markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
As a rule of thumbRemember that the pooling operation decreases the size of the image, and we lose information.However, the number of features generally increases and we get more features extracted from the images.The choices of hyperparams bother us sometimes, because DL has a lot of trial and error involved, we can choose the - learning rate- number of layers- number of neurons per layer - feature size - feature number - pooling size - stride On a side note, if you use strided convolution layers, they will decrease the size of the image as wellIf we have images with different sizes as inputs; for example; H1 x W1 x 3 and H2 x W2 x 3, then the output will be flatten-ed to different sizes, this won't work for DENSE layers as they do not have change-able input sizes, so we use global max pooling to make a vector of size 1 x 1 x (_Of_Feature_Maps_)
|
from tensorflow.keras.layers import Input, Conv2D, Dropout, Dense, Flatten, BatchNormalization, MaxPooling2D
from tensorflow.keras.models import Model
from tensorflow.keras.datasets import cifar10
(X_train, y_train), (X_test, y_test) = cifar10.load_data()
X_train, X_test = X_train / 255.0 , X_test / 255.0
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
y_train, y_test = y_train.flatten(), y_test.flatten()
print(y_train.shape)
print(y_test.shape)
classes = len(set(y_train))
print(classes)
input_shape = X_train[0].shape
i_layer = Input(shape = input_shape)
h_layer = Conv2D(32, (3,3),activation='relu', padding='same')(i_layer)
h_layer = BatchNormalization()(h_layer)
h_layer = Conv2D(64, (3,3), activation='relu', padding='same')(h_layer)
h_layer = BatchNormalization()(h_layer)
h_layer = Conv2D(128, (3,3), activation='relu', padding='same')(h_layer)
h_layer = BatchNormalization()(h_layer)
h_layer = MaxPooling2D((2,2))(h_layer)
h_layer = Conv2D(128, (3,3), activation='relu', padding='same')(h_layer)
h_layer = BatchNormalization()(h_layer)
h_layer = MaxPooling2D((2,2))(h_layer)
h_layer = Flatten()(h_layer)
h_layer = Dropout(0.5)(h_layer)
h_layer = Dense(512, activation='relu')(h_layer)
h_layer = Dropout(0.5)(h_layer)
o_layer = Dense(classes, activation='softmax')(h_layer)
model = Model(i_layer, o_layer)
model.compile(optimizer='adam',
loss = 'sparse_categorical_crossentropy',
metrics = ['accuracy'])
report = model.fit(X_train, y_train, validation_data=(X_test, y_test), epochs=50)
y_pred = model.predict(X_test).argmax(axis=1)
# only for sparse categorical crossentropy
evaluation_tf(report, y_test, y_pred, classes)
misshits = np.where(y_pred!=y_test)[0]
print("total Mishits = " + str(len(misshits)))
index = np.random.choice(misshits)
plt.imshow(X_test[index])
plt.title("Predicted = " + str(labels[y_pred[index]]) + ", Real = " + str(labels[y_test[index]]))
|
total Mishits = 1715
|
MIT
|
Tensorflow_2X_Notebooks/Demo26_CNN_CIFAR10_DataAugmentation.ipynb
|
mahnooranjum/Tensorflow_DeepLearning
|
LET'S ADD SOME DATA AUGMENTATION FROM KERAS taken from https://keras.io/api/preprocessing/image/
|
batch_size = 32
data_generator = tf.keras.preprocessing.image.ImageDataGenerator(width_shift_range = 0.1,
height_shift_range = 0.1,
horizontal_flip=True)
model_dg = Model(i_layer, o_layer)
model_dg.compile(optimizer='adam',
loss = 'sparse_categorical_crossentropy',
metrics = ['accuracy'])
train_data_generator = data_generator.flow(X_train, y_train, batch_size)
spe = X_train.shape[0] // batch_size
report = model_dg.fit_generator(train_data_generator, validation_data=(X_test, y_test), steps_per_epoch=spe, epochs=50)
y_pred = model.predict(X_test).argmax(axis=1)
# only for sparse categorical crossentropy
evaluation_tf(report, y_test, y_pred, classes)
misshits = np.where(y_pred!=y_test)[0]
print("total Mishits = " + str(len(misshits)))
index = np.random.choice(misshits)
plt.imshow(X_test[index])
plt.title("Predicted = " + str(labels[y_pred[index]]) + ", Real = " + str(labels[y_test[index]]))
|
total Mishits = 1193
|
MIT
|
Tensorflow_2X_Notebooks/Demo26_CNN_CIFAR10_DataAugmentation.ipynb
|
mahnooranjum/Tensorflow_DeepLearning
|
Google Apps Workspace Imports
|
%matplotlib inline
import os
import pandas as pd
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Load Dataset
|
apps_df = pd.read_csv('googleplaystore.csv', index_col = 0)
reviews_df = pd.read_csv('googleplaystore_user_reviews.csv', index_col = 0)
apps_df.head()
apps_df.shape
apps_df.describe()
reviews_df.head()
reviews_df.shape
reviews_df.describe()
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
**Remove empty reviews**
|
reviews_df = reviews_df.dropna(axis=0, how='all')
apps_reviews_df = pd.merge(apps_df, reviews_df, on='App', how='inner')
apps_reviews_df.head()
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Remove 1.9 Category \*Because it doesn't make sense*
|
apps_df = apps_df[apps_df['Category'] != '1.9']
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Change underscores to spaces
|
apps_df['Category'] = apps_df['Category'].str.replace('_', ' ')
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Categories
|
categories = apps_df['Category'].unique()
categories
apps_df['Reviews'] = pd.to_numeric(apps_df['Reviews'])
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Remove dollar signs
|
apps_df['Price'] = pd.to_numeric(apps_df['Price'].str.replace('$', ''))
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Standardize App size to MB
|
# apps_df['Size'] = pd.to_numeric(apps_df['Size'].str.replace('M', ''))
def convert_to_M(s):
if 'k' in s:
return str(float(s[:-1])/1000)
if 'M' in s:
return s[:-1]
return np.nan
apps_df['Size'] = apps_df['Size'].apply(convert_to_M)
apps_df['Size'] = pd.to_numeric(apps_df['Size'])
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Fill varying app sizes to the average app size of all the apps
|
apps_df['Size'] = apps_df['Size'].fillna(apps_df['Size'].mean())
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Insights Top Apps per CategoryOnly taking into account those with reviews greater than the median
|
n = 3
temp_apps_df = apps_df.reset_index()
print("Median Ratings: %.0f" % temp_apps_df['Reviews'].median())
temp_apps_df[temp_apps_df['Reviews'] > temp_apps_df['Reviews'].median()].sort_values('Rating', ascending=False).groupby('Category').head(n).reset_index(drop=True).sort_values("Category").set_index("App")
|
Median Ratings: 2094
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Free vs Paid
|
apps_df.groupby('Type').agg('size').plot.bar()
sns.jointplot(apps_df['Price'], apps_df['Rating'])
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
App Size (in MB) vs Rating
|
sns.jointplot(apps_df['Size'], apps_df['Rating'])
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Distribution of Apps per PriceIf it's not free, it's an outlier.
|
plt.figure(figsize=(18,6))
ax = sns.boxplot(x='Category', y='Price', data=apps_df, orient='v')
ax.set_xticklabels(ax.get_xticklabels(),rotation=60)
plt.show()
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Most Expensive AppsPossibly implies that you can't price an app above 400$ in Google App Store
|
apps_df.sort_values('Price', ascending=False)[['Category', 'Price', 'Installs']].head(15)
|
_____no_output_____
|
CC-BY-3.0
|
.ipynb_checkpoints/Google Apps Workspace-checkpoint.ipynb
|
henrylaynesa/kaggle-google-apps
|
Geolocalizacion de dataset de escuelas argentinas
|
#Importar librerias
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
|
_____no_output_____
|
MIT
|
source/codes/2_Geolocalizacion.ipynb
|
matuteiglesias/tutorial-datos-argentinos
|
Preparacion de data
|
# Vamos a cargar un padron de escuelas de Argentina
# Estos son los nombres de columna
cols = ['Jurisdicción','CUE Anexo','Nombre','Sector','Estado','Ámbito','Domicilio','CP','Teléfono','Código Localidad','Localidad','Departamento','E-mail','Ed. Común','Ed. Especial','Ed. de Jóvenes y Adultos','Ed. Artística','Ed. Hospitalaria Domiciliaria','Ed. Intercultural Bilingüe','Ed. Contexto de Encierro','Jardín maternal','Jardín de infantes','Primaria','Secundaria','Secundaria Técnica (INET)','Superior no Universitario','Superior No Universitario (INET)']
# Leer csv, remplazar las 'X' con True y los '' (NaN) con False
escuelas = pd.read_csv('../../datos/escuelas_arg.csv', names=cols).fillna(False).replace('X', True)
# Construir la columna 'dpto_link' con los codigos indetificatorios de partidos como los que teniamos
escuelas['dpto_link'] = escuelas['C\xc3\xb3digo Localidad'].astype(str).str.zfill(8).str[:5]
