hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
4a12697bd20ecf05dd79c37739ad90c58d3b3bec
| 30,134 |
ipynb
|
Jupyter Notebook
|
notebooks/workshop-6/variational_inference.ipynb
|
Imperial-College-Data-Science-Society/workshops
|
8ce86f9f70ab47644e1f2e988da3bac41c0b9625
|
[
"MIT"
] | 17 |
2020-08-05T03:17:50.000Z
|
2021-02-06T05:07:28.000Z
|
notebooks/workshop-6/variational_inference.ipynb
|
Imperial-College-Data-Science-Society/Workshops
|
8ce86f9f70ab47644e1f2e988da3bac41c0b9625
|
[
"MIT"
] | 4 |
2021-01-31T09:23:26.000Z
|
2022-03-12T00:50:02.000Z
|
notebooks/workshop-6/variational_inference.ipynb
|
Imperial-College-Data-Science-Society/workshops
|
8ce86f9f70ab47644e1f2e988da3bac41c0b9625
|
[
"MIT"
] | 12 |
2020-10-15T17:16:58.000Z
|
2021-03-04T18:27:39.000Z
| 65.084233 | 5,712 | 0.702496 |
[
[
[
"# Variational Inference and Learning in the Big Data Regime\n\nMany real-world modelling solutions require fitting models with large numbers of data-points and parameters, which is made convenient recently through software implementing automatic differentiation, but also require uncertainty quantification. Variational inference is a generic family of tools that reformulates (Bayesian) model inference into an optimisation problem, thereby making use of modern software tools but also having the ability to give model uncertainty. This talk will motivate how variational inference works and what the state-of-the-art methods are. We will also accompany the theory with implementations on some simple probabilistic models, such as variational autoencoders (VAE). If time-permitting, we will briefly talk about some of the recent frontiers of variational inference, namely normalising flows and Stein Variational Gradient Descent.\n\n \n\n💻 Content covered:\n\nCurrent inference methods: maximum likelihood and Markov chain Monte Carlo\n\nInformation theory and KL divergence\n\nMean field variational inference\n\nBayesian linear regression\n\nMonte Carlo variational inference (MCVI), reparameterisation trick and law of the unconscious statistician (LOTUS)\n\nExample software implementations: VAE\n\n👾 This lecture will be held online on Microsoft Teams.\n\n🔴The event will be recorded and will be publicly available.\n\n🎉 Attendance is FREE for members! Whether you are a student at Imperial College or not, sign up to be a member at www.icdss.club/joinus\n\n⭐️ We encourage participants of this workshop to have looked at our previous sessions on YouTube. Prerequisites: basic understanding of Bayesian statistics\n\n📖 A schedule of our lecture series is currently available",
"_____no_output_____"
],
[
"## Background\n\n- Variational Inference: A Review for Statisticians: https://www.tandfonline.com/doi/full/10.1080/01621459.2017.1285773\n- Auto-Encoding Variational Bayes: https://arxiv.org/pdf/1312.6114.pdf\n- http://yingzhenli.net/home/en/approximateinference\n- https://github.com/ethanluoyc/pytorch-vae\n\nConsider crop yields $y$ and we have a likelihood $p(y|z)$ where $z$ are latent parameters. Suppose $z$ has some prior distribution $p(z)$, then the posterior distribution is\n$$\np(z|y) \\propto p(y|z)p(z) := \\tilde{p}(z|y).\n$$\n\nWe then want to be able to compute quantities $\\mathbb{E}_{z\\sim p(z|y)}[h(Z)]$, for certain functions $h$ e.g. $h(z)=z$ for the posterior mean of $Z$.\n\nWe could compute $p(z|y$) analytically if we have nice priors (conjugate priors), but this is usually not the case for most models e.g. Autoencoders with latent parameters or certain Gaussian mixture models. \n\nMarkov chain Monte Carlo (MCMC) allows us to obtain samples from $z\\sim p(z|y)$ using samplers (e.g. Hamiltonian Monte Carlo (HMC) or Metropolis-Hastings), but it could be very expensive and prohibits it from being used for the big data setting.",
"_____no_output_____"
],
[
"### Variational Inference\n\nVariational Inference (VI)/Variational Bayes/Variational Approximation turns this problem into an optimisation problem. We now seek $q(z)$ in a space of functions $\\mathcal{Q}$, instead of computing the exact $p(z|y)$, in which\n\n$$KL(q(z) || p(z|y)) = \\int \\log\\frac{q(z)}{p(z|y)} q(z) dq$$\n\nis minimised. This KL denotes the KL-divergence, which is a divergence measure that looks at how close 2 distributions are to one-another. It is:\n\n- Non-negative\n- Is equal to 0 if and only if $q(z) = p(z|y)$\n- Note: $KL(q(z)||p(z|y)) \\neq KL(p(z|y) || q(z))$. Minimising $KL(p(z|y) || q(z))$ is the objective of Expectation Propagation, which is another method for approximating posterior distributions.\n\nNote that maximum likelihood estimation (MLE) is done by maximising the log-likelihood, which is the same as minimising the KL divergence:\n\n$$\n\\text{argmin}_{\\theta} KL(\\hat{p}(y|\\theta^*) || p(y|\\theta)) = \\text{argmin}_{\\theta} \\frac{1}{n}\\sum_{i=1}^n \\log \\frac{p(y_i|\\hat{\\theta})}{p(y_i|\\theta)} = \\text{argmin}_{\\theta} \\frac{1}{n}\\sum_{i=1}^n \\log \\frac{1}{p(y_i|\\theta)} = \\text{argmax}_{\\theta} \\frac{1}{n}\\sum_{i=1}^n \\log p(y_i|\\theta).\n$$\n\n**Evidence Lower-Bound**\n\nSuppose I pose a family of posteriors $q(z)$, then\n\n\\begin{align*}\nKL(q(z) || p(z|y)) = \\int \\log\\frac{q(z)}{p(z|y)} q(z) dq &= \\mathbb{E}_{z\\sim q(z)}[\\log q(z)] - \\mathbb{E}_{z\\sim q(z)}[\\log p(z|y)] \\\\\n&= \\mathbb{E}_{z\\sim q(z)}[\\log q(z)] - \\mathbb{E}_{z\\sim q(z)}[\\log p(z,y)] + \\log p(y) \\\\\n&= \\mathbb{E}_{z\\sim q(z)}[\\log q(z)] - \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)] - \\mathbb{E}_{z\\sim q(z)}[p(z)] + \\log p(y) \\\\\n&=\\log p(y) + \\mathbb{E}_{z\\sim q(z)}[\\log \\frac{q(z)}{p(z)}] - \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)] \\\\\n&= \\log p(y) + KL(q(z) || p(z)) - \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)].\n\\end{align*}\n\nSince the left term is positive and $\\log p(y)$ is fixed, it is sufficient to minimise:\n\n$$\nKL(q(z) || p(z)) - \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)].\n$$\n\nThe evidence lower-bound is $ELBO(q) = \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)] - KL(q(z) || p(z))$, which is maximised.\n",
"_____no_output_____"
],
[
"### Mean-Field Variational Inference\n\nAs fancy as it sounds, it just means specifying a family of posteriors $\\mathcal{Q}$ such that \n\n$$\nq(z) = \\prod_{j=1}^m q_j(z_j),\n$$\nwhere $m$ is the number of parameters.\n\n**Coordinate Ascent Variational Inference (CAVI)**\nBlei et al. (2017)\n\n\nLet's look at an example (Li (2021)):\n$$\ny|x \\sim \\mathcal{N}(y; x^\\intercal\\theta, \\sigma^2),\\qquad \\theta\\sim\\mathcal{N}(\\theta; \\mu_0, \\Gamma_0^{-1}).\n$$\nThis has an analytical solution\n$$\np(\\theta|\\mathcal{D}) = \\mathcal{N}(\\theta; \\mu,\\Gamma^{-1})\n$$\nwith\n\\begin{align*}\n\\Gamma &= \\Gamma_0 + \\frac{1}{\\sigma^2}X^\\intercal X \\\\\n\\mu &= \\frac{1}{\\sigma^2}(X^\\intercal X + \\Gamma_0)^{-1}X^Ty,\n\\end{align*}\n\nwhere $X=(x_1,\\ldots,x_n)^\\intercal$ and $y=(y_1,\\ldots,y_n)^\\intercal$. **Let's try CAVI**:\n\n\\begin{align*}\n\\log q_1(\\theta_1) =& \\int q_2(\\theta_2) \\log \\tilde{p}(\\theta_1, \\theta_2) d\\theta_2\\\\\n=& \\int -\\frac{1}{2}\\left[(\\theta_1-\\mu_1)^2\\Gamma_{11} + 2(\\theta_1-\\mu_1)\\Gamma_{12}(\\theta_2-\\mu_2) \\right]q_2(\\theta_2) d\\theta_2 + const \\\\\n=& -\\frac{1}{2}\\left[(\\theta_1-\\mu_1)^2\\Gamma_{11} + 2(\\theta_1-\\mu_1)\\Gamma_{12}(\\mathbb{E}_{\\theta_2\\sim q_2}[\\theta_2]-\\mu_2) \\right] + const,\n\\end{align*}\nwhich is Gaussian with mean and variance\n$$\n\\tilde{\\mu}_1 = \\mu_1 - \\Gamma_{11}^{-1}\\Gamma_{12}(\\mathbb{E}_{q_2}[\\theta_2] - \\mu_2),\\qquad \\tilde{\\gamma}_2^{-1} = \\Gamma_{11}.\n$$\nSimilarly, you can obtain a similar expression for $q_2(\\theta_2)$. For CAVI to convergence, it can be shown that $(\\tilde{\\mu}_1, \\tilde{\\mu}_2)^\\intercal = \\mu$, giving\n\n$$\n\\tilde{\\mu}_1 = \\mu_1, \\qquad \\tilde{\\mu}_2 = \\mu_2.\n$$\n\nIn this case, CAVI gives a Gaussian posteriors.",
"_____no_output_____"
],
[
"### Monte Carlo Variational Inference (MCVI)\n\nFor big data situations, the variational expectation term can be (1) very expensive and (2) is not available in closed form. We can also add some more complexity to the posterior instead of just having a mean-field approximation. Recall the bound:\n\n$$\n\\mathcal{L}(q; p) = KL(q(z) || p(z)) - \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)].\n$$\n\nMCVI calculates the variational expectation using Monte Carlo integration\n$$\n\\mathbb{E}_{z\\sim q(z)}[\\log p(y_i|z)] \\approx \\frac{1}{M}\\sum_{j=1}^M \\log p(y_i|z^j),\\qquad z^j\\sim q(z).\n$$\nEven better, we can calculate this using mini-batches:\n$$\n\\sum_{i=1}^n\\mathbb{E}_{z\\sim q(z)}[\\log p(y_i|z)] = \\mathbb{E}_{S\\sim \\{1,\\ldots,n\\}}\\left[\\frac{n}{|S|}\\sum_{i\\in S} \\mathbb{E}_q[\\log p(y_i|z)] \\right],\n$$\n\nwhere the inner expectation can be calculated as before. Now, to minimise $\\mathcal{L}(q; p)$, we differentiate with respect to the parameters, let's call it $\\theta$. Therefore, we need\n\n\\begin{align*}\n\\nabla_\\theta \\mathcal{L}(q; p) =& \\nabla_\\theta\\left[KL(q(z) || p(z)) - \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)] \\right] \\\\\n=& \\nabla_\\theta \\left[ \\frac{1}{M}\\sum_{j=1}^M \\log\\frac{q(z^j)}{p(z^j)} \\right] - \\nabla_\\theta\\left[\\mathbb{E}_{S\\sim \\{1,\\ldots,n\\}}\\left[\\frac{n}{|S|}\\sum_{i\\in S} \\frac{1}{M}\\sum_{j=1}^M \\log p(y_i|z^j)\\right] \\right],\n\\end{align*}\n\nwhere $z^j\\sim q(z)$. We can get rid of the expectation with respect to the mini-batches and get a nice approximation for the bound for each batch $S$.\n\n**Reparameterisation Trick/Law of the Unconcious Statistician (LOTUS)**\nLOTUS basically refers to the identity:\n\n$$\nE_X[f(X)] = \\int f(x) p(x) dx = \\int f(g(\\epsilon)) p(\\epsilon) dx = E_\\epsilon[f(g(\\epsilon))]\n$$\n\nfor $x=g(\\epsilon)$, via the inverse function theorem and the change of variable theorem. The reparameterisation trick thus makes it easier to compute the bound by allowing us to sample from a simpler distribution $p(\\epsilon)$ to get $q(z)$:\n\n\\begin{align*}\n\\nabla_\\theta \\mathcal{L}(q; p) =& \\nabla_\\theta\\left[KL(q(z) || p(z)) - \\mathbb{E}_{z\\sim q(z)}[\\log p(y|z)] \\right] \\\\\n=& \\nabla_\\theta\\left[KL(q(z) || p(z)) - \\mathbb{E}_{\\epsilon}[\\log p(y|g_\\theta(\\epsilon))] \\right]\\\\\n=& \\nabla_\\theta KL(q(z) || p(z)) - \\mathbb{E}_{\\epsilon}[\\nabla_g \\log p(y|g_\\theta(\\epsilon)) \\times \\nabla_\\theta g_\\theta(\\epsilon)].\n\\end{align*}\n\nThen repeat using the same MCVI integration method to approximate the variational expectation. In practice, we can also use automatic differentiation to calculate the gradients.\n\n\n**Example: Variational Autoencoders (VAEs)**\n\nModel (Taken from https://lilianweng.github.io/lil-log/2018/08/12/from-autoencoder-to-beta-vae.html)\n\n\n\n**(1)**\nThe decoder represents the likelihood $p(y|z)$, where $y$ is an image. In the upcoming example, we have\n\n$$\n\\log p(y|z) = \\log N(y; f_\\theta(z), I) \\equiv ||y - f_\\theta(z)||_2^2,\n$$\n\nthe MSE loss.\n\n**(2)**\nThe prior is $z\\sim \\mathcal{N}(0, I)$. \n\n**(3)**\nAs you will see in many applications, they people only use 1 sample to calculate the variational expectation. i.e. taking $M=1$.\n\n**(4)**\nThe variational distribution that we are going for is $$q(z|y) = N(g_\\phi(y)[0], g_\\phi(y)[1] I),$$\nwhere the variational distribution is parameterised by the encoder network.\n\n**(5)**\nWe note that we can actually analytically compute the KL divergence as they are 2 Gaussians (proceed to Wikipedia for the formula...)\n",
"_____no_output_____"
],
[
"## Experiments",
"_____no_output_____"
]
],
[
[
"# from https://github.com/ethanluoyc/pytorch-vae/blob/master/vae.py\n\nimport torch\nfrom torch.autograd import Variable\nimport numpy as np\nimport torch.nn.functional as F\nimport torchvision\nfrom torchvision import transforms\nimport torch.optim as optim\nfrom torch import nn\nimport matplotlib.pyplot as plt\nfrom six.moves import urllib\nopener = urllib.request.build_opener()\nopener.addheaders = [('User-agent', 'Mozilla/5.0')]\nurllib.request.install_opener(opener)\n\nclass Normal(object):\n def __init__(self, mu, sigma, log_sigma, v=None, r=None):\n self.mu = mu\n self.sigma = sigma # either stdev diagonal itself, or stdev diagonal from decomposition\n self.logsigma = log_sigma\n dim = mu.get_shape()\n if v is None:\n v = torch.FloatTensor(*dim)\n if r is None:\n r = torch.FloatTensor(*dim)\n self.v = v\n self.r = r\n\n\nclass Encoder(torch.nn.Module):\n def __init__(self, D_in, H, D_out):\n super(Encoder, self).__init__()\n self.linear1 = torch.nn.Linear(D_in, H)\n self.linear2 = torch.nn.Linear(H, D_out)\n\n def forward(self, x):\n x = F.relu(self.linear1(x))\n return F.relu(self.linear2(x))\n\n\nclass Decoder(torch.nn.Module):\n def __init__(self, D_in, H, D_out):\n super(Decoder, self).__init__()\n self.linear1 = torch.nn.Linear(D_in, H)\n self.linear2 = torch.nn.Linear(H, D_out)\n\n def forward(self, x):\n x = F.relu(self.linear1(x))\n return F.relu(self.linear2(x))\n\n\nclass VAE(torch.nn.Module):\n latent_dim = 8\n\n def __init__(self, encoder, decoder):\n super(VAE, self).__init__()\n self.encoder = encoder\n self.decoder = decoder\n self._enc_mu = torch.nn.Linear(100, 8)\n self._enc_log_sigma = torch.nn.Linear(100, 8)\n\n def _sample_latent(self, h_enc):\n \"\"\"\n Return the latent normal sample z ~ N(mu, sigma^2)\n \"\"\"\n mu = self._enc_mu(h_enc)\n log_sigma = self._enc_log_sigma(h_enc)\n sigma = torch.exp(log_sigma)\n std_z = torch.from_numpy(np.random.normal(0, 1, size=sigma.size())).float()\n\n self.z_mean = mu\n self.z_sigma = sigma\n\n return mu + sigma * Variable(std_z, requires_grad=False) # Reparameterization trick\n\n def forward(self, state):\n h_enc = self.encoder(state)\n z = self._sample_latent(h_enc)\n return self.decoder(z)\n\n\ndef latent_loss(z_mean, z_stddev):\n mean_sq = z_mean * z_mean\n stddev_sq = z_stddev * z_stddev\n return 0.5 * torch.mean(mean_sq + stddev_sq - torch.log(stddev_sq) - 1)\n\n\n\ninput_dim = 28 * 28\nbatch_size = 32\n\ntransform = transforms.Compose(\n [transforms.ToTensor()])\nmnist = torchvision.datasets.MNIST('./', download=True, transform=transform)\n\ndataloader = torch.utils.data.DataLoader(mnist, batch_size=batch_size,\n shuffle=True, num_workers=2)\n\nprint('Number of samples: ', len(mnist))\n\nencoder = Encoder(input_dim, 100, 100)\ndecoder = Decoder(8, 100, input_dim)\nvae = VAE(encoder, decoder)\n\ncriterion = nn.MSELoss()\n\noptimizer = optim.Adam(vae.parameters(), lr=0.001)\nl = None\nfor epoch in range(5):\n for i, data in enumerate(dataloader, 0):\n inputs, classes = data\n inputs, classes = Variable(inputs.resize_(batch_size, input_dim)), Variable(classes)\n optimizer.zero_grad()\n dec = vae(inputs)\n ll = latent_loss(vae.z_mean, vae.z_sigma)\n loss = criterion(dec, inputs) + ll\n loss.backward()\n optimizer.step()\n l = loss.item()\n print(epoch, l)\n\nplt.imshow(vae(inputs).data[0].numpy().reshape(28, 28), cmap='gray')\nplt.show(block=True)",
"Number of samples: 60000\n0 0.07181154936552048\n1 0.07108696550130844\n2 0.0702378898859024\n3 0.06724850833415985\n4 0.07055552303791046\n"
],
[
"plt.imshow(inputs[0].numpy().reshape(28, 28), cmap='gray')",
"_____no_output_____"
]
],
[
[
"### Normalising Flows\n\nUsing a \"nice\" class of diffeomorphisms, one can obtain diagonal Jacobians from the diffeomorphisms, we apply the change of variables formula:\n\\begin{align*}\nq(z_L) = q(z) \\prod_{l=1}^L |\\det(\\nabla_{z_{l-1}} T_l(z_{l-1}))|^{-1}\n\\end{align*}",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a1271caef0ec888281eb787c403207f855f889a
| 12,058 |
ipynb
|
Jupyter Notebook
|
jupyter/paddlepaddle/face_mask_detection_paddlepaddle_zh.ipynb
|
dandansamax/djl
|
3c4262de4c60922a2f7ce22fd7c686cff62c24f8
|
[
"Apache-2.0"
] | null | null | null |
jupyter/paddlepaddle/face_mask_detection_paddlepaddle_zh.ipynb
|
dandansamax/djl
|
3c4262de4c60922a2f7ce22fd7c686cff62c24f8
|
[
"Apache-2.0"
] | null | null | null |
jupyter/paddlepaddle/face_mask_detection_paddlepaddle_zh.ipynb
|
dandansamax/djl
|
3c4262de4c60922a2f7ce22fd7c686cff62c24f8
|
[
"Apache-2.0"
] | null | null | null | 34.15864 | 202 | 0.560541 |
[
[
[
"# 用飛槳+ DJL 實作人臉口罩辨識\n在這個教學中我們將會展示利用 PaddleHub 下載預訓練好的 PaddlePaddle 模型並針對範例照片做人臉口罩辨識。這個範例總共會分成兩個步驟:\n\n- 用臉部檢測模型識別圖片中的人臉(無論是否有戴口罩) \n- 確認圖片中的臉是否有戴口罩\n\n這兩個步驟會包含使用兩個 Paddle 模型,我們會在接下來的內容介紹兩個模型對應需要做的前後處理邏輯\n\n## 導入相關環境依賴及子類別\n在這個例子中的前處理飛槳深度學習引擎需要搭配 DJL 混合模式進行深度學習推理,原因是引擎本身沒有包含 NDArray 操作,因此需要藉用其他引擎的 NDArray 操作能力來完成。這邊我們導入 PyTorch 來做協同的前處理工作:",
"_____no_output_____"
]
],
[
[
"// %mavenRepo snapshots https://oss.sonatype.org/content/repositories/snapshots/\n\n%maven ai.djl:api:0.17.0\n%maven ai.djl.paddlepaddle:paddlepaddle-model-zoo:0.17.0\n%maven org.slf4j:slf4j-simple:1.7.32\n\n// second engine to do preprocessing and postprocessing\n%maven ai.djl.pytorch:pytorch-engine:0.17.0",
"_____no_output_____"
],
[
"import ai.djl.*;\nimport ai.djl.inference.*;\nimport ai.djl.modality.*;\nimport ai.djl.modality.cv.*;\nimport ai.djl.modality.cv.output.*;\nimport ai.djl.modality.cv.transform.*;\nimport ai.djl.modality.cv.translator.*;\nimport ai.djl.modality.cv.util.*;\nimport ai.djl.ndarray.*;\nimport ai.djl.ndarray.types.Shape;\nimport ai.djl.repository.zoo.*;\nimport ai.djl.translate.*;\n\nimport java.io.*;\nimport java.nio.file.*;\nimport java.util.*;",
"_____no_output_____"
]
],
[
[
"## 臉部偵測模型\n現在我們可以開始處理第一個模型,在將圖片輸入臉部檢測模型前我們必須先做一些預處理:\n•\t調整圖片尺寸: 以特定比例縮小圖片\n•\t用一個數值對縮小後圖片正規化\n對開發者來說好消息是,DJL 提供了 Translator 介面來幫助開發做這樣的預處理. 一個比較粗略的 Translator 架構如下:\n\n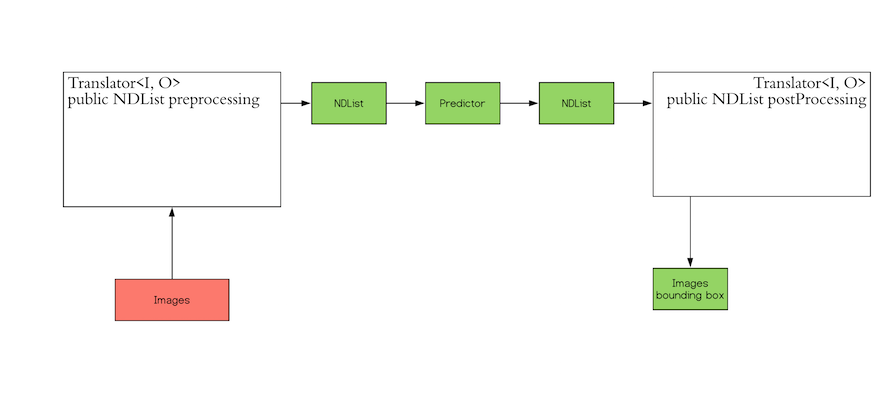\n\n在接下來的段落,我們會利用一個 FaceTranslator 子類別實作來完成工作\n### 預處理\n在這個階段我們會讀取一張圖片並且對其做一些事先的預處理,讓我們先示範讀取一張圖片:",
"_____no_output_____"
]
],
[
[
"String url = \"https://raw.githubusercontent.com/PaddlePaddle/PaddleHub/release/v1.5/demo/mask_detection/python/images/mask.jpg\";\nImage img = ImageFactory.getInstance().fromUrl(url);\nimg.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"接著,讓我們試著對圖片做一些預處理的轉換:",
"_____no_output_____"
]
],
[
[
"NDList processImageInput(NDManager manager, Image input, float shrink) {\n NDArray array = input.toNDArray(manager);\n Shape shape = array.getShape();\n array = NDImageUtils.resize(\n array, (int) (shape.get(1) * shrink), (int) (shape.get(0) * shrink));\n array = array.transpose(2, 0, 1).flip(0); // HWC -> CHW BGR -> RGB\n NDArray mean = manager.create(new float[] {104f, 117f, 123f}, new Shape(3, 1, 1));\n array = array.sub(mean).mul(0.007843f); // normalization\n array = array.expandDims(0); // make batch dimension\n return new NDList(array);\n}\n\nprocessImageInput(NDManager.newBaseManager(), img, 0.5f);",
"_____no_output_____"
]
],
[
[
"如上述所見,我們已經把圖片轉成如下尺寸的 NDArray: (披量, 通道(RGB), 高度, 寬度). 這是物件檢測模型輸入的格式\n### 後處理\n當我們做後處理時, 模型輸出的格式是 (number_of_boxes, (class_id, probability, xmin, ymin, xmax, ymax)). 我們可以將其存入預先建立好的 DJL 子類別 DetectedObjects 以便做後續操作. 我們假設有一組推論後的輸出是 ((1, 0.99, 0.2, 0.4, 0.5, 0.8)) 並且試著把人像框顯示在圖片上",
"_____no_output_____"
]
],
[
[
"DetectedObjects processImageOutput(NDList list, List<String> className, float threshold) {\n NDArray result = list.singletonOrThrow();\n float[] probabilities = result.get(\":,1\").toFloatArray();\n List<String> names = new ArrayList<>();\n List<Double> prob = new ArrayList<>();\n List<BoundingBox> boxes = new ArrayList<>();\n for (int i = 0; i < probabilities.length; i++) {\n if (probabilities[i] >= threshold) {\n float[] array = result.get(i).toFloatArray();\n names.add(className.get((int) array[0]));\n prob.add((double) probabilities[i]);\n boxes.add(\n new Rectangle(\n array[2], array[3], array[4] - array[2], array[5] - array[3]));\n }\n }\n return new DetectedObjects(names, prob, boxes);\n}\n\nNDArray tempOutput = NDManager.newBaseManager().create(new float[]{1f, 0.99f, 0.1f, 0.1f, 0.2f, 0.2f}, new Shape(1, 6));\nDetectedObjects testBox = processImageOutput(new NDList(tempOutput), Arrays.asList(\"Not Face\", \"Face\"), 0.7f);\nImage newImage = img.duplicate();\nnewImage.drawBoundingBoxes(testBox);\nnewImage.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"### 生成一個翻譯器並執行推理任務\n透過這個步驟,你會理解 DJL 中的前後處理如何運作,現在讓我們把前數的幾個步驟串在一起並對真實圖片進行操作:",
"_____no_output_____"
]
],
[
[
"class FaceTranslator implements NoBatchifyTranslator<Image, DetectedObjects> {\n\n private float shrink;\n private float threshold;\n private List<String> className;\n\n FaceTranslator(float shrink, float threshold) {\n this.shrink = shrink;\n this.threshold = threshold;\n className = Arrays.asList(\"Not Face\", \"Face\");\n }\n\n @Override\n public DetectedObjects processOutput(TranslatorContext ctx, NDList list) {\n return processImageOutput(list, className, threshold);\n }\n\n @Override\n public NDList processInput(TranslatorContext ctx, Image input) {\n return processImageInput(ctx.getNDManager(), input, shrink);\n }\n}",
"_____no_output_____"
]
],
[
[
"要執行這個人臉檢測推理,我們必須先從 DJL 的 Paddle Model Zoo 讀取模型,在讀取模型之前我們必須指定好 `Crieteria` . `Crieteria` 是用來確認要從哪邊讀取模型而後執行 `Translator` 來進行模型導入. 接著,我們只要利用 `Predictor` 就可以開始進行推論",
"_____no_output_____"
]
],
[
[
"Criteria<Image, DetectedObjects> criteria = Criteria.builder()\n .setTypes(Image.class, DetectedObjects.class)\n .optModelUrls(\"djl://ai.djl.paddlepaddle/face_detection/0.0.1/mask_detection\")\n .optFilter(\"flavor\", \"server\")\n .optTranslator(new FaceTranslator(0.5f, 0.7f))\n .build();\n \nvar model = criteria.loadModel();\nvar predictor = model.newPredictor();\n\nDetectedObjects inferenceResult = predictor.predict(img);\nnewImage = img.duplicate();\nnewImage.drawBoundingBoxes(inferenceResult);\nnewImage.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"如圖片所示,這個推論服務已經可以正確的辨識出圖片中的三張人臉\n## 口罩分類模型\n一旦有了圖片的座標,我們就可以將圖片裁剪到適當大小並且將其傳給口罩分類模型做後續的推論\n### 圖片裁剪\n圖中方框位置的數值範圍從0到1, 只要將這個數值乘上圖片的長寬我們就可以將方框對應到圖片中的準確位置. 為了使裁剪後的圖片有更好的精確度,我們將圖片裁剪成方形,讓我們示範一下:",
"_____no_output_____"
]
],
[
[
"int[] extendSquare(\n double xmin, double ymin, double width, double height, double percentage) {\n double centerx = xmin + width / 2;\n double centery = ymin + height / 2;\n double maxDist = Math.max(width / 2, height / 2) * (1 + percentage);\n return new int[] {\n (int) (centerx - maxDist), (int) (centery - maxDist), (int) (2 * maxDist)\n };\n}\n\nImage getSubImage(Image img, BoundingBox box) {\n Rectangle rect = box.getBounds();\n int width = img.getWidth();\n int height = img.getHeight();\n int[] squareBox =\n extendSquare(\n rect.getX() * width,\n rect.getY() * height,\n rect.getWidth() * width,\n rect.getHeight() * height,\n 0.18);\n return img.getSubImage(squareBox[0], squareBox[1], squareBox[2], squareBox[2]);\n}\n\nList<DetectedObjects.DetectedObject> faces = inferenceResult.items();\ngetSubImage(img, faces.get(2).getBoundingBox()).getWrappedImage();",
"_____no_output_____"
]
],
[
[
"### 事先準備 Translator 並讀取模型\n在使用臉部檢測模型的時候,我們可以利用 DJL 預先建好的 `ImageClassificationTranslator` 並且加上一些轉換。這個 Translator 提供了一些基礎的圖片翻譯處理並且同時包含一些進階的標準化圖片處理。以這個例子來說, 我們不需要額外建立新的 `Translator` 而使用預先建立的就可以",
"_____no_output_____"
]
],
[
[
"var criteria = Criteria.builder()\n .setTypes(Image.class, Classifications.class)\n .optModelUrls(\"djl://ai.djl.paddlepaddle/mask_classification/0.0.1/mask_classification\")\n .optFilter(\"flavor\", \"server\")\n .optTranslator(\n ImageClassificationTranslator.builder()\n .addTransform(new Resize(128, 128))\n .addTransform(new ToTensor()) // HWC -> CHW div(255)\n .addTransform(\n new Normalize(\n new float[] {0.5f, 0.5f, 0.5f},\n new float[] {1.0f, 1.0f, 1.0f}))\n .addTransform(nd -> nd.flip(0)) // RGB -> GBR\n .build())\n .build();\n\nvar classifyModel = criteria.loadModel();\nvar classifier = classifyModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"### 執行推論任務\n最後,要完成一個口罩識別的任務,我們只需要將上述的步驟合在一起即可。我們先將圖片做裁剪後並對其做上述的推論操作,結束之後再生成一個新的分類子類別 `DetectedObjects`:",
"_____no_output_____"
]
],
[
[
"List<String> names = new ArrayList<>();\nList<Double> prob = new ArrayList<>();\nList<BoundingBox> rect = new ArrayList<>();\nfor (DetectedObjects.DetectedObject face : faces) {\n Image subImg = getSubImage(img, face.getBoundingBox());\n Classifications classifications = classifier.predict(subImg);\n names.add(classifications.best().getClassName());\n prob.add(face.getProbability());\n rect.add(face.getBoundingBox());\n}\n\nnewImage = img.duplicate();\nnewImage.drawBoundingBoxes(new DetectedObjects(names, prob, rect));\nnewImage.getWrappedImage();",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a12777909877ab21d1c749a9f29b34afcfe9129
| 7,332 |
ipynb
|
Jupyter Notebook
|
Numpy-Assignment-2.ipynb
|
Jazz-hash/Python-Raw-Codes
|
d43cb8ec9261167037550bd59ee79e53eb9d7abf
|
[
"Apache-2.0"
] | null | null | null |
Numpy-Assignment-2.ipynb
|
Jazz-hash/Python-Raw-Codes
|
d43cb8ec9261167037550bd59ee79e53eb9d7abf
|
[
"Apache-2.0"
] | null | null | null |
Numpy-Assignment-2.ipynb
|
Jazz-hash/Python-Raw-Codes
|
d43cb8ec9261167037550bd59ee79e53eb9d7abf
|
[
"Apache-2.0"
] | null | null | null | 32.017467 | 257 | 0.30401 |
[
[
[
"import numpy as np\nimport os\nfrom PIL import Image\nimport glob\ni = 0\nrootDir = '.'\nimageDir = 'images'\nresizedImageDir = 'ResizedImages'\nimageList = []\nresizedImageList = []\nfor dirName, subDirList, fileList in os.walk(rootDir):\n if imageDir in dirName:\n print('Found Directory: %s' % dirName)\n for file in glob.glob(r\"{}/*.png\".format(imageDir)):\n print('\\t%s' %file)\n image = Image.open(file)\n imageList.append(image)\n resized_image = image.resize((200,150))\n resized_image = np.array(resized_image)\n resizedImageList.append(resized_image)\n break",
"Found Directory: .\\images\n\timages\\Screenshot-1.png\n\timages\\Screenshot-2.png\n\timages\\Screenshot-3.png\n"
],
[
"print(imageList)\nprint(resizedImageList)",
"[<PIL.PngImagePlugin.PngImageFile image mode=RGBA size=1366x768 at 0x2046903E358>, <PIL.PngImagePlugin.PngImageFile image mode=RGBA size=1366x768 at 0x20469128358>, <PIL.PngImagePlugin.PngImageFile image mode=RGBA size=1366x768 at 0x20469128780>]\n[array([[[ 48, 9, 12, 255],\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255],\n ...,\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255]],\n\n [[ 48, 9, 12, 255],\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255],\n ...,\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255]],\n\n [[ 48, 9, 12, 255],\n [ 18, 97, 146, 255],\n [ 7, 103, 161, 255],\n ...,\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255],\n [ 48, 9, 12, 255]],\n\n ...,\n\n [[ 30, 6, 8, 255],\n [255, 255, 255, 255],\n [255, 255, 255, 255],\n ...,\n [ 28, 4, 6, 255],\n [ 29, 5, 7, 255],\n [ 31, 7, 9, 255]],\n\n [[ 30, 6, 8, 255],\n [ 29, 5, 7, 255],\n [ 30, 6, 8, 255],\n ...,\n [ 31, 7, 9, 255],\n [ 30, 6, 8, 255],\n [ 29, 5, 7, 255]],\n\n [[ 28, 4, 6, 255],\n [ 29, 5, 7, 255],\n [ 29, 5, 7, 255],\n ...,\n [ 30, 6, 8, 255],\n [ 29, 5, 7, 255],\n [ 29, 5, 7, 255]]], dtype=uint8), array([[[ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n ...,\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255]],\n\n [[ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n ...,\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255]],\n\n [[ 44, 20, 22, 255],\n [ 16, 99, 149, 255],\n [ 7, 105, 163, 255],\n ...,\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255]],\n\n ...,\n\n [[ 30, 6, 8, 255],\n [255, 255, 255, 255],\n [255, 255, 255, 255],\n ...,\n [ 28, 4, 6, 255],\n [ 29, 5, 7, 255],\n [ 31, 7, 9, 255]],\n\n [[ 30, 6, 8, 255],\n [ 29, 5, 7, 255],\n [ 30, 6, 8, 255],\n ...,\n [ 31, 7, 9, 255],\n [ 30, 6, 8, 255],\n [ 29, 5, 7, 255]],\n\n [[ 28, 4, 6, 255],\n [ 29, 5, 7, 255],\n [ 29, 5, 7, 255],\n ...,\n [ 30, 6, 8, 255],\n [ 29, 5, 7, 255],\n [ 29, 5, 7, 255]]], dtype=uint8), array([[[ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n ...,\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255]],\n\n [[ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n ...,\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255]],\n\n [[ 44, 20, 22, 255],\n [ 16, 99, 149, 255],\n [ 7, 105, 163, 255],\n ...,\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255],\n [ 44, 20, 22, 255]],\n\n ...,\n\n [[ 30, 6, 8, 255],\n [255, 255, 255, 255],\n [255, 255, 255, 255],\n ...,\n [ 28, 4, 6, 255],\n [ 29, 5, 7, 255],\n [ 31, 7, 9, 255]],\n\n [[ 30, 6, 8, 255],\n [ 29, 5, 7, 255],\n [ 30, 6, 8, 255],\n ...,\n [ 31, 7, 9, 255],\n [ 30, 6, 8, 255],\n [ 29, 5, 7, 255]],\n\n [[ 28, 4, 6, 255],\n [ 29, 5, 7, 255],\n [ 29, 5, 7, 255],\n ...,\n [ 30, 6, 8, 255],\n [ 29, 5, 7, 255],\n [ 29, 5, 7, 255]]], dtype=uint8)]\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4a128342fa71f007e28422cc0439776cb803ef10
| 287,990 |
ipynb
|
Jupyter Notebook
|
_notebooks/2022-05-06-Residual-Networks.ipynb
|
geon-youn/DunGeon
|
70792a1042630fbf114fe43263b851d0b57d5b18
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2022-05-06-Residual-Networks.ipynb
|
geon-youn/DunGeon
|
70792a1042630fbf114fe43263b851d0b57d5b18
|
[
"Apache-2.0"
] | null | null | null |
_notebooks/2022-05-06-Residual-Networks.ipynb
|
geon-youn/DunGeon
|
70792a1042630fbf114fe43263b851d0b57d5b18
|
[
"Apache-2.0"
] | null | null | null | 143.63591 | 189,145 | 0.821397 |
[
[
[
"# \"Understanding Residual Networks\"\n> \"Probably about residuals, right?\"\n\n- comments: true\n- categories: [vision]",
"_____no_output_____"
]
],
[
[
"#hide\n!pip install -Uqq fastai>=2.0.0 graphviz ipywidgets matplotlib nbdev>=0.2.12 pandas scikit_learn azure-cognitiveservices-search-imagesearch sentencepiece",
"_____no_output_____"
],
[
"#hide\nfrom google.colab import drive\ndrive.mount('/content/gdrive/', force_remount=True)",
"Mounted at /content/gdrive/\n"
]
],
[
[
"## Introduction",
"_____no_output_____"
],
[
"We've covered the basics of CNNs with the MNIST data set where we trained a model to recognize handwritten digits. Today, we'll be moving towards residual networks that allow us to train CNNs with more layers. \n\nSince we've already got a good result with the MNIST data set, we'll move onto the Imagenette data set, which is a smaller version of the ImageNet data set. \n\nWe train with a smaller version so that we can make small changes without having to wait long periods of time for the model to train.",
"_____no_output_____"
]
],
[
[
"#hide\nfrom fastai.vision.all import *",
"_____no_output_____"
],
[
"def get_dls(url, presize, resize):\n path = untar_data(url)\n return DataBlock((ImageBlock, CategoryBlock), get_items=get_image_files,\n splitter=GrandparentSplitter(valid_name='val'),\n get_y=parent_label, item_tfms=Resize(presize),\n batch_tfms=[*aug_transforms(min_scale=0.5, size=resize),\n Normalize.from_stats(*imagenet_stats)]\n ).dataloaders(path, bs=128)",
"_____no_output_____"
],
[
"#hide_output\ndls = get_dls(URLs.IMAGENETTE_160, 160, 128)",
"_____no_output_____"
],
[
"dls.show_batch(max_n=4)",
"_____no_output_____"
]
],
[
[
"## Adding flexibility through fully convolutional neural networks",
"_____no_output_____"
],
[
"First, we'll first change how our model works. With the MNIST data set, we have images of shape 28 $\\times$ 28. If we added a few more layers with a stride of 1, then we'd get more layers. But, how do we do classification for images with sizes that aren't 28 $\\times$ 28? And, what do we do if we want to have additional layers of different strides?\n\nIn reducing the last two dimensions of our output from each layer (the height and width) through strides, we get two problems:\n- with larger images, we need a lot more stride 2 layers; and\n- the model won't work for images of different shape than our training images. \n\nThe latter can be solved through resizing, but do we really want to resize images to 28 $\\times$ 28? We might be losing a lot of information for more complex tasks. \n\nSo, we solve it through *fully convolutional networks*, which uses a trick of taking the average of activations along a convolutional grid. In other words, we take the average of the last two axes from the final layer like so:",
"_____no_output_____"
]
],
[
[
"def avg_pool(x):\n return x.mean((2, 3))",
"_____no_output_____"
]
],
[
[
"Remember that our final layer has a shape of `n_batches * n_channels * height * width`. So, taking the average of the last two axes gives us a new tensor of shape `n_batches * n_channels * 1` from which we flatten to get `n_batches * n_channels`. \n\nUnlike our last approach of striding until our last two axes are 1 $\\times$ 1, we can have a final layer with any value for our last two axes and still end up with 1 $\\times$ 1 after average pooling. In other words, through average pooling, we can have a CNN that can have as many layers as we want, with images of any shape during inference. \n\nOverall, a fully convolutional network has a number of convolutional layers that can be of any stride, an adaptive average pooling layer, a flatten layer, and then a linear layer. An adaptive average pooling layer allows us to specify the shape of the output, but we want one value so we pass in `1`:",
"_____no_output_____"
]
],
[
[
"def block(ni, nf):\n return ConvLayer(ni, nf, stride=2)\n\ndef get_model():\n return nn.Sequential(\n block(3, 16),\n block(16, 32),\n block(32, 64),\n block(64, 128),\n block(128, 256),\n nn.AdaptiveAvgPool2d(1), \n Flatten(),\n nn.Linear(256, dls.c)\n )",
"_____no_output_____"
]
],
[
[
"The [`ConvLayer`](https://docs.fast.ai/layers.html#ConvLayer) is fastai's version of our `conv` layer from the [last blog](https://geon-youn.github.io/DunGeon/vision/2022/04/30/Convolutional-Neural-Networks.html), which includes the convolutional layer, the activation function, and batch normalization, but also adds more functionalities. \n\n---\n\n*Activation function or nonlinearity?* In the last blog, I defined ReLU as a nonlinearity, because it is a nonlinearity. However, it can also be called an activation function since it's taking the activations from the convolutional layer to output new activations. An activation function is just that: a function between two linear layers. \n\n---\n\nIn retrospect, our simple CNN for the MNIST data set looked something like this:",
"_____no_output_____"
]
],
[
[
"def get_simple_cnn():\n return nn.Sequential(\n block(3, 16),\n block(16, 32),\n block(32, 64),\n block(64, 128),\n block(128, 10),\n Flatten()\n )",
"_____no_output_____"
]
],
[
[
"In a fully convolutional network we have what we had before in a CNN, except that we pool the last two axes from our final convolutional layer into a unit axis, flatten the output to get rid of the unit axis, then use a linear layer to get `dls.c` output channels. `dls.c` returns how many unique labels there are for our data set. \n\nSo, why didn't we just use fully convolutional networks from the beginning? Well, fully convolutional networks take an image, cut them into pieces, shake them all about, do the hokey pokey, and decide, on average, what the image should be classified as. However, we were dealing with an optical character recognition (OCR) problem. With OCR, it doesn't make sense to cut a character into pieces and decide, on averge, what character it is. \n\nThat doesn't mean fully convolutional networks are useless; they're bad for OCR problems, but they're good for problems where the objects-to-be-classified don't have a specific orientation or size. \n\nLet's try training a fully convolutional network on the Imagenette data set:",
"_____no_output_____"
]
],
[
[
"def get_learner(model):\n return Learner(dls, model, loss_func=nn.CrossEntropyLoss(), \n metrics=accuracy).to_fp16()",
"_____no_output_____"
],
[
"learn = get_learner(get_model())",
"_____no_output_____"
],
[
"learn.fit_one_cycle(5, 3e-3)",
"_____no_output_____"
]
],
[
[
"## How resnet came about",
"_____no_output_____"
],
[
"Now we're ready to add more layers through a fully convolutional network. But, how does it turn out if we just add more layers? Not that promising. \n\n<figure>\n <img src='https://inotgo.com/imagesLocal/202103/24/20210324084503876z_0.png' alt='Comparisons of small and large layer CNNs'>\n <figcaption>Comparison of small and large layer CNNs through error rate on training and test sets.</figcaption>\n</figure>\n\nYou'd expect a model with more layers to have an easier time predicting the correct label. However, [the founders](https://arxiv.org/abs/1512.03385) of resnet found different results when training and comparing the results of a 20- and 56-layer CNN: the 56-layer model was doing worse than the 20-layer model in both training and test sets. Interestingly, the lower performance isn't caused by overfitting because the same pattern persists between the training and test errors.\n\nHowever, shouldn't we be able to add 36 identity layers (that output the same activations) to the 20-layer model to achieve a 56-layer model that has the same results as the 20-layer model? For some reason, SGD isn't able to find that kind of model. \n\nSo, here comes *residuals*. Instead of each layer being the output of `F(x)` where `F` is a layer and `x` is the input, what if we had `x + F(x)`? In essense, we want each layer to learn well. We could go straight to `H(x)`, but we can have it learn `H(x)` by learning `F(x) = H(x) - x`, which turns out to `H(x) = x + F(x)`. \n\nReturning to the idea of identity layers, `F` contains a batchnorm which performs $\\gamma y + \\beta$ with the output `y`. We could have $\\gamma$ equal to 0, which turns `x + F(x)` to `x + 0`, which is equivalent to `x`.\n\nTherefore, we could start with a good 20-layer model, initialize 36 layers on top of it with $\\gamma$ initialized to 0, then fine-tune the entire model. \n\nHowever, instead of starting with a trained model, we have something like this:\n\n<figure>\n <img src='https://www.researchgate.net/publication/331195671/figure/fig1/AS:727865238753280@1550548004386/ResNet-module-adapted-from-1.ppm' alt='Diagram of a resnet block'>\n <figcaption>The \"resnet\" layer</figcaption>\n</figure>\n\nIn a residual network layer, we have an input `x` that passes through two convolutional layers `f` and `g`, where `F(x) = g(f(x))`. The right arrow is the *identity branch* or *skip connection* that gives us the identity part of the equation: `x + F(x)`, whereby `F(x)` is the residual. \n\nSo instead of adding on these resnet layers to a 20-layer model, we just initialize a model with these layers. Then, we initialize them randomly per usual and train them with SGD. Skip connections enable SGD to optimize the model even though there's more layers. \n\nWhy resnet works so well compared to just adding more layers is like how weight decay works so well: we're training to minimize residuals. \n\nAt each layer of a residual network, we're training the residuals given by the function `F` since that's the only part with trainable parameters; however, the output of each layer is `x + F(x)`. So, if the desired output is `H(x)` where `H(x) = x + F(x)`, we're asking the model to predict the *residual* `F(x) = H(x) - x`, which is the difference between the desired output and the given input. Therefore at each optimization step, we're minimizing the error (residual). Hence a residual network is good at learning the difference between doing nothing and doing something (by going through the two weight layers). \n\nSo, a residual network block looks like this:",
"_____no_output_____"
]
],
[
[
"class ResBlock(Module):\n def __init__(self, ni, nf):\n self.convs = nn.Sequential(\n ConvLayer(ni, nf),\n ConvLayer(nf, nf, norm_type=NormType.BatchZero)\n )\n\n def forward(self, x):\n return x + self.convs(x)",
"_____no_output_____"
]
],
[
[
"By passing `NormType.BatchZero` to the second convolutional layer, we initialize the $\\gamma$s in the batchnorm equation to 0. \n\n",
"_____no_output_____"
],
[
"## How to use a resblock in practice",
"_____no_output_____"
],
[
"Although we could begin training with the `ResBlock`, how would `x + F(x)` work if `x` and `F(x)` are of different shapes? \n\nWell, how could they become different shapes? When `ni != nf` or we use a stride that's not 1. Remember that `x` has the shape `n_batch * n_channels * height * width`. If we have `ni != nf`, then `n_channels` is different for `x` and `F(x)`. Similarly, if `stride != 1`, then `height` and `width` would be different for `x` and `F(x)`. \n\nInstead of accepting these restrictions, we can resize `x` to become the same shape as `F(x)`. First, we appply average pooling to change `height` and `width`, then apply a 1 $\\times$ 1 convolution (a convolution layer with a kernel size of 1 $\\times$ 1) with `ni` in-channels and `nf` out-channels to change `n_channels`. \n\nThrough changing our code, we have:",
"_____no_output_____"
]
],
[
[
"def _conv_block(ni, nf, stride):\n return nn.Sequential(\n ConvLayer(ni, nf, stride=stride),\n ConvLayer(nf, nf, act_cls=None, norm_type=NormType.BatchZero)\n )",
"_____no_output_____"
],
[
"class ResBlock(Module):\n def __init__(self, ni, nf, stride=1):\n self.convs = _conv_block(ni, nf, stride)\n # noop is `lamda x: x`, short for \"no operation\"\n self.idconv = noop if ni == nf else ConvLayer(ni, nf, 1, act_cls=None)\n self.pool = noop if stride == 1 else nn.AvgPool2d(stride, ceil_mode=True)\n\n def forward(self, x):\n # we can apply ReLU for both x and F(x) since we specified \n # no activation functions for the last layer of conv and idconvs\n return F.relu(self.idconv(self.pool(x)) + self.convs(x))",
"_____no_output_____"
]
],
[
[
"In our new `ResBlock`, one pass will lead to `H(G(x)) + F(x)` where `F` is two convolutional layers and `G` is a pooling layer followed by `H`, a convolutional layer. We have a pooling layer to make the height and width of `x` the same as `F(x)` and the convolutional layer to make `G(x)` have the same out-channels as `F(x)`. Then, `H(G(x))` and `F(x)` will have the same shape all the time, allowing them to be added. Overall, a resnet block is 2 layers deep.",
"_____no_output_____"
]
],
[
[
"def block(ni, nf):\n return ResBlock(ni, nf, stride=2)",
"_____no_output_____"
]
],
[
[
"So, let's try retraining with the new model:",
"_____no_output_____"
]
],
[
[
"learn = get_learner(get_model())\nlearn.fit_one_cycle(5, 3e-3)",
"_____no_output_____"
]
],
[
[
"We didn't get much of an accuracy boost, but that's because we still have the same number of layers. Let's double the layers:",
"_____no_output_____"
]
],
[
[
"def block(ni, nf):\n return nn.Sequential(ResBlock(ni, nf, stride=2), ResBlock(nf, nf))",
"_____no_output_____"
],
[
"learn = get_learner(get_model())\nlearn.fit_one_cycle(5, 3e-3)",
"_____no_output_____"
]
],
[
[
"## Implementing a resnet",
"_____no_output_____"
],
[
"The first step to go from a resblock to a resnet is to improve the stem of the model.\n\nThe first few layers of the model are called its *stem*. Through practice, [some researchers](https://arxiv.org/abs/1812.01187) found improvements by beginning the model with a few convolutional layers followed by a max pooling layer. A max pooling layer, unlike average pooling, takes the maximum instead of the average.\n\nThe new stem, which we prepend the the model looks like this:",
"_____no_output_____"
]
],
[
[
"def _resnet_stem(*sizes):\n return [\n ConvLayer(sizes[i], sizes[i + 1], 3, stride=2 if i == 0 else 1) \n for i in range(len(sizes) - 1)\n ] + [nn.MaxPool2d(kernel_size=3, stride=2, padding=1)]",
"_____no_output_____"
]
],
[
[
"Keeping the model simple in the beginning helps with training because with CNNs, the vast majority of *computations*, not parameters, occur in the beginning where images have large dimensions.",
"_____no_output_____"
]
],
[
[
"_resnet_stem(3, 32, 32, 64)",
"_____no_output_____"
]
],
[
[
"These same researchers found additional \"tricks\" to substantially improve the model: using **four** groups of resnet blocks with channels of 64, 128, 256, and 512. Each group starts with a stride of 2 except for the first one since it's after the max pooling layer from the stem. \n\nBelow is *the* resnet:",
"_____no_output_____"
]
],
[
[
"class ResNet(nn.Sequential):\n def __init__(self, n_out, layers, expansion=1):\n stem = _resnet_stem(3, 32, 32, 64)\n self.channels = [64, 64, 128, 256, 512]\n for i in range(1, 5): self.channels[i] *= expansion\n blocks = [self._make_layer(*o) for o in enumerate(layers)]\n super().__init__(*stem, *blocks, \n nn.AdaptiveAvgPool2d(1), Flatten(),\n nn.Linear(self.channels[-1], n_out))\n \n def _make_layer(self, idx, n_layers):\n stride = 1 if idx == 0 else 2\n ni, nf = self.channels[idx:idx + 2]\n return nn.Sequential(*[\n ResBlock(ni if i == 0 else nf, nf, stride if i == 0 else 1)\n for i in range(n_layers)\n ])",
"_____no_output_____"
]
],
[
[
"The \"total\" number of layers in a resnet is double sum of the layers passed in as `layers` in instantiating `ResNet` plus 2 from `stem` and `nn.Linear`. We consider `stem` as one layer because of the max pooling layer at the end. \n\nSo, a resnet-18 is:",
"_____no_output_____"
]
],
[
[
"#collapse_output\nrn18 = ResNet(dls.c, [2, 2, 2, 2])\nrn18",
"_____no_output_____"
]
],
[
[
"Where it has 18 layers since we pass in `[2, 2, 2, 2]` for `layers`, which sums of 8 and double that is `16` (we double since each `ResBlock` is 2 layers deep). Then, we add 2 from `stem` and `nn.Linear` to get 18 \"total\" layers. \n\nBy increasing the number of layers to 18, we see a significant increase in our model's accuracy:",
"_____no_output_____"
]
],
[
[
"learn = get_learner(rn18)\nlearn.fit_one_cycle(5, 3e-3)",
"_____no_output_____"
]
],
[
[
"## Adding more layers to resnet-18",
"_____no_output_____"
],
[
"As you increase the number of layers in a resnet, the number of parameters increases substantially. Thus, to avoid running out of GPU memory, we can apply *bottlenecking*, which alters the convolutional layer in `ResBlock` for `F` in `x + F(x)` as follows:\n\n<figure>\n <img src='https://miro.medium.com/max/588/0*9tCUFp28oQGOK6bE.jpg' alt='Bottleneck layer'>\n</figure>\n\nPreviously, we stacked two convolutions with a kernel size of 3. However, a bottleneck layer has a convolution with a kernel size of 1, which decreases the channels by a factor of 4, then a convolution with a kernel size of 3 that maintains the same number of channels, and then a convolution with a kernel size of 1 that increases the number of channels by a factor of 4 to return the original dimension. It's coined *bottleneck* because we start with, in this case, a 256-channel image that's \"bottlenecked\" by the first convolution into 64 channels, and then enlarged to 256 channels by the last convolution.\n\nAlthough we'll have another layer, it performs faster than the two convolutions with a kernel size of 3 because convolutions with a kernel size of 1 are much faster. \n\nThrough bottleneck layers, we can have more channels in more-or-less the same amount of time. Additionally, we'll have fewer parameters since we replace a 3 $\\times$ 3 kernel layer with two 1 $\\times$ 1 kernel layers, although one has 4 more out-channels. Overall, we have a difference of 4$:$9. \n\nIn code, the bottleneck layer looks like this:",
"_____no_output_____"
]
],
[
[
"def _conv_block(ni, nf, stride):\n return nn.Sequential(\n ConvLayer(ni, nf // 4, stride=1),\n ConvLayer(nf // 4, nf // 4, stride=stride),\n ConvLayer(nf // 4, nf, stride=1, act_cls=None, norm_type=NormType.BatchZero)\n )",
"_____no_output_____"
]
],
[
[
"Then, when we make our resnet model, we'll have to pass in 4 for our `expansion` to account for the decreasing factor of 4 in our bottleneck layer. ",
"_____no_output_____"
]
],
[
[
"rn50 = ResNet(dls.c, [3, 4, 6, 3], 4)",
"_____no_output_____"
],
[
"learn = get_learner(rn50)\nlearn.fit_one_cycle(20, 3e-3)",
"_____no_output_____"
]
],
[
[
"## MNIST: to CNN or resnet?",
"_____no_output_____"
],
[
"In the previous blog, we achieved a 99.2% accuracy with just the CNN. Let's try training a resnet-50 with the MNIST data set and see what we can get:",
"_____no_output_____"
]
],
[
[
"def get_dls(url, bs=512):\n path = untar_data(url)\n return DataBlock(\n blocks=(ImageBlock(cls=PILImageBW), CategoryBlock),\n get_items=get_image_files,\n splitter=GrandparentSplitter('training', 'testing'), \n get_y=parent_label,\n batch_tfms=Normalize()\n ).dataloaders(path, bs=bs)",
"_____no_output_____"
],
[
"#hide_output\ndls = get_dls(URLs.MNIST)",
"_____no_output_____"
],
[
"dls.show_batch(max_n=4)",
"_____no_output_____"
]
],
[
[
"We have to change the first input into `_resnet_stem` to 1 from 3 since the MNIST data set is a greyscale image.",
"_____no_output_____"
]
],
[
[
"class ResNet(nn.Sequential):\n def __init__(self, n_out, layers, expansion=1):\n stem = _resnet_stem(1, 32, 32, 64)\n self.channels = [64, 64, 128, 256, 512]\n for i in range(1, 5): self.channels[i] *= expansion\n blocks = [self._make_layer(*o) for o in enumerate(layers)]\n super().__init__(*stem, *blocks, \n nn.AdaptiveAvgPool2d(1), Flatten(),\n nn.Linear(self.channels[-1], n_out))\n \n def _make_layer(self, idx, n_layers):\n stride = 1 if idx == 0 else 2\n ni, nf = self.channels[idx:idx + 2]\n return nn.Sequential(*[\n ResBlock(ni if i == 0 else nf, nf, stride if i == 0 else 1)\n for i in range(n_layers)\n ])",
"_____no_output_____"
],
[
"rn50 = ResNet(dls.c, [3, 4, 6, 3], 4)",
"_____no_output_____"
],
[
"learn = get_learner(rn50)",
"_____no_output_____"
],
[
"lr = learn.lr_find().valley",
"_____no_output_____"
],
[
"learn.fit_one_cycle(20, lr)",
"_____no_output_____"
]
],
[
[
"After training the model by 20 epochs, we can confidently say that it's not always the best idea to start with a more complex architecture that takes much longer to train and gives a worse accuracy. \n\nIn addition, I mentioned before that resnets aren't the best for OCR problems like digit recognition since we're slicing the image and deciding, on average, what the digit is. \n\nIn this scenario, a regular CNN would be a better choice than a resnet. ",
"_____no_output_____"
],
[
"## Conclusion",
"_____no_output_____"
],
[
"In this blog, we covered residual networks, which allow us to train CNNs with more layers by having the model learn indirectly through residuals. It's as if before, we were just training the weights of the activations, when in reality, we also needed bias to have the models learn efficiently. To visualize, imagine the following:\n\nBefore, we had\n```\nweights * x + bias\n```\nas our activations, where `x` is our input. But, that just gives us activations. Through residual networks, we have\n```\n(weights * x + bias) + residuals\n```\nwhich adds on another parameter that we can train. Through exploring, we found that we'd need some way to make `(weights * x + bias)` have the same shape as `residuals` to allow them to be added. Thus, we formed\n```\nreshape * (weights * x + bias) + residuals\n```\nTherefore, a residual network *fixes* a CNN to be more like how we train neural networks since the above can be simplified to:\n```\nweights * x + bias\n```\n\nThen, to use this new kind of layer, we had to use fully convolutional networks that allow us to have input of any shape. Finally, we looked at a bag of tricks like stems, groups, and bottleneck layers that allow us to train RNNs more efficiently and have many more layers.\n\nAt this point, we've covered all the main architectures for training great models with computer vision (CNN and resnet), natural language processing (AWD-LSTM), tabular data (random forests and neural networks), and collaborative filtering (probabilitic matrix factorization and neural networks). \n\nFrom now on, we'll be looking at the foundations of deep learning and fastai through modifying the mentioned architectures, building better optimization functions than the standard SGD, exactly what PyTorch is doing for us, how to visualize what the model is learning, and a deeper look into what fastai is doing for us through its `Learner` class. ",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4a129330fb0ae33fad42abdccc4b5bedaa0674ad
| 19,103 |
ipynb
|
Jupyter Notebook
|
Assignment_Day_2.ipynb
|
VishnuM24/LetsUpgrade-Python
|
b1d1e74df9f6bbebab155e460e45335cd0f9888a
|
[
"Apache-2.0"
] | null | null | null |
Assignment_Day_2.ipynb
|
VishnuM24/LetsUpgrade-Python
|
b1d1e74df9f6bbebab155e460e45335cd0f9888a
|
[
"Apache-2.0"
] | null | null | null |
Assignment_Day_2.ipynb
|
VishnuM24/LetsUpgrade-Python
|
b1d1e74df9f6bbebab155e460e45335cd0f9888a
|
[
"Apache-2.0"
] | null | null | null | 23.789539 | 243 | 0.403968 |
[
[
[
"<a href=\"https://colab.research.google.com/github/VishnuM24/LetsUpgrade-Python/blob/master/Assignment_Day_2.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# - DAY 2 -",
"_____no_output_____"
],
[
"##**QUESTION 1 :**\n\nList & its default methods",
"_____no_output_____"
]
],
[
[
"vowels = [ 'a', 'e' ]",
"_____no_output_____"
],
[
"# Method 1 - extend()\nvow2 = ['i', 'o', 'u']\nvowels.extend( vow2 )\nprint( vowels )",
"['a', 'e', 'i', 'o', 'u']\n"
],
[
"# Method 2 - append()\nvowels.append( 'y' )\nprint( vowels )",
"['a', 'e', 'i', 'o', 'u', 'y']\n"
],
[
"# Method 3 - insert()\nvowels.insert( 2, 'x' )\nprint(vowels)",
"['a', 'e', 'x', 'i', 'o', 'u', 'y']\n"
],
[
"# Method 4 - pop()\nvowels.pop(-1)\nprint(vowels)",
"['a', 'e', 'x', 'i', 'o', 'u']\n"
],
[
"# Method 5 - remove()\nvowels.remove('x')\nprint(vowels)",
"['a', 'e', 'i', 'o', 'u']\n"
]
],
[
[
"## **Question 2:**\n\nDictionary & its default methods",
"_____no_output_____"
]
],
[
[
"dailyFood = {\n 'breakfast' : 'dosa',\n 'lunch' : 'rice',\n 'dinner' : 'chapathi'\n}",
"_____no_output_____"
],
[
"# Method 1 - keys()\nkeys_list = dailyFood.keys()\nprint( keys_list )",
"dict_keys(['breakfast', 'lunch', 'dinner'])\n"
],
[
"# Method 2 - items()\ndailyfood_list = dailyFood.items()\nprint( dailyfood_list )",
"dict_items([('breakfast', 'dosa'), ('lunch', 'rice'), ('dinner', 'chapathi')])\n"
],
[
"# Method 3 - update()\ndailyFood.update( { 'evening_snack' : 'biscuits' } )\nprint( dailyfood_list )",
"dict_items([('breakfast', 'dosa'), ('lunch', 'rice'), ('dinner', 'chapathi'), ('evening_snack', 'biscuits')])\n"
],
[
"# Method 4 - get()\nfood_lunch = dailyFood.get('lunch')\nprint( food_lunch )",
"rice\n"
],
[
"# Method 5 - pop()\ndailyFood.pop( 'evening_snack' )\nprint( dailyFood )",
"{'breakfast': 'dosa', 'lunch': 'rice', 'dinner': 'chapathi'}\n"
]
],
[
[
"## **Question 3:**\n\nSets & its default methods",
"_____no_output_____"
]
],
[
[
"language_set = { 'c', 'python', 'java' }",
"_____no_output_____"
],
[
"# Method 1 - add()\nlanguage_set.add( 'PHP' )\nprint( language_set )",
"{'python', 'PHP', 'c', 'java'}\n"
],
[
"# Method 2 - union()\nprogramming_set = language_set.union( { 'c#', 'cpp', 'Go', 'R' } )\nprint( programming_set )",
"{'python', 'PHP', 'java', 'R', 'c#', 'c', 'cpp', 'Go'}\n"
],
[
"# Method 3 - discard()\nprogramming_set.discard( 'PHP' )\nprint( programming_set )",
"{'python', 'java', 'R', 'c#', 'c', 'cpp', 'Go'}\n"
],
[
"# Method 4 - issubset()\nunder_programming = language_set.issubset( programming_set )\nprint( under_programming )",
"False\n"
],
[
"# Method 5 - update()\nprogramming_set.update( { 'html', 'css', 'javascript' } )\nprint( programming_set )",
"{'python', 'javascript', 'html', 'java', 'R', 'c#', 'css', 'c', 'cpp', 'Go'}\n"
]
],
[
[
"## **Question 4:**\n\nTuple & its default methods",
"_____no_output_____"
]
],
[
[
"oct_values = ( '000', '001' , '010' , '011' , '100', '101', '110', '111' )",
"_____no_output_____"
],
[
"# Method 1 - count()\nrepeat_3 = oct_values.count( '011' )\nprint( repeat_3 )",
"1\n"
],
[
"# Method 2 - index()\nindx = oct_values.index( '101' )\nprint( indx )",
"5\n"
],
[
"oct_values[6]",
"_____no_output_____"
]
],
[
[
"## **Question 5:**\n\nStrings & its default methods",
"_____no_output_____"
]
],
[
[
"intro_template = 'Hi This is <NAME> from <PLACE>'\n",
"_____no_output_____"
],
[
"# Method 1 - replace()\nmy_template = intro_template.replace( '<NAME>', 'Vishnu' )\nmy_template = my_template.replace( '<PLACE>', 'Kerala' )\n\nprint( my_template )",
"Hi This is Vishnu from Kerala\n"
],
[
"# Method 2 - split()\nwords = my_template.split(' ')\nprint( words )",
"['Hi', 'This', 'is', 'Vishnu', 'from', 'Kerala']\n"
],
[
"# Method 3 - find()\nx = my_template.find('Vishnu')\nprint( 'My name starts at position',x )",
"My name starts at position 11\n"
],
[
"# Method 4 - upper()\ncaps_template = my_template.upper()\nprint( caps_template )",
"HI THIS IS VISHNU FROM KERALA\n"
],
[
"# Method 5 - isnumeric()\nisNumber = my_template.isnumeric()\nprint( isNumber )\n\ntemp_string = '2412'\nisNo = temp_string.isnumeric()\nprint( isNo )",
"False\nTrue\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a129a1ce0ae589c15398b44b1c1bb8ad57981b7
| 5,402 |
ipynb
|
Jupyter Notebook
|
Movie recommendation system.ipynb
|
i-m-ram/movie-recommendation-system
|
9ab1d27f877765299d0079fa703da3c5587dd703
|
[
"MIT"
] | null | null | null |
Movie recommendation system.ipynb
|
i-m-ram/movie-recommendation-system
|
9ab1d27f877765299d0079fa703da3c5587dd703
|
[
"MIT"
] | null | null | null |
Movie recommendation system.ipynb
|
i-m-ram/movie-recommendation-system
|
9ab1d27f877765299d0079fa703da3c5587dd703
|
[
"MIT"
] | null | null | null | 31.776471 | 151 | 0.52351 |
[
[
[
"import pandas as pd\nimport numpy as np\n\ncredit_df=pd.read_csv(r'E:\\Projects\\Movie recommendation System\\DataSet\\tmdb_5000_credits.csv')\nmovie_df=pd.read_csv(r'E:\\Projects\\Movie recommendation System\\DataSet\\tmdb_5000_movies.csv')\n\n#movie_df.head()\n\n#credit_df.head()\n\ncredit_df.columns=['id','title','cast','crew']\n\nmovie_df=movie_df.merge(credit_df,on='id')\n\n#movie_df.head()\n\n#movie_df.describe()\n\n#movie_df.info()\n\nfrom ast import literal_eval\nfeatures = [\"cast\", \"crew\", \"keywords\", \"genres\"]\nfor feature in features:\n movie_df[feature] = movie_df[feature].apply(literal_eval)\n#movie_df[features].head(10)\n\n#movie_df['cast'].head()\n\n\n\ndef get_director(x):\n for i in x:\n if i[\"job\"] == \"Director\":\n return i[\"name\"]\n return np.nan\n\ndef get_list(x):\n if isinstance(x, list):\n names = [i[\"name\"] for i in x]\n if len(names) > 3:\n names = names[:3]\n return names\n return []\n\nmovie_df['Director']=movie_df['crew'].apply(get_director)\n\nfeatures = [\"cast\", \"keywords\", \"genres\"]\nfor feature in features:\n movie_df[feature] = movie_df[feature].apply(get_list)\n\nmovie_df['title']=movie_df['original_title']\n\n#movie_df[['title', 'cast', 'Director', 'keywords', 'genres']].head()\n\ndef clean_data(row):\n if isinstance(row, list):\n return [str.lower(i.replace(\" \", \"\")) for i in row if isinstance(i,str)]\n else:\n if isinstance(row, str):\n return str.lower(row.replace(\" \", \"\"))\n else:\n return \"\"\nfeatures = ['cast', 'keywords', 'Director', 'genres']\nfor feature in features:\n movie_df[feature] = movie_df[feature].apply(clean_data)\n\n#movie_df['cast'].head()\n\ndef create_soup(features):\n return ' '.join(features['keywords']) + ' ' + ' '.join(features['cast']) + ' ' + features['Director'] + ' ' + ' '.join(features['genres'])\n\nmovie_df['soup']=movie_df.apply(create_soup, axis=1)\n#movie_df['soup'].head()\n\n\nfrom sklearn.feature_extraction.text import CountVectorizer\nfrom sklearn.metrics.pairwise import cosine_similarity\n\ncount_vectorizer = CountVectorizer(stop_words='english')\ncount_matrix=count_vectorizer.fit_transform(movie_df['soup'])\n#print(count_matrix.shape)\ncosine_sim2 = cosine_similarity(count_matrix, count_matrix) \n#print(cosine_sim2.shape)\n\nmovie_df = movie_df.reset_index()\nindices = pd.Series(movie_df.index, index=movie_df[\"title\"]).drop_duplicates()\n#print(indices.head())\n\ndef get_recommendations(title, cosine_sim):\n idx = indices[title]\n similarity_scores = list(enumerate(cosine_sim[idx]))\n similarity_scores= sorted(similarity_scores, key=lambda x: x[1], reverse=True)\n similarity_scores= similarity_scores[1:11]\n # (a, b) where a is id of movie, b is similarity_scores\n movie_indices = [ind[0] for ind in similarity_scores]\n movies = movie_df[\"title\"].iloc[movie_indices]\n return movies\n\n#print(get_recommendations(\"Iron Man 2\", cosine_sim2))\nMovie_name=str(input('Enter Movie Name'))\nget_recommendations(Movie_name, cosine_sim2)\n",
"Enter Movie NameIron Man\n"
]
]
] |
[
"code"
] |
[
[
"code"
]
] |
4a12aaf71a4731f35b182f57673746dc3056b805
| 25,890 |
ipynb
|
Jupyter Notebook
|
Curso3.ipynb
|
camilogutierrez/MachineLearning
|
ec129ac09ae86f94b4535523bdacf800295cea8c
|
[
"MIT"
] | 2 |
2020-05-08T21:18:08.000Z
|
2020-07-18T22:13:22.000Z
|
Curso3.ipynb
|
camilogutierrez/MachineLearning
|
ec129ac09ae86f94b4535523bdacf800295cea8c
|
[
"MIT"
] | null | null | null |
Curso3.ipynb
|
camilogutierrez/MachineLearning
|
ec129ac09ae86f94b4535523bdacf800295cea8c
|
[
"MIT"
] | null | null | null | 61.937799 | 941 | 0.687524 |
[
[
[
"# ML Strategy \n* Collect more data\n* Collect more diverse trainign set\n* Train algorithm longer with gradient descetn\n* Try adam isntead of gradient descent\n* Try bigger networks\n* Try smaller networks\n* Try dropout\n* Add L2 regularizatión\n* Network architecture\n* Network archicteture \n - Activvation\n - \\# hidden units\n \n# Orthogonalization \nFor a supervised learning system to do well, you usually need to tune the knobs of your system to make sure that four things hold true. \n1. **Fit training set well on cost function** First, is that you usually have to make sure that you're at least doing well on the training set. So performance on the training set needs to pass some acceptability assessment. For some applications, this might mean doing comparably to human level performance. But this will depend on your application, and we'll talk more about comparing to human level performance next week.\n2. **Fit dev set well on cost function**\n3. **Fit test set well on cost function**\n3. **Performs well in real world**\n\nel priemro se soluccion con bigget network, the optiization algorithm\nel segundo con regularization o con un bigger traingin set\net tres con bigger dev set\ny el cuarto cambiando el dev set o la cost function\n\n The exact details of what's precision and recall don't matter too much for this example. But briefly, the definition of precision is, of the examples that your classifier recognizes as cats,\nPlay video starting at 1 minute 23 seconds and follow transcript1:23\nWhat percentage actually are cats?\nPlay video starting at 1 minute 32 seconds and follow transcript1:32\nSo if classifier A has 95% precision, this means that when classifier A says something is a cat, there's a 95% chance it really is a cat. And recall is, of all the images that really are cats, what percentage were correctly recognized by your classifier? So what percentage of actual cats, Are correctly recognized?\n\n<img align='center' src='images/metric.PNG' width='650'/>",
"_____no_output_____"
],
[
"* I often recommend that you set up a single real number evaluation metric for your problem. Let's look at an example.\n\nprecision: the examples that your classifier recognizes as cats, What percentage actually are cats\no if classifier A has 95% precision, this means that when classifier A says something is a cat, there's a 95% chance it really is a cat.\n\nrecall: of all the images that really are cats, what percentage were correctly recognized by your classifier? So what percentage of actual cats, Are correctly recognized? So if classifier A is 90% recall, this means that of all of the images in, say, your dev sets that really are cats, classifier A accurately pulled out 90% of them. \n\n\ntrade-off between precision and recall\n\n\nThe problem with using precision recall as your evaluation metric is that if classifier A does better on recall, which it does here, the classifier B does better on precision, then you're not sure which classifier is better.\n\nyou just have to find a new evaluation metric that combines precision and recall.\n \nIn the machine learning literature, the standard way to combine precision and recall is something called an F1 score. Think as average of precision (P) and recall\n\n\n$$F1 = \\frac{2}{\\frac{1}{P}+\\frac{1}{R}}$$ Harmonic mean of precition P and Recall R\n\nwhat I recommend in this example is, in addition to tracking your performance in the four different geographies, to also compute the average. And assuming that average performance is a reasonable single real number evaluation metric, by computing the average, you can quickly tell that it looks like algorithm C has a lowest average error.\n\n---\n\n**Satisficing and Optimizing metric**\n\nTo summarize, if there are multiple things you care about by say there's one as the optimizing metric that you want to do as well as possible on and one or more as satisficing metrics were you'll be satisfice. Almost it does better than some threshold you can now have an almost automatic way of quickly looking at multiple core size and picking the, quote, best one. Now these evaluation matrix must be evaluated or calculated on a training set or a development set or maybe on the test set. So one of the things you also need to do is set up training, dev or development, as well as test sets. In the next video, I want to share with you some guidelines for how to set up training, dev, and test sets. So let's go on to the next.\n\ncost = accuracy - 0.5 * running time\n\nmaximize accuracy but subject \n\nthat maximizes accuracy but subject to that the running time, that is the time it takes to classify an image, that that has to be less than or equal to 100 milliseconds. \n\nthat running time is what we call a satisficing metric\n\nSo in this case accuracy is the optimizing metric and a number of false positives every 24 hours is the satisficing metric\n\n**Train/dev/test distributions**\n\nThe way you set up your training dev, or development sets and test sets, can have a huge impact on how rapidly you or your team can make progress on building machine learning application.\n\n* Dev set So, that dev set is also called the development set, or sometimes called the hold out cross validation set. And, workflow in machine learning is that you try a lot of ideas, train up different models on the training set, and then use the dev set to evaluate the different ideas and pick one. And, keep iterating to improve dev set performance until, finally, you have one clause that you're happy with that you then evaluate on your test set.\n* choose a dev set and test set to reflect data you expect to get in future and consider important to do well on. And, in particular, the dev set and the test set here, should come from the same distribution. So, whatever type of data you expect to get in the future, and once you do well on, try to get data that looks like that. ",
"_____no_output_____"
],
[
"## Size of Dev set\n\n* So if you had a hundred examples in total, these 70/30 or 60/20/20 rule of thumb would be pretty reasonable. If you had thousand examples, maybe if you had ten thousand examples, these heuristics are not unreasonable.\n\n* say you have a million training examples. it might be quite reasonable to set up your data so that you have 98% in the training set, 1% dev, and 1% test.\n\n\n## Size of test set\n\n* Set your test set to be enough to give high confidence in the overall performance of your system.\n\n* Maybe all you need is a train and dev set, And I think, not having a test set might be okay\n\n* I do find it reassuring to have a separate test set you can use to get an unbiased estimate of how I was doing before you shift it, but if you have a very large dev set so that you think you won't overfit the dev set too bad\n\nSo to summarize, in the era of big data, I think the old rule of thumb of a 70/30 is that, that no longer applies. And the trend has been to use more data for training and less for dev and test, especially when you have a very large data sets. And the rule of thumb is really to try to set the dev set to big enough for its purpose, which helps you evaluate different ideas and pick this up from AOP better. And the purpose of test set is to help you evaluate your final cost buys. You just have to set your test set big enough for that purpose, and that could be much less than 30% of the data. So, I hope that gives some guidance or some suggestions on how to set up your dev and test sets in the Deep Learning era. Next, it turns out that sometimes, part way through a machine learning problem, you might want to change your evaluation metric, or change your dev and test sets. Let's talk about it when you might want to do that.\n\n---",
"_____no_output_____"
],
[
"### When to change dev/test sets and metrics\n\nYou've seen how set to have a dev set and evaluation metric is like placing a target somewhere for your team to aim at. \n\n**Orthogonalization for better performance**\n1. PLace target\n\n$$Error = \\frac{1}{\\sum_i w^{(i)}}\\sum_i w^{(i)}L\\{Y_{pred}^{(i)}, y^{(i)} \\}$$\n\n$w^{(i)}$ = 1 if $x^{(i)}$ is non-porn \n$w^{(i)}$ = 10 if $x^{(i)}$ is porn\n\n*Orthogonalization for cat picture: anti-porn*\n* So far weve only discussed how to define a metric to evaluate classifiers (Place the target)\n* Worry separately about how to do wel on this metric\n\n---\n\nBayes optimal error. Best posible error. That can't never being surpass.\n\n*Why compare to human-level performance*\nHumans are quite good a lot of task. So long as ML is worse than human, you can:\n* get labeled data from humans\n* Gain insight from manual error analysis. Why did a person get this right\n* Better analysis of bias/variances\n\n# Avoidable Bias\n\n* o the fact that there's a huge gap between how well your algorithm does on your training set versus how humans do shows that your algorithm isn't even fitting the training set well. So in terms of tools to reduce bias or variance, in this case I would say focus on reducing bias. So you want to do things like train a bigger neural network or run training set longer, just try to do better on the training set\n\n* Avoidable bias: difference between bayes and training error. You don't actually want to get below Bayes error.\n\n* Variance: Difference betweem training error and dev error.\n\n* Focus on either bias or variance reduction techniques\n\n<img align='center' src='images/AvoidableBIAS.PNG' width='650'/>",
"_____no_output_____"
],
[
"# Understanding Human Level Performance\n\n**Proxy for Bayes Error**\"\n\nSo to recap, having an estimate of human-level performance gives you an estimate of Bayes error. And this allows you to more quickly make decisions as to whether you should focus on trying to reduce a bias or trying to reduce the variance of your algorithm.\nSuppose:\n\n### Surpassing human-level performance\n\n* once you've surpassed this 0.5% threshold, your options, your ways of making progress on the machine learning problem are just less clear. It doesn't mean you can't make progress, you might still be able to make significant progress, but some of the tools you have for pointing you in a clear direction just don't work as well\n\n*Problems where ML significantly surpasses human -level performance*\n* Online advertising\n* Product recommendations\n* Logistic (predicting transit time)\n* Loan approvals\n\n1. All this examples are actually learning from structured data. Where you might have a databse of waht has users clicked on. This are not actual perception problems. These are not computer vision\n\n\n2. today there are speech recognition systems that can surpass human-level performance. And there are also some computer vision, some image recognition tasks, where computers have surpassed human-level performance\n\n3. Medical\n\n ECGs, skin cancer, narrow radiology task\n \n## Improving your model performance\n\n**supervised learning algorithm to work well**\n1. You can fit the training set pretty well \n - LOW Avoidable bias\n - Problem is solved by training a bigger network or training longer\n2. The Training set performance generalizes pretty well to the dev/test set\n - Variance\n - Problem is solved regularization or getting more training data that could help you generalize better to dev set dat\n \n**Steps.**\n\n1. looking at the difference between your training error and your proxy for Bayes error and just gives you a sense of the avoidable bias. In other words, just how much better do you think you should be trying to do on your training set. \n2. And then look at the difference between your dev error and your training error as an estimate of how much of a variance problem you have. In other words, how much harder you should be working to make your performance generalized from the training set to the dev set that it wasn't trained on explicitly.\n\n<img align='left' src='images/ruleofthumb.PNG' width='650'/>",
"_____no_output_____"
]
],
[
[
"[9]*7",
"_____no_output_____"
]
],
[
[
"# Carrying out error analysis\n\nIf you're trying to get a learning algorithm to do a task that humans can do. And if your learning algorithm is not yet at the performance of a human. Then manually examining mistakes that your algorithm is making, can give you insights into what to do next. This process is called error analysis. \n\nIn machine learning, sometimes we call this the ceiling on performance. Which just means, what's in the best case? How well could working on the dog problem help you?\n\n error analysis, can save you a lot of time. In terms of deciding what's the most important, or what's the most promising direction to focus on.\n \n In this slide, we'll describe using error analysis to evaluate whether or not a single idea, dogs in this case, is worth working on.\n \n*Look at dev examples to evaluate ideas*\n\nError analysis:\n* Get ~100 mislabeled dev set examples\n* count up how many are dogs\n\n5/100\n\n\"Ceiling\" upper bound on how much you could improve performance \n\nIn other case 50/100 are dogs, it is worth spending time on the dog problem.\n\n\n*Evaluate multiple idea in parallel*\n\nIdeas for cat detection:\n* Fix pictures of dogs being recognized as cats\n* Fix great cats (lions, panther, ) being misrecognized\n* Improve performance on blurry images\n \n \n# Cleaning up incorrectly labeled data\n\n**deep learning algorithms are quite robust to random errors in the training set.**\n\n* They are less robust to systematic errors.\n\nSo for example, if your labeler consistently labels white dogs as cats, then that is a problem because your classifier will learn to classify all white colored dogs as cats\n\n\n**here are a few additional guidelines or principles to consider**\n\n* f you're going in to fix something on the dev set, I would apply the same process to the test set to make sure that they continue to come from the same distribution\n\n* It's super important that your dev and test sets come from the same distribution.\n\n# Build your first system quickly, then iterate\n\n\nIf you're working on a brand new machine learning application, one of the piece of advice I often give people is that, I think you should build your first system quickly and then iterate. your main goal is to build something that works, as opposed to if your main goal is to invent a new machine learning algorithm which is a different goal, then your main goal is to get something that works really well. I'd encourage you to build something quick and dirty. Use that to do bias/variance analysis, use that to do error analysis and use the results of those analysis to help you prioritize where to go next.\n\n* Set up dev/set and metric\n* Build initial system quickly\n* Use bias/variance analysis & error analysis to prioritize next steps\n\n# Training and testing on different distributions\net. So in this video, you've seen a couple examples of when allowing your training set data to come from a different distribution than your dev and test set allows you to have much more training data. And in these examples, it will cause your learning algorithm to perform better. Now one question you might ask is, should you always use all the data you have? The answer is subtle, it is not always yes.\n\n\n# Bias and Variance with mismatched data distributions\n\nPreviously we had set up some training sets and some dev sets and some test sets as follows. And the dev and test sets have the same distribution, but the training sets will have some different distribution. What we're going to do is randomly shuffle the training sets and then carve out just a piece of the training set to be the training-dev set. So just as the dev and test set have the same distribution, the training set and the training-dev set, also have the same distribution.\n\n**Key QUantities**\n- HUman Level error\n- Train set error\n- Train dev - set error\n- Dev error\n\n\n<img align='center' src='images/biasmismatch.PNG' width='400'/>\n\n\n**More general formulation**\n\nSo what we've seen is that by using training data that can come from a different distribution as a dev and test set, this could give you a lot more data and therefore help the performance of your learning algorithm. But instead of just having bias and variance as two potential problems, you now have this third potential problem, data mismatch. So what if you perform error analysis and conclude that data mismatch is a huge source of error, how do you go about addressing that? It turns out that unfortunately there are super systematic ways to address data mismatch, but there are a few things you can try that could help. Let's take a look at them in the next video.\n\n<img align='center' src='images/data_mismatch.PNG' width='700'/>\n",
"_____no_output_____"
],
[
"# Addressing data mismatch\n\nIf your training set comes from a different distribution, than your dev and test set, and if error analysis shows you that you have a data mismatch problem, what can you do?\n\n1. Carry out manual error analysis and try to understand the differences between the training set and the dev/test sets. To avoid overfitting the test set, technically for error analysis, you should manually only look at a dev set and not at the test set \n2. try to collect more data similar to your dev and test sets. \n\nSo, to summarize, if you think you have a data mismatch problem, I recommend you do error analysis, or look at the training set, or look at the dev set to try this figure out, to try to gain insight into how these two distributions of data might differ. And then see if you can find some ways to get more training data that looks a bit more like your dev set. One of the ways we talked about is artificial data synthesis. And artificial data synthesis does work. In speech recognition, I've seen artificial data synthesis significantly boost the performance of what were already very good speech recognition system. So, it can work very well. But, if you're using artificial data synthesis, just be cautious and bear in mind whether or not you might be accidentally simulating data only from a tiny subset of the space of all possible examples. So, that's it for how to deal with data mismatch.\n\n# Transfer learning\n\nBut if you have a lot of data, then maybe you can retrain all the parameters in the network. And if you retrain all the parameters in the neural network, then this initial phase of training on image recognition is sometimes called pre-training, because you're using image recognitions data to pre-initialize or really pre-train the weights of the neural network. And then if you are updating all the weights afterwards, then training on the radiology data sometimes that's called fine tuning.\n\n- Pre-training\n- FIne tuning\n\nAnd the reason this can be helpful is that a lot of the low level features such as detecting edges, detecting curves, detecting positive objects. Learning from that, from a very large image recognition data set, might help your learning algorithm do better in radiology diagnosis. It's just learned a lot about the structure and the nature of how images look like and some of that knowledge will be useful. So having learned to recognize images, it might have learned enough about you know, just what parts of different images look like, that that knowledge about lines, dots, curves, and so on, maybe small parts of objects, that knowledge could help your radiology diagnosis network learn a bit faster or learn with less data\n\nyou're transferring from a problem with a lot of data to a problem with relatively little data. \n\n**When transfer learning makes sense**\n\n* Task A and B have the same input X\n* You have a lot more data for Task A than Task B\n* Low level features from A could be helpful for learning B",
"_____no_output_____"
],
[
"# Multi-task learning\n\n\nSo whereas in transfer learning, you have a sequential process where you learn from task A and then transfer that to task B. In multi-task learning, you start off simultaneously, trying to have one neural network do several things at the same time. And then each of these task helps hopefully all of the other task. Let's look at an example.\n\n So to summarize, multi-task learning enables you to train one neural network to do many tasks and this can give you better performance than if you were to do the tasks in isolation. Now one note of caution, in practice I see that transfer learning is used much more often than multi-task learning. So I do see a lot of tasks where if you want to solve a machine learning problem but you have a relatively small data set, then transfer learning can really help. Where if you find a related problem but you have a much bigger data set, you can train in your neural network from there and then transfer it to the problem where we have very low data. So transfer learning is used a lot today. There are some applications of transfer multi-task learning as well, but multi-task learning I think is used much less often than transfer learning. And maybe the one exception is computer vision object detection,\n \n <img align='center' src='images/multi.PNG' width='900'/>",
"_____no_output_____"
],
[
"# What is end-to-end deep learning?\n\nBriefly, there have been some data processing systems, or learning systems that require multiple stages of processing. And what end-to-end deep learning does, is it can take all those multiple stages, and replace it usually with just a single neural network.\n\nExample: Face recognition\n\nfirst crop the face, \nThen train ML to recognize the person.\n\nIt is not a good approach to train ML to images where people is approaching to the camera\n\n**Pros**\n\n* Let the data speak\n* Less hand-designing of components needed\n\n**Cons**\n* May need large amount of data\n* Excludes potentially useful hand-designed components\n\n\n## Whether to use end-to-end deep learning\n\n\n* Use DL to learn individual components\n* when applying supervised learning you should carefully choose what types of X to Y mappings you want to learn depending on what task you can get data fo",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a12be756c54dc346034b15291a28eecbcd79109
| 11,594 |
ipynb
|
Jupyter Notebook
|
usr/pir/Test 1-10/TE1_thinkpy_exam.ipynb
|
Bugnon/oc-2018
|
7961de5ba9923512bd50c579c37f1dadf070b692
|
[
"MIT"
] | 3 |
2018-09-20T12:16:48.000Z
|
2019-06-21T08:32:17.000Z
|
usr/pir/Test 1-10/TE1_thinkpy_exam.ipynb
|
Bugnon/oc-2018
|
7961de5ba9923512bd50c579c37f1dadf070b692
|
[
"MIT"
] | null | null | null |
usr/pir/Test 1-10/TE1_thinkpy_exam.ipynb
|
Bugnon/oc-2018
|
7961de5ba9923512bd50c579c37f1dadf070b692
|
[
"MIT"
] | 2 |
2018-09-20T11:55:05.000Z
|
2019-09-01T19:40:13.000Z
| 28.072639 | 764 | 0.50069 |
[
[
[
"# Travail Ecrit - Python\n* Gymnase du Bugnon, site de l'Ours\n* OC informatique\n* Sujet : chapitres 1-10 du livre *Pensez en Python*\n* Mirko Pirona\n* Date : jeudi 13 novembre 2018",
"_____no_output_____"
],
[
"## **Exercice : expression arithmétique** \nInitialisez les variables `(a, b, c, x)` avec les valeurs `(2, 3, 4, 5)`. \nCalculez l'expression \n\n$$y = a x^2 + b x +c$$\n\net imprimez le résultat.",
"_____no_output_____"
]
],
[
[
"a=2\nb=3\nc=4\nx=5\ny=a*x^2+b*x+c\nprint(y)",
"_____no_output_____"
]
],
[
[
"## **Exercice : fonction surface** \nImportez le module `math`, \ndéfinissez une fonction `surface(r)` qui calcule $s = \\pi r^2$, \naffichez avec un texte descriptif le résultat pour `r=5` ",
"_____no_output_____"
]
],
[
[
"import math\ndef surface(r):\n s=math.pi*r^2\n print(s, '=', 'la surface de rayon r')",
"_____no_output_____"
]
],
[
[
"## **Exercice : formule quadratique** \nLa solution d'une formule quadratique de forme\n\n$$ a x^2 + b x +c = 0 $$\n\ndépend du terme $\\Delta = b^2 - 4 a c$\n* Si $\\Delta < 0$ il n'y a pas de solution\n* Si $\\Delta = 0$ il y a une solution : $x = \\frac{-b}{2 a}$\n* Si $\\Delta > 0$ il y a deux solutions : \n$x_1 = \\frac{-b +\\sqrt\\Delta}{2 a}$ and $x_2 = \\frac{-b -\\sqrt\\Delta}{2 a}$\n\nDéfinissez une fonction `quadratique(a, b, c)` qui retourne la solution à l'équation quadratique dans les 3 cas: `None`, `x`, `[x1, x2]`\n.\n\nMontrez la solution pour `quadratique(1, 2, 3)`, `quadratique(1, 2, 1)` et `quadratique(1, 2, -1)`",
"_____no_output_____"
]
],
[
[
"def quadratique(a, b, c):\nt=b^2-4a*c\n if b^2-4a*c < 0:\n print (None)\n elif b^2-4a*c = 0:\n x=(-b/(2a))\n print(x)\n else b^2-4*a*c > 0:\n q=(-b+(math.sqrt(t)))/(2*a)\n s=(-b-(math.sqrt(t)))/(2*a)\n print(q, s)\n \n \n \nprint(quadratique(1, 2, 3))\nprint(quadratique(1, 2, 1))\nprint(quadratique(1, 2, -1))",
"_____no_output_____"
]
],
[
[
"## **Exercice : capitalize** \nCréez une fonction `capitalize(c)` qui transforme une lettre en majuscule si c'est une minuscule, ou la laisse inchangée autrement.",
"_____no_output_____"
]
],
[
[
"def capitalize(c):\n if type (c) ==str:\n \n if c is str(c.upper()):\n return\n else c is not str(c.upper()):\n a=c.upper()\n print(a)\n \n\ncapitalize('a'), capitalize('B'), capitalize('3')",
"_____no_output_____"
]
],
[
[
"## **Exercice : capitalize words** \nCréez une fonction `capitalize_words(s)` qui transforme la première lettre de tous les mots en majuscule.",
"_____no_output_____"
]
],
[
[
"def capitalize_words(s):\n for i in s:\n x=s.upper()\n print(x)\n \n\ncapitalize_words('hello world, how are you?')",
"HELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\nHELLO WORLD, HOW ARE YOU?\n"
]
],
[
[
"## **Exercice : tranches** \nExpliquez ce que font les 6 opérateurs de **tranches** ci-dessous.",
"_____no_output_____"
]
],
[
[
"s=['a', 'b', 'c', 'd', 'e',]\n\ns[::2]\ns[:2]\ns[::-1]\n\n\n# s[2] - Cela affiche le troisième élément d'une liste\n# s[:2] - Cela affiche le 2 premier élément d'une liste\n# s[::2] - Cela prend un élément sur 2 d'une liste et l'affiche\n\n# s[-1] -Cela prend le dernier élément d'une liste\n# s[:-1] -Cela prend tout sauf le dernier élément\n# s[::-1] -Cela affiche la liste à l'envers",
"_____no_output_____"
]
],
[
[
"## **Exercice : longueur spécifique** \nLe fichier `words.text` contient 58110 mots en anglais. \nAffichez les $n=5$ premiers mots qui ont une longueur de $m=10$ et affichez leur nombre total.",
"_____no_output_____"
]
],
[
[
"fin = open('words.txt')\nfor n in fin:\n if len(n) < 10:\n break\n else len(n) >10:\n print(n)\nn = 5\nm = 10\n",
"_____no_output_____"
]
],
[
[
"## **Exercice : répétition**. \nAffchez les $m=5$ premiers mots qui sont composé de deux parties répétées (par exemple **bonbon**).",
"_____no_output_____"
]
],
[
[
"fin = open('words.txt')\n\nfor m in fin:\n if m[0:((len(m)/2)-1)] == m[((len(m)/2)-1):(len(m)-1)]\n print(i)\nm = 5\n",
"_____no_output_____"
]
],
[
[
"## **Exercice : minimum** \nCréez une fonction `min(L)` qui retourne le minimum d'une liste et l'index de sa position sous forme de list `[val, pos]`\n.",
"_____no_output_____"
]
],
[
[
"def min(L):\n x=len(L)\n print(L[-1], x)\n \n \nL = [1, 3, 34, -4, -2, 100]\nmin(L)",
"100 6\n"
]
],
[
[
"## **Exercice : moyenne** \nEcrivez une fonction `mean(L)` qui retourne la moyenne d'une liste.\n.",
"_____no_output_____"
]
],
[
[
"def mean(L):\n x=sum(L)\n print(x/2)\n \nL = [1, 3, 34, -4, -2, 100]\nmean(L)",
"66.0\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a12da32f9212afd009d00cb3f237012e6aefd81
| 29,044 |
ipynb
|
Jupyter Notebook
|
notebooks/development/05_Pyomo_Examples.ipynb
|
gschivley/ND-Pyomo-Cookbook
|
7bddccd14eb3044e9d3e7c46999e1c9c29541e16
|
[
"Apache-2.0"
] | null | null | null |
notebooks/development/05_Pyomo_Examples.ipynb
|
gschivley/ND-Pyomo-Cookbook
|
7bddccd14eb3044e9d3e7c46999e1c9c29541e16
|
[
"Apache-2.0"
] | null | null | null |
notebooks/development/05_Pyomo_Examples.ipynb
|
gschivley/ND-Pyomo-Cookbook
|
7bddccd14eb3044e9d3e7c46999e1c9c29541e16
|
[
"Apache-2.0"
] | 2 |
2018-10-14T16:40:40.000Z
|
2021-07-20T12:02:29.000Z
| 51.043937 | 1,906 | 0.578536 |
[
[
[
"# Pyomo Examples",
"_____no_output_____"
],
[
"## Example 19.3: Linear Programming Refinery",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\nPRODUCTS = ['Gasoline', 'Kerosine', 'Fuel Oil', 'Residual']\nFEEDS = ['Crude 1', 'Crude 2']\n\nproducts = pd.DataFrame(index=PRODUCTS)\nproducts['Price'] = [72, 48, 42, 20]\nproducts['Max Production'] = [24000, 2000, 6000, 100000]\n\ncrudes = pd.DataFrame(index=FEEDS)\ncrudes['Processing Cost'] = [1.00, 2.00]\ncrudes['Feed Costs'] = [48, 30]\n\nyields = pd.DataFrame(index=PRODUCTS)\nyields['Crude 1'] = [0.80, 0.05, 0.10, 0.05]\nyields['Crude 2'] = [0.44, 0.10, 0.36, 0.10]\n\nprint('\\n', products)\nprint('\\n', crudes)\nprint('\\n', yields)",
"\n Price Max Production\nGasoline 72 24000\nKerosine 48 2000\nFuel Oil 42 6000\nResidual 20 100000\n\n Processing Cost Feed Costs\nCrude 1 1.0 48\nCrude 2 2.0 30\n\n Crude 1 Crude 2\nGasoline 0.80 0.44\nKerosine 0.05 0.10\nFuel Oil 0.10 0.36\nResidual 0.05 0.10\n"
],
[
"price = {}\nprice['Gasoline'] = 72\nprice['Kerosine'] = 48\nprice\n",
"_____no_output_____"
],
[
"products.loc['Gasoline','Price']",
"_____no_output_____"
],
[
"# model formulation\nmodel = ConcreteModel()\n\n# variables\nmodel.x = Var(FEEDS, domain=NonNegativeReals)\nmodel.y = Var(PRODUCTS, domain=NonNegativeReals)\n\n# objective\nincome = sum(products.ix[p, 'Price'] * model.y[p] for p in PRODUCTS)\nraw_materials_cost = sum(crudes.ix[f,'Feed Costs'] * model.x[f] for f in FEEDS)",
"/Users/jeff/anaconda3/lib/python3.6/site-packages/ipykernel_launcher.py:10: DeprecationWarning: \n.ix is deprecated. Please use\n.loc for label based indexing or\n.iloc for positional indexing\n\nSee the documentation here:\nhttp://pandas.pydata.org/pandas-docs/stable/indexing.html#ix-indexer-is-deprecated\n # Remove the CWD from sys.path while we load stuff.\n"
],
[
"processing_cost = sum(processing_costs[f] * model.x[f] for f in FEEDS)\nprofit = income - raw_materials_cost - processing_cost\nmodel.objective = Objective(expr = profit, sense=maximize)\n\n# constraints\nmodel.constraints = ConstraintList()\nfor p in PRODUCTS:\n model.constraints.add(0 <= model.y[p] <= max_production[p])\nfor p in PRODUCTS:\n model.constraints.add(model.y[p] == sum(model.x[f] * product_yield[(f,p)] for f in FEEDS))\n\nsolver = SolverFactory('glpk')\nsolver.solve(model)\nmodel.pprint()",
"_____no_output_____"
],
[
"from pyomo.environ import *\nimport numpy as np\n\n# problem data\nFEEDS = ['Crude #1', 'Crude #2']\nPRODUCTS = ['Gasoline', 'Kerosine', 'Fuel Oil', 'Residual']\n\n# feed costs\nfeed_costs = {'Crude #1': 48,\n 'Crude #2': 30}\n\n# processing costs\nprocessing_costs = {'Crude #1': 1.00,\n 'Crude #2': 2.00}\n\n# yield data\nproduct_yield = \nproduct_yield = {('Crude #1', 'Gasoline'): 0.80,\n ('Crude #1', 'Kerosine'): 0.05,\n ('Crude #1', 'Fuel Oil'): 0.10,\n ('Crude #1', 'Residual'): 0.05,\n ('Crude #2', 'Gasoline'): 0.44,\n ('Crude #2', 'Kerosine'): 0.10,\n ('Crude #2', 'Fuel Oil'): 0.36,\n ('Crude #2', 'Residual'): 0.10}\n\n# product sales prices\nsales_price = {'Gasoline': 72,\n 'Kerosine': 48,\n 'Fuel Oil': 42,\n 'Residual': 20}\n\n# production limits\nmax_production = {'Gasoline': 24000,\n 'Kerosine': 2000,\n 'Fuel Oil': 6000,\n 'Residual': 100000}\n\n# model formulation\nmodel = ConcreteModel()\n\n# variables\nmodel.x = Var(FEEDS, domain=NonNegativeReals)\nmodel.y = Var(PRODUCTS, domain=NonNegativeReals)\n\n# objective\nincome = sum(sales_price[p] * model.y[p] for p in PRODUCTS)\nraw_materials_cost = sum(feed_costs[f] * model.x[f] for f in FEEDS)\nprocessing_cost = sum(processing_costs[f] * model.x[f] for f in FEEDS)\n\nprofit = income - raw_materials_cost - processing_cost\nmodel.objective = Objective(expr = profit, sense=maximize)\n\n# constraints\nmodel.constraints = ConstraintList()\nfor p in PRODUCTS:\n model.constraints.add(0 <= model.y[p] <= max_production[p])\nfor p in PRODUCTS:\n model.constraints.add(model.y[p] == sum(model.x[f] * product_yield[(f,p)] for f in FEEDS))\n\nsolver = SolverFactory('glpk')\nsolver.solve(model)\nmodel.pprint()",
"3 Set Declarations\n constraints_index : Dim=0, Dimen=1, Size=8, Domain=None, Ordered=False, Bounds=None\n [1, 2, 3, 4, 5, 6, 7, 8]\n x_index : Dim=0, Dimen=1, Size=2, Domain=None, Ordered=False, Bounds=None\n ['Crude #1', 'Crude #2']\n y_index : Dim=0, Dimen=1, Size=4, Domain=None, Ordered=False, Bounds=None\n ['Fuel Oil', 'Gasoline', 'Kerosine', 'Residual']\n\n2 Var Declarations\n x : Size=2, Index=x_index\n Key : Lower : Value : Upper : Fixed : Stale : Domain\n Crude #1 : 0 : 26206.8965517241 : None : False : False : NonNegativeReals\n Crude #2 : 0 : 6896.55172413793 : None : False : False : NonNegativeReals\n y : Size=4, Index=y_index\n Key : Lower : Value : Upper : Fixed : Stale : Domain\n Fuel Oil : 0 : 5103.44827586207 : None : False : False : NonNegativeReals\n Gasoline : 0 : 24000.0 : None : False : False : NonNegativeReals\n Kerosine : 0 : 2000.0 : None : False : False : NonNegativeReals\n Residual : 0 : 2000.0 : None : False : False : NonNegativeReals\n\n1 Objective Declarations\n objective : Size=1, Index=None, Active=True\n Key : Active : Sense : Expression\n None : True : maximize : 72*y[Gasoline] + 48*y[Kerosine] + 42*y[Fuel Oil] + 20*y[Residual] - 48*x[Crude #1] - 30*x[Crude #2] - x[Crude #1] - 2.0*x[Crude #2]\n\n1 Constraint Declarations\n constraints : Size=8, Index=constraints_index, Active=True\n Key : Lower : Body : Upper : Active\n 1 : 0.0 : y[Gasoline] : 24000.0 : True\n 2 : 0.0 : y[Kerosine] : 2000.0 : True\n 3 : 0.0 : y[Fuel Oil] : 6000.0 : True\n 4 : 0.0 : y[Residual] : 100000.0 : True\n 5 : 0.0 : y[Gasoline] - 0.8*x[Crude #1] - 0.44*x[Crude #2] : 0.0 : True\n 6 : 0.0 : y[Kerosine] - 0.05*x[Crude #1] - 0.1*x[Crude #2] : 0.0 : True\n 7 : 0.0 : y[Fuel Oil] - 0.1*x[Crude #1] - 0.36*x[Crude #2] : 0.0 : True\n 8 : 0.0 : y[Residual] - 0.05*x[Crude #1] - 0.1*x[Crude #2] : 0.0 : True\n\n7 Declarations: x_index x y_index y objective constraints_index constraints\n"
],
[
"profit()",
"_____no_output_____"
],
[
"income()",
"_____no_output_____"
],
[
"raw_materials_cost()",
"_____no_output_____"
],
[
"processing_cost()",
"_____no_output_____"
]
],
[
[
"## Example: Making Change\n\nOne of the important modeling features of Pyomo is the ability to index variables and constraints. The",
"_____no_output_____"
]
],
[
[
"from pyomo.environ import *\n\ndef make_change(amount, coins):\n model = ConcreteModel()\n model.C = Set(initialize=coins.keys())\n model.x = Var(model.C, domain=NonNegativeIntegers)\n model.n = Objective(expr = sum(model.x[c] for c in model.C), sense=minimize)\n model.v = Constraint(expr = sum(model.x[c]*coins[c] for c in model.C) == amount)\n SolverFactory('glpk').solve(model)\n return {c: int(model.x[c]()) for c in model.C} \n \n# problem data\ncoins = {\n 'penny' : 1,\n 'nickel' : 5,\n 'dime' : 10,\n 'quarter': 25,\n}\n \nmake_change(93, coins)",
"_____no_output_____"
]
],
[
[
"## Example: Linear Production Model with Constraints with Duals",
"_____no_output_____"
]
],
[
[
"from pyomo.environ import *\nmodel = ConcreteModel()\n\n# for access to dual solution for constraints\nmodel.dual = Suffix(direction=Suffix.IMPORT)\n\n# declare decision variables\nmodel.x = Var(domain=NonNegativeReals)\nmodel.y = Var(domain=NonNegativeReals)\n\n# declare objective\nmodel.profit = Objective(expr = 40*model.x + 30*model.y, sense = maximize)\n\n# declare constraints\nmodel.demand = Constraint(expr = model.x <= 40)\nmodel.laborA = Constraint(expr = model.x + model.y <= 80)\nmodel.laborB = Constraint(expr = 2*model.x + model.y <= 100)\n\n# solve\nSolverFactory('glpk').solve(model).write()",
"# ==========================================================\n# = Solver Results =\n# ==========================================================\n# ----------------------------------------------------------\n# Problem Information\n# ----------------------------------------------------------\nProblem: \n- Name: unknown\n Lower bound: 2600.0\n Upper bound: 2600.0\n Number of objectives: 1\n Number of constraints: 4\n Number of variables: 3\n Number of nonzeros: 6\n Sense: maximize\n# ----------------------------------------------------------\n# Solver Information\n# ----------------------------------------------------------\nSolver: \n- Status: ok\n Termination condition: optimal\n Statistics: \n Branch and bound: \n Number of bounded subproblems: 0\n Number of created subproblems: 0\n Error rc: 0\n Time: 0.01401972770690918\n# ----------------------------------------------------------\n# Solution Information\n# ----------------------------------------------------------\nSolution: \n- number of solutions: 0\n number of solutions displayed: 0\n"
],
[
"str = \" {0:7.2f} {1:7.2f} {2:7.2f} {3:7.2f}\"\n\nprint(\"Constraint value lslack uslack dual\")\nfor c in [model.demand, model.laborA, model.laborB]:\n print(c, str.format(c(), c.lslack(), c.uslack(), model.dual[c]))",
"Constraint value lslack uslack dual\ndemand 20.00 -inf 20.00 0.00\nlaborA 80.00 -inf 0.00 20.00\nlaborB 100.00 -inf 0.00 10.00\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a12ecbde595cec78d6edb4f03ae790eac308899
| 30,925 |
ipynb
|
Jupyter Notebook
|
homework01/homework_test_modules.ipynb
|
yaPhilya/Practical_DL
|
4d39caef62e3679baa9c57a83cc42f191e9fe335
|
[
"MIT"
] | null | null | null |
homework01/homework_test_modules.ipynb
|
yaPhilya/Practical_DL
|
4d39caef62e3679baa9c57a83cc42f191e9fe335
|
[
"MIT"
] | null | null | null |
homework01/homework_test_modules.ipynb
|
yaPhilya/Practical_DL
|
4d39caef62e3679baa9c57a83cc42f191e9fe335
|
[
"MIT"
] | null | null | null | 53.411054 | 123 | 0.553727 |
[
[
[
"%run homework_modules.ipynb",
"_____no_output_____"
],
[
"import torch\nfrom torch.autograd import Variable\nimport numpy\nimport unittest",
"_____no_output_____"
],
[
"class TestLayers(unittest.TestCase):\n def test_Linear(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in, n_out = 2, 3, 4\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.Linear(n_in, n_out)\n custom_layer = Linear(n_in, n_out)\n custom_layer.W = torch_layer.weight.data.numpy()\n custom_layer.b = torch_layer.bias.data.numpy()\n\n layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(-10, 10, (batch_size, n_out)).astype(np.float32)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n \n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n\n # 3. check layer parameters grad\n custom_layer.accGradParameters(layer_input, next_layer_grad)\n weight_grad = custom_layer.gradW\n bias_grad = custom_layer.gradb\n torch_weight_grad = torch_layer.weight.grad.data.numpy()\n torch_bias_grad = torch_layer.bias.grad.data.numpy()\n self.assertTrue(np.allclose(torch_weight_grad, weight_grad, atol=1e-6))\n self.assertTrue(np.allclose(torch_bias_grad, bias_grad, atol=1e-6))\n\n def test_SoftMax(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.Softmax(dim=1)\n custom_layer = SoftMax()\n\n layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.random((batch_size, n_in)).astype(np.float32)\n next_layer_grad /= next_layer_grad.sum(axis=-1, keepdims=True)\n next_layer_grad = next_layer_grad.clip(1e-5,1.)\n next_layer_grad = 1. / next_layer_grad\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-5))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))\n \n def test_LogSoftMax(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.LogSoftmax(dim=1)\n custom_layer = LogSoftMax()\n\n layer_input = np.random.uniform(-10, 10, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.random((batch_size, n_in)).astype(np.float32)\n next_layer_grad /= next_layer_grad.sum(axis=-1, keepdims=True)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n\n def test_BatchNormalization(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 32, 16\n for _ in range(100):\n # layers initialization\n slope = np.random.uniform(0.01, 0.05)\n alpha = 0.9\n custom_layer = BatchNormalization(alpha)\n custom_layer.train()\n torch_layer = torch.nn.BatchNorm1d(n_in, eps=custom_layer.EPS, momentum=1.-alpha, affine=False)\n custom_layer.moving_mean = torch_layer.running_mean.numpy().copy()\n custom_layer.moving_variance = torch_layer.running_var.numpy().copy()\n\n layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n\n\n # please, don't increase `atol` parameter, it's garanteed that you can implement batch norm layer\n # with tolerance 1e-5\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))\n\n # 3. check moving mean\n self.assertTrue(np.allclose(custom_layer.moving_mean, torch_layer.running_mean.numpy()))\n # we don't check moving_variance because pytorch uses slightly different formula for it:\n # it computes moving average for unbiased variance (i.e var*N/(N-1))\n #self.assertTrue(np.allclose(custom_layer.moving_variance, torch_layer.running_var.numpy()))\n\n # 4. check evaluation mode\n custom_layer.moving_variance = torch_layer.running_var.numpy().copy()\n custom_layer.evaluate()\n custom_layer_output = custom_layer.updateOutput(layer_input)\n torch_layer.eval()\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n \n def test_Sequential(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n alpha = 0.9\n torch_layer = torch.nn.BatchNorm1d(n_in, eps=BatchNormalization.EPS, momentum=1.-alpha, affine=True)\n torch_layer.bias.data = torch.from_numpy(np.random.random(n_in).astype(np.float32))\n custom_layer = Sequential()\n bn_layer = BatchNormalization(alpha)\n bn_layer.moving_mean = torch_layer.running_mean.numpy().copy()\n bn_layer.moving_variance = torch_layer.running_var.numpy().copy()\n custom_layer.add(bn_layer)\n scaling_layer = ChannelwiseScaling(n_in)\n scaling_layer.gamma = torch_layer.weight.data.numpy()\n scaling_layer.beta = torch_layer.bias.data.numpy()\n custom_layer.add(scaling_layer)\n custom_layer.train()\n\n layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.backward(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-5))\n\n # 3. check layer parameters grad\n weight_grad, bias_grad = custom_layer.getGradParameters()[1]\n torch_weight_grad = torch_layer.weight.grad.data.numpy()\n torch_bias_grad = torch_layer.bias.grad.data.numpy()\n self.assertTrue(np.allclose(torch_weight_grad, weight_grad, atol=1e-6))\n self.assertTrue(np.allclose(torch_bias_grad, bias_grad, atol=1e-6))\n\n def test_Dropout(self):\n np.random.seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n p = np.random.uniform(0.3, 0.7)\n layer = Dropout(p)\n layer.train()\n\n layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n\n # 1. check layer output\n layer_output = layer.updateOutput(layer_input)\n self.assertTrue(np.all(np.logical_or(np.isclose(layer_output, 0), \n np.isclose(layer_output*(1.-p), layer_input))))\n\n # 2. check layer input grad\n layer_grad = layer.updateGradInput(layer_input, next_layer_grad)\n self.assertTrue(np.all(np.logical_or(np.isclose(layer_grad, 0), \n np.isclose(layer_grad*(1.-p), next_layer_grad))))\n\n # 3. check evaluation mode\n layer.evaluate()\n layer_output = layer.updateOutput(layer_input)\n self.assertTrue(np.allclose(layer_output, layer_input))\n\n # 4. check mask\n p = 0.0\n layer = Dropout(p)\n layer.train()\n layer_output = layer.updateOutput(layer_input)\n self.assertTrue(np.allclose(layer_output, layer_input))\n\n p = 0.5\n layer = Dropout(p)\n layer.train()\n layer_input = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)\n layer_output = layer.updateOutput(layer_input)\n zeroed_elem_mask = np.isclose(layer_output, 0)\n layer_grad = layer.updateGradInput(layer_input, next_layer_grad) \n self.assertTrue(np.all(zeroed_elem_mask == np.isclose(layer_grad, 0)))\n\n # 5. dropout mask should be generated independently for every input matrix element, not for row/column\n batch_size, n_in = 1000, 1\n p = 0.8\n layer = Dropout(p)\n layer.train()\n\n layer_input = np.random.uniform(5, 10, (batch_size, n_in)).astype(np.float32)\n layer_output = layer.updateOutput(layer_input)\n self.assertTrue(np.sum(np.isclose(layer_output, 0)) != layer_input.size)\n\n layer_input = layer_input.T\n layer_output = layer.updateOutput(layer_input)\n self.assertTrue(np.sum(np.isclose(layer_output, 0)) != layer_input.size)\n def test_LeakyReLU(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n slope = np.random.uniform(0.01, 0.05)\n torch_layer = torch.nn.LeakyReLU(slope)\n custom_layer = LeakyReLU(slope)\n\n layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n\n def test_ELU(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n alpha = 1.0\n torch_layer = torch.nn.ELU(alpha)\n custom_layer = ELU(alpha)\n\n layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n\n def test_SoftPlus(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.Softplus()\n custom_layer = SoftPlus()\n\n layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n next_layer_grad = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n\n def test_ClassNLLCriterionUnstable(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.NLLLoss()\n custom_layer = ClassNLLCriterionUnstable()\n\n layer_input = np.random.uniform(0, 1, (batch_size, n_in)).astype(np.float32)\n layer_input /= layer_input.sum(axis=-1, keepdims=True)\n layer_input = layer_input.clip(custom_layer.EPS, 1. - custom_layer.EPS) # unifies input\n target_labels = np.random.choice(n_in, batch_size)\n target = np.zeros((batch_size, n_in), np.float32)\n target[np.arange(batch_size), target_labels] = 1 # one-hot encoding\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input, target)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(torch.log(layer_input_var), \n Variable(torch.from_numpy(target_labels), requires_grad=False))\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, target)\n torch_layer_output_var.backward()\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n\n def test_ClassNLLCriterion(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 4\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.NLLLoss()\n custom_layer = ClassNLLCriterion()\n\n layer_input = np.random.uniform(-5, 5, (batch_size, n_in)).astype(np.float32)\n layer_input = torch.nn.LogSoftmax(dim=1)(Variable(torch.from_numpy(layer_input))).data.numpy()\n target_labels = np.random.choice(n_in, batch_size)\n target = np.zeros((batch_size, n_in), np.float32)\n target[np.arange(batch_size), target_labels] = 1 # one-hot encoding\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input, target)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var, \n Variable(torch.from_numpy(target_labels), requires_grad=False))\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n\n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, target)\n torch_layer_output_var.backward()\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n \n def test_adam_optimizer(self):\n state = {} \n config = {'learning_rate': 1e-3, 'beta1': 0.9, 'beta2':0.999, 'epsilon':1e-8}\n variables = [[np.arange(10).astype(np.float64)]]\n gradients = [[np.arange(10).astype(np.float64)]]\n adam_optimizer(variables, gradients, config, state)\n self.assertTrue(np.allclose(state['m'][0], np.array([0. , 0.1, 0.2, 0.3, 0.4, 0.5, \n 0.6, 0.7, 0.8, 0.9])))\n self.assertTrue(np.allclose(state['v'][0], np.array([0., 0.001, 0.004, 0.009, 0.016, 0.025, \n 0.036, 0.049, 0.064, 0.081])))\n self.assertTrue(state['t'] == 1)\n self.assertTrue(np.allclose(variables[0][0], np.array([0., 0.999, 1.999, 2.999, 3.999, 4.999, \n 5.999, 6.999, 7.999, 8.999])))\n adam_optimizer(variables, gradients, config, state)\n self.assertTrue(np.allclose(state['m'][0], np.array([0., 0.19, 0.38, 0.57, 0.76, 0.95, 1.14, \n 1.33, 1.52, 1.71])))\n self.assertTrue(np.allclose(state['v'][0], np.array([0., 0.001999, 0.007996, 0.017991, \n 0.031984, 0.049975, 0.071964, 0.097951, \n 0.127936, 0.161919])))\n self.assertTrue(state['t'] == 2)\n self.assertTrue(np.allclose(variables[0][0], np.array([0., 0.998, 1.998, 2.998, 3.998, 4.998, \n 5.998, 6.998, 7.998, 8.998])))\n \nsuite = unittest.TestLoader().loadTestsFromTestCase(TestLayers)\nunittest.TextTestRunner(verbosity=2).run(suite)",
"test_BatchNormalization (__main__.TestLayers) ... ok\ntest_ClassNLLCriterion (__main__.TestLayers) ... ok\ntest_ClassNLLCriterionUnstable (__main__.TestLayers) ... ok\ntest_Dropout (__main__.TestLayers) ... ok\ntest_ELU (__main__.TestLayers) ... ok\ntest_LeakyReLU (__main__.TestLayers) ... ok\ntest_Linear (__main__.TestLayers) ... ok\ntest_LogSoftMax (__main__.TestLayers) ... ok\ntest_Sequential (__main__.TestLayers) ... ok\ntest_SoftMax (__main__.TestLayers) ... ok\ntest_SoftPlus (__main__.TestLayers) ... ok\ntest_adam_optimizer (__main__.TestLayers) ... ok\n\n----------------------------------------------------------------------\nRan 12 tests in 0.469s\n\nOK\n"
],
[
"class TestAdvancedLayers(unittest.TestCase):\n def test_Conv2d(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in, n_out = 2, 3, 4\n h,w = 5,6\n kern_size = 3\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.Conv2d(n_in, n_out, kern_size, padding=1)\n custom_layer = Conv2d(n_in, n_out, kern_size)\n custom_layer.W = torch_layer.weight.data.numpy() # [n_out, n_in, kern, kern]\n custom_layer.b = torch_layer.bias.data.numpy()\n\n layer_input = np.random.uniform(-1, 1, (batch_size, n_in, h,w)).astype(np.float32)\n next_layer_grad = np.random.uniform(-1, 1, (batch_size, n_out, h, w)).astype(np.float32)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n \n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n \n # 3. check layer parameters grad\n custom_layer.accGradParameters(layer_input, next_layer_grad)\n weight_grad = custom_layer.gradW\n bias_grad = custom_layer.gradb\n torch_weight_grad = torch_layer.weight.grad.data.numpy()\n torch_bias_grad = torch_layer.bias.grad.data.numpy()\n #m = ~np.isclose(torch_weight_grad, weight_grad, atol=1e-5)\n self.assertTrue(np.allclose(torch_weight_grad, weight_grad, atol=1e-6, ))\n self.assertTrue(np.allclose(torch_bias_grad, bias_grad, atol=1e-6))\n \n def test_MaxPool2d(self):\n np.random.seed(42)\n torch.manual_seed(42)\n\n batch_size, n_in = 2, 3\n h,w = 4,6\n kern_size = 2\n for _ in range(100):\n # layers initialization\n torch_layer = torch.nn.MaxPool2d(kern_size)\n custom_layer = MaxPool2d(kern_size)\n\n layer_input = np.random.uniform(-10, 10, (batch_size, n_in, h,w)).astype(np.float32)\n next_layer_grad = np.random.uniform(-10, 10, (batch_size, n_in, \n h // kern_size, w // kern_size)).astype(np.float32)\n\n # 1. check layer output\n custom_layer_output = custom_layer.updateOutput(layer_input)\n layer_input_var = Variable(torch.from_numpy(layer_input), requires_grad=True)\n torch_layer_output_var = torch_layer(layer_input_var)\n self.assertTrue(np.allclose(torch_layer_output_var.data.numpy(), custom_layer_output, atol=1e-6))\n \n # 2. check layer input grad\n custom_layer_grad = custom_layer.updateGradInput(layer_input, next_layer_grad)\n torch_layer_output_var.backward(torch.from_numpy(next_layer_grad))\n torch_layer_grad_var = layer_input_var.grad\n self.assertTrue(np.allclose(torch_layer_grad_var.data.numpy(), custom_layer_grad, atol=1e-6))\n\n\nsuite = unittest.TestLoader().loadTestsFromTestCase(TestAdvancedLayers)\nunittest.TextTestRunner(verbosity=2).run(suite)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a12ecd1a9800fa7b1191f6baa67c0f02c1f836b
| 30,100 |
ipynb
|
Jupyter Notebook
|
TRN_Notebooks/ChIP_A17_KOall_bias100_TFmRNA_TFmRNA.ipynb
|
simonsfoundation/Th17_TRN_Networks
|
5b2c9427e1651b42ac913ab45dff5318bd33480b
|
[
"Apache-2.0"
] | 1 |
2019-03-06T19:37:57.000Z
|
2019-03-06T19:37:57.000Z
|
TRN_Notebooks/ChIP_A17_KOall_bias100_TFmRNA_TFmRNA.ipynb
|
simonsfoundation/Th17_TRN_Networks
|
5b2c9427e1651b42ac913ab45dff5318bd33480b
|
[
"Apache-2.0"
] | null | null | null |
TRN_Notebooks/ChIP_A17_KOall_bias100_TFmRNA_TFmRNA.ipynb
|
simonsfoundation/Th17_TRN_Networks
|
5b2c9427e1651b42ac913ab45dff5318bd33480b
|
[
"Apache-2.0"
] | 1 |
2022-02-24T22:53:03.000Z
|
2022-02-24T22:53:03.000Z
| 41.632089 | 137 | 0.414485 |
[
[
[
"# Visualization of the KO+ChIP Gold Standard from:\n# Miraldi et al. (2018) \"Leveraging chromatin accessibility for transcriptional regulatory network inference in Th17 Cells\"\n\n# TO START: In the menu above, choose \"Cell\" --> \"Run All\", and network + heatmap will load\n# NOTE: Default limits networks to TF-TF edges in top 1 TF / gene model (.93 quantile), to see the full \n# network hit \"restore\" (in the drop-down menu in cell below) and set threshold to 0 and hit \"threshold\"\n# You can search for gene names in the search box below the network (hit \"Match\"), and find regulators (\"targeted by\")\n# Change \"canvas\" to \"SVG\" (drop-down menu in cell below) to enable drag interactions with nodes & labels\n# Change \"SVG\" to \"canvas\" to speed up layout operations\n# More info about jp_gene_viz and user interface instructions are available on Github: \n# https://github.com/simonsfoundation/jp_gene_viz/blob/master/doc/dNetwork%20widget%20overview.ipynb\n\n# directory containing gene expression data and network folder\ndirectory = \".\"\n# folder containing networks\nnetPath = 'Networks'\n# network file name\nnetworkFile = 'ChIP_A17_KOall_bias100_TFmRNA_sp.tsv'\n# title for network figure\nnetTitle = 'ChIP/ATAC(Th17)+KO, bias = 100_TFmRNA, TFA = TF mRNA'\n# name of gene expression file\nexpressionFile = 'Th0_Th17_48hTh.txt'\n# column of gene expression file to color network nodes\nrnaSampleOfInt = 'Th17(48h)'\n# edge cutoff -- for Inferelator TRNs, corresponds to signed quantile (rank of edges in 15 TFs / gene models), \n# increase from 0 --> 1 to get more significant edges (e.g., .33 would correspond to edges only in 10 TFs / gene \n# models)\nedgeCutoff = .93",
"_____no_output_____"
],
[
"import sys\nif \"..\" not in sys.path:\n sys.path.append(\"..\")\nfrom jp_gene_viz import dNetwork\ndNetwork.load_javascript_support()\n# from jp_gene_viz import multiple_network\nfrom jp_gene_viz import LExpression\nLExpression.load_javascript_support()",
"_____no_output_____"
],
[
"# Load network linked to gene expression data\nL = LExpression.LinkedExpressionNetwork()\nL.show() ",
"_____no_output_____"
],
[
"# Load Network and Heatmap\nL.load_network(directory + '/' + netPath + '/' + networkFile)\nL.load_heatmap(directory + '/' + expressionFile)\nN = L.network\nN.set_title(netTitle)\nN.threshhold_slider.value = edgeCutoff\nN.apply_click(None)\nN.draw()\n# Add labels to nodes\nN.labels_button.value=True\n# Limit to TFs only, remove unconnected TFs, choose and set network layout\nN.restore_click()\nN.tf_only_click()\nN.connected_only_click()\nN.layout_dropdown.value = 'fruchterman_reingold'\nN.layout_click()\n\n# Interact with Heatmap\n# Limit genes in heatmap to network genes\nL.gene_click(None) \n# Z-score heatmap values\nL.expression.transform_dropdown.value = 'Z score' \nL.expression.apply_transform() \n# Choose a column in the heatmap (e.g., 48h Th17) to color nodes\nL.expression.col = rnaSampleOfInt\nL.condition_click(None)\n\n# Switch SVG layout to get line colors, then switch back to faster canvas mode\nN.force_svg(None)",
"('Reading network', './Networks/ChIP_A17_KOall_bias100_TFmRNA_sp.tsv')\n('Loading saved layout', './Networks/ChIP_A17_KOall_bias100_TFmRNA_sp.tsv.layout.json')\nOmitting edges, using canvas, and fast layout default because the network is large\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a12f4790332d4bd1123a6ef8e088e068a62c5dd
| 540,027 |
ipynb
|
Jupyter Notebook
|
4_Mar.ipynb
|
sayyidyofa/nescient-notebook
|
f1a7464d943546f717f1af1b1f62f41266291283
|
[
"MIT"
] | null | null | null |
4_Mar.ipynb
|
sayyidyofa/nescient-notebook
|
f1a7464d943546f717f1af1b1f62f41266291283
|
[
"MIT"
] | null | null | null |
4_Mar.ipynb
|
sayyidyofa/nescient-notebook
|
f1a7464d943546f717f1af1b1f62f41266291283
|
[
"MIT"
] | null | null | null | 636.824292 | 50,208 | 0.941999 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nplt.style.use('default')",
"_____no_output_____"
]
],
[
[
"### Data Awal\nData berasal dari dataset DARPA 2000. DDoS attack terjadi pada 2 jam 6 menit 15 detik setelah trafik mulai direkam",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"../data/darpa2000-all.csv\")\ndel df[\"Info\"]\ndel df[\"Length\"]\ndf.head()",
"_____no_output_____"
],
[
"#df.info()\n\ndf[\"T\"] = pd.to_timedelta(df[\"Time\"], unit='s')\ndf = df.set_index(\"T\")\n",
"_____no_output_____"
],
[
"# hitung source unique\nunique_source_count = df[['Source']].resample('3s').nunique()\n\n# Hitung dest unique\nunique_destination_count = df[['Destination']].resample('3s').nunique()\n\n# Hitung proto unique\nunique_protocol_count = df[['Protocol']].resample('3s').nunique()\n\n# Hitung jumlah packet\npackets_count = df[['No.']].resample('3s').count()",
"_____no_output_____"
]
],
[
[
"### Parameter Algoritma\n - A1 = Number of packets\n - A2 = Number of Unique Source IP\n - A3 = (A2) divided by (Number of Unique Dest. IP)\n - A4 = (A2) divided by (Number of Unique Protocol)\n - Beta\n - T = time interval (resampling)\n - K = Window Size",
"_____no_output_____"
]
],
[
[
"df[['Source']].resample('3s').nunique().to_csv('USIP.csv')\ndf[['Destination']].resample('3s').nunique().to_csv('UDIP.csv')\ndf[['Protocol']].resample('3s').nunique().to_csv('UPR.csv')\ndf[['No.']].resample('3s').nunique().to_csv('Packets.csv')",
"_____no_output_____"
]
],
[
[
"### Plot Data setelah resampling",
"_____no_output_____"
]
],
[
[
"# Make figure with 4 rows of plots\n#fig, plots = plt.subplots(4, 1)\n#fig.suptitle('DARPA 2000 Internal Tuesday, 7 March 2000, from 9:25 AM to 12:35 PM', fontsize=16)\nplt.plot(unique_source_count.index.seconds, unique_source_count.Source)\nplt.title(\"Unique Source IP Address Count every 3 seconds\")\nplt.xlabel(\"Time (seconds)\")\nplt.ylabel(\"Unique Source IP Count\")\nplt.show()\n\nplt.plot(unique_destination_count.index.seconds, unique_destination_count.Destination)\nplt.title(\"Unique Destination IP Address Count every 3 seconds\")\nplt.xlabel(\"Time (seconds)\")\nplt.ylabel(\"Unique Destination IP Count\")\nplt.show()\n\nplt.plot(unique_protocol_count.index.seconds, unique_protocol_count.Protocol)\nplt.title(\"Unique Protocol Count every 3 seconds\")\nplt.xlabel(\"Time (seconds)\")\nplt.ylabel(\"Unique Source IP Count\")\nplt.show()\n\nplt.plot(packets_count.index.seconds, packets_count['No.'])\nplt.title(\"Number of packets every 3 seconds\")\nplt.xlabel(\"Time (seconds)\")\nplt.ylabel(\"Number of packets\")\nplt.show()\n\n#fig.tight_layout()",
"_____no_output_____"
]
],
[
[
"### Plot A1, A2, A3, A4",
"_____no_output_____"
]
],
[
[
"##ye = packets_count.iloc[2426:2526, 0].to_list()\n##print(ye)\n#print(packets_count.loc[packets_count['No.'].idxmax()])\n# data => 38215\n# second => 7575 => 2525\n# Timedelta => 0 days 02:06:15\n\nA1_all = packets_count\nA2_all = unique_source_count\nA3_all = unique_source_count['Source'].divide(unique_destination_count['Destination']).to_frame()\nA4_all = unique_source_count['Source'].divide(unique_protocol_count['Protocol']).to_frame()\n\nplt.plot(A1_all, '*')\n#plt.plot(A1_all.iloc[:, 0].to_list())\nplt.title(\"A1 value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"A1\")\nplt.show()\n\nplt.plot(A2_all, '*')\n#plt.plot(A2_all.iloc[:, 0].to_list())\nplt.title(\"A2 value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"A2\")\nplt.show()\n\nplt.plot(A3_all, '*')\n#plt.plot(A3_all.iloc[:, 0].to_list())\nplt.title(\"A3 value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"A3\")\nplt.show()\n\nplt.plot(A4_all, '*')\n#plt.plot(A4_all.iloc[:, 0].to_list())\nplt.title(\"A4 value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"A4\")\nplt.show()\n",
"_____no_output_____"
],
[
"A_all = (A1_all, A2_all, A3_all, A4_all)\nThreshold_all = []\nN_all = []\nbeta_all = []\n\nj_to_end = range(len(A1_all.iloc[:, 0].to_list()))\n\nfor A in A_all:\n # Init values\n K = 1\n beta = 1.5\n j = 0\n T = 3\n current_threshold_array = list()\n current_N_array = list()\n current_beta_array = list()\n\n current_moving_A = A.iloc[j: j+K, 0].to_list()\n current_moving_mean = np.mean(current_moving_A)\n current_moving_variance = np.var(current_moving_A)\n current_threshold = (current_moving_mean + current_moving_variance) * beta\n\n current_threshold_array.append([j, current_threshold])\n current_beta_array.append([j, beta])\n\n #while j <= K*T-1 and j < len(A1_all.iloc[:, 0].to_list()):\n while j < len(A1_all.iloc[:, 0].to_list()):\n if j < 0 and j % K*T == 0:\n beta = 1.5\n current_moving_A = A.iloc[j: j+K, 0].to_list()\n current_moving_mean = np.mean(current_moving_A)\n current_moving_variance = np.var(current_moving_A)\n current_threshold = (current_moving_mean + current_moving_variance) * beta\n\n current_threshold_array.append([j, current_threshold])\n current_beta_array.append([j, beta])\n else:\n if A.iloc[j:j+1, 0].to_list()[0] > current_threshold:\n current_N_array.append([j, True])\n else:\n current_N_array.append([j, False])\n j = j + 1\n current_j = j\n previous_j = j - 1\n previous_moving_mean = np.mean(A.iloc[previous_j: previous_j+K, 0].to_list())\n current_moving_mean = np.mean(A.iloc[current_j: current_j+K, 0].to_list())\n if current_moving_mean > 2 * previous_moving_mean:\n beta = beta + 0.5\n current_threshold = (current_moving_mean + np.var(A.iloc[current_j: current_j+K, 0].to_list())) * beta\n else:\n beta = beta - 0.5\n if beta < 1.0:\n beta = 1\n current_threshold = (current_moving_mean + np.var(A.iloc[current_j: current_j+K, 0].to_list())) / beta\n current_threshold_array.append([current_j, current_threshold])\n current_beta_array.append([j, beta])\n\n Threshold_all.append(current_threshold_array)\n N_all.append(current_N_array)\n beta_all.append(current_beta_array)\n\n\n\n#print(beta_all)\n#print(Threshold_all)\n#print(N_all)",
"/home/yoffa/codebase/PycharmProjects/darpa2k/venv/lib/python3.8/site-packages/numpy/core/fromnumeric.py:3419: RuntimeWarning: Mean of empty slice.\n return _methods._mean(a, axis=axis, dtype=dtype,\n/home/yoffa/codebase/PycharmProjects/darpa2k/venv/lib/python3.8/site-packages/numpy/core/_methods.py:188: RuntimeWarning: invalid value encountered in double_scalars\n ret = ret.dtype.type(ret / rcount)\n/home/yoffa/codebase/PycharmProjects/darpa2k/venv/lib/python3.8/site-packages/numpy/core/fromnumeric.py:3702: RuntimeWarning: Degrees of freedom <= 0 for slice\n return _methods._var(a, axis=axis, dtype=dtype, out=out, ddof=ddof,\n/home/yoffa/codebase/PycharmProjects/darpa2k/venv/lib/python3.8/site-packages/numpy/core/_methods.py:221: RuntimeWarning: invalid value encountered in true_divide\n arrmean = um.true_divide(arrmean, div, out=arrmean, casting='unsafe',\n/home/yoffa/codebase/PycharmProjects/darpa2k/venv/lib/python3.8/site-packages/numpy/core/_methods.py:253: RuntimeWarning: invalid value encountered in double_scalars\n ret = ret.dtype.type(ret / rcount)\n"
]
],
[
[
"### Nilai Threshold berubah tiap waktu",
"_____no_output_____"
]
],
[
[
"beta1y = list()\nbeta2y = list()\nbeta3y = list()\nbeta4y = list()\n\nfor betaXY in Threshold_all[0]:\n beta1y.append(betaXY[1])\n\nfor betaXY in Threshold_all[1]:\n beta2y.append(betaXY[1])\n\nfor betaXY in Threshold_all[2]:\n beta3y.append(betaXY[1])\n\nfor betaXY in Threshold_all[3]:\n beta4y.append(betaXY[1])\n\nplt.plot(beta1y, '*')\n#plt.plot(A1_all.iloc[:, 0].to_list())\nplt.title(\"A1 Threshold value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Threshold value\")\nplt.show()\n\nplt.plot(beta2y, '*')\n#plt.plot(A2_all.iloc[:, 0].to_list())\nplt.title(\"A2 Threshold value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Threshold value\")\nplt.show()\n\nplt.plot(beta3y, '*')\n#plt.plot(A3_all.iloc[:, 0].to_list())\nplt.title(\"A3 Threshold value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Threshold value\")\nplt.show()\n\nplt.plot(beta4y, '*')\n#plt.plot(A4_all.iloc[:, 0].to_list())\nplt.title(\"A4 Threshold value over time\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Threshold value\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Nature of Network tiap waktu\nJika 1 maka deteksi positif, jika 0 maka normal",
"_____no_output_____"
]
],
[
[
"beta1y = list()\nbeta2y = list()\nbeta3y = list()\nbeta4y = list()\n\nfor betaXY in N_all[0]:\n beta1y.append(betaXY[1])\n\nfor betaXY in N_all[1]:\n beta2y.append(betaXY[1])\n\nfor betaXY in N_all[2]:\n beta3y.append(betaXY[1])\n\nfor betaXY in N_all[3]:\n beta4y.append(betaXY[1])\n\nplt.plot(beta1y, '*')\n#plt.plot(A1_all.iloc[:, 0].to_list())\nplt.title(\"A1 Network Nature\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Nature of Network\")\nplt.show()\n\nplt.plot(beta2y, '*')\n#plt.plot(A2_all.iloc[:, 0].to_list())\nplt.title(\"A2 Network Nature\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Nature of Network\")\nplt.show()\n\nplt.plot(beta3y, '*')\n#plt.plot(A3_all.iloc[:, 0].to_list())\nplt.title(\"A3 Network Nature\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Nature of Network\")\nplt.show()\n\nplt.plot(beta4y, '*')\n#plt.plot(A4_all.iloc[:, 0].to_list())\nplt.title(\"A4 Network Nature\")\nplt.xlabel(\"Time (3secs interval)\")\nplt.ylabel(\"Nature of Network\")\nplt.show()\n",
"_____no_output_____"
],
[
"N_for_testing = []\n\nfor idx in j_to_end:\n if N_all[0][idx][1] is True and N_all[1][idx][1] is True and N_all[2][idx][1] is True and N_all[3][idx][1] is True:\n N_for_testing.append(1)\n else:\n N_for_testing.append(0)\n",
"_____no_output_____"
],
[
"A1_all['real'] = 0\nA1_all.head()\n\npckts = packets_count.iloc[2525:2528, 0].index\npckts",
"_____no_output_____"
],
[
"A1_all.at['0 days 02:06:15', 'real'] = 1\nA1_all.at['0 days 02:06:18', 'real'] = 1\nA1_all.at['0 days 02:06:21', 'real'] = 1",
"_____no_output_____"
],
[
"y_real = A1_all.iloc[:, 1].to_list()",
"_____no_output_____"
],
[
"y_pred = N_for_testing",
"_____no_output_____"
],
[
"def perf_measure(y_actual, y_hat):\n TP = 0\n FP = 0\n TN = 0\n FN = 0\n\n for i in range(len(y_hat)):\n if y_actual[i]==y_hat[i]==1:\n TP += 1\n if y_hat[i]==1 and y_actual[i]!=y_hat[i]:\n FP += 1\n if y_actual[i]==y_hat[i]==0:\n TN += 1\n if y_hat[i]==0 and y_actual[i]!=y_hat[i]:\n FN += 1\n\n return TP, FP, TN, FN\n\ndef sensitivity(true_positive, false_negative):\n return true_positive / (true_positive + false_negative)\n\ndef accuracy(true_positive, false_positive, true_negative, false_negative):\n return (true_positive + true_negative) / (true_positive + false_positive + true_negative + false_negative)\n\ndef specificity(true_negative, false_positive):\n return true_negative / (true_negative + false_positive)\n\ndef precision(true_positive, false_positive):\n return true_positive / (true_positive + false_positive)",
"_____no_output_____"
],
[
"tp, fp, tn, fn = perf_measure(y_real, y_pred)\n\nprint(\n \"\\ntrue positive: {}\".format(tp),\n \"\\nfalse positive: {}\".format(fp),\n \"\\ntrue negative: {}\".format(tn),\n \"\\nfalse negative: {}\".format(fn)\n)\n\nprint(\"\\n\")\n\nprint(\"Sensitivity (TPR): {}\".format(sensitivity(tp, fn)))\nprint(\"Accuracy: {}\".format(accuracy(tp, fp, tn, fn)))\nprint(\"Specificity (TNR): {}\".format(specificity(tn, fp)))\nprint(\"Precision: {}\".format(precision(tp, fp)))",
"\ntrue positive: 1 \nfalse positive: 0 \ntrue negative: 3882 \nfalse negative: 2\n\n\nSensitivity (TPR): 0.3333333333333333\nAccuracy: 0.9994851994851995\nSpecificity (TNR): 1.0\nPrecision: 1.0\n"
],
[
"david_j = [98, 99.5, 99.6]\nmine = [33.33, 99.94, 100]\ngenerated_result = [50, 89.15, 90.12]\n\nindex = np.arange(3)\nbar_width = 0.3\n\nfig, ax = plt.subplots()\noriginal = ax.bar(index, david_j, bar_width,\n label=\"Original (DARPA 2000)\")\n\nimplementation = ax.bar(index+bar_width, mine,\n bar_width, label=\"Implementation (DARPA 2000)\")\n\ngenerated = ax.bar(index+2*bar_width, generated_result, bar_width, label=\"Implementation (Generated Dataset)\")\n\nax.set_xlabel('Evaluation Result')\nax.set_ylabel('Percentage')\nax.set_title('Evaluation Result Comparison')\nax.set_xticks(index + bar_width / 2)\nax.set_xticklabels([\"TPR\", \"Accuracy\", \"TNR\"])\nax.legend()\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a12fa26d2276214e3696566c6581f5bc6e68091
| 14,206 |
ipynb
|
Jupyter Notebook
|
08-Workshop-Overview/.ipynb_checkpoints/Workshop_Overview-checkpoint.ipynb
|
lknelson/DH-Institute-2017
|
d20246758d6da88dfedbad2e75933ad4ef370930
|
[
"BSD-2-Clause"
] | 3 |
2017-07-31T18:56:53.000Z
|
2021-03-12T14:58:00.000Z
|
08-Workshop-Overview/.ipynb_checkpoints/Workshop_Overview-checkpoint.ipynb
|
lknelson/DH-Institute-2017
|
d20246758d6da88dfedbad2e75933ad4ef370930
|
[
"BSD-2-Clause"
] | null | null | null |
08-Workshop-Overview/.ipynb_checkpoints/Workshop_Overview-checkpoint.ipynb
|
lknelson/DH-Institute-2017
|
d20246758d6da88dfedbad2e75933ad4ef370930
|
[
"BSD-2-Clause"
] | 3 |
2017-09-01T22:19:22.000Z
|
2018-07-01T09:45:46.000Z
| 34.067146 | 170 | 0.617063 |
[
[
[
" \n# <center> #DHBSI 2016: Computational Text Analysis </center>\n\n## <center> Laura Nelson <br/> <em>Postdoctoral Fellow | Digital Humanities @ Berkeley | Berkeley Institute for Data Science </em> </center>\n\n## <center> Teddy Roland <br/> <em> Coordinator, Digital Humanities @ Berkeley <br/> Lecturer, UC Berkeley </em> </center>",
"_____no_output_____"
],
[
"# <center> Summary </center>\n## <center> Text Analysis Demystified </center>\n### <center> It's Just Counting! <br/> </center>\n",
"_____no_output_____"
],
[
"## <center> The Dark Side of DH: An Invitation\n",
"_____no_output_____"
],
[
"## <center> Text Analysis in Research </center>\n",
"_____no_output_____"
],
[
"## <center> Lessons </center>\n### <center> Our workshop included 5 days and 7 lessons to learn how counting, sometimes creative counting, can amplify and augment close readings of text </center>",
"_____no_output_____"
],
[
"# Lesson 1: Introduction to Natural Language Processing",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk import word_tokenize\nfrom nltk.corpus import stopwords\nimport string\npunctuations = list(string.punctuation)\n\n#read the two text files from your hard drive, assign first mystery text to variable 'text1' and second mystery text to variable 'text2'\ntext1 = open('../01-Intro-to-NLP/text1.txt').read()\ntext2 = open('../01-Intro-to-NLP/text2.txt').read()\n\n###word frequencies\n\n#tokenize texts\ntext1_tokens = word_tokenize(text1)\ntext2_tokens = word_tokenize(text2)\n\n#pre-process for word frequency\n#lowercase\ntext1_tokens_lc = [word.lower() for word in text1_tokens]\ntext2_tokens_lc = [word.lower() for word in text2_tokens]\n\n#remove stopwords\ntext1_tokens_clean = [word for word in text1_tokens_lc if word not in stopwords.words('english')]\ntext2_tokens_clean = [word for word in text2_tokens_lc if word not in stopwords.words('english')]\n\n#remove punctuation using the list of punctuation from the string pacage\ntext1_tokens_clean = [word for word in text1_tokens_clean if word not in punctuations]\ntext2_tokens_clean = [word for word in text2_tokens_clean if word not in punctuations]\n\n#frequency distribution\ntext1_word_frequency = nltk.FreqDist(text1_tokens_clean)\ntext2_word_frequency = nltk.FreqDist(text2_tokens_clean)\n\n\n\nprint(\"Frequent Words for Text1\")\nprint(\"________________________\")\nfor word in text1_word_frequency.most_common(20):\n print(word[0])\nprint()\nprint(\"Frequent Words for Text2\")\nprint(\"________________________\")\nfor word in text2_word_frequency.most_common(20):\n print(word[0])\n \n \n### Can you guess the novel from most frequent words?",
"_____no_output_____"
]
],
[
[
"# Lesson 2: Basics of Python",
"_____no_output_____"
]
],
[
[
"# Nothing to see here, folks",
"_____no_output_____"
]
],
[
[
"# Lesson 3: Operationalizing",
"_____no_output_____"
]
],
[
[
"import pandas\ndialogue_df = pandas.read_csv('../03-Operationalizing/antigone_dialogue.csv', index_col=0)\ndialogue_tokens = [character.split() for character in dialogue_df['DIALOGUE']]\ndialogue_len = [len(tokens) for tokens in dialogue_tokens]\ndialogue_df['WORDS_SPOKEN'] = dialogue_len\ndialogue_df = dialogue_df.sort_values('WORDS_SPOKEN', ascending = False)\n# Let's visualize!\n\n# Tells Jupyter to produce images in notebook\n% pylab inline\n\n# Makes images look good\nstyle.use('ggplot')\ndialogue_df['WORDS_SPOKEN'].plot(kind='bar')\n\n###Who is the main protagonist? Maybe not Antigone?",
"_____no_output_____"
]
],
[
[
"# Lesson 4: Discriminating Words",
"_____no_output_____"
]
],
[
[
"from sklearn.feature_extraction.text import TfidfVectorizer\n\ndf = pandas.read_csv(\"../04-Discriminating-Words/BDHSI2016_music_reviews.csv\", sep = '\\t')\n\ntfidfvec = TfidfVectorizer()\n#create the dtm, but with cells weigthed by the tf-idf score.\ndtm_tfidf_df = pandas.DataFrame(tfidfvec.fit_transform(df.body).toarray(), columns=tfidfvec.get_feature_names(), index = df.index)\n\ndf_genre = df['genre'].to_frame()\nmerged_df = df_genre.join(dtm_tfidf_df, how = 'right', lsuffix='_x')\n\n#pull out the reviews for three genres, Rap, Alternative/Indie Rock, and Jazz\ndtm_rap = merged_df[merged_df['genre_x']==\"Rap\"]\ndtm_indie = merged_df[merged_df['genre_x']==\"Alternative/Indie Rock\"]\ndtm_jazz = merged_df[merged_df['genre_x']==\"Jazz\"]\n\n#print the words with the highest tf-idf scores for each genre\nprint(\"Rap Words\")\nprint(dtm_rap.max(numeric_only=True).sort_values(ascending=False)[0:20])\nprint()\nprint(\"Indie Words\")\nprint(dtm_indie.max(numeric_only=True).sort_values(ascending=False)[0:20])\nprint()\nprint(\"Jazz Words\")\nprint(dtm_jazz.max(numeric_only=True).sort_values(ascending=False)[0:20])\n\n###What words are distinct to reviews of Rap albums, Indie albums, and Jazz albums?\n##Notice the word weights for the Rap albums compared to others. Are these reviews more different than other reviews?",
"_____no_output_____"
]
],
[
[
"# Lesson 5: Sentiment Analysis using the Dictionary Method",
"_____no_output_____"
]
],
[
[
"pos_sent = open(\"../05-Dictionary-Method/positive_words.txt\").read()\nneg_sent = open(\"../05-Dictionary-Method/negative_words.txt\").read()\n\npositive_words=pos_sent.split('\\n')\nnegative_words=neg_sent.split('\\n')\n\ntext1_pos = [word for word in text1_tokens_clean if word in positive_words]\ntext2_pos = [word for word in text2_tokens_clean if word in positive_words]\n\ntext1_neg = [word for word in text1_tokens if word in negative_words]\ntext2_neg = [word for word in text2_tokens if word in negative_words]\n\nprint(\"Postive words in Melville\")\nprint(len(text1_pos)/len(text1_tokens))\nprint()\nprint(\"Negative words in Melville\")\nprint(len(text1_neg)/len(text1_tokens))\nprint()\nprint(\"Postive words in Austen\")\nprint(len(text2_pos)/len(text2_tokens))\nprint()\nprint(\"Negative words in Austen\")\nprint(len(text2_neg)/len(text2_tokens))\n\n## Who is more postive, Melville or Austen?\n## Melville has a similar precentage of postive and negative words (a whale is a whale, neither good nor bad)\n## Austen is decidedly more positive than negative (it's the gentleman thing to do)",
"_____no_output_____"
]
],
[
[
"# Lesson 6: Literary Distinction",
"_____no_output_____"
]
],
[
[
"from sklearn.naive_bayes import MultinomialNB\nfrom sklearn.feature_extraction.text import CountVectorizer\nimport os\n\nreview_path = '../06-Literary Distinction (Probably)/poems/reviewed/'\nrandom_path = '../06-Literary Distinction (Probably)/poems/random/'\nreview_files = os.listdir(review_path)\nrandom_files = os.listdir(random_path)\nreview_texts = [open(review_path+file_name).read() for file_name in review_files]\nrandom_texts = [open(random_path+file_name).read() for file_name in random_files]\nall_texts = review_texts + random_texts\nall_file_names = review_files + random_files\nall_labels = ['reviewed'] * len(review_texts) + ['random'] * len(random_texts)\ncv = CountVectorizer(stop_words = 'english', min_df=180, binary = True, max_features = None)\ndtm = cv.fit_transform(all_texts).toarray()\nnb = MultinomialNB()\nnb.fit(dtm, all_labels)\n\ndickinson_canonic = \"\"\"Because I could not stop for Death – \nHe kindly stopped for me – \nThe Carriage held but just Ourselves – \nAnd Immortality.\n\nWe slowly drove – He knew no haste\nAnd I had put away\nMy labor and my leisure too,\nFor His Civility – \n\nWe passed the School, where Children strove\nAt Recess – in the Ring – \nWe passed the Fields of Gazing Grain – \nWe passed the Setting Sun – \n\nOr rather – He passed us – \nThe Dews drew quivering and chill – \nFor only Gossamer, my Gown – \nMy Tippet – only Tulle – \n\nWe paused before a House that seemed\nA Swelling of the Ground – \nThe Roof was scarcely visible – \nThe Cornice – in the Ground – \n\nSince then – ‘tis Centuries – and yet\nFeels shorter than the Day\nI first surmised the Horses’ Heads \nWere toward Eternity – \"\"\"\n\n\nanthem_patriotic = \"\"\"O! say can you see, by the dawn's early light,\nWhat so proudly we hailed at the twilight's last gleaming,\nWhose broad stripes and bright stars through the perilous fight,\nO'er the ramparts we watched, were so gallantly streaming?\nAnd the rockets' red glare, the bombs bursting in air,\nGave proof through the night that our flag was still there;\nO! say does that star-spangled banner yet wave\nO'er the land of the free and the home of the brave?\"\"\"\n\nunknown_dtm = cv.transform([dickinson_canonic,anthem_patriotic]).toarray()\nnb.predict(unknown_dtm)\n\n## Can a computer predict whether a poem would be considered 'presitgious'?",
"_____no_output_____"
]
],
[
[
"# Lesson 6: Topic Modeling",
"_____no_output_____"
]
],
[
[
"import gensim\nimport pandas\nfrom nltk.corpus import stopwords, words\n\nmetadata_df = pandas.read_csv('../07-Topic Modeling/txtlab_Novel150_English.csv')\nfiction_path = '../07-Topic Modeling/txtalb_Novel150_English/'\nnovel_list = [open(fiction_path+file_name).read() for file_name in metadata_df['filename']]\nnovel_tokens_list = [novel.lower().split() for novel in novel_list]\ndictionary = gensim.corpora.dictionary.Dictionary(novel_tokens_list)\nproper_names = [word.lower() for word in words.words() if word.istitle()]\nnoise_tokens = [word for word in dictionary.values() if word.isalpha()==False or len(word)<=2]\nbad_words = stopwords.words('english') + proper_names + noise_tokens\nstop_ids = [_id for _id, count in dictionary.doc2bow(bad_words)]\ndictionary.filter_tokens(bad_ids = stop_ids)\ndictionary.filter_extremes(no_below = 40)\ncorpus = [dictionary.doc2bow(text) for text in novel_tokens_list]\nlda_model = gensim.models.LdaModel(corpus, num_topics=25, alpha='auto', id2word=dictionary, iterations=2500, passes = 4)\nlist_of_doctopics = [lda_model.get_document_topics(text, minimum_probability=0) for text in corpus]\nlist_of_probabilities = [[probability for label,probability in distribution] for distribution in list_of_doctopics]\nproba_distro_df = pandas.DataFrame(list_of_probabilities)\nmetadata_df = pandas.concat([metadata_df, pandas.DataFrame(list_of_probabilities)], axis=1)\nannual_means_df = metadata_df.groupby('date').mean()",
"_____no_output_____"
],
[
"annual_means_df[8].plot(kind='bar', figsize=(8,8))",
"_____no_output_____"
],
[
"lda_model.show_topic(8)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a13167a09fd010cc290954c54bfc804b859e0d0
| 4,429 |
ipynb
|
Jupyter Notebook
|
Radicality/radicality.ipynb
|
Niederb/political_science
|
d99900809c01483bf669d2dc9171ccd07465a311
|
[
"MIT"
] | null | null | null |
Radicality/radicality.ipynb
|
Niederb/political_science
|
d99900809c01483bf669d2dc9171ccd07465a311
|
[
"MIT"
] | null | null | null |
Radicality/radicality.ipynb
|
Niederb/political_science
|
d99900809c01483bf669d2dc9171ccd07465a311
|
[
"MIT"
] | null | null | null | 27.339506 | 184 | 0.516369 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport plotly.plotly as py\nimport plotly.graph_objs as go\n\ndf = pd.read_csv(\"https://raw.githubusercontent.com/Niederb/political_science/master/gewinner-kantone/alle-volksabstimmungen-resultate.csv\", sep=\";\")#, encoding=\"ascii\")\n\nstimmberechtigte = pd.read_csv(\"https://raw.githubusercontent.com/Niederb/political_science/master/gewinner-kantone/stimmberechtigte.csv\", sep=\";\")\n\ndata = df[list(df.columns[2:-1])] #df.ix[:,[2:5]]#'Schweiz':]\n\ndata_mat = data.as_matrix()\nschweiz = data_mat[:, 0]\nkantone = data_mat[:, 1:]\nschweiz_thresholded = schweiz > 50\nkantone_thresholded = kantone > 50\n\nkorrekt = schweiz_thresholded == kantone_thresholded.T\nkorrekt = korrekt.T\nsuccess_rate = np.mean(korrekt, 0)\n\nradicality = np.mean(np.abs(kantone - 50.0), 0)\n\nstimmberechtigte_2 = np.mean(stimmberechtigte.as_matrix(), 0)\nkantonsnamen = list(stimmberechtigte)\n",
"_____no_output_____"
],
[
"trace = go.Scatter(\n x = radicality,\n y = stimmberechtigte_2,\n mode = 'markers',\n text=kantonsnamen,\n marker = dict(\n size = 20,\n color = 'rgba(255, 161, 0, 1.0)',\n line = dict(\n width = 2,\n )\n ),\n)\nlayout = go.Layout(\n hovermode = 'closest',\n title='Radikalität vs Stimmberechtigte',\n xaxis=dict(title='Radikalität'),\n yaxis=dict(title='Anzahl Stimmberechtigte (Durchschnitt)')\n )\ndata = [trace] \nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig, filename='radicality-size')",
"_____no_output_____"
],
[
"trace = go.Scatter(\n x = radicality,\n y = success_rate,\n mode = 'markers',\n text=kantonsnamen,\n marker = dict(\n size = 20,\n color = 'rgba(255, 161, 0, 1.0)',\n line = dict(\n width = 2,\n )\n ),\n)\nlayout = go.Layout(\n hovermode = 'closest',\n title='Radikalität vs Erfolgsrate',\n xaxis=dict(title='Radikalität'),\n yaxis=dict(title='Erfolgsrate in %'),\n )\ndata = [trace] \nfig = go.Figure(data=data, layout=layout)\npy.iplot(fig, filename='radicality-success')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
4a131a6a2455d1ac556c0256240f0850f90f6f73
| 71,218 |
ipynb
|
Jupyter Notebook
|
SQL/My training examples/.ipynb_checkpoints/SQL Basics Working with Databases-checkpoint.ipynb
|
VertexyLV/RCS_Data_Analysis_Python
|
c706de12c8d793e41c2a1cc5ea2b2bf479c6590c
|
[
"MIT"
] | null | null | null |
SQL/My training examples/.ipynb_checkpoints/SQL Basics Working with Databases-checkpoint.ipynb
|
VertexyLV/RCS_Data_Analysis_Python
|
c706de12c8d793e41c2a1cc5ea2b2bf479c6590c
|
[
"MIT"
] | null | null | null |
SQL/My training examples/.ipynb_checkpoints/SQL Basics Working with Databases-checkpoint.ipynb
|
VertexyLV/RCS_Data_Analysis_Python
|
c706de12c8d793e41c2a1cc5ea2b2bf479c6590c
|
[
"MIT"
] | null | null | null | 38.496216 | 89 | 0.341023 |
[
[
[
"Tutorial Source: https://www.dataquest.io/blog/sql-basics/",
"_____no_output_____"
]
],
[
[
"import sqlite3\nimport pandas as pd",
"_____no_output_____"
],
[
"db = sqlite3.connect('hubway.db')",
"_____no_output_____"
],
[
"cursor = db.cursor()\ncursor.execute(\"SELECT name FROM sqlite_master WHERE type='table';\")\nprint(cursor.fetchall())",
"[('trips',), ('stations',)]\n"
],
[
"def run_query(query):\n return pd.read_sql_query(query, db)",
"_____no_output_____"
],
[
"trips = run_query(\"SELECT * FROM trips\")\ntrips",
"_____no_output_____"
],
[
"stations = run_query(\"SELECT * FROM stations\")\nstations",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a1328f59559e87189265cee1ab1e6d238d7cf9e
| 19,688 |
ipynb
|
Jupyter Notebook
|
FINAL/2. Data Preprocessing/Preprocessing_Topic Monitoring(FB)_Chien.ipynb
|
o0oBluePhoenixo0o/AbbVie2017
|
356583487455ba2616457f8a59ca741321c0b154
|
[
"MIT"
] | 2 |
2020-01-30T03:38:06.000Z
|
2020-12-15T00:31:56.000Z
|
FINAL/2. Data Preprocessing/Preprocessing_Topic Monitoring(FB)_Chien.ipynb
|
o0oBluePhoenixo0o/AbbVie2017
|
356583487455ba2616457f8a59ca741321c0b154
|
[
"MIT"
] | null | null | null |
FINAL/2. Data Preprocessing/Preprocessing_Topic Monitoring(FB)_Chien.ipynb
|
o0oBluePhoenixo0o/AbbVie2017
|
356583487455ba2616457f8a59ca741321c0b154
|
[
"MIT"
] | 7 |
2017-03-25T13:56:41.000Z
|
2017-09-03T14:03:02.000Z
| 43.175439 | 148 | 0.487099 |
[
[
[
"# Data Preprocessing for Topic Monitoring(Facebook)",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport re\nimport csv\nfrom langdetect import detect\nimport nltk\n# nltk.download('punkt')\n# nltk.download('maxent_treebank_pos_tagger')\n# nltk.download('wordnet')\n# nltk.download('averaged_perceptron_tagger')\n# nltk.download('stopwords')\nfrom nltk.corpus import stopwords\nfrom nltk import wordpunct_tokenize\nfrom IPython.display import Image\nfrom IPython.display import display",
"_____no_output_____"
],
[
"### Load the Crawled Facebook Dataset\n# Remove duplicates, NA, sorted time\ndisease = pd.read_csv('Final_utf16.csv', encoding = 'utf-16LE', sep=',',\n dtype={\"key\": object, \"id.x\": object,\"like_count.x\": float, \"from_id.x\":float,\n \"from_name.x\":object, \"message.x\":object, \"created_time.x\":object, \"type\":object,\n \"link\":object, \"story\":object, \"comments_count.x\":float,\"shares_count\":float,\n \"love_count\":float, \"haha_count\":float, \"wow_count\":float, \"sad_count\": float,\n \"angry_count\":float, \"join_id\":object, \"from_id.y\":float, \"from_name.y\":object,\n \"message.y\":object, \"created_time.y\":object, \"likes_count.y\":float, \n \"comments_count.y\": float, \"id.y\":object})\ndf = pd.DataFrame(disease, columns=['key', 'created_time.x', 'id.x','message.x' , 'id.y', 'message.y'])\ndf.columns = ['key', 'created_time.x', 'id.x','message.x' , 'id.y', 'message.y']\nrm_duplicates = df.drop_duplicates(subset=['message.x', 'message.y'])\ndtime = rm_duplicates.sort_values(['created_time.x'])\ndtime.index = range(len(dtime))\ndlang = dtime\ndlang = dlang[dlang['key']!='johnson & johnson']\ndlang = dlang[dlang['key']!='johnson&johnson']\ndlang.index = range(len(dlang))\ndisplay(dlang.head(3))\nprint(len(dlang))",
"_____no_output_____"
],
[
"# Detect the text language by majority vote\ndef calculate_languages_ratios(text):\n languages_ratios = {}\n tokens = wordpunct_tokenize(text)\n words = [word.lower() for word in tokens]\n for language in stopwords.fileids():\n stopwords_set = set(stopwords.words(language))\n words_set = set(words)\n common_elements = words_set.intersection(stopwords_set)\n languages_ratios[language] = len(common_elements)\n return languages_ratios\n \ndef detect_language(text):\n ratios = calculate_languages_ratios(text)\n most_rated_language = max(ratios, key=ratios.get)\n return most_rated_language",
"_____no_output_____"
]
],
[
[
"# Final Preprocessing",
