hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
cbc7152763804834896c66e30547c38b5d9950be
| 8,559 |
ipynb
|
Jupyter Notebook
|
quantum-with-qiskit/Q64_Phase_Kickback_Solutions.ipynb
|
jahadtariq/Quantum-Computing
|
77df163bfc9ec72035f8b3392d450da59710d4a3
|
[
"Apache-2.0",
"CC-BY-4.0"
] | 1 |
2021-07-27T13:39:00.000Z
|
2021-07-27T13:39:00.000Z
|
quantum-with-qiskit/Q64_Phase_Kickback_Solutions.ipynb
|
jahadtariq/Quantum-Computing
|
77df163bfc9ec72035f8b3392d450da59710d4a3
|
[
"Apache-2.0",
"CC-BY-4.0"
] | null | null | null |
quantum-with-qiskit/Q64_Phase_Kickback_Solutions.ipynb
|
jahadtariq/Quantum-Computing
|
77df163bfc9ec72035f8b3392d450da59710d4a3
|
[
"Apache-2.0",
"CC-BY-4.0"
] | 1 |
2021-09-14T09:24:48.000Z
|
2021-09-14T09:24:48.000Z
| 35.077869 | 310 | 0.497722 |
[
[
[
"<a href=\"https://qworld.net\" target=\"_blank\" align=\"left\"><img src=\"../qworld/images/header.jpg\" align=\"left\"></a>\n$ \\newcommand{\\bra}[1]{\\langle #1|} $\n$ \\newcommand{\\ket}[1]{|#1\\rangle} $\n$ \\newcommand{\\braket}[2]{\\langle #1|#2\\rangle} $\n$ \\newcommand{\\dot}[2]{ #1 \\cdot #2} $\n$ \\newcommand{\\biginner}[2]{\\left\\langle #1,#2\\right\\rangle} $\n$ \\newcommand{\\mymatrix}[2]{\\left( \\begin{array}{#1} #2\\end{array} \\right)} $\n$ \\newcommand{\\myvector}[1]{\\mymatrix{c}{#1}} $\n$ \\newcommand{\\myrvector}[1]{\\mymatrix{r}{#1}} $\n$ \\newcommand{\\mypar}[1]{\\left( #1 \\right)} $\n$ \\newcommand{\\mybigpar}[1]{ \\Big( #1 \\Big)} $\n$ \\newcommand{\\sqrttwo}{\\frac{1}{\\sqrt{2}}} $\n$ \\newcommand{\\dsqrttwo}{\\dfrac{1}{\\sqrt{2}}} $\n$ \\newcommand{\\onehalf}{\\frac{1}{2}} $\n$ \\newcommand{\\donehalf}{\\dfrac{1}{2}} $\n$ \\newcommand{\\hadamard}{ \\mymatrix{rr}{ \\sqrttwo & \\sqrttwo \\\\ \\sqrttwo & -\\sqrttwo }} $\n$ \\newcommand{\\vzero}{\\myvector{1\\\\0}} $\n$ \\newcommand{\\vone}{\\myvector{0\\\\1}} $\n$ \\newcommand{\\stateplus}{\\myvector{ \\sqrttwo \\\\ \\sqrttwo } } $\n$ \\newcommand{\\stateminus}{ \\myrvector{ \\sqrttwo \\\\ -\\sqrttwo } } $\n$ \\newcommand{\\myarray}[2]{ \\begin{array}{#1}#2\\end{array}} $\n$ \\newcommand{\\X}{ \\mymatrix{cc}{0 & 1 \\\\ 1 & 0} } $\n$ \\newcommand{\\I}{ \\mymatrix{rr}{1 & 0 \\\\ 0 & 1} } $\n$ \\newcommand{\\Z}{ \\mymatrix{rr}{1 & 0 \\\\ 0 & -1} } $\n$ \\newcommand{\\Htwo}{ \\mymatrix{rrrr}{ \\frac{1}{2} & \\frac{1}{2} & \\frac{1}{2} & \\frac{1}{2} \\\\ \\frac{1}{2} & -\\frac{1}{2} & \\frac{1}{2} & -\\frac{1}{2} \\\\ \\frac{1}{2} & \\frac{1}{2} & -\\frac{1}{2} & -\\frac{1}{2} \\\\ \\frac{1}{2} & -\\frac{1}{2} & -\\frac{1}{2} & \\frac{1}{2} } } $\n$ \\newcommand{\\CNOT}{ \\mymatrix{cccc}{1 & 0 & 0 & 0 \\\\ 0 & 1 & 0 & 0 \\\\ 0 & 0 & 0 & 1 \\\\ 0 & 0 & 1 & 0} } $\n$ \\newcommand{\\norm}[1]{ \\left\\lVert #1 \\right\\rVert } $\n$ \\newcommand{\\pstate}[1]{ \\lceil \\mspace{-1mu} #1 \\mspace{-1.5mu} \\rfloor } $\n$ \\newcommand{\\greenbit}[1] {\\mathbf{{\\color{green}#1}}} $\n$ \\newcommand{\\bluebit}[1] {\\mathbf{{\\color{blue}#1}}} $\n$ \\newcommand{\\redbit}[1] {\\mathbf{{\\color{red}#1}}} $\n$ \\newcommand{\\brownbit}[1] {\\mathbf{{\\color{brown}#1}}} $\n$ \\newcommand{\\blackbit}[1] {\\mathbf{{\\color{black}#1}}} $",
"_____no_output_____"
],
[
"<font style=\"font-size:28px;\" align=\"left\"><b> <font color=\"blue\"> Solutions for </font>Phase Kickback </b></font>\n<br>\n_prepared by Abuzer Yakaryilmaz_\n<br><br>",
"_____no_output_____"
],
[
"<a id=\"task1\"></a>\n<h3> Task 1</h3>\n\nCreate a quantum circuit with two qubits, say $ q[1] $ and $ q[0] $ in the reading order of Qiskit.\n\nWe start in quantum state $ \\ket{01} $:\n- set the state of $ q[1] $ to $ \\ket{0} $, and\n- set the state of $ q[0] $ to $ \\ket{1} $.\n\nApply Hadamard to both qubits.\n\nApply CNOT operator, where the controller qubit is $ q[1] $ and the target qubit is $ q[0] $.\n\nApply Hadamard to both qubits.\n\nMeasure the outcomes.",
"_____no_output_____"
],
[
"<h3> Solution </h3>",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\nq = QuantumRegister(2,\"q\") # quantum register with 2 qubits\nc = ClassicalRegister(2,\"c\") # classical register with 2 bits\n\nqc = QuantumCircuit(q,c) # quantum circuit with quantum and classical registers\n\n# the up qubit is in |0>\n\n# set the down qubit to |1>\nqc.x(q[0]) # apply x-gate (NOT operator)\n\nqc.barrier()\n\n# apply Hadamard to both qubits.\nqc.h(q[0])\nqc.h(q[1])\n\n# apply CNOT operator, where the controller qubit is the up qubit and the target qubit is the down qubit.\nqc.cx(1,0)\n\n# apply Hadamard to both qubits.\nqc.h(q[0])\nqc.h(q[1])\n\n# measure both qubits\nqc.measure(q,c)\n\n# draw the circuit in Qiskit reading order\ndisplay(qc.draw(output='mpl',reverse_bits=True))\n\n# execute the circuit 100 times in the local simulator\njob = execute(qc,Aer.get_backend('qasm_simulator'),shots=100)\ncounts = job.result().get_counts(qc)\nprint(counts)",
"_____no_output_____"
]
],
[
[
"<a id=\"task2\"></a>\n<h3> Task 2 </h3>\n\nCreate a circuit with 7 qubits, say $ q[6],\\ldots,q[0] $ in the reading order of Qiskit.\n\nSet the states of the top six qubits to $ \\ket{0} $.\n\nSet the state of the bottom qubit to $ \\ket{1} $.\n\nApply Hadamard operators to all qubits.\n\nApply CNOT operator ($q[1]$,$q[0]$) \n<br>\nApply CNOT operator ($q[4]$,$q[0]$) \n<br>\nApply CNOT operator ($q[5]$,$q[0]$) \n\nApply Hadamard operators to all qubits.\n\nMeasure all qubits. \n\nFor each CNOT operator, is there a phase-kickback effect?",
"_____no_output_____"
],
[
"<h3> Solution </h3>",
"_____no_output_____"
]
],
[
[
"# import all necessary objects and methods for quantum circuits\nfrom qiskit import QuantumRegister, ClassicalRegister, QuantumCircuit, execute, Aer\n\n# Create a circuit with 7 qubits.\nq = QuantumRegister(7,\"q\") # quantum register with 7 qubits\nc = ClassicalRegister(7) # classical register with 7 bits\n\nqc = QuantumCircuit(q,c) # quantum circuit with quantum and classical registers\n\n# the top six qubits are already in |0>\n\n# set the bottom qubit to |1>\nqc.x(0) # apply x-gate (NOT operator)\n\n# define a barrier\nqc.barrier()\n\n# apply Hadamard to all qubits.\nfor i in range(7):\n qc.h(q[i])\n\n # define a barrier\nqc.barrier()\n\n\n# apply CNOT operator (q[1],q[0]) \n# apply CNOT operator (q[4],q[0]) \n# apply CNOT operator (q[5],q[0]) \nqc.cx(q[1],q[0])\nqc.cx(q[4],q[0])\nqc.cx(q[5],q[0])\n\n# define a barrier\nqc.barrier()\n\n\n# apply Hadamard to all qubits.\nfor i in range(7):\n qc.h(q[i])\n\n# define a barrier\nqc.barrier()\n\n# measure all qubits\nqc.measure(q,c)\n\n# draw the circuit in Qiskit reading order\ndisplay(qc.draw(output='mpl',reverse_bits=True))\n\n# execute the circuit 100 times in the local simulator\njob = execute(qc,Aer.get_backend('qasm_simulator'),shots=100)\ncounts = job.result().get_counts(qc)\nprint(counts)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbc72226d18b169dfae1025fae980b0c7ea1e8fc
| 5,570 |
ipynb
|
Jupyter Notebook
|
image_augmentation.ipynb
|
siyue-zhang/dark-image-enhancement-Pix2Pix
|
5fbf447c14735e6dda7d8679c1f0a5a7e012cbab
|
[
"MIT"
] | 1 |
2021-11-17T05:41:24.000Z
|
2021-11-17T05:41:24.000Z
|
image_augmentation.ipynb
|
siyue-zhang/dark-image-enhancement-Pix2Pix
|
5fbf447c14735e6dda7d8679c1f0a5a7e012cbab
|
[
"MIT"
] | null | null | null |
image_augmentation.ipynb
|
siyue-zhang/dark-image-enhancement-Pix2Pix
|
5fbf447c14735e6dda7d8679c1f0a5a7e012cbab
|
[
"MIT"
] | null | null | null | 29.162304 | 92 | 0.515978 |
[
[
[
"import numpy as np\nimport torch\n\nuse_cuda = torch.cuda.is_available()\ndevice = torch.device(\"cuda:0\" if use_cuda else \"cpu\")\ndevice",
"_____no_output_____"
],
[
"import torchvision\nfrom torchvision import models\nfrom torchvision import transforms\nimport os\nimport glob\nfrom PIL import Image\nfrom torch.utils.data import Dataset, DataLoader\nimport matplotlib.pyplot as plt\n\nfrom torchvision import models\nfrom random import randint\n\n# tensor -> PIL image\nunloader = transforms.ToPILImage()\n\n# flip = transforms.RandomHorizontalFlip(p=1)",
"_____no_output_____"
],
[
"class ToyDataset(Dataset):\n def __init__(self, dark_img_dir, light_img_dir):\n self.dark_img_dir = dark_img_dir\n self.light_img_dir = light_img_dir\n self.n_dark = len(os.listdir(self.dark_img_dir))\n self.n_light = len(os.listdir(self.light_img_dir))\n\n def __len__(self): \n return min(self.n_dark, self.n_light)\n\n def __getitem__(self, idx):\n \n filename = os.listdir(self.light_img_dir)[idx]\n \n light_img_path = f\"{self.light_img_dir}{filename}\"\n light = Image.open(light_img_path).convert(\"RGB\")\n \n dark_img_path = f\"{self.dark_img_dir}{filename}\"\n dark = Image.open(dark_img_path).convert(\"RGB\") \n \n# if random()>0.5:\n# light = transforms.functional.rotate(light, 30)\n# dark = transforms.functional.rotate(dark, 30)\n# if random()>0.5:\n# light = transforms.functional.rotate(light, 330)\n# dark = transforms.functional.rotate(dark, 330)\n# if random()>0.5:\n# light = flip(light)\n# dark = flip(dark)\n\n s = randint(600, 700)\n transform = transforms.Compose([\n transforms.Resize(s),\n transforms.CenterCrop(512),\n transforms.ToTensor(),\n ])\n \n light = transform(light)\n dark = transform(dark) \n\n return dark, light\n\nbatch_size = 1\n \ntrain_dark_dir = f\"./data/train/dark/\"\ntrain_light_dir = f\"./data/train/light/\"\ntraining_set = ToyDataset(train_dark_dir,train_light_dir)\ntraining_generator = DataLoader(training_set, batch_size=batch_size, shuffle=True)\n\nval_dark_dir = f\"./data/test/dark/\"\nval_light_dir = f\"./data/test/light/\"\nvalidation_set = ToyDataset(val_dark_dir, val_light_dir)\nvalidation_generator = DataLoader(validation_set, batch_size=batch_size, shuffle=True)",
"_____no_output_____"
],
[
"# generate training images\nn = 1\ncycle = 5\ndark_save_path = \"./data_augment/train/dark/\"\nlight_save_path = \"./data_augment/train/light/\"\nfor i in range(cycle):\n for item in training_generator:\n dark, light = item\n dark = unloader(dark[0,])\n light = unloader(light[0,])\n dark.save(dark_save_path+f\"{n}.jpg\")\n light.save(light_save_path+f\"{n}.jpg\")\n n += 1\n",
"_____no_output_____"
],
[
"# generate testing images\nn = 1\ncycle = 1\ndark_save_path = \"./data_augment/test/dark/\"\nlight_save_path = \"./data_augment/test/light/\"\nfor i in range(cycle):\n for item in validation_generator:\n dark, light = item\n dark = unloader(dark[0,])\n light = unloader(light[0,])\n dark.save(dark_save_path+f\"{n}.jpg\")\n light.save(light_save_path+f\"{n}.jpg\")\n n += 1",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbc72aabc73a2f841de39846a6b03624709c24cb
| 5,887 |
ipynb
|
Jupyter Notebook
|
exercises/02_power_flow_and_scenario_data.ipynb
|
hmaschke/pandapower-1
|
2e93969050d3d468ce57f73d358e97fabc6e5141
|
[
"BSD-3-Clause"
] | null | null | null |
exercises/02_power_flow_and_scenario_data.ipynb
|
hmaschke/pandapower-1
|
2e93969050d3d468ce57f73d358e97fabc6e5141
|
[
"BSD-3-Clause"
] | null | null | null |
exercises/02_power_flow_and_scenario_data.ipynb
|
hmaschke/pandapower-1
|
2e93969050d3d468ce57f73d358e97fabc6e5141
|
[
"BSD-3-Clause"
] | null | null | null | 23.930894 | 155 | 0.565653 |
[
[
[
"## Exercise 2 - Running a power flow calculation and adding scenario data for electric vehicles to the grid\n\n**The goals for this exercise are:**\n\n- load the grid model from exercise 1\n- run a power flow calculation\n- display transformer, line and bus results\n- determine maximum line loading and minimum bus voltage\n- create 65 loads with random power demands between 0 and 11 kW\n- each load represents an 11 kW charging point for electric vehicles\n- connect these loads to random buses to model a future scenario for the example grid\n- run a power flow calculation again and compare the results before and after connecting the charging points to the grid\n\n**Helpful ressources for this exercise:**\n\n- https://github.com/e2nIEE/pandapower/blob/master/tutorials/minimal_example.ipynb\n- https://github.com/e2nIEE/pandapower/blob/develop/tutorials/create_simple.ipynb\n- https://github.com/e2nIEE/pandapower/blob/develop/tutorials/powerflow.ipynb ",
"_____no_output_____"
],
[
"### Step 1 - load the grid model of exercise 1 from the json file\n\nhint: use pp.from_json(FILENAME.json). You need the import the pandapower module again.",
"_____no_output_____"
],
[
"### Step 2 - run a power flow calculation",
"_____no_output_____"
],
[
"### Step 3 - display the transformer results",
"_____no_output_____"
],
[
"### Step 4 - display the line results",
"_____no_output_____"
],
[
"### Step 5 - display the bus results",
"_____no_output_____"
],
[
"### Step 6 - display the maximum line loading\n\nhint: you can determine the maximum value of a column by running net.TABLE_NAME.COLUMN_NAME.max()",
"_____no_output_____"
],
[
"### Step 7 - display the minimum bus voltage\n\nhint: you can determine the minimum value of a column by running net.TABLE_NAME.COLUMN_NAME.min()",
"_____no_output_____"
],
[
"### Step 8 - create 65 loads with random power demands between 0 and 11 kW and connect them to random buses\n\nhint: you just need to fill in the \"create load\" command in the for loop.",
"_____no_output_____"
]
],
[
[
"# just run this cell to create the list of 50 random power demand values\nimport numpy as np\nnp.random.seed(0)\np_mw_values = list(np.random.randint(0, 12, 65)/1000)\nprint(p_mw_values)",
"_____no_output_____"
],
[
"for p_mw in p_mw_values:\n bus = np.random.randint(2,7,1)[0] # chooses a bus index between 2 and 6\n load = #<replace this by a create_load command. Set the parameters bus=bus p_mw=p_mw and name=\"charging_point\">\nnet.load",
"_____no_output_____"
]
],
[
[
"### Step 9 - run a power flow calculation again, to get the new results for the grid with charging points",
"_____no_output_____"
],
[
"### Step 10 - determine the transformer loading, maximum line loading and minimum bus voltage and compare them to the results without charging points",
"_____no_output_____"
],
[
"### Step 11 - save the grid model as a json file with a new name\n\nhint: use the method pp.to_json(net, \"FILENAME.json\").",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbc74c9ee5eaae96ab62b189e93448a7fad5e365
| 41,890 |
ipynb
|
Jupyter Notebook
|
chapter_04_multilayer-perceptrons/6_numerical-stability-and-init.ipynb
|
lvyufeng/d2l-mindspore
|
e104754e381708d29fd3bb5364ea368a15776e9f
|
[
"MIT"
] | 19 |
2021-10-31T09:25:31.000Z
|
2022-03-30T02:07:51.000Z
|
chapter_04_multilayer-perceptrons/6_numerical-stability-and-init.ipynb
|
lvyufeng/d2l-mindspore
|
e104754e381708d29fd3bb5364ea368a15776e9f
|
[
"MIT"
] | null | null | null |
chapter_04_multilayer-perceptrons/6_numerical-stability-and-init.ipynb
|
lvyufeng/d2l-mindspore
|
e104754e381708d29fd3bb5364ea368a15776e9f
|
[
"MIT"
] | 1 |
2022-01-30T07:48:55.000Z
|
2022-01-30T07:48:55.000Z
| 36.617133 | 295 | 0.454404 |
[
[
[
"# 数值稳定性和模型初始化\n\n梯度消失",
"_____no_output_____"
]
],
[
[
"import sys\nsys.path.append('..')",
"_____no_output_____"
],
[
"%matplotlib inline\nimport mindspore\nimport mindspore.numpy as mnp\nimport mindspore.ops as ops\nfrom d2l import mindspore as d2l\n\ndef sigmoid(x):\n return ops.Sigmoid()(x)\n\nx = mnp.arange(-8.0, 8.0, 0.1)\ny = sigmoid(x)\ngrad_all = ops.GradOperation(get_all=True)\nx_grad = grad_all(sigmoid)(x)[0]\n\nd2l.plot(x.asnumpy(), [y.asnumpy(), x_grad.asnumpy()],\n legend=['sigmoid', 'gradient'], figsize=(4.5, 2.5))",
"[WARNING] OPTIMIZER(3870984,7f87f7fe4740,python):2021-11-05-00:20:32.730.325 [mindspore/ccsrc/frontend/optimizer/ad/dfunctor.cc:803] GetPrimalUser] J operation has no relevant primal call in the same graph. Func graph: 1_after_grad.2, J user: 1_after_grad.2:sigmoid{[0]: 3, [1]: args0}\n"
]
],
[
[
"梯度爆炸",
"_____no_output_____"
]
],
[
[
"from mindspore import Tensor\n\nM = ops.normal((4, 4), Tensor(0), Tensor(1))\nprint('一个矩阵 \\n',M)\nfor i in range(100):\n M = ops.matmul(M, ops.normal((4,4), Tensor(0), Tensor(1)))\n\nprint('乘以100个矩阵后\\n', M)",
"一个矩阵 \n [[-1.3180416 0.21586044 -1.6110967 -0.6184063 ]\n [ 0.36339304 -0.36989677 -0.6256188 -0.4508158 ]\n [-0.12890556 0.03007882 1.1935389 -0.5326446 ]\n [-0.09897626 -1.9187572 0.89836234 -1.4689225 ]]\n乘以100个矩阵后\n [[ 4.8074875e+24 -2.7623282e+24 -1.3187021e+24 1.2426934e+25]\n [-1.4034659e+24 8.0641556e+23 3.8497296e+23 -3.6278361e+24]\n [-3.0235626e+24 1.7373048e+24 8.2936814e+23 -7.8156434e+24]\n [-9.1700892e+24 5.2690288e+24 2.5153709e+24 -2.3703872e+25]]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc74da021276bfa445891aef0be7090b6efe5b9
| 21,085 |
ipynb
|
Jupyter Notebook
|
code_listings/03.04-Missing-Values.ipynb
|
cesar-rocha/PythonDataScienceHandbook
|
96c7f75d49b26a35bf76307bca86533061731859
|
[
"MIT"
] | 1 |
2021-06-02T19:42:47.000Z
|
2021-06-02T19:42:47.000Z
|
code_listings/03.04-Missing-Values.ipynb
|
matt-staton/PythonDataScienceHandbook
|
96c7f75d49b26a35bf76307bca86533061731859
|
[
"MIT"
] | null | null | null |
code_listings/03.04-Missing-Values.ipynb
|
matt-staton/PythonDataScienceHandbook
|
96c7f75d49b26a35bf76307bca86533061731859
|
[
"MIT"
] | 1 |
2019-06-14T13:38:46.000Z
|
2019-06-14T13:38:46.000Z
| 22.055439 | 1,810 | 0.39336 |
[
[
[
"# Handling Missing Data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd",
"_____no_output_____"
],
[
"vals1 = np.array([1, None, 3, 4])\nvals1",
"_____no_output_____"
],
[
"for dtype in ['object', 'int']:\n print(\"dtype =\", dtype)\n %timeit np.arange(1E6, dtype=dtype).sum()\n print()",
"dtype = object\n10 loops, best of 3: 78.2 ms per loop\n\ndtype = int\n100 loops, best of 3: 3.06 ms per loop\n\n"
],
[
"vals1.sum()",
"_____no_output_____"
],
[
"vals2 = np.array([1, np.nan, 3, 4]) \nvals2.dtype",
"_____no_output_____"
],
[
"1 + np.nan",
"_____no_output_____"
],
[
"0 * np.nan",
"_____no_output_____"
],
[
"vals2.sum(), vals2.min(), vals2.max()",
"_____no_output_____"
],
[
"np.nansum(vals2), np.nanmin(vals2), np.nanmax(vals2)",
"_____no_output_____"
],
[
"pd.Series([1, np.nan, 2, None])",
"_____no_output_____"
],
[
"x = pd.Series(range(2), dtype=int)\nx",
"_____no_output_____"
],
[
"x[0] = None\nx",
"_____no_output_____"
],
[
"data = pd.Series([1, np.nan, 'hello', None])",
"_____no_output_____"
],
[
"data.isnull()",
"_____no_output_____"
],
[
"data[data.notnull()]",
"_____no_output_____"
],
[
"data.dropna()",
"_____no_output_____"
],
[
"df = pd.DataFrame([[1, np.nan, 2],\n [2, 3, 5],\n [np.nan, 4, 6]])\ndf",
"_____no_output_____"
],
[
"df.dropna()",
"_____no_output_____"
],
[
"df.dropna(axis='columns')",
"_____no_output_____"
],
[
"df[3] = np.nan\ndf",
"_____no_output_____"
],
[
"df.dropna(axis='columns', how='all')",
"_____no_output_____"
],
[
"df.dropna(axis='rows', thresh=3)",
"_____no_output_____"
],
[
"data = pd.Series([1, np.nan, 2, None, 3], index=list('abcde'))\ndata",
"_____no_output_____"
],
[
"data.fillna(0)",
"_____no_output_____"
],
[
"# forward-fill\ndata.fillna(method='ffill')",
"_____no_output_____"
],
[
"# back-fill\ndata.fillna(method='bfill')",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.fillna(method='ffill', axis=1)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc755c2f5022587b0aab8844ddc44f82e209960
| 5,124 |
ipynb
|
Jupyter Notebook
|
results/pyhpc_benchmarks_colab.ipynb
|
ashwinvis/pyhpc-benchmarks
|
650ecc650e394df829944ffcf09e9d646ec69691
|
[
"Unlicense"
] | null | null | null |
results/pyhpc_benchmarks_colab.ipynb
|
ashwinvis/pyhpc-benchmarks
|
650ecc650e394df829944ffcf09e9d646ec69691
|
[
"Unlicense"
] | null | null | null |
results/pyhpc_benchmarks_colab.ipynb
|
ashwinvis/pyhpc-benchmarks
|
650ecc650e394df829944ffcf09e9d646ec69691
|
[
"Unlicense"
] | null | null | null | 22.473684 | 141 | 0.466432 |
[
[
[
"#pyhpc-benchmarks @ Google Colab\n\nTo run all benchmarks, you need to switch the runtime type to match the corresponding section (CPU, TPU, GPU).",
"_____no_output_____"
]
],
[
[
"!rm -rf pyhpc-benchmarks; git clone https://github.com/dionhaefner/pyhpc-benchmarks.git",
"_____no_output_____"
],
[
"%cd pyhpc-benchmarks",
"_____no_output_____"
],
[
"# check CPU model\n!lscpu |grep 'Model name'",
"_____no_output_____"
]
],
[
[
"## CPU",
"_____no_output_____"
]
],
[
[
"!pip install -U -q numba aesara",
"_____no_output_____"
],
[
"!taskset -c 0 python run.py benchmarks/equation_of_state/",
"_____no_output_____"
],
[
"!taskset -c 0 python run.py benchmarks/isoneutral_mixing/",
"_____no_output_____"
],
[
"!taskset -c 0 python run.py benchmarks/turbulent_kinetic_energy/",
"_____no_output_____"
]
],
[
[
"## TPU\n\nMake sure to set accelerator to \"TPU\" before executing this.",
"_____no_output_____"
]
],
[
[
"import jax.tools.colab_tpu\njax.tools.colab_tpu.setup_tpu()",
"_____no_output_____"
],
[
"!python run.py benchmarks/equation_of_state -b jax -b numpy --device tpu",
"_____no_output_____"
],
[
"!python run.py benchmarks/isoneutral_mixing -b jax -b numpy --device tpu",
"_____no_output_____"
],
[
"!python run.py benchmarks/turbulent_kinetic_energy -b jax -b numpy --device tpu",
"_____no_output_____"
]
],
[
[
"## GPU\n\nMake sure to set accelerator to \"GPU\" before executing this.",
"_____no_output_____"
]
],
[
[
"# get GPU model\n!nvidia-smi -L",
"_____no_output_____"
],
[
"!for backend in jax tensorflow pytorch cupy; do python run.py benchmarks/equation_of_state/ --device gpu -b $backend -b numpy; done",
"_____no_output_____"
],
[
"!for backend in jax pytorch cupy; do python run.py benchmarks/isoneutral_mixing/ --device gpu -b $backend -b numpy; done",
"_____no_output_____"
],
[
"!for backend in jax pytorch; do python run.py benchmarks/turbulent_kinetic_energy/ --device gpu -b $backend -b numpy; done",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbc757f9c352919a71ebf5e357958747c8b47dd4
| 3,262 |
ipynb
|
Jupyter Notebook
|
doc/src/UsingEKFTricksWithSPKF.ipynb
|
shkolnick-kun/yakf
|
772a0ca19a08ddce788f8438e1b38cedc8a1b2d3
|
[
"Apache-2.0"
] | 15 |
2021-04-11T01:04:11.000Z
|
2022-02-06T18:57:47.000Z
|
doc/src/UsingEKFTricksWithSPKF.ipynb
|
shkolnick-kun/yakf
|
772a0ca19a08ddce788f8438e1b38cedc8a1b2d3
|
[
"Apache-2.0"
] | null | null | null |
doc/src/UsingEKFTricksWithSPKF.ipynb
|
shkolnick-kun/yakf
|
772a0ca19a08ddce788f8438e1b38cedc8a1b2d3
|
[
"Apache-2.0"
] | 1 |
2021-04-12T07:44:53.000Z
|
2021-04-12T07:44:53.000Z
| 37.494253 | 244 | 0.530963 |
[
[
[
"# Using EKF tweaks with Sigma Point Kalman Filters\n\nIn Sigma Point Kalman Filters (SPKF, see [**[Merwe2004]**](#merwe)) Weighted Statistical Linear Regression technique is used to approximate nonlinear process and measurement functions:\n\n$\\mathbf{y} = g(\\mathbf{x}) = \\mathbf{A} \\mathbf{x} + \\mathbf{b} + \\mathbf{e}$,\n\n$\\mathbf{P}_{ee} = \\mathbf{P}_{yy} - \\mathbf{A} \\mathbf{P}_{xx} \\mathbf{A}^{\\top}$\n\nwhere: \n\n$\\mathbf{e}$ is an approximation error, \n\n$\\mathbf{A} = \\mathbf{P}_{xy}^{\\top} \\mathbf{P}_{xx}^{-1}$, \n\n$\\mathbf{b} = \\mathbf{\\bar{y}} - \\mathbf{A} \\mathbf{\\bar{x}}$,\n\n$\\mathbf{P}_{xx} = \\displaystyle\\sum_{i} {w}_{ci} \\left( \\mathbf{\\chi}_{i} - \\mathbf{\\bar{x}} \\right) \\left( \\mathbf{\\chi}_{i} - \\mathbf{\\bar{x}} \\right)$,\n\n$\\mathbf{P}_{yy} = \\displaystyle\\sum_{i} {w}_{ci} \\left( \\mathbf{\\gamma}_{i} - \\mathbf{\\bar{y}} \\right) \\left( \\mathbf{\\gamma}_{i} - \\mathbf{\\bar{y}} \\right)$,\n\n$\\mathbf{P}_{xy} = \\displaystyle\\sum_{i} {w}_{ci} \\left( \\mathbf{\\chi}_{i} - \\mathbf{\\bar{x}} \\right) \\left( \\mathbf{\\gamma}_{i} - \\mathbf{\\bar{y}} \\right)$,\n\n$\\mathbf{\\gamma}_{i} = g(\\mathbf{\\chi}_{i})$\n\n$\\mathbf{\\bar{x}} = \\displaystyle\\sum_{i} {w}_{mi} \\mathbf{\\chi}_{i}$,\n\n$\\mathbf{\\bar{y}} = \\displaystyle\\sum_{i} {w}_{mi} \\mathbf{\\gamma}_{i}$,\n\n${w}_{ci}$ are covariation weghts, ${w}_{mi}$ are mean weights.\n\nThis means that approximation errors of measurements may be treated as a part of additive noise and so we show that in SPKF we can use the following approximation of $\\mathbf{S}_{k}$:\n\n$\\mathbf{S}_{k} = \\mathbf{H}_{k} \\mathbf{P}_{k|k-1} \\mathbf{H}_{k}^{\\top} + \\mathbf{\\tilde{R}}_{k}$, \n\nwhere \n\n$\\mathbf{H}_{k} = \\mathbf{P}_{xz, k}^{\\top} \\mathbf{P}_{xx, k}^{-1}$,\n\n$\\mathbf{\\tilde{R}}_{k} = \\mathbf{R}_{k} + \\mathbf{P}_{ee, k}$,\n\nthis enables us to use some EKF tricks such as adaptive correction or generalized linear models with SPKF.",
"_____no_output_____"
],
[
"# References\n\n<a name=\"merwe\"></a>**\\[Merwe2004\\]** R. van der Merwe, \"Sigma-Point Kalman Filters for ProbabilisticInference in Dynamic State-Space Models\", PhD Thesis, OGI School of Science & Engineering, Oregon Health & Science University, USA",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown"
]
] |
cbc759d475d073d5407df1b3d469cf921b875512
| 11,710 |
ipynb
|
Jupyter Notebook
|
Notebooks/Imports.ipynb
|
theandygross/TCGA_differential_expression
|
a9e006aed526a4ae6afc768fad5dbdc421a320cd
|
[
"MIT"
] | 5 |
2017-01-15T18:53:14.000Z
|
2018-08-24T04:46:48.000Z
|
Notebooks/Imports.ipynb
|
theandygross/TCGA_differential_expression
|
a9e006aed526a4ae6afc768fad5dbdc421a320cd
|
[
"MIT"
] | null | null | null |
Notebooks/Imports.ipynb
|
theandygross/TCGA_differential_expression
|
a9e006aed526a4ae6afc768fad5dbdc421a320cd
|
[
"MIT"
] | 4 |
2018-02-20T10:31:08.000Z
|
2022-03-30T22:49:07.000Z
| 25.074946 | 236 | 0.514261 |
[
[
[
"# Global Imports",
"_____no_output_____"
]
],
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nfrom matplotlib.pyplot import subplots",
"_____no_output_____"
]
],
[
[
"### External Package Imports",
"_____no_output_____"
]
],
[
[
"import os as os\nimport pickle as pickle\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"### Module Imports",
"_____no_output_____"
],
[
"Here I am using a few of my own packages, they are availible on Github under [__theandygross__](https://github.com/theandygross) and should all be instalable by <code>python setup.py</code>.",
"_____no_output_____"
]
],
[
[
"from Stats.Scipy import *\nfrom Stats.Survival import *\n\nfrom Helpers.Pandas import *\nfrom Helpers.LinAlg import *\n\nfrom Figures.FigureHelpers import *\nfrom Figures.Pandas import *\nfrom Figures.Boxplots import *\nfrom Figures.Regression import *\n#from Figures.Survival import draw_survival_curve, survival_and_stats\n#from Figures.Survival import draw_survival_curves\n#from Figures.Survival import survival_stat_plot",
"_____no_output_____"
],
[
"import Data.Firehose as FH\nfrom Data.Containers import get_run",
"_____no_output_____"
]
],
[
[
"### Import Global Parameters \n* These need to be changed before you will be able to sucessfully run this code",
"_____no_output_____"
]
],
[
[
"import NotebookImport\nfrom Global_Parameters import *",
"_____no_output_____"
]
],
[
[
"### Tweaking Display Parameters",
"_____no_output_____"
]
],
[
[
"pd.set_option('precision', 3)\npd.set_option('display.width', 300)\nplt.rcParams['font.size'] = 12",
"_____no_output_____"
],
[
"'''Color schemes for paper taken from http://colorbrewer2.org/'''\ncolors = plt.rcParams['axes.color_cycle']\ncolors_st = ['#CA0020', '#F4A582', '#92C5DE', '#0571B0']\ncolors_th = ['#E66101', '#FDB863', '#B2ABD2', '#5E3C99']",
"_____no_output_____"
],
[
"import seaborn as sns\nsns.set_context('paper',font_scale=1.5)\nsns.set_style('white')",
"_____no_output_____"
]
],
[
[
"### Read in All of the Expression Data",
"_____no_output_____"
],
[
"This reads in data that was pre-processed in the [./Preprocessing/init_RNA](../Notebooks/init_RNA.ipynb) notebook.",
"_____no_output_____"
]
],
[
[
"codes = pd.read_hdf(RNA_SUBREAD_STORE, 'codes')\nmatched_tn = pd.read_hdf(RNA_SUBREAD_STORE, 'matched_tn')\nrna_df = pd.read_hdf(RNA_SUBREAD_STORE, 'all_rna')",
"_____no_output_____"
],
[
"data_portal = pd.read_hdf(RNA_STORE, 'matched_tn')\ngenes = data_portal.index.intersection(matched_tn.index)\npts = data_portal.columns.intersection(matched_tn.columns)\nrna_df = rna_df.ix[genes]\nmatched_tn = matched_tn.ix[genes, pts]",
"_____no_output_____"
]
],
[
[
"### Read in Gene-Sets for GSEA",
"_____no_output_____"
]
],
[
[
"from Data.Annotations import unstack_geneset_csv\n\ngene_sets = unstack_geneset_csv(GENE_SETS)\ngene_sets = gene_sets.ix[rna_df.index].fillna(0)",
"_____no_output_____"
]
],
[
[
"Initialize function for calling model-based gene set enrichment",
"_____no_output_____"
]
],
[
[
"from rpy2 import robjects\nfrom rpy2.robjects import pandas2ri\npandas2ri.activate()\n\nmgsa = robjects.packages.importr('mgsa')",
"_____no_output_____"
],
[
"gs_r = robjects.ListVector({i: robjects.StrVector(list(ti(g>0))) for i,g in \n gene_sets.iteritems()})\ndef run_mgsa(vec):\n v = robjects.r.c(*ti(vec))\n r = mgsa.mgsa(v, gs_r)\n res = pandas2ri.ri2pandas(mgsa.setsResults(r))\n return res",
"_____no_output_____"
]
],
[
[
"### Function Tweaks ",
"_____no_output_____"
],
[
"Running the binomial test across 450k probes in the same test space, we rerun the same test a lot. Here I memoize the function to cache results and not recompute them. This eats up a couple GB of memory but should be reasonable.",
"_____no_output_____"
]
],
[
[
"from scipy.stats import binom_test\n\ndef memoize(f):\n memo = {}\n def helper(x,y,z):\n if (x,y,z) not in memo: \n memo[(x,y,z)] = f(x,y,z)\n return memo[(x,y,z)]\n return helper\n\nbinom_test_mem = memoize(binom_test)\n\ndef binomial_test_screen(df, fc=1.5, p=.5):\n \"\"\"\n Run a binomial test on a DataFrame.\n\n df:\n DataFrame of measurements. Should have a multi-index with\n subjects on the first level and tissue type ('01' or '11')\n on the second level.\n fc:\n Fold-chance cutoff to use\n \"\"\"\n a, b = df.xs('01', 1, 1), df.xs('11', 1, 1)\n dx = a - b\n dx = dx[dx.abs() > np.log2(fc)]\n n = dx.count(1)\n counts = (dx > 0).sum(1)\n cn = pd.concat([counts, n], 1)\n cn = cn[cn.sum(1) > 0]\n b_test = cn.apply(lambda s: binom_test_mem(s[0], s[1], p), axis=1)\n dist = (1.*cn[0] / cn[1])\n tab = pd.concat([cn[0], cn[1], dist, b_test],\n keys=['num_ox', 'num_dx', 'frac', 'p'],\n axis=1)\n return tab",
"_____no_output_____"
]
],
[
[
"Added linewidth and number of bins arguments. This should get pushed eventually. ",
"_____no_output_____"
]
],
[
[
"def draw_dist(vec, split=None, ax=None, legend=True, colors=None, lw=2, bins=300):\n \"\"\"\n Draw a smooth distribution from data with an optional splitting factor.\n \"\"\"\n _, ax = init_ax(ax)\n if split is None:\n split = pd.Series('s', index=vec.index)\n colors = {'s': colors} if colors is not None else None\n for l,v in vec.groupby(split):\n if colors is None:\n smooth_dist(v, bins=bins).plot(label=l, lw=lw, ax=ax)\n else:\n smooth_dist(v, bins=bins).plot(label=l, lw=lw, ax=ax, color=colors[l])\n if legend and len(split.unique()) > 1:\n ax.legend(loc='upper left', frameon=False)",
"_____no_output_____"
]
],
[
[
"Some helper functions for fast calculation of odds ratios on matricies.",
"_____no_output_____"
]
],
[
[
"def odds_ratio_df(a,b):\n a = a.astype(int)\n b = b.astype(int)\n flip = lambda v: (v == 0).astype(int)\n\n a11 = (a.add(b) == 2).sum(axis=1)\n a10 = (a.add(flip(b)) == 2).sum(axis=1)\n a01 = (flip(a).add(b) == 2).sum(axis=1)\n a00 = (flip(a).add(flip(b)) == 2).sum(axis=1)\n odds_ratio = (1.*a11 * a00) / (1.*a10 * a01)\n df = pd.concat([a00, a01, a10, a11], axis=1,\n keys=['00','01','10','11'])\n return odds_ratio, df\n\ndef fet(s):\n odds, p = stats.fisher_exact([[s['00'],s['01']],\n [s['10'],s['11']]])\n return p",
"_____no_output_____"
]
],
[
[
"#### filter_pathway_hits",
"_____no_output_____"
]
],
[
[
"def filter_pathway_hits(hits, gs, cutoff=.00001):\n '''\n Takes a vector of p-values and a DataFrame of binary defined gene-sets. \n Uses the ordering defined by hits to do a greedy filtering on the gene sets. \n '''\n l = [hits.index[0]]\n for gg in hits.index:\n flag = 0\n for g2 in l:\n if gg in l:\n flag = 1\n break\n elif (chi2_cont_test(gs[gg], gs[g2])['p'] < cutoff):\n flag = 1\n break\n if flag == 0:\n l.append(gg)\n hits_filtered = hits.ix[l]\n return hits_filtered",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc75d89a95b62e0d8943b1553dd3660594a889b
| 21,248 |
ipynb
|
Jupyter Notebook
|
03-pipelines/kale/taxi-cab-classification_[TF]/taxicab_pipeline.ipynb
|
MartijnVanAndel/kubeflow_examples
|
68329ab47499dc10e55bb33827315113b8bb2a73
|
[
"Apache-2.0"
] | null | null | null |
03-pipelines/kale/taxi-cab-classification_[TF]/taxicab_pipeline.ipynb
|
MartijnVanAndel/kubeflow_examples
|
68329ab47499dc10e55bb33827315113b8bb2a73
|
[
"Apache-2.0"
] | 22 |
2020-02-20T15:27:01.000Z
|
2022-02-10T01:26:52.000Z
|
03-pipelines/kale/taxi-cab-classification_[TF]/.ipynb_checkpoints/taxicab_pipeline-checkpoint.ipynb
|
MartijnVanAndel/kubeflow_examples
|
68329ab47499dc10e55bb33827315113b8bb2a73
|
[
"Apache-2.0"
] | null | null | null | 34.947368 | 242 | 0.582643 |
[
[
[
"## Taxi Cab Classification (prior to TF2)",
"_____no_output_____"
],
[
"This notebook presents a simplified version of Kubeflow's *taxi cab clasification* pipeline, built upon TFX components.\n\nHere all the pipeline components are stripped down to their core to showcase how to run it in a self-contained local Juyter Noteobok.\n\nAdditionally, the pipeline has been upgraded to work with Python3 and all major libraries (Tensorflow, Tensorflow Transform, Tensorflow Model Analysis, Tensorflow Data Validation, Apache Beam) have been bumped to their latests versions.",
"_____no_output_____"
]
],
[
[
"!pip install tensorflow==1.15.0 --user",
"_____no_output_____"
],
[
"!pip install apache_beam tensorflow_transform tensorflow_model_analysis tensorflow_data_validation --user",
"_____no_output_____"
]
],
[
[
"You may have to restart the workbook after installing these packages",
"_____no_output_____"
]
],
[
[
"import os\nimport shutil\nimport logging\nimport apache_beam as beam\nimport tensorflow as tf\nimport tensorflow_transform as tft\nimport tensorflow_model_analysis as tfma\nimport tensorflow_data_validation as tfdv\n\nfrom apache_beam.io import textio\nfrom apache_beam.io import tfrecordio\n\nfrom tensorflow_transform.beam import impl as beam_impl\nfrom tensorflow_transform.beam.tft_beam_io import transform_fn_io\nfrom tensorflow_transform.coders.csv_coder import CsvCoder\nfrom tensorflow_transform.coders.example_proto_coder import ExampleProtoCoder\nfrom tensorflow_transform.tf_metadata import dataset_metadata\nfrom tensorflow_transform.tf_metadata import metadata_io",
"_____no_output_____"
],
[
"DATA_DIR = 'data/'\nTRAIN_DATA = os.path.join(DATA_DIR, 'taxi-cab-classification/train.csv')\nEVALUATION_DATA = os.path.join(DATA_DIR, 'taxi-cab-classification/eval.csv')\n\n# Categorical features are assumed to each have a maximum value in the dataset.\nMAX_CATEGORICAL_FEATURE_VALUES = [24, 31, 12]\nCATEGORICAL_FEATURE_KEYS = ['trip_start_hour', 'trip_start_day', 'trip_start_month']\n\nDENSE_FLOAT_FEATURE_KEYS = ['trip_miles', 'fare', 'trip_seconds']\n\n# Number of buckets used by tf.transform for encoding each feature.\nFEATURE_BUCKET_COUNT = 10\n\nBUCKET_FEATURE_KEYS = ['pickup_latitude', 'pickup_longitude', 'dropoff_latitude', 'dropoff_longitude']\n\n# Number of vocabulary terms used for encoding VOCAB_FEATURES by tf.transform\nVOCAB_SIZE = 1000\n\n# Count of out-of-vocab buckets in which unrecognized VOCAB_FEATURES are hashed.\nOOV_SIZE = 10\n\nVOCAB_FEATURE_KEYS = ['pickup_census_tract', 'dropoff_census_tract', 'payment_type', 'company',\n 'pickup_community_area', 'dropoff_community_area']\n\n# allow nan values in these features.\nOPTIONAL_FEATURES = ['dropoff_latitude', 'dropoff_longitude', 'pickup_census_tract', 'dropoff_census_tract',\n 'company', 'trip_seconds', 'dropoff_community_area']\n\nLABEL_KEY = 'tips'\nFARE_KEY = 'fare'",
"_____no_output_____"
],
[
"# training parameters\nEPOCHS = 1\nSTEPS = 3\nBATCH_SIZE = 32\nHIDDEN_LAYER_SIZE = '1500'\nLEARNING_RATE = 0.1",
"_____no_output_____"
],
[
"tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.INFO)\n# tf.get_logger().setLevel(logging.ERROR)",
"_____no_output_____"
]
],
[
[
"#### Data Validation",
"_____no_output_____"
],
[
"For an overview of the TFDV functions: https://www.tensorflow.org/tfx/tutorials/data_validation/chicago_taxi",
"_____no_output_____"
]
],
[
[
"vldn_output = os.path.join(DATA_DIR, 'validation')\n\n# TODO: Understand why this was used in the conversion to the output json\n# key columns: list of the names for columns that should be treated as unique keys.\nkey_columns = ['trip_start_timestamp']\n\n# read the first line of the cvs to have and ordered list of column names \n# (the Schema will scrable the features)\nwith open(TRAIN_DATA) as f:\n column_names = f.readline().strip().split(',')\n\nstats = tfdv.generate_statistics_from_csv(data_location=TRAIN_DATA)\nschema = tfdv.infer_schema(stats)\n\neval_stats = tfdv.generate_statistics_from_csv(data_location=EVALUATION_DATA)\nanomalies = tfdv.validate_statistics(eval_stats, schema)\n\n# Log anomalies\nfor feature_name, anomaly_info in anomalies.anomaly_info.items():\n logging.getLogger().error(\n 'Anomaly in feature \"{}\": {}'.format(\n feature_name, anomaly_info.description))\n \n# show inferred schema\ntfdv.display_schema(schema=schema)",
"_____no_output_____"
],
[
"# Resolve anomalies\ncompany = tfdv.get_feature(schema, 'company')\ncompany.distribution_constraints.min_domain_mass = 0.9\n\n# Add new value to the domain of feature payment_type.\npayment_type_domain = tfdv.get_domain(schema, 'payment_type')\npayment_type_domain.value.append('Prcard')\n\n# Validate eval stats after updating the schema \nupdated_anomalies = tfdv.validate_statistics(eval_stats, schema)\ntfdv.display_anomalies(updated_anomalies)",
"_____no_output_____"
]
],
[
[
"#### Data Transformation",
"_____no_output_____"
],
[
"For an overview of the TFT functions: https://www.tensorflow.org/tfx/tutorials/transform/simple",
"_____no_output_____"
]
],
[
[
"def to_dense(tensor):\n \"\"\"Takes as input a SparseTensor and return a Tensor with correct default value\n Args:\n tensor: tf.SparseTensor\n Returns:\n tf.Tensor with default value\n \"\"\"\n if not isinstance(tensor, tf.sparse.SparseTensor):\n return tensor\n if tensor.dtype == tf.string:\n default_value = ''\n elif tensor.dtype == tf.float32:\n default_value = 0.0\n elif tensor.dtype == tf.int32:\n default_value = 0\n else:\n raise ValueError(f\"Tensor type not recognized: {tensor.dtype}\")\n\n return tf.squeeze(tf.sparse_to_dense(tensor.indices,\n [tensor.dense_shape[0], 1],\n tensor.values, default_value=default_value), axis=1)\n # TODO: Update to below version\n # return tf.squeeze(tf.sparse.to_dense(tensor, default_value=default_value), axis=1)\n\n\ndef preprocess_fn(inputs):\n \"\"\"tf.transform's callback function for preprocessing inputs.\n Args:\n inputs: map from feature keys to raw not-yet-transformed features.\n Returns:\n Map from string feature key to transformed feature operations.\n \"\"\"\n outputs = {}\n for key in DENSE_FLOAT_FEATURE_KEYS:\n # Preserve this feature as a dense float, setting nan's to the mean.\n outputs[key] = tft.scale_to_z_score(to_dense(inputs[key]))\n\n for key in VOCAB_FEATURE_KEYS:\n # Build a vocabulary for this feature.\n if inputs[key].dtype == tf.string:\n vocab_tensor = to_dense(inputs[key])\n else:\n vocab_tensor = tf.as_string(to_dense(inputs[key]))\n outputs[key] = tft.compute_and_apply_vocabulary(\n vocab_tensor, vocab_filename='vocab_' + key,\n top_k=VOCAB_SIZE, num_oov_buckets=OOV_SIZE)\n\n for key in BUCKET_FEATURE_KEYS:\n outputs[key] = tft.bucketize(to_dense(inputs[key]), FEATURE_BUCKET_COUNT)\n\n for key in CATEGORICAL_FEATURE_KEYS:\n outputs[key] = tf.cast(to_dense(inputs[key]), tf.int64)\n\n taxi_fare = to_dense(inputs[FARE_KEY])\n taxi_tip = to_dense(inputs[LABEL_KEY])\n # Test if the tip was > 20% of the fare.\n tip_threshold = tf.multiply(taxi_fare, tf.constant(0.2))\n outputs[LABEL_KEY] = tf.logical_and(\n tf.logical_not(tf.math.is_nan(taxi_fare)),\n tf.greater(taxi_tip, tip_threshold))\n\n for key in outputs:\n if outputs[key].dtype == tf.bool:\n outputs[key] = tft.compute_and_apply_vocabulary(tf.as_string(outputs[key]),\n vocab_filename='vocab_' + key)\n \n return outputs",
"_____no_output_____"
],
[
"trns_output = os.path.join(DATA_DIR, \"transformed\")\nif os.path.exists(trns_output):\n shutil.rmtree(trns_output)\n\ntft_input_metadata = dataset_metadata.DatasetMetadata(schema)\n\nrunner = 'DirectRunner'\nwith beam.Pipeline(runner, options=None) as p:\n with beam_impl.Context(temp_dir=os.path.join(trns_output, 'tmp')):\n converter = CsvCoder(column_names, tft_input_metadata.schema)\n\n # READ TRAIN DATA\n train_data = (\n p\n | 'ReadTrainData' >> textio.ReadFromText(TRAIN_DATA, skip_header_lines=1)\n | 'DecodeTrainData' >> beam.Map(converter.decode))\n\n # TRANSFORM TRAIN DATA (and get transform_fn function)\n transformed_dataset, transform_fn = (\n (train_data, tft_input_metadata) | beam_impl.AnalyzeAndTransformDataset(preprocess_fn))\n transformed_data, transformed_metadata = transformed_dataset\n\n # SAVE TRANSFORMED TRAIN DATA\n _ = transformed_data | 'WriteTrainData' >> tfrecordio.WriteToTFRecord(\n os.path.join(trns_output, 'train'),\n coder=ExampleProtoCoder(transformed_metadata.schema))\n\n # READ EVAL DATA\n eval_data = (\n p\n | 'ReadEvalData' >> textio.ReadFromText(EVALUATION_DATA, skip_header_lines=1)\n | 'DecodeEvalData' >> beam.Map(converter.decode))\n\n # TRANSFORM EVAL DATA (using previously created transform_fn function)\n eval_dataset = (eval_data, tft_input_metadata)\n transformed_eval_data, transformed_metadata = (\n (eval_dataset, transform_fn) | beam_impl.TransformDataset())\n\n # SAVE EVAL DATA\n _ = transformed_eval_data | 'WriteEvalData' >> tfrecordio.WriteToTFRecord(\n os.path.join(trns_output, 'eval'),\n coder=ExampleProtoCoder(transformed_metadata.schema))\n\n # SAVE transform_fn FUNCTION FOR LATER USE\n # TODO: check out what is the transform function (transform_fn) that came from previous step\n _ = (transform_fn | 'WriteTransformFn' >> transform_fn_io.WriteTransformFn(trns_output))\n\n # SAVE TRANSFORMED METADATA\n metadata_io.write_metadata(\n metadata=tft_input_metadata,\n path=os.path.join(trns_output, 'metadata'))",
"_____no_output_____"
]
],
[
[
"#### Train",
"_____no_output_____"
],
[
"Estimator API: https://www.tensorflow.org/guide/premade_estimators",
"_____no_output_____"
]
],
[
[
"def training_input_fn(transformed_output, transformed_examples, batch_size, target_name):\n \"\"\"\n Args:\n transformed_output: tft.TFTransformOutput\n transformed_examples: Base filename of examples\n batch_size: Batch size.\n target_name: name of the target column.\n Returns:\n The input function for training or eval.\n \"\"\"\n dataset = tf.data.experimental.make_batched_features_dataset(\n file_pattern=transformed_examples,\n batch_size=batch_size,\n features=transformed_output.transformed_feature_spec(),\n reader=tf.data.TFRecordDataset,\n shuffle=True)\n transformed_features = dataset.make_one_shot_iterator().get_next()\n transformed_labels = transformed_features.pop(target_name)\n return transformed_features, transformed_labels\n\n\ndef get_feature_columns():\n \"\"\"Callback that returns a list of feature columns for building a tf.estimator.\n Returns:\n A list of tf.feature_column.\n \"\"\"\n return (\n [tf.feature_column.numeric_column(key, shape=()) for key in DENSE_FLOAT_FEATURE_KEYS] +\n [tf.feature_column.indicator_column(tf.feature_column.categorical_column_with_identity(key, num_buckets=VOCAB_SIZE + OOV_SIZE)) for key in VOCAB_FEATURE_KEYS] +\n [tf.feature_column.indicator_column(tf.feature_column.categorical_column_with_identity(key, num_buckets=FEATURE_BUCKET_COUNT, default_value=0)) for key in BUCKET_FEATURE_KEYS] +\n [tf.feature_column.indicator_column(tf.feature_column.categorical_column_with_identity(key, num_buckets=num_buckets, default_value=0)) for key, num_buckets in zip(CATEGORICAL_FEATURE_KEYS, MAX_CATEGORICAL_FEATURE_VALUES)]\n )",
"_____no_output_____"
],
[
"training_output = os.path.join(DATA_DIR, \"training\")\nif os.path.exists(training_output):\n shutil.rmtree(training_output)\n\nhidden_layer_size = [int(x.strip()) for x in HIDDEN_LAYER_SIZE.split(',')]\n\ntf_transform_output = tft.TFTransformOutput(trns_output)\n\n# Set how often to run checkpointing in terms of steps.\nconfig = tf.estimator.RunConfig(save_checkpoints_steps=1000)\nn_classes = tf_transform_output.vocabulary_size_by_name(\"vocab_\" + LABEL_KEY)\n# Create estimator\nestimator = tf.estimator.DNNClassifier(\n feature_columns=get_feature_columns(),\n hidden_units=hidden_layer_size,\n n_classes=n_classes,\n config=config,\n model_dir=training_output)\n\n# TODO: Simplify all this: https://www.tensorflow.org/guide/premade_estimators",
"_____no_output_____"
],
[
"estimator.train(input_fn=lambda: training_input_fn(\n tf_transform_output, \n os.path.join(trns_output, 'train' + '*'),\n BATCH_SIZE, \n \"tips\"), \n steps=STEPS)",
"_____no_output_____"
],
[
"eval_result = estimator.evaluate(input_fn=lambda: training_input_fn(\n tf_transform_output, \n os.path.join(trns_output, 'eval' + '*'),\n BATCH_SIZE, \n \"tips\"), \n steps=50)\n\nprint(eval_result)",
"_____no_output_____"
]
],
[
[
"#### Model Analysis",
"_____no_output_____"
],
[
"TF Model Analysis docs: https://www.tensorflow.org/tfx/model_analysis/get_started",
"_____no_output_____"
]
],
[
[
"# TODO: Implement model load and params analysis\n\ndef eval_input_receiver_fn(transformed_output):\n \"\"\"Build everything needed for the tf-model-analysis to run the model.\n Args:\n transformed_output: tft.TFTransformOutput\n Returns:\n EvalInputReceiver function, which contains:\n - Tensorflow graph which parses raw untranformed features, applies the\n tf-transform preprocessing operators.\n - Set of raw, untransformed features.\n - Label against which predictions will be compared.\n \"\"\"\n serialized_tf_example = tf.compat.v1.placeholder(\n dtype=tf.string, shape=[None], name='input_example_tensor')\n features = tf.io.parse_example(serialized_tf_example, transformed_output.raw_feature_spec())\n transformed_features = transformed_output.transform_raw_features(features)\n receiver_tensors = {'examples': serialized_tf_example}\n return tfma.export.EvalInputReceiver(\n features=transformed_features,\n receiver_tensors=receiver_tensors,\n labels=transformed_features[LABEL_KEY])\n\n# EXPORT MODEL\neval_model_dir = os.path.join(training_output, 'tfma_eval_model_dir')\ntfma.export.export_eval_savedmodel(\n estimator=estimator,\n export_dir_base=eval_model_dir,\n eval_input_receiver_fn=(lambda: eval_input_receiver_fn(tf_transform_output)))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
] |
cbc76a56eb82437d6c13e82ee7d03c8b4a745bed
| 52,198 |
ipynb
|
Jupyter Notebook
|
notebooks/Advanced_Usage.ipynb
|
vishalbelsare/skorch
|
9ac71e2acf8b814b65ee84ee6696e7441737e820
|
[
"BSD-3-Clause"
] | 2,748 |
2019-03-19T11:43:01.000Z
|
2022-03-31T13:55:28.000Z
|
notebooks/Advanced_Usage.ipynb
|
vishalbelsare/skorch
|
9ac71e2acf8b814b65ee84ee6696e7441737e820
|
[
"BSD-3-Clause"
] | 392 |
2019-03-19T11:17:04.000Z
|
2022-03-29T21:36:53.000Z
|
notebooks/Advanced_Usage.ipynb
|
vishalbelsare/skorch
|
9ac71e2acf8b814b65ee84ee6696e7441737e820
|
[
"BSD-3-Clause"
] | 197 |
2019-03-27T09:18:25.000Z
|
2022-03-27T00:15:23.000Z
| 33.120558 | 494 | 0.53912 |
[
[
[
"# Advanced usage",
"_____no_output_____"
],
[
"This notebook shows some more advanced features of `skorch`. More examples will be added with time.\n\n<table align=\"left\"><td>\n<a target=\"_blank\" href=\"https://colab.research.google.com/github/skorch-dev/skorch/blob/master/notebooks/Advanced_Usage.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a> \n</td><td>\n<a target=\"_blank\" href=\"https://github.com/skorch-dev/skorch/blob/master/notebooks/Advanced_Usage.ipynb\"><img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a></td></table>",
"_____no_output_____"
],
[
"### Table of contents",
"_____no_output_____"
],
[
"* [Setup](#Setup)\n* [Callbacks](#Callbacks)\n * [Writing your own callback](#Writing-a-custom-callback)\n * [Accessing callback parameters](#Accessing-callback-parameters)\n* [Working with different data types](#Working-with-different-data-types)\n * [Working with datasets](#Working-with-Datasets)\n * [Working with dicts](#Working-with-dicts)\n* [Multiple return values](#Multiple-return-values-from-forward)\n * [Implementing a simple autoencoder](#Implementing-a-simple-autoencoder)\n * [Training the autoencoder](#Training-the-autoencoder)\n * [Extracting the decoder and the encoder output](#Extracting-the-decoder-and-the-encoder-output)",
"_____no_output_____"
]
],
[
[
"! [ ! -z \"$COLAB_GPU\" ] && pip install torch skorch",
"_____no_output_____"
],
[
"import torch\nfrom torch import nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"torch.manual_seed(0)\ntorch.cuda.manual_seed(0)",
"_____no_output_____"
]
],
[
[
"## Setup",
"_____no_output_____"
],
[
"### A toy binary classification task",
"_____no_output_____"
],
[
"We load a toy classification task from `sklearn`.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.datasets import make_classification",
"_____no_output_____"
],
[
"np.random.seed(0)",
"_____no_output_____"
],
[
"X, y = make_classification(1000, 20, n_informative=10, random_state=0)\nX, y = X.astype(np.float32), y.astype(np.int64)",
"_____no_output_____"
],
[
"X.shape, y.shape, y.mean()",
"_____no_output_____"
]
],
[
[
"### Definition of the `pytorch` classification `module`",
"_____no_output_____"
],
[
"We define a vanilla neural network with two hidden layers. The output layer should have 2 output units since there are two classes. In addition, it should have a softmax nonlinearity, because later, when calling `predict_proba`, the output from the `forward` call will be used.",
"_____no_output_____"
]
],
[
[
"from skorch import NeuralNetClassifier",
"_____no_output_____"
],
[
"class ClassifierModule(nn.Module):\n def __init__(\n self,\n num_units=10,\n nonlin=F.relu,\n dropout=0.5,\n ):\n super(ClassifierModule, self).__init__()\n self.num_units = num_units\n self.nonlin = nonlin\n self.dropout = dropout\n\n self.dense0 = nn.Linear(20, num_units)\n self.nonlin = nonlin\n self.dropout = nn.Dropout(dropout)\n self.dense1 = nn.Linear(num_units, 10)\n self.output = nn.Linear(10, 2)\n\n def forward(self, X, **kwargs):\n X = self.nonlin(self.dense0(X))\n X = self.dropout(X)\n X = F.relu(self.dense1(X))\n X = F.softmax(self.output(X), dim=-1)\n return X",
"_____no_output_____"
]
],
[
[
"## Callbacks",
"_____no_output_____"
],
[
"Callbacks are a powerful and flexible way to customize the behavior of your neural network. They are all called at specific points during the model training, e.g. when training starts, or after each batch. Have a look at the `skorch.callbacks` module to see the callbacks that are already implemented.",
"_____no_output_____"
],
[
"### Writing a custom callback",
"_____no_output_____"
],
[
"Although `skorch` comes with a handful of useful callbacks, you may find that you would like to write your own callbacks. Doing so is straightforward, just remember these rules:\n* They should inherit from `skorch.callbacks.Callback`.\n* They should implement at least one of the `on_`-methods provided by the parent class (e.g. `on_batch_begin` or `on_epoch_end`).\n* As argument, the `on_`-methods first get the `NeuralNet` instance, and, where appropriate, the local data (e.g. the data from the current batch). The method should also have `**kwargs` in the signature for potentially unused arguments.\n* *Optional*: If you have attributes that should be reset when the model is re-initialized, those attributes should be set in the `initialize` method.",
"_____no_output_____"
],
[
"Here is an example of a callback that remembers at which epoch the validation accuracy reached a certain value. Then, when training is finished, it calls a mock Twitter API and tweets that epoch. We proceed as follows:\n* We set the desired minimum accuracy during `__init__`.\n* We set the critical epoch during `initialize`.\n* After each epoch, if the critical accuracy has not yet been reached, we check if it was reached.\n* When training finishes, we send a tweet informing us whether our training was successful or not.",
"_____no_output_____"
]
],
[
[
"from skorch.callbacks import Callback\n\n\ndef tweet(msg):\n print(\"~\" * 60)\n print(\"*tweet*\", msg, \"#skorch #pytorch\")\n print(\"~\" * 60)\n\n\nclass AccuracyTweet(Callback):\n def __init__(self, min_accuracy):\n self.min_accuracy = min_accuracy\n\n def initialize(self):\n self.critical_epoch_ = -1\n\n def on_epoch_end(self, net, **kwargs):\n if self.critical_epoch_ > -1:\n return\n # look at the validation accuracy of the last epoch\n if net.history[-1, 'valid_acc'] >= self.min_accuracy:\n self.critical_epoch_ = len(net.history)\n\n def on_train_end(self, net, **kwargs):\n if self.critical_epoch_ < 0:\n msg = \"Accuracy never reached {} :(\".format(self.min_accuracy)\n else:\n msg = \"Accuracy reached {} at epoch {}!!!\".format(\n self.min_accuracy, self.critical_epoch_)\n\n tweet(msg)",
"_____no_output_____"
]
],
[
[
"Now we initialize a `NeuralNetClassifier` and pass your new callback in a list to the `callbacks` argument. After that, we train the model and see what happens.",
"_____no_output_____"
]
],
[
[
"net = NeuralNetClassifier(\n ClassifierModule,\n max_epochs=15,\n lr=0.02,\n warm_start=True,\n callbacks=[AccuracyTweet(min_accuracy=0.7)],\n)",
"_____no_output_____"
],
[
"net.fit(X, y)",
" epoch train_loss valid_acc valid_loss dur\n------- ------------ ----------- ------------ ------\n 1 \u001b[36m0.6954\u001b[0m \u001b[32m0.6000\u001b[0m \u001b[35m0.6844\u001b[0m 0.0176\n 2 \u001b[36m0.6802\u001b[0m 0.5950 \u001b[35m0.6817\u001b[0m 0.0150\n 3 0.6839 0.6000 \u001b[35m0.6792\u001b[0m 0.0178\n 4 \u001b[36m0.6753\u001b[0m 0.5900 \u001b[35m0.6767\u001b[0m 0.0140\n 5 0.6769 0.5950 \u001b[35m0.6742\u001b[0m 0.0172\n 6 0.6774 \u001b[32m0.6050\u001b[0m \u001b[35m0.6720\u001b[0m 0.0166\n 7 \u001b[36m0.6693\u001b[0m \u001b[32m0.6250\u001b[0m \u001b[35m0.6695\u001b[0m 0.0134\n 8 0.6694 \u001b[32m0.6300\u001b[0m \u001b[35m0.6672\u001b[0m 0.0168\n 9 0.6703 \u001b[32m0.6400\u001b[0m \u001b[35m0.6652\u001b[0m 0.0177\n 10 \u001b[36m0.6523\u001b[0m \u001b[32m0.6550\u001b[0m \u001b[35m0.6623\u001b[0m 0.0151\n 11 0.6641 \u001b[32m0.6650\u001b[0m \u001b[35m0.6603\u001b[0m 0.0134\n 12 0.6524 0.6650 \u001b[35m0.6582\u001b[0m 0.0138\n 13 \u001b[36m0.6506\u001b[0m \u001b[32m0.6700\u001b[0m \u001b[35m0.6553\u001b[0m 0.0126\n 14 \u001b[36m0.6489\u001b[0m 0.6650 \u001b[35m0.6527\u001b[0m 0.0132\n 15 0.6505 \u001b[32m0.6750\u001b[0m \u001b[35m0.6502\u001b[0m 0.0133\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n*tweet* Accuracy never reached 0.7 :( #skorch #pytorch\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n"
]
],
[
[
"Oh no, our model never reached a validation accuracy of 0.7. Let's train some more (this is possible because we set `warm_start=True`):",
"_____no_output_____"
]
],
[
[
"net.fit(X, y)",
" 16 \u001b[36m0.6473\u001b[0m 0.6750 \u001b[35m0.6474\u001b[0m 0.0175\n 17 \u001b[36m0.6431\u001b[0m \u001b[32m0.6800\u001b[0m \u001b[35m0.6443\u001b[0m 0.0185\n 18 0.6461 \u001b[32m0.6900\u001b[0m \u001b[35m0.6418\u001b[0m 0.0162\n 19 \u001b[36m0.6430\u001b[0m 0.6850 \u001b[35m0.6392\u001b[0m 0.0131\n 20 \u001b[36m0.6364\u001b[0m \u001b[32m0.6950\u001b[0m \u001b[35m0.6366\u001b[0m 0.0146\n 21 \u001b[36m0.6266\u001b[0m \u001b[32m0.7000\u001b[0m \u001b[35m0.6334\u001b[0m 0.0149\n 22 0.6316 0.7000 \u001b[35m0.6308\u001b[0m 0.0151\n 23 \u001b[36m0.6231\u001b[0m 0.7000 \u001b[35m0.6277\u001b[0m 0.0128\n 24 \u001b[36m0.6094\u001b[0m 0.7000 \u001b[35m0.6242\u001b[0m 0.0160\n 25 0.6250 \u001b[32m0.7050\u001b[0m \u001b[35m0.6215\u001b[0m 0.0130\n 26 0.6180 \u001b[32m0.7150\u001b[0m \u001b[35m0.6187\u001b[0m 0.0139\n 27 0.6186 0.7150 \u001b[35m0.6159\u001b[0m 0.0169\n 28 0.6144 0.7150 \u001b[35m0.6134\u001b[0m 0.0171\n 29 \u001b[36m0.5993\u001b[0m 0.7150 \u001b[35m0.6100\u001b[0m 0.0147\n 30 \u001b[36m0.5976\u001b[0m 0.7150 \u001b[35m0.6071\u001b[0m 0.0138\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n*tweet* Accuracy reached 0.7 at epoch 21!!! #skorch #pytorch\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n"
],
[
"assert net.history[-1, 'valid_acc'] >= 0.7",
"_____no_output_____"
]
],
[
[
"Finally, the validation score exceeded 0.7. Hooray!",
"_____no_output_____"
],
[
"### Accessing callback parameters",
"_____no_output_____"
],
[
"Say you would like to use a learning rate schedule with your neural net, but you don't know what parameters are best for that schedule. Wouldn't it be nice if you could find those parameters with a grid search? With `skorch`, this is possible. Below, we show how to access the parameters of your callbacks.",
"_____no_output_____"
],
[
"To simplify the access to your callback parameters, it is best if you give your callback a name. This is achieved by passing the `callbacks` parameter a list of *name*, *callback* tuples, such as:\n\n callbacks=[\n ('scheduler', LearningRateScheduler)),\n ...\n ],\n \nThis way, you can access your callbacks using the double underscore semantics (as, for instance, in an `sklearn` `Pipeline`):\n\n callbacks__scheduler__epoch=50,\n\nSo if you would like to perform a grid search on, say, the number of units in the hidden layer and the learning rate schedule, it could look something like this:\n\n param_grid = {\n 'module__num_units': [50, 100, 150],\n 'callbacks__scheduler__epoch': [10, 50, 100],\n }\n \n*Note*: If you would like to refresh your knowledge on grid search, look [here](http://scikit-learn.org/stable/modules/grid_search.html#grid-search), [here](http://scikit-learn.org/stable/auto_examples/model_selection/grid_search_text_feature_extraction.html), or in the *Basic_Usage* notebok.",
"_____no_output_____"
],
[
"Below, we show how accessing the callback parameters works our `AccuracyTweet` callback:",
"_____no_output_____"
]
],
[
[
"net = NeuralNetClassifier(\n ClassifierModule,\n max_epochs=10,\n lr=0.1,\n warm_start=True,\n callbacks=[\n ('tweet', AccuracyTweet(min_accuracy=0.7)),\n ],\n callbacks__tweet__min_accuracy=0.6,\n)",
"_____no_output_____"
],
[
"net.fit(X, y)",
" epoch train_loss valid_acc valid_loss dur\n------- ------------ ----------- ------------ ------\n 1 \u001b[36m0.6932\u001b[0m \u001b[32m0.5950\u001b[0m \u001b[35m0.6749\u001b[0m 0.0161\n 2 \u001b[36m0.6686\u001b[0m \u001b[32m0.6550\u001b[0m \u001b[35m0.6613\u001b[0m 0.0160\n 3 \u001b[36m0.6641\u001b[0m 0.6450 \u001b[35m0.6487\u001b[0m 0.0168\n 4 \u001b[36m0.6438\u001b[0m \u001b[32m0.6600\u001b[0m \u001b[35m0.6354\u001b[0m 0.0171\n 5 \u001b[36m0.6293\u001b[0m \u001b[32m0.7000\u001b[0m \u001b[35m0.6190\u001b[0m 0.0171\n 6 \u001b[36m0.6091\u001b[0m \u001b[32m0.7300\u001b[0m \u001b[35m0.6040\u001b[0m 0.0147\n 7 \u001b[36m0.5872\u001b[0m \u001b[32m0.7500\u001b[0m \u001b[35m0.5868\u001b[0m 0.0130\n 8 \u001b[36m0.5820\u001b[0m \u001b[32m0.7600\u001b[0m \u001b[35m0.5736\u001b[0m 0.0138\n 9 \u001b[36m0.5778\u001b[0m \u001b[32m0.7850\u001b[0m \u001b[35m0.5595\u001b[0m 0.0129\n 10 \u001b[36m0.5626\u001b[0m 0.7750 \u001b[35m0.5484\u001b[0m 0.0131\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n*tweet* Accuracy reached 0.6 at epoch 2!!! #skorch #pytorch\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n"
]
],
[
[
"As you can see, by passing `callbacks__tweet__min_accuracy=0.6`, we changed that parameter. The same can be achieved by calling the `set_params` method with the corresponding arguments:",
"_____no_output_____"
]
],
[
[
"net.set_params(callbacks__tweet__min_accuracy=0.75)",
"_____no_output_____"
],
[
"net.fit(X, y)",
" epoch train_loss valid_acc valid_loss dur\n------- ------------ ----------- ------------ ------\n 11 \u001b[36m0.5513\u001b[0m 0.7750 \u001b[35m0.5405\u001b[0m 0.0136\n 12 0.5612 0.7800 \u001b[35m0.5361\u001b[0m 0.0133\n 13 \u001b[36m0.5473\u001b[0m \u001b[32m0.7950\u001b[0m \u001b[35m0.5303\u001b[0m 0.0159\n 14 \u001b[36m0.5304\u001b[0m 0.7900 \u001b[35m0.5241\u001b[0m 0.0162\n 15 \u001b[36m0.5088\u001b[0m 0.7850 \u001b[35m0.5198\u001b[0m 0.0170\n 16 0.5373 0.7800 \u001b[35m0.5168\u001b[0m 0.0170\n 17 0.5377 0.7750 0.5179 0.0169\n 18 0.5257 0.7700 0.5177 0.0171\n 19 0.5150 0.7700 \u001b[35m0.5132\u001b[0m 0.0169\n 20 0.5136 0.7450 \u001b[35m0.5116\u001b[0m 0.0167\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n*tweet* Accuracy reached 0.75 at epoch 11!!! #skorch #pytorch\n~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~\n"
]
],
[
[
"## Working with different data types",
"_____no_output_____"
],
[
"### Working with `Dataset`s",
"_____no_output_____"
],
[
"We encourage you to not pass `Dataset`s to `net.fit` but to let skorch handle `Dataset`s internally. Nonetheless, there are situations where passing `Dataset`s to `net.fit` is hard to avoid (e.g. if you want to load the data lazily during the training). This is supported by skorch but may have some unwanted side-effects relating to sklearn. For instance, `Dataset`s cannot split into train and validation in a stratified fashion without explicit knowledge of the classification targets.",
"_____no_output_____"
],
[
"Below we show what happens when you try to fit with `Dataset` and the stratified split fails:",
"_____no_output_____"
]
],
[
[
"class MyDataset(torch.utils.data.Dataset):\n def __init__(self, X, y):\n self.X = X\n self.y = y\n \n assert len(X) == len(y)\n\n def __len__(self):\n return len(self.X)\n\n def __getitem__(self, i):\n return self.X[i], self.y[i]",
"_____no_output_____"
],
[
"X, y = make_classification(1000, 20, n_informative=10, random_state=0)\nX, y = X.astype(np.float32), y.astype(np.int64)\ndataset = MyDataset(X, y)",
"_____no_output_____"
],
[
"net = NeuralNetClassifier(ClassifierModule)",
"_____no_output_____"
],
[
"try:\n net.fit(dataset, y=None)\nexcept ValueError as e:\n print(\"Error:\", e)",
"Error: Stratified CV requires explicitly passing a suitable y.\n"
],
[
"net.train_split.stratified",
"_____no_output_____"
]
],
[
[
"As you can see, the stratified split fails since `y` is not known. There are two solutions to this:\n\n* turn off stratified splitting ( `net.train_split.stratified=False`) \n* pass `y` explicitly (if possible), even if it is implicitely contained in the `Dataset`\n\nThe second solution is shown below:",
"_____no_output_____"
]
],
[
[
"net.fit(dataset, y=y)",
"Re-initializing module.\nRe-initializing optimizer.\n epoch train_loss valid_acc valid_loss dur\n------- ------------ ----------- ------------ ------\n 1 \u001b[36m0.6938\u001b[0m \u001b[32m0.4650\u001b[0m \u001b[35m0.6984\u001b[0m 0.0154\n 2 0.6975 0.4650 \u001b[35m0.6977\u001b[0m 0.0141\n 3 0.6938 0.4600 \u001b[35m0.6970\u001b[0m 0.0130\n 4 \u001b[36m0.6923\u001b[0m \u001b[32m0.4700\u001b[0m \u001b[35m0.6964\u001b[0m 0.0137\n 5 \u001b[36m0.6921\u001b[0m \u001b[32m0.4800\u001b[0m \u001b[35m0.6959\u001b[0m 0.0135\n 6 \u001b[36m0.6878\u001b[0m \u001b[32m0.5000\u001b[0m \u001b[35m0.6954\u001b[0m 0.0138\n 7 0.6901 0.4950 \u001b[35m0.6948\u001b[0m 0.0130\n 8 0.6884 0.4900 \u001b[35m0.6944\u001b[0m 0.0137\n 9 0.6896 0.4900 \u001b[35m0.6940\u001b[0m 0.0130\n 10 \u001b[36m0.6870\u001b[0m 0.4850 \u001b[35m0.6936\u001b[0m 0.0130\n"
]
],
[
[
"### Working with dicts",
"_____no_output_____"
],
[
"#### The standard case",
"_____no_output_____"
],
[
"skorch has built-in support for dictionaries as data containers. Here we show a somewhat contrived example of how to use dicts, but it should get the point across. First we create data and put it into a dictionary `X_dict` with two keys `X0` and `X1`:",
"_____no_output_____"
]
],
[
[
"X, y = make_classification(1000, 20, n_informative=10, random_state=0)\nX, y = X.astype(np.float32), y.astype(np.int64)\nX0, X1 = X[:, :10], X[:, 10:]\nX_dict = {'X0': X0, 'X1': X1}",
"_____no_output_____"
]
],
[
[
"When skorch passes the dict to the pytorch module, it will pass the data as keyword arguments to the forward call. That means that we should accept the two keys `XO` and `X1` in the forward method, as shown below:",
"_____no_output_____"
]
],
[
[
"class ClassifierWithDict(nn.Module):\n def __init__(\n self,\n num_units0=50,\n num_units1=50,\n nonlin=F.relu,\n dropout=0.5,\n ):\n super(ClassifierWithDict, self).__init__()\n self.num_units0 = num_units0\n self.num_units1 = num_units1\n self.nonlin = nonlin\n self.dropout = dropout\n\n self.dense0 = nn.Linear(10, num_units0)\n self.dense1 = nn.Linear(10, num_units1)\n self.nonlin = nonlin\n self.dropout = nn.Dropout(dropout)\n self.output = nn.Linear(num_units0 + num_units1, 2)\n\n # NOTE: We accept X0 and X1, the keys from the dict, as arguments\n def forward(self, X0, X1, **kwargs):\n X0 = self.nonlin(self.dense0(X0))\n X0 = self.dropout(X0)\n\n X1 = self.nonlin(self.dense1(X1))\n X1 = self.dropout(X1)\n\n X = torch.cat((X0, X1), dim=1)\n X = F.relu(X)\n X = F.softmax(self.output(X), dim=-1)\n return X",
"_____no_output_____"
]
],
[
[
"As long as we keep this in mind, we are good to go.",
"_____no_output_____"
]
],
[
[
"net = NeuralNetClassifier(ClassifierWithDict, verbose=0)",
"_____no_output_____"
],
[
"net.fit(X_dict, y)",
"_____no_output_____"
]
],
[
[
"#### Working with sklearn `Pipeline` and `GridSearchCV`",
"_____no_output_____"
]
],
[
[
"from sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import FunctionTransformer\nfrom sklearn.model_selection import GridSearchCV",
"_____no_output_____"
]
],
[
[
"sklearn makes the assumption that incoming data should be numpy/sparse arrays or something similar. This clashes with the use of dictionaries. Unfortunately, it is sometimes impossible to work around that for now (for instance using skorch with `BaggingClassifier`). Other times, there are possibilities.\n\nWhen we have a preprocessing pipeline that involves `FunctionTransformer`, we have to pass the parameter `validate=False` (which is the default value now) so that sklearn allows the dictionary to pass through. Everything else works:",
"_____no_output_____"
]
],
[
[
"pipe = Pipeline([\n ('do-nothing', FunctionTransformer(validate=False)),\n ('net', net),\n])",
"_____no_output_____"
],
[
"pipe.fit(X_dict, y)",
"_____no_output_____"
]
],
[
[
"When trying a grid or randomized search, it is not that easy to pass a dict. If we try, we will get an error:",
"_____no_output_____"
]
],
[
[
"param_grid = {\n 'net__module__num_units0': [10, 25, 50], \n 'net__module__num_units1': [10, 25, 50],\n 'net__lr': [0.01, 0.1],\n}",
"_____no_output_____"
],
[
"grid_search = GridSearchCV(pipe, param_grid, scoring='accuracy', verbose=1, cv=3)",
"_____no_output_____"
],
[
"try:\n grid_search.fit(X_dict, y)\nexcept Exception as e:\n print(e)",
"Found input variables with inconsistent numbers of samples: [2, 1000]\n"
]
],
[
[
"The error above occurs because sklearn gets the length of the input data, which is 2 for the dict, and believes that is inconsistent with the length of the target (1000). \n\nTo get around that, skorch provides a helper class called `SliceDict`. It allows us to wrap our dictionaries so that they also behave like a numpy array:",
"_____no_output_____"
]
],
[
[
"from skorch.helper import SliceDict",
"_____no_output_____"
],
[
"X_slice_dict = SliceDict(X0=X0, X1=X1) # X_slice_dict = SliceDict(**X_dict) would also work",
"_____no_output_____"
]
],
[
[
"The SliceDict shows the correct length, shape, and is sliceable across values:",
"_____no_output_____"
]
],
[
[
"print(\"Length of dict: {}, length of SliceDict: {}\".format(len(X_dict), len(X_slice_dict)))\nprint(\"Shape of SliceDict: {}\".format(X_slice_dict.shape))",
"Length of dict: 2, length of SliceDict: 1000\nShape of SliceDict: (1000,)\n"
],
[
"print(\"Slicing the SliceDict slices across values: {}\".format(X_slice_dict[:2]))",
"Slicing the SliceDict slices across values: SliceDict(**{'X0': array([[-0.9658346 , -2.1890705 , 0.16985609, 0.8138456 , -3.375209 ,\n -2.1430597 , -0.39585084, 2.9419577 , -2.1910605 , 1.2443967 ],\n [-0.454767 , 4.339768 , -0.48572844, -4.88433 , -2.8836503 ,\n 2.6097205 , -1.952876 , -0.09192174, 0.07970932, -0.08938338]],\n dtype=float32), 'X1': array([[ 0.04351204, -0.5150961 , -0.86073655, -1.1097169 , 0.31839254,\n -0.8231973 , -1.056304 , -0.89645284, 0.3759244 , -1.0849651 ],\n [-0.60726726, -1.0674309 , 0.48804346, -0.50230557, 0.55743027,\n 1.01592 , -1.9953582 , 2.9030426 , -0.9739298 , 2.1753323 ]],\n dtype=float32)})\n"
]
],
[
[
"With this, we can call `GridSearchCV` just as expected:",
"_____no_output_____"
]
],
[
[
"grid_search.fit(X_slice_dict, y)",
"Fitting 3 folds for each of 18 candidates, totalling 54 fits\n"
],
[
"grid_search.best_score_, grid_search.best_params_",
"_____no_output_____"
]
],
[
[
"## Multiple return values from `forward`",
"_____no_output_____"
],
[
"Often, we want our `Module.forward` method to return more than just one value. There can be several reasons for this. Maybe, the criterion requires not one but several outputs. Or perhaps we want to inspect intermediate values to learn more about our model (say inspecting attention in a sequence-to-sequence model). Fortunately, `skorch` makes it easy to achieve this. In the following, we demonstrate how to handle multiple outputs from the `Module`.",
"_____no_output_____"
],
[
"To demonstrate this, we implement a very simple autoencoder. It consists of an encoder that reduces our input of 20 units to 5 units using two linear layers, and a decoder that tries to reconstruct the original input, again using two linear layers.",
"_____no_output_____"
],
[
"### Implementing a simple autoencoder",
"_____no_output_____"
]
],
[
[
"from skorch import NeuralNetRegressor",
"_____no_output_____"
],
[
"class Encoder(nn.Module):\n def __init__(self, num_units=5):\n super().__init__()\n self.num_units = num_units\n \n self.encode = nn.Sequential(\n nn.Linear(20, 10),\n nn.ReLU(),\n nn.Linear(10, self.num_units),\n nn.ReLU(),\n )\n \n def forward(self, X):\n encoded = self.encode(X)\n return encoded",
"_____no_output_____"
],
[
"class Decoder(nn.Module):\n def __init__(self, num_units):\n super().__init__()\n self.num_units = num_units\n \n self.decode = nn.Sequential(\n nn.Linear(self.num_units, 10),\n nn.ReLU(),\n nn.Linear(10, 20),\n )\n \n def forward(self, X):\n decoded = self.decode(X)\n return decoded",
"_____no_output_____"
]
],
[
[
"The autoencoder module below actually returns a tuple of two values, the decoded input and the encoded input. This way, we cannot only use the decoded input to calculate the normal loss but also have access to the encoded state.",
"_____no_output_____"
]
],
[
[
"class AutoEncoder(nn.Module):\n def __init__(self, num_units):\n super().__init__()\n self.num_units = num_units\n\n self.encoder = Encoder(num_units=self.num_units)\n self.decoder = Decoder(num_units=self.num_units)\n \n def forward(self, X):\n encoded = self.encoder(X)\n decoded = self.decoder(encoded)\n return decoded, encoded # <- return a tuple of two values",
"_____no_output_____"
]
],
[
[
"Since the module's `forward` method returns two values, we have to adjust our objective to do the right thing with those values. If we don't do this, the criterion wouldn't know what to do with the two values and would raise an error.\n\nOne strategy would be to only use the decoded state for the loss and discard the encoded state. For this demonstration, we have a different plan: We would like the encoded state to be sparse. Therefore, we add an L1 loss of the encoded state to the reconstruction loss. This way, the net will try to reconstruct the input as accurately as possible while keeping the encoded state as sparse as possible.\n\nTo implement this, the right method to override is called `get_loss`, which is where `skorch` computes and returns the loss. It gets the prediction (our tuple) and the target as input, as well as other arguments and keywords that we pass through. We create a subclass of `NeuralNetRegressor` that overrides said method and implements our idea for the loss.",
"_____no_output_____"
]
],
[
[
"class AutoEncoderNet(NeuralNetRegressor):\n def get_loss(self, y_pred, y_true, *args, **kwargs):\n decoded, encoded = y_pred # <- unpack the tuple that was returned by `forward`\n loss_reconstruction = super().get_loss(decoded, y_true, *args, **kwargs)\n loss_l1 = 1e-3 * torch.abs(encoded).sum()\n return loss_reconstruction + loss_l1",
"_____no_output_____"
]
],
[
[
"*Note*: Alternatively, we could have used an unaltered `NeuralNetRegressor` but implement a custom criterion that is responsible for unpacking the tuple and computing the loss.",
"_____no_output_____"
],
[
"### Training the autoencoder",
"_____no_output_____"
],
[
"Now that everything is ready, we train the model as usual. We initialize our net subclass with the `AutoEncoder` module and call the `fit` method with `X` both as input and as target (since we want to reconstruct the original data):",
"_____no_output_____"
]
],
[
[
"net = AutoEncoderNet(\n AutoEncoder,\n module__num_units=5,\n lr=0.3,\n)",
"_____no_output_____"
],
[
"net.fit(X, X)",
" epoch train_loss valid_loss dur\n------- ------------ ------------ ------\n 1 \u001b[36m3.8328\u001b[0m \u001b[32m3.7855\u001b[0m 0.0233\n 2 \u001b[36m3.6989\u001b[0m \u001b[32m3.7111\u001b[0m 0.0244\n 3 \u001b[36m3.6417\u001b[0m \u001b[32m3.6707\u001b[0m 0.0259\n 4 \u001b[36m3.6101\u001b[0m \u001b[32m3.6463\u001b[0m 0.0209\n 5 \u001b[36m3.5914\u001b[0m \u001b[32m3.6310\u001b[0m 0.0226\n 6 \u001b[36m3.5799\u001b[0m \u001b[32m3.6212\u001b[0m 0.0242\n 7 \u001b[36m3.5725\u001b[0m \u001b[32m3.6144\u001b[0m 0.0307\n 8 \u001b[36m3.5672\u001b[0m \u001b[32m3.6090\u001b[0m 0.0347\n 9 \u001b[36m3.5627\u001b[0m \u001b[32m3.6036\u001b[0m 0.0239\n 10 \u001b[36m3.5570\u001b[0m \u001b[32m3.5963\u001b[0m 0.0264\n"
]
],
[
[
"Voilà, the model was trained using our custom loss function that makes use of both predicted values.",
"_____no_output_____"
],
[
"### Extracting the decoder and the encoder output",
"_____no_output_____"
],
[
"Sometimes, we may wish to inspect all the values returned by the `foward` method of the module. There are several ways to achieve this. In theory, we can always access the module directly by using the `net.module_` attribute. However, this is unwieldy, since this completely shortcuts the prediction loop, which takes care of important steps like casting `numpy` arrays to `pytorch` tensors and batching.\n\nAlso, we cannot use the `predict` method on the net. This method will only return the first output from the forward method, in this case the decoded state. The reason for this is that `predict` is part of the `sklearn` API, which requires there to be only one output. This is shown below:",
"_____no_output_____"
]
],
[
[
"y_pred = net.predict(X)\ny_pred.shape # only the decoded state is returned",
"_____no_output_____"
]
],
[
[
"However, the net itself provides two methods to retrieve all outputs. The first one is the `net.forward` method, which retrieves *all* the predicted batches from the `Module.forward` and concatenates them. Use this to retrieve the complete decoded and encoded state:",
"_____no_output_____"
]
],
[
[
"decoded_pred, encoded_pred = net.forward(X)\ndecoded_pred.shape, encoded_pred.shape",
"_____no_output_____"
]
],
[
[
"The other method is called `net.forward_iter`. It is similar to `net.forward` but instead of collecting all the batches, this method is lazy and only yields one batch at a time. This can be especially useful if the output doesn't fit into memory:",
"_____no_output_____"
]
],
[
[
"for decoded_pred, encoded_pred in net.forward_iter(X):\n # do something with each batch\n break\ndecoded_pred.shape, encoded_pred.shape",
"_____no_output_____"
]
],
[
[
"Finally, let's make sure that our initial goal of having a sparse encoded state was met. We check how many activities are close to zero:",
"_____no_output_____"
]
],
[
[
"torch.isclose(encoded_pred, torch.zeros_like(encoded_pred)).float().mean()",
"_____no_output_____"
]
],
[
[
"As we had hoped, the encoded state is quite sparse, with the majority of outpus being 0.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbc76c53b38fbc2a875e08ce477ac89339296154
| 378,364 |
ipynb
|
Jupyter Notebook
|
code/03-basic_image_manipulation_solutions.ipynb
|
ostanley/SDC-BIDS-fMRI-1
|
a279cdac204fcd5646d27f15a50f2f922e7b7c88
|
[
"CC-BY-4.0"
] | null | null | null |
code/03-basic_image_manipulation_solutions.ipynb
|
ostanley/SDC-BIDS-fMRI-1
|
a279cdac204fcd5646d27f15a50f2f922e7b7c88
|
[
"CC-BY-4.0"
] | null | null | null |
code/03-basic_image_manipulation_solutions.ipynb
|
ostanley/SDC-BIDS-fMRI-1
|
a279cdac204fcd5646d27f15a50f2f922e7b7c88
|
[
"CC-BY-4.0"
] | null | null | null | 729.025048 | 150,136 | 0.937597 |
[
[
[
"### Introduction to Nilearn and image manipulation\n\nThe goal of this notebook is to help get you comfortable with manipulating functional and anatomical images using nilearn. We'll be using the techniques we learned here in our final analysis...\n\n#### Content:\n1. Basic Image Operations and Masking\n2. Resampling data to work across modalities (T1/FUNC)",
"_____no_output_____"
]
],
[
[
"import os\nimport matplotlib.pyplot as plt\nfrom nilearn import image as img\nfrom nilearn import plotting as plot\nfrom bids import BIDSLayout\n\n#for inline visualization in jupyter notebook\n%matplotlib inline ",
"_____no_output_____"
]
],
[
[
"As we've done in the past we've imported <code>image as img</code>. However, we've also imported <code>plotting as plot</code> from <code>nilearn</code>. This will allow us to easily visualize our neuroimaging data!\n\nFirst let’s grab some data from where we downloaded our FMRIPREP outputs using PyBIDS:",
"_____no_output_____"
]
],
[
[
"#Base directory for fmriprep output\nfmriprep_dir = '../data/ds000030/derivatives/fmriprep/'\nlayout= BIDSLayout(fmriprep_dir, validate=False)\nT1w_files = layout.get(subject='10788', datatype='anat', suffix='preproc')\nbrainmask_files = layout.get(subject='10788', datatype='anat', suffix='brainmask')",
"_____no_output_____"
],
[
"#Display preprocessed files inside of anatomy folder\nfor f in T1w_files:\n print(f.path)",
"/mnt/tigrlab/projects/jjeyachandra/scwg2018_python_neuroimaging/data/ds000030/derivatives/fmriprep/sub-10171/anat/sub-10171_T1w_preproc.nii.gz\n/mnt/tigrlab/projects/jjeyachandra/scwg2018_python_neuroimaging/data/ds000030/derivatives/fmriprep/sub-10171/anat/sub-10171_T1w_space-MNI152NLin2009cAsym_preproc.nii.gz\n"
]
],
[
[
"## Basic Image Operations\n\nIn this section we're going to deal with the following files:\n\n1. <code>sub-10171_T1w_preproc.nii.gz</code> - the T1 image in native space\n2. <code>sub-10171_T1w_brainmask.nii.gz</code> - a mask with 1's representing the brain and 0's elsewhere.",
"_____no_output_____"
]
],
[
[
"t1 = T1w_files[0].path\nbm = brainmask_files[0].path\n\nt1_img = img.load_img(t1)\nbm_img = img.load_img(bm)",
"_____no_output_____"
]
],
[
[
"First we'll do what you've been waiting to do - plot our MR image! This can be easily achieved using Nilearn's <code>plotting</code> module as follows:",
"_____no_output_____"
]
],
[
[
"plot.plot_anat(t1_img)",
"/projects/jjeyachandra/scwg2018_python_neuroimaging/scwg2018_nilearn/lib/python3.6/site-packages/scipy/ndimage/measurements.py:272: DeprecationWarning: In future, it will be an error for 'np.bool_' scalars to be interpreted as an index\n return _nd_image.find_objects(input, max_label)\n"
]
],
[
[
"Try viewing the mask as well!",
"_____no_output_____"
]
],
[
[
"#View the mask image\nplot.plot_anat(bm_img)",
"/projects/jjeyachandra/scwg2018_python_neuroimaging/scwg2018_nilearn/lib/python3.6/site-packages/scipy/ndimage/measurements.py:272: DeprecationWarning: In future, it will be an error for 'np.bool_' scalars to be interpreted as an index\n return _nd_image.find_objects(input, max_label)\n"
]
],
[
[
"### Arithmetic Operations\n\nLet’s start performing some image operations. The simplest operations we can perform is element-wise, what this means is that we want to perform some sort of mathematical operation on each voxel of the MR image. Since voxels are represented in a 3D array, this is equivalent to performing an operation on each element (i,j,k) of a 3D array. Let’s try inverting the image, that is, flip the colour scale such that all blacks appear white and vice-versa. To do this, we’ll use the method\n\n<code>img.math_img(formula, **imgs)</code> Where:\n\n- <code>formula</code> is a mathematical expression such as 'a+1'\n- </code>**imgs</code> is a set of key-value pairs linking variable names to images. For example a=T1\n\nIn order to invert the image, we can simply flip the sign which will set the most positive elements (white) to the most negative elements (black), and the least positives elements (black) to the least negative elements (white). This effectively flips the colour-scale:",
"_____no_output_____"
]
],
[
[
"invert_img = img.math_img('-a', a=t1_img)\nplot.plot_anat(invert_img)",
"/projects/jjeyachandra/scwg2018_python_neuroimaging/scwg2018_nilearn/lib/python3.6/site-packages/scipy/ndimage/measurements.py:272: DeprecationWarning: In future, it will be an error for 'np.bool_' scalars to be interpreted as an index\n return _nd_image.find_objects(input, max_label)\n"
]
],
[
[
"Alternatively we don't need to first load in our <code>t1_img</code> using <code>img.load_img</code>. Instead we can feed in a path to <code>img.math_img</code>:\n\n~~~\ninvert_img = img.math_img('-a', a=t1)\nplot.plot_anat(invert_img)\n~~~\n\nThis will yield the same result!",
"_____no_output_____"
],
[
"### Applying a Mask\nLet’s extend this idea of applying operations to each element of an image to multiple images. Instead of specifying just one image like the following:\n\n<code>img.math_img('a+1',a=img_a)</code>\n\nWe can specify multiple images by tacking on additional variables:\n\n<code>img.math_img('a+b', a=img_a, b=img_b)</code>\n\nThe key requirement here is that when dealing with multiple images, that the size of the images must be the same. The reason being is that we’re deaing with element-wise operations. That means that some voxel (i,j,k) in img_a is being paired with some voxel (i,j,k) in <code>img_b</code> when performing operations. So every voxel in <code>img_a</code> must have some pair with a voxel in <code>img_b</code>; sizes must be the same.\n\nWe can take advantage of this property when masking our data using multiplication. Masking works by multipling a raw image (our <code>T1</code>), with some mask image (our <code>bm</code>). Whichever voxel (i,j,k) has a value of 0 in the mask multiplies with voxel (i,j,k) in the raw image resulting in a product of 0. Conversely, any voxel (i,j,k) in the mask with a value of 1 multiplies with voxel (i,j,k) in the raw image resulting in the same value. Let’s try this out in practice and see what the result is:",
"_____no_output_____"
]
],
[
[
"masked_t1 = img.math_img('a*b', a=t1, b=bm)\nplot.plot_anat(masked_t1)",
"/projects/jjeyachandra/scwg2018_python_neuroimaging/scwg2018_nilearn/lib/python3.6/site-packages/nibabel/nifti1.py:1682: DeprecationWarning: np.asscalar(a) is deprecated since NumPy v1.16, use a.item() instead\n magic = np.asscalar(hdr['magic'])\n/projects/jjeyachandra/scwg2018_python_neuroimaging/scwg2018_nilearn/lib/python3.6/site-packages/scipy/ndimage/measurements.py:272: DeprecationWarning: In future, it will be an error for 'np.bool_' scalars to be interpreted as an index\n return _nd_image.find_objects(input, max_label)\n"
]
],
[
[
"#### Exercise!\nTry applying the mask such that the brain is removed, but the rest of the head is intact!\n\n*Hint*:\n\nRemember that a mask is composed of 0's and 1's, where parts of the data labelled 1 are regions to keep, and parts of the data that are 0, are to throw away.\n\nYou can do this in 2 steps:\n\n1. Switch the 0's and 1's using an equation (simple addition/substraction) or condition (like x == 0). \n2. Apply the mask",
"_____no_output_____"
]
],
[
[
"inverted_mask = img.math_img('1-x', x=bm)\nplot.plot_anat(inverted_mask)",
"_____no_output_____"
],
[
"inverted_mask_t1 = img.math_img('a*b', a=t1, b=inverted_mask)\nplot.plot_anat(inverted_mask_t1)",
"_____no_output_____"
]
],
[
[
"### Slicing\n\nRecall that our data matrix is organized in the following manner: \n\n<img src=\"./static/images/numpy_arrays.png\" alt=\"Drawing\" align=\"middle\" width=\"500px\"/>",
"_____no_output_____"
],
[
"Slicing does exactly what it seems to imply. Given our 3D volume, we can pull out a 2D subset (called a \"slice\"). Here's an example of slicing moving from left to right via an animation:\n\n<img src=\"https://upload.wikimedia.org/wikipedia/commons/5/56/Parasagittal_MRI_of_human_head_in_patient_with_benign_familial_macrocephaly_prior_to_brain_injury_%28ANIMATED%29.gif\"/>\n\nWhat you see here is a series of 2D images that start from the left, and move toward the right. Each frame of this GIF is a slice - a 2D subset of a 3D volume. Slicing can be useful for cases in which you'd want to loop through each MR slice and perform a computation; importantly in functional imaging data slicing is useful for pulling out timepoints as we'll see later!\n\n***\nSourced from: https://en.wikipedia.org/wiki/Neuroimaging#/media/File:Parasagittal_MRI_of_human_head_in_patient_with_benign_familial_macrocephaly_prior_to_brain_injury_(ANIMATED).gif\n\n***",
"_____no_output_____"
],
[
"Slicing is done easily on an image file using the attribute <code>.slicer</code> of a Nilearn <code>image</code> object. For example we can grab the $10^{\\text{th}}$ slice along the x axis as follows:",
"_____no_output_____"
]
],
[
[
"x_slice = t1_img.slicer[10:11,:,:]",
"_____no_output_____"
]
],
[
[
"The statement $10:11$ is intentional and is required by <code>.slicer</code>. Alternatively we can slice along the x-axis using the data matrix itself:",
"_____no_output_____"
]
],
[
[
"t1_data = t1_img.get_data()\nx_slice = t1_data[10,:,:]",
"_____no_output_____"
]
],
[
[
"This will yield the same result as above. Notice that when using the <code>t1_data</code> array we can just specify which slice to grab instead of using <code>:</code>. We can use slicing in order to modify visualizations. For example, when viewing the T1 image, we may want to specify at which slice we'd like to view the image. This can be done by specifying which coordinates to *cut* the image at:",
"_____no_output_____"
]
],
[
[
"plot.plot_anat(t1_img,cut_coords=(50,30,70))",
"_____no_output_____"
]
],
[
[
"The <code>cut_coords</code> option specifies 3 numbers:\n- The first number says cut the X coordinate at slice 50 and display (sagittal view in this case!)\n- The second number says cut the Y coordinate at slice 30 and display (coronal view)\n- The third number says cut the Z coordinate at slice 70 and display (axial view)\n\nRemember <code>plot.plot_anat</code> yields 3 images, therefore <code>cut_coords</code> allows you to display where to take cross-sections of the brain from different perspectives (axial, sagittal, coronal)",
"_____no_output_____"
],
[
"***\n\nThis covers the basics of image manipulation using T1 images. To review in this section we covered:\n\n- Basic image arithmetic\n- Visualization\n- Slicing\n\nIn the next section we will cover how to integrate additional modalities (functional data) to what we've done so far using <code>Nilearn</code>. Then we can start using what we've learned in order to perform analysis and visualization!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbc76cb0825c84e89fdc76919950dbdaaa6bd810
| 279,429 |
ipynb
|
Jupyter Notebook
|
experiments/tl_3/jitter_1/oracle.run1.framed-cores_wisig/trials/2/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null |
experiments/tl_3/jitter_1/oracle.run1.framed-cores_wisig/trials/2/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null |
experiments/tl_3/jitter_1/oracle.run1.framed-cores_wisig/trials/2/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null | 109.194607 | 77,264 | 0.690955 |
[
[
[
"# Transfer Learning Template",
"_____no_output_____"
]
],
[
[
"%load_ext autoreload\n%autoreload 2\n%matplotlib inline\n\n \nimport os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom torch.optim import Adam\nfrom easydict import EasyDict\nimport matplotlib.pyplot as plt\n\nfrom steves_models.steves_ptn import Steves_Prototypical_Network\n\nfrom steves_utils.lazy_iterable_wrapper import Lazy_Iterable_Wrapper\nfrom steves_utils.iterable_aggregator import Iterable_Aggregator\nfrom steves_utils.ptn_train_eval_test_jig import PTN_Train_Eval_Test_Jig\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.torch_utils import get_dataset_metrics, ptn_confusion_by_domain_over_dataloader\nfrom steves_utils.utils_v2 import (per_domain_accuracy_from_confusion, get_datasets_base_path)\nfrom steves_utils.PTN.utils import independent_accuracy_assesment\n\nfrom torch.utils.data import DataLoader\n\nfrom steves_utils.stratified_dataset.episodic_accessor import Episodic_Accessor_Factory\n\nfrom steves_utils.ptn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.transforms import get_chained_transform",
"_____no_output_____"
]
],
[
[
"# Allowed Parameters\nThese are allowed parameters, not defaults\nEach of these values need to be present in the injected parameters (the notebook will raise an exception if they are not present)\n\nPapermill uses the cell tag \"parameters\" to inject the real parameters below this cell.\nEnable tags to see what I mean",
"_____no_output_____"
]
],
[
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"seed\",\n \"dataset_seed\",\n \"n_shot\",\n \"n_query\",\n \"n_way\",\n \"train_k_factor\",\n \"val_k_factor\",\n \"test_k_factor\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"x_net\",\n \"datasets\",\n \"torch_default_dtype\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"x_shape\",\n}",
"_____no_output_____"
],
[
"from steves_utils.CORES.utils import (\n ALL_NODES,\n ALL_NODES_MINIMUM_1000_EXAMPLES,\n ALL_DAYS\n)\n\nfrom steves_utils.ORACLE.utils_v2 import (\n ALL_DISTANCES_FEET_NARROWED,\n ALL_RUNS,\n ALL_SERIAL_NUMBERS,\n)\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"STANDALONE PTN\"\nstandalone_parameters[\"lr\"] = 0.001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"dataset_seed\"] = 1337\n\nstandalone_parameters[\"n_way\"] = 8\nstandalone_parameters[\"n_shot\"] = 3\nstandalone_parameters[\"n_query\"] = 2\nstandalone_parameters[\"train_k_factor\"] = 1\nstandalone_parameters[\"val_k_factor\"] = 2\nstandalone_parameters[\"test_k_factor\"] = 2\n\n\nstandalone_parameters[\"n_epoch\"] = 50\n\nstandalone_parameters[\"patience\"] = 10\nstandalone_parameters[\"criteria_for_best\"] = \"source_loss\"\n\nstandalone_parameters[\"datasets\"] = [\n {\n \"labels\": ALL_SERIAL_NUMBERS,\n \"domains\": ALL_DISTANCES_FEET_NARROWED,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"unit_mag\", \"minus_two\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE_\"\n },\n {\n \"labels\": ALL_NODES,\n \"domains\": ALL_DAYS,\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"unit_power\", \"times_zero\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"CORES_\"\n } \n]\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\n\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n]\n\n# Parameters relevant to results\n# These parameters will basically never need to change\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"\n\n\n\n\n",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"tl_3-jitter1:oracle.run1.framed -> cores+wisig\",\n \"device\": \"cuda\",\n \"lr\": 0.001,\n \"seed\": 1337,\n \"dataset_seed\": 1337,\n \"n_shot\": 3,\n \"n_query\": 2,\n \"train_k_factor\": 3,\n \"val_k_factor\": 2,\n \"test_k_factor\": 2,\n \"torch_default_dtype\": \"torch.float32\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_loss\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 256]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 20480, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 256}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"n_way\": 16,\n \"datasets\": [\n {\n \"labels\": [\n \"1-10.\",\n \"1-11.\",\n \"1-15.\",\n \"1-16.\",\n \"1-17.\",\n \"1-18.\",\n \"1-19.\",\n \"10-4.\",\n \"10-7.\",\n \"11-1.\",\n \"11-14.\",\n \"11-17.\",\n \"11-20.\",\n \"11-7.\",\n \"13-20.\",\n \"13-8.\",\n \"14-10.\",\n \"14-11.\",\n \"14-14.\",\n \"14-7.\",\n \"15-1.\",\n \"15-20.\",\n \"16-1.\",\n \"16-16.\",\n \"17-10.\",\n \"17-11.\",\n \"17-2.\",\n \"19-1.\",\n \"19-16.\",\n \"19-19.\",\n \"19-20.\",\n \"19-3.\",\n \"2-10.\",\n \"2-11.\",\n \"2-17.\",\n \"2-18.\",\n \"2-20.\",\n \"2-3.\",\n \"2-4.\",\n \"2-5.\",\n \"2-6.\",\n \"2-7.\",\n \"2-8.\",\n \"3-13.\",\n \"3-18.\",\n \"3-3.\",\n \"4-1.\",\n \"4-10.\",\n \"4-11.\",\n \"4-19.\",\n \"5-5.\",\n \"6-15.\",\n \"7-10.\",\n \"7-14.\",\n \"8-18.\",\n \"8-20.\",\n \"8-3.\",\n \"8-8.\",\n ],\n \"domains\": [1, 2, 3, 4, 5],\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": \"/mnt/wd500GB/CSC500/csc500-main/datasets/cores.stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"jitter_256_1\", \"take_200\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"C_A_\",\n },\n {\n \"labels\": [\n \"1-10\",\n \"1-12\",\n \"1-14\",\n \"1-16\",\n \"1-18\",\n \"1-19\",\n \"1-8\",\n \"10-11\",\n \"10-17\",\n \"10-4\",\n \"10-7\",\n \"11-1\",\n \"11-10\",\n \"11-19\",\n \"11-20\",\n \"11-4\",\n \"11-7\",\n \"12-19\",\n \"12-20\",\n \"12-7\",\n \"13-14\",\n \"13-18\",\n \"13-19\",\n \"13-20\",\n \"13-3\",\n \"13-7\",\n \"14-10\",\n \"14-11\",\n \"14-12\",\n \"14-13\",\n \"14-14\",\n \"14-19\",\n \"14-20\",\n \"14-7\",\n \"14-8\",\n \"14-9\",\n \"15-1\",\n \"15-19\",\n \"15-6\",\n \"16-1\",\n \"16-16\",\n \"16-19\",\n \"16-20\",\n \"17-10\",\n \"17-11\",\n \"18-1\",\n \"18-10\",\n \"18-11\",\n \"18-12\",\n \"18-13\",\n \"18-14\",\n \"18-15\",\n \"18-16\",\n \"18-17\",\n \"18-19\",\n \"18-2\",\n \"18-20\",\n \"18-4\",\n \"18-5\",\n \"18-7\",\n \"18-8\",\n \"18-9\",\n \"19-1\",\n \"19-10\",\n \"19-11\",\n \"19-12\",\n \"19-13\",\n \"19-14\",\n \"19-15\",\n \"19-19\",\n \"19-2\",\n \"19-20\",\n \"19-3\",\n \"19-4\",\n \"19-6\",\n \"19-7\",\n \"19-8\",\n \"19-9\",\n \"2-1\",\n \"2-13\",\n \"2-15\",\n \"2-3\",\n \"2-4\",\n \"2-5\",\n \"2-6\",\n \"2-7\",\n \"2-8\",\n \"20-1\",\n \"20-12\",\n \"20-14\",\n \"20-15\",\n \"20-16\",\n \"20-18\",\n \"20-19\",\n \"20-20\",\n \"20-3\",\n \"20-4\",\n \"20-5\",\n \"20-7\",\n \"20-8\",\n \"3-1\",\n \"3-13\",\n \"3-18\",\n \"3-2\",\n \"3-8\",\n \"4-1\",\n \"4-10\",\n \"4-11\",\n \"5-1\",\n \"5-5\",\n \"6-1\",\n \"6-15\",\n \"6-6\",\n \"7-10\",\n \"7-11\",\n \"7-12\",\n \"7-13\",\n \"7-14\",\n \"7-7\",\n \"7-8\",\n \"7-9\",\n \"8-1\",\n \"8-13\",\n \"8-14\",\n \"8-18\",\n \"8-20\",\n \"8-3\",\n \"8-8\",\n \"9-1\",\n \"9-7\",\n ],\n \"domains\": [1, 2, 3, 4],\n \"num_examples_per_domain_per_label\": 100,\n \"pickle_path\": \"/mnt/wd500GB/CSC500/csc500-main/datasets/wisig.node3-19.stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"target\",\n \"x_transforms\": [\"jitter_256_1\", \"take_200\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"W_A_\",\n },\n {\n \"labels\": [\n \"3123D52\",\n \"3123D65\",\n \"3123D79\",\n \"3123D80\",\n \"3123D54\",\n \"3123D70\",\n \"3123D7B\",\n \"3123D89\",\n \"3123D58\",\n \"3123D76\",\n \"3123D7D\",\n \"3123EFE\",\n \"3123D64\",\n \"3123D78\",\n \"3123D7E\",\n \"3124E4A\",\n ],\n \"domains\": [32, 38, 8, 44, 14, 50, 20, 26],\n \"num_examples_per_domain_per_label\": 2000,\n \"pickle_path\": \"/mnt/wd500GB/CSC500/csc500-main/datasets/oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\",\n \"source_or_target_dataset\": \"source\",\n \"x_transforms\": [\"jitter_256_1\", \"take_200\", \"resample_20Msps_to_25Msps\"],\n \"episode_transforms\": [],\n \"domain_prefix\": \"ORACLE.run1_\",\n },\n ],\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nif \"x_shape\" not in p:\n p.x_shape = [2,256] # Default to this if we dont supply x_shape\n\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"###########################################\n# The stratified datasets honor this\n###########################################\ntorch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"p.domains_source = []\np.domains_target = []\n\n\ntrain_original_source = []\nval_original_source = []\ntest_original_source = []\n\ntrain_original_target = []\nval_original_target = []\ntest_original_target = []",
"_____no_output_____"
],
[
"# global_x_transform_func = lambda x: normalize(x.to(torch.get_default_dtype()), \"unit_power\") # unit_power, unit_mag\n# global_x_transform_func = lambda x: normalize(x, \"unit_power\") # unit_power, unit_mag",
"_____no_output_____"
],
[
"def add_dataset(\n labels,\n domains,\n pickle_path,\n x_transforms,\n episode_transforms,\n domain_prefix,\n num_examples_per_domain_per_label,\n source_or_target_dataset:str,\n iterator_seed=p.seed,\n dataset_seed=p.dataset_seed,\n n_shot=p.n_shot,\n n_way=p.n_way,\n n_query=p.n_query,\n train_val_test_k_factors=(p.train_k_factor,p.val_k_factor,p.test_k_factor),\n):\n \n if x_transforms == []: x_transform = None\n else: x_transform = get_chained_transform(x_transforms)\n \n if episode_transforms == []: episode_transform = None\n else: raise Exception(\"episode_transforms not implemented\")\n \n episode_transform = lambda tup, _prefix=domain_prefix: (_prefix + str(tup[0]), tup[1])\n\n\n eaf = Episodic_Accessor_Factory(\n labels=labels,\n domains=domains,\n num_examples_per_domain_per_label=num_examples_per_domain_per_label,\n iterator_seed=iterator_seed,\n dataset_seed=dataset_seed,\n n_shot=n_shot,\n n_way=n_way,\n n_query=n_query,\n train_val_test_k_factors=train_val_test_k_factors,\n pickle_path=pickle_path,\n x_transform_func=x_transform,\n )\n\n train, val, test = eaf.get_train(), eaf.get_val(), eaf.get_test()\n train = Lazy_Iterable_Wrapper(train, episode_transform)\n val = Lazy_Iterable_Wrapper(val, episode_transform)\n test = Lazy_Iterable_Wrapper(test, episode_transform)\n\n if source_or_target_dataset==\"source\":\n train_original_source.append(train)\n val_original_source.append(val)\n test_original_source.append(test)\n\n p.domains_source.extend(\n [domain_prefix + str(u) for u in domains]\n )\n elif source_or_target_dataset==\"target\":\n train_original_target.append(train)\n val_original_target.append(val)\n test_original_target.append(test)\n p.domains_target.extend(\n [domain_prefix + str(u) for u in domains]\n )\n else:\n raise Exception(f\"invalid source_or_target_dataset: {source_or_target_dataset}\")\n ",
"_____no_output_____"
],
[
"for ds in p.datasets:\n add_dataset(**ds)",
"_____no_output_____"
],
[
"# from steves_utils.CORES.utils import (\n# ALL_NODES,\n# ALL_NODES_MINIMUM_1000_EXAMPLES,\n# ALL_DAYS\n# )\n\n# add_dataset(\n# labels=ALL_NODES,\n# domains = ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"cores.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"cores_{u}\"\n# )",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle1_{u}\"\n# )\n",
"_____no_output_____"
],
[
"# from steves_utils.ORACLE.utils_v2 import (\n# ALL_DISTANCES_FEET,\n# ALL_RUNS,\n# ALL_SERIAL_NUMBERS,\n# )\n\n\n# add_dataset(\n# labels=ALL_SERIAL_NUMBERS,\n# domains = list(set(ALL_DISTANCES_FEET) - {2,62,56}),\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"source\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"oracle2_{u}\"\n# )",
"_____no_output_____"
],
[
"# add_dataset(\n# labels=list(range(19)),\n# domains = [0,1,2],\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"metehan.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"met_{u}\"\n# )",
"_____no_output_____"
],
[
"# # from steves_utils.wisig.utils import (\n# # ALL_NODES_MINIMUM_100_EXAMPLES,\n# # ALL_NODES_MINIMUM_500_EXAMPLES,\n# # ALL_NODES_MINIMUM_1000_EXAMPLES,\n# # ALL_DAYS\n# # )\n\n# import steves_utils.wisig.utils as wisig\n\n\n# add_dataset(\n# labels=wisig.ALL_NODES_MINIMUM_100_EXAMPLES,\n# domains = wisig.ALL_DAYS,\n# num_examples_per_domain_per_label=100,\n# pickle_path=os.path.join(get_datasets_base_path(), \"wisig.node3-19.stratified_ds.2022A.pkl\"),\n# source_or_target_dataset=\"target\",\n# x_transform_func=global_x_transform_func,\n# domain_modifier=lambda u: f\"wisig_{u}\"\n# )",
"_____no_output_____"
],
[
"###################################\n# Build the dataset\n###################################\ntrain_original_source = Iterable_Aggregator(train_original_source, p.seed)\nval_original_source = Iterable_Aggregator(val_original_source, p.seed)\ntest_original_source = Iterable_Aggregator(test_original_source, p.seed)\n\n\ntrain_original_target = Iterable_Aggregator(train_original_target, p.seed)\nval_original_target = Iterable_Aggregator(val_original_target, p.seed)\ntest_original_target = Iterable_Aggregator(test_original_target, p.seed)\n\n# For CNN We only use X and Y. And we only train on the source.\n# Properly form the data using a transform lambda and Lazy_Iterable_Wrapper. Finally wrap them in a dataloader\n\ntransform_lambda = lambda ex: ex[1] # Original is (<domain>, <episode>) so we strip down to episode only\n\ntrain_processed_source = Lazy_Iterable_Wrapper(train_original_source, transform_lambda)\nval_processed_source = Lazy_Iterable_Wrapper(val_original_source, transform_lambda)\ntest_processed_source = Lazy_Iterable_Wrapper(test_original_source, transform_lambda)\n\ntrain_processed_target = Lazy_Iterable_Wrapper(train_original_target, transform_lambda)\nval_processed_target = Lazy_Iterable_Wrapper(val_original_target, transform_lambda)\ntest_processed_target = Lazy_Iterable_Wrapper(test_original_target, transform_lambda)\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"from steves_utils.transforms import get_average_magnitude, get_average_power\n\nprint(set([u for u,_ in val_original_source]))\nprint(set([u for u,_ in val_original_target]))\n\ns_x, s_y, q_x, q_y, _ = next(iter(train_processed_source))\nprint(s_x)\n\n# for ds in [\n# train_processed_source,\n# val_processed_source,\n# test_processed_source,\n# train_processed_target,\n# val_processed_target,\n# test_processed_target\n# ]:\n# for s_x, s_y, q_x, q_y, _ in ds:\n# for X in (s_x, q_x):\n# for x in X:\n# assert np.isclose(get_average_magnitude(x.numpy()), 1.0)\n# assert np.isclose(get_average_power(x.numpy()), 1.0)\n ",
"{'ORACLE.run1_26', 'ORACLE.run1_8', 'ORACLE.run1_32', 'ORACLE.run1_50', 'ORACLE.run1_38', 'ORACLE.run1_14', 'ORACLE.run1_20', 'ORACLE.run1_44'}\n"
],
[
"###################################\n# Build the model\n###################################\n# easfsl only wants a tuple for the shape\nmodel = Steves_Prototypical_Network(x_net, device=p.device, x_shape=tuple(p.x_shape))\noptimizer = Adam(params=model.parameters(), lr=p.lr)",
"(2, 256)\n"
],
[
"###################################\n# train\n###################################\njig = PTN_Train_Eval_Test_Jig(model, p.BEST_MODEL_PATH, p.device)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n patience=p.patience,\n optimizer=optimizer,\n criteria_for_best=p.criteria_for_best,\n)",
"epoch: 1, [batch: 1 / 6720], examples_per_second: 49.9724, train_label_loss: 2.7147, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"###################################\n# Evaluate the model\n###################################\nsource_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = Iterable_Aggregator((datasets.source.original.val,datasets.target.original.val))\n\nconfusion = ptn_confusion_by_domain_over_dataloader(model, p.device, val_dl)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": dict(p),\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"ptn\"),\n}",
"_____no_output_____"
],
[
"ax = get_loss_curve(experiment)\nplt.show()",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.7804361979166666 Target Test Label Accuracy: 0.6711258561643836\nSource Val Label Accuracy: 0.7753255208333333 Target Val Label Accuracy: 0.6774400684931506\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc780db15169bc56c23bfe0ecc5258c91ebcf80
| 51,874 |
ipynb
|
Jupyter Notebook
|
Shape interpolation.ipynb
|
javidcf/surfmorph
|
876bf9666cc6c47a52d0d7f899bfa181274809a3
|
[
"MIT"
] | 1 |
2018-05-31T20:03:14.000Z
|
2018-05-31T20:03:14.000Z
|
Shape interpolation.ipynb
|
javidcf/surfmorph
|
876bf9666cc6c47a52d0d7f899bfa181274809a3
|
[
"MIT"
] | null | null | null |
Shape interpolation.ipynb
|
javidcf/surfmorph
|
876bf9666cc6c47a52d0d7f899bfa181274809a3
|
[
"MIT"
] | 1 |
2021-01-07T08:46:28.000Z
|
2021-01-07T08:46:28.000Z
| 129.361596 | 32,651 | 0.752631 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbc7833e77c33387995cc16696d9bcd095c967ca
| 5,228 |
ipynb
|
Jupyter Notebook
|
2019-01-10-ESRF/notebooks/01.3.cookiecutter.ipynb
|
jtpio/quantstack-talks
|
092f93ddb9901cb614f428e13a0b1b1e3ffcc0ec
|
[
"BSD-3-Clause"
] | 82 |
2017-04-14T20:18:55.000Z
|
2021-12-25T23:38:52.000Z
|
2019-01-10-ESRF/notebooks/01.3.cookiecutter.ipynb
|
jtpio/quantstack-talks
|
092f93ddb9901cb614f428e13a0b1b1e3ffcc0ec
|
[
"BSD-3-Clause"
] | 3 |
2017-04-07T18:37:21.000Z
|
2020-07-11T09:37:53.000Z
|
2019-01-10-ESRF/notebooks/01.3.cookiecutter.ipynb
|
jtpio/quantstack-talks
|
092f93ddb9901cb614f428e13a0b1b1e3ffcc0ec
|
[
"BSD-3-Clause"
] | 59 |
2017-04-07T11:16:56.000Z
|
2022-03-25T14:48:55.000Z
| 21.966387 | 207 | 0.555471 |
[
[
[
"# A `CookieCutter` template project\n\n - Takes care of all the boilerplate for creating a new custom widget project\n - Based on Audrey Roy Greenfeld's (@audreyr) cookiecutter project.\n - URL of the project: https://github.com/jupyter/widget-cookiecutter\n \n<img src=\"./images/cookiecutter.jpg\" style=\"width:200px\"></img>\n\n```\npip install cookiecutter\ncookiecutter https://github.com/jupyter/widget-cookiecutter.git\n```\n\nThe cookiecutter project is meant to help custom widget authors get started with the packaging and the distribution of Jupyter interactive widgets.\n\nIt produces a project for a Jupyter interactive widget library following the current best practices for using interactive widgets. An implementation for a placeholder \"Hello World\" widget is provided",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown"
]
] |
cbc7954a7dc084c4761ba3846418795105efafea
| 27,876 |
ipynb
|
Jupyter Notebook
|
ML_Models/Classification/KNearestNeighbor/KNN_first_try.ipynb
|
lakshit2808/Machine-Learning-Notes
|
1b7760c2626c36a7f62c5a474e9fdadb76cb023b
|
[
"MIT"
] | 2 |
2021-09-04T17:13:48.000Z
|
2021-09-04T17:13:50.000Z
|
ML_Models/Classification/KNearestNeighbor/KNN_first_try.ipynb
|
lakshit2808/Machine-Learning-Notes
|
1b7760c2626c36a7f62c5a474e9fdadb76cb023b
|
[
"MIT"
] | null | null | null |
ML_Models/Classification/KNearestNeighbor/KNN_first_try.ipynb
|
lakshit2808/Machine-Learning-Notes
|
1b7760c2626c36a7f62c5a474e9fdadb76cb023b
|
[
"MIT"
] | 1 |
2021-11-23T19:45:02.000Z
|
2021-11-23T19:45:02.000Z
| 40.283237 | 6,414 | 0.536232 |
[
[
[
"<a href=\"https://colab.research.google.com/github/lakshit2808/Machine-Learning-Notes/blob/master/ML_Models/Classification/KNearestNeighbor/KNN_first_try.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# K-Nearest Neighbor\n**K-Nearest Neighbors** is an algorithm for supervised learning. Where the data is 'trained' with data points corresponding to their classification. Once a point is to be predicted, it takes into account the 'K' nearest points to it to determine it's classification.\n",
"_____no_output_____"
],
[
"### Here's an visualization of the K-Nearest Neighbors algorithm.\n\n<img src=\"https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-ML0101EN-SkillsNetwork/labs/Module%203/images/KNN_Diagram.png\">\n",
"_____no_output_____"
],
[
"In this case, we have data points of Class A and B. We want to predict what the star (test data point) is. If we consider a k value of 3 (3 nearest data points) we will obtain a prediction of Class B. Yet if we consider a k value of 6, we will obtain a prediction of Class A.<br><br>\nIn this sense, it is important to consider the value of k. But hopefully from this diagram, you should get a sense of what the K-Nearest Neighbors algorithm is. It considers the 'K' Nearest Neighbors (points) when it predicts the classification of the test point.\n\n",
"_____no_output_____"
],
[
"## 1. Importing Libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np \nimport matplotlib.pyplot as plt\nfrom sklearn import preprocessing",
"_____no_output_____"
]
],
[
[
"## 2. Reading Data",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('teleCust.csv')\ndf.head()",
"_____no_output_____"
]
],
[
[
"## 3. Data Visualization and Analysis\n#### Let’s see how many of each class is in our data set\n",
"_____no_output_____"
]
],
[
[
"df['custcat'].value_counts()",
"_____no_output_____"
]
],
[
[
"The target field, called **custcat**, has four possible values that correspond to the four customer groups, as follows:\n 1. Basic Service\n 2. E-Service\n 3. Plus Service\n 4. Total Service\n",
"_____no_output_____"
]
],
[
[
"df.hist(column='income' , bins=50)",
"_____no_output_____"
]
],
[
[
"### Feature Set\nLet's Define a feature set: X",
"_____no_output_____"
]
],
[
[
"df.columns",
"_____no_output_____"
]
],
[
[
"To use scikit-learn library, we have to convert the Pandas data frame to a Numpy array:\n",
"_____no_output_____"
]
],
[
[
"X = df[['region', 'tenure', 'age', 'marital', 'address', 'income', 'ed',\n 'employ', 'retire', 'gender', 'reside']].values\nX[0:5]",
"_____no_output_____"
]
],
[
[
"What are our labels?",
"_____no_output_____"
]
],
[
[
"y = df['custcat'].values\ny[0:5]",
"_____no_output_____"
]
],
[
[
"### Normalize Data\nNormalization in this case essentially means standardization. Standardization is the process of transforming data based on the mean and standard deviation for the whole set. Thus, transformed data refers to a standard distribution with a mean of 0 and a variance of 1.<br><br>\nData Standardization give data zero mean and unit variance, it is good practice, especially for algorithms such as KNN which is based on distance of cases:\n",
"_____no_output_____"
]
],
[
[
"X = preprocessing.StandardScaler().fit(X).transform(X.astype(float))\nX[0:5]",
"_____no_output_____"
]
],
[
[
"## 4. Train/Test Split ",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\nX_train , X_test , y_train , y_test = train_test_split(X , y , test_size= 0.2 , random_state = 4)\n\nprint ('Train set:', X_train.shape, y_train.shape)\nprint ('Test set:', X_test.shape, y_test.shape)",
"Train set: (800, 11) (800,)\nTest set: (200, 11) (200,)\n"
]
],
[
[
"## 5. Classification(KNN)",
"_____no_output_____"
]
],
[
[
"from sklearn.neighbors import KNeighborsClassifier",
"_____no_output_____"
]
],
[
[
"### Training\nLets start the algorithm with k=4 for now:",
"_____no_output_____"
]
],
[
[
"all_acc = []\n\nfor i in range(1, 100):\n KNN = KNeighborsClassifier(n_neighbors=i).fit(X_train , y_train)\n all_acc.append(accuracy_score(y_test , KNN.predict(X_test)))\n \nbest_acc = max(all_acc)\nbest_k = all_acc.index(best_acc) + 1\n\nKNN = KNeighborsClassifier(n_neighbors=best_k).fit(X_train , y_train)",
"_____no_output_____"
]
],
[
[
"### Prediction",
"_____no_output_____"
]
],
[
[
"y_ = KNN.predict(X_test)\ny_[0:5]",
"_____no_output_____"
]
],
[
[
"## 6. Accuracy Evaluation \nIn multilabel classification, **accuracy classification score** is a function that computes subset accuracy. This function is equal to the jaccard_score function. Essentially, it calculates how closely the actual labels and predicted labels are matched in the test set.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import accuracy_score\nprint('Train Set Accuracy: {}'.format(accuracy_score(y_train , KNN.predict(X_train))))\nprint('Ttest Set Accuracy: {}'.format(accuracy_score(y_test , KNN.predict(X_test))))",
"Train Set Accuracy: 0.4275\nTtest Set Accuracy: 0.41\n"
]
],
[
[
"#### What about other K?\n\nK in KNN, is the number of nearest neighbors to examine. It is supposed to be specified by the User. So, how can we choose right value for K?\nThe general solution is to reserve a part of your data for testing the accuracy of the model. Then chose k =1, use the training part for modeling, and calculate the accuracy of prediction using all samples in your test set. Repeat this process, increasing the k, and see which k is the best for your model.\n\nWe can calculate the accuracy of KNN for different K.\n",
"_____no_output_____"
]
],
[
[
"all_acc = []\n\nfor i in range(1, 100):\n KNN = KNeighborsClassifier(n_neighbors=i).fit(X_train , y_train)\n all_acc.append(accuracy_score(y_test , KNN.predict(X_test)))\n \nbest_acc = max(all_acc)\nbest_k = all_acc.index(best_acc) + 1\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc7a8feb38ffbb7299a04ea22ebed2447325142
| 29,417 |
ipynb
|
Jupyter Notebook
|
data/.ipynb_checkpoints/Ben_EDA-checkpoint.ipynb
|
bwright8/vis
|
4b640be619e2bbf24dc2d78b71483286c34a8ca0
|
[
"MIT"
] | 1 |
2021-09-15T01:01:43.000Z
|
2021-09-15T01:01:43.000Z
|
data/.ipynb_checkpoints/Ben_EDA-checkpoint.ipynb
|
bwright8/vis
|
4b640be619e2bbf24dc2d78b71483286c34a8ca0
|
[
"MIT"
] | null | null | null |
data/.ipynb_checkpoints/Ben_EDA-checkpoint.ipynb
|
bwright8/vis
|
4b640be619e2bbf24dc2d78b71483286c34a8ca0
|
[
"MIT"
] | 1 |
2021-11-05T21:35:57.000Z
|
2021-11-05T21:35:57.000Z
| 32.255482 | 151 | 0.360608 |
[
[
[
"import pandas as pd\nimport sklearn\nimport numpy as np\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.tree import DecisionTreeClassifier",
"_____no_output_____"
],
[
"appRecord = pd.read_csv(\"application_record.csv\");\ncreditRecord = pd.read_csv(\"credit_record.csv\");",
"_____no_output_____"
],
[
"appRecord.head()",
"_____no_output_____"
],
[
"creditRecord.head()",
"_____no_output_____"
],
[
"df = pd.merge(\n appRecord,\n creditRecord,\n how=\"inner\"\n)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"y = df['STATUS'].map({\"0\":0,\"1\":1,'2':2,'3':3,'4':4,'5':5,\"C\":-1,\"X\":-2})",
"_____no_output_____"
],
[
"from sklearn.preprocessing import LabelEncoder,OneHotEncoder\nlabel_encoder =LabelEncoder();\n",
"_____no_output_____"
],
[
"X = df.copy().drop(['ID','MONTHS_BALANCE','STATUS','OCCUPATION_TYPE'],axis=1)\nX.head()",
"_____no_output_____"
],
[
"for col in ['CODE_GENDER','FLAG_OWN_CAR','FLAG_OWN_REALTY','NAME_INCOME_TYPE','NAME_EDUCATION_TYPE','NAME_FAMILY_STATUS','NAME_HOUSING_TYPE']:\n X[col] = label_encoder.fit_transform(X[col])\n",
"_____no_output_____"
],
[
"X.columns",
"_____no_output_____"
],
[
"# as a test, make a forest model with just a single feature. \nclf = RandomForestClassifier(max_depth=5, random_state=0);\nclf.fit(X, y)",
"_____no_output_____"
],
[
"# export as dot file\n# on the command line, type dot tree.dot -Tpng -o tree.png to conver into png. \nfrom sklearn.tree import export_graphviz;\nimport os;\nlocalFilePath = \"C:/Users/yun91/Documents/GitHub/vis/data\"\nexport_graphviz(\n clf.estimators_[0],\n out_file = \"tree2.dot\",\n rounded = True,\n filled = True) ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc7c4e3672009c9c405476986956d9bcd4fcc67
| 6,570 |
ipynb
|
Jupyter Notebook
|
notebooks/envs/mountain_car_continuous_v1.ipynb
|
miquelramirez/width-lookaheads-python
|
122442136d8254be44f9dbcae365327e0f669bfa
|
[
"MIT"
] | 1 |
2019-09-10T20:36:48.000Z
|
2019-09-10T20:36:48.000Z
|
notebooks/envs/mountain_car_continuous_v1.ipynb
|
miquelramirez/width-lookaheads-python
|
122442136d8254be44f9dbcae365327e0f669bfa
|
[
"MIT"
] | null | null | null |
notebooks/envs/mountain_car_continuous_v1.ipynb
|
miquelramirez/width-lookaheads-python
|
122442136d8254be44f9dbcae365327e0f669bfa
|
[
"MIT"
] | 1 |
2019-09-10T20:36:52.000Z
|
2019-09-10T20:36:52.000Z
| 25.564202 | 249 | 0.547793 |
[
[
[
"import numpy as np\nimport gym\nimport matplotlib.pyplot as plt\n\nimport sys\nsys.path.append('../..')\n\nimport wizluk.envs\nfrom wizluk.policies import ContinuousZeroPolicy, ContinuousRandomPolicy",
"/usr/local/lib/python3.5/dist-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
]
],
[
[
"## Initialisation",
"_____no_output_____"
]
],
[
[
"env = gym.make('MountainCarContinuous-v1')",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n"
]
],
[
[
"## Parameters for Baselines",
"_____no_output_____"
]
],
[
[
"H = 500 # Horizon\nN = 10 # Number of Rollouts",
"_____no_output_____"
]
],
[
[
"### Zero Policy",
"_____no_output_____"
]
],
[
[
"env = gym.make('MountainCarContinuous-v1')\nzero_baseline = np.zeros((N,1))\n\nenv.seed(1337)\nzero_pi = ContinuousZeroPolicy()\n\nfor k in range(N):\n x = env.reset()\n for s in range(H):\n u = zero_pi.get_action(env, x)\n x, reward, done, info = env.step(u)\n zero_baseline[k] += reward\n if done : break\n\nzero_baseline_cost = np.mean(zero_baseline)\nprint(\"Zero baseline cost: {}\".format(zero_baseline_cost))",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\nZero baseline cost: -190.0490333454432\n"
]
],
[
[
"#### Demonstration",
"_____no_output_____"
]
],
[
[
"env = gym.make('MountainCarContinuous-v1')\nenv.seed(1337)\nzero_pi = ContinuousZeroPolicy()\n\nx = env.reset()\nfor s in range(H):\n env.render()\n u = zero_pi.get_action(env, x)\n x, reward, done, info = env.step(u)\n if done : break\nenv.close()",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n"
]
],
[
[
"### Random Policy",
"_____no_output_____"
]
],
[
[
"env = gym.make('MountainCarContinuous-v1')\nnp.random.seed(1337)\nenv.seed(1337)\n\nrandom_baseline = np.zeros((N,1))\nrandom_pi = ContinuousRandomPolicy()\n\nfor k in range(N):\n x = env.reset()\n for s in range(H):\n u = random_pi.get_action(env, x)\n x, reward, done, info = env.step(u)\n random_baseline[k] += reward\n if done : break\n\nrandom_baseline_cost = np.mean(random_baseline)\nprint(\"Random baseline cost: {}\".format(random_baseline_cost))",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\nRandom baseline cost: -195.41120864317648\n"
]
],
[
[
"#### Demonstration",
"_____no_output_____"
]
],
[
[
"env = gym.make('MountainCarContinuous-v1')\nenv.seed(1337)\nrandom_pi = ContinuousZeroPolicy()\n\nx = env.reset()\nfor s in range(H):\n env.render()\n u = random_pi.get_action(env, x)\n x, reward, done, info = env.step(u)\n if done : break\nenv.close()",
"\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n\u001b[33mWARN: gym.spaces.Box autodetected dtype as <class 'numpy.float32'>. Please provide explicit dtype.\u001b[0m\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc7c4e7b5b4d0b7cafeec0991e41b3686959f2c
| 79,394 |
ipynb
|
Jupyter Notebook
|
examples/Analysis/FigHI-Comparing-Modeling_Choices.ipynb
|
BiocomputeLab/FLAIR
|
9d0f439a8d692cc16c59f46092c472ce8a8b768d
|
[
"MIT"
] | null | null | null |
examples/Analysis/FigHI-Comparing-Modeling_Choices.ipynb
|
BiocomputeLab/FLAIR
|
9d0f439a8d692cc16c59f46092c472ce8a8b768d
|
[
"MIT"
] | null | null | null |
examples/Analysis/FigHI-Comparing-Modeling_Choices.ipynb
|
BiocomputeLab/FLAIR
|
9d0f439a8d692cc16c59f46092c472ce8a8b768d
|
[
"MIT"
] | 1 |
2020-12-22T12:50:37.000Z
|
2020-12-22T12:50:37.000Z
| 201.507614 | 38,620 | 0.894211 |
[
[
[
"import numpy as np\nimport pandas as pd\nimport random\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%config InlineBackend.figure_format = 'retina'\nfrom IPython.core.display import display, HTML\ndisplay(HTML(\"<style>.container {width:100% !important;}</style>\")) \nfrom scipy.special import gamma, factorial,digamma\nfrom matplotlib import cm\nfrom matplotlib.colors import ListedColormap, LinearSegmentedColormap\n\nimport matplotlib.gridspec as gridspec\n\nimport sys\nsys.path.append(r'/Users/ys18223/Documents/GitHub/FLAIR_BiocomputeLab')\n",
"_____no_output_____"
]
],
[
[
"# Fig H",
"_____no_output_____"
]
],
[
[
"#Input MAPE performance accross simulations when estimating the mean fluorescence\nd_mean = {'MAPE': [100*i for i in ([0.1674891 , 0.14371818, 0.12273398, \n 0.16679492, 0.13970324, 0.1015513 ,\n 0.16319497, 0.12743953, 0.06931147]+[0.51141972, 0.51385324, 0.51403695,\n 0.52769436, 0.51004928, 0.51341036, \n 0.53446 , 0.52250617, 0.5075517 ])]+[15.29211367, 14.14405139, 14.05101411]+[12.61702118, 10.50428435, 9.82247402]+[10.31754068, 7.2084087 , 4.77361639]+[16.35151345, 16.9359747 , 17.78217523]+[14.38362791, 14.93895699, 15.7100954 ]+[13.14528142, 13.4672431 , 14.25780018], 'distribution': ['Gamma']*18+['Lognormal']*18,'inference':['ML']*9+['MOM']*9+['ML']*9+['MOM']*9}\ndf_mean = pd.DataFrame(data=d_mean)\ndf_mean.head()\n",
"_____no_output_____"
],
[
"# Create the figure\nfig = plt.figure(figsize=(11.7,8.3))\ngs = gridspec.GridSpec(1, 1)\nax = plt.subplot(gs[0])\nmy_pal = {\"ML\": \"#2463A3\", \"MOM\": \"#B5520E\"}\nax=sns.violinplot(x=\"distribution\", y=\"MAPE\", hue=\"inference\",\n data=df_mean, palette=my_pal)\n\n\nax.set_ylabel('MAPE (mean) %') \nax.set_xlabel('')\n# my_pal = ['#2463A3', '#B5520E','#2463A3', '#B5520E']\n# INF=['ML','MOM','ML','MOM']\n# color_dict = dict(zip(INF, my_pal ))\n\n# for i in range(0,4):\n# mybox = ax.artists[i]\n# mybox.set_facecolor(color_dict[INF[i]])\n\n#plt.legend(frameon=False,fontsize=12)\nax.get_legend().remove()\n\nsns.despine()\n\nwidth=3.54\nheight=3.54\nfig.set_size_inches(width, height)\n\nplt.subplots_adjust(hspace=.0 , wspace=.00, left=.15, right=.95, top=.95, bottom=.13)\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Fig I",
"_____no_output_____"
]
],
[
[
"#Input MAPE performance accross simulations when estimating the mean variance\nd_var = {'MAPE': [56.51961891, 50.47877742, 46.13735704,\n 56.41471139, 48.30979619, 39.03006257,\n 56.08137685, 44.53477141, 27.01354216]+[287.74453306, 298.1863082 , 298.21313797,299.7961364 , 300.44014621, 311.36703739,\n 324.08161946, 323.83104867, 327.57942772]+[67.89211699, 64.24130949, 63.92732816]+[60.43748406, 50.92945822, 46.84127056]+[54.94239969, 39.2380389 , 24.5262507 ]+[195.21194215, 232.21351093, 238.5230456 ]+[219.98637949, 221.72468045, 217.98143615]+[226.76576441, 196.59937264, 221.02871965], 'distribution': ['Gamma']*18+['Lognormal']*18,'inference':['ML']*9+['MOM']*9+['ML']*9+['MOM']*9}\ndf_var = pd.DataFrame(data=d_var)\ndf_var.head()",
"_____no_output_____"
],
[
"# Create the figure\nfig = plt.figure(figsize=(11.7,8.3))\ngs = gridspec.GridSpec(1, 1)\nax = plt.subplot(gs[0])\nmy_pal = {\"ML\": \"#2463A3\", \"MOM\": \"#B5520E\"}\nax=sns.violinplot(x=\"distribution\", y=\"MAPE\", hue=\"inference\",\n data=df_var, palette=my_pal)\nax.set_ylabel('MAPE (standard deviation) %') \nax.set_xlabel('')\nax.get_legend().remove()\nsns.despine()\n\nwidth=3.54\nheight=3.54\nfig.set_size_inches(width, height)\n\nplt.subplots_adjust(hspace=.0 , wspace=.00, left=.15, right=.95, top=.95, bottom=.13)\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbc7c5444360b67c3e0d2fd5b1c216325b468f94
| 164,335 |
ipynb
|
Jupyter Notebook
|
S5_Logistic_Regr_JCS.ipynb
|
juancas9812/Inteligencia-Artificial-2020-3
|
bd626a79d9f866a0dab30c93b8cb6e85561a5036
|
[
"MIT"
] | null | null | null |
S5_Logistic_Regr_JCS.ipynb
|
juancas9812/Inteligencia-Artificial-2020-3
|
bd626a79d9f866a0dab30c93b8cb6e85561a5036
|
[
"MIT"
] | null | null | null |
S5_Logistic_Regr_JCS.ipynb
|
juancas9812/Inteligencia-Artificial-2020-3
|
bd626a79d9f866a0dab30c93b8cb6e85561a5036
|
[
"MIT"
] | null | null | null | 299.881387 | 131,812 | 0.92477 |
[
[
[
"<a href=\"https://colab.research.google.com/github/juancas9812/Inteligencia-Artificial-2020-3/blob/master/S5_Logistic_Regr_JCS.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Pontificia Universidad Javeriana\n# Inteligencia Artificial 2020-30\nTarea Semana 5\n\nJuan Camilo Sarmiento Peñuela",
"_____no_output_____"
],
[
"**1.** Implementar (adecuar) los dos métodos descritos en: https://ml-cheatsheet.readthedocs.io/en/latest/logistic_regression.html#id13 Con los datos en el csv en teams en la carpeta semana 5 (datos_multivariados.csv). Puede descargar el código también en el github del autor, pero por favor leer primero el link de arriba. https://github.com/bfortuner/ml-glossary \n\n* Primer método - Regresión Logística Binaria:\n\n\n> Primero se importan los módulos que se van a utilizar, los cuales son numpy para poder hacer uso de vectores y matrices, pandas para obtener los datos, y matplotlib para los graficos. Luego, se copian las funciones que se encuentran en el primer link, y se ajustan para que funcionen con el interprete de python 3, dado que estan escritas para python 2.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def sigmoid(z):\n return 1.0 / (1 + np.exp(-z))",
"_____no_output_____"
],
[
"def predict(features, weights):\n z = np.dot(features,weights)\n return sigmoid(z)",
"_____no_output_____"
],
[
"def cost_function(features, labels, weights):\n '''\n Using Mean Absolute Error\n\n Features:(100,3)\n Labels: (100,1)\n Weights:(3,1)\n Returns 1D matrix of predictions\n Cost = (labels*log(predictions) + (1-labels)*log(1-predictions) ) / len(labels)\n '''\n observations = len(labels)\n\n predictions = predict(features, weights)\n\n #Take the error when label=1\n class1_cost = -labels*np.log(predictions)\n\n #Take the error when label=0\n class2_cost = (1-labels)*np.log(1-predictions)\n\n #Take the sum of both costs\n cost = class1_cost - class2_cost\n\n #Take the average cost\n cost = cost.sum() / observations\n\n return cost",
"_____no_output_____"
],
[
"def update_weights(features, labels, weights, lr):\n '''\n Vectorized Gradient Descent\n\n Features:(200, 3)\n Labels: (200, 1)\n Weights:(3, 1)\n '''\n N = len(features)\n\n #1 - Get Predictions\n predictions = predict(features, weights)\n\n #2 Transpose features from (200, 3) to (3, 200)\n # So we can multiply w the (200,1) cost matrix.\n # Returns a (3,1) matrix holding 3 partial derivatives --\n # one for each feature -- representing the aggregate\n # slope of the cost function across all observations\n gradient = np.dot(features.T, predictions - labels)\n\n #3 Take the average cost derivative for each feature\n gradient /= N\n\n #4 - Multiply the gradient by our learning rate\n gradient *= lr\n\n #5 - Subtract from our weights to minimize cost\n weights -= gradient\n\n return weights",
"_____no_output_____"
],
[
"def decision_boundary(prob):\n return 1 if prob >= .5 else 0",
"_____no_output_____"
],
[
"def classify(predictions):\n '''\n input - N element array of predictions between 0 and 1\n output - N element array of 0s (False) and 1s (True)\n '''\n decision_boundary_vec = np.vectorize(decision_boundary)\n return decision_boundary_vec(predictions).flatten()",
"_____no_output_____"
],
[
"def train(features, labels, weights, lr, iters):\n cost_history = []\n\n for i in range(iters):\n weights = update_weights(features, labels, weights, lr)\n\n #Calculate error for auditing purposes\n cost = cost_function(features, labels, weights)\n cost_history.append(cost)\n\n\n return weights, cost_history",
"_____no_output_____"
],
[
"def accuracy(predicted_labels, actual_labels):\n diff = predicted_labels - actual_labels\n return 1.0 - (float(np.count_nonzero(diff)) / len(diff))",
"_____no_output_____"
],
[
"def plot_decision_boundary(trues, falses):\n fig = plt.figure()\n ax = fig.add_subplot(111)\n\n no_of_preds = len(trues) + len(falses)\n\n ax.scatter([i for i in range(len(trues))], trues, s=25, c='b', marker=\"o\", label='Trues')\n ax.scatter([i for i in range(len(falses))], falses, s=25, c='r', marker=\"s\", label='Falses')\n\n plt.legend(loc='upper right');\n ax.set_title(\"Decision Boundary\")\n ax.set_xlabel('N/2')\n ax.set_ylabel('Predicted Probability')\n plt.axhline(.5, color='black')\n plt.show()",
"_____no_output_____"
]
],
[
[
">Luego de tener todas las funciones, se procede a leer los datos del archivo data_classification.csv, y se implementan las funciones",
"_____no_output_____"
]
],
[
[
"data=pd.read_csv(\"data_classification.csv\", sep=';',header=0)\narreglo=data.values\nYCOL=2\nXb=arreglo[:,0:YCOL]\nXa=np.ones((len(Xb),1))\nX=np.concatenate((Xa,Xb),axis=1)\nY=arreglo[:,YCOL]\nYCOL2=5\nthetas=np.array([-12.5, 1.3, 1])\nNum_Iter=2500\nalpha=0.5\ncinit=112\n\nfeatures=X\nlabels=Y\n\nfor alpha in np.arange(0.4,0.5,0.001):\n weights, cost=train(X, Y, thetas, alpha, Num_Iter)\n if cost[-1]<cinit:\n final_w=weights\n cinit=cost[-1]\n alphaopt=alpha\n\nprint(final_w)\nprint(cinit)\nprint(alphaopt)\n\ncount_true=0\ncount_false=0\nacum=predict(X,final_w) \nprob=classify(acum)\ntrues=[]\nfalses=[]\nfor i in np.arange(0,acum.size):\n if prob[i]==Y[i]:\n trues.append(acum[i]) \n \n else:\n falses.append(acum[i]) \n\nplot_decision_boundary(trues,falses)",
"[-16.1602695 1.74538164 1.48650834]\n0.21700791641301476\n0.4980000000000001\n"
]
],
[
[
"* Segundo Método - Regresión logística multiclase:\n\n> Para este método se nececita importar algunos modulos de scikit-learn, debido a que se usa este modulo para la implementación de este método",
"_____no_output_____"
]
],
[
[
"import sklearn\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
]
],
[
[
"> Ahora se hace la implementación:",
"_____no_output_____"
]
],
[
[
"# Normalize grades to values between 0 and 1 for more efficient computation\nnormalized_range = sklearn.preprocessing.MinMaxScaler(feature_range=(-1,1))\n\n# Extract Features + Labels\nlabels.shape = (100,) #scikit expects this\nfeatures = normalized_range.fit_transform(features)\n\n# Create Test/Train\nfeatures_train,features_test,labels_train,labels_test = train_test_split(features,labels,test_size=0.4,random_state=0)\n\n# Scikit Logistic Regression\nscikit_log_reg = LogisticRegression()\nscikit_log_reg.fit(features_train,labels_train)\n\n#Score is Mean Accuracy\nscikit_score = scikit_log_reg.predict(features_test)\nprecision=accuracy(scikit_score,labels_test)*100\nprint('Scikit score: ', precision)\n\n#Our Mean Accuracy\nprobabilities = predict(features, weights).flatten()\nclassifications = classify(probabilities)\nour_acc = accuracy(classifications,labels.flatten())\nprint('Our score: ',our_acc)",
"Scikit score: 90.0\nOur score: 0.55\n"
]
],
[
[
"**2.** Correr el ejemplo multiclase al final de: https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html , comentar el uso y entradas de cada función que hagan uso de sklearn\n\n\n",
"_____no_output_____"
]
],
[
[
"#Se importa el dataset de Iris que se encuentra en en sklearn.datasets\nfrom sklearn.datasets import load_iris \n\n#Se importa la función para realizar la regresión logística. Se encuentra en \n#sklearn.linear_model\nfrom sklearn.linear_model import LogisticRegression\n\n\nX, y = load_iris(return_X_y=True) #Se cargan las etiquetas del dataset\nclf = LogisticRegression(random_state=0).fit(X, y) #Se entrena el modelo de la regresión logística\nclf.predict(X[:2, :]) #Hace la predicción sobre cuál etiqueta pertenece cuál caracteristica\nclf.predict_proba(X[:2, :]) #Retorna la matriz de probabilidades\nclf.score(X, y) #Se calcula el valor de la exactitud obtenida por la regresión",
"/usr/local/lib/python3.6/dist-packages/sklearn/linear_model/_logistic.py:940: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)\n"
]
],
[
[
"**3.** Encontrar la derivada de J para un theta cualquiera de la Regresión Logística. Usarla para confirmar las ecuaciones de descenso del gradiente en las presentaciones.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
" \n\n---\n\n\n",
"_____no_output_____"
],
[
"# Conclusiones",
"_____no_output_____"
],
[
"* Se encontraron problemas al intentar importar y usar los datos. Por esto se recomienda ver el tipo de dato y en caso de ser posible la forma, ya que se va a trabajar con matrices y vectores.\n\n* Asimismo, es importante revisar el tipo de dato que se requiere para utilizar ciertos métodos para las clases que utilizan los diferentes módulos.\n\n* En el caso de la regresión logística binaria, como se estaba usando el descenso del gradiente, se encuentra que al cambiar los valores de los thetas, se converge siempre a los mismos valores de los pesos y del alpha, los cuales son [-16.1602695 1.74538164 1.48650834] y 0.4980000000000001 respectivamente",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbc7c7623e406d01f30a44b717243abb37ccdb0d
| 16,720 |
ipynb
|
Jupyter Notebook
|
docs/source/guide/ipynb/hilbertspace_legacy.ipynb
|
scqubits/scqubits-doc
|
d19f165b0bf484559a3caf57b8c3896ee7e43d68
|
[
"BSD-3-Clause"
] | 5 |
2021-03-16T21:15:28.000Z
|
2022-01-20T12:05:35.000Z
|
docs/source/guide/ipynb/hilbertspace_legacy.ipynb
|
scqubits/scqubits-doc
|
d19f165b0bf484559a3caf57b8c3896ee7e43d68
|
[
"BSD-3-Clause"
] | 4 |
2021-02-20T17:25:40.000Z
|
2022-01-27T08:12:23.000Z
|
docs/source/guide/ipynb/hilbertspace_legacy.ipynb
|
scqubits/scqubits-doc
|
d19f165b0bf484559a3caf57b8c3896ee7e43d68
|
[
"BSD-3-Clause"
] | 5 |
2021-02-15T20:30:03.000Z
|
2021-09-09T19:01:20.000Z
| 35.274262 | 1,074 | 0.535347 |
[
[
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'svg'\n\nimport scqubits as scq\nimport scqubits.legacy.sweep_plotting as splot\nfrom scqubits import HilbertSpace, InteractionTerm, ParameterSweep\n\nimport numpy as np",
"_____no_output_____"
]
],
[
[
".. note::\n This describes a legacy version of the `HilbertSpace` class which is deprecated with scqubits v1.4.\n\n\n# Composite Hilbert Spaces, QuTiP Interface",
"_____no_output_____"
],
[
"The `HilbertSpace` class provides data structures and methods for handling composite Hilbert spaces which may consist of multiple qubits or qubits and oscillators coupled to each other. To harness the power of QuTiP, a toolbox for studying stationary and dynamical properties of closed and open quantum systems (and much more), `HilbertSpace` provides a convenient interface: it generates `qutip.qobj` objects which are then directly handled by QuTiP.",
"_____no_output_____"
],
[
"## Example: two transmons coupled to a harmonic mode\n\nTransmon qubits can be capacitively coupled to a common harmonic mode, realized by an LC oscillator or a transmission-line resonator. The Hamiltonian describing such a composite system is given by:\n\\begin{equation}\nH=H_\\text{tmon,1} + H_\\text{tmon,2} + \\omega_r a^\\dagger a + \\sum_{j=1,2}g_j n_j(a+a^\\dagger),\n\\end{equation}\nwhere $j=1,2$ enumerates the two transmon qubits, $\\omega_r$ is the (angular) frequency of the resonator. Furthermore, $n_j$ is the charge number operator for qubit $j$, and $g_j$ is the coupling strength between qubit $j$ and the resonator.\n\n### Create Hilbert space components\n\nThe first step consists of creating the objects describing the individual building blocks of the full Hilbert space. Here, these will be the two transmons and one oscillator:",
"_____no_output_____"
]
],
[
[
"tmon1 = scq.Transmon(\n EJ=40.0,\n EC=0.2,\n ng=0.3,\n ncut=40,\n truncated_dim=4 # after diagonalization, we will keep 3 levels\n)\n\ntmon2 = scq.Transmon(\n EJ=15.0,\n EC=0.15,\n ng=0.0,\n ncut=30,\n truncated_dim=4\n)\n\nresonator = scq.Oscillator(\n E_osc=4.5,\n truncated_dim=4 # up to 3 photons (0,1,2,3)\n)",
"_____no_output_____"
]
],
[
[
"The system objects are next grouped into a Python list, and in this form used for the initialization of a `HilbertSpace` object. Once created, a print call to this object outputs a summary of the composite Hilbert space.",
"_____no_output_____"
]
],
[
[
"hilbertspace = scq.HilbertSpace([tmon1, tmon2, resonator])\nprint(hilbertspace)",
"====== HilbertSpace object ======\n\nTRANSMON\n ———— PARAMETERS ————\nEJ\t: 40.0\nEC\t: 0.2\nng\t: 0.3\nncut\t: 40\ntruncated_dim\t: 4\nHilbert space dimension\t: 81\n\nTRANSMON\n ———— PARAMETERS ————\nEJ\t: 15.0\nEC\t: 0.15\nng\t: 0.0\nncut\t: 30\ntruncated_dim\t: 4\nHilbert space dimension\t: 61\n\nOSCILLATOR\n ———— PARAMETERS ————\nE_osc\t: 4.5\ntruncated_dim\t: 4\nHilbert space dimension\t: 4\n\n"
]
],
[
[
"One useful method of the `HilbertSpace` class is `.bare_hamiltonian()`. This yields the bare Hamiltonian of the non-interacting subsystems, expressed as a `qutip.Qobj`:",
"_____no_output_____"
]
],
[
[
"bare_hamiltonian = hilbertspace.bare_hamiltonian()\nbare_hamiltonian",
"_____no_output_____"
]
],
[
[
"### Set up the interaction between subsystems",
"_____no_output_____"
],
[
"The pairwise interactions between subsystems are assumed to have the general form \n\n$V=\\sum_{i\\not= j} g_{ij} A_i B_j$, \n\nwhere $g_{ij}$ parametrizes the interaction strength between subsystems $i$ and $j$. The operator content of the coupling is given by the two coupling operators $A_i$, $B_j$, which are operators in the two respective subsystems.\nThis structure is captured by setting up an `InteractionTerm` object:",
"_____no_output_____"
]
],
[
[
"g1 = 0.1 # coupling resonator-CPB1 (without charge matrix elements)\ng2 = 0.2 # coupling resonator-CPB2 (without charge matrix elements)\n\ninteraction1 = InteractionTerm(\n hilbertspace = hilbertspace,\n g_strength = g1,\n op1 = tmon1.n_operator(),\n subsys1 = tmon1,\n op2 = resonator.creation_operator() + resonator.annihilation_operator(),\n subsys2 =resonator\n)\n\ninteraction2 = InteractionTerm(\n hilbertspace = hilbertspace,\n g_strength = g2,\n op1 = tmon2.n_operator(),\n subsys1 = tmon2,\n op2 = resonator.creation_operator() + resonator.annihilation_operator(),\n subsys2 = resonator\n)",
"_____no_output_____"
]
],
[
[
"Each `InteractionTerm` object is initialized by specifying\n1. the Hilbert space object to which it will belong\n2. the interaction strength coefficient $g_{ij}$\n3. `op1`, `op2`: the subsystem operators $A_i$, $B_j$ (these should be operators within the subsystems' respective Hilbert spaces only)\n4. `subsys1`: the subsystem objects to which `op1` and `op2` belong\n\nNote: interaction Hamiltonians of the alternative form $V=g_{ij}A_i B_j^\\dagger + g_{ij}^* A_i^\\dagger B_J$ (a typical form when performing rotating-wave approximation) can be specified by setting `op1` to $A_i$ and `op2` to $B_j^\\dagger$, and providing the additional keyword parameter `add_hc = True`.\n\nNow, collect all interaction terms in a list, and insert into the HilbertSpace object.",
"_____no_output_____"
]
],
[
[
"interaction_list = [interaction1, interaction2]\nhilbertspace.interaction_list = interaction_list",
"_____no_output_____"
]
],
[
[
"With the interactions specified, the full Hamiltonian of the coupled system can be obtained via the method `.hamiltonian()`. Again, this conveniently results in a `qubit.Qobj` operator:",
"_____no_output_____"
]
],
[
[
"dressed_hamiltonian = hilbertspace.hamiltonian()\ndressed_hamiltonian",
"_____no_output_____"
]
],
[
[
"### Obtaining the eigenspectrum via QuTiP\n\nSince the Hamiltonian obtained this way is a proper `qutip.qobj`, all QuTiP routines are now available. In the first case, we are still making use of the scqubit `HilbertSpace.eigensys()` method. In the second, case, we use QuTiP's method `.eigenenergies()`:",
"_____no_output_____"
]
],
[
[
"evals, evecs = hilbertspace.eigensys(evals_count=4)\nprint(evals)",
"[-48.97770317 -45.02707241 -44.36656205 -41.18438832]\n"
],
[
"dressed_hamiltonian = hilbertspace.hamiltonian()\ndressed_hamiltonian.eigenenergies()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbc7c97b0b8f1fd1fd1ffaaaab0170ec8e9d44cd
| 12,484 |
ipynb
|
Jupyter Notebook
|
rdd-basics/rdd-basics.ipynb
|
sergio11/spark-py-notebooks
|
3c0542b40e21e7d5b8af8e66ed3b738b5d623810
|
[
"Apache-2.0"
] | 1 |
2020-08-17T21:13:19.000Z
|
2020-08-17T21:13:19.000Z
|
rdd-basics/rdd-basics.ipynb
|
sergio11/spark-py-notebooks
|
3c0542b40e21e7d5b8af8e66ed3b738b5d623810
|
[
"Apache-2.0"
] | null | null | null |
rdd-basics/rdd-basics.ipynb
|
sergio11/spark-py-notebooks
|
3c0542b40e21e7d5b8af8e66ed3b738b5d623810
|
[
"Apache-2.0"
] | null | null | null | 28.965197 | 1,126 | 0.544617 |
[
[
[
"# RDD basics\n\nThis notebook will introduce **three basic but essential Spark operations**. Two of them are the transformations map and filter. The other is the action collect. At the same time we will introduce the concept of persistence in Spark.",
"_____no_output_____"
],
[
"## Getting the data and creating the RDD\n\nWe will use the reduced dataset (10 percent) provided for the KDD Cup 1999, containing nearly half million network interactions. The file is provided as a Gzip file that we will download locally.",
"_____no_output_____"
]
],
[
[
"import urllib\nf = urllib.urlretrieve (\"http://kdd.ics.uci.edu/databases/kddcup99/kddcup.data_10_percent.gz\", \"kddcup.data_10_percent.gz\")",
"_____no_output_____"
]
],
[
[
"Now we can use this file to create our RDD.",
"_____no_output_____"
]
],
[
[
"data_file = \"./kddcup.data_10_percent.gz\"\nraw_data = sc.textFile(data_file)",
"_____no_output_____"
]
],
[
[
"## The filter transformation\n\nThis transformation can be applied to RDDs in order to keep just elements that satisfy a certain condition. More concretely, a function is evaluated on every element in the original RDD. The new resulting RDD will contain just those elements that make the function return True.\nFor example, imagine we want to count how many normal. interactions we have in our dataset. We can filter our raw_data RDD as follows.",
"_____no_output_____"
]
],
[
[
"normal_raw_data = raw_data.filter(lambda x: 'normal.' in x)",
"_____no_output_____"
]
],
[
[
"Now we can count how many elements we have in the new RDD.",
"_____no_output_____"
]
],
[
[
"from time import time\nt0 = time()\nnormal_count = normal_raw_data.count()\ntt = time() - t0\nprint \"There are {} 'normal' interactions\".format(normal_count)\nprint \"Count completed in {} seconds\".format(round(tt,3))",
"There are 97278 'normal' interactions\nCount completed in 13.545 seconds\n"
]
],
[
[
"The **real calculations** (distributed) in Spark **occur when we execute actions and not transformations.** In this case counting is the action that we execute in the RDD. We can apply as many transformations as we would like in a RDD and no computation will take place until we call the first action which, in this case, takes a few seconds to complete.\n",
"_____no_output_____"
],
[
"## The map transformation\n\n\nBy using the map transformation in Spark, we can apply a function to every element in our RDD. **Python's lambdas are specially expressive for this particular.**\n\nIn this case we want to read our data file as a CSV formatted one. We can do this by applying a lambda function to each element in the RDD as follows.",
"_____no_output_____"
]
],
[
[
"from pprint import pprint\ncsv_data = raw_data.map(lambda x: x.split(\",\"))\nt0 = time()\nhead_rows = csv_data.take(5)\ntt = time() - t0\nprint \"Parse completed in {} seconds\".format(round(tt,3))\npprint(head_rows[0])",
"Parse completed in 0.406 seconds\n[u'0',\n u'tcp',\n u'http',\n u'SF',\n u'181',\n u'5450',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'1',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'8',\n u'8',\n u'0.00',\n u'0.00',\n u'0.00',\n u'0.00',\n u'1.00',\n u'0.00',\n u'0.00',\n u'9',\n u'9',\n u'1.00',\n u'0.00',\n u'0.11',\n u'0.00',\n u'0.00',\n u'0.00',\n u'0.00',\n u'0.00',\n u'normal.']\n"
]
],
[
[
"Again, **all action happens once we call the first Spark action** (i.e. take in this case). What if we take a lot of elements instead of just the first few?",
"_____no_output_____"
]
],
[
[
"t0 = time()\nhead_rows = csv_data.take(100000)\ntt = time() - t0\nprint \"Parse completed in {} seconds\".format(round(tt,3))",
"Parse completed in 24.343 seconds\n"
]
],
[
[
"We can see that it takes longer. The map function is applied now in a distributed way to a lot of elements on the RDD, hence the longer execution time.",
"_____no_output_____"
],
[
"## Using map and predefined functions\n\n\nOf course we can use predefined functions with map. Imagine we want to have each element in the RDD as a key-value pair where the key is the tag (e.g. normal) and the value is the whole list of elements that represents the row in the CSV formatted file. We could proceed as follows.",
"_____no_output_____"
]
],
[
[
"def parse_interaction(line):\n elems = line.split(\",\")\n tag = elems[41]\n return (tag, elems)\n\nkey_csv_data = raw_data.map(parse_interaction)\nhead_rows = key_csv_data.take(5)\npprint(head_rows[0])",
"(u'normal.',\n [u'0',\n u'tcp',\n u'http',\n u'SF',\n u'181',\n u'5450',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'1',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'0',\n u'8',\n u'8',\n u'0.00',\n u'0.00',\n u'0.00',\n u'0.00',\n u'1.00',\n u'0.00',\n u'0.00',\n u'9',\n u'9',\n u'1.00',\n u'0.00',\n u'0.11',\n u'0.00',\n u'0.00',\n u'0.00',\n u'0.00',\n u'0.00',\n u'normal.'])\n"
]
],
[
[
"## The collect action\n\n**Basically it will get all the elements in the RDD into memory for us to work with them.** For this reason it has to be used with care, specially when working with large RDDs.\n\nAn example using our raw data.",
"_____no_output_____"
]
],
[
[
"t0 = time()\nall_raw_data = raw_data.collect()\ntt = time() - t0\nprint \"Data collected in {} seconds\".format(round(tt,3))",
"_____no_output_____"
]
],
[
[
"Every Spark worker node that has a fragment of the RDD has to be coordinated in order to retrieve its part, and then reduce everything together.",
"_____no_output_____"
],
[
"As a last example combining all the previous, we want to collect all the normal interactions as key-value pairs.",
"_____no_output_____"
]
],
[
[
"# get data from file\ndata_file = \"./kddcup.data_10_percent.gz\"\nraw_data = sc.textFile(data_file)\n\n# parse into key-value pairs\nkey_csv_data = raw_data.map(parse_interaction)\n\n# filter normal key interactions\nnormal_key_interactions = key_csv_data.filter(lambda x: x[0] == \"normal.\")\n\n# collect all\nt0 = time()\nall_normal = normal_key_interactions.collect()\ntt = time() - t0\nnormal_count = len(all_normal)\nprint \"Data collected in {} seconds\".format(round(tt,3))\nprint \"There are {} 'normal' interactions\".format(normal_count)",
"Data collected in 43.913 seconds\nThere are 97278 'normal' interactions\n"
]
],
[
[
"This count matches with the previous count for normal interactions. The new procedure is more time consuming. This is because we retrieve all the data with collect and then use Python's len on the resulting list. Before we were just counting the total number of elements in the RDD by using count.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbc7ccb7e7a3baed513e17fa2a4687a66a3f5a95
| 14,759 |
ipynb
|
Jupyter Notebook
|
part02-machine-learning/2_6_svm.ipynb
|
edgardeng/machine-learning-pytorch
|
24a060894f5226b5ef20cc311db72f1adc037548
|
[
"MIT"
] | null | null | null |
part02-machine-learning/2_6_svm.ipynb
|
edgardeng/machine-learning-pytorch
|
24a060894f5226b5ef20cc311db72f1adc037548
|
[
"MIT"
] | null | null | null |
part02-machine-learning/2_6_svm.ipynb
|
edgardeng/machine-learning-pytorch
|
24a060894f5226b5ef20cc311db72f1adc037548
|
[
"MIT"
] | null | null | null | 33.696347 | 321 | 0.536689 |
[
[
[
"## 支持向量机 (support vector machines, SVM)\n\n> 支持向量机(support vector machines,SVM)是一种二分类模型,它将实例的特征向量映射为空间中的一些点,SVM 的目的就是想要画出一条线,以 “最好地” 区分这两类点,以至如果以后有了新的点,这条线也能做出很好的分类。SVM 适合中小型数据样本、非线性、高维的分类问题\n\nSVM学习的基本想法是\n 求解能够正确划分训练数据集并且几何间隔最大的分离超平面\n\n对于线性可分的数据集来说,这样的超平面有无穷多个(即感知机),但是几何间隔最大的分离超平面却是唯一的。\n\nAdvantages 优势:\n * Effective in high dimensional spaces. 在高维空间中有效。\n * Still effective in cases where number of dimensions is greater than the number of samples. 在尺寸数大于样本数的情况下仍然有效。\n * Uses a subset of training points in the decision function (called support vectors), so it is also memory efficient.\n 在决策函数中使用训练点的子集(称为支持向量),因此它也具有记忆效率。\n * Versatile: different Kernel functions can be specified for the decision function. Common kernels are provided, but it is also possible to specify custom kernels.\n 通用:可以为决策函数指定不同的核函数。提供了通用内核,但也可以指定自定义内核。\n\ndisadvantages 缺点:\n\n * If the number of features is much greater than the number of samples, avoid over-fitting in choosing Kernel functions and regularization term is crucial.\n 当特征个数远大于样本个数时,在选择核函数和正则化项时应避免过拟合。\n\n * SVMs do not directly provide probability estimates, these are calculated using an expensive five-fold cross-validation (see Scores and probabilities, below).\n 支持向量机不直接提供概率估计,这些是使用昂贵的五倍交叉验证计算的(见下面的分数和概率)。\n\n\n[支持向量机](https://blog.csdn.net/qq_31347869/article/details/88071930)\n[sklearn文档-svm](https://scikit-learn.org/dev/modules/svm.html#svm)",
"_____no_output_____"
],
[
"The sklearn.svm module includes Support Vector Machine algorithms.\n\n|Estimators | description |\n|:---- |:---- |\n| svm.LinearSVC([penalty, loss, dual, tol, C, …]) | Linear Support Vector Classification. |\n| svm.LinearSVR(*[, epsilon, tol, C, loss, …]) | Linear Support Vector Regression. |\n| svm.NuSVC(*[, nu, kernel, degree, gamma, …]) | Nu-Support Vector Classification. |\n| svm.NuSVR(*[, nu, C, kernel, degree, gamma, …]) | Nu Support Vector Regression. |\n| svm.OneClassSVM(*[, kernel, degree, gamma, …]) | Unsupervised Outlier Detection. |\n| svm.SVC(*[, C, kernel, degree, gamma, …]) | C-Support Vector Classification. |\n| svm.SVR(*[, kernel, degree, gamma, coef0, …]) | Epsilon-Support Vector Regression. |",
"_____no_output_____"
],
[
"### Classification 使用支持向量机做分类任务\n\nSVC, NuSVC and LinearSVC are classes capable of performing binary and multi-class classification on a dataset.",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVC\nimport numpy as np\n\nX = np.random.randint(0,10,(50,2))\ny = (X[:,0] + X[:,1]) // 10\nclf = SVC()\nclf.fit(X, y)\nclf.predict([[2., 7.],[3,9]]) # 结果会不一样",
"_____no_output_____"
],
[
"print(__doc__)\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom sklearn import svm, datasets\n\ndef make_meshgrid(x, y, h=.02):\n \"\"\"Create a mesh of points to plot in\n\n Parameters\n ----------\n x: data to base x-axis meshgrid on\n y: data to base y-axis meshgrid on\n h: stepsize for meshgrid, optional\n\n Returns\n -------\n xx, yy : ndarray\n \"\"\"\n x_min, x_max = x.min() - 1, x.max() + 1\n y_min, y_max = y.min() - 1, y.max() + 1\n xx, yy = np.meshgrid(np.arange(x_min, x_max, h),\n np.arange(y_min, y_max, h))\n return xx, yy\n\n\ndef plot_contours(ax, clf, xx, yy, **params):\n \"\"\"Plot the decision boundaries for a classifier.\n\n Parameters\n ----------\n ax: matplotlib axes object\n clf: a classifier\n xx: meshgrid ndarray\n yy: meshgrid ndarray\n params: dictionary of params to pass to contourf, optional\n \"\"\"\n Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])\n Z = Z.reshape(xx.shape)\n out = ax.contourf(xx, yy, Z, **params)\n return out\n\n\n# import some data to play with\niris = datasets.load_iris()\n# Take the first two features. We could avoid this by using a two-dim dataset\nX = iris.data[:, :2]\ny = iris.target\n\n# we create an instance of SVM and fit out data. We do not scale our\n# data since we want to plot the support vectors\nC = 1.0 # SVM regularization parameter\nmodels = (svm.SVC(kernel='linear', C=C),\n svm.LinearSVC(C=C, max_iter=10000),\n svm.SVC(kernel='rbf', gamma=0.7, C=C),\n svm.SVC(kernel='poly', degree=3, gamma='auto', C=C))\nmodels = (clf.fit(X, y) for clf in models)\n\n# title for the plots\ntitles = ('SVC with linear kernel',\n 'LinearSVC (linear kernel)',\n 'SVC with RBF kernel',\n 'SVC with polynomial (degree 3) kernel')\n\n# Set-up 2x2 grid for plotting.\nfig, sub = plt.subplots(2, 2)\nplt.subplots_adjust(wspace=0.4, hspace=0.4)\n\nX0, X1 = X[:, 0], X[:, 1]\nxx, yy = make_meshgrid(X0, X1)\n\nfor clf, title, ax in zip(models, titles, sub.flatten()):\n plot_contours(ax, clf, xx, yy,\n cmap=plt.cm.coolwarm, alpha=0.8)\n ax.scatter(X0, X1, c=y, cmap=plt.cm.coolwarm, s=20, edgecolors='k')\n ax.set_xlim(xx.min(), xx.max())\n ax.set_ylim(yy.min(), yy.max())\n ax.set_xlabel('Sepal length')\n ax.set_ylabel('Sepal width')\n ax.set_xticks(())\n ax.set_yticks(())\n ax.set_title(title)\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Regression\nThere are three different implementations of Support Vector Regression: SVR, NuSVR and LinearSVR. LinearSVR provides a faster implementation than SVR but only considers the linear kernel, while NuSVR implements a slightly different formulation than SVR and LinearSVR. See Implementation details for further details.",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import SVR\nX = [[0, 0], [2, 2]]\ny = [0.5, 2.5]\nregr = SVR()\nregr.fit(X, y)\nregr.predict([[1, 1]])",
"_____no_output_____"
]
],
[
[
"Unsupervised Outlier Detection.\n\nEstimate the support of a high-dimensional distribution.\n\nOneClassSVM is based on libsvm.",
"_____no_output_____"
]
],
[
[
"from sklearn.svm import OneClassSVM\nX = [[0], [0.44], [0.45], [0.46], [1]]\nclf = OneClassSVM(gamma='auto')\nclf.fit(X)\nresult = clf.predict(X)\nprint(result)\nscores = clf.score_samples(X)\nprint(scores)",
"_____no_output_____"
]
],
[
[
"### 使用SVM做异常检测算法\n\nComparing anomaly detection algorithms for outlier detection on toy datasets\n\n[refrence](https://scikit-learn.org/dev/auto_examples/miscellaneous/plot_anomaly_comparison.htm)",
"_____no_output_____"
]
],
[
[
"# Author: Alexandre Gramfort <[email protected]>\n# Albert Thomas <[email protected]>\n# License: BSD 3 clause\n\nimport time\n\nimport numpy as np\nimport matplotlib\nimport matplotlib.pyplot as plt\nimport sklearn\nprint(sklearn.__version__)",
"_____no_output_____"
],
[
"from sklearn.datasets import make_moons, make_blobs\nfrom sklearn.covariance import EllipticEnvelope\nfrom sklearn.ensemble import IsolationForest\nfrom sklearn.neighbors import LocalOutlierFactor\nfrom sklearn.svm import OneClassSVM\nfrom sklearn.kernel_approximation import Nystroem\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.linear_model import SGDOneClassSVM\n\nprint(__doc__)\n\nmatplotlib.rcParams['contour.negative_linestyle'] = 'solid'\n\n# Example settings\nn_samples = 300\noutliers_fraction = 0.15\nn_outliers = int(outliers_fraction * n_samples)\nn_inliers = n_samples - n_outliers\n\n# define outlier/anomaly detection methods to be compared.\n# the SGDOneClassSVM must be used in a pipeline with a kernel approximation\n# to give similar results to the OneClassSVM\nanomaly_algorithms = [\n (\"Robust covariance\", EllipticEnvelope(contamination=outliers_fraction)),\n (\"One-Class SVM\", OneClassSVM(nu=outliers_fraction, kernel=\"rbf\",\n gamma=0.1)),\n (\"One-Class SVM (SGD)\", make_pipeline(\n Nystroem(gamma=0.1, random_state=42, n_components=150),\n SGDOneClassSVM(nu=outliers_fraction, shuffle=True,\n fit_intercept=True, random_state=42, tol=1e-6)\n )),\n (\"Isolation Forest\", IsolationForest(contamination=outliers_fraction,\n random_state=42)),\n (\"Local Outlier Factor\", LocalOutlierFactor(\n n_neighbors=35, contamination=outliers_fraction))]\n\n# Define datasets\nblobs_params = dict(random_state=0, n_samples=n_inliers, n_features=2)\ndatasets = [\n make_blobs(centers=[[0, 0], [0, 0]], cluster_std=0.5,\n **blobs_params)[0],\n make_blobs(centers=[[2, 2], [-2, -2]], cluster_std=[0.5, 0.5],\n **blobs_params)[0],\n make_blobs(centers=[[2, 2], [-2, -2]], cluster_std=[1.5, .3],\n **blobs_params)[0],\n 4. * (make_moons(n_samples=n_samples, noise=.05, random_state=0)[0] -\n np.array([0.5, 0.25])),\n 14. * (np.random.RandomState(42).rand(n_samples, 2) - 0.5)]\n\n# Compare given classifiers under given settings\nxx, yy = np.meshgrid(np.linspace(-7, 7, 150),\n np.linspace(-7, 7, 150))\n\nplt.figure(figsize=(len(anomaly_algorithms) * 2 + 4, 12.5))\nplt.subplots_adjust(left=.02, right=.98, bottom=.001, top=.96, wspace=.05,\n hspace=.01)\n\nplot_num = 1\nrng = np.random.RandomState(42)\n\nfor i_dataset, X in enumerate(datasets):\n # Add outliers\n X = np.concatenate([X, rng.uniform(low=-6, high=6, size=(n_outliers, 2))],\n axis=0)\n\n for name, algorithm in anomaly_algorithms:\n t0 = time.time()\n algorithm.fit(X)\n t1 = time.time()\n plt.subplot(len(datasets), len(anomaly_algorithms), plot_num)\n if i_dataset == 0:\n plt.title(name, size=18)\n\n # fit the data and tag outliers\n if name == \"Local Outlier Factor\":\n y_pred = algorithm.fit_predict(X)\n else:\n y_pred = algorithm.fit(X).predict(X)\n\n # plot the levels lines and the points\n if name != \"Local Outlier Factor\": # LOF does not implement predict\n Z = algorithm.predict(np.c_[xx.ravel(), yy.ravel()])\n Z = Z.reshape(xx.shape)\n plt.contour(xx, yy, Z, levels=[0], linewidths=2, colors='black')\n\n colors = np.array(['#377eb8', '#ff7f00'])\n plt.scatter(X[:, 0], X[:, 1], s=10, color=colors[(y_pred + 1) // 2])\n\n plt.xlim(-7, 7)\n plt.ylim(-7, 7)\n plt.xticks(())\n plt.yticks(())\n plt.text(.99, .01, ('%.2fs' % (t1 - t0)).lstrip('0'),\n transform=plt.gca().transAxes, size=15,\n horizontalalignment='right')\n plot_num += 1\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbc7dc05ccd00127863590a5e51db6cd11553ae9
| 35,758 |
ipynb
|
Jupyter Notebook
|
assignments/2021-07-10/Sophie_assingment_013_marked.ipynb
|
qqiang00/ElementaryMathPython
|
