with disable_autodiff_subgraph_inlining():\n x = torch.rand([2, 2]).requires_grad_()\n y = 0\n outputs = func(x, y, profile_and_replay=True)\n outputs_ref = torch.unbind(x, dim=y)\n self.assertEqual(outputs, outputs_ref)\n self.assertAutodiffNode(func.graph_for(x, y), True, ['aten::unbind'], [])\n\n grad = torch.autograd.grad(_sum_of_list(outputs), x)\n grad_ref = torch.autograd.grad(_sum_of_list(outputs_ref), x)\n self.assertEqual(grad, grad_ref)\n\n\n @unittest.skipIf(GRAPH_EXECUTOR == ProfilingMode.PROFILING,\n \"Profiling executor fails to recognize that tensors in a list require gradients\")\n def test_meshgrid(self):\n with enable_profiling_mode_for_profiling_tests():\n @torch.jit.script\n def func(a):\n # type: (List[Tensor]) -> List[Tensor]\n return torch.meshgrid(a)\n with disable_autodiff_subgraph_inlining():\n a = torch.tensor([1.0, 2, 3]).requires_grad_()\n b = torch.tensor([1.0, 2, 3, 4]).requires_grad_()\n inputs = [a, b]\n\n outputs_ref = torch.meshgrid(inputs)\n outputs = func(inputs, profile_and_replay=True)\n self.assertEqual(outputs, outputs_ref)\n\n if GRAPH_EXECUTOR != ProfilingMode.SIMPLE:\n self.assertAutodiffNode(func.graph_for(inputs), True, ['aten::meshgrid'], [])\n\n grads = torch.autograd.grad(_sum_of_list(outputs), inputs)\n grads_ref = torch.autograd.grad(_sum_of_list(outputs_ref), inputs)\n self.assertEqual(grads, grads_ref)\n\n def test_tensor_len(self):\n def func(x):\n return len(x)\n\n self.checkScript(func, [torch.ones(4, 5, 6)])\n\n def test_func_call(self):\n def add(a, b):\n return a + b\n\n def mul(a, x):\n return a * x\n\n def func(alpha, beta, x, y):\n return add(mul(alpha, x), mul(beta, y))\n\n alpha = torch.rand(1, dtype=torch.float, requires_grad=True)\n beta = torch.rand(1, dtype=torch.float, requires_grad=True)\n x = torch.rand(3, dtype=torch.float, requires_grad=True)\n y = torch.rand(3, dtype=torch.float, requires_grad=True)\n\n # NOTE: cannot optimize yet because broadcasts are not inserted before the fuser runs\n self.checkScript(func, [alpha, beta, x, y], optimize=False)\n\n @unittest.skip(\"bailouts are being deprecated\")\n def test_profiling_graph_executor(self):\n @torch.jit.script\n def def_in_one_branch(x, z):\n # type: (Tensor, bool) -> float\n y = x\n if z is False:\n y = x + 1\n\n return y.sum()\n\n a = torch.rand(2, 3)\n\n with enable_profiling_mode_for_profiling_tests():\n # check prim::profile are inserted\n profiled_graph_str = str(def_in_one_branch.graph_for(a, True))\n FileCheck().check_count(\"prim::profile\", 4).run(profiled_graph_str)\n # this call is optimized for\n # the given shape of (2, 3)\n def_in_one_branch(a, False)\n # change shape to (3)\n # so we go down a bailout path\n a = torch.ones(3)\n # check prim::BailOuts are inserted\n bailout_graph_str = str(def_in_one_branch.graph_for(a, True))\n FileCheck().check_count(\"prim::BailOut\", 3).run(bailout_graph_str)\n # this triggers all 3 bailouts\n self.assertEqual(def_in_one_branch(a, False), 6.0)\n # this triggers 2 bailouts\n self.assertEqual(def_in_one_branch(a, True), 3.0)\n\n @unittest.skip(\"bailouts are being deprecated\")\n def test_maxpool_guard_elimination(self):\n @torch.jit.script\n def my_maxpool(x):\n return F.max_pool1d(x, kernel_size=[1]) + torch.ones([32, 32, 32])\n\n a = torch.rand(32, 32, 32)\n\n with enable_profiling_mode_for_profiling_tests():\n my_maxpool(a)\n bailout_graph_str = str(my_maxpool.graph_for(a))\n FileCheck().check_count(\"prim::BailOut\", 1).run(bailout_graph_str)\n\n @unittest.skip(\"bailouts are being deprecated\")\n def test_slice_guard_elimination(self):\n @torch.jit.script\n def my_slice(x):\n return x[0:16:2] + x[0:16:2]\n\n a = torch.rand(32, 4)\n\n with enable_profiling_mode_for_profiling_tests():\n my_slice(a)\n bailout_graph_str = str(my_slice.graph_for(a))\n FileCheck().check_count(\"prim::BailOut\", 1).run(bailout_graph_str)\n\n @unittest.skip(\"bailouts are being deprecated\")\n def test_unsqueeze_guard_elimination(self):\n @torch.jit.script\n def my_unsqueeze(x):\n return torch.unsqueeze(x, 0) + torch.unsqueeze(x, 0)\n\n a = torch.rand(32, 4)\n\n with enable_profiling_mode_for_profiling_tests():\n my_unsqueeze(a)\n bailout_graph_str = str(my_unsqueeze.graph_for(a))\n FileCheck().check_count(\"prim::BailOut\", 2).run(bailout_graph_str)\n\n def test_resize_input_ops(self):\n # resize_ and resize_as resize the input tensor. because our shape analysis\n # is flow invariant, we set any Tensor that can alias a resized Tensor\n # to the base Tensor Type, without size information.\n\n # testing that value which is an input of a graph gets handled\n def out_op_graph_input():\n @torch.jit.script\n def test(x, y, z):\n torch.mul(x, y, out=z)\n return z\n\n graph = _propagate_shapes(test.graph,\n (torch.zeros(2, 1), torch.zeros(1, 2), torch.zeros(1, 1, 1)), False)\n self.assertTrue(next(graph.outputs()).type() == TensorType.get())\n out_op_graph_input()\n\n def test_resize():\n @torch.jit.script\n def test(x):\n after_resize_alias = torch.zeros([2])\n for _i in range(5):\n b = x + 1\n f = [1]\n before_resize_alias = b.sub_(1)\n # for i in range(10):\n f.append(1)\n b.resize_(f)\n after_resize_alias = b.add_(1)\n return after_resize_alias\n\n self.run_pass('constant_propagation', test.graph)\n g = _propagate_shapes(test.graph, (torch.zeros(1, 1),), False)\n resize_node = g.findNode(\"aten::resize_\")\n # first input and output of b.resize_ is b\n self.assertTrue(next(resize_node.inputs()).type() == TensorType.get())\n self.assertTrue(next(resize_node.outputs()).type() == TensorType.get())\n\n # correctly propagates to b alias set\n before_resize = g.findNode(\"aten::sub_\")\n self.assertTrue(next(before_resize.outputs()).type() == TensorType.get())\n\n after_resize = g.findNode(\"aten::add_\")\n self.assertTrue(next(after_resize.outputs()).type() == TensorType.get())\n\n test_resize()\n\n def test_resize_as():\n @torch.jit.script\n def test(x):\n b = torch.zeros([2, 2])\n b.resize_as_(x)\n return b\n\n g = test.graph\n self.run_pass('constant_propagation', g)\n g = _propagate_shapes(test.graph, (torch.zeros(1, 1),), False)\n\n # x doesn't alias a resized op so it shouldn't be set to base Tensor type\n self.assertTrue(next(g.inputs()).type() != TensorType.get())\n # return is resized\n self.assertTrue(next(g.outputs()).type() == TensorType.get())\n\n test_resize_as()\n\n def test_uninitialized(self):\n graph_str = \"\"\"graph():\n %1 : int = prim::Uninitialized()\n %2 : int = prim::Constant[value=1]()\n %3 : int = aten::add(%1, %2)\n return (%3)\n \"\"\"\n g = parse_ir(graph_str)\n m = self.createFunctionFromGraph(g)\n self.getExportImportCopy(m)\n with self.assertRaisesRegex(RuntimeError, \"isInt\"):\n m()\n\n\n @unittest.skipIf(GRAPH_EXECUTOR == ProfilingMode.SIMPLE, \"Simple Executor doesn't use requires_grad information\")\n @unittest.skipIf(GRAPH_EXECUTOR == ProfilingMode.PROFILING, \"Peeling is now disabled\")\n def test_requires_grad_loop(self):\n @torch.jit.script\n def test(x, y, z):\n # type: (Tensor, Tensor, int) -> Tensor\n for _ in range(z):\n x = y\n return x\n\n # x requires grad, y does not\n # testing that requires grad analysis correctly exits, with its input\n # to the loop (x) requiring grad and its output to the loop not requiring grad\n # and the output of the node conservatively setting grad to true\n\n inps = (torch.tensor(1.0, requires_grad=True), torch.tensor(1), 10)\n test(*inps, profile_and_replay=True)\n\n graph = test.graph_for(*inps)\n loop = graph.findNode(\"prim::Loop\")\n loop_body = next(loop.blocks())\n loop_inputs = list(loop_body.inputs())\n loop_outputs = list(loop_body.outputs())\n\n if GRAPH_EXECUTOR == ProfilingMode.PROFILING:\n # TODO: simplify this test as it's very sensitive\n # the optimized graph will have 3 loops\n # the original loop is peeled\n # peeled loop also gets unrolled\n index_of_x_in_peeled_unrolled_loop = -2\n self.assertTrue(loop_inputs[index_of_x_in_peeled_unrolled_loop].requires_grad())\n bailouts_in_outer_block = graph.findAllNodes(\"prim::BailOut\", False)\n last_bailout_index_on_loops_output = -1\n self.assertFalse(bailouts_in_outer_block[last_bailout_index_on_loops_output].output().requires_grad())\n else:\n self.assertTrue(loop_inputs[1].requires_grad())\n self.assertTrue(loop.output().requires_grad())\n self.assertFalse(loop_outputs[1].requires_grad())\n\n def test_view_shape_prop(self):\n cu = torch.jit.CompilationUnit('''\n def test_view_shape_prop(a):\n return a.view(size=[-1])\n ''')\n inputs = [torch.zeros(10, 10)]\n outputs = torch.zeros(100)\n\n real_outs = cu.test_view_shape_prop(*inputs)\n self.assertEqual(real_outs, outputs)\n\n def test_view_listconstruct_shape_prop(self):\n def fn(x):\n B = x.size(0)\n C = x.size(1)\n T = x.size(2)\n return x.view(T, B, C)\n\n x = torch.randn(3, 1, 5, requires_grad=True)\n fn = torch.jit.script(fn)\n graph = _propagate_shapes(fn.graph, (x,), False)\n self.assertTrue(next(graph.outputs()).type().scalarType() == 'Double')\n\n def test_shape_prop_promotion(self):\n @torch.jit.script\n def fn(x, y):\n return x + y\n\n x, y = torch.rand(3, 4, dtype=torch.float), torch.rand(3, 4, dtype=torch.double)\n graph = _propagate_shapes(fn.graph, (x, y), False)\n FileCheck().check('Double(*, *, device=cpu) = aten::add').run(graph)\n\n def test_shape_prop_promote_scalar_arg(self):\n @torch.jit.script\n def fn(x):\n return math.pi + x\n\n x = torch.zeros(3, 4, dtype=torch.long)\n graph = _propagate_shapes(fn.graph, (x,), False)\n default = torch.get_default_dtype()\n if(default == torch.float):\n FileCheck().check('Float(*, *, requires_grad=0, device=cpu) = aten::add').run(graph)\n else:\n FileCheck().check('Double(*, *, requires_grad=0, device=cpu) = aten::add').run(graph)\n\n def test_integral_shape_inference(self):\n cu = torch.jit.CompilationUnit('''\n def test_integral_shape_inference(a):\n return a * a\n ''')\n inputs = [torch.ones(10, 10, dtype=torch.long)]\n outputs = torch.ones(10, 10)\n\n # TODO(#38095): Replace assertEqualIgnoreType. See issue #38095\n self.assertEqualIgnoreType(cu.test_integral_shape_inference(*inputs), outputs)\n\n @unittest.skipIf(RUN_CUDA, 'This tests the CPU fuser')\n @unittest.skipIf(IS_SANDCASTLE, \"NYI: fuser support for Sandcastle\")\n @enable_cpu_fuser\n def test_batchnorm_fuser_cpu(self):\n code = '''\n graph(%3 : Tensor,\n %7 : Tensor,\n %12 : Float(*, *),\n %13 : Tensor,\n %25 : Tensor):\n %23 : int = prim::Constant[value=1]()\n %22 : float = prim::Constant[value=1e-05]()\n %26 : Tensor = aten::sqrt(%25)\n %24 : Tensor = aten::add(%26, %22, %23)\n %20 : Tensor = aten::reciprocal(%24)\n %norm_invstd : Tensor = aten::mul(%20, %23)\n %15 : Tensor = aten::sub(%12, %13, %23)\n %11 : Tensor = aten::mul(%15, %norm_invstd)\n %8 : Tensor = aten::mul(%11, %7)\n %5 : Tensor = aten::add(%8, %3, %23)\n %1 : Float(*, *) = aten::relu(%5)\n return (%1)\n '''\n\n graph = parse_ir(code)\n inputs = 5 * [torch.rand(26, 2048, dtype=torch.float)]\n code = torch._C._jit_fuser_get_fused_kernel_code(graph, inputs)\n FileCheck().check('sqrtf').run(code)\n\n @slowTest\n @unittest.skipIf(RUN_CUDA, 'This tests the CPU fuser')\n @unittest.skipIf(IS_SANDCASTLE, \"NYI: fuser support for Sandcastle\")\n @enable_cpu_fuser\n def test_fuser_double_float_codegen(self):\n fns = ['log', 'log10', 'log1p', 'log2', 'lgamma', 'exp', 'expm1', 'erf',\n 'erfc', 'cos', 'acos', 'cosh', 'sin', 'asin', 'sinh', 'tan',\n 'atan', 'tanh', 'sqrt', 'ceil', 'floor', 'round', 'trunc',\n 'frac']\n\n def lookup_c_equivalent_fn(aten_fn):\n if aten_fn == 'min':\n return 'fmin'\n elif aten_fn == 'max':\n return 'fmax'\n else:\n return aten_fn\n\n def test_dispatch(op, expects, dtype, binary=False):\n if dtype == torch.double:\n dtype_str = 'Double'\n elif dtype == torch.float:\n dtype_str = 'Float'\n else:\n raise RuntimeError('Unknown dtype')\n\n if binary:\n code = '''\n graph(%3 : Tensor, %4 : Tensor):\n %2 : {dtype}(*, *) = aten::{op}(%3, %4)\n %1 : {dtype}(*, *) = aten::relu(%2)\n return (%1)\n '''.format(op=op, dtype=dtype_str)\n else:\n code = '''\n graph(%3 : Tensor):\n %2 : {dtype}(*, *) = aten::{op}(%3)\n %1 : {dtype}(*, *) = aten::relu(%2)\n return (%1)\n '''.format(op=op, dtype=dtype_str)\n\n graph = parse_ir(code)\n inputs = (2 if binary else 1) * [torch.rand(26, 2048, dtype=dtype)]\n code = torch._C._jit_fuser_get_fused_kernel_code(graph, inputs)\n FileCheck().check(expects).run(code)\n\n for fn in fns:\n test_dispatch(fn, lookup_c_equivalent_fn(fn) + '(', torch.double)\n test_dispatch(fn, lookup_c_equivalent_fn(fn) + 'f(', torch.float)\n\n binary_fns = ['min', 'max', 'pow']\n for fn in binary_fns:\n test_dispatch(fn, lookup_c_equivalent_fn(fn) + '(', torch.double, binary=True)\n test_dispatch(fn, lookup_c_equivalent_fn(fn) + 'f(', torch.float, binary=True)\n\n @unittest.skipIf(RUN_CUDA, 'This tests the CPU fuser')\n @unittest.skipIf(IS_SANDCASTLE, \"NYI: fuser support for Sandcastle\")\n @enable_cpu_fuser\n def test_fuser_double_literal_precision(self):\n code = '''\n graph(%2 : Float(*, *)):\n %4 : int = prim::Constant[value=1]()\n %3 : float = prim::Constant[value=1.282549830161864]()\n %5 : Float(*, *) = aten::add(%2, %3, %4)\n %1 : Float(*, *) = aten::relu(%5)\n return (%1)\n '''\n\n graph = parse_ir(code)\n code = torch._C._jit_fuser_get_fused_kernel_code(graph, [torch.rand(3, 4)])\n FileCheck().check('1.282549830161864').run(code)\n\n def test_fuser_multiple_blocks(self):\n cu = torch.jit.CompilationUnit('''\n def test_fuser_multiple_blocks(this, that, theother, meme):\n i = 0\n while i < 20:\n this = torch.cat([this, meme], dim=0)\n that = torch.cat([that, meme], dim=0)\n theother = torch.cat([theother, meme], dim=0)\n i = i + 1\n return this, that, theother\n ''')\n\n inputs = [torch.ones(0, 10, 10)] * 3\n inputs += [torch.ones(1, 10, 10)]\n outputs = [torch.ones(20, 10, 10)] * 3\n\n self.assertEqual(cu.test_fuser_multiple_blocks(*inputs), outputs)\n\n def test_dropout_script(self):\n\n eg = torch.zeros(1, 2, 3, requires_grad=True)\n\n @_trace(eg)\n def foo(x):\n x = torch.neg(x)\n return F.dropout(x)\n\n class MyDrop(nn.Module):\n def forward(self, x):\n return foo(x)\n\n f = io.BytesIO()\n with warnings.catch_warnings(record=True):\n torch.onnx.export(MyDrop(), (eg,), f, verbose=False)\n\n @unittest.skip(\"RuntimeError: VariableType::ID() not implemented\")\n def test_cast(self):\n script = '''\n def to_int(x):\n return int(x)\n '''\n x = Variable(torch.FloatTensor([1.1, 2.3]), requires_grad=True)\n out = Variable(torch.IntTensor([1, 2]), requires_grad=True)\n self.checkScript(script, [x], optimize=True, outputs=[out], func='to_int')\n\n def test_str_cast(self):\n @torch.jit.script\n def to_str(x):\n # type: (int) -> str\n return str((x, x))\n\n self.assertEqual(\"(1, 1)\", to_str(1))\n\n def test_int_cast(self):\n @torch.jit.script\n def to_int(x):\n # type: (str) -> int\n return int(x)\n\n self.assertEqual(5, to_int('5'))\n self.assertEqual(-5, to_int('-5'))\n self.assertEqual(2147483647, to_int('2147483647'))\n self.assertEqual(-2147483648, to_int('-2147483648'))\n\n with self.assertRaisesRegex(RuntimeError, \"invalid literal for int()\"):\n to_int('0x20')\n\n with self.assertRaisesRegex(RuntimeError, \"invalid literal for int()\"):\n to_int('0b0001')\n\n def test_python_frontend(self):\n def fn(x, y, z):\n q = None\n q = x + y - z.sigmoid()\n print(q)\n w = -z\n if not x and not y and z:\n m = x if not z else y\n while x < y > z:\n q = x\n assert 1 == 1, \"hello\"\n return x\n\n ast = torch.jit.frontend.get_jit_def(fn, fn.__name__)\n self.assertExpected(str(ast))\n\n def test_python_frontend_source_range(self):\n def fn():\n raise Exception(\"hello\")\n ast = torch.jit.frontend.get_jit_def(fn, fn.__name__)\n FileCheck().check(\"SourceRange at:\") \\\n .check(\"def fn():\") \\\n .check(\"~~~~~~~~~\") \\\n .check('raise Exception(\"hello\")') \\\n .check('~~~~~~~~~~~~~~~~~ <--- HERE') \\\n .run(str(ast.range()))\n\n def test_python_frontend_py3(self):\n def fn():\n raise Exception(\"hello\")\n ast = torch.jit.frontend.get_jit_def(fn, fn.__name__)\n self.assertExpected(str(ast))\n\n def _make_scalar_vars(self, arr, dtype):\n return [torch.tensor(val, dtype=dtype) for val in arr]\n\n\n def test_string_print(self):\n def func(a):\n print(a, \"a\" 'b' '''c''' \"\"\"d\"\"\", 2, 1.5)\n return a\n\n inputs = self._make_scalar_vars([1], torch.int64)\n self.checkScript(func, inputs, capture_output=True)\n\n def test_while(self):\n def func(a, b, max):\n while bool(a < max):\n a = a + 1\n b = b + 1\n c = a + b\n return c\n\n inputs = self._make_scalar_vars([1, 1, 10], torch.int64)\n self.checkScript(func, inputs, optimize=True)\n\n def test_fibb(self):\n def func(lim):\n first = 1\n second = 1\n i = 1\n somenum = 5\n dontmutateme = 3\n third = 0\n while bool(i < lim):\n third = first + second\n first = second\n second = third\n j = 0\n while j < 10:\n somenum = somenum * 2\n j = j + 1\n i = i + j\n i = i + dontmutateme\n\n st = second + third\n fs = first + second\n return third, st, fs\n\n inputs = self._make_scalar_vars([10], torch.int64)\n self.checkScript(func, inputs, optimize=True)\n\n def test_fibb_totally_better(self):\n def fib(x):\n # type: (int) -> int\n prev = 1\n v = 1\n for i in range(0, x):\n save = v\n v = v + prev\n prev = save\n return v\n\n self.checkScript(fib, (10,))\n\n def test_if(self):\n def func(a, b):\n # type: (int, int) -> int\n d = 3\n if bool(a > 10):\n a = 3 + d\n else:\n b = 3 + d\n d = 4\n c = a + b\n return c\n\n inputs = self._make_scalar_vars([1, -1], torch.int64)\n self.checkScript(func, inputs, optimize=True)\n\n def test_if_for_in_range(self):\n def func(a, b):\n # type: (int, int) -> int\n d = 3\n for _ in range(20):\n if bool(a > 10):\n a = 3 + d\n else:\n b = 3 + d\n d = 4\n c = a + b\n return d\n inputs = self._make_scalar_vars([1, -1], torch.int64)\n self.checkScript(func, inputs, optimize=True)\n\n def test_if_noelse(self):\n def func(a, b):\n if bool(a > 10):\n a = 3 + b\n c = a + b\n return c\n\n inputs = self._make_scalar_vars([-1, 1], torch.int64)\n self.checkScript(func, inputs, optimize=True)\n\n def test_if_is_none_dispatch(self):\n\n @torch.jit.script\n def test_lhs_none_rhs_none():\n # LHS, RHS both alwaysNone, dispatch always_none_branch\n # only emit one prim::Constant\n if None is None:\n return 1\n elif None is not None:\n return 2\n else:\n return 3\n\n self.assertTrue(str(test_lhs_none_rhs_none.graph).count(': int = prim::Constant') == 1)\n\n @torch.jit.script\n def test_lhs_opt_rhs_none(lhs=None):\n # type: (Optional[Tensor]) -> int\n # LHS maybeNone: emit normal if stmt that contains 3 constants\n if lhs is not None:\n return 2\n elif lhs is None:\n return 1\n else:\n return 3\n\n self.assertTrue(str(test_lhs_opt_rhs_none.graph).count(': int = prim::Constant') == 3)\n\n @torch.jit.script\n def test_lhs_none_rhs_opt(rhs=None):\n # type: (Optional[Tensor]) -> int\n # RHS maybeNone, emit normal if stmt that contains 3 constants\n if None is rhs:\n return 1\n elif None is not rhs:\n return 2\n else:\n return 3\n\n self.assertTrue(str(test_lhs_opt_rhs_none.graph).count(': int = prim::Constant') == 3)\n\n @torch.jit.script\n def test_lhs_never_rhs_none(lhs):\n # LHS neverNone, RHS alwaysNone dispatch never_none_branch\n # only emit one prim::Constant\n if lhs is None:\n return 1\n elif lhs is not None:\n return 2\n else:\n return 3\n\n self.assertTrue(str(test_lhs_never_rhs_none.graph).count(': int = prim::Constant') == 1)\n\n @torch.jit.script\n def test_lhs_none_rhs_never(rhs):\n # LHS alwaysNone, RHS neverNone dispatch never_none_branch\n # only emit one prim::Constant\n if None is rhs:\n return 1\n elif None is not rhs:\n return 2\n else:\n return 3\n\n self.assertTrue(str(test_lhs_none_rhs_never.graph).count(': int = prim::Constant') == 1)\n\n @torch.jit.script\n def test_bool_arith_and(lhs):\n if lhs is None and lhs is not None:\n return 1\n else:\n return 2\n self.assertEqual(test_bool_arith_and(torch.zeros(3)), 2)\n self.assertTrue(str(test_bool_arith_and.graph).count('if') == 0)\n\n @torch.jit.script\n def test_bool_arith_or(lhs):\n if lhs is None or lhs is not None:\n return 1\n else:\n return 2\n self.assertEqual(test_bool_arith_or(torch.zeros(3)), 1)\n self.assertTrue(str(test_bool_arith_or.graph).count('if') == 0)\n\n\n @torch.jit.script\n def test_bool_arith_not(lhs):\n if not (lhs is None):\n return 1\n else:\n return 2\n self.assertEqual(test_bool_arith_not(torch.zeros(3)), 1)\n self.assertTrue(str(test_bool_arith_not.graph).count('if') == 0)\n\n\n def test_conditional_casting(self):\n def test_bool_cast_tensor(x):\n if x:\n return 1\n else:\n return 0\n\n for make_one_dim in [True, False]:\n for inp_val in [0.1, 0.0, -0.0, -0.1, -1, 0, 1]:\n inp_val = [inp_val] if make_one_dim else inp_val\n self.checkScript(test_bool_cast_tensor, (torch.tensor(inp_val),))\n\n self.checkScriptRaisesRegex(test_bool_cast_tensor, (torch.tensor([1, 1]),), Exception,\n \"Boolean value of Tensor with more than one value\")\n\n def test_not_cast(x):\n if not x:\n return 1\n else:\n return 0\n\n self.checkScript(test_not_cast, (torch.tensor(1),))\n self.checkScript(test_not_cast, (torch.tensor(0),))\n\n with self.assertRaisesRegex(RuntimeError, r\"Could not cast value of type Tuple\\[Tensor, Tensor\\]\"): # noqa: W605\n @torch.jit.script\n def test_mult(x, y):\n return not(x, y)\n\n def test_cast_int(x):\n # type: (int) -> int\n if x:\n return 1\n else:\n return 0\n self.checkScript(test_cast_int, (1,))\n self.checkScript(test_cast_int, (0,))\n self.checkScript(test_cast_int, (-1,))\n\n def test_cast_float(x):\n # type: (float) -> int\n if x:\n return 1\n else:\n return 0\n self.checkScript(test_cast_float, (1.,))\n self.checkScript(test_cast_float, (0.,))\n self.checkScript(test_cast_float, (-1.,))\n\n with self.assertRaisesRegex(RuntimeError, r\"Could not cast value of type Tuple\\[int, int\\] to bool\"): # noqa: W605\n\n @torch.jit.script\n def test_bad_conditional(x):\n if (1, 2): # noqa: F634\n return\n else:\n return 0\n\n def test_while_nonexistent_value(self):\n with self.assertRaisesRegex(RuntimeError, \"undefined value x\"):\n torch.jit.CompilationUnit('''\n def test_while(a, b):\n while bool(a < 10):\n a = a + x\n b = b + 1\n return a + b\n ''')\n\n def test_while_nonexistent_cond_value(self):\n with self.assertRaisesRegex(RuntimeError, \"undefined value x\"):\n torch.jit.CompilationUnit('''\n def test_while(a, b):\n while a < x:\n a = a + 1\n b = b + 1\n return a + b\n ''')\n\n @torch.jit.script\n def test_ternary(x):\n # type: (Optional[int]) -> int\n x = x if x is not None else 2\n return x\n\n @torch.jit.script\n def test_not_none(x):\n # type: (Optional[int]) -> None\n if x is not None:\n print(x + 1)\n\n @torch.jit.script\n def test_and(x, y):\n # type: (Optional[int], Optional[int]) -> None\n if x is not None and y is not None:\n print(x + y)\n\n @torch.jit.script\n def test_not(x, y):\n # type: (Optional[int], Optional[int]) -> None\n if not (x is not None and y is not None):\n pass\n else:\n print(x + y)\n\n @torch.jit.script\n def test_bool_expression(x):\n # type: (Optional[int]) -> None\n if x is not None and x < 2:\n print(x + 1)\n\n @torch.jit.script\n def test_nested_bool_expression(x, y):\n # type: (Optional[int], Optional[int]) -> int\n if x is not None and x < 2 and y is not None:\n x = x + y\n else:\n x = 5\n return x + 2\n\n @torch.jit.script\n def test_or(x, y):\n # type: (Optional[int], Optional[int]) -> None\n if y is None or x is None:\n pass\n else:\n print(x + y)\n\n # backwards compatibility\n @torch.jit.script\n def test_manual_unwrap_opt(x):\n # type: (Optional[int]) -> int\n if x is None:\n x = 1\n else:\n x = torch.jit._unwrap_optional(x)\n return x # noqa: T484\n\n with self.assertRaisesRegex(RuntimeError, \"Arguments for call are not valid\"):\n @torch.jit.script\n def or_error(x, y):\n # type: (Optional[int], Optional[int]) -> None\n if x is None or y is None:\n print(x + y) # noqa: T484\n\n with self.assertRaisesRegex(RuntimeError, \"Arguments for call are not valid\"):\n @torch.jit.script\n def and_error(x, y):\n # type: (Optional[int], Optional[int]) -> None\n if x is None and y is None:\n pass\n else:\n print(x + y) # noqa: T484\n\n with self.assertRaisesRegex(RuntimeError, \"Arguments for call are not valid\"):\n @torch.jit.script\n def named_var(x):\n # type: (Optional[int]) -> None\n x_none = x is not None\n if x_none:\n print(x + 1) # noqa: T484\n\n with self.assertRaisesRegex(RuntimeError, \"Arguments for call are not valid\"):\n @torch.jit.script\n def named_var_and(x, y):\n # type: (Optional[int], Optional[int]) -> None\n x_none = x is not None\n if y is not None and x_none:\n print(x + y) # noqa: T484\n\n def test_assertion_optional_refinement(self):\n @torch.jit.script\n def test(x, y):\n # type: (Optional[int], Optional[int]) -> int\n assert x is not None and y is not None\n return x + y\n\n self.assertEqual(test(2, 2), 4)\n with self.assertRaisesRegex(Exception, \"\"):\n test(1, None)\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY, \"the current version of Profiler doesn't profile/specialize Optionals\")\n def test_optional_tensor(self):\n @torch.jit.script\n def fn(x, y):\n # type: (Optional[Tensor], int) -> int\n if x is None:\n return y\n else:\n return 0\n\n res = fn(None, 1)\n self.assertEqual(res, 1)\n g = torch.jit.last_executed_optimized_graph()\n first_input = next(g.inputs())\n # check if input is disconnected\n self.assertEqual(first_input.type().kind(), 'OptionalType')\n self.assertEqual(first_input.uses(), [])\n t = torch.ones(1)\n res = fn(t, 1)\n self.assertEqual(res, 0)\n g = torch.jit.last_executed_optimized_graph()\n self.assertEqual(next(g.inputs()).type().kind(), 'TensorType')\n\n @torch.jit.script\n def fn(x, y, b):\n # type: (Optional[Tensor], Tensor, bool) -> Tensor\n if b:\n res = y\n else:\n res = torch.jit._unwrap_optional(x)\n return res\n\n t2 = torch.zeros(1)\n res = fn(t, t2, True)\n self.assertEqual(res, t2)\n with self.assertRaisesRegex(RuntimeError, \"Unwrapping null optional\"):\n res = fn(None, t2, False)\n res = fn(None, t2, True)\n g = torch.jit.last_executed_optimized_graph()\n self.assertIn(next(g.outputs()).type().str(), (\"Tensor\", \"Tensor(requires_grad=1)\"))\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY, \"the current version of Profiler doesn't profile/specialize Optionals\")\n def test_optional_list(self):\n @torch.jit.script\n def fn(x, y):\n # type: (Optional[List[int]], int) -> int\n if x is None:\n return y\n else:\n res = 0\n for d in x:\n res += d\n return res\n\n res = fn(None, 1)\n self.assertEqual(res, 1)\n g = torch.jit.last_executed_optimized_graph()\n first_input = next(g.inputs())\n # check if input is disconnected\n self.assertEqual(first_input.type().kind(), 'OptionalType')\n self.assertEqual(first_input.uses(), [])\n l = [2, 3]\n res = fn(l, 1)\n self.assertEqual(res, 5)\n g = torch.jit.last_executed_optimized_graph()\n self.assertEqual(next(g.inputs()).type().kind(), 'ListType')\n\n @torch.jit.script\n def fn(x, y, b):\n # type: (Optional[List[int]], List[int], bool) -> List[int]\n if b:\n l = torch.jit._unwrap_optional(x)\n else:\n l = y\n return l\n\n l2 = [0, 1]\n res = fn(l, l2, True)\n self.assertEqual(res, l)\n with self.assertRaisesRegex(RuntimeError, \"Unwrapping null optional\"):\n res = fn(None, l2, True)\n res = fn(None, l2, False)\n g = torch.jit.last_executed_optimized_graph()\n self.assertEqual(next(g.outputs()).type().str(), \"int[]\")\n\n def test_alias_covariant_type_containers(self):\n @torch.jit.script\n def foo(x):\n # type: (bool)\n if x:\n a = (None,)\n else:\n a = ([],)\n return a\n\n @torch.jit.script\n def foo2(x, li):\n # type: (bool, Tuple[Optional[List[Tensor]]])\n if x:\n li = (None,)\n return li\n\n def test_while_write_outer_then_read(self):\n def func(a, b):\n while bool(a < 10):\n a = a + 1\n b = a + 1\n return a + b\n\n inputs = self._make_scalar_vars([42, 1337], torch.int64)\n self.checkScript(func, inputs, optimize=True)\n\n def test_while_nest_if(self):\n def func(a, b):\n # type: (int, int) -> int\n c = 0\n while a < 10:\n a = a + 1\n b = b + 1\n if a > b:\n c = -a\n else:\n c = -b\n return c + 1\n\n inputs = self._make_scalar_vars([-1234, 4321], torch.int64)\n self.checkScript(func, inputs, optimize=True)\n\n def test_divmod(self):\n def func_int(a, b):\n # type: (int, int) -> Tuple[int, int]\n return divmod(a, b)\n\n def func_float(a, b):\n # type: (float, float) -> Tuple[float, float]\n return divmod(a, b)\n\n def func_int_float(a, b):\n # type: (int, float) -> Tuple[float, float]\n return divmod(a, b)\n\n def func_float_int(a, b):\n # type: (float, int) -> Tuple[float, float]\n return divmod(a, b)\n\n def divmod_test_iterator(func, num, den):\n for i in num:\n for j in den:\n self.checkScript(func, (i, j), frames_up=2)\n\n num_int = [1024, -1024]\n den_int = [10, -10]\n num_float = [5.3, -5.3]\n den_float = [2.0, -2.0]\n divmod_test_iterator(func_int, num_int, den_int)\n divmod_test_iterator(func_float, num_float, den_float)\n divmod_test_iterator(func_int_float, num_int, den_float)\n divmod_test_iterator(func_float_int, num_float, den_int)\n\n with self.assertRaisesRegex(RuntimeError, \"ZeroDivisionError: integer division or modulo by zero\"):\n cu = torch.jit.CompilationUnit(dedent(inspect.getsource(func_int)))\n cu.func_int(1024, 0)\n with self.assertRaisesRegex(RuntimeError, \"ZeroDivisionError: float divmod()\"):\n cu = torch.jit.CompilationUnit(dedent(inspect.getsource(func_float)))\n cu.func_float(5.3, 0.0)\n with self.assertRaisesRegex(RuntimeError, \"ZeroDivisionError: float divmod()\"):\n cu = torch.jit.CompilationUnit(dedent(inspect.getsource(func_int_float)))\n cu.func_int_float(1024, 0.0)\n with self.assertRaisesRegex(RuntimeError, \"ZeroDivisionError: float divmod()\"):\n cu = torch.jit.CompilationUnit(dedent(inspect.getsource(func_float_int)))\n cu.func_float_int(5.3, 0)\n\n def test_math_ops(self):\n def checkMathWrap(func_name, num_args=1, is_float=True, **args):\n if is_float:\n checkMath(func_name, num_args, True, **args)\n checkMath(func_name, num_args, False, **args)\n else:\n checkMath(func_name, num_args, is_float, **args)\n\n inf = float(\"inf\")\n NaN = float(\"nan\")\n mx_int = 2**31 - 1\n mn_int = -2**31\n float_vals = ([inf, NaN, 0.0, 1.0, 2.2, -1.0, -0.0, -2.2, -inf, 1, 0, 2] +\n [10.0 ** i for i in range(5)] + [-(10.0 ** i) for i in range(5)])\n int_vals = list(range(-5, 5, 1)) + [mx_int + 5, mx_int * 2, mn_int - 5, mn_int * 2]\n\n def checkMath(func_name, num_args, is_float=True, ret_type=\"float\", debug=False, vals=None, args_type=None):\n funcs_template = dedent('''\n def func(a, b):\n # type: {args_type} -> {ret_type}\n return math.{func}({args})\n ''')\n if num_args == 1:\n args = \"a\"\n elif num_args == 2:\n args = \"a, b\"\n else:\n raise RuntimeError(\"Test doesn't support more than 2 arguments\")\n if args_type is None:\n args_type = \"(float, float)\" if is_float else \"(int, int)\"\n funcs_str = funcs_template.format(func=func_name, args=args, args_type=args_type, ret_type=ret_type)\n scope = {}\n execWrapper(funcs_str, globals(), scope)\n cu = torch.jit.CompilationUnit(funcs_str)\n f_script = cu.func\n f = scope['func']\n\n if vals is None:\n vals = float_vals if is_float else int_vals\n vals = [(i, j) for i in vals for j in vals]\n\n for a, b in vals:\n res_python = None\n res_script = None\n try:\n res_python = f(a, b)\n except Exception as e:\n res_python = e\n try:\n res_script = f_script(a, b)\n except Exception as e:\n res_script = e\n if debug:\n print(\"in: \", a, b)\n print(\"out: \", res_python, res_script)\n # We can't use assertEqual because of a couple of differences:\n # 1. nan == nan should return true\n # 2. When python functions throw an exception, we usually want to silently ignore them.\n # (ie: We want to return `nan` for math.sqrt(-5))\n if res_python != res_script:\n if isinstance(res_python, Exception):\n continue\n\n if type(res_python) == type(res_script):\n if isinstance(res_python, tuple) and (math.isnan(res_python[0]) == math.isnan(res_script[0])):\n continue\n if isinstance(res_python, float) and math.isnan(res_python) and math.isnan(res_script):\n continue\n msg = (\"Failed on {func_name} with inputs {a} {b}. Python: {res_python}, Script: {res_script}\"\n .format(func_name=func_name, a=a, b=b, res_python=res_python, res_script=res_script))\n self.assertEqual(res_python, res_script, msg=msg, atol=(1e-4) * max(abs(res_python), res_script), rtol=0)\n\n unary_float_ops = [\"log\", \"log1p\", \"log10\", \"exp\", \"sqrt\", \"gamma\", \"lgamma\", \"erf\",\n \"erfc\", \"expm1\", \"fabs\", \"acos\", \"asin\", \"atan\", \"cos\", \"sin\", \"tan\",\n \"asinh\", \"atanh\", \"acosh\", \"sinh\", \"cosh\", \"tanh\", \"degrees\", \"radians\"]\n binary_float_ops = [\"atan2\", \"fmod\", \"copysign\"]\n for op in unary_float_ops:\n checkMathWrap(op, 1)\n for op in binary_float_ops:\n checkMathWrap(op, 2)\n\n checkMath(\"modf\", 1, ret_type=\"Tuple[float, float]\")\n checkMath(\"frexp\", 1, ret_type=\"Tuple[float, int]\")\n checkMath(\"isnan\", 1, ret_type=\"bool\")\n checkMath(\"isinf\", 1, ret_type=\"bool\")\n checkMath(\"ldexp\", 2, is_float=False, ret_type=\"float\", args_type=\"(float, int)\",\n vals=[(i, j) for i in float_vals for j in range(-10, 10)])\n checkMath(\"pow\", 2, is_float=False, ret_type=\"float\")\n checkMath(\"pow\", 2, is_float=True, ret_type=\"float\")\n checkMathWrap(\"floor\", ret_type=\"int\")\n checkMathWrap(\"ceil\", ret_type=\"int\")\n checkMathWrap(\"gcd\", 2, is_float=False, ret_type=\"int\")\n checkMath(\"isfinite\", 1, ret_type=\"bool\")\n if PY37:\n checkMathWrap(\"remainder\", 2)\n checkMathWrap(\"factorial\", 1, is_float=False, ret_type=\"int\", vals=[(i, 0) for i in range(-2, 10)])\n\n def test_if_nest_while(self):\n def func(a, b):\n # type: (int, int) -> int\n c = 0\n if a > b:\n while a > b:\n b = b + 1\n c = -b\n return c\n\n inputs = self._make_scalar_vars([4321, 1234], torch.int64)\n self.checkScript(func, inputs)\n\n def test_script_optional_none(self):\n def none_stmt(x):\n output = None\n output = x\n return output\n\n def none_args(x):\n # type: (Optional[Tensor]) -> Optional[Tensor]\n return None\n\n self.checkScript(none_stmt, [torch.arange(0, 2)], optimize=True)\n self.checkScript(none_args, [None], optimize=True)\n\n # test undefined tensor None as default param\n def test_script_optional_tensor_none(x=None):\n # type: (Optional[Tensor]) -> Tensor\n res = torch.zeros(1, dtype=torch.int8)\n if x is None:\n res = res + 1\n else:\n res = x\n return res\n\n fn = test_script_optional_tensor_none\n scripted_fn = torch.jit.script(fn)\n self.assertEqual(fn(), scripted_fn())\n self.assertEqual(fn(torch.zeros(1)), scripted_fn(torch.zeros(1)))\n\n # test typical None as default param\n def test_script_optional_other_none(x=None):\n # type: (Optional[float]) -> float\n res = 2.0\n if x is None:\n res = res + 1.0\n else:\n res = x\n return res\n\n fn = test_script_optional_other_none\n scripted_fn = torch.jit.script(fn)\n self.assertEqual(fn(), scripted_fn())\n self.assertEqual(fn(1.0), scripted_fn(1.0))\n\n def test_script_clamp_none(self):\n def test_script_clamp_max_none(x):\n return torch.clamp(x, min=2, max=None)\n\n def test_script_clamp_max(x):\n return torch.clamp(x, max=2)\n\n def test_script_clamp_min_none(x):\n return torch.clamp(x, min=None, max=2)\n\n def test_script_clamp_min(x):\n return torch.clamp(x, min=2)\n\n input = [torch.arange(0, 3)]\n self.checkScript(test_script_clamp_max_none, input, optimize=True)\n self.checkScript(test_script_clamp_max, input, optimize=True)\n self.checkScript(test_script_clamp_min_none, input, optimize=True)\n self.checkScript(test_script_clamp_min, input, optimize=True)\n\n def test_script_bool_constant(self):\n def test_script_bool_constant():\n a = True\n return a\n self.checkScript(test_script_bool_constant, [])\n\n def test_ternary(self):\n def func(a, b):\n c = 3\n c = a + b if bool(a > 3) else b\n return c\n\n inputs_true = self._make_scalar_vars([5, 2], torch.int64)\n inputs_false = self._make_scalar_vars([1, 0], torch.int64)\n self.checkScript(func, inputs_true, optimize=True)\n self.checkScript(func, inputs_false, optimize=True)\n\n def test_ternary_module_type_hint(self):\n class M1(torch.nn.Module):\n def forward(self) -> Any:\n return 'out' if self.training else {}\n\n class M2(torch.nn.Module):\n def forward(self) -> Any:\n out: Any = 'out' if self.training else {}\n return out\n\n class M3(torch.nn.Module):\n def forward(self) -> Optional[int]:\n return None if self.training else 1\n\n for module in [M1, M2, M3]:\n self.checkModule(module().train(), ())\n self.checkModule(module().eval(), ())\n\n def test_ternary_static_if(self):\n # Test for True branch when condition variable\n # is annotated as Final\n class M1(torch.nn.Module):\n flag: torch.jit.Final[bool]\n\n def __init__(self):\n super().__init__()\n self.flag = True\n\n def forward(self) -> torch.Tensor:\n return torch.ones(3) if self.flag else {}\n\n # Test for True branch when condition variable\n # is annotated as Final\n class M2(torch.nn.Module):\n flag: torch.jit.Final[bool]\n\n def __init__(self):\n super().__init__()\n self.flag = False\n\n def forward(self) -> torch.Tensor:\n return {} if self.flag else torch.ones(3)\n\n model1 = M1()\n model2 = M2()\n script_model_1 = torch.jit.script(model1)\n script_model_2 = torch.jit.script(model2)\n self.assertEqual(model1.forward(), script_model_1.forward())\n self.assertEqual(model2.forward(), script_model_2.forward())\n\n def test_print(self):\n def func(x, y):\n q = (x + y).sigmoid()\n print(q, 1, 2, [1, 2], [1.0, 2.0])\n w = -q\n return w * w\n\n x = torch.arange(4., requires_grad=True)\n y = torch.arange(0., 8, 2, requires_grad=True)\n self.checkScript(func, [x, y], optimize=True, capture_output=True)\n\n def test_format(self):\n def func(x):\n print(\"{}, I'm a {}\".format(\"Hello\", \"test\"))\n print(\"format blank\".format())\n print(\"stuff before {}\".format(\"hi\"))\n print(\"{} stuff after\".format(\"hi\"))\n return x + 1\n\n x = torch.arange(4., requires_grad=True)\n self.checkScript(func, [x], optimize=True, capture_output=True)\n\n def test_logical_short_circuit(self):\n @torch.jit.script\n def testNoThrows(t):\n c1 = 1\n if (False and bool(t[1])) or (True or bool(t[1])):\n c1 = 0\n return c1\n\n FileCheck().check_not(\"prim::If\").run(testNoThrows.graph)\n self.assertEqual(0, testNoThrows(torch.randn(0)))\n self.assertEqual(0, testNoThrows(torch.randn([2, 3])))\n\n @torch.jit.script\n def throwsOr(t):\n c0 = False or bool(t[1])\n print(c0)\n\n @torch.jit.script\n def throwsAnd(t):\n c0 = True and bool(t[1])\n print(c0)\n\n t = torch.randn(0)\n with self.assertRaisesRegex(RuntimeError, \"index 1 out of range for tensor of size\"):\n throwsOr(t)\n with self.assertRaisesRegex(RuntimeError, \"index 1 out of range for tensor of size\"):\n throwsAnd(t)\n\n def test_type_cast(self):\n template = dedent('''\n def func(v):\n # type: ({from_type}) -> {to_type}\n return {to_type}(v)\n ''')\n\n def check_cast(from_type, to_type, value, raises=False):\n code = template.format(from_type=from_type, to_type=to_type)\n self.checkScript(code, (value,))\n\n check_cast('int', 'float', 1)\n check_cast('int', 'bool', 1)\n check_cast('int', 'bool', 0)\n\n check_cast('float', 'int', 1.)\n check_cast('float', 'bool', 1.)\n check_cast('float', 'bool', 0.)\n\n check_cast('bool', 'int', True)\n check_cast('bool', 'float', True)\n\n def test_multiple_assignment(self):\n def outer_func(x):\n return x * 2, x + 2\n\n @torch.jit.script\n def func(x):\n y, z = outer_func(x)\n return y + z\n\n x = torch.arange(4)\n self.assertEqual(func(x), x * 2 + x + 2)\n\n def test_literals(self):\n def func(a):\n return a.view(size=[1, 2, 3])\n\n a = torch.randn(6)\n self.checkScript(func, [a], optimize=True)\n\n def test_return(self):\n def no_return(a):\n a + 1\n\n def void_return(a):\n return\n\n def one_return(a):\n return a + 1.\n\n def multiple_returns(a):\n return a * 1., a * 2., a * 3.\n\n a = torch.randn(1, dtype=torch.float)\n self.checkScript(no_return, [a], optimize=True)\n self.checkScript(void_return, [a], optimize=True)\n self.checkScript(one_return, [a], optimize=True)\n self.checkScript(multiple_returns, [a], optimize=True)\n\n with self.assertRaisesRegex(RuntimeError, \"does not return along all paths\"):\n torch.jit.CompilationUnit('''\n def no_return_bad_annotation(a):\n # type: (Tensor) -> Tensor\n a + 1\n ''')\n\n def test_error(self):\n @torch.jit.script\n def foo(a):\n return a.t()\n s = Variable(torch.rand(5, 5, 5))\n # XXX: this should stay quiet in stay propagation and only fail in the interpreter\n with self.assertRaisesRegex(RuntimeError, \"failed in the TorchScript interpreter\"):\n foo(s)\n\n @torch.jit.script\n def bar(c, b):\n return c + b\n\n with self.assertRaisesRegex(RuntimeError, \"failed in the TorchScript interpreter\"):\n bar(Variable(torch.rand(10), requires_grad=True), Variable(torch.rand(9), requires_grad=True))\n\n def test_error_stacktrace(self):\n @torch.jit.script\n def baz(c, b):\n return c + b\n\n @torch.jit.script\n def foo(c, b):\n return baz(c, b)\n\n @torch.jit.script\n def bar(c, b):\n return foo(c, b)\n\n with self.assertRaises(RuntimeError) as cm:\n bar(torch.rand(10), torch.rand(9))\n FileCheck().check(\"The following operation failed in the TorchScript interpreter\") \\\n .check(\"Traceback\") \\\n .check(\"in foo\").check(\"in baz\").run(str(cm.exception))\n\n def test_error_stacktrace_interface(self):\n @torch.jit.script\n def baz(c, b):\n return c + b\n\n @torch.jit.script\n def foo(c, b):\n return baz(c, b)\n\n @torch.jit.script\n def bar(c, b):\n return foo(c, b)\n\n @torch.jit.script\n class Bar(object):\n def one(self, x, y):\n return bar(x, y)\n\n @torch.jit.interface\n class IFace(object):\n def one(self, x, y):\n # type: (Tensor, Tensor) -> Tensor\n pass\n\n make_global(IFace)\n\n @torch.jit.script\n def as_interface(x):\n # type: (IFace) -> IFace\n return x\n\n f = as_interface(Bar())\n\n with self.assertRaises(RuntimeError) as cm:\n x = f.one(torch.rand(10), torch.rand(9))\n bar(torch.rand(10), torch.rand(9))\n FileCheck().check(\"The following operation failed in the TorchScript interpreter\") \\\n .check(\"Traceback\") \\\n .check(\"in foo\").check(\"in baz\").run(str(cm.exception))\n\n def test_operator_precedence(self):\n def double(x):\n # type: (int) -> int\n return 2 * x\n\n def complicated_arithmetic_operation():\n # TODO we need to test exponent operator '**' and bitwise not\n # operator '~' once they are properly supported.