"_____no_output_____"
],
[
"In this section, preprocessing is implemented into following steps.<br>\n\n| Preprocessing Steps| Packages | Notes |\n|------------------- |-----------------------------|-------------------------------------|\n| Language Detection | Self-defined function, nktk |Check the language of each post |\n| Remove Stopwords | nltk.corpus |Remove stopwords of detected language|\n| Remove Url | Regular expression | |\n| Remove Punctuation | string.punctuation | | \n| Lemmatizing | nltk.stem |Lemmatize words in Noun and Verb |\n| Part of Speech(POS)| nltk.pos_tag |Preserve Noun, Adverb and Adjective |\n| Tokenize | split |Unigram |\n| Remove NA | pandas | |\n| Drop Duplicates | pandas | |",
"_____no_output_____"
]
],
[
[
"import gensim\nfrom gensim import corpora, models, similarities\nfrom nltk.corpus import stopwords\nfrom sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer\nfrom nltk.stem import WordNetLemmatizer\nimport string\nimport time\nimport os\nexclude = set(string.punctuation)\nlemma = WordNetLemmatizer()\n\n# Create a new csv file to store the result after data preprocessing\nwith open('facebook_preprocessing.csv', 'w', encoding = 'UTF-8', newline = '') as csvfile:\n column = [['key', 'created_time.x', 'id.x', 'message.x', 'id.y', 'message.y',\n 'lang.x', 're_message.x', 'lang.y', 're_message.y']]\n writer = csv.writer(csvfile)\n writer.writerows(column)\n\n# Data preprocessing steps\nfor i in range(len(dlang['message.x'])): \n features = []\n features.append(dlang['key'][i])\n features.append(dlang['created_time.x'][i])\n features.append(dlang['id.x'][i])\n features.append(dlang['message.x'][i])\n features.append(dlang['id.y'][i])\n features.append(dlang['message.y'][i])\n if(str(dlang['message.x'][i]) == \"nan\"):\n features.append('english')\n features.append(dlang['message.x'][i])\n else:\n lang = detect_language(dlang['message.x'][i])\n features.append(lang)\n stop = set(stopwords.words(lang))\n reurl = re.sub(r\"http\\S+\", \"\", str(dlang['message.x'][i]))\n tokens = ' '.join(re.findall(r\"[\\w']+\", reurl)).lower().split()\n x = [''.join(c for c in s if c not in string.punctuation) for s in tokens]\n x = ' '.join(x)\n stop_free = \" \".join([i for i in x.lower().split() if i not in stop])\n punc_free = ''.join(ch for ch in stop_free if ch not in exclude)\n normalized = \" \".join(lemma.lemmatize(word,pos = 'n') for word in punc_free.split())\n normalized = \" \".join(lemma.lemmatize(word,pos = 'v') for word in normalized.split())\n word = \" \".join(word for word in normalized.split() if len(word)>3)\n postag = nltk.pos_tag(word.split())\n irlist = [',','.',':','#',';','CD','WRB','RB','PRP','...',')','(','-','``','@']\n poslist = ['NN','NNP','NNS','RB','RBR','RBS','JJ','JJR','JJS']\n wordlist = ['co', 'https', 'http','rt','com','amp','fe0f','www','ve','dont',\"i'm\",\"it's\",'isnt','âźă','âąă','âł_','kf4pdwe64k']\n adjandn = [word for word,pos in postag if pos in poslist and word not in wordlist and len(word)>3]\n stop = set(stopwords.words(lang))\n wordlist = [i for i in adjandn if i not in stop]\n features.append(' '.join(wordlist))\n if(str(dlang['message.y'][i]) == \"nan\"):\n features.append('english')\n features.append(dlang['message.y'][i])\n else:\n lang = detect_language(dlang['message.y'][i])\n features.append(lang)\n stop = set(stopwords.words(lang))\n reurl = re.sub(r\"http\\S+\", \"\", str(dlang['message.y'][i]))\n tokens = ' '.join(re.findall(r\"[\\w']+\", reurl)).lower().split()\n x = [''.join(c for c in s if c not in string.punctuation) for s in tokens]\n x = ' '.join(x)\n stop_free = \" \".join([i for i in x.lower().split() if i not in stop])\n punc_free = ''.join(ch for ch in stop_free if ch not in exclude)\n normalized = \" \".join(lemma.lemmatize(word,pos='n') for word in punc_free.split())\n normalized = \" \".join(lemma.lemmatize(word,pos='v') for word in normalized.split())\n word = \" \".join(word for word in normalized.split() if len(word)>3)\n postag = nltk.pos_tag(word.split())\n irlist = [',','.',':','#',';','CD','WRB','RB','PRP','...',')','(','-','``','@']\n poslist = ['NN','NNP','NNS','RB','RBR','RBS','JJ','JJR','JJS']\n wordlist = ['co', 'https', 'http','rt','com','amp','fe0f','www','ve','dont',\"i'm\",\"it's\",'isnt','âźă','âąă','âł_','kf4pdwe64k']\n adjandn = [word for word,pos in postag if pos in poslist and word not in wordlist and len(word)>3]\n stop = set(stopwords.words(lang))\n wordlist = [i for i in adjandn if i not in stop]\n features.append(' '.join(wordlist))\n with open('facebook_preprocessing.csv', 'a', encoding='UTF-8', newline='') as csvfile:\n writer = csv.writer(csvfile)\n writer.writerows([features])\n \ndf_postncomment = pd.read_csv('facebook_preprocessing.csv', encoding = 'UTF-8', sep = ',')\nrm_na = df_postncomment[pd.notnull(df_postncomment['re_message.x'])]\nrm_na.index = range(len(rm_na))\ndfinal_fb = pd.DataFrame(\n rm_na,\n columns = ['key', 'created_time.x', 'id.x', 'message.x', 'id.y', 'message.y', \n 'lang.x', 're_message.x', 'lang.y', 're_message.y'])\ndfinal_fb.to_csv(\n 'final_facebook_preprocessing.csv',\n encoding = 'UTF-8',\n columns = ['key', 'created_time.x', 'id.x', 'message.x', 'id.y', 'message.y',\n 'lang.x', 're_message.x', 'lang.y', 're_message.y'])\nos.remove('facebook_preprocessing.csv')\n#print(rm_na['re_message.x'][8])",
"_____no_output_____"
],
[
"test = pd.read_csv('final_facebook_preprocessing.csv', encoding = 'UTF-8', sep = ',', index_col = 0)\ndisplay(test.head(3))\nprint(len(test))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
4a133737aedf013305c2c58a3fba26c857f6d6e0
| 137,285 |
ipynb
|
Jupyter Notebook
|
Convert MATLAB Face Regions to Numpy.ipynb
|
vtsatskin/Hot-Car-Data-Collection
|
0a057ee987dffccd1939f771cb8e16d5b629dc81
|
[
"MIT"
] | null | null | null |
Convert MATLAB Face Regions to Numpy.ipynb
|
vtsatskin/Hot-Car-Data-Collection
|
0a057ee987dffccd1939f771cb8e16d5b629dc81
|
[
"MIT"
] | null | null | null |
Convert MATLAB Face Regions to Numpy.ipynb
|
vtsatskin/Hot-Car-Data-Collection
|
0a057ee987dffccd1939f771cb8e16d5b629dc81
|
[
"MIT"
] | null | null | null | 909.172185 | 91,628 | 0.946607 |
[
[
[
"import scipy.io as sio\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom skimage import data\n%matplotlib inline",
"_____no_output_____"
],
[
"mat_contents = sio.loadmat('positive_instances.mat')\nflattened_face_regions = [(r[0][0], r[1][0]) for r in mat_contents['positiveInstances'][0]]\n\n# Remove absolute path\nface_regions = [(x[0].replace('/Users/vt/Code/Personal/hot-car-data-collection/', ''), x[1]) for x in flattened_face_regions]\nnp.save('face_regions.npy', np.array(face_regions, dtype=object))",
"_____no_output_____"
]
],
[
[
"Try loading saved file to see if it works",
"_____no_output_____"
]
],
[
[
"saved_face_regions = np.load('face_regions.npy')\nrow = saved_face_regions[0]\nimage = data.imread(row[0])\nplt.imshow(image)",
"_____no_output_____"
]
],
[
[
"Show the face region of intee",
"_____no_output_____"
]
],
[
[
"y = row[1][0]\nx = row[1][1]\nheight = row[1][2]\nwidth = row[1][3]\nplt.imshow(image[x:x+width,y:y+height])",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a133931911205d0fe0b813838711446a31fbc07
| 12,112 |
ipynb
|
Jupyter Notebook
|
exps/conll2003 BERTBiLSTMAttnNCRF base BERT.ipynb
|
learnerhouse/ner-bert
|
606328a27a7313b6c22b78590e06618ad77402cd
|
[
"MIT"
] | 391 |
2018-12-21T01:00:18.000Z
|
2022-03-13T02:05:15.000Z
|
exps/conll2003 BERTBiLSTMAttnNCRF base BERT.ipynb
|
king-menin/nert-bert
|
b75a903c35acbd36ff5f26c525e3596294f36815
|
[
"MIT"
] | 30 |
2019-01-10T12:58:54.000Z
|
2022-01-15T15:51:14.000Z
|
exps/conll2003 BERTBiLSTMAttnNCRF base BERT.ipynb
|
king-menin/nert-bert
|
b75a903c35acbd36ff5f26c525e3596294f36815
|
[
"MIT"
] | 101 |
2018-12-21T07:40:31.000Z
|
2022-03-10T17:47:41.000Z
| 22.597015 | 197 | 0.522457 |
[
[
[
"%load_ext autoreload\n%autoreload 2\n\n\nimport sys\nimport warnings\n\n\nwarnings.filterwarnings(\"ignore\")\nsys.path.append(\"../\")",
"_____no_output_____"
],
[
"from modules.data.conll2003.prc import conll2003_preprocess",
"_____no_output_____"
],
[
"data_dir = \"/home/eartemov/ae/work/conll2003/\"",
"_____no_output_____"
],
[
"conll2003_preprocess(data_dir)",
"_____no_output_____"
]
],
[
[
"## IO markup",
"_____no_output_____"
],
[
"### Train",
"_____no_output_____"
]
],
[
[
"from modules.data import bert_data",
"_____no_output_____"
],
[
"data = bert_data.LearnData.create(\n train_df_path=\"/home/eartemov/ae/work/conll2003/eng.train.train.csv\",\n valid_df_path=\"/home/eartemov/ae/work/conll2003/eng.testa.dev.csv\",\n idx2labels_path=\"/home/eartemov/ae/work/conll2003/idx2labels5.txt\",\n clear_cache=True,\n model_name=\"bert-base-cased\"\n)",
"The pre-trained model you are loading is a cased model but you have not set `do_lower_case` to False. We are setting `do_lower_case=False` for you but you may want to check this behavior.\n"
],
[
"from modules.models.bert_models import BERTBiLSTMAttnNCRF",
"_____no_output_____"
],
[
"model = BERTBiLSTMAttnNCRF.create(\n len(data.train_ds.idx2label), model_name=\"bert-base-cased\",\n lstm_dropout=0., crf_dropout=0.3, nbest=len(data.train_ds.idx2label))",
"build CRF...\n"
],
[
"from modules.train.train import NerLearner",
"_____no_output_____"
],
[
"num_epochs = 100",
"_____no_output_____"
],
[
"learner = NerLearner(\n model, data, \"/home/eartemov/ae/work/models/conll2003-BERTBiLSTMAttnNCRF-base-IO.cpt\",\n t_total=num_epochs * len(data.train_dl))",
"_____no_output_____"
],
[
"model.get_n_trainable_params()",
"_____no_output_____"
],
[
"learner.fit(epochs=num_epochs)",
"_____no_output_____"
]
],
[
[
"### Predict",
"_____no_output_____"
]
],
[
[
"from modules.data.bert_data import get_data_loader_for_predict",
"_____no_output_____"
],
[
"dl = get_data_loader_for_predict(data, df_path=data.valid_ds.config[\"df_path\"])",
"_____no_output_____"
],
[
"preds = learner.predict(dl)",
"_____no_output_____"
],
[
"from sklearn_crfsuite.metrics import flat_classification_report",
"_____no_output_____"
],
[
"from modules.analyze_utils.utils import bert_labels2tokens, voting_choicer\nfrom modules.analyze_utils.plot_metrics import get_bert_span_report",
"_____no_output_____"
],
[
"pred_tokens, pred_labels = bert_labels2tokens(dl, preds)\ntrue_tokens, true_labels = bert_labels2tokens(dl, [x.bert_labels for x in dl.dataset])",
"_____no_output_____"
],
[
"assert pred_tokens == true_tokens\ntokens_report = flat_classification_report(true_labels, pred_labels, labels=data.train_ds.idx2label[4:], digits=4)",
"_____no_output_____"
],
[
"print(tokens_report)",
" precision recall f1-score support\n\n I_ORG 0.9552 0.9461 0.9506 2005\n I_O 0.9966 0.9973 0.9970 41651\n I_MISC 0.9343 0.8956 0.9146 1255\n I_PER 0.9846 0.9863 0.9854 2848\n I_LOC 0.9727 0.9636 0.9681 1922\n\n micro avg 0.9918 0.9907 0.9913 49681\n macro avg 0.9687 0.9578 0.9631 49681\nweighted avg 0.9917 0.9907 0.9912 49681\n\n"
]
],
[
[
"### Test",
"_____no_output_____"
]
],
[
[
"from modules.data.bert_data import get_data_loader_for_predict",
"_____no_output_____"
],
[
"dl = get_data_loader_for_predict(data, df_path=\"/home/eartemov/ae/work/conll2003/eng.testb.dev.csv\")",
"_____no_output_____"
],
[
"preds = learner.predict(dl)",
"_____no_output_____"
],
[
"from sklearn_crfsuite.metrics import flat_classification_report",
"_____no_output_____"
],
[
"from modules.analyze_utils.utils import bert_labels2tokens, voting_choicer\nfrom modules.analyze_utils.plot_metrics import get_bert_span_report",
"_____no_output_____"
],
[
"pred_tokens, pred_labels = bert_labels2tokens(dl, preds)\ntrue_tokens, true_labels = bert_labels2tokens(dl, [x.bert_labels for x in dl.dataset])",
"_____no_output_____"
],
[
"assert pred_tokens == true_tokens\ntokens_report = flat_classification_report(true_labels, pred_labels, labels=data.train_ds.idx2label[4:], digits=4)",
"_____no_output_____"
],
[
"print(tokens_report)",
" precision recall f1-score support\n\n I_ORG 0.8891 0.9174 0.9030 2360\n I_O 0.9952 0.9910 0.9931 37492\n I_MISC 0.8519 0.8154 0.8332 910\n I_PER 0.9729 0.9754 0.9741 2683\n I_LOC 0.9217 0.9207 0.9212 1815\n\n micro avg 0.9824 0.9798 0.9811 45260\n macro avg 0.9261 0.9240 0.9249 45260\nweighted avg 0.9825 0.9798 0.9811 45260\n\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a133e04198e8d10da47d22ffb7e4c0d8f1ce49a
| 107,084 |
ipynb
|
Jupyter Notebook
|
Ext_twitter.ipynb
|
ferramiCircus9/tweet_kafka
|
23650b115c0015d61c899c0bf629bce56e1354dc
|
[
"MIT"
] | null | null | null |
Ext_twitter.ipynb
|
ferramiCircus9/tweet_kafka
|
23650b115c0015d61c899c0bf629bce56e1354dc
|
[
"MIT"
] | null | null | null |
Ext_twitter.ipynb
|
ferramiCircus9/tweet_kafka
|
23650b115c0015d61c899c0bf629bce56e1354dc
|
[
"MIT"
] | null | null | null | 48.454299 | 150 | 0.329078 |
[
[
[
"# Codigo Para Twitter por streaming",
"_____no_output_____"
]
],
[
[
"#Import the necessary methods from tweepy library\nfrom tweepy.streaming import StreamListener\nfrom tweepy import OAuthHandler\nfrom tweepy import Stream\nimport json\nimport time",
"_____no_output_____"
],
[
"access_token = \"227835837-WD07ixlyOeLqkeywbnMYzk5dnebJjd1pA4sKpOjl\"\naccess_token_secret = \"6utbaX2ab3UrpL4PpfSx6ToCuuQZgZ5zDDqKQq2albTLL\"\nconsumer_key = \"dwazigqjw1ZIVtx2jKGGsw2wb\"\nconsumer_secret = \"Lyy1wItpyPTfPWIBJ5d2qrA250s3iydzwKaCubVAamOTOecK2A\"",
"_____no_output_____"
],
[
"class StdOutListener(StreamListener):\n\n def on_data(self, data):\n\n json_load = json.loads(data)\n print(data.encode())\n\n return True\n\n def on_error(self, status):\n print (status)\n\n\nif __name__ == '__main__':\n\n #This handles Twitter authetification and the connection to Twitter Streaming API\n l = StdOutListener()\n auth = OAuthHandler(consumer_key, consumer_secret)\n auth.set_access_token(access_token, access_token_secret)\n stream = Stream(auth, l)\n\n #This line filter Twitter Streams to capture data by the keywords: 'python', 'javascript', 'ruby'\nstream.filter(track=['Naruto'])\n\n",
"_____no_output_____"
]
],
[
[
"# Codigo para Descargar Historico de Twitter",
"_____no_output_____"
]
],
[
[
"import json \nimport tweepy \nfrom tweepy import Stream \nfrom tweepy.streaming import StreamListener \nfrom tweepy import OAuthHandler \nimport pandas as pd \nimport pickle \nimport sys\n \n \n",
"_____no_output_____"
],
[
"##es = Elasticsearch([{'host': 'localhost', 'port': 9200}])\n\n#Variables para poner las credenciales para la API\nconsumer_key = \"dwazigqjw1ZIVtx2jKGGsw2wb\"\nconsumer_secret = \"Lyy1wItpyPTfPWIBJ5d2qrA250s3iydzwKaCubVAamOTOecK2A\" \naccess_token = \"227835837-WD07ixlyOeLqkeywbnMYzk5dnebJjd1pA4sKpOjl\" \naccess_secret = \"6utbaX2ab3UrpL4PpfSx6ToCuuQZgZ5zDDqKQq2albTLL\" \n \nauth = OAuthHandler(consumer_key, consumer_secret) \nauth.set_access_token(access_token, access_secret) \n \napi = tweepy.API(auth)\ndatos_tweets={} \ndatos_tweets.setdefault('date',[]) \ndatos_tweets.setdefault('texts',[]) \ndatos_tweets.setdefault('user_id',[]) \ndatos_tweets.setdefault('retweet_count',[]) \ndatos_tweet_df=pd.DataFrame()\nargs=['','Soen','2021-03-12T00:00:00','2021-03-19T24:59:59']##sys.argv\nprint(args[1])\n \n#print(place_id) \nfor tweet in tweepy.Cursor(api.search, q=args[1], lang=\"es\", since=args[2], until=args[3]).items():\n if not hasattr(tweet,'retweeted_status'):\n datos_tweet_df=datos_tweet_df.append(pd.json_normalize(tweet._json))\n\ndatos_tweet_df=datos_tweet_df.reset_index(drop=True)\n",
"Soen\n"
],
[
"datos_tweet_df=datos_tweet_df.reset_index(drop=True)\ndatos_tweet_df",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a133f928e1ec0c6cdcd31eea5654c67c15e3608
| 167,898 |
ipynb
|
Jupyter Notebook
|
windTurbinePower.ipynb
|
JinaKim77/Project-2020_Emerging-Technologies
|
6dfcca5d38e251f7f25bce159540f34d7b9852b4
|
[
"MIT"
] | null | null | null |
windTurbinePower.ipynb
|
JinaKim77/Project-2020_Emerging-Technologies
|
6dfcca5d38e251f7f25bce159540f34d7b9852b4
|
[
"MIT"
] | null | null | null |
windTurbinePower.ipynb
|
JinaKim77/Project-2020_Emerging-Technologies
|
6dfcca5d38e251f7f25bce159540f34d7b9852b4
|
[
"MIT"
] | null | null | null | 112.758899 | 46,700 | 0.794238 |
[
[
[
"## Emerging Technologies\n\n\n#### Student name : Jina Kim G00353420\n\n***",
"_____no_output_____"
],
[
"#### Jupyter notebook that trains a model using the data set. (powerproduction.csv)",
"_____no_output_____"
],
[
"***\n#### References\n\nKeras Fundamentals for Deep Learning. [1]\n\nTraining & evaluation from tf.data Datasets. [2]\n\n***\n\n[1] Keras Fundamentals for Deep Learning; guru99; https://www.guru99.com/keras-tutorial.html\n\n[2] Training & evaluation from tf.data Datasets; Tensorflow; https://www.tensorflow.org/guide/keras/train_and_evaluate\n\n\n***",
"_____no_output_____"
]
],
[
[
"# Neural networks.\nimport tensorflow.keras as kr\nfrom tensorflow.keras import layers\nfrom tensorflow.python.keras.layers import Dense\nfrom tensorflow.keras.layers.experimental import preprocessing\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.python.keras.models import Model\n\n# Numerical arrays\nimport numpy as np\n\n# Data frames.\nimport pandas as pd\n\n# Plotting\nimport matplotlib.pyplot as plt\n\nfrom sklearn import linear_model",
"_____no_output_____"
],
[
"# Plot style.\nplt.style.use(\"ggplot\")\n\n# Plot size.\nplt.rcParams['figure.figsize'] = [14, 8]",
"_____no_output_____"
],
[
"# Dataset from csv file\ntrain = pd.read_csv(\"powerproduction.csv\")",
"_____no_output_____"
],
[
"print(\"TensorFlow version: {}\".format(tf.__version__))\nprint(\"Eager execution: {}\".format(tf.executing_eagerly()))",
"TensorFlow version: 2.3.0\nEager execution: True\n"
],
[
"print(\"Local copy of the dataset file: {}\".format(train))",
"Local copy of the dataset file: speed power\n0 0.000 0.0\n1 0.125 0.0\n2 0.150 0.0\n3 0.225 0.0\n4 0.275 0.0\n.. ... ...\n495 24.775 0.0\n496 24.850 0.0\n497 24.875 0.0\n498 24.950 0.0\n499 25.000 0.0\n\n[500 rows x 2 columns]\n"
],
[
"# turn dataset into dataframe\ndata = pd.DataFrame(train)",
"_____no_output_____"
],
[
"#####################\n### plot the datasel\n#####################\n\ndata.plot.scatter(x='speed', y='power',marker='o', color=\"blue\")",
"_____no_output_____"
],
[
"# Create a new neural network.\nm = kr.models.Sequential()\n\n# Add a single neuron in a single layer, initialised with weight 1 and bias 0, with sigmoid activation.\nm.add(Dense(50, input_shape=(1,), activation='sigmoid', kernel_initializer=\"glorot_uniform\", bias_initializer=\"glorot_uniform\"))\nm.add(Dense(1, activation='linear', kernel_initializer=\"glorot_uniform\", bias_initializer=\"glorot_uniform\"))\n\n# Compile the model.\nm.compile(kr.optimizers.Adam(lr=0.001), loss='mean_squared_error', metrics=['accuracy'])\n",
"_____no_output_____"
],
[
"# Model configuration\nno_epochs = 200\nbatch_size = 10\n\n# Start the training process\nmodel_one = m.fit(data['speed'], data['power'], epochs=no_epochs, batch_size=batch_size)\n\n# Show a summary of the model. Check the number of trainable parameters\n# Call its summary() method to display its contents\nprint(m.summary())",
"Epoch 1/200\n50/50 [==============================] - 0s 2ms/step - loss: 3836.7590 - accuracy: 0.0000e+00\nEpoch 2/200\n50/50 [==============================] - 0s 2ms/step - loss: 3640.8345 - accuracy: 0.0000e+00\nEpoch 3/200\n50/50 [==============================] - 0s 3ms/step - loss: 3413.5239 - accuracy: 0.0000e+00\nEpoch 4/200\n50/50 [==============================] - 0s 2ms/step - loss: 3180.1841 - accuracy: 0.0000e+00\nEpoch 5/200\n50/50 [==============================] - 0s 2ms/step - loss: 2991.1218 - accuracy: 0.0000e+00\nEpoch 6/200\n50/50 [==============================] - 0s 3ms/step - loss: 2838.3728 - accuracy: 0.0000e+00\nEpoch 7/200\n50/50 [==============================] - 0s 3ms/step - loss: 2700.5078 - accuracy: 0.0000e+00\nEpoch 8/200\n50/50 [==============================] - 0s 3ms/step - loss: 2577.8142 - accuracy: 0.0000e+00\nEpoch 9/200\n50/50 [==============================] - 0s 3ms/step - loss: 2469.1423 - accuracy: 0.0000e+00\nEpoch 10/200\n50/50 [==============================] - 0s 3ms/step - loss: 2359.7017 - accuracy: 0.0000e+00\nEpoch 11/200\n50/50 [==============================] - 0s 3ms/step - loss: 2256.6609 - accuracy: 0.0000e+00\nEpoch 12/200\n50/50 [==============================] - 0s 3ms/step - loss: 2165.9109 - accuracy: 0.0000e+00\nEpoch 13/200\n50/50 [==============================] - 0s 3ms/step - loss: 2079.3809 - accuracy: 0.0000e+00\nEpoch 14/200\n50/50 [==============================] - 0s 3ms/step - loss: 1999.2170 - accuracy: 0.0000e+00\nEpoch 15/200\n50/50 [==============================] - 0s 3ms/step - loss: 1921.4972 - accuracy: 0.0000e+00\nEpoch 16/200\n50/50 [==============================] - 0s 3ms/step - loss: 1848.0189 - accuracy: 0.0000e+00\nEpoch 17/200\n50/50 [==============================] - 0s 3ms/step - loss: 1778.4877 - accuracy: 0.0000e+00\nEpoch 18/200\n50/50 [==============================] - 0s 3ms/step - loss: 1713.7604 - accuracy: 0.0000e+00\nEpoch 19/200\n50/50 [==============================] - 0s 3ms/step - loss: 1651.2332 - accuracy: 0.0000e+00\nEpoch 20/200\n50/50 [==============================] - 0s 3ms/step - loss: 1593.0621 - accuracy: 0.0000e+00\nEpoch 21/200\n50/50 [==============================] - 0s 4ms/step - loss: 1537.6427 - accuracy: 0.0000e+00\nEpoch 22/200\n50/50 [==============================] - 0s 3ms/step - loss: 1483.9500 - accuracy: 0.0000e+00\nEpoch 23/200\n50/50 [==============================] - 0s 3ms/step - loss: 1433.1809 - accuracy: 0.0000e+00\nEpoch 24/200\n50/50 [==============================] - 0s 3ms/step - loss: 1384.1909 - accuracy: 0.0000e+00\nEpoch 25/200\n50/50 [==============================] - 0s 3ms/step - loss: 1337.5046 - accuracy: 0.0000e+00\nEpoch 26/200\n50/50 [==============================] - 0s 4ms/step - loss: 1291.7905 - accuracy: 0.0000e+00\nEpoch 27/200\n50/50 [==============================] - 0s 4ms/step - loss: 1248.7562 - accuracy: 0.0000e+00\nEpoch 28/200\n50/50 [==============================] - 0s 4ms/step - loss: 1206.1709 - accuracy: 0.0000e+00\nEpoch 29/200\n50/50 [==============================] - 0s 3ms/step - loss: 1165.7269 - accuracy: 0.0000e+00\nEpoch 30/200\n50/50 [==============================] - 0s 3ms/step - loss: 1126.3158 - accuracy: 0.0000e+00\nEpoch 31/200\n50/50 [==============================] - 0s 3ms/step - loss: 1088.7427 - accuracy: 0.0000e+00\nEpoch 32/200\n50/50 [==============================] - 0s 4ms/step - loss: 1052.0702 - accuracy: 0.0000e+00\nEpoch 33/200\n50/50 [==============================] - 0s 4ms/step - loss: 1017.2763 - accuracy: 0.0000e+00\nEpoch 34/200\n50/50 [==============================] - 0s 3ms/step - loss: 983.2903 - accuracy: 0.0000e+00\nEpoch 35/200\n50/50 [==============================] - 0s 3ms/step - loss: 951.2313 - accuracy: 0.0000e+00\nEpoch 36/200\n50/50 [==============================] - 0s 3ms/step - loss: 919.7587 - accuracy: 0.0000e+00\nEpoch 37/200\n50/50 [==============================] - 0s 3ms/step - loss: 889.5878 - accuracy: 0.0000e+00\nEpoch 38/200\n50/50 [==============================] - 0s 3ms/step - loss: 861.1971 - accuracy: 0.0000e+00\nEpoch 39/200\n50/50 [==============================] - 0s 3ms/step - loss: 834.3897 - accuracy: 0.0000e+00\nEpoch 40/200\n50/50 [==============================] - 0s 3ms/step - loss: 806.9496 - accuracy: 0.0000e+00\nEpoch 41/200\n50/50 [==============================] - 0s 4ms/step - loss: 781.2003 - accuracy: 0.0000e+00\nEpoch 42/200\n50/50 [==============================] - 0s 3ms/step - loss: 757.5896 - accuracy: 0.0000e+00\nEpoch 43/200\n50/50 [==============================] - 0s 3ms/step - loss: 733.6001 - accuracy: 0.0060\nEpoch 44/200\n50/50 [==============================] - 0s 3ms/step - loss: 710.6216 - accuracy: 0.0080\nEpoch 45/200\n50/50 [==============================] - 0s 3ms/step - loss: 689.9627 - accuracy: 0.0100\nEpoch 46/200\n50/50 [==============================] - ETA: 0s - loss: 671.6881 - accuracy: 0.01 - 0s 3ms/step - loss: 668.9572 - accuracy: 0.0120\nEpoch 47/200\n50/50 [==============================] - 0s 3ms/step - loss: 649.4540 - accuracy: 0.0140\nEpoch 48/200\n50/50 [==============================] - 0s 3ms/step - loss: 630.7135 - accuracy: 0.0140\nEpoch 49/200\n50/50 [==============================] - 0s 2ms/step - loss: 613.0167 - accuracy: 0.0180\nEpoch 50/200\n50/50 [==============================] - 0s 2ms/step - loss: 596.0219 - accuracy: 0.0200\nEpoch 51/200\n50/50 [==============================] - 0s 4ms/step - loss: 580.3693 - accuracy: 0.0220\nEpoch 52/200\n50/50 [==============================] - 0s 3ms/step - loss: 564.1965 - accuracy: 0.0240\nEpoch 53/200\n50/50 [==============================] - 0s 3ms/step - loss: 549.8567 - accuracy: 0.0260\nEpoch 54/200\n50/50 [==============================] - 0s 3ms/step - loss: 535.6124 - accuracy: 0.0280\nEpoch 55/200\n50/50 [==============================] - 0s 3ms/step - loss: 522.8572 - accuracy: 0.0300\nEpoch 56/200\n50/50 [==============================] - 0s 3ms/step - loss: 510.3208 - accuracy: 0.0320\nEpoch 57/200\n50/50 [==============================] - 0s 3ms/step - loss: 498.0914 - accuracy: 0.0340\nEpoch 58/200\n50/50 [==============================] - 0s 3ms/step - loss: 486.9695 - accuracy: 0.0340\nEpoch 59/200\n50/50 [==============================] - 0s 3ms/step - loss: 477.3657 - accuracy: 0.0340\nEpoch 60/200\n50/50 [==============================] - 0s 3ms/step - loss: 466.8929 - accuracy: 0.0340\nEpoch 61/200\n50/50 [==============================] - 0s 3ms/step - loss: 457.2222 - accuracy: 0.0340\nEpoch 62/200\n50/50 [==============================] - 0s 3ms/step - loss: 448.9042 - accuracy: 0.0340\nEpoch 63/200\n50/50 [==============================] - 0s 3ms/step - loss: 439.8732 - accuracy: 0.0340\nEpoch 64/200\n50/50 [==============================] - 0s 2ms/step - loss: 432.2128 - accuracy: 0.0340\nEpoch 65/200\n50/50 [==============================] - 0s 2ms/step - loss: 424.8849 - accuracy: 0.0340\nEpoch 66/200\n50/50 [==============================] - 0s 2ms/step - loss: 417.7518 - accuracy: 0.0360\nEpoch 67/200\n50/50 [==============================] - 0s 2ms/step - loss: 411.5956 - accuracy: 0.0360\nEpoch 68/200\n50/50 [==============================] - 0s 2ms/step - loss: 405.1595 - accuracy: 0.0360\nEpoch 69/200\n50/50 [==============================] - 0s 2ms/step - loss: 399.1534 - accuracy: 0.0360\nEpoch 70/200\n50/50 [==============================] - 0s 2ms/step - loss: 394.5169 - accuracy: 0.0360\nEpoch 71/200\n50/50 [==============================] - 0s 2ms/step - loss: 388.7432 - accuracy: 0.0420\nEpoch 72/200\n50/50 [==============================] - 0s 2ms/step - loss: 384.4977 - accuracy: 0.0400\nEpoch 73/200\n50/50 [==============================] - 0s 2ms/step - loss: 379.7592 - accuracy: 0.0420\nEpoch 74/200\n50/50 [==============================] - 0s 2ms/step - loss: 376.3634 - accuracy: 0.0440\nEpoch 75/200\n50/50 [==============================] - 0s 2ms/step - loss: 371.9331 - accuracy: 0.0440\nEpoch 76/200\n50/50 [==============================] - 0s 2ms/step - loss: 368.4671 - accuracy: 0.0480\nEpoch 77/200\n50/50 [==============================] - 0s 2ms/step - loss: 364.6064 - accuracy: 0.0480\nEpoch 78/200\n50/50 [==============================] - 0s 2ms/step - loss: 361.6483 - accuracy: 0.0480\nEpoch 79/200\n"
],
[
"#The returned \"history\" object holds a record of the loss values and metric values during training:\nmodel_one.history",
"_____no_output_____"
],
[
"# evaluate the model on the test data via evaluate()\n\n# Evaluate the model on the test data using `evaluate`\nprint(\"Evaluate on test data\")\nresults = m.evaluate(data['speed'], data['power'], batch_size=128)\nprint(\"test loss, test acc:\", results)",
"Evaluate on test data\n4/4 [==============================] - 0s 2ms/step - loss: 278.8987 - accuracy: 0.0480\ntest loss, test acc: [278.8986511230469, 0.04800000041723251]\n"
],
[
"# prediction to data frame\nprediction = pd.DataFrame(m.predict(data['speed']), columns=['speed'])\n\n# output prediction\nprediction",
"_____no_output_____"
],
[
"###################\n### save the model\n###################\n\nm.save(\"my_h5_model.h5\")\nprint(\"saving model prediction data...\")",
"saving model prediction data...\n"
],
[
"# Model accuracy into data frame\nAccuracy = pd.DataFrame(model_one.history['accuracy'], columns=['Average of accuracy is'])\n\n# output mean of model accuracy \nAccuracy.mean()",
"_____no_output_____"
],
[
"# Model accuracy into data frame\nAccuracy = pd.DataFrame(model_one.history['accuracy'], columns=['maximum of accuracy is'])\n\n# output mean of model accuracy \nAccuracy.max()",
"_____no_output_____"
],
[
"# Model losses into data frame\nLoss = pd.DataFrame(model_one.history['loss'], columns=['Average of loss is'])\n\n# output minium loss ofrom model\nLoss.mean()",
"_____no_output_____"
],
[
"# Model losses into data frame\nLoss = pd.DataFrame(model_one.history['loss'], columns=['Minium Loss is'])\n\n# output minium loss ofrom model\nLoss.min()",
"_____no_output_____"
],
[
"# Visualize the loss function over time\nfig, axes = plt.subplots(2, sharex=True, figsize=(14, 10))\nfig.suptitle('Training Metrics', fontsize=14)\n\n##########################################\n### summarize history for lost & accuracy\n##########################################\n\n# plot the losses of model \naxes[0].set_ylabel(\"Loss\", fontsize=10)\naxes[0].set_xlabel(\"Epoch\", fontsize=10)\naxes[0].set_title(\"= Model Losses =\", fontsize=13)\naxes[0].plot(model_one.history['loss'],color=\"red\")\nplt.legend(['loss'], loc='upper left')\n\n# plot accuracy of model\naxes[1].set_ylabel(\"Accuracy\", fontsize=10)\naxes[1].set_xlabel(\"Epoch\", fontsize=10)\naxes[1].set_title(\"= Model Accuracy =\", fontsize=13)\naxes[1].plot(model_one.history['accuracy'],color=\"green\")\nplt.legend(['accuracy'], loc='upper left')\n\nplt.show()\n\n# As you can see, the losses are dropped significantly and the accuracy is high.",
"_____no_output_____"
],
[
"#############################\n### plot model for comparing\n#############################\n\nplt.title('Comparison of Original Data and Predicted Data')\nplt.plot(data['speed'], data['power'], 'v', color=\"green\", markersize=2, label='Original Data')\nplt.plot(data['speed'], m.predict(data['speed']), '+', color=\"red\", markersize=2, label='Predicted Data')\nplt.legend();",
"_____no_output_____"
]
],
[
[
"***",
"_____no_output_____"
]
],
[
[
"########################################################################################\n### A program that trains a model that predicts wind turbine power based on wind speed.\n########################################################################################\n\n# load the dataset into memory\ntraining_dataset = pd.read_csv(\"powerproduction.csv\")\n\n# create a model using the linear regression algorithm\n# and train it with the data from our csv\nregression_model = linear_model.LinearRegression()\nprint (\"Training model...\")\n\n# model training\nregression_model.fit(training_dataset[['speed']], training_dataset.power) \nprint (\"Model trained!\")\n\n# ask user to enter an speed and calculate\n# its power using our model\ninput_speed = float(input(\"Enter speed: \"))\nproped_power = regression_model.predict([[input_speed]])\n\nprint (\"Proped power:\", round(proped_power[0], 3))\nprint (\"Done\")",
"Training model...\nModel trained!\nEnter speed: 8\nProped power: 25.441\nDone\n"
]
],
[
[
"***\n#### End of Model 1",
"_____no_output_____"
],
[
"## Done",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a1374fe0422d4905ee65b6ef9af0a605708d4d9
| 12,360 |
ipynb
|
Jupyter Notebook
|
Handwritten Text Recognition/Line Detection From Text/predict_lines_from_folder.ipynb
|
Psarpei/Handwritten-Text-Recognition
|
be8f12092e385f3e117ae79b08fb06d0681f67e3
|
[
"MIT"
] | 15 |
2020-05-03T20:47:59.000Z
|
2021-11-19T22:48:35.000Z
|
Handwritten Text Recognition/Line Detection From Text/predict_lines_from_folder.ipynb
|
Psarpei/Handwritten-Text-Recognition
|
be8f12092e385f3e117ae79b08fb06d0681f67e3
|
[
"MIT"
] | 1 |
2021-11-10T13:49:21.000Z
|
2021-11-10T13:49:21.000Z
|
Handwritten Text Recognition/Line Detection From Text/predict_lines_from_folder.ipynb
|
Psarpei/Handwritten-Text-Recognition
|
be8f12092e385f3e117ae79b08fb06d0681f67e3
|
[
"MIT"
] | 2 |
2021-12-21T17:15:02.000Z
|
2022-03-15T11:13:32.000Z
| 36.352941 | 146 | 0.445874 |
[
[
[
"import torch\nfrom torchvision import transforms\nimport torch.nn.functional as F\nimport torch.nn as nn\n\nfrom PIL import Image\nimport imageio\nimport os\n\nfrom google.colab import drive",
"_____no_output_____"
],
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Drive already mounted at /content/drive; to attempt to forcibly remount, call drive.mount(\"/content/drive\", force_remount=True).\n"
],
[
"class YOLO(nn.Module):\n def __init__(self, img_width, row_size):\n super(YOLO, self).__init__()\n self.row_size = row_size\n self.conv1 = nn.Conv2d(1, 16, 7, stride=2)\n self.mp1 = nn.MaxPool2d(2, 2)\n self.conv2 = nn.Conv2d(16, 32, (3, 3), stride=1)\n self.mp2 = nn.MaxPool2d(2, 2)\n self.conv3 = nn.Conv2d(32, 64, (3, 3), stride=1)\n self.mp3 = nn.MaxPool2d(2, 2)\n\n self.fc1 = nn.Linear(64*53*36, 4096)\n self.fc2 = nn.Linear(4096, row_size * 5)\n self.dropout = nn.Dropout()\n\n def forward(self, x):\n # Conv + ReLU + max pooling for two layers\n x = F.relu(self.conv1(x))\n x = self.mp1(x)\n x = F.relu(self.conv2(x))\n x = self.mp2(x)\n x = F.relu(self.conv3(x))\n x = self.mp3(x)\n x = x.view(-1, 64*53*36)\n x = F.relu(self.dropout(self.fc1(x)))\n x = self.fc2(x)\n x = x.view(-1, self.row_size, 5)\n x = torch.sigmoid(x)\n return x",
"_____no_output_____"
],
[
"def calc_x_y(row, tensor):\n \"\"\"calc coordinates\"\"\"\n\n x = tensor[1] * 619\n y = tensor[2] * (885 / 50) + row * (885 / 50)\n width = tensor[3] * 619\n height = tensor[4] * 885\n return torch.FloatTensor([1, x, y, width, height])",
"_____no_output_____"
],
[
"def calc_box(tensor):\n \"\"\"calc box for output line\"\"\"\n x1 = max(0, tensor[1] - 0.5 * tensor[3])\n y1 = max(0, tensor[2] - 0.5 * tensor[4])\n x2 = min(619, tensor[1] + 0.5 * tensor[3])\n y2 = min(885, tensor[2] + 0.5 * tensor[4])\n\n box = [x1, y1, x2, y2]\n return box",
"_____no_output_____"
],
[
"def non_maximum_suppression(tensor, percent):\n \"\"\"choose predicted lines by highest propability. \n Lines who overlap a actual choosen line by percent or higher will delete.\"\"\"\n \n for j in range(tensor.size(1)):\n if(tensor[j,0].item() < 0.5):\n tensor[j,0] = torch.tensor(0)\n found = []\n while(True):\n maximum = 0\n index = 0\n for j in range(tensor.size(1)):\n if(tensor[j,0].item() > maximum and j not in found):\n maximum = tensor[j,0].item()\n index = j\n\n if(maximum == 0):\n break\n\n found.append(index)\n tensor[index,0] = torch.tensor(1)\n \n for j in range(tensor.size(1)):\n if(j != index and tensor[j,0] >= 0.5):\n x_y_max = calc_x_y(index, tensor[index])\n x_y_other = calc_x_y(j, tensor[j])\n box1 = calc_box(x_y_max)\n box2 = calc_box(x_y_other)\n if(calc_iou(box1, box2) > percent):\n tensor[j,0] = 0",
"_____no_output_____"
],
[
"imgs_path = \"drive/My Drive/data_small/forms/forms_train_small/\"\nimgs_paths = os.listdir(imgs_path)\nweight_path = \"drive/My Drive/evaluation_small/weights_small.pt\"\npredict_path = \"drive/My Drive/testlines_predicted_small/\"\n\ntransform = transforms.Compose([transforms.Resize((885, 619)),\n transforms.ToTensor()])\n\n# set a boolean flag that indicates whether a cuda capable GPU is available\nis_gpu = torch.cuda.is_available()\nprint(\"GPU is available:\", is_gpu)\nprint(\"If you are receiving False, try setting your runtime to GPU\")\n\n# set the device to cuda if a GPU is available\ndevice = torch.device(\"cuda\" if is_gpu else \"cpu\")\nmodel = torch.load(weight_path)\n\nprint(model)",
"GPU is available: True\nIf you are receiving False, try setting your runtime to GPU\nYOLO(\n (conv1): Conv2d(1, 16, kernel_size=(7, 7), stride=(2, 2))\n (mp1): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (conv2): Conv2d(16, 32, kernel_size=(3, 3), stride=(1, 1))\n (mp2): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (conv3): Conv2d(32, 64, kernel_size=(3, 3), stride=(1, 1))\n (mp3): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)\n (fc1): Linear(in_features=122112, out_features=4096, bias=True)\n (fc2): Linear(in_features=4096, out_features=250, bias=True)\n (dropout): Dropout(p=0.5, inplace=False)\n)\n"
],
[
"def predict_lines(model,imgs_path, predict_path):\n \"\"\" predict images to lines from image path to predict_path\"\"\"\n img_count = 0\n for path in imgs_paths:\n count = 0\n img_tensor = transform(Image.open(imgs_path + path))\n output = model(torch.stack([img_tensor]).to(device))[0]\n # find right boxes\n non_maximum_suppression(output, 0.5)\n img = imageio.imread(imgs_path + path)\n yscale = round(img.shape[0] / 885)\n xscale = round(img.shape[1] / 619)\n print(xscale, xscale)\n for i in range(50):\n if(output[i][0] > 0.5):\n print(output[i])\n box = calc_box(calc_x_y(i, output[i]))\n x1 = (int(box[0])) * xscale\n x2 = (int(box[2])) * xscale\n y1 = (int(box[1])) * yscale\n y2 = (int(box[3])) * yscale\n print(box)\n imageio.imwrite(predict_path + \"pic\" + str(img_count) + \"line\" + str(count) + '.jpg', img[y1:y2, x1:x2])\n count += 1\n img_count += 1",
"_____no_output_____"
],
[
"predict_lines(model, imgs_path, predict_path)",
"4 4\ntensor([0.9624, 0.5155, 0.2458, 0.7337, 0.0322], device='cuda:0',\n grad_fn=<SelectBackward>)\n[tensor(92.0245), tensor(184.8153), tensor(546.1631), tensor(213.2853)]\ntensor([0.9611, 0.5311, 0.7832, 0.7651, 0.0283], device='cuda:0',\n grad_fn=<SelectBackward>)\n[tensor(91.9314), tensor(231.4322), tensor(565.5098), tensor(256.4917)]\ntensor([0.9774, 0.5564, 0.3270, 0.7862, 0.0289], device='cuda:0',\n grad_fn=<SelectBackward>)\n[tensor(101.0668), tensor(276.1984), tensor(587.7156), tensor(301.7772)]\ntensor([0.9356, 0.5376, 0.7916, 0.7765, 0.0302], device='cuda:0',\n grad_fn=<SelectBackward>)\n[tensor(92.4154), tensor(319.2509), tensor(573.0753), tensor(345.9702)]\ntensor([0.9861, 0.5391, 0.4071, 0.7786, 0.0331], device='cuda:0',\n grad_fn=<SelectBackward>)\n[tensor(92.7140), tensor(364.2780), tensor(574.6712), tensor(393.5328)]\ntensor([0.9241, 0.5441, 0.8382, 0.7767, 0.0290], device='cuda:0',\n grad_fn=<SelectBackward>)\n[tensor(96.4169), tensor(409.0951), tensor(577.1865), tensor(434.7762)]\ntensor([0.9694, 0.5316, 0.4824, 0.7791, 0.0314], device='cuda:0',\n grad_fn=<SelectBackward>)\n[tensor(87.9115), tensor(454.8552), tensor(570.1580), tensor(482.6223)]\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a1396ce56198a503ef5594567ba9751a7b5796e
| 70,508 |
ipynb
|
Jupyter Notebook
|
RESTful Endpoint Lab.ipynb
|
sakanmfec/CPDDVHOL4_Lab
|
28bbd5a54a9272aaa6f494d2711d6e9744f44777
|
[
"Apache-2.0"
] | 1 |
2022-01-11T13:26:24.000Z
|
2022-01-11T13:26:24.000Z
|
RESTful Endpoint Lab.ipynb
|
sakanmfec/CPDDVHOL4_Lab
|
28bbd5a54a9272aaa6f494d2711d6e9744f44777
|
[
"Apache-2.0"
] | null | null | null |
RESTful Endpoint Lab.ipynb
|
sakanmfec/CPDDVHOL4_Lab
|
28bbd5a54a9272aaa6f494d2711d6e9744f44777
|
[
"Apache-2.0"
] | 3 |
2021-12-11T18:01:44.000Z
|
2022-03-29T16:58:11.000Z
| 30.222032 | 540 | 0.558901 |
[
[
[
"# Db2 11.5.4 RESTful Programming\nThe following notebook is a brief example of how to use the Db2 11.5.4 RESTful Endpoint service to extend the capabilies of Db2.\n\nProgrammers can create Representational State Transfer (REST) endpoints that can be used to interact with Db2.\n\nEach endpoint is associated with a single SQL statement. Authenticated users of web, mobile, or cloud applications can use these REST endpoints from any REST HTTP client without having to install any Db2 drivers.\n\nThe Db2 REST server accepts an HTTP request, processes the request body, and returns results in JavaScript Object Notation (JSON).\n\nThe Db2 REST server is pre-installed and running on Docker on host3 (10.0.0.4) in the Demonstration cluster. As a programmer you can communicate with the service on port 50050. Your welcome note includes the external port you can use to interact with the Db2 RESTful Endpoint service directly.\n\nYou can find more information about this service at: https://www.ibm.com/support/producthub/db2/docs/content/SSEPGG_11.5.0/com.ibm.db2.luw.admin.rest.doc/doc/c_rest.html.",
"_____no_output_____"
],
[
"### Finding the Db2 RESTful Endpoint Service API Documentation\nIf you are running this notebook from a browser running inside the Cloud Pak for Data cluster, click: http://10.0.0.4:50050/docs If you are running this from a browser from your own desktop, check your welcome note for the address of the Db2 RESTful Service at port 50050.",
"_____no_output_____"
],
[
"## Getting Started\nBefore you can start submitting SQL or creating your own services you need to complete a few setup steps. ",
"_____no_output_____"
],
[
"### Import the required programming libraries\nThe requests library is the minimum required by Python to construct RESTful service calls. The Pandas library is used to format and manipulate JSON result sets as tables. The urllib3 library is used to manage secure https requests. ",
"_____no_output_____"
]
],
[
[
"import requests\nimport pandas as pd\nimport urllib3",
"_____no_output_____"
]
],
[
[
"### Create the Header File required for getting an authetication token\nWe have to provide the location of the RESTful service for our calls.\nThe RESTful call to the Db2 RESTful Endpoint service is contructed and transmitted as JSON. The first part of the JSON structure is the headers that define the content tyoe of the request.",
"_____no_output_____"
]
],
[
[
"headers = {\n \"content-type\": \"application/json\"\n}",
"_____no_output_____"
]
],
[
[
"### Define the RESTful Host\nThe next part defines where the request is sent to. It provides the location of the RESTful service for our calls.",
"_____no_output_____"
]
],
[
[
"Db2RESTful = \"https://10.0.0.201:32115\"",
"_____no_output_____"
]
],
[
[
"### API Authentication Service\nEach service has its own path in the RESTful call. For authentication we need to point to the `v1/auth` service.",
"_____no_output_____"
]
],
[
[
"API_Auth = \"/v1/auth\"",
"_____no_output_____"
]
],
[
[
"### Database Connection Information\nTo authenticate to the RESTful service you must provide the connection information for the database along with the userid and password that you are using to authenticate with. You can also provide an expiry time so that the access token that gets returned will be invalidated after that time period.",
"_____no_output_____"
]
],
[
[
"body = {\n \"dbParms\": {\n \"dbHost\": \"10.0.0.201\",\n \"dbName\": \"BLUDB\",\n \"dbPort\": 31684,\n \"isSSLConnection\": False,\n \"username\": \"admin\",\n \"password\": \"password\"\n },\n \"expiryTime\": \"8760h\"\n}",
"_____no_output_____"
]
],
[
[
"### Disabling HTTPS Warnings\n",
"_____no_output_____"
]
],
[
[
"urllib3.disable_warnings()",
"_____no_output_____"
]
],
[
[
"### Retrieving an Access Token\nWhen communicating with the RESTful service, you must provide the name of the service that you want to interact with. In this case the authentication service is */v1/auth*. ",
"_____no_output_____"
]
],
[
[
"try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_Auth), verify=False, headers=headers, json=body)\n print (response)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
]
],
[
[
"A response code of 200 means that the authentication worked properly, otherwise the error that was generated is printed. The response includes a connection token that is reused throughout the rest of this lab. It ensures secure a connection without requiring that you reenter a userid and password with each request.",
"_____no_output_____"
]
],
[
[
"if (response.status_code == 200):\n token = response.json()[\"token\"]\n print(\"Token: {}\".format(token))\nelse:\n print(response.json()[\"errors\"])",
"_____no_output_____"
]
],
[
[
"### Creating a standard reusable JSON header\nThe standard header for all subsequent calls will use this format. It includes the access token.",
"_____no_output_____"
]
],
[
[
"headers = {\n \"authorization\": f\"{token}\",\n \"content-type\": \"application/json\"\n}",
"_____no_output_____"
]
],
[
[
"## Executing an SQL Statement\nBefore you try creating your own customer service endpoint, you can try using some of the built in services. These let you submit SQL statements in a variety of ways. \n\nExecuting SQL requires a different service endpoint. In this case we will use \"/services/execsql\"",
"_____no_output_____"
]
],
[
[
"API_execsql = \"/v1/services/execsql\"",
"_____no_output_____"
]
],
[
[
"In this example the code requests that the RESTful function waits until the command is complete.",
"_____no_output_____"
]
],
[
[
"sql = \\\n\"\"\"\nSELECT AC.\"TAIL_NUMBER\", AC.\"MANUFACTURER\", AC.\"MODEL\", OT.\"FLIGHTDATE\", OT.\"UNIQUECARRIER\", OT.\"AIRLINEID\", OT.\"CARRIER\", OT.\"TAILNUM\", OT.\"FLIGHTNUM\", OT.\"ORIGINAIRPORTID\", OT.\"ORIGINAIRPORTSEQID\", OT.\"ORIGINCITYNAME\", OT.\"ORIGINSTATE\", OT.\"DESTAIRPORTID\", OT.\"DESTCITYNAME\", OT.\"DESTSTATE\", OT.\"DEPTIME\", OT.\"DEPDELAY\", OT.\"TAXIOUT\", OT.\"WHEELSOFF\", OT.\"WHEELSON\", OT.\"TAXIIN\", OT.\"ARRTIME\", OT.\"ARRDELAY\", OT.\"ARRDELAYMINUTES\", OT.\"CANCELLED\", OT.\"AIRTIME\", OT.\"DISTANCE\"\n FROM \"ONTIME\".\"ONTIME\" OT, \"ONTIME\".\"AIRCRAFT\" AC \n WHERE AC.\"TAIL_NUMBER\" = OT.TAILNUM\n AND ORIGINSTATE = 'NJ'\n AND DESTSTATE = 'CA'\n AND AC.MANUFACTURER = 'Boeing' \n AND AC.MODEL LIKE 'B737%'\n AND OT.TAXIOUT > 30\n AND OT.DISTANCE > 2000\n AND OT.DEPDELAY > 300\n ORDER BY OT.ARRDELAY;\n\"\"\"",
"_____no_output_____"
],
[
"body = {\n \"isQuery\": True,\n \"sqlStatement\": sql,\n \"sync\": True\n}\nprint(body)",
"_____no_output_____"
],
[
"def runStatement(sql, isQuery) :\n body = {\n \"isQuery\": isQuery,\n \"sqlStatement\": sql,\n \"sync\": True\n }\n try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_execsql), verify=False, headers=headers, json=body)\n return response\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"response = runStatement(sql, True)",
"_____no_output_____"
]
],
[
[
"If the successful call returns a **200** response code.",
"_____no_output_____"
]
],
[
[
"print(response)",
"_____no_output_____"
]
],
[
[
"Now that you know the call is a success, you can retrieve the json in the result set.",
"_____no_output_____"
]
],
[
[
"print(response.json()[\"resultSet\"])",
"_____no_output_____"
]
],
[
[
"To format the results, use a Pandas Dataframe class to convert the json result set into a table. Dataframes can be used to further manipulate results in Python.",
"_____no_output_____"
]
],
[
[
"display(pd.DataFrame(response.json()['resultSet']))",
"_____no_output_____"
]
],
[
[
"## Use Parameters in a SQL Statement\nSimple parameter passing is also available through the execsql service. In this case we are passing the employee number into the query to retrieve the full employee record. Try substituting different employee numbers and run the REST call again. For example, you can change \"000010\" to \"000020\", or \"000030\".",
"_____no_output_____"
]
],
[
[
"sqlparm = \\\n\"\"\"\nSELECT AC.\"TAIL_NUMBER\", AC.\"MANUFACTURER\", AC.\"MODEL\", OT.\"FLIGHTDATE\", OT.\"UNIQUECARRIER\", OT.\"AIRLINEID\", OT.\"CARRIER\", OT.\"TAILNUM\", OT.\"FLIGHTNUM\", OT.\"ORIGINAIRPORTID\", OT.\"ORIGINAIRPORTSEQID\", OT.\"ORIGINCITYNAME\", OT.\"ORIGINSTATE\", OT.\"DESTAIRPORTID\", OT.\"DESTCITYNAME\", OT.\"DESTSTATE\", OT.\"DEPTIME\", OT.\"DEPDELAY\", OT.\"TAXIOUT\", OT.\"WHEELSOFF\", OT.\"WHEELSON\", OT.\"TAXIIN\", OT.\"ARRTIME\", OT.\"ARRDELAY\", OT.\"ARRDELAYMINUTES\", OT.\"CANCELLED\", OT.\"AIRTIME\", OT.\"DISTANCE\"\n FROM \"ONTIME\".\"ONTIME\" OT, \"ONTIME\".\"AIRCRAFT\" AC \n WHERE AC.\"TAIL_NUMBER\" = OT.TAILNUM\n AND ORIGINSTATE = 'NJ'\n AND DESTSTATE = 'CA'\n AND AC.MANUFACTURER = 'Boeing' \n AND AC.MODEL LIKE 'B737%'\n AND OT.TAXIOUT > 30\n AND OT.DISTANCE > 2000\n AND OT.DEPDELAY > ?\n ORDER BY OT.ARRDELAY;\n\"\"\"\n\nbody = {\n \"isQuery\": True,\n \"parameters\" : {\n \"1\" : 300\n },\n \"sqlStatement\": sqlparm,\n \"sync\": True\n}",
"_____no_output_____"
],
[
"try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_execsql), verify=False, headers=headers, json=body)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"print(response)",
"_____no_output_____"
],
[
"response.json()[\"resultSet\"]",
"_____no_output_____"
],
[
"display(pd.DataFrame(response.json()['resultSet']))",
"_____no_output_____"
]
],
[
[
"## Generate a Call and don't wait for the results\nIf you know that your statement will take a long time to return a result, you can check back later. Turn **sync** off to avoid waiting.",
"_____no_output_____"
]
],
[
[
"sql = \\\n\"\"\"\nSELECT AC.\"TAIL_NUMBER\", AC.\"MANUFACTURER\", AC.\"MODEL\", OT.\"FLIGHTDATE\", OT.\"UNIQUECARRIER\", OT.\"AIRLINEID\", OT.\"CARRIER\", OT.\"TAILNUM\", OT.\"FLIGHTNUM\", OT.\"ORIGINAIRPORTID\", OT.\"ORIGINAIRPORTSEQID\", OT.\"ORIGINCITYNAME\", OT.\"ORIGINSTATE\", OT.\"DESTAIRPORTID\", OT.\"DESTCITYNAME\", OT.\"DESTSTATE\", OT.\"DEPTIME\", OT.\"DEPDELAY\", OT.\"TAXIOUT\", OT.\"WHEELSOFF\", OT.\"WHEELSON\", OT.\"TAXIIN\", OT.\"ARRTIME\", OT.\"ARRDELAY\", OT.\"ARRDELAYMINUTES\", OT.\"CANCELLED\", OT.\"AIRTIME\", OT.\"DISTANCE\"\n FROM \"ONTIME\".\"ONTIME\" OT, \"ONTIME\".\"AIRCRAFT\" AC \n WHERE AC.\"TAIL_NUMBER\" = OT.TAILNUM\n AND ORIGINSTATE = 'NJ'\n AND DESTSTATE = 'CA'\n AND AC.MANUFACTURER = 'Boeing' \n AND AC.MODEL LIKE 'B737%'\n AND OT.TAXIOUT > 30\n AND OT.DISTANCE > 2000\n AND OT.DEPDELAY > 300\n ORDER BY OT.ARRDELAY;\n\"\"\"\n\nbody = {\n \"isQuery\": True,\n \"sqlStatement\": sql,\n \"sync\": False\n}",
"_____no_output_____"
],
[
"try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_execsql), verify=False, headers=headers, json=body)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"print(response)",
"_____no_output_____"
]
],
[
[
"Retrieve the job id, so that you can retrieve the results later.",
"_____no_output_____"
]
],
[
[
"job_id = response.json()[\"id\"]",
"_____no_output_____"
],
[
"print(job_id)",
"_____no_output_____"
]
],
[
[
"## Retrieve Result set using Job ID\nThe service API needs to be appended with the Job ID.",
"_____no_output_____"
]
],
[
[
"API_get = \"/v1/services/\"",
"_____no_output_____"
]
],
[
[
"We can limit the number of rows that we return at a time. Setting the limit to zero means all of the rows are to be returned.",
"_____no_output_____"
]
],
[
[
"body = {\n \"limit\": 0\n}",
"_____no_output_____"
]
],
[
[
"Get the results.",
"_____no_output_____"
]
],
[
[
"try:\n response = requests.get(\"{}{}{}\".format(Db2RESTful,API_get,job_id), verify=False, headers=headers, json=body)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"print(response)",
"_____no_output_____"
]
],
[
[
"Retrieve the results.",
"_____no_output_____"
]
],
[
[
"display(pd.DataFrame(response.json()['resultSet']))",
"_____no_output_____"
]
],
[
[
"Now that you have some experience with the built in SQL service, you can try creating your own endpoint service. ",
"_____no_output_____"
],
[
"## Using RESTful Endpoint Services\nThe most common way of interacting with the service is to fully encapsulate an SQL statement, including any parameters, in a unique RESTful service. This creates a secure separation between the database service and the RESTful programming service. It also allows you to create versions of the same service to make maintenance and evolution of programming models simple and predictable.",
"_____no_output_____"
],
[
"### Setup the Meta Data Tables and Stored Procedures to manage Endpoint Services\nBefore you can start defining and running your own RESTful Endpoint services you need call the service to create the table and stored procedures in the database you are using. ",
"_____no_output_____"
]
],
[
[
"API_makerest = \"/v1/metadata/setup\"",
"_____no_output_____"
]
],
[
[
"You can specify the schema that the new table and stored procedures will be created in. In this example we will use **DB2REST**",
"_____no_output_____"
]
],
[
[
"body = {\n \"schema\": \"DB2REST\"\n}",
"_____no_output_____"
],
[
"try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_makerest), verify=False, headers=headers, json=body)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
]
],
[
[
"If the process is successful the service returns a 201 status code.",
"_____no_output_____"
]
],
[
[
"if (response.status_code == 201):\n print(response.reason)\nelse:\n print(response.json())",
"_____no_output_____"
]
],
[
[
"### Create a RESTful Service\nNow that the RESTful Service metadata is created in your database, you can create your first service. In this example you will pass an employee numb er, a 6 character string, to the service. It will return the department number of the employee.",
"_____no_output_____"
]
],
[
[
"API_makerest = \"/v1/services\"",
"_____no_output_____"
]
],
[
[
"The first step is to define the SQL that we want in the RESTful call. Parameters are identified using an ampersand \"@\". Notice that our SQL is nicely formatted to make this notebook easier to ready. However when creating a service it is good practice to remove the line break characters from your SQL statement. ",
"_____no_output_____"
]
],
[
[
"sql = \\\n\"\"\"\nSELECT COUNT(AC.\"TAIL_NUMBER\") FROM \"ONTIME\".\"ONTIME\" OT, \"ONTIME\".\"AIRCRAFT\" AC \n WHERE AC.\"TAIL_NUMBER\" = OT.TAILNUM\n AND ORIGINSTATE = @STATE\n AND DESTSTATE = 'CA'\n AND AC.MANUFACTURER = 'Boeing' \n AND AC.MODEL LIKE 'B737%'\n AND OT.TAXIOUT > 30\n AND OT.DISTANCE > 2000\n AND OT.DEPDELAY > @DELAY\n FETCH FIRST 5 ROWS ONLY\n\"\"\"\nsql = sql.replace(\"\\n\",\"\")",
"_____no_output_____"
]
],
[
[
"The next step is defining the json body to send along with the REST call.",
"_____no_output_____"
]
],
[
[
"body = {\"isQuery\": True,\n \"parameters\": [\n {\n \"datatype\": \"CHAR(2)\",\n \"name\": \"@STATE\"\n },\n {\n \"datatype\": \"INT\",\n \"name\": \"@DELAY\" \n }\n ],\n \"schema\": \"DEMO\",\n \"serviceDescription\": \"Delay\",\n \"serviceName\": \"delay\",\n \"sqlStatement\": sql,\n \"version\": \"1.0\"\n}",
"_____no_output_____"
]
],
[
[
"Now submit the full RESTful call to create the new service. ",
"_____no_output_____"
]
],
[
[
"try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_makerest), verify=False, headers=headers, json=body)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"print(response)",
"_____no_output_____"
]
],
[
[
"### Call the new RESTful Service\nNow you can call the RESTful service. In this case we will pass the stock symbol CAT. But like in the previous example you can try rerunning the service call with different stock symbols.",
"_____no_output_____"
]
],
[
[
"API_runrest = \"/v1/services/delay/1.0\"",
"_____no_output_____"
],
[
"body = {\n \"parameters\": {\n \"@STATE\": \"NY\",\"@DELAY\":\"300\"\n },\n \"sync\": True\n}",
"_____no_output_____"
],
[
"try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_runrest), verify=False, headers=headers, json=body)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"print(\"{}{}\".format(Db2RESTful,API_runrest))",
"_____no_output_____"
],
[
"print(response)",
"_____no_output_____"
],
[
"print(response.json())",
"_____no_output_____"
]
],
[
[
"You can retrieve the result set, convert it into a Dataframe and display the table.",
"_____no_output_____"
]
],
[
[
"display(pd.DataFrame(response.json()['resultSet']))",
"_____no_output_____"
]
],
[
[
"## Loop through the new call\nNow you can call the RESTful service with different values.",
"_____no_output_____"
]
],
[
[
"API_runrest = \"/v1/services/delay/1.0\"",
"_____no_output_____"
],
[
"repeat = 2\nfor x in range(0, repeat):\n for state in (\"OH\", \"NJ\", \"NY\", \"FL\", \"MI\"):\n\n body = {\n \"parameters\": {\n \"@STATE\": state,\"@DELAY\": \"240\"\n },\n \"sync\": True\n }\n try:\n response = requests.post(\"{}{}\".format(Db2RESTful,API_runrest), verify=False, headers=headers, json=body)\n print(state + \": \" + str(response.json()['resultSet']))\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
]
],
[
[
"## Managing Your Services \nThere are several service calls you can use to help manage the Db2 RESTful Endpoint service. ",
"_____no_output_____"
],
[
"## List Available Services\nYou can also list all the user defined services you have access to",
"_____no_output_____"
]
],
[
[
"API_listrest = \"/v1/services\"",
"_____no_output_____"
],
[
"try:\n response = requests.get(\"{}{}\".format(Db2RESTful,API_listrest), verify=False, headers=headers)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"print(response.json())",
"_____no_output_____"
],
[
"display(pd.DataFrame(response.json()['Db2Services']))",
"_____no_output_____"
]
],
[
[
"## Get Service Details\nYou can also get the details of a service",
"_____no_output_____"
]
],
[
[
"API_getDetails = \"/v1/services/delay/3.0\"",
"_____no_output_____"
],
[
"try:\n response = requests.get(\"{}{}\".format(Db2RESTful,API_getDetails), verify=False, headers=headers)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"json = response.json()\nprint(json)",
"_____no_output_____"
]
],
[
[
"You can format the result to make it easier to ready. For example, here are the input and outputs.",
"_____no_output_____"
]
],
[
[
"display(pd.DataFrame(json['inputParameters']))\ndisplay(pd.DataFrame(json['resultSetFields']))",
"_____no_output_____"
]
],
[
[
"## Delete a Service\nA single call is also available to delete a service",
"_____no_output_____"
]
],
[
[
"API_deleteService = \"/v1/services\"\nService = \"/delay\"\nVersion = \"/1.0\"",
"_____no_output_____"
],
[
"try:\n response = requests.delete(\"{}{}{}{}\".format(Db2RESTful,API_deleteService,Service,Version), verify=False, headers=headers)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"print (response)",
"_____no_output_____"
]
],
[
[
"## Get Service Logs\nYou can also easily download the Db2 RESTful Endpoint service logs. ",
"_____no_output_____"
]
],
[
[
"API_listrest = \"/v1/logs\"",
"_____no_output_____"
],
[
"try:\n response = requests.get(\"{}{}\".format(Db2RESTful,API_listrest), verify=False, headers=headers)\nexcept Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))",
"_____no_output_____"
],
[
"if (response.status_code == 200):\n myFile = response.content\n open('/tmp/logs.zip', 'wb').write(myFile)\n print(\"Downloaded\",len(myFile),\"bytes.\")\nelse:\n print(response.json())",
"_____no_output_____"
]
],
[
[
"To see the content of the logs, open the Files browser on machine host3 (10.0.0.4). Navigate to the **/tmp** directory and unzip the logs file. ",
"_____no_output_____"
],
[
"## Using the Db2 REST Class\nFor your convenience, everything in the lessons above has been included into a Db2REST Python Class. You can add or use this code as part of your own Jupyter notebooks to make working with the Db2 RESTful Endpoint service quick and easy. \n\nThere are also lots of examples in the following lesson on how to use the class.",
"_____no_output_____"
]
],
[
[
"# Run the Db2REST Class library\n# Used to construct and reuse an Autentication Key\n# Used to construct RESTAPI URLs and JSON payloads\nimport json\nimport requests\nimport pandas as pd\n\nclass Db2REST():\n \n def __init__(self, RESTServiceURL):\n self.headers = {\"content-type\": \"application/json\"}\n self.RESTServiceURL = RESTServiceURL\n self.version = \"/v1\"\n self.API_auth = self.version + \"/auth\"\n self.API_makerest = self.version + \"/metadata/setup\"\n self.API_services = self.version + \"/services/\" \n self.API_version = self.version + \"/version/\" \n self.API_execsql = self.API_services + \"execsql\"\n self.API_monitor = self.API_services + \"monitor\" \n \n self.Verify = False\n \n import urllib3\n urllib3.disable_warnings()\n \n def connectDatabase(self, dbHost, dbName, dbPort, isSSLConnection, dbUsername, dbPassword, expiryTime=\"300m\"):\n self.dbHost = dbHost\n self.dbName = dbName\n self.dbPort = dbPort\n self.isSSLConnection = isSSLConnection\n self.dbusername = dbUsername\n self.dbpassword = dbPassword \n self.connectionBody = {\n \"dbParms\": {\n \"dbHost\": dbHost,\n \"dbName\": dbName,\n \"dbPort\": dbPort,\n \"isSSLConnection\": isSSLConnection,\n \"username\": dbUsername,\n \"password\": dbPassword\n },\n \"expiryTime\": expiryTime\n }\n try:\n response = requests.post(\"{}{}\".format(self.RESTServiceURL,self.API_auth), verify=self.Verify, headers=self.headers, json=self.connectionBody)\n print (response)\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n if (response.status_code == 200):\n self.token = response.json()[\"token\"]\n print(\"Successfully connected and retrieved access token\")\n else:\n print(response)\n print(response.json())\n print(response.json()[\"errors\"])\n \n self.headers = {\n \"authorization\": f\"{self.token}\",\n \"content-type\": \"application/json\"\n }\n \n def getConnection(self):\n return self.connectionBody\n \n def getService(self):\n return self.RESTServiceURL\n \n def getToken(self):\n return(\"Token: {}\".format(self.token))\n \n def getVersion(self):\n try:\n print(\"{}{}\".format(self.RESTServiceURL,self.API_version))\n response = requests.get(\"{}{}\".format(self.RESTServiceURL,self.API_version),verify=self.Verify)\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n if (response.status_code == 200):\n return response.json()['version']\n else:\n print(response)\n print(response.json()['errors'][0]['more_info']) \n \n def runStatement(self, sql, isQuery=True, sync=True, parameters={}):\n body = {\n \"isQuery\": isQuery,\n \"sqlStatement\": sql,\n \"sync\": sync,\n \"parameters\": parameters\n }\n \n try:\n response = requests.post(\"{}{}\".format(self.RESTServiceURL,self.API_execsql), verify=self.Verify, headers=self.headers, json=body)\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n if (response.status_code == 200):\n return pd.DataFrame(response.json()['resultSet'])\n elif (response.status_code == 202):\n return response.json()[\"id\"]\n else:\n print(response.json()['errors'][0]['more_info'])\n \n def getResult(self, job_id, limit=0):\n body = {\"limit\": limit}\n \n try:\n response = requests.get(\"{}{}{}\".format(self.RESTServiceURL,self.API_services,job_id), verify=self.Verify, headers=self.headers, json=body)\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n if (response.status_code == 200):\n json = response.json() \n if (json['jobStatus'] == 2):\n return json['jobStatusDescription']\n elif (json['jobStatus'] == 3):\n return pd.DataFrame(json['resultSet']) \n elif (json['jobStatus'] == 4):\n return pd.DataFrame(json['resultSet']) \n else: \n return json\n elif (response.status_code == 404):\n print(response.json()['errors']) \n elif (response.status_code == 500):\n print(response.json()['errors'][0]['more_info']) \n else:\n print(response.json())\n \n def createServiceMetadata(self, serviceSchema=\"Db2REST\"):\n self.serviceSchema = serviceSchema\n body = {\"schema\": self.serviceSchema}\n try:\n response = requests.post(\"{}{}\".format(self.RESTServiceURL,self.API_makerest), verify=self.Verify, headers=self.headers, json=body)\n if (response.status_code == 201):\n print(response.reason)\n else:\n print(response.json())\n \n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n\n def listServices(self):\n try:\n response = requests.get(\"{}{}\".format(self.RESTServiceURL,self.API_services), verify=self.Verify, headers=self.headers)\n return pd.DataFrame(response.json()['Db2Services'])\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n def getServiceDetails(self, serviceName, version):\n try:\n response = requests.get(\"{}{}{}{}\".format(self.RESTServiceURL,self.API_services,\"/\" + serviceName,\"/\" + version), verify=self.Verify, headers=self.headers)\n print(response.status_code)\n if (response.status_code == 200):\n description = response.json()\n print(\"Input parameters:\")\n print(description[\"inputParameters\"])\n print(\"Result format:\")\n print(description[\"resultSetFields\"])\n else:\n print(response.json()) \n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e))) \n \n def createService(self, schema, serviceDescription, serviceName, sql, version, parameters=False, isQuery=True):\n if (parameters==False):\n body = {\"isQuery\": isQuery,\n \"schema\": schema,\n \"serviceDescription\": serviceDescription,\n \"serviceName\": serviceName,\n \"sqlStatement\": sql.replace(\"\\n\",\"\"),\n \"version\": version\n } \n else: \n body = {\"isQuery\": isQuery,\n \"schema\": schema,\n \"serviceDescription\": serviceDescription,\n \"serviceName\": serviceName,\n \"sqlStatement\": sql.replace(\"\\n\",\"\"),\n \"version\": version,\n \"parameters\": parameters\n } \n \n try:\n response = requests.post(\"{}{}\".format(self.RESTServiceURL,self.API_services), verify=self.Verify, headers=self.headers, json=body)\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n if (response.status_code == 201):\n print(\"Service: \" + serviceName + \" Version: \" + version + \" created\")\n else:\n print(response.json()) \n \n def deleteService(self, serviceName, version):\n try:\n response = requests.delete(\"{}{}{}{}\".format(self.RESTServiceURL,self.API_services,\"/\" + serviceName,\"/\" + version), verify=self.Verify, headers=self.headers)\n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n if (response.status_code == 204):\n print(\"Service: \" + serviceName + \" Version: \" + version + \" deleted\")\n else:\n print(response.json()) \n \n def callService(self, serviceName, version, parameters, sync=True):\n body = {\n \"parameters\": parameters,\n \"sync\": sync\n }\n try:\n response = requests.post(\"{}{}{}{}\".format(self.RESTServiceURL,self.API_services,\"/\" + serviceName,\"/\" + version), verify=self.Verify, headers=self.headers, json=body)\n if (response.status_code == 200):\n return pd.DataFrame(response.json()['resultSet'])\n elif (response.status_code == 202):\n return response.json()[\"id\"]\n else:\n print(response.json()['errors'][0]['more_info'])\n \n except Exception as e:\n if (repr(e) == \"KeyError('more_info',)\"): \n print(\"Service not found\")\n else: \n print(\"Unable to call RESTful service. Error={}\".format(repr(e)))\n \n def monitorJobs(self):\n try:\n response = requests.get(\"{}{}\".format(self.RESTServiceURL,self.API_monitor), verify=self.Verify, headers=self.headers)\n if (response.status_code == 200):\n return pd.DataFrame(response.json()['MonitorServices'])\n else:\n print(response.json()) \n except Exception as e:\n print(\"Unable to call RESTful service. Error={}\".format(repr(e))) ",
"_____no_output_____"
]
],
[
[
"### Setting up a Db2 RESTful Endpoint Service Class instance\nTo use the class first create an instance of the class. The cell below creates an object called **Db2RESTService** from the **Db2REST** class. The first call to the object is **getVersion** to confirm the version of the RESTful Endpoint Service you are connected to.",
"_____no_output_____"
],
[
"#### Connecting to the service to the database\nUnless your service is already bound to a single database, the call below connects it to a single Db2 database. You can run this command again to connect to a different database from the same RESTful Endpoint service. ",
"_____no_output_____"
]
],
[
[
"Db2RESTService = Db2REST(\"https://10.0.0.201:31315\")",
"_____no_output_____"
],
[
"print(\"Db2 RESTful Endpoint Service Version: \" + Db2RESTService.getVersion())",
"_____no_output_____"
]
],
[
[
"#### Connect to Db2 OLTP",
"_____no_output_____"
]
],
[
[
"Db2RESTService.connectDatabase(\"10.0.0.201\", \"STOCKS\", 32443, False, \"admin\", \"CP4DDataFabric\")",
"_____no_output_____"
]
],
[
[
"#### Connect to DV",
"_____no_output_____"
]
],
[
[
"Db2RESTService.connectDatabase(\"10.0.0.201\", \"BIGSQL\", 31193, False, \"admin\", \"CP4DDataFabric\")",
"_____no_output_____"
]
],
[
[
"#### Confirming the service settings\nOnce the connection to the RESTful Endpoint Service and Db2 is established you can always check your settings using the following calls.",
"_____no_output_____"
]
],
[
[
"print(Db2RESTService.getService())",
"_____no_output_____"
],
[
"print(Db2RESTService.getConnection())",
"_____no_output_____"
],
[
"print(Db2RESTService.getToken())",
"_____no_output_____"
]
],
[
[
"### Running SQL Through the Service\nYou can run an SQL Statement through the RESTful service as a simple text string.\n\nLet's start by defining the SQL to run:",
"_____no_output_____"
]
],
[
[
"sql = \\\n\"\"\"\nSELECT AC.\"TAIL_NUMBER\", AC.\"MANUFACTURER\", AC.\"MODEL\", OT.\"FLIGHTDATE\", OT.\"UNIQUECARRIER\", OT.\"AIRLINEID\", OT.\"CARRIER\", OT.\"TAILNUM\", OT.\"FLIGHTNUM\", OT.\"ORIGINAIRPORTID\", OT.\"ORIGINAIRPORTSEQID\", OT.\"ORIGINCITYNAME\", OT.\"ORIGINSTATE\", OT.\"DESTAIRPORTID\", OT.\"DESTCITYNAME\", OT.\"DESTSTATE\", OT.\"DEPTIME\", OT.\"DEPDELAY\", OT.\"TAXIOUT\", OT.\"WHEELSOFF\", OT.\"WHEELSON\", OT.\"TAXIIN\", OT.\"ARRTIME\", OT.\"ARRDELAY\", OT.\"ARRDELAYMINUTES\", OT.\"CANCELLED\", OT.\"AIRTIME\", OT.\"DISTANCE\"\n FROM \"ONTIME\".\"ONTIME\" OT, \"ONTIME\".\"AIRCRAFT\" AC \n WHERE AC.\"TAIL_NUMBER\" = OT.TAILNUM\n AND ORIGINSTATE = 'NJ'\n AND DESTSTATE = 'CA'\n AND AC.MANUFACTURER = 'Boeing' \n AND AC.MODEL LIKE 'B737%'\n AND OT.TAXIOUT > 30\n AND OT.DISTANCE > 2000\n AND OT.DEPDELAY > 300\n ORDER BY OT.DEPDELAY DESC\n FETCH FIRST 5 ROWS ONLY;\n\"\"\"",
"_____no_output_____"
]
],
[
[
"Now a single call to the **runStatement** routine runs the SQL synchronously and returns the result as a DataFrame",
"_____no_output_____"
]
],
[
[
"sql = \"SELECT * FROM SYSCAT.TABLES\"\nresult = (Db2RESTService.runStatement(sql))\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"You can also run the statement asynchronously so you don't have to wait for the result. In this case the result is the statement identifier that you can use to check the statement status.",
"_____no_output_____"
]
],
[
[
"statementID = (Db2RESTService.runStatement(sql, sync=False))\ndisplay(statementID)",
"_____no_output_____"
]
],
[
[
"If you have several statements running at the same time you can check to see their status with the **monitorStatus** routine and see where they are in the service queue. ",
"_____no_output_____"
]
],
[
[
"services = Db2RESTService.monitorJobs()\ndisplay(services)",
"_____no_output_____"
]
],
[
[
"You can try to get the results of the statment by passing the statement identifier into the getResults routine. If the statement has finished running it will return a result set as a DataFrame. It is still running, a message is returned.",
"_____no_output_____"
]
],
[
[
"result = (Db2RESTService.getResult(statementID))\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"#### Passing Parameters when running SQL Statements\nYou can also define a single SQL statement with ? parameters and call that statement with different values using the same **runStatement** routine. ",
"_____no_output_____"
]
],
[
[
"sqlparm = \\\n\"\"\"\nSELECT AC.\"TAIL_NUMBER\", AC.\"MANUFACTURER\", AC.\"MODEL\", OT.\"FLIGHTDATE\", OT.\"UNIQUECARRIER\", OT.\"AIRLINEID\", OT.\"CARRIER\", OT.\"TAILNUM\", OT.\"FLIGHTNUM\", OT.\"ORIGINAIRPORTID\", OT.\"ORIGINAIRPORTSEQID\", OT.\"ORIGINCITYNAME\", OT.\"ORIGINSTATE\", OT.\"DESTAIRPORTID\", OT.\"DESTCITYNAME\", OT.\"DESTSTATE\", OT.\"DEPTIME\", OT.\"DEPDELAY\", OT.\"TAXIOUT\", OT.\"WHEELSOFF\", OT.\"WHEELSON\", OT.\"TAXIIN\", OT.\"ARRTIME\", OT.\"ARRDELAY\", OT.\"ARRDELAYMINUTES\", OT.\"CANCELLED\", OT.\"AIRTIME\", OT.\"DISTANCE\"\n FROM \"ONTIME\".\"ONTIME\" OT, \"ONTIME\".\"AIRCRAFT\" AC \n WHERE AC.\"TAIL_NUMBER\" = OT.TAILNUM\n AND ORIGINSTATE = ?\n AND DESTSTATE = ?\n AND AC.MANUFACTURER = 'Boeing' \n AND AC.MODEL LIKE 'B737%'\n AND OT.TAXIOUT > 30\n AND OT.DISTANCE > 2000\n AND OT.DEPDELAY > ?\n ORDER BY OT.DEPDELAY DESC\n FETCH FIRST 10 ROWS ONLY;\n\"\"\"",
"_____no_output_____"
],
[
"result = Db2RESTService.runStatement(sqlparm,parameters={\"1\": 'NY', \"2\": 'CA', \"3\" : 300})\ndisplay(result)",
"_____no_output_____"
],
[
"result = Db2RESTService.runStatement(sqlparm,parameters={\"1\": 'NJ', \"2\": 'CA', \"3\" : 200})\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"#### Limiting Results\nYou also have full control of how many rows in an answer set to return. Run the following statement using **sync=False**",
"_____no_output_____"
]
],
[
[
"statementID = Db2RESTService.runStatement(sqlparm, sync=False, parameters={\"1\": 'NJ', \"2\": 'CA', \"3\" : 200})\ndisplay(statementID)\nresult = (Db2RESTService.getResult(statementID))\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"This time the **getResult** routine include a parameter to limit the result set to 5 rows. ",
"_____no_output_____"
]
],
[
[
"result = (Db2RESTService.getResult(statementID, limit=5))\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"The next cell retrieves the remaining rows.",
"_____no_output_____"
]
],
[
[
"result = (Db2RESTService.getResult(statementID))\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"After all the rows have been returned the job history is removed. If you try to retrieve the results for this statement now the service won't find it.",
"_____no_output_____"
]
],
[
[
"result = (Db2RESTService.getResult(statementID))\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"### Creating and Running Endpoint Services\nIf the MetaData tables have not already been created in your database you can use the following call to create the MetaData in the schema of your choice. In this case **DB2REST**.",
"_____no_output_____"
]
],
[
[
"Db2RESTService.createServiceMetadata(\"DB2REST\")",
"_____no_output_____"
]
],
[
[
"Let's start by defining the SQL statement. It can include parameters that have to be idenfied with an amersand \"@\".",
"_____no_output_____"
]
],
[
[
"sql = \\\n\"\"\"\nSELECT COUNT(AC.\"TAIL_NUMBER\") FROM \"ONTIME\".\"ONTIME\" OT, \"ONTIME\".\"AIRCRAFT\" AC \n WHERE AC.\"TAIL_NUMBER\" = OT.TAILNUM\n AND ORIGINSTATE = @STATE\n AND DESTSTATE = 'CA'\n AND AC.MANUFACTURER = 'Boeing' \n AND AC.MODEL LIKE 'B737%'\n AND OT.TAXIOUT > 30\n AND OT.DISTANCE > 2000\n AND OT.DEPDELAY > @DELAY\n FETCH FIRST 5 ROWS ONLY\n\"\"\"",
"_____no_output_____"
]
],
[
[
"Now we can create the service, including the two parameters, using the **createService** routine. ",
"_____no_output_____"
]
],
[
[
"parameters = [{\"datatype\": \"CHAR(2)\",\"name\": \"@STATE\"},{\"datatype\": \"INT\",\"name\": \"@DELAY\"}]\nschema = 'DEMO'\nserviceDescription = 'Delay'\nserviceName = 'delay'\nversion = '1.0'\n\nDb2RESTService.createService(schema, serviceDescription, serviceName, sql, version, parameters)",
"_____no_output_____"
]
],
[
[
"A call to the **listServices** routine confirms that you have created the new service. ",
"_____no_output_____"
]
],
[
[
"services = Db2RESTService.listServices()\ndisplay(services)",
"_____no_output_____"
]
],
[
[
"You can also see the details for any service using the **getServiceDetails** routine.",
"_____no_output_____"
]
],
[
[
"details = Db2RESTService.getServiceDetails(\"delay\",\"1.0\")\ndisplay(details)",
"_____no_output_____"
]
],
[
[
"You can all the new service using the **callService** routine. The parameters are passed into call using an array of values. By default the call is synchronous so you have to wait for the results. ",
"_____no_output_____"
]
],
[
[
"serviceName = 'delay'\nversion = '1.0'\nparameters = {\"@STATE\": \"NJ\",\"@DELAY\":\"200\"}\nresult = Db2RESTService.callService(serviceName, version, parameters)\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"You can also call the service asychronously, just like we did with SQL statements earlier. Notice the additional parameter **sync=False**. Since the cell below immediately checks the status of the job you can see it has been queued. ",
"_____no_output_____"
]
],
[
[
"serviceName = 'delay'\nversion = '1.0'\nparameters = {\"@STATE\": \"NJ\",\"@DELAY\":\"200\"}\nstatementID = Db2RESTService.callService(serviceName, version, parameters, sync=False)\ndisplay(statementID) \ndisplay(Db2RESTService.monitorJobs())",
"_____no_output_____"
]
],
[
[
"Run **monitorJobs** again to confirm that the endpoint service has completed the request.",
"_____no_output_____"
]
],
[
[
"services = Db2RESTService.monitorJobs()\ndisplay(services)",
"_____no_output_____"
]
],
[
[
"And retrieve the result set.",
"_____no_output_____"
]
],
[
[
"result = (Db2RESTService.getResult(statementID))\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"You can also delete an existing endpoint service with a call to the **deleteService** routine.",
"_____no_output_____"
]
],
[
[
"serviceName = 'delay'\nversion = '1.0'\nDb2RESTService.deleteService(serviceName, version)",
"_____no_output_____"
]
],
[
[
"#### Using a service to query the Catalog\nYou can also think about creating services to explore the database catalog. For example, here is a service that accepts a schema as an input parameter and returns a list of tables in the schema. ",
"_____no_output_____"
]
],
[
[
"sql = \\\n\"\"\"\nSELECT TABSCHEMA, TABNAME, ALTER_TIME FROM SYSCAT.TABLES WHERE TABSCHEMA = @SCHEMA\n\"\"\"\n\nparameters = [{\"datatype\": \"VARCHAR(64)\",\"name\": \"@SCHEMA\"}]\nschema = 'DEMO'\nserviceDescription = 'Tables'\nserviceName = 'tables'\nversion = '1.0'\n\nDb2RESTService.createService(schema, serviceDescription, serviceName, sql, version, parameters)",
"_____no_output_____"
],
[
"serviceName = 'tables'\nversion = '1.0'\nresult = Db2RESTService.callService(serviceName, version, parameters = {\"@SCHEMA\": \"SYSCAT\"}, sync=True)\ndisplay(result)",
"_____no_output_____"
]
],
[
[
"### Incorporating the Db2 RESTFul Endpoint Class into your Python scipts\nThe Db2 RESTful Endpoint Class is available on GIT at https://github.com/Db2-DTE-POC/CPDDVHOL4/blob/main/RESTfulEndpointServiceClass402.ipynb. You can download a copy into your own Python library and add **%run db2restendpoint.ipynb** to your own Python notebook. You can also include the following two lines which will automatically download a copy of the library from GIT and run the Class code. ",
"_____no_output_____"
]
],
[
[
"!wget -O db2restendpoint.ipynb https://raw.githubusercontent.com/Db2-DTE-POC/CPDDVHOL4/main/RESTfulEndpointServiceClass402.ipynb\n%run db2restendpoint.ipynb ",
"_____no_output_____"
],
[
"Db2RESTService = Db2REST(\"https://10.0.0.201:31315\")",
"_____no_output_____"
],
[
"print(\"Db2 RESTful Endpoint Service Version: \" + Db2RESTService.getVersion())",
"_____no_output_____"
]
],
[
[
"## What's Next\nTry experimenting. Create your own services. You can find out more at: https://www.ibm.com/support/producthub/db2/docs/content/SSEPGG_11.5.0/com.ibm.db2.luw.admin.rest.doc/doc/c_rest.html.\n\nAlso check out the OpenAPI specification for the service. It includes coding examples in Python, CURL and JavaScript. \n\nIf you are running this notebook from a browser running inside the Cloud Pak for Data cluster, click: http://10.0.0.4:50050/docs If you are running this from a browser from your own desktop, check your welcome note for the address of the Db2 RESTful Service at port 50050 and add **docs** to the end of the URL.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a1396ea37a7bcc8c64940576a48f0046e1df18c
| 213,443 |
ipynb
|
Jupyter Notebook
|
docs/t5.ipynb
|
dumpmemory/transformer-deploy
|
36993d8dd53c7440e49dce36c332fa4cc08cf9fb
|
[
"Apache-2.0"
] | 698 |
2021-11-22T17:42:40.000Z
|
2022-03-31T11:16:08.000Z
|
docs/t5.ipynb
|
dumpmemory/transformer-deploy
|
36993d8dd53c7440e49dce36c332fa4cc08cf9fb
|
[
"Apache-2.0"
] | 38 |
2021-11-23T13:45:04.000Z
|
2022-03-31T10:36:45.000Z
|
docs/t5.ipynb
|
dumpmemory/transformer-deploy
|
36993d8dd53c7440e49dce36c332fa4cc08cf9fb
|
[
"Apache-2.0"
] | 58 |
2021-11-24T11:46:21.000Z
|
2022-03-29T08:45:16.000Z
| 113.775586 | 113,708 | 0.826445 |
[
[
[
"# Inference acceleration of `T5` for large batch size / long sequence length / > large models\n\nEvery week or so, a new impressive few shots learning work taking advantage of autoregressive models is released by some team around the world. \nStill `LLM` inference is rarely discussed and few projects are focusing on this aspect. \nIn this notebook, we describe our take to significantly improve autoregressive model latency.\n\nWe plan to intensively test large autoregressive models, so we want something:\n\n* which **scales**: the improvement exists on small and large models, for short and long sequences, in greedy and beam search;\n * This is very important in a few shots learning where sequences are most of the time hundreds or thousands tokens long and beam search is used to improve text quality.\n* that has **no hidden cost**: no big increase in memory usage, no degradation in quality of generated text, support state-of-the-art decoding algorithms;\n* that is **generic**: works for any transformer based architecture, and not specific to an inference engine;\n* that is **easy to maintain**: no hard-coded behaviors or other technical debt if it doesn't bring a clear advantage.\n\nTo be clear, **we are not targeting the best performance ever but the right trade off** (for us at least) between simplicity to use/maintain and acceptable latency.\n\n## The challenge\n\nIn most situations, performing inference with `Onnx Runtime` or `TensorRT` usually bring large improvement over `Pytorch` implementations.\nIt's very true with `transformer` based models.\n\nThe main reason is that these tools will perform `kernel fusions` (merging several operations into a single one) and therefore reduce the number of memory bounded operations. Sometimes they also replace some operations by a much faster approximation.\nIn the very specific case of autoregressive languages, things are a bit more complicated.\n\nOn most `Pytorch` implementations of these models, there is a `cache` of `K` and `V` values.\nLet's remind us that in attention blocks, each token is projected on 3 matrices called `Query`, `Key`, and `Value`.\nThen, those projections will be used to compute a representation of each token which takes into account the information from the related other tokens of the sequence.\n\nAs autoregressive models generate the sequence one token at a time, they should recompute final representation of all past tokens for each new token to generate.\nBecause each token can only attend to the past, the result of these computations never changes; therefore one simple trick to reduce latency is to just memorize them and reuse them later, avoiding lots of computation.\n\nOut of the box, the cache mechanism can't be exported to `Onnx` from `Hugging Face` models (and all other `Pytorch` implementations we are aware of).\nThe reason is that those models are not `torchscript` scripting compliant (it requires `Pytorch` code to follow some [restrictive rules](https://pytorch.org/docs/stable/jit_builtin_functions.html)).\nBecause of that, `Onnx` export is done through `tracing` which erases any control flow instructions (including the `If` instruction to enable or not a cache).\n\n## Existing solutions\n\nSome interesting solutions targeting inference latency that we have considered and/or tested:\n\n* [TensorRT](https://developer.nvidia.com/blog/optimizing-t5-and-gpt-2-for-real-time-inference-with-`TensorRT`/), which targets `GPU`, heavily optimizes the computation graph, making `T5` inference very fast (they report X10 speedup on `small-T5`). The trick is that it doesn't use any cache (see below for more details), so it's very fast on short sequence and small models, as it avoids many memory bounded operations by redoing full computation again and again... but as several users have already found ([1](https://github.com/NVIDIA/TensorRT/issues/1807), [2](https://github.com/NVIDIA/TensorRT/issues/1642), [3](https://github.com/NVIDIA/TensorRT/issues/1799), [4](https://github.com/NVIDIA/TensorRT/issues/1845), ...), this approach doesn't scale when the computation intensity increases, i.e., when base or large models are used instead of a small one, when generation is done on moderately long sequence of few hundred of tokens or if beam search is used instead of a greedy search;\n* [FastT5](https://github.com/Ki6an/fastT5), which targets `CPU`, exports 2 versions of the decoder, one with cache and one without. You need the `no cache` version to compute the first token and the first `past state` tensors (aka the cached tensors), and for all the other tokens you use the `cache` version of the computation graph. Basically, it makes the memory foot print 2 times bigger as all weights are duplicated. As generative models tend to be huge, they work around the memory issue by using dynamic `int-8` quantization, the final memory foot print of the decoders is now the same as `Hugging Face` in `FP16`... but 1/ dynamic quantization only works on `CPU`, and 2/ according to several reports dynamic quantization degrades significantly generative model output, to a point where it may make them useless ([1](https://github.com/huggingface/transformers/issues/2466#issuecomment-572781378), [2](https://github.com/huggingface/transformers/issues/2466#issuecomment-982710520), and [here](https://github.com/microsoft/onnxruntime/issues/6549#issuecomment-1016948837) you can find a report in the `GPT-2` context from a Microsoft engineer: \"*int8 quantization are not recommended due to accuracy loss*\").\n* [Onnx Runtime T5 export tool](https://github.com/microsoft/onnxruntime/tree/master/onnxruntime/python/tools/transformers/models/t5) targets both `GPU` and `CPU`. It works in a similar way than `FastT5`: `decoder` module is exported 2 times. Like `FastT5`, the memory footprint of the decoder part is doubled (this time there is no `int-8` quantization).\n* [FasterTransformer](https://github.com/NVIDIA/FasterTransformer/blob/main/docs/t5_guide.md#translation-process) targets `GPU` and is a mix of `Pytorch` and `CUDA`/`C++` dedicated code. The performance boost is huge on `T5`, they report a 10X speedup like `TensorRT`. However, it may significantly decrease the accuracy of the model ([here](https://github.com/NVIDIA/FasterTransformer/blob/main/docs/t5_guide.md#translation-process) when sampling is enabled, it reduces BLEU score of translation task by 8 points, the cause may be a bug in the decoding algorithm or an approximation a bit too aggressive) plus the speedup is computed on a [translation task](https://github.com/NVIDIA/FasterTransformer/blob/main/examples/pytorch/decoding/utils/translation/test.en) where sequences are 25 tokens long on average. In our experience, improvement on very short sequences tends to decrease by large margins on longer sequences. It seems to us that their objectives are different from ours.\n\nWith the existing solutions, you need to choose one or two items of the following:\n\n* double decoder memory footprint;\n* be slower than `Hugging Face` for moderately long sequence length / beam search;\n* degrade output quality.\n\n## Our approach\n\nOur approach to make autoregressive `transformer` based models 2X faster than `Hugging Face` `Pytorch` implementation (the base line) is based on 3 key ingredients:\n\n* storing 2 computation graphs in a single `Onnx` file: this let us have both cache and no cache support without having any duplicated weights,\n* `zero copy` to retrieve output from `Onnx Runtime`: we built over our past work to connect in the most efficient way `Pytorch` tensors (used in the decoding part) and `Onnx Runtime`. Our previous work was to avoid `host` <-> `GPU` tensor copy, but it still required a `GPU` <-> `GPU`. It is now part of the official `Onnx Runtime` documentation (apparently [thanks of our project](https://github.com/microsoft/onnxruntime/pull/10651)!). This time we found out a way to directly expose the internal state of `Onnx Runtime` through a `Pytorch` tensor in zero copy way. Combined with cache mechanism, this is responsible for most of the speedup we have obtained.\n* a generic tool to convert any model (whatever the architecture) to `FP16` without any risk of having out of range values or rounding to zero: `FP16` is still the way to reduce memory footprint of a model. The main issue is that some nodes may output values outside of `FP16` range or round others to zero, resulting in `NaN` output; moreover, very small values may be rounded to zero which is an issue for log and div operations. We have built a tool which detect those nodes so we can keep their precision in `FP32`. It's quite important to reduce memory footprint of these models, not just because they tend to be huge, but also because past states (that we cache) and internal buffers can be even bigger than the weights of the model itself.\n\n## Results\n\nAs demonstrated at the end of this notebook, **we are able to provide a X2 speedup** whatever the batch size, the sequence length or the model size.\n\n> For `TensorRT` we have our own implementation of our approach described above which helps to provide similar latency to `Onnx Runtime`. It's in a Python script in the same folder as this notebook. We had to work around a documented limitation. Because of that the code is slightly more complex and we wanted to keep this notebook easy to follow.\n",
"_____no_output_____"
]
],
[
[
"! nvidia-smi",
"Mon May 23 21:22:55 2022 \r\n+-----------------------------------------------------------------------------+\r\n| NVIDIA-SMI 515.43.04 Driver Version: 515.43.04 CUDA Version: 11.7 |\r\n|-------------------------------+----------------------+----------------------+\r\n| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |\r\n| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |\r\n| | | MIG M. |\r\n|===============================+======================+======================|\r\n| 0 NVIDIA GeForce ... On | 00000000:03:00.0 On | N/A |\r\n| 39% 47C P8 41W / 350W | 263MiB / 24576MiB | 0% Default |\r\n| | | N/A |\r\n+-------------------------------+----------------------+----------------------+\r\n \r\n+-----------------------------------------------------------------------------+\r\n| Processes: |\r\n| GPU GI CI PID Type Process name GPU Memory |\r\n| ID ID Usage |\r\n|=============================================================================|\r\n| 0 N/A N/A 2366 G /usr/lib/xorg/Xorg 160MiB |\r\n| 0 N/A N/A 6173 G ...ome-remote-desktop-daemon 4MiB |\r\n| 0 N/A N/A 8174 G /usr/bin/gnome-shell 44MiB |\r\n| 0 N/A N/A 8918 G ...on/Bin/AgentConnectix.bin 4MiB |\r\n| 0 N/A N/A 393472 G ...176700596390776551,131072 47MiB |\r\n+-----------------------------------------------------------------------------+\r\n"
]
],
[
[
"## `Onnx Runtime` compilation\n\nVersion 1.11.1 of `Onnx Runtime` and older have a bug which makes them much slower when most inputs are used by subgraphs of an `If` node.\nUnfortunately, it's exactly what will do below, so we need to compile our own version of `Onnx Runtime` until the version 1.12 is released (in June 2022).\nCode below has been tested on Ubuntu 22.04 and supposes that your machine has `CUDA` 11.4 installed.\nIf not, use the Docker image of this library.\n\nWe use a specific commit of `Onnx Runtime` with a better management of `If`/`Else`/`Then` `Onnx` nodes:\n\n```shell\ngit clone --recursive https://github.com/Microsoft/onnxruntime\ncd onnxruntime\ngit checkout -b fix_if 81d78706feb1dc923f3e43f7ba8ac30b55f5b19b\nCUDACXX=/usr/local/cuda-11.4/bin/nvcc ./build.sh \\\n --config Release \\\n --build_wheel \\\n --parallel \\\n --use_cuda \\\n --cuda_home /usr/local/cuda-11.4 \\\n --cudnn_home /usr/lib/x86_\n -linux-gnu/ \\\n --skip_test\n# pip install ...\n# other required dependencies\n# pip install nvtx seaborn\n```\n\nOn our machine, it takes around 20 minutes.\n\n> to clear previous compilation, delete content of `./build` folder",
"_____no_output_____"
]
],
[
[
"import json\nimport random\nfrom transformer_deploy.backends.ort_utils import get_keep_fp32_nodes\nfrom transformer_deploy.backends.ort_utils import convert_fp16\nimport time\nfrom typing import Callable, Dict, Optional, List\nimport matplotlib.pylab as plt\nfrom onnxruntime import IOBinding\nimport numpy as np\nimport onnx\nimport torch\nfrom pathlib import Path\nfrom typing import Tuple\nfrom onnx import GraphProto, ModelProto, helper\nfrom torch.nn import Linear\nfrom transformers import AutoModelForSeq2SeqLM, AutoTokenizer, PretrainedConfig, T5ForConditionalGeneration, TensorType\nfrom transformers.generation_utils import GenerationMixin\nfrom transformers.modeling_outputs import BaseModelOutputWithPastAndCrossAttentions, Seq2SeqLMOutput\nfrom transformers.models.t5.modeling_t5 import T5Stack\nfrom nvtx import nvtx\nfrom copy import copy\n\nfrom transformer_deploy.backends.ort_utils import create_model_for_provider, inference_onnx_binding\nfrom transformer_deploy.backends.pytorch_utils import convert_to_onnx\nimport seaborn as sns\nimport operator\nfrom collections import defaultdict\nimport gc",
"_____no_output_____"
]
],
[
[
"## Loading `Hugging Face` model / tokenizer\n\nBelow we load the model and set global variables of this notebook.",
"_____no_output_____"
]
],
[
[
"np.random.seed(123)\ntorch.random.manual_seed(123)\n# other possible values: t5-small, t5-base, t5-large. t5-3b should work when ORT library is fixed\nmodel_name = \"t5-large\"\ntokenizer = AutoTokenizer.from_pretrained(model_name)\ninput_ids: torch.Tensor = tokenizer(\n \"translate English to French: This model is now very fast!\", return_tensors=TensorType.PYTORCH\n).input_ids\ninput_ids = input_ids.type(torch.int32).to(\"cuda\")\npytorch_model: T5ForConditionalGeneration = AutoModelForSeq2SeqLM.from_pretrained(model_name)\npytorch_model = pytorch_model.eval()\npytorch_model = pytorch_model.cuda()\npytorch_model.config.use_cache = True # not really needed, just to make things obvious\nnum_layers = pytorch_model.config.num_layers\n# tolerance between Onnx FP16 and Pytorch FP32.\n# Rounding errors increase with number of layers: 1e-1 for t5-small, 5e-1 for large, 3 for 3b. 11b not tested.\n# Do not impact final quality\nfp16_default_tolerance = 5e-1\n\n\ndef are_equal(a: torch.Tensor, b: torch.Tensor, atol: float = fp16_default_tolerance) -> None:\n assert np.allclose(a=a.detach().cpu().numpy(), b=b.detach().cpu().numpy(), atol=atol), f\"{a}\\n\\nVS\\n\\n{b}\"\n\n\ndef save_onnx(proto: onnx.ModelProto, model_path: str) -> None:\n # protobuff doesn't support files > 2Gb, in this case, weights are stored in another binary file\n save_external_data: bool = proto.ByteSize() > 2 * 1024**3\n filename = Path(model_path).name\n onnx.save_model(\n proto=proto,\n f=model_path,\n save_as_external_data=save_external_data,\n all_tensors_to_one_file=True,\n location=filename + \".data\",\n )\n\n\ndef prepare_folder(path: str) -> Tuple[str, str]:\n p = Path(path)\n p.mkdir(parents=True, exist_ok=True)\n [item.unlink() for item in Path(path).glob(\"*\") if item.is_file()]\n return path + \"/model.onnx\", path + \"/model_fp16.onnx\"\n\n\n# create/clean folders where each model will be stored.\n# as multiple files will be saved for T5-3B and 11B, we use different folders for the encoder and the decoders.\nencoder_model_path, encoder_fp16_model_path = prepare_folder(path=\"./test-enc\")\ndec_cache_model_path, dec_cache_fp16_model_path = prepare_folder(path=\"./test-dec-cache\")\ndec_no_cache_model_path, dec_no_cache_fp16_model_path = prepare_folder(path=\"./test-dec-no-cache\")\n_, dec_if_fp16_model_path = prepare_folder(path=\"./test-dec-if\")\n\n# some outputs to compare with\nout_enc: BaseModelOutputWithPastAndCrossAttentions = pytorch_model.encoder(input_ids=input_ids)\nout_full: Seq2SeqLMOutput = pytorch_model(input_ids=input_ids, decoder_input_ids=input_ids)",
"/home/geantvert/.local/share/virtualenvs/fast_transformer/lib/python3.9/site-packages/transformers/models/t5/tokenization_t5_fast.py:155: FutureWarning: This tokenizer was incorrectly instantiated with a model max length of 512 which will be corrected in Transformers v5.\nFor now, this behavior is kept to avoid breaking backwards compatibility when padding/encoding with `truncation is True`.\n- Be aware that you SHOULD NOT rely on t5-large automatically truncating your input to 512 when padding/encoding.\n- If you want to encode/pad to sequences longer than 512 you can either instantiate this tokenizer with `model_max_length` or pass `max_length` when encoding/padding.\n- To avoid this warning, please instantiate this tokenizer with `model_max_length` set to your preferred value.\n warnings.warn(\n"
]
],
[
[
"# Export to Onnx\n\nFirst step is to export the model to `Onnx` graph.\n`T5` is made of 2 parts, an `encoder` and a `decoder`.\n\n## Export encoder part\n\nThe `encoder` part export doesn't imply any specific challenge.\nWe use export function built for `Bert` like model, exported model is in `FP16`.",
"_____no_output_____"
]
],
[
[
"pytorch_model = pytorch_model.to(\"cuda\")\n\nconvert_to_onnx(\n model_pytorch=pytorch_model.encoder,\n output_path=encoder_model_path,\n inputs_pytorch={\"input_ids\": input_ids},\n var_output_seq=True,\n quantization=False,\n)",
"_____no_output_____"
]
],
[
[
"## Conversion to mixed precision\n\n### Why mixed precision?\n\nAs `T5` can have up to 11 billion parameters, it requires lots of computation, and even more important, it takes lots of space in device memory.\nWe convert the `encoder` to half precision.\n\nIf we blindly convert the whole graph to `FP16`, we will have 2 issues:\n* `overflow`: some nodes, like exponential nodes, will try to output values out of the `FP16` range, at the end you get some `NaN`.\n* `underflow`: values very close to 0 will be rounded to 0, which may be an issue for some operations like `Div` and `Log` .\n\n### The challenge\n\nMixed precision is done out of the box by `Pytorch` and follow some strict rules described in https://pytorch.org/docs/stable/amp.html\n\nThose rules are generic and quite conservative. Many nodes will be kept in `FP32` even if their output is always in the `FP16` range.\n\nOther approaches we have found:\n* `Onnx Runtime T5` [demo](https://github.com/microsoft/onnxruntime/blob/master/onnxruntime/python/tools/transformers/models/t5/t5_helper.py): provide a list of operations to keep in `FP32` (Pow, ReduceMean, Add, Sqrt, Div, Mul, Softmax, Relu). We have found this approach to need more an more tweaking on larger networks and encoder part (decoder part seems simpler to manage, https://github.com/microsoft/onnxruntime/issues/11119);\n* `TensorRT T5` [demo](https://github.com/NVIDIA/TensorRT/tree/main/demo/HuggingFace/notebooks): provide the exact pattern of nodes to keep in `FP32`. This approach is much more effective, but imply lots of code to describe the patterns and may not generalize well, basically what works for a `base` model may not work for 11 billion parameters model. And it does not scale to other architectures without adaptations, for a library like `transformer-deploy`, it would lead to unmaintainable technical debt.\n\n\n### Our approach\n\nWe have chosen an architecture agnostic approach: we inject random input sequences and audit the output of each computation graph node; finally, we make a list of all nodes that have output values out of the `FP16` range /close to zero values and perform some cleaning (to avoid unnecessary casting).\n\nWe have chosen to use random values only for the `input_ids` field as the search space is limited: positive integers lower than the vocabulary size.\nYou can also decide to send real data from a dataset you want to work on.\n\nTo finish, we provide the list of nodes to keep in `FP32` to the conversion function.",
"_____no_output_____"
]
],
[
[
"def get_random_input_encoder() -> Dict[str, torch.Tensor]:\n max_seq = 512\n seq_len = random.randint(a=1, b=max_seq)\n batch = max_seq // seq_len\n random_input_ids = torch.randint(\n low=0, high=tokenizer.vocab_size, size=(batch, seq_len), dtype=torch.int32, device=\"cuda\"\n )\n inputs = {\"input_ids\": random_input_ids}\n return inputs\n\n\nkeep_fp32_encoder = get_keep_fp32_nodes(onnx_model_path=encoder_model_path, get_input=get_random_input_encoder)\nassert len(keep_fp32_encoder) > 0\nenc_model_onnx = convert_fp16(onnx_model=encoder_model_path, nodes_to_exclude=keep_fp32_encoder)\nsave_onnx(proto=enc_model_onnx, model_path=encoder_fp16_model_path)\n\ndel enc_model_onnx\ntorch.cuda.empty_cache()\ngc.collect()",
"_____no_output_____"
],
[
"print(f\"20 first nodes to keep in FP32 (total {len(keep_fp32_encoder)}):\")\nkeep_fp32_encoder[:20]",
"20 first nodes to keep in FP32 (total 1247):\n"
]
],
[
[
"Compare the output of the `Onnx` `FP16` model with `Pytorch` one",
"_____no_output_____"
]
],
[
[
"enc_fp16_onnx = create_model_for_provider(encoder_fp16_model_path, \"CUDAExecutionProvider\")\nenc_fp16_onnx_binding: IOBinding = enc_fp16_onnx.io_binding()\nenc_onnx_out = inference_onnx_binding(\n model_onnx=enc_fp16_onnx,\n binding=enc_fp16_onnx_binding,\n inputs={\"input_ids\": input_ids},\n device=input_ids.device.type,\n)[\"output\"]\nare_equal(a=enc_onnx_out, b=out_enc.last_hidden_state)",
"_____no_output_____"
]
],
[
[
"## Export decoder\n\nThe decoder export part is more challenging:\n\n* we first need to wrap it in a `Pytorch` model to add the final layer so it's output provide scores for each vocabulary token and can be directly used by the `Hugging Face` `decoding` algorithm\n* then, we need to manipulate the `Onnx` graph to add support of `Key`/`Value` cache\n\nThe second point is the key ingredient of the observed acceleration of `Onnx` vs `Hugging Face` inference.\n\n\n### Wrapper to include some post-processing on the decoder output\n\nThe post-processing is mainly a projection of the decoder output on a matrix with one of its dimensions equal to model vocabulary size, so we have scores for each possible token.",
"_____no_output_____"
]
],
[
[
"class ExportT5(torch.nn.Module):\n def __init__(self, decoder: T5Stack, lm_head: Linear):\n super(ExportT5, self).__init__()\n self.decoder = decoder\n self.lm_head = lm_head\n\n def forward(self, input_ids: torch.Tensor, encoder_hidden_states: torch.Tensor, past_key_values: Tuple = None):\n out_dec = self.decoder.forward(\n input_ids=input_ids, encoder_hidden_states=encoder_hidden_states, past_key_values=past_key_values\n )\n # Rescale output before projecting on vocab\n out_dec[\"last_hidden_state\"] = out_dec[\"last_hidden_state\"] * (pytorch_model.model_dim**-0.5)\n out_dec[\"last_hidden_state\"] = self.lm_head(out_dec[\"last_hidden_state\"])\n return out_dec\n\n\npytorch_model.cuda()\nmodel_decoder = ExportT5(decoder=pytorch_model.decoder, lm_head=pytorch_model.lm_head).eval()\nout_model_export: torch.Tensor = model_decoder(input_ids=input_ids, encoder_hidden_states=out_enc.last_hidden_state)\n\nare_equal(a=out_model_export[\"last_hidden_state\"], b=out_full.logits)",
"_____no_output_____"
]
],
[
[
"### Export decoder part to `Onnx`\n\nBelow we export 2 versions of the decoder, one without cache support and one with it.\n\nModel inputs with past states (cache support):",
"_____no_output_____"
]
],
[
[
"model_decoder.cuda()\n# decoder output one step before\nout_dec_pytorch = model_decoder(input_ids=input_ids[:, :-1], encoder_hidden_states=out_enc.last_hidden_state)\n\nmodel_inputs = {\n \"input_ids\": input_ids[:, -1:].type(torch.int32),\n \"encoder_hidden_states\": out_enc.last_hidden_state,\n \"past_key_values\": out_dec_pytorch.past_key_values,\n}\n\ninput_names = [\"input_ids\", \"encoder_hidden_states\"]\n\nfor i in range(num_layers):\n input_names.append(f\"past_key_values.{i}.decoder.key\")\n input_names.append(f\"past_key_values.{i}.decoder.value\")\n input_names.append(f\"past_key_values.{i}.encoder.key\")\n input_names.append(f\"past_key_values.{i}.encoder.value\")\n\noutput_names = [\"logits\"]\n\nfor i in range(num_layers):\n output_names.append(f\"present.{i}.decoder.key\")\n output_names.append(f\"present.{i}.decoder.value\")\n output_names.append(f\"present.{i}.encoder.key\")\n output_names.append(f\"present.{i}.encoder.value\")\n\ndynamic_axis = {\n \"input_ids\": {0: \"batch\", 1: \"encoder_sequence\"},\n \"encoder_hidden_states\": {0: \"batch\", 1: \"encoder_sequence\"},\n \"logits\": {0: \"batch\", 1: \"decoder_sequence\"},\n}\n\n\nfor i in range(num_layers):\n dynamic_axis[f\"past_key_values.{i}.decoder.key\"] = {0: \"batch\", 2: \"past_decoder_sequence\"}\n dynamic_axis[f\"past_key_values.{i}.decoder.value\"] = {0: \"batch\", 2: \"past_decoder_sequence\"}\n dynamic_axis[f\"past_key_values.{i}.encoder.key\"] = {0: \"batch\", 2: \"encoder_sequence_length\"}\n dynamic_axis[f\"past_key_values.{i}.encoder.value\"] = {0: \"batch\", 2: \"encoder_sequence_length\"}\n\n dynamic_axis[f\"present.{i}.decoder.key\"] = {0: \"batch\", 2: \"decoder_sequence\"}\n dynamic_axis[f\"present.{i}.decoder.value\"] = {0: \"batch\", 2: \"decoder_sequence\"}\n dynamic_axis[f\"present.{i}.encoder.key\"] = {0: \"batch\", 2: \"encoder_sequence_length\"}\n dynamic_axis[f\"present.{i}.encoder.value\"] = {0: \"batch\", 2: \"encoder_sequence_length\"}",
"_____no_output_____"
]
],
[
[
"Export of the model with cache support:",
"_____no_output_____"
]
],
[
[
"with torch.no_grad():\n pytorch_model.config.return_dict = True\n pytorch_model.eval()\n\n # export can works with named args but the dict containing named args as to be last element of the args tuple\n torch.onnx.export(\n model_decoder,\n (model_inputs,),\n f=dec_cache_model_path,\n input_names=input_names,\n output_names=output_names,\n dynamic_axes=dynamic_axis,\n do_constant_folding=True,\n opset_version=13,\n )",
"/home/geantvert/.local/share/virtualenvs/fast_transformer/lib/python3.9/site-packages/transformers/modeling_utils.py:668: TracerWarning: Converting a tensor to a Python boolean might cause the trace to be incorrect. We can't record the data flow of Python values, so this value will be treated as a constant in the future. This means that the trace might not generalize to other inputs!\n if causal_mask.shape[1] < attention_mask.shape[1]:\nIn-place op on output of tensor.shape. See https://pytorch.org/docs/master/onnx.html#avoid-inplace-operations-when-using-tensor-shape-in-tracing-mode\nIn-place op on output of tensor.shape. See https://pytorch.org/docs/master/onnx.html#avoid-inplace-operations-when-using-tensor-shape-in-tracing-mode\n"
]
],
[
[
"Export of the model computing Key/Values for the whole sequence (we basically just remove past states from the input, the `Pytorch` code will recompute them):",
"_____no_output_____"
]
],
[
[
"model_inputs_no_cache = {\n \"input_ids\": input_ids,\n \"encoder_hidden_states\": out_enc.last_hidden_state,\n}\n\nwith torch.no_grad():\n pytorch_model.config.return_dict = True\n pytorch_model.eval()\n\n # export can works with named args but the dict containing named args as to be last element of the args tuple\n torch.onnx.export(\n model_decoder,\n (model_inputs_no_cache,),\n f=dec_no_cache_model_path,\n input_names=list(model_inputs_no_cache.keys()),\n output_names=output_names,\n dynamic_axes={k: v for k, v in dynamic_axis.items() if \"past_key_values\" not in k},\n do_constant_folding=True,\n opset_version=13,\n )\n_ = pytorch_model.cpu() # free cuda memory\ntorch.cuda.empty_cache()",
"_____no_output_____"
]
],
[
[
"## Conversion to mixed precision\n\nDecoder module has different kinds of inputs, `input_ids` but also some float tensors.\nIt would a bit more complicated to generate random values for those tensors: in theory it can be of any value in the FP32 range, but because of how models are initialized and trained, most of them are close to 0.\n\nTo avoid too much guessing, we have decided to just take the output of the real model being fed with random `input_ids`.\n",
"_____no_output_____"
]
],
[
[
"def get_random_input_no_cache() -> Dict[str, torch.Tensor]:\n inputs = get_random_input_encoder()\n encoder_hidden_states = inference_onnx_binding(\n model_onnx=enc_fp16_onnx,\n binding=enc_fp16_onnx_binding,\n inputs=inputs,\n device=\"cuda\",\n clone_tensor=False,\n )[\"output\"]\n # it will serve as input of a FP32 model\n inputs[\"encoder_hidden_states\"] = encoder_hidden_states.type(torch.float32)\n return inputs\n\n\nkeep_fp32_no_cache = get_keep_fp32_nodes(onnx_model_path=dec_no_cache_model_path, get_input=get_random_input_no_cache)\n\nonnx_model_no_cache_fp16 = convert_fp16(onnx_model=dec_no_cache_model_path, nodes_to_exclude=keep_fp32_no_cache)\nsave_onnx(proto=onnx_model_no_cache_fp16, model_path=dec_no_cache_fp16_model_path)",
"_____no_output_____"
],
[
"print(f\"20 first nodes to keep in FP32 (total {len(keep_fp32_no_cache)}):\")\nkeep_fp32_no_cache[:20]",
"20 first nodes to keep in FP32 (total 2340):\n"
],
[
"dec_no_cache_ort_model = create_model_for_provider(dec_no_cache_model_path, \"CUDAExecutionProvider\")\n\n# use info from tokenizer size and max shape provided through the command line\ndef get_random_input_cache() -> Dict[str, torch.Tensor]:\n inputs = get_random_input_no_cache()\n dec_past_states = inference_onnx_binding(\n model_onnx=dec_no_cache_ort_model,\n inputs=inputs,\n device=\"cuda\",\n clone_tensor=False,\n )\n for k, v in dec_past_states.items():\n if k == \"logits\":\n continue\n new_k = k.replace(\"present\", \"past_key_values\")\n inputs[new_k] = v\n batch, _ = inputs[\"input_ids\"].shape\n complement = torch.randint(low=0, high=tokenizer.vocab_size, size=(batch, 1), dtype=torch.int32, device=\"cuda\")\n inputs[\"input_ids\"] = torch.concat(tensors=[inputs[\"input_ids\"], complement], dim=1)\n return inputs\n\n\nkeep_fp32_cache = get_keep_fp32_nodes(onnx_model_path=dec_cache_model_path, get_input=get_random_input_cache)\ndel dec_no_cache_ort_model # free cuda memory\ntorch.cuda.empty_cache()\ngc.collect()\n\nonnx_model_cache_fp16 = convert_fp16(onnx_model=dec_cache_model_path, nodes_to_exclude=keep_fp32_cache)\nsave_onnx(proto=onnx_model_cache_fp16, model_path=dec_cache_fp16_model_path)",
"_____no_output_____"
],
[
"print(f\"20 first nodes to keep in FP32 (total {len(keep_fp32_cache)}):\")\nkeep_fp32_cache[:20]",
"20 first nodes to keep in FP32 (total 2240):\n"
]
],
[
[
"## Merge `Onnx` computation graph to deduplicate weights\n\nFinally, we will merge the 2 decoders together.\nThe idea is simple:\n\n* we prefix the node / edge names of one of them to avoid naming collision\n* we deduplicate the weights (the same weight matrix will have different names in the 2 models)\n* we join the 2 computation graphs through an `If` node\n* we generate the `Onnx` file\n\nThe new model will take a new input, `enable_cache`. When it contains a `True` value, computation graph with cache support is used.\n\n> code below is written to be easy to read, but could be made much faster to run",
"_____no_output_____"
]
],
[
[
"prefix = \"cache_node_\"\nmapping_initializer_cache_to_no_cache = dict()\n\n# search for not-duplicated weights, called initializer in Onnx\nto_add = list()\nfor node_cache in onnx_model_cache_fp16.graph.initializer:\n found = False\n for node_no_cache in onnx_model_no_cache_fp16.graph.initializer:\n if node_cache.raw_data == node_no_cache.raw_data:\n found = True\n mapping_initializer_cache_to_no_cache[node_cache.name] = node_no_cache.name\n break\n if not found:\n node_cache.name = prefix + node_cache.name\n to_add.append(node_cache)\n mapping_initializer_cache_to_no_cache[node_cache.name] = node_cache.name\n\nonnx_model_no_cache_fp16.graph.initializer.extend(to_add)\n# I/O model names should not be prefixed\nmodel_io_names = [n.name for n in list(onnx_model_cache_fp16.graph.input) + list(onnx_model_cache_fp16.graph.output)]\n\n# replace pointers to duplicated weights to their deduplicated version\nfor node in onnx_model_cache_fp16.graph.node:\n for index, input_name in enumerate(node.input):\n if input_name in model_io_names:\n continue\n node.input[index] = mapping_initializer_cache_to_no_cache.get(input_name, prefix + input_name)\n for index, output_name in enumerate(node.output):\n if output_name in model_io_names:\n continue\n node.output[index] = prefix + output_name\n node.name = prefix + node.name\nmodel_io_names = [n.name for n in list(onnx_model_cache_fp16.graph.input) + list(onnx_model_cache_fp16.graph.output)]\n\n# prefix nodes to avoid naming collision\nprefix = \"init_\"\ncache = dict()\nfor node in onnx_model_no_cache_fp16.graph.initializer:\n if node.name in model_io_names:\n new_name = prefix + node.name\n cache[node.name] = new_name\n node.name = new_name\n\nfor node in onnx_model_no_cache_fp16.graph.node:\n for input_index, n in enumerate(node.input):\n node.input[input_index] = cache.get(n, n)\n\n# mandatory for subgraph in if/else node\nassert len(onnx_model_cache_fp16.graph.output) == len(\n onnx_model_no_cache_fp16.graph.output\n), f\"{len(onnx_model_cache_fp16.graph.output)} vs {len(onnx_model_no_cache_fp16.graph.output)}\"\n\n# build a computation graph with cache support\ngraph_cache: onnx.GraphProto = onnx.helper.make_graph(\n nodes=list(onnx_model_cache_fp16.graph.node),\n name=\"graph-cache\",\n inputs=[],\n outputs=list(onnx_model_cache_fp16.graph.output),\n initializer=[],\n)\n\n# build a computation which doesn't need past states to run\ngraph_no_cache: onnx.GraphProto = onnx.helper.make_graph(\n nodes=list(onnx_model_no_cache_fp16.graph.node),\n name=\"graph-no-cache\",\n inputs=[],\n outputs=list(onnx_model_no_cache_fp16.graph.output),\n initializer=[],\n)\n\n# a new input to decide if we use past state or not\nenable_cache_input = onnx.helper.make_tensor_value_info(name=\"enable_cache\", elem_type=onnx.TensorProto.BOOL, shape=[1])\n\nif_node = onnx.helper.make_node(\n op_type=\"If\",\n inputs=[\"enable_cache\"],\n outputs=[o.name for o in list(onnx_model_no_cache_fp16.graph.output)],\n then_branch=graph_cache,\n else_branch=graph_no_cache,\n)\n\n# final model which can disable its cache\nif_graph_def: GraphProto = helper.make_graph(\n nodes=[if_node],\n name=\"if-model\",\n inputs=list(onnx_model_cache_fp16.graph.input) + [enable_cache_input],\n outputs=list(onnx_model_no_cache_fp16.graph.output),\n initializer=list(onnx_model_no_cache_fp16.graph.initializer),\n)\n\n# serialization and cleaning\nmodel_if: ModelProto = helper.make_model(\n if_graph_def, producer_name=\"onnx-example\", opset_imports=[helper.make_opsetid(onnx.defs.ONNX_DOMAIN, 13)]\n)\nsave_onnx(proto=model_if, model_path=dec_if_fp16_model_path)\ndel model_if\ntorch.cuda.empty_cache()\ngc.collect()",
"_____no_output_____"
]
],
[
[
"### Check `Onnx` decoder output\n\nCompare `Onnx` output with and without cache, plus compare with `Pytorch` output.",
"_____no_output_____"
]
],
[
[
"pytorch_model = pytorch_model.cuda()\nmodel_decoder = model_decoder.cuda()\ninput_ids = input_ids.cuda()\npytorch_model = pytorch_model.eval()\nmodel_decoder = model_decoder.eval()\ndec_onnx = create_model_for_provider(dec_if_fp16_model_path, \"CUDAExecutionProvider\", log_severity=3)\ndec_onnx_binding: IOBinding = dec_onnx.io_binding()",
"_____no_output_____"
]
],
[
[
"## Zero copy output\n\nBelow, we check that the new model output is similar to the ones from `Pytorch`.\n\nWe use our new implementation of inference call. \nThe idea is the following:\n\n* we ask `Onnx Runtime` to output a pointer to the `CUDA` array containing the result of the inference;\n* we use `Cupy` API to wrap the array and provide information regarding tensor shape and type. `Cupy` doesn't own the data;\n* we use `Dlpack` support to convert the `Cupy` tensor to `Pytorch`, another zero copy process.\n\nThis pipeline is unsafe, as the content of the tensor may change or disappear silently: only `Onnx Runtime` has the control of the array containing the data. It will happen at the next inference call. Because we know that during the text generation we discard each output before recalling `Onnx Runtime`, it works well in our case.\n\nA second benefit of this approach is that we do not have anymore to guess the output shape. \nBefore using this approach, to avoid the output to be stored on host memory (RAM) which made inference slower, we had to provide `Onnx Runtime` with a pointer to `Pytorch` tensor with the right size. As the size change with the sequence length (so it changes for each generated token), we had to store the logic to guess the size somewhere in the code. The new approach frees us from this burden.",
"_____no_output_____"
]
],
[
[
"pytorch_model = pytorch_model.half()\nwith torch.inference_mode():\n out_enc_pytorch: BaseModelOutputWithPastAndCrossAttentions = pytorch_model.encoder(input_ids=input_ids)\n previous_step_pytorch: BaseModelOutputWithPastAndCrossAttentions = model_decoder(\n input_ids=input_ids[:, :-1], encoder_hidden_states=out_enc_pytorch.last_hidden_state\n )\n out_dec_pytorch: BaseModelOutputWithPastAndCrossAttentions = model_decoder(\n input_ids=input_ids, encoder_hidden_states=out_enc_pytorch.last_hidden_state\n )",
"_____no_output_____"
],
[
"def decoder_pytorch_inference(decoder_input_ids: torch.Tensor, encoder_hidden_states: torch.Tensor, **_):\n with torch.inference_mode():\n return model_decoder(input_ids=decoder_input_ids, encoder_hidden_states=encoder_hidden_states)\n\n\ndef decoder_onnx_inference(\n decoder_input_ids: torch.Tensor,\n encoder_hidden_states: torch.Tensor,\n enable_cache: torch.Tensor,\n past_key_values: Optional[torch.Tensor],\n):\n inputs_onnx_dict = {\n \"input_ids\": decoder_input_ids,\n \"encoder_hidden_states\": encoder_hidden_states,\n \"enable_cache\": enable_cache,\n }\n\n if past_key_values is not None:\n for index, (k_dec, v_dec, k_enc, v_enc) in enumerate(past_key_values):\n inputs_onnx_dict[f\"past_key_values.{index}.decoder.key\"] = k_dec\n inputs_onnx_dict[f\"past_key_values.{index}.decoder.value\"] = v_dec\n inputs_onnx_dict[f\"past_key_values.{index}.encoder.key\"] = k_enc\n inputs_onnx_dict[f\"past_key_values.{index}.encoder.value\"] = v_enc\n\n result_dict = inference_onnx_binding(\n model_onnx=dec_onnx,\n inputs=inputs_onnx_dict,\n binding=dec_onnx_binding, # recycle the binding\n device=decoder_input_ids.device.type,\n clone_tensor=False, # no memory copy -> best perf and lowest memory footprint!\n )\n past_states = list()\n for index in range(pytorch_model.config.num_layers):\n kv = (\n result_dict[f\"present.{index}.decoder.key\"],\n result_dict[f\"present.{index}.decoder.value\"],\n result_dict[f\"present.{index}.encoder.key\"],\n result_dict[f\"present.{index}.encoder.value\"],\n )\n past_states.append(kv)\n return BaseModelOutputWithPastAndCrossAttentions(\n last_hidden_state=result_dict[\"logits\"],\n past_key_values=past_states,\n )\n\n\nout_dec_onnx_no_cache = decoder_onnx_inference(\n decoder_input_ids=input_ids,\n encoder_hidden_states=out_enc_pytorch.last_hidden_state,\n enable_cache=torch.tensor([False], device=\"cuda\", dtype=torch.bool),\n past_key_values=None,\n)\nare_equal(a=out_dec_onnx_no_cache.last_hidden_state[:, -1:, :], b=out_dec_pytorch.last_hidden_state[:, -1:, :])\n\n# check that past states are identical between Onnx and Pytorch\nassert len(out_dec_onnx_no_cache.past_key_values) == len(out_dec_pytorch.past_key_values)\nfor (o_dec_k, o_dev_v, o_enc_k, o_enc_v), (p_dec_k, p_dev_v, p_enc_k, p_enc_v) in zip(\n out_dec_onnx_no_cache.past_key_values, out_dec_pytorch.past_key_values\n):\n are_equal(a=o_dec_k, b=p_dec_k)\n are_equal(a=o_dev_v, b=p_dev_v)\n are_equal(a=o_enc_k, b=p_enc_k)\n are_equal(a=o_enc_v, b=p_enc_v)",
"_____no_output_____"
],
[
"out_dec_onnx_cache = decoder_onnx_inference(\n decoder_input_ids=input_ids[:, -1:],\n encoder_hidden_states=out_enc_pytorch.last_hidden_state,\n enable_cache=torch.tensor([True], device=\"cuda\", dtype=torch.bool),\n past_key_values=previous_step_pytorch.past_key_values,\n)\n\nare_equal(a=out_dec_onnx_cache.last_hidden_state[:, -1:, :], b=out_dec_pytorch.last_hidden_state[:, -1:, :])\n\n# check that past states are identical between Onnx and Pytorch\nassert len(out_dec_onnx_cache.past_key_values) == len(out_dec_pytorch.past_key_values)\nfor (o_dec_k, o_dev_v, o_enc_k, o_enc_v), (p_dec_k, p_dev_v, p_enc_k, p_enc_v) in zip(\n out_dec_onnx_cache.past_key_values, out_dec_pytorch.past_key_values\n):\n are_equal(a=o_dec_k, b=p_dec_k)\n are_equal(a=o_dev_v, b=p_dev_v)\n are_equal(a=o_enc_k, b=p_enc_k)\n are_equal(a=o_enc_v, b=p_enc_v)",
"_____no_output_____"
]
],
[
[
"## Benchmarks!\n\nFinally, we will compare the performances of 4 setup in end-to-end scenarii:\n\n* `Pytorch`\n* `Pytorch` + cache\n* `Onnx`\n* `Onnx` + cache\n\nFor the comparison, we first do a sanity check by just generating a short sequence (we already have checked that output tensors are OK).\n\nThen we force each model to generate:\n\n* 256 tokens + batch size 1 (similar to `TensorRT` demo)\n* 1000 tokens + batch size 4\n",
"_____no_output_____"
]
],
[
[
"def encoder_onnx_inference(input_ids: torch.Tensor, **_) -> BaseModelOutputWithPastAndCrossAttentions:\n last_hidden_state = inference_onnx_binding(\n model_onnx=enc_fp16_onnx, # noqa: F821\n inputs={\"input_ids\": input_ids},\n device=input_ids.device.type,\n binding=enc_fp16_onnx_binding,\n )[\"output\"]\n return BaseModelOutputWithPastAndCrossAttentions(last_hidden_state=last_hidden_state.type(torch.float16))\n\n\ndef encoder_pytorch_inference(input_ids, **_) -> BaseModelOutputWithPastAndCrossAttentions:\n with torch.inference_mode():\n res = pytorch_model.encoder(input_ids=input_ids).type(torch.float16)\n return res\n\n\n# https://github.com/NVIDIA/TensorRT/blob/main/demo/HuggingFace/T5/export.py\nclass ExtT5(torch.nn.Module, GenerationMixin):\n def __init__(self, config: PretrainedConfig, device: torch.device, encoder_func: Callable, decoder_func: Callable):\n super(ExtT5, self).__init__()\n self.main_input_name = \"input_ids\" # https://github.com/huggingface/transformers/pull/14803\n self.config: PretrainedConfig = config\n self.device: torch.device = device\n\n self.encoder_func = encoder_func\n self.decoder_func = decoder_func\n self.use_cache = True\n self.timings = list()\n\n def get_encoder(self):\n return self.encoder_func\n\n def get_decoder(self):\n return self.decoder_func\n\n def set_cache(self, enable: bool) -> None:\n self.use_cache = enable\n\n # from transformers library (modeling_t5.py)\n def _reorder_cache(self, past, beam_idx):\n reordered_decoder_past = ()\n for layer_past_states in past:\n # get the correct batch idx from layer past batch dim\n # batch dim of `past` is at 2nd position\n reordered_layer_past_states = ()\n for layer_past_state in layer_past_states:\n # need to set correct `past` for each of the four key / value states\n reordered_layer_past_states = reordered_layer_past_states + (\n layer_past_state.index_select(0, beam_idx),\n )\n\n assert reordered_layer_past_states[0].shape == layer_past_states[0].shape\n assert len(reordered_layer_past_states) == len(layer_past_states)\n\n reordered_decoder_past = reordered_decoder_past + (reordered_layer_past_states,)\n return reordered_decoder_past\n\n def prepare_inputs_for_generation(self, input_ids, past=None, use_cache=None, **kwargs) -> Dict[str, torch.Tensor]:\n params = {\n \"encoder_hidden_states\": kwargs[\"encoder_outputs\"][\"last_hidden_state\"],\n }\n if past is None: # this is the 1st inferred token\n self.timings = list()\n if not self.use_cache:\n past = None\n if past is None:\n params[self.main_input_name] = input_ids\n params[\"enable_cache\"] = torch.tensor([False], device=\"cuda\", dtype=torch.bool)\n else:\n params[self.main_input_name] = input_ids[:, -1:]\n params[\"enable_cache\"] = torch.tensor([True], device=\"cuda\", dtype=torch.bool)\n params[\"past_key_values\"] = past\n\n return params\n\n def forward(\n self,\n input_ids: torch.Tensor,\n encoder_hidden_states: torch.Tensor,\n enable_cache: torch.Tensor,\n past_key_values: Optional[torch.Tensor] = None,\n **_,\n ):\n start_timer = time.monotonic()\n dec_output = self.get_decoder()(\n decoder_input_ids=input_ids,\n encoder_hidden_states=encoder_hidden_states,\n enable_cache=enable_cache,\n past_key_values=past_key_values,\n )\n self.timings.append(time.monotonic() - start_timer)\n return Seq2SeqLMOutput(logits=dec_output.last_hidden_state, past_key_values=dec_output.past_key_values)\n\n\nmodel_gen = (\n ExtT5(\n config=pytorch_model.config,\n device=pytorch_model.device,\n encoder_func=encoder_onnx_inference, # encoder_pytorch_inference\n decoder_func=decoder_onnx_inference, # decoder_pytorch_inference\n )\n .cuda()\n .eval()\n)\n\ntorch.cuda.synchronize()\nwith torch.inference_mode():\n print(\"Onnx:\")\n print(\n tokenizer.decode(\n model_gen.generate(\n inputs=input_ids,\n min_length=3,\n max_length=60,\n num_beams=4,\n no_repeat_ngram_size=2,\n )[0],\n skip_special_tokens=True,\n )\n )\n print(\"Pytorch:\")\n print(\n tokenizer.decode(\n pytorch_model.generate(\n input_ids=input_ids,\n min_length=3,\n max_length=60,\n num_beams=4,\n no_repeat_ngram_size=2,\n )[0],\n skip_special_tokens=True,\n )\n )",
"Onnx:\nCe modèle est maintenant très rapide!\nPytorch:\nCe modèle est maintenant très rapide!\n"
],
[
"def print_timings(name: str, total: float, inference: float):\n percent_inference = 100 * inference / total\n print(f\"{name}: {total:.1f}, including inference: {inference:.1f} ({percent_inference:.1f}%)\")\n\n\nall_timings: Dict[str, Dict[str, List[float]]] = dict()\nfor seq_len, num_beam in [(256, 1), (1000, 4)]:\n timings = dict()\n\n print(f\"seq len: {seq_len} / # beam (batch size): {num_beam}\")\n task = \"Onnx\"\n with nvtx.annotate(\n task, color=\"red\"\n ): # nvtx is for Nvidia nsight profiler, you can remove the line or install the library\n model_gen.set_cache(enable=False)\n # warmup\n model_gen.generate(inputs=input_ids, max_length=10, num_beams=num_beam, min_length=10)\n start = time.monotonic()\n model_gen.generate(inputs=input_ids, max_length=seq_len, num_beams=num_beam, min_length=seq_len)\n total_time = time.monotonic() - start\n print_timings(name=task, total=total_time, inference=sum(model_gen.timings))\n timings[f\"{task}\"] = model_gen.timings\n\n task = \"Onnx + cache\"\n with nvtx.annotate(task, color=\"red\"):\n model_gen.set_cache(enable=True)\n # warmup\n model_gen.generate(inputs=input_ids, max_length=10, num_beams=num_beam, min_length=10)\n start = time.monotonic()\n model_gen.generate(inputs=input_ids, max_length=seq_len, num_beams=num_beam, min_length=seq_len)\n total_time = time.monotonic() - start\n print_timings(name=task, total=total_time, inference=sum(model_gen.timings))\n timings[f\"{task}\"] = model_gen.timings\n\n # monckey patching of forward function to add a timer per generated token\n old_fw = pytorch_model.forward\n timing_pytorch = list()\n\n def new_fw(self, *args, **kwargs):\n timer_start = time.monotonic()\n res = old_fw(self, *args, **kwargs)\n torch.cuda.synchronize() # makes timings correct without having significant impact on e2e latency\n total_time = time.monotonic() - timer_start\n timing_pytorch.append(total_time)\n return res\n\n task = \"Pytorch\"\n with nvtx.annotate(task, color=\"orange\"):\n pytorch_model.config.use_cache = False\n with torch.inference_mode():\n with torch.cuda.amp.autocast():\n # warmup\n pytorch_model.generate(inputs=input_ids, max_length=10, num_beams=num_beam, min_length=10)\n pytorch_model.forward = new_fw.__get__(pytorch_model)\n start = time.monotonic()\n pytorch_model.generate(inputs=input_ids, max_length=seq_len, num_beams=num_beam, min_length=seq_len)\n total_time = time.monotonic() - start\n pytorch_model.forward = old_fw\n inference_time = np.sum(timing_pytorch)\n print_timings(name=\"Pytorch\", total=total_time, inference=inference_time)\n timing_pytorch_no_cache = copy(timing_pytorch)\n timings[f\"{task}\"] = copy(timing_pytorch)\n timing_pytorch.clear()\n torch.cuda.empty_cache()\n\n task = \"Pytorch + cache\"\n with nvtx.annotate(\"Pytorch + cache\", color=\"green\"):\n pytorch_model.config.use_cache = True\n with torch.inference_mode():\n with torch.cuda.amp.autocast():\n # warmup\n pytorch_model.generate(inputs=input_ids, max_length=10, num_beams=num_beam, min_length=10)\n pytorch_model.forward = new_fw.__get__(pytorch_model)\n start = time.monotonic()\n pytorch_model.generate(inputs=input_ids, max_length=seq_len, num_beams=num_beam, min_length=seq_len)\n total_time = time.monotonic() - start\n pytorch_model.forward = old_fw\n print_timings(name=\"Pytorch + cache\", total=total_time, inference=sum(timing_pytorch))\n timings[f\"{task}\"] = copy(timing_pytorch)\n timing_pytorch.clear()\n all_timings[f\"{seq_len} / {num_beam}\"] = timings\n torch.cuda.empty_cache()",
"seq len: 256 / # beam (batch size): 1\nOnnx: 3.9, including inference: 3.8 (96.9%)\nOnnx + cache: 3.3, including inference: 3.1 (96.3%)\nPytorch: 9.8, including inference: 9.7 (99.0%)\nPytorch + cache: 8.2, including inference: 8.1 (98.7%)\nseq len: 1000 / # beam (batch size): 4\nOnnx: 99.4, including inference: 97.1 (97.7%)\nOnnx + cache: 17.7, including inference: 15.6 (87.8%)\nPytorch: 96.3, including inference: 95.5 (99.1%)\nPytorch + cache: 37.0, including inference: 35.0 (94.6%)\n"
]
],
[
[
"## Benchmark analysis\n\nBelow, we plot for each setup (short and long sequence):\n\n* the time spent on each token generation\n* the full time to generate the sequence (for each length)\n\nWe can see that for short sequence and batch size of 1, cache or not, latency appears to be stable.\nHowever, for longer sequences, we can see that the no cache approach (being `Pytorch` or `Onnx` based) doesn't scale well, and at some point, `Onnx` is even slower than `Hugging Face` code with cache support.\n\nOn the other side, `Onnx` timings are mostly stable whatever the sequence length which is quite remarkable.\nIt's because we are working one token at a time and converted a quadratic complexity in the attention layer into a linear one.",
"_____no_output_____"
]
],
[
[
"sns.set_style(\"darkgrid\") # darkgrid, whitegrid, dark, white and ticks\nplt.rc(\"axes\", titlesize=15) # fontsize of the axes title\nplt.rc(\"axes\", labelsize=14) # fontsize of the x and y labels\nplt.rc(\"xtick\", labelsize=13) # fontsize of the tick labels\nplt.rc(\"ytick\", labelsize=13) # fontsize of the tick labels\nplt.rc(\"legend\", fontsize=15) # legend fontsize\nplt.rc(\"font\", size=13) # controls default text sizes\n\ncolors = sns.color_palette(\"deep\")\nfig = plt.figure(constrained_layout=True, figsize=(12, 8))\nsubfigs = fig.subfigures(nrows=2, ncols=1)\n\nfig.supxlabel(\"seq len (# tokens)\")\nfig.supylabel(\"latency (s)\")\nfig.suptitle(f\"Small seq len and greedy search on {model_name} don't tell the whole (inference) story...\")\nfor row, (plot_name, timings) in enumerate(all_timings.items()):\n subfigs[row].suptitle(f\"setup #{1+row}: {plot_name} (seq len / beam search)\")\n axs = subfigs[row].subplots(nrows=1, ncols=2)\n for col, accumulated in enumerate([False, True]):\n plot_axis = axs[col]\n for index, (k, v) in enumerate(timings.items()):\n axis = range(len(v))\n color = colors[index]\n v = np.array(v)\n # remove extreme values\n p99 = np.percentile(v, 99)\n v[v > p99] = p99\n v = np.cumsum(v) if accumulated else v\n plot_axis.scatter(axis, v, label=k, s=2)\n\n title = f\"latency for the full sequence\" if accumulated else f\"latency for each token\"\n plot_axis.title.set_text(title)\n\n# legend deduplication\nhandles, labels = plt.gca().get_legend_handles_labels()\nby_label = dict(zip(labels, handles))\nfig.legend(by_label.values(), by_label.keys(), bbox_to_anchor=(1, 1), loc=\"upper left\", markerscale=5)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Profiling model at the kernel level\n\nBelow we reload the decoder model with `Onnx Runtime` kernel profiling enabled.\nIt will help us to understand on which part of the computation graph the GPU spends its time. \n\nThe number of events that `Onnx Runtime` can save is limited to [1 million](https://github.com/microsoft/onnxruntime/blob/a4b5fa334aa939fb159bdc571ed3d56ca8d31fc7/onnxruntime/core/common/profiler.cc#L10).\nIt is not an issue as we have seen that timings per token are mostly stable, so having only n first token information don't change anything.\n\nThe main information it gives us is that 30% of the time is spent on matrix multiplication when caching is used. \nThe rest of the time is spent on mostly memory bound operations:\n* element-wise operations which require little computation (`add`, `mul`, `div`, etc.)\n* copy pasting tensors `GPU` <-> `GPU` with little transformation in between (`transpose`, `concat`, `cast`, etc.)\n\nIt matches the information provided by both `nvidia-smi` and `Nvidia Nsight` (the GPU profiler from Nvidia): the GPU is under utilized. \nThat's why we think that a tool like `TensorRT` which will perform aggressive kernel fusion, reducing time spent on memory bounded operations, should be a good fit for autoregressive models.\n\n> there is a nice opportunity to increase the speedup by reducing the number of casting operations. We keep this work for the future.",
"_____no_output_____"
]
],
[
[
"dec_onnx = create_model_for_provider(\n dec_if_fp16_model_path, \"CUDAExecutionProvider\", enable_profiling=True, log_severity=3\n)\ndec_onnx_binding: IOBinding = dec_onnx.io_binding()\n_ = model_gen.generate(inputs=input_ids, max_length=10, num_beams=4, min_length=10)\nprofile_name = dec_onnx.end_profiling()\n\nwith open(profile_name) as f:\n content = json.load(f)\n\nop_timings = defaultdict(lambda: 0)\nfor c in content:\n if \"op_name\" not in c[\"args\"]:\n continue\n op_name = c[\"args\"][\"op_name\"]\n if op_name == \"If\":\n continue # subgraph\n time_taken = c[\"dur\"]\n op_timings[op_name] += time_taken\n\nop_timings_filter = dict(sorted(op_timings.items(), key=operator.itemgetter(1), reverse=True)[:10])\ntotal_kernel_timing = sum(op_timings.values())\nop_timings_percent = {k: 100 * v / total_kernel_timing for k, v in op_timings_filter.items()}\n\nplt.barh(list(op_timings_percent.keys()), list(op_timings_percent.values()))\nplt.title(\"Time spent per kernel\\n(top 10 kernels)\")\nplt.xlabel(\"% total inference time\")\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a13a1866dc910f1a26e2a2012ea0831c279b3fd
| 13,922 |
ipynb
|
Jupyter Notebook
|
A4.ipynb
|
CISC-372/Notebook
|
fe548df2928ec730ed1a72012a589278d2d11154
|
[
"MIT"
] | 4 |
2020-01-16T16:23:51.000Z
|
2021-01-12T09:23:38.000Z
|
A4.ipynb
|
CISC-372/Notebook
|
fe548df2928ec730ed1a72012a589278d2d11154
|
[
"MIT"
] | null | null | null |
A4.ipynb
|
CISC-372/Notebook
|
fe548df2928ec730ed1a72012a589278d2d11154
|
[
"MIT"
] | 13 |
2020-02-05T13:55:02.000Z
|
2022-03-17T08:20:57.000Z
| 33.873479 | 146 | 0.387444 |
[
[
[
"!wget -q https://github.com/CISC-372/Notebook/releases/download/a4/test.csv\n!wget -q https://github.com/CISC-372/Notebook/releases/download/a4/train.csv",
"_____no_output_____"
],
[
"# comment your understanding of each function \nimport pandas as pd\nimport csv\n\n\nxy_train_df = pd.read_csv('train.csv')\nx_test_df = pd.read_csv('test.csv', index_col='id')\n\n\nxy_train_df['length'] = xy_train_df.apply(lambda x: len(x.review), axis=1)\nxy_train_df = xy_train_df.sort_values('length')\nxy_train_df",
"_____no_output_____"
],
[
"# comment your understanding of each function and each parameter below:\n\nfrom tensorflow.keras.preprocessing.text import Tokenizer\nfrom tensorflow.keras.preprocessing.sequence import pad_sequences\nfrom sklearn.model_selection import train_test_split\n\nvocab_size = 10000\nmax_len = 256\n\nxy_train, xy_validation = train_test_split(\n xy_train_df, test_size=0.2)\n\n# build vocabulary from training set\ntokenizer = Tokenizer(num_words=vocab_size)\ntokenizer.fit_on_texts(xy_train.review)\n\n\ndef _preprocess(texts):\n return pad_sequences(\n tokenizer.texts_to_sequences(texts),\n maxlen=max_len, \n padding='post'\n )\n\n\nx_train = _preprocess(xy_train.review)\ny_train = xy_train.rating\n\nx_valid = _preprocess(xy_validation.review)\ny_valid = xy_validation.rating\n\nx_test = _preprocess(x_test_df.review)\n\nprint(x_train.shape)\nprint(x_valid.shape)\nprint(x_test.shape)",
"(4978, 256)\n(1245, 256)\n(2667, 256)\n"
],
[
"from __future__ import absolute_import, division, print_function, unicode_literals\nimport collections\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow import keras\n\n\nimport tensorflow as tf\nfrom tensorflow.keras.optimizers import Adam\n\n# comment your understanding of each line and \n# the output shape of each line below. for each dimensionality, explains its \n# meaning. (e.g. None is the batch size)\n\nx = keras.Input((max_len))\n\nembeded = keras.layers.Embedding(vocab_size, 20)(x)\n\naveraged = tf.reduce_mean(embeded, axis=1)\n\npred = keras.layers.Dense(1, activation=tf.nn.sigmoid)(averaged)\n\nmodel = keras.Model(x, pred)\n\nmodel.compile(\n optimizer=Adam(clipnorm=4.),\n loss='binary_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"history = model.fit(x_train,\n y_train,\n epochs=5,\n batch_size=64,\n validation_data=(x_valid, y_valid),\n verbose=1)",
"Epoch 1/5\n78/78 [==============================] - 2s 10ms/step - loss: 0.6611 - accuracy: 0.7026 - val_loss: 0.5413 - val_accuracy: 0.8691\nEpoch 2/5\n78/78 [==============================] - 0s 6ms/step - loss: 0.5025 - accuracy: 0.8742 - val_loss: 0.4223 - val_accuracy: 0.8691\nEpoch 3/5\n78/78 [==============================] - 0s 6ms/step - loss: 0.3925 - accuracy: 0.8821 - val_loss: 0.3885 - val_accuracy: 0.8691\nEpoch 4/5\n78/78 [==============================] - 0s 6ms/step - loss: 0.3847 - accuracy: 0.8693 - val_loss: 0.3837 - val_accuracy: 0.8691\nEpoch 5/5\n78/78 [==============================] - 0s 6ms/step - loss: 0.3564 - accuracy: 0.8825 - val_loss: 0.3820 - val_accuracy: 0.8691\n"
],
[
"model.evaluate(x_valid, y_valid)",
"39/39 [==============================] - 0s 2ms/step - loss: 0.3820 - accuracy: 0.8691\n"
],
[
"def predict_class(_dataset):\n classes = model.predict(_dataset) > 0.5\n return np.squeeze(classes * 1) \n\ny_predict = predict_class(x_valid)\n\nfrom sklearn.metrics import f1_score\nfrom sklearn.metrics import confusion_matrix\n\nprint(f1_score(y_valid, y_predict, average='micro'))",
"0.8690763052208834\n"
],
[
"# submission\npd.DataFrame(\n {'id': x_test_df.index,\n 'rating': predict_class(x_test)}).to_csv('sample_submission.csv', index=False)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a13a36bc2f758c0fda9680155578d642dd5860b
| 367,985 |
ipynb
|
Jupyter Notebook
|
nbs/04_writing_prompt_supervised_baseline_training.ipynb
|
anshradh/trl
|
946f3ca14133bc9f16d9fb1b832a41764b3e8e0c
|
[
"Apache-2.0"
] | null | null | null |
nbs/04_writing_prompt_supervised_baseline_training.ipynb
|
anshradh/trl
|
946f3ca14133bc9f16d9fb1b832a41764b3e8e0c
|
[
"Apache-2.0"
] | null | null | null |
nbs/04_writing_prompt_supervised_baseline_training.ipynb
|
anshradh/trl
|
946f3ca14133bc9f16d9fb1b832a41764b3e8e0c
|
[
"Apache-2.0"
] | null | null | null | 47.945928 | 9,127 | 0.561952 |
[
[
[
"<a href=\"https://colab.research.google.com/github/anshradh/trl_custom/blob/test/04_writing_prompt_supervised_baseline_training.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"#Writing Prompt Response Supervised Learning Baseline\nWe fine-tune a language model to respond to reddit writing prompts using standard supervised learning. This is also known as behavioral cloning or imitation learning in RL.",
"_____no_output_____"
],
[
"## Prerequisites",
"_____no_output_____"
]
],
[
[
"## Install needed libraries and log into huggingface\n!pip install datasets\n!pip install transformers\n!pip install accelerate\n!pip install huggingface_hub\n!apt install git-lfs\nfrom huggingface_hub import notebook_login\nnotebook_login()",
"Login successful\nYour token has been saved to /root/.huggingface/token\n\u001b[1m\u001b[31mAuthenticated through git-credential store but this isn't the helper defined on your machine.\nYou might have to re-authenticate when pushing to the Hugging Face Hub. Run the following command in your terminal in case you want to set this credential helper as the default\n\ngit config --global credential.helper store\u001b[0m\n"
],
[
"import torch\nfrom tqdm.auto import tqdm\nimport numpy as np\nfrom torch.utils.data import DataLoader\nimport pandas as pd\nimport numpy as np\nimport torch.nn.functional as F\nfrom torch.optim import Adam\nimport torch\nimport collections\nimport random\ntqdm.pandas()\n\nfrom datasets import load_dataset, ClassLabel, load_metric, concatenate_datasets\n\nfrom transformers import AutoModel, AutoTokenizer\nfrom transformers import top_k_top_p_filtering\nfrom torch import nn\nfrom torch.nn import Identity\nimport torch.nn.functional as F\nimport torch\n\nfrom transformers import AutoModelForCausalLM, DataCollatorWithPadding, AdamW, get_scheduler\n\nfrom accelerate import Accelerator",
"_____no_output_____"
]
],
[
[
"## Load and preprocess dataset",
"_____no_output_____"
]
],
[
[
"## Load dataset from huggingface\nprompt_response_dataset = load_dataset(\"rewardsignal/reddit_writing_prompts\", data_files=\"prompt_responses_full.csv\", split='train[:80%]')",
"Using custom data configuration rewardsignal--reddit_writing_prompts-dd5d2a64487ab606\n"
],
[
"## We tokenize the dataset and standardize the prompts and responses\n# tokenizer_name = input()\ntokenizer_name = 'distilgpt2'\ntokenizer = AutoTokenizer.from_pretrained(tokenizer_name, use_fast=True)\nprompt_prefix = \"Writing Prompt: \"\nresponse_prefix = \"Response: \"\n\ndef preprocess_text_function(examples):\n examples[\"prompt\"] = [prompt.replace('[WP] ', prompt_prefix) for prompt in examples[\"prompt\"]]\n examples[\"response\"] = [response_prefix + response for response in examples[\"response\"]]\n return tokenizer(examples['prompt'], examples['response'], truncation=True)\n\ntokenized_prompt_response_dataset = prompt_response_dataset.map(preprocess_text_function, batched=True, remove_columns=['Unnamed: 0', 'prompt_id', 'prompt', 'prompt_score', 'prompt_created_utc', 'response_id', 'response', 'response_score', 'response_created_utc', 'num_responses', 'response_children', 'score_bin', 'response_rank'], num_proc=4)\n",
"_____no_output_____"
],
[
"## We group the prompts and responses together into continuous text blocks to simplify training\nblock_size = 512\ndef group_texts(examples):\n # Concatenate all texts.\n concatenated_examples = {k: sum(examples[k], []) for k in examples.keys()}\n total_length = len(concatenated_examples[list(examples.keys())[0]])\n # We drop the small remainder, we could add padding if the model supported it instead of this drop, you can\n # customize this part to your needs.\n total_length = (total_length // block_size) * block_size\n # Split by chunks of max_len.\n result = {\n k: [t[i : i + block_size] for i in range(0, total_length, block_size)]\n for k, t in concatenated_examples.items()\n }\n result[\"labels\"] = result[\"input_ids\"].copy()\n return result\ntokenized_prompt_response_dataset = tokenized_prompt_response_dataset.map(group_texts, batched=True, batch_size = 1000, num_proc = 4)\ntokenized_prompt_response_dataset.set_format(\"torch\")",
" "
]
],
[
[
"## Prepare for training",
"_____no_output_____"
]
],
[
[
"## Split into training and evaluation datasets\nsupervised_train_dataset = tokenized_prompt_response_dataset.shuffle(seed=42).select(range(4*len(tokenized_prompt_response_dataset)//5))\nsupervised_eval_dataset = tokenized_prompt_response_dataset.shuffle(seed=42).select(range(4*len(tokenized_prompt_response_dataset)//5, len(tokenized_prompt_response_dataset)))",
"Loading cached shuffled indices for dataset at /root/.cache/huggingface/datasets/csv/rewardsignal--reddit_writing_prompts-dd5d2a64487ab606/0.0.0/433e0ccc46f9880962cc2b12065189766fbb2bee57a221866138fb9203c83519/cache-fd487d54ba07f17d.arrow\n"
],
[
"## We prepare for supervised fine-tuning on the dataset\n# supervised_model_name = input()\nsupervised_model_name = 'distilgpt2'\ntokenizer.pad_token = tokenizer.eos_token\ndata_collator = DataCollatorWithPadding(tokenizer=tokenizer)\nsupervised_model = AutoModelForCausalLM.from_pretrained(supervised_model_name, num_labels=2)\n\ntrain_dataloader = DataLoader(\n supervised_train_dataset, shuffle=True, batch_size=4, collate_fn=data_collator\n)\neval_dataloader = DataLoader(\n supervised_eval_dataset, batch_size=4, collate_fn=data_collator\n)\n\noptimizer = AdamW(supervised_model.parameters(), lr=3e-5)\naccelerator = Accelerator()\ntrain_dataloader, eval_dataloader, supervised_model, optimizer = accelerator.prepare(train_dataloader, eval_dataloader, supervised_model, optimizer)\nnum_epochs = 1\nnum_training_steps = num_epochs * len(train_dataloader)\nlr_scheduler = get_scheduler(\n \"linear\",\n optimizer=optimizer,\n num_warmup_steps=0,\n num_training_steps=num_training_steps,\n)\n\nprogress_bar = tqdm(range(num_training_steps))",
"_____no_output_____"
]
],
[
[
"## Training Loop",
"_____no_output_____"
]
],
[
[
"## Training loop for fine-tuning\nsupervised_model.train()\nfor epoch in range(num_epochs):\n for batch in train_dataloader:\n outputs = supervised_model(**batch)\n loss = outputs.loss\n accelerator.backward(loss)\n optimizer.step()\n lr_scheduler.step()\n optimizer.zero_grad()\n progress_bar.update(1)",
"_____no_output_____"
]
],
[
[
"## Evaluate Outputs",
"_____no_output_____"
]
],
[
[
"## Look at 10 batch outputs from the evaluation dataset\ndevice = torch.device(\"cuda\") if torch.cuda.is_available() else torch.device(\"cpu\")\nsupervised_model.to(device)\ncount = 0\nfor batch in eval_dataloader:\n count += 1\n batch = {k: v.to(device) for k, v in batch.items()}\n with torch.no_grad():\n outputs = supervised_model.generate(**batch, max_length=512, min_length = 200)\n print(tokenizer.batch_decode(outputs, max_length = 512))\n if count == 10: break",
"Setting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\nInput length of input_ids is 512, but ``max_length`` is set to 512. This can lead to unexpected behavior. You should consider increasing ``config.max_length`` or ``max_length``.\nSetting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\nInput length of input_ids is 512, but ``max_length`` is set to 512. This can lead to unexpected behavior. You should consider increasing ``config.max_length`` or ``max_length``.\nSetting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\nInput length of input_ids is 512, but ``max_length`` is set to 512. This can lead to unexpected behavior. You should consider increasing ``config.max_length`` or ``max_length``.\nSetting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\nInput length of input_ids is 512, but ``max_length`` is set to 512. This can lead to unexpected behavior. You should consider increasing ``config.max_length`` or ``max_length``.\nSetting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\nInput length of input_ids is 512, but ``max_length`` is set to 512. This can lead to unexpected behavior. You should consider increasing ``config.max_length`` or ``max_length``.\nSetting `pad_token_id` to `eos_token_id`:50256 for open-end generation.\nInput length of input_ids is 512, but ``max_length`` is set to 512. This can lead to unexpected behavior. You should consider increasing ``config.max_length`` or ``max_length``.\n"