7fa723335ca342b867d51776d1f009066c50092d
|
[
"MIT"
] | null | null | null |
assignments/2021-07-10/Sophie_assingment_013_marked.ipynb
|
qqiang00/ElementaryMathPython
|
7fa723335ca342b867d51776d1f009066c50092d
|
[
"MIT"
] | null | null | null |
assignments/2021-07-10/Sophie_assingment_013_marked.ipynb
|
qqiang00/ElementaryMathPython
|
7fa723335ca342b867d51776d1f009066c50092d
|
[
"MIT"
] | null | null | null | 32.185419 | 341 | 0.508502 |
[
[
[
"## 第13讲 认识和绘制数轴",
"_____no_output_____"
],
[
"### Problem 问题描述",
"_____no_output_____"
],
[
"在长宽分别为600和400像素的绘图区域绘制如下图所示的一条标有刻度、水平方向的带箭头指示方向的数轴。其中数轴的左右两端距离绘图区域左右边界均为20像素,相邻刻度的距离为50像素,刻度线的长度为20像素,表示刻度线数值的数字在刻度线的正下方且底端距离数轴20个像素。\n\n<img src=\"figures/L013_axis.png\" width=\"600px\"/>",
"_____no_output_____"
],
[
"### Math Background 数学背景\n\n1. 数轴的构成\n2. 数轴原点,正负数在数轴轴上的位置比较\n3. 每一个数在数轴上都有一个点相对应,两个数的差在数轴上表示的是这两个数对应的两个点之间的距离。",
"_____no_output_____"
],
[
"### Prerequisites 预备知识",
"_____no_output_____"
],
[
"#### 1. `write`方法可以在绘图区书写字符串",
"_____no_output_____"
]
],
[
[
"from turtle import setup, reset, pu, pd, bye, left, right, fd, bk, screensize\nfrom turtle import goto, seth, write, ht, st, home\nwidth, height = 600, 400 # 窗口的宽度和高度(单位为:像素)\nsetup(width, height, 0, 0)",
"_____no_output_____"
]
],
[
[
"比较提起画笔和放下画笔时下面的代码执行的效果有什么不同",
"_____no_output_____"
]
],
[
[
"reset()\npu()\nwrite(\"Sophie\", move=True, align=\"center\")",
"_____no_output_____"
],
[
"reset()\npd()\nwrite(\"Tony\", move=True, align=\"center\", font=(\"Arial\", 30, \"normal\"))",
"_____no_output_____"
],
[
"reset()\npd()\nwrite(\"Sophie\", move=False, align=\"center\", font=(\"Arial\", 30, \"normal\"))",
"_____no_output_____"
]
],
[
[
"#### 2. `tuple`元组数据类型\n\npos_x 是一个tuple类型的变量",
"_____no_output_____"
]
],
[
[
"pos_x = (30, 20) # pos_x 是一个tuple类型的变量",
"_____no_output_____"
]
],
[
[
"可以使用索引来获取tuple类型变量的元素",
"_____no_output_____"
]
],
[
[
"print(pos_x[0], pos_x[1]) # 可以使用索引来获取tuple类型变量的元素",
"_____no_output_____"
]
],
[
[
"可以使用len()方法来获取tuple类型数据的元素个数",
"_____no_output_____"
]
],
[
[
"len(pos_x) # 可以使用len()方法来获取tuple类型数据的元素个数",
"_____no_output_____"
]
],
[
[
" 不可以更改tuple类型变量里某一个元素的值。例如执行下面的代码将发生错误\n ```python\npos_x[0] = 40\n```\n\n```text\n---------------------------------------------------------------------------\nTypeError Traceback (most recent call last)\n<ipython-input-24-d852e9299be9> in <module>\n----> 1 pos_x[0] = 40 \n\nTypeError: 'tuple' object does not support item assignment\n```",
"_____no_output_____"
]
],
[
[
"# pos_x[0] = 40 # 不可以更改tuple类型变量里某一个元素的值",
"_____no_output_____"
]
],
[
[
"可以给整个tuple类型变量赋予一个新的tuple值",
"_____no_output_____"
]
],
[
[
"pos_x = (50, 30) # 可以给整个tuple类型变量赋予一个新的tuple值",
"_____no_output_____"
]
],
[
[
"`tuple`型的变量里的元素的类型可以互不相同",
"_____no_output_____"
]
],
[
[
"sophie = (\"Sophie\", 11, \"Female\", \"Grade4\") # tuple型的变量里的元素的类型可以互不相同\nprint(sophie)",
"_____no_output_____"
]
],
[
[
"#### 3. 理解同一个方法在接受不同的参数值时执行结果的比较\n\n对比输出的文字和海龟位置,观察下面的两条`write`方法在接受不同的`align`值或时效果有什么不同。",
"_____no_output_____"
]
],
[
[
"reset()\npu()\nwrite(\"Jason\", align=\"left\", font=(\"Arial\", 30, \"normal\"))",
"_____no_output_____"
],
[
"reset()\npu()\nwrite(\"Jason\", align=\"center\", font=(\"Arial\", 30, \"normal\"))",
"_____no_output_____"
]
],
[
[
"#### 4. 对比输出的文字,观察下面的几条`write`方法在接受不同的`font`值时效果有什么不同。\n\n参数`font`是一个`tuple`类型的变量",
"_____no_output_____"
]
],
[
[
"reset()\npu()\nwrite(\"Jason\", font=(\"Arial\", 30, \"normal\"))",
"_____no_output_____"
],
[
"reset()\npu()\nwrite(\"Jason\", font=(\"Arial\", 50, \"normal\"))",
"_____no_output_____"
],
[
"reset()\npu()\nwrite(\"Jason\", font=(\"Times New Roman\", 50, \"normal\"))",
"_____no_output_____"
],
[
"reset()\npu()\nwrite(\"Jason\", font=(\"Arial\", 50, \"italic\"))",
"_____no_output_____"
],
[
"# no reset() here 这里没有reset()\npu()\nbk(200) # 后退200\nwrite(\"Jason\", font=(\"Arial\", 50, \"underline\"))",
"_____no_output_____"
]
],
[
[
"#### 5. 区分`()`何时表示元组数据何时表示方法接受的参数\n\n看`()`前面有没有紧跟一个方法名, 下面这行代码声明了一个元组型变量,变量名为`jason`。\n\n```python\njason = (\"Jason\", (\"Arial\", 50, \"Italic\")) # \n```\n\n下面这行代码是在执行一个名为`jason`的方法\n```python\njason(\"Jason\", (\"Arial\", 50, \"Italic\")) \n```",
"_____no_output_____"
],
[
"#### 6. 练习\n\n编写下面的代码,更改变量`name`的值为你的名字,观察代码执行的效果。",
"_____no_output_____"
]
],
[
[
"# 如果没有导入绘图库相关方法以及执行setup方法,请解除下面几行代码的注释\n# from turtle import setup, reset, pu, pd, bye, left, right, fd, bk, screensize\n# from turtle import goto, seth, write, ht, st, home\n# width, height = 600, 400 # 窗口的宽度和高度(单位为:像素)\n# setup(600, 400, 0, 0)\nreset()\npu()\nht()\nname = \"Qiang\"\ntext = \"My name is {}.\\nNice to meet you.\".format(name)\nwrite(text, align=\"center\", font=(\"Arial\", 30, \"italic\"))",
"_____no_output_____"
]
],
[
[
"### Solution 编程求解",
"_____no_output_____"
]
],
[
[
"from turtle import setup, reset, pu, pd, bye, left, right, fd, bk, screensize\nfrom turtle import goto, seth, write, ht, st, home\nwidth, height = 600, 400 # 窗口的宽度和高度(单位为:像素)\nsetup(width, height, 0, 0)",
"_____no_output_____"
],
[
"origin = (0, 0) # 原点的位置\npadding = 20 # 数轴端点距离绘图区边界距离\nmax_x = width/2 - padding # x轴最大值\nshow_arrow = True # 是否显示箭头\nmark_interval = 50 # 刻度线间距\nmark_line_length = 10 # 刻度线高度\ntext_offset = 20 # 坐标值距离坐标线的距离\n\nmark_degree = 90 # 坐标刻度与坐标轴夹角\narrow_length = 100 # 箭头长度\narrow_degree = 30 # 箭头与坐标轴夹角\ndelta_x = 1 # 每次坐标值变化的幅度",
"_____no_output_____"
],
[
"# Solution1: without using goto() 第一种方法:不使用goto()\n\nreset() # 重置绘图区 # this puts turtle in the midile and it erase all for the page.\nmin_x = -1 * max_x # 根据坐标轴允许的最大值,获取该坐标轴允许的最小值 \npu() # 提起画笔,暂停绘图\nhome() # Move turtle to the origin – coordinates (0,0) 移动小海龟至初始位置\n # and set its heading to its start-orientation 并设置朝向为初始朝向\nbk(max_x) # backward max_x\npd() # 落下画笔,准备绘图\n\n# draw mark 绘制刻度线\ncur_x, last_x = min_x, min_x # 海龟当前位置和最近一次绘图后停留的位置\nwhile cur_x <= max_x: # 循环\n if cur_x % mark_interval == 0: # 海龟的位置是相邻刻度间隔长度的整数倍\n length_move = cur_x - last_x # 计算海龟应该前进的长度\n pd() # 落下画笔,准备绘图\n fd(length_move) # 海龟前进(绘制一小段)\n left(mark_degree) # 向左转90度,海龟朝正上方,准备绘制刻度线\n fd(mark_line_length) # 绘制刻度线\n pu() # 把画笔提起暂停绘图\n bk(mark_line_length + text_offset) # 后退(向下)一段长度\n text = str(int(cur_x // mark_interval))\n # 准备刻度值字符串(由整型数据转换而来)\n write(text, align=\"center\") # 在当前位置以居中的形式书写文字字符串\n fd(text_offset) # 前进(向上)一小段长度\n right(mark_degree) # 向右转90度,海龟次朝向右侧\n last_x = cur_x # 记录海龟当前位置,为下次绘图的起点\n \n cur_x += delta_x # 当前位置增加一小段长度(个单位距离:像素)\n\npd() # 落下画笔,准备绘制\nfd(max_x - last_x) # 绘制最后一个刻度线到数轴最大x值这一小段\n\nif show_arrow: # 如果需要绘制箭头\n right(arrow_degree) # 向右转,海龟朝向右侧偏下\n bk(arrow_length) # 后退一定距离,绘制箭头一边\n fd(arrow_length) # 回到max_x位置\n left(arrow_degree * 2) # 向左转,海龟朝向右侧偏上\n bk(arrow_length) # 后退一定距离,绘制箭头另一边\n\nht() # 隐藏海龟 ",
"_____no_output_____"
],
[
"# Solution2: using goto() 第二种方法:使用goto()\nreset()\nmin_x = -1 * max_x\n\n# draw line\npu() # 提起画笔,暂停绘图\nhome() # Move turtle to the origin – coordinates (0,0) 移动小海龟至初始位置\n # and set its heading to its start-orientation 并设置朝向为初始朝向\ngoto(min_x, 0) # go to the left end of the line 移动海龟到坐标轴直线的最左端\npd() # 落下画笔,准备绘图\ngoto(max_x, 0) # go to the right end of the line 移动海龟到坐标轴直线的最右段\n\n# draw mark 绘制刻度线\ncur_x = min_x # cur_x is min_x \nwhile cur_x <= max_x:\n if cur_x % mark_interval == 0:\n pu() # pen up\n goto(cur_x, 0) # go to cur_x fof x and 0 for y\n pd() # pen down\n goto(cur_x, mark_line_length) # 绘制刻度线\n \n pu() # pen up \n goto(cur_x, -text_offset) # go to cur_x for x nd -text_offset for y.\n pd() # pen down\n text = str(int(cur_x//mark_interval)) # text is str(int(cur_x//mark_interval))\n write(text, align=\"center\") # 书写刻度值\n \n cur_x += delta_x # cur_x is delta_x + delta_x\n \nif show_arrow: # if you need to draw arrows\n arrow_x, arrow_y = max_x - 10, -5\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, arrow_y) # go to arrow_x for x and arrow_y for y\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, -arrow_y) # go to arrow_x for x and arrow_y for y\nht() # hide turtle",
"_____no_output_____"
],
[
"reset()\nif show_arrow: # if you need to draw arrows\n arrow_x, arrow_y = max_x - 100, -50\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, arrow_y) # go to arrow_x for x and arrow_y for y\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, -arrow_y)# go to arrow_x for x and arrow_y for \n goto(max_x, 0)",
"_____no_output_____"
],
[
"if show_arrow: # 如果需要绘制箭头\n right(arrow_degree) # 向右转,海龟朝向右侧偏下\n bk(arrow_length) # 后退一定距离,绘制箭头一边\n fd(arrow_length) # 回到max_x位置\n left(arrow_degree * 2) # 向左转,海龟朝向右侧偏上\n bk(arrow_length) # 后退一定距离,绘制箭头另一边\n # longer\n\nht() # 隐藏海龟 ",
"_____no_output_____"
],
[
"bye()",
"_____no_output_____"
]
],
[
[
"### Summary 知识点小结",
"_____no_output_____"
],
[
"1. turtle绘图库里的新方法`write`可以在绘图区海龟的当前位置书写文字;\n2. 新的数据类型:`tuple`元组数据类型,它与`list`数据类型非常类似,但也有区别;\n3. 在执行一个方法时,方法名后面的小括号`()`内可以接受一个或多个不同的数据,这些数据成为该方法可以接受的参数。方法接受的参数的值不一样,执行该方法最后得到的结果也通常不同;\n4. 复习格式化字符串的`format`方法;\n5. 复习`while`循环,并将`while`过程中循环应用到绘图过程中;\n6. 复习操作符`//`和`%`。",
"_____no_output_____"
],
[
"### 计算机小知识",
"_____no_output_____"
],
[
"像素,字体`font`",
"_____no_output_____"
],
[
"### Assignments 作业",
"_____no_output_____"
],
[
"1. 仔细阅读本讲示例中给出的两种绘制坐标轴方法,回答下面的问题: \n Read carefully the two solutions demonstrated in the lecture, answer the following questions:\n 1. 给第二种方法中的每一行代码添加注释 \n Add comments for every code line of the second solution to tell the meaning of each code line.\n 2. 比较并说出两种方法在绘制坐标轴的差别\n Compare the two solutions and tell the difference of them in drawing the axis.\n 3. 两种方法绘制出来的箭头一模一样吗?为什么? \n Are the arrows drawn by the two solutoins exactly same? why?",
"_____no_output_____"
],
[
"(B. the first draws a little part of the line and then it draws a mark line\n the second draws the line first and then it going back to draw the line marks)\n (C. no because the arrow drawn by the second has a bigger. )\n",
"_____no_output_____"
],
[
"2. 编程绘制如下图所示的水平坐标轴。所用的刻度间距、刻度线长度等排版指标军与本讲示例相同。其中,与本讲示例不同的是: \n By programming, draw horizontal axies as the following figure shows. Most of the parameters, including the marker interval, marker length, etc, have the same value as in the lecture. However, there are still some significant differences, which are:\n 1. 将表示0刻度坐标值的文字“0”的位置向右移动距离10,刻度线仍保持与相邻的刻度线等距离不变; Move the text \"0\", which indicating the value 0 on the axis, 10 pixels right to its original position. Keep the mark line where it is.\n 2. 在箭头的下方添加字母\"x\",字母\"x\"使用的字体是\"Arial\",字号大小为10,风格为“斜体”。Add a letter \"x\" under the arrow at the right end of the axis, use font \"Arial\", size 10, and \"italic\" to write the \"x\"\n 3. 当调整绘图区域的大小为宽为800像素时,你的代码应该仅需要更新绘图区的宽度而不改变其他地方就能直接调整数轴长度和刻度的显示。When the width of drawing area changed to 800 pixels from 600 pixels, your codes should only need to change the value of `width` while keep others unchanged to draw the axis with new length and markers.\n\n<img src=\"figures/L013_assignment1.png\" />\n\n<img src=\"figures/L013_assignment1_2.png\" />",
"_____no_output_____"
]
],
[
[
"from turtle import setup, reset, pu, pd, bye, left, right, fd, bk, screensize\nfrom turtle import goto, seth, write, ht, st, home, speed\nwidth, height = 400, 500 # 窗口的宽度和高度(单位为:像素)\nsetup(width, height, 0, 0)\norigin = (0, 0) # 原点的位置\npadding = 20 # 数轴端点距离绘图区边界距离\nmax_x = width/2 - padding # x轴最大值\nshow_arrow = True # 是否显示箭头\nmark_interval = 50 # 刻度线间距\nmark_line_length = 10 # 刻度线高度\ntext_offset = 20 # 坐标值距离坐标线的距离\n\nmark_degree = 90 # 坐标刻度与坐标轴夹角\narrow_length = 10 # 箭头长度\narrow_degree = 30 # 箭头与坐标轴夹角\ndelta_x = 1 \n\norigin = (0, 0) # 原点的位置\npadding = 20 # 数轴端点距离绘图区边界距离\nmax_x = width/2 - padding # x轴最大值\nshow_arrow = True # 是否显示箭头\nmark_interval = 50 # 刻度线间距\nmark_line_length = 10 # 刻度线高度\ntext_offset = 20 # 坐标值距离坐标线的距离\n\nmark_degree = 90 # 坐标刻度与坐标轴夹角\narrow_length = 10 # 箭头长度\narrow_degree = 30 # 箭头与坐标轴夹角\ndelta_x = 1 # 每次坐标值变化的幅度",
"_____no_output_____"
],
[
"reset()\n\n# Solution2: using goto() 第二种方法:使用goto()\nreset()\nmin_x = -1 * max_x\n\n# draw line\npu() # 提起画笔,暂停绘图\nhome() # Move turtle to the origin – coordinates (0,0) 移动小海龟至初始位置\n # and set its heading to its start-orientation 并设置朝向为初始朝向\ngoto(min_x, 0) # go to the left end of the line 移动海龟到坐标轴直线的最左端\npd() # 落下画笔,准备绘图\ngoto(max_x, 0) # go to the right end of the line 移动海龟到坐标轴直线的最右段\n\n# draw mark 绘制刻度线\ncur_x = min_x # cur_x is min_x \nwhile cur_x <= max_x:\n if cur_x % mark_interval == 0:\n pu() # pen up\n goto(cur_x, 0) # go to cur_x fof x and 0 for y\n pd() # pen down\n goto(cur_x, mark_line_length) # 绘制刻度线\n \n pu() # pen up \n goto(cur_x, -text_offset) # go to cur_x for x nd -text_offset for y.\n pd() # pen down\n if cur_x == 0:\n pu()\n fd(10)\n pd()\n \n text = str(int(cur_x//mark_interval)) # text is str(int(cur_x//mark_interval))\n write(text, align=\"center\") # 书写刻度值\n \n cur_x += delta_x # cur_x is delta_x + delta_x\n \nif show_arrow: # if you need to draw arrows\n arrow_x, arrow_y = max_x - 10, -5\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, arrow_y) # go to arrow_x for x and arrow_y for y\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, -arrow_y) # go to arrow_x for x and arrow_y for y\npu()\ngoto(max_x, 0)\nright(90)\nfd(text_offset)\nwrite(\"x\", move=False, align=\"center\", font=(\"Arial\", 10, \"italic\"))\n\n\nht() # hide turtle\n\nht()",
"_____no_output_____"
],
[
"st()\ngoto(arrow_x, -arrow_y) # go to arrow_x for x and arrow_y for y",
"_____no_output_____"
]
],
[
[
"3. 编程绘制一条如下图所示的垂直方向上的坐标轴。要求:By programming, draw an ertical axis as the following figure shows. Requirement:\n\n 1. 该图所是的坐标轴基本上是把水平方向的坐标轴围绕这坐标原点向左侧旋转90度得到;\n The axis can basically be considered as a 90 degree of anti-closewise rotation of the horizontal axis illustrated in the lecture with original zero point as the rotation center;\n 2. 大部分控制数轴风格的参数值与示例中的一样,下列除外:但是刻度线位于坐标轴的右侧,刻度值位于坐标轴的左侧。Most of the parameters controlling the style of the axis are same as introduced in the lecture, except: the marker lines are located on right side of the axis line, and the marker values are on the left side;\n 3. 隐藏表示0刻度坐标值的文字“0”以及对应的刻度线; Hide the marker line and the marker value for origin point;\n 4. 在箭头的左侧添加字母\"y\",字母\"y\"使用的字体是\"Arial\",字号大小为10,风格为“斜体”。Add the letter \"y\" on left side of the axis end, the font for \"y\" is \"Arial\", size is 10, and style is \"italic\";\n 5. 如果绘图区的高度发生改变不再是400像素,你的代码应仅需要修改一处就能重新绘制出填满大部分(保留上下个20像素高的间隙)绘图区高度的数轴。If the height of drawing area is changed to any other value other than 400 pixels, your codes should only need to change one place in order to draw the new vertical axis that fullfill the most height of the draw area (keep 20 pixels paddings for both ends).\n\n<img src=\"figures/L013_assignment3.png\" style=\"align:center\" height=\"400px\"/>",
"_____no_output_____"
]
],
[
[
"reset()\n\n#TODO: Add your own codes here 在这里添加你自己的代码\n# Solution2: using goto() 第二种方法:使用goto()\nmin_x = -1 * max_x\n\n# draw line\npu() # 提起画笔,暂停绘图\nhome() # Move turtle to the origin – coordinates (0,0) 移动小海龟至初始位置\n # and set its heading to its start-orientation 并设置朝向为初始朝向\ngoto(0, min_x) # go to the left end of the line 移动海龟到坐标轴直线的最左端\npd() # 落下画笔,准备绘图\ngoto(0, max_x) # go to the right end of the line 移动海龟到坐标轴直线的最右段\n\n# draw mark 绘制刻度线\ncur_x = min_x # cur_x is min_x \nwhile cur_x <= max_x:\n if cur_x % mark_interval == 0:\n pu() # pen up\n goto(cur_x, 0) # go to cur_x fof x and 0 for y\n pd() # pen down\n goto(cur_x, mark_line_length) # 绘制刻度线\n \n pu() # pen up \n goto(cur_x, -text_offset) # go to cur_x for x nd -text_offset for y.\n pd() # pen down\n if cur_x == 0:\n pass\n \n else:\n text = str(int(cur_x//mark_interval)) # text is str(int(cur_x//mark_interval))\n write(text, align=\"center\") # 书写刻度值\n \n cur_x += delta_x # cur_x is delta_x + delta_x\n \nif show_arrow: # if you need to draw arrows\n arrow_x, arrow_y = max_x - 10, -5\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, arrow_y) # go to arrow_x for x and arrow_y for y\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, -arrow_y) # go to arrow_x for x and arrow_y for y\npu()\ngoto(max_x, 0)\nright(90)\nfd(text_offset)\nwrite(\"x\", move=False, align=\"center\", font=(\"Arial\", 10, \"italic\"))\n\n\nht() # hide turtle\n\nht()",
"_____no_output_____"
],
[
"reset()\npd()\nst()\nspeed(2)\n\nmin_x = -1 * max_x # 根据坐标轴允许的最大值,获取该坐标轴允许的最小值 \npu() # 提起画笔,暂停绘图\nhome() # Move turtle to the origin – coordinates (0,0) 移动小海龟至初始位置\nright(90) # and set its heading to its start-orientation 并设置朝向为初始朝向\nfd(max_x) # forward max_x\npd() # 落下画笔,准备绘图\n\n# draw mark 绘制刻度线\ncur_x, last_x = min_x, min_x # 海龟当前位置和最近一次绘图后停留的位置\nwhile cur_x <= max_x: # 循环\n \n if cur_x % mark_interval == 0: # 海龟的位置是相邻刻度间隔长度的整数倍\n length_move = cur_x - last_x # 计算海龟应该前进的长度\n pd() # 落下画笔,准备绘图\n bk(length_move) # 海龟前进(绘制一小段)\n left(mark_degree) # 向左转90度,海龟朝正上方,准备绘制刻度线\n fd(mark_line_length) # 绘制刻度线\n pu() # 把画笔提起暂停绘图\n bk(mark_line_length + text_offset) # 后退(向下)一段长度\n text = str(int(cur_x // mark_interval))# 准备刻度值字符串(由整型数据转换而来)\n \n if cur_x == 0:\n fd(text_offset)\n right(90)\n else:\n write(text, align=\"center\") # 在当前位置以居中的形式书写文字字符串\n fd(text_offset) # 前进(向上)一小段长度\n right(mark_degree) # 向右转90度,海龟次朝向右侧\n last_x = cur_x # 记录海龟当前位置,为下次绘图的起点\n \n cur_x += delta_x # 当前位置增加一小段长度(个单位距离:像素)\n\npd() # 落下画笔,准备绘制\nfd(max_x - last_x) # 绘制最后一个刻度线到数轴最大x值这一小段\n\nif show_arrow: # 如果需要绘制箭头\n bk(60)\n right(arrow_degree) # 向右转,海龟朝向右侧偏下\n fd(arrow_length) # 后退一定距离,绘制箭头一边\n bk(arrow_length) # 回到max_x位置\n left(arrow_degree * 2) # 向左转,海龟朝向右侧偏上\n fd(arrow_length) # 后退一定距离,绘制箭头另一边\n pu()\n right(120)\n fd(20)\n write(\"y\", move=False, align=\"left\", font=(\"Arial\", 15, \"italic\")) # 在当前位置以居中的形式书写文字字符串\nht() # 隐藏海龟 ",
"_____no_output_____"
],
[
"st()\nhome()",
"_____no_output_____"
]
],
[
[
"4. 编程绘制一条如下图所示的水平坐标轴。与本讲示例不同的是:By programming, draw a horizontal axis with major and minor marker lines as shown in the figure. Most of the parameters that control the style of the aixs remain same as introduced in the lecture, except:\n 1. 在刻度线的内部再绘制9条段的次要刻度线,这样原来相邻的两条刻度线被等间距的分为10个等分,每个等分对应的长度为5;Add 9 minor marker lines within two major marker lines so that every major marker interval is divided into 10 equal minor marker intervals, each 5 pixles length;\n 2. 与原来刻度线的宽度为10不同,次要刻度线的宽度为6; the length of the minor marker line is 6 pixels, keep the length of the major marker line 10 pixels unchanged;\n 3. (困难,可选做)在左右两侧整数刻度之外的区域**不要**绘制次要刻度线;(Difficult, Optional) Do **NOT** add minor maker lines on the parts where the position is smaller than the minimal major marker value or larger than the maximal major marker value;\n 4. 将表示0刻度坐标值的文字“0”的位置向右移动距离10,刻度线仍保持与相邻的刻度线等距离不变; Move the text \"0\", which indicating the value 0 on the axis, 10 pixels right to its original position. Keep the mark line where it is;\n 5. 在箭头的下方添加字母\"x\",字母\"x\"使用的字体是\"Arial\",字号大小为10,风格为“斜体”。Add a letter \"x\" under the arrow at the right end of the axis, use font \"Arial\", size 10, and \"italic\" to write the \"x\";\n 6. 当调整绘图区域的大小为宽为800像素时,你的代码应该仅需要更新绘图区的宽度而不改变其他地方就能直接调整数轴长度和刻度的显示。When the width of drawing area changed to 800 pixels from 600 pixels, your codes should only need to change the value of `width` while keep others unchanged to draw the axis with new length and markers.\n \n<img src=\"figures/L013_assignment4.png\" style=\"align:center\" height=\"400px\"/>",
"_____no_output_____"
]
],
[
[
"origin = (0, 0) # 原点的位置\npadding = 20 # 数轴端点距离绘图区边界距离\nmax_x = width/2 - padding # x轴最大值\nshow_arrow = True # 是否显示箭头\nmark_interval = 50 # 刻度线间距\nmark_line_length = 10 # 刻度线高度\ntext_offset = 20 # 坐标值距离坐标线的距离\nminor_mark_line_interval = 5\nminor_mark_line_length = 6\nminor_mark_degree = 90\n\nmark_degree = 90 # 坐标刻度与坐标轴夹角\narrow_length = 100 # 箭头长度\narrow_degree = 30 # 箭头与坐标轴夹角\ndelta_x = 1 ",
"_____no_output_____"
],
[
"reset()\n\n#TODO: Add your own codes here 在这里添加你自己的代码\n\nreset()\nmin_x = -1 * max_x\nminor_line_drawn_per_mark = 0\n\n# draw line\npu() # 提起画笔,暂停绘图\nhome() # Move turtle to the origin – coordinates (0,0) 移动小海龟至初始位置\n # and set its heading to its start-orientation 并设置朝向为初始朝向\ngoto(min_x, 0) # go to the left end of the line 移动海龟到坐标轴直线的最左端\npd() # 落下画笔,准备绘图\ngoto(max_x, 0) # go to the right end of the line 移动海龟到坐标轴直线的最右段\n\n# draw mark 绘制刻度线\ncur_x = min_x # cur_x is min_x \nwhile cur_x <= max_x: # while cur_x is still in the line\n if minor_line_drawn_per_mark == 9: #if\n \n minor_line_drawn_per_mark = 0 # set minor_line_drawn_per_mark to 0 at the start of each while loop\n pu() # pen up\n goto(cur_x, 0) # go to cur_x for x and 0 for y\n pd() # pen down\n goto(cur_x, mark_line_length) # 绘制刻度线\n pu() # pen up \n goto(cur_x, -text_offset) # go to cur_x for x nd -text_offset for y.\n text = str(int(cur_x//mark_interval)) # text is str(int(cur_x//mark_interval))\n write(text, align=\"center\")\n pd() # pen down \n cur_x += delta_x # cur_x is delta_x + delta_x\n\n else:\n\n pu() # pen up\n goto(cur_x, 0) # go to cur_x for x and 0 for y\n pd() # pen down\n goto(cur_x, minor_mark_line_length) # 绘制刻度线\n minor_line_drawn_per_mark += 1\n \n \n cur_x += minor_mark_line_interval\n\n \n \nif show_arrow: # if you need to draw arrows\n arrow_x, arrow_y = max_x - 10, -5\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, arrow_y) # go to arrow_x for x and arrow_y for y\n pu() # pen up\n goto(max_x, 0) # go to max_x for x and 0 for y\n pd() # pen down\n goto(arrow_x, -arrow_y) # go to arrow_x for x and arrow_y for y\npu()\ngoto(max_x, 0)\nright(90)\nfd(text_offset)\nwrite(\"x\", move=False, align=\"center\", font=(\"Arial\", 10, \"italic\"))\n\n\nht() # hide turtle\n\n",
"_____no_output_____"
]
],
[
[
"<span style=\"color:#ff0000; font-size:300%\"><u>Good</u></span> ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbc7f2b16bbac9d141676c857958200b1b990794
| 4,893 |
ipynb
|
Jupyter Notebook
|
docs/notebooks/thermal_speed.ipynb
|
davemus/PlasmaPy
|
6f163e07fb7fa3276a7b09e0f8fb1b0cf3bb3791
|
[
"MIT",
"BSD-2-Clause-Patent",
"BSD-2-Clause",
"BSD-3-Clause"
] | null | null | null |
docs/notebooks/thermal_speed.ipynb
|
davemus/PlasmaPy
|
6f163e07fb7fa3276a7b09e0f8fb1b0cf3bb3791
|
[
"MIT",
"BSD-2-Clause-Patent",
"BSD-2-Clause",
"BSD-3-Clause"
] | null | null | null |
docs/notebooks/thermal_speed.ipynb
|
davemus/PlasmaPy
|
6f163e07fb7fa3276a7b09e0f8fb1b0cf3bb3791
|
[
"MIT",
"BSD-2-Clause-Patent",
"BSD-2-Clause",
"BSD-3-Clause"
] | null | null | null | 30.58125 | 285 | 0.547517 |
[
[
[
"# Thermal Speed",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom astropy import units as u\n\nfrom plasmapy.formulary import (\n Maxwellian_speed_1D,\n Maxwellian_speed_2D,\n Maxwellian_speed_3D,\n)\nfrom plasmapy.formulary.parameters import thermal_speed",
"_____no_output_____"
]
],
[
[
"The thermal_speed function can be used to calculate the thermal velocity for a Maxwellian velocity distribution. There are three common definitions of the thermal velocity, which can be selected using the \"method\" keyword, which are defined for a 3D velocity distribution as\n\n\n- 'most_probable' <br>\n$v_{th} = \\sqrt{\\frac{2 k_B T}{m}}$\n\n- 'rms' <br>\n$v_{th} = \\sqrt{\\frac{3 k_B T}{m}}$\n\n- 'mean_magnitude' <br>\n$v_{th} = \\sqrt{\\frac{8 k_B T}{m\\pi}}$\n\nThe differences between these velocities can be seen by plotitng them on a 3D Maxwellian speed distribution",
"_____no_output_____"
]
],
[
[
"T = 1e5 * u.K\nspeeds = np.linspace(0, 8e6, num=600) * u.m / u.s\n\npdf_3D = Maxwellian_speed_3D(speeds, T=T, particle=\"e-\")\n\nfig, ax = plt.subplots(figsize=(4, 3))\n\nv_most_prob = thermal_speed(T=T, particle=\"e-\", method=\"most_probable\", ndim=3)\nv_rms = thermal_speed(T=T, particle=\"e-\", method=\"rms\", ndim=3)\nv_mean_magnitude = thermal_speed(T=T, particle=\"e-\", method=\"mean_magnitude\", ndim=3)\n\nax.plot(speeds / v_rms, pdf_3D, color=\"black\", label=\"Maxwellian\")\n\nax.axvline(x=v_most_prob / v_rms, color=\"blue\", label=\"Most Probable\")\nax.axvline(x=v_rms / v_rms, color=\"green\", label=\"RMS\")\nax.axvline(x=v_mean_magnitude / v_rms, color=\"red\", label=\"Mean Magnitude\")\n\nax.set_xlim(-0.1, 3)\nax.set_ylim(0, None)\nax.set_title(\"3D\")\nax.set_xlabel(\"|v|/|v$_{rms}|$\")\nax.set_ylabel(\"f(|v|)\")",
"_____no_output_____"
]
],
[
[
"Similar speeds are defined for 1D and 2D distributions. The differences between these definitions can be illustrated by plotting them on their respective Maxwellian speed distributions.",
"_____no_output_____"
]
],
[
[
"pdf_1D = Maxwellian_speed_1D(speeds, T=T, particle=\"e-\")\npdf_2D = Maxwellian_speed_2D(speeds, T=T, particle=\"e-\")\n\ndim = [1, 2, 3]\npdfs = [pdf_1D, pdf_2D, pdf_3D]\n\nplt.tight_layout()\nfig, ax = plt.subplots(ncols=3, figsize=(10, 3))\n\nfor n, pdf in enumerate(pdfs):\n ndim = n + 1\n v_most_prob = thermal_speed(T=T, particle=\"e-\", method=\"most_probable\", ndim=ndim)\n v_rms = thermal_speed(T=T, particle=\"e-\", method=\"rms\", ndim=ndim)\n v_mean_magnitude = thermal_speed(\n T=T, particle=\"e-\", method=\"mean_magnitude\", ndim=ndim\n )\n\n ax[n].plot(speeds / v_rms, pdf, color=\"black\", label=\"Maxwellian\")\n\n ax[n].axvline(x=v_most_prob / v_rms, color=\"blue\", label=\"Most Probable\")\n ax[n].axvline(x=v_rms / v_rms, color=\"green\", label=\"RMS\")\n ax[n].axvline(x=v_mean_magnitude / v_rms, color=\"red\", label=\"Mean Magnitude\")\n\n ax[n].set_xlim(-0.1, 3)\n ax[n].set_ylim(0, None)\n ax[n].set_title(\"{:d}D\".format(ndim))\n ax[n].set_xlabel(\"|v|/|v$_{rms}|$\")\n ax[n].set_ylabel(\"f(|v|)\")\n\n\nax[2].legend(bbox_to_anchor=(1.9, 0.8), loc=\"upper right\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc7f3643c2694d01891652f7567d285d3fcac57
| 9,291 |
ipynb
|
Jupyter Notebook
|
db2_for_machine_learning_samples/notebooks/Db2 Sample For Scikit-Learn.ipynb
|
gregstager/db2-samples
|
18ca92c4d2fbe93ac8a2a86e189577d13669cae0
|
[
"Apache-2.0"
] | 54 |
2019-08-02T13:15:07.000Z
|
2022-03-21T17:36:48.000Z
|
db2_for_machine_learning_samples/notebooks/Db2 Sample For Scikit-Learn.ipynb
|
gregstager/db2-samples
|
18ca92c4d2fbe93ac8a2a86e189577d13669cae0
|
[
"Apache-2.0"
] | 13 |
2019-07-26T13:51:16.000Z
|
2022-03-25T21:43:52.000Z
|
db2_for_machine_learning_samples/notebooks/Db2 Sample For Scikit-Learn.ipynb
|
gregstager/db2-samples
|
18ca92c4d2fbe93ac8a2a86e189577d13669cae0
|
[
"Apache-2.0"
] | 75 |
2019-07-20T04:53:24.000Z
|
2022-03-23T20:56:55.000Z
| 27.488166 | 502 | 0.583468 |
[
[
[
"#### Copyright IBM All Rights Reserved.\n#### SPDX-License-Identifier: Apache-2.0",
"_____no_output_____"
],
[
"# Db2 Sample For Scikit-Learn\n\nIn this code sample, we will show how to use the Db2 Python driver to import data from our Db2 database. Then, we will use that data to create a machine learning model with scikit-learn.\n\nMany wine connoisseurs love to taste different wines from all over the world. Mostly importantly, they want to know how the quality differs between each wine based on the ingredients. Some of them also want to be able to predict the quality before even tasting it. In this notebook, we will be using a dataset that has collected certain attributes of many wine bottles that determines the quality of the wine. Using this dataset, we will help our wine connoisseurs predict the quality of wine.\n\nThis notebook will demonstrate how to use Db2 as a data source for creating machine learning models.\n\nPrerequisites:\n1. Python 3.6 and above\n2. Db2 on Cloud instance (using free-tier option)\n3. Data already loaded in your Db2 instance\n4. Have Db2 connection credentials on hand\n\nWe will be importing two libraries- `ibm_db` and `ibm_dbi`. `ibm_db` is a library with low-level functions that will directly connect to our db2 database. To make things easier for you, we will be using `ibm-dbi`, which communicates with `ibm-db` and gives us an easy interface to interact with our data and import our data as a pandas dataframe. \n\nFor this example, we will be using the [winequality-red dataset](../data/winequality-red.csv), which we have loaded into our Db2 instance.\n\nNOTE: Running this notebook within a docker container. If `!easy_install ibm_db` doesn't work on your normally on jupter notebook, you may need to also run this notebook within a docker container as well.",
"_____no_output_____"
],
[
"## 1. Import Data\nLet's first install and import all the libraries needed for this notebook. Most important we will be installing and importing the db2 python driver `ibm_db`.",
"_____no_output_____"
]
],
[
[
"!pip install sklearn\n!easy_install ibm_db",
"_____no_output_____"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline\n\n# The two python ibm db2 drivers we need\nimport ibm_db\nimport ibm_db_dbi",
"_____no_output_____"
]
],
[
[
"Now let's import our data from our data source using the python db2 driver.",
"_____no_output_____"
]
],
[
[
"# replace only <> credentials\ndsn = \"DRIVER={{IBM DB2 ODBC DRIVER}};\" + \\\n \"DATABASE=<DATABASE NAME>;\" + \\\n \"HOSTNAME=<HOSTNMAE>;\" + \\\n \"PORT=50000;\" + \\\n \"PROTOCOL=TCPIP;\" + \\\n \"UID=<USERNAME>;\" + \\\n \"PWD=<PWD>;\"\nhdbc = ibm_db.connect(dsn, \"\", \"\")\nhdbi = ibm_db_dbi.Connection(hdbc)\n\nsql = 'SELECT * FROM <SCHEMA NAME>.<TABLE NAME>'\n\nwine = pandas.read_sql(sql,hdbi)\n#wine = pd.read_csv('../data/winequality-red.csv', sep=';') ",
"_____no_output_____"
],
[
"wine.head()",
"_____no_output_____"
]
],
[
[
"## 2. Data Exploration",
"_____no_output_____"
],
[
"In this step, we are going to try and explore our data inorder to gain insight. We hope to be able to make some assumptions of our data before we start modeling.",
"_____no_output_____"
]
],
[
[
"wine.describe()",
"_____no_output_____"
],
[
"# Minimum price of the data\nminimum_price = np.amin(wine['quality'])\n\n# Maximum price of the data\nmaximum_price = np.amax(wine['quality'])\n\n# Mean price of the data\nmean_price = np.mean(wine['quality'])\n\n# Median price of the data\nmedian_price = np.median(wine['quality'])\n\n# Standard deviation of prices of the data\nstd_price = np.std(wine['quality'])\n\n# Show the calculated statistics\nprint(\"Statistics for housing dataset:\\n\")\nprint(\"Minimum quality: {}\".format(minimum_price)) \nprint(\"Maximum quality: {}\".format(maximum_price))\nprint(\"Mean quality: {}\".format(mean_price))\nprint(\"Median quality {}\".format(median_price))\nprint(\"Standard deviation of quality: {}\".format(std_price))",
"_____no_output_____"
],
[
"wine.corr()",
"_____no_output_____"
],
[
"corr_matrix = wine.corr()\ncorr_matrix[\"quality\"].sort_values(ascending=False)",
"_____no_output_____"
]
],
[
[
"## 3. Data Visualization",
"_____no_output_____"
]
],
[
[
"wine.hist(bins=50, figsize=(30,25))\nplt.show()",
"_____no_output_____"
],
[
"boxplot = wine.boxplot(column=['quality'])",
"_____no_output_____"
]
],
[
[
"## 4. Creating Machine Learning Model",
"_____no_output_____"
],
[
"Now that we have cleaned and explored our data. We are ready to build our model that will predict the attribute `quality`. ",
"_____no_output_____"
]
],
[
[
"wine_value = wine['quality']\nwine_attributes = wine.drop(['quality'], axis=1)",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\n\n# Let us scale our data first \nsc = StandardScaler()\nwine_attributes = sc.fit_transform(wine_attributes)",
"_____no_output_____"
],
[
"from sklearn.decomposition import PCA\n\n# Apply PCA to our data\npca = PCA(n_components=8)\nx_pca = pca.fit_transform(wine_attributes)",
"_____no_output_____"
]
],
[
[
"We need to split our data into train and test data.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split\n\n# Split our data into test and train data\nx_train, x_test, y_train, y_test = train_test_split( wine_attributes,wine_value, test_size = 0.25)",
"_____no_output_____"
]
],
[
[
"We will be using Logistic Regression to model our data",
"_____no_output_____"
]
],
[
[
"from sklearn.linear_model import LogisticRegression\nfrom sklearn.metrics import confusion_matrix, accuracy_score\n\nlr = LogisticRegression()\n\n# Train our model\nlr.fit(x_train, y_train)\n\n# Predict using our trained model and our test data\nlr_predict = lr.predict(x_test)",
"_____no_output_____"
],
[
"# Print confusion matrix and accuracy score\nlr_conf_matrix = confusion_matrix(y_test, lr_predict)\nlr_acc_score = accuracy_score(y_test, lr_predict)\nprint(lr_conf_matrix)\nprint(lr_acc_score*100)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbc804ea3d3f4c0d35433f6e6a53b26b600390a2
| 8,883 |
ipynb
|
Jupyter Notebook
|
2020_week_1/Taylor_problem_3.23.ipynb
|
furnstahl/5300-notebooks
|
c3551bf2d8de22dec11d9035aa1ea78de1355cad
|
[
"MIT"
] | 3 |
2019-12-16T22:01:54.000Z
|
2020-12-11T18:07:58.000Z
|
2020_week_1/Taylor_problem_3.23.ipynb
|
furnstahl/5300-notebooks
|
c3551bf2d8de22dec11d9035aa1ea78de1355cad
|
[
"MIT"
] | null | null | null |
2020_week_1/Taylor_problem_3.23.ipynb
|
furnstahl/5300-notebooks
|
c3551bf2d8de22dec11d9035aa1ea78de1355cad
|
[
"MIT"
] | 7 |
2019-01-28T19:01:02.000Z
|
2021-04-15T22:45:23.000Z
| 31.059441 | 452 | 0.547 |
[
[
[
"# Taylor problem 3.23\n\nlast revised: 04-Jan-2020 by Dick Furnstahl [[email protected]]\n",
"_____no_output_____"
],
[
"**This notebook is almost ready to go, except that the initial conditions and $\\Delta v$ are different from the problem statement and there is no statement to print the figure. Fix these and you're done!**",
"_____no_output_____"
],
[
"This is a conservation of momentum problem, which in the end lets us determine the trajectories of the two masses before and after the explosion. How should we visualize that the center-of-mass of the pieces continues to follow the original parabolic path?",
"_____no_output_____"
],
[
"Plan:\n1. Plot the original trajectory, also continued past the explosion time.\n2. Plot the two trajectories after the explosion.\n3. For some specified times of the latter two trajectories, connect the points and indicate the center of mass.\n\nThe implementation here could certainly be improved! Please make suggestions (and develop improved versions).",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"First define some functions we think we will need. The formulas are based on our paper-and-pencil work.",
"_____no_output_____"
],
[
"The trajectory starting from $t=0$ is:\n\n$\n\\begin{align}\n x(t) &= x_0 + v_{x0} t \\\\\n y(t) &= y_0 + v_{y0} t - \\frac{1}{2} g t^2\n\\end{align}\n$",
"_____no_output_____"
]
],
[
[
"def trajectory(x0, y0, vx0, vy0, t_pts, g=9.8):\n \"\"\"Calculate the x(t) and y(t) trajectories for an array of times,\n which must start with t=0.\n \"\"\"\n return x0 + vx0*t_pts, y0 + vy0*t_pts - g*t_pts**2/2. ",
"_____no_output_____"
]
],
[
[
"The velocity at the final time $t_f$ is:\n\n$\n\\begin{align}\n v_{x}(t) &= v_{x0} \\\\\n v_{y}(t) &= v_{y0} - g t_f\n\\end{align}\n$",
"_____no_output_____"
]
],
[
[
"def final_velocity(vx0, vy0, t_pts, g=9.8):\n \"\"\"Calculate the vx(t) and vy(t) at the end of an array of times t_pts\"\"\"\n return vx0, vy0 - g*t_pts[-1] # -1 gives the last element",
"_____no_output_____"
]
],
[
[
"The center of mass of two particles at $(x_1, y_1)$ and $(x_2, y_2)$ is:\n\n$\n\\begin{align}\n x_{cm} &= \\frac{1}{2}(x_1 + x_2) \\\\\n y_{cm} &= \\frac{1}{2}(y_1 + y_2)\n\\end{align}\n$",
"_____no_output_____"
]
],
[
[
"def com_position(x1, y1, x2, y2): \n \"\"\"Find the center-of-mass (com) position given two positions (x,y).\"\"\"\n return (x1 + x2)/2., (y1 + y2)/2.",
"_____no_output_____"
]
],
[
[
"**1. Calculate and plot the original trajectory up to the explosion.**",
"_____no_output_____"
]
],
[
[
"# initial conditions\nx0_before, y0_before = [0., 0.] # put the origin at the starting point\nvx0_before, vy0_before = [6., 3.] # given in the problem statement\ng = 1. # as recommended\n\n# Array of times to calculate the trajectory up to the explosion at t=4\nt_pts_before = np.array([0., 1., 2., 3., 4.])\nx_before, y_before = trajectory(x0_before, y0_before,\n vx0_before, vy0_before,\n t_pts_before, g)",
"_____no_output_____"
],
[
"fig = plt.figure()\nax = fig.add_subplot(1,1,1)\nax.plot(x_before, y_before, 'ro-')\n\nax.set_xlabel('x')\nax.set_ylabel('y')",
"_____no_output_____"
]
],
[
[
"Does it make sense so far? Note that we could use more intermediate points to make a more correct curve (rather than the piecewise straight lines) but this is fine at least for a first pass.",
"_____no_output_____"
],
[
"**2. Calculate and plot the two trajectories after the explosion.**\n\nFor the second part of the trajectory, we reset our clock to $t=0$ because that is how our trajectory function is constructed. We'll need initial positions and velocities of the pieces just after the explosion. These are the final position of the combined piece before the explosion and the final velocity plus and minus $\\Delta \\mathbf{v}$. We are told $\\Delta \\mathbf{v}$. We have to figure out the final velocity before the explosion.",
"_____no_output_____"
]
],
[
[
"delta_v = np.array([2., 1.]) # change in velociy of one piece\n\n# reset time to 0 for calculating trajectories\nt_pts_after = np.array([0., 1., 2., 3., 4., 5.])\n# Also could have used np.arange(0.,6.,1.)\n\nx0_after = x_before[-1] # -1 here means the last element of the array\ny0_after = y_before[-1]\nvxcm0_after, vycm0_after = final_velocity(vx0_before, vy0_before,\n t_pts_before, g) \n# The _1 and _2 refer to the two pieces after the explosinon\nvx0_after_1 = vxcm0_after + delta_v[0]\nvy0_after_1 = vycm0_after + delta_v[1]\nvx0_after_2 = vxcm0_after - delta_v[0]\nvy0_after_2 = vycm0_after - delta_v[1]\n\n# Given the initial conditions after the explosion, we calculate trajectories\nx_after_1, y_after_1 = trajectory(x0_after, y0_after,\n vx0_after_1, vy0_after_1,\n t_pts_after, g)\nx_after_2, y_after_2 = trajectory(x0_after, y0_after,\n vx0_after_2, vy0_after_2,\n t_pts_after, g)\n# This is the center-of-mass trajectory\nxcm_after, ycm_after = trajectory(x0_after, y0_after,\n vxcm0_after, vycm0_after,\n t_pts_after, g)\n\n# These are calculated points of the center-of-mass\nxcm_pts, ycm_pts = com_position(x_after_1, y_after_1, x_after_2, y_after_2)",
"_____no_output_____"
],
[
"fig = plt.figure()\nax = fig.add_subplot(1,1,1)\nax.plot(x_before, y_before, 'ro-', label='before explosion')\nax.plot(x_after_1, y_after_1, 'go-', label='piece 1 after')\nax.plot(x_after_2, y_after_2, 'bo-', label='piece 2 after')\nax.plot(xcm_after, ycm_after, 'r--', label='original trajectory')\nax.plot(xcm_pts, ycm_pts, 'o', color='black', label='center-of-mass of 1 and 2')\nfor i in range(len(t_pts_after)):\n ax.plot([x_after_1[i], x_after_2[i]],\n [y_after_1[i], y_after_2[i]],\n 'k--'\n )\nax.set_xlabel('x')\nax.set_ylabel('y')\nax.legend();",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbc8068a6690a2104b7fe5c7228d32d14312c9a8
| 3,967 |
ipynb
|
Jupyter Notebook
|
Analysis-master-corrwith-self.ipynb
|
ajaymota/jax
|
25bea073cf2e3b8b9764ae8a76dba3e752fc55e6
|
[
"ECL-2.0",
"Apache-2.0"
] | null | null | null |
Analysis-master-corrwith-self.ipynb
|
ajaymota/jax
|
25bea073cf2e3b8b9764ae8a76dba3e752fc55e6
|
[
"ECL-2.0",
"Apache-2.0"
] | null | null | null |
Analysis-master-corrwith-self.ipynb
|
ajaymota/jax
|
25bea073cf2e3b8b9764ae8a76dba3e752fc55e6
|
[
"ECL-2.0",
"Apache-2.0"
] | null | null | null | 33.905983 | 111 | 0.554575 |
[
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"# Distances",
"_____no_output_____"
]
],
[
[
"activator = \"sgmd\"\n\nmnist_sgmd = []\nhands_sgmd = []\nfashn_sgmd = []\n\nfor i in range(0, 10):\n dataset = 'mnist'\n df_cnn_relu0_1 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n dataset = 'handsign_mnist'\n df_cnn_relu0_2 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n dataset = 'fashion_mnist'\n df_cnn_relu0_3 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n\n max_norm = np.linalg.norm(df_cnn_relu0_1.corrwith(df_cnn_relu0_1))\n \n mnist_sgmd.append(np.linalg.norm(df_cnn_relu0_1.corrwith(df_cnn_relu0_2)) / max_norm)\n hands_sgmd.append(np.linalg.norm(df_cnn_relu0_2.corrwith(df_cnn_relu0_3)) / max_norm)\n fashn_sgmd.append(np.linalg.norm(df_cnn_relu0_3.corrwith(df_cnn_relu0_1)) / max_norm)\n\nactivator = \"tanh\"\n\nmnist_tanh = []\nhands_tanh = []\nfashn_tanh = []\n\nfor i in range(0, 10):\n dataset = 'mnist'\n df_cnn_relu0_1 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n dataset = 'handsign_mnist'\n df_cnn_relu0_2 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n dataset = 'fashion_mnist'\n df_cnn_relu0_3 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n\n max_norm = np.linalg.norm(df_cnn_relu0_1.corrwith(df_cnn_relu0_1))\n \n mnist_tanh.append(np.linalg.norm(df_cnn_relu0_1.corrwith(df_cnn_relu0_2)) / max_norm)\n hands_tanh.append(np.linalg.norm(df_cnn_relu0_2.corrwith(df_cnn_relu0_3)) / max_norm)\n fashn_tanh.append(np.linalg.norm(df_cnn_relu0_3.corrwith(df_cnn_relu0_1)) / max_norm)\n \nactivator = \"relu\"\n\nmnist_relu = []\nhands_relu = []\nfashn_relu = []\n\nfor i in range(0, 10):\n dataset = 'mnist'\n df_cnn_relu0_1 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n dataset = 'handsign_mnist'\n df_cnn_relu0_2 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n dataset = 'fashion_mnist'\n df_cnn_relu0_3 = pd.read_csv(dataset + \"/results/\" + activator + \"/cnn_K\" + str(i) + \".csv\")\n\n max_norm = np.linalg.norm(df_cnn_relu0_1.corrwith(df_cnn_relu0_1))\n \n mnist_relu.append(np.linalg.norm(df_cnn_relu0_1.corrwith(df_cnn_relu0_2)) / max_norm)\n hands_relu.append(np.linalg.norm(df_cnn_relu0_2.corrwith(df_cnn_relu0_3)) / max_norm)\n fashn_relu.append(np.linalg.norm(df_cnn_relu0_3.corrwith(df_cnn_relu0_1)) / max_norm)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc806a88b30e6aa39640d360f1d69645ac0c3e4
| 73,248 |
ipynb
|
Jupyter Notebook
|
04其他模型教程/4.03 BERT-NER-MSRA.ipynb
|
DrDavidS/basic_Machine_Learning
|
d6f6538a13ed68543569f595fa833e6d220beedd
|
[
"MIT"
] | 15 |
2019-09-12T01:04:44.000Z
|
2022-01-01T02:30:51.000Z
|
04其他模型教程/4.03 BERT-NER-MSRA.ipynb
|
DrDavidS/basic_Machine_Learning
|
d6f6538a13ed68543569f595fa833e6d220beedd
|
[
"MIT"
] | null | null | null |
04其他模型教程/4.03 BERT-NER-MSRA.ipynb
|
DrDavidS/basic_Machine_Learning
|
d6f6538a13ed68543569f595fa833e6d220beedd
|
[
"MIT"
] | 11 |
2019-10-19T03:12:03.000Z
|
2021-01-07T05:14:01.000Z
| 36.80804 | 450 | 0.456927 |
[
[
[
"import os\nfrom pprint import pprint\n\nimport torch\nimport torch.nn as nn\nfrom transformers import BertForTokenClassification, BertTokenizer\nfrom transformers import AdamW\nfrom torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler\nfrom sklearn.model_selection import train_test_split\nimport numpy as np\nfrom tqdm.notebook import tqdm",
"_____no_output_____"
]
],
[
[
"## 读取MSRA实体识别数据集",
"_____no_output_____"
]
],
[
[
"file = \"../datasets/dh_msra.txt\"",
"_____no_output_____"
]
],
[
[
"## 检查GPU情况",
"_____no_output_____"
]
],
[
[
"# GPUcheck\n\nprint(\"CUDA Available: \", torch.cuda.is_available())\nn_gpu = torch.cuda.device_count()\n\nif torch.cuda.is_available():\n print(\"GPU numbers: \", n_gpu)\n print(\"device_name: \", torch.cuda.get_device_name(0))\n device = torch.device(\"cuda:0\") # 注意选择\n torch.cuda.set_device(0) \n print(f\"当前设备:{torch.cuda.current_device()}\")\nelse :\n device = torch.device(\"cpu\")\n print(f\"当前设备:{device}\")",
"CUDA Available: True\nGPU numbers: 1\ndevice_name: GeForce RTX 3090\n当前设备:0\n"
]
],
[
[
"## 配置参数\n\n规范化配置参数,方便使用。",
"_____no_output_____"
]
],
[
[
"class Config(object):\n \"\"\"配置参数\"\"\"\n def __init__(self):\n self.model_name = 'Bert_NER.bin'\n self.bert_path = './bert-chinese/'\n self.ner_file = '../datasets/dh_msra.txt'\n \n self.num_classes = 10 # 类别数(按需修改),这里有10种实体类型\n self.hidden_size = 768 # 隐藏层输出维度\n self.hidden_dropout_prob = 0.1 # dropout比例\n self.batch_size = 128 # mini-batch大小\n self.max_len = 103 # 句子的最长padding长度\n \n self.epochs = 3 # epoch数\n self.learning_rate = 2e-5 # 学习率 \n\n self.save_path = './saved_model/' # 模型训练结果保存路径\n \n # self.fp16 = False\n # self.fp16_opt_level = 'O1'\n # self.gradient_accumulation_steps = 1\n # self.warmup_ratio = 0.06\n # self.warmup_steps = 0\n # self.max_grad_norm = 1.0\n # self.adam_epsilon = 1e-8\n # self.class_list = class_list # 类别名单\n # self.require_improvement = 1000 # 若超过1000batch效果还没提升,则提前结束训练\n \nconfig = Config()",
"_____no_output_____"
],
[
"all_sentences_separate = []\nall_letter_labels = []\nlabel_set = set()\n\nwith open(config.ner_file, encoding=\"utf-8\") as f:\n single_sentence = []\n single_sentence_labels = []\n for s in f.readlines():\n if s != \"\\n\":\n word, label = s.split(\"\\t\")\n label = label.strip(\"\\n\")\n single_sentence.append(word)\n single_sentence_labels.append(label)\n label_set.add(label)\n elif s == \"\\n\":\n all_sentences_separate.append(single_sentence)\n all_letter_labels.append(single_sentence_labels)\n single_sentence = []\n single_sentence_labels = []",
"_____no_output_____"
],
[
"print(all_sentences_separate[0:2])\nprint(all_letter_labels[0:2])\n\nprint(f\"\\n所有的标签:{label_set}\")",
"[['当', '希', '望', '工', '程', '救', '助', '的', '百', '万', '儿', '童', '成', '长', '起', '来', ',', '科', '教', '兴', '国', '蔚', '然', '成', '风', '时', ',', '今', '天', '有', '收', '藏', '价', '值', '的', '书', '你', '没', '买', ',', '明', '日', '就', '叫', '你', '悔', '不', '当', '初', '!'], ['藏', '书', '本', '来', '就', '是', '所', '有', '传', '统', '收', '藏', '门', '类', '中', '的', '第', '一', '大', '户', ',', '只', '是', '我', '们', '结', '束', '温', '饱', '的', '时', '间', '太', '短', '而', '已', '。']]\n[['O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O'], ['O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O']]\n\n所有的标签:{'I-PER', 'B-PER', 'O', 'I-ORG', 'B-LOC', 'B-ORG', 'I-LOC'}\n"
],
[
"# 构建 tag 到 索引 的字典\ntag_to_ix = {\"B-LOC\": 0,\n \"I-LOC\": 1, \n \"B-ORG\": 2, \n \"I-ORG\": 3,\n \"B-PER\": 4,\n \"I-PER\": 5,\n \"O\": 6,\n \"[CLS]\":7,\n \"[SEP]\":8,\n \"[PAD]\":9}\n\nix_to_tag = {0:\"B-LOC\", \n 1:\"I-LOC\", \n 2:\"B-ORG\", \n 3:\"I-ORG\",\n 4:\"B-PER\",\n 5:\"I-PER\",\n 6:\"O\",\n 7:\"[CLS]\",\n 8:\"[SEP]\",\n 9:\"[PAD]\"}",
"_____no_output_____"
]
],
[
[
"## 数据示例\n\n这里简单查看一些数据例子,其中很多都是数字6。\n\n数字6说明是 O 类型的实体。",
"_____no_output_____"
]
],
[
[
"all_sentences = [] # 句子\n\nfor one_sentence in all_sentences_separate:\n sentence = \"\".join(one_sentence)\n all_sentences.append(sentence)\n\nprint(all_sentences[0:2])",
"['当希望工程救助的百万儿童成长起来,科教兴国蔚然成风时,今天有收藏价值的书你没买,明日就叫你悔不当初!', '藏书本来就是所有传统收藏门类中的第一大户,只是我们结束温饱的时间太短而已。']\n"
],
[
"all_labels = [] # labels\nfor letter_labels in all_letter_labels:\n labels = [tag_to_ix[t] for t in letter_labels]\n all_labels.append(labels)\n\nprint(all_labels[0:2])\nprint(len(all_labels[0]))",
"[[6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6], [6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6]]\n50\n"
],
[
"print(len(all_labels))",
"55289\n"
]
],
[
[
"### input数据准备",
"_____no_output_____"
]
],
[
[
"# word2token\ntokenizer = BertTokenizer.from_pretrained('./bert-chinese/', do_lower_case=True)\n\n# 新版代码,一次性处理好输入\nencoding = tokenizer(all_sentences, \n return_tensors='pt', # pt 指 pytorch,tf 就是 tensorflow \n padding='max_length', # padding 到 max_length\n truncation=True, # 激活并控制截断\n max_length=config.max_len)\n\ninput_ids = encoding['input_ids']",
"_____no_output_____"
],
[
"# 这句话的input_ids\nprint(f\"Tokenize 前的第一句话:\\n{all_sentences[0]}\\n\")\nprint(f\"Tokenize + Padding 后的第一句话: \\n{input_ids[0]}\")",
"Tokenize 前的第一句话:\n当希望工程救助的百万儿童成长起来,科教兴国蔚然成风时,今天有收藏价值的书你没买,明日就叫你悔不当初!\n\nTokenize + Padding 后的第一句话: \ntensor([ 101, 2496, 2361, 3307, 2339, 4923, 3131, 1221, 4638, 4636, 674, 1036,\n 4997, 2768, 7270, 6629, 3341, 8024, 4906, 3136, 1069, 1744, 5917, 4197,\n 2768, 7599, 3198, 8024, 791, 1921, 3300, 3119, 5966, 817, 966, 4638,\n 741, 872, 3766, 743, 8024, 3209, 3189, 2218, 1373, 872, 2637, 679,\n 2496, 1159, 8013, 102, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0])\n"
],
[
"# 新版代码\nattention_masks = encoding['attention_mask']\ntoken_type_ids = encoding['token_type_ids']",
"_____no_output_____"
],
[
"# 第一句话的 attention_masks\nprint(attention_masks[0])",
"tensor([1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,\n 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,\n 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0])\n"
]
],
[
[
"## 准备labels\n\n由于我们的input_ids是带有`[CLS]`和`[SEP]`的,所以在准备label的同时也要考虑这些情况。",
"_____no_output_____"
]
],
[
[
"# [3] 代表 O 实体\nfor label in all_labels:\n label.insert(len(label), 8) # [SEP]\n label.insert(0, 7) # [CLS]\n if config.max_len > len(label) -1:\n for i in range(config.max_len - len(label)): #+2的原因是扣除多出来的CLS和SEP\n label.append(9) # [PAD]",
"_____no_output_____"
],
[
"print(len(all_labels[0]))\nprint(all_labels[0])",
"103\n[7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9]\n"
],
[
"# 统计最长的段落\nmax_len_label = 0\nmax_len_text = 0\n\nfor label in all_labels:\n if len(label) > max_len_text:\n max_len_label = len(label)\nprint(max_len_label)\n \nfor one_input in input_ids:\n if len(one_input) > max_len_text:\n max_len_text = len(one_input) \n \nprint(max_len_text)",
"103\n103\n"
]
],
[
[
"## 切分训练和测试集",
"_____no_output_____"
]
],
[
[
"# train-test-split\ntrain_inputs, validation_inputs, train_labels, validation_labels = train_test_split(input_ids, \n all_labels, \n random_state=2021, \n test_size=0.1)\ntrain_masks, validation_masks, _, _ = train_test_split(attention_masks, \n input_ids,\n random_state=2021, \n test_size=0.1)",
"_____no_output_____"
],
[
"print(len(train_inputs))\nprint(len(validation_inputs))\n\nprint(train_inputs[0])\nprint(validation_inputs[0])",
"49760\n5529\ntensor([ 101, 5632, 5356, 510, 5632, 2193, 510, 5632, 4028, 8024, 2961, 5298,\n 1469, 4028, 1139, 6963, 3221, 1164, 4500, 5688, 969, 3189, 1469, 689,\n 865, 3198, 7313, 511, 102, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0])\ntensor([ 101, 1728, 2349, 1239, 3172, 1788, 1469, 809, 5682, 1154, 1146, 3637,\n 898, 3191, 8024, 677, 3299, 1159, 4638, 840, 3142, 833, 6379, 679,\n 3614, 5445, 3141, 8024, 1400, 3341, 5401, 1744, 2456, 6379, 4638, 1290,\n 4670, 7561, 833, 6379, 1348, 1728, 809, 2600, 4415, 1079, 1849, 2225,\n 762, 5529, 2867, 5318, 680, 833, 5445, 5522, 3647, 5592, 704, 511,\n 102, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,\n 0, 0, 0, 0, 0, 0, 0])\n"
]
],
[
[
"这里把输入的labels变为tensor形式。",
"_____no_output_____"
]
],
[
[
"train_labels = torch.tensor(train_labels).clone().detach()\nvalidation_labels = torch.tensor(validation_labels).clone().detach()",
"_____no_output_____"
],
[
"print(train_labels[0])\n\nprint(len(train_labels))\nprint(len(train_inputs))",
"tensor([7, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6,\n 6, 6, 6, 6, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9,\n 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9,\n 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9, 9,\n 9, 9, 9, 9, 9, 9, 9])\n49760\n49760\n"
],
[
"# dataloader\n\n# 形成训练数据集\ntrain_data = TensorDataset(train_inputs, train_masks, train_labels) \n# 随机采样\ntrain_sampler = RandomSampler(train_data) \n# 读取数据\ntrain_dataloader = DataLoader(train_data, sampler=train_sampler, batch_size=config.batch_size)\n\n\n# 形成验证数据集\nvalidation_data = TensorDataset(validation_inputs, validation_masks, validation_labels)\n# 随机采样\nvalidation_sampler = SequentialSampler(validation_data)\n# 读取数据\nvalidation_dataloader = DataLoader(validation_data, sampler=validation_sampler, batch_size=config.batch_size)",
"_____no_output_____"
],
[
"model = BertForTokenClassification.from_pretrained(config.bert_path, num_labels=config.num_classes)\nmodel.cuda()\n\n# 注意:\n# 在新版的 Transformers 中会给出警告\n# 原因是我们导入的预训练参数权重是不包含模型最终的线性层权重的\n# 不过我们本来就是要“微调”它,所以这个情况是符合期望的",
"Some weights of the model checkpoint at ./bert-chinese/ were not used when initializing BertForTokenClassification: ['cls.predictions.bias', 'cls.predictions.transform.dense.weight', 'cls.predictions.transform.dense.bias', 'cls.predictions.decoder.weight', 'cls.seq_relationship.weight', 'cls.seq_relationship.bias', 'cls.predictions.transform.LayerNorm.weight', 'cls.predictions.transform.LayerNorm.bias']\n- This IS expected if you are initializing BertForTokenClassification from the checkpoint of a model trained on another task or with another architecture (e.g. initializing a BertForSequenceClassification model from a BertForPreTraining model).\n- This IS NOT expected if you are initializing BertForTokenClassification from the checkpoint of a model that you expect to be exactly identical (initializing a BertForSequenceClassification model from a BertForSequenceClassification model).\nSome weights of BertForTokenClassification were not initialized from the model checkpoint at ./bert-chinese/ and are newly initialized: ['classifier.weight', 'classifier.bias']\nYou should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.\n"
],
[
"# BERT fine-tuning parameters\nparam_optimizer = list(model.named_parameters())\nno_decay = ['bias', 'LayerNorm.weight']\n\n# 权重衰减\noptimizer_grouped_parameters = [\n {'params': [p for n, p in param_optimizer if not any(nd in n for nd in no_decay)], \n 'weight_decay': 0.01},\n {'params': [p for n, p in param_optimizer if any(nd in n for nd in no_decay)], \n 'weight_decay': 0.0}]",
"_____no_output_____"
],
[
"# 优化器\noptimizer = AdamW(optimizer_grouped_parameters,\n lr=5e-5)",
"_____no_output_____"
],
[
"# 保存loss\ntrain_loss_set = []\n",
"_____no_output_____"
],
[
"# BERT training loop\nfor _ in range(config.epochs): \n ## 训练\n print(f\"当前epoch: {_}\")\n # 开启训练模式\n model.train()\n tr_loss = 0 # train loss\n nb_tr_examples, nb_tr_steps = 0, 0\n # Train the data for one epoch\n for step, batch in tqdm(enumerate(train_dataloader)):\n # 把batch放入GPU\n batch = tuple(t.to(device) for t in batch)\n # 解包batch\n b_input_ids, b_input_mask, b_labels = batch\n # 梯度归零\n optimizer.zero_grad()\n # 前向传播loss计算\n output = model(input_ids=b_input_ids, \n attention_mask=b_input_mask, \n labels=b_labels) \n loss = output[0]\n # print(loss)\n # 反向传播\n loss.backward()\n # Update parameters and take a step using the computed gradient\n # 更新模型参数\n optimizer.step()\n # Update tracking variables\n tr_loss += loss.item()\n nb_tr_examples += b_input_ids.size(0)\n nb_tr_steps += 1\n \n print(f\"当前 epoch 的 Train loss: {tr_loss/nb_tr_steps}\")",
"当前epoch: 0\n"
],
[
"# 验证状态\nmodel.eval()\n\n# 建立变量\neval_loss, eval_accuracy = 0, 0\nnb_eval_steps, nb_eval_examples = 0, 0\n# Evaluate data for one epoch",
"_____no_output_____"
],
[
"# 验证集的读取也要batch\nfor batch in tqdm(validation_dataloader):\n # 元组打包放进GPU\n batch = tuple(t.to(device) for t in batch)\n # 解开元组\n b_input_ids, b_input_mask, b_labels = batch\n # 预测\n with torch.no_grad():\n # segment embeddings,如果没有就是全0,表示单句\n # position embeddings,[0,句子长度-1]\n outputs = model(input_ids=b_input_ids, \n attention_mask=b_input_mask,\n token_type_ids=None,\n position_ids=None) \n \n # print(logits[0])\n # Move logits and labels to CPU\n scores = outputs[0].detach().cpu().numpy() # 每个字的标签的概率\n pred_flat = np.argmax(scores[0], axis=1).flatten()\n label_ids = b_labels.to('cpu').numpy() # 真实labels\n # print(logits, label_ids)",
"_____no_output_____"
],
[
"# 保存模型\n# They can then be reloaded using `from_pretrained()`\n# 创建文件夹\nif not os.path.exists(config.save_path):\n os.makedirs(config.save_path)\n print(\"文件夹不存在,创建文件夹!\")\nelse:\n pass\n\n\noutput_dir = config.save_path\nmodel_to_save = model.module if hasattr(model, 'module') else model # Take care of distributed/parallel training\n\n\n# Good practice: save your training arguments together with the trained model\ntorch.save(model_to_save.state_dict(), os.path.join(output_dir, config.model_name))",
"文件夹不存在,创建文件夹!\n"
],
[
"# 读取模型\n# Load a trained model and vocabulary that you have fine-tuned\noutput_dir = config.save_path\nmodel = BertForTokenClassification.from_pretrained(output_dir)\ntokenizer = BertTokenizer.from_pretrained(output_dir)\nmodel.to(device)",
"_____no_output_____"
],
[
"# 单句测试\n\n# test_sententce = \"在北京市朝阳区的一家网吧,我亲眼看见卢本伟和孙笑川一起开挂。\"\ntest_sententce = \"史源源的房子租在滨江区南环路税友大厦附近。\"",
"_____no_output_____"
],
[
"# 构建 tag 到 索引 的字典\ntag_to_ix = {\"B-LOC\": 0,\n \"I-LOC\": 1, \n \"B-ORG\": 2, \n \"I-ORG\": 3,\n \"B-PER\": 4,\n \"I-PER\": 5,\n \"O\": 6,\n \"[CLS]\":7,\n \"[SEP]\":8,\n \"[PAD]\":9}\n\nix_to_tag = {0:\"B-LOC\", \n 1:\"I-LOC\", \n 2:\"B-ORG\", \n 3:\"I-ORG\",\n 4:\"B-PER\",\n 5:\"I-PER\",\n 6:\"O\",\n 7:\"[CLS]\",\n 8:\"[SEP]\",\n 9:\"[PAD]\"}",
"_____no_output_____"
],
[
"encoding = tokenizer(test_sententce, \n return_tensors='pt', # pt 指 pytorch,tf 就是 tensorflow \n padding=True, # padding到最长的那句话\n truncation=True, # 激活并控制截断\n max_length=50)\n\ntest_input_ids = encoding['input_ids']\n# 创建attention masks\ntest_attention_masks = encoding['attention_mask']",
"_____no_output_____"
],
[
"# 形成验证数据集\n# 为了通用,这里还是用了 DataLoader 的形式\ntest_data = TensorDataset(test_input_ids, test_attention_masks)\n# 随机采样\ntest_sampler = SequentialSampler(test_data)\n# 读取数据\ntest_dataloader = DataLoader(test_data, sampler=test_sampler, batch_size=config.batch_size)",
"_____no_output_____"
],
[
"# 验证状态\nmodel.eval()\n\n# 建立变量\neval_loss, eval_accuracy = 0, 0\nnb_eval_steps, nb_eval_examples = 0, 0\n# Evaluate data for one epoch",
"_____no_output_____"
],
[
"# 验证集的读取也要batch\nfor batch in tqdm(test_dataloader):\n # 元组打包放进GPU\n batch = tuple(t.to(device) for t in batch)\n # 解开元组\n b_input_ids, b_input_mask = batch\n # 预测\n with torch.no_grad():\n # segment embeddings,如果没有就是全0,表示单句\n # position embeddings,[0,句子长度-1]\n outputs = model(input_ids=b_input_ids, \n attention_mask=None,\n token_type_ids=None,\n position_ids=None) \n \n # Move logits and labels to CPU\n scores = outputs[0].detach().cpu().numpy() # 每个字的标签的概率\n pred_flat = np.argmax(scores[0], axis=1).flatten()\n # label_ids = b_labels.to('cpu').numpy() # 真实labels\n print(pred_flat) # 预测值",
"_____no_output_____"
],
[
"pre_labels = [ix_to_tag[n] for n in pred_flat]\nprint(f\"测试句子: {test_sententce}\")\nprint(len(test_sententce))\nprint(pre_labels)",
"测试句子: 史源源的房子租在滨江区南环路税友大厦附近。\n21\n['[CLS]', 'B-PER', 'I-PER', 'I-PER', 'O', 'O', 'O', 'O', 'O', 'B-LOC', 'I-LOC', 'I-LOC', 'B-LOC', 'I-LOC', 'I-LOC', 'B-LOC', 'I-LOC', 'I-LOC', 'I-LOC', 'O', 'O', 'O', '[SEP]']\n"
],
[
"pre_labels_cut = pre_labels[0:len(test_sententce)+2]\npre_labels_cut",
"_____no_output_____"
],
[
"person = [] # 临时栈\npersons = []\n\nlocation = []\nlocations = []\n\n\nfor i in range(len(pre_labels_cut) - 1):\n # Person\n # 单字情况\n if pre_labels[i] == 'B-PER' and pre_labels[i+1] != 'I-PER' and len(location) == 0:\n person.append(i) \n persons.append(person)\n person = [] # 清空\n continue \n \n # 非单字\n # 如果前面有连着的 PER 实体 \n if pre_labels[i] == 'B-PER'and pre_labels[i+1] == 'I-PER' and len(person) != 0:\n person.append(i)\n \n # 如果前面没有连着的 B-PER 实体\n elif pre_labels[i] == 'B-PER'and pre_labels[i+1] == 'I-PER' and len(location) == 0:\n person.append(i) # 加入新的 B-PER\n elif pre_labels[i] != 'I-PER' and len(person) != 0:\n persons.append(person) # 临时栈内容放入正式栈\n person = [] # 清空临时栈\n elif pre_labels[i] == 'I-PER' and len(person) != 0:\n person.append(i)\n else: # 极少数情况会有 I-PER 开头的,不理\n pass\n\n # Location\n # 单字情况\n if pre_labels[i] == 'B-LOC' and pre_labels[i+1] != 'I-LOC' and len(location) == 0:\n location.append(i) \n locations.append(location)\n location = [] # 清空\n continue\n \n # 非单字\n # 如果前面有连着的 LOC 实体\n \n if pre_labels[i] == 'B-LOC' and pre_labels[i+1] == 'I-LOC' and len(location) != 0:\n locations.append(location)\n location = [] # 清空栈\n location.append(i) # 加入新的 B-LOC\n \n # 如果前面没有连着的 B-LOC 实体\n elif pre_labels[i] == 'B-LOC' and pre_labels[i+1] == 'I-LOC' and len(location) == 0:\n location.append(i) # 加入新的 B-LOC\n elif pre_labels[i] == 'I-LOC' and len(location) != 0:\n location.append(i)\n # 结尾\n elif pre_labels[i] != 'I-LOC' and len(location) != 0:\n locations.append(location) # 临时栈内容放入正式栈\n location = [] # 清空临时栈\n else: # 极少数情况会有 I-LOC 开头的,不理\n pass\n \nprint(persons)\nprint(locations)",
"[[1, 2, 3]]\n[[9, 10, 11], [12, 13, 14], [15, 16, 17, 18]]\n"
],
[
"# 从文字中提取\n# 人物\nNER_PER = []\nfor word_idx in persons:\n ONE_PER = []\n for letter_idx in word_idx: \n ONE_PER.append(test_sententce[letter_idx - 1])\n NER_PER.append(ONE_PER)\n\nNER_PER_COMBINE = []\nfor w in NER_PER:\n PER = \"\".join(w)\n NER_PER_COMBINE.append(PER)\n \n# 地点\n\nNER_LOC = []\nfor word_idx in locations:\n ONE_LOC = []\n for letter_idx in word_idx: \n # print(letter_idx)\n # print(test_sententce[letter_idx])\n ONE_LOC.append(test_sententce[letter_idx - 1])\n NER_LOC.append(ONE_LOC)\n\nNER_LOC_COMBINE = []\nfor w in NER_LOC:\n LOC = \"\".join(w)\n NER_LOC_COMBINE.append(LOC)\n\n# 组织",
"_____no_output_____"
],
[
"print(f\"当前句子:{test_sententce}\\n\")\nprint(f\" 人物:{NER_PER_COMBINE}\\n\")\nprint(f\" 地点:{NER_LOC_COMBINE}\\n\")",
"当前句子:史源源的房子租在滨江区南环路税友大厦附近。\n\n 人物:['史源源']\n\n 地点:['滨江区', '南环路', '税友大厦']\n\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc80d6dc46c9e586d2b1f990aebaee51dd655dd
| 300,923 |
ipynb
|
Jupyter Notebook
|
cloudseg/notebooks/colab_training.ipynb
|
elrichgro/irccam-pmodwrc
|
3377233c1a012ff087c22805e3b35c802564f325
|
[
"MIT"
] | 1 |
2021-12-16T22:55:24.000Z
|
2021-12-16T22:55:24.000Z
|
cloudseg/notebooks/colab_training.ipynb
|
elrichgro/irccam-pmodwrc
|
3377233c1a012ff087c22805e3b35c802564f325
|
[
"MIT"
] | 13 |
2020-12-11T16:17:03.000Z
|
2021-03-18T14:12:56.000Z
|
cloudseg/notebooks/colab_training.ipynb
|
elrichgro/irccam-pmodwrc
|
3377233c1a012ff087c22805e3b35c802564f325
|
[
"MIT"
] | 1 |
2022-01-23T20:50:39.000Z
|
2022-01-23T20:50:39.000Z
| 90.475947 | 468 | 0.583917 |
[
[
[
"<a href=\"https://colab.research.google.com/github/elrichgro/irccam-pmodwrc/blob/main/cloudseg/notebooks/colab_training.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"## Get code, mount drive",
"_____no_output_____"
]
],
[
[
"# Get code\n%cd /content\n!rm -rf irccam-pmodwrc/\n!git clone https://github.com/elrichgro/irccam-pmodwrc.git",
"/content\nCloning into 'irccam-pmodwrc'...\nremote: Enumerating objects: 387, done.\u001b[K\nremote: Counting objects: 100% (387/387), done.\u001b[K\nremote: Compressing objects: 100% (267/267), done.\u001b[K\nremote: Total 574 (delta 246), reused 241 (delta 116), pack-reused 187\u001b[K\nReceiving objects: 100% (574/574), 102.01 MiB | 16.57 MiB/s, done.\nResolving deltas: 100% (334/334), done.\n"
],
[
"# Mount data\nfrom google.colab import drive\ndrive.mount('/content/drive', force_remount=True)",
"Mounted at /content/drive\n"
]
],
[
[
"## Dependencies and data copy",
"_____no_output_____"
]
],
[
[
"# Install dependencies\n# %cd /content/irccam-pmodwrc/\n# !pip install test-tube\n# !pip install -r requirements.txt\nrequirements = \"\"\"\ntorch\ntorchvision\ntqdm\nh5py\njupyterlab\nopencv-contrib-python\nopencv-python\npytorch-lightning==1.0.5\nfuture==0.17.1\nscipy\nscikit-learn\nastral\ntest-tube\n\"\"\"\n\nwith open('requirements.txt', 'w') as f:\n f.write(requirements)\n\n!pip install -r requirements.txt",
"_____no_output_____"
],
[
"## Copy data\n!rm -rf /content/data\n!mkdir /content/data\n!time cp /content/drive/My\\ Drive/dsl/datasets/main_single_label/*.h5 /content/data\n!time cp /content/drive/My\\ Drive/dsl/datasets/main_single_label/*.txt /content/data",
"\nreal\t6m3.175s\nuser\t0m0.127s\nsys\t0m18.984s\n\nreal\t0m1.093s\nuser\t0m0.002s\nsys\t0m0.005s\n"
]
],
[
[
"## Train model",
"_____no_output_____"
]
],
[
[
"# Train model\n%cd /content/irccam-pmodwrc\nimport json \nconfig = {\n \"batch_size\": 4,\n \"batch_size_val\": 8,\n \"num_epochs\": 16,\n \"model_name\": \"unet\",\n \"dataset_root\": r\"/content/data\",\n \"learning_rate\": 0.01,\n \"experiment_name\": \"colab_test\",\n \"dataset_class\": \"hdf5\",\n \"log_dir\": \"/content/drive/My Drive/dsl/training_logs\",\n \"use_clear_sky\": True\n}\nwith open(\"config.json\", \"w\") as f:\n json.dump(config, f)\n\n!PYTHONPATH=$PYTHONPATH:/content/irccam-pmodwrc python cloudseg/training/train.py -c config.json",
"/content/irccam-pmodwrc\n/content/irccam-pmodwrc/cloudseg/datasets/cloud_dataset.py:105: H5pyDeprecationWarning: The default file mode will change to 'r' (read-only) in h5py 3.0. To suppress this warning, pass the mode you need to h5py.File(), or set the global default h5.get_config().default_file_mode, or set the environment variable H5PY_DEFAULT_READONLY=1. Available modes are: 'r', 'r+', 'w', 'w-'/'x', 'a'. See the docs for details.\n with h5py.File(file_path) as h5_file:\nGPU available: True, used: True\nTPU available: False, using: 0 TPU cores\nLOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]\n2020-12-10 15:39:28.839668: I tensorflow/stream_executor/platform/default/dso_loader.cc:48] Successfully opened dynamic library libcudart.so.10.1\n\n | Name | Type | Params\n--------------------------------------------------------\n0 | model | UNet | 17 M \n1 | cross_entropy_loss | CrossEntropyLoss | 0 \nValidation sanity check: 0it [00:00, ?it/s]/content/irccam-pmodwrc/cloudseg/datasets/cloud_dataset.py:118: H5pyDeprecationWarning: The default file mode will change to 'r' (read-only) in h5py 3.0. To suppress this warning, pass the mode you need to h5py.File(), or set the global default h5.get_config().default_file_mode, or set the environment variable H5PY_DEFAULT_READONLY=1. Available modes are: 'r', 'r+', 'w', 'w-'/'x', 'a'. See the docs for details.\n with h5py.File(self.files[file_index]) as h5_file:\nEpoch 0: 87% 1207/1395 [04:37<00:43, 4.35it/s, loss=0.380, v_num=0]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 0: 87% 1208/1395 [04:37<00:42, 4.35it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1209/1395 [04:37<00:42, 4.35it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1210/1395 [04:37<00:42, 4.35it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1211/1395 [04:38<00:42, 4.35it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1212/1395 [04:38<00:42, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1213/1395 [04:38<00:41, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1214/1395 [04:38<00:41, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1215/1395 [04:38<00:41, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1216/1395 [04:39<00:41, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1217/1395 [04:39<00:40, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1218/1395 [04:39<00:40, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1219/1395 [04:39<00:40, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 87% 1220/1395 [04:39<00:40, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1221/1395 [04:40<00:39, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1222/1395 [04:40<00:39, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1223/1395 [04:40<00:39, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1224/1395 [04:40<00:39, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1225/1395 [04:40<00:38, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1226/1395 [04:41<00:38, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1227/1395 [04:41<00:38, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1228/1395 [04:41<00:38, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1229/1395 [04:41<00:38, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1230/1395 [04:41<00:37, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1231/1395 [04:42<00:37, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1232/1395 [04:42<00:37, 4.36it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1233/1395 [04:42<00:37, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 88% 1234/1395 [04:42<00:36, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1235/1395 [04:42<00:36, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1236/1395 [04:43<00:36, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1237/1395 [04:43<00:36, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1238/1395 [04:43<00:35, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1239/1395 [04:43<00:35, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1240/1395 [04:43<00:35, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1241/1395 [04:44<00:35, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1242/1395 [04:44<00:35, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1243/1395 [04:44<00:34, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1244/1395 [04:44<00:34, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1245/1395 [04:44<00:34, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1246/1395 [04:45<00:34, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1247/1395 [04:45<00:33, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 89% 1248/1395 [04:45<00:33, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1249/1395 [04:45<00:33, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1250/1395 [04:45<00:33, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1251/1395 [04:45<00:32, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1252/1395 [04:46<00:32, 4.37it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1253/1395 [04:46<00:32, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1254/1395 [04:46<00:32, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1255/1395 [04:46<00:31, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1256/1395 [04:46<00:31, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1257/1395 [04:47<00:31, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1258/1395 [04:47<00:31, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1259/1395 [04:47<00:31, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1260/1395 [04:47<00:30, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1261/1395 [04:47<00:30, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 90% 1262/1395 [04:48<00:30, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1263/1395 [04:48<00:30, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1264/1395 [04:48<00:29, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1265/1395 [04:48<00:29, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1266/1395 [04:48<00:29, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1267/1395 [04:49<00:29, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1268/1395 [04:49<00:28, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1269/1395 [04:49<00:28, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1270/1395 [04:49<00:28, 4.38it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1271/1395 [04:49<00:28, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1272/1395 [04:50<00:28, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1273/1395 [04:50<00:27, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1274/1395 [04:50<00:27, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1275/1395 [04:50<00:27, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 91% 1276/1395 [04:50<00:27, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1277/1395 [04:50<00:26, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1278/1395 [04:51<00:26, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1279/1395 [04:51<00:26, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1280/1395 [04:51<00:26, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1281/1395 [04:51<00:25, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1282/1395 [04:51<00:25, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1283/1395 [04:52<00:25, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1284/1395 [04:52<00:25, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1285/1395 [04:52<00:25, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1286/1395 [04:52<00:24, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1287/1395 [04:52<00:24, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1288/1395 [04:53<00:24, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1289/1395 [04:53<00:24, 4.39it/s, loss=0.380, v_num=0]\nEpoch 0: 92% 1290/1395 [04:53<00:23, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1291/1395 [04:53<00:23, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1292/1395 [04:53<00:23, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1293/1395 [04:54<00:23, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1294/1395 [04:54<00:22, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1295/1395 [04:54<00:22, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1296/1395 [04:54<00:22, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1297/1395 [04:54<00:22, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1298/1395 [04:55<00:22, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1299/1395 [04:55<00:21, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1300/1395 [04:55<00:21, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1301/1395 [04:55<00:21, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1302/1395 [04:55<00:21, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1303/1395 [04:56<00:20, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 93% 1304/1395 [04:56<00:20, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1305/1395 [04:56<00:20, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1306/1395 [04:56<00:20, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1307/1395 [04:56<00:19, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1308/1395 [04:56<00:19, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1309/1395 [04:57<00:19, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1310/1395 [04:57<00:19, 4.40it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1311/1395 [04:57<00:19, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1312/1395 [04:57<00:18, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1313/1395 [04:57<00:18, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1314/1395 [04:58<00:18, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1315/1395 [04:58<00:18, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1316/1395 [04:58<00:17, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1317/1395 [04:58<00:17, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 94% 1318/1395 [04:58<00:17, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1319/1395 [04:59<00:17, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1320/1395 [04:59<00:17, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1321/1395 [04:59<00:16, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1322/1395 [04:59<00:16, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1323/1395 [04:59<00:16, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1324/1395 [05:00<00:16, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1325/1395 [05:00<00:15, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1326/1395 [05:00<00:15, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1327/1395 [05:00<00:15, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1328/1395 [05:00<00:15, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1329/1395 [05:01<00:14, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1330/1395 [05:01<00:14, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1331/1395 [05:01<00:14, 4.41it/s, loss=0.380, v_num=0]\nEpoch 0: 95% 1332/1395 [05:01<00:14, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1333/1395 [05:01<00:14, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1334/1395 [05:02<00:13, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1335/1395 [05:02<00:13, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1336/1395 [05:02<00:13, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1337/1395 [05:02<00:13, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1338/1395 [05:02<00:12, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1339/1395 [05:03<00:12, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1340/1395 [05:03<00:12, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1341/1395 [05:03<00:12, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1342/1395 [05:03<00:11, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1343/1395 [05:03<00:11, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1344/1395 [05:03<00:11, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1345/1395 [05:04<00:11, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 96% 1346/1395 [05:04<00:11, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1347/1395 [05:04<00:10, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1348/1395 [05:04<00:10, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1349/1395 [05:04<00:10, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1350/1395 [05:05<00:10, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1351/1395 [05:05<00:09, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1352/1395 [05:05<00:09, 4.42it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1353/1395 [05:05<00:09, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1354/1395 [05:05<00:09, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1355/1395 [05:06<00:09, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1356/1395 [05:06<00:08, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1357/1395 [05:06<00:08, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1358/1395 [05:06<00:08, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1359/1395 [05:06<00:08, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 97% 1360/1395 [05:07<00:07, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1361/1395 [05:07<00:07, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1362/1395 [05:07<00:07, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1363/1395 [05:07<00:07, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1364/1395 [05:07<00:06, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1365/1395 [05:08<00:06, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1366/1395 [05:08<00:06, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1367/1395 [05:08<00:06, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1368/1395 [05:08<00:06, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1369/1395 [05:08<00:05, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1370/1395 [05:09<00:05, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1371/1395 [05:09<00:05, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1372/1395 [05:09<00:05, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1373/1395 [05:09<00:04, 4.43it/s, loss=0.380, v_num=0]\nEpoch 0: 98% 1374/1395 [05:09<00:04, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1375/1395 [05:09<00:04, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1376/1395 [05:10<00:04, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1377/1395 [05:10<00:04, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1378/1395 [05:10<00:03, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1379/1395 [05:10<00:03, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1380/1395 [05:10<00:03, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1381/1395 [05:11<00:03, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1382/1395 [05:11<00:02, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1383/1395 [05:11<00:02, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1384/1395 [05:11<00:02, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1385/1395 [05:11<00:02, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1386/1395 [05:12<00:02, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1387/1395 [05:12<00:01, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 99% 1388/1395 [05:12<00:01, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 100% 1389/1395 [05:12<00:01, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 100% 1390/1395 [05:12<00:01, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 100% 1391/1395 [05:13<00:00, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 100% 1392/1395 [05:13<00:00, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 100% 1393/1395 [05:13<00:00, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 100% 1394/1395 [05:13<00:00, 4.44it/s, loss=0.380, v_num=0]\nEpoch 0: 100% 1395/1395 [05:13<00:00, 4.44it/s, loss=0.380, v_num=0]Epoch 0: val_iou reached 0.56132 (best 0.56132), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=00-val_iou=0.56.ckpt as top 1\nEpoch 0: 100% 1395/1395 [05:16<00:00, 4.40it/s, loss=0.380, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1207/1395 [04:50<00:45, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 1: 87% 1208/1395 [04:50<00:44, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1209/1395 [04:50<00:44, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1210/1395 [04:50<00:44, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1211/1395 [04:51<00:44, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1212/1395 [04:51<00:43, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1213/1395 [04:51<00:43, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1214/1395 [04:51<00:43, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1215/1395 [04:51<00:43, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1216/1395 [04:52<00:42, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1217/1395 [04:52<00:42, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1218/1395 [04:52<00:42, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1219/1395 [04:52<00:42, 4.16it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 87% 1220/1395 [04:52<00:42, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1221/1395 [04:53<00:41, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1222/1395 [04:53<00:41, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1223/1395 [04:53<00:41, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1224/1395 [04:53<00:41, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1225/1395 [04:53<00:40, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1226/1395 [04:54<00:40, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1227/1395 [04:54<00:40, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1228/1395 [04:54<00:40, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1229/1395 [04:54<00:39, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1230/1395 [04:54<00:39, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1231/1395 [04:55<00:39, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1232/1395 [04:55<00:39, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1233/1395 [04:55<00:38, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 88% 1234/1395 [04:55<00:38, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1235/1395 [04:55<00:38, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1236/1395 [04:56<00:38, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1237/1395 [04:56<00:37, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1238/1395 [04:56<00:37, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1239/1395 [04:56<00:37, 4.17it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1240/1395 [04:57<00:37, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1241/1395 [04:57<00:36, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1242/1395 [04:57<00:36, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1243/1395 [04:57<00:36, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1244/1395 [04:57<00:36, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1245/1395 [04:58<00:35, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1246/1395 [04:58<00:35, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1247/1395 [04:58<00:35, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 89% 1248/1395 [04:58<00:35, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1249/1395 [04:58<00:34, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1250/1395 [04:59<00:34, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1251/1395 [04:59<00:34, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1252/1395 [04:59<00:34, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1253/1395 [04:59<00:33, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1254/1395 [04:59<00:33, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1255/1395 [05:00<00:33, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1256/1395 [05:00<00:33, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1257/1395 [05:00<00:32, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1258/1395 [05:00<00:32, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1259/1395 [05:00<00:32, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1260/1395 [05:01<00:32, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1261/1395 [05:01<00:32, 4.18it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 90% 1262/1395 [05:01<00:31, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1263/1395 [05:01<00:31, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1264/1395 [05:01<00:31, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1265/1395 [05:02<00:31, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1266/1395 [05:02<00:30, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1267/1395 [05:02<00:30, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1268/1395 [05:02<00:30, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1269/1395 [05:02<00:30, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1270/1395 [05:03<00:29, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1271/1395 [05:03<00:29, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1272/1395 [05:03<00:29, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1273/1395 [05:03<00:29, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1274/1395 [05:03<00:28, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1275/1395 [05:04<00:28, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 91% 1276/1395 [05:04<00:28, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1277/1395 [05:04<00:28, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1278/1395 [05:04<00:27, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1279/1395 [05:04<00:27, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1280/1395 [05:05<00:27, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1281/1395 [05:05<00:27, 4.19it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1282/1395 [05:05<00:26, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1283/1395 [05:05<00:26, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1284/1395 [05:05<00:26, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1285/1395 [05:06<00:26, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1286/1395 [05:06<00:25, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1287/1395 [05:06<00:25, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1288/1395 [05:06<00:25, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1289/1395 [05:06<00:25, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 92% 1290/1395 [05:07<00:25, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1291/1395 [05:07<00:24, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1292/1395 [05:07<00:24, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1293/1395 [05:07<00:24, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1294/1395 [05:07<00:24, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1295/1395 [05:08<00:23, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1296/1395 [05:08<00:23, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1297/1395 [05:08<00:23, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1298/1395 [05:08<00:23, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1299/1395 [05:09<00:22, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1300/1395 [05:09<00:22, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1301/1395 [05:09<00:22, 4.20it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1302/1395 [05:09<00:22, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1303/1395 [05:09<00:21, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 93% 1304/1395 [05:10<00:21, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1305/1395 [05:10<00:21, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1306/1395 [05:10<00:21, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1307/1395 [05:10<00:20, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1308/1395 [05:10<00:20, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1309/1395 [05:11<00:20, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1310/1395 [05:11<00:20, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1311/1395 [05:11<00:19, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1312/1395 [05:11<00:19, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1313/1395 [05:11<00:19, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1314/1395 [05:12<00:19, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1315/1395 [05:12<00:18, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1316/1395 [05:12<00:18, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1317/1395 [05:12<00:18, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 94% 1318/1395 [05:12<00:18, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1319/1395 [05:13<00:18, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1320/1395 [05:13<00:17, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1321/1395 [05:13<00:17, 4.21it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1322/1395 [05:13<00:17, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1323/1395 [05:13<00:17, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1324/1395 [05:14<00:16, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1325/1395 [05:14<00:16, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1326/1395 [05:14<00:16, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1327/1395 [05:14<00:16, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1328/1395 [05:14<00:15, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1329/1395 [05:15<00:15, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1330/1395 [05:15<00:15, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1331/1395 [05:15<00:15, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 95% 1332/1395 [05:15<00:14, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1333/1395 [05:15<00:14, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1334/1395 [05:16<00:14, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1335/1395 [05:16<00:14, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1336/1395 [05:16<00:13, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1337/1395 [05:16<00:13, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1338/1395 [05:16<00:13, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1339/1395 [05:17<00:13, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1340/1395 [05:17<00:13, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1341/1395 [05:17<00:12, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1342/1395 [05:17<00:12, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1343/1395 [05:17<00:12, 4.22it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1344/1395 [05:18<00:12, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1345/1395 [05:18<00:11, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 96% 1346/1395 [05:18<00:11, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1347/1395 [05:18<00:11, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1348/1395 [05:18<00:11, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1349/1395 [05:19<00:10, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1350/1395 [05:19<00:10, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1351/1395 [05:19<00:10, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1352/1395 [05:19<00:10, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1353/1395 [05:19<00:09, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1354/1395 [05:20<00:09, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1355/1395 [05:20<00:09, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1356/1395 [05:20<00:09, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1357/1395 [05:20<00:08, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1358/1395 [05:20<00:08, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1359/1395 [05:21<00:08, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 97% 1360/1395 [05:21<00:08, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1361/1395 [05:21<00:08, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1362/1395 [05:21<00:07, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1363/1395 [05:21<00:07, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1364/1395 [05:22<00:07, 4.23it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1365/1395 [05:22<00:07, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1366/1395 [05:22<00:06, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1367/1395 [05:22<00:06, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1368/1395 [05:22<00:06, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1369/1395 [05:23<00:06, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1370/1395 [05:23<00:05, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1371/1395 [05:23<00:05, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1372/1395 [05:23<00:05, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1373/1395 [05:23<00:05, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 98% 1374/1395 [05:24<00:04, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1375/1395 [05:24<00:04, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1376/1395 [05:24<00:04, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1377/1395 [05:24<00:04, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1378/1395 [05:24<00:04, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1379/1395 [05:25<00:03, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1380/1395 [05:25<00:03, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1381/1395 [05:25<00:03, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1382/1395 [05:25<00:03, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1383/1395 [05:25<00:02, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1384/1395 [05:26<00:02, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1385/1395 [05:26<00:02, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1386/1395 [05:26<00:02, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1387/1395 [05:26<00:01, 4.24it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 99% 1388/1395 [05:26<00:01, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 100% 1389/1395 [05:27<00:01, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 100% 1390/1395 [05:27<00:01, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 100% 1391/1395 [05:27<00:00, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 100% 1392/1395 [05:27<00:00, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 100% 1393/1395 [05:27<00:00, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 100% 1394/1395 [05:28<00:00, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]\nEpoch 1: 100% 1395/1395 [05:28<00:00, 4.25it/s, loss=0.342, v_num=0, val_iou=0.561]Epoch 1: val_iou reached 0.57565 (best 0.57565), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=01-val_iou=0.58.ckpt as top 1\nEpoch 1: 100% 1395/1395 [05:31<00:00, 4.21it/s, loss=0.342, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1207/1395 [04:45<00:44, 4.23it/s, loss=0.265, v_num=0, val_iou=0.576]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 2: 87% 1208/1395 [04:45<00:44, 4.23it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1209/1395 [04:45<00:43, 4.23it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1210/1395 [04:45<00:43, 4.23it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1211/1395 [04:46<00:43, 4.23it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1212/1395 [04:46<00:43, 4.23it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1213/1395 [04:46<00:42, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1214/1395 [04:46<00:42, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1215/1395 [04:46<00:42, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1216/1395 [04:46<00:42, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1217/1395 [04:47<00:42, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1218/1395 [04:47<00:41, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1219/1395 [04:47<00:41, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 87% 1220/1395 [04:47<00:41, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1221/1395 [04:47<00:41, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1222/1395 [04:48<00:40, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1223/1395 [04:48<00:40, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1224/1395 [04:48<00:40, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1225/1395 [04:48<00:40, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1226/1395 [04:48<00:39, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1227/1395 [04:49<00:39, 4.24it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1228/1395 [04:49<00:39, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1229/1395 [04:49<00:39, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1230/1395 [04:49<00:38, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1231/1395 [04:49<00:38, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1232/1395 [04:50<00:38, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1233/1395 [04:50<00:38, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 88% 1234/1395 [04:50<00:37, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1235/1395 [04:50<00:37, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1236/1395 [04:50<00:37, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1237/1395 [04:51<00:37, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1238/1395 [04:51<00:36, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1239/1395 [04:51<00:36, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1240/1395 [04:51<00:36, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1241/1395 [04:51<00:36, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1242/1395 [04:51<00:35, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1243/1395 [04:52<00:35, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1244/1395 [04:52<00:35, 4.25it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1245/1395 [04:52<00:35, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1246/1395 [04:52<00:35, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1247/1395 [04:52<00:34, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 89% 1248/1395 [04:53<00:34, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1249/1395 [04:53<00:34, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1250/1395 [04:53<00:34, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1251/1395 [04:53<00:33, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1252/1395 [04:53<00:33, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1253/1395 [04:54<00:33, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1254/1395 [04:54<00:33, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1255/1395 [04:54<00:32, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1256/1395 [04:54<00:32, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1257/1395 [04:54<00:32, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1258/1395 [04:55<00:32, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1259/1395 [04:55<00:31, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1260/1395 [04:55<00:31, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1261/1395 [04:55<00:31, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 90% 1262/1395 [04:55<00:31, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1263/1395 [04:56<00:30, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1264/1395 [04:56<00:30, 4.26it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1265/1395 [04:56<00:30, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1266/1395 [04:56<00:30, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1267/1395 [04:56<00:30, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1268/1395 [04:57<00:29, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1269/1395 [04:57<00:29, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1270/1395 [04:57<00:29, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1271/1395 [04:57<00:29, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1272/1395 [04:57<00:28, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1273/1395 [04:58<00:28, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1274/1395 [04:58<00:28, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1275/1395 [04:58<00:28, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 91% 1276/1395 [04:58<00:27, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1277/1395 [04:58<00:27, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1278/1395 [04:59<00:27, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1279/1395 [04:59<00:27, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1280/1395 [04:59<00:26, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1281/1395 [04:59<00:26, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1282/1395 [04:59<00:26, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1283/1395 [05:00<00:26, 4.27it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1284/1395 [05:00<00:25, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1285/1395 [05:00<00:25, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1286/1395 [05:00<00:25, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1287/1395 [05:00<00:25, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1288/1395 [05:01<00:25, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1289/1395 [05:01<00:24, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 92% 1290/1395 [05:01<00:24, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1291/1395 [05:01<00:24, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1292/1395 [05:01<00:24, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1293/1395 [05:02<00:23, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1294/1395 [05:02<00:23, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1295/1395 [05:02<00:23, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1296/1395 [05:02<00:23, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1297/1395 [05:02<00:22, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1298/1395 [05:03<00:22, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1299/1395 [05:03<00:22, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1300/1395 [05:03<00:22, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1301/1395 [05:03<00:21, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1302/1395 [05:03<00:21, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1303/1395 [05:04<00:21, 4.28it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 93% 1304/1395 [05:04<00:21, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1305/1395 [05:04<00:21, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1306/1395 [05:04<00:20, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1307/1395 [05:04<00:20, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1308/1395 [05:05<00:20, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1309/1395 [05:05<00:20, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1310/1395 [05:05<00:19, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1311/1395 [05:05<00:19, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1312/1395 [05:05<00:19, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1313/1395 [05:06<00:19, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1314/1395 [05:06<00:18, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1315/1395 [05:06<00:18, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1316/1395 [05:06<00:18, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1317/1395 [05:06<00:18, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 94% 1318/1395 [05:07<00:17, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1319/1395 [05:07<00:17, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1320/1395 [05:07<00:17, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1321/1395 [05:07<00:17, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1322/1395 [05:07<00:17, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1323/1395 [05:08<00:16, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1324/1395 [05:08<00:16, 4.29it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1325/1395 [05:08<00:16, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1326/1395 [05:08<00:16, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1327/1395 [05:08<00:15, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1328/1395 [05:09<00:15, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1329/1395 [05:09<00:15, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1330/1395 [05:09<00:15, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1331/1395 [05:09<00:14, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 95% 1332/1395 [05:09<00:14, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1333/1395 [05:10<00:14, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1334/1395 [05:10<00:14, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1335/1395 [05:10<00:13, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1336/1395 [05:10<00:13, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1337/1395 [05:10<00:13, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1338/1395 [05:11<00:13, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1339/1395 [05:11<00:13, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1340/1395 [05:11<00:12, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1341/1395 [05:11<00:12, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1342/1395 [05:11<00:12, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1343/1395 [05:12<00:12, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1344/1395 [05:12<00:11, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1345/1395 [05:12<00:11, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 96% 1346/1395 [05:12<00:11, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1347/1395 [05:12<00:11, 4.30it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1348/1395 [05:13<00:10, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1349/1395 [05:13<00:10, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1350/1395 [05:13<00:10, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1351/1395 [05:13<00:10, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1352/1395 [05:13<00:09, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1353/1395 [05:14<00:09, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1354/1395 [05:14<00:09, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1355/1395 [05:14<00:09, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1356/1395 [05:14<00:09, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1357/1395 [05:14<00:08, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1358/1395 [05:15<00:08, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1359/1395 [05:15<00:08, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 97% 1360/1395 [05:15<00:08, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1361/1395 [05:15<00:07, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1362/1395 [05:15<00:07, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1363/1395 [05:16<00:07, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1364/1395 [05:16<00:07, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1365/1395 [05:16<00:06, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1366/1395 [05:16<00:06, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1367/1395 [05:16<00:06, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1368/1395 [05:17<00:06, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1369/1395 [05:17<00:06, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1370/1395 [05:17<00:05, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1371/1395 [05:17<00:05, 4.31it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1372/1395 [05:17<00:05, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1373/1395 [05:18<00:05, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 98% 1374/1395 [05:18<00:04, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1375/1395 [05:18<00:04, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1376/1395 [05:18<00:04, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1377/1395 [05:18<00:04, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1378/1395 [05:19<00:03, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1379/1395 [05:19<00:03, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1380/1395 [05:19<00:03, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1381/1395 [05:19<00:03, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1382/1395 [05:19<00:03, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1383/1395 [05:20<00:02, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1384/1395 [05:20<00:02, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1385/1395 [05:20<00:02, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1386/1395 [05:20<00:02, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1387/1395 [05:21<00:01, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 99% 1388/1395 [05:21<00:01, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 100% 1389/1395 [05:21<00:01, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 100% 1390/1395 [05:21<00:01, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 100% 1391/1395 [05:21<00:00, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 100% 1392/1395 [05:22<00:00, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 100% 1393/1395 [05:22<00:00, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 100% 1394/1395 [05:22<00:00, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]\nEpoch 2: 100% 1395/1395 [05:22<00:00, 4.32it/s, loss=0.265, v_num=0, val_iou=0.576]Epoch 2: val_iou reached 0.66263 (best 0.66263), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=02-val_iou=0.66.ckpt as top 1\nEpoch 2: 100% 1395/1395 [05:25<00:00, 4.28it/s, loss=0.265, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1207/1395 [04:38<00:43, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 3: 87% 1208/1395 [04:38<00:43, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1209/1395 [04:39<00:42, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1210/1395 [04:39<00:42, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1211/1395 [04:39<00:42, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1212/1395 [04:39<00:42, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1213/1395 [04:39<00:42, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1214/1395 [04:40<00:41, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1215/1395 [04:40<00:41, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1216/1395 [04:40<00:41, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1217/1395 [04:40<00:41, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1218/1395 [04:41<00:40, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1219/1395 [04:41<00:40, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 87% 1220/1395 [04:41<00:40, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1221/1395 [04:41<00:40, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1222/1395 [04:41<00:39, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1223/1395 [04:42<00:39, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1224/1395 [04:42<00:39, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1225/1395 [04:42<00:39, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1226/1395 [04:42<00:38, 4.33it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1227/1395 [04:43<00:38, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1228/1395 [04:43<00:38, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1229/1395 [04:43<00:38, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1230/1395 [04:43<00:38, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1231/1395 [04:43<00:37, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1232/1395 [04:44<00:37, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1233/1395 [04:44<00:37, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 88% 1234/1395 [04:44<00:37, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1235/1395 [04:44<00:36, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1236/1395 [04:44<00:36, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1237/1395 [04:45<00:36, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1238/1395 [04:45<00:36, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1239/1395 [04:45<00:35, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1240/1395 [04:45<00:35, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1241/1395 [04:46<00:35, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1242/1395 [04:46<00:35, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1243/1395 [04:46<00:35, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1244/1395 [04:46<00:34, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1245/1395 [04:46<00:34, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1246/1395 [04:47<00:34, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1247/1395 [04:47<00:34, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 89% 1248/1395 [04:47<00:33, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1249/1395 [04:47<00:33, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1250/1395 [04:47<00:33, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1251/1395 [04:48<00:33, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1252/1395 [04:48<00:32, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1253/1395 [04:48<00:32, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1254/1395 [04:48<00:32, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1255/1395 [04:49<00:32, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1256/1395 [04:49<00:32, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1257/1395 [04:49<00:31, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1258/1395 [04:49<00:31, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1259/1395 [04:49<00:31, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1260/1395 [04:50<00:31, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1261/1395 [04:50<00:30, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 90% 1262/1395 [04:50<00:30, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1263/1395 [04:50<00:30, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1264/1395 [04:50<00:30, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1265/1395 [04:51<00:29, 4.34it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1266/1395 [04:51<00:29, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1267/1395 [04:51<00:29, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1268/1395 [04:51<00:29, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1269/1395 [04:51<00:28, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1270/1395 [04:52<00:28, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1271/1395 [04:52<00:28, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1272/1395 [04:52<00:28, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1273/1395 [04:52<00:28, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1274/1395 [04:53<00:27, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1275/1395 [04:53<00:27, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 91% 1276/1395 [04:53<00:27, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1277/1395 [04:53<00:27, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1278/1395 [04:53<00:26, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1279/1395 [04:54<00:26, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1280/1395 [04:54<00:26, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1281/1395 [04:54<00:26, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1282/1395 [04:54<00:25, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1283/1395 [04:54<00:25, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1284/1395 [04:55<00:25, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1285/1395 [04:55<00:25, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1286/1395 [04:55<00:25, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1287/1395 [04:55<00:24, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1288/1395 [04:55<00:24, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1289/1395 [04:56<00:24, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 92% 1290/1395 [04:56<00:24, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1291/1395 [04:56<00:23, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1292/1395 [04:56<00:23, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1293/1395 [04:57<00:23, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1294/1395 [04:57<00:23, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1295/1395 [04:57<00:22, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1296/1395 [04:57<00:22, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1297/1395 [04:57<00:22, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1298/1395 [04:58<00:22, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1299/1395 [04:58<00:22, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1300/1395 [04:58<00:21, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1301/1395 [04:58<00:21, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1302/1395 [04:58<00:21, 4.35it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1303/1395 [04:59<00:21, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 93% 1304/1395 [04:59<00:20, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1305/1395 [04:59<00:20, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1306/1395 [04:59<00:20, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1307/1395 [05:00<00:20, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1308/1395 [05:00<00:19, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1309/1395 [05:00<00:19, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1310/1395 [05:00<00:19, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1311/1395 [05:00<00:19, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1312/1395 [05:01<00:19, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1313/1395 [05:01<00:18, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1314/1395 [05:01<00:18, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1315/1395 [05:01<00:18, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1316/1395 [05:01<00:18, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1317/1395 [05:02<00:17, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 94% 1318/1395 [05:02<00:17, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1319/1395 [05:02<00:17, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1320/1395 [05:02<00:17, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1321/1395 [05:03<00:16, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1322/1395 [05:03<00:16, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1323/1395 [05:03<00:16, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1324/1395 [05:03<00:16, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1325/1395 [05:03<00:16, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1326/1395 [05:04<00:15, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1327/1395 [05:04<00:15, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1328/1395 [05:04<00:15, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1329/1395 [05:04<00:15, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1330/1395 [05:05<00:14, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1331/1395 [05:05<00:14, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 95% 1332/1395 [05:05<00:14, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1333/1395 [05:05<00:14, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1334/1395 [05:05<00:13, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1335/1395 [05:06<00:13, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1336/1395 [05:06<00:13, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1337/1395 [05:06<00:13, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1338/1395 [05:06<00:13, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1339/1395 [05:07<00:12, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1340/1395 [05:07<00:12, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1341/1395 [05:07<00:12, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1342/1395 [05:07<00:12, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1343/1395 [05:07<00:11, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1344/1395 [05:08<00:11, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1345/1395 [05:08<00:11, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 96% 1346/1395 [05:08<00:11, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1347/1395 [05:08<00:11, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1348/1395 [05:09<00:10, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1349/1395 [05:09<00:10, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1350/1395 [05:09<00:10, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1351/1395 [05:09<00:10, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1352/1395 [05:09<00:09, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1353/1395 [05:10<00:09, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1354/1395 [05:10<00:09, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1355/1395 [05:10<00:09, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1356/1395 [05:10<00:08, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1357/1395 [05:11<00:08, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1358/1395 [05:11<00:08, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1359/1395 [05:11<00:08, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 97% 1360/1395 [05:11<00:08, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1361/1395 [05:11<00:07, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1362/1395 [05:12<00:07, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1363/1395 [05:12<00:07, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1364/1395 [05:12<00:07, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1365/1395 [05:12<00:06, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1366/1395 [05:13<00:06, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1367/1395 [05:13<00:06, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1368/1395 [05:13<00:06, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1369/1395 [05:13<00:05, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1370/1395 [05:13<00:05, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1371/1395 [05:14<00:05, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1372/1395 [05:14<00:05, 4.36it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1373/1395 [05:14<00:05, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 98% 1374/1395 [05:14<00:04, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1375/1395 [05:14<00:04, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1376/1395 [05:15<00:04, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1377/1395 [05:15<00:04, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1378/1395 [05:15<00:03, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1379/1395 [05:15<00:03, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1380/1395 [05:16<00:03, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1381/1395 [05:16<00:03, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1382/1395 [05:16<00:02, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1383/1395 [05:16<00:02, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1384/1395 [05:16<00:02, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1385/1395 [05:17<00:02, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1386/1395 [05:17<00:02, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1387/1395 [05:17<00:01, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 99% 1388/1395 [05:17<00:01, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 100% 1389/1395 [05:17<00:01, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 100% 1390/1395 [05:18<00:01, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 100% 1391/1395 [05:18<00:00, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 100% 1392/1395 [05:18<00:00, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 100% 1393/1395 [05:18<00:00, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 100% 1394/1395 [05:19<00:00, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]\nEpoch 3: 100% 1395/1395 [05:19<00:00, 4.37it/s, loss=0.260, v_num=0, val_iou=0.663]Epoch 3: val_iou was not in top 1\nEpoch 3: 100% 1395/1395 [05:20<00:00, 4.35it/s, loss=0.260, v_num=0, val_iou=0.63] \nEpoch 4: 87% 1207/1395 [04:30<00:42, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 4: 87% 1208/1395 [04:31<00:41, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1209/1395 [04:31<00:41, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1210/1395 [04:31<00:41, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1211/1395 [04:31<00:41, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1212/1395 [04:32<00:41, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1213/1395 [04:32<00:40, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1214/1395 [04:32<00:40, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1215/1395 [04:32<00:40, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1216/1395 [04:32<00:40, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1217/1395 [04:33<00:39, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1218/1395 [04:33<00:39, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1219/1395 [04:33<00:39, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 87% 1220/1395 [04:33<00:39, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1221/1395 [04:33<00:39, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1222/1395 [04:34<00:38, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1223/1395 [04:34<00:38, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1224/1395 [04:34<00:38, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1225/1395 [04:34<00:38, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1226/1395 [04:35<00:37, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1227/1395 [04:35<00:37, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1228/1395 [04:35<00:37, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1229/1395 [04:35<00:37, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1230/1395 [04:35<00:37, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1231/1395 [04:36<00:36, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1232/1395 [04:36<00:36, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1233/1395 [04:36<00:36, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 88% 1234/1395 [04:36<00:36, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1235/1395 [04:36<00:35, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1236/1395 [04:37<00:35, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1237/1395 [04:37<00:35, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1238/1395 [04:37<00:35, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1239/1395 [04:37<00:34, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1240/1395 [04:37<00:34, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1241/1395 [04:38<00:34, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1242/1395 [04:38<00:34, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1243/1395 [04:38<00:34, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1244/1395 [04:38<00:33, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1245/1395 [04:39<00:33, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1246/1395 [04:39<00:33, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1247/1395 [04:39<00:33, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 89% 1248/1395 [04:39<00:32, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1249/1395 [04:39<00:32, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1250/1395 [04:40<00:32, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1251/1395 [04:40<00:32, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1252/1395 [04:40<00:32, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1253/1395 [04:40<00:31, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1254/1395 [04:40<00:31, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1255/1395 [04:41<00:31, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1256/1395 [04:41<00:31, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1257/1395 [04:41<00:30, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1258/1395 [04:41<00:30, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1259/1395 [04:42<00:30, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1260/1395 [04:42<00:30, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1261/1395 [04:42<00:30, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 90% 1262/1395 [04:42<00:29, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1263/1395 [04:42<00:29, 4.46it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1264/1395 [04:43<00:29, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1265/1395 [04:43<00:29, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1266/1395 [04:43<00:28, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1267/1395 [04:43<00:28, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1268/1395 [04:43<00:28, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1269/1395 [04:44<00:28, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1270/1395 [04:44<00:27, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1271/1395 [04:44<00:27, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1272/1395 [04:44<00:27, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1273/1395 [04:44<00:27, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1274/1395 [04:45<00:27, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1275/1395 [04:45<00:26, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 91% 1276/1395 [04:45<00:26, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1277/1395 [04:45<00:26, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1278/1395 [04:46<00:26, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1279/1395 [04:46<00:25, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1280/1395 [04:46<00:25, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1281/1395 [04:46<00:25, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1282/1395 [04:46<00:25, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1283/1395 [04:47<00:25, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1284/1395 [04:47<00:24, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1285/1395 [04:47<00:24, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1286/1395 [04:47<00:24, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1287/1395 [04:47<00:24, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1288/1395 [04:48<00:23, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1289/1395 [04:48<00:23, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 92% 1290/1395 [04:48<00:23, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1291/1395 [04:48<00:23, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1292/1395 [04:48<00:23, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1293/1395 [04:49<00:22, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1294/1395 [04:49<00:22, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1295/1395 [04:49<00:22, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1296/1395 [04:49<00:22, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1297/1395 [04:50<00:21, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1298/1395 [04:50<00:21, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1299/1395 [04:50<00:21, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1300/1395 [04:50<00:21, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1301/1395 [04:50<00:21, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1302/1395 [04:51<00:20, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1303/1395 [04:51<00:20, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 93% 1304/1395 [04:51<00:20, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1305/1395 [04:51<00:20, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1306/1395 [04:51<00:19, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1307/1395 [04:52<00:19, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1308/1395 [04:52<00:19, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1309/1395 [04:52<00:19, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1310/1395 [04:52<00:18, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1311/1395 [04:53<00:18, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1312/1395 [04:53<00:18, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1313/1395 [04:53<00:18, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1314/1395 [04:53<00:18, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1315/1395 [04:53<00:17, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1316/1395 [04:54<00:17, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1317/1395 [04:54<00:17, 4.47it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 94% 1318/1395 [04:54<00:17, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1319/1395 [04:54<00:16, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1320/1395 [04:54<00:16, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1321/1395 [04:55<00:16, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1322/1395 [04:55<00:16, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1323/1395 [04:55<00:16, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1324/1395 [04:55<00:15, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1325/1395 [04:56<00:15, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1326/1395 [04:56<00:15, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1327/1395 [04:56<00:15, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1328/1395 [04:56<00:14, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1329/1395 [04:56<00:14, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1330/1395 [04:57<00:14, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1331/1395 [04:57<00:14, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 95% 1332/1395 [04:57<00:14, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1333/1395 [04:57<00:13, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1334/1395 [04:57<00:13, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1335/1395 [04:58<00:13, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1336/1395 [04:58<00:13, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1337/1395 [04:58<00:12, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1338/1395 [04:58<00:12, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1339/1395 [04:58<00:12, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1340/1395 [04:59<00:12, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1341/1395 [04:59<00:12, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1342/1395 [04:59<00:11, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1343/1395 [04:59<00:11, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1344/1395 [05:00<00:11, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1345/1395 [05:00<00:11, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 96% 1346/1395 [05:00<00:10, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1347/1395 [05:00<00:10, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1348/1395 [05:00<00:10, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1349/1395 [05:01<00:10, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1350/1395 [05:01<00:10, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1351/1395 [05:01<00:09, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1352/1395 [05:01<00:09, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1353/1395 [05:01<00:09, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1354/1395 [05:02<00:09, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1355/1395 [05:02<00:08, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1356/1395 [05:02<00:08, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1357/1395 [05:02<00:08, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1358/1395 [05:03<00:08, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1359/1395 [05:03<00:08, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 97% 1360/1395 [05:03<00:07, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1361/1395 [05:03<00:07, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1362/1395 [05:03<00:07, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1363/1395 [05:04<00:07, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1364/1395 [05:04<00:06, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1365/1395 [05:04<00:06, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1366/1395 [05:04<00:06, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1367/1395 [05:04<00:06, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1368/1395 [05:05<00:06, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1369/1395 [05:05<00:05, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1370/1395 [05:05<00:05, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1371/1395 [05:05<00:05, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1372/1395 [05:06<00:05, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1373/1395 [05:06<00:04, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 98% 1374/1395 [05:06<00:04, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1375/1395 [05:06<00:04, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1376/1395 [05:06<00:04, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1377/1395 [05:07<00:04, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1378/1395 [05:07<00:03, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1379/1395 [05:07<00:03, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1380/1395 [05:07<00:03, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1381/1395 [05:07<00:03, 4.48it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1382/1395 [05:08<00:02, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1383/1395 [05:08<00:02, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1384/1395 [05:08<00:02, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1385/1395 [05:08<00:02, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1386/1395 [05:08<00:02, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1387/1395 [05:09<00:01, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 99% 1388/1395 [05:09<00:01, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 100% 1389/1395 [05:09<00:01, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 100% 1390/1395 [05:09<00:01, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 100% 1391/1395 [05:10<00:00, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 100% 1392/1395 [05:10<00:00, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 100% 1393/1395 [05:10<00:00, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 100% 1394/1395 [05:10<00:00, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]\nEpoch 4: 100% 1395/1395 [05:10<00:00, 4.49it/s, loss=0.285, v_num=0, val_iou=0.63]Epoch 4: val_iou was not in top 1\nEpoch 4: 100% 1395/1395 [05:12<00:00, 4.47it/s, loss=0.285, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1207/1395 [04:33<00:42, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 5: 87% 1208/1395 [04:33<00:42, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1209/1395 [04:33<00:42, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1210/1395 [04:34<00:41, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1211/1395 [04:34<00:41, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1212/1395 [04:34<00:41, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1213/1395 [04:34<00:41, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1214/1395 [04:34<00:40, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1215/1395 [04:35<00:40, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1216/1395 [04:35<00:40, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1217/1395 [04:35<00:40, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1218/1395 [04:35<00:40, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1219/1395 [04:35<00:39, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 87% 1220/1395 [04:36<00:39, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1221/1395 [04:36<00:39, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1222/1395 [04:36<00:39, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1223/1395 [04:36<00:38, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1224/1395 [04:37<00:38, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1225/1395 [04:37<00:38, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1226/1395 [04:37<00:38, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1227/1395 [04:37<00:38, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1228/1395 [04:37<00:37, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1229/1395 [04:38<00:37, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1230/1395 [04:38<00:37, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1231/1395 [04:38<00:37, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1232/1395 [04:38<00:36, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1233/1395 [04:38<00:36, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 88% 1234/1395 [04:39<00:36, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1235/1395 [04:39<00:36, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1236/1395 [04:39<00:35, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1237/1395 [04:39<00:35, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1238/1395 [04:40<00:35, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1239/1395 [04:40<00:35, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1240/1395 [04:40<00:35, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1241/1395 [04:40<00:34, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1242/1395 [04:40<00:34, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1243/1395 [04:41<00:34, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1244/1395 [04:41<00:34, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1245/1395 [04:41<00:33, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1246/1395 [04:41<00:33, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1247/1395 [04:41<00:33, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 89% 1248/1395 [04:42<00:33, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1249/1395 [04:42<00:33, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1250/1395 [04:42<00:32, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1251/1395 [04:42<00:32, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1252/1395 [04:43<00:32, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1253/1395 [04:43<00:32, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1254/1395 [04:43<00:31, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1255/1395 [04:43<00:31, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1256/1395 [04:43<00:31, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1257/1395 [04:44<00:31, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1258/1395 [04:44<00:30, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1259/1395 [04:44<00:30, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1260/1395 [04:44<00:30, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1261/1395 [04:44<00:30, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 90% 1262/1395 [04:45<00:30, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1263/1395 [04:45<00:29, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1264/1395 [04:45<00:29, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1265/1395 [04:45<00:29, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1266/1395 [04:46<00:29, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1267/1395 [04:46<00:28, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1268/1395 [04:46<00:28, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1269/1395 [04:46<00:28, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1270/1395 [04:46<00:28, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1271/1395 [04:47<00:28, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1272/1395 [04:47<00:27, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1273/1395 [04:47<00:27, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1274/1395 [04:47<00:27, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1275/1395 [04:48<00:27, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 91% 1276/1395 [04:48<00:26, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1277/1395 [04:48<00:26, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1278/1395 [04:48<00:26, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1279/1395 [04:48<00:26, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1280/1395 [04:49<00:25, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1281/1395 [04:49<00:25, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1282/1395 [04:49<00:25, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1283/1395 [04:49<00:25, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1284/1395 [04:50<00:25, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1285/1395 [04:50<00:24, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1286/1395 [04:50<00:24, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1287/1395 [04:50<00:24, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1288/1395 [04:50<00:24, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1289/1395 [04:51<00:23, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 92% 1290/1395 [04:51<00:23, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1291/1395 [04:51<00:23, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1292/1395 [04:51<00:23, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1293/1395 [04:52<00:23, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1294/1395 [04:52<00:22, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1295/1395 [04:52<00:22, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1296/1395 [04:52<00:22, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1297/1395 [04:53<00:22, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1298/1395 [04:53<00:21, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1299/1395 [04:53<00:21, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1300/1395 [04:53<00:21, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1301/1395 [04:53<00:21, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1302/1395 [04:54<00:21, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1303/1395 [04:54<00:20, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 93% 1304/1395 [04:54<00:20, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1305/1395 [04:54<00:20, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1306/1395 [04:55<00:20, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1307/1395 [04:55<00:19, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1308/1395 [04:55<00:19, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1309/1395 [04:55<00:19, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1310/1395 [04:56<00:19, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1311/1395 [04:56<00:18, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1312/1395 [04:56<00:18, 4.43it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1313/1395 [04:56<00:18, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1314/1395 [04:56<00:18, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1315/1395 [04:57<00:18, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1316/1395 [04:57<00:17, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1317/1395 [04:57<00:17, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 94% 1318/1395 [04:57<00:17, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1319/1395 [04:58<00:17, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1320/1395 [04:58<00:16, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1321/1395 [04:58<00:16, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1322/1395 [04:58<00:16, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1323/1395 [04:59<00:16, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1324/1395 [04:59<00:16, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1325/1395 [04:59<00:15, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1326/1395 [04:59<00:15, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1327/1395 [04:59<00:15, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1328/1395 [05:00<00:15, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1329/1395 [05:00<00:14, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1330/1395 [05:00<00:14, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1331/1395 [05:00<00:14, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 95% 1332/1395 [05:01<00:14, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1333/1395 [05:01<00:14, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1334/1395 [05:01<00:13, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1335/1395 [05:01<00:13, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1336/1395 [05:02<00:13, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1337/1395 [05:02<00:13, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1338/1395 [05:02<00:12, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1339/1395 [05:02<00:12, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1340/1395 [05:02<00:12, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1341/1395 [05:03<00:12, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1342/1395 [05:03<00:11, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1343/1395 [05:03<00:11, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1344/1395 [05:03<00:11, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1345/1395 [05:04<00:11, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 96% 1346/1395 [05:04<00:11, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1347/1395 [05:04<00:10, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1348/1395 [05:04<00:10, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1349/1395 [05:04<00:10, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1350/1395 [05:05<00:10, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1351/1395 [05:05<00:09, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1352/1395 [05:05<00:09, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1353/1395 [05:05<00:09, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1354/1395 [05:06<00:09, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1355/1395 [05:06<00:09, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1356/1395 [05:06<00:08, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1357/1395 [05:06<00:08, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1358/1395 [05:06<00:08, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1359/1395 [05:07<00:08, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 97% 1360/1395 [05:07<00:07, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1361/1395 [05:07<00:07, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1362/1395 [05:07<00:07, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1363/1395 [05:08<00:07, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1364/1395 [05:08<00:07, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1365/1395 [05:08<00:06, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1366/1395 [05:08<00:06, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1367/1395 [05:09<00:06, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1368/1395 [05:09<00:06, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1369/1395 [05:09<00:05, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1370/1395 [05:09<00:05, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1371/1395 [05:09<00:05, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1372/1395 [05:10<00:05, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1373/1395 [05:10<00:04, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 98% 1374/1395 [05:10<00:04, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1375/1395 [05:10<00:04, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1376/1395 [05:11<00:04, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1377/1395 [05:11<00:04, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1378/1395 [05:11<00:03, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1379/1395 [05:11<00:03, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1380/1395 [05:11<00:03, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1381/1395 [05:12<00:03, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1382/1395 [05:12<00:02, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1383/1395 [05:12<00:02, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1384/1395 [05:12<00:02, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1385/1395 [05:13<00:02, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1386/1395 [05:13<00:02, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1387/1395 [05:13<00:01, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 99% 1388/1395 [05:13<00:01, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 100% 1389/1395 [05:13<00:01, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 100% 1390/1395 [05:14<00:01, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 100% 1391/1395 [05:14<00:00, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 100% 1392/1395 [05:14<00:00, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 100% 1393/1395 [05:14<00:00, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 100% 1394/1395 [05:15<00:00, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]\nEpoch 5: 100% 1395/1395 [05:15<00:00, 4.42it/s, loss=0.279, v_num=0, val_iou=0.649]Epoch 5: val_iou reached 0.67269 (best 0.67269), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=05-val_iou=0.67.ckpt as top 1\nEpoch 5: 100% 1395/1395 [05:18<00:00, 4.38it/s, loss=0.279, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1207/1395 [04:35<00:42, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 6: 87% 1208/1395 [04:35<00:42, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1209/1395 [04:35<00:42, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1210/1395 [04:35<00:42, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1211/1395 [04:36<00:41, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1212/1395 [04:36<00:41, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1213/1395 [04:36<00:41, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1214/1395 [04:36<00:41, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1215/1395 [04:36<00:41, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1216/1395 [04:37<00:40, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1217/1395 [04:37<00:40, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1218/1395 [04:37<00:40, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1219/1395 [04:37<00:40, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 87% 1220/1395 [04:38<00:39, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1221/1395 [04:38<00:39, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1222/1395 [04:38<00:39, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1223/1395 [04:38<00:39, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1224/1395 [04:38<00:38, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1225/1395 [04:39<00:38, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1226/1395 [04:39<00:38, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1227/1395 [04:39<00:38, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1228/1395 [04:39<00:38, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1229/1395 [04:39<00:37, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1230/1395 [04:40<00:37, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1231/1395 [04:40<00:37, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1232/1395 [04:40<00:37, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1233/1395 [04:40<00:36, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 88% 1234/1395 [04:41<00:36, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1235/1395 [04:41<00:36, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1236/1395 [04:41<00:36, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1237/1395 [04:41<00:35, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1238/1395 [04:41<00:35, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1239/1395 [04:42<00:35, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1240/1395 [04:42<00:35, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1241/1395 [04:42<00:35, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1242/1395 [04:42<00:34, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1243/1395 [04:43<00:34, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1244/1395 [04:43<00:34, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1245/1395 [04:43<00:34, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1246/1395 [04:43<00:33, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1247/1395 [04:43<00:33, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 89% 1248/1395 [04:44<00:33, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1249/1395 [04:44<00:33, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1250/1395 [04:44<00:33, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1251/1395 [04:44<00:32, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1252/1395 [04:45<00:32, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1253/1395 [04:45<00:32, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1254/1395 [04:45<00:32, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1255/1395 [04:45<00:31, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1256/1395 [04:45<00:31, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1257/1395 [04:46<00:31, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1258/1395 [04:46<00:31, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1259/1395 [04:46<00:30, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1260/1395 [04:46<00:30, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1261/1395 [04:47<00:30, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 90% 1262/1395 [04:47<00:30, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1263/1395 [04:47<00:30, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1264/1395 [04:47<00:29, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1265/1395 [04:47<00:29, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1266/1395 [04:48<00:29, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1267/1395 [04:48<00:29, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1268/1395 [04:48<00:28, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1269/1395 [04:48<00:28, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1270/1395 [04:48<00:28, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1271/1395 [04:49<00:28, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1272/1395 [04:49<00:27, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1273/1395 [04:49<00:27, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1274/1395 [04:49<00:27, 4.39it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1275/1395 [04:50<00:27, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 91% 1276/1395 [04:50<00:27, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1277/1395 [04:50<00:26, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1278/1395 [04:50<00:26, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1279/1395 [04:50<00:26, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1280/1395 [04:51<00:26, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1281/1395 [04:51<00:25, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1282/1395 [04:51<00:25, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1283/1395 [04:51<00:25, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1284/1395 [04:52<00:25, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1285/1395 [04:52<00:25, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1286/1395 [04:52<00:24, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1287/1395 [04:52<00:24, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1288/1395 [04:52<00:24, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1289/1395 [04:53<00:24, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 92% 1290/1395 [04:53<00:23, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1291/1395 [04:53<00:23, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1292/1395 [04:53<00:23, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1293/1395 [04:54<00:23, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1294/1395 [04:54<00:22, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1295/1395 [04:54<00:22, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1296/1395 [04:54<00:22, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1297/1395 [04:54<00:22, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1298/1395 [04:55<00:22, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1299/1395 [04:55<00:21, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1300/1395 [04:55<00:21, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1301/1395 [04:55<00:21, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1302/1395 [04:55<00:21, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1303/1395 [04:56<00:20, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 93% 1304/1395 [04:56<00:20, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1305/1395 [04:56<00:20, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1306/1395 [04:56<00:20, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1307/1395 [04:57<00:20, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1308/1395 [04:57<00:19, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1309/1395 [04:57<00:19, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1310/1395 [04:57<00:19, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1311/1395 [04:57<00:19, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1312/1395 [04:58<00:18, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1313/1395 [04:58<00:18, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1314/1395 [04:58<00:18, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1315/1395 [04:58<00:18, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1316/1395 [04:59<00:17, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1317/1395 [04:59<00:17, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 94% 1318/1395 [04:59<00:17, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1319/1395 [04:59<00:17, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1320/1395 [04:59<00:17, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1321/1395 [05:00<00:16, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1322/1395 [05:00<00:16, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1323/1395 [05:00<00:16, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1324/1395 [05:00<00:16, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1325/1395 [05:00<00:15, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1326/1395 [05:01<00:15, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1327/1395 [05:01<00:15, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1328/1395 [05:01<00:15, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1329/1395 [05:01<00:14, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1330/1395 [05:02<00:14, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1331/1395 [05:02<00:14, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 95% 1332/1395 [05:02<00:14, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1333/1395 [05:02<00:14, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1334/1395 [05:02<00:13, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1335/1395 [05:03<00:13, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1336/1395 [05:03<00:13, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1337/1395 [05:03<00:13, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1338/1395 [05:03<00:12, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1339/1395 [05:04<00:12, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1340/1395 [05:04<00:12, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1341/1395 [05:04<00:12, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1342/1395 [05:04<00:12, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1343/1395 [05:04<00:11, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1344/1395 [05:05<00:11, 4.40it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1345/1395 [05:05<00:11, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 96% 1346/1395 [05:05<00:11, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1347/1395 [05:05<00:10, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1348/1395 [05:05<00:10, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1349/1395 [05:06<00:10, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1350/1395 [05:06<00:10, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1351/1395 [05:06<00:09, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1352/1395 [05:06<00:09, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1353/1395 [05:07<00:09, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1354/1395 [05:07<00:09, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1355/1395 [05:07<00:09, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1356/1395 [05:07<00:08, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1357/1395 [05:07<00:08, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1358/1395 [05:08<00:08, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1359/1395 [05:08<00:08, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 97% 1360/1395 [05:08<00:07, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1361/1395 [05:08<00:07, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1362/1395 [05:08<00:07, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1363/1395 [05:09<00:07, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1364/1395 [05:09<00:07, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1365/1395 [05:09<00:06, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1366/1395 [05:09<00:06, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1367/1395 [05:10<00:06, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1368/1395 [05:10<00:06, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1369/1395 [05:10<00:05, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1370/1395 [05:10<00:05, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1371/1395 [05:10<00:05, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1372/1395 [05:11<00:05, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1373/1395 [05:11<00:04, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 98% 1374/1395 [05:11<00:04, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1375/1395 [05:11<00:04, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1376/1395 [05:12<00:04, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1377/1395 [05:12<00:04, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1378/1395 [05:12<00:03, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1379/1395 [05:12<00:03, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1380/1395 [05:12<00:03, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1381/1395 [05:13<00:03, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1382/1395 [05:13<00:02, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1383/1395 [05:13<00:02, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1384/1395 [05:13<00:02, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1385/1395 [05:13<00:02, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1386/1395 [05:14<00:02, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1387/1395 [05:14<00:01, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 99% 1388/1395 [05:14<00:01, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 100% 1389/1395 [05:14<00:01, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 100% 1390/1395 [05:15<00:01, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 100% 1391/1395 [05:15<00:00, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 100% 1392/1395 [05:15<00:00, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 100% 1393/1395 [05:15<00:00, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 100% 1394/1395 [05:15<00:00, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]\nEpoch 6: 100% 1395/1395 [05:16<00:00, 4.41it/s, loss=0.218, v_num=0, val_iou=0.673]Epoch 6: val_iou was not in top 1\nEpoch 6: 100% 1395/1395 [05:17<00:00, 4.39it/s, loss=0.218, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1207/1395 [04:31<00:42, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 7: 87% 1208/1395 [04:31<00:42, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1209/1395 [04:31<00:41, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1210/1395 [04:31<00:41, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1211/1395 [04:32<00:41, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1212/1395 [04:32<00:41, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1213/1395 [04:32<00:40, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1214/1395 [04:32<00:40, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1215/1395 [04:33<00:40, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1216/1395 [04:33<00:40, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1217/1395 [04:33<00:39, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1218/1395 [04:33<00:39, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1219/1395 [04:33<00:39, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 87% 1220/1395 [04:34<00:39, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1221/1395 [04:34<00:39, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1222/1395 [04:34<00:38, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1223/1395 [04:34<00:38, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1224/1395 [04:34<00:38, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1225/1395 [04:35<00:38, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1226/1395 [04:35<00:37, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1227/1395 [04:35<00:37, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1228/1395 [04:35<00:37, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1229/1395 [04:36<00:37, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1230/1395 [04:36<00:37, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1231/1395 [04:36<00:36, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1232/1395 [04:36<00:36, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1233/1395 [04:36<00:36, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 88% 1234/1395 [04:37<00:36, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1235/1395 [04:37<00:35, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1236/1395 [04:37<00:35, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1237/1395 [04:37<00:35, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1238/1395 [04:38<00:35, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1239/1395 [04:38<00:35, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1240/1395 [04:38<00:34, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1241/1395 [04:38<00:34, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1242/1395 [04:38<00:34, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1243/1395 [04:39<00:34, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1244/1395 [04:39<00:33, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1245/1395 [04:39<00:33, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1246/1395 [04:39<00:33, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1247/1395 [04:40<00:33, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 89% 1248/1395 [04:40<00:33, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1249/1395 [04:40<00:32, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1250/1395 [04:40<00:32, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1251/1395 [04:40<00:32, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1252/1395 [04:41<00:32, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1253/1395 [04:41<00:31, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1254/1395 [04:41<00:31, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1255/1395 [04:41<00:31, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1256/1395 [04:42<00:31, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1257/1395 [04:42<00:30, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1258/1395 [04:42<00:30, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1259/1395 [04:42<00:30, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1260/1395 [04:42<00:30, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1261/1395 [04:43<00:30, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 90% 1262/1395 [04:43<00:29, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1263/1395 [04:43<00:29, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1264/1395 [04:43<00:29, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1265/1395 [04:44<00:29, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1266/1395 [04:44<00:28, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1267/1395 [04:44<00:28, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1268/1395 [04:44<00:28, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1269/1395 [04:44<00:28, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1270/1395 [04:45<00:28, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1271/1395 [04:45<00:27, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1272/1395 [04:45<00:27, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1273/1395 [04:45<00:27, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1274/1395 [04:45<00:27, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1275/1395 [04:46<00:26, 4.45it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 91% 1276/1395 [04:46<00:26, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1277/1395 [04:46<00:26, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1278/1395 [04:46<00:26, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1279/1395 [04:47<00:26, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1280/1395 [04:47<00:25, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1281/1395 [04:47<00:25, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1282/1395 [04:47<00:25, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1283/1395 [04:47<00:25, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1284/1395 [04:48<00:24, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1285/1395 [04:48<00:24, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1286/1395 [04:48<00:24, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1287/1395 [04:48<00:24, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1288/1395 [04:49<00:24, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1289/1395 [04:49<00:23, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 92% 1290/1395 [04:49<00:23, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1291/1395 [04:49<00:23, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1292/1395 [04:49<00:23, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1293/1395 [04:50<00:22, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1294/1395 [04:50<00:22, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1295/1395 [04:50<00:22, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1296/1395 [04:50<00:22, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1297/1395 [04:50<00:21, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1298/1395 [04:51<00:21, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1299/1395 [04:51<00:21, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1300/1395 [04:51<00:21, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1301/1395 [04:51<00:21, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1302/1395 [04:52<00:20, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1303/1395 [04:52<00:20, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 93% 1304/1395 [04:52<00:20, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1305/1395 [04:52<00:20, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1306/1395 [04:52<00:19, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1307/1395 [04:53<00:19, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1308/1395 [04:53<00:19, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1309/1395 [04:53<00:19, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1310/1395 [04:53<00:19, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1311/1395 [04:53<00:18, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1312/1395 [04:54<00:18, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1313/1395 [04:54<00:18, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1314/1395 [04:54<00:18, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1315/1395 [04:54<00:17, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1316/1395 [04:55<00:17, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1317/1395 [04:55<00:17, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 94% 1318/1395 [04:55<00:17, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1319/1395 [04:55<00:17, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1320/1395 [04:55<00:16, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1321/1395 [04:56<00:16, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1322/1395 [04:56<00:16, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1323/1395 [04:56<00:16, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1324/1395 [04:56<00:15, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1325/1395 [04:57<00:15, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1326/1395 [04:57<00:15, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1327/1395 [04:57<00:15, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1328/1395 [04:57<00:15, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1329/1395 [04:57<00:14, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1330/1395 [04:58<00:14, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1331/1395 [04:58<00:14, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 95% 1332/1395 [04:58<00:14, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1333/1395 [04:58<00:13, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1334/1395 [04:58<00:13, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1335/1395 [04:59<00:13, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1336/1395 [04:59<00:13, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1337/1395 [04:59<00:12, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1338/1395 [04:59<00:12, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1339/1395 [04:59<00:12, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1340/1395 [05:00<00:12, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1341/1395 [05:00<00:12, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1342/1395 [05:00<00:11, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1343/1395 [05:00<00:11, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1344/1395 [05:01<00:11, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1345/1395 [05:01<00:11, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 96% 1346/1395 [05:01<00:10, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1347/1395 [05:01<00:10, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1348/1395 [05:01<00:10, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1349/1395 [05:02<00:10, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1350/1395 [05:02<00:10, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1351/1395 [05:02<00:09, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1352/1395 [05:02<00:09, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1353/1395 [05:03<00:09, 4.46it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1354/1395 [05:03<00:09, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1355/1395 [05:03<00:08, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1356/1395 [05:03<00:08, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1357/1395 [05:03<00:08, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1358/1395 [05:04<00:08, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1359/1395 [05:04<00:08, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 97% 1360/1395 [05:04<00:07, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1361/1395 [05:04<00:07, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1362/1395 [05:04<00:07, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1363/1395 [05:05<00:07, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1364/1395 [05:05<00:06, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1365/1395 [05:05<00:06, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1366/1395 [05:05<00:06, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1367/1395 [05:06<00:06, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1368/1395 [05:06<00:06, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1369/1395 [05:06<00:05, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1370/1395 [05:06<00:05, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1371/1395 [05:06<00:05, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1372/1395 [05:07<00:05, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1373/1395 [05:07<00:04, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 98% 1374/1395 [05:07<00:04, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1375/1395 [05:07<00:04, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1376/1395 [05:07<00:04, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1377/1395 [05:08<00:04, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1378/1395 [05:08<00:03, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1379/1395 [05:08<00:03, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1380/1395 [05:08<00:03, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1381/1395 [05:09<00:03, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1382/1395 [05:09<00:02, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1383/1395 [05:09<00:02, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1384/1395 [05:09<00:02, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1385/1395 [05:09<00:02, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1386/1395 [05:10<00:02, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1387/1395 [05:10<00:01, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 99% 1388/1395 [05:10<00:01, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 100% 1389/1395 [05:10<00:01, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 100% 1390/1395 [05:10<00:01, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 100% 1391/1395 [05:11<00:00, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 100% 1392/1395 [05:11<00:00, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 100% 1393/1395 [05:11<00:00, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 100% 1394/1395 [05:11<00:00, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]\nEpoch 7: 100% 1395/1395 [05:12<00:00, 4.47it/s, loss=0.242, v_num=0, val_iou=0.622]Epoch 7: val_iou reached 0.68488 (best 0.68488), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=07-val_iou=0.68.ckpt as top 1\nEpoch 7: 100% 1395/1395 [05:15<00:00, 4.43it/s, loss=0.242, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1207/1395 [04:35<00:42, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 8: 87% 1208/1395 [04:36<00:42, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1209/1395 [04:36<00:42, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1210/1395 [04:36<00:42, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1211/1395 [04:36<00:42, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1212/1395 [04:37<00:41, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1213/1395 [04:37<00:41, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1214/1395 [04:37<00:41, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1215/1395 [04:37<00:41, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1216/1395 [04:37<00:40, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1217/1395 [04:38<00:40, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1218/1395 [04:38<00:40, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1219/1395 [04:38<00:40, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 87% 1220/1395 [04:38<00:39, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1221/1395 [04:39<00:39, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1222/1395 [04:39<00:39, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1223/1395 [04:39<00:39, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1224/1395 [04:39<00:39, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1225/1395 [04:40<00:38, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1226/1395 [04:40<00:38, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1227/1395 [04:40<00:38, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1228/1395 [04:40<00:38, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1229/1395 [04:40<00:37, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1230/1395 [04:41<00:37, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1231/1395 [04:41<00:37, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1232/1395 [04:41<00:37, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1233/1395 [04:41<00:37, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 88% 1234/1395 [04:42<00:36, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1235/1395 [04:42<00:36, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1236/1395 [04:42<00:36, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1237/1395 [04:42<00:36, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1238/1395 [04:43<00:35, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1239/1395 [04:43<00:35, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1240/1395 [04:43<00:35, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1241/1395 [04:43<00:35, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1242/1395 [04:43<00:34, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1243/1395 [04:44<00:34, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1244/1395 [04:44<00:34, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1245/1395 [04:44<00:34, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1246/1395 [04:44<00:34, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1247/1395 [04:45<00:33, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 89% 1248/1395 [04:45<00:33, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1249/1395 [04:45<00:33, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1250/1395 [04:45<00:33, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1251/1395 [04:45<00:32, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1252/1395 [04:46<00:32, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1253/1395 [04:46<00:32, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1254/1395 [04:46<00:32, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1255/1395 [04:46<00:32, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1256/1395 [04:47<00:31, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1257/1395 [04:47<00:31, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1258/1395 [04:47<00:31, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1259/1395 [04:47<00:31, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1260/1395 [04:48<00:30, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1261/1395 [04:48<00:30, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 90% 1262/1395 [04:48<00:30, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1263/1395 [04:48<00:30, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1264/1395 [04:48<00:29, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1265/1395 [04:49<00:29, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1266/1395 [04:49<00:29, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1267/1395 [04:49<00:29, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1268/1395 [04:49<00:29, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1269/1395 [04:50<00:28, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1270/1395 [04:50<00:28, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1271/1395 [04:50<00:28, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1272/1395 [04:50<00:28, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1273/1395 [04:50<00:27, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1274/1395 [04:51<00:27, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1275/1395 [04:51<00:27, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 91% 1276/1395 [04:51<00:27, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1277/1395 [04:51<00:26, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1278/1395 [04:52<00:26, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1279/1395 [04:52<00:26, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1280/1395 [04:52<00:26, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1281/1395 [04:52<00:26, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1282/1395 [04:52<00:25, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1283/1395 [04:53<00:25, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1284/1395 [04:53<00:25, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1285/1395 [04:53<00:25, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1286/1395 [04:53<00:24, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1287/1395 [04:54<00:24, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1288/1395 [04:54<00:24, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1289/1395 [04:54<00:24, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 92% 1290/1395 [04:54<00:23, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1291/1395 [04:55<00:23, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1292/1395 [04:55<00:23, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1293/1395 [04:55<00:23, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1294/1395 [04:55<00:23, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1295/1395 [04:55<00:22, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1296/1395 [04:56<00:22, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1297/1395 [04:56<00:22, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1298/1395 [04:56<00:22, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1299/1395 [04:56<00:21, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1300/1395 [04:57<00:21, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1301/1395 [04:57<00:21, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1302/1395 [04:57<00:21, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1303/1395 [04:57<00:21, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 93% 1304/1395 [04:57<00:20, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1305/1395 [04:58<00:20, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1306/1395 [04:58<00:20, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1307/1395 [04:58<00:20, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1308/1395 [04:58<00:19, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1309/1395 [04:59<00:19, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1310/1395 [04:59<00:19, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1311/1395 [04:59<00:19, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1312/1395 [04:59<00:18, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1313/1395 [05:00<00:18, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1314/1395 [05:00<00:18, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1315/1395 [05:00<00:18, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1316/1395 [05:00<00:18, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1317/1395 [05:00<00:17, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 94% 1318/1395 [05:01<00:17, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1319/1395 [05:01<00:17, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1320/1395 [05:01<00:17, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1321/1395 [05:01<00:16, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1322/1395 [05:02<00:16, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1323/1395 [05:02<00:16, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1324/1395 [05:02<00:16, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1325/1395 [05:02<00:15, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1326/1395 [05:03<00:15, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1327/1395 [05:03<00:15, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1328/1395 [05:03<00:15, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1329/1395 [05:03<00:15, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1330/1395 [05:03<00:14, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1331/1395 [05:04<00:14, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 95% 1332/1395 [05:04<00:14, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1333/1395 [05:04<00:14, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1334/1395 [05:04<00:13, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1335/1395 [05:05<00:13, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1336/1395 [05:05<00:13, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1337/1395 [05:05<00:13, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1338/1395 [05:05<00:13, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1339/1395 [05:05<00:12, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1340/1395 [05:06<00:12, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1341/1395 [05:06<00:12, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1342/1395 [05:06<00:12, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1343/1395 [05:06<00:11, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1344/1395 [05:07<00:11, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1345/1395 [05:07<00:11, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 96% 1346/1395 [05:07<00:11, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1347/1395 [05:07<00:10, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1348/1395 [05:08<00:10, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1349/1395 [05:08<00:10, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1350/1395 [05:08<00:10, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1351/1395 [05:08<00:10, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1352/1395 [05:08<00:09, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1353/1395 [05:09<00:09, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1354/1395 [05:09<00:09, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1355/1395 [05:09<00:09, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1356/1395 [05:09<00:08, 4.38it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1357/1395 [05:10<00:08, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1358/1395 [05:10<00:08, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1359/1395 [05:10<00:08, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 97% 1360/1395 [05:10<00:08, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1361/1395 [05:11<00:07, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1362/1395 [05:11<00:07, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1363/1395 [05:11<00:07, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1364/1395 [05:11<00:07, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1365/1395 [05:12<00:06, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1366/1395 [05:12<00:06, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1367/1395 [05:12<00:06, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1368/1395 [05:12<00:06, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1369/1395 [05:13<00:05, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1370/1395 [05:13<00:05, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1371/1395 [05:13<00:05, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1372/1395 [05:13<00:05, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1373/1395 [05:14<00:05, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 98% 1374/1395 [05:14<00:04, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1375/1395 [05:14<00:04, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1376/1395 [05:14<00:04, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1377/1395 [05:14<00:04, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1378/1395 [05:15<00:03, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1379/1395 [05:15<00:03, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1380/1395 [05:15<00:03, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1381/1395 [05:15<00:03, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1382/1395 [05:16<00:02, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1383/1395 [05:16<00:02, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1384/1395 [05:16<00:02, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1385/1395 [05:16<00:02, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1386/1395 [05:17<00:02, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1387/1395 [05:17<00:01, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 99% 1388/1395 [05:17<00:01, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 100% 1389/1395 [05:17<00:01, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 100% 1390/1395 [05:18<00:01, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 100% 1391/1395 [05:18<00:00, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 100% 1392/1395 [05:18<00:00, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 100% 1393/1395 [05:18<00:00, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 100% 1394/1395 [05:19<00:00, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 8: 100% 1395/1395 [05:19<00:00, 4.37it/s, loss=0.172, v_num=0, val_iou=0.685]Epoch 8: val_iou was not in top 1\nEpoch 8: 100% 1395/1395 [05:20<00:00, 4.35it/s, loss=0.172, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1207/1395 [04:37<00:43, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 9: 87% 1208/1395 [04:37<00:42, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1209/1395 [04:37<00:42, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1210/1395 [04:37<00:42, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1211/1395 [04:38<00:42, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1212/1395 [04:38<00:42, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1213/1395 [04:38<00:41, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1214/1395 [04:38<00:41, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1215/1395 [04:39<00:41, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1216/1395 [04:39<00:41, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1217/1395 [04:39<00:40, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1218/1395 [04:39<00:40, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1219/1395 [04:39<00:40, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 87% 1220/1395 [04:40<00:40, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1221/1395 [04:40<00:39, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1222/1395 [04:40<00:39, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1223/1395 [04:40<00:39, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1224/1395 [04:41<00:39, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1225/1395 [04:41<00:39, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1226/1395 [04:41<00:38, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1227/1395 [04:41<00:38, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1228/1395 [04:42<00:38, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1229/1395 [04:42<00:38, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1230/1395 [04:42<00:37, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1231/1395 [04:42<00:37, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1232/1395 [04:42<00:37, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1233/1395 [04:43<00:37, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 88% 1234/1395 [04:43<00:36, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1235/1395 [04:43<00:36, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1236/1395 [04:43<00:36, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1237/1395 [04:44<00:36, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1238/1395 [04:44<00:36, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1239/1395 [04:44<00:35, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1240/1395 [04:44<00:35, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1241/1395 [04:44<00:35, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1242/1395 [04:45<00:35, 4.35it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1243/1395 [04:45<00:34, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1244/1395 [04:45<00:34, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1245/1395 [04:45<00:34, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1246/1395 [04:46<00:34, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1247/1395 [04:46<00:33, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 89% 1248/1395 [04:46<00:33, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1249/1395 [04:46<00:33, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1250/1395 [04:47<00:33, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1251/1395 [04:47<00:33, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1252/1395 [04:47<00:32, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1253/1395 [04:47<00:32, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1254/1395 [04:47<00:32, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1255/1395 [04:48<00:32, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1256/1395 [04:48<00:31, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1257/1395 [04:48<00:31, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1258/1395 [04:48<00:31, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1259/1395 [04:49<00:31, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1260/1395 [04:49<00:30, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1261/1395 [04:49<00:30, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 90% 1262/1395 [04:49<00:30, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1263/1395 [04:49<00:30, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1264/1395 [04:50<00:30, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1265/1395 [04:50<00:29, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1266/1395 [04:50<00:29, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1267/1395 [04:50<00:29, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1268/1395 [04:51<00:29, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1269/1395 [04:51<00:28, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1270/1395 [04:51<00:28, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1271/1395 [04:51<00:28, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1272/1395 [04:52<00:28, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1273/1395 [04:52<00:28, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1274/1395 [04:52<00:27, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1275/1395 [04:52<00:27, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 91% 1276/1395 [04:52<00:27, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1277/1395 [04:53<00:27, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1278/1395 [04:53<00:26, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1279/1395 [04:53<00:26, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1280/1395 [04:53<00:26, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1281/1395 [04:54<00:26, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1282/1395 [04:54<00:25, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1283/1395 [04:54<00:25, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1284/1395 [04:54<00:25, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1285/1395 [04:54<00:25, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1286/1395 [04:55<00:25, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1287/1395 [04:55<00:24, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1288/1395 [04:55<00:24, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1289/1395 [04:55<00:24, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 92% 1290/1395 [04:56<00:24, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1291/1395 [04:56<00:23, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1292/1395 [04:56<00:23, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1293/1395 [04:56<00:23, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1294/1395 [04:57<00:23, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1295/1395 [04:57<00:22, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1296/1395 [04:57<00:22, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1297/1395 [04:57<00:22, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1298/1395 [04:57<00:22, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1299/1395 [04:58<00:22, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1300/1395 [04:58<00:21, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1301/1395 [04:58<00:21, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1302/1395 [04:58<00:21, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1303/1395 [04:59<00:21, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 93% 1304/1395 [04:59<00:20, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1305/1395 [04:59<00:20, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1306/1395 [04:59<00:20, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1307/1395 [04:59<00:20, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1308/1395 [05:00<00:19, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1309/1395 [05:00<00:19, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1310/1395 [05:00<00:19, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1311/1395 [05:00<00:19, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1312/1395 [05:01<00:19, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1313/1395 [05:01<00:18, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1314/1395 [05:01<00:18, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1315/1395 [05:01<00:18, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1316/1395 [05:02<00:18, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1317/1395 [05:02<00:17, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 94% 1318/1395 [05:02<00:17, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1319/1395 [05:02<00:17, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1320/1395 [05:02<00:17, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1321/1395 [05:03<00:16, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1322/1395 [05:03<00:16, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1323/1395 [05:03<00:16, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1324/1395 [05:03<00:16, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1325/1395 [05:04<00:16, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1326/1395 [05:04<00:15, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1327/1395 [05:04<00:15, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1328/1395 [05:04<00:15, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1329/1395 [05:05<00:15, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1330/1395 [05:05<00:14, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1331/1395 [05:05<00:14, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 95% 1332/1395 [05:05<00:14, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1333/1395 [05:05<00:14, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1334/1395 [05:06<00:14, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1335/1395 [05:06<00:13, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1336/1395 [05:06<00:13, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1337/1395 [05:06<00:13, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1338/1395 [05:07<00:13, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1339/1395 [05:07<00:12, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1340/1395 [05:07<00:12, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1341/1395 [05:07<00:12, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1342/1395 [05:08<00:12, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1343/1395 [05:08<00:11, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1344/1395 [05:08<00:11, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1345/1395 [05:08<00:11, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 96% 1346/1395 [05:08<00:11, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1347/1395 [05:09<00:11, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1348/1395 [05:09<00:10, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1349/1395 [05:09<00:10, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1350/1395 [05:09<00:10, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1351/1395 [05:10<00:10, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1352/1395 [05:10<00:09, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1353/1395 [05:10<00:09, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1354/1395 [05:10<00:09, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1355/1395 [05:11<00:09, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1356/1395 [05:11<00:08, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1357/1395 [05:11<00:08, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1358/1395 [05:11<00:08, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1359/1395 [05:11<00:08, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 97% 1360/1395 [05:12<00:08, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1361/1395 [05:12<00:07, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1362/1395 [05:12<00:07, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1363/1395 [05:12<00:07, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1364/1395 [05:13<00:07, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1365/1395 [05:13<00:06, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1366/1395 [05:13<00:06, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1367/1395 [05:13<00:06, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1368/1395 [05:13<00:06, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1369/1395 [05:14<00:05, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1370/1395 [05:14<00:05, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1371/1395 [05:14<00:05, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1372/1395 [05:14<00:05, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1373/1395 [05:15<00:05, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 98% 1374/1395 [05:15<00:04, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1375/1395 [05:15<00:04, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1376/1395 [05:15<00:04, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1377/1395 [05:15<00:04, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1378/1395 [05:16<00:03, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1379/1395 [05:16<00:03, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1380/1395 [05:16<00:03, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1381/1395 [05:16<00:03, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1382/1395 [05:17<00:02, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1383/1395 [05:17<00:02, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1384/1395 [05:17<00:02, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1385/1395 [05:17<00:02, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1386/1395 [05:18<00:02, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1387/1395 [05:18<00:01, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 99% 1388/1395 [05:18<00:01, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 100% 1389/1395 [05:18<00:01, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 100% 1390/1395 [05:18<00:01, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 100% 1391/1395 [05:19<00:00, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 100% 1392/1395 [05:19<00:00, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 100% 1393/1395 [05:19<00:00, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 100% 1394/1395 [05:19<00:00, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]\nEpoch 9: 100% 1395/1395 [05:20<00:00, 4.36it/s, loss=0.211, v_num=0, val_iou=0.685]Epoch 9: val_iou reached 0.69248 (best 0.69248), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=09-val_iou=0.69.ckpt as top 1\nEpoch 9: 100% 1395/1395 [05:22<00:00, 4.32it/s, loss=0.211, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1207/1395 [04:38<00:43, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 10: 87% 1208/1395 [04:38<00:43, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1209/1395 [04:39<00:42, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1210/1395 [04:39<00:42, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1211/1395 [04:39<00:42, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1212/1395 [04:39<00:42, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1213/1395 [04:39<00:41, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1214/1395 [04:40<00:41, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1215/1395 [04:40<00:41, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1216/1395 [04:40<00:41, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1217/1395 [04:40<00:41, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1218/1395 [04:41<00:40, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1219/1395 [04:41<00:40, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 87% 1220/1395 [04:41<00:40, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1221/1395 [04:41<00:40, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1222/1395 [04:41<00:39, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1223/1395 [04:42<00:39, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1224/1395 [04:42<00:39, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1225/1395 [04:42<00:39, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1226/1395 [04:42<00:38, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1227/1395 [04:43<00:38, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1228/1395 [04:43<00:38, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1229/1395 [04:43<00:38, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1230/1395 [04:43<00:38, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1231/1395 [04:44<00:37, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1232/1395 [04:44<00:37, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1233/1395 [04:44<00:37, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 88% 1234/1395 [04:44<00:37, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1235/1395 [04:44<00:36, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1236/1395 [04:45<00:36, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1237/1395 [04:45<00:36, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1238/1395 [04:45<00:36, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1239/1395 [04:45<00:35, 4.33it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1240/1395 [04:46<00:35, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1241/1395 [04:46<00:35, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1242/1395 [04:46<00:35, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1243/1395 [04:46<00:35, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1244/1395 [04:46<00:34, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1245/1395 [04:47<00:34, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1246/1395 [04:47<00:34, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1247/1395 [04:47<00:34, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 89% 1248/1395 [04:47<00:33, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1249/1395 [04:48<00:33, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1250/1395 [04:48<00:33, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1251/1395 [04:48<00:33, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1252/1395 [04:48<00:32, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1253/1395 [04:48<00:32, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1254/1395 [04:49<00:32, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1255/1395 [04:49<00:32, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1256/1395 [04:49<00:32, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1257/1395 [04:49<00:31, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1258/1395 [04:50<00:31, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1259/1395 [04:50<00:31, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1260/1395 [04:50<00:31, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1261/1395 [04:50<00:30, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 90% 1262/1395 [04:50<00:30, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1263/1395 [04:51<00:30, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1264/1395 [04:51<00:30, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1265/1395 [04:51<00:29, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1266/1395 [04:51<00:29, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1267/1395 [04:52<00:29, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1268/1395 [04:52<00:29, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1269/1395 [04:52<00:29, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1270/1395 [04:52<00:28, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1271/1395 [04:53<00:28, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1272/1395 [04:53<00:28, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1273/1395 [04:53<00:28, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1274/1395 [04:53<00:27, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1275/1395 [04:53<00:27, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 91% 1276/1395 [04:54<00:27, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1277/1395 [04:54<00:27, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1278/1395 [04:54<00:26, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1279/1395 [04:54<00:26, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1280/1395 [04:55<00:26, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1281/1395 [04:55<00:26, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1282/1395 [04:55<00:26, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1283/1395 [04:55<00:25, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1284/1395 [04:55<00:25, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1285/1395 [04:56<00:25, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1286/1395 [04:56<00:25, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1287/1395 [04:56<00:24, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1288/1395 [04:56<00:24, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1289/1395 [04:57<00:24, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 92% 1290/1395 [04:57<00:24, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1291/1395 [04:57<00:23, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1292/1395 [04:57<00:23, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1293/1395 [04:57<00:23, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1294/1395 [04:58<00:23, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1295/1395 [04:58<00:23, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1296/1395 [04:58<00:22, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1297/1395 [04:58<00:22, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1298/1395 [04:59<00:22, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1299/1395 [04:59<00:22, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1300/1395 [04:59<00:21, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1301/1395 [04:59<00:21, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1302/1395 [05:00<00:21, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1303/1395 [05:00<00:21, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 93% 1304/1395 [05:00<00:20, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1305/1395 [05:00<00:20, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1306/1395 [05:00<00:20, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1307/1395 [05:01<00:20, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1308/1395 [05:01<00:20, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1309/1395 [05:01<00:19, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1310/1395 [05:01<00:19, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1311/1395 [05:02<00:19, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1312/1395 [05:02<00:19, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1313/1395 [05:02<00:18, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1314/1395 [05:02<00:18, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1315/1395 [05:02<00:18, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1316/1395 [05:03<00:18, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1317/1395 [05:03<00:17, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 94% 1318/1395 [05:03<00:17, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1319/1395 [05:03<00:17, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1320/1395 [05:04<00:17, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1321/1395 [05:04<00:17, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1322/1395 [05:04<00:16, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1323/1395 [05:04<00:16, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1324/1395 [05:05<00:16, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1325/1395 [05:05<00:16, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1326/1395 [05:05<00:15, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1327/1395 [05:05<00:15, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1328/1395 [05:05<00:15, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1329/1395 [05:06<00:15, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1330/1395 [05:06<00:14, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1331/1395 [05:06<00:14, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 95% 1332/1395 [05:06<00:14, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1333/1395 [05:07<00:14, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1334/1395 [05:07<00:14, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1335/1395 [05:07<00:13, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1336/1395 [05:07<00:13, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1337/1395 [05:08<00:13, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1338/1395 [05:08<00:13, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1339/1395 [05:08<00:12, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1340/1395 [05:08<00:12, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1341/1395 [05:08<00:12, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1342/1395 [05:09<00:12, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1343/1395 [05:09<00:11, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1344/1395 [05:09<00:11, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1345/1395 [05:09<00:11, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 96% 1346/1395 [05:10<00:11, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1347/1395 [05:10<00:11, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1348/1395 [05:10<00:10, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1349/1395 [05:10<00:10, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1350/1395 [05:10<00:10, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1351/1395 [05:11<00:10, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1352/1395 [05:11<00:09, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1353/1395 [05:11<00:09, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1354/1395 [05:11<00:09, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1355/1395 [05:12<00:09, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1356/1395 [05:12<00:08, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1357/1395 [05:12<00:08, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1358/1395 [05:12<00:08, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1359/1395 [05:12<00:08, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 97% 1360/1395 [05:13<00:08, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1361/1395 [05:13<00:07, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1362/1395 [05:13<00:07, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1363/1395 [05:13<00:07, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1364/1395 [05:14<00:07, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1365/1395 [05:14<00:06, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1366/1395 [05:14<00:06, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1367/1395 [05:14<00:06, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1368/1395 [05:15<00:06, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1369/1395 [05:15<00:05, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1370/1395 [05:15<00:05, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1371/1395 [05:15<00:05, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1372/1395 [05:15<00:05, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1373/1395 [05:16<00:05, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 98% 1374/1395 [05:16<00:04, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1375/1395 [05:16<00:04, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1376/1395 [05:16<00:04, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1377/1395 [05:17<00:04, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1378/1395 [05:17<00:03, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1379/1395 [05:17<00:03, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1380/1395 [05:17<00:03, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1381/1395 [05:17<00:03, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1382/1395 [05:18<00:02, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1383/1395 [05:18<00:02, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1384/1395 [05:18<00:02, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1385/1395 [05:18<00:02, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1386/1395 [05:19<00:02, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1387/1395 [05:19<00:01, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 99% 1388/1395 [05:19<00:01, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 100% 1389/1395 [05:19<00:01, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 100% 1390/1395 [05:19<00:01, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 100% 1391/1395 [05:20<00:00, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 100% 1392/1395 [05:20<00:00, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 100% 1393/1395 [05:20<00:00, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 100% 1394/1395 [05:20<00:00, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]\nEpoch 10: 100% 1395/1395 [05:21<00:00, 4.34it/s, loss=0.256, v_num=0, val_iou=0.692]Epoch 10: val_iou was not in top 1\nEpoch 10: 100% 1395/1395 [05:22<00:00, 4.32it/s, loss=0.256, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1207/1395 [04:34<00:42, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 11: 87% 1208/1395 [04:34<00:42, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1209/1395 [04:34<00:42, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1210/1395 [04:35<00:42, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1211/1395 [04:35<00:41, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1212/1395 [04:35<00:41, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1213/1395 [04:35<00:41, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1214/1395 [04:35<00:41, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1215/1395 [04:36<00:40, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1216/1395 [04:36<00:40, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1217/1395 [04:36<00:40, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1218/1395 [04:36<00:40, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1219/1395 [04:37<00:39, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 87% 1220/1395 [04:37<00:39, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1221/1395 [04:37<00:39, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1222/1395 [04:37<00:39, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1223/1395 [04:37<00:39, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1224/1395 [04:38<00:38, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1225/1395 [04:38<00:38, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1226/1395 [04:38<00:38, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1227/1395 [04:38<00:38, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1228/1395 [04:39<00:37, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1229/1395 [04:39<00:37, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1230/1395 [04:39<00:37, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1231/1395 [04:39<00:37, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1232/1395 [04:39<00:37, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1233/1395 [04:40<00:36, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 88% 1234/1395 [04:40<00:36, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1235/1395 [04:40<00:36, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1236/1395 [04:40<00:36, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1237/1395 [04:41<00:35, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1238/1395 [04:41<00:35, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1239/1395 [04:41<00:35, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1240/1395 [04:41<00:35, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1241/1395 [04:42<00:34, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1242/1395 [04:42<00:34, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1243/1395 [04:42<00:34, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1244/1395 [04:42<00:34, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1245/1395 [04:42<00:34, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1246/1395 [04:43<00:33, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1247/1395 [04:43<00:33, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 89% 1248/1395 [04:43<00:33, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1249/1395 [04:43<00:33, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1250/1395 [04:44<00:32, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1251/1395 [04:44<00:32, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1252/1395 [04:44<00:32, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1253/1395 [04:44<00:32, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1254/1395 [04:45<00:32, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1255/1395 [04:45<00:31, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1256/1395 [04:45<00:31, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1257/1395 [04:45<00:31, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1258/1395 [04:46<00:31, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1259/1395 [04:46<00:30, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1260/1395 [04:46<00:30, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1261/1395 [04:46<00:30, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 90% 1262/1395 [04:47<00:30, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1263/1395 [04:47<00:30, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1264/1395 [04:47<00:29, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1265/1395 [04:47<00:29, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1266/1395 [04:47<00:29, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1267/1395 [04:48<00:29, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1268/1395 [04:48<00:28, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1269/1395 [04:48<00:28, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1270/1395 [04:48<00:28, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1271/1395 [04:49<00:28, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1272/1395 [04:49<00:27, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1273/1395 [04:49<00:27, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1274/1395 [04:49<00:27, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1275/1395 [04:49<00:27, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 91% 1276/1395 [04:50<00:27, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1277/1395 [04:50<00:26, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1278/1395 [04:50<00:26, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1279/1395 [04:50<00:26, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1280/1395 [04:51<00:26, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1281/1395 [04:51<00:25, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1282/1395 [04:51<00:25, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1283/1395 [04:51<00:25, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1284/1395 [04:52<00:25, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1285/1395 [04:52<00:25, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1286/1395 [04:52<00:24, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1287/1395 [04:52<00:24, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1288/1395 [04:53<00:24, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1289/1395 [04:53<00:24, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 92% 1290/1395 [04:53<00:23, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1291/1395 [04:53<00:23, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1292/1395 [04:53<00:23, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1293/1395 [04:54<00:23, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1294/1395 [04:54<00:22, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1295/1395 [04:54<00:22, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1296/1395 [04:54<00:22, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1297/1395 [04:55<00:22, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1298/1395 [04:55<00:22, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1299/1395 [04:55<00:21, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1300/1395 [04:55<00:21, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1301/1395 [04:56<00:21, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1302/1395 [04:56<00:21, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1303/1395 [04:56<00:20, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 93% 1304/1395 [04:56<00:20, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1305/1395 [04:56<00:20, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1306/1395 [04:57<00:20, 4.40it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1307/1395 [04:57<00:20, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1308/1395 [04:57<00:19, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1309/1395 [04:57<00:19, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1310/1395 [04:58<00:19, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1311/1395 [04:58<00:19, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1312/1395 [04:58<00:18, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1313/1395 [04:58<00:18, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1314/1395 [04:59<00:18, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1315/1395 [04:59<00:18, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1316/1395 [04:59<00:17, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1317/1395 [04:59<00:17, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 94% 1318/1395 [04:59<00:17, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1319/1395 [05:00<00:17, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1320/1395 [05:00<00:17, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1321/1395 [05:00<00:16, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1322/1395 [05:00<00:16, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1323/1395 [05:01<00:16, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1324/1395 [05:01<00:16, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1325/1395 [05:01<00:15, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1326/1395 [05:01<00:15, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1327/1395 [05:02<00:15, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1328/1395 [05:02<00:15, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1329/1395 [05:02<00:15, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1330/1395 [05:02<00:14, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1331/1395 [05:02<00:14, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 95% 1332/1395 [05:03<00:14, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1333/1395 [05:03<00:14, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1334/1395 [05:03<00:13, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1335/1395 [05:03<00:13, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1336/1395 [05:04<00:13, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1337/1395 [05:04<00:13, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1338/1395 [05:04<00:12, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1339/1395 [05:04<00:12, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1340/1395 [05:05<00:12, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1341/1395 [05:05<00:12, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1342/1395 [05:05<00:12, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1343/1395 [05:05<00:11, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1344/1395 [05:05<00:11, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1345/1395 [05:06<00:11, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 96% 1346/1395 [05:06<00:11, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1347/1395 [05:06<00:10, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1348/1395 [05:06<00:10, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1349/1395 [05:07<00:10, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1350/1395 [05:07<00:10, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1351/1395 [05:07<00:10, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1352/1395 [05:07<00:09, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1353/1395 [05:08<00:09, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1354/1395 [05:08<00:09, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1355/1395 [05:08<00:09, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1356/1395 [05:08<00:08, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1357/1395 [05:09<00:08, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1358/1395 [05:09<00:08, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1359/1395 [05:09<00:08, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 97% 1360/1395 [05:09<00:07, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1361/1395 [05:09<00:07, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1362/1395 [05:10<00:07, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1363/1395 [05:10<00:07, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1364/1395 [05:10<00:07, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1365/1395 [05:10<00:06, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1366/1395 [05:11<00:06, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1367/1395 [05:11<00:06, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1368/1395 [05:11<00:06, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1369/1395 [05:11<00:05, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1370/1395 [05:12<00:05, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1371/1395 [05:12<00:05, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1372/1395 [05:12<00:05, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1373/1395 [05:12<00:05, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 98% 1374/1395 [05:12<00:04, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1375/1395 [05:13<00:04, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1376/1395 [05:13<00:04, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1377/1395 [05:13<00:04, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1378/1395 [05:13<00:03, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1379/1395 [05:14<00:03, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1380/1395 [05:14<00:03, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1381/1395 [05:14<00:03, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1382/1395 [05:14<00:02, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1383/1395 [05:15<00:02, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1384/1395 [05:15<00:02, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1385/1395 [05:15<00:02, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1386/1395 [05:15<00:02, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1387/1395 [05:15<00:01, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 99% 1388/1395 [05:16<00:01, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 100% 1389/1395 [05:16<00:01, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 100% 1390/1395 [05:16<00:01, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 100% 1391/1395 [05:16<00:00, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 100% 1392/1395 [05:17<00:00, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 100% 1393/1395 [05:17<00:00, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 100% 1394/1395 [05:17<00:00, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]\nEpoch 11: 100% 1395/1395 [05:17<00:00, 4.39it/s, loss=0.228, v_num=0, val_iou=0.684]Epoch 11: val_iou reached 0.69357 (best 0.69357), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=11-val_iou=0.69.ckpt as top 1\nEpoch 11: 100% 1395/1395 [05:20<00:00, 4.35it/s, loss=0.228, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1207/1395 [04:34<00:42, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 12: 87% 1208/1395 [04:35<00:42, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1209/1395 [04:35<00:42, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1210/1395 [04:35<00:42, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1211/1395 [04:35<00:41, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1212/1395 [04:36<00:41, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1213/1395 [04:36<00:41, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1214/1395 [04:36<00:41, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1215/1395 [04:36<00:41, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1216/1395 [04:37<00:40, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1217/1395 [04:37<00:40, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1218/1395 [04:37<00:40, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1219/1395 [04:37<00:40, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 87% 1220/1395 [04:38<00:39, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1221/1395 [04:38<00:39, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1222/1395 [04:38<00:39, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1223/1395 [04:38<00:39, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1224/1395 [04:38<00:38, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1225/1395 [04:39<00:38, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1226/1395 [04:39<00:38, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1227/1395 [04:39<00:38, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1228/1395 [04:39<00:38, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1229/1395 [04:40<00:37, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1230/1395 [04:40<00:37, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1231/1395 [04:40<00:37, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1232/1395 [04:40<00:37, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1233/1395 [04:41<00:36, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 88% 1234/1395 [04:41<00:36, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1235/1395 [04:41<00:36, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1236/1395 [04:41<00:36, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1237/1395 [04:42<00:36, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1238/1395 [04:42<00:35, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1239/1395 [04:42<00:35, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1240/1395 [04:42<00:35, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1241/1395 [04:42<00:35, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1242/1395 [04:43<00:34, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1243/1395 [04:43<00:34, 4.39it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1244/1395 [04:43<00:34, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1245/1395 [04:43<00:34, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1246/1395 [04:44<00:33, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1247/1395 [04:44<00:33, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 89% 1248/1395 [04:44<00:33, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1249/1395 [04:44<00:33, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1250/1395 [04:45<00:33, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1251/1395 [04:45<00:32, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1252/1395 [04:45<00:32, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1253/1395 [04:45<00:32, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1254/1395 [04:46<00:32, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1255/1395 [04:46<00:31, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1256/1395 [04:46<00:31, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1257/1395 [04:46<00:31, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1258/1395 [04:47<00:31, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1259/1395 [04:47<00:31, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1260/1395 [04:47<00:30, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1261/1395 [04:47<00:30, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 90% 1262/1395 [04:47<00:30, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1263/1395 [04:48<00:30, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1264/1395 [04:48<00:29, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1265/1395 [04:48<00:29, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1266/1395 [04:48<00:29, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1267/1395 [04:49<00:29, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1268/1395 [04:49<00:28, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1269/1395 [04:49<00:28, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1270/1395 [04:49<00:28, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1271/1395 [04:50<00:28, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1272/1395 [04:50<00:28, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1273/1395 [04:50<00:27, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1274/1395 [04:50<00:27, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1275/1395 [04:51<00:27, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 91% 1276/1395 [04:51<00:27, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1277/1395 [04:51<00:26, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1278/1395 [04:51<00:26, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1279/1395 [04:51<00:26, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1280/1395 [04:52<00:26, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1281/1395 [04:52<00:26, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1282/1395 [04:52<00:25, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1283/1395 [04:52<00:25, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1284/1395 [04:53<00:25, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1285/1395 [04:53<00:25, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1286/1395 [04:53<00:24, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1287/1395 [04:53<00:24, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1288/1395 [04:54<00:24, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1289/1395 [04:54<00:24, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 92% 1290/1395 [04:54<00:23, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1291/1395 [04:54<00:23, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1292/1395 [04:55<00:23, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1293/1395 [04:55<00:23, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1294/1395 [04:55<00:23, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1295/1395 [04:55<00:22, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1296/1395 [04:55<00:22, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1297/1395 [04:56<00:22, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1298/1395 [04:56<00:22, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1299/1395 [04:56<00:21, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1300/1395 [04:56<00:21, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1301/1395 [04:57<00:21, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1302/1395 [04:57<00:21, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1303/1395 [04:57<00:21, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 93% 1304/1395 [04:57<00:20, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1305/1395 [04:58<00:20, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1306/1395 [04:58<00:20, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1307/1395 [04:58<00:20, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1308/1395 [04:58<00:19, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1309/1395 [04:59<00:19, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1310/1395 [04:59<00:19, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1311/1395 [04:59<00:19, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1312/1395 [04:59<00:18, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1313/1395 [05:00<00:18, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1314/1395 [05:00<00:18, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1315/1395 [05:00<00:18, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1316/1395 [05:00<00:18, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1317/1395 [05:00<00:17, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 94% 1318/1395 [05:01<00:17, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1319/1395 [05:01<00:17, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1320/1395 [05:01<00:17, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1321/1395 [05:01<00:16, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1322/1395 [05:02<00:16, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1323/1395 [05:02<00:16, 4.38it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1324/1395 [05:02<00:16, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1325/1395 [05:02<00:16, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1326/1395 [05:03<00:15, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1327/1395 [05:03<00:15, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1328/1395 [05:03<00:15, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1329/1395 [05:03<00:15, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1330/1395 [05:04<00:14, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1331/1395 [05:04<00:14, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 95% 1332/1395 [05:04<00:14, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1333/1395 [05:04<00:14, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1334/1395 [05:05<00:13, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1335/1395 [05:05<00:13, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1336/1395 [05:05<00:13, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1337/1395 [05:05<00:13, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1338/1395 [05:05<00:13, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1339/1395 [05:06<00:12, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1340/1395 [05:06<00:12, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1341/1395 [05:06<00:12, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1342/1395 [05:06<00:12, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1343/1395 [05:07<00:11, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1344/1395 [05:07<00:11, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1345/1395 [05:07<00:11, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 96% 1346/1395 [05:07<00:11, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1347/1395 [05:08<00:10, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1348/1395 [05:08<00:10, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1349/1395 [05:08<00:10, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1350/1395 [05:08<00:10, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1351/1395 [05:09<00:10, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1352/1395 [05:09<00:09, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1353/1395 [05:09<00:09, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1354/1395 [05:09<00:09, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1355/1395 [05:09<00:09, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1356/1395 [05:10<00:08, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1357/1395 [05:10<00:08, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1358/1395 [05:10<00:08, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1359/1395 [05:10<00:08, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 97% 1360/1395 [05:11<00:08, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1361/1395 [05:11<00:07, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1362/1395 [05:11<00:07, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1363/1395 [05:11<00:07, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1364/1395 [05:12<00:07, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1365/1395 [05:12<00:06, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1366/1395 [05:12<00:06, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1367/1395 [05:12<00:06, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1368/1395 [05:13<00:06, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1369/1395 [05:13<00:05, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1370/1395 [05:13<00:05, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1371/1395 [05:13<00:05, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1372/1395 [05:13<00:05, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1373/1395 [05:14<00:05, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 98% 1374/1395 [05:14<00:04, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1375/1395 [05:14<00:04, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1376/1395 [05:14<00:04, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1377/1395 [05:15<00:04, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1378/1395 [05:15<00:03, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1379/1395 [05:15<00:03, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1380/1395 [05:15<00:03, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1381/1395 [05:16<00:03, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1382/1395 [05:16<00:02, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1383/1395 [05:16<00:02, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1384/1395 [05:16<00:02, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1385/1395 [05:17<00:02, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1386/1395 [05:17<00:02, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1387/1395 [05:17<00:01, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 99% 1388/1395 [05:17<00:01, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 100% 1389/1395 [05:18<00:01, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 100% 1390/1395 [05:18<00:01, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 100% 1391/1395 [05:18<00:00, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 100% 1392/1395 [05:18<00:00, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 100% 1393/1395 [05:18<00:00, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 100% 1394/1395 [05:19<00:00, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]\nEpoch 12: 100% 1395/1395 [05:19<00:00, 4.37it/s, loss=0.202, v_num=0, val_iou=0.694]Epoch 12: val_iou was not in top 1\nEpoch 12: 100% 1395/1395 [05:20<00:00, 4.35it/s, loss=0.202, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1207/1395 [04:44<00:44, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 13: 87% 1208/1395 [04:44<00:44, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1209/1395 [04:44<00:43, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1210/1395 [04:44<00:43, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1211/1395 [04:44<00:43, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1212/1395 [04:45<00:43, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1213/1395 [04:45<00:42, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1214/1395 [04:45<00:42, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1215/1395 [04:45<00:42, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1216/1395 [04:46<00:42, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1217/1395 [04:46<00:41, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1218/1395 [04:46<00:41, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1219/1395 [04:46<00:41, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 87% 1220/1395 [04:47<00:41, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1221/1395 [04:47<00:40, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1222/1395 [04:47<00:40, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1223/1395 [04:47<00:40, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1224/1395 [04:48<00:40, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1225/1395 [04:48<00:40, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1226/1395 [04:48<00:39, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1227/1395 [04:48<00:39, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1228/1395 [04:48<00:39, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1229/1395 [04:49<00:39, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1230/1395 [04:49<00:38, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1231/1395 [04:49<00:38, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1232/1395 [04:49<00:38, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1233/1395 [04:50<00:38, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 88% 1234/1395 [04:50<00:37, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1235/1395 [04:50<00:37, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1236/1395 [04:50<00:37, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1237/1395 [04:51<00:37, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1238/1395 [04:51<00:36, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1239/1395 [04:51<00:36, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1240/1395 [04:51<00:36, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1241/1395 [04:52<00:36, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1242/1395 [04:52<00:36, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1243/1395 [04:52<00:35, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1244/1395 [04:52<00:35, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1245/1395 [04:53<00:35, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1246/1395 [04:53<00:35, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1247/1395 [04:53<00:34, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 89% 1248/1395 [04:53<00:34, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1249/1395 [04:53<00:34, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1250/1395 [04:54<00:34, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1251/1395 [04:54<00:33, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1252/1395 [04:54<00:33, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1253/1395 [04:54<00:33, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1254/1395 [04:55<00:33, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1255/1395 [04:55<00:32, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1256/1395 [04:55<00:32, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1257/1395 [04:55<00:32, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1258/1395 [04:56<00:32, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1259/1395 [04:56<00:32, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1260/1395 [04:56<00:31, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1261/1395 [04:56<00:31, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 90% 1262/1395 [04:57<00:31, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1263/1395 [04:57<00:31, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1264/1395 [04:57<00:30, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1265/1395 [04:57<00:30, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1266/1395 [04:57<00:30, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1267/1395 [04:58<00:30, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1268/1395 [04:58<00:29, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1269/1395 [04:58<00:29, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1270/1395 [04:58<00:29, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1271/1395 [04:59<00:29, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1272/1395 [04:59<00:28, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1273/1395 [04:59<00:28, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1274/1395 [04:59<00:28, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1275/1395 [05:00<00:28, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 91% 1276/1395 [05:00<00:28, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1277/1395 [05:00<00:27, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1278/1395 [05:00<00:27, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1279/1395 [05:01<00:27, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1280/1395 [05:01<00:27, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1281/1395 [05:01<00:26, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1282/1395 [05:01<00:26, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1283/1395 [05:02<00:26, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1284/1395 [05:02<00:26, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1285/1395 [05:02<00:25, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1286/1395 [05:02<00:25, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1287/1395 [05:02<00:25, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1288/1395 [05:03<00:25, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1289/1395 [05:03<00:24, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 92% 1290/1395 [05:03<00:24, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1291/1395 [05:03<00:24, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1292/1395 [05:04<00:24, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1293/1395 [05:04<00:24, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1294/1395 [05:04<00:23, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1295/1395 [05:04<00:23, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1296/1395 [05:05<00:23, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1297/1395 [05:05<00:23, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1298/1395 [05:05<00:22, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1299/1395 [05:05<00:22, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1300/1395 [05:06<00:22, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1301/1395 [05:06<00:22, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1302/1395 [05:06<00:21, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1303/1395 [05:06<00:21, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 93% 1304/1395 [05:07<00:21, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1305/1395 [05:07<00:21, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1306/1395 [05:07<00:20, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1307/1395 [05:07<00:20, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1308/1395 [05:07<00:20, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1309/1395 [05:08<00:20, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1310/1395 [05:08<00:20, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1311/1395 [05:08<00:19, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1312/1395 [05:08<00:19, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1313/1395 [05:09<00:19, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1314/1395 [05:09<00:19, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1315/1395 [05:09<00:18, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1316/1395 [05:09<00:18, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1317/1395 [05:10<00:18, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 94% 1318/1395 [05:10<00:18, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1319/1395 [05:10<00:17, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1320/1395 [05:10<00:17, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1321/1395 [05:11<00:17, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1322/1395 [05:11<00:17, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1323/1395 [05:11<00:16, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1324/1395 [05:11<00:16, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1325/1395 [05:11<00:16, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1326/1395 [05:12<00:16, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1327/1395 [05:12<00:16, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1328/1395 [05:12<00:15, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1329/1395 [05:12<00:15, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1330/1395 [05:13<00:15, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1331/1395 [05:13<00:15, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 95% 1332/1395 [05:13<00:14, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1333/1395 [05:13<00:14, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1334/1395 [05:14<00:14, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1335/1395 [05:14<00:14, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1336/1395 [05:14<00:13, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1337/1395 [05:14<00:13, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1338/1395 [05:15<00:13, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1339/1395 [05:15<00:13, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1340/1395 [05:15<00:12, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1341/1395 [05:15<00:12, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1342/1395 [05:16<00:12, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1343/1395 [05:16<00:12, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1344/1395 [05:16<00:12, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1345/1395 [05:16<00:11, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 96% 1346/1395 [05:16<00:11, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1347/1395 [05:17<00:11, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1348/1395 [05:17<00:11, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1349/1395 [05:17<00:10, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1350/1395 [05:17<00:10, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1351/1395 [05:18<00:10, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1352/1395 [05:18<00:10, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1353/1395 [05:18<00:09, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1354/1395 [05:18<00:09, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1355/1395 [05:19<00:09, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1356/1395 [05:19<00:09, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1357/1395 [05:19<00:08, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1358/1395 [05:19<00:08, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1359/1395 [05:20<00:08, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 97% 1360/1395 [05:20<00:08, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1361/1395 [05:20<00:08, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1362/1395 [05:20<00:07, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1363/1395 [05:20<00:07, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1364/1395 [05:21<00:07, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1365/1395 [05:21<00:07, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1366/1395 [05:21<00:06, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1367/1395 [05:21<00:06, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1368/1395 [05:22<00:06, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1369/1395 [05:22<00:06, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1370/1395 [05:22<00:05, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1371/1395 [05:22<00:05, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1372/1395 [05:23<00:05, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1373/1395 [05:23<00:05, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 98% 1374/1395 [05:23<00:04, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1375/1395 [05:23<00:04, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1376/1395 [05:24<00:04, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1377/1395 [05:24<00:04, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1378/1395 [05:24<00:04, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1379/1395 [05:24<00:03, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1380/1395 [05:24<00:03, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1381/1395 [05:25<00:03, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1382/1395 [05:25<00:03, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1383/1395 [05:25<00:02, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1384/1395 [05:25<00:02, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1385/1395 [05:26<00:02, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1386/1395 [05:26<00:02, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1387/1395 [05:26<00:01, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 99% 1388/1395 [05:26<00:01, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 100% 1389/1395 [05:27<00:01, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 100% 1390/1395 [05:27<00:01, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 100% 1391/1395 [05:27<00:00, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 100% 1392/1395 [05:27<00:00, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 100% 1393/1395 [05:28<00:00, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 100% 1394/1395 [05:28<00:00, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]\nEpoch 13: 100% 1395/1395 [05:28<00:00, 4.25it/s, loss=0.201, v_num=0, val_iou=0.648]Epoch 13: val_iou reached 0.70197 (best 0.70197), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=13-val_iou=0.70.ckpt as top 1\nEpoch 13: 100% 1395/1395 [05:31<00:00, 4.21it/s, loss=0.201, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1207/1395 [04:41<00:43, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 14: 87% 1208/1395 [04:42<00:43, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1209/1395 [04:42<00:43, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1210/1395 [04:42<00:43, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1211/1395 [04:42<00:42, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1212/1395 [04:42<00:42, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1213/1395 [04:43<00:42, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1214/1395 [04:43<00:42, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1215/1395 [04:43<00:42, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1216/1395 [04:43<00:41, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1217/1395 [04:44<00:41, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1218/1395 [04:44<00:41, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1219/1395 [04:44<00:41, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 87% 1220/1395 [04:44<00:40, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1221/1395 [04:45<00:40, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1222/1395 [04:45<00:40, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1223/1395 [04:45<00:40, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1224/1395 [04:45<00:39, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1225/1395 [04:45<00:39, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1226/1395 [04:46<00:39, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1227/1395 [04:46<00:39, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1228/1395 [04:46<00:38, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1229/1395 [04:46<00:38, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1230/1395 [04:47<00:38, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1231/1395 [04:47<00:38, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1232/1395 [04:47<00:38, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1233/1395 [04:47<00:37, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 88% 1234/1395 [04:48<00:37, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1235/1395 [04:48<00:37, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1236/1395 [04:48<00:37, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1237/1395 [04:48<00:36, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1238/1395 [04:49<00:36, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1239/1395 [04:49<00:36, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1240/1395 [04:49<00:36, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1241/1395 [04:49<00:35, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1242/1395 [04:50<00:35, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1243/1395 [04:50<00:35, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1244/1395 [04:50<00:35, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1245/1395 [04:50<00:35, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1246/1395 [04:50<00:34, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1247/1395 [04:51<00:34, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 89% 1248/1395 [04:51<00:34, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1249/1395 [04:51<00:34, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1250/1395 [04:51<00:33, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1251/1395 [04:52<00:33, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1252/1395 [04:52<00:33, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1253/1395 [04:52<00:33, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1254/1395 [04:52<00:32, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1255/1395 [04:53<00:32, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1256/1395 [04:53<00:32, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1257/1395 [04:53<00:32, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1258/1395 [04:53<00:31, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1259/1395 [04:54<00:31, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1260/1395 [04:54<00:31, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1261/1395 [04:54<00:31, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 90% 1262/1395 [04:54<00:31, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1263/1395 [04:54<00:30, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1264/1395 [04:55<00:30, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1265/1395 [04:55<00:30, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1266/1395 [04:55<00:30, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1267/1395 [04:55<00:29, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1268/1395 [04:56<00:29, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1269/1395 [04:56<00:29, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1270/1395 [04:56<00:29, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1271/1395 [04:56<00:28, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1272/1395 [04:57<00:28, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1273/1395 [04:57<00:28, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1274/1395 [04:57<00:28, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1275/1395 [04:57<00:28, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 91% 1276/1395 [04:58<00:27, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1277/1395 [04:58<00:27, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1278/1395 [04:58<00:27, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1279/1395 [04:58<00:27, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1280/1395 [04:58<00:26, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1281/1395 [04:59<00:26, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1282/1395 [04:59<00:26, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1283/1395 [04:59<00:26, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1284/1395 [04:59<00:25, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1285/1395 [05:00<00:25, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1286/1395 [05:00<00:25, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1287/1395 [05:00<00:25, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1288/1395 [05:00<00:24, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1289/1395 [05:01<00:24, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 92% 1290/1395 [05:01<00:24, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1291/1395 [05:01<00:24, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1292/1395 [05:01<00:24, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1293/1395 [05:01<00:23, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1294/1395 [05:02<00:23, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1295/1395 [05:02<00:23, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1296/1395 [05:02<00:23, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1297/1395 [05:02<00:22, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1298/1395 [05:03<00:22, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1299/1395 [05:03<00:22, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1300/1395 [05:03<00:22, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1301/1395 [05:03<00:21, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1302/1395 [05:04<00:21, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1303/1395 [05:04<00:21, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 93% 1304/1395 [05:04<00:21, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1305/1395 [05:04<00:21, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1306/1395 [05:04<00:20, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1307/1395 [05:05<00:20, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1308/1395 [05:05<00:20, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1309/1395 [05:05<00:20, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1310/1395 [05:05<00:19, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1311/1395 [05:06<00:19, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1312/1395 [05:06<00:19, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1313/1395 [05:06<00:19, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1314/1395 [05:06<00:18, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1315/1395 [05:07<00:18, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1316/1395 [05:07<00:18, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1317/1395 [05:07<00:18, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 94% 1318/1395 [05:07<00:17, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1319/1395 [05:08<00:17, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1320/1395 [05:08<00:17, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1321/1395 [05:08<00:17, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1322/1395 [05:08<00:17, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1323/1395 [05:08<00:16, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1324/1395 [05:09<00:16, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1325/1395 [05:09<00:16, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1326/1395 [05:09<00:16, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1327/1395 [05:09<00:15, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1328/1395 [05:10<00:15, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1329/1395 [05:10<00:15, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1330/1395 [05:10<00:15, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1331/1395 [05:10<00:14, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 95% 1332/1395 [05:11<00:14, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1333/1395 [05:11<00:14, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1334/1395 [05:11<00:14, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1335/1395 [05:11<00:14, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1336/1395 [05:11<00:13, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1337/1395 [05:12<00:13, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1338/1395 [05:12<00:13, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1339/1395 [05:12<00:13, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1340/1395 [05:12<00:12, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1341/1395 [05:13<00:12, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1342/1395 [05:13<00:12, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1343/1395 [05:13<00:12, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1344/1395 [05:13<00:11, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1345/1395 [05:14<00:11, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 96% 1346/1395 [05:14<00:11, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1347/1395 [05:14<00:11, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1348/1395 [05:14<00:10, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1349/1395 [05:15<00:10, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1350/1395 [05:15<00:10, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1351/1395 [05:15<00:10, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1352/1395 [05:15<00:10, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1353/1395 [05:15<00:09, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1354/1395 [05:16<00:09, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1355/1395 [05:16<00:09, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1356/1395 [05:16<00:09, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1357/1395 [05:16<00:08, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1358/1395 [05:17<00:08, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1359/1395 [05:17<00:08, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 97% 1360/1395 [05:17<00:08, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1361/1395 [05:17<00:07, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1362/1395 [05:18<00:07, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1363/1395 [05:18<00:07, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1364/1395 [05:18<00:07, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1365/1395 [05:18<00:07, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1366/1395 [05:18<00:06, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1367/1395 [05:19<00:06, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1368/1395 [05:19<00:06, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1369/1395 [05:19<00:06, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1370/1395 [05:19<00:05, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1371/1395 [05:20<00:05, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1372/1395 [05:20<00:05, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1373/1395 [05:20<00:05, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 98% 1374/1395 [05:20<00:04, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1375/1395 [05:21<00:04, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1376/1395 [05:21<00:04, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1377/1395 [05:21<00:04, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1378/1395 [05:21<00:03, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1379/1395 [05:22<00:03, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1380/1395 [05:22<00:03, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1381/1395 [05:22<00:03, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1382/1395 [05:22<00:03, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1383/1395 [05:22<00:02, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1384/1395 [05:23<00:02, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1385/1395 [05:23<00:02, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1386/1395 [05:23<00:02, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1387/1395 [05:23<00:01, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 99% 1388/1395 [05:24<00:01, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 100% 1389/1395 [05:24<00:01, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 100% 1390/1395 [05:24<00:01, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 100% 1391/1395 [05:24<00:00, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 100% 1392/1395 [05:25<00:00, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 100% 1393/1395 [05:25<00:00, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 100% 1394/1395 [05:25<00:00, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]\nEpoch 14: 100% 1395/1395 [05:25<00:00, 4.28it/s, loss=0.190, v_num=0, val_iou=0.702]Epoch 14: val_iou reached 0.70413 (best 0.70413), saving model to /content/drive/My Drive/dsl/training_logs/20201210153924-colab_test/tube_logs/version_0/checkpoints/best-epoch=14-val_iou=0.70.ckpt as top 1\nEpoch 14: 100% 1395/1395 [05:28<00:00, 4.25it/s, loss=0.190, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1207/1395 [04:44<00:44, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nValidating: 0it [00:00, ?it/s]\u001b[A\nEpoch 15: 87% 1208/1395 [04:44<00:44, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1209/1395 [04:44<00:43, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1210/1395 [04:45<00:43, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1211/1395 [04:45<00:43, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1212/1395 [04:45<00:43, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1213/1395 [04:45<00:42, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1214/1395 [04:45<00:42, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1215/1395 [04:46<00:42, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1216/1395 [04:46<00:42, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1217/1395 [04:46<00:41, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1218/1395 [04:46<00:41, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1219/1395 [04:47<00:41, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 87% 1220/1395 [04:47<00:41, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1221/1395 [04:47<00:40, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1222/1395 [04:47<00:40, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1223/1395 [04:48<00:40, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1224/1395 [04:48<00:40, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1225/1395 [04:48<00:40, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1226/1395 [04:48<00:39, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1227/1395 [04:49<00:39, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1228/1395 [04:49<00:39, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1229/1395 [04:49<00:39, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1230/1395 [04:49<00:38, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1231/1395 [04:49<00:38, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1232/1395 [04:50<00:38, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1233/1395 [04:50<00:38, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 88% 1234/1395 [04:50<00:37, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1235/1395 [04:50<00:37, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1236/1395 [04:51<00:37, 4.24it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1237/1395 [04:51<00:37, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1238/1395 [04:51<00:36, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1239/1395 [04:51<00:36, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1240/1395 [04:52<00:36, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1241/1395 [04:52<00:36, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1242/1395 [04:52<00:36, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1243/1395 [04:52<00:35, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1244/1395 [04:53<00:35, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1245/1395 [04:53<00:35, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1246/1395 [04:53<00:35, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1247/1395 [04:53<00:34, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 89% 1248/1395 [04:53<00:34, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1249/1395 [04:54<00:34, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1250/1395 [04:54<00:34, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1251/1395 [04:54<00:33, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1252/1395 [04:54<00:33, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1253/1395 [04:55<00:33, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1254/1395 [04:55<00:33, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1255/1395 [04:55<00:32, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1256/1395 [04:55<00:32, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1257/1395 [04:56<00:32, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1258/1395 [04:56<00:32, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1259/1395 [04:56<00:32, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1260/1395 [04:56<00:31, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1261/1395 [04:57<00:31, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 90% 1262/1395 [04:57<00:31, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1263/1395 [04:57<00:31, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1264/1395 [04:57<00:30, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1265/1395 [04:57<00:30, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1266/1395 [04:58<00:30, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1267/1395 [04:58<00:30, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1268/1395 [04:58<00:29, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1269/1395 [04:58<00:29, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1270/1395 [04:59<00:29, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1271/1395 [04:59<00:29, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1272/1395 [04:59<00:28, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1273/1395 [04:59<00:28, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1274/1395 [05:00<00:28, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1275/1395 [05:00<00:28, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 91% 1276/1395 [05:00<00:28, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1277/1395 [05:00<00:27, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1278/1395 [05:00<00:27, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1279/1395 [05:01<00:27, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1280/1395 [05:01<00:27, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1281/1395 [05:01<00:26, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1282/1395 [05:01<00:26, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1283/1395 [05:02<00:26, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1284/1395 [05:02<00:26, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1285/1395 [05:02<00:25, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1286/1395 [05:02<00:25, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1287/1395 [05:03<00:25, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1288/1395 [05:03<00:25, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1289/1395 [05:03<00:24, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 92% 1290/1395 [05:03<00:24, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1291/1395 [05:04<00:24, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1292/1395 [05:04<00:24, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1293/1395 [05:04<00:24, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1294/1395 [05:04<00:23, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1295/1395 [05:04<00:23, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1296/1395 [05:05<00:23, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1297/1395 [05:05<00:23, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1298/1395 [05:05<00:22, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1299/1395 [05:05<00:22, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1300/1395 [05:06<00:22, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1301/1395 [05:06<00:22, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1302/1395 [05:06<00:21, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1303/1395 [05:06<00:21, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 93% 1304/1395 [05:07<00:21, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1305/1395 [05:07<00:21, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1306/1395 [05:07<00:20, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1307/1395 [05:07<00:20, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1308/1395 [05:07<00:20, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1309/1395 [05:08<00:20, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1310/1395 [05:08<00:20, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1311/1395 [05:08<00:19, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1312/1395 [05:08<00:19, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1313/1395 [05:09<00:19, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1314/1395 [05:09<00:19, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1315/1395 [05:09<00:18, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1316/1395 [05:09<00:18, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1317/1395 [05:10<00:18, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 94% 1318/1395 [05:10<00:18, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1319/1395 [05:10<00:17, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1320/1395 [05:10<00:17, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1321/1395 [05:11<00:17, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1322/1395 [05:11<00:17, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1323/1395 [05:11<00:16, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1324/1395 [05:11<00:16, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1325/1395 [05:11<00:16, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1326/1395 [05:12<00:16, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1327/1395 [05:12<00:16, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1328/1395 [05:12<00:15, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1329/1395 [05:12<00:15, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1330/1395 [05:13<00:15, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1331/1395 [05:13<00:15, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 95% 1332/1395 [05:13<00:14, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1333/1395 [05:13<00:14, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1334/1395 [05:14<00:14, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1335/1395 [05:14<00:14, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1336/1395 [05:14<00:13, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1337/1395 [05:14<00:13, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1338/1395 [05:14<00:13, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1339/1395 [05:15<00:13, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1340/1395 [05:15<00:12, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1341/1395 [05:15<00:12, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1342/1395 [05:15<00:12, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1343/1395 [05:16<00:12, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1344/1395 [05:16<00:12, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1345/1395 [05:16<00:11, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 96% 1346/1395 [05:16<00:11, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1347/1395 [05:17<00:11, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1348/1395 [05:17<00:11, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1349/1395 [05:17<00:10, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1350/1395 [05:17<00:10, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1351/1395 [05:18<00:10, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1352/1395 [05:18<00:10, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1353/1395 [05:18<00:09, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1354/1395 [05:18<00:09, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1355/1395 [05:18<00:09, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1356/1395 [05:19<00:09, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1357/1395 [05:19<00:08, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1358/1395 [05:19<00:08, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1359/1395 [05:19<00:08, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 97% 1360/1395 [05:20<00:08, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1361/1395 [05:20<00:08, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1362/1395 [05:20<00:07, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1363/1395 [05:20<00:07, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1364/1395 [05:21<00:07, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1365/1395 [05:21<00:07, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1366/1395 [05:21<00:06, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1367/1395 [05:21<00:06, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1368/1395 [05:22<00:06, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1369/1395 [05:22<00:06, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1370/1395 [05:22<00:05, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1371/1395 [05:22<00:05, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1372/1395 [05:22<00:05, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1373/1395 [05:23<00:05, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 98% 1374/1395 [05:23<00:04, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1375/1395 [05:23<00:04, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1376/1395 [05:23<00:04, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1377/1395 [05:24<00:04, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1378/1395 [05:24<00:04, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1379/1395 [05:24<00:03, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1380/1395 [05:24<00:03, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1381/1395 [05:25<00:03, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1382/1395 [05:25<00:03, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1383/1395 [05:25<00:02, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1384/1395 [05:25<00:02, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1385/1395 [05:26<00:02, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1386/1395 [05:26<00:02, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1387/1395 [05:26<00:01, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 99% 1388/1395 [05:26<00:01, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 100% 1389/1395 [05:26<00:01, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 100% 1390/1395 [05:27<00:01, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 100% 1391/1395 [05:27<00:00, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 100% 1392/1395 [05:27<00:00, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 100% 1393/1395 [05:27<00:00, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 100% 1394/1395 [05:28<00:00, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]\nEpoch 15: 100% 1395/1395 [05:28<00:00, 4.25it/s, loss=0.195, v_num=0, val_iou=0.704]Epoch 15: val_iou was not in top 1\nEpoch 15: 100% 1395/1395 [05:29<00:00, 4.23it/s, loss=0.195, v_num=0, val_iou=0.703]\n \u001b[ASaving latest checkpoint...\nEpoch 15: 100% 1395/1395 [05:29<00:00, 4.23it/s, loss=0.195, v_num=0, val_iou=0.703]\nTesting: 100% 202/202 [00:24<00:00, 8.16it/s]--------------------------------------------------------------------------------\nDATALOADER:0 TEST RESULTS\n{'test_loss': tensor(0.2589, device='cuda:0'),\n 'train_loss': tensor(0.1162, device='cuda:0'),\n 'val_iou': tensor(0.7035, device='cuda:0'),\n 'val_loss': tensor(0.2552, device='cuda:0')}\n--------------------------------------------------------------------------------\nTesting: 100% 202/202 [00:25<00:00, 7.99it/s]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc80fff74eee4a69bcfe0212fe527c4328eecd9
| 1,885 |
ipynb
|
Jupyter Notebook
|
FantasyPL-Flask/Untitled1.ipynb
|
AlexBrady/FYP-FlaskPredictions
|
46eef7be1567da6c52bd540d026417ceb433665a
|
[
"MIT"
] | null | null | null |
FantasyPL-Flask/Untitled1.ipynb
|
AlexBrady/FYP-FlaskPredictions
|
46eef7be1567da6c52bd540d026417ceb433665a
|
[
"MIT"
] | null | null | null |
FantasyPL-Flask/Untitled1.ipynb
|
AlexBrady/FYP-FlaskPredictions
|
46eef7be1567da6c52bd540d026417ceb433665a
|
[
"MIT"
] | null | null | null | 27.318841 | 332 | 0.531565 |
[
[
[
"from data_cleaning import clean_defender_data\nfrom flask import *\nimport pandas as pd\napp = Flask(__name__)\n\n\[email protected](\"/tables\")\ndef show_tables():\n DefenderDF = clean_defender_data()\n DefenderDF.set_index(['player_id'], inplace=True)\n DefenderDF.index.name=None\n return render_template('defenderDF.html',tables=[DefenderDF.to_html(classes='defenders')],\n\n titles = ['na', 'Defenders'])\n\nif __name__ == \"__main__\":\n app.run(debug=True)\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbc8230a0f097912382861fa7aef36b0acb6f097
| 18,337 |
ipynb
|
Jupyter Notebook
|
notebooks/Dstripes/Basic/convolutional/VAE/DstripesVAE_Convolutional_reconst_psnr.ipynb
|
Fidan13/Generative_Models
|
2c700da53210a16f75c468ba521061106afa6982
|
[
"MIT"
] | null | null | null |
notebooks/Dstripes/Basic/convolutional/VAE/DstripesVAE_Convolutional_reconst_psnr.ipynb
|
Fidan13/Generative_Models
|
2c700da53210a16f75c468ba521061106afa6982
|
[
"MIT"
] | null | null | null |
notebooks/Dstripes/Basic/convolutional/VAE/DstripesVAE_Convolutional_reconst_psnr.ipynb
|
Fidan13/Generative_Models
|
2c700da53210a16f75c468ba521061106afa6982
|
[
"MIT"
] | null | null | null | 22.226667 | 138 | 0.554289 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbc827cabada81682fc6a89bb5a35ff398e72da6
| 144,103 |
ipynb
|
Jupyter Notebook
|
ONNXclassifier/onnxClassify.ipynb
|
econdesousa/portfolio
|
0d3d6d8112a8d1e36bec194d81331c7ce2b1cced
|
[
"BSD-3-Clause"
] | null | null | null |
ONNXclassifier/onnxClassify.ipynb
|
econdesousa/portfolio
|
0d3d6d8112a8d1e36bec194d81331c7ce2b1cced
|
[
"BSD-3-Clause"
] | null | null | null |
ONNXclassifier/onnxClassify.ipynb
|
econdesousa/portfolio
|
0d3d6d8112a8d1e36bec194d81331c7ce2b1cced
|
[
"BSD-3-Clause"
] | null | null | null | 357.575682 | 130,794 | 0.928509 |
[
[
[
"<a id='StartingPoint'></a>\n# ONNX classification example\n\nSharing DL models between frameworks or programming languages is possible with Open Neural Network Exchange (ONNX for short).\n\nThis notebook starts from an onnx model exported from MATLAB and uses it in Python.\nOn MATLAB a GoogleNet model pre-trained on ImageNet was loaded and saved to onnx file format through a one-line command [exportONNXNetwork(net,filename)](https://www.mathworks.com/help/deeplearning/ref/exportonnxnetwork.html).\n\nThe model is then loaded here, as well as the data to evaluate (some images retrieved from google).\nImages are preprocessed to the desired format np.array with shape (BatchSize, numChannels, width, heigh), and the model is applied to classify the images and get the probabilities of the classifications.",
"_____no_output_____"
],
[
"## Input Variables\n\nThe why for the existence of each variable is explained [bellow](#ModelDef), near the model definition.",
"_____no_output_____"
]
],
[
[
"%%time\n# model vars\nmodelPath = 'D:/onnxStartingCode/model/preTrainedImageNetGooglenet.onnx'\nlabelsPath = 'D:/onnxStartingCode/model/labels.csv'\nhasHeader = 1\n\n#data vars\nimage_folder = 'D:/onnxStartingCode/ImageFolder/'\nEXT = (\"jfif\",\"jpg\")",
"Wall time: 0 ns\n"
]
],
[
[
"## Import Modules\n\nLet's start by importing all the needed modules\n\n[back to top](#StartingPoint)",
"_____no_output_____"
]
],
[
[
"%%time\nimport onnx\nimport numpy as np\nfrom PIL import Image\nimport os as os\nimport matplotlib.pyplot as plt\nfrom onnxruntime import InferenceSession\nimport csv",
"Wall time: 416 ms\n"
]
],
[
[
"<a id='ModelDef'></a>\n## Define Model and Data functions\n\nWe need to define functions to retrieve the Classifier and Data array.\n\nTo load the model we need the path to the file that stores it and the pat to the file that stores the labels. Finnaly, the parameter hasHeader defines the way the firs row of the labelsFile is treated, as header or ar a label.\nThe labelsPath is required here becuase the model here used does not contain label information, so an external csv file needs to be read.\n\n[back to top](#StartingPoint)",
"_____no_output_____"
]
],
[
[
"%%time\ndef loadmodel(modelPath,labelsPath,hasHeader):\n\n # define network\n # load and check the model\n # load the inference module\n onnx.checker.check_model(modelPath)\n sess = InferenceSession(modelPath)\n \n # Determine the name of the input and output layers\n inname = [input.name for input in sess.get_inputs()]\n outname = [output.name for output in sess.get_outputs()]\n \n \n # auxiliary function to load labels file\n def extractLabels( filename , hasHeader ):\n file = open(filename)\n csvreader = csv.reader(file)\n if (hasHeader>0):\n header = next(csvreader)\n #print(header)\n\n rows = []\n for row in csvreader:\n rows.append(row)\n #print(rows)\n file.close()\n return rows\n \n \n # Get labels\n labels = extractLabels(labelsPath,hasHeader)\n\n\n # Extract information on the inputSize =(width, heigh) and numChannels = 3(RGB) or 1(Grayscale) \n for inp in sess.get_inputs():\n inputSize = inp.shape\n \n numChannels = inputSize[1]\n inputSize = inputSize[2:4]\n \n return sess,inname,outname,numChannels,inputSize,labels\n\n\ndef getData(image_folder,EXT,inputSize):\n \n def getImagesFromFolder(EXT):\n imageList = os.listdir(image_folder)\n if (not(isinstance(EXT, list)) and not(isinstance(EXT,tuple))):\n ext = [EXT]\n\n fullFilePath = [os.path.join(image_folder, f) \n for ext in EXT for f in imageList if os.path.isfile(os.path.join(image_folder, f)) & f.endswith(ext)]\n\n return fullFilePath\n\n def imFile2npArray(imFile,inputSize):\n data = np.array([\n np.array(\n Image.open(fname).resize(inputSize),\n dtype=np.int64) \n for fname in fullFilePath\n ])\n\n X=data.transpose(0,3,1,2)\n X = X.astype(np.float32)\n return X, data\n \n fullFilePath = getImagesFromFolder(EXT)\n X, data = imFile2npArray(fullFilePath,inputSize)\n \n \n return X,data,fullFilePath\n\n",
"Wall time: 0 ns\n"
]
],
[
[
"## Run loading functions to get model and data\n\n* get full filename of all files in a gives directory that end with a given ext (might be an array of EXTENSIONS)\n* load data into numpy arrays for future use:\n * to plot data has to have shape = (x,y,3)\n * the model here presented requires data with shape (3,x,y)\n * two data arrays are then exported, data for ploting and X for classification\n \n[back to top](#StartingPoint)",
"_____no_output_____"
]
],
[
[
"%%time\n# run code\nsess,inname,outname,numChannels,inputSize,labels = loadmodel(modelPath,labelsPath,hasHeader)\nX,data,fullFilePath = getData(image_folder,EXT,inputSize)\n\n\nprint(\"inputSize: \" + str(inputSize))\nprint(\"numChannels: \" + str(numChannels))\nprint(\"inputName: \", inname[0])\nprint(\"outputName: \", outname[0])",
"_____no_output_____"
]
],
[
[
"## Define a functions to load all data\n\n1. get full filename of all files in a gives directory that end with a given ext (might be an arrat of EXT)\n2. load data into numpy arrays for future use:\n * to plot data has to have shape = (x,y,3)\n * the model here presented requires data with shape (3,x,y)\n * two data arrays are then exported, data for ploting and X for classification\n\n[back to top](#StartingPoint)",
"_____no_output_____"
],
[
"## Classification\n\n[back to top](#StartingPoint)",
"_____no_output_____"
]
],
[
[
"%%time\n#data_output = sess.run(outname, {inname: X[0]})\nout = sess.run(None, {inname[0]: X})\nout=np.asarray(out[0])\n\nprint(out.shape)\n\nIND = []\nPROB= []\nfor i in range(out.shape[0]):\n ind=np.where(out[i] == np.amax(out[i]))\n IND.append(ind[0][0])\n PROB.append(out[i,ind[0][0]])\n\nl = [labels[ind] for ind in IND]\nprint([labels[ind] for ind in IND])\nprint(IND)\nprint(PROB)\n",
"(5, 1000)\n[['tabby'], ['pug'], ['daisy'], ['tank'], ['tabby']]\n[281, 254, 985, 847, 281]\n[0.45442542, 0.96363264, 0.99984944, 0.6701486, 0.27902907]\nWall time: 87.9 ms\n"
]
],
[
[
"## Plot some examples\n\n[back to top](#StartingPoint)",
"_____no_output_____"
]
],
[
[
"%%time\nplt.figure(figsize=(10,10))\nif data.shape[0]>=6:\n nPlots=6\n subArray=[2,3]\nelse:\n nPlots=data.shape[0]\n subArray = [1, nPlots]\n \nfor i in range(nPlots):\n plt.subplot(subArray[0],subArray[1],i+1)\n plt.imshow(data[i])\n plt.axis('off')\n plt.title(l[i][0] + ' --- ' + str(round(100*PROB[i])) + '%')\n \nplt.show()\n",
"_____no_output_____"
],
[
"[back to top](#StartingPoint)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbc832f3961916e482070710f75a89b5dab53a56
| 24,398 |
ipynb
|
Jupyter Notebook
|
Exploration_Flux/5_Flux_script.ipynb
|
QuarkNet-HEP/CosmicExplorationBooks
|
215bf86fe73ca1af7a0a567b9e1bdf735ddf32c8
|
[
"MIT"
] | 1 |
2018-10-26T19:08:46.000Z
|
2018-10-26T19:08:46.000Z
|
Analysis/script_Flux.ipynb
|
QuarkNet-HEP/cosmic-ray-notebooks
|
8e80242a85581d43205f08a6350e88690923ef32
|
[
"MIT"
] | 1 |
2019-06-25T20:45:28.000Z
|
2019-06-26T13:30:21.000Z
|
Analysis/script_Flux.ipynb
|
QuarkNet-HEP/cosmic-ray-notebooks
|
8e80242a85581d43205f08a6350e88690923ef32
|
[
"MIT"
] | 1 |
2019-06-25T20:42:39.000Z
|
2019-06-25T20:42:39.000Z
| 35.617518 | 575 | 0.617387 |
[
[
[
"# Flux.pl",
"_____no_output_____"
],
[
"The `Flux.pl` Perl script takes four input parameters:\n\n`Flux.pl [input file] [output file] [bin width (s)] [geometry base directory]`\n\nor, as invoked from the command line,\n\n`$ perl ./perl/Flux.pl [input file] [output file] [bin width (s)] [geometry directory]`",
"_____no_output_____"
],
[
"## Input Parameters",
"_____no_output_____"
],
[
"* `[input file]`\n\n`Flux.pl` expects the first non-comment line of the input file to begin with a string of the form `<DAQ ID>.<channel>`. This is satisfied by threshold and wire delay files, as well as the outputs of data transformation scripts like `Sort.pl` and `Combine.pl` if their inputs are of the appropriate form.\n\nIf the input file doesn't meet this condition, `Flux.pl` -- specifically, the `all_geo_info{}` subroutine of `CommonSubs.pl` -- won't be able to load the appropriate geometry files and execution will fail (see the `[geometry directory]` parameter below).",
"_____no_output_____"
],
[
"* `[output file]`\n\nThis is what the output file will be named.",
"_____no_output_____"
],
[
"* `[binWidth]`\n\nIn physical terms, cosmic ray _flux_ is the number of incident rays per unit area per unit time. The `[binWidth]` parameter determines the \"per unit time\" portion of this quantity. `Flux.pl` will sort the events in its input data into bins of the given time interval, returning the number of events per unit area recorded within each bin.",
"_____no_output_____"
],
[
"* `[geometry directory]`\n\nWith `[binWidth]` handling the \"per unit time\" portion of the flux calculation, the geometry file associated with each detector handles the \"per unit area\". \n\n`Flux.pl` expects geometry files to be stored in a directory structure of the form\n\n```\ngeo/\n├── 6119/\n│ └── 6119.geo\n├── 6148/\n│ └── 6148.geo\n└── 6203/\n └── 6203.geo\n```\n\nwhere each DAQ has its own subdirectory whose name is the DAQ ID, and each such subdirectory has a geometry file whose name is given by the DAQ ID with the `.geo` extension. The command-line argument in this case is `geo/`, the parent directory. With this as the base directory, `Flux.pl` determines what geometry file to load by looking for the DAQ ID in the first line of data. This is why, as noted above, the first non-comment line of `[input file]` must begin with `<DAQ ID>.<channel>`.",