\n list = [0, 1, 2, 3]\n result = list[1:3][0] + double(4) + (-3 + 8) * 6 // 2 % 4 << 2 + 1 >> 1 | 23 & 16 + 3 ^ 4\n return result\n\n self.checkScript(complicated_arithmetic_operation, ())\n\n def test_in_operator_with_two_strings(self):\n def fn() -> bool:\n return \"a\" in \"abcd\"\n self.checkScript(fn, ())\n\n def test_bitwise_ops(self):\n\n def int_test():\n return 2 & 3, 2 ^ 3, 2 | 3, 2 << 3, 2 >> 3\n\n self.checkScript(int_test, ())\n\n def bool_test(x, y):\n # type: (bool, bool) -> Tuple[bool, bool, bool]\n return x & y, x ^ y, x | y\n\n self.checkScript(bool_test, (True, False))\n self.checkScript(bool_test, (True, True))\n\n def tensor_test(x, y):\n return x & y, x ^ y, x | y\n\n def tensor_with_int_test(x, y):\n # type: (Tensor, int) -> Tuple[Tensor, Tensor]\n return x << y, x >> y\n\n x = torch.tensor(2)\n y = torch.tensor(3)\n\n self.checkScript(tensor_test, (x, y))\n self.checkScript(tensor_with_int_test, (x, 2))\n\n def not_test(x):\n return ~x\n\n self.checkScript(not_test, (torch.tensor([2, 4]), ))\n\n def test_all(self):\n @torch.jit.script\n def test_all_tensor(x):\n return all(x)\n self.assertFalse(test_all_tensor(torch.tensor([1, 0, 3], dtype=torch.uint8)))\n self.assertTrue(test_all_tensor(torch.tensor([3.14, 3, 99], dtype=torch.uint8)))\n self.assertTrue(test_all_tensor(torch.tensor([True, True], dtype=torch.uint8)))\n self.assertFalse(test_all_tensor(torch.tensor([True, False], dtype=torch.uint8)))\n\n @torch.jit.script\n def test_all_bool_list(x):\n # type: (List[bool]) -> bool\n return all(x)\n self.assertTrue(test_all_bool_list([True, True]))\n self.assertTrue(test_all_bool_list([True, 1]))\n self.assertFalse(test_all_bool_list([True, False]))\n self.assertFalse(test_all_bool_list([True, 0]))\n self.assertFalse(test_all_bool_list([False, 0]))\n self.assertTrue(test_all_bool_list([]))\n\n @torch.jit.script\n def test_all_int_list(x):\n # type: (List[int]) -> bool\n return all(x)\n self.assertTrue(test_all_int_list([3, 6]))\n self.assertFalse(test_all_int_list([2, 0]))\n\n @torch.jit.script\n def test_all_float_list(x):\n # type: (List[float]) -> bool\n return all(x)\n self.assertTrue(test_all_float_list([3.14, 8.1]))\n self.assertFalse(test_all_float_list([3.14, 0, 8.9]))\n\n\n def test_number_math(self):\n ops_template = dedent('''\n def func():\n return {scalar1} {op} {scalar2}\n ''')\n ops = ['+', '-', '*', '%', '<', '<=', '>', '>=', '==', '!=', '//']\n funcs_template = dedent('''\n def func():\n return {func}({scalar1}, {scalar2})\n ''')\n funcs = ['min', 'max']\n scalars = ['7', '2', '3', '-3', '3.14', '0.125', '-0.5', '2.0', '-2.0']\n scalar_pairs = [(scalar1, scalar2) for scalar1 in scalars for scalar2 in scalars]\n\n def run_test(code):\n scope = {}\n execWrapper(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n\n self.assertEqual(cu.func(), scope['func']())\n\n for scalar1, scalar2 in scalar_pairs:\n for op in ops:\n code = ops_template.format(op=op, scalar1=scalar1, scalar2=scalar2)\n run_test(code)\n for func in funcs:\n code = funcs_template.format(func=func, scalar1=scalar1, scalar2=scalar2)\n run_test(code)\n\n # test Scalar overloads\n for scalar1, scalar2 in scalar_pairs:\n item1 = 'torch.tensor(' + scalar1 + ').item()'\n item2 = 'torch.tensor(' + scalar2 + ').item()'\n for op in ops:\n code = ops_template.format(op=op, scalar1=item1, scalar2=scalar2)\n run_test(code)\n code = ops_template.format(op=op, scalar1=scalar1, scalar2=item2)\n run_test(code)\n code = ops_template.format(op=op, scalar1=item1, scalar2=item2)\n run_test(code)\n for func in funcs:\n code = funcs_template.format(func=func, scalar1=item1, scalar2=scalar2)\n run_test(code)\n code = funcs_template.format(func=func, scalar1=scalar1, scalar2=item2)\n run_test(code)\n code = funcs_template.format(func=func, scalar1=item1, scalar2=item2)\n run_test(code)\n\n def test_number_abs(self):\n def func1(x):\n # type: (float) -> float\n return abs(x)\n\n def func2(x):\n # type: (int) -> int\n return abs(x)\n\n def func3(x):\n return abs(x)\n\n self.checkScript(func1, (-3.14,))\n self.checkScript(func1, (3.14,))\n self.checkScript(func2, (-10,))\n self.checkScript(func2, (10,))\n self.checkScript(func3, (torch.tensor([-5, -10, -20]),))\n self.checkScript(func3, (torch.tensor([5, 10, 20]),))\n self.checkScript(func3, (torch.tensor([-5, 10, -20]),))\n\n def test_number_div(self):\n self.assertEqual(div_int_future(), torch.jit.script(div_int_future)())\n self.checkScript(div_float_future, ())\n\n self.checkScript(div_int_nofuture, ())\n self.checkScript(div_float_nofuture, ())\n\n def test_floor_div(self):\n @torch.jit.script\n def foo(a, b):\n # type: (int, int) -> int\n return a // b\n for i in range(-8, 8):\n for j in range(-8, 8):\n if j != 0:\n self.assertEqual(foo(i, j), i // j)\n else:\n with self.assertRaisesRegex(RuntimeError, 'division by 0'):\n foo(i, j)\n\n # Testing bitwise shorthand aug assignment\n def test_bool_augassign_bitwise_or(self):\n def func(a: bool, b: bool) -> bool:\n a |= b\n return a\n\n self.checkScript(func, (True, False), optimize=True)\n self.checkScript(func, (True, True), optimize=True)\n self.checkScript(func, (False, False), optimize=True)\n self.checkScript(func, (False, True), optimize=True)\n\n def test_bool_augassign_bitwise_and(self):\n def func(a: bool, b: bool) -> bool:\n a &= b\n return a\n\n self.checkScript(func, (True, False), optimize=True)\n self.checkScript(func, (True, True), optimize=True)\n self.checkScript(func, (False, False), optimize=True)\n self.checkScript(func, (False, True), optimize=True)\n\n def test_bool_augassign_bitwise_xor(self):\n def func(a: bool, b: bool) -> bool:\n a ^= b\n return a\n\n self.checkScript(func, (True, False), optimize=True)\n self.checkScript(func, (True, True), optimize=True)\n self.checkScript(func, (False, False), optimize=True)\n self.checkScript(func, (False, True), optimize=True)\n\n def test_number_augassign_bitwise_lshift(self):\n def func() -> int:\n z = 8\n z <<= 2\n return z\n\n self.checkScript(func, (), optimize=True)\n\n def test_number_augassign_bitwise_rshift(self):\n def func() -> int:\n z = 8\n z >>= 2\n return z\n\n self.checkScript(func, (), optimize=True)\n\n def test_number_augassign_bitwise_pow(self):\n def func() -> float:\n z = 8\n z **= 2\n return z\n\n self.checkScript(func, (), optimize=True)\n\n def test_number_augassign(self):\n def func():\n z = 1\n z += 2\n return z\n\n self.checkScript(func, (), optimize=True)\n\n def test_nested_select_assign(self):\n class SubSubModule(torch.nn.Module):\n def __init__(self):\n super(SubSubModule, self).__init__()\n self.abc = 11\n\n def forward(self, x):\n return self.abc\n\n class SubModule(torch.nn.Module):\n def __init__(self):\n super(SubModule, self).__init__()\n self.a = 11\n self.nested = SubSubModule()\n\n def forward(self, x):\n return self.a\n\n class TestModule(torch.nn.Module):\n def __init__(self):\n super(TestModule, self).__init__()\n self.sub = SubModule()\n self.hi = 1\n\n def forward(self):\n self.hi = 5\n self.sub.a = 1\n self.sub.nested.abc = 5\n return self.sub.a * 20 + self.sub.nested.abc * 3 + self.hi\n\n self.checkModule(TestModule(), ())\n\n def test_number_neg(self):\n # int -> int\n def func1():\n return -8\n\n # float -> float\n def func2():\n return -3.14\n\n self.checkScript(func1, (), optimize=True)\n self.checkScript(func2, (), optimize=True)\n\n def test_compare_two_bool_inputs(self):\n def compare_eq(a: bool, b: bool):\n return a == b\n\n def compare_ne(a: bool, b: bool):\n return a != b\n\n scripted_fn_eq = torch.jit.script(compare_eq)\n scripted_fn_ne = torch.jit.script(compare_ne)\n self.assertEqual(scripted_fn_eq(True, False), compare_eq(True, False))\n self.assertEqual(scripted_fn_eq(False, True), compare_eq(False, True))\n self.assertEqual(scripted_fn_eq(True, True), compare_eq(True, True))\n self.assertEqual(scripted_fn_eq(False, False), compare_eq(False, False))\n\n self.assertEqual(scripted_fn_ne(True, False), compare_ne(True, False))\n self.assertEqual(scripted_fn_ne(False, True), compare_ne(False, True))\n self.assertEqual(scripted_fn_ne(True, True), compare_ne(True, True))\n self.assertEqual(scripted_fn_ne(False, False), compare_ne(False, False))\n\n\n def _test_tensor_number_math(self, device='cpu'):\n template = dedent('''\n def func(t):\n return {lhs} {op} {rhs}\n ''')\n\n def test(op, tensor, const, swap_args, template=template):\n args = ('t', const)\n if swap_args:\n args = (const, 't')\n\n code = template.format(lhs=args[0], rhs=args[1], op=op)\n scope = {}\n execWrapper(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n message = 'with code `{} {} {}` and t={}'.format(args[0], op, args[1], tensor)\n res1 = cu.func(tensor)\n res2 = scope['func'](tensor)\n self.assertEqual(res1, res2, msg=message + \"\\nres1=\" + str(res1) + \"\\nres2=\" + str(res2))\n self.assertEqual(res1.dtype, res2.dtype, msg=message + \"\\nres1=\" + str(res1) + \"\\nres2=\" + str(res2))\n\n var_int = [2, -2]\n var_float = [1.4321, -1.2]\n\n ops = ['+', '-', '*', '%', '<', '<=', '>', '>=', '==', '!=', '/']\n\n float_tensor = torch.randn(5, 5, device=device)\n double_tensor = torch.randn(5, 5, dtype=torch.double, device=device)\n long_tensor = torch.randint(-5, 5, (5, 5), dtype=torch.long, device=device)\n long_tensor[long_tensor == 0] = 2\n\n tensors = [float_tensor, double_tensor, long_tensor]\n consts = var_int + var_float\n\n for op, tensor, const, swap_args in product(ops, tensors, consts, [True, False]):\n # FIXME: things like 2 / long_tensor are not implemented correctly\n # Look in torch/_tensor.py to see how pytorch implements it.\n if op == '/' and tensor.data_ptr() == long_tensor.data_ptr():\n continue\n\n # % operator does not take: const % tensor\n if op == '%' and swap_args is True:\n continue\n\n test(op, tensor, const, swap_args)\n\n def test_tensor_number_math(self):\n self._test_tensor_number_math()\n\n def test_torch_tensor_bad_input(self):\n with self.assertRaisesRegex(RuntimeError, \"must be of ints, floats, \"\n \"or bools, got None\"):\n @torch.jit.script\n def test():\n return torch.tensor([None])\n test()\n\n with self.assertRaisesRegex(RuntimeError, r\"Empty lists default to List\\[Tensor\\]\"):\n @torch.jit.script\n def tmp():\n return torch.tensor([])\n tmp()\n\n @torch.jit.script\n def foo():\n return torch.tensor([[2, 2], [1]])\n with self.assertRaisesRegex(RuntimeError, \"Expected sequence of length\"):\n foo()\n\n @suppress_warnings\n def test_torch_tensor_as_tensor_empty_list(self):\n tensor_template = dedent('''\n def func():\n empty_list = torch.jit.annotate(List[int], [])\n ten1 = torch.{tensor_op}({input})\n return ten1\n ''')\n ops = ['tensor', 'as_tensor']\n inputs = ['empty_list', '[empty_list, empty_list]', '[[[empty_list]]]']\n\n for op in ops:\n for inp in inputs:\n code = tensor_template.format(tensor_op=op, input=inp)\n scope = {}\n exec(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n t1 = cu.func()\n t2 = scope['func']()\n if inp == 'empty_list':\n # torchscript returns int tensor, python returns float tensor\n self.assertNotEqual(t1.dtype, t2.dtype)\n\n # TODO(#38095): Replace assertEqualIgnoreType. See issue #38095\n self.assertEqualIgnoreType(t1, t2)\n self.assertEqual(t1.device, t2.device)\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY, \"Simple Executor doesn't have any shapes to propagate\")\n def test_tensor_as_tensor_shape_prop(self):\n tensor_template = dedent('''\n def func():\n return torch.{tensor_op}({input})\n ''')\n ops = ['tensor', 'as_tensor']\n inputs = ['[1]', '[False]', '[2.5]', '0.5', '1', 'False', '[[1]]', 'torch.jit.annotate(List[List[int]], [])']\n expected_shape = [\"Long(*, device=cpu)\", \"Bool(*, device=cpu)\",\n \"Double(*, device=cpu)\", \"Double(device=cpu)\",\n \"Long(device=cpu)\", \"Bool(device=cpu)\", \"Long(*, *, device=cpu)\"]\n\n for op in ops:\n for inp, expect in zip(inputs, expected_shape):\n code = tensor_template.format(tensor_op=op, input=inp)\n scope = {}\n exec(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n torch._C._jit_pass_complete_shape_analysis(cu.func.graph, (), False)\n FileCheck().check(expect).check(\"aten::{tensor_op}\".format(tensor_op=op)).run(cu.func.graph)\n\n @torch.jit.script\n def test_dtype(inp_dtype: torch.dtype):\n a = torch.tensor(1.0, dtype=torch.float, requires_grad=True)\n return a, torch.tensor(1.0, dtype=inp_dtype)\n\n if GRAPH_EXECUTOR == ProfilingMode.PROFILING:\n g = test_dtype.graph_for(5, profile_and_replay=True)\n # both should have completed shapes\n FileCheck().check(\"Tensor = aten::tensor\").check(\"Float(device=cpu) = prim::BailOut\") \\\n .check(\"Tensor = aten::tensor\").check(\"Half(device=cpu) = prim::BailOut\").run(g)\n else:\n g = test_dtype.graph_for(5)\n # first should have type set second should not\n FileCheck().check(\"Float(requires_grad=1, device=cpu) = aten::tensor\") \\\n .check(\"Tensor(requires_grad=0) = aten::tensor\").run(g)\n\n @torch.jit.script\n def test_as_tensor_tensor_input(input):\n a = torch.as_tensor(input, dtype=input.dtype)\n return a, torch.as_tensor(input, dtype=torch.float)\n\n if GRAPH_EXECUTOR == ProfilingMode.PROFILING:\n g = test_as_tensor_tensor_input.graph_for(torch.ones(3, 4), profile_and_replay=True)\n FileCheck().check(\"Tensor = aten::as_tensor\").check(\"Float(3, 4) = prim::BailOut\") \\\n .check(\"Tensor = aten::as_tensor\").check(\"Float(3, 4) = prim::BailOut\").run(g)\n else:\n g = test_as_tensor_tensor_input.graph_for(torch.ones(3, 4))\n FileCheck().check(\"Tensor = aten::as_tensor\").check(\"Float(*, *, requires_grad=0, device=cpu) = aten::as_tensor\").run(g)\n\n\n def test_tensor_requires_grad(self):\n @torch.jit.script\n def test(b):\n # type: (bool) -> Tuple[Tensor, Tensor, Tensor]\n a = torch.tensor(1., requires_grad=b)\n b = torch.tensor(1., requires_grad=True)\n c = torch.tensor(1., requires_grad=False)\n return a, b, c\n\n g = test.graph_for(True)\n out = next(g.outputs())\n out_inp = list(out.node().inputs())\n\n self.assertTrue(out_inp[0].requires_grad())\n self.assertTrue(out_inp[1].requires_grad())\n self.assertFalse(out_inp[2].requires_grad())\n\n def test_grad_from_script(self):\n def test():\n a = torch.tensor(2.5, requires_grad=True)\n b = a * 2\n return a, b\n\n a, b = test()\n b.backward()\n\n a_script, b_script = torch.jit.script(test)()\n b_script.backward()\n self.assertEqual(a.grad, a_script.grad)\n\n def test_torch_tensor_as_tensor(self):\n tensor_template = dedent('''\n def func():\n li = {list_create}\n ten1 = torch.{tensor_op}(li {options})\n return ten1\n ''')\n\n lists = [\"2.5\", \"4\", \"True\", \"False\", \"[2]\", \"[-.5]\", \"[False, True, False]\", \"[2, 2]\", \"(1, 1)\",\n \"torch.jit.annotate(List[List[int]], [])\",\n \"torch.jit.annotate(List[int], [])\", \"[2.5, 2.5]\", \"[[2], [2]]\", \"[[-.5], [2.2]]\", \"[[False], [True]]\"]\n\n dtypes = [\"\", \", dtype=torch.float\", \", dtype=torch.double\", \", dtype=torch.half\",\n \", dtype=torch.uint8\", \", dtype=torch.int8\", \", dtype=torch.short\",\n \", dtype=torch.int\", \", dtype=torch.long\", \", dtype=torch.cfloat\",\n \", dtype=torch.cdouble\"]\n\n ops = ['tensor', 'as_tensor']\n devices = ['', \", device='cpu'\"]\n if RUN_CUDA:\n devices.append(\", device='cuda'\")\n\n option_pairs = [dtype + device for dtype in dtypes for device in devices]\n for op in ops:\n for li in lists:\n for option in option_pairs:\n # tensor from empty list is type float in python and annotated type in torchscript\n if \"annotate\" in li and \"dtype\" not in option:\n continue\n code = tensor_template.format(list_create=li, tensor_op=op, options=option)\n scope = {}\n exec(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n t1 = cu.func()\n t2 = scope['func']()\n if t1.dtype == torch.float16: # equality NYI for half tensor\n self.assertTrue(str(t1) == str(t2))\n else:\n self.assertEqual(t1, t2)\n self.assertEqual(t1.dtype, t2.dtype)\n self.assertEqual(t1.device, t2.device)\n\n def test_as_tensor_tensor_input(input):\n # type: (Tensor) -> Tuple[Tensor, Tensor, Tensor]\n return torch.as_tensor(input, dtype=torch.cfloat), torch.as_tensor(input, dtype=torch.float), \\\n torch.as_tensor(input, dtype=torch.int32)\n\n inp = torch.randn(3, 4, dtype=torch.cfloat)\n self.checkScript(test_as_tensor_tensor_input, (inp,))\n\n def test_torch_tensor_dtype(self):\n def foo(s: float):\n return torch.tensor(s), torch.tensor([s, s])\n\n # need to clear function cache so we re run shape analysis\n with set_default_dtype(torch.double):\n self.assertEqual(torch.jit.script(foo)(1.), foo(1.), exact_dtype=True)\n if GRAPH_EXECUTOR == ProfilingMode.LEGACY:\n FileCheck().check(\"Double\").check_same(\"aten::tensor\").run(torch.jit.last_executed_optimized_graph())\n with set_default_dtype(torch.float):\n del torch.jit._state._jit_caching_layer[foo]\n self.assertEqual(torch.jit.script(foo)(1.), foo(1.), exact_dtype=True)\n if GRAPH_EXECUTOR == ProfilingMode.LEGACY:\n FileCheck().check(\"Float\").check_same(\"aten::tensor\").run(torch.jit.last_executed_optimized_graph())\n with set_default_dtype(torch.half):\n del torch.jit._state._jit_caching_layer[foo]\n self.assertEqual(torch.jit.script(foo)(1.), foo(1.), exact_dtype=True)\n if GRAPH_EXECUTOR == ProfilingMode.LEGACY:\n FileCheck().check(\"Half\").check_same(\"aten::tensor\").run(torch.jit.last_executed_optimized_graph())\n\n def test_shape_analysis_grad_property(self):\n @torch.jit.script\n def foo(x):\n return torch.sub(x, torch.tanh(x))\n\n torch._C._jit_pass_complete_shape_analysis(foo.graph, (torch.tensor([0.39]),), False)\n\n # requires_grad property shouldn't be accidentally set by shape analysis\n self.assertTrue(foo.graph.findNode(\"aten::sub\").output().requiresGrad() is None)\n\n def test_empty_like_memory_format_bc(self):\n def f(x):\n # type: (Tensor) -> Tensor\n return torch.zeros_like(x, memory_format=None)\n\n scripted_f = torch.jit.script(f)\n x = torch.rand(3, 4)\n self.assertEqual(scripted_f(x), f(x))\n\n def test_multiline_string_dedents(self):\n def foo() -> None:\n multiline_string_dedent_1 = \"\"\"\nThis is a string dedent \"\"\"\n multiline_string_dedent_2 = \"\"\" This is a\n string dedent \"\"\"\n multiline_string_dedent_3 = \"\"\"\n This is a string\ndedent \"\"\"\n multiline_string_dedent_4 = \"\"\" This is a string dedent \"\"\"\n\n scripted_foo = torch.jit.script(foo)\n self.assertEqual(scripted_foo(), foo())\n\n def test_class_with_comment_at_lower_indentation(self):\n class Foo(torch.nn.Module):\n def forward(self, x):\n x = torch.neg(x)\n # This comment is at the wrong indent\n return x\n\n torch.jit.script(Foo())\n\n # adapted from test in test_torch\n def test_tensor_to(self):\n template = dedent('''\n def func(t):\n cuda = \"{cuda}\"\n device = \"{device}\"\n non_blocking = {non_blocking}\n return {to_str}\n ''')\n\n def s(t, to_str, non_blocking=None, device=None, cuda=None):\n device = device if device is not None else str(t.device)\n non_blocking = non_blocking if non_blocking is not None else False\n cuda = \"cuda\" if cuda is None else cuda\n code = template.format(to_str=to_str, device=device, non_blocking=non_blocking, cuda=cuda)\n scope = {}\n cu = torch.jit.CompilationUnit(code)\n return cu.func(t, profile_and_replay=True)\n\n def test_copy_behavior(t, non_blocking=False):\n self.assertIs(t, s(t, 't.to(t, non_blocking=non_blocking)', non_blocking))\n self.assertIs(t, s(t, 't.to(t.dtype, non_blocking=non_blocking)', non_blocking))\n self.assertIs(t, s(t, 't.to(torch.empty_like(t), non_blocking=non_blocking)', non_blocking))\n self.assertIsNot(t, s(t, 't.to(t, non_blocking=non_blocking, copy=True)', non_blocking))\n self.assertIsNot(t, s(t, 't.to(t.dtype, non_blocking=non_blocking, copy=True)', non_blocking))\n self.assertIsNot(t, s(t, 't.to(torch.empty_like(t), non_blocking=non_blocking, copy=True)', non_blocking))\n\n devices = [t.device]\n if t.device.type == 'cuda':\n if t.device.index == -1:\n devices.append('cuda:{}'.format(torch.cuda.current_device()))\n elif t.device.index == torch.cuda.current_device():\n devices.append('cuda')\n for device in devices:\n self.assertIs(t, s(t, 't.to(device, non_blocking=non_blocking)', non_blocking, device))\n self.assertIs(t, s(t, 't.to(device, t.dtype, non_blocking=non_blocking)', non_blocking, device))\n self.assertIsNot(t, s(t, 't.to(device, non_blocking=non_blocking, copy=True)', non_blocking, device))\n self.assertIsNot(t, s(t, 't.to(device, t.dtype, non_blocking=non_blocking, copy=True)',\n non_blocking, device))\n\n t = torch.tensor(5)\n test_copy_behavior(t)\n\n self.assertEqual(t.device, s(t, \"t.to('cpu')\").device)\n self.assertEqual(t.device, s(t, \"t.to('cpu', dtype=torch.float32)\").device)\n self.assertIs(torch.float32, s(t, \"t.to('cpu', dtype=torch.float32)\").dtype)\n self.assertEqual(t.device, s(t, \"t.to(torch.float32)\").device)\n self.assertIs(torch.float32, s(t, \"t.to(dtype=torch.float32)\").dtype)\n self.assertEqual(t.data_ptr(), s(t, \"t.to('cpu')\").data_ptr())\n self.assertEqual(t.data_ptr(), s(t, \"t.to(dtype=t.dtype, device=t.device, copy=False)\").data_ptr())\n self.assertEqual(t.data_ptr(), s(t, \"t.to('cpu', copy=False)\").data_ptr())\n self.assertNotEqual(t.data_ptr(), s(t, \"t.to('cpu', copy=True)\").data_ptr())\n\n a = torch.tensor(5)\n if torch.cuda.is_available():\n for non_blocking in [True, False]:\n for cuda in ['cuda', 'cuda:0' if torch.cuda.device_count() == 1 else 'cuda:1']:\n b = torch.tensor(5., device=cuda)\n test_copy_behavior(b, non_blocking)\n self.assertEqual(b.device, s(b, \"t.to(cuda, non_blocking=non_blocking).device\", cuda=cuda))\n self.assertEqual(a.device, s(b, \"t.to('cpu', non_blocking=non_blocking).device\"))\n self.assertEqual(b.device, s(b, \"t.to(cuda, non_blocking=non_blocking).device\", cuda=cuda))\n self.assertIs(torch.int32, s(b, \"t.to('cpu', dtype=torch.int32, non_blocking=non_blocking)\").dtype)\n self.assertEqual(a.device, s(b, \"t.to('cpu', dtype=torch.int32, non_blocking=non_blocking)\").device)\n self.assertIs(torch.int32, s(b, \"t.to(dtype=torch.int32)\").dtype)\n self.assertEqual(b.device, s(b, \"t.to(dtype=torch.int32)\").device)\n\n # Test AD: aten::to(Tensor self, int dtype, bool non_blocking, bool copy) -> Tensor\n t = torch.tensor(5).float().requires_grad_()\n out_ref = t.to(torch.float32)\n out = s(t, \"t.to(torch.float32)\")\n self.assertEqual(out_ref, out)\n\n grad_ref = torch.autograd.grad(out_ref.sum(), t)\n grad = torch.autograd.grad(out.sum(), t)\n self.assertEqual(grad_ref, grad)\n\n # Test AD: aten::to(Tensor self, Device? device, int? dtype, bool non_blocking, bool copy) -> Tensor\n out_ref = t.to('cpu')\n out = s(t, \"t.to('cpu')\")\n self.assertEqual(out_ref, out)\n\n grad_ref = torch.autograd.grad(out_ref.sum(), t)\n grad = torch.autograd.grad(out.sum(), t)\n self.assertEqual(grad_ref, grad)\n\n # Test AD: aten::to(Tensor self, Tensor other, bool non_blocking, bool copy) -> Tensor\n @torch.jit.script\n def func2(t, t_ref):\n return t.to(t_ref)\n\n with disable_autodiff_subgraph_inlining():\n t_ref = torch.tensor(4).double()\n out_ref = t.to(t_ref)\n out = func2(t, t_ref)\n grad_ref = torch.autograd.grad(out_ref.sum(), t)\n grad = torch.autograd.grad(out.sum(), t)\n self.assertEqual(grad_ref, grad)\n\n @unittest.skipIf(not RUN_CUDA, \"No CUDA\")\n def test_tensor_number_math_cuda(self):\n self._test_tensor_number_math(device='cuda')\n\n def test_not(self):\n # test not operator in python\n # TODO: add more tests when bool conversions ready\n def test_not_op(a):\n return not bool(a > 1)\n\n self.checkScript(test_not_op, (torch.tensor(2), ), optimize=True)\n\n def test_is_isnot(self):\n # test is and is not operator in python\n template = dedent('''\n def func():\n # type: () -> bool\n return {lhs} {op} {rhs}\n ''')\n\n def test(op, args):\n code = template.format(lhs=args[0], rhs=args[1], op=op)\n scope = {}\n execWrapper(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n self.assertEqual(\n cu.func(),\n scope['func'](),\n msg=\"Failed with op: {}, lhs: {}, rhs: {}\"\n .format(op, args[0], args[1])\n )\n\n ops = ['is', 'is not']\n type_literals = [True, False, None, [1, 1], 1, 2, .5, 1.5]\n\n # do literals product to try any types combinations\n for op, lhs, rhs in product(ops, type_literals, type_literals):\n test(op, [lhs, rhs])\n\n def test_isinstance_refinement(self):\n @torch.jit.script\n def foo(a):\n # type: (Optional[int]) -> int\n if isinstance(a, int):\n return a + 3\n else:\n return 4\n self.assertEqual(foo(4), 7)\n self.assertEqual(foo(None), 4)\n\n @torch.jit.script\n def foo2(a, b):\n # type: (Optional[int], Optional[int]) -> int\n if not isinstance(a, int) or not isinstance(b, int):\n return 0\n else:\n return a + b\n self.assertEqual(foo2(3, 4), 7)\n self.assertEqual(foo2(None, 4), 0)\n self.assertEqual(foo2(4, None), 0)\n\n @torch.jit.script\n def any_refinement(a, b):\n # type: (Any, Any) -> int\n if isinstance(a, int) and isinstance(b, int):\n return a + b\n return 0\n\n self.assertEqual(any_refinement(3, 4), 7)\n self.assertEqual(any_refinement(3, \"hi\"), 0)\n\n @torch.jit.script\n def any_refinement2(a):\n # type: (Any) -> Tensor\n if isinstance(a, Tensor):\n return a\n return torch.tensor(3)\n\n self.assertEqual(any_refinement2(3), torch.tensor(3))\n self.assertEqual(any_refinement2(torch.tensor(5)), torch.tensor(5))\n\n @unittest.skipIf(GRAPH_EXECUTOR == ProfilingMode.LEGACY, \"bug persists in deprecated executor\")\n def test_unspecialized_any_binding(self):\n # any binding will infer the type, if it infers\n # a specialized tensor type `x` Dict type will fail isinstance check\n\n @torch.jit.script\n def foo(x: Any):\n assert isinstance(x, Dict[str, torch.Tensor])\n\n foo({\"1\": torch.tensor(3)})\n with self.assertRaises(Exception):\n foo(2)\n\n def test_isinstance(self):\n # test isinstance operator for static type checking\n template = dedent('''\n def func(x):\n # type: ({type_hint}) -> bool\n return isinstance(x, {typ})\n ''')\n\n def test(inp, typ, type_hint):\n code = template.format(typ=typ, type_hint=type_hint)\n scope = {}\n execWrapper(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n self.assertEqual(\n cu.func(inp),\n scope['func'](inp),\n msg=\"Failed with typ: {}\"\n .format(typ)\n )\n\n inputs = [True, 1, 1.0, torch.tensor(1), [1, 2], (1.0,), [1, 2], 1]\n type_literals = ['bool', 'int', 'float', 'torch.Tensor', 'list', 'tuple',\n '(list, tuple)', '(int, float, bool)']\n type_annotations = ['bool', 'int', 'float', 'Tensor', 'List[int]', 'Tuple[float]',\n 'List[int]', 'int']\n\n # do zipping to try different types\n for inp, typ, type_hint in zip(inputs, type_literals, type_annotations):\n test(inp, typ, type_hint)\n\n # test optional isinstance check\n @torch.jit.script\n def opt_func(x):\n # type: (Optional[int]) -> bool\n return isinstance(x, int)\n self.assertTrue(opt_func(3))\n self.assertFalse(opt_func(None))\n\n def test_dropout_eval(self):\n class ScriptedConv2d(torch.jit.ScriptModule):\n def __init__(self, in_channels, out_channels, **kwargs):\n super(ScriptedConv2d, self).__init__()\n self.conv = nn.Conv2d(in_channels, out_channels, bias=False, **kwargs)\n self.bn = nn.BatchNorm2d(out_channels, eps=0.001)\n\n @torch.jit.script_method\n def forward(self, x):\n x = self.conv(x)\n x = self.bn(x)\n return F.relu(x, inplace=True)\n\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n self.Conv2d_1a_3x3 = ScriptedConv2d(3, 32, kernel_size=3, stride=2)\n\n @torch.jit.script_method\n def forward(self, x):\n x = self.Conv2d_1a_3x3(x)\n return F.dropout(x, training=self.training)\n\n class EagerConv2d(torch.nn.Module):\n def __init__(self, in_channels, out_channels, **kwargs):\n super(EagerConv2d, self).__init__()\n self.conv = nn.Conv2d(in_channels, out_channels, bias=False, **kwargs)\n self.bn = nn.BatchNorm2d(out_channels, eps=0.001)\n\n def forward(self, x):\n x = self.conv(x)\n x = self.bn(x)\n return F.relu(x, inplace=True)\n\n class EagerMod(torch.nn.Module):\n def __init__(self):\n super(EagerMod, self).__init__()\n self.Conv2d_1a_3x3 = EagerConv2d(3, 32, kernel_size=3, stride=2)\n\n def forward(self, x):\n x = self.Conv2d_1a_3x3(x)\n return F.dropout(x, training=self.training)\n\n script_input = torch.rand(4, 3, 299, 299)\n eager_input = script_input.clone()\n\n with freeze_rng_state():\n script_mod = ScriptMod()\n script_mod.eval()\n script_output = script_mod(script_input)\n\n with freeze_rng_state():\n eager_mod = EagerMod()\n eager_mod.eval()\n eager_output = eager_mod(eager_input)\n\n self.assertEqual(script_output, eager_output)\n\n with freeze_rng_state():\n script_mod = ScriptMod()\n script_mod.train()\n script_output = script_mod(script_input)\n\n with freeze_rng_state():\n eager_mod = EagerMod()\n eager_mod.train()\n eager_output = eager_mod(eager_input)\n\n self.assertEqual(script_output, eager_output)\n\n def test_nested_breaks(self):\n def no_bool_loop_outputs(g):\n # testing that the \"did exit\" transform values are not loop block\n # outputs (and thus not affecting one loop from another)\n loops = g.findAllNodes(\"prim::Loop\")\n for loop in loops:\n for out in loop.outputs():\n self.assertTrue(out.type() != BoolType.get())\n\n def test(y):\n # type: (int)\n ret = 0\n tensor = torch.tensor(0)\n while int(tensor.add_(1)) < 4:\n if y == 1:\n continue\n for i in range(y):\n continue\n ret += 1\n ret += 1\n return ret, int(tensor)\n\n self.assertEqual(torch.jit.script(test)(1), test(1))\n self.assertEqual(torch.jit.script(test)(2), test(2))\n no_bool_loop_outputs(torch.jit.script(test).graph)\n\n def foo():\n y = torch.tensor(0)\n z = 0\n while int(y.add_(1)) < 20:\n if int(y) < 10:\n for i in range(6):\n if i == 3:\n continue\n else:\n if i > 3:\n break\n z += 2\n if int(y) == 18:\n break\n if int(y) == 15:\n continue\n z += 1\n return int(y), z\n\n no_bool_loop_outputs(torch.jit.script(foo).graph)\n self.checkScript(foo, ())\n\n def test_nested_two():\n i = 0\n k = 0\n while i < 5:\n for j in range(5):\n k += 1\n if j == 3:\n continue\n i += 1\n k += 1\n if i == 4:\n break\n return i, k\n\n self.checkScript(test_nested_two, ())\n no_bool_loop_outputs(torch.jit.script(test_nested_two).graph)\n\n def test_breaks_continues(self):\n def foo_continue(cond):\n # type: (int)\n j = 1\n for i in range(5):\n if i == cond:\n continue\n j += 1\n return j\n\n def foo_break(cond):\n # type: (int)\n j = 1\n for i in range(5):\n if i == cond:\n break\n j += 1\n return j\n\n for i in range(1, 4):\n self.checkScript(foo_continue, (i,))\n self.checkScript(foo_break, (i,))\n\n def test_refine_outside_loop():\n if 1 == 1:\n x = None\n else:\n x = 1\n i = 0\n j = 0\n while (x is None or torch.jit._unwrap_optional(x) > 3):\n if i < 3:\n if i < 3:\n x = torch.jit.annotate(Optional[int], None)\n i += 1\n continue\n x = 1\n else:\n x = 1 if x is None else x\n x = x + 1\n j = x + x\n\n return x, j\n\n self.checkScript(test_refine_outside_loop, ())\n\n def assign_after_break(y):\n # type: (int)\n x = 0\n for i in range(y):\n x = y * 2 + i\n break\n x = 4\n return x\n\n self.checkScript(assign_after_break, (1,))\n self.checkScript(assign_after_break, (2,))\n self.checkScript(assign_after_break, (3,))\n\n def assign_after_break_nested(y):\n # type: (int)\n x = 0\n for i in range(y):\n if y == 1:\n x = 5\n break\n assert 1 == 2\n else:\n x = x + 1\n break\n assert 1 == 2\n x = -30\n assert 1 == 2\n return x\n\n self.checkScript(assign_after_break_nested, (1,))\n self.checkScript(assign_after_break_nested, (2,))\n self.checkScript(assign_after_break_nested, (3,))\n\n def may_break(y):\n # type: (int)\n x = 0\n for i in range(y):\n if y == 1:\n x = 5\n else:\n x = x + 1\n break\n x = -30\n return x\n\n self.checkScript(may_break, (1,))\n self.checkScript(may_break, (2,))\n self.checkScript(may_break, (3,))\n\n def test(x, y):\n # type: (int, int)\n a = 1\n while (x > 0):\n if y == 3:\n for i in range(y):\n a += (1 % (i + 1))\n x -= 1\n if x == 3:\n a = x * 3\n break\n if x < 3:\n if x == 1:\n a -= 2\n x -= 1\n break\n a -= 1\n x -= 3\n return a, x\n\n self.checkScript(test, (10, 3))\n self.checkScript(test, (10, 2))\n self.checkScript(test, (3, 2))\n self.checkScript(test, (5, 3))\n self.checkScript(test, (2, 3))\n\n def test_delete_after_break(x):\n # type: (int)\n a = 1\n b = 1\n for i in range(x):\n a = i * 3\n break\n b = i * 5\n return a, b\n\n self.checkScript(test_delete_after_break, (0,))\n self.checkScript(test_delete_after_break, (1,))\n\n def test_will_break_after_guard(x):\n # type: (int)\n a = 1\n for i in range(x):\n if i == 4:\n a = 3\n break\n a -= 1\n break\n assert 1 == 2\n a -= -100\n return a\n\n self.checkScript(test_will_break_after_guard, (0,))\n self.checkScript(test_will_break_after_guard, (2,))\n self.checkScript(test_will_break_after_guard, (4,))\n\n def test_varexit(cond):\n # type: (int)\n m = 0\n for i in range(3):\n if cond == 2:\n if cond == 2:\n m = 2\n break\n k = 1\n else:\n k = 2\n m += k\n return m\n\n # use of k tests the pathway where we have to insert unitialized\n self.checkScript(test_varexit, (3,))\n self.checkScript(test_varexit, (2,))\n\n def test_break_true():\n i = 0\n while True:\n i += 1\n if i == 3:\n break\n while False:\n i += 1\n return i\n\n self.checkScript(test_break_true, ())\n\n def test_break_continue_error(self):\n with self.assertRaisesRegex(RuntimeError, \"Syntax\"):\n cu = torch.jit.CompilationUnit('''\n def other_func(a):\n break\n ''')\n\n with self.assertRaisesRegex(RuntimeError, \"Syntax\"):\n cu = torch.jit.CompilationUnit('''\n def other_func(a):\n for i in range(5):\n def foo():\n break\n ''')\n\n with self.assertRaisesRegex(RuntimeError, \"do not support break or continue inside\"):\n @torch.jit.script\n def foo(x):\n i = 0\n for a in (1, \"2\", 1.5):\n b = a\n if x:\n break\n return b\n\n def test_python_call(self):\n def pyfunc(a):\n return a * 3.0\n\n cu = torch.jit.CompilationUnit('''\n def other_func(a):\n return a + a\n\n def test_call_python(a):\n b = pyfunc(a)\n b = other_func(b)\n i = 0\n step = 1\n while i < 10:\n b = pyfunc(b)\n if bool(b > 3.0):\n b = pyfunc(b)\n i = 11\n return b\n ''')\n inputs = self._make_scalar_vars([1], torch.float)\n outputs = self._make_scalar_vars([54], torch.float)\n\n self.assertEqual(cu.test_call_python(*inputs), outputs[0])\n\n def test_python_call_failure(self):\n with self.assertRaisesRegex(RuntimeError, \"undefined value pyfunc2\"):\n def pyfunc(a):\n return a * 3.0\n\n cu = torch.jit.CompilationUnit('''\n def other_func(a):\n return a + a\n\n def test_call_python(a):\n b = pyfunc(a)\n b = other_func(b)\n i = 0\n step = 1\n while i < 10:\n b = pyfunc2(b)\n if b > 3.0:\n b = pyfunc(b)\n i = 11\n return b\n ''')\n inputs = self._make_scalar_vars([1], torch.float)\n outputs = self._make_scalar_vars([54], torch.float)\n\n self.assertEqual(cu.test_call_python(*inputs), outputs)\n\n def test_type_call_in_script(self):\n @torch.jit.script\n def fn(x):\n return type(x)\n\n with self.assertRaisesRegex(RuntimeError, \"value of type _TensorMeta\"):\n fn(torch.tensor(.5))\n\n def test_python_call_annotation(self):\n def pyfunc(a):\n return a * 3.0\n\n @torch.jit.script\n def foo(a):\n return pyfunc(a) + pyfunc(a)\n\n inputs = self._make_scalar_vars([1], torch.float)\n outputs = self._make_scalar_vars([6], torch.float)\n self.assertEqual(foo(*inputs), outputs[0])\n\n def test_python_call_annoytation_failure(self):\n with self.assertRaisesRegex(RuntimeError, \"undefined value pyfunc2\"):\n def pyfunc(a):\n return a * 3.0\n\n @torch.jit.script\n def foo(a):\n return pyfunc2(a) + pyfunc(a)\n\n inputs = self._make_scalar_vars([1], torch.float)\n outputs = self._make_scalar_vars([6], torch.float)\n\n self.assertEqual(foo(*inputs), outputs[0])\n\n def test_desugar_module(self):\n import torch.nn.functional as F\n\n def fn(x, slope):\n a = torch.abs(x)\n b = torch.nn.functional.prelu(x, slope)\n c = F.prelu(x, slope)\n return a, b, c\n\n x = torch.arange(-3., 4)\n slope = torch.tensor([0.5])\n self.checkScript(fn, [x, slope], optimize=True)\n\n def test_script_docstring(self):\n @torch.jit.script\n def with_docstring(x):\n \"\"\"test str\"\"\"\n y = x\n \"\"\"y is the same as x\"\"\"\n return y\n self.assertEqual(with_docstring.__doc__, 'test str')\n\n def test_script_method_docstring(self):\n class A(torch.jit.ScriptModule):\n @torch.jit.script_method\n def with_docstring(self, x):\n \"\"\"test str\"\"\"\n y = x\n \"\"\"y is the same as x\"\"\"\n return y\n a = A()\n self.assertEqual(a.with_docstring.__doc__, 'test str')\n\n def test_script_module(self):\n class M1(torch.jit.ScriptModule):\n def __init__(self):\n super(M1, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class PModule(nn.Module):\n def __init__(self):\n super(PModule, self).