# Tenemos los radios censales del AMBA, que creamos en el notebook anterior. Creemos los 'dpto_link' del AMBA.
radios_censales_AMBA = pd.read_csv('../../datos/AMBA_datos', dtype=object)
dpto_links_AMBA = (radios_censales_AMBA['prov'] + radios_censales_AMBA['depto']).unique()
# Filtramos las escuelas AMBA
escuelas_AMBA = escuelas.loc[escuelas['dpto_link'].isin(dpto_links_AMBA)]
escuelas_AMBA = pd.concat([escuelas_AMBA, escuelas.loc[escuelas['Jurisdicci\xc3\xb3n'] == 'Ciudad de Buenos Aires']])
# Filtramos secundaria estatal
escuelas_AMBA_secundaria_estatal = escuelas_AMBA.loc[escuelas_AMBA['Secundaria'] & (escuelas_AMBA[u'Sector'] == 'Estatal')]
escuelas_AMBA_secundaria_estatal.reset_index(inplace=True, drop=True)
|
_____no_output_____
|
MIT
|
source/codes/2_Geolocalizacion.ipynb
|
matuteiglesias/tutorial-datos-argentinos
|
Columnas de 'Address'
|
# Creamos un campo que llamamos 'Address', uniendo domicilio, localidad, departamento, jurisdiccion, y ', Argentina'
escuelas_AMBA_secundaria_estatal['Address'] = \
escuelas_AMBA_secundaria_estatal['Domicilio'].astype(str) + ', ' + \
escuelas_AMBA_secundaria_estatal['Localidad'].astype(str) + ', ' + \
escuelas_AMBA_secundaria_estatal['Departamento'].astype(str) + ', ' + \
escuelas_AMBA_secundaria_estatal['Jurisdicci\xc3\xb3n'].astype(str) +', Argentina'
pd.set_option('display.max_colwidth', -1)
import re
def filtrar_entre_calles(string):
"""
Removes substring between 'E/' and next field (delimited by ','). Case insensitive.
example:
>>> out = filtrar_entre_calles('LASCANO E/ ROMA E ISLAS MALVINAS 6213, ISIDRO CASANOVA')
>>> print out
'LASCANO 6213, ISIDRO CASANOVA'
"""
s = string.lower()
try:
m = re.search("\d", s)
start = s.index( 'e/' )
# end = s.index( last, start )
end = m.start()
return string[:start] + string[end:]
except:
return string
def filtrar_barrio(string, n = 3):
"""
Leaves only n most aggregate fields and the address.
example:
>>> out = filtrar_entre_calles('LASCANO 6213, ISIDRO CASANOVA, LA MATANZA, Buenos Aires, Argentina')
>>> print out
'LASCANO 6213, LA MATANZA, Buenos Aires, Argentina'
"""
try:
coma_partido_jurisdiccion = [m.start() for m in re.finditer(',', string)][-n]
coma_direccion = [m.start() for m in re.finditer(',', string)][0]
s = string[:coma_direccion][::-1]
if "n/s" in s.lower():
start = s.lower().index('n/s')
cut = len(s) - len('n/s') - start
else:
m = re.search("\d", s)
cut = len(s) - m.start(0)
return string[:cut] + string[coma_partido_jurisdiccion:]
except AttributeError:
return string
escuelas_AMBA_secundaria_estatal['Address_2'] = escuelas_AMBA_secundaria_estatal['Address'].apply(filtrar_entre_calles)
escuelas_AMBA_secundaria_estatal['Address_3'] = escuelas_AMBA_secundaria_estatal['Address_2'].apply(filtrar_barrio)
escuelas_AMBA_secundaria_estatal.to_csv('../../datos/escuelas_AMBA_secundaria_estatal.csv', index = False)
|
_____no_output_____
|
MIT
|
source/codes/2_Geolocalizacion.ipynb
|
matuteiglesias/tutorial-datos-argentinos
|
Geolocalizacion
|
import json
import time
import urllib
import urllib2
def geolocate(inp, API_key = None, BACKOFF_TIME = 30):
# See https://developers.google.com/maps/documentation/timezone/get-api-key
# with open('googleMapsAPIkey.txt', 'r') as myfile:
# maps_key = myfile.read().replace('\n', '')
base_url = 'https://maps.googleapis.com/maps/api/geocode/json'
# This joins the parts of the URL together into one string.
url = base_url + '?' + urllib.urlencode({
'address': "%s" % (inp),
'key': API_key,
})
try:
# Get the API response.
response = str(urllib2.urlopen(url).read())
except IOError:
pass # Fall through to the retry loop.
else:
# If we didn't get an IOError then parse the result.
result = json.loads(response.replace('\\n', ''))
if result['status'] == 'OK':
return result['results'][0]
elif result['status'] != 'UNKNOWN_ERROR':
# Many API errors cannot be fixed by a retry, e.g. INVALID_REQUEST or
# ZERO_RESULTS. There is no point retrying these requests.
# raise Exception(result['error_message'])
return None
# If we're over the API limit, backoff for a while and try again later.
elif result['status'] == 'OVER_QUERY_LIMIT':
print "Hit Query Limit! Backing off for "+str(BACKOFF_TIME)+" minutes..."
time.sleep(BACKOFF_TIME * 60) # sleep for 30 minutes
geocoded = False
def set_geolocation_values(df, loc):
df.set_value(i,'lng', loc['geometry']['location']['lng'])
df.set_value(i,'lat', loc['geometry']['location']['lat'])
df.set_value(i, 'id', loc['place_id'])
dataframe = escuelas_AMBA_secundaria_estatal
col, col_2, col_3 = 'Address', 'Address_2', 'Address_3'
API_key = 'AIzaSyDjBFMZlNTyds2Sfihu2D5LTKupKDBpf6c'
for i, row in dataframe.iterrows():
loc = geolocate(row[col], API_key)
if loc:
set_geolocation_values(dataframe, loc)
else:
loc = geolocate(row[col_2], API_key)
if loc:
set_geolocation_values(dataframe, loc)
else:
loc = geolocate(row[col_3], API_key)
if loc:
set_geolocation_values(dataframe, loc)
if i%50 == 0:
print 'processed row '+str(i)
dataframe.to_csv('../../datos/esc_sec_AMBA_geoloc.csv', index = False, encoding = 'utf8')
# esc_sec_AMBA_geoloc_1200 = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_1200.csv', encoding = 'utf8')
# esc_sec_AMBA_geoloc_480_1200 = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_480_1200.csv', encoding = 'utf8')
# esc_sec_AMBA_geoloc = pd.read_csv('../../datos/esc_sec_AMBA_geoloc.csv', encoding = 'utf8')
# esc_sec_AMBA_geoloc_900_1200 = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_900_1200.csv', encoding = 'utf8')
# pd.concat([esc_sec_AMBA_geoloc[:480],esc_sec_AMBA_geoloc_480_1200[:420],esc_sec_AMBA_geoloc_900_1200, esc_sec_AMBA_geoloc_1200]).to_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', index = False, encoding = 'utf8')
print len(pd.read_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', encoding = 'utf8').dropna())
print len(pd.read_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', encoding = 'utf8'))
1840/2066.
import numpy as np
df = pd.read_csv('../../datos/esc_sec_AMBA_geoloc_full.csv', encoding = 'utf8')
index = df['lat'].index[df['lat'].apply(np.isnan)]
plt.hist(index, 100)
# plt.xlim(900, 1300)
plt.show()
df.iloc[np.where(pd.isnull(df['lat']))][['Nombre','Address', 'Address_2', 'Address_3']].to_csv('../../datos/no_result_addresses.csv', index = False, encoding = 'utf8')
|
_____no_output_____
|
MIT
|
source/codes/2_Geolocalizacion.ipynb
|
matuteiglesias/tutorial-datos-argentinos
|
Now You Code 1: Hello 2 WaysWrite a Python program which prompts you to input your first name and then your last name. It should then print your name two ways First Last and Last, First. For example:```What is your first name? MichaelWhat is your last name? FudgeHello, Michael FudgeOr should I say Fudge, Michael``` Step 1: Problem AnalysisInputs:first namelast nameOutputs:Hello, first name, last nameOr should I say last name first nameAlgorithm (Steps in Program):input first name input last name print line one with embedded variables print line two with embedded variables
|
first_name = input('What is your first name? ')
last_name = input('What is your last name? ')
print('Hello,' , first_name , last_name)
print('Or should I say' , last_name , first_name)
|
What is your first name? Puzzanghera
What is your last name? Hope
Hello, Puzzanghera Hope
Or should I say Hope Puzzanghera
|
MIT
|
content/lessons/02/Now-You-Code/NYC1-Hello-2-Ways.ipynb
|
hvpuzzan-su/intern-project
|
Combining DataFrames with pandasIn many "real world" situations, the data that we want to use come in multiplefiles. We often need to combine these files into a single DataFrame to analyzethe data. The pandas package provides [various methods for combiningDataFrames](http://pandas.pydata.org/pandas-docs/stable/merging.html) including`merge` and `concat`.To work through the examples below, we first need to load the species andsurveys files into pandas DataFrames. In iPython: Take note that the `read_csv` method we used can take some additional options whichwe didn't use previously. Many functions in python have a set of options thatcan be set by the user if needed. In this case, we have told Pandas to assignempty values in our CSV to NaN `keep_default_na=False, na_values=[""]`.[More about all of the read_csv options here.](http://pandas.pydata.org/pandas-docs/dev/generated/pandas.io.parsers.read_csv.html) Concatenating DataFramesWe can use the `concat` function in Pandas to append either columns or rows fromone DataFrame to another. Let's grab two subsets of our data to see how thisworks.