"_____no_output_____"
],
[
"## Flux Input Files",
"_____no_output_____"
],
[
"As we mentioned above, threshold files have the appropriate first-line structure to allow `Flux.pl` to access geometry data for them. So what does `Flux.pl` do when acting on a threshold file?",
"_____no_output_____"
],
[
"We'll test it using the threshold files `files/6148.2016.0109.0.thresh` and `files/6119.2016.0104.1.thresh` as input. First, take a look at the files themselves so we know what the input looks like:",
"_____no_output_____"
]
],
[
[
"!head -10 files/6148.2016.0109.0.thresh",
"#$md5\r\n#md5_hex(0)\r\n#ID.CHANNEL, Julian Day, RISING EDGE(sec), FALLING EDGE(sec), TIME OVER THRESHOLD (nanosec), RISING EDGE(INT), FALLING EDGE(INT)\r\n6148.4\t2457396\t0.5006992493422453\t0.5006992493424479\t17.51\t4326041514317000\t4326041514318750\r\n6148.3\t2457396\t0.5006992493422887\t0.5006992493424768\t16.25\t4326041514317375\t4326041514319000\r\n6148.2\t2457396\t0.5007005963399161\t0.5007005963400029\t7.49\t4326053152376876\t4326053152377625\r\n6148.3\t2457396\t0.5007005963401910\t0.5007005963404514\t22.49\t4326053152379250\t4326053152381500\r\n6148.4\t2457396\t0.5007005963401765\t0.5007005963404658\t25.00\t4326053152379125\t4326053152381624\r\n6148.1\t2457396\t0.5014987243978154\t0.5014987243980903\t23.75\t4332948978797125\t4332948978799500\r\n6148.2\t2457396\t0.5014987243980759\t0.5014987243982495\t15.00\t4332948978799376\t4332948978800875\r\n"
],
[
"!wc -l files/6148.2016.0109.0.thresh",
"6703 files/6148.2016.0109.0.thresh\r\n"
],
[
"!head -10 files/6119.2016.0104.1.thresh",
"#$md5\r\n#md5_hex(0)\r\n#ID.CHANNEL, Julian Day, RISING EDGE(sec), FALLING EDGE(sec), TIME OVER THRESHOLD (nanosec), RISING EDGE(INT), FALLING EDGE(INT)\r\n6119.1\t2457392\t0.3721863017828993\t0.3721863017831598\t22.50\t3215689647404250\t3215689647406500\r\n6119.3\t2457392\t0.3721863017829138\t0.3721863017831598\t21.25\t3215689647404375\t3215689647406500\r\n6119.2\t2457392\t0.3721885846820747\t0.3721885846822772\t17.50\t3215709371653125\t3215709371654875\r\n6119.4\t2457392\t0.3721885846820747\t0.3721885846822917\t18.75\t3215709371653125\t3215709371655000\r\n6119.4\t2457392\t0.3721901866161603\t0.3721901866163773\t18.75\t3215723212363625\t3215723212365500\r\n6119.1\t2457392\t0.3721901866161748\t0.3721901866164496\t23.75\t3215723212363750\t3215723212366125\r\n6119.1\t2457392\t0.3721903650327546\t0.3721903650329427\t16.25\t3215724753883000\t3215724753884625\r\n"
],
[
"!wc -l files/6119.2016.0104.1.thresh",
"181008 files/6119.2016.0104.1.thresh\r\n"
]
],
[
[
"(remember, `wc -l` returns a count of the number of lines in the file). These look like fairly standard threshold files. Now we'll see what `Flux.pl` does with them.",
"_____no_output_____"
],
[
"## The Parsl Flux App",
"_____no_output_____"
],
[
"For convenience, we'll wrap the UNIX command-line invocation of the `Flux.pl` script in a Parsl App, which will make it easier to work with from within the Jupyter Notebook environment.",
"_____no_output_____"
]
],
[
[
"# The prep work:\nimport parsl\nfrom parsl.config import Config\nfrom parsl.executors.threads import ThreadPoolExecutor\nfrom parsl.app.app import bash_app,python_app\nfrom parsl import File\n\nconfig = Config(\n executors=[ThreadPoolExecutor()],\n lazy_errors=True\n)\nparsl.load(config)",
"_____no_output_____"
],
[
"# The App:\n@bash_app\ndef Flux(inputs=[], outputs=[], binWidth='600', geoDir='geo/', stdout='stdout.txt', stderr='stderr.txt'):\n return 'perl ./perl/Flux.pl %s %s %s %s' % (inputs[0], outputs[0], binWidth, geoDir)",
"_____no_output_____"
]
],
[
[
"_Edit stuff below to use the App_",
"_____no_output_____"
],
[
"## Flux Output",
"_____no_output_____"
],
[
"Below is the output generated by `Flux.pl` using the threshold files `6148.2016.0109.0.thresh` and `6119.2016.0104.1.thresh` (separately) as input:",
"_____no_output_____"
],
[
"```\n$ perl ./perl/Flux.pl files/6148.2016.0109.0.thresh outputs/ThreshFluxOut6148_01 600 geo/\n$ head -15 outputs/ThreshFluxOut6148_01 \n#cf12d07ed2dfe4e4c0d52eb663dd9956\n#md5_hex(1536259294 1530469616 files/6148.2016.0109.0.thresh outputs/ThreshFluxOut6148_01 600 geo/)\n01/09/2016 00:06:00 59.416172 8.760437\n01/09/2016 00:16:00 63.291139 9.041591\n01/09/2016 00:26:00 71.041075 9.579177\n01/09/2016 00:36:00 50.374580 8.066389\n01/09/2016 00:46:00 55.541204 8.469954\n01/09/2016 00:56:00 73.624386 9.751788\n01/09/2016 01:06:00 42.624645 7.419998\n01/09/2016 01:16:00 54.249548 8.370887\n01/09/2016 01:26:00 45.207957 7.641539\n01/09/2016 01:36:00 42.624645 7.419998\n01/09/2016 01:46:00 65.874451 9.224268\n01/09/2016 01:56:00 59.416172 8.760437\n01/09/2016 02:06:00 94.290881 11.035913\n```",
"_____no_output_____"
],
[
"```\n$ perl ./perl/Flux.pl files/6119.2016.0104.1.thresh outputs/ThreshFluxOut6119_01 600 geo/\n$ head -15 outputs/ThreshFluxOut6119_01 \n#84d0f02f26edb8f59da2d4011a27389d\n#md5_hex(1536259294 1528996902 files/6119.2016.0104.1.thresh outputs/ThreshFluxOut6119_01 600 geo/)\n01/04/2016 21:00:56 12496.770860 127.049313\n01/04/2016 21:10:56 12580.728494 127.475379\n01/04/2016 21:20:56 12929.475588 129.230157\n01/04/2016 21:30:56 12620.769827 127.678079\n01/04/2016 21:40:56 12893.309222 129.049289\n01/04/2016 21:50:56 12859.726169 128.881113\n01/04/2016 22:00:56 12782.226815 128.492174\n01/04/2016 22:10:56 12520.020666 127.167443\n01/04/2016 22:20:56 12779.643503 128.479189\n01/04/2016 22:30:56 12746.060449 128.310265\n01/04/2016 22:40:56 12609.144924 127.619264\n01/04/2016 22:50:56 12372.771894 126.417419\n01/04/2016 23:00:56 12698.269181 128.069490\n```",
"_____no_output_____"
],
[
"`Flux.pl` seems to give reasonable output with a threshold file as input, provided the DAQ has a geometry file that's up to standards. Can we interpret the output? Despite the lack of a header line, some reasonable inferences will make it clear.",
"_____no_output_____"
],
[
"The first column is clearly the date that the data was taken, and in both cases it agrees with the date indicated by the threshold file's filename.\n\nThe second column is clearly time-of-day values, but what do they mean? We might be tempted to think of them as the full-second portion of cosmic ray event times, but we note in both cases that they occur in a regular pattern of exactly every ten minutes. Of course, that happens to be exactly what we selected as the `binWidth` parameter, 600s = 10min. These are the time bins into which the cosmic ray event data is organized.",
"_____no_output_____"
],
[
"Since we're calculating flux -- muon strikes per unit area per unit time -- we expect the flux count itself to be included in the data, and in fact this is what the third column is, in units of $events/m^2/min$. Note that the \"$/min$\" part is *always* a part of the units of the third column, no matter what the size of the time bins we selected.",
"_____no_output_____"
],
[
"Finally, when doing science, having a measurement means having uncertainty. The fourth column is the obligatory statistical uncertainty in the flux.",
"_____no_output_____"
],
[
"## An exercise in statistical uncertainty",
"_____no_output_____"
],
[
"The general formula for flux $\\Phi$ is\n\n$$\\Phi = \\frac{N}{AT}$$\n\nwhere $N$ is the number of incident events, $A$ is the cross-sectional area over which the flux is measured, and $T$ is the time interval over which the flux is measured.\n\nBy the rule of quadrature for propagating uncertainties,\n\n$$\\frac{\\delta \\Phi}{\\Phi} \\approx \\frac{\\delta N}{N} + \\frac{\\delta A}{A} + \\frac{\\delta T}{T}$$",
"_____no_output_____"
],
[
"Here, $N$ is the raw count of muon hits in the detector, an integer with a standard statistical uncertainty of $\\sqrt{N}$.",
"_____no_output_____"
],
[
"In our present analysis, errors in the bin width and detector area are negligible compared to the statistical fluctuation of cosmic ray muons. Thus, we'll take $\\delta A \\approx \\delta T \\approx 0$ to leave",
"_____no_output_____"
],
[
"$$\\delta \\Phi \\approx \\frac{\\delta N}{N} \\Phi = \\frac{\\Phi}{\\sqrt{N}}$$",
"_____no_output_____"
],
[
"Rearranging this a bit, we find that we should be able to calculate the exact number of muon strikes for each time bin as\n\n$$N \\approx \\left(\\frac{\\Phi}{\\delta\\Phi}\\right)^2.$$\n\nLet's see what happens when we apply this to the data output from `Flux.pl`. For the 6148 data with `binWidth=600`, we find",
"_____no_output_____"
],
[
"```\nDate Time Phi dPhi (Phi/dPhi)^2\n01/09/16\t12:06:00 AM\t59.416172\t8.760437\t45.999996082\n01/09/16\t12:16:00 AM\t63.291139\t9.041591\t49.0000030968\n01/09/16\t12:26:00 AM\t71.041075\t9.579177\t54.9999953935\n01/09/16\t12:36:00 AM\t50.37458\t 8.066389\t38.9999951081\n01/09/16\t12:46:00 AM\t55.541204\t8.469954\t43.0000020769\n01/09/16\t12:56:00 AM\t73.624386\t9.751788\t57.000001784\n01/09/16\t01:06:00 AM\t42.624645\t7.419998\t33.0000025577\n01/09/16\t01:16:00 AM\t54.249548\t8.370887\t41.999999903\n01/09/16\t01:26:00 AM\t45.207957\t7.641539\t35.0000040418\n01/09/16\t01:36:00 AM\t42.624645\t7.419998\t33.0000025577\n01/09/16\t01:46:00 AM\t65.874451\t9.224268\t51.00000197\n01/09/16\t01:56:00 AM\t59.416172\t8.760437\t45.999996082\n01/09/16\t02:06:00 AM\t94.290881\t11.035913 72.9999984439\n```",
"_____no_output_____"
],
[
"The numbers we come up with are in fact integers to an excellent approximation!",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"### Exercise 1",
"_____no_output_____"
],
[
"**A)** Using the data table above, round the `(Phi/dPhi)^2` column to the nearest integer, calling it `N`. With $\\delta N = \\sqrt{N}$, calculate $\\frac{\\delta N}{N}$ for each row in the data.",
"_____no_output_____"
],
[
"**B)** Using your knowledge of the cosmic ray muon detector, estimate the uncertainty $\\delta A$ in the detector area $A$ and the uncertainty $\\delta T$ in the time bin $T$ given as the input `binWidth` parameter. Calculate $\\frac{\\delta A}{A}$ and $\\frac{\\delta T}{T}$ for this analysis.",
"_____no_output_____"
],
[
"**C)** Considering the results of **A)** and **B)**, do you think our previous assumption that $\\frac{\\delta A}{A} \\approx 0$ and $\\frac{\\delta T}{T} \\approx 0$ compared to $\\frac{\\delta N}{N}$ is justified?",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"### Additional Exercises",
"_____no_output_____"
],
[
"* Do the number of counts $N$ in one `binWidth=600s` bin match the sum of counts in the ten corresponding `binWidth=60s` bins?\n\n* Considering raw counts, do you think the \"zero\" bins in the above analyses are natural fluctuations in cosmic ray muon strikes?\n\n* Do the flux values shown above reasonably agree with the known average CR muon flux at sea level? If \"no,\" what effects do you think might account for the difference?",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"We can dig more information out of the `Flux.pl` output by returning to the definition of flux\n\n$$\\Phi = \\frac{N}{AT}.$$\n\nNow that we know $N$ for each data point, and given that we know the bin width $T$ because we set it for the entire analysis, we should be able to calculate the area of the detector as\n\n$$A = \\frac{N}{\\Phi T}$$\n\nOne important comment: `Flux.pl` gives flux values in units of `events/m^2/min` - note the use of minutes instead of seconds. When substituting a numerical value for $T$, we must convert the command line parameter `binWidth=600` from $600s$ to $10min$.\n\nWhen we perform this calculation, we find consistent values for $A$:",
"_____no_output_____"
],
[
"```\nDate Time Phi dPhi N=(Phi/dPhi)^2 A=N/Phi T\n01/09/16\t12:06:00 AM\t59.416172\t8.760437\t45.999996082\t 0.0774199928\n01/09/16\t12:16:00 AM\t63.291139\t9.041591\t49.0000030968\t0.0774200052\n01/09/16\t12:26:00 AM\t71.041075\t9.579177\t54.9999953935\t0.0774199931\n01/09/16\t12:36:00 AM\t50.37458\t 8.066389\t38.9999951081\t0.0774199906\n01/09/16\t12:46:00 AM\t55.541204\t8.469954\t43.0000020769\t0.0774200035\n01/09/16\t12:56:00 AM\t73.624386\t9.751788\t57.000001784\t 0.0774200029\n01/09/16\t01:06:00 AM\t42.624645\t7.419998\t33.0000025577\t0.0774200056\n01/09/16\t01:16:00 AM\t54.249548\t8.370887\t41.999999903\t 0.0774199997\n01/09/16\t01:26:00 AM\t45.207957\t7.641539\t35.0000040418\t0.0774200083\n01/09/16\t01:36:00 AM\t42.624645\t7.419998\t33.0000025577\t0.0774200056\n01/09/16\t01:46:00 AM\t65.874451\t9.224268\t51.00000197\t 0.077420003\n01/09/16\t01:56:00 AM\t59.416172\t8.760437\t45.999996082\t 0.0774199928\n01/09/16\t02:06:00 AM\t94.290881\t11.035913 72.9999984439\t0.0774199983\n```",
"_____no_output_____"
],
[
"In fact, the area of one standard 6000-series QuarkNet CRMD detector panel is $0.07742m^2$.",
"_____no_output_____"
],
[
"It's important to note that we're reversing only the calculations, not the physics! That is, we find $A=0.07742m^2$ because that's the value stored in the `6248.geo` file, not because we're able to determine the actual area of the detector panel from the `Flux.pl` output data using physical principles.",
"_____no_output_____"
],
[
"## Testing binWidth",
"_____no_output_____"
],
[
"To verify that the third-column flux values behave as expected, we can run a quick check by manipulating the `binWidth` parameter. We'll run `Flux.pl` on the above two threshold files again, but this time we'll reduce `binWidth` by a factor of 10:",
"_____no_output_____"
],
[
"```\n$ perl ./perl/Flux.pl files/6148.2016.0109.0.thresh outputs/ThreshFluxOut6148_02 60 geo/\n```",
"_____no_output_____"
]
],
[
[
"!head -15 outputs/ThreshFluxOut6148_02 ",
"#d28fbf9f1f5e4939813797ac0d28f3db\r\n#md5_hex(1536259294 1530469616 files/6148.2016.0109.0.thresh outputs/ThreshFluxOut6148_02 60 geo/)\r\n01/09/2016 00:01:30 64.582795 28.882304\r\n01/09/2016 00:02:30 77.499354 31.638979\r\n01/09/2016 00:13:30 25.833118 18.266773\r\n01/09/2016 00:14:30 25.833118 18.266773\r\n01/09/2016 00:15:30 142.082149 42.839380\r\n01/09/2016 00:16:30 116.249031 38.749677\r\n01/09/2016 00:17:30 77.499354 31.638979\r\n01/09/2016 00:18:30 90.415913 34.174003\r\n01/09/2016 00:19:30 51.666236 25.833118\r\n01/09/2016 00:23:30 103.332472 36.533546\r\n01/09/2016 00:24:30 64.582795 28.882304\r\n01/09/2016 00:25:30 103.332472 36.533546\r\n01/09/2016 00:26:30 90.415913 34.174003\r\n"
]
],
[
[
"```\n$ perl ./perl/Flux.pl files/6119.2016.0104.1.thresh outputs/ThreshFluxOut6119_02 60 geo/\n```",
"_____no_output_____"
]
],
[
[
"!head -15 outputs/ThreshFluxOut6119_02 ",
"#d20cb8a6a91adb6dd45998a811d75401\r\n#md5_hex(1536259294 1528996902 files/6119.2016.0104.1.thresh outputs/ThreshFluxOut6119_02 60 geo/)\r\n01/04/2016 20:56:26 12580.728494 403.112543\r\n01/04/2016 20:57:26 12399.896668 400.204944\r\n01/04/2016 20:58:26 12942.392147 408.865714\r\n01/04/2016 20:59:26 12735.727202 405.588181\r\n01/04/2016 21:00:26 11883.234306 391.778633\r\n01/04/2016 21:01:26 12231.981400 397.485987\r\n01/04/2016 21:02:26 12076.982692 394.959567\r\n01/04/2016 21:03:26 12593.645053 403.319426\r\n01/04/2016 21:04:26 12903.642470 408.253181\r\n01/04/2016 21:05:26 12619.478171 403.732875\r\n01/04/2016 21:06:26 12671.144407 404.558506\r\n01/04/2016 21:07:26 12929.475588 408.661638\r\n01/04/2016 21:08:26 13097.390855 411.306725\r\n"
]
],
[
[
"In the case of the 6148 data, our new fine-grained binning reveals some sparsity in the first several minutes of the data, as all of the bins between the `2:30` bin and the `13:30` bin are empty of muon events (and therefore not reported). What happened here? It's difficult to say -- under normal statistical variations, it's possible that there were simply no recorded events during these bins. It's also possible that the experimenter adjusted the level of physical shielding around the detector during these times, or had a cable unplugged while troubleshooting.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbc836fcf4f37e5fbc7875936d51f8172a0481ec
| 26,679 |
ipynb
|
Jupyter Notebook
|
ETL_Deliverable1_function_test.ipynb
|
AKumar1-lab/Movies_ETL
|
b35679108c8d054cb91c30c60767c30a856fab9a
|
[
"MIT"
] | null | null | null |
ETL_Deliverable1_function_test.ipynb
|
AKumar1-lab/Movies_ETL
|
b35679108c8d054cb91c30c60767c30a856fab9a
|
[
"MIT"
] | null | null | null |
ETL_Deliverable1_function_test.ipynb
|
AKumar1-lab/Movies_ETL
|
b35679108c8d054cb91c30c60767c30a856fab9a
|
[
"MIT"
] | null | null | null | 37.896307 | 95 | 0.389145 |
[
[
[
"import json\nimport pandas as pd\nimport numpy as np\nimport time\nimport re\n#import psycopg2\n#import psycopg2-binary\nfrom sqlalchemy import create_engine\nfrom config import db_password",
"_____no_output_____"
],
[
"# 1. Create a function that takes in three arguments;\n# Wikipedia data, Kaggle metadata, and MovieLens rating data (from Kaggle)\ndef extract_transform_load():\n file_dir = \"./Resources\"\n# 2. Read in the kaggle metadata and MovieLens ratings CSV files as Pandas DataFrames.\n kaggle_file = pd.read_csv(f'{file_dir}/movies_metadata.csv', low_memory=False)\n ratings_file = pd.read_csv(f'{file_dir}/ratings.csv')\n# 3. Open the read the Wikipedia data JSON file.\n with open(f'{file_dir}/wikipedia-movies.json', mode='r') as file:\n wiki_movies_raw = json.load(file)\n# 4. Read in the raw wiki movie data as a Pandas DataFrame.\n wiki_movies_df = pd.DataFrame(wiki_movies_raw)\n# 5. Return the three DataFrames\n return wiki_movies_df, kaggle_file, ratings_file",
"_____no_output_____"
],
[
"# 6 Create the path to your file directory and variables for the three files. \nfile_dir = \"./Resources\"\n# Wikipedia data\nwiki_file = f'{file_dir}/wikipedia-movies.json'\n# Kaggle metadata\nkaggle_file = f'{file_dir}/movies_metadata.csv'\n# MovieLens rating data.\nratings_file = f'{file_dir}/ratings.csv'",
"_____no_output_____"
],
[
"# 7. Set the three variables in Step 6 equal to the function created in Step 1.\nwiki_file, kaggle_file, ratings_file = extract_transform_load()",
"_____no_output_____"
],
[
"# 8. Set the DataFrames from the return statement equal to the file names in Step 6. \n#wiki_movies_df = wiki_file\n#import pandas as pd\nwiki_movies_df = pd.DataFrame(wiki_file)",
"_____no_output_____"
],
[
"#kaggle_metadata = kaggle_file\nkaggle_metadata = pd.DataFrame(kaggle_file)\n\n#ratings = ratings_file\nratings = pd.DataFrame(ratings_file)",
"_____no_output_____"
],
[
"# 9. Check the wiki_movies_df DataFrame.\nwiki_movies_df.head(5)",
"_____no_output_____"
],
[
"# 10. Check the kaggle_metadata DataFrame.\nkaggle_metadata.head()",
"_____no_output_____"
],
[
"# 11. Check the ratings DataFrame.\nratings.head()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc868ae8abbe51003e0feccedf7daaf017ba250
| 14,520 |
ipynb
|
Jupyter Notebook
|
examples/nlp/ipynb/lstm_seq2seq.ipynb
|
wariua/keras-io-ko
|
b89fa9c34af006aa3584dd765fe78f36374246a7
|
[
"Apache-2.0"
] | 1,542 |
2020-05-06T20:23:07.000Z
|
2022-03-31T15:25:03.000Z
|
examples/nlp/ipynb/lstm_seq2seq.ipynb
|
wariua/keras-io-ko
|
b89fa9c34af006aa3584dd765fe78f36374246a7
|
[
"Apache-2.0"
] | 625 |
2020-05-07T10:21:15.000Z
|
2022-03-31T17:19:35.000Z
|
examples/nlp/ipynb/lstm_seq2seq.ipynb
|
wariua/keras-io-ko
|
b89fa9c34af006aa3584dd765fe78f36374246a7
|
[
"Apache-2.0"
] | 1,616 |
2020-05-07T06:28:33.000Z
|
2022-03-31T13:35:35.000Z
| 34.326241 | 95 | 0.594146 |
[
[
[
"# Character-level recurrent sequence-to-sequence model\n\n**Author:** [fchollet](https://twitter.com/fchollet)<br>\n**Date created:** 2017/09/29<br>\n**Last modified:** 2020/04/26<br>\n**Description:** Character-level recurrent sequence-to-sequence model.",
"_____no_output_____"
],
[
"## Introduction\n\nThis example demonstrates how to implement a basic character-level\nrecurrent sequence-to-sequence model. We apply it to translating\nshort English sentences into short French sentences,\ncharacter-by-character. Note that it is fairly unusual to\ndo character-level machine translation, as word-level\nmodels are more common in this domain.\n\n**Summary of the algorithm**\n\n- We start with input sequences from a domain (e.g. English sentences)\n and corresponding target sequences from another domain\n (e.g. French sentences).\n- An encoder LSTM turns input sequences to 2 state vectors\n (we keep the last LSTM state and discard the outputs).\n- A decoder LSTM is trained to turn the target sequences into\n the same sequence but offset by one timestep in the future,\n a training process called \"teacher forcing\" in this context.\n It uses as initial state the state vectors from the encoder.\n Effectively, the decoder learns to generate `targets[t+1...]`\n given `targets[...t]`, conditioned on the input sequence.\n- In inference mode, when we want to decode unknown input sequences, we:\n - Encode the input sequence into state vectors\n - Start with a target sequence of size 1\n (just the start-of-sequence character)\n - Feed the state vectors and 1-char target sequence\n to the decoder to produce predictions for the next character\n - Sample the next character using these predictions\n (we simply use argmax).\n - Append the sampled character to the target sequence\n - Repeat until we generate the end-of-sequence character or we\n hit the character limit.\n",
"_____no_output_____"
],
[
"## Setup\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\n",
"_____no_output_____"
]
],
[
[
"## Download the data\n",
"_____no_output_____"
]
],
[
[
"!!curl -O http://www.manythings.org/anki/fra-eng.zip\n!!unzip fra-eng.zip\n",
"_____no_output_____"
]
],
[
[
"## Configuration\n",
"_____no_output_____"
]
],
[
[
"batch_size = 64 # Batch size for training.\nepochs = 100 # Number of epochs to train for.\nlatent_dim = 256 # Latent dimensionality of the encoding space.\nnum_samples = 10000 # Number of samples to train on.\n# Path to the data txt file on disk.\ndata_path = \"fra.txt\"\n",
"_____no_output_____"
]
],
[
[
"## Prepare the data\n",
"_____no_output_____"
]
],
[
[
"# Vectorize the data.\ninput_texts = []\ntarget_texts = []\ninput_characters = set()\ntarget_characters = set()\nwith open(data_path, \"r\", encoding=\"utf-8\") as f:\n lines = f.read().split(\"\\n\")\nfor line in lines[: min(num_samples, len(lines) - 1)]:\n input_text, target_text, _ = line.split(\"\\t\")\n # We use \"tab\" as the \"start sequence\" character\n # for the targets, and \"\\n\" as \"end sequence\" character.\n target_text = \"\\t\" + target_text + \"\\n\"\n input_texts.append(input_text)\n target_texts.append(target_text)\n for char in input_text:\n if char not in input_characters:\n input_characters.add(char)\n for char in target_text:\n if char not in target_characters:\n target_characters.add(char)\n\ninput_characters = sorted(list(input_characters))\ntarget_characters = sorted(list(target_characters))\nnum_encoder_tokens = len(input_characters)\nnum_decoder_tokens = len(target_characters)\nmax_encoder_seq_length = max([len(txt) for txt in input_texts])\nmax_decoder_seq_length = max([len(txt) for txt in target_texts])\n\nprint(\"Number of samples:\", len(input_texts))\nprint(\"Number of unique input tokens:\", num_encoder_tokens)\nprint(\"Number of unique output tokens:\", num_decoder_tokens)\nprint(\"Max sequence length for inputs:\", max_encoder_seq_length)\nprint(\"Max sequence length for outputs:\", max_decoder_seq_length)\n\ninput_token_index = dict([(char, i) for i, char in enumerate(input_characters)])\ntarget_token_index = dict([(char, i) for i, char in enumerate(target_characters)])\n\nencoder_input_data = np.zeros(\n (len(input_texts), max_encoder_seq_length, num_encoder_tokens), dtype=\"float32\"\n)\ndecoder_input_data = np.zeros(\n (len(input_texts), max_decoder_seq_length, num_decoder_tokens), dtype=\"float32\"\n)\ndecoder_target_data = np.zeros(\n (len(input_texts), max_decoder_seq_length, num_decoder_tokens), dtype=\"float32\"\n)\n\nfor i, (input_text, target_text) in enumerate(zip(input_texts, target_texts)):\n for t, char in enumerate(input_text):\n encoder_input_data[i, t, input_token_index[char]] = 1.0\n encoder_input_data[i, t + 1 :, input_token_index[\" \"]] = 1.0\n for t, char in enumerate(target_text):\n # decoder_target_data is ahead of decoder_input_data by one timestep\n decoder_input_data[i, t, target_token_index[char]] = 1.0\n if t > 0:\n # decoder_target_data will be ahead by one timestep\n # and will not include the start character.\n decoder_target_data[i, t - 1, target_token_index[char]] = 1.0\n decoder_input_data[i, t + 1 :, target_token_index[\" \"]] = 1.0\n decoder_target_data[i, t:, target_token_index[\" \"]] = 1.0\n",
"_____no_output_____"
]
],
[
[
"## Build the model\n",
"_____no_output_____"
]
],
[
[
"# Define an input sequence and process it.\nencoder_inputs = keras.Input(shape=(None, num_encoder_tokens))\nencoder = keras.layers.LSTM(latent_dim, return_state=True)\nencoder_outputs, state_h, state_c = encoder(encoder_inputs)\n\n# We discard `encoder_outputs` and only keep the states.\nencoder_states = [state_h, state_c]\n\n# Set up the decoder, using `encoder_states` as initial state.\ndecoder_inputs = keras.Input(shape=(None, num_decoder_tokens))\n\n# We set up our decoder to return full output sequences,\n# and to return internal states as well. We don't use the\n# return states in the training model, but we will use them in inference.\ndecoder_lstm = keras.layers.LSTM(latent_dim, return_sequences=True, return_state=True)\ndecoder_outputs, _, _ = decoder_lstm(decoder_inputs, initial_state=encoder_states)\ndecoder_dense = keras.layers.Dense(num_decoder_tokens, activation=\"softmax\")\ndecoder_outputs = decoder_dense(decoder_outputs)\n\n# Define the model that will turn\n# `encoder_input_data` & `decoder_input_data` into `decoder_target_data`\nmodel = keras.Model([encoder_inputs, decoder_inputs], decoder_outputs)\n",
"_____no_output_____"
]
],
[
[
"## Train the model\n",
"_____no_output_____"
]
],
[
[
"model.compile(\n optimizer=\"rmsprop\", loss=\"categorical_crossentropy\", metrics=[\"accuracy\"]\n)\nmodel.fit(\n [encoder_input_data, decoder_input_data],\n decoder_target_data,\n batch_size=batch_size,\n epochs=epochs,\n validation_split=0.2,\n)\n# Save model\nmodel.save(\"s2s\")\n",
"_____no_output_____"
]
],
[
[
"## Run inference (sampling)\n\n1. encode input and retrieve initial decoder state\n2. run one step of decoder with this initial state\nand a \"start of sequence\" token as target.\nOutput will be the next target token.\n3. Repeat with the current target token and current states\n",
"_____no_output_____"
]
],
[
[
"# Define sampling models\n# Restore the model and construct the encoder and decoder.\nmodel = keras.models.load_model(\"s2s\")\n\nencoder_inputs = model.input[0] # input_1\nencoder_outputs, state_h_enc, state_c_enc = model.layers[2].output # lstm_1\nencoder_states = [state_h_enc, state_c_enc]\nencoder_model = keras.Model(encoder_inputs, encoder_states)\n\ndecoder_inputs = model.input[1] # input_2\ndecoder_state_input_h = keras.Input(shape=(latent_dim,))\ndecoder_state_input_c = keras.Input(shape=(latent_dim,))\ndecoder_states_inputs = [decoder_state_input_h, decoder_state_input_c]\ndecoder_lstm = model.layers[3]\ndecoder_outputs, state_h_dec, state_c_dec = decoder_lstm(\n decoder_inputs, initial_state=decoder_states_inputs\n)\ndecoder_states = [state_h_dec, state_c_dec]\ndecoder_dense = model.layers[4]\ndecoder_outputs = decoder_dense(decoder_outputs)\ndecoder_model = keras.Model(\n [decoder_inputs] + decoder_states_inputs, [decoder_outputs] + decoder_states\n)\n\n# Reverse-lookup token index to decode sequences back to\n# something readable.\nreverse_input_char_index = dict((i, char) for char, i in input_token_index.items())\nreverse_target_char_index = dict((i, char) for char, i in target_token_index.items())\n\n\ndef decode_sequence(input_seq):\n # Encode the input as state vectors.\n states_value = encoder_model.predict(input_seq)\n\n # Generate empty target sequence of length 1.\n target_seq = np.zeros((1, 1, num_decoder_tokens))\n # Populate the first character of target sequence with the start character.\n target_seq[0, 0, target_token_index[\"\\t\"]] = 1.0\n\n # Sampling loop for a batch of sequences\n # (to simplify, here we assume a batch of size 1).\n stop_condition = False\n decoded_sentence = \"\"\n while not stop_condition:\n output_tokens, h, c = decoder_model.predict([target_seq] + states_value)\n\n # Sample a token\n sampled_token_index = np.argmax(output_tokens[0, -1, :])\n sampled_char = reverse_target_char_index[sampled_token_index]\n decoded_sentence += sampled_char\n\n # Exit condition: either hit max length\n # or find stop character.\n if sampled_char == \"\\n\" or len(decoded_sentence) > max_decoder_seq_length:\n stop_condition = True\n\n # Update the target sequence (of length 1).\n target_seq = np.zeros((1, 1, num_decoder_tokens))\n target_seq[0, 0, sampled_token_index] = 1.0\n\n # Update states\n states_value = [h, c]\n return decoded_sentence\n\n",
"_____no_output_____"
]
],
[
[
"You can now generate decoded sentences as such:\n",
"_____no_output_____"
]
],
[
[
"for seq_index in range(20):\n # Take one sequence (part of the training set)\n # for trying out decoding.\n input_seq = encoder_input_data[seq_index : seq_index + 1]\n decoded_sentence = decode_sequence(input_seq)\n print(\"-\")\n print(\"Input sentence:\", input_texts[seq_index])\n print(\"Decoded sentence:\", decoded_sentence)\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc87baa98779ef6b444297918a998dc5b7a2ea5
| 14,868 |
ipynb
|
Jupyter Notebook
|
campus_data_mining/reference/5/BRP_1VS2.ipynb
|
Yeolnim/big-data-competition
|
de7065d774d52885d76a2d4cfac165d06c2520b8
|
[
"MIT"
] | 1 |
2020-11-10T09:25:33.000Z
|
2020-11-10T09:25:33.000Z
|
campus_data_mining/reference/5/BRP_1VS2.ipynb
|
Yeolnim/big-data-competition
|
de7065d774d52885d76a2d4cfac165d06c2520b8
|
[
"MIT"
] | null | null | null |
campus_data_mining/reference/5/BRP_1VS2.ipynb
|
Yeolnim/big-data-competition
|
de7065d774d52885d76a2d4cfac165d06c2520b8
|
[
"MIT"
] | null | null | null | 37.640506 | 540 | 0.512846 |
[
[
[
"import numpy as np\nimport tensorflow as tf\nimport os\nimport random\nfrom collections import defaultdict\nimport pandas as pd\nimport time\n\ndef load_data_train():\n user_movie = defaultdict(set)\n data=pd.read_csv('BRP_datas\\\\BRP_common_user_book\\\\common_user_book_19_1VS2.csv')\n num_user=len(pd.unique(data['user_id']))\n num_book=len(pd.unique(data['book_id']))\n print('训练集借阅记录数:{}'.format(data.shape[0]))\n for row,val in data.iterrows():\n u = int(val['user_id'])\n i = int(val['book_id'])\n user_movie[u].add(i) #\n\n print(\"num_user:\", num_user)\n print(\"num_book\", num_book)\n\n return num_user, num_book, user_movie\n\n\ndef load_data_test():\n user_movie = defaultdict(set)\n data=pd.read_csv('BRP_datas\\\\BRP_common_user_book\\\\common_user_book_19_2VS1.csv')\n num_user=len(pd.unique(data['user_id']))\n num_book=len(pd.unique(data['book_id']))\n print('测试集借阅记录数:{}'.format(data.shape[0]))\n for row,val in data.iterrows():\n u = int(val['user_id'])\n i = int(val['book_id'])\n user_movie[u].add(i)\n\n print(\"num_user:\", num_user)\n print(\"num_book\", num_book)\n\n return num_user, num_book, user_movie\n\ndef generate_test(user_movie_pair_test):\n \"\"\"\n 对每一个用户u,在user_movie_pair_test中随机找到他借阅过的一本书,保存在user_ratings_test,\n 后面构造训练集和测试集需要用到。\n \"\"\"\n user_test = dict()\n for u,i_list in user_movie_pair_test.items():\n user_test[u] = random.sample(user_movie_pair_test[u],1)[0]\n return user_test\n\n\ndef generate_train_batch(user_movie_pair_train,item_count,batch_size=50):\n t = []\n for b in range(batch_size):\n u = random.sample(user_movie_pair_train.keys(),1)[0]\n i = random.sample(user_movie_pair_train[u],1)[0]\n j = random.randint(0,item_count)\n while j in user_movie_pair_train[u]:\n j = random.randint(0,item_count)\n\n t.append([u,i,j])\n\n return np.asarray(t)\n\n\ndef generate_test_batch(user_ratings_test,user_movie_pair_test,item_count):\n \"\"\"\n 对于每个用户u,它的评分图书i是我们在user_ratings_test中随机抽取的,它的j是用户u所有没有借阅过的图书集合,\n 比如用户u有1000本书没有借阅,那么这里该用户的测试集样本就有1000个\n \"\"\"\n for u in user_movie_pair_test.keys():\n t = []\n i = user_ratings_test[u]\n for j in range(0,item_count):\n if not(j in user_movie_pair_test[u]):\n t.append([u,i,j])\n yield np.asarray(t)\n\n\ndef bpr_mf(user_count,item_count,hidden_dim):\n u = tf.placeholder(tf.int32,[None])\n i = tf.placeholder(tf.int32,[None])\n j = tf.placeholder(tf.int32,[None])\n\n user_emb_w = tf.get_variable(\"user_emb_w\", [user_count+1, hidden_dim],\n initializer=tf.random_normal_initializer(0, 0.1))\n item_emb_w = tf.get_variable(\"item_emb_w\", [item_count+1, hidden_dim],\n initializer=tf.random_normal_initializer(0, 0.1))\n\n u_emb = tf.nn.embedding_lookup(user_emb_w, u)\n i_emb = tf.nn.embedding_lookup(item_emb_w, i)\n j_emb = tf.nn.embedding_lookup(item_emb_w, j)\n\n\n x = tf.reduce_sum(tf.multiply(u_emb,(i_emb-j_emb)),1,keep_dims=True)\n\n mf_auc = tf.reduce_mean(tf.to_float(x>0))\n\n l2_norm = tf.add_n([\n tf.reduce_sum(tf.multiply(u_emb, u_emb)),\n tf.reduce_sum(tf.multiply(i_emb, i_emb)),\n tf.reduce_sum(tf.multiply(j_emb, j_emb))\n ])\n\n regulation_rate = 0.0001\n bprloss = regulation_rate * l2_norm - tf.reduce_mean(tf.log(tf.sigmoid(x)))\n\n train_op = tf.train.GradientDescentOptimizer(0.01).minimize(bprloss)\n return u, i, j, mf_auc, bprloss, train_op\n\nstart=time.clock()\nuser_count,item_count,user_movie_pair_train = load_data_train()\ntest_user_count,test_item_count,user_movie_pair_test = load_data_test()\nuser_ratings_test = generate_test(user_movie_pair_test)\n\nprint('user_ratings_test的值为:{}'.format(user_ratings_test))\nwith tf.Session() as sess:\n u,i,j,mf_auc,bprloss,train_op = bpr_mf(user_count,item_count,20)\n sess.run(tf.global_variables_initializer())\n\n for epoch in range(1,6):\n print('epoch的值为{}'.format(epoch))\n _batch_bprloss = 0\n for k in range(1,5000):\n\n uij = generate_train_batch(user_movie_pair_train,item_count)\n _bprloss,_train_op = sess.run([bprloss,train_op],\n feed_dict={u:uij[:,0],i:uij[:,1],j:uij[:,2]})\n\n _batch_bprloss += _bprloss\n\n print(\"epoch:\",epoch)\n print(\"bpr_loss:\",_batch_bprloss / k)\n print(\"_train_op\")\n\n user_count = 0\n _auc_sum = 0.0\n\n for t_uij in generate_test_batch(user_ratings_test,user_movie_pair_test,item_count):\n _auc, _test_bprloss = sess.run([mf_auc, bprloss],\n feed_dict={u: t_uij[:, 0], i: t_uij[:, 1], j: t_uij[:, 2]}\n )\n user_count += 1\n _auc_sum += _auc\n print(\"test_loss: \", _test_bprloss, \"test_auc: \", _auc_sum / user_count)\n print(\"\")\n variable_names = [v.name for v in tf.trainable_variables()]\n values = sess.run(variable_names)\n for k, v in zip(variable_names, values):\n print(\"Variable: \", k)\n print(\"Shape: \", v.shape)\n print(v)\n\n\nsession1 = tf.Session()\nu1_all = tf.matmul(values[0], values[1],transpose_b=True)\nresult_1 = session1.run(u1_all)\nprint (result_1)\n\n\np = np.squeeze(result_1)\n# np.argsort(p),将元素从小到大排列,提取对应的索引。找到了索引就是找到了书\nind = np.argsort(p)[:,-5:]\nprint('top5对应的索引为{}'.format(ind))\n\nnum=0\nall_num_user_item=0\nfor ii in range(len(user_movie_pair_test)):\n num_user_item=0\n for jj in user_movie_pair_test[ii]:\n num_user_item+=1\n if jj in (ind[ii]):\n num+=1\n all_num_user_item+=num_user_item\nprint('num的值为:{}'.format(num))\nprint('用户的数目为{}'.format(len(user_movie_pair_test)))\nprint('用户喜欢的物品的数目为:{}'.format(all_num_user_item))\nprint('召回率为{}'.format(num/all_num_user_item))\nprint('准确率为{}'.format(num/(len(user_movie_pair_test)*5)))\nduration=time.clock()-start\nprint('耗费时间:{}'.format(duration))",
"训练集借阅记录数:88\nnum_user: 66\nnum_book 88\n测试集借阅记录数:89\nnum_user: 66\nnum_book 88\nuser_ratings_test的值为:{37: 40, 60: 28, 58: 60, 49: 85, 36: 76, 25: 15, 47: 41, 16: 77, 23: 65, 27: 23, 0: 47, 31: 62, 48: 61, 30: 31, 43: 50, 28: 5, 7: 38, 34: 22, 21: 52, 29: 58, 26: 7, 50: 69, 10: 57, 40: 59, 39: 8, 18: 21, 32: 11, 59: 83, 65: 13, 1: 33, 6: 20, 44: 64, 63: 4, 19: 37, 55: 17, 17: 24, 54: 55, 42: 81, 56: 2, 57: 12, 11: 34, 61: 26, 53: 29, 64: 27, 2: 18, 41: 82, 35: 0, 12: 45, 46: 87, 15: 53, 51: 9, 52: 42, 20: 1, 9: 66, 22: 43, 13: 25, 62: 70, 3: 6, 45: 86, 8: 39, 4: 51, 24: 79, 38: 48, 5: 49, 14: 44, 33: 32}\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbc884aeafc0ae453a2771f5c407e53e15854d1e
| 6,413 |
ipynb
|
Jupyter Notebook
|
quiz/Quiz0_Solutions.ipynb
|
cliburn/bios-823-2020
|
842dde98b28899bb52b315fc2df10813629183bb
|
[
"MIT"
] | 13 |
2020-08-17T20:59:59.000Z
|
2021-09-27T16:30:59.000Z
|
quiz/Quiz0_Solutions.ipynb
|
cliburn/bios-823-2020
|
842dde98b28899bb52b315fc2df10813629183bb
|
[
"MIT"
] | null | null | null |
quiz/Quiz0_Solutions.ipynb
|
cliburn/bios-823-2020
|
842dde98b28899bb52b315fc2df10813629183bb
|
[
"MIT"
] | 11 |
2020-08-17T21:35:22.000Z
|
2021-09-19T16:05:45.000Z
| 24.291667 | 234 | 0.551848 |
[
[
[
"**Note**: There are multiple ways to solve these problems in SQL. Your solution may be quite different from mine and still be correct.",
"_____no_output_____"
],
[
"**1**. Connect to the SQLite3 database at `data/faculty.db` in the `notebooks` folder using the `sqlite` package or `ipython-sql` magic functions. Inspect the `sql` creation statement for each tables so you know their structure.",
"_____no_output_____"
]
],
[
[
"%load_ext sql",
"_____no_output_____"
],
[
"%sql sqlite:///../notebooks/data/faculty.db",
"_____no_output_____"
],
[
"%%sql\n\nSELECT sql FROM sqlite_master WHERE type='table';",
"_____no_output_____"
]
],
[
[
"2. Find the youngest and oldest faculty member(s) of each gender.",
"_____no_output_____"
]
],
[
[
"%%sql\n\nSELECT min(age), max(age) FROM person",
"_____no_output_____"
],
[
"%%sql\n\nSELECT first, last, age, gender\nFROM person \nINNER JOIN gender \n ON person.gender_id = gender.gender_id\nWHERE age IN (SELECT min(age) FROM person) AND gender = 'Male'\nUNION\nSELECT first, last, age, gender\nFROM person \nINNER JOIN gender \n ON person.gender_id = gender.gender_id\nWHERE age IN (SELECT min(age) FROM person) AND gender = 'Female'\nUNION\nSELECT first, last, age, gender\nFROM person \nINNER JOIN gender \n ON person.gender_id = gender.gender_id\nWHERE age IN (SELECT max(age) FROM person) AND gender = 'Male'\nUNION\nSELECT first, last, age, gender\nFROM person \nINNER JOIN gender \n ON person.gender_id = gender.gender_id\nWHERE age IN (SELECT max(age) FROM person) AND gender = 'Female'\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"3. Find the median age of the faculty members who know Python.\n\nAs SQLite3 does not provide a median function, you can create a User Defined Function (UDF) to do this. See [documentation](https://docs.python.org/2/library/sqlite3.html#sqlite3.Connection.create_function).",
"_____no_output_____"
]
],
[
[
"import statistics",
"_____no_output_____"
],
[
"class Median:\n def __init__(self):\n self.acc = []\n\n def step(self, value):\n self.acc.append(value)\n\n def finalize(self):\n return statistics.median(self.acc)",
"_____no_output_____"
],
[
"import sqlite3\ncon = sqlite3.connect('../notebooks/data/faculty.db')\ncon.create_aggregate(\"Median\", 1, Median)",
"_____no_output_____"
],
[
"cr = con.cursor()\ncr.execute('SELECT median(age) FROM person')\ncr.fetchall()",
"_____no_output_____"
]
],
[
[
"4. Arrange countries by the average age of faculty in descending order. Countries are only included in the table if there are at least 3 faculty members from that country.",
"_____no_output_____"
]
],
[
[
"%%sql\n\nSELECT country, count(country), avg(age)\nFROM person\nINNER JOIN country\n ON person.country_id = country.country_id\nGROUP BY country\nHAVING count(*) > 3\nORDER BY age DESC\nLIMIT 3",
"_____no_output_____"
]
],
[
[
"5. Which country has the highest average body mass index (BMII) among the faculty? Recall that BMI is weight (kg) / (height (m))^2.",
"_____no_output_____"
]
],
[
[
"%%sql\n\nSELECT country, avg(weight / (height*height)) as avg_bmi\nFROM person\nINNER JOIN country\n ON person.country_id = country.country_id\nGROUP BY country\nORDER BY avg_bmi DESC\nLIMIT 3",
"_____no_output_____"
]
],
[
[
"6. Do obese faculty (BMI > 30) know more languages on average than non-obese faculty?",
"_____no_output_____"
]
],
[
[
"%%sql\n\nSELECT is_obese, avg(language)\nFROM (\n SELECT \n weight / (height*height) > 30 AS is_obese, \n count(language_name) AS language\n FROM person\n INNER JOIN person_language\n ON person.person_id = person_language.person_id\n INNER JOIN language\n ON person_language.language_id = language.language_id\n GROUP BY person.person_id\n)\nGROUP BY is_obese",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc893957328d81baa4f7e770207b2916c88961f
| 410,835 |
ipynb
|
Jupyter Notebook
|
notebooks/EDA.ipynb
|
dinabandhu50/FREQUENT_OPIATE_PRESCRIBER
|
3a434be2b15a987301bf165a6978c6fe24ad43d5
|
[
"MIT"
] | 1 |
2020-09-30T12:34:40.000Z
|
2020-09-30T12:34:40.000Z
|
notebooks/EDA.ipynb
|
dinabandhu50/FREQUENT_OPIATE_PRESCRIBER
|
3a434be2b15a987301bf165a6978c6fe24ad43d5
|
[
"MIT"
] | null | null | null |
notebooks/EDA.ipynb
|
dinabandhu50/FREQUENT_OPIATE_PRESCRIBER
|
3a434be2b15a987301bf165a6978c6fe24ad43d5
|
[
"MIT"
] | null | null | null | 148.102019 | 194,948 | 0.834547 |
[
[
[
"# Frequent opiate prescriber",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport numpy as np \nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nimport preprocessors as pp\n\nsns.set(style=\"darkgrid\")",
"_____no_output_____"
],
[
"data = pd.read_csv('../data/prescriber-info.csv')",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
]
],
[
[
"## Variable Separation",
"_____no_output_____"
]
],
[
[
"uniq_cols = ['NPI']",
"_____no_output_____"
],
[
"cat_cols = list(data.columns[1:5])\ncat_cols",
"_____no_output_____"
],
[
"num_cols = list(data.columns[5:-1])\n# print(num_cols)",
"_____no_output_____"
],
[
"target = [data.columns[-1]]\ntarget",
"_____no_output_____"
]
],
[
[
"## Categorical Variable Analysis & EDA",
"_____no_output_____"
],
[
"### Missing values",
"_____no_output_____"
]
],
[
[
"# chcking for missing values\ndata[cat_cols].isnull().sum()",
"_____no_output_____"
],
[
"# checking for missing value percentage\ndata[cat_cols].isnull().sum()/data.shape[0] *100",
"_____no_output_____"
],
[
"# checking for null value in drugs column\ndata[num_cols].isnull().sum().sum()",
"_____no_output_____"
],
[
"data['NPI'].nunique()",
"_____no_output_____"
]
],
[
[
"Remarks:\n\n1. We dont need `NPI` column it has all unique values.\n2. The `Credentials` column has missing values ~3% of total.\n<!-- 3. All the `med_clos` are sparse in nature -->",
"_____no_output_____"
],
[
"### Basic plots",
"_____no_output_____"
]
],
[
[
"data[num_cols].iloc[:,2].value_counts()",
"_____no_output_____"
],
[
"cat_cols",
"_____no_output_____"
],
[
"for item in cat_cols[1:]:\n print('-'*25)\n print(data[item].value_counts())\n",
"-------------------------\nCA 2562\nNY 1956\nFL 1570\nTX 1500\nPA 1211\nIL 1002\nOH 981\nMI 872\nNC 778\nMA 725\nNJ 649\nGA 613\nWA 578\nVA 568\nTN 552\nIN 533\nMD 509\nAZ 509\nWI 498\nMO 483\nMN 448\nCO 393\nSC 390\nCT 388\nKY 367\nLA 354\nAL 344\nOR 344\nOK 281\nPR 231\nIA 225\nAR 216\nKS 203\nWV 199\nMS 193\nNM 166\nUT 162\nNV 155\nME 147\nNE 137\nID 133\nNH 119\nRI 117\nDE 91\nHI 91\nSD 83\nDC 79\nMT 77\nND 66\nVT 65\nAK 39\nWY 38\nVI 3\nAE 2\nZZ 2\nGU 2\nAA 1\nName: State, dtype: int64\n-------------------------\nMD 7034\nM.D. 6772\nDDS 1145\nD.O. 866\nPA-C 845\n ... \nD.D.S., F.A.G.D. 1\nM,D 1\nBDS, DDS 1\nM.D., F.A.A.F.P. 1\nRN, CS, MS(N),FNP 1\nName: Credentials, Length: 887, dtype: int64\n-------------------------\nInternal Medicine 3194\nFamily Practice 2975\nDentist 2800\nNurse Practitioner 2512\nPhysician Assistant 1839\n ... \nMilitary Health Care Provider 1\nHealth Maintenance Organization 1\nMidwife 1\nPharmacy Technician 1\nSlide Preparation Facility 1\nName: Specialty, Length: 109, dtype: int64\n"
],
[
"cat_cols",
"_____no_output_____"
],
[
"# Gender analysis",
"_____no_output_____"
],
[
"plt.figure(figsize=(7,5))\nsns.countplot(data=data,x='Gender')\nplt.title('Count plot of Gender column')\nplt.show()",
"_____no_output_____"
],
[
"# State column",
"_____no_output_____"
],
[
"plt.figure(figsize=(15,5))\nsns.countplot(data=data,x='State')\nplt.title('Count plot of State column')\nplt.show()",
"_____no_output_____"
],
[
"# lets check out `Speciality` column",
"_____no_output_____"
],
[
"data['Specialty'].nunique()",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,5))\nsns.countplot(data=data,x='Specialty')\nplt.title('Count plot of Specialty column')\nplt.xticks(rotation=90)\nplt.show()",
"_____no_output_____"
],
[
"data['Specialty'].value_counts()[:20]",
"_____no_output_____"
],
[
"# filling missing values with mean",
"_____no_output_____"
]
],
[
[
"In `credentials` we can do lot more\n\n1. The credentals column have multiple occupation in the same row.\n2. \\[PHD, MD\\] and \\[MD, PHD\\] are treated differently.\n3. P,A, is treated different from P.A and PA\n4. MD ----- M.D. , M.D, M D, MD\\` \n5. This column is a mess",
"_____no_output_____"
]
],
[
[
"cat_cols",
"_____no_output_____"
]
],
[
[
"Remarks:\n\n1. We don't need `Credentials` column which is a real mess, the `Specialty` column has the same information as of `Credentials`. \n\n2. Cat Features to remove - `NPI`, `Credentials` \n\n3. Cat Features to keep - `Gender`, `State`, `Speciality` \n\n4. Cat encoder pipeline - \n 1. Gender - normal 1/0 encoding using category_encoders\n 2. State - Frequency encoding using category_encoders\n 3. Speciality - Frequency encoding ",
"_____no_output_____"
],
[
"### Numerical Variable Analysis & Engineering",
"_____no_output_____"
]
],
[
[
"for item in num_cols:\n print('-'*25)\n print(f'frequency - {data[item].nunique()}')\n",
"-------------------------\nfrequency - 208\n-------------------------\nfrequency - 123\n-------------------------\nfrequency - 81\n-------------------------\nfrequency - 230\n-------------------------\nfrequency - 72\n-------------------------\nfrequency - 268\n-------------------------\nfrequency - 255\n-------------------------\nfrequency - 420\n-------------------------\nfrequency - 145\n-------------------------\nfrequency - 148\n-------------------------\nfrequency - 699\n-------------------------\nfrequency - 158\n-------------------------\nfrequency - 158\n-------------------------\nfrequency - 106\n-------------------------\nfrequency - 107\n-------------------------\nfrequency - 367\n-------------------------\nfrequency - 660\n-------------------------\nfrequency - 130\n-------------------------\nfrequency - 198\n-------------------------\nfrequency - 181\n-------------------------\nfrequency - 168\n-------------------------\nfrequency - 186\n-------------------------\nfrequency - 113\n-------------------------\nfrequency - 86\n-------------------------\nfrequency - 214\n-------------------------\nfrequency - 95\n-------------------------\nfrequency - 133\n-------------------------\nfrequency - 94\n-------------------------\nfrequency - 128\n-------------------------\nfrequency - 155\n-------------------------\nfrequency - 159\n-------------------------\nfrequency - 142\n-------------------------\nfrequency - 100\n-------------------------\nfrequency - 198\n-------------------------\nfrequency - 129\n-------------------------\nfrequency - 57\n-------------------------\nfrequency - 433\n-------------------------\nfrequency - 90\n-------------------------\nfrequency - 158\n-------------------------\nfrequency - 157\n-------------------------\nfrequency - 110\n-------------------------\nfrequency - 119\n-------------------------\nfrequency - 92\n-------------------------\nfrequency - 206\n-------------------------\nfrequency - 331\n-------------------------\nfrequency - 69\n-------------------------\nfrequency - 152\n-------------------------\nfrequency - 333\n-------------------------\nfrequency - 207\n-------------------------\nfrequency - 434\n-------------------------\nfrequency - 94\n-------------------------\nfrequency - 102\n-------------------------\nfrequency - 88\n-------------------------\nfrequency - 295\n-------------------------\nfrequency - 142\n-------------------------\nfrequency - 115\n-------------------------\nfrequency - 184\n-------------------------\nfrequency - 139\n-------------------------\nfrequency - 100\n-------------------------\nfrequency - 148\n-------------------------\nfrequency - 139\n-------------------------\nfrequency - 104\n-------------------------\nfrequency - 124\n-------------------------\nfrequency - 71\n-------------------------\nfrequency - 71\n-------------------------\nfrequency - 184\n-------------------------\nfrequency - 52\n-------------------------\nfrequency - 196\n-------------------------\nfrequency - 152\n-------------------------\nfrequency - 354\n-------------------------\nfrequency - 183\n-------------------------\nfrequency - 165\n-------------------------\nfrequency - 77\n-------------------------\nfrequency - 109\n-------------------------\nfrequency - 246\n-------------------------\nfrequency - 222\n-------------------------\nfrequency - 271\n-------------------------\nfrequency - 116\n-------------------------\nfrequency - 137\n-------------------------\nfrequency - 226\n-------------------------\nfrequency - 75\n-------------------------\nfrequency - 237\n-------------------------\nfrequency - 200\n-------------------------\nfrequency - 254\n-------------------------\nfrequency - 62\n-------------------------\nfrequency - 91\n-------------------------\nfrequency - 222\n-------------------------\nfrequency - 288\n-------------------------\nfrequency - 625\n-------------------------\nfrequency - 496\n-------------------------\nfrequency - 118\n-------------------------\nfrequency - 274\n-------------------------\nfrequency - 233\n-------------------------\nfrequency - 122\n-------------------------\nfrequency - 101\n-------------------------\nfrequency - 85\n-------------------------\nfrequency - 132\n-------------------------\nfrequency - 148\n-------------------------\nfrequency - 201\n-------------------------\nfrequency - 401\n-------------------------\nfrequency - 681\n-------------------------\nfrequency - 87\n-------------------------\nfrequency - 107\n-------------------------\nfrequency - 80\n-------------------------\nfrequency - 74\n-------------------------\nfrequency - 187\n-------------------------\nfrequency - 135\n-------------------------\nfrequency - 85\n-------------------------\nfrequency - 124\n-------------------------\nfrequency - 264\n-------------------------\nfrequency - 113\n-------------------------\nfrequency - 97\n-------------------------\nfrequency - 196\n-------------------------\nfrequency - 136\n-------------------------\nfrequency - 93\n-------------------------\nfrequency - 80\n-------------------------\nfrequency - 155\n-------------------------\nfrequency - 98\n-------------------------\nfrequency - 114\n-------------------------\nfrequency - 218\n-------------------------\nfrequency - 99\n-------------------------\nfrequency - 191\n-------------------------\nfrequency - 198\n-------------------------\nfrequency - 361\n-------------------------\nfrequency - 102\n-------------------------\nfrequency - 102\n-------------------------\nfrequency - 209\n-------------------------\nfrequency - 156\n-------------------------\nfrequency - 761\n-------------------------\nfrequency - 117\n-------------------------\nfrequency - 700\n-------------------------\nfrequency - 260\n-------------------------\nfrequency - 105\n-------------------------\nfrequency - 329\n-------------------------\nfrequency - 248\n-------------------------\nfrequency - 452\n-------------------------\nfrequency - 252\n-------------------------\nfrequency - 98\n-------------------------\nfrequency - 219\n-------------------------\nfrequency - 187\n-------------------------\nfrequency - 121\n-------------------------\nfrequency - 295\n-------------------------\nfrequency - 536\n-------------------------\nfrequency - 222\n-------------------------\nfrequency - 142\n-------------------------\nfrequency - 82\n-------------------------\nfrequency - 183\n-------------------------\nfrequency - 136\n-------------------------\nfrequency - 107\n-------------------------\nfrequency - 82\n-------------------------\nfrequency - 443\n-------------------------\nfrequency - 529\n-------------------------\nfrequency - 73\n-------------------------\nfrequency - 263\n-------------------------\nfrequency - 271\n-------------------------\nfrequency - 92\n-------------------------\nfrequency - 205\n-------------------------\nfrequency - 116\n-------------------------\nfrequency - 106\n-------------------------\nfrequency - 282\n-------------------------\nfrequency - 183\n-------------------------\nfrequency - 148\n-------------------------\nfrequency - 90\n-------------------------\nfrequency - 267\n-------------------------\nfrequency - 76\n-------------------------\nfrequency - 76\n-------------------------\nfrequency - 158\n-------------------------\nfrequency - 87\n-------------------------\nfrequency - 132\n-------------------------\nfrequency - 104\n-------------------------\nfrequency - 125\n-------------------------\nfrequency - 134\n-------------------------\nfrequency - 99\n-------------------------\nfrequency - 222\n-------------------------\nfrequency - 682\n-------------------------\nfrequency - 89\n-------------------------\nfrequency - 51\n-------------------------\nfrequency - 74\n-------------------------\nfrequency - 117\n-------------------------\nfrequency - 125\n-------------------------\nfrequency - 145\n-------------------------\nfrequency - 312\n-------------------------\nfrequency - 292\n-------------------------\nfrequency - 161\n-------------------------\nfrequency - 350\n-------------------------\nfrequency - 176\n-------------------------\nfrequency - 83\n-------------------------\nfrequency - 107\n-------------------------\nfrequency - 155\n-------------------------\nfrequency - 208\n-------------------------\nfrequency - 428\n-------------------------\nfrequency - 119\n-------------------------\nfrequency - 112\n-------------------------\nfrequency - 436\n-------------------------\nfrequency - 328\n-------------------------\nfrequency - 95\n-------------------------\nfrequency - 98\n-------------------------\nfrequency - 252\n-------------------------\nfrequency - 104\n-------------------------\nfrequency - 93\n-------------------------\nfrequency - 61\n-------------------------\nfrequency - 314\n-------------------------\nfrequency - 100\n-------------------------\nfrequency - 78\n-------------------------\nfrequency - 152\n-------------------------\nfrequency - 122\n-------------------------\nfrequency - 288\n-------------------------\nfrequency - 143\n-------------------------\nfrequency - 280\n-------------------------\nfrequency - 149\n-------------------------\nfrequency - 121\n-------------------------\nfrequency - 342\n-------------------------\nfrequency - 690\n-------------------------\nfrequency - 117\n-------------------------\nfrequency - 246\n-------------------------\nfrequency - 205\n-------------------------\nfrequency - 107\n-------------------------\nfrequency - 139\n-------------------------\nfrequency - 63\n-------------------------\nfrequency - 172\n-------------------------\nfrequency - 262\n-------------------------\nfrequency - 414\n-------------------------\nfrequency - 199\n-------------------------\nfrequency - 126\n-------------------------\nfrequency - 206\n-------------------------\nfrequency - 188\n-------------------------\n"
],
[
"print(f'Min \\t Average \\t Max \\t Prob>0')\nfor item in num_cols:\n print('-'*40)\n prob = sum(data[item] > 0) / data[item].shape[0]\n print(f'{data[item].min()}\\t{data[item].mean(): .4f} \\t{data[item].max()} \\t {prob:.4f}')",
"Min \t Average \t Max \t Prob>0\n----------------------------------------\n0\t 3.1572 \t770 \t 0.0637\n----------------------------------------\n0\t 2.3704 \t644 \t 0.0853\n----------------------------------------\n0\t 1.0537 \t356 \t 0.0467\n----------------------------------------\n0\t 7.0410 \t1105 \t 0.1614\n----------------------------------------\n0\t 0.7084 \t275 \t 0.0317\n----------------------------------------\n0\t 8.9628 \t2431 \t 0.1728\n----------------------------------------\n0\t 9.3057 \t790 \t 0.1818\n----------------------------------------\n0\t 16.1281 \t5716 \t 0.2061\n----------------------------------------\n0\t 2.3948 \t596 \t 0.0664\n----------------------------------------\n0\t 4.3472 \t430 \t 0.1373\n----------------------------------------\n0\t 44.3146 \t2429 \t 0.3250\n----------------------------------------\n0\t 2.8432 \t665 \t 0.0798\n----------------------------------------\n0\t 6.0060 \t617 \t 0.2028\n----------------------------------------\n0\t 2.7837 \t511 \t 0.1116\n----------------------------------------\n0\t 0.9858 \t1072 \t 0.0285\n----------------------------------------\n0\t 15.7880 \t1873 \t 0.2276\n----------------------------------------\n0\t 39.5564 \t3665 \t 0.3074\n----------------------------------------\n0\t 1.5150 \t1122 \t 0.0442\n----------------------------------------\n0\t 7.5719 \t780 \t 0.2087\n----------------------------------------\n0\t 3.3026 \t611 \t 0.0886\n----------------------------------------\n0\t 3.0960 \t477 \t 0.0937\n----------------------------------------\n0\t 3.7726 \t624 \t 0.0997\n----------------------------------------\n0\t 1.6150 \t494 \t 0.0584\n----------------------------------------\n0\t 0.9588 \t359 \t 0.0386\n----------------------------------------\n0\t 2.7067 \t1800 \t 0.0442\n----------------------------------------\n0\t 0.8848 \t309 \t 0.0305\n----------------------------------------\n0\t 1.2786 \t698 \t 0.0276\n----------------------------------------\n0\t 1.1218 \t517 \t 0.0393\n----------------------------------------\n0\t 2.1952 \t306 \t 0.0765\n----------------------------------------\n0\t 2.9993 \t882 \t 0.0894\n----------------------------------------\n0\t 2.8691 \t434 \t 0.0832\n----------------------------------------\n0\t 2.3500 \t992 \t 0.0680\n----------------------------------------\n0\t 1.2618 \t324 \t 0.0472\n----------------------------------------\n0\t 2.9498 \t1242 \t 0.0576\n----------------------------------------\n0\t 1.4513 \t955 \t 0.0409\n----------------------------------------\n0\t 0.6372 \t122 \t 0.0311\n----------------------------------------\n0\t 16.6003 \t1623 \t 0.2249\n----------------------------------------\n0\t 0.8456 \t270 \t 0.0288\n----------------------------------------\n0\t 3.4250 \t412 \t 0.1054\n----------------------------------------\n0\t 4.2962 \t571 \t 0.1518\n----------------------------------------\n0\t 1.1457 \t464 \t 0.0382\n----------------------------------------\n0\t 1.3663 \t492 \t 0.0442\n----------------------------------------\n0\t 0.9022 \t331 \t 0.0343\n----------------------------------------\n0\t 7.2761 \t1260 \t 0.2100\n----------------------------------------\n0\t 12.4999 \t1380 \t 0.2101\n----------------------------------------\n0\t 1.0438 \t190 \t 0.0518\n----------------------------------------\n0\t 1.7283 \t590 \t 0.0438\n----------------------------------------\n0\t 11.2784 \t4762 \t 0.1837\n----------------------------------------\n0\t 5.4953 \t1078 \t 0.1368\n----------------------------------------\n0\t 18.1012 \t1757 \t 0.2347\n----------------------------------------\n0\t 1.2238 \t392 \t 0.0461\n----------------------------------------\n0\t 1.2981 \t271 \t 0.0486\n----------------------------------------\n0\t 1.3856 \t157 \t 0.0581\n----------------------------------------\n0\t 10.7466 \t1742 \t 0.1862\n----------------------------------------\n0\t 3.3464 \t774 \t 0.1114\n----------------------------------------\n0\t 1.5742 \t382 \t 0.0515\n----------------------------------------\n0\t 4.4573 \t2297 \t 0.1207\n----------------------------------------\n0\t 2.0304 \t679 \t 0.0600\n----------------------------------------\n0\t 1.3038 \t440 \t 0.0484\n----------------------------------------\n0\t 2.7456 \t311 \t 0.0835\n----------------------------------------\n0\t 2.3570 \t465 \t 0.0733\n----------------------------------------\n0\t 1.8271 \t300 \t 0.0685\n----------------------------------------\n0\t 2.5110 \t326 \t 0.0861\n----------------------------------------\n0\t 1.0293 \t371 \t 0.0480\n----------------------------------------\n0\t 0.8081 \t356 \t 0.0356\n----------------------------------------\n0\t 4.5984 \t1144 \t 0.1221\n----------------------------------------\n0\t 0.5467 \t116 \t 0.0277\n----------------------------------------\n0\t 3.0597 \t939 \t 0.0667\n----------------------------------------\n0\t 1.9886 \t513 \t 0.0481\n----------------------------------------\n0\t 9.9764 \t1813 \t 0.1515\n----------------------------------------\n0\t 1.8335 \t858 \t 0.0279\n----------------------------------------\n0\t 3.3642 \t1323 \t 0.0926\n----------------------------------------\n0\t 0.7875 \t765 \t 0.0323\n----------------------------------------\n0\t 2.2835 \t310 \t 0.0878\n----------------------------------------\n0\t 7.8734 \t1204 \t 0.1691\n----------------------------------------\n0\t 5.0352 \t1099 \t 0.1202\n----------------------------------------\n0\t 8.1249 \t1059 \t 0.1644\n----------------------------------------\n0\t 1.8532 \t705 \t 0.0624\n----------------------------------------\n0\t 1.5391 \t896 \t 0.0365\n----------------------------------------\n0\t 4.6361 \t1224 \t 0.1103\n----------------------------------------\n0\t 0.6960 \t413 \t 0.0282\n----------------------------------------\n0\t 6.0914 \t717 \t 0.1272\n----------------------------------------\n0\t 3.4177 \t1292 \t 0.0786\n----------------------------------------\n0\t 5.4874 \t1696 \t 0.1064\n----------------------------------------\n0\t 0.6587 \t187 \t 0.0329\n----------------------------------------\n0\t 1.8199 \t201 \t 0.0798\n----------------------------------------\n0\t 7.2282 \t1363 \t 0.1694\n----------------------------------------\n0\t 12.1769 \t2313 \t 0.2239\n----------------------------------------\n0\t 33.1264 \t3181 \t 0.3004\n----------------------------------------\n0\t 26.1796 \t1805 \t 0.3144\n----------------------------------------\n0\t 2.0210 \t955 \t 0.0704\n----------------------------------------\n0\t 6.8652 \t940 \t 0.1180\n----------------------------------------\n0\t 6.3118 \t922 \t 0.1366\n----------------------------------------\n0\t 2.0847 \t239 \t 0.0700\n----------------------------------------\n0\t 1.2196 \t268 \t 0.0463\n----------------------------------------\n0\t 1.2848 \t254 \t 0.0531\n----------------------------------------\n0\t 1.2414 \t409 \t 0.0321\n----------------------------------------\n0\t 2.1901 \t508 \t 0.0650\n----------------------------------------\n0\t 4.4926 \t722 \t 0.1113\n----------------------------------------\n0\t 21.4727 \t1134 \t 0.2735\n----------------------------------------\n0\t 38.3233 \t5249 \t 0.4018\n----------------------------------------\n0\t 0.7806 \t287 \t 0.0310\n----------------------------------------\n0\t 0.9271 \t532 \t 0.0289\n----------------------------------------\n0\t 1.0490 \t165 \t 0.0473\n----------------------------------------\n0\t 0.6863 \t273 \t 0.0299\n----------------------------------------\n0\t 5.4752 \t829 \t 0.1585\n----------------------------------------\n0\t 2.1718 \t576 \t 0.0658\n----------------------------------------\n0\t 0.7488 \t233 \t 0.0269\n----------------------------------------\n0\t 1.5318 \t761 \t 0.0483\n----------------------------------------\n0\t 6.9781 \t861 \t 0.1372\n----------------------------------------\n0\t 1.0879 \t681 \t 0.0314\n----------------------------------------\n0\t 1.0062 \t471 \t 0.0372\n----------------------------------------\n0\t 4.9468 \t755 \t 0.1167\n----------------------------------------\n0\t 1.6740 \t1337 \t 0.0480\n----------------------------------------\n0\t 1.1896 \t205 \t 0.0486\n----------------------------------------\n0\t 0.9702 \t152 \t 0.0403\n----------------------------------------\n0\t 3.2020 \t406 \t 0.0930\n----------------------------------------\n0\t 1.2828 \t434 \t 0.0481\n----------------------------------------\n0\t 1.4040 \t390 \t 0.0454\n----------------------------------------\n0\t 3.3603 \t974 \t 0.0705\n----------------------------------------\n0\t 1.8493 \t238 \t 0.0727\n----------------------------------------\n0\t 4.6093 \t534 \t 0.1178\n----------------------------------------\n0\t 4.9841 \t728 \t 0.1246\n----------------------------------------\n0\t 7.6578 \t2858 \t 0.0649\n----------------------------------------\n0\t 1.0975 \t476 \t 0.0368\n----------------------------------------\n0\t 1.4439 \t207 \t 0.0519\n----------------------------------------\n0\t 3.5700 \t705 \t 0.0806\n----------------------------------------\n0\t 4.1746 \t447 \t 0.1351\n----------------------------------------\n0\t 45.7890 \t3300 \t 0.2975\n----------------------------------------\n0\t 1.5334 \t626 \t 0.0512\n----------------------------------------\n0\t 47.0497 \t1926 \t 0.3421\n----------------------------------------\n0\t 8.8335 \t754 \t 0.1728\n----------------------------------------\n0\t 0.9206 \t444 \t 0.0272\n----------------------------------------\n0\t 11.5414 \t1011 \t 0.1958\n----------------------------------------\n0\t 6.7295 \t1003 \t 0.1378\n----------------------------------------\n0\t 22.1488 \t2503 \t 0.2572\n----------------------------------------\n0\t 6.7808 \t4194 \t 0.1378\n----------------------------------------\n0\t 1.0840 \t655 \t 0.0370\n----------------------------------------\n0\t 2.6667 \t3605 \t 0.0323\n----------------------------------------\n0\t 4.7552 \t642 \t 0.1296\n----------------------------------------\n0\t 2.2614 \t625 \t 0.0781\n----------------------------------------\n"
],
[
"print(f'Maximun of all maxs - {data[num_cols].max().max()}')\nprint(f'Average of all maxs - {data[num_cols].max().mean()}')\nprint(f'Minimun of all maxs - {data[num_cols].max().min()}')\nprint(f'Maximun of all mins - {data[num_cols].min().max()}')\nprint(f'Minimun of all mins - {data[num_cols].min().min()}')",
"Maximun of all maxs - 6029\nAverage of all maxs - 961.464\nMinimun of all maxs - 92\nMaximun of all mins - 0\nMinimun of all mins - 0\n"
],
[
"sns.distplot(data[num_cols[0]]);",
"/home/db/anaconda3/envs/fop/lib/python3.8/site-packages/seaborn/distributions.py:2551: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"sns.boxplot(data = data, x = num_cols[0],orient=\"v\");",
"/home/db/anaconda3/envs/fop/lib/python3.8/site-packages/seaborn/_core.py:1303: UserWarning: Vertical orientation ignored with only `x` specified.\n warnings.warn(single_var_warning.format(\"Vertical\", \"x\"))\n"
]
],
[
[
"Problem:\n\n1. All the continuous cols have large number of zeros, and other values are counting value.\n2. The solutions I stumble accross are - `Two-part-models(twopm)`, `hurdle models` and `zero inflated poisson models(ZIP)`\n3. These models thinks the target variable has lots of zero and the non-zero values are not 1, if they had been 1s and 0s we could use a a classification model but they are like 0s mostly and if not zeros they are continuous variable like 100,120, 234, 898, etc.\n4. In our case our feature variable has lots of zeros.",
"_____no_output_____"
]
],
[
[
"data[data[num_cols[0]] > 0][num_cols[0]]",
"_____no_output_____"
],
[
"temp = 245\nsns.distplot(data[data[num_cols[temp]] > 0][num_cols[temp]]);",
"/home/db/anaconda3/envs/fop/lib/python3.8/site-packages/seaborn/distributions.py:2551: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"temp = 5\nsns.distplot(np.log(data[data[num_cols[temp]] > 0][num_cols[temp]]));",
"/home/db/anaconda3/envs/fop/lib/python3.8/site-packages/seaborn/distributions.py:2551: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"from sklearn.preprocessing import power_transform\n\ntemp = 5\n# data_without_0 = data[data[num_cols[temp]] > 0][num_cols[temp]]\ndata_without_0 = data[num_cols[temp]]\ndata_0 = np.array(data_without_0).reshape(-1,1)\ndata_0_trans = power_transform(data_0, method='yeo-johnson')\nsns.distplot(data_0_trans);",
"/home/db/anaconda3/envs/fop/lib/python3.8/site-packages/seaborn/distributions.py:2551: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"temp = 5\n# data_without_0 = data[data[num_cols[temp]] > 0][num_cols[temp]]\ndata_without_0 = data[num_cols[temp]]\ndata_0 = np.array(data_without_0).reshape(-1,1)\ndata_0_trans = power_transform(data_0+1, method='box-cox')\n# data_0_trans = np.log(data_0 + 1 )\n# data_0\nsns.distplot(data_0_trans);",
"/home/db/anaconda3/envs/fop/lib/python3.8/site-packages/seaborn/distributions.py:2551: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).\n warnings.warn(msg, FutureWarning)\n"
],
[
"from sklearn.decomposition import PCA",
"_____no_output_____"
],
[
"pca = PCA(n_components=0.8,svd_solver='full')\n# pca = PCA(n_components='mle',svd_solver='full')",
"_____no_output_____"
],
[
"pca.fit(data[num_cols])",
"_____no_output_____"
],
[
"pca_var_ratio = pca.explained_variance_ratio_\npca_var_ratio",
"_____no_output_____"
],
[
"len(pca_var_ratio)",
"_____no_output_____"
],
[
"plt.plot(pca_var_ratio[:],'-*');",
"_____no_output_____"
],
[
"sum(pca_var_ratio[:10])",
"_____no_output_____"
],
[
"data[num_cols].sample(2)",
"_____no_output_____"
],
[
"pca.transform(data[num_cols].sample(1))",
"_____no_output_____"
],
[
"pca2 = pp.PCATransformer(cols=num_cols,n_components=0.8)",
"_____no_output_____"
],
[
"pca2.fit(data)",
"/home/db/anaconda3/envs/fop/lib/python3.8/site-packages/sklearn/base.py:209: FutureWarning: From version 0.24, get_params will raise an AttributeError if a parameter cannot be retrieved as an instance attribute. Previously it would return None.\n warnings.warn('From version 0.24, get_params will raise an '\n"
],
[
"pca2.transform(data[num_cols].sample(1))",
"_____no_output_____"
]
],
[
[
"### Train test split and data saving",
"_____no_output_____"
]
],
[
[
"# train test split\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X = data.drop(target,axis=1)\ny = data[target]\nX_train, X_test, y_train, y_test = train_test_split(X,y, test_size=0.20, random_state=1)",
"_____no_output_____"
],
[
"pd.concat([X_train,y_train],axis=1).to_csv('../data/train.csv',index=False)\npd.concat([X_test,y_test],axis=1).to_csv('../data/test.csv',index=False)",
"_____no_output_____"
]
],
[
[
"## Data Engineering",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import LabelBinarizer",
"_____no_output_____"
],
[
"lbin = LabelBinarizer()\nlbin.fit(X_train['Gender'])",
"_____no_output_____"
],
[
"gen_tra = lbin.transform(X_train['Gender'])\ngen_tra",
"_____no_output_____"
],
[
"X_train[num_cols[:5]].info();",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 20000 entries, 6655 to 235\nData columns (total 5 columns):\n # Column Non-Null Count Dtype\n--- ------ -------------- -----\n 0 ABILIFY 20000 non-null int64\n 1 ACETAMINOPHEN.CODEINE 20000 non-null int64\n 2 ACYCLOVIR 20000 non-null int64\n 3 ADVAIR.DISKUS 20000 non-null int64\n 4 AGGRENOX 20000 non-null int64\ndtypes: int64(5)\nmemory usage: 937.5 KB\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbc8a7e37dac2e4b84a779c78529f3ed7d0a104d
| 5,611 |
ipynb
|
Jupyter Notebook
|
sentiment_classification.ipynb
|
jbyte23/diploma
|
3d0f0584820a69413cb5671d0cd74299af169cfb
|
[
"MIT"
] | null | null | null |
sentiment_classification.ipynb
|
jbyte23/diploma
|
3d0f0584820a69413cb5671d0cd74299af169cfb
|
[
"MIT"
] | null | null | null |
sentiment_classification.ipynb
|
jbyte23/diploma
|
3d0f0584820a69413cb5671d0cd74299af169cfb
|
[
"MIT"
] | null | null | null | 5,611 | 5,611 | 0.646052 |
[
[
[
"# Priprava okolja",
"_____no_output_____"
]
],
[
[
"!pip install transformers\n!pip install sentencepiece",
"_____no_output_____"
],
[
"import csv\nimport torch\nfrom torch import nn\nfrom transformers import AutoTokenizer, AutoModel\nimport pandas as pd\nfrom google.colab import drive\n\nimport transformers\nimport json\nfrom tqdm import tqdm\nfrom torch.utils.data import Dataset, DataLoader\n\nRANDOM_SEED = 42\n\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")",
"_____no_output_____"
]
],
[
[
"# Pomožni razredi in funkcije",
"_____no_output_____"
]
],
[
[
"class SentimentClassifier(nn.Module):\n\n def __init__(self, n_classes):\n super(SentimentClassifier, self).__init__()\n self.model = AutoModel.from_pretrained('EMBEDDIA/sloberta')\n self.pre_classifier = torch.nn.Linear(768, 768)\n self.dropout = torch.nn.Dropout(0.2)\n self.classifier = nn.Linear(self.model.config.hidden_size, n_classes)\n \n\n def forward(self, input_ids, attention_mask):\n output = self.model(\n input_ids=input_ids, \n attention_mask=attention_mask\n )\n last_hidden_state = output[0]\n pooler = last_hidden_state[:, 0, :]\n pooler = self.dropout(pooler)\n pooler = self.pre_classifier(pooler)\n pooler = torch.nn.ReLU()(pooler)\n pooler = self.dropout(pooler)\n output = self.classifier(pooler)\n return output\n\n\nclass ArticleTestDataset(torch.utils.data.Dataset):\n def __init__(self, dataframe, tokenizer, max_len):\n self.tokenizer = tokenizer\n self.df = dataframe\n self.text = dataframe.body\n self.max_len = max_len\n\n def __getitem__(self, idx):\n text = str(self.text[idx])\n\n inputs = tokenizer.encode_plus(\n text,\n None,\n add_special_tokens=True,\n padding='max_length',\n truncation=True,\n max_length=self.max_len,\n return_attention_mask=True,\n return_token_type_ids=True\n )\n \n input_ids = inputs['input_ids']\n attention_mask = inputs['attention_mask']\n\n return {\n 'text': text,\n 'input_ids': torch.tensor(input_ids, dtype=torch.long),\n 'attention_mask': torch.tensor(attention_mask, dtype=torch.long),\n }\n\n def __len__(self):\n return len(self.text)\n\n\ndef get_predictions(model, data_loader):\n model = model.eval()\n predictions = []\n\n data_iterator = tqdm(data_loader, desc=\"Iteration\")\n with torch.no_grad():\n for step, d in enumerate(data_iterator):\n input_ids = d[\"input_ids\"].to(device)\n attention_mask = d[\"attention_mask\"].to(device)\n outputs = model(\n input_ids=input_ids,\n attention_mask=attention_mask\n )\n _, preds = torch.max(outputs, dim=1)\n predictions.extend(preds)\n \n predictions = torch.stack(predictions).cpu()\n \n return predictions\n",
"_____no_output_____"
]
],
[
[
"# MAIN",
"_____no_output_____"
]
],
[
[
"model_path = '/content/drive/MyDrive/Diploma/best_model_state_latest.bin'",
"_____no_output_____"
],
[
"MAX_LEN = 512\nBATCH_SIZE = 8\n\ntest_params = {'batch_size': BATCH_SIZE,\n 'shuffle': False,\n 'num_workers': 0\n }\n\ntokenizer = AutoTokenizer.from_pretrained('EMBEDDIA/sloberta', use_fast=False)\nmodel = SentimentClassifier(3)\nmodel.load_state_dict(torch.load(model_path))\nmodel = model.to(device)",
"_____no_output_____"
],
[
"# V naslednji vrstici lako spremenite vrednost. Možne vrednosti so:\n# \"2019_slovenija_sentiment\",\n# \"2019_svet_sentiment\",\n# \"2020_korona_sentiment\",\n# \"2020_svet_sentiment\",\n# \"2020_slovenska_politika_sentiment\",\nfile_name = '2019_slovenija_sentiment'\nfilepath = f'/content/drive/MyDrive/Diploma/data/{file_name}.pkl'\ndata = pd.read_pickle(filepath)\n\ndataloader = DataLoader(ArticleTestDataset(data, tokenizer, MAX_LEN), **test_params)",
"_____no_output_____"
],
[
"preds = get_predictions(model, dataloader)\ndata['sentiment'] = preds",
"_____no_output_____"
],
[
"# data.to_pickle(filepath)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbc8a94556a135973d3ffac0ca8a48239a8e15bc
| 36,421 |
ipynb
|
Jupyter Notebook
|
hm-ensembling.ipynb
|
ManashJKonwar/Kaggle-HM-Recommender
|
abd6e7f040b6560ded1f7f67fe68dff40aa58ff7
|
[
"MIT"
] | null | null | null |
hm-ensembling.ipynb
|
ManashJKonwar/Kaggle-HM-Recommender
|
abd6e7f040b6560ded1f7f67fe68dff40aa58ff7
|
[
"MIT"
] | null | null | null |
hm-ensembling.ipynb
|
ManashJKonwar/Kaggle-HM-Recommender
|
abd6e7f040b6560ded1f7f67fe68dff40aa58ff7
|
[
"MIT"
] | null | null | null | 41.200226 | 175 | 0.545729 |
[
[
[
"import os\nimport numpy as np\nimport pandas as pd\nimport gc",
"_____no_output_____"
]
],
[
[
"# To ensemble I used submissions from 8 public notebooks:\n* LB: 0.0225 - https://www.kaggle.com/lunapandachan/h-m-trending-products-weekly-add-test/notebook\n* LB: 0.0217 - https://www.kaggle.com/tarique7/hnm-exponential-decay-with-alternate-items/notebook\n* LB: 0.0221 - https://www.kaggle.com/astrung/lstm-sequential-modelwith-item-features-tutorial\n* LB: 0.0224 - https://www.kaggle.com/code/hirotakanogami/h-m-eda-customer-clustering-by-kmeans\n* LB: 0.0220 - https://www.kaggle.com/code/hengzheng/time-is-our-best-friend-v2/notebook\n* LB: 0.0227 - https://www.kaggle.com/code/hechtjp/h-m-eda-rule-base-by-customer-age\n* LB: 0.0231 - https://www.kaggle.com/code/ebn7amdi/trending/notebook?scriptVersionId=90980162\n* LB: 0.0225 - https://www.kaggle.com/code/mayukh18/svd-model-reranking-implicit-to-explicit-feedback",
"_____no_output_____"
]
],
[
[
"sub0 = pd.read_csv('../input/hm-00231-solution/submission.csv').sort_values('customer_id').reset_index(drop=True) # 0.0231\nsub1 = pd.read_csv('../input/handmbestperforming/h-m-trending-products-weekly-add-test.csv').sort_values('customer_id').reset_index(drop=True) # 0.0225\nsub2 = pd.read_csv('../input/handmbestperforming/hnm-exponential-decay-with-alternate-items.csv').sort_values('customer_id').reset_index(drop=True) # 0.0217\nsub3 = pd.read_csv('../input/handmbestperforming/lstm-sequential-modelwith-item-features-tutorial.csv').sort_values('customer_id').reset_index(drop=True) # 0.0221\nsub4 = pd.read_csv('../input/hm-00224-solution/submission.csv').sort_values('customer_id').reset_index(drop=True) # 0.0224\nsub5 = pd.read_csv('../input/handmbestperforming/time-is-our-best-friend-v2.csv').sort_values('customer_id').reset_index(drop=True) # 0.0220\nsub6 = pd.read_csv('../input/handmbestperforming/rule-based-by-customer-age.csv').sort_values('customer_id').reset_index(drop=True) # 0.0227\nsub7 = pd.read_csv('../input/h-m-faster-trending-products-weekly/submission.csv').sort_values('customer_id').reset_index(drop=True) # 0.0231\nsub8 = pd.read_csv('../input/h-m-framework-for-partitioned-validation/submission.csv').sort_values('customer_id').reset_index(drop=True) # 0.0225",
"_____no_output_____"
],
[
"sub0.columns = ['customer_id', 'prediction0']\nsub0['prediction1'] = sub1['prediction']\nsub0['prediction2'] = sub2['prediction']\nsub0['prediction3'] = sub3['prediction']\nsub0['prediction4'] = sub4['prediction']\nsub0['prediction5'] = sub5['prediction']\nsub0['prediction6'] = sub6['prediction']\nsub0['prediction7'] = sub7['prediction'].astype(str)\n\ndel sub1, sub2, sub3, sub4, sub5, sub6, sub7\ngc.collect()\nsub0.head()",
"_____no_output_____"
],
[
"def cust_blend(dt, W = [1,1,1,1,1,1,1,1]):\n #Create a list of all model predictions\n REC = []\n\n # Second Try\n REC.append(dt['prediction0'].split())\n REC.append(dt['prediction1'].split())\n REC.append(dt['prediction2'].split())\n REC.append(dt['prediction3'].split())\n REC.append(dt['prediction4'].split())\n REC.append(dt['prediction5'].split())\n REC.append(dt['prediction6'].split())\n REC.append(dt['prediction7'].split())\n\n #Create a dictionary of items recommended.\n #Assign a weight according the order of appearance and multiply by global weights\n res = {}\n for M in range(len(REC)):\n for n, v in enumerate(REC[M]):\n if v in res:\n res[v] += (W[M]/(n+1))\n else:\n res[v] = (W[M]/(n+1))\n\n # Sort dictionary by item weights\n res = list(dict(sorted(res.items(), key=lambda item: -item[1])).keys())\n\n # Return the top 12 items only\n return ' '.join(res[:12])\n\nsub0['prediction'] = sub0.apply(cust_blend, W = [1.05, 0.78, 0.86, 0.85, 0.68, 0.64, 0.70, 0.24], axis=1)\n\nsub0.head()",
"_____no_output_____"
]
],
[
[
"# Make a submission",
"_____no_output_____"
]
],
[
[
"del sub0['prediction0']\ndel sub0['prediction1']\ndel sub0['prediction2']\ndel sub0['prediction3']\ndel sub0['prediction4']\ndel sub0['prediction5']\ndel sub0['prediction6']\ndel sub0['prediction7']\ngc.collect()",
"_____no_output_____"
],
[
"sub1 = pd.read_csv('../input/h-m-framework-for-partitioned-validation/submission.csv').sort_values('customer_id').reset_index(drop=True)\nsub1['prediction'] = sub1['prediction'].astype(str)\n\nsub0.columns = ['customer_id', 'prediction0']\nsub0['prediction1'] = sub1['prediction']\n\ndel sub1\ngc.collect()",
"_____no_output_____"
],
[
"def cust_blend(dt, W = [1,1,1,1,1]):\n #Global ensemble weights\n #W = [1.15,0.95,0.85]\n\n #Create a list of all model predictions\n REC = []\n\n # Second Try\n REC.append(dt['prediction0'].split())\n REC.append(dt['prediction1'].split())\n\n #Create a dictionary of items recommended.\n #Assign a weight according the order of appearance and multiply by global weights\n res = {}\n for M in range(len(REC)):\n for n, v in enumerate(REC[M]):\n if v in res:\n res[v] += (W[M]/(n+1))\n else:\n res[v] = (W[M]/(n+1))\n\n # Sort dictionary by item weights\n res = list(dict(sorted(res.items(), key=lambda item: -item[1])).keys())\n\n # Return the top 12 items only\n return ' '.join(res[:12])",
"_____no_output_____"
],
[
"sub0['prediction'] = sub0.apply(cust_blend, W = [1.20, 0.85], axis=1)",
"_____no_output_____"
],
[
"del sub0['prediction0']\ndel sub0['prediction1']\n\nsub0.head()",
"_____no_output_____"
],
[
"sub0.to_csv('submission.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc8b82703f8c2a38f000d9b9cb558353b950ee7
| 19,948 |
ipynb
|
Jupyter Notebook
|
notebook/Unit8-2-GCC_DLL.ipynb
|
hervey-su/home
|
655b9e7b8180592742a132832795170a00debb47
|
[
"MIT"
] | 2 |
2019-03-13T15:34:42.000Z
|
2019-03-13T15:34:47.000Z
|
notebook/Unit8-2-GCC_DLL.ipynb
|
hervey-su/home
|
655b9e7b8180592742a132832795170a00debb47
|
[
"MIT"
] | null | null | null |
notebook/Unit8-2-GCC_DLL.ipynb
|
hervey-su/home
|
655b9e7b8180592742a132832795170a00debb47
|
[
"MIT"
] | null | null | null | 25.314721 | 386 | 0.509926 |
[
[
[
"# The Shared Library with GCC\n\nWhen your program is linked against a shared library, only a small table is created in the executable. Before the executable starts running, **the operating system loads the machine code needed for the external functions** - a process known as **dynamic linking.** \n\n \n* Dynamic linking makes executable files smaller and saves disk space, because `one` copy of a **library** can be **shared** between `multiple` programs. \n\n\n* Furthermore, most operating systems allows one copy of a shared library in memory to be used by all running programs, thus, saving memory. \n\n\n* The shared library codes can be upgraded without the need to recompile your program.\n\n\nA **shared library** has file extension of \n\n * **`.so`** (shared objects) in `Linux(Unixes)`\n \n \n * **`.dll** (dynamic link library) in `Windows`. \n",
"_____no_output_____"
],
[
"## 1: Building the shared library\n\nThe shared library we will build consist of a single source file: `SumArray.c/h`\n\nWe will compile the C file with `Position Independent Code( PIC )` into a shared library。\n\nGCC assumes that all libraries \n \n* `start` with `lib`\n\n* `end` with `.dll`(windows) or `.so`(Linux),\n\nso, we should name the shared library begin with `lib prefix` and the `.so/.dll` extensions.\n\n* libSumArray.dll(Windows)\n\n* libSumArray.so(Linux)\n",
"_____no_output_____"
],
[
"\n#### Under Windows",
"_____no_output_____"
]
],
[
[
"!gcc -c -O3 -Wall -fPIC -o ./demo/bin/SumArray.o ./demo/src/SumArray.c\n!gcc -shared -o ./demo/bin/libSumArray.dll ./demo/bin/SumArray.o",
"_____no_output_____"
],
[
"!dir .\\demo\\bin\\libSumArray.*",
"_____no_output_____"
]
],
[
[
"#### under Linux",
"_____no_output_____"
]
],
[
[
"!gcc -c -O3 -Wall -fPIC -o ./demo/obj/SumArray.o ./demo/gcc/SumArray.c\n!gcc -shared -o ./cdemo/bin/libSumArray ./demo/obj/SumArray.o ",
"_____no_output_____"
],
[
"!ls ./demo/bin/libSumArray.*",
"_____no_output_____"
]
],
[
[
"\n* `-c`: compile into object file with default name : funs.o.\n\n By default, the object file has the same name as the source file with extension of \".o\" \n \n \n* `-O3`: Optimize yet more.\n\n turns on all optimizations specified by -O2 and also turns on the -finline-functions, -fweb, -frename-registers and -funswitch-loops optionsturns on all optimizations \n \n \n* `-Wall`: prints \"all\" compiler's warning message. \n\n This option should always be used, in order to generate better code.\n\n\n* **`-fPIC`** : stands for `Position Independent Code`(位置无关代码)\n \n the generated machine code is `not dependent` on being located at a `specific address` in order to `work`.\n \n Position-independent code can be `executed` at `any memory address`\n \n \n* **-shared:** creating a shared library\n",
"_____no_output_____"
]
],
[
[
"%%file ./demo/makefile-SumArray-dll\n\nCC=gcc\nCFLAGS=-O3 -Wall -fPIC \n\nSRCDIR= ./demo/src/\nOBJDIR= ./demo/obj/\nBINDIR= ./demo/bin/\n\nall: libdll\n\nlibdll: obj\n\t $(CC) -shared -o $(BINDIR)libSumArray.dll $(OBJDIR)SumArray.o\n\t del .\\demo\\obj\\SumArray.o\n \nobj: ./demo/src/SumArray.c\n\t $(CC) -c $(CFLAGS) -o $(OBJDIR)SumArray.o $(SRCDIR)SumArray.c\n \nclean:\n\t del .\\demo\\src\\libSumArray.dll",
"_____no_output_____"
],
[
"!make -f ./demo/makefile-SumArray-dll",
"_____no_output_____"
],
[
"!dir .\\demo\\bin\\libSum*.dll",
"_____no_output_____"
]
],
[
[
"#### Under Linux",
"_____no_output_____"
]
],
[
[
"%%file ./code/makefile-SumArray-so\n\n\nCC=gcc\nCFLAGS=-O3 -Wall -fPIC\n\nSRCDIR= ./demo/src/\nOBJDIR= ./demo/obj/\nBINDIR= ./demo/bin/\n\nall: libdll\n\nlibdll: obj\n\t $(CC) -shared -o $(BINDIR)libSumArray.dll $(OBJDIR)SumArray.o\n\t rm -f ./demo/obj/SumArray.o\n \nobj: ./demo/src/SumArray.c\n\t $(CC) -c $(CFLAGS) -o $(OBJDIR)SumArray.o $(SRCDIR)SumArray.c\n \nclean:\n\t rm -f ./demo/src/libSumArray.dll\n",
"_____no_output_____"
],
[
"!make -f ./code/makefile-SumArray-so",
"_____no_output_____"
],
[
"!ls ./code/bin/libSum*.so",
"_____no_output_____"
]
],
[
[
"## 2 Building a client executable ",
"_____no_output_____"
],
[
"### Header Files and Libraries \n\n* `Header File`: When compiling the program, the **compiler** needs the **header** files to compile the source codes;\n\n* `libraries`: the **linker** needs the **libraries** to resolve external references from other object files or libraries. \n\nThe `compiler` and `linker` will not find the `headers/libraries` unless you set **the appropriate options**\n\n* **1 Searching for Header Files**\n\n **`-Idir`:** The include-paths are specified via **-Idir** option (`uppercase` 'I' followed by the directory path or environment variable **CPATH**). \n \n \n* **2 Searching for libraries Files**\n\n **`-Ldir`**: The library-path is specified via **-Ldir** option (`uppercase` 'L' followed by the directory path(or environment variable **LIBRARY_PATH**). \n\n\n* **3 Linking the library**\n\n **`-llibname`**: Link with the library name **without** the `lib` prefix and the `.so/.dll` extensions.\n \n Windows\n ```bash\n -I./demo/src/ -L./demo/bin/ -lSumArray\n ```\n Linux\n ```bash\n -I./demo/src/ -L./demo/bin/ -lSumArray -Wl,-rpath=./demo/bin/ \n ```\n\n * **`-Wl,option`**\n\n Pass option as an option to the **linker**. If option contains `commas`, it is split into multiple options at the commas. You can use this syntax to pass an argument to the option. For example, -Wl,-Map,output.map passes -Map output.map to the linker. When using the GNU linker, you can also get the same effect with `-Wl,-Map=output.map'.\n \n * **`-rpath=dir`** \n\n **Add a directory to the runtime library search path**. This is used when linking an ELF executable with shared objects. All -rpath arguments are concatenated and passed to the runtime linker, which uses them to locate shared objects at runtime. The -rpath option is also used when locating shared objects which are needed by shared objects explicitly included in the link;\n \n",
"_____no_output_____"
],
[
"---\nThe following source code `\"mainSum.c\"` demonstrates calling the DLL's functions: \n\n**NOTE:** mainSum.c is the same code in multi-source example",
"_____no_output_____"
]
],
[
[
"%%file ./demo/src/mainSum.c\n\n#include <stdio.h> \n#include \"SumArray.h\"\n\nint main() {\n \n int a1[] = {8, 4, 5, 3, 2};\n printf(\"sum is %d\\n\", sum(a1, 5)); // sum is 22\n return 0;\n}",
"_____no_output_____"
]
],
[
[
"#### Windows",
"_____no_output_____"
]
],
[
[
"!gcc -c -o ./demo/obj/mainSum.o ./demo/src/mainSum.c \n!gcc -o ./demo/bin/mainSum ./demo/obj/mainSum.o -I./demo/src/ -L./demo/bin/ -lSumArray",
"_____no_output_____"
],
[
"!.\\demo\\bin\\mainSum",
"_____no_output_____"
]
],
[
[
"#### Linux",
"_____no_output_____"
]
],
[
[
"!gcc -c -o ./demo/obj/mainSum.o ./demo/obj/mainSum.c \n!gcc -o ./demo/bin/mainSum ./demo/obj/mainSum.o -I./demo/obj/ -L./demo/bin/ -lSumArray -Wl,-rpath=./demo/bin/",
"_____no_output_____"
],
[
"!ldd ./demo/bin/mainSum",
"_____no_output_____"
],
[
"!./code/demo/mainSum",
"_____no_output_____"
]
],
[
[
"#### Under Windows",
"_____no_output_____"
]
],
[
[
"%%file ./demo/makefile-call-dll\n\nSRCDIR= ./demo/src/\nOBJDIR= ./demo/obj/\nBINDIR= ./demo/bin/\n\nall: mainexe\n\nclean:\n\tdel .\\demo\\bin\\mainSum.exe\n\nmainexe: sumobj $(SRCDIR)SumArray.h \n\tgcc -o $(BINDIR)mainSum.exe $(OBJDIR)mainSum.o -I$(SRCDIR) -L$(BINDIR) -lSumArray\n\tdel .\\demo\\obj\\mainSum.o\n\nsumobj: $(SRCDIR)mainSum.c \n\tgcc -c -o $(OBJDIR)mainSum.o $(SRCDIR)mainSum.c ",
"_____no_output_____"
],
[
"!make -f ./demo/makefile-call-dll",
"_____no_output_____"
],
[
"!.\\demo\\bin\\mainSum",
"_____no_output_____"
]
],
[
[
"#### Under Linux",
"_____no_output_____"
]
],
[
[
"%%file ./demo/makefile-call-so\n\n\nSRCDIR= ./demo/src/\nOBJDIR= ./demo/obj/\nBINDIR= ./demo/bin/\n\nall: main\n\nclean:\n\trm -f ./demo/bin/mainSum.exe\n\nmain: sumobj $(SRCDIR)SumArray.h \n\tgcc -o $(BINDIR)mainSum.exe $(OBJDIR)mainSum.o -I$(SRCDIR) -L$(BINDIR) -lSumArray -Wl,-rpath=./code/bin/ \n\trm -f ./demo/obj/mainSum.o\n\nsumobj: $(SRCDIR)mainSum.c \n\tgcc -c -o $(OBJDIR)mainSum.o $(SRCDIR)mainSum.c \n ",
"_____no_output_____"
],
[
"!make -f ./demo/makefile-call-so",
"_____no_output_____"
],
[
"!./demo/bin/mainSum",
"_____no_output_____"
]
],
[
[
"## 3 Building a `shared library` with `multi-source` files\n\nThe shared library we will build consist of a multi-source files\n\n* funs.c/h\n\n* SumArray.c/h",
"_____no_output_____"
]
],
[
[
"%%file ./demo/src/funs.h\n\n#ifndef FUNS_H\n#define FUNS_H\n\ndouble dprod(double *x, int n);\nint factorial(int n);\n\n#endif",
"_____no_output_____"
],
[
"%%file ./demo/src/funs.c\n\n#include \"funs.h\"\n\n// x[0]*x[1]*...*x[n-1]\ndouble dprod(double *x, int n)\n{\n double y = 1.0;\n for (int i = 0; i < n; i++)\n {\n y *= x[i];\n }\n return y;\n}\n\n// The factorial of a positive integer n, denoted by n!, is the product of all positive integers less than or equal to n. \n// For example,5!=5*4*3*2*1=120\n// The value of 0! is 1 \nint factorial(int n)\n{\n if (n == 0 ) {\n return 1;\n }\n else \n {\n return n * factorial(n - 1);\n }\n}",
"_____no_output_____"
]
],
[
[
"#### Building `funs.c` and `SumArray.c` into libmultifuns.dll",
"_____no_output_____"
]
],
[
[
"!gcc -c -O3 -Wall -fPIC -o ./demo/obj/funs.o ./demo/src/funs.c \n!gcc -c -O3 -Wall -fPIC -o ./demo/obj/SumArray.o ./demo/src/SumArray.c\n!gcc -shared -o ./demo/bin/libmultifuns.dll ./demo/obj/funs.o ./demo/obj/SumArray.o",
"_____no_output_____"
],
[
"!dir .\\demo\\bin\\libmulti*.dll",
"_____no_output_____"
]
],
[
[
"#### Building with makefile\n",
"_____no_output_____"
]
],
[
[
"%%file ./demo/makefile-libmultifun\n\nCC=gcc\nCFLAGS=-O3 -Wall -fPIC\n\nSRCDIR= ./demo/src/\nOBJDIR= ./demo/obj/\nBINDIR= ./demo/bin/\n\nall: libmultifuns.dll\n\nlibmultifuns.dll: multifunsobj\n\t $(CC) -shared -o $(BINDIR)libmultifuns.dll $(OBJDIR)funs.o $(OBJDIR)SumArray.o\n\t del .\\demo\\obj\\funs.o .\\demo\\obj\\SumArray.o\n \nmultifunsobj: $(SRCDIR)funs.c $(SRCDIR)SumArray.c\n\t$(CC) -c $(CFLAGS) -o $(OBJDIR)SumArray.o $(SRCDIR)SumArray.c\n\t$(CC) -c $(CFLAGS) -o $(OBJDIR)funs.o $(SRCDIR)funs.c \n \n \nclean:\n\t del .\\demo\\bin\\libmultifuns.dll",
"_____no_output_____"
],
[
"!make -f ./demo/makefile-libmultifun",
"_____no_output_____"
]
],
[
[
"The result is a compiled shared library **`libmultifuns.dll`**",
"_____no_output_____"
],
[
"##### makefile-libmultifun - more vars",
"_____no_output_____"
]
],
[
[
"%%file ./code/makefile-libmultifun\n\nCC=gcc\nCFLAGS=-O3 -Wall -fPIC \n\nSRCDIR= ./demo/src/\nOBJDIR= ./demo/obj/\nBINDIR= ./demo/bin/\n\nINC = -I$(SRCDIR) \n\nSRCS= $(SRCDIR)funs.c \\\n $(SRCDIR)SumArray.c \n\nall: libmultifuns.dll\n\nlibmultifuns.dll: multifunsobj\n\t $(CC) -shared -o $(BINDIR)libmultifuns.dll funs.o SumArray.o\n\t del funs.o SumArray.o\n \nmultifunsobj: \n\t $(CC) -c $(CFLAGS) $(INC) $(SRCS) \n \nclean:\n\t del .\\demo\\bin\\libmultifuns.dll",
"_____no_output_____"
],
[
"!make -f ./code/makefile-libmultifun",
"_____no_output_____"
]
],
[
[
"##### Building a client executable \n\nThe following source code `\"mainMultifuns.c\"` demonstrates calling the DLL's functions:\n",
"_____no_output_____"
]
],
[
[
"%%file ./demo/src/mainMultifuns.c\n\n#include <stdio.h> \n#include \"SumArray.h\"\n#include \"funs.h\"\n\nint main() {\n \n int a1[] = {8, 4, 5, 3, 2};\n printf(\"sum is %d\\n\", sum(a1, 5)); // sum is 22\n \n double a2[] = {8.0, 4.0, 5.0, 3.0, 2.0};\n printf(\"dprod is %f\\n\", dprod(a2, 5)); // dprod is 960\n \n int n =5;\n printf(\"the factorial of %d is %d\\n\",n,factorial(n)); // 5!=120\n return 0;\n}",
"_____no_output_____"
],
[
"!gcc -c -o ./demo/obj/mainMultifuns.o ./demo/src/mainMultifuns.c \n!gcc -o ./demo/bin/mainMultifuns ./demo/obj/mainMultifuns.o -I./demo/src/ -L./demo/bin/ -lmultifuns",
"_____no_output_____"
],
[
"!.\\demo\\bin\\mainMultifuns",
"_____no_output_____"
]
],
[
[
"## Reference\n\n* GCC (GNU compilers) http://gcc.gnu.org\n\n * GCC Manual http://gcc.gnu.org/onlinedocs\n\n * An Introduction to GCC http://www.network-theory.co.uk/docs/gccintro/index.html.\n\n * GCC and Make:Compiling, Linking and Building C/C++ Applications http://www3.ntu.edu.sg/home/ehchua/programming/cpp/gcc_make.html\n\n * MinGW-W64 (GCC) Compiler Suite: http://www.mingw-w64.org/doku.php\n\n\n* C/C++ for VS Code https://code.visualstudio.com/docs/languages/cpp\n\n* C/C++ Preprocessor Directives http://www.cplusplus.com/doc/tutorial/preprocessor/\n\n\n* What is a DLL and How Do I Create or Use One? http://www.mingw.org/wiki/DLL\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbc8c4337e3585c0cc93964eaec6a9a79a995ba9
| 8,211 |
ipynb
|
Jupyter Notebook
|
notebooks/UniProt_refined_query.ipynb
|
Lean-Mean-Protein-Machine-Learning/LMPM
|
e163d5e7ea634e6acc43aff56605c0da002265c1
|
[
"MIT"
] | 1 |
2021-03-18T00:18:15.000Z
|
2021-03-18T00:18:15.000Z
|
notebooks/UniProt_refined_query.ipynb
|
Lean-Mean-Protein-Machine-Learning/LMPM
|
e163d5e7ea634e6acc43aff56605c0da002265c1
|
[
"MIT"
] | null | null | null |
notebooks/UniProt_refined_query.ipynb
|
Lean-Mean-Protein-Machine-Learning/LMPM
|
e163d5e7ea634e6acc43aff56605c0da002265c1
|
[
"MIT"
] | 4 |
2021-02-05T06:34:42.000Z
|
2021-03-16T20:16:42.000Z
| 35.240343 | 288 | 0.601267 |
[
[
[
"# Searching the UniProt database and saving fastas:\n\nThis notebook is really just to demonstrate how Andrew finds the sequences for the datasets. <br>\n\nIf you do call it from within our github repository, you'll probably want to add the fastas to the `.gitignore` file.",
"_____no_output_____"
]
],
[
[
"# Import bioservices module, to run remote UniProt queries\n# (will probably need to pip install this to use)\nfrom bioservices import UniProt\n",
"_____no_output_____"
]
],
[
[
"## Connecting to UniProt using bioservices:\n",
"_____no_output_____"
]
],
[
[
"service = UniProt() \nfasta_path = 'refined_query_fastas/' #optional file organization param",
"_____no_output_____"
]
],
[
[
"## Query with signal_peptide",
"_____no_output_____"
]
],
[
[
"def data_saving_function_with_SP(organism,save_path=''):\n \n secreted_query = f'(((organism:{organism} OR host:{organism}) annotation:(\"signal peptide\") keyword:secreted) NOT annotation:(type:transmem)) AND reviewed:yes'\n secreted_result = service.search(secreted_query, frmt=\"fasta\")\n secreted_outfile = f'{save_path}{organism}_secreted_SP_new.fasta'\n with open(secreted_outfile, 'a') as ofh:\n ofh.write(secreted_result)\n \n\n cytoplasm_query = f'(((organism:{organism} OR host:{organism}) locations:(location:cytoplasm)) NOT (annotation:(type:transmem) OR annotation:(\"signal peptide\"))) AND reviewed:yes'\n cytoplasm_result = service.search(cytoplasm_query, frmt=\"fasta\")\n cytoplasm_outfile = f'{save_path}{organism}_cytoplasm_SP_new.fasta'\n with open(cytoplasm_outfile, 'a') as ofh:\n ofh.write(cytoplasm_result)\n \n membrane_query = f'(((organism:{organism} OR host:{organism}) annotation:(type:transmem)) annotation:(\"signal peptide\")) AND reviewed:yes'\n membrane_result = service.search(membrane_query, frmt=\"fasta\")\n membrane_outfile = f'{save_path}{organism}_membrane_SP_new.fasta'\n with open(membrane_outfile, 'a') as ofh:\n ofh.write(membrane_result)\n \n\n ",
"_____no_output_____"
],
[
"data_saving_function_with_SP('human',fasta_path)",
"_____no_output_____"
],
[
"data_saving_function_with_SP('escherichia',fasta_path)",
"_____no_output_____"
]
],
[
[
"## Query without signal_peptide",
"_____no_output_____"
]
],
[
[
"def data_saving_function_without_SP(organism,save_path=''):\n# maybe new: \n\n secreted_query = f'(((organism:{organism} OR host:{organism}) AND (keyword:secreted OR goa:(\"extracellular region [5576]\"))) NOT (annotation:(type:transmem) OR goa:(\"membrane [16020]\") OR locations:(location:cytoplasm) OR goa:(\"cytoplasm [5737]\") )) AND reviewed:yes'\n secreted_result = service.search(secreted_query, frmt=\"fasta\")\n secreted_outfile = f'{save_path}{organism}_secreted_noSP_new_new.fasta'\n with open(secreted_outfile, 'a') as ofh:\n ofh.write(secreted_result)\n \n cytoplasm_query = f'(((organism:{organism} OR host:{organism}) AND (locations:(location:cytoplasm) OR goa:(\"cytoplasm [5737]\")) ) NOT (annotation:(type:transmem) OR goa:(\"membrane [16020]\") OR keyword:secreted OR goa:(\"extracellular region [5576]\") )) AND reviewed:yes'\n cytoplasm_result = service.search(cytoplasm_query, frmt=\"fasta\")\n cytoplasm_outfile = f'{save_path}{organism}_cytoplasm_noSP_new_new.fasta'\n with open(cytoplasm_outfile, 'a') as ofh:\n ofh.write(cytoplasm_result)\n \n\n membrane_query= f'(((organism:{organism} OR host:{organism}) AND ( annotation:(type:transmem) OR goa:(\"membrane [16020]\") )) NOT ( keyword:secreted OR goa:(\"extracellular region [5576]\") OR locations:(location:cytoplasm) OR goa:(\"cytoplasm [5737]\") )) AND reviewed:yes'\n membrane_result = service.search(membrane_query, frmt=\"fasta\")\n membrane_outfile = f'{save_path}{organism}_membrane_noSP_new_new.fasta'\n with open(membrane_outfile, 'a') as ofh:\n ofh.write(membrane_result)\n \n\n ",
"_____no_output_____"
],
[
"data_saving_function_without_SP('human',fasta_path)",
"_____no_output_____"
],
[
"data_saving_function_without_SP('yeast',fasta_path)",
"_____no_output_____"
],
[
"data_saving_function_without_SP('escherichia',fasta_path)",
"_____no_output_____"
]
],
[
[
"## Query ALL SHIT (warning: do not do unless you have lots of free time and computer memory)",
"_____no_output_____"
]
],
[
[
"def data_saving_function_without_SP_full_uniprot(save_path=''):\n# maybe new: \n secreted_query = f'((keyword:secreted OR goa:(\"extracellular region [5576]\")) NOT (annotation:(type:transmem) OR goa:(\"membrane [16020]\") OR locations:(location:cytoplasm) OR goa:(\"cytoplasm [5737]\") )) AND reviewed:yes'\n secreted_result = service.search(secreted_query, frmt=\"fasta\")\n secreted_outfile = f'{save_path}all_secreted_noSP_new_new.fasta'\n with open(secreted_outfile, 'a') as ofh:\n ofh.write(secreted_result)\n \n cytoplasm_query = f'(( locations:(location:cytoplasm) OR goa:(\"cytoplasm [5737]\") ) NOT (annotation:(type:transmem) OR goa:(\"membrane [16020]\") OR keyword:secreted OR goa:(\"extracellular region [5576]\") )) AND reviewed:yes'\n cytoplasm_result = service.search(cytoplasm_query, frmt=\"fasta\")\n cytoplasm_outfile = f'{save_path}all_cytoplasm_noSP_new_new.fasta'\n with open(cytoplasm_outfile, 'a') as ofh:\n ofh.write(cytoplasm_result)\n \n membrane_query= f'(( annotation:(type:transmem) OR goa:(\"membrane [16020]\") ) NOT ( keyword:secreted OR goa:(\"extracellular region [5576]\") OR locations:(location:cytoplasm) OR goa:(\"cytoplasm [5737]\") )) AND reviewed:yes'\n membrane_result = service.search(membrane_query, frmt=\"fasta\")\n membrane_outfile = f'{save_path}all_membrane_noSP_new_new.fasta'\n with open(membrane_outfile, 'a') as ofh:\n ofh.write(membrane_result)\n \n\n ",
"_____no_output_____"
],
[
"data_saving_function_without_SP_full_uniprot(fasta_path)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbc8d455e4f6f95b2859c83c1b559b3ec2a13ce3
| 886,417 |
ipynb
|
Jupyter Notebook
|
dmu26/dmu26_XID+PACS_COSMOS/validation_report-PACS_red.ipynb
|
djbsmith/dmu_products
|
4a6e1496a759782057c87ab5a65763282f61c497
|
[
"MIT"
] | null | null | null |
dmu26/dmu26_XID+PACS_COSMOS/validation_report-PACS_red.ipynb
|
djbsmith/dmu_products
|
4a6e1496a759782057c87ab5a65763282f61c497
|
[
"MIT"
] | null | null | null |
dmu26/dmu26_XID+PACS_COSMOS/validation_report-PACS_red.ipynb
|
djbsmith/dmu_products
|
4a6e1496a759782057c87ab5a65763282f61c497
|
[
"MIT"
] | null | null | null | 1,446.030995 | 567,666 | 0.945922 |
[
[
[
"# Validation report for dmu26_XID+PACS_COSMOS_20170303",
"_____no_output_____"
],
[
"The data product dmu26_XID+PACS_COSMOS_20170303, contains three files:\n\n1. dmu26_XID+PACS_COSMOS_20170303.fits: The catalogue file\n2. dmu26_XID+PACS_COSMOS_20170303_Bayes_pval_PACS100.fits: The Bayesian pvalue map\n3. dmu26_XID+PACS_COSMOS_20170303_Bayes_pval_PACS160.fits: The Bayesian pvalue map",
"_____no_output_____"
],
[
"## Catalogue Validation\nValidation of the catalogue should cover the following as a minimum:\n\n* Compare XID+ Fluxes with previous catalogues\n* Check for sources with poor convergence (i.e. $\\hat{R}$ >1.2 and $n_{eff}$ <40)\n* Check for sources with strange error (i.e. small upper limit and large lower limit, which would be indicating prior is limiting flux)\n* Check for sources that return prior (i.e. probably very large flux and large error)\n* Check background estimate is similar across neighbouring tiles (will vary depending on depth of prior list)\n\n",
"_____no_output_____"
]
],
[
[
"from astropy.table import Table\nimport numpy as np\nimport pylab as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"table=Table.read('/Users/williamp/validation/cosmos/PACS/dmu26_XID+PACS_COSMOS_20170303.fits', format='fits')",
"WARNING: UnitsWarning: 'degrees' did not parse as fits unit: At col 0, Unit 'degrees' not supported by the FITS standard. [astropy.units.core]\nWARNING:astropy:UnitsWarning: 'degrees' did not parse as fits unit: At col 0, Unit 'degrees' not supported by the FITS standard. \n"
],
[
"table[:10].show_in_notebook()",
"_____no_output_____"
],
[
"import seaborn as sns",
":0: FutureWarning: IPython widgets are experimental and may change in the future.\n"
]
],
[
[
"### Comparison to previous catalogues\nUsing COSMOS2015 catalogue and matching to closest PACS objects within 1''",
"_____no_output_____"
]
],
[
[
"#table.sort('help_id')\n\nCOSMOS2015 = Table.read('/Users/williamp/validation/cosmos/PACS/COSMOS2015_Laigle+v1.1_wHELPids_PACS_red.fits')\nCOSMOS2015.sort('HELP_ID')",
"_____no_output_____"
],
[
"plt.scatter(np.log10(COSMOS2015['F_PACS_160']), np.log10(COSMOS2015['FLUX_160']))\nplt.xlabel('$\\log_{10}S_{160 \\mathrm{\\mu m}, XID+}$')\nplt.ylabel('$\\log_{10}S_{160 \\mathrm{\\mu m}, COSMOS2015}$')\nplt.show()\n\nplot=sns.jointplot(x=np.log10(COSMOS2015['F_PACS_160']), y=np.log10(COSMOS2015['FLUX_160']), xlim=(-1,3), kind='hex')\nplot.set_axis_labels('$\\log_{10}S_{160 \\mathrm{\\mu m}, XID+}$', '$\\log_{10}S_{160 \\mathrm{\\mu m}, COSMOS2015}$')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Agreement is reasonable good. Lower flux objects can be given lower flux density by XID+ as would be expecetd. High flux density objects have a convergance in XID+ and COSMOS2015 as would be expected.",
"_____no_output_____"
],
[
"### Convergence Statistics\ne.g. How many of the objects satisfy critera? \n(note Some of the $\\hat{R}$ values are NaN. This is a PyStan bug. They are most likely 1.0",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(20,10))\nplt.subplot(1,2,1)\nRhat=plt.hist(np.isfinite(table['Rhat_PACS_160']), bins=np.arange(0.9,1.2,0.01))\nplt.xlabel(r'$\\hat{R}$')\nplt.subplot(1,2,2)\nneff=plt.hist(table['n_eff_PACS_160'])\nplt.yscale('log')\nplt.xlabel(r'$n_{eff.}$')",
"_____no_output_____"
],
[
"numRhat = 0\nnumNeff = 0\n\nfor i in range(0, len(table)):\n if table['Rhat_PACS_160'][i] > 1.2 and np.isfinite(table['Rhat_PACS_160']):\n numRhat += 1\n if table['n_eff_PACS_160'][i] < 40:\n numNeff += 1\n \nprint(str(numRhat)+' objects have $\\hat{R}$ > 1.2')\nprint(str(numNeff)+' objects have n$_{eff}$ < 40')",
"0 objects have $\\hat{R}$ > 1.2\n0 objects have n$_{eff}$ < 40\n"
]
],
[
[
"All objects have good $\\hat{R}$ and n$_{eff}$ values",
"_____no_output_____"
],
[
"### Skewness",
"_____no_output_____"
]
],
[
[
"plot=sns.jointplot(x=np.log10(table['F_PACS_160']), y=(table['FErr_PACS_160_u']-table['F_PACS_160'])/(table['F_PACS_160']-table['FErr_PACS_160_l*1000.0']), xlim=(-1,2), kind='hex')\nplot.set_axis_labels(r'$\\log_{10}S_{160 \\mathrm{\\mu m}}$ ', r'$(84^{th}-50^{th})/(50^{th}-16^{th})$ percentiles')\n",
"_____no_output_____"
]
],
[
[
"### Sources where posterior=prior\nSuggest looking at size of errors to diagnose. How many appear to be returning prior? Where are they on Bayesian P value map? Does it make sense why?",
"_____no_output_____"
],
[
"The lower errors appear to be very, very small. The catalogue appears to have them multiplied by 1000 and they are still 10\\,000 times smaller than the upper errors.",
"_____no_output_____"
],
[
"### Background value\nAre all the background values similar? For those that aren't is it obvious why? (e.g. edge of map, extended source not fitted well etc)",
"_____no_output_____"
]
],
[
[
"plt.hist(table['Bkg_PACS_160'])\nplt.xlabel(r'Background (MJy/sr)')\nplt.show()",
"_____no_output_____"
]
],
[
[
"The background seems to have quite a large scatter but roughly consistent around ~-1.0",
"_____no_output_____"
],
[
"-------------\n## Bayesian P value map\nThe Bayesian P value map can be thought of as a more robust residual map. They provide the probability our model would obtain the pixel data value, having inferred our model on the data. The probabilitiy value is expressed in terms of a sigma value. \n\n* a value of < -2 indicates that our model cannot explain why there is so little flux in the map\n* a value of > -2 indicates our model cannot explain why there is so much flux in the map \n* a value of ~ 0 indicates a good fit\n\nPlotting the distribution of the Bayesian P value map can indicate whether the map has been been fit well in general. If the distribution is centered on 0 and roughly symmetric then it is a good fit.\n\nSuggested validation checks: \n\n* Check distribution is reasonable\n* Check for strange areas in map",
"_____no_output_____"
]
],
[
[
"import aplpy\nfrom astropy.io import fits",
"_____no_output_____"
],
[
"hdulist=fits.open('/Users/williamp/validation/cosmos/PACS/dmu26_XID+PACS_COSMOS_20170303_Bayes_pval_PACS160.fits')",
"_____no_output_____"
],
[
"plt.figure()\nBayes_hist=plt.hist(np.isfinite(hdulist[1].data), bins=np.arange(-6,6.1,0.05))\nplt.xlabel(r'$\\sigma$ value')",
"_____no_output_____"
],
[
"plt.show()",
"_____no_output_____"
]
],
[
[
"#Checking the Positions\nUsing the PACS map to check where the high flux objects and objects with highish bakground are.",
"_____no_output_____"
]
],
[
[
"hdulist = fits.open('/Users/williamp/dev/XID_plus/input/cosmosKs/pep_COSMOS_red_Map.DR1.fits')\n\nhdulist[1].header['CTYPE1'] = 'RA'\nhdulist[1].header['CTYPE2'] = 'DEC'",
"_____no_output_____"
]
],
[
[
"PACS_red map is in greyscale, object positions are in blue, objects with fluxes > 1e3 are in red, objects with background values > 1.25 MJy/sr are in yellow, objects with P-values > 0.5 are in green.",
"_____no_output_____"
]
],
[
[
"vmin=0.0001\nvmax=0.02\nfig = plt.figure(figsize=(30,10))\npltut = aplpy.FITSFigure(hdulist[1], figure=fig)\n\npltut.show_colorscale(vmin=vmin,vmax=vmax,cmap='Greys',stretch='log')\n\npltut.show_circles(table['RA'],table['Dec'], radius=0.0025, color='b')\n\nbRA = []\nbDec = []\nfor i in range(0, len(table)):\n if table['F_PACS_160'][i] > 1e3:\n bRA.append(table['RA'][i])\n bDec.append(table['Dec'][i])\nif len(bRA) > 0:\n pltut.show_circles(bRA, bDec, radius=0.005, color='r')\n\nbgRA = []\nbgDec = []\nfor i in range(0, len(table)):\n if table['Bkg_PACS_160'][i] > 1.25:\n bgRA.append(table['RA'][i])\n bgDec.append(table['Dec'][i])\nif len(bgRA) > 0:\n pltut.show_circles(bgRA, bgDec, radius=0.005, color='y')\n\npRA = []\npDec = []\nfor i in range(0, len(table)):\n if table['Pval_res_160'][i] > 0.5:\n pRA.append(table['RA'][i])\n pDec.append(table['Dec'][i])\nif len(pRA) > 0:\n pltut.show_circles(pRA, pDec, radius=0.005, color='g')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"The high flux value objects (red) appear to mainly cluster arround the edges of the masked regions, so may just be artefacts of bad masking.\n\nThe high background objects (yellow) appear to be a property of individual tiles as they form squares on the image above. These tiles are on the edge of the map and the high background is probably a result of this.\n\nThe high P-value objects (green) appear arround brighter objects (but not all bright objects) which makes sense; bright blended objects are harder to deblend than non-blended objects. They also often coincide with the high background tiles.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbc8d60a9477ec23117d2f16a814372e4efd0f3f
| 4,017 |
ipynb
|
Jupyter Notebook
|
006_Python_Sets_Methods/015_Python_Set_symmetric_difference_update().ipynb
|
Maxc390/python-discrete-structures
|
f7bc7d7caed16f357cc712630b562182355e072b
|
[
"MIT"
] | 175 |
2021-06-28T03:51:13.000Z
|
2022-03-25T06:29:14.000Z
|
006_Python_Sets_Methods/015_Python_Set_symmetric_difference_update().ipynb
|
Wangcx225/02_Python_Datatypes
|
57828db90aa8d960c62612ed33bb985483442e8a
|
[
"MIT"
] | null | null | null |
006_Python_Sets_Methods/015_Python_Set_symmetric_difference_update().ipynb
|
Wangcx225/02_Python_Datatypes
|
57828db90aa8d960c62612ed33bb985483442e8a
|
[
"MIT"
] | 164 |
2021-06-28T03:54:15.000Z
|
2022-03-25T08:08:53.000Z
| 23.910714 | 200 | 0.546179 |
[
[
[
"<small><small><i>\nAll the IPython Notebooks in this lecture series by Dr. Milan Parmar are available @ **[GitHub](https://github.com/milaan9/02_Python_Datatypes/tree/main/006_Python_Sets_Methods)**\n</i></small></small>",
"_____no_output_____"
],
[
"# Python Set `symmetric_difference_update()`\n\nThe **`symmetric_difference_update()`** method finds the symmetric difference of two sets and updates the set calling it.\n\nThe symmetric difference of two sets **`A`** and **`B`** is the set of elements that are in either **`A`** or **`B`**, but not in their intersection.\n\n<div>\n<img src=\"img/symmetric_difference_update.png\" width=\"250\"/>\n</div>",
"_____no_output_____"
],
[
"**Syntax**:\n\n```python\nA.symmetric_difference_update(B)\n```",
"_____no_output_____"
],
[
"## Return Value from `symmetric_difference_update()`\n\nThe **`symmetric_difference_update()`** returns None (returns nothing). Rather, it updates the set calling it.",
"_____no_output_____"
]
],
[
[
"# Example: Working of symmetric_difference_update()\n\nA = {'a', 'c', 'd'}\nB = {'c', 'd', 'e' }\n\nresult = A.symmetric_difference_update(B)\n\nprint('A =', A)\nprint('B =', B)\nprint('result =', result)",
"A = {'a', 'e'}\nB = {'d', 'e', 'c'}\nresult = None\n"
]
],
[
[
"Here, the set **`A`** is updated with the symmetric difference of set **`A`** and **`B`**. However, the set **`B`** is unchanged.",
"_____no_output_____"
],
[
"**Recommended Reading:** **[Python Set symmetric_difference()](https://github.com/milaan9/02_Python_Datatypes/blob/main/006_Python_Sets_Methods/014_Python_Set_symmetric_difference%28%29.ipynb)**",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbc8e0b2ca86d4a4904ffe9d3adc368309f77b29
| 124,787 |
ipynb
|
Jupyter Notebook
|
content/07/2/.ipynb_checkpoints/RNA World-checkpoint.ipynb
|
edur409/ASTROBIOLOGY200
|
868d02a10b4be5f71935325c43e89b02567e4e26
|
[
"MIT"
] | null | null | null |
content/07/2/.ipynb_checkpoints/RNA World-checkpoint.ipynb
|
edur409/ASTROBIOLOGY200
|
868d02a10b4be5f71935325c43e89b02567e4e26
|
[
"MIT"
] | null | null | null |
content/07/2/.ipynb_checkpoints/RNA World-checkpoint.ipynb
|
edur409/ASTROBIOLOGY200
|
868d02a10b4be5f71935325c43e89b02567e4e26
|
[
"MIT"
] | null | null | null | 89.77482 | 25,196 | 0.805965 |
[
[
[
"# RNA World Hypothesis\n\nRNA is a simpler cousin of DNA. As you may know, RNA is widely thought to be the first self replicating life-form to arise perhaps around 4 billion years ago. One of the strongest arguments for this theory is that RNA is able to carry information in its nucleotides like DNA, and like protein, it is able to adopt higher order structures to catalyze reactions, such as self replication. So it is likely, and there is growing evidence that this is the case, that the first form of replicating life was RNA. And because of this dual property of RNA as a hereditary information vessel as well as a structural/functional element we can use RNA molecules to build very nice population models. \n\nSo in this section, we'll be walking you through building genetic populations, simulating their evolution, and using statistics and other mathematical tools for understanding key properties of populations.",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\n\n# Youtube\nHTML('<iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/K1xnYFCZ9Yg\" frameborder=\"0\" allow=\"accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture\" allowfullscreen></iframe>')",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\IPython\\core\\display.py:694: UserWarning: Consider using IPython.display.IFrame instead\n warnings.warn(\"Consider using IPython.display.IFrame instead\")\n"
],
[
"#HTML('<iframe width=\"784\" height=\"441\" src=\"https://scaleofuniverse.com/\" /iframe>')",
"_____no_output_____"
]
],
[
[
"## Population Evolution in *an* RNA World\n\nIn order to study the evolution of a population, we first need a model of a population. And even before that, we need to define what we mean by *population*. Populations can be defined on many levels and with many diffferent criteria. For our purposes, we will simply say that a population is a set of individuals sharing a common environment. And because this is population *genetics* we can think of individuals as entities comprising of specific genes or chromosomes. \n\nSo where do we get a population from? As you may have discussed in previous workshops, there are very large datasets containing sequencing information from different populations. So we could download one of these datasets and perform some analysis on it. But I find this can be dry and tedious. So why download data when we can simply create our own?\n\nIn this section we're going to be creating and studying our own \"artificial\" populations to illustrate some important population genetics concepts and methodologies. Not only will this help you learn population genetics, but you will get a lot more programming practice than if we were to simply parse data files and go from there. \n\nMore specifically, we're going to build our own RNA world.\n\n### Building an RNA population\n\nAs we saw earlier, RNA has the nice property of posessing a strong mapping between information carrying (sequence) and function (structure). This is analogous to what is known in evolutionary terms as a genotype and a phenotype. With these properties, we have everything we need to model a population, and simulate its evolution.\n\n#### RNA sequence-structure\n\nWe can think of the genotype as a sequence $s$ consisting of letters/nucleotides from the alphabet $\\{U,A,C,G\\}$. The corresponding phenotype $\\omega$ is the secondary structure of $s$ which can be thought of as a pairing between nucleotides in the primary sequence that give rise to a 2D architecture. Because it has been shown that the function of many biomolecules, including RNA, is driven by structure this gives us a good proxy for phenotype. \n\nBelow is an example of what an RNA secondary structure, or pairing, looks like.",
"_____no_output_____"
]
],
[
[
"### 1\n\nfrom IPython.display import Image\n#This will load an image of an RNA secondary structure\nImage(url='https://viennarna.github.io/forgi/_images/1y26_ss.png')\n",
"_____no_output_____"
],
[
"from IPython.display import HTML\n\n# Youtube\nHTML('<iframe width=\"560\" height=\"315\" src=\"https://www.youtube.com/embed/JQByjprj_mA\" frameborder=\"0\" allow=\"accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture\" allowfullscreen></iframe>')\n#import matplotlib.pyplot as plt\n#import forgi.visual.mplotlib as fvm\n#import forgi\n#cg = forgi.load_rna(\"1y26.fx\", allow_many=False)\n#fvm.plot_rna(cg, text_kwargs={\"fontweight\":\"black\"}, lighten=0.7,\n# backbone_kwargs={\"linewidth\":3})\n#plt.show()",
"_____no_output_____"
]
],
[
[
"As you can see, unpaired positions are forming loop-like structures, and paired positions are forming stem-like structures. It is this spatial arrangement of nucleotides that drives RNA's function. Therefore, another sequence that adopts a similar shape, is likely to behave in a similar manner. Another thing to notice is that, although in reality this is often not the case, in general we only allow pairs between $\\{C,G\\}$ and $\\{A, U\\}$ nucleotides, most modern approaches allow for non-canonical pairings and you will find some examples of this in the above structure.\n\n*How do we go from a sequence to a structure?*\n\nSo a secondary structure is just a list of pairings between positions. How do we get the optimal pairing?\n\nThe algorithm we're going to be using in our simulations is known as the Nussinov Algorithm. The Nussinov algorithm is one of the first and simplest attempts at predicting RNA structure. Because bonds tend to stabilize RNA, the algorithm tries to maximize the number of pairs in the structure and return that as its solution. Current approaches achieve more accurate solutions by using energy models based one experimental values to then obtain a structure that minimizes free energy. But since we're not really concerned with the accuracy of our predictions, Nussinov is a good entry point. Furthermore, the main algorithmic concepts are the same between Nussinov and state of the art RNA structure prediction algorithms. I implemented the algorithm in a separate file called `fold.py` that we can import and use its functions. I'm not going to go into detail here on how the algorithm works because it is beyond the scope of this workshop but there is a bonus exercise at the end if you're curious.\n\nYou can predict a secondary structure by calling `nussinov()` with a sequence string and it will return a tuple in the form `(structure, pairs)`.",
"_____no_output_____"
]
],
[
[
"### 2\nimport numpy as np\nfrom fold import nussinov #Codes by Carlos G. Oliver (https://github.com/cgoliver)\n\nsequence_to_fold = \"ACCCGAUGUUAUAUAUACCU\"\nstruc = nussinov(sequence_to_fold)\nprint(\">test\") #creates the structure of a .fx file for \"forna\"\nprint(sequence_to_fold)\nprint(struc[0])\n#Check the molecule at: http://rna.tbi.univie.ac.at/forna/\n# Paste the text below in the webpage and see the structure\n#>test\n#ACCCGAUGUUAUAUAUACCU\n#(...(..(((....).))))",
">test\nACCCGAUGUUAUAUAUACCU\n(...(..(((....).))))\n"
]
],
[
[
"You will see a funny dot-bracket string in the output. This is a representation of the structure of an RNA. Quite simply, a matching parir of parentheses (open and close) correspond to the nucleotides at those positions being paried. Whereas, a dot means that that position is unpaired in the structure. Feel free to play around with the input sequence to get a better understanding of the notation.\n\nIf you want to visually check the sequence, go to: [forna](http://rna.tbi.univie.ac.at/forna/forna.html) and copy the text above with the sequence and its structure in the **Add Molecule** button. The webpage is embedded below.\n\nSo that's enough about RNA structure prediction. Let's move on to building our populations.",
"_____no_output_____"
]
],
[
[
"HTML('<iframe width=\"784\" height=\"441\" src=\"http://rna.tbi.univie.ac.at/forna/forna.html\" /iframe>')",
"_____no_output_____"
],
[
"print(np.random.choice(5, 3, p=[0.1, 0, 0.6, 0.3, 0],replace=True))",
"[2 2 2]\n"
]
],
[
[
"### Fitness of a sequence: Target Structure\n\nNow that we have a good way of getting a phenotype (secondary structure), we need a way to evaluate the fitness of that phenotype. If we think in real life terms, fitness is the ability of a genotype to replicate into the next generation. If you have a gene carrying a mutation that causes some kind of disease, your fitness is decreased and you have a lower chance of contributing offspring to the next generation. On a molecular level the same concept applies. A molecule needs to accomplish a certain function, i.e. bind to some other molecule or send some kind of signal. And as we've seen before, the most important factor that determines how well it can carry out this function is its structure. So we can imagine that a certain structure, we can call this a 'target' structure, is required in order to accomplish a certain function. So a sequence that folds correctly to a target structure is seen as having a greater fitness than one that does not. Since we've encoded structures as simple dot-bracket strings, we can easily compare structures and thus evaluate the fitness between a given structure and the target, or 'correct' structure. \n\nThere are many ways to compare structures $w_{1}$ and $w_{2}$, but we're going to use one of the simplest ways, which is base-pair distance. This is just the number of pairs in $w_{1}$ that are not in $w_{2}$. Again, this is beyond the scope of this workshop so I'll just give you the code for it and if you would like to know more you can ask me.",
"_____no_output_____"
]
],
[
[
"### 3\n\n#ss_to_bp() and bp_distance() by Vladimir Reinharz.\ndef ss_to_bp(ss):\n bps = set()\n l = []\n for i, x in enumerate(ss):\n if x == '(':\n l.append(i)\n elif x == ')':\n bps.add((l.pop(), i))\n return bps\n\ndef bp_distance(w1, w2):\n \"\"\"\n return base pair distance between structures w1 and w1. \n w1 and w1 are lists of tuples representing pairing indices.\n \"\"\"\n return len(set(w1).symmetric_difference(set(w2)))\n\n#let's fold two sequences\nw1 = nussinov(\"CCAAAAGG\")\nw2 = nussinov(\"ACAAAAGA\")\n\nprint(w1)\nprint(w2)\n\n#give the list of pairs to bp_distance and see what the distance is.\nprint(bp_distance(w1[-1], w2[-1]))",
"('((....))', [(0, 7), (1, 6)])\n('.(....).', [(1, 6)])\n1\n"
]
],
[
[
"## Defining a cell\n\nThe cell we will define here is a simple organism with two copies of an RNA gene, each with its own structure. This simple definition of a cell will help us create populations to play around in our evolutionary reactor.",
"_____no_output_____"
]
],
[
[
"### 4\nclass Cell:\n def __init__(self, seq_1, struc_1, seq_2, struc_2):\n self.sequence_1 = seq_1\n self.sequence_2 = seq_2\n self.structure_1 = struc_1\n self.structure_2 = struc_2\n \n#for now just try initializing a Cell with made up sequences and structures\ncell = Cell(\"AACCCCUU\", \"((.....))\", \"GGAAAACA\", \"(....).\")\nprint(cell.sequence_1, cell.structure_2, cell.sequence_1, cell.structure_2)",
"AACCCCUU (....). AACCCCUU (....).\n"
]
],
[
[
"## Populations of Cells\n\nNow we've defined a 'Cell'. Since a population is a collection of individuals our populations will naturally consist of **lists** of 'Cell' objects, each with their own sequences. Here we initialize all the Cells with random sequences and add them to the 'population' list.",
"_____no_output_____"
]
],
[
[
"### 5\nimport random\n\ndef populate(target, pop_size):\n '''Creates a population of cells (pop_size) with a number of random RNA nucleotides (AUCG)\n matching the length of the target structure'''\n population = []\n\n for i in range(pop_size):\n #get a random sequence to start with\n sequence = \"\".join([random.choice(\"AUCG\") for _ in range(len(target))])\n #use nussinov to get the secondary structure for the sequence\n structure = nussinov(sequence)\n #add a new Cell object to the population list\n new_cell = Cell(sequence, structure, sequence, structure)\n new_cell.id = i\n new_cell.parent = i\n population.append(new_cell)\n \n return population",
"_____no_output_____"
]
],
[
[
"Try creating a new population and printing the first 10 sequences and structures (in dot-bracket) on the first chromosome!",
"_____no_output_____"
]
],
[
[
"### 6\ntarget = \"(((....)))\"\npop = populate(target, pop_size=300)\nfor p in pop[:10]:\n print(p.id, p.sequence_1, p.structure_1[0], p.sequence_2, p.structure_2[0])\n \nfor p in pop[-10:]:#for p in pop[:10]:#for p in pop[-10:]:#for p in pop[:10]:\n print(p.id, p.sequence_1, p.structure_1[0], p.sequence_2, p.structure_2[0])",
"0 ACUUUGCACC ..(....).. ACUUUGCACC ..(....)..\n1 GACUGUGAAA ...(....). GACUGUGAAA ...(....).\n2 UGUCUGCAAG ((....)).. UGUCUGCAAG ((....))..\n3 AGGUGCCCAC .((....).) AGGUGCCCAC .((....).)\n4 GUCCUAGUGA .((.....)) GUCCUAGUGA .((.....))\n5 AUAGAGACAG .(....)... AUAGAGACAG .(....)...\n6 CCGCGUGGCC ((....)).. CCGCGUGGCC ((....))..\n7 GAACACCUUA .((....)). GAACACCUUA .((....)).\n8 CUUAAUAUCC .(....)... CUUAAUAUCC .(....)...\n9 CCGUCUGGUA ((....)).. CCGUCUGGUA ((....))..\n290 CGGGGUACUG ((.....).) CGGGGUACUG ((.....).)\n291 AGAACUGACA (....).... AGAACUGACA (....)....\n292 AACUGUAAAA (....).... AACUGUAAAA (....)....\n293 CAGGCACCUA .((....)). CAGGCACCUA .((....)).\n294 AUGCAACGAC .(......). AUGCAACGAC .(......).\n295 GUCACAGGCA (.(....)). GUCACAGGCA (.(....)).\n296 GUACGAGGCU (.......). GUACGAGGCU (.......).\n297 AUUACUAUAA ((....)).. AUUACUAUAA ((....))..\n298 UUCACUCUGC ..(.....). UUCACUCUGC ..(.....).\n299 GGAUAACUGA (.....)... GGAUAACUGA (.....)...\n"
]
],
[
[
"## The Fitness of a Cell \n\n\nOnce we are able to create populations of cells, we need a way of asssessing their individual fitness. In our model, a *Cell* is an object that contains two sequences of RNA, something analogous to having two copies of a gene in each chromosome. \n\nWe could simply loop through each *Cell* in the population and check the base pair distance to the target structure we defined. However, this approach of using base-pair distance is not the best for defining fitness. There are two reasons for this: \n\n1. We want fitness to represent a *probability* that a cell will reproduce (pass its genes to the next generation), and base pair distance is, in our case, an integer value.\n2. We want this probability to be a *relative* measure of fitness. That is, we want the fitness to be proportional to how good a cell is with respect to all others in the population. This touches on an important principle in evolution where we only need to be 'better' than the rest of the population (the competition) and not good in some absolute measure. For instance, if you and I are being chased by a predator. In order to survive, I only need to be faster than you, and not necessarily some absolute level of fitness.\n\nIn order to get a probability (number between 0 and 1) we use the following equation to define the fitness of a structure $\\omega$ on a target structure $T$:\n\n$$P(\\omega, T) = N^{-1} exp(\\frac{-\\beta \\texttt{dist}(\\omega, T)}{\\texttt{len}(\\omega)})$$\n\n$$N = \\sum_{i \\in Pop}{P(\\omega_i, T})$$\n\nHere, the $N$ is what gives us the 'relative' measure because we divide the fitness of the Cell by the sum of the fitness of every other Cell. \n\nLet's take a quick look at how this function behaves if we plot different base pair distance values.\n\nWhat is the effect of the parameter $\\beta$? Try plotting the same function but with different values of $\\beta$.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nimport math\n#import seaborn as sns\n\ntarget_length = 50\nbeta = -3\n\nplt.plot([math.exp(beta * (bp_dist / float(target_length))) for bp_dist in range(target_length)])\nplt.xlabel(\"Base pair distance to target structure\")\nplt.ylabel(\"P(w, T)\")",
"_____no_output_____"
]
],
[
[
"As you can see, it's a very simple function that evaluates to 1 (highest fitness) if the base pair distance is 0, and decreases as the structures get further and further away from the target. I didn't include the $N$ in the plotting as it will be a bit more annoying to compute, but it is simply a scaling factor so the shape and main idea won't be different.\n\nNow we can use this function to get a fitness value for each Cell in our population.",
"_____no_output_____"
]
],
[
[
"### 7\n\ndef compute_fitness(population, target, beta=-3):\n \"\"\"\n Assigns a fitness and bp_distance value to each cell in the population.\n \"\"\"\n #store the fitness values of each cell\n tot = []\n #iterate through each cell\n for cell in population:\n \n #calculate the bp_distance of each chromosome using the cell's structure\n bp_distance_1 = bp_distance(cell.structure_1[-1], ss_to_bp(target))\n bp_distance_2 = bp_distance(cell.structure_2[-1], ss_to_bp(target))\n \n #use the bp_distances and the above fitness equation to calculate the fitness of each chromosome\n fitness_1 = math.exp((beta * bp_distance_1 / float(len(cell.sequence_1))))\n fitness_2 = math.exp((beta * bp_distance_2 / float(len(cell.sequence_2))))\n\n #get the fitness of the whole cell by multiplying the fitnesses of each chromosome\n cell.fitness = fitness_1 * fitness_2\n \n #store the bp_distance of each chromosome.\n cell.bp_distance_1 = bp_distance_1\n cell.bp_distance_2 = bp_distance_2\n \n \n #add the cell's fitness value to the list of all fitness values (used for normalization later)\n tot.append(cell.fitness)\n\n #normalization factor is sum of all fitness values in population\n norm = np.sum(tot)\n #divide all fitness values by the normalization factor.\n for cell in population:\n cell.fitness = cell.fitness / norm\n\n return None\n\ncompute_fitness(pop, target)\nfor cell in pop[:10]:\n print(cell.fitness, cell.bp_distance_1, cell.bp_distance_2)",
"0.005730832462563744 2 2\n0.002575029013933113 4 4\n0.0017260935671627514 5 5\n0.003841491880128266 3 3\n0.0017260935671627514 5 5\n0.002575029013933113 4 4\n0.0017260935671627514 5 5\n0.008549397405696933 1 1\n0.002575029013933113 4 4\n0.0017260935671627514 5 5\n"
]
],
[
[
"## Introducing diversity: Mutations\n\nEvolution would go nowhere without random mutations. While mutations are technically just random errors in the copying of genetic material, they are essential in the process of evolution. This is because they introduce novel diversity to populatons, which with a low frequency can be beneficial. And when a beneficial mutation arises (i.e. a mutation that increases fitness, or replication probability) it quickly takes over the population and the populatioin as a whole has a higher fitness.\n\nImplementing mutations in our model will be quite straightforward. Since mutations happen at the genotype/sequence level, we simply have to iterate through our strings of nucleotides (sequences) and randomly introduce changes.",
"_____no_output_____"
]
],
[
[
"def mutate(sequence, mutation_rate=0.001):\n \"\"\"Takes a sequence and mutates bases with probability mutation_rate\"\"\"\n \n #start an empty string to store the mutated sequence\n new_sequence = \"\"\n #boolean storing whether or not the sequence got mutated\n mutated = False\n #go through every bp in the sequence\n for bp in sequence:\n #generate a random number between 0 and 1\n r = random.random()\n #if r is below mutation rate, introduce a mutation\n if r < mutation_rate:\n #add a randomly sampled nucleotide to the new sequence\n new_sequence = new_sequence + random.choice(\"aucg\")\n mutated = True\n else:\n #if the mutation condition did not get met, copy the current bp to the new sequence\n new_sequence = new_sequence + bp\n \n return (new_sequence, mutated)\n\nsequence_to_mutate = 'AAAAGGAGUGUGUAUGU'\nprint(sequence_to_mutate)\nprint(mutate(sequence_to_mutate, mutation_rate=0.5))",
"AAAAGGAGUGUGUAUGU\n('AgAcccuguGgGUaucc', True)\n"
]
],
[
[
"## Selection\n\nThe final process in this evolution model is selection. Once you have populations with a diverse range of fitnesses, we need to select the fittest individuals and let them replicate and contribute offspring to the next generation. In real populations this is just the process of reproduction. If you're fit enough you will be likely to reproduce more than another individual who is not as well suited to the environment.\n\nIn order to represent this process in our model, we will use the fitness values that we assigned to each Cell earlier and use that to select replicating Cells. This is equivalent to sampling from a population with the sampling being weighted by the fitness of each Cell. Thankfully, `numpy.random.choice` comes to the rescue here. Once we have sampled enough Cells to build our next generation, we introduce mutations and compute the fitness values of the new generation.",
"_____no_output_____"
]
],
[
[
"def selection(population, target, mutation_rate=0.001, beta=-3):\n \"\"\"\n Returns a new population with offspring of the input population\n \"\"\"\n\n #select the sequences that will be 'parents' and contribute to the next generation\n parents = np.random.choice(population, len(population), p=[rna.fitness for rna in population], replace=True)\n\n #build the next generation using the parents list\n next_generation = [] \n for i, p in enumerate(parents):\n new_cell = Cell(p.sequence_1, p.structure_1, p.sequence_2, p.structure_2)\n new_cell.id = i\n new_cell.parent = p.id\n \n next_generation.append(new_cell)\n\n #introduce mutations in next_generation sequeneces and re-fold when a mutation occurs\n for rna in next_generation: \n mutated_sequence_1, mutated_1 = mutate(rna.sequence_1, mutation_rate=mutation_rate)\n mutated_sequence_2, mutated_2 = mutate(rna.sequence_2, mutation_rate=mutation_rate)\n \n if mutated_1:\n rna.sequence_1 = mutated_sequence_1\n rna.structure_1 = nussinov(mutated_sequence_1)\n if mutated_2:\n rna.sequence_2 = mutated_sequence_2\n rna.structure_2 = nussinov(mutated_sequence_2)\n else:\n continue\n\n #update fitness values for the new generation\n compute_fitness(next_generation, target, beta=beta)\n\n return next_generation\n\nnext_gen = selection(pop, target)\nfor cell in next_gen[:10]:\n print(cell.sequence_1)",
"GUCCAUUGGG\nGUACGAGGCU\nGCACCGAGUC\nAGACCCUUUA\nCUGGCUCUAA\nUAGGAGCGUC\nACGCUAUUGU\nCAGGCACCUA\nCUGCCCUCUG\nCAGGAAGCUC\n"
]
],
[
[
"## Gathering information on our populations\n\nHere we simply store some statistics (in a dictionary) on the population at each generation such as the average base pair distance and the average fitness of the populations. No coding to do here, it's not a very interesting function but feel free to give it a look.",
"_____no_output_____"
]
],
[
[
"def record_stats(pop, population_stats):\n \"\"\"\n Takes a population list and a dictionary and updates it with stats on the population.\n \"\"\"\n generation_bp_distance_1 = [rna.bp_distance_1 for rna in pop]\n generation_bp_distance_2 = [rna.bp_distance_2 for rna in pop]\n\n mean_bp_distance_1 = np.mean(generation_bp_distance_1)\n mean_bp_distance_2 = np.mean(generation_bp_distance_2)\n \n mean_fitness = np.mean([rna.fitness for rna in pop])\n\n\n population_stats.setdefault('mean_bp_distance_1', []).append(mean_bp_distance_1)\n population_stats.setdefault('mean_bp_distance_2', []).append(mean_bp_distance_2)\n \n population_stats.setdefault('mean_fitness', []).append(mean_fitness)\n \n return None",
"_____no_output_____"
]
],
[
[
"## And finally.... evolution\n\nWe can put all the above parts together in a simple function that does the following:\n\n1. start a new population and compute its fitness\n2. repeat the following for the desired number of generations:\n 1. record statistics on population\n 2. perform selection+mutation\n 3. store new population\n\nAnd that's it! We have an evolutionary reactor!",
"_____no_output_____"
]
],
[
[
"def evolve(target, generations=20, pop_size=100, mutation_rate=0.001, beta=-2):\n \"\"\"\n Takes target structure and sets up initial population, performs selection and iterates for desired generations.\n \"\"\"\n #store list of all populations throughotu generations [[cells from generation 1], [cells from gen. 2]...]\n populations = []\n #start a dictionary that will hold some stats on the populations.\n population_stats = {}\n \n #get a starting population\n initial_population = populate(target, pop_size=pop_size)\n #compute fitness of initial population\n compute_fitness(initial_population, target)\n\n #set current_generation to initial population.\n current_generation = initial_population\n\n #iterate the selection process over the desired number of generations\n for i in range(generations):\n\n #let's get some stats on the structures in the populations \n record_stats(current_generation, population_stats)\n \n #add the current generation to our list of populations.\n populations.append(current_generation)\n\n #select the next generation\n new_gen = selection(current_generation, target, mutation_rate=mutation_rate, beta=beta)\n #set current generation to be the generation we just obtained.\n current_generation = new_gen \n \n return (populations, population_stats)",
"_____no_output_____"
]
],
[
[
"Try a run of the `evolve()` function.",
"_____no_output_____"
]
],
[
[
"target = \"(((....)))\"\npops, pops_stats = evolve(target, generations=20, pop_size=300, mutation_rate=0.001, beta=-2)\n\n#Print the first 10 sequences of the population\nfor p in pop[:10]:\n print(p.id, p.sequence_1, p.structure_1[0], p.sequence_2, p.structure_2[0])\n\n#Print the last 10 sequences of the population \nfor p in pop[-10:]:\n print(p.id, p.sequence_1, p.structure_1[0], p.sequence_2, p.structure_2[0])",
"0 ACUUUGCACC ..(....).. ACUUUGCACC ..(....)..\n1 GACUGUGAAA ...(....). GACUGUGAAA ...(....).\n2 UGUCUGCAAG ((....)).. UGUCUGCAAG ((....))..\n3 AGGUGCCCAC .((....).) AGGUGCCCAC .((....).)\n4 GUCCUAGUGA .((.....)) GUCCUAGUGA .((.....))\n5 AUAGAGACAG .(....)... AUAGAGACAG .(....)...\n6 CCGCGUGGCC ((....)).. CCGCGUGGCC ((....))..\n7 GAACACCUUA .((....)). GAACACCUUA .((....)).\n8 CUUAAUAUCC .(....)... CUUAAUAUCC .(....)...\n9 CCGUCUGGUA ((....)).. CCGUCUGGUA ((....))..\n290 CGGGGUACUG ((.....).) CGGGGUACUG ((.....).)\n291 AGAACUGACA (....).... AGAACUGACA (....)....\n292 AACUGUAAAA (....).... AACUGUAAAA (....)....\n293 CAGGCACCUA .((....)). CAGGCACCUA .((....)).\n294 AUGCAACGAC .(......). AUGCAACGAC .(......).\n295 GUCACAGGCA (.(....)). GUCACAGGCA (.(....)).\n296 GUACGAGGCU (.......). GUACGAGGCU (.......).\n297 AUUACUAUAA ((....)).. AUUACUAUAA ((....))..\n298 UUCACUCUGC ..(.....). UUCACUCUGC ..(.....).\n299 GGAUAACUGA (.....)... GGAUAACUGA (.....)...\n"
]
],
[
[
"Let's see if it actually worked by plotting the average base pair distance as a function of generations for both genes in each cell. We should expect a gradual decrease as the populations get closer to the target structure.",
"_____no_output_____"
]
],
[
[
"def evo_plot(pops_stats):\n \"\"\"\n Plot base pair distance for each chromosome over generations.\n \"\"\"\n plt.figure('Mean base pair Distance',figsize=(10,5))\n for m in ['mean_bp_distance_1', 'mean_bp_distance_2']:\n plt.plot(pops_stats[m], label=m)\n plt.legend()\n plt.xlabel(\"Generations\")\n plt.ylabel(\"Mean Base Pair Distance\")\n \nevo_plot(pops_stats)",
"_____no_output_____"
]
],
[
[
"Let's see the structure of random cells from each generation. Run the code below and copy the output in the RNA folding webpage. Compare the base-pair distance plot with the structures.\n\nQuestions:\n- Do you notice some simmilarities from a particular generation onwards? Compare your observations to the plot with the Mean Base Pair Distance.\n- What could trigger another evolutionary jump? \n\n\n",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\nHTML('<iframe width=\"784\" height=\"441\" src=\"http://rna.tbi.univie.ac.at/forna/forna.html\" /iframe>')",
"_____no_output_____"
],
[
"#Select a random RNA sequence from each generation to check its folding structure\nfrom random import randrange\n#print(randrange(999))\ngenerations=20\npop_size=300\n#Print some random cells from each generation \n#pops[generation][cell in that generation].{quality to retrieve}\nfor gen in range(0,generations):\n cid=randrange(pop_size)\n print('>Gen'+np.str(gen+1)+'_Cell_'+np.str(pops[gen][cid].id)+'')\n print(pops[gen][2].sequence_1)\n print(''+np.str(pops[gen][2].structure_1[0])+'\\n')",
">Gen1_Cell_207\nAUUAACAUAA\n((....))..\n\n>Gen2_Cell_83\nCUGUCUCCUA\n.((....).)\n\n>Gen3_Cell_45\nCCGCGCCAAG\n(........)\n\n>Gen4_Cell_165\nGCCAUACAGC\n((......))\n\n>Gen5_Cell_227\nGCCAUACAGC\n((......))\n\n>Gen6_Cell_112\nAUUGGGAAAU\n(((....)))\n\n>Gen7_Cell_218\nGUCaCGUCAC\n((......))\n\n>Gen8_Cell_101\nCGGCUUUUCG\n((......))\n\n>Gen9_Cell_27\nGGAAUCUUCC\n(((....)))\n\n>Gen10_Cell_61\nAUUGGGAAAU\n(((....)))\n\n>Gen11_Cell_40\nCUAAUAUUAG\n(((....)))\n\n>Gen12_Cell_195\nCUAAUAUUAG\n(((....)))\n\n>Gen13_Cell_135\nAUUGGGAAAU\n(((....)))\n\n>Gen14_Cell_206\nAUUGGGAAAU\n(((....)))\n\n>Gen15_Cell_136\nGGAAUCUUCC\n(((....)))\n\n>Gen16_Cell_149\nAUUGGGAAAU\n(((....)))\n\n>Gen17_Cell_10\nCUAAUAUUAG\n(((....)))\n\n>Gen18_Cell_262\nAUUGGGAAAU\n(((....)))\n\n>Gen19_Cell_233\nAUUGGGAAAU\n(((....)))\n\n>Gen20_Cell_92\nAUUGGGAAAU\n(((....)))\n\n"
]
],
[
[
"You should see a nice drop in base pair distance! Another way of visualizing this is by plotting a histogram of the base pair distance of all Cells in the initial population versus the final population.",
"_____no_output_____"
]
],
[
[
"def bp_distance_distributions(pops):\n \"\"\"\n Plots histograms of base pair distance in initial and final populations.\n \"\"\"\n #plot bp_distance_1 for rnas in first population\n g = sns.distplot([rna.bp_distance_1 for rna in pops[0]], label='initial population')\n #plot bp_distance_1 for rnas in first population\n g = sns.distplot([rna.bp_distance_1 for rna in pops[-1]], label='final population')\n g.set(xlabel='Mean Base Pair Distance')\n g.legend()\nbp_distance_distributions(pops)",
"_____no_output_____"
]
],
[
[
"## Introducing mating to the model\n\nThe populations we generated evolved asexually. This means that individuals do not mate or exchange genetic information. So to make our simulation a bit more interesting let's let the Cells mate. This is going to require a few small changes in the `selection()` function. Previously, when we selected sequences to go into the next generation we just let them provide one offspring which was a copy of itself and introduced mutations. Now instead of choosing one Cell at a time, we will randomly choose two 'parents' that will mate. When they mate, each parent will contribute one of its chromosomes to the child. We'll repeat this process until we have filled the next generation.",
"_____no_output_____"
]
],
[
[
"def selection_with_mating(population, target, mutation_rate=0.001, beta=-2):\n next_generation = []\n \n counter = 0\n while len(next_generation) < len(population):\n #select two parents based on their fitness\n parents_pair = np.random.choice(population, 2, p=[rna.fitness for rna in population], replace=False)\n \n #take the sequence and structure from the first parent's first chromosome and give it to the child\n child_chrom_1 = (parents_pair[0].sequence_1, parents_pair[0].structure_1)\n\n #do the same for the child's second chromosome and the second parent.\n child_chrom_2 = (parents_pair[1].sequence_2, parents_pair[1].structure_2)\n\n\n #initialize the new child Cell with the new chromosomes.\n child_cell = Cell(child_chrom_1[0], child_chrom_1[1], child_chrom_2[0], child_chrom_2[1])\n\n #give the child and id and store who its parents are\n child_cell.id = counter\n child_cell.parent_1 = parents_pair[0].id\n child_cell.parent_2 = parents_pair[1].id\n\n #add the child to the new generation\n next_generation.append(child_cell)\n \n counter = counter + 1\n \n \n #introduce mutations in next_generation sequeneces and re-fold when a mutation occurs (same as before)\n for rna in next_generation: \n mutated_sequence_1, mutated_1 = mutate(rna.sequence_1, mutation_rate=mutation_rate)\n mutated_sequence_2, mutated_2 = mutate(rna.sequence_2, mutation_rate=mutation_rate)\n\n if mutated_1:\n rna.sequence_1 = mutated_sequence_1\n rna.structure_1 = nussinov(mutated_sequence_1)\n if mutated_2:\n rna.sequence_2 = mutated_sequence_2\n rna.structure_2 = nussinov(mutated_sequence_2)\n else:\n continue\n\n #update fitness values for the new generation\n compute_fitness(next_generation, target, beta=beta)\n\n return next_generation \n\n#run a small test to make sure it works\nnext_gen = selection_with_mating(pop, target)\nfor cell in next_gen[:10]:\n print(cell.sequence_1)",
"CGAAGUACCU\nACGAGUUUGC\nCAGGUACACG\nCACCGGGAUG\nCCUACACAGG\nACCCCGCUUA\nGUUUAUGUGC\nAUGGAGUUCA\nUUUGGCGGUA\nCCUACACAGG\n"
]
],
[
[
"Now we just have to update our `evolution()` function to call the new `selection_with_mating()` function.",
"_____no_output_____"
]
],
[
[
"def evolve_with_mating(target, generations=10, pop_size=100, mutation_rate=0.001, beta=-2):\n populations = []\n population_stats = {}\n \n initial_population = populate(target, pop_size=pop_size)\n compute_fitness(initial_population, target)\n \n current_generation = initial_population\n\n #iterate the selection process over the desired number of generations\n for i in range(generations):\n #let's get some stats on the structures in the populations \n record_stats(current_generation, population_stats)\n \n #add the current generation to our list of populations.\n populations.append(current_generation)\n\n #select the next generation, but this time with mutations\n new_gen = selection_with_mating(current_generation, target, mutation_rate=mutation_rate, beta=beta)\n current_generation = new_gen \n \n return (populations, population_stats)",
"_____no_output_____"
]
],
[
[
"Try out the new evolution model!",
"_____no_output_____"
]
],
[
[
"pops_mating, pops_stats_mating = evolve_with_mating(\"(((....)))\", generations=20, pop_size=1000, beta=-2)\n\nevo_plot(pops_stats_mating)",
"_____no_output_____"
],
[
"#Select a random RNA sequence from each generation to check its folding structure\nfrom random import randrange\n#print(randrange(999))\ngenerations=20\npop_size=1000\n#Print some random cells from each generation \n#pops[generation][cell in that generation].{quality to retrieve}\nfor gen in range(0,generations):\n cid=randrange(pop_size)\n print('>Gen'+np.str(gen+1)+'_Cell_'+np.str(pops_mating[gen][cid].id)+'')\n print(pops_mating[gen][2].sequence_1)\n print(''+np.str(pops_mating[gen][2].structure_1[0])+'\\n')",
">Gen1_Cell_826\nCUCCGCUGAC\n.((....)).\n\n>Gen2_Cell_944\nCCCACAGACA\n(.....)...\n\n>Gen3_Cell_616\nAGUUCUCGGU\n((....)..)\n\n>Gen4_Cell_958\nCAAUAAGUUA\n.((....)).\n\n>Gen5_Cell_606\nCCACCUUGAG\n((.....).)\n\n>Gen6_Cell_993\nCGAUUCCUCA\n.((....)).\n\n>Gen7_Cell_50\nUAUCUGAAUA\n(((....)))\n\n>Gen8_Cell_115\nGCUAUUCAGA\n.((....)).\n\n>Gen9_Cell_983\nACAACCCUCU\n(.(....).)\n\n>Gen10_Cell_531\nAUCGAUGGAC\n.((....)).\n\n>Gen11_Cell_998\nGCACAUGUGC\n(((....)))\n\n>Gen12_Cell_80\nCUCGCACGAG\n(((....)))\n\n>Gen13_Cell_255\nACAACCCUCU\n(.(....).)\n\n>Gen14_Cell_24\nGUUUUCGAAC\n(((....)))\n\n>Gen15_Cell_904\nCAAUAGUUUG\n(((....)))\n\n>Gen16_Cell_229\nUAUCUGAAUA\n(((....)))\n\n>Gen17_Cell_522\nGUUAAGUUAC\n((......))\n\n>Gen18_Cell_608\nAUGCCGUCCU\n(.(....).)\n\n>Gen19_Cell_752\nUAUCUGAAUA\n(((....)))\n\n>Gen20_Cell_862\nCUCGCACGAG\n(((....)))\n\n"
]
],
[
[
"# Acknowledgements\n\nThe computational codes of this notebook were originally created by [Carlos G. Oliver](https://github.com/cgoliver), and adapted by Evert Durán for ASTRO200.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbc8e20169337846e96cd69556da0241d1be391d
| 93,588 |
ipynb
|
Jupyter Notebook
|
samples/gaussian_mixture.ipynb
|
yokonami/prml_practice
|
9d5d160df32d3d98a2092d14cb2a7130f69ac0fd
|
[
"MIT"
] | null | null | null |
samples/gaussian_mixture.ipynb
|
yokonami/prml_practice
|
9d5d160df32d3d98a2092d14cb2a7130f69ac0fd
|
[
"MIT"
] | null | null | null |
samples/gaussian_mixture.ipynb
|
yokonami/prml_practice
|
9d5d160df32d3d98a2092d14cb2a7130f69ac0fd
|
[
"MIT"
] | 1 |
2019-11-15T02:32:43.000Z
|
2019-11-15T02:32:43.000Z
| 519.933333 | 89,908 | 0.946916 |
[
[
[
"import random\nimport numpy as np\nimport sys\nsys.path.append('../')",
"_____no_output_____"
],
[
"from gaussian_mixture_em import GaussianMixtureEM",
"_____no_output_____"
],
[
"from sklearn.datasets import load_iris\ndata = load_iris()\ndata.keys()\n\ndata['data'].shape, data['target'].shape",
"_____no_output_____"
],
[
"n_class = len(np.unique(data['target']))\nn_class",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\nscaler = StandardScaler()\nX = scaler.fit_transform(data['data'])",
"_____no_output_____"
],
[
"model = GaussianMixtureEM(n_class, max_iter=300)\nmodel.fit(X, data['target'])\npred = model.predict(X)",
"_____no_output_____"
],
[
"print(model.pi)",
"[0.22955384 0.33328799 0.43715817]\n"
],
[
"import matplotlib\nimport matplotlib.pyplot as plt\nfrom scipy.stats import multivariate_normal\nmatplotlib.rcParams['figure.figsize'] = (21, 4)\nax = 0\nay = 2\n\nmu = model.mu\ncov = model.cov\nrnk = model.rnk\nfor k in range(n_class):\n plt.subplot(1, n_class, k+1)\n plt.scatter(X[:, ax], X[:, ay], c=rnk[:, k])\n plt.scatter(mu[:, ax], mu[:, ay], c='b', marker='x', s=100)\n plt.scatter(mu[k, ax], mu[k, ay], c='r', marker='x', s=100)\n \n rv = multivariate_normal(mu[k, [ax, ay]], cov[k, [ax, ay], [ax, ay]])\n x, y = np.mgrid[-3:3:.01, -3:3:.01]\n pos = np.empty(x.shape + (2,))\n pos[:, :, 0] = x; pos[:, :, 1] = y\n plt.contour(x, y, rv.pdf(pos))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc8e85d887e4edfe48397f262edfe192f1d293c
| 22,836 |
ipynb
|
Jupyter Notebook
|
Phase - 1/Session4/4_Architectures in CNN.ipynb
|
yash1996/DL_curated_intuition
|
ae6d966bea93a0ece50dd7a2aa316ccde8c6d6b2
|
[
"CC0-1.0"
] | 33 |
2020-05-10T13:13:27.000Z
|
2021-04-22T08:40:55.000Z
|
Phase - 1/Session4/4_Architectures in CNN.ipynb
|
yash1996/DL_curated_intuition
|
ae6d966bea93a0ece50dd7a2aa316ccde8c6d6b2
|
[
"CC0-1.0"
] | 9 |
2020-09-26T00:39:54.000Z
|
2022-03-12T00:14:11.000Z
|
Phase - 1/Session4/4_Architectures in CNN.ipynb
|
nikhilpradeep/deep
|
d55562100fe3804e55ea1cf1637a669da69baec3
|
[
"CC0-1.0"
] | 18 |
2020-06-07T12:58:21.000Z
|
2022-02-21T17:18:25.000Z
| 53.858491 | 607 | 0.681906 |
[
[
[
"# Introduction\nIn this post,we will talk about some of the most important papers that have been published over the last 5 years and discuss why they’re so important.We will go through different CNN Architectures (LeNet to DenseNet) showcasing the advancements in general network architecture that made these architectures top the ILSVRC results.",
"_____no_output_____"
],
[
"# What is ImageNet\n\n[ImageNet](http://www.image-net.org/)\n\nImageNet is formally a project aimed at (manually) labeling and categorizing images into almost 22,000 separate object categories for the purpose of computer vision research.\n\nHowever, when we hear the term “ImageNet” in the context of deep learning and Convolutional Neural Networks, we are likely referring to the ImageNet Large Scale Visual Recognition Challenge, or ILSVRC for short.\n\nThe ImageNet project runs an annual software contest, the ImageNet Large Scale Visual Recognition Challenge (ILSVRC), where software programs compete to correctly classify and detect objects and scenes.\n\nThe goal of this image classification challenge is to train a model that can correctly classify an input image into 1,000 separate object categories.\n\nModels are trained on ~1.2 million training images with another 50,000 images for validation and 100,000 images for testing.\n\nThese 1,000 image categories represent object classes that we encounter in our day-to-day lives, such as species of dogs, cats, various household objects, vehicle types, and much more. You can find the full list of object categories in the ILSVRC challenge\n\nWhen it comes to image classification, the **ImageNet** challenge is the de facto benchmark for computer vision classification algorithms — and the leaderboard for this challenge has been dominated by Convolutional Neural Networks and deep learning techniques since 2012.\n",
"_____no_output_____"
],
[
"# LeNet-5(1998)\n\n[Gradient Based Learning Applied to Document Recognition](http://yann.lecun.com/exdb/publis/pdf/lecun-01a.pdf)\n\n1. A pioneering 7-level convolutional network by LeCun that classifies digits,\n2. Found its application by several banks to recognise hand-written numbers on checks (cheques) \n3. These numbers were digitized in 32x32 pixel greyscale which acted as an input images. \n4. The ability to process higher resolution images requires larger and more convolutional layers, so this technique is constrained by the availability of computing resources.\n\n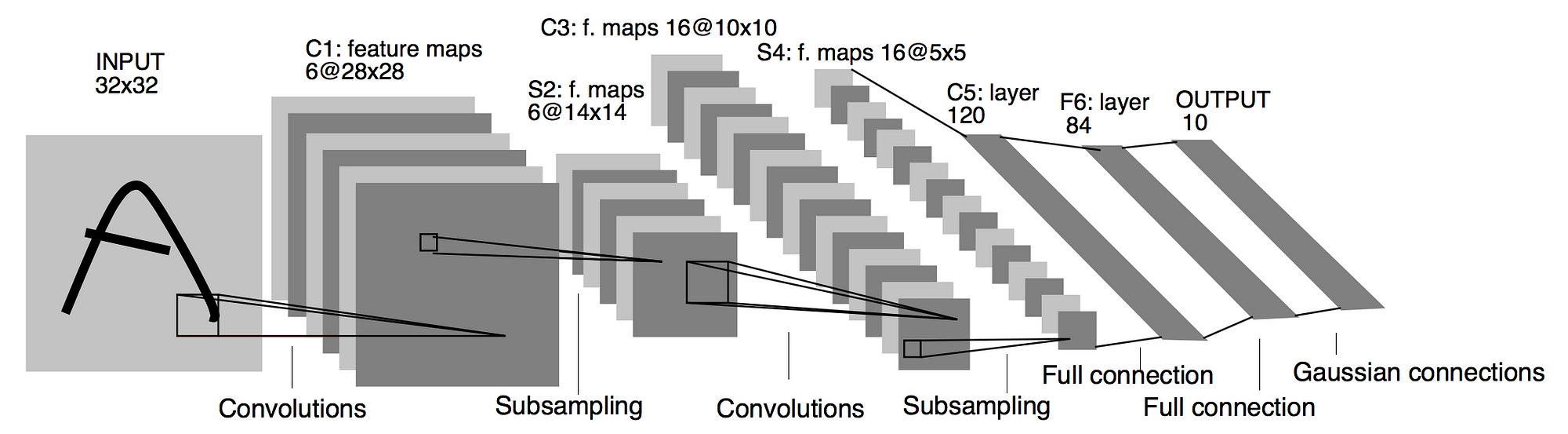",
"_____no_output_____"
],
[
"# AlexNet(2012)\n\n[ImageNet Classification with Deep Convolutional Networks](https://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf)\n\n1. One of the most influential publications in the field by Alex Krizhevsky, Ilya Sutskever, and Geoffrey Hinton that started the revolution of CNN in Computer Vision.This was the first time a model performed so well on a historically difficult ImageNet dataset.\n2. The network consisted 11x11, 5x5,3x3, convolutions and made up of 5 conv layers, max-pooling layers, dropout layers, and 3 fully connected layers.\n3. Used ReLU for the nonlinearity functions (Found to decrease training time as ReLUs are several times faster than the conventional tanh function) and used SGD with momentum for training.\n4. Used data augmentation techniques that consisted of image translations, horizontal reflections, and patch extractions.\n5. Implemented dropout layers in order to combat the problem of overfitting to the training data.\n6. Trained the model using batch stochastic gradient descent, with specific values for momentum and weight decay.\n7. AlexNet was trained for 6 days simultaneously on two Nvidia Geforce GTX 580 GPUs which is the reason for why their network is split into two pipelines.\n8. AlexNet significantly outperformed all the prior competitors and won the challenge by reducing the top-5 error from 26% to 15.3%\n\n ",
"_____no_output_____"
],
[
"# ZFNet(2013)\n\n[Visualizing and Understanding Convolutional Neural Networks](https://cs.nyu.edu/~fergus/papers/zeilerECCV2014.pdf)\n<br>\nThis architecture was more of a fine tuning to the previous AlexNet structure by tweaking the hyper-parameters of AlexNet while maintaining the same structure but still developed some very keys ideas about improving performance.Few minor modifications done were the following:\n1. AlexNet trained on 15 million images, while ZF Net trained on only 1.3 million images.\n2. Instead of using 11x11 sized filters in the first layer (which is what AlexNet implemented), ZF Net used filters of size 7x7 and a decreased stride value. The reasoning behind this modification is that a smaller filter size in the first conv layer helps retain a lot of original pixel information in the input volume. A filtering of size 11x11 proved to be skipping a lot of relevant information, especially as this is the first conv layer.\n3. As the network grows, we also see a rise in the number of filters used.\n4. Used ReLUs for their activation functions, cross-entropy loss for the error function, and trained using batch stochastic gradient descent.\n5. Trained on a GTX 580 GPU for twelve days.\n6. Developed a visualization technique named **Deconvolutional Network**, which helps to examine different feature activations and their relation to the input space. Called **deconvnet** because it maps features to pixels (the opposite of what a convolutional layer does).\n7. It achieved a top-5 error rate of 14.8%\n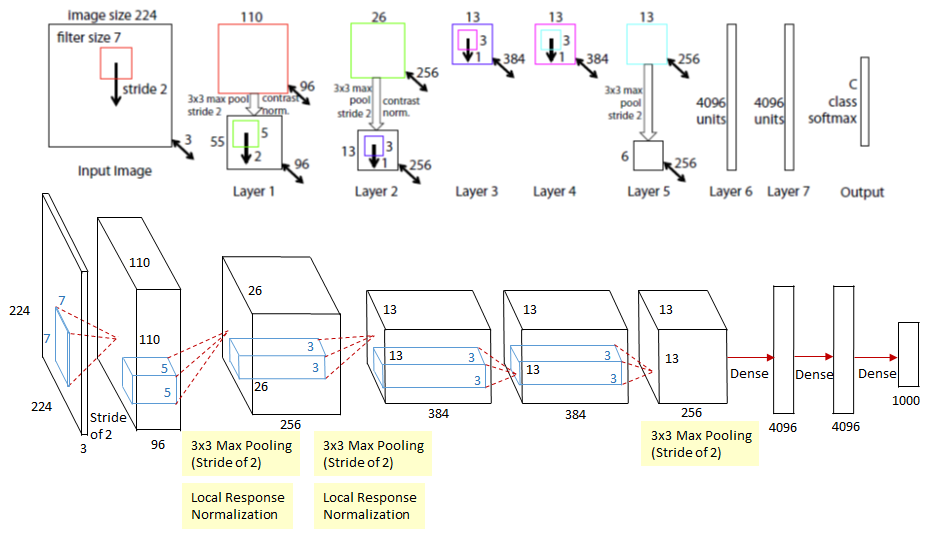",