__init__()\n self.a = nn.Parameter(torch.randn(2, 3))\n\n def forward(self, a):\n return self.a.mm(a)\n\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n # test submodule\n self.sub = M1()\n self.sub2 = PModule()\n # test parameters\n self.weight = nn.Parameter(torch.randn(2, 3))\n self.bias = nn.Parameter(torch.randn(2))\n # test defining a method from a string\n self.define(\"\"\"\n def hi(self, a):\n return self.weight.mm(a)\n \"\"\")\n # test script methods\n\n @torch.jit.script_method\n def doit(self, input):\n # test use of parameter\n return self.weight.mm(input)\n\n @torch.jit.script_method\n def doit2(self, input):\n return self.weight.mm(input)\n\n @torch.jit.script_method\n def forward(self, input):\n a = self.doit(input)\n b = self.doit2(input)\n c = self.hi(input)\n d = self.sub2(input)\n return a + b + self.bias + self.sub(a) + c + d\n with torch.jit.optimized_execution(False):\n m2 = M2()\n input = torch.randn(3, 2)\n a = m2.weight.mm(input)\n b = m2.weight.mm(input)\n c = m2.weight.mm(input)\n d = m2.sub2.a.mm(input)\n ref = a + b + m2.bias + m2.sub.weight + a + c + d\n self.assertEqual(ref, m2.forward(input))\n m2.weight = nn.Parameter(torch.zeros_like(m2.weight))\n m2.bias = nn.Parameter(torch.zeros_like(m2.bias))\n m2.sub.weight = nn.Parameter(torch.zeros_like(m2.sub.weight))\n m2.sub2.a.data.zero_()\n self.assertEqual(torch.zeros(2, 2), m2.forward(torch.randn(3, 2)))\n\n def test_irparser(self):\n graph_str = \"\"\"graph(%0 : Double(5, 5)):\n # CHECK: aten::relu\n %1 : Double(5, 5) = aten::relu(%0)\n return (%1)\n \"\"\"\n FileCheck().run(graph_str, parse_ir(graph_str))\n\n def test_is_after_use(self):\n def sorted_input_use(g):\n uses = list(next(g.inputs()).uses())\n return sorted(uses, key=functools.cmp_to_key(type(uses[0]).isAfter))\n\n @torch.jit.script\n def foo(x):\n a = x + 1\n return (x, x, a)\n\n uses_sorted = sorted_input_use(foo.graph)\n # sorts last use to the end\n self.assertFalse(uses_sorted[0].isAfter(uses_sorted[1]))\n self.assertTrue(uses_sorted[0].user.kind() == \"aten::add\")\n self.assertEqual(uses_sorted[1].offset, 0)\n\n @torch.jit.script\n def foo(x, cond: bool):\n if cond:\n return x + 3\n else:\n return x - 3\n\n uses_sorted = sorted_input_use(foo.graph)\n self.assertTrue(uses_sorted[0].user.kind() == \"aten::add\")\n self.assertTrue(uses_sorted[1].user.kind() == \"aten::sub\")\n\n @torch.jit.script\n def foo(x, cond: bool, cond2: bool):\n if cond:\n return x + 3\n elif cond2 :\n return x - 3\n\n return x / 3\n\n graph1 = foo.graph\n\n @torch.jit.script\n def foo(x, cond: bool, cond2: bool):\n if cond:\n return x + 3\n else:\n if cond2 :\n return x - 3\n return x / 3\n\n graph2 = foo.graph\n\n for graph in [graph1, graph2]:\n uses_sorted = sorted_input_use(graph)\n self.assertTrue(uses_sorted[0].user.kind() == \"aten::add\")\n self.assertTrue(uses_sorted[1].user.kind() == \"aten::sub\")\n self.assertTrue(uses_sorted[2].user.kind() == \"aten::div\")\n\n def test_canonicalize_control_outputs(self):\n def test_all_outputs(g):\n ifs = g.findAllNodes(\"prim::If\")\n loops = g.findAllNodes(\"prim::Loop\")\n\n def contained_blocks(node):\n return len(node.findAllNodes(\"prim::If\")) * 2 + len(node.findAllNodes(\"prim::Loop\"))\n for node in ifs + loops:\n outs = list(node.outputs())\n out_name = [x.debugName() for x in outs]\n if len(out_name) == 0:\n continue\n fc = FileCheck()\n # find the last output, then all subsequent uses\n fc.check(out_name[-1] + \" : \")\n # skip past node body\n for i in range(contained_blocks(node)):\n fc.check(\"->\")\n if (node.kind() == \"prim::If\"):\n fc.check(\"->\").check(\"->\").check(\"\\n\")\n else:\n fc.check(\"->\").check(\"\\n\")\n # the canonical order is the same order as the first use\n # appears in text\n for name in out_name:\n fc.check(name)\n fc.run(g)\n\n @torch.jit.script\n def test(x):\n # type: (bool) -> Tuple[int, int]\n b = 2\n a = 1\n if x:\n a = 1\n b = 2\n x = False\n if x:\n b = a\n else:\n a = b\n\n return a, b\n test_all_outputs(test.graph)\n\n @torch.jit.script\n def test2(x):\n # type: (bool) -> Tuple[int, int]\n b = 2\n a = 1\n if x:\n a = 1\n b = 2\n x = False\n if x:\n print(a)\n else:\n if x:\n print(b)\n\n return a, b\n test_all_outputs(test2.graph)\n\n @torch.jit.script\n def test_loop(x, iter):\n # type: (bool, int) -> (None)\n a = 1\n b = 2\n c = 3\n for i in range(iter):\n a = 4\n b = 5\n c = 6\n x = True\n print(c)\n if x:\n print(a, b)\n test_all_outputs(test_loop.graph)\n\n @torch.jit.script\n def loop_unused(iter):\n # type: (int) -> (None)\n a = 1\n b = 2\n c = 3\n for i in range(iter):\n c = c + 1\n b = b + 1\n a = a + 1\n print(a, b)\n print(c)\n\n # c is used, then unused should be ordered by alphabetical\n FileCheck().check(r\"%c : int, %a : int, %b : int\").run(loop_unused.graph)\n\n def test_filecheck(self):\n def test_check():\n file = \"232\"\n FileCheck().check(\"2\").check(\"3\").check(\"2\").run(file)\n FileCheck().check(\"232\").run(file)\n\n with self.assertRaisesRegex(RuntimeError, 'Expected to find \"22\"'):\n FileCheck().check(\"22\").run(file)\n with self.assertRaisesRegex(RuntimeError, \"CHECK: 3\"):\n FileCheck().check(\"3\").check(\"3\").run(file)\n\n test_check()\n\n def test_check_count():\n file = \"22222\"\n FileCheck().check_count(\"2\", 5).run(file)\n FileCheck().check_count(\"22\", 2).run(file)\n FileCheck().check_count(\"222\", 1).run(file)\n\n with self.assertRaisesRegex(RuntimeError, 'Expected to not find'):\n FileCheck().check_count(\"2\", 4, exactly=True).run(file)\n\n with self.assertRaisesRegex(RuntimeError, 'Expected to find \"22\"'):\n FileCheck().check_count(\"22\", 3).run(file)\n\n with self.assertRaisesRegex(RuntimeError, \"CHECK-COUNT-6: 2\"):\n FileCheck().check_count(\"2\", 6).run(file)\n\n test_check_count()\n\n def test_check_same():\n file = \"22\\n33\"\n FileCheck().check_same(\"22\").run(file)\n\n with self.assertRaisesRegex(RuntimeError, \"Expected to not find\"):\n FileCheck().check_same(\"33\").run(file)\n\n file = \"22 1 3\"\n\n FileCheck().check(\"2\").check_same(\"3\").run(file)\n FileCheck().check_count(\"2\", 2).check_same(\"3\").run(file)\n\n test_check_same()\n\n def test_check_next():\n file = \"\\n1\\n2\\n3\"\n FileCheck().check(\"1\").check_next(\"2\").check_next(\"3\").run(file)\n FileCheck().check_next(\"1\").check_next(\"2\").check_next(\"3\").run(file)\n\n with self.assertRaisesRegex(RuntimeError, \"Expected to find\"):\n FileCheck().check(\"1\").check_next(\"2\").run(\"12\")\n\n with self.assertRaisesRegex(RuntimeError, \"Expected to not find\"):\n FileCheck().check(\"1\").check_next(\"2\").run(\"1\\n\\n2\")\n\n test_check_next()\n\n def test_check_dag():\n fc = FileCheck().check_dag(\"1\").check_dag(\"2\").check_not(\"2\")\n fc.run(\"12\")\n fc.run(\"21\")\n\n fc = FileCheck()\n fc.check_not(\"3\").check_dag(\"1\").check_dag(\"2\").check_not(\"3\")\n fc.run(\"1 3 2\")\n fc.run(\"2 3 1\")\n\n fc = FileCheck().check_dag(\"1\").check_dag(\"2\").check(\"3\")\n with self.assertRaisesRegex(RuntimeError, 'Expected to find \"3\" but did not find it'):\n fc.run(\"1 3 2\")\n\n test_check_dag()\n\n def test_check_not():\n FileCheck().check_not(\"2\").check(\"1\").run(\"12\")\n FileCheck().check(\"2\").check_not(\"2\").run(\"12\")\n\n with self.assertRaisesRegex(RuntimeError, 'Expected to not find \"2\"'):\n FileCheck().check_not(\"2\").check(\"1\").run(\"21\")\n\n with self.assertRaisesRegex(RuntimeError, 'Expected to not find \"1\"'):\n FileCheck().check(\"2\").check_not(\"1\").run(\"21\")\n\n # checks with distinct range matchings\n fb = FileCheck().check_count(\"2\", 2).check_count(\"2\", 2).check_not(\"2\")\n with self.assertRaisesRegex(RuntimeError, 'Expected to not find \"2\"'):\n fb.run(\"22 2 22\")\n\n fb = FileCheck().check_count(\"2\", 2).check_not(\"1\").check_count(\"2\", 2)\n with self.assertRaisesRegex(RuntimeError, 'Expected to not find \"1\"'):\n fb.run(\"22 1 22\")\n\n def _dtype_to_jit_name(self, dtype):\n if(dtype == torch.float32):\n return \"Float\"\n if(dtype == torch.float64):\n return \"Double\"\n if(dtype == torch.int64):\n return \"Long\"\n if(dtype == torch.int32):\n return \"Int\"\n if(dtype == torch.bool):\n return \"Bool\"\n raise RuntimeError('dtype not handled')\n\n def _dtype_to_expect(self, dtype, dim=0):\n param = ', '.join(['*'] * dim + ['device=cpu'])\n param = '(' + param + ')'\n jit_type = self._dtype_to_jit_name(dtype)\n if dim >= 0:\n return jit_type + param\n # special case representing wrapped number\n else:\n return jit_type.lower()\n\n\n def _test_dtype_op_shape(self, ops, args, input_dims=1):\n if input_dims < 1:\n raise RuntimeError(\"input dims must be at least 1\")\n dtypes = [torch.float32, torch.float64, torch.int64, torch.int32]\n str_args = ', '.join([str(arg) for arg in args]) + (', ' if len(args) else '')\n tensor_data = ('[' * input_dims) + '1, 2, 3' + (input_dims * ']')\n template = dedent('''\n def func():\n return {return_line}\n ''')\n\n for op in ops:\n for dtype in (dtypes + [None]):\n for tensor_type in dtypes:\n # a couple of ops aren't implemented for non-floating types\n if(not tensor_type.is_floating_point or (dtype is not None and not dtype.is_floating_point)):\n if op in ['mean', 'softmax', 'log_softmax']:\n continue\n return_line = \"torch.tensor({}, dtype={}).{}({}dtype={})\".format(tensor_data, tensor_type, op, str_args, dtype)\n # uncomment for debugging a failed test:\n # print(\"testing {}\".format(return_line))\n code = template.format(return_line=return_line)\n scope = {}\n exec(code, globals(), scope)\n cu = torch.jit.CompilationUnit(code)\n graph = cu.func.graph\n torch._C._jit_pass_complete_shape_analysis(graph, (), False)\n input_array = [1, 2, 3]\n for _ in range(1, input_dims):\n input_array = [input_array]\n t = torch.tensor(input_array, dtype=tensor_type)\n attr = getattr(t, op)\n kwargs = {'dtype': dtype}\n result = attr(*args, **kwargs)\n expect = self._dtype_to_expect(result.dtype, result.dim())\n FileCheck().check(\"aten::tensor\").check(expect).run(graph)\n\n def test_dtype_op_shape(self):\n ops = ['prod']\n self._test_dtype_op_shape(ops, args=[])\n self._test_dtype_op_shape(ops, args=[0, False])\n self._test_dtype_op_shape(ops, args=[0, False])\n self._test_dtype_op_shape(ops, args=[0, True])\n\n def test_dtype_op_shape2(self):\n ops = ['cumprod', 'cumsum', 'softmax', 'log_softmax']\n self._test_dtype_op_shape(ops, args=[0])\n\n self._test_dtype_op_shape(ops, args=[1], input_dims=4)\n\n\n def _test_binary_op_shape(self, ops, input_dims=1):\n\n dtypes = [torch.float32, torch.float64, torch.int64, torch.int32, torch.bool]\n\n if input_dims == 0:\n shape = '1'\n else:\n shape = '[' + ('1,' * 4) + ']'\n for _ in range(1, input_dims):\n shape = '[' + \",\".join([shape] * 4) + ']'\n\n template = dedent('''\n def func():\n arg1 = {}\n arg2 = {}\n return torch.{}(arg1, arg2)\n ''')\n\n args = []\n for dtype in dtypes:\n args = args + [\"torch.tensor({}, dtype={})\".format(shape, dtype)]\n args = args + [1, 1.5]\n\n def isBool(arg):\n return type(arg) == bool or (type(arg) == str and \"torch.bool\" in arg)\n\n for op in ops:\n for first_arg in args:\n for second_arg in args:\n # subtract not supported for bool\n if (op == 'sub' or op == 'div') and (isBool(first_arg) or isBool(second_arg)):\n continue\n # div is not implemented correctly for mixed-type or int params\n if (op == 'div' and (type(first_arg) != type(second_arg) or\n isinstance(first_arg, int) or\n (isinstance(first_arg, str) and 'int' in first_arg))):\n continue\n return_line = \"torch.{}({}, {})\".format(op, first_arg, second_arg)\n # uncomment for debugging a failed test:\n # print(\"testing {}\".format(return_line))\n code = template.format(first_arg, second_arg, op)\n scope = {}\n exec(code, globals(), scope)\n non_jit_result = scope['func']()\n\n cu = torch.jit.CompilationUnit(code)\n graph = cu.func.graph\n torch._C._jit_pass_complete_shape_analysis(graph, (), False)\n # use dim=-1 to represent a python/jit scalar.\n dim = -1 if type(first_arg) != str and type(second_arg) != str else non_jit_result.dim()\n dtype = non_jit_result.dtype\n # jit only supports int/float scalars.\n if dim < 0:\n if dtype == torch.int64:\n dtype = torch.int32\n if dtype == torch.float64:\n dtype = torch.float32\n expect = self._dtype_to_expect(dtype, dim)\n jit_output = next(graph.outputs())\n\n check = FileCheck()\n check.check(expect).run(str(jit_output))\n\n def test_binary_op_shape(self):\n self._test_binary_op_shape(['mul', 'div', 'add', 'sub'], 0)\n self._test_binary_op_shape(['mul', 'div', 'add', 'sub'], 3)\n\n def test_no_dtype_shape(self):\n\n @torch.jit.script\n def foo(x):\n scalar_number = x.item()\n return x.add(scalar_number)\n\n @torch.jit.script\n def foo2(x):\n scalar_number = x.item()\n return torch.tensor(1).add(scalar_number)\n\n t = torch.tensor(5)\n g = foo.graph_for(t)\n type = next(g.outputs())\n self.assertTrue(type.type() == torch._C.TensorType.get())\n g2 = foo2.graph_for(t)\n type = next(g.outputs())\n self.assertTrue(type.type() == torch._C.TensorType.get())\n\n\n def test_filecheck_parse(self):\n def test_check():\n file = \"\"\"\n # CHECK: 2\n # CHECK: 3\n # CHECK: 2\n 232\n \"\"\"\n FileCheck().run(checks_file=file, test_file=file)\n file = \"\"\"\n # CHECK: 232\n 232\n \"\"\"\n FileCheck().run(file, \"232\")\n with self.assertRaisesRegex(RuntimeError, 'Expected to find \"232\"'):\n FileCheck().run(file, \"22\")\n with self.assertRaisesRegex(RuntimeError, 'Expected to find \"22\"'):\n FileCheck().run(\"# CHECK: 22\", \"23\")\n test_check()\n\n def test_check_count():\n file = \"22222\"\n FileCheck().run(\"# CHECK-COUNT-5: 2\", file)\n FileCheck().run(\"# CHECK-COUNT-EXACTLY-5: 2\", file)\n FileCheck().run(\"# CHECK-COUNT-2: 22\", file)\n FileCheck().run(\"# CHECK-COUNT-1: 222\", file)\n\n with self.assertRaisesRegex(RuntimeError, 'Expected to not find'):\n FileCheck().run(\"# CHECK-COUNT-EXACTLY-2: 2\", file)\n test_check_count()\n\n def test_check_same():\n file = \"22\\n33\"\n FileCheck().run(\"# CHECK-SAME: 22\", file)\n\n with self.assertRaisesRegex(RuntimeError, \"Expected to not find\"):\n FileCheck().run(\"# CHECK-SAME: 33\", file)\n\n file = \"22 1 3\"\n\n FileCheck().run(\"# CHECK: 2\\n # CHECK-SAME: 3\", file)\n FileCheck().run(\"# CHECK-COUNT-2: 2\\n # CHECK-SAME: 3\", file)\n test_check_same()\n\n def test_bad_input():\n with self.assertRaisesRegex(RuntimeError, \"Check for bad input\"):\n FileCheck().run(\"\", \"1\")\n\n with self.assertRaisesRegex(RuntimeError, \"Could not parse check\"):\n FileCheck().run(\"# CHECK1\", \"\")\n\n test_bad_input()\n\n def test_script_module_call_noscript(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.value = 1\n\n @torch.jit.ignore\n def foo(self):\n return torch.ones(2, 2) + self.value\n\n @torch.jit.script_method\n def forward(self, input):\n return input + self.foo()\n\n with torch.jit.optimized_execution(False):\n m = M()\n input = torch.randn(2, 2)\n o = m(input)\n self.assertEqual(o, input + torch.ones(2, 2) + 1)\n # check that we can change python attributes\n # and that those changes are picked up in script methods\n m.value = 2\n o = m(input)\n self.assertEqual(o, input + torch.ones(2, 2) + 2)\n\n def test_script_module_nochange_submodule(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.sub = nn.Linear(5, 5)\n\n @torch.jit.script_method\n def forward(self, input):\n return self.sub(input)\n with torch.jit.optimized_execution(False):\n m = M()\n input = torch.randn(1, 5, 5)\n o = m(input)\n self.assertEqual(o, m.sub(input))\n with self.assertRaisesRegex(RuntimeError, \"Cannot re-assign\"):\n m.sub = nn.Linear(5, 5)\n\n def test_module_apis(self):\n class Sub(torch.nn.Module):\n def __init__(self):\n super(Sub, self).__init__()\n\n def forward(self, thing):\n return thing - 2\n\n class Double(torch.nn.Module):\n def __init__(self):\n super(Double, self).__init__()\n\n def forward(self, thing):\n return thing * 2\n\n class MyMod(torch.nn.Module):\n def __init__(self):\n super(MyMod, self).__init__()\n self.mod = (Sub())\n self.mod2 = (Sub())\n self.mod3 = nn.Sequential(nn.Sequential(Sub()))\n self.mod4 = nn.Sequential(Sub(), Double())\n\n @torch.jit.export\n def method(self, x, x1, y, y1):\n mod_names = \"\"\n for name, mod in self.named_modules():\n mod_names = mod_names + \" \" + name\n x = mod(x)\n\n children_names = \"\"\n for name, mod in self.named_children():\n children_names = children_names + \" \" + name\n x1 = mod(x1)\n\n for mod in self.modules():\n y = mod(y)\n\n for mod in self.children():\n y1 = mod(y1)\n\n return mod_names, children_names, x, x1, y, y1\n\n def forward(self, x):\n return x + 2\n\n mod = torch.jit.script(MyMod())\n inps = tuple([torch.tensor(i) for i in range(1, 5)])\n self.assertEqual(mod.method(*inps), MyMod().method(*inps))\n\n def test_script_module_const(self):\n class M(torch.jit.ScriptModule):\n\n __constants__ = ['b', 'i', 'c', 's']\n\n def __init__(self):\n super(M, self).__init__()\n self.b = False\n self.i = 1\n self.c = 3.5\n self.s = [\"hello\"]\n\n @torch.jit.script_method\n def forward(self):\n return self.b, self.i, self.c\n\n with torch.jit.optimized_execution(False):\n m = M()\n o0, o1, o2 = m()\n self.assertEqual(o0, 0)\n self.assertEqual(o1, 1)\n self.assertEqual(o2, 3.5)\n\n def test_script_module_fail_exist(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x):\n return x + self.whatisgoingon\n with self.assertRaisesRegex(RuntimeError, \"Module 'M' has no attribute\"):\n M()\n\n @unittest.skip(\"[module dedupe] currently NoneType refinement on optional attributes doesn't work.\")\n def test_script_module_none_exist_fail(self):\n class M(torch.jit.ScriptModule):\n def __init__(self, my_optional):\n super(M, self).__init__()\n self.my_optional = my_optional\n\n @torch.jit.script_method\n def forward(self, x):\n if self.my_optional is not None:\n return torch.neg(x) + self.my_optional\n return torch.neg(x)\n with self.assertRaisesRegex(RuntimeError, \"has no attribute 'my_optional'\"):\n x = torch.rand(3, 4)\n fb = M(None)\n fb(x)\n\n def test_script_module_invalid_consts(self):\n class Foo(torch.jit.ScriptModule):\n __constants__ = ['invalid']\n\n def __init__(self):\n super(Foo, self).__init__()\n self.invalid = [nn.Linear(3, 4)]\n\n with self.assertRaisesRegex(\n TypeError,\n \"Linear' object in attribute 'Foo.invalid' is not a valid constant\"):\n Foo()\n\n class Foo2(torch.jit.ScriptModule):\n __constants__ = ['invalid']\n\n def __init__(self):\n super(Foo2, self).__init__()\n self.invalid = type(1)\n\n with self.assertRaisesRegex(TypeError, \"not a valid constant\"):\n Foo2()\n\n class Foo3(torch.jit.ScriptModule):\n __constants__ = ['invalid']\n\n def __init__(self):\n super(Foo3, self).__init__()\n self.invalid = (3, 4, {})\n\n with self.assertRaisesRegex(TypeError, \"not a valid constant\"):\n Foo3()\n\n class Foo4(torch.jit.ScriptModule):\n __constants__ = ['invalid']\n\n def __init__(self):\n super(Foo4, self).__init__()\n self.invalid = np.int64(5)\n\n # verify that we capture human understandable class name\n with self.assertRaisesRegex(TypeError, \"numpy.int64\"):\n Foo4()\n\n def test_script_module_param_buffer_mutation(self):\n # TODO: add param mutation test case after JIT support it\n class ModuleBufferMutate(torch.jit.ScriptModule):\n def __init__(self):\n super(ModuleBufferMutate, self).__init__()\n self.register_buffer('running_var', torch.tensor(0, dtype=torch.long))\n\n @torch.jit.script_method\n def forward(self):\n if self.training:\n self.running_var += 1\n return self.running_var\n\n with torch.jit.optimized_execution(False):\n m = ModuleBufferMutate()\n self.assertEqual(m(), 1)\n m.eval()\n self.assertEqual(m(), 1)\n\n def test_script_module_for(self):\n class M(torch.jit.ScriptModule):\n __constants__ = ['b']\n\n def __init__(self):\n super(M, self).__init__()\n self.b = [1, 2, 3, 4]\n\n @torch.jit.script_method\n def forward(self):\n sum = 0\n for i in self.b:\n sum += i\n return sum\n\n with torch.jit.optimized_execution(False):\n m = M()\n self.assertEqual(m(), 10)\n\n def test_override_magic(self):\n class OverrideMagic(nn.Module):\n def __init__(self):\n super(OverrideMagic, self).__init__()\n\n @torch.jit.export\n def __len__(self):\n return 10\n\n mod = OverrideMagic()\n self.assertEqual(len(mod), len(torch.jit.script(mod)))\n\n class OverrideMagicSeq(nn.Sequential):\n def __init__(self):\n super(OverrideMagicSeq, self).__init__()\n\n @torch.jit.export\n def __len__(self):\n return 10\n\n mod = OverrideMagicSeq()\n self.assertEqual(len(mod), len(torch.jit.script(mod)))\n self.assertTrue(torch.jit.script(mod))\n\n def test_script_module_for2(self):\n class Sub(torch.jit.ScriptModule):\n def __init__(self):\n super(Sub, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.mods = nn.ModuleList([Sub() for i in range(10)])\n\n @torch.jit.script_method\n def forward(self, v):\n for m in self.mods:\n v = m(v)\n return v\n\n with torch.jit.optimized_execution(False):\n i = torch.empty(2)\n m = M()\n o = m(i)\n v = i\n for sub in m.mods:\n v = sub(v)\n self.assertEqual(o, v)\n with self.assertRaisesRegex(Exception, \"object is not iterable\"):\n print(list(m))\n\n def test_attr_qscheme_script(self):\n class Foo(torch.nn.Module):\n def __init__(self):\n super(Foo, self).__init__()\n self.qscheme = torch.per_tensor_affine\n\n def forward(self):\n if self.qscheme == torch.per_tensor_symmetric:\n return 3\n else:\n return 4\n\n f = Foo()\n scripted = torch.jit.script(f)\n self.assertEqual(f(), scripted())\n\n def test_script_module_const_submodule_fail(self):\n class Sub(torch.jit.ScriptModule):\n def __init__(self):\n super(Sub, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.mods = [Sub() for _ in range(10)]\n\n @torch.jit.script_method\n def forward(self):\n for _ in self.mods:\n print(1)\n return 4\n\n with self.assertRaisesRegex(RuntimeError, \"has no attribute 'mods'\"):\n M()\n\n class DerivedStateModule(torch.jit.ScriptModule):\n def __init__(self):\n super(TestScript.DerivedStateModule, self).__init__()\n self.param = torch.nn.Parameter(torch.ones(3, 4, dtype=torch.float))\n self.register_buffer('derived', torch.neg(self.param).detach().clone())\n\n # This is a flag so we can test that the pack method was called\n self.register_buffer('pack_called', torch.zeros(1, dtype=torch.long))\n # This is a flag so we can test that the unpack method was called\n self.register_buffer('unpack_called', torch.zeros(1, dtype=torch.long))\n\n @torch.jit.script_method\n def _pack(self):\n self.pack_called.set_(torch.ones(1, dtype=torch.long))\n self.derived.set_(torch.rand(1, dtype=torch.float).detach())\n\n @torch.jit.script_method\n def _unpack(self):\n self.unpack_called.set_(torch.ones(1, dtype=torch.long))\n self.derived.set_(torch.neg(self.param).detach())\n\n @torch.jit.script_method\n def forward(self, x):\n return x + self.derived\n\n def test_pack_unpack_state(self):\n sm = TestScript.DerivedStateModule()\n x = torch.rand(3, 4, dtype=torch.float)\n torch.testing.assert_allclose(sm(x), x + torch.neg(torch.ones(3, 4, dtype=torch.float)))\n\n # Test save path\n self.assertFalse(sm.pack_called.item())\n self.assertFalse(sm.unpack_called.item())\n imported = self.getExportImportCopyWithPacking(sm)\n # ensure pack was called before serialization\n self.assertTrue(sm.pack_called.item())\n # ensure unpack was called after serialization so as to leave the module in an initialized state\n self.assertTrue(sm.unpack_called.item())\n\n torch.testing.assert_allclose(sm.derived, torch.neg(sm.param))\n\n # Test load paths\n self.assertTrue(imported.unpack_called.item())\n torch.testing.assert_allclose(imported(x), x + torch.neg(torch.ones(3, 4, dtype=torch.float)))\n\n @unittest.skipIf(not TEST_MKL, \"PyTorch is built without MKL support\")\n def test_torch_functional(self):\n def stft(input, n_fft):\n # type: (Tensor, int) -> Tensor\n return torch.stft(input, n_fft, return_complex=True)\n\n inps = (torch.randn(10), 7)\n self.assertEqual(stft(*inps), torch.jit.script(stft)(*inps))\n\n def istft(input, n_fft):\n # type: (Tensor, int) -> Tensor\n return torch.istft(input, n_fft)\n\n inps2 = (stft(*inps), inps[1])\n self.assertEqual(istft(*inps2), torch.jit.script(istft)(*inps2))\n\n def lu(x):\n # type: (Tensor) -> Tuple[Tensor, Tensor]\n return torch.lu(x)\n\n self.checkScript(lu, (torch.randn(2, 3, 3),))\n\n def lu_infos(x):\n # type: (Tensor) -> Tuple[Tensor, Tensor, Tensor]\n return torch.lu(x, get_infos=True)\n\n self.checkScript(lu_infos, (torch.randn(2, 3, 3),))\n\n def lu_unpack(x):\n A_LU, pivots = torch.lu(x)\n return torch.lu_unpack(A_LU, pivots)\n\n for shape in ((3, 3), (5, 3, 3), (7, 3, 5, 5), (7, 5, 3, 3, 3)):\n a = torch.randn(*shape)\n self.checkScript(lu_unpack, (a,))\n\n def cdist_fn():\n a = torch.tensor([[0.9041, 0.0196], [-0.3108, -2.4423], [-0.4821, 1.059]])\n b = torch.tensor([[-2.1763, -0.4713], [-0.6986, 1.3702]])\n return torch.cdist(a, b, compute_mode=\"use_mm_for_euclid_dist\")\n\n self.checkScript(cdist_fn, ())\n\n def norm():\n c = torch.tensor([[1, 2, 3], [-1, 1, 4]], dtype=torch.float)\n return torch.norm(c, p=\"fro\"), torch.norm(c, p=\"nuc\"), torch.norm(c), torch.norm(c, p=.5)\n\n self.checkScript(norm, ())\n\n def torch_unique(dim: Optional[int]):\n ten = torch.unique(torch.tensor([[1, 3], [2, 3]], dtype=torch.long))\n a = torch.unique(ten, dim=dim)\n b = torch.unique(ten, return_counts=True, dim=dim)\n c = torch.unique(ten, return_inverse=True, dim=dim)\n d = torch.unique(ten, return_counts=True, return_inverse=True, dim=dim)\n return a, b, c, d\n\n self.checkScript(torch_unique, (None,))\n self.checkScript(torch_unique, (0,))\n\n def torch_unique_consecutive(dim: Optional[int]):\n ten = torch.unique(torch.tensor([[1, 3], [3, 2], [3, 2], [2, 3]], dtype=torch.long))\n a = torch.unique_consecutive(ten, dim=dim)\n b = torch.unique_consecutive(ten, return_counts=True, dim=dim)\n c = torch.unique_consecutive(ten, return_inverse=True, dim=dim)\n d = torch.unique_consecutive(ten, return_counts=True, return_inverse=True, dim=dim)\n return a, b, c, d\n\n self.checkScript(torch_unique_consecutive, (None,))\n self.checkScript(torch_unique_consecutive, (0,))\n\n def test_torch_functional_tensordot_int(self):\n def tensordot_dims_int(a: torch.Tensor, b: torch.Tensor, dims: int):\n return torch.tensordot(a, b, dims=dims)\n\n a = torch.arange(120.).reshape(2, 3, 4, 5)\n b = torch.arange(840.).reshape(4, 5, 6, 7)\n dims = 2\n self.checkScript(tensordot_dims_int, (a, b, dims))\n\n def test_torch_functional_tensordot_tensor(self):\n def tensordot_dims_tensor(a: torch.Tensor, b: torch.Tensor, dims: torch.Tensor):\n return torch.tensordot(a, b, dims=dims)\n\n a = torch.arange(120.).reshape(2, 3, 4, 5)\n b = torch.arange(840.).reshape(4, 5, 6, 7)\n dims = torch.tensor([2])\n self.checkScript(tensordot_dims_tensor, (a, b, dims))\n\n a = torch.arange(60.).reshape(3, 4, 5)\n b = torch.arange(24.).reshape(4, 3, 2)\n dims = torch.tensor([[1, 0], [0, 1]], dtype=torch.long)\n self.checkScript(tensordot_dims_tensor, (a, b, dims))\n\n def test_torch_functional_tensordot_list(self):\n def tensordot_dims_list(a: torch.Tensor, b: torch.Tensor, dims: List[List[int]]):\n return torch.tensordot(a, b, dims=dims)\n\n a = torch.arange(60.).reshape(3, 4, 5)\n b = torch.arange(24.).reshape(4, 3, 2)\n dims = [[1, 0], [0, 1]]\n self.checkScript(tensordot_dims_list, (a, b, dims))\n\n def test_torch_functional_tensordot_tuple(self):\n def tensordot_dims_tuple(a: torch.Tensor, b: torch.Tensor, dims: Tuple[List[int], List[int]]):\n return torch.tensordot(a, b, dims=dims)\n\n a = torch.arange(60.).reshape(3, 4, 5)\n b = torch.arange(24.).reshape(4, 3, 2)\n dims = ([1, 0], [0, 1])\n self.checkScript(tensordot_dims_tuple, (a, b, dims))\n\n def test_missing_getstate(self):\n class Foo(torch.nn.Module):\n def __init__(self):\n super(Foo, self).__init__()\n self.x = 1\n\n def forward(self, x):\n return x * self.x\n\n @torch.jit.export\n def __setstate__(self, state):\n self.x = state[0]\n self.training = state[1]\n\n with self.assertRaisesRegex(RuntimeError, \"getstate\"):\n scripted = torch.jit.script(Foo())\n\n def test_inlining_cleanup(self):\n def foo(x):\n return F.linear(x, x)\n\n @torch.jit.script\n def fee(x):\n return foo(x)\n\n # inlining optimizations should have cleaned up linear if statement\n self.run_pass(\"inline\", fee.graph)\n FileCheck().check_not(\"prim::If\").run(fee.graph)\n\n def test_pack_unpack_nested(self):\n class SubSubMod(torch.jit.ScriptModule):\n def __init__(self):\n super(SubSubMod, self).__init__()\n self.register_buffer('buf', torch.ones(3, 4) * 3)\n\n @torch.jit.script_method\n def _pack(self):\n self.buf.set_(torch.zeros(1, dtype=torch.double))\n\n @torch.jit.script_method\n def _unpack(self):\n self.buf.set_(torch.ones(3, 4, dtype=torch.double) * 3)\n\n @torch.jit.script_method\n def forward(self, x):\n return x + self.buf\n\n class SubMod(torch.jit.ScriptModule):\n def __init__(self):\n super(SubMod, self).__init__()\n self.register_buffer('buf', torch.ones(3, 4) * 2)\n self.ssm = SubSubMod()\n\n @torch.jit.script_method\n def _pack(self):\n self.buf.set_(torch.zeros(1, dtype=torch.double))\n\n @torch.jit.script_method\n def _unpack(self):\n self.buf.set_(torch.ones(3, 4, dtype=torch.double) * 2)\n\n @torch.jit.script_method\n def forward(self, x):\n return self.ssm(x + self.buf)\n\n class Mod(torch.jit.ScriptModule):\n def __init__(self):\n super(Mod, self).__init__()\n self.submod = SubMod()\n self.register_buffer('buf', torch.ones(3, 4) * 1)\n\n @torch.jit.script_method\n def _pack(self):\n self.buf.set_(torch.zeros(1, dtype=torch.double))\n\n @torch.jit.script_method\n def _unpack(self):\n self.buf.set_(torch.ones(3, 4, dtype=torch.double))\n\n @torch.jit.script_method\n def forward(self, x):\n return self.submod(x + self.buf)\n\n m = Mod()\n torch.testing.assert_allclose(m(torch.zeros(3, 4)), torch.ones(3, 4) * 6)\n m.apply(lambda s: s._pack())\n torch.testing.assert_allclose(m(torch.zeros(3, 4)), torch.zeros(3, 4))\n m.apply(lambda s: s._unpack())\n torch.testing.assert_allclose(m(torch.zeros(3, 4)), torch.ones(3, 4) * 6)\n\n def test_torch_any(self):\n def fn(x):\n return torch.any(x)\n\n def fn1(x, dim: int):\n return torch.any(x, dim)\n\n self.checkScript(fn, (torch.randn(3, 4), ))\n self.checkScript(fn, (torch.empty(3), ))\n self.checkScript(fn, (torch.empty(1), ))\n self.checkScript(fn, (torch.ones(3, 4),))\n self.checkScript(fn, (torch.zeros(5, 7, 1),))\n self.checkScript(fn1, (torch.empty(3, 4), -2))\n self.checkScript(fn1, (torch.randn(3, 8), 1))\n self.checkScript(fn1, (torch.zeros(3, 6, 9), -3))\n self.checkScript(fn1, (torch.empty(5), 0))\n\n def test_any(self):\n def fn(x: List[int]):\n return any(x)\n\n def fn1(x: List[float]):\n return any(x)\n\n def fn2(x: List[bool]):\n return any(x)\n\n def fn3(x: List[str]):\n return any(x)\n\n self.checkScript(fn, ([0, 0, 0, 0], ))\n self.checkScript(fn, ([0, 3, 0], ))\n self.checkScript(fn, ([], ))\n self.checkScript(fn1, ([1.0, 2.0, 3.0], ))\n self.checkScript(fn1, ([0.0, 0.0, 0.0], ))\n self.checkScript(fn1, ([0, 0, 0], ))\n self.checkScript(fn1, ([], ))\n self.checkScript(fn2, ([True, False, False], ))\n self.checkScript(fn2, ([False, False, False], ))\n self.checkScript(fn2, ([True, True, True, True], ))\n self.checkScript(fn2, ([], ))\n self.checkScript(fn3, ([\"\", \"\", \"\"], ))\n self.checkScript(fn3, ([\"\", \"\", \"\", \"-1\"], ))\n self.checkScript(fn3, ([], ))\n\n def test_script_module_not_tuple(self):\n class M(torch.jit.ScriptModule):\n __constants__ = ['mods']\n\n def __init__(self):\n super(M, self).__init__()\n self.mods = 1\n\n @torch.jit.script_method\n def forward(self, v):\n for m in self.mods:\n print(m)\n return v\n with self.assertRaisesRegex(RuntimeError, \"'int' object is not iterable\"):\n M()\n\n def test_attr_module_constants(self):\n class M2(torch.jit.ScriptModule):\n def __init__(self, mod_list):\n super(M2, self).__init__()\n self.mods = mod_list\n\n @torch.jit.script_method\n def forward(self, x):\n return self.mods.forward(x)\n\n with torch.jit.optimized_execution(False):\n m = M2(nn.Sequential(nn.ReLU()))\n self.assertExportImportModule(m, (torch.randn(2, 2),))\n\n def test_script_sequential_for(self):\n class Sub(torch.jit.ScriptModule):\n def __init__(self):\n super(Sub, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.mods = nn.Sequential(Sub(), Sub(), Sub())\n\n @torch.jit.script_method\n def forward(self, v):\n for m in self.mods:\n v = m(v)\n return v\n\n @torch.jit.script_method\n def forward2(self, v):\n return self.mods(v)\n\n with torch.jit.optimized_execution(False):\n i = torch.empty(2)\n m = M()\n o = m(i)\n v = i\n for sub in m.mods._modules.values():\n v = sub(v)\n self.assertEqual(o, v)\n\n o2 = m.forward2(i)\n self.assertEqual(o2, v)\n\n def test_script_sequential_sliced_iteration(self):\n class seq_mod(nn.Module):\n def __init__(self):\n super(seq_mod, self).__init__()\n self.layers = [nn.ReLU(), nn.ReLU(), nn.ReLU()]\n self.layers = nn.Sequential(*self.layers)\n\n def forward(self, input):\n x = self.layers[0].forward(input)\n for layer in self.layers[1:3]:\n x = layer.forward(x)\n for layer in self.layers[2:]:\n x = layer.forward(x)\n return x\n\n seq = seq_mod()\n self.checkModule(seq, [torch.tensor([-2, 1, -1, 2])])\n\n def test_script_sequential_orderdict(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.mods = nn.Sequential(OrderedDict([\n (\"conv\", nn.Conv2d(1, 20, 5)),\n (\"relu\", nn.ReLU())\n ]))\n\n @torch.jit.script_method\n def forward(self, input):\n return self.mods(input)\n\n m = M()\n self.assertTrue('mods.conv.weight' in m.state_dict().keys())\n\n def test_script_sequential_multi_output_fail(self):\n class Sub(torch.jit.ScriptModule):\n def __init__(self):\n super(Sub, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class ReturnMulti(torch.jit.ScriptModule):\n def __init__(self):\n super(ReturnMulti, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x):\n return x, x, x\n\n class HaveSequential(torch.jit.ScriptModule):\n def __init__(self):\n super(HaveSequential, self).__init__()\n self.someseq = nn.Sequential(\n Sub(),\n ReturnMulti(),\n Sub()\n )\n\n @torch.jit.script_method\n def forward(self, x):\n return self.someseq(x)\n\n with self.assertRaisesRegex(RuntimeError, \"(Tensor, Tensor, Tensor)\"):\n with torch.jit.optimized_execution(False):\n hs = HaveSequential()\n i = torch.empty(2)\n hs(i)\n\n @_tmp_donotuse_dont_inline_everything\n def test_script_sequential_in_mod_list(self):\n class Sub(torch.jit.ScriptModule):\n def __init__(self):\n super(Sub, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.mods = nn.ModuleList([Sub(), nn.Sequential(Sub(), nn.Sequential(Sub(), Sub()), Sub())])\n\n @torch.jit.script_method\n def forward(self, v):\n for mod in self.mods:\n v = mod(v)\n return v\n\n m = M()\n graph = str(m.graph)\n self.assertTrue(graph.count(\"prim::CallMethod\") == 2)\n self.assertTrue(\"python\" not in graph)\n\n @_tmp_donotuse_dont_inline_everything\n def test_script_nested_mod_list(self):\n class Sub(torch.jit.ScriptModule):\n def __init__(self):\n super(Sub, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.mods = nn.ModuleList([nn.ModuleList([Sub()]), nn.Sequential(Sub()), nn.ModuleList([Sub(), Sub()])])\n\n @torch.jit.script_method\n def forward(self, v):\n for mod in self.mods:\n for m in mod:\n v = m(v)\n return v\n\n m = M()\n graph = str(m.graph)\n self.assertTrue(graph.count(\"prim::CallMethod\") == 4)\n self.assertTrue(\"python\" not in graph)\n\n def test_constant_as_attr(self):\n class M(torch.jit.ScriptModule):\n __constants__ = ['dim']\n\n def __init__(self):\n super(M, self).__init__()\n self.dim = 1\n\n @torch.jit.script_method\n def forward(self, v):\n return torch.cat([v, v, v], dim=self.dim)\n v = torch.zeros(1, 1)\n with torch.jit.optimized_execution(False):\n self.assertEqual(torch.cat([v, v, v], dim=1), M()(v))\n\n class StarTestSumStarred(torch.nn.Module):\n def __init__(self):\n super(TestScript.StarTestSumStarred, self).__init__()\n\n def forward(self, *inputs):\n output = inputs[0]\n for i in range(1, len(inputs)):\n output += inputs[i]\n return output\n\n class StarTestReturnThree(torch.nn.Module):\n def __init__(self):\n super(TestScript.StarTestReturnThree, self).__init__()\n\n def forward(self, rep):\n return rep, rep, rep\n\n def test_script_star_expr(self):\n\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n self.m = torch.jit.trace(TestScript.StarTestSumStarred(),\n (torch.ones(4, 3), torch.ones(4, 3), torch.ones(4, 3)))\n self.g = torch.jit.trace(TestScript.StarTestReturnThree(), torch.ones(4, 3))\n\n @torch.jit.script_method\n def forward(self, rep):\n tup = self.g(rep)\n return self.m(*tup)\n\n m = M2()\n self.assertEqual(m(torch.zeros(4, 3)), 3 * torch.zeros(4, 3))\n\n def test_script_star_expr_string(self):\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n self.m = torch.jit.trace(TestScript.StarTestSumStarred(),\n (torch.ones(4, 3), torch.ones(4, 3), torch.ones(4, 3)))\n self.g = torch.jit.trace(TestScript.StarTestReturnThree(), torch.ones(4, 3))\n\n self.define('''\n def forward(self, rep):\n tup = self.g(rep)\n return self.m(*tup)\n ''')\n\n m = M2()\n self.assertEqual(m(torch.zeros(4, 3)), 3 * torch.zeros(4, 3))\n\n class StarTestSumAndReturnThree(torch.nn.Module):\n def __init__(self):\n super(TestScript.StarTestSumAndReturnThree, self).__init__()\n\n def forward(self, *inputs):\n output = inputs[0]\n for i in range(1, len(inputs)):\n output += inputs[i]\n return output, output, output\n\n def test_script_star_assign(self):\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n self.g = torch.jit.trace(TestScript.StarTestSumAndReturnThree(), torch.ones(4, 3))\n self.define('''\n def forward(self, rep):\n head, *tail = self.g(rep)\n return head\n ''')\n\n m = M2()\n self.assertEqual(m(torch.zeros(4, 3)), 3 * torch.zeros(4, 3))\n\n def test_script_module_star_assign2(self):\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n self.g = torch.jit.trace(\n TestScript.StarTestSumAndReturnThree(),\n (torch.ones(4, 3), torch.ones(4, 3), torch.ones(4, 3)),\n _force_outplace=True)\n self.define('''\n def forward(self, rep):\n *head, tail = self.g(rep, rep, rep)\n return tail\n ''')\n\n m = M2()\n self.assertEqual(m(torch.ones(4, 3)), 3 * torch.ones(4, 3))\n\n def test_script_module_star_assign2_inplace(self):\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n self.g = torch.jit.trace(\n TestScript.StarTestSumAndReturnThree(),\n (torch.ones(4, 3), torch.ones(4, 3), torch.ones(4, 3)),\n _force_outplace=False)\n self.define('''\n def forward(self, rep):\n *head, tail = self.g(rep, rep, rep)\n return tail\n ''')\n\n m = M2()\n # since forward() makes three aliases to the input `rep` before passing\n # it to StarTestSumAndReturnThree(), in-place behavior will be different\n # than the above out of place.\n self.assertEqual(m(torch.ones(4, 3)), 4 * torch.ones(4, 3))\n\n def test_script_module_star_assign_fail_pythonop(self):\n\n with self.assertRaisesRegex(RuntimeError, \"cannot be used as a tuple\"):\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n\n @torch.jit.ignore\n def myfunc():\n return torch.zeros(1, 2, 3), torch.zeros(1, 2, 3)\n\n self.define('''\n def forward(self, rep):\n a, *b = myfunc()\n return a\n ''')\n\n m = M2()\n m(torch.zeros(4, 3))\n\n def test_script_module_star_assign_fail_builtin(self):\n with self.assertRaisesRegex(RuntimeError, \"cannot be used as a tuple\"):\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n\n self.define('''\n def forward(self, rep):\n a, *b = torch.neg(rep)\n return a\n ''')\n\n m = M2()\n m(torch.zeros(4, 3))\n\n @skipIfCompiledWithoutNumpy\n def test_pack_padded_pad_packed_trace(self):\n from torch.nn.utils.rnn import pack_padded_sequence, pad_packed_sequence\n T, B, C = 3, 5, 7\n\n class PadPackedWrapper(torch.nn.Module):\n def __init__(self):\n super(PadPackedWrapper, self).__init__()\n\n def forward(self, x, seq_lens):\n x = pack_padded_sequence(x, seq_lens)\n x, _ = pad_packed_sequence(x)\n return x\n\n x = np.ones((T, B, C))\n seq_lens = np.array([3, 3, 2, 2, 1], dtype=np.int32)\n # set padding value so we can test equivalence\n for b in range(B):\n if seq_lens[b] < T:\n x[seq_lens[b]:, b, :] = 0\n seq_lens = torch.from_numpy(seq_lens)\n x = torch.autograd.Variable(torch.from_numpy(x), requires_grad=True)\n\n m = PadPackedWrapper()\n m_traced = torch.jit.trace(m, (x, seq_lens,))\n\n y = m(x, seq_lens)\n loss = torch.sum(y)\n loss.backward()\n grad = x.grad.clone()\n x.grad.zero_()\n\n y_traced = m_traced(x, seq_lens)\n loss_traced = torch.sum(y_traced)\n loss_traced.backward()\n grad_traced = x.grad.clone()\n\n self.assertEqual(y_traced, x)\n self.assertEqual(y_traced, y)\n self.assertEqual(grad, grad_traced)\n\n f = io.BytesIO()\n torch.onnx._export(m, (x, seq_lens), f, verbose=False)\n\n def test_script_pack_padded_sequence(self):\n from torch.nn.utils.rnn import pack_padded_sequence, pad_packed_sequence\n\n def pack_padded_pad_packed_script(x, seq_lens):\n x = pack_padded_sequence(x, seq_lens)\n x, lengths = pad_packed_sequence(x)\n return x, lengths\n\n T, B, C = 3, 5, 7\n x = torch.ones((T, B, C))\n seq_lens = torch.tensor([3, 3, 2, 2, 1])\n # set padding value so we can test equivalence\n for b in range(B):\n if seq_lens[b] < T:\n x[seq_lens[b]:, b, :] = 0\n\n eager_seq, eager_lengths = pack_padded_pad_packed_script(x, seq_lens)\n with torch._jit_internal._disable_emit_hooks():\n scripted_pack_padded_seq = torch.jit.script(pack_padded_pad_packed_script)\n script_seq, script_lengths = scripted_pack_padded_seq(x, seq_lens)\n self.assertEqual(eager_seq, script_seq)\n self.assertEqual(eager_lengths, script_lengths)\n\n class ExperimentalLSTM(torch.nn.Module):\n def __init__(self, input_dim, hidden_dim):\n super().__init__()\n\n def forward(self, input):\n # type: (Tensor)\n packed = pack_padded_sequence(\n input=input, lengths=torch.tensor([1, 2]), enforce_sorted=False\n )\n output, lengths = pad_packed_sequence(\n sequence=packed, total_length=2\n )\n # lengths is flipped, so is output\n return output[0]\n\n lstm = ExperimentalLSTM(input_dim=2, hidden_dim=2)\n\n with torch._jit_internal._disable_emit_hooks():\n self.checkModule(lstm, [torch.ones(2, 2)])\n\n def test_script_pad_sequence_pack_sequence(self):\n from torch.nn.utils.rnn import pad_sequence, pack_sequence, pad_packed_sequence\n\n def pad_sequence_func(tensor_list, batch_first=False, padding_value=0.0):\n # type: (List[Tensor], bool, float) -> Tensor\n return pad_sequence(tensor_list, batch_first, padding_value)\n\n def pack_sequence_func(tensor_list, enforce_sorted=True):\n # type: (List[Tensor], bool) -> Tensor\n return pad_packed_sequence(pack_sequence(tensor_list, enforce_sorted))[0]\n\n ones3 = torch.ones(3, 5)\n ones4 = torch.ones(4, 5)\n ones5 = torch.ones(5, 5)\n tensor1 = torch.tensor([1, 2, 3])\n tensor2 = torch.tensor([4, 5])\n tensor3 = torch.tensor([6])\n with torch._jit_internal._disable_emit_hooks():\n self.checkScript(pad_sequence_func,\n ([ones3, ones4, ones5],))\n self.checkScript(pad_sequence_func,\n ([ones3, ones4, ones5], True))\n self.checkScript(pad_sequence_func,\n ([ones3, ones4, ones5], True, 2.5))\n self.checkScript(pack_sequence_func,\n ([tensor1, tensor2, tensor3],))\n self.checkScript(pack_sequence_func,\n ([tensor1, tensor2, tensor3], False))\n\n def test_script_get_tracing_state(self):\n def test_if_tracing(x):\n if torch._C._get_tracing_state():\n return x + 1\n else:\n return x - 1\n\n inp = torch.randn(3, 3)\n self.checkScript(test_if_tracing, (inp,))\n\n def test_script_is_tracing(self):\n def test_is_tracing(x):\n if torch.jit.is_tracing():\n return x + 1\n else:\n return x - 1\n\n inp = torch.randn(3, 3)\n self.checkScript(test_is_tracing, (inp,))\n\n def test_is_scripting(self):\n def foo():\n return torch.jit.is_scripting()\n\n self.assertFalse(foo())\n scripted = torch.jit.script(foo)\n self.assertTrue(scripted())\n\n def test_script_outputs(self):\n with self.assertRaisesRegex(RuntimeError, \"cannot be used as a tuple\"):\n @torch.jit.script\n def foo(a):\n c, d = a + a\n return c + d\n\n @torch.jit.script\n def return3():\n return 1, 2, 3\n\n with self.assertRaisesRegex(RuntimeError, \"too many values to unpack\"):\n @torch.jit.script\n def bind2():\n a, b = return3()\n print(a)\n print(b)\n\n @unittest.skipIf(not RUN_CUDA, \"requires CUDA\")\n def test_script_get_device_cuda(self):\n @torch.jit.script\n def foo(a):\n return a.get_device()\n\n v = torch.randn(1, device='cuda')\n self.assertEqual(foo(v), 0)\n\n def test_script_chunk(self):\n @torch.jit.script\n def foo(a):\n b, c = torch.chunk(a, dim=0, chunks=2)\n return b\n v = torch.rand(10, 3)\n self.assertEqual(torch.chunk(v, dim=0, chunks=2)[0], foo(v))\n\n def test_script_copy(self):\n class M(torch.nn.Module):\n __annotations__ = {\n \"val\": Optional[torch.Tensor]\n }\n\n def __init__(self):\n super(M, self).__init__()\n self.val = None\n\n def some_method(self):\n return 3\n\n def forward(self, x):\n # type: (Tensor) -> Tensor\n self.val = x + self.some_method()\n return x\n\n m = torch.jit.script(M())\n # test copy\n copy.copy(m)\n copy.deepcopy(m)\n\n def test_script_forward_method_replacement(self):\n # We want to support the use case of attaching a different `forward` method\n class LowLevelModule(torch.nn.Module):\n def __init__(self):\n super(LowLevelModule, self).__init__()\n\n def forward(self, input: torch.Tensor):\n # Generic forward dispatch\n return self.forward_pytorch(input) * 2\n\n class TestModule(LowLevelModule):\n def __init__(self):\n super(TestModule, self).__init__()\n # Replace the forward method\n self.forward = types.MethodType(LowLevelModule.forward, self)\n\n def forward_pytorch(self, input: torch.Tensor):\n return torch.tensor(123)\n\n def forward(self, input: torch.Tensor):\n # Should not use this forward method\n raise AssertionError(\"This method should not be used\")\n return self.forward_pytorch(input)\n\n m = TestModule()\n self.assertEqual(m(torch.tensor(1)), torch.tensor(246))\n\n m_scripted = torch.jit.script(m)\n self.assertEqual(m_scripted(torch.tensor(1)), torch.tensor(246))\n\n # Suppression: ONNX warns when exporting RNNs because of potential batch size mismatch.