|
# read in first 10 lines of surveys table
# grab the last 10 rows
# reset the index values to the second dataframe appends properly
# drop=True option avoids adding new index column with old index values
|
_____no_output_____
|
CC-BY-4.0
|
_episodes_pynb/04-merging-data_clean.ipynb
|
scw-ss/-2018-06-27-cfmehu-python-ecology-lesson
|
When we concatenate DataFrames, we need to specify the axis. `axis=0` tellsPandas to stack the second DataFrame under the first one. It will automaticallydetect whether the column names are the same and will stack accordingly.`axis=1` will stack the columns in the second DataFrame to the RIGHT of thefirst DataFrame. To stack the data vertically, we need to make sure we have thesame columns and associated column format in both datasets. When we stackhorizonally, we want to make sure what we are doing makes sense (ie the data arerelated in some way).
|
# stack the DataFrames on top of each other
# place the DataFrames side by side
|
_____no_output_____
|
CC-BY-4.0
|
_episodes_pynb/04-merging-data_clean.ipynb
|
scw-ss/-2018-06-27-cfmehu-python-ecology-lesson
|
Row Index Values and ConcatHave a look at the `vertical_stack` dataframe? Notice anything unusual?The row indexes for the two data frames `survey_sub` and `survey_sub_last10`have been repeated. We can reindex the new dataframe using the `reset_index()` method. Writing Out Data to CSVWe can use the `to_csv` command to do export a DataFrame in CSV format. Note that the codebelow will by default save the data into the current working directory. We cansave it to a different folder by adding the foldername and a slash to the file`vertical_stack.to_csv('foldername/out.csv')`. We use the 'index=False' so thatpandas doesn't include the index number for each line.
|
# Write DataFrame to CSV
|
_____no_output_____
|
CC-BY-4.0
|
_episodes_pynb/04-merging-data_clean.ipynb
|
scw-ss/-2018-06-27-cfmehu-python-ecology-lesson
|
Check out your working directory to make sure the CSV wrote out properly, andthat you can open it! If you want, try to bring it back into python to make sureit imports properly.
|
# for kicks read our output back into python and make sure all looks good
|
_____no_output_____
|
CC-BY-4.0
|
_episodes_pynb/04-merging-data_clean.ipynb
|
scw-ss/-2018-06-27-cfmehu-python-ecology-lesson
|
> Challenge - Combine Data>> In the data folder, there are two survey data files: `survey2001.csv` and> `survey2002.csv`. Read the data into python and combine the files to make one> new data frame. Create a plot of average plot weight by year grouped by sex.> Export your results as a CSV and make sure it reads back into python properly. Joining DataFramesWhen we concatenated our DataFrames we simply added them to each other -stacking them either vertically or side by side. Another way to combineDataFrames is to use columns in each dataset that contain common values (acommon unique id). Combining DataFrames using a common field is called"joining". The columns containing the common values are called "join key(s)".Joining DataFrames in this way is often useful when one DataFrame is a "lookuptable" containing additional data that we want to include in the other.NOTE: This process of joining tables is similar to what we do with tables in anSQL database.For example, the `species.csv` file that we've been working with is a lookuptable. This table contains the genus, species and taxa code for 55 species. Thespecies code is unique for each line. These species are identified in our surveydata as well using the unique species code. Rather than adding 3 more columnsfor the genus, species and taxa to each of the 35,549 line Survey data table, wecan maintain the shorter table with the species information. When we want toaccess that information, we can create a query that joins the additional columnsof information to the Survey data.Storing data in this way has many benefits including:1. It ensures consistency in the spelling of species attributes (genus, species and taxa) given each species is only entered once. Imagine the possibilities for spelling errors when entering the genus and species thousands of times!2. It also makes it easy for us to make changes to the species information once without having to find each instance of it in the larger survey data.3. It optimizes the size of our data. Joining Two DataFramesTo better understand joins, let's grab the first 10 lines of our data as asubset to work with. We'll use the `.head` method to do this. We'll also readin a subset of the species table.
|
# read in first 10 lines of surveys table
# import a small subset of the species data designed for this part of the lesson.
# It is stored in the data folder.
|
_____no_output_____
|
CC-BY-4.0
|
_episodes_pynb/04-merging-data_clean.ipynb
|
scw-ss/-2018-06-27-cfmehu-python-ecology-lesson
|
Chapter 13: Analyzing sound waves with Fourier Series Helper functions
|
import matplotlib.pyplot as plt
def plot_function(f,xmin,xmax,**kwargs):
ts = np.linspace(xmin,xmax,1000)
plt.plot(ts,[f(t) for t in ts],**kwargs)
def plot_sequence(points,max=100,line=False,**kwargs):
if line:
plt.plot(range(0,max),points[0:max],**kwargs)
else:
plt.scatter(range(0,max),points[0:max],**kwargs)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.1 Playing sound waves in Python 13.1.1 Producing our first sound
|
import pygame, pygame.sndarray
pygame.mixer.init(frequency=44100, size=-16, channels=1)
import numpy as np
arr = np.random.randint(-32768, 32767, size=44100)
arr
plot_sequence(arr)
plot_sequence(arr,line=True,max=441)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
**CAUTION: May play a loud sound!!!**
|
sound = pygame.sndarray.make_sound(arr)
sound.play()
arr = np.random.randint(-10000, 10000, size=44100)
sound = pygame.sndarray.make_sound(arr)
sound.play()
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.1.2 Playing a musical note
|
form = np.repeat([10000,-10000],50) #<1>
plot_sequence(form)
arr = np.tile(form,441)
plot_sequence(arr,line=True,max=1000)
sound = pygame.sndarray.make_sound(arr)
sound.play()
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.1.3 Exercises **Exercise:** Our musical note “A” was a pattern that repeated 441 times in a second. Create a similar pattern that repeats 350 times in one second, which will produce the musical note “F”. **Solution:**
|
form = np.repeat([10000,-10000],63)
arr = np.tile(form,350)
sound = pygame.sndarray.make_sound(arr)
sound.play()
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.2 Turning a sinusoidal wave into a sound 13.2.1 Making audio from sinusoidal functions
|
from math import sin,cos,pi
plot_function(sin,0,4*pi)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.2.2 Changing the frequency of a sinusoid
|
def make_sinusoid(frequency,amplitude):
def f(t): #<1>
return amplitude * sin(2*pi*frequency*t) #<2>
return f
plot_function(make_sinusoid(5,4),0,1)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.2.3 Sampling and playing the sound wave
|
sinusoid = make_sinusoid(441,8000)
np.arange(0,1,0.1)
np.arange(0,1,1/44100)
def sample(f,start,end,count): #<1>
mapf = np.vectorize(f) #<2>
ts = np.arange(start,end,(end-start)/count) #<3>
values = mapf(ts) #<4>
return values.astype(np.int16) #<5>
sinusoid = make_sinusoid(441,8000)
arr = sample(sinusoid, 0, 1, 44100)
sound = pygame.sndarray.make_sound(arr)
sound.play()
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.2.4 Exercises **Exercise:** Plot the tangent function $\tan(t) = \sin(t)/\cos(t).$ What is its period? **Solution:** The period is $\pi$.
|
from math import tan
plot_function(tan,0,5*pi)
plt.ylim(-10,10) #<1>
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
**Exercise:** Find the value of $k$ such that $\cos(kt)$ has a frequency of 5. Plot the resulting function $\cos(kt)$ from zero to one and show that it repeats itself 5 times. **Solution:**
|
plot_function(lambda t: cos(10*pi*t),0,1)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.3 Combining sound waves to make new ones 13.3.1 Adding sampled sound waves to build a chord
|
np.array([1,2,3]) + np.array([4,5,6])
sample1 = sample(make_sinusoid(441,8000),0,1,44100)
sample2 = sample(make_sinusoid(551,8000),0,1,44100)
sound1 = pygame.sndarray.make_sound(sample1)
sound2 = pygame.sndarray.make_sound(sample2)
sound1.play()
sound2.play()
chord = pygame.sndarray.make_sound(sample1 + sample2)
chord.play()
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.3.2 Picturing the sum of two sound waves
|
plot_sequence(sample1,max=400)
plot_sequence(sample2,max=400)
plot_sequence(sample1+sample2,max=400)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.3.3 Building a linear combination of sinusoids
|
def const(n):
return 1
def fourier_series(a0,a,b):
def result(t):
cos_terms = [an*cos(2*pi*(n+1)*t) for (n,an) in enumerate(a)] #<1>
sin_terms = [bn*sin(2*pi*(n+1)*t) for (n,bn) in enumerate(b)] #<2>
return a0*const(t) + sum(cos_terms) + sum(sin_terms) #<3>
return result
f = fourier_series(0,[0,0,0,0,0],[0,0,0,1,1])
plot_function(f,0,1)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.3.4 Building a familiar function with sinusoids
|
f1 = fourier_series(0,[],[4/pi])
f3 = fourier_series(0,[],[4/pi,0,4/(3*pi)])
plot_function(f1,0,1)
plot_function(f3,0,1)
b = [4/(n * pi) if n%2 != 0 else 0 for n in range(1,10)] #<1>
f = fourier_series(0,[],b)
plot_function(f,0,1)
b = [4/(n * pi) if n%2 != 0 else 0 for n in range(1,20)]
f = fourier_series(0,[],b)
plot_function(f,0,1)
b = [4/(n * pi) if n%2 != 0 else 0 for n in range(1,100)]
f = fourier_series(0,[],b)
plot_function(f,0,1)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.3.5 Exercises **Mini-project:** Create a manipulated version of the square wave Fourier series so that is frequency is 441 Hz, sample it, and confirm that it doesn’t just look like the square wave -- it sounds like the square wave as well. **Solution:** Here's a quick idea of how to do this with the function `f` you just built.