"_____no_output_____"
],
[
"# VggNet(2014)\n\n[VERY DEEP CONVOLUTIONAL NETWORKS FOR LARGE-SCALE IMAGE RECOGNITION](https://arxiv.org/pdf/1409.1556v6.pdf)\n\nThis architecture is well konwn for **Simplicity and depth**.. VGGNet is very appealing because of its very uniform architecture.They proposed 6 different variations of VggNet however 16 layer with all 3x3 convolution produced the best result.\n\nFew things to note:\n1. The use of only 3x3 sized filters is quite different from AlexNet’s 11x11 filters in the first layer and ZF Net’s 7x7 filters. The authors’ reasoning is that the combination of two 3x3 conv layers has an effective receptive field of 5x5. This in turn simulates a larger filter while keeping the benefits of smaller filter sizes. One of the benefits is a decrease in the number of parameters. Also, with two conv layers, we’re able to use two ReLU layers instead of one.\n2. 3 conv layers back to back have an effective receptive field of 7x7.\n3. As the spatial size of the input volumes at each layer decrease (result of the conv and pool layers), the depth of the volumes increase due to the increased number of filters as you go down the network.\n4. Interesting to notice that the number of filters doubles after each maxpool layer. This reinforces the idea of shrinking spatial dimensions, but growing depth.\n5. Worked well on both image classification and localization tasks. The authors used a form of localization as regression (see page 10 of the paper for all details).\n6. Built model with the Caffe toolbox.\n7. Used scale jittering as one data augmentation technique during training.\n8. Used ReLU layers after each conv layer and trained with batch gradient descent.\n9. Trained on 4 Nvidia Titan Black GPUs for two to three weeks.\n10. It achieved a top-5 error rate of 7.3% \n\n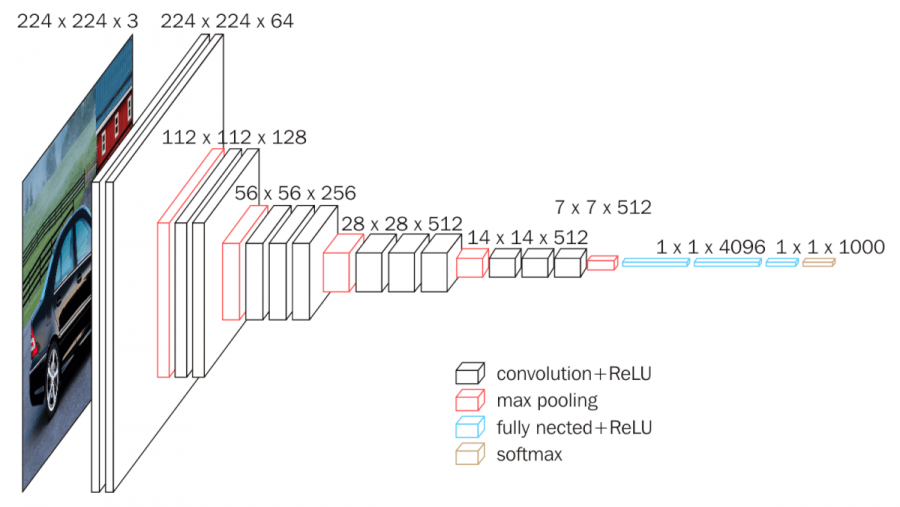\n\n\n\n**In Standard ConvNet, input image goes through multiple convolution and obtain high-level features.**",
"_____no_output_____"
],
[
"After Inception V1 ,the author proposed a number of upgrades which increased the accuracy and reduced the computational complexity.This lead to many new upgrades resulting in different versions of Inception Network :\n1. Inception v2\n2. Inception V3",
"_____no_output_____"
],
[
"# Inception Network (GoogleNet)(2014)\n[Going Deeper with Convolutions](https://www.cv-foundation.org/openaccess/content_cvpr_2015/papers/Szegedy_Going_Deeper_With_2015_CVPR_paper.pdf)\n\nPrior to this, most popular CNNs just stacked convolution layers deeper and deeper, hoping to get better performance,however **Inception Network** was one of the first CNN architectures that really strayed from the general approach of simply stacking conv and pooling layers on top of each other in a sequential structure and came up with the **Inception Module**.The Inception network was complex. It used a lot of tricks to push performance; both in terms of speed and accuracy. Its constant evolution lead to the creation of several versions of the network. The popular versions are as follows:\n\n1. Inception v1.\n2. Inception v2 and Inception v3.\n3. Inception v4 and Inception-ResNet.\n<br>\n\nEach version is an iterative improvement over the previous one.Let us go ahead and explore them one by one\n\n\n\n",
"_____no_output_____"
],
[
"## Inception V1\n[Inception v1](https://arxiv.org/pdf/1409.4842v1.pdf)\n\n\n**Problems this network tried to solve:**\n1. **What is the right kernel size for convolution**\n<br>\nA larger kernel is preferred for information that is distributed more globally, and a smaller kernel is preferred for information that is distributed more locally.\n<br>\n**Ans-** Filters with multiple sizes.The network essentially would get a bit “wider” rather than “deeper”\n<br>\n<br>\n3. **How to stack convolution which can be less computationally expensive**\n<BR>\nStacking them naively computationally expensive.\n<br>\n**Ans-**Limit the number of input channels by adding an extra 1x1 convolution before the 3x3 and 5x5 convolutions\n<br>\n<br>\n2. **How to avoid overfitting in a very deep network**\n<br>\nVery deep networks are prone to overfitting. It also hard to pass gradient updates through the entire network.\n<br>\n**Ans-**Introduce two auxiliary classifiers (The purple boxes in the image). They essentially applied softmax to the outputs of two of the inception modules, and computed an auxiliary loss over the same labels. The total loss function is a weighted sum of the auxiliary loss and the real loss.\n\nThe total loss used by the inception net during training.\n<br>\n **total_loss = real_loss + 0.3 * aux_loss_1 + 0.3 * aux_loss_2**\n<br>\n<br>\n\n\n\n**Points to note**\n\n1. Used 9 Inception modules in the whole architecture, with over 100 layers in total! Now that is deep…\n2. No use of fully connected layers! They use an average pool instead, to go from a 7x7x1024 volume to a 1x1x1024 volume. This saves a huge number of parameters.\n3. Uses 12x fewer parameters than AlexNet.\n4. Trained on “a few high-end GPUs within a week”.\n5. It achieved a top-5 error rate of 6.67% ",
"_____no_output_____"
],
[
"## Inception V2\n[Rethinking the Inception Architecture for Computer Vision](https://arxiv.org/pdf/1512.00567v3.pdf)\n\n\nUpgrades were targeted towards:\n1. Reducing representational bottleneck by replacing 5x5 convolution to two 3x3 convolution operations which further improves computational speed\n<br>\nThe intuition was that, neural networks perform better when convolutions didn’t alter the dimensions of the input drastically. Reducing the dimensions too much may cause loss of information, known as a **“representational bottleneck”**\n<br>\n\n2. Using smart factorization method where they factorize convolutions of filter size nxn to a combination of 1xn and nx1 convolutions.\n<br>\nFor example, a 3x3 convolution is equivalent to first performing a 1x3 convolution, and then performing a 3x1 convolution on its output. They found this method to be 33% more cheaper than the single 3x3 convolution.\n\n",
"_____no_output_____"
],
[
"# ResNet(2015)\n[Deep Residual Learning for Image Recognition](https://arxiv.org/pdf/1512.03385.pdf)\n\n**In ResNet, identity mapping is proposed to promote the gradient propagation. Element-wise addition is used. It can be viewed as algorithms with a state passed from one ResNet module to another one.**\n\n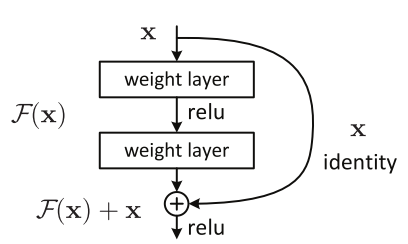\n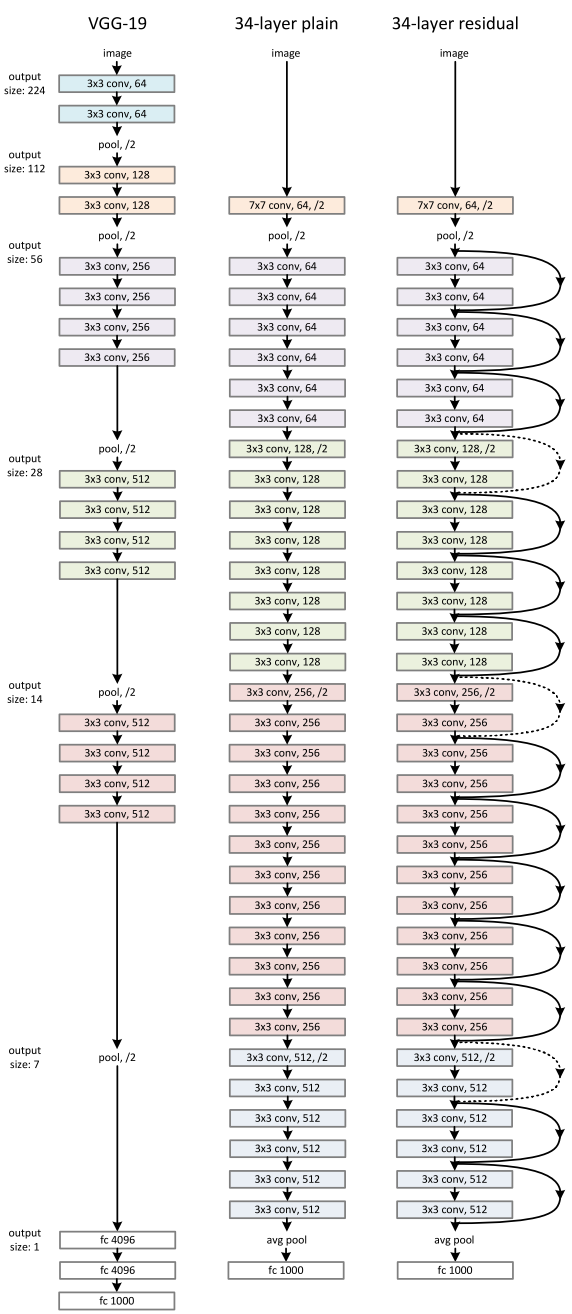",
"_____no_output_____"
],
[
"# ResNet-Wide\n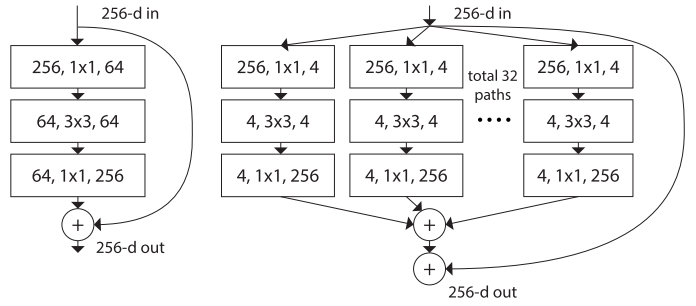\nleft: a building block of [2], right: a building block of ResNeXt with cardinality = 32",
"_____no_output_____"
],
[
"# DenseNet(2017)\n\n[Densely Connected Convolutional Networks](https://arxiv.org/pdf/1608.06993v3.pdf)\n<br>\nIt is a logical extension to ResNet.\n\n**From the paper:**\nRecent work has shown that convolutional networks can be substantially deeper, more accurate, and efficient to train if they contain shorter connections between layers close to the input and those close to the output. In this paper, we embrace this observation and introduce the Dense Convolutional Network (DenseNet), which connects each layer to every other layer in a feed-forward fashion.\n\n**DenseNet Architecture**\n\n\nLet us explore different componenets of the network\n<br>\n<br>\n**1. Dense Block**\n<br>\nFeature map sizes are the same within the dense block so that they can be concatenated together easily.\n\n\n**In DenseNet, each layer obtains additional inputs from all preceding layers and passes on its own feature-maps to all subsequent layers. Concatenation is used. Each layer is receiving a “collective knowledge” from all preceding layers.**\n\n\nSince each layer receives feature maps from all preceding layers, network can be thinner and compact, i.e. number of channels can be fewer. The growth rate k is the additional number of channels for each layer.\n\nThe paper proposed different ways to implement DenseNet with/without B/C by adding some variations in the Dense block to further reduce the complexity,size and to bring more compression in the architecture.\n\n 1. Dense Block (DenseNet)\n - Batch Norm (BN)\n - ReLU\n - 3×3 Convolution \n 2. Dense Block(DenseNet B)\n - Batch Norm (BN)\n - ReLU\n - 1×1 Convolution\n - Batch Norm (BN)\n - ReLU\n - 3×3 Convolution\n 3. Dense Block(DenseNet C)\n - If a dense block contains m feature-maps, The transition layer generate $\\theta $ output feature maps, where $\\theta \\leq \\theata \\leq$ is referred to as the compression factor.\n - $\\theta$=0.5 was used in the experiemnt which reduced the number of feature maps by 50%.\n \n 4. Dense Block(DenseNet BC)\n - Combination of Densenet B and Densenet C\n<br>\n**2. Trasition Layer**\n<br>\nThe layers between two adjacent blocks are referred to as transition layers where the following operations are done to change feature-map sizes:\n - 1×1 Convolution\n - 2×2 Average pooling \n\n\n**Points to Note:**\n1. it requires fewer parameters than traditional convolutional networks\n2. Traditional convolutional networks with L layers have L connections — one between each layer and its subsequent layer — our network has L(L+1)/ 2 direct connections.\n3. Improved flow of information and gradients throughout the network, which makes them easy to train\n4. They alleviate the vanishing-gradient problem, strengthen feature propagation, encourage feature reuse, and substantially reduce the number of parameters.\n5. Concatenating feature maps instead of summing learned by different layers increases variation in the input of subsequent layers and improves efficiency. This constitutes a major difference between DenseNets and ResNets.\n6. It achieved a top-5 error rate of 6.66% ",
"_____no_output_____"
],
[
"# MobileNet",
"_____no_output_____"
],
[
"## Spatial Seperable Convolution\n\n\n**Divides a kernel into two, smaller kernels**\n\n\n\n**Instead of doing one convolution with 9 multiplications(parameters), we do two convolutions with 3 multiplications(parameters) each (6 in total) to achieve the same effect**\n\n\n\n\n**With less multiplications, computational complexity goes down, and the network is able to run faster.**\n\nThis was used in an architecture called [Effnet](https://arxiv.org/pdf/1801.06434v1.pdf) showing promising results.\n\nThe main issue with the spatial separable convolution is that not all kernels can be “separated” into two, smaller kernels. This becomes particularly bothersome during training, since of all the possible kernels the network could have adopted, it can only end up using one of the tiny portion that can be separated into two smaller kernels.\n\n\n\n## Depthwise Convolution\n\n\nSay we need to increase the number of channels from 16 to 32 using 3x3 kernel.\n<br>\n\n**Normal Convolution**\n<br>\nTotal No of Parameters = 3 x 3 x 16 x 32 = 4608\n\n\n\n**Depthwise Convolution**\n\n1. DepthWise Convolution = 16 x [3 x 3 x 1]\n2. PointWise Convolution = 32 x [1 x 1 x 16]\n\nTotal Number of Parameters = 656\n\n\n**Mobile net uses depthwise seperable convolution to reduce the number of parameters**",
"_____no_output_____"
],
[
"# References\n\n[Standford CS231n Lecture Notes](http://cs231n.stanford.edu/slides/2017/cs231n_2017_lecture9.pdf)\n<br>\n[The 9 Deep Learning Papers You Need To Know About](https://adeshpande3.github.io/adeshpande3.github.io/The-9-Deep-Learning-Papers-You-Need-To-Know-About.html)\n<br>\n[CNN Architectures](https://medium.com/@sidereal/cnns-architectures-lenet-alexnet-vgg-googlenet-resnet-and-more-666091488df5)\n<br>\n[Lets Keep It Simple](https://arxiv.org/pdf/1608.06037.pdf)\n<br>\n[CNN Architectures Keras](https://www.pyimagesearch.com/2017/03/20/imagenet-vggnet-resnet-inception-xception-keras/)\n<br>\n[Inception Versions](https://towardsdatascience.com/a-simple-guide-to-the-versions-of-the-inception-network-7fc52b863202)\n<br>\n[DenseNet Review](https://towardsdatascience.com/review-densenet-image-classification-b6631a8ef803)\n<br>\n[DenseNet](https://towardsdatascience.com/densenet-2810936aeebb)\n<br>\n[ResNet](http://teleported.in/posts/decoding-resnet-architecture/)\n<br>\n[ResNet Versions](https://towardsdatascience.com/an-overview-of-resnet-and-its-variants-5281e2f56035)\n<br>\n[Depthwise Convolution](https://towardsdatascience.com/a-basic-introduction-to-separable-convolutions-b99ec3102728)",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbc8ebc9f5208abaf7bbd95ad364e1ff21879f08
| 2,386 |
ipynb
|
Jupyter Notebook
|
DownloadOpenStreetCam.ipynb
|
brandon-castaing-ucb/Wall-E
|
dc3e5864a823c29bf6c5bf20bb80af7d00dd8f13
|
[
"MIT"
] | 1 |
2020-01-20T20:30:48.000Z
|
2020-01-20T20:30:48.000Z
|
DownloadOpenStreetCam.ipynb
|
brandon-castaing-ucb/Wall-E
|
dc3e5864a823c29bf6c5bf20bb80af7d00dd8f13
|
[
"MIT"
] | 2 |
2020-03-18T02:02:46.000Z
|
2020-03-29T03:46:23.000Z
|
DownloadOpenStreetCam.ipynb
|
brandon-castaing-ucb/Wall-E
|
dc3e5864a823c29bf6c5bf20bb80af7d00dd8f13
|
[
"MIT"
] | null | null | null | 27.744186 | 130 | 0.477368 |
[
[
[
"!pip uninstall -y s3fs\n!pip install s3fs==0.4.0\n\nimport s3fs\nassert(s3fs.__version__ == \"0.4.0\")\nbucket_name = \"ucb-mids-wall-e-andy-test\"\n\nfs = s3fs.S3FileSystem(anon=False, key='', secret='')\nprint(fs.ls(bucket_name))\n\nimport sys\nimport urllib.request\n",
"_____no_output_____"
],
[
"for fname in fs.ls(bucket_name + \"/OpenStreetCam/openstreetcam_data_raw/\"):\n with fs.open(fname, 'rb') as f_in:\n for raw_line in f_in:\n line = raw_line.decode(\"utf-8\")\n split_line = line.split(\"\\t\")\n track_id, image_id, url = split_line[0], split_line[1], split_line[-1]\n target_name = bucket_name + \"/OpenStreetCam/openstreetcam_data_images/{}_{}.jpg\".format(track_id, image_id)\n print(target_name)\n\n if fs.exists(target_name):\n print(\"skipped\")\n continue\n \n f = urllib.request.urlopen(url, timeout=1800)\n imbytes = f.read()\n \n if not imbytes is None:\n fs.touch(target_name)\n f_out = fs.open(target_name, \"wb\")\n f_out.write(imbytes)\n f_out.close()\n print(\"downloaded\")\n \n f_in.close()\n \n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbc90a6291947dee2f25657a4757033b86f3544f
| 776,198 |
ipynb
|
Jupyter Notebook
|
notebooks/4.3-mbml_kf_final.ipynb
|
Peymankor/MBML_Final_Project
|
1d989b07773f1b8e9a5dd11379845959d62e8780
|
[
"MIT"
] | 1 |
2020-05-21T18:02:06.000Z
|
2020-05-21T18:02:06.000Z
|
notebooks/4.3-mbml_kf_final.ipynb
|
Peymankor/MBML_Final_Project
|
1d989b07773f1b8e9a5dd11379845959d62e8780
|
[
"MIT"
] | null | null | null |
notebooks/4.3-mbml_kf_final.ipynb
|
Peymankor/MBML_Final_Project
|
1d989b07773f1b8e9a5dd11379845959d62e8780
|
[
"MIT"
] | 1 |
2020-05-09T21:52:21.000Z
|
2020-05-09T21:52:21.000Z
| 344.517532 | 46,552 | 0.924711 |
[
[
[
"## Probalistic Confirmed COVID19 Cases- Denmark",
"_____no_output_____"
],
[
"**Jorge: remember to reexecute the cell with the photo.**",
"_____no_output_____"
],
[
"### Table of contents\n[Initialization](#Initialization) \n[Data Importing and Processing](#Data-Importing-and-Processing)\n1. [Kalman Filter Modeling: Case of Denmark Data](#1.-Kalman-Filter-Modeling:-Case-of-Denmark-Data) \n 1.1. [Model with the vector c fixed as [0, 1]](#1.1.-Kalman-Filter-Model-vector-c-fixed-as-[0,-1]) \n 1.2. [Model with the vector c as a random variable with prior](#1.2.-Kalman-Filter-with-the-vector-c-as-a-random-variable-with-prior) \n 1.3. [Model without input (2 hidden variables)](#1.3.-Kalman-Filter-without-Input) \n2. [Kalman Filter Modeling: Case of Norway Data](#2.-Kalman-Filter-Modeling:-Case-of-Norway-Data) \n 2.1. [Model with the vector c fixed as [0, 1]](#2.1.-Kalman-Filter-Model-vector-c-fixed-as-[0,-1]) \n 2.2. [Model with the vector c as a random variable with prior](#2.2.-Kalman-Filter-with-the-vector-c-as-a-random-variable-with-prior) \n 2.3. [Model without input (2 hidden variables)](#2.3.-Kalman-Filter-without-Input) \n3. [Kalman Filter Modeling: Case of Sweden Data](#Kalman-Filter-Modeling:-Case-of-Sweden-Data) \n 3.1. [Model with the vector c fixed as [0, 1]](#3.1.-Kalman-Filter-Model-vector-c-fixed-as-[0,-1]) \n 3.2. [Model with the vector c as a random variable with prior](#3.2.-Kalman-Filter-with-the-vector-c-as-a-random-variable-with-prior) \n 3.3. [Model without input (2 hidden variables)](#3.3.-Kalman-Filter-without-Input) ",
"_____no_output_____"
],
[
"## Initialization",
"_____no_output_____"
]
],
[
[
"from os.path import join, pardir\n\nimport jax\nimport jax.numpy as jnp\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport numpyro\nimport numpyro.distributions as dist\nimport pandas as pd\nimport seaborn as sns\n\nfrom jax import lax, random, vmap\nfrom jax.scipy.special import logsumexp\nfrom numpyro import handlers\nfrom numpyro.infer import MCMC, NUTS\nfrom sklearn.preprocessing import StandardScaler\n\nnp.random.seed(2103)",
"_____no_output_____"
],
[
"ROOT = pardir\nDATA = join(ROOT, \"data\", \"raw\")\n\n# random seed\nnp.random.seed(42)\n\n#plot style\nplt.style.use('ggplot')\n\n%matplotlib inline\nplt.rcParams['figure.figsize'] = (16, 10)",
"_____no_output_____"
]
],
[
[
"## Data Importing and Processing",
"_____no_output_____"
],
[
"The data in this case are the confirmed cases of the COVID-19 and the the mobility data (from Google) for three specific countries: Denmark, Sweden and Norway.\n",
"_____no_output_____"
]
],
[
[
"adress = join(ROOT, \"data\", \"processed\")\ndata = pd.read_csv(join(adress, 'data_three_mob_cov.csv'),parse_dates=['Date'])\ndata.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 268 entries, 0 to 267\nData columns (total 9 columns):\nDate 268 non-null datetime64[ns]\nCountry 268 non-null object\nConfirmed 268 non-null int64\nret_rec_ch 268 non-null int64\ngr_ph_ch 265 non-null float64\npar_cha 268 non-null int64\ntra_sta_ch 268 non-null int64\nwor_ch 268 non-null int64\nres_ch 268 non-null int64\ndtypes: datetime64[ns](1), float64(1), int64(6), object(1)\nmemory usage: 19.0+ KB\n"
],
[
"data.head(5)",
"_____no_output_____"
]
],
[
[
"Handy functions to split the data, train the models and plot the results.",
"_____no_output_____"
]
],
[
[
"def split_forecast(df, n_train=65):\n \"\"\"Split dataframe `df` as training, test and input mobility data.\"\"\"\n # just take the first 4 mobility features\n X = df.iloc[:, 3:7].values.astype(np.float_)\n # confirmed cases\n y = df.iloc[:,2].values.astype(np.float_)\n\n idx_train = [*range(0,n_train)]\n idx_test = [*range(n_train, len(y))]\n\n y_train = y[:n_train]\n y_test = y[n_train:]\n\n return X, y_train, y_test\n\n\ndef train_kf(model, data, n_train, n_test, num_samples=9000, num_warmup=3000, **kwargs):\n \"\"\"Train a Kalman Filter model.\"\"\"\n rng_key = random.PRNGKey(0)\n rng_key, rng_key_ = random.split(rng_key)\n\n nuts_kernel = NUTS(model=model)\n # burn-in is still too much in comparison with the samples\n mcmc = MCMC(\n nuts_kernel, num_samples=num_samples, num_warmup=num_warmup, num_chains=1\n )\n mcmc.run(rng_key_, T=n_train, T_forecast=n_test, obs=data, **kwargs)\n return mcmc\n\n\ndef get_samples(mcmc):\n \"\"\"Get samples from variables in MCMC.\"\"\"\n return {k: v for k, v in mcmc.get_samples().items()}\n\n\ndef plot_samples(hmc_samples, nodes, dist=True):\n \"\"\"Plot samples from the variables in `nodes`.\"\"\"\n for node in nodes:\n if len(hmc_samples[node].shape) > 1:\n n_vars = hmc_samples[node].shape[1]\n for i in range(n_vars):\n plt.figure(figsize=(4, 3))\n if dist:\n sns.distplot(hmc_samples[node][:, i], label=node + \"%d\" % i)\n else:\n plt.plot(hmc_samples[node][:, i], label=node + \"%d\" % i)\n plt.legend()\n plt.show()\n else:\n plt.figure(figsize=(4, 3))\n if dist:\n sns.distplot(hmc_samples[node], label=node)\n else:\n plt.plot(hmc_samples[node], label=node)\n plt.legend()\n plt.show()\n\n \ndef plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test):\n \"\"\"Plot the results of forecasting (dimension are different).\"\"\"\n y_hat = hmc_samples[\"y_pred\"].mean(axis=0)\n y_std = hmc_samples[\"y_pred\"].std(axis=0)\n y_pred_025 = y_hat - 1.96 * y_std\n y_pred_975 = y_hat + 1.96 * y_std\n plt.plot(idx_train, y_train, \"b-\")\n plt.plot(idx_test, y_test, \"bx\")\n plt.plot(idx_test[:-1], y_hat, \"r-\")\n plt.plot(idx_test[:-1], y_pred_025, \"r--\")\n plt.plot(idx_test[:-1], y_pred_975, \"r--\")\n plt.fill_between(idx_test[:-1], y_pred_025, y_pred_975, alpha=0.3)\n plt.legend(\n [\n \"true (train)\",\n \"true (test)\",\n \"forecast\",\n \"forecast + stddev\",\n \"forecast - stddev\",\n ]\n )\n plt.show()\n\nn_train = 65 # number of points to train\nn_test = 20 # number of points to forecast\nidx_train = [*range(0,n_train)]\nidx_test = [*range(n_train, n_train+n_test)]",
"_____no_output_____"
]
],
[
[
"## 1. Kalman Filter Modeling: Case of Denmark Data",
"_____no_output_____"
]
],
[
[
"data_dk=data[data['Country'] == \"Denmark\"]\ndata_dk.head(5)",
"_____no_output_____"
],
[
"print(\"The length of the full dataset for Denmark is:\" + \" \" )\nprint(len(data_dk))",
"The length of the full dataset for Denmark is: \n85\n"
]
],
[
[
"Prepare input of the models (we are using numpyro so the inputs are numpy arrays).",
"_____no_output_____"
]
],
[
[
"X, y_train, y_test = split_forecast(data_dk)",
"_____no_output_____"
]
],
[
[
"### 1.1. Kalman Filter Model vector c fixed as [0, 1]",
"_____no_output_____"
],
[
"First model: the sampling distribution is replaced by one fixed variable $c$.",
"_____no_output_____"
]
],
[
[
"def f(carry, input_t):\n x_t, noise_t = input_t\n W, beta, z_prev, tau = carry\n z_t = beta * z_prev + W @ x_t + noise_t\n z_prev = z_t\n return (W, beta, z_prev, tau), z_t\n\n\ndef model_wo_c(T, T_forecast, x, obs=None):\n \"\"\"Define KF with inputs and fixed sampling dist.\"\"\"\n # Define priors over beta, tau, sigma, z_1\n W = numpyro.sample(\n name=\"W\", fn=dist.Normal(loc=jnp.zeros((2, 4)), scale=jnp.ones((2, 4)))\n )\n beta = numpyro.sample(\n name=\"beta\", fn=dist.Normal(loc=jnp.zeros(2), scale=jnp.ones(2))\n )\n tau = numpyro.sample(name=\"tau\", fn=dist.HalfCauchy(scale=jnp.ones(2)))\n sigma = numpyro.sample(name=\"sigma\", fn=dist.HalfCauchy(scale=0.1))\n z_prev = numpyro.sample(\n name=\"z_1\", fn=dist.Normal(loc=jnp.zeros(2), scale=jnp.ones(2))\n )\n # Define LKJ prior\n L_Omega = numpyro.sample(\"L_Omega\", dist.LKJCholesky(2, 10.0))\n Sigma_lower = jnp.matmul(\n jnp.diag(jnp.sqrt(tau)), L_Omega\n ) # lower cholesky factor of the covariance matrix\n noises = numpyro.sample(\n \"noises\",\n fn=dist.MultivariateNormal(loc=jnp.zeros(2), scale_tril=Sigma_lower),\n sample_shape=(T + T_forecast - 2,),\n )\n # Propagate the dynamics forward using jax.lax.scan\n carry = (W, beta, z_prev, tau)\n z_collection = [z_prev]\n carry, zs_exp = lax.scan(f, carry, (x, noises), T + T_forecast - 2)\n z_collection = jnp.concatenate((jnp.array(z_collection), zs_exp), axis=0)\n\n obs_mean = z_collection[:T, 1]\n pred_mean = z_collection[T:, 1]\n\n # Sample the observed y (y_obs)\n numpyro.sample(name=\"y_obs\", fn=dist.Normal(loc=obs_mean, scale=sigma), obs=obs)\n numpyro.sample(name=\"y_pred\", fn=dist.Normal(loc=pred_mean, scale=sigma), obs=None)",
"_____no_output_____"
],
[
"mcmc = train_kf(model_wo_c, y_train, n_train, n_test, x=X[2:])",
"sample: 100%|██████████| 12000/12000 [04:46<00:00, 41.87it/s, 1023 steps of size 7.60e-04. acc. prob=0.54]\n"
]
],
[
[
"Plots of the distribution of the samples for each variable.",
"_____no_output_____"
]
],
[
[
"hmc_samples = get_samples(mcmc)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
]
],
[
[
"Forecasting prediction, all the datapoints in the test set are within the Confidence Interval.",
"_____no_output_____"
]
],
[
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"### 1.2. Kalman Filter with the vector c as a random variable with prior",
"_____no_output_____"
],
[
"Second model: the sampling distribution is a Normal distribution $c$.",
"_____no_output_____"
]
],
[
[
"def model_w_c(T, T_forecast, x, obs=None):\n # Define priors over beta, tau, sigma, z_1 (keep the shapes in mind)\n W = numpyro.sample(\n name=\"W\", fn=dist.Normal(loc=jnp.zeros((2, 4)), scale=jnp.ones((2, 4)))\n )\n beta = numpyro.sample(\n name=\"beta\", fn=dist.Normal(loc=jnp.array([0.0, 0.0]), scale=jnp.ones(2))\n )\n tau = numpyro.sample(name=\"tau\", fn=dist.HalfCauchy(scale=jnp.array([2,2])))\n sigma = numpyro.sample(name=\"sigma\", fn=dist.HalfCauchy(scale=1))\n z_prev = numpyro.sample(\n name=\"z_1\", fn=dist.Normal(loc=jnp.zeros(2), scale=jnp.ones(2))\n )\n # Define LKJ prior\n L_Omega = numpyro.sample(\"L_Omega\", dist.LKJCholesky(2, 10.0))\n Sigma_lower = jnp.matmul(\n jnp.diag(jnp.sqrt(tau)), L_Omega\n ) # lower cholesky factor of the covariance matrix\n noises = numpyro.sample(\n \"noises\",\n fn=dist.MultivariateNormal(loc=jnp.zeros(2), scale_tril=Sigma_lower),\n sample_shape=(T + T_forecast - 2,),\n )\n # Propagate the dynamics forward using jax.lax.scan\n carry = (W, beta, z_prev, tau)\n z_collection = [z_prev]\n carry, zs_exp = lax.scan(f, carry, (x, noises), T + T_forecast - 2)\n z_collection = jnp.concatenate((jnp.array(z_collection), zs_exp), axis=0)\n\n c = numpyro.sample(\n name=\"c\", fn=dist.Normal(loc=jnp.array([[0.0], [0.0]]), scale=jnp.ones((2, 1)))\n )\n obs_mean = jnp.dot(z_collection[:T, :], c).squeeze()\n pred_mean = jnp.dot(z_collection[T:, :], c).squeeze()\n\n # Sample the observed y (y_obs)\n numpyro.sample(name=\"y_obs\", fn=dist.Normal(loc=obs_mean, scale=sigma), obs=obs)\n numpyro.sample(name=\"y_pred\", fn=dist.Normal(loc=pred_mean, scale=sigma), obs=None)",
"_____no_output_____"
],
[
"mcmc2 = train_kf(model_w_c, y_train, n_train, n_test, x=X[:-2])",
"sample: 100%|██████████| 12000/12000 [06:47<00:00, 29.46it/s, 1023 steps of size 4.92e-04. acc. prob=0.85]\n"
],
[
"hmc_samples = get_samples(mcmc2)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"### 1.3. Kalman Filter without Input",
"_____no_output_____"
],
[
"Third model: no input mobility data, **two** hidden states.",
"_____no_output_____"
]
],
[
[
"def f_s(carry, noise_t):\n \"\"\"Propagate forward the time series.\"\"\"\n beta, z_prev, tau = carry\n z_t = beta * z_prev + noise_t\n z_prev = z_t\n return (beta, z_prev, tau), z_t\n\ndef twoh_c_kf(T, T_forecast, obs=None):\n \"\"\"Define Kalman Filter with two hidden variates.\"\"\"\n # Define priors over beta, tau, sigma, z_1\n # W = numpyro.sample(name=\"W\", fn=dist.Normal(loc=jnp.zeros((2,4)), scale=jnp.ones((2,4))))\n beta = numpyro.sample(\n name=\"beta\", fn=dist.Normal(loc=jnp.array([0.0, 0.0]), scale=jnp.ones(2))\n )\n tau = numpyro.sample(name=\"tau\", fn=dist.HalfCauchy(scale=jnp.array([10,10])))\n sigma = numpyro.sample(name=\"sigma\", fn=dist.HalfCauchy(scale=5))\n z_prev = numpyro.sample(\n name=\"z_1\", fn=dist.Normal(loc=jnp.zeros(2), scale=jnp.ones(2))\n )\n # Define LKJ prior\n L_Omega = numpyro.sample(\"L_Omega\", dist.LKJCholesky(2, 10.0))\n Sigma_lower = jnp.matmul(\n jnp.diag(jnp.sqrt(tau)), L_Omega\n ) # lower cholesky factor of the covariance matrix\n noises = numpyro.sample(\n \"noises\",\n fn=dist.MultivariateNormal(loc=jnp.zeros(2), scale_tril=Sigma_lower),\n sample_shape=(T + T_forecast - 2,),\n )\n # Propagate the dynamics forward using jax.lax.scan\n carry = (beta, z_prev, tau)\n z_collection = [z_prev]\n carry, zs_exp = lax.scan(f_s, carry, noises, T + T_forecast - 2)\n z_collection = jnp.concatenate((jnp.array(z_collection), zs_exp), axis=0)\n\n c = numpyro.sample(\n name=\"c\", fn=dist.Normal(loc=jnp.array([[0.0], [0.0]]), scale=jnp.ones((2, 1)))\n )\n obs_mean = jnp.dot(z_collection[:T, :], c).squeeze()\n pred_mean = jnp.dot(z_collection[T:, :], c).squeeze()\n\n # Sample the observed y (y_obs)\n numpyro.sample(name=\"y_obs\", fn=dist.Normal(loc=obs_mean, scale=sigma), obs=obs)\n numpyro.sample(name=\"y_pred\", fn=dist.Normal(loc=pred_mean, scale=sigma), obs=None)",
"_____no_output_____"
],
[
"mcmc3 = train_kf(twoh_c_kf, y_train, n_train, n_test, num_samples=12000, num_warmup=5000)",
"sample: 100%|██████████| 17000/17000 [06:55<00:00, 40.92it/s, 1023 steps of size 2.57e-03. acc. prob=0.61] \n"
],
[
"hmc_samples = get_samples(mcmc3)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"## 2. Kalman Filter Modeling: Case of Norway Data",
"_____no_output_____"
]
],
[
[
"data_no=data[data['Country'] == \"Norway\"]\ndata_no.head(5)",
"_____no_output_____"
],
[
"print(\"The length of the full dataset for Norway is:\" + \" \" )\nprint(len(data_no))",
"The length of the full dataset for Norway is: \n86\n"
],
[
"n_train = 66 # number of points to train\nn_test = 20 # number of points to forecast\nidx_train = [*range(0,n_train)]\nidx_test = [*range(n_train, n_train+n_test)]",
"_____no_output_____"
],
[
"X, y_train, y_test = split_forecast(data_no, n_train)",
"_____no_output_____"
]
],
[
[
"### 2.1. Kalman Filter Model vector c fixed as [0, 1]",
"_____no_output_____"
]
],
[
[
"mcmc_no = train_kf(model_wo_c, y_train, n_train, n_test, x=X[:-2])",
"_____no_output_____"
],
[
"hmc_samples = get_samples(mcmc_no)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"### 2.2. Kalman Filter with the vector c as a random variable with prior",
"_____no_output_____"
]
],
[
[
"mcmc2_no = train_kf(model_w_c, y_train, n_train, n_test, x=X[:-2])",
"_____no_output_____"
],
[
"hmc_samples = get_samples(mcmc2_no)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"### 2.3. Kalman Filter without Input",
"_____no_output_____"
]
],
[
[
"mcmc3_no = train_kf(twoh_c_kf, y_train, n_train, n_test)",
"sample: 100%|██████████| 12000/12000 [05:02<00:00, 39.63it/s, 1023 steps of size 8.29e-05. acc. prob=0.43]\n"
],
[
"hmc_samples = get_samples(mcmc3_no)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"## 3. Kalman Filter Modeling: Case of Sweden Data",
"_____no_output_____"
]
],
[
[
"data_sw=data[data['Country'] == \"Sweden\"]\ndata_sw.head(5)",
"_____no_output_____"
],
[
"print(\"The length of the full dataset for Sweden is:\" + \" \" )\nprint(len(data_sw))",
"The length of the full dataset for Sweden is: \n97\n"
],
[
"n_train = 75 # number of points to train\nn_test = 22 # number of points to forecast\nidx_train = [*range(0,n_train)]\nidx_test = [*range(n_train, n_train+n_test)]",
"_____no_output_____"
],
[
"X, y_train, y_test = split_forecast(data_sw, n_train)",
"_____no_output_____"
]
],
[
[
"### 3.1. Kalman Filter Model vector c fixed as [0, 1]",
"_____no_output_____"
]
],
[
[
"mcmc_sw = train_kf(model_wo_c, y_train, n_train, n_test, x=X[:-2])",
"sample: 100%|██████████| 12000/12000 [03:07<00:00, 64.08it/s, 7 steps of size 9.13e-03. acc. prob=0.37] \n"
],
[
"hmc_samples = get_samples(mcmc_sw)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"### 3.2. Kalman Filter with the vector c as a random variable with prior",
"_____no_output_____"
]
],
[
[
"mcmc2_sw = train_kf(model_w_c, y_train, n_train, n_test, x=X[:-2])",
"sample: 100%|██████████| 12000/12000 [07:50<00:00, 25.49it/s, 1023 steps of size 3.14e-04. acc. prob=0.75]\n"
],
[
"hmc_samples = get_samples(mcmc2_sw)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"### 3.3. Kalman Filter without Input",
"_____no_output_____"
]
],
[
[
"mcmc3_sw = train_kf(twoh_c_kf, y_train, n_train, n_test)",
"sample: 100%|██████████| 12000/12000 [04:13<00:00, 47.42it/s, 106 steps of size 3.37e-03. acc. prob=0.37] \n"
],
[
"hmc_samples = get_samples(mcmc3_sw)\nplot_samples(hmc_samples, [\"beta\", \"tau\", \"sigma\"])",
"_____no_output_____"
],
[
"plot_forecast(hmc_samples, idx_train, idx_test, y_train, y_test)",
"_____no_output_____"
]
],
[
[
"Save results to rerun the plotting functions.",
"_____no_output_____"
]
],
[
[
"import pickle",
"_____no_output_____"
],
[
"MODELS = join(ROOT, \"models\")\nfor i, mc in enumerate([mcmc3_no, mcmc_sw, mcmc2_sw, mcmc3_sw]):\n with open(join(MODELS, f\"hmc_ok_{i}.pickle\"), \"wb\") as f:\n pickle.dump(get_samples(mc),f)",
"_____no_output_____"
]
],
[
[
"## Gaussian Process",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbc90d4431c030809b35195fd8a27772c954f9ca
| 73,550 |
ipynb
|
Jupyter Notebook
|
Covid19_death_prediction.ipynb
|
Janani-harshu/Machine_Learning_Projects
|
3fed52a49c62105336165c1fe2b20ca1ab9362a9
|
[
"MIT"
] | null | null | null |
Covid19_death_prediction.ipynb
|
Janani-harshu/Machine_Learning_Projects
|
3fed52a49c62105336165c1fe2b20ca1ab9362a9
|
[
"MIT"
] | null | null | null |
Covid19_death_prediction.ipynb
|
Janani-harshu/Machine_Learning_Projects
|
3fed52a49c62105336165c1fe2b20ca1ab9362a9
|
[
"MIT"
] | null | null | null | 150.102041 | 24,047 | 0.551353 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Janani-harshu/Machine_Learning_Projects/blob/main/Covid19_death_prediction.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"Covid-19 is one of the deadliest viruses you’ve ever heard. Mutations in covid-19 make it either more deadly or more infectious. We have seen a lot of deaths from covid-19 while there is a higher wave of cases. We can use historical data on covid-19 cases and deaths to predict the number of deaths in the future.\nIn this notebook, I will take you through the task of Covid-19 deaths prediction with machine learning using Python.",
"_____no_output_____"
],
[
"## Covid-19 Deaths Prediction (Case Study)\nYou are given a dataset of Covid-19 in India from 30 January 2020 to 18 January 2022. The dataset contains information about the daily confirmed cases and deaths. Below are all the columns of the dataset:\n\nDate: Contains the date of the record\n\nDate_YMD: Contains date in Year-Month-Day Format\n\nDaily Confirmed: Contains the daily confirmed cases of Covid-19\n\nDaily Deceased: Contains the daily deaths due to Covid-19\n\nYou need to use this historical data of covid-19 cases and deaths to predict the number of deaths for the next 30 days",
"_____no_output_____"
]
],
[
[
"# Importing the libraries \nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"data = pd.read_csv(\"COVID19 data for overall INDIA.csv\")\nprint(data.head())",
" Date Date_YMD Daily Confirmed Daily Deceased\n0 30 January 2020 2020-01-30 1 0\n1 31 January 2020 2020-01-31 0 0\n2 1 February 2020 2020-02-01 0 0\n3 2 February 2020 2020-02-02 1 0\n4 3 February 2020 2020-02-03 1 0\n"
],
[
"data.isnull().sum()",
"_____no_output_____"
],
[
"data = data.drop(\"Date\", axis=1)",
"_____no_output_____"
],
[
"# Daily confirmed cases of Covid-19\nimport plotly.express as px\nfig = px.box(data, x='Date_YMD', y='Daily Confirmed')\nfig.show()",
"_____no_output_____"
]
],
[
[
"## Covid-19 Death Rate Analysis\nNow let’s visualize the death rate due to Covid-19:",
"_____no_output_____"
]
],
[
[
"cases = data[\"Daily Confirmed\"].sum()\ndeceased = data[\"Daily Deceased\"].sum()\n\nlabels = [\"Confirmed\", \"Deceased\"]\nvalues = [cases, deceased]\n\nfig = px.funnel_area(data, values=values, \n names=labels, \n title='Daily Confirmed Cases vs Daily Deaths')\nfig.show()",
"_____no_output_____"
],
[
"# calculate the death rate of Covid-19:\ndeath_rate = (data[\"Daily Deceased\"].sum() / data[\"Daily Confirmed\"].sum()) * 100\nprint(death_rate)",
"1.2840580507834722\n"
],
[
"# daily deaths of covid-19:\nimport plotly.express as px\nfig = px.box(data, x='Date_YMD', y='Daily Deceased')\nfig.show()",
"_____no_output_____"
],
[
"!pip install AutoTS",
"_____no_output_____"
]
],
[
[
"## Covid-19 Deaths Prediction Model\nNow let’s move to the task of covid-19 deaths prediction for the next 30 days. Here I will be using the AutoTS library, which is one of the best Automatic Machine Learning libraries for Time Series Analysis.",
"_____no_output_____"
]
],
[
[
"from autots import AutoTS\n\nmodel = AutoTS(forecast_length=30, frequency='infer', ensemble='simple')\nmodel = model.fit(data, date_col=\"Date_YMD\", value_col='Daily Deceased', id_col=None)",
"[Parallel(n_jobs=-2)]: Done 5000 out of 5000 | elapsed: 8.0s finished\n"
],
[
"# Predict covid-19 deaths with machine learning for the next 30 days:\nprediction = model.predict()\nforecast = prediction.forecast\nprint(forecast)",
" Daily Deceased\n2022-01-19 293.988778\n2022-01-20 278.076363\n2022-01-21 309.494481\n2022-01-22 300.350164\n2022-01-23 277.788000\n2022-01-24 278.432313\n2022-01-25 309.712306\n2022-01-26 310.099204\n2022-01-27 284.818877\n2022-01-28 260.745164\n2022-01-29 290.874466\n2022-01-30 278.761165\n2022-01-31 247.638798\n2022-02-01 278.680893\n2022-02-02 265.598129\n2022-02-03 272.476395\n2022-02-04 274.777953\n2022-02-05 302.135985\n2022-02-06 290.745774\n2022-02-07 283.601135\n2022-02-08 285.481865\n2022-02-09 308.135826\n2022-02-10 275.678988\n2022-02-11 286.125338\n2022-02-12 287.022266\n2022-02-13 314.892784\n2022-02-14 299.079899\n2022-02-15 310.061025\n2022-02-16 266.635495\n2022-02-17 313.559686\n"
]
],
[
[
"So this is how we can predict covid-19 deaths with machine learning using the Python programming language. We can use the historical data of covid-19 cases and deaths to predict the number of deaths in future. You can implement the same method for predicting covid-19 deaths and waves on the latest dataset",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbc92aa077185c08c3e4e792f814e696ae34fc27
| 191,009 |
ipynb
|
Jupyter Notebook
|
Model backlog/Train/120-jigsaw-fold1-xlm-roberta-ratio-1-exp-3-epochs.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | 4 |
2020-06-23T02:31:07.000Z
|
2020-07-04T11:50:08.000Z
|
Model backlog/Train/120-jigsaw-fold1-xlm-roberta-ratio-1-exp-3-epochs.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | null | null | null |
Model backlog/Train/120-jigsaw-fold1-xlm-roberta-ratio-1-exp-3-epochs.ipynb
|
dimitreOliveira/Jigsaw-Multilingual-Toxic-Comment-Classification
|
44422e6aeeff227e22dbb5c05101322e9d4aabbe
|
[
"MIT"
] | null | null | null | 134.418719 | 105,964 | 0.835128 |
[
[
[
"## Dependencies",
"_____no_output_____"
]
],
[
[
"import json, warnings, shutil, glob\nfrom jigsaw_utility_scripts import *\nfrom scripts_step_lr_schedulers import *\nfrom transformers import TFXLMRobertaModel, XLMRobertaConfig\nfrom tensorflow.keras.models import Model\nfrom tensorflow.keras import optimizers, metrics, losses, layers\n\nSEED = 0\nseed_everything(SEED)\nwarnings.filterwarnings(\"ignore\")\npd.set_option('max_colwidth', 120)\npd.set_option('display.float_format', lambda x: '%.4f' % x)",
"\u001b[34m\u001b[1mwandb\u001b[0m: \u001b[33mWARNING\u001b[0m W&B installed but not logged in. Run `wandb login` or set the WANDB_API_KEY env variable.\n"
]
],
[
[
"## TPU configuration",
"_____no_output_____"
]
],
[
[
"strategy, tpu = set_up_strategy()\nprint(\"REPLICAS: \", strategy.num_replicas_in_sync)\nAUTO = tf.data.experimental.AUTOTUNE",
"Running on TPU grpc://10.0.0.2:8470\nREPLICAS: 8\n"
]
],
[
[
"# Load data",
"_____no_output_____"
]
],
[
[
"database_base_path = '/kaggle/input/jigsaw-data-split-roberta-192-ratio-1-clean-polish/'\nk_fold = pd.read_csv(database_base_path + '5-fold.csv')\nvalid_df = pd.read_csv(\"/kaggle/input/jigsaw-multilingual-toxic-comment-classification/validation.csv\", \n usecols=['comment_text', 'toxic', 'lang'])\n\nprint('Train samples: %d' % len(k_fold))\ndisplay(k_fold.head())\nprint('Validation samples: %d' % len(valid_df))\ndisplay(valid_df.head())\n\nbase_data_path = 'fold_1/'\nfold_n = 1\n# Unzip files\n!tar -xf /kaggle/input/jigsaw-data-split-roberta-192-ratio-1-clean-polish/fold_1.tar.gz",
"Train samples: 267220\n"
]
],
[
[
"# Model parameters",
"_____no_output_____"
]
],
[
[
"base_path = '/kaggle/input/jigsaw-transformers/XLM-RoBERTa/'\n\nconfig = {\n \"MAX_LEN\": 192,\n \"BATCH_SIZE\": 128,\n \"EPOCHS\": 3,\n \"LEARNING_RATE\": 1e-5, \n \"ES_PATIENCE\": None,\n \"base_model_path\": base_path + 'tf-xlm-roberta-large-tf_model.h5',\n \"config_path\": base_path + 'xlm-roberta-large-config.json'\n}\n\nwith open('config.json', 'w') as json_file:\n json.dump(json.loads(json.dumps(config)), json_file)\n \nconfig",
"_____no_output_____"
]
],
[
[
"## Learning rate schedule",
"_____no_output_____"
]
],
[
[
"lr_min = 1e-7\nlr_start = 0\nlr_max = config['LEARNING_RATE']\nstep_size = (len(k_fold[k_fold[f'fold_{fold_n}'] == 'train']) * 2) // config['BATCH_SIZE']\ntotal_steps = config['EPOCHS'] * step_size\nhold_max_steps = 0\nwarmup_steps = step_size * 1\ndecay = .9998\n\nrng = [i for i in range(0, total_steps, config['BATCH_SIZE'])]\ny = [exponential_schedule_with_warmup(tf.cast(x, tf.float32), warmup_steps, hold_max_steps, \n lr_start, lr_max, lr_min, decay) for x in rng]\n\nsns.set(style=\"whitegrid\")\nfig, ax = plt.subplots(figsize=(20, 6))\nplt.plot(rng, y)\nprint(\"Learning rate schedule: {:.3g} to {:.3g} to {:.3g}\".format(y[0], max(y), y[-1]))",
"Learning rate schedule: 0 to 9.96e-06 to 2.65e-06\n"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"module_config = XLMRobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)\n\ndef model_fn(MAX_LEN):\n input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')\n attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')\n \n base_model = TFXLMRobertaModel.from_pretrained(config['base_model_path'], config=module_config)\n last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})\n cls_token = last_hidden_state[:, 0, :]\n \n output = layers.Dense(1, activation='sigmoid', name='output')(cls_token)\n \n model = Model(inputs=[input_ids, attention_mask], outputs=output)\n \n return model",
"_____no_output_____"
]
],
[
[
"# Train",
"_____no_output_____"
]
],
[
[
"# Load data\nx_train = np.load(base_data_path + 'x_train.npy')\ny_train = np.load(base_data_path + 'y_train_int.npy').reshape(x_train.shape[1], 1).astype(np.float32)\nx_valid = np.load(base_data_path + 'x_valid.npy')\ny_valid = np.load(base_data_path + 'y_valid_int.npy').reshape(x_valid.shape[1], 1).astype(np.float32)\nx_valid_ml = np.load(database_base_path + 'x_valid.npy')\ny_valid_ml = np.load(database_base_path + 'y_valid.npy').reshape(x_valid_ml.shape[1], 1).astype(np.float32)\n\n#################### ADD TAIL ####################\nx_train_tail = np.load(base_data_path + 'x_train_tail.npy')\ny_train_tail = np.load(base_data_path + 'y_train_int_tail.npy').reshape(x_train_tail.shape[1], 1).astype(np.float32)\nx_train = np.hstack([x_train, x_train_tail])\ny_train = np.vstack([y_train, y_train_tail])\n\nstep_size = x_train.shape[1] // config['BATCH_SIZE']\nvalid_step_size = x_valid_ml.shape[1] // config['BATCH_SIZE']\nvalid_2_step_size = x_valid.shape[1] // config['BATCH_SIZE']\n\n# Build TF datasets\ntrain_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_train, y_train, config['BATCH_SIZE'], AUTO, seed=SEED))\nvalid_dist_ds = strategy.experimental_distribute_dataset(get_validation_dataset(x_valid_ml, y_valid_ml, config['BATCH_SIZE'], AUTO, repeated=True, seed=SEED))\nvalid_2_dist_ds = strategy.experimental_distribute_dataset(get_validation_dataset(x_valid, y_valid, config['BATCH_SIZE'], AUTO, repeated=True, seed=SEED))\ntrain_data_iter = iter(train_dist_ds)\nvalid_data_iter = iter(valid_dist_ds)\nvalid_2_data_iter = iter(valid_2_dist_ds)",
"_____no_output_____"
],
[
"# Step functions\[email protected]\ndef train_step(data_iter):\n def train_step_fn(x, y):\n with tf.GradientTape() as tape:\n probabilities = model(x, training=True)\n loss = loss_fn(y, probabilities)\n grads = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n train_auc.update_state(y, probabilities)\n train_loss.update_state(loss)\n for _ in tf.range(step_size):\n strategy.experimental_run_v2(train_step_fn, next(data_iter))\n\[email protected]\ndef valid_step(data_iter):\n def valid_step_fn(x, y):\n probabilities = model(x, training=False)\n loss = loss_fn(y, probabilities)\n valid_auc.update_state(y, probabilities)\n valid_loss.update_state(loss)\n for _ in tf.range(valid_step_size):\n strategy.experimental_run_v2(valid_step_fn, next(data_iter))\n\[email protected]\ndef valid_2_step(data_iter):\n def valid_step_fn(x, y):\n probabilities = model(x, training=False)\n loss = loss_fn(y, probabilities)\n valid_2_auc.update_state(y, probabilities)\n valid_2_loss.update_state(loss)\n for _ in tf.range(valid_2_step_size):\n strategy.experimental_run_v2(valid_step_fn, next(data_iter))",
"_____no_output_____"
],
[
"# Train model\nwith strategy.scope():\n model = model_fn(config['MAX_LEN'])\n \n lr = lambda: exponential_schedule_with_warmup(tf.cast(optimizer.iterations, tf.float32), \n warmup_steps=warmup_steps, lr_start=lr_start, \n lr_max=lr_max, decay=decay)\n \n optimizer = optimizers.Adam(learning_rate=lr)\n loss_fn = losses.binary_crossentropy\n train_auc = metrics.AUC()\n valid_auc = metrics.AUC()\n valid_2_auc = metrics.AUC()\n train_loss = metrics.Sum()\n valid_loss = metrics.Sum()\n valid_2_loss = metrics.Sum()\n\nmetrics_dict = {'loss': train_loss, 'auc': train_auc, \n 'val_loss': valid_loss, 'val_auc': valid_auc, \n 'val_2_loss': valid_2_loss, 'val_2_auc': valid_2_auc}\n\nhistory = custom_fit_2(model, metrics_dict, train_step, valid_step, valid_2_step, train_data_iter, \n valid_data_iter, valid_2_data_iter, step_size, valid_step_size, valid_2_step_size, \n config['BATCH_SIZE'], config['EPOCHS'], config['ES_PATIENCE'], save_last=False)\n# model.save_weights('model.h5')\n\n# Make predictions\n# x_train = np.load(base_data_path + 'x_train.npy')\n# x_valid = np.load(base_data_path + 'x_valid.npy')\nx_valid_ml_eval = np.load(database_base_path + 'x_valid.npy')\n\n# train_preds = model.predict(get_test_dataset(x_train, config['BATCH_SIZE'], AUTO))\n# valid_preds = model.predict(get_test_dataset(x_valid, config['BATCH_SIZE'], AUTO))\nvalid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))\n\n# k_fold.loc[k_fold[f'fold_{fold_n}'] == 'train', f'pred_{fold_n}'] = np.round(train_preds)\n# k_fold.loc[k_fold[f'fold_{fold_n}'] == 'validation', f'pred_{fold_n}'] = np.round(valid_preds)\nvalid_df[f'pred_{fold_n}'] = valid_ml_preds",
"Train for 3340 steps, validate for 62 steps, validate_2 for 417 steps\n\nEPOCH 1/3\ntime: 1074.3s loss: 0.3107 auc: 0.9400 val_loss: 0.3254 val_auc: 0.9244 val_2_loss: 0.2137 val_2_auc: 0.9749\n\nEPOCH 2/3\ntime: 918.5s loss: 0.1801 auc: 0.9791 val_loss: 0.3355 val_auc: 0.9229 val_2_loss: 0.2184 val_2_auc: 0.9761\n\nEPOCH 3/3\ntime: 918.4s loss: 0.1327 auc: 0.9881 val_loss: 0.3875 val_auc: 0.9116 val_2_loss: 0.2594 val_2_auc: 0.9714\nTraining finished\n"
],
[
"# Fine-tune on validation set\n#################### ADD TAIL ####################\nx_valid_ml_tail = np.hstack([x_valid_ml, np.load(database_base_path + 'x_valid_tail.npy')])\ny_valid_ml_tail = np.vstack([y_valid_ml, y_valid_ml])\n\nvalid_step_size_tail = x_valid_ml_tail.shape[1] // config['BATCH_SIZE']\n\n# Build TF datasets\ntrain_ml_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_valid_ml_tail, y_valid_ml_tail, config['BATCH_SIZE'], AUTO, seed=SEED))\ntrain_ml_data_iter = iter(train_ml_dist_ds)",
"_____no_output_____"
],
[
"# Step functions\[email protected]\ndef train_ml_step(data_iter):\n def train_step_fn(x, y):\n with tf.GradientTape() as tape:\n probabilities = model(x, training=True)\n loss = loss_fn(y, probabilities)\n grads = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n train_auc.update_state(y, probabilities)\n train_loss.update_state(loss)\n for _ in tf.range(valid_step_size_tail):\n strategy.experimental_run_v2(train_step_fn, next(data_iter))",
"_____no_output_____"
],
[
"# Fine-tune on validation set\nhistory_ml = custom_fit_2(model, metrics_dict, train_ml_step, valid_step, valid_2_step, train_ml_data_iter, \n valid_data_iter, valid_2_data_iter, valid_step_size_tail, valid_step_size, valid_2_step_size, \n config['BATCH_SIZE'], 2, config['ES_PATIENCE'], save_last=False)\n\n# Join history\nfor key in history_ml.keys():\n history[key] += history_ml[key]\n \nmodel.save_weights('model.h5')\n\n# Make predictions\nvalid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))\nvalid_df[f'pred_ml_{fold_n}'] = valid_ml_preds\n\n### Delete data dir\nshutil.rmtree(base_data_path)",
"Train for 125 steps, validate for 62 steps, validate_2 for 417 steps\n\nEPOCH 1/2\ntime: 163.6s loss: 0.2160 auc: 0.9393 val_loss: 0.1695 val_auc: 0.9699 val_2_loss: 0.2167 val_2_auc: 0.9713\n\nEPOCH 2/2\ntime: 69.7s loss: 0.1598 auc: 0.9675 val_loss: 0.1141 val_auc: 0.9858 val_2_loss: 0.2452 val_2_auc: 0.9694\nTraining finished\n"
]
],
[
[
"## Model loss graph",
"_____no_output_____"
]
],
[
[
"plot_metrics_2(history)",
"_____no_output_____"
]
],
[
[
"# Model evaluation",
"_____no_output_____"
]
],
[
[
"# display(evaluate_model_single_fold(k_fold, fold_n, label_col='toxic_int').style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Confusion matrix",
"_____no_output_____"
]
],
[
[
"# train_set = k_fold[k_fold[f'fold_{fold_n}'] == 'train']\n# validation_set = k_fold[k_fold[f'fold_{fold_n}'] == 'validation'] \n# plot_confusion_matrix(train_set['toxic_int'], train_set[f'pred_{fold_n}'], \n# validation_set['toxic_int'], validation_set[f'pred_{fold_n}'])",
"_____no_output_____"
]
],
[
[
"# Model evaluation by language",
"_____no_output_____"
]
],
[
[
"display(evaluate_model_single_fold_lang(valid_df, fold_n).style.applymap(color_map))\n# ML fine-tunned preds\ndisplay(evaluate_model_single_fold_lang(valid_df, fold_n, pred_col='pred_ml').style.applymap(color_map))",
"_____no_output_____"
]
],
[
[
"# Visualize predictions",
"_____no_output_____"
]
],
[
[
"print('English validation set')\ndisplay(k_fold[['comment_text', 'toxic'] + [c for c in k_fold.columns if c.startswith('pred')]].head(10))\n\nprint('Multilingual validation set')\ndisplay(valid_df[['comment_text', 'toxic'] + [c for c in valid_df.columns if c.startswith('pred')]].head(10))",
"English validation set\n"
]
],
[
[
"# Test set predictions",
"_____no_output_____"
]
],
[
[
"x_test = np.load(database_base_path + 'x_test.npy')\ntest_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE'], AUTO))",
"_____no_output_____"
],
[
"submission = pd.read_csv('/kaggle/input/jigsaw-multilingual-toxic-comment-classification/sample_submission.csv')\nsubmission['toxic'] = test_preds\nsubmission.to_csv('submission.csv', index=False)\n\ndisplay(submission.describe())\ndisplay(submission.head(10))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbc935b9ea9ff5d11b6052c4a7be344a9f08a8b7
| 18,934 |
ipynb
|
Jupyter Notebook
|
Data_Pipeline.ipynb
|
CLandauGWU/group_e
|
af9b8ca619e9afeb0b276c1a83f5cd84310e927a
|
[
"MIT"
] | 1 |
2018-07-24T15:16:22.000Z
|
2018-07-24T15:16:22.000Z
|
Data_Pipeline.ipynb
|
CLandauGWU/group_e
|
af9b8ca619e9afeb0b276c1a83f5cd84310e927a
|
[
"MIT"
] | 2 |
2017-10-25T18:43:03.000Z
|
2017-11-08T22:58:23.000Z
|
Data_Pipeline.ipynb
|
CLandauGWU/group_e
|
af9b8ca619e9afeb0b276c1a83f5cd84310e927a
|
[
"MIT"
] | 2 |
2017-10-19T22:44:21.000Z
|
2018-01-10T13:45:18.000Z
| 38.79918 | 478 | 0.507553 |
[
[
[
"import pandas as pd\nimport numpy as np\n\nimport matplotlib as plt\n\nfrom shapely.geometry import Point, Polygon\n\nfrom sklearn.linear_model import LogisticRegression\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import KFold\n\nimport zipfile\nimport requests\nimport os\nimport shutil\n\n\n\nfrom downloading_funcs import addr_shape, down_extract_zip\nfrom supp_funcs import *\nimport lnks\n\nimport warnings #DANGER: I triggered a ton of warnings.\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"import geopandas as gpd\n\n%matplotlib inline",
"_____no_output_____"
],
[
"#Load the BBL list\nBBL12_17CSV = ['https://hub.arcgis.com/datasets/82ab09c9541b4eb8ba4b537e131998ce_22.csv', 'https://hub.arcgis.com/datasets/4c4d6b4defdf4561b737a594b6f2b0dd_23.csv', 'https://hub.arcgis.com/datasets/d7aa6d3a3fdc42c4b354b9e90da443b7_1.csv', 'https://hub.arcgis.com/datasets/a8434614d90e416b80fbdfe2cb2901d8_2.csv', 'https://hub.arcgis.com/datasets/714d5f8b06914b8596b34b181439e702_36.csv', 'https://hub.arcgis.com/datasets/c4368a66ce65455595a211d530facc54_3.csv',]",
"_____no_output_____"
],
[
"def data_pipeline(shapetype, bbl_links, supplement=None,\n dex=None, ts_lst_range=None):\n #A pipeline for group_e dataframe operations\n \n \n #Test inputs --------------------------------------------------------------\n if supplement:\n assert isinstance(supplement, list)\n assert isinstance(bbl_links, list)\n if ts_lst_range:\n assert isinstance(ts_lst_range, list)\n assert len(ts_lst_range) == 2 #Must be list of format [start-yr, end-yr]\n \n #We'll need our addresspoints and our shapefile\n if not dex:\n dex = addr_shape(shapetype)\n \n #We need a list of time_unit_of_analysis\n if ts_lst_range:\n ts_lst = [x+(i/100) for i in range(1,13,1) for x in range(1980, 2025)]\n ts_lst = [x for x in ts_lst if \n x >= ts_lst_range[0] and x <= ts_lst_range[1]]\n ts_lst = sorted(ts_lst)\n if not ts_lst_range:\n ts_lst = [x+(i/100) for i in range(1,13,1) for x in range(2012, 2017)]\n ts_lst = sorted(ts_lst)\n \n #Now we need to stack our BBL data ----------------------------------------\n \n #Begin by forming an empty DF \n bbl_df = pd.DataFrame()\n for i in list(range(2012, 2018)):\n bblpth = './data/bbls/Basic_Business_License_in_'+str(i)+'.csv' #Messy hack\n #TODO: generalize bblpth above\n bbl = pd.read_csv(bblpth, low_memory=False)\n col_len = len(bbl.columns)\n bbl_df = bbl_df.append(bbl)\n if len(bbl.columns) != col_len:\n print('Column Mismatch!')\n del bbl\n \n bbl_df.LICENSE_START_DATE = pd.to_datetime(\n bbl_df.LICENSE_START_DATE)\n \n bbl_df.LICENSE_EXPIRATION_DATE = pd.to_datetime(\n bbl_df.LICENSE_EXPIRATION_DATE)\n \n bbl_df.LICENSE_ISSUE_DATE = pd.to_datetime(\n bbl_df.LICENSE_ISSUE_DATE)\n\n \n bbl_df.sort_values('LICENSE_START_DATE')\n \n #Set up our time unit of analysis\n bbl_df['month'] = 0\n bbl_df['endMonth'] = 0\n bbl_df['issueMonth'] = 0\n \n bbl_df['month'] = bbl_df['LICENSE_START_DATE'].dt.year + (\n bbl_df['LICENSE_START_DATE'].dt.month/100\n )\n bbl_df['endMonth'] = bbl_df['LICENSE_EXPIRATION_DATE'].dt.year + (\n bbl_df['LICENSE_EXPIRATION_DATE'].dt.month/100\n )\n bbl_df['issueMonth'] = bbl_df['LICENSE_ISSUE_DATE'].dt.year + (\n bbl_df['LICENSE_ISSUE_DATE'].dt.month/100\n )\n bbl_df.endMonth.fillna(max(ts_lst))\n bbl_df['endMonth'][bbl_df['endMonth'] > max(ts_lst)] = max(ts_lst)\n \n #Sort on month\n bbl_df = bbl_df.dropna(subset=['month'])\n bbl_df = bbl_df.set_index(['MARADDRESSREPOSITORYID','month'])\n bbl_df = bbl_df.sort_index(ascending=True)\n bbl_df.reset_index(inplace=True)\n \n \n bbl_df = bbl_df[bbl_df['MARADDRESSREPOSITORYID'] >= 0]\n \n bbl_df = bbl_df.dropna(subset=['LICENSESTATUS', 'issueMonth', 'endMonth',\n 'MARADDRESSREPOSITORYID','month', \n 'LONGITUDE', 'LATITUDE'\n ])\n \n #Now that we have the BBL data, let's create our flag and points data -----\n \n #This is the addresspoints, passed from the dex param\n addr_df = dex[0]\n \n #Zip the latlongs\n addr_df['geometry'] = [\n Point(xy) for xy in zip(\n addr_df.LONGITUDE.apply(float), addr_df.LATITUDE.apply(float)\n )\n ]\n \n addr_df['Points'] = addr_df['geometry'] #Duplicate, so raw retains points\n \n addr_df['dummy_counter'] = 1 #Always one, always dropped before export\n \n crs='EPSG:4326' #Convenience assignment of crs\n \n #Now we're stacking for each month ----------------------------------------\n \n \n out_gdf = pd.DataFrame() #Empty storage df\n for i in ts_lst: #iterate through the list of months\n print('Month '+ str(i))\n strmfile_pth = str(\n './data/strm_file/' + str(i) +'_' + shapetype + '.csv')\n if os.path.exists(strmfile_pth):\n print('Skipping, ' + str(i) + ' stream file path already exists:')\n print(strmfile_pth)\n continue\n\n #dex[1] is the designated shapefile passed from the dex param, \n #and should match the shapetype defined in that param\n \n #Copy of the dex[1] shapefile\n shp_gdf = dex[1]\n \n #Active BBL in month i\n bbl_df['inRange'] = 0\n bbl_df['inRange'][(bbl_df.endMonth > i) & (bbl_df.month <= i)] = 1\n \n #Issued BBL in month i\n bbl_df['isuFlag'] = 0\n bbl_df['isuFlag'][bbl_df.issueMonth == i] = 1\n \n #Merge BBL and MAR datasets -------------------------------------------\n addr = pd.merge(addr_df, bbl_df, how='left', \n left_on='ADDRESS_ID', right_on='MARADDRESSREPOSITORYID')\n addr = gpd.GeoDataFrame(addr, crs=crs, geometry=addr.geometry)\n \n shp_gdf.crs = addr.crs\n \n raw = gpd.sjoin(shp_gdf, addr, how='left', op='intersects')\n \n #A simple percent of buildings with active flags per shape,\n #and call it a 'utilization index'\n numer = raw.groupby('NAME').sum()\n numer = numer.inRange\n denom = raw.groupby('NAME').sum()\n denom = denom.dummy_counter\n issue = raw.groupby('NAME').sum()\n issue = issue.isuFlag\n \n flags = []\n \n utl_inx = pd.DataFrame(numer/denom)\n \n utl_inx.columns = [\n 'Util_Indx_BBL'\n ]\n flags.append(utl_inx)\n \n #This is number of buildings with an active BBL in month i\n bbl_count = pd.DataFrame(numer)\n \n bbl_count.columns = [\n 'countBBL'\n ]\n flags.append(bbl_count)\n \n #This is number of buildings that were issued a BBL in month i\n isu_count = pd.DataFrame(issue)\n isu_count.columns = [\n 'countIssued'\n ]\n flags.append(isu_count)\n \n for flag in flags:\n flag.crs = shp_gdf.crs\n\n shp_gdf = shp_gdf.merge(flag,\n how=\"left\", left_on='NAME', right_index=True)\n shp_gdf['month'] = i\n \n \n \n #Head will be the list of retained columns\n head = ['NAME', 'Util_Indx_BBL',\n 'countBBL', 'countIssued',\n 'month', 'geometry']\n shp_gdf = shp_gdf[head]\n \n print('Merging...')\n if supplement: #this is where your code will be fed into the pipeline.\n \n #To include time unit of analysis, pass 'i=i' as the last\n #item in your args list over on lnks.py, and the for-loop\n #will catch that. Else, it will pass your last item as an arg.\n \n #Ping CDL if you need to pass a func with more args and we\n #can extend this.\n \n for supp_func in supplement:\n if len(supp_func) == 2:\n if supp_func[1] == 'i=i':\n shp_gdf = supp_func[0](shp_gdf, raw, i=i)\n if supp_func[1] != 'i=i':\n shp_gdf = supp_func[0](shp_gdf, raw, supp_func[1])\n \n if len(supp_func) == 3:\n if supp_func[2] == 'i=i':\n shp_gdf = supp_func[0](shp_gdf, raw, supp_func[1], i=i)\n if supp_func[2] != 'i=i':\n shp_gdf = supp_func[0](shp_gdf, raw, supp_func[1],\n supp_func[2])\n if len(supp_func) == 4:\n if supp_func[3] == 'i=i':\n shp_gdf = supp_func[0](shp_gdf, raw, supp_func[1],\n supp_func[2], i=i)\n if supp_func[3] != 'i=i':\n shp_gdf = supp_func[0](shp_gdf, raw, supp_func[1],\n supp_func[2], supp_func[3])\n print(str(supp_func[0]) + ' is done.')\n \n \n if not os.path.exists(strmfile_pth):\n shp_gdf = shp_gdf.drop('geometry', axis=1)\n \n #Save, also verify re-read works\n shp_gdf.to_csv(strmfile_pth, encoding='utf-8', index=False)\n shp_gdf = pd.read_csv(strmfile_pth, encoding='utf-8', \n engine='python')\n del shp_gdf, addr, utl_inx, numer, denom, issue, raw #Save me some memory please!\n #if i != 2016.12:\n # del raw\n print('Merged month:', i)\n print()\n \n #Done iterating through months here....\n pth = './data/strm_file/' #path of the streamfiles\n for file in os.listdir(pth):\n try:\n filepth = str(os.path.join(pth, file))\n print([os.path.getsize(filepth), filepth])\n fl = pd.read_csv(filepth, encoding='utf-8', engine='python') #read the stream file\n out_gdf = out_gdf.append(fl) #This does the stacking\n del fl\n except IsADirectoryError:\n continue\n out_gdf.to_csv('./data/' + shapetype + '_out.csv') #Save\n #shutil.rmtree('./data/strm_file/')\n \n print('Done!')\n return [bbl_df, addr_df, out_gdf] #Remove this later, for testing now\n ",
"_____no_output_____"
],
[
"dex = addr_shape('anc')",
"_____no_output_____"
],
[
"sets = data_pipeline('anc', BBL12_17CSV, supplement=lnks.supplm, dex=dex, ts_lst_range=None)",
"Month 2012.01\nSkipping, 2012.01 stream file path already exists:\n./data/strm_file/2012.01_anc.csv\nMonth 2012.02\nSkipping, 2012.02 stream file path already exists:\n./data/strm_file/2012.02_anc.csv\nMonth 2012.03\nSkipping, 2012.03 stream file path already exists:\n./data/strm_file/2012.03_anc.csv\nMonth 2012.04\nSkipping, 2012.04 stream file path already exists:\n./data/strm_file/2012.04_anc.csv\nMonth 2012.05\nSkipping, 2012.05 stream file path already exists:\n./data/strm_file/2012.05_anc.csv\nMonth 2012.06\nSkipping, 2012.06 stream file path already exists:\n./data/strm_file/2012.06_anc.csv\nMonth 2012.07\nSkipping, 2012.07 stream file path already exists:\n./data/strm_file/2012.07_anc.csv\nMonth 2012.08\nSkipping, 2012.08 stream file path already exists:\n./data/strm_file/2012.08_anc.csv\nMonth 2012.09\nSkipping, 2012.09 stream file path already exists:\n./data/strm_file/2012.09_anc.csv\nMonth 2012.1\nSkipping, 2012.1 stream file path already exists:\n./data/strm_file/2012.1_anc.csv\nMonth 2012.11\nSkipping, 2012.11 stream file path already exists:\n./data/strm_file/2012.11_anc.csv\nMonth 2012.12\nSkipping, 2012.12 stream file path already exists:\n./data/strm_file/2012.12_anc.csv\nMonth 2013.01\nSkipping, 2013.01 stream file path already exists:\n./data/strm_file/2013.01_anc.csv\nMonth 2013.02\nSkipping, 2013.02 stream file path already exists:\n./data/strm_file/2013.02_anc.csv\nMonth 2013.03\nSkipping, 2013.03 stream file path already exists:\n./data/strm_file/2013.03_anc.csv\nMonth 2013.04\nSkipping, 2013.04 stream file path already exists:\n./data/strm_file/2013.04_anc.csv\nMonth 2013.05\nSkipping, 2013.05 stream file path already exists:\n./data/strm_file/2013.05_anc.csv\nMonth 2013.06\nSkipping, 2013.06 stream file path already exists:\n./data/strm_file/2013.06_anc.csv\nMonth 2013.07\nSkipping, 2013.07 stream file path already exists:\n./data/strm_file/2013.07_anc.csv\nMonth 2013.08\nSkipping, 2013.08 stream file path already exists:\n./data/strm_file/2013.08_anc.csv\nMonth 2013.09\nSkipping, 2013.09 stream file path already exists:\n./data/strm_file/2013.09_anc.csv\nMonth 2013.1\nSkipping, 2013.1 stream file path already exists:\n./data/strm_file/2013.1_anc.csv\nMonth 2013.11\nSkipping, 2013.11 stream file path already exists:\n./data/strm_file/2013.11_anc.csv\nMonth 2013.12\nSkipping, 2013.12 stream file path already exists:\n./data/strm_file/2013.12_anc.csv\nMonth 2014.01\nSkipping, 2014.01 stream file path already exists:\n./data/strm_file/2014.01_anc.csv\nMonth 2014.02\nSkipping, 2014.02 stream file path already exists:\n./data/strm_file/2014.02_anc.csv\nMonth 2014.03\nSkipping, 2014.03 stream file path already exists:\n./data/strm_file/2014.03_anc.csv\nMonth 2014.04\nSkipping, 2014.04 stream file path already exists:\n./data/strm_file/2014.04_anc.csv\nMonth 2014.05\nSkipping, 2014.05 stream file path already exists:\n./data/strm_file/2014.05_anc.csv\nMonth 2014.06\nSkipping, 2014.06 stream file path already exists:\n./data/strm_file/2014.06_anc.csv\nMonth 2014.07\nMerging...\n<function ITSPExtract at 0x7fee9bea9ea0> is done.\n<function clim_ingest at 0x7fee9bea9e18> is done.\n<function oecdGdpQs at 0x7fee9bea9d08> is done.\n"
],
[
"sets[2].columns #Our number of rows equals our number of shapes * number of months",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc93b41ad6fe99e6032de2517a537994fcef8d2
| 1,048,248 |
ipynb
|
Jupyter Notebook
|
python/money_making_model.ipynb
|
mlisovyi/dfine_hack2018
|
fcab07535643faae9c4600584cf9b2fbf233d01d
|
[
"MIT"
] | null | null | null |
python/money_making_model.ipynb
|
mlisovyi/dfine_hack2018
|
fcab07535643faae9c4600584cf9b2fbf233d01d
|
[
"MIT"
] | null | null | null |
python/money_making_model.ipynb
|
mlisovyi/dfine_hack2018
|
fcab07535643faae9c4600584cf9b2fbf233d01d
|
[
"MIT"
] | 2 |
2018-10-19T13:47:41.000Z
|
2018-10-19T13:52:45.000Z
| 554.922181 | 178,972 | 0.826352 |
[
[
[
"# Important installation",
"_____no_output_____"
],
[
"This notebook requires unusual packages: `LightGBM`, `SHAP` and `LIME`. \nFor installation, do:\n\n`conda install lightgbm lime shap`\n## Initial classical imports",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\nimport pandas as pd\nimport warnings\nwarnings.simplefilter(action='ignore', category=Warning)\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Read in data\nThe stock data first:",
"_____no_output_____"
]
],
[
[
"dfs = {}\n\nfor ticker in ['BLK', 'GS', 'MS']:\n dfs[ticker] = pd.read_pickle('stock_{}.pickle'.format(ticker)).set_index('Date')",
"_____no_output_____"
]
],
[
[
"The media data:",
"_____no_output_____"
]
],
[
[
"df_media = pd.read_csv('MediaAttention_Mini.csv', parse_dates=[0], index_col='Time')",
"_____no_output_____"
],
[
"df_media.info()",
"<class 'pandas.core.frame.DataFrame'>\nDatetimeIndex: 4556 entries, 2005-07-22 to 2018-01-10\nData columns (total 23 columns):\nMorgan_Stanley_count 4556 non-null int64\nGoldman_Sachs_count 4556 non-null int64\nBlackRock_count 4556 non-null int64\npositive_count 4556 non-null int64\nnegative_count 4556 non-null int64\nMorgan_Stanley_positive_count 4556 non-null int64\nMorgan_Stanley_negative_count 4556 non-null int64\nGoldman_Sachs_positive_count 4556 non-null int64\nGoldman_Sachs_negative_count 4556 non-null int64\nBlackRock_positive_count 4556 non-null int64\nBlackRock_negative_count 4556 non-null int64\ntotal_msg_count 4556 non-null int64\nMorgan_Stanley_frac 3439 non-null float64\nGoldman_Sachs_frac 3439 non-null float64\nBlackRock_frac 3439 non-null float64\npositive_frac 3439 non-null float64\nnegative_frac 3439 non-null float64\nMorgan_Stanley_positive_frac 3439 non-null float64\nMorgan_Stanley_negative_frac 3439 non-null float64\nGoldman_Sachs_positive_frac 3439 non-null float64\nGoldman_Sachs_negative_frac 3439 non-null float64\nBlackRock_positive_frac 3439 non-null float64\nBlackRock_negative_frac 3439 non-null float64\ndtypes: float64(11), int64(12)\nmemory usage: 854.2 KB\n"
],
[
"import glob\ndf_media_long = pd.DataFrame()\nfor fle in glob.glob('MediaAttentionLong*.csv'):\n dummy = pd.read_csv(fle, parse_dates=[0], index_col='Time')\n df_media_long = pd.concat([df_media_long,dummy])\n \ndf_media_long = df_media_long['2007':'2013']\ndf_media_long = df_media_long.resample('1D').sum()\ndf_media_long.shape",
"_____no_output_____"
],
[
"#df_media_long= pd.read_csv('MediaAttentionLong_2008.csv', parse_dates=[0], index_col='Time')",
"_____no_output_____"
],
[
"df_media_long.columns = ['{}_Long'.format(c) for c in df_media_long.columns]",
"_____no_output_____"
],
[
"df_media_long.loc['2008',:].info()",
"<class 'pandas.core.frame.DataFrame'>\nDatetimeIndex: 366 entries, 2008-01-01 to 2008-12-31\nFreq: D\nData columns (total 23 columns):\nMorgan_Stanley_count_Long 366 non-null int64\nGoldman_Sachs_count_Long 366 non-null int64\nBlackRock_count_Long 366 non-null int64\npositive_count_Long 366 non-null int64\nnegative_count_Long 366 non-null int64\nMorgan_Stanley_positive_count_Long 366 non-null int64\nMorgan_Stanley_negative_count_Long 366 non-null int64\nGoldman_Sachs_positive_count_Long 366 non-null int64\nGoldman_Sachs_negative_count_Long 366 non-null int64\nBlackRock_positive_count_Long 366 non-null int64\nBlackRock_negative_count_Long 366 non-null int64\ntotal_msg_count_Long 366 non-null int64\nMorgan_Stanley_frac_Long 366 non-null float64\nGoldman_Sachs_frac_Long 366 non-null float64\nBlackRock_frac_Long 366 non-null float64\npositive_frac_Long 366 non-null float64\nnegative_frac_Long 366 non-null float64\nMorgan_Stanley_positive_frac_Long 366 non-null float64\nMorgan_Stanley_negative_frac_Long 366 non-null float64\nGoldman_Sachs_positive_frac_Long 366 non-null float64\nGoldman_Sachs_negative_frac_Long 366 non-null float64\nBlackRock_positive_frac_Long 366 non-null float64\nBlackRock_negative_frac_Long 366 non-null float64\ndtypes: float64(11), int64(12)\nmemory usage: 68.6 KB\n"
],
[
"for ticker in dfs:\n dfs[ticker] = dfs[ticker].merge(df_media, how='inner', left_index=True, right_index=True)\n dfs[ticker] = dfs[ticker].merge(df_media_long, how='inner', left_index=True, right_index=True)",
"_____no_output_____"
],
[
"dfs['BLK'].head(10)",
"_____no_output_____"
]
],
[
[
"# Model",
"_____no_output_____"
]
],
[
[
"from sklearn.ensemble import RandomForestClassifier\nfrom sklearn.metrics import accuracy_score, recall_score, roc_auc_score, make_scorer\nfrom sklearn.model_selection import cross_val_score, KFold",
"/home/mlisovyi/Drive/anaconda3/envs/dfine_py36/lib/python3.6/site-packages/sklearn/ensemble/weight_boosting.py:29: DeprecationWarning: numpy.core.umath_tests is an internal NumPy module and should not be imported. It will be removed in a future NumPy release.\n from numpy.core.umath_tests import inner1d\n"
],
[
"model = RandomForestClassifier(n_estimators=100, max_depth=4, random_state=314, n_jobs=4)",
"_____no_output_____"
],
[
"for ticker in dfs:\n X = dfs[ticker].drop(['Label', 'Close'], axis=1).fillna(-1)\n y = dfs[ticker].loc[:,'Label']\n scores = cross_val_score(model, X, y, \n scoring=make_scorer(accuracy_score), \n cv=KFold(10, shuffle=True, random_state=314), \n n_jobs=1\n )\n print('{} prediction performance in accuracy = {:.3f}+-{:.3f}'.format(ticker,\n np.mean(scores),\n np.std(scores)\n ))",
"BLK prediction performance in accuracy = 0.500+-0.041\nGS prediction performance in accuracy = 0.514+-0.037\nMS prediction performance in accuracy = 0.515+-0.030\n"
],
[
"plt.figure(figsize=(15,12))\nsns.heatmap(dfs['BLK'].corr(), vmin=-0.2, vmax=0.2)",
"_____no_output_____"
]
],
[
[
"### Train a model on all data",
"_____no_output_____"
]
],
[
[
" df = pd.concat(list(dfs.values()), axis=0)\n X = df.drop(['Label', 'Close'], axis=1).fillna(-1)\n y = df.loc[:,'Label']\n scores = cross_val_score(model, X, y, \n scoring=make_scorer(accuracy_score), \n cv=KFold(10, shuffle=True, random_state=314), \n n_jobs=1\n )\n print('{} prediction performance in accuracy = {:.3f}+-{:.3f}'.format('ALL',\n np.mean(scores),\n np.std(scores)\n ))",
"ALL prediction performance in accuracy = 0.587+-0.015\n"
],
[
"scores",
"_____no_output_____"
],
[
"mdl = model.fit(X,y)",
"_____no_output_____"
],
[
"import shap\nshap.initjs()",
"_____no_output_____"
],
[
"explainer=shap.TreeExplainer(mdl)\nshap_values = explainer.shap_values(X)",
"_____no_output_____"
],
[
"shap.summary_plot(shap_values, X, plot_type=\"bar\")",
"_____no_output_____"
]
],
[
[
"## LightGBM",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import StratifiedKFold, KFold, GroupKFold\nfrom sklearn.base import clone, ClassifierMixin, RegressorMixin\nimport lightgbm as lgb\n\n\ndef train_single_model(clf_, X_, y_, random_state_=314, opt_parameters_={}, fit_params_={}):\n '''\n A wrapper to train a model with particular parameters\n '''\n c = clone(clf_)\n c.set_params(**opt_parameters_)\n c.set_params(random_state=random_state_)\n return c.fit(X_, y_, **fit_params_)\n\ndef train_model_in_CV(model, X, y, metric, metric_args={},\n model_name='xmodel',\n seed=31416, n=5,\n opt_parameters_={}, fit_params_={},\n verbose=True,\n groups=None, y_eval=None):\n # the list of classifiers for voting ensable\n clfs = []\n # performance \n perf_eval = {'score_i_oof': 0,\n 'score_i_ave': 0,\n 'score_i_std': 0,\n 'score_i': []\n }\n # full-sample oof prediction\n y_full_oof = pd.Series(np.zeros(shape=(y.shape[0],)), \n index=y.index)\n \n sample_weight=None\n if 'sample_weight' in metric_args:\n sample_weight=metric_args['sample_weight']\n \n index_weight=None\n if 'index_weight' in metric_args:\n index_weight=metric_args['index_weight']\n del metric_args['index_weight']\n \n doSqrt=False\n if 'sqrt' in metric_args:\n doSqrt=True\n del metric_args['sqrt']\n\n if groups is None:\n cv = KFold(n, shuffle=True, random_state=seed) #Stratified\n else:\n cv = GroupKFold(n)\n # The out-of-fold (oof) prediction for the k-1 sample in the outer CV loop\n y_oof = pd.Series(np.zeros(shape=(X.shape[0],)), \n index=X.index)\n scores = []\n clfs = []\n\n for n_fold, (trn_idx, val_idx) in enumerate(cv.split(X, (y!=0).astype(np.int8), groups=groups)):\n X_trn, y_trn = X.iloc[trn_idx], y.iloc[trn_idx]\n X_val, y_val = X.iloc[val_idx], y.iloc[val_idx]\n \n if 'LGBMRanker' in type(model).__name__ and groups is not None:\n G_trn, G_val = groups.iloc[trn_idx], groups.iloc[val_idx] \n\n if fit_params_:\n # use _stp data for early stopping\n fit_params_[\"eval_set\"] = [(X_trn,y_trn), (X_val,y_val)]\n fit_params_['verbose'] = verbose\n if index_weight is not None:\n fit_params_[\"sample_weight\"] = y_trn.index.map(index_weight).values\n fit_params_[\"eval_sample_weight\"] = [None, y_val.index.map(index_weight).values]\n if 'LGBMRanker' in type(model).__name__ and groups is not None:\n fit_params_['group'] = G_trn.groupby(G_trn, sort=False).count()\n fit_params_['eval_group'] = [G_trn.groupby(G_trn, sort=False).count(),\n G_val.groupby(G_val, sort=False).count()]\n\n #display(y_trn.head())\n clf = train_single_model(model, X_trn, y_trn, 314+n_fold, opt_parameters_, fit_params_)\n\n clfs.append(('{}{}'.format(model_name,n_fold), clf))\n # oof predictions\n if isinstance(clf, RegressorMixin):\n y_oof.iloc[val_idx] = clf.predict(X_val)\n elif isinstance(clf, ClassifierMixin) and metric.__name__=='roc_auc_score':\n y_oof.iloc[val_idx] = clf.predict_proba(X_val)[:,1]\n else:\n y_oof.iloc[val_idx] = clf.predict(X_val)\n # prepare weights for evaluation\n if sample_weight is not None:\n metric_args['sample_weight'] = y_val.map(sample_weight)\n elif index_weight is not None:\n metric_args['sample_weight'] = y_val.index.map(index_weight).values\n # prepare target values\n y_true_tmp = y_val if 'LGBMRanker' not in type(model).__name__ and y_eval is None else y_eval.iloc[val_idx]\n y_pred_tmp = y_oof.iloc[val_idx] if y_eval is None else y_oof.iloc[val_idx] \n #store evaluated metric\n scores.append(metric(y_true_tmp, y_pred_tmp, **metric_args))\n #cleanup\n del X_trn, y_trn, X_val, y_val, y_true_tmp, y_pred_tmp\n\n # Store performance info for this CV\n if sample_weight is not None:\n metric_args['sample_weight'] = y_oof.map(sample_weight)\n elif index_weight is not None:\n metric_args['sample_weight'] = y_oof.index.map(index_weight).values\n perf_eval['score_i_oof'] = metric(y, y_oof, **metric_args)\n perf_eval['score_i'] = scores\n \n if doSqrt:\n for k in perf_eval.keys():\n if 'score' in k:\n perf_eval[k] = np.sqrt(perf_eval[k])\n scores = np.sqrt(scores)\n \n perf_eval['score_i_ave'] = np.mean(scores)\n perf_eval['score_i_std'] = np.std(scores)\n\n return clfs, perf_eval, y_oof\n\ndef print_perf_clf(name, perf_eval):\n print('Performance of the model:') \n print('Mean(Val) score inner {} Classifier: {:.4f}+-{:.4f}'.format(name, \n perf_eval['score_i_ave'],\n perf_eval['score_i_std']\n ))\n print('Min/max scores on folds: {:.4f} / {:.4f}'.format(np.min(perf_eval['score_i']),\n np.max(perf_eval['score_i'])))\n print('OOF score inner {} Classifier: {:.4f}'.format(name, perf_eval['score_i_oof']))\n print('Scores in individual folds: {}'.format(perf_eval['score_i']))\n\n\n\n",
"_____no_output_____"
],
[
"mdl_inputs = {\n 'lgbm1_reg': (lgb.LGBMClassifier(max_depth=-1, min_child_samples=400, random_state=314, silent=True, metric='None', \n n_jobs=4, n_estimators=1000, learning_rate=0.1),\n {'colsample_bytree': 0.9, 'min_child_weight': 10.0, 'num_leaves': 20, 'reg_alpha': 1, 'subsample': 0.9}, \n {\"early_stopping_rounds\":20, \n \"eval_metric\" : 'binary_logloss',\n 'eval_names': ['train', 'early_stop'],\n 'verbose': False, \n #'callbacks': [lgb.reset_parameter(learning_rate=learning_rate_decay_power)],\n },#'categorical_feature': 'auto'},\n y,\n None\n ),\n 'rf1': (\n RandomForestClassifier(n_estimators=100, max_depth=4, random_state=314, n_jobs=4),\n {},\n {},\n y,\n None\n )\n}",
"_____no_output_____"
],
[
"%%time\nmdls = {}\nresults = {}\ny_oofs = {}\nfor name, (mdl, mdl_pars, fit_pars, y_, g_) in mdl_inputs.items():\n print('--------------- {} -----------'.format(name))\n mdl_, perf_eval_, y_oof_ = train_model_in_CV(mdl, X, y_, accuracy_score, \n metric_args={},\n model_name=name, \n opt_parameters_=mdl_pars,\n fit_params_=fit_pars, \n n=10, seed=3146,\n verbose=500, \n groups=g_)\n results[name] = perf_eval_\n mdls[name] = mdl_\n y_oofs[name] = y_oof_\n print_perf_clf(name, perf_eval_)\n\n",
"--------------- lgbm1_reg -----------\nTraining until validation scores don't improve for 20 rounds.\n[500]\ttrain's binary_logloss: 0.421587\tearly_stop's binary_logloss: 0.606957\nEarly stopping, best iteration is:\n[567]\ttrain's binary_logloss: 0.405288\tearly_stop's binary_logloss: 0.60054\nint8 float64\nTraining until validation scores don't improve for 20 rounds.\n"
]
],
[
[
"# Train LGBM model on a simple rain/val/test split (70/15/15)",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"X_1, X_tst, y_1, y_tst = train_test_split(X, y, test_size=0.15, shuffle=True, random_state=314)\nX_trn, X_val, y_trn, y_val = train_test_split(X_1, y_1, test_size=0.15, shuffle=True, random_state=31)",
"_____no_output_____"
],
[
"mdl = lgb.LGBMClassifier(max_depth=-1, min_child_samples=400, random_state=314,\n silent=True, metric='None', \n n_jobs=4, n_estimators=1000, learning_rate=0.1,\n **{'colsample_bytree': 0.9, 'min_child_weight': 10.0, 'num_leaves': 20, 'reg_alpha': 1, 'subsample': 0.9}\n )",
"_____no_output_____"
],
[
"mdl.fit(X_trn, y_trn, \n **{\"early_stopping_rounds\":20, \n \"eval_metric\" : 'binary_logloss',\n 'eval_set': [(X_trn, y_trn), (X_val, y_val)],\n 'eval_names': ['train', 'early_stop'],\n 'verbose': 100\n })",
"Training until validation scores don't improve for 20 rounds.\n[100]\ttrain's binary_logloss: 0.601553\tearly_stop's binary_logloss: 0.662331\n[200]\ttrain's binary_logloss: 0.550266\tearly_stop's binary_logloss: 0.645603\n[300]\ttrain's binary_logloss: 0.510233\tearly_stop's binary_logloss: 0.630478\n[400]\ttrain's binary_logloss: 0.479222\tearly_stop's binary_logloss: 0.621539\n[500]\ttrain's binary_logloss: 0.452081\tearly_stop's binary_logloss: 0.612112\n[600]\ttrain's binary_logloss: 0.429007\tearly_stop's binary_logloss: 0.602617\nEarly stopping, best iteration is:\n[604]\ttrain's binary_logloss: 0.428126\tearly_stop's binary_logloss: 0.602573\n"
],
[
"print('Accuracy score on train/validation/test samples is: {:.3f}/{:.3f}/{:.3f}'.format(accuracy_score(y_trn, mdl.predict(X_trn)),\n accuracy_score(y_val, mdl.predict(X_val)),\n accuracy_score(y_tst, mdl.predict(X_tst))\n ))",
"Accuracy score on train/validation/test samples is: 0.849/0.680/0.662\n"
]
],
[
[
"## Do LGBM model exlanation\n### SHAP",
"_____no_output_____"
]
],
[
[
"explainer=shap.TreeExplainer(mdl)\nshap_values = explainer.shap_values(X_tst)\nshap.summary_plot(shap_values, X_tst, plot_type=\"bar\")",
"_____no_output_____"
]
],
[
[
"_To understand how a single feature effects the output of the model we can plot **the SHAP value of that feature vs. the value of the feature** for all the examples in a dataset. Since SHAP values represent a feature's responsibility for a change in the model output, **the plot below represents the change in predicted label as either of chosen variables changes**._",
"_____no_output_____"
]
],
[
[
"for f in ['negative_frac', 'BlackRock_count_Long', 'positive_count', 'diff_7d']:\n shap.dependence_plot(f, shap_values, X_tst)",
"_____no_output_____"
]
],
[
[
"### LIME",
"_____no_output_____"
]
],
[
[
"import lime\nfrom lime.lime_tabular import LimeTabularExplainer",
"_____no_output_____"
],
[
"explainer = LimeTabularExplainer(X_trn.values, \n feature_names=X_trn.columns, \n class_names=['Down','Up'], \n verbose=False, \n mode='classification')",
"_____no_output_____"
],
[
"exp= []\nfor i in range(5):\n e = explainer.explain_instance(X_trn.iloc[i,:].values, mdl.predict_proba)\n _ = e.as_pyplot_figure(label=1)\n #exp.append(e)",
"_____no_output_____"
],
[
"import pickle",
"_____no_output_____"
],
[
"with open('model_lighgbm.pkl', 'wb') as fout:\n pickle.dump(mdl, fout)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbc944800a312555771b81d5f067e311ae5f52e0
| 6,330 |
ipynb
|
Jupyter Notebook
|
CE019_Lab5/019_lab5_linear_regression.ipynb
|
neel4888/019_Neel
|
2d4458d796ed64a592fc32aef23750608b20c4f8
|
[
"Apache-2.0"
] | null | null | null |
CE019_Lab5/019_lab5_linear_regression.ipynb
|
neel4888/019_Neel
|
2d4458d796ed64a592fc32aef23750608b20c4f8
|
[
"Apache-2.0"
] | null | null | null |
CE019_Lab5/019_lab5_linear_regression.ipynb
|
neel4888/019_Neel
|
2d4458d796ed64a592fc32aef23750608b20c4f8
|
[
"Apache-2.0"
] | null | null | null | 6,330 | 6,330 | 0.624961 |
[
[
[
"# Exercise 1: Try Linear Regression just using numpy (Without Tensorflow/Pytorch or other torch library). ",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"# Input (temp, rainfall, humidity)\ninputs = np.array([[73, 67, 43], \n [91, 88, 64], \n [87, 134, 58], \n [102, 43, 37], \n [69, 96, 70]], dtype='float32')",
"_____no_output_____"
],
[
"# Target (apples)\ntargets = np.array([[56], \n [81], \n [119], \n [22], \n [103]], dtype='float32')",
"_____no_output_____"
],
[
"print(\"Inputs:\",inputs)\nprint(\"Targets:\",targets)",
"Inputs: [[ 73. 67. 43.]\n [ 91. 88. 64.]\n [ 87. 134. 58.]\n [102. 43. 37.]\n [ 69. 96. 70.]]\nTargets: [[ 56.]\n [ 81.]\n [119.]\n [ 22.]\n [103.]]\n"
],
[
"from numpy import random\n\nw = random.rand(1, 3)\nb=random.rand(1,1)\nprint(w)\nprint(b)",
"[[0.00535657 0.3576928 0.27095887]]\n[[0.98186623]]\n"
],
[
"# Define the model\n# @=FOR DOT PRODUCT\ndef model(x):\n return x @ w.transpose()+b",
"_____no_output_____"
],
[
"# Generate predictions\npred=model(inputs)\nprint(pred)",
"[[36.98954494]\n [50.28764826]\n [65.09433768]\n [26.93450507]\n [54.65709929]]\n"
],
[
"# Compare with targets\nprint(targets)",
"[[ 56.]\n [ 81.]\n [119.]\n [ 22.]\n [103.]]\n"
]
],
[
[
"## Loss Function\n\nWe can compare the predictions with the actual targets, using the following method: \n* Calculate the difference between the two matrices (`preds` and `targets`).\n* Square all elements of the difference matrix to remove negative values.\n* Calculate the average of the elements in the resulting matrix.\n\nThe result is a single number, known as the **mean squared error** (MSE).",
"_____no_output_____"
]
],
[
[
"# MSE loss\ndef mse(t1,t2):\n diff=(t1-t2)**2\n _sum_=np.sum(diff) \n return _sum_/diff.size\n",
"_____no_output_____"
],
[
"# Compute loss\nloss=mse(pred,targets)\nprint(loss)\nl_rate=1e-4",
"1314.3703541168882\n"
]
],
[
[
"## Gradient Descent Algorithm",
"_____no_output_____"
]
],
[
[
"def grad_desc(w,b):\n m=len(targets)\n b_de=(l_rate * ((1/m) * np.sum(model(inputs) - targets)))\n for i in range(len(w)):\n w_de=(l_rate * ((1/m) * np.sum(model(inputs) - targets)))\n return w_de,b_de",
"_____no_output_____"
]
],
[
[
"## Training for 200 epochs",
"_____no_output_____"
]
],
[
[
"# Train for 200 epochs\nfor i in range(200):\n pred=model(inputs)\n loss=mse(pred,targets)\n w_de,b_de=grad_desc(w,b)\n w=w-w_de\n b=b-b_de",
"_____no_output_____"
],
[
"print(model(inputs)) ",
"[[ 60.99553615]\n [ 82.12168008]\n [101.62519387]\n [ 50.81002894]\n [ 85.44739237]]\n"
],
[
"print(targets)",
"[[ 56.]\n [ 81.]\n [119.]\n [ 22.]\n [103.]]\n"
],
[
"loss=mse(pred,targets)\nprint(loss)",
"293.241851057183\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbc948ce5891ca345b83350194865b61fe0ce92e
| 129 |
ipynb
|
Jupyter Notebook
|
01-Lesson-Plans/11-Classification/3/Activities/04-Stu_Do_More_Loans/Solved/more_loans.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 1 |
2021-04-13T07:14:34.000Z
|
2021-04-13T07:14:34.000Z
|
01-Lesson-Plans/11-Classification/3/Activities/04-Stu_Do_More_Loans/Solved/more_loans.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 2 |
2021-06-02T03:14:19.000Z
|
2022-02-11T23:21:24.000Z
|
01-Lesson-Plans/11-Classification/3/Activities/04-Stu_Do_More_Loans/Solved/more_loans.ipynb
|
tatianegercina/FinTech
|
b40687aa362d78674e223eb15ecf14bc59f90b62
|
[
"ADSL"
] | 1 |
2021-05-07T13:26:50.000Z
|
2021-05-07T13:26:50.000Z
| 32.25 | 75 | 0.883721 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbc950e1afd15da39b25938144ab1aa71fc7085d
| 23,489 |
ipynb
|
Jupyter Notebook
|
02-lesson/07-groupby.ipynb
|
chilperic/scipy-2017-tutorial-pandas
|
6b52344ad58d6dfa4aaaad6327bb5054fb2a5b93
|
[
"MIT"
] | 164 |
2017-06-27T19:20:26.000Z
|
2022-01-09T03:31:02.000Z
|
02-lesson/07-groupby.ipynb
|
chilperic/scipy-2017-tutorial-pandas
|
6b52344ad58d6dfa4aaaad6327bb5054fb2a5b93
|
[
"MIT"
] | 2 |
2017-11-28T17:03:33.000Z
|
2018-03-13T15:23:04.000Z
|
02-lesson/07-groupby.ipynb
|
chilperic/scipy-2017-tutorial-pandas
|
6b52344ad58d6dfa4aaaad6327bb5054fb2a5b93
|
[
"MIT"
] | 167 |
2017-06-29T21:16:19.000Z
|
2021-10-01T22:53:29.000Z
| 27.281069 | 141 | 0.371834 |
[
[
[
"import pandas as pd",
"_____no_output_____"
],
[
"gapminder = pd.read_csv('../data/gapminder.tsv', delimiter='\\t')",
"_____no_output_____"
],
[
"gapminder.head()",
"_____no_output_____"
],
[
"gapminder.groupby('year')['lifeExp'].mean()",
"_____no_output_____"
],
[
"y1952 = gapminder.loc[gapminder['year'] == 1952, :]",
"_____no_output_____"
],
[
"l = [1, 2, 3, 4, 5]",
"_____no_output_____"
],
[
"l[:]",
"_____no_output_____"
],
[
"y1952['lifeExp'].mean()",
"_____no_output_____"
],
[
"gapminder.groupby('year')['lifeExp'].describe()",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"gapminder.groupby('continent')['lifeExp'].agg(np.mean)",
"_____no_output_____"
],
[
"gapminder.groupby('continent')['lifeExp'].aggregate(np.std)",
"_____no_output_____"
],
[
"def my_mean(values):\n n = len(values)\n s = np.sum(values)\n return s / n",
"_____no_output_____"
],
[
"gapminder.groupby('continent')['lifeExp'].aggregate(my_mean)",
"_____no_output_____"
],
[
"gapminder.groupby('continent')['lifeExp'].aggregate([\n np.count_nonzero,\n np.mean,\n np.std\n])",
"_____no_output_____"
],
[
"gapminder.groupby('continent')['lifeExp'].aggregate({\n 'ncount': np.count_nonzero,\n 'mean': np.mean,\n 'std': np.std\n})",
"/home/dchen/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:4: FutureWarning: using a dict on a Series for aggregation\nis deprecated and will be removed in a future version\n after removing the cwd from sys.path.\n"
],
[
"gapminder.groupby('continent')['lifeExp'].aggregate([\n np.count_nonzero,\n np.mean,\n np.std]).\\\n rename(columns = {'count_nonzero': 'count',\n 'mean': 'avg',\n 'std': 'std_dev'}).\\\n reset_index()",
"_____no_output_____"
],
[
"|",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc9522a67500265ed57ab9217ae688dbb595c20
| 168,441 |
ipynb
|
Jupyter Notebook
|
notebooks/14_Advanced_RNNs.ipynb
|
zrsmith92/practicalAI
|
6856bfcb25db3b9725ee2e53697b6cb989f80d5a
|
[
"MIT"
] | 45 |
2020-05-29T05:03:20.000Z
|
2021-11-28T11:43:37.000Z
|
notebooks/14_Advanced_RNNs.ipynb
|
Amore-HDU/practicalAI
|
afe66e56ec0f984782fe5a0987a50f25f55153cc
|
[
"MIT"
] | 9 |
2021-03-18T21:44:20.000Z
|
2022-03-11T23:37:17.000Z
|
notebooks/14_Advanced_RNNs.ipynb
|
Amore-HDU/practicalAI
|
afe66e56ec0f984782fe5a0987a50f25f55153cc
|
[
"MIT"
] | 30 |
2020-05-21T09:04:06.000Z
|
2021-10-16T14:52:40.000Z
| 57.764403 | 41,582 | 0.638888 |
[
[
[
"# Advanced RNNs",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/GokuMohandas/practicalAI/master/images/logo.png\" width=150>\n\nIn this notebook we're going to cover some advanced topics related to RNNs.\n\n1. Conditioned hidden state\n2. Char-level embeddings\n3. Encoder and decoder\n4. Attentional mechanisms\n5. Implementation\n\n\n",
"_____no_output_____"
],
[
"# Set up",
"_____no_output_____"
]
],
[
[
"# Load PyTorch library\n!pip3 install torch",
"Requirement already satisfied: torch in /usr/local/lib/python3.6/dist-packages (1.0.0)\n"
],
[
"import os\nfrom argparse import Namespace\nimport collections\nimport copy\nimport json\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport re\nimport torch",
"_____no_output_____"
],
[
"# Set Numpy and PyTorch seeds\ndef set_seeds(seed, cuda):\n np.random.seed(seed)\n torch.manual_seed(seed)\n if cuda:\n torch.cuda.manual_seed_all(seed)\n \n# Creating directories\ndef create_dirs(dirpath):\n if not os.path.exists(dirpath):\n os.makedirs(dirpath)",
"_____no_output_____"
],
[
"# Arguments\nargs = Namespace(\n seed=1234,\n cuda=True,\n batch_size=4,\n condition_vocab_size=3, # vocabulary for condition possibilities\n embedding_dim=100,\n rnn_hidden_dim=100,\n hidden_dim=100,\n num_layers=1,\n bidirectional=False,\n)\n\n# Set seeds\nset_seeds(seed=args.seed, cuda=args.cuda)\n\n# Check CUDA\nif not torch.cuda.is_available():\n args.cuda = False\nargs.device = torch.device(\"cuda\" if args.cuda else \"cpu\")\nprint(\"Using CUDA: {}\".format(args.cuda))",
"Using CUDA: True\n"
]
],
[
[
"# Conditioned RNNs",
"_____no_output_____"
],
[
"Conditioning an RNN is to add extra information that will be helpful towards a prediction. We can encode (embed it) this information and feed it along with the sequential input into our model. For example, suppose in our document classificaiton example in the previous notebook, we knew the publisher of each news article (NYTimes, ESPN, etc.). We could have encoded that information to help with the prediction. There are several different ways of creating a conditioned RNN.\n\n**Note**: If the conditioning information is novel for each input in the sequence, just concatenate it along with each time step's input.",
"_____no_output_____"
],
[
"1. Make the initial hidden state the encoded information instead of using the initial zerod hidden state. Make sure that the size of the encoded information is the same as the hidden state for the RNN.\n",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/GokuMohandas/practicalAI/master/images/conditioned_rnn1.png\" width=400>",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"# Condition\ncondition = torch.LongTensor([0, 2, 1, 2]) # batch size of 4 with a vocab size of 3\ncondition_embeddings = nn.Embedding(\n embedding_dim=args.embedding_dim, # should be same as RNN hidden dim\n num_embeddings=args.condition_vocab_size) # of unique conditions\n\n# Initialize hidden state\nnum_directions = 1\nif args.bidirectional:\n num_directions = 2\n \n# If using multiple layers and directions, the hidden state needs to match that size\nhidden_t = condition_embeddings(condition).unsqueeze(0).repeat(\n args.num_layers * num_directions, 1, 1).to(args.device) # initial state to RNN\nprint (hidden_t.size())\n\n# Feed into RNN\n# y_out, _ = self.rnn(x_embedded, hidden_t)\n",
"torch.Size([1, 4, 100])\n"
]
],
[
[
"2. Concatenate the encoded information with the hidden state at each time step. Do not replace the hidden state because the RNN needs that to learn. ",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/GokuMohandas/practicalAI/master/images/conditioned_rnn2.png\" width=400>",
"_____no_output_____"
]
],
[
[
"# Initialize hidden state\nhidden_t = torch.zeros((args.num_layers * num_directions, args.batch_size, args.rnn_hidden_dim))\nprint (hidden_t.size())",
"torch.Size([1, 4, 100])\n"
],
[
"def concat_condition(condition_embeddings, condition, hidden_t, num_layers, num_directions):\n condition_t = condition_embeddings(condition).unsqueeze(0).repeat(\n num_layers * num_directions, 1, 1)\n hidden_t = torch.cat([hidden_t, condition_t], 2)\n return hidden_t",
"_____no_output_____"
],
[
"# Loop through the inputs time steps\nhiddens = []\nseq_size = 1\nfor t in range(seq_size):\n hidden_t = concat_condition(condition_embeddings, condition, hidden_t, \n args.num_layers, num_directions).to(args.device)\n print (hidden_t.size())\n \n # Feed into RNN\n # hidden_t = rnn_cell(x_in[t], hidden_t)\n ...",
"torch.Size([1, 4, 200])\n"
]
],
[
[
"# Char-level embeddings",
"_____no_output_____"
],
[
"Our conv operations will have inputs that are words in a sentence represented at the character level| $\\in \\mathbb{R}^{NXSXWXE}$ and outputs are embeddings for each word (based on convlutions applied at the character level.) \n\n**Word embeddings**: capture the temporal correlations among\nadjacent tokens so that similar words have similar representations. Ex. \"New Jersey\" is close to \"NJ\" is close to \"Garden State\", etc.\n\n**Char embeddings**: create representations that map words at a character level. Ex. \"toy\" and \"toys\" will be close to each other.",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/GokuMohandas/practicalAI/master/images/char_embeddings.png\" width=450>",
"_____no_output_____"
]
],
[
[
"# Arguments\nargs = Namespace(\n seed=1234,\n cuda=False,\n shuffle=True,\n batch_size=64,\n vocab_size=20, # vocabulary\n seq_size=10, # max length of each sentence\n word_size=15, # max length of each word\n embedding_dim=100,\n num_filters=100, # filters per size\n)",
"_____no_output_____"
],
[
"class Model(nn.Module):\n def __init__(self, embedding_dim, num_embeddings, num_input_channels, \n num_output_channels, padding_idx):\n super(Model, self).__init__()\n \n # Char-level embedding\n self.embeddings = nn.Embedding(embedding_dim=embedding_dim,\n num_embeddings=num_embeddings,\n padding_idx=padding_idx)\n \n # Conv weights\n self.conv = nn.ModuleList([nn.Conv1d(num_input_channels, num_output_channels, \n kernel_size=f) for f in [2,3,4]])\n\n def forward(self, x, channel_first=False, apply_softmax=False):\n \n # x: (N, seq_len, word_len)\n input_shape = x.size()\n batch_size, seq_len, word_len = input_shape\n x = x.view(-1, word_len) # (N*seq_len, word_len)\n \n # Embedding\n x = self.embeddings(x) # (N*seq_len, word_len, embedding_dim)\n \n # Rearrange input so num_input_channels is in dim 1 (N, embedding_dim, word_len)\n if not channel_first:\n x = x.transpose(1, 2)\n \n # Convolution\n z = [F.relu(conv(x)) for conv in self.conv]\n \n # Pooling\n z = [F.max_pool1d(zz, zz.size(2)).squeeze(2) for zz in z] \n z = [zz.view(batch_size, seq_len, -1) for zz in z] # (N, seq_len, embedding_dim)\n \n # Concat to get char-level embeddings\n z = torch.cat(z, 2) # join conv outputs\n \n return z",
"_____no_output_____"
],
[
"# Input\ninput_size = (args.batch_size, args.seq_size, args.word_size)\nx_in = torch.randint(low=0, high=args.vocab_size, size=input_size).long()\nprint (x_in.size())",
"torch.Size([64, 10, 15])\n"
],
[
"# Initial char-level embedding model\nmodel = Model(embedding_dim=args.embedding_dim, \n num_embeddings=args.vocab_size, \n num_input_channels=args.embedding_dim, \n num_output_channels=args.num_filters,\n padding_idx=0)\nprint (model.named_modules)",
"<bound method Module.named_modules of Model(\n (embeddings): Embedding(20, 100, padding_idx=0)\n (conv): ModuleList(\n (0): Conv1d(100, 100, kernel_size=(2,), stride=(1,))\n (1): Conv1d(100, 100, kernel_size=(3,), stride=(1,))\n (2): Conv1d(100, 100, kernel_size=(4,), stride=(1,))\n )\n)>\n"
],
[
"# Forward pass to get char-level embeddings\nz = model(x_in)\nprint (z.size())",
"torch.Size([64, 10, 300])\n"
]
],
[
[
"There are several different ways you can use these char-level embeddings:\n\n1. Concat char-level embeddings with word-level embeddings, since we have an embedding for each word (at a char-level) and then feed it into an RNN. \n2. You can feed the char-level embeddings into an RNN to processes them.",
"_____no_output_____"
],
[
"# Encoder and decoder",
"_____no_output_____"
],
[
"So far we've used RNNs to `encode` a sequential input and generate hidden states. We use these hidden states to `decode` the predictions. So far, the encoder was an RNN and the decoder was just a few fully connected layers followed by a softmax layer (for classification). But the encoder and decoder can assume other architectures as well. For example, the decoder could be an RNN that processes the hidden state outputs from the encoder RNN. ",
"_____no_output_____"
]
],
[
[
"# Arguments\nargs = Namespace(\n batch_size=64,\n embedding_dim=100,\n rnn_hidden_dim=100,\n hidden_dim=100,\n num_layers=1,\n bidirectional=False,\n dropout=0.1,\n)",
"_____no_output_____"
],
[
"class Encoder(nn.Module):\n def __init__(self, embedding_dim, num_embeddings, rnn_hidden_dim, \n num_layers, bidirectional, padding_idx=0):\n super(Encoder, self).__init__()\n \n # Embeddings\n self.word_embeddings = nn.Embedding(embedding_dim=embedding_dim,\n num_embeddings=num_embeddings,\n padding_idx=padding_idx)\n \n # GRU weights\n self.gru = nn.GRU(input_size=embedding_dim, hidden_size=rnn_hidden_dim, \n num_layers=num_layers, batch_first=True, \n bidirectional=bidirectional)\n\n def forward(self, x_in, x_lengths):\n \n # Word level embeddings\n z_word = self.word_embeddings(x_in)\n \n # Feed into RNN\n out, h_n = self.gru(z)\n \n # Gather the last relevant hidden state\n out = gather_last_relevant_hidden(out, x_lengths)\n \n return out",
"_____no_output_____"
],
[
"class Decoder(nn.Module):\n def __init__(self, rnn_hidden_dim, hidden_dim, output_dim, dropout_p):\n super(Decoder, self).__init__()\n \n # FC weights\n self.dropout = nn.Dropout(dropout_p)\n self.fc1 = nn.Linear(rnn_hidden_dim, hidden_dim)\n self.fc2 = nn.Linear(hidden_dim, output_dim)\n\n def forward(self, encoder_output, apply_softmax=False):\n \n # FC layers\n z = self.dropout(encoder_output)\n z = self.fc1(z)\n z = self.dropout(z)\n y_pred = self.fc2(z)\n\n if apply_softmax:\n y_pred = F.softmax(y_pred, dim=1)\n return y_pred",
"_____no_output_____"
],
[
"class Model(nn.Module):\n def __init__(self, embedding_dim, num_embeddings, rnn_hidden_dim, \n hidden_dim, num_layers, bidirectional, output_dim, dropout_p, \n padding_idx=0):\n super(Model, self).__init__()\n self.encoder = Encoder(embedding_dim, num_embeddings, rnn_hidden_dim, \n num_layers, bidirectional, padding_idx=0)\n self.decoder = Decoder(rnn_hidden_dim, hidden_dim, output_dim, dropout_p)\n \n def forward(self, x_in, x_lengths, apply_softmax=False):\n encoder_outputs = self.encoder(x_in, x_lengths)\n y_pred = self.decoder(encoder_outputs, apply_softmax)\n return y_pred",
"_____no_output_____"
],
[
"model = Model(embedding_dim=args.embedding_dim, num_embeddings=1000, \n rnn_hidden_dim=args.rnn_hidden_dim, hidden_dim=args.hidden_dim, \n num_layers=args.num_layers, bidirectional=args.bidirectional, \n output_dim=4, dropout_p=args.dropout, padding_idx=0)\nprint (model.named_parameters)",
"<bound method Module.named_parameters of Model(\n (encoder): Encoder(\n (word_embeddings): Embedding(1000, 100, padding_idx=0)\n (gru): GRU(100, 100, batch_first=True)\n )\n (decoder): Decoder(\n (dropout): Dropout(p=0.1)\n (fc1): Linear(in_features=100, out_features=100, bias=True)\n (fc2): Linear(in_features=100, out_features=4, bias=True)\n )\n)>\n"
]
],
[
[
"# Attentional mechanisms",
"_____no_output_____"
],
[
"When processing an input sequence with an RNN, recall that at each time step we process the input and the hidden state at that time step. For many use cases, it's advantageous to have access to the inputs at all time steps and pay selective attention to the them at each time step. For example, in machine translation, it's advantageous to have access to all the words when translating to another language because translations aren't necessarily word for word. ",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/GokuMohandas/practicalAI/master/images/attention1.jpg\" width=650>",
"_____no_output_____"
],
[
"Attention can sound a bit confusing so let's see what happens at each time step. At time step j, the model has processed inputs $x_0, x_1, x_2, ..., x_j$ and has generted hidden states $h_0, h_1, h_2, ..., h_j$. The idea is to use all the processed hidden states to make the prediction and not just the most recent one. There are several approaches to how we can do this.\n\nWith **soft attention**, we learn a vector of floating points (probabilities) to multiply with the hidden states to create the context vector.\n\nEx. [0.1, 0.3, 0.1, 0.4, 0.1]\n\nWith **hard attention**, we can learn a binary vector to multiply with the hidden states to create the context vector. \n\nEx. [0, 0, 0, 1, 0]",
"_____no_output_____"
],
[
"We're going to focus on soft attention because it's more widley used and we can visualize how much of each hidden state helps with the prediction, which is great for interpretability. ",
"_____no_output_____"
],
[
"<img src=\"https://raw.githubusercontent.com/GokuMohandas/practicalAI/master/images/attention2.jpg\" width=650>",
"_____no_output_____"
],
[
"We're going to implement attention in the document classification task below.",
"_____no_output_____"
],
[
"# Document classification with RNNs",
"_____no_output_____"
],
[
"We're going to implement the same document classification task as in the previous notebook but we're going to use an attentional interface for interpretability.\n\n**Why not machine translation?** Normally, machine translation is the go-to example for demonstrating attention but it's not really practical. How many situations can you think of that require a seq to generate another sequence? Instead we're going to apply attention with our document classification example to see which input tokens are more influential towards predicting the genre.",
"_____no_output_____"
],
[
"## Set up",
"_____no_output_____"
]
],
[
[
"from argparse import Namespace\nimport collections\nimport copy\nimport json\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport re\nimport torch",
"_____no_output_____"
],
[
"def set_seeds(seed, cuda):\n np.random.seed(seed)\n torch.manual_seed(seed)\n if cuda:\n torch.cuda.manual_seed_all(seed)\n \n# Creating directories\ndef create_dirs(dirpath):\n if not os.path.exists(dirpath):\n os.makedirs(dirpath)",
"_____no_output_____"
],
[
"args = Namespace(\n seed=1234,\n cuda=True,\n shuffle=True,\n data_file=\"news.csv\",\n split_data_file=\"split_news.csv\",\n vectorizer_file=\"vectorizer.json\",\n model_state_file=\"model.pth\",\n save_dir=\"news\",\n train_size=0.7,\n val_size=0.15,\n test_size=0.15,\n pretrained_embeddings=None,\n cutoff=25,\n num_epochs=5,\n early_stopping_criteria=5,\n learning_rate=1e-3,\n batch_size=128,\n embedding_dim=100,\n kernels=[3,5],\n num_filters=100,\n rnn_hidden_dim=128,\n hidden_dim=200,\n num_layers=1,\n bidirectional=False,\n dropout_p=0.25,\n)\n\n# Set seeds\nset_seeds(seed=args.seed, cuda=args.cuda)\n\n# Create save dir\ncreate_dirs(args.save_dir)\n\n# Expand filepaths\nargs.vectorizer_file = os.path.join(args.save_dir, args.vectorizer_file)\nargs.model_state_file = os.path.join(args.save_dir, args.model_state_file)\n\n# Check CUDA\nif not torch.cuda.is_available():\n args.cuda = False\nargs.device = torch.device(\"cuda\" if args.cuda else \"cpu\")\nprint(\"Using CUDA: {}\".format(args.cuda))",
"Using CUDA: True\n"
]
],
[
[
"## Data",
"_____no_output_____"
]
],
[
[
"import urllib",
"_____no_output_____"
],
[
"url = \"https://raw.githubusercontent.com/GokuMohandas/practicalAI/master/data/news.csv\"\nresponse = urllib.request.urlopen(url)\nhtml = response.read()\nwith open(args.data_file, 'wb') as fp:\n fp.write(html)",
"_____no_output_____"
],
[
"df = pd.read_csv(args.data_file, header=0)\ndf.head()",
"_____no_output_____"
],
[
"by_category = collections.defaultdict(list)\nfor _, row in df.iterrows():\n by_category[row.category].append(row.to_dict())\nfor category in by_category:\n print (\"{0}: {1}\".format(category, len(by_category[category])))",
"Business: 30000\nSci/Tech: 30000\nSports: 30000\nWorld: 30000\n"
],
[
"final_list = []\nfor _, item_list in sorted(by_category.items()):\n if args.shuffle:\n np.random.shuffle(item_list)\n n = len(item_list)\n n_train = int(args.train_size*n)\n n_val = int(args.val_size*n)\n n_test = int(args.test_size*n)\n\n # Give data point a split attribute\n for item in item_list[:n_train]:\n item['split'] = 'train'\n for item in item_list[n_train:n_train+n_val]:\n item['split'] = 'val'\n for item in item_list[n_train+n_val:]:\n item['split'] = 'test' \n\n # Add to final list\n final_list.extend(item_list)",
"_____no_output_____"
],
[
"split_df = pd.DataFrame(final_list)\nsplit_df[\"split\"].value_counts()",
"_____no_output_____"
],
[
"def preprocess_text(text):\n text = ' '.join(word.lower() for word in text.split(\" \"))\n text = re.sub(r\"([.,!?])\", r\" \\1 \", text)\n text = re.sub(r\"[^a-zA-Z.,!?]+\", r\" \", text)\n text = text.strip()\n return text\n \nsplit_df.title = split_df.title.apply(preprocess_text)",
"_____no_output_____"
],
[
"split_df.to_csv(args.split_data_file, index=False)\nsplit_df.head()",
"_____no_output_____"
]
],
[
[
"## Vocabulary",
"_____no_output_____"
]
],
[
[
"class Vocabulary(object):\n def __init__(self, token_to_idx=None):\n\n # Token to index\n if token_to_idx is None:\n token_to_idx = {}\n self.token_to_idx = token_to_idx\n\n # Index to token\n self.idx_to_token = {idx: token \\\n for token, idx in self.token_to_idx.items()}\n\n def to_serializable(self):\n return {'token_to_idx': self.token_to_idx}\n\n @classmethod\n def from_serializable(cls, contents):\n return cls(**contents)\n\n def add_token(self, token):\n if token in self.token_to_idx:\n index = self.token_to_idx[token]\n else:\n index = len(self.token_to_idx)\n self.token_to_idx[token] = index\n self.idx_to_token[index] = token\n return index\n\n def add_tokens(self, tokens):\n return [self.add_token[token] for token in tokens]\n\n def lookup_token(self, token):\n return self.token_to_idx[token]\n\n def lookup_index(self, index):\n if index not in self.idx_to_token:\n raise KeyError(\"the index (%d) is not in the Vocabulary\" % index)\n return self.idx_to_token[index]\n\n def __str__(self):\n return \"<Vocabulary(size=%d)>\" % len(self)\n\n def __len__(self):\n return len(self.token_to_idx)",
"_____no_output_____"
],
[
"# Vocabulary instance\ncategory_vocab = Vocabulary()\nfor index, row in df.iterrows():\n category_vocab.add_token(row.category)\nprint (category_vocab) # __str__\nprint (len(category_vocab)) # __len__\nindex = category_vocab.lookup_token(\"Business\")\nprint (index)\nprint (category_vocab.lookup_index(index))",
"<Vocabulary(size=4)>\n4\n0\nBusiness\n"
]
],
[
[
"## Sequence vocabulary",
"_____no_output_____"
],
[
"Next, we're going to create our Vocabulary classes for the article's title, which is a sequence of words.",
"_____no_output_____"
]
],
[
[
"from collections import Counter\nimport string",
"_____no_output_____"
],
[
"class SequenceVocabulary(Vocabulary):\n def __init__(self, token_to_idx=None, unk_token=\"<UNK>\",\n mask_token=\"<MASK>\", begin_seq_token=\"<BEGIN>\",\n end_seq_token=\"<END>\"):\n\n super(SequenceVocabulary, self).__init__(token_to_idx)\n\n self.mask_token = mask_token\n self.unk_token = unk_token\n self.begin_seq_token = begin_seq_token\n self.end_seq_token = end_seq_token\n\n self.mask_index = self.add_token(self.mask_token)\n self.unk_index = self.add_token(self.unk_token)\n self.begin_seq_index = self.add_token(self.begin_seq_token)\n self.end_seq_index = self.add_token(self.end_seq_token)\n \n # Index to token\n self.idx_to_token = {idx: token \\\n for token, idx in self.token_to_idx.items()}\n\n def to_serializable(self):\n contents = super(SequenceVocabulary, self).to_serializable()\n contents.update({'unk_token': self.unk_token,\n 'mask_token': self.mask_token,\n 'begin_seq_token': self.begin_seq_token,\n 'end_seq_token': self.end_seq_token})\n return contents\n\n def lookup_token(self, token):\n return self.token_to_idx.get(token, self.unk_index)\n \n def lookup_index(self, index):\n if index not in self.idx_to_token:\n raise KeyError(\"the index (%d) is not in the SequenceVocabulary\" % index)\n return self.idx_to_token[index]\n \n def __str__(self):\n return \"<SequenceVocabulary(size=%d)>\" % len(self.token_to_idx)\n\n def __len__(self):\n return len(self.token_to_idx)\n",
"_____no_output_____"
],
[
"# Get word counts\nword_counts = Counter()\nfor title in split_df.title:\n for token in title.split(\" \"):\n if token not in string.punctuation:\n word_counts[token] += 1\n\n# Create SequenceVocabulary instance\ntitle_word_vocab = SequenceVocabulary()\nfor word, word_count in word_counts.items():\n if word_count >= args.cutoff:\n title_word_vocab.add_token(word)\nprint (title_word_vocab) # __str__\nprint (len(title_word_vocab)) # __len__\nindex = title_word_vocab.lookup_token(\"general\")\nprint (index)\nprint (title_word_vocab.lookup_index(index))",
"<SequenceVocabulary(size=4400)>\n4400\n4\ngeneral\n"
]
],
[
[
"We're also going to create an instance fo SequenceVocabulary that processes the input on a character level.",
"_____no_output_____"
]
],
[
[
"# Create SequenceVocabulary instance\ntitle_char_vocab = SequenceVocabulary()\nfor title in split_df.title:\n for token in title:\n title_char_vocab.add_token(token)\nprint (title_char_vocab) # __str__\nprint (len(title_char_vocab)) # __len__\nindex = title_char_vocab.lookup_token(\"g\")\nprint (index)\nprint (title_char_vocab.lookup_index(index))",
"<SequenceVocabulary(size=35)>\n35\n4\ng\n"
]
],
[
[
"## Vectorizer",
"_____no_output_____"
],
[
"Something new that we introduce in this Vectorizer is calculating the length of our input sequence. We will use this later on to extract the last relevant hidden state for each input sequence.",
"_____no_output_____"
]
],
[
[
"class NewsVectorizer(object):\n def __init__(self, title_word_vocab, title_char_vocab, category_vocab):\n self.title_word_vocab = title_word_vocab\n self.title_char_vocab = title_char_vocab\n self.category_vocab = category_vocab\n\n def vectorize(self, title):\n \n # Word-level vectorization\n word_indices = [self.title_word_vocab.lookup_token(token) for token in title.split(\" \")]\n word_indices = [self.title_word_vocab.begin_seq_index] + word_indices + \\\n [self.title_word_vocab.end_seq_index]\n title_length = len(word_indices)\n word_vector = np.zeros(title_length, dtype=np.int64)\n word_vector[:len(word_indices)] = word_indices\n \n # Char-level vectorization\n word_length = max([len(word) for word in title.split(\" \")])\n char_vector = np.zeros((len(word_vector), word_length), dtype=np.int64)\n char_vector[0, :] = self.title_word_vocab.mask_index # <BEGIN>\n char_vector[-1, :] = self.title_word_vocab.mask_index # <END>\n for i, word in enumerate(title.split(\" \")):\n char_vector[i+1,:len(word)] = [title_char_vocab.lookup_token(char) \\\n for char in word] # i+1 b/c of <BEGIN> token\n \n return word_vector, char_vector, len(word_indices)\n \n def unvectorize_word_vector(self, word_vector):\n tokens = [self.title_word_vocab.lookup_index(index) for index in word_vector]\n title = \" \".join(token for token in tokens)\n return title\n \n def unvectorize_char_vector(self, char_vector):\n title = \"\"\n for word_vector in char_vector:\n for index in word_vector:\n if index == self.title_char_vocab.mask_index:\n break\n title += self.title_char_vocab.lookup_index(index)\n title += \" \"\n return title\n \n @classmethod\n def from_dataframe(cls, df, cutoff):\n \n # Create class vocab\n category_vocab = Vocabulary() \n for category in sorted(set(df.category)):\n category_vocab.add_token(category)\n\n # Get word counts\n word_counts = Counter()\n for title in df.title:\n for token in title.split(\" \"):\n word_counts[token] += 1\n \n # Create title vocab (word level)\n title_word_vocab = SequenceVocabulary()\n for word, word_count in word_counts.items():\n if word_count >= cutoff:\n title_word_vocab.add_token(word)\n \n # Create title vocab (char level)\n title_char_vocab = SequenceVocabulary()\n for title in df.title:\n for token in title:\n title_char_vocab.add_token(token)\n \n return cls(title_word_vocab, title_char_vocab, category_vocab)\n\n @classmethod\n def from_serializable(cls, contents):\n title_word_vocab = SequenceVocabulary.from_serializable(contents['title_word_vocab'])\n title_char_vocab = SequenceVocabulary.from_serializable(contents['title_char_vocab'])\n category_vocab = Vocabulary.from_serializable(contents['category_vocab'])\n return cls(title_word_vocab=title_word_vocab, \n title_char_vocab=title_char_vocab, \n category_vocab=category_vocab)\n \n def to_serializable(self):\n return {'title_word_vocab': self.title_word_vocab.to_serializable(),\n 'title_char_vocab': self.title_char_vocab.to_serializable(),\n 'category_vocab': self.category_vocab.to_serializable()}",
"_____no_output_____"
],
[
"# Vectorizer instance\nvectorizer = NewsVectorizer.from_dataframe(split_df, cutoff=args.cutoff)\nprint (vectorizer.title_word_vocab)\nprint (vectorizer.title_char_vocab)\nprint (vectorizer.category_vocab)\nword_vector, char_vector, title_length = vectorizer.vectorize(preprocess_text(\n \"Roger Federer wins the Wimbledon tennis tournament.\"))\nprint (\"word_vector:\", np.shape(word_vector))\nprint (\"char_vector:\", np.shape(char_vector))\nprint (\"title_length:\", title_length)\nprint (word_vector)\nprint (char_vector)\nprint (vectorizer.unvectorize_word_vector(word_vector))\nprint (vectorizer.unvectorize_char_vector(char_vector))",
"<SequenceVocabulary(size=4404)>\n<SequenceVocabulary(size=35)>\n<Vocabulary(size=4)>\nword_vector: (10,)\nchar_vector: (10, 10)\ntitle_length: 10\n[ 2 1 4151 1231 25 1 2392 4076 38 3]\n[[ 0 0 0 0 0 0 0 0 0 0]\n [ 7 15 4 5 7 0 0 0 0 0]\n [21 5 18 5 7 5 7 0 0 0]\n [26 13 6 16 0 0 0 0 0 0]\n [12 17 5 0 0 0 0 0 0 0]\n [26 13 23 25 9 5 18 15 6 0]\n [12 5 6 6 13 16 0 0 0 0]\n [12 15 20 7 6 8 23 5 6 12]\n [30 0 0 0 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]]\n<BEGIN> <UNK> federer wins the <UNK> tennis tournament . <END>\n roger federer wins the wimbledon tennis tournament . \n"
]
],
[
[
"## Dataset",
"_____no_output_____"
]
],
[
[
"from torch.utils.data import Dataset, DataLoader",
"_____no_output_____"
],
[
"class NewsDataset(Dataset):\n def __init__(self, df, vectorizer):\n self.df = df\n self.vectorizer = vectorizer\n\n # Data splits\n self.train_df = self.df[self.df.split=='train']\n self.train_size = len(self.train_df)\n self.val_df = self.df[self.df.split=='val']\n self.val_size = len(self.val_df)\n self.test_df = self.df[self.df.split=='test']\n self.test_size = len(self.test_df)\n self.lookup_dict = {'train': (self.train_df, self.train_size), \n 'val': (self.val_df, self.val_size),\n 'test': (self.test_df, self.test_size)}\n self.set_split('train')\n\n # Class weights (for imbalances)\n class_counts = df.category.value_counts().to_dict()\n def sort_key(item):\n return self.vectorizer.category_vocab.lookup_token(item[0])\n sorted_counts = sorted(class_counts.items(), key=sort_key)\n frequencies = [count for _, count in sorted_counts]\n self.class_weights = 1.0 / torch.tensor(frequencies, dtype=torch.float32)\n\n @classmethod\n def load_dataset_and_make_vectorizer(cls, split_data_file, cutoff):\n df = pd.read_csv(split_data_file, header=0)\n train_df = df[df.split=='train']\n return cls(df, NewsVectorizer.from_dataframe(train_df, cutoff))\n\n @classmethod\n def load_dataset_and_load_vectorizer(cls, split_data_file, vectorizer_filepath):\n df = pd.read_csv(split_data_file, header=0)\n vectorizer = cls.load_vectorizer_only(vectorizer_filepath)\n return cls(df, vectorizer)\n\n def load_vectorizer_only(vectorizer_filepath):\n with open(vectorizer_filepath) as fp:\n return NewsVectorizer.from_serializable(json.load(fp))\n\n def save_vectorizer(self, vectorizer_filepath):\n with open(vectorizer_filepath, \"w\") as fp:\n json.dump(self.vectorizer.to_serializable(), fp)\n\n def set_split(self, split=\"train\"):\n self.target_split = split\n self.target_df, self.target_size = self.lookup_dict[split]\n\n def __str__(self):\n return \"<Dataset(split={0}, size={1})\".format(\n self.target_split, self.target_size)\n\n def __len__(self):\n return self.target_size\n\n def __getitem__(self, index):\n row = self.target_df.iloc[index]\n title_word_vector, title_char_vector, title_length = \\\n self.vectorizer.vectorize(row.title)\n category_index = self.vectorizer.category_vocab.lookup_token(row.category)\n return {'title_word_vector': title_word_vector, \n 'title_char_vector': title_char_vector, \n 'title_length': title_length, \n 'category': category_index}\n\n def get_num_batches(self, batch_size):\n return len(self) // batch_size\n\n def generate_batches(self, batch_size, collate_fn, shuffle=True, \n drop_last=False, device=\"cpu\"):\n dataloader = DataLoader(dataset=self, batch_size=batch_size,\n collate_fn=collate_fn, shuffle=shuffle, \n drop_last=drop_last)\n for data_dict in dataloader:\n out_data_dict = {}\n for name, tensor in data_dict.items():\n out_data_dict[name] = data_dict[name].to(device)\n yield out_data_dict",
"_____no_output_____"
],
[
"# Dataset instance\ndataset = NewsDataset.load_dataset_and_make_vectorizer(args.split_data_file,\n args.cutoff)\nprint (dataset) # __str__\ninput_ = dataset[10] # __getitem__\nprint (input_['title_word_vector'])\nprint (input_['title_char_vector'])\nprint (input_['title_length'])\nprint (input_['category'])\nprint (dataset.vectorizer.unvectorize_word_vector(input_['title_word_vector']))\nprint (dataset.vectorizer.unvectorize_char_vector(input_['title_char_vector']))\nprint (dataset.class_weights)",
"<Dataset(split=train, size=84000)\n[ 2 51 1 52 53 26 54 3]\n[[ 0 0 0 0 0 0 0 0 0 0]\n [18 5 9 12 8 0 0 0 0 0]\n [18 15 18 4 5 16 0 0 0 0]\n [25 8 6 27 7 20 14 12 11 22]\n [26 13 12 17 0 0 0 0 0 0]\n [ 9 8 25 15 7 0 0 0 0 0]\n [18 5 8 9 0 0 0 0 0 0]\n [ 0 0 0 0 0 0 0 0 0 0]]\n8\n0\n<BEGIN> delta <UNK> bankruptcy with labor deal <END>\n delta dodges bankruptcy with labor deal \ntensor([3.3333e-05, 3.3333e-05, 3.3333e-05, 3.3333e-05])\n"
]
],
[
[
"## Model",
"_____no_output_____"
],
[
"embed → encoder → attend → predict",
"_____no_output_____"
]
],
[
[
"import torch.nn as nn\nimport torch.nn.functional as F",
"_____no_output_____"
],
[
"class NewsEncoder(nn.Module):\n def __init__(self, embedding_dim, num_word_embeddings, num_char_embeddings,\n kernels, num_input_channels, num_output_channels, \n rnn_hidden_dim, num_layers, bidirectional, \n word_padding_idx=0, char_padding_idx=0):\n super(NewsEncoder, self).__init__()\n \n self.num_layers = num_layers\n self.bidirectional = bidirectional\n \n # Embeddings\n self.word_embeddings = nn.Embedding(embedding_dim=embedding_dim,\n num_embeddings=num_word_embeddings,\n padding_idx=word_padding_idx)\n self.char_embeddings = nn.Embedding(embedding_dim=embedding_dim,\n num_embeddings=num_char_embeddings,\n padding_idx=char_padding_idx)\n \n # Conv weights\n self.conv = nn.ModuleList([nn.Conv1d(num_input_channels, \n num_output_channels, \n kernel_size=f) for f in kernels])\n \n \n # GRU weights\n self.gru = nn.GRU(input_size=embedding_dim*(len(kernels)+1), \n hidden_size=rnn_hidden_dim, num_layers=num_layers, \n batch_first=True, bidirectional=bidirectional)\n \n def initialize_hidden_state(self, batch_size, rnn_hidden_dim, device):\n \"\"\"Modify this to condition the RNN.\"\"\"\n num_directions = 1\n if self.bidirectional:\n num_directions = 2\n hidden_t = torch.zeros(self.num_layers * num_directions, \n batch_size, rnn_hidden_dim).to(device)\n \n def get_char_level_embeddings(self, x):\n # x: (N, seq_len, word_len)\n input_shape = x.size()\n batch_size, seq_len, word_len = input_shape\n x = x.view(-1, word_len) # (N*seq_len, word_len)\n \n # Embedding\n x = self.char_embeddings(x) # (N*seq_len, word_len, embedding_dim)\n \n # Rearrange input so num_input_channels is in dim 1 (N, embedding_dim, word_len)\n x = x.transpose(1, 2)\n \n # Convolution\n z = [F.relu(conv(x)) for conv in self.conv]\n \n # Pooling\n z = [F.max_pool1d(zz, zz.size(2)).squeeze(2) for zz in z] \n z = [zz.view(batch_size, seq_len, -1) for zz in z] # (N, seq_len, embedding_dim)\n \n # Concat to get char-level embeddings\n z = torch.cat(z, 2) # join conv outputs\n \n return z\n \n def forward(self, x_word, x_char, x_lengths, device):\n \"\"\"\n x_word: word level representation (N, seq_size)\n x_char: char level representation (N, seq_size, word_len)\n \"\"\"\n \n # Word level embeddings\n z_word = self.word_embeddings(x_word)\n \n # Char level embeddings\n z_char = self.get_char_level_embeddings(x=x_char)\n \n # Concatenate\n z = torch.cat([z_word, z_char], 2)\n \n # Feed into RNN\n initial_h = self.initialize_hidden_state(\n batch_size=z.size(0), rnn_hidden_dim=self.gru.hidden_size,\n device=device)\n out, h_n = self.gru(z, initial_h)\n \n return out",
"_____no_output_____"
],
[
"class NewsDecoder(nn.Module):\n def __init__(self, rnn_hidden_dim, hidden_dim, output_dim, dropout_p):\n super(NewsDecoder, self).__init__()\n \n # Attention FC layer\n self.fc_attn = nn.Linear(rnn_hidden_dim, rnn_hidden_dim)\n self.v = nn.Parameter(torch.rand(rnn_hidden_dim))\n \n # FC weights\n self.dropout = nn.Dropout(dropout_p)\n self.fc1 = nn.Linear(rnn_hidden_dim, hidden_dim)\n self.fc2 = nn.Linear(hidden_dim, output_dim)\n\n def forward(self, encoder_outputs, apply_softmax=False):\n \n # Attention\n z = torch.tanh(self.fc_attn(encoder_outputs))\n z = z.transpose(2,1) # [B*H*T]\n v = self.v.repeat(encoder_outputs.size(0),1).unsqueeze(1) #[B*1*H]\n z = torch.bmm(v,z).squeeze(1) # [B*T]\n attn_scores = F.softmax(z, dim=1)\n context = torch.matmul(encoder_outputs.transpose(-2, -1), \n attn_scores.unsqueeze(dim=2)).squeeze()\n if len(context.size()) == 1:\n context = context.unsqueeze(0)\n \n # FC layers\n z = self.dropout(context)\n z = self.fc1(z)\n z = self.dropout(z)\n y_pred = self.fc2(z)\n\n if apply_softmax:\n y_pred = F.softmax(y_pred, dim=1)\n return attn_scores, y_pred",
"_____no_output_____"
],
[
"class NewsModel(nn.Module):\n def __init__(self, embedding_dim, num_word_embeddings, num_char_embeddings,\n kernels, num_input_channels, num_output_channels, \n rnn_hidden_dim, hidden_dim, output_dim, num_layers, \n bidirectional, dropout_p, word_padding_idx, char_padding_idx):\n super(NewsModel, self).__init__()\n self.encoder = NewsEncoder(embedding_dim, num_word_embeddings,\n num_char_embeddings, kernels, \n num_input_channels, num_output_channels, \n rnn_hidden_dim, num_layers, bidirectional, \n word_padding_idx, char_padding_idx)\n self.decoder = NewsDecoder(rnn_hidden_dim, hidden_dim, output_dim, \n dropout_p)\n \n def forward(self, x_word, x_char, x_lengths, device, apply_softmax=False):\n encoder_outputs = self.encoder(x_word, x_char, x_lengths, device)\n y_pred = self.decoder(encoder_outputs, apply_softmax)\n return y_pred",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"import torch.optim as optim",
"_____no_output_____"
],
[
"class Trainer(object):\n def __init__(self, dataset, model, model_state_file, save_dir, device, \n shuffle, num_epochs, batch_size, learning_rate, \n early_stopping_criteria):\n self.dataset = dataset\n self.class_weights = dataset.class_weights.to(device)\n self.device = device\n self.model = model.to(device)\n self.save_dir = save_dir\n self.device = device\n self.shuffle = shuffle\n self.num_epochs = num_epochs\n self.batch_size = batch_size\n self.loss_func = nn.CrossEntropyLoss(self.class_weights)\n self.optimizer = optim.Adam(self.model.parameters(), lr=learning_rate)\n self.scheduler = optim.lr_scheduler.ReduceLROnPlateau(\n optimizer=self.optimizer, mode='min', factor=0.5, patience=1)\n self.train_state = {\n 'stop_early': False, \n 'early_stopping_step': 0,\n 'early_stopping_best_val': 1e8,\n 'early_stopping_criteria': early_stopping_criteria,\n 'learning_rate': learning_rate,\n 'epoch_index': 0,\n 'train_loss': [],\n 'train_acc': [],\n 'val_loss': [],\n 'val_acc': [],\n 'test_loss': -1,\n 'test_acc': -1,\n 'model_filename': model_state_file}\n \n def update_train_state(self):\n\n # Verbose\n print (\"[EPOCH]: {0:02d} | [LR]: {1} | [TRAIN LOSS]: {2:.2f} | [TRAIN ACC]: {3:.1f}% | [VAL LOSS]: {4:.2f} | [VAL ACC]: {5:.1f}%\".format(\n self.train_state['epoch_index'], self.train_state['learning_rate'], \n self.train_state['train_loss'][-1], self.train_state['train_acc'][-1], \n self.train_state['val_loss'][-1], self.train_state['val_acc'][-1]))\n\n # Save one model at least\n if self.train_state['epoch_index'] == 0:\n torch.save(self.model.state_dict(), self.train_state['model_filename'])\n self.train_state['stop_early'] = False\n\n # Save model if performance improved\n elif self.train_state['epoch_index'] >= 1:\n loss_tm1, loss_t = self.train_state['val_loss'][-2:]\n\n # If loss worsened\n if loss_t >= self.train_state['early_stopping_best_val']:\n # Update step\n self.train_state['early_stopping_step'] += 1\n\n # Loss decreased\n else:\n # Save the best model\n if loss_t < self.train_state['early_stopping_best_val']:\n torch.save(self.model.state_dict(), self.train_state['model_filename'])\n\n # Reset early stopping step\n self.train_state['early_stopping_step'] = 0\n\n # Stop early ?\n self.train_state['stop_early'] = self.train_state['early_stopping_step'] \\\n >= self.train_state['early_stopping_criteria']\n return self.train_state\n \n def compute_accuracy(self, y_pred, y_target):\n _, y_pred_indices = y_pred.max(dim=1)\n n_correct = torch.eq(y_pred_indices, y_target).sum().item()\n return n_correct / len(y_pred_indices) * 100\n \n def pad_word_seq(self, seq, length):\n vector = np.zeros(length, dtype=np.int64)\n vector[:len(seq)] = seq\n vector[len(seq):] = self.dataset.vectorizer.title_word_vocab.mask_index\n return vector\n \n def pad_char_seq(self, seq, seq_length, word_length):\n vector = np.zeros((seq_length, word_length), dtype=np.int64)\n vector.fill(self.dataset.vectorizer.title_char_vocab.mask_index)\n for i in range(len(seq)):\n char_padding = np.zeros(word_length-len(seq[i]), dtype=np.int64)\n vector[i] = np.concatenate((seq[i], char_padding), axis=None)\n return vector\n \n def collate_fn(self, batch):\n \n # Make a deep copy\n batch_copy = copy.deepcopy(batch)\n processed_batch = {\"title_word_vector\": [], \"title_char_vector\": [], \n \"title_length\": [], \"category\": []}\n \n # Max lengths\n get_seq_length = lambda sample: len(sample[\"title_word_vector\"])\n get_word_length = lambda sample: len(sample[\"title_char_vector\"][0])\n max_seq_length = max(map(get_seq_length, batch))\n max_word_length = max(map(get_word_length, batch))\n\n\n # Pad\n for i, sample in enumerate(batch_copy):\n padded_word_seq = self.pad_word_seq(\n sample[\"title_word_vector\"], max_seq_length)\n padded_char_seq = self.pad_char_seq(\n sample[\"title_char_vector\"], max_seq_length, max_word_length)\n processed_batch[\"title_word_vector\"].append(padded_word_seq)\n processed_batch[\"title_char_vector\"].append(padded_char_seq)\n processed_batch[\"title_length\"].append(sample[\"title_length\"])\n processed_batch[\"category\"].append(sample[\"category\"])\n \n # Convert to appropriate tensor types\n processed_batch[\"title_word_vector\"] = torch.LongTensor(\n processed_batch[\"title_word_vector\"])\n processed_batch[\"title_char_vector\"] = torch.LongTensor(\n processed_batch[\"title_char_vector\"])\n processed_batch[\"title_length\"] = torch.LongTensor(\n processed_batch[\"title_length\"])\n processed_batch[\"category\"] = torch.LongTensor(\n processed_batch[\"category\"])\n \n return processed_batch \n \n def run_train_loop(self):\n for epoch_index in range(self.num_epochs):\n self.train_state['epoch_index'] = epoch_index\n \n # Iterate over train dataset\n\n # initialize batch generator, set loss and acc to 0, set train mode on\n self.dataset.set_split('train')\n batch_generator = self.dataset.generate_batches(\n batch_size=self.batch_size, collate_fn=self.collate_fn, \n shuffle=self.shuffle, device=self.device)\n running_loss = 0.0\n running_acc = 0.0\n self.model.train()\n\n for batch_index, batch_dict in enumerate(batch_generator):\n # zero the gradients\n self.optimizer.zero_grad()\n \n # compute the output\n _, y_pred = self.model(x_word=batch_dict['title_word_vector'],\n x_char=batch_dict['title_char_vector'],\n x_lengths=batch_dict['title_length'],\n device=self.device)\n \n # compute the loss\n loss = self.loss_func(y_pred, batch_dict['category'])\n loss_t = loss.item()\n running_loss += (loss_t - running_loss) / (batch_index + 1)\n\n # compute gradients using loss\n loss.backward()\n\n # use optimizer to take a gradient step\n self.optimizer.step()\n \n # compute the accuracy\n acc_t = self.compute_accuracy(y_pred, batch_dict['category'])\n running_acc += (acc_t - running_acc) / (batch_index + 1)\n\n self.train_state['train_loss'].append(running_loss)\n self.train_state['train_acc'].append(running_acc)\n\n # Iterate over val dataset\n\n # initialize batch generator, set loss and acc to 0, set eval mode on\n self.dataset.set_split('val')\n batch_generator = self.dataset.generate_batches(\n batch_size=self.batch_size, collate_fn=self.collate_fn, \n shuffle=self.shuffle, device=self.device)\n running_loss = 0.\n running_acc = 0.\n self.model.eval()\n\n for batch_index, batch_dict in enumerate(batch_generator):\n\n # compute the output\n _, y_pred = self.model(x_word=batch_dict['title_word_vector'],\n x_char=batch_dict['title_char_vector'],\n x_lengths=batch_dict['title_length'],\n device=self.device)\n\n # compute the loss\n loss = self.loss_func(y_pred, batch_dict['category'])\n loss_t = loss.to(\"cpu\").item()\n running_loss += (loss_t - running_loss) / (batch_index + 1)\n\n # compute the accuracy\n acc_t = self.compute_accuracy(y_pred, batch_dict['category'])\n running_acc += (acc_t - running_acc) / (batch_index + 1)\n\n self.train_state['val_loss'].append(running_loss)\n self.train_state['val_acc'].append(running_acc)\n\n self.train_state = self.update_train_state()\n self.scheduler.step(self.train_state['val_loss'][-1])\n if self.train_state['stop_early']:\n break\n \n def run_test_loop(self):\n # initialize batch generator, set loss and acc to 0, set eval mode on\n self.dataset.set_split('test')\n batch_generator = self.dataset.generate_batches(\n batch_size=self.batch_size, collate_fn=self.collate_fn, \n shuffle=self.shuffle, device=self.device)\n running_loss = 0.0\n running_acc = 0.0\n self.model.eval()\n\n for batch_index, batch_dict in enumerate(batch_generator):\n # compute the output\n _, y_pred = self.model(x_word=batch_dict['title_word_vector'],\n x_char=batch_dict['title_char_vector'],\n x_lengths=batch_dict['title_length'],\n device=self.device)\n\n # compute the loss\n loss = self.loss_func(y_pred, batch_dict['category'])\n loss_t = loss.item()\n running_loss += (loss_t - running_loss) / (batch_index + 1)\n\n # compute the accuracy\n acc_t = self.compute_accuracy(y_pred, batch_dict['category'])\n running_acc += (acc_t - running_acc) / (batch_index + 1)\n\n self.train_state['test_loss'] = running_loss\n self.train_state['test_acc'] = running_acc\n \n def plot_performance(self):\n # Figure size\n plt.figure(figsize=(15,5))\n\n # Plot Loss\n plt.subplot(1, 2, 1)\n plt.title(\"Loss\")\n plt.plot(trainer.train_state[\"train_loss\"], label=\"train\")\n plt.plot(trainer.train_state[\"val_loss\"], label=\"val\")\n plt.legend(loc='upper right')\n\n # Plot Accuracy\n plt.subplot(1, 2, 2)\n plt.title(\"Accuracy\")\n plt.plot(trainer.train_state[\"train_acc\"], label=\"train\")\n plt.plot(trainer.train_state[\"val_acc\"], label=\"val\")\n plt.legend(loc='lower right')\n\n # Save figure\n plt.savefig(os.path.join(self.save_dir, \"performance.png\"))\n\n # Show plots\n plt.show()\n \n def save_train_state(self):\n with open(os.path.join(self.save_dir, \"train_state.json\"), \"w\") as fp:\n json.dump(self.train_state, fp)",
"_____no_output_____"
],
[
"# Initialization\ndataset = NewsDataset.load_dataset_and_make_vectorizer(args.split_data_file,\n args.cutoff)\ndataset.save_vectorizer(args.vectorizer_file)\nvectorizer = dataset.vectorizer\nmodel = NewsModel(embedding_dim=args.embedding_dim, \n num_word_embeddings=len(vectorizer.title_word_vocab), \n num_char_embeddings=len(vectorizer.title_char_vocab),\n kernels=args.kernels,\n num_input_channels=args.embedding_dim,\n num_output_channels=args.num_filters,\n rnn_hidden_dim=args.rnn_hidden_dim,\n hidden_dim=args.hidden_dim,\n output_dim=len(vectorizer.category_vocab),\n num_layers=args.num_layers,\n bidirectional=args.bidirectional,\n dropout_p=args.dropout_p, \n word_padding_idx=vectorizer.title_word_vocab.mask_index,\n char_padding_idx=vectorizer.title_char_vocab.mask_index)\nprint (model.named_modules)",
"<bound method Module.named_modules of NewsModel(\n (encoder): NewsEncoder(\n (word_embeddings): Embedding(3406, 100, padding_idx=0)\n (char_embeddings): Embedding(35, 100, padding_idx=0)\n (conv): ModuleList(\n (0): Conv1d(100, 100, kernel_size=(3,), stride=(1,))\n (1): Conv1d(100, 100, kernel_size=(5,), stride=(1,))\n )\n (gru): GRU(300, 128, batch_first=True)\n )\n (decoder): NewsDecoder(\n (fc_attn): Linear(in_features=128, out_features=128, bias=True)\n (dropout): Dropout(p=0.25)\n (fc1): Linear(in_features=128, out_features=200, bias=True)\n (fc2): Linear(in_features=200, out_features=4, bias=True)\n )\n)>\n"
],
[
"# Train\ntrainer = Trainer(dataset=dataset, model=model, \n model_state_file=args.model_state_file, \n save_dir=args.save_dir, device=args.device,\n shuffle=args.shuffle, num_epochs=args.num_epochs, \n batch_size=args.batch_size, learning_rate=args.learning_rate, \n early_stopping_criteria=args.early_stopping_criteria)\ntrainer.run_train_loop()",
"[EPOCH]: 00 | [LR]: 0.001 | [TRAIN LOSS]: 0.78 | [TRAIN ACC]: 68.6% | [VAL LOSS]: 0.58 | [VAL ACC]: 78.5%\n[EPOCH]: 01 | [LR]: 0.001 | [TRAIN LOSS]: 0.50 | [TRAIN ACC]: 82.0% | [VAL LOSS]: 0.48 | [VAL ACC]: 83.2%\n[EPOCH]: 02 | [LR]: 0.001 | [TRAIN LOSS]: 0.43 | [TRAIN ACC]: 84.6% | [VAL LOSS]: 0.47 | [VAL ACC]: 83.1%\n[EPOCH]: 03 | [LR]: 0.001 | [TRAIN LOSS]: 0.39 | [TRAIN ACC]: 86.2% | [VAL LOSS]: 0.46 | [VAL ACC]: 83.7%\n[EPOCH]: 04 | [LR]: 0.001 | [TRAIN LOSS]: 0.35 | [TRAIN ACC]: 87.4% | [VAL LOSS]: 0.44 | [VAL ACC]: 84.2%\n"
],
[
"# Plot performance\ntrainer.plot_performance()",
"_____no_output_____"
],
[
"# Test performance\ntrainer.run_test_loop()\nprint(\"Test loss: {0:.2f}\".format(trainer.train_state['test_loss']))\nprint(\"Test Accuracy: {0:.1f}%\".format(trainer.train_state['test_acc']))",
"Test loss: 0.44\nTest Accuracy: 84.4%\n"
],
[
"# Save all results\ntrainer.save_train_state()",
"_____no_output_____"
]
],
[
[
"## Inference",
"_____no_output_____"
]
],
[
[
"class Inference(object):\n def __init__(self, model, vectorizer):\n self.model = model\n self.vectorizer = vectorizer\n \n def predict_category(self, title):\n # Vectorize\n word_vector, char_vector, title_length = self.vectorizer.vectorize(title)\n title_word_vector = torch.tensor(word_vector).unsqueeze(0)\n title_char_vector = torch.tensor(char_vector).unsqueeze(0)\n title_length = torch.tensor([title_length]).long() \n \n # Forward pass\n self.model.eval()\n attn_scores, y_pred = self.model(x_word=title_word_vector, \n x_char=title_char_vector,\n x_lengths=title_length, \n device=\"cpu\",\n apply_softmax=True)\n\n # Top category\n y_prob, indices = y_pred.max(dim=1)\n index = indices.item()\n\n # Predicted category\n category = vectorizer.category_vocab.lookup_index(index)\n probability = y_prob.item()\n return {'category': category, 'probability': probability, \n 'attn_scores': attn_scores}\n \n def predict_top_k(self, title, k):\n # Vectorize\n word_vector, char_vector, title_length = self.vectorizer.vectorize(title)\n title_word_vector = torch.tensor(word_vector).unsqueeze(0)\n title_char_vector = torch.tensor(char_vector).unsqueeze(0)\n title_length = torch.tensor([title_length]).long()\n \n # Forward pass\n self.model.eval()\n _, y_pred = self.model(x_word=title_word_vector,\n x_char=title_char_vector,\n x_lengths=title_length, \n device=\"cpu\",\n apply_softmax=True)\n \n # Top k categories\n y_prob, indices = torch.topk(y_pred, k=k)\n probabilities = y_prob.detach().numpy()[0]\n indices = indices.detach().numpy()[0]\n\n # Results\n results = []\n for probability, index in zip(probabilities, indices):\n category = self.vectorizer.category_vocab.lookup_index(index)\n results.append({'category': category, 'probability': probability})\n\n return results",
"_____no_output_____"
],
[
"# Load the model\ndataset = NewsDataset.load_dataset_and_load_vectorizer(\n args.split_data_file, args.vectorizer_file)\nvectorizer = dataset.vectorizer\nmodel = NewsModel(embedding_dim=args.embedding_dim, \n num_word_embeddings=len(vectorizer.title_word_vocab), \n num_char_embeddings=len(vectorizer.title_char_vocab),\n kernels=args.kernels,\n num_input_channels=args.embedding_dim,\n num_output_channels=args.num_filters,\n rnn_hidden_dim=args.rnn_hidden_dim,\n hidden_dim=args.hidden_dim,\n output_dim=len(vectorizer.category_vocab),\n num_layers=args.num_layers,\n bidirectional=args.bidirectional,\n dropout_p=args.dropout_p, \n word_padding_idx=vectorizer.title_word_vocab.mask_index,\n char_padding_idx=vectorizer.title_char_vocab.mask_index)\nmodel.load_state_dict(torch.load(args.model_state_file))\nmodel = model.to(\"cpu\")\nprint (model.named_modules)",
"<bound method Module.named_modules of NewsModel(\n (encoder): NewsEncoder(\n (word_embeddings): Embedding(3406, 100, padding_idx=0)\n (char_embeddings): Embedding(35, 100, padding_idx=0)\n (conv): ModuleList(\n (0): Conv1d(100, 100, kernel_size=(3,), stride=(1,))\n (1): Conv1d(100, 100, kernel_size=(5,), stride=(1,))\n )\n (gru): GRU(300, 128, batch_first=True)\n )\n (decoder): NewsDecoder(\n (fc_attn): Linear(in_features=128, out_features=128, bias=True)\n (dropout): Dropout(p=0.25)\n (fc1): Linear(in_features=128, out_features=200, bias=True)\n (fc2): Linear(in_features=200, out_features=4, bias=True)\n )\n)>\n"
],
[
"# Inference\ninference = Inference(model=model, vectorizer=vectorizer)\ntitle = input(\"Enter a title to classify: \")\nprediction = inference.predict_category(preprocess_text(title))\nprint(\"{} → {} (p={:0.2f})\".format(title, prediction['category'], \n prediction['probability']))",
"Enter a title to classify: Sale of Apple's new iphone are skyrocketing.\nSale of Apple's new iphone are skyrocketing. → Sci/Tech (p=0.86)\n"
],
[
"# Top-k inference\ntop_k = inference.predict_top_k(preprocess_text(title), k=len(vectorizer.category_vocab))\nprint (\"{}: \".format(title))\nfor result in top_k:\n print (\"{} (p={:0.2f})\".format(result['category'], \n result['probability']))",
"Sale of Apple's new iphone are skyrocketing.: \nSci/Tech (p=0.86)\nBusiness (p=0.12)\nWorld (p=0.01)\nSports (p=0.00)\n"
]
],
[
[
"# Interpretability",
"_____no_output_____"
],
[
"We can inspect the probability vector that is generated at each time step to visualize the importance of each of the previous hidden states towards a particular time step's prediction. ",
"_____no_output_____"
]
],
[
[
"import seaborn as sns\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"attn_matrix = prediction['attn_scores'].detach().numpy()\nax = sns.heatmap(attn_matrix, linewidths=2, square=True)\ntokens = [\"<BEGIN>\"]+preprocess_text(title).split(\" \")+[\"<END>\"]\nax.set_xticklabels(tokens, rotation=45)\nax.set_xlabel(\"Token\")\nax.set_ylabel(\"Importance\\n\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"# TODO",
"_____no_output_____"
],
[
"- attn visualization isn't always great\n- bleu score\n- ngram-overlap\n- perplexity\n- beamsearch\n- hierarchical softmax\n- hierarchical attention\n- Transformer networks\n- attention interpretability is hit/miss\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbc95e375e1da0d0de3300002aceec961afeff52
| 78,707 |
ipynb
|
Jupyter Notebook
|
mouseNiu2020_SVM_supporting.ipynb
|
ventolab/HGDA
|
baacdf627f1c5fdd4712db1c98d94ab175e33fdf
|
[
"MIT"
] | null | null | null |
mouseNiu2020_SVM_supporting.ipynb
|
ventolab/HGDA
|
baacdf627f1c5fdd4712db1c98d94ab175e33fdf
|
[
"MIT"
] | null | null | null |
mouseNiu2020_SVM_supporting.ipynb
|
ventolab/HGDA
|
baacdf627f1c5fdd4712db1c98d94ab175e33fdf
|
[
"MIT"
] | null | null | null | 57.324836 | 29,860 | 0.708679 |
[
[
[
"### Supervised Machine Learning Models for Cross Species comparison of supporting cells",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport scanpy as sc\nimport matplotlib.pyplot as plt\nimport os\nimport sys\nimport anndata\n\n\ndef MovePlots(plotpattern, subplotdir):\n os.system('mkdir -p '+str(sc.settings.figdir)+'/'+subplotdir)\n os.system('mv '+str(sc.settings.figdir)+'/*'+plotpattern+'** '+str(sc.settings.figdir)+'/'+subplotdir)\n\n\nsc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)\nsc.settings.figdir = '/home/jovyan/Gonads/Flat_SupportVectorMachine_Fetal/SVM/training/'\nsc.logging.print_versions()\nsc.settings.set_figure_params(dpi=80) # low dpi (dots per inch) yields small inline figures\n\nsys.executable",
"WARNING: If you miss a compact list, please try `print_header`!\n"
]
],
[
[
"**Load our fetal samples**",
"_____no_output_____"
]
],
[
[
"human = sc.read('/nfs/team292/lg18/with_valentina/FCA-M5-annotatedCluster4Seurat.h5ad')\nhuman = human[[i in ['female'] for i in human.obs['sex']]]\nhuman.obs['stage'].value_counts()",
"/home/jovyan/my-conda-envs/scanpy_env/lib/python3.8/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.\n res = method(*args, **kwargs)\n"
]
],
[
[
"**Take fine grained annotations from Luz on supporting cells**",
"_____no_output_____"
]
],
[
[
"supporting = pd.read_csv('/nfs/team292/lg18/with_valentina/supporting_nocycling_annotation.csv', index_col = 0)\nprint(supporting['annotated_clusters'].value_counts())\nsupporting = supporting[supporting['annotated_clusters'].isin(['coelEpi', 'sLGR5', 'sPAX8b', 'preGC_III_Notch', 'preGC_II', \n 'preGC_II_hypoxia', 'preGC_I_OSR1', 'sKITLG',\n 'ovarianSurf'])]\nmapping = supporting['annotated_clusters'].to_dict()\nhuman.obs['supporting_clusters'] = human.obs_names.map(mapping)\n\n# Remove doublets as well as NaNs corresponding to cells from enriched samples\nhuman.obs['supporting_clusters'] = human.obs['supporting_clusters'].astype(str)\nhuman = human[[i not in ['nan'] for i in human.obs['supporting_clusters']]]\nhuman.obs['supporting_clusters'].value_counts(dropna = False)",
"Trying to set attribute `.obs` of view, copying.\n"
],
[
"### Join sub-states of preGC_II and preGC_III\njoined = {'coelEpi' : 'coelEpi', 'sLGR5' : 'sLGR5', 'sPAX8b' : 'sPAX8b', 'preGC_III_Notch' : 'preGC_III', 'preGC_II' : 'preGC_II', \n 'preGC_II_hypoxia' : 'preGC_II', 'preGC_I_OSR1' : 'preGC_I_OSR1', 'sKITLG' : 'sKITLG',\n 'ovarianSurf' : 'ovarianSurf'}\nhuman.obs['supporting_clusters'] = human.obs['supporting_clusters'].map(joined)\nhuman.obs['supporting_clusters'].value_counts(dropna = False)",
"Trying to set attribute `.obs` of view, copying.\n"
]
],
[
[
"**Intersect genes present in all fetal gonads scRNAseq datasets of human and mouse**",
"_____no_output_____"
],
[
"Mouse ovary",
"_____no_output_____"
]
],
[
[
"mouse = sc.read(\"/nfs/team292/vl6/Mouse_Niu2020/supporting_mesothelial.h5ad\")\nmouse = anndata.AnnData(X= mouse.raw.X, var=mouse.raw.var, obs=mouse.obs)\nmouse ",
"_____no_output_____"
]
],
[
[
"Extract the genes from all datasets",
"_____no_output_____"
]
],
[
[
"human_genes = human.var_names.to_list()\nmouse_genes = mouse.var_names.to_list()",
"_____no_output_____"
],
[
"from functools import reduce\ninters = reduce(np.intersect1d, (human_genes, mouse_genes))\nlen(inters)",
"_____no_output_____"
],
[
"cell_cycle_genes = [x.strip() for x in open(file='/nfs/users/nfs_v/vl6/regev_lab_cell_cycle_genes.txt')]\ncell_cycle_genes = [x for x in cell_cycle_genes if x in list(inters)]\ninters = [x for x in list(inters) if x not in cell_cycle_genes]\nlen(inters)",
"_____no_output_____"
]
],
[
[
"**Subset fetal data to keep only these genes**",
"_____no_output_____"
]
],
[
[
"human = human[:, list(inters)]\nhuman",
"_____no_output_____"
]
],
[
[
"**Downsample more frequent classes**",
"_____no_output_____"
]
],
[
[
"myindex = human.obs['supporting_clusters'].value_counts().index \nmyvalues = human.obs['supporting_clusters'].value_counts().values\nclusters = pd.Series(myvalues, index = myindex)\nclusters.values",
"_____no_output_____"
],
[
"import random\nfrom itertools import chain\n\n# Find clusters with > n cells\nn = 1500\ncl2downsample = clusters.index[ clusters.values > n ]\n\n# save all barcode ids from small clusters\nholder = []\nholder.append( human.obs_names[[ i not in cl2downsample for i in human.obs['supporting_clusters'] ]] ) \n\n# randomly sample n cells in the cl2downsample\nfor cl in cl2downsample:\n print(cl)\n cl_sample = human[[ i == cl for i in human.obs['supporting_clusters'] ]].obs_names\n # n = int(round(len(cl_sample)/2, 0))\n cl_downsample = random.sample(set(cl_sample), n )\n holder.append(cl_downsample)\n \n# samples to include\nsamples = list(chain(*holder))\n\n# Filter adata_count\nhuman = human[[ i in samples for i in human.obs_names ]]\nhuman.X.shape",
"preGC_II\npreGC_I_OSR1\novarianSurf\npreGC_III\nsLGR5\ncoelEpi\n"
]
],
[
[
"**Preprocess the data**",
"_____no_output_____"
]
],
[
[
"# Per cell normalization\nsc.pp.normalize_per_cell(human, counts_per_cell_after=1e4)\n# Log transformation \nsc.pp.log1p(human)",
"normalizing by total count per cell\nTrying to set attribute `.obs` of view, copying.\n finished (0:00:01): normalized adata.X and added 'n_counts', counts per cell before normalization (adata.obs)\n"
],
[
"# Filter HVGs --> Select top 300 highly variable genes that will serve as features to the machine learning models \nsc.pp.highly_variable_genes(human, n_top_genes = 300)\nhighly_variable_genes = human.var[\"highly_variable\"]\nhuman = human[:, highly_variable_genes]",
"If you pass `n_top_genes`, all cutoffs are ignored.\nextracting highly variable genes\n finished (0:00:02)\n--> added\n 'highly_variable', boolean vector (adata.var)\n 'means', float vector (adata.var)\n 'dispersions', float vector (adata.var)\n 'dispersions_norm', float vector (adata.var)\n/home/jovyan/my-conda-envs/scanpy_env/lib/python3.8/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.\n res = method(*args, **kwargs)\n"
],
[
"# Scale\nsc.pp.scale(human, max_value=10)\nprint('Total number of cells: {:d}'.format(human.n_obs))\nprint('Total number of genes: {:d}'.format(human.n_vars))",
"/home/jovyan/my-conda-envs/scanpy_env/lib/python3.8/site-packages/scanpy/preprocessing/_simple.py:810: UserWarning: Revieved a view of an AnnData. Making a copy.\n view_to_actual(adata)\n... as `zero_center=True`, sparse input is densified and may lead to large memory consumption\n"
]
],
[
[
"**Import libraries**",
"_____no_output_____"
]
],
[
[
"# Required libraries regardless of the model you choose\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import confusion_matrix, classification_report, accuracy_score\nfrom sklearn.model_selection import GridSearchCV, RandomizedSearchCV\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.decomposition import PCA\nfrom sklearn.pipeline import Pipeline\n\n# Library for Logistic Regression\nfrom sklearn.linear_model import LogisticRegression\n\n# Library for Random Forest \nfrom sklearn.ensemble import RandomForestClassifier\n\n# Library for Support Vector Machine \nfrom sklearn.svm import SVC",
"_____no_output_____"
],
[
"print(\"Loading data\")\nX = np.array(human.X) # Fetching the count matrix to use as input to the model \nprint(type(X), X.shape)",
"Loading data\n<class 'numpy.ndarray'> (11422, 300)\n"
],
[
"# Choose output variable, meaning the labels you want to predict \ny = list(human.obs.supporting_clusters.astype('str'))",
"_____no_output_____"
],
[
"# Split the training dataset into train and test sets \nX_train, X_test, y_train, y_test = train_test_split(\n X,\n y,\n test_size=0.25, # This can be changed, though it makes sense to use 25-30% of the data for test\n random_state=1234,\n )",
"_____no_output_____"
]
],
[
[
"**Option 1: Logistic Regression classifier**",
"_____no_output_____"
]
],
[
[
"# Instantiate a Logistic Regression Classifier and specify L2 regularization\nlr = LogisticRegression(penalty='l2', multi_class=\"multinomial\", max_iter = 2000)\n\n# Instantiate a PCA object\npca = PCA()\n\n# Create pipeline object\npipe = Pipeline(steps=[('pca', pca), ('LogReg', lr)])\n\nprint('Hyperparameter tuning with exhaustive grid search')\n\n# Choose a grid of hyperparameters values (these are arbitrary but reasonable as I took reference values from the documentation)\nparams_lr = {'LogReg__C' : [0.001, 0.01, 0.1, 1, 10, 100], 'LogReg__solver' : [\"lbfgs\", 'newton-cg', 'sag'], \n 'pca__n_components' : [0.7, 0.8, 0.9]}\n\n# Use grid search cross validation to span the hyperparameter space and choose the best \ngrid_lr = RandomizedSearchCV(estimator = pipe, param_distributions = params_lr, cv = 5, n_jobs = -1)\n\n# Fit the model to the training set of the training data\ngrid_lr.fit(X_train, y_train)\n\n# Report the best parameters\nprint(\"Best CV params\", grid_lr.best_params_)\n\n# Report the best hyperparameters and the corresponding score\nprint(\"Softmax training accuracy:\", grid_lr.score(X_train, y_train))\nprint(\"Softmax test accuracy:\", grid_lr.score(X_test, y_test))",
"Hyperparameter tuning with exhaustive grid search\nBest CV params {'pca__n_components': 0.9, 'LogReg__solver': 'sag', 'LogReg__C': 0.01}\nSoftmax training accuracy: 0.9324071912211067\nSoftmax test accuracy: 0.9103641456582633\n"
]
],
[
[
"**Option 2: Support Vector Machine classifier**",
"_____no_output_____"
]
],
[
[
"# Instantiate an RBF Support Vector Machine\nsvm = SVC(kernel = \"rbf\", probability = True)\n\n# Instantiate a PCA \npca = PCA()\n\n# Create pipeline object\npipe = Pipeline(steps=[('pca', pca), ('SVC', svm)])\n\nprint('Hyperparameter tuning with exhaustive grid search')\n\n# Choose a grid of hyperparameters values (these are arbitrary but reasonable as I took reference values from the documentation)\nparams_svm = {'SVC__C':[0.1, 1, 10, 100], 'SVC__gamma':[0.001, 0.01, 0.1], 'pca__n_components': [0.7, 0.8, 0.9]}\n\n# Use grid search cross validation to span the hyperparameter space and choose the best \ngrid_svm = RandomizedSearchCV(pipe, param_distributions = params_svm, cv=5, verbose =1, n_jobs = -1)\n\n# Fit the model to the training set of the training data\ngrid_svm.fit(X_train, y_train)\n\n# Report the best hyperparameters and the corresponding score\nprint(\"Best CV params\", grid_svm.best_params_)\nprint(\"Best CV accuracy\", grid_svm.best_score_)",
"Hyperparameter tuning with exhaustive grid search\nFitting 5 folds for each of 10 candidates, totalling 50 fits\nBest CV params {'pca__n_components': 0.7, 'SVC__gamma': 0.01, 'SVC__C': 1}\nBest CV accuracy 0.9096435998722106\n"
]
],
[
[
"**Option 3: Random Forest classifier**",
"_____no_output_____"
]
],
[
[
"# Instantiate a Random Forest Classifier \nSEED = 123\nrf = RandomForestClassifier(random_state = SEED) # set a seed to ensure reproducibility of results\nprint(rf.get_params()) # Look at the hyperparameters that can be tuned \n\n# Instantiate a PCA object\npca = PCA()\n\n# Create pipeline object\npipe = Pipeline(steps=[('pca', pca), ('RF', rf)])\n\nprint('Hyperparameter tuning with exhaustive grid search')\n\n# Choose a grid of hyperparameters values (these are arbitrary but reasonable as I took reference values from the documentation)\nparams_rf = {\"RF__n_estimators\": [50, 100, 200, 300], 'RF__min_samples_leaf': [1, 5], 'RF__min_samples_split': [2, 5, 10], \n 'pca__n_components' : [0.7, 0.8,0.9]}\n\n# Use grid search cross validation to span the hyperparameter space and choose the best \ngrid_rf = RandomizedSearchCV(estimator = pipe, param_distributions = params_rf, cv = 5, n_jobs = -1)\n\n# Fit the model to the training set of the training data\ngrid_rf.fit(X_train, y_train)\n\n# Report the best hyperparameters and the corresponding score\nprint(\"Best CV params\", grid_rf.best_params_)\nprint(\"Best CV accuracy\", grid_rf.best_score_)",
"{'bootstrap': True, 'ccp_alpha': 0.0, 'class_weight': None, 'criterion': 'gini', 'max_depth': None, 'max_features': 'auto', 'max_leaf_nodes': None, 'max_samples': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'n_estimators': 100, 'n_jobs': None, 'oob_score': False, 'random_state': 123, 'verbose': 0, 'warm_start': False}\nHyperparameter tuning with exhaustive grid search\nBest CV params {'pca__n_components': 0.7, 'RF__n_estimators': 300, 'RF__min_samples_split': 5, 'RF__min_samples_leaf': 1}\nBest CV accuracy 0.8991372175572753\n"
]
],
[
[
"All 3 models return an object (which I called *grid_lr*, *grid_rf*, *grid_svm*, respectively) that has an attribute called **.best_estimator_** which holds the model with the best hyperparameters that was found using grid search cross validation. This is the model that you will use to make predictions.",
"_____no_output_____"
],
[
"**Evaluating the model's performance on the test set of the training data**",
"_____no_output_____"
]
],
[
[
"predicted_labels = grid_svm.best_estimator_.predict(X_test) # Here as an example I am using the support vector machine model\nreport_rf = classification_report(y_test, predicted_labels)\nprint(report_rf)\nprint(\"Accuracy:\", accuracy_score(y_test, predicted_labels))",
" precision recall f1-score support\n\n coelEpi 0.96 0.95 0.95 366\n ovarianSurf 0.93 0.89 0.91 366\n preGC_II 0.84 0.89 0.86 368\n preGC_III 0.99 0.96 0.98 385\npreGC_I_OSR1 0.88 0.84 0.86 392\n sKITLG 0.88 0.85 0.86 275\n sLGR5 0.89 0.92 0.91 369\n sPAX8b 0.95 0.99 0.97 335\n\n accuracy 0.91 2856\n macro avg 0.91 0.91 0.91 2856\nweighted avg 0.91 0.91 0.91 2856\n\nAccuracy: 0.9142156862745098\n"
],
[
"cnf_matrix = confusion_matrix(y_test, predicted_labels)\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline\n\nclass_names=[0,1] # name of classes\nfig, ax = plt.subplots()\ntick_marks = np.arange(len(class_names))\nplt.xticks(tick_marks, class_names)\nplt.yticks(tick_marks, class_names)\n# create heatmap\nsns.heatmap(pd.DataFrame(cnf_matrix), annot=True, cmap=\"YlGnBu\" ,fmt='g')\nax.xaxis.set_label_position(\"top\")\nplt.tight_layout()\nplt.title('Confusion matrix', y=1.1)\nplt.ylabel('Actual label')\nplt.xlabel('Predicted label')",
"_____no_output_____"
],
[
"print(\"Accuracy:\", accuracy_score(y_test, predicted_labels))",
"Accuracy: 0.9142156862745098\n"
],
[
"grid_svm.best_estimator_.feature_names = list(human.var_names)",
"_____no_output_____"
]
],
[
[
"**Predict cell types in the mouse data**",
"_____no_output_____"
]
],
[
[
"def process_and_subset_data(adata, genes):\n # save the log transformed counts as raw \n adata.raw = adata.copy()\n # Per cell normalization\n sc.pp.normalize_per_cell(adata, counts_per_cell_after=1e4)\n # Log transformation \n sc.pp.log1p(adata)\n # Subset data\n adata = adata[:, list(genes)]\n # Scale\n sc.pp.scale(adata, max_value=10)\n return adata",
"_____no_output_____"
],
[
"def process_data(adata):\n # Per cell normalization\n sc.pp.normalize_per_cell(adata, counts_per_cell_after=1e4)\n # Log transformation \n sc.pp.log1p(adata)\n # Scale\n sc.pp.scale(adata, max_value=10)",
"_____no_output_____"
],
[
"import scipy\ndef make_single_predictions(adata, classifier): \n #if scipy.sparse.issparse(adata.X):\n #adata.X = adata.X.toarray()\n adata_X = np.array(adata.X)\n print(type(adata_X), adata_X.shape)\n adata_preds = classifier.predict(adata_X)\n adata.obs['human_classifier_supporting'] = adata_preds\n print(adata.obs.human_classifier_supporting.value_counts(dropna = False))",
"_____no_output_____"
],
[
"def make_correspondence(classifier):\n corr = {}\n for i in range(0,len(classifier.classes_)):\n corr[i] = classifier.classes_[i]\n return corr",
"_____no_output_____"
],
[
"def make_probability_predictions(adata, classifier):\n adata_X = np.array(adata.X)\n print(type(adata_X), adata_X.shape)\n proba_preds = classifier.predict_proba(adata_X)\n df_probs = pd.DataFrame(np.column_stack(list(zip(*proba_preds))))\n corr = make_correspondence(classifier)\n for index in df_probs.columns.values:\n celltype = corr[index]\n adata.obs['prob_'+celltype] = df_probs[index].to_list()",
"_____no_output_____"
]
],
[
[
"Mouse ovary (Niu et al., 2020)",
"_____no_output_____"
]
],
[
[
"mouse = process_and_subset_data(mouse, grid_svm.best_estimator_.feature_names)\n\nmake_single_predictions(mouse, grid_svm.best_estimator_)",
"normalizing by total count per cell\n/home/jovyan/my-conda-envs/scanpy_env/lib/python3.8/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.\n res = method(*args, **kwargs)\n finished (0:00:01): normalized adata.X and added 'n_counts', counts per cell before normalization (adata.obs)\n/home/jovyan/my-conda-envs/scanpy_env/lib/python3.8/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.\n res = method(*args, **kwargs)\n/home/jovyan/my-conda-envs/scanpy_env/lib/python3.8/site-packages/scanpy/preprocessing/_simple.py:810: UserWarning: Revieved a view of an AnnData. Making a copy.\n view_to_actual(adata)\n... as `zero_center=True`, sparse input is densified and may lead to large memory consumption\n"