\n @suppress_warnings\n @skipIfCompiledWithoutNumpy\n def test_rnn_trace_override(self):\n from torch.nn.utils.rnn import pack_padded_sequence, pad_packed_sequence\n num_layers = 3\n T, B, C = 11, 5, 7\n\n class RNNTraceWrapper(torch.nn.Module):\n def __init__(self, cell_type):\n super(RNNTraceWrapper, self).__init__()\n if cell_type == 'RNN':\n self.rnn = torch.nn.RNN(input_size=C, hidden_size=C, num_layers=num_layers)\n elif cell_type == 'LSTM':\n self.rnn = torch.nn.LSTM(input_size=C, hidden_size=C, num_layers=num_layers)\n elif cell_type == 'GRU':\n self.rnn = torch.nn.GRU(input_size=C, hidden_size=C, num_layers=num_layers)\n\n def forward(self, x, seq_lens):\n x = pack_padded_sequence(x, seq_lens)\n x, _ = self.rnn(x)\n x, _ = pad_packed_sequence(x)\n return x\n\n for cell_type in ['RNN', 'LSTM', 'GRU']:\n x = torch.ones(T, B, C, requires_grad=True)\n seq_lens = torch.from_numpy(np.array([11, 3, 2, 2, 1], dtype=np.int32))\n\n m = RNNTraceWrapper(cell_type)\n m_traced = torch.jit.trace(m, (x, seq_lens,))\n\n y = m(x, seq_lens)\n loss = torch.sum(y)\n loss.backward()\n grad = x.grad.clone()\n x.grad.zero_()\n\n y_traced = m_traced(x, seq_lens)\n loss_traced = torch.sum(y_traced)\n loss_traced.backward()\n grad_traced = x.grad.clone()\n\n self.assertEqual(y_traced, y)\n self.assertEqual(grad, grad_traced)\n\n f = io.BytesIO()\n torch.onnx._export(m, (x, seq_lens), f, verbose=False)\n\n def test_python_call_non_tensor(self):\n def foo(a, b, c):\n # type: (Tensor, int, Tuple[Tensor, int]) -> Tuple[int, Tensor]\n d, e = c\n return b + e, a + d\n\n @torch.jit.script\n def bar():\n x = torch.ones(3, 4)\n a, b = foo(x, 3, (x, 3))\n return a, b\n\n self.assertEqual((6, torch.ones(3, 4) + 1), bar())\n\n def test_python_call_non_tensor_wrong(self):\n with self.assertRaisesRegex(RuntimeError, r\"but instead got value of type tuple\"):\n @torch.jit.ignore\n def foo():\n # type: () -> Tensor\n return ((3, 4),) # noqa: T484\n\n @torch.jit.script\n def bar():\n return foo()\n\n bar()\n\n def test_if_different_type(self):\n with self.assertRaisesRegex(RuntimeError, \"Type mismatch: c0 is set to type int \"\n \"in the true branch and type float in the false branch:\"):\n @torch.jit.script\n def diff_type_used():\n if 1 == 2:\n c0 = 1\n else:\n c0 = 1.0\n return c0\n\n with self.assertRaisesRegex(RuntimeError, \"Variable 'c0' previously has type float\"):\n @torch.jit.script\n def diff_existing_type(x):\n c0 = 1.0\n if 1 == 2:\n c0 = 1\n print(x)\n return x\n\n @torch.jit.script\n def diff_type_unused():\n if 1 == 1:\n c0 = 1\n print(c0)\n else:\n c0 = 1.0\n print(c0)\n return 1\n\n def test_if_not_defined_error(self):\n with self.assertRaisesRegex(RuntimeError, \"c0 is not defined in the false branch\"):\n @torch.jit.script\n def test():\n if 1 == 1:\n c0 = 1\n return c0\n with self.assertRaisesRegex(RuntimeError, \"c0 is not defined in the true branch\"):\n @torch.jit.script\n def test2():\n if 1 == 1:\n pass\n else:\n c0 = 1\n return c0\n\n def test_if_list_cat(self):\n # testing that different length lists don't throw error on cat in shape prop\n @torch.jit.script\n def test_list(x):\n if bool(x.sum() < 1):\n c = [x, x]\n else:\n c = [x, x, x]\n return torch.cat(c)\n\n b = torch.zeros(2, 4)\n _propagate_shapes(test_list.graph, (b,), False)\n\n def test_if_supertype(self):\n @torch.jit.script\n def tensor_unifying(x, y, z):\n # testing dynamic is appropriately set for y and z\n if bool(x):\n x, y, z = x + 1, y, z\n else:\n x, y, z = x + 1, x, y\n\n return x, y, z\n\n a = torch.zeros(2, 2, dtype=torch.float)\n b = torch.zeros(2, 4, dtype=torch.long)\n c = torch.zeros(2, 4, dtype=torch.float)\n\n graph = _propagate_shapes(tensor_unifying.graph, (a, b, c), False)\n if_outputs = list(graph.findNode(\"prim::If\").outputs())\n self.assertTrue(if_outputs[0].type().str() == \"Float(*, *, requires_grad=0, device=cpu)\")\n self.assertTrue(if_outputs[1].type().str() == \"Tensor(*, *, requires_grad=0, device=cpu)\")\n self.assertTrue(if_outputs[2].type().str() == \"Tensor(*, *, requires_grad=0, device=cpu)\")\n\n def test_list_unify(self):\n # allowing a unififed int?[] would cause a runtime error b/c\n # the index operation expects int?[] to be a generic list,\n # but in the true branch the IValue will be a int list\n with self.assertRaisesRegex(RuntimeError, \"int[] in the true branch and type None[]\"):\n @torch.jit.script\n def list_optional_fails(x):\n # type: (bool) -> Optional[int]\n if x:\n y = [1]\n else:\n y = [None] # noqa: T484\n return y[0]\n\n @torch.jit.script\n def list_tensors(x):\n # type: (bool) -> Tuple[Tensor, List[Tensor]]\n if x:\n a = torch.zeros([1, 1])\n y = [a]\n else:\n a = torch.zeros([1, 2])\n y = [a]\n return a, y\n\n self.run_pass('constant_propagation', list_tensors.graph)\n m = self.createFunctionFromGraph(list_tensors.graph)\n # testing that tensor type of lists is unified\n self.getExportImportCopy(m)\n\n @_inline_everything\n def test_import_constants_not_specialized(self):\n class Mod(torch.nn.Module):\n def forward(self, x):\n return torch.cat(2 * [x], dim=0)\n\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self, mod):\n super(ScriptMod, self).__init__()\n x = torch.zeros(1, 3)\n mod_fn = lambda : mod(x) # noqa: E731\n self.mod = torch.jit.trace(mod_fn, tuple())\n\n @torch.jit.script_method\n def forward(self):\n return self.mod()\n\n cm = ScriptMod(Mod())\n # specialized tensor in graph\n FileCheck().check(\"Double(1, 3, strides=[3, 1], requires_grad=0, device=cpu)\").run(cm.forward.graph)\n buffer = io.BytesIO()\n torch.jit.save(cm, buffer)\n buffer.seek(0)\n # when tensor is loaded as constant it isnt specialized\n cm_load = torch.jit.load(buffer)\n FileCheck().check_not(\"Double(1, 3)\").run(cm_load.forward.graph)\n\n def test_type_annotations_repeated_list(self):\n @torch.jit.script\n def float_fn(x, y):\n # type: (float, BroadcastingList3[float]) -> List[float]\n return y\n self.assertEqual(float_fn(2.0, 1.0), float_fn(2.0, [1.0, 1.0, 1.0]))\n self.assertEqual(float_fn(2.0, 1.0), float_fn(2.0, (1.0, 1.0, 1.0)))\n\n @torch.jit.script\n def float_fn_call():\n print(float_fn(1.0, 1.0))\n print(float_fn(1.0, (1.0, 1.0, 1.0)))\n\n @torch.jit.script\n def int_fn(x):\n # type: (BroadcastingList3[int]) -> List[int]\n return x\n self.assertEqual(int_fn(1), int_fn([1, 1, 1]))\n self.assertEqual(int_fn(1), int_fn((1, 1, 1)))\n\n @torch.jit.script\n def int_fn_call():\n print(int_fn(1))\n print(int_fn((1, 1, 1)))\n\n with self.assertRaisesRegex(RuntimeError, \"must be a positive integer:\"):\n @torch.jit.script # noqa: T484\n def fn(x):\n # type: (BroadcastingListx[int]) -> List[int] # noqa: T484\n return x\n\n # using CU so that flake8 error on int[2] is not raised (noqa not working)\n with self.assertRaisesRegex(RuntimeError, \"Unknown type constructor\"):\n cu = torch.jit.CompilationUnit('''\n def nested(x, y):\n # type: (int, Tuple[int, int[2]]) -> List[int]\n return x # noqa: T484\n ''')\n\n @torch.jit.script\n def f(x: BroadcastingList2[int]):\n return x\n\n out = f(1)\n self.assertTrue(isinstance(out[0], int))\n self.assertEqual(out, [1, 1])\n\n def test_ntuple_builtins(self):\n from torch.nn.modules.utils import _single, _pair, _triple, _quadruple\n\n def test_ints():\n return _single(1), _pair(2), _triple(3), _quadruple(4)\n\n def test_floats():\n return _single(1), _pair(2.1), _triple(3.1), _quadruple(4.1)\n\n self.checkScript(test_ints, ())\n self.checkScript(test_floats, ())\n\n def test_embedding_renorm_grad_error(self):\n # Testing that the builtin call to embedding_renorm_ correctly throws\n # Error when .backward() is called on its input\n\n def embedding_norm(input, embedding_matrix, max_norm):\n F.embedding(input, embedding_matrix, max_norm=0.01)\n\n @torch.jit.script\n def embedding_norm_script(input, embedding_matrix, max_norm):\n # type: (Tensor, Tensor, float) -> None\n F.embedding(input, embedding_matrix, max_norm=0.01)\n\n for _ in [embedding_norm, embedding_norm_script]:\n input = torch.tensor([[1, 2, 4, 5], [4, 3, 2, 9]])\n embedding_matrix = torch.randn(10, 3)\n\n var1 = torch.randn(10, 3, requires_grad=True)\n var2 = var1.detach().requires_grad_()\n output1 = var1 * embedding_matrix\n output2 = var2 * embedding_matrix\n\n output1.sum().backward()\n\n ignore = F.embedding(input, embedding_matrix, max_norm=0.01)\n with self.assertRaisesRegex(RuntimeError, \"modified\"):\n output2.sum().backward()\n\n def test_type_annotations(self):\n def fn(x, y):\n # type: (Tensor, Tensor) -> Tuple[Tensor, Tensor, Tensor]\n return x, x * 2, x * 3\n\n with self.assertRaisesRegex(RuntimeError, r\"need 4 values .* found only 3\"):\n @torch.jit.script\n def script_fn(x):\n x, y, z, w = fn(x, x)\n\n with self.assertRaisesRegex(RuntimeError, r\"too many values .* need 2 but found 3\"):\n @torch.jit.script\n def script_fn2(x):\n x, y = fn(x, x)\n\n def fn_unpack(x):\n y, z, w = fn(x, x)\n return y\n\n def fn_index(x):\n q = fn(x, x)\n return x\n\n def fn_string(str, strpair):\n # type: (str, Tuple[str, str]) -> Tuple[str, int, str, str]\n str1, str2 = strpair\n return str, 2, str1, str2\n\n x = torch.ones(2, 2)\n self.checkScript(fn_unpack, (x,), optimize=True)\n self.checkScript(fn_index, (x,), optimize=True)\n self.checkScript(fn_string, (\"1\", (\"3\", \"4\")), optimize=True)\n\n def test_type_annotations_varargs(self):\n @torch.jit.ignore\n def fn_varargs(x, *args):\n return args[0] if args else x\n\n def fn1(x, y, z):\n return fn_varargs(x)\n\n def fn2(x, y, z):\n return fn_varargs(x, y)\n\n def fn3(x, y, z):\n return fn_varargs(x, y, z)\n\n x, y, z = [torch.randn(2, 2) for _ in range(3)]\n self.checkScript(fn1, (x, y, z), optimize=True)\n self.checkScript(fn2, (x, y, z), optimize=True)\n self.checkScript(fn3, (x, y, z), optimize=True)\n\n def test_type_annotation_py3(self):\n code = dedent(\"\"\"\n import torch\n from torch import Tensor\n from typing import Tuple\n\n def fn(x : torch.Tensor, y : Tensor, z) -> Tuple[Tensor, Tensor, Tensor]:\n return (x, y + z, z)\n \"\"\")\n\n with tempfile.TemporaryDirectory() as tmp_dir:\n script_path = os.path.join(tmp_dir, 'script.py')\n with open(script_path, 'w') as f:\n f.write(code)\n fn = get_fn('test_type_annotation_py3', script_path)\n fn = torch.jit.ignore(fn)\n\n with self.assertRaisesRegex(RuntimeError, r\"Expected a value of type 'Tensor' for argument\"\n r\" 'x' but instead found type 'Tuple\\[Tensor,\"):\n @torch.jit.script\n def bad_fn(x):\n x, y = fn((x, x), x, x)\n return y\n\n with self.assertRaisesRegex(RuntimeError, r\"too many values .* need 2 but found 3\"):\n @torch.jit.script\n def bad_fn2(x):\n x, y = fn(x, x, x)\n return y\n\n with self.assertRaisesRegex(RuntimeError, r\"need 4 values .* found only 3\"):\n @torch.jit.script\n def bad_fn3(x):\n x, y, z, w = fn(x, x, x)\n return y\n\n def good_fn(x):\n y, z, w = fn(x, x, x)\n return y, z, w\n\n self.checkScript(good_fn, (torch.ones(2, 2),), optimize=True)\n\n def test_type_annotation_module(self):\n class BaseModule(torch.jit.ScriptModule):\n @torch.jit.ignore\n def foo(self, x):\n # type: (Tensor) -> Tensor\n return x + 1\n\n @torch.jit.ignore\n def bar(self, x, y):\n # type: (Tensor, Tensor) -> Tuple[Tensor, Tensor]\n return x + y, y\n\n @torch.jit.ignore\n def baz(self, x, y):\n return x\n\n class ModuleTooMany(BaseModule):\n @torch.jit.script_method\n def method(self, x):\n return self.foo(x, x)\n\n class ModuleTooFew(BaseModule):\n @torch.jit.script_method\n def method(self, x):\n return self.bar(x)\n\n class ModuleTooManyAssign(BaseModule):\n @torch.jit.script_method\n def method(self, x):\n y, z, w = self.bar(x, x)\n return x\n\n class ModuleDefault(BaseModule):\n @torch.jit.script_method\n def method(self, x):\n y = self.baz(x)\n return x\n\n with self.assertRaisesRegex(RuntimeError, \"Expected at most 2 arguments but found 3\"):\n ModuleTooMany()\n with self.assertRaisesRegex(RuntimeError, \"Argument y not provided\"):\n ModuleTooFew()\n with self.assertRaisesRegex(RuntimeError, \"need 3 values .* found only 2\"):\n ModuleTooManyAssign()\n with self.assertRaisesRegex(RuntimeError, \"Argument y not provided.\"):\n ModuleDefault()\n\n def test_type_inferred_from_empty_annotation(self):\n \"\"\"\n Test that the type inferred from an empty or missing annotation is Torch.Tensor wtih `inferred=true`\n \"\"\"\n @torch.jit.script\n def fn(x):\n return x\n\n graph = fn.graph\n n = next(graph.inputs())\n self.assertTrue(n.type() == torch._C.TensorType.getInferred())\n\n with self.assertRaisesRegex(RuntimeError, \"Inferred \\'x\\' to be of type \\'Tensor\"):\n fn(1)\n\n def test_script_define_order(self):\n class M(torch.jit.ScriptModule):\n\n @torch.jit.script_method\n def call_foo(self, input):\n return self.foo(input)\n\n @torch.jit.script_method\n def foo(self, input):\n return input + 1\n m = M()\n self.assertEqual(2, m.call_foo(torch.ones((), dtype=torch.int64)))\n\n def test_script_define_order_recursive_fail(self):\n class M(torch.jit.ScriptModule):\n\n @torch.jit.script_method\n def call_foo(self, input):\n return self.foo(input)\n\n @torch.jit.script_method\n def foo(self, input):\n self.call_foo(input)\n\n with self.assertRaisesRegex(RuntimeError, 'called recursively'):\n M()\n\n def test_script_kwargs_fn_call(self):\n class M(torch.jit.ScriptModule):\n\n @torch.jit.script_method\n def call_foo(self, input):\n return self.foo(input=input, bar=1)\n\n @torch.jit.script_method\n def foo(self, bar, input):\n # type: (int, Tensor) -> Tensor\n return input + bar\n m = M()\n self.assertEqual(2, m.call_foo(torch.ones((), dtype=torch.int64)))\n\n def test_if_define(self):\n @torch.jit.script\n def foo(a):\n if bool(a == 0):\n b = 1\n else:\n b = 0\n return b + 1\n\n @torch.jit.script\n def foo2(a):\n b = 0\n if bool(a == 0):\n b = 1\n return b + 1\n\n @torch.jit.script\n def foo3(a):\n b = 1\n if bool(a == 0):\n c = 4\n else:\n b = 0\n return b + 1\n\n a = torch.ones(1, dtype=torch.long)\n b = torch.zeros(1, dtype=torch.long)\n self.assertEqual(1, foo(a))\n self.assertEqual(2, foo(b))\n self.assertEqual(1, foo2(a))\n self.assertEqual(2, foo2(b))\n self.assertEqual(1, foo3(a))\n self.assertEqual(2, foo3(b))\n\n def test_script_module_export_submodule(self):\n class M1(torch.jit.ScriptModule):\n def __init__(self):\n super(M1, self).__init__()\n self.weight = nn.Parameter(torch.randn(2))\n\n @torch.jit.script_method\n def forward(self, thing):\n return self.weight + thing\n\n class M2(torch.jit.ScriptModule):\n def __init__(self):\n super(M2, self).__init__()\n # test submodule\n self.sub = M1()\n self.weight = nn.Parameter(torch.randn(2, 3))\n self.bias = nn.Parameter(torch.randn(2))\n self.define(\"\"\"\n def hi(self, a):\n return self.weight.mm(a)\n \"\"\")\n\n @torch.jit.script_method\n def doit(self, input):\n return self.weight.mm(input)\n\n @torch.jit.script_method\n def doit2(self, input):\n return self.weight.mm(input)\n\n @torch.jit.script_method\n def doit3(self, input):\n return input + torch.ones([1], dtype=torch.double)\n\n @torch.jit.script_method\n def forward(self, input):\n a = self.doit(input)\n b = self.doit2(input)\n c = self.hi(input)\n return a + b + self.bias + c\n\n with torch.jit.optimized_execution(False):\n m_orig = M2()\n m_import = self.getExportImportCopy(m_orig)\n\n input = torch.randn(3, 2)\n self.assertEqual(m_orig.doit(input), m_import.doit(input))\n self.assertEqual(m_orig.hi(input), m_import.hi(input))\n self.assertEqual(m_orig.doit3(input), m_import.doit3(input))\n self.assertEqual(m_orig.forward(input), m_import.forward(input))\n\n @slowTest\n def test_compile_module_with_constant(self):\n class Double(nn.Module):\n def __init__(self, downsample=None):\n super(Double, self).__init__()\n\n def forward(self, input):\n return input * 2\n\n class Mod(nn.Module):\n __constants__ = ['downsample']\n\n def __init__(self, downsample=None):\n super(Mod, self).__init__()\n self.downsample = downsample\n\n def forward(self, input):\n if self.downsample is not None:\n return self.downsample(input)\n return input\n\n none_mod = torch.jit.script(Mod(None))\n double_mod = torch.jit.script(Mod(Double()))\n self.assertEqual(none_mod(torch.tensor(1)), torch.tensor(1))\n self.assertEqual(double_mod(torch.tensor(1)), torch.tensor(1) * 2)\n\n def test_script_module_export_tensor_type(self):\n class M(torch.jit.ScriptModule):\n def __init__(self, type):\n super(M, self).__init__()\n self.param = torch.nn.Parameter(torch.zeros((5, 5), dtype=type).random_())\n\n @torch.jit.script_method\n def foo(self):\n return self.param\n\n with torch.jit.optimized_execution(False):\n for type in [torch.float, torch.double]:\n m_orig = M(type)\n m_import = self.getExportImportCopy(m_orig)\n # check to make sure the storage wasn't resized\n self.assertTrue(m_orig.param.storage().size() == 25)\n self.assertEqual(m_orig.foo(), m_import.foo())\n self.assertTrue(m_orig.foo().dtype == m_import.foo().dtype)\n\n @unittest.skipIf(not RUN_CUDA, \"testing cuda tensors require CUDA\")\n def test_script_module_export_tensor_cuda(self):\n class M(torch.jit.ScriptModule):\n\n def __init__(self):\n super(M, self).__init__()\n self.param = torch.nn.Parameter(torch.zeros((5, 5), device='cuda:0').random_())\n\n @torch.jit.script_method\n def foo(self):\n return self.param\n\n m_orig = M()\n m_import = self.getExportImportCopy(m_orig)\n # check to make sure the storage wasn't resized\n self.assertTrue(m_orig.param.storage().size() == 25)\n self.assertTrue(m_import.foo().device == torch.device('cuda:0'))\n self.assertEqual(m_orig.foo(), m_import.foo())\n self.assertTrue(m_orig.foo().dtype == m_import.foo().dtype)\n\n def test_script_module_export_blocks(self):\n class M(torch.jit.ScriptModule):\n def __init__(self, n, m):\n super(M, self).__init__()\n self.weight = torch.nn.Parameter(torch.rand(n, m))\n\n @torch.jit.script_method\n def forward(self, input):\n if bool(input.sum() > 0):\n output = self.weight.mv(input)\n else:\n output = self.weight + input\n return output\n\n m_orig = M(200, 200)\n m_import = self.getExportImportCopy(m_orig)\n\n t = torch.rand(200)\n self.assertEqual(m_orig(t), m_import(t))\n\n def test_script_module_export_shared_storage(self):\n class M(torch.jit.ScriptModule):\n\n def __init__(self):\n super(M, self).__init__()\n self.param1 = torch.nn.Parameter(torch.rand(5, 5))\n self.param2 = torch.nn.Parameter(self.param1[3])\n self.param3 = torch.nn.Parameter(torch.rand(5, 5))\n self.param4 = torch.nn.Parameter(torch.rand(11, 5)[1:6])\n\n @torch.jit.script_method\n def foo(self):\n return self.param1 + self.param2 + self.param3 + self.param4\n\n with torch.jit.optimized_execution(False):\n m_orig = M()\n m_import = self.getExportImportCopy(m_orig)\n\n self.assertEqual(m_orig.foo(), m_import.foo())\n\n self.assertTrue(m_import.param1.storage().data_ptr() == m_import.param2.storage().data_ptr())\n self.assertTrue(m_import.param1.storage().data_ptr() != m_import.param3.storage().data_ptr())\n\n def test_sequential_intermediary_types(self):\n class A(torch.nn.Module):\n def __init__(self):\n super(A, self).__init__()\n\n def forward(self, x):\n return x + 3\n\n class B(torch.nn.Module):\n def __init__(self):\n super(B, self).__init__()\n\n def forward(self, x):\n return {\"1\": x}\n\n class C(torch.nn.Module):\n def __init__(self):\n super(C, self).__init__()\n self.foo = torch.nn.Sequential(A(), B())\n\n def forward(self, x):\n return self.foo(x)\n\n self.checkModule(C(), (torch.tensor(1),))\n\n def test_ellipsis_const_mid(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[2, Ellipsis, 0:4, 4:8].size()\n\n dummy = torch.zeros(8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_ellipsis_const_mid_select(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[2, Ellipsis, 4, 4, 4:8, 2].size()\n\n dummy = torch.zeros(8, 8, 8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_ellipsis_const_start(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[Ellipsis, 0:4, 4:8].size()\n dummy = torch.zeros(8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_ellipsis_const_end(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[0:4, 2, Ellipsis].size()\n dummy = torch.zeros(8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_ellipsis_mid(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[2, ..., 0:4, 4:8].size()\n\n dummy = torch.zeros(8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_ellipsis_mid_select(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[2, ..., 4, 4, 4:8, 2].size()\n\n dummy = torch.zeros(8, 8, 8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_ellipsis_start(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[..., 0:4, 4:8].size()\n dummy = torch.zeros(8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_ellipsis_end(self):\n def ellipsize(x):\n # type: (Tensor) -> List[int]\n return x[0:4, 2, ...].size()\n dummy = torch.zeros(8, 8, 8, 8, 8)\n self.checkScript(ellipsize, (dummy,), optimize=True)\n\n def test_torch_manual_seed(self):\n with freeze_rng_state():\n def test():\n torch.manual_seed(2)\n return torch.rand(1)\n\n script = torch.jit.script(test)\n self.assertEqual(test(), script())\n graph = script.graph_for()\n FileCheck().check(\"aten::manual_seed\").run(graph)\n\n def test_index_select_shape_prop(self):\n\n @torch.jit.script\n def foo(x, y):\n return torch.index_select(x, index=y, dim=1)\n\n a = torch.zeros(2, 2)\n b = torch.zeros(4, dtype=torch.long)\n torch._C._jit_pass_complete_shape_analysis(foo.graph, (a, b), False)\n FileCheck().check(\"Double(2, 4, strides=[4, 1], requires_grad=0, device=cpu)\").run(str(foo.graph))\n\n def test_shape_analysis_loop(self):\n def foo(a, b, x):\n c = a\n # on the first iteration of the loop it appears that\n # c should have a expand to the size of b\n # but on the second+ iterations, there is no broadcast and the\n # sizes are different.\n # previously this would cause the compiler to (1) enter an infinite\n # loop trying to compute the shape, and (2) insert invalid\n # broadcasts.\n # this test ensure we don't regress on these issues\n for _ in range(2):\n a = c + b\n c = x\n b = x\n return a\n\n self.checkScript(foo, (torch.zeros(1), torch.zeros(4), torch.zeros(5)), optimize=False)\n\n def test_intlist_args(self):\n def func_1(x):\n return torch.nn.functional.adaptive_avg_pool1d(x, 1)\n\n def func_2(x):\n return torch.nn.functional.adaptive_avg_pool1d(x, output_size=1)\n\n def func_3(x):\n return torch.nn.functional.adaptive_avg_pool1d(x, output_size=[1])\n\n x = torch.randn(8, 8, 8)\n self.checkScript(func_1, [x], optimize=True)\n self.checkScript(func_2, [x], optimize=True)\n self.checkScript(func_3, [x], optimize=True)\n\n def test_wrong_implicit_expand(self):\n\n @_trace(torch.zeros(3), torch.zeros(1))\n def foo(a, b):\n return a + b\n\n a = torch.rand(4)\n b = torch.rand(4)\n self.assertEqual(a + b, foo(a, b))\n\n def test_builtin_args_fails(self):\n\n with self.assertRaisesRegex(RuntimeError, 'Argument self not provided'):\n @torch.jit.script\n def f1(a):\n torch.sum(foo=4)\n\n with self.assertRaisesRegex(RuntimeError, 'specified twice'):\n @torch.jit.script\n def f2(a):\n torch.sum(a, self=a)\n\n with self.assertRaisesRegex(RuntimeError, 'not provided'):\n @torch.jit.script\n def f3(a):\n torch.sum(dim=4)\n\n with self.assertRaisesRegex(RuntimeError, 'for argument \\'tensors\\' but instead found type \\'Tensor'):\n @torch.jit.script\n def f4(a):\n torch.cat(a)\n\n with self.assertRaisesRegex(RuntimeError, r'argument \\'tensors\\' but instead found type \\'List\\[int\\]'):\n @torch.jit.script\n def f5(a):\n torch.cat([3])\n\n with self.assertRaisesRegex(RuntimeError, 'Lists must contain only a single type'):\n @torch.jit.script\n def f6(a):\n a.expand(size=[3, [4]])\n\n def test_builtin_args(self):\n\n def t0(a):\n # default arg dim\n return torch.cat([a, a])\n\n self.checkScript(t0, (torch.zeros(1, 1),))\n\n def t1(a):\n # keywords out of order\n return torch.cat(dim=1, tensors=[a, a])\n\n self.checkScript(t1, (torch.zeros(1, 1, 2),))\n\n def t2(a):\n # mix const/non-const attributes\n if 1 == 1:\n b = 1\n else:\n b = 0\n return torch.sum(a, dim=b, keepdim=False)\n\n self.checkScript(t2, (torch.zeros(1, 1, 2),))\n\n def test_parser_type_annotations(self):\n cu = torch.jit.CompilationUnit('''\n def foo(x : Tensor, y : Tuple[Tuple[Tensor, Tensor], Tensor]) -> Tuple[Tensor, Tensor]:\n return x, x\n ''')\n\n self.assertExpected(str(cu.foo.schema))\n\n def test_parser_type_annotations_comment(self):\n cu = torch.jit.CompilationUnit('''\n def foo(x, y):\n # type: (Tensor, Tuple[Tuple[Tensor, Tensor], Tensor]) -> Tuple[Tensor, Tensor]\n return x, x\n ''')\n\n self.assertExpected(str(cu.foo.schema))\n\n def test_parser_type_annotations_unknown_type(self):\n with self.assertRaisesRegex(RuntimeError, \"Unknown type name 'Foo'\"):\n cu = torch.jit.CompilationUnit('''\n def foo(x : Tensor, y : Tuple[Tuple[Foo, Tensor], Tensor]) -> Tuple[Tensor, Tensor]:\n return x, x\n ''')\n\n def test_parser_type_annotations_subscript_non_ident(self):\n with self.assertRaisesRegex(RuntimeError, r'Subscripted type must be a type identifier'):\n cu = torch.jit.CompilationUnit('''\n def foo(x : Tensor, y : Tuple[Tensor, Tensor][Tensor]) -> Tuple[Tensor, Tensor]:\n return x, x\n ''')\n\n def test_parser_type_annotations_subscript_tensor(self):\n with self.assertRaisesRegex(RuntimeError, r'Unknown type constructor Tensor'):\n cu = torch.jit.CompilationUnit('''\n def foo(x : Tensor, y : Tensor[Tensor, Tensor]) -> Tuple[Tensor, Tensor]:\n return x, x\n ''')\n\n def test_parser_type_annotations_incompatible_expression(self):\n with self.assertRaisesRegex(RuntimeError, r'Expression of type \\+ cannot be used in a type expression'):\n cu = torch.jit.CompilationUnit('''\n def foo(x : Tensor, y : Tuple[3 + 4, Tensor]) -> Tuple[Tensor, Tensor]:\n return x, x\n ''')\n\n def test_gather_dynamic_index(self):\n def t(x):\n gather1 = x[0]\n idx = 0 + 1\n gather2 = x[idx]\n return gather1 + gather2\n\n self.checkScript(t, (torch.zeros(3, 2, 3),))\n\n def test_torch_ignore_conversion_to_none(self):\n class A(torch.nn.Module):\n def __init__(self):\n super(A, self).__init__()\n\n @torch.jit.ignore\n def ignored(self, a: int) -> None:\n l: int = len([2 for i in range(a) if i > 2])\n return\n\n def forward(self) -> int:\n a: int = 4\n b: int = 5\n self.ignored(a)\n return a + b\n\n class B(torch.nn.Module):\n def __init__(self):\n super(B, self).__init__()\n\n @torch.jit.ignore\n def ignored(self, a: int):\n l: int = len([2 for i in range(a) if i > 2])\n return\n\n def forward(self) -> int:\n a: int = 4\n b: int = 5\n self.ignored(a)\n return a + b\n\n modelA = torch.jit.script(A())\n self.assertEqual(modelA(), 9)\n\n with self.assertRaisesRegexWithHighlight(RuntimeError, \"expected value of type Tensor\", \"self.ignored\"):\n modelB = torch.jit.script(B())\n modelB()\n\n def test_addmm_grad(self):\n \"\"\" This test checks several things:\n 1. An expand node was inserted before the addmm operating on the\n bias term.\n 2. The fused form of addmm appears in the ultimate graph that's\n executed.\n 3. A sum op was emitted for accumulating gradients along the 0th\n (expanded) dimension of the bias term.\n 4. The correct symbolic representation for the backward pass of the\n mm operator was emitted (x.t() -> mm)\n\n TODO: we should actually check these conditions once we have a way\n to dump the GraphExecutor state. Namely the processed forward graph\n and the backward graph.\n \"\"\"\n @torch.jit.script\n def addmm_grad_test(b, x, w):\n return torch.addmm(b, x, w)\n\n # Initialize param and input values\n w_init = torch.rand(2, 5)\n b_init = torch.rand(5)\n x = torch.rand(3, 2)\n\n # Clone trainable params\n b = b_init.clone()\n b.requires_grad_()\n w = w_init.clone()\n w.requires_grad_()\n\n # Test symbolic differentiation\n y = addmm_grad_test(b, x, w)\n y.sum().backward()\n\n # clone params for autograd reference\n b_ref = b_init.clone()\n b_ref.requires_grad_()\n w_ref = w_init.clone()\n w_ref.requires_grad_()\n y_ref = torch.addmm(b_ref, x, w_ref)\n y_ref.sum().backward()\n\n self.assertEqual(w.grad, w_ref.grad)\n self.assertEqual(b.grad, b_ref.grad)\n\n def test_zeros(self):\n class M(torch.jit.ScriptModule):\n __constants__ = ['d']\n\n def __init__(self):\n super(M, self).__init__()\n self.d = torch.device('cpu')\n\n @torch.jit.script_method\n def create(self):\n return torch.zeros([1, 1, 2], dtype=torch.float, device=self.d, layout=torch.strided)\n\n r = M().create()\n self.assertEqual(r.dtype, torch.float)\n self.assertEqual(torch.zeros([1, 1, 2], dtype=torch.float), r)\n\n def fn():\n return torch.zeros((1, 2, 3))\n\n self.checkScript(fn, ())\n\n def test_vararg_zeros(self):\n def foo():\n return torch.zeros(3, 4, 5, dtype=torch.int)\n\n self.checkScript(foo, ())\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.LEGACY, \"the original version of test_rand\")\n def test_rand(self):\n def test_rand():\n a = torch.rand([3, 4])\n return a + 1.0 - a\n\n self.checkScript(test_rand, ())\n fn = torch.jit.script(test_rand)\n out = fn()\n self.assertEqual(out.dtype, torch.double)\n g = fn.graph_for()\n # Testing shape analysis correctly setting type\n if GRAPH_EXECUTOR != ProfilingMode.SIMPLE:\n FileCheck().check(\"Double(*, *, requires_grad=0, device=cpu)\") \\\n .check_not(\"Float(*, *, requires_grad=0, device=cpu)\").run(g)\n\n @torch.jit.script\n def randint():\n return torch.randint(0, 5, [1, 2])\n out = randint()\n self.assertEqual(out.dtype, torch.double)\n # although the type should be int here, testing that the runtime dtype\n # and shape analysis dtype is the same.\n if GRAPH_EXECUTOR != ProfilingMode.SIMPLE:\n FileCheck().check(\"Double(*, *, requires_grad=0, device=cpu)\") \\\n .check_not(\"Float(*, *, requires_grad=0, device=cpu)\").run(randint.graph_for())\n\n def test_linear_grad(self):\n with enable_profiling_mode_for_profiling_tests():\n def t(x: torch.Tensor, w: torch.Tensor, b: Optional[torch.Tensor]):\n return torch.nn.functional.linear(x, w, b)\n\n x_init = torch.randn(4, 2)\n w_init = torch.randn(3, 2)\n b_init = torch.randn(3)\n grad = torch.randn(4, 3)\n\n with disable_autodiff_subgraph_inlining():\n # script module\n jit_t = torch.jit.script(t)\n\n x = x_init.detach().requires_grad_()\n w = w_init.detach().requires_grad_()\n b = b_init.detach().requires_grad_()\n x_ref = x_init.detach().requires_grad_()\n w_ref = w_init.detach().requires_grad_()\n b_ref = b_init.detach().requires_grad_()\n\n # profiling/optimization runs\n jit_o = jit_t(x, w, b)\n jit_o.backward(grad)\n jit_o = jit_t(x, w, b)\n jit_o.backward(grad)\n\n x.grad.zero_()\n w.grad.zero_()\n b.grad.zero_()\n jit_o = jit_t(x, w, b)\n jit_o.backward(grad)\n o = t(x_ref, w_ref, b_ref)\n o.backward(grad)\n\n self.assertEqual(jit_o, o)\n self.assertEqual(x.grad, x_ref.grad)\n self.assertEqual(w.grad, w_ref.grad)\n self.assertEqual(b.grad, b_ref.grad)\n\n x.grad.zero_()\n w.grad.zero_()\n x_ref.grad.zero_()\n w_ref.grad.zero_()\n jit_o = jit_t(x, w, None)\n jit_o.backward(grad)\n o = t(x_ref, w_ref, None)\n o.backward(grad)\n\n self.assertEqual(jit_o, o)\n self.assertEqual(x.grad, x_ref.grad)\n self.assertEqual(w.grad, w_ref.grad)\n\n @unittest.skipIf(GRAPH_EXECUTOR != ProfilingMode.PROFILING, \"the profiling version of test_rand\")\n def test_rand_profiling(self):\n def test_rand():\n a = torch.rand([3, 4])\n return a + 1.0 - a\n\n # Testing shape analysis correctly setting type\n with enable_profiling_mode_for_profiling_tests():\n with num_profiled_runs(1):\n fn = torch.jit.script(test_rand)\n out = fn()\n graph_str = torch.jit.last_executed_optimized_graph()\n self.assertEqual(out.dtype, torch.double)\n FileCheck().check(\"Double(3, 4, strides=[4, 1], requires_grad=0, device=cpu)\") \\\n .check_not(\"Float(3, 4, strides=[4, 1], requires_grad=0, device=cpu)\").run(graph_str)\n\n # fn = self.checkScript(test_rand, ())\n # out = fn()\n # self.assertEqual(out.dtype, torch.double)\n\n @torch.jit.script\n def randint():\n return torch.randint(0, 5, [1, 2])\n\n # although the type should be int here, testing that the runtime dtype\n # and shape analysis dtype is the same.\n with enable_profiling_mode_for_profiling_tests():\n with num_profiled_runs(1):\n out = randint()\n graph_str = torch.jit.last_executed_optimized_graph()\n self.assertEqual(out.dtype, torch.double)\n FileCheck().check(\"profiled_type=Double(1, 2, strides=[2, 1], requires_grad=0, device=cpu)\").run(graph_str)\n\n\n def test_erase_number_types(self):\n def func(a):\n b = 7 + 1 + 3\n c = a + b\n c += b\n return c\n\n graph = torch.jit.script(func).graph\n FileCheck().check(\"int = prim::Constant\").check(\"aten::add_\").run(str(graph))\n self.run_pass('remove_inplace_ops', graph)\n self.run_pass('erase_number_types', graph)\n FileCheck().check_not(\"int = prim::Constant\").check_not(\"aten::add_\").run(str(graph))\n\n def test_remove_dropout(self):\n weight_0_shape = (20, 5)\n weight_1_shape = (20, 20)\n input_shape = (10, 5)\n\n class M(torch.nn.Module):\n def __init__(self):\n super(M, self).__init__()\n self.weight_0 = torch.nn.Parameter(torch.rand(weight_0_shape))\n self.weight_1 = torch.nn.Parameter(torch.rand(weight_1_shape))\n\n def forward(self, x):\n o = F.linear(x, self.weight_0)\n o = F.dropout(o, training=self.training)\n o = F.linear(o, self.weight_1)\n return o\n\n data = torch.rand(input_shape)\n m = M()\n m = torch.jit.script(m)\n with self.assertRaisesRegex(RuntimeError, r'Dropout removal module in training mode is not yet supported'):\n torch._C._jit_pass_remove_dropout(m._c)\n m.eval()\n ref_res = m(data)\n # Need to inline otherwise we see instances of Function.\n # We would have to use torch.linear/dropout to get around it otherwise.\n from torch.jit._recursive import wrap_cpp_module\n m = wrap_cpp_module(torch._C._freeze_module(m._c))\n torch._C._jit_pass_remove_dropout(m._c)\n res = m(data)\n FileCheck().check_not(\"aten::dropout\").run(str(m.graph))\n torch.testing.assert_allclose(ref_res, res, rtol=1e-2, atol=1e-3)\n\n def test_unfold_zero_dim(self):\n def fn(x):\n return x.unfold(0, 1, 1)\n\n graph = torch.jit.script(fn).graph\n torch._C._jit_pass_complete_shape_analysis(graph, (torch.tensor(0.39),), False)\n out_dims = fn(torch.tensor(0.3923)).ndim\n self.assertEqual(graph.findNode(\"aten::unfold\").output().type().dim(), out_dims)\n\n def test_mm_batching(self):\n\n with enable_profiling_mode_for_profiling_tests():\n lstm_cell = torch.jit.script(LSTMCellS)\n\n def lstm(x, hx, cx, w_ih, w_hh, b_ih, b_hh):\n for i in range(x.size(0)):\n hx, cx = lstm_cell(x[i], hx, cx, w_ih, w_hh, b_ih, b_hh)\n return hx\n\n slstm = torch.jit.script(lstm)\n\n inputs = get_lstm_inputs('cpu', training=True, seq_length=10)\n slstm(*inputs, profile_and_replay=True).sum().backward(retain_graph=True)\n if GRAPH_EXECUTOR == ProfilingMode.PROFILING:\n slstm(*inputs, profile_and_replay=True).sum().backward()\n\n fw_graph = slstm.graph_for(*inputs)\n if GRAPH_EXECUTOR == ProfilingMode.LEGACY:\n bw_graph = backward_graph(slstm, diff_graph_idx=0)\n self.assertTrue('prim::MMBatchSide' in str(fw_graph))\n self.assertTrue('prim::MMTreeReduce' in str(bw_graph))\n\n sout = slstm(*inputs)\n out = lstm(*inputs)\n self.assertEqual(sout, out)\n self.assertEqual(torch.autograd.grad(sout.sum(), inputs),\n torch.autograd.grad(out.sum(), inputs))\n\n def test_loop_unrolling(self):\n def fn(x):\n y = 0\n for i in range(int(x)):\n y -= i\n return y\n\n graph = torch.jit.script(fn).graph\n self.run_pass('loop_unrolling', graph)\n unroll_factor = 8\n FileCheck().check(\"prim::Loop\").check_count(\"aten::sub\", unroll_factor) \\\n .check(\"prim::Loop\").check(\"aten::sub\").run(str(graph))\n self.checkScript(fn, (torch.tensor(10),))\n\n def test_loop_unrolling_const(self):\n def fn():\n y = 0\n for _ in range(10):\n y -= 1\n return y\n\n def fn2():\n y = 0\n for i in range(10):\n y -= i\n return y\n\n def check(fn, name):\n graph = torch.jit.script(fn).graph\n self.run_pass('loop_unrolling', graph)\n # entirely unrolled\n FileCheck().check_not(\"prim::Loop'\").run(str(graph))\n self.checkScript(fn, ())\n\n check(fn, 'add_const')\n check(fn2, 'add_iter')\n\n def test_loop_unrolling_nested(self):\n def fn(x):\n y = 0\n for _ in range(10):\n for j in range(int(x)):\n y -= j\n return y\n\n graph = torch.jit.script(fn).graph\n self.run_pass('loop_unrolling', graph)\n # inner loop with 8 subs followed by loop epilogue\n unroll_factor = 8\n FileCheck().check(\"prim::Loop\").check(\"prim::Loop\").check_count('aten::sub', unroll_factor) \\\n .check(\"prim::Loop\").check(\"aten::sub\").run(str(graph))\n self.checkScript(fn, (torch.tensor(10),))\n\n def test_loop_unroll_unused_counter(self):\n def fn(x):\n y = 0\n for _ in range(int(x)):\n y -= 1\n return y\n\n graph = torch.jit.script(fn).graph\n self.run_pass('loop_unrolling', graph)\n FileCheck().check(\"prim::Loop\").check_not(\"aten::add\").check(\"return\") \\\n .run(str(graph))\n\n def test_loop_unroll_negative(self):\n def fn(x):\n y = 0\n for _ in range(int(x)):\n y += 1\n return y\n\n self.checkScript(fn, (torch.tensor(-20),))\n self.checkScript(fn, (torch.tensor(-2),))\n self.checkScript(fn, (torch.tensor(-1),))\n self.checkScript(fn, (torch.tensor(0),))\n self.checkScript(fn, (torch.tensor(1),))\n self.checkScript(fn, (torch.tensor(2),))\n\n def test_where(self):\n def fn(x, y):\n return torch.where(x > 0.0, x, y)\n\n self.checkScript(fn, (torch.randn(3, 2, dtype=torch.float), torch.ones(3, 2, dtype=torch.float)))\n\n def test_where_method(self):\n def fn(x, y):\n return x.where(x > 0.0, y)\n\n self.checkScript(fn, (torch.randn(3, 2, dtype=torch.float), torch.ones(3, 2, dtype=torch.float)))\n\n def test_reassign_module_lhs(self):\n with self.assertRaisesRegex(RuntimeError, 'Cannot re-assign \\'self\\''):\n class ReassignSelfLHS(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, x):\n for _ in range(20):\n self = x\n return self\n\n ReassignSelfLHS()\n\n def test_reassign_module_rhs(self):\n with self.assertRaisesRegex(RuntimeError, 'Cannot re-assign \\'x\\' to a value of type module'):\n class ReassignSelfRHS(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, x):\n for _ in range(20):\n x = self\n return self\n\n ReassignSelfRHS()\n\n def test_unknown_builtin(self):\n with self.assertRaisesRegex(RuntimeError, 'object has no attribute or method'):\n @torch.jit.script\n def unknown_builtin(x):\n return x.splork(3)\n\n def test_return_tuple(self):\n def return_tuple(x):\n a = (x, x)\n return a, x\n self.checkScript(return_tuple, (torch.rand(4),))\n\n def test_add_tuple_optional(self):\n def foo(input: Tuple[torch.Tensor, Optional[torch.Tensor], Optional[torch.Tensor]]) -> Optional[torch.Tensor]:\n changed_input = input[0] + 1\n value: Tuple[torch.Tensor, Optional[torch.Tensor], Optional[torch.Tensor]] = (changed_input,) + input[1:]\n return value[2]\n inp: Tuple[torch.Tensor, Optional[torch.Tensor], Optional[torch.Tensor]] = (torch.rand(4), None, None)\n self.checkScript(foo, (inp,))\n\n def test_add_tuple_non_optional(self):\n def foo(input: Tuple[torch.Tensor, torch.Tensor, torch.Tensor]) -> torch.Tensor:\n changed_input = input[0] + 1\n value: Tuple[torch.Tensor, torch.Tensor, torch.Tensor] = (changed_input,) + input[1:]\n return torch.sum(value[2]) + 4\n inp: Tuple[torch.Tensor, torch.Tensor, torch.Tensor] = (torch.rand(4), torch.rand(4), torch.rand(4))\n self.checkScript(foo, (inp,))\n\n def test_add_tuple_different_types(self):\n def foo(a: Tuple[int, float], b: Tuple[int]) -> int:\n c: Tuple[int, float, int] = a + b\n d: Tuple[int, float, int, int] = c + b\n return d[3] + 1\n a = (1, 2.0)\n b = (3,)\n self.checkScript(foo, (a, b))\n\n def test_add_tuple_same_types(self):\n def foo(a: Tuple[int, int], b: Tuple[int, int, int]) -> int:\n c: Tuple[int, int, int, int, int] = a + b\n d: Tuple[int, int, int, int, int, int, int, int] = c + b\n return d[6] - 2\n a = (1, 2)\n b = (3, 4, 5)\n self.checkScript(foo, (a, b))\n\n def test_method_no_self(self):\n with self.assertRaisesRegex(RuntimeError, 'methods must have a self argument'):\n class MethodNoSelf(torch.jit.ScriptModule):\n @torch.jit.script_method # noqa: B902\n def forward(): # noqa: B902\n return torch.zeros(3, 4)\n\n MethodNoSelf()\n\n def test_return_stmt_not_at_end(self):\n def return_stmt(x):\n if bool(x > 3):\n return x + 3\n else:\n return x\n self.checkScript(return_stmt, (torch.rand(1),))\n\n def test_for_in_range(self):\n def fn():\n c = 0\n for i in range(100):\n c += i\n return c\n self.checkScript(fn, ())\n\n def test_for_in_range_dynamic(self):\n def fn():\n c = 0\n for i in range(100):\n acc = 0\n for j in range(i):\n acc += j\n c += acc\n return c\n self.checkScript(fn, (), optimize=False)\n\n def test_for_in_range_ast(self):\n def test_script_for_in_range_ast():\n c = 0\n for i in range(100):\n acc = 0\n for j in range(i):\n acc += j\n c += acc\n return c\n\n self.checkScript(test_script_for_in_range_ast, ())\n\n def test_for_in_range_if_ast(self):\n @torch.jit.script\n def test_script_for_in_range_if_ast(x):\n output = x\n for i in range(20):\n if i == 0:\n output = x.unsqueeze(0)\n else:\n output = torch.cat((output, x.unsqueeze(0)), dim=0)\n return output\n inputs = self._make_scalar_vars([0], torch.int64)\n\n self.assertEqual(test_script_for_in_range_if_ast(*inputs).shape[0], 20)\n\n def test_for_in_range_start_end(self):\n def fn():\n x = 0\n for i in range(7, 100):\n x += i\n return x\n self.checkScript(fn, ())\n\n def test_for_in_range_start_end_step(self):\n def fn(start, end, step):\n # type: (int, int, int) -> int\n x = 0\n for i in range(start, end, step):\n x += i\n return x\n\n self.checkScript(fn, (7, 100, 7))\n self.checkScript(fn, (7, 100, -7))\n self.checkScript(fn, (2, -11, -3))\n self.checkScript(fn, (2, -11, 3))\n self.checkScript(fn, (2, 10, 3))\n self.checkScript(fn, (-2, -10, -10))\n\n def test_for_in_range_zero_step(self):\n @torch.jit.script\n def fn():\n x = 0\n for i in range(2, -11, 0):\n x += i\n return x\n\n with self.assertRaisesRegex(RuntimeError, \"must not be zero\"):\n fn()\n\n def test_range_args(self):\n with self.assertRaisesRegex(RuntimeError, r'range expected at least 1 arguments, got 0'):\n @torch.jit.script\n def range_no_arg(x):\n for _ in range():\n x += 1\n return x\n with self.assertRaisesRegex(RuntimeError, r'found float'):\n @torch.jit.script\n def range_non_float():\n for i in range(.5):\n print(i)\n\n def test_parse_empty_tuple_annotation(self):\n cu = torch.jit.CompilationUnit('''\n def foo(x : Tuple[()]) -> Tuple[()]:\n return x\n ''')\n\n foo_code = cu.find_function('foo').code\n FileCheck().check(\"Tuple[()]\").check(\"Tuple[()]\").run(foo_code)\n\n def test_parse_empty_tuple_annotation_element_error(self):\n with self.assertRaisesRegex(\n RuntimeError, 'Tuple literal in Tuple type annotation must not have any elements'):\n cu = torch.jit.CompilationUnit('''\n def foo(x : Tuple[(int,)]) -> Tuple[(int,)]:\n return x\n ''')\n\n def test_parse_none_type_annotation(self):\n cu = torch.jit.CompilationUnit('''\n def foo(x : NoneType) -> NoneType:\n return x\n ''')\n\n foo_code = cu.find_function('foo').code\n FileCheck().check(\": NoneType\").check(\"-> NoneType\").run(foo_code)\n\n def test_empty_tuple_str(self):\n empty_tuple_type = torch._C.TupleType([])\n g = {'Tuple' : typing.Tuple}\n python_type = eval(empty_tuple_type.annotation_str, g)\n assert python_type is typing.Tuple[()]\n\n def test_none_type_str(self):\n none_type = torch._C.NoneType.get()\n g = {'NoneType' : type(None)}\n python_type = eval(none_type.annotation_str, g)\n assert python_type is type(None)\n\n def test_zip_enumerate_modulelist(self):\n class Sub(torch.nn.Module):\n def __init__(self):\n super(Sub, self).