],
[
"## Upload model to huggingface hub\nsupervised_model.push_to_hub(tokenizer_name + \"_supervised_model_final\", use_temp_dir=True)",
"/usr/local/lib/python3.7/dist-packages/huggingface_hub/utils/_deprecation.py:43: FutureWarning: Pass token='distilgpt2_supervised_model_final' as keyword args. From version 0.7 passing these as positional arguments will result in an error\n FutureWarning,\n/usr/local/lib/python3.7/dist-packages/huggingface_hub/hf_api.py:599: FutureWarning: `create_repo` now takes `token` as an optional positional argument. Be sure to adapt your code!\n FutureWarning,\nCloning https://huggingface.co/anshr/distilgpt2_supervised_model_final into local empty directory.\n"
]
],
[
[
"## Results and Discussion\nOverall, this step was pretty straightforward and should provide us a good supervised baseline to apply RL on top of.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a13afce37f5247660d63d0efa183fb1e6724b58
| 701,753 |
ipynb
|
Jupyter Notebook
|
02. Sentiment Analysis Network/Sentiment_Classification_Projects.ipynb
|
darkmatter18/Udacity-Deep-Learning-Nanodegree
|
20e0dea11b670a86882dd34e4f591ac724159f80
|
[
"MIT"
] | null | null | null |
02. Sentiment Analysis Network/Sentiment_Classification_Projects.ipynb
|
darkmatter18/Udacity-Deep-Learning-Nanodegree
|
20e0dea11b670a86882dd34e4f591ac724159f80
|
[
"MIT"
] | 1 |
2020-05-01T13:08:55.000Z
|
2020-05-01T13:10:59.000Z
|
02. Sentiment Analysis Network/Sentiment_Classification_Projects.ipynb
|
darkmatter18/Udacity-Deep-Learning-Nanodegree
|
20e0dea11b670a86882dd34e4f591ac724159f80
|
[
"MIT"
] | 3 |
2020-05-11T21:06:09.000Z
|
2021-01-09T05:58:04.000Z
| 79.070761 | 46,924 | 0.718897 |
[
[
[
"# Sentiment Classification & How To \"Frame Problems\" for a Neural Network\n\nby Andrew Trask\n\n- **Twitter**: @iamtrask\n- **Blog**: http://iamtrask.github.io",
"_____no_output_____"
],
[
"### What You Should Already Know\n\n- neural networks, forward and back-propagation\n- stochastic gradient descent\n- mean squared error\n- and train/test splits\n\n### Where to Get Help if You Need it\n- Re-watch previous Udacity Lectures\n- Leverage the recommended Course Reading Material - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) (Check inside your classroom for a discount code)\n- Shoot me a tweet @iamtrask\n\n\n### Tutorial Outline:\n\n- Intro: The Importance of \"Framing a Problem\" (this lesson)\n\n- [Curate a Dataset](#lesson_1)\n- [Developing a \"Predictive Theory\"](#lesson_2)\n- [**PROJECT 1**: Quick Theory Validation](#project_1)\n\n\n- [Transforming Text to Numbers](#lesson_3)\n- [**PROJECT 2**: Creating the Input/Output Data](#project_2)\n\n\n- Putting it all together in a Neural Network (video only - nothing in notebook)\n- [**PROJECT 3**: Building our Neural Network](#project_3)\n\n\n- [Understanding Neural Noise](#lesson_4)\n- [**PROJECT 4**: Making Learning Faster by Reducing Noise](#project_4)\n\n\n- [Analyzing Inefficiencies in our Network](#lesson_5)\n- [**PROJECT 5**: Making our Network Train and Run Faster](#project_5)\n\n\n- [Further Noise Reduction](#lesson_6)\n- [**PROJECT 6**: Reducing Noise by Strategically Reducing the Vocabulary](#project_6)\n\n\n- [Analysis: What's going on in the weights?](#lesson_7)",
"_____no_output_____"
],
[
"# Lesson: Curate a Dataset<a id='lesson_1'></a>\nThe cells from here until Project 1 include code Andrew shows in the videos leading up to mini project 1. We've included them so you can run the code along with the videos without having to type in everything.",
"_____no_output_____"
]
],
[
[
"def pretty_print_review_and_label(i):\n print(labels[i] + \"\\t:\\t\" + reviews[i][:80] + \"...\")\n\ng = open('reviews.txt','r') # What we know!\nreviews = list(map(lambda x:x[:-1],g.readlines()))\ng.close()\n\ng = open('labels.txt','r') # What we WANT to know!\nlabels = list(map(lambda x:x[:-1].upper(),g.readlines()))\ng.close()",
"_____no_output_____"
]
],
[
[
"**Note:** The data in `reviews.txt` we're using has already been preprocessed a bit and contains only lower case characters. If we were working from raw data, where we didn't know it was all lower case, we would want to add a step here to convert it. That's so we treat different variations of the same word, like `The`, `the`, and `THE`, all the same way.",
"_____no_output_____"
]
],
[
[
"len(reviews)",
"_____no_output_____"
],
[
"reviews[0]",
"_____no_output_____"
],
[
"labels[0]",
"_____no_output_____"
]
],
[
[
"# Lesson: Develop a Predictive Theory<a id='lesson_2'></a>",
"_____no_output_____"
]
],
[
[
"print(\"labels.txt \\t : \\t reviews.txt\\n\")\npretty_print_review_and_label(2137)\npretty_print_review_and_label(12816)\npretty_print_review_and_label(6267)\npretty_print_review_and_label(21934)\npretty_print_review_and_label(5297)\npretty_print_review_and_label(4998)",
"labels.txt \t : \t reviews.txt\n\nNEGATIVE\t:\tthis movie is terrible but it has some good effects . ...\nPOSITIVE\t:\tadrian pasdar is excellent is this film . he makes a fascinating woman . ...\nNEGATIVE\t:\tcomment this movie is impossible . is terrible very improbable bad interpretat...\nPOSITIVE\t:\texcellent episode movie ala pulp fiction . days suicides . it doesnt get more...\nNEGATIVE\t:\tif you haven t seen this it s terrible . it is pure trash . i saw this about ...\nPOSITIVE\t:\tthis schiffer guy is a real genius the movie is of excellent quality and both e...\n"
]
],
[
[
"# Project 1: Quick Theory Validation<a id='project_1'></a>\n\nThere are multiple ways to implement these projects, but in order to get your code closer to what Andrew shows in his solutions, we've provided some hints and starter code throughout this notebook.\n\nYou'll find the [Counter](https://docs.python.org/2/library/collections.html#collections.Counter) class to be useful in this exercise, as well as the [numpy](https://docs.scipy.org/doc/numpy/reference/) library.",
"_____no_output_____"
]
],
[
[
"from collections import Counter\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"We'll create three `Counter` objects, one for words from postive reviews, one for words from negative reviews, and one for all the words.",
"_____no_output_____"
]
],
[
[
"# Create three Counter objects to store positive, negative and total counts\npositive_counts = Counter()\nnegative_counts = Counter()\ntotal_counts = Counter()",
"_____no_output_____"
]
],
[
[
"**TODO:** Examine all the reviews. For each word in a positive review, increase the count for that word in both your positive counter and the total words counter; likewise, for each word in a negative review, increase the count for that word in both your negative counter and the total words counter.\n\n**Note:** Throughout these projects, you should use `split(' ')` to divide a piece of text (such as a review) into individual words. If you use `split()` instead, you'll get slightly different results than what the videos and solutions show.",
"_____no_output_____"
]
],
[
[
"# TODO: Loop over all the words in all the reviews and increment the counts in the appropriate counter objects\nfor review, label in zip(reviews, labels):\n r = review.split(' ')\n if label == 'POSITIVE':\n for i in r:\n positive_counts[i] += 1\n if label == 'NEGATIVE':\n for i in r:\n negative_counts[i] += 1\n \n for i in r:\n total_counts[i] += 1",
"_____no_output_____"
]
],
[
[
"Run the following two cells to list the words used in positive reviews and negative reviews, respectively, ordered from most to least commonly used. ",
"_____no_output_____"
]
],
[
[
"# Examine the counts of the most common words in positive reviews\npositive_counts.most_common()",
"_____no_output_____"
],
[
"# Examine the counts of the most common words in negative reviews\nnegative_counts.most_common()",
"_____no_output_____"
]
],
[
[
"As you can see, common words like \"the\" appear very often in both positive and negative reviews. Instead of finding the most common words in positive or negative reviews, what you really want are the words found in positive reviews more often than in negative reviews, and vice versa. To accomplish this, you'll need to calculate the **ratios** of word usage between positive and negative reviews.\n\n**TODO:** Check all the words you've seen and calculate the ratio of postive to negative uses and store that ratio in `pos_neg_ratios`. \n>Hint: the positive-to-negative ratio for a given word can be calculated with `positive_counts[word] / float(negative_counts[word]+1)`. Notice the `+1` in the denominator – that ensures we don't divide by zero for words that are only seen in positive reviews.",
"_____no_output_____"
]
],
[
[
"# Create Counter object to store positive/negative ratios\npos_neg_ratios = Counter()\n\n# TODO: Calculate the ratios of positive and negative uses of the most common words\n# Consider words to be \"common\" if they've been used at least 100 times\nfor term, count in list(total_counts.most_common()):\n if count > 10 :\n ratio = positive_counts[term] / float(negative_counts[term]+1)\n pos_neg_ratios[term] = ratio",
"_____no_output_____"
]
],
[
[
"Examine the ratios you've calculated for a few words:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 1.0607993145235326\nPos-to-neg ratio for 'amazing' = 4.022813688212928\nPos-to-neg ratio for 'terrible' = 0.17744252873563218\n"
]
],
[
[
"Looking closely at the values you just calculated, we see the following:\n\n* Words that you would expect to see more often in positive reviews – like \"amazing\" – have a ratio greater than 1. The more skewed a word is toward postive, the farther from 1 its positive-to-negative ratio will be.\n* Words that you would expect to see more often in negative reviews – like \"terrible\" – have positive values that are less than 1. The more skewed a word is toward negative, the closer to zero its positive-to-negative ratio will be.\n* Neutral words, which don't really convey any sentiment because you would expect to see them in all sorts of reviews – like \"the\" – have values very close to 1. A perfectly neutral word – one that was used in exactly the same number of positive reviews as negative reviews – would be almost exactly 1. The `+1` we suggested you add to the denominator slightly biases words toward negative, but it won't matter because it will be a tiny bias and later we'll be ignoring words that are too close to neutral anyway.\n\nOk, the ratios tell us which words are used more often in postive or negative reviews, but the specific values we've calculated are a bit difficult to work with. A very positive word like \"amazing\" has a value above 4, whereas a very negative word like \"terrible\" has a value around 0.18. Those values aren't easy to compare for a couple of reasons:\n\n* Right now, 1 is considered neutral, but the absolute value of the postive-to-negative rations of very postive words is larger than the absolute value of the ratios for the very negative words. So there is no way to directly compare two numbers and see if one word conveys the same magnitude of positive sentiment as another word conveys negative sentiment. So we should center all the values around netural so the absolute value fro neutral of the postive-to-negative ratio for a word would indicate how much sentiment (positive or negative) that word conveys.\n* When comparing absolute values it's easier to do that around zero than one. \n\nTo fix these issues, we'll convert all of our ratios to new values using logarithms.\n\n**TODO:** Go through all the ratios you calculated and convert them to logarithms. (i.e. use `np.log(ratio)`)\n\nIn the end, extremely positive and extremely negative words will have positive-to-negative ratios with similar magnitudes but opposite signs.",
"_____no_output_____"
]
],
[
[
"# TODO: Convert ratios to logs\nfor term, count in pos_neg_ratios.most_common():\n if count > 1:\n pos_neg_ratios[term] = np.log(count)\n else:\n pos_neg_ratios[term] = - np.log((1 / (count + 0.01)))",
"_____no_output_____"
]
],
[
[
"Examine the new ratios you've calculated for the same words from before:",
"_____no_output_____"
]
],
[
[
"print(\"Pos-to-neg ratio for 'the' = {}\".format(pos_neg_ratios[\"the\"]))\nprint(\"Pos-to-neg ratio for 'amazing' = {}\".format(pos_neg_ratios[\"amazing\"]))\nprint(\"Pos-to-neg ratio for 'terrible' = {}\".format(pos_neg_ratios[\"terrible\"]))",
"Pos-to-neg ratio for 'the' = 0.05902269426102881\nPos-to-neg ratio for 'amazing' = 1.3919815802404802\nPos-to-neg ratio for 'terrible' = -1.6742829939664696\n"
]
],
[
[
"If everything worked, now you should see neutral words with values close to zero. In this case, \"the\" is near zero but slightly positive, so it was probably used in more positive reviews than negative reviews. But look at \"amazing\"'s ratio - it's above `1`, showing it is clearly a word with positive sentiment. And \"terrible\" has a similar score, but in the opposite direction, so it's below `-1`. It's now clear that both of these words are associated with specific, opposing sentiments.\n\nNow run the following cells to see more ratios. \n\nThe first cell displays all the words, ordered by how associated they are with postive reviews. (Your notebook will most likely truncate the output so you won't actually see *all* the words in the list.)\n\nThe second cell displays the 30 words most associated with negative reviews by reversing the order of the first list and then looking at the first 30 words. (If you want the second cell to display all the words, ordered by how associated they are with negative reviews, you could just write `reversed(pos_neg_ratios.most_common())`.)\n\nYou should continue to see values similar to the earlier ones we checked – neutral words will be close to `0`, words will get more positive as their ratios approach and go above `1`, and words will get more negative as their ratios approach and go below `-1`. That's why we decided to use the logs instead of the raw ratios.",
"_____no_output_____"
]
],
[
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]\n\n# Note: Above is the code Andrew uses in his solution video, \n# so we've included it here to avoid confusion.\n# If you explore the documentation for the Counter class, \n# you will see you could also find the 30 least common\n# words like this: pos_neg_ratios.most_common()[:-31:-1]",
"_____no_output_____"
]
],
[
[
"# End of Project 1. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n\n# Transforming Text into Numbers<a id='lesson_3'></a>\nThe cells here include code Andrew shows in the next video. We've included it so you can run the code along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\n\nreview = \"This was a horrible, terrible movie.\"\n\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"review = \"The movie was excellent\"\n\nImage(filename='sentiment_network_pos.png')",
"_____no_output_____"
]
],
[
[
"# Project 2: Creating the Input/Output Data<a id='project_2'></a>\n\n**TODO:** Create a [set](https://docs.python.org/3/tutorial/datastructures.html#sets) named `vocab` that contains every word in the vocabulary.",
"_____no_output_____"
]
],
[
[
"# TODO: Create set named \"vocab\" containing all of the words from all of the reviews\nvocab = total_counts.keys()",
"_____no_output_____"
]
],
[
[
"Run the following cell to check your vocabulary size. If everything worked correctly, it should print **74074**",
"_____no_output_____"
]
],
[
[
"vocab_size = len(vocab)\nprint(vocab_size)",
"74074\n"
]
],
[
[
"Take a look at the following image. It represents the layers of the neural network you'll be building throughout this notebook. `layer_0` is the input layer, `layer_1` is a hidden layer, and `layer_2` is the output layer.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network_2.png')",
"_____no_output_____"
]
],
[
[
"**TODO:** Create a numpy array called `layer_0` and initialize it to all zeros. You will find the [zeros](https://docs.scipy.org/doc/numpy/reference/generated/numpy.zeros.html) function particularly helpful here. Be sure you create `layer_0` as a 2-dimensional matrix with 1 row and `vocab_size` columns. ",
"_____no_output_____"
]
],
[
[
"# TODO: Create layer_0 matrix with dimensions 1 by vocab_size, initially filled with zeros\nlayer_0 = np.zeros((1, len(total_counts)))",
"_____no_output_____"
]
],
[
[
"Run the following cell. It should display `(1, 74074)`",
"_____no_output_____"
]
],
[
[
"layer_0.shape",
"_____no_output_____"
],
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
]
],
[
[
"`layer_0` contains one entry for every word in the vocabulary, as shown in the above image. We need to make sure we know the index of each word, so run the following cell to create a lookup table that stores the index of every word.",
"_____no_output_____"
]
],
[
[
"# Create a dictionary of words in the vocabulary mapped to index positions\n# (to be used in layer_0)\nword2index = {}\nfor i,word in enumerate(vocab):\n word2index[word] = i\n \n# display the map of words to indices\nword2index",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `update_input_layer`. It should count \n how many times each word is used in the given review, and then store\n those counts at the appropriate indices inside `layer_0`.",
"_____no_output_____"
]
],
[
[
"def update_input_layer(review):\n \"\"\" Modify the global layer_0 to represent the vector form of review.\n The element at a given index of layer_0 should represent\n how many times the given word occurs in the review.\n Args:\n review(string) - the string of the review\n Returns:\n None\n \"\"\"\n global layer_0\n # clear out previous state by resetting the layer to be all 0s\n layer_0 *= 0\n for term in review.split(' '):\n layer_0[:,word2index[term]] = 1\n # TODO: count how many times each word is used in the given review and store the results in layer_0 ",
"_____no_output_____"
]
],
[
[
"Run the following cell to test updating the input layer with the first review. The indices assigned may not be the same as in the solution, but hopefully you'll see some non-zero values in `layer_0`. ",
"_____no_output_____"
]
],
[
[
"update_input_layer(reviews[0])\nlayer_0",
"_____no_output_____"
]
],
[
[
"**TODO:** Complete the implementation of `get_target_for_labels`. It should return `0` or `1`, \n depending on whether the given label is `NEGATIVE` or `POSITIVE`, respectively.",
"_____no_output_____"
]
],
[
[
"def get_target_for_label(label):\n \"\"\"Convert a label to `0` or `1`.\n Args:\n label(string) - Either \"POSITIVE\" or \"NEGATIVE\".\n Returns:\n `0` or `1`.\n \"\"\"\n # TODO: Your code here\n return 1 if label == 'POSITIVE' else 0",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out`'POSITIVE'` and `1`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[0]",
"_____no_output_____"
],
[
"get_target_for_label(labels[0])",
"_____no_output_____"
]
],
[
[
"Run the following two cells. They should print out `'NEGATIVE'` and `0`, respectively.",
"_____no_output_____"
]
],
[
[
"labels[1]",
"_____no_output_____"
],
[
"get_target_for_label(labels[1])",
"_____no_output_____"
]
],
[
[
"# End of Project 2. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Project 3: Building a Neural Network<a id='project_3'></a>",
"_____no_output_____"
],
[
"**TODO:** We've included the framework of a class called `SentimentNetork`. Implement all of the items marked `TODO` in the code. These include doing the following:\n- Create a basic neural network much like the networks you've seen in earlier lessons and in Project 1, with an input layer, a hidden layer, and an output layer. \n- Do **not** add a non-linearity in the hidden layer. That is, do not use an activation function when calculating the hidden layer outputs.\n- Re-use the code from earlier in this notebook to create the training data (see `TODO`s in the code)\n- Implement the `pre_process_data` function to create the vocabulary for our training data generating functions\n- Ensure `train` trains over the entire corpus",
"_____no_output_____"
],
[
"### Where to Get Help if You Need it\n- Re-watch earlier Udacity lectures\n- Chapters 3-5 - [Grokking Deep Learning](https://www.manning.com/books/grokking-deep-learning) - (Check inside your classroom for a discount code)",
"_____no_output_____"
]
],
[
[
"import time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews, labels, hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n review_vocab = set()\n # TODO: populate review_vocab with all of the words in the given reviews\n # Remember to split reviews into individual words \n # using \"split(' ')\" instead of \"split()\".\n for review in reviews:\n for r in review.split(' '):\n review_vocab.add(r)\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n label_vocab = set()\n # TODO: populate label_vocab with all of the words in the given labels.\n # There is no need to split the labels because each one is a single word.\n for label in labels:\n label_vocab.add(label)\n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n # TODO: populate self.word2index with indices for all the words in self.review_vocab\n # like you saw earlier in the notebook\n for idx, word in enumerate(self.review_vocab):\n self.word2index[word] = idx\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n # TODO: do the same thing you did for self.word2index and self.review_vocab, \n # but for self.label2index and self.label_vocab instead\n for idx, label in enumerate(self.label_vocab):\n self.label2index[label] = idx\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Store the number of nodes in input, hidden, and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n \n # TODO: initialize self.weights_0_1 as a matrix of zeros. These are the weights between\n # the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((input_nodes, hidden_nodes))\n \n # TODO: initialize self.weights_1_2 as a matrix of random values. \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # TODO: Create the input layer, a two-dimensional matrix with shape \n # 1 x input_nodes, with all values initialized to zero\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n # TODO: You can copy most of the code you wrote for update_input_layer \n # earlier in this notebook. \n #\n # However, MAKE SURE YOU CHANGE ALL VARIABLES TO REFERENCE\n # THE VERSIONS STORED IN THIS OBJECT, NOT THE GLOBAL OBJECTS.\n # For example, replace \"layer_0 *= 0\" with \"self.layer_0 *= 0\"\n self.layer_0 *= 0\n \n for term in review.split(' '):\n if(term in self.word2index.keys()):\n self.layer_0[:,self.word2index[term]] = 1\n \n def get_target_for_label(self,label):\n # TODO: Copy the code you wrote for get_target_for_label \n # earlier in this notebook. \n return 1 if label == 'POSITIVE' else 0\n \n def sigmoid(self,x):\n # TODO: Return the result of calculating the sigmoid activation function\n # shown in the lectures\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n # TODO: Return the derivative of the sigmoid activation function, \n # where \"output\" is the original output from the sigmoid fucntion \n return output * (1 - output)\n\n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n \n # Remember when we started for printing time statistics\n start = time.time()\n\n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # TODO: Get the next review and its correct label\n self.update_input_layer(training_reviews[i])\n label = self.get_target_for_label(training_labels[i])\n # TODO: Implement the forward pass through the network. \n # That means use the given review to update the input layer, \n # then calculate values for the hidden layer,\n # and finally calculate the output layer.\n # \n # Do not use an activation function for the hidden layer,\n # but use the sigmoid activation function for the output layer.\n out_0_1 = np.dot(self.layer_0, self.weights_0_1)\n out_1_2 = self.sigmoid(np.dot(out_0_1, self.weights_1_2))\n \n # TODO: Implement the back propagation pass here. \n # That means calculate the error for the forward pass's prediction\n # and update the weights in the network according to their\n # contributions toward the error, as calculated via the\n # gradient descent and back propagation algorithms you \n # learned in class.\n error = out_1_2 - label\n \n layel_2_error = error * self.sigmoid_output_2_derivative(out_1_2)\n \n layer_1_error = np.dot(layel_2_error, self.weights_1_2.T)\n \n self.weights_1_2 -= np.dot(out_1_2, layel_2_error) * self.learning_rate\n self.weights_0_1 -= np.dot(self.layer_0.T, layer_1_error) * self.learning_rate\n \n # TODO: Keep track of correct predictions. To determine if the prediction was\n # correct, check that the absolute value of the output error \n # is less than 0.5. If so, add one to the correct_so_far count.\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n \n if(out_1_2 >= 0.5 and label == 1):\n correct_so_far += 1\n elif(out_1_2 < 0.5 and label == 0):\n correct_so_far += 1\n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # TODO: Run a forward pass through the network, like you did in the\n # \"train\" function. That means use the given review to \n # update the input layer, then calculate values for the hidden layer,\n # and finally calculate the output layer.\n #\n # Note: The review passed into this function for prediction \n # might come from anywhere, so you should convert it \n # to lower case prior to using it.\n \n # TODO: The output layer should now contain a prediction. \n # Return `POSITIVE` for predictions greater-than-or-equal-to `0.5`, \n # and `NEGATIVE` otherwise.\n self.update_input_layer(review)\n \n out_0_1 = np.dot(self.layer_0, self.weights_0_1)\n out_1_2 = self.sigmoid(np.dot(out_0_1, self.weights_1_2))\n \n if out_1_2 > 0.5:\n return 'POSITIVE'\n else:\n return 'NEGATIVE'\n",
"_____no_output_____"
]
],
[
[
"Run the following cell to create a `SentimentNetwork` that will train on all but the last 1000 reviews (we're saving those for testing). Here we use a learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)",
"_____no_output_____"
]
],
[
[
"Run the following cell to test the network's performance against the last 1000 reviews (the ones we held out from our training set). \n\n**We have not trained the model yet, so the results should be about 50% as it will just be guessing and there are only two possible values to choose from.**",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"\rProgress:0.0% Speed(reviews/sec):0.0 #Correct:0 #Tested:1 Testing Accuracy:0.0%\rProgress:0.1% Speed(reviews/sec):333.7 #Correct:1 #Tested:2 Testing Accuracy:50.0%\rProgress:0.2% Speed(reviews/sec):500.1 #Correct:1 #Tested:3 Testing Accuracy:33.3%\rProgress:0.3% Speed(reviews/sec):600.1 #Correct:2 #Tested:4 Testing Accuracy:50.0%\rProgress:0.4% Speed(reviews/sec):666.7 #Correct:2 #Tested:5 Testing Accuracy:40.0%\rProgress:0.5% Speed(reviews/sec):833.4 #Correct:3 #Tested:6 Testing Accuracy:50.0%\rProgress:0.6% Speed(reviews/sec):750.2 #Correct:3 #Tested:7 Testing Accuracy:42.8%\rProgress:0.7% Speed(reviews/sec):778.0 #Correct:4 #Tested:8 Testing Accuracy:50.0%\rProgress:0.8% Speed(reviews/sec):800.3 #Correct:4 #Tested:9 Testing Accuracy:44.4%\rProgress:0.9% Speed(reviews/sec):750.2 #Correct:5 #Tested:10 Testing Accuracy:50.0%\rProgress:1.0% Speed(reviews/sec):769.5 #Correct:5 #Tested:11 Testing Accuracy:45.4%\rProgress:1.1% Speed(reviews/sec):785.9 #Correct:6 #Tested:12 Testing Accuracy:50.0%\rProgress:1.2% Speed(reviews/sec):800.2 #Correct:6 #Tested:13 Testing Accuracy:46.1%\rProgress:1.3% Speed(reviews/sec):812.7 #Correct:7 #Tested:14 Testing Accuracy:50.0%\rProgress:1.4% Speed(reviews/sec):823.7 #Correct:7 #Tested:15 Testing Accuracy:46.6%\rProgress:1.5% Speed(reviews/sec):833.5 #Correct:8 #Tested:16 Testing Accuracy:50.0%\rProgress:1.6% Speed(reviews/sec):842.2 #Correct:8 #Tested:17 Testing Accuracy:47.0%\rProgress:1.7% Speed(reviews/sec):850.1 #Correct:9 #Tested:18 Testing Accuracy:50.0%\rProgress:1.8% Speed(reviews/sec):857.2 #Correct:9 #Tested:19 Testing Accuracy:47.3%\rProgress:1.9% Speed(reviews/sec):904.8 #Correct:10 #Tested:20 Testing Accuracy:50.0%\rProgress:2.0% Speed(reviews/sec):909.2 #Correct:10 #Tested:21 Testing Accuracy:47.6%\rProgress:2.1% Speed(reviews/sec):913.2 #Correct:11 #Tested:22 Testing Accuracy:50.0%\rProgress:2.2% Speed(reviews/sec):880.0 #Correct:11 #Tested:23 Testing Accuracy:47.8%\rProgress:2.3% Speed(reviews/sec):884.6 #Correct:12 #Tested:24 Testing Accuracy:50.0%\rProgress:2.4% Speed(reviews/sec):857.2 #Correct:12 #Tested:25 Testing Accuracy:48.0%\rProgress:2.5% Speed(reviews/sec):862.0 #Correct:13 #Tested:26 Testing Accuracy:50.0%\rProgress:2.6% Speed(reviews/sec):866.7 #Correct:13 #Tested:27 Testing Accuracy:48.1%\rProgress:2.7% Speed(reviews/sec):843.9 #Correct:14 #Tested:28 Testing Accuracy:50.0%\rProgress:2.8% Speed(reviews/sec):823.5 #Correct:14 #Tested:29 Testing Accuracy:48.2%\rProgress:2.9% Speed(reviews/sec):828.6 #Correct:15 #Tested:30 Testing Accuracy:50.0%\rProgress:3.0% Speed(reviews/sec):833.4 #Correct:15 #Tested:31 Testing Accuracy:48.3%\rProgress:3.1% Speed(reviews/sec):837.9 #Correct:16 #Tested:32 Testing Accuracy:50.0%\rProgress:3.2% Speed(reviews/sec):842.1 #Correct:16 #Tested:33 Testing Accuracy:48.4%\rProgress:3.3% Speed(reviews/sec):846.2 #Correct:17 #Tested:34 Testing Accuracy:50.0%\rProgress:3.4% Speed(reviews/sec):829.3 #Correct:17 #Tested:35 Testing Accuracy:48.5%\rProgress:3.5% Speed(reviews/sec):833.4 #Correct:18 #Tested:36 Testing Accuracy:50.0%\rProgress:3.6% Speed(reviews/sec):837.3 #Correct:18 #Tested:37 Testing Accuracy:48.6%\rProgress:3.7% Speed(reviews/sec):841.0 #Correct:19 #Tested:38 Testing Accuracy:50.0%\rProgress:3.8% Speed(reviews/sec):844.5 #Correct:19 #Tested:39 Testing Accuracy:48.7%\rProgress:3.9% Speed(reviews/sec):829.7 #Correct:20 #Tested:40 Testing Accuracy:50.0%\rProgress:4.0% Speed(reviews/sec):833.2 #Correct:20 #Tested:41 Testing Accuracy:48.7%\rProgress:4.1% Speed(reviews/sec):854.1 #Correct:21 #Tested:42 Testing Accuracy:50.0%\rProgress:4.2% Speed(reviews/sec):840.0 #Correct:21 #Tested:43 Testing Accuracy:48.8%\rProgress:4.3% Speed(reviews/sec):843.2 #Correct:22 #Tested:44 Testing Accuracy:50.0%\rProgress:4.4% Speed(reviews/sec):846.2 #Correct:22 #Tested:45 Testing Accuracy:48.8%\rProgress:4.5% Speed(reviews/sec):833.4 #Correct:23 #Tested:46 Testing Accuracy:50.0%\rProgress:4.6% Speed(reviews/sec):836.4 #Correct:23 #Tested:47 Testing Accuracy:48.9%\rProgress:4.7% Speed(reviews/sec):854.6 #Correct:24 #Tested:48 Testing Accuracy:50.0%\rProgress:4.8% Speed(reviews/sec):857.2 #Correct:24 #Tested:49 Testing Accuracy:48.9%\rProgress:4.9% Speed(reviews/sec):859.7 #Correct:25 #Tested:50 Testing Accuracy:50.0%\rProgress:5.0% Speed(reviews/sec):862.1 #Correct:25 #Tested:51 Testing Accuracy:49.0%\rProgress:5.1% Speed(reviews/sec):879.3 #Correct:26 #Tested:52 Testing Accuracy:50.0%\rProgress:5.2% Speed(reviews/sec):866.6 #Correct:26 #Tested:53 Testing Accuracy:49.0%\rProgress:5.3% Speed(reviews/sec):883.3 #Correct:27 #Tested:54 Testing Accuracy:50.0%\rProgress:5.4% Speed(reviews/sec):885.2 #Correct:27 #Tested:55 Testing Accuracy:49.0%\rProgress:5.5% Speed(reviews/sec):887.1 #Correct:28 #Tested:56 Testing Accuracy:50.0%\rProgress:5.6% Speed(reviews/sec):861.5 #Correct:28 #Tested:57 Testing Accuracy:49.1%\rProgress:5.7% Speed(reviews/sec):863.6 #Correct:29 #Tested:58 Testing Accuracy:50.0%\rProgress:5.8% Speed(reviews/sec):852.5 #Correct:29 #Tested:59 Testing Accuracy:49.1%\rProgress:5.9% Speed(reviews/sec):855.1 #Correct:30 #Tested:60 Testing Accuracy:50.0%\rProgress:6.0% Speed(reviews/sec):857.2 #Correct:30 #Tested:61 Testing Accuracy:49.1%\rProgress:6.1% Speed(reviews/sec):847.2 #Correct:31 #Tested:62 Testing Accuracy:50.0%\rProgress:6.2% Speed(reviews/sec):849.3 #Correct:31 #Tested:63 Testing Accuracy:49.2%\rProgress:6.3% Speed(reviews/sec):863.0 #Correct:32 #Tested:64 Testing Accuracy:50.0%\rProgress:6.4% Speed(reviews/sec):853.3 #Correct:32 #Tested:65 Testing Accuracy:49.2%\rProgress:6.5% Speed(reviews/sec):855.3 #Correct:33 #Tested:66 Testing Accuracy:50.0%\rProgress:6.6% Speed(reviews/sec):857.2 #Correct:33 #Tested:67 Testing Accuracy:49.2%\rProgress:6.7% Speed(reviews/sec):859.0 #Correct:34 #Tested:68 Testing Accuracy:50.0%\rProgress:6.8% Speed(reviews/sec):860.8 #Correct:34 #Tested:69 Testing Accuracy:49.2%\rProgress:6.9% Speed(reviews/sec):862.5 #Correct:35 #Tested:70 Testing Accuracy:50.0%\rProgress:7.0% Speed(reviews/sec):864.2 #Correct:35 #Tested:71 Testing Accuracy:49.2%\rProgress:7.1% Speed(reviews/sec):876.6 #Correct:36 #Tested:72 Testing Accuracy:50.0%\rProgress:7.2% Speed(reviews/sec):878.1 #Correct:36 #Tested:73 Testing Accuracy:49.3%\rProgress:7.3% Speed(reviews/sec):879.5 #Correct:37 #Tested:74 Testing Accuracy:50.0%\rProgress:7.4% Speed(reviews/sec):870.2 #Correct:37 #Tested:75 Testing Accuracy:49.3%\rProgress:7.5% Speed(reviews/sec):872.0 #Correct:38 #Tested:76 Testing Accuracy:50.0%\rProgress:7.6% Speed(reviews/sec):873.5 #Correct:38 #Tested:77 Testing Accuracy:49.3%\rProgress:7.7% Speed(reviews/sec):875.0 #Correct:39 #Tested:78 Testing Accuracy:50.0%\rProgress:7.8% Speed(reviews/sec):876.4 #Correct:39 #Tested:79 Testing Accuracy:49.3%\rProgress:7.9% Speed(reviews/sec):877.8 #Correct:40 #Tested:80 Testing Accuracy:50.0%\rProgress:8.0% Speed(reviews/sec):879.1 #Correct:40 #Tested:81 Testing Accuracy:49.3%\rProgress:8.1% Speed(reviews/sec):880.4 #Correct:41 #Tested:82 Testing Accuracy:50.0%\rProgress:8.2% Speed(reviews/sec):872.3 #Correct:41 #Tested:83 Testing Accuracy:49.3%\rProgress:8.3% Speed(reviews/sec):873.6 #Correct:42 #Tested:84 Testing Accuracy:50.0%\rProgress:8.4% Speed(reviews/sec):874.7 #Correct:42 #Tested:85 Testing Accuracy:49.4%\rProgress:8.5% Speed(reviews/sec):876.3 #Correct:43 #Tested:86 Testing Accuracy:50.0%\rProgress:8.6% Speed(reviews/sec):877.5 #Correct:43 #Tested:87 Testing Accuracy:49.4%\rProgress:8.7% Speed(reviews/sec):878.8 #Correct:44 #Tested:88 Testing Accuracy:50.0%\rProgress:8.8% Speed(reviews/sec):880.0 #Correct:44 #Tested:89 Testing Accuracy:49.4%\rProgress:8.9% Speed(reviews/sec):872.5 #Correct:45 #Tested:90 Testing Accuracy:50.0%\rProgress:9.0% Speed(reviews/sec):873.8 #Correct:45 #Tested:91 Testing Accuracy:49.4%\rProgress:9.1% Speed(reviews/sec):883.5 #Correct:46 #Tested:92 Testing Accuracy:50.0%\rProgress:9.2% Speed(reviews/sec):884.6 #Correct:46 #Tested:93 Testing Accuracy:49.4%\rProgress:9.3% Speed(reviews/sec):885.7 #Correct:47 #Tested:94 Testing Accuracy:50.0%\rProgress:9.4% Speed(reviews/sec):886.8 #Correct:47 #Tested:95 Testing Accuracy:49.4%\rProgress:9.5% Speed(reviews/sec):887.9 #Correct:48 #Tested:96 Testing Accuracy:50.0%\rProgress:9.6% Speed(reviews/sec):888.9 #Correct:48 #Tested:97 Testing Accuracy:49.4%\rProgress:9.7% Speed(reviews/sec):889.9 #Correct:49 #Tested:98 Testing Accuracy:50.0%\rProgress:9.8% Speed(reviews/sec):890.9 #Correct:49 #Tested:99 Testing Accuracy:49.4%\rProgress:9.9% Speed(reviews/sec):891.9 #Correct:50 #Tested:100 Testing Accuracy:50.0%\rProgress:10.0% Speed(reviews/sec):892.9 #Correct:50 #Tested:101 Testing Accuracy:49.5%\rProgress:10.1% Speed(reviews/sec):893.8 #Correct:51 #Tested:102 Testing Accuracy:50.0%\rProgress:10.2% Speed(reviews/sec):894.7 #Correct:51 #Tested:103 Testing Accuracy:49.5%\rProgress:10.3% Speed(reviews/sec):895.6 #Correct:52 #Tested:104 Testing Accuracy:50.0%\rProgress:10.4% Speed(reviews/sec):896.5 #Correct:52 #Tested:105 Testing Accuracy:49.5%\rProgress:10.5% Speed(reviews/sec):905.2 #Correct:53 #Tested:106 Testing Accuracy:50.0%\rProgress:10.6% Speed(reviews/sec):898.3 #Correct:53 #Tested:107 Testing Accuracy:49.5%\rProgress:10.7% Speed(reviews/sec):906.8 #Correct:54 #Tested:108 Testing Accuracy:50.0%\rProgress:10.8% Speed(reviews/sec):907.6 #Correct:54 #Tested:109 Testing Accuracy:49.5%\rProgress:10.9% Speed(reviews/sec):908.3 #Correct:55 #Tested:110 Testing Accuracy:50.0%\rProgress:11.0% Speed(reviews/sec):909.1 #Correct:55 #Tested:111 Testing Accuracy:49.5%\rProgress:11.1% Speed(reviews/sec):909.8 #Correct:56 #Tested:112 Testing Accuracy:50.0%\rProgress:11.2% Speed(reviews/sec):910.6 #Correct:56 #Tested:113 Testing Accuracy:49.5%\rProgress:11.3% Speed(reviews/sec):911.3 #Correct:57 #Tested:114 Testing Accuracy:50.0%\rProgress:11.4% Speed(reviews/sec):912.0 #Correct:57 #Tested:115 Testing Accuracy:49.5%\rProgress:11.5% Speed(reviews/sec):912.7 #Correct:58 #Tested:116 Testing Accuracy:50.0%\rProgress:11.6% Speed(reviews/sec):920.6 #Correct:58 #Tested:117 Testing Accuracy:49.5%\rProgress:11.7% Speed(reviews/sec):921.3 #Correct:59 #Tested:118 Testing Accuracy:50.0%\rProgress:11.8% Speed(reviews/sec):921.9 #Correct:59 #Tested:119 Testing Accuracy:49.5%\rProgress:11.9% Speed(reviews/sec):922.5 #Correct:60 #Tested:120 Testing Accuracy:50.0%\rProgress:12.0% Speed(reviews/sec):930.2 #Correct:60 #Tested:121 Testing Accuracy:49.5%\rProgress:12.1% Speed(reviews/sec):930.8 #Correct:61 #Tested:122 Testing Accuracy:50.0%\rProgress:12.2% Speed(reviews/sec):938.5 #Correct:61 #Tested:123 Testing Accuracy:49.5%\rProgress:12.3% Speed(reviews/sec):938.9 #Correct:62 #Tested:124 Testing Accuracy:50.0%\rProgress:12.4% Speed(reviews/sec):939.4 #Correct:62 #Tested:125 Testing Accuracy:49.6%\rProgress:12.5% Speed(reviews/sec):939.8 #Correct:63 #Tested:126 Testing Accuracy:50.0%\rProgress:12.6% Speed(reviews/sec):947.4 #Correct:63 #Tested:127 Testing Accuracy:49.6%\rProgress:12.7% Speed(reviews/sec):947.7 #Correct:64 #Tested:128 Testing Accuracy:50.0%\rProgress:12.8% Speed(reviews/sec):948.1 #Correct:64 #Tested:129 Testing Accuracy:49.6%\rProgress:12.9% Speed(reviews/sec):948.5 #Correct:65 #Tested:130 Testing Accuracy:50.0%\rProgress:13.0% Speed(reviews/sec):948.9 #Correct:65 #Tested:131 Testing Accuracy:49.6%\rProgress:13.1% Speed(reviews/sec):949.3 #Correct:66 #Tested:132 Testing Accuracy:50.0%\rProgress:13.2% Speed(reviews/sec):949.6 #Correct:66 #Tested:133 Testing Accuracy:49.6%\rProgress:13.3% Speed(reviews/sec):956.8 #Correct:67 #Tested:134 Testing Accuracy:50.0%\rProgress:13.4% Speed(reviews/sec):957.1 #Correct:67 #Tested:135 Testing Accuracy:49.6%\rProgress:13.5% Speed(reviews/sec):957.4 #Correct:68 #Tested:136 Testing Accuracy:50.0%\rProgress:13.6% Speed(reviews/sec):957.7 #Correct:68 #Tested:137 Testing Accuracy:49.6%\rProgress:13.7% Speed(reviews/sec):958.0 #Correct:69 #Tested:138 Testing Accuracy:50.0%\rProgress:13.8% Speed(reviews/sec):958.3 #Correct:69 #Tested:139 Testing Accuracy:49.6%\rProgress:13.9% Speed(reviews/sec):958.6 #Correct:70 #Tested:140 Testing Accuracy:50.0%\rProgress:14.0% Speed(reviews/sec):958.9 #Correct:70 #Tested:141 Testing Accuracy:49.6%\rProgress:14.1% Speed(reviews/sec):965.7 #Correct:71 #Tested:142 Testing Accuracy:50.0%\rProgress:14.2% Speed(reviews/sec):966.0 #Correct:71 #Tested:143 Testing Accuracy:49.6%\rProgress:14.3% Speed(reviews/sec):966.2 #Correct:72 #Tested:144 Testing Accuracy:50.0%\rProgress:14.4% Speed(reviews/sec):966.4 #Correct:72 #Tested:145 Testing Accuracy:49.6%\rProgress:14.5% Speed(reviews/sec):966.6 #Correct:73 #Tested:146 Testing Accuracy:50.0%\rProgress:14.6% Speed(reviews/sec):966.9 #Correct:73 #Tested:147 Testing Accuracy:49.6%\rProgress:14.7% Speed(reviews/sec):967.1 #Correct:74 #Tested:148 Testing Accuracy:50.0%\rProgress:14.8% Speed(reviews/sec):967.3 #Correct:74 #Tested:149 Testing Accuracy:49.6%\rProgress:14.9% Speed(reviews/sec):967.5 #Correct:75 #Tested:150 Testing Accuracy:50.0%\rProgress:15.0% Speed(reviews/sec):974.0 #Correct:75 #Tested:151 Testing Accuracy:49.6%\rProgress:15.1% Speed(reviews/sec):974.2 #Correct:76 #Tested:152 Testing Accuracy:50.0%\rProgress:15.2% Speed(reviews/sec):974.3 #Correct:76 #Tested:153 Testing Accuracy:49.6%\rProgress:15.3% Speed(reviews/sec):974.5 #Correct:77 #Tested:154 Testing Accuracy:50.0%\rProgress:15.4% Speed(reviews/sec):980.9 #Correct:77 #Tested:155 Testing Accuracy:49.6%\rProgress:15.5% Speed(reviews/sec):981.0 #Correct:78 #Tested:156 Testing Accuracy:50.0%\rProgress:15.6% Speed(reviews/sec):981.1 #Correct:78 #Tested:157 Testing Accuracy:49.6%\rProgress:15.7% Speed(reviews/sec):981.2 #Correct:79 #Tested:158 Testing Accuracy:50.0%\rProgress:15.8% Speed(reviews/sec):987.5 #Correct:79 #Tested:159 Testing Accuracy:49.6%\rProgress:15.9% Speed(reviews/sec):987.6 #Correct:80 #Tested:160 Testing Accuracy:50.0%\rProgress:16.0% Speed(reviews/sec):987.6 #Correct:80 #Tested:161 Testing Accuracy:49.6%\rProgress:16.1% Speed(reviews/sec):993.8 #Correct:81 #Tested:162 Testing Accuracy:50.0%\rProgress:16.2% Speed(reviews/sec):993.9 #Correct:81 #Tested:163 Testing Accuracy:49.6%\rProgress:16.3% Speed(reviews/sec):1000. #Correct:82 #Tested:164 Testing Accuracy:50.0%\rProgress:16.4% Speed(reviews/sec):1000. #Correct:82 #Tested:165 Testing Accuracy:49.6%\rProgress:16.5% Speed(reviews/sec):1000. #Correct:83 #Tested:166 Testing Accuracy:50.0%\rProgress:16.6% Speed(reviews/sec):1006. #Correct:83 #Tested:167 Testing Accuracy:49.7%\rProgress:16.7% Speed(reviews/sec):1006. #Correct:84 #Tested:168 Testing Accuracy:50.0%\rProgress:16.8% Speed(reviews/sec):1006. #Correct:84 #Tested:169 Testing Accuracy:49.7%\rProgress:16.9% Speed(reviews/sec):1005. #Correct:85 #Tested:170 Testing Accuracy:50.0%\rProgress:17.0% Speed(reviews/sec):1005. #Correct:85 #Tested:171 Testing Accuracy:49.7%\rProgress:17.1% Speed(reviews/sec):1005. #Correct:86 #Tested:172 Testing Accuracy:50.0%\rProgress:17.2% Speed(reviews/sec):1005. #Correct:86 #Tested:173 Testing Accuracy:49.7%\rProgress:17.3% Speed(reviews/sec):1011. #Correct:87 #Tested:174 Testing Accuracy:50.0%\rProgress:17.4% Speed(reviews/sec):1011. #Correct:87 #Tested:175 Testing Accuracy:49.7%\rProgress:17.5% Speed(reviews/sec):1011. #Correct:88 #Tested:176 Testing Accuracy:50.0%\rProgress:17.6% Speed(reviews/sec):1011. #Correct:88 #Tested:177 Testing Accuracy:49.7%\rProgress:17.7% Speed(reviews/sec):1011. #Correct:89 #Tested:178 Testing Accuracy:50.0%\rProgress:17.8% Speed(reviews/sec):1011. #Correct:89 #Tested:179 Testing Accuracy:49.7%\rProgress:17.9% Speed(reviews/sec):1011. #Correct:90 #Tested:180 Testing Accuracy:50.0%\rProgress:18.0% Speed(reviews/sec):1011. #Correct:90 #Tested:181 Testing Accuracy:49.7%\rProgress:18.1% Speed(reviews/sec):1016. #Correct:91 #Tested:182 Testing Accuracy:50.0%\rProgress:18.2% Speed(reviews/sec):1016. #Correct:91 #Tested:183 Testing Accuracy:49.7%\rProgress:18.3% Speed(reviews/sec):1016. #Correct:92 #Tested:184 Testing Accuracy:50.0%\rProgress:18.4% Speed(reviews/sec):1016. #Correct:92 #Tested:185 Testing Accuracy:49.7%\rProgress:18.5% Speed(reviews/sec):1016. #Correct:93 #Tested:186 Testing Accuracy:50.0%\rProgress:18.6% Speed(reviews/sec):1016. #Correct:93 #Tested:187 Testing Accuracy:49.7%\rProgress:18.7% Speed(reviews/sec):1016. #Correct:94 #Tested:188 Testing Accuracy:50.0%\rProgress:18.8% Speed(reviews/sec):1016. #Correct:94 #Tested:189 Testing Accuracy:49.7%\rProgress:18.9% Speed(reviews/sec):1016. #Correct:95 #Tested:190 Testing Accuracy:50.0%\rProgress:19.0% Speed(reviews/sec):1021. #Correct:95 #Tested:191 Testing Accuracy:49.7%\rProgress:19.1% Speed(reviews/sec):1021. #Correct:96 #Tested:192 Testing Accuracy:50.0%\rProgress:19.2% Speed(reviews/sec):1021. #Correct:96 #Tested:193 Testing Accuracy:49.7%"
]
],
[
[
"Run the following cell to actually train the network. During training, it will display the model's accuracy repeatedly as it trains so you can see how well it's doing.",
"_____no_output_____"
]
],
[
[
"mlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):155.3 #Correct:1750 #Trained:2501 Training Accuracy:69.9%\nProgress:20.8% Speed(reviews/sec):155.6 #Correct:3591 #Trained:5001 Training Accuracy:71.8%\nProgress:31.2% Speed(reviews/sec):155.5 #Correct:5530 #Trained:7501 Training Accuracy:73.7%\nProgress:41.6% Speed(reviews/sec):155.6 #Correct:7478 #Trained:10001 Training Accuracy:74.7%\nProgress:52.0% Speed(reviews/sec):155.2 #Correct:9409 #Trained:12501 Training Accuracy:75.2%\nProgress:62.5% Speed(reviews/sec):154.9 #Correct:11349 #Trained:15001 Training Accuracy:75.6%\nProgress:72.9% Speed(reviews/sec):154.7 #Correct:13256 #Trained:17501 Training Accuracy:75.7%\nProgress:83.3% Speed(reviews/sec):153.0 #Correct:15195 #Trained:20001 Training Accuracy:75.9%\nProgress:93.7% Speed(reviews/sec):152.6 #Correct:17034 #Trained:22501 Training Accuracy:75.7%\nProgress:99.9% Speed(reviews/sec):152.6 #Correct:18220 #Trained:24000 Training Accuracy:75.9%"
]
],
[
[
"That most likely didn't train very well. Part of the reason may be because the learning rate is too high. Run the following cell to recreate the network with a smaller learning rate, `0.01`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):143.1 #Correct:1961 #Trained:2501 Training Accuracy:78.4%\nProgress:20.8% Speed(reviews/sec):141.0 #Correct:4001 #Trained:5001 Training Accuracy:80.0%\nProgress:31.2% Speed(reviews/sec):142.7 #Correct:6121 #Trained:7501 Training Accuracy:81.6%\nProgress:41.6% Speed(reviews/sec):137.3 #Correct:8272 #Trained:10001 Training Accuracy:82.7%\nProgress:52.0% Speed(reviews/sec):136.6 #Correct:10432 #Trained:12501 Training Accuracy:83.4%\nProgress:62.5% Speed(reviews/sec):135.8 #Correct:12566 #Trained:15001 Training Accuracy:83.7%\nProgress:72.9% Speed(reviews/sec):133.6 #Correct:14671 #Trained:17501 Training Accuracy:83.8%\nProgress:83.3% Speed(reviews/sec):121.7 #Correct:16833 #Trained:20001 Training Accuracy:84.1%\nProgress:93.7% Speed(reviews/sec):123.6 #Correct:19013 #Trained:22501 Training Accuracy:84.4%\nProgress:99.9% Speed(reviews/sec):125.0 #Correct:20333 #Trained:24000 Training Accuracy:84.7%"
]
],
[
[
"That probably wasn't much different. Run the following cell to recreate the network one more time with an even smaller learning rate, `0.001`, and then train the new network.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.001)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):156.9 #Correct:1941 #Trained:2501 Training Accuracy:77.6%\nProgress:20.8% Speed(reviews/sec):156.8 #Correct:3988 #Trained:5001 Training Accuracy:79.7%\nProgress:31.2% Speed(reviews/sec):156.4 #Correct:6085 #Trained:7501 Training Accuracy:81.1%\nProgress:41.6% Speed(reviews/sec):156.1 #Correct:8206 #Trained:10001 Training Accuracy:82.0%\nProgress:52.0% Speed(reviews/sec):155.7 #Correct:10337 #Trained:12501 Training Accuracy:82.6%\nProgress:62.5% Speed(reviews/sec):155.5 #Correct:12422 #Trained:15001 Training Accuracy:82.8%\nProgress:72.9% Speed(reviews/sec):155.3 #Correct:14528 #Trained:17501 Training Accuracy:83.0%\nProgress:83.3% Speed(reviews/sec):155.1 #Correct:16700 #Trained:20001 Training Accuracy:83.4%\nProgress:93.7% Speed(reviews/sec):154.7 #Correct:18860 #Trained:22501 Training Accuracy:83.8%\nProgress:99.9% Speed(reviews/sec):154.6 #Correct:20175 #Trained:24000 Training Accuracy:84.0%"
]
],
[
[
"With a learning rate of `0.001`, the network should finally have started to improve during training. It's still not very good, but it shows that this solution has potential. We will improve it in the next lesson.",
"_____no_output_____"
],
[
"# End of Project 3. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Understanding Neural Noise<a id='lesson_4'></a>\n\nThe following cells include includes the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image\nImage(filename='sentiment_network.png')",
"_____no_output_____"
],
[
"def update_input_layer(review):\n \n global layer_0\n \n # clear out previous state, reset the layer to be all 0s\n layer_0 *= 0\n for word in review.split(\" \"):\n layer_0[0][word2index[word]] += 1\n\nupdate_input_layer(reviews[0])",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"review_counter = Counter()",
"_____no_output_____"
],
[
"for word in reviews[0].split(\" \"):\n review_counter[word] += 1",
"_____no_output_____"
],
[
"review_counter.most_common()",
"_____no_output_____"
]
],
[
[
"# Project 4: Reducing Noise in Our Input Data<a id='project_4'></a>\n\n**TODO:** Attempt to reduce the noise in the input data like Andrew did in the previous video. Specifically, do the following:\n* Copy the `SentimentNetwork` class you created earlier into the following cell.\n* Modify `update_input_layer` so it does not count how many times each word is used, but rather just stores whether or not a word was used. ",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Projet 3 lesson\n# -Modify it to reduce noise, like in the video \nimport time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews, labels, hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n review_vocab = set()\n # TODO: populate review_vocab with all of the words in the given reviews\n # Remember to split reviews into individual words \n # using \"split(' ')\" instead of \"split()\".\n for review in reviews:\n for r in review.split(' '):\n review_vocab.add(r)\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n label_vocab = set()\n # TODO: populate label_vocab with all of the words in the given labels.\n # There is no need to split the labels because each one is a single word.\n for label in labels:\n label_vocab.add(label)\n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n # TODO: populate self.word2index with indices for all the words in self.review_vocab\n # like you saw earlier in the notebook\n for idx, word in enumerate(self.review_vocab):\n self.word2index[word] = idx\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n # TODO: do the same thing you did for self.word2index and self.review_vocab, \n # but for self.label2index and self.label_vocab instead\n for idx, label in enumerate(self.label_vocab):\n self.label2index[label] = idx\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Store the number of nodes in input, hidden, and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n \n # TODO: initialize self.weights_0_1 as a matrix of zeros. These are the weights between\n # the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((input_nodes, hidden_nodes))\n \n # TODO: initialize self.weights_1_2 as a matrix of random values. \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # TODO: Create the input layer, a two-dimensional matrix with shape \n # 1 x input_nodes, with all values initialized to zero\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n # TODO: You can copy most of the code you wrote for update_input_layer \n # earlier in this notebook. \n #\n # However, MAKE SURE YOU CHANGE ALL VARIABLES TO REFERENCE\n # THE VERSIONS STORED IN THIS OBJECT, NOT THE GLOBAL OBJECTS.\n # For example, replace \"layer_0 *= 0\" with \"self.layer_0 *= 0\"\n self.layer_0 *= 0\n \n for term in review.split(' '):\n if(term in self.word2index.keys()):\n self.layer_0[:,self.word2index[term]] = 1\n \n def get_target_for_label(self,label):\n # TODO: Copy the code you wrote for get_target_for_label \n # earlier in this notebook. \n return 1 if label == 'POSITIVE' else 0\n \n def sigmoid(self,x):\n # TODO: Return the result of calculating the sigmoid activation function\n # shown in the lectures\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n # TODO: Return the derivative of the sigmoid activation function, \n # where \"output\" is the original output from the sigmoid fucntion \n return output * (1 - output)\n\n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n \n # Remember when we started for printing time statistics\n start = time.time()\n\n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # TODO: Get the next review and its correct label\n self.update_input_layer(training_reviews[i])\n label = self.get_target_for_label(training_labels[i])\n # TODO: Implement the forward pass through the network. \n # That means use the given review to update the input layer, \n # then calculate values for the hidden layer,\n # and finally calculate the output layer.\n # \n # Do not use an activation function for the hidden layer,\n # but use the sigmoid activation function for the output layer.\n out_0_1 = np.dot(self.layer_0, self.weights_0_1)\n out_1_2 = self.sigmoid(np.dot(out_0_1, self.weights_1_2))\n \n # TODO: Implement the back propagation pass here. \n # That means calculate the error for the forward pass's prediction\n # and update the weights in the network according to their\n # contributions toward the error, as calculated via the\n # gradient descent and back propagation algorithms you \n # learned in class.\n error = out_1_2 - label\n \n layel_2_error = error * self.sigmoid_output_2_derivative(out_1_2)\n \n layer_1_error = np.dot(layel_2_error, self.weights_1_2.T)\n \n self.weights_1_2 -= np.dot(out_1_2, layel_2_error) * self.learning_rate\n self.weights_0_1 -= np.dot(self.layer_0.T, layer_1_error) * self.learning_rate\n \n # TODO: Keep track of correct predictions. To determine if the prediction was\n # correct, check that the absolute value of the output error \n # is less than 0.5. If so, add one to the correct_so_far count.\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n \n if(out_1_2 >= 0.5 and label == 1):\n correct_so_far += 1\n elif(out_1_2 < 0.5 and label == 0):\n correct_so_far += 1\n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # TODO: Run a forward pass through the network, like you did in the\n # \"train\" function. That means use the given review to \n # update the input layer, then calculate values for the hidden layer,\n # and finally calculate the output layer.\n #\n # Note: The review passed into this function for prediction \n # might come from anywhere, so you should convert it \n # to lower case prior to using it.\n \n # TODO: The output layer should now contain a prediction. \n # Return `POSITIVE` for predictions greater-than-or-equal-to `0.5`, \n # and `NEGATIVE` otherwise.\n self.update_input_layer(review)\n \n out_0_1 = np.dot(self.layer_0, self.weights_0_1)\n out_1_2 = self.sigmoid(np.dot(out_0_1, self.weights_1_2))\n \n if out_1_2 > 0.5:\n return 'POSITIVE'\n else:\n return 'NEGATIVE'\n",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it. Notice we've gone back to the higher learning rate of `0.1`.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"Progress:0.0% Speed(reviews/sec):0.0 #Correct:1 #Trained:1 Training Accuracy:100.%\nProgress:10.4% Speed(reviews/sec):133.2 #Correct:1750 #Trained:2501 Training Accuracy:69.9%\nProgress:20.8% Speed(reviews/sec):117.0 #Correct:3591 #Trained:5001 Training Accuracy:71.8%\nProgress:31.2% Speed(reviews/sec):114.9 #Correct:5530 #Trained:7501 Training Accuracy:73.7%\nProgress:41.6% Speed(reviews/sec):115.9 #Correct:7478 #Trained:10001 Training Accuracy:74.7%\nProgress:52.0% Speed(reviews/sec):118.7 #Correct:9409 #Trained:12501 Training Accuracy:75.2%\nProgress:62.5% Speed(reviews/sec):123.4 #Correct:11349 #Trained:15001 Training Accuracy:75.6%\nProgress:72.9% Speed(reviews/sec):126.9 #Correct:13256 #Trained:17501 Training Accuracy:75.7%\nProgress:83.3% Speed(reviews/sec):128.8 #Correct:15195 #Trained:20001 Training Accuracy:75.9%\nProgress:93.7% Speed(reviews/sec):130.2 #Correct:17034 #Trained:22501 Training Accuracy:75.7%\nProgress:99.9% Speed(reviews/sec):131.3 #Correct:18220 #Trained:24000 Training Accuracy:75.9%"
]
],
[
[
"That should have trained much better than the earlier attempts. It's still not wonderful, but it should have improved dramatically. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"\rProgress:0.0% Speed(reviews/sec):0 #Correct:1 #Tested:1 Testing Accuracy:100.%\rProgress:0.1% Speed(reviews/sec):506.8 #Correct:1 #Tested:2 Testing Accuracy:50.0%\rProgress:0.2% Speed(reviews/sec):1013. #Correct:2 #Tested:3 Testing Accuracy:66.6%\rProgress:0.3% Speed(reviews/sec):755.5 #Correct:3 #Tested:4 Testing Accuracy:75.0%\rProgress:0.4% Speed(reviews/sec):1007. #Correct:4 #Tested:5 Testing Accuracy:80.0%\rProgress:0.5% Speed(reviews/sec):837.6 #Correct:5 #Tested:6 Testing Accuracy:83.3%\rProgress:0.6% Speed(reviews/sec):860.5 #Correct:6 #Tested:7 Testing Accuracy:85.7%\rProgress:0.7% Speed(reviews/sec):878.4 #Correct:7 #Tested:8 Testing Accuracy:87.5%\rProgress:0.8% Speed(reviews/sec):891.9 #Correct:8 #Tested:9 Testing Accuracy:88.8%\rProgress:0.9% Speed(reviews/sec):902.8 #Correct:9 #Tested:10 Testing Accuracy:90.0%\rProgress:1.0% Speed(reviews/sec):911.7 #Correct:10 #Tested:11 Testing Accuracy:90.9%\rProgress:1.1% Speed(reviews/sec):919.2 #Correct:11 #Tested:12 Testing Accuracy:91.6%\rProgress:1.2% Speed(reviews/sec):925.4 #Correct:12 #Tested:13 Testing Accuracy:92.3%\rProgress:1.3% Speed(reviews/sec):930.7 #Correct:13 #Tested:14 Testing Accuracy:92.8%\rProgress:1.4% Speed(reviews/sec):935.1 #Correct:14 #Tested:15 Testing Accuracy:93.3%\rProgress:1.5% Speed(reviews/sec):939.3 #Correct:15 #Tested:16 Testing Accuracy:93.7%\rProgress:1.6% Speed(reviews/sec):942.9 #Correct:16 #Tested:17 Testing Accuracy:94.1%\rProgress:1.7% Speed(reviews/sec):946.1 #Correct:17 #Tested:18 Testing Accuracy:94.4%\rProgress:1.8% Speed(reviews/sec):948.9 #Correct:18 #Tested:19 Testing Accuracy:94.7%\rProgress:1.9% Speed(reviews/sec):906.0 #Correct:18 #Tested:20 Testing Accuracy:90.0%\rProgress:2.0% Speed(reviews/sec):910.3 #Correct:19 #Tested:21 Testing Accuracy:90.4%\rProgress:2.1% Speed(reviews/sec):914.2 #Correct:20 #Tested:22 Testing Accuracy:90.9%\rProgress:2.2% Speed(reviews/sec):917.9 #Correct:21 #Tested:23 Testing Accuracy:91.3%\rProgress:2.3% Speed(reviews/sec):921.2 #Correct:22 #Tested:24 Testing Accuracy:91.6%\rProgress:2.4% Speed(reviews/sec):889.9 #Correct:23 #Tested:25 Testing Accuracy:92.0%\rProgress:2.5% Speed(reviews/sec):893.8 #Correct:24 #Tested:26 Testing Accuracy:92.3%\rProgress:2.6% Speed(reviews/sec):897.5 #Correct:25 #Tested:27 Testing Accuracy:92.5%\rProgress:2.7% Speed(reviews/sec):871.9 #Correct:26 #Tested:28 Testing Accuracy:92.8%\rProgress:2.8% Speed(reviews/sec):849.2 #Correct:27 #Tested:29 Testing Accuracy:93.1%\rProgress:2.9% Speed(reviews/sec):853.6 #Correct:28 #Tested:30 Testing Accuracy:93.3%\rProgress:3.0% Speed(reviews/sec):834.0 #Correct:29 #Tested:31 Testing Accuracy:93.5%\rProgress:3.1% Speed(reviews/sec):861.8 #Correct:30 #Tested:32 Testing Accuracy:93.7%\rProgress:3.2% Speed(reviews/sec):865.6 #Correct:31 #Tested:33 Testing Accuracy:93.9%\rProgress:3.3% Speed(reviews/sec):869.0 #Correct:31 #Tested:34 Testing Accuracy:91.1%\rProgress:3.4% Speed(reviews/sec):872.4 #Correct:32 #Tested:35 Testing Accuracy:91.4%\rProgress:3.5% Speed(reviews/sec):875.5 #Correct:32 #Tested:36 Testing Accuracy:88.8%\rProgress:3.6% Speed(reviews/sec):878.6 #Correct:33 #Tested:37 Testing Accuracy:89.1%\rProgress:3.7% Speed(reviews/sec):903.0 #Correct:33 #Tested:38 Testing Accuracy:86.8%\rProgress:3.8% Speed(reviews/sec):905.3 #Correct:34 #Tested:39 Testing Accuracy:87.1%\rProgress:3.9% Speed(reviews/sec):907.6 #Correct:34 #Tested:40 Testing Accuracy:85.0%\rProgress:4.0% Speed(reviews/sec):909.7 #Correct:35 #Tested:41 Testing Accuracy:85.3%\rProgress:4.1% Speed(reviews/sec):911.6 #Correct:36 #Tested:42 Testing Accuracy:85.7%\rProgress:4.2% Speed(reviews/sec):913.6 #Correct:37 #Tested:43 Testing Accuracy:86.0%\rProgress:4.3% Speed(reviews/sec):915.4 #Correct:38 #Tested:44 Testing Accuracy:86.3%\rProgress:4.4% Speed(reviews/sec):898.5 #Correct:39 #Tested:45 Testing Accuracy:86.6%\rProgress:4.5% Speed(reviews/sec):865.8 #Correct:39 #Tested:46 Testing Accuracy:84.7%\rProgress:4.6% Speed(reviews/sec):868.4 #Correct:40 #Tested:47 Testing Accuracy:85.1%\rProgress:4.7% Speed(reviews/sec):870.8 #Correct:41 #Tested:48 Testing Accuracy:85.4%\rProgress:4.8% Speed(reviews/sec):857.5 #Correct:42 #Tested:49 Testing Accuracy:85.7%\rProgress:4.9% Speed(reviews/sec):844.9 #Correct:43 #Tested:50 Testing Accuracy:86.0%\rProgress:5.0% Speed(reviews/sec):847.8 #Correct:44 #Tested:51 Testing Accuracy:86.2%\rProgress:5.1% Speed(reviews/sec):850.4 #Correct:45 #Tested:52 Testing Accuracy:86.5%\rProgress:5.2% Speed(reviews/sec):852.9 #Correct:46 #Tested:53 Testing Accuracy:86.7%\rProgress:5.3% Speed(reviews/sec):841.7 #Correct:47 #Tested:54 Testing Accuracy:87.0%\rProgress:5.4% Speed(reviews/sec):857.5 #Correct:48 #Tested:55 Testing Accuracy:87.2%\rProgress:5.5% Speed(reviews/sec):859.8 #Correct:49 #Tested:56 Testing Accuracy:87.5%\rProgress:5.6% Speed(reviews/sec):848.9 #Correct:50 #Tested:57 Testing Accuracy:87.7%\rProgress:5.7% Speed(reviews/sec):864.0 #Correct:51 #Tested:58 Testing Accuracy:87.9%\rProgress:5.8% Speed(reviews/sec):866.0 #Correct:51 #Tested:59 Testing Accuracy:86.4%\rProgress:5.9% Speed(reviews/sec):868.0 #Correct:52 #Tested:60 Testing Accuracy:86.6%\rProgress:6.0% Speed(reviews/sec):857.4 #Correct:53 #Tested:61 Testing Accuracy:86.8%\rProgress:6.1% Speed(reviews/sec):859.5 #Correct:54 #Tested:62 Testing Accuracy:87.0%\rProgress:6.2% Speed(reviews/sec):861.5 #Correct:55 #Tested:63 Testing Accuracy:87.3%\rProgress:6.3% Speed(reviews/sec):863.4 #Correct:56 #Tested:64 Testing Accuracy:87.5%\rProgress:6.4% Speed(reviews/sec):853.7 #Correct:57 #Tested:65 Testing Accuracy:87.6%\rProgress:6.5% Speed(reviews/sec):855.6 #Correct:57 #Tested:66 Testing Accuracy:86.3%\rProgress:6.6% Speed(reviews/sec):868.7 #Correct:58 #Tested:67 Testing Accuracy:86.5%\rProgress:6.7% Speed(reviews/sec):859.3 #Correct:59 #Tested:68 Testing Accuracy:86.7%\rProgress:6.8% Speed(reviews/sec):861.0 #Correct:60 #Tested:69 Testing Accuracy:86.9%\rProgress:6.9% Speed(reviews/sec):862.7 #Correct:61 #Tested:70 Testing Accuracy:87.1%\rProgress:7.0% Speed(reviews/sec):864.5 #Correct:61 #Tested:71 Testing Accuracy:85.9%\rProgress:7.1% Speed(reviews/sec):866.1 #Correct:61 #Tested:72 Testing Accuracy:84.7%\rProgress:7.2% Speed(reviews/sec):867.8 #Correct:62 #Tested:73 Testing Accuracy:84.9%\rProgress:7.3% Speed(reviews/sec):869.3 #Correct:63 #Tested:74 Testing Accuracy:85.1%\rProgress:7.4% Speed(reviews/sec):870.9 #Correct:64 #Tested:75 Testing Accuracy:85.3%\rProgress:7.5% Speed(reviews/sec):872.4 #Correct:65 #Tested:76 Testing Accuracy:85.5%\rProgress:7.6% Speed(reviews/sec):863.9 #Correct:66 #Tested:77 Testing Accuracy:85.7%\rProgress:7.7% Speed(reviews/sec):865.3 #Correct:67 #Tested:78 Testing Accuracy:85.8%\rProgress:7.8% Speed(reviews/sec):866.9 #Correct:68 #Tested:79 Testing Accuracy:86.0%\rProgress:7.9% Speed(reviews/sec):868.4 #Correct:68 #Tested:80 Testing Accuracy:85.0%\rProgress:8.0% Speed(reviews/sec):879.4 #Correct:69 #Tested:81 Testing Accuracy:85.1%\rProgress:8.1% Speed(reviews/sec):871.2 #Correct:69 #Tested:82 Testing Accuracy:84.1%\rProgress:8.2% Speed(reviews/sec):872.6 #Correct:70 #Tested:83 Testing Accuracy:84.3%\rProgress:8.3% Speed(reviews/sec):873.9 #Correct:70 #Tested:84 Testing Accuracy:83.3%\rProgress:8.4% Speed(reviews/sec):884.5 #Correct:71 #Tested:85 Testing Accuracy:83.5%\rProgress:8.5% Speed(reviews/sec):885.7 #Correct:72 #Tested:86 Testing Accuracy:83.7%\rProgress:8.6% Speed(reviews/sec):886.8 #Correct:73 #Tested:87 Testing Accuracy:83.9%\rProgress:8.7% Speed(reviews/sec):897.1 #Correct:73 #Tested:88 Testing Accuracy:82.9%\rProgress:8.8% Speed(reviews/sec):898.2 #Correct:74 #Tested:89 Testing Accuracy:83.1%\rProgress:8.9% Speed(reviews/sec):899.2 #Correct:75 #Tested:90 Testing Accuracy:83.3%\rProgress:9.0% Speed(reviews/sec):900.2 #Correct:76 #Tested:91 Testing Accuracy:83.5%\rProgress:9.1% Speed(reviews/sec):901.2 #Correct:77 #Tested:92 Testing Accuracy:83.6%\rProgress:9.2% Speed(reviews/sec):893.2 #Correct:78 #Tested:93 Testing Accuracy:83.8%\rProgress:9.3% Speed(reviews/sec):894.5 #Correct:79 #Tested:94 Testing Accuracy:84.0%\rProgress:9.4% Speed(reviews/sec):895.5 #Correct:80 #Tested:95 Testing Accuracy:84.2%\rProgress:9.5% Speed(reviews/sec):896.5 #Correct:81 #Tested:96 Testing Accuracy:84.3%\rProgress:9.6% Speed(reviews/sec):905.9 #Correct:82 #Tested:97 Testing Accuracy:84.5%\rProgress:9.7% Speed(reviews/sec):898.4 #Correct:83 #Tested:98 Testing Accuracy:84.6%\rProgress:9.8% Speed(reviews/sec):907.6 #Correct:84 #Tested:99 Testing Accuracy:84.8%\rProgress:9.9% Speed(reviews/sec):908.5 #Correct:85 #Tested:100 Testing Accuracy:85.0%\rProgress:10.0% Speed(reviews/sec):909.3 #Correct:86 #Tested:101 Testing Accuracy:85.1%\rProgress:10.1% Speed(reviews/sec):910.1 #Correct:87 #Tested:102 Testing Accuracy:85.2%\rProgress:10.2% Speed(reviews/sec):910.9 #Correct:88 #Tested:103 Testing Accuracy:85.4%\rProgress:10.3% Speed(reviews/sec):911.7 #Correct:88 #Tested:104 Testing Accuracy:84.6%\rProgress:10.4% Speed(reviews/sec):912.5 #Correct:89 #Tested:105 Testing Accuracy:84.7%\rProgress:10.5% Speed(reviews/sec):921.3 #Correct:90 #Tested:106 Testing Accuracy:84.9%\rProgress:10.6% Speed(reviews/sec):922.0 #Correct:91 #Tested:107 Testing Accuracy:85.0%\rProgress:10.7% Speed(reviews/sec):922.6 #Correct:91 #Tested:108 Testing Accuracy:84.2%\rProgress:10.8% Speed(reviews/sec):923.3 #Correct:92 #Tested:109 Testing Accuracy:84.4%\rProgress:10.9% Speed(reviews/sec):923.9 #Correct:92 #Tested:110 Testing Accuracy:83.6%\rProgress:11.0% Speed(reviews/sec):924.6 #Correct:93 #Tested:111 Testing Accuracy:83.7%\rProgress:11.1% Speed(reviews/sec):925.2 #Correct:93 #Tested:112 Testing Accuracy:83.0%\rProgress:11.2% Speed(reviews/sec):925.8 #Correct:94 #Tested:113 Testing Accuracy:83.1%\rProgress:11.3% Speed(reviews/sec):926.4 #Correct:95 #Tested:114 Testing Accuracy:83.3%\rProgress:11.4% Speed(reviews/sec):927.0 #Correct:96 #Tested:115 Testing Accuracy:83.4%\rProgress:11.5% Speed(reviews/sec):935.2 #Correct:96 #Tested:116 Testing Accuracy:82.7%\rProgress:11.6% Speed(reviews/sec):935.7 #Correct:96 #Tested:117 Testing Accuracy:82.0%\rProgress:11.7% Speed(reviews/sec):936.2 #Correct:97 #Tested:118 Testing Accuracy:82.2%\rProgress:11.8% Speed(reviews/sec):944.2 #Correct:98 #Tested:119 Testing Accuracy:82.3%\rProgress:11.9% Speed(reviews/sec):944.6 #Correct:99 #Tested:120 Testing Accuracy:82.5%\rProgress:12.0% Speed(reviews/sec):945.1 #Correct:100 #Tested:121 Testing Accuracy:82.6%\rProgress:12.1% Speed(reviews/sec):952.9 #Correct:101 #Tested:122 Testing Accuracy:82.7%\rProgress:12.2% Speed(reviews/sec):953.3 #Correct:102 #Tested:123 Testing Accuracy:82.9%\rProgress:12.3% Speed(reviews/sec):946.3 #Correct:103 #Tested:124 Testing Accuracy:83.0%\rProgress:12.4% Speed(reviews/sec):954.0 #Correct:104 #Tested:125 Testing Accuracy:83.2%\rProgress:12.5% Speed(reviews/sec):954.4 #Correct:104 #Tested:126 Testing Accuracy:82.5%\rProgress:12.6% Speed(reviews/sec):954.7 #Correct:105 #Tested:127 Testing Accuracy:82.6%\rProgress:12.7% Speed(reviews/sec):962.3 #Correct:106 #Tested:128 Testing Accuracy:82.8%\rProgress:12.8% Speed(reviews/sec):962.6 #Correct:107 #Tested:129 Testing Accuracy:82.9%\rProgress:12.9% Speed(reviews/sec):962.9 #Correct:108 #Tested:130 Testing Accuracy:83.0%\rProgress:13.0% Speed(reviews/sec):970.3 #Correct:108 #Tested:131 Testing Accuracy:82.4%\rProgress:13.1% Speed(reviews/sec):970.5 #Correct:109 #Tested:132 Testing Accuracy:82.5%\rProgress:13.2% Speed(reviews/sec):970.8 #Correct:110 #Tested:133 Testing Accuracy:82.7%\rProgress:13.3% Speed(reviews/sec):978.1 #Correct:111 #Tested:134 Testing Accuracy:82.8%\rProgress:13.4% Speed(reviews/sec):978.3 #Correct:112 #Tested:135 Testing Accuracy:82.9%\rProgress:13.5% Speed(reviews/sec):978.4 #Correct:113 #Tested:136 Testing Accuracy:83.0%\rProgress:13.6% Speed(reviews/sec):985.7 #Correct:114 #Tested:137 Testing Accuracy:83.2%\rProgress:13.7% Speed(reviews/sec):985.8 #Correct:115 #Tested:138 Testing Accuracy:83.3%\rProgress:13.8% Speed(reviews/sec):993.0 #Correct:116 #Tested:139 Testing Accuracy:83.4%\rProgress:13.9% Speed(reviews/sec):993.0 #Correct:117 #Tested:140 Testing Accuracy:83.5%\rProgress:14.0% Speed(reviews/sec):993.1 #Correct:118 #Tested:141 Testing Accuracy:83.6%\rProgress:14.1% Speed(reviews/sec):1000. #Correct:118 #Tested:142 Testing Accuracy:83.0%\rProgress:14.2% Speed(reviews/sec):1000. #Correct:119 #Tested:143 Testing Accuracy:83.2%\rProgress:14.3% Speed(reviews/sec):1000. #Correct:120 #Tested:144 Testing Accuracy:83.3%\rProgress:14.4% Speed(reviews/sec):1007. #Correct:121 #Tested:145 Testing Accuracy:83.4%\rProgress:14.5% Speed(reviews/sec):1007. #Correct:122 #Tested:146 Testing Accuracy:83.5%\rProgress:14.6% Speed(reviews/sec):1007. #Correct:123 #Tested:147 Testing Accuracy:83.6%\rProgress:14.7% Speed(reviews/sec):1014. #Correct:124 #Tested:148 Testing Accuracy:83.7%\rProgress:14.8% Speed(reviews/sec):1013. #Correct:125 #Tested:149 Testing Accuracy:83.8%\rProgress:14.9% Speed(reviews/sec):1013. #Correct:126 #Tested:150 Testing Accuracy:84.0%\rProgress:15.0% Speed(reviews/sec):1020. #Correct:127 #Tested:151 Testing Accuracy:84.1%\rProgress:15.1% Speed(reviews/sec):1020. #Correct:127 #Tested:152 Testing Accuracy:83.5%\rProgress:15.2% Speed(reviews/sec):1020. #Correct:128 #Tested:153 Testing Accuracy:83.6%\rProgress:15.3% Speed(reviews/sec):1020. #Correct:129 #Tested:154 Testing Accuracy:83.7%\rProgress:15.4% Speed(reviews/sec):1026. #Correct:130 #Tested:155 Testing Accuracy:83.8%\rProgress:15.5% Speed(reviews/sec):1026. #Correct:131 #Tested:156 Testing Accuracy:83.9%\rProgress:15.6% Speed(reviews/sec):1026. #Correct:132 #Tested:157 Testing Accuracy:84.0%\rProgress:15.7% Speed(reviews/sec):1026. #Correct:133 #Tested:158 Testing Accuracy:84.1%\rProgress:15.8% Speed(reviews/sec):1032. #Correct:133 #Tested:159 Testing Accuracy:83.6%\rProgress:15.9% Speed(reviews/sec):1032. #Correct:134 #Tested:160 Testing Accuracy:83.7%\rProgress:16.0% Speed(reviews/sec):1032. #Correct:134 #Tested:161 Testing Accuracy:83.2%\rProgress:16.1% Speed(reviews/sec):1038. #Correct:134 #Tested:162 Testing Accuracy:82.7%\rProgress:16.2% Speed(reviews/sec):1038. #Correct:135 #Tested:163 Testing Accuracy:82.8%\rProgress:16.3% Speed(reviews/sec):1038. #Correct:136 #Tested:164 Testing Accuracy:82.9%\rProgress:16.4% Speed(reviews/sec):1044. #Correct:137 #Tested:165 Testing Accuracy:83.0%\rProgress:16.5% Speed(reviews/sec):1044. #Correct:138 #Tested:166 Testing Accuracy:83.1%\rProgress:16.6% Speed(reviews/sec):1044. #Correct:138 #Tested:167 Testing Accuracy:82.6%\rProgress:16.7% Speed(reviews/sec):1050. #Correct:139 #Tested:168 Testing Accuracy:82.7%\rProgress:16.8% Speed(reviews/sec):1050. #Correct:140 #Tested:169 Testing Accuracy:82.8%\rProgress:16.9% Speed(reviews/sec):1049. #Correct:141 #Tested:170 Testing Accuracy:82.9%\rProgress:17.0% Speed(reviews/sec):1056. #Correct:141 #Tested:171 Testing Accuracy:82.4%\rProgress:17.1% Speed(reviews/sec):1055. #Correct:142 #Tested:172 Testing Accuracy:82.5%\rProgress:17.2% Speed(reviews/sec):1055. #Correct:143 #Tested:173 Testing Accuracy:82.6%\rProgress:17.3% Speed(reviews/sec):1055. #Correct:143 #Tested:174 Testing Accuracy:82.1%\rProgress:17.4% Speed(reviews/sec):1054. #Correct:144 #Tested:175 Testing Accuracy:82.2%\rProgress:17.5% Speed(reviews/sec):1054. #Correct:145 #Tested:176 Testing Accuracy:82.3%\rProgress:17.6% Speed(reviews/sec):1054. #Correct:146 #Tested:177 Testing Accuracy:82.4%\rProgress:17.7% Speed(reviews/sec):1053. #Correct:147 #Tested:178 Testing Accuracy:82.5%\rProgress:17.8% Speed(reviews/sec):1053. #Correct:148 #Tested:179 Testing Accuracy:82.6%\rProgress:17.9% Speed(reviews/sec):1059. #Correct:149 #Tested:180 Testing Accuracy:82.7%\rProgress:18.0% Speed(reviews/sec):1059. #Correct:150 #Tested:181 Testing Accuracy:82.8%\rProgress:18.1% Speed(reviews/sec):1058. #Correct:150 #Tested:182 Testing Accuracy:82.4%\rProgress:18.2% Speed(reviews/sec):1058. #Correct:151 #Tested:183 Testing Accuracy:82.5%\rProgress:18.3% Speed(reviews/sec):1058. #Correct:152 #Tested:184 Testing Accuracy:82.6%\rProgress:18.4% Speed(reviews/sec):1057. #Correct:153 #Tested:185 Testing Accuracy:82.7%\rProgress:18.5% Speed(reviews/sec):1057. #Correct:154 #Tested:186 Testing Accuracy:82.7%\rProgress:18.6% Speed(reviews/sec):1063. #Correct:155 #Tested:187 Testing Accuracy:82.8%\rProgress:18.7% Speed(reviews/sec):1062. #Correct:156 #Tested:188 Testing Accuracy:82.9%\rProgress:18.8% Speed(reviews/sec):1062. #Correct:157 #Tested:189 Testing Accuracy:83.0%\rProgress:18.9% Speed(reviews/sec):1061. #Correct:158 #Tested:190 Testing Accuracy:83.1%"
]
],
[
[
"# End of Project 4. \n## Andrew's solution was actually in the previous video, so rewatch that video if you had any problems with that project. Then continue on to the next lesson.\n# Analyzing Inefficiencies in our Network<a id='lesson_5'></a>\nThe following cells include the code Andrew shows in the next video. We've included it here so you can run the cells along with the video without having to type in everything.",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"layer_0 = np.zeros(10)",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"layer_0[4] = 1\nlayer_0[9] = 1",
"_____no_output_____"
],
[
"layer_0",
"_____no_output_____"
],
[
"weights_0_1 = np.random.randn(10,5)",
"_____no_output_____"
],
[
"layer_0.dot(weights_0_1)",
"_____no_output_____"
],
[
"indices = [4,9]",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (1 * weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"layer_1 = np.zeros(5)",
"_____no_output_____"
],
[
"for index in indices:\n layer_1 += (weights_0_1[index])",
"_____no_output_____"
],
[
"layer_1",
"_____no_output_____"
]
],
[
[
"# Project 5: Making our Network More Efficient<a id='project_5'></a>\n**TODO:** Make the `SentimentNetwork` class more efficient by eliminating unnecessary multiplications and additions that occur during forward and backward propagation. To do that, you can do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Remove the `update_input_layer` function - you will not need it in this version.\n* Modify `init_network`:\n>* You no longer need a separate input layer, so remove any mention of `self.layer_0`\n>* You will be dealing with the old hidden layer more directly, so create `self.layer_1`, a two-dimensional matrix with shape 1 x hidden_nodes, with all values initialized to zero\n* Modify `train`:\n>* Change the name of the input parameter `training_reviews` to `training_reviews_raw`. This will help with the next step.\n>* At the beginning of the function, you'll want to preprocess your reviews to convert them to a list of indices (from `word2index`) that are actually used in the review. This is equivalent to what you saw in the video when Andrew set specific indices to 1. Your code should create a local `list` variable named `training_reviews` that should contain a `list` for each review in `training_reviews_raw`. Those lists should contain the indices for words found in the review.\n>* Remove call to `update_input_layer`\n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* In the forward pass, replace the code that updates `layer_1` with new logic that only adds the weights for the indices used in the review.\n>* When updating `weights_0_1`, only update the individual weights that were used in the forward pass.\n* Modify `run`:\n>* Remove call to `update_input_layer` \n>* Use `self`'s `layer_1` instead of a local `layer_1` object.\n>* Much like you did in `train`, you will need to pre-process the `review` so you can work with word indices, then update `layer_1` by adding weights for the indices used in the review.",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Project 4 lesson\n# -Modify it according to the above instructions \nimport time\nimport sys\nimport numpy as np\n\n# Encapsulate our neural network in a class\nclass SentimentNetwork:\n def __init__(self, reviews, labels, hidden_nodes = 10, learning_rate = 0.1):\n \"\"\"Create a SentimenNetwork with the given settings\n Args:\n reviews(list) - List of reviews used for training\n labels(list) - List of POSITIVE/NEGATIVE labels associated with the given reviews\n hidden_nodes(int) - Number of nodes to create in the hidden layer\n learning_rate(float) - Learning rate to use while training\n \n \"\"\"\n # Assign a seed to our random number generator to ensure we get\n # reproducable results during development \n np.random.seed(1)\n\n # process the reviews and their associated labels so that everything\n # is ready for training\n self.pre_process_data(reviews, labels)\n \n # Build the network to have the number of hidden nodes and the learning rate that\n # were passed into this initializer. Make the same number of input nodes as\n # there are vocabulary words and create a single output node.\n self.init_network(len(self.review_vocab),hidden_nodes, 1, learning_rate)\n\n def pre_process_data(self, reviews, labels):\n \n review_vocab = set()\n # TODO: populate review_vocab with all of the words in the given reviews\n # Remember to split reviews into individual words \n # using \"split(' ')\" instead of \"split()\".\n for review in reviews:\n for r in review.split(' '):\n review_vocab.add(r)\n # Convert the vocabulary set to a list so we can access words via indices\n self.review_vocab = list(review_vocab)\n \n label_vocab = set()\n # TODO: populate label_vocab with all of the words in the given labels.\n # There is no need to split the labels because each one is a single word.\n for label in labels:\n label_vocab.add(label)\n # Convert the label vocabulary set to a list so we can access labels via indices\n self.label_vocab = list(label_vocab)\n \n # Store the sizes of the review and label vocabularies.\n self.review_vocab_size = len(self.review_vocab)\n self.label_vocab_size = len(self.label_vocab)\n \n # Create a dictionary of words in the vocabulary mapped to index positions\n self.word2index = {}\n # TODO: populate self.word2index with indices for all the words in self.review_vocab\n # like you saw earlier in the notebook\n for idx, word in enumerate(self.review_vocab):\n self.word2index[word] = idx\n \n # Create a dictionary of labels mapped to index positions\n self.label2index = {}\n # TODO: do the same thing you did for self.word2index and self.review_vocab, \n # but for self.label2index and self.label_vocab instead\n for idx, label in enumerate(self.label_vocab):\n self.label2index[label] = idx\n \n def init_network(self, input_nodes, hidden_nodes, output_nodes, learning_rate):\n # Store the number of nodes in input, hidden, and output layers.\n self.input_nodes = input_nodes\n self.hidden_nodes = hidden_nodes\n self.output_nodes = output_nodes\n\n # Store the learning rate\n self.learning_rate = learning_rate\n\n # Initialize weights\n \n # TODO: initialize self.weights_0_1 as a matrix of zeros. These are the weights between\n # the input layer and the hidden layer.\n self.weights_0_1 = np.zeros((input_nodes, hidden_nodes))\n \n # TODO: initialize self.weights_1_2 as a matrix of random values. \n # These are the weights between the hidden layer and the output layer.\n self.weights_1_2 = np.random.normal(0.0, self.output_nodes**-0.5, \n (self.hidden_nodes, self.output_nodes))\n \n # TODO: Create the input layer, a two-dimensional matrix with shape \n # 1 x input_nodes, with all values initialized to zero\n self.layer_0 = np.zeros((1,input_nodes))\n \n \n def update_input_layer(self,review):\n # TODO: You can copy most of the code you wrote for update_input_layer \n # earlier in this notebook. \n #\n # However, MAKE SURE YOU CHANGE ALL VARIABLES TO REFERENCE\n # THE VERSIONS STORED IN THIS OBJECT, NOT THE GLOBAL OBJECTS.\n # For example, replace \"layer_0 *= 0\" with \"self.layer_0 *= 0\"\n self.layer_0 *= 0\n \n for term in review.split(' '):\n if(term in self.word2index.keys()):\n self.layer_0[:,self.word2index[term]] = 1\n \n def get_target_for_label(self,label):\n # TODO: Copy the code you wrote for get_target_for_label \n # earlier in this notebook. \n return 1 if label == 'POSITIVE' else 0\n \n def sigmoid(self,x):\n # TODO: Return the result of calculating the sigmoid activation function\n # shown in the lectures\n return 1 / (1 + np.exp(-x))\n \n def sigmoid_output_2_derivative(self,output):\n # TODO: Return the derivative of the sigmoid activation function, \n # where \"output\" is the original output from the sigmoid fucntion \n return output * (1 - output)\n\n def train(self, training_reviews, training_labels):\n \n # make sure out we have a matching number of reviews and labels\n assert(len(training_reviews) == len(training_labels))\n \n # Keep track of correct predictions to display accuracy during training \n correct_so_far = 0\n \n # Remember when we started for printing time statistics\n start = time.time()\n\n # loop through all the given reviews and run a forward and backward pass,\n # updating weights for every item\n for i in range(len(training_reviews)):\n \n # TODO: Get the next review and its correct label\n self.update_input_layer(training_reviews[i])\n label = self.get_target_for_label(training_labels[i])\n # TODO: Implement the forward pass through the network. \n # That means use the given review to update the input layer, \n # then calculate values for the hidden layer,\n # and finally calculate the output layer.\n # \n # Do not use an activation function for the hidden layer,\n # but use the sigmoid activation function for the output layer.\n out_0_1 = np.dot(self.layer_0, self.weights_0_1)\n out_1_2 = self.sigmoid(np.dot(out_0_1, self.weights_1_2))\n \n # TODO: Implement the back propagation pass here. \n # That means calculate the error for the forward pass's prediction\n # and update the weights in the network according to their\n # contributions toward the error, as calculated via the\n # gradient descent and back propagation algorithms you \n # learned in class.\n error = out_1_2 - label\n \n layel_2_error = error * self.sigmoid_output_2_derivative(out_1_2)\n \n layer_1_error = np.dot(layel_2_error, self.weights_1_2.T)\n \n self.weights_1_2 -= np.dot(out_1_2, layel_2_error) * self.learning_rate\n self.weights_0_1 -= np.dot(self.layer_0.T, layer_1_error) * self.learning_rate\n \n # TODO: Keep track of correct predictions. To determine if the prediction was\n # correct, check that the absolute value of the output error \n # is less than 0.5. If so, add one to the correct_so_far count.\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the training process. \n \n if(out_1_2 >= 0.5 and label == 1):\n correct_so_far += 1\n elif(out_1_2 < 0.5 and label == 0):\n correct_so_far += 1\n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(training_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct_so_far) + \" #Trained:\" + str(i+1) \\\n + \" Training Accuracy:\" + str(correct_so_far * 100 / float(i+1))[:4] + \"%\")\n if(i % 2500 == 0):\n print(\"\")\n \n def test(self, testing_reviews, testing_labels):\n \"\"\"\n Attempts to predict the labels for the given testing_reviews,\n and uses the test_labels to calculate the accuracy of those predictions.\n \"\"\"\n \n # keep track of how many correct predictions we make\n correct = 0\n\n # we'll time how many predictions per second we make\n start = time.time()\n\n # Loop through each of the given reviews and call run to predict\n # its label. \n for i in range(len(testing_reviews)):\n pred = self.run(testing_reviews[i])\n if(pred == testing_labels[i]):\n correct += 1\n \n # For debug purposes, print out our prediction accuracy and speed \n # throughout the prediction process. \n\n elapsed_time = float(time.time() - start)\n reviews_per_second = i / elapsed_time if elapsed_time > 0 else 0\n \n sys.stdout.write(\"\\rProgress:\" + str(100 * i/float(len(testing_reviews)))[:4] \\\n + \"% Speed(reviews/sec):\" + str(reviews_per_second)[0:5] \\\n + \" #Correct:\" + str(correct) + \" #Tested:\" + str(i+1) \\\n + \" Testing Accuracy:\" + str(correct * 100 / float(i+1))[:4] + \"%\")\n \n def run(self, review):\n \"\"\"\n Returns a POSITIVE or NEGATIVE prediction for the given review.\n \"\"\"\n # TODO: Run a forward pass through the network, like you did in the\n # \"train\" function. That means use the given review to \n # update the input layer, then calculate values for the hidden layer,\n # and finally calculate the output layer.\n #\n # Note: The review passed into this function for prediction \n # might come from anywhere, so you should convert it \n # to lower case prior to using it.\n \n # TODO: The output layer should now contain a prediction. \n # Return `POSITIVE` for predictions greater-than-or-equal-to `0.5`, \n # and `NEGATIVE` otherwise.\n self.update_input_layer(review)\n \n out_0_1 = np.dot(self.layer_0, self.weights_0_1)\n out_1_2 = self.sigmoid(np.dot(out_0_1, self.weights_1_2))\n \n if out_1_2 > 0.5:\n return 'POSITIVE'\n else:\n return 'NEGATIVE'\n",
"_____no_output_____"
]
],
[
[
"Run the following cell to recreate the network and train it once again.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000], learning_rate=0.1)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"That should have trained much better than the earlier attempts. Run the following cell to test your model with 1000 predictions.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"# End of Project 5. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.\n# Further Noise Reduction<a id='lesson_6'></a>",
"_____no_output_____"
]
],
[
[
"Image(filename='sentiment_network_sparse_2.png')",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"POSITIVE\" label\npos_neg_ratios.most_common()",
"_____no_output_____"
],
[
"# words most frequently seen in a review with a \"NEGATIVE\" label\nlist(reversed(pos_neg_ratios.most_common()))[0:30]",
"_____no_output_____"
],
[
"from bokeh.models import ColumnDataSource, LabelSet\nfrom bokeh.plotting import figure, show, output_file\nfrom bokeh.io import output_notebook\noutput_notebook()",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],pos_neg_ratios.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"Word Positive/Negative Affinity Distribution\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"_____no_output_____"
],
[
"frequency_frequency = Counter()\n\nfor word, cnt in total_counts.most_common():\n frequency_frequency[cnt] += 1",
"_____no_output_____"
],
[
"hist, edges = np.histogram(list(map(lambda x:x[1],frequency_frequency.most_common())), density=True, bins=100, normed=True)\n\np = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"The frequency distribution of the words in our corpus\")\np.quad(top=hist, bottom=0, left=edges[:-1], right=edges[1:], line_color=\"#555555\")\nshow(p)",
"_____no_output_____"
]
],
[
[
"# Project 6: Reducing Noise by Strategically Reducing the Vocabulary<a id='project_6'></a>\n\n**TODO:** Improve `SentimentNetwork`'s performance by reducing more noise in the vocabulary. Specifically, do the following:\n* Copy the `SentimentNetwork` class from the previous project into the following cell.\n* Modify `pre_process_data`:\n>* Add two additional parameters: `min_count` and `polarity_cutoff`\n>* Calculate the positive-to-negative ratios of words used in the reviews. (You can use code you've written elsewhere in the notebook, but we are moving it into the class like we did with other helper code earlier.)\n>* Andrew's solution only calculates a postive-to-negative ratio for words that occur at least 50 times. This keeps the network from attributing too much sentiment to rarer words. You can choose to add this to your solution if you would like. \n>* Change so words are only added to the vocabulary if they occur in the vocabulary more than `min_count` times.\n>* Change so words are only added to the vocabulary if the absolute value of their postive-to-negative ratio is at least `polarity_cutoff`\n* Modify `__init__`:\n>* Add the same two parameters (`min_count` and `polarity_cutoff`) and use them when you call `pre_process_data`",
"_____no_output_____"
]
],
[
[
"# TODO: -Copy the SentimentNetwork class from Project 5 lesson\n# -Modify it according to the above instructions ",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a small polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.05,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"And run the following cell to test it's performance. It should be ",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"Run the following cell to train your network with a much larger polarity cutoff.",
"_____no_output_____"
]
],
[
[
"mlp = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=20,polarity_cutoff=0.8,learning_rate=0.01)\nmlp.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
]
],
[
[
"And run the following cell to test it's performance.",
"_____no_output_____"
]
],
[
[
"mlp.test(reviews[-1000:],labels[-1000:])",
"_____no_output_____"
]
],
[
[
"# End of Project 6. \n## Watch the next video to see Andrew's solution, then continue on to the next lesson.",
"_____no_output_____"
],
[
"# Analysis: What's Going on in the Weights?<a id='lesson_7'></a>",
"_____no_output_____"
]
],
[
[
"mlp_full = SentimentNetwork(reviews[:-1000],labels[:-1000],min_count=0,polarity_cutoff=0,learning_rate=0.01)",
"_____no_output_____"
],
[
"mlp_full.train(reviews[:-1000],labels[:-1000])",
"_____no_output_____"
],
[
"Image(filename='sentiment_network_sparse.png')",
"_____no_output_____"
],
[
"def get_most_similar_words(focus = \"horrible\"):\n most_similar = Counter()\n\n for word in mlp_full.word2index.keys():\n most_similar[word] = np.dot(mlp_full.weights_0_1[mlp_full.word2index[word]],mlp_full.weights_0_1[mlp_full.word2index[focus]])\n \n return most_similar.most_common()",
"_____no_output_____"
],
[
"get_most_similar_words(\"excellent\")",
"_____no_output_____"
],
[
"get_most_similar_words(\"terrible\")",
"_____no_output_____"
],
[
"import matplotlib.colors as colors\n\nwords_to_visualize = list()\nfor word, ratio in pos_neg_ratios.most_common(500):\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)\n \nfor word, ratio in list(reversed(pos_neg_ratios.most_common()))[0:500]:\n if(word in mlp_full.word2index.keys()):\n words_to_visualize.append(word)",
"_____no_output_____"
],
[
"pos = 0\nneg = 0\n\ncolors_list = list()\nvectors_list = list()\nfor word in words_to_visualize:\n if word in pos_neg_ratios.keys():\n vectors_list.append(mlp_full.weights_0_1[mlp_full.word2index[word]])\n if(pos_neg_ratios[word] > 0):\n pos+=1\n colors_list.append(\"#00ff00\")\n else:\n neg+=1\n colors_list.append(\"#000000\")",
"_____no_output_____"
],
[
"from sklearn.manifold import TSNE\ntsne = TSNE(n_components=2, random_state=0)\nwords_top_ted_tsne = tsne.fit_transform(vectors_list)",
"_____no_output_____"
],
[
"p = figure(tools=\"pan,wheel_zoom,reset,save\",\n toolbar_location=\"above\",\n title=\"vector T-SNE for most polarized words\")\n\nsource = ColumnDataSource(data=dict(x1=words_top_ted_tsne[:,0],\n x2=words_top_ted_tsne[:,1],\n names=words_to_visualize,\n color=colors_list))\n\np.scatter(x=\"x1\", y=\"x2\", size=8, source=source, fill_color=\"color\")\n\nword_labels = LabelSet(x=\"x1\", y=\"x2\", text=\"names\", y_offset=6,\n text_font_size=\"8pt\", text_color=\"#555555\",\n source=source, text_align='center')\np.add_layout(word_labels)\n\nshow(p)\n\n# green indicates positive words, black indicates negative words",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a13b55a886114b1f03033b3906a8308040923e2
| 235,927 |
ipynb
|
Jupyter Notebook
|
Google play store user reviews.ipynb
|
nipun1992/Google-play-store-user-reviews-sentiments
|
171d2d1e407429957e5fc7df14bbd042d4430b07
|
[
"CC-BY-3.0"
] | null | null | null |
Google play store user reviews.ipynb
|
nipun1992/Google-play-store-user-reviews-sentiments
|
171d2d1e407429957e5fc7df14bbd042d4430b07
|
[
"CC-BY-3.0"
] | null | null | null |
Google play store user reviews.ipynb
|
nipun1992/Google-play-store-user-reviews-sentiments
|
171d2d1e407429957e5fc7df14bbd042d4430b07
|
[
"CC-BY-3.0"
] | 1 |
2020-12-29T22:35:48.000Z
|
2020-12-29T22:35:48.000Z
| 55.420954 | 578 | 0.558673 |
[
[
[
"#### Importing the libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"#### Reading the csv as dataframe",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('googleplaystore_user_reviews.csv')",
"_____no_output_____"
],
[
"df.head(5)",
"_____no_output_____"
]
],
[
[
"*We will perform sentiment analysis on Google Play store user reviews*",
"_____no_output_____"
],
[
"#### Renaming the column",
"_____no_output_____"
]
],
[
[
"df = df.rename(columns={'Translated_Review':'Reviews'})",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
]
],
[
[
"*The column **Translated_Review** has been renamed to **Reviews**.*",
"_____no_output_____"
],
[
"#### Checking information of the dataset",
"_____no_output_____"
]
],
[
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 64295 entries, 0 to 64294\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 App 64295 non-null object \n 1 Reviews 37427 non-null object \n 2 Sentiment 37432 non-null object \n 3 Sentiment_Polarity 37432 non-null float64\n 4 Sentiment_Subjectivity 37432 non-null float64\ndtypes: float64(2), object(3)\nmemory usage: 2.5+ MB\n"
]
],
[
[
"*There are total of 64295 rocords and 5 columns in the dataset*",
"_____no_output_____"
]
],
[
[
"df.isnull().sum()",
"_____no_output_____"
]
],
[
[
"*There are 26868 missing values in **Reviews** column and 26863 missing values in **Sentiment**, **Sentiment_Polarity** and **Sentiment_Subjectivity** columns of the dataset.* ",
"_____no_output_____"
],
[
"#### Checking for duplicate records",
"_____no_output_____"
]
],
[
[
"df.duplicated(subset=None, keep='first').sum()",
"_____no_output_____"
]
],
[
[
"*There are 33616 duplicate rows*",
"_____no_output_____"
]
],
[
[
"df.shape",
"_____no_output_____"
],
[
"df = df[df.duplicated(df.columns.tolist(), keep='first')==False]",
"_____no_output_____"
],
[
"df.duplicated(subset=None, keep='first').sum()",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
]
],
[
[
"*The duplicate rows have been removed*",
"_____no_output_____"
]
],
[
[
"df.isnull().sum()",
"_____no_output_____"
]
],
[
[
"#### Handling missing values in Reviews column",
"_____no_output_____"
]
],
[
[
"df[(df['Reviews'].isnull())].loc[:, 'App']",
"_____no_output_____"
]
],
[
[
"*The above displays the rows for which there are missing values in **Reviews** column*",
"_____no_output_____"
]
],
[
[
"df[(df['Reviews'].isnull())].loc[:, 'App'].unique()",
"_____no_output_____"
]
],
[
[
"*The above displays the unique Apps for which there are missing values in **Reviews** column*",
"_____no_output_____"
]
],
[
[
"'''\ndef apps_missing_reviews(dataset):\n for values in dataset['App'].unique().tolist():\n if dataset[dataset['App']==values].App.count() != dataset[dataset['App']==values].Reviews.count():\n print(values, \" : \", dataset[dataset['App']==values].App.count(), \" : \", dataset[dataset['App']==values].Reviews.count())\n \napps_missing_reviews(df)\n'''",
"_____no_output_____"
]
],
[
[
"*The above displays the Apps for which there are 1 or more missing values in the **Reviews** column*",
"_____no_output_____"
]
],
[
[
"'''\ndef apps_no_missing_reviews(dataset):\n for values in dataset['App'].unique().tolist():\n #print(values)\n if dataset[dataset['App']==values].App.count() == dataset[dataset['App']==values].Reviews.count():\n print(values, \" : \", dataset[dataset['App']==values].App.count(), \" : \", dataset[dataset['App']==values].Reviews.count())\n \napps_no_missing_reviews(df)\n'''",
"_____no_output_____"
]
],
[
[
"*The above displays the Apps for which there are no missing values in the **Reviews** column*",
"_____no_output_____"
],
[
"#### Dropping rows having missing values in the Reviews column",
"_____no_output_____"
]
],
[
[
"df = df[df['Reviews'].isnull()==False]",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
]
],
[
[
"*There are no more missing values in the dataset*",
"_____no_output_____"
]
],
[
[
"df.to_csv('file1.csv', index=False)",
"_____no_output_____"
],
[
"df = pd.read_csv('file1.csv')",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df.isnull().sum()",
"_____no_output_____"
]
],
[
[
"#### Cleaning the punctuation marks",
"_____no_output_____"
]
],
[
[
"import re\nimport string",
"_____no_output_____"
],
[
"#df['Reviews'].unique().tolist()",
"_____no_output_____"
],
[
"def remove_punctuation(text):\n no_punct = \"\".join([c for c in text if c not in string.punctuation])\n return no_punct\n \ndf['Reviews'] = df['Reviews'].apply(lambda x: remove_punctuation(x))",
"_____no_output_____"
],
[
"def cleaning(dataset, characters):\n dataset['Reviews'] = dataset['Reviews'].str.lower()\n dataset['Reviews'] = dataset['Reviews'].str.replace('*', 'i')\n dataset['Reviews'] = [re.sub('♥️|❤|\\d', '', e) for e in df['Reviews']]\n dataset['Reviews'] = [re.sub('\\s+', ' ', e) for e in df['Reviews']]\n for ch in characters:\n dataset['Reviews'] = dataset['Reviews'].str.replace(ch, '')\n return dataset\n \n \n \nchar=['☆', '✌', '—']\ndf = cleaning(df, char)",
"_____no_output_____"
],
[
"df.Reviews.unique().tolist()",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"*The punctuation marks have been cleaned in the dataset*",
"_____no_output_____"
],
[
"#### Tokenizing the Reviews",
"_____no_output_____"
]
],
[
[
"from nltk.tokenize import word_tokenize\ndf['Reviews'].dropna(inplace=True)\ndf['Reviews'] = df['Reviews'].apply(word_tokenize)",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"*The Reviews have been tokenized i.e. split into tokens/pieces*",
"_____no_output_____"
],
[
"#### Removing the stop words from Reviews",
"_____no_output_____"
]
],
[
[
"#nltk.download('stopwords')\nfrom nltk.corpus import stopwords",
"_____no_output_____"
],
[
"stopwords = stopwords.words('english')",
"_____no_output_____"
],
[
"def remove_stopwords(text): \n words = [w for w in text if w not in stopwords]\n return words\n\n\ndf['Reviews'] = df['Reviews'].apply(lambda x: remove_stopwords(x))",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
]
],
[
[
"*The sopwords have been removed from Reviews*",
"_____no_output_____"
],
[
"#### Lemmatization of Reviews",
"_____no_output_____"
]
],
[
[
"from nltk.stem import WordNetLemmatizer\nlemmatizer = WordNetLemmatizer()\n\ndef word_lemmatizer(text):\n lem_text = [lemmatizer.lemmatize(i) for i in text]\n return lem_text\n\n\ndf['Reviews'] = df['Reviews'].apply(lambda x: word_lemmatizer(x))",
"_____no_output_____"
],
[
"df['Reviews'].head(10)",
"_____no_output_____"
]
],
[
[
"#### Determining sentiment polarity and subjectivity using TextBlob library",
"_____no_output_____"
]
],
[
[
"from textblob import TextBlob",
"_____no_output_____"
],
[
"pol = lambda x: TextBlob(x).sentiment.polarity\nsub = lambda x: TextBlob(x).sentiment.subjectivity",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df['Reviews'] = df['Reviews'].apply(lambda x: \" \".join(x))",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"df['Predicted_Polarity'] = df['Reviews'].apply(pol)\ndf['Predicted_Subjectivity'] = df['Reviews'].apply(sub)",
"_____no_output_____"
],
[
"df.head(10)",
"_____no_output_____"
],
[
"df[df['App'] == 'Housing-Real Estate & Property']",
"_____no_output_____"
]
],
[
[
"#### Summary\n\n*The actual and the predicted values of snetiment polarity and subjectivity are very close to each other for the respective applications.*",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a13c3ac9308aef9a3c339b9778a4820abe126a2
| 382,735 |
ipynb
|
Jupyter Notebook
|
8-US-airports-case-study-instructor.ipynb
|
lzblack/Network-Analysis-Made-Simple
|
5b4c73da0a5935b87e7dee6d328bcf404008e1ac
|
[
"MIT"
] | 2 |
2021-01-09T15:57:26.000Z
|
2021-11-29T01:44:21.000Z
|
8-US-airports-case-study-instructor.ipynb
|
lzblack/Network-Analysis-Made-Simple
|
5b4c73da0a5935b87e7dee6d328bcf404008e1ac
|
[
"MIT"
] | 5 |
2019-11-15T02:00:26.000Z
|
2021-01-06T04:26:40.000Z
|
Network-Analysis-Made-Simple/8-US-airports-case-study-instructor.ipynb
|
sunny2309/scipy_conf_notebooks
|
30a85d5137db95e01461ad21519bc1bdf294044b
|
[
"MIT"
] | null | null | null | 69.995428 | 23,608 | 0.717763 |
[
[
[
"## Exploratory analysis of the US Airport Dataset\n\nThis dataset contains data for 25 years[1995-2015] of flights between various US airports and metadata about these routes. Taken from Bureau of Transportation Statistics, United States Department of Transportation.\n\nLet's see what can we make out of this!",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport networkx as nx\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport warnings\nwarnings.filterwarnings('ignore')\n\npass_air_data = pd.read_csv('datasets/passengers.csv')",
"_____no_output_____"
]
],
[
[
"In the `pass_air_data` dataframe we have the information of number of people that fly every year on a particular route on the list of airlines that fly that route.",
"_____no_output_____"
]
],
[
[
"pass_air_data.head()",
"_____no_output_____"
],
[
"# Create a MultiDiGraph from this dataset\n\npassenger_graph = nx.from_pandas_edgelist(pass_air_data, source='ORIGIN', target='DEST', edge_attr=['YEAR', 'PASSENGERS', 'UNIQUE_CARRIER_NAME'], create_using=nx.MultiDiGraph())",
"_____no_output_____"
]
],
[
[
"### Cleveland to Chicago, how many people fly this route?",
"_____no_output_____"
]
],
[
[
"passenger_graph['CLE']['ORD'][25]",
"_____no_output_____"
],
[
"temp = [(i['YEAR'], i['PASSENGERS'])for i in dict(passenger_graph['CLE']['ORD']).values()]\nx, y = zip(*temp)\nplt.plot(x, y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Exercise\n\nFind the busiest route in 1990 and in 2015 according to number of passengers, and plot the time series of number of passengers on these routes.\n\nYou can use the DataFrame instead of working with the network. It will be faster ;)\n[5 mins]",
"_____no_output_____"
]
],
[
[
"temp = pass_air_data.groupby(['YEAR'])['PASSENGERS'].transform(max) == pass_air_data['PASSENGERS']",
"_____no_output_____"
],
[
"pass_air_data[temp][pass_air_data.YEAR.isin([1990, 2015])]",
"_____no_output_____"
],
[
"pass_air_data[(pass_air_data['ORIGIN'] == 'LAX') & (pass_air_data['DEST'] == 'HNL')].plot('YEAR', 'PASSENGERS')",
"_____no_output_____"
],
[
"pass_air_data[(pass_air_data['ORIGIN'] == 'LAX') & (pass_air_data['DEST'] == 'SFO')].plot('YEAR', 'PASSENGERS')",
"_____no_output_____"
]
],
[
[
"So let's have a look at the important nodes in this network, i.e. important airports in this network. We'll use pagerank, betweenness centrality and degree centrality.",
"_____no_output_____"
]
],
[
[
"# nx.pagerank(passenger_graph)",
"_____no_output_____"
],
[
"def year_network(G, year):\n temp_g = nx.DiGraph()\n for i in G.edges(data=True):\n if i[2]['YEAR'] == year:\n temp_g.add_edge(i[0], i[1], weight=i[2]['PASSENGERS'])\n return temp_g",
"_____no_output_____"
],
[
"pass_2015 = year_network(passenger_graph, 2015)",
"_____no_output_____"
],
[
"len(pass_2015)",
"_____no_output_____"
],
[
"len(pass_2015.edges())",
"_____no_output_____"
],
[
"# Load in the GPS coordinates of all the airports\nlat_long = pd.read_csv('datasets/GlobalAirportDatabase.txt', delimiter=':', header=None)",
"_____no_output_____"
],
[
"lat_long[lat_long[1].isin(list(pass_2015.nodes()))]",
"_____no_output_____"
],
[
"pos_dict = {}\nfor airport in lat_long[lat_long[1].isin(list(pass_2015.nodes()))].iterrows():\n pos_dict[airport[1][1]] = (airport[1][15], airport[1][14]) ",
"_____no_output_____"
],
[
"pos_dict",
"_____no_output_____"
]
],
[
[
"## Exercise\n\nUsing the position dictionary `pos_dict` create a plot of the airports, only the nodes not the edges.\n\n- As we don't have coordinates for all the airports we have to create a subgraph first.\n- Use `nx.subgraph(Graph, iterable of nodes)` to create the subgraph\n- Use `nx.draw_networkx_nodes(G, pos)` to map the nodes. \n\nor \n\n- Just use a scatter plot :)",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(20, 9))\nG = nx.subgraph(pass_2015, pos_dict.keys())\nnx.draw_networkx_nodes(G, pos=pos_dict, node_size=10, alpha=0.6, node_color='b')\n# nx.draw_networkx_edges(G, pos=pos_dict, width=0.1, arrows=False)\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(20, 9))\nx = [i[0] for i in pos_dict.values()]\ny = [i[1] for i in pos_dict.values()]\nplt.scatter(x, y)",
"_____no_output_____"
]
],
[
[
"### What about degree distribution of this network?",
"_____no_output_____"
]
],
[
[
"plt.hist(list(nx.degree_centrality(pass_2015).values()))\nplt.show()",
"_____no_output_____"
]
],
[
[
"Let's plot a log log plot to get a better overview of this.",
"_____no_output_____"
]
],
[
[
"d = {}\nfor i, j in dict(nx.degree(pass_2015)).items():\n if j in d:\n d[j] += 1\n else:\n d[j] = 1\nx = np.log2(list((d.keys())))\ny = np.log2(list(d.values()))\nplt.scatter(x, y, alpha=0.4)\nplt.show()",
"_____no_output_____"
]
],
[
[
"### Directed Graphs\n\n",
"_____no_output_____"
]
],
[
[
"G = nx.DiGraph()\n\nG.add_edge(1, 2, weight=1)\n\n# print(G.edges())\n# G[1][2]\n# G[2][1]\n# G.is_directed()\n# type(G)",
"_____no_output_____"
],
[
"G.add_edges_from([(1, 2), (3, 2), (4, 2), (5, 2), (6, 2), (7, 2)])\nnx.draw_circular(G, with_labels=True)",
"_____no_output_____"
],
[
"G.in_degree()",
"_____no_output_____"
],
[
"nx.pagerank(G)",
"_____no_output_____"
],
[
"G.add_edge(5, 6)\nnx.draw_circular(G, with_labels=True)",
"_____no_output_____"
],
[
"nx.pagerank(G)",
"_____no_output_____"
],
[
"G.add_edge(2, 8)\nnx.draw_circular(G, with_labels=True)",
"_____no_output_____"
],
[
"nx.pagerank(G)",
"_____no_output_____"
]
],
[
[
"### Moving back to Airports",
"_____no_output_____"
]
],
[
[
"sorted(nx.pagerank(pass_2015, weight=None).items(), key=lambda x:x[1], reverse=True)[:10]",
"_____no_output_____"
],
[
"sorted(nx.betweenness_centrality(pass_2015).items(), key=lambda x:x[1], reverse=True)[0:10]",
"_____no_output_____"
],
[
"sorted(nx.degree_centrality(pass_2015).items(), key=lambda x:x[1], reverse=True)[0:10]",
"_____no_output_____"
]
],
[
[
"'ANC' is the airport code of Anchorage airport, a place in Alaska, and according to pagerank and betweenness centrality it is the most important airport in this network Isn't that weird? Thoughts?\n\nrelated blog post: https://toreopsahl.com/2011/08/12/why-anchorage-is-not-that-important-binary-ties-and-sample-selection/\n\nLet's look at weighted version, i.e taking into account the number of people flying to these places.",
"_____no_output_____"
]
],
[
[
"sorted(nx.betweenness_centrality(pass_2015, weight='weight').items(), key=lambda x:x[1], reverse=True)[0:10]",
"_____no_output_____"
],
[
"sorted(nx.pagerank(pass_2015, weight='weight').items(), key=lambda x:x[1], reverse=True)[0:10]",
"_____no_output_____"
]
],
[
[
"## How reachable is this network?\n\nWe calculate the average shortest path length of this network, it gives us an idea about the number of jumps we need to make around the network to go from one airport to any other airport in this network.",
"_____no_output_____"
]
],
[
[
"# nx.average_shortest_path_length(pass_2015)",
"_____no_output_____"
]
],
[
[
"Wait, What??? This network is not connected. That seems like a really stupid thing to do.",
"_____no_output_____"
]
],
[
[
"list(nx.weakly_connected_components(pass_2015))",
"_____no_output_____"
]
],
[
[
"### SPB, SSB, AIK anyone?",
"_____no_output_____"
]
],
[
[
"pass_air_data[(pass_air_data['YEAR'] == 2015) & (pass_air_data['ORIGIN'] == 'AIK')]",
"_____no_output_____"
],
[
"pass_2015.remove_nodes_from(['SPB', 'SSB', 'AIK'])",
"_____no_output_____"
],
[
"nx.is_weakly_connected(pass_2015)",
"_____no_output_____"
],
[
"nx.is_strongly_connected(pass_2015)",
"_____no_output_____"
]
],
[
[
"### Strongly vs weakly connected graphs.",
"_____no_output_____"
]
],
[
[
"G = nx.DiGraph()\nG.add_edge(1, 2)\nG.add_edge(2, 3)\nG.add_edge(3, 1)\nnx.draw(G)",
"_____no_output_____"
],
[
"G.add_edge(3, 4)\nnx.draw(G)",
"_____no_output_____"
],
[
"nx.is_strongly_connected(G)",
"_____no_output_____"
],
[
"list(nx.strongly_connected_components(pass_2015))",
"_____no_output_____"
],
[
"pass_air_data[(pass_air_data['YEAR'] == 2015) & (pass_air_data['DEST'] == 'TSP')]",
"_____no_output_____"
],
[
"pass_2015_strong = max(nx.strongly_connected_component_subgraphs(pass_2015), key=len)",
"_____no_output_____"
],
[
"len(pass_2015_strong)",
"_____no_output_____"
],
[
"nx.average_shortest_path_length(pass_2015_strong)",
"_____no_output_____"
]
],
[
[
"#### Exercise! (Actually this is a game :D)\n\nHow can we decrease the avg shortest path length of this network?\n\nThink of an effective way to add new edges to decrease the avg shortest path length.\nLet's see if we can come up with a nice way to do this, and the one who gets the highest decrease wins!!!\n\nThe rules are simple:\n- You can't add more than 2% of the current edges( ~500 edges)\n\n[10 mins]",
"_____no_output_____"
]
],
[
[
"sort_degree = sorted(nx.degree_centrality(pass_2015_strong).items(), key=lambda x:x[1], reverse=True)\ntop_count = 0\nfor n, v in sort_degree:\n count = 0\n for node, val in sort_degree:\n if node != n:\n if node not in pass_2015_strong.adj[n]:\n pass_2015_strong.add_edge(n, node)\n count += 1\n if count == 25:\n break\n top_count += 1\n if top_count == 20:\n break",
"_____no_output_____"
],
[
"nx.average_shortest_path_length(pass_2015_strong)",
"_____no_output_____"
]
],
[
[
"### What about airlines? Can we find airline specific reachability?",
"_____no_output_____"
]
],
[
[
"passenger_graph['JFK']['SFO'][25]",
"_____no_output_____"
],
[
"def str_to_list(a):\n return a[1:-1].split(', ')",
"_____no_output_____"
],
[
"for i in str_to_list(passenger_graph['JFK']['SFO'][25]['UNIQUE_CARRIER_NAME']):\n print(i)",
"'Delta Air Lines Inc.'\n'Virgin America'\n'American Airlines Inc.'\n'Sun Country Airlines d/b/a MN Airlines'\n'JetBlue Airways'\n'Vision Airlines'\n'United Air Lines Inc.'\n"
],
[
"%%time\nfor origin, dest in passenger_graph.edges():\n for key in passenger_graph[origin][dest]:\n passenger_graph[origin][dest][key]['airlines'] = str_to_list(passenger_graph[origin][dest][key]['UNIQUE_CARRIER_NAME'])",
"CPU times: user 52.9 s, sys: 722 ms, total: 53.6 s\nWall time: 56.3 s\n"
]
],
[
[
"### Exercise\n\nPlay around with United Airlines network.\n\n- Extract a network for United Airlines flights from the metagraph `passenger_graph` for the year 2015\n- Make sure it's a weighted network, where weight is the number of passengers.\n- Find the number of airports and connections in this network\n- Find the most important airport, according to PageRank and degree centrality.",
"_____no_output_____"
]
],
[
[
"united_network = nx.DiGraph()\nfor origin, dest in passenger_graph.edges():\n if 25 in passenger_graph[origin][dest]:\n if \"'United Air Lines Inc.'\" in passenger_graph[origin][dest][25]['airlines']:\n united_network.add_edge(origin, dest, weight=passenger_graph[origin][dest][25]['PASSENGERS'])",
"_____no_output_____"
],
[
"len(united_network)",
"_____no_output_____"
],
[
"len(united_network.edges())",
"_____no_output_____"
],
[
"sorted(nx.pagerank(united_network, weight='weight').items(), key=lambda x:x[1], reverse=True)[0:10]",
"_____no_output_____"
],
[
"sorted(nx.degree_centrality(united_network).items(), key=lambda x:x[1], reverse=True)[0:10]",
"_____no_output_____"
]
],
[
[
"### Exercise\n\nWe are in Cleveland so what should we do?\n\nObviously we will make a time series of number of passengers flying out of Cleveland with United Airlines over the years.\n\nThere are 2 ways of doing it.\n- Create a new multidigraph specifically for this exercise.\n\nOR\n\n- exploit the `pass_air_data` dataframe.",
"_____no_output_____"
]
],
[
[
"pass_air_data[(pass_air_data.ORIGIN == 'CLE') &\n (pass_air_data.UNIQUE_CARRIER_NAME.str.contains('United Air Lines Inc.'))\n ].groupby('YEAR')['PASSENGERS'].sum().plot()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a1408166b9283b6cb5aedc4dcfa720234110d3f
| 3,042 |
ipynb
|
Jupyter Notebook
|
test/ipynb/python/TableDisplayWIthIndex.ipynb
|
ssadedin/beakerx
|
34479b07d2dfdf1404692692f483faf0251632c3
|
[
"Apache-2.0"
] | 1,491 |
2017-03-30T03:05:05.000Z
|
2022-03-27T04:26:02.000Z
|
test/ipynb/python/TableDisplayWIthIndex.ipynb
|
ssadedin/beakerx
|
34479b07d2dfdf1404692692f483faf0251632c3
|
[
"Apache-2.0"
] | 3,268 |
2015-01-01T00:10:26.000Z
|
2017-05-05T18:59:41.000Z
|
test/ipynb/python/TableDisplayWIthIndex.ipynb
|
ssadedin/beakerx
|
34479b07d2dfdf1404692692f483faf0251632c3
|
[
"Apache-2.0"
] | 287 |
2017-04-03T01:30:06.000Z
|
2022-03-17T06:09:15.000Z
| 24.532258 | 94 | 0.540763 |
[
[
[
"from beakerx import *\nfrom beakerx.object import beakerx\n\nimport pandas as pd\ndf = pd.read_csv('../resources/data/interest-rates-small-with_index.csv')\ndf",
"_____no_output_____"
],
[
"from beakerx import *\nfrom beakerx.object import beakerx\n\nimport pandas as pd\ndf = pd.read_csv('../resources/data/interest-rates-small-with_index.csv',index_col=0)\ndf",
"_____no_output_____"
],
[
"from beakerx import *\nfrom beakerx.object import beakerx\n\nimport pandas as pd\n\ns1 = pd.Series([0, 1, 2], index=pd.date_range('2000', freq='D', periods=3))\ns2 = pd.Series([3, 4, 5], index=pd.date_range('2000', freq='D', periods=3))\ns3 = pd.Series([6, 7, 8], index=pd.date_range('2000', freq='D', periods=3))\n\ndf = pd.concat([s1, s2, s3], axis=1)\ndf",
"_____no_output_____"
],
[
"from beakerx import *\nfrom beakerx.object import beakerx\n\nimport pandas as pd\n\ns1 = pd.Series([0, 1, 2], index=pd.date_range('2000', freq='D', periods=3))\ns2 = pd.Series([3, 4, 5], index=pd.date_range('2000', freq='D', periods=3))\ns3 = pd.Series([6, 7, 8], index=pd.date_range('2000', freq='D', periods=3))\n\ndf = pd.concat([s1, s2, s3], axis=1)\n\ndef color_provider(row, column, table):\n row = table.values[row]\n val = row[column]\n if val == 4:\n return Color.GREEN\n return Color.BLACK\n \ntable = TableDisplay(df)\ntable.setStringFormatForTimes(TimeUnit.DAYS)\ntable.setFontColorProvider(color_provider)\n\ntable",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a141819f85f3a3054555142ba724248202d8255
| 9,236 |
ipynb
|
Jupyter Notebook
|
notebooks/01_preprocessing.ipynb
|
IKNL/intro-ds-students
|
81c9685d4065f24fa9b4c64f70d1da05d35028d2
|
[
"MIT"
] | null | null | null |
notebooks/01_preprocessing.ipynb
|
IKNL/intro-ds-students
|
81c9685d4065f24fa9b4c64f70d1da05d35028d2
|
[
"MIT"
] | null | null | null |
notebooks/01_preprocessing.ipynb
|
IKNL/intro-ds-students
|
81c9685d4065f24fa9b4c64f70d1da05d35028d2
|
[
"MIT"
] | null | null | null | 26.238636 | 161 | 0.534106 |
[
[
[
"Before we begin, we will change a few settings to make the notebook look a bit prettier",
"_____no_output_____"
]
],
[
[
"%%html\n<style> body {font-family: \"Calibri\", cursive, sans-serif;} </style>",
"_____no_output_____"
]
],
[
[
"<img src=\"https://github.com/IKNL/guidelines/blob/master/resources/logos/iknl_nl.png?raw=true\" width=200 align=\"right\">\n\n# 01 - Pre-processing\n\nPerform some pre-processing on the (synthetic) data\n\n**Important**: include all your imports here and here only.",
"_____no_output_____"
],
[
"## 1.1 Read the data\n**Tip**: take a look at [`pandas read_csv`](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html)",
"_____no_output_____"
],
[
"## 1.2 Rename the columns\nRename the columns of the original `DataFrame` as follows:\n\n| Original name | New name |\n|---------------|----------|\n| `rn` | `nkr_id` |\n| `zid` | `disease_id` |\n| `eid` | `episode_id` |\n| `gesl` | `gender` |\n| `gebdat` | `birth_date` |\n| `incdat` | `incidence_date` |\n| `topo` | `topo` |\n| `sublok` | `sublocalization` |\n| `topog` | `topography` |\n| `later` | `lateralization` |\n| `morf` | `morphology` |\n| `gedrag` | `behaviour` |\n| `diffgr` | `grade` |\n| `tumorsoort` | `tumor_type` |\n| `basisd` | `diagnosis_basis` |\n| `ct` | `t_clin` |\n| `cn` | `n_clin` |\n| `cm` | `m_clin` |\n| `pt` | `t_patho` |\n| `pn` | `n_patho` |\n| `pm` | `m_patho` |\n| `stadiumc` | `stage_clin` |\n| `stadium` | `stage_patho` |\n| `vitdat` | `contact_date` |\n| `ovldat` | `death_date` |\n\nThis will make your code much easier to read (and more accessible to \nnon-Dutch speakers as well 😉).\n\n**Tip**: take a look at [`pandas rename`](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.rename.html)\n\nAs you get familiarized with the data, be sure to complete the table found in `../data/README.md`.",
"_____no_output_____"
],
[
"## 1.3 Check data integrity\nData are never perfect. Let's see how imperfect this dataset is.\n\nFirst, generate a visualization of the missing values.\n\n**Tip**: take a look at the [`missingno` package](https://github.com/ResidentMario/missingno)",
"_____no_output_____"
],
[
"How many missing values (in absolute numbers and as a percentage)\nare there for each column? Generate a bar plot showing this.",
"_____no_output_____"
],
[
"What methods do you know for dealing with missing values? \nWhat are the advantages and disadvantages of each of them?",
"_____no_output_____"
],
[
"* ...",
"_____no_output_____"
],
[
"For now, we will keep it simple. Drop all the rows that contain missing data. How many records were lost because of this??",
"_____no_output_____"
],
[
"*In theory*, `topography` should be the concatenation of `topo` and\n`sublocalization`. For what number/percentage of records\nis this actually true?",
"_____no_output_____"
],
[
"## 1.4 Feature engineering\nLet's make a small feature engineering. In all cases, make sure that you \nonly add these new features (i.e., columns) only if they aren't already present \nin the `DataFrame`. \n\n* Create a new column called `age_incidence`, which \ncontains the age of the patient at the moment of diagnosis. Use the \ninformation contained in the columns `incidence_year` and `birth_date`. ",
"_____no_output_____"
],
[
"* Create two new columns called `stage_clin_num` and `stage_patho_num`. \nEach of them will have the numerical part of the data contained\nin the columns `stage_clin` and `state_patho`, respectively.",
"_____no_output_____"
],
[
"* Create q new column called `death`, which has a `False` if the patient\nis still alive or a `True` if the patient already passed away.",
"_____no_output_____"
],
[
"## 1.5 Decode features\nMany features in the NKR are encoded. While this will be useful later on,\nit can also make the data cryptic at this point.\n\nDecode the column `gender` as indicated in the following table\n\n| Original value | New value |\n|----------------|-----------------|\n| 1 | `male` |\n| 2 | `female` |\n| 3 | `hermaphrodite` |\n",
"_____no_output_____"
],
[
"## 1.6 Save the data\nLastly, save the modified DataFrame as `df_preprocessing.csv` in the\n`data_processed` directory. \n\n**Tip**: take a look at [`pandas to_csv`](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.to_csv.html)",
"_____no_output_____"
],
[
"Make sure that you generate a new section in the data `README.md` corresponding to this new data file. Remember to include information of the new features.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a141d969fc28f49832fe32cbf2a34c3bcf96bd8
| 8,827 |
ipynb
|
Jupyter Notebook
|
2016/ferran/day9.ipynb
|
bbglab/adventofcode
|
65b6d8331d10f229b59232882d60024b08d69294
|
[
"MIT"
] | null | null | null |
2016/ferran/day9.ipynb
|
bbglab/adventofcode
|
65b6d8331d10f229b59232882d60024b08d69294
|
[
"MIT"
] | null | null | null |
2016/ferran/day9.ipynb
|
bbglab/adventofcode
|
65b6d8331d10f229b59232882d60024b08d69294
|
[
"MIT"
] | 3 |
2016-12-02T09:20:42.000Z
|
2021-12-01T13:31:07.000Z
| 18.313278 | 118 | 0.455647 |
[
[
[
"# Chellenge 9\n\n## Challenge 9.1",
"_____no_output_____"
]
],
[
[
"myinput = '/home/fmuinos/projects/adventofcode/2016/ferran/inputs/input9.txt'",
"_____no_output_____"
],
[
"import re\n\nMARKER = re.compile('\\(\\d+x\\d+\\)')\n\ndef parse_line(myinput):\n with open(myinput, 'rt') as f:\n return next(f).rstrip()\n\ndef next_marker(mystr):\n marker = next(MARKER.finditer(mystr))\n return marker.group(), marker.start()",
"_____no_output_____"
]
],
[
[
"### by Brute Force",
"_____no_output_____"
]
],
[
[
"def unzip(mystr):\n try:\n m, p = next_marker(mystr)\n a, b = map(int, m[1:-1].split('x'))\n prefix = ''\n if p > 0:\n prefix = mystr[:p]\n upper_index = p + len(m) + a\n if upper_index > len(mystr):\n upper_index = len(mystr)\n return prefix + mystr[p + len(m): upper_index] * b + unzip(mystr[upper_index:])\n except:\n return mystr",
"_____no_output_____"
]
],
[
[
"### by Length Computation",
"_____no_output_____"
]
],
[
[
"def unzip_length(mystr):\n if MARKER.findall(mystr):\n m, p = next_marker(mystr)\n a, b = map(int, m[1:-1].split('x'))\n return p + a * b + unzip_length(mystr[p + len(m) + a:])\n else:\n return len(mystr)",
"_____no_output_____"
]
],
[
[
"### Testing",
"_____no_output_____"
]
],
[
[
"def test(mystr):\n l = unzip_length(mystr)\n s = unzip(mystr)\n print(s[:100] + '...' * (len(s) >= 100))\n print(len(s) == l)\n print('length = {}'.format(l))",
"_____no_output_____"
],
[
"test('ADVENT')",
"ADVENT\nTrue\nlength = 6\n"
],
[
"test('A(1x5)BC')",
"ABBBBBC\nTrue\nlength = 7\n"
],
[
"test('(3x3)XYZ')",
"XYZXYZXYZ\nTrue\nlength = 9\n"
],
[
"test('(6x1)(1x3)A')",
"(1x3)A\nTrue\nlength = 6\n"
],
[
"test('X(8x2)(3x3)ABCY')",
"X(3x3)ABC(3x3)ABCY\nTrue\nlength = 18\n"
]
],
[
[
"### Results",
"_____no_output_____"
]
],
[
[
"mystr = parse_line(myinput)",
"_____no_output_____"
],
[
"len(unzip(mystr))",
"_____no_output_____"
],
[
"unzip_length(mystr)",
"_____no_output_____"
]
],
[
[
"## Challenge 9.2",
"_____no_output_____"
],
[
"### by Brute Force (not to be used with *myinput*)",
"_____no_output_____"
]
],
[
[
"def deep_unzip(mystr):\n while MARKER.findall(mystr):\n mystr = unzip(mystr)\n return mystr",
"_____no_output_____"
]
],
[
[
"### by Length Computation",
"_____no_output_____"
]
],
[
[
"def deep_unzip_length(mystr):\n if MARKER.findall(mystr):\n m, p = next_marker(mystr)\n a, b = map(int, m[1:-1].split('x'))\n return p + b * deep_unzip_length(mystr[p+len(m): p+len(m)+a]) + deep_unzip_length(mystr[p+len(m)+a:])\n else:\n return len(mystr)",
"_____no_output_____"
]
],
[
[
"### Testing",
"_____no_output_____"
]
],
[
[
"def deep_test(mystr):\n l = deep_unzip_length(mystr)\n s = deep_unzip(mystr)\n print(s[:100] + '...' * (len(s) > 100))\n print(len(s) == l)\n print('length = {}'.format(l))",
"_____no_output_____"
],
[
"deep_test('(6x1)(1x3)A')",
"AAA\nTrue\nlength = 3\n"
],
[
"deep_test('X(8x2)(3x3)ABCY')",
"XABCABCABCABCABCABCY\nTrue\nlength = 20\n"
],
[
"deep_test('(27x12)(20x12)(13x14)(7x10)(1x12)A')",
"AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA...\nTrue\nlength = 241920\n"
]
],
[
[
"### Results",
"_____no_output_____"
]
],
[
[
"mystr = parse_line(myinput)\ndeep_unzip_length(mystr)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a14245a4029e95264ddde0f9d773475011dc743
| 35,863 |
ipynb
|
Jupyter Notebook
|
Lesson 3 - Introduction to Databases and SQL.ipynb
|
ZaharaMC/Accelerated_Intro_to_CompBio_Part_2
|
06deb66bfaaf51fd18314ed0d4cade8be4501cbe
|
[
"CC-BY-4.0"
] | null | null | null |
Lesson 3 - Introduction to Databases and SQL.ipynb
|
ZaharaMC/Accelerated_Intro_to_CompBio_Part_2
|
06deb66bfaaf51fd18314ed0d4cade8be4501cbe
|
[
"CC-BY-4.0"
] | null | null | null |
Lesson 3 - Introduction to Databases and SQL.ipynb
|
ZaharaMC/Accelerated_Intro_to_CompBio_Part_2
|
06deb66bfaaf51fd18314ed0d4cade8be4501cbe
|
[
"CC-BY-4.0"
] | 1 |
2021-03-01T02:31:16.000Z
|
2021-03-01T02:31:16.000Z
| 35.056696 | 573 | 0.576276 |
[
[
[
"## Before we begin\n\nWe need to install the mysql client software. My apologies that I forgot to install it in the virtual machine this year!\n\nopen a Terminal window and type:\n\n\n sudo apt-get update\n sudo apt-get install mysql-client\n \nThis will install the client software we need to connect to our Docker mySQL database.\n",
"_____no_output_____"
],
[
"# Introduction to Databases and Structured Query Language (SQL)\n\nAs Data Scientists, you will frequently want to store data in an organized, structured manner that allows you to do complex queries. Because you are good Data Scientists, [**you do not use Excel!!!**](https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-5-80)\n\nIn this course, we will only discuss **Relational Databases**, because those are the most common in bioinformatics. (There are other kinds!!). So when I say \"database\" I mean \"relational database\".\n\nDatabases are used to store information in a manner that, when used properly, is:\n a) highly structured\n b) constrained (i.e. detects errors)\n c) transactional (i.e. can undo a command if it discovers a problem)\n d) indexed (for speed of search)\n e) searchable\n \nThe core concept of a database is a **Table**. Tables contain one particular \"kind\" of information (e.g. a Table could represent a Student, a University, a Book, or a portion of a Clinical Record.\n\nTables contain **Rows** and **Columns** where, generally, every column represents a \"feature\" of that information (e.g. a Student table might have **[\"name\", \"gender\", \"studentID\", \"age\"]** as its columns/features). Every row represents an \"individual\", and their values for each feature (e.g. a Row in a Student table might have **[\"Mark Wilkinson\", \"M\", \"163483\", \"35\"]** as its values.\n\nA Database may have many Tables that represent various kinds of related information. For example, a library database might have a Books table, a Publishers table, and a Locations table. A Book has a Publisher, and a Location, so the tables need to be connected to one another. This is achieved using **keys**. Generally, every row (individual) in a table has a unique identifier (generally a number), and this is called its **key**. Because it is unique, it is possible to refer unambiguously to that individual record.\n\nI think the easiest way to learn about databases and SQL is to start building one! We will use the MySQL Docker Container that we created in the previous lesson. We are going to create a Germplasm database (seed stocks). It will contain information about the seed (its amount, its harvest date, its location), the germplasm (its species, the allele it carries), and about the genetics related to that allele (the gene_id, the gene name, the protein name, and a link to the GenBank record)\n\n(if that Docker Container isn't running, please **docker start course-mysql** now!)\n\n**Note: This Jupyter Notebook is running the Python kernel. This allows us to use some nice tools in Python (the sql extension and SqlMagic) that provide access to the mysql database server from inside of the Notebook. You don't need to know any Python to do this. Note also that you can do exactly the same commands in your Terminal window.**\n\nTo connect to the MySQL Docker Container from your terminal window, type:\n\n mysql -h 127.0.0.1 -P 3306 --protocol=tcp -u root -p\n \n(then enter your password 'root' to access the database)\n \n<pre>\n\n\n</pre>\n# SQL\n\nStructured Query Language is a way to interact with a database server. It is used to create, delete, edit, fill, and query tables and their contents. \n\nFirst, we will learn the SQL commands that allow us to explore the database server, and create new databases and tables.. Later, we will use SQL to put information into those tables. Finally, we will use SQL to query those tables.\n",
"_____no_output_____"
],
[
"## Python SQL Extension\n\nThe commands below are used to connect to the MySQL server in our Docker Container. You need to execute them ONCE. In every subsequent Juputer code window, you will have access to the database.\n\nall SQL commands are preceded by \n\n %sql \n \n(**only in the Python extension! Not in your terminal window!**)\n\nall SQL commands end with a \";\"",
"_____no_output_____"
]
],
[
[
"# UNCOMMENT THIS SECTION IF YOU ARE RUNNING THE DOCKER IMAGE OF THIS COURSE\n#import os\n#os.system(\"echo jovyan | sudo -S service mysql start\")\n#%load_ext sql\n#%sql mysql+pymysql://[email protected]:3306/mysql\n#%sql DROP DATABASE IF EXISTS germplasm;\n\n# ===============================================================================================\n\n# COMMENT THIS SECTION IF YOU ARE RUNNING THE DOCKER IMAGE OF THIS COURSE\n%load_ext sql\n#%config SqlMagic.autocommit=False\n%sql mysql+pymysql://root:[email protected]:3306/mysql\n#%sql mysql+pymysql://[email protected]/homo_sapiens_core_92_38\n#%sql DROP DATABASE IF EXISTS germplasm;",
"_____no_output_____"
]
],
[
[
"## show databases\n\n**show databases** is the command to see what databases exist in the server. The ones you see now are the default databases that MySQL uses to organize itself. _**DO NOT TOUCH THESE DATABASES**_ **EVER EVER EVER EVER**",
"_____no_output_____"
]
],
[
[
"%sql show databases;\n\n",
"_____no_output_____"
]
],
[
[
"## create database\n\nThe command to create a database is **create database** (surprise! ;-) )\n\nWe will create a database called \"germplasm\"\n\n\n",
"_____no_output_____"
]
],
[
[
"%sql create database germplasm;\n%sql show databases\n",
"_____no_output_____"
]
],
[
[
"## use database_name\n\nthe **use** command tells the server which database you want to interact with. Here we will use the database we just created",
"_____no_output_____"
]
],
[
[
"%sql use germplasm;\n\n",
"_____no_output_____"
]
],
[
[
"## show tables\n\nThe show tables command shows what tables the database contains (right now, none!)",
"_____no_output_____"
]
],
[
[
"%sql show tables;",
"_____no_output_____"
]
],
[
[
"# Planning your data structure\n\nThis is the hard part. What does our data \"look like\" in a well-structured, relational format?\n\nStarting simply:\n\n<center>stock table</center>\n\n amount | date | location \n --- | --- | --- \n 5 | 10/5/2013 | Room 2234 \n 9.8 | 12/1/2015 | Room 998 \n\n\n-----------------------------\n\n\n<center>germplasm table</center>\n\n taxonid | allele\n --- | --- \n 4150 | def-1\n 3701 | ap3\n \n--------------------------------\n\n<center>gene table</center>\n\n gene | gene_name | embl\n --- | --- | --- \n DEF | Deficiens | https://www.ebi.ac.uk/ena/data/view/AB516402\n AP3 | Apetala3 | https://www.ebi.ac.uk/ena/data/view/AF056541\n \n \n \n",
"_____no_output_____"
],
[
"## add indexes\n\nIt is usually a good idea to have an index column on every table, so let's add that first:\n\n\n<center>stock table</center>\n\nid | amount | date | location \n--- | --- | --- | --- \n1 | 5 | 10/5/2013 | Room 2234 \n2 | 9.8 | 12/1/2015 | Room 998 \n\n\n-----------------------------\n\n\n<center>germplasm table</center>\n\nid | taxonid | allele\n--- | --- | --- \n1 | 4150 | def-1\n2 | 3701 | ap3\n \n--------------------------------\n\n<center>gene table</center>\n\nid | gene | gene_name | embl\n--- | --- | --- | --- \n1 | DEF | Deficiens | https://www.ebi.ac.uk/ena/data/view/AB516402\n2 | AP3 | Apetala3 | https://www.ebi.ac.uk/ena/data/view/AF056541\n \n",
"_____no_output_____"
],
[
"## find linkages\n\n* Every germplasm has a stock record. This is a 1:1 relationship.\n* Every germplasm represents a specific gene. This is a 1:1 relationship\n\nSo every germplasm must point to the index of a stock, and also to the index of a gene\n\nAdding that into our tables we have:\n\n\n\n<center>stock table</center>\n\nid | amount | date | location \n--- | --- | --- | --- \n1 | 5 | 10/5/2013 | Room 2234 \n2 | 9.8 | 12/1/2015 | Room 998 \n\n\n-----------------------------\n\n\n<center>germplasm table</center>\n\nid | taxonid | allele | stock_id | genetics_id\n--- | --- | --- | --- | ---\n1 | 4150 | def-1 | 2 | 1\n2 | 3701 | ap3 | 1 | 2\n \n--------------------------------\n\n<center>gene table</center>\n\nid | gene | gene_name | embl\n--- | --- | --- | --- \n1 | DEF | Deficiens | https://www.ebi.ac.uk/ena/data/view/AB516402\n2 | AP3 | Apetala3 | https://www.ebi.ac.uk/ena/data/view/AF056541\n \n",
"_____no_output_____"
],
[
"## data types in MySQL\n\nI will not discuss [all MySQL Datatypes](https://dev.mysql.com/doc/refman/5.7/en/data-types.html), but we will look at only the ones we need. We need:\n\n* Integers (type INTEGER)\n* Floating point (type FLOAT)\n* Date (type DATE [in yyyy-mm-dd format](https://dev.mysql.com/doc/refman/5.7/en/datetime.html) )\n* Characters (small, variable-length --> type [VARCHAR(x)](https://dev.mysql.com/doc/refman/5.7/en/char.html) )\n\n<pre>\n\n\n</pre>\n## create table \n\ntables are created using the **create table** command (surprise!)\n\nThe [syntax of create table](https://dev.mysql.com/doc/refman/5.7/en/create-table.html) can be quite complicated, but we are only going to do the most simple examples.\n\n create table table_name (column_name column_definition, column_name column_definition, ........)\n \ncolumn definitions include the data-type, and other options like if it is allowed to be null(blank), or if it should be treated as an \"index\" column.\n\nExamples are easier to understand than words... so here are our table definitions:\n\n ",
"_____no_output_____"
]
],
[
[
"#%sql drop table stock\n%sql CREATE TABLE stock(id INTEGER NOT NULL AUTO_INCREMENT PRIMARY KEY, amount FLOAT NOT NULL, date DATE NOT NULL, location VARCHAR(20) NOT NULL);\n%sql DESCRIBE stock\n",
"_____no_output_____"
],
[
"#%sql drop table germplasm\n%sql CREATE TABLE germplasm(id INTEGER NOT NULL AUTO_INCREMENT PRIMARY KEY, taxonid INTEGER NOT NULL, allele VARCHAR(10) NOT NULL, stock_id INTEGER NOT NULL, gene_id INTEGER NOT NULL);\n%sql DESCRIBE germplasm\n",
"_____no_output_____"
],
[
"#%sql drop table gene\n%sql CREATE TABLE gene(id INTEGER NOT NULL AUTO_INCREMENT PRIMARY KEY, gene VARCHAR(10) NOT NULL, gene_name VARCHAR(30) NOT NULL, embl VARCHAR(70) NOT NULL);\n%sql DESCRIBE gene",
"_____no_output_____"
],
[
"%sql show tables;",
"_____no_output_____"
]
],
[
[
"## loading data\n\nThere are many ways to import data into MySQL. If you have data in another (identical) MySQL database, you can \"dump\" the data, and then import it directly. If you have tab or comma-delimited (tsv, csv) you can **sometimes** import it directly from these formats. You can also enter data using SQL itself. This is usually the safest way, when you have to keep multiple tables synchronized (as we do, since the germplasm table is \"linked to\" the other two tables)\n\n## insert into\n\nThe command to load data is:\n\n insert into table_name (field1, field2, field3) values (value1, value2, value3)\n \nNow... what data do we need to add, in what order?\n\nThe germplasm table needs the ID number from both the gene table and the stock table, so we cannot enter the germplasm information first. We must therefore enter the gene and stock data first.\n",
"_____no_output_____"
]
],
[
[
"# NOTE - we DO NOT put data into the \"id\" column! This column is auto_increment, so it \"magically\" creates its own value\n%sql INSERT INTO gene (gene, gene_name, embl) VALUES ('DEF', \"Deficiens\", 'https://www.ebi.ac.uk/ena/data/view/AB516402');\n%sql INSERT INTO gene (gene, gene_name, embl) VALUES ('AP3', \"Apetala3\", 'https://www.ebi.ac.uk/ena/data/view/AF056541');\n",
"_____no_output_____"
],
[
"%sql SELECT last_insert_id(); # just to show you that this function exists!",
"_____no_output_____"
],
[
"%sql INSERT INTO stock(amount, date, location) VALUES (5, '2013-05-10', 'Room 2234');\n%sql INSERT INTO stock(amount, date, location) VALUES (9.8, '2015-01-12', 'Room 998');\n",
"_____no_output_____"
]
],
[
[
"#### Almost ready! \n\nWe now need to know the index numbers from the stock and gene databases that correspond to the data for the germplasm table. For this, we need to learn another function: **select**\n\n## Select statements\n\n**Select** is the command used to query the database. We will look in more detail later, but now all you need to know is that the most basic structure is:\n\n select * from table_name\n \n",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM stock; # notice that the id number was automatically generated\n",
"_____no_output_____"
],
[
"%sql SELECT * FROM gene;",
"_____no_output_____"
]
],
[
[
"<pre>\n\n\n</pre>\n\nJust a reminder, our germplasm data is:\n\nid | \ttaxonid | allele | \tstock_id |\tgene_id\n--- | --- | --- | --- | --- |\n1 \t | 4150 \t|def-1 \t\n2 \t| 3701 |\tap3 \t\n\n\nWe need to connect the *germplasm* table **gene_id** to the appropriate **id** from the *gene* table. i.e.\n\n def-1 allele ---> DEF gene (id = 1)\n ap3 allele ---> AP3 gene (id = 2)\n \nWe need to connect the *germplasm* table **stock_id** to the appropriate **id** from the *stock* table. i.e.\n\n def-1 allele ---> Room 998 (id = 2)\n ap3 allele ---> Room 2234 (id = 1)\n\nNow we are ready to do our (\"manual\") insert of data into the *germplasm* table:\n",
"_____no_output_____"
]
],
[
[
"%sql INSERT INTO germplasm (taxonid, allele, stock_id, gene_id) VALUES (4150, 'def-1', 2, 1 );\n%sql INSERT INTO germplasm (taxonid, allele, stock_id, gene_id) VALUES (3701, 'ap3', 1, 2 );\n",
"_____no_output_____"
],
[
"%sql SELECT * FROM germplasm;",
"_____no_output_____"
]
],
[
[
"\n## SQL UPDATE & SQL WHERE\n\nImagine that we are going to plant some seed from our def-1 germplasm. We need to update the *stock* record to show that there is now less seed available. We do this using an [UPDATE statement](https://www.techonthenet.com/mysql/update.php). UPDATE is used to change the values of a particular column or set of columns. But we don't want to change **all** of the values in that column, we only want to change the values for the DEF stock. For that, we need a WHERE clause. \n\nWHERE allows you to set the conditions for an update. The general form is:\n\n UPDATE table_name SET column = value WHERE column = value;\n \nWe will sow 1g of seed from DEF (stock.id = 2) (note that I am now starting to use the MySQL syntax for referring to *table*.**column** - the tablename followed by a \".\" followed by the column name).\n\nThe simplest UPDATE statement is:\n",
"_____no_output_____"
]
],
[
[
"%sql UPDATE stock SET amount = 8.8 WHERE id = 2;\n%sql SELECT * FROM stock;",
"_____no_output_____"
]
],
[
[
"\n<pre>\n\n</pre>\nThis simple solution is not very \"friendly\"... you are asking the database user to already know what the remaining amount is! It would be better if we simply reduced the amount by 1g. \n\nThat is done using in-line equations, like this:\n\n",
"_____no_output_____"
]
],
[
[
"%sql UPDATE stock SET amount = amount-1 WHERE id = 2;\n%sql SELECT * FROM stock;",
"_____no_output_____"
]
],
[
[
"<pre>\n\n\n</pre>\n## Using indexes and 'joining' tables\n\nThe UPDATE we did is still not very friendly! My stock table does not have any information about what gene or allele is in that stock, so we have to **know** that the stock record is stock.id=2. This is bad!\n\nIt would be better if we could say \"plant 1 gram of the stock that represents gene record DEF\", but that information exists in two different tables. How do we join tables?\n\nThis is the main purpose of the \"id\" column. Note that, when we defined that column, we said that it is \"auto_increment, not null, primary key\", meaning that every record must have an id, and every id must be unique (NOTE: an auto-increment id *should never be manually modified/added*!!! <span style=\"color:red;\">You Have Been Warned!!!</span>). Being a 'primary key' means that this column was intended to be the \"pointer\" from other tables in the database (like our germplasm table, that points to the id of the stock, and the id of the gene, tables)\n\nWhen using UPDATE with multiple tables, we must name all of the tables, and then make the connection between them in the \"where\" clause, using *table*.**column** notation. \n\nThe update clause below shows how this is done (a \"\\\" character means that the command continues on the next line):\n",
"_____no_output_____"
]
],
[
[
"%sql UPDATE stock, germplasm SET stock.amount = stock.amount-1 \\ \n WHERE \\\n stock.id = germplasm.stock_id \\\n AND \\\n germplasm.allele = 'def-1'; \n \n%sql SELECT * FROM stock;",
"_____no_output_____"
]
],
[
[
"<pre>\n\n</pre>\n# Challenges for you!\n\n1. (hard) when we plant our seeds, we should update both the quantity, and the date. What does that UPDATE statement look like?\n\n2. (very hard!) when we plant our seed, instead of using the allele designation (def-1) I want to use the gene designation (DEF). This query spans **all three tables**. What does the UPDATE statement look like?\n\n<span style=\"visibility:hidden;\">\n Challenge 1\n %sql UPDATE stock,germplasm SET stock.amount=stock.amount-1, stock.date=\"2018-09-06\" WHERE \\\n stock.id = germplasm.stock_id AND \\\n germplasm.allele='def-1';\n Challenge2\n%sql UPDATE stock,germplasm,gene SET stock.amount=stock.amount-0.2, stock.date=\"2018-09-06\" WHERE \\\n stock.id = germplasm.stock_id AND \\\n gene.id = germplasm.gene_id AND \\\n gene.gene='DEF'; \n</span>",
"_____no_output_____"
]
],
[
[
"# challenge 1\n",
"_____no_output_____"
],
[
"# challenge 2\n",
"_____no_output_____"
]
],
[
[
"<pre>\n\n\n</pre>\n\n# SELECT queries\n\nQuerying the data is the most common operation on a database. You have seen simple SELECT queries, but now we will look at more complex ones.\n\nThe general structure is:\n\n SELECT table1.column1, ... FROM table1, ... WHERE condition1 [AND|OR] condition2....\n \nYou probably understand this enough to show you the query that will show you all of the data:",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM gene, stock, germplasm WHERE \\\n germplasm.stock_id = stock.id AND \\\n germplasm.gene_id = gene.id;",
"_____no_output_____"
]
],
[
[
"\n# Dealing with missing records - JOIN clauses\n\n**Credit for the Venn diagrams used in this section goes to [Flatiron School](https://learn.co/) and are linked from their tutorial on [JOINs in SQL](https://learn.co/lessons/sql-complex-joins-readme) published under the [CC-BY-NC 4.0 license](https://creativecommons.org/licenses/by-nc-sa/4.0/)**\n\nYour first database will probably be complete, and perfect! You will be very proud of it! ;-)\n\nOver time, things will happen. Records will be deleted, and records will be added where there is incomplete information, for example, a germplasm stock record where the gene is unknown. You should think about these situations, because there are NO RULES telling you what you should do! You have to make a decision - a *policy* for your database - and you should follow that policy like a religion!\n\nFor example:\n\n* If there is no known gene for a given germplasm, what does in the stock.allele column? What goes in the stock.gene_id column? What goes in the gene table? Discuss....\n* If a stock is fully planted - no more seeds - what do you do? Should you delete the record? If you do, then your germplasm table is linked through the stock_id to a stock that no longer exists. If you gather more seed in the future, is that the same stock? (answer: NO, it is not!!!).... so do you update the existing stock record to say there is now 10g of seed? \n* Remember, you are trapped! In your table definition you declared all columns to be \"NOT NULL\", meaning that if the row exists, there must be a value for each column in the row! What do you do if there isn't a value to put into that column? \n * zero? \n * What if you change the column definition to allow NULL?\n * What does NULL mean? What does zero mean? \n * How does software respond to NULL or zero values? (you don't know this yet, but we can talk about it)\n\n\nFor our database, I am going to suggest this policy:\n1. if we don't know the allele, we put \"unknown\" in the allele column\n2. We put '0' into the gene_id column (auto_increment starts with 1 in the gene table, so a 0 will match nothing!) \n3. We DO NOT add a gene record at all.\n\nLet's add a record like this one to our database:\n",
"_____no_output_____"
]
],
[
[
"%sql INSERT INTO stock(amount, date, location) VALUES (23, '2018-05-12', 'Room 289');\n%sql INSERT INTO germplasm (taxonid, allele, stock_id, gene_id) VALUES (4150, 'unknown', LAST_INSERT_ID(), 0 );\n# note that I am using LAST_INSERT_ID to capture the auto_increment value from the stock table insert\n# this ensures that the germplasm and stock tables are 'synchronized'\n%sql SELECT * FROM germplasm;\n#%sql SELECT * FROM gene;",
"_____no_output_____"
]
],
[
[
"<pre>\n\n</pre>\nThat looks good! ...but we have just created a problem! gene_id=0 doesn't exist in the gene table, so what happens with our beautiful SELECT query that we just created above?\n\n",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM gene, germplasm WHERE \\\n germplasm.gene_id = gene.id;",
"_____no_output_____"
]
],
[
[
"\n### OH CRAP!!!! We lost our data!!\n\nOur \"unknown\" germplasm has disappeared!! Or has it?\n\nThe problem is that stock.gene_id = gene.id failed for the \"unknown\" record, and so it isn't reflected in the output from the query. THIS IS BAD, if (for example) you were trying to take an inventory of all germplasm stocks you had! \n\nHow do we solve this? The answer is to use SQL's \"JOIN\" instruction.\n\nThere are four kinds of JOIN: INNER, LEFT OUTER, RIGHT OUTER, and FULL OUTER.\n\nThe join we are doing with our current SELECT query is an INNER join. Using a Venn diagram, the query looks like this:\n\n<a href='https://learn.co/lessons/sql-complex-joins-readme'><img src='http://readme-pics.s3.amazonaws.com/Inner%20Join%20Venn%20Diagram.png' width=300px/></a>\n\n\nEffectively, the intersection where BOTH the 'left' (gene.id) and 'right' (germplasm.id) are true.\n\nYou can duplicate this behavior using the INNER JOIN instruction. The syntax is a little bit different - look:\n",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM gene INNER JOIN germplasm ON \\\n germplasm.gene_id = gene.id;",
"_____no_output_____"
]
],
[
[
"<pre>\n\n\n</pre>\nWhat we want is a query that allows one side to be \"missing/absent/NULL\", but the other side to exist.\n\nPerhaps we need a \"LEFT JOIN\"?\n\n ... gene LEFT JOIN germplasm ...\n\n\nAgain, in this situation, \"LEFT\" means the table on the Left side of the SQL JOIN statement (gene)\n\nAs a Venn diagram, Left joins look like this (ignore the :\n\n<a href='https://learn.co/lessons/sql-complex-joins-readme'><img src='http://readme-pics.s3.amazonaws.com/Left%20Outer%20Join%20Venn%20Diagram.png' width=300px/></a>\n\nWhat it means is that, **in addition to the perfect matches at the intersection**, the record on the left (the gene record) should be included in the result set, **even if it doesn't match** with a germplasm record (on the right). Is that the solution to our problem?\n",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM gene LEFT JOIN germplasm ON \\\n germplasm.gene_id = gene.id;",
"_____no_output_____"
]
],
[
[
"## PFFFFFF!! No, that was not the solution\n\nWhy?\n\nWhat about a RIGHT JOIN?\n\n ... gene RIGHT JOIN germplasm ...\n\n<a href='https://learn.co/lessons/sql-complex-joins-readme'><img src='http://readme-pics.s3.amazonaws.com/Right%20Outer%20Join%20Venn%20Diagram.png' width=300px/></a>\n\n\n\nAgain, in this situation, \"RIGHT\" means the table on the Right side of the SQL JOIN statement (germplasm). So the germplasm record should be included in the result set, even if a gene record does not exist. ...that sounds much more likely to be correct! \n",