|
arr = sample(lambda t: 10000* f(441*t), 0, 1, 44100)
sound = pygame.sndarray.make_sound(arr)
sound.play()
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.4 Decomposing a sound wave into its Fourier Series 13.4.1 Finding vector components with an inner product 13.4.2 Defining an inner product for periodic functions
|
def inner_product(f,g,N=1000):
dt = 1/N #<1>
return 2*sum([f(t)*g(t)*dt for t in np.arange(0,1,dt)]) #<2>
def s(n): #<1>
def f(t):
return sin(2*pi*n*t)
return f
def c(n): #<2>
def f(t):
return cos(2*pi*n*t)
return f
inner_product(s(1),c(1))
inner_product(s(1),s(2))
inner_product(c(3),s(10))
inner_product(s(1),s(1))
inner_product(c(1),c(1))
inner_product(c(3),c(3))
from math import sqrt
def const(n):
return 1 /sqrt(2)
inner_product(const,s(1))
inner_product(const,c(1))
inner_product(const,const)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.4.3 Writing a function to find Fourier coefficients **note** we have a new `const` function so `fourier_series` will behave differently
|
def fourier_series(a0,a,b):
def result(t):
cos_terms = [an*cos(2*pi*(n+1)*t) for (n,an) in enumerate(a)] #<1>
sin_terms = [bn*sin(2*pi*(n+1)*t) for (n,bn) in enumerate(b)] #<2>
return a0*const(t) + sum(cos_terms) + sum(sin_terms) #<3>
return result
def fourier_coefficients(f,N):
a0 = inner_product(f,const) #<1>
an = [inner_product(f,c(n)) for n in range(1,N+1)] #<2>
bn = [inner_product(f,s(n)) for n in range(1,N+1)] #<3>
return a0, an, bn
f = fourier_series(0,[2,3,4],[5,6,7])
fourier_coefficients(f,3)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.4.4 Finding the Fourier coefficients for the square wave
|
def square(t):
return 1 if (t%1) < 0.5 else -1
a0, a, b = fourier_coefficients(square,10)
b[0], 4/pi
b[2], 4/(3*pi)
b[4], 4/(5*pi)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
4.5 Fourier coefficients for other waveforms
|
def sawtooth(t):
return t%1
plot_function(sawtooth,0,5)
approx = fourier_series(*fourier_coefficients(sawtooth,10))
plot_function(sawtooth,0,5)
plot_function(approx,0,5)
def speedbumps(t):
if abs(t%1 - 0.5) > 0.25:
return 0
else:
return sqrt(0.25*0.25 - (t%1 - 0.5)**2)
approx = fourier_series(*fourier_coefficients(speedbumps,10))
plot_function(speedbumps,0,5)
plot_function(approx,0,5)
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
13.4.6 Exercises **Mini project:** Play a sawtooth wave at 441 Hz and compare it with the square and sinusoidal waves you played at that frequency. **Solution:**
|
def modified_sawtooth(t):
return 8000 * sawtooth(441*t)
arr = sample(modified_sawtooth,0,1,44100)
sound = pygame.sndarray.make_sound(arr)
sound.play()
|
_____no_output_____
|
MIT
|
Chapter 13/Chapter13Walkthrough.ipynb
|
ZhangXinNan/MathForProgrammers
|
info クレンジング1. 欠損値があった場合、基礎分析の結果に基づいて値埋めか行の削除を行なっている1. 表記揺れがあった場合、漏れなく修正している1. 水準数が多く、なおかつまとめられそうな質的変数があった場合に、論理的な基準に基づいて値をまとめている 特徴量エンジニアリング1. 質的変数を量的変数(加減乗除して意味のある数値)に変換している1. 量的変数を基礎分析の結果をもとに変換している1. 量的変数のスケーリングを行っている1. 元データを素に、有用であると考えられるような特徴を少なくとも1は生成している init
|
import numpy as np
import pandas as pd
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
load
|
path_data = "../data/"
path_raw = path_data + "raw/"
path_mid = path_data + "mid/"
path_clns = path_data + "clns/"
cats = pd.read_csv(path_mid+"cats.csv", index_col=0)
nums = pd.read_csv(path_mid+"nums.csv", index_col=0)
bools = pd.read_csv(path_mid+"bools.csv", index_col=0)
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
clns fillna embarked
|
cats["embarked"] = cats["embarked"].fillna("S")
cats["embarked"].isna().any()
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
age
|
from sklearn.linear_model import LogisticRegression, LinearRegression
from sklearn.metrics import roc_auc_score
from sklearn.preprocessing import StandardScaler
fill_mean = nums[["age", "survived"]]
fill_mean = fill_mean[~fill_mean.survived.isna()]
fill_mean["age"] = fill_mean.age.fillna(fill_mean.age.mean())
fill_median = nums[["age", "survived"]]
fill_median = fill_median[~fill_median.survived.isna()]
fill_median["age"] = fill_median.age.fillna(fill_median.age.median())
# 線形回帰で値埋め
def zscore(x):
m = x.mean()
s = x.std(ddof=1)
return (x-m)/s
X = pd.get_dummies(cats, drop_first=True)
z = zscore(nums.drop(["age", "survived"], 1))
X = X.join(z)
y = nums.age
is_na = y.isna()
y = np.log1p(y)
rgs = LinearRegression()
rgs.fit(X[~is_na], y[~is_na])
pred = rgs.predict(X[is_na])
pred = np.exp(pred)-1
base = X[~is_na]
base["age"] = np.exp(y[~is_na])-1
fill = X[is_na]
fill["age"] = pred
fill_linear = pd.concat([base, fill]).join(nums[["survived"]])
fill_linear = fill_linear[["age", "survived"]]
fill_linear = fill_linear[~fill_linear.survived.isna()]
def check_auc(data):
X, y = data[["age"]], data["survived"]
clf = LogisticRegression()
clf.fit(X, y)
proba = clf.predict_proba(X)[:,1]
auc = roc_auc_score(y_true=y, y_score=proba)
print(auc)
check_auc(fill_mean)
check_auc(fill_median)
check_auc(fill_linear)
nums["age"] = nums.fillna(nums.age.mean())
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
チェックポイント1: 欠損値があった場合、基礎分析の結果に基づいて値埋めか行の削除を行なっている チェックポイント2: 表記揺れがあった場合、漏れなく修正している- 表記揺れはない union-value
|
cats_union = cats.copy()
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
embarked
|
cats_union["embarked"] = cats_union.embarked.replace(["Q", "S"], "QorS")
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
family-size
|
family = nums.parch + nums.sibsp
parch = nums.parch.apply(lambda x: "4+" if x >= 4 else x)
sibsp = nums.sibsp.apply(lambda x: "4+" if x >= 4 else x)
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
チェックポイント3: 水準数が多く、なおかつまとめられそうな質的変数があった場合に、論理的な基準に基づいて値をまとめている feature engineering onehot-encoding
|
pd.get_dummies(cats_union, drop_first=True).to_csv(path_clns+"onehot_cats.csv")
pd.get_dummies(pd.concat([parch, sibsp], axis=1), drop_first=True).to_csv(path_clns+"onehot_parch_sibsp.csv")
pd.get_dummies(family, drop_first=True, prefix="family-size").to_csv(path_clns+"onehot_familysize.csv")
(bools*1).to_csv(path_clns+"onehot_bools.csv")
nums[["survived"]].to_csv(path_clns+"y.csv")
is_child = (nums.age <= 7)*1
is_child.name = "is_child"
is_child.to_frame().to_csv(path_clns+"onehot_ischild.csv")
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
チェックポイント4: 元データを素に、有用であると考えられるような特徴を少なくとも1は生成している target-encoding
|
def tgt_encoding(data, y):
data = data.copy()
idname = data.index.name
data = data.reset_index()
train = data.dropna()
for x in set(data)-set([idname, y]):
dfg = train.groupby(x)[y].mean()
dfg = dfg.to_frame()
data = data.merge(dfg, on=x, suffixes=["", "_%s_tgt"%x], how="left")
data = data.set_index(idname)
data = data.filter(regex="_tgt")
return data
y = "survived"
data = cats.join(nums[[y]])
data = tgt_encoding(data, y)
data.to_csv(path_clns+"tgt_cats.csv")
data = bools.join(nums[[y]])
data = tgt_encoding(data, y)
data.to_csv(path_clns+"tgt_bools.csv")
data = pd.concat([parch, sibsp], axis=1).join(nums[[y]])
data = tgt_encoding(data, y)
data.to_csv(path_clns+"tgt_parch_sibsp.csv")
data = family.to_frame()
data.columns = ["familysize"]
data = data.join(nums[[y]])
data = tgt_encoding(data, y)
data.to_csv(path_clns+"tgt_familysize.csv")
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
チェックポイント1: 質的変数を量的変数(加減乗除して意味のある数値)に変換している log, zscore
|
def zscore(x):
m = x.mean()
s = x.std(ddof=1)
return (x-m)/s
nums_tgt = nums[["age", "fare"]]
z = zscore(nums_tgt)
z.to_csv(path_clns+"num_z.csv")
z = zscore(np.log1p(nums_tgt))
z.to_csv(path_clns+"num_logz.csv")
|
_____no_output_____
|
MIT
|
notebooks/2.PreProcessing.ipynb
|
mnm-analytics/titanic
|
FloPy shapefile export demoThe goal of this notebook is to demonstrate ways to export model information to shapefiles.This example will cover:* basic exporting of information for a model, individual package, or dataset* custom exporting of combined data from different packages* general exporting and importing of geographic data from other sources
|
import sys
import os
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import pandas as pd
# run installed version of flopy or add local path
try:
import flopy
except:
fpth = os.path.abspath(os.path.join('..', '..'))