__init__()\n\n def forward(self, thing):\n return thing - 2\n\n class Double(torch.nn.Module):\n def __init__(self):\n super(Double, self).__init__()\n\n def forward(self, thing):\n return thing * 2\n\n # zipping over two\n class ZipModLists(torch.nn.Module):\n def __init__(self, mods, mods2):\n super(ZipModLists, self).__init__()\n self.mods = mods\n self.mods2 = mods2\n\n def forward(self, x):\n iter = 0\n for mod1, mod2 in zip(self.mods, self.mods2):\n x = mod2(mod1(x))\n iter += 1\n return x, iter\n\n class ZipWithValues(torch.nn.Module):\n __constants__ = ['tup_larger', 'tup_smaller']\n\n def __init__(self, mods, mods2):\n super(ZipWithValues, self).__init__()\n self.mods = mods\n self.mods2 = mods2\n self.tup_larger = list(range(len(mods2) + 1))\n self.tup_smaller = list(range(max(len(mods2) + 1, 1)))\n\n def forward(self, x):\n iter = 0\n x2 = x\n for val, mod1, mod2 in zip(self.tup_larger, self.mods, self.mods2):\n x = mod2(mod1(x)) + val\n iter += 1\n for val, mod1, mod2 in zip(self.tup_smaller, self.mods, self.mods2):\n x2 = mod2(mod1(x2)) + val\n iter += 1\n return x, iter\n\n mods = nn.ModuleList([Double()]), nn.ModuleList([Double(), Sub(), Sub()]), nn.ModuleList([Sub(), Double()])\n for i in range(len(mods)):\n for j in range(len(mods)):\n mod = ZipModLists(mods[i], mods[j])\n self.checkModule(mod, (torch.tensor(.5),))\n mod2 = ZipWithValues(mods[i], mods[j])\n self.checkModule(mod2, (torch.tensor(.5),))\n\n\n def test_enumerate_modlist_range(self):\n class Double(torch.nn.Module):\n def forward(self, thing):\n return thing * 2\n\n class Mod(torch.nn.Module):\n def __init__(self):\n super(Mod, self).__init__()\n self.mods = nn.ModuleList([Double(), Double()])\n\n def forward(self, x):\n x2 = x\n iter = 0\n for val, mod in enumerate(self.mods):\n x2 = mod(x2) * val\n iter += 1\n return iter, x, x2\n\n self.checkModule(Mod(), (torch.tensor(.5),))\n\n # variable length, modulelist\n class Mod2(Mod):\n def forward(self, x):\n for val, mod in zip(range(int(x)), self.mods):\n x = mod(x) * val\n return x\n\n with self.assertRaisesRegex(Exception, \"that does not have a statically determinable length\"):\n torch.jit.script(Mod2())\n\n # modulelist, variable length\n class Mod3(Mod):\n def forward(self, x):\n for val, mod in zip(self.mods, range(int(x))):\n x = mod(x) * val\n return x\n\n with self.assertRaisesRegex(Exception, \"that does not have a statically determinable length\"):\n torch.jit.script(Mod3())\n\n def test_for_in_enumerate(self):\n def fn(x):\n # type: (List[int]) -> int\n sum = 0\n for (i, v) in enumerate(x):\n sum += i * v\n\n return sum\n\n self.checkScript(fn, ([1, 2, 3, 4, 5],))\n\n def fn_enumerate_start_index(x):\n # type: (List[int]) -> int\n sum = 0\n for (i, v) in enumerate(x, start=1):\n sum += i * v\n\n return sum\n\n self.checkScript(fn, ([1, 2, 3, 4, 5],))\n\n def fn_nested_enumerate(x):\n # type: (List[int]) -> int\n sum = 0\n for (i, (j, v)) in enumerate(enumerate(x)):\n sum += i * j * v\n\n return sum\n\n self.checkScript(fn, ([1, 2, 3, 4, 5],))\n\n with self.assertRaisesRegex(RuntimeError, r'enumerate expected at least 1 arguments, got 0'):\n @torch.jit.script\n def enumerate_no_arg(x):\n # type: (List[int]) -> int\n sum = 0\n for _ in enumerate():\n sum += 1\n\n return sum\n\n with self.assertRaisesRegex(RuntimeError, r'enumerate expected at most 2 arguments, got 3'):\n @torch.jit.script\n def enumerate_too_many_args(x):\n # type: (List[int]) -> int\n sum = 0\n for _ in enumerate(x, x, x):\n sum += 1\n\n return sum\n\n def test_list_comprehension_modulelist(self):\n class Inner(torch.nn.Module):\n def forward(self, x):\n return x + 10\n\n class M(torch.nn.Module):\n def __init__(self, mod_list):\n super(M, self).__init__()\n self.module_list = mod_list\n\n def forward(self, x):\n out = torch.jit.annotate(List[Tensor], [mod(x) for mod in self.module_list])\n return out\n\n mod = M(nn.ModuleList([Inner(), Inner()]))\n self.checkModule(mod, (torch.tensor(3),))\n\n mod = M(nn.ModuleList([]))\n torch.jit.script(mod)\n\n class M2(M):\n def __init__(self, mod_list):\n super(M2, self).__init__(mod_list)\n\n def forward(self, x):\n out = [mod(x) for mod in self.module_list]\n return out\n\n mod = M2(nn.ModuleList([Inner(), Inner()]))\n self.checkModule(mod, (torch.tensor(3),))\n\n mod = M2(nn.ModuleList([]))\n # defaults to List of Tensor for empty modulelist\n self.assertEqual(torch.jit.script(mod)(torch.tensor(.5)), [])\n\n def bad_type_annotation():\n out = torch.jit.annotate(int, [x for x in [1, 2, 3]]) # noqa: C416\n return out\n\n with self.assertRaisesRegex(Exception, \"Expected list type annotation\"):\n torch.jit.script(bad_type_annotation)\n\n def test_list_comprehension_variable_write(self):\n # i in comprehension doesn't write to function scope\n def foo():\n i = 1\n x = [i if i != 5 else 3 for i in range(7)] # noqa: C416\n return i, x\n\n self.assertEqual(foo(), torch.jit.script(foo)())\n\n def test_for_in_zip(self):\n def fn(x, y):\n # type: (List[int], List[int]) -> int\n sum = 0\n for (i, j) in zip(x, y):\n sum += i * j\n\n return sum\n\n self.checkScript(fn, ([1, 2, 3, 4, 5], [2, 3, 4, 5, 6]))\n\n def fn_multi_inputs(x, y, z):\n # type: (List[int], List[int], List[int]) -> int\n sum = 0\n for (i, j, k) in zip(x, y, z):\n sum += i * j * k\n\n return sum\n\n self.checkScript(fn_multi_inputs, ([1, 2, 3, 4], [2, 3, 4, 5], [3, 4, 5, 6]))\n\n def fn_nested_zip(x, y, z):\n # type: (List[int], List[int], List[int]) -> int\n sum = 0\n for (i, (j, k)) in zip(x, zip(y, z)):\n sum += i * j * k\n\n return sum\n\n self.checkScript(fn_multi_inputs, ([1, 2, 3, 4], [2, 3, 4, 5], [3, 4, 5, 6]))\n\n with self.assertRaisesRegex(RuntimeError, r'zip expected at least 1 arguments, got 0'):\n @torch.jit.script\n def zip_no_arg(x):\n # type: (List[int]) -> int\n sum = 0\n for _ in zip():\n sum += 1\n\n return sum\n\n with self.assertRaisesRegex(RuntimeError, r'too many values to unpack: need 2 but found 3'):\n @torch.jit.script\n def fn_nested_zip_wrong_target_assign(x, y, z):\n # type: (List[int], List[int], List[int]) -> int\n sum = 0\n for (i, (j, k)) in zip(x, y, z):\n sum += i * j * k\n\n return sum\n\n def test_for_in_zip_enumerate(self):\n def fn_zip_enumerate(x, y):\n # type: (List[int], List[int]) -> int\n sum = 0\n for (i, (j, v), k) in zip(x, enumerate(y), range(0, 100)):\n sum += i * j * v * k\n\n return sum\n\n self.checkScript(fn_zip_enumerate, ([1, 2, 3, 4], [2, 3, 4, 5]))\n\n def fn_enumerate_zip(x, y):\n # type: (List[int], List[int]) -> int\n sum = 0\n for (i, (j, v)) in enumerate(zip(x, y)):\n sum += i * j * v\n\n return sum\n\n self.checkScript(fn_enumerate_zip, ([1, 2, 3, 4], [2, 3, 4, 5]))\n\n def test_for_in_tensors(self):\n def test_sizes(x):\n sumz = 0\n for s in x:\n sumz += 1\n return sumz\n self.checkScript(test_sizes, (torch.rand(5, 4, 3, 2, 1),))\n self.checkScript(test_sizes, (torch.rand(777),))\n self.checkScript(test_sizes, (torch.rand(0),))\n\n def test_for_in_tensors_rank0(self):\n with self.assertRaisesRegex(RuntimeError, \"of a 0-d tensor\"):\n @torch.jit.script\n def test_sizes(x):\n sumz = 0\n for s in x:\n sumz += 1\n return sumz\n\n test_sizes(torch.tensor(1))\n\n def test_for_in_tensors_fail_scalar(self):\n with self.assertRaisesRegex(RuntimeError, \"'float' object is not iterable\"):\n @torch.jit.script\n def test_sizes(x):\n # type: (float) -> int\n sumz = 0\n for s in x:\n sumz += 1\n return sumz\n\n test_sizes(0.0)\n\n def test_for_in_tensors_nested(self):\n def test_sizes(x):\n sumz = 0\n for n in x:\n for t in n:\n sumz += 1\n return sumz\n\n self.checkScript(test_sizes, (torch.rand(5, 4, 3, 2, 1),))\n\n # to avoid defining sum_list in multiple tests\n def get_sum_list_fn(self):\n def sum_list(a):\n # type: (List[int]) -> int\n sum = 0\n for i in a:\n sum += i\n\n return sum\n\n return sum_list\n\n def test_sum_list_diff_elms(self):\n self.checkScript(self.get_sum_list_fn(), ([1, 2, 3, 4, 5],))\n\n def test_sum_list_empty(self):\n self.checkScript(self.get_sum_list_fn(), ([],))\n\n def test_sum_list_one(self):\n self.checkScript(self.get_sum_list_fn(), ([1],))\n\n def test_sum_list_literal(self):\n\n def sum_list():\n # type: () -> int\n sum = 0\n for i in [1, 2, 3, 4, 5]:\n sum += i\n\n return sum\n\n self.checkScript(sum_list, ())\n\n def test_sum_list_wrong_type(self):\n\n with self.assertRaisesRegex(RuntimeError, \"'int' object is not iterable\"):\n @torch.jit.script\n def sum_list(a):\n # type: (int) -> int\n sum = 0\n for i in a: # noqa: T484\n sum += i\n\n return sum\n\n sum_list(1)\n\n def test_list_iterables(self):\n with self.assertRaisesRegex(RuntimeError, 'List of iterables is not supported currently'):\n cu = torch.jit.CompilationUnit('''\n def list_iterables(x):\n for i, j in [2, 3, 4], [5, 6, 7]:\n x += i\n x += j\n return x\n ''')\n\n def test_for_in_string(self):\n def test_strings(x):\n # type: (str) -> str\n reverse = \"\"\n for c in x:\n reverse = c + reverse\n return reverse\n\n self.checkScript(test_strings, (\"hello\",))\n self.checkScript(test_strings, (\"\",))\n\n def test_list_strings(x):\n # type: (List[str]) -> str\n result = \"\"\n for sub_str in x:\n result += sub_str\n return result\n\n self.checkScript(test_list_strings, ([\"hello\", \"world\"],))\n self.checkScript(test_list_strings, ([\"hello\", \" \", \"world\", \"\"],))\n\n def test_for_in_dict(self):\n def test_dicts(x):\n # type: (Dict[str, int]) -> int\n sum = 0\n for key in x:\n sum += x[key]\n return sum\n\n self.checkScript(test_dicts, ({\"a\": 1, \"b\": 2, \"c\": 3},))\n\n def test_dict_keys_values(x):\n # type: (Dict[str, int]) -> Tuple[str, int]\n key_str = \"\"\n sum = 0\n for key in x.keys():\n key_str += key\n for val in x.values():\n sum += val\n return key_str, sum\n\n self.checkScript(test_dicts, ({\"a\": 1, \"b\": 2, \"c\": 3},))\n\n def test_for_tuple_unpack(self):\n def for_tuple_unpack(x, y):\n for i, j in [[3, 4], [5, 6], [7, 8]]:\n x += i\n y += j\n return x, y\n\n self.checkScript(for_tuple_unpack, (torch.tensor(3), torch.tensor(5)))\n\n def nested_tuple_unpack(x, y):\n # type: (List[int], List[int]) -> int\n sum = 0\n for i, (j, k), v in zip(x, enumerate(x), y):\n sum += i + j + k + v\n return sum\n\n self.checkScript(nested_tuple_unpack, ([1, 3, 5], [2, 4, 6]))\n\n def test_for_tuple_assign(self):\n def test_simple_assign(x):\n # type: (Tuple[int, float]) -> float\n sum = 0.0\n for a in x:\n sum += float(a)\n return sum\n\n self.checkScript(test_simple_assign, ((1, 2.5),))\n\n def test_tuple_assign(x):\n # type: (Tuple[Tuple[int, int], Tuple[int, int]]) -> int\n sum = 0\n for a in x:\n sum += a[0]\n sum += a[1]\n return sum\n\n self.checkScript(test_tuple_assign, (((1, 2), (4, 7)), ))\n\n def test_single_starred_lhs(self):\n with self.assertRaisesRegex(RuntimeError, 'A Starred expression may only appear on the lhs within the presence'\n ' of another non-starred expression'):\n cu = torch.jit.CompilationUnit('''\n def single_starred_lhs(x):\n a = (x, x, x)\n *b, = a\n return b\n ''')\n\n def test_singleton_tuple_unpack(self):\n def foo(a):\n b, = (a,)\n return b + 1\n self.checkScript(foo, (torch.rand(3),))\n\n def test_tuple_assignments(self):\n def var_tuple_assign(x, y):\n # type: (Tuple[Tensor, Tensor], Tensor) -> Tensor\n (a, b), c = x, y\n return a + b + c\n\n tuple_inputs = (torch.randn(1, 4), torch.randn(3, 4))\n self.checkScript(var_tuple_assign, (tuple_inputs, torch.randn(3, 4)))\n\n def nested_tuple_assign(x, y, z):\n # type: (int, Tuple[int, Tuple[int, int]], Tuple[int, int]) -> int\n a, (b, (c, d)), (e, f) = x, y, z\n return a + b + c + d + e + f\n\n self.checkScript(nested_tuple_assign, ((1, (2, (3, 4)), (5, 6))))\n\n def subscript_tuple_assign(a, x, i):\n # type: (List[int], Tensor, int) -> Tuple[int, Tensor, int]\n a[i], (x[i], b) = 1, (2, 3)\n return a[i] + 1, x + 5, b\n\n self.checkScript(subscript_tuple_assign, ([12, 7, 9, 11], torch.tensor((3, 13, 17)), 0))\n\n def star_tuple_assign():\n # type: () -> Tuple[int, int, Tuple[int, int], Tuple[int, int]]\n a, (b, *c), *d = 1, (2, 3, 4), 5, 6\n return a, b, c, d\n\n self.checkScript(star_tuple_assign, ())\n\n def subscript_tuple_augmented_assign(a):\n # type: (Tuple[int, int]) -> Tuple[int, int]\n a[0] += 1\n return a\n\n with self.assertRaisesRegex(RuntimeError, 'does not support augmented assign'):\n scripted_aug_assign = torch.jit.script(subscript_tuple_augmented_assign)\n\n class AttrTupleAssignmentTestClass:\n def __init__(self, a: int, b: int):\n self.a = a\n self.b = b\n\n def set_ab(self, a: int, b: int):\n self.a, self.b = (a, b)\n\n def get(self) -> Tuple[int, int]:\n return (self.a, self.b)\n\n make_global(AttrTupleAssignmentTestClass)\n\n @torch.jit.script\n def attr_tuple_assignment(o: AttrTupleAssignmentTestClass, a: int, b: int):\n o.set_ab(a, b)\n return o\n\n o = AttrTupleAssignmentTestClass(1, 2)\n self.assertEqual(attr_tuple_assignment(o, 3, 4).get(), (3, 4))\n\n def test_multiple_assign(self):\n def test():\n a = b, c = d, f = (1, 1)\n\n # side effect\n ten = torch.tensor(1)\n ten1 = ten2 = ten.add_(1)\n\n # ordering\n x = 1\n y = 3\n x, y = y, x + y\n\n return a, b, c, d, f, ten, ten1, ten2, x, y\n\n self.checkScript(test, ())\n\n def test_multi_reduction(self):\n with self.assertRaisesRegex(\n RuntimeError,\n 'augmented assignment can only have one LHS expression'):\n cu = torch.jit.CompilationUnit('''\n def multi_reduction(x):\n a, b += x\n return a, b\n ''')\n\n def test_invalid_call_arguments(self):\n with self.assertRaisesRegex(RuntimeError, 'but instead found type '):\n @torch.jit.script\n def invalid_call_arguments(x):\n return torch.unsqueeze(3, 4, 5, 6, 7, 8)\n\n def test_invalid_lhs_assignment(self):\n with self.assertRaisesRegex(RuntimeError, 'unexpected expression'):\n cu = torch.jit.CompilationUnit('''\n def invalid_lhs_assignment(x):\n x + 1 = x\n return x\n ''')\n\n def test_multi_starred_expr_lhs(self):\n with self.assertRaisesRegex(RuntimeError, 'Only one starred expression is allowed on the lhs'):\n cu = torch.jit.CompilationUnit('''\n def multi_starred_expr_lhs():\n a, *b, *c = [1, 2, 3, 4, 5, 6]\n return a\n ''')\n\n def test_pack_tuple_into_non_var(self):\n with self.assertRaisesRegex(RuntimeError, 'Cannot pack a tuple into a non-variable'):\n cu = torch.jit.CompilationUnit('''\n def pack_tuple_into_non_var(x):\n a, *1 = (3, 4, 5)\n return x\n ''')\n\n def test_print_kwargs(self):\n with self.assertRaisesRegex(RuntimeError, 'print doesn\\'t accept any keyword arguments'):\n cu = torch.jit.CompilationUnit('''\n def print_kwargs(x):\n print(x, flush=True)\n return x\n ''')\n\n def test_builtin_use_as_value(self):\n with self.assertRaisesRegex(RuntimeError, 'builtin cannot be used as a value'):\n @torch.jit.script\n def builtin_use_as_value(x):\n return x.unsqueeze\n\n def test_wrong_use_as_tuple(self):\n with self.assertRaisesRegex(RuntimeError, 'cannot be used as a tuple'):\n def test_fn():\n return 3\n\n @torch.jit.script\n def wrong_use_as_tuple(self):\n a, b = test_fn\n return a\n\n def test_wrong_attr_lookup(self):\n with self.assertRaisesRegex(RuntimeError, 'attribute lookup is not defined on builtin'):\n @torch.jit.script\n def wrong_attr_lookup(self, x):\n a = x.unsqueeze.myattr\n return a\n\n def test_wrong_use_as_callable(self):\n with self.assertRaisesRegex(RuntimeError, 'cannot call a value'):\n @torch.jit.script\n def wrong_use_as_callable(x):\n return x(3, 4, 5)\n\n def test_python_val_doesnt_have_attr(self):\n with self.assertRaisesRegex(RuntimeError, 'object has no attribute abcd'):\n\n @torch.jit.script\n def python_val_doesnt_have_attr():\n # this has to be a module otherwise attr lookup would not be\n # allowed in the first place\n return shutil.abcd\n\n def test_wrong_module_attr_lookup(self):\n with self.assertRaisesRegex(RuntimeError, 'python value of type \\'type\\' cannot be used as a value'):\n import io\n\n @torch.jit.script\n def wrong_module_attr_lookup():\n return io.BytesIO\n\n def test_wrong_method_call_inputs(self):\n with self.assertRaisesRegex(RuntimeError, 'Argument y not provided'):\n class SomeModule(torch.jit.ScriptModule):\n\n @torch.jit.script_method\n def foo(self, x, y):\n return x\n\n @torch.jit.script_method\n def forward(self, x, y):\n return self.foo(x)\n SomeModule()\n\n def test_single_starred_expr_for_loop(self):\n with self.assertRaisesRegex(RuntimeError, 'A Starred expression may only appear'):\n cu = torch.jit.CompilationUnit('''\n def test():\n x = 0\n for *a in [1, 2, 3]:\n x = x + 1\n return x\n ''')\n\n def test_call_ge(self):\n with self.assertRaisesRegex(RuntimeError, 'Expected at most 1 arguments but found 3'):\n @_trace(torch.zeros(1, 2, 3))\n def foo(x):\n return x\n\n @torch.jit.script\n def test_fn():\n return foo(torch.full([1], 1), torch.full([1], 2), torch.full([1], 3))\n\n def test_wrong_return_type(self):\n with self.assertRaisesRegex(RuntimeError, 'but instead got value of type tuple'):\n @torch.jit.ignore\n def somefunc():\n # type: () -> Tuple[Tuple[Tensor, Tensor]]\n return torch.zeros(3, 4), torch.zeros(4, 5) # noqa: T484\n\n @torch.jit.script\n def wrong_return_type():\n return somefunc()\n wrong_return_type()\n\n # Tests for calling between different front-end modes\n def test_call_python_fn_from_tracing_fn(self):\n def python_fn(x):\n return torch.neg(x)\n\n @_trace(torch.rand(3, 4))\n def traced_fn(x):\n return python_fn(x) + 1\n\n # The neg op in the python function should be properly inlined to the\n # graph\n FileCheck().check(\"aten::neg\").run(str(traced_fn.graph))\n\n def test_call_python_mod_from_tracing_fn(self):\n class PythonMod(torch.nn.Module):\n def __init__(self):\n super(PythonMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 3), requires_grad=False)\n\n def forward(self, x):\n return torch.mm(x, self.param)\n\n pm = PythonMod()\n\n @_trace(torch.rand(3, 4))\n def traced_fn(x):\n return pm(x) + 1.0\n\n # Note: the parameter self.param from the Python module is inlined\n # into the graph\n self.assertTrue(len(list(traced_fn.graph.inputs())) == 1)\n FileCheck().check(\"aten::mm\").check(\"aten::add\").run(str(traced_fn.graph))\n\n @_tmp_donotuse_dont_inline_everything\n def test_call_traced_fn_from_tracing_fn(self):\n @_trace(torch.rand(3, 4))\n def traced_fn1(x):\n return torch.neg(x)\n\n @_trace(torch.rand(3, 4))\n def traced_fn(x):\n return traced_fn1(x) + 1\n\n FileCheck().check(\"traced_fn\").check(\"prim::CallFunction\").check(\"aten::add\") \\\n .run(str(traced_fn.graph))\n\n @unittest.skip(\"error in first class mode\")\n def test_call_traced_mod_from_tracing_fn(self):\n class TracedModule(torch.nn.Module):\n def __init__(self):\n super(TracedModule, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 3), requires_grad=False)\n\n def forward(self, x):\n return torch.mm(x, self.param)\n\n tm = torch.jit.trace(TracedModule(), torch.rand(3, 4))\n\n with self.assertRaisesRegex(RuntimeError, \"must be registered as submodules\"):\n @_trace(torch.rand(3, 4))\n def traced_fn(x):\n return tm(x) + 1.0\n\n @_tmp_donotuse_dont_inline_everything\n def test_call_script_fn_from_tracing_fn(self):\n @torch.jit.script\n def script_fn(x):\n return torch.neg(x)\n\n @_trace(torch.rand(3, 4))\n def traced_fn(x):\n return script_fn(x) + 1\n\n FileCheck().check(\"prim::CallFunction\").check(\"aten::add\").run(str(traced_fn.graph))\n\n @unittest.skip(\"error in first class mode\")\n def test_call_script_mod_from_tracing_fn(self):\n with self.assertRaisesRegex(RuntimeError, \"must be registered as submodules\"):\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(3, 4), requires_grad=False)\n\n @torch.jit.script_method\n def forward(self, x):\n for _i in range(4):\n x += self.param\n return x\n\n sm = ScriptMod()\n\n @_trace(torch.rand(3, 4))\n def traced_fn(x):\n return sm(x) + 1.0\n\n\n def test_call_python_fn_from_traced_module(self):\n def python_fn(x):\n return torch.neg(x)\n\n class TracedModule(torch.nn.Module):\n def __init__(self):\n super(TracedModule, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 3))\n\n def forward(self, x):\n return torch.mm(python_fn(x), self.param)\n\n tm = torch.jit.trace(TracedModule(), torch.rand(3, 4))\n\n # Note: parameter self.param from the traced module should appear as\n # an input to the graph and the neg op from the Python function should\n # be properly inlined\n self.assertTrue(len(list(tm.graph.inputs())) == 2)\n FileCheck().check(\"aten::neg\").check(\"aten::mm\").run(str(tm.graph))\n\n def test_call_python_mod_from_traced_module(self):\n class PythonModule(torch.nn.Module):\n def __init__(self):\n super(PythonModule, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(5, 7))\n\n def forward(self, x):\n return torch.mm(x, self.param)\n\n class TracedModule(torch.nn.Module):\n def __init__(self):\n super(TracedModule, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 5))\n self.mod = PythonModule()\n\n def forward(self, x):\n return self.mod(torch.mm(x, self.param)) + 1.0\n\n tm = torch.jit.trace(TracedModule(), torch.rand(3, 4))\n\n FileCheck().check_not(\"value=<Tensor>\").check(\"aten::mm\")\\\n .check(\"prim::CallMethod[name=\\\"forward\\\"]\").check(\"aten::add\") \\\n .run(str(tm.graph))\n FileCheck().check(\"aten::mm\").run(str(tm.mod.graph))\n\n def test_op_dtype(self):\n\n def check_equal_and_dtype(a, b):\n self.assertEqual(a, b)\n self.assertEqual(a.dtype, b.dtype)\n\n def fn():\n a = torch.arange(10)\n b = torch.arange(10, dtype=torch.float)\n c = torch.arange(1, 10, 2)\n d = torch.arange(1, 10, 2, dtype=torch.float)\n e = torch.arange(1, 10., 2)\n f = torch.arange(1, 10., 2, dtype=torch.float)\n return a, b, c, d, e, f\n\n scripted_fn = torch.jit.script(fn)\n eager_out = fn()\n script_out = scripted_fn()\n for a, b in zip(eager_out, script_out):\n check_equal_and_dtype(a, b)\n\n def test_floordiv(self):\n funcs_template = dedent('''\n def fn():\n ten = {a_construct}\n ten_or_scalar = {b_construct}\n return ten // ten_or_scalar, torch.floor_divide(ten, ten_or_scalar)\n ''')\n\n lhs = [\"torch.tensor([5.5, 3.2])\", \"torch.tensor([2, 2])\", \"torch.tensor([3, 2])\"]\n rhs = [\"1.5\", \"2\", \"4\", \"1.1\"] + lhs\n for tensor in lhs:\n for tensor_or_scalar in rhs:\n funcs_str = funcs_template.format(a_construct=tensor, b_construct=tensor_or_scalar)\n scope = {}\n execWrapper(funcs_str, globals(), scope)\n cu = torch.jit.CompilationUnit(funcs_str)\n f_script = cu.fn\n f = scope['fn']\n with self.assertWarnsOnceRegex(UserWarning, \"floor_divide\"):\n self.assertEqual(f_script(), f())\n\n def test_call_python_fn_from_script_fn(self):\n @torch.jit.ignore\n def python_fn(x):\n return torch.neg(x)\n\n @torch.jit.script\n def script_fn(x):\n return python_fn(x) + 1\n\n # Note: the call to python_fn appears as `^python_fn()` and is called\n # as a PythonOp in the interpreter\n a = torch.tensor(1)\n self.assertEqual(script_fn(a), torch.tensor(0))\n FileCheck().check(\"python_fn\").run(str(script_fn.graph))\n\n def test_call_python_mod_from_script_fn(self):\n class PythonModule(torch.nn.Module):\n def __init__(self):\n super(PythonModule, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(5, 7))\n\n def forward(self, x):\n return torch.mm(x, self.param)\n\n pm = PythonModule()\n\n @torch.jit.script\n def script_fn(x):\n return pm(x) + 1\n\n # Note: call to pm(x) appears as ^<python_value>() in the trace.\n # Parameters are NOT inlined.\n FileCheck().check(\"python_value\").check(\"aten::add\").run(str(script_fn.graph))\n\n @_tmp_donotuse_dont_inline_everything\n def test_call_script_fn_from_script_fn(self):\n @torch.jit.script\n def script_fn1(x):\n return torch.neg(x)\n\n @torch.jit.script\n def script_fn(x):\n return script_fn1(x) + 1\n\n FileCheck().check(\"prim::CallFunction\").run(str(script_fn.graph))\n\n def test_call_script_mod_from_script_fn(self):\n with self.assertRaisesRegex(RuntimeError, \"Cannot call a ScriptModule that is not a submodule of the caller\"):\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n\n @torch.jit.script_method\n def forward(self, x):\n return torch.mm(x, torch.zeros([4, 3]))\n\n sm = ScriptMod()\n\n @torch.jit.script\n def script_fn(x):\n return sm(x) + 1\n\n def test_call_python_fn_from_script_module(self):\n @torch.jit.ignore\n def python_fn(x):\n return torch.neg(x)\n\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 3))\n\n @torch.jit.script_method\n def forward(self, x):\n return python_fn(torch.mm(x, self.param))\n\n sm = ScriptMod()\n FileCheck().check(\"aten::mm\").check(\"python_fn\") \\\n .run(str(sm.forward.graph))\n\n def test_call_python_mod_from_script_module(self):\n class PythonMod(torch.nn.Module):\n def __init__(self):\n super(PythonMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(3, 5))\n\n @torch.jit.ignore\n def forward(self, x):\n return torch.mm(x, self.param)\n\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 3))\n self.pm = PythonMod()\n\n @torch.jit.script_method\n def forward(self, x):\n return self.pm(torch.mm(x, self.param))\n\n sm = ScriptMod()\n # Note: the call into PythonMod appears as ^forward(). Parameters\n # are NOT inlined\n FileCheck().check(\"aten::mm\").check(\"forward\").run(str(sm.graph))\n\n @_tmp_donotuse_dont_inline_everything\n def test_call_script_fn_from_script_module(self):\n @torch.jit.script\n def script_fn(x):\n return torch.neg(x)\n\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 3))\n\n @torch.jit.script_method\n def forward(self, x):\n return script_fn(torch.mm(x, self.param))\n\n sm = ScriptMod()\n graph = (sm.forward.graph)\n FileCheck().check(\"aten::mm\").check(\"prim::CallFunction\").run(str(graph))\n\n @_tmp_donotuse_dont_inline_everything\n def test_call_script_mod_from_script_module(self):\n class ScriptMod1(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod1, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(3, 5))\n\n @torch.jit.script_method\n def forward(self, x):\n return torch.mm(x, self.param)\n\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(4, 3))\n self.tm = ScriptMod1()\n\n @torch.jit.script_method\n def forward(self, x):\n return self.tm(torch.mm(x, self.param))\n\n sm = ScriptMod()\n # Note: the parameters from both modules should appear in the flattened\n # input list to the graph. The mm op from ScriptMod1 should be properly\n # inlined\n # 3 % values in graph input lists, two mms in body\n FileCheck().check_count('%', 3).check(\":\").check_count(\"mm\", 1).check(\"prim::CallMethod\").run(str(sm.graph))\n\n def test_module_with_params_called_fails(self):\n with self.assertRaisesRegex(RuntimeError, \"Cannot call a ScriptModule that is not a submodule of the caller\"):\n class ScriptMod(torch.jit.ScriptModule):\n def __init__(self):\n super(ScriptMod, self).__init__()\n self.param = torch.nn.Parameter(torch.rand(3, 3))\n\n @torch.jit.script_method\n def forward(self, x):\n return torch.mm(x, self.param)\n\n sm = ScriptMod()\n\n @torch.jit.script\n def some_func(x):\n return sm(x)\n\n def test_tuple_index_to_list(self):\n def test_non_constant_input(a):\n # type: (bool) -> int\n if a:\n b = 1\n else:\n b = 0\n c = (0, 1)\n return c[b]\n\n self.checkScript(test_non_constant_input, (True,))\n self.checkScript(test_non_constant_input, (False,))\n\n with self.assertRaisesRegex(RuntimeError, \"because we cannot resolve the output type\"):\n @torch.jit.script\n def test_non_constant_input(a):\n # type: (bool) -> None\n if a:\n b = 1\n else:\n b = 0\n c = (0, 1.1)\n print(c[b])\n\n def test_tuple_indexing(self):\n def tuple_index(a):\n if bool(a):\n b = (1, 2)\n else:\n b = (0, 2)\n return b[-2], b[1]\n\n self.checkScript(tuple_index, (torch.tensor([0]),))\n self.checkScript(tuple_index, (torch.tensor([1]),))\n self.checkScript(tuple_index, (torch.tensor([1]),), optimize=True)\n tuple_comp = torch.jit.script(tuple_index)\n FileCheck().check_count(\"TupleIndex\", 2, exactly=True).run(str(tuple_comp.graph))\n\n with self.assertRaisesRegex(RuntimeError, \"index must be an integer\"):\n @torch.jit.script\n def test_indexing_float():\n c = (1, 2)\n return c[0.1]\n\n def test_indexing_out_of_bounds_pos():\n c = (1, 2)\n return c[2]\n\n self.checkScriptRaisesRegex(test_indexing_out_of_bounds_pos, (), Exception,\n \"out of range\")\n\n def test_indexing_out_of_bounds_neg():\n c = (1, 2)\n return c[-3]\n\n self.checkScriptRaisesRegex(test_indexing_out_of_bounds_pos, (), Exception,\n \"out of range\")\n\n def negative_index():\n tup = (1, 2, 3, 4)\n return tup[-1]\n\n self.checkScript(negative_index, [])\n\n def really_negative_index():\n tup = (1, 2, 3, 4)\n return tup[-100]\n\n self.checkScriptRaisesRegex(really_negative_index, [], Exception, \"index out of range\")\n\n def negative_slice():\n tup = (1, 2, 3, 4)\n return tup[-3:4]\n\n self.checkScript(negative_slice, [])\n\n def really_slice_out_of_bounds():\n tup = (1, 2, 3, 4)\n return tup[-300:4000]\n\n self.checkScript(really_slice_out_of_bounds, [])\n\n def test_namedtuple_attr(self):\n def f(x):\n return x.max(dim=1).indices + torch.max(x, dim=1).indices\n\n self.checkScript(f, (torch.rand(20, 20, 20),), optimize=True)\n\n with self.assertRaisesRegex(RuntimeError, \"object has no attribute or method\"):\n @torch.jit.script\n def g1(x):\n return x.max(dim=1).unknown_symbol\n\n with self.assertRaisesRegex(RuntimeError, \"object has no attribute or method\"):\n @torch.jit.script\n def g2(x):\n print((x, x, x).__doc__)\n return x\n\n def test_tuple_len(self):\n @torch.jit.script\n def foo():\n return len((1, \"str\", None))\n\n self.assertEqual(foo(), 3)\n\n @torch.jit.script\n def test_indexing_end_out_of_bounds():\n c = (1, 2)\n return c[2:10]\n\n self.assertEqual(test_indexing_end_out_of_bounds(), ())\n\n def test_lower_nested_tuples(self):\n @torch.jit.script\n def test():\n return ((1, 2), 3)\n\n self.run_pass('constant_propagation', test.graph)\n FileCheck().check(\"prim::Constant\").check_not(\"TupleConstruct\").run(test.graph)\n # fails if a tuple can't be lowered\n self.run_pass('lower_all_tuples', test.graph)\n\n def test_unwrap_optional_builtin(self):\n def test(x):\n # type: (Optional[int]) -> int\n x = torch.jit._unwrap_optional(x)\n x = x + x # noqa: T484\n return x\n\n self.checkScript(test, (3,))\n\n with self.assertRaisesRegex(AssertionError, \"Unwrapping null optional\"):\n test(None)\n\n test_script = torch.jit.script(test)\n with self.assertRaisesRegex(RuntimeError, \"Unwrapping null optional\"):\n test_script(None)\n\n @torch.jit.script\n def test_test():\n return torch.jit._unwrap_optional(1)\n\n with self.assertRaisesRegex(RuntimeError, r\"could not be inferred from actual type None\"):\n @torch.jit.script\n def test_no_type():\n # type: () -> int\n return torch.jit._unwrap_optional(None)\n\n def test_indexing_error(self):\n with self.assertRaisesRegex(RuntimeError, \"'int' object is not subscriptable\"):\n @torch.jit.script\n def test_wrong_type():\n a = 8\n return a[0]\n\n def test_unsupported_builtin_error(self):\n with self.assertRaisesRegex(RuntimeError,\n \"Python builtin <built-in function hypot> is currently\"):\n @torch.jit.script\n def test_unsupported(a):\n return math.hypot(a, 2.0)\n\n def test_annotated_script_fn(self):\n @torch.jit.script\n def foo(x, y, z):\n # type: (Tensor, Tuple[Tensor, Tensor, Tensor], Tuple[Tensor, Tuple[Tensor, Tensor]]) -> Tensor\n return x\n\n self.assertExpected(str(foo.schema))\n\n def test_annotated_script_method(self):\n class SM(torch.jit.ScriptModule):\n @torch.jit.script_method\n def forward(self, x, y):\n # type: (Tuple[Tensor, Tensor], Tensor) -> Tuple[Tensor, Tensor, Tensor]\n return y, y, y\n\n sm = SM()\n\n self.assertExpectedStripMangled(str(sm.forward.schema))\n\n def test_annotated_script_fn_return_mismatch(self):\n with self.assertRaisesRegex(RuntimeError, \"but is actually of type\"):\n @torch.jit.script\n def return_tup(x):\n # type: (Tensor) -> Tuple[Tuple[Tensor, Tensor], Tensor]\n return x, x # noqa: T484\n\n def test_annotated_script_fn_arg_mismatch(self):\n with self.assertRaisesRegex(RuntimeError, r\"Arguments for call are not valid\"):\n @torch.jit.script\n def tuple_arg(x):\n # type: (Tuple[Tensor, Tensor]) -> Tensor\n return x + 1 # noqa: T484\n\n def test_script_non_tensor_args_outputs(self):\n @torch.jit.script\n def fn(x, y):\n # type: (Tensor, float) -> float\n return float((x + y).sum())\n\n x = torch.ones(2, 2)\n z = fn(x, 1)\n self.assertIsInstance(z, float)\n self.assertEqual(z, 8.)\n\n @unittest.skip('https://github.com/pytorch/pytorch/issues/9595')\n def test_inline_and_run_annotated_script_fn(self):\n @torch.jit.script\n def to_inline(x, y):\n # type: (Tuple[Tensor, Tensor], Tensor) -> Tensor\n return y\n\n @torch.jit.script\n def some_func(x):\n return to_inline((x, x), x)\n\n x = torch.rand(3, 4)\n self.assertEqual(some_func(x), x)\n\n def test_file_format_serialization(self):\n filename = tempfile.mktemp()\n writer = torch._C.PyTorchFileWriter(filename)\n buffers = [os.urandom(size) for size in [random.randint(1, 100) for i in range(20)]]\n offsets = []\n for i, buf in enumerate(buffers):\n writer.write_record(str(i), buf, len(buf))\n offsets.append(i)\n serialized_offsets = pickle.dumps(offsets)\n writer.write_record(\"meta\", serialized_offsets, len(serialized_offsets))\n writer.write_end_of_file()\n\n reader = torch._C.PyTorchFileReader(filename)\n serialized_offsets_read = reader.get_record(\"meta\")\n parsed_serialized_offsets = pickle.loads(serialized_offsets)\n\n for i, offset in enumerate(parsed_serialized_offsets):\n data = reader.get_record(str(offset))\n assert(data == buffers[i])\n\n # for each type, the input type annotation and corresponding return type annotation\n def type_input_return_pairs(self):\n return [\n ('Tensor', 'Tensor'),\n ('torch.Tensor', 'Tensor'),\n ('str', 'str'),\n ('int', 'int'),\n ('bool', 'bool'),\n ('BroadcastingList3[float]', 'List[float]'),\n ('BroadcastingList2[int]', 'List[int]'),\n ('List[int]', 'List[int]'),\n ('Optional[int]', 'Optional[int]'),\n ]\n\n # replacing code input & return type pair\n def format_code(self, code, pair):\n return code.format(input=pair[0], output=pair[1])\n\n # ***** Type annotation tests ****\n # Test combinations of:\n # {String frontend, Python AST Frontend}\n # {Python 3-style type annotations, MyPy-style type comments}\n # {Script method, Script function}\n\n # String frontend , Python 3-style type annotations , Script function\n def test_annot_string_py3_fn(self):\n code = '''\n def foo(x : {input}, y : Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]:\n return x, x\n '''\n test_str = []\n for pair in self.type_input_return_pairs():\n cu = torch.jit.CompilationUnit(self.format_code(code, pair))\n test_str.append(str(cu.foo.schema))\n self.assertExpected(\"\\n\".join(test_str))\n\n # String frontend , Python 3-style type annotations , Script method\n def test_annot_string_py3_method(self):\n class TestModule(torch.jit.ScriptModule):\n def __init__(self):\n super(TestModule, self).__init__()\n\n code = '''\n def foo(self, x : {input}, y : Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]:\n return x, x\n '''\n test_str = []\n for pair in self.type_input_return_pairs():\n # clear the class registry as we will be defining foo multiple times\n jit_utils.clear_class_registry()\n tm = TestModule()\n tm.define(self.format_code(code, pair))\n test_str.append(str(tm.foo.schema))\n self.assertExpectedStripMangled(\"\\n\".join(test_str))\n\n # String frontend , MyPy-style type comments , Script function\n def test_annot_string_mypy_fn(self):\n code = '''\n def foo(x, y):\n # type: ({input}, Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]\n return x, x\n '''\n test_str = []\n for pair in self.type_input_return_pairs():\n cu = torch.jit.CompilationUnit(self.format_code(code, pair))\n test_str.append(str(cu.foo.schema))\n self.assertExpectedStripMangled(\"\\n\".join(test_str))\n\n # String frontend , MyPy-style type comments , Script method\n def test_annot_string_mypy_method(self):\n class TestModule(torch.jit.ScriptModule):\n def __init__(self):\n super(TestModule, self).__init__()\n\n code = '''\n def foo(self, x, y):\n # type: ({input}, Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]\n return x, x\n '''\n\n test_str = []\n for pair in self.type_input_return_pairs():\n # clear the class registry as we will be defining foo multiple times\n jit_utils.clear_class_registry()\n tm = TestModule()\n tm.define(self.format_code(code, pair))\n test_str.append(str(tm.foo.schema))\n self.assertExpectedStripMangled(\"\\n\".join(test_str))\n\n # Python AST Frontend , Python 3-style type annotations , Script function\n def test_annot_ast_py3_fn(self):\n code = dedent('''\n from typing import Tuple, List, Optional\n from torch import Tensor\n from torch.jit.annotations import BroadcastingList2, BroadcastingList3\n import torch\n @torch.jit.script\n def foo(x : {input}, y : Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]:\n return x, x\n ''')\n test_str = []\n for pair in self.type_input_return_pairs():\n fn = jit_utils._get_py3_code(self.format_code(code, pair), 'foo')\n test_str.append(str(fn.schema))\n self.assertExpectedStripMangled(\"\\n\".join(test_str))\n\n def test_multiline_annot_ast_py3_fn(self):\n code = dedent('''\n from typing import Tuple, List, Optional\n from torch import Tensor\n from torch.jit.annotations import BroadcastingList2, BroadcastingList3\n import torch\n @torch.jit.script\n def foo(x, # type: {input}\n y # type: Tuple[Tensor, Tensor]\n ):\n # type: (...) -> Tuple[{output}, {output}]\n return x, x\n ''')\n test_str = []\n\n for pair in self.type_input_return_pairs():\n fn = jit_utils._get_py3_code(self.format_code(code, pair), 'foo')\n args = fn.schema.arguments\n returns = fn.schema.returns\n self.assertEqual(str(args[0].type), pair[1])\n self.assertEqual(str(args[1].type), \"Tuple[Tensor, Tensor]\")\n self.assertEqual(str(returns[0].type), \"Tuple[{}, {}]\".format(pair[1], pair[1]))\n\n def test_bad_multiline_annotations(self):\n with self.assertRaisesRegex(RuntimeError, \"Return type line\"):\n @torch.jit.script\n def bad_type_line(a, # type: Tensor\n b, # type: Tensor\n c # type: Tensor\n ):\n # type: (int, int, int) -> Tensor\n # type: bad type line # noqa: F723\n\n return a + b + c\n\n with self.assertRaisesRegex(RuntimeError, \"Return type line\"):\n @torch.jit.script\n def bad_return_line(a, # type: Tensor\n b,\n c # type: Tensor\n ):\n # type: (int, int, int) -> Tensor\n return a + b + c\n\n # TODO: this should be supported but is difficult to parse\n with self.assertRaisesRegex(RuntimeError, \"Number of type annotations\"):\n @torch.jit.script\n def missing_type(a, # type: Tensor\n b,\n c # type: Tensor\n ):\n # type: (...) -> Tensor\n return a + b + c\n\n # Python AST Frontend , Python 3-style type annotations , Script method\n def test_annot_ast_py3_method(self):\n code = dedent('''\n from typing import Tuple, List, Optional\n from torch import Tensor\n from torch.jit.annotations import BroadcastingList2, \\\\\n BroadcastingList3\n import torch\n class FooModule(torch.jit.ScriptModule):\n @torch.jit.script_method\n def foo(self, x : {input}, y : Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]:\n return x, x\n instance = FooModule()\n ''')\n\n test_str = []\n for pair in self.type_input_return_pairs():\n fn = jit_utils._get_py3_code(self.format_code(code, pair), 'instance')\n test_str.append(str(fn.foo.schema))\n self.assertExpectedStripMangled(\"\\n\".join(test_str))\n\n # Python AST Frontend , MyPy-style type comments , Script function\n def test_annot_ast_mypy_fn(self):\n code = dedent('''\n import torch\n @torch.jit.script\n def foo(x, y):\n # type: ({input}, Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]\n return x, x\n ''')\n\n test_str = []\n for pair in self.type_input_return_pairs():\n fn = jit_utils._get_py3_code(self.format_code(code, pair), 'foo')\n test_str.append(str(fn.schema))\n self.assertExpected(\"\\n\".join(test_str))\n\n # Python AST Frontend , MyPy-style type comments , Script method\n def test_annot_ast_mypy_method(self):\n code = dedent('''\n import torch\n class FooModule(torch.jit.ScriptModule):\n @torch.jit.script_method\n def foo(self, x, y):\n # type: ({input}, Tuple[Tensor, Tensor]) -> Tuple[{output}, {output}]\n return x, x\n instance = FooModule()\n ''')\n\n test_str = []\n for pair in self.type_input_return_pairs():\n fn = jit_utils._get_py3_code(self.format_code(code, pair), 'instance')\n test_str.append(str(fn.foo.schema))\n self.assertExpectedStripMangled(\"\\n\".join(test_str))\n\n # Tests that \"# type: ignore[*]\" is supported in type lines and is\n # properly ignored.