"_____no_output_____"
]
],
[
[
"%sql SELECT * FROM gene RIGHT JOIN germplasm ON \\\n germplasm.gene_id = gene.id;",
"_____no_output_____"
]
],
[
[
"### Voila!!\n\n\n",
"_____no_output_____"
],
[
"\n## Your turn\n\n1) Create another record, where in this case, there is no **stock**, but there is a germplasm and a gene record. \n2) Create the JOIN query between germplasm and stock that includes all germplasm records\n",
"_____no_output_____"
],
[
"<pre>\n\n\n</pre>\n# Other SELECT \"magic\"\n\nYou can do many other useful things with SELECTS, such as:\n\n## COUNT()\n\nIf you want to count the number of records returned from a query, use the **COUNT() AS your_name** function:\n",
"_____no_output_____"
]
],
[
[
"%sql SELECT COUNT(*) AS \"Number Of Matches\" FROM gene RIGHT JOIN germplasm ON \\\n germplasm.gene_id = gene.id;",
"_____no_output_____"
]
],
[
[
"## SUM(), AVG(), MAX()\n\nYou can do mathematical functions on results also, for example, you can take the SUM of a column - how much seed do we have in total? \n\n(look carefully at this query! It's quite complicated!):\n",
"_____no_output_____"
]
],
[
[
"%sql SELECT SUM(stock.amount) FROM gene RIGHT JOIN germplasm ON \\\n germplasm.gene_id = gene.id \\\n INNER JOIN stock ON germplasm.stock_id = stock.id;\n",
"_____no_output_____"
]
],
[
[
"<pre>\n\n</pre>\nOr you could take the **average AVG()** of a column - what is the average quantity of seed we have?\n",
"_____no_output_____"
]
],
[
[
"%sql SELECT AVG(stock.amount) FROM gene RIGHT JOIN germplasm ON \\\n germplasm.gene_id = gene.id \\\n INNER JOIN stock ON germplasm.stock_id = stock.id;\n",
"_____no_output_____"
]
],
[
[
"<pre>\n\n</pre>\nOr you could take the **max MAX()** value of a column - what is the largest quantity of seed we have in our stocks?",
"_____no_output_____"
]
],
[
[
"%sql SELECT MAX(stock.amount) FROM gene RIGHT JOIN germplasm ON \\\n germplasm.gene_id = gene.id \\\n INNER JOIN stock ON germplasm.stock_id = stock.id;\n",
"_____no_output_____"
]
],
[
[
"## ORDER BY\n\nYou can put your results in a specific order:\n\n",
"_____no_output_____"
]
],
[
[
"%sql SELECT gene.gene_name, stock.amount FROM gene RIGHT JOIN germplasm ON \\\n germplasm.gene_id = gene.id \\\n INNER JOIN stock ON germplasm.stock_id = stock.id \\\n ORDER BY stock.amount DESC; # change this to ASC\n",
"_____no_output_____"
]
],
[
[
"<pre>\n\n\n</pre>\n## Conclusion\n\n1) Databases are a very powerful way to store structured information - far far better than Excel Spreadsheets! \n2) It will take you years (literally, years!) to become an expert in MySQL! We have only explored the most common functions here.",
"_____no_output_____"
]
],
[
[
"%sql drop database germplasm;",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a144180b830f5a3c7b14b69c329aea2d169903a
| 7,684 |
ipynb
|
Jupyter Notebook
|
pages/gallery/500hPa_Vorticity_Advection.ipynb
|
LProx2020/python-training
|
fee59beab788f923be491e16451544c055a754ff
|
[
"BSD-3-Clause"
] | 1 |
2020-01-18T20:19:53.000Z
|
2020-01-18T20:19:53.000Z
|
pages/gallery/500hPa_Vorticity_Advection.ipynb
|
LProx2020/python-training
|
fee59beab788f923be491e16451544c055a754ff
|
[
"BSD-3-Clause"
] | null | null | null |
pages/gallery/500hPa_Vorticity_Advection.ipynb
|
LProx2020/python-training
|
fee59beab788f923be491e16451544c055a754ff
|
[
"BSD-3-Clause"
] | 1 |
2019-12-16T05:46:07.000Z
|
2019-12-16T05:46:07.000Z
| 33.85022 | 104 | 0.551926 |
[
[
[
"500 hPa Vorticity Advection\n===========================\n\nPlot an 500-hPa map with calculating vorticity advection using MetPy calculations.\n\nBeyond just plotting 500-hPa level data, this uses calculations from `metpy.calc` to find\nthe vorticity and vorticity advection. Currently, this needs an extra helper function to\ncalculate the distance between lat/lon grid points.",
"_____no_output_____"
],
[
"Imports",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\n\nimport cartopy.crs as ccrs\nimport cartopy.feature as cfeature\nimport matplotlib.gridspec as gridspec\nimport matplotlib.pyplot as plt\nimport metpy.calc as mpcalc\nimport numpy as np\nimport scipy.ndimage as ndimage\n\nfrom metpy.units import units\nfrom netCDF4 import num2date\nfrom siphon.catalog import TDSCatalog",
"_____no_output_____"
]
],
[
[
"Data Aquisition\n---------------",
"_____no_output_____"
]
],
[
[
"dt = datetime(2016, 4, 16, 18)\n\n# Assemble our URL to the THREDDS Data Server catalog,\n# and access our desired dataset within via NCSS\nbase_url = 'https://www.ncei.noaa.gov/thredds/catalog/model-namanl-old/'\ncat = TDSCatalog(f'{base_url}{dt:%Y%m}/{dt:%Y%m%d}/catalog.xml')\nncss = cat.datasets[f'namanl_218_{dt:%Y%m%d}_{dt:%H}00_000.grb'].subset()\n\n# Query for Latest GFS Run\nquery = ncss.query()\n\nquery.time(dt)\nquery.accept('netcdf')\nquery.variables('Geopotential_height_isobaric',\n 'u-component_of_wind_isobaric',\n 'v-component_of_wind_isobaric')\nquery.add_lonlat()\n\n# Obtain our queried data\nds = ncss.get_data(query)\n\nlon = ds.variables['lon'][:]\nlat = ds.variables['lat'][:]\n\ntimes = ds.variables[ds.variables['Geopotential_height_isobaric'].dimensions[0]]\nvtime = num2date(times[:].squeeze(), units=times.units)\n\nlev_500 = np.where(ds.variables['isobaric'][:] == 500)[0][0]\n\nhght_500 = ds.variables['Geopotential_height_isobaric'][0, lev_500, :, :]\nhght_500 = ndimage.gaussian_filter(hght_500, sigma=3, order=0) * units.meter\n\nuwnd_500 = units('m/s') * ds.variables['u-component_of_wind_isobaric'][0, lev_500, :, :]\nvwnd_500 = units('m/s') * ds.variables['v-component_of_wind_isobaric'][0, lev_500, :, :]",
"_____no_output_____"
]
],
[
[
"Begin Data Calculations\n-----------------------",
"_____no_output_____"
]
],
[
[
"dx, dy = mpcalc.lat_lon_grid_deltas(lon, lat)\n\nf = mpcalc.coriolis_parameter(np.deg2rad(lat)).to(units('1/sec'))\n\navor = mpcalc.vorticity(uwnd_500, vwnd_500, dx, dy, dim_order='yx') + f\n\navor = ndimage.gaussian_filter(avor, sigma=3, order=0) * units('1/s')\n\nvort_adv = mpcalc.advection(avor, [uwnd_500, vwnd_500], (dx, dy), dim_order='yx') * 1e9",
"_____no_output_____"
]
],
[
[
"Map Creation\n------------",
"_____no_output_____"
]
],
[
[
"# Set up Coordinate System for Plot and Transforms\ndproj = ds.variables['LambertConformal_Projection']\nglobe = ccrs.Globe(ellipse='sphere', semimajor_axis=dproj.earth_radius,\n semiminor_axis=dproj.earth_radius)\ndatacrs = ccrs.LambertConformal(central_latitude=dproj.latitude_of_projection_origin,\n central_longitude=dproj.longitude_of_central_meridian,\n standard_parallels=[dproj.standard_parallel],\n globe=globe)\nplotcrs = ccrs.LambertConformal(central_latitude=45., central_longitude=-100.,\n standard_parallels=[30, 60])\n\nfig = plt.figure(1, figsize=(14., 12))\ngs = gridspec.GridSpec(2, 1, height_ratios=[1, .02], bottom=.07, top=.99,\n hspace=0.01, wspace=0.01)\nax = plt.subplot(gs[0], projection=plotcrs)\n\n# Plot Titles\nplt.title(r'500-hPa Heights (m), AVOR$*10^5$ ($s^{-1}$), AVOR Adv$*10^8$ ($s^{-2}$)',\n loc='left')\nplt.title(f'VALID: {vtime}', loc='right')\n\n# Plot Background\nax.set_extent([235., 290., 20., 58.], ccrs.PlateCarree())\nax.coastlines('50m', edgecolor='black', linewidth=0.75)\nax.add_feature(cfeature.STATES, linewidth=.5)\n\n# Plot Height Contours\nclev500 = np.arange(5100, 6061, 60)\ncs = ax.contour(lon, lat, hght_500.m, clev500, colors='black', linewidths=1.0,\n linestyles='solid', transform=ccrs.PlateCarree())\nplt.clabel(cs, fontsize=10, inline=1, inline_spacing=10, fmt='%i',\n rightside_up=True, use_clabeltext=True)\n\n# Plot Absolute Vorticity Contours\nclevvort500 = np.arange(-9, 50, 5)\ncs2 = ax.contour(lon, lat, avor*10**5, clevvort500, colors='grey',\n linewidths=1.25, linestyles='dashed', transform=ccrs.PlateCarree())\nplt.clabel(cs2, fontsize=10, inline=1, inline_spacing=10, fmt='%i',\n rightside_up=True, use_clabeltext=True)\n\n# Plot Colorfill of Vorticity Advection\nclev_avoradv = np.arange(-30, 31, 5)\ncf = ax.contourf(lon, lat, vort_adv.m, clev_avoradv[clev_avoradv != 0], extend='both',\n cmap='bwr', transform=ccrs.PlateCarree())\ncax = plt.subplot(gs[1])\ncb = plt.colorbar(cf, cax=cax, orientation='horizontal', extendrect='True', ticks=clev_avoradv)\ncb.set_label(r'$1/s^2$', size='large')\n\n# Plot Wind Barbs\n# Transform Vectors and plot wind barbs.\nax.barbs(lon, lat, uwnd_500.m, vwnd_500.m, length=6, regrid_shape=20,\n pivot='middle', transform=ccrs.PlateCarree())",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a1442575d343eb2e7240c2005077215bc3cdaba
| 16,439 |
ipynb
|
Jupyter Notebook
|
databases/snowflake/batch_prediction_api/snow_batch_prediction_api_query_s3.ipynb
|
joemeree/ai_engineering
|
7dba6c808f27d895afc86cf119e876384106fb3c
|
[
"Apache-2.0"
] | 9 |
2021-04-08T17:45:46.000Z
|
2022-02-28T09:43:44.000Z
|
databases/snowflake/batch_prediction_api/snow_batch_prediction_api_query_s3.ipynb
|
joemeree/ai_engineering
|
7dba6c808f27d895afc86cf119e876384106fb3c
|
[
"Apache-2.0"
] | 13 |
2020-08-14T15:17:00.000Z
|
2022-02-27T20:12:44.000Z
|
databases/snowflake/batch_prediction_api/snow_batch_prediction_api_query_s3.ipynb
|
joemeree/ai_engineering
|
7dba6c808f27d895afc86cf119e876384106fb3c
|
[
"Apache-2.0"
] | 3 |
2020-08-14T12:47:28.000Z
|
2022-03-23T18:58:13.000Z
| 29.30303 | 152 | 0.530142 |
[
[
[
"\"\"\"\nSnowflake Batch Prediction API Snowflake S3 scoring job\n\nv1.0 Mike Taveirne (doyouevendata) 3/21/2020\n\"\"\"",
"_____no_output_____"
],
[
"import pandas as pd\nimport requests\nimport time\nfrom pandas.io.json import json_normalize\nimport snowflake.connector\n\nimport my_creds\n#from imp import reload\n#reload(my_creds)",
"_____no_output_____"
],
[
"# datarobot parameters\nAPI_KEY = my_creds.API_KEY\nUSERNAME = my_creds.USERNAME\nDEPLOYMENT_ID = my_creds.DEPLOYMENT_ID\nDATAROBOT_KEY = my_creds.DATAROBOT_KEY\n# replace with the load balancer for your prediction instance(s)\nDR_PREDICTION_HOST = my_creds.DR_PREDICTION_HOST\nDR_APP_HOST = 'https://app.datarobot.com'\n\nDR_MODELING_HEADERS = {'Content-Type': 'application/json', 'Authorization': 'token %s' % API_KEY}",
"_____no_output_____"
],
[
"# snowflake parameters\nSNOW_ACCOUNT = my_creds.SNOW_ACCOUNT\nSNOW_USER = my_creds.SNOW_USER\nSNOW_PASS = my_creds.SNOW_PASS\nSNOW_DB = 'TITANIC'\nSNOW_SCHEMA = 'PUBLIC'\n\n# ETL parameters\nJOB_NAME = 'pass_scoring'",
"_____no_output_____"
]
],
[
[
"### Retrieve or Create S3 Credentials",
"_____no_output_____"
]
],
[
[
"# get a saved credential set, return None if not found\ndef dr_get_catalog_credentials(name, cred_type):\n if cred_type not in ['basic', 's3']:\n print('credentials type must be: basic, s3 - value passed was {ct}'.format(ct=cred_type))\n return None\n \n credentials_id = None\n\n response = requests.get(\n DR_APP_HOST + '/api/v2/credentials/',\n headers=DR_MODELING_HEADERS,\n )\n\n if response.status_code == 200:\n\n df = pd.io.json.json_normalize(response.json()['data'])[['credentialId', 'name', 'credentialType']]\n\n if df[(df['name'] == name) & (df['credentialType'] == cred_type)]['credentialId'].size > 0:\n credentials_id = df[(df['name'] == name) & (df['credentialType'] == cred_type)]['credentialId'].iloc[0]\n \n else:\n\n print('Request failed; http error {code}: {content}'.format(code=response.status_code, content=response.content))\n\n return credentials_id",
"_____no_output_____"
],
[
"# create credentials set\ndef dr_create_catalog_credentials(name, cred_type, user, password, token=None):\n if cred_type not in ['basic', 's3']:\n print('credentials type must be: basic, s3 - value passed was {ct}'.format(ct=cred_type))\n return None\n \n if cred_type == 'basic': \n json = {\n \"credentialType\": cred_type,\n \"user\": user,\n \"password\": password,\n \"name\": name\n }\n elif cred_type == 's3' and token != None: \n json = {\n \"credentialType\": cred_type,\n \"awsAccessKeyId\": user,\n \"awsSecretAccessKey\": password,\n \"awsSessionToken\": token,\n \"name\": name\n }\n elif cred_type == 's3' and token == None: \n json = {\n \"credentialType\": cred_type,\n \"awsAccessKeyId\": user,\n \"awsSecretAccessKey\": password,\n \"name\": name\n }\n \n response = requests.post(\n url = DR_APP_HOST + '/api/v2/credentials/',\n headers=DR_MODELING_HEADERS,\n json=json\n )\n \n if response.status_code == 201:\n\n return response.json()['credentialId']\n \n else:\n\n print('Request failed; http error {code}: {content}'.format(code=response.status_code, content=response.content))\n",
"_____no_output_____"
],
[
"# get or create a credential set\ndef dr_get_or_create_catalog_credentials(name, cred_type, user, password, token=None):\n cred_id = dr_get_catalog_credentials(name, cred_type)\n \n if cred_id == None:\n return dr_create_catalog_credentials(name, cred_type, user, password, token=None)\n else:\n return cred_id",
"_____no_output_____"
],
[
"credentials_id = dr_get_or_create_catalog_credentials('s3_community', \n 's3', my_creds.SNOW_USER, my_creds.SNOW_PASS)",
"_____no_output_____"
]
],
[
[
"### Extract Data to S3 via Snowflake",
"_____no_output_____"
]
],
[
[
"# create a connection\nctx = snowflake.connector.connect(\n user=SNOW_USER,\n password=SNOW_PASS,\n account=SNOW_ACCOUNT,\n database=SNOW_DB,\n schema=SNOW_SCHEMA,\n protocol='https'\n)\n\n# create a cursor\ncur = ctx.cursor()\n\n# execute sql to get start/end timestamps to use\nsql = \"select last_ts_scored_through, current_timestamp::TIMESTAMP_NTZ cur_ts \" \\\n \"from etl_history \" \\\n \"where job_nm = '{job}' \" \\\n \"order by last_ts_scored_through desc \" \\\n \"limit 1 \".format(job=JOB_NAME)\ncur.execute(sql)\n\n# fetch results into dataframe\ndf = cur.fetch_pandas_all()\nstart_ts = df['LAST_TS_SCORED_THROUGH'][0]\nend_ts = df['CUR_TS'][0]\n\n# execute sql to dump data into a single file in S3 stage bucket\n# AWS single file snowflake limit 5 GB\nsql = \"COPY INTO @S3_SUPPORT/titanic/community/\" + JOB_NAME + \".csv \" \\\n \"from \" \\\n \"( \" \\\n \" select passengerid, pclass, name, sex, age, sibsp, parch, ticket, fare, cabin, embarked \" \\\n \" from passengers_500k_ts \" \\\n \" where nvl(updt_ts, crt_ts) >= '{start}' \" \\\n \" and nvl(updt_ts, crt_ts) < '{end}' \" \\\n \") \" \\\n \"file_format = (format_name='default_csv' compression='none') header=true overwrite=true single=true;\".format(start=start_ts, end=end_ts)\ncur.execute(sql)",
"_____no_output_____"
]
],
[
[
"### Create DataRobot Session and Running Batch Prediction API Job",
"_____no_output_____"
]
],
[
[
"session = requests.Session()\nsession.headers = {\n 'Authorization': 'Bearer {}'.format(API_KEY)\n}",
"_____no_output_____"
],
[
"INPUT_FILE = 's3://'+ my_creds.S3_BUCKET + '/titanic/community/' + JOB_NAME + '.csv'\nOUTPUT_FILE = 's3://'+ my_creds.S3_BUCKET + '/titanic/community/' + JOB_NAME + '_scored.csv'\n\njob_details = {\n 'deploymentId': DEPLOYMENT_ID,\n 'passthroughColumns': ['PASSENGERID'],\n 'numConcurrent': 4,\n \"predictionInstance\" : {\n \"hostName\": DR_PREDICTION_HOST,\n \"datarobotKey\": DATAROBOT_KEY\n },\n 'intakeSettings': {\n 'type': 's3',\n 'url': INPUT_FILE,\n 'credentialId': credentials_id\n },\n 'outputSettings': {\n 'type': 's3',\n 'url': OUTPUT_FILE,\n 'credentialId': credentials_id\n }\n}",
"_____no_output_____"
],
[
"response = session.post(\n DR_APP_HOST + '/api/v2/batchPredictions',\n json=job_details\n )",
"_____no_output_____"
]
],
[
[
"### Monitor S3 Scoring Status and Return Control Upon Completion",
"_____no_output_____"
]
],
[
[
"if response.status_code == 202:\n \n job = response.json()\n print('queued batch job: {}'.format(job['links']['self']))\n\n while job['status'] == 'INITIALIZING':\n time.sleep(3)\n response = session.get(job['links']['self'])\n response.raise_for_status()\n job = response.json()\n \n print('completed INITIALIZING')\n \n if job['status'] == 'RUNNING':\n\n while job['status'] == 'RUNNING':\n time.sleep(3)\n response = session.get(job['links']['self'])\n response.raise_for_status()\n job = response.json()\n \n print('completed RUNNING')\n print('status is now {status}'.format(status=job['status']))\n \n if job['status'] != 'COMPLETED':\n for i in job['logs']:\n print(i)\n \nelse:\n \n print('Job submission failed; http error {code}: {content}'.format(code=response.status_code, content=response.content))",
"queued batch job: https://app.datarobot.com/api/v2/batchPredictions/1234567891234567893/\ncompleted INITIALIZING\ncompleted RUNNING\nstatus is now COMPLETED\n"
]
],
[
[
"### Truncate and Reload STG Staging Table with Results",
"_____no_output_____"
]
],
[
[
"# multi-statement executions\n# https://docs.snowflake.com/en/user-guide/python-connector-api.html#execute_string\n\n# truncate and load STG schema table with scored results\nsql = \"truncate titanic.stg.PASSENGERS_SCORED_BATCH_API; \" \\\n \" copy into titanic.stg.PASSENGERS_SCORED_BATCH_API from @S3_SUPPORT/titanic/community/\" + JOB_NAME + \"_scored.csv\" \\\n \" FILE_FORMAT = 'DEFAULT_CSV' ON_ERROR = 'ABORT_STATEMENT' PURGE = FALSE;\"\nctx.execute_string(sql)",
"_____no_output_____"
]
],
[
[
"### Update Presentation Target Table With Results",
"_____no_output_____"
]
],
[
[
"# update target presentation table and ETL history table in transaction\n\nsql = \\\n \"begin; \" \\\n \"update titanic.public.passengers_500k_ts trg \" \\\n \"set trg.survival = src.survived_1_prediction \" \\\n \"from titanic.stg.PASSENGERS_SCORED_BATCH_API src \" \\\n \"where src.passengerid = trg.passengerid; \" \\\n \"insert into etl_history values ('{job}', '{run_through_ts}'); \" \\\n \"commit; \".format(job=JOB_NAME, run_through_ts=end_ts)\nctx.execute_string(sql)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a14581348260d9ebe00881ff3dd32c79506f67c
| 10,931 |
ipynb
|
Jupyter Notebook
|
MR_left_t4.ipynb
|
geraldfj/multiregressor
|
780d46c3299f550fde3de098b86ec7028932db59
|
[
"Apache-2.0"
] | null | null | null |
MR_left_t4.ipynb
|
geraldfj/multiregressor
|
780d46c3299f550fde3de098b86ec7028932db59
|
[
"Apache-2.0"
] | null | null | null |
MR_left_t4.ipynb
|
geraldfj/multiregressor
|
780d46c3299f550fde3de098b86ec7028932db59
|
[
"Apache-2.0"
] | null | null | null | 30.196133 | 145 | 0.514409 |
[
[
[
"import pandas as pd\nimport numpy as np\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn import ensemble\nfrom sklearn.utils import shuffle\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn.metrics import r2_score\nfrom sklearn.metrics import accuracy_score\nfrom sklearn import metrics\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.multioutput import MultiOutputRegressor\nfrom sklearn.ensemble import RandomForestRegressor\nfrom matplotlib import pyplot as plt\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.decomposition.pca import PCA\nfrom sklearn.preprocessing import scale",
"_____no_output_____"
],
[
"def find_nearest(array,value):\n idx = (np.abs(array-value)).argmin()\n return array[idx]",
"_____no_output_____"
],
[
"def refine_data(arr1,arr2,arr3):\n refined_data = []\n for i in range(arr1.shape[0]):\n rt = find_nearest(arr1[i],arr3[i][0])\n refined_data.append([rt,arr2[i][arr1[i].tolist().index(rt)]])\n \n refined_data = np.array(refined_data)\n return refined_data",
"_____no_output_____"
],
[
"def get_metrics(arr1,arr2):\n mse = mean_squared_error(arr1,arr2)\n r2 = r2_score(arr1,arr2)\n return mse,r2",
"_____no_output_____"
],
[
"d1 = pd.read_csv('X_data_t3',sep = ',')\nd2 = pd.read_csv('y_data',sep = ',')\nd3 = pd.concat([d1,d2],axis=1)\n\n#data = d3.loc[(d3['maxRT_ab'] < 1e6) & (d3['maxRT_ab'] > 3e4)]\ndata = d3.loc[(d3['maxRT_ab'] > 3e4)]\n#data = d3.loc[(d3['maxRT_ab'] < 1e6)]\n#d3 = shuffle(d3)\n\n#X = np.array(d3[['maxRT_t','x_start_t','diff_start','diff_end']])\n#X = np.array(d3[['maxRT_ab','maxRT_baseline','x_start_ab','x_end_ab']])\n#X = np.array(d3[['maxRT_t','maxRT_ab','maxRT_baseline','x_start_ab','x_end_ab']])\nX = np.array(d3[['rt','maxRT_t','maxRT_ab','maxRT_base','x_start_t','x_start_ab','diff_start','x_end_t','x_end_ab','diff_end','width']])\n\n#X = np.array(d3[['rt','maxRT_ab','x_start_t','x_end_t','width']])\n\n\ny = np.array(d3[['y_left_t','y_left_ab']])\n\n\ntime = np.array(pd.read_csv('time',sep = ',',header = None).dropna(axis = 'columns').round(3))\nabundance = np.array(pd.read_csv('abundance',sep = ',',header = None).dropna(axis = 'columns'))\nbaseline = np.array(pd.read_csv('baseline',sep = ',',header = None).dropna(axis = 'columns'))\n#time = time[0:420]\n#abundance = abundance[0:420]\n#baseline = baseline[0:420]",
"_____no_output_____"
],
[
"np.amax(d3['maxRT_ab']), np.amin(d3['maxRT_ab']), X.shape",
"_____no_output_____"
],
[
"X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.2,random_state=42)",
"_____no_output_____"
],
[
"scaler_train = MinMaxScaler(feature_range=(0,1))\n#scaler_train = StandardScaler()\nscaler_train.fit(X_train)\nX_train =scaler_train.transform(X_train)\n\nscaler_test = MinMaxScaler(feature_range=(0,1))\n#scaler_test = StandardScaler()\nscaler_test.fit(X_test)\nX_test = scaler_test.transform(X_test)",
"_____no_output_____"
],
[
"n_estimators = 1000\nmax_depth = 1000\nrandom_state = 1000\nite = 1\n\n#mse_t,r2_t = [[]for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]\n#mse_ab,r2_ab = [[]for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]\n\n\nparams = {'n_estimators': n_estimators, 'max_depth': max_depth, 'min_samples_split': 20,\n 'n_jobs': -1, 'random_state':random_state}\n \nclf = RandomForestRegressor(**params)\n\n\nscores = cross_val_score(clf, X_train, y_train, cv=5)\nclf.fit(X_train,y_train)\ny_predict = clf.predict(X_test).round(3)\n \nmse_1,r2_1 = get_metrics(y_test[:,0],y_predict[:,0])\nmse_2,r2_2 = get_metrics(y_test[:,1],y_predict[:,1])\nprint(mse_1,r2_1,mse_2,r2_2)\n \n \n \n",
"0.0002529294117647059 -0.0052749823140381125 15299224.448273474 0.5448056130182723\n"
],
[
"y_test[:,0]",
"_____no_output_____"
],
[
"y_predict[:,0]",
"_____no_output_____"
],
[
"from sklearn.decomposition.pca import PCA\nfrom sklearn.preprocessing import scale\npca = PCA()\nX_reduced = pca.fit_transform(X_train)\nscores = cross_val_score(clf, X_reduced, y_train, cv=5)\nprint(scores)\nclf.fit(X_reduced,y_train)\ny_predict = clf.predict(X_test).round(3)\n \n \nmse_1,r2_1 = get_metrics(y_test[:,0],y_predict[:,0])\nmse_2,r2_2 = get_metrics(y_test[:,1],y_predict[:,1])\n \n \nprint(r2_1,r2_2)\nprint(mse_1,mse_2)\n",
"[0.73976687 0.75249782 0.67413778 0.72655178 0.36610512]\n-0.07639565062882814 -5.2099801026128905\n0.00027082352941176503 208719356.22747636\n"
],
[
"y_predict[:,0]",
"_____no_output_____"
],
[
"np.cumsum(np.round(pca.explained_variance_ratio_, decimals=4)*100)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a145d35c2ea78cff13365ab66a11c76916b74fe
| 5,875 |
ipynb
|
Jupyter Notebook
|
Week7BeamShear/.ipynb_checkpoints/ENGR213Lec11-checkpoint.ipynb
|
smithrockmaker/ENGR213
|
96e1c9da5aa02d280dd602194a5c7ace55cbd71b
|
[
"MIT"
] | null | null | null |
Week7BeamShear/.ipynb_checkpoints/ENGR213Lec11-checkpoint.ipynb
|
smithrockmaker/ENGR213
|
96e1c9da5aa02d280dd602194a5c7ace55cbd71b
|
[
"MIT"
] | null | null | null |
Week7BeamShear/.ipynb_checkpoints/ENGR213Lec11-checkpoint.ipynb
|
smithrockmaker/ENGR213
|
96e1c9da5aa02d280dd602194a5c7ace55cbd71b
|
[
"MIT"
] | null | null | null | 52.927928 | 653 | 0.679149 |
[
[
[
"## ENGR 213 Lecture Notes\n\n### Lecture 10\n#### Shear Stress:\n\nNow that we have some sense of shear and moment diagrams and how they relate to the normal stresses across the face of a cross section of a beam it's time to add another layer to our thinking. An important characteristic of the normal stress across the cross section of a beam is that it is **NOT** constant. That means that there are different normal forces between adjacent longitudinal slices of the beam. This results in what are called **longitudinal shear** forces that can also lead to failure of the beam. Be sure to distinguish the longitudinal shear forces from transverse shear forces which are perpendicular to the axis of the beam.\n\n<img src=\"https://raw.githubusercontent.com/smithrockmaker/ENGR213/main/images/beamBendingVisual.jpg\" width=\"600\"/>",
"_____no_output_____"
],
[
"### Longitudinal Shear: Conceptual\n\nIf you think of a beam as being made up of a lot of thin layers of the material the concept of transverse shear is relatively apparent. Take a stack of paper and flex the stack (bending a beam) and notice that the ends of the paper stick out relative to each other. This illustrates that the individual layers of paper have slipped (sheared) relative to each other. If the stack of paper were a solid block then the individual layers would be prevented from slipping leading to shear stress between the layers. A simplified illustration is shown below.\n\n<img src=\"https://raw.githubusercontent.com/smithrockmaker/ENGR213/master/images/beamLongitudinalShear.jpg\" width=\"600\"/>\n",
"_____no_output_____"
],
[
"### Shear Formula\n\nThe derivation of the shear formula is pretty cool but quite complex and I'm not sure how much it adds to our understanding. Like many of the formulae we have developed it involves considering a small cross section of the beam and slicing it parallel to the axis of beam. Then it's just a 'simple' matter of setting the sum of the forces and moments to 0 and solving for the shear stress.\n\nThe **shear formula** is deceptively simple. Knowing meaning of each term and how to determine it is the challenge.\n\n.$$\\large \\tau_{longitudinal} = \\tau = \\frac{V\\: Q}{I\\: t}$$\n\nV: V is the transverse shear force at the particular cross section we are considering. Note that this means that any point on the shear diagram where V = 0 there are no longitudinal shear stresses anywhere along that cross section.\n\nI: I is the 2nd moment of inertia around the neutral axis of the cross section of the beam. This is the same moment of inertia we used in the flexure formula for normal stresses in the beam. Nothing new here.\n\nt: t is the width of the shear surface. In general this is the width of the beam at the location of interest. Note that because it is in the denominator of the expression the shear stress is higher where the beam is narrower, typically on the vertical web.\n\nQ: Q is the first moment of the area above the shear surface we are interested in. Consider the graphic below...\n\n<img src=\"https://raw.githubusercontent.com/smithrockmaker/ENGR213/master/images/beamQDef.png\" width=\"300\"/>\n\nFor simple geometries $Q = y_s A_s$ while more generally...\n\n.$$\\large Q = \\int_{y_{shear}}^{y_{max}} dA = y_s A_s$$\n\nNotice that Q is **NOT** 0 for the neutral axis because $y_s$ is the distance from then neutral axis to the centroid of the area above the shear surface. In fact, Q reaches it's maximum value at the neutral axis (an interesting proof to consider). \n",
"_____no_output_____"
],
[
"#### Conceptually:\n\nAn important conceptual feature is that the shear stress is generally a maximum on the neutral axis. This is precisely the opposite of the normal stress in a beam. \n\n#### Beam Design:\n\nI don't have a great personal sense of the relationship between the shear strength of materials used in beams vs the normal strength. The fact that it is generally prudent to put holes through floor joists (a wooden I beam) at the neutral axis is a suggestion that shear stresses are not the primary concern (see image below). \n\n<img src=\"https://raw.githubusercontent.com/smithrockmaker/ENGR213/master/images/TJIPenetrations.png\" width=\"600\"/>\n\nA clear counterexample are wooden beams made up of laminates of smaller stock (called glulams) where the shear stresses on the glue joints can lead to delamination of the beams. Here's a related example that suggests shear failure though it might be more complex.\n\n<img src=\"https://raw.githubusercontent.com/smithrockmaker/ENGR213/master/images/beamDelamination.jpg\" width=\"600\"/>",
"_____no_output_____"
]
]
] |
[
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
]
] |
4a145fac2af9b7c82bf8898c3f64b4e9d8f91abe
| 109,490 |
ipynb
|
Jupyter Notebook
|
intro-to-pytorch/Part 5 - Inference and Validation.ipynb
|
jetsonai11/Udacity_Deep_Learning_Nanodegree_2020
|
7193092812f0fcde975a93180e547081f50766c4
|
[
"MIT"
] | 2 |
2020-05-09T01:52:34.000Z
|
2020-05-12T08:06:58.000Z
|
intro-to-pytorch/Part 5 - Inference and Validation.ipynb
|
jetsonai11/Udacity_Deep_Learning_Nanodegree_2020
|
7193092812f0fcde975a93180e547081f50766c4
|
[
"MIT"
] | 3 |
2020-11-13T18:49:26.000Z
|
2022-02-10T01:53:18.000Z
|
intro-to-pytorch/Part 5 - Inference and Validation.ipynb
|
jetsonai11/Udacity_Deep_Learning_Nanodegree_2020
|
7193092812f0fcde975a93180e547081f50766c4
|
[
"MIT"
] | null | null | null | 107.554028 | 51,224 | 0.825436 |
[
[
[
"# Inference and Validation\n\nNow that you have a trained network, you can use it for making predictions. This is typically called **inference**, a term borrowed from statistics. However, neural networks have a tendency to perform *too well* on the training data and aren't able to generalize to data that hasn't been seen before. This is called **overfitting** and it impairs inference performance. To test for overfitting while training, we measure the performance on data not in the training set called the **validation** set. We avoid overfitting through regularization such as dropout while monitoring the validation performance during training. In this notebook, I'll show you how to do this in PyTorch. \n\nAs usual, let's start by loading the dataset through torchvision. You'll learn more about torchvision and loading data in a later part. This time we'll be taking advantage of the test set which you can get by setting `train=False` here:\n\n```python\ntestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)\n```\n\nThe test set contains images just like the training set. Typically you'll see 10-20% of the original dataset held out for testing and validation with the rest being used for training.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torchvision import datasets, transforms\n\n# Define a transform to normalize the data\ntransform = transforms.Compose([transforms.ToTensor(),\n transforms.Normalize((0.5,), (0.5,))])\n# Download and load the training data\ntrainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)\ntrainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)\n\n# Download and load the test data\ntestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)\ntestloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True)",
"_____no_output_____"
]
],
[
[
"Here I'll create a model like normal, using the same one from my solution for part 4.",
"_____no_output_____"
]
],
[
[
"from torch import nn, optim\nimport torch.nn.functional as F\n\nclass Classifier(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(784, 256)\n self.fc2 = nn.Linear(256, 128)\n self.fc3 = nn.Linear(128, 64)\n self.fc4 = nn.Linear(64, 10)\n \n def forward(self, x):\n # make sure input tensor is flattened\n x = x.view(x.shape[0], -1)\n \n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = F.relu(self.fc3(x))\n x = F.log_softmax(self.fc4(x), dim=1)\n \n return x",
"_____no_output_____"
]
],
[
[
"The goal of validation is to measure the model's performance on data that isn't part of the training set. Performance here is up to the developer to define though. Typically this is just accuracy, the percentage of classes the network predicted correctly. Other options are [precision and recall](https://en.wikipedia.org/wiki/Precision_and_recall#Definition_(classification_context)) and top-5 error rate. We'll focus on accuracy here. First I'll do a forward pass with one batch from the test set.",
"_____no_output_____"
]
],
[
[
"model = Classifier()\n\nimages, labels = next(iter(testloader))\n# Get the class probabilities\nps = torch.exp(model(images))\n# Make sure the shape is appropriate, we should get 10 class probabilities for 64 examples\nprint(ps.shape)",
"torch.Size([64, 10])\n"
]
],
[
[
"With the probabilities, we can get the most likely class using the `ps.topk` method. This returns the $k$ highest values. Since we just want the most likely class, we can use `ps.topk(1)`. This returns a tuple of the top-$k$ values and the top-$k$ indices. If the highest value is the fifth element, we'll get back 4 as the index.",
"_____no_output_____"
]
],
[
[
"top_p, top_class = ps.topk(1, dim=1)\n# Look at the most likely classes for the first 10 examples\nprint(top_class[:10,:])",
"tensor([[2],\n [2],\n [2],\n [3],\n [3],\n [2],\n [2],\n [2],\n [3],\n [2]])\n"
]
],
[
[
"Now we can check if the predicted classes match the labels. This is simple to do by equating `top_class` and `labels`, but we have to be careful of the shapes. Here `top_class` is a 2D tensor with shape `(64, 1)` while `labels` is 1D with shape `(64)`. To get the equality to work out the way we want, `top_class` and `labels` must have the same shape.\n\nIf we do\n\n```python\nequals = top_class == labels\n```\n\n`equals` will have shape `(64, 64)`, try it yourself. What it's doing is comparing the one element in each row of `top_class` with each element in `labels` which returns 64 True/False boolean values for each row.",
"_____no_output_____"
]
],
[
[
"equals = top_class == labels.view(top_class.shape)",
"_____no_output_____"
],
[
"equals",
"_____no_output_____"
]
],
[
[
"Now we need to calculate the percentage of correct predictions. `equals` has binary values, either 0 or 1. This means that if we just sum up all the values and divide by the number of values, we get the percentage of correct predictions. This is the same operation as taking the mean, so we can get the accuracy with a call to `torch.mean`. If only it was that simple. If you try `torch.mean(equals)`, you'll get an error\n\n```\nRuntimeError: mean is not implemented for type torch.ByteTensor\n```\n\nThis happens because `equals` has type `torch.ByteTensor` but `torch.mean` isn't implemented for tensors with that type. So we'll need to convert `equals` to a float tensor. Note that when we take `torch.mean` it returns a scalar tensor, to get the actual value as a float we'll need to do `accuracy.item()`.",
"_____no_output_____"
]
],
[
[
"accuracy = torch.mean(equals.type(torch.FloatTensor))\nprint(f'Accuracy: {accuracy.item()*100}%')\n",
"Accuracy: 9.375%\n"
]
],
[
[
"The network is untrained so it's making random guesses and we should see an accuracy around 10%. Now let's train our network and include our validation pass so we can measure how well the network is performing on the test set. Since we're not updating our parameters in the validation pass, we can speed up our code by turning off gradients using `torch.no_grad()`:\n\n```python\n# turn off gradients\nwith torch.no_grad():\n # validation pass here\n for images, labels in testloader:\n ...\n```\n\n>**Exercise:** Implement the validation loop below and print out the total accuracy after the loop. You can largely copy and paste the code from above, but I suggest typing it in because writing it out yourself is essential for building the skill. In general you'll always learn more by typing it rather than copy-pasting. You should be able to get an accuracy above 80%.",
"_____no_output_____"
]
],
[
[
"model = Classifier()\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.parameters(), lr=0.003)\n\nepochs = 30\nsteps = 0\n\ntrain_losses, test_losses = [], []\nfor e in range(epochs):\n running_loss = 0\n for images, labels in trainloader:\n images = images.view(images.shape[0], -1)\n optimizer.zero_grad()\n \n log_ps = model(images)\n loss = criterion(log_ps, labels)\n loss.backward()\n optimizer.step()\n \n running_loss += loss.item()\n \n else:\n test_loss = 0\n accuracy = 0\n ## TODO: Implement the validation pass and print out the validation accuracy\n with torch.no_grad():\n for images, labels in testloader:\n images = images.view(images.shape[0], -1)\n log_ps_test = model(images)\n test_loss += criterion(log_ps_test, labels)\n \n output = torch.exp(log_ps_test)\n top_p, top_class = output.topk(1, dim=1)\n equals = top_class == labels.view(top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor))\n \n train_losses.append(running_loss/len(trainloader))\n test_losses.append(test_loss/len(testloader))\n\n print(\"Epoch: {}/{}.. \".format(e+1, epochs),\n \"Training Loss: {:.3f}.. \".format(running_loss/len(trainloader)),\n \"Test Loss: {:.3f}.. \".format(test_loss/len(testloader)),\n \"Test Accuracy: {:.3f}\".format(accuracy/len(testloader)))",
"Epoch: 1/30.. Training Loss: 0.515.. Test Loss: 0.451.. Test Accuracy: 0.831\nEpoch: 2/30.. Training Loss: 0.388.. Test Loss: 0.412.. Test Accuracy: 0.851\nEpoch: 3/30.. Training Loss: 0.353.. Test Loss: 0.389.. Test Accuracy: 0.858\nEpoch: 4/30.. Training Loss: 0.330.. Test Loss: 0.376.. Test Accuracy: 0.866\nEpoch: 5/30.. Training Loss: 0.314.. Test Loss: 0.392.. Test Accuracy: 0.861\nEpoch: 6/30.. Training Loss: 0.297.. Test Loss: 0.358.. Test Accuracy: 0.873\nEpoch: 7/30.. Training Loss: 0.289.. Test Loss: 0.402.. Test Accuracy: 0.867\nEpoch: 8/30.. Training Loss: 0.280.. Test Loss: 0.357.. Test Accuracy: 0.873\nEpoch: 9/30.. Training Loss: 0.269.. Test Loss: 0.384.. Test Accuracy: 0.867\nEpoch: 10/30.. Training Loss: 0.265.. Test Loss: 0.365.. Test Accuracy: 0.872\nEpoch: 11/30.. Training Loss: 0.253.. Test Loss: 0.368.. Test Accuracy: 0.877\nEpoch: 12/30.. Training Loss: 0.248.. Test Loss: 0.394.. Test Accuracy: 0.870\nEpoch: 13/30.. Training Loss: 0.245.. Test Loss: 0.381.. Test Accuracy: 0.876\nEpoch: 14/30.. Training Loss: 0.240.. Test Loss: 0.399.. Test Accuracy: 0.872\nEpoch: 15/30.. Training Loss: 0.235.. Test Loss: 0.398.. Test Accuracy: 0.877\nEpoch: 16/30.. Training Loss: 0.229.. Test Loss: 0.378.. Test Accuracy: 0.879\nEpoch: 17/30.. Training Loss: 0.223.. Test Loss: 0.427.. Test Accuracy: 0.866\nEpoch: 18/30.. Training Loss: 0.224.. Test Loss: 0.362.. Test Accuracy: 0.888\nEpoch: 19/30.. Training Loss: 0.221.. Test Loss: 0.379.. Test Accuracy: 0.882\nEpoch: 20/30.. Training Loss: 0.215.. Test Loss: 0.381.. Test Accuracy: 0.884\nEpoch: 21/30.. Training Loss: 0.208.. Test Loss: 0.392.. Test Accuracy: 0.885\nEpoch: 22/30.. Training Loss: 0.207.. Test Loss: 0.406.. Test Accuracy: 0.881\nEpoch: 23/30.. Training Loss: 0.204.. Test Loss: 0.411.. Test Accuracy: 0.879\nEpoch: 24/30.. Training Loss: 0.193.. Test Loss: 0.412.. Test Accuracy: 0.880\nEpoch: 25/30.. Training Loss: 0.197.. Test Loss: 0.405.. Test Accuracy: 0.880\nEpoch: 26/30.. Training Loss: 0.194.. Test Loss: 0.402.. Test Accuracy: 0.880\nEpoch: 27/30.. Training Loss: 0.189.. Test Loss: 0.416.. Test Accuracy: 0.884\nEpoch: 28/30.. Training Loss: 0.187.. Test Loss: 0.408.. Test Accuracy: 0.883\nEpoch: 29/30.. Training Loss: 0.185.. Test Loss: 0.408.. Test Accuracy: 0.885\nEpoch: 30/30.. Training Loss: 0.187.. Test Loss: 0.416.. Test Accuracy: 0.886\n"
]
],
[
[
"## Overfitting\n\nIf we look at the training and validation losses as we train the network, we can see a phenomenon known as overfitting.\n\n<img src='assets/overfitting.png' width=450px>\n\nThe network learns the training set better and better, resulting in lower training losses. However, it starts having problems generalizing to data outside the training set leading to the validation loss increasing. The ultimate goal of any deep learning model is to make predictions on new data, so we should strive to get the lowest validation loss possible. One option is to use the version of the model with the lowest validation loss, here the one around 8-10 training epochs. This strategy is called *early-stopping*. In practice, you'd save the model frequently as you're training then later choose the model with the lowest validation loss.\n\nThe most common method to reduce overfitting (outside of early-stopping) is *dropout*, where we randomly drop input units. This forces the network to share information between weights, increasing it's ability to generalize to new data. Adding dropout in PyTorch is straightforward using the [`nn.Dropout`](https://pytorch.org/docs/stable/nn.html#torch.nn.Dropout) module.\n\n```python\nclass Classifier(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(784, 256)\n self.fc2 = nn.Linear(256, 128)\n self.fc3 = nn.Linear(128, 64)\n self.fc4 = nn.Linear(64, 10)\n \n # Dropout module with 0.2 drop probability\n self.dropout = nn.Dropout(p=0.2)\n \n def forward(self, x):\n # make sure input tensor is flattened\n x = x.view(x.shape[0], -1)\n \n # Now with dropout\n x = self.dropout(F.relu(self.fc1(x)))\n x = self.dropout(F.relu(self.fc2(x)))\n x = self.dropout(F.relu(self.fc3(x)))\n \n # output so no dropout here\n x = F.log_softmax(self.fc4(x), dim=1)\n \n return x\n```\n\nDuring training we want to use dropout to prevent overfitting, but during inference we want to use the entire network. So, we need to turn off dropout during validation, testing, and whenever we're using the network to make predictions. To do this, you use `model.eval()`. This sets the model to evaluation mode where the dropout probability is 0. You can turn dropout back on by setting the model to train mode with `model.train()`. In general, the pattern for the validation loop will look like this, where you turn off gradients, set the model to evaluation mode, calculate the validation loss and metric, then set the model back to train mode.\n\n```python\n# turn off gradients\nwith torch.no_grad():\n \n # set model to evaluation mode\n model.eval()\n \n # validation pass here\n for images, labels in testloader:\n ...\n\n# set model back to train mode\nmodel.train()\n```",
"_____no_output_____"
],
[
"> **Exercise:** Add dropout to your model and train it on Fashion-MNIST again. See if you can get a lower validation loss or higher accuracy.",
"_____no_output_____"
]
],
[
[
"## TODO: Define your model with dropout added\nfrom torch import nn\nimport torch.nn.functional as F\n\nclass Classifier(nn.Module):\n def __init__(self):\n super().__init__()\n self.fc1 = nn.Linear(784,256)\n self.fc2 = nn.Linear(256,128)\n self.fc3 = nn.Linear(128,64)\n self.fc4 = nn.Linear(64,10)\n \n self.dropout = nn.Dropout(p=0.2)\n \n def forward(self, x):\n x = x.view(x.shape[0], -1)\n x = self.dropout(F.relu(self.fc1(x)))\n x = self.dropout(F.relu(self.fc2(x)))\n x = self.dropout(F.relu(self.fc3(x)))\n x = F.log_softmax(self.fc4(x),dim = 1)\n \n return x",
"_____no_output_____"
],
[
"## TODO: Train your model with dropout, and monitor the training progress with the validation loss and accuracy\nfrom torch import optim\nfrom tqdm import tqdm\n\nmodel = Classifier()\ncriterion = nn.NLLLoss()\noptimizer = optim.Adam(model.parameters(), lr = 0.005)\n\nepochs = 30\nsteps = 0\n\ntrain_losses, test_losses = [], []\n\nfor e in tqdm(range(epochs)):\n running_loss = 0\n for images, labels in trainloader:\n optimizer.zero_grad()\n \n log_ps = model(images)\n loss = criterion(log_ps, labels)\n loss.backward()\n optimizer.step()\n \n running_loss += loss.item()\n else:\n test_loss = 0\n accuracy = 0\n with torch.no_grad():\n model.eval()\n for images, labels in testloader:\n log_ps_test = model(images)\n test_loss += criterion(log_ps_test, labels)\n \n output = torch.exp(log_ps_test)\n top_p, top_class = output.topk(1, dim = 1)\n equals = top_class == labels.view(top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor))\n model.train()\n \n train_losses.append(running_loss/len(trainloader))\n test_losses.append(test_loss/len(testloader))\n \n print(\"Epoch: {}/{}.. \".format(e+1, epochs),\n \"Training Loss: {:.3f}.. \".format(running_loss/len(trainloader)),\n \"Test Loss: {:.3f}.. \".format(test_loss/len(testloader)),\n \"Test Accuracy: {:.3f}\".format(accuracy/len(testloader)))\n ",
" 3%|▎ | 1/30 [00:24<11:53, 24.61s/it]"
],
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"plt.plot(train_losses, label='Training loss')\nplt.plot(test_losses, label='Validation loss')\nplt.legend(frameon=False)",
"_____no_output_____"
]
],
[
[
"## Inference\n\nNow that the model is trained, we can use it for inference. We've done this before, but now we need to remember to set the model in inference mode with `model.eval()`. You'll also want to turn off autograd with the `torch.no_grad()` context.",
"_____no_output_____"
]
],
[
[
"# Import helper module (should be in the repo)\nimport helper\n\n# Test out your network!\n\nmodel.eval()\n\ndataiter = iter(testloader)\nimages, labels = dataiter.next()\nimg = images[0]\n# Convert 2D image to 1D vector\nimg = img.view(1, 784)\n\n# Calculate the class probabilities (softmax) for img\nwith torch.no_grad():\n output = model.forward(img)\n\nps = torch.exp(output)\n\n# Plot the image and probabilities\nhelper.view_classify(img.view(1, 28, 28), ps, version='Fashion')",
"_____no_output_____"
],
[
"import numpy as np\nA = np.random.randn(100,100)\nA.reshape(100*100, -1)\nA.shape",
"_____no_output_____"
]
],
[
[
"## Next Up!\n\nIn the next part, I'll show you how to save your trained models. In general, you won't want to train a model everytime you need it. Instead, you'll train once, save it, then load the model when you want to train more or use if for inference.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a14698f3ece131995b7f99410150e6280497a5f
| 62,079 |
ipynb
|
Jupyter Notebook
|
Introduction to Python/Week 1/w1-python-datatypes.ipynb
|
drkndl/PHY-546
|
2b088885d9709400f8080a89b4240bb8a27a506a
|
[
"MIT"
] | null | null | null |
Introduction to Python/Week 1/w1-python-datatypes.ipynb
|
drkndl/PHY-546
|
2b088885d9709400f8080a89b4240bb8a27a506a
|
[
"MIT"
] | null | null | null |
Introduction to Python/Week 1/w1-python-datatypes.ipynb
|
drkndl/PHY-546
|
2b088885d9709400f8080a89b4240bb8a27a506a
|
[
"MIT"
] | null | null | null | 29.282547 | 490 | 0.506403 |
[
[
[
"from __future__ import print_function",
"_____no_output_____"
]
],
[
[
"# Jupyter",
"_____no_output_____"
],
[
"We'll be using Jupyter for all of our examples -- this allows us to run python in a web-based notebook, keeping a history of input and output, along with text and images.\n\nFor Jupyter help, visit:\nhttps://jupyter.readthedocs.io/en/latest/content-quickstart.html",
"_____no_output_____"
],
[
"We interact with python by typing into _cells_ in the notebook. By default, a cell is a _code_ cell, which means that you can enter any valid python code into it and run it. Another important type of cell is a _markdown_ cell. This lets you put text, with different formatting (italics, bold, etc) that describes what the notebook is doing.\n\nYou can change the cell type via the menu at the top, or using the shortcuts:\n\n * ctrl-m m : mark down cell\n * ctrl-m y : code cell",
"_____no_output_____"
],
[
"Some useful short-cuts:\n\n * shift+enter = run cell and jump to the next (creating a new cell if there is no other new one)\n * ctrl+enter = run cell-in place\n * alt+enter = run cell and insert a new one below\n\nctrl+m h lists other commands",
"_____no_output_____"
],
[
"A \"markdown cell\" enables you to typeset LaTeX equations right in your notebook. Just put them in <span>$</span> or <span>$$</span>:\n\n$$\\frac{\\partial \\rho}{\\partial t} + \\nabla \\cdot (\\rho U) = 0$$",
"_____no_output_____"
],
[
"<div style=\"background-color: palevioletred; color: white; padding: 10px;\">\n**Important**: when you work through a notebook, everything you did in previous cells is still in memory and _known_ by python, so you can refer to functions and variables that were previously defined. Even if you go up to the top of a notebook and insert a cell, all the information done earlier in your notebook session is still defined -- it doesn't matter where physically you are in the notebook. If you want to reset things, you can use the options under the _Kernel_ menu.\n</div>",
"_____no_output_____"
],
[
"<div style=\"background-color:yellow; padding: 10px\"><h3><span class=\"fa fa-flash\"></span> Quick Exercise:</h3></div>\n\nCreate a new cell below this one. Make sure that it is a _code_ cell, and enter the following code and run it:\n```\nprint(\"Hello, World\")\n```\n<hr>",
"_____no_output_____"
]
],
[
[
"print(\"Hello, World\")",
"Hello, World\n"
]
],
[
[
"`print()` is a _function_ in python that takes arguments (in the `()`) and outputs to the screen. You can print multiple quantities at once like:",
"_____no_output_____"
]
],
[
[
"print(1, 2, 3)",
"1 2 3\n"
]
],
[
[
"# Basic Datatypes\n\nNow we'll look at some of the basic datatypes in python -- these are analogous to what you will find in most programming languages, including numbers (integers and floating point), and strings.\n\nSome examples come from the python tutorial:\nhttp://docs.python.org/3/tutorial/",
"_____no_output_____"
],
[
"## integers",
"_____no_output_____"
],
[
"Integers are numbers without a decimal point. They can be positive or negative. Most programming languages use a finite-amount of memory to store a single integer, but in python will expand the amount of memory as necessary to store large integers.\n\nThe basic operators, `+`, `-`, `*`, and `/` work with integers",
"_____no_output_____"
]
],
[
[
"2+2+3",
"_____no_output_____"
],
[
"2*-4",
"_____no_output_____"
]
],
[
[
"Note: integer division is one place where python 2 and python 3 different\n \nIn python 3.x, dividing 2 integers results in a float. In python 2.x, dividing 2 integers results in an integer. The latter is consistent with many strongly-typed programming languages (like Fortran or C), since the data-type of the result is the same as the inputs, but the former is more inline with our expectations",
"_____no_output_____"
]
],
[
[
"1/2",
"_____no_output_____"
]
],
[
[
"To get an integer result, we can use the // operator.",
"_____no_output_____"
]
],
[
[
"1//2",
"_____no_output_____"
]
],
[
[
"Python is a _dynamically-typed language_—this means that we do not need to declare the datatype of a variable before initializing it. \n\nHere we'll create a variable (think of it as a descriptive label that can refer to some piece of data). The `=` operator assigns a value to a variable. ",
"_____no_output_____"
]
],
[
[
"a = 1\nb = 2",
"_____no_output_____"
]
],
[
[
"Functions operate on variables and return a result. Here, `print()` will output to the screen.",
"_____no_output_____"
]
],
[
[
"print(a+b)",
"3\n"
],
[
"print(a*b)",
"2\n"
]
],
[
[
"Note that variable names are case sensitive, so a and A are different",
"_____no_output_____"
]
],
[
[
"A = 2048",
"_____no_output_____"
],
[
"print(a, A)",
"1 2048\n"
]
],
[
[
"Here we initialize 3 variable all to `0`, but these are still distinct variables, so we can change one without affecting the others.",
"_____no_output_____"
]
],
[
[
"x = y = z = 0",
"_____no_output_____"
],
[
"print(x, y, z)",
"0 0 0\n"
],
[
"z = 1",
"_____no_output_____"
],
[
"print(x, y, z)",
"0 0 1\n"
]
],
[
[
"Python has some built in help (and Jupyter/ipython has even more)",
"_____no_output_____"
]
],
[
[
"help(x)",
"Help on int object:\n\nclass int(object)\n | int([x]) -> integer\n | int(x, base=10) -> integer\n | \n | Convert a number or string to an integer, or return 0 if no arguments\n | are given. If x is a number, return x.__int__(). For floating point\n | numbers, this truncates towards zero.\n | \n | If x is not a number or if base is given, then x must be a string,\n | bytes, or bytearray instance representing an integer literal in the\n | given base. The literal can be preceded by '+' or '-' and be surrounded\n | by whitespace. The base defaults to 10. Valid bases are 0 and 2-36.\n | Base 0 means to interpret the base from the string as an integer literal.\n | >>> int('0b100', base=0)\n | 4\n | \n | Methods defined here:\n | \n | __abs__(self, /)\n | abs(self)\n | \n | __add__(self, value, /)\n | Return self+value.\n | \n | __and__(self, value, /)\n | Return self&value.\n | \n | __bool__(self, /)\n | self != 0\n | \n | __ceil__(...)\n | Ceiling of an Integral returns itself.\n | \n | __divmod__(self, value, /)\n | Return divmod(self, value).\n | \n | __eq__(self, value, /)\n | Return self==value.\n | \n | __float__(self, /)\n | float(self)\n | \n | __floor__(...)\n | Flooring an Integral returns itself.\n | \n | __floordiv__(self, value, /)\n | Return self//value.\n | \n | __format__(self, format_spec, /)\n | Default object formatter.\n | \n | __ge__(self, value, /)\n | Return self>=value.\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __getnewargs__(self, /)\n | \n | __gt__(self, value, /)\n | Return self>value.\n | \n | __hash__(self, /)\n | Return hash(self).\n | \n | __index__(self, /)\n | Return self converted to an integer, if self is suitable for use as an index into a list.\n | \n | __int__(self, /)\n | int(self)\n | \n | __invert__(self, /)\n | ~self\n | \n | __le__(self, value, /)\n | Return self<=value.\n | \n | __lshift__(self, value, /)\n | Return self<<value.\n | \n | __lt__(self, value, /)\n | Return self<value.\n | \n | __mod__(self, value, /)\n | Return self%value.\n | \n | __mul__(self, value, /)\n | Return self*value.\n | \n | __ne__(self, value, /)\n | Return self!=value.\n | \n | __neg__(self, /)\n | -self\n | \n | __or__(self, value, /)\n | Return self|value.\n | \n | __pos__(self, /)\n | +self\n | \n | __pow__(self, value, mod=None, /)\n | Return pow(self, value, mod).\n | \n | __radd__(self, value, /)\n | Return value+self.\n | \n | __rand__(self, value, /)\n | Return value&self.\n | \n | __rdivmod__(self, value, /)\n | Return divmod(value, self).\n | \n | __repr__(self, /)\n | Return repr(self).\n | \n | __rfloordiv__(self, value, /)\n | Return value//self.\n | \n | __rlshift__(self, value, /)\n | Return value<<self.\n | \n | __rmod__(self, value, /)\n | Return value%self.\n | \n | __rmul__(self, value, /)\n | Return value*self.\n | \n | __ror__(self, value, /)\n | Return value|self.\n | \n | __round__(...)\n | Rounding an Integral returns itself.\n | Rounding with an ndigits argument also returns an integer.\n | \n | __rpow__(self, value, mod=None, /)\n | Return pow(value, self, mod).\n | \n | __rrshift__(self, value, /)\n | Return value>>self.\n | \n | __rshift__(self, value, /)\n | Return self>>value.\n | \n | __rsub__(self, value, /)\n | Return value-self.\n | \n | __rtruediv__(self, value, /)\n | Return value/self.\n | \n | __rxor__(self, value, /)\n | Return value^self.\n | \n | __sizeof__(self, /)\n | Returns size in memory, in bytes.\n | \n | __str__(self, /)\n | Return str(self).\n | \n | __sub__(self, value, /)\n | Return self-value.\n | \n | __truediv__(self, value, /)\n | Return self/value.\n | \n | __trunc__(...)\n | Truncating an Integral returns itself.\n | \n | __xor__(self, value, /)\n | Return self^value.\n | \n | bit_length(self, /)\n | Number of bits necessary to represent self in binary.\n | \n | >>> bin(37)\n | '0b100101'\n | >>> (37).bit_length()\n | 6\n | \n | conjugate(...)\n | Returns self, the complex conjugate of any int.\n | \n | to_bytes(self, /, length, byteorder, *, signed=False)\n | Return an array of bytes representing an integer.\n | \n | length\n | Length of bytes object to use. An OverflowError is raised if the\n | integer is not representable with the given number of bytes.\n | byteorder\n | The byte order used to represent the integer. If byteorder is 'big',\n | the most significant byte is at the beginning of the byte array. If\n | byteorder is 'little', the most significant byte is at the end of the\n | byte array. To request the native byte order of the host system, use\n | `sys.byteorder' as the byte order value.\n | signed\n | Determines whether two's complement is used to represent the integer.\n | If signed is False and a negative integer is given, an OverflowError\n | is raised.\n | \n | ----------------------------------------------------------------------\n | Class methods defined here:\n | \n | from_bytes(bytes, byteorder, *, signed=False) from builtins.type\n | Return the integer represented by the given array of bytes.\n | \n | bytes\n | Holds the array of bytes to convert. The argument must either\n | support the buffer protocol or be an iterable object producing bytes.\n | Bytes and bytearray are examples of built-in objects that support the\n | buffer protocol.\n | byteorder\n | The byte order used to represent the integer. If byteorder is 'big',\n | the most significant byte is at the beginning of the byte array. If\n | byteorder is 'little', the most significant byte is at the end of the\n | byte array. To request the native byte order of the host system, use\n | `sys.byteorder' as the byte order value.\n | signed\n | Indicates whether two's complement is used to represent the integer.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n | \n | ----------------------------------------------------------------------\n | Data descriptors defined here:\n | \n | denominator\n | the denominator of a rational number in lowest terms\n | \n | imag\n | the imaginary part of a complex number\n | \n | numerator\n | the numerator of a rational number in lowest terms\n | \n | real\n | the real part of a complex number\n\n"
],
[
"x?",
"_____no_output_____"
]
],
[
[
"Another function, `type()` returns the data type of a variable",
"_____no_output_____"
]
],
[
[
"print(type(x))",
"<class 'int'>\n"
]
],
[
[
"Note in languages like Fortran and C, you specify the amount of memory an integer can take (usually 2 or 4 bytes). This puts a restriction on the largest size integer that can be represented. Python will adapt the size of the integer so you don't *overflow*",
"_____no_output_____"
]
],
[
[
"a = 12345678901234567890123456789012345123456789012345678901234567890\nprint(a)\nprint(a.bit_length())\nprint(type(a))",
"12345678901234567890123456789012345123456789012345678901234567890\n213\n<class 'int'>\n"
]
],
[
[
"## floating point",
"_____no_output_____"
],
[
"when operating with both floating point and integers, the result is promoted to a float. This is true of both python 2.x and 3.x",
"_____no_output_____"
]
],
[
[
"1./2",
"_____no_output_____"
]
],
[
[
"but note the special integer division operator",
"_____no_output_____"
]
],
[
[
"1.//2",
"_____no_output_____"
]
],
[
[
"It is important to understand that since there are infinitely many real numbers between any two bounds, on a computer we have to approximate this by a finite number. There is an IEEE standard for floating point that pretty much all languages and processors follow. \n\nThe means two things\n\n* not every real number will have an exact representation in floating point\n* there is a finite precision to numbers -- below this we lose track of differences (this is usually called *roundoff* error)\n\nOn our course website, I posted a link to a paper, _What every computer scientist should know about floating-point arithmetic_ -- this is a great reference on understanding how a computer stores numbers.\n\nConsider the following expression, for example:",
"_____no_output_____"
]
],
[
[
"0.3/0.1 - 3",
"_____no_output_____"
]
],
[
[
"Here's another example: The number 0.1 cannot be exactly represented on a computer. In our print, we use a format specifier (the stuff inside of the {}) to ask for more precision to be shown:",
"_____no_output_____"
]
],
[
[
"a = 0.1\nprint(\"{:1.25}\".format(a))",
"0.1000000000000000055511151\n"
]
],
[
[
"we can ask python to report the limits on floating point",
"_____no_output_____"
]
],
[
[
"import sys\nprint(sys.float_info)",
"sys.float_info(max=1.7976931348623157e+308, max_exp=1024, max_10_exp=308, min=2.2250738585072014e-308, min_exp=-1021, min_10_exp=-307, dig=15, mant_dig=53, epsilon=2.220446049250313e-16, radix=2, rounds=1)\n"
]
],
[
[
"Note that this says that we can only store numbers between 2.2250738585072014e-308 and 1.7976931348623157e+308\n\nWe also see that the precision is 2.220446049250313e-16 (this is commonly called _machine epsilon_). To see this, consider adding a small number to 1.0. We'll use the equality operator (`==`) to test if two numbers are equal:",
"_____no_output_____"
],
[
"<div style=\"background-color:yellow; padding: 10px\"><h3><span class=\"fa fa-flash\"></span> Quick Exercise:</h3></div>\n\nDefine two variables, $a = 1$, and $e = 10^{-16}$.\n\nNow define a third variable, `b = a + e`\n\nWe can use the python `==` operator to test for equality. What do you expect `b == a` to return? run it an see if it agrees with your guess.\n<hr>",
"_____no_output_____"
]
],
[
[
"a = 1\ne = 10**-16\nb = a+e\nif b==a:\n print(True)\nelse:\n print(False)",
"True\n"
]
],
[
[
"## modules\n\nThe core python language is extended by a standard library that provides additional functionality. These added pieces are in the form of modules that we can _import_ into our python session (or program).\n\nThe `math` module provides functions that do the basic mathematical operations as well as provide constants (note there is a separate `cmath` module for complex numbers).\n\nIn python, you `import` a module. The functions are then defined in a separate _namespace_—this is a separate region that defines names and variables, etc. A variable in one namespace can have the same name as a variable in a different namespace, and they don't clash. You use the \"`.`\" operator to access a member of a namespace.\n\nBy default, when you type stuff into the python interpreter or here in the Jupyter notebook, or in a script, it is in its own default namespace, and you don't need to prefix any of the variables with a namespace indicator.",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
]
],
[
[
"`math` provides the value of pi",
"_____no_output_____"
]
],
[
[
"print(math.pi)",
"3.141592653589793\n"
]
],
[
[
"This is distinct from any variable `pi` we might define here",
"_____no_output_____"
]
],
[
[
"pi = 3",
"_____no_output_____"
],
[
"print(pi, math.pi)",
"3 3.141592653589793\n"
]
],
[
[
"Note here that `pi` and `math.pi` are distinct from one another—they are in different namespaces.",
"_____no_output_____"
],
[
"### floating point operations",
"_____no_output_____"
],
[
"The same operators, `+`, `-`, `*`, `/` work are usual for floating point numbers. To raise an number to a power, we use the `**` operator (this is the same as Fortran)",
"_____no_output_____"
]
],
[
[
"R = 2.0",
"_____no_output_____"
],
[
"print(math.pi*R**2)",
"12.566370614359172\n"
]
],
[
[
"operator precedence follows that of most languages. See\n\nhttps://docs.python.org/3/reference/expressions.html#operator-precedence\n \nin order of precedence:\n* quantites in `()`\n* slicing, calls, subscripts\n* exponentiation (`**`)\n* `+x`, `-x`, `~x`\n* `*`, `@`, `/`, `//`, `%`\n* `+`, `-`\n\n(after this are bitwise operations and comparisons)\n\nParantheses can be used to override the precedence.",
"_____no_output_____"
],
[
"<div style=\"background-color:yellow; padding: 10px\"><h3><span class=\"fa fa-flash\"></span> Quick Exercise:</h3></div>\n\nConsider the following expressions. Using the ideas of precedence, think about what value will result, then try it out in the cell below to see if you were right.\n\n * `1 + 3*2**2`\n * `1 + (3*2)**2`\n * `2**3**2`\n\n<hr>",
"_____no_output_____"
]
],
[
[
"\"\"\"\nMy guesses: \n1 + 3*2**2 = 13\n1 + (3*2)**2 = 37\n2**3**2 = 64?\n\"\"\"\n\nprint(1 + 3*2**2)\nprint(1 + (3*2)**2)\nprint(2**3**2)\n\n#Okay, so if precedence is same, order is right to left. Whoops!",
"13\n37\n512\n"
]
],
[
[
"The math module provides a lot of the standard math functions we might want to use.\n\nFor the trig functions, the expectation is that the argument to the function is in radians—you can use `math.radians()` to convert from degrees to radians, ex:",
"_____no_output_____"
]
],
[
[
"print(math.cos(math.radians(45)))",
"0.7071067811865476\n"
]
],
[
[
"Notice that in that statement we are feeding the output of one function (`math.radians()`) into a second function, `math.cos()`\n\nWhen in doubt, as for help to discover all of the things a module provides:",
"_____no_output_____"
]
],
[
[
"help(math.sin)",
"Help on built-in function sin in module math:\n\nsin(x, /)\n Return the sine of x (measured in radians).\n\n"
]
],
[
[
"## complex numbers",
"_____no_output_____"
],
[
"python uses '`j`' to denote the imaginary unit",
"_____no_output_____"
]
],
[
[
"print(1.0 + 2j)",
"(1+2j)\n"
],
[
"a = 1j\nb = 3.0 + 2.0j\nprint(a + b)\nprint(a*b)",
"(3+3j)\n(-2+3j)\n"
]
],
[
[
"we can use `abs()` to get the magnitude and separately get the real or imaginary parts ",
"_____no_output_____"
]
],
[
[
"print(abs(b))\nprint(a.real)\nprint(a.imag)",
"3.605551275463989\n0.0\n1.0\n"
]
],
[
[
"## strings",
"_____no_output_____"
],
[
"python doesn't care if you use single or double quotes for strings:",
"_____no_output_____"
]
],
[
[
"a = \"this is my string\"\nb = 'another string'",
"_____no_output_____"
],
[
"print(a)\nprint(b)",
"this is my string\nanother string\n"
]
],
[
[
"Many of the usual mathematical operators are defined for strings as well. For example to concatenate or duplicate:",
"_____no_output_____"
]
],
[
[
"print(a+b)",
"this is my stringanother string\n"
],
[
"print(a + \". \" + b)",
"this is my string. another string\n"
],
[
"print(a*2)",
"this is my stringthis is my string\n"
]
],
[
[
"There are several escape codes that are interpreted in strings. These start with a backwards-slash, `\\`. E.g., you can use `\\n` for new line",
"_____no_output_____"
]
],
[
[
"a = a + \"\\n\"\nprint(a)",
"this is my string\n\n"
]
],
[
[
"<div style=\"background-color:yellow; padding: 10px\"><h3><span class=\"fa fa-flash\"></span> Quick Exercise:</h3></div>\n\nThe `input()` function can be used to ask the user for input.\n\n * Use `help(input)` to see how it works. \n * Write code to ask for input and store the result in a variable. `input()` will return a string.\n\n * Use the `float()` function to convert a number entered as input to a floating point variable. \n * Check to see if the conversion worked using the `type()` function.\n<hr>",
"_____no_output_____"
]
],
[
[
"help(input)\nnum = input()\nnum = float(num)\nprint(type(num))",
"Help on method raw_input in module ipykernel.kernelbase:\n\nraw_input(prompt='') method of ipykernel.ipkernel.IPythonKernel instance\n Forward raw_input to frontends\n \n Raises\n ------\n StdinNotImplentedError if active frontend doesn't support stdin.\n\n5\n<class 'float'>\n"
]
],
[
[
"\"\"\" can enclose multiline strings. This is useful for docstrings at the start of functions (more on that later...)",
"_____no_output_____"
]
],
[
[
"c = \"\"\"\nLorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor \nincididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis \nnostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. \nDuis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore \neu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt \nin culpa qui officia deserunt mollit anim id est laborum.\"\"\"",
"_____no_output_____"
],
[
"print(c)",
"\nLorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor \nincididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis \nnostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. \nDuis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore \neu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt \nin culpa qui officia deserunt mollit anim id est laborum.\n"
]
],
[
[
"a raw string does not replace escape sequences (like \\n). Just put a `r` before the first quote:",
"_____no_output_____"
]
],
[
[
"d = r\"this is a raw string\\n\"\nprint(d)",
"this is a raw string\\n\n"
]
],
[
[
"slicing is used to access a portion of a string.\n\nslicing a string can seem a bit counterintuitive if you are coming from Fortran. The trick is to think of the index as representing the left edge of a character in the string. When we do arrays later, the same will apply.\n\nAlso note that python (like C) uses 0-based indexing\n\nNegative indices count from the right.",
"_____no_output_____"
]
],
[
[
"a = \"this is my string\"\nprint(a)\nprint(a[5:7])\nprint(a[0])\nprint(d)\nprint(d[-2])",
"this is my string\nis\nt\nthis is a raw string\\n\n\\\n"
]
],
[
[
"<div style=\"background-color:yellow; padding: 10px\"><h3><span class=\"fa fa-flash\"></span> Quick Exercise:</h3></div>\n\nStrings have a lot of _methods_ (functions that know how to work with a particular datatype, in this case strings). A useful method is `.find()`. For a string `a`,\n`a.find(s)` will return the index of the first occurrence of `s`.\n\nFor our string `c` above, find the first `.` (identifying the first full sentence), and print out just the first sentence in `c` using this result\n\n<hr>",
"_____no_output_____"
]
],
[
[
"index = c.find('.')\nfirst = c[:index+1]\nprint(index, first)",
"125 \nLorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor \nincididunt ut labore et dolore magna aliqua.\n"
]
],
[
[
"there are also a number of methods and functions that work with strings. Here are some examples:",
"_____no_output_____"
]
],
[
[
"print(a.replace(\"this\", \"that\"))\nprint(len(a))\nprint(a.strip()) # Also notice that strip removes the \\n\nprint(a.strip()[-1])",
"that is my string\n17\nthis is my string\ng\n"
]
],
[
[
"Note that our original string, `a`, has not changed. In python, strings are *immutable*. Operations on strings return a new string.",
"_____no_output_____"
]
],
[
[
"print(a)",
"this is my string\n"
],
[
"print(type(a))",
"<class 'str'>\n"
]
],
[
[
"As usual, ask for help to learn more:",
"_____no_output_____"
]
],
[
[
"help(str)",
"Help on class str in module builtins:\n\nclass str(object)\n | str(object='') -> str\n | str(bytes_or_buffer[, encoding[, errors]]) -> str\n | \n | Create a new string object from the given object. If encoding or\n | errors is specified, then the object must expose a data buffer\n | that will be decoded using the given encoding and error handler.\n | Otherwise, returns the result of object.__str__() (if defined)\n | or repr(object).\n | encoding defaults to sys.getdefaultencoding().\n | errors defaults to 'strict'.\n | \n | Methods defined here:\n | \n | __add__(self, value, /)\n | Return self+value.\n | \n | __contains__(self, key, /)\n | Return key in self.\n | \n | __eq__(self, value, /)\n | Return self==value.\n | \n | __format__(self, format_spec, /)\n | Return a formatted version of the string as described by format_spec.\n | \n | __ge__(self, value, /)\n | Return self>=value.\n | \n | __getattribute__(self, name, /)\n | Return getattr(self, name).\n | \n | __getitem__(self, key, /)\n | Return self[key].\n | \n | __getnewargs__(...)\n | \n | __gt__(self, value, /)\n | Return self>value.\n | \n | __hash__(self, /)\n | Return hash(self).\n | \n | __iter__(self, /)\n | Implement iter(self).\n | \n | __le__(self, value, /)\n | Return self<=value.\n | \n | __len__(self, /)\n | Return len(self).\n | \n | __lt__(self, value, /)\n | Return self<value.\n | \n | __mod__(self, value, /)\n | Return self%value.\n | \n | __mul__(self, value, /)\n | Return self*value.\n | \n | __ne__(self, value, /)\n | Return self!=value.\n | \n | __repr__(self, /)\n | Return repr(self).\n | \n | __rmod__(self, value, /)\n | Return value%self.\n | \n | __rmul__(self, value, /)\n | Return value*self.\n | \n | __sizeof__(self, /)\n | Return the size of the string in memory, in bytes.\n | \n | __str__(self, /)\n | Return str(self).\n | \n | capitalize(self, /)\n | Return a capitalized version of the string.\n | \n | More specifically, make the first character have upper case and the rest lower\n | case.\n | \n | casefold(self, /)\n | Return a version of the string suitable for caseless comparisons.\n | \n | center(self, width, fillchar=' ', /)\n | Return a centered string of length width.\n | \n | Padding is done using the specified fill character (default is a space).\n | \n | count(...)\n | S.count(sub[, start[, end]]) -> int\n | \n | Return the number of non-overlapping occurrences of substring sub in\n | string S[start:end]. Optional arguments start and end are\n | interpreted as in slice notation.\n | \n | encode(self, /, encoding='utf-8', errors='strict')\n | Encode the string using the codec registered for encoding.\n | \n | encoding\n | The encoding in which to encode the string.\n | errors\n | The error handling scheme to use for encoding errors.\n | The default is 'strict' meaning that encoding errors raise a\n | UnicodeEncodeError. Other possible values are 'ignore', 'replace' and\n | 'xmlcharrefreplace' as well as any other name registered with\n | codecs.register_error that can handle UnicodeEncodeErrors.\n | \n | endswith(...)\n | S.endswith(suffix[, start[, end]]) -> bool\n | \n | Return True if S ends with the specified suffix, False otherwise.\n | With optional start, test S beginning at that position.\n | With optional end, stop comparing S at that position.\n | suffix can also be a tuple of strings to try.\n | \n | expandtabs(self, /, tabsize=8)\n | Return a copy where all tab characters are expanded using spaces.\n | \n | If tabsize is not given, a tab size of 8 characters is assumed.\n | \n | find(...)\n | S.find(sub[, start[, end]]) -> int\n | \n | Return the lowest index in S where substring sub is found,\n | such that sub is contained within S[start:end]. Optional\n | arguments start and end are interpreted as in slice notation.\n | \n | Return -1 on failure.\n | \n | format(...)\n | S.format(*args, **kwargs) -> str\n | \n | Return a formatted version of S, using substitutions from args and kwargs.\n | The substitutions are identified by braces ('{' and '}').\n | \n | format_map(...)\n | S.format_map(mapping) -> str\n | \n | Return a formatted version of S, using substitutions from mapping.\n | The substitutions are identified by braces ('{' and '}').\n | \n | index(...)\n | S.index(sub[, start[, end]]) -> int\n | \n | Return the lowest index in S where substring sub is found, \n | such that sub is contained within S[start:end]. Optional\n | arguments start and end are interpreted as in slice notation.\n | \n | Raises ValueError when the substring is not found.\n | \n | isalnum(self, /)\n | Return True if the string is an alpha-numeric string, False otherwise.\n | \n | A string is alpha-numeric if all characters in the string are alpha-numeric and\n | there is at least one character in the string.\n | \n | isalpha(self, /)\n | Return True if the string is an alphabetic string, False otherwise.\n | \n | A string is alphabetic if all characters in the string are alphabetic and there\n | is at least one character in the string.\n | \n | isascii(self, /)\n | Return True if all characters in the string are ASCII, False otherwise.\n | \n | ASCII characters have code points in the range U+0000-U+007F.\n | Empty string is ASCII too.\n | \n | isdecimal(self, /)\n | Return True if the string is a decimal string, False otherwise.\n | \n | A string is a decimal string if all characters in the string are decimal and\n | there is at least one character in the string.\n | \n | isdigit(self, /)\n | Return True if the string is a digit string, False otherwise.\n | \n | A string is a digit string if all characters in the string are digits and there\n | is at least one character in the string.\n | \n | isidentifier(self, /)\n | Return True if the string is a valid Python identifier, False otherwise.\n | \n | Use keyword.iskeyword() to test for reserved identifiers such as \"def\" and\n | \"class\".\n | \n | islower(self, /)\n | Return True if the string is a lowercase string, False otherwise.\n | \n | A string is lowercase if all cased characters in the string are lowercase and\n | there is at least one cased character in the string.\n | \n | isnumeric(self, /)\n | Return True if the string is a numeric string, False otherwise.\n | \n | A string is numeric if all characters in the string are numeric and there is at\n | least one character in the string.\n | \n | isprintable(self, /)\n | Return True if the string is printable, False otherwise.\n | \n | A string is printable if all of its characters are considered printable in\n | repr() or if it is empty.\n | \n | isspace(self, /)\n | Return True if the string is a whitespace string, False otherwise.\n | \n | A string is whitespace if all characters in the string are whitespace and there\n | is at least one character in the string.\n | \n | istitle(self, /)\n | Return True if the string is a title-cased string, False otherwise.\n | \n | In a title-cased string, upper- and title-case characters may only\n | follow uncased characters and lowercase characters only cased ones.\n | \n | isupper(self, /)\n | Return True if the string is an uppercase string, False otherwise.\n | \n | A string is uppercase if all cased characters in the string are uppercase and\n | there is at least one cased character in the string.\n | \n | join(self, iterable, /)\n | Concatenate any number of strings.\n | \n | The string whose method is called is inserted in between each given string.\n | The result is returned as a new string.\n | \n | Example: '.'.join(['ab', 'pq', 'rs']) -> 'ab.pq.rs'\n | \n | ljust(self, width, fillchar=' ', /)\n | Return a left-justified string of length width.\n | \n | Padding is done using the specified fill character (default is a space).\n | \n | lower(self, /)\n | Return a copy of the string converted to lowercase.\n | \n | lstrip(self, chars=None, /)\n | Return a copy of the string with leading whitespace removed.\n | \n | If chars is given and not None, remove characters in chars instead.\n | \n | partition(self, sep, /)\n | Partition the string into three parts using the given separator.\n | \n | This will search for the separator in the string. If the separator is found,\n | returns a 3-tuple containing the part before the separator, the separator\n | itself, and the part after it.\n | \n | If the separator is not found, returns a 3-tuple containing the original string\n | and two empty strings.\n | \n | replace(self, old, new, count=-1, /)\n | Return a copy with all occurrences of substring old replaced by new.\n | \n | count\n | Maximum number of occurrences to replace.\n | -1 (the default value) means replace all occurrences.\n | \n | If the optional argument count is given, only the first count occurrences are\n | replaced.\n | \n | rfind(...)\n | S.rfind(sub[, start[, end]]) -> int\n | \n | Return the highest index in S where substring sub is found,\n | such that sub is contained within S[start:end]. Optional\n | arguments start and end are interpreted as in slice notation.\n | \n | Return -1 on failure.\n | \n | rindex(...)\n | S.rindex(sub[, start[, end]]) -> int\n | \n | Return the highest index in S where substring sub is found,\n | such that sub is contained within S[start:end]. Optional\n | arguments start and end are interpreted as in slice notation.\n | \n | Raises ValueError when the substring is not found.\n | \n | rjust(self, width, fillchar=' ', /)\n | Return a right-justified string of length width.\n | \n | Padding is done using the specified fill character (default is a space).\n | \n | rpartition(self, sep, /)\n | Partition the string into three parts using the given separator.\n | \n | This will search for the separator in the string, starting at the end. If\n | the separator is found, returns a 3-tuple containing the part before the\n | separator, the separator itself, and the part after it.\n | \n | If the separator is not found, returns a 3-tuple containing two empty strings\n | and the original string.\n | \n | rsplit(self, /, sep=None, maxsplit=-1)\n | Return a list of the words in the string, using sep as the delimiter string.\n | \n | sep\n | The delimiter according which to split the string.\n | None (the default value) means split according to any whitespace,\n | and discard empty strings from the result.\n | maxsplit\n | Maximum number of splits to do.\n | -1 (the default value) means no limit.\n | \n | Splits are done starting at the end of the string and working to the front.\n | \n | rstrip(self, chars=None, /)\n | Return a copy of the string with trailing whitespace removed.\n | \n | If chars is given and not None, remove characters in chars instead.\n | \n | split(self, /, sep=None, maxsplit=-1)\n | Return a list of the words in the string, using sep as the delimiter string.\n | \n | sep\n | The delimiter according which to split the string.\n | None (the default value) means split according to any whitespace,\n | and discard empty strings from the result.\n | maxsplit\n | Maximum number of splits to do.\n | -1 (the default value) means no limit.\n | \n | splitlines(self, /, keepends=False)\n | Return a list of the lines in the string, breaking at line boundaries.\n | \n | Line breaks are not included in the resulting list unless keepends is given and\n | true.\n | \n | startswith(...)\n | S.startswith(prefix[, start[, end]]) -> bool\n | \n | Return True if S starts with the specified prefix, False otherwise.\n | With optional start, test S beginning at that position.\n | With optional end, stop comparing S at that position.\n | prefix can also be a tuple of strings to try.\n | \n | strip(self, chars=None, /)\n | Return a copy of the string with leading and trailing whitespace removed.\n | \n | If chars is given and not None, remove characters in chars instead.\n | \n | swapcase(self, /)\n | Convert uppercase characters to lowercase and lowercase characters to uppercase.\n | \n | title(self, /)\n | Return a version of the string where each word is titlecased.\n | \n | More specifically, words start with uppercased characters and all remaining\n | cased characters have lower case.\n | \n | translate(self, table, /)\n | Replace each character in the string using the given translation table.\n | \n | table\n | Translation table, which must be a mapping of Unicode ordinals to\n | Unicode ordinals, strings, or None.\n | \n | The table must implement lookup/indexing via __getitem__, for instance a\n | dictionary or list. If this operation raises LookupError, the character is\n | left untouched. Characters mapped to None are deleted.\n | \n | upper(self, /)\n | Return a copy of the string converted to uppercase.\n | \n | zfill(self, width, /)\n | Pad a numeric string with zeros on the left, to fill a field of the given width.\n | \n | The string is never truncated.\n | \n | ----------------------------------------------------------------------\n | Static methods defined here:\n | \n | __new__(*args, **kwargs) from builtins.type\n | Create and return a new object. See help(type) for accurate signature.\n | \n | maketrans(x, y=None, z=None, /)\n | Return a translation table usable for str.translate().\n | \n | If there is only one argument, it must be a dictionary mapping Unicode\n | ordinals (integers) or characters to Unicode ordinals, strings or None.\n | Character keys will be then converted to ordinals.\n | If there are two arguments, they must be strings of equal length, and\n | in the resulting dictionary, each character in x will be mapped to the\n | character at the same position in y. If there is a third argument, it\n | must be a string, whose characters will be mapped to None in the result.\n\n"
]
],
[
[
"We can format strings when we are printing to insert quantities in particular places in the string. A `{}` serves as a placeholder for a quantity and is replaced using the `.format()` method:",
"_____no_output_____"
]
],
[
[
"a = 1\nb = 2.0\nc = \"test\"\nprint(\"a = {}; b = {}; c = {}\".format(a, b, c))\nprint(\"a = {}\".format(a))",
"a = 1; b = 2.0; c = test\na = 1\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a146ba83b5da2d4613845e3e68204387b961e57
| 33,627 |
ipynb
|
Jupyter Notebook
|
Classification using TF-IDF & Count Vectorizer.ipynb
|
Nimori/Classification_Models-NLP
|
64d6b539d7bcf0505a3cc96573d93bbff5d50bc7
|
[
"MIT"
] | null | null | null |
Classification using TF-IDF & Count Vectorizer.ipynb
|
Nimori/Classification_Models-NLP
|
64d6b539d7bcf0505a3cc96573d93bbff5d50bc7
|
[
"MIT"
] | null | null | null |
Classification using TF-IDF & Count Vectorizer.ipynb
|
Nimori/Classification_Models-NLP
|
64d6b539d7bcf0505a3cc96573d93bbff5d50bc7
|
[
"MIT"
] | null | null | null | 33.660661 | 237 | 0.487317 |
[
[
[
"from sklearn import model_selection, preprocessing, linear_model, naive_bayes, metrics, svm\nfrom sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer\nfrom sklearn import decomposition, ensemble\n\n#import textblob\nfrom keras.preprocessing import text, sequence\nfrom keras import layers, models, optimizers\n\nimport numpy as np\nimport pandas as pd\nimport string\n\nimport nltk\nnltk.download('punkt')",
"[nltk_data] Downloading package punkt to /home/nimori/nltk_data...\n[nltk_data] Unzipping tokenizers/punkt.zip.\n"
],
[
"# create a dataframe using texts and lables\n\ndf= pd.read_csv('data_1000.csv')\ndf2= pd.DataFrame()\ndf2['text']= df['post_description_en']\ndf2['labels']= df['categories']",
"_____no_output_____"
],
[
"df2['labels']= df2['labels'].replace('Journalist, Writing And Translation', 'Government Works')",
"_____no_output_____"
],
[
"df2.groupby('labels').count()",
"_____no_output_____"
],
[
"df2= df2[(df2.labels!='Agriculture And Farming')& (df2.labels!='Artist')& (df2.labels!='Brokering / Agent')& (df2.labels!='Labor')& (df2.labels!='Plumber')& (df2.labels!='Sanitation Works')& (df2.labels!='Travel And Transport')]\n#We remove all those categories whose count is in single digits",
"_____no_output_____"
],
[
"df2.groupby('labels').count()",
"_____no_output_____"
],
[
"df3= pd.read_csv('report_04-11.csv')\n#len(df3)",
"_____no_output_____"
],
[
"df_04= pd.DataFrame()\ndf_04['text']= df3['translation']\ndf_04['labels']= df3['classifier']",
"_____no_output_____"
],
[
"df_04['labels']= df_04['labels'].apply(lambda x: x.lower())\ndf_04['text']= df_04['text'].apply(lambda x: x.lower())",
"_____no_output_____"
],
[
"def text_cleaning(a):\n list2 = nltk.word_tokenize(a) \n a_lemma = ' '.join([lemmatizer.lemmatize(words) for words in list2])\n rem_punc= [char for char in a_lemma if char not in string.punctuation]\n rem_punc= ''.join(rem_punc)\n return [word for word in rem_punc.split() if word.lower() not in stopwords.words('english')]",
"_____no_output_____"
],
[
"from nltk.stem import WordNetLemmatizer \nnltk.download('wordnet')\nlemmatizer = WordNetLemmatizer()",
"[nltk_data] Downloading package wordnet to /home/nimori/nltk_data...\n[nltk_data] Unzipping corpora/wordnet.zip.\n"
],
[
"import nltk\nfrom nltk import word_tokenize\nfrom nltk.corpus import stopwords\n\nstopWords= set(stopwords.words('english'))",
"_____no_output_____"
],
[
"df_04['text_cleaned']= df_04.iloc[:,0].apply(text_cleaning)",
"_____no_output_____"
],
[
"for i,row in df_04.iterrows():\n row['text_cleaned']= ' '.join([str(elem) for elem in row['text_cleaned']])",
"_____no_output_____"
],
[
"df_04",
"_____no_output_____"
],
[
"df2['text_cleaned']= df2.iloc[:,0].apply(text_cleaning)",
"_____no_output_____"
],
[
"for i,row in df2.iterrows():\n row['text_cleaned']= ' '.join([str(elem) for elem in row['text_cleaned']])",
"_____no_output_____"
],
[
"df2['labels']= df2['labels'].apply(lambda x: x.lower())\ndf2['text']= df2['text'].apply(lambda x: x.lower())\ndf2['text_cleaned']= df2['text_cleaned'].apply(lambda x: x.lower())",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"# split the dataset into training and validation datasets \ntrain_x, valid_x, train_y, valid_y = model_selection.train_test_split(df2['text_cleaned'], df2['labels'], test_size=0.25, random_state=0, stratify= df2['labels'])\nfrom sklearn.model_selection import StratifiedShuffleSplit\n# sss = StratifiedShuffleSplit(n_splits=5, test_size=0.25, random_state=0)\n# sss.get_n_splits(X, y)\n# for train_index, test_index in sss.split(X, y):\n# #print(\"TRAIN:\", train_index, \"TEST:\", test_index)\n# train_x, valid_x = X[train_index], X[test_index]\n# train_y, valid_y = y[train_index], y[test_index]",
"_____no_output_____"
],
[
"x_test= df_04.text_cleaned\ny_test= df_04.labels",
"_____no_output_____"
],
[
"# label encode the target variable \nencoder = preprocessing.LabelEncoder()\ntrain_y = encoder.fit_transform(train_y)\nvalid_y = encoder.fit_transform(valid_y)\ny_test = encoder.fit_transform(y_test)",
"_____no_output_____"
],
[
"# create a count vectorizer object \ncount_vect = CountVectorizer(analyzer='word', token_pattern=r'\\w{1,}')\n#count_vect.fit(df2['text_cleaned'])\ncount_vect.fit(train_x)\n\n# transform the training and validation data using count vectorizer object\nxtrain_count = count_vect.transform(train_x)\nxvalid_count = count_vect.transform(valid_x)\nxtest_count = count_vect.transform(x_test)",
"_____no_output_____"
],
[
"# word level tf-idf\ntfidf_vect = TfidfVectorizer(analyzer='word', token_pattern=r'\\w{1,}', max_features=5000)\ntfidf_vect.fit(df2['text'])\nxtrain_tfidf = tfidf_vect.transform(train_x)\nxvalid_tfidf = tfidf_vect.transform(valid_x)\nxtest_tfidf = tfidf_vect.transform(x_test)\n\n# ngram level tf-idf \ntfidf_vect_ngram = TfidfVectorizer(analyzer='word', token_pattern=r'\\w{1,}', ngram_range=(2,3), max_features=5000)\ntfidf_vect_ngram.fit(df2['text'])\nxtrain_tfidf_ngram = tfidf_vect_ngram.transform(train_x)\nxvalid_tfidf_ngram = tfidf_vect_ngram.transform(valid_x)\nxtest_tfidf_ngram = tfidf_vect_ngram.transform(x_test)\n\n# characters level tf-idf\ntfidf_vect_ngram_chars = TfidfVectorizer(analyzer='char', token_pattern=r'\\w{1,}', ngram_range=(2,3), max_features=5000)\ntfidf_vect_ngram_chars.fit(df2['text'])\nxtrain_tfidf_ngram_chars = tfidf_vect_ngram_chars.transform(train_x) \nxvalid_tfidf_ngram_chars = tfidf_vect_ngram_chars.transform(valid_x) \nxtest_tfidf_ngram_chars = tfidf_vect_ngram_chars.transform(x_test)",
"/home/nimori/venv_envs/tf24/lib/python3.8/site-packages/sklearn/feature_extraction/text.py:506: UserWarning: The parameter 'token_pattern' will not be used since 'analyzer' != 'word'\n warnings.warn(\"The parameter 'token_pattern' will not be used\"\n"
],
[
"# load the pre-trained word-embedding vectors \nembeddings_index = {}\nfor i, line in enumerate(open('wiki-news-300d-1M.vec')):\n values = line.split()\n embeddings_index[values[0]] = np.asarray(values[1:], dtype='float32')\n\n# create a tokenizer \ntoken = text.Tokenizer()\ntoken.fit_on_texts(df2['text'])\nword_index = token.word_index\n\n# convert text to sequence of tokens and pad them to ensure equal length vectors \ntrain_seq_x = sequence.pad_sequences(token.texts_to_sequences(train_x), maxlen=70)\nvalid_seq_x = sequence.pad_sequences(token.texts_to_sequences(valid_x), maxlen=70)\n\n# create token-embedding mapping\nembedding_matrix = np.zeros((len(word_index) + 1, 300))\nfor word, i in word_index.items():\n embedding_vector = embeddings_index.get(word)\n if embedding_vector is not None:\n embedding_matrix[i] = embedding_vector",
"_____no_output_____"
],
[
"def train_model(classifier, feature_vector_train, label, feature_vector_valid, is_neural_net=False):\n # fit the training dataset on the classifier\n classifier.fit(feature_vector_train, label)\n \n # predict the labels on validation dataset\n predictions = classifier.predict(feature_vector_valid)\n \n if is_neural_net:\n predictions = predictions.argmax(axis=-1)\n \n return metrics.accuracy_score(predictions, valid_y)",
"_____no_output_____"
],
[
"# Naive Bayes on Count Vectors\naccuracy = train_model(naive_bayes.MultinomialNB(), xtrain_count, train_y, xvalid_count)\nprint(\"NB, Count Vectors: \", accuracy)\n\n# Naive Bayes on Word Level TF IDF Vectors\naccuracy = train_model(naive_bayes.MultinomialNB(), xtrain_tfidf, train_y, xvalid_tfidf)\nprint(\"NB, WordLevel TF-IDF: \", accuracy)\n\n# Naive Bayes on Ngram Level TF IDF Vectors\naccuracy = train_model(naive_bayes.MultinomialNB(), xtrain_tfidf_ngram, train_y, xvalid_tfidf_ngram)\nprint(\"NB, N-Gram Vectors: \", accuracy)\n\n# Naive Bayes on Character Level TF IDF Vectors\naccuracy = train_model(naive_bayes.MultinomialNB(), xtrain_tfidf_ngram_chars, train_y, xvalid_tfidf_ngram_chars)\nprint(\"NB, CharLevel Vectors: \", accuracy)",
"NB, Count Vectors: 0.6341463414634146\nNB, WordLevel TF-IDF: 0.5934959349593496\nNB, N-Gram Vectors: 0.5813008130081301\nNB, CharLevel Vectors: 0.491869918699187\n"
],
[
"# Linear Classifier on Count Vectors\naccuracy = train_model(linear_model.LogisticRegression(), xtrain_count, train_y, xvalid_count)\nprint (\"LR, Count Vectors: \", accuracy)\n\n# Linear Classifier on Word Level TF IDF Vectors\naccuracy = train_model(linear_model.LogisticRegression(), xtrain_tfidf, train_y, xvalid_tfidf)\nprint(\"LR, WordLevel TF-IDF: \", accuracy)\n\n# Linear Classifier on Ngram Level TF IDF Vectors\naccuracy = train_model(linear_model.LogisticRegression(), xtrain_tfidf_ngram, train_y, xvalid_tfidf_ngram)\nprint(\"LR, N-Gram Vectors: \", accuracy)\n\n# Linear Classifier on Character Level TF IDF Vectors\naccuracy = train_model(linear_model.LogisticRegression(), xtrain_tfidf_ngram_chars, train_y, xvalid_tfidf_ngram_chars)\nprint(\"LR, CharLevel Vectors: \", accuracy)",
"LR, Count Vectors: 0.6626016260162602\nLR, WordLevel TF-IDF: 0.6219512195121951\nLR, N-Gram Vectors: 0.5813008130081301\nLR, CharLevel Vectors: 0.6463414634146342\n"
],
[
"# SVM on Ngram Level TF IDF Vectors\naccuracy = train_model(svm.SVC(), xtrain_tfidf_ngram, train_y, xvalid_tfidf_ngram)\nprint(\"SVM, N-Gram Vectors: \", accuracy)",
"SVM, N-Gram Vectors: 0.5772357723577236\n"
],
[
"# RF on Count Vectors\naccuracy = train_model(ensemble.RandomForestClassifier(), xtrain_count, train_y, xvalid_count)\nprint(\"RF, Count Vectors: \", accuracy)\n\n# RF on Word Level TF IDF Vectors\naccuracy = train_model(ensemble.RandomForestClassifier(), xtrain_tfidf, train_y, xvalid_tfidf)\nprint(\"RF, WordLevel TF-IDF: \", accuracy)",
"RF, Count Vectors: 0.6382113821138211\nRF, WordLevel TF-IDF: 0.6504065040650406\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a146baa8ac933f969ce71f8f471930a1cd3b5ea
| 9,880 |
ipynb
|
Jupyter Notebook
|
aquaponics-master/notebooks/On Test.ipynb
|
JunwenLIU/Aquaponics_system_Simulink
|
5048012730815f3e8bb5ccb254c7e1d4be8a84d4
|
[
"Apache-2.0"
] | null | null | null |
aquaponics-master/notebooks/On Test.ipynb
|
JunwenLIU/Aquaponics_system_Simulink
|
5048012730815f3e8bb5ccb254c7e1d4be8a84d4
|
[
"Apache-2.0"
] | null | null | null |
aquaponics-master/notebooks/On Test.ipynb
|
JunwenLIU/Aquaponics_system_Simulink
|
5048012730815f3e8bb5ccb254c7e1d4be8a84d4
|
[
"Apache-2.0"
] | null | null | null | 95.92233 | 7,788 | 0.869737 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"planting_day = 10\nharvest_day = 20\n\ndef switch(left, right, on, loc, k):\n sigma = 1 / (1 + np.exp(-k * (on - loc)))\n return (1 - sigma) * left + sigma * right\n\ndef _on(t, k):\n on_harvest = switch(1, 0, t, harvest_day, k)\n return switch(0, on_harvest, t, planting_day, k)",
"_____no_output_____"
],
[
"k = 100\nts = np.arange(0, 30, 0.1)\nons = [_on(t, k) for t in ts]\n\nplt.plot(ts, ons)",
"/home/nwoodbury/.virtualenvs/aquaponics/lib/python3.5/site-packages/ipykernel_launcher.py:5: RuntimeWarning: overflow encountered in exp\n \"\"\"\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code"
]
] |
4a148424c1b33f8b93cddb31192c15da986d9338
| 424,616 |
ipynb
|
Jupyter Notebook
|
NASA_GISS.ipynb
|
waterupto/EnvHydrology
|
e04f1f0e1de1ce2faa998ccb79d893badc924d37
|
[
"MIT"
] | 1 |
2022-02-14T07:52:12.000Z
|
2022-02-14T07:52:12.000Z
|
NASA_GISS.ipynb
|
waterupto/EnvHydrology
|
e04f1f0e1de1ce2faa998ccb79d893badc924d37
|
[
"MIT"
] | null | null | null |
NASA_GISS.ipynb
|
waterupto/EnvHydrology
|
e04f1f0e1de1ce2faa998ccb79d893badc924d37
|
[
"MIT"
] | 2 |
2021-11-09T14:04:12.000Z
|
2021-11-20T06:08:16.000Z
| 207.230844 | 122,090 | 0.740182 |
[
[
[
"**Import library**",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport calendar\nfrom datetime import datetime\nimport time\n# Standard plotly imports\nimport plotly.express as px\nimport plotly.graph_objects as go\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set_context(\"paper\", font_scale=1.3)\nsns.set_style('white')\n# stats\nfrom statsmodels.tsa.statespace.sarimax import SARIMAX\nfrom random import random\nfrom statsmodels.tsa.stattools import adfuller\n#Prophet\nfrom fbprophet import Prophet\n# SKLEARN\nfrom sklearn.metrics import mean_squared_error",
"_____no_output_____"
]
],
[
[
"**Import data**",
"_____no_output_____"
]
],
[
[
"# Read in the raw temperature dataset\nraw_global = pd.read_csv('GLB.Ts+dSST.csv', skiprows=1)\nraw_global = raw_global.iloc[:,:13]",
"_____no_output_____"
],
[
"raw_global.head()",
"_____no_output_____"
],
[
"raw_global.tail()",
"_____no_output_____"
]
],
[
[
"**Data Preprocessing**",
"_____no_output_____"
]
],
[
[
"def clean_value(raw_value):\n try:\n return float(raw_value)\n except:\n return np.NaN",
"_____no_output_____"
],
[
"def preprocess_data(raw):\n data_horizon = pd.date_range(start='1/1/1880', end='12/31/2019', freq='M')\n data = pd.DataFrame(data_horizon, columns=['Date'])\n #extract temperature data\n temp_list = []\n for idx in range(raw.shape[0]):\n temp_list.extend(raw.iloc[idx,1:])\n data['Temp'] = temp_list\n #clean value\n data['Temp'] = data['Temp'].apply(lambda x: clean_value(x))\n data.fillna(method='ffill', inplace=True)\n return data",
"_____no_output_____"
],
[
"global_t = preprocess_data(raw_global)",
"_____no_output_____"
],
[
"global_t.head()",
"_____no_output_____"
],
[
"global_t.tail()",
"_____no_output_____"
]
],
[
[
"**Data Visualization**",
"_____no_output_____"
]
],
[
[
"fig = px.line(global_t, x=\"Date\", y=\"Temp\", title='Global-mean monthly Combined Land-Surface Air and Sea-Surface Water Temperature Anomalies')\nfig.show()",
"_____no_output_____"
],
[
"fig = px.line(global_t.resample('A', on='Date').mean().reset_index(), x=\"Date\", y=\"Temp\", title='Global-mean yearly Combined Land-Surface Air and Sea-Surface Water Temperature Anomalies')\nfig.show()",
"_____no_output_____"
]
],
[
[
"Test stationarity",
"_____no_output_____"
]
],
[
[
"def test_stationarity(timeseries):\n rolmean = timeseries.rolling(window=30).mean()\n rolstd = timeseries.rolling(window=30).std()\n \n plt.figure(figsize=(14,5))\n sns.despine(left=True)\n orig = plt.plot(timeseries, color='blue',label='Original')\n mean = plt.plot(rolmean, color='red', label='Rolling Mean')\n std = plt.plot(rolstd, color='black', label = 'Rolling Std')\n\n plt.legend(loc='best'); plt.title('Rolling Mean & Standard Deviation')\n plt.show()\n \n print ('<Results of Dickey-Fuller Test>')\n dftest = adfuller(timeseries, autolag='AIC')\n dfoutput = pd.Series(dftest[0:4],\n index=['Test Statistic','p-value','#Lags Used','Number of Observations Used'])\n for key,value in dftest[4].items():\n dfoutput['Critical Value (%s)'%key] = value\n print(dfoutput)",
"_____no_output_____"
],
[
"test_stationarity(global_t.Temp.dropna())",
"_____no_output_____"
]
],
[
[
"since the p-value > 0.05, we accept the null hypothesis (H0), the data has a unit root and is non-stationary.",
"_____no_output_____"
],
[
"**Time Series Prediction - SARIMA**",
"_____no_output_____"
],
[
"The Seasonal Autoregressive Integrated Moving Average (SARIMA) method models the next step in the sequence as a linear function of the differenced observations, errors, differenced seasonal observations, and seasonal errors at prior time steps.\n\nIt combines the ARIMA model with the ability to perform the same autoregression, differencing, and moving average modeling at the seasonal level.\n\nThe notation for the model involves specifying the order for the AR(p), I(d), and MA(q) models as parameters to an ARIMA function and AR(P), I(D), MA(Q) and m parameters at the seasonal level, e.g. SARIMA(p, d, q)(P, D, Q)m where “m” is the number of time steps in each season (the seasonal period). A SARIMA model can be used to develop AR, MA, ARMA and ARIMA models.\n\nThe method is suitable for univariate time series with trend and/or seasonal components.",
"_____no_output_____"
]
],
[
[
"def plot(y_true,y_pred):\n # Plot\n fig = go.Figure()\n x = global_t['Date'][global_t.shape[0]-len(y_true):]\n fig.add_trace(go.Scatter(x=x, y=y_true, mode='lines', name='actual'))\n fig.add_trace(go.Scatter(x=x, y=y_pred, mode='lines', name='predicted'))\n # Edit the layout\n fig.update_layout(title='Southern Hemisphere-mean Temperature: Predicted v.s. Actual',\n xaxis_title='Month',\n yaxis_title='Temperature')\n fig.show()",
"_____no_output_____"
],
[
"def SARIMA_prediction(temp_data):\n y_true = []\n y_pred = []\n temperature = temp_data['Temp'].tolist()\n train = temperature[:-336]\n test = temperature[len(train):]\n #predict the latest 336 values (20% of data)\n for idx in range(len(test)):\n true_val = test[idx]\n if len(y_pred)>0:\n record = train+y_pred\n else:\n record = train\n # fit model\n model = SARIMAX(record, order=(1, 1, 1), seasonal_order=(1, 1, 1, 1))\n model_fit = model.fit(disp=False,low_memory=True)\n # make predictions \n yhat = model_fit.predict(len(record), len(record))\n # save value\n y_true.append(true_val)\n y_pred.extend(yhat)\n print(mean_squared_error(y_true, y_pred))\n plot(y_true,y_pred)",
"_____no_output_____"
],
[
"start_time = time.time()\nSARIMA_prediction(global_t)",
"/usr/local/lib/python3.6/dist-packages/statsmodels/base/model.py:512: ConvergenceWarning:\n\nMaximum Likelihood optimization failed to converge. Check mle_retvals\n\n"
],
[
"print(\"--- %s seconds ---\" % (time.time() - start_time))",
"--- 256.6636118888855 seconds ---\n"
]
],
[
[
"**Time Series Prediction - Prophet**",
"_____no_output_____"
],
[
"Prophet is a procedure for forecasting time series data based on an additive model where non-linear trends are fit with yearly, weekly, and daily seasonality, plus holiday effects. It works best with time series that have strong seasonal effects and several seasons of historical data. Prophet is robust to missing data and shifts in the trend, and typically handles outliers well.",
"_____no_output_____"
]
],
[
[
"def prophet_prediction(temp_data):\n #removing the last 336 values (10 years)\n df = temp_data.iloc[:-336]\n df = df.rename(columns={'Date':'ds', 'Temp':'y'})\n #load prophet model\n model = Prophet(weekly_seasonality=True)\n model.fit(df)\n #prediction\n future = model.make_future_dataframe(periods=336, freq = 'm')\n forecast = model.predict(future)\n model.plot(forecast)\n return forecast",
"_____no_output_____"
],
[
"start_time = time.time()\nprophet_forecast = prophet_prediction(global_t)",
"INFO:numexpr.utils:NumExpr defaulting to 2 threads.\nINFO:fbprophet:Disabling daily seasonality. Run prophet with daily_seasonality=True to override this.\n"
],
[
"print(\"--- %s seconds ---\" % (time.time() - start_time))",
"--- 6.097748279571533 seconds ---\n"
],
[
"prophet_forecast_last = prophet_forecast.iloc[prophet_forecast.shape[0]-336:]\nglobal_t_last = global_t.iloc[global_t.shape[0]-336:]",
"_____no_output_____"
],
[
"mean_squared_error(global_t_last.Temp, prophet_forecast_last.yhat)",
"_____no_output_____"
]
],
[
[
"**Time series prediction - LSTM**",
"_____no_output_____"
]
],
[
[
"from keras.models import Sequential\nfrom keras.layers.recurrent import LSTM\nfrom keras.layers.core import Dense, Activation, Dropout\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.metrics import mean_squared_error\nfrom sklearn.utils import shuffle\nfrom keras.callbacks import EarlyStopping\nearlyStop=EarlyStopping(monitor=\"val_loss\",verbose=2,mode='min',patience=5)",
"_____no_output_____"
]
],
[
[
"**Data preparation**",
"_____no_output_____"
]
],
[
[
"temp_raw = np.array(global_t.Temp.astype(\"float32\")).reshape(-1,1)\n# Apply the MinMax scaler from sklearn to normalize data in the (0, 1) interval.\nscaler = MinMaxScaler(feature_range = (0, 1))\ntemp_LSTM = scaler.fit_transform(temp_raw)",
"_____no_output_____"
],
[
"# Train test split - Using 80% of data for training, 20% for validation.\nratio = 0.6\ntrain_size = int(len(temp_LSTM) * ratio)\nval_size = int(len(temp_LSTM) * 0.2)\ntest_size = len(temp_LSTM) - train_size - val_size\ntrain, val, test = temp_LSTM[0:train_size, :], temp_LSTM[train_size:train_size+val_size, :], temp_LSTM[train_size+val_size:len(temp_LSTM), :]\nprint(\"Number of entries (training set, val set, test set): \" + str((len(train), len(val), len(test))))",
"Number of entries (training set, val set, test set): (1008, 336, 336)\n"
],
[
"def create_dataset(dataset):\n window_size = 1\n data_X, data_Y = [], []\n for i in range(len(dataset) - window_size - 1):\n a = dataset[i:(i + window_size), 0]\n data_X.append(a)\n data_Y.append(dataset[i + window_size, 0])\n return(np.array(data_X), np.array(data_Y))",
"_____no_output_____"
],
[
"# Create test and training sets for one-step-ahead regression.\ntrain_X, train_Y = create_dataset(train)\nval_X, val_Y = create_dataset(val)\ntest_X, test_Y = create_dataset(test)",
"_____no_output_____"
],
[
"# Reshape the input data into appropriate form for Keras.\ntrain_X = np.reshape(train_X, (train_X.shape[0], 1,train_X.shape[1]))\nval_X = np.reshape(val_X, (val_X.shape[0], 1,val_X.shape[1]))\ntest_X = np.reshape(test_X, (test_X.shape[0], 1,test_X.shape[1]))\nprint(\"Training data for Keras shape:\")\nprint(train_X.shape)",
"Training data for Keras shape:\n(1006, 1, 1)\n"
]
],
[
[
"**LSTM Model**",
"_____no_output_____"
],
[
"The LSTM architecture here consists of:\n\n- One input layer.\n- One LSTM layer of 4 blocks.\n- One Dense layer to produce a single output.\n- Use MSE as loss function.",
"_____no_output_____"
]
],
[
[
"def LSTM_modelone(train_X, train_Y, window_size):\n model = Sequential()\n model.add(LSTM(4, \n input_shape = (1, window_size)))\n model.add(Dense(1))\n model.compile(loss = \"mean_squared_error\", \n optimizer = \"adam\")\n model.fit(train_X, \n train_Y, \n epochs = 100, \n batch_size = 10, \n verbose = 2,\n validation_data=(val_X,val_Y),callbacks=[earlyStop])\n \n return model",
"_____no_output_____"
],
[
"start_time = time.time()\nLSTM_model1 = LSTM_modelone(train_X, train_Y, window_size=1)",
"Train on 1006 samples, validate on 334 samples\nEpoch 1/100\n - 1s - loss: 0.0328 - val_loss: 0.0362\nEpoch 2/100\n - 1s - loss: 0.0063 - val_loss: 0.0143\nEpoch 3/100\n - 1s - loss: 0.0044 - val_loss: 0.0116\nEpoch 4/100\n - 1s - loss: 0.0043 - val_loss: 0.0116\nEpoch 5/100\n - 1s - loss: 0.0042 - val_loss: 0.0108\nEpoch 6/100\n - 1s - loss: 0.0040 - val_loss: 0.0101\nEpoch 7/100\n - 1s - loss: 0.0039 - val_loss: 0.0093\nEpoch 8/100\n - 1s - loss: 0.0037 - val_loss: 0.0086\nEpoch 9/100\n - 1s - loss: 0.0036 - val_loss: 0.0085\nEpoch 10/100\n - 1s - loss: 0.0034 - val_loss: 0.0067\nEpoch 11/100\n - 1s - loss: 0.0033 - val_loss: 0.0064\nEpoch 12/100\n - 1s - loss: 0.0032 - val_loss: 0.0060\nEpoch 13/100\n - 1s - loss: 0.0031 - val_loss: 0.0059\nEpoch 14/100\n - 1s - loss: 0.0030 - val_loss: 0.0051\nEpoch 15/100\n - 1s - loss: 0.0029 - val_loss: 0.0050\nEpoch 16/100\n - 1s - loss: 0.0028 - val_loss: 0.0044\nEpoch 17/100\n - 1s - loss: 0.0028 - val_loss: 0.0041\nEpoch 18/100\n - 1s - loss: 0.0028 - val_loss: 0.0037\nEpoch 19/100\n - 1s - loss: 0.0028 - val_loss: 0.0034\nEpoch 20/100\n - 1s - loss: 0.0027 - val_loss: 0.0036\nEpoch 21/100\n - 1s - loss: 0.0027 - val_loss: 0.0036\nEpoch 22/100\n - 1s - loss: 0.0027 - val_loss: 0.0038\nEpoch 00022: early stopping\n"
],
[
"print(\"--- %s seconds ---\" % (time.time() - start_time))",
"--- 14.512033939361572 seconds ---\n"
],
[
"def predict_and_score(model, X, Y):\n # Make predictions on the original scale of the data.\n pred = scaler.inverse_transform(model.predict(X))\n # Prepare Y data to also be on the original scale for interpretability.\n orig_data = scaler.inverse_transform([Y])\n # Calculate RMSE.\n score = mean_squared_error(orig_data[0], pred[:, 0])\n return score",
"_____no_output_____"
],
[
"print(\"Test data score: %.3f MSE\" % predict_and_score(LSTM_model1,test_X, test_Y))",
"Test data score: 0.047 MSE\n"
]
],
[
[
"The second model architecture is slightly more complex. Its elements are:\n\n - Define the LSTM with 100 neurons in the first hidden layer and 1 neuron in the output layer\n - Dropout 20%.\n - Use the MSE loss function and the efficient Adam version of stochastic gradient descent.\n - The model will be fit for 50 training epochs with a batch size of 5.",
"_____no_output_____"
]
],
[
[
"def LSTM_modeltwo(train_X, train_Y):\n model = Sequential()\n model.add(LSTM(50, input_shape=(train_X.shape[1], train_X.shape[2])))\n model.add(Dropout(0.2))\n model.add(Dense(1))\n model.compile(loss='mean_squared_error', optimizer='adam')\n print(model.summary())\n\n model.fit(train_X, train_Y, epochs=50, batch_size=5, verbose=2, shuffle=False, validation_data=(val_X,val_Y),callbacks=[earlyStop])\n\n return model",
"_____no_output_____"
],
[
"start_time = time.time()\nLSTM_model2 = LSTM_modeltwo(train_X, train_Y)",
"Model: \"sequential_37\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nlstm_14 (LSTM) (None, 50) 10400 \n_________________________________________________________________\ndropout_30 (Dropout) (None, 50) 0 \n_________________________________________________________________\ndense_71 (Dense) (None, 1) 51 \n=================================================================\nTotal params: 10,451\nTrainable params: 10,451\nNon-trainable params: 0\n_________________________________________________________________\nNone\nTrain on 1006 samples, validate on 334 samples\nEpoch 1/50\n - 5s - loss: 0.0143 - val_loss: 0.0074\nEpoch 2/50\n - 1s - loss: 0.0046 - val_loss: 0.0065\nEpoch 3/50\n - 1s - loss: 0.0042 - val_loss: 0.0062\nEpoch 4/50\n - 1s - loss: 0.0039 - val_loss: 0.0051\nEpoch 5/50\n - 1s - loss: 0.0037 - val_loss: 0.0043\nEpoch 6/50\n - 1s - loss: 0.0036 - val_loss: 0.0039\nEpoch 7/50\n - 1s - loss: 0.0035 - val_loss: 0.0036\nEpoch 8/50\n - 1s - loss: 0.0034 - val_loss: 0.0034\nEpoch 9/50\n - 1s - loss: 0.0034 - val_loss: 0.0034\nEpoch 10/50\n - 1s - loss: 0.0033 - val_loss: 0.0030\nEpoch 11/50\n - 1s - loss: 0.0032 - val_loss: 0.0029\nEpoch 12/50\n - 1s - loss: 0.0032 - val_loss: 0.0029\nEpoch 13/50\n - 1s - loss: 0.0031 - val_loss: 0.0030\nEpoch 14/50\n - 1s - loss: 0.0031 - val_loss: 0.0031\nEpoch 15/50\n - 1s - loss: 0.0030 - val_loss: 0.0029\nEpoch 16/50\n - 1s - loss: 0.0030 - val_loss: 0.0029\nEpoch 00016: early stopping\n"
],
[
"print(\"--- %s seconds ---\" % (time.time() - start_time))",
"--- 24.736785173416138 seconds ---\n"
],
[
"print(\"Test data score: %.3f MSE\" % predict_and_score(LSTM_model2,test_X, test_Y))",
"Test data score: 0.042 MSE\n"
],
[
"def predict_and_plot(model, X, Y):\n # Make predictions on the original scale of the data.\n pred = scaler.inverse_transform(model.predict(X))\n # Prepare Y data to also be on the original scale for interpretability.\n orig_data = scaler.inverse_transform([Y])\n # Plot\n fig = go.Figure()\n x = global_t['Date'][global_t.shape[0]-len(orig_data[0]):]\n fig.add_trace(go.Scatter(x=x, y=orig_data[0], mode='lines', name='actual'))\n fig.add_trace(go.Scatter(x=x, y=pred[:, 0], mode='lines', name='predicted'))\n # Edit the layout\n fig.update_layout(title='Global Temperature: Predicted v.s. Actual',\n xaxis_title='Month',\n yaxis_title='Temperature')\n fig.show()",
"_____no_output_____"
],
[
"predict_and_plot(LSTM_model2,test_X, test_Y)",
"_____no_output_____"
]
],
[
[
"**MLP Model**",
"_____no_output_____"
]
],
[
[
"def MLP_model(train_X, train_Y):\n model = Sequential()\n model.add(Dense(100, input_shape=(1,)))\n model.add(Activation('relu'))\n model.add(Dropout(0.25))\n model.add(Dense(50))\n model.add(Activation('relu'))\n model.add(Dense(1))\n model.add(Activation('linear'))\n model.compile(optimizer='adam', loss='mse')\n print(model.summary())\n model.fit(train_X, train_Y, epochs=50, batch_size=10, verbose=2, shuffle=False, validation_data=(val_X,val_Y),callbacks=[earlyStop])\n return model",
"_____no_output_____"
],
[
"start_time = time.time()\nMLP_model_result = MLP_model(train_X, train_Y)",
"Model: \"sequential_32\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_65 (Dense) (None, 100) 200 \n_________________________________________________________________\nactivation_52 (Activation) (None, 100) 0 \n_________________________________________________________________\ndropout_26 (Dropout) (None, 100) 0 \n_________________________________________________________________\ndense_66 (Dense) (None, 50) 5050 \n_________________________________________________________________\nactivation_53 (Activation) (None, 50) 0 \n_________________________________________________________________\ndense_67 (Dense) (None, 1) 51 \n_________________________________________________________________\nactivation_54 (Activation) (None, 1) 0 \n=================================================================\nTotal params: 5,301\nTrainable params: 5,301\nNon-trainable params: 0\n_________________________________________________________________\nNone\nTrain on 1006 samples, validate on 334 samples\nEpoch 1/50\n - 3s - loss: 0.0150 - val_loss: 0.0052\nEpoch 2/50\n - 0s - loss: 0.0039 - val_loss: 0.0054\nEpoch 3/50\n - 0s - loss: 0.0036 - val_loss: 0.0042\nEpoch 4/50\n - 0s - loss: 0.0034 - val_loss: 0.0037\nEpoch 5/50\n - 0s - loss: 0.0033 - val_loss: 0.0035\nEpoch 6/50\n - 0s - loss: 0.0033 - val_loss: 0.0035\nEpoch 7/50\n - 0s - loss: 0.0030 - val_loss: 0.0034\nEpoch 8/50\n - 0s - loss: 0.0031 - val_loss: 0.0032\nEpoch 9/50\n - 0s - loss: 0.0032 - val_loss: 0.0032\nEpoch 10/50\n - 0s - loss: 0.0031 - val_loss: 0.0032\nEpoch 11/50\n - 0s - loss: 0.0031 - val_loss: 0.0031\nEpoch 12/50\n - 0s - loss: 0.0031 - val_loss: 0.0030\nEpoch 13/50\n - 0s - loss: 0.0031 - val_loss: 0.0031\nEpoch 14/50\n - 0s - loss: 0.0031 - val_loss: 0.0032\nEpoch 15/50\n - 0s - loss: 0.0031 - val_loss: 0.0031\nEpoch 16/50\n - 0s - loss: 0.0031 - val_loss: 0.0032\nEpoch 17/50\n - 0s - loss: 0.0031 - val_loss: 0.0032\nEpoch 00017: early stopping\n"
],
[
"print(\"--- %s seconds ---\" % (time.time() - start_time))",
"--- 8.819153308868408 seconds ---\n"
],
[
"print(\"Test data score: %.3f MSE\" % predict_and_score(MLP_model_result,test_X, test_Y))",
"Test data score: 0.059 MSE\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a1488b04dd96bb2fa8ba278be925c7914e40871
| 33,835 |
ipynb
|
Jupyter Notebook
|
eppy_resi_modelling.ipynb
|
oisindoherty3/energyplus-archetypes
|
01e12b6fc5bd2654a04cecae8dffa31799d15108
|
[
"MIT"
] | null | null | null |
eppy_resi_modelling.ipynb
|
oisindoherty3/energyplus-archetypes
|
01e12b6fc5bd2654a04cecae8dffa31799d15108
|
[
"MIT"
] | null | null | null |
eppy_resi_modelling.ipynb
|
oisindoherty3/energyplus-archetypes
|
01e12b6fc5bd2654a04cecae8dffa31799d15108
|
[
"MIT"
] | null | null | null | 35.503673 | 1,523 | 0.519285 |
[
[
[
"from eppy import modeleditor\nfrom eppy.modeleditor import IDF\nimport pandas as pd",
"_____no_output_____"
],
[
"pip install pandas",
"Requirement already satisfied: pandas in /usr/local/lib/python3.7/site-packages (1.2.0)\nRequirement already satisfied: numpy>=1.16.5 in /usr/local/lib/python3.7/site-packages (from pandas) (1.19.5)\nRequirement already satisfied: pytz>=2017.3 in /usr/local/lib/python3.7/site-packages (from pandas) (2020.5)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.7/site-packages (from pandas) (2.8.1)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.7/site-packages (from python-dateutil>=2.7.3->pandas) (1.15.0)\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"IDF.setiddname('/usr/local/bin/Energy+.idd')",
"_____no_output_____"
],
[
"idf = IDF('/idf/det_pre/detatched_pre.idf')\nidf.epw = \"idf/det_pre/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/det_pre\")",
"\r\n/usr/local/bin/energyplus --weather /idf/det_pre/IRL_Dublin.039690_IWEC.epw --output-directory /idf/det_pre --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/det_pre/eplusmtr.csv\")\npeak_demand_joule_det_pre = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/det_pre/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_det_pre = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_det_pre = df3.iloc[16,1]\nann_heat_demand_kwh_det_pre = df3.iloc[16,5]\nprint(peak_demand_joule_det_pre, ann_energy_demand_kwh_det_pre, ann_elec_demand_kwh_det_pre, ann_heat_demand_kwh_det_pre)",
"8584366.459649652 26343.66 6439.15 19904.51\n"
],
[
"del idf\nidf = IDF('/idf/det_post/detatched_post.idf')\nidf.epw = \"idf/det_post/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/det_post\")",
"\r\n/usr/local/bin/energyplus --weather /idf/det_post/IRL_Dublin.039690_IWEC.epw --output-directory /idf/det_post --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/det_post/eplusmtr.csv\")\npeak_demand_joule_det_post = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/det_post/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_det_post = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_det_post = df3.iloc[16,1]\nann_heat_demand_kwh_det_post = df3.iloc[16,5]\nprint(peak_demand_joule_det_post, ann_energy_demand_kwh_det_post, ann_elec_demand_kwh_det_post, ann_heat_demand_kwh_det_post)",
"7240166.818522849 16097.65 5171.22 10926.43\n"
],
[
"del idf\nidf = IDF('/idf/semi_d_pre/semi_d_pre.idf')\nidf.epw = \"idf/semi_d_pre/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/semid_pre\")",
"\r\n/usr/local/bin/energyplus --weather /idf/semid_pre/IRL_Dublin.039690_IWEC.epw --output-directory /idf/semid_pre --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/semi_d_pre/eplusmtr.csv\")\npeak_demand_joule_semid_pre = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/semi_d_pre/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_semid_pre = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_semid_pre = df3.iloc[16,1]\nann_heat_demand_kwh_semid_pre = df3.iloc[16,5]\nprint(peak_demand_joule_semid_pre, ann_energy_demand_kwh_semid_pre, ann_elec_demand_kwh_semid_pre, ann_heat_demand_kwh_semid_pre)",
"6177061.326873895 25113.73 5149.22 19964.51\n"
],
[
"del idf\nidf = IDF('/idf/semi_d_post/semi_d_post.idf')\nidf.epw = \"idf/semi_d_post/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/semid_post\")",
"\r\n/usr/local/bin/energyplus --weather /idf/semid_post/IRL_Dublin.039690_IWEC.epw --output-directory /idf/semid_post --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/semi_d_post/eplusmtr.csv\")\npeak_demand_joule_semid_post = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/semi_d_post/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_semid_post = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_semid_post = df3.iloc[16,1]\nann_heat_demand_kwh_semid_post = df3.iloc[16,5]\nprint(peak_demand_joule_semid_post, ann_energy_demand_kwh_semid_post, ann_elec_demand_kwh_semid_post, ann_heat_demand_kwh_semid_post)",
"4723727.966424771 9736.54 3804.43 5932.10\n"
],
[
"del idf\nidf = IDF('/idf/terr_pre/terraced_pre.idf')\nidf.epw = \"idf/terr_pre/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/terr_pre\")",
"\r\n/usr/local/bin/energyplus --weather /idf/terr_pre/IRL_Dublin.039690_IWEC.epw --output-directory /idf/terr_pre --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/terr_pre/eplusmtr.csv\")\npeak_demand_joule_terr_pre = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/terr_pre/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_terr_pre = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_terr_pre = df3.iloc[16,1]\nann_heat_demand_kwh_terr_pre = df3.iloc[16,5]\nprint(peak_demand_joule_terr_pre, ann_energy_demand_kwh_terr_pre, ann_elec_demand_kwh_terr_pre, ann_heat_demand_kwh_terr_pre)",
"3783938.4093833286 15126.49 2531.93 12594.56\n"
],
[
"del idf\nidf = IDF('/idf/terr_post/terraced_post.idf')\nidf.epw = \"idf/terr_post/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/terr_post\")",
"\r\n/usr/local/bin/energyplus --weather /idf/terr_post/IRL_Dublin.039690_IWEC.epw --output-directory /idf/terr_post --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/terr_post/eplusmtr.csv\")\npeak_demand_joule_terr_post = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/terr_post/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_terr_post = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_terr_post = df3.iloc[16,1]\nann_heat_demand_kwh_terr_post = df3.iloc[16,5]\nprint(peak_demand_joule_terr_post, ann_energy_demand_kwh_terr_post, ann_elec_demand_kwh_terr_post, ann_heat_demand_kwh_terr_post)",
"2869567.332309684 4082.00 1913.10 2168.90\n"
],
[
"del idf\nidf = IDF('/idf/apt_pre/mid_apt_pre.idf')\nidf.epw = \"idf/apt_pre/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/apt_pre\")",
"\r\n/usr/local/bin/energyplus --weather /idf/apt_pre/IRL_Dublin.039690_IWEC.epw --output-directory /idf/apt_pre --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/mid_apt_pre/eplusmtr.csv\")\npeak_demand_joule_mid_apt_pre = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/mid_apt_pre/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_mid_apt_pre = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_mid_apt_pre = df3.iloc[16,1]\nann_heat_demand_kwh_mid_apt_pre = df3.iloc[16,5]\nprint(peak_demand_joule_mid_apt_pre, ann_energy_demand_kwh_mid_apt_pre, ann_elec_demand_kwh_mid_apt_pre, ann_heat_demand_kwh_mid_apt_pre)",
"6265227.970799999 7929.74 1381.93 6547.81\n"
],
[
"del idf\nidf = IDF('/idf/mid_apt_post/mid_apt_post.idf')\nidf.epw = \"idf/mid_apt_post/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/mid_apt_post\")",
"_____no_output_____"
],
[
"df = pd.read_csv(\"idf/mid_apt_post/eplusmtr.csv\")\npeak_demand_joule_mid_apt_post = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/mid_apt_post/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_mid_apt_post = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_mid_apt_post = df3.iloc[16,1]\nann_heat_demand_kwh_mid_apt_post = df3.iloc[16,5]\nprint(peak_demand_joule_mid_apt_post, ann_energy_demand_kwh_mid_apt_post, ann_elec_demand_kwh_mid_apt_post, ann_heat_demand_kwh_mid_apt_post)",
"1036469.4732 3821.79 984.32 2837.47\n"
],
[
"del idf\nidf = IDF('/idf/top_apt_pre/top_apt_pre.idf')\nidf.epw = \"idf/top_apt_pre/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/top_apt_pre\")",
"\r\n/usr/local/bin/energyplus --weather /idf/top_apt_pre/IRL_Dublin.039690_IWEC.epw --output-directory /idf/top_apt_pre --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/top_apt_pre/eplusmtr.csv\")\npeak_demand_joule_top_apt_pre = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/top_apt_pre/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_top_apt_pre = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_top_apt_pre = df3.iloc[16,1]\nann_heat_demand_kwh_top_apt_pre = df3.iloc[16,5]\nprint(peak_demand_joule_top_apt_pre, ann_energy_demand_kwh_top_apt_pre, ann_elec_demand_kwh_top_apt_pre, ann_heat_demand_kwh_top_apt_pre)",
"6627898.778400001 16025.90 2030.69 13995.20\n"
],
[
"del idf\nidf = IDF('/idf/top_apt_post/top_apt_post.idf')\nidf.epw = \"idf/top_apt_post/IRL_Dublin.039690_IWEC.epw\"\nidf.run(expandobjects=True, readvars=True, output_directory=\"/idf/top_apt_post\")",
"\r\n/usr/local/bin/energyplus --weather /idf/top_apt_post/IRL_Dublin.039690_IWEC.epw --output-directory /idf/top_apt_post --idd /usr/local/bin/Energy+.idd --expandobjects --readvars /in.idf\r\n\n"
],
[
"df = pd.read_csv(\"idf/top_apt_post/eplusmtr.csv\")\npeak_demand_joule_top_apt_post = df[\"Electricity:Facility [J](Hourly)\"].max()\ndf2 = pd.read_html(\"idf/top_apt_post/eplustbl.htm\")\ndf_e = pd.DataFrame(df2[0])\nann_energy_demand_kwh_top_apt_post = df_e.iloc[1,1]\ndf3 = pd.DataFrame(df2[3])\nann_elec_demand_kwh_top_apt_post = df3.iloc[16,1]\nann_heat_demand_kwh_top_apt_post = df3.iloc[16,5]\nprint(peak_demand_joule_top_apt_post, ann_energy_demand_kwh_top_apt_post, ann_elec_demand_kwh_top_apt_post, ann_heat_demand_kwh_top_apt_post)",
"5803007.378400002 11013.22 1508.87 9504.34\n"
],
[
"peak_demand_joule_top_apt_post",
"_____no_output_____"
],
[
"peak_data = [['Detatched housepre', peak_demand_joule_det_pre, ann_energy_demand_kwh_det_pre, ann_elec_demand_kwh_det_pre, ann_heat_demand_kwh_det_pre], ['Detatched housepost', peak_demand_joule_det_post, ann_energy_demand_kwh_det_post, ann_elec_demand_kwh_det_post, ann_heat_demand_kwh_det_post], ['Semi detatched housepre', peak_demand_joule_semid_pre, ann_energy_demand_kwh_semid_pre, ann_elec_demand_kwh_semid_pre, ann_heat_demand_kwh_semid_pre],['Semi detatched housepost', peak_demand_joule_semid_post, ann_energy_demand_kwh_semid_post, ann_elec_demand_kwh_semid_post, ann_heat_demand_kwh_semid_post],['Terraced housepre', peak_demand_joule_terr_pre, ann_energy_demand_kwh_terr_pre, ann_elec_demand_kwh_terr_pre, ann_heat_demand_kwh_terr_pre], ['Terraced housepost', peak_demand_joule_terr_post, ann_energy_demand_kwh_terr_post, ann_elec_demand_kwh_terr_post, ann_heat_demand_kwh_terr_post], ['Apartmentpre', peak_demand_joule_mid_apt_pre, ann_energy_demand_kwh_mid_apt_pre, ann_elec_demand_kwh_mid_apt_pre, ann_heat_demand_kwh_mid_apt_pre],['Apartmentpost', peak_demand_joule_mid_apt_post, ann_energy_demand_kwh_mid_apt_post, ann_elec_demand_kwh_mid_apt_post, ann_heat_demand_kwh_mid_apt_post],['Top floor apt.pre', peak_demand_joule_top_apt_pre, ann_energy_demand_kwh_top_apt_pre, ann_elec_demand_kwh_top_apt_pre, ann_heat_demand_kwh_top_apt_pre],['Top floor apt.post', peak_demand_joule_top_apt_post, ann_energy_demand_kwh_top_apt_post, ann_elec_demand_kwh_top_apt_post, ann_heat_demand_kwh_top_apt_post], ] ",
"_____no_output_____"
],
[
"df_peaks = pd.DataFrame(peak_data, columns = ['dwelling_type','peak_hourly_elec_demand(J)', \"annual_energy_demand_kwh\", \"annual_elec_demand_kwh\", \"annual_heat_demand_kwh\"]) ",
"_____no_output_____"
],
[
"df_peaks",
"_____no_output_____"
],
[
"df_peaks[\"peak_elec_demand(kW)\"] = df_peaks[\"peak_hourly_elec_demand(J)\"]/3600000",
"_____no_output_____"
],
[
"df_peaks[\"peak_elec_demand(kVA)\"] = df_peaks[\"peak_elec_demand(kW)\"]*0.85",
"_____no_output_____"
],
[
"df_peaks",
"_____no_output_____"
],
[
"df_peaks.to_csv(\"/idf/resi_modelling/energy_demand_by_building_type.csv\")",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a1492c1690628cdf12ae389a162ca6cdde4b5dc
| 92,175 |
ipynb
|
Jupyter Notebook
|
Section_2_Federated_Learning_Final_Project.ipynb
|
ewotawa/secure_private_ai
|
f2b1e92ba7abba2a50b7a5f42dbdf30b88d9d1ae
|
[
"OLDAP-2.6"
] | 2 |
2019-08-13T04:09:57.000Z
|
2021-07-09T18:47:11.000Z
|
Section_2_Federated_Learning_Final_Project.ipynb
|
ewotawa/secure_private_ai
|
f2b1e92ba7abba2a50b7a5f42dbdf30b88d9d1ae
|
[
"OLDAP-2.6"
] | null | null | null |
Section_2_Federated_Learning_Final_Project.ipynb
|
ewotawa/secure_private_ai
|
f2b1e92ba7abba2a50b7a5f42dbdf30b88d9d1ae
|
[
"OLDAP-2.6"
] | null | null | null | 61.903962 | 364 | 0.535731 |
[
[
[
"<a href=\"https://colab.research.google.com/github/ewotawa/secure_private_ai/blob/master/Section_2_Federated_Learning_Final_Project.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Federated Learning Final Project\n\n## Overview\n* See <a href=\"https://classroom.udacity.com/nanodegrees/nd185/parts/3fe1bb10-68d7-4d84-9c99-9539dedffad5/modules/28d685f0-0cb1-4f94-a8ea-2e16614ab421/lessons/c8fe481d-81ea-41be-8206-06d2deeb8575/concepts/a5fb4b4c-e38a-48de-b2a7-4e853c62acbe\">video</a> for additional details. \n* Do Federated Learning where the central server is not trusted with the raw gradients. \n* In the final project notebook, you'll receive a dataset. \n* Train on the dataset using Federated Learning. \n* The gradients should not come up to the server in raw form. \n* Instead, use the new .move() command to move all of the gradients to one of the workers, sum them up there, and then bring that batch up to the central server and then bring that batch up \n* Idea: the central server never actually sees the raw gradient for any person. \n* We'll look at secure aggregation in course 3. \n* For now, do a larger-scale Federated Learning case where you handle the gradients in a special way.\n\n## Approach\n* Use the method illustrated in the \"DEEP LEARNING\" article referenced below. Update the code such that the MNIST model trains locally. Updated for my personal code style preferences.\n* Per conversation in the SPAIC Slack channel, use of a federated data loader approach trains the model and keeps the disaggregated gradients off of the local machine. The aggregate model returns when model.get() is called.\n* Contacted the team at OpenMined. They confirmed that PySyft currently does not work with GPUs, although updates are in progress. (7/18/2019).\n\n## References\n* <a href = \"https://blog.openmined.org/upgrade-to-federated-learning-in-10-lines/\">DEEP LEARNING -> FEDERATED LEARNING IN 10 LINES OF PYTORCH + PYSYFT</a>\n* <a href =\"https://github.com/udacity/private-ai/pull/10\">added data for Federated Learning project</a>\n* <a href=\"https://github.com/OpenMined/PySyft/blob/master/examples/tutorials/Part%206%20-%20Federated%20Learning%20on%20MNIST%20using%20a%20CNN.ipynb\">Part 6 - Federated Learning on MNIST using a CNN.ipynb</a>\n* <a href=\"https://docs.google.com/spreadsheets/d/1x-QQK-3Wn86bvSbNTf2_p2FXVCqiic2QwjcArQEuQlg/edit#gid=0\">Slack Channel's reference sheet </a>\n* <a href=\"https://github.com/ucalyptus/Federated-Learning/blob/master/Federated%20Learning.ipynb\">Federated Learning Example from Slack Channel reference sheet</a>",
"_____no_output_____"
],
[
"### Install libraries and dependencies",
"_____no_output_____"
]
],
[
[
"!pip install syft\n\nimport syft as sy",
"Collecting syft\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/38/2e/16bdefc78eb089e1efa9704c33b8f76f035a30dc935bedd7cbb22f6dabaa/syft-0.1.21a1-py3-none-any.whl (219kB)\n\u001b[K |████████████████████████████████| 225kB 2.8MB/s \n\u001b[?25hCollecting zstd>=1.4.0.0 (from syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/8e/27/1ea8086d37424e83ab692015cc8dd7d5e37cf791e339633a40dc828dfb74/zstd-1.4.0.0.tar.gz (450kB)\n\u001b[K |████████████████████████████████| 450kB 28.4MB/s \n\u001b[?25hRequirement already satisfied: Flask>=1.0.2 in /usr/local/lib/python3.6/dist-packages (from syft) (1.1.1)\nCollecting msgpack>=0.6.1 (from syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/92/7e/ae9e91c1bb8d846efafd1f353476e3fd7309778b582d2fb4cea4cc15b9a2/msgpack-0.6.1-cp36-cp36m-manylinux1_x86_64.whl (248kB)\n\u001b[K |████████████████████████████████| 256kB 41.2MB/s \n\u001b[?25hRequirement already satisfied: tblib>=1.4.0 in /usr/local/lib/python3.6/dist-packages (from syft) (1.4.0)\nRequirement already satisfied: torch>=1.1 in /usr/local/lib/python3.6/dist-packages (from syft) (1.1.0)\nCollecting flask-socketio>=3.3.2 (from syft)\n Downloading https://files.pythonhosted.org/packages/4b/68/fe4806d3a0a5909d274367eb9b3b87262906c1515024f46c2443a36a0c82/Flask_SocketIO-4.1.0-py2.py3-none-any.whl\nRequirement already satisfied: torchvision>=0.3.0 in /usr/local/lib/python3.6/dist-packages (from syft) (0.3.0)\nRequirement already satisfied: scikit-learn>=0.21.0 in /usr/local/lib/python3.6/dist-packages (from syft) (0.21.2)\nRequirement already satisfied: numpy>=1.14.0 in /usr/local/lib/python3.6/dist-packages (from syft) (1.16.4)\nCollecting lz4>=2.1.6 (from syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/0a/c6/96bbb3525a63ebc53ea700cc7d37ab9045542d33b4d262d0f0408ad9bbf2/lz4-2.1.10-cp36-cp36m-manylinux1_x86_64.whl (385kB)\n\u001b[K |████████████████████████████████| 389kB 41.2MB/s \n\u001b[?25hCollecting tf-encrypted>=0.5.4 (from syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/55/ff/7dbd5fc77fcec0df1798268a6b72a2ab0150b854761bc39c77d566798f0b/tf_encrypted-0.5.7-py3-none-manylinux1_x86_64.whl (2.1MB)\n\u001b[K |████████████████████████████████| 2.1MB 35.3MB/s \n\u001b[?25hCollecting websocket-client>=0.56.0 (from syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/29/19/44753eab1fdb50770ac69605527e8859468f3c0fd7dc5a76dd9c4dbd7906/websocket_client-0.56.0-py2.py3-none-any.whl (200kB)\n\u001b[K |████████████████████████████████| 204kB 41.8MB/s \n\u001b[?25hCollecting websockets>=7.0 (from syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/61/5e/2fe6afbb796c6ac5c006460b5503cd674d33706660337f2dbff10d4aa12d/websockets-8.0-cp36-cp36m-manylinux1_x86_64.whl (72kB)\n\u001b[K |████████████████████████████████| 81kB 27.7MB/s \n\u001b[?25hRequirement already satisfied: itsdangerous>=0.24 in /usr/local/lib/python3.6/dist-packages (from Flask>=1.0.2->syft) (1.1.0)\nRequirement already satisfied: Jinja2>=2.10.1 in /usr/local/lib/python3.6/dist-packages (from Flask>=1.0.2->syft) (2.10.1)\nRequirement already satisfied: Werkzeug>=0.15 in /usr/local/lib/python3.6/dist-packages (from Flask>=1.0.2->syft) (0.15.4)\nRequirement already satisfied: click>=5.1 in /usr/local/lib/python3.6/dist-packages (from Flask>=1.0.2->syft) (7.0)\nCollecting python-socketio>=2.1.0 (from flask-socketio>=3.3.2->syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/26/1b/57e860a86f2a01be86ae1dacfa0cd8c4dfbfcd4593322268b61b5a07b564/python_socketio-4.2.0-py2.py3-none-any.whl (46kB)\n\u001b[K |████████████████████████████████| 51kB 19.5MB/s \n\u001b[?25hRequirement already satisfied: pillow>=4.1.1 in /usr/local/lib/python3.6/dist-packages (from torchvision>=0.3.0->syft) (4.3.0)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from torchvision>=0.3.0->syft) (1.12.0)\nRequirement already satisfied: scipy>=0.17.0 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.21.0->syft) (1.3.0)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.21.0->syft) (0.13.2)\nCollecting pyyaml>=5.1 (from tf-encrypted>=0.5.4->syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/a3/65/837fefac7475963d1eccf4aa684c23b95aa6c1d033a2c5965ccb11e22623/PyYAML-5.1.1.tar.gz (274kB)\n\u001b[K |████████████████████████████████| 276kB 21.3MB/s \n\u001b[?25hRequirement already satisfied: tensorflow<2,>=1.12.0 in /usr/local/lib/python3.6/dist-packages (from tf-encrypted>=0.5.4->syft) (1.14.0)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.6/dist-packages (from Jinja2>=2.10.1->Flask>=1.0.2->syft) (1.1.1)\nCollecting python-engineio>=3.8.0 (from python-socketio>=2.1.0->flask-socketio>=3.3.2->syft)\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/bd/b8/0fc389ca5c445051b37b17802f80bbf1b51c1e3b48b772ee608efbb90583/python_engineio-3.8.2.post1-py2.py3-none-any.whl (119kB)\n\u001b[K |████████████████████████████████| 122kB 39.3MB/s \n\u001b[?25hRequirement already satisfied: olefile in /usr/local/lib/python3.6/dist-packages (from pillow>=4.1.1->torchvision>=0.3.0->syft) (0.46)\nRequirement already satisfied: keras-preprocessing>=1.0.5 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (1.1.0)\nRequirement already satisfied: termcolor>=1.1.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (1.1.0)\nRequirement already satisfied: protobuf>=3.6.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (3.7.1)\nRequirement already satisfied: google-pasta>=0.1.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (0.1.7)\nRequirement already satisfied: astor>=0.6.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (0.8.0)\nRequirement already satisfied: wheel>=0.26 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (0.33.4)\nRequirement already satisfied: absl-py>=0.7.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (0.7.1)\nRequirement already satisfied: tensorflow-estimator<1.15.0rc0,>=1.14.0rc0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (1.14.0)\nRequirement already satisfied: keras-applications>=1.0.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (1.0.8)\nRequirement already satisfied: grpcio>=1.8.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (1.15.0)\nRequirement already satisfied: gast>=0.2.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (0.2.2)\nRequirement already satisfied: wrapt>=1.11.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (1.11.2)\nRequirement already satisfied: tensorboard<1.15.0,>=1.14.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (1.14.0)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from protobuf>=3.6.1->tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (41.0.1)\nRequirement already satisfied: h5py in /usr/local/lib/python3.6/dist-packages (from keras-applications>=1.0.6->tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (2.8.0)\nRequirement already satisfied: markdown>=2.6.8 in /usr/local/lib/python3.6/dist-packages (from tensorboard<1.15.0,>=1.14.0->tensorflow<2,>=1.12.0->tf-encrypted>=0.5.4->syft) (3.1.1)\nBuilding wheels for collected packages: zstd, pyyaml\n Building wheel for zstd (setup.py) ... \u001b[?25l\u001b[?25hdone\n Stored in directory: /root/.cache/pip/wheels/ad/9a/f4/3105b5209674ac77fcca7fede95184c62a95df0196888e0e76\n Building wheel for pyyaml (setup.py) ... \u001b[?25l\u001b[?25hdone\n Stored in directory: /root/.cache/pip/wheels/16/27/a1/775c62ddea7bfa62324fd1f65847ed31c55dadb6051481ba3f\nSuccessfully built zstd pyyaml\nInstalling collected packages: zstd, msgpack, python-engineio, python-socketio, flask-socketio, lz4, pyyaml, tf-encrypted, websocket-client, websockets, syft\n Found existing installation: msgpack 0.5.6\n Uninstalling msgpack-0.5.6:\n Successfully uninstalled msgpack-0.5.6\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\nSuccessfully installed flask-socketio-4.1.0 lz4-2.1.10 msgpack-0.6.1 python-engineio-3.8.2.post1 python-socketio-4.2.0 pyyaml-5.1.1 syft-0.1.21a1 tf-encrypted-0.5.7 websocket-client-0.56.0 websockets-8.0 zstd-1.4.0.0\n"
],
[
"!pip install torch\n!pip install torchvision\n\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nimport torchvision\nfrom torchvision import datasets, transforms\n\nimport numpy as np",
"Requirement already satisfied: torch in /usr/local/lib/python3.6/dist-packages (1.1.0)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torch) (1.16.4)\nRequirement already satisfied: torchvision in /usr/local/lib/python3.6/dist-packages (0.3.0)\nRequirement already satisfied: pillow>=4.1.1 in /usr/local/lib/python3.6/dist-packages (from torchvision) (4.3.0)\nRequirement already satisfied: torch>=1.1.0 in /usr/local/lib/python3.6/dist-packages (from torchvision) (1.1.0)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from torchvision) (1.12.0)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from torchvision) (1.16.4)\nRequirement already satisfied: olefile in /usr/local/lib/python3.6/dist-packages (from pillow>=4.1.1->torchvision) (0.46)\n"
],
[
"hook = sy.TorchHook(torch) # <-- NEW: hook PyTorch ie add extra functionalities to support Federated Learning\nvw00 = sy.VirtualWorker(hook, id=\"vw00\")\nvw01 = sy.VirtualWorker(hook, id=\"vw01\")\n\naggr = sy.VirtualWorker(hook, id=\"aggr\")",
"_____no_output_____"
],
[
"class Arguments():\n def __init__(self):\n self.batch_size = 64\n self.test_batch_size = 1000\n self.epochs = 10\n self.lr = 0.01\n self.momentum = 0.5\n self.no_cuda = False\n self.seed = 1\n self.log_interval = 10\n self.save_model = False\n\nargs = Arguments()\n\nuse_cuda = not args.no_cuda and torch.cuda.is_available()\n\ntorch.manual_seed(args.seed)\n\ndevice = torch.device(\"cuda\" if use_cuda else \"cpu\")\n\nkwargs = {'num_workers': 1, 'pin_memory': True} if use_cuda else {}",
"_____no_output_____"
],
[
"# Note: removed **kwargs from end of federated_train_loader and test_loader definitions.\n\ntransform = transform=transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))\n ])\n\nfederated_train_loader = sy.FederatedDataLoader(datasets.MNIST('../data', train=True, download=True, transform=transform).federate((vw00, vw01)), \n batch_size=args.batch_size, shuffle=True)\n\ntest_loader = torch.utils.data.DataLoader(datasets.MNIST('../data', train=False, transform=transform), \n batch_size=args.test_batch_size, shuffle=True)",
"\r0it [00:00, ?it/s]"
],
[
"class Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n self.conv1 = nn.Conv2d(1, 20, 5, 1)\n self.conv2 = nn.Conv2d(20, 50, 5, 1)\n self.fc1 = nn.Linear(4*4*50, 500)\n self.fc2 = nn.Linear(500, 10)\n\n def forward(self, x):\n x = F.relu(self.conv1(x))\n x = F.max_pool2d(x, 2, 2)\n x = F.relu(self.conv2(x))\n x = F.max_pool2d(x, 2, 2)\n x = x.view(-1, 4*4*50)\n x = F.relu(self.fc1(x))\n x = self.fc2(x)\n return F.log_softmax(x, dim=1)",
"_____no_output_____"
],
[
"def train(args, model, device, train_loader, optimizer, epoch):\n model.train()\n for batch_idx, (data, target) in enumerate(federated_train_loader): # <-- now it is a distributed dataset\n model.send(data.location) # <-- NEW: send the model to the right location\n # data, target = data.to(device), target.to(device)\n optimizer.zero_grad()\n output = model(data)\n loss = F.nll_loss(output, target)\n loss.backward()\n optimizer.step()\n model.get() # <-- NEW: get the model back\n if batch_idx % args.log_interval == 0:\n loss = loss.get() # <-- NEW: get the loss back\n print('Train Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n epoch, batch_idx * args.batch_size, len(train_loader) * args.batch_size, #batch_idx * len(data), len(train_loader.dataset),\n 100. * batch_idx / len(train_loader), loss.item()))\n ",
"_____no_output_____"
],
[
"def test(args, model, device, test_loader):\n model.eval()\n test_loss = 0\n correct = 0\n with torch.no_grad():\n for data, target in test_loader:\n # data, target = data.to(device), target.to(device)\n output = model(data)\n test_loss += F.nll_loss(output, target, reduction='sum').item() # sum up batch loss\n pred = output.argmax(1, keepdim=True) # get the index of the max log-probability \n correct += pred.eq(target.view_as(pred)).sum().item()\n\n test_loss /= len(test_loader.dataset)\n\n print('\\nTest set: Average loss: {:.4f}, Accuracy: {}/{} ({:.0f}%)\\n'.format(\n test_loss, correct, len(test_loader.dataset),\n 100. * correct / len(test_loader.dataset)))",
"_____no_output_____"
],
[
"# model = Net().to(device)\nmodel = Net()\noptimizer = optim.SGD(model.parameters(), lr=args.lr) # TODO momentum is not supported at the moment\n\nfor epoch in range(1, args.epochs + 1):\n train(args, model, device, federated_train_loader, optimizer, epoch)\n test(args, model, device, test_loader)\n\nif (args.save_model):\n torch.save(model.state_dict(), \"mnist_cnn.pt\")",