],
[
"make_probability_predictions(mouse, grid_svm.best_estimator_)",
"<class 'numpy.ndarray'> (17686, 300)\n"
],
[
"mouse",
"_____no_output_____"
],
[
"mouse.write('/nfs/team292/vl6/Mouse_Niu2020/supporting_cells_with_human_preds.h5ad')",
"... storing 'human_classifier_supporting' as categorical\n"
],
[
"mouse = sc.read('/nfs/team292/vl6/Mouse_Niu2020/supporting_cells_with_human_preds.h5ad')\nmouse",
"_____no_output_____"
],
[
"mouse_predictions = mouse.obs[['prob_coelEpi', 'prob_ovarianSurf', 'prob_preGC_II', 'prob_preGC_III', 'prob_preGC_I_OSR1', 'prob_sKITLG', 'prob_sLGR5', 'prob_sPAX8b']]",
"_____no_output_____"
],
[
"mouse_predictions.columns = ['prob_coelEpi', 'prob_ovarianSurf', 'prob_preGC-II', 'prob_preGC-II-late', 'prob_preGC-I', \n 'prob_sKITLG', 'prob_sLGR5', 'prob_sPAX8b']",
"_____no_output_____"
],
[
"mouse_predictions.head()",
"_____no_output_____"
],
[
"mouse_predictions.to_csv('/nfs/team292/vl6/Mouse_Niu2020/mouse_Niu2020_supporting_predictions.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc965f0ffee8f4a5b29c19e5b23b7096ee86167
| 7,394 |
ipynb
|
Jupyter Notebook
|
01 - Python para Zumbis/PPZ_03.ipynb
|
GabrielTrentino/Python_Basico
|
f13f6448c275c14896337d2018b04cbf5a54efd3
|
[
"MIT"
] | null | null | null |
01 - Python para Zumbis/PPZ_03.ipynb
|
GabrielTrentino/Python_Basico
|
f13f6448c275c14896337d2018b04cbf5a54efd3
|
[
"MIT"
] | null | null | null |
01 - Python para Zumbis/PPZ_03.ipynb
|
GabrielTrentino/Python_Basico
|
f13f6448c275c14896337d2018b04cbf5a54efd3
|
[
"MIT"
] | null | null | null | 28.658915 | 390 | 0.449013 |
[
[
[
"# Lista de Exercício 03:",
"_____no_output_____"
],
[
"## Q01: Faça um programa que peça uma nota, entre zero e dez. Mostre uma mensagem caso o valor seja inválido e continue pedindo até que o usuário informe um valor válido.",
"_____no_output_____"
]
],
[
[
"while True:\n try:\n nota = int(input('Digite uma nota de 0 a 10: '))\n if (nota >= 0) & (nota <= 10):\n print(\"Ok, validado.\")\n break\n else:\n print('Valor não valido')\n except:\n print('Digite um número valido')",
"Digite uma nota de 0 a 10: gfd\nDigite um número valido\nDigite uma nota de 0 a 10: 39\nValor não valido\nDigite uma nota de 0 a 10: 3\nOk, validado.\n"
]
],
[
[
"## Q02 - Faça um programa que leia um nome de usuário e a sua senha e não aceite a senha igual ao nome do usuário, mostrando uma mensagem de erro e voltando a pedir as informações.",
"_____no_output_____"
]
],
[
[
"while True:\n user = input('Digite seu usario: ')\n senha = input('Digite sua senha: ')\n if user == senha:\n print('Usuario e senhas iguais, digite novamente.')\n else:\n print('Validado')\n break",
"Digite seu usario: 5626\nDigite sua senha: 5626\nUsuario e senhas iguais, digite novamente.\nDigite seu usario: gabriel\nDigite sua senha: 123\nValidado\n"
]
],
[
[
"## Q03 - Supondo que a população de um país A seja da ordem de 80000 habitantes com uma taxa anual de crescimento de 3% e que a população de B seja 200000 habitantes com uma taxa de crescimento de 1.5%. Faça um programa que calcule e escreva o número de anos necessários para que a população do país A ultrapasse ou iguale a população do país B, mantidas as taxas de crescimento\n",
"_____no_output_____"
]
],
[
[
"pop_A = 8000\npop_B = 200000\nano = 0\nwhile True:\n ano += 1\n pop_A = pop_A*1.03\n pop_B = pop_B*1.015\n if pop_A > pop_B:\n break\nprint('Em {} anos a Cidade A terá mais habitantes que a cidade B'.format(ano))",
"Em 220 anos a Cidade A terá mais habitantes que a cidade B\n"
]
],
[
[
"## Q04 - A seqüência de Fibonacci é a seguinte: 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, ... Sua regra de formação é simples: os dois primeiros elementos são 1; a partir de então, cada elemento é a soma dos dois anteriores. Faça um algoritmo que leia um número inteiro calcule o seu número de Fibonacci. F1 = 1, F2 = 1, F3 = 2, etc.\n",
"_____no_output_____"
]
],
[
[
"num = int(input('Digite o número: '))\nfib = []\nfor i in range(num):\n if (i==0) | (i==1):\n fib.append(1)\n print(fib)\n else:\n fib.append(fib[-2]+fib[-1])\n print(fib)\nprint(fib[-1])",
"Digite o número: 9\n[1]\n[1, 1]\n[1, 1, 2]\n[1, 1, 2, 3]\n[1, 1, 2, 3, 5]\n[1, 1, 2, 3, 5, 8]\n[1, 1, 2, 3, 5, 8, 13]\n[1, 1, 2, 3, 5, 8, 13, 21]\n[1, 1, 2, 3, 5, 8, 13, 21, 34]\n34\n"
]
],
[
[
"## Q05 - Dados dois números inteiros positivos, determinar o máximo divisor comum entre eles usando o algoritmo de Euclides. ",
"_____no_output_____"
]
],
[
[
"num1 = int(input('Digite o primeiro número: '))\nnum2 = int(input('Digite o segundo número: '))\n\ndividendo = num1\ndivisor = num2\n\nwhile True:\n resto = dividendo % divisor\n if resto == 0:\n break\n else:\n dividendo = divisor\n divisor = resto\n \nprint(\"O Mdc de {} com {} é {}\".format(num1, num2, divisor))",
"Digite o primeiro número: 32\nDigite o segundo número: 12\nO Mdc de 32 com 12 é 4\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc9855a0ade06d4d43266769f12853982e3d043
| 137,452 |
ipynb
|
Jupyter Notebook
|
examples/models/nodejs_mnist/nodejs_mnist.ipynb
|
MarcoGorelli/seldon-core
|
fdab6ecc718d5c20aa2df3b90592c3b4a410bd0e
|
[
"Apache-2.0"
] | null | null | null |
examples/models/nodejs_mnist/nodejs_mnist.ipynb
|
MarcoGorelli/seldon-core
|
fdab6ecc718d5c20aa2df3b90592c3b4a410bd0e
|
[
"Apache-2.0"
] | null | null | null |
examples/models/nodejs_mnist/nodejs_mnist.ipynb
|
MarcoGorelli/seldon-core
|
fdab6ecc718d5c20aa2df3b90592c3b4a410bd0e
|
[
"Apache-2.0"
] | null | null | null | 40.714455 | 9,220 | 0.319464 |
[
[
[
"# Nodejs MNIST model Deployment\n\n * Wrap a nodejs tensorflow model for use as a prediction microservice in seldon-core\n * Run locally on Docker to test\n \n## Dependencies\n\n * ```pip install seldon-core```\n * [Helm](https://github.com/kubernetes/helm)\n * [Minikube](https://github.com/kubernetes/minikube)\n * [S2I](https://github.com/openshift/source-to-image)\n * node (version>=8.11.0)\n * npm\n\n## Train locally using npm commands\n This model takes in mnist images of size 28x28x1 as input and outputs an array of size 10 with prediction of each digits from 0-9",
"_____no_output_____"
]
],
[
[
"!make train && make clean_build",
"npm install\n\u001b[K\u001b[?25h \u001b[27m\u001b[90m......\u001b[0m] \\ refresh-package-json:argparse: \u001b[32;40mtiming\u001b[0m \u001b[35maction:finalize\u001b[0m Co\u001b[0m\u001b[K\n> @tensorflow/[email protected] install /home/clive/work/seldon-core/fork-seldon-core/examples/models/nodejs_mnist/node_modules/@tensorflow/tfjs-node\n> node scripts/install.js\n\n* Downloading libtensorflow\n\u001b[1G[ ] 1180235/bps 0% 16.7s\u001b[0K\u001b[1G[ ] 1875303/bps 0% 10.5s\u001b[0K\u001b[1G[ ] 2741939/bps 1% 7.2s\u001b[0K\u001b[1G[ ] 3032508/bps 1% 6.5s\u001b[0K\u001b[1G[= ] 4188852/bps 2% 4.6s\u001b[0K\u001b[1G[= ] 4196020/bps 2% 4.6s\u001b[0K\u001b[1G[= ] 3919906/bps 3% 4.9s\u001b[0K\u001b[1G[= ] 4554618/bps 3% 4.2s\u001b[0K\u001b[1G[= ] 5345319/bps 4% 3.5s\u001b[0K\u001b[1G[== ] 5635676/bps 5% 3.3s\u001b[0K\u001b[1G[== ] 7673917/bps 7% 2.4s\u001b[0K\u001b[1G[=== ] 8695962/bps 9% 2.1s\u001b[0K\u001b[1G[=== ] 8116991/bps 10% 2.2s\u001b[0K\u001b[1G[==== ] 8513097/bps 12% 2.0s\u001b[0K\u001b[1G[==== ] 8410939/bps 13% 2.1s\u001b[0K\u001b[1G[==== ] 8673246/bps 14% 2.0s\u001b[0K\u001b[1G[===== ] 9525507/bps 16% 1.7s\u001b[0K\u001b[1G[===== ] 10068442/bps 18% 1.6s\u001b[0K\u001b[1G[====== ] 10564293/bps 20% 1.5s\u001b[0K\u001b[1G[====== ] 10311254/bps 20% 1.5s\u001b[0K\u001b[1G[====== ] 10324710/bps 21% 1.5s\u001b[0K\u001b[1G[======= ] 10551756/bps 23% 1.4s\u001b[0K\u001b[1G[======= ] 10651449/bps 24% 1.4s\u001b[0K\u001b[1G[======= ] 10507130/bps 24% 1.4s\u001b[0K\u001b[1G[======== ] 11083294/bps 27% 1.3s\u001b[0K\u001b[1G[========= ] 10975382/bps 29% 1.3s\u001b[0K\u001b[1G[========= ] 11084185/bps 30% 1.2s\u001b[0K\u001b[1G[========= ] 11108293/bps 31% 1.2s\u001b[0K\u001b[1G[========== ] 11150222/bps 33% 1.2s\u001b[0K\u001b[1G[========== ] 11199892/bps 33% 1.2s\u001b[0K\u001b[1G[========== ] 11255565/bps 35% 1.1s\u001b[0K\u001b[1G[=========== ] 11298406/bps 36% 1.1s\u001b[0K\u001b[1G[=========== ] 11279950/bps 37% 1.1s\u001b[0K\u001b[1G[=========== ] 11335154/bps 38% 1.1s\u001b[0K\u001b[1G[============ ] 11461561/bps 39% 1.0s\u001b[0K\u001b[1G[============ ] 11322851/bps 40% 1.0s\u001b[0K\u001b[1G[============ ] 11328628/bps 41% 1.0s\u001b[0K\u001b[1G[============= ] 11569110/bps 43% 1.0s\u001b[0K\u001b[1G[============= ] 11394723/bps 43% 1.0s\u001b[0K\u001b[1G[============= ] 11119219/bps 44% 1.0s\u001b[0K\u001b[1G[============== ] 11200476/bps 46% 1.0s\u001b[0K\u001b[1G[============== ] 11327456/bps 47% 0.9s\u001b[0K\u001b[1G[============== ] 11185786/bps 48% 0.9s\u001b[0K\u001b[1G[=============== ] 11092156/bps 49% 0.9s\u001b[0K\u001b[1G[=============== ] 11057818/bps 50% 0.9s\u001b[0K\u001b[1G[=============== ] 11169269/bps 51% 0.9s\u001b[0K\u001b[1G[=============== ] 11017133/bps 52% 0.9s\u001b[0K\u001b[1G[================ ] 11178032/bps 53% 0.8s\u001b[0K\u001b[1G[================ ] 11042508/bps 53% 0.8s\u001b[0K\u001b[1G[================ ] 11034122/bps 55% 0.8s\u001b[0K\u001b[1G[================= ] 11136514/bps 56% 0.8s\u001b[0K\u001b[1G[================= ] 11187099/bps 57% 0.8s\u001b[0K\u001b[1G[================== ] 11267984/bps 59% 0.7s\u001b[0K\u001b[1G[================== ] 11277706/bps 60% 0.7s\u001b[0K\u001b[1G[================== ] 11120431/bps 61% 0.7s\u001b[0K\u001b[1G[=================== ] 11001318/bps 62% 0.7s\u001b[0K\u001b[1G[=================== ] 10961331/bps 63% 0.7s\u001b[0K\u001b[1G[=================== ] 11194304/bps 65% 0.6s\u001b[0K\u001b[1G[==================== ] 11293962/bps 66% 0.6s\u001b[0K\u001b[1G[==================== ] 11124598/bps 67% 0.6s\u001b[0K\u001b[1G[===================== ] 11200894/bps 69% 0.5s\u001b[0K\u001b[1G[===================== ] 11152804/bps 70% 0.5s\u001b[0K\u001b[1G[===================== ] 11284431/bps 72% 0.5s\u001b[0K\u001b[1G[====================== ] 11148271/bps 72% 0.5s\u001b[0K\u001b[1G[====================== ] 11149697/bps 73% 0.5s\u001b[0K\u001b[1G[====================== ] 11250762/bps 75% 0.4s\u001b[0K\u001b[1G[======================= ] 11291676/bps 76% 0.4s\u001b[0K\u001b[1G[======================= ] 11344624/bps 78% 0.4s\u001b[0K\u001b[1G[======================= ] 11224529/bps 78% 0.4s\u001b[0K\u001b[1G[======================== ] 11293257/bps 80% 0.3s\u001b[0K\u001b[1G[======================== ] 11269785/bps 81% 0.3s\u001b[0K\u001b[1G[========================= ] 11268995/bps 82% 0.3s\u001b[0K\u001b[1G[========================= ] 11256269/bps 83% 0.3s\u001b[0K\u001b[1G[========================= ] 11148678/bps 83% 0.3s\u001b[0K\u001b[1G[========================= ] 11177869/bps 85% 0.3s\u001b[0K\u001b[1G[========================== ] 11138957/bps 85% 0.3s\u001b[0K\u001b[1G[========================== ] 11235963/bps 87% 0.2s\u001b[0K\u001b[1G[========================== ] 11239572/bps 88% 0.2s\u001b[0K\u001b[1G[=========================== ] 11305796/bps 90% 0.2s\u001b[0K\u001b[1G[=========================== ] 11334843/bps 91% 0.2s\u001b[0K\u001b[1G[============================ ] 11302114/bps 92% 0.1s\u001b[0K\u001b[1G[============================ ] 11233882/bps 93% 0.1s\u001b[0K\u001b[1G[============================ ] 11240417/bps 94% 0.1s\u001b[0K\u001b[1G[============================ ] 11171803/bps 94% 0.1s\u001b[0K\u001b[1G[============================ ] 11133717/bps 95% 0.1s\u001b[0K\u001b[1G[============================= ] 11128391/bps 96% 0.1s\u001b[0K\u001b[1G[============================= ] 11139209/bps 97% 0.1s\u001b[0K\u001b[1G[============================= ] 11140363/bps 98% 0.0s\u001b[0K\u001b[1G[============================= ] 11116375/bps 98% 0.0s\u001b[0K\u001b[1G[==============================] 11154472/bps 100% 0.0s\u001b[0K\u001b[1G[==============================] 11158360/bps 100% 0.0s\u001b[0K\n* Building TensorFlow Node.js bindings\n\n> [email protected] postinstall /home/clive/work/seldon-core/fork-seldon-core/examples/models/nodejs_mnist/node_modules/protobufjs\n> node scripts/postinstall\n\n\u001b[37;40mnpm\u001b[0m \u001b[0m\u001b[34;40mnotice\u001b[0m\u001b[35m\u001b[0m created a lockfile as package-lock.json. You should commit this file.\n\u001b[0m\u001b[37;40mnpm\u001b[0m \u001b[0m\u001b[30;43mWARN\u001b[0m\u001b[35m\u001b[0m [email protected] No repository field.\n\u001b[0m\u001b[37;40mnpm\u001b[0m \u001b[0m\u001b[30;43mWARN\u001b[0m\u001b[35m\u001b[0m [email protected] No license field.\n\u001b[0m\nadded 50 packages from 58 contributors and audited 64 packages in 8.793s\nfound \u001b[92m0\u001b[0m vulnerabilities\n\n\u001b[K\u001b[?25hnpm start \u001b[27m\u001b[90m.....\u001b[0m] - prepare:nodejs_mnist: \u001b[7msill\u001b[0m \u001b[35minstall\u001b[0m printInstalled\u001b[0m\u001b[K\n\n> [email protected] start /home/clive/work/seldon-core/fork-seldon-core/examples/models/nodejs_mnist\n> node train.js --epoch 1 --batch_size 128 --model_save_path /\n\n2019-04-16 15:13:12.477002: I tensorflow/core/platform/cpu_feature_guard.cc:141] Your CPU supports instructions that this TensorFlow binary was not compiled to use: SSE4.1 SSE4.2 AVX AVX2 FMA\n1 128 '/home/clive/work/seldon-core/fork-seldon-core/examples/models/nodejs_mnist/'\n * Downloading from: https://storage.googleapis.com/cvdf-datasets/mnist/train-images-idx3-ubyte.gz\n * Downloading from: https://storage.googleapis.com/cvdf-datasets/mnist/train-labels-idx1-ubyte.gz\n * Downloading from: https://storage.googleapis.com/cvdf-datasets/mnist/t10k-images-idx3-ubyte.gz\n * Downloading from: https://storage.googleapis.com/cvdf-datasets/mnist/t10k-labels-idx1-ubyte.gz\n_________________________________________________________________\nLayer (type) Output shape Param # \n=================================================================\nconv2d_Conv2D1 (Conv2D) [null,26,26,32] 320 \n_________________________________________________________________\nconv2d_Conv2D2 (Conv2D) [null,24,24,32] 9248 \n_________________________________________________________________\nmax_pooling2d_MaxPooling2D1 [null,12,12,32] 0 \n_________________________________________________________________\nconv2d_Conv2D3 (Conv2D) [null,10,10,64] 18496 \n_________________________________________________________________\nconv2d_Conv2D4 (Conv2D) [null,8,8,64] 36928 \n_________________________________________________________________\nmax_pooling2d_MaxPooling2D2 [null,4,4,64] 0 \n_________________________________________________________________\nflatten_Flatten1 (Flatten) [null,1024] 0 \n_________________________________________________________________\ndropout_Dropout1 (Dropout) [null,1024] 0 \n_________________________________________________________________\ndense_Dense1 (Dense) [null,512] 524800 \n_________________________________________________________________\ndropout_Dropout2 (Dropout) [null,512] 0 \n_________________________________________________________________\ndense_Dense2 (Dense) [null,10] 5130 \n=================================================================\nTotal params: 594922\nTrainable params: 594922\nNon-trainable params: 0\n_________________________________________________________________\n2019-04-16 15:13:14.898522: W tensorflow/core/framework/allocator.cc:108] Allocation of 188160000 exceeds 10% of system memory.\n"
]
],
[
[
"Training creates a model.json file and a weights.bin file which is utilized for prediction",
"_____no_output_____"
],
[
"## Prediction using REST API on the docker container",
"_____no_output_____"
]
],
[
[
"!s2i build . seldonio/seldon-core-s2i-nodejs:0.2-SNAPSHOT node-s2i-mnist-model:0.1",
"---> Installing application source...\n---> Installing dependencies ...\n\n> @tensorflow/[email protected] install /microservice/model/node_modules/@tensorflow/tfjs-node\n> node scripts/install.js\n\n* Downloading libtensorflow\n\n* Building TensorFlow Node.js bindings\n\n> [email protected] postinstall /microservice/model/node_modules/protobufjs\n> node scripts/postinstall\n\nnpm notice created a lockfile as package-lock.json. You should commit this file.\nnpm WARN [email protected] No repository field.\nnpm WARN [email protected] No license field.\n\nadded 50 packages from 58 contributors and audited 64 packages in 16.212s\nfound 0 vulnerabilities\n\nBuild completed successfully\n"
],
[
"!docker run --name \"nodejs_mnist_predictor\" -d --rm -p 5000:5000 node-s2i-mnist-model:0.1",
"a41526a3110245adad542b85a8ac0c5822e35d0cb0530f3cffe46c10e421cfff\r\n"
]
],
[
[
"Send some random features that conform to the contract",
"_____no_output_____"
]
],
[
[
"!seldon-core-tester contract.json 0.0.0.0 5000 -p -t",
"----------------------------------------\r\nSENDING NEW REQUEST:\r\n\r\n[[[[0.109]\r\n [0.536]\r\n [0.906]\r\n [0.169]\r\n [0.342]\r\n [0.662]\r\n [0.834]\r\n [0.724]\r\n [0.466]\r\n [0.199]\r\n [0.503]\r\n [0.502]\r\n [0.899]\r\n [0.761]\r\n [0.155]\r\n [0.105]\r\n [0.225]\r\n [0.296]\r\n [0.198]\r\n [0.917]\r\n [0.483]\r\n [0.881]\r\n [0.283]\r\n [0.919]\r\n [0.817]\r\n [0.509]\r\n [0.08 ]\r\n [0.965]]\r\n\r\n [[0.564]\r\n [0.931]\r\n [0.953]\r\n [0.001]\r\n [0.858]\r\n [0.332]\r\n [0.942]\r\n [0.699]\r\n [0.95 ]\r\n [0.486]\r\n [0.876]\r\n [0.727]\r\n [0.163]\r\n [0.535]\r\n [0.881]\r\n [0.937]\r\n [0.369]\r\n [0.556]\r\n [0.985]\r\n [0.574]\r\n [0.022]\r\n [0.835]\r\n [0.174]\r\n [0.615]\r\n [0.041]\r\n [0.916]\r\n [0.619]\r\n [0.342]]\r\n\r\n [[0.874]\r\n [0.522]\r\n [0.911]\r\n [0.163]\r\n [0.341]\r\n [0.233]\r\n [0.626]\r\n [0.528]\r\n [0.194]\r\n [0.566]\r\n [0.644]\r\n [0.897]\r\n [0.659]\r\n [0.065]\r\n [0.383]\r\n [0.778]\r\n [0.746]\r\n [0.492]\r\n [0.496]\r\n [0.628]\r\n [0.245]\r\n [0.07 ]\r\n [0.647]\r\n [0.159]\r\n [0.97 ]\r\n [0.407]\r\n [0.395]\r\n [0.4 ]]\r\n\r\n [[0.466]\r\n [0.914]\r\n [0.617]\r\n [0.202]\r\n [0.285]\r\n [0.183]\r\n [0.137]\r\n [0.952]\r\n [0.608]\r\n [0.434]\r\n [0.89 ]\r\n [0.293]\r\n [0.555]\r\n [0.034]\r\n [0.988]\r\n [0.548]\r\n [0.743]\r\n [0.066]\r\n [0.683]\r\n [0.205]\r\n [0.739]\r\n [0.205]\r\n [0.35 ]\r\n [0.297]\r\n [0.918]\r\n [0.928]\r\n [0.562]\r\n [0.747]]\r\n\r\n [[0.143]\r\n [0.686]\r\n [0.432]\r\n [0.701]\r\n [0.243]\r\n [0.503]\r\n [0.469]\r\n [0.553]\r\n [0.206]\r\n [0.444]\r\n [0.065]\r\n [0.711]\r\n [0.614]\r\n [0.722]\r\n [0.885]\r\n [0.754]\r\n [0.594]\r\n [0.124]\r\n [0.455]\r\n [0.276]\r\n [0.363]\r\n [0.125]\r\n [0.41 ]\r\n [0.078]\r\n [0.902]\r\n [0.321]\r\n [0.279]\r\n [0.164]]\r\n\r\n [[0.903]\r\n [0.556]\r\n [0.266]\r\n [0.927]\r\n [0.958]\r\n [0.2 ]\r\n [0.095]\r\n [0.761]\r\n [0.419]\r\n [0.047]\r\n [0.055]\r\n [0.831]\r\n [0.527]\r\n [0.404]\r\n [0.512]\r\n [0.653]\r\n [0.652]\r\n [0.325]\r\n [0.456]\r\n [0.681]\r\n [0.791]\r\n [0.601]\r\n [0.514]\r\n [0.255]\r\n [0.415]\r\n [0.831]\r\n [0.394]\r\n [0.02 ]]\r\n\r\n [[0.904]\r\n [0.89 ]\r\n [0.793]\r\n [0.342]\r\n [0.125]\r\n [0.039]\r\n [0.423]\r\n [0.714]\r\n [0.546]\r\n [0.299]\r\n [0.902]\r\n [0.717]\r\n [0.507]\r\n [0.355]\r\n [0.174]\r\n [0.836]\r\n [0.473]\r\n [0.635]\r\n [0.887]\r\n [0.755]\r\n [0.62 ]\r\n [0.508]\r\n [0.479]\r\n [0.915]\r\n [0.1 ]\r\n [0.665]\r\n [0.148]\r\n [0.421]]\r\n\r\n [[0.724]\r\n [0.336]\r\n [0.596]\r\n [0.432]\r\n [0.848]\r\n [0.737]\r\n [0.971]\r\n [0.408]\r\n [0.395]\r\n [0.371]\r\n [0.588]\r\n [0.778]\r\n [0.428]\r\n [0.687]\r\n [0.534]\r\n [0.427]\r\n [0.723]\r\n [0.161]\r\n [0.532]\r\n [0.739]\r\n [0.855]\r\n [0.37 ]\r\n [0.512]\r\n [0.862]\r\n [0.061]\r\n [0.817]\r\n [0.768]\r\n [0.735]]\r\n\r\n [[0.464]\r\n [0.604]\r\n [0.541]\r\n [0.349]\r\n [0.102]\r\n [0.704]\r\n [0.24 ]\r\n [0.38 ]\r\n [0.884]\r\n [0.131]\r\n [0.944]\r\n [0.494]\r\n [0.794]\r\n [0.934]\r\n [0.81 ]\r\n [0.015]\r\n [0.612]\r\n [0.399]\r\n [0.484]\r\n [0.18 ]\r\n [0.596]\r\n [0.347]\r\n [0.582]\r\n [0.47 ]\r\n [0.759]\r\n [0.232]\r\n [0.827]\r\n [0.635]]\r\n\r\n [[0.362]\r\n [0.976]\r\n [0.588]\r\n [0.466]\r\n [0.371]\r\n [0.139]\r\n [0.201]\r\n [0.927]\r\n [0.998]\r\n [0.063]\r\n [0.439]\r\n [0.483]\r\n [0.07 ]\r\n [0.478]\r\n [0.827]\r\n [0.77 ]\r\n [0.971]\r\n [0.318]\r\n [0.42 ]\r\n [0.835]\r\n [0.751]\r\n [0.84 ]\r\n [0.838]\r\n [0.654]\r\n [0.204]\r\n [0.153]\r\n [0.441]\r\n [0.015]]\r\n\r\n [[0.489]\r\n [0.252]\r\n [0. ]\r\n [0.417]\r\n [0.92 ]\r\n [0.442]\r\n [0.307]\r\n [0.073]\r\n [0.784]\r\n [0.481]\r\n [0.154]\r\n [0.334]\r\n [0.276]\r\n [0.163]\r\n [0.156]\r\n [0.145]\r\n [0.348]\r\n [0.356]\r\n [0.276]\r\n [0.68 ]\r\n [0.431]\r\n [0.238]\r\n [0.404]\r\n [0.284]\r\n [0.848]\r\n [0.865]\r\n [0.052]\r\n [0.433]]\r\n\r\n [[0.19 ]\r\n [0.266]\r\n [0.835]\r\n [0.469]\r\n [0.808]\r\n [0.189]\r\n [0.424]\r\n [0.127]\r\n [0.766]\r\n [0.638]\r\n [0.283]\r\n [0.494]\r\n [0.459]\r\n [0.299]\r\n [0.764]\r\n [0.368]\r\n [0.248]\r\n [0.28 ]\r\n [0.328]\r\n [0.356]\r\n [0.083]\r\n [0.067]\r\n [0.616]\r\n [0.994]\r\n [0.683]\r\n [0.975]\r\n [0.96 ]\r\n [0.706]]\r\n\r\n [[0.198]\r\n [0.879]\r\n [0.205]\r\n [0.14 ]\r\n [0.251]\r\n [0.814]\r\n [0.808]\r\n [0.61 ]\r\n [0.762]\r\n [0.847]\r\n [0.115]\r\n [0.894]\r\n [0.043]\r\n [0.342]\r\n [0.967]\r\n [0.269]\r\n [0.507]\r\n [0.072]\r\n [0.126]\r\n [0.091]\r\n [0.023]\r\n [0.917]\r\n [0.984]\r\n [0.007]\r\n [0.668]\r\n [0.992]\r\n [0.846]\r\n [0.062]]\r\n\r\n [[0.786]\r\n [0.823]\r\n [0.279]\r\n [0.888]\r\n [0.02 ]\r\n [0.12 ]\r\n [0.56 ]\r\n [0.383]\r\n [0.08 ]\r\n [0.013]\r\n [0.525]\r\n [0.078]\r\n [0.012]\r\n [0.242]\r\n [0.162]\r\n [0.088]\r\n [0.359]\r\n [0.276]\r\n [0.968]\r\n [0.219]\r\n [0.441]\r\n [0.957]\r\n [0.954]\r\n [0.779]\r\n [0.419]\r\n [0.109]\r\n [0.312]\r\n [0.708]]\r\n\r\n [[0.993]\r\n [0.588]\r\n [0.856]\r\n [0.924]\r\n [0.43 ]\r\n [0.725]\r\n [0.285]\r\n [0.961]\r\n [0.251]\r\n [0.631]\r\n [0.458]\r\n [0.416]\r\n [0.478]\r\n [0.872]\r\n [0.833]\r\n [0.285]\r\n [0.28 ]\r\n [0.669]\r\n [0.589]\r\n [0.238]\r\n [0.467]\r\n [0.706]\r\n [0.067]\r\n [0.665]\r\n [0.397]\r\n [0.021]\r\n [0.597]\r\n [0.76 ]]\r\n\r\n [[0.057]\r\n [0.769]\r\n [0.938]\r\n [0.901]\r\n [0.288]\r\n [0.145]\r\n [0.305]\r\n [0.275]\r\n [0.222]\r\n [0.04 ]\r\n [0.84 ]\r\n [0.734]\r\n [0.329]\r\n [0.106]\r\n [0.879]\r\n [0.876]\r\n [0.245]\r\n [0.179]\r\n [0.892]\r\n [0.731]\r\n [0.08 ]\r\n [0.307]\r\n [0.368]\r\n [0.668]\r\n [0.673]\r\n [0.867]\r\n [0.187]\r\n [0.583]]\r\n\r\n [[0.393]\r\n [0.241]\r\n [0.192]\r\n [0.787]\r\n [0.469]\r\n [0.128]\r\n [0.006]\r\n [0.271]\r\n [0.013]\r\n [0.79 ]\r\n [0.447]\r\n [0.557]\r\n [0.844]\r\n [0.303]\r\n [0.628]\r\n [0.339]\r\n [0.413]\r\n [0.441]\r\n [0.538]\r\n [0.235]\r\n [0.241]\r\n [0.229]\r\n [0.228]\r\n [0.374]\r\n [0.433]\r\n [0.838]\r\n [0.922]\r\n [0.24 ]]\r\n\r\n [[0.903]\r\n [0.151]\r\n [0.391]\r\n [0.079]\r\n [0.574]\r\n [0.94 ]\r\n [0.742]\r\n [0.677]\r\n [0.622]\r\n [0.969]\r\n [0.9 ]\r\n [0.785]\r\n [0.271]\r\n [0.373]\r\n [0.253]\r\n [0.03 ]\r\n [0.771]\r\n [0.717]\r\n [0.483]\r\n [0.004]\r\n [0.939]\r\n [0.301]\r\n [0.208]\r\n [0.081]\r\n [0.282]\r\n [0.288]\r\n [0.253]\r\n [0.556]]\r\n\r\n [[0.324]\r\n [0.683]\r\n [0.038]\r\n [0.487]\r\n [0.537]\r\n [0.955]\r\n [0.901]\r\n [0.862]\r\n [0.846]\r\n [0.544]\r\n [0.844]\r\n [0.451]\r\n [0.234]\r\n [0.34 ]\r\n [0.739]\r\n [0.18 ]\r\n [0.958]\r\n [0.674]\r\n [0.443]\r\n [0.389]\r\n [0.762]\r\n [0.836]\r\n [0.868]\r\n [0.437]\r\n [0.976]\r\n [0.478]\r\n [0.018]\r\n [0.971]]\r\n\r\n [[0.062]\r\n [0.232]\r\n [0.729]\r\n [0.697]\r\n [0.257]\r\n [0.599]\r\n [0.503]\r\n [0.49 ]\r\n [0.046]\r\n [0.781]\r\n [0.222]\r\n [0.566]\r\n [0.329]\r\n [0.379]\r\n [0.951]\r\n [0.421]\r\n [0.57 ]\r\n [0.333]\r\n [0.18 ]\r\n [0.96 ]\r\n [0.156]\r\n [0.116]\r\n [0.112]\r\n [0.063]\r\n [0.348]\r\n [0.878]\r\n [0.251]\r\n [0.041]]\r\n\r\n [[0.535]\r\n [0.085]\r\n [0.914]\r\n [0.981]\r\n [0.593]\r\n [0.098]\r\n [0.129]\r\n [0.803]\r\n [0.457]\r\n [0.177]\r\n [0.062]\r\n [0.26 ]\r\n [0.074]\r\n [0.865]\r\n [0.092]\r\n [0.607]\r\n [0.725]\r\n [0.402]\r\n [0.486]\r\n [0.392]\r\n [0.983]\r\n [0.715]\r\n [0.511]\r\n [0.334]\r\n [0.595]\r\n [0.875]\r\n [0.553]\r\n [0.619]]\r\n\r\n [[0.336]\r\n [0.758]\r\n [0.818]\r\n [0.907]\r\n [0.41 ]\r\n [0.397]\r\n [0.013]\r\n [0.874]\r\n [0.312]\r\n [0.242]\r\n [0.939]\r\n [0.318]\r\n [0.985]\r\n [0.135]\r\n [0.115]\r\n [0.92 ]\r\n [0.223]\r\n [0.166]\r\n [0.737]\r\n [0.023]\r\n [0.588]\r\n [0.995]\r\n [0.57 ]\r\n [0.376]\r\n [0.367]\r\n [0.278]\r\n [0.387]\r\n [0.406]]\r\n\r\n [[0.783]\r\n [0.672]\r\n [0.019]\r\n [0.94 ]\r\n [0.839]\r\n [0.976]\r\n [0.604]\r\n [0.692]\r\n [0.859]\r\n [0.383]\r\n [0.601]\r\n [0.247]\r\n [0.983]\r\n [0.84 ]\r\n [0.94 ]\r\n [0.227]\r\n [0.395]\r\n [0.424]\r\n [0.342]\r\n [0.01 ]\r\n [0.2 ]\r\n [0.077]\r\n [0.047]\r\n [0.024]\r\n [0.678]\r\n [0.862]\r\n [0.958]\r\n [0.209]]\r\n\r\n [[0.484]\r\n [0.186]\r\n [0.686]\r\n [0.531]\r\n [0.487]\r\n [0.642]\r\n [0.717]\r\n [0.178]\r\n [0.537]\r\n [0.681]\r\n [0.895]\r\n [0.645]\r\n [0.365]\r\n [0.943]\r\n [0.786]\r\n [0.346]\r\n [0.505]\r\n [0.576]\r\n [0.224]\r\n [0.984]\r\n [0.823]\r\n [0.361]\r\n [0.7 ]\r\n [0.111]\r\n [0.651]\r\n [0.986]\r\n [0.083]\r\n [0.367]]\r\n\r\n [[0.858]\r\n [0.919]\r\n [0.071]\r\n [0.45 ]\r\n [0.41 ]\r\n [0.162]\r\n [0.586]\r\n [0.842]\r\n [0.667]\r\n [0.902]\r\n [0.97 ]\r\n [0.606]\r\n [0.873]\r\n [0.804]\r\n [0.403]\r\n [0.805]\r\n [0.386]\r\n [0.311]\r\n [0.282]\r\n [0.913]\r\n [0.528]\r\n [0.118]\r\n [0.851]\r\n [0.371]\r\n [0.784]\r\n [0.736]\r\n [0.053]\r\n [0.819]]\r\n\r\n [[0.457]\r\n [0.274]\r\n [0.906]\r\n [0.006]\r\n [0.008]\r\n [0.772]\r\n [0.657]\r\n [0.84 ]\r\n [0.022]\r\n [0.08 ]\r\n [0.643]\r\n [0.808]\r\n [0.732]\r\n [0.681]\r\n [0.778]\r\n [0.664]\r\n [0.248]\r\n [0.254]\r\n [0.066]\r\n [0.527]\r\n [0.126]\r\n [0.297]\r\n [0.891]\r\n [0.836]\r\n [0.276]\r\n [0.478]\r\n [0.96 ]\r\n [0.263]]\r\n\r\n [[0.841]\r\n [0.094]\r\n [0.367]\r\n [0.519]\r\n [0.811]\r\n [0.803]\r\n [0.63 ]\r\n [0.704]\r\n [0.92 ]\r\n [0.963]\r\n [0.497]\r\n [0.833]\r\n [0.606]\r\n [0.136]\r\n [0.209]\r\n [0.191]\r\n [0.764]\r\n [0.143]\r\n [0.22 ]\r\n [0.787]\r\n [0.365]\r\n [0.428]\r\n [0.952]\r\n [0.994]\r\n [0.287]\r\n [0.164]\r\n [0.096]\r\n [0.413]]\r\n\r\n [[0.014]\r\n [0.904]\r\n [0.188]\r\n [0.77 ]\r\n [0.85 ]\r\n [0.858]\r\n [0.674]\r\n [0.679]\r\n [0.377]\r\n [0.581]\r\n [0.286]\r\n [0.742]\r\n [0.862]\r\n [0.113]\r\n [0.4 ]\r\n [0.047]\r\n [0.53 ]\r\n [0.958]\r\n [0.673]\r\n [0.499]\r\n [0.504]\r\n [0.041]\r\n [0.011]\r\n [0.252]\r\n [0.781]\r\n [0.142]\r\n [0.783]\r\n [0.933]]]]\r\n"
],
[
"!docker rm nodejs_mnist_predictor --force",
"nodejs_mnist_predictor\r\n"
]
],
[
[
"## Prediction using GRPC API on the docker container",
"_____no_output_____"
]
],
[
[
"!s2i build -E ./.s2i/environment_grpc . seldonio/seldon-core-s2i-nodejs:0.2-SNAPSHOT node-s2i-mnist-model:0.2",
"---> Installing application source...\n---> Installing dependencies ...\n\n> @tensorflow/[email protected] install /microservice/model/node_modules/@tensorflow/tfjs-node\n> node scripts/install.js\n\n* Downloading libtensorflow\n\n* Building TensorFlow Node.js bindings\n\n> [email protected] postinstall /microservice/model/node_modules/protobufjs\n> node scripts/postinstall\n\nnpm notice created a lockfile as package-lock.json. You should commit this file.\nnpm WARN [email protected] No repository field.\nnpm WARN [email protected] No license field.\n\nadded 50 packages from 57 contributors and audited 64 packages in 8.297s\nfound 0 vulnerabilities\n\nBuild completed successfully\n"
],
[
"!docker run --name \"nodejs_mnist_predictor\" -d --rm -p 5000:5000 node-s2i-mnist-model:0.2",
"7059dc3cd930b287c0eff2649034e338ddbaff4ce868c9fc695428bd426c04bd\r\n"
]
],
[
[
"Send some random features that conform to the contract",
"_____no_output_____"
]
],
[
[
"!seldon-core-tester contract.json 0.0.0.0 5000 -p -t --grpc",
"----------------------------------------\nSENDING NEW REQUEST:\nRECEIVED RESPONSE:\nSuccess:True message:\nRequest:\ndata {\n tensor {\n shape: 1\n shape: 28\n shape: 28\n shape: 1\n values: 0.401\n values: 0.774\n values: 0.944\n values: 0.816\n values: 0.069\n values: 0.403\n values: 0.572\n values: 0.724\n values: 0.972\n values: 0.433\n values: 0.697\n values: 0.868\n values: 0.523\n values: 0.937\n values: 0.06\n values: 0.921\n values: 0.726\n values: 0.336\n values: 0.635\n values: 0.242\n values: 0.648\n values: 0.104\n values: 0.615\n values: 0.873\n values: 0.224\n values: 0.88\n values: 0.99\n values: 0.92\n values: 0.234\n values: 0.723\n values: 0.462\n values: 0.346\n values: 0.034\n values: 0.037\n values: 0.426\n values: 0.656\n values: 0.24\n values: 0.375\n values: 0.818\n values: 0.998\n values: 0.129\n values: 0.108\n values: 0.1\n values: 0.633\n values: 0.518\n values: 0.742\n values: 0.248\n values: 0.671\n values: 0.898\n values: 0.59\n values: 0.414\n values: 0.749\n values: 0.639\n values: 0.974\n values: 0.218\n values: 0.021\n values: 0.82\n values: 0.944\n values: 0.249\n values: 0.678\n values: 0.638\n values: 0.427\n values: 0.64\n values: 0.94\n values: 0.976\n values: 0.151\n values: 0.056\n values: 0.439\n values: 0.955\n values: 0.604\n values: 0.968\n values: 0.192\n values: 0.036\n values: 0.279\n values: 0.947\n values: 0.955\n values: 0.93\n values: 0.707\n values: 0.579\n values: 0.373\n values: 0.636\n values: 0.628\n values: 0.052\n values: 0.755\n values: 0.111\n values: 0.03\n values: 0.429\n values: 0.585\n values: 0.658\n values: 0.661\n values: 0.732\n values: 0.705\n values: 0.358\n values: 0.8\n values: 0.13\n values: 0.471\n values: 0.477\n values: 0.267\n values: 0.511\n values: 0.913\n values: 0.497\n values: 0.242\n values: 0.869\n values: 0.171\n values: 0.774\n values: 0.42\n values: 0.778\n values: 0.253\n values: 0.309\n values: 0.552\n values: 0.233\n values: 0.199\n values: 0.941\n values: 0.731\n values: 0.917\n values: 0.435\n values: 0.26\n values: 0.578\n values: 0.365\n values: 0.261\n values: 0.348\n values: 0.725\n values: 0.428\n values: 0.987\n values: 0.157\n values: 0.048\n values: 0.457\n values: 0.529\n values: 0.883\n values: 0.999\n values: 0.763\n values: 0.575\n values: 0.519\n values: 0.979\n values: 0.979\n values: 0.408\n values: 0.096\n values: 0.352\n values: 0.21\n values: 0.977\n values: 0.669\n values: 0.826\n values: 0.466\n values: 0.081\n values: 0.612\n values: 0.421\n values: 0.972\n values: 0.407\n values: 0.288\n values: 0.725\n values: 0.153\n values: 0.508\n values: 0.989\n values: 0.718\n values: 0.797\n values: 0.973\n values: 0.968\n values: 0.729\n values: 0.229\n values: 0.44\n values: 0.842\n values: 0.971\n values: 0.472\n values: 0.238\n values: 0.611\n values: 0.68\n values: 0.36\n values: 0.16\n values: 0.905\n values: 0.869\n values: 0.545\n values: 0.09\n values: 0.837\n values: 0.699\n values: 0.113\n values: 0.735\n values: 0.201\n values: 0.241\n values: 0.527\n values: 0.227\n values: 0.702\n values: 0.026\n values: 0.532\n values: 0.93\n values: 0.982\n values: 0.446\n values: 0.103\n values: 0.926\n values: 0.335\n values: 0.591\n values: 0.013\n values: 0.107\n values: 0.973\n values: 0.97\n values: 0.476\n values: 0.038\n values: 0.501\n values: 0.633\n values: 0.075\n values: 0.965\n values: 0.161\n values: 0.383\n values: 0.336\n values: 0.415\n values: 0.482\n values: 0.822\n values: 0.36\n values: 0.452\n values: 0.913\n values: 0.346\n values: 0.873\n values: 0.282\n values: 0.225\n values: 0.924\n values: 0.458\n values: 0.79\n values: 0.861\n values: 0.813\n values: 0.762\n values: 0.394\n values: 0.893\n values: 0.038\n values: 0.327\n values: 0.397\n values: 0.296\n values: 0.681\n values: 0.68\n values: 0.088\n values: 0.058\n values: 0.942\n values: 0.483\n values: 0.689\n values: 0.09\n values: 0.217\n values: 0.263\n values: 0.872\n values: 0.407\n values: 0.322\n values: 0.875\n values: 0.019\n values: 0.796\n values: 0.492\n values: 0.71\n values: 0.122\n values: 0.725\n values: 0.164\n values: 0.555\n values: 0.421\n values: 0.577\n values: 0.011\n values: 0.583\n values: 0.802\n values: 0.008\n values: 0.928\n values: 0.41\n values: 0.903\n values: 0.062\n values: 0.918\n values: 0.468\n values: 0.372\n values: 0.504\n values: 0.69\n values: 0.572\n values: 0.058\n values: 0.761\n values: 0.05\n values: 0.475\n values: 0.686\n values: 0.696\n values: 0.963\n values: 0.864\n values: 0.746\n values: 0.1\n values: 0.024\n values: 0.71\n values: 0.289\n values: 0.826\n values: 0.819\n values: 0.508\n values: 0.303\n values: 0.231\n values: 0.17\n values: 0.156\n values: 0.935\n values: 0.01\n values: 0.01\n values: 0.305\n values: 0.071\n values: 0.248\n values: 0.379\n values: 0.219\n values: 0.597\n values: 0.977\n values: 0.916\n values: 0.401\n values: 0.19\n values: 0.679\n values: 0.791\n values: 0.397\n values: 0.763\n values: 0.961\n values: 0.109\n values: 0.062\n values: 0.655\n values: 0.924\n values: 0.497\n values: 0.955\n values: 0.921\n values: 0.201\n values: 0.837\n values: 0.689\n values: 0.417\n values: 0.667\n values: 0.463\n values: 0.19\n values: 0.414\n values: 0.525\n values: 0.905\n values: 0.585\n values: 0.191\n values: 0.997\n values: 0.477\n values: 0.214\n values: 0.081\n values: 0.106\n values: 0.568\n values: 0.978\n values: 0.88\n values: 0.202\n values: 0.766\n values: 0.16\n values: 0.422\n values: 0.07\n values: 0.87\n values: 0.211\n values: 0.074\n values: 0.944\n values: 0.304\n values: 0.383\n values: 0.793\n values: 0.984\n values: 0.735\n values: 0.669\n values: 0.308\n values: 0.73\n values: 0.418\n values: 0.099\n values: 0.637\n values: 0.493\n values: 0.705\n values: 0.307\n values: 0.966\n values: 0.532\n values: 0.867\n values: 0.665\n values: 0.601\n values: 0.443\n values: 0.589\n values: 0.296\n values: 0.256\n values: 0.524\n values: 0.898\n values: 0.816\n values: 0.001\n values: 0.673\n values: 0.156\n values: 0.302\n values: 0.979\n values: 0.722\n values: 0.479\n values: 0.635\n values: 0.479\n values: 0.879\n values: 0.394\n values: 0.078\n values: 0.354\n values: 0.741\n values: 0.182\n values: 0.796\n values: 0.792\n values: 0.59\n values: 0.305\n values: 0.056\n values: 0.536\n values: 0.041\n values: 0.487\n values: 0.679\n values: 0.491\n values: 0.586\n values: 0.358\n values: 0.444\n values: 0.967\n values: 0.739\n values: 0.556\n values: 0.959\n values: 0.214\n values: 0.919\n values: 0.636\n values: 0.734\n values: 0.171\n values: 0.96\n values: 0.193\n values: 0.402\n values: 0.896\n values: 0.694\n values: 0.523\n values: 0.431\n values: 0.305\n values: 0.624\n values: 0.892\n values: 0.252\n values: 0.422\n values: 0.11\n values: 0.573\n values: 0.989\n values: 0.548\n values: 0.097\n values: 0.829\n values: 0.547\n values: 0.337\n values: 0.334\n values: 0.769\n values: 0.76\n values: 0.539\n values: 0.269\n values: 0.54\n values: 0.424\n values: 0.491\n values: 0.488\n values: 0.029\n values: 0.9\n values: 0.635\n values: 0.934\n values: 0.926\n values: 0.291\n values: 0.556\n values: 0.361\n values: 0.235\n values: 0.588\n values: 0.389\n values: 0.753\n values: 0.748\n values: 0.117\n values: 0.166\n values: 0.382\n values: 0.163\n values: 0.43\n values: 0.595\n values: 0.067\n values: 0.908\n values: 0.217\n values: 0.717\n values: 0.721\n values: 0.858\n values: 0.363\n values: 0.568\n values: 0.32\n values: 0.071\n values: 0.099\n values: 0.91\n values: 0.774\n values: 0.67\n values: 0.308\n values: 0.616\n values: 0.109\n values: 0.339\n values: 0.311\n values: 0.023\n values: 0.368\n values: 0.462\n values: 0.158\n values: 0.965\n values: 0.338\n values: 0.374\n values: 0.048\n values: 0.195\n values: 0.972\n values: 0.57\n values: 0.623\n values: 0.898\n values: 0.244\n values: 0.457\n values: 0.845\n values: 0.381\n values: 0.89\n values: 0.814\n values: 0.713\n values: 0.309\n values: 0.73\n values: 0.739\n values: 0.596\n values: 0.556\n values: 0.712\n values: 0.843\n values: 0.568\n values: 0.386\n values: 0.095\n values: 0.041\n values: 0.551\n values: 0.556\n values: 0.446\n values: 0.049\n values: 0.21\n values: 0.537\n values: 0.685\n values: 0.225\n values: 0.84\n values: 0.034\n values: 0.703\n values: 0.725\n values: 0.542\n values: 0.338\n values: 0.628\n values: 0.022\n values: 0.338\n values: 0.568\n values: 0.528\n values: 0.475\n values: 0.911\n values: 0.312\n values: 0.898\n values: 0.362\n values: 0.68\n values: 0.111\n values: 0.77\n values: 0.505\n values: 0.847\n values: 0.468\n values: 0.086\n values: 0.127\n values: 0.775\n values: 0.758\n values: 0.423\n values: 0.442\n values: 0.721\n values: 0.181\n values: 0.733\n values: 0.308\n values: 0.705\n values: 0.147\n values: 0.42\n values: 0.148\n values: 0.576\n values: 0.048\n values: 0.018\n values: 0.225\n values: 0.231\n values: 0.942\n values: 0.196\n values: 0.968\n values: 0.515\n values: 0.856\n values: 0.461\n values: 0.935\n values: 0.92\n values: 0.574\n values: 0.693\n values: 0.87\n values: 0.36\n values: 0.533\n values: 0.749\n values: 0.908\n values: 0.019\n values: 0.796\n values: 0.462\n values: 0.603\n values: 0.81\n values: 0.076\n values: 0.612\n values: 0.542\n values: 0.08\n values: 0.859\n values: 0.218\n values: 0.127\n values: 0.998\n values: 0.824\n values: 0.712\n values: 0.216\n values: 0.788\n values: 0.327\n values: 0.786\n values: 0.651\n values: 0.329\n values: 0.413\n values: 0.071\n values: 0.444\n values: 0.005\n values: 0.89\n values: 0.152\n values: 0.186\n values: 0.359\n values: 0.527\n values: 0.202\n values: 0.308\n values: 0.161\n values: 0.642\n values: 0.272\n values: 0.662\n values: 0.075\n values: 0.948\n values: 0.104\n values: 0.949\n values: 0.613\n values: 0.231\n values: 0.342\n values: 0.011\n values: 0.664\n values: 0.792\n values: 0.939\n values: 0.659\n values: 0.023\n values: 0.498\n values: 0.154\n values: 0.318\n values: 0.973\n values: 0.822\n values: 0.828\n values: 0.118\n values: 0.602\n values: 0.712\n values: 0.076\n values: 0.178\n values: 0.311\n values: 0.747\n values: 0.116\n values: 0.11\n values: 0.633\n values: 0.38\n values: 0.153\n values: 0.202\n values: 0.522\n values: 0.632\n values: 0.602\n values: 0.205\n values: 0.288\n values: 0.878\n values: 0.948\n values: 0.745\n values: 0.019\n values: 0.542\n values: 0.531\n values: 0.611\n values: 0.188\n values: 0.416\n values: 0.861\n values: 0.329\n values: 0.438\n values: 0.434\n values: 0.983\n values: 0.065\n values: 0.496\n values: 0.657\n values: 0.142\n values: 0.181\n values: 0.685\n values: 0.635\n values: 0.767\n values: 0.336\n values: 0.914\n values: 0.135\n values: 0.99\n values: 0.34\n values: 0.101\n values: 0.138\n values: 0.905\n values: 0.056\n values: 0.487\n values: 0.627\n values: 0.109\n values: 0.06\n values: 0.089\n values: 0.761\n values: 0.043\n values: 0.229\n values: 0.253\n values: 0.379\n values: 0.541\n values: 0.756\n values: 0.654\n values: 0.654\n values: 0.83\n values: 0.598\n values: 0.349\n values: 0.689\n values: 0.606\n values: 0.335\n values: 0.139\n values: 0.67\n values: 0.641\n values: 0.907\n values: 0.563\n values: 0.188\n values: 0.43\n values: 0.412\n values: 0.635\n values: 0.051\n values: 0.445\n values: 0.086\n values: 0.157\n values: 0.482\n values: 0.699\n values: 0.805\n values: 0.849\n values: 0.812\n values: 0.381\n values: 0.459\n values: 0.974\n values: 0.257\n values: 0.879\n values: 0.42\n values: 0.008\n values: 0.931\n values: 0.028\n values: 0.968\n values: 0.891\n values: 0.408\n values: 0.253\n values: 0.174\n values: 0.608\n values: 0.847\n values: 0.908\n values: 0.488\n values: 0.287\n values: 0.656\n values: 0.013\n values: 0.377\n values: 0.638\n values: 0.851\n values: 0.631\n values: 0.726\n values: 0.528\n values: 0.405\n values: 0.315\n values: 0.629\n values: 0.812\n values: 0.734\n values: 0.01\n values: 0.417\n values: 0.731\n values: 0.615\n values: 0.384\n values: 0.902\n values: 0.16\n values: 0.703\n values: 0.24\n values: 0.493\n values: 0.899\n values: 0.047\n values: 0.41\n values: 0.576\n values: 0.581\n values: 0.425\n values: 0.483\n values: 0.592\n values: 0.817\n values: 0.675\n values: 0.49\n values: 0.033\n values: 0.672\n values: 0.638\n values: 0.173\n values: 0.504\n values: 0.829\n values: 0.445\n values: 0.369\n values: 0.54\n values: 0.146\n values: 0.332\n values: 0.716\n values: 0.485\n values: 0.741\n values: 0.427\n values: 0.607\n values: 0.437\n values: 0.082\n values: 0.302\n values: 0.177\n values: 0.22\n values: 0.795\n values: 0.129\n values: 0.834\n values: 0.905\n values: 0.04\n values: 0.193\n }\n}\n\nResponse:\ndata {\n names: \"t:0\"\n names: \"t:1\"\n names: \"t:2\"\n names: \"t:3\"\n names: \"t:4\"\n names: \"t:5\"\n names: \"t:6\"\n names: \"t:7\"\n names: \"t:8\"\n names: \"t:9\"\n tensor {\n shape: 1\n shape: 10\n values: 0.016527308151125908\n values: 0.03343753144145012\n values: 0.13515257835388184\n values: 0.2837943434715271\n values: 0.046536825597286224\n values: 0.04741676151752472\n values: 0.015463653951883316\n values: 0.01107808854430914\n values: 0.37488991022109985\n values: 0.03570308908820152\n }\n}\n\n\n"
],
[
"!docker rm nodejs_mnist_predictor --force",
"nodejs_mnist_predictor\r\n"
]
],
[
[
"## Test using Minikube\n\n**Due to a [minikube/s2i issue](https://github.com/SeldonIO/seldon-core/issues/253) you will need [s2i >= 1.1.13](https://github.com/openshift/source-to-image/releases/tag/v1.1.13)**",
"_____no_output_____"
]
],
[
[
"!minikube start --memory 4096",
"😄 minikube v0.34.1 on linux (amd64)\n🔥 Creating virtualbox VM (CPUs=2, Memory=4096MB, Disk=20000MB) ...\n📶 \"minikube\" IP address is 192.168.99.100\n🐳 Configuring Docker as the container runtime ...\n✨ Preparing Kubernetes environment ...\n🚜 Pulling images required by Kubernetes v1.13.3 ...\n🚀 Launching Kubernetes v1.13.3 using kubeadm ... \n🔑 Configuring cluster permissions ...\n🤔 Verifying component health .....\n💗 kubectl is now configured to use \"minikube\"\n🏄 Done! Thank you for using minikube!\n"
],
[
"!kubectl create clusterrolebinding kube-system-cluster-admin --clusterrole=cluster-admin --serviceaccount=kube-system:default",
"clusterrolebinding.rbac.authorization.k8s.io/kube-system-cluster-admin created\r\n"
],
[
"!helm init",
"$HELM_HOME has been configured at /home/clive/.helm.\n\nTiller (the Helm server-side component) has been installed into your Kubernetes Cluster.\n\nPlease note: by default, Tiller is deployed with an insecure 'allow unauthenticated users' policy.\nTo prevent this, run `helm init` with the --tiller-tls-verify flag.\nFor more information on securing your installation see: https://docs.helm.sh/using_helm/#securing-your-helm-installation\nHappy Helming!\n"
],
[
"!kubectl rollout status deploy/tiller-deploy -n kube-system",
"Waiting for deployment \"tiller-deploy\" rollout to finish: 0 of 1 updated replicas are available...\ndeployment \"tiller-deploy\" successfully rolled out\n"
],
[
"!helm install ../../../helm-charts/seldon-core-operator --name seldon-core --set usageMetrics.enabled=true --namespace seldon-system",
"NAME: seldon-core\nLAST DEPLOYED: Tue Apr 16 15:29:41 2019\nNAMESPACE: seldon-system\nSTATUS: DEPLOYED\n\nRESOURCES:\n==> v1/ClusterRoleBinding\nNAME AGE\nseldon-operator-manager-rolebinding 1s\n\n==> v1/Service\nNAME TYPE CLUSTER-IP EXTERNAL-IP PORT(S) AGE\nseldon-operator-controller-manager-service ClusterIP 10.107.173.100 <none> 443/TCP 1s\n\n==> v1beta1/Deployment\nNAME DESIRED CURRENT UP-TO-DATE AVAILABLE AGE\nseldon-spartakus-volunteer 1 0 0 0 1s\n\n==> v1beta1/ClusterRole\nNAME AGE\nseldon-spartakus-volunteer 1s\n\n==> v1beta1/ClusterRoleBinding\nNAME AGE\nseldon-spartakus-volunteer 1s\n\n==> v1/Secret\nNAME TYPE DATA AGE\nseldon-operator-webhook-server-secret Opaque 0 1s\n\n==> v1/ConfigMap\nNAME DATA AGE\nseldon-spartakus-config 3 1s\n\n==> v1beta1/CustomResourceDefinition\nNAME AGE\nseldondeployments.machinelearning.seldon.io 1s\n\n==> v1/Pod(related)\nNAME READY STATUS RESTARTS AGE\nseldon-operator-controller-manager-0 0/1 ContainerCreating 0 1s\n\n==> v1/ClusterRole\nNAME AGE\nseldon-operator-manager-role 1s\n\n==> v1/StatefulSet\nNAME DESIRED CURRENT AGE\nseldon-operator-controller-manager 1 1 1s\n\n==> v1/ServiceAccount\nNAME SECRETS AGE\nseldon-spartakus-volunteer 1 1s\n\n\nNOTES:\nNOTES: TODO\n\n\n"
],
[
"!kubectl rollout status statefulset.apps/seldon-operator-controller-manager -n seldon-system",
"partitioned roll out complete: 1 new pods have been updated...\r\n"
]
],
[
[
"## Setup Ingress\nPlease note: There are reported gRPC issues with ambassador (see https://github.com/SeldonIO/seldon-core/issues/473).",
"_____no_output_____"
]
],
[
[
"!helm install stable/ambassador --name ambassador --set crds.keep=false",
"NAME: ambassador\nLAST DEPLOYED: Tue Apr 16 15:30:27 2019\nNAMESPACE: seldon\nSTATUS: DEPLOYED\n\nRESOURCES:\n==> v1/Pod(related)\nNAME READY STATUS RESTARTS AGE\nambassador-5b89d44544-b2snz 0/1 ContainerCreating 0 0s\nambassador-5b89d44544-ktd9h 0/1 ContainerCreating 0 0s\nambassador-5b89d44544-x8tcg 0/1 ContainerCreating 0 0s\n\n==> v1/ServiceAccount\nNAME SECRETS AGE\nambassador 1 0s\n\n==> v1beta1/ClusterRole\nNAME AGE\nambassador 0s\n\n==> v1beta1/ClusterRoleBinding\nNAME AGE\nambassador 0s\n\n==> v1/Service\nNAME TYPE CLUSTER-IP EXTERNAL-IP PORT(S) AGE\nambassador-admins ClusterIP 10.98.139.122 <none> 8877/TCP 0s\nambassador LoadBalancer 10.98.135.113 <pending> 80:31109/TCP,443:30421/TCP 0s\n\n==> v1/Deployment\nNAME DESIRED CURRENT UP-TO-DATE AVAILABLE AGE\nambassador 3 3 3 0 0s\n\n\nNOTES:\nCongratuations! You've successfully installed Ambassador.\n\nFor help, visit our Slack at https://d6e.co/slack or view the documentation online at https://www.getambassador.io.\n\nTo get the IP address of Ambassador, run the following commands:\nNOTE: It may take a few minutes for the LoadBalancer IP to be available.\n You can watch the status of by running 'kubectl get svc -w --namespace seldon ambassador'\n\n On GKE/Azure:\n export SERVICE_IP=$(kubectl get svc --namespace seldon ambassador -o jsonpath='{.status.loadBalancer.ingress[0].ip}')\n\n On AWS:\n export SERVICE_IP=$(kubectl get svc --namespace seldon ambassador -o jsonpath='{.status.loadBalancer.ingress[0].hostname}')\n\n echo http://$SERVICE_IP:\n\n"
],
[
"!kubectl rollout status deployment.apps/ambassador",
"Waiting for deployment \"ambassador\" rollout to finish: 0 of 3 updated replicas are available...\nWaiting for deployment \"ambassador\" rollout to finish: 1 of 3 updated replicas are available...\nWaiting for deployment \"ambassador\" rollout to finish: 2 of 3 updated replicas are available...\ndeployment \"ambassador\" successfully rolled out\n"
],
[
"!eval $(minikube docker-env) && s2i build . seldonio/seldon-core-s2i-nodejs:0.2-SNAPSHOT node-s2i-mnist-model:0.1",
"---> Installing application source...\n---> Installing dependencies ...\n\n> @tensorflow/[email protected] install /microservice/model/node_modules/@tensorflow/tfjs-node\n> node scripts/install.js\n\n* Downloading libtensorflow\n\n* Building TensorFlow Node.js bindings\n\n> [email protected] postinstall /microservice/model/node_modules/protobufjs\n> node scripts/postinstall\n\nnpm notice created a lockfile as package-lock.json. You should commit this file.\nnpm WARN [email protected] No repository field.\nnpm WARN [email protected] No license field.\n\nadded 50 packages from 58 contributors and audited 64 packages in 15.755s\nfound 0 vulnerabilities\n\nBuild completed successfully\n"
],
[
"!kubectl create -f nodejs_mnist_deployment.json",
"seldondeployment.machinelearning.seldon.io/seldon-deployment-example created\r\n"
],
[
"!kubectl rollout status deploy/nodejs-mnist-deployment-nodejs-mnist-predictor-5aa9edd",
"Waiting for deployment \"nodejs-mnist-deployment-nodejs-mnist-predictor-5aa9edd\" rollout to finish: 0 of 1 updated replicas are available...\ndeployment \"nodejs-mnist-deployment-nodejs-mnist-predictor-5aa9edd\" successfully rolled out\n"
],
[
"!seldon-core-api-tester contract.json `minikube ip` `kubectl get svc ambassador -o jsonpath='{.spec.ports[0].nodePort}'` \\\n seldon-deployment-example --namespace seldon -p",
"----------------------------------------\r\nSENDING NEW REQUEST:\r\n\r\n[[[[0.743]\r\n [0.335]\r\n [0.726]\r\n [0.051]\r\n [0.954]\r\n [0.716]\r\n [0.094]\r\n [0.447]\r\n [0.524]\r\n [0.431]\r\n [0.749]\r\n [0.034]\r\n [0.607]\r\n [0.342]\r\n [0.035]\r\n [0.744]\r\n [0.838]\r\n [0.29 ]\r\n [0.311]\r\n [0.091]\r\n [0.177]\r\n [0.049]\r\n [0.43 ]\r\n [0.469]\r\n [0.766]\r\n [0.137]\r\n [0.517]\r\n [0.814]]\r\n\r\n [[0.066]\r\n [0.168]\r\n [0.69 ]\r\n [0.733]\r\n [0.658]\r\n [0.838]\r\n [0.867]\r\n [0.485]\r\n [0.093]\r\n [0.405]\r\n [0.809]\r\n [0.592]\r\n [0.574]\r\n [0.449]\r\n [0.776]\r\n [0.988]\r\n [0.428]\r\n [0.912]\r\n [0.351]\r\n [0.406]\r\n [0.919]\r\n [0.832]\r\n [0.639]\r\n [0.342]\r\n [0.778]\r\n [0.31 ]\r\n [0.476]\r\n [0.828]]\r\n\r\n [[0.99 ]\r\n [0.076]\r\n [0.238]\r\n [0.941]\r\n [0.442]\r\n [0.998]\r\n [0.591]\r\n [0.486]\r\n [0.631]\r\n [0.312]\r\n [0.679]\r\n [0.214]\r\n [0.79 ]\r\n [0.207]\r\n [0.965]\r\n [0.912]\r\n [0.247]\r\n [0.851]\r\n [0.761]\r\n [0.833]\r\n [0.875]\r\n [0.284]\r\n [0.352]\r\n [0.884]\r\n [0.517]\r\n [0.547]\r\n [0.487]\r\n [0.13 ]]\r\n\r\n [[0.395]\r\n [0.259]\r\n [0.773]\r\n [0.935]\r\n [0.686]\r\n [0.526]\r\n [0.319]\r\n [0.254]\r\n [0.771]\r\n [0.428]\r\n [0.621]\r\n [0.546]\r\n [0.167]\r\n [0.678]\r\n [0.364]\r\n [0.173]\r\n [0.834]\r\n [0.298]\r\n [0.069]\r\n [0.427]\r\n [0.98 ]\r\n [0.53 ]\r\n [0.422]\r\n [0.303]\r\n [0.828]\r\n [0.857]\r\n [0.258]\r\n [0.837]]\r\n\r\n [[0.204]\r\n [0.755]\r\n [0.256]\r\n [0.964]\r\n [0.774]\r\n [0.533]\r\n [0.692]\r\n [0.152]\r\n [0.491]\r\n [0.913]\r\n [0.161]\r\n [0.909]\r\n [0.278]\r\n [0.436]\r\n [0.642]\r\n [0.598]\r\n [0.094]\r\n [0.39 ]\r\n [0.706]\r\n [0.882]\r\n [0.906]\r\n [0.402]\r\n [0.653]\r\n [0.792]\r\n [0.458]\r\n [0.106]\r\n [0.601]\r\n [0.867]]\r\n\r\n [[0.548]\r\n [0.711]\r\n [0.079]\r\n [0.998]\r\n [0.812]\r\n [0.547]\r\n [0.582]\r\n [0.086]\r\n [0.336]\r\n [0.223]\r\n [0.037]\r\n [0.945]\r\n [0.816]\r\n [0.354]\r\n [0.991]\r\n [0.817]\r\n [0.41 ]\r\n [0.591]\r\n [0.138]\r\n [0.485]\r\n [0.348]\r\n [0.503]\r\n [0.626]\r\n [0.037]\r\n [0.862]\r\n [0.258]\r\n [0.128]\r\n [0.828]]\r\n\r\n [[0.483]\r\n [0.789]\r\n [0.076]\r\n [0.618]\r\n [0.162]\r\n [0.201]\r\n [0.927]\r\n [0.436]\r\n [0.609]\r\n [0.123]\r\n [0.599]\r\n [0.038]\r\n [0.836]\r\n [0.506]\r\n [0.727]\r\n [0.783]\r\n [0.522]\r\n [0.925]\r\n [0.628]\r\n [0.938]\r\n [0.388]\r\n [0.07 ]\r\n [0.262]\r\n [0.163]\r\n [0.295]\r\n [0.632]\r\n [0.331]\r\n [0.129]]\r\n\r\n [[0.704]\r\n [0.126]\r\n [0.011]\r\n [0.974]\r\n [0.22 ]\r\n [0.693]\r\n [0.798]\r\n [0.579]\r\n [0.074]\r\n [0.76 ]\r\n [0.482]\r\n [0.651]\r\n [0.85 ]\r\n [0.972]\r\n [0.911]\r\n [0.297]\r\n [0.036]\r\n [0.71 ]\r\n [0.854]\r\n [0.489]\r\n [0.211]\r\n [0.778]\r\n [0.374]\r\n [0.454]\r\n [0.717]\r\n [0.142]\r\n [0.378]\r\n [0.087]]\r\n\r\n [[0.487]\r\n [0.761]\r\n [0.703]\r\n [0.202]\r\n [0.948]\r\n [0.699]\r\n [0.946]\r\n [0.444]\r\n [0.229]\r\n [0.612]\r\n [0.659]\r\n [0.45 ]\r\n [0.554]\r\n [0.13 ]\r\n [0.97 ]\r\n [0.215]\r\n [0.519]\r\n [0.627]\r\n [0.519]\r\n [0.948]\r\n [0.212]\r\n [0.087]\r\n [0.704]\r\n [0.853]\r\n [0.931]\r\n [0.484]\r\n [0.218]\r\n [0.998]]\r\n\r\n [[0.376]\r\n [0.14 ]\r\n [0.053]\r\n [0.322]\r\n [0.542]\r\n [0.357]\r\n [0.858]\r\n [0.903]\r\n [0.292]\r\n [0.812]\r\n [0.436]\r\n [0.049]\r\n [0.629]\r\n [0.679]\r\n [0.968]\r\n [0.974]\r\n [0.842]\r\n [0.936]\r\n [0.855]\r\n [0.371]\r\n [0.808]\r\n [0.243]\r\n [0.355]\r\n [0.221]\r\n [0.141]\r\n [0.972]\r\n [0.104]\r\n [0.729]]\r\n\r\n [[0.843]\r\n [0.73 ]\r\n [0.963]\r\n [0.455]\r\n [0.357]\r\n [0.34 ]\r\n [0.251]\r\n [0.939]\r\n [0.118]\r\n [0.237]\r\n [0.926]\r\n [0.853]\r\n [0.832]\r\n [0.59 ]\r\n [0.132]\r\n [0.55 ]\r\n [0.792]\r\n [0.974]\r\n [0.025]\r\n [0.417]\r\n [0.517]\r\n [0.083]\r\n [0.152]\r\n [0.01 ]\r\n [0.782]\r\n [0.036]\r\n [0.022]\r\n [0.244]]\r\n\r\n [[0.207]\r\n [0.481]\r\n [0.655]\r\n [0.173]\r\n [0.721]\r\n [0.283]\r\n [0.163]\r\n [0.418]\r\n [0.402]\r\n [0.006]\r\n [0.974]\r\n [0.953]\r\n [0.989]\r\n [0.952]\r\n [0.895]\r\n [0.112]\r\n [0.886]\r\n [0.226]\r\n [0.394]\r\n [0.566]\r\n [0.728]\r\n [0.959]\r\n [0.452]\r\n [0.296]\r\n [0.662]\r\n [0.372]\r\n [0.872]\r\n [0.443]]\r\n\r\n [[0.579]\r\n [0.698]\r\n [0.936]\r\n [0.521]\r\n [0.126]\r\n [0.216]\r\n [0.36 ]\r\n [0.208]\r\n [0.709]\r\n [0.747]\r\n [0.178]\r\n [0.201]\r\n [0.875]\r\n [0.518]\r\n [0.173]\r\n [0.453]\r\n [0.293]\r\n [0.571]\r\n [0.448]\r\n [0.16 ]\r\n [0.096]\r\n [0.572]\r\n [0.837]\r\n [0.276]\r\n [0.597]\r\n [0.151]\r\n [0.56 ]\r\n [0.653]]\r\n\r\n [[0.905]\r\n [0.3 ]\r\n [0.38 ]\r\n [0.8 ]\r\n [0.475]\r\n [0.52 ]\r\n [0.982]\r\n [0.077]\r\n [0.691]\r\n [0.204]\r\n [0.931]\r\n [0.573]\r\n [0.875]\r\n [0.084]\r\n [0.112]\r\n [0.312]\r\n [0.422]\r\n [0.393]\r\n [0.98 ]\r\n [0.145]\r\n [0.734]\r\n [0.21 ]\r\n [0.741]\r\n [0.666]\r\n [0.925]\r\n [0.987]\r\n [0.583]\r\n [0.84 ]]\r\n\r\n [[0.036]\r\n [0.916]\r\n [0.312]\r\n [0.495]\r\n [0.701]\r\n [0.547]\r\n [0.483]\r\n [0.231]\r\n [0.218]\r\n [0.912]\r\n [0.561]\r\n [0.881]\r\n [0.039]\r\n [0.969]\r\n [0.147]\r\n [0.615]\r\n [0.582]\r\n [0.554]\r\n [0.715]\r\n [0.607]\r\n [0.972]\r\n [0.898]\r\n [0.923]\r\n [0.08 ]\r\n [0.511]\r\n [0.111]\r\n [0.446]\r\n [0.984]]\r\n\r\n [[0.646]\r\n [0.753]\r\n [0.474]\r\n [0.689]\r\n [0.697]\r\n [0.093]\r\n [0.372]\r\n [0.584]\r\n [0.55 ]\r\n [0.768]\r\n [0.964]\r\n [0.414]\r\n [0.129]\r\n [0.021]\r\n [0.635]\r\n [0.766]\r\n [0.588]\r\n [0.454]\r\n [0.927]\r\n [0.898]\r\n [0.613]\r\n [0.992]\r\n [0.736]\r\n [0.888]\r\n [0.329]\r\n [0.931]\r\n [0.043]\r\n [0.12 ]]\r\n\r\n [[0.514]\r\n [0.151]\r\n [0.128]\r\n [0.178]\r\n [0.809]\r\n [0.338]\r\n [0.104]\r\n [0.454]\r\n [0.523]\r\n [0.391]\r\n [0.005]\r\n [0.045]\r\n [0.837]\r\n [0.023]\r\n [0.483]\r\n [0.148]\r\n [0.13 ]\r\n [0.457]\r\n [0.517]\r\n [0.344]\r\n [0.94 ]\r\n [0.461]\r\n [0.98 ]\r\n [0.368]\r\n [0.453]\r\n [0.71 ]\r\n [0.65 ]\r\n [0.838]]\r\n\r\n [[0.691]\r\n [0.838]\r\n [0.411]\r\n [0.18 ]\r\n [0.769]\r\n [0.928]\r\n [0.898]\r\n [0.237]\r\n [0.134]\r\n [0.892]\r\n [0.428]\r\n [0.977]\r\n [0.295]\r\n [0.917]\r\n [0.374]\r\n [0.494]\r\n [0.913]\r\n [0.889]\r\n [0.635]\r\n [0.633]\r\n [0.452]\r\n [0.76 ]\r\n [0.809]\r\n [0.593]\r\n [0.001]\r\n [0.708]\r\n [0.546]\r\n [0.826]]\r\n\r\n [[0.011]\r\n [0.106]\r\n [0.125]\r\n [0.169]\r\n [0.569]\r\n [0.88 ]\r\n [0.078]\r\n [0.221]\r\n [0.43 ]\r\n [0.703]\r\n [0.172]\r\n [0.228]\r\n [0.33 ]\r\n [0.829]\r\n [0.798]\r\n [0.759]\r\n [0.124]\r\n [0.137]\r\n [0.222]\r\n [0.547]\r\n [0.112]\r\n [0.304]\r\n [0.074]\r\n [0.344]\r\n [0.786]\r\n [0.569]\r\n [0.288]\r\n [0.192]]\r\n\r\n [[0.205]\r\n [0.676]\r\n [0.823]\r\n [0.66 ]\r\n [0.019]\r\n [0.135]\r\n [0.962]\r\n [0.68 ]\r\n [0.19 ]\r\n [0.238]\r\n [0.403]\r\n [0.228]\r\n [0.98 ]\r\n [0.419]\r\n [0.307]\r\n [0.726]\r\n [0.634]\r\n [0.732]\r\n [0.45 ]\r\n [0.757]\r\n [0.53 ]\r\n [0.869]\r\n [0.283]\r\n [0.143]\r\n [0.805]\r\n [0.232]\r\n [0.525]\r\n [0.55 ]]\r\n\r\n [[0.712]\r\n [0.512]\r\n [0.908]\r\n [0.655]\r\n [0.136]\r\n [0.133]\r\n [0.403]\r\n [0.518]\r\n [0.172]\r\n [0.984]\r\n [0.776]\r\n [0.06 ]\r\n [0.784]\r\n [0.46 ]\r\n [0.281]\r\n [0.698]\r\n [0.209]\r\n [0.862]\r\n [0.37 ]\r\n [0.98 ]\r\n [0.328]\r\n [0.195]\r\n [0.786]\r\n [0.222]\r\n [0.741]\r\n [0.524]\r\n [0.297]\r\n [0.021]]\r\n\r\n [[0.923]\r\n [0.54 ]\r\n [0.257]\r\n [0.1 ]\r\n [0.793]\r\n [0.992]\r\n [0.106]\r\n [0.435]\r\n [0.094]\r\n [0.53 ]\r\n [0.457]\r\n [0.15 ]\r\n [0.284]\r\n [0.019]\r\n [0.811]\r\n [0.835]\r\n [0.422]\r\n [0.112]\r\n [0.16 ]\r\n [0.314]\r\n [0.581]\r\n [0.238]\r\n [0.136]\r\n [0.118]\r\n [0.069]\r\n [0.826]\r\n [0.779]\r\n [0.35 ]]\r\n\r\n [[0.709]\r\n [0.162]\r\n [0.412]\r\n [0.893]\r\n [0.03 ]\r\n [0.818]\r\n [0.195]\r\n [0.523]\r\n [0.547]\r\n [0.168]\r\n [0.718]\r\n [0.294]\r\n [0.507]\r\n [0.405]\r\n [0.36 ]\r\n [0.483]\r\n [0.06 ]\r\n [0.9 ]\r\n [0.276]\r\n [0.877]\r\n [0.363]\r\n [0.197]\r\n [0.063]\r\n [0.076]\r\n [0.759]\r\n [0.427]\r\n [0.339]\r\n [0.942]]\r\n\r\n [[0.058]\r\n [0.93 ]\r\n [0.589]\r\n [0.873]\r\n [0.165]\r\n [0.784]\r\n [0.552]\r\n [0.648]\r\n [0.491]\r\n [0.291]\r\n [0.39 ]\r\n [0.966]\r\n [0.967]\r\n [0.652]\r\n [0.982]\r\n [0.474]\r\n [0.703]\r\n [0.869]\r\n [0.799]\r\n [0.102]\r\n [0.267]\r\n [0.106]\r\n [0.188]\r\n [0.568]\r\n [0.474]\r\n [0.69 ]\r\n [0.156]\r\n [0.855]]\r\n\r\n [[0.551]\r\n [0.414]\r\n [0.006]\r\n [0.677]\r\n [0.719]\r\n [0.958]\r\n [0.964]\r\n [0.422]\r\n [0.179]\r\n [0.798]\r\n [0.358]\r\n [0.966]\r\n [0.774]\r\n [0.519]\r\n [0.067]\r\n [0.717]\r\n [0.188]\r\n [0.587]\r\n [0.268]\r\n [0.804]\r\n [0.918]\r\n [0.818]\r\n [0.887]\r\n [0.377]\r\n [0.863]\r\n [0.882]\r\n [0.547]\r\n [0.455]]\r\n\r\n [[0.278]\r\n [0.23 ]\r\n [0.245]\r\n [0.029]\r\n [0.697]\r\n [0.623]\r\n [0.41 ]\r\n [0.385]\r\n [0.595]\r\n [0.043]\r\n [0.372]\r\n [0.076]\r\n [0.085]\r\n [0.565]\r\n [0.946]\r\n [0.461]\r\n [0.193]\r\n [0.138]\r\n [0.259]\r\n [0.042]\r\n [0.706]\r\n [0.087]\r\n [0.662]\r\n [0.76 ]\r\n [0.28 ]\r\n [0.827]\r\n [0.121]\r\n [0.903]]\r\n\r\n [[0.599]\r\n [0.265]\r\n [0.632]\r\n [0.924]\r\n [0.739]\r\n [0.816]\r\n [0.374]\r\n [0.822]\r\n [0.672]\r\n [0.349]\r\n [0.245]\r\n [0.071]\r\n [0.799]\r\n [0.73 ]\r\n [0.342]\r\n [0.196]\r\n [0.749]\r\n [0.987]\r\n [0.419]\r\n [0.667]\r\n [0.28 ]\r\n [0.526]\r\n [0.298]\r\n [0.241]\r\n [0.071]\r\n [0.544]\r\n [0.079]\r\n [0.126]]\r\n\r\n [[0.376]\r\n [0.599]\r\n [0.283]\r\n [0.94 ]\r\n [0.4 ]\r\n [0.318]\r\n [0.721]\r\n [0.594]\r\n [0.869]\r\n [0.025]\r\n [0.107]\r\n [0.626]\r\n [0.89 ]\r\n [0.792]\r\n [0.22 ]\r\n [0.33 ]\r\n [0.188]\r\n [0.651]\r\n [0.657]\r\n [0.991]\r\n [0.355]\r\n [0.854]\r\n [0.107]\r\n [0.802]\r\n [0.594]\r\n [0.125]\r\n [0.197]\r\n [0.995]]]]\r\n"
],
[
"!minikube delete",
"🔥 Deleting \"minikube\" from virtualbox ...\n💔 The \"minikube\" cluster has been deleted.\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc9886f9a9a75a6c9499b682cd44f1d281831ba
| 156,172 |
ipynb
|
Jupyter Notebook
|
tutorials/W1D5_DimensionalityReduction/student/W1D5_Tutorial3.ipynb
|
himahuja/course-content
|
f3e17aedf722c818708b83b213a267682a238194
|
[
"CC-BY-4.0"
] | 1 |
2020-08-04T10:11:55.000Z
|
2020-08-04T10:11:55.000Z
|
tutorials/W1D5_DimensionalityReduction/student/W1D5_Tutorial3.ipynb
|
modi471/course-content
|
a1a213918f46c87ccb22b798c5a6c1ecb2d86368
|
[
"CC-BY-4.0"
] | null | null | null |
tutorials/W1D5_DimensionalityReduction/student/W1D5_Tutorial3.ipynb
|
modi471/course-content
|
a1a213918f46c87ccb22b798c5a6c1ecb2d86368
|
[
"CC-BY-4.0"
] | 1 |
2021-03-29T21:08:26.000Z
|
2021-03-29T21:08:26.000Z
| 99.031072 | 44,900 | 0.826665 |
[
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W1D5_DimensionalityReduction/student/W1D5_Tutorial3.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Neuromatch Academy: Week 1, Day 5, Tutorial 3\n# Dimensionality Reduction and reconstruction\n\n__Content creators:__ Alex Cayco Gajic, John Murray\n\n__Content reviewers:__ Roozbeh Farhoudi, Matt Krause, Spiros Chavlis, Richard Gao, Michael Waskom",
"_____no_output_____"
],
[
"---\n# Tutorial Objectives\n\nIn this notebook we'll learn to apply PCA for dimensionality reduction, using a classic dataset that is often used to benchmark machine learning algorithms: MNIST. We'll also learn how to use PCA for reconstruction and denoising.\n\nOverview:\n- Perform PCA on MNIST\n- Calculate the variance explained\n- Reconstruct data with different numbers of PCs\n- (Bonus) Examine denoising using PCA\n\nYou can learn more about MNIST dataset [here](https://en.wikipedia.org/wiki/MNIST_database).",
"_____no_output_____"
]
],
[
[
"# @title Video 1: PCA for dimensionality reduction\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"oO0bbInoO_0\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=oO0bbInoO_0\n"
]
],
[
[
"---\n# Setup\nRun these cells to get the tutorial started.",
"_____no_output_____"
]
],
[
[
"# Imports\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"# @title Figure Settings\nimport ipywidgets as widgets # interactive display\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/nma.mplstyle\")",
"_____no_output_____"
],
[
"# @title Helper Functions\n\n\ndef plot_variance_explained(variance_explained):\n \"\"\"\n Plots eigenvalues.\n\n Args:\n variance_explained (numpy array of floats) : Vector of variance explained\n for each PC\n\n Returns:\n Nothing.\n\n \"\"\"\n\n plt.figure()\n plt.plot(np.arange(1, len(variance_explained) + 1), variance_explained,\n '--k')\n plt.xlabel('Number of components')\n plt.ylabel('Variance explained')\n plt.show()\n\n\ndef plot_MNIST_reconstruction(X, X_reconstructed):\n \"\"\"\n Plots 9 images in the MNIST dataset side-by-side with the reconstructed\n images.\n\n Args:\n X (numpy array of floats) : Data matrix each column\n corresponds to a different\n random variable\n X_reconstructed (numpy array of floats) : Data matrix each column\n corresponds to a different\n random variable\n\n Returns:\n Nothing.\n \"\"\"\n\n plt.figure()\n ax = plt.subplot(121)\n k = 0\n for k1 in range(3):\n for k2 in range(3):\n k = k + 1\n plt.imshow(np.reshape(X[k, :], (28, 28)),\n extent=[(k1 + 1) * 28, k1 * 28, (k2 + 1) * 28, k2 * 28],\n vmin=0, vmax=255)\n plt.xlim((3 * 28, 0))\n plt.ylim((3 * 28, 0))\n plt.tick_params(axis='both', which='both', bottom=False, top=False,\n labelbottom=False)\n ax.set_xticks([])\n ax.set_yticks([])\n plt.title('Data')\n plt.clim([0, 250])\n ax = plt.subplot(122)\n k = 0\n for k1 in range(3):\n for k2 in range(3):\n k = k + 1\n plt.imshow(np.reshape(np.real(X_reconstructed[k, :]), (28, 28)),\n extent=[(k1 + 1) * 28, k1 * 28, (k2 + 1) * 28, k2 * 28],\n vmin=0, vmax=255)\n plt.xlim((3 * 28, 0))\n plt.ylim((3 * 28, 0))\n plt.tick_params(axis='both', which='both', bottom=False, top=False,\n labelbottom=False)\n ax.set_xticks([])\n ax.set_yticks([])\n plt.clim([0, 250])\n plt.title('Reconstructed')\n plt.tight_layout()\n\n\ndef plot_MNIST_sample(X):\n \"\"\"\n Plots 9 images in the MNIST dataset.\n\n Args:\n X (numpy array of floats) : Data matrix each column corresponds to a\n different random variable\n\n Returns:\n Nothing.\n\n \"\"\"\n\n fig, ax = plt.subplots()\n k = 0\n for k1 in range(3):\n for k2 in range(3):\n k = k + 1\n plt.imshow(np.reshape(X[k, :], (28, 28)),\n extent=[(k1 + 1) * 28, k1 * 28, (k2+1) * 28, k2 * 28],\n vmin=0, vmax=255)\n plt.xlim((3 * 28, 0))\n plt.ylim((3 * 28, 0))\n plt.tick_params(axis='both', which='both', bottom=False, top=False,\n labelbottom=False)\n plt.clim([0, 250])\n ax.set_xticks([])\n ax.set_yticks([])\n plt.show()\n\n\ndef plot_MNIST_weights(weights):\n \"\"\"\n Visualize PCA basis vector weights for MNIST. Red = positive weights,\n blue = negative weights, white = zero weight.\n\n Args:\n weights (numpy array of floats) : PCA basis vector\n\n Returns:\n Nothing.\n \"\"\"\n\n fig, ax = plt.subplots()\n cmap = plt.cm.get_cmap('seismic')\n plt.imshow(np.real(np.reshape(weights, (28, 28))), cmap=cmap)\n plt.tick_params(axis='both', which='both', bottom=False, top=False,\n labelbottom=False)\n plt.clim(-.15, .15)\n plt.colorbar(ticks=[-.15, -.1, -.05, 0, .05, .1, .15])\n ax.set_xticks([])\n ax.set_yticks([])\n plt.show()\n\n\ndef add_noise(X, frac_noisy_pixels):\n \"\"\"\n Randomly corrupts a fraction of the pixels by setting them to random values.\n\n Args:\n X (numpy array of floats) : Data matrix\n frac_noisy_pixels (scalar) : Fraction of noisy pixels\n\n Returns:\n (numpy array of floats) : Data matrix + noise\n\n \"\"\"\n\n X_noisy = np.reshape(X, (X.shape[0] * X.shape[1]))\n N_noise_ixs = int(X_noisy.shape[0] * frac_noisy_pixels)\n noise_ixs = np.random.choice(X_noisy.shape[0], size=N_noise_ixs,\n replace=False)\n X_noisy[noise_ixs] = np.random.uniform(0, 255, noise_ixs.shape)\n X_noisy = np.reshape(X_noisy, (X.shape[0], X.shape[1]))\n\n return X_noisy\n\n\ndef change_of_basis(X, W):\n \"\"\"\n Projects data onto a new basis.\n\n Args:\n X (numpy array of floats) : Data matrix each column corresponding to a\n different random variable\n W (numpy array of floats) : new orthonormal basis columns correspond to\n basis vectors\n\n Returns:\n (numpy array of floats) : Data matrix expressed in new basis\n \"\"\"\n\n Y = np.matmul(X, W)\n\n return Y\n\n\ndef get_sample_cov_matrix(X):\n \"\"\"\n Returns the sample covariance matrix of data X.\n\n Args:\n X (numpy array of floats) : Data matrix each column corresponds to a\n different random variable\n\n Returns:\n (numpy array of floats) : Covariance matrix\n\"\"\"\n\n X = X - np.mean(X, 0)\n cov_matrix = 1 / X.shape[0] * np.matmul(X.T, X)\n return cov_matrix\n\n\ndef sort_evals_descending(evals, evectors):\n \"\"\"\n Sorts eigenvalues and eigenvectors in decreasing order. Also aligns first two\n eigenvectors to be in first two quadrants (if 2D).\n\n Args:\n evals (numpy array of floats) : Vector of eigenvalues\n evectors (numpy array of floats) : Corresponding matrix of eigenvectors\n each column corresponds to a different\n eigenvalue\n\n Returns:\n (numpy array of floats) : Vector of eigenvalues after sorting\n (numpy array of floats) : Matrix of eigenvectors after sorting\n \"\"\"\n\n index = np.flip(np.argsort(evals))\n evals = evals[index]\n evectors = evectors[:, index]\n if evals.shape[0] == 2:\n if np.arccos(np.matmul(evectors[:, 0],\n 1 / np.sqrt(2) * np.array([1, 1]))) > np.pi / 2:\n evectors[:, 0] = -evectors[:, 0]\n if np.arccos(np.matmul(evectors[:, 1],\n 1 / np.sqrt(2)*np.array([-1, 1]))) > np.pi / 2:\n evectors[:, 1] = -evectors[:, 1]\n\n return evals, evectors\n\n\ndef pca(X):\n \"\"\"\n Performs PCA on multivariate data. Eigenvalues are sorted in decreasing order\n\n Args:\n X (numpy array of floats) : Data matrix each column corresponds to a\n different random variable\n\n Returns:\n (numpy array of floats) : Data projected onto the new basis\n (numpy array of floats) : Vector of eigenvalues\n (numpy array of floats) : Corresponding matrix of eigenvectors\n\n \"\"\"\n\n X = X - np.mean(X, 0)\n cov_matrix = get_sample_cov_matrix(X)\n evals, evectors = np.linalg.eigh(cov_matrix)\n evals, evectors = sort_evals_descending(evals, evectors)\n score = change_of_basis(X, evectors)\n\n return score, evectors, evals\n\n\ndef plot_eigenvalues(evals, limit=True):\n \"\"\"\n Plots eigenvalues.\n\n Args:\n (numpy array of floats) : Vector of eigenvalues\n\n Returns:\n Nothing.\n\n \"\"\"\n\n plt.figure()\n plt.plot(np.arange(1, len(evals) + 1), evals, 'o-k')\n plt.xlabel('Component')\n plt.ylabel('Eigenvalue')\n plt.title('Scree plot')\n if limit:\n plt.show()",
"_____no_output_____"
]
],
[
[
"---\n# Section 1: Perform PCA on MNIST\n\nThe MNIST dataset consists of a 70,000 images of individual handwritten digits. Each image is a 28x28 pixel grayscale image. For convenience, each 28x28 pixel image is often unravelled into a single 784 (=28*28) element vector, so that the whole dataset is represented as a 70,000 x 784 matrix. Each row represents a different image, and each column represents a different pixel.\n \nEnter the following cell to load the MNIST dataset and plot the first nine images.",
"_____no_output_____"
]
],
[
[
"from sklearn.datasets import fetch_openml\nmnist = fetch_openml(name='mnist_784')\nX = mnist.data\nplot_MNIST_sample(X)",
"_____no_output_____"
]
],
[
[
"The MNIST dataset has an extrinsic dimensionality of 784, much higher than the 2-dimensional examples used in the previous tutorials! To make sense of this data, we'll use dimensionality reduction. But first, we need to determine the intrinsic dimensionality $K$ of the data. One way to do this is to look for an \"elbow\" in the scree plot, to determine which eigenvalues are signficant.",
"_____no_output_____"
],
[
"## Exercise 1: Scree plot of MNIST\n\nIn this exercise you will examine the scree plot in the MNIST dataset.\n\n**Steps:**\n- Perform PCA on the dataset and examine the scree plot. \n- When do the eigenvalues appear (by eye) to reach zero? (**Hint:** use `plt.xlim` to zoom into a section of the plot).\n",
"_____no_output_____"
]
],
[
[
"help(pca)\nhelp(plot_eigenvalues)",
"Help on function pca in module __main__:\n\npca(X)\n Performs PCA on multivariate data. Eigenvalues are sorted in decreasing order\n \n Args:\n X (numpy array of floats) : Data matrix each column corresponds to a\n different random variable\n \n Returns:\n (numpy array of floats) : Data projected onto the new basis\n (numpy array of floats) : Vector of eigenvalues\n (numpy array of floats) : Corresponding matrix of eigenvectors\n\nHelp on function plot_eigenvalues in module __main__:\n\nplot_eigenvalues(evals, limit=True)\n Plots eigenvalues.\n \n Args:\n (numpy array of floats) : Vector of eigenvalues\n \n Returns:\n Nothing.\n\n"
],
[
"#################################################\n## TO DO for students: perform PCA and plot the eigenvalues\n#################################################\n\n# perform PCA\n# score, evectors, evals = ...\n# plot the eigenvalues\n# plot_eigenvalues(evals, limit=False)\n# plt.xlim(...) # limit x-axis up to 100 for zooming",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D5_DimensionalityReduction/solutions/W1D5_Tutorial3_Solution_a876e927.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=558 height=414 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D5_DimensionalityReduction/static/W1D5_Tutorial3_Solution_a876e927_0.png>\n\n",
"_____no_output_____"
],
[
"---\n# Section 2: Calculate the variance explained\n\nThe scree plot suggests that most of the eigenvalues are near zero, with fewer than 100 having large values. Another common way to determine the intrinsic dimensionality is by considering the variance explained. This can be examined with a cumulative plot of the fraction of the total variance explained by the top $K$ components, i.e.,\n\n\\begin{equation}\n\\text{var explained} = \\frac{\\sum_{i=1}^K \\lambda_i}{\\sum_{i=1}^N \\lambda_i}\n\\end{equation}\n\nThe intrinsic dimensionality is often quantified by the $K$ necessary to explain a large proportion of the total variance of the data (often a defined threshold, e.g., 90%).",
"_____no_output_____"
],
[
"## Exercise 2: Plot the explained variance\n\nIn this exercise you will plot the explained variance.\n\n**Steps:**\n- Fill in the function below to calculate the fraction variance explained as a function of the number of principal componenets. **Hint:** use `np.cumsum`.\n- Plot the variance explained using `plot_variance_explained`.\n\n**Questions:**\n- How many principal components are required to explain 90% of the variance?\n- How does the intrinsic dimensionality of this dataset compare to its extrinsic dimensionality?\n",
"_____no_output_____"
]
],
[
[
"help(plot_variance_explained)",
"Help on function plot_variance_explained in module __main__:\n\nplot_variance_explained(variance_explained)\n Plots eigenvalues.\n \n Args:\n variance_explained (numpy array of floats) : Vector of variance explained\n for each PC\n \n Returns:\n Nothing.\n\n"
],
[
"def get_variance_explained(evals):\n \"\"\"\n Calculates variance explained from the eigenvalues.\n\n Args:\n evals (numpy array of floats) : Vector of eigenvalues\n\n Returns:\n (numpy array of floats) : Vector of variance explained\n\n \"\"\"\n\n #################################################\n ## TO DO for students: calculate the explained variance using the equation\n ## from Section 2.\n # Comment once you've filled in the function\n raise NotImplementedError(\"Student excercise: calculate explaine variance!\")\n #################################################\n\n # cumulatively sum the eigenvalues\n csum = ...\n # normalize by the sum of eigenvalues\n variance_explained = ...\n\n return variance_explained\n\n\n#################################################\n## TO DO for students: call the function and plot the variance explained\n#################################################\n\n# calculate the variance explained\nvariance_explained = ...\n\n# Uncomment to plot the variance explained\n# plot_variance_explained(variance_explained)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D5_DimensionalityReduction/solutions/W1D5_Tutorial3_Solution_0f5f51b9.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D5_DimensionalityReduction/static/W1D5_Tutorial3_Solution_0f5f51b9_0.png>\n\n",
"_____no_output_____"
],
[
"---\n# Section 3: Reconstruct data with different numbers of PCs\n",
"_____no_output_____"
]
],
[
[
"# @title Video 2: Data Reconstruction\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"ZCUhW26AdBQ\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"Video available at https://youtube.com/watch?v=ZCUhW26AdBQ\n"
]
],
[
[
"Now we have seen that the top 100 or so principal components of the data can explain most of the variance. We can use this fact to perform *dimensionality reduction*, i.e., by storing the data using only 100 components rather than the samples of all 784 pixels. Remarkably, we will be able to reconstruct much of the structure of the data using only the top 100 components. To see this, recall that to perform PCA we projected the data $\\bf X$ onto the eigenvectors of the covariance matrix:\n\\begin{equation}\n\\bf S = X W\n\\end{equation}\nSince $\\bf W$ is an orthogonal matrix, ${\\bf W}^{-1} = {\\bf W}^T$. So by multiplying by ${\\bf W}^T$ on each side we can rewrite this equation as \n\\begin{equation}\n{\\bf X = S W}^T.\n\\end{equation}\nThis now gives us a way to reconstruct the data matrix from the scores and loadings. To reconstruct the data from a low-dimensional approximation, we just have to truncate these matrices. Let's call ${\\bf S}_{1:K}$ and ${\\bf W}_{1:K}$ as keeping only the first $K$ columns of this matrix. Then our reconstruction is:\n\\begin{equation}\n{\\bf \\hat X = S}_{1:K} ({\\bf W}_{1:K})^T.\n\\end{equation}\n",
"_____no_output_____"
],
[
"## Exercise 3: Data reconstruction\n\nFill in the function below to reconstruct the data using different numbers of principal components. \n\n**Steps:**\n\n* Fill in the following function to reconstruct the data based on the weights and scores. Don't forget to add the mean!\n* Make sure your function works by reconstructing the data with all $K=784$ components. The two images should look identical.",
"_____no_output_____"
]
],
[
[
"help(plot_MNIST_reconstruction)",
"Help on function plot_MNIST_reconstruction in module __main__:\n\nplot_MNIST_reconstruction(X, X_reconstructed)\n Plots 9 images in the MNIST dataset side-by-side with the reconstructed\n images.\n \n Args:\n X (numpy array of floats) : Data matrix each column\n corresponds to a different\n random variable\n X_reconstructed (numpy array of floats) : Data matrix each column\n corresponds to a different\n random variable\n \n Returns:\n Nothing.\n\n"
],
[
"def reconstruct_data(score, evectors, X_mean, K):\n \"\"\"\n Reconstruct the data based on the top K components.\n\n Args:\n score (numpy array of floats) : Score matrix\n evectors (numpy array of floats) : Matrix of eigenvectors\n X_mean (numpy array of floats) : Vector corresponding to data mean\n K (scalar) : Number of components to include\n\n Returns:\n (numpy array of floats) : Matrix of reconstructed data\n\n \"\"\"\n\n #################################################\n ## TO DO for students: Reconstruct the original data in X_reconstructed\n # Comment once you've filled in the function\n raise NotImplementedError(\"Student excercise: reconstructing data function!\")\n #################################################\n\n # Reconstruct the data from the score and eigenvectors\n # Don't forget to add the mean!!\n X_reconstructed = ...\n\n return X_reconstructed\n\n\nK = 784\n\n#################################################\n## TO DO for students: Calculate the mean and call the function, then plot\n## the original and the recostructed data\n#################################################\n\n# Reconstruct the data based on all components\nX_mean = ...\nX_reconstructed = ...\n\n# Plot the data and reconstruction\n# plot_MNIST_reconstruction(X, X_reconstructed)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D5_DimensionalityReduction/solutions/W1D5_Tutorial3_Solution_e3395916.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=557 height=289 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D5_DimensionalityReduction/static/W1D5_Tutorial3_Solution_e3395916_0.png>\n\n",
"_____no_output_____"
],
[
"## Interactive Demo: Reconstruct the data matrix using different numbers of PCs\n\nNow run the code below and experiment with the slider to reconstruct the data matrix using different numbers of principal components.\n\n**Steps**\n* How many principal components are necessary to reconstruct the numbers (by eye)? How does this relate to the intrinsic dimensionality of the data?\n* Do you see any information in the data with only a single principal component?",
"_____no_output_____"
]
],
[
[
"# @title\n\n# @markdown Make sure you execute this cell to enable the widget!\n\n\ndef refresh(K=100):\n X_reconstructed = reconstruct_data(score, evectors, X_mean, K)\n plot_MNIST_reconstruction(X, X_reconstructed)\n plt.title('Reconstructed, K={}'.format(K))\n\n\n_ = widgets.interact(refresh, K=(1, 784, 10))",
"_____no_output_____"
]
],
[
[
"## Exercise 4: Visualization of the weights\n\nNext, let's take a closer look at the first principal component by visualizing its corresponding weights. \n\n**Steps:**\n\n* Enter `plot_MNIST_weights` to visualize the weights of the first basis vector.\n* What structure do you see? Which pixels have a strong positive weighting? Which have a strong negative weighting? What kinds of images would this basis vector differentiate?\n* Try visualizing the second and third basis vectors. Do you see any structure? What about the 100th basis vector? 500th? 700th?",
"_____no_output_____"
]
],
[
[
"help(plot_MNIST_weights)",
"Help on function plot_MNIST_weights in module __main__:\n\nplot_MNIST_weights(weights)\n Visualize PCA basis vector weights for MNIST. Red = positive weights,\n blue = negative weights, white = zero weight.\n \n Args:\n weights (numpy array of floats) : PCA basis vector\n \n Returns:\n Nothing.\n\n"
],
[
"#################################################\n## TO DO for students: plot the weights calling the plot_MNIST_weights function\n#################################################\n\n# Plot the weights of the first principal component\n# plot_MNIST_weights(...)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D5_DimensionalityReduction/solutions/W1D5_Tutorial3_Solution_f358e413.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=499 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D5_DimensionalityReduction/static/W1D5_Tutorial3_Solution_f358e413_0.png>\n\n",
"_____no_output_____"
],
[
"---\n# Summary\n* In this tutorial, we learned how to use PCA for dimensionality reduction by selecting the top principal components. This can be useful as the intrinsic dimensionality ($K$) is often less than the extrinsic dimensionality ($N$) in neural data. $K$ can be inferred by choosing the number of eigenvalues necessary to capture some fraction of the variance.\n* We also learned how to reconstruct an approximation of the original data using the top $K$ principal components. In fact, an alternate formulation of PCA is to find the $K$ dimensional space that minimizes the reconstruction error.\n* Noise tends to inflate the apparent intrinsic dimensionality, however the higher components reflect noise rather than new structure in the data. PCA can be used for denoising data by removing noisy higher components.\n* In MNIST, the weights corresponding to the first principal component appear to discriminate between a 0 and 1. We will discuss the implications of this for data visualization in the following tutorial.",
"_____no_output_____"
],
[
"---\n# Bonus: Examine denoising using PCA\n\nIn this lecture, we saw that PCA finds an optimal low-dimensional basis to minimize the reconstruction error. Because of this property, PCA can be useful for denoising corrupted samples of the data.",
"_____no_output_____"
],
[
"## Exercise 5: Add noise to the data\nIn this exercise you will add salt-and-pepper noise to the original data and see how that affects the eigenvalues. \n\n**Steps:**\n- Use the function `add_noise` to add noise to 20% of the pixels.\n- Then, perform PCA and plot the variance explained. How many principal components are required to explain 90% of the variance? How does this compare to the original data? \n",
"_____no_output_____"
]
],
[
[
"help(add_noise)",
"Help on function add_noise in module __main__:\n\nadd_noise(X, frac_noisy_pixels)\n Randomly corrupts a fraction of the pixels by setting them to random values.\n \n Args:\n X (numpy array of floats) : Data matrix\n frac_noisy_pixels (scalar) : Fraction of noisy pixels\n \n Returns:\n (numpy array of floats) : Data matrix + noise\n\n"
],
[
"###################################################################\n# Insert your code here to:\n# Add noise to the data\n# Plot noise-corrupted data\n# Perform PCA on the noisy data\n# Calculate and plot the variance explained\n###################################################################\nnp.random.seed(2020) # set random seed\nX_noisy = ...\n# score_noisy, evectors_noisy, evals_noisy = ...\n# variance_explained_noisy = ...\n# plot_MNIST_sample(X_noisy)\n# plot_variance_explained(variance_explained_noisy)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D5_DimensionalityReduction/solutions/W1D5_Tutorial3_Solution_d4a41b8c.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=424 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D5_DimensionalityReduction/static/W1D5_Tutorial3_Solution_d4a41b8c_0.png>\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D5_DimensionalityReduction/static/W1D5_Tutorial3_Solution_d4a41b8c_1.png>\n\n",
"_____no_output_____"
],
[
"## Exercise 6: Denoising\n\nNext, use PCA to perform denoising by projecting the noise-corrupted data onto the basis vectors found from the original dataset. By taking the top K components of this projection, we can reduce noise in dimensions orthogonal to the K-dimensional latent space. \n\n**Steps:**\n- Subtract the mean of the noise-corrupted data.\n- Project the data onto the basis found with the original dataset (`evectors`, not `evectors_noisy`) and take the top $K$ components. \n- Reconstruct the data as normal, using the top 50 components. \n- Play around with the amount of noise and K to build intuition.\n",
"_____no_output_____"
]
],
[
[
"###################################################################\n# Insert your code here to:\n# Subtract the mean of the noise-corrupted data\n# Project onto the original basis vectors evectors\n# Reconstruct the data using the top 50 components\n# Plot the result\n###################################################################\n\nX_noisy_mean = ...\nprojX_noisy = ...\nX_reconstructed = ...\n# plot_MNIST_reconstruction(X_noisy, X_reconstructed)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D5_DimensionalityReduction/solutions/W1D5_Tutorial3_Solution_e3ee8262.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=557 height=289 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D5_DimensionalityReduction/static/W1D5_Tutorial3_Solution_e3ee8262_0.png>\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbc98f93ffef560b19bd817d9e7ed47033d15beb
| 13,898 |
ipynb
|
Jupyter Notebook
|
docs/source/notebooks/tutorial/3-Projection-Join-Sort.ipynb
|
andrewseidl/ibis
|
1468b8c4f96d9d58f6fa147a2579b0d9e5796186
|
[
"Apache-2.0"
] | null | null | null |
docs/source/notebooks/tutorial/3-Projection-Join-Sort.ipynb
|
andrewseidl/ibis
|
1468b8c4f96d9d58f6fa147a2579b0d9e5796186
|
[
"Apache-2.0"
] | 6 |
2017-05-18T19:49:09.000Z
|
2019-03-27T15:37:14.000Z
|
docs/source/notebooks/tutorial/3-Projection-Join-Sort.ipynb
|
andrewseidl/ibis
|
1468b8c4f96d9d58f6fa147a2579b0d9e5796186
|
[
"Apache-2.0"
] | 1 |
2019-07-18T02:24:16.000Z
|
2019-07-18T02:24:16.000Z
| 26.986408 | 345 | 0.574039 |
[
[
[
"# Projection, Joining, and Sorting",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"import ibis\nimport os\nhdfs_port = os.environ.get('IBIS_WEBHDFS_PORT', 50070)\nhdfs = ibis.hdfs_connect(host='quickstart.cloudera', port=hdfs_port)\ncon = ibis.impala.connect(host='quickstart.cloudera', database='ibis_testing',\n hdfs_client=hdfs)\nprint('Hello!')",
"_____no_output_____"
]
],
[
[
"## Projections: adding/selecting columns\n\nProjections are the general way for adding new columns to tables, or selecting or removing existing ones.",
"_____no_output_____"
]
],
[
[
"table = con.table('functional_alltypes')\ntable.limit(5)",
"_____no_output_____"
]
],
[
[
"First, the basics: selecting columns:",
"_____no_output_____"
]
],
[
[
"proj = table['bool_col', 'int_col', 'double_col']\n\nproj.limit(5)",
"_____no_output_____"
]
],
[
[
"You can make a list of columns you want, too, and pass that:",
"_____no_output_____"
]
],
[
[
"to_select = ['bool_col', 'int_col']\ntable[to_select].limit(5)",
"_____no_output_____"
]
],
[
[
"You can also use the explicit `projection` or `select` functions",
"_____no_output_____"
]
],
[
[
"table.select(['int_col', 'double_col']).limit(5)",
"_____no_output_____"
]
],
[
[
"We can add new columns by using named column expressions",
"_____no_output_____"
]
],
[
[
"bigger_expr = (table.int_col * 2).name('bigger_ints')\nproj2 = table['int_col', bigger_expr]\nproj2.limit(5)",
"_____no_output_____"
]
],
[
[
"Adding columns is a shortcut for projection. In Ibis, adding columns always produces a new table reference",
"_____no_output_____"
]
],
[
[
"table2 = table.add_column(bigger_expr)\ntable2.limit(5)",
"_____no_output_____"
]
],
[
[
"In more complicated projections involving joins, we may need to refer to all of the columns in a table at once. This is how `add_column` works. We just pass the whole table in the projection:",
"_____no_output_____"
]
],
[
[
"table.select([table, bigger_expr]).limit(5)",
"_____no_output_____"
]
],
[
[
"\nTo use constants in projections, we have to use a special `ibis.literal` function",
"_____no_output_____"
]
],
[
[
"foo_constant = ibis.literal(5).name('foo')\ntable.select([table.bigint_col, foo_constant]).limit(5)",
"_____no_output_____"
]
],
[
[
"## Joins\n\nIbis attempts to provide good support for all the standard relational joins supported by Impala, Hive, and other relational databases.\n\n- inner, outer, left, right joins\n- semi and anti-joins\n\nTo illustrate the joins we'll use the TPC-H tables for now",
"_____no_output_____"
]
],
[
[
"region = con.table('tpch_region')\nnation = con.table('tpch_nation')\ncustomer = con.table('tpch_customer')\nlineitem = con.table('tpch_lineitem')",
"_____no_output_____"
]
],
[
[
"`region` and `nation` are connected by their respective `regionkey` columns",
"_____no_output_____"
]
],
[
[
"join_expr = region.r_regionkey == nation.n_regionkey\njoined = region.inner_join(nation, join_expr)",
"_____no_output_____"
]
],
[
[
"If you have multiple join conditions, either compose them yourself (like filters) or pass a list to the join function\n\n join_exprs = [cond1, cond2, cond3]\n joined = table1.inner_join(table2, join_exprs)",
"_____no_output_____"
],
[
"Once you've joined tables, you don't necessarily have anything yet. I'll put it in big letters\n\n### Joins are declarations of intent\n\nAfter calling the join function (which validates the join condition, of course), you may perform any number of other operations:\n\n- Aggregation\n- Projection\n- Filtering\n\nand so forth. Most importantly, depending on your schemas, the joined tables may include overlapping column names that could create a conflict if not addressed directly. Some other systems, like pandas, handle this by applying suffixes to the overlapping column names and computing the fully joined tables immediately. We don't do this.\n\nSo, with the above data, suppose we just want the region name and all the nation table data. We can then make a projection on the joined reference:",
"_____no_output_____"
]
],
[
[
"table_ref = joined[nation, region.r_name.name('region')]\ntable_ref.columns",
"_____no_output_____"
],
[
"table_ref.limit(5)",
"_____no_output_____"
],
[
"agged = table_ref.aggregate([table_ref.n_name.count().name('nrows')], by=['region'])\nagged",
"_____no_output_____"
]
],
[
[
"Things like `group_by` work with unmaterialized joins, too, as you would hope. ",
"_____no_output_____"
]
],
[
[
"joined.group_by(region.r_name).size()",
"_____no_output_____"
]
],
[
[
"### Explicit join materialization\n\nIf you're lucky enough to have two table schemas with no overlapping column names (lucky you!), the join can be *materialized* without having to perform some other relational algebra operation:\n\n joined = a.inner_join(b, join_expr).materialize()\n \nNote that this is equivalent to doing\n\n joined = a.join(b, join_expr)[a, b]\n \ni.e., joining and then selecting all columns from both joined tables. If there is a name overlap, just like with the equivalent projection, there will be an immediate error.",
"_____no_output_____"
],
[
"### Writing down join keys\n\nIn addition to having explicit comparison expressions as join keys, you can also write down column names, or use expressions referencing the joined tables, e.g.:\n\n joined = a.join(b, [('a_key1', 'b_key2')])\n \n joined2 = a.join(b, [(left_expr, right_expr)])\n\n joined3 = a.join(b, ['common_key'])\n\nThese will be compared for equality when performing the join; if you want non-equality conditions in the join, you will have to form those yourself.",
"_____no_output_____"
],
[
"### Join referential nuances\n\nThere's nothing to stop you from doing many joins in succession, and, in fact, with complex schemas it will be to your advantage to build the joined table references for your analysis first, then reuse the objects as you go:\n\n joined_ref = (a.join(b, a.key1 == b.key2)\n .join(c, [a.key3 == c.key4, b.key5 == c.key6]))\n\nNote that, at least right now, you need to provide explicit comparison expressions (or tuples of column references) referencing the joined tables.",
"_____no_output_____"
],
[
"### Aggregating joined table with metrics involving more than one base reference\n\nLet's consider the case similar to the SQL query\n\n SELECT a.key, sum(a.foo - b.bar) AS metric\n FROM a\n JOIN b\n ON a.key = b.key\n GROUP BY 1\n \nI'll use a somewhat contrived example using the data we already have to show you what this looks like. Take the `functional.alltypes` table, and suppose we want to compute the **mean absolute deviation (MAD) from the hourly mean of the double_col**. Silly, I know, but bear with me.\n\nFirst, the hourly mean:",
"_____no_output_____"
]
],
[
[
"table = con.table('functional_alltypes')\n\nhour_dim = table.timestamp_col.hour().name('hour')\n\nhourly_mean = (table.group_by(hour_dim)\n .aggregate([table.double_col.mean().name('avg_double')]))\nhourly_mean",
"_____no_output_____"
]
],
[
[
"Okay, great, now how about the MAD? The only trick here is that we can form an aggregate metric from the two tables, and we then have to join it later. Ibis **will not** figure out how to join the tables automatically for us. ",
"_____no_output_____"
]
],
[
[
"mad = (table.double_col - hourly_mean.avg_double).abs().mean().name('MAD')",
"_____no_output_____"
]
],
[
[
"This metric is only valid if used in the context of `table` joined with `hourly_mean`, so let's do that. Writing down the join condition is simply a matter of writing:",
"_____no_output_____"
]
],
[
[
"join_expr = hour_dim == hourly_mean.hour",
"_____no_output_____"
]
],
[
[
"Now let's compute the MAD grouped by `string_col`",
"_____no_output_____"
]
],
[
[
"result = (table.inner_join(hourly_mean, join_expr)\n .group_by(table.string_col)\n .aggregate([mad]))\nresult",
"_____no_output_____"
]
],
[
[
"## Sorting\n\nSorting tables works similarly to the SQL `ORDER BY` clause. We use the `sort_by` function and pass one of the following:\n\n- Column names\n- Column expressions\n- One of these, with a False (descending order) or True (ascending order) qualifier\n\nSo, to sort by `total` in ascending order we write:\n\n table.sort_by('total')\n\nor by `key` then by `total` in descending order\n\n table.sort_by(['key', ('total', False)])\n \nFor descending sort order, there is a convenience function `desc` which can wrap sort keys\n\n from ibis import desc\n table.sort_by(['key', desc(table.total)])",
"_____no_output_____"
],
[
"Here's a concrete example involving filters, custom grouping dimension, and sorting",
"_____no_output_____"
]
],
[
[
"table = con.table('functional_alltypes')\n\nkeys = ['string_col', (table.bigint_col > 40).ifelse('high', 'low').name('bigint_tier')]\nmetrics = [table.double_col.sum().name('total')]\n\nagged = (table\n .filter(table.int_col < 8)\n .group_by(keys)\n .aggregate(metrics))\n\nsorted_agged = agged.sort_by(['bigint_tier', ('total', False)])\nsorted_agged",
"_____no_output_____"
]
],
[
[
"For sorting in descending order, you can use the special `ibis.desc` function:",
"_____no_output_____"
]
],
[
[
"agged.sort_by(ibis.desc('total'))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc999ca918b5b55b154ca088701ee1d81b812a7
| 20,223 |
ipynb
|
Jupyter Notebook
|
Tutorials/Boston Housing - XGBoost (Hyperparameter Tuning) - High Level.ipynb
|
Zomma2/AWS-deployment-notebooks-
|
b7be9909be16d99a9954e72aa7951eb951068956
|
[
"MIT"
] | null | null | null |
Tutorials/Boston Housing - XGBoost (Hyperparameter Tuning) - High Level.ipynb
|
Zomma2/AWS-deployment-notebooks-
|
b7be9909be16d99a9954e72aa7951eb951068956
|
[
"MIT"
] | null | null | null |
Tutorials/Boston Housing - XGBoost (Hyperparameter Tuning) - High Level.ipynb
|
Zomma2/AWS-deployment-notebooks-
|
b7be9909be16d99a9954e72aa7951eb951068956
|
[
"MIT"
] | null | null | null | 41.020284 | 551 | 0.623795 |
[
[
[
"# Predicting Boston Housing Prices\n\n## Using XGBoost in SageMaker (Hyperparameter Tuning)\n\n_Deep Learning Nanodegree Program | Deployment_\n\n---\n\nAs an introduction to using SageMaker's High Level Python API for hyperparameter tuning, we will look again at the [Boston Housing Dataset](https://www.cs.toronto.edu/~delve/data/boston/bostonDetail.html) to predict the median value of a home in the area of Boston Mass.\n\nThe documentation for the high level API can be found on the [ReadTheDocs page](http://sagemaker.readthedocs.io/en/latest/)\n\n## General Outline\n\nTypically, when using a notebook instance with SageMaker, you will proceed through the following steps. Of course, not every step will need to be done with each project. Also, there is quite a lot of room for variation in many of the steps, as you will see throughout these lessons.\n\n1. Download or otherwise retrieve the data.\n2. Process / Prepare the data.\n3. Upload the processed data to S3.\n4. Train a chosen model.\n5. Test the trained model (typically using a batch transform job).\n6. Deploy the trained model.\n7. Use the deployed model.\n\nIn this notebook we will only be covering steps 1 through 5 as we are only interested in creating a tuned model and testing its performance.",
"_____no_output_____"
],
[
"## Step 0: Setting up the notebook\n\nWe begin by setting up all of the necessary bits required to run our notebook. To start that means loading all of the Python modules we will need.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport os\n\nimport numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\n\nfrom sklearn.datasets import load_boston\nimport sklearn.model_selection",
"_____no_output_____"
]
],
[
[
"In addition to the modules above, we need to import the various bits of SageMaker that we will be using. ",
"_____no_output_____"
]
],
[
[
"import sagemaker\nfrom sagemaker import get_execution_role\nfrom sagemaker.amazon.amazon_estimator import get_image_uri\nfrom sagemaker.predictor import csv_serializer\n\n# This is an object that represents the SageMaker session that we are currently operating in. This\n# object contains some useful information that we will need to access later such as our region.\nsession = sagemaker.Session()\n\n# This is an object that represents the IAM role that we are currently assigned. When we construct\n# and launch the training job later we will need to tell it what IAM role it should have. Since our\n# use case is relatively simple we will simply assign the training job the role we currently have.\nrole = get_execution_role()",
"_____no_output_____"
]
],
[
[
"## Step 1: Downloading the data\n\nFortunately, this dataset can be retrieved using sklearn and so this step is relatively straightforward.",
"_____no_output_____"
]
],
[
[
"boston = load_boston()",
"_____no_output_____"
]
],
[
[
"## Step 2: Preparing and splitting the data\n\nGiven that this is clean tabular data, we don't need to do any processing. However, we do need to split the rows in the dataset up into train, test and validation sets.",
"_____no_output_____"
]
],
[
[
"# First we package up the input data and the target variable (the median value) as pandas dataframes. This\n# will make saving the data to a file a little easier later on.\n\nX_bos_pd = pd.DataFrame(boston.data, columns=boston.feature_names)\nY_bos_pd = pd.DataFrame(boston.target)\n\n# We split the dataset into 2/3 training and 1/3 testing sets.\nX_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X_bos_pd, Y_bos_pd, test_size=0.33)\n\n# Then we split the training set further into 2/3 training and 1/3 validation sets.\nX_train, X_val, Y_train, Y_val = sklearn.model_selection.train_test_split(X_train, Y_train, test_size=0.33)",
"_____no_output_____"
]
],
[
[
"## Step 3: Uploading the data files to S3\n\nWhen a training job is constructed using SageMaker, a container is executed which performs the training operation. This container is given access to data that is stored in S3. This means that we need to upload the data we want to use for training to S3. In addition, when we perform a batch transform job, SageMaker expects the input data to be stored on S3. We can use the SageMaker API to do this and hide some of the details.\n\n### Save the data locally\n\nFirst we need to create the test, train and validation csv files which we will then upload to S3.",
"_____no_output_____"
]
],
[
[
"# This is our local data directory. We need to make sure that it exists.\ndata_dir = '../data/boston'\nif not os.path.exists(data_dir):\n os.makedirs(data_dir)",
"_____no_output_____"
],
[
"# We use pandas to save our test, train and validation data to csv files. Note that we make sure not to include header\n# information or an index as this is required by the built in algorithms provided by Amazon. Also, for the train and\n# validation data, it is assumed that the first entry in each row is the target variable.\n\nX_test.to_csv(os.path.join(data_dir, 'test.csv'), header=False, index=False)\n\npd.concat([Y_val, X_val], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)\npd.concat([Y_train, X_train], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)",
"_____no_output_____"
]
],
[
[
"### Upload to S3\n\nSince we are currently running inside of a SageMaker session, we can use the object which represents this session to upload our data to the 'default' S3 bucket. Note that it is good practice to provide a custom prefix (essentially an S3 folder) to make sure that you don't accidentally interfere with data uploaded from some other notebook or project.",
"_____no_output_____"
]
],
[
[
"prefix = 'boston-xgboost-tuning-HL'\n\ntest_location = session.upload_data(os.path.join(data_dir, 'test.csv'), key_prefix=prefix)\nval_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)\ntrain_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)",
"_____no_output_____"
]
],
[
[
"## Step 4: Train the XGBoost model\n\nNow that we have the training and validation data uploaded to S3, we can construct our XGBoost model and train it. Unlike in the previous notebooks, instead of training a single model, we will use SageMaker's hyperparameter tuning functionality to train multiple models and use the one that performs the best on the validation set.\n\nTo begin with, as in the previous approaches, we will need to construct an estimator object.",
"_____no_output_____"
]
],
[
[
"# As stated above, we use this utility method to construct the image name for the training container.\ncontainer = get_image_uri(session.boto_region_name, 'xgboost')\n\n# Now that we know which container to use, we can construct the estimator object.\nxgb = sagemaker.estimator.Estimator(container, # The name of the training container\n role, # The IAM role to use (our current role in this case)\n train_instance_count=1, # The number of instances to use for training\n train_instance_type='ml.m4.xlarge', # The type of instance ot use for training\n output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),\n # Where to save the output (the model artifacts)\n sagemaker_session=session) # The current SageMaker session",
"_____no_output_____"
]
],
[
[
"Before beginning the hyperparameter tuning, we should make sure to set any model specific hyperparameters that we wish to have default values. There are quite a few that can be set when using the XGBoost algorithm, below are just a few of them. If you would like to change the hyperparameters below or modify additional ones you can find additional information on the [XGBoost hyperparameter page](https://docs.aws.amazon.com/sagemaker/latest/dg/xgboost_hyperparameters.html)",
"_____no_output_____"
]
],
[
[
"xgb.set_hyperparameters(max_depth=5,\n eta=0.2,\n gamma=4,\n min_child_weight=6,\n subsample=0.8,\n objective='reg:linear',\n early_stopping_rounds=10,\n num_round=200)",
"_____no_output_____"
]
],
[
[
"Now that we have our estimator object completely set up, it is time to create the hyperparameter tuner. To do this we need to construct a new object which contains each of the parameters we want SageMaker to tune. In this case, we wish to find the best values for the `max_depth`, `eta`, `min_child_weight`, `subsample`, and `gamma` parameters. Note that for each parameter that we want SageMaker to tune we need to specify both the *type* of the parameter and the *range* of values that parameter may take on.\n\nIn addition, we specify the *number* of models to construct (`max_jobs`) and the number of those that can be trained in parallel (`max_parallel_jobs`). In the cell below we have chosen to train `20` models, of which we ask that SageMaker train `3` at a time in parallel. Note that this results in a total of `20` training jobs being executed which can take some time, in this case almost a half hour. With more complicated models this can take even longer so be aware!",
"_____no_output_____"
]
],
[
[
"from sagemaker.tuner import IntegerParameter, ContinuousParameter, HyperparameterTuner\n\nxgb_hyperparameter_tuner = HyperparameterTuner(estimator = xgb, # The estimator object to use as the basis for the training jobs.\n objective_metric_name = 'validation:rmse', # The metric used to compare trained models.\n objective_type = 'Minimize', # Whether we wish to minimize or maximize the metric.\n max_jobs = 20, # The total number of models to train\n max_parallel_jobs = 3, # The number of models to train in parallel\n hyperparameter_ranges = {\n 'max_depth': IntegerParameter(3, 12),\n 'eta' : ContinuousParameter(0.05, 0.5),\n 'min_child_weight': IntegerParameter(2, 8),\n 'subsample': ContinuousParameter(0.5, 0.9),\n 'gamma': ContinuousParameter(0, 10),\n })",
"_____no_output_____"
]
],
[
[
"Now that we have our hyperparameter tuner object completely set up, it is time to train it. To do this we make sure that SageMaker knows our input data is in csv format and then execute the `fit` method.",
"_____no_output_____"
]
],
[
[
"# This is a wrapper around the location of our train and validation data, to make sure that SageMaker\n# knows our data is in csv format.\ns3_input_train = sagemaker.s3_input(s3_data=train_location, content_type='csv')\ns3_input_validation = sagemaker.s3_input(s3_data=val_location, content_type='csv')\n\nxgb_hyperparameter_tuner.fit({'train': s3_input_train, 'validation': s3_input_validation})",
"_____no_output_____"
]
],
[
[
"As in many of the examples we have seen so far, the `fit()` method takes care of setting up and fitting a number of different models, each with different hyperparameters. If we wish to wait for this process to finish, we can call the `wait()` method.",
"_____no_output_____"
]
],
[
[
"xgb_hyperparameter_tuner.wait()",
"_____no_output_____"
]
],
[
[
"Once the hyperamater tuner has finished, we can retrieve information about the best performing model. ",
"_____no_output_____"
]
],
[
[
"xgb_hyperparameter_tuner.best_training_job()",
"_____no_output_____"
]
],
[
[
"In addition, since we'd like to set up a batch transform job to test the best model, we can construct a new estimator object from the results of the best training job. The `xgb_attached` object below can now be used as though we constructed an estimator with the best performing hyperparameters and then fit it to our training data.",
"_____no_output_____"
]
],
[
[
"xgb_attached = sagemaker.estimator.Estimator.attach(xgb_hyperparameter_tuner.best_training_job())",
"_____no_output_____"
]
],
[
[
"## Step 5: Test the model\n\nNow that we have our best performing model, we can test it. To do this we will use the batch transform functionality. To start with, we need to build a transformer object from our fit model.",
"_____no_output_____"
]
],
[
[
"xgb_transformer = xgb_attached.transformer(instance_count = 1, instance_type = 'ml.m4.xlarge')",
"_____no_output_____"
]
],
[
[
"Next we ask SageMaker to begin a batch transform job using our trained model and applying it to the test data we previous stored in S3. We need to make sure to provide SageMaker with the type of data that we are providing to our model, in our case `text/csv`, so that it knows how to serialize our data. In addition, we need to make sure to let SageMaker know how to split our data up into chunks if the entire data set happens to be too large to send to our model all at once.\n\nNote that when we ask SageMaker to do this it will execute the batch transform job in the background. Since we need to wait for the results of this job before we can continue, we use the `wait()` method. An added benefit of this is that we get some output from our batch transform job which lets us know if anything went wrong.",
"_____no_output_____"
]
],
[
[
"xgb_transformer.transform(test_location, content_type='text/csv', split_type='Line')",
"_____no_output_____"
],
[
"xgb_transformer.wait()",
"_____no_output_____"
]
],
[
[
"Now that the batch transform job has finished, the resulting output is stored on S3. Since we wish to analyze the output inside of our notebook we can use a bit of notebook magic to copy the output file from its S3 location and save it locally.",
"_____no_output_____"
]
],
[
[
"!aws s3 cp --recursive $xgb_transformer.output_path $data_dir",
"_____no_output_____"
]
],
[
[
"To see how well our model works we can create a simple scatter plot between the predicted and actual values. If the model was completely accurate the resulting scatter plot would look like the line $x=y$. As we can see, our model seems to have done okay but there is room for improvement.",
"_____no_output_____"
]
],
[
[
"Y_pred = pd.read_csv(os.path.join(data_dir, 'test.csv.out'), header=None)",
"_____no_output_____"
],
[
"plt.scatter(Y_test, Y_pred)\nplt.xlabel(\"Median Price\")\nplt.ylabel(\"Predicted Price\")\nplt.title(\"Median Price vs Predicted Price\")",
"_____no_output_____"
]
],
[
[
"## Optional: Clean up\n\nThe default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.",
"_____no_output_____"
]
],
[
[
"# First we will remove all of the files contained in the data_dir directory\n!rm $data_dir/*\n\n# And then we delete the directory itself\n!rmdir $data_dir",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc99d9b45ef93d505411d00ced62290571e3b64
| 4,957 |
ipynb
|
Jupyter Notebook
|
component-library/input/input-url.ipynb
|
Dani7B/claimed
|
037bfdeed98d6cb3e86db2d9160e21b9fa20ce44
|
[
"Apache-2.0"
] | null | null | null |
component-library/input/input-url.ipynb
|
Dani7B/claimed
|
037bfdeed98d6cb3e86db2d9160e21b9fa20ce44
|
[
"Apache-2.0"
] | null | null | null |
component-library/input/input-url.ipynb
|
Dani7B/claimed
|
037bfdeed98d6cb3e86db2d9160e21b9fa20ce44
|
[
"Apache-2.0"
] | null | null | null | 23.492891 | 109 | 0.526125 |
[
[
[
"# Input URL",
"_____no_output_____"
],
[
"This component reads a file from a HTTP(s) source via wget",
"_____no_output_____"
]
],
[
[
"!pip install wget==3.2",
"_____no_output_____"
],
[
"import logging\nimport os\nimport re\nimport sys\nimport wget",
"_____no_output_____"
],
[
"# path and file name for output\noutput_data_csv = os.environ.get('output_data_csv', 'data.csv')\n\n# url of souce\nurl = os.environ.get('url')\n\n# temporal data storage for local execution\ndata_dir = os.environ.get('data_dir', '../../data/')",
"_____no_output_____"
],
[
"parameters = list(\n map(lambda s: re.sub('$', '\"', s),\n map(\n lambda s: s.replace('=', '=\"'),\n filter(\n lambda s: s.find('=') > -1 and bool(re.match(r'[A-Za-z0-9_]*=[.\\/A-Za-z0-9]*', s)),\n sys.argv\n )\n )))\n\nfor parameter in parameters:\n logging.warning('Parameter: ' + parameter)\n exec(parameter)",
"_____no_output_____"
],
[
"destination = os.path.join(data_dir, output_data_csv)\nos.remove(destination) if os.path.exists(destination) else None\nwget.download(url, out=destination)",
"_____no_output_____"
],
[
"print('Data written successfully')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc9a7e2a062dd2dc2b982116fe7963956e9078a
| 393,866 |
ipynb
|
Jupyter Notebook
|
M1 Python For Data Science/Week_3_Python_For_Data_Science_Data_Visulization_With_Python/PDS_UberDriveProject_Question.ipynb
|
fborrasumh/greatlearning-pgp-dsba
|
2aff5e00f8d6a60e1d819b970901492af703de85
|
[
"MIT"
] | 1 |
2021-12-04T12:11:50.000Z
|
2021-12-04T12:11:50.000Z
|
M1 Python For Data Science/Week_3_Python_For_Data_Science_Data_Visulization_With_Python/PDS_UberDriveProject_Question.ipynb
|
fborrasumh/greatlearning-pgp-dsba
|
2aff5e00f8d6a60e1d819b970901492af703de85
|
[
"MIT"
] | null | null | null |
M1 Python For Data Science/Week_3_Python_For_Data_Science_Data_Visulization_With_Python/PDS_UberDriveProject_Question.ipynb
|
fborrasumh/greatlearning-pgp-dsba
|
2aff5e00f8d6a60e1d819b970901492af703de85
|
[
"MIT"
] | 1 |
2022-03-20T07:01:46.000Z
|
2022-03-20T07:01:46.000Z
| 411.133612 | 303,608 | 0.633078 |
[
[
[
"**Instructions:** \n1. **For all questions after 10th, Please only use the data specified in the note given just below the question**\n2. **You need to add answers in the same file i.e. PDS_UberDriveProject_Questions.ipynb' and rename that file as 'Name_Date.ipynb'.You can mention the date on which you will be uploading/submitting the file.For e.g. if you plan to submit your assignment on 31-March, you can rename the file as 'STUDENTNAME_31-Mar-2020'**",
"_____no_output_____"
],
[
"# Load the necessary libraries. Import and load the dataset with a name uber_drives .",
"_____no_output_____"
]
],
[
[
"import pandas as pd \nimport numpy as np",
"_____no_output_____"
],
[
"# Get the Data\ndata_uber_driver = pd.read_csv('uberdrive-1.csv')",
"_____no_output_____"
]
],
[
[
"## Q1. Show the last 10 records of the dataset. (2 point)",
"_____no_output_____"
]
],
[
[
"data_uber_driver.tail(10)",
"_____no_output_____"
]
],
[
[
"## Q2. Show the first 10 records of the dataset. (2 points)",
"_____no_output_____"
]
],
[
[
"data_uber_driver.head(10)",
"_____no_output_____"
]
],
[
[
"## Q3. Show the dimension(number of rows and columns) of the dataset. (2 points)",
"_____no_output_____"
]
],
[
[
"data_uber_driver.shape",
"_____no_output_____"
]
],
[
[
"## Q4. Show the size (Total number of elements) of the dataset. (2 points)",
"_____no_output_____"
]
],
[
[
"data_uber_driver.size",
"_____no_output_____"
]
],
[
[
"## Q5. Print the information about all the variables of the data set. (2 points)",
"_____no_output_____"
]
],
[
[
"data_uber_driver.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1155 entries, 0 to 1154\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 START_DATE* 1155 non-null object \n 1 END_DATE* 1155 non-null object \n 2 CATEGORY* 1155 non-null object \n 3 START* 1155 non-null object \n 4 STOP* 1155 non-null object \n 5 MILES* 1155 non-null float64\n 6 PURPOSE* 653 non-null object \ndtypes: float64(1), object(6)\nmemory usage: 63.3+ KB\n"
]
],
[
[
"## Q6. Check for missing values. (2 points) - Note: Output should be boolean only.",
"_____no_output_____"
]
],
[
[
"data_uber_driver.isna()",
"_____no_output_____"
]
],
[
[
"## Q7. How many missing values are present? (2 points)",
"_____no_output_____"
]
],
[
[
"data_uber_driver.isna().sum().sum()",
"_____no_output_____"
]
],
[
[
"## Q8. Get the summary of the original data. (2 points). Hint:Outcome will contain only numerical column.",
"_____no_output_____"
]
],
[
[
"data_uber_driver.describe()",
"_____no_output_____"
]
],
[
[
"\n\n## Q9. Drop the missing values and store the data in a new dataframe (name it\"df\") (2-points)\n\n### Note: Dataframe \"df\" will not contain any missing value",
"_____no_output_____"
]
],
[
[
"df = data_uber_driver.dropna()",
"_____no_output_____"
]
],
[
[
"## Q10. Check the information of the dataframe(df). (2 points)",
"_____no_output_____"
]
],
[
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 653 entries, 0 to 1154\nData columns (total 7 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 START_DATE* 653 non-null object \n 1 END_DATE* 653 non-null object \n 2 CATEGORY* 653 non-null object \n 3 START* 653 non-null object \n 4 STOP* 653 non-null object \n 5 MILES* 653 non-null float64\n 6 PURPOSE* 653 non-null object \ndtypes: float64(1), object(6)\nmemory usage: 40.8+ KB\n"
]
],
[
[
"## Q11. Get the unique start destinations. (2 points)\n### Note: This question is based on the dataframe with no 'NA' values\n### Hint- You need to print the unique destination place names in this and not the count.",
"_____no_output_____"
]
],
[
[
"df['START*'].unique()",
"_____no_output_____"
]
],
[
[
"## Q12. What is the total number of unique start destinations? (2 points)\n### Note: Use the original dataframe without dropping 'NA' values",
"_____no_output_____"
]
],
[
[
"data_uber_driver['START*'].nunique()",
"_____no_output_____"
]
],
[
[
"## Q13. Print the total number of unique stop destinations. (2 points)\n### Note: Use the original dataframe without dropping 'NA' values.",
"_____no_output_____"
]
],
[
[
"data_uber_driver['STOP*'].unique().size",
"_____no_output_____"
]
],
[
[
"## Q14. Print all the Uber trips that has the starting point of San Francisco. (2 points)\n### Note: Use the original dataframe without dropping the 'NA' values.\n\n### Hint: Use the loc function",
"_____no_output_____"
]
],
[
[
"data_uber_driver[data_uber_driver['START*']=='San Francisco']",
"_____no_output_____"
]
],
[
[
"## Q15. What is the most popular starting point for the Uber drivers? (2 points)\n### Note: Use the original dataframe without dropping the 'NA' values.\n\n### Hint:Popular means the place that is visited the most",
"_____no_output_____"
]
],
[
[
"data_uber_driver['START*'].value_counts().idxmax()",
"_____no_output_____"
]
],
[
[
"## Q16. What is the most popular dropping point for the Uber drivers? (2 points)\n### Note: Use the original dataframe without dropping the 'NA' values.\n\n### Hint: Popular means the place that is visited the most",
"_____no_output_____"
]
],
[
[
"data_uber_driver['STOP*'].value_counts().idxmax()",
"_____no_output_____"
]
],
[
[
"## Q17. List the most frequent route taken by Uber drivers. (3 points)\n### Note: This question is based on the new dataframe with no 'na' values.\n### Hint-Print the most frequent route taken by Uber drivers (Route= combination of START & END points present in the Data set).",
"_____no_output_____"
]
],
[
[
"df.groupby(['START*', 'STOP*']).size().sort_values(ascending=False)",
"_____no_output_____"
]
],
[
[
"## Q18. Print all types of purposes for the trip in an array. (3 points)\n### Note: This question is based on the new dataframe with no 'NA' values.",
"_____no_output_____"
]
],
[
[
"df['PURPOSE*']",
"_____no_output_____"
]
],
[
[
"## Q19. Plot a bar graph of Purpose vs Miles(Distance). (3 points)\n### Note: Use the original dataframe without dropping the 'NA' values.\n### Hint:You have to plot total/sum miles per purpose",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfig = plt.figure(figsize=(19,5))\nax = fig.add_axes([0,0,1,1])\n# replacing na 'PURPOSE' values \ndata_uber_driver[\"PURPOSE*\"].fillna(\"NO_PURPOSE_PROVIDED\", inplace = True) \n# replacing na 'MILES' values \ndata_uber_driver[\"MILES*\"].fillna(\"NO_MILES_PROVIDED\", inplace = True) \nax.bar(data_uber_driver['PURPOSE*'],data_uber_driver['MILES*'])\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Q20. Print a dataframe of Purposes and the distance travelled for that particular Purpose. (3 points)\n### Note: Use the original dataframe without dropping \"NA\" values",
"_____no_output_____"
]
],
[
[
"data_uber_driver.groupby(by=[\"PURPOSE*\"]).sum()",
"_____no_output_____"
]
],
[
[
"## Q21. Plot number of trips vs Category of trips. (3 points)\n### Note: Use the original dataframe without dropping the 'NA' values.\n### Hint : You can make a countplot or barplot.",
"_____no_output_____"
]
],
[
[
"# import seaborn as sns\n# sns.countplot(x='CATEGORY*',data=data_uber_driver)\ndata_uber_driver['CATEGORY*'].value_counts().plot(kind='bar',figsize=(19,7),color='red');",
"_____no_output_____"
]
],
[
[
"## Q22. What is proportion of trips that is Business and what is the proportion of trips that is Personal? (3 points)\n\n### Note:Use the original dataframe without dropping the 'NA' values. The proportion calculation is with respect to the 'miles' variable.\n### Hint:Out of the category of trips, you need to find percentage wise how many are business and how many are personal on the basis of miles per category.",
"_____no_output_____"
]
],
[
[
"data_uber_driver['CATEGORY*'].value_counts(normalize=True)*100",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc9a8cb41ff128a5d0bdba09fb3ae4935840366
| 6,284 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/prepare-image-checkpoint.ipynb
|
cjporteo/thoughtful-ai
|
e3a2563db283aeea7f259f5ffeee2eda07efb856
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/prepare-image-checkpoint.ipynb
|
cjporteo/thoughtful-ai
|
e3a2563db283aeea7f259f5ffeee2eda07efb856
|
[
"MIT"
] | 8 |
2020-11-13T18:55:08.000Z
|
2022-03-12T00:36:10.000Z
|
.ipynb_checkpoints/prepare-image-checkpoint.ipynb
|
cjporteo/thoughtful-ai
|
e3a2563db283aeea7f259f5ffeee2eda07efb856
|
[
"MIT"
] | null | null | null | 32.729167 | 104 | 0.407702 |
[
[
[
"import process_output\nfrom PIL import Image, ImageEnhance, ImageFilter\nimport requests\nfrom io import BytesIO\nimport imgkit\nimport json",
"_____no_output_____"
],
[
"def get_unsplash_url(client_id, query, orientation):\n \n root = 'https://api.unsplash.com/'\n path = 'photos/random/?client_id={}&query={}&orientation={}'\n search_url = root + path.format(client_id, query, orientation)\n \n api_response = requests.get(search_url)\n data = api_response.json()\n api_response.close()\n #print(json.dumps(data, indent=4, sort_keys=True))\n \n return data['urls']['regular']\n\nclient_id = 'L-CxZwGQjlKToJ1xdSiBCnj1gAyUJ0nBLKYqaQOXOAg'\nquery = 'nature dark'\norientation = 'landscape'\n\nimage_url = get_unsplash_url(client_id, query, orientation)",
"_____no_output_____"
],
[
"quote_text = process_output.get_quote()\n\nimage_response = requests.get(image_url)\nimg = Image.open(BytesIO(image_response.content))\nimage_response.close()\n\n# resize down until either a desired width or height is acheived, then crop the other dimension\n# to acheive a non-distorted version of the image with desired dimensions\ndef resize_crop(im, desired_width=800, desired_height=600):\n \n width, height = im.size\n if width/height > desired_width/desired_height:\n im.thumbnail((width, desired_height))\n else:\n im.thumbnail((desired_width, height))\n \n width, height = im.size\n box = [0, 0, width, height] # left, upper, right, lower\n if width > desired_width:\n box[0] = width/2 - desired_width/2\n box[2] = width/2 + desired_width/2\n if height > desired_height:\n box[1] = height/2 - desired_height/2\n box[3] = height/2 + desired_height/2\n \n im = im.crop(box=box)\n return im\n\ndef reduce_color(im, desired_color=0.5):\n converter = ImageEnhance.Color(im)\n im = converter.enhance(desired_color)\n return im\n\ndef gaussian_blur(im, radius=2):\n im = im.filter(ImageFilter.GaussianBlur(radius=radius))\n return im\n\nimg = resize_crop(img)\nimg = reduce_color(img)\n#img = gaussian_blur(img)\nimg.save('backdrop.jpg')",
"_____no_output_____"
],
[
"html_doc = None\n\nwith open('image_template.html', 'r') as f:\n html_doc = f.read()\n\nhtml_doc = html_doc.replace('dynamictext', quote_text)\n\n#print(len(quote_text))\n\ndef get_font_size(text):\n size = len(text)\n if size < 40:\n return '44'\n if size < 75:\n return '36'\n return '30'\n\nhtml_doc = html_doc.replace('dynamicfontsize', get_font_size(quote_text))\n\nwith open('image_out.html', 'w') as f:\n f.write(html_doc)",
"_____no_output_____"
],
[
"imgkit.from_file('image_out.html', 'image_out.jpg', options={'width' : 800,\n 'height' : 600,\n 'quality' : 100,\n 'encoding' : 'utf-8'\n })",
"Loading page (1/2)\n[> ] 0%\r[======> ] 10%\r[==========> ] 17%\r[====================> ] 34%\r[=========================> ] 43%\r[=========================> ] 43%\r[====================================> ] 60%\r[========================================> ] 68%\r[=============================================> ] 75%\r[=================================================> ] 82%\r[===================================================> ] 85%\r[============================================================] 100%\rRendering (2/2) \n[> ] 0%\r[===============> ] 25%\r[============================================================] 100%\rDone \n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
cbc9aa44fc7b7c8bd6ea8068bc79eb8f44b3ff6e
| 3,317 |
ipynb
|
Jupyter Notebook
|
Interfaces.ipynb
|
Marcosddf/programacaojava
|
52ba3148721c6a9b09af0f3acc764680221c8826
|
[
"Unlicense"
] | 2 |
2020-10-28T21:24:07.000Z
|
2021-03-09T20:29:52.000Z
|
Interfaces.ipynb
|
Marcosddf/programacaojava
|
52ba3148721c6a9b09af0f3acc764680221c8826
|
[
"Unlicense"
] | 4 |
2020-03-24T18:17:03.000Z
|
2021-02-02T22:32:31.000Z
|
Interfaces.ipynb
|
Marcosddf/programacaojava
|
52ba3148721c6a9b09af0f3acc764680221c8826
|
[
"Unlicense"
] | 1 |
2021-06-12T15:26:25.000Z
|
2021-06-12T15:26:25.000Z
| 28.350427 | 228 | 0.529997 |
[
[
[
"## Interfaces\n\nUma interface é um tipo abstrato que é usado para especificar comportamento das classes, agindo como um protocolo. \n\nAs interfaces podem declarar apenas assinaturas de métodos que serão implementados.\n\nUma classe **implementa** uma ou mais interfaces. Caso uma classe implemente uma interface, todos os métodos declarados na interface deverão ser implementados pela classe.\n\nAbaixo o exemplo da [https://github.com/Marcosddf/programacaojava/blob/master/Calculadora.ipynb](calculadora polimórfica) desenvolvido com interfaces. \n\nNeste exemplo, a classe *Soma* também implementa a interface *imprime*, então esta deve obrigatoriamente implementar este método. \n\nNa última linha da execução, há uma chamada sobre a variavel *soma*, do tipo *IOperacao*, porém com um *cast* para *IImprime*. Isto só é possível pois o objeto instanciado é do tipo Soma, que implementa as 2 interfaces.\n\n",
"_____no_output_____"
]
],
[
[
"interface IOperacao {\n public int executa (int a, int b);\n}\n\ninterface IImprime {\n void imprime();\n}\n\nclass Soma implements IOperacao, IImprime {\n int valor;\n public int executa (int a, int b){\n valor = a + b;\n return valor;\n }\n public void imprime (){\n System.out.println(\"impressão do valor calculado previamente: \"+valor);\n }\n}\n\nclass Subtracao implements IOperacao {\n public int executa (int a, int b){\n return a - b;\n } \n}\n\n\nclass Calculadora {\n int calcula (IOperacao opt, int a, int b){\n return opt.executa(a,b);\n }\n}\n\nclass Programa {\n public static void main (){\n Calculadora calc = new Calculadora ();\n \n IOperacao soma = new Soma();\n IOperacao subtracao = new Subtracao();\n \n System.out.println( calc.calcula (soma,2,3) );\n \n System.out.println( calc.calcula (subtracao,2,3) );\n \n ((IImprime)soma).imprime(); \n \n \n }\n}\n\nPrograma.main();",
"5\n-1\nimpressão do valor calculado previamente: 5\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
cbc9cacb11712a1236146d21aef5704427c45373
| 12,085 |
ipynb
|
Jupyter Notebook
|
tasks/robust-scaler/Experiment.ipynb
|
vitoryeso/tasks
|
5f530a0fff6b0312ce9a68f4b160be42a4b98044
|
[
"MIT",
"Apache-2.0",
"BSD-3-Clause"
] | 2 |
2021-02-16T12:39:57.000Z
|
2021-07-21T11:36:39.000Z
|
tasks/robust-scaler/Experiment.ipynb
|
vitoryeso/tasks
|
5f530a0fff6b0312ce9a68f4b160be42a4b98044
|
[
"MIT",
"Apache-2.0",
"BSD-3-Clause"
] | 20 |
2020-10-26T18:05:27.000Z
|
2021-11-30T19:05:22.000Z
|
tasks/robust-scaler/Experiment.ipynb
|
vitoryeso/tasks
|
5f530a0fff6b0312ce9a68f4b160be42a4b98044
|
[
"MIT",
"Apache-2.0",
"BSD-3-Clause"
] | 7 |
2020-10-13T18:12:22.000Z
|
2021-08-13T19:16:21.000Z
| 40.827703 | 1,828 | 0.683409 |
[
[
[
"# Robust Scaler - Experimento\n\nEste é um componante que dimensiona atributos usando estatísticas robustas para outliers. Este Scaler remove a mediana e dimensiona os dados de acordo com o intervalo quantil (o padrão é Amplitude interquartil). Amplitude interquartil é o intervalo entre o 1º quartil (25º quantil) e o 3º quartil (75º quantil). Faz uso da implementação do [Scikit-learn](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.RobustScaler.html). <br>\nScikit-learn é uma biblioteca open source de machine learning que suporta apredizado supervisionado e não supervisionado. Também provê várias ferramentas para montagem de modelo, pré-processamento de dados, seleção e avaliação de modelos, e muitos outros utilitários.",
"_____no_output_____"
],
[
"## Declaração de parâmetros e hiperparâmetros\n\nDeclare parâmetros com o botão <img src=\"data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAABQAAAAUCAYAAACNiR0NAAABhWlDQ1BJQ0MgcHJvZmlsZQAAKJF9kT1Iw0AcxV9TtaIVBzuIOASpThb8QhylikWwUNoKrTqYXPohNGlIUlwcBdeCgx+LVQcXZ10dXAVB8APEydFJ0UVK/F9SaBHjwXE/3t173L0DhFqJqWbbGKBqlpGMRcVMdkUMvKID3QhiCOMSM/V4aiENz/F1Dx9f7yI8y/vcn6NHyZkM8InEs0w3LOJ14ulNS+e8TxxiRUkhPiceNeiCxI9cl11+41xwWOCZISOdnCMOEYuFFpZbmBUNlXiKOKyoGuULGZcVzluc1VKFNe7JXxjMacsprtMcRAyLiCMBETIq2EAJFiK0aqSYSNJ+1MM/4PgT5JLJtQFGjnmUoUJy/OB/8LtbMz854SYFo0D7i21/DAOBXaBete3vY9uunwD+Z+BKa/rLNWDmk/RqUwsfAb3bwMV1U5P3gMsdoP9JlwzJkfw0hXweeD+jb8oCfbdA16rbW2Mfpw9AmrpaugEODoGRAmWveby7s7W3f880+vsBocZyukMJsmwAAAAGYktHRAD/AP8A/6C9p5MAAAAJcEhZcwAADdcAAA3XAUIom3gAAAAHdElNRQfkBgsMIwnXL7c0AAACDUlEQVQ4y92UP4gTQRTGf29zJxhJZ2NxbMBKziYWlmJ/ile44Nlkd+dIYWFzItiNgoIEtFaTzF5Ac/inE/urtLWxsMqmUOwCEpt1Zmw2xxKi53XitPO9H9978+aDf/3IUQvSNG0450Yi0jXG7C/eB0cFeu9viciGiDyNoqh2KFBrHSilWstgnU7nFLBTgl+ur6/7PwK11kGe5z3n3Hul1MaiuCgKDZwALHA7z/Oe1jpYCtRaB+PxuA8kQM1aW68Kt7e3zwBp6a5b1ibj8bhfhQYVZwMRiQHrvW9nWfaqCrTWPgRWvPdvsiy7IyLXgEJE4slk8nw+T5nDgDbwE9gyxryuwpRSF5xz+0BhrT07HA4/AyRJchUYASvAbhiGaRVWLIMBYq3tAojIszkMoNRulbXtPM8HwV/sXSQi54HvQRDcO0wfhGGYArvAKjAq2wAgiqJj3vsHpbtur9f7Vi2utLx60LLW2hljEuBJOYu9OI6vAzQajRvAaeBLURSPlsBelA+VhWGYaq3dwaZvbm6+m06noYicE5ErrVbrK3AXqHvvd4bD4Ye5No7jSERGwKr3Pms2m0pr7Rb30DWbTQWYcnFvAieBT7PZbFB1V6vVfpQaU4UtDQetdTCZTC557/eA48BlY8zbRZ1SqrW2tvaxCvtt2iRJ0i9/xb4x5uJRwmNlaaaJ3AfqIvKY/+78Av++6uiSZhYMAAAAAElFTkSuQmCC\" /> na barra de ferramentas.<br>\nA variável `dataset` possui o caminho para leitura do arquivos importados na tarefa de \"Upload de dados\".<br>\nVocê também pode importar arquivos com o botão <img src=\"data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAABQAAAAUCAYAAACNiR0NAAABhWlDQ1BJQ0MgcHJvZmlsZQAAKJF9kT1Iw0AcxV9TtaIVBzuIOASpThb8QhylikWwUNoKrTqYXPohNGlIUlwcBdeCgx+LVQcXZ10dXAVB8APEydFJ0UVK/F9SaBHjwXE/3t173L0DhFqJqWbbGKBqlpGMRcVMdkUMvKID3QhiCOMSM/V4aiENz/F1Dx9f7yI8y/vcn6NHyZkM8InEs0w3LOJ14ulNS+e8TxxiRUkhPiceNeiCxI9cl11+41xwWOCZISOdnCMOEYuFFpZbmBUNlXiKOKyoGuULGZcVzluc1VKFNe7JXxjMacsprtMcRAyLiCMBETIq2EAJFiK0aqSYSNJ+1MM/4PgT5JLJtQFGjnmUoUJy/OB/8LtbMz854SYFo0D7i21/DAOBXaBete3vY9uunwD+Z+BKa/rLNWDmk/RqUwsfAb3bwMV1U5P3gMsdoP9JlwzJkfw0hXweeD+jb8oCfbdA16rbW2Mfpw9AmrpaugEODoGRAmWveby7s7W3f880+vsBocZyukMJsmwAAAAGYktHRAD/AP8A/6C9p5MAAAAJcEhZcwAADdcAAA3XAUIom3gAAAAHdElNRQfkBgsOBy6ASTeXAAAC/0lEQVQ4y5WUT2gcdRTHP29m99B23Uiq6dZisgoWCxVJW0oL9dqLfyhCvGWY2YUBI95MsXgwFISirQcLhS5hfgk5CF3wJIhFI7aHNsL2VFZFik1jS1qkiZKdTTKZ3/MyDWuz0fQLc/m99/vMvDfv+4RMlUrlkKqeAAaBAWAP8DSgwJ/AXRG5rao/WWsvTU5O3qKLBMD3fSMiPluXFZEPoyj67PGAMzw83PeEMABHVT/oGpiamnoAmCcEWhH5tFsgF4bh9oWFhfeKxeJ5a+0JVT0oImWgBPQCKfAQuAvcBq67rltX1b+6ApMkKRcKhe9V9QLwbavV+qRer692Sx4ZGSnEcXw0TdP3gSrQswGYz+d/S5IkVtXTwOlCoZAGQXAfmAdagAvsAErtdnuXiDy6+023l7qNRsMODg5+CawBzwB9wFPA7mx8ns/KL2Tl3xCRz5eWlkabzebahrHxPG+v4zgnc7ncufHx8Z+Hhoa29fT0lNM03Q30ikiqqg+ttX/EcTy3WTvWgdVqtddaOw/kgXvADHBHROZVNRaRvKruUNU+EdkPfGWM+WJTYOaSt1T1LPDS/4zLWWPMaLVaPWytrYvIaBRFl/4F9H2/JCKvGmMu+76/X0QOqGoZKDmOs1NV28AicMsYc97zvFdc1/0hG6kEeNsY83UnsCwivwM3VfU7YEZE7lhr74tIK8tbnJiYWPY8b6/ruleAXR0ftQy8boyZXi85CIIICDYpc2ZgYODY3NzcHmvt1eyvP64lETkeRdE1yZyixWLx5U2c8q4x5mIQBE1g33/0d3FlZeXFR06ZttZesNZejuO4q1NE5CPgWVV9E3ij47wB1IDlJEn+ljAM86urq7+KyAtZTgqsO0VV247jnOnv7/9xbGzMViqVMVX9uANYj6LonfVtU6vVkjRNj6jqGeCXzGrPAQeA10TkuKpOz87ONrayhnIA2Qo7BZwKw3B7kiRloKSqO13Xja21C47jPNgysFO1Wi0GmtmzQap6DWgD24A1Vb3SGf8Hfstmz1CuXEIAAAAASUVORK5CYII=\" /> na barra de ferramentas.",
"_____no_output_____"
]
],
[
[
"# parâmetros\ndataset = \"/tmp/data/iris.csv\" #@param {type:\"string\"}\ntarget = None #@param {type:\"feature\", label:\"Atributo alvo\", description: \"Esse valor será utilizado para garantir que o alvo não seja removido.\"}\n\nwith_centering = True #@param {type:\"boolean\", label:\"Centralização\", description:\"Centralizar os dados antes de dimensionar. Ocorre exceção quando usado com matrizes esparsas\"}\nwith_scaling = True #@param {type:\"boolean\", label:\"Dimensionamento\", description:\"Dimensionar os dados para um intervalo interquartil\"}",
"_____no_output_____"
]
],
[
[
"## Acesso ao conjunto de dados\n\nO conjunto de dados utilizado nesta etapa será o mesmo carregado através da plataforma.<br>\nO tipo da variável retornada depende do arquivo de origem:\n- [pandas.DataFrame](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.html) para CSV e compressed CSV: .csv .csv.zip .csv.gz .csv.bz2 .csv.xz\n- [Binary IO stream](https://docs.python.org/3/library/io.html#binary-i-o) para outros tipos de arquivo: .jpg .wav .zip .h5 .parquet etc",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\ndf = pd.read_csv(dataset)",
"_____no_output_____"
],
[
"has_target = True if target is not None and target in df.columns else False",
"_____no_output_____"
],
[
"X = df.copy()\n\nif has_target:\n X = df.drop(target, axis=1)\n y = df[target]",
"_____no_output_____"
]
],
[
[
"## Acesso aos metadados do conjunto de dados\n\nUtiliza a função `stat_dataset` do [SDK da PlatIAgro](https://platiagro.github.io/sdk/) para carregar metadados. <br>\nPor exemplo, arquivos CSV possuem `metadata['featuretypes']` para cada coluna no conjunto de dados (ex: categorical, numerical, or datetime).",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom platiagro import stat_dataset\n\nmetadata = stat_dataset(name=dataset)\nfeaturetypes = metadata[\"featuretypes\"]\n\ncolumns = df.columns.to_numpy()\nfeaturetypes = np.array(featuretypes)\n\nif has_target:\n target_index = np.argwhere(columns == target)\n columns = np.delete(columns, target_index)\n featuretypes = np.delete(featuretypes, target_index)",
"_____no_output_____"
]
],
[
[
"## Configuração dos atributos",
"_____no_output_____"
]
],
[
[
"from platiagro.featuretypes import NUMERICAL\n\n# Selects the indexes of numerical\nnumerical_indexes = np.where(featuretypes == NUMERICAL)[0]\nnon_numerical_indexes = np.where(~(featuretypes == NUMERICAL))[0]\n\n# After the step of the make_column_transformer,\n# numerical features are grouped in the beggining of the array\nnumerical_indexes_after_first_step = np.arange(len(numerical_indexes))",
"_____no_output_____"
]
],
[
[
"## Treina um modelo usando sklearn.preprocessing.RobustScaler",
"_____no_output_____"
]
],
[
[
"from sklearn.compose import make_column_transformer\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.preprocessing import RobustScaler\n\npipeline = Pipeline(\n steps=[\n (\n \"imputer\",\n make_column_transformer(\n (SimpleImputer(), numerical_indexes), remainder=\"passthrough\"\n ),\n ),\n (\n \"robust_scaler\",\n make_column_transformer(\n (\n RobustScaler(\n with_centering=with_centering, with_scaling=with_scaling\n ),\n numerical_indexes_after_first_step,\n ),\n remainder=\"passthrough\",\n ),\n ),\n ]\n)\n\n# Train model and transform dataset\nX = pipeline.fit_transform(X)\n\n# Put numerical features in the lowest indexes\nfeatures_after_pipeline = np.concatenate(\n (columns[numerical_indexes], columns[non_numerical_indexes])\n)\n",
"_____no_output_____"
],
[
"# Put data back in a pandas.DataFrame\ndf = pd.DataFrame(data=X, columns=features_after_pipeline)\n\nif has_target:\n df[target] = y",
"_____no_output_____"
]
],
[
[
"## Cria visualização do resultado\n\nCria visualização do resultado como uma planilha.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom platiagro.plotting import plot_data_table\n\nax = plot_data_table(df)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Salva alterações no conjunto de dados\n\nO conjunto de dados será salvo (e sobrescrito com as respectivas mudanças) localmente, no container da experimentação, utilizando a função `pandas.DataFrame.to_csv`.<br>",
"_____no_output_____"
]
],
[
[
"# save dataset changes\ndf.to_csv(dataset, index=False)",
"_____no_output_____"
]
],
[
[
"## Salva resultados da tarefa \n\nA plataforma guarda o conteúdo de `/tmp/data/` para as tarefas subsequentes.",
"_____no_output_____"
]
],
[
[
"from joblib import dump\n\nartifacts = {\n \"pipeline\": pipeline,\n \"columns\": columns,\n \"features_after_pipeline\": features_after_pipeline,\n}\n\ndump(artifacts, \"/tmp/data/robust-scaler.joblib\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc9d5c54284d3bcd50328367c0e4609288c8892
| 136,945 |
ipynb
|
Jupyter Notebook
|
src/05_desafio/05_missao05.ipynb
|
ralsouza/python_fundamentos
|
f3ccf4b036036327394aac181659eaf463768e9d
|
[
"MIT"
] | 1 |
2019-07-29T02:43:25.000Z
|
2019-07-29T02:43:25.000Z
|
src/05_desafio/05_missao05.ipynb
|
ralsouza/python_fundamentos
|
f3ccf4b036036327394aac181659eaf463768e9d
|
[
"MIT"
] | null | null | null |
src/05_desafio/05_missao05.ipynb
|
ralsouza/python_fundamentos
|
f3ccf4b036036327394aac181659eaf463768e9d
|
[
"MIT"
] | null | null | null | 61.575989 | 15,620 | 0.622622 |
[
[
[
"<a href=\"https://colab.research.google.com/github/ralsouza/python_fundamentos/blob/master/src/05_desafio/05_missao05.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"## **Missão: Analisar o Comportamento de Compra de Consumidores.**\n### Nível de Dificuldade: Alto\n\nVocê recebeu a tarefa de analisar os dados de compras de um web site! Os dados estão no formato JSON e disponíveis junto com este notebook.\n\nNo site, cada usuário efetua login usando sua conta pessoal e pode adquirir produtos à medida que navega pela lista de produtos oferecidos. Cada produto possui um valor de venda. Dados de idade e sexo de cada usuário foram coletados e estão fornecidos no arquivo JSON.\n\nSeu trabalho é entregar uma análise de comportamento de compra dos consumidores. Esse é um tipo de atividade comum realizado por Cientistas de Dados e o resultado deste trabalho pode ser usado, por exemplo, para alimentar um modelo de Machine Learning e fazer previsões sobre comportamentos futuros.\n\nMas nesta missão você vai analisar o comportamento de compra dos consumidores usando o pacote Pandas da linguagem Python e seu relatório final deve incluir cada um dos seguintes itens:\n\n\n**Contagem de Consumidores**\n* Número total de consumidores\n\n**Análise Geral de Compras**\n* Número de itens exclusivos\n* Preço médio de compra\n* Número total de compras\n* Rendimento total (Valor Total)\n\n**Informações Demográficas Por Gênero**\n* Porcentagem e contagem de compradores masculinos\n* Porcentagem e contagem de compradores do sexo feminino\n* Porcentagem e contagem de outros / não divulgados\n\n**Análise de Compras Por Gênero**\n* Número de compras\n* Preço médio de compra\n* Valor Total de Compra\n* Compras for faixa etária\n\n**Identifique os 5 principais compradores pelo valor total de compra e, em seguida, liste (em uma tabela):**\n* Login\n* Número de compras\n* Preço médio de compra\n* Valor Total de Compra\n* Itens mais populares\n\n**Identifique os 5 itens mais populares por contagem de compras e, em seguida, liste (em uma tabela):**\n* ID do item\n* Nome do item\n* Número de compras\n* Preço Médio do item\n* Valor Total de Compra\n* Itens mais lucrativos\n\n**Identifique os 5 itens mais lucrativos pelo valor total de compra e, em seguida, liste (em uma tabela):**\n* ID do item\n* Nome do item\n* Número de compras\n* Preço Médio do item\n* Valor Total de Compra\n\n**Como considerações finais:**\n* Seu script deve funcionar para o conjunto de dados fornecido.\n* Você deve usar a Biblioteca Pandas e o Jupyter Notebook.\n\n",
"_____no_output_____"
]
],
[
[
"# Imports\nimport pandas as pd\nimport numpy as np\n\n# Load file from Drive\nfrom google.colab import drive\ndrive.mount('/content/drive')\n\n# Load file to Dataframe\nload_file = \"/content/drive/My Drive/dados_compras.json\"\npurchase_file = pd.read_json(load_file, orient = \"records\")",
"_____no_output_____"
]
],
[
[
"## **1. Análise Exploratória**",
"_____no_output_____"
],
[
"### **1.1 Checagem das primeiras linhas**",
"_____no_output_____"
]
],
[
[
"# Nota-se que os logins se repetem.\npurchase_file.sort_values('Login')",
"_____no_output_____"
]
],
[
[
"### **1.2 Checagem dos tipos dos dados**",
"_____no_output_____"
]
],
[
[
"purchase_file.dtypes",
"_____no_output_____"
]
],
[
[
"### **1.3 Checagem de valores nulos**",
"_____no_output_____"
]
],
[
[
"purchase_file.isnull().sum().sort_values(ascending = False)",
"_____no_output_____"
]
],
[
[
"### **1.4 Checagem de valores zero**",
"_____no_output_____"
]
],
[
[
"(purchase_file == 0).sum()",
"_____no_output_____"
]
],
[
[
"### **1.5 Distribuição de idades**\nO público mais representativo desta amostra encontra-se entre 19 há 26 anos de idade.",
"_____no_output_____"
]
],
[
[
"plt.hist(purchase_file['Idade'], histtype='bar', rwidth=0.8)\n\nplt.title('Distribuição de vendas por idade')\nplt.xlabel('Idade')\nplt.ylabel('Quantidade de compradores')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"### **1.6 Distribuição dos valores**\nA maioria das vendas são dos produtos de `R$ 2,30`, `R$ 3,40` e `R$ 4,20`.",
"_____no_output_____"
]
],
[
[
"plt.hist(purchase_file['Valor'], histtype='bar', rwidth=0.8)\n\nplt.title('Distribuição por Valores')\nplt.xlabel('Reais R$')\nplt.ylabel('Quantidade de vendas')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## **2. Informações Sobre os Consumidores**\n* Número total de consumidores",
"_____no_output_____"
]
],
[
[
"# Contar a quantidade de logins, removendo as linhas com dados duplicados.\n\ntotal_consumidores = purchase_file['Login'].drop_duplicates().count()\nprint('O total de consumidores na amostra são: {}'.format(total_consumidores))",
"O total de consumidores na amostra são: 573\n"
]
],
[
[
"## **3. Análise Geral de Compras**\n* Número de itens exclusivos\n* Preço médio de compra\n* Número total de compras\n* Rendimento total (Valor Total)",
"_____no_output_____"
]
],
[
[
"# Número de itens exclusivos\nitens_exclusivos = purchase_file['Item ID'].drop_duplicates().count()\n\npreco_medio = np.average(purchase_file['Valor'])\n\ntotal_compras = purchase_file['Nome do Item'].count()\n\nvalor_total = np.sum(purchase_file['Valor'])\n\nanalise_geral = pd.DataFrame({\n 'Itens Exclusivos':[itens_exclusivos],\n 'Preço Médio (R$)':[np.round(preco_medio, decimals=2)],\n 'Qtd. Compras':[total_compras],\n 'Valor Total (R$)':[valor_total]\n})\n\nanalise_geral",
"_____no_output_____"
]
],
[
[
"## **4. Análise Demográfica por Genêro**\n* Porcentagem e contagem de compradores masculinos\n* Porcentagem e contagem de compradores do sexo feminino\n* Porcentagem e contagem de outros / não divulgados",
"_____no_output_____"
]
],
[
[
"# Selecionar os dados únicos do compradores para deduplicação\ninfo_compradores = purchase_file.loc[:,['Login','Sexo','Idade']]\n\n# Deduplicar os dados\ninfo_compradores = info_compradores.drop_duplicates()",
"_____no_output_____"
],
[
"# Quantidade de compradores por genêro\nqtd_compradores = info_compradores['Sexo'].value_counts()\n\n# Percentual de compradores por genêro\nperc_compradores = round(info_compradores['Sexo'].value_counts(normalize=True) * 100, 2)\n\n# Armazenar dados no Dataframe\nanalise_demografica = pd.DataFrame(\n {'Percentual':perc_compradores,\n 'Qtd. Compradores':qtd_compradores\n }\n)",
"_____no_output_____"
],
[
"# Impressão da tabela\nanalise_demografica",
"_____no_output_____"
],
[
"plot = analise_demografica['Percentual'].plot(kind='pie',\n title='Percentual de Compras por Genêro',\n autopct='%.2f')",
"_____no_output_____"
],
[
"plot = analise_demografica['Qtd. Compradores'].plot(kind='barh',\n title='Quantidade de Compradores por Genêro')\n# Add labels\nfor i in plot.patches:\n plot.text(i.get_width()+.1, i.get_y()+.31, \\\n str(round((i.get_width()), 2)), fontsize=10)",
"_____no_output_____"
]
],
[
[
"## **5. Análise de Compras Por Gênero**\n\n* Número de compras\n* Preço médio de compra\n* Valor Total de Compra\n* Compras for faixa etária\n",
"_____no_output_____"
]
],
[
[
"# Número de compras por genêro\nnro_compras_gen = purchase_file['Sexo'].value_counts()\n\n# Preço médio de compra por genêro\nmedia_compras_gen = round(purchase_file.groupby('Sexo')['Valor'].mean(), 2)\n\n# Total de compras por genêro\ntotal_compras_gen = purchase_file.groupby('Sexo')['Valor'].sum()\n\nanalise_compras = pd.DataFrame(\n {'Qtd. de Compras':nro_compras_gen,\n 'Preço Médio (R$)':media_compras_gen,\n 'Total Compras (R$)':total_compras_gen}\n)",
"_____no_output_____"
],
[
"# Impressão da tabela\nanalise_compras",
"_____no_output_____"
],
[
"# Usar dataframe deduplicado\n\ninfo_compradores",
"_____no_output_____"
],
[
"# Compras por faixa etária\nage_bins = [0, 9.99, 14.99, 19.99, 24.99, 29.99, 34.99, 39.99, 999]\nseg_idade = ['Menor de 10', '10-14', '15-19', '20-24', '25-29', '30-34', '35-39', 'Maior de 39']\n\ninfo_compradores['Intervalo Idades'] = pd.cut(info_compradores['Idade'], age_bins, labels=seg_idade)",
"_____no_output_____"
],
[
"df_hist_compras = pd.DataFrame(info_compradores['Intervalo Idades'].value_counts(), index=seg_idade)\n\nhist = df_hist_compras.plot(kind='bar', legend=False)\n\nhist.set_title('Compras for faixa etária', fontsize=15)\nhist.set_ylabel('Frequência')\nhist.set_xlabel('Faixas de Idades')",
"_____no_output_____"
]
],
[
[
"## **6. Consumidores Mais Populares (Top 5)**\nIdentifique os 5 principais compradores pelo valor total de compra e, em seguida, liste (em uma tabela):\n\n* Login\n* Número de compras\n* Preço médio de compra\n* Valor Total de Compra\n* Itens mais populares",
"_____no_output_____"
]
],
[
[
"consumidores_populares = purchase_file[['Login','Nome do Item','Valor']]",
"_____no_output_____"
],
[
"consumidores_populares.head(5)",
"_____no_output_____"
],
[
"top_por_compras = consumidores_populares.groupby(['Login']).count()['Nome do Item']\ntop_por_valor_medio = round(consumidores_populares.groupby('Login').mean()['Valor'], 2)\ntop_por_valor_total = consumidores_populares.groupby('Login').sum()['Valor']\n\ntop_consumidores = pd.DataFrame({'Número de Compras': top_por_compras,\n 'Preço Médio(R$)': top_por_valor_medio,\n 'Valor Total(R$)': top_por_valor_total}) \\\n .sort_values(by=['Valor Total(R$)'], ascending=False) \\\n .head(5)\n\ntop_itens = consumidores_populares['Nome do Item'].value_counts().head(5)",
"_____no_output_____"
],
[
"top_consumidores",
"_____no_output_____"
],
[
"itens_populares = pd.DataFrame(consumidores_populares['Nome do Item'].value_counts().head(5))\nitens_populares",
"_____no_output_____"
]
],
[
[
"## **7. Itens Mais Populares**\nIdentifique os 5 itens mais populares **por contagem de compras** e, em seguida, liste (em uma tabela):\n* ID do item\n* Nome do item\n* Número de compras\n* Preço Médio do item\n* Valor Total de Compra\n* Itens mais lucrativos",
"_____no_output_____"
]
],
[
[
"itens_populares = purchase_file[['Item ID','Nome do Item','Valor']]",
"_____no_output_____"
],
[
"num_compras = itens_populares.groupby('Nome do Item').count()['Item ID']\nmedia_preco = round(itens_populares.groupby('Nome do Item').mean()['Valor'], 2)\ntotal_preco = itens_populares.groupby('Nome do Item').sum()['Valor']\n\ndf_itens_populares = pd.DataFrame({\n 'Numero de Compras': num_compras,\n 'Preço Médio do Item': media_preco,\n 'Valor Total da Compra': total_preco})\n\ndf_itens_populares.sort_values(by=['Numero de Compras'], ascending=False).head(5)",
"_____no_output_____"
]
],
[
[
"## **8. Itens Mais Lucrativos**\nIdentifique os 5 itens mais lucrativos pelo **valor total de compra** e, em seguida, liste (em uma tabela):\n* ID do item\n* Nome do item\n* Número de compras\n* Preço Médio do item\n* Valor Total de Compra",
"_____no_output_____"
]
],
[
[
"itens_lucrativos = purchase_file[['Item ID','Nome do Item','Valor']]",
"_____no_output_____"
],
[
"itens_lucrativos.head(5)",
"_____no_output_____"
],
[
"qtd_compras = itens_lucrativos.groupby(['Nome do Item']).count()['Valor']\navg_compras = itens_lucrativos.groupby(['Nome do Item']).mean()['Valor']\nsum_compras = itens_lucrativos.groupby(['Nome do Item']).sum()['Valor']",
"_____no_output_____"
],
[
"df_itens_lucrativos = pd.DataFrame({\n 'Número de Compras': qtd_compras,\n 'Preço Médio do Item (R$)': round(avg_compras, 2),\n 'Valor Total de Compra (R$)': sum_compras\n})",
"_____no_output_____"
],
[
"df_itens_lucrativos.sort_values(by='Valor Total de Compra (R$)', ascending=False).head(5)",
"_____no_output_____"
],
[
"itens_lucrativos.sort_values('Nome do Item')",
"_____no_output_____"
],
[
"itens_lucrativos.sort_values(by='Nome do Item')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbc9d7fee7fc3bbc62a8bdb681beec97d8a694e4
| 171,617 |
ipynb
|
Jupyter Notebook
|
notebooks/wine_classifier.ipynb
|
herr-kistler/python-nlp
|
30d329adb2d796647a3edb7f8b3427d9ace08530
|
[
"MIT"
] | null | null | null |
notebooks/wine_classifier.ipynb
|
herr-kistler/python-nlp
|
30d329adb2d796647a3edb7f8b3427d9ace08530
|
[
"MIT"
] | null | null | null |
notebooks/wine_classifier.ipynb
|
herr-kistler/python-nlp
|
30d329adb2d796647a3edb7f8b3427d9ace08530
|
[
"MIT"
] | null | null | null | 62.338177 | 22,002 | 0.64789 |
[
[
[
"WINE CLASSIFIER",
"_____no_output_____"
]
],
[
[
"# Imports\n\nfrom io import StringIO\nimport pandas as pd\nimport spacy\nfrom cytoolz import *\nimport numpy as np\nfrom IPython.display import display\nimport seaborn as sns\n\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.feature_selection import chi2\nfrom sklearn.svm import LinearSVC\nfrom sklearn.naive_bayes import MultinomialNB\nfrom sklearn.model_selection import *\nfrom sklearn.linear_model import *\nfrom sklearn.dummy import *\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.feature_extraction.text import *\nfrom sklearn.metrics import *\nfrom sklearn.decomposition import *\nfrom sklearn import metrics\n\n%precision 4\n%matplotlib inline",
"_____no_output_____"
],
[
"nlp = spacy.load('en', disable=['tagger', 'ner', 'parser'])",
"_____no_output_____"
],
[
"#1. Prepare Data\n\ndf = pd.read_msgpack('http://bulba.sdsu.edu/wine.dat')\n#df.head()\n\n#about 40,000 rows in full msgpack\n#sample created to increase speed, but remove sample definition for increased accuracy!\ndf = df.sample(4000)\ndf = df[pd.notnull(df['review_text'])]\ndf = df[pd.notnull(df['wine_variant'])]\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 4000 entries, 137 to 16668\nData columns (total 2 columns):\nwine_variant 4000 non-null object\nreview_text 4000 non-null object\ndtypes: object(2)\nmemory usage: 93.8+ KB\n"
],
[
"#Create 'category_id' column for LinearSVC use\n\ndf['category_id'] = df['wine_variant'].factorize()[0]\ncategory_id_df = df[['wine_variant', 'category_id']].drop_duplicates().sort_values('category_id')\ncategory_to_id = dict(category_id_df.values)\nid_to_category = dict(category_id_df[['category_id', 'wine_variant']].values)\n\n\n#Create a tokenized column for Logistical Regression use\n\ndef tokenize(text):\n return [tok.orth_ for tok in nlp.tokenizer(text)]\n\ndf['tokens'] = df['review_text'].apply(tokenize)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"#Check to see sample sizes for each variant, ensurring result accuracy\n\nimport matplotlib.pyplot as plt\n\nfig = plt.figure(figsize=(10,8))\ndf.groupby('wine_variant').review_text.count().plot.bar(ylim=0)\nplt.show()",
"_____no_output_____"
],
[
"#2. BASELINE\n\nfolds = StratifiedKFold(shuffle = True, \n n_splits = 10, \n random_state = 10)\n\nsum(df['wine_variant'] == True), len(df)",
"_____no_output_____"
],
[
"baseline = make_pipeline(CountVectorizer(analyzer = identity), \n DummyClassifier('most_frequent'))\n\nbase_score = cross_val_score(baseline, \n df['tokens'], \n df['wine_variant'], \n cv=folds, \n n_jobs = -1)\n\nbase_score.mean(), base_score.std()",
"_____no_output_____"
],
[
"#3. SIMPLE LOGISTIC REGRESSION CLASSIFIER\n\nlr = make_pipeline(CountVectorizer(analyzer = identity), \n LogisticRegression())\n\nparams = {'logisticregression__C': [0.01, 0.1, 1.0],\n 'countvectorizer__min_df': [1, 2],\n 'countvectorizer__max_df': [0.25, 0.5]}\n\ngrid_search = GridSearchCV(lr, \n params, \n n_jobs = -1, \n verbose = 1, \n return_train_score = True)\n\ngrid_search.fit(df['tokens'], df['wine_variant'])",
"Fitting 3 folds for each of 12 candidates, totalling 36 fits\n"
],
[
"grid_search.best_params_",
"_____no_output_____"
],
[
"lr.set_params(**grid_search.best_params_)\n\nlr_score = cross_val_score(lr, \n df['tokens'], \n df['wine_variant'], \n cv = folds, \n n_jobs = -1)\n\nlr_score.mean(), lr_score.std()",
"_____no_output_____"
],
[
"grid = pd.DataFrame(grid_search.cv_results_, dtype = float)\n\ngrid.plot.line('param_countvectorizer__max_df', 'mean_test_score')",
"_____no_output_____"
],
[
"#4. BEST CLASSIFIER -- found through n_gram correlation \n\nbest = make_pipeline(CountVectorizer(analyzer = identity), \n TfidfTransformer(), \n LinearSVC())\n\nparams_best = {'tfidftransformer__norm': ['l2', None],\n 'tfidftransformer__use_idf': [True, False],\n 'tfidftransformer__sublinear_tf': [True, False], \n 'linearsvc__penalty': ['l2'],\n 'linearsvc__C': [0.01, 0.1, 1.0],\n 'countvectorizer__min_df': [1, 2, 3],\n 'countvectorizer__max_df': [0.1, 0.5, 1.0]}\n\nbest_grid_search = GridSearchCV(best, \n params_best, \n n_jobs = -1, \n verbose = 1, \n return_train_score = True)\n\nbest_grid_search.fit(df['tokens'], df['wine_variant'])",
"Fitting 3 folds for each of 216 candidates, totalling 648 fits\n"
],
[
"best_grid_search.best_params_",
"_____no_output_____"
],
[
"#Set hyperparameters for best model\nbest.set_params(**best_grid_search.best_params_)\n\nbest_score = cross_val_score(best, \n df['tokens'], \n df['wine_variant'], \n cv = folds, \n n_jobs = -1)\n\nbest_score.mean(), best_score.std()\n#Result score is slightly higher than using LR model, and std is slightly less",
"_____no_output_____"
],
[
"best_grid = pd.DataFrame(best_grid_search.cv_results_, dtype = float)\n\nbest_grid.plot.line('param_countvectorizer__max_df', 'mean_test_score')",
"_____no_output_____"
],
[
"#5. Error Analysis & Discussion\n\n#Inspect feautues\n\ntfidf = TfidfVectorizer(sublinear_tf = True, \n min_df = 1, \n norm = 'l2', \n encoding = 'latin-1', \n ngram_range = (1, 3), \n stop_words = 'english')\n\nfeatures = tfidf.fit_transform(df.review_text).toarray()\n\nlabels = df.category_id\n\nfeatures.shape",
"_____no_output_____"
],
[
"# Display the n_grams with highest correlation for each variant\nN = 5\nfor wine_variant, category_id in sorted(category_to_id.items()):\n # chi squared determines the correlation of each ngram to each variant, taking into account sample size\n features_chi2 = chi2(features, \n labels == category_id)\n indices = np.argsort(features_chi2[0])\n feature_names = np.array(tfidf.get_feature_names())[indices]\n unigrams = [v for v in feature_names if len(v.split(' ')) == 1]\n bigrams = [v for v in feature_names if len(v.split(' ')) == 2]\n trigrams = [v for v in feature_names if len(v.split(' ')) == 3]\n print(\"# '{}':\".format(wine_variant))\n print(\" . Most correlated unigrams:\\n . {}\".format('\\n . '.join(unigrams[-N:])))\n print(\" . Most correlated bigrams:\\n . {}\".format('\\n . '.join(bigrams[-N:])))\n print(\" . Most correlated trigrams:\\n . {}\".format('\\n . '.join(trigrams[-N:])))\n \n\n#The ngrams below appear more accurate and unique to each of the different variants.",
"# 'Cabernet Sauvignon':\n . Most correlated unigrams:\n . black\n . dark\n . tannins\n . cassis\n . cab\n . Most correlated bigrams:\n . dark fruits\n . cali cab\n . black currant\n . dark fruit\n . napa cab\n . Most correlated trigrams:\n . fruit smooth tannins\n . decanted 45 minutes\n . classic napa cab\n . drinking great right\n . decanted 90 minutes\n# 'Chardonnay':\n . Most correlated unigrams:\n . buttery\n . apple\n . chard\n . butter\n . chardonnay\n . Most correlated bigrams:\n . yellow color\n . golden color\n . white burgundy\n . gold color\n . apple pear\n . Most correlated trigrams:\n . nose tasty tart\n . mineral nose tasty\n . medium gold color\n . lemon mineral palate\n . golden yellow color\n# 'Merlot':\n . Most correlated unigrams:\n . plum\n . chocolate\n . merlots\n . cherry\n . merlot\n . Most correlated bigrams:\n . red fruit\n . great merlot\n . sauvignon blanc\n . good merlot\n . nice merlot\n . Most correlated trigrams:\n . smooth lingering finish\n . red black fruit\n . black cherry fruit\n . ruby red color\n . red color nose\n# 'Sauvignon Blanc':\n . Most correlated unigrams:\n . sauvignon\n . sb\n . grass\n . blanc\n . grapefruit\n . Most correlated bigrams:\n . cut grass\n . new zealand\n . nz sb\n . sauv blanc\n . sauvignon blanc\n . Most correlated trigrams:\n . nice sauvignon blanc\n . grapefruit lemon grass\n . new zealand sauvignon\n . tropical fruit flavors\n . fresh cut grass\n"
],
[
"# Heatmap & Confusion Matrix to display accuracies of predictions with LinearSVC\n\nmodel = LinearSVC()\n\nX_train, X_test, y_train, y_test, indices_train, indices_test = train_test_split(features, \n labels, \n df.index, \n test_size = 0.33, \n random_state = 10)\nmodel.fit(X_train, y_train)\ny_pred = model.predict(X_test)\n\nconf_mat = confusion_matrix(y_test, y_pred)\nfig, ax = plt.subplots(figsize = (10,8))\nsns.heatmap(conf_mat, \n annot = True, \n fmt = 'd',\n xticklabels = category_id_df.wine_variant.values, \n yticklabels = category_id_df.wine_variant.values)\nplt.ylabel('Actual')\nplt.xlabel('Predicted')\nplt.show()",
"_____no_output_____"
],
[
"\n#WRONG RESULT EXAMPLES FOR LINEAR SVC CLASSIFIER\n\nfor predicted in category_id_df.category_id:\n for actual in category_id_df.category_id:\n if predicted != actual and conf_mat[actual, predicted] >= 6:\n print(\"'{}' predicted as '{}' : {} examples.\".format(id_to_category[actual], \n id_to_category[predicted], \n conf_mat[actual, predicted]))\n display(df.loc[indices_test[(y_test == actual) & (y_pred == predicted)]][['wine_variant', 'review_text']])\n print('')",
"'Chardonnay' predicted as 'Cabernet Sauvignon' : 9 examples.\n"
],
[
"model.fit(features, labels)",
"_____no_output_____"
],
[