\n def test_mypy_type_ignore(self):\n @torch.jit.script\n def foo(x): # type: ignore\n return x\n\n @torch.jit.script\n def bar(x): # type: ignore[no-redef]\n return x\n\n def test_method_casts_script(self):\n cast_types = [\n 'byte', 'char', 'double', 'float', 'int', 'long', 'short'\n ]\n\n for cast_type in cast_types:\n cu = torch.jit.CompilationUnit('''\n def cast_to(x):\n return x.{cast_type}()\n '''.format(cast_type=cast_type))\n\n x = torch.rand(3, 4, 5) * 128\n cu_result = cu.cast_to(x)\n reference = getattr(x, cast_type)()\n self.assertEqual(cu_result, reference)\n\n def test_string_frontend_elif(self):\n code = '''\n def func(niter):\n # type: (int)\n rv = 0\n for i in range(niter):\n if i % 3 == 0 and i % 5 == 0:\n rv += 35\n elif i % 3 == 0:\n rv += 3\n elif i % 5 == 0:\n rv += 5\n else:\n rv += i\n return rv\n '''\n\n self.checkScript(dedent(code), (101,))\n\n def test_pyop_exception_message(self):\n class Foo(torch.jit.ScriptModule):\n def __init__(self):\n super(Foo, self).__init__()\n self.conv = nn.Conv2d(1, 10, kernel_size=5)\n\n @torch.jit.script_method\n def forward(self, x):\n return self.conv(x)\n foo = Foo()\n # testing that the correct error message propagates\n with self.assertRaisesRegex(RuntimeError, \"Expected 4-dimensional input for 4-dimensional weight\"):\n foo(torch.ones([123])) # wrong size\n\n def test_builtin_error_messsage(self):\n with self.assertRaisesRegex(RuntimeError, \"Arguments for call are not valid\"):\n @torch.jit.script\n def close_match(x):\n return x.masked_fill(True)\n\n with self.assertRaisesRegex(RuntimeError, \"This op may not exist or may not be currently \"\n \"supported in TorchScript\"):\n @torch.jit.script\n def unknown_op(x):\n torch.set_anomaly_enabled(True)\n return x\n\n def test_exceptions(self):\n cu = torch.jit.CompilationUnit('''\n def foo(cond):\n if bool(cond):\n raise ValueError(3)\n return 1\n ''')\n\n cu.foo(torch.tensor(0))\n with self.assertRaisesRegex(torch.jit.Error, \"3\"):\n cu.foo(torch.tensor(1))\n\n def foo(cond):\n a = 3\n if bool(cond):\n raise ArbitraryError(a, \"hi\")\n if 1 == 2:\n raise ArbitraryError\n return a\n\n with self.assertRaisesRegex(RuntimeError, \"undefined value ArbitraryError\"):\n torch.jit.script(foo)\n\n def exception_as_value():\n a = Exception()\n print(a)\n\n with self.assertRaisesRegex(RuntimeError, \"cannot be used as a value\"):\n torch.jit.script(exception_as_value)\n\n @torch.jit.script\n def foo_no_decl_always_throws():\n raise RuntimeError(\"Hi\")\n\n # function that has no declared type but always throws set to None\n output_type = next(foo_no_decl_always_throws.graph.outputs()).type()\n self.assertTrue(str(output_type) == \"NoneType\")\n\n @torch.jit.script\n def foo_decl_always_throws():\n # type: () -> Tensor\n raise Exception(\"Hi\")\n\n output_type = next(foo_decl_always_throws.graph.outputs()).type()\n self.assertTrue(str(output_type) == \"Tensor\")\n\n def foo():\n raise 3 + 4\n\n with self.assertRaisesRegex(RuntimeError, \"must derive from BaseException\"):\n torch.jit.script(foo)\n\n # a escapes scope\n @torch.jit.script\n def foo():\n if 1 == 1:\n a = 1\n else:\n if 1 == 1:\n raise Exception(\"Hi\")\n else:\n raise Exception(\"Hi\")\n return a\n self.assertEqual(foo(), 1)\n\n @torch.jit.script\n def tuple_fn():\n raise RuntimeError(\"hello\", \"goodbye\")\n\n with self.assertRaisesRegex(torch.jit.Error, \"hello, goodbye\"):\n tuple_fn()\n\n @torch.jit.script\n def no_message():\n raise RuntimeError\n\n with self.assertRaisesRegex(torch.jit.Error, \"RuntimeError\"):\n no_message()\n\n def test_assertions(self):\n cu = torch.jit.CompilationUnit('''\n def foo(cond):\n assert bool(cond), \"hi\"\n return 0\n ''')\n\n cu.foo(torch.tensor(1))\n with self.assertRaisesRegex(torch.jit.Error, \"AssertionError: hi\"):\n cu.foo(torch.tensor(0))\n\n @torch.jit.script\n def foo(cond):\n assert bool(cond), \"hi\"\n\n foo(torch.tensor(1))\n # we don't currently validate the name of the exception\n with self.assertRaisesRegex(torch.jit.Error, \"AssertionError: hi\"):\n foo(torch.tensor(0))\n\n def test_python_op_exception(self):\n @torch.jit.ignore\n def python_op(x):\n raise Exception(\"bad!\")\n\n @torch.jit.script\n def fn(x):\n return python_op(x)\n\n with self.assertRaisesRegex(RuntimeError, \"operation failed in the TorchScript interpreter\"):\n fn(torch.tensor(4))\n\n def test_dict_expansion_raises_error(self):\n def fn(self):\n d = {\"foo\": 1, \"bar\": 2, \"baz\": 3}\n return {**d}\n\n with self.assertRaisesRegex(torch.jit.frontend.NotSupportedError,\n \"Dict expansion \"):\n torch.jit.script(fn)\n\n def test_module_parameters_and_buffers(self):\n weights = torch.randn(10, 10)\n bias = torch.randn(10)\n weights2 = torch.randn(10, 10)\n bias2 = torch.randn(10)\n\n class TestLinear(torch.nn.Module):\n def __init__(self, in_features, out_features):\n super(TestLinear, self).__init__()\n self.in_features = in_features\n self.out_features = out_features\n self.weight = torch.nn.Parameter(torch.empty(out_features, in_features))\n self.bias = torch.nn.Parameter(torch.empty(out_features))\n self.register_buffer('counter', torch.ones(out_features))\n self.reset_parameters()\n\n def reset_parameters(self):\n torch.nn.init.kaiming_uniform_(self.weight, a=math.sqrt(5))\n if self.bias is not None:\n fan_in, _ = torch.nn.init._calculate_fan_in_and_fan_out(self.weight)\n bound = 1 / math.sqrt(fan_in)\n torch.nn.init.uniform_(self.bias, -bound, bound)\n\n def forward(self, input):\n return F.linear(input, self.weight, self.bias) + self.counter\n\n # Initialize a ScriptModule that uses the weak module above multiple times\n class Strong(torch.jit.ScriptModule):\n def __init__(self):\n super(Strong, self).__init__()\n self.fc1 = TestLinear(10, 10)\n self.fc1.weight = torch.nn.Parameter(weights)\n self.fc1.bias = torch.nn.Parameter(bias)\n self.fc2 = TestLinear(10, 10)\n self.fc2.weight = torch.nn.Parameter(weights2)\n self.fc2.bias = torch.nn.Parameter(bias2)\n\n @torch.jit.script_method\n def forward(self, x):\n return x + self.fc1(x) + self.fc1(x) + self.fc2(x)\n\n strong_mod = Strong()\n\n # Run same calculation as module\n inp = torch.ones(10)\n lin = torch.nn.Linear(10, 10)\n lin.weight = torch.nn.Parameter(weights)\n lin.bias = torch.nn.Parameter(bias)\n lin2 = torch.nn.Linear(10, 10)\n lin2.weight = torch.nn.Parameter(weights2)\n lin2.bias = torch.nn.Parameter(bias2)\n expected_result = inp + (lin(inp) + torch.ones(10)) * 2 + lin2(inp) + torch.ones(10)\n\n self.assertEqual(strong_mod(inp), expected_result)\n self.assertExportImportModule(strong_mod, (inp,))\n\n def test_module_copying(self):\n class Submodule(torch.nn.Module):\n def __init__(self):\n super(Submodule, self).__init__()\n\n def forward(self, x):\n return x + 100\n\n class Weak(torch.nn.Module):\n def __init__(self, in_features, out_features):\n super(Weak, self).__init__()\n self.weight = torch.nn.Parameter(torch.ones(out_features, in_features))\n self.bias = torch.nn.Parameter(torch.ones(out_features))\n self.register_buffer(\"buffer\", torch.ones(out_features))\n self.submodule = Submodule()\n\n def forward(self, x):\n return F.linear(x, self.weight, self.bias) \\\n + self.buffer + self.submodule(x)\n\n class Strong(torch.jit.ScriptModule):\n def __init__(self, weak):\n super(Strong, self).__init__()\n self.weak = weak\n\n @torch.jit.script_method\n def forward(self, x):\n return self.weak(x)\n\n inp = torch.ones(5, 5) * 5\n weak_mod = Weak(5, 5)\n strong_mod = Strong(weak_mod)\n\n self.assertTrue(isinstance(strong_mod.weak, torch.jit.ScriptModule))\n self.assertFalse(isinstance(weak_mod, torch.jit.ScriptModule))\n\n self.assertIs(strong_mod.weak.weight, weak_mod.weight)\n self.assertIs(strong_mod.weak.buffer, weak_mod.buffer)\n # strong_mod.weak.submodule has been recursively scripted\n self.assertIsNot(strong_mod.weak.submodule, weak_mod.submodule)\n\n weak_mod.weight.data += torch.ones(5, 5) * 100\n self.assertTrue(strong_mod(inp).allclose(weak_mod(inp)))\n\n # Re-assignment is not tracked\n weak_mod.weight = torch.nn.Parameter(torch.ones(5, 5) * 100)\n self.assertFalse(strong_mod(inp).allclose(weak_mod(inp)))\n\n def test_backend_cudnn_enabled(self):\n # Only test that this compiles\n @torch.jit.script\n def fn(x):\n if torch.backends.cudnn.enabled:\n x = x + 2\n else:\n x = x + 3\n return x\n\n def test_inplace_add(self):\n\n def foo(a, b):\n c = a + b\n c.add_(b)\n return c\n self.checkScript(foo, (torch.rand(3), torch.rand(3)))\n\n def test_add_out(self):\n def foo(a, b):\n c = a + b\n e = 2 * a\n torch.add(c, b, out=e)\n return e\n self.checkScript(foo, (torch.rand(3), torch.rand(3)))\n\n def test_tuple_error_msg(self):\n def fn(t: Any):\n if isinstance(t, tuple):\n a, b = t\n return a + b\n with self.assertRaisesRegexWithHighlight(RuntimeError, \"Provided tuple is not fully defined/refined\", \"t\"):\n s = torch.jit.script(fn)\n\n def test_augmented_assign(self):\n def foo(a, b):\n a += b\n a -= b\n a /= b\n a *= b\n return a, b\n self.checkScript(foo, (torch.rand(3), torch.rand(3)))\n\n def test_ignored_props(self):\n class A(nn.Module):\n __jit_ignored_attributes__ = [\"ignored\", \"ignored_return_val\"]\n\n def __init__(self):\n super().__init__()\n\n @property\n def ignored(self):\n raise ValueError(\"shouldn't be called\")\n\n @property\n def ignored_return_val(self):\n return 1\n\n @torch.jit.ignore\n def call(self):\n return self.ignored_return_val\n\n f = torch.jit.script(A())\n # jank way to test if there is no error\n self.assertTrue(isinstance(f, torch.jit.ScriptModule))\n self.assertTrue(isinstance(f.call(), property))\n\n\n def test_pass(self):\n def foo(x):\n # type: (bool) -> int\n for _i in range(3):\n pass\n if x:\n pass\n else:\n pass\n return 3\n\n self.checkScript(foo, (True,))\n\n def test_lhs_indexing(self):\n def foo(a, b):\n a = a.clone()\n a[0] = b\n return a\n self.checkScript(foo, (torch.rand(2, 3), torch.rand(3)))\n\n def test_lhs_advanced_indexing_assignment(self):\n def foo(x, y):\n a = torch.exp(x)\n b = x == 1\n a[b] = y[b]\n return a\n self.checkScript(foo, (torch.ones(4, 3), torch.ones(4, 3)))\n\n def test_lhs_advanced_indexing_augmented_assignment(self):\n def foo(x, y):\n a = torch.exp(x)\n b = x == 1\n a[b] += y[b]\n return a\n self.checkScript(foo, (torch.ones(4, 3), torch.ones(4, 3)))\n\n def test_lhs_indexing_list(self):\n def foo(a, b):\n ls = [a]\n ls[0] = b\n return ls\n self.checkScript(foo, (torch.rand(2, 3), torch.rand(3)))\n\n def test_inplace_copy_script(self):\n def foo(x):\n a = torch.rand(3, 4)\n a.copy_(x)\n return a\n self.checkScript(foo, (torch.rand(3, 4),))\n\n def test_lhs_indexing_increment(self):\n def foo(a, b):\n a[0] += b\n return a\n self.checkScript(foo, (torch.rand(2, 3), torch.rand(3)))\n\n def test_lhs_indexing_increment_list(self):\n def foo(a, b):\n a = a.clone()\n ls = [a, b]\n ls[0] += b\n return ls\n self.checkScript(foo, (torch.rand(2, 3), torch.rand(3)))\n\n def test_lhs_indexing_increment_list_prim(self):\n def foo():\n ls = [1, 2, 3]\n ls[0] += 5\n return ls\n self.checkScript(foo, ())\n\n def test_lhs_indexing_multi(self):\n def foo(a, b):\n a = a.clone()\n foo, a[0], bar = (1, b, 3)\n return foo, a, bar\n self.checkScript(foo, (torch.rand(2, 3), torch.rand(3)))\n\n def test_bool_dispatch(self):\n with torch._jit_internal._disable_emit_hooks(): # TODO: Python print broadcasting list\n def kwarg_false(x):\n # type: (Tensor) -> Tensor\n return F.max_pool1d(x, 1, 1, return_indices=False)\n self.checkScript(kwarg_false, (torch.randn(3, 3, 3),))\n\n def kwarg_true(x):\n # type: (Tensor) -> Tuple[Tensor, Tensor]\n return F.max_pool1d(x, 1, 1, return_indices=True)\n self.checkScript(kwarg_true, (torch.randn(3, 3, 3),))\n\n def full_kwarg_false(x):\n # type: (Tensor) -> Tensor\n return F.max_pool1d(x, 1, 1, ceil_mode=False, return_indices=False)\n self.checkScript(full_kwarg_false, (torch.randn(3, 3, 3),))\n\n def full_kwarg_true(x):\n # type: (Tensor) -> Tuple[Tensor, Tensor]\n return F.max_pool1d(x, 1, 1, ceil_mode=False, return_indices=True)\n self.checkScript(full_kwarg_true, (torch.randn(3, 3, 3),))\n\n def use_default(x):\n # type: (Tensor) -> Tensor\n return F.max_pool1d(x, 1, 1)\n self.checkScript(use_default, (torch.randn(3, 3, 3),))\n\n def arg_false(x):\n # type: (Tensor) -> Tensor\n return F.max_pool1d(x, 1, 1, 0, 1, False, False)\n self.checkScript(arg_false, (torch.randn(3, 3, 3),))\n\n def arg_true(x):\n # type: (Tensor) -> Tuple[Tensor, Tensor]\n return F.max_pool1d(x, 1, 1, 0, 1, False, True)\n self.checkScript(arg_true, (torch.randn(3, 3, 3),))\n\n def test_infer_size(self):\n from torch._C import _infer_size\n\n def fn(x, y):\n # type: (Tensor, Tensor) -> List[int]\n return _infer_size(x.size(), y.size())\n\n self.checkScript(fn, (torch.ones(2, 4, 2), torch.ones(2, 4, 2)))\n\n def test_hash(self):\n def tester(fn, inputs):\n for x in inputs:\n for y in inputs:\n if x == y:\n self.assertEqual(fn(x), fn(y))\n else:\n self.assertNotEqual(fn(x), fn(y))\n\n @torch.jit.script\n def int_hash(x):\n # type: (int) -> int\n return hash(x)\n\n @torch.jit.script\n def float_hash(x):\n # type: (float) -> int\n return hash(x)\n\n @torch.jit.script\n def str_hash(x):\n # type: (str) -> int\n return hash(x)\n\n tester(int_hash, (20, 21, 22))\n tester(float_hash, (20.0, 21.00001, 22.443))\n tester(str_hash, (\"\", \"hello\", \"a\"))\n\n def test_id(self):\n with self.assertRaisesRegex(RuntimeError, \"Expected a value\"):\n @torch.jit.script\n def test_id_scalars():\n return id(2) == id(None)\n\n @torch.jit.script\n class FooTest(object):\n def __init__(self, x):\n self.foo = x\n\n def getFooTest(self):\n return self.foo\n\n @torch.jit.script\n def test_id_class_types():\n obj1 = FooTest(torch.tensor(3))\n obj2 = FooTest(torch.tensor(2))\n assert obj1 is not obj2\n assert id(obj1) != id(obj2)\n assert id(obj1) != id(None)\n return True\n\n self.assertTrue(test_id_class_types())\n\n def test_mutable_dce(self):\n @torch.jit.script\n def foo():\n a = torch.rand(2, 3)\n a += torch.rand(2, 3)\n b = torch.rand(2, 3)\n b += torch.rand(2, 3)\n # b should be cleaned up but not a\n return a\n\n FileCheck().check_count(\"aten::rand\", 2, exactly=True) \\\n .check_count(\"aten::add\", 1, exactly=True).run(str(foo.graph))\n\n def test_mutable_dce_block(self):\n @torch.jit.script\n def foo():\n a = torch.rand(2, 3)\n a += torch.rand(2, 3)\n b = torch.rand(2, 3)\n if bool(a > torch.zeros(2, 3)):\n b += torch.rand(2, 3)\n a += torch.rand(2, 3)\n # a should be cleaned up but not b\n return b\n\n FileCheck().check(\"prim::If\").check_count(\"aten::rand\", 1, exactly=True) \\\n .run(str(foo.graph))\n\n def test_mutable_dce_graph_input(self):\n @torch.jit.script\n def foo(a):\n a += torch.rand(2, 3)\n # shouldn't clean up `a` even though it's not used in the output\n\n FileCheck().check(\"aten::rand\").check(\"aten::add\").run(str(foo.graph))\n\n def test_mutable_dce_list(self):\n @torch.jit.script\n def foo(a):\n l = []\n l.append(a)\n c = l[0]\n b = torch.rand(2, 3)\n c += torch.rand(2, 3)\n return b\n\n # c does not get cleaned up because there is a wildcard + mutation\n FileCheck().check_count(\"aten::rand\", 2, exactly=True).run(str(foo.graph))\n\n def test_mutable_dce_loop(self):\n @torch.jit.script\n def foo(a):\n l = []\n l.append(a)\n i = 0\n b = torch.rand(2, 3)\n while i < 1:\n dead = torch.rand(2, 3)\n c = l[0]\n c += torch.rand(2, 3)\n i += 1\n return b\n\n FileCheck().check(\"prim::Loop\").check_not(\"aten::rand\").check(\"aten::__getitem__\") \\\n .check_count(\"aten::rand\", 1, exactly=True).run(str(foo.graph))\n\n def test_mutable_dce_indirect_wildcards(self):\n def fn():\n x = torch.ones(2, 3)\n x_1 = x.view(-1)\n l = []\n l.append(x_1)\n x_view = l[0]\n x.add_(torch.ones(2, 3))\n return x_view\n self.checkScript(fn, ())\n\n def test_mutable_dce_indirect_wildcard_write(self):\n def fn():\n indexes = torch.jit.annotate(List[Tensor], [])\n word_ids = torch.zeros(10, dtype=torch.int32)\n word_ids[1] = 1\n indexes.append(word_ids)\n\n return word_ids\n self.checkScript(fn, ())\n\n def test_mutable_dce_wildcards(self):\n def fn():\n x = torch.ones(2, 3)\n l = []\n l.append(x)\n x_view = l[0]\n x.add_(torch.ones(2, 3))\n return x_view\n\n self.checkScript(fn, (), profiling=ProfilingMode.SIMPLE)\n\n def test_cpp_function_tensor_str(self):\n x = torch.randn(2, 2)\n scale = torch.randn(2, 2, requires_grad=True)\n shift = torch.randn(2, 2, requires_grad=True)\n\n @torch.jit.script\n def fn(x, scale, shift):\n return scale * x + shift\n\n with self.capture_stdout() as captured:\n print(fn(x, scale, shift))\n\n def test_string_index(self):\n def fn(x):\n # type: (str)\n return x[2], x[-1]\n\n self.checkScript(fn, (\"abcde\",))\n\n def test_ord(self):\n def fn(x):\n # type: (str) -> int\n return ord(x)\n\n self.checkScript(fn, (\"h\"))\n self.checkScript(fn, (\"y\"))\n\n def index_str_to_tensor(s):\n # type: (str) -> int\n return torch.tensor(ord(s))\n\n s = u'\\u00a3'.encode('utf8')[:1]\n self.checkScript(index_str_to_tensor, (s,))\n\n def test_chr(self):\n def fn(x):\n # type: (int) -> str\n return chr(x)\n\n self.checkScript(fn, (1,))\n self.checkScript(fn, (97,))\n\n def test_round(self):\n def round_float(x):\n # type: (float) -> float\n return round(x)\n\n def round_int(x):\n # type: (int) -> float\n return round(x)\n\n self.checkScript(round_float, (1.5,))\n self.checkScript(round_int, (2,))\n\n def test_convert_base(self):\n def test_hex(x):\n # type: (int) -> str\n return hex(x)\n\n def test_oct(x):\n # type: (int) -> str\n return oct(x)\n\n def test_bin(x):\n # type: (int) -> str\n return bin(x)\n\n numbers = [-1000, -10, 0, 1, 10, 2343]\n for n in numbers:\n self.checkScript(test_bin, (n,))\n self.checkScript(test_oct, (n,))\n self.checkScript(test_hex, (n,))\n\n @unittest.skipIf(IS_WINDOWS or IS_SANDCASTLE, \"NYI: TemporaryFileName support for Windows or Sandcastle\")\n def test_get_set_state(self):\n class Root(torch.jit.ScriptModule):\n __constants__ = ['number']\n\n def __init__(self, number):\n super(Root, self).__init__()\n self.register_buffer('buffer1', torch.ones(2, 2))\n self.register_buffer('buffer2', torch.ones(2, 2))\n self.number = number\n\n @torch.jit.script_method\n def __getstate__(self):\n return (self.buffer1, self.buffer2, 74, self.training)\n\n @torch.jit.script_method\n def __setstate__(self, state):\n self.buffer1 = state[0] + 10\n self.buffer2 = state[1] + 10\n self.training = state[3]\n\n class M(torch.jit.ScriptModule):\n __constants__ = ['number']\n\n def __init__(self, number, submodule):\n super(M, self).__init__()\n self.register_buffer('buffer1', torch.ones(2, 2))\n self.register_buffer('buffer2', torch.ones(2, 2))\n self.number = number\n self.submodule = submodule\n\n @torch.jit.script_method\n def __getstate__(self):\n return (self.buffer1, self.buffer2, 74, self.submodule, self.training)\n\n @torch.jit.script_method\n def __setstate__(self, state):\n self.buffer1 = state[0] + 10\n self.buffer2 = state[1] + 10\n self.submodule = state[3]\n self.training = state[4]\n\n with TemporaryFileName() as fname:\n m = M(23, submodule=Root(99))\n m.save(fname)\n loaded = torch.jit.load(fname)\n\n # Check original module\n self.assertEqual(m.buffer1, torch.ones(2, 2))\n self.assertEqual(m.buffer2, torch.ones(2, 2))\n\n # Check top level module\n self.assertEqual(loaded.buffer1, torch.ones(2, 2) + 10)\n self.assertEqual(loaded.buffer2, torch.ones(2, 2) + 10)\n\n # Check submodule\n self.assertEqual(loaded.submodule.buffer1, torch.ones(2, 2) + 10)\n self.assertEqual(loaded.submodule.buffer2, torch.ones(2, 2) + 10)\n\n # Check simpler module\n class NoArgState(torch.nn.Module):\n def __init__(self):\n super(NoArgState, self).__init__()\n self.register_buffer('buffer1', torch.ones(2, 2))\n self.register_buffer('buffer2', torch.ones(2, 2))\n\n def forward(self):\n pass\n\n @torch.jit.export\n def __getstate__(self):\n return 5, self.training\n\n @torch.jit.export\n def __setstate__(self, state):\n self.buffer1 = torch.ones(2, 2) + state[0]\n self.buffer2 = torch.ones(2, 2) + 10\n self.training = state[1]\n\n with TemporaryFileName() as fname:\n m = torch.jit.script(NoArgState())\n m.save(fname)\n loaded = torch.jit.load(fname)\n self.assertEqual(loaded.buffer1, torch.ones(2, 2) + 5)\n self.assertEqual(loaded.buffer2, torch.ones(2, 2) + 10)\n\n\n\n def test_string_slicing(self):\n def fn1(x):\n # type: (str) -> str\n return x[1:3]\n\n def fn2(x):\n # type: (str) -> str\n return x[-1:3]\n\n def fn3(x):\n # type: (str) -> str\n return x[3:1]\n\n def fn4(x):\n # type: (str) -> str\n return x[3:100]\n\n self.checkScript(fn1, (\"abcdefghi\",))\n self.checkScript(fn2, (\"abcdefghi\",))\n self.checkScript(fn3, (\"abcdefghi\",))\n self.checkScript(fn4, (\"abcdefghi\",))\n\n def test_early_return_closure(self):\n code = dedent('''\n def tanh(self):\n output = torch.tanh(self)\n def backward(grad_output):\n pass\n return output, backward\n ''')\n cu = torch.jit.CompilationUnit(code)\n g = cu.tanh.graph\n FileCheck().check_count(\"prim::Closure_0\", 2).check(\"NoneType = prim::Constant\") \\\n .check_next(\"return\").run(g)\n\n code = dedent('''\n def tanh(self):\n output = torch.tanh(self)\n def backward(grad_output):\n a = 1\n if output:\n return 1\n else:\n a = 2\n return a\n return output, backward\n ''')\n cu = torch.jit.CompilationUnit(code)\n g = cu.tanh.graph\n FileCheck().check_count(\"prim::Closure_0\", 2).check(\"int = prim::If\") \\\n .run(g)\n\n code = dedent('''\n def loop_in_closure(self):\n output = torch.tanh(self)\n def backward(grad_output):\n for i in range(3):\n return 1\n return 4\n return output, backward\n ''')\n cu = torch.jit.CompilationUnit(code)\n fc = FileCheck()\n fc.check(\"prim::Closure\").check(\"(Tensor, NoneType) = prim::TupleConstruct\")\n # Loop then two if's added in exit transform\n fc.check(\"prim::Closure\").check(\"prim::Loop\").check_count(\"prim::If\", 2)\n fc.run(cu.loop_in_closure.graph)\n\n code = dedent('''\n def tanh(self):\n output = torch.tanh(self)\n def backward(grad_output):\n if 1 == 1:\n return 1\n else:\n return 1.\n return output, backward\n ''')\n with self.assertRaisesRegex(RuntimeError, \"returned a value of type int but\"):\n cu = torch.jit.CompilationUnit(code)\n\n @_inline_everything\n def test_early_return_fork_join(self):\n @torch.jit.script\n def foo(x):\n if x.dim() == 2:\n return torch.neg(x), x\n else:\n return torch.neg(x), x + 1\n\n x = torch.rand(3, 4)\n\n @torch.jit.script\n def wait_script(x):\n fut = torch.jit._fork(foo, x)\n y_hat = foo(x)\n y = torch.jit._wait(fut)\n return y, y_hat\n\n FileCheck().check(\"with prim::fork\").check(\"prim::If\").check(\"return\")\\\n .run(wait_script.graph)\n\n def test_early_return_type_refinement(self):\n @torch.jit.script\n def test(x):\n # type: (Optional[int]) -> int\n if x is None:\n return 1\n else:\n return x\n self.assertEqual(test(None), 1)\n self.assertEqual(test(2), 2)\n\n def test_exceptions_with_control_flow(self):\n def test_num_ifs(func, num_ifs):\n g = torch.jit.script(func).graph\n FileCheck().check_count(\"prim::If\", num_ifs, exactly=True).run(g)\n\n def no_guard_ifs_added(x):\n # type: (int) -> int\n if x == 1:\n return 1\n else:\n if x == 2:\n raise RuntimeError(\"hi\")\n else:\n raise RuntimeError(\"hi\")\n\n self.checkScript(no_guard_ifs_added, (1,))\n self.checkScriptRaisesRegex(no_guard_ifs_added, (2,), Exception, \"\")\n test_num_ifs(no_guard_ifs_added, 2)\n\n # FUNCTION LOOKS LIKE:\n # graph(%x.1 : int):\n # %7 : str = prim::Constant[value=\"Exception\"]()\n # %2 : int = prim::Constant[value=1]()\n # %5 : int = prim::Constant[value=2]()\n # %19 : int = prim::Uninitialized()\n # %3 : bool = aten::eq(%x.1, %2)\n # %20 : int = prim::If(%3)\n # block0():\n # -> (%2)\n # block1():\n # %6 : bool = aten::eq(%x.1, %5)\n # = prim::If(%6)\n # block0():\n # = prim::RaiseException(%7)\n # -> ()\n # block1():\n # = prim::RaiseException(%7)\n # -> ()\n # -> (%19)\n # return (%20)\n\n def no_ifs_added(x):\n # type: (int) -> int\n if x < 0:\n raise RuntimeError(\"hi\")\n return x\n\n self.checkScript(no_ifs_added, (1,))\n self.checkScriptRaisesRegex(no_ifs_added, (-2,), Exception, \"\")\n test_num_ifs(no_ifs_added, 1)\n\n def test_if_might(x):\n # type: (int)\n if x > 0:\n if x == 1:\n return 1\n else:\n a = 2\n else:\n raise RuntimeError(\"hi\")\n return a + 2\n\n self.checkScript(test_if_might, (1,))\n self.checkScript(test_if_might, (3,))\n self.checkScriptRaisesRegex(no_ifs_added, (-2,), Exception, \"\")\n test_num_ifs(test_if_might, 3) # one if added to guard a + 2\n\n def test_loop_no_escape(x):\n # type: (int)\n if x >= 0:\n for i in range(x):\n raise RuntimeError(\"hi\")\n else:\n return 5\n return x + 3\n\n self.checkScript(test_loop_no_escape, (0,))\n self.checkScript(test_loop_no_escape, (-1,))\n self.checkScriptRaisesRegex(test_loop_no_escape, (1,), Exception, \"\")\n\n # if guard gets optimized away\n test_num_ifs(test_loop_no_escape, 1)\n\n def test_loop_exception_with_continue(x):\n # type: (int)\n i = 0\n for i in range(5):\n if i == x:\n raise RuntimeError(\"hi\")\n else:\n continue\n print(i)\n return i + 5\n\n self.checkScript(test_loop_exception_with_continue, (-1,))\n self.checkScriptRaisesRegex(test_loop_exception_with_continue, (1,), Exception, \"\")\n test_num_ifs(test_loop_exception_with_continue, 1) # no ifs added to guard print\n\n\n def test_exception_exits_closure(self):\n code = dedent('''\n def no_return_func(self):\n # type: (Tensor) -> Tensor\n output = torch.tanh(self)\n def backward(grad_output):\n raise RuntimeError(\"Hi\")\n ''')\n with self.assertRaisesRegex(RuntimeError, \"does not return along all\"):\n cu = torch.jit.CompilationUnit(code)\n\n code = dedent('''\n def test_exit_pair_reset(x):\n # type: (int) -> int\n if x > 0:\n a = 0\n def backward(grad_output):\n raise RuntimeError(\"Hi\")\n a = a + 1\n else:\n return x\n return a + 1\n ''')\n func = torch.jit.CompilationUnit(code).test_exit_pair_reset\n self.assertEqual(func(1,), 2)\n self.assertEqual(func(-1,), -1)\n # final a + 1 gets inlined into the first branch and optimized away\n FileCheck().check_count(\"prim::If\", 1, exactly=True).run(func.graph)\n\n def test_non_final_return(self):\n def simple(x):\n if bool(x > 3):\n return x + 1\n else:\n return x + 2\n raise RuntimeError(\"nope\")\n\n def nest(x):\n x = x + 1\n if bool(x > 3):\n if bool(x > 4):\n x += 1\n return x + 1\n else:\n return x + 2\n\n def early_ret(x):\n x = x + 1\n if bool(x > 3):\n return x + 1\n x = x + 1\n return x + 2\n\n def nest_early_ret(x):\n x = x + 1\n if bool(x > 3):\n if bool(x > 4):\n return x + 2\n return x + 1\n x = x + 1\n return x + 2\n\n def not_early_ret(x):\n s = \"\"\n if bool(x > 3):\n if bool(x > 4):\n return 1, s\n s += \"foo\"\n else:\n s += \"5\"\n s += \"hi\"\n return 7, s\n\n def not_total_ret(x):\n s = \"\"\n if bool(x > 3):\n if bool(x > 4):\n return 1, s\n else:\n return 2, s\n else:\n s += \"5\"\n return 7, s\n\n for i in range(3):\n for func in [simple, nest, early_ret, nest_early_ret, not_early_ret,\n not_total_ret]:\n self.checkScript(func, (torch.tensor(2.5 + i),))\n\n def vars_used_after_ret(x):\n # type: (int) -> int\n if x == 0:\n return x\n else:\n y = 2\n z = 3\n return x + y * z\n\n self.checkScript(vars_used_after_ret, (1,))\n self.checkScript(vars_used_after_ret, (0,))\n\n def complicated(x):\n # type: (int) -> int\n if x:\n if x == 2:\n return 1\n assert 1 == 2\n else:\n if x == 3:\n return 2\n assert 1 == 2\n else:\n a = 2\n b = 3\n else:\n a = 4\n b = 1\n return a + b\n assert 1 == 2\n\n for i in range(4):\n self.checkScript(complicated, (i,))\n\n def test_partial_returns(self):\n with self.assertRaisesRegex(RuntimeError, \"does not return along all\"):\n @torch.jit.script\n def no_ret():\n # type: () -> int\n pass\n\n with self.assertRaisesRegex(RuntimeError, \"does not return along all\"):\n @torch.jit.script\n def partial(x):\n # type: (Tensor) -> int\n if x:\n return 1\n\n with self.assertRaisesRegex(RuntimeError, \"does not return along all\"):\n @torch.jit.script\n def typed_none():\n # type: () -> Optional[int]\n pass\n\n @torch.jit.script\n def none_ret():\n pass\n\n self.assertIs(none_ret(), None)\n FileCheck().check(\": None\").run(none_ret.graph)\n\n def test_early_returns_loops(self):\n def nest_while_ret(x):\n # type: (int) -> int\n y = 4\n while x < 4:\n if x < 3:\n return y\n else:\n y = y + 1\n break\n y = y + 2\n y = y + 1\n return y\n\n self.checkScript(nest_while_ret, (2,))\n self.checkScript(nest_while_ret, (3,))\n self.checkScript(nest_while_ret, (4,))\n\n def loop_ret(x, y):\n # type: (int, int) -> (int)\n i = 0\n for i in range(x):\n if x == y:\n return x + y\n i = i + y\n i = i - 1\n return i\n\n self.checkScript(loop_ret, (3, 3))\n self.checkScript(loop_ret, (2, 3))\n self.checkScript(loop_ret, (3, 1))\n\n def test_will_ret(y):\n # type: (int) -> int\n for i in range(y):\n return 2\n return 1\n\n self.checkScript(test_will_ret, (0,))\n self.checkScript(test_will_ret, (1,))\n\n def test_loop_nest_ret(y):\n # type: (int) -> int\n for i in range(y):\n for i in range(y - 2):\n return 10\n return 5\n return 0\n\n self.checkScript(test_loop_nest_ret, (0,))\n self.checkScript(test_loop_nest_ret, (1,))\n self.checkScript(test_loop_nest_ret, (2,))\n\n def test_nn_init(self):\n tests = (\n ('constant_', (lambda: (torch.ones(2, 2), 2.5)), \"Tensor, float\"),\n ('ones_', (lambda: (torch.ones(2, 2),)), \"Tensor\"),\n ('zeros_', (lambda: (torch.ones(2, 2),)), \"Tensor\"),\n ('uniform_', (lambda: (torch.ones(2, 2),)), \"Tensor\"),\n ('normal_', (lambda: (torch.ones(2, 2),)), \"Tensor\"),\n ('xavier_normal_', (lambda: (torch.ones(2, 2),)), \"Tensor\"),\n ('xavier_uniform_', (lambda: (torch.ones(2, 2),)), \"Tensor\"),\n )\n\n for name, args_fn, type_str in tests:\n # Build test code\n arg_str = ', '.join([chr(i + ord('a')) for i in range(len(args_fn()))])\n\n code = dedent('''\n def test({arg_str}):\n # type: ({type_str})\n return torch.nn.init.{name}({arg_str})\n ''').format(arg_str=arg_str, type_str=type_str, name=name)\n cu = torch.jit.CompilationUnit(code)\n\n # Compare functions\n init_fn = getattr(torch.nn.init, name)\n script_out = self.runAndSaveRNG(cu.test, args_fn())\n eager_out = self.runAndSaveRNG(init_fn, args_fn())\n self.assertEqual(script_out, eager_out)\n\n FileCheck().check_not(\"prim::PythonOp\").run(cu.test.graph)\n\n def test_early_return_rewrite(self):\n def test_foo(x: bool):\n if x:\n return 1\n return 2\n\n self.checkScript(test_foo, (True,))\n self.checkScript(test_foo, (False,))\n FileCheck().check_count(\"prim::If\", 1, exactly=True).run(torch.jit.script(test_foo).graph)\n\n def test_multiple(x: int):\n if x == 5:\n return x * x\n else:\n y = 2 * x\n\n z = y * 2\n if z == 8:\n return 1\n\n if z != 16:\n z = z - 2\n abc = 4\n else:\n return 3\n\n z = z * abc\n return z * z * z\n\n self.checkScript(test_multiple, (5,))\n self.checkScript(test_multiple, (2,))\n self.checkScript(test_multiple, (4,))\n self.checkScript(test_multiple, (3,))\n self.checkScript(test_multiple, (10,))\n\n graph = torch.jit.script(test_multiple).graph\n FileCheck().check_count(\"prim::If\", 3, exactly=True).run(graph)\n\n def test_is_scripting_metacompile(self):\n @torch.jit.script\n def foo():\n if torch.jit.is_scripting():\n return 1\n else:\n print(\"hello\") + 2 # will not be compiled\n\n self.assertEqual(foo(), 1)\n\n def test_boolean_literal_constant_metacompile(self):\n class Mod(torch.nn.Module):\n __constants__ = ['val']\n\n def __init__(self, val):\n super(Mod, self).__init__()\n self.val = val\n\n def forward(self):\n if self.val:\n return 1\n else:\n return \"2\"\n\n self.checkModule(Mod(True), ())\n self.checkModule(Mod(False), ())\n\n @torch.jit.script\n def foo():\n if True:\n return 1\n else:\n return \"2\"\n\n self.assertEqual(foo(), 1)\n\n def test_assert_is_scripting_metacompile(self):\n def foo():\n assert not torch.jit.is_scripting(), \"TestErrorMsg\"\n print(\"hello\") + 2 # will not be compiled\n\n f = torch.jit.script(foo)\n with self.assertRaisesRegex(torch.jit.Error, \"TestErrorMsg\"):\n f()\n\n def test_isinstance_metacompile(self):\n @torch.jit.script\n def test_primitive_type(x):\n # type: (int) -> int\n if isinstance(x, int):\n return x + 1\n else:\n return x - 1\n\n self.assertEqual(test_primitive_type(1), 2)\n with self.assertRaisesRegex(Exception, \"Expected a value of type\"):\n test_primitive_type(1.5)\n\n _MyNamedTuple = namedtuple('_MyNamedTuple', ['value'])\n\n @torch.jit.script\n def test_non_primitive_types(x):\n # type: (_MyNamedTuple) -> Tensor\n if isinstance(1, _MyNamedTuple):\n return 10\n\n if isinstance(x, _MyNamedTuple):\n return x.value + 1\n else:\n return 1\n\n out = test_non_primitive_types(_MyNamedTuple(value=torch.tensor(5.0)))\n self.assertEqual(out, torch.tensor(6.0))\n\n def test_namedtuple_type_inference(self):\n _AnnotatedNamedTuple = NamedTuple('_NamedTupleAnnotated', [('value', int)])\n _UnannotatedNamedTuple = namedtuple('_NamedTupleUnAnnotated', ['value'])\n\n def test_check_named_tuple_value():\n named_tuple = _AnnotatedNamedTuple(1)\n return named_tuple.value\n\n self.checkScript(test_check_named_tuple_value, ())\n\n def test_error():\n return _UnannotatedNamedTuple(1)\n\n with self.assertRaisesRegex(RuntimeError, r\"Expected a value of type \\'Tensor \\(inferred\\)\\' \"\n r\"for argument \\'value\\' but instead found type \\'int\\'.\"):\n torch.jit.script(test_error)\n\n def test_namedtuple_default_values_simple_type(self):\n\n class Point(NamedTuple):\n x: Optional[int] = None\n y: int = 2\n\n make_global(Point)\n\n class M(torch.nn.Module):\n def __init__(self):\n super(M, self).__init__()\n\n def forward(self, point: Point):\n return point\n\n p = Point(x=3, y=2)\n\n self.checkModule(M(), (p,))\n self.checkModule(M(), (Point(),))\n\n m = torch.jit.script(M())\n\n FileCheck().check(r\"NamedTuple(x : int? = None, y : int = 2))\") \\\n .run(m.graph)\n\n def test_namedtuple_default_values_missing(self):\n\n class Point(NamedTuple):\n x: Optional[int]\n y: int\n z: int = 3\n\n make_global(Point)\n\n class M(torch.nn.Module):\n def __init__(self):\n super(M, self).__init__()\n\n def forward(self, point: Point):\n return point\n\n p1 = Point(x=3, y=2)\n p2 = Point(x=3, y=2, z=1)\n\n self.checkModule(M(), (p1,))\n self.checkModule(M(), (p2,))\n\n m = torch.jit.script(M())\n\n FileCheck().check(r\"NamedTuple(x : int?, y : int, z : int = 3))\") \\\n .run(m.graph)\n\n def test_namedtuple_default_values_container_type(self):\n\n class Point(NamedTuple):\n x: Optional[List[int]] = None\n y: List[int] = [1, 2, 3]\n z: Optional[Dict[str, int]] = {\"a\": 1}\n\n make_global(Point)\n\n class M(torch.nn.Module):\n def __init__(self):\n super(M, self).__init__()\n\n def forward(self, point: Point):\n return point\n\n p = Point(x=[4, 5, 6], y=[3, 2, 1], z={\"b\": 2})\n\n self.checkModule(M(), (p,))\n self.checkModule(M(), (Point(),))\n\n m = torch.jit.script(M())\n\n first_line = r\"NamedTuple(x : int[]? = None, y : int[] = \" \\\n r\"[1, 2, 3], z : Dict(str, int)? = {a: 1}))\"\n\n FileCheck().check(first_line) \\\n .run(m.graph)\n\n def test_namedtuple_default_values_Tensor_type(self):\n\n class Point(NamedTuple):\n x: torch.Tensor = torch.rand(2, 3)\n\n make_global(Point)\n\n class M(torch.nn.Module):\n def __init__(self):\n super(M, self).__init__()\n\n def forward(self, point: Point):\n return point\n\n p = Point(x=torch.rand(2, 3))\n\n with self.assertRaisesRegex(RuntimeError, \"Tensors are not \"\n \"supported as default NamedTuple \"\n \"fields\"):\n m = torch.jit.script(M())\n m(p)\n\n @unittest.skipIf(sys.version_info < (3, 7, 0), \"defaults keyword added in Python 3.8\")\n def test_namedtuple_default_values_using_factory_constructor(self):\n Pair = namedtuple(\"Pair\", [\"x\", \"y\"], defaults=(1, 2))\n\n make_global(Pair)\n\n @torch.jit.script\n def fn(x: Pair) -> Pair:\n return x\n\n # TODO: We can't use `checkScript` with the NamedTuple factory\n # constructor. Using the factory constructor with TorchScript\n # TorchScript creates an anonymous `NamedTuple` class instead of\n # preserving the actual name. For example, the actual generated\n # signature in this case is:\n # graph(%x.1 : NamedTuple(x : Tensor, y : Tensor))\n # It looks like similar test cases have had this issue as well\n # (see: `test_namedtuple_python`).\n FileCheck().check(r\"NamedTuple(x : Tensor = 1, y : Tensor = 2))\") \\\n .check_next(r\"return (%x.1)\") \\\n .run(fn.graph)\n\n def test_isinstance_dynamic(self):\n @torch.jit.script\n def foo(a):\n # type: (Optional[List[int]]) -> int\n b = 0\n if isinstance(a, (int, (float,), list, str)):\n b += 1\n if isinstance(a, (int, str)):\n b += 1\n if isinstance(a, List[int]):\n b += 1\n return b\n self.assertEqual(foo([3, 4]), 2)\n self.assertEqual(foo(None), 0)\n\n def test_function_overloads(self):\n # TODO: pyflakes currently does not compose @overload annotation with other\n # decorators. This is fixed on master but not on version 2.1.1.\n # Next version update remove noqa and add @typing.overload annotation\n\n @torch.jit._overload # noqa: F811\n def test_simple(x1): # noqa: F811\n # type: (int) -> int\n pass\n\n @torch.jit._overload # noqa: F811\n def test_simple(x1): # noqa: F811\n # type: (float) -> float\n pass\n\n def test_simple(x1): # noqa: F811\n return x1\n\n def invoke_function():\n return test_simple(1.0), test_simple(.5)\n\n self.checkScript(invoke_function, ())\n\n # testing that the functions are cached\n compiled_fns_1 = torch.jit._script._get_overloads(test_simple)\n compiled_fns_2 = torch.jit._script._get_overloads(test_simple)\n for a, b in zip(compiled_fns_1, compiled_fns_2):\n self.assertIs(a.graph, b.graph)\n\n old_func = test_simple\n\n # testing that new functions added work with caching\n @torch.jit._overload # noqa: F811\n def test_simple(x1): # noqa: F811\n # type: (str) -> str\n pass\n\n @torch.jit.script\n def my_func():\n return old_func(\"hi\")\n\n # testing new function same qualified name\n @torch.jit._overload # noqa: F811\n def test_simple(a, b): # noqa: F811\n # type: (int, int) -> int\n pass\n\n def test_simple(a, b):\n return a + b\n\n @torch.jit.script\n def fn():\n return test_simple(3, 4)\n\n self.assertEqual(fn(), 7)\n\n # currently we take the default values have to be specified in the\n # overload as well - TODO take them from implementation and apply\n # where the type is valid.\n @torch.jit._overload # noqa: F811\n def identity(x1): # noqa: F811\n # type: (str) -> str\n pass\n\n @torch.jit._overload # noqa: F811\n def identity(x1): # noqa: F811\n # type: (float) -> float\n pass\n\n def identity(x1=1.0): # noqa: F811\n return x1\n\n def invoke():\n return identity(), identity(.5), identity(\"hi\")\n\n self.checkScript(invoke, ())\n\n def schema_match_failure():\n return identity((1, 2))\n\n thrown = False\n try:\n torch.jit.script(schema_match_failure)\n except Exception as e:\n thrown = True\n self.assertTrue(r\"of type 'str'\" in str(e) and r\"of type 'float\" in str(e))\n self.assertTrue(thrown)\n\n with self.assertRaisesRegex(Exception, \"cannot be directly compiled\"):\n torch.jit.script(identity)\n\n @torch.jit._overload # noqa: F811\n def impl_compile_failure(x, y): # noqa: F811\n # type: (str, str) -> (str)\n pass\n\n @torch.jit._overload # noqa: F811\n def impl_compile_failure(x, y): # noqa: F811\n # type: (int, int) -> (int)\n pass\n\n def impl_compile_failure(x, y): # noqa: F811\n return x - y\n\n def test():\n impl_compile_failure(\"one\", \"two\")\n\n\n with self.assertRaisesRegex(Exception, \"Arguments for call are not valid\"):\n torch.jit.script(test)\n\n @torch.jit._overload # noqa: F811\n def good_overload(x=1): # noqa: F811\n # type: (int) -> (int)\n pass\n\n def good_overload(x=1): # noqa: F811\n return x\n\n @torch.jit.script\n def foo():\n return good_overload()\n\n self.assertEqual(foo(), 1)\n\n\n with self.assertRaisesRegex(Exception, \"must equal to the default parameter\"):\n @torch.jit._overload # noqa: F811\n def bad_default_on_overload(x, y=2): # noqa: F811\n # type: (int, int) -> (int)\n pass\n\n def bad_default_on_overload(x, y=1): # noqa: F811\n # type: (int, int) -> (int)\n pass\n\n @torch.jit.script\n def test():\n return bad_default_on_overload(1, 2)\n\n @torch.jit._overload # noqa: F811\n def diff_default(x): # noqa: F811\n # type: (int) -> int\n pass\n\n @torch.jit._overload # noqa: F811\n def diff_default(x): # noqa: F811\n # type: (str) -> str\n pass\n\n def diff_default(x=\"hi\"): # noqa: F811\n return x\n\n def test():\n return diff_default(), diff_default(2), diff_default(\"abc\")\n\n self.assertEqual(test(), torch.jit.script(test)())\n\n @torch.jit._overload # noqa: F811\n def diff_num_params(x): # noqa: F811\n # type: (float) -> float\n pass\n\n @torch.jit._overload # noqa: F811\n def diff_num_params(x, y): # noqa: F811\n # type: (int, int) -> int\n pass\n\n def diff_num_params(x, y=2, z=3): # noqa: F811\n # type: (Union[float, int], int, int)\n return x + y + z\n\n def test():\n return diff_num_params(1.0), diff_num_params(1, 2), diff_num_params(1), diff_num_params(1, 2, 3)\n\n self.assertEqual(test(), torch.jit.script(test)())\n\n @torch.jit._overload # noqa: F811\n def diff_num_params_no_annot():\n # type: () -> int\n pass\n\n def diff_num_params_no_annot(x=1): # noqa: F811\n return x\n\n def test():\n return diff_num_params_no_annot(1.0)\n\n with self.assertRaisesRegex(Exception, \"Parameters not specified\"):\n torch.jit.script(test)\n\n def test_script_method_torch_function_overload(self):\n class MyCustomTensor(torch.Tensor):\n pass\n\n class MyCustomModule(torch.nn.Module):\n def forward(self, x):\n return torch.relu(x)\n\n scripted_mod = torch.jit.script(MyCustomModule())\n t = torch.tensor([3.0])\n ref_out = scripted_mod(t)\n\n t_custom = MyCustomTensor([3.0])\n out1 = scripted_mod(t_custom)\n self.assertEqual(out1, ref_out)\n\n out2 = scripted_mod.forward(t_custom)\n self.assertEqual(out2, ref_out)\n\n def test_function_overloading_isinstance(self):\n @torch.jit._overload # noqa: F811\n def my_conv(x, y): # noqa: F811\n # type: (float, str) -> (float)\n pass\n\n @torch.jit._overload # noqa: F811\n def my_conv(x, y): # noqa: F811\n # type: (float, float) -> (float)\n pass\n\n def my_conv(x, y=2.0): # noqa: F811\n if isinstance(y, str):\n if y == \"hi\":\n return 4.0 - x\n else:\n return 5.0 - x\n else:\n return 2.0 + x\n\n def test_uses():\n return my_conv(1.5), my_conv(1.5, \"hi\"), my_conv(1.5, 5.0)\n\n self.checkScript(test_uses, ())\n\n def test_method_overloading(self):\n class Over(torch.nn.Module):\n def __init__(self):\n super(Over, self).