"Train Epoch: 1 [0/60032 (0%)]\tLoss: 2.305818\nTrain Epoch: 1 [640/60032 (1%)]\tLoss: 2.281389\nTrain Epoch: 1 [1280/60032 (2%)]\tLoss: 2.249339\nTrain Epoch: 1 [1920/60032 (3%)]\tLoss: 2.207941\nTrain Epoch: 1 [2560/60032 (4%)]\tLoss: 2.154252\nTrain Epoch: 1 [3200/60032 (5%)]\tLoss: 2.107017\nTrain Epoch: 1 [3840/60032 (6%)]\tLoss: 2.000264\nTrain Epoch: 1 [4480/60032 (7%)]\tLoss: 1.924190\nTrain Epoch: 1 [5120/60032 (9%)]\tLoss: 1.751883\nTrain Epoch: 1 [5760/60032 (10%)]\tLoss: 1.495690\nTrain Epoch: 1 [6400/60032 (11%)]\tLoss: 1.385576\nTrain Epoch: 1 [7040/60032 (12%)]\tLoss: 1.188629\nTrain Epoch: 1 [7680/60032 (13%)]\tLoss: 1.049302\nTrain Epoch: 1 [8320/60032 (14%)]\tLoss: 0.858565\nTrain Epoch: 1 [8960/60032 (15%)]\tLoss: 0.622664\nTrain Epoch: 1 [9600/60032 (16%)]\tLoss: 0.820056\nTrain Epoch: 1 [10240/60032 (17%)]\tLoss: 0.529858\nTrain Epoch: 1 [10880/60032 (18%)]\tLoss: 0.546309\nTrain Epoch: 1 [11520/60032 (19%)]\tLoss: 0.677349\nTrain Epoch: 1 [12160/60032 (20%)]\tLoss: 0.372410\nTrain Epoch: 1 [12800/60032 (21%)]\tLoss: 0.512144\nTrain Epoch: 1 [13440/60032 (22%)]\tLoss: 0.506294\nTrain Epoch: 1 [14080/60032 (23%)]\tLoss: 0.308307\nTrain Epoch: 1 [14720/60032 (25%)]\tLoss: 0.615366\nTrain Epoch: 1 [15360/60032 (26%)]\tLoss: 0.390446\nTrain Epoch: 1 [16000/60032 (27%)]\tLoss: 0.305409\nTrain Epoch: 1 [16640/60032 (28%)]\tLoss: 0.376919\nTrain Epoch: 1 [17280/60032 (29%)]\tLoss: 0.372103\nTrain Epoch: 1 [17920/60032 (30%)]\tLoss: 0.406021\nTrain Epoch: 1 [18560/60032 (31%)]\tLoss: 0.533358\nTrain Epoch: 1 [19200/60032 (32%)]\tLoss: 0.312826\nTrain Epoch: 1 [19840/60032 (33%)]\tLoss: 0.273819\nTrain Epoch: 1 [20480/60032 (34%)]\tLoss: 0.303732\nTrain Epoch: 1 [21120/60032 (35%)]\tLoss: 0.394617\nTrain Epoch: 1 [21760/60032 (36%)]\tLoss: 0.269441\nTrain Epoch: 1 [22400/60032 (37%)]\tLoss: 0.321690\nTrain Epoch: 1 [23040/60032 (38%)]\tLoss: 0.299298\nTrain Epoch: 1 [23680/60032 (39%)]\tLoss: 0.262346\nTrain Epoch: 1 [24320/60032 (41%)]\tLoss: 0.212261\nTrain Epoch: 1 [24960/60032 (42%)]\tLoss: 0.360453\nTrain Epoch: 1 [25600/60032 (43%)]\tLoss: 0.434933\nTrain Epoch: 1 [26240/60032 (44%)]\tLoss: 0.271046\nTrain Epoch: 1 [26880/60032 (45%)]\tLoss: 0.176902\nTrain Epoch: 1 [27520/60032 (46%)]\tLoss: 0.257633\nTrain Epoch: 1 [28160/60032 (47%)]\tLoss: 0.180958\nTrain Epoch: 1 [28800/60032 (48%)]\tLoss: 0.386504\nTrain Epoch: 1 [29440/60032 (49%)]\tLoss: 0.332910\nTrain Epoch: 1 [30080/60032 (50%)]\tLoss: 0.669243\nTrain Epoch: 1 [30720/60032 (51%)]\tLoss: 0.144631\nTrain Epoch: 1 [31360/60032 (52%)]\tLoss: 0.232713\nTrain Epoch: 1 [32000/60032 (53%)]\tLoss: 0.305170\nTrain Epoch: 1 [32640/60032 (54%)]\tLoss: 0.218618\nTrain Epoch: 1 [33280/60032 (55%)]\tLoss: 0.225743\nTrain Epoch: 1 [33920/60032 (57%)]\tLoss: 0.237649\nTrain Epoch: 1 [34560/60032 (58%)]\tLoss: 0.151107\nTrain Epoch: 1 [35200/60032 (59%)]\tLoss: 0.290875\nTrain Epoch: 1 [35840/60032 (60%)]\tLoss: 0.221027\nTrain Epoch: 1 [36480/60032 (61%)]\tLoss: 0.318646\nTrain Epoch: 1 [37120/60032 (62%)]\tLoss: 0.274984\nTrain Epoch: 1 [37760/60032 (63%)]\tLoss: 0.320128\nTrain Epoch: 1 [38400/60032 (64%)]\tLoss: 0.206872\nTrain Epoch: 1 [39040/60032 (65%)]\tLoss: 0.194813\nTrain Epoch: 1 [39680/60032 (66%)]\tLoss: 0.426123\nTrain Epoch: 1 [40320/60032 (67%)]\tLoss: 0.178330\nTrain Epoch: 1 [40960/60032 (68%)]\tLoss: 0.210610\nTrain Epoch: 1 [41600/60032 (69%)]\tLoss: 0.203684\nTrain Epoch: 1 [42240/60032 (70%)]\tLoss: 0.183487\nTrain Epoch: 1 [42880/60032 (71%)]\tLoss: 0.109545\nTrain Epoch: 1 [43520/60032 (72%)]\tLoss: 0.214975\nTrain Epoch: 1 [44160/60032 (74%)]\tLoss: 0.349123\nTrain Epoch: 1 [44800/60032 (75%)]\tLoss: 0.228906\nTrain Epoch: 1 [45440/60032 (76%)]\tLoss: 0.359266\nTrain Epoch: 1 [46080/60032 (77%)]\tLoss: 0.123895\nTrain Epoch: 1 [46720/60032 (78%)]\tLoss: 0.212147\nTrain Epoch: 1 [47360/60032 (79%)]\tLoss: 0.241777\nTrain Epoch: 1 [48000/60032 (80%)]\tLoss: 0.259264\nTrain Epoch: 1 [48640/60032 (81%)]\tLoss: 0.263612\nTrain Epoch: 1 [49280/60032 (82%)]\tLoss: 0.281947\nTrain Epoch: 1 [49920/60032 (83%)]\tLoss: 0.173935\nTrain Epoch: 1 [50560/60032 (84%)]\tLoss: 0.149824\nTrain Epoch: 1 [51200/60032 (85%)]\tLoss: 0.110589\nTrain Epoch: 1 [51840/60032 (86%)]\tLoss: 0.211629\nTrain Epoch: 1 [52480/60032 (87%)]\tLoss: 0.304069\nTrain Epoch: 1 [53120/60032 (88%)]\tLoss: 0.195024\nTrain Epoch: 1 [53760/60032 (90%)]\tLoss: 0.376578\nTrain Epoch: 1 [54400/60032 (91%)]\tLoss: 0.175444\nTrain Epoch: 1 [55040/60032 (92%)]\tLoss: 0.126718\nTrain Epoch: 1 [55680/60032 (93%)]\tLoss: 0.164699\nTrain Epoch: 1 [56320/60032 (94%)]\tLoss: 0.150968\nTrain Epoch: 1 [56960/60032 (95%)]\tLoss: 0.089778\nTrain Epoch: 1 [57600/60032 (96%)]\tLoss: 0.175772\nTrain Epoch: 1 [58240/60032 (97%)]\tLoss: 0.251346\nTrain Epoch: 1 [58880/60032 (98%)]\tLoss: 0.166730\nTrain Epoch: 1 [59520/60032 (99%)]\tLoss: 0.173113\n\nTest set: Average loss: 0.1714, Accuracy: 9488/10000 (95%)\n\nTrain Epoch: 2 [0/60032 (0%)]\tLoss: 0.136982\nTrain Epoch: 2 [640/60032 (1%)]\tLoss: 0.131242\nTrain Epoch: 2 [1280/60032 (2%)]\tLoss: 0.145176\nTrain Epoch: 2 [1920/60032 (3%)]\tLoss: 0.119726\nTrain Epoch: 2 [2560/60032 (4%)]\tLoss: 0.200899\nTrain Epoch: 2 [3200/60032 (5%)]\tLoss: 0.284986\nTrain Epoch: 2 [3840/60032 (6%)]\tLoss: 0.121851\nTrain Epoch: 2 [4480/60032 (7%)]\tLoss: 0.144987\nTrain Epoch: 2 [5120/60032 (9%)]\tLoss: 0.142260\nTrain Epoch: 2 [5760/60032 (10%)]\tLoss: 0.112734\nTrain Epoch: 2 [6400/60032 (11%)]\tLoss: 0.189147\nTrain Epoch: 2 [7040/60032 (12%)]\tLoss: 0.172275\nTrain Epoch: 2 [7680/60032 (13%)]\tLoss: 0.156510\nTrain Epoch: 2 [8320/60032 (14%)]\tLoss: 0.071181\nTrain Epoch: 2 [8960/60032 (15%)]\tLoss: 0.146095\nTrain Epoch: 2 [9600/60032 (16%)]\tLoss: 0.044013\nTrain Epoch: 2 [10240/60032 (17%)]\tLoss: 0.172235\nTrain Epoch: 2 [10880/60032 (18%)]\tLoss: 0.295077\nTrain Epoch: 2 [11520/60032 (19%)]\tLoss: 0.193669\nTrain Epoch: 2 [12160/60032 (20%)]\tLoss: 0.150169\nTrain Epoch: 2 [12800/60032 (21%)]\tLoss: 0.081198\nTrain Epoch: 2 [13440/60032 (22%)]\tLoss: 0.121817\nTrain Epoch: 2 [14080/60032 (23%)]\tLoss: 0.277597\nTrain Epoch: 2 [14720/60032 (25%)]\tLoss: 0.126618\nTrain Epoch: 2 [15360/60032 (26%)]\tLoss: 0.171404\nTrain Epoch: 2 [16000/60032 (27%)]\tLoss: 0.196170\nTrain Epoch: 2 [16640/60032 (28%)]\tLoss: 0.162388\nTrain Epoch: 2 [17280/60032 (29%)]\tLoss: 0.163096\nTrain Epoch: 2 [17920/60032 (30%)]\tLoss: 0.095635\nTrain Epoch: 2 [18560/60032 (31%)]\tLoss: 0.150551\nTrain Epoch: 2 [19200/60032 (32%)]\tLoss: 0.055939\nTrain Epoch: 2 [19840/60032 (33%)]\tLoss: 0.087581\nTrain Epoch: 2 [20480/60032 (34%)]\tLoss: 0.277243\nTrain Epoch: 2 [21120/60032 (35%)]\tLoss: 0.162521\nTrain Epoch: 2 [21760/60032 (36%)]\tLoss: 0.268383\nTrain Epoch: 2 [22400/60032 (37%)]\tLoss: 0.133994\nTrain Epoch: 2 [23040/60032 (38%)]\tLoss: 0.364789\nTrain Epoch: 2 [23680/60032 (39%)]\tLoss: 0.083243\nTrain Epoch: 2 [24320/60032 (41%)]\tLoss: 0.151107\nTrain Epoch: 2 [24960/60032 (42%)]\tLoss: 0.268702\nTrain Epoch: 2 [25600/60032 (43%)]\tLoss: 0.031650\nTrain Epoch: 2 [26240/60032 (44%)]\tLoss: 0.096981\nTrain Epoch: 2 [26880/60032 (45%)]\tLoss: 0.112181\nTrain Epoch: 2 [27520/60032 (46%)]\tLoss: 0.103714\nTrain Epoch: 2 [28160/60032 (47%)]\tLoss: 0.133328\nTrain Epoch: 2 [28800/60032 (48%)]\tLoss: 0.138665\nTrain Epoch: 2 [29440/60032 (49%)]\tLoss: 0.041654\nTrain Epoch: 2 [30080/60032 (50%)]\tLoss: 0.107013\nTrain Epoch: 2 [30720/60032 (51%)]\tLoss: 0.137944\nTrain Epoch: 2 [31360/60032 (52%)]\tLoss: 0.078677\nTrain Epoch: 2 [32000/60032 (53%)]\tLoss: 0.116954\nTrain Epoch: 2 [32640/60032 (54%)]\tLoss: 0.176856\nTrain Epoch: 2 [33280/60032 (55%)]\tLoss: 0.030993\nTrain Epoch: 2 [33920/60032 (57%)]\tLoss: 0.129508\nTrain Epoch: 2 [34560/60032 (58%)]\tLoss: 0.183357\nTrain Epoch: 2 [35200/60032 (59%)]\tLoss: 0.111668\nTrain Epoch: 2 [35840/60032 (60%)]\tLoss: 0.024881\nTrain Epoch: 2 [36480/60032 (61%)]\tLoss: 0.063785\nTrain Epoch: 2 [37120/60032 (62%)]\tLoss: 0.075077\nTrain Epoch: 2 [37760/60032 (63%)]\tLoss: 0.144636\nTrain Epoch: 2 [38400/60032 (64%)]\tLoss: 0.050517\nTrain Epoch: 2 [39040/60032 (65%)]\tLoss: 0.121883\nTrain Epoch: 2 [39680/60032 (66%)]\tLoss: 0.166800\nTrain Epoch: 2 [40320/60032 (67%)]\tLoss: 0.135950\nTrain Epoch: 2 [40960/60032 (68%)]\tLoss: 0.065102\nTrain Epoch: 2 [41600/60032 (69%)]\tLoss: 0.055656\nTrain Epoch: 2 [42240/60032 (70%)]\tLoss: 0.119884\nTrain Epoch: 2 [42880/60032 (71%)]\tLoss: 0.129070\nTrain Epoch: 2 [43520/60032 (72%)]\tLoss: 0.080352\nTrain Epoch: 2 [44160/60032 (74%)]\tLoss: 0.053117\nTrain Epoch: 2 [44800/60032 (75%)]\tLoss: 0.129221\nTrain Epoch: 2 [45440/60032 (76%)]\tLoss: 0.026495\nTrain Epoch: 2 [46080/60032 (77%)]\tLoss: 0.205546\nTrain Epoch: 2 [46720/60032 (78%)]\tLoss: 0.293277\nTrain Epoch: 2 [47360/60032 (79%)]\tLoss: 0.132079\nTrain Epoch: 2 [48000/60032 (80%)]\tLoss: 0.083449\nTrain Epoch: 2 [48640/60032 (81%)]\tLoss: 0.093549\nTrain Epoch: 2 [49280/60032 (82%)]\tLoss: 0.032769\nTrain Epoch: 2 [49920/60032 (83%)]\tLoss: 0.120211\nTrain Epoch: 2 [50560/60032 (84%)]\tLoss: 0.091749\nTrain Epoch: 2 [51200/60032 (85%)]\tLoss: 0.159344\nTrain Epoch: 2 [51840/60032 (86%)]\tLoss: 0.115711\nTrain Epoch: 2 [52480/60032 (87%)]\tLoss: 0.036213\nTrain Epoch: 2 [53120/60032 (88%)]\tLoss: 0.084314\nTrain Epoch: 2 [53760/60032 (90%)]\tLoss: 0.085498\nTrain Epoch: 2 [54400/60032 (91%)]\tLoss: 0.051328\nTrain Epoch: 2 [55040/60032 (92%)]\tLoss: 0.087684\nTrain Epoch: 2 [55680/60032 (93%)]\tLoss: 0.092397\nTrain Epoch: 2 [56320/60032 (94%)]\tLoss: 0.116120\nTrain Epoch: 2 [56960/60032 (95%)]\tLoss: 0.025299\nTrain Epoch: 2 [57600/60032 (96%)]\tLoss: 0.050432\nTrain Epoch: 2 [58240/60032 (97%)]\tLoss: 0.073126\nTrain Epoch: 2 [58880/60032 (98%)]\tLoss: 0.054885\nTrain Epoch: 2 [59520/60032 (99%)]\tLoss: 0.065184\n\nTest set: Average loss: 0.0881, Accuracy: 9739/10000 (97%)\n\nTrain Epoch: 3 [0/60032 (0%)]\tLoss: 0.196212\nTrain Epoch: 3 [640/60032 (1%)]\tLoss: 0.027683\nTrain Epoch: 3 [1280/60032 (2%)]\tLoss: 0.049790\nTrain Epoch: 3 [1920/60032 (3%)]\tLoss: 0.038210\nTrain Epoch: 3 [2560/60032 (4%)]\tLoss: 0.062873\nTrain Epoch: 3 [3200/60032 (5%)]\tLoss: 0.096338\nTrain Epoch: 3 [3840/60032 (6%)]\tLoss: 0.038556\nTrain Epoch: 3 [4480/60032 (7%)]\tLoss: 0.091497\nTrain Epoch: 3 [5120/60032 (9%)]\tLoss: 0.186206\nTrain Epoch: 3 [5760/60032 (10%)]\tLoss: 0.097394\nTrain Epoch: 3 [6400/60032 (11%)]\tLoss: 0.092199\nTrain Epoch: 3 [7040/60032 (12%)]\tLoss: 0.158342\nTrain Epoch: 3 [7680/60032 (13%)]\tLoss: 0.149393\nTrain Epoch: 3 [8320/60032 (14%)]\tLoss: 0.108441\nTrain Epoch: 3 [8960/60032 (15%)]\tLoss: 0.076324\nTrain Epoch: 3 [9600/60032 (16%)]\tLoss: 0.082731\nTrain Epoch: 3 [10240/60032 (17%)]\tLoss: 0.074252\nTrain Epoch: 3 [10880/60032 (18%)]\tLoss: 0.047882\nTrain Epoch: 3 [11520/60032 (19%)]\tLoss: 0.121217\nTrain Epoch: 3 [12160/60032 (20%)]\tLoss: 0.071879\nTrain Epoch: 3 [12800/60032 (21%)]\tLoss: 0.086744\nTrain Epoch: 3 [13440/60032 (22%)]\tLoss: 0.036404\nTrain Epoch: 3 [14080/60032 (23%)]\tLoss: 0.118092\nTrain Epoch: 3 [14720/60032 (25%)]\tLoss: 0.128600\nTrain Epoch: 3 [15360/60032 (26%)]\tLoss: 0.128535\nTrain Epoch: 3 [16000/60032 (27%)]\tLoss: 0.122295\nTrain Epoch: 3 [16640/60032 (28%)]\tLoss: 0.111799\nTrain Epoch: 3 [17280/60032 (29%)]\tLoss: 0.140395\nTrain Epoch: 3 [17920/60032 (30%)]\tLoss: 0.051356\nTrain Epoch: 3 [18560/60032 (31%)]\tLoss: 0.062547\nTrain Epoch: 3 [19200/60032 (32%)]\tLoss: 0.082309\nTrain Epoch: 3 [19840/60032 (33%)]\tLoss: 0.056778\nTrain Epoch: 3 [20480/60032 (34%)]\tLoss: 0.045520\nTrain Epoch: 3 [21120/60032 (35%)]\tLoss: 0.051373\nTrain Epoch: 3 [21760/60032 (36%)]\tLoss: 0.090650\nTrain Epoch: 3 [22400/60032 (37%)]\tLoss: 0.036855\nTrain Epoch: 3 [23040/60032 (38%)]\tLoss: 0.022794\nTrain Epoch: 3 [23680/60032 (39%)]\tLoss: 0.137001\nTrain Epoch: 3 [24320/60032 (41%)]\tLoss: 0.072350\nTrain Epoch: 3 [24960/60032 (42%)]\tLoss: 0.092482\nTrain Epoch: 3 [25600/60032 (43%)]\tLoss: 0.021641\nTrain Epoch: 3 [26240/60032 (44%)]\tLoss: 0.039240\nTrain Epoch: 3 [26880/60032 (45%)]\tLoss: 0.077412\nTrain Epoch: 3 [27520/60032 (46%)]\tLoss: 0.035526\nTrain Epoch: 3 [28160/60032 (47%)]\tLoss: 0.049093\nTrain Epoch: 3 [28800/60032 (48%)]\tLoss: 0.019213\nTrain Epoch: 3 [29440/60032 (49%)]\tLoss: 0.115915\nTrain Epoch: 3 [30080/60032 (50%)]\tLoss: 0.022377\nTrain Epoch: 3 [30720/60032 (51%)]\tLoss: 0.049881\nTrain Epoch: 3 [31360/60032 (52%)]\tLoss: 0.215625\nTrain Epoch: 3 [32000/60032 (53%)]\tLoss: 0.114067\nTrain Epoch: 3 [32640/60032 (54%)]\tLoss: 0.075174\nTrain Epoch: 3 [33280/60032 (55%)]\tLoss: 0.025694\nTrain Epoch: 3 [33920/60032 (57%)]\tLoss: 0.096331\nTrain Epoch: 3 [34560/60032 (58%)]\tLoss: 0.037031\nTrain Epoch: 3 [35200/60032 (59%)]\tLoss: 0.123958\nTrain Epoch: 3 [35840/60032 (60%)]\tLoss: 0.111565\nTrain Epoch: 3 [36480/60032 (61%)]\tLoss: 0.040589\nTrain Epoch: 3 [37120/60032 (62%)]\tLoss: 0.041095\nTrain Epoch: 3 [37760/60032 (63%)]\tLoss: 0.081622\nTrain Epoch: 3 [38400/60032 (64%)]\tLoss: 0.057721\nTrain Epoch: 3 [39040/60032 (65%)]\tLoss: 0.046868\nTrain Epoch: 3 [39680/60032 (66%)]\tLoss: 0.094902\nTrain Epoch: 3 [40320/60032 (67%)]\tLoss: 0.080666\nTrain Epoch: 3 [40960/60032 (68%)]\tLoss: 0.117332\nTrain Epoch: 3 [41600/60032 (69%)]\tLoss: 0.112522\nTrain Epoch: 3 [42240/60032 (70%)]\tLoss: 0.107774\nTrain Epoch: 3 [42880/60032 (71%)]\tLoss: 0.060894\nTrain Epoch: 3 [43520/60032 (72%)]\tLoss: 0.058937\nTrain Epoch: 3 [44160/60032 (74%)]\tLoss: 0.142228\nTrain Epoch: 3 [44800/60032 (75%)]\tLoss: 0.171978\nTrain Epoch: 3 [45440/60032 (76%)]\tLoss: 0.093110\nTrain Epoch: 3 [46080/60032 (77%)]\tLoss: 0.049810\nTrain Epoch: 3 [46720/60032 (78%)]\tLoss: 0.055311\nTrain Epoch: 3 [47360/60032 (79%)]\tLoss: 0.053296\nTrain Epoch: 3 [48000/60032 (80%)]\tLoss: 0.080236\nTrain Epoch: 3 [48640/60032 (81%)]\tLoss: 0.033535\nTrain Epoch: 3 [49280/60032 (82%)]\tLoss: 0.021829\nTrain Epoch: 3 [49920/60032 (83%)]\tLoss: 0.057428\nTrain Epoch: 3 [50560/60032 (84%)]\tLoss: 0.040245\nTrain Epoch: 3 [51200/60032 (85%)]\tLoss: 0.143099\nTrain Epoch: 3 [51840/60032 (86%)]\tLoss: 0.215343\nTrain Epoch: 3 [52480/60032 (87%)]\tLoss: 0.157364\nTrain Epoch: 3 [53120/60032 (88%)]\tLoss: 0.048516\nTrain Epoch: 3 [53760/60032 (90%)]\tLoss: 0.094795\nTrain Epoch: 3 [54400/60032 (91%)]\tLoss: 0.064886\nTrain Epoch: 3 [55040/60032 (92%)]\tLoss: 0.083711\nTrain Epoch: 3 [55680/60032 (93%)]\tLoss: 0.091166\nTrain Epoch: 3 [56320/60032 (94%)]\tLoss: 0.133744\nTrain Epoch: 3 [56960/60032 (95%)]\tLoss: 0.160619\nTrain Epoch: 3 [57600/60032 (96%)]\tLoss: 0.027220\nTrain Epoch: 3 [58240/60032 (97%)]\tLoss: 0.017269\nTrain Epoch: 3 [58880/60032 (98%)]\tLoss: 0.061630\nTrain Epoch: 3 [59520/60032 (99%)]\tLoss: 0.037820\n\nTest set: Average loss: 0.0815, Accuracy: 9750/10000 (98%)\n\nTrain Epoch: 4 [0/60032 (0%)]\tLoss: 0.034388\nTrain Epoch: 4 [640/60032 (1%)]\tLoss: 0.008091\nTrain Epoch: 4 [1280/60032 (2%)]\tLoss: 0.039770\nTrain Epoch: 4 [1920/60032 (3%)]\tLoss: 0.132797\nTrain Epoch: 4 [2560/60032 (4%)]\tLoss: 0.046503\nTrain Epoch: 4 [3200/60032 (5%)]\tLoss: 0.079162\nTrain Epoch: 4 [3840/60032 (6%)]\tLoss: 0.052506\nTrain Epoch: 4 [4480/60032 (7%)]\tLoss: 0.112856\nTrain Epoch: 4 [5120/60032 (9%)]\tLoss: 0.047054\nTrain Epoch: 4 [5760/60032 (10%)]\tLoss: 0.077705\nTrain Epoch: 4 [6400/60032 (11%)]\tLoss: 0.035396\nTrain Epoch: 4 [7040/60032 (12%)]\tLoss: 0.058383\nTrain Epoch: 4 [7680/60032 (13%)]\tLoss: 0.032164\nTrain Epoch: 4 [8320/60032 (14%)]\tLoss: 0.058356\nTrain Epoch: 4 [8960/60032 (15%)]\tLoss: 0.091825\nTrain Epoch: 4 [9600/60032 (16%)]\tLoss: 0.088521\nTrain Epoch: 4 [10240/60032 (17%)]\tLoss: 0.019547\nTrain Epoch: 4 [10880/60032 (18%)]\tLoss: 0.058892\nTrain Epoch: 4 [11520/60032 (19%)]\tLoss: 0.059722\nTrain Epoch: 4 [12160/60032 (20%)]\tLoss: 0.175013\nTrain Epoch: 4 [12800/60032 (21%)]\tLoss: 0.085420\nTrain Epoch: 4 [13440/60032 (22%)]\tLoss: 0.085398\nTrain Epoch: 4 [14080/60032 (23%)]\tLoss: 0.066407\nTrain Epoch: 4 [14720/60032 (25%)]\tLoss: 0.086988\nTrain Epoch: 4 [15360/60032 (26%)]\tLoss: 0.094427\nTrain Epoch: 4 [16000/60032 (27%)]\tLoss: 0.030238\nTrain Epoch: 4 [16640/60032 (28%)]\tLoss: 0.045739\nTrain Epoch: 4 [17280/60032 (29%)]\tLoss: 0.022443\nTrain Epoch: 4 [17920/60032 (30%)]\tLoss: 0.022746\nTrain Epoch: 4 [18560/60032 (31%)]\tLoss: 0.073060\nTrain Epoch: 4 [19200/60032 (32%)]\tLoss: 0.065248\nTrain Epoch: 4 [19840/60032 (33%)]\tLoss: 0.027066\nTrain Epoch: 4 [20480/60032 (34%)]\tLoss: 0.014112\nTrain Epoch: 4 [21120/60032 (35%)]\tLoss: 0.031931\nTrain Epoch: 4 [21760/60032 (36%)]\tLoss: 0.047272\nTrain Epoch: 4 [22400/60032 (37%)]\tLoss: 0.116918\nTrain Epoch: 4 [23040/60032 (38%)]\tLoss: 0.159644\nTrain Epoch: 4 [23680/60032 (39%)]\tLoss: 0.059512\nTrain Epoch: 4 [24320/60032 (41%)]\tLoss: 0.077351\nTrain Epoch: 4 [24960/60032 (42%)]\tLoss: 0.238766\nTrain Epoch: 4 [25600/60032 (43%)]\tLoss: 0.098239\nTrain Epoch: 4 [26240/60032 (44%)]\tLoss: 0.041562\nTrain Epoch: 4 [26880/60032 (45%)]\tLoss: 0.023171\nTrain Epoch: 4 [27520/60032 (46%)]\tLoss: 0.042277\nTrain Epoch: 4 [28160/60032 (47%)]\tLoss: 0.037537\nTrain Epoch: 4 [28800/60032 (48%)]\tLoss: 0.054304\nTrain Epoch: 4 [29440/60032 (49%)]\tLoss: 0.037641\nTrain Epoch: 4 [30080/60032 (50%)]\tLoss: 0.045417\nTrain Epoch: 4 [30720/60032 (51%)]\tLoss: 0.032511\nTrain Epoch: 4 [31360/60032 (52%)]\tLoss: 0.081783\nTrain Epoch: 4 [32000/60032 (53%)]\tLoss: 0.117212\nTrain Epoch: 4 [32640/60032 (54%)]\tLoss: 0.044074\nTrain Epoch: 4 [33280/60032 (55%)]\tLoss: 0.084898\nTrain Epoch: 4 [33920/60032 (57%)]\tLoss: 0.128583\nTrain Epoch: 4 [34560/60032 (58%)]\tLoss: 0.029901\nTrain Epoch: 4 [35200/60032 (59%)]\tLoss: 0.035927\nTrain Epoch: 4 [35840/60032 (60%)]\tLoss: 0.097918\nTrain Epoch: 4 [36480/60032 (61%)]\tLoss: 0.025856\nTrain Epoch: 4 [37120/60032 (62%)]\tLoss: 0.070445\nTrain Epoch: 4 [37760/60032 (63%)]\tLoss: 0.018995\nTrain Epoch: 4 [38400/60032 (64%)]\tLoss: 0.077177\nTrain Epoch: 4 [39040/60032 (65%)]\tLoss: 0.044427\nTrain Epoch: 4 [39680/60032 (66%)]\tLoss: 0.110785\nTrain Epoch: 4 [40320/60032 (67%)]\tLoss: 0.118569\nTrain Epoch: 4 [40960/60032 (68%)]\tLoss: 0.148673\nTrain Epoch: 4 [41600/60032 (69%)]\tLoss: 0.052738\nTrain Epoch: 4 [42240/60032 (70%)]\tLoss: 0.016911\nTrain Epoch: 4 [42880/60032 (71%)]\tLoss: 0.045542\nTrain Epoch: 4 [43520/60032 (72%)]\tLoss: 0.044842\nTrain Epoch: 4 [44160/60032 (74%)]\tLoss: 0.045369\nTrain Epoch: 4 [44800/60032 (75%)]\tLoss: 0.010688\nTrain Epoch: 4 [45440/60032 (76%)]\tLoss: 0.015408\nTrain Epoch: 4 [46080/60032 (77%)]\tLoss: 0.017765\nTrain Epoch: 4 [46720/60032 (78%)]\tLoss: 0.074281\nTrain Epoch: 4 [47360/60032 (79%)]\tLoss: 0.081282\nTrain Epoch: 4 [48000/60032 (80%)]\tLoss: 0.026275\nTrain Epoch: 4 [48640/60032 (81%)]\tLoss: 0.019319\nTrain Epoch: 4 [49280/60032 (82%)]\tLoss: 0.166881\nTrain Epoch: 4 [49920/60032 (83%)]\tLoss: 0.093638\nTrain Epoch: 4 [50560/60032 (84%)]\tLoss: 0.044577\nTrain Epoch: 4 [51200/60032 (85%)]\tLoss: 0.072052\nTrain Epoch: 4 [51840/60032 (86%)]\tLoss: 0.040266\nTrain Epoch: 4 [52480/60032 (87%)]\tLoss: 0.156111\nTrain Epoch: 4 [53120/60032 (88%)]\tLoss: 0.089448\nTrain Epoch: 4 [53760/60032 (90%)]\tLoss: 0.142163\nTrain Epoch: 4 [54400/60032 (91%)]\tLoss: 0.045538\nTrain Epoch: 4 [55040/60032 (92%)]\tLoss: 0.068240\nTrain Epoch: 4 [55680/60032 (93%)]\tLoss: 0.051040\nTrain Epoch: 4 [56320/60032 (94%)]\tLoss: 0.113153\nTrain Epoch: 4 [56960/60032 (95%)]\tLoss: 0.093316\nTrain Epoch: 4 [57600/60032 (96%)]\tLoss: 0.063761\nTrain Epoch: 4 [58240/60032 (97%)]\tLoss: 0.107417\nTrain Epoch: 4 [58880/60032 (98%)]\tLoss: 0.045168\nTrain Epoch: 4 [59520/60032 (99%)]\tLoss: 0.016856\n\nTest set: Average loss: 0.0568, Accuracy: 9814/10000 (98%)\n\nTrain Epoch: 5 [0/60032 (0%)]\tLoss: 0.020153\nTrain Epoch: 5 [640/60032 (1%)]\tLoss: 0.027963\nTrain Epoch: 5 [1280/60032 (2%)]\tLoss: 0.177104\nTrain Epoch: 5 [1920/60032 (3%)]\tLoss: 0.026191\nTrain Epoch: 5 [2560/60032 (4%)]\tLoss: 0.023304\nTrain Epoch: 5 [3200/60032 (5%)]\tLoss: 0.018702\nTrain Epoch: 5 [3840/60032 (6%)]\tLoss: 0.026195\nTrain Epoch: 5 [4480/60032 (7%)]\tLoss: 0.046623\nTrain Epoch: 5 [5120/60032 (9%)]\tLoss: 0.114872\nTrain Epoch: 5 [5760/60032 (10%)]\tLoss: 0.108763\nTrain Epoch: 5 [6400/60032 (11%)]\tLoss: 0.016490\nTrain Epoch: 5 [7040/60032 (12%)]\tLoss: 0.048616\nTrain Epoch: 5 [7680/60032 (13%)]\tLoss: 0.010754\nTrain Epoch: 5 [8320/60032 (14%)]\tLoss: 0.106226\nTrain Epoch: 5 [8960/60032 (15%)]\tLoss: 0.087185\nTrain Epoch: 5 [9600/60032 (16%)]\tLoss: 0.085100\nTrain Epoch: 5 [10240/60032 (17%)]\tLoss: 0.112376\nTrain Epoch: 5 [10880/60032 (18%)]\tLoss: 0.087230\nTrain Epoch: 5 [11520/60032 (19%)]\tLoss: 0.060482\nTrain Epoch: 5 [12160/60032 (20%)]\tLoss: 0.027084\nTrain Epoch: 5 [12800/60032 (21%)]\tLoss: 0.022883\nTrain Epoch: 5 [13440/60032 (22%)]\tLoss: 0.031407\nTrain Epoch: 5 [14080/60032 (23%)]\tLoss: 0.220639\nTrain Epoch: 5 [14720/60032 (25%)]\tLoss: 0.049089\nTrain Epoch: 5 [15360/60032 (26%)]\tLoss: 0.084462\nTrain Epoch: 5 [16000/60032 (27%)]\tLoss: 0.015038\nTrain Epoch: 5 [16640/60032 (28%)]\tLoss: 0.037550\nTrain Epoch: 5 [17280/60032 (29%)]\tLoss: 0.028653\nTrain Epoch: 5 [17920/60032 (30%)]\tLoss: 0.028285\nTrain Epoch: 5 [18560/60032 (31%)]\tLoss: 0.150356\nTrain Epoch: 5 [19200/60032 (32%)]\tLoss: 0.036568\nTrain Epoch: 5 [19840/60032 (33%)]\tLoss: 0.018198\nTrain Epoch: 5 [20480/60032 (34%)]\tLoss: 0.218620\nTrain Epoch: 5 [21120/60032 (35%)]\tLoss: 0.031804\nTrain Epoch: 5 [21760/60032 (36%)]\tLoss: 0.025349\nTrain Epoch: 5 [22400/60032 (37%)]\tLoss: 0.090573\nTrain Epoch: 5 [23040/60032 (38%)]\tLoss: 0.065329\nTrain Epoch: 5 [23680/60032 (39%)]\tLoss: 0.032602\nTrain Epoch: 5 [24320/60032 (41%)]\tLoss: 0.059301\nTrain Epoch: 5 [24960/60032 (42%)]\tLoss: 0.082533\nTrain Epoch: 5 [25600/60032 (43%)]\tLoss: 0.177886\nTrain Epoch: 5 [26240/60032 (44%)]\tLoss: 0.041511\nTrain Epoch: 5 [26880/60032 (45%)]\tLoss: 0.051170\nTrain Epoch: 5 [27520/60032 (46%)]\tLoss: 0.059319\nTrain Epoch: 5 [28160/60032 (47%)]\tLoss: 0.175344\nTrain Epoch: 5 [28800/60032 (48%)]\tLoss: 0.034557\nTrain Epoch: 5 [29440/60032 (49%)]\tLoss: 0.028588\nTrain Epoch: 5 [30080/60032 (50%)]\tLoss: 0.119612\nTrain Epoch: 5 [30720/60032 (51%)]\tLoss: 0.090046\nTrain Epoch: 5 [31360/60032 (52%)]\tLoss: 0.046450\nTrain Epoch: 5 [32000/60032 (53%)]\tLoss: 0.073137\nTrain Epoch: 5 [32640/60032 (54%)]\tLoss: 0.083834\nTrain Epoch: 5 [33280/60032 (55%)]\tLoss: 0.038151\nTrain Epoch: 5 [33920/60032 (57%)]\tLoss: 0.110557\nTrain Epoch: 5 [34560/60032 (58%)]\tLoss: 0.045256\nTrain Epoch: 5 [35200/60032 (59%)]\tLoss: 0.194613\nTrain Epoch: 5 [35840/60032 (60%)]\tLoss: 0.009550\nTrain Epoch: 5 [36480/60032 (61%)]\tLoss: 0.057306\nTrain Epoch: 5 [37120/60032 (62%)]\tLoss: 0.026960\nTrain Epoch: 5 [37760/60032 (63%)]\tLoss: 0.116204\nTrain Epoch: 5 [38400/60032 (64%)]\tLoss: 0.033455\nTrain Epoch: 5 [39040/60032 (65%)]\tLoss: 0.024697\nTrain Epoch: 5 [39680/60032 (66%)]\tLoss: 0.090211\nTrain Epoch: 5 [40320/60032 (67%)]\tLoss: 0.071277\nTrain Epoch: 5 [40960/60032 (68%)]\tLoss: 0.009953\nTrain Epoch: 5 [41600/60032 (69%)]\tLoss: 0.039897\nTrain Epoch: 5 [42240/60032 (70%)]\tLoss: 0.052740\nTrain Epoch: 5 [42880/60032 (71%)]\tLoss: 0.133722\nTrain Epoch: 5 [43520/60032 (72%)]\tLoss: 0.045977\nTrain Epoch: 5 [44160/60032 (74%)]\tLoss: 0.051510\nTrain Epoch: 5 [44800/60032 (75%)]\tLoss: 0.045732\nTrain Epoch: 5 [45440/60032 (76%)]\tLoss: 0.016773\nTrain Epoch: 5 [46080/60032 (77%)]\tLoss: 0.062058\nTrain Epoch: 5 [46720/60032 (78%)]\tLoss: 0.034382\nTrain Epoch: 5 [47360/60032 (79%)]\tLoss: 0.035736\nTrain Epoch: 5 [48000/60032 (80%)]\tLoss: 0.115613\nTrain Epoch: 5 [48640/60032 (81%)]\tLoss: 0.112691\nTrain Epoch: 5 [49280/60032 (82%)]\tLoss: 0.014247\nTrain Epoch: 5 [49920/60032 (83%)]\tLoss: 0.024166\nTrain Epoch: 5 [50560/60032 (84%)]\tLoss: 0.185810\nTrain Epoch: 5 [51200/60032 (85%)]\tLoss: 0.059340\nTrain Epoch: 5 [51840/60032 (86%)]\tLoss: 0.058731\nTrain Epoch: 5 [52480/60032 (87%)]\tLoss: 0.055707\nTrain Epoch: 5 [53120/60032 (88%)]\tLoss: 0.068775\nTrain Epoch: 5 [53760/60032 (90%)]\tLoss: 0.018685\nTrain Epoch: 5 [54400/60032 (91%)]\tLoss: 0.013157\nTrain Epoch: 5 [55040/60032 (92%)]\tLoss: 0.030340\nTrain Epoch: 5 [55680/60032 (93%)]\tLoss: 0.054379\nTrain Epoch: 5 [56320/60032 (94%)]\tLoss: 0.108384\nTrain Epoch: 5 [56960/60032 (95%)]\tLoss: 0.052427\nTrain Epoch: 5 [57600/60032 (96%)]\tLoss: 0.020344\nTrain Epoch: 5 [58240/60032 (97%)]\tLoss: 0.009642\nTrain Epoch: 5 [58880/60032 (98%)]\tLoss: 0.028303\nTrain Epoch: 5 [59520/60032 (99%)]\tLoss: 0.087671\n\nTest set: Average loss: 0.0533, Accuracy: 9836/10000 (98%)\n\nTrain Epoch: 6 [0/60032 (0%)]\tLoss: 0.025014\nTrain Epoch: 6 [640/60032 (1%)]\tLoss: 0.028698\nTrain Epoch: 6 [1280/60032 (2%)]\tLoss: 0.064421\nTrain Epoch: 6 [1920/60032 (3%)]\tLoss: 0.043281\nTrain Epoch: 6 [2560/60032 (4%)]\tLoss: 0.016751\nTrain Epoch: 6 [3200/60032 (5%)]\tLoss: 0.102817\nTrain Epoch: 6 [3840/60032 (6%)]\tLoss: 0.025380\nTrain Epoch: 6 [4480/60032 (7%)]\tLoss: 0.062252\nTrain Epoch: 6 [5120/60032 (9%)]\tLoss: 0.029176\nTrain Epoch: 6 [5760/60032 (10%)]\tLoss: 0.049006\nTrain Epoch: 6 [6400/60032 (11%)]\tLoss: 0.020854\nTrain Epoch: 6 [7040/60032 (12%)]\tLoss: 0.053753\nTrain Epoch: 6 [7680/60032 (13%)]\tLoss: 0.073204\nTrain Epoch: 6 [8320/60032 (14%)]\tLoss: 0.092593\nTrain Epoch: 6 [8960/60032 (15%)]\tLoss: 0.045797\nTrain Epoch: 6 [9600/60032 (16%)]\tLoss: 0.067838\nTrain Epoch: 6 [10240/60032 (17%)]\tLoss: 0.006947\nTrain Epoch: 6 [10880/60032 (18%)]\tLoss: 0.012980\nTrain Epoch: 6 [11520/60032 (19%)]\tLoss: 0.029988\nTrain Epoch: 6 [12160/60032 (20%)]\tLoss: 0.016550\nTrain Epoch: 6 [12800/60032 (21%)]\tLoss: 0.030582\nTrain Epoch: 6 [13440/60032 (22%)]\tLoss: 0.069579\nTrain Epoch: 6 [14080/60032 (23%)]\tLoss: 0.029539\nTrain Epoch: 6 [14720/60032 (25%)]\tLoss: 0.044889\nTrain Epoch: 6 [15360/60032 (26%)]\tLoss: 0.178108\nTrain Epoch: 6 [16000/60032 (27%)]\tLoss: 0.027673\nTrain Epoch: 6 [16640/60032 (28%)]\tLoss: 0.047966\nTrain Epoch: 6 [17280/60032 (29%)]\tLoss: 0.038432\nTrain Epoch: 6 [17920/60032 (30%)]\tLoss: 0.102820\nTrain Epoch: 6 [18560/60032 (31%)]\tLoss: 0.150295\nTrain Epoch: 6 [19200/60032 (32%)]\tLoss: 0.044012\nTrain Epoch: 6 [19840/60032 (33%)]\tLoss: 0.011486\nTrain Epoch: 6 [20480/60032 (34%)]\tLoss: 0.006812\nTrain Epoch: 6 [21120/60032 (35%)]\tLoss: 0.016451\nTrain Epoch: 6 [21760/60032 (36%)]\tLoss: 0.014458\nTrain Epoch: 6 [22400/60032 (37%)]\tLoss: 0.053117\nTrain Epoch: 6 [23040/60032 (38%)]\tLoss: 0.040178\nTrain Epoch: 6 [23680/60032 (39%)]\tLoss: 0.023645\nTrain Epoch: 6 [24320/60032 (41%)]\tLoss: 0.043930\nTrain Epoch: 6 [24960/60032 (42%)]\tLoss: 0.019009\nTrain Epoch: 6 [25600/60032 (43%)]\tLoss: 0.052494\nTrain Epoch: 6 [26240/60032 (44%)]\tLoss: 0.010974\nTrain Epoch: 6 [26880/60032 (45%)]\tLoss: 0.141218\nTrain Epoch: 6 [27520/60032 (46%)]\tLoss: 0.042667\nTrain Epoch: 6 [28160/60032 (47%)]\tLoss: 0.081838\nTrain Epoch: 6 [28800/60032 (48%)]\tLoss: 0.019254\nTrain Epoch: 6 [29440/60032 (49%)]\tLoss: 0.050912\nTrain Epoch: 6 [30080/60032 (50%)]\tLoss: 0.020576\nTrain Epoch: 6 [30720/60032 (51%)]\tLoss: 0.043702\nTrain Epoch: 6 [31360/60032 (52%)]\tLoss: 0.037019\nTrain Epoch: 6 [32000/60032 (53%)]\tLoss: 0.054425\nTrain Epoch: 6 [32640/60032 (54%)]\tLoss: 0.061798\nTrain Epoch: 6 [33280/60032 (55%)]\tLoss: 0.036123\nTrain Epoch: 6 [33920/60032 (57%)]\tLoss: 0.044808\nTrain Epoch: 6 [34560/60032 (58%)]\tLoss: 0.053852\nTrain Epoch: 6 [35200/60032 (59%)]\tLoss: 0.005029\nTrain Epoch: 6 [35840/60032 (60%)]\tLoss: 0.007349\nTrain Epoch: 6 [36480/60032 (61%)]\tLoss: 0.091853\nTrain Epoch: 6 [37120/60032 (62%)]\tLoss: 0.016010\nTrain Epoch: 6 [37760/60032 (63%)]\tLoss: 0.069505\nTrain Epoch: 6 [38400/60032 (64%)]\tLoss: 0.021467\nTrain Epoch: 6 [39040/60032 (65%)]\tLoss: 0.027197\nTrain Epoch: 6 [39680/60032 (66%)]\tLoss: 0.073208\nTrain Epoch: 6 [40320/60032 (67%)]\tLoss: 0.007481\nTrain Epoch: 6 [40960/60032 (68%)]\tLoss: 0.176389\nTrain Epoch: 6 [41600/60032 (69%)]\tLoss: 0.057661\nTrain Epoch: 6 [42240/60032 (70%)]\tLoss: 0.031201\nTrain Epoch: 6 [42880/60032 (71%)]\tLoss: 0.020894\nTrain Epoch: 6 [43520/60032 (72%)]\tLoss: 0.090538\nTrain Epoch: 6 [44160/60032 (74%)]\tLoss: 0.029354\nTrain Epoch: 6 [44800/60032 (75%)]\tLoss: 0.021749\nTrain Epoch: 6 [45440/60032 (76%)]\tLoss: 0.008537\nTrain Epoch: 6 [46080/60032 (77%)]\tLoss: 0.060737\nTrain Epoch: 6 [46720/60032 (78%)]\tLoss: 0.055961\nTrain Epoch: 6 [47360/60032 (79%)]\tLoss: 0.207721\nTrain Epoch: 6 [48000/60032 (80%)]\tLoss: 0.016195\nTrain Epoch: 6 [48640/60032 (81%)]\tLoss: 0.046007\nTrain Epoch: 6 [49280/60032 (82%)]\tLoss: 0.008330\nTrain Epoch: 6 [49920/60032 (83%)]\tLoss: 0.018175\nTrain Epoch: 6 [50560/60032 (84%)]\tLoss: 0.026913\nTrain Epoch: 6 [51200/60032 (85%)]\tLoss: 0.062922\nTrain Epoch: 6 [51840/60032 (86%)]\tLoss: 0.114062\nTrain Epoch: 6 [52480/60032 (87%)]\tLoss: 0.033819\nTrain Epoch: 6 [53120/60032 (88%)]\tLoss: 0.116976\nTrain Epoch: 6 [53760/60032 (90%)]\tLoss: 0.041320\nTrain Epoch: 6 [54400/60032 (91%)]\tLoss: 0.010514\nTrain Epoch: 6 [55040/60032 (92%)]\tLoss: 0.019040\nTrain Epoch: 6 [55680/60032 (93%)]\tLoss: 0.004178\nTrain Epoch: 6 [56320/60032 (94%)]\tLoss: 0.023471\nTrain Epoch: 6 [56960/60032 (95%)]\tLoss: 0.022724\nTrain Epoch: 6 [57600/60032 (96%)]\tLoss: 0.058093\nTrain Epoch: 6 [58240/60032 (97%)]\tLoss: 0.039848\nTrain Epoch: 6 [58880/60032 (98%)]\tLoss: 0.043645\nTrain Epoch: 6 [59520/60032 (99%)]\tLoss: 0.075563\n\nTest set: Average loss: 0.0475, Accuracy: 9853/10000 (99%)\n\nTrain Epoch: 7 [0/60032 (0%)]\tLoss: 0.146034\nTrain Epoch: 7 [640/60032 (1%)]\tLoss: 0.008706\nTrain Epoch: 7 [1280/60032 (2%)]\tLoss: 0.130339\nTrain Epoch: 7 [1920/60032 (3%)]\tLoss: 0.022218\nTrain Epoch: 7 [2560/60032 (4%)]\tLoss: 0.004462\nTrain Epoch: 7 [3200/60032 (5%)]\tLoss: 0.023724\nTrain Epoch: 7 [3840/60032 (6%)]\tLoss: 0.008415\nTrain Epoch: 7 [4480/60032 (7%)]\tLoss: 0.056233\nTrain Epoch: 7 [5120/60032 (9%)]\tLoss: 0.164741\nTrain Epoch: 7 [5760/60032 (10%)]\tLoss: 0.037281\nTrain Epoch: 7 [6400/60032 (11%)]\tLoss: 0.036307\nTrain Epoch: 7 [7040/60032 (12%)]\tLoss: 0.017701\nTrain Epoch: 7 [7680/60032 (13%)]\tLoss: 0.008410\nTrain Epoch: 7 [8320/60032 (14%)]\tLoss: 0.022548\nTrain Epoch: 7 [8960/60032 (15%)]\tLoss: 0.026380\nTrain Epoch: 7 [9600/60032 (16%)]\tLoss: 0.024258\nTrain Epoch: 7 [10240/60032 (17%)]\tLoss: 0.011242\nTrain Epoch: 7 [10880/60032 (18%)]\tLoss: 0.041713\nTrain Epoch: 7 [11520/60032 (19%)]\tLoss: 0.016677\nTrain Epoch: 7 [12160/60032 (20%)]\tLoss: 0.024791\nTrain Epoch: 7 [12800/60032 (21%)]\tLoss: 0.071365\nTrain Epoch: 7 [13440/60032 (22%)]\tLoss: 0.008650\nTrain Epoch: 7 [14080/60032 (23%)]\tLoss: 0.129404\nTrain Epoch: 7 [14720/60032 (25%)]\tLoss: 0.018914\nTrain Epoch: 7 [15360/60032 (26%)]\tLoss: 0.055319\nTrain Epoch: 7 [16000/60032 (27%)]\tLoss: 0.039595\nTrain Epoch: 7 [16640/60032 (28%)]\tLoss: 0.015238\nTrain Epoch: 7 [17280/60032 (29%)]\tLoss: 0.011442\nTrain Epoch: 7 [17920/60032 (30%)]\tLoss: 0.026230\nTrain Epoch: 7 [18560/60032 (31%)]\tLoss: 0.007713\nTrain Epoch: 7 [19200/60032 (32%)]\tLoss: 0.074849\nTrain Epoch: 7 [19840/60032 (33%)]\tLoss: 0.109825\nTrain Epoch: 7 [20480/60032 (34%)]\tLoss: 0.075948\nTrain Epoch: 7 [21120/60032 (35%)]\tLoss: 0.052406\nTrain Epoch: 7 [21760/60032 (36%)]\tLoss: 0.006732\nTrain Epoch: 7 [22400/60032 (37%)]\tLoss: 0.036843\nTrain Epoch: 7 [23040/60032 (38%)]\tLoss: 0.026656\nTrain Epoch: 7 [23680/60032 (39%)]\tLoss: 0.096858\nTrain Epoch: 7 [24320/60032 (41%)]\tLoss: 0.044328\nTrain Epoch: 7 [24960/60032 (42%)]\tLoss: 0.022555\nTrain Epoch: 7 [25600/60032 (43%)]\tLoss: 0.079736\nTrain Epoch: 7 [26240/60032 (44%)]\tLoss: 0.008342\nTrain Epoch: 7 [26880/60032 (45%)]\tLoss: 0.012412\nTrain Epoch: 7 [27520/60032 (46%)]\tLoss: 0.022173\nTrain Epoch: 7 [28160/60032 (47%)]\tLoss: 0.028194\nTrain Epoch: 7 [28800/60032 (48%)]\tLoss: 0.086265\nTrain Epoch: 7 [29440/60032 (49%)]\tLoss: 0.051854\nTrain Epoch: 7 [30080/60032 (50%)]\tLoss: 0.044413\nTrain Epoch: 7 [30720/60032 (51%)]\tLoss: 0.011174\nTrain Epoch: 7 [31360/60032 (52%)]\tLoss: 0.010169\nTrain Epoch: 7 [32000/60032 (53%)]\tLoss: 0.004514\nTrain Epoch: 7 [32640/60032 (54%)]\tLoss: 0.024473\nTrain Epoch: 7 [33280/60032 (55%)]\tLoss: 0.005404\nTrain Epoch: 7 [33920/60032 (57%)]\tLoss: 0.021634\nTrain Epoch: 7 [34560/60032 (58%)]\tLoss: 0.018731\nTrain Epoch: 7 [35200/60032 (59%)]\tLoss: 0.109460\nTrain Epoch: 7 [35840/60032 (60%)]\tLoss: 0.074583\nTrain Epoch: 7 [36480/60032 (61%)]\tLoss: 0.082778\nTrain Epoch: 7 [37120/60032 (62%)]\tLoss: 0.029774\nTrain Epoch: 7 [37760/60032 (63%)]\tLoss: 0.143449\nTrain Epoch: 7 [38400/60032 (64%)]\tLoss: 0.019281\nTrain Epoch: 7 [39040/60032 (65%)]\tLoss: 0.033991\nTrain Epoch: 7 [39680/60032 (66%)]\tLoss: 0.014584\nTrain Epoch: 7 [40320/60032 (67%)]\tLoss: 0.038756\nTrain Epoch: 7 [40960/60032 (68%)]\tLoss: 0.034623\nTrain Epoch: 7 [41600/60032 (69%)]\tLoss: 0.043312\nTrain Epoch: 7 [42240/60032 (70%)]\tLoss: 0.105817\nTrain Epoch: 7 [42880/60032 (71%)]\tLoss: 0.020727\nTrain Epoch: 7 [43520/60032 (72%)]\tLoss: 0.120223\nTrain Epoch: 7 [44160/60032 (74%)]\tLoss: 0.027469\nTrain Epoch: 7 [44800/60032 (75%)]\tLoss: 0.009150\nTrain Epoch: 7 [45440/60032 (76%)]\tLoss: 0.008214\nTrain Epoch: 7 [46080/60032 (77%)]\tLoss: 0.083218\nTrain Epoch: 7 [46720/60032 (78%)]\tLoss: 0.055152\nTrain Epoch: 7 [47360/60032 (79%)]\tLoss: 0.038762\nTrain Epoch: 7 [48000/60032 (80%)]\tLoss: 0.007858\nTrain Epoch: 7 [48640/60032 (81%)]\tLoss: 0.004001\nTrain Epoch: 7 [49280/60032 (82%)]\tLoss: 0.070956\nTrain Epoch: 7 [49920/60032 (83%)]\tLoss: 0.051868\nTrain Epoch: 7 [50560/60032 (84%)]\tLoss: 0.065362\nTrain Epoch: 7 [51200/60032 (85%)]\tLoss: 0.059228\nTrain Epoch: 7 [51840/60032 (86%)]\tLoss: 0.039857\nTrain Epoch: 7 [52480/60032 (87%)]\tLoss: 0.112386\nTrain Epoch: 7 [53120/60032 (88%)]\tLoss: 0.050384\nTrain Epoch: 7 [53760/60032 (90%)]\tLoss: 0.043221\nTrain Epoch: 7 [54400/60032 (91%)]\tLoss: 0.009287\nTrain Epoch: 7 [55040/60032 (92%)]\tLoss: 0.025575\nTrain Epoch: 7 [55680/60032 (93%)]\tLoss: 0.009971\nTrain Epoch: 7 [56320/60032 (94%)]\tLoss: 0.004897\nTrain Epoch: 7 [56960/60032 (95%)]\tLoss: 0.113037\nTrain Epoch: 7 [57600/60032 (96%)]\tLoss: 0.116349\nTrain Epoch: 7 [58240/60032 (97%)]\tLoss: 0.194649\nTrain Epoch: 7 [58880/60032 (98%)]\tLoss: 0.020764\nTrain Epoch: 7 [59520/60032 (99%)]\tLoss: 0.016565\n\nTest set: Average loss: 0.0439, Accuracy: 9858/10000 (99%)\n\nTrain Epoch: 8 [0/60032 (0%)]\tLoss: 0.033885\nTrain Epoch: 8 [640/60032 (1%)]\tLoss: 0.191931\nTrain Epoch: 8 [1280/60032 (2%)]\tLoss: 0.042910\nTrain Epoch: 8 [1920/60032 (3%)]\tLoss: 0.054583\nTrain Epoch: 8 [2560/60032 (4%)]\tLoss: 0.041497\nTrain Epoch: 8 [3200/60032 (5%)]\tLoss: 0.009083\nTrain Epoch: 8 [3840/60032 (6%)]\tLoss: 0.010484\nTrain Epoch: 8 [4480/60032 (7%)]\tLoss: 0.049081\nTrain Epoch: 8 [5120/60032 (9%)]\tLoss: 0.014216\nTrain Epoch: 8 [5760/60032 (10%)]\tLoss: 0.003393\nTrain Epoch: 8 [6400/60032 (11%)]\tLoss: 0.015030\nTrain Epoch: 8 [7040/60032 (12%)]\tLoss: 0.076544\nTrain Epoch: 8 [7680/60032 (13%)]\tLoss: 0.018571\nTrain Epoch: 8 [8320/60032 (14%)]\tLoss: 0.044120\nTrain Epoch: 8 [8960/60032 (15%)]\tLoss: 0.017073\nTrain Epoch: 8 [9600/60032 (16%)]\tLoss: 0.017347\nTrain Epoch: 8 [10240/60032 (17%)]\tLoss: 0.005205\nTrain Epoch: 8 [10880/60032 (18%)]\tLoss: 0.048844\nTrain Epoch: 8 [11520/60032 (19%)]\tLoss: 0.015564\nTrain Epoch: 8 [12160/60032 (20%)]\tLoss: 0.024815\nTrain Epoch: 8 [12800/60032 (21%)]\tLoss: 0.071916\nTrain Epoch: 8 [13440/60032 (22%)]\tLoss: 0.041922\nTrain Epoch: 8 [14080/60032 (23%)]\tLoss: 0.012463\nTrain Epoch: 8 [14720/60032 (25%)]\tLoss: 0.041193\nTrain Epoch: 8 [15360/60032 (26%)]\tLoss: 0.004859\nTrain Epoch: 8 [16000/60032 (27%)]\tLoss: 0.080889\nTrain Epoch: 8 [16640/60032 (28%)]\tLoss: 0.058989\nTrain Epoch: 8 [17280/60032 (29%)]\tLoss: 0.072190\nTrain Epoch: 8 [17920/60032 (30%)]\tLoss: 0.084213\nTrain Epoch: 8 [18560/60032 (31%)]\tLoss: 0.040844\nTrain Epoch: 8 [19200/60032 (32%)]\tLoss: 0.045398\nTrain Epoch: 8 [19840/60032 (33%)]\tLoss: 0.031646\nTrain Epoch: 8 [20480/60032 (34%)]\tLoss: 0.065922\nTrain Epoch: 8 [21120/60032 (35%)]\tLoss: 0.028012\nTrain Epoch: 8 [21760/60032 (36%)]\tLoss: 0.051286\nTrain Epoch: 8 [22400/60032 (37%)]\tLoss: 0.130897\nTrain Epoch: 8 [23040/60032 (38%)]\tLoss: 0.009301\nTrain Epoch: 8 [23680/60032 (39%)]\tLoss: 0.025238\nTrain Epoch: 8 [24320/60032 (41%)]\tLoss: 0.065967\nTrain Epoch: 8 [24960/60032 (42%)]\tLoss: 0.008086\nTrain Epoch: 8 [25600/60032 (43%)]\tLoss: 0.030019\nTrain Epoch: 8 [26240/60032 (44%)]\tLoss: 0.113557\nTrain Epoch: 8 [26880/60032 (45%)]\tLoss: 0.008590\nTrain Epoch: 8 [27520/60032 (46%)]\tLoss: 0.027831\nTrain Epoch: 8 [28160/60032 (47%)]\tLoss: 0.053981\nTrain Epoch: 8 [28800/60032 (48%)]\tLoss: 0.090621\nTrain Epoch: 8 [29440/60032 (49%)]\tLoss: 0.016549\nTrain Epoch: 8 [30080/60032 (50%)]\tLoss: 0.040018\nTrain Epoch: 8 [30720/60032 (51%)]\tLoss: 0.004209\nTrain Epoch: 8 [31360/60032 (52%)]\tLoss: 0.004295\nTrain Epoch: 8 [32000/60032 (53%)]\tLoss: 0.047793\nTrain Epoch: 8 [32640/60032 (54%)]\tLoss: 0.051148\nTrain Epoch: 8 [33280/60032 (55%)]\tLoss: 0.034580\nTrain Epoch: 8 [33920/60032 (57%)]\tLoss: 0.062458\nTrain Epoch: 8 [34560/60032 (58%)]\tLoss: 0.012093\nTrain Epoch: 8 [35200/60032 (59%)]\tLoss: 0.136102\nTrain Epoch: 8 [35840/60032 (60%)]\tLoss: 0.003715\nTrain Epoch: 8 [36480/60032 (61%)]\tLoss: 0.086116\nTrain Epoch: 8 [37120/60032 (62%)]\tLoss: 0.006770\nTrain Epoch: 8 [37760/60032 (63%)]\tLoss: 0.006999\nTrain Epoch: 8 [38400/60032 (64%)]\tLoss: 0.127277\nTrain Epoch: 8 [39040/60032 (65%)]\tLoss: 0.214078\nTrain Epoch: 8 [39680/60032 (66%)]\tLoss: 0.040323\nTrain Epoch: 8 [40320/60032 (67%)]\tLoss: 0.041333\nTrain Epoch: 8 [40960/60032 (68%)]\tLoss: 0.026706\nTrain Epoch: 8 [41600/60032 (69%)]\tLoss: 0.021022\nTrain Epoch: 8 [42240/60032 (70%)]\tLoss: 0.042266\nTrain Epoch: 8 [42880/60032 (71%)]\tLoss: 0.030710\nTrain Epoch: 8 [43520/60032 (72%)]\tLoss: 0.007389\nTrain Epoch: 8 [44160/60032 (74%)]\tLoss: 0.017878\nTrain Epoch: 8 [44800/60032 (75%)]\tLoss: 0.041308\nTrain Epoch: 8 [45440/60032 (76%)]\tLoss: 0.051370\nTrain Epoch: 8 [46080/60032 (77%)]\tLoss: 0.075144\nTrain Epoch: 8 [46720/60032 (78%)]\tLoss: 0.004884\nTrain Epoch: 8 [47360/60032 (79%)]\tLoss: 0.012424\nTrain Epoch: 8 [48000/60032 (80%)]\tLoss: 0.023663\nTrain Epoch: 8 [48640/60032 (81%)]\tLoss: 0.161230\nTrain Epoch: 8 [49280/60032 (82%)]\tLoss: 0.027597\nTrain Epoch: 8 [49920/60032 (83%)]\tLoss: 0.039839\nTrain Epoch: 8 [50560/60032 (84%)]\tLoss: 0.043766\nTrain Epoch: 8 [51200/60032 (85%)]\tLoss: 0.038147\nTrain Epoch: 8 [51840/60032 (86%)]\tLoss: 0.018665\nTrain Epoch: 8 [52480/60032 (87%)]\tLoss: 0.006185\nTrain Epoch: 8 [53120/60032 (88%)]\tLoss: 0.004579\nTrain Epoch: 8 [53760/60032 (90%)]\tLoss: 0.007689\nTrain Epoch: 8 [54400/60032 (91%)]\tLoss: 0.001980\nTrain Epoch: 8 [55040/60032 (92%)]\tLoss: 0.016289\nTrain Epoch: 8 [55680/60032 (93%)]\tLoss: 0.056811\nTrain Epoch: 8 [56320/60032 (94%)]\tLoss: 0.008514\nTrain Epoch: 8 [56960/60032 (95%)]\tLoss: 0.041729\nTrain Epoch: 8 [57600/60032 (96%)]\tLoss: 0.075201\nTrain Epoch: 8 [58240/60032 (97%)]\tLoss: 0.011030\nTrain Epoch: 8 [58880/60032 (98%)]\tLoss: 0.110500\nTrain Epoch: 8 [59520/60032 (99%)]\tLoss: 0.015867\n\nTest set: Average loss: 0.0392, Accuracy: 9874/10000 (99%)\n\nTrain Epoch: 9 [0/60032 (0%)]\tLoss: 0.122474\nTrain Epoch: 9 [640/60032 (1%)]\tLoss: 0.020601\nTrain Epoch: 9 [1280/60032 (2%)]\tLoss: 0.011366\nTrain Epoch: 9 [1920/60032 (3%)]\tLoss: 0.022259\nTrain Epoch: 9 [2560/60032 (4%)]\tLoss: 0.053344\nTrain Epoch: 9 [3200/60032 (5%)]\tLoss: 0.025076\nTrain Epoch: 9 [3840/60032 (6%)]\tLoss: 0.013174\nTrain Epoch: 9 [4480/60032 (7%)]\tLoss: 0.012291\nTrain Epoch: 9 [5120/60032 (9%)]\tLoss: 0.125409\nTrain Epoch: 9 [5760/60032 (10%)]\tLoss: 0.019490\nTrain Epoch: 9 [6400/60032 (11%)]\tLoss: 0.018156\nTrain Epoch: 9 [7040/60032 (12%)]\tLoss: 0.020821\nTrain Epoch: 9 [7680/60032 (13%)]\tLoss: 0.128987\nTrain Epoch: 9 [8320/60032 (14%)]\tLoss: 0.016785\nTrain Epoch: 9 [8960/60032 (15%)]\tLoss: 0.041369\nTrain Epoch: 9 [9600/60032 (16%)]\tLoss: 0.083387\nTrain Epoch: 9 [10240/60032 (17%)]\tLoss: 0.046702\nTrain Epoch: 9 [10880/60032 (18%)]\tLoss: 0.067936\nTrain Epoch: 9 [11520/60032 (19%)]\tLoss: 0.004627\nTrain Epoch: 9 [12160/60032 (20%)]\tLoss: 0.007573\nTrain Epoch: 9 [12800/60032 (21%)]\tLoss: 0.010968\nTrain Epoch: 9 [13440/60032 (22%)]\tLoss: 0.012924\nTrain Epoch: 9 [14080/60032 (23%)]\tLoss: 0.077692\nTrain Epoch: 9 [14720/60032 (25%)]\tLoss: 0.109353\nTrain Epoch: 9 [15360/60032 (26%)]\tLoss: 0.048015\nTrain Epoch: 9 [16000/60032 (27%)]\tLoss: 0.055696\nTrain Epoch: 9 [16640/60032 (28%)]\tLoss: 0.056478\nTrain Epoch: 9 [17280/60032 (29%)]\tLoss: 0.066137\nTrain Epoch: 9 [17920/60032 (30%)]\tLoss: 0.042421\nTrain Epoch: 9 [18560/60032 (31%)]\tLoss: 0.036250\nTrain Epoch: 9 [19200/60032 (32%)]\tLoss: 0.010536\nTrain Epoch: 9 [19840/60032 (33%)]\tLoss: 0.004409\nTrain Epoch: 9 [20480/60032 (34%)]\tLoss: 0.094395\nTrain Epoch: 9 [21120/60032 (35%)]\tLoss: 0.065235\nTrain Epoch: 9 [21760/60032 (36%)]\tLoss: 0.008797\nTrain Epoch: 9 [22400/60032 (37%)]\tLoss: 0.063747\nTrain Epoch: 9 [23040/60032 (38%)]\tLoss: 0.023818\nTrain Epoch: 9 [23680/60032 (39%)]\tLoss: 0.010220\nTrain Epoch: 9 [24320/60032 (41%)]\tLoss: 0.013939\nTrain Epoch: 9 [24960/60032 (42%)]\tLoss: 0.002819\nTrain Epoch: 9 [25600/60032 (43%)]\tLoss: 0.015462\nTrain Epoch: 9 [26240/60032 (44%)]\tLoss: 0.049887\nTrain Epoch: 9 [26880/60032 (45%)]\tLoss: 0.055040\nTrain Epoch: 9 [27520/60032 (46%)]\tLoss: 0.046757\nTrain Epoch: 9 [28160/60032 (47%)]\tLoss: 0.009853\nTrain Epoch: 9 [28800/60032 (48%)]\tLoss: 0.045440\nTrain Epoch: 9 [29440/60032 (49%)]\tLoss: 0.034183\nTrain Epoch: 9 [30080/60032 (50%)]\tLoss: 0.004226\nTrain Epoch: 9 [30720/60032 (51%)]\tLoss: 0.043186\nTrain Epoch: 9 [31360/60032 (52%)]\tLoss: 0.033284\nTrain Epoch: 9 [32000/60032 (53%)]\tLoss: 0.010189\nTrain Epoch: 9 [32640/60032 (54%)]\tLoss: 0.020675\nTrain Epoch: 9 [33280/60032 (55%)]\tLoss: 0.031353\nTrain Epoch: 9 [33920/60032 (57%)]\tLoss: 0.021329\nTrain Epoch: 9 [34560/60032 (58%)]\tLoss: 0.012174\nTrain Epoch: 9 [35200/60032 (59%)]\tLoss: 0.117923\nTrain Epoch: 9 [35840/60032 (60%)]\tLoss: 0.050959\nTrain Epoch: 9 [36480/60032 (61%)]\tLoss: 0.034033\nTrain Epoch: 9 [37120/60032 (62%)]\tLoss: 0.016778\nTrain Epoch: 9 [37760/60032 (63%)]\tLoss: 0.025700\nTrain Epoch: 9 [38400/60032 (64%)]\tLoss: 0.106904\nTrain Epoch: 9 [39040/60032 (65%)]\tLoss: 0.004782\nTrain Epoch: 9 [39680/60032 (66%)]\tLoss: 0.040262\nTrain Epoch: 9 [40320/60032 (67%)]\tLoss: 0.053806\nTrain Epoch: 9 [40960/60032 (68%)]\tLoss: 0.036670\nTrain Epoch: 9 [41600/60032 (69%)]\tLoss: 0.001931\nTrain Epoch: 9 [42240/60032 (70%)]\tLoss: 0.079998\nTrain Epoch: 9 [42880/60032 (71%)]\tLoss: 0.005327\nTrain Epoch: 9 [43520/60032 (72%)]\tLoss: 0.052708\nTrain Epoch: 9 [44160/60032 (74%)]\tLoss: 0.044313\nTrain Epoch: 9 [44800/60032 (75%)]\tLoss: 0.032287\nTrain Epoch: 9 [45440/60032 (76%)]\tLoss: 0.057284\nTrain Epoch: 9 [46080/60032 (77%)]\tLoss: 0.017856\nTrain Epoch: 9 [46720/60032 (78%)]\tLoss: 0.061742\nTrain Epoch: 9 [47360/60032 (79%)]\tLoss: 0.159292\nTrain Epoch: 9 [48000/60032 (80%)]\tLoss: 0.040139\nTrain Epoch: 9 [48640/60032 (81%)]\tLoss: 0.039376\nTrain Epoch: 9 [49280/60032 (82%)]\tLoss: 0.013785\nTrain Epoch: 9 [49920/60032 (83%)]\tLoss: 0.045515\nTrain Epoch: 9 [50560/60032 (84%)]\tLoss: 0.010760\nTrain Epoch: 9 [51200/60032 (85%)]\tLoss: 0.021212\nTrain Epoch: 9 [51840/60032 (86%)]\tLoss: 0.016506\nTrain Epoch: 9 [52480/60032 (87%)]\tLoss: 0.027196\nTrain Epoch: 9 [53120/60032 (88%)]\tLoss: 0.172877\nTrain Epoch: 9 [53760/60032 (90%)]\tLoss: 0.004599\nTrain Epoch: 9 [54400/60032 (91%)]\tLoss: 0.014329\nTrain Epoch: 9 [55040/60032 (92%)]\tLoss: 0.013661\nTrain Epoch: 9 [55680/60032 (93%)]\tLoss: 0.041459\nTrain Epoch: 9 [56320/60032 (94%)]\tLoss: 0.014859\nTrain Epoch: 9 [56960/60032 (95%)]\tLoss: 0.049032\nTrain Epoch: 9 [57600/60032 (96%)]\tLoss: 0.035813\nTrain Epoch: 9 [58240/60032 (97%)]\tLoss: 0.032728\nTrain Epoch: 9 [58880/60032 (98%)]\tLoss: 0.077139\nTrain Epoch: 9 [59520/60032 (99%)]\tLoss: 0.072986\n\nTest set: Average loss: 0.0427, Accuracy: 9854/10000 (99%)\n\nTrain Epoch: 10 [0/60032 (0%)]\tLoss: 0.011451\nTrain Epoch: 10 [640/60032 (1%)]\tLoss: 0.021752\nTrain Epoch: 10 [1280/60032 (2%)]\tLoss: 0.064347\nTrain Epoch: 10 [1920/60032 (3%)]\tLoss: 0.011675\nTrain Epoch: 10 [2560/60032 (4%)]\tLoss: 0.002058\nTrain Epoch: 10 [3200/60032 (5%)]\tLoss: 0.042565\nTrain Epoch: 10 [3840/60032 (6%)]\tLoss: 0.007678\nTrain Epoch: 10 [4480/60032 (7%)]\tLoss: 0.002436\nTrain Epoch: 10 [5120/60032 (9%)]\tLoss: 0.013937\nTrain Epoch: 10 [5760/60032 (10%)]\tLoss: 0.008509\nTrain Epoch: 10 [6400/60032 (11%)]\tLoss: 0.022384\nTrain Epoch: 10 [7040/60032 (12%)]\tLoss: 0.057672\nTrain Epoch: 10 [7680/60032 (13%)]\tLoss: 0.022134\nTrain Epoch: 10 [8320/60032 (14%)]\tLoss: 0.005648\nTrain Epoch: 10 [8960/60032 (15%)]\tLoss: 0.006835\nTrain Epoch: 10 [9600/60032 (16%)]\tLoss: 0.007762\nTrain Epoch: 10 [10240/60032 (17%)]\tLoss: 0.040670\nTrain Epoch: 10 [10880/60032 (18%)]\tLoss: 0.020584\nTrain Epoch: 10 [11520/60032 (19%)]\tLoss: 0.033016\nTrain Epoch: 10 [12160/60032 (20%)]\tLoss: 0.030611\nTrain Epoch: 10 [12800/60032 (21%)]\tLoss: 0.024301\nTrain Epoch: 10 [13440/60032 (22%)]\tLoss: 0.019356\nTrain Epoch: 10 [14080/60032 (23%)]\tLoss: 0.028614\nTrain Epoch: 10 [14720/60032 (25%)]\tLoss: 0.120555\nTrain Epoch: 10 [15360/60032 (26%)]\tLoss: 0.005400\nTrain Epoch: 10 [16000/60032 (27%)]\tLoss: 0.102693\nTrain Epoch: 10 [16640/60032 (28%)]\tLoss: 0.031609\nTrain Epoch: 10 [17280/60032 (29%)]\tLoss: 0.012520\nTrain Epoch: 10 [17920/60032 (30%)]\tLoss: 0.026162\nTrain Epoch: 10 [18560/60032 (31%)]\tLoss: 0.045229\nTrain Epoch: 10 [19200/60032 (32%)]\tLoss: 0.025283\nTrain Epoch: 10 [19840/60032 (33%)]\tLoss: 0.013817\nTrain Epoch: 10 [20480/60032 (34%)]\tLoss: 0.181028\nTrain Epoch: 10 [21120/60032 (35%)]\tLoss: 0.007453\nTrain Epoch: 10 [21760/60032 (36%)]\tLoss: 0.024282\nTrain Epoch: 10 [22400/60032 (37%)]\tLoss: 0.028244\nTrain Epoch: 10 [23040/60032 (38%)]\tLoss: 0.018918\nTrain Epoch: 10 [23680/60032 (39%)]\tLoss: 0.013389\nTrain Epoch: 10 [24320/60032 (41%)]\tLoss: 0.007258\nTrain Epoch: 10 [24960/60032 (42%)]\tLoss: 0.021336\nTrain Epoch: 10 [25600/60032 (43%)]\tLoss: 0.097925\nTrain Epoch: 10 [26240/60032 (44%)]\tLoss: 0.009022\nTrain Epoch: 10 [26880/60032 (45%)]\tLoss: 0.019054\nTrain Epoch: 10 [27520/60032 (46%)]\tLoss: 0.038322\nTrain Epoch: 10 [28160/60032 (47%)]\tLoss: 0.123178\nTrain Epoch: 10 [28800/60032 (48%)]\tLoss: 0.063764\nTrain Epoch: 10 [29440/60032 (49%)]\tLoss: 0.046642\nTrain Epoch: 10 [30080/60032 (50%)]\tLoss: 0.058700\nTrain Epoch: 10 [30720/60032 (51%)]\tLoss: 0.031819\nTrain Epoch: 10 [31360/60032 (52%)]\tLoss: 0.081197\nTrain Epoch: 10 [32000/60032 (53%)]\tLoss: 0.001933\nTrain Epoch: 10 [32640/60032 (54%)]\tLoss: 0.021334\nTrain Epoch: 10 [33280/60032 (55%)]\tLoss: 0.034316\nTrain Epoch: 10 [33920/60032 (57%)]\tLoss: 0.008230\nTrain Epoch: 10 [34560/60032 (58%)]\tLoss: 0.036735\nTrain Epoch: 10 [35200/60032 (59%)]\tLoss: 0.020100\nTrain Epoch: 10 [35840/60032 (60%)]\tLoss: 0.082057\nTrain Epoch: 10 [36480/60032 (61%)]\tLoss: 0.011593\nTrain Epoch: 10 [37120/60032 (62%)]\tLoss: 0.061417\nTrain Epoch: 10 [37760/60032 (63%)]\tLoss: 0.003418\nTrain Epoch: 10 [38400/60032 (64%)]\tLoss: 0.005231\nTrain Epoch: 10 [39040/60032 (65%)]\tLoss: 0.010868\nTrain Epoch: 10 [39680/60032 (66%)]\tLoss: 0.023426\nTrain Epoch: 10 [40320/60032 (67%)]\tLoss: 0.034223\nTrain Epoch: 10 [40960/60032 (68%)]\tLoss: 0.022992\nTrain Epoch: 10 [41600/60032 (69%)]\tLoss: 0.070264\nTrain Epoch: 10 [42240/60032 (70%)]\tLoss: 0.026406\nTrain Epoch: 10 [42880/60032 (71%)]\tLoss: 0.011131\nTrain Epoch: 10 [43520/60032 (72%)]\tLoss: 0.078265\nTrain Epoch: 10 [44160/60032 (74%)]\tLoss: 0.044495\nTrain Epoch: 10 [44800/60032 (75%)]\tLoss: 0.019916\nTrain Epoch: 10 [45440/60032 (76%)]\tLoss: 0.015379\nTrain Epoch: 10 [46080/60032 (77%)]\tLoss: 0.003633\nTrain Epoch: 10 [46720/60032 (78%)]\tLoss: 0.010158\nTrain Epoch: 10 [47360/60032 (79%)]\tLoss: 0.053545\nTrain Epoch: 10 [48000/60032 (80%)]\tLoss: 0.028536\nTrain Epoch: 10 [48640/60032 (81%)]\tLoss: 0.016899\nTrain Epoch: 10 [49280/60032 (82%)]\tLoss: 0.034963\nTrain Epoch: 10 [49920/60032 (83%)]\tLoss: 0.027169\nTrain Epoch: 10 [50560/60032 (84%)]\tLoss: 0.020548\nTrain Epoch: 10 [51200/60032 (85%)]\tLoss: 0.021499\nTrain Epoch: 10 [51840/60032 (86%)]\tLoss: 0.009159\nTrain Epoch: 10 [52480/60032 (87%)]\tLoss: 0.004679\nTrain Epoch: 10 [53120/60032 (88%)]\tLoss: 0.240532\nTrain Epoch: 10 [53760/60032 (90%)]\tLoss: 0.001164\nTrain Epoch: 10 [54400/60032 (91%)]\tLoss: 0.008152\nTrain Epoch: 10 [55040/60032 (92%)]\tLoss: 0.082201\nTrain Epoch: 10 [55680/60032 (93%)]\tLoss: 0.017729\nTrain Epoch: 10 [56320/60032 (94%)]\tLoss: 0.053357\nTrain Epoch: 10 [56960/60032 (95%)]\tLoss: 0.009189\nTrain Epoch: 10 [57600/60032 (96%)]\tLoss: 0.031754\nTrain Epoch: 10 [58240/60032 (97%)]\tLoss: 0.023230\nTrain Epoch: 10 [58880/60032 (98%)]\tLoss: 0.011324\nTrain Epoch: 10 [59520/60032 (99%)]\tLoss: 0.152542\n\nTest set: Average loss: 0.0330, Accuracy: 9889/10000 (99%)\n\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a149c2d12696d4e9ecbd756597b5ce957359c87
| 88,536 |
ipynb
|
Jupyter Notebook
|
notebooks/1.2-dgb-initial-data-exploration.ipynb
|
Burton-David/KickstarterClassificationProject
|
470bab8233aadb2b4e5ddd2135307e59ec37ebbd
|
[
"MIT"
] | null | null | null |
notebooks/1.2-dgb-initial-data-exploration.ipynb
|
Burton-David/KickstarterClassificationProject
|
470bab8233aadb2b4e5ddd2135307e59ec37ebbd
|
[
"MIT"
] | null | null | null |
notebooks/1.2-dgb-initial-data-exploration.ipynb
|
Burton-David/KickstarterClassificationProject
|
470bab8233aadb2b4e5ddd2135307e59ec37ebbd
|
[
"MIT"
] | null | null | null | 66.319101 | 17,756 | 0.696124 |
[
[
[
"## Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport missingno as miss\nimport os\nimport glob\nfrom pathlib import Path\nfrom datetime import datetime\nimport pandas as pd\nimport seaborn as sns\nimport matplotlib.pyplot as plt\nimport matplotlib.cm as cm\nimport sklearn\nfrom xgboost import XGBClassifier\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Importing Data",
"_____no_output_____"
]
],
[
[
"print(f\"Current directory: {Path.cwd()}\")\nprint(f\"Home directory: {Path.home()}\")",
"Current directory: /Users/davidburton/DS/M3/KickstarterSuccessClassifier/notebooks\nHome directory: /Users/davidburton\n"
],
[
"path = \"/Users/davidburton/DS/M3/KickstarterSuccessClassifier/data/interim/ks-projects-201801.csv\"\npath_processed = \"/Users/davidburton/DS/M3/KickstarterSuccessClassifier/data/processed/ks-projects-201801.csv\"\npath_processed2 = \"../data/processed/explored.csv\"",
"_____no_output_____"
],
[
"df = pd.read_csv(path,\n encoding = \"ISO-8859-1\")\ndf.head(2)",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.isnull().any()",
"_____no_output_____"
],
[
"# Primer\n# What I want to predict\ntarget = \"outcome\"\n\n# The Features or Attributes\nfeatures = df.drop(target,1).columns",
"_____no_output_____"
],
[
"features_by_dtype = {}\n\nfor f in features:\n dtype = str(df[f].dtype)\n if dtype not in features_by_dtype.keys():\n features_by_dtype[dtype] = [f]\n else:\n features_by_dtype[dtype] += [f]\n \nfor k in features_by_dtype.keys():\n string = \"%s: %s\" % (k , len(features_by_dtype[k]))\n print(string)",
"object: 9\nint64: 9\n"
],
[
"keys = iter(features_by_dtype.keys())",
"_____no_output_____"
],
[
"k = next(keys)\ndtype_list = features_by_dtype[k]\nfor d in dtype_list:\n string = \"%s: %s\" % (d,len(df[d].unique()))\n print(string)\n",
"_____no_output_____"
],
[
"sns.pairplot(df)",
"_____no_output_____"
],
[
"g = sns.pairplot(df[[\"backers\", \"pledged\", \"goal\", \"deadline_dayofweek\",\n \"deadline_weekofyear\", \"launched_dayofweek\", \n \"launched_weekofyear\", \"launch_hourofday\", \"duration_days\"]],\n diag_kind=\"hist\")\n \nfor ax in g.axes.flat: \n plt.setp(ax.get_xticklabels(), rotation=45)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df['outcome'].value_counts().plot(kind='box');",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"df.drop(['name'], axis = 1, inplace=True)",
"_____no_output_____"
],
[
"df.category.value_counts()",
"_____no_output_____"
],
[
"df.corr(method='spearman')",
"_____no_output_____"
],
[
"df.corr(method='pearson')",
"_____no_output_____"
],
[
"df.groupby('outcome').nunique()\n# less backers means more likely to fail\n# longer campaigns more likely to fail ",
"_____no_output_____"
],
[
"df.category.unique()",
"_____no_output_____"
],
[
"#create a category dictionary\ncategory_dict = {}\ncategory_list = ['Publishing', 'Film & Video', 'Music', 'Food', 'Crafts', 'Games',\n 'Design', 'Comics', 'Fashion', 'Theater', 'Art', 'Photography',\n 'Technology', 'Dance', 'Journalism']",
"_____no_output_____"
],
[
"df.category.value_counts(normalize=True).plot(kind = 'pie', figsize=(14,7))",
"_____no_output_____"
],
[
"\ndf_cat = df.category.value_counts()\ndf_cat = pd.DataFrame(df_cat)\ndf_cat = df_cat.rename(columns={'category':'count'})\ndf_cat['category'] = df_cat.index\nax = sns.barplot(x=\"category\" , y= \"count\", data= df_cat, palette='rainbow')\nax.set_xlabel(\"Category\", fontweight='bold')\nax.set_ylabel(\"Number of Projects\", fontweight='bold')\nplt.xticks(rotation=90);\n",
"_____no_output_____"
]
],
[
[
"Start doing some modeling",
"_____no_output_____"
]
],
[
[
"#encode\ndf.dtypes",
"_____no_output_____"
],
[
"#convert objects into datetime where appropriate \ndf.launch_time = pd.to_datetime(df.launch_time)",
"_____no_output_____"
],
[
"df.launched = pd.to_datetime(df.launched)",
"_____no_output_____"
],
[
"df.deadline = pd.to_datetime(df.deadline)",
"_____no_output_____"
],
[
"#encode\ndf_dummies = pd.get_dummies(df[['sub_category', 'category', \n 'currency', 'country']], drop_first=True)\n",
"_____no_output_____"
],
[
"df = df_dummies.merge(df, left_index=True, right_index=True)",
"_____no_output_____"
],
[
"#drop original categorical object features\ndf = df.drop(['sub_category', 'category', \n 'currency', 'country'],1)",
"_____no_output_____"
],
[
"df.shape",
"_____no_output_____"
],
[
"df.head(2)",
"_____no_output_____"
],
[
"df.columns",
"_____no_output_____"
],
[
"#https://elitedatascience.com/python-machine-learning-tutorial-scikit-learn\nimport numpy as np\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\nfrom sklearn import preprocessing\n\n#families\n#random forest\nfrom sklearn.ensemble import RandomForestRegressor\nfrom sklearn.ensemble import RandomForestClassifier\n\n#cross-validation pipeline\nfrom sklearn.pipeline import make_pipeline\nfrom sklearn.model_selection import GridSearchCV\n\n#evaluation metrics\nfrom sklearn.metrics import mean_squared_error, r2_score\n\n#for saving models\nfrom sklearn.externals import joblib",
"_____no_output_____"
],
[
"#keep only feature known from begining of campaign\nX = df.drop(['outcome', 'backers', 'pledged','duration','deadline',\n 'launched', 'launch_time'],1)\n#Target Variable\ny = df.outcome",
"_____no_output_____"
],
[
"X=X.astype(int);",
"_____no_output_____"
],
[
"y.unique()",
"_____no_output_____"
],
[
"y.replace('failed',value=0,inplace=True)",
"_____no_output_____"
],
[
"y.replace('successful',value=1,inplace=True)",
"_____no_output_____"
],
[
"y.value_counts(normalize=True)",
"_____no_output_____"
],
[
"y = y.astype(int)",
"_____no_output_____"
],
[
"#split data into train and test sets\nX_train, X_test, y_train, y_test = train_test_split(X, y, \n test_size=0.2, \n random_state=123, \n stratify=y)\n",
"_____no_output_____"
]
],
[
[
"Fit the transformer on the training set (saving the means and standard deviations)<br>\nApply the transformer to the training set (scaling the training data)<br>\nApply the transformer to the test set (using the same means and standard deviations)<br>\n",
"_____no_output_____"
]
],
[
[
"# fit the transformer\nscaler = preprocessing.StandardScaler().fit(X_train)\n",
"/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n"
],
[
"# sanity check\nX_train_scaled = scaler.transform(X_train)\n \n",
"/anaconda3/envs/learn-env/lib/python3.6/site-packages/ipykernel_launcher.py:2: DataConversionWarning: Data with input dtype uint8, int64 were all converted to float64 by StandardScaler.\n \n"
],
[
"%%time\nX_train_scaled.mean(axis=0)[:10]",
"CPU times: user 138 ms, sys: 375 ms, total: 513 ms\nWall time: 514 ms\n"
],
[
"X_train_scaled.std()",
"_____no_output_____"
],
[
"#worked on train set, use same method to scale on test data\nX_test_scaled = scaler.transform(X_test)",
"/anaconda3/envs/learn-env/lib/python3.6/site-packages/ipykernel_launcher.py:2: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n \n"
],
[
"# sanity check\nX_test_scaled.mean(axis=0)[:10]",
"_____no_output_____"
],
[
"X_test_scaled.std()",
"_____no_output_____"
],
[
"#easier method\n#pipleine with preprocessing and model\npipeline = make_pipeline(preprocessing.StandardScaler(), \n RandomForestClassifier(n_estimators=100))",
"_____no_output_____"
]
],
[
[
"The above is a modeling pipeline that first transforms the data using StandardScaler() and then fits a model using a random forest regressor.",
"_____no_output_____"
]
],
[
[
"# figuring out hyperparameters \npipeline.get_params()",
"_____no_output_____"
],
[
"#define hyper parameters to tune\nhyperparameters = { 'randomforestclassifier__max_features' : ['auto', 'sqrt', 'log2'],\n 'randomforestclassifier__max_depth': [None, 5, 3, 1]}",
"_____no_output_____"
]
],
[
[
"Cross-validation<br>\n1. Split data into k equal parts, or \"folds\" (typically k=10).<br>\n2. Preprocess k-1 training folds.<br>\n3. Train model on k-1 folds (e.g. the first 9 folds).<br>\n4. Preprocess the hold-out fold using the same transformations from step(2)<br>\n5. Evaluate it on the remaining \"hold-out\" fold (e.g. the 10th fold).<br>\n6. Perform steps (2) and (3) k times, each time holding out a different fold.<br>\n7. Aggregate the performance across all k folds. This is the performance metric.<br>",
"_____no_output_____"
]
],
[
[
"%%time\n#cross-validation with pipeline\nclf = GridSearchCV(pipeline, hyperparameters, cv=10)\n",
"CPU times: user 32 µs, sys: 29 µs, total: 61 µs\nWall time: 66 µs\n"
],
[
"%%time\n# Fit and tune model\n#clf.fit(X_train, y_train)",
"/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/pipeline.py:511: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n Xt = transform.transform(Xt)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/preprocessing/data.py:625: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.partial_fit(X, y)\n/anaconda3/envs/learn-env/lib/python3.6/site-packages/sklearn/base.py:465: DataConversionWarning: Data with input dtype int64 were all converted to float64 by StandardScaler.\n return self.fit(X, y, **fit_params).transform(X)\n"
],
[
"rf = RandomForestClassifier(random_state = 42)\nfrom pprint import pprint\n# Look at parameters used by our current forest\nprint('Parameters currently in use:\\n')\npprint(rf.get_params())",
"Parameters currently in use:\n\n{'bootstrap': True,\n 'class_weight': None,\n 'criterion': 'gini',\n 'max_depth': None,\n 'max_features': 'auto',\n 'max_leaf_nodes': None,\n 'min_impurity_decrease': 0.0,\n 'min_impurity_split': None,\n 'min_samples_leaf': 1,\n 'min_samples_split': 2,\n 'min_weight_fraction_leaf': 0.0,\n 'n_estimators': 'warn',\n 'n_jobs': None,\n 'oob_score': False,\n 'random_state': 42,\n 'verbose': 0,\n 'warm_start': False}\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a14a34a88d484400b3c74356fb7fb39b3376fb4
| 30,788 |
ipynb
|
Jupyter Notebook
|
notebooks/mnist/00_pretrain_model.ipynb
|
Yu-Group/adaptive-wavelets
|
e67f726e741d83c94c3aee3ed97a772db4ce0bb3
|
[
"MIT"
] | 22 |
2021-02-13T05:22:13.000Z
|
2022-03-07T09:55:55.000Z
|
notebooks/mnist/00_pretrain_model.ipynb
|
Yu-Group/adaptive-wavelets
|
e67f726e741d83c94c3aee3ed97a772db4ce0bb3
|
[
"MIT"
] | null | null | null |
notebooks/mnist/00_pretrain_model.ipynb
|
Yu-Group/adaptive-wavelets
|
e67f726e741d83c94c3aee3ed97a772db4ce0bb3
|
[
"MIT"
] | 5 |
2021-12-11T13:43:19.000Z
|
2022-03-19T07:07:37.000Z
| 96.81761 | 11,952 | 0.842374 |
[
[
[
"% load_ext autoreload\n% autoreload 2\n% matplotlib inline\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport torch\n\ndevice = 'cuda' if torch.cuda.is_available() else 'cpu'\nimport os, sys\n\nopj = os.path.join\nfrom tqdm import tqdm\n\nfrom ex_mnist import p\nfrom dset import get_dataloader\n\nsys.path.append('../../src/models')\nfrom models import CNN, FFN",
"_____no_output_____"
],
[
"# load data\ntrain_loader, test_loader = get_dataloader(p.data_path,\n batch_size=p.batch_size)\n\n# import models\ncnn = CNN().to(device)\nffn = FFN().to(device)",
"_____no_output_____"
]
],
[
[
"# train cnn",
"_____no_output_____"
]
],
[
[
"optimizer = torch.optim.Adam(cnn.parameters(), lr=0.001)\ncriterion = torch.nn.CrossEntropyLoss()\nnum_epochs = 50\n\ntrain_losses = []\n\nfor epoch in range(num_epochs):\n epoch_loss = 0.\n for batch_idx, (data, y) in enumerate(train_loader):\n data = data.to(device)\n y = y.to(device)\n # zero grad\n optimizer.zero_grad()\n output = cnn(data)\n loss = criterion(output, y)\n\n # backward\n loss.backward()\n # update step\n optimizer.step()\n\n iter_loss = loss.item()\n epoch_loss += iter_loss\n\n print('\\rTrain Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n epoch, batch_idx * len(data), len(train_loader.dataset),\n 100. * batch_idx / len(train_loader), iter_loss), end='')\n\n mean_epoch_loss = epoch_loss / (batch_idx + 1)\n train_losses.append(mean_epoch_loss)\n\n# save model\ntorch.save(cnn.state_dict(), opj(p.model_path, 'CNN.pth'))\n",
"Train Epoch: 49 [59900/60000 (100%)]\tLoss: 0.004418"
],
[
"plt.plot(train_losses)",
"_____no_output_____"
]
],
[
[
"# train ffn",
"_____no_output_____"
]
],
[
[
"optimizer = torch.optim.Adam(ffn.parameters(), lr=0.001)\ncriterion = torch.nn.CrossEntropyLoss()\nnum_epochs = 50\n\ntrain_losses = []\n\nfor epoch in range(num_epochs):\n epoch_loss = 0.\n for batch_idx, (data, y) in enumerate(train_loader):\n data = data.to(device)\n y = y.to(device)\n # zero grad\n optimizer.zero_grad()\n output = ffn(data)\n loss = criterion(output, y)\n\n # backward\n loss.backward()\n # update step\n optimizer.step()\n\n iter_loss = loss.item()\n epoch_loss += iter_loss\n\n print('\\rTrain Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}'.format(\n epoch, batch_idx * len(data), len(train_loader.dataset),\n 100. * batch_idx / len(train_loader), iter_loss), end='')\n\n mean_epoch_loss = epoch_loss / (batch_idx + 1)\n train_losses.append(mean_epoch_loss)\n\n# save model\ntorch.save(ffn.state_dict(), opj(p.model_path, 'FFN.pth'))\n",
"Train Epoch: 49 [59900/60000 (100%)]\tLoss: 0.000006"
],
[
"plt.plot(train_losses)",
"_____no_output_____"
]
],
[
[
"# model prediction",
"_____no_output_____"
]
],
[
[
"# check prediction\nm = len(test_loader.dataset)\nbatch_size = test_loader.batch_size\n\ny_pred_cnn = np.zeros(m)\ny_pred_ffn = np.zeros(m)\ny_true = np.zeros(m)\nwith torch.no_grad():\n for batch_idx, (data, y) in tqdm(enumerate(test_loader, 0), total=int(np.ceil(m / batch_size))):\n data = data.to(device)\n # cnn prediction\n outputs_cnn = cnn(data)\n _, y_pred = torch.max(outputs_cnn.data, 1)\n y_pred_cnn[batch_idx * batch_size:(batch_idx + 1) * batch_size] = y_pred.cpu().numpy()\n\n # ffn prediction\n outputs_ffn = ffn(data)\n _, y_pred = torch.max(outputs_ffn.data, 1)\n y_pred_ffn[batch_idx * batch_size:(batch_idx + 1) * batch_size] = y_pred.cpu().numpy()\n\n # labels\n y_true[batch_idx * batch_size:(batch_idx + 1) * batch_size] = y.numpy()\n\nprint(\"CNN accuracy {:.5f}% FFN accuracy {:.5f}%\".format((y_true == y_pred_cnn).sum() / m * 100,\n (y_true == y_pred_ffn).sum() / m * 100))",
"100%|██████████| 100/100 [00:02<00:00, 33.85it/s]"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a14c0b693ceab7e25eb08553b0a7e52eff9ddd1
| 8,298 |
ipynb
|
Jupyter Notebook
|
d2l-en/chapter_deep-learning-computation/read-write.ipynb
|
mru4913/Dive-into-Deep-Learning
|
bcd16ac602f011292bd1d5540ef3833cd3fd7c72
|
[
"MIT"
] | null | null | null |
d2l-en/chapter_deep-learning-computation/read-write.ipynb
|
mru4913/Dive-into-Deep-Learning
|
bcd16ac602f011292bd1d5540ef3833cd3fd7c72
|
[
"MIT"
] | null | null | null |
d2l-en/chapter_deep-learning-computation/read-write.ipynb
|
mru4913/Dive-into-Deep-Learning
|
bcd16ac602f011292bd1d5540ef3833cd3fd7c72
|
[
"MIT"
] | null | null | null | 28.912892 | 738 | 0.566884 |
[
[
[
"# File I/O\n\nSo far we discussed how to process data, how to build, train and test deep learning models. However, at some point we are likely happy with what we obtained and we want to save the results for later use and distribution. Likewise, when running a long training process it is best practice to save intermediate results (checkpointing) to ensure that we don't lose several days worth of computation when tripping over the power cord of our server. At the same time, we might want to load a pretrained model (e.g. we might have word embeddings for English and use it for our fancy spam classifier). For all of these cases we need to load and store both individual weight vectors and entire models. This section addresses both issues.\n\n## NDArray\n\nIn its simplest form, we can directly use the `save` and `load` functions to store and read NDArrays separately. This works just as expected.",
"_____no_output_____"
]
],
[
[
"from mxnet import nd\nfrom mxnet.gluon import nn\n\nx = nd.arange(4)\nnd.save('x-file', x)",
"_____no_output_____"
]
],
[
[
"Then, we read the data from the stored file back into memory.",
"_____no_output_____"
]
],
[
[
"x2 = nd.load('x-file')\nx2",
"_____no_output_____"
]
],
[
[
"We can also store a list of NDArrays and read them back into memory.",
"_____no_output_____"
]
],
[
[
"y = nd.zeros(4)\nnd.save('x-files', [x, y])\nx2, y2 = nd.load('x-files')\n(x2, y2)",
"_____no_output_____"
]
],
[
[
"We can even write and read a dictionary that maps from a string to an NDArray. This is convenient, for instance when we want to read or write all the weights in a model.",
"_____no_output_____"
]
],
[
[
"mydict = {'x': x, 'y': y}\nnd.save('mydict', mydict)\nmydict2 = nd.load('mydict')\nmydict2",
"_____no_output_____"
]
],
[
[
"## Gluon Model Parameters\n\nSaving individual weight vectors (or other NDArray tensors) is useful but it\ngets very tedious if we want to save (and later load) an entire model. After\nall, we might have hundreds of parameter groups sprinkled throughout. Writing a\nscript that collects all the terms and matches them to an architecture is quite\nsome work. For this reason Gluon provides built-in functionality to load and\nsave entire networks rather than just single weight vectors. An important detail\nto note is that this saves model *parameters* and not the entire model. I.e. if\nwe have a 3 layer MLP we need to specify the *architecture* separately. The\nreason for this is that the models themselves can contain arbitrary code, hence\nthey cannot be serialized quite so easily (there is a way to do this for\ncompiled models - please refer to the [MXNet documentation](http://www.mxnet.io)\nfor the technical details on it). The result is that in order to reinstate a\nmodel we need to generate the architecture in code and then load the parameters\nfrom disk. The deferred initialization (:numref:`chapter_deferred_init`) is quite advantageous here since we can simply define a model without the need to put actual values in place. Let's start with our favorite MLP.",
"_____no_output_____"
]
],
[
[
"class MLP(nn.Block):\n def __init__(self, **kwargs):\n super(MLP, self).__init__(**kwargs)\n self.hidden = nn.Dense(256, activation='relu')\n self.output = nn.Dense(10)\n\n def forward(self, x):\n return self.output(self.hidden(x))\n\nnet = MLP()\nnet.initialize()\nx = nd.random.uniform(shape=(2, 20))\ny = net(x)",
"_____no_output_____"
]
],
[
[
"Next, we store the parameters of the model as a file with the name 'mlp.params'.",
"_____no_output_____"
]
],
[
[
"net.save_parameters('mlp.params')",
"_____no_output_____"
]
],
[
[
"To check whether we are able to recover the model we instantiate a clone of the original MLP model. Unlike the random initialization of model parameters, here we read the parameters stored in the file directly.",
"_____no_output_____"
]
],
[
[
"clone = MLP()\nclone.load_parameters('mlp.params')",
"_____no_output_____"
]
],
[
[
"Since both instances have the same model parameters, the computation result of the same input `x` should be the same. Let's verify this.",
"_____no_output_____"
]
],
[
[
"yclone = clone(x)\nyclone == y",
"_____no_output_____"
]
],
[
[
"## Summary\n\n* The `save` and `load` functions can be used to perform File I/O for NDArray objects.\n* The `load_parameters` and `save_parameters` functions allow us to save entire sets of parameters for a network in Gluon.\n* Saving the architecture has to be done in code rather than in parameters.\n\n## Exercises\n\n1. Even if there is no need to deploy trained models to a different device, what are the practical benefits of storing model parameters?\n1. Assume that we want to reuse only parts of a network to be incorporated into a network of a *different* architecture. How would you go about using, say the first two layers from a previous network in a new network.\n1. How would you go about saving network architecture and parameters? What restrictions would you impose on the architecture?\n\n## Scan the QR Code to [Discuss](https://discuss.mxnet.io/t/2329)\n\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a14c77c22eb09e015590c1a1692cd4c1c7580c1
| 13,740 |
ipynb
|
Jupyter Notebook
|
17 JSON/Twitter/Twitter Konversationen als Netzwerk.ipynb
|
MAZ-CAS-DDJ/kurs_19_20
|
143b90f2295cb790615564408595ba16c9ae05de
|
[
"MIT"
] | 6 |
2019-08-06T14:53:34.000Z
|
2020-10-16T19:44:16.000Z
|
17 JSON/Twitter/Twitter Konversationen als Netzwerk.ipynb
|
MAZ-CAS-DDJ/kurs_19_20
|
143b90f2295cb790615564408595ba16c9ae05de
|
[
"MIT"
] | 1 |
2020-06-25T09:46:58.000Z
|
2020-06-25T09:46:58.000Z
|
17 JSON/Twitter/Twitter Konversationen als Netzwerk.ipynb
|
MAZ-CAS-DDJ/kurs_19_20
|
143b90f2295cb790615564408595ba16c9ae05de
|
[
"MIT"
] | 2 |
2019-09-16T13:05:51.000Z
|
2019-09-27T09:07:49.000Z
| 37.438692 | 319 | 0.522416 |
[
[
[
"# Twitter Konversationen zu einem Thema als Netzwerk untersuchen\n\n- Aus Twitter-Daten kann man besonders gut Netzwerke basteln.\n- Dabei können wir frei definieren,wann eigentlich ein Nutzer mit einem anderen verbunden ist. Die gebräuchlichsten Definitionen sind:\n 1. Nutzer A retweetet Nutzer B (RT plotti was für ein super tweet)\n 2. Nutzer A erwähnt Nutzer B (Ich geh das so die Straße lang und seh @plotti)\n 3. Nutzer A schreibt Nutzer B (@plotti was geht heute)\n 4. (Nutzer A folgt Nutzer B (Leider um die Struktur einer Konversationen nicht sooo hilfreich. Außerdem muss man über Twarc recht viele User sammeln um diese Information zu erhalten, es geht aber.))",
"_____no_output_____"
],
[
"# Daten Sammeln über Twarc",
"_____no_output_____"
],
[
"- https://github.com/DocNow/twarc\n- Twarc: A command line tool (and Python library) for archiving Twitter JSON\n- Sehr praktisch um Tweets zu einem Stichwort zu sammeln. \n- Man muss eine Twiter app beantragen :(\n- ```pip install twarc```\n- ```twarc configure```\n",
"_____no_output_____"
],
[
"## Daten Sammeln ",
"_____no_output_____"
],
[
"```twarc search zürich > zürich.json```",
"_____no_output_____"
]
],
[
[
"import sys\nimport json\nimport re\nimport numpy as np\nfrom datetime import datetime\nimport pandas as pd \nimport networkx as nx\n\ntweetfile = 'zürich.json'",
"_____no_output_____"
]
],
[
[
"# 1. Kanten erzeugen durch Retweets\n- Personen retweeten sich und deswegen erzeugen wir eine Kante zwischen ihnen.",
"_____no_output_____"
]
],
[
[
"# 1. Export edges from Retweets\n\nfh = open(tweetfile, 'r')\n\nuserdata = pd.DataFrame(columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count' ))\nedges = pd.DataFrame(columns=('Source','Target','Time', \"Strength\"))\n\nfor line in fh:\n try:\n tweet = json.loads(line)\n except:\n continue\n if 'retweeted_status' not in tweet:\n continue\n \n userdata = userdata.append(pd.DataFrame([[tweet['user']['id_str'],\n tweet['user']['screen_name'],\n tweet['user']['created_at'],\n tweet['user']['profile_image_url_https'],\n tweet['user']['followers_count'],\n tweet['user']['friends_count']]], columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count')), ignore_index=True)\n userdata = userdata.append(pd.DataFrame([[tweet['retweeted_status']['user']['id_str'],\n tweet['retweeted_status']['user']['screen_name'],\n tweet['retweeted_status']['user']['created_at'],\n tweet['retweeted_status']['user']['profile_image_url_https'],\n tweet['retweeted_status']['user']['followers_count'],\n tweet['retweeted_status']['user']['friends_count']]], columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count')), ignore_index=True) \n edges = edges.append(pd.DataFrame([[tweet['user']['id_str'],\n tweet['retweeted_status']['user']['id_str'],\n str(datetime.strptime(tweet['created_at'], '%a %b %d %H:%M:%S +0000 %Y')),1]]\n , columns=('Source','Target',\"Time\",'Strength')), ignore_index=True) ",
"_____no_output_____"
],
[
"userdata.head()",
"_____no_output_____"
],
[
"edges.head()",
"_____no_output_____"
]
],
[
[
"# 2. Kanten erzeugen durch Mentions\n- Personen erwähnen sich und deshalb erzeugen wir eine Kante zwischen den Personen. ",
"_____no_output_____"
]
],
[
[
"fh = open(tweetfile, 'r')\n\nuserdata = pd.DataFrame(columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count' ))\nedges = pd.DataFrame(columns=('Source','Target','Strength'))\n\nfor line in fh:\n try:\n tweet = json.loads(line)\n except:\n continue\n if len(tweet['entities']['user_mentions']) == 0:\n continue\n \n for mention in tweet['entities']['user_mentions']:\n userdata = userdata.append(pd.DataFrame([[tweet['user']['id_str'],\n tweet['user']['screen_name'],\n tweet['user']['created_at'],\n tweet['user']['profile_image_url_https'],\n tweet['user']['followers_count'],\n tweet['user']['friends_count']]], columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count')), ignore_index=True)\n if len(userdata[userdata['Id'].str.contains(mention['id_str'])]) == 0:\n userdata = userdata.append(pd.DataFrame([[tweet['user']['id_str'],\n tweet['user']['screen_name'],\n np.nan,\n np.nan,\n np.nan,\n np.nan]], columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count')), ignore_index=True)\n edges = edges.append(pd.DataFrame([[tweet['user']['id_str'],\n mention['id_str'],\n str(datetime.strptime(tweet['created_at'], '%a %b %d %H:%M:%S +0000 %Y'))]]\n , columns=('Source','Target','Strength')), ignore_index=True) ",
"_____no_output_____"
]
],
[
[
"# 3. Kanten erzeugen durch gemeinsame Kommunikation\n- Personen diskutieren miteinander und deshalb erzeugen wir eine Kante zwischen ihnen. ",
"_____no_output_____"
]
],
[
[
"\nfh = open(tweetfile, 'r')\n\nuserdata = pd.DataFrame(columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count' ))\nedges = pd.DataFrame(columns=('Source','Target','Strength'))\n\nfor line in fh:\n try:\n tweet = json.loads(line)\n except:\n continue\n if tweet['in_reply_to_user_id_str'] is None:\n continue\n\n userdata = userdata.append(pd.DataFrame([[tweet['user']['id_str'],\n tweet['user']['screen_name'],\n tweet['user']['created_at'],\n tweet['user']['profile_image_url_https'],\n tweet['user']['followers_count'],\n tweet['user']['friends_count']]], columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count')), ignore_index=True)\n if len(userdata[userdata['Id'].str.contains(tweet['in_reply_to_user_id_str'])]) == 0:\n userdata = userdata.append(pd.DataFrame([[tweet['in_reply_to_user_id_str'],\n tweet['in_reply_to_screen_name'],\n np.nan,\n np.nan,\n np.nan,\n np.nan]], columns=('Id','Label','user_created_at','profile_image','followers_count','friends_count')), ignore_index=True)\n edges = edges.append(pd.DataFrame([[tweet['user']['id_str'],\n tweet['in_reply_to_user_id_str'],\n str(datetime.strptime(tweet['created_at'], '%a %b %d %H:%M:%S +0000 %Y'))]]\n , columns=('Source','Target','Strength')), ignore_index=True)",
"_____no_output_____"
]
],
[
[
"# Nur jene Kanten behalten die eine gewisse Stärke haben.",
"_____no_output_____"
]
],
[
[
"strengthLevel = 3 # Network connection strength level: the number of times in total each of the tweeters responded to or mentioned the other.\n # If you have 1 as the level, then all tweeters who mentioned or replied to another at least once will be displayed. But if you have 5, only those who have mentioned or responded to a particular tweeter at least 5 times will be displayed, which means that only the strongest bonds are shown.\n\nedges2 = edges.groupby(['Source','Target'])['Strength'].count()\nedges2 = edges2.reset_index()\nedges2 = edges2[edges2['Strength'] >= strengthLevel]",
"_____no_output_____"
],
[
"len(edges2)",
"_____no_output_____"
]
],
[
[
"# Daten als Gephi Netzwerk Exportieren",
"_____no_output_____"
]
],
[
[
"def robust_decode(bs):\n '''Takes a byte string as param and convert it into a unicode one.\nFirst tries UTF8, and fallback to Latin1 if it fails'''\n cr = None\n cr = bs.decode('ascii', 'ignore').encode('ascii')\n return cr",
"_____no_output_____"
],
[
"import sys\nreload(sys)\nsys.setdefaultencoding('utf8')\nuserdata = userdata.sort_values(['Id','followers_count'], ascending=[True, False])\nuserdata = userdata.drop_duplicates(['Id'], keep='first') \n\nids = edges2['Source'].append(edges2['Target']).to_frame()\nids.columns = ['Id']\nids = ids.drop_duplicates()\n\nnodes = pd.merge(ids, userdata, on='Id', how='left')\nnodes = nodes.dropna()\nnodes[\"Label\"] = nodes[\"Label\"].astype(str)\nnodes[\"Id\"] = nodes[\"Id\"].astype(str)\n\n\nG = nx.DiGraph(name=\"zürich\")\n\nfor i, row in nodes.iterrows():\n G.add_node(robust_decode(row[\"Id\"]), label=robust_decode(row[\"Label\"]))\n\nfor i, row in edges2.iterrows():\n G.add_edge(robust_decode(row[\"Source\"]),robust_decode(row[\"Target\"]),weight=row[\"Strength\"])\n\nnx.write_gexf(G,\"Zürich.gexf\")",
"_____no_output_____"
]
],
[
[
"# Alternativ als csv speichern für Kumu.io",
"_____no_output_____"
]
],
[
[
"# Export nodes from the edges and add node attributes for both Sources and Targets.\nuserdata = userdata.sort_values(['Id','followers_count'], ascending=[True, False])\nuserdata = userdata.drop_duplicates(['Id'], keep='first') \n\nids = edges2['Source'].append(edges2['Target']).to_frame()\nids.columns = ['Id']\nids = ids.drop_duplicates()\n\nnodes = pd.merge(ids, userdata, on='Id', how='left')\n\n# change column names for Kumu import (Run this when using Kumu)\nnodes.columns = ['Id', 'Label', 'Date', 'Image', 'followers_count', 'friends_count']\nedges2.columns = ['From','To','Strength']\n\n# Export nodes and edges to csv files\nnodes.to_csv('nodes.csv', encoding='utf-8', index=False)\nedges2.to_csv('edges.csv', encoding='utf-8', index=False)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a14d0a4f43d85f189c5093f3d781ef6927e44d7
| 70,550 |
ipynb
|
Jupyter Notebook
|
Supervised Learning/Regression Algorithms/Simple Linear Regression/Simple_Linear_Regression.ipynb
|
Jawwad-Fida/Machine-Learning-Algorithms
|
c326cd83850b771b979b8dfcbca6a54c508b035a
|
[
"MIT"
] | 1 |
2021-07-07T07:44:20.000Z
|
2021-07-07T07:44:20.000Z
|
Supervised Learning/Regression Algorithms/Simple Linear Regression/Simple_Linear_Regression.ipynb
|
Jawwad-Fida/Machine-Learning-Algorithms
|
c326cd83850b771b979b8dfcbca6a54c508b035a
|
[
"MIT"
] | null | null | null |
Supervised Learning/Regression Algorithms/Simple Linear Regression/Simple_Linear_Regression.ipynb
|
Jawwad-Fida/Machine-Learning-Algorithms
|
c326cd83850b771b979b8dfcbca6a54c508b035a
|
[
"MIT"
] | null | null | null | 108.538462 | 28,498 | 0.819391 |
[
[
[
"%matplotlib inline \nimport matplotlib.pyplot as plt # for plotting\nimport numpy as np # for matrix and vector computations\nimport pandas as pd\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"### Debugging\n\n* Python array indices start from zero\n* Vector/matrix operations work only with numpy arrays.Inspect matrix operations to make sure that you are adding and multiplying matrices of compatible dimensions. Printing the dimensions of numpy arrays using the shape property will help you debug.\n* If you want to do matrix multiplication, you need to use the dot function in numpy. For, example if A and B are two numpy matrices, then the matrix operation AB is np.dot(A, B)",
"_____no_output_____"
],
[
"## Return a 5x5 Identity Matrix",
"_____no_output_____"
]
],
[
[
"A = np.eye(5) # using eye()\nA",
"_____no_output_____"
]
],
[
[
"Implement linear regression with one variable to predict profits for a food truck. Suppose you are the CEO of a restaurant franchise and are considering different cities for opening a new outlet. The chain already has trucks in various cities and you have data for profits and populations from the cities. You would like to use this data to help you select which city to expand to next.\n\nThe file Data/ex1data1.txt contains the dataset for our linear regression problem. The first column is the population of a city (in 10,000s) and the second column is the profit of a food truck in that city (in $10,000s). A negative value for profit indicates a loss.\n",
"_____no_output_____"
],
[
"## 1) Load the dataset",
"_____no_output_____"
]
],
[
[
"# Load the dataset\ndata = np.loadtxt('ex1data1.txt',delimiter=',')\nX = data[:,0]\ny = data[:,1]\n\n# X and y are matrices ",
"_____no_output_____"
],
[
"m = y.size # number of training samples\nm",
"_____no_output_____"
],
[
"X.shape, y.shape, X.ndim, y.ndim",
"_____no_output_____"
]
],
[
[
"## 2) Plotting the Data\n\nBefore starting on any task, it is often useful to understand the data by visualizing it. For this dataset has only two properties to plot (profit and population).",
"_____no_output_____"
]
],
[
[
"\"\"\"\n Plots the data points x and y into a new figure. Plots the data \n points and gives the figure axes labels of population and profit.\n \n Parameters\n ----------\n x : array_like\n Data point values for x-axis.\n\n y : array_like\n Data point values for y-axis. Note x and y should have the same size.\n \n ----\n You can use the 'ro' option with plot to have the markers\n appear as red circles. Furthermore, you can make the markers larger by\n using plot(..., 'ro', ms=10), where `ms` refers to marker size. You \n can also set the marker edge color using the `mec` property.\n \"\"\"",
"_____no_output_____"
],
[
"def plotData(x,y):\n fig = plt.figure(figsize=(8,6))\n plt.plot(x,y,'ro',ms=10,mec='k')\n plt.xlabel('Profit in $10,000')\n plt.ylabel('Population of a city in 10,000')",
"_____no_output_____"
],
[
"plotData(X,y)",
"_____no_output_____"
]
],
[
[
"## 3) Gradient Descent\n\nFit the linear regression parameters $\\theta$ to the dataset using gradient descent.",
"_____no_output_____"
],
[
"<a id=\"section2\"></a>\n### 3.1 Update Equations\n\nThe objective of linear regression is to minimize the cost function $J(\\theta)$\n\n$$ J(\\theta) = \\frac{1}{2m} \\sum_{i=1}^m \\left( h_{\\theta}(x^{(i)}) - y^{(i)}\\right)^2$$\n\nwhere the hypothesis $h_\\theta(x)$ is given by the linear model\n$$ h_\\theta(x) = \\theta^Tx = \\theta_0 + \\theta_1 x_1$$\n\nRecall that the parameters of your model are the $\\theta_j$ values. These are\nthe values you will adjust to minimize cost $J(\\theta)$. One way to do this is to\nuse the **batch gradient descent algorithm**. In batch gradient descent, each\niteration performs the update\n\n$$ \\theta_j = \\theta_j - \\alpha \\frac{1}{m} \\sum_{i=1}^m \\left( h_\\theta(x^{(i)}) - y^{(i)}\\right)x_j^{(i)} \\qquad \\text{simultaneously update } \\theta_j \\text{ for all } j$$\n\nWith each step of gradient descent, your parameters $\\theta_j$ come closer to the optimal values that will achieve the lowest cost J($\\theta$).\n\n<div class=\"alert alert-block alert-warning\">\n**Implementation Note:** We store each sample as a row in the the $X$ matrix in Python `numpy`. To take into account the intercept term ($\\theta_0$), we add an additional first column to $X$ and set it to all ones. This allows us to treat $\\theta_0$ as simply another 'feature'.\n</div>\n",
"_____no_output_____"
]
],
[
[
"# initially X contains features x1,x2. Add x0 = 1, so X will now contain the features x0,x1,x2\n\n#### Add a column of ones to X. The numpy function stack() joins arrays along a given axis. \n\n# The first axis (axis=0) refers to rows (training samples), and second axis (axis=1) refers to columns (features).\n\nX = np.stack([np.ones(m),X],axis=1) # This cell is executed only once!",
"_____no_output_____"
]
],
[
[
"<a id=\"section2\"></a>\n### 3.2 Computing the cost $J(\\theta)$\n\nAs you perform gradient descent to minimize the cost function $J(\\theta)$, it is helpful to monitor the convergence by computing the cost. Implement a function to calculate $J(\\theta)$ so you can check the convergence of your gradient descent implementation. \n\nRemember that the variables $X$ and $y$ are not scalar values. $X$ is a matrix whose rows represent the samples from the training set (feature) and $y$ (label) is a vector whose each element represent the value at a given row of $X$.\n<a id=\"computeCost\"></a>",
"_____no_output_____"
]
],
[
[
"\"\"\"\n Compute cost for linear regression. Computes the cost of using theta as the\n parameter for linear regression to fit the data points in X and y.\n \n Parameters\n ----------\n X : array_like\n The input dataset of shape (m x n+1) dimesnions, where m is the number of samples,\n and n is the number of features. We assume a vector of one's already \n appended to the features so we have n+1 columns.\n \n y : array_like\n The values of the function at each data point. This is a vector of\n shape (m, ) i.e. (mx1) dimensions\n \n theta : array_like\n The parameters for the hypothesis/regression function. This is a vector of \n shape (n+1, ) i.e. (n+1)x1 dimensions.\n \n Returns\n -------\n J : float - The value of the regression cost function.\n \"\"\"",
"_____no_output_____"
],
[
"def computeCost(X,y,theta):\n m = y.size # no. of training samples\n J = 0\n\n h = np.dot(X,theta) # X and theta are matrices\n J = (1/(2 * m)) * np.sum(np.square(np.dot(X, theta) - y))\n\n return J\n",
"_____no_output_____"
],
[
"# take random values of theta0 and theta1\n\nJ = computeCost(X,y ,theta=np.array([0.0,0.0])) # two values for theta0 and theta1\nprint(f\"With theta = [0, 0] \\nCost computed = {J:.2f}\")\nprint()\n\nJ = computeCost(X,y ,theta=np.array([-1,2])) \nprint(f\"With theta = [-1, 2] \\nCost computed = {J:.2f}\")",
"With theta = [0, 0] \nCost computed = 32.07\n\nWith theta = [-1, 2] \nCost computed = 54.24\n"
]
],
[
[
"<a id=\"section3\"></a>\n### 3.3 Gradient descent\n\nComplete a function which Implements gradient descent. Update $\\theta$ with each iteration of the loop. \n\nAs you program, make sure you understand what you are trying to optimize and what is being updated. Keep in mind that the cost $J(\\theta)$ is parameterized by the vector $\\theta$, not $X$ and $y$. That is, we minimize the value of $J(\\theta)$ by changing the values of the vector $\\theta$, not by changing $X$ or $y$. \n\nA good way to verify that gradient descent is working correctly is to look at the value of $J(\\theta)$ and check that it is decreasing with each step. \n",
"_____no_output_____"
]
],
[
[
" \"\"\"\n Performs gradient descent to learn `theta`. Updates theta by taking `num_iters`\n gradient steps with learning rate `alpha`.\n \n Parameters\n ----------\n X : array_like\n The input dataset of shape (m x n+1).\n \n y : array_like\n Value at given features. A vector of shape (m, ), i.e. (mx1) dimensions\n \n theta : array_like\n Initial values for the linear regression parameters. \n A vector of shape (n+1, ), i.e. (n+1)x1 dimensions\n \n alpha : float\n The learning rate.\n \n num_iters : int\n The number of iterations for gradient descent. \n \n Returns\n -------\n theta : array_like\n The learned linear regression parameters. A vector of shape (n+1, ). This is the optimal theta\n for which J is minimum\n \n J_history : list\n A python list for the values of the cost function after each iteration.\n \n Instructions\n ------------\n Peform a single gradient step on the parameter vector theta.\n\n While debugging, it can be useful to print out the values of \n the cost function (computeCost) and gradient here.\n \"\"\"",
"_____no_output_____"
],
[
"def gradient_descent(X,y,theta,alpha,num_iters):\n m = y.size # or y.shape[0] # number of training samples\n\n # make a copy of theta, to avoid changing the original array, since numpy arrays are passed by reference to functions\n theta = theta.copy()\n\n J_history = [] # Use a python list to store cost in every iteration\n\n for i in range(num_iters):\n theta = theta - (alpha/m) * (np.dot(X,theta) - y).dot(X)\n # print(theta)\n\n # save the cost J in every iteration\n min_cost = computeCost(X,y,theta)\n J_history.append(min_cost)\n # print(J_history[i])\n\n return theta, J_history # theta will return 2 values --> theta0, theta1",
"_____no_output_____"
],
[
"# randomly initialize fitting parameters\ntheta = np.zeros(2)\n\n# some gradient descent settings\niterations = 1500\nalpha = 0.01\n\ntheta, J_history = gradient_descent(X,y,theta,alpha,iterations)\nprint('Theta found by gradient descent: {:.4f}, {:.4f}'.format(*theta)) # adds theta to empty string",
"Theta found by gradient descent: -3.6303, 1.1664\n"
]
],
[
[
"## 4) Plot the linear fit",
"_____no_output_____"
]
],
[
[
"plotData(X[:,1],y) # plot the samples - excluding x1=0 (0th column)\n\n# Linear regression line/hypothesis line of best fit --> y = h(x) = theta0 + theta1*X\n# x is feature except x1=0,y is entire equation\nplt.plot(X[:,1],np.dot(X,theta),ls='-') \n\nplt.legend(['Training Data','Linear Regression']); # x is training data, y is linear regression line",
"_____no_output_____"
]
],
[
[
"## 5) Predict some values",
"_____no_output_____"
]
],
[
[
"# we now have the optimal theta\n\n# Predict values for population sizes of 35,000 and 70,000\n\n# Note that the first argument to the `numpy` function `dot` is a python list. \n# `numpy` can internally convert **valid** python lists to numpy arrays when explicitly provided as arguments to `numpy` functions.\n\n# profit x in 10,000 and population y in 10,000, so 3.5 --> 350000, 1 -> 10000\npredict1 = np.dot([1,3.5],theta)\nprint(f\"For population = 35,000, we predict a profit of {predict1 * 10000:.2f}\")\n\npredict2 = np.dot([1,7],theta)\nprint(f\"For population = 35,000, we predict a profit of {predict2 * 10000:.2f}\")",
"For population = 35,000, we predict a profit of 4519.77\nFor population = 35,000, we predict a profit of 45342.45\n"
],
[
"",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a14d21d030676aabbd1f3831d1d5e7672eaa040
| 6,844 |
ipynb
|
Jupyter Notebook
|
1-Basic Data Processing and Visualization/Wk2/Reading CSV & JSON Files2.ipynb
|
garynth41/Python-Data-Products-for-Predictive-Analytics-Specialization
|
41934a56cf33ffe9231082d281e89173eee59b41
|
[
"MIT"
] | 2 |
2020-10-15T05:13:09.000Z
|
2022-02-05T05:52:14.000Z
|
1-Basic Data Processing and Visualization/Wk2/Reading CSV & JSON Files2.ipynb
|
garynth41/Python-Data-Products-for-Predictive-Analytics-Specialization
|
41934a56cf33ffe9231082d281e89173eee59b41
|
[
"MIT"
] | null | null | null |
1-Basic Data Processing and Visualization/Wk2/Reading CSV & JSON Files2.ipynb
|
garynth41/Python-Data-Products-for-Predictive-Analytics-Specialization
|
41934a56cf33ffe9231082d281e89173eee59b41
|
[
"MIT"
] | 1 |
2021-03-14T15:29:13.000Z
|
2021-03-14T15:29:13.000Z
| 22.966443 | 439 | 0.526447 |
[
[
[
"# <center> Basic Data Processing and Visualization</center> \n<center> Week 2 \n<center> Reading CSV & JSON Files</center>",
"_____no_output_____"
],
[
"# Code:eval() Function",
"_____no_output_____"
]
],
[
[
"import csv\nimport ast",
"_____no_output_____"
],
[
"path = \"datasets/review.json\"",
"_____no_output_____"
],
[
"f = open(path)",
"_____no_output_____"
],
[
"line = f.readline()",
"_____no_output_____"
],
[
"line # print the first line of the yelp review dataset",
"_____no_output_____"
],
[
"d = eval(line)",
"_____no_output_____"
],
[
"d",
"_____no_output_____"
],
[
"d['user_id']",
"_____no_output_____"
],
[
"# here python treats an arbitrary string as a bit of python code, which is nice, but could be \n# dangerous.\neval(\"2 + 4\") ",
"_____no_output_____"
]
],
[
[
"#### To prevent the execution of arbitrary code, you should use, ast or json library",
"_____no_output_____"
]
],
[
[
"ast.literal_eval(line)",
"_____no_output_____"
]
],
[
[
"Both eval() and ast.literal_eval() evaluate strings containing Python code in the current Python environment, but ast.literal_eval() restricts its evaluation to a subset of valid Python datatypes, while eval() does not.",
"_____no_output_____"
],
[
"#### ast and json libraries",
"_____no_output_____"
]
],
[
[
"import json",
"_____no_output_____"
],
[
"json.loads(line)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
4a14e0428a446e03e66aba7bdf9d8abe90751ca4
| 8,967 |
ipynb
|
Jupyter Notebook
|
01Pytorch/Custom_Dataset_sol.ipynb
|
aonekoda/pytorch
|
1433ec75404bf0823e1499ba0a24293b6ff5ab37
|
[
"MIT"
] | null | null | null |
01Pytorch/Custom_Dataset_sol.ipynb
|
aonekoda/pytorch
|
1433ec75404bf0823e1499ba0a24293b6ff5ab37
|
[
"MIT"
] | null | null | null |
01Pytorch/Custom_Dataset_sol.ipynb
|
aonekoda/pytorch
|
1433ec75404bf0823e1499ba0a24293b6ff5ab37
|
[
"MIT"
] | null | null | null | 24.433243 | 117 | 0.489573 |
[
[
[
"# 과제\n\n데이터셋을 좀 더 쉽게 다룰 수 있도록 유용한 도구로서 torch.utils.data.Dataset과 torch.utils.data.DataLoader를 제공 한다. \n 이를 사용하면 미니 배치 학습, 데이터 셔플(shuffle), 병렬 처리까지 간단히 수행할 수 있다. 기본적인 사용 방법은 Dataset을 정의하고, 이를 DataLoader에 전달하는 것이다\n \n# 커스텀 데이터셋(Custom Dataset) \ntorch.utils.data.Dataset을 상속받아 직접 커스텀 데이터셋(Custom Dataset)을 만들 수 있다. \nDataset을 상속받아 다음 메소드들을 오버라이드 하여 커스텀 데이터셋을 생성해 보자. \n커스텀 데이터셋을 만들 때, 일단 가장 기본적인 뼈대는 아래와 같다. \n```\nclass CustomDataset(torch.utils.data.Dataset): \n def __init__(self):\n\n def __len__(self):\n\n def __getitem__(self, idx): \n```",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport torch\nfrom torch.utils.data import Dataset",
"_____no_output_____"
]
],
[
[
"### Diabetes dataset\nDiabetes dataset은 총 442명의 당뇨병 환자에 대한 자료이다. \nage, sex, body mass index, average blood pressure, 6개의 혈청값으로 이루어져 있다. \n442명의 당뇨병 환자를 대상으로한 검사 결과를 나타내는 데이터이다.\n\n- 타겟 데이터 : 1년 뒤 측정한 당뇨병의 진행률\n\n- 특징 데이터 (이 데이터셋의 특징 데이터는 모두 정규화된 값이다.)\n\n - Age\n - Sex\n - Body mass index\n - Average blood pressure\n - S1\n - S2\n - S3\n - S4\n - S5\n - S6\n",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('diabetes.csv')\ndf.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 442 entries, 0 to 441\nData columns (total 11 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 age 442 non-null float64\n 1 sex 442 non-null float64\n 2 bmi 442 non-null float64\n 3 bp 442 non-null float64\n 4 s1 442 non-null float64\n 5 s2 442 non-null float64\n 6 s3 442 non-null float64\n 7 s4 442 non-null float64\n 8 s5 442 non-null float64\n 9 s6 442 non-null float64\n 10 target 442 non-null float64\ndtypes: float64(11)\nmemory usage: 38.1 KB\n"
]
],
[
[
"### 문제 1\n다음을 참조하여 `diabetes.csv` 파일을 읽고 custom dataset으로 생성해 보시오.\n\n* len(dataset)을 했을 때 데이터셋의 크기를 리턴할 len\n* dataset[i]을 했을 때 i번째 샘플을 가져오도록 하는 인덱싱을 위한 get_item",
"_____no_output_____"
],
[
"# TODO : 다음의 코드를 완성하시오.\nclass CustomDataset(torch.utils.data.Dataset): \n def __init__(self):\n # 여기에 코드를 작성하시오.\n def __len__(self):\n # 여기에 코드를 작성하시오.\n def __getitem__(self, idx): \n # 여기에 코드를 작성하시오.",
"_____no_output_____"
]
],
[
[
"class CustomDataset(Dataset):\n def __init__(self):\n data = np.loadtxt('diabetes.csv', delimiter=',', dtype=np.float32, skiprows=1)\n self.x_data = torch.from_numpy(data[:, :-1]) # age,sex,bmi,bp,s1,s2,s3,s4,s5,s6\n self.y_data = torch.from_numpy(data[:, -1]).view(-1,1) # target\n def __getitem__(self, idx):\n x = self.x_data[idx]\n y = self.y_data[idx]\n return x, y\n def __len__(self):\n return len(self.x_data)",
"_____no_output_____"
]
],
[
[
"### 문제 2\ndataset을 생성하고 `__getitem__()` 속성을 사용하여 데이터를 조회해 보시오.",
"_____no_output_____"
]
],
[
[
"dataset = CustomDataset()\n\ndataset.__getitem__([0])",
"_____no_output_____"
]
],
[
[
"### 문제 3\n읽어들인 데이셋을 신경망 모형의 입력에 사용할 수 있도록 `DataLoader` 를 사용하시오. \n적절한 batch_size를 설정하시오.",
"_____no_output_____"
]
],
[
[
"from torch.utils.data import DataLoader\n\ndataloader = DataLoader(dataset, batch_size=10, shuffle=True)\n\nx, y = iter(dataloader).next()\n\nprint(x.shape, y.shape)",
"torch.Size([10, 10]) torch.Size([10, 1])\n"
]
],
[
[
"### 문제 4\ndiabetes를 예측하는 신경망 모형을 생성하시오.",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch import nn\n\nmodel = nn.Sequential(nn.Linear(10, 1))\nprint(model)",
"Sequential(\n (0): Linear(in_features=10, out_features=1, bias=True)\n)\n"
]
],
[
[
"### 문제 5\n\nloss 함수와 optimizer를 설정하시오. learning_rate를 적절히 선택하시오.",
"_____no_output_____"
]
],
[
[
"criterion = torch.nn.MSELoss()\noptimizer = torch.optim.SGD(model.parameters(), lr=0.0001) ",
"_____no_output_____"
]
],
[
[
"### 문제 6\n모형을 훈련하시오. epoch의 횟수를 적절히 선택하시오.",
"_____no_output_____"
]
],
[
[
"epochs = 1000\n\nfor epoch in range(epochs):\n running_loss = 0\n for data, target in dataloader:\n optimizer.zero_grad()\n pred = model(data)\n loss = criterion(pred, target)\n loss.backward()\n optimizer.step()\n running_loss += loss.item()\n else:\n if epoch % 100 == 0 :\n print(f\"Training loss: {running_loss/len(dataloader)}\")",
"Training loss: 28609.126323784723\nTraining loss: 9504.185909016927\nTraining loss: 6431.6843044704865\nTraining loss: 5825.768329196506\nTraining loss: 5974.438216145833\nTraining loss: 5753.489246961805\nTraining loss: 5752.729806857639\nTraining loss: 5658.602563476563\nTraining loss: 5636.333854166666\nTraining loss: 5628.204378255208\n"
]
],
[
[
"### 문제 7\n```\nnew_var = torch.FloatTensor([[ 0.0381, 0.0507, 0.0617, 0.0219, -0.0442, -0.0348, \n -0.0434, -0.0026, 0.0199, -0.0176]])\n```\n새로운 데이터로 diabets를 예측해 보시오.",
"_____no_output_____"
]
],
[
[
"new_var = torch.FloatTensor([[ 0.0381, 0.0507, 0.0617, 0.0219, -0.0442, -0.0348, \n -0.0434, -0.0026, 0.0199, -0.0176]])\nwith torch.no_grad():\n y_hat = model(new_var)\n \nprint(y_hat)",
"tensor([[153.9861]])\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a14ff7b2c2115e482176518c6784ff4bdb9f0bb
| 66,790 |
ipynb
|
Jupyter Notebook
|
homeworks/D006/Day_006_outliers_detection.ipynb
|
peteryuX/100Day-ML-Marathon
|
cd61add6fa91ef117429eb1300cbdd96682d2d43
|
[
"MIT"
] | 6 |
2019-05-19T05:53:07.000Z
|
2020-04-18T05:02:13.000Z
|
homeworks/D006/Day_006_outliers_detection.ipynb
|
peteryuX/100Day-ML-Marathon
|
cd61add6fa91ef117429eb1300cbdd96682d2d43
|
[
"MIT"
] | null | null | null |
homeworks/D006/Day_006_outliers_detection.ipynb
|
peteryuX/100Day-ML-Marathon
|
cd61add6fa91ef117429eb1300cbdd96682d2d43
|
[
"MIT"
] | 1 |
2019-11-20T14:33:12.000Z
|
2019-11-20T14:33:12.000Z
| 101.969466 | 22,482 | 0.790133 |
[
[
[
"# 檢視 Outliers\n### 為何會有 outliers, 常見的 outlier 原因\n* 未知值,隨意填補 (約定俗成的代入),如年齡常見 0,999\n* 可能的錯誤紀錄/手誤/系統性錯誤,如某本書在某筆訂單的銷售量 = 1000 本",
"_____no_output_____"
]
],
[
[
"# Import 需要的套件\nimport os\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\n%matplotlib inline\n\n# 設定 data_path\ndir_data = './data/'",
"_____no_output_____"
],
[
"f_app = os.path.join(dir_data, 'application_train.csv')\nprint('Path of read in data: %s' % (f_app))\napp_train = pd.read_csv(f_app)\napp_train.head()",
"Path of read in data: ./data/application_train.csv\n"
]
],
[
[
"## 開始檢視不同欄位是否有異常值",
"_____no_output_____"
]
],
[
[
"# DAYS_BIRTH: 客戶申請貸款時的年齡\n(app_train['DAYS_BIRTH'] / (-365)).describe()",
"_____no_output_____"
],
[
"# DAYS_EMPLOYED: 申請貸款前,申請人已在現職工作的時間\n(app_train['DAYS_EMPLOYED'] / 365).describe()\nplt.hist(app_train['DAYS_EMPLOYED'])\nplt.show()\napp_train['DAYS_EMPLOYED'].value_counts()",
"_____no_output_____"
]
],
[
[
"### 從上面的圖與數值可以看出, 365243 顯然是個奇怪的數值",
"_____no_output_____"
]
],
[
[
"anom = app_train[app_train['DAYS_EMPLOYED'] == 365243]\nnon_anom = app_train[app_train['DAYS_EMPLOYED'] != 365243]\nprint('The non-anomalies default on %0.2f%% of loans' % (100 * non_anom['TARGET'].mean()))\nprint('The anomalies default on %0.2f%% of loans' % (100 * anom['TARGET'].mean()))\nprint('There are %d anomalous days of employment' % len(anom))",
"The non-anomalies default on 8.66% of loans\nThe anomalies default on 5.40% of loans\nThere are 55374 anomalous days of employment\n"
],
[
"# 新增一個欄位: DAYS_EMPLOYED_ANOM 來標記 DAYS_EMPLOYED 是否異常\napp_train['DAYS_EMPLOYED_ANOM'] = app_train[\"DAYS_EMPLOYED\"] == 365243\nprint(app_train['DAYS_EMPLOYED_ANOM'].value_counts())\n\n# 這邊我們用 nan 將異常值取代\napp_train['DAYS_EMPLOYED'].replace({365243: np.nan}, inplace = True)\n\napp_train['DAYS_EMPLOYED'].plot.hist(title = 'Days Employment Histogram');\nplt.xlabel('Days Employment');",
"False 252137\nTrue 55374\nName: DAYS_EMPLOYED_ANOM, dtype: int64\n"
],
[
"# 檢查 OWN_CAR_AGE: 貸款人的車齡\nplt.hist(app_train[~app_train.OWN_CAR_AGE.isnull()]['OWN_CAR_AGE'])\nplt.show()\napp_train['OWN_CAR_AGE'].value_counts()",
"_____no_output_____"
],
[
"app_train[app_train['OWN_CAR_AGE'] > 50]['OWN_CAR_AGE'].value_counts()",
"_____no_output_____"
]
],
[
[
"### 從上面我們可以發現車齡為 64, 65 的人特別多,是否合理?\n* 記得,這沒有正確答案 - 但我們總是可以給他們一些標記,讓最後的模型來決定",
"_____no_output_____"
]
],
[
[
"print(\"Target of OWN_CAR_AGE >= 50: %.2f%%\" % (app_train[app_train['OWN_CAR_AGE'] >= 50 ]['TARGET'].mean() * 100 ))\nprint(\"Target of OWN_CAR_AGE < 50: %.2f%%\" % (app_train[app_train['OWN_CAR_AGE'] < 50]['TARGET'].mean() * 100))\n\napp_train['OWN_CAR_AGE_ANOM'] = app_train['OWN_CAR_AGE'] >= 50",
"Target of OWN_CAR_AGE >= 50: 8.36%\nTarget of OWN_CAR_AGE < 50: 7.21%\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a1507cc797c285340b0fd73d30ada4f5bd2220f
| 593,536 |
ipynb
|
Jupyter Notebook
|
Model Chart/model.ipynb
|
vmoraesfl/infectious_disease_modelling
|
74d0d5ac4d7e71ee22f22a63850af9bcf09be12f
|
[
"MIT"
] | 1 |
2020-06-09T02:24:28.000Z
|
2020-06-09T02:24:28.000Z
|
Model Chart/model.ipynb
|
vmoraesfl/FightThePandemicProject
|
74d0d5ac4d7e71ee22f22a63850af9bcf09be12f
|
[
"MIT"
] | null | null | null |
Model Chart/model.ipynb
|
vmoraesfl/FightThePandemicProject
|
74d0d5ac4d7e71ee22f22a63850af9bcf09be12f
|
[
"MIT"
] | 1 |
2020-06-02T21:32:44.000Z
|
2020-06-02T21:32:44.000Z
| 528.527159 | 32,402 | 0.782276 |
[
[
[
"<a href=\"https://colab.research.google.com/github/hf2000510/infectious_disease_modelling/blob/master/part_two.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"Make sure to open in Colab to see the plots!",
"_____no_output_____"
],
[
"### Importing the libraries",
"_____no_output_____"
]
],
[
[
"from scipy.integrate import odeint\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline \n!pip install mpld3\nimport mpld3\nmpld3.enable_notebook()",
"_____no_output_____"
]
],
[
[
"### Plot Function",
"_____no_output_____"
]
],
[
[
"def plotseird(t, S, E, I, R, D=None, L=None, R0=None, Alpha=None, CFR=None):\n f, ax = plt.subplots(1,1,figsize=(10,4))\n ax.plot(t, S, 'b', alpha=0.7, linewidth=2, label='Susceptible')\n ax.plot(t, E, 'y', alpha=0.7, linewidth=2, label='Exposed')\n ax.plot(t, I, 'r', alpha=0.7, linewidth=2, label='Infected')\n ax.plot(t, R, 'g', alpha=0.7, linewidth=2, label='Recovered')\n if D is not None:\n ax.plot(t, D, 'k', alpha=0.7, linewidth=2, label='Dead')\n ax.plot(t, S+E+I+R+D, 'c--', alpha=0.7, linewidth=2, label='Total')\n else:\n ax.plot(t, S+E+I+R, 'c--', alpha=0.7, linewidth=2, label='Total')\n\n ax.set_xlabel('Time (days)')\n\n ax.yaxis.set_tick_params(length=0)\n ax.xaxis.set_tick_params(length=0)\n ax.grid(b=True, which='major', c='w', lw=2, ls='-')\n legend = ax.legend(borderpad=2.0)\n legend.get_frame().set_alpha(0.5)\n for spine in ('top', 'right', 'bottom', 'left'):\n ax.spines[spine].set_visible(False)\n if L is not None:\n plt.title(\"Lockdown after {} days\".format(L))\n plt.show();\n\n if R0 is not None or CFR is not None:\n f = plt.figure(figsize=(12,4))\n \n if R0 is not None:\n # sp1\n ax1 = f.add_subplot(121)\n ax1.plot(t, R0, 'b--', alpha=0.7, linewidth=2, label='R_0')\n\n ax1.set_xlabel('Time (days)')\n ax1.title.set_text('R_0 over time')\n # ax.set_ylabel('Number (1000s)')\n # ax.set_ylim(0,1.2)\n ax1.yaxis.set_tick_params(length=0)\n ax1.xaxis.set_tick_params(length=0)\n ax1.grid(b=True, which='major', c='w', lw=2, ls='-')\n legend = ax1.legend()\n legend.get_frame().set_alpha(0.5)\n for spine in ('top', 'right', 'bottom', 'left'):\n ax.spines[spine].set_visible(False)\n\n if Alpha is not None:\n # sp2\n ax2 = f.add_subplot(122)\n ax2.plot(t, Alpha, 'r--', alpha=0.7, linewidth=2, label='alpha')\n\n ax2.set_xlabel('Time (days)')\n ax2.title.set_text('fatality rate over time')\n # ax.set_ylabel('Number (1000s)')\n # ax.set_ylim(0,1.2)\n ax2.yaxis.set_tick_params(length=0)\n ax2.xaxis.set_tick_params(length=0)\n ax2.grid(b=True, which='major', c='w', lw=2, ls='-')\n legend = ax2.legend()\n legend.get_frame().set_alpha(0.5)\n for spine in ('top', 'right', 'bottom', 'left'):\n ax.spines[spine].set_visible(False)\n\n plt.show();",
"_____no_output_____"
]
],
[
[
"## Basic SIR Equations",
"_____no_output_____"
]
],
[
[
"def deriv(y, t, N, beta, gamma):\n S, I, R = y\n dSdt = -beta * S * I / N\n dIdt = beta * S * I / N - gamma * I\n dRdt = gamma * I\n return dSdt, dIdt, dRdt",
"_____no_output_____"
]
],
[
[
"## The Exposed-Compartment",
"_____no_output_____"
]
],
[
[
"def deriv(y, t, N, beta, gamma, delta):\n S, E, I, R = y\n dSdt = -beta * S * I / N\n dEdt = beta * S * I / N - delta * E\n dIdt = delta * E - gamma * I\n dRdt = gamma * I\n return dSdt, dEdt, dIdt, dRdt",
"_____no_output_____"
]
],
[
[
"### Variables that we define:",
"_____no_output_____"
]
],
[
[
"N = 1_000_000 # total population\nD = 4.0 # infections lasts four days\ngamma = 1.0 / D\ndelta = 1.0 / 5.0 # incubation period of five days\nR_0 = 5.0\nbeta = R_0 * gamma # R_0 = beta / gamma, so beta = R_0 * gamma\nS0, E0, I0, R0 = N-1, 1, 0, 0 # initial conditions: one exposed",
"_____no_output_____"
],
[
"t = np.linspace(0, 99, 100) # Grid of time points (in days)\ny0 = S0, E0, I0, R0 # Initial conditions vector\n\n# Integrate the SIR equations over the time grid, t.\nret = odeint(deriv, y0, t, args=(N, beta, gamma, delta))\nS, E, I, R = ret.T",
"_____no_output_____"
]
],
[
[
"### Plot the result:",
"_____no_output_____"
]
],
[
[
"plotseird(t, S, E, I, R)",
"_____no_output_____"
]
],
[
[
"## Programming the Dead-Compartment",
"_____no_output_____"
]
],
[
[
"def deriv(y, t, N, beta, gamma, delta, alpha, rho):\n S, E, I, R, D = y\n dSdt = -beta * S * I / N\n dEdt = beta * S * I / N - delta * E\n dIdt = delta * E - (1 - alpha) * gamma * I - alpha * rho * I\n dRdt = (1 - alpha) * gamma * I\n dDdt = alpha * rho * I\n return dSdt, dEdt, dIdt, dRdt, dDdt",
"_____no_output_____"
]
],
[
[
"### New variables:",
"_____no_output_____"
]
],
[
[
"N = 1_000_000\nD = 4.0 # infections lasts four days\ngamma = 1.0 / D\ndelta = 1.0 / 5.0 # incubation period of five days\nR_0 = 5.0\nbeta = R_0 * gamma # R_0 = beta / gamma, so beta = R_0 * gamma\nalpha = 0.2 # 20% death rate\nrho = 1/9 # 9 days from infection until death\nS0, E0, I0, R0, D0 = N-1, 1, 0, 0, 0 # initial conditions: one exposed",
"_____no_output_____"
],
[
"t = np.linspace(0, 99, 100) # Grid of time points (in days)\ny0 = S0, E0, I0, R0, D0 # Initial conditions vector\n\n# Integrate the SIR equations over the time grid, t.\nret = odeint(deriv, y0, t, args=(N, beta, gamma, delta, alpha, rho))\nS, E, I, R, D = ret.T",
"_____no_output_____"
]
],
[
[
"### Plot the result:",
"_____no_output_____"
]
],
[
[
"plotseird(t, S, E, I, R, D)",
"_____no_output_____"
]
],
[
[
"## Time-Dependent $R_{0}$",
"_____no_output_____"
],
[
"### Simple Approach: Single Lockdown",
"_____no_output_____"
]
],
[
[
"def deriv(y, t, N, beta, gamma, delta, alpha, rho):\n S, E, I, R, D = y\n dSdt = -beta(t) * S * I / N\n dEdt = beta(t) * S * I / N - delta * E\n dIdt = delta * E - (1 - alpha) * gamma * I - alpha * rho * I\n dRdt = (1 - alpha) * gamma * I\n dDdt = alpha * rho * I\n return dSdt, dEdt, dIdt, dRdt, dDdt",
"_____no_output_____"
],
[
"L = 40",
"_____no_output_____"
],
[
"N = 1_000_000\nD = 4.0 # infections lasts four days\ngamma = 1.0 / D\ndelta = 1.0 / 5.0 # incubation period of five days\ndef R_0(t):\n return 5.0 if t < L else 0.9\ndef beta(t):\n return R_0(t) * gamma\n\nalpha = 0.2 # 20% death rate\nrho = 1/9 # 9 days from infection until death\nS0, E0, I0, R0, D0 = N-1, 1, 0, 0, 0 # initial conditions: one exposed",
"_____no_output_____"
],
[
"t = np.linspace(0, 99, 100) # Grid of time points (in days)\ny0 = S0, E0, I0, R0, D0 # Initial conditions vector\n\n# Integrate the SIR equations over the time grid, t.\nret = odeint(deriv, y0, t, args=(N, beta, gamma, delta, alpha, rho))\nS, E, I, R, D = ret.T",
"_____no_output_____"
]
],
[
[
"### Plot the result:",
"_____no_output_____"
]
],
[
[
"plotseird(t, S, E, I, R, D, L)",
"_____no_output_____"
]
],
[
[
"### Advanced Approach: logistic $R_{0}$",
"_____no_output_____"
]
],
[
[
"### we will use the logistic R in our model, because R probably never “jumps” from one value to another. Rather, it continuously changes. ",
"_____no_output_____"
],
[
"def deriv(y, t, N, beta, gamma, delta, alpha, rho):\n S, E, I, R, D = y\n dSdt = -beta(t) * S * I / N\n dEdt = beta(t) * S * I / N - delta * E\n dIdt = delta * E - (1 - alpha) * gamma * I - alpha * rho * I\n dRdt = (1 - alpha) * gamma * I\n dDdt = alpha * rho * I\n return dSdt, dEdt, dIdt, dRdt, dDdt",
"_____no_output_____"
],
[
"N = 1_000_000\nD = 4.0 # infections lasts four days\ngamma = 1.0 / D\ndelta = 1.0 / 5.0 # incubation period of five days\n\nR_0_start, k, x0, R_0_end = 5.0, 0.5, 50, 0.5\n\ndef logistic_R_0(t):\n return (R_0_start-R_0_end) / (1 + np.exp(-k*(-t+x0))) + R_0_end\n\ndef beta(t):\n return logistic_R_0(t) * gamma\n\nalpha = 0.2 # 20% death rate\nrho = 1/9 # 9 days from infection until death\nS0, E0, I0, R0, D0 = N-1, 1, 0, 0, 0 # initial conditions: one exposed",
"_____no_output_____"
],
[
"t = np.linspace(0, 99, 100) # Grid of time points (in days)\ny0 = S0, E0, I0, R0, D0 # Initial conditions vector\n\n# Integrate the SIR equations over the time grid, t.\nret = odeint(deriv, y0, t, args=(N, beta, gamma, delta, alpha, rho))\nS, E, I, R, D = ret.T\nR0_over_time = [logistic_R_0(i) for i in range(len(t))] # to plot R_0 over time: get function values",
"_____no_output_____"
]
],
[
[
"### Plot the result:",
"_____no_output_____"
]
],
[
[
"plotseird(t, S, E, I, R, D, R0=R0_over_time)",
"_____no_output_____"
]
],
[
[
"## Resource- and Age-Dependent Fatality Rate",
"_____no_output_____"
]
],
[
[
"def deriv(y, t, N, beta, gamma, delta, alpha_opt, rho):\n S, E, I, R, D = y\n def alpha(t):\n return s * I/N + alpha_opt\n\n dSdt = -beta(t) * S * I / N\n dEdt = beta(t) * S * I / N - delta * E\n dIdt = delta * E - (1 - alpha(t)) * gamma * I - alpha(t) * rho * I\n dRdt = (1 - alpha(t)) * gamma * I\n dDdt = alpha(t) * rho * I\n return dSdt, dEdt, dIdt, dRdt, dDdt",
"_____no_output_____"
],
[
"### New variables:",
"_____no_output_____"
],
[
"N = 1_000_000\nD = 4.0 # infections lasts four days\ngamma = 1.0 / D\ndelta = 1.0 / 5.0 # incubation period of five days\n\nR_0_start, k, x0, R_0_end = 5.0, 0.5, 50, 0.5\n\ndef logistic_R_0(t):\n return (R_0_start-R_0_end) / (1 + np.exp(-k*(-t+x0))) + R_0_end\n\ndef beta(t):\n return logistic_R_0(t) * gamma\n\nalpha_by_agegroup = {\"0-29\": 0.01, \"30-59\": 0.05, \"60-89\": 0.2, \"89+\": 0.3}\nproportion_of_agegroup = {\"0-29\": 0.1, \"30-59\": 0.3, \"60-89\": 0.4, \"89+\": 0.2}\ns = 0.01\nalpha_opt = sum(alpha_by_agegroup[i] * proportion_of_agegroup[i] for i in list(alpha_by_agegroup.keys()))\n\nrho = 1/9 # 9 days from infection until death\nS0, E0, I0, R0, D0 = N-1, 1, 0, 0, 0 # initial conditions: one exposed",
"_____no_output_____"
],
[
"t = np.linspace(0, 99, 100) # Grid of time points (in days)\ny0 = S0, E0, I0, R0, D0 # Initial conditions vector\n\n# Integrate the SIR equations over the time grid, t.\nret = odeint(deriv, y0, t, args=(N, beta, gamma, delta, alpha_opt, rho))\nS, E, I, R, D = ret.T\nR0_over_time = [logistic_R_0(i) for i in range(len(t))] # to plot R_0 over time: get function values\nAlpha_over_time = [s * I[i]/N + alpha_opt for i in range(len(t))] # to plot alpha over time",
"_____no_output_____"
]
],
[
[
"### Plot the result:",
"_____no_output_____"
]
],
[
[
"plotseird(t, S, E, I, R, D, R0=R0_over_time, Alpha=Alpha_over_time)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a15132fc1dc6c7cc35c9beb121423ece79978b5
| 120,469 |
ipynb
|
Jupyter Notebook
|
NHL_data_shape_notebooks/Approach_3_bulk_Leung_data_process/Merge_Leung_clean_up_v5.ipynb
|
joeyamosjohns/nhl_prediction_Aug_2021
|
3b3f24cc7f0eff0aca5b7f1d8f85a90b02dbd52b
|
[
"MIT"
] | null | null | null |
NHL_data_shape_notebooks/Approach_3_bulk_Leung_data_process/Merge_Leung_clean_up_v5.ipynb
|
joeyamosjohns/nhl_prediction_Aug_2021
|
3b3f24cc7f0eff0aca5b7f1d8f85a90b02dbd52b
|
[
"MIT"
] | null | null | null |
NHL_data_shape_notebooks/Approach_3_bulk_Leung_data_process/Merge_Leung_clean_up_v5.ipynb
|
joeyamosjohns/nhl_prediction_Aug_2021
|
3b3f24cc7f0eff0aca5b7f1d8f85a90b02dbd52b
|
[
"MIT"
] | null | null | null | 32.76285 | 297 | 0.434361 |
[
[
[
"\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"##bring in Leung data (data is from Leung's Git Repo on betting on nhl 2018); he scraped it from nhl.com\n\n\npathjj = '/Users/joejohns/' \ndata_path = 'data_bootcamp/GitHub/final_project_nhl_prediction/Data/Leung_Data_Results/nhl_data_Leung.csv'\nresults_path = 'data_bootcamp/GitHub/final_project_nhl_prediction/Data/Leung_Data_Results/results_Leung.csv'\nL_data = pd.read_csv(pathjj+data_path) #stats CF etc ... one line every data and team so num_dates*30? rows\nL_res = pd.read_csv(pathjj+results_path) #results win, loss etc\n#df_data.dropna(inplace = True)",
"_____no_output_____"
],
[
"#My old data sets (next two cells)\n\n\nKaggle_path = \"/Users/joejohns/data_bootcamp/GitHub/final_project_nhl_prediction/Data/Kaggle_Data_Ellis/\"\nmp_path = \"/Users/joejohns/data_bootcamp/GitHub/final_project_nhl_prediction/Data/Money_Puck_Data/\"\nbetting_path = \"/Users/joejohns/data_bootcamp/GitHub/final_project_nhl_prediction/Data/Betting_Data/\"\n#data_bootcamp/GitHub/final_project_nhl_prediction/Data/Betting_Data/nhl odds 2007-08.xlsx\n#data_bootcamp/GitHub/final_project_nhl_prediction/Data/Kaggle_Data_Ellis/\n#data_bootcamp/GitHub/final_project_nhl_prediction/Data/Money_Puck_Data/\n\n\n##Kaggle files\n\ndf_game = pd.read_csv(Kaggle_path+'game.csv')\ndf_game_team_stats = pd.read_csv(Kaggle_path+'game_teams_stats.csv')\n\ndf_team_info = pd.read_csv(Kaggle_path+'team_info.csv')\n",
"_____no_output_____"
],
[
"##? not sure what this is ... \nfrom numpy.core.numeric import True_\n\n\n##import all the files \n\n##file paths\n\nKaggle_path = \"/Users/joejohns/data_bootcamp/GitHub/final_project_nhl_prediction/Data/Kaggle_Data_Ellis/\"\nmp_path = \"/Users/joejohns/data_bootcamp/GitHub/final_project_nhl_prediction/Data/Money_Puck_Data/\"\nbetting_path = \"/Users/joejohns/data_bootcamp/GitHub/final_project_nhl_prediction/Data/Betting_Data/\"\n#data_bootcamp/GitHub/final_project_nhl_prediction/Data/Betting_Data/nhl odds 2007-08.xlsx\n#data_bootcamp/GitHub/final_project_nhl_prediction/Data/Kaggle_Data_Ellis/\n#data_bootcamp/GitHub/final_project_nhl_prediction/Data/Money_Puck_Data/\n\n\n##Kaggle files\n\ndf_game = pd.read_csv(Kaggle_path+'game.csv')\ndf_game_team_stats = pd.read_csv(Kaggle_path+'game_teams_stats.csv')\ndf_game_skater_stats = pd.read_csv(Kaggle_path+'game_skater_stats.csv')\ndf_game_goalie_stats = pd.read_csv(Kaggle_path+'game_goalie_stats.csv')\n\n##more subtle Kaggle features:\ndf_game_scratches = pd.read_csv(Kaggle_path+'game_scratches.csv')\ndf_game_officials = pd.read_csv(Kaggle_path+'game_officials.csv')\ndf_team_info = pd.read_csv(Kaggle_path+'team_info.csv')\n\n## grab all the moneypuck data \n\ndf_mp_teams = pd.read_csv(mp_path+'all_teams.csv')\n\n\n## grab all betting data \ndf1 = pd.read_excel(io = betting_path+'nhl odds 2007-08.xlsx')\ndf2 = pd.read_excel(io = betting_path+'nhl odds 2008-09.xlsx')\ndf3 = pd.read_excel(io = betting_path+'nhl odds 2009-10.xlsx')\ndf4 = pd.read_excel(io = betting_path+'nhl odds 2010-11.xlsx')\ndf5 = pd.read_excel(io = betting_path+'nhl odds 2011-12.xlsx')\ndf6 = pd.read_excel(io = betting_path+'nhl odds 2012-13.xlsx')\ndf7 = pd.read_excel(io = betting_path+'nhl odds 2013-14.xlsx')\ndf8 = pd.read_excel(io = betting_path+'nhl odds 2014-15.xlsx')\ndf9 = pd.read_excel(io = betting_path+'nhl odds 2015-16.xlsx')\ndf10 = pd.read_excel(io = betting_path+'nhl odds 2016-17.xlsx')\ndf11 = pd.read_excel(io = betting_path+'nhl odds 2017-18.xlsx')\ndf12 = pd.read_excel(io = betting_path+'nhl odds 2018-19.xlsx')\ndf13 = pd.read_excel(io = betting_path+'nhl odds 2019-20.xlsx')\n\n\n\n\ndf1['season'] = 20072008\ndf2['season'] = 20082009\ndf3['season'] = 20092010\ndf4['season'] = 20102011\ndf5['season'] = 20112012\ndf6['season'] = 20122013\ndf7['season'] = 20132014\ndf8['season'] = 20142015\ndf9['season'] = 20152016\ndf10['season'] = 20162017\ndf11['season'] = 20172018\ndf12['season'] = 20182019\ndf13['season'] = 20192020\n\n##omit 20072008 \ndf_betting = pd.concat([df2, df3, df4, df5, df6, df7, df8, df9, df10, df11, df12, df13])",
"_____no_output_____"
],
[
"\ndef fav_pred(x):\n if x < 0:\n return 1\n if x>0:\n return 0\n if x==0:\n print(\"O odds detected, nan returned\")\n return np.NaN\nv_fav_pred = np.vectorize(fav_pred)\n\ndef implied_proba(odds):\n if odds > 0: \n return 100/(odds+100) #bet 100 to get 100+odds; profit = odds\n if odds < 0:\n return (-odds)/(-odds + 100) #bet |odds| to get 100+|odds|; profit = 100\n \n \n\ndf= df_betting.copy()\n\ndf",
"_____no_output_____"
]
],
[
[
"##Note Bene: In L_data or L_res team id wrong! not franchise id either ... ",
"_____no_output_____"
]
],
[
[
"## cleaning tool\n\n\ndef perc_null(X):\n \n total = X.isnull().sum().sort_values(ascending=False)\n data_types = X.dtypes\n percent = (X.isnull().sum()/X.isnull().count()).sort_values(ascending=False)\n\n missing_data = pd.concat([total, data_types, percent], axis=1, keys=['Total','Type' ,'Percent'])\n return missing_data\n",
"_____no_output_____"
],
[
"##fixing dates and season label \n\n\ndef to_string(n):\n s = str(n)\n if len(s) ==1:\n return \"0\"+s\n else:\n return s\n\n#makes mp_date for L_data '2014-1-16' --> 20140116 (integer)\n \ndef L_to_mp_date(s):\n ymd = s.split('-')\n y = ymd[0]\n m = ymd[1]\n d = ymd[2]\n if len(m) ==1:\n m = '0'+m\n if len(d) == 1:\n d = '0'+d\n return int(y+m+d)\n\n#test '2014-1-16'.split('-')\n#make_mp_date('2014-10-16') \n \n \n##for assigning dates in df_game\n#takes in date_time date and returns integer yyyymmdd eg 20080904\n##note this is df_mp format\n\ndef game_add_mp_date(date_time):\n d = date_time.day\n m = date_time.month\n y = date_time.year\n dd = to_string(d)\n mm = to_string(m)\n yyyy = str(y)\n return(int(yyyy+mm+dd))\n\n##test \ndate_time1 = pd.to_datetime('2016-10-19T00:30:00Z')\ndate_time2 = pd.to_datetime('2008-09-19T00:30:00Z')\n\nprint(\"two tests for game_add_mp_date:\")\nprint(game_add_mp_date(date_time1) == 20161019)\nprint(game_add_mp_date(date_time2)==20080919)\nprint(\" \")\n##for assigning dates to df_betting\n#takes in ingegers date = 913 or 1026 season = 20082009 \n# returns returns integer yyyymmdd eg 20080904\n#note this is df_mp format\n\ndef bet_add_mp_date(date, season):\n d = str(date)\n s = str(season)\n if len(d) == 3:\n d = \"0\"+d \n if (900 < date):\n y= s[:4]\n else:\n y = s[4:]\n return int(y+d)\n\n\n##fix season format for df_mp\n##takes in integer 2008 and returns 20082009\n##note: I have verified in df_mp for two seasons 2008 labels 20082009\n\ndef fix_mp_season(n):\n return int(str(n)+str(n+1))\n\ndef L_add_season(mp_date):\n st = str(mp_date)\n y = st[0:4]\n ym1 = str(int(y)-1) \n yp1 = str(int(y)+1)\n \n m = st[4:6]\n d = st[6:8]\n if 9 <= int(m) <= 12:\n return int(y+yp1)\n if 1 <= int(m) <= 8:\n return int(ym1+y)\n\n##df_game_team_stats does not have 'season' or 'won' so we add that \n\ndef add_season_from_id(game_id):\n id = str(game_id)\n y = int(id[0:4])\n yn = y+1\n return int(str(y)+str(yn))\n\n\n\n##test\nprint(\"three tests for bed_add_mp_date:\")\nprint(bet_add_mp_date(912, 20082009) == 20080912)\nprint(bet_add_mp_date(1025,20102011) == 20101025)\nprint(bet_add_mp_date(227,20102011) == 20110227)\nprint(\" \")\n\n##tests\nprint(\"two tests for fix_mp_season:\")\nprint(fix_mp_season(2008) == 20082009)\nprint(fix_mp_season(2019) == 20192020)\nprint(\" \")\n\nprint(\"three tests for L_add_season:\")\nprint(L_add_season('20140119')==20132014)\nprint(L_add_season('20140912')==20142015)\n\n \nprint(\" \")\n\n\nprint(\"three tests for L_to_mp_season:\")\nprint(L_to_mp_date('2014-10-16')==20141016)\nprint(L_to_mp_date('2014-1-16')==20140116)\nprint(L_to_mp_date('2014-1-6')==20140106)\n \nprint(\" \")\n\nprint('test for add_season_from_id')\n#test \nprint(add_season_from_id(2019021081) == 20192020)\n",
"two tests for game_add_mp_date:\nTrue\nTrue\n \nthree tests for bed_add_mp_date:\nTrue\nTrue\nTrue\n \ntwo tests for fix_mp_season:\nTrue\nTrue\n \nthree tests for L_add_season:\nTrue\nTrue\n \nthree tests for L_to_mp_season:\nTrue\nTrue\nTrue\n \ntest for add_season_from_id\nTrue\n"
],
[
"##fix team names\n##Leung dropped all the ATL games so I should do same \n# issues CAL MON and WAS\n\nL_to_nhl_dic = {'ANA': 'ANA',\n 'ARI': 'ARI',\n 'ATL': 'ATL',\n 'BOS': 'BOS',\n 'BUF': 'BUF',\n 'CAR': 'CAR',\n 'CBJ': 'CBJ',\n 'CAL': 'CGY',\n 'CHI': 'CHI',\n 'COL': 'COL',\n 'DAL': 'DAL',\n 'DET': 'DET',\n 'EDM': 'EDM',\n 'FLA': 'FLA',\n 'LAK': 'LAK',\n 'MIN': 'MIN',\n 'MON': 'MTL',\n 'NJD': 'NJD',\n 'NSH': 'NSH',\n 'NYI': 'NYI',\n 'NYR': 'NYR',\n 'OTT': 'OTT',\n 'PHI': 'PHI',\n 'PIT': 'PIT',\n 'SJS': 'SJS',\n 'STL': 'STL',\n 'TBL': 'TBL',\n 'TOR': 'TOR',\n 'VAN': 'VAN',\n 'VGK': 'VGK',\n 'WPG': 'WPG',\n 'WAS': 'WSH'}\n\n\nmp_to_nhl_dic = {'ANA': 'ANA',\n 'ARI': 'ARI',\n 'ATL': 'ATL',\n 'BOS': 'BOS',\n 'BUF': 'BUF',\n 'CAR': 'CAR',\n 'CBJ': 'CBJ',\n 'CGY': 'CGY',\n 'CHI': 'CHI',\n 'COL': 'COL',\n 'DAL': 'DAL',\n 'DET': 'DET',\n 'EDM': 'EDM',\n 'FLA': 'FLA',\n 'L.A': 'LAK',\n 'MIN': 'MIN',\n 'MTL': 'MTL',\n 'N.J': 'NJD',\n 'NSH': 'NSH',\n 'NYI': 'NYI',\n 'NYR': 'NYR',\n 'OTT': 'OTT',\n 'PHI': 'PHI',\n 'PIT': 'PIT',\n 'S.J': 'SJS',\n 'STL': 'STL',\n 'T.B': 'TBL',\n 'TOR': 'TOR',\n 'VAN': 'VAN',\n 'VGK': 'VGK',\n 'WPG': 'WPG',\n 'WSH': 'WSH'}\n\nbet_to_nhl_dic = {'Anaheim': 'ANA',\n 'Arizona': 'ARI',\n 'Arizonas': 'ARI',\n 'Atlanta': 'ATL',\n 'Boston': 'BOS',\n 'Buffalo': 'BUF',\n 'Calgary': 'CGY',\n 'Carolina': 'CAR',\n 'Chicago': 'CHI',\n 'Colorado': 'COL',\n 'Columbus': 'CBJ',\n 'Dallas': 'DAL',\n 'Detroit': 'DET',\n 'Edmonton': 'EDM',\n 'Florida': 'FLA',\n 'LosAngeles': 'LAK',\n 'Minnesota': 'MIN',\n 'Montreal': 'MTL',\n 'NYIslanders': 'NYI',\n 'NYRangers': 'NYR',\n 'Nashville': 'NSH',\n 'NewJersey': 'NJD',\n 'Ottawa': 'OTT',\n 'Philadelphia': 'PHI',\n 'Phoenix': 'ARI',\n 'Pittsburgh': 'PIT',\n 'SanJose': 'SJS',\n 'St.Louis': 'STL',\n 'Tampa': 'TBL',\n 'TampaBay': 'TBL',\n 'Toronto': 'TOR',\n 'Vancouver': 'VAN',\n 'Vegas': 'VGK',\n 'Washington': 'WSH',\n 'Winnipeg': 'WPG',\n 'WinnipegJets': 'WPG'}\n\ngame_to_nhl_dic = {1: 'NJD',\n 4: 'PHI',\n 26: 'LAK',\n 14: 'TBL',\n 6: 'BOS',\n 3: 'NYR',\n 5: 'PIT',\n 17: 'DET',\n 28: 'SJS',\n 18: 'NSH',\n 23: 'VAN',\n 16: 'CHI',\n 9: 'OTT',\n 8: 'MTL',\n 30: 'MIN',\n 15: 'WSH',\n 19: 'STL',\n 24: 'ANA',\n 27: 'ARI',\n 2: 'NYI',\n 10: 'TOR',\n 13: 'FLA',\n 7: 'BUF',\n 20: 'CGY',\n 21: 'COL',\n 25: 'DAL',\n 29: 'CBJ',\n 52: 'WPG',\n 22: 'EDM',\n 54: 'VGK',\n 12: 'CAR',\n 53: 'ARI',\n 11: 'ATL'}\n\ndef L_to_nhl(abbrevName):\n return L_to_nhl_dic[abbrevName]\n\n\ndef bet_to_nhl(Team):\n return bet_to_nhl_dic[Team]\n\ndef mp_to_nhl(team):\n return mp_to_nhl_dic[team]\n\ndef game_to_nhl(team_id):\n return game_to_nhl_dic[team_id]\n\n##simple tests:\nprint(set(bet_to_nhl_dic.values()) == set(mp_to_nhl_dic.values()) )\nprint(set(bet_to_nhl_dic.values() )== set(game_to_nhl_dic.values()) )\nprint(set(mp_to_nhl_dic.values()) == set(game_to_nhl_dic.values()) )\nprint(set(mp_to_nhl_dic.values()) == set(L_to_nhl_dic.values()) )\nprint(set(game_to_nhl_dic.values()))\nprint(set(bet_to_nhl_dic.values()))\nprint(set(mp_to_nhl_dic.values()))\n#print(set(mp_to_nhl_dic.values()) == set(L_data['teamAbbreviation'])) #these are different bec no ATL in L_data\nlen(set(mp_to_nhl_dic.values()))",
"True\nTrue\nTrue\nTrue\n{'BUF', 'CGY', 'DAL', 'COL', 'CHI', 'MTL', 'PIT', 'STL', 'NJD', 'PHI', 'OTT', 'ATL', 'CBJ', 'LAK', 'NYR', 'DET', 'ARI', 'CAR', 'MIN', 'NSH', 'FLA', 'TOR', 'SJS', 'EDM', 'BOS', 'VAN', 'NYI', 'VGK', 'ANA', 'WPG', 'TBL', 'WSH'}\n{'CGY', 'DAL', 'COL', 'CHI', 'MTL', 'PIT', 'STL', 'NJD', 'PHI', 'OTT', 'ATL', 'CBJ', 'LAK', 'NYR', 'DET', 'ARI', 'CAR', 'MIN', 'NSH', 'FLA', 'WSH', 'TOR', 'EDM', 'SJS', 'BOS', 'VAN', 'NYI', 'VGK', 'ANA', 'WPG', 'TBL', 'BUF'}\n{'CGY', 'DAL', 'COL', 'CHI', 'MTL', 'PIT', 'STL', 'NJD', 'PHI', 'OTT', 'ATL', 'CBJ', 'LAK', 'NYR', 'DET', 'ARI', 'CAR', 'MIN', 'NSH', 'FLA', 'WSH', 'TOR', 'EDM', 'SJS', 'BOS', 'VAN', 'NYI', 'VGK', 'ANA', 'WPG', 'TBL', 'BUF'}\n"
],
[
"##assign game_id to df_betting (two ways)\n\n##these two functions \n# assign game_id to df_betting ... using two different look up tables, df_game and df_mp_teams\n##note the following error codes are assigned if a game is not found or found more than once \n# if a home game is empty: 80 #8 is for h\n# if a home game has >= 2 entries: 82\n# if an away game is empty: 10 #1 is for a\n# if an away game has >= 12 entries: 82\n\n#df_betting['mp_game_id'] = np.vectorize(mp_to_bet_add_game_id, excluded={0})(df_mp_teams, df_betting['mp_date'], df_betting['nhl_name'], df_betting['VH'] ) \n\n\n \ndef game_to_bet_add_game_id(df_game_team_stats, mp_date, nhl_name, VH):##put VH in there! duh\n if VH == 'H':\n ## this h and (also a) is an array of values ... it may be empty; we expect exactly one element; might have >=2 or 0 if something is wrong\n h = df_game.loc[(df_game_team_stats['mp_date'] == mp_date)&(df_game_team_stats['nhl_name'] == nhl_name)&(df_game_team_stats['HoA'] == 'home'), :]['game_id'].values\n if len(h) ==1:\n return h[0]\n elif len(h)==0: #and mp_date < 2010000:\n return 80\n print(mp_date)\n elif len(h) > 1:\n return 82\n if VH == 'V':\n a = df_game.loc[(df_game_team_stats['mp_date'] == mp_date)&(df_game_team_stats['nhl_name'] == nhl_name)&(df_game_team_stats['HoA'] == 'away'), :]['game_id']\n if len(a) ==1:\n return a[0]\n elif len(a)==0:\n return 220\n elif len(a) > 1:\n return 222\n\n\n\ndef mp_to_bet_add_game_id(df_mp_teams, mp_date, nhl_name, VH):##put VH in there! duh\n if VH == 'H':\n h = df_mp_teams.loc[(df_mp_teams['mp_date'] == mp_date)&(df_mp_teams['nhl_name'] == nhl_name)&(df_mp_teams['home_or_away'] == 'HOME'), :]['game_id'].values\n if len(h) ==1:\n return h[0]\n elif len(h)==0: \n return 80 \n print(mp_date)\n elif len(h) > 1:\n return 82\n if VH == 'V':\n a = df_mp_teams.loc[(df_mp_teams['mp_date'] == mp_date)&(df_mp_teams['nhl_name'] == nhl_name)&(df_mp_teams['home_or_away'] == 'AWAY'), :]['game_id'].values\n if len(a) ==1:\n return a[0]\n elif len(a)==0:\n return 10 \n elif len(a) > 1:\n return 12\n\n if VH == 'N':\n n = df_mp_teams.loc[(df_mp_teams['mp_date'] == mp_date)&(df_mp_teams['nhl_name'] == nhl_name), :]['game_id'].values\n if len(n) ==1:\n return n[0]\n elif len(n)==0:\n return 130 \n elif len(n) > 1:\n return 132\n \n else:\n return 26\n \n\ndef mp_to_bet_add_game_id_no_VH(df_mp_teams, mp_date, nhl_name):##put VH in there! duh\n ha = df_mp_teams.loc[(df_mp_teams['mp_date'] == mp_date)&(df_mp_teams['nhl_name'] == nhl_name), :]['game_id'].values\n if len(ha) ==1:\n return ha[0]\n elif len(ha)==0: \n return 180 \n elif len(ha) > 1:\n return 182\n else:\n return 26\n\n",
"_____no_output_____"
],
[
"#cleaning up df's before merge ...\n\ndf_betting = df_betting.loc[:, ['Date', 'season','VH', 'Team', 'Open']].copy()\ndf_mp_teams.rename(columns={\"teamId\": \"team_id\", 'gameDate': 'mp_date', 'gameId':'game_id' }, inplace = True)\n##note there are other column names which differ, but team_id, mp_date, game_id are key for joining so we \n##make it consistent\n\ndf_mp_teams_big = df_mp_teams.copy()\n##only has the rows for all situations (divide num rows by 5)\n\n#restrict situation to all \ndf_mp_teams = df_mp_teams.loc[(df_mp_teams['situation'] == 'all'), :] #&(df_mp_teams['playoffGame']==0), :].copy()\n\n##restrict types to all situations later before merge \n\ndf_game_team_stats.drop_duplicates(inplace = True)\ndf_game_team_stats = df_game_team_stats[~df_game_team_stats['team_id'].isin([87,88,89,90])].copy()\ndf_betting = df_betting[~(df_betting['VH'] == 'N')].copy() ",
"_____no_output_____"
],
[
"##add season and won to df_game_team_stats (need later to re-do part of missing Leung data )\n\ndf_game_team_stats['season'] = np.vectorize(add_season_from_id)(df_game_team_stats['game_id']).copy()\ndf_game_team_stats['won'] = df_game_team_stats['won'].apply(int).copy()\n\n#change 2008 to 20082009 for df_mp_teams\n\ndf_mp_teams['season'] = np.vectorize(fix_mp_season)(df_mp_teams['season']).copy()\n",
"_____no_output_____"
],
[
"# fix these in L_data: 'endDate', 'teamAbbreviation'\nL_data['mp_date'] = np.vectorize(L_to_mp_date)(L_data['endDate']).copy() \nL_data['nhl_name'] = np.vectorize(L_to_nhl)(L_data['teamAbbreviation']).copy() \n\nL_data['season'] = np.vectorize(L_add_season)(L_data['mp_date'])\n#L_data.columns",
"_____no_output_____"
],
[
"##fix some labels in df_betting and df_mp_teams\n\ndf_betting['mp_date'] = np.vectorize(bet_add_mp_date)(df_betting['Date'], df_betting['season']).copy()\n\nHA_dic = { 'H': 'home', 'V': 'away', 'HOME': 'home', 'AWAY':'away'}\ndef add_HoA(h_or_a): # h_or_a can be either of the formats used by mp or betting\n return HA_dic[h_or_a]\n\ndf_betting['HoA'] = df_betting['VH'].apply(add_HoA).copy()\n\ndf_mp_teams['HoA'] = df_mp_teams['home_or_away'].apply(add_HoA).copy()\n\n\n\n\ndf_betting['nhl_name'] = df_betting['Team'].apply(bet_to_nhl).copy()\ndf_game_team_stats['nhl_name'] = df_game_team_stats['team_id'].apply(game_to_nhl).copy()\ndf_mp_teams['nhl_name'] = df_mp_teams['name'].apply(mp_to_nhl).copy()\n\ndf_betting['game_id'] = np.vectorize(mp_to_bet_add_game_id_no_VH, excluded={0})(df_mp_teams, df_betting['mp_date'], df_betting['nhl_name'] ).copy() ",
"_____no_output_____"
],
[
"\n\n##get ready to restrict seasons; I reduced it to 20182019 for Leung\nseasons = []\nfor n in range(2008,2019):\n seasons.append(int(str(n)+str(n+1)))\n\n#check seasons look ok\nprint(seasons)\n",
"[20082009, 20092010, 20102011, 20112012, 20122013, 20132014, 20142015, 20152016, 20162017, 20172018, 20182019]\n"
],
[
"#restrict seasons \n\nL_data = L_data.loc[L_data['season'].isin(seasons), :].copy()\n\ndf_betting = df_betting.loc[df_betting['season'].isin(seasons), :].copy()\n#df_game_team_stats = df_game_team_stats.loc[df_game_team_stats['season'].isin(seasons), :].copy()\ndf_mp_teams = df_mp_teams.loc[df_mp_teams['season'].isin(seasons), :].copy()\n#df_mp_teams = df_mp_teams.loc[df_mp_teams['season'].isin(seasons), :].copy()\n\n\n## the index is no longer consecutive so we reset:\n\ndf_betting.reset_index(drop = True, inplace = True)\n#df_game_team_stats.reset_index(drop = True, inplace = True)\ndf_mp_teams.reset_index(drop = True, inplace = True)\nL_data.reset_index(drop = True, inplace = True)\n\n\n##! restrict df_game_team_stats later using the game_id",
"_____no_output_____"
]
],
[
[
"df_temp = df_results.dropna(inplace = False).copy()\n\nA = [set(df_betting['game_id']) , set(df_game_team_stats['game_id']) , set(df_results['game_id']) ] \n\nfor i in range(3):\n j = i + 1 %3 \n print(i, len(A[i])) \n print(i, j, 'inters: ', len( set.intersection(A[i], A[j]) )\n print(\"all: \": len( set.intersection(A[0],A[1],A[2]))",
"_____no_output_____"
]
],
[
[
"##only doing first merge now ... \n\ncommon_game_ids = set.intersection( set(df_betting['game_id']) , set(df_game_team_stats['game_id']) ) #, set(df_results['game_id']) ) \n\nA = [set(df_betting['game_id']) , set(df_game_team_stats['game_id'])] \n\nprint(len(A[0]))\nprint(len(A[1]))\nprint(\"all: \", len( set.intersection(A[0],A[1]) ))\n ",
"14064\n23730\nall: 13887\n"
],
[
" \ndf_betting = df_betting.loc[df_betting['game_id'].isin(common_game_ids), :].copy()\ndf_game_team_stats = df_game_team_stats.loc[df_game_team_stats['game_id'].isin(common_game_ids), :].copy()\n#df_mp_teams = df_mp_teams.loc[df_mp_teams['game_id'].isin(common_game_ids), :].copy()",
"_____no_output_____"
],
[
"##check consistency of nhl_names, HoA ...\n\n#df_mp_teams.sort_values(by = ['game_id', 'nhl_name'], inplace = True)\ndf_game_team_stats.sort_values(by = ['game_id', 'nhl_name'], inplace = True)\ndf_betting.sort_values(by = ['game_id', 'nhl_name'], inplace = True)\n\ndf_betting.reset_index(drop = True, inplace = True)\ndf_game_team_stats.reset_index(drop = True, inplace = True)\n#df_mp_teams.reset_index(drop = True, inplace = True)",
"_____no_output_____"
],
[
"##The big merge! Note: I am not using X_merge2 in this note book, but it is important on prev version\n\n\nX_merge = pd.merge(df_betting, df_game_team_stats, on = ['game_id', 'nhl_name'], how = 'inner', suffixes = ('_bet', '_gm_stats'))\n\n#X_merge2 = pd.merge(X_merge, df_mp_teams, on = ['game_id', 'nhl_name', 'mp_date', 'season'], how = 'inner', suffixes = ('_merge', '_mp_teams'))\n\n#print(2*len(common_game_ids), X_merge2.shape[0])",
"_____no_output_____"
],
[
"#drop ATL because Leung's data did\n\nX_merge = X_merge.loc[ (X_merge['nhl_name'] !='ATL'), :].copy()\n\nX_merge.rename(columns = {'season_bet':'season'}, inplace=True)\nX_merge.rename(columns = {'HoA_gm_stats':'HoA'}, inplace=True)\n#X_merge_L3 = pd.merge(X_merge, L_data, on = ['nhl_name', 'mp_date', 'season'], how = 'left', suffixes = ('_merge', '_L'))\n\n##using this as 'HoA' results in less data loss ",
"_____no_output_____"
],
[
"##convenient function for turning Vegas odds (-200 = fav, 120 = underdog) into game predictions win 1, lose 0\ndef Vegas_pred(Open):\n if Open < 0:\n return 1 \n elif Open > 0:\n return 0\nVegas_pred(-200) ",
"_____no_output_____"
],
[
"X_merge['Open'].isnull().sum()",
"_____no_output_____"
],
[
"np.vectorize(Vegas_pred)(np.array([2,-2]))",
"_____no_output_____"
],
[
"X_merge['Vegas_pred_won'] = X_merge['Open'].apply(Vegas_pred).copy()",
"_____no_output_____"
],
[
"X_merge = X_merge.loc[~(X_merge['Open'] == 0), :].copy()",
"_____no_output_____"
],
[
"X_merge['won'] = X_merge['won'].apply(int).copy()",
"_____no_output_____"
],
[
"X_merge['won'] = X_merge['won'].apply(int)\n",
"_____no_output_____"
],
[
"X_merge['won'] == X_merge['Vegas_pred_won']",
"_____no_output_____"
],
[
"##Here we start creating a new df called df_results ... it will have similar form to df_game\n##and is suitable fora attaching Leung's data ... (I am not using df_game because I'm scared of it's GMT. dates\n##which did work sometimes properly (I got tired of it so never checked super thoroughly)\n\n#From X_merge use the game_id as a seed to make new df in the form of df_game ...(df_game is annoying bc of date)\n\n\n##these are the games we will use:\nlist_game_ids = sorted(set(X_merge['game_id']))\nprint(len(sorted(set(X_merge['game_id']))))\n\ndf_results = pd.DataFrame({'game_id':list_game_ids})\n",
"13886\n"
],
[
"\n###This is adapted from Leung's code.\n\n\ndef value(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n elif len(frame[key]) >=2:\n return 3.14\n else: \n return frame.iloc[0][key].astype(float)\n\ndef intValue(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n elif len(frame[key]) >=2:\n return 314\n else: \n return int(frame.iloc[0][key])\n \ndef strValue(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n elif len(frame[key]) >=2:\n return \"314\"\n else: \n return frame.iloc[0][key]\n \ndef get_home_results(game_id):\n return X_merge.loc[(X_merge['game_id'] == game_id)&(X_merge['HoA'] == 'home'), :]\n\n\ndef get_away_results(game_id):\n return X_merge.loc[(X_merge['game_id'] == game_id)&(X_merge['HoA'] == 'away'), :]\n\n \ndef add_results(row):\n home = get_home_results(row['game_id']) #can use home \n away = get_away_results(row['game_id'])\n \n row['mp_date'] = intValue(home, 'mp_date')\n row['season'] = intValue(home, 'season')\n \n row['home_team'] = strValue(home, 'nhl_name')\n row['away_team'] = strValue(away, 'nhl_name')\n \n \n row['home_odds'] = value(home, 'Open')\n row['away_odds'] = value(away, 'Open')\n \n row['home_goals'] = value(home, 'goals')\n row['away_goals'] = value(away, 'goals')\n \n row['home_win'] = intValue(home, 'won') #can use home\n row['settled_in'] = strValue(home, 'settled_in')\n \n return row\n\ndf_results = df_results.apply(add_results, axis=1).copy()\n",
"_____no_output_____"
],
[
"df_results.shape",
"_____no_output_____"
],
[
"(df_results ==\"314\").sum()",
"_____no_output_____"
],
[
"perc_null(df_results)",
"_____no_output_____"
]
],
[
[
"\n###This is adapted from Leung's code.\n\n##Here we taking data from L_data and attaching to our new df_results (one row for each game id in X_merge)\n##we do not use X_merge because it has two rows for every game id (H and A)\n\n \n##get results info from X_merge: \n\n##game_id restricts season ... check\n\n\ndef get_home_results(game_id):\n return X_merge.loc[(X_merge['game_id'] == game_id)&(X_merge['HoA'] == 'home'), :]\n\n\ndef get_away_results(game_id):\n return X_merge.loc[(X_merge['game_id'] == game_id)&(X_merge['HoA'] == 'away'), :]\n\n#below row will come from df_result (game_id ) and from will be the either home info or away info from X_merge\n \ndef value(frame, key):\n if len(frame[key]) ==0:\n # print( \"There is only one value for some feature in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n elif len(frame[key]) >=2:\n #print( \"A feature value appears more than once in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n else: \n return frame.iloc[0][key].astype(float)\n\ndef intValue(frame, key):\n if len(frame[key]) ==0:\n #print( \"There emight be a missing (only 1 not 2?) value for some feature in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n elif len(frame[key]) >=2:\n #print( \"A feature value appears more than once in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n else: \n return int(frame.iloc[0][key])\n \ndef strValue(frame, key):\n if len(frame[key]) ==0:\n #print( \"There might be a missing (only 1 not 2?) value for some feature in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n elif len(frame[key]) >=2:\n #print( \"A feature value appears more than once in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n else: \n return frame.iloc[0][key]\n \n \ndef add_results(row):\n home = get_home_results(row['game_id'])\n away = get_away_results(row['game_id'])\n \n id rows (to go with game_id)\n \n row['mp_date'] = intValue(home, 'mp_date') #we can use the home one for these 2\n row['season'] = intValue(home, 'season')\n \n row['home_team'] = strValue(home, 'nhl_name')\n row['away_team'] = strValue(away, 'nhl_name')\n \n #odds (treat as a target ?)\n row['home_odds'] = value(home, 'Open')\n row['away_odds'] = value(away, 'Open')\n \n #target vars\n row['home_goals'] = value(home, 'goals')\n row['away_goals'] = value(away, 'goals')\n row['goal_difference']=value(home, 'goals')-value(away, 'goals') #keep same name as before\n row['home_win'] = intValue(home, 'won')\n row['settled_in'] = strValue(home, 'settled_in')\n \n return row\n",
"_____no_output_____"
]
],
[
[
"X_merge.columns",
"_____no_output_____"
],
[
"\ndf_results = df_results.apply(add_results, axis=1).copy()\n\n\nperc_null(df_results)\n\n##\n##df_results.dropna(inplace=True)",
"_____no_output_____"
],
[
"##get_L_stats is called getStats in Leung's code\n\ndef get_L_stats(nhl_name, mp_date): #season is restriced because date is fixed as date -1 \n return L_data.loc[(L_data['nhl_name'] == nhl_name)& (L_data['mp_date'] == mp_date -1), :] #(!) -1 VERY important so you don't leak!\n\n#the L_data stats are cumulative up to <= date ... so we grab cumulative stats up to mp_date = mp_date -1\n\n##note the mp_date-1 !! this is what gives us the *previous day's'* cumulative stats for that te\n##if the frame is empty value() will return NaN; this should happen for all first games for each team. \n \n\n##value is defined above when we created df_results from X_merge\n \ndef add_stats(row):\n home = get_L_stats(row['home_team'], row['mp_date'])\n away = get_L_stats(row['away_team'], row['mp_date'])\n\n row['CF%'] = value(home, 'cF%') - value(away, 'cF%')\n corsiShooting = (value(home, 'gF') / value(home, 'cF')) - (value(away, 'gF') / value(away, 'cF'))\n row['CSh%'] = corsiShooting * 100\n corsiSave = (1 - (value(home, 'gA') / value(home, 'cA'))) - (1 - (value(away, 'gA') / value(away, 'cA')))\n row['CSv%'] = corsiSave * 100\n row['FF%'] = value(home, 'fF%') - value(away, 'fF%')\n fenwickShooting = (value(home, 'gF') / value(home, 'fF')) - (value(away, 'gF') / value(away, 'fF'))\n row['FSh%'] = fenwickShooting * 100\n fenwickSave = (1 - (value(home, 'gA') / value(home, 'fA'))) - (1 - (value(away, 'gA') / value(away, 'fA')))\n row['FSv%'] = fenwickSave * 100\n row['GDiff'] = (value(home, 'gF60') - value(home, 'gA60')) - (value(away, 'gF60') - value(away, 'gA60'))\n row['GF%'] = value(home, 'gF%') - value(away, 'gF%')\n row['PDO'] = value(home, 'pDO') - value(away, 'pDO')\n row['PENDiff'] = (value(home, 'pEND') - value(home, 'pENT')) - (value(away, 'pEND') - value(away, 'pENT'))\n row['SF%'] = value(home, 'sF%') - value(away, 'sF%')\n row['SDiff'] = (value(home, 'sF60') - value(home, 'sA60')) - (value(away, 'sF60') - value(away, 'sA60'))\n shootingPercentage = (value(home, 'gF') / value(home, 'sF')) - (value(away, 'gF') / value(away, 'sF'))\n row['Sh%'] = shootingPercentage * 100\n row['Sv'] = value(home, 'sv%') - value(away, 'sv%')\n \n #L_data is missing GP, W, FOW% \n #row['FOW%'] = value(home, 'FOW%') - value(away, 'FOW%')\n #winPercentage = (intValue(home, 'W') / intValue(home, 'GP')) - (intValue(away, 'W') / intValue(away, 'GP'))\n #row['W%'] = np.float64(winPercentage)\n return row\n",
"_____no_output_____"
],
[
"##test for code \nL_data.loc[(L_data['nhl_name'] == 'LAK')& (L_data['mp_date'] == 20151112 -1), :]['cF%']\n",
"_____no_output_____"
],
[
"#test getStats:\nget_L_stats('LAK', 20151112).iloc[0]['cF'].astype(float)\n\n#test empty frame if no game ... \nlen(get_L_stats('LAK', 20121112)['cF'])",
"_____no_output_____"
],
[
"\ndf_results_L_data = df_results.apply(add_stats, axis =1).copy() ##new df combines X_merge (results) and L_data\n\n#check on diff between X_merge and L_data games:\n#print(L_data.groupby(['mp_date, 'nhl_team']) ... hard to do grup by b/c team appears on all dates (whether played or not)\n ",
"_____no_output_____"
],
[
" \nperc_null(df_results_L_data)\n",
"_____no_output_____"
],
[
"# one of the games have 0 odds ... we will remove those\ndf_results_L_data= df_results_L_data.loc[ (df_results_L_data['home_odds'] != 0), :].copy()",
"_____no_output_____"
],
[
"df_results_L_data.shape\n\nperc_null(df_results_L_data2)\n",
"_____no_output_____"
],
[
"df_results_L_data2 = df_results_L_data.dropna(inplace=False) #drop na first 25%",
"_____no_output_____"
],
[
"df_results_L_data2",
"_____no_output_____"
],
[
"df_mp_teams.loc[(df_mp_teams['season'] == 20162017)&(df_mp_teams['nhl_name'] == 'LAK')&(df_mp_teams['mp_date'] <= 20141015), : ]['team'].sum()\n\n#df_LA = df_mp_teams.loc[(df_mp_teams['season'] == 20082009)&(df_mp_teams['nhl_name'] == 'LAK')&(df_mp_teams['mp_date'] <= 20090230), : ]\n#df_LA2 = df_game_team_stats.loc[(df_game_team_stats['season'] == 20082009)&(df_game_team_stats['nhl_name'] == 'LAK')&(df_game_team_stats['game_id'] <= 2008020919), :]\n",
"_____no_output_____"
],
[
"##from old notebook ... aha ... the counyt and sum are getting messed up by null values I think\n \n ##add stats part two ... using mp \ndef get_mp_stats(season, nhl_name, mp_date): #note each game has two rows, with each team listed ONCE under nhl_name; date<= mp_date\n return df_mp_teams.loc[(df_mp_teams['season'] == season)&(df_mp_teams['nhl_name'] == nhl_name)&(df_mp_teams['mp_date'] <= mp_date-1), : ]\n\n#game_id is used as date:\ndef get_game_stats(season, nhl_name, game_id): #note each game has two rows, with each team listed ONCE under nhl_name; date<= mp_date\n return df_game_team_stats.loc[(df_game_team_stats['season'] == season)&(df_game_team_stats['nhl_name'] == nhl_name)&(df_game_team_stats['game_id'] <= game_id-1), :]\n \n \ndef do_sum(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n else: \n return frame[key].sum() ##! remove the .iloc[0] you want the whole frame now to count or sum\n\n\ndef do_count(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n elif len(frame[key]) >=2:\n return 3.14\n else: \n return frame[key].count()\n \n \ndef add_missing_stats(row):\n mp_stats_home = get_mp_stats(row['season'], row['home_team'], row['mp_date'])\n mp_stats_away = get_mp_stats(row['season'], row['away_team'], row['mp_date'])\n game_stats_home = get_game_stats(row['season'], row['home_team'], row['game_id'])\n game_stats_away = get_game_stats(row['season'], row['away_team'], row['game_id'])\n \n GP_home = do_count(mp_stats_home, 'game_id') #game_id is just to count\n GP_away = do_count(mp_stats_away, 'game_id')\n W_home = do_sum(game_stats_home, 'won') \n W_away = do_sum(game_stats_away, 'won') \n Wper_home = W_home/GP_home\n Wper_away = W_away/GP_away\n \n row['W%'] = Wper_home - Wper_away \n \n FOL_home= do_sum(mp_stats_home,'faceOffsWonAgainst')\n FOL_away= do_sum(mp_stats_away,'faceOffsWonAgainst')\n FOW_home = do_sum(mp_stats_home,'faceOffsWonFor')\n FOW_away = do_sum(mp_stats_away,'faceOffsWonFor')\n FOper_home = FOW_home/(FOW_home+FOL_home)\n FOper_away = FOW_away/(FOW_away+FOL_away)\n \n row['FOW%'] = FOper_home-FOper_away\n ",
"_____no_output_____"
],
[
"##a test for the code \n\ndf_LA = df_mp_teams.loc[(df_mp_teams['season'] == 20082009)&(df_mp_teams['nhl_name'] == 'LAK')&(df_mp_teams['mp_date'] <= 20090230), : ]\ndf_LA2 = df_game_team_stats.loc[(df_game_team_stats['season'] == 20082009)&(df_game_team_stats['nhl_name'] == 'LAK')&(df_game_team_stats['game_id'] <= 2008020919), :]\n\nprint(df_LA['game_id'].count())\nprint(df_LA2['game_id'].count())\n#check game_id works as well as date:\nlist(df_LA2['game_id']) == list(df_LA['game_id']) #using game_id as date will work within a season",
"61\n61\n"
],
[
"#try dropping na first\n\ndf_results_L_data3 = df_results_L_data2.apply(add_missing_stats, axis =1).copy()\n",
"_____no_output_____"
],
[
"#perc_null(df_mp_teams)\ndf_results_L_data3",
"_____no_output_____"
]
],
[
[
" ##add stats part two ... using mp to add FOW% and W% ... Leung's data is missing the entries 'GP' etc needed to do those\n \ndef get_mp_stats(season, nhl_name, mp_date): #note each game has two rows, with each team listed ONCE under nhl_name; date<= mp_date-1\n return df_mp_teams.loc[(df_mp_teams['season'] == season)&(df_mp_teams['nhl_name'] == nhl_name)&(df_mp_teams['mp_date'] <= mp_date-1), : ]\n\n##note the mp_date -1 (!)\n\n#game_id is used as date (cell above checks this works)\ndef get_game_stats(season, nhl_name, game_id): #note each game has two rows, with each team listed ONCE under nhl_name; date<= mp_date-1\n return df_game_team_stats.loc[(df_game_team_stats['season'] == season)&(df_game_team_stats['nhl_name'] == nhl_name)&(df_game_team_stats['game_id'] <= game_id-1), :]\n \n##note the game_id -1 (!) \n \ndef do_sum(frame, key):\n if len(frame[key]) ==0:\n print( \"There emight be a missing (only 1 not 2?) value for some feature in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n elif len(frame[key]) >=2:\n print( \"A feature value appears more than once in X_merge for this game_id: \", frame['game_id'] )\n return np.NaN\n else: \n return frame[key].sum() ##! remove the .iloc[0] you want the whole frame now to count or sum\n\n\ndef do_count(frame, key):\n if len(frame[key]) ==0:\n print( \"Thermight be a missing (only 1 not 2?) value for some feature in df_game_stats for this game_id: \", frame['game_id'] )\n return np.NaN\n elif len(frame[key]) >=2:\n print( \"A feature value appears more than once in df_game_stats for this game_id: \", frame['game_id'] )\n return np.NaN\n else: \n return frame[key].count()\n \n#will be applied to rows of df_results_L_data \ndef add_missing_stats(row):\n mp_stats_home = get_mp_stats(row['season'], row['home_team'], row['mp_date'])\n mp_stats_away = get_mp_stats(row['season'], row['away_team'], row['mp_date'])\n game_stats_home = get_game_stats(row['season'], row['home_team'], row['game_id'])\n game_stats_away = get_game_stats(row['season'], row['away_team'], row['game_id'])\n \n GP_home = do_count(mp_stats_home, 'game_id') #game_id is just to count\n GP_away = do_count(mp_stats_away, 'game_id')\n W_home = do_sum(game_stats_home, 'won') \n W_away = do_sum(game_stats_away, 'won') \n Wper_home = W_home/GP_home\n Wper_away = W_away/GP_home\n \n row['W%'] = Wper_home - Wper_away \n \n FOL_home= do_sum(mp_stats_home,'faceOffsWonAgainst')\n FOL_away= do_sum(mp_stats_away,'faceOffsWonAgainst')\n FOW_home = do_sum(mp_stats_home,'faceOffsWonFor')\n FOW_away = do_sum(mp_stats_away,'faceOffsWonFor')\n FOper_home = FOW_home/(FOW_home+FOL_home)\n FOper_away = FOW_away/(FOW_away+FOL_away)\n \n \n row['FOW%'] = FOper_home-FOper_away\n \n",
"_____no_output_____"
]
],
[
[
"##should be about same NaN as df_result, around 29%?\n\n\ndf_results_L_data3 = df_results_L_data2.apply(add_missing_stats, axis =1).copy()\n\n",
"_____no_output_____"
],
[
"df_results_L_data3",
"_____no_output_____"
],
[
"perc_null(df_results_L_data3)",
"_____no_output_____"
],
[
"df_results_L_data.columns",
"_____no_output_____"
]
],
[
[
"##from Git\n\n##add stats part two ... using mp \ndef get_mp_stats(season, nhl_name, mp_date): #note each game has two rows, with each team listed ONCE under nhl_name; date<= mp_date\n return df_mp_teams.loc[(df_mp_teams['season'] == season)&(df_mp_teams['nhl_name'] == nhl_name)&(df_mp_teams['mp_date'] <= mp_date), : ]\n\n#game_id is used as date:\ndef get_game_stats(season, nhl_name, game_id): #note each game has two rows, with each team listed ONCE under nhl_name; date<= mp_date\n return df_game_team_stats.loc[(df_game_team_stats['season'] == season)&(df_game_team_stats['nhl_name'] == nhl_name)&(df_game_team_stats['game_id'] <= game_id), :]\n \n \ndef do_sum(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n elif len(frame[key]) >=2:\n return 3.14\n else: \n return frame[key].sum() ##! remove the .iloc[0] you want the whole frame now to count or sum\n\n\ndef do_count(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n elif len(frame[key]) >=2:\n return 3.14\n else: \n return frame[key].count()\n \n \ndef add_missing_stats(row):\n mp_stats_home = get_mp_stats(row['season'], row['home_team'], row['mp_date'])\n mp_stats_away = get_mp_stats(row['season'], row['away_team'], row['mp_date'])\n game_stats_home = get_game_stats(row['season'], row['home_team'], row['game_id'])\n game_stats_away = get_game_stats(row['season'], row['away_team'], row['game_id'])\n \n GP_home = do_count(mp_stats_home, 'game_id') #game_id is just to count\n GP_away = do_count(mp_stats_away, 'game_id')\n W_home = do_sum(game_stats_home, 'won') \n W_away = do_sum(game_stats_away, 'won') \n Wper_home = W_home/GP_home\n Wper_away = W_away/GP_home\n \n row['W%'] = Wper_home - Wper_away \n \n FOL_home= do_sum(mp_stats_home,'faceOffsWonAgainst')\n FOL_away= do_sum(mp_stats_away,'faceOffsWonAgainst')\n FOW_home = do_sum(mp_stats_home,'faceOffsWonFor')\n FOW_away = do_sum(mp_stats_away,'faceOffsWonFor')\n FOper_home = FOW_home/(FOW_home+FOL_home)\n FOper_away = FOW_away/(FOW_away+FOL_away)\n \n row['FOW%'] = FOper_home-FOper_away\n \n\n\ndf_results_L_data5 = df_results_L_data.head(60).apply(add_missing_stats, axis =1).copy()",