"N = 5\nfor wine_variant, category_id in sorted(category_to_id.items()):\n indices = np.argsort(model.coef_[category_id])\n feature_names = np.array(tfidf.get_feature_names())[indices]\n unigrams = [v for v in reversed(feature_names) if len(v.split(' ')) == 1][:N]\n bigrams = [v for v in reversed(feature_names) if len(v.split(' ')) == 2][:N]\n trigrams = [v for v in reversed(feature_names) if len(v.split(' ')) == 3][:N]\n print(\"# '{}':\".format(wine_variant))\n print(\" . Top unigrams:\\n . {}\".format('\\n . '.join(unigrams)))\n print(\" . Top bigrams:\\n . {}\".format('\\n . '.join(bigrams)))\n print(\" . Top trigrams:\\n . {}\".format('\\n . '.join(trigrams)))",
"# 'Cabernet Sauvignon':\n . Top unigrams:\n . cab\n . cassis\n . tannins\n . cabernet\n . dark\n . Top bigrams:\n . dark fruit\n . black currant\n . dark fruits\n . napa cab\n . cali cab\n . Top trigrams:\n . oak oak oak\n . medium bodied light\n . medium bodied wine\n . drinking great right\n . afraid waited long\n# 'Chardonnay':\n . Top unigrams:\n . chardonnay\n . chard\n . butter\n . oak\n . buttery\n . Top bigrams:\n . golden color\n . tropical fruit\n . gold color\n . white burgundy\n . yellow color\n . Top trigrams:\n . light gold color\n . nice citrus nose\n . golden yellow color\n . tropical fruit notes\n . lot oak point\n# 'Merlot':\n . Top unigrams:\n . merlot\n . cherry\n . chocolate\n . merlots\n . plum\n . Top bigrams:\n . red fruit\n . nice balance\n . good merlot\n . berry fruit\n . wine really\n . Top trigrams:\n . color nice nose\n . red black fruit\n . reduction tart currant\n . aromas dark fruit\n . deep dark purple\n# 'Sauvignon Blanc':\n . Top unigrams:\n . grapefruit\n . sb\n . blanc\n . grass\n . sauvignon\n . Top bigrams:\n . sauvignon blanc\n . sauv blanc\n . new zealand\n . passion fruit\n . nz sb\n . Top trigrams:\n . tropical fruit flavors\n . second calmed little\n . night better second\n . better second calmed\n . little bit acidity\n"
],
[
"# Scores for each variant using a LinearSVC classifier with Tfid\n\nprint(metrics.classification_report(y_test, \n y_pred, \n target_names=df['wine_variant'].unique()))",
" precision recall f1-score support\n\nCabernet Sauvignon 0.73 0.74 0.74 351\n Chardonnay 0.85 0.85 0.85 350\n Sauvignon Blanc 0.90 0.83 0.86 304\n Merlot 0.67 0.70 0.69 315\n\n avg / total 0.79 0.78 0.79 1320\n\n"
]
],
[
[
"My best classifier (Linear SVC + tfid) struggles with classifying reviews in which the reviewer states key words that the actual wine variant lacks, whether the reviewer is uneducated or they are saying the identifying phrase in a negative (or lacking) manner. Also, generic reviews that do not include significantly unique characteristics will struggle, mainly due to the fact that wine shares a lot of characteristics between variants, but certain of those characteristics are more heavily present (on average) with specific variants. \n\nI have learned that classification can be done from string comparisions, statistics (logs), indexes, and various other measurable variables/attributes. The best way to ensure that classification is most successful is to combine the various models, depending from situation to situation. This wine prediction strongly supports the idea of vector and phrase classifications for multi-classes, leading to a need for identifying the distinguishing qualities of each class. Even though there are many variables and similarities between the variants, I found the beginning predictions to be quite easy and efficient. The main errors came from trying to distingusih merlot from cabernet, which does make sense since they are the closest related wines as far as shared features. As more and more data is collected, the dictionary of unique words grows in number and certainty in making a correct prediction. I do believe that a score better than 90% is achievable after subpar and uneducated reveiws are removed, tokenization further cleans reamining punctuation errors, and continued training data is supplied to the program to increase its correlation certainties. Also, to increase the accuracy, it would be advantageous to have the phrases with a negative or quantifying variable treated separate from the usage count. By recognizing the negative and quantifying variables as an un-splittable part of its modifying phrase, we ensure that certain features, mainly in the unigram category, aren't wrongly weighting that feauture for that variant. An example of this dilemma can be seen when comparing things like 'red' and 'not red,' which obviously mean a distinct difference, but if 'not' is allowed to separate from 'red,' non-red wine variants could get too much weight on the word 'red,' creating a higher chance for inaccuracies. ",
"_____no_output_____"
]
],
[
[
"\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbc9da360cbe2305009aabf337249f0a904dcf77
| 179,911 |
ipynb
|
Jupyter Notebook
|
criminology-portugal/Criminology in Portugal - 2011.ipynb
|
rubenandrebarreiro/studying-crimonology-worldwide
|
4a863eb1b829529fb55dd843a88b95513239fa90
|
[
"MIT"
] | 1 |
2019-07-20T21:36:07.000Z
|
2019-07-20T21:36:07.000Z
|
criminology-portugal/Criminology in Portugal - 2011.ipynb
|
rubenandrebarreiro/studying-crimonology-worldwide
|
4a863eb1b829529fb55dd843a88b95513239fa90
|
[
"MIT"
] | null | null | null |
criminology-portugal/Criminology in Portugal - 2011.ipynb
|
rubenandrebarreiro/studying-crimonology-worldwide
|
4a863eb1b829529fb55dd843a88b95513239fa90
|
[
"MIT"
] | null | null | null | 106.89899 | 30,700 | 0.749621 |
[
[
[
"# Criminology in Portugal (2011)\n\n## Introduction\n\n> In this _study case_, it will be analysed the **_crimes occurred_** in **_Portugal_**, during the civil year of **_2011_**. It will analysed all the _categories_ or _natures_ of this **_crimes_**, _building some statistics and making some filtering of data related to them_.\n\n> It will be applied some _filtering_ and made some _analysis_ on the data related to **_Portugal_** as _country_, like the following:\n* _Crimes by **Nature/Category**_\n* _Crimes by **Geographical Zone**_\n* _Crimes by **Region/City** (only considered th 5 more populated **regions/cities** in **Portugal**)_\n* _Conclusions_\n\n> It will be applied some _filtering_ and made some _analysis_ on the data related to the **_5 biggest/more populated regions/cities_** (_Metropolitan Area of Lisbon_, _North_, _Center_, _Metropolitan Area of Porto_, and _Algarve_) of **_Portugal_**, like the following:\n* **_Metropolitan Area of Lisbon_**\n * _Crimes by **Nature/Category**_\n * _Crimes by **Locality/Village**_\n * _Conclusions_\n* **_North_**\n * _Crimes by **Nature/Category**_\n * _Crimes by **Locality/Village**_\n * _Conclusions_\n* **_Center_**\n * _Crimes by **Nature/Category**_\n * _Crimes by **Locality/Village**_\n * _Conclusions_\n* **_Metropolitan Area of Porto_**\n * _Crimes by **Nature/Category**_\n * _Crimes by **Locality/Village**_\n * _Conclusions_\n* **_Algarve_**\n * _Crimes by **Nature/Category**_\n * _Crimes by **Locality/Village**_\n * _Conclusions_",
"_____no_output_____"
]
],
[
[
"# Importing pandas library\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\n\n%matplotlib inline",
"_____no_output_____"
],
[
"crimes_by_geozone_2011 = pd.read_csv(\"datasets/ine.pt/2011/dataset-crimes-portugal-2011-by-geozone-2.csv\" , header=1)\n\ncrimes_by_geozone_2011 = crimes_by_geozone_2011.rename(columns={'Unnamed: 0': 'Zona Geográfica'})\n\ncrimes_by_geozone_2011 = crimes_by_geozone_2011.set_index(\"Zona Geográfica\", drop = True)",
"_____no_output_____"
]
],
[
[
"#### Data Available in the Dataset\n\n> All the data available and used for this _study case_ can be found in the following _hyperlink_:\n* [dataset-crimes-portugal-2011-by-geozone-2.csv](datasets/ine.pt/2011/dataset-crimes-portugal-2011-by-geozone-2.csv)\n\n##### Note:\n> If you pretend to see all the data available and used for this _study case_, uncomment the following line.",
"_____no_output_____"
]
],
[
[
"# Just for debug\n#crimes_by_geozone_2011",
"_____no_output_____"
]
],
[
[
"## Starting the Study Case",
"_____no_output_____"
],
[
"### Criminology in **_Metropolitan Area of Lisbon_** (**_2011_**)",
"_____no_output_____"
],
[
"#### Analysing the **_crimes occurred_** in **_Metropolitan Area of Lisbon_**, during **_2011_**",
"_____no_output_____"
],
[
"* The total of **_crime occurrences_** in **_Metropolitan Area of Lisbon_**, during **_2011_**, filtered by _category or nature of the crime_:",
"_____no_output_____"
]
],
[
[
"crimes_lisbon_2011 = crimes_by_geozone_2011.loc[\"170: Área Metropolitana de Lisboa\", : ]\n\ncrimes_lisbon_2011",
"_____no_output_____"
]
],
[
[
"* The total number of **_crime occurrences_** in **_Metropolitan Area of Lisbon_**, during **_2011_**, filtered by _category or nature of the crime_ (_organised as a Data Frame_):",
"_____no_output_____"
]
],
[
[
"crimes_lisbon_2011 = pd.DataFrame(crimes_lisbon_2011).T\n\ncrimes_lisbon_2011 = crimes_lisbon_2011.iloc[:,1:8]\n\ncrimes_lisbon_2011",
"_____no_output_____"
],
[
"# Just for debug\n#crimes_lisbon_2011.columns",
"_____no_output_____"
],
[
"crimes_lisbon_2011.values[0,3] = 0\n\ncrimes_lisbon_2011.values[0,6] = 0\n\ncrimes_lisbon_2011.values[0,0] = int(crimes_lisbon_2011.values[0,0])\ncrimes_lisbon_2011.values[0,1] = int(float(crimes_lisbon_2011.values[0,1]))\ncrimes_lisbon_2011.values[0,2] = int(crimes_lisbon_2011.values[0,2])\ncrimes_lisbon_2011.values[0,4] = int(float(crimes_lisbon_2011.values[0,4]))\ncrimes_lisbon_2011.values[0,5] = int(float(crimes_lisbon_2011.values[0,5]))\n\n# Just for debug\n#crimes_lisbon_2011.values",
"_____no_output_____"
],
[
"# Just for debug\n#crimes_lisbon_2011",
"_____no_output_____"
]
],
[
[
"* The total number of **_crime occurrences_** in **_Metropolitan Area of Lisbon_**, during **_2011_**, filtered by _category or nature of the crime_ (_organised as a Data Frame_ and excluding some _redundant fields and data_):",
"_____no_output_____"
]
],
[
[
"del crimes_lisbon_2011['Crimes contra a identidade cultural e integridade pessoal']\ndel crimes_lisbon_2011['Crimes contra animais de companhia']\n\ncrimes_lisbon_2011",
"_____no_output_____"
],
[
"crimes_lisbon_2011_categories = crimes_lisbon_2011.columns.tolist()\n\n# Just for debug\n#crimes_lisbon_2011_categories",
"_____no_output_____"
],
[
"crimes_lisbon_2011_values = crimes_lisbon_2011.values[0].tolist()\n\n# Just for debug\n#crimes_lisbon_2011_values",
"_____no_output_____"
]
],
[
[
"* A _plot_ of a representation of the total of **_crime occurrences_** in **_Metropolitan Area of Lisbon_**, during **_2011_**, filtered by _category or nature of the crime_ (_organised as a Data Frame_ and excluding some _redundant fields and data_):",
"_____no_output_____"
]
],
[
[
"plt.bar(crimes_lisbon_2011_categories, crimes_lisbon_2011_values)\n\nplt.xticks(crimes_lisbon_2011_categories, rotation='vertical')\n\nplt.xlabel('\\nCrime Category/Nature\\n')\nplt.ylabel('\\nNum. Occurrences\\n')\n\nplt.title('Crimes in Metropolitan Area of Lisbon, during 2011 (by Crime Category/Nature) - Bars Chart\\n')\n\nprint('\\n')\n\nplt.show()",
"\n\n"
],
[
"plt.pie(crimes_lisbon_2011_values, labels=crimes_lisbon_2011_categories, autopct='%.2f%%')\n\nplt.title('Crimes in Metropolitan Area of Lisbon, during 2011 (by Crime Category/Nature) - Pie Chart\\n\\n')\nplt.axis('equal')\n\nprint('\\n')\n\nplt.show()",
"\n\n"
]
],
[
[
"* The total number of **_crime occurrences_** in all the **_localities/villages_** of the **_Metropolitan Area of Lisbon_**, during **_2011_**, filtered by _category or nature of the crime_ (_organised as a Data Frame_):",
"_____no_output_____"
]
],
[
[
"crimes_lisbon_2011_by_locality = crimes_by_geozone_2011.loc[\"1701502: Alcochete\":\"1701114: Vila Franca de Xira\", : ]\n\ncrimes_lisbon_2011_by_locality",
"_____no_output_____"
]
],
[
[
"* The total number of **_crime occurrences_** in all the **_localities/villages_** of the **_Metropolitan Area of Lisbon_**, during **_2011_**, filtered by _category or nature of the crime_ (_organised as a Data Frame_ and excluding some _redundant fields and data_):",
"_____no_output_____"
]
],
[
[
"del crimes_lisbon_2011_by_locality['Crimes de homicídio voluntário consumado']\ndel crimes_lisbon_2011_by_locality['Crimes contra a identidade cultural e integridade pessoal']\ndel crimes_lisbon_2011_by_locality['Crimes contra animais de companhia']\n\ncrimes_lisbon_2011_by_locality",
"_____no_output_____"
],
[
"top_6_crimes_lisbon_2011_by_locality = crimes_lisbon_2011_by_locality.sort_values(by='Total', ascending=False).head(6)\n\ntop_6_crimes_lisbon_2011_by_locality",
"_____no_output_____"
],
[
"top_6_crimes_lisbon_2011_by_locality_total = top_6_crimes_lisbon_2011_by_locality.loc[:,\"Total\"]\n\ntop_6_crimes_lisbon_2011_by_locality_total",
"_____no_output_____"
],
[
"top_6_crimes_lisbon_2011_by_locality_total = pd.DataFrame(top_6_crimes_lisbon_2011_by_locality_total).T\n\ntop_6_crimes_lisbon_2011_by_locality_total = top_6_crimes_lisbon_2011_by_locality_total.iloc[:,0:6]\n\ntop_6_crimes_lisbon_2011_by_locality_total",
"_____no_output_____"
],
[
"top_6_crimes_lisbon_2011_by_locality_total_localities = top_6_crimes_lisbon_2011_by_locality_total.columns.tolist()\n\n# Just for debug\n#top_6_crimes_lisbon_2011_by_locality_total_localities",
"_____no_output_____"
],
[
"top_6_crimes_lisbon_2011_by_locality_total_values = top_6_crimes_lisbon_2011_by_locality_total.values[0].tolist()\n\n# Just for debug\n#top_6_crimes_lisbon_2011_by_locality_total_values",
"_____no_output_____"
],
[
"plt.bar(top_6_crimes_lisbon_2011_by_locality_total_localities, top_6_crimes_lisbon_2011_by_locality_total_values)\n\nplt.xticks(top_6_crimes_lisbon_2011_by_locality_total_localities, rotation='vertical')\n\nplt.xlabel('\\nLocality/Village')\nplt.ylabel('\\nNum. Occurrences\\n')\n\nplt.title('Crimes in Metropolitan Area of Lisbon, during 2011 (by Locality/Village in Top 6) - Bars Chart\\n')\n\nprint('\\n')\n\nplt.show()",
"\n\n"
],
[
"plt.pie(top_6_crimes_lisbon_2011_by_locality_total_values, labels=top_6_crimes_lisbon_2011_by_locality_total_localities, autopct='%.2f%%')\n\nplt.title('Crimes in Metropolitan Area of Lisbon, during 2011 (by Locality/Village in Top 6) - Pie Chart\\n\\n')\nplt.axis('equal')\n\nprint('\\n')\n\nplt.show()",
"\n\n"
]
],
[
[
"#### Conclusion of the **_crimes occurred_** in **_Metropolitan Area of Lisbon_**, during **_2011_**",
"_____no_output_____"
],
[
"* After studying all the perspectives about the **_crimes occurred_** in **_Metropolitan Area of Lisbon_**, during **_2011_**, it's possible to conclude the following:\n\n * a) The most of the **_crimes_** occurred was against:\n\n > 1) The **_country's patrimony_** (**68.52%**)\n\n > 2) The **_people_**, at general (**20.35%**)\n\n > 3) The **_life in society_** (**9.32%**)\n\n\n",
"_____no_output_____"
],
[
"Thank you, and I hope you enjoy it!\n\nSincerely,\n> Rúben André Barreiro.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
cbc9f2c4ea9b821e8f57ba4e8dcfc9145e645fa7
| 1,910 |
ipynb
|
Jupyter Notebook
|
Keras_XOR_.ipynb
|
sunilkami/AI-ML-DL-NN
|
f503dc051b8b90e0a0faf453aca9cc380ea81d8d
|
[
"MIT"
] | null | null | null |
Keras_XOR_.ipynb
|
sunilkami/AI-ML-DL-NN
|
f503dc051b8b90e0a0faf453aca9cc380ea81d8d
|
[
"MIT"
] | null | null | null |
Keras_XOR_.ipynb
|
sunilkami/AI-ML-DL-NN
|
f503dc051b8b90e0a0faf453aca9cc380ea81d8d
|
[
"MIT"
] | 1 |
2021-05-29T05:52:56.000Z
|
2021-05-29T05:52:56.000Z
| 30.31746 | 241 | 0.512042 |
[
[
[
"<a href=\"https://colab.research.google.com/github/ShubhInfotech-Bhilai/AI-ML-DL-NN/blob/master/Keras_XOR_.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom keras.models import Sequential\nfrom keras.layers.core import Dense\n\n# the four different states of the XOR gate\ntraining_data = np.array([[0,0],[0,1],[1,0],[1,1]], \"float32\")\n\n# the four expected results in the same order\ntarget_data = np.array([[0],[1],[1],[0]], \"float32\")\n\nmodel = Sequential()\nmodel.add(Dense(16, input_dim=2, activation='relu'))\nmodel.add(Dense(1, activation='sigmoid'))\n\nmodel.compile(loss='mean_squared_error',\n optimizer='adam',\n metrics=['binary_accuracy'])\n\nmodel.fit(training_data, target_data, epochs=500, verbose=2)\n\nprint( model.predict(training_data).round())",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
]
] |
cbc9f343cbef2d52650e11332d96d3c77a9a9475
| 16,067 |
ipynb
|
Jupyter Notebook
|
Old-k20-model/kitchen20/training_script.ipynb
|
kevinmicha/kitchen20
|
20290451fa3ddc5b45f212b8d9f8cd4076e57047
|
[
"Apache-2.0"
] | null | null | null |
Old-k20-model/kitchen20/training_script.ipynb
|
kevinmicha/kitchen20
|
20290451fa3ddc5b45f212b8d9f8cd4076e57047
|
[
"Apache-2.0"
] | 8 |
2021-11-25T11:00:44.000Z
|
2022-03-24T08:28:53.000Z
|
Old-k20-model/kitchen20/training_script.ipynb
|
kevinmicha/kitchen20
|
20290451fa3ddc5b45f212b8d9f8cd4076e57047
|
[
"Apache-2.0"
] | 1 |
2022-03-24T08:05:33.000Z
|
2022-03-24T08:05:33.000Z
| 16,067 | 16,067 | 0.691044 |
[
[
[
"!pip install torch torchvision\n!pip install wavio\n!pip install sounddevice",
"Requirement already satisfied: torch in /usr/local/lib/python3.7/dist-packages (1.10.0+cu111)\nRequirement already satisfied: torchvision in /usr/local/lib/python3.7/dist-packages (0.11.1+cu111)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from torch) (3.10.0.2)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from torchvision) (1.19.5)\nRequirement already satisfied: pillow!=8.3.0,>=5.3.0 in /usr/local/lib/python3.7/dist-packages (from torchvision) (7.1.2)\nRequirement already satisfied: wavio in /usr/local/lib/python3.7/dist-packages (0.0.4)\nRequirement already satisfied: numpy>=1.6.0 in /usr/local/lib/python3.7/dist-packages (from wavio) (1.19.5)\nRequirement already satisfied: sounddevice in /usr/local/lib/python3.7/dist-packages (0.4.3)\nRequirement already satisfied: CFFI>=1.0 in /usr/local/lib/python3.7/dist-packages (from sounddevice) (1.15.0)\nRequirement already satisfied: pycparser in /usr/local/lib/python3.7/dist-packages (from CFFI>=1.0->sounddevice) (2.21)\n"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')\n\n!ls \"/content/drive/My Drive/IMT Atlantique/Projet 3A /master/kitchen20\"\n%cd /content/drive/My Drive/IMT Atlantique/Projet 3A /master/kitchen20",
"Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount(\"/content/drive\", force_remount=True).\n audioUtils.py\t\t envnet.pyc\t kitch_env\t\t transforms.py\n config.ini\t\t esc.py\t __pycache__\t\t utils.py\n'Copie de kitchen20.py' __init__.py\t training.py\n envnet.py\t\t kitchen20.py\t training_script.ipynb\n/content/drive/My Drive/IMT Atlantique/Projet 3A /master/kitchen20\n"
],
[
"from envnet import EnvNet\nfrom kitchen20 import Kitchen20\nfrom torch.utils.data import DataLoader\nimport torch.nn as nn\nimport utils as U\nimport torch\n\n\n# Model\nmodel = EnvNet(20, True)\nmodel.cuda()\ncriterion = nn.CrossEntropyLoss()\noptimizer = torch.optim.SGD(model.parameters(), lr=1e-2)\n\n\n# Dataset\nbatchSize = 32\ninputLength = 48000\ntransforms = []\ntransforms += [U.random_scale(1.25)] # Strong augment\ntransforms += [U.padding(inputLength // 2)] # Padding\ntransforms += [U.random_crop(inputLength)] # Random crop\ntransforms += [U.normalize(float(2 ** 16 / 2))] # 16 bit signed\ntransforms += [U.random_flip()] # Random +-\n\ntrainData = Kitchen20(root='../',\n transforms=transforms,\n folds=[1,2,3,4,5,6,7,8],\n overwrite=False,\n audio_rate=44100,\n use_bc_learning=False)\ntrainIter = DataLoader(trainData, batch_size=batchSize,\n shuffle=True, num_workers=2)\n\n\ninputLength = 64000\ntransforms = []\ntransforms += [U.padding(inputLength // 2)] # Padding\ntransforms += [U.random_crop(inputLength)] # Random crop\ntransforms += [U.normalize(float(2 ** 16 / 2))] # 16 bit signed\ntransforms += [U.random_flip()] # Random +-\n\nvalData = Kitchen20(root='../',\n transforms=transforms,\n folds=[9,],\n audio_rate=44100,\n overwrite=False,\n use_bc_learning=False)\nvalIter = DataLoader(valData, batch_size=batchSize,\n shuffle=True, num_workers=2)\n\n\nfor epoch in range(600):\n tAcc = tLoss = 0\n vAcc = vLoss = 0\n for x, y in trainIter: # Train epoch\n model.train()\n x = x[:, None, None, :]\n x = x.to('cuda:0')\n y = y.to('cuda:0')\n # Forward pass: Compute predicted y by passing x to the model\n y_pred = model(x)\n y_pred = y_pred[:, :, 0, 0]\n\n # Compute and print loss\n loss = criterion(y_pred, y.long())\n acc = (y_pred.argmax(dim=1).long() == y.long()).sum()\n\n # Zero gradients, perform a backward pass, and update the weights.\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n\n tLoss += loss.item()\n tAcc += acc.item()/len(trainData)\n\n for x, y in valIter: # Test epoch\n model.eval()\n x = x[:, None, None, :]\n x = x.to('cuda:0')\n y = y.to('cuda:0')\n # Forward pass: Compute predicted y by passing x to the model\n y_pred = model(x)\n y_pred = y_pred[:, :, 0, 0]\n loss = criterion(y_pred, y.long())\n acc = (y_pred.argmax(dim=1).long() == y.long()).sum()\n vLoss += loss.item()\n vAcc += acc.item()/len(valData)\n\n\n # loss = loss / len(dataset)\n # acc = acc / float(len(dataset))\n print('epoch {} -- train: {}/{} -- val:{}/{}'.format(\n epoch, tAcc, tLoss, vAcc, vLoss))",
"epoch 0 -- train: 0.12181303116147313/97.97092843055725 -- val:0.0/3.187875509262085\nepoch 1 -- train: 0.15675165250236073/94.00973343849182 -- val:0.0/3.1697351932525635\nepoch 2 -- train: 0.2181303116147309/91.95808124542236 -- val:0.0/3.1046066284179688\nepoch 3 -- train: 0.2341831916902738/88.9431459903717 -- val:0.0/3.0460920333862305\nepoch 4 -- train: 0.22096317280453262/88.35655999183655 -- val:0.0/3.2392897605895996\nepoch 5 -- train: 0.22474032105760147/87.95760774612427 -- val:0.0/3.060206651687622\nepoch 6 -- train: 0.25212464589235123/86.58423566818237 -- val:0.09090909090909091/3.020418643951416\nepoch 7 -- train: 0.23984891406987724/85.94967555999756 -- val:0.0/3.058861494064331\nepoch 8 -- train: 0.2587346553352219/85.23419046401978 -- val:0.09090909090909091/2.9845855236053467\nepoch 9 -- train: 0.28045325779036834/83.57538199424744 -- val:0.09090909090909091/3.025662422180176\nepoch 10 -- train: 0.2672332389046271/83.15415334701538 -- val:0.18181818181818182/3.059786081314087\nepoch 11 -- train: 0.2832861189801699/83.64054203033447 -- val:0.0/3.4704513549804688\nepoch 12 -- train: 0.28989612842304063/83.41219472885132 -- val:0.09090909090909091/2.872743844985962\nepoch 13 -- train: 0.3002832861189801/82.20750951766968 -- val:0.0/3.416393280029297\nepoch 14 -- train: 0.29745042492917845/82.02136540412903 -- val:0.18181818181818182/2.902827501296997\nepoch 15 -- train: 0.32200188857412654/81.13914394378662 -- val:0.09090909090909091/3.028014898300171\nepoch 16 -- train: 0.30878186968838534/80.96466517448425 -- val:0.18181818181818182/2.9827158451080322\nepoch 17 -- train: 0.3021718602455147/80.38183355331421 -- val:0.0/3.171856641769409\nepoch 18 -- train: 0.31255901794145424/80.28044271469116 -- val:0.0/3.180346727371216\nepoch 19 -- train: 0.2898961284230407/80.6487455368042 -- val:0.0/3.0260045528411865\nepoch 20 -- train: 0.32577903682719556/78.67753052711487 -- val:0.0/3.132235050201416\nepoch 21 -- train: 0.31916902738432484/79.15378284454346 -- val:0.09090909090909091/2.8326916694641113\nepoch 22 -- train: 0.33899905571293676/78.86282205581665 -- val:0.18181818181818182/2.7451322078704834\nepoch 23 -- train: 0.32011331444759195/78.66623330116272 -- val:0.18181818181818182/2.7057619094848633\nepoch 24 -- train: 0.32861189801699714/78.31696283817291 -- val:0.09090909090909091/2.9312524795532227\nepoch 25 -- train: 0.3305004721435317/78.01028180122375 -- val:0.18181818181818182/2.904731035232544\nepoch 26 -- train: 0.3673276676109538/76.05531537532806 -- val:0.18181818181818182/2.7527639865875244\nepoch 27 -- train: 0.3418319169027384/76.73610639572144 -- val:0.0/2.7431089878082275\nepoch 28 -- train: 0.35410764872521255/75.75055360794067 -- val:0.36363636363636365/2.462319850921631\nepoch 29 -- train: 0.35221907459867796/76.41767501831055 -- val:0.2727272727272727/2.4759721755981445\nepoch 30 -- train: 0.3512747875354108/76.37492430210114 -- val:0.09090909090909091/2.699296474456787\nepoch 31 -- train: 0.35599622285174704/76.33455348014832 -- val:0.18181818181818182/2.843254804611206\nepoch 32 -- train: 0.38149197355996217/75.07489621639252 -- val:0.18181818181818182/2.711885452270508\nepoch 33 -- train: 0.35410764872521255/74.97323966026306 -- val:0.0/2.840402841567993\nepoch 34 -- train: 0.38149197355996217/74.21288025379181 -- val:0.36363636363636365/2.2271344661712646\nepoch 35 -- train: 0.3692162417374882/75.43889880180359 -- val:0.2727272727272727/2.7112581729888916\nepoch 36 -- train: 0.380547686496695/74.40289449691772 -- val:0.45454545454545453/2.50362491607666\nepoch 37 -- train: 0.37960339943342775/73.41236245632172 -- val:0.36363636363636365/2.729421377182007\nepoch 38 -- train: 0.3729933899905571/73.33772647380829 -- val:0.18181818181818182/2.4002199172973633\nepoch 39 -- train: 0.380547686496695/73.58163237571716 -- val:0.36363636363636365/2.7423362731933594\nepoch 40 -- train: 0.34560906515580736/74.29612827301025 -- val:0.45454545454545453/2.0393266677856445\nepoch 41 -- train: 0.3748819641170917/73.52354204654694 -- val:0.2727272727272727/2.309627056121826\nepoch 42 -- train: 0.3616619452313503/73.46115028858185 -- val:0.2727272727272727/2.657866954803467\nepoch 43 -- train: 0.38715769593956556/72.54269027709961 -- val:0.45454545454545453/2.145785093307495\nepoch 44 -- train: 0.3644948064211521/72.95265483856201 -- val:0.18181818181818182/3.055500030517578\nepoch 45 -- train: 0.3786591123701604/72.68001461029053 -- val:0.18181818181818182/2.714223861694336\nepoch 46 -- train: 0.37488196411709157/71.68151068687439 -- val:0.0/2.781144618988037\nepoch 47 -- train: 0.38243626062322944/71.73811388015747 -- val:0.2727272727272727/2.5556812286376953\nepoch 48 -- train: 0.37110481586402255/72.43550848960876 -- val:0.09090909090909091/2.6619157791137695\nepoch 49 -- train: 0.376770538243626/72.64651453495026 -- val:0.09090909090909091/2.947298526763916\nepoch 50 -- train: 0.39565627950897075/71.53214168548584 -- val:0.09090909090909091/2.930819511413574\nepoch 51 -- train: 0.3852691218130313/70.45468926429749 -- val:0.18181818181818182/3.5568060874938965\nepoch 52 -- train: 0.3862134088762984/72.1697770357132 -- val:0.2727272727272727/2.5803451538085938\nepoch 53 -- train: 0.3777148253068934/71.39509272575378 -- val:0.7272727272727273/1.8067984580993652\nepoch 54 -- train: 0.37204910292728993/70.99451231956482 -- val:0.2727272727272727/2.3066396713256836\nepoch 55 -- train: 0.37393767705382436/71.57057964801788 -- val:0.36363636363636365/2.7376952171325684\nepoch 56 -- train: 0.37488196411709157/71.01202774047852 -- val:0.09090909090909091/2.968686103820801\nepoch 57 -- train: 0.3805476864966949/71.35291814804077 -- val:0.09090909090909091/2.7508857250213623\nepoch 58 -- train: 0.38715769593956567/71.20707619190216 -- val:0.18181818181818182/2.469641923904419\nepoch 59 -- train: 0.38621340887629835/69.27867317199707 -- val:0.18181818181818182/2.637190580368042\nepoch 60 -- train: 0.4013220018885741/69.73752450942993 -- val:0.09090909090909091/2.668821334838867\nepoch 61 -- train: 0.43437204910292715/68.56845200061798 -- val:0.09090909090909091/2.8345232009887695\nepoch 62 -- train: 0.4211520302171861/68.3389595746994 -- val:0.2727272727272727/3.145677328109741\nepoch 63 -- train: 0.4003777148253068/69.42544376850128 -- val:0.36363636363636365/2.1436607837677\nepoch 64 -- train: 0.4126534466477809/67.94003248214722 -- val:0.2727272727272727/2.5086171627044678\nepoch 65 -- train: 0.40887629839471207/68.02043533325195 -- val:0.45454545454545453/2.0190937519073486\nepoch 66 -- train: 0.408876298394712/67.94542229175568 -- val:0.2727272727272727/2.1100358963012695\nepoch 67 -- train: 0.39848914069877245/68.31405460834503 -- val:0.09090909090909091/2.693007469177246\nepoch 68 -- train: 0.396600566572238/68.59649908542633 -- val:0.0/3.0592544078826904\n"
],
[
"testData = Kitchen20(root='../',\n transforms=transforms,\n folds=[10,],\n audio_rate=44100,\n overwrite=False,\n use_bc_learning=False)\ntestIter = DataLoader(testData, batch_size=1,\n shuffle=True, num_workers=2)",
"_____no_output_____"
],
[
"testAcc = 0\n \nfor x, y in testIter: # Test epoch\n model.eval()\n x = x[:, None, None, :]\n x = x.to('cuda:0')\n y = y.to('cuda:0')\n # Forward pass: Compute predicted y by passing x to the model\n y_test = model(x)\n y_test = y_test[:, :, 0, 0]\n print(y_test)\n print(y_test.argmax(dim=1))\n #loss = criterion(y_pred, y.long())\n acc = (y_test.argmax(dim=1).long() == y.long()).sum()\n #vLoss += loss.item()\n testAcc += acc.item()/len(testData)",
"_____no_output_____"
],
[
"testAcc",
"_____no_output_____"
],
[
"len(testData)",
"_____no_output_____"
],
[
"y",
"_____no_output_____"
],
[
"y_pred.argmax(dim=1).long()",
"_____no_output_____"
],
[
"import numpy as np\n\ndata = np.load('../audio/44100.npz', allow_pickle=True)\nlst = data.files\n\nfor item in lst:\n print(item)\n print(data[item])",
"_____no_output_____"
],
[
"len(trainData)",
"_____no_output_____"
],
[
"acc.item()",
"_____no_output_____"
],
[
"for i in range(10):\n print(len(data[data.files[i]].item()['sounds']))",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbca089a3a5954c6e09ea704fa3440efedaa7e4a
| 275,865 |
ipynb
|
Jupyter Notebook
|
ensemble_flight.ipynb
|
SandyGuru/TeamFunFinalProject
|
0bcbdb32e7212423f9f94df489026041f00c8bbd
|
[
"Apache-2.0"
] | null | null | null |
ensemble_flight.ipynb
|
SandyGuru/TeamFunFinalProject
|
0bcbdb32e7212423f9f94df489026041f00c8bbd
|
[
"Apache-2.0"
] | null | null | null |
ensemble_flight.ipynb
|
SandyGuru/TeamFunFinalProject
|
0bcbdb32e7212423f9f94df489026041f00c8bbd
|
[
"Apache-2.0"
] | null | null | null | 694.874055 | 264,284 | 0.947877 |
[
[
[
"from sklearn import *\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.datasets import load_iris\nfrom sklearn.ensemble import (RandomForestClassifier, ExtraTreesClassifier,\n AdaBoostClassifier)\nfrom sklearn.tree import DecisionTreeClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.feature_selection import RFE\n\n\n\n%pylab",
"Using matplotlib backend: Qt5Agg\nPopulating the interactive namespace from numpy and matplotlib\n"
],
[
"import numpy as np\n%matplotlib inline\nimport matplotlib.pyplot as plt\nimport seaborn as sns #for graphics and figure styling\nimport pandas as pd\nfrom matplotlib.colors import ListedColormap",
"_____no_output_____"
],
[
"data = pd.read_csv('E:/Stony Brook/AMS560/Data/FlightDelay2018.csv')\ndata=data[1:50000]\ndata.DepDelayMinutes.fillna(1)\ndata.DepDelayMinutes[data.DepDelayMinutes!=0]=1",
"D:\\Anaconda\\lib\\site-packages\\IPython\\core\\interactiveshell.py:2728: DtypeWarning: Columns (77,84) have mixed types. Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"from collections import defaultdict\na=0\nb=0\nmissing=defaultdict(int)\nfor col in data:\n for i in data[col].isnull():\n if i:\n a+=1\n b+=1\n #print('Missing data in',col,'is',a/b*100,'%')\n missing[col]=a/b*100\n a=0\n b=0\nmissing['Year']\n\nfor col in data:\n if missing[col]>5:\n data=data.drop(col, axis=1)",
"_____no_output_____"
],
[
"data=data.drop('TailNum',axis=1)\nenc = LabelEncoder()\ndata = data.apply(LabelEncoder().fit_transform)",
"_____no_output_____"
],
[
"depDelayColumn = data.DepDelayMinutes\ndata = data.drop('DepDelayMinutes', axis=1)\ndata = data.drop('DepDelay', axis=1)\ndata = data.drop(['CRSDepTime','DepTime','DepartureDelayGroups'], axis=1)\n\n",
"_____no_output_____"
],
[
"data_train, data_test, y_train, y_test = train_test_split(data, depDelayColumn, test_size=.3)\nscaler = StandardScaler().fit(data)\nstandard_data_test = scaler.transform(data_test)\n\nscaler = StandardScaler().fit(data_train)\nstandard_data = scaler.transform(data_train)",
"_____no_output_____"
],
[
"#Using the Random Forest Classifier on our Data, with depth 3.\ndepth=3;\nn_features=5;\ncensusIDM = RandomForestClassifier(max_depth=depth, random_state=0)\nfrfe = RFE(censusIDM, n_features_to_select=n_features)\nfrfe.fit(data_train, y_train)\nprint(frfe.ranking_)\nfrfe.score(data_test, y_test)",
"[46 45 44 12 20 1 19 15 14 10 13 25 26 11 22 29 28 21 9 30 32 33 36 38\n 40 42 43 41 1 16 3 2 5 8 7 4 1 1 6 1 17 18 23 24 27 31 34 35\n 37 39]\n"
],
[
"feature_to_select=[0]*n_features\nj=0\nfor i in range(len(frfe.ranking_)):\n if frfe.ranking_[i]==1:\n feature_to_select[j]=i\n j=j+1\nprint(feature_to_select)",
"[5, 28, 36, 37, 39]\n"
],
[
"data.columns[36]",
"_____no_output_____"
],
[
"# Parameters\nn_classes = 2\nn_estimators = 30\ncmap = plt.cm.RdYlBu\nplot_step = 0.02 # fine step width for decision surface contours\nplot_step_coarser = 0.5 # step widths for coarse classifier guesses\nRANDOM_SEED = 13 # fix the seed on each iteration\n\nfig=plt.figure(figsize=[15,5])\nplt.subplot(1,3, 1)\n\nf1=[36,28]\nf2=[5,36]\nf3=[5,28]\n\n#X=standardized_test_data[:,[0,4]];\nX=standard_data[:,f1];\ny=y_train\n\nfrfe.fit(X, y)\nprint(frfe.score(standard_data_test[:,f1], y_test))\n\nmean = X.mean(axis=0)\nstd = X.std(axis=0)\nX = (X - mean) / std\n\n # Now plot the decision boundary using a fine mesh as input to a\n # filled contour plot\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nxx, yy = np.meshgrid(np.arange(x_min, x_max, plot_step),\n np.arange(y_min, y_max, plot_step))\n\nZ = frfe.predict(np.c_[xx.ravel(), yy.ravel()])\nZ = Z.reshape(xx.shape)\ncs = plt.contourf(xx, yy, Z, cmap=cmap)\n \nxx_coarser, yy_coarser = np.meshgrid(\n np.arange(x_min, x_max, plot_step_coarser),\n np.arange(y_min, y_max, plot_step_coarser))\nZ_points_coarser = frfe.predict(np.c_[xx_coarser.ravel(),yy_coarser.ravel()]).reshape(xx_coarser.shape)\ncs_points = plt.scatter(xx_coarser, yy_coarser, s=15,c=Z_points_coarser, cmap=cmap, edgecolors=\"none\")\n\n # Plot the training points, these are clustered together and have a\n # black outline\nplt.scatter(X[:, 0], X[:, 1], c=y,\n cmap=ListedColormap(['r', 'y', 'b']),\n edgecolor='k', s=20)\nxlabel('ArrDelay')\nylabel('DepDel15')\n\n\nplt.subplot(1,3,2)\nX=standard_data[:,f2];\n\nfrfe.fit(X, y)\nprint(frfe.score(standard_data_test[:,f2], y_test))\n\nmean = X.mean(axis=0)\nstd = X.std(axis=0)\nX = (X - mean) / std\n\n # Now plot the decision boundary using a fine mesh as input to a\n # filled contour plot\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nxx, yy = np.meshgrid(np.arange(x_min, x_max, plot_step),\n np.arange(y_min, y_max, plot_step))\n\nZ = frfe.predict(np.c_[xx.ravel(), yy.ravel()])\nZ = Z.reshape(xx.shape)\ncs = plt.contourf(xx, yy, Z, cmap=cmap)\n \nxx_coarser, yy_coarser = np.meshgrid(\n np.arange(x_min, x_max, plot_step_coarser),\n np.arange(y_min, y_max, plot_step_coarser))\nZ_points_coarser = frfe.predict(np.c_[xx_coarser.ravel(),yy_coarser.ravel()]).reshape(xx_coarser.shape)\ncs_points = plt.scatter(xx_coarser, yy_coarser, s=15,c=Z_points_coarser, cmap=cmap, edgecolors=\"none\")\n\n # Plot the training points, these are clustered together and have a\n # black outline\nplt.scatter(X[:, 0], X[:, 1], c=y,\n cmap=ListedColormap(['r', 'y', 'b']),\n edgecolor='k', s=20)\nxlabel('DayOfWeek')\nylabel('ArrDelay')\n\n\nplt.subplot(1,3,3)\nX=standard_data[:,f3];\n\nfrfe.fit(X, y)\nprint(frfe.score(standard_data_test[:,f3], y_test))\n\nmean = X.mean(axis=0)\nstd = X.std(axis=0)\nX = (X - mean) / std\n\n # Now plot the decision boundary using a fine mesh as input to a\n # filled contour plot\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nxx, yy = np.meshgrid(np.arange(x_min, x_max, plot_step),\n np.arange(y_min, y_max, plot_step))\n\nZ = frfe.predict(np.c_[xx.ravel(), yy.ravel()])\nZ = Z.reshape(xx.shape)\ncs = plt.contourf(xx, yy, Z, cmap=cmap)\n \nxx_coarser, yy_coarser = np.meshgrid(\n np.arange(x_min, x_max, plot_step_coarser),\n np.arange(y_min, y_max, plot_step_coarser))\nZ_points_coarser = frfe.predict(np.c_[xx_coarser.ravel(),yy_coarser.ravel()]).reshape(xx_coarser.shape)\ncs_points = plt.scatter(xx_coarser, yy_coarser, s=15,c=Z_points_coarser, cmap=cmap, edgecolors=\"none\")\n\n # Plot the training points, these are clustered together and have a\n # black outline\nplt.scatter(X[:, 0], X[:, 1], c=y,\n cmap=ListedColormap(['r', 'y', 'b']),\n edgecolor='k', s=20)\nxlabel('DayOfWeek')\nylabel('DepDel15')\n\nplt.suptitle('RandomForestTree model on feature subsets ');\n\n#fig.savefig('RandomForest.pdf',dpi=200)",
"0.8458666666666667\n0.8132666666666667\n0.8464666666666667\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbca0a00a7cbddda1a9c15e5f1d572eb7dc912d2
| 14,059 |
ipynb
|
Jupyter Notebook
|
employee data analysis.ipynb
|
DHANIDHI/SQL-challenge
|
721fcbb1a5e70e5574e1c6ae707b52f103745e50
|
[
"ADSL"
] | null | null | null |
employee data analysis.ipynb
|
DHANIDHI/SQL-challenge
|
721fcbb1a5e70e5574e1c6ae707b52f103745e50
|
[
"ADSL"
] | null | null | null |
employee data analysis.ipynb
|
DHANIDHI/SQL-challenge
|
721fcbb1a5e70e5574e1c6ae707b52f103745e50
|
[
"ADSL"
] | null | null | null | 55.350394 | 1,876 | 0.605022 |
[
[
[
"from sqlalchemy import create_engine\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np\nfrom config import DB_USER, DB_PASS",
"_____no_output_____"
],
[
"engine = create_engine(f\"postgresql://{DB_USER}:{DB_PASS}@localhost/employee_db\")\nconn = engine.connect()",
"_____no_output_____"
],
[
"salaries = pd.read_sql(\"SELECT * FROM salaries\", conn)\nsalaries.head()",
"_____no_output_____"
],
[
"titles = pd.read_sql(\"SELECT * FROM titles\", conn)\ntitles.head()",
"_____no_output_____"
],
[
"merged = pd.merge(salaries, titles, on=\"emp_no\", how=\"inner\")\nmerged.head()",
"_____no_output_____"
],
[
"grouped = merged.groupby(\"title\").mean()\ngrouped",
"_____no_output_____"
],
[
"title_salary_df = grouped.drop(columns = \"emp_no\")\ntitle_salary_df",
"_____no_output_____"
],
[
"title_salary_df = title_salary_df.reset_index()\ntitle_salary_df",
"_____no_output_____"
],
[
"x_axis = title_salary_df[\"title\"]\nticks = np.arange(len(x_axis))\ny_axis = title_salary_df[\"salary\"]\n \nplt.bar(x_axis, y_axis, align=\"center\", alpha=1.0, color=[\"pink\", \"b\", \"r\", \"orange\", \"y\", \"b\", \"g\"])\n\nplt.xticks(ticks, x_axis, rotation=\"vertical\")\n\nplt.ylabel(\"Salaries ($)\")\nplt.xlabel(\"Employee Title\")\nplt.title(\"Average Employee Salary by Title\")\n\nplt.savefig(\"../Images/avg_salary_by_title.png\")\n\nplt.show()\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbca28f72de2914f9dd0c86887bb0185ad10e9db
| 181 |
ipynb
|
Jupyter Notebook
|
.ipynb_checkpoints/Untitled0-checkpoint.ipynb
|
andreqi/coh-metrix-experimetation
|
c259799a82226486516d6bcb1532d3fb45306c80
|
[
"MIT"
] | 1 |
2019-06-04T13:55:26.000Z
|
2019-06-04T13:55:26.000Z
|
.ipynb_checkpoints/Untitled0-checkpoint.ipynb
|
andreqi/coh-metrix-experimetation
|
c259799a82226486516d6bcb1532d3fb45306c80
|
[
"MIT"
] | null | null | null |
.ipynb_checkpoints/Untitled0-checkpoint.ipynb
|
andreqi/coh-metrix-experimetation
|
c259799a82226486516d6bcb1532d3fb45306c80
|
[
"MIT"
] | null | null | null | 20.111111 | 88 | 0.690608 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
cbca2ddc66b8f3696e4226be547b4ead1abe0964
| 17,281 |
ipynb
|
Jupyter Notebook
|
docs/qcvv/xeb_theory.ipynb
|
anonymousr007/Cirq
|
fae0d85f79440e046ef365b58d86605ce35d4626
|
[
"Apache-2.0"
] | 3,326 |
2018-07-18T23:17:21.000Z
|
2022-03-29T22:28:24.000Z
|
docs/qcvv/xeb_theory.ipynb
|
anonymousr007/Cirq
|
fae0d85f79440e046ef365b58d86605ce35d4626
|
[
"Apache-2.0"
] | 3,443 |
2018-07-18T21:07:28.000Z
|
2022-03-31T20:23:21.000Z
|
docs/qcvv/xeb_theory.ipynb
|
anonymousr007/Cirq
|
fae0d85f79440e046ef365b58d86605ce35d4626
|
[
"Apache-2.0"
] | 865 |
2018-07-18T23:30:24.000Z
|
2022-03-30T11:43:23.000Z
| 31.650183 | 385 | 0.559632 |
[
[
[
"##### Copyright 2021 The Cirq Developers",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://quantumai.google/cirq/qcvv/xeb_theory>\"><img src=\"https://quantumai.google/site-assets/images/buttons/quantumai_logo_1x.png\" />View on QuantumAI</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/quantumlib/Cirq/blob/master/docs/qcvv/xeb_theory.ipynb\"><img src=\"https://quantumai.google/site-assets/images/buttons/colab_logo_1x.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/quantumlib/Cirq/blob/master/docs/qcvv/xeb_theory.ipynb\"><img src=\"https://quantumai.google/site-assets/images/buttons/github_logo_1x.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/Cirq/docs/qcvv/xeb_theory.ipynb\"><img src=\"https://quantumai.google/site-assets/images/buttons/download_icon_1x.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
]
],
[
[
"try:\n import cirq\nexcept ImportError:\n print(\"installing cirq...\")\n !pip install --quiet cirq\n print(\"installed cirq.\")",
"_____no_output_____"
]
],
[
[
"# Cross Entropy Benchmarking Theory\n\nCross entropy benchmarking uses the properties of random quantum programs to determine the fidelity of a wide variety of circuits. When applied to circuits with many qubits, XEB can characterize the performance of a large device. When applied to deep, two-qubit circuits it can be used to accurately characterize a two-qubit interaction potentially leading to better calibration.",
"_____no_output_____"
]
],
[
[
"# Standard imports\nimport numpy as np\nimport cirq\n\nfrom cirq.contrib.svg import SVGCircuit",
"_____no_output_____"
]
],
[
[
"## The action of random circuits with noise\nAn XEB experiment collects data from the execution of random circuits\nsubject to noise. The effect of applying a random circuit with unitary $U$ is\nmodeled as $U$ followed by a depolarizing channel. The result is that the\ninitial state $|𝜓⟩$ is mapped to a density matrix $ρ_U$ as follows:\n\n$$\n |𝜓⟩ → ρ_U = f |𝜓_U⟩⟨𝜓_U| + (1 - f) I / D\n$$\n\nwhere $|𝜓_U⟩ = U|𝜓⟩$, $D$ is the dimension of the Hilbert space, $I / D$ is the\nmaximally mixed state, and $f$ is the fidelity with which the circuit is\napplied.\n\nFor this model to be accurate, we require $U$ to be a random circuit that scrambles errors. In practice, we use a particular circuit ansatz consisting of random single-qubit rotations interleaved with entangling gates.",
"_____no_output_____"
],
[
"### Possible single-qubit rotations\nThese 8*8 possible rotations are chosen randomly when constructing the circuit.\n\nGeometrically, we choose 8 axes in the XY plane to perform a quarter-turn (pi/2 rotation) around. This is followed by a rotation around the Z axis of 8 different magnitudes.",
"_____no_output_____"
]
],
[
[
"exponents = np.linspace(0, 7/4, 8)\nexponents",
"_____no_output_____"
],
[
"import itertools\nSINGLE_QUBIT_GATES = [\n cirq.PhasedXZGate(x_exponent=0.5, z_exponent=z, axis_phase_exponent=a)\n for a, z in itertools.product(exponents, repeat=2)\n]\nSINGLE_QUBIT_GATES[:10], '...'",
"_____no_output_____"
]
],
[
[
"### Random circuit\n\nWe use `random_rotations_between_two_qubit_circuit` to generate a random two-qubit circuit. Note that we provide the possible single-qubit rotations from above and declare that our two-qubit operation is the $\\sqrt{i\\mathrm{SWAP}}$ gate.",
"_____no_output_____"
]
],
[
[
"import cirq_google as cg\nfrom cirq.experiments import random_quantum_circuit_generation as rqcg\n\nq0, q1 = cirq.LineQubit.range(2)\ncircuit = rqcg.random_rotations_between_two_qubit_circuit(\n q0, q1, \n depth=4, \n two_qubit_op_factory=lambda a, b, _: cirq.SQRT_ISWAP(a, b), \n single_qubit_gates=SINGLE_QUBIT_GATES\n)\nSVGCircuit(circuit)",
"_____no_output_____"
]
],
[
[
"## Estimating fidelity\n\nLet $O_U$ be an observable that is diagonal in the computational\nbasis. Then the expectation value of $O_U$ on $ρ_U$ is given by\n\n$$\n Tr(ρ_U O_U) = f ⟨𝜓_U|O_U|𝜓_U⟩ + (1 - f) Tr(O_U / D).\n$$\n\nThis equation shows how $f$ can be estimated, since $Tr(ρ_U O_U)$ can be\nestimated from experimental data, and $⟨𝜓_U|O_U|𝜓_U⟩$ and $Tr(O_U / D)$ can be\ncomputed.\n\nLet $e_U = ⟨𝜓_U|O_U|𝜓_U⟩$, $u_U = Tr(O_U / D)$, and $m_U$ denote the experimental\nestimate of $Tr(ρ_U O_U)$. We can write the following linear equation (equivalent to the\nexpression above):\n\n$$\n m_U = f e_U + (1-f) u_U \\\\\n m_U - u_U = f (e_U - u_U)\n$$",
"_____no_output_____"
]
],
[
[
"# Make long circuits (which we will truncate)\nMAX_DEPTH = 100\nN_CIRCUITS = 10\ncircuits = [\n rqcg.random_rotations_between_two_qubit_circuit(\n q0, q1, \n depth=MAX_DEPTH, \n two_qubit_op_factory=lambda a, b, _: cirq.SQRT_ISWAP(a, b), \n single_qubit_gates=SINGLE_QUBIT_GATES)\n for _ in range(N_CIRCUITS)\n]",
"_____no_output_____"
],
[
"# We will truncate to these lengths\ncycle_depths = np.arange(1, MAX_DEPTH + 1, 9)\ncycle_depths",
"_____no_output_____"
]
],
[
[
"### Execute circuits\nCross entropy benchmarking requires sampled bitstrings from the device being benchmarked *as well as* the true probabilities from a noiseless simulation. We find these quantities for all `(cycle_depth, circuit)` permutations.",
"_____no_output_____"
]
],
[
[
"pure_sim = cirq.Simulator()\n\n# Pauli Error. If there is an error, it is either X, Y, or Z\n# with probability E_PAULI / 3\nE_PAULI = 5e-3\nnoisy_sim = cirq.DensityMatrixSimulator(noise=cirq.depolarize(E_PAULI))\n\n# These two qubit circuits have 2^2 = 4 probabilities\nDIM = 4\n\nrecords = []\nfor cycle_depth in cycle_depths:\n for circuit_i, circuit in enumerate(circuits):\n \n # Truncate the long circuit to the requested cycle_depth\n circuit_depth = cycle_depth * 2 + 1\n assert circuit_depth <= len(circuit)\n trunc_circuit = circuit[:circuit_depth]\n\n # Pure-state simulation\n psi = pure_sim.simulate(trunc_circuit)\n psi = psi.final_state_vector\n pure_probs = np.abs(psi)**2\n\n # Noisy execution\n meas_circuit = trunc_circuit + cirq.measure(q0, q1)\n sampled_inds = noisy_sim.sample(meas_circuit, repetitions=10_000).values[:,0]\n sampled_probs = np.bincount(sampled_inds, minlength=DIM) / len(sampled_inds)\n\n # Save the results\n records += [{\n 'circuit_i': circuit_i,\n 'cycle_depth': cycle_depth,\n 'circuit_depth': circuit_depth,\n 'pure_probs': pure_probs,\n 'sampled_probs': sampled_probs,\n }]\n print('.', end='', flush=True)",
"_____no_output_____"
]
],
[
[
"## What's the observable\n\nWhat is $O_U$? Let's define it to be the observable that gives the sum of all probabilities, i.e.\n\n$$\n O_U |x \\rangle = p(x) |x \\rangle\n$$\n\nfor any bitstring $x$. We can use this to derive expressions for our quantities of interest.\n\n$$\ne_U = \\langle \\psi_U | O_U | \\psi_U \\rangle \\\\\n = \\sum_x a_x^* \\langle x | O_U | x \\rangle a_x \\\\\n = \\sum_x p(x) \\langle x | O_U | x \\rangle \\\\\n = \\sum_x p(x) p(x)\n$$\n\n$e_U$ is simply the sum of squared ideal probabilities. $u_U$ is a normalizing factor that only depends on the operator. Since this operator has the true probabilities in the definition, they show up here anyways.\n\n$$\nu_U = \\mathrm{Tr}[O_U / D] \\\\\n = 1/D \\sum_x \\langle x | O_U | x \\rangle \\\\\n = 1/D \\sum_x p(x)\n$$\n\nFor the measured values, we use the definition of an expectation value\n$$\n\\langle f(x) \\rangle_\\rho = \\sum_x p(x) f(x)\n$$\nIt becomes notationally confusing because remember: our operator on basis states returns the ideal probability of that basis state $p(x)$. The probability of observing a measured basis state is estimated from samples and denoted $p_\\mathrm{est}(x)$ here.\n\n$$\nm_U = \\mathrm{Tr}[\\rho_U O_U] \\\\\n = \\langle O_U \\rangle_{\\rho_U} = \\sum_{x} p_\\mathrm{est}(x) p(x)\n$$",
"_____no_output_____"
]
],
[
[
"for record in records:\n e_u = np.sum(record['pure_probs']**2)\n u_u = np.sum(record['pure_probs']) / DIM\n m_u = np.sum(record['pure_probs'] * record['sampled_probs'])\n record.update(\n e_u=e_u,\n u_u=u_u,\n m_u=m_u, \n )",
"_____no_output_____"
]
],
[
[
"Remember:\n\n$$\n m_U - u_U = f (e_U - u_U)\n$$\n\nWe estimate f by performing least squares\nminimization of the sum of squared residuals\n\n$$\n \\sum_U \\left(f (e_U - u_U) - (m_U - u_U)\\right)^2\n$$\n\nover different random circuits. The solution to the\nleast squares problem is given by\n\n$$\n f = (∑_U (m_U - u_U) * (e_U - u_U)) / (∑_U (e_U - u_U)^2)\n$$",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndf = pd.DataFrame(records)\ndf['y'] = df['m_u'] - df['u_u']\ndf['x'] = df['e_u'] - df['u_u']\n\ndf['numerator'] = df['x'] * df['y']\ndf['denominator'] = df['x'] ** 2\ndf.head()",
"_____no_output_____"
]
],
[
[
"### Fit\n\nWe'll plot the linear relationship and least-squares fit while we transform the raw DataFrame into one containing fidelities.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nfrom matplotlib import pyplot as plt\n\n# Color by cycle depth\nimport seaborn as sns\ncolors = sns.cubehelix_palette(n_colors=len(cycle_depths)) \ncolors = {k: colors[i] for i, k in enumerate(cycle_depths)}\n\n_lines = []\ndef per_cycle_depth(df):\n fid_lsq = df['numerator'].sum() / df['denominator'].sum()\n \n cycle_depth = df.name \n xx = np.linspace(0, df['x'].max())\n l, = plt.plot(xx, fid_lsq*xx, color=colors[cycle_depth])\n plt.scatter(df['x'], df['y'], color=colors[cycle_depth])\n \n global _lines\n _lines += [l] # for legend\n return pd.Series({'fidelity': fid_lsq})\n\nfids = df.groupby('cycle_depth').apply(per_cycle_depth).reset_index()\nplt.xlabel(r'$e_U - u_U$', fontsize=18)\nplt.ylabel(r'$m_U - u_U$', fontsize=18)\n_lines = np.asarray(_lines)\nplt.legend(_lines[[0,-1]], cycle_depths[[0,-1]], loc='best', title='Cycle depth')\nplt.tight_layout()",
"_____no_output_____"
]
],
[
[
"### Fidelities",
"_____no_output_____"
]
],
[
[
"plt.plot(\n fids['cycle_depth'], \n fids['fidelity'],\n marker='o',\n label='Least Squares')\n\nxx = np.linspace(0, fids['cycle_depth'].max())\n\n# In XEB, we extract the depolarizing fidelity, which is\n# related to (but not equal to) the Pauli error.\n# For the latter, an error involves doing X, Y, or Z with E_PAULI/3\n# but for the former, an error involves doing I, X, Y, or Z with e_depol/4\ne_depol = E_PAULI / (1 - 1/DIM**2)\n\n# The additional factor of four in the exponent is because each layer\n# involves two moments of two qubits (so each layer has four applications\n# of a single-qubit single-moment depolarizing channel).\nplt.plot(xx, (1-e_depol)**(4*xx), label=r'$(1-\\mathrm{e\\_depol})^{4d}$')\n\nplt.ylabel('Circuit fidelity', fontsize=18)\nplt.xlabel('Cycle Depth $d$', fontsize=18)\nplt.legend(loc='best')\nplt.yscale('log')\nplt.tight_layout()",
"_____no_output_____"
],
[
"from cirq.experiments.xeb_fitting import fit_exponential_decays\n\n# Ordinarily, we'd use this function to fit curves for multiple pairs.\n# We add our qubit pair as a column.\nfids['pair'] = [(q0, q1)] * len(fids)\n\nfit_df = fit_exponential_decays(fids)\nfit_row = fit_df.iloc[0]\nprint(f\"Noise model fidelity: {(1-e_depol)**4:.3e}\")\nprint(f\"XEB layer fidelity: {fit_row['layer_fid']:.3e} +- {fit_row['layer_fid_std']:.2e}\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbca318d3c2260f5d56ac878e2e255febe03c433
| 80,045 |
ipynb
|
Jupyter Notebook
|
AKAZE_code.ipynb
|
Shubham0Rajput/Feature-Detection-with-AKAZE
|
ca738535689292e235f7b5252758dafdba4fc961
|
[
"MIT"
] | 2 |
2020-09-12T06:38:34.000Z
|
2020-09-12T08:29:35.000Z
|
AKAZE_code.ipynb
|
Shubham0Rajput/Feature-Detection-with-AKAZE
|
ca738535689292e235f7b5252758dafdba4fc961
|
[
"MIT"
] | null | null | null |
AKAZE_code.ipynb
|
Shubham0Rajput/Feature-Detection-with-AKAZE
|
ca738535689292e235f7b5252758dafdba4fc961
|
[
"MIT"
] | null | null | null | 381.166667 | 72,938 | 0.91997 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Shubham0Rajput/Feature-Detection-with-AKAZE/blob/master/AKAZE_code.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"#IMPORT FILES\nimport matplotlib.pyplot as plt\nimport cv2\n#matplotlib inline\n\n#MOUNTIING DRIVE\nfrom google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"from __future__ import print_function\nimport cv2 as cv\nimport numpy as np\nimport argparse\nfrom math import sqrt\nimport matplotlib.pyplot as plt\n\n\nimge1 = cv.imread('/content/drive/My Drive/e2.jpg') \nimg1 = cv.cvtColor(imge1, cv.COLOR_BGR2GRAY) # queryImage\n\nimge2 = cv.imread('/content/drive/My Drive/e1.jpg') \nimg2 = cv.cvtColor(imge2, cv.COLOR_BGR2GRAY) # trainImage\n\nif img1 is None or img2 is None:\n print('Could not open or find the images!')\n exit(0)\n\nfs = cv.FileStorage('/content/drive/My Drive/H1to3p.xml', cv.FILE_STORAGE_READ)\nhomography = fs.getFirstTopLevelNode().mat()\n\n\n## [AKAZE]\nakaze = cv.AKAZE_create()\nkpts1, desc1 = akaze.detectAndCompute(img1, None)\nkpts2, desc2 = akaze.detectAndCompute(img2, None)\n## [AKAZE]\n\n## [2-nn matching]\nmatcher = cv.DescriptorMatcher_create(cv.DescriptorMatcher_BRUTEFORCE_HAMMING)\nnn_matches = matcher.knnMatch(desc1, desc2, 2)\n## [2-nn matching]\n\n## [ratio test filtering]\nmatched1 = []\nmatched2 = []\nnn_match_ratio = 0.8 # Nearest neighbor matching ratio\nfor m, n in nn_matches:\n if m.distance < nn_match_ratio * n.distance:\n matched1.append(kpts1[m.queryIdx])\n matched2.append(kpts2[m.trainIdx])\n\n## [homography check]\ninliers1 = []\ninliers2 = []\ngood_matches = []\ninlier_threshold = 2.5 # Distance threshold to identify inliers with homography check\nfor i, m in enumerate(matched1):\n col = np.ones((3,1), dtype=np.float64)\n col[0:2,0] = m.pt\n\n col = np.dot(homography, col)\n col /= col[2,0]\n dist = sqrt(pow(col[0,0] - matched2[i].pt[0], 2) +\\\n pow(col[1,0] - matched2[i].pt[1], 2))\n\n if dist > inlier_threshold:\n good_matches.append(cv.DMatch(len(inliers1), len(inliers2), 0))\n inliers1.append(matched1[i])\n inliers2.append(matched2[i])\n## [homography check]\n\n## [draw final matches]\nres = np.empty((max(img1.shape[0], img2.shape[0]), img1.shape[1]+img2.shape[1], 3), dtype=np.uint8)\nimg0 = cv.drawMatches(img1, inliers1, img2, inliers2, good_matches, res)\n#img0 = cv.drawMatchesKnn(img1,inliers1,img2,inliers2,res,None,flags=cv.DrawMatchesFlags_NOT_DRAW_SINGLE_POINTS)\ncv.imwrite(\"akaze_result.png\", res)\n\ninlier_ratio = len(inliers1) / float(len(matched1))\nprint('A-KAZE Matching Results')\nprint('*******************************')\nprint('# Keypoints 1: \\t', len(kpts1))\nprint('# Keypoints 2: \\t', len(kpts2))\nprint('# Matches: \\t', len(matched1))\nprint('# Inliers: \\t', len(inliers1))\nprint('# Inliers Ratio: \\t', inlier_ratio)\nprint('# Dist: \\t', dist)\n\nplt.imshow(img0),plt.show()\n## [draw final matches]",
"A-KAZE Matching Results\n*******************************\n# Keypoints 1: \t 402\n# Keypoints 2: \t 572\n# Matches: \t 65\n# Inliers: \t 65\n# Inliers Ratio: \t 1.0\n# Dist: \t 147.93780754404938\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbca348102ba93774fe993848a0b4ae1d98ea4a8
| 73,771 |
ipynb
|
Jupyter Notebook
|
Programming for Data Analysis Project 2018.ipynb
|
patmcdonald/ProgData52465_project
|
a341f26126eec5eb4e42e46a861eaaaee4a86a99
|
[
"MIT"
] | null | null | null |
Programming for Data Analysis Project 2018.ipynb
|
patmcdonald/ProgData52465_project
|
a341f26126eec5eb4e42e46a861eaaaee4a86a99
|
[
"MIT"
] | null | null | null |
Programming for Data Analysis Project 2018.ipynb
|
patmcdonald/ProgData52465_project
|
a341f26126eec5eb4e42e46a861eaaaee4a86a99
|
[
"MIT"
] | null | null | null | 41.374649 | 1,818 | 0.434114 |
[
[
[
"## Programming for Data Analysis Project 2018\n\n### Patrick McDonald G00281051\n\n#### Problem statement\n\nFor this project you must create a data set by simulating a real-world phenomenon of your choosing. You may pick any phenomenon you wish – you might pick one that is of interest to you in your personal or professional life. Then, rather than collect data related to the phenomenon, you should model and synthesise such data using Python. We suggest you use the numpy.random package for this purpose.\n\nSpecifically, in this project you should:\n\n1. Choose a real-world phenomenon that can be measured and for which you could collect at least one-hundred data points across at least four different variables.\n2. Investigate the types of variables involved, their likely distributions, and their relationships with each other.\n3. Synthesise/simulate a data set as closely matching their properties as possible.\n4. Detail your research and implement the simulation in a Jupyter notebook – the data set itself can simply be displayed in an output cell within the notebook.\n",
"_____no_output_____"
],
[
"### 1. Choose a real-world phenomenon that can be measured and for which you could collect at least one-hundred data points across at least four different variables.\n\nFor the purpose of this project, I shall extract some wave buoy data from the [M6 weather buoy](http://www.marine.ie/Home/site-area/data-services/real-time-observations/irish-weather-buoy-network) off the westcoast of Ireland. I surf occassionally, and many surfers, like myself; use weather buoy data in order to predict when there will be decent waves to surf. There are many online resources that provide such information, but I thought this may be an enjoyable exploration of raw data that is used everyday, worldwide.",
"_____no_output_____"
]
],
[
[
"# Import libraries\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n%matplotlib inline\n\n# Downloaded hly62095.csv from https://data.gov.ie/dataset/hourly-data-for-buoy-m6 \n# Opened dataset in VSCode. It contains the label legend, so I have skipped these rows.\n# I also only want to utilise 4 relevant columns of data, I'll use the 'usecols' arguement:\n# https://realpython.com/python-data-cleaning-numpy-pandas/#dropping-columns-in-a-dataframe\n\ndf = pd.read_csv(\"hly62095.csv\", skiprows = 19, low_memory = False, usecols= ['date', 'dir', 'per', 'wavht'])\n\n# Change the date column to a Pythonic datetime - \n# reference: https://github.com/ianmcloughlin/jupyter-teaching-notebooks/raw/master/time-series.ipynb\n\ndf['datetime'] = pd.to_datetime(df['date'])",
"_____no_output_____"
]
],
[
[
"Downloaded hly62095.csv from https://data.gov.ie/dataset/hourly-data-for-buoy-m6. Opened dataset in VSCode. It contains the label legend, so I have skipped these rows 1-19:\n\n###Label legend\n\n```\n1. Station Name: M6\n2. Station Height: 0 M \n3. Latitude:52.990 ,Longitude: -15.870\n4. \n5. \n6. date:\t - Date and Time (utc)\n7. temp: \t - Air Temperature (C)\t\n8. rhum:\t - Relative Humidity (%)\n9. windsp:\t - Mean Wind Speed (kt)\n10. dir:\t - Mean Wind \tDirection (degrees)\n11. gust:\t - Maximum Gust (kt)\n12. msl:\t - Mean Sea Level Pressure (hPa)\n13. seatp:\t - Sea Temperature (C)\n14. per:\t - Significant Wave Period (seconds)\n15. wavht:\t - Significant Wave Height (m)\n16. mxwav: \t - Individual Maximum Wave Height(m)\n17. wvdir: - Wave Direction (degrees)\n18. ind: - Indicator \n19. \n20. date,temp,rhum,wdsp,dir,gust,msl,seatp,per,wavht,mxwave,wvdir\n21. 25-sep-2006 09:00,15.2, ,8.000,240.000, ,1007.2,15.4,6.000,1.5, , \n22. 25-sep-2006 10:00,15.2, ,8.000,220.000, ,1008.0,15.4,6.000,1.5, ,......... \n\n```",
"_____no_output_____"
]
],
[
[
"# View DataFrame\ndf",
"_____no_output_____"
]
],
[
[
"There are a significant missing datapoints, and its a large sample, with 94248 rows. I'm going to explore this further, and extract the relevant data for September 2018. This will give me enough data to explore and simulate for this project.\nFirst, I'll describe the datatypes in the set.",
"_____no_output_____"
]
],
[
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"I want to view the data for September 2018. So I'll extract the relevant datapoints from this dataset.",
"_____no_output_____"
]
],
[
[
"# Create a datetime index for a data frame.\n\n# Adapted from: https://pandas.pydata.org/pandas-docs/stable/timeseries.html \n# https://pandas.pydata.org/pandas-docs/stable/generated/pandas.date_range.html\n\n\nrng = pd.date_range(start='1-sep-2018', periods=30, freq='D')",
"_____no_output_____"
],
[
"rng",
"_____no_output_____"
]
],
[
[
"I'm using 4 variables from the dataset. These are;\n\n1. date: - Date and Time (utc)\n2. dir:\t - Mean Wind \tDirection (degrees)\n3. per:\t - Significant Wave Period (seconds) - This is important for quality waves!\n4. wavht: - Significant Wave Height (m)",
"_____no_output_____"
]
],
[
[
"df.head(10)",
"_____no_output_____"
]
],
[
[
"Next, I'll display the data columns from 1st September 2018. Since I've already removed rows 1-19, the row label numbers have been modified by pandas. In this case - I worked backwards to find the right label number.\n\nI'm going to name this smaller dataframe 'wavedata'.",
"_____no_output_____"
]
],
[
[
"wavedata = df.loc['93530':]",
"_____no_output_____"
],
[
"wavedata",
"_____no_output_____"
]
],
[
[
"I'm now going to check for Null values in the data.",
"_____no_output_____"
]
],
[
[
"# checking for null values\n# There are no null values in this dataframe, according to pandas!\nwavedata.isnull().sum()",
"_____no_output_____"
]
],
[
[
"### 2. Investigate the types of variables involved, their likely distributions, and their relationships with each other.\n\nNow for some exploratory data analysis.",
"_____no_output_____"
]
],
[
[
"# Check datatypes\ndf.dtypes",
"_____no_output_____"
]
],
[
[
"#### Explore distributions",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nplt.figure(figsize=(26, 10))\n\nplot = sns.scatterplot(x=\"datetime\", y=\"wavht\", hue='per', data=wavedata)",
"_____no_output_____"
]
],
[
[
"It looks like I've a problem with this data! It's mostly objects, not floats.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbca54f39e0aeb71dfa00833868b121ef96b8e22
| 16,231 |
ipynb
|
Jupyter Notebook
|
Materials/Enums.ipynb
|
FMakhnach/csharp-materials
|
6b936c34656170270ca496d5c77225351688955e
|
[
"MIT"
] | null | null | null |
Materials/Enums.ipynb
|
FMakhnach/csharp-materials
|
6b936c34656170270ca496d5c77225351688955e
|
[
"MIT"
] | null | null | null |
Materials/Enums.ipynb
|
FMakhnach/csharp-materials
|
6b936c34656170270ca496d5c77225351688955e
|
[
"MIT"
] | null | null | null | 22.143247 | 378 | 0.505083 |
[
[
[
"# Перечислимые типы (enums)",
"_____no_output_____"
],
[
"## 1. Базовые возможности",
"_____no_output_____"
]
],
[
[
"enum Color\n{\n White, // 0\n Red, // 1\n Green, // 2\n Blue, // 3\n Orange, // 4\n}",
"_____no_output_____"
],
[
"Color white = Color.White; \nConsole.WriteLine(white); // White\n\nColor red = (Color)1; // Так можно приводить к типу перечисления \nConsole.WriteLine(red); // Red\n\nColor unknown = (Color)42; // Нет ошибки!\nConsole.WriteLine(unknown); // 42",
"White\nRed\n42\n"
],
[
"Color green = Enum.Parse<Color>(\"Green\");\ngreen.ToString()",
"_____no_output_____"
],
[
"Enum.TryParse<Color>(\"Blue\", out Color blue);\nblue.ToString()",
"_____no_output_____"
],
[
"// Посмотрим, какими типами можно задавать перечисления\nenum Dummy : object {}",
"_____no_output_____"
]
],
[
[
"## 2. Приведение перечислимых типов",
"_____no_output_____"
]
],
[
[
"enum Fruit\n{\n Melon, // 0\n Tomato, // 1\n Apple, // 2\n Blueberry, // 3\n Orange, // 4\n}",
"_____no_output_____"
],
[
"Fruit orange = Color.Orange; // Безопасность типов -> ошибка",
"_____no_output_____"
],
[
"Fruit tomato = (Fruit)Color.Red; // А вот так уже можно\nConsole.WriteLine(tomato);",
"Tomato\r\n"
],
[
"Color unknownColor = (Color)42;\nFruit unknownFruit = (Fruit)unknownColor;\nConsole.WriteLine(unknownFruit);",
"42\r\n"
],
[
"// Любой enum имеет следующую цепочку наследования: MyEnum <- System.Enum <- System.ValueType <- System.Object\n\nEnum enumEnum = Color.Blue;\nValueType enumValueType = Color.Blue;\nobject enumObj = Color.Blue; // BOXING\n\nConsole.WriteLine($\"{enumEnum}, {enumValueType}, {enumObj}\");",
"Blue, Blue, Blue\r\n"
]
],
[
[
"## 3. Использование одного целочисленного значения для нескольких enum значений",
"_____no_output_____"
]
],
[
[
"public enum Subject\n{\n Programming = 0,\n DiscreteMath = 1,\n Algebra = 2,\n Calculus = 3,\n Economics = 4,\n\n MostDifficultSubject = Algebra,\n MostUsefulSubject = Programming,\n // MostHatefulSubject = Programming\n}",
"_____no_output_____"
],
[
"Console.WriteLine(Subject.Programming);\nConsole.WriteLine(Subject.MostUsefulSubject);\nConsole.WriteLine((Subject)0);\n\nConsole.WriteLine(Subject.Programming == Subject.MostUsefulSubject)",
"Programming\nProgramming\nProgramming\nTrue\n"
],
[
"Console.WriteLine(Subject.Algebra);\nConsole.WriteLine(Subject.MostDifficultSubject);\nConsole.WriteLine((Subject)2);\n\nConsole.WriteLine(Subject.Algebra == Subject.MostDifficultSubject)",
"Algebra\nAlgebra\nAlgebra\nTrue\n"
]
],
[
[
"## 4. Рефлексия перечислимых типов",
"_____no_output_____"
],
[
"Статический метод Enum.GetUnderlyingType возвращает целочисленный тип для енама",
"_____no_output_____"
]
],
[
[
"Enum.GetUnderlyingType(typeof(Subject))",
"_____no_output_____"
],
[
"Enum.GetUnderlyingType(typeof(Subject))",
"_____no_output_____"
]
],
[
[
"В типе System.Type также есть метод GetEnumUnderlyingType",
"_____no_output_____"
]
],
[
[
"typeof(Subject).GetEnumUnderlyingType()",
"_____no_output_____"
]
],
[
[
"Который работает только с объектами-типами енамов",
"_____no_output_____"
]
],
[
[
"typeof(short).GetEnumUnderlyingType()",
"_____no_output_____"
]
],
[
[
"Можно получить все значения енама c помощью Enum.GetValues(Type)",
"_____no_output_____"
]
],
[
[
"var enumValues = Enum.GetValues(typeof(Subject)); // Аналог: typeof(Subject).GetEnumValues();\nforeach(var value in enumValues){\n Console.WriteLine(value);\n}",
"Programming\nProgramming\nDiscreteMath\nAlgebra\nAlgebra\nCalculus\nEconomics\n"
],
[
"Enum.GetNames(typeof(Subject)) // Аналог: typeof(Subject).GetEnumNames()",
"_____no_output_____"
]
],
[
[
"Проверка, есть ли в енаме соответствующее значение.",
"_____no_output_____"
]
],
[
[
"Enum.IsDefined(typeof(Subject), 3)",
"_____no_output_____"
],
[
"Enum.IsDefined(typeof(Subject), 42)",
"_____no_output_____"
]
],
[
[
"## 5. Битовые флаги",
"_____no_output_____"
]
],
[
[
"[Flags]\nenum FilePermission : byte\n{\n None = 0b00000000,\n\n Read = 0b00000001,\n Write = 0b00000010,\n Execute = 0b00000100,\n Rename = 0b00001000,\n Move = 0b00010000,\n Delete = 0b00100000,\n\n User = Read | Execute,\n ReadWrite = Read | Write,\n Admin = Read | Write | Execute | Rename | Move | Delete\n}",
"_____no_output_____"
]
],
[
[
"[Про FlagsAttribute](https://docs.microsoft.com/ru-ru/dotnet/api/system.flagsattribute?view=net-5.0)",
"_____no_output_____"
]
],
[
[
"FilePermission permission = FilePermission.User;\npermission.HasFlag(FilePermission.Read)",
"_____no_output_____"
]
],
[
[
"Пример использования:\n```\nvoid RenameFile(File file, User user)\n{\n if (!user.Permission.HasFlag(FilePermission.Rename)) {\n throw new SomeException(\"you can't.\")\n }\n ...\n}\n```",
"_____no_output_____"
]
],
[
[
"for (int i = 0; i <= 16; ++i) {\n FilePermission fp = (FilePermission)i;\n Console.WriteLine(fp.ToString(\"G\"));\n}",
"None\nRead\nWrite\nReadWrite\nExecute\nUser\nWrite, Execute\nWrite, User\nRename\nRead, Rename\nWrite, Rename\nReadWrite, Rename\nExecute, Rename\nUser, Rename\nWrite, Execute, Rename\nWrite, User, Rename\nMove\n"
]
],
[
[
"Пример из стандартной библиотеки: System.AttributeTargets",
"_____no_output_____"
]
],
[
[
"[Flags, Serializable]\npublic enum AttributeTargets {\n Assembly = 0x0001,\n Module = 0x0002,\n Class = 0x0004,\n Struct = 0x0008,\n Enum = 0x0010,\n Constructor = 0x0020,\n Method = 0x0040,\n Property = 0x0080,\n Field = 0x0100,\n Event = 0x0200,\n Interface = 0x0400,\n Parameter = 0x0800,\n Delegate = 0x1000,\n ReturnValue = 0x2000,\n GenericParameter = 0x4000,\n All = Assembly | Module | Class | Struct | Enum |\n Constructor | Method | Property | Field | Event |\n Interface | Parameter | Delegate | ReturnValue |\n GenericParameter\n}",
"_____no_output_____"
]
],
[
[
"## 6. Методы расширения для enum",
"_____no_output_____"
],
[
"Перечислениям можно \"добавлять функциональность\" с помощью методов расширения",
"_____no_output_____"
]
],
[
[
"//public static class EnumExtentions\n//{\n public static int GetMark(this Subject subject)\n {\n return subject switch \n {\n Subject.Programming => 8,\n Subject.DiscreteMath => 10,\n Subject.Algebra => 5,\n Subject.Calculus => 7,\n Subject.Economics => 6,\n _ => 0,\n };\n }\n//}",
"_____no_output_____"
],
[
"Subject prog = Subject.Programming;\nprog.GetMark()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
cbca5ca7f6d9fba60012c9d3eba03250179d3854
| 45,130 |
ipynb
|
Jupyter Notebook
|
Data Collecion with Web Scraping.ipynb
|
fa-901/ibm-capstone
|
0ebaac17dc03e09f348e625d475f45c202fc98dc
|
[
"MIT"
] | null | null | null |
Data Collecion with Web Scraping.ipynb
|
fa-901/ibm-capstone
|
0ebaac17dc03e09f348e625d475f45c202fc98dc
|
[
"MIT"
] | null | null | null |
Data Collecion with Web Scraping.ipynb
|
fa-901/ibm-capstone
|
0ebaac17dc03e09f348e625d475f45c202fc98dc
|
[
"MIT"
] | null | null | null | 119.076517 | 20,069 | 0.61961 |
[
[
[
"<center>\n <img src=\"https://gitlab.com/ibm/skills-network/courses/placeholder101/-/raw/master/labs/module%201/images/IDSNlogo.png\" width=\"300\" alt=\"cognitiveclass.ai logo\" />\n</center>\n",
"_____no_output_____"
],
[
"# **Space X Falcon 9 First Stage Landing Prediction**\n",
"_____no_output_____"
],
[
"## Web scraping Falcon 9 and Falcon Heavy Launches Records from Wikipedia\n",
"_____no_output_____"
],
[
"Estimated time needed: **40** minutes\n",
"_____no_output_____"
],
[
"In this lab, you will be performing web scraping to collect Falcon 9 historical launch records from a Wikipedia page titled `List of Falcon 9 and Falcon Heavy launches`\n\n[https://en.wikipedia.org/wiki/List_of_Falcon\\_9\\_and_Falcon_Heavy_launches](https://en.wikipedia.org/wiki/List_of_Falcon\\_9\\_and_Falcon_Heavy_launches?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDS0321ENSkillsNetwork26802033-2021-01-01)\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"Falcon 9 first stage will land successfully\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"Several examples of an unsuccessful landing are shown here:\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"More specifically, the launch records are stored in a HTML table shown below:\n",
"_____no_output_____"
],
[
"\n",
"_____no_output_____"
],
[
"## Objectives\n\nWeb scrap Falcon 9 launch records with `BeautifulSoup`:\n\n* Extract a Falcon 9 launch records HTML table from Wikipedia\n* Parse the table and convert it into a Pandas data frame\n",
"_____no_output_____"
],
[
"First let's import required packages for this lab\n",
"_____no_output_____"
]
],
[
[
"!pip3 install beautifulsoup4\n!pip3 install requests",
"Requirement already satisfied: beautifulsoup4 in /opt/conda/envs/Python-3.8-main/lib/python3.8/site-packages (4.9.3)\nRequirement already satisfied: soupsieve>1.2 in /opt/conda/envs/Python-3.8-main/lib/python3.8/site-packages (from beautifulsoup4) (2.2.1)\nRequirement already satisfied: requests in /opt/conda/envs/Python-3.8-main/lib/python3.8/site-packages (2.25.1)\nRequirement already satisfied: certifi>=2017.4.17 in /opt/conda/envs/Python-3.8-main/lib/python3.8/site-packages (from requests) (2021.10.8)\nRequirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/conda/envs/Python-3.8-main/lib/python3.8/site-packages (from requests) (1.26.6)\nRequirement already satisfied: idna<3,>=2.5 in /opt/conda/envs/Python-3.8-main/lib/python3.8/site-packages (from requests) (2.8)\nRequirement already satisfied: chardet<5,>=3.0.2 in /opt/conda/envs/Python-3.8-main/lib/python3.8/site-packages (from requests) (3.0.4)\n"
],
[
"import sys\n\nimport requests\nfrom bs4 import BeautifulSoup\nimport re\nimport unicodedata\nimport pandas as pd",
"_____no_output_____"
]
],
[
[
"and we will provide some helper functions for you to process web scraped HTML table\n",
"_____no_output_____"
]
],
[
[
"def date_time(table_cells):\n \"\"\"\n This function returns the data and time from the HTML table cell\n Input: the element of a table data cell extracts extra row\n \"\"\"\n return [data_time.strip() for data_time in list(table_cells.strings)][0:2]\n\ndef booster_version(table_cells):\n \"\"\"\n This function returns the booster version from the HTML table cell \n Input: the element of a table data cell extracts extra row\n \"\"\"\n out=''.join([booster_version for i,booster_version in enumerate( table_cells.strings) if i%2==0][0:-1])\n return out\n\ndef landing_status(table_cells):\n \"\"\"\n This function returns the landing status from the HTML table cell \n Input: the element of a table data cell extracts extra row\n \"\"\"\n out=[i for i in table_cells.strings][0]\n return out\n\n\ndef get_mass(table_cells):\n mass=unicodedata.normalize(\"NFKD\", table_cells.text).strip()\n if mass:\n mass.find(\"kg\")\n new_mass=mass[0:mass.find(\"kg\")+2]\n else:\n new_mass=0\n return new_mass\n\n\ndef extract_column_from_header(row):\n \"\"\"\n This function returns the landing status from the HTML table cell \n Input: the element of a table data cell extracts extra row\n \"\"\"\n if (row.br):\n row.br.extract()\n if row.a:\n row.a.extract()\n if row.sup:\n row.sup.extract()\n \n colunm_name = ' '.join(row.contents)\n \n # Filter the digit and empty names\n if not(colunm_name.strip().isdigit()):\n colunm_name = colunm_name.strip()\n return colunm_name \n",
"_____no_output_____"
]
],
[
[
"To keep the lab tasks consistent, you will be asked to scrape the data from a snapshot of the `List of Falcon 9 and Falcon Heavy launches` Wikipage updated on\n`9th June 2021`\n",
"_____no_output_____"
]
],
[
[
"static_url = \"https://en.wikipedia.org/w/index.php?title=List_of_Falcon_9_and_Falcon_Heavy_launches&oldid=1027686922\"",
"_____no_output_____"
]
],
[
[
"Next, request the HTML page from the above URL and get a `response` object\n",
"_____no_output_____"
],
[
"### TASK 1: Request the Falcon9 Launch Wiki page from its URL\n",
"_____no_output_____"
],
[
"First, let's perform an HTTP GET method to request the Falcon9 Launch HTML page, as an HTTP response.\n",
"_____no_output_____"
]
],
[
[
"# use requests.get() method with the provided static_url\n# assign the response to a object\nresponse = requests.get(static_url)",
"_____no_output_____"
]
],
[
[
"Create a `BeautifulSoup` object from the HTML `response`\n",
"_____no_output_____"
]
],
[
[
"# Use BeautifulSoup() to create a BeautifulSoup object from a response text content\nsoup = BeautifulSoup(response.text)",
"_____no_output_____"
]
],
[
[
"Print the page title to verify if the `BeautifulSoup` object was created properly\n",
"_____no_output_____"
]
],
[
[
"# Use soup.title attribute\nprint(soup.title)",
"<title>List of Falcon 9 and Falcon Heavy launches - Wikipedia</title>\n"
]
],
[
[
"### TASK 2: Extract all column/variable names from the HTML table header\n",
"_____no_output_____"
],
[
"Next, we want to collect all relevant column names from the HTML table header\n",
"_____no_output_____"
],
[
"Let's try to find all tables on the wiki page first. If you need to refresh your memory about `BeautifulSoup`, please check the external reference link towards the end of this lab\n",
"_____no_output_____"
]
],
[
[
"# Use the find_all function in the BeautifulSoup object, with element type `table`\n# Assign the result to a list called `html_tables`\nhtml_tables = soup.find_all('table')",
"_____no_output_____"
]
],
[
[
"Starting from the third table is our target table contains the actual launch records.\n",
"_____no_output_____"
]
],
[
[
"# Let's print the third table and check its content\nfirst_launch_table = html_tables[2]\nprint(first_launch_table)",
"<table class=\"wikitable plainrowheaders collapsible\" style=\"width: 100%;\">\n<tbody><tr>\n<th scope=\"col\">Flight No.\n</th>\n<th scope=\"col\">Date and<br/>time (<a href=\"/wiki/Coordinated_Universal_Time\" title=\"Coordinated Universal Time\">UTC</a>)\n</th>\n<th scope=\"col\"><a href=\"/wiki/List_of_Falcon_9_first-stage_boosters\" title=\"List of Falcon 9 first-stage boosters\">Version,<br/>Booster</a> <sup class=\"reference\" id=\"cite_ref-booster_11-0\"><a href=\"#cite_note-booster-11\">[b]</a></sup>\n</th>\n<th scope=\"col\">Launch site\n</th>\n<th scope=\"col\">Payload<sup class=\"reference\" id=\"cite_ref-Dragon_12-0\"><a href=\"#cite_note-Dragon-12\">[c]</a></sup>\n</th>\n<th scope=\"col\">Payload mass\n</th>\n<th scope=\"col\">Orbit\n</th>\n<th scope=\"col\">Customer\n</th>\n<th scope=\"col\">Launch<br/>outcome\n</th>\n<th scope=\"col\"><a href=\"/wiki/Falcon_9_first-stage_landing_tests\" title=\"Falcon 9 first-stage landing tests\">Booster<br/>landing</a>\n</th></tr>\n<tr>\n<th rowspan=\"2\" scope=\"row\" style=\"text-align:center;\">1\n</th>\n<td>4 June 2010,<br/>18:45\n</td>\n<td><a href=\"/wiki/Falcon_9_v1.0\" title=\"Falcon 9 v1.0\">F9 v1.0</a><sup class=\"reference\" id=\"cite_ref-MuskMay2012_13-0\"><a href=\"#cite_note-MuskMay2012-13\">[7]</a></sup><br/>B0003.1<sup class=\"reference\" id=\"cite_ref-block_numbers_14-0\"><a href=\"#cite_note-block_numbers-14\">[8]</a></sup>\n</td>\n<td><a href=\"/wiki/Cape_Canaveral_Space_Force_Station\" title=\"Cape Canaveral Space Force Station\">CCAFS</a>,<br/><a href=\"/wiki/Cape_Canaveral_Space_Launch_Complex_40\" title=\"Cape Canaveral Space Launch Complex 40\">SLC-40</a>\n</td>\n<td><a href=\"/wiki/Dragon_Spacecraft_Qualification_Unit\" title=\"Dragon Spacecraft Qualification Unit\">Dragon Spacecraft Qualification Unit</a>\n</td>\n<td>\n</td>\n<td><a href=\"/wiki/Low_Earth_orbit\" title=\"Low Earth orbit\">LEO</a>\n</td>\n<td><a href=\"/wiki/SpaceX\" title=\"SpaceX\">SpaceX</a>\n</td>\n<td class=\"table-success\" style=\"background: LightGreen; color: black; vertical-align: middle; text-align: center;\">Success\n</td>\n<td class=\"table-failure\" style=\"background: #ffbbbb; color: black; vertical-align: middle; text-align: center;\">Failure<sup class=\"reference\" id=\"cite_ref-ns20110930_15-0\"><a href=\"#cite_note-ns20110930-15\">[9]</a></sup><sup class=\"reference\" id=\"cite_ref-16\"><a href=\"#cite_note-16\">[10]</a></sup><br/><small>(parachute)</small>\n</td></tr>\n<tr>\n<td colspan=\"9\">First flight of Falcon 9 v1.0.<sup class=\"reference\" id=\"cite_ref-sfn20100604_17-0\"><a href=\"#cite_note-sfn20100604-17\">[11]</a></sup> Used a boilerplate version of Dragon capsule which was not designed to separate from the second stage.<small>(<a href=\"#First_flight_of_Falcon_9\">more details below</a>)</small> Attempted to recover the first stage by parachuting it into the ocean, but it burned up on reentry, before the parachutes even deployed.<sup class=\"reference\" id=\"cite_ref-parachute_18-0\"><a href=\"#cite_note-parachute-18\">[12]</a></sup>\n</td></tr>\n<tr>\n<th rowspan=\"2\" scope=\"row\" style=\"text-align:center;\">2\n</th>\n<td>8 December 2010,<br/>15:43<sup class=\"reference\" id=\"cite_ref-spaceflightnow_Clark_Launch_Report_19-0\"><a href=\"#cite_note-spaceflightnow_Clark_Launch_Report-19\">[13]</a></sup>\n</td>\n<td><a href=\"/wiki/Falcon_9_v1.0\" title=\"Falcon 9 v1.0\">F9 v1.0</a><sup class=\"reference\" id=\"cite_ref-MuskMay2012_13-1\"><a href=\"#cite_note-MuskMay2012-13\">[7]</a></sup><br/>B0004.1<sup class=\"reference\" id=\"cite_ref-block_numbers_14-1\"><a href=\"#cite_note-block_numbers-14\">[8]</a></sup>\n</td>\n<td><a href=\"/wiki/Cape_Canaveral_Space_Force_Station\" title=\"Cape Canaveral Space Force Station\">CCAFS</a>,<br/><a href=\"/wiki/Cape_Canaveral_Space_Launch_Complex_40\" title=\"Cape Canaveral Space Launch Complex 40\">SLC-40</a>\n</td>\n<td><a href=\"/wiki/SpaceX_Dragon\" title=\"SpaceX Dragon\">Dragon</a> <a class=\"mw-redirect\" href=\"/wiki/COTS_Demo_Flight_1\" title=\"COTS Demo Flight 1\">demo flight C1</a><br/>(Dragon C101)\n</td>\n<td>\n</td>\n<td><a href=\"/wiki/Low_Earth_orbit\" title=\"Low Earth orbit\">LEO</a> (<a href=\"/wiki/International_Space_Station\" title=\"International Space Station\">ISS</a>)\n</td>\n<td><div class=\"plainlist\">\n<ul><li><a href=\"/wiki/NASA\" title=\"NASA\">NASA</a> (<a href=\"/wiki/Commercial_Orbital_Transportation_Services\" title=\"Commercial Orbital Transportation Services\">COTS</a>)</li>\n<li><a href=\"/wiki/National_Reconnaissance_Office\" title=\"National Reconnaissance Office\">NRO</a></li></ul>\n</div>\n</td>\n<td class=\"table-success\" style=\"background: LightGreen; color: black; vertical-align: middle; text-align: center;\">Success<sup class=\"reference\" id=\"cite_ref-ns20110930_15-1\"><a href=\"#cite_note-ns20110930-15\">[9]</a></sup>\n</td>\n<td class=\"table-failure\" style=\"background: #ffbbbb; color: black; vertical-align: middle; text-align: center;\">Failure<sup class=\"reference\" id=\"cite_ref-ns20110930_15-2\"><a href=\"#cite_note-ns20110930-15\">[9]</a></sup><sup class=\"reference\" id=\"cite_ref-20\"><a href=\"#cite_note-20\">[14]</a></sup><br/><small>(parachute)</small>\n</td></tr>\n<tr>\n<td colspan=\"9\">Maiden flight of <a class=\"mw-redirect\" href=\"/wiki/Dragon_capsule\" title=\"Dragon capsule\">Dragon capsule</a>, consisting of over 3 hours of testing thruster maneuvering and reentry.<sup class=\"reference\" id=\"cite_ref-spaceflightnow_Clark_unleashing_Dragon_21-0\"><a href=\"#cite_note-spaceflightnow_Clark_unleashing_Dragon-21\">[15]</a></sup> Attempted to recover the first stage by parachuting it into the ocean, but it disintegrated upon reentry, before the parachutes were deployed.<sup class=\"reference\" id=\"cite_ref-parachute_18-1\"><a href=\"#cite_note-parachute-18\">[12]</a></sup> <small>(<a href=\"#COTS_demo_missions\">more details below</a>)</small> It also included two <a href=\"/wiki/CubeSat\" title=\"CubeSat\">CubeSats</a>,<sup class=\"reference\" id=\"cite_ref-NRO_Taps_Boeing_for_Next_Batch_of_CubeSats_22-0\"><a href=\"#cite_note-NRO_Taps_Boeing_for_Next_Batch_of_CubeSats-22\">[16]</a></sup> and a wheel of <a href=\"/wiki/Brou%C3%A8re\" title=\"Brouère\">Brouère</a> cheese.\n</td></tr>\n<tr>\n<th rowspan=\"2\" scope=\"row\" style=\"text-align:center;\">3\n</th>\n<td>22 May 2012,<br/>07:44<sup class=\"reference\" id=\"cite_ref-BBC_new_era_23-0\"><a href=\"#cite_note-BBC_new_era-23\">[17]</a></sup>\n</td>\n<td><a href=\"/wiki/Falcon_9_v1.0\" title=\"Falcon 9 v1.0\">F9 v1.0</a><sup class=\"reference\" id=\"cite_ref-MuskMay2012_13-2\"><a href=\"#cite_note-MuskMay2012-13\">[7]</a></sup><br/>B0005.1<sup class=\"reference\" id=\"cite_ref-block_numbers_14-2\"><a href=\"#cite_note-block_numbers-14\">[8]</a></sup>\n</td>\n<td><a href=\"/wiki/Cape_Canaveral_Space_Force_Station\" title=\"Cape Canaveral Space Force Station\">CCAFS</a>,<br/><a href=\"/wiki/Cape_Canaveral_Space_Launch_Complex_40\" title=\"Cape Canaveral Space Launch Complex 40\">SLC-40</a>\n</td>\n<td><a href=\"/wiki/SpaceX_Dragon\" title=\"SpaceX Dragon\">Dragon</a> <a class=\"mw-redirect\" href=\"/wiki/Dragon_C2%2B\" title=\"Dragon C2+\">demo flight C2+</a><sup class=\"reference\" id=\"cite_ref-C2_24-0\"><a href=\"#cite_note-C2-24\">[18]</a></sup><br/>(Dragon C102)\n</td>\n<td>525 kg (1,157 lb)<sup class=\"reference\" id=\"cite_ref-25\"><a href=\"#cite_note-25\">[19]</a></sup>\n</td>\n<td><a href=\"/wiki/Low_Earth_orbit\" title=\"Low Earth orbit\">LEO</a> (<a href=\"/wiki/International_Space_Station\" title=\"International Space Station\">ISS</a>)\n</td>\n<td><a href=\"/wiki/NASA\" title=\"NASA\">NASA</a> (<a href=\"/wiki/Commercial_Orbital_Transportation_Services\" title=\"Commercial Orbital Transportation Services\">COTS</a>)\n</td>\n<td class=\"table-success\" style=\"background: LightGreen; color: black; vertical-align: middle; text-align: center;\">Success<sup class=\"reference\" id=\"cite_ref-26\"><a href=\"#cite_note-26\">[20]</a></sup>\n</td>\n<td class=\"table-noAttempt\" style=\"background: #ececec; color: black; vertical-align: middle; white-space: nowrap; text-align: center;\">No attempt\n</td></tr>\n<tr>\n<td colspan=\"9\">Dragon spacecraft demonstrated a series of tests before it was allowed to approach the <a href=\"/wiki/International_Space_Station\" title=\"International Space Station\">International Space Station</a>. Two days later, it became the first commercial spacecraft to board the ISS.<sup class=\"reference\" id=\"cite_ref-BBC_new_era_23-1\"><a href=\"#cite_note-BBC_new_era-23\">[17]</a></sup> <small>(<a href=\"#COTS_demo_missions\">more details below</a>)</small>\n</td></tr>\n<tr>\n<th rowspan=\"3\" scope=\"row\" style=\"text-align:center;\">4\n</th>\n<td rowspan=\"2\">8 October 2012,<br/>00:35<sup class=\"reference\" id=\"cite_ref-SFN_LLog_27-0\"><a href=\"#cite_note-SFN_LLog-27\">[21]</a></sup>\n</td>\n<td rowspan=\"2\"><a href=\"/wiki/Falcon_9_v1.0\" title=\"Falcon 9 v1.0\">F9 v1.0</a><sup class=\"reference\" id=\"cite_ref-MuskMay2012_13-3\"><a href=\"#cite_note-MuskMay2012-13\">[7]</a></sup><br/>B0006.1<sup class=\"reference\" id=\"cite_ref-block_numbers_14-3\"><a href=\"#cite_note-block_numbers-14\">[8]</a></sup>\n</td>\n<td rowspan=\"2\"><a href=\"/wiki/Cape_Canaveral_Space_Force_Station\" title=\"Cape Canaveral Space Force Station\">CCAFS</a>,<br/><a href=\"/wiki/Cape_Canaveral_Space_Launch_Complex_40\" title=\"Cape Canaveral Space Launch Complex 40\">SLC-40</a>\n</td>\n<td><a href=\"/wiki/SpaceX_CRS-1\" title=\"SpaceX CRS-1\">SpaceX CRS-1</a><sup class=\"reference\" id=\"cite_ref-sxManifest20120925_28-0\"><a href=\"#cite_note-sxManifest20120925-28\">[22]</a></sup><br/>(Dragon C103)\n</td>\n<td>4,700 kg (10,400 lb)\n</td>\n<td><a href=\"/wiki/Low_Earth_orbit\" title=\"Low Earth orbit\">LEO</a> (<a href=\"/wiki/International_Space_Station\" title=\"International Space Station\">ISS</a>)\n</td>\n<td><a href=\"/wiki/NASA\" title=\"NASA\">NASA</a> (<a href=\"/wiki/Commercial_Resupply_Services\" title=\"Commercial Resupply Services\">CRS</a>)\n</td>\n<td class=\"table-success\" style=\"background: LightGreen; color: black; vertical-align: middle; text-align: center;\">Success\n</td>\n<td rowspan=\"2\" style=\"background:#ececec; text-align:center;\"><span class=\"nowrap\">No attempt</span>\n</td></tr>\n<tr>\n<td><a href=\"/wiki/Orbcomm_(satellite)\" title=\"Orbcomm (satellite)\">Orbcomm-OG2</a><sup class=\"reference\" id=\"cite_ref-Orbcomm_29-0\"><a href=\"#cite_note-Orbcomm-29\">[23]</a></sup>\n</td>\n<td>172 kg (379 lb)<sup class=\"reference\" id=\"cite_ref-gunter-og2_30-0\"><a href=\"#cite_note-gunter-og2-30\">[24]</a></sup>\n</td>\n<td><a href=\"/wiki/Low_Earth_orbit\" title=\"Low Earth orbit\">LEO</a>\n</td>\n<td><a href=\"/wiki/Orbcomm\" title=\"Orbcomm\">Orbcomm</a>\n</td>\n<td class=\"table-partial\" style=\"background: wheat; color: black; vertical-align: middle; text-align: center;\">Partial failure<sup class=\"reference\" id=\"cite_ref-nyt-20121030_31-0\"><a href=\"#cite_note-nyt-20121030-31\">[25]</a></sup>\n</td></tr>\n<tr>\n<td colspan=\"9\">CRS-1 was successful, but the <a href=\"/wiki/Secondary_payload\" title=\"Secondary payload\">secondary payload</a> was inserted into an abnormally low orbit and subsequently lost. This was due to one of the nine <a href=\"/wiki/SpaceX_Merlin\" title=\"SpaceX Merlin\">Merlin engines</a> shutting down during the launch, and NASA declining a second reignition, as per <a href=\"/wiki/International_Space_Station\" title=\"International Space Station\">ISS</a> visiting vehicle safety rules, the primary payload owner is contractually allowed to decline a second reignition. NASA stated that this was because SpaceX could not guarantee a high enough likelihood of the second stage completing the second burn successfully which was required to avoid any risk of secondary payload's collision with the ISS.<sup class=\"reference\" id=\"cite_ref-OrbcommTotalLoss_32-0\"><a href=\"#cite_note-OrbcommTotalLoss-32\">[26]</a></sup><sup class=\"reference\" id=\"cite_ref-sn20121011_33-0\"><a href=\"#cite_note-sn20121011-33\">[27]</a></sup><sup class=\"reference\" id=\"cite_ref-34\"><a href=\"#cite_note-34\">[28]</a></sup>\n</td></tr>\n<tr>\n<th rowspan=\"2\" scope=\"row\" style=\"text-align:center;\">5\n</th>\n<td>1 March 2013,<br/>15:10\n</td>\n<td><a href=\"/wiki/Falcon_9_v1.0\" title=\"Falcon 9 v1.0\">F9 v1.0</a><sup class=\"reference\" id=\"cite_ref-MuskMay2012_13-4\"><a href=\"#cite_note-MuskMay2012-13\">[7]</a></sup><br/>B0007.1<sup class=\"reference\" id=\"cite_ref-block_numbers_14-4\"><a href=\"#cite_note-block_numbers-14\">[8]</a></sup>\n</td>\n<td><a href=\"/wiki/Cape_Canaveral_Space_Force_Station\" title=\"Cape Canaveral Space Force Station\">CCAFS</a>,<br/><a href=\"/wiki/Cape_Canaveral_Space_Launch_Complex_40\" title=\"Cape Canaveral Space Launch Complex 40\">SLC-40</a>\n</td>\n<td><a href=\"/wiki/SpaceX_CRS-2\" title=\"SpaceX CRS-2\">SpaceX CRS-2</a><sup class=\"reference\" id=\"cite_ref-sxManifest20120925_28-1\"><a href=\"#cite_note-sxManifest20120925-28\">[22]</a></sup><br/>(Dragon C104)\n</td>\n<td>4,877 kg (10,752 lb)\n</td>\n<td><a href=\"/wiki/Low_Earth_orbit\" title=\"Low Earth orbit\">LEO</a> (<a class=\"mw-redirect\" href=\"/wiki/ISS\" title=\"ISS\">ISS</a>)\n</td>\n<td><a href=\"/wiki/NASA\" title=\"NASA\">NASA</a> (<a href=\"/wiki/Commercial_Resupply_Services\" title=\"Commercial Resupply Services\">CRS</a>)\n</td>\n<td class=\"table-success\" style=\"background: LightGreen; color: black; vertical-align: middle; text-align: center;\">Success\n</td>\n<td class=\"table-noAttempt\" style=\"background: #ececec; color: black; vertical-align: middle; white-space: nowrap; text-align: center;\">No attempt\n</td></tr>\n<tr>\n<td colspan=\"9\">Last launch of the original Falcon 9 v1.0 <a href=\"/wiki/Launch_vehicle\" title=\"Launch vehicle\">launch vehicle</a>, first use of the unpressurized trunk section of Dragon.<sup class=\"reference\" id=\"cite_ref-sxf9_20110321_35-0\"><a href=\"#cite_note-sxf9_20110321-35\">[29]</a></sup>\n</td></tr>\n<tr>\n<th rowspan=\"2\" scope=\"row\" style=\"text-align:center;\">6\n</th>\n<td>29 September 2013,<br/>16:00<sup class=\"reference\" id=\"cite_ref-pa20130930_36-0\"><a href=\"#cite_note-pa20130930-36\">[30]</a></sup>\n</td>\n<td><a href=\"/wiki/Falcon_9_v1.1\" title=\"Falcon 9 v1.1\">F9 v1.1</a><sup class=\"reference\" id=\"cite_ref-MuskMay2012_13-5\"><a href=\"#cite_note-MuskMay2012-13\">[7]</a></sup><br/>B1003<sup class=\"reference\" id=\"cite_ref-block_numbers_14-5\"><a href=\"#cite_note-block_numbers-14\">[8]</a></sup>\n</td>\n<td><a class=\"mw-redirect\" href=\"/wiki/Vandenberg_Air_Force_Base\" title=\"Vandenberg Air Force Base\">VAFB</a>,<br/><a href=\"/wiki/Vandenberg_Space_Launch_Complex_4\" title=\"Vandenberg Space Launch Complex 4\">SLC-4E</a>\n</td>\n<td><a href=\"/wiki/CASSIOPE\" title=\"CASSIOPE\">CASSIOPE</a><sup class=\"reference\" id=\"cite_ref-sxManifest20120925_28-2\"><a href=\"#cite_note-sxManifest20120925-28\">[22]</a></sup><sup class=\"reference\" id=\"cite_ref-CASSIOPE_MDA_37-0\"><a href=\"#cite_note-CASSIOPE_MDA-37\">[31]</a></sup>\n</td>\n<td>500 kg (1,100 lb)\n</td>\n<td><a href=\"/wiki/Polar_orbit\" title=\"Polar orbit\">Polar orbit</a> <a href=\"/wiki/Low_Earth_orbit\" title=\"Low Earth orbit\">LEO</a>\n</td>\n<td><a href=\"/wiki/Maxar_Technologies\" title=\"Maxar Technologies\">MDA</a>\n</td>\n<td class=\"table-success\" style=\"background: LightGreen; color: black; vertical-align: middle; text-align: center;\">Success<sup class=\"reference\" id=\"cite_ref-pa20130930_36-1\"><a href=\"#cite_note-pa20130930-36\">[30]</a></sup>\n</td>\n<td class=\"table-no2\" style=\"background: #ffdddd; color: black; vertical-align: middle; text-align: center;\">Uncontrolled<br/><small>(ocean)</small><sup class=\"reference\" id=\"cite_ref-ocean_landing_38-0\"><a href=\"#cite_note-ocean_landing-38\">[d]</a></sup>\n</td></tr>\n<tr>\n<td colspan=\"9\">First commercial mission with a private customer, first launch from Vandenberg, and demonstration flight of Falcon 9 v1.1 with an improved 13-tonne to LEO capacity.<sup class=\"reference\" id=\"cite_ref-sxf9_20110321_35-1\"><a href=\"#cite_note-sxf9_20110321-35\">[29]</a></sup> After separation from the second stage carrying Canadian commercial and scientific satellites, the first stage booster performed a controlled reentry,<sup class=\"reference\" id=\"cite_ref-39\"><a href=\"#cite_note-39\">[32]</a></sup> and an <a href=\"/wiki/Falcon_9_first-stage_landing_tests\" title=\"Falcon 9 first-stage landing tests\">ocean touchdown test</a> for the first time. This provided good test data, even though the booster started rolling as it neared the ocean, leading to the shutdown of the central engine as the roll depleted it of fuel, resulting in a hard impact with the ocean.<sup class=\"reference\" id=\"cite_ref-pa20130930_36-2\"><a href=\"#cite_note-pa20130930-36\">[30]</a></sup> This was the first known attempt of a rocket engine being lit to perform a supersonic retro propulsion, and allowed SpaceX to enter a public-private partnership with <a href=\"/wiki/NASA\" title=\"NASA\">NASA</a> and its Mars entry, descent, and landing technologies research projects.<sup class=\"reference\" id=\"cite_ref-40\"><a href=\"#cite_note-40\">[33]</a></sup> <small>(<a href=\"#Maiden_flight_of_v1.1\">more details below</a>)</small>\n</td></tr>\n<tr>\n<th rowspan=\"2\" scope=\"row\" style=\"text-align:center;\">7\n</th>\n<td>3 December 2013,<br/>22:41<sup class=\"reference\" id=\"cite_ref-sfn_wwls20130624_41-0\"><a href=\"#cite_note-sfn_wwls20130624-41\">[34]</a></sup>\n</td>\n<td><a href=\"/wiki/Falcon_9_v1.1\" title=\"Falcon 9 v1.1\">F9 v1.1</a><br/>B1004\n</td>\n<td><a href=\"/wiki/Cape_Canaveral_Space_Force_Station\" title=\"Cape Canaveral Space Force Station\">CCAFS</a>,<br/><a href=\"/wiki/Cape_Canaveral_Space_Launch_Complex_40\" title=\"Cape Canaveral Space Launch Complex 40\">SLC-40</a>\n</td>\n<td><a href=\"/wiki/SES-8\" title=\"SES-8\">SES-8</a><sup class=\"reference\" id=\"cite_ref-sxManifest20120925_28-3\"><a href=\"#cite_note-sxManifest20120925-28\">[22]</a></sup><sup class=\"reference\" id=\"cite_ref-spx-pr_42-0\"><a href=\"#cite_note-spx-pr-42\">[35]</a></sup><sup class=\"reference\" id=\"cite_ref-aw20110323_43-0\"><a href=\"#cite_note-aw20110323-43\">[36]</a></sup>\n</td>\n<td>3,170 kg (6,990 lb)\n</td>\n<td><a href=\"/wiki/Geostationary_transfer_orbit\" title=\"Geostationary transfer orbit\">GTO</a>\n</td>\n<td><a href=\"/wiki/SES_S.A.\" title=\"SES S.A.\">SES</a>\n</td>\n<td class=\"table-success\" style=\"background: LightGreen; color: black; vertical-align: middle; text-align: center;\">Success<sup class=\"reference\" id=\"cite_ref-SNMissionStatus7_44-0\"><a href=\"#cite_note-SNMissionStatus7-44\">[37]</a></sup>\n</td>\n<td class=\"table-noAttempt\" style=\"background: #ececec; color: black; vertical-align: middle; white-space: nowrap; text-align: center;\">No attempt<br/><sup class=\"reference\" id=\"cite_ref-sf10120131203_45-0\"><a href=\"#cite_note-sf10120131203-45\">[38]</a></sup>\n</td></tr>\n<tr>\n<td colspan=\"9\">First <a href=\"/wiki/Geostationary_transfer_orbit\" title=\"Geostationary transfer orbit\">Geostationary transfer orbit</a> (GTO) launch for Falcon 9,<sup class=\"reference\" id=\"cite_ref-spx-pr_42-1\"><a href=\"#cite_note-spx-pr-42\">[35]</a></sup> and first successful reignition of the second stage.<sup class=\"reference\" id=\"cite_ref-46\"><a href=\"#cite_note-46\">[39]</a></sup> SES-8 was inserted into a <a href=\"/wiki/Geostationary_transfer_orbit\" title=\"Geostationary transfer orbit\">Super-Synchronous Transfer Orbit</a> of 79,341 km (49,300 mi) in apogee with an <a href=\"/wiki/Orbital_inclination\" title=\"Orbital inclination\">inclination</a> of 20.55° to the <a href=\"/wiki/Equator\" title=\"Equator\">equator</a>.\n</td></tr></tbody></table>\n"
]
],
[
[
"You should able to see the columns names embedded in the table header elements `<th>` as follows:\n",
"_____no_output_____"
],
[
"```\n<tr>\n<th scope=\"col\">Flight No.\n</th>\n<th scope=\"col\">Date and<br/>time (<a href=\"/wiki/Coordinated_Universal_Time\" title=\"Coordinated Universal Time\">UTC</a>)\n</th>\n<th scope=\"col\"><a href=\"/wiki/List_of_Falcon_9_first-stage_boosters\" title=\"List of Falcon 9 first-stage boosters\">Version,<br/>Booster</a> <sup class=\"reference\" id=\"cite_ref-booster_11-0\"><a href=\"#cite_note-booster-11\">[b]</a></sup>\n</th>\n<th scope=\"col\">Launch site\n</th>\n<th scope=\"col\">Payload<sup class=\"reference\" id=\"cite_ref-Dragon_12-0\"><a href=\"#cite_note-Dragon-12\">[c]</a></sup>\n</th>\n<th scope=\"col\">Payload mass\n</th>\n<th scope=\"col\">Orbit\n</th>\n<th scope=\"col\">Customer\n</th>\n<th scope=\"col\">Launch<br/>outcome\n</th>\n<th scope=\"col\"><a href=\"/wiki/Falcon_9_first-stage_landing_tests\" title=\"Falcon 9 first-stage landing tests\">Booster<br/>landing</a>\n</th></tr>\n```\n",
"_____no_output_____"
],
[
"Next, we just need to iterate through the `<th>` elements and apply the provided `extract_column_from_header()` to extract column name one by one\n",
"_____no_output_____"
]
],
[
[
"column_names = []\n\nth = first_launch_table.find_all('th')\nfor name in th:\n name = extract_column_from_header(name)\n if name is not None and len(name) > 0:\n column_names.append(name)\n\n# Apply find_all() function with `th` element on first_launch_table\n# Iterate each th element and apply the provided extract_column_from_header() to get a column name\n# Append the Non-empty column name (`if name is not None and len(name) > 0`) into a list called column_names\n",
"_____no_output_____"
]
],
[
[
"Check the extracted column names\n",
"_____no_output_____"
]
],
[
[
"print(column_names)",
"['Flight No.', 'Date and time ( )', 'Launch site', 'Payload', 'Payload mass', 'Orbit', 'Customer', 'Launch outcome']\n"
]
],
[
[
"## TASK 3: Create a data frame by parsing the launch HTML tables\n",
"_____no_output_____"
],
[
"We will create an empty dictionary with keys from the extracted column names in the previous task. Later, this dictionary will be converted into a Pandas dataframe\n",
"_____no_output_____"
]
],
[
[
"launch_dict= dict.fromkeys(column_names)\n\n# Remove an irrelvant column\ndel launch_dict['Date and time ( )']\n\n# Let's initial the launch_dict with each value to be an empty list\nlaunch_dict['Flight No.'] = []\nlaunch_dict['Launch site'] = []\nlaunch_dict['Payload'] = []\nlaunch_dict['Payload mass'] = []\nlaunch_dict['Orbit'] = []\nlaunch_dict['Customer'] = []\nlaunch_dict['Launch outcome'] = []\n# Added some new columns\nlaunch_dict['Version Booster']=[]\nlaunch_dict['Booster landing']=[]\nlaunch_dict['Date']=[]\nlaunch_dict['Time']=[]",
"_____no_output_____"
]
],
[
[
"Next, we just need to fill up the `launch_dict` with launch records extracted from table rows.\n",
"_____no_output_____"
],
[
"Usually, HTML tables in Wiki pages are likely to contain unexpected annotations and other types of noises, such as reference links `B0004.1[8]`, missing values `N/A [e]`, inconsistent formatting, etc.\n",
"_____no_output_____"
],
[
"To simplify the parsing process, we have provided an incomplete code snippet below to help you to fill up the `launch_dict`. Please complete the following code snippet with TODOs or you can choose to write your own logic to parse all launch tables:\n",
"_____no_output_____"
]
],
[
[
"extracted_row = 0\n#Extract each table \nfor table_number,table in enumerate(soup.find_all('table',\"wikitable plainrowheaders collapsible\")):\n # get table row \n for rows in table.find_all(\"tr\"):\n #check to see if first table heading is as number corresponding to launch a number \n if rows.th:\n if rows.th.string:\n flight_number=rows.th.string.strip()\n flag=flight_number.isdigit()\n else:\n flag=False\n #get table element \n row=rows.find_all('td')\n #if it is number save cells in a dictonary \n if flag:\n extracted_row += 1\n # Flight Number value\n # TODO: Append the flight_number into launch_dict with key `Flight No.`\n #print(flight_number)\n datatimelist=date_time(row[0])\n \n # Date value\n # TODO: Append the date into launch_dict with key `Date`\n date = datatimelist[0].strip(',')\n #print(date)\n \n # Time value\n # TODO: Append the time into launch_dict with key `Time`\n time = datatimelist[1]\n #print(time)\n \n # Booster version\n # TODO: Append the bv into launch_dict with key `Version Booster`\n bv=booster_version(row[1])\n if not(bv):\n bv=row[1].a.string\n print(bv)\n \n # Launch Site\n # TODO: Append the bv into launch_dict with key `Launch Site`\n launch_site = row[2].a.string\n #print(launch_site)\n \n # Payload\n # TODO: Append the payload into launch_dict with key `Payload`\n payload = row[3].a.string\n #print(payload)\n \n # Payload Mass\n # TODO: Append the payload_mass into launch_dict with key `Payload mass`\n payload_mass = get_mass(row[4])\n #print(payload)\n \n # Orbit\n # TODO: Append the orbit into launch_dict with key `Orbit`\n orbit = row[5].a.string\n #print(orbit)\n \n # Customer\n # TODO: Append the customer into launch_dict with key `Customer`\n customer = row[6].a.string\n #print(customer)\n \n # Launch outcome\n # TODO: Append the launch_outcome into launch_dict with key `Launch outcome`\n launch_outcome = list(row[7].strings)[0]\n #print(launch_outcome)\n \n # Booster landing\n # TODO: Append the launch_outcome into launch_dict with key `Booster landing`\n booster_landing = landing_status(row[8])\n #print(booster_landing)\n ",
"F9 v1.0B0003.1\nF9 v1.0B0004.1\nF9 v1.0B0005.1\nF9 v1.0B0006.1\nF9 v1.0B0007.1\nF9 v1.1B1003\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 v1.1\nF9 FT\nF9 v1.1\nF9 FT\nF9 FT\nF9 FT\nF9 FT\nF9 FT\nF9 FT\nF9 FT\nF9 FT\nF9 FT\nF9 FT\nF9 FT♺\nF9 FT\nF9 FT\nF9 FT\nF9 FTB1029.2\nF9 FT\nF9 FT\nF9 B4\nF9 FT\nF9 B4\nF9 B4\nF9 FTB1031.2\nF9 B4\nF9 FTB1035.2\nF9 FTB1036.2\nF9 B4\nF9 FTB1032.2\nF9 FTB1038.2\nF9 B4\nF9 B4B1041.2\nF9 B4B1039.2\nF9 B4\nF9 B5B1046.1\nF9 B4B1043.2\nF9 B4B1040.2\nF9 B4B1045.2\nF9 B5\nF9 B5B1048\nF9 B5B1046.2\nF9 B5\nF9 B5B1048.2\nF9 B5B1047.2\nF9 B5B1046.3\nF9 B5\nF9 B5\nF9 B5B1049.2\nF9 B5B1048.3\nF9 B5[268]\nF9 B5\nF9 B5B1049.3\nF9 B5B1051.2\nF9 B5B1056.2\nF9 B5B1047.3\nF9 B5\nF9 B5\nF9 B5B1056.3\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5B1058.2\nF9 B5\nF9 B5B1049.6\nF9 B5\nF9 B5B1060.2\nF9 B5B1058.3\nF9 B5B1051.6\nF9 B5\nF9 B5\nF9 B5\nF9 B5\nF9 B5 ♺\nF9 B5 ♺\nF9 B5 ♺\nF9 B5 ♺\nF9 B5\nF9 B5B1051.8\nF9 B5B1058.5\n"
]
],
[
[
"After you have fill in the parsed launch record values into `launch_dict`, you can create a dataframe from it.\n",
"_____no_output_____"
]
],
[
[
"df=pd.DataFrame(launch_dict)",
"_____no_output_____"
]
],
[
[
"We can now export it to a <b>CSV</b> for the next section, but to make the answers consistent and in case you have difficulties finishing this lab.\n\nFollowing labs will be using a provided dataset to make each lab independent.\n",
"_____no_output_____"
],
[
"<code>df.to_csv('spacex_web_scraped.csv', index=False)</code>\n",
"_____no_output_____"
],
[
"## Authors\n",
"_____no_output_____"
],
[
"<a href=\"https://www.linkedin.com/in/yan-luo-96288783/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDS0321ENSkillsNetwork26802033-2021-01-01\">Yan Luo</a>\n",
"_____no_output_____"
],
[
"<a href=\"https://www.linkedin.com/in/nayefaboutayoun/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDS0321ENSkillsNetwork26802033-2021-01-01\">Nayef Abou Tayoun</a>\n",
"_____no_output_____"
],
[
"## Change Log\n",
"_____no_output_____"
],
[
"| Date (YYYY-MM-DD) | Version | Changed By | Change Description |\n| ----------------- | ------- | ---------- | --------------------------- |\n| 2021-06-09 | 1.0 | Yan Luo | Tasks updates |\n| 2020-11-10 | 1.0 | Nayef | Created the initial version |\n",
"_____no_output_____"
],
[
"Copyright © 2021 IBM Corporation. All rights reserved.\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbca64866d868ad4656b732ee1cee937fc4f13b3
| 214,328 |
ipynb
|
Jupyter Notebook
|
docs/load-entities.ipynb
|
DevconX/Malaya
|
a2e7030f0911d65c9c1c72d38bc3e7c53b8e06fc
|
[
"MIT"
] | 39 |
2018-03-12T04:26:42.000Z
|
2018-12-05T03:53:45.000Z
|
docs/load-entities.ipynb
|
DevconX/Malaya
|
a2e7030f0911d65c9c1c72d38bc3e7c53b8e06fc
|
[
"MIT"
] | 12 |
2018-10-01T07:28:23.000Z
|
2018-12-10T01:59:25.000Z
|
example/entities/load-entities.ipynb
|
DevconX/Malaya
|
a2e7030f0911d65c9c1c72d38bc3e7c53b8e06fc
|
[
"MIT"
] | 16 |
2018-03-16T05:46:12.000Z
|
2018-12-10T04:15:07.000Z
| 85.971921 | 75,480 | 0.770478 |
[
[
[
"# Entities Recognition",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\">\n\nThis tutorial is available as an IPython notebook at [Malaya/example/entities](https://github.com/huseinzol05/Malaya/tree/master/example/entities).\n \n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-warning\">\n\nThis module only trained on standard language structure, so it is not save to use it for local language structure.\n \n</div>",
"_____no_output_____"
]
],
[
[
"%%time\nimport malaya",
"CPU times: user 6.33 s, sys: 1.4 s, total: 7.73 s\nWall time: 9.25 s\n"
]
],
[
[
"### Models accuracy\n\nWe use `sklearn.metrics.classification_report` for accuracy reporting, check at https://malaya.readthedocs.io/en/latest/models-accuracy.html#entities-recognition and https://malaya.readthedocs.io/en/latest/models-accuracy.html#entities-recognition-ontonotes5",
"_____no_output_____"
],
[
"### Describe supported entities",
"_____no_output_____"
]
],
[
[
"import pandas as pd\npd.set_option('display.max_colwidth', -1)\nmalaya.entity.describe()",
"_____no_output_____"
]
],
[
[
"### Describe supported Ontonotes 5 entities",
"_____no_output_____"
]
],
[
[
"malaya.entity.describe_ontonotes5()",
"_____no_output_____"
]
],
[
[
"### List available Transformer NER models",
"_____no_output_____"
]
],
[
[
"malaya.entity.available_transformer()",
"INFO:root:tested on 20% test set.\n"
]
],
[
[
"### List available Transformer NER Ontonotes 5 models",
"_____no_output_____"
]
],
[
[
"malaya.entity.available_transformer_ontonotes5()",
"INFO:root:tested on 20% test set.\n"
],
[
"string = 'KUALA LUMPUR: Sempena sambutan Aidilfitri minggu depan, Perdana Menteri Tun Dr Mahathir Mohamad dan Menteri Pengangkutan Anthony Loke Siew Fook menitipkan pesanan khas kepada orang ramai yang mahu pulang ke kampung halaman masing-masing. Dalam video pendek terbitan Jabatan Keselamatan Jalan Raya (JKJR) itu, Dr Mahathir menasihati mereka supaya berhenti berehat dan tidur sebentar sekiranya mengantuk ketika memandu.'\nstring1 = 'memperkenalkan Husein, dia sangat comel, berumur 25 tahun, bangsa melayu, agama islam, tinggal di cyberjaya malaysia, bercakap bahasa melayu, semua membaca buku undang-undang kewangan, dengar laju Siti Nurhaliza - Seluruh Cinta sambil makan ayam goreng KFC'",
"_____no_output_____"
]
],
[
[
"### Load Transformer model\n\n```python\ndef transformer(model: str = 'xlnet', quantized: bool = False, **kwargs):\n \"\"\"\n Load Transformer Entity Tagging model trained on Malaya Entity, transfer learning Transformer + CRF.\n\n Parameters\n ----------\n model : str, optional (default='bert')\n Model architecture supported. Allowed values:\n\n * ``'bert'`` - Google BERT BASE parameters.\n * ``'tiny-bert'`` - Google BERT TINY parameters.\n * ``'albert'`` - Google ALBERT BASE parameters.\n * ``'tiny-albert'`` - Google ALBERT TINY parameters.\n * ``'xlnet'`` - Google XLNET BASE parameters.\n * ``'alxlnet'`` - Malaya ALXLNET BASE parameters.\n * ``'fastformer'`` - FastFormer BASE parameters.\n * ``'tiny-fastformer'`` - FastFormer TINY parameters.\n\n quantized : bool, optional (default=False)\n if True, will load 8-bit quantized model.\n Quantized model not necessary faster, totally depends on the machine.\n\n Returns\n -------\n result: model\n List of model classes:\n\n * if `bert` in model, will return `malaya.model.bert.TaggingBERT`.\n * if `xlnet` in model, will return `malaya.model.xlnet.TaggingXLNET`.\n * if `fastformer` in model, will return `malaya.model.fastformer.TaggingFastFormer`.\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"model = malaya.entity.transformer(model = 'alxlnet')",
"INFO:root:running entity/alxlnet using device /device:CPU:0\n"
]
],
[
[
"#### Load Quantized model\n\nTo load 8-bit quantized model, simply pass `quantized = True`, default is `False`.\n\nWe can expect slightly accuracy drop from quantized model, and not necessary faster than normal 32-bit float model, totally depends on machine.",
"_____no_output_____"
]
],
[
[
"quantized_model = malaya.entity.transformer(model = 'alxlnet', quantized = True)",
"WARNING:root:Load quantized model will cause accuracy drop.\nINFO:root:running entity/alxlnet-quantized using device /device:CPU:0\n"
]
],
[
[
"#### Predict\n\n```python\ndef predict(self, string: str):\n \"\"\"\n Tag a string.\n\n Parameters\n ----------\n string : str\n\n Returns\n -------\n result: Tuple[str, str]\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"model.predict(string)",
"_____no_output_____"
],
[
"model.predict(string1)",
"_____no_output_____"
],
[
"quantized_model.predict(string)",
"_____no_output_____"
],
[
"quantized_model.predict(string1)",
"_____no_output_____"
]
],
[
[
"#### Group similar tags\n\n```python\ndef analyze(self, string: str):\n \"\"\"\n Analyze a string.\n\n Parameters\n ----------\n string : str\n\n Returns\n -------\n result: {'words': List[str], 'tags': [{'text': 'text', 'type': 'location', 'score': 1.0, 'beginOffset': 0, 'endOffset': 1}]}\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"model.analyze(string)",
"_____no_output_____"
],
[
"model.analyze(string1)",
"_____no_output_____"
]
],
[
[
"#### Vectorize\n\nLet say you want to visualize word level in lower dimension, you can use `model.vectorize`,\n\n```python\ndef vectorize(self, string: str):\n \"\"\"\n vectorize a string.\n\n Parameters\n ----------\n string: List[str]\n\n Returns\n -------\n result: np.array\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"strings = [string, \n 'Husein baca buku Perlembagaan yang berharga 3k ringgit dekat kfc sungai petani minggu lepas, 2 ptg 2 oktober 2019 , suhu 32 celcius, sambil makan ayam goreng dan milo o ais',\n 'contact Husein at [email protected]',\n 'tolong tempahkan meja makan makan nasi dagang dan jus apple, milo tarik esok dekat Restoran Sebulek']",
"_____no_output_____"
],
[
"r = [quantized_model.vectorize(string) for string in strings]",
"_____no_output_____"
],
[
"x, y = [], []\nfor row in r:\n x.extend([i[0] for i in row])\n y.extend([i[1] for i in row])",
"_____no_output_____"
],
[
"from sklearn.manifold import TSNE\nimport matplotlib.pyplot as plt\n\ntsne = TSNE().fit_transform(y)\ntsne.shape",
"_____no_output_____"
],
[
"plt.figure(figsize = (7, 7))\nplt.scatter(tsne[:, 0], tsne[:, 1])\nlabels = x\nfor label, x, y in zip(\n labels, tsne[:, 0], tsne[:, 1]\n):\n label = (\n '%s, %.3f' % (label[0], label[1])\n if isinstance(label, list)\n else label\n )\n plt.annotate(\n label,\n xy = (x, y),\n xytext = (0, 0),\n textcoords = 'offset points',\n )",
"_____no_output_____"
]
],
[
[
"Pretty good, the model able to know cluster similar entities.",
"_____no_output_____"
],
[
"### Load Transformer Ontonotes 5 model\n\n```python\ndef transformer_ontonotes5(\n model: str = 'xlnet', quantized: bool = False, **kwargs\n):\n \"\"\"\n Load Transformer Entity Tagging model trained on Ontonotes 5 Bahasa, transfer learning Transformer + CRF.\n\n Parameters\n ----------\n model : str, optional (default='bert')\n Model architecture supported. Allowed values:\n\n * ``'bert'`` - Google BERT BASE parameters.\n * ``'tiny-bert'`` - Google BERT TINY parameters.\n * ``'albert'`` - Google ALBERT BASE parameters.\n * ``'tiny-albert'`` - Google ALBERT TINY parameters.\n * ``'xlnet'`` - Google XLNET BASE parameters.\n * ``'alxlnet'`` - Malaya ALXLNET BASE parameters.\n * ``'fastformer'`` - FastFormer BASE parameters.\n * ``'tiny-fastformer'`` - FastFormer TINY parameters.\n\n quantized : bool, optional (default=False)\n if True, will load 8-bit quantized model.\n Quantized model not necessary faster, totally depends on the machine.\n\n Returns\n -------\n result: model\n List of model classes:\n\n * if `bert` in model, will return `malaya.model.bert.TaggingBERT`.\n * if `xlnet` in model, will return `malaya.model.xlnet.TaggingXLNET`.\n * if `fastformer` in model, will return `malaya.model.fastformer.TaggingFastFormer`.\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"albert = malaya.entity.transformer_ontonotes5(model = 'albert')",
"INFO:root:running entity-ontonotes5/albert using device /device:CPU:0\n"
],
[
"alxlnet = malaya.entity.transformer_ontonotes5(model = 'alxlnet')",
"INFO:root:running entity-ontonotes5/alxlnet using device /device:CPU:0\n"
]
],
[
[
"#### Load Quantized model\n\nTo load 8-bit quantized model, simply pass `quantized = True`, default is `False`.\n\nWe can expect slightly accuracy drop from quantized model, and not necessary faster than normal 32-bit float model, totally depends on machine.",
"_____no_output_____"
]
],
[
[
"quantized_albert = malaya.entity.transformer_ontonotes5(model = 'albert', quantized = True)",
"WARNING:root:Load quantized model will cause accuracy drop.\nINFO:root:running entity-ontonotes5/albert-quantized using device /device:CPU:0\n"
],
[
"quantized_alxlnet = malaya.entity.transformer_ontonotes5(model = 'alxlnet', quantized = True)",
"WARNING:root:Load quantized model will cause accuracy drop.\nINFO:root:running entity-ontonotes5/alxlnet-quantized using device /device:CPU:0\n"
]
],
[
[
"#### Predict\n\n```python\ndef predict(self, string: str):\n \"\"\"\n Tag a string.\n\n Parameters\n ----------\n string : str\n\n Returns\n -------\n result: Tuple[str, str]\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"albert.predict(string)",
"_____no_output_____"
],
[
"alxlnet.predict(string)",
"_____no_output_____"
],
[
"albert.predict(string1)",
"_____no_output_____"
],
[
"alxlnet.predict(string1)",
"_____no_output_____"
],
[
"quantized_albert.predict(string)",
"_____no_output_____"
],
[
"quantized_alxlnet.predict(string1)",
"_____no_output_____"
]
],
[
[
"#### Group similar tags\n\n```python\ndef analyze(self, string: str):\n \"\"\"\n Analyze a string.\n\n Parameters\n ----------\n string : str\n\n Returns\n -------\n result: {'words': List[str], 'tags': [{'text': 'text', 'type': 'location', 'score': 1.0, 'beginOffset': 0, 'endOffset': 1}]}\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"alxlnet.analyze(string1)",
"_____no_output_____"
]
],
[
[
"#### Vectorize\n\nLet say you want to visualize word level in lower dimension, you can use `model.vectorize`,\n\n```python\ndef vectorize(self, string: str):\n \"\"\"\n vectorize a string.\n\n Parameters\n ----------\n string: List[str]\n\n Returns\n -------\n result: np.array\n \"\"\"\n```",
"_____no_output_____"
]
],
[
[
"strings = [string, string1]\nr = [quantized_model.vectorize(string) for string in strings]",
"_____no_output_____"
],
[
"x, y = [], []\nfor row in r:\n x.extend([i[0] for i in row])\n y.extend([i[1] for i in row])",
"_____no_output_____"
],
[
"tsne = TSNE().fit_transform(y)\ntsne.shape",
"_____no_output_____"
],
[
"plt.figure(figsize = (7, 7))\nplt.scatter(tsne[:, 0], tsne[:, 1])\nlabels = x\nfor label, x, y in zip(\n labels, tsne[:, 0], tsne[:, 1]\n):\n label = (\n '%s, %.3f' % (label[0], label[1])\n if isinstance(label, list)\n else label\n )\n plt.annotate(\n label,\n xy = (x, y),\n xytext = (0, 0),\n textcoords = 'offset points',\n )",
"_____no_output_____"
]
],
[
[
"Pretty good, the model able to know cluster similar entities.",
"_____no_output_____"
],
[
"### Load general Malaya entity model\n\nThis model able to classify,\n\n1. date\n2. money\n3. temperature\n4. distance\n5. volume\n6. duration\n7. phone\n8. email\n9. url\n10. time\n11. datetime\n12. local and generic foods, can check available rules in malaya.texts._food\n13. local and generic drinks, can check available rules in malaya.texts._food\n\nWe can insert BERT or any deep learning model by passing `malaya.entity.general_entity(model = model)`, as long the model has `predict` method and return `[(string, label), (string, label)]`. This is an optional.",
"_____no_output_____"
]
],
[
[
"entity = malaya.entity.general_entity(model = model)",
"_____no_output_____"
],
[
"entity.predict('Husein baca buku Perlembagaan yang berharga 3k ringgit dekat kfc sungai petani minggu lepas, 2 ptg 2 oktober 2019 , suhu 32 celcius, sambil makan ayam goreng dan milo o ais')",
"_____no_output_____"
],
[
"entity.predict('contact Husein at [email protected]')",
"_____no_output_____"
],
[
"entity.predict('tolong tempahkan meja makan makan nasi dagang dan jus apple, milo tarik esok dekat Restoran Sebulek')",
"_____no_output_____"
]
],
[
[
"### Voting stack model",
"_____no_output_____"
]
],
[
[
"malaya.stack.voting_stack([albert, alxlnet, alxlnet], string1)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
cbca68300d14adf63188c2d4b0966bbe05ce98cf
| 70,403 |
ipynb
|
Jupyter Notebook
|
FCM.ipynb
|
Phantom-Ren/PR_TH
|
e7ffbbddb51c2852ae7d37cdecec40aaf706c8f2
|
[
"Apache-2.0"
] | null | null | null |
FCM.ipynb
|
Phantom-Ren/PR_TH
|
e7ffbbddb51c2852ae7d37cdecec40aaf706c8f2
|
[
"Apache-2.0"
] | null | null | null |
FCM.ipynb
|
Phantom-Ren/PR_TH
|
e7ffbbddb51c2852ae7d37cdecec40aaf706c8f2
|
[
"Apache-2.0"
] | null | null | null | 117.730769 | 33,658 | 0.834325 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Phantom-Ren/PR_TH/blob/master/FCM.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"<center>\n\n# 模式识别·第七次作业·模糊聚类(Fussy C Means)\n\n#### 纪泽西 17375338\n\n#### Last Modified:26th,April,2020\n\n</center>\n\n<table align=\"center\">\n <td align=\"center\"><a target=\"_blank\" href=\"https://colab.research.google.com/github/Phantom-Ren/PR_TH/blob/master/FCM.ipynb\"> \n <img src=\"http://introtodeeplearning.com/images/colab/colab.png?v2.0\" style=\"padding-bottom:5px;\" /><br>Run in Google Colab</a></td>\n</table>\n",
"_____no_output_____"
],
[
"## Part1: 导入库文件及数据集\n\n#### 如需在其他环境运行需改变数据集所在路径",
"_____no_output_____"
]
],
[
[
" !pip install -U scikit-fuzzy",
"Collecting scikit-fuzzy\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/6c/f0/5eb5dbe0fd8dfe7d4651a8f4e591a196623a22b9e5339101e559695b4f6c/scikit-fuzzy-0.4.2.tar.gz (993kB)\n\u001b[K |████████████████████████████████| 1.0MB 157kB/s \n\u001b[?25hRequirement already satisfied, skipping upgrade: numpy>=1.6.0 in /usr/local/lib/python3.6/dist-packages (from scikit-fuzzy) (1.18.3)\nRequirement already satisfied, skipping upgrade: scipy>=0.9.0 in /usr/local/lib/python3.6/dist-packages (from scikit-fuzzy) (1.4.1)\nRequirement already satisfied, skipping upgrade: networkx>=1.9.0 in /usr/local/lib/python3.6/dist-packages (from scikit-fuzzy) (2.4)\nRequirement already satisfied, skipping upgrade: decorator>=4.3.0 in /usr/local/lib/python3.6/dist-packages (from networkx>=1.9.0->scikit-fuzzy) (4.4.2)\nBuilding wheels for collected packages: scikit-fuzzy\n Building wheel for scikit-fuzzy (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for scikit-fuzzy: filename=scikit_fuzzy-0.4.2-cp36-none-any.whl size=894070 sha256=e15ba9b4e39e9dfbc5cce9735fa101b6c76bdf502ba8a21f3e5e56915783fc82\n Stored in directory: /root/.cache/pip/wheels/b9/4e/77/da79b16f64ef1738d95486e2731eea09d73e90a72465096600\nSuccessfully built scikit-fuzzy\nInstalling collected packages: scikit-fuzzy\nSuccessfully installed scikit-fuzzy-0.4.2\n"
],
[
"%tensorflow_version 2.x\nimport tensorflow as tf\nimport sklearn\nfrom sklearn.metrics import confusion_matrix\nfrom skfuzzy.cluster import cmeans\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.model_selection import cross_val_score\n\nimport glob\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set()\nfrom time import *\n\nimport os\nimport scipy.io as sio\n\n%cd /content/drive/My Drive/Pattern Recognition/Dataset/cell_dataset",
"/usr/local/lib/python3.6/dist-packages/statsmodels/tools/_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead.\n import pandas.util.testing as tm\n"
],
[
"x_train = np.load(\"x_train.npy\")\ny_train = np.load(\"y_train.npy\")\nx_test = np.load(\"x_test.npy\")\ny_test = np.load(\"y_test.npy\")",
"_____no_output_____"
],
[
"print(x_train.shape,x_test.shape)\nprint(np.unique(y_test))\nprint(np.bincount(y_test.astype(int)))",
"(14536, 51, 51) (6229, 51, 51)\n[0. 1. 2.]\n[ 683 2439 3107]\n"
]
],
[
[
"## Part2:数据预处理",
"_____no_output_____"
]
],
[
[
"x_train = x_train.reshape(x_train.shape[0],-1)\nx_test = x_test.reshape(x_test.shape[0],-1)\nx_train = x_train/255.0\nx_test = x_test/255.0\nprint(x_train.shape,x_test.shape)",
"(14536, 2601) (6229, 2601)\n"
]
],
[
[
"## Part3:模型建立\n",
"_____no_output_____"
],
[
"由于skfuzzy模块内提到对于高维特征数据,cmeans聚类可能存在问题,故使用[第五次作业:细胞聚类](https://colab.research.google.com/github/Phantom-Ren/PR_TH/blob/master/细胞聚类.ipynb)中使用的AutoEncoder进行特征降维。",
"_____no_output_____"
]
],
[
[
"encoding_dim = 10",
"_____no_output_____"
],
[
"encoder = tf.keras.models.Sequential([\n tf.keras.layers.Dense(128,activation='relu') , \n tf.keras.layers.Dense(32,activation='relu') ,\n tf.keras.layers.Dense(8,activation='relu') ,\n tf.keras.layers.Dense(encoding_dim) \n])\n\ndecoder = tf.keras.models.Sequential([\n tf.keras.layers.Dense(8,activation='relu') , \n tf.keras.layers.Dense(32,activation='relu') ,\n tf.keras.layers.Dense(128,activation='relu') ,\n tf.keras.layers.Dense(2601,activation='sigmoid') \n])\n\nAE = tf.keras.models.Sequential([\n encoder, \n decoder\n])",
"_____no_output_____"
],
[
"AE.compile(optimizer='adam',loss='binary_crossentropy')",
"_____no_output_____"
],
[
"AE.fit(x_train,x_train,epochs=10,batch_size=256)",
"Epoch 1/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.2308\nEpoch 2/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0672\nEpoch 3/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0623\nEpoch 4/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0592\nEpoch 5/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0581\nEpoch 6/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0577\nEpoch 7/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0576\nEpoch 8/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0574\nEpoch 9/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0573\nEpoch 10/10\n57/57 [==============================] - 2s 37ms/step - loss: 0.0572\n"
],
[
"x_encoded = encoder.predict(x_train)\nx_encoded_test = encoder.predict(x_test)",
"_____no_output_____"
],
[
"x_encoded_t = x_encoded.T\nprint(x_encoded_t.shape)",
"(10, 14536)\n"
],
[
"st=time()\ncenter, u, u0, d, jm, p, fpc = cmeans(x_encoded_t, m=2, c=8, error=0.0005, maxiter=1000)\net=time()\nprint('Time Usage:',et-st,'s')\nprint('Numbers of iterations used:',p)\nfor i in u:\n yhat = np.argmax(u, axis=0)",
"Time Usage: 9.911698341369629 s\nNumbers of iterations used: 410\n"
],
[
"print(center)\nprint(center.shape)",
"[[ -3.44933402 -8.44556914 -16.94252588 -8.74556024 -12.61217008\n -9.7781175 18.60453291 2.95457345 3.22953149 5.69484494]\n [-11.97403419 -16.07650986 -26.56396647 -16.34790619 -21.81834829\n -15.61480001 34.48161701 8.90122248 11.26525733 12.4596491 ]\n [ -7.22867202 -12.76481299 -23.26152398 -12.80788411 -18.18223393\n -13.88773779 27.39076737 5.79463356 6.88977848 9.47686693]\n [ -0.72018174 -3.36455579 -8.15082003 -4.31916584 -5.82211945\n -4.75698858 9.04330793 1.32119156 0.68767281 2.14246547]\n [-23.36955657 -21.53172434 -25.71895429 -23.30417817 -25.17116461\n -11.51322674 46.47580771 14.77245746 22.20161965 14.57876323]\n [-29.09174015 -26.21911058 -19.85356267 -29.59542209 -25.20324848\n -3.53020116 53.2928569 16.38369228 28.97699993 11.33743856]\n [-24.79542182 -22.26765841 -15.34363889 -25.22701977 -20.6935394\n -1.61246474 44.78221591 13.69816692 24.88078417 8.79516102]\n [-17.60714073 -18.62241444 -27.38277968 -19.41323712 -23.98275131\n -14.91679002 40.47605938 12.07882916 16.45713245 14.44150938]]\n(8, 10)\n"
],
[
"from sklearn.metrics import fowlkes_mallows_score\ndef draw_confusionmatrix(ytest, yhat):\n plt.figure(figsize=(10,7))\n cm = confusion_matrix(ytest, yhat)\n ax = sns.heatmap(cm, annot=True, fmt=\"d\")\n plt.ylabel('True label')\n plt.xlabel('Predicted label')\n\n acc = accuracy_score(ytest, yhat)\n score_f=fowlkes_mallows_score(ytest,yhat)\n print(f\"Sum Axis-1 as Classification accuracy: {acc}\")\n print('F-Score:',score_f)",
"_____no_output_____"
],
[
"draw_confusionmatrix(y_train,yhat)",
"Sum Axis-1 as Classification accuracy: 0.19888552559163455\nF-Score: 0.3129005083679034\n"
],
[
"temp=[2,2,2,2,1,0,1,1]\ny_hat1=np.zeros(14536)\nfor i in range(0,14536):\n y_hat1[i] = temp[yhat[i]]\ndraw_confusionmatrix(y_train,y_hat1) ",
"Sum Axis-1 as Classification accuracy: 0.6651073197578427\nF-Score: 0.5610603391756352\n"
]
],
[
[
"将结果与Kmeans聚类相比,发现结果有较大提升(61%->67%)。但相对有监督学习方法,结果仍不尽如人意。",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
cbca70a6a575d61aef635e19e7f8593244fc70dc
| 22,713 |
ipynb
|
Jupyter Notebook
|
examples/pygrid/homomorphic-encryption/Tutorial_2_TenSEAL_Syft_Data_Owner.ipynb
|
H4LL/PySyft
|
baaeec792e90919f0b27f583cbecc96d61b33fd6
|
[
"Apache-2.0"
] | null | null | null |
examples/pygrid/homomorphic-encryption/Tutorial_2_TenSEAL_Syft_Data_Owner.ipynb
|
H4LL/PySyft
|
baaeec792e90919f0b27f583cbecc96d61b33fd6
|
[
"Apache-2.0"
] | null | null | null |
examples/pygrid/homomorphic-encryption/Tutorial_2_TenSEAL_Syft_Data_Owner.ipynb
|
H4LL/PySyft
|
baaeec792e90919f0b27f583cbecc96d61b33fd6
|
[
"Apache-2.0"
] | null | null | null | 40.777379 | 6,316 | 0.631885 |
[
[
[
"<img src=\"https://github.com/OpenMined/design-assets/raw/master/logos/OM/horizontal-primary-light.png\" alt=\"he-black-box\" width=\"600\"/>\n\n\n# Homomorphic Encryption using Duet: Data Owner\n## Tutorial 2: Encrypted image evaluation\n\n\nWelcome!\nThis tutorial will show you how to evaluate Encrypted images using Duet and TenSEAL. This notebook illustrates the Data Owner view on the operations.\n\nWe recommend going through Tutorial 0 and 1 before trying this one.",
"_____no_output_____"
],
[
"### Setup\n\nAll modules are imported here, make sure everything is installed by running the cell below.",
"_____no_output_____"
]
],
[
[
"import os\nimport requests\n\nimport syft as sy\nimport tenseal as ts\nfrom torchvision import transforms\nfrom random import randint\nimport numpy as np\nfrom PIL import Image\nfrom matplotlib.pyplot import imshow\nimport torch\nfrom syft.grid.client.client import connect\nfrom syft.grid.client.grid_connection import GridHTTPConnection\nfrom syft.core.node.domain.client import DomainClient\nsy.load_lib(\"tenseal\")",
"_____no_output_____"
]
],
[
[
"## Connect to PyGrid\n\nConnect to PyGrid Domain server.",
"_____no_output_____"
]
],
[
[
"client = connect(\n url=\"http://localhost:5000\", # Domain Address\n credentials={\"email\":\"[email protected]\", \"password\":\"pwd123\"},\n conn_type= GridHTTPConnection, # HTTP Connection Protocol\n client_type=DomainClient) # Domain Client type",
"_____no_output_____"
]
],
[
[
"### <img src=\"https://github.com/OpenMined/design-assets/raw/master/logos/OM/mark-primary-light.png\" alt=\"he-black-box\" width=\"100\"/> Checkpoint 1 : Now STOP and run the Data Scientist notebook until the same checkpoint.",
"_____no_output_____"
],
[
"### Data Owner helpers",
"_____no_output_____"
]
],
[
[
"# Create the TenSEAL security context\ndef create_ctx():\n \"\"\"Helper for creating the CKKS context.\n CKKS params:\n - Polynomial degree: 8192.\n - Coefficient modulus size: [40, 21, 21, 21, 21, 21, 21, 40].\n - Scale: 2 ** 21.\n - The setup requires the Galois keys for evaluating the convolutions.\n \"\"\"\n poly_mod_degree = 8192\n coeff_mod_bit_sizes = [40, 21, 21, 21, 21, 21, 21, 40]\n ctx = ts.context(ts.SCHEME_TYPE.CKKS, poly_mod_degree, -1, coeff_mod_bit_sizes)\n ctx.global_scale = 2 ** 21\n ctx.generate_galois_keys()\n return ctx\n\ndef download_images():\n try:\n os.mkdir(\"data/mnist-samples\")\n except BaseException as e:\n pass\n\n url = \"https://raw.githubusercontent.com/OpenMined/TenSEAL/master/tutorials/data/mnist-samples/img_{}.jpg\"\n path = \"data/mnist-samples/img_{}.jpg\"\n \n for idx in range(6):\n img_url = url.format(idx)\n img_path = path.format(idx)\n r = requests.get(img_url)\n\n with open(img_path, 'wb') as f:\n f.write(r.content)\n \n# Sample an image\ndef load_input():\n download_images()\n \n transform = transforms.Compose(\n [transforms.ToTensor(), transforms.Normalize((0.1307,), (0.3081,))]\n )\n idx = randint(1, 5)\n img_name = \"data/mnist-samples/img_{}.jpg\".format(idx)\n img = Image.open(img_name)\n return transform(img).view(28, 28).tolist(), img\n\n# Helper for encoding the image\ndef prepare_input(ctx, plain_input):\n enc_input, windows_nb = ts.im2col_encoding(ctx, plain_input, 7, 7, 3)\n assert windows_nb == 64\n return enc_input\n",
"_____no_output_____"
]
],
[
[
"### Prepare the context",
"_____no_output_____"
]
],
[
[
"context = create_ctx()",
"_____no_output_____"
]
],
[
[
"### Sample and encrypt an image",
"_____no_output_____"
]
],
[
[
"image, orig = load_input()\n\nencrypted_image = prepare_input(context, image)\n\nprint(\"Encrypted image \", encrypted_image)\nprint(\"Original image \")\nimshow(np.asarray(orig))",
"Encrypted image <tenseal.tensors.ckksvector.CKKSVector object at 0x7f80db007220>\nOriginal image \n"
],
[
"ctx_ptr = context.send(client, searchable=True, tags=[\"context\"])\nenc_image_ptr = encrypted_image.send(client, searchable=True, tags=[\"enc_image\"])",
"_____no_output_____"
],
[
"client.store.pandas",
"_____no_output_____"
]
],
[
[
"### <img src=\"https://github.com/OpenMined/design-assets/raw/master/logos/OM/mark-primary-light.png\" alt=\"he-black-box\" width=\"100\"/> Checkpoint 2 : Now STOP and run the Data Scientist notebook until the same checkpoint.",
"_____no_output_____"
],
[
"### Approve the requests",
"_____no_output_____"
]
],
[
[
"client.requests.pandas",
"_____no_output_____"
],
[
"client.requests[0].accept()\nclient.requests[0].accept()",
"_____no_output_____"
],
[
"client.requests.pandas",
"_____no_output_____"
]
],
[
[
"### <img src=\"https://github.com/OpenMined/design-assets/raw/master/logos/OM/mark-primary-light.png\" alt=\"he-black-box\" width=\"100\"/> Checkpoint 3 : Now STOP and run the Data Scientist notebook until the same checkpoint.",
"_____no_output_____"
],
[
"### Retrieve and decrypt the evaluation result",
"_____no_output_____"
]
],
[
[
"result = client.store[\"result\"].get(delete_obj=False)\nresult.link_context(context)\n\nresult = result.decrypt()",
"_____no_output_____"
]
],
[
[
"### Run the activation and retrieve the label",
"_____no_output_____"
]
],
[
[
"probs = torch.softmax(torch.tensor(result), 0)\nlabel_max = torch.argmax(probs)\n\nprint(\"Maximum probability for label {}\".format(label_max))",
"Maximum probability for label 9\n"
]
],
[
[
"### <img src=\"https://github.com/OpenMined/design-assets/raw/master/logos/OM/mark-primary-light.png\" alt=\"he-black-box\" width=\"100\"/> Checkpoint 4 : Well done!",
"_____no_output_____"
],
[
"# Congratulations!!! - Time to Join the Community!\n\nCongratulations on completing this notebook tutorial! If you enjoyed this and would like to join the movement toward privacy preserving, decentralized ownership of AI and the AI supply chain (data), you can do so in the following ways!\n\n### Star PySyft and TenSEAL on GitHub\n\nThe easiest way to help our community is just by starring the Repos! This helps raise awareness of the cool tools we're building.\n\n- [Star PySyft](https://github.com/OpenMined/PySyft)\n- [Star TenSEAL](https://github.com/OpenMined/TenSEAL)\n\n### Join our Slack!\n\nThe best way to keep up to date on the latest advancements is to join our community! You can do so by filling out the form at [http://slack.openmined.org](http://slack.openmined.org). #lib_tenseal and #code_tenseal are the main channels for the TenSEAL project.\n\n### Donate\n\nIf you don't have time to contribute to our codebase, but would still like to lend support, you can also become a Backer on our Open Collective. All donations go toward our web hosting and other community expenses such as hackathons and meetups!\n\n[OpenMined's Open Collective Page](https://opencollective.com/openmined)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbca718a7417bfd005a52fab136fb67a243834f9
| 10,388 |
ipynb
|
Jupyter Notebook
|
machineLearning/02_shallows/03_regression.ipynb
|
csc-training/geocomputing
|
1e8043c864fb663526d1c15cfd3bb390a1379181
|
[
"CC-BY-4.0"
] | 25 |
2017-10-11T06:54:48.000Z
|
2022-03-14T07:27:07.000Z
|
machineLearning/02_shallows/03_regression.ipynb
|
csc-training/geocomputing
|
1e8043c864fb663526d1c15cfd3bb390a1379181
|
[
"CC-BY-4.0"
] | 5 |
2020-04-15T08:02:10.000Z
|
2022-03-21T10:34:03.000Z
|
machineLearning/02_shallows/03_regression.ipynb
|
csc-training/geocomputing
|
1e8043c864fb663526d1c15cfd3bb390a1379181
|
[
"CC-BY-4.0"
] | 17 |
2017-11-02T13:17:38.000Z
|
2022-03-28T12:54:46.000Z
| 29.344633 | 157 | 0.603485 |
[
[
[
"# Shallow regression for vector data\n\nThis script reads zip code data produced by **vectorDataPreparations** and creates different machine learning models for\npredicting the average zip code income from population and spatial variables.\n\nIt assesses the model accuracy with a test dataset but also predicts the number to all zip codes and writes it to a geopackage\nfor closer inspection",
"_____no_output_____"
],
[
"# 1. Read the data",
"_____no_output_____"
]
],
[
[
"import time\nimport geopandas as gpd\nimport pandas as pd\nfrom math import sqrt\nimport os\nimport matplotlib.pyplot as plt\n\nfrom sklearn.ensemble import GradientBoostingRegressor, RandomForestRegressor, BaggingRegressor,ExtraTreesRegressor, AdaBoostRegressor\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import mean_squared_error, mean_absolute_error,r2_score",
"_____no_output_____"
]
],
[
[
"### 1.1 Input and output file paths ",
"_____no_output_____"
]
],
[
[
"paavo_data = \"../data/paavo\"\n\n### Relative path to the zip code geopackage file that was prepared by vectorDataPreparations.py\ninput_geopackage_path = os.path.join(paavo_data,\"zip_code_data_after_preparation.gpkg\")\n\n### Output file. You can change the name to identify different regression models\noutput_geopackage_path = os.path.join(paavo_data,\"median_income_per_zipcode_shallow_model.gpkg\")",
"_____no_output_____"
]
],
[
[
"### 1.2 Read the input data to a Geopandas dataframe",
"_____no_output_____"
]
],
[
[
"original_gdf = gpd.read_file(input_geopackage_path)\noriginal_gdf.head()",
"_____no_output_____"
]
],
[
[
"# 2. Train the model \n\nHere we try training different models. We encourage you to dive into the documentation of different models a bit and try different parameters. \n\nWhich one is the best model? Can you figure out how to improve it even more?",
"_____no_output_____"
],
[
"### 2.1 Split the dataset to train and test datasets",
"_____no_output_____"
]
],
[
[
"### Split the gdf to x (the predictor attributes) and y (the attribute to be predicted)\ny = original_gdf['hr_mtu'] # Average income\n\n### Remove geometry and textual fields\nx = original_gdf.drop(['geometry','postinumer','nimi','hr_mtu'],axis=1)\n\n### Split the both datasets to train (80%) and test (20%) datasets\nx_train, x_test, y_train, y_test = train_test_split(x, y, test_size=.2, random_state=42)\n",
"_____no_output_____"
]
],
[
[
"### 2.2 These are the functions used for training, estimating and predicting.",
"_____no_output_____"
]
],
[
[
"def trainModel(x_train, y_train, model):\n start_time = time.time() \n print(model)\n model.fit(x_train,y_train)\n print('Model training took: ', round((time.time() - start_time), 2), ' seconds')\n return model\n\ndef estimateModel(x_test,y_test, model):\n ### Predict the unemployed number to the test dataset\n prediction = model.predict(x_test)\n \n ### Assess the accuracy of the model with root mean squared error, mean absolute error and coefficient of determination r2\n rmse = sqrt(mean_squared_error(y_test, prediction))\n mae = mean_absolute_error(y_test, prediction)\n r2 = r2_score(y_test, prediction)\n\n print(f\"\\nMODEL ACCURACY METRICS WITH TEST DATASET: \\n\" +\n f\"\\t Root mean squared error: {round(rmse)} \\n\" +\n f\"\\t Mean absolute error: {round(mae)} \\n\" +\n f\"\\t Coefficient of determination: {round(r2,4)} \\n\")",
"_____no_output_____"
]
],
[
[
"### 2.3 Run different models",
"_____no_output_____"
],
[
"### Gradient Boosting Regressor\n* https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.GradientBoostingRegressor.html\n* https://scikit-learn.org/stable/modules/ensemble.html#regression",
"_____no_output_____"
]
],
[
[
"model = GradientBoostingRegressor(n_estimators=30, learning_rate=0.1,verbose=1)\nmodel_name = \"Gradient Boosting Regressor\"\ntrainModel(x_train, y_train,model)\nestimateModel(x_test,y_test, model)",
"_____no_output_____"
]
],
[
[
"### Random Forest Regressor\n\n* https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestRegressor.html\n* https://scikit-learn.org/stable/modules/ensemble.html#forest",
"_____no_output_____"
]
],
[
[
"model = RandomForestRegressor(n_estimators=30,verbose=1)\nmodel_name = \"Random Forest Regressor\"\ntrainModel(x_train, y_train,model)\nestimateModel(x_test,y_test, model)",
"_____no_output_____"
]
],
[
[
"### Extra Trees Regressor\n\n* https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.ExtraTreesRegressor.html",
"_____no_output_____"
]
],
[
[
"model = ExtraTreesRegressor(n_estimators=30,verbose=1)\nmodel_name = \"Extra Trees Regressor\"\ntrainModel(x_train, y_train,model)\nestimateModel(x_test,y_test, model)",
"_____no_output_____"
]
],
[
[
"### Bagging Regressor\n\n* https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.BaggingRegressor.html\n* https://scikit-learn.org/stable/modules/ensemble.html#bagging",
"_____no_output_____"
]
],
[
[
"model = BaggingRegressor(n_estimators=30,verbose=1)\nmodel_name = \"Bagging Regressor\"\ntrainModel(x_train, y_train,model)\nestimateModel(x_test,y_test, model)",
"_____no_output_____"
]
],
[
[
"### AdaBoost Regressor\n\n* https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.AdaBoostRegressor.html\n* https://scikit-learn.org/stable/modules/ensemble.html#adaboost",
"_____no_output_____"
]
],
[
[
"model = AdaBoostRegressor(n_estimators=30)\nmodel_name = \"AdaBoost Regressor\"\ntrainModel(x_train, y_train,model)\nestimateModel(x_test,y_test, model)",
"_____no_output_____"
]
],
[
[
"# 3. Predict average income to all zip codes\nHere we predict the average income to the whole dataset. Prediction is done with the model you have stored in the model variable - the one you ran last",
"_____no_output_____"
]
],
[
[
"### Print chosen model (the one you ran last)\nprint(model)\n\n### Drop the not-used columns from original_gdf as done before model training.\nx = original_gdf.drop(['geometry','postinumer','nimi','hr_mtu'],axis=1)\n\n### Predict the median income with already trained model\nprediction = model.predict(x)\n\n### Join the predictions to the original geodataframe and pick only interesting columns for results\noriginal_gdf['predicted_hr_mtu'] = prediction.round(0)\noriginal_gdf['difference'] = original_gdf['predicted_hr_mtu'] - original_gdf['hr_mtu']\nresulting_gdf = original_gdf[['postinumer','nimi','hr_mtu','predicted_hr_mtu','difference','geometry']]",
"_____no_output_____"
],
[
"fig, ax = plt.subplots(figsize=(20, 10))\nax.set_title(\"Predicted average income by zip code \" + model_name, fontsize=25)\nax.set_axis_off()\nresulting_gdf.plot(column='predicted_hr_mtu', ax=ax, legend=True, cmap=\"magma\")",
"_____no_output_____"
]
],
[
[
"# 4. EXERCISE: Calculate the difference between real and predicted incomes\n\nCalculate the difference of real and predicted income amounts by zip code level and plot a map of it\n\n* **original_gdf** is the original dataframe\n* **resulting_gdf** is the predicted one",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbca781e7623fa3a8a9e463e41c62ba01efb3662
| 17,519 |
ipynb
|
Jupyter Notebook
|
notebooks/rnn_sentiment_classifier.ipynb
|
tromgy/deep-learning-illustrated
|
e9dc671e555bcdf7e9501606b207d1015981733a
|
[
"MIT"
] | 504 |
2019-04-29T09:28:04.000Z
|
2022-03-25T15:42:41.000Z
|
notebooks/rnn_sentiment_classifier.ipynb
|
tromgy/deep-learning-illustrated
|
e9dc671e555bcdf7e9501606b207d1015981733a
|
[
"MIT"
] | 8 |
2019-06-17T18:30:10.000Z
|
2021-06-07T00:23:59.000Z
|
notebooks/rnn_sentiment_classifier.ipynb
|
tromgy/deep-learning-illustrated
|
e9dc671e555bcdf7e9501606b207d1015981733a
|
[
"MIT"
] | 262 |
2019-03-20T07:22:35.000Z
|
2022-03-28T01:56:07.000Z
| 46.967828 | 6,960 | 0.697585 |
[
[
[
"# RNN Sentiment Classifier",
"_____no_output_____"
],
[
"In this notebook, we use an RNN to classify IMDB movie reviews by their sentiment.",
"_____no_output_____"
],
[
"[](https://colab.research.google.com/github/the-deep-learners/deep-learning-illustrated/blob/master/notebooks/rnn_sentiment_classifier.ipynb)",
"_____no_output_____"
],
[
"#### Load dependencies",
"_____no_output_____"
]
],
[
[
"import keras\nfrom keras.datasets import imdb\nfrom keras.preprocessing.sequence import pad_sequences\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Embedding, SpatialDropout1D\nfrom keras.layers import SimpleRNN # new! \nfrom keras.callbacks import ModelCheckpoint\nimport os\nfrom sklearn.metrics import roc_auc_score \nimport matplotlib.pyplot as plt \n%matplotlib inline",
"Using TensorFlow backend.\n"
]
],
[
[
"#### Set hyperparameters",
"_____no_output_____"
]
],
[
[
"# output directory name:\noutput_dir = 'model_output/rnn'\n\n# training:\nepochs = 16 # way more!\nbatch_size = 128\n\n# vector-space embedding: \nn_dim = 64 \nn_unique_words = 10000 \nmax_review_length = 100 # lowered due to vanishing gradient over time\npad_type = trunc_type = 'pre'\ndrop_embed = 0.2 \n\n# RNN layer architecture:\nn_rnn = 256 \ndrop_rnn = 0.2\n\n# dense layer architecture: \n# n_dense = 256\n# dropout = 0.2",
"_____no_output_____"
]
],
[
[
"#### Load data",
"_____no_output_____"
]
],
[
[
"(x_train, y_train), (x_valid, y_valid) = imdb.load_data(num_words=n_unique_words) # removed n_words_to_skip",
"_____no_output_____"
]
],
[
[
"#### Preprocess data",
"_____no_output_____"
]
],
[
[
"x_train = pad_sequences(x_train, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)\nx_valid = pad_sequences(x_valid, maxlen=max_review_length, padding=pad_type, truncating=trunc_type, value=0)",
"_____no_output_____"
]
],
[
[
"#### Design neural network architecture",
"_____no_output_____"
]
],
[
[
"model = Sequential()\nmodel.add(Embedding(n_unique_words, n_dim, input_length=max_review_length)) \nmodel.add(SpatialDropout1D(drop_embed))\nmodel.add(SimpleRNN(n_rnn, dropout=drop_rnn))\n# model.add(Dense(n_dense, activation='relu')) # typically don't see top dense layer in NLP like in \n# model.add(Dropout(dropout))\nmodel.add(Dense(1, activation='sigmoid'))",
"_____no_output_____"
],
[
"model.summary() ",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_1 (Embedding) (None, 100, 64) 640000 \n_________________________________________________________________\nspatial_dropout1d_1 (Spatial (None, 100, 64) 0 \n_________________________________________________________________\nsimple_rnn_1 (SimpleRNN) (None, 256) 82176 \n_________________________________________________________________\ndense_1 (Dense) (None, 1) 257 \n=================================================================\nTotal params: 722,433\nTrainable params: 722,433\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"#### Configure model",
"_____no_output_____"
]
],
[
[
"model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy'])",
"_____no_output_____"
],
[
"modelcheckpoint = ModelCheckpoint(filepath=output_dir+\"/weights.{epoch:02d}.hdf5\")\nif not os.path.exists(output_dir):\n os.makedirs(output_dir)",
"_____no_output_____"
]
],
[
[
"#### Train!",
"_____no_output_____"
]
],
[
[
"model.fit(x_train, y_train, batch_size=batch_size, epochs=epochs, verbose=1, validation_data=(x_valid, y_valid), callbacks=[modelcheckpoint])",
"Train on 25000 samples, validate on 25000 samples\nEpoch 1/16\n25000/25000 [==============================] - 18s 709us/step - loss: 0.7023 - acc: 0.5132 - val_loss: 0.6993 - val_acc: 0.5311\nEpoch 2/16\n25000/25000 [==============================] - 17s 689us/step - loss: 0.6865 - acc: 0.5422 - val_loss: 0.6649 - val_acc: 0.5922\nEpoch 3/16\n25000/25000 [==============================] - 17s 682us/step - loss: 0.5905 - acc: 0.6776 - val_loss: 0.5816 - val_acc: 0.6835\nEpoch 4/16\n25000/25000 [==============================] - 17s 682us/step - loss: 0.5716 - acc: 0.6966 - val_loss: 0.5647 - val_acc: 0.7055\nEpoch 5/16\n25000/25000 [==============================] - 17s 680us/step - loss: 0.5848 - acc: 0.6982 - val_loss: 0.5329 - val_acc: 0.7458\nEpoch 6/16\n25000/25000 [==============================] - 17s 678us/step - loss: 0.5263 - acc: 0.7399 - val_loss: 0.6334 - val_acc: 0.6453\nEpoch 7/16\n25000/25000 [==============================] - 17s 681us/step - loss: 0.4484 - acc: 0.7962 - val_loss: 0.5039 - val_acc: 0.7758\nEpoch 8/16\n25000/25000 [==============================] - 17s 678us/step - loss: 0.5540 - acc: 0.7086 - val_loss: 0.6503 - val_acc: 0.6119\nEpoch 9/16\n25000/25000 [==============================] - 17s 677us/step - loss: 0.5298 - acc: 0.7416 - val_loss: 0.6444 - val_acc: 0.7170\nEpoch 10/16\n25000/25000 [==============================] - 17s 675us/step - loss: 0.5070 - acc: 0.7624 - val_loss: 0.5628 - val_acc: 0.7406\nEpoch 11/16\n25000/25000 [==============================] - 17s 675us/step - loss: 0.4657 - acc: 0.7842 - val_loss: 0.5673 - val_acc: 0.7324\nEpoch 12/16\n25000/25000 [==============================] - 17s 672us/step - loss: 0.4607 - acc: 0.7895 - val_loss: 0.6675 - val_acc: 0.5698\nEpoch 13/16\n25000/25000 [==============================] - 17s 671us/step - loss: 0.4289 - acc: 0.8077 - val_loss: 0.5532 - val_acc: 0.7346\nEpoch 14/16\n25000/25000 [==============================] - 17s 673us/step - loss: 0.6410 - acc: 0.6558 - val_loss: 0.7551 - val_acc: 0.6234\nEpoch 15/16\n25000/25000 [==============================] - 17s 675us/step - loss: 0.5058 - acc: 0.7543 - val_loss: 0.5916 - val_acc: 0.7038\nEpoch 16/16\n25000/25000 [==============================] - 17s 672us/step - loss: 0.4012 - acc: 0.8309 - val_loss: 0.5920 - val_acc: 0.7510\n"
]
],
[
[
"#### Evaluate",
"_____no_output_____"
]
],
[
[
"model.load_weights(output_dir+\"/weights.07.hdf5\") ",
"_____no_output_____"
],
[
"y_hat = model.predict_proba(x_valid)",
"_____no_output_____"
],
[
"plt.hist(y_hat)\n_ = plt.axvline(x=0.5, color='orange')",
"_____no_output_____"
],
[
"\"{:0.2f}\".format(roc_auc_score(y_valid, y_hat)*100.0)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
] |
cbca7827750a56620434f9e239b9a97bafb95d5d
| 886,941 |
ipynb
|
Jupyter Notebook
|
notebooks/.ipynb_checkpoints/Intro_02_JupyterNotebooks-checkpoint.ipynb
|
python4oceanography/ocean_python_tutorial
|
7215b3c740ab8fb4b177a8ae764411c273cc7a06
|
[
"Apache-2.0"
] | 12 |
2019-09-01T19:00:25.000Z
|
2021-12-01T00:45:49.000Z
|
notebooks/.ipynb_checkpoints/Intro_02_JupyterNotebooks-checkpoint.ipynb
|
ognancy4life/ocean_python_tutorial
|
7215b3c740ab8fb4b177a8ae764411c273cc7a06
|
[
"Apache-2.0"
] | 1 |
2021-07-20T02:02:58.000Z
|
2021-07-20T02:02:58.000Z
|
notebooks/.ipynb_checkpoints/Intro_02_JupyterNotebooks-checkpoint.ipynb
|
ognancy4life/ocean_python_tutorial
|
7215b3c740ab8fb4b177a8ae764411c273cc7a06
|
[
"Apache-2.0"
] | 6 |
2019-09-14T19:10:52.000Z
|
2021-06-25T20:08:47.000Z
| 2,390.67655 | 754,468 | 0.9613 |
[
[
[
"# Intro to Jupyter Notebooks\n\n",
"_____no_output_____"
],
[
"### `Jupyter` is a project for developing open-source software\n### `Jupyter Notebooks` is a `web` application to create scripts\n### `Jupyter Lab` is the new generation of web user interface for Jypyter",
"_____no_output_____"
],
[
"### But it is more than that\n#### It lets you insert and save text, equations & visualizations ... in the same page!\n",
"_____no_output_____"
],
[
"***",
"_____no_output_____"
],
[
"# Notebook dashboard\nWhen you launch the Jupyter notebook server in your computer, you would see a dashboard like this:\n\n\n",
"_____no_output_____"
],
[
"# Saving your own script\nAll scripts we are showing here today are running online & we will make changes through the workshop. To keep your modified script for further reference, you will need to save a copy on your own computer at the end.\n\n<div class=\"alert alert-block alert-info\">\n <b>Try it out! </b>\n <br><br>\n Go to <b>File</b> in the top menu -> Download As -> Notebook </div>\n<br>\nAny changes made online, even if saved (not downloaded) will be lost once the binder connection is closed.",
"_____no_output_____"
],
[
"***",
"_____no_output_____"
],
[
"## Two type of cells\n### `Code` Cells: execute code\n### `Markdown` Cells: show formated text\n\nThere are two ways to change the type on a cell:\n- Cliking on the scroll-down menu on the top\n\n- using the shortcut `Esc-y` for code and `Esc-m` for markdown types\n<br>\n<div class=\"alert alert-block alert-info\"><b>Try it out! </b>\n <bR>\n <br>- Click on the next cell\n <br>- Change the type using the scroll-down menu & select <b>Code</b>\n <br>- Change it back to <b>Markdown</b>\n</div>",
"_____no_output_____"
],
[
"## This is a simple operation\ny = 4 + 6\nprint(y)",
"_____no_output_____"
],
[
"## <i>Note the change in format of the first line & the text color in the second line</i>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\"><b>Try it out!</br>\n <br><br>In the next cell:\n <br>- Double-Click on the next cell\n <br>- Press <b> Esc</b> (note to blue color of the left border)\n <br>- Type <b>y</b> to change it to <b>Code</b> type \n <br>- Use <b>m</b> to change it back to <b>Markdown</b> type\n</div>",
"_____no_output_____"
]
],
[
[
"# This is a simple operation\ny = 4 + 6 \nprint(y)",
"10\n"
]
],
[
[
"***",
"_____no_output_____"
],
[
"# To execute commands\n\n## - `Shift-Enter` : executes cell & advance to next\n## - `Control-enter` : executes cell & stay in the same cell\n\n<div class=\"alert alert-block alert-info\"><b>Try it out!</b>\n <br>\n <br>In the previous cell:\n <br>- Double-Click on the previous cell\n <br>- Use <b>Shift-Enter</b> to execute\n <br>- Double-Click on the in the previous cell again\n <br>- This time use <b>Control-Enter</b> to execute\n <br>\n <br>- Now change the type to <b>Code</b> & execute the cell\n</div>",
"_____no_output_____"
],
[
"## You could also execute the entire script use the `Run` tab in the top menu\n\n",
"_____no_output_____"
],
[
"## Or even the entire script from the `Cell` menu at the top",
"_____no_output_____"
],
[
"***",
"_____no_output_____"
],
[
"## Other commands\n### From the icon menu: \n### Save, Add Cell, Cut Cell, Copy Cell, Paste Cell, Move Cell Up, Move Cell Down\n\n\n\n### or the drop down menu 'command palette'",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\"><b>Try them out!</b>",
"_____no_output_____"
],
[
"## Now, the keyboard shortcuts\n#### First press `Esc`, then:\n - `s` : save changes\n <br>\n - `a`, `b` : create cell above and below\n <br>\n - `dd` : delete cell\n <br>\n - `x`, `c`, `v` : cut, copy and paste cell\n <br>\n - `z` undo last change",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n <b> Let's practice!</b>\n <br>\n <br>- Create a cell bellow with <b>Esc-b</b>, and click on it\n <br>- Type print('Hello world!') and execute it using <b>Control-Enter</b>\n <br>- Copy-paste the cell to make a duplicate by typing <b>Esc-c</b> & <b>Esc-v</b>\n <br>- Cut the first cell using <b>Esc-x</b>\n</div>",
"_____no_output_____"
],
[
"## And the last one: adding line numbers\n- `Esc-l` : in Jupyter Notebooks\n- `Esc-Shift-l`: in Jupyter Lab\n\n<div class=\"alert alert-block alert-info\">\n<b>Try it out!</b>\n<br><br>\n - Try it in a code cell\n <br>- And now try it in the markdown cell\n</div>",
"_____no_output_____"
]
],
[
[
"y = 5\nprint(y + 4)\nx = 8\nprint(y*x)",
"_____no_output_____"
]
],
[
[
"***",
"_____no_output_____"
],
[
"## Last note about the `Kernel`\n#### That little program that is running in the background & let you run your notebook\n<div class=\"alert alert-block alert-danger\">\nOnce in a while the <b>kernel</b> will die or your program will get stucked, & like everything else in the computer world.... you'll have to restart it.\n</div>\n\n### You can do this by going to the `Kernel` menu -> Restart, & then you'll have to run all your cells (or at least the ones above the one you're working on (use `Cell` menu -> Run all Above.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
cbca7d75a482438f605dee18f097513fc420a84c
| 81,831 |
ipynb
|
Jupyter Notebook
|
qiskit/advanced/aqua/optimization/vehicle_routing.ipynb
|
dgrinko/qiskit-iqx-tutorials
|
51144fe48b29b0bc6e4e4033d0dfad84907099c7
|
[
"Apache-2.0"
] | null | null | null |
qiskit/advanced/aqua/optimization/vehicle_routing.ipynb
|
dgrinko/qiskit-iqx-tutorials
|
51144fe48b29b0bc6e4e4033d0dfad84907099c7
|
[
"Apache-2.0"
] | null | null | null |
qiskit/advanced/aqua/optimization/vehicle_routing.ipynb
|
dgrinko/qiskit-iqx-tutorials
|
51144fe48b29b0bc6e4e4033d0dfad84907099c7
|
[
"Apache-2.0"
] | null | null | null | 73.854693 | 21,748 | 0.748494 |
[
[
[
"",
"_____no_output_____"
],
[