sys.path.append(fpth)
import flopy
print(sys.version)
print('numpy version: {}'.format(np.__version__))
print('matplotlib version: {}'.format(mpl.__version__))
print('flopy version: {}'.format(flopy.__version__))
# set the output directory
outdir = os.path.join('temp', 'shapefile_export')
if not os.path.isdir(outdir):
os.makedirs(outdir)
# load an existing model
model_ws = "../data/freyberg"
m = flopy.modflow.Modflow.load("freyberg.nam", model_ws=model_ws, verbose=False,
check=False, exe_name="mfnwt")
m.get_package_list()
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
set the model coordinate informationthe coordinate information where the grid is located in a projected coordinate system (e.g. UTM)
|
grid = m.modelgrid
grid.set_coord_info(xoff=273170, yoff=5088657, epsg=26916)
grid.extent
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
Declarative export using attached `.export()` methods Export the whole model to a single shapefile
|
fname = '{}/model.shp'.format(outdir)
m.export(fname)
ax = plt.subplot(1, 1, 1, aspect='equal')
extents = grid.extent
pc = flopy.plot.plot_shapefile(fname, ax=ax, edgecolor='k', facecolor='none')
ax.set_xlim(extents[0], extents[1])
ax.set_ylim(extents[2], extents[3])
ax.set_title(fname);
fname = '{}/wel.shp'.format(outdir)
m.wel.export(fname)
|
wrote temp/shapefile_export/wel.shp
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
Export a package to a shapefile Export a FloPy list or array object
|
m.lpf.hk
fname = '{}/hk.shp'.format(outdir)
m.lpf.hk.export('{}/hk.shp'.format(outdir))
ax = plt.subplot(1, 1, 1, aspect='equal')
extents = grid.extent
a = m.lpf.hk.array.ravel()
pc = flopy.plot.plot_shapefile(fname, ax=ax, a=a)
ax.set_xlim(extents[0], extents[1])
ax.set_ylim(extents[2], extents[3])
ax.set_title(fname);
m.riv.stress_period_data
m.riv.stress_period_data.export('{}/riv_spd.shp'.format(outdir))
|
wrote temp/shapefile_export/riv_spd.shp
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
MfList.export() exports the whole grid by default, regardless of the locations of the boundary cells`sparse=True` only exports the boundary cells in the MfList
|
m.riv.stress_period_data.export('{}/riv_spd.shp'.format(outdir), sparse=True)
m.wel.stress_period_data.export('{}/wel_spd.shp'.format(outdir), sparse=True)
|
wrote temp/shapefile_export/wel_spd.shp
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
Ad-hoc exporting using `recarray2shp`* The main idea is to create a recarray with all of the attribute information, and a list of geometry features (one feature per row in the recarray)* each geometry feature is an instance of the `Point`, `LineString` or `Polygon` classes in `flopy.utils.geometry`. The shapefile format requires all the features to be of the same type.* We will use pandas dataframes for these examples because they are easy to work with, and then convert them to recarrays prior to exporting.
|
from flopy.export.shapefile_utils import recarray2shp
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
combining data from different packageswrite a shapefile of RIV and WEL package cells
|
wellspd = pd.DataFrame(m.wel.stress_period_data[0])
rivspd = pd.DataFrame(m.riv.stress_period_data[0])
spd = wellspd.append(rivspd)
spd.head()
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
create a list of Polygon features from the cell vertices stored in the SpatialReference object
|
from flopy.utils.geometry import Polygon
vertices = []
for row, col in zip(spd.i, spd.j):
vertices.append(grid.get_cell_vertices(row, col))
polygons = [Polygon(vrt) for vrt in vertices]
polygons
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
write the shapefile
|
fname = '{}/bcs.shp'.format(outdir)
recarray2shp(spd.to_records(), geoms=polygons,
shpname=fname,
epsg=grid.epsg)
ax = plt.subplot(1, 1, 1, aspect='equal')
extents = grid.extent
pc = flopy.plot.plot_shapefile(fname, ax=ax)
ax.set_xlim(extents[0], extents[1])
ax.set_ylim(extents[2], extents[3])
ax.set_title(fname);
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
exporting other dataSuppose we have some well data with actual locations that we want to export to a shapefile
|
welldata = pd.DataFrame({'wellID': np.arange(0, 10),
'q': np.random.randn(10)*100 - 1000,
'x_utm': np.random.rand(10)*1000 + grid.yoffset,
'y_utm': grid.xoffset - np.random.rand(10)*3000})
welldata.head()
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
convert the x, y coorindates to point features and then export
|
from flopy.utils.geometry import Point
geoms = [Point(x, y) for x, y in zip(welldata.x_utm, welldata.y_utm)]
fname = '{}/wel_data.shp'.format(outdir)
recarray2shp(welldata.to_records(), geoms=geoms,
shpname=fname,
epsg=grid.epsg)
ax = plt.subplot(1, 1, 1, aspect='equal')
extents = grid.extent
pc = flopy.plot.plot_shapefile(fname, ax=ax, radius=25)
ax.set_xlim(extents[0], extents[1])
ax.set_ylim(extents[2], extents[3])
ax.set_title(fname);
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
Adding attribute data to an existing shapefileSuppose we have a GIS coverage representing the river in the riv package
|
from flopy.utils.geometry import LineString
### make up a linestring shapefile of the river reaches
i, j = m.riv.stress_period_data[0].i, m.riv.stress_period_data[0].j
x0 = grid.xyzcellcenters[0][i[0], j[0]]
x1 = grid.xyzcellcenters[0][i[-1], j[-1]]
y0 = grid.xyzcellcenters[1][i[0], j[0]]
y1 = grid.xyzcellcenters[1][i[-1], j[-1]]
x = np.linspace(x0, x1, m.nrow+1)
y = np.linspace(y0, y1, m.nrow+1)
l0 = zip(list(zip(x[:-1], y[:-1])), list(zip(x[1:], y[1:])))
lines = [LineString(l) for l in l0]
rivdata = pd.DataFrame(m.riv.stress_period_data[0])
rivdata['reach'] = np.arange(len(lines))
lines_shapefile = '{}/riv_reaches.shp'.format(outdir)
recarray2shp(rivdata.to_records(index=False), geoms=lines,
shpname=lines_shapefile,
epsg=grid.epsg)
ax = plt.subplot(1, 1, 1, aspect='equal')
extents = grid.extent
pc = flopy.plot.plot_shapefile(lines_shapefile, ax=ax, radius=25)
ax.set_xlim(extents[0], extents[1])
ax.set_ylim(extents[2], extents[3])
ax.set_title(lines_shapefile);
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
read in the GIS coverage using `shp2recarray``shp2recarray` reads a shapefile into a numpy record array, which can easily be converted to a DataFrame
|
from flopy.export.shapefile_utils import shp2recarray
linesdata = shp2recarray(lines_shapefile)
linesdata = pd.DataFrame(linesdata)
linesdata.head()
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
Suppose we have some flow information that we read in from the cell budget file
|
# make up some fluxes between the river and aquifer at each reach
q = np.random.randn(len(linesdata))+1
q
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
Add reachs fluxes and cumulative flow to lines DataFrame
|
linesdata['qreach'] = q
linesdata['qstream'] = np.cumsum(q)
recarray2shp(linesdata.drop('geometry', axis=1).to_records(),
geoms=linesdata.geometry,
shpname=lines_shapefile,
epsg=grid.epsg)
ax = plt.subplot(1, 1, 1, aspect='equal')
extents = grid.extent
pc = flopy.plot.plot_shapefile(lines_shapefile, ax=ax, radius=25)
ax.set_xlim(extents[0], extents[1])
ax.set_ylim(extents[2], extents[3])
ax.set_title(lines_shapefile);
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
Overriding the model's modelgrid with a user supplied modelgridIn some cases it may be necessary to override the model's modelgrid instance with a seperate modelgrid. An example of this is if the model discretization is in feet and the user would like it projected in meters. Exporting can be accomplished by supplying a modelgrid as a `kwarg` in any of the `export()` methods within flopy. Below is an example:
|
mg0 = m.modelgrid
# build a new modelgrid instance with discretization in meters
modelgrid = flopy.discretization.StructuredGrid(delc=mg0.delc * 0.3048, delr=mg0.delr * 0.3048,
top= mg0.top, botm=mg0.botm, idomain=mg0.idomain,
xoff=mg0.xoffset * 0.3048, yoff=mg0.yoffset * 0.3048)
# exporting an entire model
m.export('{}/freyberg.shp'.format(outdir), modelgrid=modelgrid)
|
wrote temp/shapefile_export/freyberg.shp
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
And for a specific parameter the method is the same
|
fname = '{}/hk.shp'.format(outdir)
m.lpf.hk.export(fname, modelgrid=modelgrid)
ax = plt.subplot(1, 1, 1, aspect='equal')
extents = modelgrid.extent
a = m.lpf.hk.array.ravel()
pc = flopy.plot.plot_shapefile(fname, ax=ax, a=a)