"_____no_output_____"
]
],
[
[
"df_results_L_data5 ",
"_____no_output_____"
],
[
"##constructing FavoriteW% .. measures how accurately the betting company predicted games in past for both home team and away team\n\n\n#fav_stats = df_results_L_data.copy() #we create an auxiliary stats repo ... so it doesn't get all tangled up\n\nfav_stats = X_merge.copy() #renamed fav_stats as convenience; X_merge has a row for every team and pred_Vegas_won =0,1\n\ndef get_fav_stats(nhl_name, mp_date, season):\n return X_merge.loc[(fav_stats['nhl_name'] == season)&(fav_stats['nhl_name'] == nhl_name)& (fav_stats['mp_date'] <= mp_date -1), :] #(!) -1 VERY important so you don't leak!\n\n\n \n##note the mp_date-1 we still need this because fav% is based on vegas pred plus actual results\n##if the frame is empty value() will return NaN; this should happen for all first games for each team. \n\n\n \ndef do_sum(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n else: \n return frame[key].sum() ##! remove the .iloc[0] you want the whole frame now to count or sum\n\n\ndef do_count(frame, key):\n if len(frame[key]) ==0:\n return np.NaN\n elif len(frame[key]) >=2:\n return 3.14\n else: \n return frame[key].count()\n\ndef count_when_same(frame, key1, key2): #homeFav\n if len(frame[key1])==0 or len(frame[key1])==0:\n #print( \"There might be 0 values for some feature in X_merge for this game_id: \", str(key1), str(key2), frame['game_id'] )\n return np.NaN\n elif len(frame[key1])!=len(frame[key1]): \n #print( \"There might be different values for some feature in X_merge for this game_id: \", str(key1), str(key2), frame['game_id'] )\n return np.NaN\n else: \n return (frame[key1] == frame[key2]).sum()\n \n #def count_when_diff(frame, key1, key2): #homeFav\n# return(frame[key1] != frame[key2]).sum()\n\n#rows of df_results_L_data\ndef add_fav(row):\n home = get_fav_stats(row['home_team'], row['mp_date'], row['season']) #stats on *all* previous (stricly previous) games of home team (H or A games)\n away = get_fav_stats(row['away_team'], row['mp_date'], row['season'])\n \n GP_home = do_count(home, 'game_id') #game_id is just to count\n GP_away = do_count(away, 'game_id')\n home_win_pred_corr = count_when_same(home, 'Vegas_pred_won', 'won')\n away_win_pred_corr = count_when_same(away, 'Vegas_pred_won', 'won')\n home_fav = home_win_pred_corr/GP_home #this measures how accurate Vegas is with this home team in all games strictly passed (H or A) \n away_fav = away_win_pred_corr/GP_away #same for away team\n FavPerc = home_fav*away_fav \n \n row['FavoritesW%'] = FavPerc ## the product measures the overall accuracy of Vegas for both teams eg (0.9)(0.9) = 0.81, 0.9*0.5 = 0.45\n \n\n \n",
"_____no_output_____"
],
[
"##tests \nget_fav_stats('LAK', 20151112, 20152016 ) \n",
"_____no_output_____"
],
[
" \ndf_results_L_data4 = df_results_L_data2.head(20).apply(add_fav, axis =1).copy()\n\ndf_results_L_data4 \n\n\n#30% missing ... I'll take it!\n",
"_____no_output_____"
],
[
"df_results_L_data4\n\n#df_results_L_data.shape\n",
"_____no_output_____"
],
[
"\n##!! I think I drop a lot here ... 30% ish (because Leung's data seems to be missing about 30% of games)\ndata_LJ = df_results_L_data.dropna(inplace=False).copy()\n#df_results_L_data.shape",
"_____no_output_____"
],
[
"\ndata_LJ.to_csv('/Users/joejohns/data_bootcamp/GitHub/final_project_nhl_prediction/Data/Shaped_Data/data_LJ.csv')",
"_____no_output_____"
],
[
"##exrta investigation\n\ndf_temp = df_results.dropna(inplace = False).copy()\n\nA = [set(df_betting['game_id']) , set(df_game_team_stats['game_id']) , set(df_results['game_id']) ] \n\nfor i in range(3):\n j = i + 1 %3 \n print(i, len(A[i])) \n print(i, j, 'inters: ', len( set.intersection(A[i], A[j]) )\n print(\"all: \": len( set.intersection(A[0],A[1],A[2]))",
"_____no_output_____"
],
[
"common_game_ids = set.intersection( set(df_betting['game_id']) , set(df_game_team_stats['game_id']) , set(df_results['game_id']) ) \n \ndf_betting = df_betting.loc[df_betting['game_id'].isin(common_game_ids), :].copy()\ndf_game_team_stats = df_game_team_stats.loc[df_game_team_stats['game_id'].isin(common_game_ids), :].copy()\ndf_mp_teams = df_mp_teams.loc[df_mp_teams['game_id'].isin(common_game_ids), :].copy()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a15149165e6dcf048931a1fd40e1a911204bec5
| 14,087 |
ipynb
|
Jupyter Notebook
|
projects/day15/day15.ipynb
|
dmcauslan/advent-of-code-2021
|
30af2bed6b2d0b8aa269bbc5283642a81f3adcae
|
[
"BSD-3-Clause"
] | null | null | null |
projects/day15/day15.ipynb
|
dmcauslan/advent-of-code-2021
|
30af2bed6b2d0b8aa269bbc5283642a81f3adcae
|
[
"BSD-3-Clause"
] | null | null | null |
projects/day15/day15.ipynb
|
dmcauslan/advent-of-code-2021
|
30af2bed6b2d0b8aa269bbc5283642a81f3adcae
|
[
"BSD-3-Clause"
] | null | null | null | 28.690428 | 133 | 0.563569 |
[
[
[
"SAMPLE_TEXT = \"\"\"\n1163751742\n1381373672\n2136511328\n3694931569\n7463417111\n1319128137\n1359912421\n3125421639\n1293138521\n2311944581\n\"\"\"\n\nSAMPLE_TEXT_2 = \"\"\"\n11637517422274862853338597396444961841755517295286\n13813736722492484783351359589446246169155735727126\n21365113283247622439435873354154698446526571955763\n36949315694715142671582625378269373648937148475914\n74634171118574528222968563933317967414442817852555\n13191281372421239248353234135946434524615754563572\n13599124212461123532357223464346833457545794456865\n31254216394236532741534764385264587549637569865174\n12931385212314249632342535174345364628545647573965\n23119445813422155692453326671356443778246755488935\n22748628533385973964449618417555172952866628316397\n24924847833513595894462461691557357271266846838237\n32476224394358733541546984465265719557637682166874\n47151426715826253782693736489371484759148259586125\n85745282229685639333179674144428178525553928963666\n24212392483532341359464345246157545635726865674683\n24611235323572234643468334575457944568656815567976\n42365327415347643852645875496375698651748671976285\n23142496323425351743453646285456475739656758684176\n34221556924533266713564437782467554889357866599146\n33859739644496184175551729528666283163977739427418\n35135958944624616915573572712668468382377957949348\n43587335415469844652657195576376821668748793277985\n58262537826937364893714847591482595861259361697236\n96856393331796741444281785255539289636664139174777\n35323413594643452461575456357268656746837976785794\n35722346434683345754579445686568155679767926678187\n53476438526458754963756986517486719762859782187396\n34253517434536462854564757396567586841767869795287\n45332667135644377824675548893578665991468977611257\n44961841755517295286662831639777394274188841538529\n46246169155735727126684683823779579493488168151459\n54698446526571955763768216687487932779859814388196\n69373648937148475914825958612593616972361472718347\n17967414442817852555392896366641391747775241285888\n46434524615754563572686567468379767857948187896815\n46833457545794456865681556797679266781878137789298\n64587549637569865174867197628597821873961893298417\n45364628545647573965675868417678697952878971816398\n56443778246755488935786659914689776112579188722368\n55172952866628316397773942741888415385299952649631\n57357271266846838237795794934881681514599279262561\n65719557637682166874879327798598143881961925499217\n71484759148259586125936169723614727183472583829458\n28178525553928963666413917477752412858886352396999\n57545635726865674683797678579481878968159298917926\n57944568656815567976792667818781377892989248891319\n75698651748671976285978218739618932984172914319528\n56475739656758684176786979528789718163989182927419\n67554889357866599146897761125791887223681299833479\n\"\"\"",
"_____no_output_____"
],
[
"from dataclasses import dataclass\nfrom typing import Dict, Set, Optional, List",
"_____no_output_____"
],
[
"def tokenize_line(line):\n return [int(i) for i in list(line)]\n\ndef parse_text(raw_text):\n return [tokenize_line(l) for l in raw_text.split(\"\\n\") if l]\n\ndef read_input():\n with open(\"input.txt\", \"rt\") as f:\n return f.read()",
"_____no_output_____"
],
[
"# Use Dijkstra's algorithm since it will definitely work.\n\n@dataclass(frozen=True)\nclass Position:\n x: int\n y: int\n\n@dataclass\nclass Cost:\n parent: Position\n cost: int\n\n def update(self, new_parent: Position, new_cost: int) -> bool:\n if new_cost < self.cost:\n self.parent = new_parent\n self.cost = new_cost\n return True\n return False\n\ndef find_cheapest_node(costs: Dict[Position, Cost], visited: Set[Position]) -> Optional[Position]:\n non_visited_costs = (i for i in costs.items() if i[0] not in visited)\n sorted_costs = sorted(non_visited_costs, key=lambda x: x[1].cost)\n if not sorted_costs:\n return None\n return sorted_costs[0][0]\n\ndef get_route(start: Position, end: Position, costs: Dict[Position, Cost]) -> List[Position]:\n result = [end]\n while True:\n parent = costs[result[-1]].parent\n result.append(parent)\n if parent == start:\n break\n result.reverse()\n return result\n\ndef get_adjacent_positions(from_position: Position, weights: Dict[Position, int], visited: Set[Position]) -> List[Position]:\n possible_positions = [\n Position(from_position.x - 1, from_position.y),\n Position(from_position.x + 1, from_position.y),\n Position(from_position.x, from_position.y - 1),\n Position(from_position.x, from_position.y + 1),\n ]\n return [p for p in possible_positions if p in weights and p not in visited]\n\ndef find_cheapest_path(weights: Dict[Position, int], start: Position, end: Position):\n costs: Dict[Position, Cost] = dict()\n visited: Set[Position] = set()\n\n if start == end:\n return 0, []\n\n visited.add(start)\n for neighbour in get_adjacent_positions(start, weights, visited):\n costs[neighbour] = Cost(start, weights[neighbour])\n\n node = find_cheapest_node(costs, visited)\n i = 0\n while node is not None:\n i += 1\n if i == 10000:\n pct_visited = len(visited) / ((end.x + 1) * (end.y + 1))\n print(pct_visited)\n i = 0\n running_total = costs[node].cost\n for neighbour in get_adjacent_positions(node, weights, visited):\n new_cost = running_total + weights[neighbour]\n if neighbour not in costs:\n costs[neighbour] = Cost(node, new_cost)\n else:\n costs[neighbour].update(node, new_cost)\n visited.add(node)\n node = find_cheapest_node(costs, visited)\n if end not in costs:\n raise ValueError(f\"No route to {end}\")\n return costs[end], get_route(start, end, costs)\n\ndef make_weights(lines) -> Dict[Position, int]:\n result = {}\n for y in range(len(lines)):\n for x in range(len(lines[0])):\n result[Position(x, y)] = lines[y][x]\n return result\n\ndef find_end(weights: Dict[Position, int]) -> Position:\n max_x = 0\n max_y = 0\n for k in weights.keys():\n max_x = max(max_x, k.x)\n max_y = max(max_y, k.y)\n return Position(max_x, max_y)\n\ndef print_weights(weights: Dict[Position, int]):\n end = find_end(weights)\n result = \"\"\n for y in range(end.y + 1):\n for x in range(end.x + 1):\n result += str(weights[Position(x, y)])\n result += '\\n'\n print(result)",
"_____no_output_____"
],
[
"weights = make_weights(parse_text(SAMPLE_TEXT))\ncost, route = find_cheapest_path(weights, Position(0, 0), find_end(weights))\ncost.cost",
"% visited 1.0\n"
],
[
"weights = make_weights(parse_text(read_input()))\ncost, route = find_cheapest_path(weights, Position(0, 0), find_end(weights))\ncost.cost",
"% visited 1.0\n"
],
[
"print_weights(make_weights(parse_text(SAMPLE_TEXT)))",
"1163751742\n1381373672\n2136511328\n3694931569\n7463417111\n1319128137\n1359912421\n3125421639\n1293138521\n2311944581\n\n"
],
[
"def multiply_weights(original_weights: Dict[Position, int], times):\n end = find_end(original_weights)\n new_max_x = (end.x + 1) * times - 1\n new_max_y = (end.y + 1) * times - 1\n result = {}\n for x in range(new_max_x + 1):\n for y in range(new_max_y + 1):\n orig_x = x % (end.x + 1)\n orig_y = y % (end.y + 1)\n multiplier = (x // (end.x + 1)) + (y // (end.y + 1))\n original_value = original_weights[Position(orig_x, orig_y)]\n new_value = original_value + multiplier\n if new_value > 9:\n new_value -= 9\n result[Position(x, y)] = new_value\n return result",
"_____no_output_____"
],
[
"multiply_weights(make_weights(parse_text(SAMPLE_TEXT)), 5) == make_weights(parse_text(SAMPLE_TEXT_2))",
"_____no_output_____"
],
[
"new_weights = multiply_weights(make_weights(parse_text(SAMPLE_TEXT)), 5)\ncost, route = find_cheapest_path(new_weights, Position(0, 0), find_end(new_weights))\ncost.cost",
"_____no_output_____"
],
[
"# Need a better solution for the big map. The approach above takes way too long\nnew_weights = multiply_weights(make_weights(parse_text(read_input())), 5)\ncost, route = find_cheapest_path(new_weights, Position(0, 0), find_end(new_weights))\ncost.cost",
"0.04\n0.08\n0.12\n0.16\n0.2\n0.24\n0.28\n0.32\n0.36\n0.4\n0.44\n0.48\n0.52\n0.56\n0.6\n0.64\n0.68\n0.72\n0.76\n0.8\n0.84\n0.88\n0.92\n0.96\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a15417e5b94b5d0cbdf74f8b02f78943e60dbd6
| 5,549 |
ipynb
|
Jupyter Notebook
|
notebooks/Heartbeat-model.ipynb
|
adrn/apogee-kepler-binaries
|
1162f4472995a044cb440bb3cffa30c5e34d78b0
|
[
"MIT"
] | null | null | null |
notebooks/Heartbeat-model.ipynb
|
adrn/apogee-kepler-binaries
|
1162f4472995a044cb440bb3cffa30c5e34d78b0
|
[
"MIT"
] | null | null | null |
notebooks/Heartbeat-model.ipynb
|
adrn/apogee-kepler-binaries
|
1162f4472995a044cb440bb3cffa30c5e34d78b0
|
[
"MIT"
] | null | null | null | 30.827778 | 99 | 0.445125 |
[
[
[
"import os\n\nfrom astropy.time import Time\nimport astropy.coordinates as coord\nimport astropy.units as u\n\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport numpy as np\nfrom tqdm.notebook import tqdm\n\nfrom twobody import TwoBodyKeplerElements, KeplerOrbit\nfrom twobody import (eccentric_anomaly_from_mean_anomaly, \n true_anomaly_from_eccentric_anomaly)",
"_____no_output_____"
],
[
"def true_anomaly(orbit, time):\n # mean anomaly\n with u.set_enabled_equivalencies(u.dimensionless_angles()):\n M = 2*np.pi * (time.tcb - orbit.t0.tcb) / orbit.P - orbit.M0\n M = M.to(u.radian)\n\n # eccentric anomaly\n E = eccentric_anomaly_from_mean_anomaly(M, orbit.e)\n\n # true anomaly\n return true_anomaly_from_eccentric_anomaly(E, orbit.e)",
"_____no_output_____"
],
[
"def hb_model(S, i, omega, f, R, a):\n num = 1 - 3*np.sin(i)**2 * np.sin(f - omega)**2\n den = (R / a) ** 3\n return S * num / den",
"_____no_output_____"
],
[
"P = 20 * u.day\ne = 0.5\nS = 1.\n\nepoch = Time(Time.now().mjd, format='mjd')\nt = epoch + np.linspace(0, P.value, 8192)",
"_____no_output_____"
],
[
"for e in [0.3, 0.5, 0.7]:\n fig, axes = plt.subplots(6, 6, figsize=(16, 16), \n sharex=True, sharey=True,\n constrained_layout=True)\n\n n = 0\n omegas = np.linspace(-90, 90, axes.shape[0]) * u.deg\n incls = np.linspace(6, 90, axes.shape[0]) * u.deg\n for omega in omegas:\n for incl in incls:\n ax = axes.flat[n]\n\n elem = TwoBodyKeplerElements(P=P, e=e, \n m1=1.*u.Msun, m2=0.25*u.Msun,\n omega=omega, i=incl,\n t0=epoch)\n orbit1 = KeplerOrbit(elem.primary)\n orbit2 = KeplerOrbit(elem.secondary)\n\n x1 = orbit1.reference_plane(t)\n x2 = orbit2.reference_plane(t)\n\n R = (x1.data.without_differentials() - x2.data.without_differentials()).norm()\n a = elem.a\n f = true_anomaly(orbit1, t)\n\n phase = ((t.mjd - t.mjd.min()) / P.to_value(u.day) + 0.5) % 1 - 0.5\n y = hb_model(S, elem.i, elem.omega, f, R, a)\n y = y[phase.argsort()]\n phase = phase[phase.argsort()]\n ax.plot(phase, y, marker='', ls='-', lw=2, color='k')\n ax.plot(phase - 1, y, marker='', ls='-', lw=2, color='k')\n ax.plot(phase + 1, y, marker='', ls='-', lw=2, color='k')\n ax.axhline(0, marker='', zorder=-100, \n color='tab:blue', alpha=0.2)\n\n # plt.setp(ax.get_xticklabels(), fontsize=8)\n # plt.setp(ax.get_yticklabels(), fontsize=8)\n ax.xaxis.set_visible(False)\n ax.yaxis.set_visible(False)\n\n n += 1\n \n ax.set_xlim(-0.75, 0.75)\n \n n = 0\n for omega in omegas:\n for incl in incls:\n ax = axes.flat[n]\n \n xlim = ax.get_xlim()\n xspan = xlim[1] - xlim[0]\n ylim = ax.get_ylim()\n yspan = ylim[1] - ylim[0]\n ax.text(xlim[0] + 0.05 * xspan, \n ylim[0] + 0.05 * yspan, \n (rf'$\\omega = {omega.value:.1f}^\\circ$' + \n f'\\n$i = {incl.value:.0f}^\\circ$'),\n ha='left', va='bottom', fontsize=12)\n \n n += 1\n\n fig.suptitle(f'$e={e:.1f}$', fontsize=16)\n fig.set_facecolor('w')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a1542371cc2b9c67d375486246c4f44d7369ae8
| 13,579 |
ipynb
|
Jupyter Notebook
|
01-Fundamentos-Programacao-DS/Scripts/Mod-01/aula_04/.ipynb_checkpoints/exemplos_resolvidos-checkpoint.ipynb
|
fgandrade/Estudos-Comunidade-DS
|
bc956d4cb6237a305584ee20813ee47bfaca375c
|
[
"MIT"
] | null | null | null |
01-Fundamentos-Programacao-DS/Scripts/Mod-01/aula_04/.ipynb_checkpoints/exemplos_resolvidos-checkpoint.ipynb
|
fgandrade/Estudos-Comunidade-DS
|
bc956d4cb6237a305584ee20813ee47bfaca375c
|
[
"MIT"
] | null | null | null |
01-Fundamentos-Programacao-DS/Scripts/Mod-01/aula_04/.ipynb_checkpoints/exemplos_resolvidos-checkpoint.ipynb
|
fgandrade/Estudos-Comunidade-DS
|
bc956d4cb6237a305584ee20813ee47bfaca375c
|
[
"MIT"
] | null | null | null | 32.563549 | 326 | 0.535975 |
[
[
[
"# Exemplo 1",
"_____no_output_____"
],
[
"## Problema",
"_____no_output_____"
],
[
"Considere um arquivo de entrada no formato CSV (comma separated values) com informações relativas a acidentes na aviação civil brasileira nos últimos 10 anos (arquivo anv.csv)\n\nAs informações estão separadas pelo caracter separador ~ e entre “” (aspas) conforme o exemplo abaixo:\n\n```javascript\n\"201106142171203\"~\"PPGXE\"~\"AEROCLUBE\"~\"AVIÃO\"~\"NEIVA INDUSTRIA AERONAUTICA\"~\"56- C\"~\"PAUL\"~\"PISTÃO\"~\"MONOMOTOR\"~\"660\"~\"LEVE\"~\"2\"~\"1962\"~\"BRASIL\"~\"BRASIL\"~\"PRI\"~\" INSTRUÇÃO\"~\"SDPW\"~\"SDPW\"~\"INDETERMINADA\"~\"UNKNOWN\"~\"VOO DE INSTRUÇÃO\"~\"SUBSTANCIAL\"~\"0\"~\"2018-07-09“\n\n\"201707111402595\"~\"PPNCG\"~\"OPERADOR PARTICULAR\"~\"AVIÃO\"~\"PIPER AIRCRAFT\"~\"PA-46- 350P\"~\"PA46\"~\"TURBOÉLICE\"~\"MONOMOTOR\"~\"1950\"~\"LEVE\"~\"6\"~\"1990\"~\"NULL\"~\"BRASIL\"~\" TPP\"~\"PARTICULAR\"~\"SBBR\"~\"SBGR\"~\"POUSO\"~\"LANDING\"~\"VOO PRIVADO\"~\"NENHUM\"~\"0\"~\"2018-07-09\"'''\n```\n\nO arquivo é composto das seguintes colunas:\n1. codigo_ocorrencia\n2. aeronave_matricula\n3. aeronave_operador_categoria \n4. aeronave_tipo_veiculo\n5. aeronave_fabricante\n6. aeronave_modelo\n7. aeronave_tipo_icao\n8. aeronave_motor_tipo\n9. aeronave_motor_quantidade \n10. aeronave_pmd \n11. aeronave_pmd_categoria \n12. aeronave_assentos \n13. aeronave_ano_fabricacao\n14. aeronave_pais_fabricante \n15. aeronave_pais_registro \n16. aeronave_registro_categoria \n17. aeronave_registro_segmento \n18. aeronave_voo_origem \n19. aeronave_voo_destino \n20. aeronave_fase_operacao \n21. aeronave_fase_operacao_icao \n22. aeronave_tipo_operacao \n23. aeronave_nivel_dano \n24. total_fatalidades\n25. aeronave_dia_extracao\n\nCrie uma função que efetue a leitura do arquivo, sem a utilização de bibliotecas externas, e processe o arquivo produzindo dois novos arquivos.\n\nOBS: Efetuar apenas uma leitura do arquivo de entrada",
"_____no_output_____"
],
[
"### Arquivo 1",
"_____no_output_____"
],
[
"O primeiro arquivo deve ter seu conteúdo em formato JSON, com o nome statistics.json, e deve possuir as as estatísticas: \n* fase de operação\n* número de total de ocorrências\n* percentual de quanto essa fase representa dentro de todos os dados\n\n\nExemplo de como deve estar o arquivo:\n```json\n[\n {\n \"fase_operacao\": \"APROXIMAÇÃO FINAL\", \n \"ocorrencias\": 234,\n \"percentual\": \"4,51%\"\n },\n {\n \"fase_operacao\": \"INDETERMINADA\", \n \"ocorrencias\": 180,\n \"percentual\": \"2,43%\"\n },\n {\n \"fase_operacao\": \"MANOBRA\", \n \"ocorrencias\": 80,\n \"percentual\": \"0,95%\"\n }\n]\n```",
"_____no_output_____"
],
[
"### Arquivo 2",
"_____no_output_____"
],
[
"Crie um arquivo de saida (formato CSV) com nome levels.csv contendo as seguintes informações:\n* operation -> aeronave_operador_categoria\n* type -> aeronave_tipo_veiculo\n* manufacturer -> aeronave_fabricante\n* engine_type aeronave_motor_tipo\n* engines -> aeronave_motor_quantidade\n* year_manufacturing -> aeronave_ano_fabricacao\n* seating -> aeronave_assentos\n* fatalities -> total_fatalidades\n\nConsiderando apenas acidentes cujo nível de dano da aeronave tenha sido LEVE ou NENHUM (coluna aeronave_nivel_dano) e que o número de fatalidades (total_fatalidades) tenha sido superior à 0 (zero)",
"_____no_output_____"
],
[
"## Análise",
"_____no_output_____"
],
[
"Receberemos um arquio de entrada no formato CSV, com o caractere separador ~ e entre “”.\n\nO arquivo possuí 25 colunas\n\nDevemos criar uma função que lê e processa o arquivo, sem a utilizaão de bibliotecas, e cria outros dois arquivos de saída.\n\nTemos que efetuar a leitura do arquivo uma única vez.",
"_____no_output_____"
],
[
"## Implicações / Proposições / Afirmações",
"_____no_output_____"
],
[
"* Receberemos um arquivo de entrada\n* O arquivo contém 10 anos de dados de acidentes de avição\n* O arquivo contém 25 colunas\n* O caractere separado do arquivo é `~` e `\"\"`\n* Devemos criar uma função que irá ler e precessar o arquivo de entrada\n* Devemos criar dois arquivo de saída: um `.json`, com o nome `statistics.json`; e outro arquivo `.csv` com o nome `levels.csv`\n* O arquivo `.json` deve ter um formato específico\n* O arquivo `.json` deve conter as seguintes informações:\n * fase de operação\n * número de total de ocorrências\n * percentual de quanto essa fase representa dentro de todos os dados\n",
"_____no_output_____"
],
[
"## Resolução e Resposta",
"_____no_output_____"
]
],
[
[
"# import das bibliotecas\nimport csv\nimport json",
"_____no_output_____"
],
[
"# Função que irá processar o Arquivo\ndef read_file(file_path):\n \"\"\"Função que irá processar o arquivo de entrada para ajustar\n as informações requisitadas de saída.\n \n Argumentos:\n file_path -- Caminho do arquivo de entrada\n \"\"\"\n \n # Estruturas que serão utilizadas para produzir as respostas\n map_of_incidents = {} # Arquivo que produzirá a saída JSON\n list_of_incidents = [] # Arquivo que produzirá a saída CSV\n count = 0 # Variável utilizada para contarmos a quantidade de elementos\n \n # Abre e processa o arquivo\n with open(file_path, 'r', encoding='utf-8') as f:\n reader = csv.DictReader(f, delimiter='~', quotechar='\"')\n \n # Itera pelas linhas do arquivo para \n for row in reader:\n count += 1 \n key = row['aeronave_fase_operacao']\n \n # Verifica se a chave ja existe no dicionário que será\n # utilizado para escrever o arquivo JSON\n \n if key in map_of_incidents:\n map_of_incidents[key] += 1 # Se existir, incrementa mais mais um acidente\n else:\n map_of_incidents[key] = 1 # Se não existir, cria a chave e adiciona um acidente\n \n # Verifica se o dano na aeronave foi leve ou nenhum E se as\n # fatalidades foram maiores que zero\n if ( \n (row['aeronave_nivel_dano'] == 'LEVE' or row['aeronave_nivel_dano'] == 'NENHUM' ) and\n (int(row['total_fatalidades']) > 0) \n ):\n # Se sim, adiciona as informações pertinentes na lista que \n # será utilizada para escrever o arquivo CSV\n list_of_incidents.append([row['aeronave_operador_categoria'],\n row['aeronave_tipo_veiculo'],\n row['aeronave_fabricante'],\n row['aeronave_motor_tipo'],\n row['aeronave_motor_quantidade'],\n row['aeronave_ano_fabricacao'],\n row['aeronave_assentos'],\n row['total_fatalidades']])\n \n # Assim que juntarmos todas as informações, vamos criar os\n # arquivos necessários\n save_json(map_of_incidents, count)\n save_csv(list_of_incidents)",
"_____no_output_____"
],
[
"# Função que irá criar o arquivo JSON\ndef save_json(data, count):\n \"\"\"Função resposável por criar o arquivo JSON\n \n Argumentos:\n data -- Dados que será escritos no arquivo\n count -- Quantidade de acidentes registrados no arquivo de entrada\n \"\"\"\n \n # Lista que será escrita dentro do arquivo JSON\n array_of_incidents = []\n \n # Itera sobre o dicionário com os dados\n for key, value in data.items():\n array_of_incidents.append(\n {\n \"fase_operacao\": key,\n \"ocorrencias\": value,\n \"percentual\": '{:.3%}'.format(value / count)\n }\n )\n \n # Cria o arquivo com os dados ajustados\n with open('statistics.json', 'w', encoding='utf8') as outfile:\n json.dump(array_of_incidents, outfile, ensure_ascii=False, indent=4) ",
"_____no_output_____"
],
[
"# Função que irá criar o arquivo CSV\ndef save_csv(data):\n \"\"\"Função responsável por criar o arquivo .csv\n \n Argumentos\n data -- dados que serão escritos no arquivo\n \"\"\"\n \n # Cabeçalho do arquivo .csv\n header = [\n 'operation',\n 'type',\n 'manufacturer',\n 'engine_type',\n 'engines',\n 'year_manufacturing',\n 'seating',\n 'fatalities'\n ]\n \n # Cria o arquivo com os dados ajustados\n with open('levels.csv', 'w', encoding='utf8', newline='') as outfile:\n w = csv.writer(outfile)\n w.writerow(header)\n w.writerows(data)",
"_____no_output_____"
],
[
"# Execução da Aplicação\nread_file('csv/anv.csv')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a154b54946aa995537e3e560dba90ddc9bbc363
| 96,222 |
ipynb
|
Jupyter Notebook
|
site/ko/swift/tutorials/python_interoperability.ipynb
|
justaverygoodboy/docs-l10n
|
8d4857750f2b5e8e6889acbb4b1e2f98ad7ce34e
|
[
"Apache-2.0"
] | null | null | null |
site/ko/swift/tutorials/python_interoperability.ipynb
|
justaverygoodboy/docs-l10n
|
8d4857750f2b5e8e6889acbb4b1e2f98ad7ce34e
|
[
"Apache-2.0"
] | null | null | null |
site/ko/swift/tutorials/python_interoperability.ipynb
|
justaverygoodboy/docs-l10n
|
8d4857750f2b5e8e6889acbb4b1e2f98ad7ce34e
|
[
"Apache-2.0"
] | null | null | null | 133.456311 | 76,010 | 0.862194 |
[
[
[
"##### Copyright 2018 The TensorFlow Authors. [Licensed under the Apache License, Version 2.0](#scrollTo=ByZjmtFgB_Y5).",
"_____no_output_____"
]
],
[
[
"// #@title Licensed under the Apache License, Version 2.0 (the \"License\"); { display-mode: \"form\" }\n// Licensed under the Apache License, Version 2.0 (the \"License\");\n// you may not use this file except in compliance with the License.\n// You may obtain a copy of the License at\n//\n// https://www.apache.org/licenses/LICENSE-2.0\n//\n// Unless required by applicable law or agreed to in writing, software\n// distributed under the License is distributed on an \"AS IS\" BASIS,\n// WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n// See the License for the specific language governing permissions and\n// limitations under the License.",
"_____no_output_____"
]
],
[
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/swift/tutorials/python_interoperability\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />TensorFlow.org에서 보기</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/ko/swift/tutorials/python_interoperability.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />구글 코랩(Colab)에서 실행하기</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/ko/swift/tutorials/python_interoperability.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />깃허브(GitHub)에서 소스 보기</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/ko/swift/tutorials/python_interoperability.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: 이 문서는 텐서플로 커뮤니티에서 번역했습니다. 커뮤니티 번역 활동의 특성상 정확한 번역과 최신 내용을 반영하기 위해 노력함에도\n불구하고 [공식 영문 문서](https://www.tensorflow.org/?hl=en)의 내용과 일치하지 않을 수 있습니다.\n이 번역에 개선할 부분이 있다면\n[tensorflow/docs-l10n](https://github.com/tensorflow/docs-l10n/) 깃헙 저장소로 풀 리퀘스트를 보내주시기 바랍니다.\n문서 번역이나 리뷰에 참여하려면\n[[email protected]](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs-ko)로\n메일을 보내주시기 바랍니다.",
"_____no_output_____"
],
[
"# 파이썬 상호 호환성\n\n텐서플로를 위한 스위프트는 파이썬과 상호 호환됩니다.\n\n스위프트에서 파이썬 모듈을 임포트해서, 스위프트와 파이썬 사이의 값을 바꾸거나 파이썬 함수를 호출할 수 있습니다.",
"_____no_output_____"
]
],
[
[
"// comment so that Colab does not interpret `#if ...` as a comment\n#if canImport(PythonKit)\n import PythonKit\n#else\n import Python\n#endif\nprint(Python.version)",
"3.6.9 (default, Nov 7 2019, 10:44:02) \r\n[GCC 8.3.0]\r\n"
]
],
[
[
"## 파이썬 버전 설정",
"_____no_output_____"
],
[
"기본적으로 `import Python`를 하면, 스위프트는 시스템 라이브러리 경로에 따라 설치된 최신 버전의 파이썬을 검색합니다. \n특정한 파이썬을 설치하려면, `PYTHON_LIBRARY` 환경변수에 설치시 제공받은 `libpython` 공유 라이브러리를 설정합니다. 예를 들어: \n\n`export PYTHON_LIBRARY=\"~/anaconda3/lib/libpython3.7m.so\"`\n\n정확한 파일명은 파이썬 환경과 플랫폼마다 다를 수 있습니다.",
"_____no_output_____"
],
[
"또는 스위프트가 시스템 라이브러리 경로에서 알맞은 파이썬 버전을 찾도록 해서 `PYTHON_VERSION` 환경변수를 설정할 수 있습니다. `PYTHON_LIBRARY`가 `PYTHON_VERSION` 보다 우선한다는 점을 유의해야 합니다.\n\n또한 코드에서 `PYTHON_VERSION` 설정과 동일한 기능을 하는 `PythonLibrary.useVersion` 함수를 호출할 수 있습니다.",
"_____no_output_____"
]
],
[
[
"// PythonLibrary.useVersion(2)\n// PythonLibrary.useVersion(3, 7)",
"_____no_output_____"
]
],
[
[
"__Note: 파이썬을 임포트한 직후, 파이썬 코드를 호출하기 전에 `PythonLibrary.useVersion`을 실행해야 합니다. 이것은 파이썬 버전을 동적으로 바꾸는 데 사용될 수 없습니다.__",
"_____no_output_____"
],
[
"[파이썬 라이브러리 로딩 과정에서 생성되는 디버그 출력](https://github.com/apple/swift/pull/20674#discussion_r235207008)을 확인하기 위해서 `PYTHON_LOADER_LOGGING=1`를 설정하세요. ",
"_____no_output_____"
],
[
"## 기초\n\n스위프트에서 `PythonObject`는 파이썬 객체를 나타냅니다.\n모든 파이썬 API는 `PythonObject` 인스턴스를 사용하거나 반환합니다.\n\n스위프트에서 기본형(숫자 및 배열처럼)은 `PythonObject`로 전환할 수 있습니다. 몇몇 경우에 (`PythonConvertible` 인수를 받는 리터럴과 함수의 경우), 변환이 암묵적으로 일어납니다. 스위프트 값을 `PythonObject`에 명시적으로 지정하려면 `PythonObject` 이니셜라이저를 사용합니다.\n\n`PythonObject`는 숫자 연산, 인덱싱, 반복을 포함한 많은 표준 연산을 정의합니다.",
"_____no_output_____"
]
],
[
[
"// 표준 스위프트 자료형을 Python으로 변환합니다.\nlet pythonInt: PythonObject = 1\nlet pythonFloat: PythonObject = 3.0\nlet pythonString: PythonObject = \"Hello Python!\"\nlet pythonRange: PythonObject = PythonObject(5..<10)\nlet pythonArray: PythonObject = [1, 2, 3, 4]\nlet pythonDict: PythonObject = [\"foo\": [0], \"bar\": [1, 2, 3]]\n\n// 파이썬 객체에 표준 연산을 수행합니다.\nprint(pythonInt + pythonFloat)\nprint(pythonString[0..<6])\nprint(pythonRange)\nprint(pythonArray[2])\nprint(pythonDict[\"bar\"])",
"4.0\r\nHello \r\nslice(5, 10, None)\r\n3\r\n[1, 2, 3]\r\n"
],
[
"// 파이썬 객체를 다시 스위프트로 변환합니다.\nlet int = Int(pythonInt)!\nlet float = Float(pythonFloat)!\nlet string = String(pythonString)!\nlet range = Range<Int>(pythonRange)!\nlet array: [Int] = Array(pythonArray)!\nlet dict: [String: [Int]] = Dictionary(pythonDict)!\n\n// 표준 연산을 수행합니다.\n// 출력은 파이썬과 동일합니다!\nprint(Float(int) + float)\nprint(string.prefix(6))\nprint(range)\nprint(array[2])\nprint(dict[\"bar\"]!)",
"4.0\r\nHello \r\n5..<10\r\n3\r\n[1, 2, 3]\r\n"
]
],
[
[
"`PythonObject`는 많은 표준 스위프트 프로토콜에 대해 적합하도록 정의합니다:\n* `Equatable`\n* `Comparable`\n* `Hashable`\n* `SignedNumeric`\n* `Strideable`\n* `MutableCollection`\n* 모든 `ExpressibleBy_Literal` 프로토콜\n\n이러한 적합성은 형안전(type-safe)하지 않다는 점에 유의해야 합니다: 호환되지 않는 `PythonObject` 인스턴스에서 프로토콜 기능을 사용하려고 할 때 충돌이 발생할 수 있습니다.",
"_____no_output_____"
]
],
[
[
"let one: PythonObject = 1\nprint(one == one)\nprint(one < one)\nprint(one + one)\n\nlet array: PythonObject = [1, 2, 3]\nfor (i, x) in array.enumerated() {\n print(i, x)\n}",
"true\r\nfalse\r\n2\r\n0 1\r\n1 2\r\n2 3\r\n"
]
],
[
[
"튜플을 파이썬에서 스위프트로 변환하려면, 정확한 튜플의 길이를 알아야 합니다.\n\n다음 인스턴스 메서드 중 하나를 호출합니다:\n- `PythonObject.tuple2`\n- `PythonObject.tuple3`\n- `PythonObject.tuple4`",
"_____no_output_____"
]
],
[
[
"let pythonTuple = Python.tuple([1, 2, 3])\nprint(pythonTuple, Python.len(pythonTuple))\n\n// 스위프트로 변환합니다.\nlet tuple = pythonTuple.tuple3\nprint(tuple)",
"(1, 2, 3) 3\r\n(1, 2, 3)\r\n"
]
],
[
[
"## 파이썬 내장 객체\n\n전역 `Python` 인터페이스를 활용해 파이썬 내장 객체에 접근합니다.",
"_____no_output_____"
]
],
[
[
"// `Python.builtins`은 모든 파이썬 내장 객체의 딕셔너리입니다.\n_ = Python.builtins\n\n// 파이썬 내장 객체를 사용합니다.\nprint(Python.type(1))\nprint(Python.len([1, 2, 3]))\nprint(Python.sum([1, 2, 3]))",
"<class 'int'>\r\n3\r\n6\r\n"
]
],
[
[
"## 파이썬 모듈 임포트\n\n`Python.import`를 사용하여 파이썬 모듈을 임포트합니다. 이것은 `Python`의 `import` 키워드처럼 동작합니다.",
"_____no_output_____"
]
],
[
[
"let np = Python.import(\"numpy\")\nprint(np)\nlet zeros = np.ones([2, 3])\nprint(zeros)",
"<module 'numpy' from '/usr/local/lib/python3.6/dist-packages/numpy/__init__.py'>\r\n[[1. 1. 1.]\r\n [1. 1. 1.]]\r\n"
]
],
[
[
"안전하게 패키지를 가져오기 위해 예외처리 함수 `Python.attemptImport`를 사용하세요.",
"_____no_output_____"
]
],
[
[
"let maybeModule = try? Python.attemptImport(\"nonexistent_module\")\nprint(maybeModule)",
"nil\r\n"
]
],
[
[
"## `numpy.ndarray`로 변환\n\n다음 스위프트 자료형은 `numpy.ndarray`로 변환할 수 있습니다:\n- `Array<Element>`\n- `ShapedArray<Scalar>`\n- `Tensor<Scalar>`\n\n`Numpy.ndarray`의 `dtype`이 `Element` 또는 `Scalar`의 일반 파라미터 타입과 호환되어야만 변환이 성공합니다.\n\n`Array`의 경우 `numpy.ndarray`가 1-D일 경우에만 `numpy`에서 변환이 성공합니다.",
"_____no_output_____"
]
],
[
[
"import TensorFlow\n\nlet numpyArray = np.ones([4], dtype: np.float32)\nprint(\"Swift type:\", type(of: numpyArray))\nprint(\"Python type:\", Python.type(numpyArray))\nprint(numpyArray.shape)",
"Swift type: PythonObject\r\nPython type: <class 'numpy.ndarray'>\r\n(4,)\r\n"
],
[
"// `numpy.ndarray`에서 스위프트 타입으로 변환하는 예제.\nlet array: [Float] = Array(numpy: numpyArray)!\nlet shapedArray = ShapedArray<Float>(numpy: numpyArray)!\nlet tensor = Tensor<Float>(numpy: numpyArray)!\n\n// 스위프트 타입에서 `numpy.ndarray`으로 변환하는 예제.\nprint(array.makeNumpyArray())\nprint(shapedArray.makeNumpyArray())\nprint(tensor.makeNumpyArray())\n\n// dtype이 다른 예제.\nlet doubleArray: [Double] = Array(numpy: np.ones([3], dtype: np.float))!\nlet intTensor = Tensor<Int32>(numpy: np.ones([2, 3], dtype: np.int32))!",
"[1. 1. 1. 1.]\r\n[1. 1. 1. 1.]\r\n[1. 1. 1. 1.]\r\n"
]
],
[
[
"## 이미지 표시\n\n파이썬 노트북에서처럼 `matplotlib`를 이용해 이미지를 결과 창에 표시할 수 있습니다.",
"_____no_output_____"
]
],
[
[
"// 주피터 노트북에 그래프를 표시하기 위한 셀입니다.\n// 다른 환경에서는 사용하지 마세요.\n%include \"EnableIPythonDisplay.swift\"\nIPythonDisplay.shell.enable_matplotlib(\"inline\")",
"_____no_output_____"
],
[
"let np = Python.import(\"numpy\")\nlet plt = Python.import(\"matplotlib.pyplot\")\n\nlet time = np.arange(0, 10, 0.01)\nlet amplitude = np.exp(-0.1 * time)\nlet position = amplitude * np.sin(3 * time)\n\nplt.figure(figsize: [15, 10])\n\nplt.plot(time, position)\nplt.plot(time, amplitude)\nplt.plot(time, -amplitude)\n\nplt.xlabel(\"Time (s)\")\nplt.ylabel(\"Position (m)\")\nplt.title(\"Oscillations\")\n\nplt.show()",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a15521ac9d1251b35210ad54d2e4bcb6193818c
| 64,903 |
ipynb
|
Jupyter Notebook
|
Assignment2_DarJuan.ipynb
|
darjuangeloys/LinearAlgebra2021
|
81a0614c50c2102a4de743531b7600124340118d
|
[
"Apache-2.0"
] | null | null | null |
Assignment2_DarJuan.ipynb
|
darjuangeloys/LinearAlgebra2021
|
81a0614c50c2102a4de743531b7600124340118d
|
[
"Apache-2.0"
] | null | null | null |
Assignment2_DarJuan.ipynb
|
darjuangeloys/LinearAlgebra2021
|
81a0614c50c2102a4de743531b7600124340118d
|
[
"Apache-2.0"
] | null | null | null | 57.538121 | 12,698 | 0.744619 |
[
[
[
"<a href=\"https://colab.research.google.com/github/darjuangeloys/LinearAlgebra2021/blob/main/Assignment2_DarJuan.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# **Welcome to Python Fundamentals!**\n\n\nIn this module, we are going to establish our skills in Python programming. In this notebook we are going to cover:\n\n\n* Variables and Data Types\n* Operations\n* Input and Output Operations\n* Logic Control\n* Iterables\n* Functions\n\n\n\n\n\n",
"_____no_output_____"
],
[
"## ***Variables and Data Types***\n\n\nVariables and data types in python as the name suggests are the values that vary. In a programming language, a variable is a memory location where you store a value. The value that you have stored may change in the future according to the specifications. A variable in python is created as soon as a value is assigned to it. [1]",
"_____no_output_____"
]
],
[
[
"x = 1\na, b = 3, -2\n",
"_____no_output_____"
],
[
"type(x)",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
],
[
[
"y = 3.0\ntype(y)",
"_____no_output_____"
],
[
"x = float(x)\ntype(x)\nx",
"_____no_output_____"
],
[
"s, t, u = \"1\", '3', 'three'\ntype(u)",
"_____no_output_____"
]
],
[
[
"## ***Operations***\n\n\nOperators are special symbols in Python that carry out arithmetic or logical computation. The value that the operator operates on is called the \"operand\" [2]",
"_____no_output_____"
],
[
"### *Arithmetic*\n\n\nArithmetic operators are used to perform mathematical operations like addition, subtraction, multiplication, etc.",
"_____no_output_____"
]
],
[
[
"w, x, y, z = 4.0, -3.0, 1, -32",
"_____no_output_____"
],
[
"### Addition\nS = w + x",
"_____no_output_____"
],
[
"### Subtraction\nD = y - z",
"_____no_output_____"
],
[
"### Multiplication\nP = w*z",
"_____no_output_____"
],
[
"### Division\nQ = y/x",
"_____no_output_____"
],
[
"### Floor Division\nQf = w//x\nQf",
"_____no_output_____"
]
],
[
[
"***What's the difference between a standard division and a floor division?***\n\n-In floor division, it returns the largest possible integer. [3]",
"_____no_output_____"
]
],
[
[
"### Exponentiation\nE = w**y",
"_____no_output_____"
],
[
"### Modulo \nmod = z%x\nmod",
"_____no_output_____"
]
],
[
[
"***What is Modulo?***\n\n\n\nIf in a floor division, it gives the largest possible integer, Modulo gives the remainder of the two divided values. [4]",
"_____no_output_____"
],
[
"### *Assignment*\n\nAssignment operators are used to assign values to variables.",
"_____no_output_____"
]
],
[
[
"A, B, C, D, E = 0, 100, 2, 1, 2",
"_____no_output_____"
],
[
"A += w",
"_____no_output_____"
],
[
"B -= x",
"_____no_output_____"
],
[
"C *= w",
"_____no_output_____"
],
[
"D /= x",
"_____no_output_____"
],
[
"E **= y",
"_____no_output_____"
]
],
[
[
"### *Comparators*\n\n\nComparators or Comparison operators are used to compare values. It returns either True or False according to the condition. ",
"_____no_output_____"
]
],
[
[
"size_1, size_2, size_3 = 1, 2.0, \"1\"\ntrue_size = 1.0;",
"_____no_output_____"
],
[
"## Equality\nsize_1 == true_size",
"_____no_output_____"
],
[
"## Non-equality\nsize_2 != true_size",
"_____no_output_____"
],
[
"## Inequality\ns1 = size_1 > size_2\ns2 = size_1 < size_2/2\ns3 = true_size >= size_1\ns4 = size_2 <= true_size",
"_____no_output_____"
]
],
[
[
"### *Logical*\nLogical operators are the and, or, not operators ",
"_____no_output_____"
]
],
[
[
"size_1 == true_size",
"_____no_output_____"
],
[
"size_1 is true_size",
"_____no_output_____"
],
[
"size_1 is not true_size",
"_____no_output_____"
],
[
"P, Q = True, False\nconj = P and Q",
"_____no_output_____"
],
[
"disj = P or Q\ndisj",
"_____no_output_____"
],
[
"nand = not(P and Q)\nnand",
"_____no_output_____"
],
[
"xor = (not P and Q) or (P and not Q)\nxor",
"_____no_output_____"
]
],
[
[
"## ***Input and Output***\nThese are functions used to connect with the user of the program. We use the print() function to output data to the standard output device and we use the input () to take the input from the user. [5]",
"_____no_output_____"
]
],
[
[
"print(\"Hello World!\")",
"Hello World!\n"
],
[
"cnt = 14000",
"_____no_output_____"
],
[
"string = \"Hello World!\"\nprint(string, \", Current COVID count is:\", cnt)\ncnt += 10000",
"_____no_output_____"
],
[
"print(f\"{string}, current count is: {cnt}\")",
"Hello World!, current count is: 64007\n"
],
[
"sem_grade = 86.25\nname = \"Lois\"\nprint(\"Hello {}, your semestral grades is: {}\". format(name, sem_grade))",
"Hello Lois, your semestral grades is: 86.25\n"
],
[
"pg, mg, fg = 0.3, 0.3, 0.4\nprint(\"The weights of your semestral grade are:\\\n\\n\\t {:.2%} for Prelims\\\n\\n\\t {:.2%} for Midterms, and\\\n\\n\\t {:.2%} for Finals.\".format(pg, mg, fg))",
"_____no_output_____"
],
[
"e = input(\"Enter a number: \")\ne",
"_____no_output_____"
],
[
"name = input(\"Enter your name: \")\npg = input(\"Enter prelim grade: \")\nmg = input(\"Enter midterm grade: \")\nfg = input(\"Enter final grade: \")\nsem_grade = None\nprint(\"Hello {}, your semestral grade is: {}\".format(name, sem_grade))",
"_____no_output_____"
]
],
[
[
"## ***Looping statements***\nLoops are powerful programming concepts supported by almost all modern programming languages. It allows a program to implement iterations, which basically means executing the same block of code two or more times depending on the specified conditions of the user or the codes in the program itself. [6]",
"_____no_output_____"
],
[
"## *While*\n\n\nA while loop is used to iterate over a block of code as long as the test expression or condition is \"true\". [7]",
"_____no_output_____"
]
],
[
[
"## while loops\ni, j = 0, 10\nwhile(i <= j):\n print(f\"{i}\\t|\\t{j}\")\n i += 1",
"0\t|\t10\n1\t|\t10\n2\t|\t10\n3\t|\t10\n4\t|\t10\n5\t|\t10\n6\t|\t10\n7\t|\t10\n8\t|\t10\n9\t|\t10\n10\t|\t10\n"
]
],
[
[
"## *For*\n\n\nThe for loop in Python is used to iterate over a sequence (list, tuple, string) or other iterable objects [8]",
"_____no_output_____"
]
],
[
[
"# for(int = 0; i<=10, i++){\n# printf(i)\n# }\n\ni = 0 \nfor i in range(11):\n print(i)",
"_____no_output_____"
],
[
"playlist = [\"Bahay Kubo\", \"Magandang Kanta\", \"Buko\"]\nprint('Now Playing:\\n')\nfor song in playlist: \n print(song)",
"Now Playing:\n\nBahay Kubo\nMagandang Kanta\nBuko\n"
]
],
[
[
"## ***Flow Control***\nFlow control is the order in which statements or the blocks of code in the program are executed at runtime based on a condition. [9]",
"_____no_output_____"
],
[
"### *Condition Statements*",
"_____no_output_____"
]
],
[
[
"num_1, num_2 = 14, 12\nif(num_1 == num_2):\n print(\"HAHA\")\nelif(num_1>num_2):\n print(\"HOHO\")\nelse:\n print(\"HUHU\")",
"_____no_output_____"
]
],
[
[
"## ***Functions***\nA function is a group of related statements that performs a specific task. It helps break our program into smaller and modular chunks, making it more organized and manageable. [10]",
"_____no_output_____"
]
],
[
[
"# void DeleteUser (int userid){\n# delete(userid);\n#}\n\ndef delete_user (userid):\n print(\"Successfully deleted user: {}\".format(userid))",
"_____no_output_____"
],
[
"userid = 2020_100100\ndelete_user(2020_100100)",
"Successfully deleted user: 2020100100\n"
],
[
"def add(addend1, addend2):\n sum = addend1 + addend2\n return sum",
"_____no_output_____"
],
[
"add(5, 4)",
"_____no_output_____"
]
],
[
[
"## **References:**\n\n[1] Variables and Data Types in Python (2021). (https://www.edureka.co/blog/variables-and-data-types-in-python/)\n\n[2] Python Operators (2021). (https://www.programiz.com/python-programming/operators)\n\n[3] Python Floor Division (2021). https://www.pythontutorial.net/advanced-python/python-floor-division/\n\n[4] Python Modulo in Practice: How to Use the % Operator (2020). (https://realpython.com/python-modulo-operator/)\n\n[5] Python Input, Output, and Import (2021). (https://www.programiz.com/python-programming/input-output-import)\n\n[6] Python Loops – For, While, Nested Loops With Examples (2021). (https://www.softwaretestinghelp.com/python/looping-in-python-for-while-nested-loops/)\n\n[7] Python while Loop (2021). (https://www.programiz.com/python-programming/while-loop)\n\n[8] Python for Loop (2021). (https://www.programiz.com/python-programming/for-loop)\n\n[9] Python Control Flow Statements and Loops (2021). (https://pynative.com/python-control-flow-statements/)\n\n[10] Python Functions (2021). (https://www.programiz.com/python-programming/function)",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a1555caf9cca5bc65043b1aeb917b3097840a7a
| 436,264 |
ipynb
|
Jupyter Notebook
|
tmp/keras_resnet.ipynb
|
angellmethod/DogFaceNet
|
83b4f58978faea9efd8cb64160c26f4f5085f931
|
[
"MIT"
] | 79 |
2019-01-08T02:35:24.000Z
|
2022-03-30T11:56:31.000Z
|
tmp/keras_resnet.ipynb
|
angellmethod/DogFaceNet
|
83b4f58978faea9efd8cb64160c26f4f5085f931
|
[
"MIT"
] | 3 |
2020-01-13T06:25:57.000Z
|
2021-08-10T01:34:05.000Z
|
tmp/keras_resnet.ipynb
|
angellmethod/DogFaceNet
|
83b4f58978faea9efd8cb64160c26f4f5085f931
|
[
"MIT"
] | 34 |
2019-07-24T12:25:13.000Z
|
2022-03-16T07:13:47.000Z
| 99.626399 | 1,816 | 0.686717 |
[
[
[
"import keras\nimport keras.layers as KL",
"Using TensorFlow backend.\n"
],
[
"mod = keras.applications.nasnet.NASNetMobile(input_shape=(224,224,3), include_top=False, weights='imagenet', input_tensor=None, pooling=None)",
"_____no_output_____"
],
[
"mod.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 224, 224, 3) 0 \n__________________________________________________________________________________________________\nstem_conv1 (Conv2D) (None, 111, 111, 32) 864 input_1[0][0] \n__________________________________________________________________________________________________\nstem_bn1 (BatchNormalization) (None, 111, 111, 32) 128 stem_conv1[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_stem_1 (Conv2D (None, 111, 111, 11) 352 activation_1[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_stem_1 (BatchNor (None, 111, 111, 11) 44 reduction_conv_1_stem_1[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 111, 111, 11) 0 reduction_bn_1_stem_1[0][0] \n__________________________________________________________________________________________________\nactivation_4 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 115, 115, 11) 0 activation_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 117, 117, 32) 0 activation_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 56, 56, 11) 396 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 56, 56, 11) 1920 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_3 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_5 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 56, 56, 11) 396 activation_3[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 56, 56, 11) 660 activation_5[0][0] \n__________________________________________________________________________________________________\nactivation_6 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 117, 117, 32) 0 activation_6[0][0] \n__________________________________________________________________________________________________\nactivation_8 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_stem_1 (Add) (None, 56, 56, 11) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 56, 56, 11) 1920 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 115, 115, 32) 0 activation_8[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 56, 56, 11) 0 reduction_add_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 56, 56, 11) 1152 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 56, 56, 11) 220 activation_10[0][0] \n__________________________________________________________________________________________________\nactivation_7 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_stem_1 (ZeroPad (None, 113, 113, 11) 0 reduction_bn_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 56, 56, 11) 660 activation_7[0][0] \n__________________________________________________________________________________________________\nactivation_9 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_stem_1 (MaxPool (None, 56, 56, 11) 0 reduction_pad_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 56, 56, 11) 396 activation_9[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 56, 56, 11) 220 activation_11[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_stem_2 (Activatio (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_stem_1 (Add) (None, 56, 56, 11) 0 reduction_left2_stem_1[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_stem_1 (Average (None, 56, 56, 11) 0 reduction_pad_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_stem_1 (Average (None, 56, 56, 11) 0 reduction_add_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_stem_1 (MaxPoo (None, 56, 56, 11) 0 reduction_pad_1_stem_1[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_1 (ZeroPadding2D (None, 112, 112, 32) 0 adjust_relu_1_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_add3_stem_1 (Add) (None, 56, 56, 11) 0 reduction_left3_stem_1[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_1 (Add) (None, 56, 56, 11) 0 reduction_add_2_stem_1[0][0] \n reduction_left4_stem_1[0][0] \n__________________________________________________________________________________________________\nreduction_add4_stem_1 (Add) (None, 56, 56, 11) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_stem_1[0][0] \n__________________________________________________________________________________________________\ncropping2d_1 (Cropping2D) (None, 111, 111, 32) 0 zero_padding2d_1[0][0] \n__________________________________________________________________________________________________\nreduction_concat_stem_1 (Concat (None, 56, 56, 44) 0 reduction_add_2_stem_1[0][0] \n reduction_add3_stem_1[0][0] \n add_1[0][0] \n reduction_add4_stem_1[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_stem_2 (Avera (None, 56, 56, 32) 0 adjust_relu_1_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_stem_2 (Avera (None, 56, 56, 32) 0 cropping2d_1[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 56, 56, 44) 0 reduction_concat_stem_1[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_stem_2 (Conv2D) (None, 56, 56, 11) 352 adjust_avg_pool_1_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_stem_2 (Conv2D) (None, 56, 56, 11) 352 adjust_avg_pool_2_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_stem_2 (Conv2D (None, 56, 56, 22) 968 activation_12[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 56, 56, 22) 0 adjust_conv_1_stem_2[0][0] \n adjust_conv_2_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_stem_2 (BatchNor (None, 56, 56, 22) 88 reduction_conv_1_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_bn_stem_2 (BatchNormaliz (None, 56, 56, 22) 88 concatenate_1[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 56, 56, 22) 0 reduction_bn_1_stem_2[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 56, 56, 22) 0 adjust_bn_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 59, 59, 22) 0 activation_13[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 61, 61, 22) 0 activation_15[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 28, 28, 22) 1034 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 28, 28, 22) 1562 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_16 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 28, 28, 22) 1034 activation_14[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 28, 28, 22) 1562 activation_16[0][0] \n__________________________________________________________________________________________________\nactivation_17 (Activation) (None, 56, 56, 22) 0 adjust_bn_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 61, 61, 22) 0 activation_17[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 56, 56, 22) 0 adjust_bn_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_stem_2 (Add) (None, 28, 28, 22) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 28, 28, 22) 1562 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 59, 59, 22) 0 activation_19[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 28, 28, 22) 0 reduction_add_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 28, 28, 22) 1034 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 28, 28, 22) 682 activation_21[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_stem_2 (ZeroPad (None, 57, 57, 22) 0 reduction_bn_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 28, 28, 22) 1562 activation_18[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_stem_2 (MaxPool (None, 28, 28, 22) 0 reduction_pad_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 28, 28, 22) 1034 activation_20[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 28, 28, 22) 682 activation_22[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_0 (Activation) (None, 56, 56, 44) 0 reduction_concat_stem_1[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_stem_2 (Add) (None, 28, 28, 22) 0 reduction_left2_stem_2[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_stem_2 (Average (None, 28, 28, 22) 0 reduction_pad_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_stem_2 (Average (None, 28, 28, 22) 0 reduction_add_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_stem_2 (MaxPoo (None, 28, 28, 22) 0 reduction_pad_1_stem_2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_2 (ZeroPadding2D (None, 57, 57, 44) 0 adjust_relu_1_0[0][0] \n__________________________________________________________________________________________________\nreduction_add3_stem_2 (Add) (None, 28, 28, 22) 0 reduction_left3_stem_2[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_2 (Add) (None, 28, 28, 22) 0 reduction_add_2_stem_2[0][0] \n reduction_left4_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_add4_stem_2 (Add) (None, 28, 28, 22) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_stem_2[0][0] \n__________________________________________________________________________________________________\ncropping2d_2 (Cropping2D) (None, 56, 56, 44) 0 zero_padding2d_2[0][0] \n__________________________________________________________________________________________________\nreduction_concat_stem_2 (Concat (None, 28, 28, 88) 0 reduction_add_2_stem_2[0][0] \n reduction_add3_stem_2[0][0] \n add_2[0][0] \n reduction_add4_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_0 (AveragePoo (None, 28, 28, 44) 0 adjust_relu_1_0[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_0 (AveragePoo (None, 28, 28, 44) 0 cropping2d_2[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_0 (Conv2D) (None, 28, 28, 22) 968 adjust_avg_pool_1_0[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_0 (Conv2D) (None, 28, 28, 22) 968 adjust_avg_pool_2_0[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 28, 28, 88) 0 reduction_concat_stem_2[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 28, 28, 44) 0 adjust_conv_1_0[0][0] \n adjust_conv_2_0[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_0 (Conv2D) (None, 28, 28, 44) 3872 activation_23[0][0] \n__________________________________________________________________________________________________\nadjust_bn_0 (BatchNormalization (None, 28, 28, 44) 176 concatenate_2[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_0 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_0[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 28, 28, 44) 0 normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 28, 28, 44) 0 normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_0 (None, 28, 28, 44) 3036 activation_24[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_26[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_0 (None, 28, 28, 44) 3036 activation_28[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_30[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_0 (None, 28, 28, 44) 2332 activation_32[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_0[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_0[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_0[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_0[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_0[0\n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_0 (None, 28, 28, 44) 3036 activation_25[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_27[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_0 (None, 28, 28, 44) 3036 activation_29[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_31[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_0 (None, 28, 28, 44) 2332 activation_33[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_0[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_0[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_0[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_0[\n__________________________________________________________________________________________________\nnormal_left3_0 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nnormal_left4_0 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nnormal_right4_0 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_0[0\n__________________________________________________________________________________________________\nnormal_add_1_0 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_0 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_0 (Add) (None, 28, 28, 44) 0 normal_left3_0[0][0] \n adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_0 (Add) (None, 28, 28, 44) 0 normal_left4_0[0][0] \n normal_right4_0[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_0 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nnormal_concat_0 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_0[0][0] \n normal_add_1_0[0][0] \n normal_add_2_0[0][0] \n normal_add_3_0[0][0] \n normal_add_4_0[0][0] \n normal_add_5_0[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 28, 28, 88) 0 reduction_concat_stem_2[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 28, 28, 264) 0 normal_concat_0[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_1 (Conv2 (None, 28, 28, 44) 3872 activation_34[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_1 (Conv2D) (None, 28, 28, 44) 11616 activation_35[0][0] \n__________________________________________________________________________________________________\nadjust_bn_1 (BatchNormalization (None, 28, 28, 44) 176 adjust_conv_projection_1[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_1 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_1[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 28, 28, 44) 0 normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nactivation_40 (Activation) (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nactivation_42 (Activation) (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nactivation_44 (Activation) (None, 28, 28, 44) 0 normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 28, 28, 44) 3036 activation_36[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_38[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 28, 28, 44) 3036 activation_40[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_42[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 28, 28, 44) 2332 activation_44[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_1[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_1[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_1[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_1[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_1[0\n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_41 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_43 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_45 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 28, 28, 44) 3036 activation_37[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_39[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 28, 28, 44) 3036 activation_41[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_43[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 28, 28, 44) 2332 activation_45[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_1[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_1[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_1[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_1[\n__________________________________________________________________________________________________\nnormal_left3_1 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nnormal_left4_1 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nnormal_right4_1 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_1[0\n__________________________________________________________________________________________________\nnormal_add_1_1 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_1 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_1 (Add) (None, 28, 28, 44) 0 normal_left3_1[0][0] \n adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_1 (Add) (None, 28, 28, 44) 0 normal_left4_1[0][0] \n normal_right4_1[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_1 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nnormal_concat_1 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_1[0][0] \n normal_add_1_1[0][0] \n normal_add_2_1[0][0] \n normal_add_3_1[0][0] \n normal_add_4_1[0][0] \n normal_add_5_1[0][0] \n__________________________________________________________________________________________________\nactivation_46 (Activation) (None, 28, 28, 264) 0 normal_concat_0[0][0] \n__________________________________________________________________________________________________\nactivation_47 (Activation) (None, 28, 28, 264) 0 normal_concat_1[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_2 (Conv2 (None, 28, 28, 44) 11616 activation_46[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_2 (Conv2D) (None, 28, 28, 44) 11616 activation_47[0][0] \n__________________________________________________________________________________________________\nadjust_bn_2 (BatchNormalization (None, 28, 28, 44) 176 adjust_conv_projection_2[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_2 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_2[0][0] \n__________________________________________________________________________________________________\nactivation_48 (Activation) (None, 28, 28, 44) 0 normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nactivation_50 (Activation) (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nactivation_52 (Activation) (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nactivation_54 (Activation) (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nactivation_56 (Activation) (None, 28, 28, 44) 0 normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_2 (None, 28, 28, 44) 3036 activation_48[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_50[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_2 (None, 28, 28, 44) 3036 activation_52[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_54[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_2 (None, 28, 28, 44) 2332 activation_56[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_2[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_2[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_2[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_2[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_2[0\n__________________________________________________________________________________________________\nactivation_49 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_51 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_53 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_55 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_57 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_2 (None, 28, 28, 44) 3036 activation_49[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_51[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_2 (None, 28, 28, 44) 3036 activation_53[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_55[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_2 (None, 28, 28, 44) 2332 activation_57[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_2[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_2[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_2[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_2[\n__________________________________________________________________________________________________\nnormal_left3_2 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nnormal_left4_2 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nnormal_right4_2 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_2[0\n__________________________________________________________________________________________________\nnormal_add_1_2 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_2 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_2 (Add) (None, 28, 28, 44) 0 normal_left3_2[0][0] \n adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_2 (Add) (None, 28, 28, 44) 0 normal_left4_2[0][0] \n normal_right4_2[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_2 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nnormal_concat_2 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_2[0][0] \n normal_add_1_2[0][0] \n normal_add_2_2[0][0] \n normal_add_3_2[0][0] \n normal_add_4_2[0][0] \n normal_add_5_2[0][0] \n__________________________________________________________________________________________________\nactivation_58 (Activation) (None, 28, 28, 264) 0 normal_concat_1[0][0] \n__________________________________________________________________________________________________\nactivation_59 (Activation) (None, 28, 28, 264) 0 normal_concat_2[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_3 (Conv2 (None, 28, 28, 44) 11616 activation_58[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_3 (Conv2D) (None, 28, 28, 44) 11616 activation_59[0][0] \n__________________________________________________________________________________________________\nadjust_bn_3 (BatchNormalization (None, 28, 28, 44) 176 adjust_conv_projection_3[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_3 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_3[0][0] \n__________________________________________________________________________________________________\nactivation_60 (Activation) (None, 28, 28, 44) 0 normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nactivation_62 (Activation) (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nactivation_64 (Activation) (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nactivation_66 (Activation) (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nactivation_68 (Activation) (None, 28, 28, 44) 0 normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_3 (None, 28, 28, 44) 3036 activation_60[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_62[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_3 (None, 28, 28, 44) 3036 activation_64[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_66[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_3 (None, 28, 28, 44) 2332 activation_68[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_3[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_3[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_3[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_3[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_3[0\n__________________________________________________________________________________________________\nactivation_61 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_63 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_65 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_67 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_69 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_3 (None, 28, 28, 44) 3036 activation_61[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_63[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_3 (None, 28, 28, 44) 3036 activation_65[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_67[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_3 (None, 28, 28, 44) 2332 activation_69[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_3[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_3[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_3[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_3[\n__________________________________________________________________________________________________\nnormal_left3_3 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nnormal_left4_3 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nnormal_right4_3 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_3[0\n__________________________________________________________________________________________________\nnormal_add_1_3 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_3 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_3 (Add) (None, 28, 28, 44) 0 normal_left3_3[0][0] \n adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_3 (Add) (None, 28, 28, 44) 0 normal_left4_3[0][0] \n normal_right4_3[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_3 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nnormal_concat_3 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_3[0][0] \n normal_add_1_3[0][0] \n normal_add_2_3[0][0] \n normal_add_3_3[0][0] \n normal_add_4_3[0][0] \n normal_add_5_3[0][0] \n__________________________________________________________________________________________________\nactivation_71 (Activation) (None, 28, 28, 264) 0 normal_concat_3[0][0] \n__________________________________________________________________________________________________\nactivation_70 (Activation) (None, 28, 28, 264) 0 normal_concat_2[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_reduce_4 (Conv (None, 28, 28, 88) 23232 activation_71[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_reduce_4 (None, 28, 28, 88) 23232 activation_70[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_reduce_4 (BatchN (None, 28, 28, 88) 352 reduction_conv_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nadjust_bn_reduce_4 (BatchNormal (None, 28, 28, 88) 352 adjust_conv_projection_reduce_4[0\n__________________________________________________________________________________________________\nactivation_72 (Activation) (None, 28, 28, 88) 0 reduction_bn_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nactivation_74 (Activation) (None, 28, 28, 88) 0 adjust_bn_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 31, 31, 88) 0 activation_72[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 33, 33, 88) 0 activation_74[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 14, 14, 88) 9944 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 14, 14, 88) 12056 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_73 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_75 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 14, 14, 88) 9944 activation_73[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 14, 14, 88) 12056 activation_75[0][0] \n__________________________________________________________________________________________________\nactivation_76 (Activation) (None, 28, 28, 88) 0 adjust_bn_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 33, 33, 88) 0 activation_76[0][0] \n__________________________________________________________________________________________________\nactivation_78 (Activation) (None, 28, 28, 88) 0 adjust_bn_reduce_4[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_reduce_4 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 14, 14, 88) 12056 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 31, 31, 88) 0 activation_78[0][0] \n__________________________________________________________________________________________________\nactivation_80 (Activation) (None, 14, 14, 88) 0 reduction_add_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 14, 14, 88) 9944 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 14, 14, 88) 8536 activation_80[0][0] \n__________________________________________________________________________________________________\nactivation_77 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_reduce_4 (ZeroP (None, 29, 29, 88) 0 reduction_bn_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 14, 14, 88) 12056 activation_77[0][0] \n__________________________________________________________________________________________________\nactivation_79 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_81 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_reduce_4 (MaxPo (None, 14, 14, 88) 0 reduction_pad_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 14, 14, 88) 9944 activation_79[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 14, 14, 88) 8536 activation_81[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_5 (Activation) (None, 28, 28, 264) 0 normal_concat_3[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_reduce_4 (Add) (None, 14, 14, 88) 0 reduction_left2_reduce_4[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_reduce_4 (Avera (None, 14, 14, 88) 0 reduction_pad_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_reduce_4 (Avera (None, 14, 14, 88) 0 reduction_add_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_reduce_4 (MaxP (None, 14, 14, 88) 0 reduction_pad_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_3 (ZeroPadding2D (None, 29, 29, 264) 0 adjust_relu_1_5[0][0] \n__________________________________________________________________________________________________\nreduction_add3_reduce_4 (Add) (None, 14, 14, 88) 0 reduction_left3_reduce_4[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_3 (Add) (None, 14, 14, 88) 0 reduction_add_2_reduce_4[0][0] \n reduction_left4_reduce_4[0][0] \n__________________________________________________________________________________________________\nreduction_add4_reduce_4 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_reduce_4[0][0] \n__________________________________________________________________________________________________\ncropping2d_3 (Cropping2D) (None, 28, 28, 264) 0 zero_padding2d_3[0][0] \n__________________________________________________________________________________________________\nreduction_concat_reduce_4 (Conc (None, 14, 14, 352) 0 reduction_add_2_reduce_4[0][0] \n reduction_add3_reduce_4[0][0] \n add_3[0][0] \n reduction_add4_reduce_4[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_5 (AveragePoo (None, 14, 14, 264) 0 adjust_relu_1_5[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_5 (AveragePoo (None, 14, 14, 264) 0 cropping2d_3[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_5 (Conv2D) (None, 14, 14, 44) 11616 adjust_avg_pool_1_5[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_5 (Conv2D) (None, 14, 14, 44) 11616 adjust_avg_pool_2_5[0][0] \n__________________________________________________________________________________________________\nactivation_82 (Activation) (None, 14, 14, 352) 0 reduction_concat_reduce_4[0][0] \n__________________________________________________________________________________________________\nconcatenate_3 (Concatenate) (None, 14, 14, 88) 0 adjust_conv_1_5[0][0] \n adjust_conv_2_5[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_5 (Conv2D) (None, 14, 14, 88) 30976 activation_82[0][0] \n__________________________________________________________________________________________________\nadjust_bn_5 (BatchNormalization (None, 14, 14, 88) 352 concatenate_3[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_5 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_5[0][0] \n__________________________________________________________________________________________________\nactivation_83 (Activation) (None, 14, 14, 88) 0 normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nactivation_85 (Activation) (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nactivation_87 (Activation) (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nactivation_89 (Activation) (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nactivation_91 (Activation) (None, 14, 14, 88) 0 normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_5 (None, 14, 14, 88) 9944 activation_83[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_85[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_5 (None, 14, 14, 88) 9944 activation_87[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_89[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_5 (None, 14, 14, 88) 8536 activation_91[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_5[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_5[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_5[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_5[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_5[0\n__________________________________________________________________________________________________\nactivation_84 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_86 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_88 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_90 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_92 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_5 (None, 14, 14, 88) 9944 activation_84[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_86[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_5 (None, 14, 14, 88) 9944 activation_88[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_90[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_5 (None, 14, 14, 88) 8536 activation_92[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_5[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_5[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_5[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_5[\n__________________________________________________________________________________________________\nnormal_left3_5 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nnormal_left4_5 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nnormal_right4_5 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_5[0\n__________________________________________________________________________________________________\nnormal_add_1_5 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_5 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_5 (Add) (None, 14, 14, 88) 0 normal_left3_5[0][0] \n adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_5 (Add) (None, 14, 14, 88) 0 normal_left4_5[0][0] \n normal_right4_5[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_5 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nnormal_concat_5 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_5[0][0] \n normal_add_1_5[0][0] \n normal_add_2_5[0][0] \n normal_add_3_5[0][0] \n normal_add_4_5[0][0] \n normal_add_5_5[0][0] \n__________________________________________________________________________________________________\nactivation_93 (Activation) (None, 14, 14, 352) 0 reduction_concat_reduce_4[0][0] \n__________________________________________________________________________________________________\nactivation_94 (Activation) (None, 14, 14, 528) 0 normal_concat_5[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_6 (Conv2 (None, 14, 14, 88) 30976 activation_93[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_6 (Conv2D) (None, 14, 14, 88) 46464 activation_94[0][0] \n__________________________________________________________________________________________________\nadjust_bn_6 (BatchNormalization (None, 14, 14, 88) 352 adjust_conv_projection_6[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_6 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_6[0][0] \n__________________________________________________________________________________________________\nactivation_95 (Activation) (None, 14, 14, 88) 0 normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nactivation_97 (Activation) (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nactivation_99 (Activation) (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nactivation_101 (Activation) (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nactivation_103 (Activation) (None, 14, 14, 88) 0 normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_6 (None, 14, 14, 88) 9944 activation_95[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_97[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_6 (None, 14, 14, 88) 9944 activation_99[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_101[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_6 (None, 14, 14, 88) 8536 activation_103[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_6[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_6[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_6[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_6[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_6[0\n__________________________________________________________________________________________________\nactivation_96 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_98 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_100 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_102 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_104 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_6 (None, 14, 14, 88) 9944 activation_96[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_98[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_6 (None, 14, 14, 88) 9944 activation_100[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_102[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_6 (None, 14, 14, 88) 8536 activation_104[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_6[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_6[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_6[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_6[\n__________________________________________________________________________________________________\nnormal_left3_6 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nnormal_left4_6 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nnormal_right4_6 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_6[0\n__________________________________________________________________________________________________\nnormal_add_1_6 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_6 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_6 (Add) (None, 14, 14, 88) 0 normal_left3_6[0][0] \n adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_6 (Add) (None, 14, 14, 88) 0 normal_left4_6[0][0] \n normal_right4_6[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_6 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nnormal_concat_6 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_6[0][0] \n normal_add_1_6[0][0] \n normal_add_2_6[0][0] \n normal_add_3_6[0][0] \n normal_add_4_6[0][0] \n normal_add_5_6[0][0] \n__________________________________________________________________________________________________\nactivation_105 (Activation) (None, 14, 14, 528) 0 normal_concat_5[0][0] \n__________________________________________________________________________________________________\nactivation_106 (Activation) (None, 14, 14, 528) 0 normal_concat_6[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_7 (Conv2 (None, 14, 14, 88) 46464 activation_105[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_7 (Conv2D) (None, 14, 14, 88) 46464 activation_106[0][0] \n__________________________________________________________________________________________________\nadjust_bn_7 (BatchNormalization (None, 14, 14, 88) 352 adjust_conv_projection_7[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_7 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_7[0][0] \n__________________________________________________________________________________________________\nactivation_107 (Activation) (None, 14, 14, 88) 0 normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nactivation_109 (Activation) (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nactivation_111 (Activation) (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nactivation_113 (Activation) (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nactivation_115 (Activation) (None, 14, 14, 88) 0 normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_7 (None, 14, 14, 88) 9944 activation_107[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_109[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_7 (None, 14, 14, 88) 9944 activation_111[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_113[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_7 (None, 14, 14, 88) 8536 activation_115[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_7[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_7[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_7[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_7[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_7[0\n__________________________________________________________________________________________________\nactivation_108 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_110 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_112 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_114 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_116 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_7 (None, 14, 14, 88) 9944 activation_108[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_110[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_7 (None, 14, 14, 88) 9944 activation_112[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_114[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_7 (None, 14, 14, 88) 8536 activation_116[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_7[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_7[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_7[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_7[\n__________________________________________________________________________________________________\nnormal_left3_7 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nnormal_left4_7 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nnormal_right4_7 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_7[0\n__________________________________________________________________________________________________\nnormal_add_1_7 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_7 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_7 (Add) (None, 14, 14, 88) 0 normal_left3_7[0][0] \n adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_7 (Add) (None, 14, 14, 88) 0 normal_left4_7[0][0] \n normal_right4_7[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_7 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nnormal_concat_7 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_7[0][0] \n normal_add_1_7[0][0] \n normal_add_2_7[0][0] \n normal_add_3_7[0][0] \n normal_add_4_7[0][0] \n normal_add_5_7[0][0] \n__________________________________________________________________________________________________\nactivation_117 (Activation) (None, 14, 14, 528) 0 normal_concat_6[0][0] \n__________________________________________________________________________________________________\nactivation_118 (Activation) (None, 14, 14, 528) 0 normal_concat_7[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_8 (Conv2 (None, 14, 14, 88) 46464 activation_117[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_8 (Conv2D) (None, 14, 14, 88) 46464 activation_118[0][0] \n__________________________________________________________________________________________________\nadjust_bn_8 (BatchNormalization (None, 14, 14, 88) 352 adjust_conv_projection_8[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_8 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_8[0][0] \n__________________________________________________________________________________________________\nactivation_119 (Activation) (None, 14, 14, 88) 0 normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nactivation_121 (Activation) (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nactivation_123 (Activation) (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nactivation_125 (Activation) (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nactivation_127 (Activation) (None, 14, 14, 88) 0 normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_8 (None, 14, 14, 88) 9944 activation_119[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_121[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_8 (None, 14, 14, 88) 9944 activation_123[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_125[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_8 (None, 14, 14, 88) 8536 activation_127[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_8[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_8[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_8[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_8[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_8[0\n__________________________________________________________________________________________________\nactivation_120 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_122 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_124 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_126 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_128 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_8 (None, 14, 14, 88) 9944 activation_120[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_122[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_8 (None, 14, 14, 88) 9944 activation_124[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_126[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_8 (None, 14, 14, 88) 8536 activation_128[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_8[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_8[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_8[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_8[\n__________________________________________________________________________________________________\nnormal_left3_8 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nnormal_left4_8 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nnormal_right4_8 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_8[0\n__________________________________________________________________________________________________\nnormal_add_1_8 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_8 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_8 (Add) (None, 14, 14, 88) 0 normal_left3_8[0][0] \n adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_8 (Add) (None, 14, 14, 88) 0 normal_left4_8[0][0] \n normal_right4_8[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_8 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nnormal_concat_8 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_8[0][0] \n normal_add_1_8[0][0] \n normal_add_2_8[0][0] \n normal_add_3_8[0][0] \n normal_add_4_8[0][0] \n normal_add_5_8[0][0] \n__________________________________________________________________________________________________\nactivation_130 (Activation) (None, 14, 14, 528) 0 normal_concat_8[0][0] \n__________________________________________________________________________________________________\nactivation_129 (Activation) (None, 14, 14, 528) 0 normal_concat_7[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_reduce_8 (Conv (None, 14, 14, 176) 92928 activation_130[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_reduce_8 (None, 14, 14, 176) 92928 activation_129[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_reduce_8 (BatchN (None, 14, 14, 176) 704 reduction_conv_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nadjust_bn_reduce_8 (BatchNormal (None, 14, 14, 176) 704 adjust_conv_projection_reduce_8[0\n__________________________________________________________________________________________________\nactivation_131 (Activation) (None, 14, 14, 176) 0 reduction_bn_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nactivation_133 (Activation) (None, 14, 14, 176) 0 adjust_bn_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 17, 17, 176) 0 activation_131[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 19, 19, 176) 0 activation_133[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 7, 7, 176) 35376 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 7, 7, 176) 39600 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_132 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_134 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 7, 7, 176) 35376 activation_132[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 7, 7, 176) 39600 activation_134[0][0] \n__________________________________________________________________________________________________\nactivation_135 (Activation) (None, 14, 14, 176) 0 adjust_bn_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 19, 19, 176) 0 activation_135[0][0] \n__________________________________________________________________________________________________\nactivation_137 (Activation) (None, 14, 14, 176) 0 adjust_bn_reduce_8[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_reduce_8 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 7, 7, 176) 39600 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 17, 17, 176) 0 activation_137[0][0] \n__________________________________________________________________________________________________\nactivation_139 (Activation) (None, 7, 7, 176) 0 reduction_add_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 7, 7, 176) 35376 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 7, 7, 176) 32560 activation_139[0][0] \n__________________________________________________________________________________________________\nactivation_136 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_reduce_8 (ZeroP (None, 15, 15, 176) 0 reduction_bn_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 7, 7, 176) 39600 activation_136[0][0] \n__________________________________________________________________________________________________\nactivation_138 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_140 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_reduce_8 (MaxPo (None, 7, 7, 176) 0 reduction_pad_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 7, 7, 176) 35376 activation_138[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 7, 7, 176) 32560 activation_140[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_9 (Activation) (None, 14, 14, 528) 0 normal_concat_8[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_reduce_8 (Add) (None, 7, 7, 176) 0 reduction_left2_reduce_8[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_reduce_8 (Avera (None, 7, 7, 176) 0 reduction_pad_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_reduce_8 (Avera (None, 7, 7, 176) 0 reduction_add_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_reduce_8 (MaxP (None, 7, 7, 176) 0 reduction_pad_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_4 (ZeroPadding2D (None, 15, 15, 528) 0 adjust_relu_1_9[0][0] \n__________________________________________________________________________________________________\nreduction_add3_reduce_8 (Add) (None, 7, 7, 176) 0 reduction_left3_reduce_8[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_4 (Add) (None, 7, 7, 176) 0 reduction_add_2_reduce_8[0][0] \n reduction_left4_reduce_8[0][0] \n__________________________________________________________________________________________________\nreduction_add4_reduce_8 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_reduce_8[0][0] \n__________________________________________________________________________________________________\ncropping2d_4 (Cropping2D) (None, 14, 14, 528) 0 zero_padding2d_4[0][0] \n__________________________________________________________________________________________________\nreduction_concat_reduce_8 (Conc (None, 7, 7, 704) 0 reduction_add_2_reduce_8[0][0] \n reduction_add3_reduce_8[0][0] \n add_4[0][0] \n reduction_add4_reduce_8[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_9 (AveragePoo (None, 7, 7, 528) 0 adjust_relu_1_9[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_9 (AveragePoo (None, 7, 7, 528) 0 cropping2d_4[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_9 (Conv2D) (None, 7, 7, 88) 46464 adjust_avg_pool_1_9[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_9 (Conv2D) (None, 7, 7, 88) 46464 adjust_avg_pool_2_9[0][0] \n__________________________________________________________________________________________________\nactivation_141 (Activation) (None, 7, 7, 704) 0 reduction_concat_reduce_8[0][0] \n__________________________________________________________________________________________________\nconcatenate_4 (Concatenate) (None, 7, 7, 176) 0 adjust_conv_1_9[0][0] \n adjust_conv_2_9[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_9 (Conv2D) (None, 7, 7, 176) 123904 activation_141[0][0] \n__________________________________________________________________________________________________\nadjust_bn_9 (BatchNormalization (None, 7, 7, 176) 704 concatenate_4[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_9 (BatchNormalizati (None, 7, 7, 176) 704 normal_conv_1_9[0][0] \n__________________________________________________________________________________________________\nactivation_142 (Activation) (None, 7, 7, 176) 0 normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nactivation_144 (Activation) (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nactivation_146 (Activation) (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nactivation_148 (Activation) (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nactivation_150 (Activation) (None, 7, 7, 176) 0 normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_9 (None, 7, 7, 176) 35376 activation_142[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_144[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_9 (None, 7, 7, 176) 35376 activation_146[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_148[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_9 (None, 7, 7, 176) 32560 activation_150[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_9[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_9[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_9[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_9[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_9[0\n__________________________________________________________________________________________________\nactivation_143 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_145 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_147 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_149 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_151 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_9 (None, 7, 7, 176) 35376 activation_143[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_145[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_9 (None, 7, 7, 176) 35376 activation_147[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_149[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_9 (None, 7, 7, 176) 32560 activation_151[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_9[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_9[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_9[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_9[\n__________________________________________________________________________________________________\nnormal_left3_9 (AveragePooling2 (None, 7, 7, 176) 0 normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nnormal_left4_9 (AveragePooling2 (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nnormal_right4_9 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_9[0\n__________________________________________________________________________________________________\nnormal_add_1_9 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_9 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_9 (Add) (None, 7, 7, 176) 0 normal_left3_9[0][0] \n adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_9 (Add) (None, 7, 7, 176) 0 normal_left4_9[0][0] \n normal_right4_9[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_9 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nnormal_concat_9 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_9[0][0] \n normal_add_1_9[0][0] \n normal_add_2_9[0][0] \n normal_add_3_9[0][0] \n normal_add_4_9[0][0] \n normal_add_5_9[0][0] \n__________________________________________________________________________________________________\nactivation_152 (Activation) (None, 7, 7, 704) 0 reduction_concat_reduce_8[0][0] \n__________________________________________________________________________________________________\nactivation_153 (Activation) (None, 7, 7, 1056) 0 normal_concat_9[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_10 (Conv (None, 7, 7, 176) 123904 activation_152[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_10 (Conv2D) (None, 7, 7, 176) 185856 activation_153[0][0] \n__________________________________________________________________________________________________\nadjust_bn_10 (BatchNormalizatio (None, 7, 7, 176) 704 adjust_conv_projection_10[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_10 (BatchNormalizat (None, 7, 7, 176) 704 normal_conv_1_10[0][0] \n__________________________________________________________________________________________________\nactivation_154 (Activation) (None, 7, 7, 176) 0 normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nactivation_156 (Activation) (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nactivation_158 (Activation) (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nactivation_160 (Activation) (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nactivation_162 (Activation) (None, 7, 7, 176) 0 normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 7, 7, 176) 35376 activation_154[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_156[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 7, 7, 176) 35376 activation_158[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_160[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 7, 7, 176) 32560 activation_162[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_10[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_10\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_10[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_10\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_10[\n__________________________________________________________________________________________________\nactivation_155 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_157 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_159 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_161 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_163 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 7, 7, 176) 35376 activation_155[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_157[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 7, 7, 176) 35376 activation_159[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_161[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 7, 7, 176) 32560 activation_163[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_10[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_10\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_10[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_10\n__________________________________________________________________________________________________\nnormal_left3_10 (AveragePooling (None, 7, 7, 176) 0 normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nnormal_left4_10 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nnormal_right4_10 (AveragePoolin (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_10[\n__________________________________________________________________________________________________\nnormal_add_1_10 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_10 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_10 (Add) (None, 7, 7, 176) 0 normal_left3_10[0][0] \n adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_10 (Add) (None, 7, 7, 176) 0 normal_left4_10[0][0] \n normal_right4_10[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_10 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nnormal_concat_10 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_10[0][0] \n normal_add_1_10[0][0] \n normal_add_2_10[0][0] \n normal_add_3_10[0][0] \n normal_add_4_10[0][0] \n normal_add_5_10[0][0] \n__________________________________________________________________________________________________\nactivation_164 (Activation) (None, 7, 7, 1056) 0 normal_concat_9[0][0] \n__________________________________________________________________________________________________\nactivation_165 (Activation) (None, 7, 7, 1056) 0 normal_concat_10[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_11 (Conv (None, 7, 7, 176) 185856 activation_164[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_11 (Conv2D) (None, 7, 7, 176) 185856 activation_165[0][0] \n__________________________________________________________________________________________________\nadjust_bn_11 (BatchNormalizatio (None, 7, 7, 176) 704 adjust_conv_projection_11[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_11 (BatchNormalizat (None, 7, 7, 176) 704 normal_conv_1_11[0][0] \n__________________________________________________________________________________________________\nactivation_166 (Activation) (None, 7, 7, 176) 0 normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nactivation_168 (Activation) (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nactivation_170 (Activation) (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nactivation_172 (Activation) (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nactivation_174 (Activation) (None, 7, 7, 176) 0 normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 7, 7, 176) 35376 activation_166[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_168[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 7, 7, 176) 35376 activation_170[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_172[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 7, 7, 176) 32560 activation_174[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_11[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_11\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_11[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_11\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_11[\n__________________________________________________________________________________________________\nactivation_167 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_169 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_171 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_173 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_175 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 7, 7, 176) 35376 activation_167[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_169[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 7, 7, 176) 35376 activation_171[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_173[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 7, 7, 176) 32560 activation_175[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_11[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_11\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_11[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_11\n__________________________________________________________________________________________________\nnormal_left3_11 (AveragePooling (None, 7, 7, 176) 0 normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nnormal_left4_11 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nnormal_right4_11 (AveragePoolin (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_11[\n__________________________________________________________________________________________________\nnormal_add_1_11 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_11 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_11 (Add) (None, 7, 7, 176) 0 normal_left3_11[0][0] \n adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_11 (Add) (None, 7, 7, 176) 0 normal_left4_11[0][0] \n normal_right4_11[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_11 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nnormal_concat_11 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_11[0][0] \n normal_add_1_11[0][0] \n normal_add_2_11[0][0] \n normal_add_3_11[0][0] \n normal_add_4_11[0][0] \n normal_add_5_11[0][0] \n__________________________________________________________________________________________________\nactivation_176 (Activation) (None, 7, 7, 1056) 0 normal_concat_10[0][0] \n__________________________________________________________________________________________________\nactivation_177 (Activation) (None, 7, 7, 1056) 0 normal_concat_11[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_12 (Conv (None, 7, 7, 176) 185856 activation_176[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_12 (Conv2D) (None, 7, 7, 176) 185856 activation_177[0][0] \n__________________________________________________________________________________________________\nadjust_bn_12 (BatchNormalizatio (None, 7, 7, 176) 704 adjust_conv_projection_12[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_12 (BatchNormalizat (None, 7, 7, 176) 704 normal_conv_1_12[0][0] \n__________________________________________________________________________________________________\nactivation_178 (Activation) (None, 7, 7, 176) 0 normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nactivation_180 (Activation) (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nactivation_182 (Activation) (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nactivation_184 (Activation) (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nactivation_186 (Activation) (None, 7, 7, 176) 0 normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 7, 7, 176) 35376 activation_178[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_180[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 7, 7, 176) 35376 activation_182[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_184[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 7, 7, 176) 32560 activation_186[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_12[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_12\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_12[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_12\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_12[\n__________________________________________________________________________________________________\nactivation_179 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_181 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_183 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_185 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_187 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 7, 7, 176) 35376 activation_179[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_181[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 7, 7, 176) 35376 activation_183[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_185[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 7, 7, 176) 32560 activation_187[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_12[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_12\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_12[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_12\n__________________________________________________________________________________________________\nnormal_left3_12 (AveragePooling (None, 7, 7, 176) 0 normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nnormal_left4_12 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nnormal_right4_12 (AveragePoolin (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_12[\n__________________________________________________________________________________________________\nnormal_add_1_12 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_12 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_12 (Add) (None, 7, 7, 176) 0 normal_left3_12[0][0] \n adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_12 (Add) (None, 7, 7, 176) 0 normal_left4_12[0][0] \n normal_right4_12[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_12 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nnormal_concat_12 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_12[0][0] \n normal_add_1_12[0][0] \n normal_add_2_12[0][0] \n normal_add_3_12[0][0] \n normal_add_4_12[0][0] \n normal_add_5_12[0][0] \n__________________________________________________________________________________________________\nactivation_188 (Activation) (None, 7, 7, 1056) 0 normal_concat_12[0][0] \n==================================================================================================\nTotal params: 4,269,716\nTrainable params: 4,232,978\nNon-trainable params: 36,738\n__________________________________________________________________________________________________\n"
],
[
"x = mod.output\nx = KL.GlobalAveragePooling2D()(x)\nx = KL.Dense(1056, activation='relu')(x)\nmodel = keras.Model(inputs=mod.input, outputs=x)\nmodel.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) (None, 224, 224, 3) 0 \n__________________________________________________________________________________________________\nstem_conv1 (Conv2D) (None, 111, 111, 32) 864 input_1[0][0] \n__________________________________________________________________________________________________\nstem_bn1 (BatchNormalization) (None, 111, 111, 32) 128 stem_conv1[0][0] \n__________________________________________________________________________________________________\nactivation_1 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_stem_1 (Conv2D (None, 111, 111, 11) 352 activation_1[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_stem_1 (BatchNor (None, 111, 111, 11) 44 reduction_conv_1_stem_1[0][0] \n__________________________________________________________________________________________________\nactivation_2 (Activation) (None, 111, 111, 11) 0 reduction_bn_1_stem_1[0][0] \n__________________________________________________________________________________________________\nactivation_4 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 115, 115, 11) 0 activation_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 117, 117, 32) 0 activation_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 56, 56, 11) 396 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 56, 56, 11) 1920 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_3 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_5 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 56, 56, 11) 396 activation_3[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 56, 56, 11) 660 activation_5[0][0] \n__________________________________________________________________________________________________\nactivation_6 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 117, 117, 32) 0 activation_6[0][0] \n__________________________________________________________________________________________________\nactivation_8 (Activation) (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_stem_1 (Add) (None, 56, 56, 11) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 56, 56, 11) 1920 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 115, 115, 32) 0 activation_8[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 56, 56, 11) 0 reduction_add_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 56, 56, 11) 1152 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 56, 56, 11) 220 activation_10[0][0] \n__________________________________________________________________________________________________\nactivation_7 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_stem_1 (ZeroPad (None, 113, 113, 11) 0 reduction_bn_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 56, 56, 11) 660 activation_7[0][0] \n__________________________________________________________________________________________________\nactivation_9 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 56, 56, 11) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_stem_1 (MaxPool (None, 56, 56, 11) 0 reduction_pad_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 56, 56, 11) 396 activation_9[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 56, 56, 11) 220 activation_11[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_stem_2 (Activatio (None, 111, 111, 32) 0 stem_bn1[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_stem_1 (Add) (None, 56, 56, 11) 0 reduction_left2_stem_1[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_stem_1 (Average (None, 56, 56, 11) 0 reduction_pad_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 56, 56, 11) 44 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_stem_1 (Average (None, 56, 56, 11) 0 reduction_add_1_stem_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 56, 56, 11) 44 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_stem_1 (MaxPoo (None, 56, 56, 11) 0 reduction_pad_1_stem_1[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_1 (ZeroPadding2D (None, 112, 112, 32) 0 adjust_relu_1_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_add3_stem_1 (Add) (None, 56, 56, 11) 0 reduction_left3_stem_1[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_1 (Add) (None, 56, 56, 11) 0 reduction_add_2_stem_1[0][0] \n reduction_left4_stem_1[0][0] \n__________________________________________________________________________________________________\nreduction_add4_stem_1 (Add) (None, 56, 56, 11) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_stem_1[0][0] \n__________________________________________________________________________________________________\ncropping2d_1 (Cropping2D) (None, 111, 111, 32) 0 zero_padding2d_1[0][0] \n__________________________________________________________________________________________________\nreduction_concat_stem_1 (Concat (None, 56, 56, 44) 0 reduction_add_2_stem_1[0][0] \n reduction_add3_stem_1[0][0] \n add_1[0][0] \n reduction_add4_stem_1[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_stem_2 (Avera (None, 56, 56, 32) 0 adjust_relu_1_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_stem_2 (Avera (None, 56, 56, 32) 0 cropping2d_1[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 56, 56, 44) 0 reduction_concat_stem_1[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_stem_2 (Conv2D) (None, 56, 56, 11) 352 adjust_avg_pool_1_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_stem_2 (Conv2D) (None, 56, 56, 11) 352 adjust_avg_pool_2_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_stem_2 (Conv2D (None, 56, 56, 22) 968 activation_12[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 56, 56, 22) 0 adjust_conv_1_stem_2[0][0] \n adjust_conv_2_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_stem_2 (BatchNor (None, 56, 56, 22) 88 reduction_conv_1_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_bn_stem_2 (BatchNormaliz (None, 56, 56, 22) 88 concatenate_1[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 56, 56, 22) 0 reduction_bn_1_stem_2[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 56, 56, 22) 0 adjust_bn_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 59, 59, 22) 0 activation_13[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 61, 61, 22) 0 activation_15[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 28, 28, 22) 1034 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 28, 28, 22) 1562 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_16 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 28, 28, 22) 1034 activation_14[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 28, 28, 22) 1562 activation_16[0][0] \n__________________________________________________________________________________________________\nactivation_17 (Activation) (None, 56, 56, 22) 0 adjust_bn_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 61, 61, 22) 0 activation_17[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 56, 56, 22) 0 adjust_bn_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_stem_2 (Add) (None, 28, 28, 22) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 28, 28, 22) 1562 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 59, 59, 22) 0 activation_19[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 28, 28, 22) 0 reduction_add_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 28, 28, 22) 1034 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 28, 28, 22) 682 activation_21[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_stem_2 (ZeroPad (None, 57, 57, 22) 0 reduction_bn_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 28, 28, 22) 1562 activation_18[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 28, 28, 22) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_stem_2 (MaxPool (None, 28, 28, 22) 0 reduction_pad_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 28, 28, 22) 1034 activation_20[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 28, 28, 22) 682 activation_22[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_0 (Activation) (None, 56, 56, 44) 0 reduction_concat_stem_1[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_stem_2 (Add) (None, 28, 28, 22) 0 reduction_left2_stem_2[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_stem_2 (Average (None, 28, 28, 22) 0 reduction_pad_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 28, 28, 22) 88 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_stem_2 (Average (None, 28, 28, 22) 0 reduction_add_1_stem_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 28, 28, 22) 88 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_stem_2 (MaxPoo (None, 28, 28, 22) 0 reduction_pad_1_stem_2[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_2 (ZeroPadding2D (None, 57, 57, 44) 0 adjust_relu_1_0[0][0] \n__________________________________________________________________________________________________\nreduction_add3_stem_2 (Add) (None, 28, 28, 22) 0 reduction_left3_stem_2[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_2 (Add) (None, 28, 28, 22) 0 reduction_add_2_stem_2[0][0] \n reduction_left4_stem_2[0][0] \n__________________________________________________________________________________________________\nreduction_add4_stem_2 (Add) (None, 28, 28, 22) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_stem_2[0][0] \n__________________________________________________________________________________________________\ncropping2d_2 (Cropping2D) (None, 56, 56, 44) 0 zero_padding2d_2[0][0] \n__________________________________________________________________________________________________\nreduction_concat_stem_2 (Concat (None, 28, 28, 88) 0 reduction_add_2_stem_2[0][0] \n reduction_add3_stem_2[0][0] \n add_2[0][0] \n reduction_add4_stem_2[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_0 (AveragePoo (None, 28, 28, 44) 0 adjust_relu_1_0[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_0 (AveragePoo (None, 28, 28, 44) 0 cropping2d_2[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_0 (Conv2D) (None, 28, 28, 22) 968 adjust_avg_pool_1_0[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_0 (Conv2D) (None, 28, 28, 22) 968 adjust_avg_pool_2_0[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 28, 28, 88) 0 reduction_concat_stem_2[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 28, 28, 44) 0 adjust_conv_1_0[0][0] \n adjust_conv_2_0[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_0 (Conv2D) (None, 28, 28, 44) 3872 activation_23[0][0] \n__________________________________________________________________________________________________\nadjust_bn_0 (BatchNormalization (None, 28, 28, 44) 176 concatenate_2[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_0 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_0[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 28, 28, 44) 0 normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 28, 28, 44) 0 normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_0 (None, 28, 28, 44) 3036 activation_24[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_26[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_0 (None, 28, 28, 44) 3036 activation_28[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_30[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_0 (None, 28, 28, 44) 2332 activation_32[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_0[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_0[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_0[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_0[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_0[0\n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_0 (None, 28, 28, 44) 3036 activation_25[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_27[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_0 (None, 28, 28, 44) 3036 activation_29[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_31[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_0 (None, 28, 28, 44) 2332 activation_33[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_0[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_0[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_0[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_0[\n__________________________________________________________________________________________________\nnormal_left3_0 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nnormal_left4_0 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nnormal_right4_0 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_0[0\n__________________________________________________________________________________________________\nnormal_add_1_0 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_0 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_0 (Add) (None, 28, 28, 44) 0 normal_left3_0[0][0] \n adjust_bn_0[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_0 (Add) (None, 28, 28, 44) 0 normal_left4_0[0][0] \n normal_right4_0[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_0 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_0[0][0] \n__________________________________________________________________________________________________\nnormal_concat_0 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_0[0][0] \n normal_add_1_0[0][0] \n normal_add_2_0[0][0] \n normal_add_3_0[0][0] \n normal_add_4_0[0][0] \n normal_add_5_0[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 28, 28, 88) 0 reduction_concat_stem_2[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 28, 28, 264) 0 normal_concat_0[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_1 (Conv2 (None, 28, 28, 44) 3872 activation_34[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_1 (Conv2D) (None, 28, 28, 44) 11616 activation_35[0][0] \n__________________________________________________________________________________________________\nadjust_bn_1 (BatchNormalization (None, 28, 28, 44) 176 adjust_conv_projection_1[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_1 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_1[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 28, 28, 44) 0 normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nactivation_40 (Activation) (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nactivation_42 (Activation) (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nactivation_44 (Activation) (None, 28, 28, 44) 0 normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 28, 28, 44) 3036 activation_36[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_38[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 28, 28, 44) 3036 activation_40[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_42[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 28, 28, 44) 2332 activation_44[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_1[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_1[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_1[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_1[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_1[0\n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_41 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_43 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_45 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 28, 28, 44) 3036 activation_37[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_39[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 28, 28, 44) 3036 activation_41[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_43[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 28, 28, 44) 2332 activation_45[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_1[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_1[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_1[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_1[\n__________________________________________________________________________________________________\nnormal_left3_1 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nnormal_left4_1 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nnormal_right4_1 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_1[0\n__________________________________________________________________________________________________\nnormal_add_1_1 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_1 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_1 (Add) (None, 28, 28, 44) 0 normal_left3_1[0][0] \n adjust_bn_1[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_1 (Add) (None, 28, 28, 44) 0 normal_left4_1[0][0] \n normal_right4_1[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_1 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_1[0][0] \n__________________________________________________________________________________________________\nnormal_concat_1 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_1[0][0] \n normal_add_1_1[0][0] \n normal_add_2_1[0][0] \n normal_add_3_1[0][0] \n normal_add_4_1[0][0] \n normal_add_5_1[0][0] \n__________________________________________________________________________________________________\nactivation_46 (Activation) (None, 28, 28, 264) 0 normal_concat_0[0][0] \n__________________________________________________________________________________________________\nactivation_47 (Activation) (None, 28, 28, 264) 0 normal_concat_1[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_2 (Conv2 (None, 28, 28, 44) 11616 activation_46[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_2 (Conv2D) (None, 28, 28, 44) 11616 activation_47[0][0] \n__________________________________________________________________________________________________\nadjust_bn_2 (BatchNormalization (None, 28, 28, 44) 176 adjust_conv_projection_2[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_2 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_2[0][0] \n__________________________________________________________________________________________________\nactivation_48 (Activation) (None, 28, 28, 44) 0 normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nactivation_50 (Activation) (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nactivation_52 (Activation) (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nactivation_54 (Activation) (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nactivation_56 (Activation) (None, 28, 28, 44) 0 normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_2 (None, 28, 28, 44) 3036 activation_48[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_50[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_2 (None, 28, 28, 44) 3036 activation_52[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_54[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_2 (None, 28, 28, 44) 2332 activation_56[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_2[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_2[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_2[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_2[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_2[0\n__________________________________________________________________________________________________\nactivation_49 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_51 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_53 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_55 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_57 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_2 (None, 28, 28, 44) 3036 activation_49[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_51[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_2 (None, 28, 28, 44) 3036 activation_53[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_55[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_2 (None, 28, 28, 44) 2332 activation_57[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_2[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_2[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_2[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_2[\n__________________________________________________________________________________________________\nnormal_left3_2 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nnormal_left4_2 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nnormal_right4_2 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_2[0\n__________________________________________________________________________________________________\nnormal_add_1_2 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_2 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_2 (Add) (None, 28, 28, 44) 0 normal_left3_2[0][0] \n adjust_bn_2[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_2 (Add) (None, 28, 28, 44) 0 normal_left4_2[0][0] \n normal_right4_2[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_2 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_2[0][0] \n__________________________________________________________________________________________________\nnormal_concat_2 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_2[0][0] \n normal_add_1_2[0][0] \n normal_add_2_2[0][0] \n normal_add_3_2[0][0] \n normal_add_4_2[0][0] \n normal_add_5_2[0][0] \n__________________________________________________________________________________________________\nactivation_58 (Activation) (None, 28, 28, 264) 0 normal_concat_1[0][0] \n__________________________________________________________________________________________________\nactivation_59 (Activation) (None, 28, 28, 264) 0 normal_concat_2[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_3 (Conv2 (None, 28, 28, 44) 11616 activation_58[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_3 (Conv2D) (None, 28, 28, 44) 11616 activation_59[0][0] \n__________________________________________________________________________________________________\nadjust_bn_3 (BatchNormalization (None, 28, 28, 44) 176 adjust_conv_projection_3[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_3 (BatchNormalizati (None, 28, 28, 44) 176 normal_conv_1_3[0][0] \n__________________________________________________________________________________________________\nactivation_60 (Activation) (None, 28, 28, 44) 0 normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nactivation_62 (Activation) (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nactivation_64 (Activation) (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nactivation_66 (Activation) (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nactivation_68 (Activation) (None, 28, 28, 44) 0 normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_3 (None, 28, 28, 44) 3036 activation_60[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 28, 28, 44) 2332 activation_62[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_3 (None, 28, 28, 44) 3036 activation_64[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 28, 28, 44) 2332 activation_66[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_3 (None, 28, 28, 44) 2332 activation_68[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left1_3[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right1_3[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left2_3[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_1_normal_right2_3[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 28, 28, 44) 176 separable_conv_1_normal_left5_3[0\n__________________________________________________________________________________________________\nactivation_61 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_63 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_65 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_67 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_69 (Activation) (None, 28, 28, 44) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_3 (None, 28, 28, 44) 3036 activation_61[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 28, 28, 44) 2332 activation_63[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_3 (None, 28, 28, 44) 3036 activation_65[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 28, 28, 44) 2332 activation_67[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_3 (None, 28, 28, 44) 2332 activation_69[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left1_3[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right1_3[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left2_3[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 28, 28, 44) 176 separable_conv_2_normal_right2_3[\n__________________________________________________________________________________________________\nnormal_left3_3 (AveragePooling2 (None, 28, 28, 44) 0 normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nnormal_left4_3 (AveragePooling2 (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nnormal_right4_3 (AveragePooling (None, 28, 28, 44) 0 adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 28, 28, 44) 176 separable_conv_2_normal_left5_3[0\n__________________________________________________________________________________________________\nnormal_add_1_3 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_3 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_3 (Add) (None, 28, 28, 44) 0 normal_left3_3[0][0] \n adjust_bn_3[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_3 (Add) (None, 28, 28, 44) 0 normal_left4_3[0][0] \n normal_right4_3[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_3 (Add) (None, 28, 28, 44) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_3[0][0] \n__________________________________________________________________________________________________\nnormal_concat_3 (Concatenate) (None, 28, 28, 264) 0 adjust_bn_3[0][0] \n normal_add_1_3[0][0] \n normal_add_2_3[0][0] \n normal_add_3_3[0][0] \n normal_add_4_3[0][0] \n normal_add_5_3[0][0] \n__________________________________________________________________________________________________\nactivation_71 (Activation) (None, 28, 28, 264) 0 normal_concat_3[0][0] \n__________________________________________________________________________________________________\nactivation_70 (Activation) (None, 28, 28, 264) 0 normal_concat_2[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_reduce_4 (Conv (None, 28, 28, 88) 23232 activation_71[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_reduce_4 (None, 28, 28, 88) 23232 activation_70[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_reduce_4 (BatchN (None, 28, 28, 88) 352 reduction_conv_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nadjust_bn_reduce_4 (BatchNormal (None, 28, 28, 88) 352 adjust_conv_projection_reduce_4[0\n__________________________________________________________________________________________________\nactivation_72 (Activation) (None, 28, 28, 88) 0 reduction_bn_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nactivation_74 (Activation) (None, 28, 28, 88) 0 adjust_bn_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 31, 31, 88) 0 activation_72[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 33, 33, 88) 0 activation_74[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 14, 14, 88) 9944 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 14, 14, 88) 12056 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_73 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_75 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 14, 14, 88) 9944 activation_73[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 14, 14, 88) 12056 activation_75[0][0] \n__________________________________________________________________________________________________\nactivation_76 (Activation) (None, 28, 28, 88) 0 adjust_bn_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 33, 33, 88) 0 activation_76[0][0] \n__________________________________________________________________________________________________\nactivation_78 (Activation) (None, 28, 28, 88) 0 adjust_bn_reduce_4[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_reduce_4 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 14, 14, 88) 12056 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 31, 31, 88) 0 activation_78[0][0] \n__________________________________________________________________________________________________\nactivation_80 (Activation) (None, 14, 14, 88) 0 reduction_add_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 14, 14, 88) 9944 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 14, 14, 88) 8536 activation_80[0][0] \n__________________________________________________________________________________________________\nactivation_77 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_reduce_4 (ZeroP (None, 29, 29, 88) 0 reduction_bn_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 14, 14, 88) 12056 activation_77[0][0] \n__________________________________________________________________________________________________\nactivation_79 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_81 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_reduce_4 (MaxPo (None, 14, 14, 88) 0 reduction_pad_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 14, 14, 88) 9944 activation_79[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 14, 14, 88) 8536 activation_81[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_5 (Activation) (None, 28, 28, 264) 0 normal_concat_3[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_reduce_4 (Add) (None, 14, 14, 88) 0 reduction_left2_reduce_4[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_reduce_4 (Avera (None, 14, 14, 88) 0 reduction_pad_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 14, 14, 88) 352 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_reduce_4 (Avera (None, 14, 14, 88) 0 reduction_add_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 14, 14, 88) 352 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_reduce_4 (MaxP (None, 14, 14, 88) 0 reduction_pad_1_reduce_4[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_3 (ZeroPadding2D (None, 29, 29, 264) 0 adjust_relu_1_5[0][0] \n__________________________________________________________________________________________________\nreduction_add3_reduce_4 (Add) (None, 14, 14, 88) 0 reduction_left3_reduce_4[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_3 (Add) (None, 14, 14, 88) 0 reduction_add_2_reduce_4[0][0] \n reduction_left4_reduce_4[0][0] \n__________________________________________________________________________________________________\nreduction_add4_reduce_4 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_reduce_4[0][0] \n__________________________________________________________________________________________________\ncropping2d_3 (Cropping2D) (None, 28, 28, 264) 0 zero_padding2d_3[0][0] \n__________________________________________________________________________________________________\nreduction_concat_reduce_4 (Conc (None, 14, 14, 352) 0 reduction_add_2_reduce_4[0][0] \n reduction_add3_reduce_4[0][0] \n add_3[0][0] \n reduction_add4_reduce_4[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_5 (AveragePoo (None, 14, 14, 264) 0 adjust_relu_1_5[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_5 (AveragePoo (None, 14, 14, 264) 0 cropping2d_3[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_5 (Conv2D) (None, 14, 14, 44) 11616 adjust_avg_pool_1_5[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_5 (Conv2D) (None, 14, 14, 44) 11616 adjust_avg_pool_2_5[0][0] \n__________________________________________________________________________________________________\nactivation_82 (Activation) (None, 14, 14, 352) 0 reduction_concat_reduce_4[0][0] \n__________________________________________________________________________________________________\nconcatenate_3 (Concatenate) (None, 14, 14, 88) 0 adjust_conv_1_5[0][0] \n adjust_conv_2_5[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_5 (Conv2D) (None, 14, 14, 88) 30976 activation_82[0][0] \n__________________________________________________________________________________________________\nadjust_bn_5 (BatchNormalization (None, 14, 14, 88) 352 concatenate_3[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_5 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_5[0][0] \n__________________________________________________________________________________________________\nactivation_83 (Activation) (None, 14, 14, 88) 0 normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nactivation_85 (Activation) (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nactivation_87 (Activation) (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nactivation_89 (Activation) (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nactivation_91 (Activation) (None, 14, 14, 88) 0 normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_5 (None, 14, 14, 88) 9944 activation_83[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_85[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_5 (None, 14, 14, 88) 9944 activation_87[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_89[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_5 (None, 14, 14, 88) 8536 activation_91[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_5[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_5[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_5[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_5[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_5[0\n__________________________________________________________________________________________________\nactivation_84 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_86 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_88 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_90 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_92 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_5 (None, 14, 14, 88) 9944 activation_84[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_86[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_5 (None, 14, 14, 88) 9944 activation_88[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_90[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_5 (None, 14, 14, 88) 8536 activation_92[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_5[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_5[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_5[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_5[\n__________________________________________________________________________________________________\nnormal_left3_5 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nnormal_left4_5 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nnormal_right4_5 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_5[0\n__________________________________________________________________________________________________\nnormal_add_1_5 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_5 