__init__()\n\n @torch.jit._overload_method # noqa: F811\n def forward(self, x): # noqa: F811\n # type: (Tuple[Tensor, Tensor]) -> Tensor\n pass\n\n @torch.jit._overload_method # noqa: F811\n def forward(self, x): # noqa: F811\n # type: (Tensor) -> Tensor\n pass\n\n def forward(self, x): # noqa: F811\n if isinstance(x, Tensor):\n return x + 20\n else:\n return x[0] + 5\n\n class S(torch.jit.ScriptModule):\n def __init__(self):\n super(S, self).__init__()\n self.weak = Over()\n\n @torch.jit.script_method\n def forward(self, x):\n return self.weak(x) + self.weak((x, x))\n\n s_mod = S()\n x = torch.ones(1)\n self.assertEqual(s_mod(x), x + 20 + 5 + x)\n\n over = Over()\n self.assertEqual(over((x, x)), x + 5)\n self.assertEqual(over((x)), x + 20)\n\n class Unannotated(torch.nn.Module):\n def __init__(self):\n super(Unannotated, self).__init__()\n\n @torch.jit._overload_method # noqa: F811\n def hello(self, x): # noqa: F811\n pass\n\n @torch.jit._overload_method # noqa: F811\n def hello(self, x): # noqa: F811\n # type: (int) -> (int)\n pass\n\n def hello(self, x): # noqa: F811\n return x + 3\n\n def forward(self):\n return self.hello(1), self.hello(.5)\n\n w = Unannotated()\n with self.assertRaisesRegex(Exception, \"explicitly add type annotations to overloaded functions\"):\n torch.jit.script(w)\n\n class CompileOverloadError(torch.nn.Module):\n def __init__(self):\n super(CompileOverloadError, self).__init__()\n\n @torch.jit._overload_method # noqa: F811\n def hello(self, x): # noqa: F811\n # type: (str) -> (int)\n pass\n\n @torch.jit._overload_method # noqa: F811\n def hello(self, x): # noqa: F811\n # type: (int) -> (int)\n pass\n\n def hello(self, x): # noqa: F811\n return x + 1\n\n def forward(self):\n return self.hello(\"hi\"), self.hello(.5)\n\n w = CompileOverloadError()\n with self.assertRaisesRegex(Exception, \"but instead found type \\'str\\'\"):\n torch.jit.script(w)\n\n # testing overload declared first, then non-overload\n with self.assertRaisesRegex(Exception, \"Overloads are not useable when a module\"):\n class W3(torch.nn.Module):\n def __init__(self):\n super(W3, self).__init__()\n\n @torch.jit._overload_method # noqa: F811\n def forward(self, x): # noqa: F811\n # type: (int) -> int\n pass\n\n @torch.jit._overload_method # noqa: F811\n def forward(self, x): # noqa: F811\n # type: (Tensor) -> Tensor\n pass\n\n def forward(self, x): # noqa: F811\n return x + 5\n\n a = W3()\n b = torch.jit.script(a)\n\n class W3(torch.nn.Module):\n def __init__(self):\n super(W3, self).__init__()\n\n def forward(self, x): # noqa: F811\n return x + 5 + 10\n\n a = W3()\n b = torch.jit.script(a)\n\n # testing non-overload declared first, then overload\n class W2(torch.nn.Module):\n def __init__(self):\n super(W2, self).__init__()\n\n def hello(self, x1, x2):\n return x1 + x2\n\n def forward(self, x):\n return self.hello(x, x)\n\n a = torch.jit.script(W2())\n self.assertEqual(a(torch.tensor(1)), torch.tensor(2))\n\n class W2(torch.nn.Module):\n def __init__(self):\n super(W2, self).__init__()\n\n @torch.jit._overload_method # noqa: F811\n def hello(self, x): # noqa: F811\n pass\n\n @torch.jit._overload_method # noqa: F811\n def hello(self, x): # noqa: F811\n # type: (int) -> (int)\n pass\n\n def hello(self, x): # noqa: F811\n return x + 5 + 10\n\n def forward(self, x):\n return self.hello(1), self.hello(x)\n\n with self.assertRaisesRegex(Exception, \"Overloads are not useable when a module\"):\n a = torch.jit.script(W2())\n\n def test_select_after_chunk(self):\n def foo(x):\n chunked = torch.chunk(x, 1)\n foo = chunked[0]\n foo.add_(5)\n return x\n\n self.checkScript(foo, [torch.rand(2, 3)])\n\n def test_nn_LSTM_with_layers(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.rnn = nn.LSTM(2, 3, 2, dropout=0)\n\n @torch.jit.script_method\n def forward(self, x, lengths, h0, c0):\n return self.rnn(x, (h0, c0))[0]\n\n class Eager(torch.nn.Module):\n def __init__(self):\n super(Eager, self).__init__()\n self.rnn = nn.LSTM(2, 3, 2, dropout=0)\n\n def forward(self, x, lengths, h0, c0):\n return self.rnn(x, (h0, c0))[0]\n\n inputs = (torch.randn(1, 1, 2), torch.LongTensor([7]), torch.randn(2, 1, 3), torch.randn(2, 1, 3))\n eager_out = self.runAndSaveRNG(lambda: Eager()(*inputs), ())[0]\n script_out = self.runAndSaveRNG(lambda: M()(*inputs), ())[0]\n\n self.assertEqual(eager_out, script_out)\n\n def test_nn_LSTM(self):\n input = torch.nn.utils.rnn.pack_sequence([torch.randn(5, 5)])\n\n class S(torch.jit.ScriptModule):\n def __init__(self):\n super(S, self).__init__()\n self.x = torch.nn.LSTM(5, 5)\n\n @torch.jit.script_method\n def forward(self, input: PackedSequence) -> Tuple[PackedSequence, Tuple[torch.Tensor, torch.Tensor]]:\n return self.x(input)\n\n eager_out = self.runAndSaveRNG(lambda x: torch.nn.LSTM(5, 5)(x), (input,))[0]\n script_out = self.runAndSaveRNG(lambda x: S()(x), (input,))[0]\n\n self.assertEqual(eager_out, script_out)\n\n def test_nn_GRU(self):\n seq_input = torch.nn.utils.rnn.pack_sequence([torch.randn(5, 5)])\n tensor_input = torch.randn(5, 5, 5)\n\n class SeqLengthGRU(torch.jit.ScriptModule):\n def __init__(self):\n super(SeqLengthGRU, self).__init__()\n self.x = torch.nn.GRU(5, 5)\n\n @torch.jit.script_method\n def forward(self, input: PackedSequence) -> Tuple[PackedSequence, torch.Tensor]:\n return self.x(input)\n\n class TensorGRU(torch.jit.ScriptModule):\n def __init__(self):\n super(TensorGRU, self).__init__()\n self.x = torch.nn.GRU(5, 5)\n\n @torch.jit.script_method\n def forward(self, input: torch.Tensor) -> Tuple[torch.Tensor, torch.Tensor]:\n return self.x(input)\n\n seq_eager_out = self.runAndSaveRNG(lambda x: torch.nn.GRU(5, 5)(x), (seq_input,))[0]\n seq_script_out = self.runAndSaveRNG(lambda x: SeqLengthGRU()(x), (seq_input,))[0]\n tensor_eager_out = self.runAndSaveRNG(lambda x: torch.nn.GRU(5, 5)(x), (tensor_input,))[0]\n tensor_script_out = self.runAndSaveRNG(lambda x: TensorGRU()(x), (tensor_input,))[0]\n\n self.assertEqual(seq_eager_out, seq_script_out)\n self.assertEqual(tensor_eager_out, tensor_script_out)\n\n def test_torchscript_memoryformat(self):\n @torch.jit.script\n def fn(x):\n return x.contiguous(memory_format=torch.channels_last)\n x = torch.randn(4, 3, 6, 6)\n y = fn(x)\n self.assertTrue(y.is_contiguous(memory_format=torch.channels_last))\n\n def test_torchscript_multi_head_attn(self):\n @torch.jit.script\n def jit_multihead_attn_forward(query, # type: Tensor\n key, # type: Tensor\n value, # type: Tensor\n embed_dim_to_check, # type: int\n num_heads, # type: int\n in_proj_weight, # type: Tensor\n in_proj_bias, # type: Tensor\n bias_k, # type: Optional[Tensor]\n bias_v, # type: Optional[Tensor]\n add_zero_attn, # type: bool\n dropout, # type: float\n out_proj_weight, # type: Tensor\n out_proj_bias, # type: Tensor\n training=True, # type: bool\n key_padding_mask=None, # type: Optional[Tensor]\n need_weights=True, # type: bool\n attn_mask=None # type: Optional[Tensor]\n ):\n # type: (...) -> Tuple[Tensor, Optional[Tensor]]\n return torch.nn.functional.multi_head_attention_forward(query, key, value,\n embed_dim_to_check, num_heads,\n in_proj_weight, in_proj_bias,\n bias_k, bias_v,\n add_zero_attn, dropout,\n out_proj_weight, out_proj_bias,\n training, key_padding_mask,\n need_weights, attn_mask)\n\n src_l = 3\n bsz = 5\n embed_size = 8\n nhead = 2\n multi_head_attn = torch.nn.MultiheadAttention(embed_size, nhead)\n query = torch.rand((src_l, bsz, embed_size))\n key = torch.rand((src_l, bsz, embed_size))\n value = torch.rand((src_l, bsz, embed_size))\n\n mask = (torch.triu(torch.ones(src_l, src_l)) == 1).transpose(0, 1)\n mask = mask.float().masked_fill(mask == 0, float('-inf')).masked_fill(mask == 1, float(0.0)).double()\n\n jit_out = jit_multihead_attn_forward(query, key, value,\n embed_size, nhead,\n multi_head_attn.in_proj_weight,\n multi_head_attn.in_proj_bias,\n multi_head_attn.bias_k, multi_head_attn.bias_v,\n multi_head_attn.add_zero_attn, multi_head_attn.dropout,\n multi_head_attn.out_proj.weight,\n multi_head_attn.out_proj.bias, attn_mask=mask)[0]\n\n py_out = torch.nn.functional.multi_head_attention_forward(query, key, value,\n embed_size, nhead,\n multi_head_attn.in_proj_weight,\n multi_head_attn.in_proj_bias,\n multi_head_attn.bias_k,\n multi_head_attn.bias_v,\n multi_head_attn.add_zero_attn,\n multi_head_attn.dropout,\n multi_head_attn.out_proj.weight,\n multi_head_attn.out_proj.bias,\n attn_mask=mask)[0]\n # print(\"rel. error: \")\n # print(jit_out / py_out - 1)\n self.assertTrue(torch.allclose(jit_out, py_out, atol=5e-4, rtol=1e-4))\n\n @unittest.skipIf(not RUN_CUDA, \"no CUDA\")\n def test_scriptmodule_multi_head_attn_cuda(self):\n\n class MyModule(torch.jit.ScriptModule):\n def __init__(self, embed_dim, num_heads):\n super(MyModule, self).__init__()\n sample_q = torch.randn(3, 2, embed_dim)\n sample_kv = torch.randn(3, 2, embed_dim)\n attention = nn.MultiheadAttention(embed_dim, num_heads)\n attention.eval()\n\n self.mod = torch.jit.trace(attention,\n (sample_q, sample_kv, sample_kv))\n\n @torch.jit.script_method\n def forward(self, q, k, v):\n return self.mod(q, k, v)\n\n embed_dim = 8\n num_heads = 2\n sl = 3\n bs = 2\n model = MyModule(embed_dim, num_heads).cuda()\n q = torch.randn(sl, bs, embed_dim, device=\"cuda\")\n kv = torch.randn(sl, bs, embed_dim, device=\"cuda\")\n\n jit_out = model(q, kv, kv)[0]\n py_out = torch.nn.functional.multi_head_attention_forward(q, kv, kv,\n embed_dim, num_heads,\n model.mod.in_proj_weight,\n model.mod.in_proj_bias,\n None, None, None, 0.0,\n model.mod.out_proj.weight,\n model.mod.out_proj.bias)[0]\n self.assertTrue(torch.allclose(jit_out, py_out, atol=5e-4, rtol=1e-4))\n\n @unittest.skipIf(not RUN_CUDA, \"no CUDA\")\n def test_scriptmodule_transformer_cuda(self):\n\n class MyModule(torch.jit.ScriptModule):\n def __init__(self, transformer, sample_q, sample_kv):\n super(MyModule, self).__init__()\n transformer.eval()\n\n self.mod = torch.jit.trace(transformer,\n (sample_q, sample_kv))\n\n @torch.jit.script_method\n def forward(self, q, k):\n return self.mod(q, k)\n\n d_model = 8\n nhead = 2\n num_encoder_layers = 2\n num_decoder_layers = 2\n dim_feedforward = 16\n bsz = 2\n seq_length = 5\n tgt_length = 3\n\n src = torch.randn(seq_length, bsz, d_model)\n tgt = torch.randn(tgt_length, bsz, d_model)\n transformer = nn.Transformer(d_model, nhead, num_encoder_layers,\n num_decoder_layers, dim_feedforward, dropout=0.0)\n model = MyModule(transformer, tgt, src)\n\n src = torch.randn(seq_length, bsz, d_model)\n tgt = torch.randn(tgt_length, bsz, d_model)\n jit_out = model(tgt, src)\n py_out = transformer(tgt, src)\n\n # print(jit_out/py_out-1)\n # print(torch.allclose(jit_out, py_out, atol=5e-4, rtol=1e-4))\n self.assertTrue(torch.allclose(jit_out, py_out, atol=5e-4, rtol=1e-4))\n\n def test_list_python_op(self):\n def python_list_op(lst):\n # type: (List[Tensor]) -> Tensor\n return lst[0]\n\n def fn(lst):\n # type: (List[Tensor]) -> Tensor\n return python_list_op(lst)\n\n self.checkScript(fn, ([torch.ones(2) + 2, torch.ones(2)],))\n\n @unittest.skipIf(not RUN_CUDA, \"no CUDA\")\n def test_weak_cuda(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.lstm = torch.nn.LSTM(5, 5)\n self.lstm.cuda()\n\n @torch.jit.script_method\n def forward(self, x):\n return self.lstm(x)\n\n m = M()\n m.cuda()\n out = m(torch.ones(5, 5, 5).cuda())\n self.assertTrue(out[0].is_cuda)\n\n def test_ignore_decorator(self):\n with warnings.catch_warnings(record=True) as warns:\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n tensor = torch.zeros(1, requires_grad=False)\n self.register_buffer('some_state', torch.nn.Parameter(tensor))\n\n @torch.jit.script_method\n def forward(self, x):\n self.ignored_code(x)\n return x\n\n @torch.jit.ignore(drop_on_export=True)\n def ignored_code(self, x):\n self.some_state = torch.tensor((100,))\n\n FileCheck().check(\"TorchScript will now drop the function\").run(str(warns[0]))\n\n # Assert ignored code is run\n m = M()\n\n m2 = self.getExportImportCopy(m)\n pp = str(m2.forward.code)\n self.assertNotIn('ignored_code', pp)\n\n with self.assertRaisesRegex(torch.jit.Error, \"annotated to be ignored and cannot be run\"):\n m2.forward(torch.ones(1))\n\n def test_ignored_as_value(self):\n class Model(nn.Module):\n def __init__(self):\n super(Model, self).__init__()\n\n @torch.jit.unused\n def tuple_ignored(self, x):\n # type: (Tensor) -> Tuple[Tensor, Tensor]\n return x, x\n\n @torch.jit.unused\n def single_val_ignored(self, x, y):\n # type: (Tensor, Tensor) -> Tensor\n return x\n\n def forward(self, x, use_ignore_path):\n # type: (Tensor, bool) -> Tuple[Tensor, Tensor]\n if 1 == 2:\n return self.tuple_ignored(x)\n if use_ignore_path:\n return self.single_val_ignored(x, x), self.single_val_ignored(x, x)\n return x, x\n\n original = Model()\n scripted = torch.jit.script(original)\n self.assertEqual(scripted(torch.tensor(.5), False), (torch.tensor(.5), torch.tensor(.5)))\n\n buffer = io.BytesIO()\n torch.jit.save(scripted, buffer)\n buffer.seek(0)\n loaded = torch.jit.load(buffer)\n\n with self.assertRaisesRegex(torch.jit.Error, \"annotated to be ignored and cannot be run\"):\n loaded(torch.tensor(.5), True)\n\n def test_module_error(self):\n class MyModule(torch.nn.Module):\n def __init__(self):\n super(MyModule, self).__init__()\n\n def forward(self, foo):\n return foo\n\n with self.assertRaisesRegex(RuntimeError, \"cannot be compiled since it inherits from nn.Module\"):\n torch.jit.script(MyModule)\n\n def test_view_write(self):\n def fn(x, y):\n l = []\n l.append(x)\n x_view = l[0]\n a = x + x\n x_view.add_(y)\n b = x + x\n return a == b\n self.checkScript(fn, (torch.rand(2, 3), torch.rand(2, 3)))\n\n def test_module_attrs(self):\n class M(torch.jit.ScriptModule):\n def __init__(self, table):\n super(M, self).__init__()\n self.table = torch.jit.Attribute(table, Dict[str, torch.Tensor])\n self.x = torch.nn.Parameter(torch.tensor([100.0]))\n\n @torch.jit.script_method\n def forward(self, key):\n # type: (str) -> Tensor\n return self.table[key] + self.x\n\n with torch._jit_internal._disable_emit_hooks():\n # TODO: re-enable module hook when Python printing of attributes is\n # supported\n m = M({char : torch.ones(1) + ord(char) - ord(\"a\") for char in \"abcdefg\"})\n # TODO(#38095): Replace assertEqualIgnoreType. See issue #38095\n self.assertEqualIgnoreType(m(\"c\"), torch.tensor([103]))\n\n def test_module_none_attrs(self):\n class MyMod(torch.jit.ScriptModule):\n def __init__(self):\n super(MyMod, self).__init__()\n self.optional_value = None\n\n @torch.jit.script_method\n def forward(self):\n return self.optional_value\n\n graph = MyMod().forward.graph\n FileCheck().check(\"prim::GetAttr\").run(graph)\n self.run_pass('peephole', graph)\n FileCheck().check_not(\"prim::GetAttr\").run(graph)\n\n def test_tensor_import_export(self):\n @torch.jit.script\n def foo(x):\n a = torch.tensor(1)\n b = torch.tensor([1, 2])\n c = [a, b]\n return c\n\n self.run_pass('constant_propagation', foo.graph)\n m = self.createFunctionFromGraph(foo.graph)\n self.getExportImportCopy(m)\n\n def get_pickle_values(self):\n return (('dict', {\"I\": \"am\", \"a test\": \"test\"}, Dict[str, str]),\n ('float', 2.3, float),\n ('int', 99, int),\n ('bool', False, bool),\n ('tuple', (1, 2, 3, 4), Tuple[int, int, int, int]),\n ('list', [(1, 2), (3, 4)], List[Tuple[int, int]]),\n ('tensor', torch.randn(2, 2), torch.Tensor),\n ('int_list', [1, 2, 3, 4], List[int]),\n ('tensor_list', [torch.ones(2, 2) + i for i in range(4)], List[torch.Tensor]),\n ('bool_list', [True, True, False, True], List[bool]),\n ('float_list', [1., 2., 3., 4.], List[float]),\n ('str_list', ['hello', 'bye'], List[str]),\n ('none', None, Optional[int]),\n ('a_device', torch.device('cpu'), torch.device),\n ('another_device', torch.device('cuda:1'), torch.device))\n\n def test_attribute_serialization(self):\n tester = self\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n for name, value, the_type in tester.get_pickle_values():\n setattr(self, name, torch.jit.Attribute(value, the_type))\n\n @torch.jit.script_method\n def forward(self):\n return (self.dict, self.float, self.int, self.bool, self.tuple,\n self.list, self.int_list, self.tensor_list, self.bool_list,\n self.float_list, self.str_list, self.none)\n\n m = M()\n imported_m = self.getExportImportCopy(m)\n self.assertEqual(m(), imported_m())\n\n def test_string_len(self):\n def fn(x):\n # type: (str) -> int\n return len(x)\n\n self.checkScript(fn, (\"\",))\n self.checkScript(fn, (\"h\",))\n self.checkScript(fn, (\"hello\",))\n\n def test_multiline_optional_future_refinement(self):\n @torch.jit.script\n def fun() -> int:\n future: Optional[\n torch.jit.Future[Tuple[torch.Tensor]]\n ] = None\n\n return 1\n self.assertEqual(fun(), 1)\n\n @unittest.skipIf(IS_WINDOWS or IS_SANDCASTLE, \"NYI: TemporaryFileName support for Windows or Sandcastle\")\n def test_attribute_unpickling(self):\n tensor = torch.randn(2, 2)\n tester = self\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n for name, value, the_type in tester.get_pickle_values():\n setattr(self, \"_\" + name, torch.jit.Attribute(value, the_type))\n\n @torch.jit.script_method\n def forward(self):\n return (self._dict, self._float, self._int, self._bool, self._tuple,\n self._list, self._int_list, self._tensor_list, self._bool_list,\n self._float_list, self._str_list, self._none)\n\n with TemporaryFileName() as fname:\n M().save(fname)\n loaded = torch.jit.load(fname)\n\n def is_tensor_value(item):\n if isinstance(item, torch.Tensor):\n return True\n if isinstance(item, list):\n return is_tensor_value(item[0])\n return False\n for name, value, the_type in self.get_pickle_values():\n if is_tensor_value(value):\n continue\n self.assertEqual(value, getattr(loaded, \"_\" + name))\n\n @unittest.skipIf(IS_WINDOWS or IS_SANDCASTLE, \"NYI: TemporaryFileName support for Windows or Sandcastle\")\n def test_old_models_bc(self):\n model = {\n 'archive/version': b'1',\n 'archive/code/archive.py':\n b'''\n op_version_set = 0\n def forward(self,\n _0: Tensor) -> Tensor:\n _1 = torch.zeros([10], dtype=6, layout=0, device=torch.device(\"cpu\"))\n result = torch.to(torch.fill_(_1, 5), dtype=6, layout=0, device=torch.device(\"cpu\"),\n non_blocking=False, copy=False)\n result2 = torch.rand([10], dtype=6, layout=0, device=torch.device(\"cpu\"))\n result3 = torch.rand_like(result2, dtype=6, layout=0, device=torch.device(\"cpu\"))\n _2 = torch.add(torch.add(result, result2, alpha=1), result3, alpha=1)\n return _2\n ''',\n 'archive/attributes.pkl': b'\\x80\\x02](e.',\n 'archive/libs.py': b'op_version_set = 0\\n',\n 'archive/model.json':\n b'''\n {\n \"protoVersion\":\"2\",\n \"mainModule\":{\n \"torchscriptArena\":{\n \"key\":\"code/archive.py\"\n },\n \"name\":\"archive\",\n \"optimize\":true\n },\n \"producerName\":\"pytorch\",\n \"producerVersion\":\"1.0\",\n \"libs\":{\n \"torchscriptArena\":{\n \"key\":\"libs.py\"\n }\n }\n }'''}\n with TemporaryFileName() as fname:\n archive_name = os.path.basename(os.path.normpath(fname))\n with zipfile.ZipFile(fname, 'w') as archive:\n for k, v in model.items():\n archive.writestr(k, v)\n\n with open(fname, \"rb\") as f:\n fn = torch.jit.load(f)\n\n x = torch.zeros(10)\n fn(x)\n\n def test_submodule_attribute_serialization(self):\n class S(torch.jit.ScriptModule):\n def __init__(self, list_data):\n super(S, self).__init__()\n self.table = torch.jit.Attribute({\"I\": \"am\", \"a test\": \"test\"}, Dict[str, str])\n self.list = torch.jit.Attribute(list_data, List[Tuple[int, int]])\n\n @torch.jit.script_method\n def forward(self):\n return (self.table, self.list)\n\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.table = torch.jit.Attribute({\"this\": \"is\", \"a different\": \"dict\"}, Dict[str, str])\n self.tensor = torch.jit.Attribute(torch.randn(2, 2), torch.Tensor)\n self.s1 = S([(1, 2)])\n self.s2 = S([(4, 5)])\n\n @torch.jit.script_method\n def forward(self):\n return (self.table, self.tensor, self.s1.table, self.s2.list, self.s1.list)\n\n m = M()\n imported_m = self.getExportImportCopy(m)\n self.assertEqual(m(), imported_m())\n\n def test_serialization_big_ints(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.int32_max = torch.jit.Attribute(2**31 - 1, int)\n self.int32_min = torch.jit.Attribute(-2**31, int)\n self.uint32_max = torch.jit.Attribute(2**32, int)\n\n self.int64_max = torch.jit.Attribute(2**63 - 1, int)\n self.int64_min = torch.jit.Attribute(-2**63, int)\n\n self.tensor = torch.nn.Parameter(torch.ones(2, 2))\n\n @torch.jit.script_method\n def forward(self, x):\n # type: (int) -> (int)\n return x + (self.int32_max + self.int32_min) + (self.int64_max + self.int64_min)\n\n m = M()\n imported = self.getExportImportCopy(m)\n self.assertEqual(m(10), imported(10))\n\n self.assertEqual(m.int32_max, imported.int32_max)\n self.assertEqual(m.int32_min, imported.int32_min)\n self.assertEqual(m.uint32_max, imported.uint32_max)\n self.assertEqual(m.int64_max, imported.int64_max)\n self.assertEqual(m.int64_min, imported.int64_min)\n\n def test_script_scope(self):\n scripted = torch.jit.script(torch.nn.functional.pad)\n\n @unittest.skipIf(IS_WINDOWS, \"NYI: TemporaryFileName on Windows\")\n def test_serialization_sharing(self):\n class M(torch.jit.ScriptModule):\n def __init__(self):\n super(M, self).__init__()\n self.list = torch.jit.Attribute([], List[str])\n\n @torch.jit.script_method\n def forward(self, key):\n # type: (str) -> List[str]\n self.list.append(key)\n self.list.append(key)\n self.list.append(key)\n return self.list\n\n # the text of the string should only appear once in the pickling\n m = M()\n s1 = \"a long string\"\n s2 = \"a different, even longer string\"\n self.assertEqual(m(s1), [s1] * 3)\n self.assertEqual(m(s2), [s1] * 3 + [s2] * 3)\n with TemporaryFileName() as fname:\n m.save(fname)\n archive_name = os.path.basename(os.path.normpath(fname))\n archive = zipfile.ZipFile(fname, 'r')\n pickled_data = archive.read(os.path.join(archive_name, 'data.pkl'))\n\n out = io.StringIO()\n pickletools.dis(pickled_data, out=out)\n disassembled = out.getvalue()\n\n FileCheck().check_count(s1, 1, exactly=True) \\\n .check_count(\"BINGET\", 2, exactly=True) \\\n .check_count(s2, 1, exactly=True) \\\n .check_count(\"BINGET\", 2, exactly=True).run(out.getvalue())\n\n def test_sys_stdout_override(self):\n @torch.jit.script\n def foo():\n print('foo')\n\n class Redirect(object):\n def __init__(self):\n self.s = ''\n\n def write(self, s):\n self.s += s\n\n old_stdout = sys.stdout\n redirect = Redirect()\n try:\n sys.stdout = redirect\n foo()\n finally:\n sys.stdout = old_stdout\n\n FileCheck().check('foo').run(redirect.s)\n\n def test_dtype_attr(self):\n class Foo(torch.nn.Module):\n def __init__(self):\n super(Foo, self).__init__()\n self.dtype = torch.zeros([]).dtype\n\n def forward(self):\n return torch.zeros(3, 4, dtype=self.dtype)\n\n f = Foo()\n torch.jit.script(f)\n\n\n def test_named_buffers_are_iterable(self):\n class MyMod(torch.nn.Module):\n def __init__(self):\n super(MyMod, self).__init__()\n self.mod = (torch.nn.ReLU())\n self.mod2 = (torch.nn.ReLU())\n self.mod3 = torch.nn.Sequential(torch.nn.Sequential(torch.nn.ReLU()))\n self.register_buffer('x', torch.zeros(3))\n self.register_buffer('y', torch.zeros(3))\n self.z = torch.zeros(3)\n\n def bleh(self):\n return self.z + 4\n\n @torch.jit.export\n def method(self):\n names = [\"\"]\n vals = []\n for name, buffer in self.named_buffers():\n names.append(name)\n vals.append(buffer + 2)\n\n return names, vals\n\n def forward(self, x):\n return x\n\n model = MyMod()\n x = torch.jit.script(model)\n z = self.getExportImportCopy(x)\n\n self.assertEqual(z.method(), x.method())\n self.assertEqual(z.method(), model.method())\n self.assertEqual(x.method(), model.method())\n names = x.method()\n for name in names:\n self.assertNotEqual('z', name)\n\n\n def test_static_if_prop(self):\n class MaybeHasAttr(torch.nn.Module):\n def __init__(self, add_attr):\n super(MaybeHasAttr, self).__init__()\n if add_attr:\n self.maybe_attr = 1\n\n def forward(self):\n if hasattr(self, \"maybe_attr\") and True:\n return self.maybe_attr\n else:\n return 0\n\n class MaybeHasAttr2(torch.nn.Module):\n def __init__(self, add_attr):\n super(MaybeHasAttr2, self).__init__()\n if add_attr:\n self.maybe_attr = 1\n\n def forward(self):\n if not hasattr(self, \"maybe_attr\") or False:\n return 0\n else:\n return self.maybe_attr\n\n torch.jit.script(MaybeHasAttr(True))\n torch.jit.script(MaybeHasAttr(False))\n torch.jit.script(MaybeHasAttr2(True))\n torch.jit.script(MaybeHasAttr2(False))\n\n class MyMod(torch.nn.Module):\n def forward(self):\n if hasattr(self, \"foo\"):\n return 1\n else:\n return 0\n\n @torch.jit.export\n def fee(self):\n return 1\n\n self.checkModule(MyMod(), ())\n\n class HasAttrMod(torch.nn.Module):\n __constants__ = [\"fee\"]\n\n def __init__(self):\n super().__init__()\n self.fee = 3\n\n def forward(self):\n a = hasattr(self, \"fee\")\n b = hasattr(self, \"foo\")\n c = hasattr(self, \"hi\")\n d = hasattr(self, \"nonexistant\")\n return (a, b, c, d)\n\n def foo(self):\n return 1\n\n @torch.jit._overload_method\n def hi(self, x: Tensor): ... # noqa: E704\n\n def hi(self, x): # noqa: F811\n return 2\n\n self.checkModule(HasAttrMod(), ())\n\n @torch.jit.script\n class FooTest(object):\n def __init__(self):\n self.x = 1\n\n def foo(self, y):\n return self.x + y\n\n def foo():\n a = FooTest()\n val1 = hasattr(a, \"foo\"), hasattr(a, \"x\"), hasattr(a, \"bla\")\n val2 = hasattr(FooTest, \"foo\"), hasattr(FooTest, \"a\")\n return val1, val2\n\n self.assertEqual(foo(), torch.jit.script(foo)())\n\n def _test_pickle_checkpoint(self, device):\n with TemporaryFileName() as fname:\n class M(torch.jit.ScriptModule):\n __constants__ = ['fname']\n\n def __init__(self, tensor):\n super(M, self).__init__()\n self.fname = fname\n self.tensor = torch.nn.Parameter(tensor)\n\n @torch.jit.script_method\n def forward(self, x):\n y = self.tensor + x\n torch.save(y, self.fname)\n return y\n\n param = torch.randn(2, 2).to(device)\n input = torch.randn(2, 2).to(device)\n m = M(param)\n m(input)\n with open(fname, \"rb\") as handle:\n loaded_tensor = torch.load(fname)\n self.assertEqual(loaded_tensor, input + param)\n\n def _test_pickle_checkpoint_views(self, device):\n with TemporaryFileName() as fname:\n class M(torch.jit.ScriptModule):\n __constants__ = ['fname']\n\n def __init__(self, tensor):\n super(M, self).__init__()\n self.fname = fname\n self.tensor = torch.nn.Parameter(tensor)\n\n @torch.jit.script_method\n def forward(self, x):\n y = self.tensor + x\n y_view = y.view(4)\n torch.save((y, y_view, y), self.fname)\n return y\n\n param = torch.randn(2, 2).to(device)\n input = torch.randn(2, 2).to(device)\n m = M(param)\n m(input)\n with open(fname, \"rb\") as handle:\n loaded_y, loaded_y_view, loaded_y_2 = torch.load(fname)\n self.assertEqual(loaded_y, input + param)\n with torch.no_grad():\n loaded_y_view[1] += 20\n # assert that loaded_y changed as well\n self.assertEqual(loaded_y.view(4), loaded_y_view)\n self.assertEqual(loaded_y_2.view(4), loaded_y_view)\n\n @unittest.skipIf(not RUN_CUDA, \"no CUDA\")\n def test_pickle_checkpoint_cuda(self):\n self._test_pickle_checkpoint('cuda')\n self._test_pickle_checkpoint_views('cuda')\n\n def test_pickle_checkpoint(self):\n self._test_pickle_checkpoint('cpu')\n self._test_pickle_checkpoint_views('cpu')\n\n def test_pickle_checkpoint_tup(self):\n @torch.jit.script\n def foo(fname):\n # type: (str) -> None\n torch.save((3, 4), fname)\n with TemporaryFileName() as name:\n foo(name)\n self.assertEqual(torch.load(name), (3, 4))\n\n def test_string_list(self):\n def fn(string):\n # type: (str) -> List[str]\n return list(string)\n\n self.checkScript(fn, (\"abcdefgh\",))\n\n def test_unicode_comments(self):\n @torch.jit.script\n def test(self, a):\n # 🤷🤷🤷🤷\n return torch.nn.functional.relu(a)\n\n def test_get_set_state_with_tensors(self):\n class M(torch.nn.Module):\n def __init__(self):\n super(M, self).__init__()\n self.tensor = torch.randn(2, 2)\n\n @torch.jit.export\n def __getstate__(self):\n return (self.tensor, self.training)\n\n @torch.jit.export\n def __setstate__(self, state):\n self.tensor = state[0]\n self.training = state[1]\n\n def forward(self, x):\n return x + self.tensor\n\n with TemporaryFileName() as fname:\n m = torch.jit.script(M())\n m.save(fname)\n loaded = torch.jit.load(fname)\n self.assertEqual(loaded.tensor, m.tensor)\n\n def test_in_for_and_comp_expr(self):\n def fn(d):\n # type: (Dict[str, int]) -> List[int]\n out = [1]\n for i in range(d[\"hi\"] if \"hi\" in d else 6):\n out.append(i)\n return out\n\n self.checkScript(fn, ({'hi': 2, 'bye': 3},))\n self.checkScript(fn, ({'bye': 3},))\n\n def test_for_else(self):\n def fn():\n c = 0\n for i in range(4):\n c += 10\n else:\n print(\"In else block of for...else\")\n\n with self.assertRaisesRegex(torch.jit.frontend.NotSupportedError, \"else branches of for loops aren't supported\"):\n torch.jit.script(fn)\n\n def test_split(self):\n def split_two(tensor):\n a, b, c = torch.split(tensor, 2, dim=1)\n return a, b, c\n x = torch.randn(3, 6)\n y = torch.randn(3, 6)\n self.checkScript(split_two, [(x + y)])\n\n def test_conv_error(self):\n @torch.jit.script\n def fn(x, y):\n return F.conv2d(x, y)\n\n try:\n fn(torch.ones(2, 2), torch.ones(4, 4))\n except RuntimeError as e:\n self.assertFalse('frame' in str(e))\n\n def test_python_op_name(self):\n import random\n\n with self.assertRaisesRegex(RuntimeError, \"randint\"):\n @torch.jit.script\n def fn():\n return random.randint()\n\n def test_dir(self):\n class M(torch.jit.ScriptModule):\n def forward(self, t):\n return t\n\n self.assertTrue('forward' in dir(M()))\n\n def test_kwarg_expansion_error(self):\n @torch.jit.ignore\n def something_else(h, i):\n pass\n\n def fn(x):\n something_else(**x)\n\n with self.assertRaisesRegex(torch.jit.frontend.NotSupportedError, \"keyword-arg expansion is not supported\"):\n torch.jit.script(fn)\n\n def test_kwargs_error_msg(self):\n def other(**kwargs):\n print(kwargs)\n\n def fn():\n return other()\n\n with self.assertRaisesRegex(torch.jit.frontend.NotSupportedError, 'variable number'):\n torch.jit.script(fn)\n\n def another_other(*args):\n print(args)\n\n def another_fn():\n return another_other()\n\n with self.assertRaisesRegex(torch.jit.frontend.NotSupportedError, 'variable number'):\n torch.jit.script(another_fn)\n\n def test_inferred_error_msg(self):\n \"\"\"\n Test that when we get a type mismatch on a function where we inferred\n the type to be tensor, a good error message is given.\n \"\"\"\n @torch.jit.script\n def foo(a):\n return a\n\n with self.assertRaisesRegex(RuntimeError, (r\"Expected a value of type \\'Tensor \\(inferred\\)\\'\"\n r\"[\\S\\s]*Inferred \\'a\\' to be of type \\'Tensor\\'\")):\n foo(1)\n\n def test_type_comments_in_body(self):\n @torch.jit.script\n def foo(a, # type: int\n b, # type: int\n ):\n # type: (...) -> int\n # type: int\n return a + b\n\n class M(torch.nn.Module):\n def __init__(self,\n a, # type: int\n b # type: int\n ):\n # type: (...) -> None\n super(M, self).__init__()\n self.a = a # type: int\n self.b = b # type: int\n\n torch.jit.script(M(2, 3))\n\n def test_module_method_reassignment(self):\n class Foo(torch.nn.Module):\n def __init__(self):\n super().__init__()\n\n def _forward(self, x):\n return x\n\n forward = _forward\n\n sm = torch.jit.script(Foo())\n input = torch.ones(2, 2)\n self.assertEqual(input, sm(input))\n\n # Tests the case where a torch.Tensor subclass (like Parameter) is used as\n # input.\n def test_script_module_tensor_subclass_argument(self):\n @torch.jit.script\n def parameter_script(x: torch.nn.Parameter):\n return x\n\n input = torch.ones(2, 2)\n self.assertEqual(input, parameter_script(input))\n\n def test_save_load_attr_error(self):\n class Inner(nn.Module):\n def __init__(self):\n super().__init__()\n\n def forward(self, x):\n return x\n\n class Wrapper(nn.Module):\n def __init__(self, inner):\n super().__init__()\n self.inner = inner\n\n def forward(self, x):\n # this attribute doesn't exist on `Inner`\n return self.inner.b(x)\n\n inner_module = torch.jit.script(Inner())\n inner_module = self.getExportImportCopy(inner_module)\n wrapped = Wrapper(inner_module)\n # This should properly complain that `self.inner` doesn't have the attribute `b`\n with self.assertRaisesRegex(RuntimeError, 'has no attribute'):\n torch.jit.script(wrapped)\n\n def test_rescripting_loaded_modules(self):\n class InnerSubmod(nn.Module):\n __constants__ = ['my_constant']\n\n def __init__(self):\n super().__init__()\n self.register_buffer(\"foo\", torch.ones(1))\n self.register_parameter(\"bar\", torch.nn.Parameter(torch.ones(1)))\n self.baz = torch.ones(1)\n self.my_constant = 1\n\n def forward(self, x):\n return x + x\n\n class Inner(nn.Module):\n def __init__(self):\n super().__init__()\n self.submod = InnerSubmod()\n\n def forward(self, x):\n return self.submod(x)\n\n class Wrapper(nn.Module):\n def __init__(self, inner):\n super().__init__()\n self.inner = inner\n\n def forward(self, x):\n # access inner elements\n ret = self.inner.submod(x) + self.inner.submod.foo + self.inner.submod.bar + self.inner.submod.baz\n ret = ret + self.inner.submod.my_constant\n return ret\n\n inner_module = torch.jit.script(Inner())\n wrapped = Wrapper(inner_module)\n self.checkModule(wrapped, torch.ones(1))\n\n inner_module_loaded = self.getExportImportCopy(inner_module)\n wrapped_loaded = Wrapper(inner_module_loaded)\n self.assertEqual(wrapped(torch.ones(1)), wrapped_loaded(torch.ones(1)))\n\n def test_interpret_graph(self):\n def fn(x):\n return x.unfold(0, 1, 1)\n\n graph_str = \"\"\"\n graph(%a : Tensor, %b : Tensor):\n %c : Tensor = aten::mul(%a, %b)\n return (%c)\n \"\"\"\n graph = parse_ir(graph_str)\n a = torch.rand(10)\n b = torch.rand(10)\n test = torch._C._jit_interpret_graph(graph, (a, b))\n ref = a * b\n self.assertEqual(test, ref)\n\n def test_signed_float_zero(self):\n\n class MyModule(torch.nn.Module):\n def __init__(self):\n super(MyModule, self).__init__()\n\n def forward(self, x):\n return torch.div(x, -0.)\n\n inp = torch.ones(1)\n self.checkModule(MyModule(), inp)\n\n# known to be failing in tracer\nEXCLUDE_TRACED = {\n # The following fail due to #12024.\n # A prim::ListConstruct is involved and the indices get traced as TensorType,\n # which always require_grad. This causes a crash in autodiff.\n 'test___getitem___adv_index',\n 'test___getitem___adv_index_beg',\n 'test___getitem___adv_index_comb',\n 'test___getitem___adv_index_dup',\n 'test___getitem___adv_index_sub',\n 'test___getitem___adv_index_sub_2',\n 'test___getitem___adv_index_sub_3',\n 'test___getitem___adv_index_var',\n\n # jit doesn't support sparse tensors.\n 'test_to_sparse',\n 'test_to_sparse_dim',\n}\n\nEXCLUDE_TYPE_CHECK = {\n # slogdet tests use itemgetter to select its only differentiable output,\n # but this happens outside of the graph we handle, so there are fewer\n # reference outputs than graph outputs.\n 'test_slogdet_1x1_neg_det',\n 'test_slogdet_1x1_pos_det',\n 'test_slogdet_distinct_singular_values',\n 'test_slogdet_neg_det',\n 'test_slogdet_pos_det',\n 'test_slogdet_symmetric',\n 'test_slogdet_symmetric_pd',\n 'test_slogdet_batched_1x1_neg_det',\n 'test_slogdet_batched_pos_det',\n 'test_slogdet_batched_symmetric',\n 'test_slogdet_batched_symmetric_pd',\n 'test_slogdet_batched_distinct_singular_values'\n}\n\n# chunk returns a list in scripting and we don't unpack the list,\n# Thus it won't be replaced by ConstantChunk and run AD.\n# It's explicitly checked in test_chunk_constant_script_ad\n# Similary for split, it's replaced by split_with_sizes in tracing,\n# but we don't have AD formula for aten::split(Tensor, int[], int),\n# an op registered in JIT so AD is not triggered in scripting.\nEXCLUDE_SCRIPT_AD_CHECK = {\n 'test_chunk',\n 'test_chunk_dim',\n 'test_chunk_dim_neg0',\n 'test_split_size_list',\n 'test_split_size_list_dim',\n 'test_split_size_list_dim_neg0',\n 'test_tensor_indices_sections',\n 'test_tensor_indices_sections_dim',\n 'test_tensor_indices_sections_dim_neg0',\n 'test_tensor_split_sections',\n 'test_tensor_split_sections_dim',\n 'test_tensor_split_sections_dim_neg0'\n}\n\nEXCLUDE_PYTHON_PRINT = {\n # no support for BroadcastingList in python printer\n 'test_nn_max_unpool1d',\n 'test_nn_max_unpool2d',\n 'test_nn_max_unpool3d',\n 'test_nn_max_pool1d',\n 'test_nn_max_pool2d',\n 'test_nn_max_pool3d',\n 'test_nn_max_pool1d_with_indices',\n}\n\nEXCLUDE_ALIAS = {\n # aliases, which may appear in method_tests but are tested elsewhere\n 'true_divide',\n\n # Disable tests for lu from common_methods_invocations.py\n # TODO(@nikitaved) Enable jit tests once autograd.Function does support scripting\n 'lu'\n}\n\n\nclass TestJitGeneratedModule(JitTestCase):\n pass\n\n\nclass TestJitGeneratedFunctional(JitTestCase):\n pass\n\n\n# UBSAN per-function exclusions don't seem to work with OpenMP pragmas,\n# and we have to disable the failing tests here instead.\nUBSAN_DISABLED_TESTS = [\n \"test___rdiv___constant\",\n \"test___rdiv___scalar_constant\",\n \"test_addcdiv\",\n \"test_addcdiv_broadcast_all\",\n \"test_addcdiv_broadcast_rhs\",\n \"test_addcdiv_scalar\",\n \"test_addcdiv_scalar_broadcast_lhs\",\n \"test_addcdiv_scalar_broadcast_rhs\",\n \"test_addcdiv_scalar_scale\",\n \"test_addcdiv_scalar_scale_broadcast_lhs\",\n \"test_addcdiv_scalar_scale_broadcast_rhs\",\n \"test_addcdiv_scale\",\n \"test_addcdiv_scale_broadcast_all\",\n \"test_addcdiv_scale_broadcast_rhs\",\n \"test_add_broadcast_all\",\n \"test_add_broadcast_lhs\",\n \"test_add_broadcast_rhs\",\n \"test_add_constant\",\n \"test_add_scalar\",\n \"test_add_scalar_broadcast_lhs\",\n \"test_add_scalar_broadcast_rhs\",\n \"test_div\",\n \"test_div_broadcast_all\",\n \"test_div_broadcast_lhs\",\n \"test_div_broadcast_rhs\",\n \"test_div_scalar\",\n \"test_div_scalar_broadcast_lhs\",\n \"test_div_scalar_broadcast_rhs\",\n \"test_rsqrt\",\n \"test_rsqrt_scalar\",\n \"test_add\",\n \"test_reciprocal\",\n \"test_reciprocal_scalar\",\n]\n\nL = 20\nM = 10\nS = 5\n\n\ndef add_nn_functional_test(name, self_size, args, variant_name='', check_ad=(), skipTestIf=(),\n output_process_fn=lambda x: x, kwargs=None):\n test_name = 'test_nn_' + name\n\n if variant_name != '':\n test_name = test_name + '_' + variant_name\n\n no_grad = variant_name == 'inplace'\n\n @suppress_warnings\n def do_test(self, name=name, args=args, test_name=test_name, check_ad=check_ad):\n torch.manual_seed(2)\n\n self_variable = create_input((self_size,))[0][0]\n\n # need to record this because methods can change the size (e.g. unsqueeze)\n args_variable, kwargs_variable = create_input(args, call_kwargs=kwargs)\n\n self_tensor = deepcopy(self_variable.data)\n args_tensor = deepcopy(unpack_variables(args_variable))\n\n if not no_grad:\n output_variable = getattr(F, name)(self_variable, *args_variable, **kwargs_variable)\n\n def fn(*inputs, **kwargs):\n return getattr(F, name)(*inputs, **kwargs)\n\n f_args_variable = (self_variable,) + args_variable\n f_args_tensor = (self_tensor,) + args_tensor\n should_autodiff_node, autodiff_nodes, fusible_nodes = normalize_check_ad(check_ad, name)\n\n if test_name not in EXCLUDE_SCRIPT:\n def run_test():\n # XXX: this test should always run with disable_autodiff_subgraph_inlining(True),\n # so that we don't regress on autodiff support.\n with disable_autodiff_subgraph_inlining():\n script_fn = create_script_fn(self, name, 'nn_functional')\n check_against_reference(self, script_fn, fn, output_process_fn,\n f_args_variable, kwargs_variable, no_grad=no_grad)\n # For tests we disabled AD subgraph inlining, make sure it's not falling back to autograd\n if (doAutodiffCheck(test_name)):\n self.assertAutodiffNode(script_fn.last_graph, should_autodiff_node, autodiff_nodes, fusible_nodes)\n\n if test_name in EXCLUDE_PYTHON_PRINT:\n with torch._jit_internal._disable_emit_hooks():\n run_test()\n else:\n run_test()\n\n post_add_test(test_name, skipTestIf, do_test, TestJitGeneratedFunctional)\n\n\ndef add_nn_module_test(*args, **kwargs):\n name = get_nn_module_name_from_kwargs(**kwargs)\n\n no_grad = False if 'no_grad' not in kwargs else kwargs['no_grad']\n\n if 'desc' in kwargs and 'eval' in kwargs['desc']:\n # eval() is not supported, so skip these tests\n return\n\n test_name = name\n if 'desc' in kwargs:\n test_name = \"{}_{}\".format(test_name, kwargs['desc'])\n test_name = 'test_nn_{}'.format(test_name)\n\n @suppress_warnings\n def do_test(self):\n if test_name in EXCLUDE_SCRIPT_MODULES:\n return\n if not kwargs.get('check_jit', True):\n raise unittest.SkipTest('module test skipped on JIT')\n\n if 'constructor' in kwargs:\n nn_module = kwargs['constructor']\n else:\n nn_module = getattr(torch.nn, name)\n\n if \"FunctionalModule\" in str(nn_module):\n return\n\n if 'constructor_args_fn' in kwargs:\n constructor_args = kwargs['constructor_args_fn']()\n else:\n constructor_args = kwargs.get('constructor_args', ())\n\n module_name = get_nn_module_name_from_kwargs(**kwargs)\n\n # Construct a script module that passes arguments through\n # to self.submodule\n def create_script_module(*args, **kwargs):\n formals, tensors, actuals = get_script_args(args)\n\n method_args = ', '.join(['self'] + actuals)\n call_args_str = ', '.join(actuals)\n call = \"self.submodule({})\".format(call_args_str)\n script = script_method_template.format(method_args, call)\n\n submodule_constants = []\n if kwargs.get('is_constant'):\n submodule_constants = ['submodule']\n\n # Create module to use the script method\n class TheModule(torch.jit.ScriptModule):\n __constants__ = submodule_constants\n\n def __init__(self):\n super(TheModule, self).__init__()\n self.submodule = nn_module(*constructor_args)\n\n def make_module(script):\n module = TheModule()\n # check __repr__\n str(module)\n module.define(script)\n return module\n\n module = make_module(script)\n self.assertExportImportModule(module, tensors)\n create_script_module.last_graph = module.graph\n mod = module(*args)\n return mod\n\n # Construct a normal nn module to stay consistent with create_script_module\n # and make use of a single global rng_state in module initialization\n def create_nn_module(*args, **kwargs):\n module = nn_module(*constructor_args)\n return module(*args)\n\n # Set up inputs from tuple of sizes or constructor fn\n dtype = torch.double\n if 'input_fn' in kwargs:\n input = kwargs['input_fn']()\n if isinstance(input, Tensor):\n input = (input,)\n\n if all(tensor.is_complex() for tensor in input):\n dtype = torch.cdouble\n else:\n input = (kwargs['input_size'],)\n\n if 'target_size' in kwargs:\n input = input + (kwargs['target_size'],)\n elif 'target_fn' in kwargs:\n if torch.is_tensor(input):\n input = (input,)\n input = input + (kwargs['target_fn'](),)\n elif 'target' in kwargs:\n input = input + (kwargs['target'],)\n\n # Extra parameters to forward()\n if 'extra_args' in kwargs:\n input = input + kwargs['extra_args']\n\n args_variable, kwargs_variable = create_input(input, dtype=dtype)\n f_args_variable = deepcopy(unpack_variables(args_variable))\n\n # TODO(issue#52052) Neither this nor no_grad should be required\n # if check_against_reference() is updated to check gradients\n # w.r.t. weights and then only check w.r.t. inputs if any\n # inputs require it.\n any_requires_grad = any(input.requires_grad for input in f_args_variable)\n\n # Check against Python module as reference\n check_against_reference(self, create_script_module, create_nn_module,\n lambda x: x, f_args_variable,\n no_grad=no_grad or not any_requires_grad)\n\n if 'slowTest' in kwargs:\n do_test = slowTest(do_test)\n\n post_add_test(test_name, (), do_test, TestJitGeneratedModule)\n\n\ndef post_add_test(test_name, skipTestIf, do_test, test_class):\n assert not hasattr(test_class, test_name), 'Two tests have the same name: ' + test_name\n\n for skip in skipTestIf:\n do_test = skip(do_test)\n\n if not (TEST_WITH_UBSAN and test_name in UBSAN_DISABLED_TESTS):\n setattr(test_class, test_name, do_test)\n\n\ndef normalize_check_ad(check_ad, name):\n # normalized check_ad is 3-element tuple: (bool, List[str], List[str])\n if len(check_ad) == 0:\n check_ad = [False, ['aten::' + name], []]\n elif len(check_ad) == 1:\n check_ad = [check_ad[0], ['aten::' + name], []]\n elif len(check_ad) == 2:\n check_ad = [check_ad[0], check_ad[1], []]\n elif len(check_ad) == 3:\n check_ad = list(check_ad)\n else:\n raise Exception('Invalid check_ad, requires (bool, str|List[str], str|List[str])')\n\n check_ad = [[t] if isinstance(t, str) else t for t in check_ad]\n\n return check_ad\n\n\nclass TestProducerVersion(unittest.TestCase):\n\n def test_version(self):\n # issue gh-32561\n self.assertTrue(torch.__version__.startswith(torch.onnx.producer_version))\n\n\nfor test in nn_functional_tests:\n add_nn_functional_test(*test)\n\nfor test in module_tests + new_module_tests + additional_module_tests:\n add_nn_module_test(**test)\n\nfor test in criterion_tests:\n test['no_grad'] = True\n add_nn_module_test(**test)\n\nif __name__ == '__main__':\n run_tests()\n import test_jit_py3\n import jit.test_module_interface\n suite = unittest.findTestCases(test_jit_py3)\n unittest.TextTestRunner().run(suite)\n suite = unittest.findTestCases(jit.test_module_interface)\n unittest.TextTestRunner().run(suite)\n"