ax.set_xlim(extents[0], extents[1])
ax.set_ylim(extents[2], extents[3])
ax.set_title(fname);
|
_____no_output_____
|
CC0-1.0
|
examples/Notebooks/flopy3_shapefile_export.ipynb
|
tomvansteijn/flopy
|
What Packages do I have Installed
|
import pip
!pip freeze
|
alabaster==0.7.10
anaconda-client==1.6.9
anaconda-navigator==1.7.0
anaconda-project==0.8.2
asn1crypto==0.24.0
astroid==1.6.1
astropy==2.0.3
attrs==17.4.0
Babel==2.5.3
backports.shutil-get-terminal-size==1.0.0
beautifulsoup4==4.6.0
bitarray==0.8.1
bkcharts==0.2
blaze==0.11.3
bleach==2.1.2
blpapi==3.9.1
bokeh==0.12.13
boto==2.48.0
Bottleneck==1.2.1
bqplot==0.11.6
certifi==2018.1.18
cffi==1.11.4
chardet==3.0.4
click==6.7
cloudpickle==0.5.2
clyent==1.2.2
colorama==0.3.9
comtypes==1.1.4
conda==4.4.10
conda-build==3.4.1
conda-verify==2.0.0
contextlib2==0.5.5
cryptography==2.1.4
cycler==0.10.0
Cython==0.27.3
cytoolz==0.9.0
dask==0.16.1
datashape==0.5.4
decorator==4.2.1
distributed==1.20.2
docutils==0.14
entrypoints==0.2.3
et-xmlfile==1.0.1
fastcache==1.0.2
filelock==2.0.13
Flask==0.12.2
Flask-Cors==3.0.3
gevent==1.2.2
glob2==0.6
greenlet==0.4.12
h5py==2.7.1
heapdict==1.0.0
html5lib==1.0.1
idna==2.6
imageio==2.2.0
imagesize==0.7.1
ipython==6.2.1
ipython-genutils==0.2.0
ipywidgets==7.1.1
isort==4.2.15
itsdangerous==0.24
jdcal==1.3
jedi==0.11.1
Jinja2==2.10
jsonschema==2.6.0
jupyter==1.0.0
jupyter-client==5.2.2
jupyter-console==5.2.0
jupyter-contrib-core==0.3.3
jupyter-contrib-nbextensions==0.5.1
jupyter-core==4.4.0
jupyter-highlight-selected-word==0.2.0
jupyter-latex-envs==1.4.6
jupyter-nbextensions-configurator==0.4.1
jupyterlab==0.31.4
jupyterlab-launcher==0.10.2
lazy-object-proxy==1.3.1
llvmlite==0.21.0
locket==0.2.0
lxml==4.1.1
MarkupSafe==1.0
matplotlib==2.1.2
mccabe==0.6.1
menuinst==1.4.11
mistune==0.8.3
mpmath==1.0.0
msgpack-python==0.5.1
multipledispatch==0.4.9
navigator-updater==0.1.0
nbconvert==5.3.1
nbformat==4.4.0
networkx==2.1
nltk==3.2.5
nose==1.3.7
notebook==5.4.0
numba==0.36.2
numexpr==2.6.4
numpy==1.14.0
numpydoc==0.7.0
odo==0.5.1
olefile==0.45.1
openpyxl==2.4.10
packaging==16.8
pandas==0.22.0
pandocfilters==1.4.2
parso==0.1.1
partd==0.3.8
path.py==10.5
pathlib2==2.3.0
patsy==0.5.0
pdblp==0.1.8
pep8==1.7.1
pickleshare==0.7.4
Pillow==5.0.0
pkginfo==1.4.1
pluggy==0.6.0
ply==3.10
prompt-toolkit==1.0.15
psutil==5.4.3
py==1.5.2
pycodestyle==2.3.1
pycosat==0.6.3
pycparser==2.18
pycrypto==2.6.1
pycurl==7.43.0.1
pyflakes==1.6.0
Pygments==2.2.0
pylint==1.8.2
pyodbc==4.0.22
pyOpenSSL==17.5.0
pyparsing==2.2.0
PySocks==1.6.7
pytest==3.3.2
python-dateutil==2.6.1
pytz==2017.3
PyWavelets==0.5.2
pywin32==222
pywinpty==0.5
PyYAML==3.12
pyzmq==16.0.3
QtAwesome==0.4.4
qtconsole==4.3.1
QtPy==1.3.1
requests==2.18.4
rope==0.10.7
ruamel-yaml==0.15.35
scikit-image==0.13.1
scikit-learn==0.19.1
scipy==1.0.0
seaborn==0.8.1
Send2Trash==1.4.2
simplegeneric==0.8.1
singledispatch==3.4.0.3
six==1.11.0
snowballstemmer==1.2.1
sortedcollections==0.5.3
sortedcontainers==1.5.9
Sphinx==1.6.6
sphinxcontrib-websupport==1.0.1
spyder==3.2.6
SQLAlchemy==1.2.1
statsmodels==0.8.0
sympy==1.1.1
tables==3.4.2
tblib==1.3.2
terminado==0.8.1
testpath==0.3.1
toolz==0.9.0
tornado==4.5.3
traitlets==4.3.2
traittypes==0.2.1
typing==3.6.2
unicodecsv==0.14.1
urllib3==1.22
wcwidth==0.1.7
webencodings==0.5.1
Werkzeug==0.14.1
widgetsnbextension==3.1.0
win-inet-pton==1.0.1
win-unicode-console==0.5
wincertstore==0.2
wrapt==1.10.11
xlrd==1.1.0
XlsxWriter==1.0.2
xlwings==0.11.5
xlwt==1.3.0
zict==0.1.3
|
BSD-2-Clause
|
Bloomberg Python Interop.ipynb
|
dmitchell28/Other-People-s-Code
|
Bloomberg Interop via python wrapper Accessing Bloomberg Desktop API directly INSTRUCTIONS FOR INSTALLING THE LIBRARY1. https://www.bloomberg.com/professional/support/api-library/- download the Python Supported Release2. go to command line and run: pip install pdblp
|
import pandas as pd
import pdblp #pandas wrapper to Bloomberg API. also xbbg & pybbg are popular
con = pdblp.BCon(debug=True, port=8194)
con.start()
con.debug = False #turn off 'verbose' mode
#historical data via 'bdh'
tickers = ['QCOM US Equity', 'AAPL US Equity', 'XIU CN Equity']
fields = ['PX_LAST', 'DX_SAF', 'VL158'] #borrow cost p/a and 3mo IVOL
start = '20200105'
end = '20200805'
response = con.bdh(tickers, fields, start, end)
type(response)
response.tail()
|
_____no_output_____
|
BSD-2-Clause
|
Bloomberg Python Interop.ipynb
|
dmitchell28/Other-People-s-Code
|
Chart Prices (same axis)
|
%matplotlib inline
response.filter(like="PX_LAST").plot()
list(response.columns) #format for columns
|
_____no_output_____
|
BSD-2-Clause
|
Bloomberg Python Interop.ipynb
|
dmitchell28/Other-People-s-Code
|
extract one Series (column) from DataFrame
|
spycloses = response[('XIU CN Equity', 'PX_LAST')]
spycloses.tail()
%matplotlib inline
spycloses.plot() #chart it
type(spycloses) #confirm dtype
spx_df = pd.DataFrame(spycloses) #confirm to dataframe with dates as index
spx_df.tail()
|
_____no_output_____
|
BSD-2-Clause
|
Bloomberg Python Interop.ipynb
|
dmitchell28/Other-People-s-Code
|
Non-Equity Examples
|
# individual datapoint using 'ref' (similar to 'bdp')
response2 = con.ref(['AUDUSD Curncy'], 'SETTLE_DT')
print(response2)
response3 = con.ref(['NDX Index'], 'VL137')
print(response3)
response3.value[0]
response = con.bsrch("FI:ytd_green")
response
|
_____no_output_____
|
BSD-2-Clause
|
Bloomberg Python Interop.ipynb
|
dmitchell28/Other-People-s-Code
|
BERT finetuning on AG_news-4 Librairy
|
# !pip install transformers==4.8.2
# !pip install datasets==1.7.0
import os
import time
import pickle
import numpy as np
import torch
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, recall_score, precision_score, f1_score
from transformers import BertTokenizer, BertTokenizerFast
from transformers import BertForSequenceClassification, AdamW
from transformers import Trainer, TrainingArguments
from transformers import EarlyStoppingCallback
from transformers.data.data_collator import DataCollatorWithPadding
from datasets import load_dataset, Dataset, concatenate_datasets
# print(torch.__version__)
# print(torch.cuda.device_count())
# print(torch.cuda.is_available())
# print(torch.cuda.get_device_name(0))
device = torch.device('cuda') if torch.cuda.is_available() else torch.device('cpu')
# if torch.cuda.is_available():
# torch.set_default_tensor_type('torch.cuda.FloatTensor')
device
|
_____no_output_____
|
MIT
|
notebooks_paper_2021/BERT_finetuning/BERT_finetune_AG_news-4.ipynb
|
PlaytikaResearch/esntorch
|
Global variables
|
BATCH_SIZE = 24
NB_EPOCHS = 4
RESULTS_FILE = '~/Results/BERT_finetune/ag_news-4_BERT_finetune_b'+str(BATCH_SIZE)+'_results.pkl'
RESULTS_PATH = '~/Results/BERT_finetune/ag_news-4_b'+str(BATCH_SIZE)+'/'
CACHE_DIR = '~/Data/huggignface/' # path of your folder
|
_____no_output_____
|
MIT
|
notebooks_paper_2021/BERT_finetuning/BERT_finetune_AG_news-4.ipynb
|
PlaytikaResearch/esntorch
|
Dataset
|
# download dataset
raw_datasets = load_dataset('ag_news', cache_dir=CACHE_DIR)
# tokenize
tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
def tokenize_function(examples):
return tokenizer(examples["text"], padding=True, truncation=True)
tokenized_datasets = raw_datasets.map(tokenize_function, batched=True)
tokenized_datasets.set_format(type='torch', columns=['input_ids', 'attention_mask', 'label'])
train_dataset = tokenized_datasets["train"].shuffle(seed=42)
train_val_datasets = train_dataset.train_test_split(train_size=0.8)
train_dataset = train_val_datasets['train'].rename_column('label', 'labels')
val_dataset = train_val_datasets['test'].rename_column('label', 'labels')
test_dataset = tokenized_datasets["test"].shuffle(seed=42).rename_column('label', 'labels')
# get number of labels
num_labels = len(set(train_dataset['labels'].tolist()))
num_labels
|
_____no_output_____
|
MIT
|
notebooks_paper_2021/BERT_finetuning/BERT_finetune_AG_news-4.ipynb
|
PlaytikaResearch/esntorch
|
Model Model
|
model = BertForSequenceClassification.from_pretrained("bert-base-uncased", num_labels=num_labels)
model.to(device)
|
_____no_output_____
|
MIT
|
notebooks_paper_2021/BERT_finetuning/BERT_finetune_AG_news-4.ipynb
|
PlaytikaResearch/esntorch
|
Training
|
training_args = TrainingArguments(
# output
output_dir=RESULTS_PATH,
# params
num_train_epochs=NB_EPOCHS, # nb of epochs
per_device_train_batch_size=BATCH_SIZE, # batch size per device during training
per_device_eval_batch_size=BATCH_SIZE, # cf. paper Sun et al.