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_5 (Add) (None, 14, 14, 88) 0 normal_left3_5[0][0] \n adjust_bn_5[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_5 (Add) (None, 14, 14, 88) 0 normal_left4_5[0][0] \n normal_right4_5[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_5 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_5[0][0] \n__________________________________________________________________________________________________\nnormal_concat_5 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_5[0][0] \n normal_add_1_5[0][0] \n normal_add_2_5[0][0] \n normal_add_3_5[0][0] \n normal_add_4_5[0][0] \n normal_add_5_5[0][0] \n__________________________________________________________________________________________________\nactivation_93 (Activation) (None, 14, 14, 352) 0 reduction_concat_reduce_4[0][0] \n__________________________________________________________________________________________________\nactivation_94 (Activation) (None, 14, 14, 528) 0 normal_concat_5[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_6 (Conv2 (None, 14, 14, 88) 30976 activation_93[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_6 (Conv2D) (None, 14, 14, 88) 46464 activation_94[0][0] \n__________________________________________________________________________________________________\nadjust_bn_6 (BatchNormalization (None, 14, 14, 88) 352 adjust_conv_projection_6[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_6 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_6[0][0] \n__________________________________________________________________________________________________\nactivation_95 (Activation) (None, 14, 14, 88) 0 normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nactivation_97 (Activation) (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nactivation_99 (Activation) (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nactivation_101 (Activation) (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nactivation_103 (Activation) (None, 14, 14, 88) 0 normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_6 (None, 14, 14, 88) 9944 activation_95[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_97[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_6 (None, 14, 14, 88) 9944 activation_99[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_101[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_6 (None, 14, 14, 88) 8536 activation_103[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_6[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_6[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_6[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_6[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_6[0\n__________________________________________________________________________________________________\nactivation_96 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_98 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_100 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_102 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_104 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_6 (None, 14, 14, 88) 9944 activation_96[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_98[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_6 (None, 14, 14, 88) 9944 activation_100[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_102[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_6 (None, 14, 14, 88) 8536 activation_104[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_6[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_6[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_6[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_6[\n__________________________________________________________________________________________________\nnormal_left3_6 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nnormal_left4_6 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nnormal_right4_6 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_6[0\n__________________________________________________________________________________________________\nnormal_add_1_6 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_6 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_6 (Add) (None, 14, 14, 88) 0 normal_left3_6[0][0] \n adjust_bn_6[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_6 (Add) (None, 14, 14, 88) 0 normal_left4_6[0][0] \n normal_right4_6[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_6 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_6[0][0] \n__________________________________________________________________________________________________\nnormal_concat_6 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_6[0][0] \n normal_add_1_6[0][0] \n normal_add_2_6[0][0] \n normal_add_3_6[0][0] \n normal_add_4_6[0][0] \n normal_add_5_6[0][0] \n__________________________________________________________________________________________________\nactivation_105 (Activation) (None, 14, 14, 528) 0 normal_concat_5[0][0] \n__________________________________________________________________________________________________\nactivation_106 (Activation) (None, 14, 14, 528) 0 normal_concat_6[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_7 (Conv2 (None, 14, 14, 88) 46464 activation_105[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_7 (Conv2D) (None, 14, 14, 88) 46464 activation_106[0][0] \n__________________________________________________________________________________________________\nadjust_bn_7 (BatchNormalization (None, 14, 14, 88) 352 adjust_conv_projection_7[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_7 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_7[0][0] \n__________________________________________________________________________________________________\nactivation_107 (Activation) (None, 14, 14, 88) 0 normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nactivation_109 (Activation) (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nactivation_111 (Activation) (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nactivation_113 (Activation) (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nactivation_115 (Activation) (None, 14, 14, 88) 0 normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_7 (None, 14, 14, 88) 9944 activation_107[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_109[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_7 (None, 14, 14, 88) 9944 activation_111[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_113[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_7 (None, 14, 14, 88) 8536 activation_115[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_7[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_7[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_7[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_7[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_7[0\n__________________________________________________________________________________________________\nactivation_108 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_110 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_112 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_114 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_116 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_7 (None, 14, 14, 88) 9944 activation_108[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_110[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_7 (None, 14, 14, 88) 9944 activation_112[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_114[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_7 (None, 14, 14, 88) 8536 activation_116[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_7[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_7[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_7[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_7[\n__________________________________________________________________________________________________\nnormal_left3_7 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nnormal_left4_7 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nnormal_right4_7 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_7[0\n__________________________________________________________________________________________________\nnormal_add_1_7 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_7 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_7 (Add) (None, 14, 14, 88) 0 normal_left3_7[0][0] \n adjust_bn_7[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_7 (Add) (None, 14, 14, 88) 0 normal_left4_7[0][0] \n normal_right4_7[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_7 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_7[0][0] \n__________________________________________________________________________________________________\nnormal_concat_7 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_7[0][0] \n normal_add_1_7[0][0] \n normal_add_2_7[0][0] \n normal_add_3_7[0][0] \n normal_add_4_7[0][0] \n normal_add_5_7[0][0] \n__________________________________________________________________________________________________\nactivation_117 (Activation) (None, 14, 14, 528) 0 normal_concat_6[0][0] \n__________________________________________________________________________________________________\nactivation_118 (Activation) (None, 14, 14, 528) 0 normal_concat_7[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_8 (Conv2 (None, 14, 14, 88) 46464 activation_117[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_8 (Conv2D) (None, 14, 14, 88) 46464 activation_118[0][0] \n__________________________________________________________________________________________________\nadjust_bn_8 (BatchNormalization (None, 14, 14, 88) 352 adjust_conv_projection_8[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_8 (BatchNormalizati (None, 14, 14, 88) 352 normal_conv_1_8[0][0] \n__________________________________________________________________________________________________\nactivation_119 (Activation) (None, 14, 14, 88) 0 normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nactivation_121 (Activation) (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nactivation_123 (Activation) (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nactivation_125 (Activation) (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nactivation_127 (Activation) (None, 14, 14, 88) 0 normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_8 (None, 14, 14, 88) 9944 activation_119[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 14, 14, 88) 8536 activation_121[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_8 (None, 14, 14, 88) 9944 activation_123[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 14, 14, 88) 8536 activation_125[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_8 (None, 14, 14, 88) 8536 activation_127[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left1_8[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right1_8[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left2_8[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_1_normal_right2_8[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 14, 14, 88) 352 separable_conv_1_normal_left5_8[0\n__________________________________________________________________________________________________\nactivation_120 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_122 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_124 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_126 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_128 (Activation) (None, 14, 14, 88) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_8 (None, 14, 14, 88) 9944 activation_120[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 14, 14, 88) 8536 activation_122[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_8 (None, 14, 14, 88) 9944 activation_124[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 14, 14, 88) 8536 activation_126[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_8 (None, 14, 14, 88) 8536 activation_128[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left1_8[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right1_8[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left2_8[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 14, 14, 88) 352 separable_conv_2_normal_right2_8[\n__________________________________________________________________________________________________\nnormal_left3_8 (AveragePooling2 (None, 14, 14, 88) 0 normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nnormal_left4_8 (AveragePooling2 (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nnormal_right4_8 (AveragePooling (None, 14, 14, 88) 0 adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 14, 14, 88) 352 separable_conv_2_normal_left5_8[0\n__________________________________________________________________________________________________\nnormal_add_1_8 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_8 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_8 (Add) (None, 14, 14, 88) 0 normal_left3_8[0][0] \n adjust_bn_8[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_8 (Add) (None, 14, 14, 88) 0 normal_left4_8[0][0] \n normal_right4_8[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_8 (Add) (None, 14, 14, 88) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_8[0][0] \n__________________________________________________________________________________________________\nnormal_concat_8 (Concatenate) (None, 14, 14, 528) 0 adjust_bn_8[0][0] \n normal_add_1_8[0][0] \n normal_add_2_8[0][0] \n normal_add_3_8[0][0] \n normal_add_4_8[0][0] \n normal_add_5_8[0][0] \n__________________________________________________________________________________________________\nactivation_130 (Activation) (None, 14, 14, 528) 0 normal_concat_8[0][0] \n__________________________________________________________________________________________________\nactivation_129 (Activation) (None, 14, 14, 528) 0 normal_concat_7[0][0] \n__________________________________________________________________________________________________\nreduction_conv_1_reduce_8 (Conv (None, 14, 14, 176) 92928 activation_130[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_reduce_8 (None, 14, 14, 176) 92928 activation_129[0][0] \n__________________________________________________________________________________________________\nreduction_bn_1_reduce_8 (BatchN (None, 14, 14, 176) 704 reduction_conv_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nadjust_bn_reduce_8 (BatchNormal (None, 14, 14, 176) 704 adjust_conv_projection_reduce_8[0\n__________________________________________________________________________________________________\nactivation_131 (Activation) (None, 14, 14, 176) 0 reduction_bn_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nactivation_133 (Activation) (None, 14, 14, 176) 0 adjust_bn_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 17, 17, 176) 0 activation_131[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 19, 19, 176) 0 activation_133[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 7, 7, 176) 35376 separable_conv_1_pad_reduction_le\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 7, 7, 176) 39600 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_1_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_1_reduction_right1\n__________________________________________________________________________________________________\nactivation_132 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nactivation_134 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 7, 7, 176) 35376 activation_132[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 7, 7, 176) 39600 activation_134[0][0] \n__________________________________________________________________________________________________\nactivation_135 (Activation) (None, 14, 14, 176) 0 adjust_bn_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_2_reduction_left1_\n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_2_reduction_right1\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 19, 19, 176) 0 activation_135[0][0] \n__________________________________________________________________________________________________\nactivation_137 (Activation) (None, 14, 14, 176) 0 adjust_bn_reduce_8[0][0] \n__________________________________________________________________________________________________\nreduction_add_1_reduce_8 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_reduction_lef\n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 7, 7, 176) 39600 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_pad_reduction_ (None, 17, 17, 176) 0 activation_137[0][0] \n__________________________________________________________________________________________________\nactivation_139 (Activation) (None, 7, 7, 176) 0 reduction_add_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_1_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_righ (None, 7, 7, 176) 35376 separable_conv_1_pad_reduction_ri\n__________________________________________________________________________________________________\nseparable_conv_1_reduction_left (None, 7, 7, 176) 32560 activation_139[0][0] \n__________________________________________________________________________________________________\nactivation_136 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_1_reduction_right3\n__________________________________________________________________________________________________\nseparable_conv_1_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_1_reduction_left4_\n__________________________________________________________________________________________________\nreduction_pad_1_reduce_8 (ZeroP (None, 15, 15, 176) 0 reduction_bn_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 7, 7, 176) 39600 activation_136[0][0] \n__________________________________________________________________________________________________\nactivation_138 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_rig\n__________________________________________________________________________________________________\nactivation_140 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_reduction_lef\n__________________________________________________________________________________________________\nreduction_left2_reduce_8 (MaxPo (None, 7, 7, 176) 0 reduction_pad_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_2_reduction_right2\n__________________________________________________________________________________________________\nseparable_conv_2_reduction_righ (None, 7, 7, 176) 35376 activation_138[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_reduction_left (None, 7, 7, 176) 32560 activation_140[0][0] \n__________________________________________________________________________________________________\nadjust_relu_1_9 (Activation) (None, 14, 14, 528) 0 normal_concat_8[0][0] \n__________________________________________________________________________________________________\nreduction_add_2_reduce_8 (Add) (None, 7, 7, 176) 0 reduction_left2_reduce_8[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nreduction_left3_reduce_8 (Avera (None, 7, 7, 176) 0 reduction_pad_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_r (None, 7, 7, 176) 704 separable_conv_2_reduction_right3\n__________________________________________________________________________________________________\nreduction_left4_reduce_8 (Avera (None, 7, 7, 176) 0 reduction_add_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_reduction_l (None, 7, 7, 176) 704 separable_conv_2_reduction_left4_\n__________________________________________________________________________________________________\nreduction_right5_reduce_8 (MaxP (None, 7, 7, 176) 0 reduction_pad_1_reduce_8[0][0] \n__________________________________________________________________________________________________\nzero_padding2d_4 (ZeroPadding2D (None, 15, 15, 528) 0 adjust_relu_1_9[0][0] \n__________________________________________________________________________________________________\nreduction_add3_reduce_8 (Add) (None, 7, 7, 176) 0 reduction_left3_reduce_8[0][0] \n separable_conv_2_bn_reduction_rig\n__________________________________________________________________________________________________\nadd_4 (Add) (None, 7, 7, 176) 0 reduction_add_2_reduce_8[0][0] \n reduction_left4_reduce_8[0][0] \n__________________________________________________________________________________________________\nreduction_add4_reduce_8 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_reduction_lef\n reduction_right5_reduce_8[0][0] \n__________________________________________________________________________________________________\ncropping2d_4 (Cropping2D) (None, 14, 14, 528) 0 zero_padding2d_4[0][0] \n__________________________________________________________________________________________________\nreduction_concat_reduce_8 (Conc (None, 7, 7, 704) 0 reduction_add_2_reduce_8[0][0] \n reduction_add3_reduce_8[0][0] \n add_4[0][0] \n reduction_add4_reduce_8[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_1_9 (AveragePoo (None, 7, 7, 528) 0 adjust_relu_1_9[0][0] \n__________________________________________________________________________________________________\nadjust_avg_pool_2_9 (AveragePoo (None, 7, 7, 528) 0 cropping2d_4[0][0] \n__________________________________________________________________________________________________\nadjust_conv_1_9 (Conv2D) (None, 7, 7, 88) 46464 adjust_avg_pool_1_9[0][0] \n__________________________________________________________________________________________________\nadjust_conv_2_9 (Conv2D) (None, 7, 7, 88) 46464 adjust_avg_pool_2_9[0][0] \n__________________________________________________________________________________________________\nactivation_141 (Activation) (None, 7, 7, 704) 0 reduction_concat_reduce_8[0][0] \n__________________________________________________________________________________________________\nconcatenate_4 (Concatenate) (None, 7, 7, 176) 0 adjust_conv_1_9[0][0] \n adjust_conv_2_9[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_9 (Conv2D) (None, 7, 7, 176) 123904 activation_141[0][0] \n__________________________________________________________________________________________________\nadjust_bn_9 (BatchNormalization (None, 7, 7, 176) 704 concatenate_4[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_9 (BatchNormalizati (None, 7, 7, 176) 704 normal_conv_1_9[0][0] \n__________________________________________________________________________________________________\nactivation_142 (Activation) (None, 7, 7, 176) 0 normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nactivation_144 (Activation) (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nactivation_146 (Activation) (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nactivation_148 (Activation) (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nactivation_150 (Activation) (None, 7, 7, 176) 0 normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_9 (None, 7, 7, 176) 35376 activation_142[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_144[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_9 (None, 7, 7, 176) 35376 activation_146[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_148[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_9 (None, 7, 7, 176) 32560 activation_150[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_9[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_9[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_9[0\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_9[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_9[0\n__________________________________________________________________________________________________\nactivation_143 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_145 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_147 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_149 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_151 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_9 (None, 7, 7, 176) 35376 activation_143[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_145[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_9 (None, 7, 7, 176) 35376 activation_147[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_149[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_9 (None, 7, 7, 176) 32560 activation_151[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_9[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_9[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_9[0\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_9[\n__________________________________________________________________________________________________\nnormal_left3_9 (AveragePooling2 (None, 7, 7, 176) 0 normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nnormal_left4_9 (AveragePooling2 (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nnormal_right4_9 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_9[0\n__________________________________________________________________________________________________\nnormal_add_1_9 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_9 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_9 (Add) (None, 7, 7, 176) 0 normal_left3_9[0][0] \n adjust_bn_9[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_9 (Add) (None, 7, 7, 176) 0 normal_left4_9[0][0] \n normal_right4_9[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_9 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_9[0][0] \n__________________________________________________________________________________________________\nnormal_concat_9 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_9[0][0] \n normal_add_1_9[0][0] \n normal_add_2_9[0][0] \n normal_add_3_9[0][0] \n normal_add_4_9[0][0] \n normal_add_5_9[0][0] \n__________________________________________________________________________________________________\nactivation_152 (Activation) (None, 7, 7, 704) 0 reduction_concat_reduce_8[0][0] \n__________________________________________________________________________________________________\nactivation_153 (Activation) (None, 7, 7, 1056) 0 normal_concat_9[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_10 (Conv (None, 7, 7, 176) 123904 activation_152[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_10 (Conv2D) (None, 7, 7, 176) 185856 activation_153[0][0] \n__________________________________________________________________________________________________\nadjust_bn_10 (BatchNormalizatio (None, 7, 7, 176) 704 adjust_conv_projection_10[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_10 (BatchNormalizat (None, 7, 7, 176) 704 normal_conv_1_10[0][0] \n__________________________________________________________________________________________________\nactivation_154 (Activation) (None, 7, 7, 176) 0 normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nactivation_156 (Activation) (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nactivation_158 (Activation) (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nactivation_160 (Activation) (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nactivation_162 (Activation) (None, 7, 7, 176) 0 normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 7, 7, 176) 35376 activation_154[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_156[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 7, 7, 176) 35376 activation_158[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_160[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 7, 7, 176) 32560 activation_162[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_10[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_10\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_10[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_10\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_10[\n__________________________________________________________________________________________________\nactivation_155 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_157 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_159 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_161 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_163 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 7, 7, 176) 35376 activation_155[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_157[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 7, 7, 176) 35376 activation_159[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_161[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 7, 7, 176) 32560 activation_163[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_10[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_10\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_10[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_10\n__________________________________________________________________________________________________\nnormal_left3_10 (AveragePooling (None, 7, 7, 176) 0 normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nnormal_left4_10 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nnormal_right4_10 (AveragePoolin (None, 7, 7, 176) 0 adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_10[\n__________________________________________________________________________________________________\nnormal_add_1_10 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_10 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_10 (Add) (None, 7, 7, 176) 0 normal_left3_10[0][0] \n adjust_bn_10[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_10 (Add) (None, 7, 7, 176) 0 normal_left4_10[0][0] \n normal_right4_10[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_10 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_10[0][0] \n__________________________________________________________________________________________________\nnormal_concat_10 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_10[0][0] \n normal_add_1_10[0][0] \n normal_add_2_10[0][0] \n normal_add_3_10[0][0] \n normal_add_4_10[0][0] \n normal_add_5_10[0][0] \n__________________________________________________________________________________________________\nactivation_164 (Activation) (None, 7, 7, 1056) 0 normal_concat_9[0][0] \n__________________________________________________________________________________________________\nactivation_165 (Activation) (None, 7, 7, 1056) 0 normal_concat_10[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_11 (Conv (None, 7, 7, 176) 185856 activation_164[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_11 (Conv2D) (None, 7, 7, 176) 185856 activation_165[0][0] \n__________________________________________________________________________________________________\nadjust_bn_11 (BatchNormalizatio (None, 7, 7, 176) 704 adjust_conv_projection_11[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_11 (BatchNormalizat (None, 7, 7, 176) 704 normal_conv_1_11[0][0] \n__________________________________________________________________________________________________\nactivation_166 (Activation) (None, 7, 7, 176) 0 normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nactivation_168 (Activation) (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nactivation_170 (Activation) (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nactivation_172 (Activation) (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nactivation_174 (Activation) (None, 7, 7, 176) 0 normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 7, 7, 176) 35376 activation_166[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_168[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 7, 7, 176) 35376 activation_170[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_172[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 7, 7, 176) 32560 activation_174[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_11[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_11\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_11[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_11\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_11[\n__________________________________________________________________________________________________\nactivation_167 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_169 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_171 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_173 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_175 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 7, 7, 176) 35376 activation_167[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_169[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 7, 7, 176) 35376 activation_171[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_173[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 7, 7, 176) 32560 activation_175[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_11[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_11\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_11[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_11\n__________________________________________________________________________________________________\nnormal_left3_11 (AveragePooling (None, 7, 7, 176) 0 normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nnormal_left4_11 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nnormal_right4_11 (AveragePoolin (None, 7, 7, 176) 0 adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_11[\n__________________________________________________________________________________________________\nnormal_add_1_11 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_11 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_11 (Add) (None, 7, 7, 176) 0 normal_left3_11[0][0] \n adjust_bn_11[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_11 (Add) (None, 7, 7, 176) 0 normal_left4_11[0][0] \n normal_right4_11[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_11 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_11[0][0] \n__________________________________________________________________________________________________\nnormal_concat_11 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_11[0][0] \n normal_add_1_11[0][0] \n normal_add_2_11[0][0] \n normal_add_3_11[0][0] \n normal_add_4_11[0][0] \n normal_add_5_11[0][0] \n__________________________________________________________________________________________________\nactivation_176 (Activation) (None, 7, 7, 1056) 0 normal_concat_10[0][0] \n__________________________________________________________________________________________________\nactivation_177 (Activation) (None, 7, 7, 1056) 0 normal_concat_11[0][0] \n__________________________________________________________________________________________________\nadjust_conv_projection_12 (Conv (None, 7, 7, 176) 185856 activation_176[0][0] \n__________________________________________________________________________________________________\nnormal_conv_1_12 (Conv2D) (None, 7, 7, 176) 185856 activation_177[0][0] \n__________________________________________________________________________________________________\nadjust_bn_12 (BatchNormalizatio (None, 7, 7, 176) 704 adjust_conv_projection_12[0][0] \n__________________________________________________________________________________________________\nnormal_bn_1_12 (BatchNormalizat (None, 7, 7, 176) 704 normal_conv_1_12[0][0] \n__________________________________________________________________________________________________\nactivation_178 (Activation) (None, 7, 7, 176) 0 normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nactivation_180 (Activation) (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nactivation_182 (Activation) (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nactivation_184 (Activation) (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nactivation_186 (Activation) (None, 7, 7, 176) 0 normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left1_1 (None, 7, 7, 176) 35376 activation_178[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right1_ (None, 7, 7, 176) 32560 activation_180[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left2_1 (None, 7, 7, 176) 35376 activation_182[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_right2_ (None, 7, 7, 176) 32560 activation_184[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_normal_left5_1 (None, 7, 7, 176) 32560 activation_186[0][0] \n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left1_12[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right1_12\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left2_12[\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_1_normal_right2_12\n__________________________________________________________________________________________________\nseparable_conv_1_bn_normal_left (None, 7, 7, 176) 704 separable_conv_1_normal_left5_12[\n__________________________________________________________________________________________________\nactivation_179 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left1_\n__________________________________________________________________________________________________\nactivation_181 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right1\n__________________________________________________________________________________________________\nactivation_183 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left2_\n__________________________________________________________________________________________________\nactivation_185 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_right2\n__________________________________________________________________________________________________\nactivation_187 (Activation) (None, 7, 7, 176) 0 separable_conv_1_bn_normal_left5_\n__________________________________________________________________________________________________\nseparable_conv_2_normal_left1_1 (None, 7, 7, 176) 35376 activation_179[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right1_ (None, 7, 7, 176) 32560 activation_181[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left2_1 (None, 7, 7, 176) 35376 activation_183[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_right2_ (None, 7, 7, 176) 32560 activation_185[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_normal_left5_1 (None, 7, 7, 176) 32560 activation_187[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left1_12[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right1_12\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left2_12[\n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_righ (None, 7, 7, 176) 704 separable_conv_2_normal_right2_12\n__________________________________________________________________________________________________\nnormal_left3_12 (AveragePooling (None, 7, 7, 176) 0 normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nnormal_left4_12 (AveragePooling (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nnormal_right4_12 (AveragePoolin (None, 7, 7, 176) 0 adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nseparable_conv_2_bn_normal_left (None, 7, 7, 176) 704 separable_conv_2_normal_left5_12[\n__________________________________________________________________________________________________\nnormal_add_1_12 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left1_\n separable_conv_2_bn_normal_right1\n__________________________________________________________________________________________________\nnormal_add_2_12 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left2_\n separable_conv_2_bn_normal_right2\n__________________________________________________________________________________________________\nnormal_add_3_12 (Add) (None, 7, 7, 176) 0 normal_left3_12[0][0] \n adjust_bn_12[0][0] \n__________________________________________________________________________________________________\nnormal_add_4_12 (Add) (None, 7, 7, 176) 0 normal_left4_12[0][0] \n normal_right4_12[0][0] \n__________________________________________________________________________________________________\nnormal_add_5_12 (Add) (None, 7, 7, 176) 0 separable_conv_2_bn_normal_left5_\n normal_bn_1_12[0][0] \n__________________________________________________________________________________________________\nnormal_concat_12 (Concatenate) (None, 7, 7, 1056) 0 adjust_bn_12[0][0] \n normal_add_1_12[0][0] \n normal_add_2_12[0][0] \n normal_add_3_12[0][0] \n normal_add_4_12[0][0] \n normal_add_5_12[0][0] \n__________________________________________________________________________________________________\nactivation_188 (Activation) (None, 7, 7, 1056) 0 normal_concat_12[0][0] \n__________________________________________________________________________________________________\nglobal_average_pooling2d_1 (Glo (None, 1056) 0 activation_188[0][0] \n__________________________________________________________________________________________________\ndense_1 (Dense) (None, 1024) 1082368 global_average_pooling2d_1[0][0] \n==================================================================================================\nTotal params: 5,352,084\nTrainable params: 5,315,346\nNon-trainable params: 36,738\n__________________________________________________________________________________________________\n"
],
[
"import tensorflow as tf\nimport numpy as np",
"_____no_output_____"
],
[
"inputs = tf.constant(np.zeros((32,224,224,3)), dtype=tf.float32)\n\nx = mod.layers[1](inputs)\nx = KL.GlobalAveragePooling2D()(x)\nx = KL.Dense(1056, activation='relu')(x)\nif training:\n x = KL.Dropout(0.5)(x)\nx = KL.Dense(128)(x)\n\nsess = tf.Session()\ninit = tf.global_variable_initializer()\nsess.run(init)\nsess.run(x)",
"_____no_output_____"
],
[
"mod.layers",
"_____no_output_____"
],
[
"input_tensor = keras.engine.Input(shape=(224, 224, 3))",
"_____no_output_____"
],
[
"input_tensor",
"_____no_output_____"
],
[
"model = keras.applications.nasnet.NASNetMobile(input_shape=(224,224,3), include_top=False, weights='imagenet', input_tensor=inputs, pooling=None)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a155c75d47c5bd1bbeaf20b4545dbdc30315410
| 44,703 |
ipynb
|
Jupyter Notebook
|
tutorials/W1D4_MachineLearning/student/W1D4_Tutorial1.ipynb
|
bgalbraith/course-content
|
3db3bbba0fee7af1def2a67e34be073c43434f4a
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null |
tutorials/W1D4_MachineLearning/student/W1D4_Tutorial1.ipynb
|
bgalbraith/course-content
|
3db3bbba0fee7af1def2a67e34be073c43434f4a
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | 1 |
2021-06-16T05:41:08.000Z
|
2021-06-16T05:41:08.000Z
|
tutorials/W1D4_MachineLearning/student/W1D4_Tutorial1.ipynb
|
bgalbraith/course-content
|
3db3bbba0fee7af1def2a67e34be073c43434f4a
|
[
"CC-BY-4.0",
"BSD-3-Clause"
] | null | null | null | 38.273116 | 502 | 0.593025 |
[
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W1D4_MachineLearning/student/W1D4_Tutorial1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Tutorial 1: GLMs for Encoding\n**Week 1, Day 4: Machine Learning (GLMs)**\n\n**By Neuromatch Academy**\n\n__Content creators:__ Pierre-Etienne H. Fiquet, Ari Benjamin, Jakob Macke\n\n__Content reviewers:__ Davide Valeriani, Alish Dipani, Michael Waskom\n\n",
"_____no_output_____"
],
[
"This is part 1 of a 2-part series about Generalized Linear Models (GLMs), which are a fundamental framework for supervised learning.\n\nIn this tutorial, the objective is to model a retinal ganglion cell spike train by fitting a temporal receptive field. First with a Linear-Gaussian GLM (also known as ordinary least-squares regression model) and then with a Poisson GLM (aka \"Linear-Nonlinear-Poisson\" model). In the next tutorial, we’ll extend to a special case of GLMs, logistic regression, and learn how to ensure good model performance.\n\nThis tutorial is designed to run with retinal ganglion cell spike train data from [Uzzell & Chichilnisky 2004](https://journals.physiology.org/doi/full/10.1152/jn.01171.2003?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed).\n\n*Acknowledgements:*\n\n- We thank EJ Chichilnisky for providing the dataset. Please note that it is provided for tutorial purposes only, and should not be distributed or used for publication without express permission from the author ([email protected]).\n- We thank Jonathan Pillow, much of this tutorial is inspired by exercises asigned in his 'Statistical Modeling and Analysis of Neural Data' class.",
"_____no_output_____"
],
[
"# Setup\n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nfrom scipy.optimize import minimize\nfrom scipy.io import loadmat",
"_____no_output_____"
],
[
"#@title Figure settings\n%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/nma.mplstyle\")",
"_____no_output_____"
],
[
"#@title Helper functions\n\ndef plot_stim_and_spikes(stim, spikes, dt, nt=120):\n \"\"\"Show time series of stim intensity and spike counts.\n\n Args:\n stim (1D array): vector of stimulus intensities\n spikes (1D array): vector of spike counts\n dt (number): duration of each time step\n nt (number): number of time steps to plot\n\n \"\"\"\n timepoints = np.arange(120)\n time = timepoints * dt\n\n f, (ax_stim, ax_spikes) = plt.subplots(\n nrows=2, sharex=True, figsize=(8, 5),\n )\n ax_stim.plot(time, stim[timepoints])\n ax_stim.set_ylabel('Stimulus intensity')\n\n ax_spikes.plot(time, spikes[timepoints])\n ax_spikes.set_xlabel('Time (s)')\n ax_spikes.set_ylabel('Number of spikes')\n\n f.tight_layout()\n\n\ndef plot_glm_matrices(X, y, nt=50):\n \"\"\"Show X and Y as heatmaps.\n\n Args:\n X (2D array): Design matrix.\n y (1D or 2D array): Target vector.\n\n \"\"\"\n from matplotlib.colors import BoundaryNorm\n from mpl_toolkits.axes_grid1 import make_axes_locatable\n Y = np.c_[y] # Ensure Y is 2D and skinny\n\n f, (ax_x, ax_y) = plt.subplots(\n ncols=2,\n figsize=(6, 8),\n sharey=True,\n gridspec_kw=dict(width_ratios=(5, 1)),\n )\n norm = BoundaryNorm([-1, -.2, .2, 1], 256)\n imx = ax_x.pcolormesh(X[:nt], cmap=\"coolwarm\", norm=norm)\n\n ax_x.set(\n title=\"X\\n(lagged stimulus)\",\n xlabel=\"Time lag (time bins)\",\n xticks=[4, 14, 24],\n xticklabels=['-20', '-10', '0'],\n ylabel=\"Time point (time bins)\",\n )\n plt.setp(ax_x.spines.values(), visible=True)\n\n divx = make_axes_locatable(ax_x)\n caxx = divx.append_axes(\"right\", size=\"5%\", pad=0.1)\n cbarx = f.colorbar(imx, cax=caxx)\n cbarx.set_ticks([-.6, 0, .6])\n cbarx.set_ticklabels(np.sort(np.unique(X)))\n\n norm = BoundaryNorm(np.arange(y.max() + 1), 256)\n imy = ax_y.pcolormesh(Y[:nt], cmap=\"magma\", norm=norm)\n ax_y.set(\n title=\"Y\\n(spike count)\",\n xticks=[]\n )\n ax_y.invert_yaxis()\n plt.setp(ax_y.spines.values(), visible=True)\n\n divy = make_axes_locatable(ax_y)\n caxy = divy.append_axes(\"right\", size=\"30%\", pad=0.1)\n cbary = f.colorbar(imy, cax=caxy)\n cbary.set_ticks(np.arange(y.max()) + .5)\n cbary.set_ticklabels(np.arange(y.max()))\n\ndef plot_spike_filter(theta, dt, **kws):\n \"\"\"Plot estimated weights based on time lag model.\n\n Args:\n theta (1D array): Filter weights, not including DC term.\n dt (number): Duration of each time bin.\n kws: Pass additional keyword arguments to plot()\n\n \"\"\"\n d = len(theta)\n t = np.arange(-d + 1, 1) * dt\n\n ax = plt.gca()\n ax.plot(t, theta, marker=\"o\", **kws)\n ax.axhline(0, color=\".2\", linestyle=\"--\", zorder=1)\n ax.set(\n xlabel=\"Time before spike (s)\",\n ylabel=\"Filter weight\",\n )\n\n\ndef plot_spikes_with_prediction(\n spikes, predicted_spikes, dt, nt=50, t0=120, **kws):\n \"\"\"Plot actual and predicted spike counts.\n\n Args:\n spikes (1D array): Vector of actual spike counts\n predicted_spikes (1D array): Vector of predicted spike counts\n dt (number): Duration of each time bin.\n nt (number): Number of time bins to plot\n t0 (number): Index of first time bin to plot.\n kws: Pass additional keyword arguments to plot()\n\n \"\"\"\n t = np.arange(t0, t0 + nt) * dt\n\n f, ax = plt.subplots()\n lines = ax.stem(t, spikes[:nt], use_line_collection=True)\n plt.setp(lines, color=\".5\")\n lines[-1].set_zorder(1)\n kws.setdefault(\"linewidth\", 3)\n yhat, = ax.plot(t, predicted_spikes[:nt], **kws)\n ax.set(\n xlabel=\"Time (s)\",\n ylabel=\"Spikes\",\n )\n ax.yaxis.set_major_locator(plt.MaxNLocator(integer=True))\n ax.legend([lines[0], yhat], [\"Spikes\", \"Predicted\"])\n\n plt.show()",
"_____no_output_____"
],
[
"#@title Data retrieval and loading\nimport os\nimport hashlib\nimport requests\n\nfname = \"RGCdata.mat\"\nurl = \"https://osf.io/mzujs/download\"\nexpected_md5 = \"1b2977453020bce5319f2608c94d38d0\"\n\nif not os.path.isfile(fname):\n try:\n r = requests.get(url)\n except requests.ConnectionError:\n print(\"!!! Failed to download data !!!\")\n else:\n if r.status_code != requests.codes.ok:\n print(\"!!! Failed to download data !!!\")\n elif hashlib.md5(r.content).hexdigest() != expected_md5:\n print(\"!!! Data download appears corrupted !!!\")\n else:\n with open(fname, \"wb\") as fid:\n fid.write(r.content)",
"_____no_output_____"
]
],
[
[
"-----\n",
"_____no_output_____"
],
[
"#Section 1: Linear-Gaussian GLM",
"_____no_output_____"
]
],
[
[
"#@title Video 1: General linear model\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"Yv89UHeSa9I\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"## Section 1.1: Load retinal ganglion cell activity data\n\nIn this exercise we use data from an experiment that presented a screen which randomly alternated between two luminance values and recorded responses from retinal ganglion cell (RGC), a type of neuron in the retina in the back of the eye. This kind of visual stimulus is called a \"full-field flicker\", and it was presented at ~120Hz (ie. the stimulus presented on the screen was refreshed about every 8ms). These same time bins were used to count the number of spikes emitted by each neuron.\n\nThe file `RGCdata.mat` contains three variablies:\n\n- `Stim`, the stimulus intensity at each time point. It is an array with shape $T \\times 1$, where $T=144051$.\n\n- `SpCounts`, the binned spike counts for 2 ON cells, and 2 OFF cells. It is a $144051 \\times 4$ array, and each column has counts for a different cell.\n\n- `dtStim`, the size of a single time bin (in seconds), which is needed for computing model output in units of spikes / s. The stimulus frame rate is given by `1 / dtStim`.\n\nBecause these data were saved in MATLAB, where everything is a matrix, we will also process the variables to more Pythonic representations (1D arrays or scalars, where appropriate) as we load the data.",
"_____no_output_____"
]
],
[
[
"data = loadmat('RGCdata.mat') # loadmat is a function in scipy.io\ndt_stim = data['dtStim'].item() # .item extracts a scalar value\n\n# Extract the stimulus intensity\nstim = data['Stim'].squeeze() # .squeeze removes dimensions with 1 element\n\n# Extract the spike counts for one cell\ncellnum = 2\nspikes = data['SpCounts'][:, cellnum]\n\n# Don't use all of the timepoints in the dataset, for speed\nkeep_timepoints = 20000\nstim = stim[:keep_timepoints]\nspikes = spikes[:keep_timepoints]",
"_____no_output_____"
]
],
[
[
"Use the `plot_stim_and_spikes` helper function to visualize the changes in stimulus intensities and spike counts over time.",
"_____no_output_____"
]
],
[
[
"plot_stim_and_spikes(stim, spikes, dt_stim)",
"_____no_output_____"
]
],
[
[
"### Exercise 1: Create design matrix\n\nOur goal is to predict the cell's activity from the stimulus intensities preceding it. That will help us understand how RGCs process information over time. To do so, we first need to create the *design matrix* for this model, which organizes the stimulus intensities in matrix form such that the $i$th row has the stimulus frames preceding timepoint $i$.\n\nIn this exercise, we will create the design matrix $X$ using $d=25$ time lags. That is, $X$ should be a $T \\times d$ matrix. $d = 25$ (about 200 ms) is a choice we're making based on our prior knowledge of the temporal window that influences RGC responses. In practice, you might not know the right duration to use.\n\nThe last entry in row `t` should correspond to the stimulus that was shown at time `t`, the entry to the left of it should contain the value that was show one time bin earlier, etc. Specifically, $X_{ij}$ will be the stimulus intensity at time $i + d - 1 - j$.\n\nNote that for the first few time bins, we have access to the recorded spike counts but not to the stimulus shown in the recent past. For simplicity we are going to assume that values of `stim` are 0 for the time lags prior to the first timepoint in the dataset. This is known as \"zero-padding\", so that the design matrix has the same number of rows as the response vectors in `spikes`.\n\nYour task is is to complete the function below to:\n\n - make a zero-padded version of the stimulus\n - initialize an empty design matrix with the correct shape\n - **fill in each row of the design matrix, using the zero-padded version of the stimulus**\n\nTo visualize your design matrix (and the corresponding vector of spike counts), we will plot a \"heatmap\", which encodes the numerical value in each position of the matrix as a color. The helper functions include some code to do this.",
"_____no_output_____"
]
],
[
[
"def make_design_matrix(stim, d=25):\n \"\"\"Create time-lag design matrix from stimulus intensity vector.\n\n Args:\n stim (1D array): Stimulus intensity at each time point.\n d (number): Number of time lags to use.\n\n Returns\n X (2D array): GLM design matrix with shape T, d\n\n \"\"\"\n\n # Create version of stimulus vector with zeros before onset\n padded_stim = np.concatenate([np.zeros(d - 1), stim])\n\n #####################################################################\n # Fill in missing code (...),\n # then remove or comment the line below to test your function\n raise NotImplementedError(\"Complete the make_design_matrix function\")\n #####################################################################\n\n\n # Construct a matrix where each row has the d frames of\n # the stimulus proceeding and including timepoint t\n T = len(...) # Total number of timepoints (hint: number of stimulus frames)\n X = np.zeros((T, d))\n for t in range(T):\n X[t] = ...\n\n return X\n\n# Uncomment and run to test your function\n# X = make_design_matrix(stim)\n# plot_glm_matrices(X, spikes, nt=50)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D4_MachineLearning/solutions/W1D4_Tutorial1_Solution_3eaecf0a.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=415 height=558 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D4_MachineLearning/static/W1D4_Tutorial1_Solution_3eaecf0a_0.png>\n\n",
"_____no_output_____"
],
[
"##Section 1.2: Fit Linear-Gaussian regression model \n\nFirst, we will use the design matrix to compute the maximum likelihood estimate for a linear-Gaussian GLM (aka \"general linear model\"). The maximum likelihood estimate of $\\theta$ in this model can be solved analytically using the equation you learned about on Day 3:\n\n$$\\hat \\theta = (X^TX)^{-1}X^Ty$$\n\nBefore we can apply this equation, we need to augment the design matrix to account for the mean of $y$, because the spike counts are all $\\geq 0$. We do this by adding a constant column of 1's to the design matrix, which will allow the model to learn an additive offset weight. We will refer to this additional weight as $b$ (for bias), although it is alternatively known as a \"DC term\" or \"intercept\".",
"_____no_output_____"
]
],
[
[
"# Build the full design matrix\ny = spikes\nconstant = np.ones_like(y)\nX = np.column_stack([constant, make_design_matrix(stim)])\n\n# Get the MLE weights for the LG model\ntheta = np.linalg.inv(X.T @ X) @ X.T @ y\ntheta_lg = theta[1:]",
"_____no_output_____"
]
],
[
[
"Plot the resulting maximum likelihood filter estimate (just the 25-element weight vector $\\theta$ on the stimulus elements, not the DC term $b$).",
"_____no_output_____"
]
],
[
[
"plot_spike_filter(theta_lg, dt_stim)",
"_____no_output_____"
]
],
[
[
"---\n\n### Exercise 2: Predict spike counts with Linear-Gaussian model\n\nNow we are going to put these pieces together and write a function that outputs a predicted spike count for each timepoint using the stimulus information.\n\nYour steps should be:\n\n- Create the complete design matrix\n- Obtain the MLE weights ($\\hat \\theta$)\n- Compute $\\hat y = X\\hat \\theta$",
"_____no_output_____"
]
],
[
[
"def predict_spike_counts_lg(stim, spikes, d=25):\n \"\"\"Compute a vector of predicted spike counts given the stimulus.\n\n Args:\n stim (1D array): Stimulus values at each timepoint\n spikes (1D array): Spike counts measured at each timepoint\n d (number): Number of time lags to use.\n\n Returns:\n yhat (1D array): Predicted spikes at each timepoint.\n\n \"\"\"\n ##########################################################################\n # Fill in missing code (...) and then comment or remove the error to test\n raise NotImplementedError(\"Complete the predict_spike_counts_lg function\")\n ##########################################################################\n\n # Create the design matrix\n y = spikes\n constant = ...\n X = ...\n\n # Get the MLE weights for the LG model\n theta = ...\n\n # Compute predicted spike counts\n yhat = X @ theta\n return yhat\n\n# Uncomment and run to test your function and plot prediction\n# predicted_counts = predict_spike_counts_lg(stim, spikes)\n# plot_spikes_with_prediction(spikes, predicted_counts, dt_stim)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D4_MachineLearning/solutions/W1D4_Tutorial1_Solution_6576a3e7.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D4_MachineLearning/static/W1D4_Tutorial1_Solution_6576a3e7_0.png>\n\n",
"_____no_output_____"
],
[
"Is this a good model? The prediction line more-or-less follows the bumps in the spikes, but it never predicts as many spikes as are actually observed. And, more troublingly, it's predicting *negative* spikes for some time points.\n\nThe Poisson GLM will help to address these failures.\n\n\n### Bonus challenge\n\nThe \"spike-triggered average\" falls out as a subcase of the linear Gaussian GLM: $\\mathrm{STA} = X^T y \\,/\\, \\textrm{sum}(y)$, where $y$ is the vector of spike counts of the neuron. In the LG GLM, the term $(X^TX)^{-1}$ corrects for potential correlation between the regressors. Because the experiment that produced these data used a white noise stimulus, there are no such correlations. Therefore the two methods are equivalent. (How would you check the statement about no correlations?)",
"_____no_output_____"
],
[
"#Section 2: Linear-Nonlinear-Poisson GLM",
"_____no_output_____"
]
],
[
[
"#@title Video 2: Generalized linear model\nfrom IPython.display import YouTubeVideo\nvideo = YouTubeVideo(id=\"wRbvwdze4uE\", width=854, height=480, fs=1)\nprint(\"Video available at https://youtube.com/watch?v=\" + video.id)\nvideo",
"_____no_output_____"
]
],
[
[
"## Section 2.1: Nonlinear optimization with `scipy.optimize`\n\nBefore diving into the Poisson GLM case, let us review the use and importance of convexity in optimization:\n- We have seen previously that in the Linear-Gaussian case, maximum likelihood parameter estimate can be computed analytically. That is great because it only takes us a single line of code!\n- Unfortunately in general there is no analytical solution to our statistical estimation problems of interest. Instead, we need to apply a nonlinear optimization algorithm to find the parameter values that minimize some *objective function*. This can be extremely tedious because there is no general way to check whether we have found *the optimal solution* or if we are just stuck in some local minimum.\n- Somewhere in between theses two extremes, the spetial case of convex objective function is of great practical importance. Indeed, such optimization problems can be solved very reliably (and usually quite rapidly too!) using some standard software.\n\nNotes:\n- a function is convex if and only if its curve lies below any chord joining two of its points\n- to learn more about optimization, you can consult the book of Stephen Boyd and Lieven Vandenberghe [Convex Optimization](https://web.stanford.edu/~boyd/cvxbook/).",
"_____no_output_____"
],
[
"Here we will use the `scipy.optimize` module, it contains a function called [`minimize`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.minimize.html) that provides a generic interface to a large number of optimization algorithms. This function expects as argument an objective function and an \"initial guess\" for the parameter values. It then returns a dictionary that includes the minimum function value, the parameters that give this minimum, and other information.\n\nLet's see how this works with a simple example. We want to minimize the function $f(x) = x^2$:",
"_____no_output_____"
]
],
[
[
"f = np.square\n\nres = minimize(f, x0=2)\nprint(\n f\"Minimum value: {res['fun']:.4g}\",\n f\"at x = {res['x']}\",\n)",
"_____no_output_____"
]
],
[
[
"When minimizing a $f(x) = x^2$, we get a minimum value of $f(x) \\approx 0$ when $x \\approx 0$. The algorithm doesn't return exactly $0$, because it stops when it gets \"close enough\" to a minimum. You can change the `tol` parameter to control how it defines \"close enough\".\n\nA point about the code bears emphasis. The first argument to `minimize` is not a number or a string but a *function*. Here, we used `np.square`. Take a moment to make sure you understand what's going on, because it's a bit unusual, and it will be important for the exercise you're going to do in a moment.\n\nIn this example, we started at $x_0 = 2$. Let's try different values for the starting point:",
"_____no_output_____"
]
],
[
[
"start_points = -1, 1.5\n\nxx = np.linspace(-2, 2, 100)\nplt.plot(xx, f(xx), color=\".2\")\nplt.xlabel(\"$x$\")\nplt.ylabel(\"$f(x)$\")\n\nfor i, x0 in enumerate(start_points):\n res = minimize(f, x0)\n plt.plot(x0, f(x0), \"o\", color=f\"C{i}\", ms=10, label=f\"Start {i}\")\n plt.plot(res[\"x\"].item(), res[\"fun\"], \"x\", c=f\"C{i}\", ms=10, mew=2, label=f\"End {i}\")\n plt.legend()",
"_____no_output_____"
]
],
[
[
"The runs started at different points (the dots), but they each ended up at roughly the same place (the cross): $f(x_\\textrm{final}) \\approx 0$. Let's see what happens if we use a different function:",
"_____no_output_____"
]
],
[
[
"g = lambda x: x / 5 + np.cos(x)\nstart_points = -.5, 1.5\n\nxx = np.linspace(-4, 4, 100)\nplt.plot(xx, g(xx), color=\".2\")\nplt.xlabel(\"$x$\")\nplt.ylabel(\"$f(x)$\")\n\nfor i, x0 in enumerate(start_points):\n res = minimize(g, x0)\n plt.plot(x0, g(x0), \"o\", color=f\"C{i}\", ms=10, label=f\"Start {i}\")\n plt.plot(res[\"x\"].item(), res[\"fun\"], \"x\", color=f\"C{i}\", ms=10, mew=2, label=f\"End {i}\")\n plt.legend()",
"_____no_output_____"
]
],
[
[
"Unlike $f(x) = x^2$, $g(x) = \\frac{x}{5} + \\cos(x)$ is not *convex*. We see that the final position of the minimization algorithm depends on the starting point, which adds a layer of comlpexity to such problems.",
"_____no_output_____"
],
[
"### Exercise 3: Fitting the Poisson GLM and prediction spikes\n\nIn this exercise, we will use [`scipy.optimize.minimize`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.optimize.minimize.html) to compute maximum likelihood estimates for the filter weights in the Poissson GLM model with an exponential nonlinearity (LNP: Linear-Nonlinear-Poisson).\n\nIn practice, this will involve filling out two functions.\n\n- The first should be an *objective function* that takes a design matrix, a spike count vector, and a vector of parameters. It should return a negative log likelihood.\n- The second function should take `stim` and `spikes`, build the design matrix and then use `minimize` internally, and return the MLE parameters.\n\nWhat should the objective function look like? We want it to return the negative log likelihood: $-\\log P(y \\mid X, \\theta).$\n\nIn the Poisson GLM,\n\n$$\n\\log P(\\mathbf{y} \\mid X, \\theta) = \\sum_t \\log P(y_t \\mid \\mathbf{x_t},\\theta),\n$$\n\nwhere\n\n$$ P(y_t \\mid \\mathbf{x_t}, \\theta) = \\frac{\\lambda_t^{y_t}\\exp(-\\lambda_t)}{y_t!} \\text{, with rate } \\lambda_t = \\exp(\\mathbf{x_t}^{\\top} \\theta).$$\n\nNow, taking the log likelihood for all the data we obtain:\n$\\log P(\\mathbf{y} \\mid X, \\theta) = \\sum_t( y_t \\log(\\lambda_t) - \\lambda_t - \\log(y_t !)).$\n\nBecause we are going to minimize the negative log likelihood with respct to the parameters $\\theta$, we can ignore the last term that does not depend on $\\theta$. For faster implementation, let us rewrite this in matrix notation:\n\n$$\\mathbf{y}^T \\log(\\mathbf{\\lambda}) - \\mathbf{1}^T \\mathbf{\\lambda} \\text{, with rate } \\mathbf{\\lambda} = \\exp(X^{\\top} \\theta)$$\n\nFinally, don't forget to add the minus sign for your function to return the negative log likelihood.",
"_____no_output_____"
]
],
[
[
"def neg_log_lik_lnp(theta, X, y):\n \"\"\"Return -loglike for the Poisson GLM model.\n\n Args:\n theta (1D array): Parameter vector.\n X (2D array): Full design matrix.\n y (1D array): Data values.\n\n Returns:\n number: Negative log likelihood.\n\n \"\"\"\n #####################################################################\n # Fill in missing code (...), then remove the error\n raise NotImplementedError(\"Complete the neg_log_lik_lnp function\")\n #####################################################################\n\n # Compute the Poisson log likeliood\n rate = np.exp(X @ theta)\n log_lik = y @ ... - ...\n\n return ...\n\n\ndef fit_lnp(stim, spikes, d=25):\n \"\"\"Obtain MLE parameters for the Poisson GLM.\n\n Args:\n stim (1D array): Stimulus values at each timepoint\n spikes (1D array): Spike counts measured at each timepoint\n d (number): Number of time lags to use.\n\n Returns:\n 1D array: MLE parameters\n\n \"\"\"\n #####################################################################\n # Fill in missing code (...), then remove the error\n raise NotImplementedError(\"Complete the fit_lnp function\")\n #####################################################################\n\n # Build the design matrix\n y = spikes\n constant = np.ones_like(y)\n X = np.column_stack([constant, make_design_matrix(stim)])\n\n # Use a random vector of weights to start (mean 0, sd .2)\n x0 = np.random.normal(0, .2, d + 1)\n\n # Find parameters that minmize the negative log likelihood function\n res = minimize(..., args=(X, y))\n\n return ...\n\n\n# Uncomment and run to test your function\n# theta_lnp = fit_lnp(stim, spikes)\n# plot_spike_filter(theta_lg[1:], dt_stim, color=\".5\", label=\"LG\")\n# plot_spike_filter(theta_lnp[1:], dt_stim, label=\"LNP\")\n# plt.legend(loc=\"upper left\");",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D4_MachineLearning/solutions/W1D4_Tutorial1_Solution_c0c87e2d.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=558 height=414 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D4_MachineLearning/static/W1D4_Tutorial1_Solution_c0c87e2d_0.png>\n\n",
"_____no_output_____"
],
[
"Plotting the LG and LNP weights together, we see that they are broadly similar, but the LNP weights are generally larger. What does that mean for the model's ability to *predict* spikes? To see that, let's finish the exercise by filling out the `predict_spike_counts_lnp` function:",
"_____no_output_____"
]
],
[
[
"def predict_spike_counts_lnp(stim, spikes, theta=None, d=25):\n \"\"\"Compute a vector of predicted spike counts given the stimulus.\n\n Args:\n stim (1D array): Stimulus values at each timepoint\n spikes (1D array): Spike counts measured at each timepoint\n theta (1D array): Filter weights; estimated if not provided.\n d (number): Number of time lags to use.\n\n Returns:\n yhat (1D array): Predicted spikes at each timepoint.\n\n \"\"\"\n ###########################################################################\n # Fill in missing code (...) and then remove the error to test\n raise NotImplementedError(\"Complete the predict_spike_counts_lnp function\")\n ###########################################################################\n\n y = spikes\n constant = np.ones_like(spikes)\n X = np.column_stack([constant, make_design_matrix(stim)])\n if theta is None: # Allow pre-cached weights, as fitting is slow\n theta = fit_lnp(X, y, d)\n\n yhat = ...\n return yhat\n\n# Uncomment and run to test predict_spike_counts_lnp\n# yhat = predict_spike_counts_lnp(stim, spikes, theta_lnp)\n# plot_spikes_with_prediction(spikes, yhat, dt_stim)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W1D4_MachineLearning/solutions/W1D4_Tutorial1_Solution_8fbc44cf.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=560 height=416 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W1D4_MachineLearning/static/W1D4_Tutorial1_Solution_8fbc44cf_0.png>\n\n",
"_____no_output_____"
],
[
"We see that the LNP model does a better job of fitting the actual spiking data. Importantly, it never predicts negative spikes!\n\n*Bonus:* Our statement that the LNP model \"does a better job\" is qualitative and based mostly on the visual appearance of the plot. But how would you make this a quantitative statement?",
"_____no_output_____"
],
[
"## Summary\n\nIn this first tutorial, we used two different models to learn something about how retinal ganglion cells respond to a flickering white noise stimulus. We learned how to construct a design matrix that we could pass to different GLMs, and we found that the Linear-Nonlinear-Poisson (LNP) model allowed us to predict spike rates better than a simple Linear-Gaussian (LG) model.\n\nIn the next tutorial, we'll extend these ideas further. We'll meet yet another GLM — logistic regression — and we'll learn how to ensure good model performance even when the number of parameters `d` is large compared to the number of data points `N`.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
] |
4a15732aa2050d9e4817c3dbfda6fef848f1337d
| 21,120 |
ipynb
|
Jupyter Notebook
|
Version 1/titanic-v-1-1.ipynb
|
insomniac-klutz/Titanic-Kaggle-EDA
|
855ceaf38724a2dd9393b41ebb2201198d6fa33d
|
[
"MIT"
] | null | null | null |
Version 1/titanic-v-1-1.ipynb
|
insomniac-klutz/Titanic-Kaggle-EDA
|
855ceaf38724a2dd9393b41ebb2201198d6fa33d
|
[
"MIT"
] | null | null | null |
Version 1/titanic-v-1-1.ipynb
|
insomniac-klutz/Titanic-Kaggle-EDA
|
855ceaf38724a2dd9393b41ebb2201198d6fa33d
|
[
"MIT"
] | null | null | null | 31.104566 | 163 | 0.433191 |
[
[
[
"# This Python 3 environment comes with many helpful analytics libraries installed\n# It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python\n# For example, here's several helpful packages to load\n\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\n\n# Input data files are available in the read-only \"../input/\" directory\n# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory\n\nimport os\nfor dirname, _, filenames in os.walk('/kaggle/input'):\n for filename in filenames:\n print(os.path.join(dirname, filename))\n\n# You can write up to 5GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using \"Save & Run All\" \n# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session",
"/kaggle/input/titanic/gender_submission.csv\n/kaggle/input/titanic/test.csv\n/kaggle/input/titanic/train.csv\n"
],
[
"train_data = pd.read_csv(\"/kaggle/input/titanic/train.csv\")\ntrain_data.head()",
"_____no_output_____"
],
[
"test_data = pd.read_csv(\"/kaggle/input/titanic/test.csv\")\ntest_data.head()",
"_____no_output_____"
],
[
"women = train_data.loc[train_data.Sex == 'female'][\"Survived\"]\nrate_women = sum(women)/len(women)\n\nprint(\"% of women who survived:\", rate_women)",
"% of women who survived: 0.7420382165605095\n"
],
[
"men = train_data.loc[train_data.Sex == 'male'][\"Survived\"]\nrate_men = sum(men)/len(men)\n\nprint(\"% of men who survived:\", rate_men)",
"% of men who survived: 0.18890814558058924\n"
],
[
"train_data.columns",
"_____no_output_____"
],
[
"leaves=[10,16,30,35,40,49,55,50]\n\ny = train_data[\"Survived\"]\n\nfeatures = [\"Pclass\", \"Sex\", \"SibSp\", \"Parch\", \"Age\",\"Fare\",\"Embarked\"]\n\nX = pd.get_dummies(train_data[features])\nX_test = pd.get_dummies(test_data[features])\n\nprint(X.info())\nprint(X_test.info())",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 891 entries, 0 to 890\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Pclass 891 non-null int64 \n 1 SibSp 891 non-null int64 \n 2 Parch 891 non-null int64 \n 3 Age 714 non-null float64\n 4 Fare 891 non-null float64\n 5 Sex_female 891 non-null uint8 \n 6 Sex_male 891 non-null uint8 \n 7 Embarked_C 891 non-null uint8 \n 8 Embarked_Q 891 non-null uint8 \n 9 Embarked_S 891 non-null uint8 \ndtypes: float64(2), int64(3), uint8(5)\nmemory usage: 39.3 KB\nNone\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 418 entries, 0 to 417\nData columns (total 10 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 Pclass 418 non-null int64 \n 1 SibSp 418 non-null int64 \n 2 Parch 418 non-null int64 \n 3 Age 332 non-null float64\n 4 Fare 417 non-null float64\n 5 Sex_female 418 non-null uint8 \n 6 Sex_male 418 non-null uint8 \n 7 Embarked_C 418 non-null uint8 \n 8 Embarked_Q 418 non-null uint8 \n 9 Embarked_S 418 non-null uint8 \ndtypes: float64(2), int64(3), uint8(5)\nmemory usage: 18.5 KB\nNone\n"
],
[
"from xgboost import XGBClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import mean_absolute_error\nfrom sklearn.impute import KNNImputer\nfrom sklearn.impute import SimpleImputer\nfrom sklearn.metrics import accuracy_score\nfrom sklearn.compose import ColumnTransformer\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.model_selection import cross_val_score\nfrom sklearn.preprocessing import OneHotEncoder",
"_____no_output_____"
],
[
"na_list=X.isna().sum() + X_test.isna().sum()\nprint(na_list)\n\nnum_cols=[col for col in X if X[col].dtype in ['int64','float64'] and na_list[col]>0]\ncat_cols=[col for col in X_test if X[col].dtype in ['object']]\n\nprint(\" Numerical Columns \",num_cols,\"\\n Categorical Columns\",cat_cols)",
"Pclass 0\nSibSp 0\nParch 0\nAge 263\nFare 1\nSex_female 0\nSex_male 0\nEmbarked_C 0\nEmbarked_Q 0\nEmbarked_S 0\ndtype: int64\n Numerical Columns ['Age', 'Fare'] \n Categorical Columns []\n"
],
[
"X_train,X_valid,y_train,y_valid=train_test_split(X,y,random_state=21)\n\nbenchmark_model=XGBClassifier(random_state=21)\n\npip_b=Pipeline(steps=[(\"benchmark_model\",benchmark_model)])\n\npip_b.fit(X_train,y_train)\npred=pip_b.predict(X_valid)\n\nacc=accuracy_score(y_valid,pred)\nprint(acc)",
"0.8340807174887892\n"
],
[
"import xgboost as xgb\nfrom sklearn.model_selection import RandomizedSearchCV\n\n# Create the parameter grid: gbm_param_grid \ngbm_param_grid = {\n 'n_estimators': range(0,400,25),\n 'max_depth': range(3,7),\n 'learning_rate': [0.4, 0.45, 0.5, 0.55, 0.6],\n 'subsample':[0.8,0.9,1],\n 'colsample_bytree': [0.6, 0.7, 0.8, 0.9, 1],\n 'n_jobs':[4]\n}\n\n# Instantiate the regressor: gbm\ngbm = XGBClassifier(random_state=21)\n\n# Perform random search: grid_mse\nxgb_random = RandomizedSearchCV(param_distributions=gbm_param_grid, \n estimator = gbm, scoring = \"accuracy\", \n verbose = 1, n_iter = 40, cv = 6,n_jobs=4)\n\n\n# Fit randomized_mse to the data\nxgb_random.fit(X, y)\n\n# Print the best parameters and lowest RMSE\nprint(\"Best parameters found: \", xgb_random.best_params_)\nprint(\"Best accuracy found: \", xgb_random.best_score_)",
"Fitting 6 folds for each of 40 candidates, totalling 240 fits\n"
],
[
"model = XGBClassifiermodel_dummy =XGBClassifier(n_estimators=75,max_depth= 3,subsample=0.9,colsample_bytree=1,learning_rate=0.45,n_jobs=4,random_state=21)\n\n#numerical_transformer = KNNImputer(n_neighbors=10)\n\n#imputed_X_test=pd.DataFrame(numerical_transformer.fit_transform(X_test))\n#imputed_X_test.columns=X_test.columns\n\n#print(imputed_X_test.info())\n\npip = Pipeline(steps=[('model',model)])\npip.fit(X, y)\npredic = pip.predict(X_test)\n\n\noutput = pd.DataFrame({'PassengerId': test_data.PassengerId, 'Survived': predic})\noutput.to_csv('my_submission.csv', index=False)\nprint(\"Your submission was successfully saved!\")",
"Your submission was successfully saved!\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a1577dc0d667dbfea5d5687fe6231dc6781beba
| 469,663 |
ipynb
|
Jupyter Notebook
|
Train_Basline_With_Gradient_Clipping.ipynb
|
ayulockin/Explore-NFNet
|
82897576e04f27cab3866e4fa9f4bee610c81658
|
[
"MIT"
] | 4 |
2021-03-05T13:51:39.000Z
|
2021-09-07T23:01:46.000Z
|
Train_Basline_With_Gradient_Clipping.ipynb
|
ayulockin/Explore-NFNet
|
82897576e04f27cab3866e4fa9f4bee610c81658
|
[
"MIT"
] | null | null | null |
Train_Basline_With_Gradient_Clipping.ipynb
|
ayulockin/Explore-NFNet
|
82897576e04f27cab3866e4fa9f4bee610c81658
|
[
"MIT"
] | 3 |
2021-07-05T07:42:24.000Z
|
2022-01-03T16:23:26.000Z
| 434.873148 | 426,914 | 0.920581 |
[
[
[
"<a href=\"https://colab.research.google.com/github/ayulockin/Explore-NFNet/blob/main/Train_Basline_With_Gradient_Clipping.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# 🧰 Setups, Installations and Imports",
"_____no_output_____"
]
],
[
[
"%%capture\r\n!pip install wandb --upgrade\r\n!pip install albumentations",
"_____no_output_____"
],
[
"!git clone https://github.com/ayulockin/Explore-NFNet",
"Cloning into 'Explore-NFNet'...\nremote: Enumerating objects: 28, done.\u001b[K\nremote: Counting objects: 100% (28/28), done.\u001b[K\nremote: Compressing objects: 100% (24/24), done.\u001b[K\nremote: Total 28 (delta 8), reused 11 (delta 2), pack-reused 0\u001b[K\nUnpacking objects: 100% (28/28), done.\n"
],
[
"import tensorflow as tf\r\nprint(tf.__version__)\r\n\r\nimport tensorflow_datasets as tfds\r\n\r\nimport sys\r\nsys.path.append(\"Explore-NFNet\")\r\nimport os\r\nimport cv2\r\nimport numpy as np\r\nfrom functools import partial\r\nimport matplotlib.pyplot as plt\r\n\r\n# Imports from the cloned repository\r\nfrom models.resnet import resnet_v1 \r\nfrom models.mini_vgg import get_mini_vgg\r\n\r\n# Augmentation related imports\r\nimport albumentations as A\r\n\r\n# Seed everything for reproducibility\r\ndef seed_everything():\r\n # Set the random seeds\r\n os.environ['TF_CUDNN_DETERMINISTIC'] = '1' \r\n np.random.seed(hash(\"improves reproducibility\") % 2**32 - 1)\r\n tf.random.set_seed(hash(\"by removing stochasticity\") % 2**32 - 1)\r\n\r\nseed_everything()\r\n\r\n# Avoid TensorFlow to allocate all the GPU at once. \r\n# Ref: https://www.tensorflow.org/guide/gpu\r\ngpus = tf.config.experimental.list_physical_devices('GPU')\r\nif gpus:\r\n try:\r\n # Currently, memory growth needs to be the same across GPUs\r\n for gpu in gpus:\r\n tf.config.experimental.set_memory_growth(gpu, True)\r\n logical_gpus = tf.config.experimental.list_logical_devices('GPU')\r\n print(len(gpus), \"Physical GPUs,\", len(logical_gpus), \"Logical GPUs\")\r\n except RuntimeError as e:\r\n # Memory growth must be set before GPUs have been initialized\r\n print(e)",
"2.4.1\n1 Physical GPUs, 1 Logical GPUs\n"
],
[
"import wandb\r\nfrom wandb.keras import WandbCallback\r\n\r\nwandb.login()",
"_____no_output_____"
],
[
"DATASET_NAME = 'cifar10'\r\nIMG_HEIGHT = 32\r\nIMG_WIDTH = 32\r\nNUM_CLASSES = 10\r\nSHUFFLE_BUFFER = 1024\r\nBATCH_SIZE = 256\r\nEPOCHS = 100\r\n\r\nAUTOTUNE = tf.data.experimental.AUTOTUNE\r\n\r\nprint(f'Global batch size is: {BATCH_SIZE}')",
"Global batch size is: 256\n"
]
],
[
[
"# ⛄ Download and Prepare Dataset",
"_____no_output_____"
]
],
[
[
"(train_ds, val_ds, test_ds), info = tfds.load(name=DATASET_NAME, \r\n split=[\"train[:85%]\", \"train[85%:]\", \"test\"], \r\n with_info=True,\r\n as_supervised=True)",
"_____no_output_____"
],
[
"@tf.function\r\ndef preprocess(image, label):\r\n # preprocess image\r\n image = tf.cast(image, tf.float32)\r\n image = image/255.0\r\n\r\n return image, label\r\n\r\n# Define the augmentation policies. Note that they are applied sequentially with some probability p.\r\ntransforms = A.Compose([\r\n A.HorizontalFlip(p=0.7),\r\n A.Rotate(limit=30, p=0.7)\r\n ])\r\n\r\n# Apply augmentation policies.\r\ndef aug_fn(image):\r\n data = {\"image\":image}\r\n aug_data = transforms(**data)\r\n aug_img = aug_data[\"image\"]\r\n\r\n return aug_img\r\n\r\[email protected]\r\ndef apply_augmentation(image, label):\r\n aug_img = tf.numpy_function(func=aug_fn, inp=[image], Tout=tf.float32)\r\n aug_img.set_shape((IMG_HEIGHT, IMG_WIDTH, 3))\r\n \r\n return aug_img, label\r\n\r\ntrain_ds = (\r\n train_ds\r\n .shuffle(SHUFFLE_BUFFER)\r\n .map(preprocess, num_parallel_calls=AUTOTUNE)\r\n .map(apply_augmentation, num_parallel_calls=AUTOTUNE)\r\n .batch(BATCH_SIZE)\r\n .prefetch(AUTOTUNE)\r\n)\r\n\r\nval_ds = (\r\n val_ds\r\n .map(preprocess, num_parallel_calls=AUTOTUNE)\r\n .batch(BATCH_SIZE)\r\n .prefetch(AUTOTUNE)\r\n)\r\n\r\ntest_ds = (\r\n test_ds\r\n .map(preprocess, num_parallel_calls=AUTOTUNE)\r\n .batch(BATCH_SIZE)\r\n .prefetch(AUTOTUNE)\r\n)",
"_____no_output_____"
],
[
"def show_batch(image_batch, label_batch):\r\n plt.figure(figsize=(10,10))\r\n for n in range(25):\r\n ax = plt.subplot(5,5,n+1)\r\n plt.imshow(image_batch[n])\r\n # plt.title(f'{np.argmax(label_batch[n].numpy())}')\r\n plt.title(f'{label_batch[n].numpy()}')\r\n plt.axis('off')\r\n \r\nimage_batch, label_batch = next(iter(train_ds))\r\nshow_batch(image_batch, label_batch)\r\n\r\nprint(image_batch.shape, label_batch.shape)",
"(256, 32, 32, 3) (256,)\n"
]
],
[
[
"# 🐤 Model",
"_____no_output_____"
]
],
[
[
"class ResNetModel(tf.keras.Model):\r\n def __init__(self, resnet):\r\n super(ResNetModel, self).__init__()\r\n self.resnet = resnet\r\n \r\n def train_step(self, data):\r\n images, labels = data\r\n\r\n with tf.GradientTape() as tape:\r\n predictions = self.resnet(images)\r\n loss = self.compiled_loss(labels, predictions)\r\n\r\n trainable_params = self.resnet.trainable_variables\r\n gradients = tape.gradient(loss, trainable_params) \r\n gradients_clipped = [tf.clip_by_norm(g, 0.01) for g in gradients]\r\n\r\n self.optimizer.apply_gradients(zip(gradients_clipped, trainable_params))\r\n\r\n self.compiled_metrics.update_state(labels, predictions)\r\n return {m.name: m.result() for m in self.metrics}\r\n\r\n def test_step(self, data):\r\n images, labels = data\r\n predictions = self.resnet(images, training=False)\r\n loss = self.compiled_loss(labels, predictions)\r\n self.compiled_metrics.update_state(labels, predictions)\r\n return {m.name: m.result() for m in self.metrics}\r\n\r\n def save_weights(self, filepath):\r\n self.resnet.save_weights(filepath=filepath, save_format=\"tf\")\r\n\r\n def call(self, inputs, *args, **kwargs):\r\n return self.resnet(inputs)\r\n\r\ntf.keras.backend.clear_session()\r\ntest_model = ResNetModel(resnet_v1((IMG_HEIGHT, IMG_WIDTH, 3), 20, num_classes=NUM_CLASSES, use_bn=False))\r\ntest_model.build((1, IMG_HEIGHT, IMG_WIDTH, 3))\r\ntest_model.summary()\r\nprint(f\"Total learnable parameters: {test_model.count_params()/1e6} M\")",
"Model: \"res_net_model\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nmodel (Functional) (None, 10) 271754 \n=================================================================\nTotal params: 271,754\nTrainable params: 271,722\nNon-trainable params: 32\n_________________________________________________________________\nTotal learnable parameters: 0.271754 M\n"
]
],
[
[
"# 📲 Callbacks",
"_____no_output_____"
]
],
[
[
"earlystopper = tf.keras.callbacks.EarlyStopping(\r\n monitor='val_loss', patience=10, verbose=0, mode='auto',\r\n restore_best_weights=True\r\n)\r\n\r\nreducelronplateau = tf.keras.callbacks.ReduceLROnPlateau(\r\n monitor=\"val_loss\", factor=0.5,\r\n patience=3, verbose=1\r\n)",
"_____no_output_____"
]
],
[
[
"# 🚋 Train with W&B",
"_____no_output_____"
]
],
[
[
"tf.keras.backend.clear_session()\r\n# Intialize model\r\nmodel = ResNetModel(resnet_v1((IMG_HEIGHT, IMG_WIDTH, 3), 20, num_classes=NUM_CLASSES, use_bn=False))\r\nmodel.compile('adam', 'sparse_categorical_crossentropy', metrics=['acc'])\r\n\r\n# Intialize W&B run\r\nrun = wandb.init(entity='ayush-thakur', project='nfnet', job_type='train-baseline')\r\n\r\n# Train model\r\nmodel.fit(train_ds,\r\n epochs=EPOCHS,\r\n validation_data=val_ds,\r\n callbacks=[WandbCallback(),\r\n reducelronplateau,\r\n earlystopper]) \r\n\r\n# Evaluate model on test set\r\nloss, acc = model.evaluate(test_ds)\r\nwandb.log({'Test Accuracy': round(acc, 3)})\r\n\r\n# Close W&B run\r\nrun.finish()",
"\u001b[34m\u001b[1mwandb\u001b[0m: Currently logged in as: \u001b[33mayush-thakur\u001b[0m (use `wandb login --relogin` to force relogin)\n"
]
],
[
[
"",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a159754ed6324237bf5b5bfaa17286cd5205c51
| 2,721 |
ipynb
|
Jupyter Notebook
|
doc/_notebooks/stubs/sarsa.ipynb
|
frederikschubert/coax
|
96a1fae4f6f527064a1601a25847204b514d5767
|
[
"MIT"
] | 81 |
2021-03-11T06:10:25.000Z
|
2022-03-18T18:00:10.000Z
|
doc/_notebooks/stubs/sarsa.ipynb
|
frederikschubert/coax
|
96a1fae4f6f527064a1601a25847204b514d5767
|
[
"MIT"
] | 12 |
2021-05-27T12:21:54.000Z
|
2022-02-07T16:12:54.000Z
|
doc/_notebooks/stubs/sarsa.ipynb
|
frederikschubert/coax
|
96a1fae4f6f527064a1601a25847204b514d5767
|
[
"MIT"
] | 2 |
2021-08-10T06:03:22.000Z
|
2022-03-14T06:05:33.000Z
| 26.417476 | 101 | 0.498346 |
[
[
[
"%pip install git+https://github.com/coax-dev/coax.git@main",
"_____no_output_____"
],
[
"import gym\nimport coax\nimport optax\nimport haiku as hk\nimport jax\nimport jax.numpy as jnp\n\n\n# pick environment\nenv = gym.make(...)\nenv = coax.wrappers.TrainMonitor(env)\n\n\ndef func_type1(S, A, is_training):\n # custom haiku function: s,a -> q(s,a)\n value = hk.Sequential([...])\n X = jax.vmap(jnp.kron)(S, A) # or jnp.concatenate((S, A), axis=-1) or whatever you like\n return value(X) # output shape: (batch_size,)\n\n\ndef func_type2(S, is_training):\n # custom haiku function: s -> q(s,.)\n value = hk.Sequential([...])\n return value(S) # output shape: (batch_size, num_actions)\n\n\n# function approximator\nfunc = ... # func_type1 or func_type2\nq = coax.Q(func, env)\npi = coax.EpsilonGreedy(q, epsilon=0.1)\n\n\n# specify how to update q-function\nsarsa = coax.td_learning.Sarsa(q, optimizer=optax.adam(0.001))\n\n\n# specify how to trace the transitions\ntracer = coax.reward_tracing.NStep(n=1, gamma=0.9)\n\n\nfor ep in range(100):\n pi.epsilon = ... # exploration schedule\n s = env.reset()\n\n for t in range(env.spec.max_episode_steps):\n a = pi(s)\n s_next, r, done, info = env.step(a)\n\n # trace rewards to create training data\n tracer.add(s, a, r, done)\n\n # update\n while tracer:\n transition_batch = tracer.pop()\n metrics = sarsa.update(transition_batch)\n env.record_metrics(metrics)\n\n if done:\n break\n\n s = s_next\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4a15b105692b6a6a975efd7b4ecd7e611c9b538a
| 851,616 |
ipynb
|
Jupyter Notebook
|
train_0722_history.ipynb
|
kanand77/unet-updated
|
54a4b7169f280ec78cedd787ffd9404f1c70645d
|
[
"MIT"
] | null | null | null |
train_0722_history.ipynb
|
kanand77/unet-updated
|
54a4b7169f280ec78cedd787ffd9404f1c70645d
|
[
"MIT"
] | null | null | null |
train_0722_history.ipynb
|
kanand77/unet-updated
|
54a4b7169f280ec78cedd787ffd9404f1c70645d
|
[
"MIT"
] | null | null | null | 1,257.926145 | 365,308 | 0.952691 |
[
[
[
"from model_2 import *\nfrom data_2 import *\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport tensorflow as tf",
"_____no_output_____"
],
[
"print(\"Num GPUs Available: \", len(tf.config.experimental.list_physical_devices('GPU')))",
"Num GPUs Available: 2\n"
],
[
"N_training = 150\nN_validate = 15\nbatch_size = 4\n\ntrain_ids = np.arange(N_training)\nprint(train_ids)\ntrain_generator = DataGenerator(train_ids, '../training_data_v1/', '../training_data_v1/', do_fft = False, batch_size = batch_size)\n\nvalidation_ids = np.arange(N_training, N_training+N_validate)\nprint(validation_ids)\nvalidation_generator = DataGenerator(validation_ids, '../training_data_v1/', '../training_data_v1/', do_fft = False, batch_size = batch_size)",
"[ 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17\n 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35\n 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53\n 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71\n 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89\n 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107\n 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125\n 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143\n 144 145 146 147 148 149]\nInitialized with 3750 total IDs\n[150 151 152 153 154 155 156 157 158 159 160 161 162 163 164]\nInitialized with 375 total IDs\n"
],
[
"with tf.device('/GPU:1'):\n model = unet(init_layers=128)\n print(model.summary())",
"Model: \"model\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 256, 256, 2) 0 \n__________________________________________________________________________________________________\nconv2d (Conv2D) (None, 256, 256, 128 2432 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_1 (Conv2D) (None, 256, 256, 128 147584 conv2d[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 128, 128, 128 0 conv2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_2 (Conv2D) (None, 128, 128, 256 295168 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nconv2d_3 (Conv2D) (None, 128, 128, 256 590080 conv2d_2[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_1 (MaxPooling2D) (None, 64, 64, 256) 0 conv2d_3[0][0] \n__________________________________________________________________________________________________\nconv2d_4 (Conv2D) (None, 64, 64, 512) 1180160 max_pooling2d_1[0][0] \n__________________________________________________________________________________________________\nconv2d_5 (Conv2D) (None, 64, 64, 512) 2359808 conv2d_4[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_2 (MaxPooling2D) (None, 32, 32, 512) 0 conv2d_5[0][0] \n__________________________________________________________________________________________________\nconv2d_6 (Conv2D) (None, 32, 32, 1024) 4719616 max_pooling2d_2[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 32, 32, 1024) 9438208 conv2d_6[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d_3 (MaxPooling2D) (None, 16, 16, 1024) 0 conv2d_7[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 16, 16, 2048) 18876416 max_pooling2d_3[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 16, 16, 2048) 37750784 conv2d_8[0][0] \n__________________________________________________________________________________________________\nup_sampling2d (UpSampling2D) (None, 32, 32, 2048) 0 conv2d_9[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 32, 32, 1024) 8389632 up_sampling2d[0][0] \n__________________________________________________________________________________________________\nconcatenate (Concatenate) (None, 32, 32, 2048) 0 conv2d_7[0][0] \n conv2d_10[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 32, 32, 1024) 18875392 concatenate[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 32, 32, 1024) 9438208 conv2d_11[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_1 (UpSampling2D) (None, 64, 64, 1024) 0 conv2d_12[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 64, 64, 512) 2097664 up_sampling2d_1[0][0] \n__________________________________________________________________________________________________\nconcatenate_1 (Concatenate) (None, 64, 64, 1024) 0 conv2d_5[0][0] \n conv2d_13[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 64, 64, 512) 4719104 concatenate_1[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 64, 64, 512) 2359808 conv2d_14[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_2 (UpSampling2D) (None, 128, 128, 512 0 conv2d_15[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 128, 128, 256 524544 up_sampling2d_2[0][0] \n__________________________________________________________________________________________________\nconcatenate_2 (Concatenate) (None, 128, 128, 512 0 conv2d_3[0][0] \n conv2d_16[0][0] \n__________________________________________________________________________________________________\nconv2d_17 (Conv2D) (None, 128, 128, 256 1179904 concatenate_2[0][0] \n__________________________________________________________________________________________________\nconv2d_18 (Conv2D) (None, 128, 128, 256 590080 conv2d_17[0][0] \n__________________________________________________________________________________________________\nup_sampling2d_3 (UpSampling2D) (None, 256, 256, 256 0 conv2d_18[0][0] \n__________________________________________________________________________________________________\nconv2d_19 (Conv2D) (None, 256, 256, 128 131200 up_sampling2d_3[0][0] \n__________________________________________________________________________________________________\nconcatenate_3 (Concatenate) (None, 256, 256, 256 0 conv2d_1[0][0] \n conv2d_19[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 256, 256, 128 295040 concatenate_3[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 256, 256, 128 147584 conv2d_20[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 256, 256, 2) 258 conv2d_21[0][0] \n==================================================================================================\nTotal params: 124,108,674\nTrainable params: 124,108,674\nNon-trainable params: 0\n__________________________________________________________________________________________________\nNone\n"
],
[
"# Load test data\ncardiac_test = np.load('../tagsim/test_data_v1.npz')\n\ntruth_data = cardiac_test['truth']\n\ntest_data = cardiac_test['test']\n\nX = np.zeros(test_data.shape + (2,))\ny = np.zeros(truth_data.shape + (2,))\n\nX[:,:,:,0] = np.real(test_data)\nX[:,:,:,1] = np.imag(test_data)\n\ny[:,:,:,0] = np.real(truth_data)\ny[:,:,:,1] = np.imag(truth_data)\n\nwith tf.device('/GPU:2'):\n res = model.predict(X)",
"_____no_output_____"
],
[
"print(X.shape)\nprint(y.shape)",
"(25, 256, 256, 2)\n(25, 256, 256, 2)\n"
],
[
"with tf.device('/GPU:1'):\n hist = model.fit_generator(train_generator, validation_data= (X,y) , epochs=12)",
"WARNING:tensorflow:From <ipython-input-7-b354daf376c5>:2: Model.fit_generator (from tensorflow.python.keras.engine.training) is deprecated and will be removed in a future version.\nInstructions for updating:\nPlease use Model.fit, which supports generators.\nEpoch 1/12\n937/937 [==============================] - 504s 538ms/step - loss: 0.0016 - mean_squared_error: 0.0031 - val_loss: 6.3768e-04 - val_mean_squared_error: 0.0013\nEpoch 2/12\n937/937 [==============================] - 501s 535ms/step - loss: 8.9586e-04 - mean_squared_error: 0.0018 - val_loss: 6.4992e-04 - val_mean_squared_error: 0.0013\nEpoch 3/12\n937/937 [==============================] - 502s 535ms/step - loss: 7.8760e-04 - mean_squared_error: 0.0016 - val_loss: 5.4855e-04 - val_mean_squared_error: 0.0011\nEpoch 4/12\n937/937 [==============================] - 501s 535ms/step - loss: 7.2125e-04 - mean_squared_error: 0.0014 - val_loss: 2.8781e-04 - val_mean_squared_error: 5.7561e-04\nEpoch 5/12\n937/937 [==============================] - 501s 535ms/step - loss: 6.8687e-04 - mean_squared_error: 0.0014 - val_loss: 3.4628e-04 - val_mean_squared_error: 6.9256e-04\nEpoch 6/12\n937/937 [==============================] - 501s 535ms/step - loss: 6.6172e-04 - mean_squared_error: 0.0013 - val_loss: 4.0902e-04 - val_mean_squared_error: 8.1805e-04\nEpoch 7/12\n937/937 [==============================] - 500s 534ms/step - loss: 6.2996e-04 - mean_squared_error: 0.0013 - val_loss: 3.9105e-04 - val_mean_squared_error: 7.8210e-04\nEpoch 8/12\n937/937 [==============================] - 501s 535ms/step - loss: 6.0791e-04 - mean_squared_error: 0.0012 - val_loss: 2.4323e-04 - val_mean_squared_error: 4.8646e-04\nEpoch 9/12\n937/937 [==============================] - 501s 534ms/step - loss: 5.7950e-04 - mean_squared_error: 0.0012 - val_loss: 4.0583e-04 - val_mean_squared_error: 8.1167e-04\nEpoch 10/12\n937/937 [==============================] - 500s 534ms/step - loss: 6.2104e-04 - mean_squared_error: 0.0012 - val_loss: 3.1062e-04 - val_mean_squared_error: 6.2125e-04\nEpoch 11/12\n937/937 [==============================] - 501s 535ms/step - loss: 5.3747e-04 - mean_squared_error: 0.0011 - val_loss: 2.5463e-04 - val_mean_squared_error: 5.0927e-04\nEpoch 12/12\n937/937 [==============================] - 501s 534ms/step - loss: 4.9947e-04 - mean_squared_error: 9.9893e-04 - val_loss: 4.4079e-04 - val_mean_squared_error: 8.8158e-04\n"
],
[
"plt.figure()\nplt.plot(hist.history['loss'], label='Training Loss')\nplt.plot(hist.history['val_loss'], label='Validation Loss')\nplt.legend()",
"_____no_output_____"
],
[
"plt.figure()\nplt.imshow(np.log(np.abs(np.fft.fftshift(np.fft.fft2(np.fft.fftshift(im_X))))))\n\nplt.figure()\nplt.imshow(np.log(np.abs(np.fft.fftshift(np.fft.fft2(np.fft.fftshift(im_res[11]))))))\n\nplt.figure()\nplt.imshow(np.log(np.abs(np.fft.fftshift(np.fft.fft2(np.fft.fftshift(im_truth[11]))))))",
"_____no_output_____"
],
[
"#Visualize magnitude of input, results, truth\n\ntt = 11\n\nim_X = X[tt,:,:,0] + 1j * X[tt,:,:,1]\nim_res = res[tt,:,:,0] + 1j * res[tt,:,:,1]\nim_truth = truth_data[tt]\n\nfig, axs = plt.subplots(1, 3, squeeze=False, figsize=(18,8))\n\naxs[0,0].imshow((np.abs(im_X)), cmap='gray')\naxs[0,1].imshow((np.abs(im_res)), cmap='gray')\naxs[0,2].imshow((np.abs(im_truth)), cmap='gray')",
"_____no_output_____"
],
[
"#Visualize phase of input, results, truth\n\ntt = 11\n\nim_X = X[tt,:,:,0] + 1j * X[tt,:,:,1]\nim_res = res[tt,:,:,0] + 1j * res[tt,:,:,1]\nim_truth = truth_data[tt]\n\nfig, axs = plt.subplots(1, 3, squeeze=False, figsize=(18,8))\n\naxs[0,0].imshow((np.angle(im_X)), cmap='gray')\naxs[0,1].imshow((np.angle(im_res)), cmap='gray')\naxs[0,2].imshow((np.angle(im_truth)), cmap='gray')",
"_____no_output_____"
],
[
"# Compute error:\n\n# Phase difference of results and truth\nim_res = res[:,:,:,0] + 1j * res[:,:,:,1]\nim_truth = truth_data\nph_diff = np.angle(im_res * np.exp(-1j * np.angle(im_truth)))\n\n\n# Plot phase difference\ntt_show = 11 # timeframe to look at\nplt.figure()\nplt.imshow(ph_diff[tt_show])\nplt.title('Phase difference')\n\n# We dont actually care about anything but the hard, I saved a mask in the test dataset\nmask = cardiac_test['mask']\nplt.figure()\nplt.imshow(mask[tt_show])\nplt.title('Mask')\n\n# Multiply results by the mask\nph_diff_mask = ph_diff * mask\nplt.figure()\nplt.imshow(ph_diff_mask[tt_show])\nplt.title('Masked Phase difference')\n\n# Convert to mm error\nke = 0.12 # This was set in simulation\nmm_diff_mask = ph_diff_mask / (2*np.pi) / ke\nplt.figure()\nplt.imshow(mm_diff_mask[tt_show])\nplt.colorbar()\nplt.title('Masked mm difference')\n\n# Calculate mean and standard deviation error, time frame specific\n# We mask differently because we don't want to incorporate all the zeros from masking\nerr_mean = []\nerr_std = []\nfor tt in range(im_res.shape[0]):\n t_diff = mm_diff_mask[tt, mask[tt]>0]\n err_mean.append(t_diff.mean())\n err_std.append(t_diff.std())\n\nprint('Time resolved mean error:', err_mean)\nprint('Time resolved stdev error:', err_std)\nx_ax = np.arange(25)\nplt.figure()\nplt.errorbar(x_ax, err_mean, yerr = err_std, fmt='o')\nplt.title('Mean error by timeframe')\nplt.xlabel(\"time frames 0 to 24\")\n\n# And the total error as a single metric\ntotal_err = mm_diff_mask[mask>0]\nprint('Total Error:', np.abs(total_err).mean(), 'mm +/-', np.abs(total_err).std(), 'mm')",
"Time resolved mean error: [0.96500105, 0.9515384, 0.95983005, 0.96939254, 0.9828257, 0.7993082, 0.51279, 0.22943455, -0.040202476, -0.112036906, -0.24206087, -0.31499285, -0.32632458, -0.32638112, -0.298206, -0.22652699, -0.149608, -0.0031538545, 0.27350715, 0.65129435, 0.9077374, 0.93075264, 0.93126166, 0.9324114, 0.9252203]\nTime resolved stdev error: [0.43198806, 0.54968226, 0.8365565, 1.1900026, 1.5250155, 1.9023885, 2.2192175, 2.427327, 2.53268, 2.5788047, 2.5980444, 2.597569, 2.5923252, 2.580618, 2.562899, 2.5258877, 2.475548, 2.392444, 2.2452936, 1.9741125, 1.5742894, 1.2147238, 0.864236, 0.55586547, 0.42180642]\nTotal Error: 1.7595571 mm +/- 1.1636698 mm\n"
]
],
[
[
"## Tuning ideas\n* Use all timeframes together\n * As channels (3D convolutions will be too expensive)\n* Operate in k-space\n* Which loss function is best\n* Operate in image space, but loss function in k-space\n* Play with activation functions, where to use batch normalization, learning rate adjustments, batch size\n* Check initial image normalization (zero mean, stdev = 1)\n\n## Implement validation, test data\n* Validate on different simulations\n* Test on the cardiac simulation\n* Eventually we will test on our acquired DENSE data (I need to prep it)",
"_____no_output_____"
]
],
[
[
"list(cardiac_test.keys())",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a15b5708d9ca52dfac65032bdf5ef40e5079f7b
| 3,695 |
ipynb
|
Jupyter Notebook
|
100days/day 70 - deadlock.ipynb
|
gopala-kr/ds-notebooks
|
bc35430ecdd851f2ceab8f2437eec4d77cb59423
|
[
"MIT"
] | 13 |
2021-03-11T00:25:22.000Z
|
2022-03-19T00:19:23.000Z
|
100days/day 70 - deadlock.ipynb
|
gopala-kr/ds-notebooks
|
bc35430ecdd851f2ceab8f2437eec4d77cb59423
|
[
"MIT"
] | 160 |
2021-04-26T19:04:15.000Z
|
2022-03-26T20:18:37.000Z
|
100days/day 70 - deadlock.ipynb
|
gopala-kr/ds-notebooks
|
bc35430ecdd851f2ceab8f2437eec4d77cb59423
|
[
"MIT"
] | 12 |
2021-04-26T19:43:01.000Z
|
2022-01-31T08:36:29.000Z
| 22.668712 | 80 | 0.481732 |
[
[
[
"from collections import defaultdict\nfrom time import sleep\nfrom threading import Thread, Lock",
"_____no_output_____"
]
],
[
[
"## algorithm",
"_____no_output_____"
]
],
[
[
"class SharedState:\n\n def __init__(self, n):\n self._lock = Lock()\n self._state = defaultdict(int)\n self._resources = [Lock() for _ in range(n)]\n\n def atomic(self, key, value=0):\n with self._lock:\n self._state[key] += value\n return self._state[key]\n\n def resource(self, i):\n return self._resources[i]\n\n def kill(self):\n resources = self._resources\n self._resources = None\n for i in resources:\n i.release()",
"_____no_output_____"
],
[
"def worker(pid, state):\n try:\n while True:\n state.atomic('waiting', 1)\n with state.resource(pid):\n state.atomic('waiting', 1)\n with state.resource(pid - 1):\n state.atomic('waiting', -2)\n state.atomic('tasks', 1)\n\n except RuntimeError:\n pass",
"_____no_output_____"
],
[
"def deadlock(n):\n state = SharedState(n)\n\n for i in range(n):\n Thread(target=worker, args=(i, state)).start()\n\n while state.atomic('waiting') < 2 * n:\n sleep(1)\n\n print(n, 'workers; deadlock after', state.atomic('tasks'), 'tasks')\n state.kill()",
"_____no_output_____"
]
],
[
[
"## run",
"_____no_output_____"
]
],
[
[
"for i in range(1, 10):\n deadlock(10 * i)",
"10 workers; deadlock after 19935 tasks\n20 workers; deadlock after 36262 tasks\n30 workers; deadlock after 77059 tasks\n40 workers; deadlock after 15107 tasks\n50 workers; deadlock after 43405 tasks\n60 workers; deadlock after 9465 tasks\n70 workers; deadlock after 78949 tasks\n80 workers; deadlock after 10051 tasks\n90 workers; deadlock after 14169 tasks\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a15c72a3543fdd2393cc6ce3b0ea2f81ca91e61
| 24,555 |
ipynb
|
Jupyter Notebook
|
make_optimal_filter_data.ipynb
|
prashanth-prakash/Spiking-rdd
|
82a6487fe7a99cc8571d40b154c3aa298313d43f
|
[
"MIT"
] | 4 |
2019-01-21T18:21:07.000Z
|
2021-04-30T16:31:12.000Z
|
make_optimal_filter_data.ipynb
|
prashanth-prakash/Spiking-rdd
|
82a6487fe7a99cc8571d40b154c3aa298313d43f
|
[
"MIT"
] | null | null | null |
make_optimal_filter_data.ipynb
|
prashanth-prakash/Spiking-rdd
|
82a6487fe7a99cc8571d40b154c3aa298313d43f
|
[
"MIT"
] | null | null | null | 116.374408 | 20,528 | 0.877988 |
[
[
[
"%pylab inline\npylab.rcParams['figure.figsize'] = (10, 6)\n\nimport numpy as np\nimport numpy.random as random\nimport matplotlib.pyplot as plt",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"a = np.load('./sweeps/param_w_N_19_nsims_1_c_0.010000_deltaT_simulations.npz')\nfn_out = './sweeps/param_w_N_19_i_10_j_10_c_0.010000_deltaT_simulation.npz'",
"_____no_output_____"
],
[
"a.keys()",
"_____no_output_____"
],
[
"vs = a['vs']",
"_____no_output_____"
],
[
"i = 10\nj = 10\nv = vs[i,j,0,0,:]",
"_____no_output_____"
],
[
"N = 10000\n#Take different deltaT sizes\ndeltaT = [2, 5, 10, 25, 50]\n\nv_out = np.zeros((N, len(deltaT)))\n\nfor idx, DT in enumerate(deltaT):\n v_r = v.reshape((-1, DT))\n v_out[:,idx] = np.max(v_r[0:N], 1)",
"_____no_output_____"
],
[
"#Save\nnp.save(fn_out, v_out)",
"_____no_output_____"
],
[
"#Generate N Gaussian pts\ngauss = random.randn(N)",
"_____no_output_____"
],
[
"np.save('./sweeps/Gaussian_1000.npz', gauss)",
"_____no_output_____"
]
],
[
[
"## Compute optimal filter in R",
"_____no_output_____"
]
],
[
[
"#Then reimport\nprespike = np.load('./optimal_filter_prespike.npy')\npostspike = np.load('./optimal_filter_postspike.npy')",
"_____no_output_____"
],
[
"plt.plot(prespike[:,0], prespike[:,1], color=(0,0,0))\nplt.plot(postspike[:,0], postspike[:,1], color=(0,0,0))\nplt.axvline(x = 1, linestyle = '--', color = (0,0,0))\nplt.xlim([0, 1.5])\nplt.savefig('./fig_3.pdf')",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
] |
4a15c8cc11bdffab0c5939659aa950328d561f62
| 1,760 |
ipynb
|
Jupyter Notebook
|
tools/add-numbers/run.ipynb
|
crosscompute/crosscompute-examples
|
b341994d9353206fe0ee6b69688e70999bc2196e
|
[
"MIT"
] | 6 |
2015-12-18T15:54:24.000Z
|
2021-05-19T17:30:39.000Z
|
tools/add-numbers/run.ipynb
|
crosscompute/crosscompute-examples
|
b341994d9353206fe0ee6b69688e70999bc2196e
|
[
"MIT"
] | 7 |
2016-04-22T16:31:08.000Z
|
2022-02-07T17:54:46.000Z
|
tools/add-numbers/run.ipynb
|
crosscompute/crosscompute-examples
|
b341994d9353206fe0ee6b69688e70999bc2196e
|
[
"MIT"
] | 7 |
2016-04-20T21:03:57.000Z
|
2022-02-04T16:45:35.000Z
| 21.463415 | 67 | 0.54375 |
[
[
[
"# Get settings\nfrom os import environ\n\ninput_folder = environ.get(\n 'CROSSCOMPUTE_INPUT_FOLDER', 'tests/integers/input')\noutput_folder = environ.get(\n 'CROSSCOMPUTE_OUTPUT_FOLDER', 'tests/integers/output')",
"_____no_output_____"
],
[
"# Load input\nimport json\nfrom os.path import join\nnumbers_path = join(input_folder, 'numbers.json')\nnumbers_dictionary = json.load(open(numbers_path, 'rt'))\nnumbers_dictionary",
"_____no_output_____"
],
[
"# Make output\na = numbers_dictionary['a']\nb = numbers_dictionary['b']\nc = a + b\nc",
"_____no_output_____"
],
[
"# Save output\nproperties_path = join(output_folder, 'properties.json')\nproperties_dictionary = {\n 'c': c,\n}\njson.dump(properties_dictionary, open(properties_path, 'wt'))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code"
]
] |
4a15cb42a82472ec34f98858d88d7e6162e3b5ca
| 79,195 |
ipynb
|
Jupyter Notebook
|
3_quality_control/expression_QC_part2.ipynb
|
kevin-rychel/modulome-workflow
|
865c8e6039382750f792ad21231f26ef36c4f2e8
|
[
"MIT"
] | 1 |
2021-11-16T12:34:14.000Z
|
2021-11-16T12:34:14.000Z
|
3_quality_control/expression_QC_part2.ipynb
|
kevin-rychel/modulome-workflow
|
865c8e6039382750f792ad21231f26ef36c4f2e8
|
[
"MIT"
] | 8 |
2021-06-14T19:06:30.000Z
|
2022-03-17T23:29:52.000Z
|
3_quality_control/expression_QC_part2.ipynb
|
kevin-rychel/modulome-workflow
|
865c8e6039382750f792ad21231f26ef36c4f2e8
|
[
"MIT"
] | 2 |
2022-01-28T22:37:28.000Z
|
2022-03-31T22:26:44.000Z
| 53.008701 | 28,016 | 0.628297 |
[
[
[
"# Expression Quality Control (Part 2)",
"_____no_output_____"
],
[
"This is a template notebook for performing the final quality control on your organism's expression data. This requires a curated metadata sheet.",
"_____no_output_____"
],
[
"## Setup ",
"_____no_output_____"
]
],
[
[
"import itertools\n\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport seaborn as sns\n\nfrom os import path\nfrom scipy import stats\nfrom tqdm.notebook import tqdm",
"_____no_output_____"
],
[
"sns.set_style('ticks')",
"_____no_output_____"
]
],
[
[
"### Inputs",
"_____no_output_____"
]
],
[
[
"logTPM_file = path.join('..','data','raw_data','log_tpm.csv') # Enter log-TPM filename here\nall_metadata_file = path.join('..','data','interim','metadata_qc_part1_all.tsv') # Enter full metadata filename here\nmetadata_file = path.join('..','data','interim','metadata_qc_part1_curated.tsv') # Enter curated metadata filename here",
"_____no_output_____"
]
],
[
[
"### Load expression data",
"_____no_output_____"
]
],
[
[
"DF_log_tpm = pd.read_csv(logTPM_file,index_col=0).fillna(0)\nprint('Number of genes:',DF_log_tpm.shape[0])\nprint('Number of samples:',DF_log_tpm.shape[1])\nDF_log_tpm.head()",
"Number of genes: 4325\nNumber of samples: 708\n"
]
],
[
[
"### Load metadata",
"_____no_output_____"
]
],
[
[
"DF_metadata = pd.read_csv(metadata_file,index_col=0,sep='\\t')\nprint('Number of samples with curated metadata:',DF_metadata.shape[0])\nDF_metadata.head()",
"Number of samples with curated metadata: 387\n"
],
[
"DF_metadata_all = pd.read_csv(all_metadata_file,index_col=0,sep='\\t')",
"_____no_output_____"
]
],
[
[
"## Remove samples due to poor metadata",
"_____no_output_____"
],
[
"After curation, some samples either did not have enough replicates or metadata to warrant inclusion in this database.",
"_____no_output_____"
]
],
[
[
"DF_metadata_passed_step4 = DF_metadata[~DF_metadata.skip.fillna(False)].copy()\nprint('New number of samples with curated metadata:',DF_metadata_passed_step4.shape[0])\nDF_metadata_passed_step4.head()",
"New number of samples with curated metadata: 292\n"
]
],
[
[
"### Check curation\nSince manual curation is error-prone, we want to make sure that all samples have labels for their project and condition. In addition, there should only be one reference condition in each project, and it should be in the project itself.\n\nAny samples that fail these checks will be printed below.",
"_____no_output_____"
]
],
[
[
"assert(DF_metadata_passed_step4.project.notnull().all())\nassert(DF_metadata_passed_step4.condition.notnull().all())\n\nfor name,group in DF_metadata_passed_step4.groupby('project'):\n ref_cond = group.reference_condition.unique()\n \n # Ensure that there is only one reference condition per project\n if not len(ref_cond) == 1:\n print('Multiple reference conditions for:, name')\n \n # Ensure the reference condition is in fact in the project\n ref_cond = ref_cond[0]\n if not ref_cond in group.condition.tolist():\n print('Reference condition not in project:', name)",
"_____no_output_____"
]
],
[
[
"Next, make a new column called ``full_name`` that gives every experimental condition a unique, human-readable identifier.",
"_____no_output_____"
]
],
[
[
"DF_metadata_passed_step4['full_name'] = DF_metadata_passed_step4['project'].str.cat(DF_metadata_passed_step4['condition'],sep=':')",
"_____no_output_____"
]
],
[
[
"### Remove samples with only one replicate",
"_____no_output_____"
],
[
"First, find sample names that have at least two replicates.",
"_____no_output_____"
]
],
[
[
"counts = DF_metadata_passed_step4.full_name.value_counts()\nkeep_samples = counts[counts >= 2].index\nprint(keep_samples[:5])",
"Index(['nusA:no_nusA', 'nusA:nusA_induced', 'biofilm_time:8h',\n 'biofilm_time:12h', 'biofilm_time:16h'],\n dtype='object')\n"
]
],
[
[
"Only keep these samples",
"_____no_output_____"
]
],
[
[
"DF_metadata_passed_step4 = DF_metadata_passed_step4[DF_metadata_passed_step4.full_name.isin(keep_samples)]\nprint('New number of samples with curated metadata:',DF_metadata_passed_step4.shape[0])\nDF_metadata_passed_step4.head()",
"New number of samples with curated metadata: 273\n"
]
],
[
[
"### Save this information to the full metadata dataframe",
"_____no_output_____"
]
],
[
[
"DF_metadata_all['passed_curation'] = DF_metadata_all.index.isin(DF_metadata_passed_step4.index)",
"_____no_output_____"
]
],
[
[
"## Check correlations between replicates",
"_____no_output_____"
],
[
"### Remove failed data from log_tpm files",
"_____no_output_____"
]
],
[
[
"DF_log_tpm = DF_log_tpm[DF_metadata_passed_step4.index]",
"_____no_output_____"
]
],
[
[
"### Compute Pearson R Score",
"_____no_output_____"
],
[
"Biological replicates should have a Pearson R correlation above 0.95. For samples with more than 2 replicates, the replicates must have R >= 0.95 with at least one other replicate or it will be dropped. The correlation threshold can be changed below:",
"_____no_output_____"
]
],
[
[
"rcutoff = 0.95",
"_____no_output_____"
]
],
[
[
"The following code computes correlations between all samples and collects correlations between replicates and non-replicates.",
"_____no_output_____"
]
],
[
[
"rep_corrs = {}\nrand_corrs = {}\n\nnum_comparisons = len(DF_metadata_passed_step4)*(len(DF_metadata_passed_step4)-1)/2\n\nfor exp1,exp2 in tqdm(itertools.combinations(DF_metadata_passed_step4.index,2),total=num_comparisons):\n if DF_metadata_passed_step4.loc[exp1,'full_name'] == DF_metadata_passed_step4.loc[exp2,'full_name']:\n rep_corrs[(exp1,exp2)] = stats.pearsonr(DF_log_tpm[exp1],DF_log_tpm[exp2])[0]\n else:\n rand_corrs[(exp1,exp2)] = stats.pearsonr(DF_log_tpm[exp1],DF_log_tpm[exp2])[0]",
"_____no_output_____"
]
],
[
[
"Correlations can be plotted on a histogram",
"_____no_output_____"
]
],
[
[
"fig,ax = plt.subplots(figsize=(5,5))\nax2 = ax.twinx()\nax2.hist(rep_corrs.values(),bins=50,range=(0.2,1),alpha=0.8,color='green',linewidth=0)\nax.hist(rand_corrs.values(),bins=50,range=(0.2,1),alpha=0.8,color='blue',linewidth=0)\nax.set_title('Pearson R correlation between experiments',fontsize=14)\nax.set_xlabel('Pearson R correlation',fontsize=14)\nax.set_ylabel('Different Conditions',fontsize=14)\nax2.set_ylabel('Known Replicates',fontsize=14)\n\nmed_corr = np.median([v for k,v in rep_corrs.items()])\nprint('Median Pearson R between replicates: {:.2f}'.format(med_corr))",
"Median Pearson R between replicates: 0.98\n"
]
],
[
[
"Remove samples without any high-correlation replicates",
"_____no_output_____"
]
],
[
[
"dissimilar = []\nfor idx, grp in DF_metadata_passed_step4.groupby('full_name'):\n ident = np.identity(len(grp))\n corrs = (DF_log_tpm[grp.index].corr() - ident).max()\n dissimilar.extend(corrs[corrs<rcutoff].index)\n\n# Save this information in both the original metadata dataframe and the new metadata dataframe\nDF_metadata_all['passed_replicate_correlations'] = ~DF_metadata_all.index.isin(dissimilar)\nDF_metadata_passed_step4['passed_replicate_correlations'] = ~DF_metadata_passed_step4.index.isin(dissimilar)",
"_____no_output_____"
],
[
"DF_metadata_final = DF_metadata_passed_step4[DF_metadata_passed_step4['passed_replicate_correlations']]\nprint('# Samples that passed replicate correlations:',len(DF_metadata_final))",
"# Samples that passed replicate correlations: 265\n"
]
],
[
[
"## Check that reference conditions still exist\nIf a reference condition was removed due to poor replicate correlations, a new reference condition needs to be defined.\n\nAgain, any samples that fail these checks will be printed below.",
"_____no_output_____"
]
],
[
[
"project_exprs = []\nfor name,group in DF_metadata_final.groupby('project'):\n \n # Get reference condition\n ref_cond = group.reference_condition.iloc[0]\n \n # Ensure the reference condition is still in the project\n if ref_cond not in group.condition.tolist():\n print('Reference condition missing from:', name)\n \n # Check that each project has at least two conditions (a reference and at least one test condition)\n if len(group.condition.unique()) <= 1:\n print('Only one condition in:', name)",
"_____no_output_____"
]
],
[
[
"If necessary, choose a new condition for failed projects and re-run notebook.",
"_____no_output_____"
],
[
"## Normalize dataset to reference conditions",
"_____no_output_____"
]
],
[
[
"DF_log_tpm_final = DF_log_tpm[DF_metadata_final.index]",
"_____no_output_____"
],
[
"project_exprs = []\nfor name,group in DF_metadata_final.groupby('project'):\n \n # Get reference condition\n ref_cond = group.reference_condition.iloc[0]\n \n # Get reference condition sample ids\n ref_samples = group[group.condition == ref_cond].index\n \n # Get reference condition expression\n ref_expr = DF_log_tpm_final[ref_samples].mean(axis=1)\n \n # Subtract reference expression from project\n project_exprs.append(DF_log_tpm_final[group.index].sub(ref_expr,axis=0))\n\nDF_log_tpm_norm = pd.concat(project_exprs,axis=1)",
"_____no_output_____"
]
],
[
[
"## Save final datasets",
"_____no_output_____"
]
],
[
[
"logTPM_qc_file = path.join('..','data','processed_data','log_tpm.csv')\nlogTPM_norm_file = path.join('..','data','processed_data','log_tpm_norm.csv')\nfinal_metadata_file = path.join('..','data','processed_data','metadata.tsv')\nfinal_metadata_all_file = path.join('..','data','interim','metadata_qc_part2_all.tsv')\n\nDF_log_tpm_final.to_csv(logTPM_qc_file)\nDF_log_tpm_norm.to_csv(logTPM_norm_file)\nDF_metadata_final.to_csv(final_metadata_file, sep='\\t')\nDF_metadata_all.to_csv(final_metadata_all_file, sep='\\t')",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a15cda3ec7576061d22dda96817b3ac9da96973
| 16,760 |
ipynb
|
Jupyter Notebook
|
examples/Hyperparams And Distributions.ipynb
|
vincent-antaki/Neuraxle
|
cef1284a261010c655f8ef02b4fca5b8bb45850c
|
[
"Apache-2.0"
] | 4 |
2019-06-24T01:06:57.000Z
|
2020-08-18T08:16:10.000Z
|
examples/Hyperparams And Distributions.ipynb
|
Tubbz-alt/Neuraxle
|
308f24248cdb242b7e2f6ec7c51daf2ee3e38834
|
[
"Apache-2.0"
] | 1 |
2020-02-07T15:08:42.000Z
|
2020-02-07T15:08:42.000Z
|
examples/Hyperparams And Distributions.ipynb
|
Tubbz-alt/Neuraxle
|
308f24248cdb242b7e2f6ec7c51daf2ee3e38834
|
[
"Apache-2.0"
] | null | null | null | 30.865562 | 381 | 0.564797 |
[
[
[
"# Hyperparams And Distributions",
"_____no_output_____"
],
[
"This page introduces the hyperparams, and distributions in Neuraxle. You can find [Hyperparams Distribution API here](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.distributions.html), and \n[Hyperparameter Samples API here](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.space.html).\n\nHyperparameter is a parameter drawn from a prior distribution. In Neuraxle, we have a few built-in distributions, and we are also compatible with scipy distributions. \n\nCreate a [Uniform Distribution](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.distributions.html#neuraxle.hyperparams.distributions.Uniform):",
"_____no_output_____"
]
],
[
[
"from neuraxle.hyperparams.distributions import Uniform\n\nhd = Uniform(\n min_included=-10, \n max_included=10, \n null_default_value=0\n)",
"_____no_output_____"
]
],
[
[
"Sample the random variable using [rvs](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.distributions.html#neuraxle.hyperparams.distributions.HyperparameterDistribution.rvs):",
"_____no_output_____"
]
],
[
[
"sample = hd.rvs()\nprint(sample)",
"-5.873595345744127\n"
]
],
[
[
"Nullify the random variable using [nullify](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.distributions.html#neuraxle.hyperparams.distributions.HyperparameterDistribution.nullify):",
"_____no_output_____"
]
],
[
[
"nullified_sample = hd.nullify()\nassert nullified_sample == hd.null_default_value",
"_____no_output_____"
]
],
[
[
"Get the probability distribution function value at `x` using [pdf](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.distributions.html#neuraxle.hyperparams.distributions.HyperparameterDistribution.pdf):",
"_____no_output_____"
]
],
[
[
"pdf = hd.pdf(1)\nprint('pdf: {}'.format(pdf))",
"pdf: 0.05\n"
]
],
[
[
"Get the cumulative probability distribution function value at `x` using [cdf](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.distributions.html#neuraxle.hyperparams.distributions.HyperparameterDistribution.cdf)",
"_____no_output_____"
]
],
[
[
"cdf = hd.cdf(1)\nprint('cdf: {}'.format(cdf))",
"cdf: 0.55\n"
]
],
[
[
"## Setting And Updating Hyperparams\n\n\nIn Neuraxle, each step has hyperparams of type [HyperparameterSamples](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.space.html#neuraxle.hyperparams.space.HyperparameterSamples), and spaces of type [HyperparameterSpace](https://www.neuraxle.org/stable/api/neuraxle.hyperparams.distributions.html#neuraxle.hyperparams.distributions.HyperparameterDistribution). \n\nConsider a simple pipeline that contains 2 MultiplyByN steps, and one PCA component inside a nested pipeline:",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\n\nfrom neuraxle.hyperparams.distributions import RandInt\nfrom neuraxle.hyperparams.space import HyperparameterSpace, HyperparameterSamples\nfrom neuraxle.pipeline import Pipeline\nfrom neuraxle.steps.numpy import MultiplyByN\n\np = Pipeline([\n ('step1', MultiplyByN(2)),\n ('step2', MultiplyByN(2)),\n Pipeline([\n PCA(n_components=4)\n ])\n])",
"_____no_output_____"
]
],
[
[
"We can set or update the hyperparams, and spaces by doing the following: ",
"_____no_output_____"
]
],
[
[
"p.set_hyperparams(HyperparameterSamples({\n 'step1__multiply_by': 42,\n 'step2__multiply_by': -10,\n 'Pipeline__PCA__n_components': 2\n}))\n\np.update_hyperparams(HyperparameterSamples({\n 'Pipeline__PCA__n_components': 3\n}))\n\np.set_hyperparams_space(HyperparameterSpace({\n 'step1__multiply_by': RandInt(42, 50),\n 'step2__multiply_by': RandInt(-10, 0),\n 'Pipeline__PCA__n_components': RandInt(2, 3)\n}))",
"_____no_output_____"
]
],
[
[
"We can sample the space of random variables:",
"_____no_output_____"
]
],
[
[
"samples = p.get_hyperparams_space().rvs()\n\nassert 42 <= samples['step1__multiply_by'] <= 50\nassert -10 <= samples['step2__multiply_by'] <= 0\nassert samples['Pipeline__PCA__n_components'] in [2, 3]",
"_____no_output_____"
]
],
[
[
"We can get all hyperparams:",
"_____no_output_____"
]
],
[
[
"samples = p.get_hyperparams()\n\nassert 42 <= samples['step1__multiply_by'] <= 50\nassert -10 <= samples['step2__multiply_by'] <= 0\nassert samples['Pipeline__PCA__n_components'] in [2, 3]\nassert p['Pipeline']['PCA'].get_wrapped_sklearn_predictor().n_components in [2, 3]",
"_____no_output_____"
]
],
[
[
"## Neuraxle Custom Distributions",
"_____no_output_____"
],
[
"## Scipy Distributions \n\nTo define a scipy distribution that is compatible with Neuraxle, you need to wrap the scipy distribution with ScipyDistributionWrapper: ",
"_____no_output_____"
]
],
[
[
"from neuraxle.hyperparams.scipy_distributions import ScipyDistributionWrapper, BaseContinuousDistribution, BaseDiscreteDistribution\nfrom scipy.integrate import quad\nfrom scipy.special import factorial\nfrom scipy.stats import rv_continuous, norm, rv_discrete, rv_histogram, truncnorm, randint\nimport numpy as np\nimport math \n\nhd = ScipyDistributionWrapper(\n scipy_distribution=randint(low=0, high=10),\n is_continuous=False,\n null_default_value=0\n)",
"_____no_output_____"
]
],
[
[
"### Discrete Distributions\nFor discrete distribution that inherit from [rv_discrete](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.rv_discrete.html#scipy.stats.rv_discrete), you only need to implement _pmf. The rest is taken care of magically by scipy. \n\nFor example, here is a discrete poisson distribution: ",
"_____no_output_____"
]
],
[
[
"class Poisson(BaseDiscreteDistribution):\n def __init__(self, min_included: float, max_included: float, null_default_value: float = None, mu=0.6):\n super().__init__(\n min_included=min_included,\n max_included=max_included,\n name='poisson',\n null_default_value=null_default_value\n )\n self.mu = mu\n\n def _pmf(self, x):\n return math.exp(-self.mu) * self.mu ** x / factorial(x)",
"_____no_output_____"
]
],
[
[
"### Continuous Distributions\n\nFor continous distribution that inherit from [rv_continuous](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.rv_continuous.html), you only need to implement _pdf function. The rest is taken care of magically by scipy. \n\nFor example, here is a continous gaussian distribution: ",
"_____no_output_____"
]
],
[
[
"class Gaussian(BaseContinuousDistribution): \n def __init__(self, min_included: int, max_included: int, null_default_value: float = None):\n self.max_included = max_included\n self.min_included = min_included\n\n BaseContinuousDistribution.__init__(\n self,\n name='gaussian',\n min_included=min_included,\n max_included=max_included,\n null_default_value=null_default_value\n )\n\n def _pdf(self, x):\n return math.exp(-x ** 2 / 2.) / np.sqrt(2.0 * np.pi)",
"_____no_output_____"
]
],
[
[
"### Custom Arguments \n\nIf you want to add more properties to calculate your distributions, just add them in self. They will be available in all of the scipy private methods you can override like _pmf, and _pdf. ",
"_____no_output_____"
]
],
[
[
"class LogNormal(BaseContinuousDistribution):\n def __init__(\n self,\n log2_space_mean: float,\n log2_space_std: float,\n hard_clip_min: float,\n hard_clip_max: float,\n null_default_value: float = None\n ):\n if null_default_value is None:\n null_default_value = hard_clip_min\n\n if hard_clip_min is None:\n hard_clip_min = np.nan\n\n if hard_clip_max is None:\n hard_clip_max = np.nan\n\n self.log2_space_mean = log2_space_mean\n self.log2_space_std = log2_space_std\n\n super().__init__(\n name='log_normal',\n min_included=hard_clip_min,\n max_included=hard_clip_max,\n null_default_value=null_default_value\n )\n\n def _pdf(self, x):\n if x <= 0:\n return 0.\n\n cdf_min = 0.\n cdf_max = 1.\n\n pdf_x = 1 / (x * math.log(2) * self.log2_space_std * math.sqrt(2 * math.pi)) * math.exp(\n -(math.log2(x) - self.log2_space_mean) ** 2 / (2 * self.log2_space_std ** 2))\n return pdf_x / (cdf_max - cdf_min)",
"_____no_output_____"
]
],
[
[
"### Scipy methods\n\nAll of the scipy distribution methods are available:",
"_____no_output_____"
]
],
[
[
"def get_many_samples_for(hd, num_trial):\n return [hd.rvs() for _ in range(num_trial)]\n\nsamples = get_many_samples_for(hd, 1000)\n\nfor s in samples:\n assert type(s) == int\n \nhd = Gaussian(min_included=0, max_included=10, null_default_value=0)\n\nassert 0.0 <= hd.rvs() <= 10.0\nassert hd.pdf(10) < 0.001\nassert hd.pdf(0) < 0.42\nassert 0.55 > hd.cdf(5.0) > 0.45\nassert hd.cdf(0) == 0.0\nassert hd.logpdf(5) == -13.418938533204672\nassert hd.logcdf(5) == -0.6931477538632531\nassert hd.sf(5) == 0.5000002866515718\nassert hd.logsf(5) == -0.693146607256966\nassert np.all(hd.ppf([0.0, 0.01, 0.05, 0.1, 1 - 0.10, 1 - 0.05, 1 - 0.01, 1.0], 10))\nassert np.isclose(hd.moment(2), 50.50000000091249)\nassert hd.stats()[0]\nassert hd.stats()[1]\nassert np.array_equal(hd.entropy(), np.array(0.7094692666023363))\nassert hd.median()\nassert hd.mean() == 5.398942280397029\nassert np.isclose(hd.std(), 4.620759921685374)\nassert np.isclose(hd.var(), 21.35142225385382)\nassert np.isclose(hd.expect(), 0.39894228040143276)\ninterval = hd.interval(alpha=[0.25, 0.50])\nassert np.all(interval[0])\nassert np.all(interval[1])\nassert hd.support() == (0, 10)",
"_____no_output_____"
]
],
[
[
"## SKLearn Hyperparams\n\nSKLearnWrapper wraps sklearn predictors so that they can be compatible with Neuraxle. When you set the hyperparams of an SKLearnWrapper, it automatically sets the params of the sklearn predictor for you: ",
"_____no_output_____"
]
],
[
[
"from neuraxle.hyperparams.distributions import Choice\nfrom neuraxle.hyperparams.distributions import RandInt\nfrom neuraxle.hyperparams.space import HyperparameterSpace\nfrom neuraxle.steps.sklearn import SKLearnWrapper\nfrom sklearn.tree import DecisionTreeClassifier\n\n\ndecision_tree_classifier = SKLearnWrapper(\n DecisionTreeClassifier(), \n HyperparameterSpace({\n 'criterion': Choice(['gini', 'entropy']), \n 'splitter': Choice(['best', 'random']),\n 'min_samples_leaf': RandInt(2, 5), \n 'min_samples_split': RandInt(1, 3) \n })\n).set_hyperparams(HyperparameterSamples({\n 'criterion': 'gini', \n 'splitter': 'best',\n 'min_samples_leaf': 3, \n 'min_samples_split': 3 \n}))",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a15d4cfe0f5fbaa73c542d672f3decca9a9929b
| 13,034 |
ipynb
|
Jupyter Notebook
|
notebooks/hpo_tensorflow_mnist.ipynb
|
randyridgley/sagemaker-tensorflow
|
c3b935f429968b8c7a1362c95be1bc945007738c
|
[
"MIT"
] | 2 |
2019-03-29T01:16:13.000Z
|
2019-05-23T02:30:23.000Z
|
notebooks/hpo_tensorflow_mnist.ipynb
|
randyridgley/sagemaker-tensorflow
|
c3b935f429968b8c7a1362c95be1bc945007738c
|
[
"MIT"
] | null | null | null |
notebooks/hpo_tensorflow_mnist.ipynb
|
randyridgley/sagemaker-tensorflow
|
c3b935f429968b8c7a1362c95be1bc945007738c
|
[
"MIT"
] | 1 |
2019-04-04T00:41:44.000Z
|
2019-04-04T00:41:44.000Z
| 39.377644 | 557 | 0.64209 |
[
[
[
"# Hyperparameter Tuning using SageMaker Tensorflow Container\n\nThis tutorial focuses on how to create a convolutional neural network model to train the [MNIST dataset](http://yann.lecun.com/exdb/mnist/) using **SageMaker TensorFlow container**. It leverages hyperparameter tuning to kick off multiple training jobs with different hyperparameter combinations, to find the one with best model training result.",
"_____no_output_____"
],
[
"## Set up the environment\nWe will set up a few things before starting the workflow. \n\n1. specify the s3 bucket and prefix where training data set and model artifacts will be stored\n2. get the execution role which will be passed to sagemaker for accessing your resources such as s3 bucket",
"_____no_output_____"
]
],
[
[
"import sagemaker\nimport project_path\nfrom lib import utils\n\nbucket = '{{s3_workshop_bucket}}}'\nprefix = 'sagemaker/DEMO-hpo-tensorflow-high' # you can customize the prefix (subfolder) here\n\nrole = sagemaker.get_execution_role() # we are using the notebook instance role for training in this example",
"_____no_output_____"
]
],
[
[
"Now we'll import the Python libraries we'll need.",
"_____no_output_____"
]
],
[
[
"import boto3\nfrom time import gmtime, strftime\nfrom sagemaker.tensorflow import TensorFlow\nfrom sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner",
"_____no_output_____"
]
],
[
[
"## Download the MNIST dataset",
"_____no_output_____"
]
],
[
[
"import utils\nfrom tensorflow.contrib.learn.python.learn.datasets import mnist\nimport tensorflow as tf\n\ndata_sets = mnist.read_data_sets('data', dtype=tf.uint8, reshape=False, validation_size=5000)\n\nutils.convert_to(data_sets.train, 'train', 'data')\nutils.convert_to(data_sets.validation, 'validation', 'data')\nutils.convert_to(data_sets.test, 'test', 'data')",
"_____no_output_____"
]
],
[
[
"## Upload the data\nWe use the ```sagemaker.Session.upload_data``` function to upload our datasets to an S3 location. The return value identifies the location -- we will use this later when we start the training job.",
"_____no_output_____"
]
],
[
[
"inputs = sagemaker.Session().upload_data(path='data', bucket=bucket, key_prefix=prefix+'/data/mnist')\nprint (inputs)",
"_____no_output_____"
]
],
[
[
"## Construct a script for distributed training \nHere is the full code for the network model:",
"_____no_output_____"
]
],
[
[
"!cat '../scripts/mnist.py'",
"_____no_output_____"
]
],
[
[
"The script here is and adaptation of the [TensorFlow MNIST example](https://github.com/tensorflow/models/tree/master/official/mnist). It provides a ```model_fn(features, labels, mode)```, which is used for training, evaluation and inference. \n\n### A regular ```model_fn```\n\nA regular **```model_fn```** follows the pattern:\n1. [defines a neural network](https://github.com/tensorflow/models/blob/master/official/mnist/mnist.py#L96)\n- [applies the ```features``` in the neural network](https://github.com/tensorflow/models/blob/master/official/mnist/mnist.py#L178)\n- [if the ```mode``` is ```PREDICT```, returns the output from the neural network](https://github.com/tensorflow/models/blob/master/official/mnist/mnist.py#L186)\n- [calculates the loss function comparing the output with the ```labels```](https://github.com/tensorflow/models/blob/master/official/mnist/mnist.py#L188)\n- [creates an optimizer and minimizes the loss function to improve the neural network](https://github.com/tensorflow/models/blob/master/official/mnist/mnist.py#L193)\n- [returns the output, optimizer and loss function](https://github.com/tensorflow/models/blob/master/official/mnist/mnist.py#L205)\n\n### Writing a ```model_fn``` for distributed training\nWhen distributed training happens, the same neural network will be sent to the multiple training instances. Each instance will predict a batch of the dataset, calculate loss and minimize the optimizer. One entire loop of this process is called **training step**.\n\n#### Syncronizing training steps\nA [global step](https://www.tensorflow.org/api_docs/python/tf/train/global_step) is a global variable shared between the instances. It necessary for distributed training, so the optimizer will keep track of the number of **training steps** between runs: \n\n```python\ntrain_op = optimizer.minimize(loss, tf.train.get_or_create_global_step())\n```\n\nThat is the only required change for distributed training!",
"_____no_output_____"
],
[
"## Set up hyperparameter tuning job\n*Note, with the default setting below, the hyperparameter tuning job can take about 30 minutes to complete.*\n\nNow we will set up the hyperparameter tuning job using SageMaker Python SDK, following below steps:\n* Create an estimator to set up the TensorFlow training job\n* Define the ranges of hyperparameters we plan to tune, in this example, we are tuning \"learning_rate\"\n* Define the objective metric for the tuning job to optimize\n* Create a hyperparameter tuner with above setting, as well as tuning resource configurations ",
"_____no_output_____"
],
[
"Similar to training a single TensorFlow job in SageMaker, we define our TensorFlow estimator passing in the TensorFlow script, IAM role, and (per job) hardware configuration.",
"_____no_output_____"
]
],
[
[
"estimator = TensorFlow(entry_point='../scripts/mnist.py',\n role=role,\n framework_version='1.11.0',\n training_steps=1000, \n evaluation_steps=100,\n train_instance_count=1,\n train_instance_type='ml.m4.xlarge',\n base_job_name='DEMO-hpo-tensorflow')",
"_____no_output_____"
]
],
[
[
"Once we've defined our estimator we can specify the hyperparameters we'd like to tune and their possible values. We have three different types of hyperparameters.\n- Categorical parameters need to take one value from a discrete set. We define this by passing the list of possible values to `CategoricalParameter(list)`\n- Continuous parameters can take any real number value between the minimum and maximum value, defined by `ContinuousParameter(min, max)`\n- Integer parameters can take any integer value between the minimum and maximum value, defined by `IntegerParameter(min, max)`\n\n*Note, if possible, it's almost always best to specify a value as the least restrictive type. For example, tuning learning rate as a continuous value between 0.01 and 0.2 is likely to yield a better result than tuning as a categorical parameter with values 0.01, 0.1, 0.15, or 0.2.*",
"_____no_output_____"
]
],
[
[
"hyperparameter_ranges = {'learning_rate': ContinuousParameter(0.01, 0.2)}",
"_____no_output_____"
]
],
[
[
"Next we'll specify the objective metric that we'd like to tune and its definition, which includes the regular expression (Regex) needed to extract that metric from the CloudWatch logs of the training job. In this particular case, our script emits loss value and we will use it as the objective metric, we also set the objective_type to be 'minimize', so that hyperparameter tuning seeks to minize the objective metric when searching for the best hyperparameter setting. By default, objective_type is set to 'maximize'.",
"_____no_output_____"
]
],
[
[
"objective_metric_name = 'loss'\nobjective_type = 'Minimize'\nmetric_definitions = [{'Name': 'loss',\n 'Regex': 'loss = ([0-9\\\\.]+)'}]",
"_____no_output_____"
]
],
[
[
"Now, we'll create a `HyperparameterTuner` object, to which we pass:\n- The TensorFlow estimator we created above\n- Our hyperparameter ranges\n- Objective metric name and definition\n- Tuning resource configurations such as Number of training jobs to run in total and how many training jobs can be run in parallel.",
"_____no_output_____"
]
],
[
[
"tuner = HyperparameterTuner(estimator,\n objective_metric_name,\n hyperparameter_ranges,\n metric_definitions,\n max_jobs=9,\n max_parallel_jobs=3,\n objective_type=objective_type)",
"_____no_output_____"
]
],
[
[
"## Launch hyperparameter tuning job\nAnd finally, we can start our hyperprameter tuning job by calling `.fit()` and passing in the S3 path to our train and test dataset.\n\nAfter the hyperprameter tuning job is created, you should be able to describe the tuning job to see its progress in the next step, and you can go to SageMaker console->Jobs to check out the progress of the progress of the hyperparameter tuning job.",
"_____no_output_____"
]
],
[
[
"tuner.fit(inputs)",
"_____no_output_____"
]
],
[
[
"Let's just run a quick check of the hyperparameter tuning jobs status to make sure it started successfully.",
"_____no_output_____"
]
],
[
[
"boto3.client('sagemaker').describe_hyper_parameter_tuning_job(\n HyperParameterTuningJobName=tuner.latest_tuning_job.job_name)['HyperParameterTuningJobStatus']",
"_____no_output_____"
]
],
[
[
"## Analyze tuning job results - after tuning job is completed\nPlease refer to \"HPO_Analyze_TuningJob_Results.ipynb\" to see example code to analyze the tuning job results.",
"_____no_output_____"
],
[
"## Deploy the best model\nNow that we have got the best model, we can deploy it to an endpoint. Please refer to other SageMaker sample notebooks or SageMaker documentation to see how to deploy a model.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a15d6baf589005e2b72ae68d0bf380e40dc62c7
| 301,781 |
ipynb
|
Jupyter Notebook
|
Your_first_neural_network.ipynb
|
jmontag43/udacity_bike_prediction
|
efe573efe00359dbf2b735c9f2aa493b48ce77ac
|
[
"Xnet",
"X11"
] | null | null | null |
Your_first_neural_network.ipynb
|
jmontag43/udacity_bike_prediction
|
efe573efe00359dbf2b735c9f2aa493b48ce77ac
|
[
"Xnet",
"X11"
] | null | null | null |
Your_first_neural_network.ipynb
|
jmontag43/udacity_bike_prediction
|
efe573efe00359dbf2b735c9f2aa493b48ce77ac
|
[
"Xnet",
"X11"
] | null | null | null | 312.402692 | 161,644 | 0.90907 |
[
[
[
"# Your first neural network\n\nIn this project, you'll build your first neural network and use it to predict daily bike rental ridership. We've provided some of the code, but left the implementation of the neural network up to you (for the most part). After you've submitted this project, feel free to explore the data and the model more.\n\n",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n%config InlineBackend.figure_format = 'retina'\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Load and prepare the data\n\nA critical step in working with neural networks is preparing the data correctly. Variables on different scales make it difficult for the network to efficiently learn the correct weights. Below, we've written the code to load and prepare the data. You'll learn more about this soon!",
"_____no_output_____"
]
],
[
[
"data_path = 'hour.csv'\n\nrides = pd.read_csv(data_path)",
"_____no_output_____"
],
[
"rides.head()",
"_____no_output_____"
]
],
[
[
"## Checking out the data\n\nThis dataset has the number of riders for each hour of each day from January 1 2011 to December 31 2012. The number of riders is split between casual and registered, summed up in the `cnt` column. You can see the first few rows of the data above.\n\nBelow is a plot showing the number of bike riders over the first 10 days or so in the data set. (Some days don't have exactly 24 entries in the data set, so it's not exactly 10 days.) You can see the hourly rentals here. This data is pretty complicated! The weekends have lower over all ridership and there are spikes when people are biking to and from work during the week. Looking at the data above, we also have information about temperature, humidity, and windspeed, all of these likely affecting the number of riders. You'll be trying to capture all this with your model.",
"_____no_output_____"
]
],
[
[
"rides[:24*10].plot(x='dteday', y='cnt')",
"_____no_output_____"
]
],
[
[
"### Dummy variables\nHere we have some categorical variables like season, weather, month. To include these in our model, we'll need to make binary dummy variables. This is simple to do with Pandas thanks to `get_dummies()`.",
"_____no_output_____"
]
],
[
[
"dummy_fields = ['season', 'weathersit', 'mnth', 'hr', 'weekday']\nfor each in dummy_fields:\n dummies = pd.get_dummies(rides[each], prefix=each, drop_first=False)\n rides = pd.concat([rides, dummies], axis=1)\n\nfields_to_drop = ['instant', 'dteday', 'season', 'weathersit', \n 'weekday', 'atemp', 'mnth', 'workingday', 'hr']\ndata = rides.drop(fields_to_drop, axis=1)\ndata.head()",
"_____no_output_____"
]
],
[
[
"### Scaling target variables\nTo make training the network easier, we'll standardize each of the continuous variables. That is, we'll shift and scale the variables such that they have zero mean and a standard deviation of 1.\n\nThe scaling factors are saved so we can go backwards when we use the network for predictions.",
"_____no_output_____"
]
],
[
[
"quant_features = ['casual', 'registered', 'cnt', 'temp', 'hum', 'windspeed']\n# Store scalings in a dictionary so we can convert back later\nscaled_features = {}\nfor each in quant_features:\n mean, std = data[each].mean(), data[each].std()\n scaled_features[each] = [mean, std]\n data.loc[:, each] = (data[each] - mean)/std",
"_____no_output_____"
]
],
[
[
"### Splitting the data into training, testing, and validation sets\n\nWe'll save the data for the last approximately 21 days to use as a test set after we've trained the network. We'll use this set to make predictions and compare them with the actual number of riders.",
"_____no_output_____"
]
],
[
[
"# Save data for approximately the last 21 days \ntest_data = data[-21*24:]\n\n# Now remove the test data from the data set \ndata = data[:-21*24]\n\n# Separate the data into features and targets\ntarget_fields = ['cnt', 'casual', 'registered']\nfeatures, targets = data.drop(target_fields, axis=1), data[target_fields]\ntest_features, test_targets = test_data.drop(target_fields, axis=1), test_data[target_fields]",
"_____no_output_____"
]
],
[
[
"We'll split the data into two sets, one for training and one for validating as the network is being trained. Since this is time series data, we'll train on historical data, then try to predict on future data (the validation set).",
"_____no_output_____"
]
],
[
[
"# Hold out the last 60 days or so of the remaining data as a validation set\ntrain_features, train_targets = features[:-60*24], targets[:-60*24]\nval_features, val_targets = features[-60*24:], targets[-60*24:]",
"_____no_output_____"
]
],
[
[
"## Time to build the network\n\nBelow you'll build your network. We've built out the structure. You'll implement both the forward pass and backwards pass through the network. You'll also set the hyperparameters: the learning rate, the number of hidden units, and the number of training passes.\n\n<img src=\"assets/neural_network.png\" width=300px>\n\nThe network has two layers, a hidden layer and an output layer. The hidden layer will use the sigmoid function for activations. The output layer has only one node and is used for the regression, the output of the node is the same as the input of the node. That is, the activation function is $f(x)=x$. A function that takes the input signal and generates an output signal, but takes into account the threshold, is called an activation function. We work through each layer of our network calculating the outputs for each neuron. All of the outputs from one layer become inputs to the neurons on the next layer. This process is called *forward propagation*.\n\nWe use the weights to propagate signals forward from the input to the output layers in a neural network. We use the weights to also propagate error backwards from the output back into the network to update our weights. This is called *backpropagation*.\n\n> **Hint:** You'll need the derivative of the output activation function ($f(x) = x$) for the backpropagation implementation. If you aren't familiar with calculus, this function is equivalent to the equation $y = x$. What is the slope of that equation? That is the derivative of $f(x)$.\n\nBelow, you have these tasks:\n1. Implement the sigmoid function to use as the activation function. Set `self.activation_function` in `__init__` to your sigmoid function.\n2. Implement the forward pass in the `train` method.\n3. Implement the backpropagation algorithm in the `train` method, including calculating the output error.\n4. Implement the forward pass in the `run` method.\n ",
"_____no_output_____"
]
],
[
[
"#############\n# In the my_answers.py file, fill out the TODO sections as specified\n#############\n\nfrom my_answers import NeuralNetwork",
"_____no_output_____"
],
[
"def MSE(y, Y):\n return np.mean((y-Y)**2)",
"_____no_output_____"
]
],
[
[
"## Unit tests\n\nRun these unit tests to check the correctness of your network implementation. This will help you be sure your network was implemented correctly befor you starting trying to train it. These tests must all be successful to pass the project.",
"_____no_output_____"
]
],
[
[
"import unittest\n\ninputs = np.array([[0.5, -0.2, 0.1]])\ntargets = np.array([[0.4]])\ntest_w_i_h = np.array([[0.1, -0.2],\n [0.4, 0.5],\n [-0.3, 0.2]])\ntest_w_h_o = np.array([[0.3],\n [-0.1]])\n\nclass TestMethods(unittest.TestCase):\n \n ##########\n # Unit tests for data loading\n ##########\n \n def test_data_path(self):\n # Test that file path to dataset has been unaltered\n self.assertTrue(data_path.lower() == 'hour.csv')\n \n def test_data_loaded(self):\n # Test that data frame loaded\n self.assertTrue(isinstance(rides, pd.DataFrame))\n \n ##########\n # Unit tests for network functionality\n ##########\n\n def test_activation(self):\n network = NeuralNetwork(3, 2, 1, 0.5)\n # Test that the activation function is a sigmoid\n self.assertTrue(np.all(network.activation_function(0.5) == 1/(1+np.exp(-0.5))))\n\n def test_train(self):\n # Test that weights are updated correctly on training\n network = NeuralNetwork(3, 2, 1, 0.5)\n network.weights_input_to_hidden = test_w_i_h.copy()\n network.weights_hidden_to_output = test_w_h_o.copy()\n \n network.train(inputs, targets)\n print(network.weights_hidden_to_output)\n self.assertTrue(np.allclose(network.weights_hidden_to_output, \n np.array([[ 0.37275328], \n [-0.03172939]])))\n self.assertTrue(np.allclose(network.weights_input_to_hidden,\n np.array([[ 0.10562014, -0.20185996], \n [0.39775194, 0.50074398], \n [-0.29887597, 0.19962801]])))\n\n def runTest(self):\n # Test correctness of run method\n network = NeuralNetwork(3, 2, 1, 0.5)\n network.weights_input_to_hidden = test_w_i_h.copy()\n network.weights_hidden_to_output = test_w_h_o.copy()\n\n self.assertTrue(np.allclose(network.run(inputs), 0.09998924))\n\nsuite = unittest.TestLoader().loadTestsFromModule(TestMethods())\nunittest.TextTestRunner().run(suite)",
"...."