"# Qiskit Aqua: Vehicle Routing\n\n## The Introduction\n\nLogistics is a major industry, with some estimates valuing it at USD 8183 billion globally in 2015. Most service providers operate a number of vehicles (e.g., trucks and container ships), a number of depots, where the vehicles are based overnight, and serve a number of client locations with each vehicle during each day. There are many optimization and control problems that consider these parameters. Computationally, the key challenge is how to design routes from depots to a number of client locations and back to the depot, so as to minimize vehicle-miles traveled, time spent, or similar objective functions. In this notebook we formalize an idealized version of the problem and showcase its solution using the quantum approximate optimization approach of Farhi, Goldstone, and Gutman (2014). \n\nThe overall workflow we demonstrate comprises:\n\n1. establish the client locations. Normally, these would be available ahead of the day of deliveries from a database. In our use case, we generate these randomly.\n\n3. compute the pair-wise distances, travel times, or similar. In our case, we consider the Euclidean distance, \"as the crow flies\", which is perhaps the simplest possible.\n\n4. compute the actual routes. This step is run twice, actually. First, we obtain a reference value by a run of a classical solver (IBM CPLEX) on the classical computer. Second, we run an alternative, hybrid algorithm partly on the quantum computer.\n\n5. visualization of the results. In our case, this is again a simplistic plot.\n\nIn the following, we first explain the model, before we proceed with the installation of the pre-requisites and the data loading.\n\n## The Model \n\nMathematically speaking, the vehicle routing problem (VRP) is a combinatorial problem, wherein the best routes from a depot to a number of clients and back to the depot are sought, given a number of available vehicles. There are a number of formulations possible, extending a number of formulations of the traveling salesman problem [Applegate et al, 2006]. Here, we present a formulation known as MTZ [Miller, Tucker, Zemlin, 1960]. \n\nLet $n$ be the number of clients (indexed as $1,\\dots,n$), and $K$ be the number of available vehicles. Let $x_{ij} = \\{0,1\\}$ be the binary decision variable which, if it is $1$, activates the segment from node $i$ to node $j$. The node index runs from $0$ to $n$, where $0$ is (by convention) the depot. There are twice as many distinct decision variables as edges. For example, in a fully connected graph, there are $n(n+1)$ binary decision variables. \n\nIf two nodes $i$ and $j$ have a link from $i$ to $j$, we write $i \\sim j$. We also denote with $\\delta(i)^+$ the set of nodes to which $i$ has a link, i.e., $j \\in \\delta(i)^+$ if and only if $i \\sim j$. Similarly, we denote with \n$\\delta(i)^-$ the set of nodes which are connected to $i$, in the sense that $j \\in \\delta(i)^-$ if and only if $j \\sim i$. \n\nIn addition, we consider continuous variables, for all nodes $i = 1,\\dots, n$, denoted $u_i$. These variables are needed in the MTZ formulation of the problem to eliminate sub-tours between clients. \n\nThe VRP can be formulated as:\n\n$$\n(VRP) \\quad f = \\min_{\\{x_{ij}\\}_{i\\sim j}\\in \\{0,1\\}, \\{u_i\\}_{i=1,\\dots,n}\\in \\mathbb{R}} \\quad \\sum_{i \\sim j} w_{ij} x_{ij}\n$$\n\nsubject to the node-visiting constraint:\n\n$$\n\\sum_{j \\in \\delta(i)^+} x_{ij} = 1, \\,\\sum_{j \\in \\delta(i)^-} x_{ji} = 1,\\, \\forall i \\in \\{1,\\dots,n\\},\n$$\n\nthe depot-visiting constraints:\n\n$$\n\\sum_{i \\in \\delta(0)^+} x_{0i} = K, \\, \\sum_{j \\in \\delta(0)^+} x_{j0} = K,\n$$\n\nand the sub-tour elimination constraints:\n\n$$\nu_i - u_j + Q x_{ij} \\leq Q-q_j, \\, \\forall i \\sim j, \\,i ,j \\neq 0, \\quad q_i \\leq u_i \\leq Q,\\, \\forall i, i \\neq 0.\n$$\n\nIn particular, \n- The cost function is linear in the cost functions and weighs the different arches based on a positive weight $w_{ij}>0$ (typically the distance between node $i$ and node $j$);\n- The first set of constraints enforce that from and to every client, only one link is allowed;\n- The second set of constraints enforce that from and to the depot, exactly $K$ links are allowed;\n- The third set of constraints enforce the sub-tour elimination constraints and are bounds on $u_i$, with $Q>q_j>0$, and $Q,q_i \\in \\mathbb{R}$.\n\n\n## Classical solution\n\nWe can solve the VRP classically, e.g., by using CPLEX. CPLEX uses a branch-and-bound-and-cut method to find an approximate solution of the VRP, which, in this formulation, is a mixed-integer linear program (MILP). For the sake of notation, we pack the decision variables in one vector as\n\n$$\n{\\bf z} = [x_{01},x_{02},\\ldots,x_{10}, x_{12},\\ldots,x_{n(n-1)}]^T,\n$$\n\nwherein ${\\bf z} \\in \\{0,1\\}^N$, with $N = n (n+1)$. So the dimension of the problem scales quadratically with the number of nodes. Let us denote the optimal solution by ${\\bf z}^*$, and the associated optimal cost $f^*$. \n\n\n## Quantum solution\n\nHere, we demonstrate an approach that combines classical and quantum computing steps, following the quantum approximate optimization approach of Farhi, Goldstone, and Gutman (2014). In particular, we use the variational quantum eigensolver (VQE). We stress that given the use of limited depth of the quantum circuits employed (variational forms), it is hard to discuss the speed-up of the algorithm, as the solution obtained is heuristic in nature. At the same time, due to the nature and importance of the target problems, it is worth investigating heuristic approaches, which may be worthwhile for some problem classes. \n\nFollowing [5], the algorithm can be summarized as follows:\n- Preparation steps: \n\t- Transform the combinatorial problem into a binary polynomial optimization problem with equality constraints only;\n\t- Map the resulting problem into an Ising Hamiltonian ($H$) for variables ${\\bf z}$ and basis $Z$, via penalty methods if necessary;\n\t- Choose the depth of the quantum circuit $m$. Note that the depth can be modified adaptively.\n\t- Choose a set of controls $\\theta$ and make a trial function $\\big|\\psi(\\boldsymbol\\theta)\\rangle$, built using a quantum circuit made of C-Phase gates and single-qubit Y rotations, parameterized by the components of $\\boldsymbol\\theta$.\n\n\n- Algorithm steps: \n\t- Evaluate $C(\\boldsymbol\\theta) = \\langle\\psi(\\boldsymbol\\theta)\\big|H\\big|\\psi(\\boldsymbol\\theta)\\rangle$ by sampling the outcome of the circuit in the Z-basis and adding the expectation values of the individual Ising terms together. In general, different control points around $\\boldsymbol\\theta$ have to be estimated, depending on the classical optimizer chosen.\n\t- Use a classical optimizer to choose a new set of controls.\n\t- Continue until $C(\\boldsymbol\\theta)$ reaches a minimum, close enough to the solution $\\boldsymbol\\theta^*$.\n\t- Use the last $\\boldsymbol\\theta$ to generate a final set of samples from the distribution $\\Big|\\langle z_i\\big|\\psi(\\boldsymbol\\theta)\\rangle\\Big|^2\\;\\forall i$ to obtain the answer.\n\n\nThere are many parameters throughout, notably the choice of the trial wavefunction. Below, we consider:\n\n$$\n\\big|\\psi(\\theta)\\rangle = [U_\\mathrm{single}(\\boldsymbol\\theta) U_\\mathrm{entangler}]^m \\big|+\\rangle\n$$\n\nwhere $U_\\mathrm{entangler}$ is a collection of C-Phase gates (fully-entangling gates), and $U_\\mathrm{single}(\\theta) = \\prod_{i=1}^N Y(\\theta_{i})$, where $N$ is the number of qubits and $m$ is the depth of the quantum circuit. \n\n\n### Construct the Ising Hamiltonian\n\nFrom $VRP$ one can construct a binary polynomial optimization with equality constraints only by considering cases in which $K=n-1$. In these cases the sub-tour elimination constraints are not necessary and the problem is only on the variable ${\\bf z}$. In particular, we can write an augmented Lagrangian as\n\n$$\n(IH) \\quad H = \\sum_{i \\sim j} w_{ij} x_{ij} + A \\sum_{i \\in \\{1,\\dots,n\\}} \\Big(\\sum_{j \\in \\delta(i)^+} x_{ij} - 1\\Big)^2 + A \\sum_{i \\in \\{1,\\dots,n\\}}\\Big(\\sum_{j \\in \\delta(i)^-} x_{ji} - 1\\Big)^2 +A \\Big(\\sum_{i \\in \\delta(0)^+} x_{0i} - K\\Big)^2 + A\\Big(\\sum_{j \\in \\delta(0)^+} x_{j0} - K\\Big)^2\n$$\n\nwhere $A$ is a big enough parameter. \n\n### From Hamiltonian to QP formulation \n\nIn the vector ${\\bf z}$, and for a complete graph ($\\delta(i)^+ = \\delta(i)^- = \\{0,1,\\dots,i-1,i+1,\\dots,n\\}$), $H$ can be written as follows.\n\n$$\n\\min_{{\\bf z}\\in \\{0,1\\}^{n(n+1)}} {\\bf w}^T {\\bf z} + A \\sum_{i \\in \\{1,\\dots,n\\}} \\Big({\\bf e}_i \\otimes {\\bf 1}_n^T {\\bf z} - 1\\Big)^2 + A \\sum_{i \\in \\{1,\\dots,n\\}}\\Big({\\bf v}_i^T {\\bf z} - 1\\Big)^2 + A \\Big(({\\bf e}_0 \\otimes {\\bf 1}_n)^T{\\bf z} - K\\Big)^2 + A\\Big({\\bf v}_0^T{\\bf z} - K\\Big)^2.\n$$\n\nThat is:\n\n$$\n\\min_{\\bf z\\in \\{0,1\\}^{n(n+1)}} \\bf z^T {\\bf Q} \\bf z + {\\bf g}^T \\bf z + c,\n$$\n\nWhere: first term:\n\n$$\n{\\bf Q} = A \\sum_{i \\in \\{0,1,\\dots,n\\}} \\Big[({\\bf e}_i \\otimes {\\bf 1}_n)({\\bf e}_i \\otimes {\\bf 1}_n)^T + {\\bf v}_i{\\bf v}_i^T \\Big] \n$$\n\nSecond term:\n\n$$\n{\\bf g} = {\\bf w} -2 A \\sum_{i \\in \\{1,\\dots,n\\}} \\Big[({\\bf e}_i \\otimes {\\bf 1}_n) + {\\bf v}_i \\Big] -2 A K \\Big[({\\bf e}_0 \\otimes {\\bf 1}_n) + {\\bf v}_0 \\Big]\n$$\n\nThird term:\n\n$$\nc = 2An +2AK^2.\n$$\n\nThe QP formulation of the Ising Hamiltonian is ready for the use of VQE. \n\n\n\n## References\n\n[1] E. Farhi, J. Goldstone, S. Gutmann e-print arXiv 1411.4028, 2014\n\n[2] https://github.com/Qiskit/qiskit-tutorial/blob/master/qiskit/aqua/optimization/maxcut_and_tsp.ipynb\n\n[3] C. E. Miller, E. W. Tucker, and R. A. Zemlin (1960). \"Integer Programming Formulations and Travelling Salesman Problems\". J. ACM. 7: 326–329. doi:10.1145/321043.321046.\n\n[4] D. L. Applegate, R. M. Bixby, V. Chvátal, and W. J. Cook (2006). The Traveling Salesman Problem. Princeton University Press, ISBN 978-0-691-12993-8.",
"_____no_output_____"
],
[
"## Initialization\n\nFirst of all we load all the packages that we need: \n - Python 3.6 or greater is required;\n - CPLEX 12.8 or greater is required for the classical computations;\n - Latest Qiskit is required for the quantum computations.",
"_____no_output_____"
]
],
[
[
"# Load the packages that are required\nimport numpy as np\nimport operator\nimport matplotlib.pyplot as plt\n\nimport sys\nif sys.version_info < (3, 6):\n raise Exception('Please use Python version 3.6 or greater.')\n\ntry:\n import cplex\n from cplex.exceptions import CplexError\nexcept: \n print(\"Warning: Cplex not found.\")\nimport math\n\n# Qiskit packages\nfrom qiskit.quantum_info import Pauli\nfrom qiskit.aqua.input import EnergyInput\nfrom qiskit.aqua import run_algorithm\nfrom qiskit.aqua.operators import WeightedPauliOperator\n\n# setup aqua logging\nimport logging\nfrom qiskit.aqua._logging import set_logging_config, build_logging_config\n#set_logging_config(build_logging_config(logging.DEBUG)) # choose INFO, DEBUG to see the log",
"Warning: Cplex not found.\n"
]
],
[
[
"We then initialize the variables",
"_____no_output_____"
]
],
[
[
"# Initialize the problem by defining the parameters\nn = 3 # number of nodes + depot (n+1)\nK = 2 # number of vehicles",
"_____no_output_____"
]
],
[
[
"We define an initializer class that randomly places the nodes in a 2-D plane and computes the distance between them. ",
"_____no_output_____"
]
],
[
[
"# Get the data\nclass Initializer():\n\n def __init__(self, n):\n self.n = n\n\n def generate_instance(self):\n\n n = self.n\n\n # np.random.seed(33)\n np.random.seed(1543)\n\n xc = (np.random.rand(n) - 0.5) * 10\n yc = (np.random.rand(n) - 0.5) * 10\n\n instance = np.zeros([n, n])\n for ii in range(0, n):\n for jj in range(ii + 1, n):\n instance[ii, jj] = (xc[ii] - xc[jj]) ** 2 + (yc[ii] - yc[jj]) ** 2\n instance[jj, ii] = instance[ii, jj]\n\n return xc, yc, instance",
"_____no_output_____"
],
[
"# Initialize the problem by randomly generating the instance\ninitializer = Initializer(n)\nxc,yc,instance = initializer.generate_instance()",
"_____no_output_____"
]
],
[
[
"## Classical solution using IBM ILOG CPLEX\n\nFor a classical solution, we use IBM ILOG CPLEX. CPLEX is able to find the exact solution of this problem. We first define a ClassicalOptimizer class that encodes the problem in a way that CPLEX can solve, and then instantiate the class and solve it. \n",
"_____no_output_____"
]
],
[
[
"class ClassicalOptimizer:\n\n def __init__(self, instance,n,K):\n\n self.instance = instance\n self.n = n # number of nodes\n self.K = K # number of vehicles\n\n\n def compute_allowed_combinations(self):\n f = math.factorial\n return f(self.n) / f(self.K) / f(self.n-self.K)\n\n\n def cplex_solution(self):\n\n # refactoring\n instance = self.instance\n n = self.n\n K = self.K\n\n my_obj = list(instance.reshape(1, n**2)[0])+[0. for x in range(0,n-1)]\n my_ub = [1 for x in range(0,n**2+n-1)]\n my_lb = [0 for x in range(0,n**2)] + [0.1 for x in range(0,n-1)]\n my_ctype = \"\".join(['I' for x in range(0,n**2)]) + \"\".join(['C' for x in range(0,n-1)])\n\n my_rhs = 2*([K] + [1 for x in range(0,n-1)]) + [1-0.1 for x in range(0,(n-1)**2-(n-1))] + [0 for x in range(0,n)]\n my_sense = \"\".join(['E' for x in range(0,2*n)]) + \"\".join(['L' for x in range(0,(n-1)**2-(n-1))])+\"\".join(['E' for x in range(0,n)])\n\n try:\n my_prob = cplex.Cplex()\n self.populatebyrow(my_prob,my_obj,my_ub,my_lb,my_ctype,my_sense,my_rhs)\n\n my_prob.solve()\n\n except CplexError as exc:\n print(exc)\n return\n\n x = my_prob.solution.get_values()\n x = np.array(x)\n cost = my_prob.solution.get_objective_value()\n\n return x,cost\n \n\n def populatebyrow(self,prob,my_obj,my_ub,my_lb,my_ctype,my_sense,my_rhs):\n\n n = self.n\n \n prob.objective.set_sense(prob.objective.sense.minimize)\n prob.variables.add(obj = my_obj, lb = my_lb, ub = my_ub, types = my_ctype)\n \n prob.set_log_stream(None)\n prob.set_error_stream(None)\n prob.set_warning_stream(None)\n prob.set_results_stream(None)\n\n rows = []\n for ii in range(0,n):\n col = [x for x in range(0+n*ii,n+n*ii)]\n coef = [1 for x in range(0,n)]\n rows.append([col, coef])\n\n for ii in range(0,n):\n col = [x for x in range(0+ii,n**2,n)]\n coef = [1 for x in range(0,n)]\n\n rows.append([col, coef])\n\n # Sub-tour elimination constraints:\n for ii in range(0, n):\n for jj in range(0,n):\n if (ii != jj)and(ii*jj>0):\n\n col = [ii+(jj*n), n**2+ii-1, n**2+jj-1]\n coef = [1, 1, -1]\n\n rows.append([col, coef])\n\n for ii in range(0,n):\n col = [(ii)*(n+1)]\n coef = [1]\n rows.append([col, coef])\n\n prob.linear_constraints.add(lin_expr=rows, senses=my_sense, rhs=my_rhs)",
"_____no_output_____"
],
[
"# Instantiate the classical optimizer class\nclassical_optimizer = ClassicalOptimizer(instance,n,K)\n\n# Print number of feasible solutions\nprint('Number of feasible solutions = ' + str(classical_optimizer.compute_allowed_combinations()))",
"Number of feasible solutions = 3.0\n"
],
[
"# Solve the problem in a classical fashion via CPLEX\nx = None\nz = None\ntry:\n x,classical_cost = classical_optimizer.cplex_solution()\n # Put the solution in the z variable\n z = [x[ii] for ii in range(n**2) if ii//n != ii%n]\n # Print the solution\n print(z)\nexcept: \n print(\"CPLEX may be missing.\")",
"CPLEX may be missing.\n"
],
[
"# Visualize the solution\ndef visualize_solution(xc, yc, x, C, n, K, title_str):\n plt.figure()\n plt.scatter(xc, yc, s=200)\n for i in range(len(xc)):\n plt.annotate(i, (xc[i] + 0.15, yc[i]), size=16, color='r')\n plt.plot(xc[0], yc[0], 'r*', ms=20)\n\n plt.grid()\n\n for ii in range(0, n ** 2):\n\n if x[ii] > 0:\n ix = ii // n\n iy = ii % n\n plt.arrow(xc[ix], yc[ix], xc[iy] - xc[ix], yc[iy] - yc[ix], length_includes_head=True, head_width=.25)\n\n plt.title(title_str+' cost = ' + str(int(C * 100) / 100.))\n plt.show() \n\nif x: visualize_solution(xc, yc, x, classical_cost, n, K, 'Classical')",
"_____no_output_____"
]
],
[
[
"If you have CPLEX, the solution shows the depot with a star and the selected routes for the vehicles with arrows. ",
"_____no_output_____"
],
[
"## Quantum solution from the ground up\n\nFor the quantum solution, we use Qiskit. \n\nFirst, we derive the solution from the ground up, using a class QuantumOptimizer that encodes the quantum approach to solve the problem and then we instantiate it and solve it. We define the following methods inside the class:\n- `binary_representation` : encodes the problem $(M)$ into a the Ising Hamiltonian QP (that's basically linear algebra);\n- `construct_hamiltonian` : constructs the Ising Hamiltonian in terms of the $Z$ basis;\n- `check_hamiltonian` : makes sure that the Ising Hamiltonian is correctly encoded in the $Z$ basis: to do this, it solves a eigenvalue-eigenvector problem for a symmetric matrix of dimension $2^N \\times 2^N$. For the problem at hand $n=3$, that is $N = 12$ seems the limit; \n- `vqe_solution` : solves the problem $(M)$ via VQE by using the SPSA solver (with default parameters);\n- `_q_solution` : internal routine to represent the solution in a usable format.\n",
"_____no_output_____"
]
],
[
[
"class QuantumOptimizer:\n\n def __init__(self, instance, n, K, max_trials=1000):\n\n self.instance = instance\n self.n = n\n self.K = K\n self.max_trials = max_trials\n\n def binary_representation(self,x_sol=0):\n\n instance = self.instance\n n = self.n\n K = self.K\n\n A = np.max(instance) * 100 # A parameter of cost function\n\n # Determine the weights w\n instance_vec = instance.reshape(n ** 2)\n w_list = [instance_vec[x] for x in range(n ** 2) if instance_vec[x] > 0]\n w = np.zeros(n * (n - 1))\n for ii in range(len(w_list)):\n w[ii] = w_list[ii]\n\n # Some variables I will use\n Id_n = np.eye(n)\n Im_n_1 = np.ones([n - 1, n - 1])\n Iv_n_1 = np.ones(n)\n Iv_n_1[0] = 0\n Iv_n = np.ones(n-1)\n neg_Iv_n_1 = np.ones(n) - Iv_n_1\n\n v = np.zeros([n, n*(n-1)])\n for ii in range(n):\n count = ii-1\n for jj in range(n*(n-1)):\n\n if jj//(n-1) == ii:\n count = ii\n\n if jj//(n-1) != ii and jj%(n-1) == count:\n v[ii][jj] = 1.\n\n vn = np.sum(v[1:], axis=0)\n\n # Q defines the interactions between variables\n Q = A*(np.kron(Id_n, Im_n_1) + np.dot(v.T, v))\n\n # g defines the contribution from the individual variables\n g = w - 2 * A * (np.kron(Iv_n_1,Iv_n) + vn.T) - \\\n 2 * A * K * (np.kron(neg_Iv_n_1, Iv_n) + v[0].T)\n\n # c is the constant offset\n c = 2 * A * (n-1) + 2 * A * (K ** 2)\n\n try:\n max(x_sol)\n # Evaluates the cost distance from a binary representation of a path\n fun = lambda x: np.dot(np.around(x), np.dot(Q, np.around(x))) + np.dot(g, np.around(x)) + c\n cost = fun(x_sol)\n except:\n cost = 0\n\n return Q,g,c,cost\n\n def construct_hamiltonian(self):\n\n instance = self.instance\n n = self.n\n K = self.K\n\n N = (n - 1) * n # number of qubits\n Q,g,c,_ = self.binary_representation()\n\n # Defining the new matrices in the Z-basis\n\n Iv = np.ones(N)\n Qz = (Q / 4)\n gz = (-g / 2 - np.dot(Iv, Q / 4) - np.dot(Q / 4, Iv))\n cz = (c + np.dot(g / 2, Iv) + np.dot(Iv, np.dot(Q / 4, Iv)))\n\n cz = cz + np.trace(Qz)\n Qz = Qz - np.diag(np.diag(Qz))\n\n # Getting the Hamiltonian in the form of a list of Pauli terms\n\n pauli_list = []\n for i in range(N):\n if gz[i] != 0:\n wp = np.zeros(N)\n vp = np.zeros(N)\n vp[i] = 1\n pauli_list.append((gz[i], Pauli(vp, wp)))\n for i in range(N):\n for j in range(i):\n if Qz[i, j] != 0:\n wp = np.zeros(N)\n vp = np.zeros(N)\n vp[i] = 1\n vp[j] = 1\n pauli_list.append((2 * Qz[i, j], Pauli(vp, wp)))\n\n pauli_list.append((cz, Pauli(np.zeros(N), np.zeros(N))))\n\n return cz, pauli_list\n\n def check_hamiltonian(self):\n\n cz, op = self.construct_hamiltonian()\n Op = WeightedPauliOperator(paulis=op)\n\n qubitOp, offset = Op, 0\n algo_input = EnergyInput(qubitOp)\n\n # Making the Hamiltonian in its full form and getting the lowest eigenvalue and eigenvector\n\n algorithm_cfg = {\n 'name': 'ExactEigensolver',\n }\n\n params = {\n 'problem': {'name': 'ising'},\n 'algorithm': algorithm_cfg\n }\n result = run_algorithm(params, algo_input)\n\n quantum_solution = self._q_solution(result['eigvecs'][0],self.n*(self.n+1))\n ground_level = result['energy'] + offset\n\n return quantum_solution, ground_level\n\n def vqe_solution(self):\n\n cz, op = self.construct_hamiltonian()\n Op = WeightedPauliOperator(paulis=op)\n\n qubitOp, offset = Op, cz\n algo_input = EnergyInput(qubitOp)\n\n\n algorithm_cfg = {\n 'name': 'VQE'\n }\n\n optimizer_cfg = {\n 'name': 'SPSA',\n 'max_trials': self.max_trials\n }\n\n var_form_cfg = {\n 'name': 'RY',\n 'depth': 5,\n 'entanglement': 'linear'\n }\n\n params = {\n 'problem': {'name': 'ising', 'random_seed': 10598},\n 'algorithm': algorithm_cfg,\n 'optimizer': optimizer_cfg,\n 'variational_form': var_form_cfg,\n 'backend': {'name': 'qasm_simulator'\n }\n }\n\n result = run_algorithm(params, algo_input)\n\n #quantum_solution = self._q_solution(result['eigvecs'][0], self.n * (self.n + 1))\n quantum_solution_dict = result['eigvecs'][0]\n\n q_s = max(quantum_solution_dict.items(), key=operator.itemgetter(1))[0]\n quantum_solution= [int(chars) for chars in q_s]\n quantum_solution = np.flip(quantum_solution, axis=0)\n\n _,_,_,level = self.binary_representation(x_sol=quantum_solution)\n return quantum_solution_dict, quantum_solution, level\n\n def _q_solution(self, v, N):\n\n index_value = [x for x in range(len(v)) if v[x] == max(v)][0]\n string_value = \"{0:b}\".format(index_value)\n\n while len(string_value)<N:\n string_value = '0'+string_value\n\n sol = list()\n for elements in string_value:\n if elements == '0':\n sol.append(0)\n else:\n sol.append(1)\n\n sol = np.flip(sol, axis=0)\n\n return sol",
"_____no_output_____"
]
],
[
[
"### Step 1\n\nInstantiate the quantum optimizer class with parameters: \n- the instance;\n- the number of nodes and vehicles `n` and `K`;\n- the number of iterations for SPSA in VQE (default 1000)",
"_____no_output_____"
]
],
[
[
"# Instantiate the quantum optimizer class with parameters: \nquantum_optimizer = QuantumOptimizer(instance, n, K, 100)",
"_____no_output_____"
]
],
[
[
"### Step 2\n\nEncode the problem as a binary formulation (IH-QP).\n\nSanity check: make sure that the binary formulation in the quantum optimizer is correct (i.e., yields the same cost given the same solution).",
"_____no_output_____"
]
],
[
[
"# Check if the binary representation is correct\ntry:\n if z:\n Q,g,c,binary_cost = quantum_optimizer.binary_representation(x_sol = z)\n print(binary_cost,classical_cost)\n if np.abs(binary_cost - classical_cost)<0.01:\n print('Binary formulation is correct')\n else: print('Error in the binary formulation')\n else:\n print('Could not verify the correctness, due to CPLEX solution being unavailable.')\n Q,g,c,binary_cost = quantum_optimizer.binary_representation()\nexcept NameError as e:\n print(\"Warning: Please run the cells above first.\")\n print(e)",
"Could not verify the correctness, due to CPLEX solution being unavailable.\n"
]
],
[
[
"### Step 3\n\nEncode the problem as an Ising Hamiltonian in the Z basis. \n\nSanity check: make sure that the formulation is correct (i.e., yields the same cost given the same solution)",
"_____no_output_____"
]
],
[
[
"ground_state, ground_level = quantum_optimizer.check_hamiltonian()\nprint(ground_state)\n\nif z:\n if np.abs(ground_level - classical_cost)<0.01:\n print('Ising Hamiltonian in Z basis is correct')\n else: print('Error in the Ising Hamiltonian formulation')",
"/Users/paul/opt/anaconda3/envs/qiskit/lib/python3.7/site-packages/qiskit/providers/models/backendconfiguration.py:337: UserWarning: `dt` and `dtm` now have units of seconds(s) rather than nanoseconds(ns).\n warnings.warn('`dt` and `dtm` now have units of seconds(s) rather '\n"
]
],
[
[
"### Step 4\n\nSolve the problem via VQE. N.B. Depending on the number of qubits, the state-vector simulation can can take a while; for example with 12 qubits, it takes more than 12 hours. Logging is useful to see what the program is doing.",
"_____no_output_____"
]
],
[
[
"quantum_dictionary, quantum_solution, quantum_cost = quantum_optimizer.vqe_solution()\n\nprint(quantum_solution, quantum_cost)",
"[1 1 1 0 1 0] 132.11148115684045\n"
]
],
[
[
"### Step 5\nVisualize the solution",
"_____no_output_____"
]
],
[
[
"# Put the solution in a way that is compatible with the classical variables\nx_quantum = np.zeros(n**2)\nkk = 0\nfor ii in range(n ** 2):\n if ii // n != ii % n:\n x_quantum[ii] = quantum_solution[kk]\n kk += 1\n\n\n# visualize the solution \nvisualize_solution(xc, yc, x_quantum, quantum_cost, n, K, 'Quantum')\n \n# and visualize the classical for comparison\nif x: visualize_solution(xc, yc, x, classical_cost, n, K, 'Classical')",
"_____no_output_____"
]
],
[
[
"The plots present the depot with a star and the selected routes for the vehicles with arrows. Note that in this particular case, we can find the optimal solution of the QP formulation, which happens to coincide with the optimal solution of the ILP.\n\nKeep in mind that VQE is an heuristic working on the QP formulation of the Ising Hamiltonian, though. For suitable choices of A, local optima of the QP formulation will be feasible solutions to the ILP. While for some small instances, as above, we can find optimal solutions of the QP formulation which coincide with optima of the ILP, finding optimal solutions of the ILP is harder than finding local optima of the QP formulation, in general, which in turn is harder than finding feasible solutions of the ILP. Even within the VQE, one may provide stronger guarantees, for specific variational forms (trial wave functions). \n\nLast but not least, you may be pleased to learn that the above has been packaged in Qiskit Aqua.",
"_____no_output_____"
]
],
[
[
"from qiskit import Aer\nfrom qiskit.aqua import QuantumInstance\nfrom qiskit.aqua import run_algorithm\nfrom qiskit.aqua.input import EnergyInput\nfrom qiskit.aqua.algorithms import VQE, QAOA, ExactEigensolver\nfrom qiskit.aqua.components.optimizers import COBYLA\nfrom qiskit.aqua.components.variational_forms import RY\nfrom qiskit.aqua.translators.ising.vehicle_routing import *\n\nqubitOp = get_vehiclerouting_qubitops(instance, n, K)\nbackend = Aer.get_backend('statevector_simulator')\nseed = 50\ncobyla = COBYLA()\ncobyla.set_options(maxiter=250)\nry = RY(qubitOp.num_qubits, depth=3, entanglement='full')\nvqe = VQE(qubitOp, ry, cobyla)\nvqe.random_seed = seed\nquantum_instance = QuantumInstance(backend=backend, seed_simulator=seed, seed_transpiler=seed)\nresult = vqe.run(quantum_instance)\n# print(result)\nx_quantum2 = get_vehiclerouting_solution(instance, n, K, result)\nprint(x_quantum2)\nquantum_cost2 = get_vehiclerouting_cost(instance, n, K, x_quantum2)\nprint(quantum_cost2)",
"[1 1 1 0 0 1]\n12434.909288240102\n"
],
[
"import qiskit.tools.jupyter\n%qiskit_version_table\n%qiskit_copyright",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbca819f317de6f5fc67c9fc297ad55ad4740daf
| 194,543 |
ipynb
|
Jupyter Notebook
|
decision_score.ipynb
|
JoseAugustoVital/Decision-Score-Marketplace
|
5c1b76cd931bd4b3852583bcdb7d9bced363ce34
|
[
"MIT"
] | null | null | null |
decision_score.ipynb
|
JoseAugustoVital/Decision-Score-Marketplace
|
5c1b76cd931bd4b3852583bcdb7d9bced363ce34
|
[
"MIT"
] | null | null | null |
decision_score.ipynb
|
JoseAugustoVital/Decision-Score-Marketplace
|
5c1b76cd931bd4b3852583bcdb7d9bced363ce34
|
[
"MIT"
] | null | null | null | 37.746023 | 695 | 0.352847 |
[
[
[
"<a href=\"https://colab.research.google.com/github/JoseAugustoVital/Decision-Score-MarketPlace/blob/main/decision_score.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# ***UNIVERSIDADE FEDERAL DO MATO GROSSO DO SUL***\n# Análise de dados para aumentar nível de satisfação dos clientes de um marketplace utilizando árvore de decisão.\n**TRABALHO 3 - INTELIGÊNCIA ARTIFICIAL 2021/1**\n__________________________________________________\n\n**Aluno:** \n\nNome: **José Augusto Lajo Vieira Vital** \n\n",
"_____no_output_____"
]
],
[
[
"# INÍCIO DO ESTUDO",
"_____no_output_____"
]
],
[
[
"**Importação do sistema operacional**",
"_____no_output_____"
]
],
[
[
"import os",
"_____no_output_____"
]
],
[
[
"**Determinação do acesso ao diretório**",
"_____no_output_____"
]
],
[
[
"os.chdir('/content/drive/MyDrive/datasets')",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount(\"/content/drive\", force_remount=True).\n"
],
[
"!pwd",
"/content/drive/My Drive/datasets\n"
],
[
"!ls",
"olist_customers_dataset.csv\t olist_products_dataset.csv\nolist_geolocation_dataset.csv\t olist_sellers_dataset.csv\nolist_order_items_dataset.csv\t product_category_name_translation.csv\nolist_order_payments_dataset.csv tabela_resulta.csv\nolist_order_reviews_dataset.csv tabela_result.csv\nolist_orders_dataset.csv\n"
]
],
[
[
"**Importação das bibliotecas Pandas e Numpy**",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"**Leitura dos tabelas .csv**",
"_____no_output_____"
]
],
[
[
"tabela_cliente = pd.read_csv('olist_customers_dataset.csv')\ntabela_localizacao = pd.read_csv('olist_geolocation_dataset.csv')\ntabela_pedido = pd.read_csv('olist_order_items_dataset.csv')\ntabela_pagamento = pd.read_csv('olist_order_payments_dataset.csv')\ntabela_review = pd.read_csv('olist_order_reviews_dataset.csv')\ntabela_entrega_pedido = pd.read_csv('olist_orders_dataset.csv')\ntabela_descricao_produto = pd.read_csv('olist_products_dataset.csv')\ntabela_vendedor = pd.read_csv('olist_sellers_dataset.csv')\ntabela_categoria_traduzido = pd.read_csv('product_category_name_translation.csv')",
"_____no_output_____"
]
],
[
[
"**Checagem dos 5 primeiros elementos de cada tabela**",
"_____no_output_____"
]
],
[
[
"tabela_cliente.head()",
"_____no_output_____"
],
[
"tabela_localizacao.head()",
"_____no_output_____"
],
[
"tabela_pedido.head()",
"_____no_output_____"
],
[
"tabela_pagamento.head()",
"_____no_output_____"
],
[
"tabela_review.head()",
"_____no_output_____"
],
[
"tabela_entrega_pedido.head()",
"_____no_output_____"
],
[
"tabela_descricao_produto.head()",
"_____no_output_____"
],
[
"tabela_vendedor.head()",
"_____no_output_____"
],
[
"tabela_categoria_traduzido.head()",
"_____no_output_____"
]
],
[
[
"**Início do processo de união das 9 tabelas disponibilizadas com a finalidade de produzir uma tabela resultante que possua os elementos mais importantes para a determinação do review_score. No primero merge realizado, unimos a tabela de clientes com as respectivas entregas dos pedidos usando o código individual de cada consumidor como parâmetro.**",
"_____no_output_____"
]
],
[
[
"pd.merge(tabela_cliente, tabela_entrega_pedido, on=[\"customer_id\"], how=\"inner\")",
"_____no_output_____"
]
],
[
[
"**Processo de união com as demais tabelas disponibilizadas**\n\n**1 - (Clientes, Entregas)**\n\n**2 - (1, Pedidos)**\n\n**3 - (2, Pagamentos)**\n\n**4 - (3, Review)**\n\n**5 - (4, Vendedor)**",
"_____no_output_____"
]
],
[
[
"test = pd.merge(tabela_cliente, tabela_entrega_pedido, on=[\"customer_id\"], how=\"inner\")\ntest = pd.merge(test, tabela_pedido, on=[\"order_id\"], how=\"inner\")\ntest = pd.merge(test, tabela_pagamento, on=[\"order_id\"], how=\"inner\")\ntest = pd.merge(test, tabela_review, on=[\"order_id\"], how=\"inner\")\ntest = pd.merge(test, tabela_vendedor, on=[\"seller_id\"], how=\"inner\")",
"_____no_output_____"
]
],
[
[
"**Tabela Resultante**\n\n**Linhas: 118315**\n\n**Colunas: 31**",
"_____no_output_____"
]
],
[
[
"test",
"_____no_output_____"
]
],
[
[
"**Segunda filtragem consiste em remover elementos que não possuem\nrelação com a variável review_score**",
"_____no_output_____"
]
],
[
[
"#test = test.drop(columns=[\"customer_unique_id\"],axis=1)\n#test = test.drop(columns=[\"customer_city\"],axis=1)\n#test = test.drop(columns=[\"customer_state\"],axis=1)\n#test = test.drop(columns=[\"order_status\"],axis=1)\n#test = test.drop(columns=[\"order_purchase_timestamp\"],axis=1)\n#test = test.drop(columns=[\"order_approved_at\"],axis=1)\n#test = test.drop(columns=[\"order_delivered_carrier_date\"],axis=1)\n#test = test.drop(columns=[\"order_delivered_customer_date\"],axis=1)\n#test = test.drop(columns=[\"order_estimated_delivery_date\"],axis=1)\n#test = test.drop(columns=[\"shipping_limit_date\"],axis=1)\n#test = test.drop(columns=[\"review_creation_date\"],axis=1)\n#test = test.drop(columns=[\"review_answer_timestamp\"],axis=1)\n#test = test.drop(columns=[\"seller_city\"],axis=1)\n#test = test.drop(columns=[\"seller_state\"],axis=1)\n#test = test.drop(columns=[\"review_comment_title\"],axis=1)\n#test = test.drop(columns=[\"review_comment_message\"],axis=1)",
"_____no_output_____"
]
],
[
[
"**Tabela Resultante após a remoção de atributos não prioritários para o nível de\nsatisfação dos clientes**\n\n**Linhas: 118315**\n\n**Colunas: 15**",
"_____no_output_____"
]
],
[
[
"test",
"_____no_output_____"
]
],
[
[
"**Inserindo cada atributo da tabela resultante em um vetor para melhor manipulação dos dados**\n",
"_____no_output_____"
]
],
[
[
"vetor_cliente = np.array(test.customer_id)\nvetor_cepcliente = np.array(test.customer_zip_code_prefix)\nvetor_pedido = np.array(test.order_id)\nvetor_idpedido = np.array(test.order_item_id)\nvetor_produto = np.array(test.product_id)\nvetor_vendedor = np.array(test.seller_id)\nvetor_preco_produto = np.array(test.price)\nvetor_frete = np.array(test.freight_value)\nvetor_parcela = np.array(test.payment_sequential)\nvetor_tipopagamento = np.array(test.payment_type)\nvetor_pay = np.array(test.payment_installments)\nvetor_valorfinal = np.array(test.payment_value)\nvetor_review = np.array(test.review_id)\nvetor_score = np.array(test.review_score)\nvetor_cepvendedor = np.array(test.seller_zip_code_prefix)",
"_____no_output_____"
]
],
[
[
"**Definindo um novo dataframe vazio**",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame()\ndf",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Definindo as colunas do novo dataframe e atribuindo para cada coluna, seu respectivo vetor de dados registrado anteriormente.**",
"_____no_output_____"
]
],
[
[
"COLUNAS = [\n 'Cliente',\n 'CEP_Cliente',\n 'Pedido',\n 'id_Pedido',\n 'Produto',\n 'Vendedor',\n 'Preco_produto',\n 'Frete',\n 'Parcela',\n 'Tipo_pagamento',\n 'Installments',\n 'Valor_total',\n 'ID_Review',\n 'CEP_Vendedor',\n 'Score'\n]\n\ndf = pd.DataFrame(columns =COLUNAS)\n\ndf.Cliente = vetor_cliente\ndf.CEP_Cliente = vetor_cepcliente\ndf.Pedido = vetor_pedido\ndf.id_Pedido = vetor_idpedido\ndf.Produto = vetor_produto\ndf.Vendedor = vetor_vendedor\ndf.Preco_produto = vetor_preco_produto\ndf.Frete = vetor_frete\ndf.Parcela = vetor_parcela\ndf.Tipo_pagamento = vetor_tipopagamento\ndf.Installments = vetor_pay\ndf.Valor_total = vetor_valorfinal\ndf.ID_Review = vetor_review\ndf.CEP_Vendedor = vetor_cepvendedor\ndf.Score = vetor_score\ndf",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Impressão da coluna de clientes.**",
"_____no_output_____"
]
],
[
[
"df.Cliente",
"_____no_output_____"
],
[
"#for index, row in df.iterrows():\n# if row['Score'] == 1:\n# df.loc[index,'Classe'] = 'Pessimo'\n# if row['Score'] == 2:\n# df.loc[index,'Classe'] = 'Ruim'\n# if row['Score'] == 3:\n# df.loc[index,'Classe'] = 'Mediano'\n# if row['Score'] == 4:\n# df.loc[index,'Classe'] = 'Bom'\n# if row['Score'] == 5:\n# df.loc[index,'Classe'] = 'Otimo'",
"_____no_output_____"
]
],
[
[
"**Informações do dataframe**\n\n**Atributos, elementos não nulos, Tipo das variáveis da coluna**",
"_____no_output_____"
]
],
[
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 118315 entries, 0 to 118314\nData columns (total 15 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Cliente 118315 non-null object \n 1 CEP_Cliente 118315 non-null int64 \n 2 Pedido 118315 non-null object \n 3 id_Pedido 118315 non-null int64 \n 4 Produto 118315 non-null object \n 5 Vendedor 118315 non-null object \n 6 Preco_produto 118315 non-null float64\n 7 Frete 118315 non-null float64\n 8 Parcela 118315 non-null int64 \n 9 Tipo_pagamento 118315 non-null object \n 10 Installments 118315 non-null int64 \n 11 Valor_total 118315 non-null float64\n 12 ID_Review 118315 non-null object \n 13 CEP_Vendedor 118315 non-null int64 \n 14 Score 118315 non-null int64 \ndtypes: float64(3), int64(6), object(6)\nmemory usage: 13.5+ MB\n"
]
],
[
[
"**Agrupando os elementos do dataframe por consumidor**",
"_____no_output_____"
]
],
[
[
"df.groupby(by='Cliente').size()",
"_____no_output_____"
]
],
[
[
"**Importando os métodos de árvore de decisão**",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier, export_graphviz\nfrom sklearn.model_selection import train_test_split\nfrom sklearn import metrics",
"_____no_output_____"
]
],
[
[
"**Para simplificar os dados e evitar criar um dummy com esse dataframe, removemos todos os elementos não-numéricos para que o modelo seja capaz de realizar a execução. Solução encontrada para simplificar os atributos do tipo \"Objeto\" para tipos numéricos.**",
"_____no_output_____"
]
],
[
[
"df['Cliente'] = df['Cliente'].str.replace(r'\\D', '')\ndf['Pedido'] = df['Pedido'].str.replace(r'\\D', '')\ndf['Produto'] = df['Produto'].str.replace(r'\\D', '')\ndf['Vendedor'] = df['Vendedor'].str.replace(r'\\D', '')\ndf['ID_Review'] = df['ID_Review'].str.replace(r'\\D', '')\ndf",
"_____no_output_____"
]
],
[
[
"**Realizamos o procedimento de remoção dos elementos não-numéricos para todas as colunas do tipo objeto com exceção do tipo de pagamento pois o tipo de pagamento se resume a poucas opções. Dessa forma, usamos a função get_dummies apenas para o tipo de pagamento**\n\n**Portanto, a coluna Tipo_pagamento se divide em quatro colunas com lógica booleana. As novas colunas são: Tipo_pagamento_boleto, \tTipo_pagamento_credit_card, Tipo_pagamento_debit_card, Tipo_pagamento_voucher**",
"_____no_output_____"
]
],
[
[
"result_df = pd.get_dummies(df, columns=[\"Tipo_pagamento\"])\n",
"_____no_output_____"
]
],
[
[
"**Resultado final do dataframe**",
"_____no_output_____"
]
],
[
[
"result_df",
"_____no_output_____"
]
],
[
[
"**Criação de um dataframe reserva para possíveis conclusões**",
"_____no_output_____"
]
],
[
[
"reserva = result_df\nreserva",
"_____no_output_____"
]
],
[
[
"**Eliminando todas as linhas com nível 4 ou nível 5 de satisfação. Dessa forma, temos um dataframe com todos os dados, e um apenas com dados classificados com nível 3,2 ou 1 ou seja, mediano, ruim ou péssimo. (elementos que apresentam nível de insatisfação interessante para análise)**",
"_____no_output_____"
]
],
[
[
"reserva = reserva.drop(reserva[reserva.Score > 3].index)\nreserva\n",
"_____no_output_____"
]
],
[
[
"**Processo de separação de treine/teste**\n\n**Proporções estabelecidas:**\n\n**70% treino**\n\n**30% teste**",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(result_df.drop('Score', axis=1), result_df['Score'], test_size=0.3)",
"_____no_output_____"
]
],
[
[
"**Número de amostras para cada processo**",
"_____no_output_____"
]
],
[
[
"X_train.shape, X_test.shape",
"_____no_output_____"
]
],
[
[
"**Número de targets para cada processo**",
"_____no_output_____"
]
],
[
[
"y_train.shape, y_test.shape",
"_____no_output_____"
]
],
[
[
"**Criação do classifcador**",
"_____no_output_____"
]
],
[
[
"cls = DecisionTreeClassifier()",
"_____no_output_____"
]
],
[
[
"**Treinamento**",
"_____no_output_____"
]
],
[
[
"cls = cls.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"**Vetor com as importancias de cada atributo para a determinação do review_score**",
"_____no_output_____"
]
],
[
[
"cls.feature_importances_",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"**Para tornar o modelo mais visual, criamos um laço para a impressão dos pesos de cada atributo para determinar o score**",
"_____no_output_____"
]
],
[
[
"for feature, importancia in zip(result_df.columns, cls.feature_importances_):\n print(\"{}:{:.1f}%\".format(feature,((importancia*100))))\n",
"Cliente:10.4%\nCEP_Cliente:11.0%\nPedido:10.4%\nid_Pedido:0.8%\nProduto:9.4%\nVendedor:8.0%\nPreco_produto:8.0%\nFrete:8.1%\nParcela:0.3%\nInstallments:3.7%\nValor_total:8.6%\nID_Review:11.4%\nCEP_Vendedor:7.7%\nScore:0.8%\nTipo_pagamento_boleto:0.8%\nTipo_pagamento_credit_card:0.2%\nTipo_pagamento_debit_card:0.3%\n"
]
],
[
[
"**Vetor com predições para checagem do aprendizado**",
"_____no_output_____"
]
],
[
[
"result = cls.predict(X_test)\nresult",
"_____no_output_____"
],
[
"result_df.Score[118310]",
"_____no_output_____"
]
],
[
[
"**Representação das métricas de precisão e médias do modelo**",
"_____no_output_____"
]
],
[
[
"from sklearn import metrics\nprint(metrics.classification_report(y_test,result))",
" precision recall f1-score support\n\n 1 0.26 0.27 0.27 4602\n 2 0.12 0.13 0.12 1193\n 3 0.15 0.17 0.16 2975\n 4 0.24 0.25 0.24 6685\n 5 0.61 0.58 0.60 20040\n\n accuracy 0.43 35495\n macro avg 0.28 0.28 0.28 35495\nweighted avg 0.44 0.43 0.44 35495\n\n"
]
],
[
[
"**Precisão total**",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import cross_val_score\nallScores = cross_val_score(cls, X_train, y_train , cv=10)\nallScores.mean()",
"_____no_output_____"
]
],
[
[
"**Treinamento utilizando o dataframe reserva. (apenas com os níveis de satisfação abaixo da média, score <=3)** ",
"_____no_output_____"
],
[
"**Split do dataframe em treino e teste (70%, 30%, respectivamente)**",
"_____no_output_____"
]
],
[
[
"X_train, X_test, y_train, y_test = train_test_split(reserva.drop('Score', axis=1), reserva['Score'], test_size=0.3)",
"_____no_output_____"
]
],
[
[
"**Quantidade de amostras do treino**",
"_____no_output_____"
]
],
[
[
"X_train.shape, X_test.shape\n",
"_____no_output_____"
]
],
[
[
"**Classificador**",
"_____no_output_____"
]
],
[
[
"clf = DecisionTreeClassifier()",
"_____no_output_____"
]
],
[
[
"**Treino**",
"_____no_output_____"
]
],
[
[
"clf = clf.fit(X_train, y_train)\n",
"_____no_output_____"
]
],
[
[
"**Importancia de cada atributo para determinar o nível de satisfação dos consumidores**",
"_____no_output_____"
]
],
[
[
"clf.feature_importances_",
"_____no_output_____"
],
[
"for feature, importancia in zip(reserva.columns, clf.feature_importances_):\n print(\"{}:{:.1f}%\".format(feature,((importancia*100))))\n ",
"Cliente:10.0%\nCEP_Cliente:11.1%\nPedido:10.9%\nid_Pedido:0.8%\nProduto:9.5%\nVendedor:8.3%\nPreco_produto:7.4%\nFrete:8.3%\nParcela:0.2%\nInstallments:3.6%\nValor_total:8.7%\nID_Review:11.6%\nCEP_Vendedor:7.6%\nScore:0.7%\nTipo_pagamento_boleto:0.7%\nTipo_pagamento_credit_card:0.2%\nTipo_pagamento_debit_card:0.3%\n"
]
],
[
[
"# ANÁLISE DOS DADOS PROCESSADOS\n\n**Com o objetivo de aumentar o review_score desse marketplace, utilizamos o algoritmo de árvore de decisão para encontrar elementos que influenciam diretamente a aceitação e satisfação dos clientes, analisando desde o processo de despacho do produto, até a qualidade do atendimento e do produto final. Dessa forma, utilizamos estratégias de filtragem. Inicialmente eliminamos os elementos que, evidentemente, não apresentavam influencia sobre a nota registrada pelo cliente. Após esse passo inicial, simplificamos o tipo dos dados para facilitar o processamento do dataframe pelo algoritmo. Após o processo de preparar os dados para o aprendizado registramos os seguintes valores:**\n\n**Considerando todos as notas (Ótimo, bom, mediano, ruim, péssimo) :**\n\nCliente:10.4%\n\nCEP_Cliente:11.0%\n\nPedido:10.4%\n\nid_Pedido:0.8%\n\nProduto:9.4%\n\nVendedor:8.0%\n\nPreco_produto:8.0%\n\nFrete:8.1%\n\nParcela:0.3%\n\nInstallments:3.7%\n\nValor_total:8.6%\n\nID_Review:11.4%\n\nCEP_Vendedor:7.7%\n\nScore:0.8%\n\nTipo_pagamento_boleto:0.8%\n\nTipo_pagamento_credit_card:0.2%\n\nTipo_pagamento_debit_card:0.3%\n\n------------------------------------\n\n**Considerando apenas avaliações com notas instatisfatórias (mediano, ruim, péssimo) :**\n\nCliente:10.0%\n\nCEP_Cliente:11.1%\n\nPedido:10.9%\n\nid_Pedido:0.8%\n\nProduto:9.5%\n\nVendedor:8.3%\n\nPreco_produto:7.4%\n\nFrete:8.3%\n\nParcela:0.2%\n\nInstallments:3.6%\n\nValor_total:8.7%\n\nID_Review:11.6%\n\nCEP_Vendedor:7.6%\n\nScore:0.7%\n\nTipo_pagamento_boleto:0.7%\n\nTipo_pagamento_credit_card:0.2%\n\nTipo_pagamento_debit_card:0.3%\n\n---------------------\n**Portanto, com base nos resultados obtidos, o dono do marketplace deve se atentar aos seguintes parâmetros de sua logística:**\n\n**1 - Relação entre CEP_Cliente, Frete e CEP_Vendedor**\n\n**Esses atributos apresentaram importancia direta na nota registrada pelo cliente, dessa forma, algumas razões devem ser consideradas:**\n\n**-Problemas com a entrega.**\n\n**-Qualidade da entrega(tempo de entrega,comprometimento do produto durante o processo de transporte).**\n\n**-Alto preço do frete para determinadas regiões.**\n\n-----------------------------\n**2 - Produto**\n\n**O atributo produto apresentou importancia direta na insatisfação dos clientes. Portanto, deve-se considerar:**\n\n**-Qualidade ruim de determinados produtos.**\n\n**-O produto entregue não apresentar as caracteristicas do produto anunciado.**\n\n**-Entrega de produtos errados, problema de logística.**\n\n---------------------------------\n\n**3 - Vendedor**\n\n**O atributo Vendedor apresentou importancia direta na insatisfação dos clientes. Portanto, deve-se considerar:**\n\n**-Qualidade duvidosa do atendimento por parte do vendedor.**\n\n**-Erro do vendedor em alguma etapa do processo em específico.**\n\n--------------------------\n\n\n# **CONCLUSÃO**\n\n\n**Portanto, é possível concluir que para o marketplace aumentar seu score, analisar problemas logísticos por parte do processo de transporte, aspectos do produto e os vendedores são os fatores mais importantes para entender o que se passa na empresa e solucionar o problema que está gerando insatisfação dos clientes desse e-commerce.**\n\n\n\n\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
cbcaa5c4d8af7df00df5b80b7ce485a633b2728e
| 20,764 |
ipynb
|
Jupyter Notebook
|
01-introduction-geospatial-data.ipynb
|
darribas/scipy2018-geospatial-data
|
7a294204eb9a2d6c1207da7f899fa98dadc27378
|
[
"BSD-3-Clause"
] | 4 |
2019-08-27T17:11:56.000Z
|
2020-05-06T14:02:10.000Z
|
01-introduction-geospatial-data.ipynb
|
SpatialMac/scipy2018-geospatial-data
|
7a294204eb9a2d6c1207da7f899fa98dadc27378
|
[
"BSD-3-Clause"
] | null | null | null |
01-introduction-geospatial-data.ipynb
|
SpatialMac/scipy2018-geospatial-data
|
7a294204eb9a2d6c1207da7f899fa98dadc27378
|
[
"BSD-3-Clause"
] | 1 |
2020-05-06T08:29:11.000Z
|
2020-05-06T08:29:11.000Z
| 26.826873 | 340 | 0.575082 |
[
[
[
"# Introduction to geospatial vector data in Python",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n\nimport pandas as pd\nimport geopandas\n\npd.options.display.max_rows = 10",
"_____no_output_____"
]
],
[
[
"## Importing geospatial data",
"_____no_output_____"
],
[
"Geospatial data is often available from specific GIS file formats or data stores, like ESRI shapefiles, GeoJSON files, geopackage files, PostGIS (PostgreSQL) database, ...\n\nWe can use the GeoPandas library to read many of those GIS file formats (relying on the `fiona` library under the hood, which is an interface to GDAL/OGR), using the `geopandas.read_file` function.\n\nFor example, let's start by reading a shapefile with all the countries of the world (adapted from http://www.naturalearthdata.com/downloads/110m-cultural-vectors/110m-admin-0-countries/, zip file is available in the `/data` directory), and inspect the data:",
"_____no_output_____"
]
],
[
[
"countries = geopandas.read_file(\"zip://./data/ne_110m_admin_0_countries.zip\")\n# or if the archive is unpacked:\n# countries = geopandas.read_file(\"data/ne_110m_admin_0_countries/ne_110m_admin_0_countries.shp\")",
"_____no_output_____"
],
[
"countries.head()",
"_____no_output_____"
],
[
"countries.plot()",
"_____no_output_____"
]
],
[
[
"What can we observe:\n\n- Using `.head()` we can see the first rows of the dataset, just like we can do with Pandas.\n- There is a 'geometry' column and the different countries are represented as polygons\n- We can use the `.plot()` method to quickly get a *basic* visualization of the data",
"_____no_output_____"
],
[
"## What's a GeoDataFrame?\n\nWe used the GeoPandas library to read in the geospatial data, and this returned us a `GeoDataFrame`:",
"_____no_output_____"
]
],
[
[
"type(countries)",
"_____no_output_____"
]
],
[
[
"A GeoDataFrame contains a tabular, geospatial dataset:\n\n* It has a **'geometry' column** that holds the geometry information (or features in GeoJSON).\n* The other columns are the **attributes** (or properties in GeoJSON) that describe each of the geometries\n\nSuch a `GeoDataFrame` is just like a pandas `DataFrame`, but with some additional functionality for working with geospatial data:\n\n* A `.geometry` attribute that always returns the column with the geometry information (returning a GeoSeries). The column name itself does not necessarily need to be 'geometry', but it will always be accessible as the `.geometry` attribute.\n* It has some extra methods for working with spatial data (area, distance, buffer, intersection, ...), which we will see in later notebooks",
"_____no_output_____"
]
],
[
[
"countries.geometry",
"_____no_output_____"
],
[
"type(countries.geometry)",
"_____no_output_____"
],
[
"countries.geometry.area",
"_____no_output_____"
]
],
[
[
"**It's still a DataFrame**, so we have all the pandas functionality available to use on the geospatial dataset, and to do data manipulations with the attributes and geometry information together.\n\nFor example, we can calculate average population number over all countries (by accessing the 'pop_est' column, and calling the `mean` method on it):",
"_____no_output_____"
]
],
[
[
"countries['pop_est'].mean()",
"_____no_output_____"
]
],
[
[
"Or, we can use boolean filtering to select a subset of the dataframe based on a condition:",
"_____no_output_____"
]
],
[
[
"africa = countries[countries['continent'] == 'Africa']",
"_____no_output_____"
],
[
"africa.plot()",
"_____no_output_____"
]
],
[
[
"---\n\n**Exercise**: create a plot of South America\n",
"_____no_output_____"
],
[
"<!--\ncountries[countries['continent'] == 'South America'].plot()\n-->",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
]
],
[
[
"countries.head()",
"_____no_output_____"
]
],
[
[
"---\n\nThe rest of the tutorial is going to assume you already know some pandas basics, but we will try to give hints for that part for those that are not familiar. \nA few resources in case you want to learn more about pandas:\n\n- Pandas docs: https://pandas.pydata.org/pandas-docs/stable/10min.html\n- Other tutorials: chapter from pandas in https://jakevdp.github.io/PythonDataScienceHandbook/, https://github.com/jorisvandenbossche/pandas-tutorial, https://github.com/TomAugspurger/pandas-head-to-tail, ...",
"_____no_output_____"
],
[
"<div class=\"alert alert-info\" style=\"font-size:120%\">\n<b>REMEMBER</b>: <br>\n\n<ul>\n <li>A `GeoDataFrame` allows to perform typical tabular data analysis together with spatial operations</li>\n <li>A `GeoDataFrame` (or *Feature Collection*) consists of:\n <ul>\n <li>**Geometries** or **features**: the spatial objects</li>\n <li>**Attributes** or **properties**: columns with information about each spatial object</li>\n </ul>\n </li>\n</ul>\n</div>",
"_____no_output_____"
],
[
"## Geometries: Points, Linestrings and Polygons\n\nSpatial **vector** data can consist of different types, and the 3 fundamental types are:\n\n* **Point** data: represents a single point in space.\n* **Line** data (\"LineString\"): represents a sequence of points that form a line.\n* **Polygon** data: represents a filled area.\n\nAnd each of them can also be combined in multi-part geometries (See https://shapely.readthedocs.io/en/stable/manual.html#geometric-objects for extensive overview).",
"_____no_output_____"
],
[
"For the example we have seen up to now, the individual geometry objects are Polygons:",
"_____no_output_____"
]
],
[
[
"print(countries.geometry[2])",
"_____no_output_____"
]
],
[
[
"Let's import some other datasets with different types of geometry objects.\n\nA dateset about cities in the world (adapted from http://www.naturalearthdata.com/downloads/110m-cultural-vectors/110m-populated-places/, zip file is available in the `/data` directory), consisting of Point data:",
"_____no_output_____"
]
],
[
[
"cities = geopandas.read_file(\"zip://./data/ne_110m_populated_places.zip\")",
"_____no_output_____"
],
[
"print(cities.geometry[0])",
"_____no_output_____"
]
],
[
[
"And a dataset of rivers in the world (from http://www.naturalearthdata.com/downloads/50m-physical-vectors/50m-rivers-lake-centerlines/, zip file is available in the `/data` directory) where each river is a (multi-)line:",
"_____no_output_____"
]
],
[
[
"rivers = geopandas.read_file(\"zip://./data/ne_50m_rivers_lake_centerlines.zip\")",
"_____no_output_____"
],
[
"print(rivers.geometry[0])",
"_____no_output_____"
]
],
[
[
"### The `shapely` library\n\nThe individual geometry objects are provided by the [`shapely`](https://shapely.readthedocs.io/en/stable/) library",
"_____no_output_____"
]
],
[
[
"type(countries.geometry[0])",
"_____no_output_____"
]
],
[
[
"To construct one ourselves:",
"_____no_output_____"
]
],
[
[
"from shapely.geometry import Point, Polygon, LineString",
"_____no_output_____"
],
[
"p = Point(1, 1)",
"_____no_output_____"
],
[
"print(p)",
"_____no_output_____"
],
[
"polygon = Polygon([(1, 1), (2,2), (2, 1)])",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-info\" style=\"font-size:120%\">\n<b>REMEMBER</b>: <br><br>\n\nSingle geometries are represented by `shapely` objects:\n\n<ul>\n <li>If you access a single geometry of a GeoDataFrame, you get a shapely geometry object</li>\n <li>Those objects have similar functionality as geopandas objects (GeoDataFrame/GeoSeries). For example:\n <ul>\n <li>`single_shapely_object.distance(other_point)` -> distance between two points</li>\n <li>`geodataframe.distance(other_point)` -> distance for each point in the geodataframe to the other point</li>\n </ul>\n </li>\n</ul>\n</div>",
"_____no_output_____"
],
[
"## Coordinate reference systems\n\nA **coordinate reference system (CRS)** determines how the two-dimensional (planar) coordinates of the geometry objects should be related to actual places on the (non-planar) earth.\n\nFor a nice in-depth explanation, see https://docs.qgis.org/2.8/en/docs/gentle_gis_introduction/coordinate_reference_systems.html",
"_____no_output_____"
],
[
"A GeoDataFrame or GeoSeries has a `.crs` attribute which holds (optionally) a description of the coordinate reference system of the geometries:",
"_____no_output_____"
]
],
[
[
"countries.crs",
"_____no_output_____"
]
],
[
[
"For the `countries` dataframe, it indicates that it used the EPSG 4326 / WGS84 lon/lat reference system, which is one of the most used. \nIt uses coordinates as latitude and longitude in degrees, as can you be seen from the x/y labels on the plot:",
"_____no_output_____"
]
],
[
[
"countries.plot()",
"_____no_output_____"
]
],
[
[
"The `.crs` attribute is given as a dictionary. In this case, it only indicates the EPSG code, but it can also contain the full \"proj4\" string (in dictionary form). \n\nUnder the hood, GeoPandas uses the `pyproj` / `proj4` libraries to deal with the re-projections.\n\nFor more information, see also http://geopandas.readthedocs.io/en/latest/projections.html.",
"_____no_output_____"
],
[
"---\n\nThere are sometimes good reasons you want to change the coordinate references system of your dataset, for example:\n\n- different sources with different crs -> need to convert to the same crs\n- distance-based operations -> if you a crs that has meter units (not degrees)\n- plotting in a certain crs (eg to preserve area)\n\nWe can convert a GeoDataFrame to another reference system using the `to_crs` function. \n\nFor example, let's convert the countries to the World Mercator projection (http://epsg.io/3395):",
"_____no_output_____"
]
],
[
[
"# remove Antartica, as the Mercator projection cannot deal with the poles\ncountries = countries[(countries['name'] != \"Antarctica\")]",
"_____no_output_____"
],
[
"countries_mercator = countries.to_crs(epsg=3395) # or .to_crs({'init': 'epsg:3395'})",
"_____no_output_____"
],
[
"countries_mercator.plot()",
"_____no_output_____"
]
],
[
[
"Note the different scale of x and y.",
"_____no_output_____"
],
[
"---\n\n**Exercise**: project the countries to [Web Mercator](http://epsg.io/3857), the CRS used by Google Maps, OpenStreetMap and most web providers.\n",
"_____no_output_____"
],
[
"<!--\ncountries.to_crs(epsg=3857)\n-->",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"## Plotting our different layers together",
"_____no_output_____"
]
],
[
[
"ax = countries.plot(edgecolor='k', facecolor='none', figsize=(15, 10))\nrivers.plot(ax=ax)\ncities.plot(ax=ax, color='red')\nax.set(xlim=(-20, 60), ylim=(-40, 40))",
"_____no_output_____"
]
],
[
[
"See the [04-more-on-visualization.ipynb](04-more-on-visualization.ipynb) notebook for more details on visualizing geospatial datasets.",
"_____no_output_____"
],
[
"---\n\n**Exercise**: replicate the figure above by coloring the countries in black and cities in yellow\n",
"_____no_output_____"
],
[
"<!--\nax = countries.plot(edgecolor='w', facecolor='k', figsize=(15, 10))\nrivers.plot(ax=ax)\ncities.plot(ax=ax, color='yellow')\nax.set(xlim=(-20, 60), ylim=(-40, 40))\n-->",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
],
[
"## A bit more on importing and creating GeoDataFrames",
"_____no_output_____"
],
[
"### Note on `fiona`\n\nUnder the hood, GeoPandas uses the [Fiona library](http://toblerity.org/fiona/) (pythonic interface to GDAL/OGR) to read and write data. GeoPandas provides a more user-friendly wrapper, which is sufficient for most use cases. But sometimes you want more control, and in that case, to read a file with fiona you can do the following:\n",
"_____no_output_____"
]
],
[
[
"import fiona\nfrom shapely.geometry import shape\n\nwith fiona.drivers():\n with fiona.open(\"data/ne_110m_admin_0_countries/ne_110m_admin_0_countries.shp\") as collection:\n for feature in collection:\n # ... do something with geometry\n geom = shape(feature['geometry'])\n # ... do something with properties\n print(feature['properties']['name'])",
"_____no_output_____"
]
],
[
[
"### Constructing a GeoDataFrame manually",
"_____no_output_____"
]
],
[
[
"geopandas.GeoDataFrame({\n 'geometry': [Point(1, 1), Point(2, 2)],\n 'attribute1': [1, 2],\n 'attribute2': [0.1, 0.2]})",
"_____no_output_____"
]
],
[
[
"### Creating a GeoDataFrame from an existing dataframe\n\nFor example, if you have lat/lon coordinates in two columns:",
"_____no_output_____"
]
],
[
[
"df = pd.DataFrame(\n {'City': ['Buenos Aires', 'Brasilia', 'Santiago', 'Bogota', 'Caracas'],\n 'Country': ['Argentina', 'Brazil', 'Chile', 'Colombia', 'Venezuela'],\n 'Latitude': [-34.58, -15.78, -33.45, 4.60, 10.48],\n 'Longitude': [-58.66, -47.91, -70.66, -74.08, -66.86]})",
"_____no_output_____"
],
[
"df['Coordinates'] = list(zip(df.Longitude, df.Latitude))",
"_____no_output_____"
],
[
"df['Coordinates'] = df['Coordinates'].apply(Point)",
"_____no_output_____"
],
[
"gdf = geopandas.GeoDataFrame(df, geometry='Coordinates')",
"_____no_output_____"
],
[
"gdf",
"_____no_output_____"
]
],
[
[
"See http://geopandas.readthedocs.io/en/latest/gallery/create_geopandas_from_pandas.html#sphx-glr-gallery-create-geopandas-from-pandas-py for full example",
"_____no_output_____"
],
[
"---\n\n**Exercise**: use [geojson.io](http://geojson.io) to mark five points, and create a `GeoDataFrame` with it. Note that coordinates will be expressed in longitude and latitude, so you'll have to set the CRS accordingly.\n",
"_____no_output_____"
],
[
"<!--\ndf = pd.DataFrame(\n {'Name': ['Hotel', 'Capitol', 'Barton Springs'],\n 'Latitude': [30.28195889019179, 30.274782936992608, 30.263728440902543],\n 'Longitude': [-97.74006128311157, -97.74038314819336, -97.77013421058655]})\ndf['Coordinates'] = list(zip(df.Longitude, df.Latitude))\ndf['Coordinates'] = df['Coordinates'].apply(Point)\ngdf = geopandas.GeoDataFrame(df, geometry='Coordinates', crs={'init': 'epsg:4326'})\n-->",
"_____no_output_____"
],
[
"---",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
cbcaa9369607a9da2ced4b309b6d6f13962a875f
| 1,270 |
ipynb
|
Jupyter Notebook
|
OpenJudge/test.ipynb
|
PKUfudawei/Data-Structure-Algorithm
|
55d6d1764e46197f661163beffd0cec51c1aecd2
|
[
"MIT"
] | null | null | null |
OpenJudge/test.ipynb
|
PKUfudawei/Data-Structure-Algorithm
|
55d6d1764e46197f661163beffd0cec51c1aecd2
|
[
"MIT"
] | null | null | null |
OpenJudge/test.ipynb
|
PKUfudawei/Data-Structure-Algorithm
|
55d6d1764e46197f661163beffd0cec51c1aecd2
|
[
"MIT"
] | null | null | null | 25.918367 | 81 | 0.472441 |
[
[
[
"N, K=map(int, input().split())\nx_max=100000\narrived=[False for i in range(x_max+1)]\narrived[N]=True\ntime=[0 for i in range(x_max+1)]\nstart_mark=0\nend_mark=0\nlocation=[0 for i in range(x_max+1)]\nlocation[0]=N\nSTOP=False\nwhile end_mark<=x_max:\n start=location[start_mark]\n start_mark+=1\n for i in range(3):\n if i==0: next_loc=start+1\n if i==1: next_loc=start-1\n if i==2: next_loc=start*2\n if next_loc>=0 and next_loc<=x_max and arrived[next_loc]==False:\n end_mark+=1\n location[end_mark]=next_loc\n arrived[next_loc]=True\n time[next_loc]=time[start]+1\n if next_loc==K:\n print(time[next_loc])\n STOP=True\n if STOP:\n break\n ",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbcabe13544047f2e6727b9c67c0b60833a35694
| 246,375 |
ipynb
|
Jupyter Notebook
|
experiments/cnn_2/wisig/trials/12/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null |
experiments/cnn_2/wisig/trials/12/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null |
experiments/cnn_2/wisig/trials/12/trial.ipynb
|
stevester94/csc500-notebooks
|
4c1b04c537fe233a75bed82913d9d84985a89177
|
[
"MIT"
] | null | null | null | 104.885057 | 75,040 | 0.768008 |
[
[
[
"import os, json, sys, time, random\nimport numpy as np\nimport torch\nfrom easydict import EasyDict\nfrom math import floor\nfrom easydict import EasyDict\n\nfrom steves_utils.vanilla_train_eval_test_jig import Vanilla_Train_Eval_Test_Jig\n\nfrom steves_utils.torch_utils import get_dataset_metrics, independent_accuracy_assesment\nfrom steves_models.configurable_vanilla import Configurable_Vanilla\nfrom steves_utils.torch_sequential_builder import build_sequential\nfrom steves_utils.lazy_map import Lazy_Map\nfrom steves_utils.sequence_aggregator import Sequence_Aggregator\n\nfrom steves_utils.stratified_dataset.traditional_accessor import Traditional_Accessor_Factory\n\nfrom steves_utils.cnn_do_report import (\n get_loss_curve,\n get_results_table,\n get_parameters_table,\n get_domain_accuracies,\n)\n\nfrom steves_utils.torch_utils import (\n confusion_by_domain_over_dataloader,\n independent_accuracy_assesment\n)\n\nfrom steves_utils.utils_v2 import (\n per_domain_accuracy_from_confusion,\n get_datasets_base_path\n)\n\n# from steves_utils.ptn_do_report import TBD",
"_____no_output_____"
],
[
"required_parameters = {\n \"experiment_name\",\n \"lr\",\n \"device\",\n \"dataset_seed\",\n \"seed\",\n \"labels\",\n \"domains_target\",\n \"domains_source\",\n \"num_examples_per_domain_per_label_source\",\n \"num_examples_per_domain_per_label_target\",\n \"batch_size\",\n \"n_epoch\",\n \"patience\",\n \"criteria_for_best\",\n \"normalize_source\",\n \"normalize_target\",\n \"x_net\",\n \"NUM_LOGS_PER_EPOCH\",\n \"BEST_MODEL_PATH\",\n \"pickle_name_source\",\n \"pickle_name_target\",\n \"torch_default_dtype\",\n}",
"_____no_output_____"
],
[
"from steves_utils.ORACLE.utils_v2 import (\n ALL_SERIAL_NUMBERS,\n ALL_DISTANCES_FEET_NARROWED,\n)\n\nstandalone_parameters = {}\nstandalone_parameters[\"experiment_name\"] = \"MANUAL CORES CNN\"\nstandalone_parameters[\"lr\"] = 0.0001\nstandalone_parameters[\"device\"] = \"cuda\"\n\nstandalone_parameters[\"dataset_seed\"] = 1337\nstandalone_parameters[\"seed\"] = 1337\nstandalone_parameters[\"labels\"] = ALL_SERIAL_NUMBERS\n\nstandalone_parameters[\"domains_source\"] = [8,32,50]\nstandalone_parameters[\"domains_target\"] = [14,20,26,38,44,]\n\nstandalone_parameters[\"num_examples_per_domain_per_label_source\"]=-1\nstandalone_parameters[\"num_examples_per_domain_per_label_target\"]=-1\n\nstandalone_parameters[\"pickle_name_source\"] = \"oracle.Run1_framed_2000Examples_stratified_ds.2022A.pkl\"\nstandalone_parameters[\"pickle_name_target\"] = \"oracle.Run2_framed_2000Examples_stratified_ds.2022A.pkl\"\n\nstandalone_parameters[\"torch_default_dtype\"] = \"torch.float32\" \n\nstandalone_parameters[\"batch_size\"]=128\n\nstandalone_parameters[\"n_epoch\"] = 3\n\nstandalone_parameters[\"patience\"] = 10\n\nstandalone_parameters[\"criteria_for_best\"] = \"target_accuracy\"\nstandalone_parameters[\"normalize_source\"] = False\nstandalone_parameters[\"normalize_target\"] = False\n\nstandalone_parameters[\"x_net\"] = [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\":[-1, 1, 2, 256]}},\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":1, \"out_channels\":256, \"kernel_size\":(1,7), \"bias\":False, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Conv2d\", \"kargs\": { \"in_channels\":256, \"out_channels\":80, \"kernel_size\":(2,7), \"bias\":True, \"padding\":(0,3), },},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\":80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 80*256, \"out_features\": 256}}, # 80 units per IQ pair\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\":256}},\n\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": len(standalone_parameters[\"labels\"])}},\n]\n\nstandalone_parameters[\"NUM_LOGS_PER_EPOCH\"] = 10\nstandalone_parameters[\"BEST_MODEL_PATH\"] = \"./best_model.pth\"",
"_____no_output_____"
],
[
"# Parameters\nparameters = {\n \"experiment_name\": \"cnn_2:wisig\",\n \"labels\": [\n \"1-10\",\n \"1-12\",\n \"1-14\",\n \"1-16\",\n \"1-18\",\n \"1-19\",\n \"1-8\",\n \"10-11\",\n \"10-17\",\n \"10-4\",\n \"10-7\",\n \"11-1\",\n \"11-10\",\n \"11-19\",\n \"11-20\",\n \"11-4\",\n \"11-7\",\n \"12-19\",\n \"12-20\",\n \"12-7\",\n \"13-14\",\n \"13-18\",\n \"13-19\",\n \"13-20\",\n \"13-3\",\n \"13-7\",\n \"14-10\",\n \"14-11\",\n \"14-12\",\n \"14-13\",\n \"14-14\",\n \"14-19\",\n \"14-20\",\n \"14-7\",\n \"14-8\",\n \"14-9\",\n \"15-1\",\n \"15-19\",\n \"15-6\",\n \"16-1\",\n \"16-16\",\n \"16-19\",\n \"16-20\",\n \"17-10\",\n \"17-11\",\n \"18-1\",\n \"18-10\",\n \"18-11\",\n \"18-12\",\n \"18-13\",\n \"18-14\",\n \"18-15\",\n \"18-16\",\n \"18-17\",\n \"18-19\",\n \"18-2\",\n \"18-20\",\n \"18-4\",\n \"18-5\",\n \"18-7\",\n \"18-8\",\n \"18-9\",\n \"19-1\",\n \"19-10\",\n \"19-11\",\n \"19-12\",\n \"19-13\",\n \"19-14\",\n \"19-15\",\n \"19-19\",\n \"19-2\",\n \"19-20\",\n \"19-3\",\n \"19-4\",\n \"19-6\",\n \"19-7\",\n \"19-8\",\n \"19-9\",\n \"2-1\",\n \"2-13\",\n \"2-15\",\n \"2-3\",\n \"2-4\",\n \"2-5\",\n \"2-6\",\n \"2-7\",\n \"2-8\",\n \"20-1\",\n \"20-12\",\n \"20-14\",\n \"20-15\",\n \"20-16\",\n \"20-18\",\n \"20-19\",\n \"20-20\",\n \"20-3\",\n \"20-4\",\n \"20-5\",\n \"20-7\",\n \"20-8\",\n \"3-1\",\n \"3-13\",\n \"3-18\",\n \"3-2\",\n \"3-8\",\n \"4-1\",\n \"4-10\",\n \"4-11\",\n \"5-1\",\n \"5-5\",\n \"6-1\",\n \"6-15\",\n \"6-6\",\n \"7-10\",\n \"7-11\",\n \"7-12\",\n \"7-13\",\n \"7-14\",\n \"7-7\",\n \"7-8\",\n \"7-9\",\n \"8-1\",\n \"8-13\",\n \"8-14\",\n \"8-18\",\n \"8-20\",\n \"8-3\",\n \"8-8\",\n \"9-1\",\n \"9-7\",\n ],\n \"domains_source\": [1, 2, 3, 4],\n \"domains_target\": [1, 2, 3, 4],\n \"pickle_name_target\": \"wisig.node3-19.stratified_ds.2022A.pkl\",\n \"pickle_name_source\": \"wisig.node3-19.stratified_ds.2022A.pkl\",\n \"device\": \"cuda\",\n \"lr\": 0.0001,\n \"batch_size\": 128,\n \"normalize_source\": False,\n \"normalize_target\": False,\n \"num_examples_per_domain_per_label_source\": -1,\n \"num_examples_per_domain_per_label_target\": -1,\n \"torch_default_dtype\": \"torch.float32\",\n \"n_epoch\": 50,\n \"patience\": 3,\n \"criteria_for_best\": \"target_accuracy\",\n \"x_net\": [\n {\"class\": \"nnReshape\", \"kargs\": {\"shape\": [-1, 1, 2, 256]}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 1,\n \"out_channels\": 256,\n \"kernel_size\": [1, 7],\n \"bias\": False,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 256}},\n {\n \"class\": \"Conv2d\",\n \"kargs\": {\n \"in_channels\": 256,\n \"out_channels\": 80,\n \"kernel_size\": [2, 7],\n \"bias\": True,\n \"padding\": [0, 3],\n },\n },\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm2d\", \"kargs\": {\"num_features\": 80}},\n {\"class\": \"Flatten\", \"kargs\": {}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 20480, \"out_features\": 256}},\n {\"class\": \"ReLU\", \"kargs\": {\"inplace\": True}},\n {\"class\": \"BatchNorm1d\", \"kargs\": {\"num_features\": 256}},\n {\"class\": \"Linear\", \"kargs\": {\"in_features\": 256, \"out_features\": 130}},\n ],\n \"NUM_LOGS_PER_EPOCH\": 10,\n \"BEST_MODEL_PATH\": \"./best_model.pth\",\n \"dataset_seed\": 7,\n \"seed\": 7,\n}\n",
"_____no_output_____"
],
[
"# Set this to True if you want to run this template directly\nSTANDALONE = False\nif STANDALONE:\n print(\"parameters not injected, running with standalone_parameters\")\n parameters = standalone_parameters\n\nif not 'parameters' in locals() and not 'parameters' in globals():\n raise Exception(\"Parameter injection failed\")\n\n#Use an easy dict for all the parameters\np = EasyDict(parameters)\n\nsupplied_keys = set(p.keys())\n\nif supplied_keys != required_parameters:\n print(\"Parameters are incorrect\")\n if len(supplied_keys - required_parameters)>0: print(\"Shouldn't have:\", str(supplied_keys - required_parameters))\n if len(required_parameters - supplied_keys)>0: print(\"Need to have:\", str(required_parameters - supplied_keys))\n raise RuntimeError(\"Parameters are incorrect\")\n\n",
"_____no_output_____"
],
[
"###################################\n# Set the RNGs and make it all deterministic\n###################################\nnp.random.seed(p.seed)\nrandom.seed(p.seed)\ntorch.manual_seed(p.seed)\n\ntorch.use_deterministic_algorithms(True) ",
"_____no_output_____"
],
[
"torch.set_default_dtype(eval(p.torch_default_dtype))",
"_____no_output_____"
],
[
"###################################\n# Build the network(s)\n# Note: It's critical to do this AFTER setting the RNG\n###################################\nx_net = build_sequential(p.x_net)",
"_____no_output_____"
],
[
"start_time_secs = time.time()",
"_____no_output_____"
],
[
"def wrap_in_dataloader(p, ds):\n return torch.utils.data.DataLoader(\n ds,\n batch_size=p.batch_size,\n shuffle=True,\n num_workers=1,\n persistent_workers=True,\n prefetch_factor=50,\n pin_memory=True\n )\n\ntaf_source = Traditional_Accessor_Factory(\n labels=p.labels,\n domains=p.domains_source,\n num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_source,\n pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name_source),\n seed=p.dataset_seed\n)\ntrain_original_source, val_original_source, test_original_source = \\\n taf_source.get_train(), taf_source.get_val(), taf_source.get_test()\n\n\ntaf_target = Traditional_Accessor_Factory(\n labels=p.labels,\n domains=p.domains_target,\n num_examples_per_domain_per_label=p.num_examples_per_domain_per_label_source,\n pickle_path=os.path.join(get_datasets_base_path(), p.pickle_name_target),\n seed=p.dataset_seed\n)\ntrain_original_target, val_original_target, test_original_target = \\\n taf_target.get_train(), taf_target.get_val(), taf_target.get_test()\n\n\n# For CNN We only use X and Y. And we only train on the source.\n# Properly form the data using a transform lambda and Lazy_Map. Finally wrap them in a dataloader\n\ntransform_lambda = lambda ex: ex[:2] # Strip the tuple to just (x,y)\n\n\ntrain_processed_source = wrap_in_dataloader(\n p,\n Lazy_Map(train_original_source, transform_lambda)\n)\nval_processed_source = wrap_in_dataloader(\n p,\n Lazy_Map(val_original_source, transform_lambda)\n)\ntest_processed_source = wrap_in_dataloader(\n p,\n Lazy_Map(test_original_source, transform_lambda)\n)\n\ntrain_processed_target = wrap_in_dataloader(\n p,\n Lazy_Map(train_original_target, transform_lambda)\n)\nval_processed_target = wrap_in_dataloader(\n p,\n Lazy_Map(val_original_target, transform_lambda)\n)\ntest_processed_target = wrap_in_dataloader(\n p,\n Lazy_Map(test_original_target, transform_lambda)\n)\n\n\n\ndatasets = EasyDict({\n \"source\": {\n \"original\": {\"train\":train_original_source, \"val\":val_original_source, \"test\":test_original_source},\n \"processed\": {\"train\":train_processed_source, \"val\":val_processed_source, \"test\":test_processed_source}\n },\n \"target\": {\n \"original\": {\"train\":train_original_target, \"val\":val_original_target, \"test\":test_original_target},\n \"processed\": {\"train\":train_processed_target, \"val\":val_processed_target, \"test\":test_processed_target}\n },\n})",
"_____no_output_____"
],
[
"ep = next(iter(test_processed_target))\nep[0].dtype",
"_____no_output_____"
],
[
"model = Configurable_Vanilla(\n x_net=x_net,\n label_loss_object=torch.nn.NLLLoss(),\n learning_rate=p.lr\n)",
"_____no_output_____"
],
[
"jig = Vanilla_Train_Eval_Test_Jig(\n model=model,\n path_to_best_model=p.BEST_MODEL_PATH,\n device=p.device,\n label_loss_object=torch.nn.NLLLoss(),\n)\n\njig.train(\n train_iterable=datasets.source.processed.train,\n source_val_iterable=datasets.source.processed.val,\n target_val_iterable=datasets.target.processed.val,\n patience=p.patience,\n num_epochs=p.n_epoch,\n num_logs_per_epoch=p.NUM_LOGS_PER_EPOCH,\n criteria_for_best=p.criteria_for_best\n)",
"epoch: 1, [batch: 1 / 1479], examples_per_second: 739.2661, train_label_loss: 4.8518, \n"
],
[
"total_experiment_time_secs = time.time() - start_time_secs",
"_____no_output_____"
],
[
"source_test_label_accuracy, source_test_label_loss = jig.test(datasets.source.processed.test)\ntarget_test_label_accuracy, target_test_label_loss = jig.test(datasets.target.processed.test)\n\nsource_val_label_accuracy, source_val_label_loss = jig.test(datasets.source.processed.val)\ntarget_val_label_accuracy, target_val_label_loss = jig.test(datasets.target.processed.val)\n\nhistory = jig.get_history()\n\ntotal_epochs_trained = len(history[\"epoch_indices\"])\n\nval_dl = wrap_in_dataloader(p, Sequence_Aggregator((datasets.source.original.val, datasets.target.original.val)))\n\nconfusion = confusion_by_domain_over_dataloader(model, p.device, val_dl, forward_uses_domain=False)\nper_domain_accuracy = per_domain_accuracy_from_confusion(confusion)\n\n# Add a key to per_domain_accuracy for if it was a source domain\nfor domain, accuracy in per_domain_accuracy.items():\n per_domain_accuracy[domain] = {\n \"accuracy\": accuracy,\n \"source?\": domain in p.domains_source\n }\n\n# Do an independent accuracy assesment JUST TO BE SURE!\n# _source_test_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.test, p.device)\n# _target_test_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.test, p.device)\n# _source_val_label_accuracy = independent_accuracy_assesment(model, datasets.source.processed.val, p.device)\n# _target_val_label_accuracy = independent_accuracy_assesment(model, datasets.target.processed.val, p.device)\n\n# assert(_source_test_label_accuracy == source_test_label_accuracy)\n# assert(_target_test_label_accuracy == target_test_label_accuracy)\n# assert(_source_val_label_accuracy == source_val_label_accuracy)\n# assert(_target_val_label_accuracy == target_val_label_accuracy)\n\n###################################\n# Write out the results\n###################################\n\nexperiment = {\n \"experiment_name\": p.experiment_name,\n \"parameters\": p,\n \"results\": {\n \"source_test_label_accuracy\": source_test_label_accuracy,\n \"source_test_label_loss\": source_test_label_loss,\n \"target_test_label_accuracy\": target_test_label_accuracy,\n \"target_test_label_loss\": target_test_label_loss,\n \"source_val_label_accuracy\": source_val_label_accuracy,\n \"source_val_label_loss\": source_val_label_loss,\n \"target_val_label_accuracy\": target_val_label_accuracy,\n \"target_val_label_loss\": target_val_label_loss,\n \"total_epochs_trained\": total_epochs_trained,\n \"total_experiment_time_secs\": total_experiment_time_secs,\n \"confusion\": confusion,\n \"per_domain_accuracy\": per_domain_accuracy,\n },\n \"history\": history,\n \"dataset_metrics\": get_dataset_metrics(datasets, \"cnn\"),\n}",
"_____no_output_____"
],
[
"get_loss_curve(experiment)",
"_____no_output_____"
],
[
"get_results_table(experiment)",
"_____no_output_____"
],
[
"get_domain_accuracies(experiment)",
"_____no_output_____"
],
[
"print(\"Source Test Label Accuracy:\", experiment[\"results\"][\"source_test_label_accuracy\"], \"Target Test Label Accuracy:\", experiment[\"results\"][\"target_test_label_accuracy\"])\nprint(\"Source Val Label Accuracy:\", experiment[\"results\"][\"source_val_label_accuracy\"], \"Target Val Label Accuracy:\", experiment[\"results\"][\"target_val_label_accuracy\"])",
"Source Test Label Accuracy: 0.9493803413600838 Target Test Label Accuracy: 0.9493803413600838\nSource Val Label Accuracy: 0.9506888690653187 Target Val Label Accuracy: 0.9506888690653187\n"
],
[
"json.dumps(experiment)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
cbcac4150b0ca3f957679e0b47c295bd6629bd81
| 81,335 |
ipynb
|
Jupyter Notebook
|
docs_src/vision.learner.ipynb
|
ksasi/fastai
|
18f6f439317b92f0aa8e4ba1a4044a43172a8c2c
|
[
"Apache-2.0"
] | 1 |
2019-06-08T22:44:43.000Z
|
2019-06-08T22:44:43.000Z
|
docs_src/vision.learner.ipynb
|
eisenjulian/fastai
|
202a81c511864e5b98acaafb3ea58a3e6ef500c0
|
[
"Apache-2.0"
] | null | null | null |
docs_src/vision.learner.ipynb
|
eisenjulian/fastai
|
202a81c511864e5b98acaafb3ea58a3e6ef500c0
|
[
"Apache-2.0"
] | null | null | null | 86.711087 | 21,504 | 0.776062 |
[
[
[
"## Computer Vision Learner",
"_____no_output_____"
],
[
"[`vision.learner`](/vision.learner.html#vision.learner) is the module that defines the [`cnn_learner`](/vision.learner.html#cnn_learner) method, to easily get a model suitable for transfer learning.",
"_____no_output_____"
]
],
[
[
"from fastai.gen_doc.nbdoc import *\nfrom fastai.vision import *\n",
"_____no_output_____"
]
],
[
[
"## Transfer learning",
"_____no_output_____"
],
[
"Transfer learning is a technique where you use a model trained on a very large dataset (usually [ImageNet](http://image-net.org/) in computer vision) and then adapt it to your own dataset. The idea is that it has learned to recognize many features on all of this data, and that you will benefit from this knowledge, especially if your dataset is small, compared to starting from a randomly initialized model. It has been proved in [this article](https://arxiv.org/abs/1805.08974) on a wide range of tasks that transfer learning nearly always give better results.\n\nIn practice, you need to change the last part of your model to be adapted to your own number of classes. Most convolutional models end with a few linear layers (a part will call head). The last convolutional layer will have analyzed features in the image that went through the model, and the job of the head is to convert those in predictions for each of our classes. In transfer learning we will keep all the convolutional layers (called the body or the backbone of the model) with their weights pretrained on ImageNet but will define a new head initialized randomly.\n\nThen we will train the model we obtain in two phases: first we freeze the body weights and only train the head (to convert those analyzed features into predictions for our own data), then we unfreeze the layers of the backbone (gradually if necessary) and fine-tune the whole model (possibly using differential learning rates).\n\nThe [`cnn_learner`](/vision.learner.html#cnn_learner) factory method helps you to automatically get a pretrained model from a given architecture with a custom head that is suitable for your data.",
"_____no_output_____"
]
],
[
[
"show_doc(cnn_learner)",
"_____no_output_____"
]
],
[
[
"This method creates a [`Learner`](/basic_train.html#Learner) object from the [`data`](/vision.data.html#vision.data) object and model inferred from it with the backbone given in `arch`. Specifically, it will cut the model defined by `arch` (randomly initialized if `pretrained` is False) at the last convolutional layer by default (or as defined in `cut`, see below) and add:\n- an [`AdaptiveConcatPool2d`](/layers.html#AdaptiveConcatPool2d) layer,\n- a [`Flatten`](/layers.html#Flatten) layer,\n- blocks of \\[[`nn.BatchNorm1d`](https://pytorch.org/docs/stable/nn.html#torch.nn.BatchNorm1d), [`nn.Dropout`](https://pytorch.org/docs/stable/nn.html#torch.nn.Dropout), [`nn.Linear`](https://pytorch.org/docs/stable/nn.html#torch.nn.Linear), [`nn.ReLU`](https://pytorch.org/docs/stable/nn.html#torch.nn.ReLU)\\] layers.\n\nThe blocks are defined by the `lin_ftrs` and `ps` arguments. Specifically, the first block will have a number of inputs inferred from the backbone `arch` and the last one will have a number of outputs equal to `data.c` (which contains the number of classes of the data) and the intermediate blocks have a number of inputs/outputs determined by `lin_frts` (of course a block has a number of inputs equal to the number of outputs of the previous block). The default is to have an intermediate hidden size of 512 (which makes two blocks `model_activation` -> 512 -> `n_classes`). If you pass a float then the final dropout layer will have the value `ps`, and the remaining will be `ps/2`. If you pass a list then the values are used for dropout probabilities directly.\n\nNote that the very last block doesn't have a [`nn.ReLU`](https://pytorch.org/docs/stable/nn.html#torch.nn.ReLU) activation, to allow you to use any final activation you want (generally included in the loss function in pytorch). Also, the backbone will be frozen if you choose `pretrained=True` (so only the head will train if you call [`fit`](/basic_train.html#fit)) so that you can immediately start phase one of training as described above.\n\nAlternatively, you can define your own `custom_head` to put on top of the backbone. If you want to specify where to split `arch` you should so in the argument `cut` which can either be the index of a specific layer (the result will not include that layer) or a function that, when passed the model, will return the backbone you want.\n\nThe final model obtained by stacking the backbone and the head (custom or defined as we saw) is then separated in groups for gradual unfreezing or differential learning rates. You can specify how to split the backbone in groups with the optional argument `split_on` (should be a function that returns those groups when given the backbone). \n\nThe `kwargs` will be passed on to [`Learner`](/basic_train.html#Learner), so you can put here anything that [`Learner`](/basic_train.html#Learner) will accept ([`metrics`](/metrics.html#metrics), `loss_func`, `opt_func`...)",
"_____no_output_____"
]
],
[
[
"path = untar_data(URLs.MNIST_SAMPLE)\ndata = ImageDataBunch.from_folder(path)",
"_____no_output_____"
],
[
"learner = cnn_learner(data, models.resnet18, metrics=[accuracy])\nlearner.fit_one_cycle(1,1e-3)",
"_____no_output_____"
],
[
"learner.save('one_epoch')",
"_____no_output_____"
],
[
"show_doc(unet_learner)",
"_____no_output_____"
]
],
[
[
"This time the model will be a [`DynamicUnet`](/vision.models.unet.html#DynamicUnet) with an encoder based on `arch` (maybe `pretrained`) that is cut depending on `split_on`. `blur_final`, `norm_type`, `blur`, `self_attention`, `y_range`, `last_cross` and `bottle` are passed to unet constructor, the `kwargs` are passed to the initialization of the [`Learner`](/basic_train.html#Learner).",
"_____no_output_____"
]
],
[
[
"jekyll_warn(\"The models created with this function won't work with pytorch `nn.DataParallel`, you have to use distributed training instead!\")",
"_____no_output_____"
]
],
[
[
"### Get predictions",
"_____no_output_____"
],
[
"Once you've actually trained your model, you may want to use it on a single image. This is done by using the following method.",
"_____no_output_____"
]
],
[
[
"show_doc(Learner.predict)",
"_____no_output_____"
],
[
"img = learner.data.train_ds[0][0]\nlearner.predict(img)",
"_____no_output_____"
]
],
[
[
"Here the predict class for our image is '3', which corresponds to a label of 0. The probabilities the model found for each class are 99.65% and 0.35% respectively, so its confidence is pretty high.\n\nNote that if you want to load your trained model and use it on inference mode with the previous function, you should export your [`Learner`](/basic_train.html#Learner).",
"_____no_output_____"
]
],
[
[
"learner.export()",
"_____no_output_____"
]
],
[
[
"And then you can load it with an empty data object that has the same internal state like this:",
"_____no_output_____"
]
],
[
[
"learn = load_learner(path)",
"_____no_output_____"
]
],
[
[
"### Customize your model",
"_____no_output_____"
],
[
"You can customize [`cnn_learner`](/vision.learner.html#cnn_learner) for your own model's default `cut` and `split_on` functions by adding them to the dictionary `model_meta`. The key should be your model and the value should be a dictionary with the keys `cut` and `split_on` (see the source code for examples). The constructor will call [`create_body`](/vision.learner.html#create_body) and [`create_head`](/vision.learner.html#create_head) for you based on `cut`; you can also call them yourself, which is particularly useful for testing.",
"_____no_output_____"
]
],
[
[
"show_doc(create_body)",
"_____no_output_____"
],
[
"show_doc(create_head, doc_string=False)",
"_____no_output_____"
]
],
[
[
"Model head that takes `nf` features, runs through `lin_ftrs`, and ends with `nc` classes. `ps` is the probability of the dropouts, as documented above in [`cnn_learner`](/vision.learner.html#cnn_learner).",
"_____no_output_____"
]
],
[
[
"show_doc(ClassificationInterpretation, title_level=3)",
"_____no_output_____"
]
],
[
[
"This provides a confusion matrix and visualization of the most incorrect images. Pass in your [`data`](/vision.data.html#vision.data), calculated `preds`, actual `y`, and your `losses`, and then use the methods below to view the model interpretation results. For instance:",
"_____no_output_____"
]
],
[
[
"learn = cnn_learner(data, models.resnet18)\nlearn.fit(1)\npreds,y,losses = learn.get_preds(with_loss=True)\ninterp = ClassificationInterpretation(learn, preds, y, losses)",
"_____no_output_____"
]
],
[
[
"The following factory method gives a more convenient way to create an instance of this class:",
"_____no_output_____"
]
],
[
[
"show_doc(ClassificationInterpretation.from_learner, full_name='from_learner')",
"_____no_output_____"
]
],
[
[
"You can also use a shortcut `learn.interpret()` to do the same.",
"_____no_output_____"
]
],
[
[
"show_doc(Learner.interpret, full_name='interpret')",
"_____no_output_____"
]
],
[
[
"Note that this shortcut is a [`Learner`](/basic_train.html#Learner) object/class method that can be called as: `learn.interpret()`.",
"_____no_output_____"
]
],
[
[
"show_doc(ClassificationInterpretation.plot_top_losses, full_name='plot_top_losses')",
"_____no_output_____"
]
],
[
[
"The `k` items are arranged as a square, so it will look best if `k` is a square number (4, 9, 16, etc). The title of each image shows: prediction, actual, loss, probability of actual class. When `heatmap` is True (by default it's True) , Grad-CAM heatmaps (http://openaccess.thecvf.com/content_ICCV_2017/papers/Selvaraju_Grad-CAM_Visual_Explanations_ICCV_2017_paper.pdf) are overlaid on each image. `plot_top_losses` should be used with single-labeled datasets. See `plot_multi_top_losses` below for a version capable of handling multi-labeled datasets.",
"_____no_output_____"
]
],
[
[
"interp.plot_top_losses(9, figsize=(7,7))",
"_____no_output_____"
],
[
"show_doc(ClassificationInterpretation.top_losses)",
"_____no_output_____"
]
],
[
[
"Returns tuple of *(losses,indices)*.",
"_____no_output_____"
]
],
[
[
"interp.top_losses(9)",
"_____no_output_____"
],
[
"show_doc(ClassificationInterpretation.plot_multi_top_losses, full_name='plot_multi_top_losses')",
"_____no_output_____"
]
],
[
[
"Similar to `plot_top_losses()` but aimed at multi-labeled datasets. It plots misclassified samples sorted by their respective loss. \nSince you can have multiple labels for a single sample, they can easily overlap in a grid plot. So it plots just one sample per row. \nNote that you can pass `save_misclassified=True` (by default it's `False`). In such case, the method will return a list containing the misclassified images which you can use to debug your model and/or tune its hyperparameters. ",
"_____no_output_____"
]
],
[
[
"show_doc(ClassificationInterpretation.plot_confusion_matrix)",
"_____no_output_____"
]
],
[
[
"If [`normalize`](/vision.data.html#normalize), plots the percentages with `norm_dec` digits. `slice_size` can be used to avoid out of memory error if your set is too big. `kwargs` are passed to `plt.figure`.",
"_____no_output_____"
]
],
[
[
"interp.plot_confusion_matrix()",
"_____no_output_____"
],
[
"show_doc(ClassificationInterpretation.confusion_matrix)",
"_____no_output_____"
],
[
"interp.confusion_matrix()",
"_____no_output_____"
],
[
"show_doc(ClassificationInterpretation.most_confused)",
"_____no_output_____"
]
],
[
[
"#### Working with large datasets",
"_____no_output_____"
],
[
"When working with large datasets, memory problems can arise when computing the confusion matrix. For example, an error can look like this:\n\n RuntimeError: $ Torch: not enough memory: you tried to allocate 64GB. Buy new RAM!\n\nIn this case it is possible to force [`ClassificationInterpretation`](/train.html#ClassificationInterpretation) to compute the confusion matrix for data slices and then aggregate the result by specifying slice_size parameter. ",
"_____no_output_____"
]
],
[
[
"interp.confusion_matrix(slice_size=10)",
"_____no_output_____"
],
[
"interp.plot_confusion_matrix(slice_size=10)",
"_____no_output_____"
],
[
"interp.most_confused(slice_size=10)",
"_____no_output_____"
]
],
[
[
"## Undocumented Methods - Methods moved below this line will intentionally be hidden",
"_____no_output_____"
],
[
"## New Methods - Please document or move to the undocumented section",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
cbcacf79e336a57f95cdd6d63d4200614fa30638
| 38,302 |
ipynb
|
Jupyter Notebook
|
02_fundamentos_pandas/notebook/11_formas_de_selecao.ipynb
|
eltonrp/formacao_Data_Science
|
278c807740d0ba0e8294937181b41076a1697622
|
[
"MIT"
] | null | null | null |
02_fundamentos_pandas/notebook/11_formas_de_selecao.ipynb
|
eltonrp/formacao_Data_Science
|
278c807740d0ba0e8294937181b41076a1697622
|
[
"MIT"
] | null | null | null |
02_fundamentos_pandas/notebook/11_formas_de_selecao.ipynb
|
eltonrp/formacao_Data_Science
|
278c807740d0ba0e8294937181b41076a1697622
|
[
"MIT"
] | null | null | null | 24.33418 | 138 | 0.343037 |
[
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"## Formas de Criar uma lista",
"_____no_output_____"
],
[
"### Usando espaço e a função split",
"_____no_output_____"
]
],
[
[
"data = ['1 2 3 4'.split(),\n '5 6 7 8 '.split(),\n '9 10 11 12'.split(),\n '13 14 15 16'.split()]\ndata",
"_____no_output_____"
]
],
[
[
"## Passando int para cada elemento",
"_____no_output_____"
],
[
"### Com a função map",
"_____no_output_____"
]
],
[
[
"# map usa os parâmetros (function sem parênteses, variável)\nfor i, l in enumerate(data):\n data[i] = list(map(int, l))\ndata",
"_____no_output_____"
]
],
[
[
"### Com for e range",
"_____no_output_____"
]
],
[
[
"for l in range(len(data)):\n for i in range(len(data[l])):\n data[l][i] = int(data[l][i])\ndata",
"_____no_output_____"
]
],
[
[
"### Com for e enumerate",
"_____no_output_____"
]
],
[
[
"for i, l in enumerate(data):\n for p, n in enumerate(data[i]):\n data[i][p] = int(n)\ndata",
"_____no_output_____"
]
],
[
[
"## Criando um Dataframe com split\n* para criar o index e as columns pode-se passar uma str e a função split\n* o split usa por padrão o espaço vazio como separador\n* se for usado outro separador, deve ser passado como parâmetro",
"_____no_output_____"
]
],
[
[
"data = [(1, 2, 3, 4),\n (5, 6, 7, 8),\n (9, 10, 11, 12),\n (13, 14, 15, 16)]\ndf = pd.DataFrame(data, 'l1 l2 l3 l4'.split(), 'c1 c2 c3 c4'.split())\ndf",
"_____no_output_____"
]
],
[
[
"## Tipos de Seleção",
"_____no_output_____"
],
[
"### Selecionar coluna\n* em outras palavras, selecionando uma series",
"_____no_output_____"
]
],
[
[
"df['c1']",
"_____no_output_____"
],
[
"type(df['c1'])",
"_____no_output_____"
]
],
[
[
"### Mais de uma coluna",
"_____no_output_____"
]
],
[
[
"df[['c3', 'c1']]\n# as colunas são apresentadas na ordem que foi passada",
"_____no_output_____"
],
[
"type(df[['c3', 'c1']])",
"_____no_output_____"
]
],
[
[
"### Seleção por linha",
"_____no_output_____"
]
],
[
[
"# a seleção por linha obedece ao padrão de fatiamento de string\n# não se usa o index sozinho, usa-se os ':'\n# e mais ':' se for usar o passo além do intervalo\n# ex => [::2] => seleciona tudo dando 2 passos\ndf[:]",
"_____no_output_____"
]
],
[
[
"### Seleção de linhas e colunas",
"_____no_output_____"
]
],
[
[
"# selecionar a partir da linha de index 1\n# e ainda, as colunas c3 e c1, nessa ordem\ndf[1:][['c3', 'c1']]",
"_____no_output_____"
]
],
[
[
"### Selecionando linhas com o loc\n* o loc permite selecionar linhas usando o label da linha",
"_____no_output_____"
]
],
[
[
"df",
"_____no_output_____"
],
[
"# pega uma linha e transforma numa series\ndf.loc['l1']",
"_____no_output_____"
],
[
"# obedece o mesmo formato para selecionar mais de uma linha\ndf.loc[['l3', 'l2']]",
"_____no_output_____"
],
[
"# para selecionar um elemento, usa-se a conotação matricial\ndf.loc['l3', 'c1']",
"_____no_output_____"
]
],
[
[
"### Selecionando com iloc\n* o iloc tem a mesma função do loc, mas usa o index da linha, não o label",
"_____no_output_____"
]
],
[
[
"# selecionando o mesmo elemento com o iloc\ndf.iloc[2, 0]",
"_____no_output_____"
]
],
[
[
"### Selecionando várias linhas e colunas com loc e iloc",
"_____no_output_____"
]
],
[
[
"df",
"_____no_output_____"
],
[
"# usando o loc\ndf.loc[['l3', 'l1'], ['c4', 'c1']]",
"_____no_output_____"
],
[
"# usando o iloc\ndf.iloc[[2, 0], [3, 0]]",
"_____no_output_____"
]
],
[
[
"## Exercícios",
"_____no_output_____"
],
[
"### Crie um DataFrame somente com os alunos reprovados e mantenha neste DataFrame apenas as colunas Nome, Sexo e Idade, nesta ordem.",
"_____no_output_____"
]
],
[
[
"alunos = pd.DataFrame({'Nome': ['Ary', 'Cátia', 'Denis', 'Beto', 'Bruna', 'Dara', 'Carlos', 'Alice'], \n 'Sexo': ['M', 'F', 'M', 'M', 'F', 'F', 'M', 'F'], \n 'Idade': [15, 27, 56, 32, 42, 21, 19, 35], \n 'Notas': [7.5, 2.5, 5.0, 10, 8.2, 7, 6, 5.6], \n 'Aprovado': [True, False, False, True, True, True, False, False]}, \n columns = ['Nome', 'Idade', 'Sexo', 'Notas', 'Aprovado'])\nalunos",
"_____no_output_____"
]
],
[
[
"### Resposta do exercício",
"_____no_output_____"
]
],
[
[
"selecao = alunos['Aprovado'] == False\nreprovados = alunos[['Nome', 'Sexo', 'Idade']][selecao]\nreprovados",
"_____no_output_____"
]
],
[
[
"### Resposta organizando por sexo, colocando em ordem alfabética e refazendo o index",
"_____no_output_____"
]
],
[
[
"reprovados = alunos['Aprovado'] == False\n# cria uma variável em que a coluna Aprovado tem valor False\nalunos_reprovados = alunos[reprovados]\n# cria um Dataframe apenas com Aprovado == False\nalunos_reprovados = alunos_reprovados[['Nome', 'Sexo', 'Idade']].sort_values(by=['Sexo', 'Nome'])\n# sobrescreve o Dataframe apenas com as colunas desejadas e com o filtro criado\nalunos_reprovados.index = range(alunos_reprovados.shape[0])\n# refaz o index com o range do tamanho do nº de linhas desse Dataframe\nalunos_reprovados",
"_____no_output_____"
]
],
[
[
"### Crie uma visualização com os três alunos mais novos",
"_____no_output_____"
]
],
[
[
"alunos",
"_____no_output_____"
],
[
"alunos.sort_values(by='Idade', inplace=True)\nalunos.iloc[:3]",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
cbcad0b927adab22fa03fa5ba9a7934f2dbba802
| 74,337 |
ipynb
|
Jupyter Notebook
|
intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Solution).ipynb
|
r-keller/pytorch-udacity-fb
|
f45f51074106c554d7096138477ccca76f702211
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Solution).ipynb
|
r-keller/pytorch-udacity-fb
|
f45f51074106c554d7096138477ccca76f702211
|
[
"MIT"
] | null | null | null |
intro-to-pytorch/Part 2 - Neural Networks in PyTorch (Solution).ipynb
|
r-keller/pytorch-udacity-fb
|
f45f51074106c554d7096138477ccca76f702211
|
[
"MIT"
] | null | null | null | 86.038194 | 16,184 | 0.781952 |
[
[
[
"# Neural networks with PyTorch\n\nDeep learning networks tend to be massive with dozens or hundreds of layers, that's where the term \"deep\" comes from. You can build one of these deep networks using only weight matrices as we did in the previous notebook, but in general it's very cumbersome and difficult to implement. PyTorch has a nice module `nn` that provides a nice way to efficiently build large neural networks.",
"_____no_output_____"
]
],
[
[
"# Import necessary packages\n\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport numpy as np\nimport torch\n\nimport helper\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"\nNow we're going to build a larger network that can solve a (formerly) difficult problem, identifying text in an image. Here we'll use the MNIST dataset which consists of greyscale handwritten digits. Each image is 28x28 pixels, you can see a sample below\n\n<img src='assets/mnist.png'>\n\nOur goal is to build a neural network that can take one of these images and predict the digit in the image.\n\nFirst up, we need to get our dataset. This is provided through the `torchvision` package. The code below will download the MNIST dataset, then create training and test datasets for us. Don't worry too much about the details here, you'll learn more about this later.",
"_____no_output_____"
]
],
[
[
"### Run this cell\n\nfrom torchvision import datasets, transforms\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5,), (0.5,)),\n ])\n# Download and load the training data\ntrainset = datasets.MNIST('~/.pytorch/MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"We have the training data loaded into `trainloader` and we make that an iterator with `iter(trainloader)`. Later, we'll use this to loop through the dataset for training, like\n\n```python\nfor image, label in trainloader:\n ## do things with images and labels\n```\n\nYou'll notice I created the `trainloader` with a batch size of 64, and `shuffle=True`. The batch size is the number of images we get in one iteration from the data loader and pass through our network, often called a *batch*. And `shuffle=True` tells it to shuffle the dataset every time we start going through the data loader again. But here I'm just grabbing the first batch so we can check out the data. We can see below that `images` is just a tensor with size `(64, 1, 28, 28)`. So, 64 images per batch, 1 color channel, and 28x28 images.",
"_____no_output_____"
]
],
[
[
"dataiter = iter(trainloader)\nimages, labels = dataiter.next()\nprint(type(images))\nprint(images.shape)\nprint(labels.shape)",
"<class 'torch.Tensor'>\ntorch.Size([64, 1, 28, 28])\ntorch.Size([64])\n"
]
],
[
[
"This is what one of the images looks like. ",
"_____no_output_____"
]
],
[
[
"plt.imshow(images[1].numpy().squeeze(), cmap='Greys_r');",
"_____no_output_____"
]
],
[
[
"First, let's try to build a simple network for this dataset using weight matrices and matrix multiplications. Then, we'll see how to do it using PyTorch's `nn` module which provides a much more convenient and powerful method for defining network architectures.\n\nThe networks you've seen so far are called *fully-connected* or *dense* networks. Each unit in one layer is connected to each unit in the next layer. In fully-connected networks, the input to each layer must be a one-dimensional vector (which can be stacked into a 2D tensor as a batch of multiple examples). However, our images are 28x28 2D tensors, so we need to convert them into 1D vectors. Thinking about sizes, we need to convert the batch of images with shape `(64, 1, 28, 28)` to a have a shape of `(64, 784)`, 784 is 28 times 28. This is typically called *flattening*, we flattened the 2D images into 1D vectors.\n\nPreviously you built a network with one output unit. Here we need 10 output units, one for each digit. We want our network to predict the digit shown in an image, so what we'll do is calculate probabilities that the image is of any one digit or class. This ends up being a discrete probability distribution over the classes (digits) that tells us the most likely class for the image. That means we need 10 output units for the 10 classes (digits). We'll see how to convert the network output into a probability distribution next.\n\n> **Exercise:** Flatten the batch of images `images`. Then build a multi-layer network with 784 input units, 256 hidden units, and 10 output units using random tensors for the weights and biases. For now, use a sigmoid activation for the hidden layer. Leave the output layer without an activation, we'll add one that gives us a probability distribution next.",
"_____no_output_____"
]
],
[
[
"## Solution\ndef activation(x):\n return 1/(1+torch.exp(-x))\n\n# Flatten the input images\ninputs = images.view(images.shape[0], -1)\n\n# Create parameters\nw1 = torch.randn(784, 256)\nb1 = torch.randn(256)\n\nw2 = torch.randn(256, 10)\nb2 = torch.randn(10)\n\nh = activation(torch.mm(inputs, w1) + b1)\n\nout = torch.mm(h, w2) + b2",
"_____no_output_____"
]
],
[
[
"Now we have 10 outputs for our network. We want to pass in an image to our network and get out a probability distribution over the classes that tells us the likely class(es) the image belongs to. Something that looks like this:\n<img src='assets/image_distribution.png' width=500px>\n\nHere we see that the probability for each class is roughly the same. This is representing an untrained network, it hasn't seen any data yet so it just returns a uniform distribution with equal probabilities for each class.\n\nTo calculate this probability distribution, we often use the [**softmax** function](https://en.wikipedia.org/wiki/Softmax_function). Mathematically this looks like\n\n$$\n\\Large \\sigma(x_i) = \\cfrac{e^{x_i}}{\\sum_k^K{e^{x_k}}}\n$$\n\nWhat this does is squish each input $x_i$ between 0 and 1 and normalizes the values to give you a proper probability distribution where the probabilites sum up to one.\n\n> **Exercise:** Implement a function `softmax` that performs the softmax calculation and returns probability distributions for each example in the batch. Note that you'll need to pay attention to the shapes when doing this. If you have a tensor `a` with shape `(64, 10)` and a tensor `b` with shape `(64,)`, doing `a/b` will give you an error because PyTorch will try to do the division across the columns (called broadcasting) but you'll get a size mismatch. The way to think about this is for each of the 64 examples, you only want to divide by one value, the sum in the denominator. So you need `b` to have a shape of `(64, 1)`. This way PyTorch will divide the 10 values in each row of `a` by the one value in each row of `b`. Pay attention to how you take the sum as well. You'll need to define the `dim` keyword in `torch.sum`. Setting `dim=0` takes the sum across the rows while `dim=1` takes the sum across the columns.",
"_____no_output_____"
]
],
[
[
"## Solution\ndef softmax(x):\n return torch.exp(x)/torch.sum(torch.exp(x), dim=1).view(-1, 1)\n\nprobabilities = softmax(out)\n\n# Does it have the right shape? Should be (64, 10)\nprint(probabilities.shape)\n# Does it sum to 1?\nprint(probabilities.sum(dim=1))",
"torch.Size([64, 10])\ntensor([ 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000, 1.0000,\n 1.0000])\n"
]
],
[
[
"## Building networks with PyTorch\n\nPyTorch provides a module `nn` that makes building networks much simpler. Here I'll show you how to build the same one as above with 784 inputs, 256 hidden units, 10 output units and a softmax output.",
"_____no_output_____"
]
],
[
[
"from torch import nn",
"_____no_output_____"
],
[
"class Network(nn.Module):\n def __init__(self):\n super().__init__()\n \n # Inputs to hidden layer linear transformation\n self.hidden = nn.Linear(784, 256)\n # Output layer, 10 units - one for each digit\n self.output = nn.Linear(256, 10)\n \n # Define sigmoid activation and softmax output \n self.sigmoid = nn.Sigmoid()\n self.softmax = nn.Softmax(dim=1)\n \n def forward(self, x):\n # Pass the input tensor through each of our operations\n x = self.hidden(x)\n x = self.sigmoid(x)\n x = self.output(x)\n x = self.softmax(x)\n \n return x",
"_____no_output_____"
]
],
[
[
"Let's go through this bit by bit.\n\n```python\nclass Network(nn.Module):\n```\n\nHere we're inheriting from `nn.Module`. Combined with `super().__init__()` this creates a class that tracks the architecture and provides a lot of useful methods and attributes. It is mandatory to inherit from `nn.Module` when you're creating a class for your network. The name of the class itself can be anything.\n\n```python\nself.hidden = nn.Linear(784, 256)\n```\n\nThis line creates a module for a linear transformation, $x\\mathbf{W} + b$, with 784 inputs and 256 outputs and assigns it to `self.hidden`. The module automatically creates the weight and bias tensors which we'll use in the `forward` method. You can access the weight and bias tensors once the network (`net`) is created with `net.hidden.weight` and `net.hidden.bias`.\n\n```python\nself.output = nn.Linear(256, 10)\n```\n\nSimilarly, this creates another linear transformation with 256 inputs and 10 outputs.\n\n```python\nself.sigmoid = nn.Sigmoid()\nself.softmax = nn.Softmax(dim=1)\n```\n\nHere I defined operations for the sigmoid activation and softmax output. Setting `dim=1` in `nn.Softmax(dim=1)` calculates softmax across the columns.\n\n```python\ndef forward(self, x):\n```\n\nPyTorch networks created with `nn.Module` must have a `forward` method defined. It takes in a tensor `x` and passes it through the operations you defined in the `__init__` method.\n\n```python\nx = self.hidden(x)\nx = self.sigmoid(x)\nx = self.output(x)\nx = self.softmax(x)\n```\n\nHere the input tensor `x` is passed through each operation a reassigned to `x`. We can see that the input tensor goes through the hidden layer, then a sigmoid function, then the output layer, and finally the softmax function. It doesn't matter what you name the variables here, as long as the inputs and outputs of the operations match the network architecture you want to build. The order in which you define things in the `__init__` method doesn't matter, but you'll need to sequence the operations correctly in the `forward` method.\n\nNow we can create a `Network` object.",
"_____no_output_____"
]
],
[
[
"# Create the network and look at it's text representation\nmodel = Network()\nmodel",
"_____no_output_____"
]
],
[
[
"You can define the network somewhat more concisely and clearly using the `torch.nn.functional` module. This is the most common way you'll see networks defined as many operations are simple element-wise functions. We normally import this module as `F`, `import torch.nn.functional as F`.",
"_____no_output_____"
]
],
[
[
"import torch.nn.functional as F\n\nclass Network(nn.Module):\n def __init__(self):\n super().__init__()\n # Inputs to hidden layer linear transformation\n self.hidden = nn.Linear(784, 256)\n # Output layer, 10 units - one for each digit\n self.output = nn.Linear(256, 10)\n \n def forward(self, x):\n # Hidden layer with sigmoid activation\n x = F.sigmoid(self.hidden(x))\n # Output layer with softmax activation\n x = F.softmax(self.output(x), dim=1)\n \n return x",
"_____no_output_____"
]
],
[
[
"### Activation functions\n\nSo far we've only been looking at the softmax activation, but in general any function can be used as an activation function. The only requirement is that for a network to approximate a non-linear function, the activation functions must be non-linear. Here are a few more examples of common activation functions: Tanh (hyperbolic tangent), and ReLU (rectified linear unit).\n\n<img src=\"assets/activation.png\" width=700px>\n\nIn practice, the ReLU function is used almost exclusively as the activation function for hidden layers.",
"_____no_output_____"
],
[
"### Your Turn to Build a Network\n\n<img src=\"assets/mlp_mnist.png\" width=600px>\n\n> **Exercise:** Create a network with 784 input units, a hidden layer with 128 units and a ReLU activation, then a hidden layer with 64 units and a ReLU activation, and finally an output layer with a softmax activation as shown above. You can use a ReLU activation with the `nn.ReLU` module or `F.relu` function.",
"_____no_output_____"
]
],
[
[
"## Solution\n\nclass Network(nn.Module):\n def __init__(self):\n super().__init__()\n # Defining the layers, 128, 64, 10 units each\n self.fc1 = nn.Linear(784, 128)\n self.fc2 = nn.Linear(128, 64)\n # Output layer, 10 units - one for each digit\n self.fc3 = nn.Linear(64, 10)\n \n def forward(self, x):\n ''' Forward pass through the network, returns the output logits '''\n \n x = self.fc1(x)\n x = F.relu(x)\n x = self.fc2(x)\n x = F.relu(x)\n x = self.fc3(x)\n x = F.softmax(x, dim=1)\n \n return x\n\nmodel = Network()\nmodel",
"_____no_output_____"
]
],
[
[
"### Initializing weights and biases\n\nThe weights and such are automatically initialized for you, but it's possible to customize how they are initialized. The weights and biases are tensors attached to the layer you defined, you can get them with `model.fc1.weight` for instance.",
"_____no_output_____"
]
],
[
[
"print(model.fc1.weight)\nprint(model.fc1.bias)",
"Parameter containing:\ntensor([[-2.3278e-02, -1.2170e-03, -1.1882e-02, ..., 3.3567e-02,\n 4.4827e-03, 1.4840e-02],\n [ 4.8464e-03, 1.9844e-02, 3.9791e-03, ..., -2.6048e-02,\n -3.5558e-02, -2.2386e-02],\n [-1.9664e-02, 8.1722e-03, 2.6729e-02, ..., -1.5122e-02,\n 2.7632e-02, -1.9567e-02],\n ...,\n [-3.3571e-02, -2.9686e-02, -2.1387e-02, ..., 3.0770e-02,\n 1.0800e-02, -6.5941e-03],\n [ 2.9749e-02, 1.2849e-02, 2.7320e-02, ..., -1.9899e-02,\n 2.7131e-02, 2.2082e-02],\n [ 1.3992e-02, -2.1520e-02, 3.1907e-02, ..., 2.2435e-02,\n 1.1370e-02, 2.1568e-02]])\nParameter containing:\ntensor(1.00000e-02 *\n [-1.3222, 2.4094, -2.1571, 3.2237, 2.5302, -1.1515, 2.6382,\n -2.3426, -3.5689, -1.0724, -2.8842, -2.9667, -0.5022, 1.1381,\n 1.2849, 3.0731, -2.0207, -2.3282, 0.3168, -2.8098, -1.0740,\n -1.8273, 1.8692, 2.9404, 0.1783, 0.9391, -0.7085, -1.2522,\n -2.7769, 0.0916, -1.4283, -0.3267, -1.6876, -1.8580, -2.8724,\n -3.5512, 3.2155, 1.5532, 0.8836, -1.2911, 1.5735, -3.0478,\n -1.3089, -2.2117, 1.5162, -0.8055, -1.3307, -2.4267, -1.2665,\n 0.8666, -2.2325, -0.4797, -0.5448, -0.6612, -0.6022, 2.6399,\n 1.4673, -1.5417, -2.9492, -2.7507, 0.6157, -0.0681, -0.8171,\n -0.3554, -0.8225, 3.3906, 3.3509, -1.4484, 3.5124, -2.6519,\n 0.9721, -2.5068, -3.4962, 3.4743, 1.1525, -2.7555, -3.1673,\n 2.2906, 2.5914, 1.5992, -1.2859, -0.5682, 2.1488, -2.0631,\n 2.6281, -2.4639, 2.2622, 2.3632, -0.1979, 0.7160, 1.7594,\n 0.0761, -2.8886, -3.5467, 2.7691, 0.8280, -2.2398, -1.4602,\n -1.3475, -1.4738, 0.6338, 3.2811, -3.0628, 2.7044, 1.2775,\n 2.8856, -3.3938, 2.7056, 0.5826, -0.6286, 1.2381, 0.7316,\n -2.4725, -1.2958, -3.1543, -0.8584, 0.5517, 2.8176, 0.0947,\n -1.6849, -1.4968, 3.1039, 1.7680, 1.1803, -1.4402, 2.5710,\n -3.3057, 1.9027])\n"
]
],
[
[
"For custom initialization, we want to modify these tensors in place. These are actually autograd *Variables*, so we need to get back the actual tensors with `model.fc1.weight.data`. Once we have the tensors, we can fill them with zeros (for biases) or random normal values.",
"_____no_output_____"
]
],
[
[
"# Set biases to all zeros\nmodel.fc1.bias.data.fill_(0)",
"_____no_output_____"
],
[
"# sample from random normal with standard dev = 0.01\nmodel.fc1.weight.data.normal_(std=0.01)",
"_____no_output_____"
]
],
[
[
"### Forward pass\n\nNow that we have a network, let's see what happens when we pass in an image.",
"_____no_output_____"
]
],
[
[
"# Grab some data \ndataiter = iter(trainloader)\nimages, labels = dataiter.next()\n\n# Resize images into a 1D vector, new shape is (batch size, color channels, image pixels) \nimages.resize_(64, 1, 784)\n# or images.resize_(images.shape[0], 1, 784) to automatically get batch size\n\n# Forward pass through the network\nimg_idx = 0\nps = model.forward(images[img_idx,:])\n\nimg = images[img_idx]\nhelper.view_classify(img.view(1, 28, 28), ps)",
"_____no_output_____"
]
],
[
[
"As you can see above, our network has basically no idea what this digit is. It's because we haven't trained it yet, all the weights are random!\n\n### Using `nn.Sequential`\n\nPyTorch provides a convenient way to build networks like this where a tensor is passed sequentially through operations, `nn.Sequential` ([documentation](https://pytorch.org/docs/master/nn.html#torch.nn.Sequential)). Using this to build the equivalent network:",
"_____no_output_____"
]
],
[
[
"# Hyperparameters for our network\ninput_size = 784\nhidden_sizes = [128, 64]\noutput_size = 10\n\n# Build a feed-forward network\nmodel = nn.Sequential(nn.Linear(input_size, hidden_sizes[0]),\n nn.ReLU(),\n nn.Linear(hidden_sizes[0], hidden_sizes[1]),\n nn.ReLU(),\n nn.Linear(hidden_sizes[1], output_size),\n nn.Softmax(dim=1))\nprint(model)\n\n# Forward pass through the network and display output\nimages, labels = next(iter(trainloader))\nimages.resize_(images.shape[0], 1, 784)\nps = model.forward(images[0,:])\nhelper.view_classify(images[0].view(1, 28, 28), ps)",
"Sequential(\n (0): Linear(in_features=784, out_features=128, bias=True)\n (1): ReLU()\n (2): Linear(in_features=128, out_features=64, bias=True)\n (3): ReLU()\n (4): Linear(in_features=64, out_features=10, bias=True)\n (5): Softmax()\n)\n"
]
],
[
[
"The operations are availble by passing in the appropriate index. For example, if you want to get first Linear operation and look at the weights, you'd use `model[0]`.",
"_____no_output_____"
]
],
[
[
"print(model[0])\nmodel[0].weight",
"Linear(in_features=784, out_features=128, bias=True)\n"
]
],
[
[
"You can also pass in an `OrderedDict` to name the individual layers and operations, instead of using incremental integers. Note that dictionary keys must be unique, so _each operation must have a different name_.",
"_____no_output_____"
]
],
[
[
"from collections import OrderedDict\nmodel = nn.Sequential(OrderedDict([\n ('fc1', nn.Linear(input_size, hidden_sizes[0])),\n ('relu1', nn.ReLU()),\n ('fc2', nn.Linear(hidden_sizes[0], hidden_sizes[1])),\n ('relu2', nn.ReLU()),\n ('output', nn.Linear(hidden_sizes[1], output_size)),\n ('softmax', nn.Softmax(dim=1))]))\nmodel",
"_____no_output_____"
]
],
[
[
"Now you can access layers either by integer or the name",
"_____no_output_____"
]
],
[
[
"print(model[0])\nprint(model.fc1)",
"Linear(in_features=784, out_features=128, bias=True)\nLinear(in_features=784, out_features=128, bias=True)\n"
]
],
[
[
"In the next notebook, we'll see how we can train a neural network to accuractly predict the numbers appearing in the MNIST images.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
cbcaf9f570651612d6e5d5a7c069f35dbe4ae73c
| 7,424 |
ipynb
|
Jupyter Notebook
|
notebooks/Schema.ipynb
|
GenomicMedLab/vrs-python
|
4978e88396c8a718576ade805c8b1332b7108496
|
[
"Apache-2.0"
] | null | null | null |
notebooks/Schema.ipynb
|
GenomicMedLab/vrs-python
|
4978e88396c8a718576ade805c8b1332b7108496
|
[
"Apache-2.0"
] | null | null | null |
notebooks/Schema.ipynb
|
GenomicMedLab/vrs-python
|
4978e88396c8a718576ade805c8b1332b7108496
|
[
"Apache-2.0"
] | null | null | null | 27.194139 | 497 | 0.560345 |
[
[
[
"# GA4GH Variation Representation Schema\n\nThis notebook demonstrates the use of the VR schema to represent variation in APOE. Objects created in this notebook are saved at the end and used by other notebooks to demonstrate other features of the VR specification.\n\n\n## APOE Variation\n\n rs7412 \n NC_000019.10:g.44908822\n NM_000041.3:c.526\n C T\n rs429358 C APOE-ε4 APOE-ε1\n NC_000019.10:g.44908684 T APOE-ε3 APOE-ε2\n NM_000041.3:c.388\n\nNote: The example currently uses only rs7412:T. Future versions of the schema will support haplotypes and genotypes, and these examples will be extended appropriately.",
"_____no_output_____"
],
[
"## Using the VR Reference Implemention\n\nSee https://github.com/ga4gh/vr-python for information about installing the reference implementation.",
"_____no_output_____"
]
],
[
[
"from ga4gh.vrs import __version__, models\n__version__",
"_____no_output_____"
]
],
[
[
"## Schema Overview\n\n<img src=\"images/schema-current.png\" width=\"75%\" alt=\"Current Schema\"/>",
"_____no_output_____"
],
[
"## Sequences\n\nThe VR Specfication expects the existence of a repository of biological sequences. At a minimum, these sequences must be indexed using whatever accessions are available. Implementations that wish to use the computed identifier mechanism should also have precomputed ga4gh sequence accessions. Either way, sequences must be referred to using [W3C Compact URIs (CURIEs)](https://w3.org/TR/curie/). In the examples below, we'll use \"refseq:NC_000019.10\" to refer to chromosome 19 from GRCh38.",
"_____no_output_____"
],
[
"## Locations\nA Location is an *abstract* object that refer to contiguous regions of biological sequences.\n\nIn the initial release of VR, the only Location is a SequenceLocation, which represents a precise interval (`SimpleInterval`) on a sequence. GA4GH VR uses interbase coordinates exclusively; therefore the 1-based residue position 44908822 is referred to using the 0-based interbase interval <44908821, 44908822>.\n\nFuture Location subclasses will provide for approximate coordinates, gene symbols, and cytogenetic bands.",
"_____no_output_____"
],
[
"#### SequenceLocation",
"_____no_output_____"
]
],
[
[
"location = models.SequenceLocation(\n sequence_id=\"refseq:NC_000019.10\",\n interval=models.SimpleInterval(start=44908821, end=44908822))",
"_____no_output_____"
],
[
"location.as_dict()",
"_____no_output_____"
]
],
[
[
"## Variation\n\n### Text Variation\n\nThe TextVariation class represents variation descriptions that cannot be parsed, or cannot be parsed yet. The primary use for this class is to allow unparsed variation to be represented within the VR framework and be associated with annotations.",
"_____no_output_____"
]
],
[
[
"variation = models.Text(definition=\"APO loss\")\nvariation.as_dict()",
"_____no_output_____"
]
],
[
[
"### Alleles\n\nAn Allele is an asserion of a state of biological sequence at a Location. In the first version of the VR Schema, the only State subclass is SequenceState, which represents the replacement of sequence. Future versions of State will enable representations of copy number variation.",
"_____no_output_____"
],
[
"### \"Simple\" sequence replacements\nThis case covers any \"ref-alt\" style variation, which includes SNVs, MNVs, del, ins, and delins.",
"_____no_output_____"
]
],
[
[
"allele = models.Allele(location=location,\n state=models.SequenceState(sequence=\"A\"))\nallele.as_dict()",
"_____no_output_____"
]
],
[
[
"----\n\n## Saving the objects\n\nObjects created in this notebook will be saved as a json file and loaded by subsequent notebooks.",
"_____no_output_____"
]
],
[
[
"import json\nfilename = \"objects.json\"",
"_____no_output_____"
],
[
"data = {\n \"alleles\": [allele.as_dict()],\n \"locations\": [location.as_dict()]\n}",
"_____no_output_____"
],
[
"json.dump(data, open(filename, \"w\"))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbcafcbda081ba558475edb465a71d5cc785f313
| 14,379 |
ipynb
|
Jupyter Notebook
|
opencv_tutorial/contour-hierarchy/contour-hierachy-bgr.ipynb
|
nooshhub/pycv-sample
|
d49c5588032b8ba27e5891d4dfa0238989ab5e64
|
[
"Apache-2.0"
] | null | null | null |
opencv_tutorial/contour-hierarchy/contour-hierachy-bgr.ipynb
|
nooshhub/pycv-sample
|
d49c5588032b8ba27e5891d4dfa0238989ab5e64
|
[
"Apache-2.0"
] | null | null | null |
opencv_tutorial/contour-hierarchy/contour-hierachy-bgr.ipynb
|
nooshhub/pycv-sample
|
d49c5588032b8ba27e5891d4dfa0238989ab5e64
|
[
"Apache-2.0"
] | 1 |
2021-06-23T07:01:44.000Z
|
2021-06-23T07:01:44.000Z
| 36.310606 | 132 | 0.512762 |
[
[
[
"import numpy as np\nimport cv2 as cv\nimport json\n\n\"\"\"缩小图像,方便看效果\nresize会损失像素,造成边缘像素模糊,不要再用于计算的原图上使用\n\"\"\" \ndef resizeImg(src):\n height, width = src.shape[:2]\n size = (int(width * 0.3), int(height * 0.3)) \n img = cv.resize(src, size, interpolation=cv.INTER_AREA)\n return img\n\n\"\"\"找出ROI,用于分割原图\n原图有四块区域,一个是地块区域,一个是颜色示例区域,一个距离标尺区域,一个南北方向区域\n理论上倒排后的最大轮廓的是地块区域\n\"\"\"\ndef findROIContours(src):\n copy = src.copy()\n gray = cv.cvtColor(copy, cv.COLOR_BGR2GRAY)\n # cv.imshow(\"gray\", gray)\n \n # 低于thresh都变为黑色,maxval是给binary用的\n # 白底 254, 255 黑底 0, 255\n threshold = cv.threshold(gray, 0, 255, cv.THRESH_BINARY)[1]\n # cv.imshow(\"threshold\", threshold)\n contours, hierarchy = cv.findContours(threshold, cv.RETR_EXTERNAL, cv.CHAIN_APPROX_SIMPLE)\n sortedCnts = sorted(contours, key = cv.contourArea, reverse=True)\n # cv.drawContours(copy, [maxCnt], -1, (255, 0, 0), 2)\n # cv.imshow(\"roi contours\", copy)\n return sortedCnts\n \n\n\n\"\"\"按照mask,截取roi\n\"\"\"\ndef getROIByContour(src, cnt):\n copy = src.copy()\n # black background\n mask = np.zeros(copy.shape[:2], np.uint8)\n mask = cv.fillConvexPoly(mask, cnt, (255,255,255)) \n# cv.imshow(\"mask\", resizeImg(mask))\n # print(mask.shape)\n # print(copy.dtype)\n roi = cv.bitwise_and(copy, copy, mask=mask)\n # cv.imshow(\"roi\", roi)\n \n # white background for none roi and fill the roi's backgroud\n mask = cv.bitwise_not(mask)\n whiteBg = np.full(copy.shape[:2], 255, dtype=np.uint8)\n whiteBg = cv.bitwise_and(whiteBg, whiteBg, mask=mask)\n whiteBg = cv.merge((whiteBg,whiteBg,whiteBg))\n# cv.imshow(\"whiteBg\", resizeImg(whiteBg))\n \n roiWithAllWhite = cv.bitwise_or(roi, whiteBg)\n return roiWithAllWhite\n\n\n\"\"\"找出所有的地块轮廓\n\"\"\"\ndef findAllBlockContours(src):\n copy = src.copy()\n contours = findExternalContours(copy)\n return contours\n\n\n\"\"\"找出颜色示例里的颜色BGR\n根据限定长,高,比例来过滤出实例颜色区域\n\"\"\"\ndef findBGRColors(cnts):\n W_RANGE = [170,180]\n H_RANGE = [75, 85]\n RATIO_RANGE = [0.40, 0.50]\n colors = []\n \n # TODO 如果可以知道颜色示例的个数可以提前统计退出循环\n for cnt in cnts:\n x,y,w,h = cv.boundingRect(cnt)\n ratio = round(h/w, 2)\n \n if ratio > RATIO_RANGE[0] and ratio < RATIO_RANGE[1] \\\n and w > W_RANGE[0] and w < W_RANGE[1] \\\n and h > H_RANGE[0] and h < H_RANGE[1]:\n # print(ratio,x,y,w,h)\n # 因为,原图色块矩形的周边和mask出来的颜色区都有模糊渐变的线\n # 无法使用cv.mean(colorRegion, meanMask)来计算实际颜色\n # 所以,取矩形的中心点的颜色最为准确\n cx,cy = round(x+w/2), round(y+h/2)\n bgr = img_white_bg[cy, cx]\n # print(bgr)\n colors.append(bgr)\n \n return colors\n\n\ndef drawnForTest(img, contours, rect=False):\n img = img.copy()\n for i in contours:\n if rect:\n x, y, w, h = cv.boundingRect(i)\n cv.rectangle(img, (x, y), (x + w, y + h), (255, 0, 0), 2)\n cv.putText(img, 'Area' + str(cv.contourArea(i)), (x+5,y+15), cv.FONT_HERSHEY_PLAIN, 1,(255,0,0), 1, cv.LINE_AA)\n else:\n cv.drawContours(img, [i], -1, (0, 255, 0), 2) \n cv.imshow(\"detect\", resizeImg(img))\n cv.waitKey(0)\n\n\n\"\"\"Find Original Contours\nFind Original Contours from source image, we only need external contour.\nArgs:\n src: source image\nReturns:\n Original contours\n\"\"\"\ndef findExternalContours(src):\n # 必须是白色背景的图片,如果是黑色背景,将黑色改成白色\n# src[np.where((src == [0,0,0]).all(axis = 2))] = [255,255,255]\n \n # preprocess, remove noise, a lot noise on the road\n gray = cv.cvtColor(src, cv.COLOR_BGR2GRAY)\n # 测试这里道理有没有必要高斯\n# blur = cv.GaussianBlur(gray, (3,3), 0)\n \n thresVal = 254\n maxVal = 255\n ret,thresh1 = cv.threshold(gray, thresVal, maxVal, cv.THRESH_BINARY)\n \n kernel = np.ones((7,7),np.uint8)\n morph = cv.morphologyEx(thresh1, cv.MORPH_CLOSE, kernel)\n \n # ??threshold怎么计算的?\n edges = cv.Canny(morph,100,200)\n # edges找出来,但是是锯齿状,会在找轮廓时形成很多点,这里加一道拉普拉斯锐化一下\n edges = cv.Laplacian(edges, -1, (3,3))\n \n contours, hierarchy = cv.findContours(edges, cv.RETR_EXTERNAL, cv.CHAIN_APPROX_SIMPLE)\n# contours, hierarchy = cv.findContours(edges, cv.RETR_EXTERNAL, cv.CHAIN_APPROX_SIMPLE)\n# cv.imshow('gray', resizeImg(gray))\n# cv.imshow('thresh1', resizeImg(thresh1))\n# cv.imshow('edges', resizeImg(edges))\n# cv.imshow('opening', opening)\n# cv.imshow('opening', resizeImg(opening))\n return contours, hierarchy \n\n\n\"\"\"根据找出的轮廓和层级关系计算地块和色块的父子关系\n\"\"\"\ndef getBlockColorTree(copy, blockCnts, hierarchy):\n # print(hierarchy)\n # hierarchy [Next, Previous, First_Child, Parent]\n currentRootIndex = -1\n rootRegions = {}\n for i,cnt in enumerate(blockCnts):\n x,y,w,h = cv.boundingRect(cnt)\n cntArea = cv.contourArea(cnt)\n if cntArea > 1000:\n continue\n cv.putText(copy, str(i), (x+5,y+15), cv.FONT_HERSHEY_PLAIN, 1,(255,0,0), 1, cv.LINE_AA)\n\n print(i, hierarchy[0][i])\n if hierarchy[0][i][3] == -1:\n # root region\n currentRootIndex = i\n if currentRootIndex == len(blockCnts):\n break\n rootRegion = {'index': i, 'contour': cv.contourArea(blockCnts[currentRootIndex]), 'childRegion': []}\n rootRegions[currentRootIndex] = rootRegion\n elif hierarchy[0][i][3] == currentRootIndex:\n rootRegions[currentRootIndex]['childRegion'].append({'index': i, 'contour': cv.contourArea(cnt)})\n\n cv.imshow(\"blockCnts with debug info\", resizeImg(copy))\n\n print(rootRegions) \n data2 = json.dumps(rootRegions, sort_keys=True, indent=4, separators=(',', ': '))\n print(data2)\n\n \n\"\"\"使用颜色来分块,并返回所有地块和色块父子关系\ndebug 只演示前三个地块的识别过程,可以通过debugFrom:debugLen来调整debug开始位置和长度\n\"\"\"\ndef findColorRegionsForAllBlocks(img_white_bg, blockCnts, debug=False, debugFrom=0, debugLen=3):\n filteredBlockCnts = [cnt for cnt in blockCnts if cv.contourArea(cnt) > 100]\n \n if debug:\n filteredBlockCnts = filteredBlockCnts[debugFrom:debugLen]\n \n for blockCnt in filteredBlockCnts:\n findColorRegionsForBlock(img_white_bg, blockCnt, debug)\n\n\"\"\"根据threshold重新计算BGR的值\n\"\"\"\ndef bgrWithThreshold(bgr, threshold): \n newBgr = []\n for x in bgr.tolist():\n if x + threshold < 0:\n newBgr.append(0)\n elif x + threshold > 255:\n newBgr.append(255)\n else:\n newBgr.append(x + threshold )\n \n return newBgr \n \n \n\"\"\"使用颜色来找出单个地块内的色块\n\"\"\"\ndef findColorRegionsForBlock(img_white_bg, blockCnt, debug=False): \n blockWithColorsDict = {'area': cv.contourArea(blockCnt), 'points':[] , 'children': []}\n \n blockRegion = getROIByContour(img_white_bg, blockCnt) \n if debug:\n cv.imshow(\"blockRegions\", np.hstack([resizeImg(img_white_bg), resizeImg(blockRegion)]))\n \n colorCnts = []\n for bgr in bgrColors:\n # 图片里的颜色可能和示例颜色不相等,适当增加点阈值来防色差\n threshold = 5\n lower = np.array(bgrWithThreshold(bgr, -threshold), dtype=\"uint8\") \n upper = np.array(bgrWithThreshold(bgr, threshold), dtype=\"uint8\") \n # 根据阈值找到对应颜色区域,黑底白块\n mask = cv.inRange(blockRegion, lower, upper) \n # cv.imshow(\"mask\", resizeImg(mask))\n \n # 过滤出于颜色匹配的色块\n nonZeroCount = cv.countNonZero(mask)\n # print('none zero count', nonZeroCount)\n if nonZeroCount == 0:\n continue\n \n contours, hierarchy = cv.findContours(mask.copy(), cv.RETR_EXTERNAL, cv.CHAIN_APPROX_SIMPLE)\n# print('external', len(contours))\n# print(hierarchy)\n colorCnts.extend(contours)\n \n \n# contours, hierarchy = cv.findContours(mask.copy(), cv.RETR_TREE, cv.CHAIN_APPROX_SIMPLE)\n# print('tree', len(contours))\n# print(hierarchy)\n\n if debug:\n # 黑白变白黑\n mask_inv = 255 - mask \n # cv.imshow(\"mask_inv\", resizeImg(mask_inv))\n # 展示图片\n output = cv.bitwise_and(blockRegion, blockRegion, mask=mask_inv) \n # cv.drawContours(output, contours, -1, (0, 0, 255), 3) \n \n cv.imshow(\"images\", np.hstack([resizeImg(blockRegion), resizeImg(output)]))\n cv.waitKey(0)\n \n # 色块在同一个地块内也可能是多个的,色块内又内嵌了色块\n # TODO 嵌套的\n # colorCnts 循环递归,不到叶子节点,所有的色块统一当作地块继续处理,这样就可以解决嵌套问题\n # 所以要先构建一个三层数据模型\n # 第一层 是一个列表 存放所有的地块\n # 第二层 是以色块为节点,色块内无嵌套,则为叶子节点,有嵌套,色块升级为地块,继续深入查找色块,直到没有内嵌,成为叶子节点\n # 第三层 是以色块为叶子节点\n colorDicts = []\n for colorCnt in contours:\n colorDict = {'area': cv.contourArea(colorCnt), 'points':[], 'color': bgr.tolist()}\n colorDicts.append(colorDict)\n blockWithColorsDict['children'].extend(colorDicts)\n \n # print(blockWithColorsDict) \n jsonData = json.dumps(blockWithColorsDict, sort_keys=True, indent=4, separators=(',', ': '))\n print(jsonData) \n \n return colorCnts\n\n \n# 用于找ROI\nimg = cv.imread('data_hierarchy2.png')\n# 用于真实计算,\n# 1. 色调区间误差,原图4识别最高,图3识别一般,需要增加threshold到5,\n# 2. 间隙与边框误差,色块面积总和与地块面积相差3000左右,应该是线的面积没计算进去\n# 3. \nimg_white_bg = cv.imread('data_hierarchy4.png')\n\n# 将图片按照轮廓排序,最大的是总地块\n# 按照原图中的比例和实际距离来分割图片,参考findBGRColors的计算方式\nsortedCnts = findROIContours(img) \n# print(len(sortedCnts[2:]))\n# drawnForTest(img_white_bg, sortedCnts[3], rect=True)\n# print(sortedCnts[3])\nprint(cv.boundingRect(sortedCnts[3]))\nprint(img_white_bg.shape)\n\n# 2401 * 3151\n# 670\npx_km_scale = 670/1000\narea_px = (2401*3151)\narea_km2 = (2401*px_km_scale*3151*px_km_scale)\nprint(area_px/area_km2)\nprint(1/(px_km_scale*px_km_scale))\n\n# 获取总地块\nrootRegion = getROIByContour(img_white_bg, sortedCnts[0])\n# cv.imshow(\"rootRegion\", resizeImg(rootRegion))\n\n# 找出色块的颜色\nbgrColors = findBGRColors(sortedCnts[1:])\n# print(bgrColors)\n\n\n# 找出地块\ncopy = rootRegion.copy()\nblockCnts, hierarchy = findAllBlockContours(copy)\n# print(len(blockCnts))\n# drawnForTest(img_white_bg, blockCnts, rect=False)\n\n# 通过颜色来检测地块内色块\n# findColorRegionsForAllBlocks(img_white_bg, blockCnts, debug=True, debugLen=1)\n# findColorRegionsForAllBlocks(img_white_bg, blockCnts)\n\n\n# 根据轮廓找地块的方法首先hierarchy的转换父子关系,还有很多小的干扰项待解决\n# getBlockColorTree(copy, blockCnts, hierarchy)\n\ncv.waitKey(0)\ncv.destroyAllWindows()\n",
"(2238, 2132, 670, 80)\n(2401, 3151, 3)\n2.227667631989307\n2.227667631989307\n"
],
[
"a = [1]\nb = [2,3]\na.extend(b)\na\n\nc=1\n-c",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
cbcb06882f3b55e1264afc290e14e6b8a6c6d9e1
| 7,189 |
ipynb
|
Jupyter Notebook
|
docs/_build/.jupyter_cache/executed/384bbed82e7d63ab2e9be1c4d93b6a79/base.ipynb
|
cancermqiao/CancerMBook
|
bd26c0e3e1f76f66b75aacf75b3cb8602715e803
|
[
"Apache-2.0"
] | null | null | null |
docs/_build/.jupyter_cache/executed/384bbed82e7d63ab2e9be1c4d93b6a79/base.ipynb
|
cancermqiao/CancerMBook
|
bd26c0e3e1f76f66b75aacf75b3cb8602715e803
|
[
"Apache-2.0"
] | null | null | null |
docs/_build/.jupyter_cache/executed/384bbed82e7d63ab2e9be1c4d93b6a79/base.ipynb
|
cancermqiao/CancerMBook
|
bd26c0e3e1f76f66b75aacf75b3cb8602715e803
|
[
"Apache-2.0"
] | null | null | null | 36.866667 | 121 | 0.564195 |
[
[
[
"\"\"\"\nEstimating the causal effect of sodium on blood pressure in a simulated example\nadapted from Luque-Fernandez et al. (2018):\n https://academic.oup.com/ije/article/48/2/640/5248195\n\"\"\"\n\nimport numpy as np\nimport pandas as pd\nfrom sklearn.linear_model import LinearRegression\n\ndef generate_data(n=1000, seed=0, beta1=1.05, alpha1=0.4, alpha2=0.3, binary_treatment=True, binary_cutoff=3.5):\n np.random.seed(seed)\n age = np.random.normal(65, 5, n)\n sodium = age / 18 + np.random.normal(size=n)\n if binary_treatment:\n if binary_cutoff is None:\n binary_cutoff = sodium.mean()\n sodium = (sodium > binary_cutoff).astype(int)\n blood_pressure = beta1 * sodium + 2 * age + np.random.normal(size=n)\n proteinuria = alpha1 * sodium + alpha2 * blood_pressure + np.random.normal(size=n)\n hypertension = (blood_pressure >= 140).astype(int) # not used, but could be used for binary outcomes\n return pd.DataFrame({'blood_pressure': blood_pressure, 'sodium': sodium,\n 'age': age, 'proteinuria': proteinuria})\n\ndef estimate_causal_effect(Xt, y, model=LinearRegression(), treatment_idx=0, regression_coef=False):\n model.fit(Xt, y)\n if regression_coef:\n return model.coef_[treatment_idx]\n else:\n Xt1 = pd.DataFrame.copy(Xt)\n Xt1[Xt.columns[treatment_idx]] = 1\n Xt0 = pd.DataFrame.copy(Xt)\n Xt0[Xt.columns[treatment_idx]] = 0\n return (model.predict(Xt1) - model.predict(Xt0)).mean()\n\nbinary_t_df = generate_data(beta1=1.05, alpha1=.4, alpha2=.3, binary_treatment=True, n=10000000)\ncontinuous_t_df = generate_data(beta1=1.05, alpha1=.4, alpha2=.3, binary_treatment=False, n=10000000)\n\nate_est_naive = None\nate_est_adjust_all = None\nate_est_adjust_age = None\n\nfor df, name in zip([binary_t_df, continuous_t_df],\n ['Binary Treatment Data', 'Continuous Treatment Data']):\n print()\n print('### {} ###'.format(name))\n print()\n\n # Adjustment formula estimates\n ate_est_naive = estimate_causal_effect(df[['sodium']], df['blood_pressure'], treatment_idx=0)\n ate_est_adjust_all = estimate_causal_effect(df[['sodium', 'age', 'proteinuria']],\n df['blood_pressure'], treatment_idx=0)\n ate_est_adjust_age = estimate_causal_effect(df[['sodium', 'age']], df['blood_pressure'])\n print('# Adjustment Formula Estimates #')\n print('Naive ATE estimate:\\t\\t\\t\\t\\t\\t\\t', ate_est_naive)\n print('ATE estimate adjusting for all covariates:\\t', ate_est_adjust_all)\n print('ATE estimate adjusting for age:\\t\\t\\t\\t', ate_est_adjust_age)\n print()\n\n # Linear regression coefficient estimates\n ate_est_naive = estimate_causal_effect(df[['sodium']], df['blood_pressure'], treatment_idx=0,\n regression_coef=True)\n ate_est_adjust_all = estimate_causal_effect(df[['sodium', 'age', 'proteinuria']],\n df['blood_pressure'], treatment_idx=0,\n regression_coef=True)\n ate_est_adjust_age = estimate_causal_effect(df[['sodium', 'age']], df['blood_pressure'],\n regression_coef=True)\n print('# Regression Coefficient Estimates #')\n print('Naive ATE estimate:\\t\\t\\t\\t\\t\\t\\t', ate_est_naive)\n print('ATE estimate adjusting for all covariates:\\t', ate_est_adjust_all)\n print('ATE estimate adjusting for age:\\t\\t\\t\\t', ate_est_adjust_age)\n print()",
"\n### Binary Treatment Data ###\n\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
cbcb0a1853c634a40484f6fb3aa9dcbb223654e3
| 15,894 |
ipynb
|
Jupyter Notebook
|
notebook/LDA.ipynb
|
devlarrywong/News-Text-Summarization-Project
|
ea8dedd06e334371fecbf20fb729c66d4ea73f45
|
[
"MIT"
] | null | null | null |
notebook/LDA.ipynb
|
devlarrywong/News-Text-Summarization-Project
|
ea8dedd06e334371fecbf20fb729c66d4ea73f45
|
[
"MIT"
] | null | null | null |
notebook/LDA.ipynb
|
devlarrywong/News-Text-Summarization-Project
|
ea8dedd06e334371fecbf20fb729c66d4ea73f45
|
[
"MIT"
] | null | null | null | 29.216912 | 4,927 | 0.592236 |
[
[
[
"# abc",
"_____no_output_____"
]
],
[
[
"doc_a = \"Brocolli is good to eat. My brother likes to eat good brocolli, but not my mother.\"\ndoc_b = \"My mother spends a lot of time driving my brother around to baseball practice.\"\ndoc_c = \"Some health experts suggest that driving may cause increased tension and blood pressure.\"\ndoc_d = \"I often feel pressure to perform well at school, but my mother never seems to drive my brother to do better.\"\ndoc_e = \"Health professionals say that brocolli is good for your health.\"\n\n# compile sample documents into a list\ndoc_set = [doc_a, doc_b, doc_c, doc_d, doc_e]",
"_____no_output_____"
]
],
[
[
"# Tokenization",
"_____no_output_____"
]
],
[
[
"from nltk.tokenize import RegexpTokenizer\ntokenizer = RegexpTokenizer(r'\\w+')",
"_____no_output_____"
],
[
"raw = doc_a.lower()\ntokens = tokenizer.tokenize(raw)\nprint(tokens)",
"['brocolli', 'is', 'good', 'to', 'eat', 'my', 'brother', 'likes', 'to', 'eat', 'good', 'brocolli', 'but', 'not', 'my', 'mother']\n"
]
],
[
[
"# Stop words",
"_____no_output_____"
]
],
[
[
"from nltk.corpus import stopwords\n# create English stop words list\nen_stop = stopwords.words('english')",
"_____no_output_____"
],
[
"# remove stop words from tokens\nstopped_tokens = [i for i in tokens if not i in en_stop]\n\nprint(stopped_tokens)",
"['brocolli', 'good', 'eat', 'brother', 'likes', 'eat', 'good', 'brocolli', 'mother']\n"
]
],
[
[
"# Stemming",
"_____no_output_____"
]
],
[
[
"from nltk.stem.porter import PorterStemmer\n\n# Create p_stemmer of class PorterStemmer\np_stemmer = PorterStemmer()",
"_____no_output_____"
],
[
"# stem token\ntexts = [p_stemmer.stem(i) for i in stopped_tokens]\nprint(texts)",
"['brocolli', 'good', 'eat', 'brother', 'like', 'eat', 'good', 'brocolli', 'mother']\n"
]
],
[
[
"# Constructing a document-term matrix",
"_____no_output_____"
]
],
[
[
"#!pip install gensim",
"_____no_output_____"
],
[
"a = []\na.append(texts)\na",
"_____no_output_____"
],
[
"from gensim import corpora, models\n\ndictionary = corpora.Dictionary(a)",
"_____no_output_____"
],
[
"corpus = [dictionary.doc2bow(text) for text in a]",
"_____no_output_____"
],
[
"print(corpus[0])",
"[(0, 2), (1, 1), (2, 2), (3, 2), (4, 1), (5, 1)]\n"
]
],
[
[
"# Applying the LDA model",
"_____no_output_____"
]
],
[
[
"ldamodel = models.ldamodel.LdaModel(corpus, num_topics=3, id2word = dictionary, passes=20)",
"_____no_output_____"
]
],
[
[
"# Examining the result",
"_____no_output_____"
]
],
[
[
"print(ldamodel.print_topics(num_topics=3, num_words=3))",
"[(0, '0.167*\"mother\" + 0.167*\"like\" + 0.167*\"brother\"'), (1, '0.212*\"brocolli\" + 0.212*\"eat\" + 0.212*\"good\"'), (2, '0.167*\"good\" + 0.167*\"mother\" + 0.167*\"like\"')]\n"
],
[
"ldamodel = models.ldamodel.LdaModel(corpus, num_topics=2, id2word = dictionary, passes=20)\n\nb = ldamodel.print_topics(num_topics=2, num_words=4)",
"_____no_output_____"
],
[
"len(b)",
"_____no_output_____"
],
[
"for i in b:\n print(i)",
"(0, '0.167*\"brocolli\" + 0.167*\"eat\" + 0.167*\"good\" + 0.167*\"like\"')\n(1, '0.209*\"good\" + 0.209*\"eat\" + 0.209*\"brocolli\" + 0.125*\"brother\"')\n"
]
],
[
[
"-----",
"_____no_output_____"
],
[
"# My Example",
"_____no_output_____"
]
],
[
[
"doc_f = \"The decision to ban lawmaker Eddie Chu Hoi-dick from running in a rural representative election was based on a shaky argument that could be struck down in court, according to leading legal scholars, who also called on Hong Kong’s courts to clarify the vagueness in election laws. Johannes Chan Man-mun, the former law dean of the University of Hong Kong, was speaking on Sunday after Chu was told he would not be allowed to run for a post as a local village’s representative. Returning officer Enoch Yuen Ka-lok pointed to Chu’s stance on Hong Kong independence and said the lawmaker had dodged his questions on his political beliefs. Yuen took this to imply that Chu supported the possibility of Hong Kong breaking with Beijing in the future. Chan, however, said Chu’s responses to the returning officer were open to interpretation. The legal scholar did not believe they met the standard of giving the election officer “cogent, clear and compelling” evidence as required by the precedent set in the case of Andy Chan Ho-tin. Andy Chan was barred from standing in a Legislative Council by-election in New Territories West in 2016 because of his political beliefs. According to Section 24 of the Rural Representative Election Ordinance, candidates are required to declare their allegiance to the Hong Kong Special Administrative Region and to state they will uphold the Basic Law, Hong Kong’s mini-constitution, when filing their application. The allegiance requirement was written into law in 2003, mirroring clauses in the rules for the Legco and district council elections, but it had never been applied by an election officer. The situation changed after separatist Andy Chan lost his election appeal in February this year, with the courts saying returning officers could ban candidates who held political views that ran contrary to the Basic Law. While the landmark ruling was concerned only with Legco elections, Johannes Chan said, after Chu’s case, returning officers for other elections could have similar powers to ban candidates from running, including in the district council elections next year. Gladys Li, the lawyer who represented Andy Chan, said the ruling would be binding on returning officers for other elections. Eric Cheung Tat-ming, another legal scholar at HKU, said Yuen had provided weak reasons for disqualifying Chu. He agreed that there will be room for Chu to launch an appeal. “The logic has become – if your interpretation of the Basic Law is different from the government’s, it means you have no intention of upholding the Basic Law,” Cheung said. He also said Hong Kong courts must clarify the vagueness in election laws and process such appeals more quickly. Stephen Fisher, the former deputy home affairs secretary who led the government’s effort to formalise rural representative elections under the ordinance, said it was “common sense” that rural representatives had to uphold allegiance to Hong Kong. “The village representatives are also elected by people, and they are empowered to identify who the indigenous villagers are,” Fisher said before Chu’s disqualification. “So it’s normal that the legal drafting [of the ordinance] follows the law on Legislative Council and district council elections.” Fisher, who would not comment on Chu’s case, said it would have been “unthinkable” for anyone back then to have imagined a candidate being disqualified for their political views. “The requirement was written there, but it was never contentious,” Fisher said. Chu was disqualified by Yuen because he had “defended independence as an option to Hongkongers” in a statement in 2016. Pressed twice by the returning officer to clarify his position, Chu would say only that he did not support Hong Kong’s independence, but added that he would support another’s right to peacefully advocate it. Johannes Chan said Chu’s political stance was open to interpretation, and the election officer could hardly fulfil the criteria for providing “cogent, clear and compelling” evidence to disqualify him. “At best, we could argue Chu’s reply to the officer was vague about self-determination – even the returning officer himself confessed Chu was only ‘implicitly’ confirming independence as an option,” he said. “But we can’t take a candidate’s silence as his stance. That would have jumped many, many steps.” The decision on Sunday would also create a “conflicting” situation over Chu's political allegiance, Chan added, since the lawmaker remained in office but was disqualified in a separate election. Both Chan and Li said how the returning officer had come to the disqualification might require clarification in any future court ruling. “It was as if they [government officials] could read your mind,” Li said. “The court still has not clarified how far back election officials can look – such as in this case, could we go back to statements Chu made two years ago?” Chan asked.\"",
"_____no_output_____"
]
],
[
[
"# Tokenization",
"_____no_output_____"
]
],
[
[
"from nltk.tokenize import RegexpTokenizer\ntokenizer = RegexpTokenizer(r'\\w+')",
"_____no_output_____"
],
[
"my_raw = doc_f.lower()\nmy_tokens = tokenizer.tokenize(my_raw)\n#print(my_tokens)",
"_____no_output_____"
]
],
[
[
"# Stop words",
"_____no_output_____"
]
],
[
[
"from nltk.corpus import stopwords\n# create English stop words list\neng_stop = stopwords.words('english')",
"_____no_output_____"
],
[
"# remove stop words from tokens\nmy_stopped_tokens = [i for i in my_tokens if not i in eng_stop]\n\n#print(my_stopped_tokens)",
"_____no_output_____"
]
],
[
[
"# Stemming",
"_____no_output_____"
]
],
[
[
"from nltk.stem.porter import PorterStemmer\n\n# Create p_stemmer of class PorterStemmer\np_stemmer = PorterStemmer()",
"_____no_output_____"
],
[
"# stem token\nmy_texts = [p_stemmer.stem(i) for i in my_stopped_tokens]\n#print(texts)",
"_____no_output_____"
],
[
"my_texts_list = []\n#my_texts_list.append(my_texts)\nmy_texts_list.append(my_stopped_tokens)\n#my_texts_list",
"_____no_output_____"
],
[
"from gensim import corpora, models\n\nmy_dictionary = corpora.Dictionary(my_texts_list)",
"_____no_output_____"
],
[
"my_corpus = [my_dictionary.doc2bow(text) for text in my_texts_list]",
"_____no_output_____"
],
[
"corpus[0]",
"_____no_output_____"
]
],
[
[
"# Applying the LDA model",
"_____no_output_____"
]
],
[
[
"my_ldamodel = models.ldamodel.LdaModel(my_corpus, num_topics=3, id2word = my_dictionary, passes=20)",
"_____no_output_____"
],
[
"result = my_ldamodel.print_topics(num_topics=3, num_words=3)",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
cbcb0b0f2f6cbb8502145353db6bd6a20ffa685f
| 69,384 |
ipynb
|
Jupyter Notebook
|
McKinsey-Hackathon/20180310_McKinseyDSHackathon.ipynb
|
FTLiao/KaggleProjects
|
9e290dfd47d7c2b2ab80205f48b338e50451a958
|
[
"MIT"
] | null | null | null |
McKinsey-Hackathon/20180310_McKinseyDSHackathon.ipynb
|
FTLiao/KaggleProjects
|
9e290dfd47d7c2b2ab80205f48b338e50451a958
|
[
"MIT"
] | 2 |
2017-12-23T21:33:57.000Z
|
2018-02-17T13:50:05.000Z
|
McKinsey-Hackathon/20180310_McKinseyDSHackathon.ipynb
|
FTLiao/KaggleProjects
|
9e290dfd47d7c2b2ab80205f48b338e50451a958
|
[
"MIT"
] | null | null | null | 32.137101 | 437 | 0.391603 |
[
[
[
"# McKinsey Data Scientist Hackathon\n\nlink: https://datahack.analyticsvidhya.com/contest/mckinsey-analytics-online-hackathon-recommendation/?utm_source=sendinblue&utm_campaign=Download_The_Dataset_McKinsey_Analytics_Online_Hackathon__Recommendation_Design_is_now_Live&utm_medium=email\n\nslack:https://analyticsvidhya.slack.com/messages/C8X88UJ5P/\n\n\n## Problem Statement ##\n\nYour client is a fast-growing mobile platform, for hosting coding challenges. They have a unique business model, where they crowdsource problems from various creators(authors). These authors create the problem and release it on the client's platform. The users then select the challenges they want to solve. The authors make money based on the level of difficulty of their problems and how many users take up their challenge.\n \nThe client, on the other hand makes money when the users can find challenges of their interest and continue to stay on the platform. Till date, the client has relied on its domain expertise, user interface and experience with user behaviour to suggest the problems a user might be interested in. You have now been appointed as the data scientist who needs to come up with the algorithm to keep the users engaged on the platform.\nThe client has provided you with history of last 10 challenges the user has solved, and you need to predict which might be the next 3 challenges the user might be interested to solve. Apply your data science skills to help the client make a big mark in their user engagements/revenue.\n\n### Data Relationships\nClient: problem platform maintainer\nCreators: problem contributors\nUsers: people who solve these problems\n\nQuestion? Given the 10 challenges the user solved, what might be the next 3 challenges user want to solve?",
"_____no_output_____"
],
[
"## Now let's first look at some raw data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport sklearn\nimport pandas\nimport seaborn",
"_____no_output_____"
],
[
"x_data = pandas.read_csv('./train_mddNHeX/train.csv')\ny_data = pandas.read_csv('./train_mddNHeX/challenge_data.csv')\nx_test = pandas.read_csv('./test.csv')\ny_sub_temp = pandas.read_csv('./sample_submission_J0OjXLi_DDt3uQN.csv')",
"_____no_output_____"
],
[
"print('shape of submission data = {}, number of users = {}'.format(y_sub_temp.shape, y_sub_temp.shape[0]/13))\ny_sub_temp.head()",
"shape of submission data = (119196, 2), number of users = 9168\n"
],
[
"print('shape of user data = {}, number of users = {}'.format(x_data.shape, x_data.shape[0]/13))\n#x_data.sort_values('user_id')\nx_data[0:20]",
"shape of user data = (903916, 4), number of users = 69532\n"
],
[
"print('shape of user test data = {}, number of users = {}'.format(x_test.shape, x_test.shape[0]/10))\n#x_test[0:15]\nx_test.head(15)\n#x_test.sort_values('user_id').head(15)",
"shape of user test data = (397320, 4), number of users = 39732\n"
],
[
"print('shape of challenge data = {}'.format(y_data.shape))\ny_data[0:10]#.tail()\n#print(y_data.loc[:,['challenge_ID','challenge_series_ID']])\n#print(y_data.groupby('challenge_series_ID'))",
"shape of challenge data = (5606, 9)\n"
]
],
[
[
"## Dirty try\n1. Need to find a feature vector for a given challenge \n - This is associated with [prog_lang, challenge_series, total submission, publish_time, auth_id, auth_org, categ]\n2. Create a preference vector for each user \n - This will be randomly initialized\n3. Use the first 10 samples from each users as ground truth for training the feature vector and the preference vector",
"_____no_output_____"
],
[
"## Prepare training data",
"_____no_output_____"
],
[
"Let's prepare the challange id as a lookup table to constuct training data\n",
"_____no_output_____"
]
],
[
[
"def str2ascii(astr):\n \"\"\"\n input: \n astr: a string\n output: \n val: a number which is sum of char's ascii.\n \"\"\"\n val = 0\n real = 0\n count_val, count_real = 0, 0\n for i in list(astr):\n num = ord(i)\n if 48<= num and num <= 57:\n real = real*10 + int(i)\n count_real += 1\n else:\n val += num\n count_val += 1\n val = val*10**count_real + real\n return val",
"_____no_output_____"
],
[
"# Retain the original copy of the y_data\nch_table = y_data\norig_y_data = y_data.copy()",
"_____no_output_____"
],
[
"print(ch_table.columns)",
"Index([u'challenge_ID', u'programming_language', u'challenge_series_ID',\n u'total_submissions', u'publish_date', u'author_ID', u'author_gender',\n u'author_org_ID', u'category_id'],\n dtype='object')\n"
],
[
"## Fill NaN with some values\nvalues = {'challenge_series_ID':'SI0000','author_ID':'AI000000','author_gender':'I'\n ,'author_org_ID':'AOI000000', 'category_id':0.0\n ,'programming_language':0,'total_submissions':0, 'publish_date':'00-00-0000'}\nch_table = y_data.fillna(value = values)\nprint(y_data.head(), ch_table.head())",
"( challenge_ID programming_language challenge_series_ID total_submissions \\\n0 CI23478 2 SI2445 37.0 \n1 CI23479 2 SI2435 48.0 \n2 CI23480 1 SI2435 15.0 \n3 CI23481 1 SI2710 236.0 \n4 CI23482 2 SI2440 137.0 \n\n publish_date author_ID author_gender author_org_ID category_id \n0 06-05-2006 AI563576 M AOI100001 NaN \n1 17-10-2002 AI563577 M AOI100002 32.0 \n2 16-10-2002 AI563578 M AOI100003 NaN \n3 19-09-2003 AI563579 M AOI100004 70.0 \n4 21-03-2002 AI563580 M AOI100005 NaN , challenge_ID programming_language challenge_series_ID total_submissions \\\n0 CI23478 2 SI2445 37.0 \n1 CI23479 2 SI2435 48.0 \n2 CI23480 1 SI2435 15.0 \n3 CI23481 1 SI2710 236.0 \n4 CI23482 2 SI2440 137.0 \n\n publish_date author_ID author_gender author_org_ID category_id \n0 06-05-2006 AI563576 M AOI100001 0.0 \n1 17-10-2002 AI563577 M AOI100002 32.0 \n2 16-10-2002 AI563578 M AOI100003 0.0 \n3 19-09-2003 AI563579 M AOI100004 70.0 \n4 21-03-2002 AI563580 M AOI100005 0.0 )\n"
],
[
"ch_table.iloc[3996]",
"_____no_output_____"
],
[
"## Change strings to some encoded values\ncolumns = ['challenge_series_ID','author_ID','author_gender','author_org_ID','publish_date']\n#print(ch_table[0:10])\nfor col in columns:\n print(col)\n #ch_table[col] = ch_table.apply(lambda x: str2ascii(x[col]),axis=1)\n ch_table[col] = ch_table[col].apply(lambda x: str2ascii(x))",
"challenge_series_ID\nauthor_ID\nauthor_gender\nauthor_org_ID\npublish_date\n"
],
[
"ch_table[0:10]\ny_data['programming_language'].describe()",
"_____no_output_____"
]
],
[
[
"### Now, we need to normalize the table",
"_____no_output_____"
]
],
[
[
"## using normalizer\nfrom sklearn import preprocessing\nnormalizer = preprocessing.Normalizer()\nmin_max_scaler = preprocessing.MinMaxScaler()",
"_____no_output_____"
],
[
"## Decrease the variance between data points in each columns\ncolumns = ch_table.columns\n#print(columns[1:],ch_table.loc[:,columns[1:]])\nch_table.loc[:,columns[1:]].head()\nminmax_ch_table = min_max_scaler.fit_transform(ch_table.loc[:,columns[1:]])\nnorm_ch_table = preprocessing.normalize(ch_table.loc[:,columns[1:]],norm='l2')\n#ch_table.loc[:,columns[1:]] = norm_ch_table",
"_____no_output_____"
],
[
"#ch_table.head()\nprint(pandas.DataFrame(minmax_ch_table, columns=columns[1:]).head(2))\nprint(pandas.DataFrame(norm_ch_table, columns=columns[1:]).head(2))",
" programming_language challenge_series_ID total_submissions publish_date \\\n0 0.5 0.852213 0.000852 0.167498 \n1 0.5 0.848728 0.001106 0.534729 \n\n author_ID author_gender author_org_ID category_id \n0 0.993856 1.0 0.98312 0.000000 \n1 0.993858 1.0 0.98313 0.105263 \n programming_language challenge_series_ID total_submissions publish_date \\\n0 2.219821e-10 0.000173 4.106670e-09 0.999591 \n1 2.217103e-10 0.000173 5.321048e-09 0.999592 \n\n author_ID author_gender author_org_ID category_id \n0 0.015379 8.546312e-09 0.024096 0.000000e+00 \n1 0.015360 8.535848e-09 0.024067 3.547365e-09 \n"
],
[
"## Finally put the scaled data back\nch_table[columns[1:]] = minmax_ch_table",
"_____no_output_____"
],
[
"ch_table.head(10)",
"_____no_output_____"
]
],
[
[
"## Great!, now we have feature vectors for every challenges\n\nNext lets prepare the ground truth matrix for users\n\nShape of y = (n_c, n_u)\n\n 1. n_c: the number of challenges\n 2. n_u: the number of users",
"_____no_output_____"
]
],
[
[
"## The ch_features contains \nch_features = ch_table.sort_values('challenge_ID')\nch_features = ch_features.loc[:,columns[1:]].values\n#ch_features.head(10)\nprint('Shape of feature (n_c, n_f) = {}'.format(ch_features.shape))",
"Shape of feature (n_c, n_f) = (5606, 8)\n"
],
[
"## Setting up the lookup table\nch_lookup = {}\ntmp = ch_table['challenge_ID'].to_dict()\nch_id_lookup=tmp\n#for key in tmp.keys\nfor key in tmp.keys():\n #print(key, tmp[key])\n ch_lookup[tmp[key]] = key\n#ch_lookup",
"_____no_output_____"
],
[
"## now lets set up a training y array with shape = (n_c, n_u)\ndef findChallengeFeatures(challenge_id, table, lookup):\n \"\"\"\n input: \n challenge_id: a string of the challenge_id\n table: pandas dataframe lookup table\n output:\n features: numpy array of features\n \"\"\"\n columns = table.columns\n return table.loc[lookup[challenge_id], columns[1:]]",
"_____no_output_____"
],
[
"%%time\nch_table.head()\nfeatureVec = findChallengeFeatures(x_data.loc[0,'challenge'],ch_table, ch_lookup)\nprint(featureVec.shape)",
"(8,)\nCPU times: user 68 ms, sys: 36.5 ms, total: 105 ms\nWall time: 117 ms\n"
],
[
"%%time\nfrom operator import itemgetter\n#myvalues = itemgetter(*mykeys)(mydict)\ncolumns = ch_table.columns.values\nusr_table = x_data\nprint(columns[1:])\nfor i in columns[1:]:\\\n usr_table[i] = np.nan\nnSamples = x_data.shape[0]\n## Finding indices\nindices = np.array([ch_lookup[i] for i in x_data.loc[:nSamples-1,'challenge']])\nprint(indices.shape)\nusr_table.loc[:nSamples-1, columns[1:]] = ch_table.loc[indices, columns[1:]].values\n#print(ch_table.loc[indices,columns[1:3]])\n#print(usr_table.loc[:nSamples-1, columns[1:]].shape)",
"['programming_language' 'challenge_series_ID' 'total_submissions'\n 'publish_date' 'author_ID' 'author_gender' 'author_org_ID' 'category_id']\n(903916,)\nCPU times: user 1.33 s, sys: 333 ms, total: 1.66 s\nWall time: 1.77 s\n"
],
[
"usr_table.head(15)\nusr_table.to_csv('train_withFeatureVec_allsamples.csv')\nch_table.to_csv('challenge_featureVecTable_allsamples.csv')",
"_____no_output_____"
]
],
[
[
"## Let's prepare the labels\n\nFirst, we need an empyty array to hold challenges",
"_____no_output_____"
]
],
[
[
"ch_emptyVec = np.zeros((ch_table.shape[0]))\nch_emptyVec.shape",
"_____no_output_____"
],
[
"x_data.head(13)",
"_____no_output_____"
],
[
"## constructing a (n_u, n_c) array for nSamples\n%%time\n\ncolumns = ch_table.columns\nnSamples = int(x_data.shape[0])\nx_train = np.zeros((nSamples, 10, len(ch_table.columns)-1)) ## (m, n_i, n_f)\ny_train = np.zeros((nSamples, ch_table.shape[0])) ## (m, n_c)\nfor i in range(nSamples/13): \n curpt = i*13\n #print(i)\n x_train[i] = x_data.loc[curpt:(curpt+9), columns[1:]] ## 0-10, 13-26\n #print(x_train[i].shape)\n #y_train[i] = ch_emptyVec\n #tmp = x_data.loc[(curpt+10):(curpt+12), 'challenge'].values\n #tmp = [ch_lookup[tmp[0]],ch_lookup[tmp[1]],ch_lookup[tmp[2]]]\n #y_train[i,tmp] = 1 ## 10-13, 26-29\n indices = [int(ch_lookup[j]) for j in x_data.loc[(curpt+10):(curpt+12), 'challenge']] \n #print(indices, np.ones(3), tmp)\n y_train[i, indices] = 1 ## 10-13, 26-29\n #break\nprint('x_train shape = {}, y_train shape = {}'.format(x_train.shape, y_train.shape))\n ",
"x_train shape = (903916, 10, 8), y_train shape = (903916, 5606)\n"
],
[
"## Flatten the array\nx_train = x_train.reshape((x_data.shape[0],-1))",
"_____no_output_____"
]
],
[
[
"## Finally lets dunmp it into a classifier",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import GaussianNB\nfrom sklearn import tree\ngnb = GaussianNB()\nclf = tree.DecisionTreeClassifier()\n",
"_____no_output_____"
],
[
"clf.fit(x_train, y_train)",
"_____no_output_____"
],
[
"## A simple NN\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Activation\n\nmodel = Sequential()\nmodel.add(Dense(64, input_dim=80))\nmodel.add(Activation('relu'))\nmodel.add(Dense(128))\nmodel.add(Activation('relu'))\nmodel.add(Dense(256))\nmodel.add(Activation('relu'))\nmodel.add(Dense(512))\nmodel.add(Activation('relu'))\nmodel.add(Dense(1024))\nmodel.add(Activation('relu'))\nmodel.add(Dense(y_train.shape[1]))\nmodel.add(Activation('softmax'))\n\n",
"Using TensorFlow backend.\n"
],
[
"model.compile(optimizer='adam',\n loss='categorical_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"model.fit(x_train, y_train, epochs=1)",
"Epoch 1/1\n903916/903916 [==============================] - 5157s 6ms/step - loss: 1.4728 - acc: 0.0011\n"
],
[
"model.save_weights('simpleNN.h5')",
"_____no_output_____"
]
],
[
[
"\n## Running out of time just gonna plug it in and submit",
"_____no_output_____"
]
],
[
[
"%%time\nnSamples = x_test.shape[0]\ncolumns = ch_table.columns\ntest_table = x_test\nprint(columns[1:]) \nfor i in columns[1:]:\n test_table[i] = np.nan\nindices = np.array([ch_lookup[i] for i in x_test.loc[:nSamples-1,'challenge']])\ntest_table.loc[:nSamples-1, columns[1:]] = ch_table.loc[indices, columns[1:]].values\nprint(indices.shape)\n",
"Index([u'programming_language', u'challenge_series_ID', u'total_submissions',\n u'publish_date', u'author_ID', u'author_gender', u'author_org_ID',\n u'category_id'],\n dtype='object')\n(397320,)\nCPU times: user 647 ms, sys: 412 ms, total: 1.06 s\nWall time: 1.66 s\n"
],
[
"%%time\ntest_table.to_csv('prepared_test_table_for_prediction.csv')",
"CPU times: user 2.92 s, sys: 274 ms, total: 3.19 s\nWall time: 3.93 s\n"
],
[
"x_submit = np.zeros((nSamples/10, 10, len(ch_table.columns)-1)) ## (m, n_i, n_f)\ny_submit = pandas.DataFrame(columns=['user_sequence','challenge'], \n data = np.empty((x_test.shape[0]/10*3,2), dtype=np.str))\n#y_submit['user_sequence']\n#y_submit.head(15)",
"_____no_output_____"
],
[
"%%time\nfor i in range(nSamples/10): \n curpt = i*10\n #print(i)\n x_submit[i] = x_test.loc[curpt:(curpt+9), columns[1:]] ## 0-10, 13-26\n pred = model.predict(x_submit[i].reshape((1,80)))\n ids = np.argsort(pred.reshape(-1))[-3:]\n #print(pred, ids, ids.shape)\n #print(pred[0,ids])\n \n outpt = i*3\n user_id = x_test.loc[curpt,'user_id']\n y_submit.iloc[outpt:outpt+3,:] = [[str(user_id)+'_11', ch_id_lookup[ids[0]]],\n [str(user_id)+'_12', ch_id_lookup[ids[1]]],\n [str(user_id)+'_13', ch_id_lookup[ids[2]]]\n ]\n #print(y_submit.iloc[outpt:outpt+3,:])\ny_submit.head()",
"CPU times: user 6min 9s, sys: 30.8 s, total: 6min 40s\nWall time: 6min 29s\n"
],
[
"y_submit.head(15)",
"_____no_output_____"
],
[
"y_submit.to_csv('ftl_submission.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.