learning_rate=2e-5, # cf. paper Sun et al.
# warmup_steps=500, # number of warmup steps for learning rate scheduler
warmup_ratio=0.1, # cf. paper Sun et al.
weight_decay=0.01, # strength of weight decay
# # eval
evaluation_strategy="steps",
eval_steps=50,
# evaluation_strategy='no', # no more evaluation, takes time
# log
logging_dir=RESULTS_PATH+'logs',
logging_strategy='steps',
logging_steps=50,
# save
# save_strategy='epoch',
# save_strategy='steps',
# load_best_model_at_end=False
load_best_model_at_end=True # cf. paper Sun et al.
)
def compute_metrics(p):
pred, labels = p
pred = np.argmax(pred, axis=1)
accuracy = accuracy_score(y_true=labels, y_pred=pred)
return {"val_accuracy": accuracy}
trainer = Trainer(
model=model,
args=training_args,
tokenizer=tokenizer,
train_dataset=train_dataset,
eval_dataset=val_dataset,
# compute_metrics=compute_metrics,
# callbacks=[EarlyStoppingCallback(early_stopping_patience=5)]
)
results = trainer.train()
training_time = results.metrics["train_runtime"]
training_time_per_epoch = training_time / training_args.num_train_epochs
training_time_per_epoch
trainer.save_model(os.path.join(RESULTS_PATH, 'best_model-0'))
|
_____no_output_____
|
MIT
|
notebooks_paper_2021/BERT_finetuning/BERT_finetune_AG_news-4.ipynb
|
PlaytikaResearch/esntorch
|
Results
|
results_d = {}
epoch = 1
ordered_files = sorted( [f for f in os.listdir(RESULTS_PATH)
if (not f.endswith("logs")) and (f.startswith("best")) # best model eval only
],
key=lambda x: int(x.split('-')[1]) )
for filename in ordered_files:
print(filename)
# load model
model_file = os.path.join(RESULTS_PATH, filename)
finetuned_model = BertForSequenceClassification.from_pretrained(model_file, num_labels=num_labels)
finetuned_model.to(device)
finetuned_model.eval()
# compute test acc
test_trainer = Trainer(finetuned_model, data_collator=DataCollatorWithPadding(tokenizer))
raw_preds, labels, _ = test_trainer.predict(test_dataset)
preds = np.argmax(raw_preds, axis=1)
test_acc = accuracy_score(y_true=labels, y_pred=preds)
# results_d[filename] = (test_acc, training_time_per_epoch*epoch)
results_d[filename] = test_acc # best model evaluation only
print((test_acc, training_time_per_epoch*epoch))
epoch += 1
results_d['training_time'] = training_time
# save results
with open(RESULTS_FILE, 'wb') as fh:
pickle.dump(results_d, fh)
# load results
with open(RESULTS_FILE, 'rb') as fh:
results_d = pickle.load(fh)
results_d
|
_____no_output_____
|
MIT
|
notebooks_paper_2021/BERT_finetuning/BERT_finetune_AG_news-4.ipynb
|
PlaytikaResearch/esntorch
|
This tutorial shows how to generate an image of handwritten digits using Deep Convolutional Generative Adversarial Network (DCGAN).Generative Adversarial Networks (GANs) are one of the most interesting fields in machine learning. The standard GAN consists of two models, a generative and a discriminator one. Two models are trained simultaneously by an adversarial process. A generative model (`the artist`) learns to generate images that look real, while the discriminator (`the art critic`) one learns to tell real images apart from the fakes.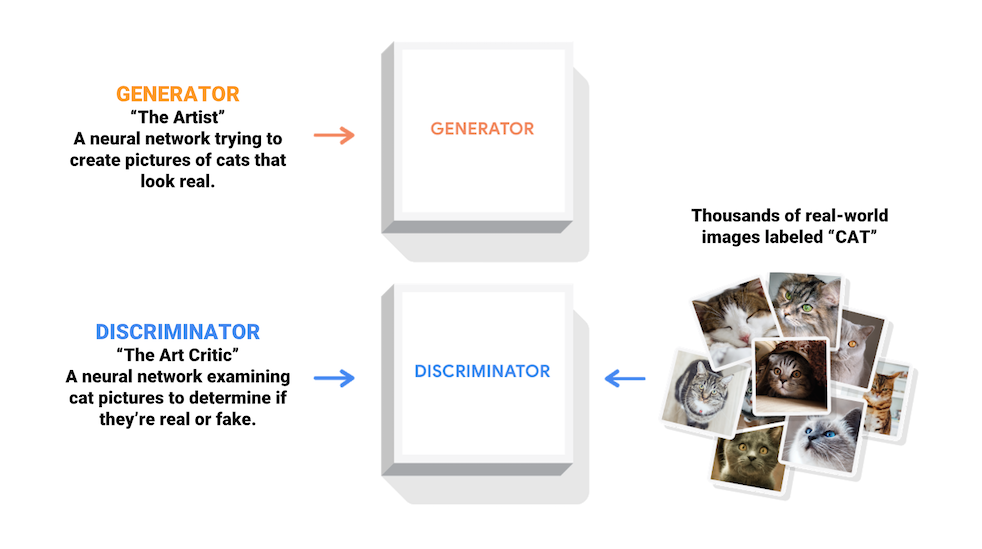Refer to Tensorflow.org (2020).During training, the generative model becomes progressively creating images that look real, and the discriminator model becomes progressively telling them apart. The whole process reaches equilibrium when the discriminator is no longer able to distinguish real images from fakes.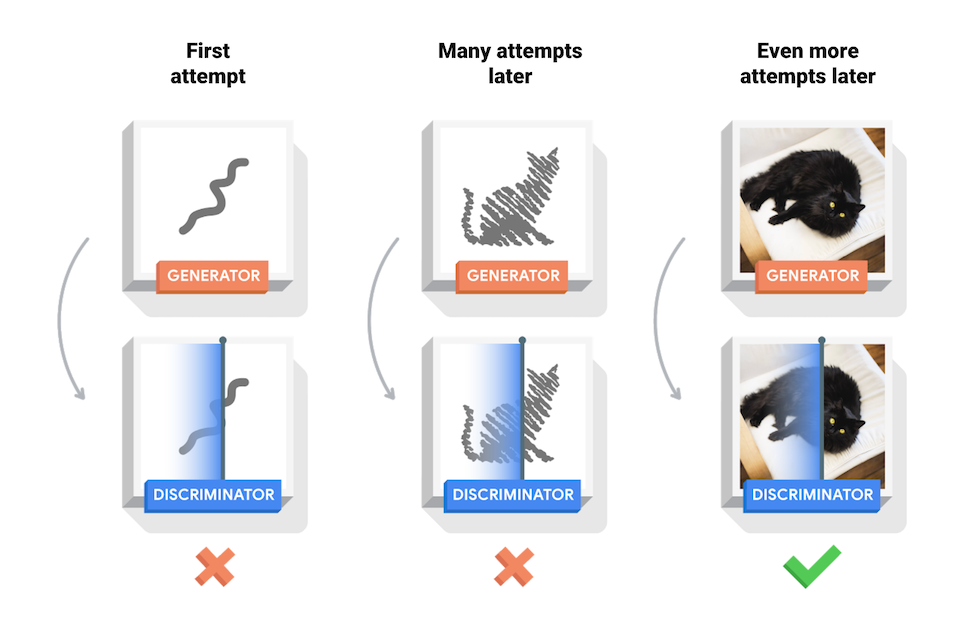Refer to Tensorflow.org (2020).In this demo, we show how to train a GAN model on MNIST and FASHION MNIST dataset.
|
!pip uninstall -y tensorflow
!pip install -q tf-nightly tfds-nightly
import glob
import tensorflow as tf
import tensorflow_datasets as tfds
import matplotlib.pyplot as plt
from tensorflow.keras.layers import Conv2D, Conv2DTranspose, Dense, Flatten, BatchNormalization, ELU, LeakyReLU, Reshape, Dropout
import numpy as np
import IPython.display as display
from IPython.display import clear_output
import os
import time
import imageio
tfds.disable_progress_bar()
print("Tensorflow Version: {}".format(tf.__version__))
print("GPU {} available.".format("is" if tf.config.experimental.list_physical_devices("GPU") else "not"))
|
Tensorflow Version: 2.4.0-dev20200706
GPU is available.