]
| [
[
"torch.BoolTensor",
"torch.jit.load",
"torch.randint",
"torch.lu_unpack",
"torch._C.PyTorchFileWriter",
"torch.testing._internal.jit_utils.get_execution_plan",
"torch.zeros",
"torch.nn.functional.dropout",
"torch.max",
"torch.nn.GRU",
"torch.neg",
"torch.testing._internal.jit_metaprogramming_utils.create_script_fn",
"torch.acos",
"torch.jit.verify",
"torch.where",
"torch._C.NoneType.get",
"torch.device",
"torch.randn",
"torch._C.TensorType.get",
"torch.equal",
"torch.nn.init._calculate_fan_in_and_fan_out",
"torch.jit.CompilationUnit",
"torch.autograd.grad",
"torch.blargh",
"torch.jit.optimized_execution",
"torch.enable_grad",
"torch.full",
"torch.jit._hide_source_ranges",
"torch._C.Graph",
"torch.random.fork_rng",
"numpy.random.choice",
"torch.nn.Conv2d",
"torch.nn.ReplicationPad1d",
"torch.cuda.current_device",
"torch.nn.Module",
"torch.nn.Linear",
"torch.exp",
"torch._C._jit_pass_canonicalize",
"torch.nn.ReflectionPad3d",
"torch.nn.BatchNorm2d",
"torch._C._jit_pass_fuse_add_relu",
"numpy.array",
"torch.testing._internal.common_utils.run_tests",
"torch._C._get_tracing_state",
"torch._C.TupleType",
"torch.nn.ReflectionPad2d",
"torch._C._jit_set_profiling_executor",
"torch.nn.modules.utils._triple",
"torch.nn.ConvTranspose3d",
"torch.any",
"torch.unbind",
"torch.nn.modules.utils._pair",
"torch.testing._internal.common_utils.TemporaryFileName",
"torch.ops.aten.add",
"torch.jit.compile",
"torch.abs",
"torch.nn.functional.prelu",
"torch.nn.RNN",
"torch.sum",
"torch.nn.utils.rnn.pad_sequence",
"torch.nn.functional.adaptive_avg_pool1d",
"torch.nn.utils.rnn.pad_packed_sequence",
"torch.set_grad_enabled",
"torch.FloatTensor",
"torch.cuda.is_available",
"torch._C._jit_debug_module_iterators",
"torch._C._jit_pass_complete_shape_analysis",
"torch.jit._fork",
"torch.addmm",
"torch.testing._internal.common_utils.set_default_dtype",
"torch.split",
"torch.nn.ReplicationPad2d",
"torch.jit._script._get_overloads",
"torch._C._jit_has_cpp_tests",
"torch.norm",
"torch.testing.assert_allclose",
"torch._C._ivalue_debug_python_object",
"torch.nn.MultiheadAttention",
"torch.autograd.backward",
"torch.einsum",
"torch._C._freeze_module",
"torch.jit.Attribute",
"torch.testing._internal.jit_metaprogramming_utils.script_method_template.format",
"torch.nn.ConstantPad2d",
"torch.testing._internal.common_utils.slowTest",
"torch.tensor",
"torch.nn.LSTMCell",
"torch.mul",
"torch.rand",
"torch.atan",
"torch._C._jit_interpret_graph",
"torch.tensordot",
"torch.LongTensor",
"torch._C._get_graph_executor_optimize",
"torch.zeros_like",
"torch.nn.functional.multi_head_attention_forward",
"torch.is_tensor",
"numpy.int64",
"torch.jit.is_tracing",
"torch.testing._internal.jit_metaprogramming_utils.get_script_args",
"torch.__version__.startswith",
"torch.cuda.device_count",
"torch.stack",
"torch.as_tensor",
"torch.nn.ReflectionPad1d",
"torch.istft",
"torch.testing._internal.common_utils.num_profiled_runs",
"torch.nn.LSTM",
"torch.manual_seed",
"torch.jit.ignore",
"torch.sub",
"torch._C._jit_pass_custom_pattern_based_rewrite_graph",
"numpy.ones",
"torch.onnx._export",
"torch.IntTensor",
"torch.sparse.FloatTensor",
"torch.meshgrid",
"torch.nn.ReLU",
"torch.stft",
"torch.nn.init.uniform_",
"torch.cat",
"torch.load",
"torch.jit._state.enable",
"torch.sparse.mm",
"torch.unique",
"torch.jit.is_scripting",
"torch.nn.functional.interpolate",
"torch.save",
"torch.mm",
"torch.ones",
"torch.jit.trace",
"torch.testing._internal.jit_utils.make_global",
"torch.add",
"torch._C.TensorType.getInferred",
"torch.from_numpy",
"torch.sparse.addmm",
"torch._C._jit_set_texpr_fuser_enabled",
"torch.testing._internal.jit_utils.clear_class_registry",
"torch.relu",
"torch.nn.functional.relu",
"torch.arange",
"torch.jit._state.disable",
"torch.DoubleTensor",
"torch.index_select",
"torch.sparse.sum",
"torch.nn.utils.rnn.pack_sequence",
"torch._C.parse_ir",
"torch.nn.Parameter",
"torch.jit._wait",
"torch.nn.ConvTranspose2d",
"torch.testing._internal.jit_utils.disable_autodiff_subgraph_inlining",
"torch.testing._internal.common_utils.freeze_rng_state",
"torch.nn.ModuleList",
"torch.nn.functional.conv2d",
"torch.unsqueeze",
"torch.testing._internal.common_methods_invocations.create_input",
"torch._jit_internal._disable_emit_hooks",
"torch.nn.Conv3d",
"torch._C._jit_run_cpp_tests",
"torch._C._jit_fuser_get_fused_kernel_code",
"torch.jit.save",
"torch.nn.functional.embedding",
"torch._C.PyTorchFileReader",
"torch._jit_internal.is_optional",
"torch._C._set_graph_executor_optimize",
"torch.jit.last_executed_optimized_graph",
"torch.chunk",
"torch._C._jit_flatten",
"torch.nn.functional.max_pool1d",
"torch.jit.script_if_tracing",
"torch.nn.ConstantPad1d",
"torch.jit.frontend.get_jit_def",
"torch.nn.ReplicationPad3d",
"torch.nn.Transformer",
"torch._C._jit_set_profiling_mode",
"torch._C._jit_pass_remove_dropout",
"torch.nn.modules.utils._quadruple",
"torch.testing.FileCheck",
"torch.tanh",
"torch.jit._get_trace_graph",
"torch.cdist",
"torch.no_grad",
"torch.complex",
"torch.allclose",
"torch.jit.script",
"torch.nn.Dropout",
"torch.nn.modules.utils._single",
"torch.serialization._is_zipfile",
"numpy.arange",
"torch._C.BoolType.get",
"torch.nn.utils.rnn.pack_padded_sequence",
"torch.testing._internal.common_utils.make_tensor",
"torch.testing._internal.common_jit.check_against_reference",
"torch.unique_consecutive",
"torch.get_default_dtype",
"torch.nn.ZeroPad2d",
"torch.nn.functional.linear",
"torch.nn.ConstantPad3d",
"torch.nn.BatchNorm1d",
"torch.sigmoid",
"torch.nn.Sequential",
"torch.testing._internal.jit_utils._trace",
"torch.empty",
"torch.div",
"torch.testing._internal.common_utils.enable_profiling_mode_for_profiling_tests",
"torch.jit.annotate",
"torch.jit.export_opnames",
"torch.lu",
"torch.nn.Conv1d",
"torch._C._propagate_shapes",
"torch.testing._internal.common_methods_invocations.unpack_variables",
"torch.set_anomaly_enabled",
"torch.jit.ScriptModule",
"torch.jit._unique_state_dict",
"torch.testing._internal.jit_metaprogramming_utils.get_nn_module_name_from_kwargs",
"torch.nn.ConvTranspose1d",
"torch.clamp",
"torch.jit._unwrap_optional",
"torch.autograd.profiler.profile",
"torch.testing._internal.test_module.future_div.div_int_future"
]
]
|
panda0881/Selectional_Preference | [
"e48c5ff3feade82fd7f6963bca8135989ce148fe"
]
| [
"Label_data_preparation.py"
]
| [
"from nltk.corpus import wordnet as wn\nfrom nltk.corpus import verbnet\nimport pandas\nimport json\nimport xml.etree.ElementTree as etree\nimport os\nimport random\n\n\ndef filter_word(input_word):\n tmp_output = ''\n for c in input_word:\n if c in 'zxcvbnmasdfghjklqwertyuiopZXCVBNMASDFGHJKLQWERTYUIOP':\n tmp_output += c\n return tmp_output\n\n\nfrequent_verbs_data = pandas.read_csv('verb_data/verb_frequency.csv')\nfrequent_verbs = list()\nfor index, row in frequent_verbs_data.iterrows():\n # test = row['Word']\n test = filter_word(row[1])\n frequent_verbs.append(test)\n # print('lalala')\n# print(frequency_data)\n\nfrequent_words_data = pandas.read_csv('verb_data/word_frequency.csv')\nfrequent_nouns = list()\nfrequent_adjectives = list()\nfor index, row in frequent_words_data.iterrows():\n part_of_speech = row[2]\n if part_of_speech == 'n':\n frequent_nouns.append(filter_word(row[1]))\n elif part_of_speech == 'j':\n frequent_adjectives.append(filter_word(row[1]))\n\n\n\n# test = wn.all_synsets(pos='v')\n# verb = list()\n# for verb_synset in wn.all_synsets(pos='v'):\n# # verb.append(verb_synset)\n# verb.append(verb_synset._name.split('.')[0])\n# print(len(verb))\n# new_verb = set(verb)\n# print(len(new_verb))\n\n# test_example = 'verb_data/new_vn/eat-39.1.xml'\n# category = test_example.split('-')[1].split('.')[0]\nverb_dict = dict()\n\nfor f_name in os.listdir('verb_data/new_vn'):\n file_name = 'verb_data/new_vn/' + f_name\n name_verb = f_name.split('-')[0]\n category = file_name.split('-')[1].split('.')[0]\n if category not in verb_dict:\n verb_dict[category] = list()\n verb_dict[category].append(name_verb)\n with open(file_name, 'r') as f:\n raw_data = f.read()\n tmp = raw_data.split('name=')\n for tmp2 in tmp[1:]:\n verb_dict[category].append(tmp2[1:].split('\"')[0])\n\nselected_all_verbs = list()\nall_verbnet_verbs = list()\nlimitation = 3\nadded_verbs = list()\n\nfor category in verb_dict:\n tmp_verbs = verb_dict[category]\n contained_verbs = list()\n for v in tmp_verbs:\n if v in frequent_verbs:\n contained_verbs.append(v)\n random.shuffle(tmp_verbs)\n contained_verbs = contained_verbs + tmp_verbs\n for v in contained_verbs[:limitation]:\n if v not in frequent_verbs:\n added_verbs.append(v)\n # if len(tmp_verbs) < limitation:\n # for verb in tmp_verbs:\n # selected_all_verbs.append(verb)\n # else:\n # selected_tmp_verbs = list()\n # for v in frequent_verbs:\n # if v in tmp_verbs:\n # selected_tmp_verbs.append(v)\n # if len(selected_tmp_verbs) >= limitation:\n # break\n # if len(selected_tmp_verbs) < limitation:\n # random.shuffle(tmp_verbs)\n # selected_tmp_verbs = selected_tmp_verbs + tmp_verbs\n # selected_tmp_verbs = selected_tmp_verbs[:limitation]\n # for v in selected_tmp_verbs:\n # selected_all_verbs.append(v)\n\nprint(len(added_verbs))\ntest_verbs = frequent_verbs + added_verbs\n# test_verbs = list()\n# for v in frequent_verbs:\n# if v in all_verbnet_verbs:\n# test_verbs.append(v)\n# print(test_verbs)\n\nwith open('selected_verbs.json', 'w') as f:\n json.dump(test_verbs, f)\n\n\nwith open('Wino_verb.json', 'r') as f:\n wino_data = json.load(f)\n#\nprint('We analyzing PDP')\nPDP_data = wino_data['PDP']\nPDP_covered_verbs = list()\nfor verb in PDP_data:\n if verb in test_verbs:\n PDP_covered_verbs.append(verb)\n\nprint('We analyzing Wino')\nWino_data = wino_data['Wino']\nWino_covered_verbs = list()\nfor verb in Wino_data:\n if verb in test_verbs:\n Wino_covered_verbs.append(verb)\n#\nprint(len(PDP_covered_verbs), len(PDP_data), len(PDP_covered_verbs)/len(PDP_data))\nprint(len(Wino_covered_verbs), len(Wino_data), len(Wino_covered_verbs)/len(Wino_data))\n\nprint('end')\n"
]
| [
[
"pandas.read_csv"
]
]
|
Remosy/iceHocekeyIRL | [
"1ffeaf8a9bd9585038629be41a2da552e0a4473b"
]
| [
"Demo/Demo_gym/envs/mujoco/humanoidstandup.py"
]
| [
"from Demo_gym.envs.mujoco import mujoco_env\nfrom Demo_gym import utils\nimport numpy as np\n\nclass HumanoidStandupEnv(mujoco_env.MujocoEnv, utils.EzPickle):\n def __init__(self):\n mujoco_env.MujocoEnv.__init__(self, 'humanoidstandup.xml', 5)\n utils.EzPickle.__init__(self)\n\n def _get_obs(self):\n data = self.sim.data\n return np.concatenate([data.qpos.flat[2:],\n data.qvel.flat,\n data.cinert.flat,\n data.cvel.flat,\n data.qfrc_actuator.flat,\n data.cfrc_ext.flat])\n\n def step(self, a):\n self.do_simulation(a, self.frame_skip)\n pos_after = self.sim.data.qpos[2]\n data = self.sim.data\n uph_cost = (pos_after - 0) / self.model.opt.timestep\n\n quad_ctrl_cost = 0.1 * np.square(data.ctrl).sum()\n quad_impact_cost = .5e-6 * np.square(data.cfrc_ext).sum()\n quad_impact_cost = min(quad_impact_cost, 10)\n reward = uph_cost - quad_ctrl_cost - quad_impact_cost + 1\n\n done = bool(False)\n return self._get_obs(), reward, done, dict(reward_linup=uph_cost, reward_quadctrl=-quad_ctrl_cost, reward_impact=-quad_impact_cost)\n\n def reset_model(self):\n c = 0.01\n self.set_state(\n self.init_qpos + self.np_random.uniform(low=-c, high=c, size=self.model.nq),\n self.init_qvel + self.np_random.uniform(low=-c, high=c, size=self.model.nv,)\n )\n return self._get_obs()\n\n def viewer_setup(self):\n self.viewer.cam.trackbodyid = 1\n self.viewer.cam.distance = self.model.stat.extent * 1.0\n self.viewer.cam.lookat[2] = 0.8925\n self.viewer.cam.elevation = -20\n"
]
| [
[
"numpy.concatenate",
"numpy.square"
]
]
|
matthijsz/etchasketch | [
"336186cb566a8716595b0de45ad9cebc3043092b"
]
| [
"pointerclass.py"
]
| [
"import cv2, random, time, imageio,os, argparse, datetime, multiprocessing, tkinter\r\nimport numpy as np\r\nfrom PIL import Image\r\nfrom scipy.spatial.distance import cdist\r\nimport PIL.Image, PIL.ImageTk\r\nfrom tkinter import filedialog\r\nfrom joblib import Parallel, delayed\r\nfrom my_gui import *\r\nfrom Google_Image import *\r\nfrom pointerclass import *\r\nfrom functions import *\r\nfrom Astar import *\r\n#Find the middle of the image\r\nclass start:\r\n def __init__(self,img,method='topleft'):\r\n if method=='middle':\r\n self.x=int(len(img[0])/2)\r\n self.y=int(len(img)/2)\r\n if method.endswith('left'):\r\n self.x=int(1)\r\n if method.endswith('right'):\r\n self.x=int(len(img[0])-1)\r\n if method.startswith('top'):\r\n self.y=int(1)\r\n if method.endswith('bottom'):\r\n self.y=int(len(img)-1)\r\n\r\n#Define the pointer that moves over\r\n\r\n\r\nclass pointer:\r\n def __init__(self, x_, y_, img, print_process=False, Directory='', Target=''): # Things stored in the pointer:\r\n self.Target = Target\r\n self.Directory = Directory\r\n self.x = x_ # X coordinate\r\n self.y = y_ # Y coordinate\r\n self.history = [] # History as list of coordinates\r\n self.visited = np.zeros(img.shape,dtype=np.uint8) # A 2D-array of cells that are visited by the pointer\r\n self.visited[self.y, self.x] = 1\r\n self.print_process = False\r\n self.imgsum = img.sum()\r\n self.stepcounter=0\r\n self.Graph = Create_graph2(img)\r\n self.r_array = np.zeros(img.shape) + 255 # for black background: self.r_array = img * 255\r\n self.g_array = np.zeros(img.shape) + 255 # for black background: self.r_array = img * 255\r\n self.b_array = np.zeros(img.shape) + 255 # for black background: self.r_array = img * 255\r\n self.CPUtimes = {'moveto': 0, 'move_one_step': 0, 'returnto_fast': 0, 'go_to_next_block': 0, 'finish': 0}\r\n if print_process:\r\n os.makedirs(Directory+'/Steps')\r\n self.print_process = True\r\n\r\n def update(self, x_, y_): # Update pointer position\r\n self.history += [[self.x, self.y]] # Append old position to the history\r\n self.x = x_ # Change current x\r\n self.y = y_ # Change current y\r\n self.visited[self.y, self.x] = 1 # Add current position to visited array\r\n self.stepcounter += 1\r\n if (self.stepcounter % 1000) == 0:\r\n visitsum = self.visited.sum()\r\n print('Filled {0}/{1}px after {2} steps '.format(visitsum, self.imgsum, self.stepcounter),end='\\r')\r\n if self.print_process:\r\n self.r_array[self.y, self.x] = 255\r\n self.g_array[self.y, self.x] = 0\r\n self.b_array[self.y, self.x] = 0\r\n self.r_array[self.history[-1:][0][1], self.history[-1:][0][0]] = 0\r\n self.b_array[self.history[-1:][0][1], self.history[-1:][0][0]] = 255\r\n img_outr = Image.fromarray(np.uint8(self.r_array))\r\n img_outg = Image.fromarray(np.uint8(self.g_array))\r\n img_outb = Image.fromarray(np.uint8(self.b_array))\r\n merged = Image.merge(\"RGB\", (img_outr, img_outg, img_outb))\r\n merged.save('{0}/Steps/step_{1}.png'.format(self.Directory, self.stepcounter))\r\n\r\n def moveto(self,x_,y_): #Move to a coordinate, unrestricted, this will also walk over white cells\r\n substart=time.time()\r\n x_dist = x_-self.x\r\n y_dist = y_-self.y\r\n for n in range(np.max([abs(x_dist), abs(y_dist)])):\r\n if not abs(x_dist) == abs(y_dist):\r\n if abs(x_dist) < abs(y_dist):\r\n self.update(self.x, self.y + 1 * get_sign(y_dist))\r\n y_dist = y_dist + (-1 * get_sign(y_dist))\r\n elif abs(x_dist) > abs(y_dist):\r\n self.update(self.x + 1 * get_sign(x_dist), self.y)\r\n x_dist = x_dist + (-1 * get_sign(x_dist))\r\n else:\r\n self.update(self.x + 1 * get_sign(x_dist), self.y + 1 * get_sign(y_dist))\r\n y_dist = y_dist + (-1 * get_sign(y_dist))\r\n x_dist = x_dist + (-1 * get_sign(x_dist))\r\n self.CPUtimes['moveto'] += time.time() - substart\r\n def move_one_step(self,img): #Move the pointer to the next black cell\r\n substart=time.time()\r\n deadend=False\r\n adjecent_ones = find_adjecent_ones(self.x,self.y,self.history,img)\r\n if len(adjecent_ones['vh']) == 0:\r\n targets = adjecent_ones['diag']\r\n else:\r\n targets = adjecent_ones['vh']\r\n if len(targets) >= 1:\r\n next_cell = targets[0]\r\n elif len(targets)==0:\r\n deadend=True\r\n if not deadend:\r\n self.CPUtimes['move_one_step'] += time.time() - substart\r\n self.update(next_cell[0],next_cell[1])\r\n return True\r\n else:\r\n self.CPUtimes['move_one_step'] += time.time() - substart\r\n return False\r\n def returnto_fast(self,img):\r\n substart=time.time()\r\n Goals = np.transpose(np.array(np.where((img - self.visited) == 1)))\r\n Goals = [(i[1], i[0]) for i in Goals]\r\n Path = bfs_shortest_path_goallist(self.Graph,(self.x,self.y),Goals)\r\n if not Path:\r\n self.CPUtimes['returnto_fast'] += time.time() - substart\r\n return False\r\n else:\r\n for i in Path:\r\n self.update(i[0],i[1])\r\n self.CPUtimes['returnto_fast'] += time.time() - substart\r\n return True\r\n\r\n def find_returnto_targets(self,img): #Find a cell the pointer can go back to that still has open ends\r\n cells_to_returnto=[]\r\n if len(self.returnto) > 0:\r\n tmp_list=self.returnto.copy()\r\n for i in tmp_list:\r\n i_targets=find_adjecent_ones(i[0],i[1],self.history,img)\r\n if (len(i_targets['diag']) != 0) or (len(i_targets['vh']) != 0):\r\n cells_to_returnto+=[i]\r\n if (len(i_targets['diag']) == 0) and (len(i_targets['vh']) == 0):\r\n if i in self.returnto:\r\n self.returnto.remove(i)\r\n if (len(i_targets['diag'])+len(i_targets['vh'])) == 1:\r\n self.returnto.remove(cells_to_returnto[-1:][0])\r\n if (len(cells_to_returnto) > 0):\r\n return cells_to_returnto[-1:][0] #Return the most recently visited one\r\n else:\r\n return False\r\n def go_back_to(self,target,graph):\r\n movesequence,cost=AStarSearch((self.x,self.y),target,graph)\r\n for i in movesequence[1:]:\r\n self.update(i[0],i[1])\r\n def go_to_next_block(self, img):\r\n img2=img\r\n img2[np.where(self.visited==1)]=1\r\n astar_graph = AStarGraph(img2)\r\n substart=time.time()\r\n print('Moving to a different block... Progress:{0}% '.format(round(((self.visited.sum() / img.sum()) * 100), 2)),end='\\r')\r\n visited_coords = np.array(np.where(self.visited == 1))\r\n not_visited_coords = np.array(np.where((img - self.visited) == 1))\r\n chunksize = 10000000\r\n if (len(visited_coords[0]) * len(not_visited_coords[0]) < chunksize):\r\n distance_matrix = cdist(np.transpose(visited_coords), np.transpose(not_visited_coords),metric='chebyshev')\r\n min_distance = distance_matrix.min()\r\n best_path_index = np.array(np.where(distance_matrix == min_distance))[:, 0]\r\n goto_visited = visited_coords[:, best_path_index[0]]\r\n goto_new = not_visited_coords[:, best_path_index[1]]\r\n goto_visited=[goto_visited[1],goto_visited[0]]\r\n goto_new=[goto_new[1],goto_new[0]]\r\n else:\r\n num_cores = multiprocessing.cpu_count() - 1\r\n results = Parallel(n_jobs=num_cores)(\r\n delayed(my_distance)(i, not_visited_coords, visited_coords) for i in range(not_visited_coords.shape[1]))\r\n overall_min = 1e8\r\n n1, v1 = [],[]\r\n for i in results:\r\n if i[0] == 1: #Sometimes the current version forgets to visit some pxs that it actually could visit\r\n v1+=[i[1]] #So save those, and return to them iteratively (using A*)\r\n n1+=[i[2]]\r\n elif i[0] < overall_min:\r\n overall_min = i[0]\r\n goto_visited = i[1]\r\n goto_new = i[2]\r\n if len(n1) > 0:\r\n n1=find_best_order(self.x,self.y,n1)\r\n for i in range(len(n1)):\r\n if self.visited[n1[i][1],n1[i][0]] != 1:\r\n print('Using A* to move back to {0},{1}... '.format(n1[i][0], n1[i][1]),end='\\n')\r\n self.go_back_to(tuple(n1[i]), astar_graph)\r\n print('Using A* to move back to {0},{1}... '.format(goto_visited[0],goto_visited[1]), end='\\n')\r\n self.go_back_to(tuple(goto_visited),astar_graph)\r\n print('Moving to a new block at {0},{1}... '.format(goto_new[0], goto_new[1]), end='\\n')\r\n self.moveto(goto_new[0],goto_new[1])\r\n print('Making new graph...{0} '.format(self.visited.sum()),end='\\n')\r\n self.Graph=Create_graph2(img,self.visited)\r\n self.CPUtimes['go_to_next_block'] += time.time() - substart\r\n\r\n def finish(self,start,img,write_instructions=True): #Wrap up: Return to starting position, and save gif and instructions\r\n substart=time.time()\r\n img2 = img\r\n img2[np.where(self.visited == 1)] = 1\r\n astar_graph = AStarGraph(img2)\r\n print('Using A* to move back to starting position {0},{1} '.format(start.x,start.y),end='\\r')\r\n self.go_back_to((start.x, start.y), astar_graph)\r\n print('Finishing up... The pointer made {0} steps '.format(len(self.history)))\r\n self.history += [[self.x, self.y]]\r\n self.r_array[np.where(self.visited==1)] = 0\r\n self.g_array[np.where(self.visited == 1)] = 0\r\n self.b_array[np.where(self.visited == 1)] = 0\r\n img_outr = Image.fromarray(np.uint8(self.r_array))\r\n img_outg = Image.fromarray(np.uint8(self.g_array))\r\n img_outb = Image.fromarray(np.uint8(self.b_array))\r\n merged = Image.merge(\"RGB\", (img_outr, img_outg, img_outb))\r\n merged.save('{0}_finalstep.png'.format(self.Target))\r\n if write_instructions:\r\n last_coord = self.history[0]\r\n with open(self.Target+'.instructions', 'w') as f:\r\n for i in range(1, len(self.history)):\r\n instruction_1 = [self.history[i][n] - last_coord[n] for n in range(2)]\r\n f.write('{0},{1}\\n'.format(instruction_1[0], instruction_1[1]))\r\n last_coord = self.history[i]\r\n self.CPUtimes['finish'] += time.time() - substart\r\n print('CPU time per classfunction:')\r\n for lab,s in self.CPUtimes.items():\r\n print(lab+':'+' '*(20-(len(lab)))+'{0}'.format(str(datetime.timedelta(seconds=s))))\r\n \r\n\r\n\r\n\r\n\r\n\r\n\r\n"
]
| [
[
"numpy.uint8",
"numpy.zeros",
"numpy.where",
"numpy.transpose"
]
]
|
AntoineGuillot2/RLFirmStrategy | [
"1ab7b59ccad2bbcad641f83dbad552e39b0f1591"
]
| [
"CNN_market_reaction/Firm.py"
]
| [
"#!/usr/bin/env python3\n# -*- coding: utf-8 -*-\n\"\"\"\nCreated on Tue Jan 23 10:47:13 2018\n\n@author: antoine\n\"\"\"\nimport numpy as np\nfrom keras.models import Model\nfrom keras.layers import Input,Dense, Conv1D, MaxPooling1D, Concatenate, Flatten\n\n\nclass Firm:\n def __init__(self,params,observation_size=12):\n print('NEW INIT')\n \n \n \n ###Initial firm parameters\n self.initial_funds=params['initial_funds']\n self.funds=self.initial_funds\n self.initial_inventory=params['initial_inventory']\n self.inventory=params['initial_inventory']\n self.max_inventory=params['max_inventory']\n self.production_queue=[0]*params['production_time']\n self.current_reward=0.\n self.WACC=params['WACC']\n self.cost=params['cost']\n self.production_time=params['production_time']\n self.epsilon_greedy=params['epsilon_greedy']\n ##Set explore rate evolution\n self.explore_rate=params['initial_explore_rate']\n self.explore_rate_decay=params['explore_rate_decay']\n self.min_explore_rate=params['min_explore_rate']\n self.explore_turns=params['explore_turns']\n self.temperature=10\n \n self.bankrupt=False\n \n ##Scaling parameters:\n self.mean_action=0\n self.std_action=1\n self.mean_obs=0\n self.std_obs=1\n self.mean_Q=0\n self.std_Q=1\n \n \n self.plot_frequency=params['plot_frequency']\n self.possible_actions=params['possible_actions']\n self.last_action=self.possible_actions[0]\n self.env_obs_size=observation_size\n self.memory_size=params['memory_size']\n self.init_event_memory()\n self.init_Q_estimator(observation_size)\n \n self.epochs=params['epochs']\n \n self.replay_memory_size=params['replay_memory_size']\n self.init_replay_memory()\n\n self.n_step=0\n self.played_games=0\n \n \n self.list_rewards=[]\n self.funds_evolution=[]\n self.list_actions=[]\n\n def get_state(self):\n return np.array([(self.funds,self.current_reward,self.inventory,*self.last_action)])\n \n \n def init_Q_estimator(self,n_input):\n self.Q_estimator_shape=(n_input,self.possible_actions.shape[1],1)\n input_actions=Input(shape=(self.possible_actions.shape[1],))\n x_actions = Dense(10,activation='relu')(input_actions)\n input_data = Input(shape=(self.memory_size,n_input+self.get_state().shape[1],))\n x_data = Conv1D(10,3,activation='relu')(input_data)\n x_data = Conv1D(10,3,activation='relu')(x_data)\n x_data = MaxPooling1D(3)(x_data)\n x_data=Flatten()(x_data)\n x=Concatenate()([x_data,x_actions])\n x=Dense(10,activation='relu')(x)\n estimated_Q = Dense(1,activation='linear')(x)\n self.Q_estimator = Model(inputs=[input_actions,input_data], outputs=estimated_Q)\n self.Q_estimator.compile(optimizer='rmsprop',\n loss='mse')\n \n def estimate_Q(self,actions,observations,rescale=True):\n actions=(actions-self.mean_action)/self.std_action\n observations=(observations-self.mean_obs)/self.std_obs\n return self.Q_estimator.predict([actions,observations])[:,0]\n \n def fit_Q(self,rescale=True):\n if rescale:\n action_memory=(self.action_memory-self.mean_action)/self.std_action\n observation_memory=(self.observation_memory-self.mean_obs)/self.std_obs\n Q_memory=(self.Q_memory-self.mean_Q)/self.std_Q\n self.Q_estimator.fit([action_memory,observation_memory],Q_memory,epochs=self.epochs,verbose=0)\n \n \n\n ###Initialize the replay memory\n def init_replay_memory(self):\n self.action_memory=np.zeros((0,self.Q_estimator_shape[1]))\n self.observation_memory=np.zeros((0,self.event_memory.shape[1],self.event_memory.shape[2]))\n self.Q_memory=np.zeros((0,self.Q_estimator_shape[2]))\n \n def update_replay_memory(self,action,state_observation,Q_value):\n self.observation_memory=np.concatenate((self.observation_memory,state_observation),0)\n self.action_memory=np.concatenate((self.action_memory,action),0)\n Q_value=np.array([(Q_value,)])\n self.Q_memory=np.concatenate((self.Q_memory,Q_value),0)\n if np.shape(self.action_memory)[0]>self.replay_memory_size:\n self.observation_memory=self.observation_memory[1:]\n self.action_memory=self.action_memory[1:]\n self.Q_memory=self.Q_memory[1:]\n \n def update_scaling(self):\n self.mean_obs=0.5*self.mean_obs+0.5*np.mean(self.observation_memory,0)\n self.std_obs=0.5*self.std_obs+0.5*np.std(self.observation_memory,0)\n \n self.mean_action=0.5*self.mean_action+0.5*np.mean(self.action_memory,0)\n self.std_action=0.5*self.std_action+0.5*np.std(self.action_memory,0)\n \n self.std_Q=0.5*self.std_Q+np.std(self.Q_memory,0)*0.5\n self.mean_Q=0.5*self.mean_Q+np.mean(self.Q_memory,0)*0.5\n \n def init_event_memory(self):\n self.event_memory=np.zeros((1,self.memory_size,self.env_obs_size+self.get_state().shape[1]))\n \n def update_event_memory(self,new_observation):\n new_event_memory=np.concatenate((new_observation.reshape((1,1,-1)),self.event_memory),1)\n self.event_memory=new_event_memory[:,:self.memory_size,:]\n \n def reset(self):\n self.bankrupt=False\n self.update_scaling()\n if self.explore_rate>self.min_explore_rate:\n self.explore_rate*=self.explore_rate_decay\n if self.temperature>0.1:\n self.temperature*=self.explore_rate_decay\n self.fit_Q()\n \n self.played_games+=1\n self.n_step=0\n print(self.played_games)\n print(self.funds)\n self.rewards=0\n self.inventory=self.initial_inventory\n self.funds=self.initial_funds\n self.init_event_memory()\n \n def compute_best_action(self,observation):\n observation=np.repeat(observation,len(self.possible_actions),0)\n values=self.estimate_Q(self.possible_actions,observation)\n return self.possible_actions[np.argmax(values)], np.max(values)\n \n def compute_action_values(self,observation):\n observation=np.repeat(observation,len(self.possible_actions),0)\n values=self.estimate_Q(self.possible_actions,observation)\n return values\n\n def act(self, observation):\n if self.funds<0:\n self.bankrupt=True\n print('Firm bankruptcy')\n self.n_step+=1\n state_observation=np.concatenate((self.get_state(),observation),1)\n self.previous_observation=state_observation\n self.update_event_memory(state_observation)\n if self.epsilon_greedy:\n if (self.played_games<self.explore_turns):\n if np.random.uniform()<self.explore_rate:\n random_action=np.random.randint(len(self.possible_actions))\n action = self.possible_actions[random_action]\n else:\n action = self.compute_best_action(self.event_memory)[0]\n else:\n action = self.compute_best_action(self.event_memory)[0]\n else:\n values=np.exp(self.compute_action_values(self.event_memory)/self.temperature)\n values=values/np.sum(values)\n action = self.possible_actions[np.random.choice(values.shape[0],p=values)]\n \n self.last_action=action\n return action\n \n def reward(self, observation, action, reward):\n self.current_reward=reward\n if self.funds<0:\n target=-1000\n\n else:\n target=reward+(1-self.WACC)*self.compute_best_action(self.event_memory)[1]\n self.update_replay_memory(action.reshape(1,-1),self.event_memory,target)\n \n def update_production_queue(self,production):\n self.production_queue=np.concatenate((self.production_queue,[production]))\n \n def produce(self):\n self.inventory+=self.production_queue[0]\n if self.inventory>=self.max_inventory:\n self.inventory=self.max_inventory\n self.production_queue=self.production_queue[1:]\n \n def get_sales(self,purchase):\n if self.inventory-purchase>=0:\n self.inventory=self.inventory-purchase\n return purchase\n else:\n self.inventory=0\n return self.inventory\n"
]
| [
[
"numpy.random.choice",
"numpy.concatenate",
"numpy.max",
"numpy.std",
"numpy.argmax",
"numpy.shape",
"numpy.mean",
"numpy.random.uniform",
"numpy.array",
"numpy.zeros",
"numpy.sum"
]
]
|
mjarrett/bikeraccoonAPI | [
"c3b15e63c4e6f32eaa6464ef86d4399733dd4919"
]
| [
"bikeraccoonAPI/query_functions.py"
]
| [
"import pandas as pd\nimport json\nimport requests\nimport datetime as dt\nimport timeout_decorator\nimport ssl\n\nimport logging\nlogger = logging.getLogger(\"Rotating Log\")\n\ndef get_station_status_url(sys_url):\n data = requests.get(sys_url).json()\n return [x for x in data['data']['en']['feeds'] if x['name']=='station_status'][0]['url'] \n\ndef get_station_info_url(sys_url):\n data = requests.get(sys_url).json()\n return [x for x in data['data']['en']['feeds'] if x['name']=='station_information'][0]['url'] \n\n\ndef get_system_info_url(sys_url):\n data = requests.get(sys_url).json()\n return [x for x in data['data']['en']['feeds'] if x['name']=='system_information'][0]['url']\n\n@timeout_decorator.timeout(30) \ndef query_system_info(sys_url):\n url = get_system_info_url(sys_url)\n\n data = requests.get(url).json()\n\n return data\n\n \n@timeout_decorator.timeout(30) \ndef query_station_status(sys_url):\n \"\"\"\n Query station_status.json\n \"\"\"\n \n url = get_station_status_url(sys_url)\n\n\n data = requests.get(url).json()\n\n try:\n df = pd.DataFrame(data['data']['stations'])\n except KeyError:\n df = pd.DataFrame(data['stations'])\n \n df = df.drop_duplicates(['station_id','last_reported'])\n try:\n df['datetime'] = data['last_updated']\n df['datetime'] = df['datetime'].map(lambda x: dt.datetime.utcfromtimestamp(x))\n except KeyError:\n df['datetime'] = dt.datetime.utcnow()\n \n df['datetime'] = df['datetime'].dt.tz_localize('UTC')\n \n df = df[['datetime','num_bikes_available','num_docks_available','is_renting','station_id']]\n\n\n return df\n\n@timeout_decorator.timeout(30) \ndef query_station_info(sys_url):\n \n \"\"\"\n Query station_information.json\n \"\"\"\n url = get_station_info_url(sys_url)\n\n data = requests.get(url).json()\n\n try:\n df = pd.DataFrame(data['data']['stations'])\n except KeyError:\n df = pd.DataFrame(data['stations'])\n return df[['name','station_id','lat','lon']]\n\n@timeout_decorator.timeout(30) \ndef query_free_bikes(sys_url):\n \n \"\"\"\n Query free_bikes.json\n \"\"\"\n \n url = get_free_bike_url(sys_url)\n\n data = requests.get(url).json()\n\n try: \n df = pd.DataFrame(data['data']['bikes'])\n except KeyError:\n df = pd.DataFrame(data['bikes'])\n \n df['bike_id'] = df['bike_id'].astype(str)\n\n try:\n df['datetime'] = data['last_updated']\n df['datetime'] = df['datetime'].map(lambda x: dt.datetime.utcfromtimestamp(x))\n except KeyError:\n df['datetime'] = dt.datetime.utcnow()\n \n df['datetime'] = df['datetime'].dt.tz_localize('UTC')\n \n \n df = df[['bike_id','lat','lon','datetime']]\n\n return df\n\n \ndef get_free_bike_url(sys_url):\n data = requests.get(sys_url).json()\n return [x for x in data['data']['en']['feeds'] if x['name']=='free_bike_status'][0]['url']"
]
| [
[
"pandas.DataFrame"
]
]
|
Ali1999-AK/heart-prediction-ML-and-Flask | [
"d14e65cd918c8da0b6a2b35594c6708fd0e3ef19"
]
| [
"Heart_D/heart.py"
]
| [
"import numpy as np\r\nimport pandas as pd\r\nfrom sklearn import *\r\nfrom sklearn.model_selection import train_test_split\r\nfrom sklearn.ensemble import RandomForestClassifier\r\nfrom sklearn.svm import SVC\r\nimport warnings\r\nimport pickle\r\nwarnings.filterwarnings(\"ignore\")\r\n\r\ndata = pd.read_csv('heart.csv')\r\nX = data[[\"thalachh\",\"oldpeak\",\"caa\",\"cp\",\"exng\",\"chol\",\"age\",\"trtbps\",\"slp\",\"sex\"]]\r\nX = np.array(X)\r\nY = data['output']\r\nY = np.array(Y)\r\nX_train,X_test,y_train,y_test = train_test_split(X,Y,test_size = 0.2,random_state = 0)\r\nclf = RandomForestClassifier(n_estimators=168,criterion='gini',max_depth=6,max_features=2,min_samples_leaf=8).fit(X_train,y_train)\r\npickle.dump(clf,open('model.pkl','wb'))\r\nmodel=pickle.load(open('model.pkl','rb'))\r\n\r\n"
]
| [
[
"numpy.array",
"pandas.read_csv",
"sklearn.model_selection.train_test_split",
"sklearn.ensemble.RandomForestClassifier"
]
]
|
tylerbinski/recordlinkage | [
"5b3230f5cff92ef58968eedc451735e972035793"
]
| [
"recordlinkage/algorithms/indexing.py"
]
| [
"\"\"\"Algorithms for indexing.\"\"\"\n\nimport numpy as np\n\nfrom recordlinkage.measures import full_index_size\n\n\ndef _map_tril_1d_on_2d(indices, dims):\n \"\"\"Map 1d indices on lower triangular matrix in 2d. \"\"\"\n\n N = (dims * dims - dims) / 2\n\n m = np.ceil(np.sqrt(2 * N))\n c = m - np.round(np.sqrt(2 * (N - indices))) - 1\n r = np.mod(indices + (c + 1) * (c + 2) / 2 - 1, m) + 1\n\n return np.array([r, c], dtype=np.int64)\n\n\ndef random_pairs_with_replacement(n, shape, random_state=None):\n \"\"\"make random record pairs\"\"\"\n\n if not isinstance(random_state, np.random.RandomState):\n random_state = np.random.RandomState(random_state)\n\n n_max = full_index_size(shape)\n\n if n_max <= 0:\n raise ValueError('n_max must be larger than 0')\n\n # make random pairs\n indices = random_state.randint(0, n_max, n)\n\n if len(shape) == 1:\n return _map_tril_1d_on_2d(indices, shape[0])\n else:\n return np.array(np.unravel_index(indices, shape))\n\n\ndef random_pairs_without_replacement(\n n, shape, random_state=None):\n \"\"\"Return record pairs for dense sample.\n\n Sample random record pairs without replacement bounded by the\n maximum number of record pairs (based on shape). This algorithm is\n efficient and fast for relative small samples.\n \"\"\"\n\n n_max = full_index_size(shape)\n\n if not isinstance(random_state, np.random.RandomState):\n random_state = np.random.RandomState(random_state)\n\n if not isinstance(n, int) or n <= 0 or n > n_max:\n raise ValueError(\"n must be a integer satisfying 0<n<=%s\" % n_max)\n\n # make a sample without replacement\n sample = random_state.choice(\n np.arange(n_max), n, replace=False)\n\n # return 2d indices\n if len(shape) == 1:\n return _map_tril_1d_on_2d(sample, shape[0])\n else:\n return np.array(np.unravel_index(sample, shape))\n\n\ndef random_pairs_without_replacement_low_memory(\n n, shape, random_state=None):\n \"\"\"Make a sample of random pairs with replacement.\n\n Sample random record pairs without replacement bounded by the\n maximum number of record pairs (based on shape). This algorithm\n consumes low memory and is fast for relatively small samples.\n \"\"\"\n\n n_max = full_index_size(shape)\n\n if not isinstance(random_state, np.random.RandomState):\n random_state = np.random.RandomState(random_state)\n\n if not isinstance(n, int) or n <= 0 or n > n_max:\n raise ValueError(\"n must be a integer satisfying 0<n<=%s\" % n_max)\n\n sample = np.array([], dtype=np.int64)\n\n # Run as long as the number of pairs is less than the requested number\n # of pairs n.\n while len(sample) < n:\n\n # The number of pairs to sample (sample twice as much record pairs\n # because the duplicates are dropped).\n n_sample_size = (n - len(sample)) * 2\n sample_sub = random_state.randint(\n n_max, \n size=n_sample_size\n )\n\n # concatenate pairs and deduplicate\n pairs_non_unique = np.append(sample, sample_sub)\n sample = np.unique(pairs_non_unique)\n\n # return 2d indices\n if len(shape) == 1:\n return _map_tril_1d_on_2d(sample[0:n], shape[0])\n else:\n return np.array(np.unravel_index(sample[0:n], shape))\n"
]
| [
[
"numpy.sqrt",
"numpy.unique",
"numpy.arange",
"numpy.append",
"numpy.mod",
"numpy.array",
"numpy.unravel_index",
"numpy.random.RandomState"
]
]
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.