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Data Preprocessing
|
def normalize(image):
img = image['image']
img = (tf.cast(img, tf.float32) - 127.5) / 127.5
return img
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
MNIST Dataset
|
raw_datasets, metadata = tfds.load(name="mnist", with_info=True)
raw_train_datasets, raw_test_datasets = raw_datasets['train'], raw_datasets['test']
raw_test_datasets, metadata
BUFFER_SIZE = 10000
BATCH_SIZE = 256
train_datasets = raw_train_datasets.map(normalize).cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE)
test_datasets = raw_test_datasets.map(normalize).batch(BATCH_SIZE)
for imgs in train_datasets.take(1):
img = imgs[0]
plt.imshow(tf.keras.preprocessing.image.array_to_img(img))
plt.axis("off")
plt.show()
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Fashion_MNIST Dataset
|
raw_datasets, metadata = tfds.load(name="fashion_mnist", with_info=True)
raw_train_datasets, raw_test_datasets = raw_datasets['train'], raw_datasets['test']
raw_train_datasets
for image in raw_train_datasets.take(1):
plt.imshow(tf.keras.preprocessing.image.array_to_img(image['image']))
plt.axis("off")
plt.title("Label: {}".format(image['label']))
plt.show()
BUFFER_SIZE = 10000
BATCH_SIZE = 256
train_datasets = raw_train_datasets.map(normalize).cache().prefetch(BUFFER_SIZE).batch(BATCH_SIZE)
test_datasets = raw_test_datasets.map(normalize).batch(BATCH_SIZE)
for imgs in train_datasets.take(1):
img = imgs[0]
plt.imshow(tf.keras.preprocessing.image.array_to_img(img))
plt.axis("off")
plt.show()
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Build the GAN Model The GeneratorThe generator uses the `tf.keras.layers.Conv2DTranspose` (upsampling) layer to produce an image from a seed input (a random noise). Start from this seed input, upsample it several times to reach the desired output (28x28x1).
|
def build_generator_model():
model = tf.keras.Sequential()
model.add(Dense(units=7 * 7 * 256, use_bias=False, input_shape=(100,)))
model.add(BatchNormalization())
model.add(LeakyReLU())
model.add(Reshape(target_shape=[7,7,256]))
assert model.output_shape == (None, 7, 7, 256)
model.add(Conv2DTranspose(filters=128, kernel_size=(5,5), strides=(1,1), padding="same", use_bias=False))
model.add(BatchNormalization())
model.add(LeakyReLU())
assert model.output_shape == (None, 7, 7, 128)
model.add(Conv2DTranspose(filters=64, kernel_size=(5,5), strides=(2,2), padding='same', use_bias=False))
model.add(BatchNormalization())
model.add(LeakyReLU())
assert model.output_shape == (None, 14, 14, 64)
model.add(Conv2DTranspose(filters=1, kernel_size=(5,5), strides=(2,2), padding='same', use_bias=False,
activation="tanh"))
assert model.output_shape == (None, 28, 28, 1)
return model
generator = build_generator_model()
generator_input = tf.random.normal(shape=[1, 100])
generator_outputs = generator(generator_input, training=False)
plt.imshow(generator_outputs[0, :, :, 0], cmap='gray')
plt.show()
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
The DiscriminatorThe discriminator is basically a CNN network.
|
def build_discriminator_model():
model = tf.keras.Sequential()
# [None, 28, 28, 64]
model.add(Conv2D(filters=64, kernel_size=(5,5), strides=(1,1), padding="same",
input_shape=[28,28,1]))
model.add(LeakyReLU())
model.add(Dropout(rate=0.3))
# [None, 14, 14, 128]
model.add(Conv2D(filters=128, kernel_size=(3,3), strides=(2,2), padding='same'))
model.add(LeakyReLU())
model.add(Dropout(rate=0.3))
model.add(Flatten())
model.add(Dense(units=1))
return model
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
The output of the discriminator was trained that the negative values are for the fake images and the positive values are for real ones.
|
discriminator = build_discriminator_model()
discriminator_outputs = discriminator(generator_outputs)
discriminator_outputs
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Define the losses and optimizersDefine the loss functions and the optimizers for both models.
|
# define the cross entropy as the helper function
cross_entropy = tf.keras.losses.BinaryCrossentropy(from_logits=True)
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Discriminator LossThe discriminator's loss quantifies how well the discriminator can tell the real images from fakes. It compares the discriminator's predictions on real images to an array of 1s, and the discriminator's predictions on fake images to an array of 0s.
|
def discriminator_loss(real_output, fake_output):
real_loss = cross_entropy(tf.ones_like(real_output), real_output)
fake_loss = cross_entropy(tf.zeros_like(fake_output), fake_output)
total_loss = real_loss + fake_loss
return total_loss
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Generator Loss The generator's loss quantifies how well the generator model can trick the discriminator model. If the generator performs well, the discriminator will classify the fake images as real (or 1). Here, we will compare the discriminator decisions on the generated images to an array of 1s.
|
def generator_loss(fake_output):
# the generator learns to make the discriminator predictions became real
# (or an array of 1s) on the fake images
return cross_entropy(tf.ones_like(fake_output), fake_output)
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Define optimizers.
|
generator_optimizer = tf.keras.optimizers.Adam(learning_rate=1e-4)
discriminator_optimizer = tf.keras.optimizers.Adam(learning_rate=1e-4)
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Save Checkpoints
|
ckpt_dir = "./gan_ckpt"
ckpt_prefix = os.path.join(ckpt_dir, "ckpt")
ckpt = tf.train.Checkpoint(generator_optimizer=generator_optimizer,
discriminator_optimizer=discriminator_optimizer,
generator=generator,
discriminator=discriminator)
ckpt
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Define the training loop
|
EPOCHS = 50
noise_dim = 100
num_generated_examples = 16
# You will reuse the seed overtime to visualize progress in the animated GIF.
seed = tf.random.normal(shape=[num_generated_examples, noise_dim])
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
In the training loop, the generator model takes the noise as the input to generate the fake images. The discriminator model takes real images and fake images to give the discriminations (or outputs) for them. Calculate the generator and discriminator losses each using the real outputs and the fake outputs. Calculate the gradients of the model trainable variables based on these losses and then apply gradients back to them.
|
@tf.function
def train_step(images):
fake_noises = tf.random.normal(shape=[BATCH_SIZE, noise_dim])
with tf.GradientTape() as disc_tape, tf.GradientTape() as gen_tape:
fake_images = generator(fake_noises, training=True)
fake_outputs = discriminator(fake_images, training=True)
real_outputs = discriminator(images, training=True)
disc_loss = discriminator_loss(real_output=real_outputs,
fake_output=fake_outputs)
gen_loss = generator_loss(fake_output=fake_outputs)
disc_gradients = disc_tape.gradient(disc_loss, discriminator.trainable_variables)
gen_gradients = gen_tape.gradient(gen_loss, generator.trainable_variables)
discriminator_optimizer.apply_gradients(zip(disc_gradients, discriminator.trainable_variables))
generator_optimizer.apply_gradients(zip(gen_gradients, generator.trainable_variables))
def generate_and_save_images(model, epoch, test_input):
"""Helps to generate the images from a fixed seed."""
predictions = model(test_input, training=False)
fig = plt.figure(figsize=(8,8))
for i in range(predictions.shape[0]):
plt.subplot(4, 4, i+1)
plt.imshow(predictions[i, :, :, 0] * 127.5 + 127.5, cmap='gray')
plt.axis("off")
plt.savefig('image_epoch_{:04d}.png'.format(epoch))
plt.show()
def train(dataset, epochs):
for epoch in range(epochs):
start = time.time()
for batch_dataset in dataset:
train_step(batch_dataset)
clear_output(wait=True)
generate_and_save_images(generator, epoch+1, seed)
if (epoch+1) % 15 == 0:
ckpt.save(file_prefix=ckpt_prefix)
print("Epoch {} in time {}.".format(epoch + 1, time.time()-start))
# after the training
clear_output(wait=True)
generate_and_save_images(generator, epoch+1, seed)
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Train the ModelCall the `train()` function to start the model training. Note, training GANs can be tricky. It's important that the generator and discriminator do not overpower each other (e.g. they train at a similar rate).
|
train(train_datasets, epochs=EPOCHS)
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Create a GIF
|
def display_image(epoch_no):
image_path = 'image_epoch_{:04d}.png'.format(epoch_no)
img = plt.imread(fname=image_path)
plt.imshow(img)
plt.margins(0)
plt.axis("off")
plt.tight_layout()
plt.show()
display_image(50)
anim_file = 'dcgan.gif'
with imageio.get_writer(anim_file, mode="I") as writer:
filenames = glob.glob('image*.png')
filenames = sorted(filenames)
for _, filename in enumerate(filenames):
image = imageio.imread(filename)
writer.append_data(image)
try:
from google.colab import files
except ImportError:
pass
else:
files.download(anim_file)
|
_____no_output_____
|
MIT
|
deep_learning/generative/TF2Keras_VanillaGAN_DCGAN.ipynb
|
jiankaiwang/sophia.ml
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.