]
],
[
[
"## Training the network\n\nHere you'll set the hyperparameters for the network. The strategy here is to find hyperparameters such that the error on the training set is low, but you're not overfitting to the data. If you train the network too long or have too many hidden nodes, it can become overly specific to the training set and will fail to generalize to the validation set. That is, the loss on the validation set will start increasing as the training set loss drops.\n\nYou'll also be using a method know as Stochastic Gradient Descent (SGD) to train the network. The idea is that for each training pass, you grab a random sample of the data instead of using the whole data set. You use many more training passes than with normal gradient descent, but each pass is much faster. This ends up training the network more efficiently. You'll learn more about SGD later.\n\n### Choose the number of iterations\nThis is the number of batches of samples from the training data we'll use to train the network. The more iterations you use, the better the model will fit the data. However, this process can have sharply diminishing returns and can waste computational resources if you use too many iterations. You want to find a number here where the network has a low training loss, and the validation loss is at a minimum. The ideal number of iterations would be a level that stops shortly after the validation loss is no longer decreasing.\n\n### Choose the learning rate\nThis scales the size of weight updates. If this is too big, the weights tend to explode and the network fails to fit the data. Normally a good choice to start at is 0.1; however, if you effectively divide the learning rate by n_records, try starting out with a learning rate of 1. In either case, if the network has problems fitting the data, try reducing the learning rate. Note that the lower the learning rate, the smaller the steps are in the weight updates and the longer it takes for the neural network to converge.\n\n### Choose the number of hidden nodes\nIn a model where all the weights are optimized, the more hidden nodes you have, the more accurate the predictions of the model will be. (A fully optimized model could have weights of zero, after all.) However, the more hidden nodes you have, the harder it will be to optimize the weights of the model, and the more likely it will be that suboptimal weights will lead to overfitting. With overfitting, the model will memorize the training data instead of learning the true pattern, and won't generalize well to unseen data. \n\nTry a few different numbers and see how it affects the performance. You can look at the losses dictionary for a metric of the network performance. If the number of hidden units is too low, then the model won't have enough space to learn and if it is too high there are too many options for the direction that the learning can take. The trick here is to find the right balance in number of hidden units you choose. You'll generally find that the best number of hidden nodes to use ends up being between the number of input and output nodes.",
"_____no_output_____"
]
],
[
[
"import sys\n\n####################\n### Set the hyperparameters in you myanswers.py file ###\n####################\n\nfrom my_answers import iterations, learning_rate, hidden_nodes, output_nodes\n\n\nN_i = train_features.shape[1]\nnetwork = NeuralNetwork(N_i, hidden_nodes, output_nodes, learning_rate)\n\nlosses = {'train':[], 'validation':[]}\nfor ii in range(iterations):\n # Go through a random batch of 128 records from the training data set\n batch = np.random.choice(train_features.index, size=128)\n X, y = train_features.ix[batch].values, train_targets.ix[batch]['cnt']\n \n network.train(X, y)\n \n # Printing out the training progress\n train_loss = MSE(network.run(train_features).T, train_targets['cnt'].values)\n val_loss = MSE(network.run(val_features).T, val_targets['cnt'].values)\n sys.stdout.write(\"\\rProgress: {:2.1f}\".format(100 * ii/float(iterations)) \\\n + \"% ... Training loss: \" + str(train_loss)[:5] \\\n + \" ... Validation loss: \" + str(val_loss)[:5])\n sys.stdout.flush()\n \n losses['train'].append(train_loss)\n losses['validation'].append(val_loss)",
"Progress: 0.3% ... Training loss: 0.907 ... Validation loss: 1.386"
],
[
"plt.plot(losses['train'], label='Training loss')\nplt.plot(losses['validation'], label='Validation loss')\nplt.legend()\n_ = plt.ylim()",
"_____no_output_____"
]
],
[
[
"## Check out your predictions\n\nHere, use the test data to view how well your network is modeling the data. If something is completely wrong here, make sure each step in your network is implemented correctly.",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(8,4))\n\nmean, std = scaled_features['cnt']\npredictions = network.run(test_features).T*std + mean\nax.plot(predictions[0], label='Prediction')\nax.plot((test_targets['cnt']*std + mean).values, label='Data')\nprint(np.average(np.absolute((test_targets['cnt']*std + mean).values - predictions[0])))\nax.set_xlim(right=len(predictions))\nax.legend()\n\ndates = pd.to_datetime(rides.ix[test_data.index]['dteday'])\ndates = dates.apply(lambda d: d.strftime('%b %d'))\nax.set_xticks(np.arange(len(dates))[12::24])\n_ = ax.set_xticklabels(dates[12::24], rotation=45)",
"51.86683374276577\n"
]
],
[
[
"## OPTIONAL: Thinking about your results(this question will not be evaluated in the rubric).\n \nAnswer these questions about your results. How well does the model predict the data? Where does it fail? Why does it fail where it does?\n\n> **Note:** You can edit the text in this cell by double clicking on it. When you want to render the text, press control + enter\n\n#### Your answer below\n\nFor normal days, it seems to predict pretty well. It follows the graph very well between peaks and valleys and is usually within 50 - 100 bikes or so. It is interesting that it predicts less than 0 bikes on occasion. It seems to fail at predicting the holidays, and I would expect that to be due to the lack of training data for this period of time.",
"_____no_output_____"
],
[
"## Submitting:\nOpen up the 'jwt' file in the first-neural-network directory (which also contains this notebook) for submission instructions",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a15f5ffe068e6a0b885950aed4bf719f2299713
| 92,066 |
ipynb
|
Jupyter Notebook
|
notebooks/Housing.ipynb
|
ystkmeg/handson-ml-practice
|
5042d9f260bfd3c0079c8a609f34cda81ff04fff
|
[
"Apache-2.0"
] | null | null | null |
notebooks/Housing.ipynb
|
ystkmeg/handson-ml-practice
|
5042d9f260bfd3c0079c8a609f34cda81ff04fff
|
[
"Apache-2.0"
] | null | null | null |
notebooks/Housing.ipynb
|
ystkmeg/handson-ml-practice
|
5042d9f260bfd3c0079c8a609f34cda81ff04fff
|
[
"Apache-2.0"
] | null | null | null | 107.805621 | 58,236 | 0.839984 |
[
[
[
"## データの構造をざっと見てみる",
"_____no_output_____"
]
],
[
[
"import os\n\nHOUSING_PATH = os.path.join('/src/datasets', 'housing')",
"_____no_output_____"
],
[
"import pandas as pd\n\ndef load_housing_data(housing_path=HOUSING_PATH):\n csv_path = os.path.join(housing_path, 'housing.csv')\n return pd.read_csv(csv_path)",
"_____no_output_____"
],
[
"housing = load_housing_data() # csvファイルを読み込む",
"_____no_output_____"
],
[
"housing.head() # headで最初の5行取得",
"_____no_output_____"
],
[
"# longiture=経度\n# latitude=緯度\n# housing_median_age=築年数の中央値\n# total_rooms=部屋数\n# total_bedrooms=寝室数\n# population=人口\n# households=世帯数\n# median_income=収入の中央値\n# median_house_value=住宅価格の中央値\n# ocean_proximity=海との位置関係",
"_____no_output_____"
],
[
"housing.info() # infoで総行数、各属性のタイプ、nullではない値の数などが取得できる",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 20640 entries, 0 to 20639\nData columns (total 10 columns):\nlongitude 20640 non-null float64\nlatitude 20640 non-null float64\nhousing_median_age 20640 non-null float64\ntotal_rooms 20640 non-null float64\ntotal_bedrooms 20433 non-null float64\npopulation 20640 non-null float64\nhouseholds 20640 non-null float64\nmedian_income 20640 non-null float64\nmedian_house_value 20640 non-null float64\nocean_proximity 20640 non-null object\ndtypes: float64(9), object(1)\nmemory usage: 1.6+ MB\n"
],
[
"# total_bedroomsはnullではない値が20,433個しかない\n# つまりtotal_bedroomsという特徴量(feature)を持たない区域が207あるということ",
"_____no_output_____"
],
[
"# ocean_proximityのタイプはobject\n# この場合はtextオブジェクト\n# 先頭5行の出力から、カテゴリを示すものだとわかる\n# value_counts()を使えば、どのようなカテゴリがあってそれぞれのカテゴリに何個の区域が含まれているかわかる",
"_____no_output_____"
],
[
"housing['ocean_proximity'].value_counts()",
"_____no_output_____"
],
[
"housing.describe() # describe()メソッドを使うと、数値属性の集計情報が表示される",
"_____no_output_____"
],
[
"# std(standard deviation)行は、標準偏差(値の散らばり具合)を示している\n# 標準偏差に関する詳しい説明 https://atarimae.biz/archives/5379",
"_____no_output_____"
],
[
"# 25%, 50%, 75%の各行は、パーセンタイル(percentile)を示している\n# パーセンタイルとは、観測値のグループのうち下から数えて指定された割合の観測値の値がどうなっているかを示す。\n# 例えば、下から数えて25%の区域のhousing_median_ageは18年、50%の区域では29年、75%の区域では37年となる。",
"_____no_output_____"
],
[
"# ヒストグラムをプロットすることも、データの感じをつかむためには効果的",
"_____no_output_____"
],
[
"%matplotlib inline \nimport matplotlib.pyplot as plt\nhousing.hist(bins=50, figsize=(20,15)) # bins = 表示するビン(棒)の数\nplt.show()",
"_____no_output_____"
],
[
"# 収入の中央値(median_income)はデータ収集時にスケーリングされたもの。上限15.0001,下限0.4999\n# 機械学習では、前処理済みの属性を使うのは、ごく普通のことであるため、データがどのように計算されたものなのかは理解しておいた方がよい。",
"_____no_output_____"
],
[
"# 築年数の中央値(housing_median_age)と住宅価格の中央値(median_house_value)も条件を切ってある。後者の場合、ターゲット属性なので、重大な問題になる。\n# 50万ドルを越えても正確な予測が必要な場合、以下の選択肢がある。\n# a. 上限を越えている区域の正しいラベルを集める。\n# b. 訓練セットからそれらの区域を取り除く(50万ドルを越える値を予測したときにシステムの評価が下がるので、テストセットからも取り除く)。",
"_____no_output_____"
],
[
"# ほとんどのヒストグラムがテールヘビー(tail heavy)になっている。\n# つまり、中央値の左側より右側が大きく広がっている。\n# このような形になると、一部の機械学習アルゴリズムはパターンを見つけにくくなることがある。\n# そういった属性については、ベル型分布に近づくように変換する。",
"_____no_output_____"
]
],
[
[
"## テストセットを作る",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef split_train_test(data, test_ratio):\n shuffled_indices = np.random.permutation(len(data))\n test_set_size = int(len(data) * test_ratio)\n test_indices = shuffled_indices[:test_set_size]\n train_indices = shuffled_indices[test_set_size:]\n return data.iloc[train_indices], data.iloc[test_indices] # pandas.DataFrame.ilocの説明 https://note.nkmk.me/python-pandas-at-iat-loc-iloc/",
"_____no_output_____"
],
[
"train_set, test_set = split_train_test(housing, 0.2)\nprint(len(train_set), 'train +', len(test_set), 'test')",
"16512 train + 4128 test\n"
],
[
"# これでテストセットを作ることはできるが、完璧ではない。\n# よく理解出来てない(p49~p50)",
"_____no_output_____"
],
[
"import hashlib\n\ndef test_set_check(identifier, test_ratio, hash):\n return hash(np.int64(identifier)).digest()[-1] < 256 * test_ratio\n\ndef split_train_test_by_id(data, test_ratio, id_column, hash=hashlib.md5):\n ids = data[id_column]\n in_test_set = ids.apply(lambda id_: test_set_check(id_, test_ratio, hash))\n return data.loc[~in_test_set], data.loc[in_test_set]",
"_____no_output_____"
],
[
"# 住宅価格データセットには、識別子の列がない。そのような場合、もっとも単純な方法は、行番号をIDにすること。",
"_____no_output_____"
],
[
"housing_with_id = housing.reset_index() # ID列の追加\ntrain_set, test_set = split_train_test_by_id(housing_with_id, 0.2, 'index')",
"_____no_output_____"
],
[
"# この場合一意の識別子を作るためにもっと安定した方法は、区域の緯度経度を組み合わせる方法。",
"_____no_output_____"
],
[
"housing_with_id['id'] = housing['longitude'] * 1000 + housing['latitude']\ntrain_set, test_set = split_train_test_by_id(housing_with_id, 0.2, 'id')",
"_____no_output_____"
],
[
"# scikit-learnには、さまざまな方法でデータセットを複数のサブセットに分割する関数がいくつもある。\n# train_set_splitは、上記split_train_testとほぼ同じことに加え、乱数生成器の種を設定するrandom_state引数や、\n# 複数のデータセットに同じ行番号を与え、同じインデックスでデータセットを分割する機能がある。\n# これはラベルのために別個のDataFrameがある場合などにとても役立つ。",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\ntrain_set, test_set = train_test_split(housing, test_size=0.2, random_state=42)",
"_____no_output_____"
],
[
"# これまでのものは、無作為なサンプリング方法。\n# データセットが十分大規模かつ属性数と相対的な割合ならいいけど、\n# そうでない場合、大きなサンプリングバイアスを持ち込む危険がある。",
"_____no_output_____"
],
[
"# 例えば、調査会社が1000人の人に電話をかけて質問をする時、電話帳で無作為に1000人の人々を拾い出すわけではない。\n# 米国の人口は女性51.3%, 男性48.7%の比率なので、サンプルでも同じ火率にするため、\n# 513人の女性と487人の男性に尋ねる",
"_____no_output_____"
],
[
"# これを層化抽出法(stractified sampling)という。\n# 人口全体の層(stratum)と呼ばれる同種の下位集団に分割し、\n# 各層から適切な数のインスタンスをサンプリング抽出し、テストセットが人口全体の代表になるようにする。\n# 純粋に無作為なサンプリングを使うと、12%の確率で、女性が49%より少なく、54%よりも多い歪んだ検証セットをサンプリングしてしまう。",
"_____no_output_____"
],
[
"# 今回のケースでは、収入の中央値は、住宅価格の中央値を予測する上で非常に重要な属性。\n# テストセットは、データセット全体のさまざまな収入カテゴリを代表するものにしたい\n# 収入の中央値は連続的な数値属性なので、収入カテゴリという属性を作る必要がある。",
"_____no_output_____"
],
[
"# 収入の中央値のヒストグラムから、収入の中央値の大半は2~5万ドルの周辺に集まっているが、\n# 一部の値は6万ドルを大きく越えている。\n# データセットの各層に十分な数のインスタンスがあることが重要\n# そうでなければ、層を重視することがバイアスになってしまう\n# つまり、層の数が大きくなり過ぎないようにしながら、各層は十分に大きくなければならない",
"_____no_output_____"
],
[
"# 以下のコードは、収入の中央値を1.5で割り(収入カテゴリの数を減らすため)、\n# ceilで端数を切り上げ(離散したカテゴリを作るため)て、\n# 5以上のカテゴリをすべて5にまとめるという方法で収入カテゴリを作る。",
"_____no_output_____"
],
[
"housing['income_cat'] = np.ceil(housing['median_income'] / 1.5)\nhousing['income_cat'].where(housing['income_cat'] < 5, 5.0, inplace=True)",
"_____no_output_____"
],
[
"housing['income_cat'].hist(bins=50, figsize=(8,6))\nplt.show()",
"_____no_output_____"
],
[
"# これらの収入カテゴリに基づき、scikit-learnのStratified ShuffleSplitクラスで層化抽出をする",
"_____no_output_____"
],
[
"from sklearn.model_selection import StratifiedShuffleSplit\n\nsplit = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)\nfor train_index, test_index in split.split(housing, housing['income_cat']):\n strat_train_set = housing.loc[train_index]\n strat_test_set = housing.loc[test_index]",
"_____no_output_____"
],
[
"strat_test_set['income_cat'].value_counts() / len(strat_test_set)",
"_____no_output_____"
],
[
"# 同じようなコードを使ってデータセット全体の収入カテゴリの割合を調べることができる。\n# 謎謎謎",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a15ff78eb79b5214f6d74d393528915827acb6c
| 124,089 |
ipynb
|
Jupyter Notebook
|
training/code/ml_assign.ipynb
|
Shetty073/car-price-prediction
|
443133014960e033755ffcf4ca4bc4e91f880a0e
|
[
"MIT"
] | null | null | null |
training/code/ml_assign.ipynb
|
Shetty073/car-price-prediction
|
443133014960e033755ffcf4ca4bc4e91f880a0e
|
[
"MIT"
] | null | null | null |
training/code/ml_assign.ipynb
|
Shetty073/car-price-prediction
|
443133014960e033755ffcf4ca4bc4e91f880a0e
|
[
"MIT"
] | null | null | null | 97.554245 | 18,105 | 0.703197 |
[
[
[
"from google.colab import drive\ndrive.mount('/content/drive')",
"Mounted at /content/drive\n"
],
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nimport numpy as np\nfrom sklearn import datasets, linear_model, preprocessing, model_selection, linear_model, metrics\nimport joblib\nimport pandas as pd",
"_____no_output_____"
],
[
"df = pd.read_csv('/content/drive/My Drive/car_data.csv')\ndf.head()",
"_____no_output_____"
],
[
"numerical_feature = [feature for feature in df.columns if df[feature].dtypes!=\"O\"]\nnumerical_feature",
"_____no_output_____"
],
[
"for feature in numerical_feature:\n sns.scatterplot(x = df[feature], y = df['Selling_Price'])\n plt.show()",
"_____no_output_____"
],
[
"df = df.drop(['Car_Name', 'Fuel_Type', 'Seller_Type', 'Transmission'], axis=1)\ndf",
"_____no_output_____"
],
[
"scaler = preprocessing.MinMaxScaler()\nscaler.fit(df)\ndataset=pd.DataFrame(scaler.transform(df),columns=df.columns)\ndataset.head()",
"_____no_output_____"
],
[
"X = df.drop(['Selling_Price'], axis=1)\ny = df['Selling_Price']\nX_train, X_test, y_train, y_test = model_selection.train_test_split(X, y, test_size=0.1, random_state=42)\nX_test",
"_____no_output_____"
],
[
"lr = linear_model.LinearRegression()\nlr.fit(X_train,y_train)",
"_____no_output_____"
],
[
"y_predLR = lr.predict(X_test)\nmetrics.r2_score(y_test, y_predLR)",
"_____no_output_____"
],
[
"joblib.dump(lr, '/content/drive/My Drive/car_price_lr.pkl')",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a162539fdcf874495a49a1e48ef9b78a0bd6ccf
| 21,780 |
ipynb
|
Jupyter Notebook
|
notebooks/development/preprocessing/make_grid.ipynb
|
kuechenrole/tidal_melting
|
b71eec6aa502e1eb0570e9fc4a9d0170aa4dc24b
|
[
"MIT"
] | 2 |
2020-08-11T09:23:27.000Z
|
2021-04-25T01:30:07.000Z
|
notebooks/development/preprocessing/make_grid.ipynb
|
kuechenrole/tidal_melting
|
b71eec6aa502e1eb0570e9fc4a9d0170aa4dc24b
|
[
"MIT"
] | 4 |
2020-03-30T20:12:46.000Z
|
2021-06-01T22:02:55.000Z
|
notebooks/development/preprocessing/make_grid.ipynb
|
kuechenrole/tidal_melting
|
b71eec6aa502e1eb0570e9fc4a9d0170aa4dc24b
|
[
"MIT"
] | 2 |
2019-09-29T16:40:30.000Z
|
2021-04-25T01:30:09.000Z
| 36.728499 | 159 | 0.552158 |
[
[
[
"import numpy as np \nimport os\nimport sys\nimport xarray as xr\nimport scipy.io as sio\nimport matplotlib.pyplot as plt\nimport datetime\n\nfrom dotenv import load_dotenv, find_dotenv\n\n# find .env automagically by walking up directories until it's found\ndotenv_path = find_dotenv()\nload_dotenv(dotenv_path)\nsrc_dir = os.environ.get('srcdir')\nsys.path.append(src_dir)\n\n# always reload modules marked with \"%aimport\"\n%load_ext autoreload\n%autoreload 1\n\n%aimport features.bathy_smoothing\nfrom features.resample_roms import resample\nfrom features.bathy_smoothing import smoothing_PlusMinus_rx0,smoothing_PositiveVolume_rx0\nfrom features.cartesian_grid_2d import haversine",
"_____no_output_____"
],
[
"#run = os.environ.get('run')\nrun ='waom10'\nmr = 10 #km\nsmooth = False\ndeepen = False",
"_____no_output_____"
],
[
"#establish the grid with grid point distances of mr/2 in km\n#we need double resolution to cover all of the staggered grid points (we subset to rho, psi, u, v points later)\n#we need an extra line of u and v points at first to calculate all dx and dy on rho points\nx,y = np.meshgrid(np.arange(-3000,3300+mr/2,mr/2),np.arange(-2700,2600+mr/2,mr/2))\n#x,y = np.meshgrid(np.arange(-4300,4300+mr/2,mr/2),np.arange(-3700,3600+mr/2,mr/2))",
"_____no_output_____"
],
[
"#load south polar stereographic projection to convert from grid point distance in m to lat/lon and back\nfrom mpl_toolkits.basemap import Basemap\nm = Basemap(projection='spstere',lon_0=0,boundinglat=-50,lat_ts=-71)\n\n#get lat/lon coordinates at all grid points by shifting the grid to the lower left corner of the map\nlon,lat=m(x*1000+m.urcrnrx/2,y*1000+m.urcrnry/2,inverse=True)\n\n#calculate curvilinear coordinate distances at rho points\ndx = haversine(lon[1::2,0:-2:2],lat[1::2,0:-2:2],lon[1::2,2::2],lat[1::2,2::2])\ndy = haversine(lon[0:-2:2,1::2],lat[0:-2:2,1::2],lon[2::2,1::2],lat[2::2,1::2])\n\n#calculate curvilinear coordinate metrices\npm = 1.0/dx\npn = 1.0/dy\n \ndndx = np.empty_like(pm)\ndmde = np.empty_like(pn)\n\ndndx[:,1:-1] = 0.5*(pn[:,2:] - pn[:,:-2])\ndmde[1:-1,:] = 0.5*(pm[2:,:] - pm[:-2,:])\n\ndndx[:,0] = 2*dndx[:,1] - dndx[:,2]\ndndx[:,-1] = 2*dndx[:,-2] - dndx[:,-3]\ndmde[0,:] = 2*dmde[1,:] - dmde[2,:]\ndmde[-1,:] = 2*dmde[-2,:] - dmde[-3,:]\n\n#subset lat and lon at rho, psi, u and v points\nlon_rho = lon[1::2,1::2]\nlat_rho = lat[1::2,1::2]\n\nlon_psi = lon[2:-1:2,2:-1:2]\nlat_psi = lat[2:-1:2,2:-1:2]\n\nlon_u = lon[1::2,2:-1:2]\nlat_u = lat[1::2,2:-1:2]\n\nlon_v = lon[2:-1:2,1::2]\nlat_v = lat[2:-1:2,1::2]",
"_____no_output_____"
],
[
"#load rtopo bed and ice topography and resample to rho points\nrtopo_path = os.path.join(os.environ.get('extdir'),'rtopo','RTopo-2.0.1_30sec_*_S30.nc')\nrtopo = xr.open_mfdataset(rtopo_path,data_vars='minimal')#.sel(latdim=np.arange(0,7501,50),londim=np.arange(0,43201,100))\n\nrt_lon,rt_lat = np.meshgrid(rtopo.lon.values,rtopo.lat.values)\nbed_raw = resample(rt_lon,rt_lat,lon_rho,lat_rho,rtopo.bedrock_topography.values)\nice_raw = resample(rt_lon,rt_lat,lon_rho,lat_rho,rtopo.ice_base_topography.values)\n\n#make a copy of the raw bathymetry\nbed = bed_raw.copy()\nice = ice_raw.copy()",
"_____no_output_____"
],
[
"#set bed minimum depth to 10 cm\nbed[bed>-0.1]= -0.1\n#set ice draft at these places to zero \nice[bed>0.1] = 0.0\n#set ice mountains to zero\nice[ice>0]= 0.0\n\n#set water column thickness to a small positive value (ROMS don't like when bed = ice draft)\nwct = (-(bed-ice)).copy()\nice[wct==0] = bed[wct==0] + 0.1",
"_____no_output_____"
],
[
"#generate a land/ocean mask depending on water column thickness\n#(distance between ice and bed or sea surface and bed)\n#wct = (-(bed-ice)).copy()\nmask = np.ones_like(wct)\nmask[wct<20] = 0",
"_____no_output_____"
],
[
"#smooth=True\n#deepen=True\nif smooth:\n #if smoothing is activated smooth wct and bed and set ice draft as bed + wct\n mask = np.ones_like(wct)\n mask[wct<=0.1] = 0\n dA = 1.0/(pn*pm) \n bed = -(smoothing_PositiveVolume_rx0(mask,-bed,0.8,dA))\n wct = smoothing_PositiveVolume_rx0(mask,wct,0.8,dA)\n ice = bed + wct\n \n #update the minimum wct points as before\n bed[bed>-0.1]= -0.1\n ice[bed>0.1] = 0.0\n ice[ice>0]= 0.0\n wct = (-(bed-ice)).copy()\n ice[wct==0] = bed[wct==0] + 0.1\n \n #update the mask\n wct = (-(bed-ice)).copy()\n mask = np.ones_like(wct)\n mask[wct<20] = 0\n \n#if deepening is activated, deepen the bed to a minimum water column thickness of 50m \nif deepen:\n shallow = (wct<50)&(wct>=20)\n bed[shallow] = ice[shallow]-50.0",
"_____no_output_____"
],
[
"#set spherical flag to 1, since we're creating a curvilinear spherical grid\nspherical_da = xr.DataArray(int(1),name='spherical',attrs={'flag_meanings': 'Cartesian spherical',\n 'flag_values': np.array([0, 1], dtype=int),\n 'long_name': 'grid type logical switch'})\n\nxl = mr*np.size(lat_rho,1)*1000\nxl_da = xr.DataArray(xl,name='xl',attrs={'long_name': 'basin length in the XI-direction', 'units': 'meter'} )\nel = mr*np.size(lon_rho,0)*1000\nel_da = xr.DataArray(el,name='el',attrs={'long_name': 'basin length in the ETA-direction', 'units': 'meter'} )\n\nangle = lon_rho/180.0*np.pi\nangle_da = xr.DataArray(angle,name='angle',dims=['eta_rho','xi_rho'],attrs={'long_name': 'angle between XI-axis and EAST', 'units': 'radians'})\n\npn_da = xr.DataArray(pn,name=\"pn\",dims=['eta_rho','xi_rho'],attrs={'long_name': 'curvilinear coordinate metric in ETA', 'units': 'meter-1'})\npm_da = xr.DataArray(pm,name='pm',dims=['eta_rho','xi_rho'],attrs={'long_name': 'curvilinear coordinate metric in XI', 'units': 'meter-1'})\n\ndmde_da = xr.DataArray(dmde,name='dmde',dims=['eta_rho','xi_rho'],attrs={'long_name': 'ETA-derivative of inverse metric factor pm', 'units': 'meter'})\ndndx_da = xr.DataArray(dndx,name='dndx',dims=['eta_rho','xi_rho'],attrs={'long_name': 'XI-derivative of inverse metric factor nm', 'units': 'meter'})\n\nf = 2*7.29e-5*np.sin(lat_rho*np.pi/180)\nf_da = xr.DataArray(f,name='f',dims=['eta_rho','xi_rho'],attrs={'long_name': 'Coriolis parameter at RHO-points', 'units': 'second-1'})\n\nh_da = xr.DataArray(-bed,name='h',dims=['eta_rho','xi_rho'],attrs={'long_name': 'model bathymetry at RHO-points', 'units': 'meter'})\nhraw_da = xr.DataArray(-bed_raw,name='hraw',dims=['eta_rho','xi_rho'],attrs={'long_name': 'Working bathymetry at RHO-points', 'units': 'meter'})\n\nzice_da = xr.DataArray(ice,name='zice',dims=['eta_rho','xi_rho'],attrs={'long_name': 'model ice draft at RHO-points', 'units': 'meter'})\n\nlon_rho_da = xr.DataArray(lon_rho,name='lon_rho',dims=['eta_rho','xi_rho'],attrs={'long_name': 'longitude of RHO-points',\n 'standard_name': 'longitude',\n 'units': 'degree_east'})\nlat_rho_da = xr.DataArray(lat_rho,name='lat_rho',dims=['eta_rho','xi_rho'],attrs={'long_name': 'latitude of RHO-points',\n 'standard_name': 'latitude',\n 'units': 'degree_north'})\nlon_psi_da = xr.DataArray(lon_psi,name='lon_psi',dims=['eta_psi','xi_psi'],attrs={'long_name': 'longitude of psi-points',\n 'standard_name': 'longitude',\n 'units': 'degree_east'})\nlat_psi_da = xr.DataArray(lat_psi,name='lat_psi',dims=['eta_psi','xi_psi'],attrs={'long_name': 'latitude of psi-points',\n 'standard_name': 'latitude',\n 'units': 'degree_north'})\nlon_u_da = xr.DataArray(lon_u,name='lon_u',dims=['eta_u','xi_u'],attrs={'long_name': 'longitude of u-points',\n 'standard_name': 'longitude',\n 'units': 'degree_east'})\nlat_u_da = xr.DataArray(lat_u,name='lat_u',dims=['eta_u','xi_u'],attrs={'long_name': 'latitude of u-points',\n 'standard_name': 'latitude',\n 'units': 'degree_north'})\nlon_v_da = xr.DataArray(lon_v,name='lon_v',dims=['eta_v','xi_v'],attrs={'long_name': 'longitude of v-points',\n 'standard_name': 'longitude',\n 'units': 'degree_east'})\nlat_v_da = xr.DataArray(lat_v,name='lat_v',dims=['eta_v','xi_v'],attrs={'long_name': 'latitude of v-points',\n 'standard_name': 'latitude',\n 'units': 'degree_north'})\n\nfrom features.mask_roms_uvp import uvp_masks\n\nmask_rho = mask.copy()\nmask_u,mask_v,mask_psi = uvp_masks(mask_rho)\n\nmask_rho_da = xr.DataArray(mask_rho,name='mask_rho',dims=['eta_rho','xi_rho'],attrs={'flag_meanings': 'land water',\n 'flag_values': np.array([ 0., 1.]),\n 'long_name': 'mask on RHO-points'})\nmask_psi_da = xr.DataArray(mask_psi,name='mask_psi',dims=['eta_psi','xi_psi'],attrs={'flag_meanings': 'land water',\n 'flag_values': np.array([ 0., 1.]),\n 'long_name': 'mask on psi-points'})\nmask_u_da = xr.DataArray(mask_u,name='mask_u',dims=['eta_u','xi_u'],attrs={'flag_meanings': 'land water',\n 'flag_values': np.array([ 0., 1.]),\n 'long_name': 'mask on u-points'})\nmask_v_da = xr.DataArray(mask_v,name='mask_v',dims=['eta_v','xi_v'],attrs={'flag_meanings': 'land water',\n 'flag_values': np.array([ 0., 1.]),\n 'long_name': 'mask on v-points'})\n\ngrd = xr.Dataset({'spherical':spherical_da,\n 'xl':xl_da,\n 'el':el_da,\n 'angle':angle_da,\n 'pm':pn_da,\n 'pn':pn_da,\n 'dndx':dndx_da,\n 'dmde':dmde_da,\n 'f':f_da,\n 'h':h_da,\n 'hraw':hraw_da,\n 'zice':zice_da,\n 'lon_rho':lon_rho_da,\n 'lat_rho':lat_rho_da,\n 'lon_psi':lon_psi_da,\n 'lat_psi':lat_psi_da,\n 'lon_u':lon_u_da,\n 'lat_u':lat_u_da,\n 'lon_v':lon_v_da,\n 'lat_v':lat_v_da,\n 'mask_rho':mask_rho_da,\n 'mask_psi':mask_psi_da,\n 'mask_u':mask_u_da,\n 'mask_v':mask_v_da,},\n attrs={'history': 'GRID file using make_grid.py, smoothing='+str(smooth)+\n ', deepening='+str(deepen)+', '+str(datetime.date.today()),\n 'type': 'ROMS grid file'})",
"_____no_output_____"
],
[
"out_path = os.path.join(os.environ.get('intdir'),'waom'+str(mr)+'_grd_raw.nc')\n#out_path = '~/raijin/short/m68/oxr581/waom10_test/waom10_grd_smooth.nc'\ngrd.to_netcdf(out_path,unlimited_dims='bath')",
"_____no_output_____"
]
],
[
[
"Below just left overs from development",
"_____no_output_____"
]
],
[
[
"#include masks psi u v in smoothing and deepening",
"_____no_output_____"
],
[
"x_earth = np.empty((rows,columns))\ny_earth = np.empty((rows,columns))\n\nx_earth[:,x_0ind] = 0\ny_earth[y_0ind,:] = 0\n\nfor column in np.arange(columns-x_0ind-1):\n dx = haversine(lon[:,x_0ind+column],lat[:,x_0ind+column],lon[:,x_0ind+column+1],lat[:,x_0ind+column+1])\n x_earth[:,x_0ind+column+1] = x_earth[:,x_0ind+column]+dx\n \nfor column in np.arange(x_0ind): \n dx = haversine(lon[:,x_0ind-column],lat[:,x_0ind-column],lon[:,x_0ind-column-1],lat[:,x_0ind-column-1])\n x_earth[:,x_0ind-column-1] = x_earth[:,x_0ind-column]-dist\n \nfor row in np.arange(rows-y_0ind-1): \n dy = haversine(lon[y_0ind+row,:],lat[y_0ind+row,:],lon[y_0ind+row+1,:],lat[y_0ind+row+1,:])\n y_earth[y_0ind+row+1,:] = y_earth[y_0ind+row,:]+dy\n\nfor row in np.arange(y_0ind):\n dy = haversine(lon[y_0ind-row,:],lat[y_0ind-row,:],lon[y_0ind-row-1,:],lat[y_0ind-row-1,:])\n y_earth[y_0ind-row-1,:] = y_earth[y_0ind-row,:]-dy\n ",
"_____no_output_____"
],
[
"def calc_angle(lon_u,lat_u,lon_v,lat_v):\n \n DTOR = np.pi/180\n \n a1 = lat_u[1:,:] - lat_u[:-1,:]\n a2 = lon_u[1:,:] - lon_u[:-1,:]\n a2[a2<-180]+=360\n a2[a2>180]-=360\n \n a2 = a2*np.cos(0.5*DTOR*(lat_u[1:,:]+lat_u[:-1,:]))\n angle = np.arctan2(a1,a2)\n \n a1 = lat_v[:,:-1] - lat_v[:,1:]\n a2 = lon_v[:,:-1] - lon_v[:,1:]\n a2[a2<-180]+=360\n a2[a2>180]-=360\n \n a2 = a2*np.cos(0.5*DTOR*(lat_v[:,:-1] + lat_v[:,1:]))\n angle = 0.5*(angle + np.arctan2(a2,-a1)) \n \n return angle\n\nangle = calc_angle(lon_u,lat_u,lon_v,lat_v)\n\nprint(np.shape(angle))\nplt.close()\nplt.contour(angle)\nplt.colorbar()\nplt.show()",
"_____no_output_____"
],
[
"x_rho_da = xr.DataArray(x_rho*1000,name='x_rho',dims=['eta_rho','xi_rho'],attrs={'long_name': 'X-location of RHO-points', 'units': 'meter'})\ny_rho_da = xr.DataArray(y_rho*1000,name='y_rho',dims=['eta_rho','xi_rho'],attrs={'long_name': 'Y-location of RHO-points', 'units': 'meter'})",
"_____no_output_____"
],
[
"x_psi_da = xr.DataArray(x_psi*1000,name='x_psi',dims=['eta_rho','xi_rho'],attrs={'long_name': 'X-location of PSI-points', 'units': 'meter'})\ny_psi_da = xr.DataArray(y_psi*1000,name='y_psi',dims=['eta_psi','xi_psi'],attrs={'long_name': 'Y-location of PSI-points', 'units': 'meter'})",
"_____no_output_____"
],
[
"x_u_da = xr.DataArray(x_u*1000,name='x_u',dims=['eta_u','xi_u'],attrs={'long_name': 'X-location of U-points', 'units': 'meter'})\ny_u_da = xr.DataArray(y_u*1000,name='y_u',dims=['eta_u','xi_u'],attrs={'long_name': 'Y-location of U-points', 'units': 'meter'})",
"_____no_output_____"
],
[
"x_v_da = xr.DataArray(x_v*1000,name='x_v',dims=['eta_v','xi_v'],attrs={'long_name': 'X-location of V-points', 'units': 'meter'})\ny_v_da = xr.DataArray(y_v*1000,name='y_v',dims=['eta_v','xi_v'],attrs={'long_name': 'Y-location of V-points', 'units': 'meter'})",
"_____no_output_____"
],
[
"%matplotlib notebook\n#set up mask_rho depending on water column thickness\nwct = (bed+ice).copy()\nmask_rho = np.ones_like(wct)\nmask_rho[wct<20] = 0\n\ndef make_neibhour_mask(mask):\n neib_xi = np.empty_like(mask)\n neib_eta = np.empty_like(mask)\n\n neib_xi[:,1:-1] = mask[:,2:]+mask[:,:-2]\n neib_xi[:,0] = neib_xi[:,1]\n neib_xi[:,-1] = neib_xi[:,-2]\n\n neib_eta[1:-1,:] = mask[2:,:]+mask[:-2,:]\n neib_eta[0,:] = neib_eta[1,:]\n neib_eta[-1,:] = neib_eta[-2,:]\n\n return neib_eta + neib_xi",
"_____no_output_____"
],
[
"#first get rid of any dubious ocean point nort of 60S\nmask_sa = mask_rho.copy()\nmask_sa[(lat_rho>-60)&(neib<=1)] = 0\n\n#neib = make_neibhour_mask(mask_sa)\n#mask_sa[(lat_rho>-60)&(neib<=3)] = 0\n\nplt.close()\nplt.pcolormesh(mask_sa)\nplt.colorbar()\nplt.show()",
"_____no_output_____"
],
[
"intdirintdirplt.close()\nplt.pcolormesh(neib)\nplt.colorbar()\nplt.show()",
"_____no_output_____"
],
[
"#then, more carefully, find and mask ocean point around antarctica that are sorounded by land\nNmodif=1\nwhile Nmodif > 0:\n mask_new = mask_tmp.copy()\n neib = make_neibhour_mask(mask_tmp)\n mask_new[(lat_rho<=-60)&(neib<=1)] = 0\n Nmodif = np.sum(mask_tmp)-np.sum(mask_new)\n print(Nmodif)\n mask_tmp = mask_new.copy()\n\nmask_tmp[(lat_rho<=-60)&(neib<=2)] =0\n\nNmodif=1\nwhile Nmodif > 0:\n mask_new = mask_tmp.copy()\n neib = make_neibhour_mask(mask_tmp)\n mask_new[(lat_rho<=-60)&(neib<=1)] = 0\n Nmodif = np.sum(mask_tmp)-np.sum(mask_new)\n print(Nmodif)\n mask_tmp = mask_new.copy()\nplt.close()\nplt.pcolormesh(mask_rho)\nplt.colorbar()\nplt.show()",
"_____no_output_____"
],
[
"%matplotlib notebook\n",
"_____no_output_____"
],
[
"#include the amundsen fix from Milan 2017\n\n#first load and resample to whole roms rho grid\nimport scipy.io as sio\namundsen_fix_path = os.path.join(os.environ.get('extdir'),'rtopo','Bathymetry_ASE_Millan_et_al_2017.nc')\namu_fix = xr.open_dataset(amundsen_fix_path)\namundsen_fix_latlon_path = os.path.join(os.environ.get('extdir'),'rtopo','lon_lat_romain_grid.mat')\namu_fix_latlon = sio.loadmat(amundsen_fix_latlon_path)\n\namu_lon = amu_fix_latlon['lone']\namu_lat = amu_fix_latlon['late']\namu_lon[amu_lon>180]-=360.0\namu = resample(amu_lon,amu_lat,lon_rho,lat_rho,amu_fix.BED.values)\n\n#then lookup indices of coorner coordinates and subset the working bathymetry just there \nfrom scipy.spatial import KDTree\n\npoints = np.column_stack((lon_rho.flatten(),lat_rho.flatten()))\n\ntree = KDTree(points)\n\ntarget = np.column_stack((amu_lon[0,0],amu_lat[0,0]))\nllcDist, llcInd = tree.query(target)\nllcInd = np.squeeze(llcInd)\n\ntarget = np.column_stack((amu_lon[-1,-1],amu_lat[-1,-1]))\nurcDist, urcInd = tree.query(target)\nurcInd = np.squeeze(urcInd)\n\n#establish grid coordinates at rho points only (needed for the amundsen fix)\nxi_rho = np.arange((np.size(x,1)-1)/2,dtype=int)\neta_rho = np.arange((np.size(x,0)-1)/2,dtype=int)\n\nxi_2d,eta_2d = np.meshgrid(xi_rho,eta_rho)\nxiEta = np.column_stack((xi_2d.flatten(),eta_2d.flatten()))\n\nxi_amu_ll,xi_amu_ur = xiEta[llcInd][0],xiEta[urcInd][0]\neta_amu_ll,eta_amu_ur = xiEta[llcInd][1],xiEta[urcInd][1]\n\nbed[eta_amu_ll:eta_amu_ur+1,xi_amu_ll:xi_amu_ur+1] = amu[eta_amu_ll:eta_amu_ur+1,xi_amu_ll:xi_amu_ur+1]\nbed=bed*-1",
"_____no_output_____"
],
[
"#have a look at the amundsen patch\nplt.close()\nplt.pcolormesh(bed[eta_amu_ll-20:eta_amu_ur+21,xi_amu_ll-20:xi_amu_ur+21])\nplt.colorbar()\nplt.show()",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"raw"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw",
"raw"
]
] |
4a162fa17c63f6705346092c3694d8ae3572e3f1
| 258,875 |
ipynb
|
Jupyter Notebook
|
Liver Segmentation.ipynb
|
jon-young/medicalimage
|
ef8dd3b7c65ab501d1d44ba9ee8f2ad442d42820
|
[
"MIT"
] | 6 |
2017-04-05T13:39:55.000Z
|
2021-12-27T21:47:14.000Z
|
Liver Segmentation.ipynb
|
jon-young/medicalimage
|
ef8dd3b7c65ab501d1d44ba9ee8f2ad442d42820
|
[
"MIT"
] | 1 |
2017-04-06T06:06:05.000Z
|
2017-04-06T06:06:05.000Z
|
Liver Segmentation.ipynb
|
jon-young/medicalimage
|
ef8dd3b7c65ab501d1d44ba9ee8f2ad442d42820
|
[
"MIT"
] | 6 |
2016-03-17T03:04:28.000Z
|
2019-10-21T08:28:43.000Z
| 324.404762 | 89,936 | 0.925987 |
[
[
[
"import matplotlib",
"_____no_output_____"
],
[
"matplotlib.use('Agg')",
"_____no_output_____"
],
[
"%matplotlib qt",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"import os",
"_____no_output_____"
],
[
"import SimpleITK as sitk",
"_____no_output_____"
],
[
"from os.path import expanduser, join",
"_____no_output_____"
],
[
"from scipy.spatial.distance import euclidean",
"_____no_output_____"
],
[
"os.chdir(join(expanduser('~'), 'Medical Imaging'))",
"_____no_output_____"
],
[
"import liversegmentation",
"_____no_output_____"
]
],
[
[
"---",
"_____no_output_____"
],
[
"# Read in DICOM images",
"_____no_output_____"
]
],
[
[
"sliceNum = 42",
"_____no_output_____"
],
[
"dicomPath = join(expanduser('~'), 'Documents', 'SlicerDICOMDatabase', 'TCIALocal', '0', 'images', '')\nreader = sitk.ImageSeriesReader()\nseriesIDread = reader.GetGDCMSeriesIDs(dicomPath)[1]\ndicomFilenames = reader.GetGDCMSeriesFileNames(dicomPath, seriesIDread)\nreader.SetFileNames(dicomFilenames)\nimgSeries = reader.Execute()\nimgSlice = imgSeries[:,:,sliceNum]",
"_____no_output_____"
]
],
[
[
"Note that the TCGA-BC-4073 patient has 2 series of images (series 9 & 10). The series IDs are:",
"_____no_output_____"
]
],
[
[
"reader.GetGDCMSeriesIDs(dicomPath)",
"_____no_output_____"
]
],
[
[
"By comparing images between OsiriX and plots of the SimpleITK images, the 2<sup>nd</sup> tuple element corresponds to series 9. ",
"_____no_output_____"
]
],
[
[
"liversegmentation.sitk_show(imgSlice)",
"_____no_output_____"
]
],
[
[
"Cast original slice to unsigned 8-bit integer so that segmentations can be overlaid on top",
"_____no_output_____"
]
],
[
[
"imgSliceUInt8 = sitk.Cast(sitk.RescaleIntensity(imgSlice), sitk.sitkUInt8)",
"_____no_output_____"
]
],
[
[
"# Filtering",
"_____no_output_____"
],
[
"## Curvature anisotropic diffusion",
"_____no_output_____"
]
],
[
[
"anisoParams = (0.06, 9.0, 5)",
"_____no_output_____"
],
[
"imgFilter = liversegmentation.anisotropic_diffusion(imgSlice, *anisoParams)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(imgFilter)",
"_____no_output_____"
]
],
[
[
"## Median filter",
"_____no_output_____"
]
],
[
[
"med = sitk.MedianImageFilter()\nmed.SetRadius(3)\nimgFilter = med.Execute(imgSlice)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(imgFilter)",
"_____no_output_____"
]
],
[
[
"# Edge potential",
"_____no_output_____"
],
[
"## Gradient magnitude recursive Gaussian",
"_____no_output_____"
]
],
[
[
"#sigma = 3.0\nsigma = 1.0\nimgGauss = liversegmentation.gradient_magnitude(imgFilter, sigma)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(imgGauss)",
"_____no_output_____"
]
],
[
[
"# Feature Image",
"_____no_output_____"
],
[
"## Sigmoid mapping",
"_____no_output_____"
]
],
[
[
"#K1, K2 = 20.0, 6.0\n#K1, K2 = 14.0, 4.0\nK1, K2 = 8.0, 2.0",
"_____no_output_____"
],
[
"imgSigmoid = liversegmentation.sigmoid_filter(imgGauss, K1, K2)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(imgSigmoid)",
"_____no_output_____"
]
],
[
[
"# Input level set",
"_____no_output_____"
],
[
"Create 2 lists, one to hold the seed coordinates and the other for the radii. The radius in the 1<sup>st</sup> index corresponds to the 1<sup>st</sup> index, and so on. ",
"_____no_output_____"
]
],
[
[
"coords = [(118, 286), (135, 254), (202, 75), (169, 89), (145, 209), (142, 147), (252, 58), (205, 119)]",
"_____no_output_____"
],
[
"radii = [10, 10, 10, 10, 10, 10, 5, 5]",
"_____no_output_____"
],
[
"seed2radius = {tuple(reversed(p[0])): p[1] for p in zip(coords, radii)}",
"_____no_output_____"
],
[
"initImg = liversegmentation.input_level_set(imgSigmoid, seed2radius)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(initImg)",
"_____no_output_____"
]
],
[
[
"Creating new level set from segmentation of downsampled image.\n\nFirst convert the segmentation result into a workable format:",
"_____no_output_____"
]
],
[
[
"binaryThresh = sitk.BinaryThresholdImageFilter()\nbinaryThresh.SetLowerThreshold(-2.3438)\nbinaryThresh.SetUpperThreshold(0.0)\nbinaryThresh.SetInsideValue(1)\nbinaryThresh.SetOutsideValue(0)\nbinaryImg = binaryThresh.Execute(imgGac2)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(binaryImg)",
"_____no_output_____"
]
],
[
[
"Add in new seeds:",
"_____no_output_____"
]
],
[
[
"coords2 = [(235, 108), (199, 188), (120, 113), (96, 140)]\nradii2 = [5, 5, 5, 5]",
"_____no_output_____"
],
[
"seed2radius2 = {tuple(reversed(p[0])): p[1] for p in zip(coords2, radii2)}",
"_____no_output_____"
]
],
[
[
"Now create new level set image:",
"_____no_output_____"
]
],
[
[
"X_1 = sitk.GetArrayFromImage(binaryImg)\n\n# create a 2nd seed matrix from the 2nd set of coordinates\nsetupImg = sitk.Image(imgSigmoid.GetSize()[0], imgSigmoid.GetSize()[1], sitk.sitkUInt8)\nX_2 = sitk.GetArrayFromImage(setupImg)\nfor i in range(X_2.shape[0]):\n for j in range(X_2.shape[1]):\n for s in seed2radius2.keys():\n if euclidean((i,j), s) <= seed2radius2[s]:\n X_2[i,j] = 1\n\nX = X_1.astype(bool) + X_2.astype(bool)\ninitImg2 = sitk.Cast(sitk.GetImageFromArray(X.astype(int)), imgSigmoid.GetPixelIDValue()) * -1 + 0.5\ninitImg2.SetSpacing(imgSigmoid.GetSpacing())\ninitImg2.SetOrigin(imgSigmoid.GetOrigin())\ninitImg2.SetDirection(imgSigmoid.GetDirection())",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(initImg2)",
"_____no_output_____"
]
],
[
[
"Add in a 3<sup>rd</sup> set of seeds:",
"_____no_output_____"
]
],
[
[
"coords3 = [(225, 177), (246, 114), (83, 229), (78, 208), (82, 183), (238, 126)]\nradii3 = [5, 10, 5, 5, 5, 15]",
"_____no_output_____"
],
[
"seed2radius3 = {tuple(reversed(p[0])): p[1] for p in zip(coords3, radii3)}",
"_____no_output_____"
],
[
"X_1 = sitk.GetArrayFromImage(binaryImg)\n\n# create a 3rd seed matrix from the 3rd set of coordinates\nsetupImg = sitk.Image(imgSigmoid.GetSize()[0], imgSigmoid.GetSize()[1], sitk.sitkUInt8)\nX_2 = sitk.GetArrayFromImage(setupImg)\nfor i in range(X_2.shape[0]):\n for j in range(X_2.shape[1]):\n for s in seed2radius3.keys():\n if euclidean((i,j), s) <= seed2radius3[s]:\n X_2[i,j] = 1\n\nX = X_1.astype(bool) + X_2.astype(bool)\ninitImg3 = sitk.Cast(sitk.GetImageFromArray(X.astype(int)), imgSigmoid.GetPixelIDValue()) * -1 + 0.5\ninitImg3.SetSpacing(imgSigmoid.GetSpacing())\ninitImg3.SetOrigin(imgSigmoid.GetOrigin())\ninitImg3.SetDirection(imgSigmoid.GetDirection())",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(initImg3)",
"_____no_output_____"
]
],
[
[
"# Segmentation",
"_____no_output_____"
],
[
"## Geodesic Active Contour",
"_____no_output_____"
]
],
[
[
"#gacParams = (1.0, 0.2, 4.5, 0.01, 250)\n#gacParams = (1.0, 0.2, 4.5, 0.01, 200)\ngacParams = (1.0, 0.2, 5.0, 0.01, 350)",
"_____no_output_____"
],
[
"imgGac3 = liversegmentation.geodesic_active_contour(initImg3, imgSigmoid, *gacParams)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(imgGac)",
"_____no_output_____"
]
],
[
[
"Display overlay of segmentation over original slice:",
"_____no_output_____"
]
],
[
[
"labelLowThresh = -2.3438\nlabelUpThresh = 0.0",
"_____no_output_____"
],
[
"binarySegImg3 = liversegmentation.binary_threshold(imgGac3, labelLowThresh, labelUpThresh)",
"_____no_output_____"
],
[
"liversegmentation.sitk_show(sitk.LabelOverlay(imgSliceUInt8, binarySegImg3, backgroundValue=255))",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a163d2119dc4df54819f0d7e6f25f1a4a6b92c3
| 1,922 |
ipynb
|
Jupyter Notebook
|
createResults.ipynb
|
dorazhao99/pycocoevalcap
|
8961b637334daa5a4fe8a823abfe70fca6afae85
|
[
"BSD-2-Clause-FreeBSD"
] | null | null | null |
createResults.ipynb
|
dorazhao99/pycocoevalcap
|
8961b637334daa5a4fe8a823abfe70fca6afae85
|
[
"BSD-2-Clause-FreeBSD"
] | null | null | null |
createResults.ipynb
|
dorazhao99/pycocoevalcap
|
8961b637334daa5a4fe8a823abfe70fca6afae85
|
[
"BSD-2-Clause-FreeBSD"
] | null | null | null | 21.120879 | 80 | 0.511967 |
[
[
[
"import json\nimport os",
"_____no_output_____"
],
[
"# read in the vis.json from ImageCaptioning\nwith open('./results/vis.json', 'r') as f:\n res = json.load(f)",
"_____no_output_____"
],
[
"# read in corresponding image ids\nwith open('./results/val.txt', 'rb') as filename:\n ids = filename.readlines()\n ids = [os.path.splitext(x)[0] for x in ids]\n ids = [int(os.path.basename(x.decode('utf-8').strip())) for x in ids] ",
"_____no_output_____"
],
[
"results = []\nfor index, r in enumerate(res):\n caption_dict = {}\n caption_dict['image_id'] = ids[index]\n caption_dict['caption'] = r['caption']\n results.append(caption_dict)",
"_____no_output_____"
],
[
"with open('./results/captions_val2017_res.json', 'w') as f:\n json.dump(results, f)",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a1648910a73538eb0371dec54260c8828e40264
| 30,959 |
ipynb
|
Jupyter Notebook
|
Lexical Dispersion Revamp.ipynb
|
burgamacha/lexicalDispersion
|
6d79089ad008c69904918dc4c672990eb38a1d15
|
[
"MIT"
] | null | null | null |
Lexical Dispersion Revamp.ipynb
|
burgamacha/lexicalDispersion
|
6d79089ad008c69904918dc4c672990eb38a1d15
|
[
"MIT"
] | null | null | null |
Lexical Dispersion Revamp.ipynb
|
burgamacha/lexicalDispersion
|
6d79089ad008c69904918dc4c672990eb38a1d15
|
[
"MIT"
] | null | null | null | 34.785393 | 231 | 0.41106 |
[
[
[
"# Plotting Lexical Dispersion - working with JSON reviews",
"_____no_output_____"
]
],
[
[
"import os\nimport pandas as pd\nimport json",
"_____no_output_____"
]
],
[
[
"## Data Munging",
"_____no_output_____"
]
],
[
[
"path = './data/690_webhose-2017-03_20170904112233'\ngood_review_folder = os.listdir(path)",
"_____no_output_____"
],
[
"good_reviews = []\nfor file in good_review_folder:\n with open(path + '/' +file, 'r') as json_file:\n data = json_file.readlines()\n good_reviews.append(list(map(json.loads, data))[0])\nprint(len(good_reviews))",
"9233\n"
],
[
"good_reviews[25]",
"_____no_output_____"
],
[
"keys = good_reviews[0].keys()\nkeys",
"_____no_output_____"
],
[
"for UON in good_reviews:\n for key in UON.keys():\n if key not in keys:\n keys.append(key)\n print('added ', key, ' to namespace')\n else:\n pass",
"_____no_output_____"
],
[
"columns = ['uuid', 'title', 'published', 'text']",
"_____no_output_____"
],
[
"review_df = pd.DataFrame.from_dict(good_reviews, orient = 'columns')",
"_____no_output_____"
],
[
"columns = ['uuid', 'title', 'published', 'text']\nreview_df[columns].head(3)",
"_____no_output_____"
]
],
[
[
"## Feature Engineering - True or False?",
"_____no_output_____"
]
],
[
[
"bed = \"|\".join(('pillow', 'bed', 'sheets', 'blankets', 'covers', 'comforter'))\ndesk = \"|\".join(('chair', 'desk', 'stationary', 'outlet', 'plug', 'plugs'))\nroom = \"|\".join(('carpet', 'wallpaper', 'paint', 'fridge', 'light', 'lights', 'curtain'))\nbathroom = \"|\".join(('towels', 'bath', 'tub', 'shower', 'mirror', 'toilet', 'soap'))",
"_____no_output_____"
],
[
"elements = [bed, desk, room, bathroom]\n\nfor element in elements:\n review_df[element[0:3]] = review_df['text'].str.contains(element)",
"_____no_output_____"
],
[
"review_df.rename(columns = {'pil': 'bed', 'cha': 'desk', 'car': 'room', 'tow': 'bathroom'}, inplace = True)",
"_____no_output_____"
],
[
"review_df.head(3)",
"_____no_output_____"
],
[
"print(review_df.bed.value_counts(), '\\n')\nprint(review_df.desk.value_counts(), '\\n')\nprint(review_df.room.value_counts(), '\\n')\nprint(review_df.bathroom.value_counts(), '\\n')",
"False 7052\nTrue 2181\nName: bed, dtype: int64 \n\nFalse 8192\nTrue 1041\nName: desk, dtype: int64 \n\nFalse 8073\nTrue 1160\nName: room, dtype: int64 \n\nFalse 6829\nTrue 2404\nName: bathroom, dtype: int64 \n\n"
]
],
[
[
"## Feature Engineering - List of Element Mentions",
"_____no_output_____"
]
],
[
[
"room_elements = ['pillow', 'bed', 'sheets', 'blankets', 'covers', 'comforter', \n 'chair', 'desk', 'stationary', 'outlet', 'plug', 'plugs',\n 'carpet', 'wallpaper', 'paint', 'fridge', 'light', 'lights', 'curtain',\n 'towels', 'bath', 'tub', 'shower', 'mirror', 'toilet', 'soap']\n\npunctuation = ',?!.\\/#@\"><[]'",
"_____no_output_____"
],
[
"def room_list(x):\n list_of_words = x.split(\" \")\n out_data = []\n for word in list_of_words:\n word = word.lower()\n if word.strip(punctuation) in room_elements:\n out_data.append(word.strip(punctuation))\n return (str(out_data))",
"_____no_output_____"
],
[
"review_df['room_list'] = review_df.text.apply(room_list)\nreview_df.room_list.value_counts()",
"_____no_output_____"
]
],
[
[
"## Feature Engineering - Final Element Mentions",
"_____no_output_____"
]
],
[
[
"import string\ndef room_item(x):\n list_of_words = x.split(\" \")\n for word in list_of_words:\n word = word.lower()\n if word.strip(punctuation) in room_elements:\n return word.strip(punctuation)\n else:\n pass",
"_____no_output_____"
],
[
"review_df['room_item'] = review_df.text.apply(room_item)\nreview_df.room_item.value_counts()",
"_____no_output_____"
],
[
"review_df.room_item.fillna('none', inplace = True)",
"_____no_output_____"
],
[
"review_df[['uuid', 'published', 'url', 'language', 'text', 'title', 'bed', 'desk', 'room', 'bathroom', 'room_list', 'room_item']].to_csv('good_review_lexical_dispersion.csv', sep = ',')",
"_____no_output_____"
],
[
"review_df.columns",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a16556576ff0aeec631d862f95eb361a2428343
| 499,424 |
ipynb
|
Jupyter Notebook
|
analysis/apa/train_scrambler_apa_target_bits_05.ipynb
|
willshi88/scrambler
|
fd77c05824fc99e6965d204c4f5baa1e3b0c4fb3
|
[
"MIT"
] | 19 |
2021-04-30T04:12:58.000Z
|
2022-03-07T19:09:32.000Z
|
analysis/apa/train_scrambler_apa_target_bits_05.ipynb
|
willshi88/scrambler
|
fd77c05824fc99e6965d204c4f5baa1e3b0c4fb3
|
[
"MIT"
] | 4 |
2021-07-02T15:07:27.000Z
|
2021-08-01T12:41:28.000Z
|
analysis/apa/train_scrambler_apa_target_bits_05.ipynb
|
willshi88/scrambler
|
fd77c05824fc99e6965d204c4f5baa1e3b0c4fb3
|
[
"MIT"
] | 4 |
2021-06-28T09:41:01.000Z
|
2022-02-28T09:13:29.000Z
| 259.710868 | 44,048 | 0.891885 |
[
[
[
"import keras\nfrom keras.models import Sequential, Model, load_model\n\nfrom keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda\nfrom keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, CuDNNLSTM, CuDNNGRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional\nfrom keras.layers import Concatenate, Reshape, Softmax, Conv2DTranspose, Embedding, Multiply\nfrom keras.callbacks import ModelCheckpoint, EarlyStopping, Callback\nfrom keras import regularizers\nfrom keras import backend as K\nfrom keras.utils.generic_utils import Progbar\nfrom keras.layers.merge import _Merge\nimport keras.losses\n\nfrom functools import partial\n\nfrom collections import defaultdict\n\nimport tensorflow as tf\nfrom tensorflow.python.framework import ops\n\nimport isolearn.keras as iso\n\nimport numpy as np\n\nimport tensorflow as tf\nimport logging\nlogging.getLogger('tensorflow').setLevel(logging.ERROR)\n\nimport pandas as pd\n\nimport os\nimport pickle\nimport numpy as np\n\nimport scipy.sparse as sp\nimport scipy.io as spio\n\nimport matplotlib.pyplot as plt\n\nimport isolearn.io as isoio\nimport isolearn.keras as isol\n\nfrom sequence_logo_helper import plot_dna_logo\n\nimport pandas as pd\n\nfrom keras.backend.tensorflow_backend import set_session\n\ndef contain_tf_gpu_mem_usage() :\n config = tf.ConfigProto()\n config.gpu_options.allow_growth = True\n sess = tf.Session(config=config)\n set_session(sess)\n\ncontain_tf_gpu_mem_usage()\n\nclass EpochVariableCallback(Callback) :\n \n def __init__(self, my_variable, my_func) :\n self.my_variable = my_variable \n self.my_func = my_func\n \n def on_epoch_begin(self, epoch, logs={}) :\n K.set_value(self.my_variable, self.my_func(K.get_value(self.my_variable), epoch))\n",
"Using TensorFlow backend.\n"
],
[
"#Define dataset/experiment name\ndataset_name = \"apa_doubledope\"\n\n#Load cached dataframe\ncached_dict = pickle.load(open('apa_doubledope_cached_set.pickle', 'rb'))\ndata_df = cached_dict['data_df']\n\nprint(\"len(data_df) = \" + str(len(data_df)) + \" (loaded)\")\n",
"len(data_df) = 34748 (loaded)\n"
],
[
"#Make generators\n\nvalid_set_size = 0.05\ntest_set_size = 0.05\n\nbatch_size = 32\n\n#Generate training and test set indexes\ndata_index = np.arange(len(data_df), dtype=np.int)\n\ntrain_index = data_index[:-int(len(data_df) * (valid_set_size + test_set_size))]\nvalid_index = data_index[train_index.shape[0]:-int(len(data_df) * test_set_size)]\ntest_index = data_index[train_index.shape[0] + valid_index.shape[0]:]\n\nprint('Training set size = ' + str(train_index.shape[0]))\nprint('Validation set size = ' + str(valid_index.shape[0]))\nprint('Test set size = ' + str(test_index.shape[0]))\n\n\ndata_gens = {\n gen_id : iso.DataGenerator(\n idx,\n {'df' : data_df},\n batch_size=batch_size,\n inputs = [\n {\n 'id' : 'seq',\n 'source_type' : 'dataframe',\n 'source' : 'df',\n 'extractor' : iso.SequenceExtractor('padded_seq', start_pos=180, end_pos=180 + 205),\n 'encoder' : iso.OneHotEncoder(seq_length=205),\n 'dim' : (1, 205, 4),\n 'sparsify' : False\n }\n ],\n outputs = [\n {\n 'id' : 'hairpin',\n 'source_type' : 'dataframe',\n 'source' : 'df',\n 'extractor' : lambda row, index: row['proximal_usage'],\n 'transformer' : lambda t: t,\n 'dim' : (1,),\n 'sparsify' : False\n }\n ],\n randomizers = [],\n shuffle = True if gen_id == 'train' else False\n ) for gen_id, idx in [('all', data_index), ('train', train_index), ('valid', valid_index), ('test', test_index)]\n}\n",
"Training set size = 31274\nValidation set size = 1737\nTest set size = 1737\n"
],
[
"#Load data matrices\n\nx_train = np.concatenate([data_gens['train'][i][0][0] for i in range(len(data_gens['train']))], axis=0)\nx_test = np.concatenate([data_gens['test'][i][0][0] for i in range(len(data_gens['test']))], axis=0)\n\ny_train = np.concatenate([data_gens['train'][i][1][0] for i in range(len(data_gens['train']))], axis=0)\ny_test = np.concatenate([data_gens['test'][i][1][0] for i in range(len(data_gens['test']))], axis=0)\n\nprint(\"x_train.shape = \" + str(x_train.shape))\nprint(\"x_test.shape = \" + str(x_test.shape))\n\nprint(\"y_train.shape = \" + str(y_train.shape))\nprint(\"y_test.shape = \" + str(y_test.shape))\n",
"x_train.shape = (31264, 1, 205, 4)\nx_test.shape = (1728, 1, 205, 4)\ny_train.shape = (31264, 1)\ny_test.shape = (1728, 1)\n"
],
[
"#Define sequence template (APA Doubledope sublibrary)\n\nsequence_template = 'CTTCCGATCTNNNNNNNNNNNNNNNNNNNNCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCTAC'\n\nsequence_mask = np.array([1 if sequence_template[j] == 'N' else 0 for j in range(len(sequence_template))])\n",
"_____no_output_____"
],
[
"#Visualize background sequence distribution\n\npseudo_count = 1.0\n\nx_mean = (np.sum(x_train, axis=(0, 1)) + pseudo_count) / (x_train.shape[0] + 4. * pseudo_count)\nx_mean_logits = np.log(x_mean / (1. - x_mean))\n\nplot_dna_logo(np.copy(x_mean), sequence_template=sequence_template, figsize=(14, 0.65), logo_height=1.0, plot_start=0, plot_end=205)\n",
"_____no_output_____"
],
[
"#Calculate mean training set conservation\n\nentropy = np.sum(x_mean * -np.log(x_mean), axis=-1) / np.log(2.0)\nconservation = 2.0 - entropy\n\nx_mean_conservation = np.sum(conservation) / np.sum(sequence_mask)\n\nprint(\"Mean conservation (bits) = \" + str(x_mean_conservation))\n",
"Mean conservation (bits) = 0.6565171060290858\n"
],
[
"#Calculate mean training set kl-divergence against background\n\nx_train_clipped = np.clip(np.copy(x_train[:, 0, :, :]), 1e-8, 1. - 1e-8)\n\nkl_divs = np.sum(x_train_clipped * np.log(x_train_clipped / np.tile(np.expand_dims(x_mean, axis=0), (x_train_clipped.shape[0], 1, 1))), axis=-1) / np.log(2.0)\n\nx_mean_kl_divs = np.sum(kl_divs * sequence_mask, axis=-1) / np.sum(sequence_mask)\nx_mean_kl_div = np.mean(x_mean_kl_divs)\n\nprint(\"Mean KL Div against background (bits) = \" + str(x_mean_kl_div))\n",
"Mean KL Div against background (bits) = 1.8729605733372379\n"
],
[
"from tensorflow.python.framework import ops\n\n#Stochastic Binarized Neuron helper functions (Tensorflow)\n#ST Estimator code adopted from https://r2rt.com/beyond-binary-ternary-and-one-hot-neurons.html\n#See Github https://github.com/spitis/\n\ndef st_sampled_softmax(logits):\n with ops.name_scope(\"STSampledSoftmax\") as namescope :\n nt_probs = tf.nn.softmax(logits)\n onehot_dim = logits.get_shape().as_list()[1]\n sampled_onehot = tf.one_hot(tf.squeeze(tf.multinomial(logits, 1), 1), onehot_dim, 1.0, 0.0)\n with tf.get_default_graph().gradient_override_map({'Ceil': 'Identity', 'Mul': 'STMul'}):\n return tf.ceil(sampled_onehot * nt_probs)\n\ndef st_hardmax_softmax(logits):\n with ops.name_scope(\"STHardmaxSoftmax\") as namescope :\n nt_probs = tf.nn.softmax(logits)\n onehot_dim = logits.get_shape().as_list()[1]\n sampled_onehot = tf.one_hot(tf.argmax(nt_probs, 1), onehot_dim, 1.0, 0.0)\n with tf.get_default_graph().gradient_override_map({'Ceil': 'Identity', 'Mul': 'STMul'}):\n return tf.ceil(sampled_onehot * nt_probs)\n\[email protected](\"STMul\")\ndef st_mul(op, grad):\n return [grad, grad]\n\n#Gumbel Distribution Sampler\ndef gumbel_softmax(logits, temperature=0.5) :\n gumbel_dist = tf.contrib.distributions.RelaxedOneHotCategorical(temperature, logits=logits)\n batch_dim = logits.get_shape().as_list()[0]\n onehot_dim = logits.get_shape().as_list()[1]\n return gumbel_dist.sample()\n",
"_____no_output_____"
],
[
"#PWM Masking and Sampling helper functions\n\ndef mask_pwm(inputs) :\n pwm, onehot_template, onehot_mask = inputs\n\n return pwm * onehot_mask + onehot_template\n\ndef sample_pwm_st(pwm_logits) :\n n_sequences = K.shape(pwm_logits)[0]\n seq_length = K.shape(pwm_logits)[2]\n\n flat_pwm = K.reshape(pwm_logits, (n_sequences * seq_length, 4))\n sampled_pwm = st_sampled_softmax(flat_pwm)\n\n return K.reshape(sampled_pwm, (n_sequences, 1, seq_length, 4))\n\ndef sample_pwm_gumbel(pwm_logits) :\n n_sequences = K.shape(pwm_logits)[0]\n seq_length = K.shape(pwm_logits)[2]\n\n flat_pwm = K.reshape(pwm_logits, (n_sequences * seq_length, 4))\n sampled_pwm = gumbel_softmax(flat_pwm, temperature=0.5)\n\n return K.reshape(sampled_pwm, (n_sequences, 1, seq_length, 4))\n\n#Generator helper functions\ndef initialize_sequence_templates(generator, sequence_templates, background_matrices) :\n\n embedding_templates = []\n embedding_masks = []\n embedding_backgrounds = []\n\n for k in range(len(sequence_templates)) :\n sequence_template = sequence_templates[k]\n onehot_template = iso.OneHotEncoder(seq_length=len(sequence_template))(sequence_template).reshape((1, len(sequence_template), 4))\n\n for j in range(len(sequence_template)) :\n if sequence_template[j] not in ['N', 'X'] :\n nt_ix = np.argmax(onehot_template[0, j, :])\n onehot_template[:, j, :] = -4.0\n onehot_template[:, j, nt_ix] = 10.0\n elif sequence_template[j] == 'X' :\n onehot_template[:, j, :] = -1.0\n\n onehot_mask = np.zeros((1, len(sequence_template), 4))\n for j in range(len(sequence_template)) :\n if sequence_template[j] == 'N' :\n onehot_mask[:, j, :] = 1.0\n\n embedding_templates.append(onehot_template.reshape(1, -1))\n embedding_masks.append(onehot_mask.reshape(1, -1))\n embedding_backgrounds.append(background_matrices[k].reshape(1, -1))\n\n embedding_templates = np.concatenate(embedding_templates, axis=0)\n embedding_masks = np.concatenate(embedding_masks, axis=0)\n embedding_backgrounds = np.concatenate(embedding_backgrounds, axis=0)\n\n generator.get_layer('template_dense').set_weights([embedding_templates])\n generator.get_layer('template_dense').trainable = False\n\n generator.get_layer('mask_dense').set_weights([embedding_masks])\n generator.get_layer('mask_dense').trainable = False\n \n generator.get_layer('background_dense').set_weights([embedding_backgrounds])\n generator.get_layer('background_dense').trainable = False\n\n#Generator construction function\ndef build_sampler(batch_size, seq_length, n_classes=1, n_samples=1, sample_mode='st') :\n\n #Initialize Reshape layer\n reshape_layer = Reshape((1, seq_length, 4))\n \n #Initialize background matrix\n onehot_background_dense = Embedding(n_classes, seq_length * 4, embeddings_initializer='zeros', name='background_dense')\n\n #Initialize template and mask matrices\n onehot_template_dense = Embedding(n_classes, seq_length * 4, embeddings_initializer='zeros', name='template_dense')\n onehot_mask_dense = Embedding(n_classes, seq_length * 4, embeddings_initializer='ones', name='mask_dense')\n\n #Initialize Templating and Masking Lambda layer\n masking_layer = Lambda(mask_pwm, output_shape = (1, seq_length, 4), name='masking_layer')\n background_layer = Lambda(lambda x: x[0] + x[1], name='background_layer')\n \n #Initialize PWM normalization layer\n pwm_layer = Softmax(axis=-1, name='pwm')\n \n #Initialize sampling layers\n sample_func = None\n if sample_mode == 'st' :\n sample_func = sample_pwm_st\n elif sample_mode == 'gumbel' :\n sample_func = sample_pwm_gumbel\n \n upsampling_layer = Lambda(lambda x: K.tile(x, [n_samples, 1, 1, 1]), name='upsampling_layer')\n sampling_layer = Lambda(sample_func, name='pwm_sampler')\n permute_layer = Lambda(lambda x: K.permute_dimensions(K.reshape(x, (n_samples, batch_size, 1, seq_length, 4)), (1, 0, 2, 3, 4)), name='permute_layer')\n \n def _sampler_func(class_input, raw_logits) :\n \n #Get Template and Mask\n onehot_background = reshape_layer(onehot_background_dense(class_input))\n onehot_template = reshape_layer(onehot_template_dense(class_input))\n onehot_mask = reshape_layer(onehot_mask_dense(class_input))\n \n #Add Template and Multiply Mask\n pwm_logits = masking_layer([background_layer([raw_logits, onehot_background]), onehot_template, onehot_mask])\n \n #Compute PWM (Nucleotide-wise Softmax)\n pwm = pwm_layer(pwm_logits)\n \n #Tile each PWM to sample from and create sample axis\n pwm_logits_upsampled = upsampling_layer(pwm_logits)\n sampled_pwm = sampling_layer(pwm_logits_upsampled)\n sampled_pwm = permute_layer(sampled_pwm)\n\n sampled_mask = permute_layer(upsampling_layer(onehot_mask))\n \n return pwm_logits, pwm, sampled_pwm, onehot_mask, sampled_mask\n \n return _sampler_func\n",
"_____no_output_____"
],
[
"#Scrambler network definition\n\ndef make_resblock(n_channels=64, window_size=8, dilation_rate=1, group_ix=0, layer_ix=0, drop_rate=0.0) :\n\n #Initialize res block layers\n batch_norm_0 = BatchNormalization(name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_batch_norm_0')\n\n relu_0 = Lambda(lambda x: K.relu(x, alpha=0.0))\n\n conv_0 = Conv2D(n_channels, (1, window_size), dilation_rate=dilation_rate, strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_conv_0')\n\n batch_norm_1 = BatchNormalization(name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_batch_norm_1')\n\n relu_1 = Lambda(lambda x: K.relu(x, alpha=0.0))\n\n conv_1 = Conv2D(n_channels, (1, window_size), dilation_rate=dilation_rate, strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_conv_1')\n\n skip_1 = Lambda(lambda x: x[0] + x[1], name='scrambler_resblock_' + str(group_ix) + '_' + str(layer_ix) + '_skip_1')\n\n drop_1 = None\n if drop_rate > 0.0 :\n drop_1 = Dropout(drop_rate)\n \n #Execute res block\n def _resblock_func(input_tensor) :\n batch_norm_0_out = batch_norm_0(input_tensor)\n relu_0_out = relu_0(batch_norm_0_out)\n conv_0_out = conv_0(relu_0_out)\n\n batch_norm_1_out = batch_norm_1(conv_0_out)\n relu_1_out = relu_1(batch_norm_1_out)\n \n if drop_rate > 0.0 :\n conv_1_out = drop_1(conv_1(relu_1_out))\n else :\n conv_1_out = conv_1(relu_1_out)\n\n skip_1_out = skip_1([conv_1_out, input_tensor])\n \n return skip_1_out\n\n return _resblock_func\n\ndef load_scrambler_network(n_groups=1, n_resblocks_per_group=4, n_channels=32, window_size=8, dilation_rates=[1], drop_rate=0.0) :\n\n #Discriminator network definition\n conv_0 = Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_conv_0')\n \n skip_convs = []\n resblock_groups = []\n for group_ix in range(n_groups) :\n \n skip_convs.append(Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_skip_conv_' + str(group_ix)))\n \n resblocks = []\n for layer_ix in range(n_resblocks_per_group) :\n resblocks.append(make_resblock(n_channels=n_channels, window_size=window_size, dilation_rate=dilation_rates[group_ix], group_ix=group_ix, layer_ix=layer_ix, drop_rate=drop_rate))\n \n resblock_groups.append(resblocks)\n\n last_block_conv = Conv2D(n_channels, (1, 1), strides=(1, 1), padding='same', activation='linear', kernel_initializer='glorot_normal', name='scrambler_last_block_conv')\n \n skip_add = Lambda(lambda x: x[0] + x[1], name='scrambler_skip_add')\n \n final_conv = Conv2D(1, (1, 1), strides=(1, 1), padding='same', activation='softplus', kernel_initializer='glorot_normal', name='scrambler_final_conv')\n \n onehot_to_logits = Lambda(lambda x: 2. * x - 1., name='scrambler_onehot_to_logits')\n \n scale_logits = Lambda(lambda x: K.tile(x[0], (1, 1, 1, 4)) * x[1], name='scrambler_logit_scale')\n \n def _scrambler_func(sequence_input) :\n conv_0_out = conv_0(sequence_input)\n\n #Connect group of res blocks\n output_tensor = conv_0_out\n\n #Res block group execution\n skip_conv_outs = []\n for group_ix in range(n_groups) :\n skip_conv_out = skip_convs[group_ix](output_tensor)\n skip_conv_outs.append(skip_conv_out)\n\n for layer_ix in range(n_resblocks_per_group) :\n output_tensor = resblock_groups[group_ix][layer_ix](output_tensor)\n \n #Last res block extr conv\n last_block_conv_out = last_block_conv(output_tensor)\n\n skip_add_out = last_block_conv_out\n for group_ix in range(n_groups) :\n skip_add_out = skip_add([skip_add_out, skip_conv_outs[group_ix]])\n\n #Final conv out\n final_conv_out = final_conv(skip_add_out)\n \n #Scale logits by importance scores\n scaled_logits = scale_logits([final_conv_out, onehot_to_logits(sequence_input)])\n \n return scaled_logits, final_conv_out\n\n return _scrambler_func\n",
"_____no_output_____"
],
[
"#Keras loss functions\n\ndef get_sigmoid_nll() :\n\n def _sigmoid_nll(y_true, y_pred) :\n\n y_pred = K.clip(y_pred, K.epsilon(), 1.0 - K.epsilon())\n\n return K.mean(-y_true * K.log(y_pred) - (1.0 - y_true) * K.log(1.0 - y_pred), axis=-1)\n \n return _sigmoid_nll\n\ndef get_kl_divergence() :\n\n def _kl_divergence(y_true, y_pred) :\n\n y_pred = K.clip(y_pred, K.epsilon(), 1.0 - K.epsilon())\n y_true = K.clip(y_true, K.epsilon(), 1.0 - K.epsilon())\n \n left_mean_kl = K.mean(y_true * K.log(y_true / y_pred) + (1.0 - y_true) * K.log((1.0 - y_true) / (1.0 - y_pred)), axis=-1)\n right_mean_kl = K.mean(y_pred * K.log(y_pred / y_true) + (1.0 - y_pred) * K.log((1.0 - y_pred) / (1.0 - y_true)), axis=-1)\n\n return left_mean_kl + right_mean_kl\n \n return _kl_divergence\n\ndef get_margin_entropy_ame_masked(pwm_start, pwm_end, pwm_background, max_bits=1.0) :\n \n def _margin_entropy_ame_masked(pwm, pwm_mask) :\n conservation = pwm[:, 0, pwm_start:pwm_end, :] * K.log(K.clip(pwm[:, 0, pwm_start:pwm_end, :], K.epsilon(), 1. - K.epsilon()) / K.constant(pwm_background[pwm_start:pwm_end, :])) / K.log(2.0)\n conservation = K.sum(conservation, axis=-1)\n \n mask = K.max(pwm_mask[:, 0, pwm_start:pwm_end, :], axis=-1)\n n_unmasked = K.sum(mask, axis=-1)\n \n mean_conservation = K.sum(conservation * mask, axis=-1) / n_unmasked\n\n margin_conservation = K.switch(mean_conservation > K.constant(max_bits, shape=(1,)), mean_conservation - K.constant(max_bits, shape=(1,)), K.zeros_like(mean_conservation))\n \n return margin_conservation\n \n return _margin_entropy_ame_masked\n\ndef get_target_entropy_sme_masked(pwm_start, pwm_end, pwm_background, target_bits=1.0) :\n \n def _target_entropy_sme_masked(pwm, pwm_mask) :\n conservation = pwm[:, 0, pwm_start:pwm_end, :] * K.log(K.clip(pwm[:, 0, pwm_start:pwm_end, :], K.epsilon(), 1. - K.epsilon()) / K.constant(pwm_background[pwm_start:pwm_end, :])) / K.log(2.0)\n conservation = K.sum(conservation, axis=-1)\n \n mask = K.max(pwm_mask[:, 0, pwm_start:pwm_end, :], axis=-1)\n n_unmasked = K.sum(mask, axis=-1)\n \n mean_conservation = K.sum(conservation * mask, axis=-1) / n_unmasked\n\n return (mean_conservation - target_bits)**2\n \n return _target_entropy_sme_masked\n\ndef get_weighted_loss(loss_coeff=1.) :\n \n def _min_pred(y_true, y_pred) :\n return loss_coeff * y_pred\n \n return _min_pred\n",
"_____no_output_____"
],
[
"#Initialize Encoder and Decoder networks\nbatch_size = 32\nseq_length = 205\nn_samples = 32\nsample_mode = 'st'\n#sample_mode = 'gumbel'\n\n#Resnet parameters\nresnet_n_groups = 1\nresnet_n_resblocks_per_group = 4\nresnet_n_channels = 32\nresnet_window_size = 8\nresnet_dilation_rates = [1]\nresnet_drop_rate = 0.25\n\n#Load scrambler\nscrambler = load_scrambler_network(\n n_groups=resnet_n_groups,\n n_resblocks_per_group=resnet_n_resblocks_per_group,\n n_channels=resnet_n_channels, window_size=resnet_window_size,\n dilation_rates=resnet_dilation_rates,\n drop_rate=resnet_drop_rate\n)\n\n#Load sampler\nsampler = build_sampler(batch_size, seq_length, n_classes=1, n_samples=n_samples, sample_mode=sample_mode)\n",
"_____no_output_____"
],
[
"#Load Predictor\npredictor_path = '../../../aparent/saved_models/aparent_plasmid_iso_cut_distalpas_all_libs_no_sampleweights_sgd.h5'\n\npredictor = load_model(predictor_path)\n\npredictor.trainable = False\npredictor.compile(optimizer=keras.optimizers.SGD(lr=0.1), loss='mean_squared_error')\n",
"/home/ubuntu/anaconda3/envs/tensorflow_p36/lib/python3.6/site-packages/keras/engine/saving.py:292: UserWarning: No training configuration found in save file: the model was *not* compiled. Compile it manually.\n warnings.warn('No training configuration found in save file: '\n"
],
[
"#Build scrambler model\nscrambler_class = Input(shape=(1,), name='scrambler_class')\nscrambler_input = Input(shape=(1, seq_length, 4), name='scrambler_input')\n\nscrambled_logits, importance_scores = scrambler(scrambler_input)\n\npwm_logits, pwm, sampled_pwm, _, _ = sampler(scrambler_class, scrambled_logits)\n\nscrambler_model = Model([scrambler_input, scrambler_class], [pwm_logits, pwm, sampled_pwm, importance_scores])\n\n#Initialize Sequence Templates and Masks\ninitialize_sequence_templates(scrambler_model, [sequence_template], [x_mean_logits])\n\nscrambler_model.compile(\n optimizer=keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999),\n loss='mean_squared_error'\n)\n",
"_____no_output_____"
],
[
"#Build Auto-scrambler pipeline\n\n#Define model inputs\nae_scrambler_class = Input(shape=(1,), name='ae_scrambler_class')\nae_scrambler_input = Input(shape=(1, seq_length, 4), name='ae_scrambler_input')\n\n#APARENT-specific tensors\naparent_lib = Input(shape=(13,), name='aparent_lib_input')\naparent_distal_pas = Input(shape=(1,), name='aparent_distal_pas_input')\n\n#Run encoder and decoder\n_, scrambled_pwm, scrambled_sample, pwm_mask, _ = sampler(ae_scrambler_class, scrambler(ae_scrambler_input)[0])\n\n#Define layer to deflate sample axis\ndeflate_scrambled_sample = Lambda(lambda x: K.reshape(x, (batch_size * n_samples, 1, seq_length, 4)), name='deflate_scrambled_sample')\n\n#Deflate sample axis\nscrambled_sample_deflated = deflate_scrambled_sample(scrambled_sample)\n\ndef _make_prediction(inputs, predictor=predictor) :\n pred_seq_in, pred_lib_in, pred_distal_pas_in = inputs\n \n pred_seq_in_perm = K.expand_dims(pred_seq_in[:, 0, ...], axis=-1)\n \n return predictor([pred_seq_in_perm, pred_lib_in, pred_distal_pas_in])[0]\n\ndef _make_prediction_scrambled(inputs, predictor=predictor, n_samples=n_samples) :\n pred_seq_in, pred_lib_in, pred_distal_pas_in = inputs\n \n pred_seq_in_perm = K.expand_dims(pred_seq_in[:, 0, ...], axis=-1)\n \n return predictor([pred_seq_in_perm, K.tile(pred_lib_in, (n_samples, 1)), K.tile(pred_distal_pas_in, (n_samples, 1))])[0]\n\n#Make reference prediction on non-scrambled input sequence\ny_pred_non_scrambled = Lambda(_make_prediction, name='make_prediction_non_scrambled')([ae_scrambler_input, aparent_lib, aparent_distal_pas])\n\n#Make prediction on scrambled sequence samples\ny_pred_scrambled_deflated = Lambda(_make_prediction_scrambled, name='make_prediction_scrambled')([scrambled_sample_deflated, aparent_lib, aparent_distal_pas])\n\n#Define layer to inflate sample axis\ninflate_scrambled_prediction = Lambda(lambda x: K.reshape(x, (batch_size, n_samples)), name='inflate_scrambled_prediction')\n\n#Inflate sample axis\ny_pred_scrambled = inflate_scrambled_prediction(y_pred_scrambled_deflated)\n\n#Cost function parameters\npwm_start = 10\npwm_end = 201\ntarget_bits = 0.5\n\n#NLL cost\nnll_loss_func = get_kl_divergence()\n\n#Conservation cost\nconservation_loss_func = get_target_entropy_sme_masked(pwm_start=pwm_start, pwm_end=pwm_end, pwm_background=x_mean, target_bits=1.8)\n\n#Entropy cost\nentropy_loss_func = get_target_entropy_sme_masked(pwm_start=pwm_start, pwm_end=pwm_end, pwm_background=x_mean, target_bits=target_bits)\n#entropy_loss_func = get_margin_entropy_ame_masked(pwm_start=pwm_start, pwm_end=pwm_end, pwm_background=x_mean, max_bits=target_bits)\n\n#Define annealing coefficient\nanneal_coeff = K.variable(1.0)\n\n#Execute NLL cost\nnll_loss = Lambda(lambda x: nll_loss_func(K.tile(x[0], (1, K.shape(x[1])[1])), x[1]), name='nll')([y_pred_non_scrambled, y_pred_scrambled])\n\n#Execute conservation cost\nconservation_loss = Lambda(lambda x: anneal_coeff * conservation_loss_func(x[0], x[1]), name='conservation')([scrambled_pwm, pwm_mask])\n\n#Execute entropy cost\nentropy_loss = Lambda(lambda x: (1. - anneal_coeff) * entropy_loss_func(x[0], x[1]), name='entropy')([scrambled_pwm, pwm_mask])\n\nloss_model = Model(\n [ae_scrambler_class, ae_scrambler_input, aparent_lib, aparent_distal_pas],\n [nll_loss, conservation_loss, entropy_loss]\n)\n\n#Initialize Sequence Templates and Masks\ninitialize_sequence_templates(loss_model, [sequence_template], [x_mean_logits])\n\nloss_model.compile(\n optimizer=keras.optimizers.Adam(lr=0.0001, beta_1=0.5, beta_2=0.9),\n loss={\n 'nll' : get_weighted_loss(loss_coeff=1.0),\n 'conservation' : get_weighted_loss(loss_coeff=1.0),\n 'entropy' : get_weighted_loss(loss_coeff=1.0)\n }\n)\n",
"_____no_output_____"
],
[
"scrambler_model.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nscrambler_input (InputLayer) (None, 1, 205, 4) 0 \n__________________________________________________________________________________________________\nscrambler_conv_0 (Conv2D) (None, 1, 205, 32) 160 scrambler_input[0][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_0_batch_no (None, 1, 205, 32) 128 scrambler_conv_0[0][0] \n__________________________________________________________________________________________________\nlambda_1 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_0_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_0_conv_0 ( (None, 1, 205, 32) 8224 lambda_1[0][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_0_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_0_conv_0[0][\n__________________________________________________________________________________________________\nlambda_2 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_0_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_0_conv_1 ( (None, 1, 205, 32) 8224 lambda_2[0][0] \n__________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_0_conv_1[0][\n__________________________________________________________________________________________________\nscrambler_resblock_0_0_skip_1 ( (None, 1, 205, 32) 0 dropout_1[0][0] \n scrambler_conv_0[0][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_1_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_0_skip_1[0][\n__________________________________________________________________________________________________\nlambda_3 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_1_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_1_conv_0 ( (None, 1, 205, 32) 8224 lambda_3[0][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_1_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_1_conv_0[0][\n__________________________________________________________________________________________________\nlambda_4 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_1_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_1_conv_1 ( (None, 1, 205, 32) 8224 lambda_4[0][0] \n__________________________________________________________________________________________________\ndropout_2 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_1_conv_1[0][\n__________________________________________________________________________________________________\nscrambler_resblock_0_1_skip_1 ( (None, 1, 205, 32) 0 dropout_2[0][0] \n scrambler_resblock_0_0_skip_1[0][\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_1_skip_1[0][\n__________________________________________________________________________________________________\nlambda_5 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_2_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_conv_0 ( (None, 1, 205, 32) 8224 lambda_5[0][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_2_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_2_conv_0[0][\n__________________________________________________________________________________________________\nlambda_6 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_2_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_conv_1 ( (None, 1, 205, 32) 8224 lambda_6[0][0] \n__________________________________________________________________________________________________\ndropout_3 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_2_conv_1[0][\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_skip_1 ( (None, 1, 205, 32) 0 dropout_3[0][0] \n scrambler_resblock_0_1_skip_1[0][\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_2_skip_1[0][\n__________________________________________________________________________________________________\nlambda_7 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_3_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_conv_0 ( (None, 1, 205, 32) 8224 lambda_7[0][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_3_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_3_conv_0[0][\n__________________________________________________________________________________________________\nlambda_8 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_3_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_conv_1 ( (None, 1, 205, 32) 8224 lambda_8[0][0] \n__________________________________________________________________________________________________\ndropout_4 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_3_conv_1[0][\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_skip_1 ( (None, 1, 205, 32) 0 dropout_4[0][0] \n scrambler_resblock_0_2_skip_1[0][\n__________________________________________________________________________________________________\nscrambler_last_block_conv (Conv (None, 1, 205, 32) 1056 scrambler_resblock_0_3_skip_1[0][\n__________________________________________________________________________________________________\nscrambler_skip_conv_0 (Conv2D) (None, 1, 205, 32) 1056 scrambler_conv_0[0][0] \n__________________________________________________________________________________________________\nscrambler_skip_add (Lambda) (None, 1, 205, 32) 0 scrambler_last_block_conv[0][0] \n scrambler_skip_conv_0[0][0] \n__________________________________________________________________________________________________\nscrambler_class (InputLayer) (None, 1) 0 \n__________________________________________________________________________________________________\nscrambler_final_conv (Conv2D) (None, 1, 205, 1) 33 scrambler_skip_add[0][0] \n__________________________________________________________________________________________________\nscrambler_onehot_to_logits (Lam (None, 1, 205, 4) 0 scrambler_input[0][0] \n__________________________________________________________________________________________________\nbackground_dense (Embedding) (None, 1, 820) 820 scrambler_class[0][0] \n__________________________________________________________________________________________________\nscrambler_logit_scale (Lambda) (None, 1, 205, 4) 0 scrambler_final_conv[0][0] \n scrambler_onehot_to_logits[0][0] \n__________________________________________________________________________________________________\nreshape_1 (Reshape) (None, 1, 205, 4) 0 background_dense[0][0] \n template_dense[0][0] \n mask_dense[0][0] \n__________________________________________________________________________________________________\ntemplate_dense (Embedding) (None, 1, 820) 820 scrambler_class[0][0] \n__________________________________________________________________________________________________\nmask_dense (Embedding) (None, 1, 820) 820 scrambler_class[0][0] \n__________________________________________________________________________________________________\nbackground_layer (Lambda) (None, 1, 205, 4) 0 scrambler_logit_scale[0][0] \n reshape_1[0][0] \n__________________________________________________________________________________________________\nmasking_layer (Lambda) (None, 1, 205, 4) 0 background_layer[0][0] \n reshape_1[1][0] \n reshape_1[2][0] \n__________________________________________________________________________________________________\nupsampling_layer (Lambda) (None, 1, 205, 4) 0 masking_layer[0][0] \n__________________________________________________________________________________________________\npwm_sampler (Lambda) (None, 1, None, 4) 0 upsampling_layer[0][0] \n__________________________________________________________________________________________________\npwm (Softmax) (None, 1, 205, 4) 0 masking_layer[0][0] \n__________________________________________________________________________________________________\npermute_layer (Lambda) (32, 32, 1, 205, 4) 0 pwm_sampler[0][0] \n==================================================================================================\nTotal params: 71,581\nTrainable params: 68,609\nNon-trainable params: 2,972\n__________________________________________________________________________________________________\n"
],
[
"loss_model.summary()",
"__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\nae_scrambler_input (InputLayer) (None, 1, 205, 4) 0 \n__________________________________________________________________________________________________\nscrambler_conv_0 (Conv2D) (None, 1, 205, 32) 160 ae_scrambler_input[0][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_0_batch_no (None, 1, 205, 32) 128 scrambler_conv_0[1][0] \n__________________________________________________________________________________________________\nlambda_1 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_0_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_0_conv_0 ( (None, 1, 205, 32) 8224 lambda_1[1][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_0_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_0_conv_0[1][\n__________________________________________________________________________________________________\nlambda_2 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_0_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_0_conv_1 ( (None, 1, 205, 32) 8224 lambda_2[1][0] \n__________________________________________________________________________________________________\ndropout_1 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_0_conv_1[1][\n__________________________________________________________________________________________________\nscrambler_resblock_0_0_skip_1 ( (None, 1, 205, 32) 0 dropout_1[1][0] \n scrambler_conv_0[1][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_1_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_0_skip_1[1][\n__________________________________________________________________________________________________\nlambda_3 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_1_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_1_conv_0 ( (None, 1, 205, 32) 8224 lambda_3[1][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_1_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_1_conv_0[1][\n__________________________________________________________________________________________________\nlambda_4 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_1_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_1_conv_1 ( (None, 1, 205, 32) 8224 lambda_4[1][0] \n__________________________________________________________________________________________________\ndropout_2 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_1_conv_1[1][\n__________________________________________________________________________________________________\nscrambler_resblock_0_1_skip_1 ( (None, 1, 205, 32) 0 dropout_2[1][0] \n scrambler_resblock_0_0_skip_1[1][\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_1_skip_1[1][\n__________________________________________________________________________________________________\nlambda_5 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_2_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_conv_0 ( (None, 1, 205, 32) 8224 lambda_5[1][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_2_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_2_conv_0[1][\n__________________________________________________________________________________________________\nlambda_6 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_2_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_conv_1 ( (None, 1, 205, 32) 8224 lambda_6[1][0] \n__________________________________________________________________________________________________\ndropout_3 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_2_conv_1[1][\n__________________________________________________________________________________________________\nscrambler_resblock_0_2_skip_1 ( (None, 1, 205, 32) 0 dropout_3[1][0] \n scrambler_resblock_0_1_skip_1[1][\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_2_skip_1[1][\n__________________________________________________________________________________________________\nlambda_7 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_3_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_conv_0 ( (None, 1, 205, 32) 8224 lambda_7[1][0] \n__________________________________________________________________________________________________\nscrambler_resblock_0_3_batch_no (None, 1, 205, 32) 128 scrambler_resblock_0_3_conv_0[1][\n__________________________________________________________________________________________________\nlambda_8 (Lambda) (None, 1, 205, 32) 0 scrambler_resblock_0_3_batch_norm\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_conv_1 ( (None, 1, 205, 32) 8224 lambda_8[1][0] \n__________________________________________________________________________________________________\ndropout_4 (Dropout) (None, 1, 205, 32) 0 scrambler_resblock_0_3_conv_1[1][\n__________________________________________________________________________________________________\nscrambler_resblock_0_3_skip_1 ( (None, 1, 205, 32) 0 dropout_4[1][0] \n scrambler_resblock_0_2_skip_1[1][\n__________________________________________________________________________________________________\nscrambler_last_block_conv (Conv (None, 1, 205, 32) 1056 scrambler_resblock_0_3_skip_1[1][\n__________________________________________________________________________________________________\nscrambler_skip_conv_0 (Conv2D) (None, 1, 205, 32) 1056 scrambler_conv_0[1][0] \n__________________________________________________________________________________________________\nscrambler_skip_add (Lambda) (None, 1, 205, 32) 0 scrambler_last_block_conv[1][0] \n scrambler_skip_conv_0[1][0] \n__________________________________________________________________________________________________\nae_scrambler_class (InputLayer) (None, 1) 0 \n__________________________________________________________________________________________________\nscrambler_final_conv (Conv2D) (None, 1, 205, 1) 33 scrambler_skip_add[1][0] \n__________________________________________________________________________________________________\nscrambler_onehot_to_logits (Lam (None, 1, 205, 4) 0 ae_scrambler_input[0][0] \n__________________________________________________________________________________________________\nbackground_dense (Embedding) (None, 1, 820) 820 ae_scrambler_class[0][0] \n__________________________________________________________________________________________________\nscrambler_logit_scale (Lambda) (None, 1, 205, 4) 0 scrambler_final_conv[1][0] \n scrambler_onehot_to_logits[1][0] \n__________________________________________________________________________________________________\nreshape_1 (Reshape) (None, 1, 205, 4) 0 background_dense[1][0] \n template_dense[1][0] \n mask_dense[1][0] \n__________________________________________________________________________________________________\ntemplate_dense (Embedding) (None, 1, 820) 820 ae_scrambler_class[0][0] \n__________________________________________________________________________________________________\nmask_dense (Embedding) (None, 1, 820) 820 ae_scrambler_class[0][0] \n__________________________________________________________________________________________________\nbackground_layer (Lambda) (None, 1, 205, 4) 0 scrambler_logit_scale[1][0] \n reshape_1[3][0] \n__________________________________________________________________________________________________\nmasking_layer (Lambda) (None, 1, 205, 4) 0 background_layer[1][0] \n reshape_1[4][0] \n reshape_1[5][0] \n__________________________________________________________________________________________________\nupsampling_layer (Lambda) (None, 1, 205, 4) 0 masking_layer[1][0] \n__________________________________________________________________________________________________\npwm_sampler (Lambda) (None, 1, None, 4) 0 upsampling_layer[2][0] \n__________________________________________________________________________________________________\npermute_layer (Lambda) (32, 32, 1, 205, 4) 0 pwm_sampler[1][0] \n__________________________________________________________________________________________________\naparent_lib_input (InputLayer) (None, 13) 0 \n__________________________________________________________________________________________________\naparent_distal_pas_input (Input (None, 1) 0 \n__________________________________________________________________________________________________\ndeflate_scrambled_sample (Lambd (1024, 1, 205, 4) 0 permute_layer[2][0] \n__________________________________________________________________________________________________\nmake_prediction_scrambled (Lamb (1024, 1) 0 deflate_scrambled_sample[0][0] \n aparent_lib_input[0][0] \n aparent_distal_pas_input[0][0] \n__________________________________________________________________________________________________\nmake_prediction_non_scrambled ( (None, 1) 0 ae_scrambler_input[0][0] \n aparent_lib_input[0][0] \n aparent_distal_pas_input[0][0] \n__________________________________________________________________________________________________\ninflate_scrambled_prediction (L (32, 32) 0 make_prediction_scrambled[0][0] \n__________________________________________________________________________________________________\npwm (Softmax) (None, 1, 205, 4) 0 masking_layer[1][0] \n__________________________________________________________________________________________________\nnll (Lambda) (32,) 0 make_prediction_non_scrambled[0][\n inflate_scrambled_prediction[0][0\n__________________________________________________________________________________________________\nconservation (Lambda) (None,) 0 pwm[1][0] \n reshape_1[5][0] \n__________________________________________________________________________________________________\nentropy (Lambda) (None,) 0 pwm[1][0] \n reshape_1[5][0] \n==================================================================================================\nTotal params: 71,581\nTrainable params: 68,609\nNon-trainable params: 2,972\n__________________________________________________________________________________________________\n"
],
[
"#Training configuration\n\n#Define number of training epochs\nn_epochs = 50\n\n#Define experiment suffix (optional)\nexperiment_suffix = \"\"\n\n#Define anneal function\ndef _anneal_func(val, epoch, n_epochs=n_epochs) :\n if epoch in [0] :\n return 1.0\n \n return 0.0\n\narchitecture_str = \"resnet_\" + str(resnet_n_groups) + \"_\" + str(resnet_n_resblocks_per_group) + \"_\" + str(resnet_n_channels) + \"_\" + str(resnet_window_size) + \"_\" + str(resnet_drop_rate).replace(\".\", \"\")\n\nmodel_name = \"autoscrambler_dataset_\" + dataset_name + \"_sample_mode_\" + sample_mode + \"_n_samples_\" + str(n_samples) + \"_\" + architecture_str + \"_n_epochs_\" + str(n_epochs) + \"_target_bits_\" + str(target_bits).replace(\".\", \"\") + experiment_suffix\n\nprint(\"Model save name = \" + model_name)\n",
"Model save name = autoscrambler_dataset_apa_doubledope_sample_mode_st_n_samples_32_resnet_1_4_32_8_025_n_epochs_50_target_bits_05\n"
],
[
"#Execute training procedure\n\ncallbacks =[\n #ModelCheckpoint(\"model_checkpoints/\" + model_name + \"_epoch_{epoch:02d}.hdf5\", monitor='val_loss', mode='min', period=10, save_weights_only=True),\n EpochVariableCallback(anneal_coeff, _anneal_func)\n]\n\ns_train = np.zeros((x_train.shape[0], 1))\ns_test = np.zeros((x_test.shape[0], 1))\n\naparent_l_train = np.zeros((x_train.shape[0], 13))\naparent_l_train[:, 4] = 1.\naparent_l_test = np.zeros((x_test.shape[0], 13))\naparent_l_test[:, 4] = 1.\n\naparent_d_train = np.ones((x_train.shape[0], 1))\naparent_d_test = np.ones((x_test.shape[0], 1))\n\n# train the autoencoder\ntrain_history = loss_model.fit(\n [s_train, x_train, aparent_l_train, aparent_d_train],\n [s_train, s_train, s_train],\n shuffle=True,\n epochs=n_epochs,\n batch_size=batch_size,\n validation_data=(\n [s_test, x_test, aparent_l_test, aparent_d_test],\n [s_test, s_test, s_test]\n ),\n callbacks=callbacks\n)\n",
"Train on 31264 samples, validate on 1728 samples\nEpoch 1/50\n31264/31264 [==============================] - 44s 1ms/step - loss: 0.0604 - nll_loss: 0.0492 - conservation_loss: 0.0112 - entropy_loss: 0.0000e+00 - val_loss: 0.0075 - val_nll_loss: 0.0063 - val_conservation_loss: 0.0011 - val_entropy_loss: 0.0000e+00\nEpoch 2/50\n31264/31264 [==============================] - 35s 1ms/step - loss: 0.3763 - nll_loss: 0.3456 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0308 - val_loss: 0.2929 - val_nll_loss: 0.2732 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0197\nEpoch 3/50\n31264/31264 [==============================] - 36s 1ms/step - loss: 0.3251 - nll_loss: 0.3082 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0169 - val_loss: 0.2711 - val_nll_loss: 0.2524 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0187\nEpoch 4/50\n31264/31264 [==============================] - 36s 1ms/step - loss: 0.3076 - nll_loss: 0.2919 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0157 - val_loss: 0.2678 - val_nll_loss: 0.2520 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0159\nEpoch 5/50\n31264/31264 [==============================] - 35s 1ms/step - loss: 0.2984 - nll_loss: 0.2835 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0149 - val_loss: 0.2573 - val_nll_loss: 0.2412 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0161\nEpoch 6/50\n31264/31264 [==============================] - 29s 921us/step - loss: 0.2895 - nll_loss: 0.2755 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0140 - val_loss: 0.2668 - val_nll_loss: 0.2454 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0214\nEpoch 7/50\n31264/31264 [==============================] - 28s 905us/step - loss: 0.2813 - nll_loss: 0.2682 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0131 - val_loss: 0.2367 - val_nll_loss: 0.2209 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0158\nEpoch 8/50\n31264/31264 [==============================] - 27s 867us/step - loss: 0.2758 - nll_loss: 0.2630 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0127 - val_loss: 0.2522 - val_nll_loss: 0.2351 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0171\nEpoch 9/50\n31264/31264 [==============================] - 27s 868us/step - loss: 0.2714 - nll_loss: 0.2592 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0121 - val_loss: 0.2367 - val_nll_loss: 0.2190 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0177\nEpoch 10/50\n31264/31264 [==============================] - 27s 868us/step - loss: 0.2673 - nll_loss: 0.2559 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0114 - val_loss: 0.2211 - val_nll_loss: 0.1978 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0233\nEpoch 11/50\n31264/31264 [==============================] - 27s 868us/step - loss: 0.2655 - nll_loss: 0.2539 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0116 - val_loss: 0.2227 - val_nll_loss: 0.2077 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0151\nEpoch 12/50\n31264/31264 [==============================] - 27s 867us/step - loss: 0.2628 - nll_loss: 0.2514 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0113 - val_loss: 0.2232 - val_nll_loss: 0.2078 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0154\nEpoch 13/50\n31264/31264 [==============================] - 27s 875us/step - loss: 0.2604 - nll_loss: 0.2494 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0109 - val_loss: 0.2178 - val_nll_loss: 0.1912 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0266\nEpoch 14/50\n31264/31264 [==============================] - 28s 889us/step - loss: 0.2570 - nll_loss: 0.2459 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0111 - val_loss: 0.2200 - val_nll_loss: 0.2049 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0151\nEpoch 15/50\n31264/31264 [==============================] - 32s 1ms/step - loss: 0.2567 - nll_loss: 0.2457 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0109 - val_loss: 0.2152 - val_nll_loss: 0.2008 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0145\nEpoch 16/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2545 - nll_loss: 0.2437 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0108 - val_loss: 0.2138 - val_nll_loss: 0.1923 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0215\nEpoch 17/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2536 - nll_loss: 0.2429 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0108 - val_loss: 0.2105 - val_nll_loss: 0.1976 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0130\nEpoch 18/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2502 - nll_loss: 0.2395 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0107 - val_loss: 0.2131 - val_nll_loss: 0.2003 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0128\nEpoch 19/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2501 - nll_loss: 0.2394 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0106 - val_loss: 0.2095 - val_nll_loss: 0.1939 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0156\nEpoch 20/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2498 - nll_loss: 0.2390 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0107 - val_loss: 0.2255 - val_nll_loss: 0.1996 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0260\nEpoch 21/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2472 - nll_loss: 0.2368 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0104 - val_loss: 0.2021 - val_nll_loss: 0.1859 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0162\nEpoch 22/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2451 - nll_loss: 0.2348 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0103 - val_loss: 0.2168 - val_nll_loss: 0.1968 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0199\nEpoch 23/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2443 - nll_loss: 0.2339 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0103 - val_loss: 0.2105 - val_nll_loss: 0.1949 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0156\nEpoch 24/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2431 - nll_loss: 0.2325 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0105 - val_loss: 0.2011 - val_nll_loss: 0.1861 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0150\nEpoch 25/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2420 - nll_loss: 0.2315 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0106 - val_loss: 0.2041 - val_nll_loss: 0.1854 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0187\nEpoch 26/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2404 - nll_loss: 0.2299 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0105 - val_loss: 0.2003 - val_nll_loss: 0.1868 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0135\nEpoch 27/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2387 - nll_loss: 0.2283 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0104 - val_loss: 0.2093 - val_nll_loss: 0.1931 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0163\nEpoch 28/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2403 - nll_loss: 0.2300 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0102 - val_loss: 0.2065 - val_nll_loss: 0.1914 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0152\nEpoch 29/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2391 - nll_loss: 0.2288 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0103 - val_loss: 0.2187 - val_nll_loss: 0.2048 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0138\nEpoch 30/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2364 - nll_loss: 0.2263 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0101 - val_loss: 0.2048 - val_nll_loss: 0.1903 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0145\nEpoch 31/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2367 - nll_loss: 0.2265 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0103 - val_loss: 0.2012 - val_nll_loss: 0.1836 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0176\nEpoch 32/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2367 - nll_loss: 0.2265 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0103 - val_loss: 0.1968 - val_nll_loss: 0.1789 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0179\nEpoch 33/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2346 - nll_loss: 0.2244 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0102 - val_loss: 0.1956 - val_nll_loss: 0.1779 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0177\nEpoch 34/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2356 - nll_loss: 0.2257 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0099 - val_loss: 0.2043 - val_nll_loss: 0.1849 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0194\nEpoch 35/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2332 - nll_loss: 0.2230 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0102 - val_loss: 0.2025 - val_nll_loss: 0.1894 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0131\nEpoch 36/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2312 - nll_loss: 0.2212 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0101 - val_loss: 0.1955 - val_nll_loss: 0.1808 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0147\nEpoch 37/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2308 - nll_loss: 0.2209 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0099 - val_loss: 0.2058 - val_nll_loss: 0.1920 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0138\nEpoch 38/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2298 - nll_loss: 0.2199 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0099 - val_loss: 0.1912 - val_nll_loss: 0.1710 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0202\nEpoch 39/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2292 - nll_loss: 0.2190 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0102 - val_loss: 0.1931 - val_nll_loss: 0.1766 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0165\nEpoch 40/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2275 - nll_loss: 0.2175 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0099 - val_loss: 0.1997 - val_nll_loss: 0.1840 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0158\nEpoch 41/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2281 - nll_loss: 0.2183 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0098 - val_loss: 0.1916 - val_nll_loss: 0.1734 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0181\nEpoch 42/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2263 - nll_loss: 0.2166 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0098 - val_loss: 0.1990 - val_nll_loss: 0.1747 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0243\nEpoch 43/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2265 - nll_loss: 0.2170 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0095 - val_loss: 0.1988 - val_nll_loss: 0.1836 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0151\nEpoch 44/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2248 - nll_loss: 0.2150 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0098 - val_loss: 0.2145 - val_nll_loss: 0.1910 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0235\nEpoch 45/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2263 - nll_loss: 0.2167 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0097 - val_loss: 0.1918 - val_nll_loss: 0.1774 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0144\nEpoch 46/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2243 - nll_loss: 0.2147 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0096 - val_loss: 0.1919 - val_nll_loss: 0.1745 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0175\nEpoch 47/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2237 - nll_loss: 0.2141 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0097 - val_loss: 0.1873 - val_nll_loss: 0.1702 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0171\nEpoch 48/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2235 - nll_loss: 0.2142 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0093 - val_loss: 0.1906 - val_nll_loss: 0.1716 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0190\nEpoch 49/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2225 - nll_loss: 0.2132 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0093 - val_loss: 0.1991 - val_nll_loss: 0.1808 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0184\nEpoch 50/50\n31264/31264 [==============================] - 34s 1ms/step - loss: 0.2220 - nll_loss: 0.2126 - conservation_loss: 0.0000e+00 - entropy_loss: 0.0094 - val_loss: 0.1905 - val_nll_loss: 0.1747 - val_conservation_loss: 0.0000e+00 - val_entropy_loss: 0.0158\n"
],
[
"\nf, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(3 * 4, 3))\n\nn_epochs_actual = len(train_history.history['nll_loss'])\n\nax1.plot(np.arange(1, n_epochs_actual + 1), train_history.history['nll_loss'], linewidth=3, color='green')\nax1.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_nll_loss'], linewidth=3, color='orange')\n\nplt.sca(ax1)\nplt.xlabel(\"Epochs\", fontsize=14)\nplt.ylabel(\"NLL\", fontsize=14)\nplt.xlim(1, n_epochs_actual)\nplt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)\nplt.yticks(fontsize=12)\n\nax2.plot(np.arange(1, n_epochs_actual + 1), train_history.history['entropy_loss'], linewidth=3, color='green')\nax2.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_entropy_loss'], linewidth=3, color='orange')\n\nplt.sca(ax2)\nplt.xlabel(\"Epochs\", fontsize=14)\nplt.ylabel(\"Entropy Loss\", fontsize=14)\nplt.xlim(1, n_epochs_actual)\nplt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)\nplt.yticks(fontsize=12)\n\nax3.plot(np.arange(1, n_epochs_actual + 1), train_history.history['conservation_loss'], linewidth=3, color='green')\nax3.plot(np.arange(1, n_epochs_actual + 1), train_history.history['val_conservation_loss'], linewidth=3, color='orange')\n\nplt.sca(ax3)\nplt.xlabel(\"Epochs\", fontsize=14)\nplt.ylabel(\"Conservation Loss\", fontsize=14)\nplt.xlim(1, n_epochs_actual)\nplt.xticks([1, n_epochs_actual], [1, n_epochs_actual], fontsize=12)\nplt.yticks(fontsize=12)\n\nplt.tight_layout()\n\nplt.show()\n",
"_____no_output_____"
],
[
"# Save model and weights\nsave_dir = 'saved_models'\n\nif not os.path.isdir(save_dir):\n os.makedirs(save_dir)\n\nmodel_path = os.path.join(save_dir, model_name + '.h5')\n\nscrambler_model.save(model_path)\nprint('Saved scrambler model at %s ' % (model_path))\n",
"Saved scrambler model at saved_models/autoscrambler_dataset_apa_doubledope_sample_mode_st_n_samples_32_resnet_1_4_32_8_025_n_epochs_50_target_bits_05.h5 \n"
],
[
"#Load models\nsave_dir = 'saved_models'\n\nif not os.path.isdir(save_dir):\n os.makedirs(save_dir)\n\nmodel_path = os.path.join(save_dir, model_name + '.h5')\nscrambler_model = load_model(model_path, custom_objects={\n 'st_sampled_softmax' : st_sampled_softmax\n})\n\nprint('Loaded scrambler model %s ' % (model_path))\n",
"Loaded scrambler model saved_models/autoscrambler_dataset_apa_doubledope_sample_mode_st_n_samples_32_resnet_1_4_32_8_025_n_epochs_50_target_bits_05.h5 \n"
],
[
"#Load models\nsave_dir = 'saved_models'\n\nif not os.path.isdir(save_dir):\n os.makedirs(save_dir)\n\nmodel_path = os.path.join(save_dir, model_name + '.h5')\nscrambler_model.load_weights(model_path, by_name=True)\n\nprint('Loaded scrambler model %s ' % (model_path))\n",
"Loaded scrambler model saved_models/autoscrambler_dataset_apa_doubledope_sample_mode_st_n_samples_32_resnet_1_4_32_8_025_n_epochs_50_target_bits_05.h5 \n"
],
[
"#Visualize a few reconstructed sequence patterns\n\nsequence_template = 'CTTCCGATCTNNNNNNNNNNNNNNNNNNNNCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNANTAAANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCTAC'\n\nsave_examples = [2, 3, 4, 5, 6, 7]\n\ns_test = np.zeros((x_test.shape[0], 1))\n\naparent_l_test = np.zeros((x_test.shape[0], 13))\naparent_l_test[:, 4] = 1.\n\naparent_d_test = np.ones((x_test.shape[0], 1))\n\n_, pwm_test, sample_test, _ = scrambler_model.predict_on_batch(x=[x_test[:32], s_test[:32]])\n\nfor plot_i in range(0, 10) :\n \n print(\"Test sequence \" + str(plot_i) + \":\")\n \n y_test_hat_ref = predictor.predict(x=[np.expand_dims(np.expand_dims(x_test[plot_i, 0, :, :], axis=0), axis=-1), aparent_l_test[:1], aparent_d_test[:1]], batch_size=1)[0][0, 0]\n y_test_hat = predictor.predict(x=[np.expand_dims(sample_test[plot_i, :, 0, :, :], axis=-1), aparent_l_test[:32], aparent_d_test[:32]], batch_size=32)[0][:10, 0].tolist()\n \n print(\" - Prediction (original) = \" + str(round(y_test_hat_ref, 2))[:4])\n print(\" - Predictions (scrambled) = \" + str([float(str(round(y_test_hat[i], 2))[:4]) for i in range(len(y_test_hat))]))\n \n save_figs = False\n if save_examples is not None and plot_i in save_examples :\n save_figs = True\n \n plot_dna_logo(x_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(14, 0.65), plot_start=0, plot_end=205, plot_sequence_template=True, save_figs=save_figs, fig_name=model_name + \"_test_ix_\" + str(plot_i))\n plot_dna_logo(pwm_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(14, 0.65), plot_start=0, plot_end=205, plot_sequence_template=True, save_figs=save_figs, fig_name=model_name + \"_test_ix_\" + str(plot_i))\n",
"Test sequence 0:\n - Prediction (original) = 0.09\n - Predictions (scrambled) = [0.03, 0.07, 0.05, 0.03, 0.09, 0.03, 0.09, 0.06, 0.04, 0.05]\n"
],
[
"#Visualize a few reconstructed images\n\ns_test = np.zeros((x_test.shape[0], 1))\n\n_, pwm_test, sample_test, importance_scores_test = scrambler_model.predict(x=[x_test, s_test], batch_size=32, verbose=True)\n",
"1728/1728 [==============================] - 2s 1ms/step\n"
],
[
"#Save predicted importance scores\n\nnp.save(model_name + \"_importance_scores_test\", importance_scores_test)\n",
"_____no_output_____"
],
[
"#Calculate original and scrambled predictions\n\naparent_l_test = np.zeros((x_test.shape[0], 13))\naparent_l_test[:, 4] = 1.\n\naparent_d_test = np.ones((x_test.shape[0], 1))\n\ny_test_hats = []\ny_test_hats_scrambled = []\nfor i in range(x_test.shape[0]) :\n \n y_test_hat_ref = predictor.predict(x=[np.expand_dims(np.expand_dims(x_test[i, 0, :, :], axis=0), axis=-1), aparent_l_test[:1], aparent_d_test[:1]], batch_size=1)[0][0, 0]\n y_test_hat = np.mean(predictor.predict(x=[np.expand_dims(sample_test[i, :, 0, :, :], axis=-1), aparent_l_test[:32], aparent_d_test[:32]], batch_size=32)[0][:, 0])\n \n y_test_hats.append(y_test_hat_ref)\n y_test_hats_scrambled.append(y_test_hat)\n\ny_test_hat = np.array(y_test_hats)\ny_test_hat_scrambled = np.array(y_test_hats_scrambled)\n",
"_____no_output_____"
],
[
"from scipy.stats import pearsonr\n\nsave_figs = True\n\nr_val, _ = pearsonr(y_test_hat, y_test_hat_scrambled)\n\nleft_kl_divs = y_test_hat * np.log(y_test_hat / y_test_hat_scrambled) + (1. - y_test_hat) * np.log((1. - y_test_hat) / (1. - y_test_hat_scrambled))\nright_kl_divs = y_test_hat_scrambled * np.log(y_test_hat_scrambled / y_test_hat) + (1. - y_test_hat_scrambled) * np.log((1. - y_test_hat_scrambled) / (1. - y_test_hat))\n\nmean_kl_div = np.mean(left_kl_divs + right_kl_divs)\n\nf = plt.figure(figsize=(4, 4))\n\nplt.scatter(y_test_hat, y_test_hat_scrambled, color='black', s=5, alpha=0.25)\n\nplt.xlim(0, 1)\nplt.ylim(0, 1)\n\nplt.xticks([0.0, 0.2, 0.4, 0.6, 0.8, 1.0], [0.0, 0.2, 0.4, 0.6, 0.8, 1.0], fontsize=14)\nplt.yticks([0.0, 0.2, 0.4, 0.6, 0.8, 1.0], [0.0, 0.2, 0.4, 0.6, 0.8, 1.0], fontsize=14)\n\nplt.xlabel(\"Original Prediction\", fontsize=14)\nplt.ylabel(\"Scrambled Prediction\", fontsize=14)\n\nplt.title(\"R^2 = \" + str(round(r_val**2, 2)) + \", KL = \" + str(round(mean_kl_div, 2)), fontsize=14)\n\nplt.tight_layout()\n\nif save_figs :\n plt.savefig(model_name + \"_test_scatter.png\", transparent=True, dpi=300)\n plt.savefig(model_name + \"_test_scatter.eps\")\n\nplt.show()\n",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a165d00d375b0cdcbc70f1dbdf4d7723b2cd3d2
| 3,559 |
ipynb
|
Jupyter Notebook
|
ap_third_semester/compact_objects/calculations/transsonic.ipynb
|
jacopok/notes
|
805ebe1be49bbd14c6b46b24055f9fc7d1cd2586
|
[
"Apache-2.0"
] | 6 |
2019-10-10T13:10:57.000Z
|
2022-01-13T14:52:50.000Z
|
ap_third_semester/compact_objects/calculations/transsonic.ipynb
|
jacopok/notes
|
805ebe1be49bbd14c6b46b24055f9fc7d1cd2586
|
[
"Apache-2.0"
] | null | null | null |
ap_third_semester/compact_objects/calculations/transsonic.ipynb
|
jacopok/notes
|
805ebe1be49bbd14c6b46b24055f9fc7d1cd2586
|
[
"Apache-2.0"
] | 3 |
2019-10-03T16:20:19.000Z
|
2021-08-06T16:11:07.000Z
| 24.376712 | 177 | 0.527114 |
[
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport astropy.units as u\nfrom astropy.constants import codata2018 as ac\nfrom astropy.visualization import quantity_support\nfrom ipywidgets import interact",
"_____no_output_____"
],
[
"rho_inf = 1*u.g / u.m**3\na_inf = 2000 * u.m / u.s\nGM = aa.GM_sun\nGamma = 4/3\n\n# not working right now",
"_____no_output_____"
],
[
"def u_ellipse(a, r, Mdot, rho_inf=rho_inf, a_inf=a_inf):\n rhs = GM / r + (a_inf**2 - a**2)/(Gamma-1) \n return(np.sqrt(2 * rhs))\n\ndef u_hyperbola(a, r, Mdot, rho_inf=rho_inf, a_inf=a_inf):\n rhs = Mdot * (a_inf / a)**(2/ (Gamma -1))/ (4 * np.pi * r**2 * rho_inf) \n return(rhs)",
"_____no_output_____"
],
[
"ass = np.linspace(1e1 * u.m/u.s, 1e8*u.m/u.s)\ndef plot(r, Mdot):\n us_e = u_ellipse(ass, r*u.m, Mdot*u.kg/u.s)\n us_h = u_hyperbola(ass, r*u.m, Mdot*u.kg/u.s)\n with(quantity_support()):\n plt.loglog(ass, us_e)\n plt.loglog(ass, us_h)",
"_____no_output_____"
],
[
"interact(plot, r=(0, 1e6, 1e4), Mdot=(1e10, 1e15, 1e13))",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a1664e6dd780a76be0439849664eecb9d6a2da9
| 51,095 |
ipynb
|
Jupyter Notebook
|
src/tools/ecos/cvxpy/examples/notebooks/WWW/l1_trend_filter.ipynb
|
riadnassiffe/Simulator
|
7d9ff09f26367d3714e3d10be3dd4a9817b8ed6b
|
[
"MIT"
] | 5 |
2017-08-31T01:37:00.000Z
|
2022-03-24T04:23:09.000Z
|
examples/notebooks/WWW/l1_trend_filter.ipynb
|
quantopian/cvxpy
|
7deee4d172470aa8f629dab7fead50467afa75ff
|
[
"Apache-2.0"
] | 11 |
2020-06-05T17:21:59.000Z
|
2021-03-22T23:20:32.000Z
|
src/tools/ecos/cvxpy/examples/notebooks/WWW/l1_trend_filter.ipynb
|
riadnassiffe/Simulator
|
7d9ff09f26367d3714e3d10be3dd4a9817b8ed6b
|
[
"MIT"
] | 6 |
2017-02-09T19:37:07.000Z
|
2021-01-07T00:17:54.000Z
| 145.985714 | 41,695 | 0.860847 |
[
[
[
"empty"
]
]
] |
[
"empty"
] |
[
[
"empty"
]
] |
4a167ddb4d572596ed7d2cc81b187753fe3143bc
| 39,559 |
ipynb
|
Jupyter Notebook
|
model_notebooks/BP/model.ipynb
|
indralab/adeft_indra
|
6f039b58b6dea5eefa529cf15afaffff2d485513
|
[
"BSD-2-Clause"
] | null | null | null |
model_notebooks/BP/model.ipynb
|
indralab/adeft_indra
|
6f039b58b6dea5eefa529cf15afaffff2d485513
|
[
"BSD-2-Clause"
] | null | null | null |
model_notebooks/BP/model.ipynb
|
indralab/adeft_indra
|
6f039b58b6dea5eefa529cf15afaffff2d485513
|
[
"BSD-2-Clause"
] | null | null | null | 37.425733 | 1,456 | 0.516317 |
[
[
[
"import os\nimport json\nimport pickle\nimport random\nfrom collections import defaultdict, Counter\n\nfrom indra.literature.adeft_tools import universal_extract_text\nfrom indra.databases.hgnc_client import get_hgnc_name, get_hgnc_id\n\nfrom adeft.discover import AdeftMiner\nfrom adeft.gui import ground_with_gui\nfrom adeft.modeling.label import AdeftLabeler\nfrom adeft.modeling.classify import AdeftClassifier\nfrom adeft.disambiguate import AdeftDisambiguator, load_disambiguator\n\n\nfrom adeft_indra.ground.ground import AdeftGrounder\nfrom adeft_indra.model_building.s3 import model_to_s3\nfrom adeft_indra.model_building.escape import escape_filename\nfrom adeft_indra.db.content import get_pmids_for_agent_text, get_pmids_for_entity, \\\n get_plaintexts_for_pmids",
"_____no_output_____"
],
[
"adeft_grounder = AdeftGrounder()",
"_____no_output_____"
],
[
"shortforms = ['BP']\nmodel_name = ':'.join(sorted(escape_filename(shortform) for shortform in shortforms))\nresults_path = os.path.abspath(os.path.join('../..', 'results', model_name))",
"_____no_output_____"
],
[
"miners = dict()\nall_texts = {}\nfor shortform in shortforms:\n pmids = get_pmids_for_agent_text(shortform)\n text_dict = get_plaintexts_for_pmids(pmids, contains=shortforms)\n text_dict = {pmid: text for pmid, text in text_dict.items() if len(text) > 5}\n miners[shortform] = AdeftMiner(shortform)\n miners[shortform].process_texts(text_dict.values())\n all_texts.update(text_dict)\n\nlongform_dict = {}\nfor shortform in shortforms:\n longforms = miners[shortform].get_longforms()\n longforms = [(longform, count, score) for longform, count, score in longforms\n if count*score > 2]\n longform_dict[shortform] = longforms\n \ncombined_longforms = Counter()\nfor longform_rows in longform_dict.values():\n combined_longforms.update({longform: count for longform, count, score\n in longform_rows})\ngrounding_map = {}\nnames = {}\nfor longform in combined_longforms:\n groundings = adeft_grounder.ground(longform)\n if groundings:\n grounding = groundings[0]['grounding']\n grounding_map[longform] = grounding\n names[grounding] = groundings[0]['name']\nlongforms, counts = zip(*combined_longforms.most_common())\npos_labels = []",
"_____no_output_____"
],
[
"list(zip(longforms, counts))",
"_____no_output_____"
],
[
"try:\n disamb = load_disambiguator(shortforms[0])\n for shortform, gm in disamb.grounding_dict.items():\n for longform, grounding in gm.items():\n grounding_map[longform] = grounding\n for grounding, name in disamb.names.items():\n names[grounding] = name\n pos_labels = disamb.pos_labels\nexcept Exception:\n pass",
"_____no_output_____"
],
[
"names",
"_____no_output_____"
],
[
"grounding_map, names, pos_labels = ground_with_gui(longforms, counts, \n grounding_map=grounding_map,\n names=names, pos_labels=pos_labels, no_browser=True, port=8891)",
"_____no_output_____"
],
[
"result = [grounding_map, names, pos_labels]",
"_____no_output_____"
],
[
"result",
"_____no_output_____"
],
[
"grounding_map, names, pos_labels = [{'b phymatum stm815': 'ungrounded',\n 'back pain': 'MESH:D001416',\n 'back propagation': 'ungrounded',\n 'backpropagation': 'ungrounded',\n 'bacterial pneumonia': 'MESH:D018410',\n 'bacterial production': 'ungrounded',\n 'bacterioplankton production': 'ungrounded',\n 'ball peptide': 'ungrounded',\n 'banana peels': 'ungrounded',\n 'band pass': 'ungrounded',\n 'barometric pressure': 'ungrounded',\n 'basal progenitor': 'ungrounded',\n 'basic protein': 'ungrounded',\n 'basilar papilla': 'ungrounded',\n 'beat period': 'ungrounded',\n 'bee pollen': 'ungrounded',\n 'beet pulp': 'ungrounded',\n 'bell s palsy': 'MESH:D020330',\n 'bench press': 'ungrounded',\n 'benzo a pyrene': 'CHEBI:CHEBI:29865',\n 'benzo alpha pyrene': 'CHEBI:CHEBI:29865',\n 'benzo α pyrene': 'CHEBI:CHEBI:29865',\n 'benzophenone': 'ungrounded',\n 'benzopyrene': 'ungrounded',\n 'benzoyl peroxide': 'ungrounded',\n 'benzphetamine': 'CHEBI:CHEBI:3044',\n 'benzpyrene': 'CHEBI:CHEBI:29865',\n 'benzylpenicillin': 'CHEBI:CHEBI:18208',\n 'bereitschaftspotential': 'ungrounded',\n 'biliopancreatic': 'ungrounded',\n 'binding potential': 'ungrounded',\n 'binding protein': 'ungrounded',\n 'biodegradable polymer': 'ungrounded',\n 'biological process': 'ungrounded',\n 'biopharmaceutical': 'ungrounded',\n 'biopolymer': 'CHEBI:CHEBI:33694',\n 'biopterin': 'CHEBI:CHEBI:15373',\n 'biparental': 'ungrounded',\n 'biphenyl': 'CHEBI:CHEBI:17097',\n 'bipolar': 'MESH:D001714',\n 'bipolar disorder': 'MESH:D001714',\n 'bipolar patients': 'MESH:D001714',\n 'bipyridyl': 'CHEBI:CHEBI:35545',\n 'birch pollen': 'ungrounded',\n 'bisection point': 'ungrounded',\n 'bisphenol': 'CHEBI:CHEBI:22901',\n 'bisphosphonate': 'MESH:D004164',\n 'black phosphorus': 'ungrounded',\n 'blast phase': 'MESH:D001752',\n 'blastic phase': 'ungrounded',\n 'blocking peptide': 'ungrounded',\n 'blood perfusion': 'MESH:D010477',\n 'blood plasma': 'MESH:D010949',\n 'blood pressure': 'MESH:D001794',\n 'blueberry juice and probiotics': 'MESH:D019936',\n 'bodily pain': 'MESH:D010146',\n 'bodipy ® 665 676': 'ungrounded',\n 'body project': 'ungrounded',\n 'bovine pericardium': 'MESH:D010496',\n 'bowenoid papulosis': 'ungrounded',\n 'boysenberry polyphenol': 'CHEBI:CHEBI:26195',\n 'brachial plexus': 'ungrounded',\n 'branch point': 'ungrounded',\n 'branchpoint': 'ungrounded',\n 'brazilian propolis': 'MESH:D011429',\n 'breaking point': 'ungrounded',\n 'breakpoint': 'ungrounded',\n 'brevipedicellus': 'UP:P46639',\n 'bromopyruvate': 'MESH:C017092',\n 'bronchopneumonia': 'MESH:D001996',\n 'bruce peninsula': 'ungrounded',\n 'buckypaper': 'ungrounded',\n 'bullous pemphigoid': 'MESH:D010391',\n 'bupropion': 'CHEBI:CHEBI:3219',\n 'burkholderia pseudomallei': 'MESH:D016957',\n 'butyl paraben': 'CHEBI:CHEBI:85122',\n 'butylparaben': 'CHEBI:CHEBI:88542',\n 'butylphenol': 'ungrounded'},\n {'MESH:D001416': 'Back Pain',\n 'MESH:D018410': 'Pneumonia, Bacterial',\n 'MESH:D020330': 'Bell Palsy',\n 'CHEBI:CHEBI:29865': 'benzo[a]pyrene',\n 'CHEBI:CHEBI:3044': 'benzphetamine',\n 'CHEBI:CHEBI:18208': 'benzylpenicillin',\n 'CHEBI:CHEBI:33694': 'biomacromolecule',\n 'CHEBI:CHEBI:15373': 'biopterin',\n 'CHEBI:CHEBI:17097': 'biphenyl',\n 'MESH:D001714': 'Bipolar Disorder',\n 'CHEBI:CHEBI:35545': 'bipyridine',\n 'CHEBI:CHEBI:22901': 'bisphenol',\n 'MESH:D004164': 'Diphosphonates',\n 'MESH:D001752': 'Blast Crisis',\n 'MESH:D010477': 'Perfusion',\n 'MESH:D010949': 'Plasma',\n 'MESH:D001794': 'Blood Pressure',\n 'MESH:D019936': 'Probiotics',\n 'MESH:D010146': 'Pain',\n 'MESH:D010496': 'Pericardium',\n 'CHEBI:CHEBI:26195': 'polyphenol',\n 'MESH:D011429': 'Propolis',\n 'UP:P46639': 'KNAT1',\n 'MESH:C017092': 'bromopyruvate',\n 'MESH:D001996': 'Bronchopneumonia',\n 'MESH:D010391': 'Pemphigoid, Bullous',\n 'CHEBI:CHEBI:3219': 'bupropion',\n 'MESH:D016957': 'Burkholderia pseudomallei',\n 'CHEBI:CHEBI:85122': 'paraben',\n 'CHEBI:CHEBI:88542': 'Butylparaben'},\n ['CHEBI:CHEBI:29865', 'MESH:D001794', 'MESH:D004164']]",
"_____no_output_____"
],
[
"excluded_longforms = ['bp']",
"_____no_output_____"
],
[
"grounding_dict = {shortform: {longform: grounding_map[longform] \n for longform, _, _ in longforms if longform in grounding_map\n and longform not in excluded_longforms}\n for shortform, longforms in longform_dict.items()}\nresult = [grounding_dict, names, pos_labels]\n\nif not os.path.exists(results_path):\n os.mkdir(results_path)\nwith open(os.path.join(results_path, f'{model_name}_preliminary_grounding_info.json'), 'w') as f:\n json.dump(result, f)",
"_____no_output_____"
],
[
"additional_entities = {}",
"_____no_output_____"
],
[
"unambiguous_agent_texts = {}",
"_____no_output_____"
],
[
"labeler = AdeftLabeler(grounding_dict)\ncorpus = labeler.build_from_texts((text, pmid) for pmid, text in all_texts.items())\nagent_text_pmid_map = defaultdict(list)\nfor text, label, id_ in corpus:\n agent_text_pmid_map[label].append(id_)\n\nentity_pmid_map = {entity: set(get_pmids_for_entity(*entity.split(':', maxsplit=1),\n major_topic=True))for entity in additional_entities}",
"_____no_output_____"
],
[
"intersection1 = []\nfor entity1, pmids1 in entity_pmid_map.items():\n for entity2, pmids2 in entity_pmid_map.items():\n intersection1.append((entity1, entity2, len(pmids1 & pmids2)))",
"_____no_output_____"
],
[
"intersection2 = []\nfor entity1, pmids1 in agent_text_pmid_map.items():\n for entity2, pmids2 in entity_pmid_map.items():\n intersection2.append((entity1, entity2, len(set(pmids1) & pmids2)))",
"_____no_output_____"
],
[
"intersection1",
"_____no_output_____"
],
[
"intersection2",
"_____no_output_____"
],
[
"all_used_pmids = set()\nfor entity, agent_texts in unambiguous_agent_texts.items():\n used_pmids = set()\n for agent_text in agent_texts[1]:\n pmids = set(get_pmids_for_agent_text(agent_text))\n new_pmids = list(pmids - all_texts.keys() - used_pmids)\n text_dict = get_plaintexts_for_pmids(new_pmids, contains=agent_texts)\n corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) >= 5])\n used_pmids.update(new_pmids)\n all_used_pmids.update(used_pmids)\n \nfor entity, pmids in entity_pmid_map.items():\n new_pmids = list(set(pmids) - all_texts.keys() - all_used_pmids)\n if len(new_pmids) > 10000:\n new_pmids = random.choices(new_pmids, k=10000)\n _, contains = additional_entities[entity]\n text_dict = get_plaintexts_for_pmids(new_pmids, contains=contains)\n corpus.extend([(text, entity, pmid) for pmid, text in text_dict.items() if len(text) >= 5])",
"_____no_output_____"
],
[
"names.update({key: value[0] for key, value in additional_entities.items()})\nnames.update({key: value[0] for key, value in unambiguous_agent_texts.items()})\npos_labels = list(set(pos_labels) | additional_entities.keys() |\n unambiguous_agent_texts.keys())",
"_____no_output_____"
],
[
"%%capture\n\nclassifier = AdeftClassifier(shortforms, pos_labels=pos_labels, random_state=1729)\nparam_grid = {'C': [100.0], 'max_features': [10000]}\ntexts, labels, pmids = zip(*corpus)\nclassifier.cv(texts, labels, param_grid, cv=5, n_jobs=5)",
"INFO: [2020-11-03 04:02:29] /adeft/PP/adeft/adeft/modeling/classify.py - Beginning grid search in parameter space:\n{'C': [100.0], 'max_features': [10000]}\nINFO: [2020-11-03 04:06:26] /adeft/PP/adeft/adeft/modeling/classify.py - Best f1 score of 0.9870412939858071 found for parameter values:\n{'logit__C': 100.0, 'tfidf__max_features': 10000}\n"
],
[
"classifier.stats",
"_____no_output_____"
],
[
"disamb = AdeftDisambiguator(classifier, grounding_dict, names)",
"_____no_output_____"
],
[
"disamb.dump(model_name, results_path)",
"_____no_output_____"
],
[
"print(disamb.info())",
"Disambiguation model for BP\n\nProduces the disambiguations:\n\tBack Pain\tMESH:D001416\n\tBell Palsy\tMESH:D020330\n\tBipolar Disorder\tMESH:D001714\n\tBlast Crisis\tMESH:D001752\n\tBlood Pressure*\tMESH:D001794\n\tBronchopneumonia\tMESH:D001996\n\tBurkholderia pseudomallei\tMESH:D016957\n\tButylparaben\tCHEBI:CHEBI:88542\n\tDiphosphonates*\tMESH:D004164\n\tKNAT1\tUP:P46639\n\tPain\tMESH:D010146\n\tPemphigoid, Bullous\tMESH:D010391\n\tPerfusion\tMESH:D010477\n\tPericardium\tMESH:D010496\n\tPlasma\tMESH:D010949\n\tPneumonia, Bacterial\tMESH:D018410\n\tProbiotics\tMESH:D019936\n\tPropolis\tMESH:D011429\n\tbenzo[a]pyrene*\tCHEBI:CHEBI:29865\n\tbenzphetamine\tCHEBI:CHEBI:3044\n\tbenzylpenicillin\tCHEBI:CHEBI:18208\n\tbiomacromolecule\tCHEBI:CHEBI:33694\n\tbiopterin\tCHEBI:CHEBI:15373\n\tbiphenyl\tCHEBI:CHEBI:17097\n\tbipyridine\tCHEBI:CHEBI:35545\n\tbisphenol\tCHEBI:CHEBI:22901\n\tbromopyruvate\tMESH:C017092\n\tbupropion\tCHEBI:CHEBI:3219\n\tparaben\tCHEBI:CHEBI:85122\n\tpolyphenol\tCHEBI:CHEBI:26195\n\nClass level metrics:\n--------------------\nGrounding \tCount\tF1 \n Blood Pressure*\t6046\t0.99204\n Ungrounded\t 373\t0.82332\n benzo[a]pyrene*\t 301\t0.90708\n Pemphigoid, Bullous\t 155\t0.93306\n Diphosphonates*\t 97\t0.92438\n Bipolar Disorder\t 70\t0.88796\n KNAT1\t 29\t0.97778\n biphenyl\t 11\t0.13333\n Pericardium\t 11\t0.56667\n Blast Crisis\t 10\t 0.64\n paraben\t 10\t0.52667\n Butylparaben\t 8\t 0.2\n bisphenol\t 7\t 0.3\n Pain\t 6\t0.73333\n Back Pain\t 6\t 0.0\n benzylpenicillin\t 5\t0.13333\n bupropion\t 4\t 0.0\n Bell Palsy\t 4\t 0.0\n bipyridine\t 3\t 0.0\n bromopyruvate\t 3\t 0.0\n Pneumonia, Bacterial\t 3\t 0.0\n Plasma\t 3\t 0.0\n biomacromolecule\t 3\t 0.0\n benzphetamine\t 2\t 0.0\n biopterin\t 2\t 0.0\n Propolis\t 2\t 0.0\n Perfusion\t 2\t 0.0\n Probiotics\t 2\t 0.0\n Bronchopneumonia\t 2\t 0.0\nBurkholderia pseudomallei\t 1\t 0.0\n polyphenol\t 1\t 0.0\n\nWeighted Metrics:\n-----------------\n\tF1 score:\t0.98704\n\tPrecision:\t0.98114\n\tRecall:\t\t0.99333\n\n* Positive labels\nSee Docstring for explanation\n\n"
],
[
"model_to_s3(disamb)",
"_____no_output_____"
],
[
"preds = [disamb.disambiguate(text) for text in all_texts.values()]",
"_____no_output_____"
],
[
"texts = [text for pred, text in zip(preds, all_texts.values()) if pred[0] == 'HGNC:10967']",
"_____no_output_____"
],
[
"texts[3]",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a16823d953c89b5c59b7e669a6ab790a8c16a8d
| 338,760 |
ipynb
|
Jupyter Notebook
|
fmnist-nn-and-cnn.ipynb
|
digs1998/Deep-Learning-Implementations
|
3f92ccad257b3c2067e44f0d7d386e6e5a5ae005
|
[
"Apache-2.0"
] | null | null | null |
fmnist-nn-and-cnn.ipynb
|
digs1998/Deep-Learning-Implementations
|
3f92ccad257b3c2067e44f0d7d386e6e5a5ae005
|
[
"Apache-2.0"
] | null | null | null |
fmnist-nn-and-cnn.ipynb
|
digs1998/Deep-Learning-Implementations
|
3f92ccad257b3c2067e44f0d7d386e6e5a5ae005
|
[
"Apache-2.0"
] | null | null | null | 323.244275 | 136,448 | 0.915297 |
[
[
[
"# Import the Fashion MNIST dataset\n\nThis guide uses the Fashion MNIST dataset, which contains 70,000 grayscale images in 10 categories. The images show individual articles of clothing at low resolution (28 × 28 pixels), as seen here:\nFashion MNIST sprite\nFigure 1. Fashion-MNIST samples (by Zalando, MIT License).\n \n<table>\n <tr><td>\n <img src=\"https://tensorflow.org/images/fashion-mnist-sprite.png\"\n alt=\"Fashion MNIST sprite\" width=\"600\">\n </td></tr>\n <tr><td align=\"center\">\n <b>Figure 1.</b> <a href=\"https://github.com/zalandoresearch/fashion-mnist\">Fashion-MNIST samples</a> (by Zalando, MIT License).<br/> \n </td></tr>\n</table>\n \n \n \n\nFashion MNIST is intended as a drop-in replacement for the classic MNIST dataset—often used as the \"Hello, World\" of machine learning programs for computer vision. The MNIST dataset contains images of handwritten digits (0, 1, 2, etc) in an identical format to the articles of clothing we'll use here.\n\nThis guide uses Fashion MNIST for variety, and because it's a slightly more challenging problem than regular MNIST. Both datasets are relatively small and are used to verify that an algorithm works as expected. They're good starting points to test and debug code.\n\nWe will use 60,000 images to train the network and 10,000 images to evaluate how accurately the network learned to classify images. You can access the Fashion MNIST directly from TensorFlow, using the Datasets API:\n",
"_____no_output_____"
],
[
"# Load the Libraries",
"_____no_output_____"
]
],
[
[
"# This Python 3 environment comes with many helpful analytics libraries installed\n# It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python\n# For example, here's several helpful packages to load\n\nimport numpy as np # linear algebra\nimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)\n\n# Input data files are available in the read-only \"../input/\" directory\n# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom sklearn.model_selection import train_test_split\n\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport os\nfor dirname, _, filenames in os.walk('/kaggle/input'):\n for filename in filenames:\n print(os.path.join(dirname, filename))\n\n# You can write up to 5GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using \"Save & Run All\" \n# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session",
"/kaggle/input/fashionmnist/t10k-labels-idx1-ubyte\n/kaggle/input/fashionmnist/train-images-idx3-ubyte\n/kaggle/input/fashionmnist/fashion-mnist_train.csv\n/kaggle/input/fashionmnist/train-labels-idx1-ubyte\n/kaggle/input/fashionmnist/t10k-images-idx3-ubyte\n/kaggle/input/fashionmnist/fashion-mnist_test.csv\n"
]
],
[
[
"# Load and Explore Dataset",
"_____no_output_____"
]
],
[
[
"train_df = pd.read_csv('../input/fashionmnist/fashion-mnist_train.csv')\ntest_df = pd.read_csv('../input/fashionmnist/fashion-mnist_test.csv')\nprint('The shape of training dataset : ', train_df.shape)\nprint('The shape of testing dataset : ', test_df.shape)",
"The shape of training dataset : (60000, 785)\nThe shape of testing dataset : (10000, 785)\n"
],
[
"train_df.head(5)",
"_____no_output_____"
]
],
[
[
"**Now we start with converting the pixel values into array format**",
"_____no_output_____"
]
],
[
[
"train = np.array(train_df, dtype = 'float32')\ntest = np.array(test_df, dtype = 'float32')",
"_____no_output_____"
],
[
"x_train = train[:,1:]/255\n\ny_train = train[:,0]\n\nx_test= test[:,1:]/255\n\ny_test=test[:,0]",
"_____no_output_____"
]
],
[
[
"**Let us split the training and test datasets**",
"_____no_output_____"
]
],
[
[
"X_train, X_validate,y_train, y_validate = train_test_split(x_train, y_train, test_size = 0.2, random_state = 5000)\nprint('The size of training data after model selection : ', X_train.shape, y_train.shape)\nprint('The size of Validation data after model selection : ', X_validate.shape, y_validate.shape)",
"The size of training data after model selection : (48000, 784) (48000,)\nThe size of Validation data after model selection : (12000, 784) (12000,)\n"
]
],
[
[
"# Data Visualization",
"_____no_output_____"
]
],
[
[
"class_names = ['T_shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat', \n 'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']\nplt.figure(figsize=(17, 17))\n\nfor i in range(25):\n plt.subplot(5, 5, i + 1)\n plt.grid(False)\n plt.imshow(x_train[i].reshape((28,28)))\n plt.colorbar()\n label_index = int(y_train[i])\n plt.title(class_names[label_index])\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Model Development",
"_____no_output_____"
],
[
"**Before we start with model layout let's do some basic things**",
"_____no_output_____"
]
],
[
[
"#Assigning image dimensions for model\nimg_rows = 28\nimg_cols = 28\nimg_shape = (img_rows, img_cols,1)\n\nX_train = X_train.reshape(X_train.shape[0],*img_shape)\nx_test = x_test.reshape(x_test.shape[0],*img_shape)\nX_validate = X_validate.reshape(X_validate.shape[0],*img_shape)\n",
"_____no_output_____"
]
],
[
[
"# Simple layered Neural Network",
"_____no_output_____"
],
[
"**Without Dropout**",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n tf.keras.layers.Flatten(input_shape = img_shape),\n tf.keras.layers.Dense(512, activation = 'relu'),\n tf.keras.layers.Dropout(0.8),\n tf.keras.layers.Dense(10, activation = 'softmax') #since we want a probability based output\n])\nmodel.compile(optimizer = 'adam',\n loss = tf.keras.losses.SparseCategoricalCrossentropy(),\n metrics = ['accuracy'])\nhistory = model.fit(X_train, y_train, epochs = 30, verbose=2, validation_data=(X_validate, y_validate))",
"Train on 48000 samples, validate on 12000 samples\nEpoch 1/30\n48000/48000 - 5s - loss: 0.7594 - accuracy: 0.7246 - val_loss: 0.4805 - val_accuracy: 0.8183\nEpoch 2/30\n48000/48000 - 4s - loss: 0.5976 - accuracy: 0.7799 - val_loss: 0.4349 - val_accuracy: 0.8416\nEpoch 3/30\n48000/48000 - 4s - loss: 0.5648 - accuracy: 0.7947 - val_loss: 0.4150 - val_accuracy: 0.8467\nEpoch 4/30\n48000/48000 - 4s - loss: 0.5487 - accuracy: 0.7990 - val_loss: 0.4215 - val_accuracy: 0.8488\nEpoch 5/30\n48000/48000 - 4s - loss: 0.5351 - accuracy: 0.8032 - val_loss: 0.4107 - val_accuracy: 0.8545\nEpoch 6/30\n48000/48000 - 4s - loss: 0.5237 - accuracy: 0.8105 - val_loss: 0.3942 - val_accuracy: 0.8582\nEpoch 7/30\n48000/48000 - 4s - loss: 0.5150 - accuracy: 0.8126 - val_loss: 0.3850 - val_accuracy: 0.8627\nEpoch 8/30\n48000/48000 - 4s - loss: 0.5066 - accuracy: 0.8150 - val_loss: 0.3895 - val_accuracy: 0.8622\nEpoch 9/30\n48000/48000 - 4s - loss: 0.4992 - accuracy: 0.8186 - val_loss: 0.3780 - val_accuracy: 0.8655\nEpoch 10/30\n48000/48000 - 4s - loss: 0.4914 - accuracy: 0.8221 - val_loss: 0.3743 - val_accuracy: 0.8660\nEpoch 11/30\n48000/48000 - 4s - loss: 0.4855 - accuracy: 0.8249 - val_loss: 0.3703 - val_accuracy: 0.8692\nEpoch 12/30\n48000/48000 - 4s - loss: 0.4793 - accuracy: 0.8238 - val_loss: 0.3770 - val_accuracy: 0.8669\nEpoch 13/30\n48000/48000 - 5s - loss: 0.4799 - accuracy: 0.8266 - val_loss: 0.3646 - val_accuracy: 0.8712\nEpoch 14/30\n48000/48000 - 4s - loss: 0.4685 - accuracy: 0.8277 - val_loss: 0.3706 - val_accuracy: 0.8679\nEpoch 15/30\n48000/48000 - 4s - loss: 0.4762 - accuracy: 0.8279 - val_loss: 0.3600 - val_accuracy: 0.8737\nEpoch 16/30\n48000/48000 - 4s - loss: 0.4688 - accuracy: 0.8297 - val_loss: 0.3600 - val_accuracy: 0.8737\nEpoch 17/30\n48000/48000 - 4s - loss: 0.4645 - accuracy: 0.8315 - val_loss: 0.3643 - val_accuracy: 0.8700\nEpoch 18/30\n48000/48000 - 4s - loss: 0.4670 - accuracy: 0.8321 - val_loss: 0.3618 - val_accuracy: 0.8726\nEpoch 19/30\n48000/48000 - 4s - loss: 0.4584 - accuracy: 0.8327 - val_loss: 0.3571 - val_accuracy: 0.8757\nEpoch 20/30\n48000/48000 - 4s - loss: 0.4596 - accuracy: 0.8334 - val_loss: 0.3717 - val_accuracy: 0.8707\nEpoch 21/30\n48000/48000 - 4s - loss: 0.4572 - accuracy: 0.8330 - val_loss: 0.3538 - val_accuracy: 0.8758\nEpoch 22/30\n48000/48000 - 4s - loss: 0.4512 - accuracy: 0.8357 - val_loss: 0.3613 - val_accuracy: 0.8719\nEpoch 23/30\n48000/48000 - 4s - loss: 0.4512 - accuracy: 0.8364 - val_loss: 0.3588 - val_accuracy: 0.8680\nEpoch 24/30\n48000/48000 - 4s - loss: 0.4520 - accuracy: 0.8374 - val_loss: 0.3582 - val_accuracy: 0.8748\nEpoch 25/30\n48000/48000 - 4s - loss: 0.4472 - accuracy: 0.8374 - val_loss: 0.3521 - val_accuracy: 0.8772\nEpoch 26/30\n48000/48000 - 4s - loss: 0.4437 - accuracy: 0.8387 - val_loss: 0.3631 - val_accuracy: 0.8721\nEpoch 27/30\n48000/48000 - 4s - loss: 0.4428 - accuracy: 0.8381 - val_loss: 0.3571 - val_accuracy: 0.8753\nEpoch 28/30\n48000/48000 - 5s - loss: 0.4439 - accuracy: 0.8385 - val_loss: 0.3510 - val_accuracy: 0.8766\nEpoch 29/30\n48000/48000 - 4s - loss: 0.4409 - accuracy: 0.8402 - val_loss: 0.3521 - val_accuracy: 0.8762\nEpoch 30/30\n48000/48000 - 4s - loss: 0.4340 - accuracy: 0.8414 - val_loss: 0.3527 - val_accuracy: 0.8773\n"
]
],
[
[
"**Lets check the plots**",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(17,17))\n\nplt.subplot(2, 2, 1)\nplt.plot(history.history['loss'], label='Loss')\nplt.plot(history.history['val_loss'], label='Validation Loss')\nplt.legend()\nplt.title('Training - Loss Function')\n\nplt.subplot(2, 2, 2)\nplt.plot(history.history['accuracy'], label='Accuracy')\nplt.plot(history.history['val_accuracy'], label='Validation Accuracy')\nplt.legend()\nplt.title('Train - Accuracy')",
"_____no_output_____"
]
],
[
[
"**With Dropout**",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n tf.keras.layers.Flatten(input_shape = img_shape),\n tf.keras.layers.Dense(512, activation = 'relu'),\n tf.keras.layers.Dropout(0.2),\n tf.keras.layers.Dense(10, activation = 'softmax') #since we want a probability based output\n])\n",
"_____no_output_____"
]
],
[
[
"**Description of model is shown below**",
"_____no_output_____"
]
],
[
[
"model.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nflatten_1 (Flatten) (None, 784) 0 \n_________________________________________________________________\ndense_2 (Dense) (None, 512) 401920 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 512) 0 \n_________________________________________________________________\ndense_3 (Dense) (None, 10) 5130 \n=================================================================\nTotal params: 407,050\nTrainable params: 407,050\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.compile(optimizer = 'adam',\n loss = tf.keras.losses.SparseCategoricalCrossentropy(),\n metrics = ['accuracy'])",
"_____no_output_____"
],
[
"history = model.fit(X_train, y_train, epochs = 30, verbose=2, validation_data=(X_validate, y_validate))",
"Train on 48000 samples, validate on 12000 samples\nEpoch 1/30\n48000/48000 - 5s - loss: 0.5187 - accuracy: 0.8151 - val_loss: 0.5125 - val_accuracy: 0.7949\nEpoch 2/30\n48000/48000 - 4s - loss: 0.3994 - accuracy: 0.8545 - val_loss: 0.3832 - val_accuracy: 0.8600\nEpoch 3/30\n48000/48000 - 4s - loss: 0.3645 - accuracy: 0.8656 - val_loss: 0.3467 - val_accuracy: 0.8741\nEpoch 4/30\n48000/48000 - 4s - loss: 0.3395 - accuracy: 0.8729 - val_loss: 0.3207 - val_accuracy: 0.8856\nEpoch 5/30\n48000/48000 - 4s - loss: 0.3230 - accuracy: 0.8803 - val_loss: 0.3330 - val_accuracy: 0.8823\nEpoch 6/30\n48000/48000 - 4s - loss: 0.3122 - accuracy: 0.8835 - val_loss: 0.3329 - val_accuracy: 0.8816\nEpoch 7/30\n48000/48000 - 4s - loss: 0.2991 - accuracy: 0.8879 - val_loss: 0.3136 - val_accuracy: 0.8864\nEpoch 8/30\n48000/48000 - 4s - loss: 0.2900 - accuracy: 0.8916 - val_loss: 0.3124 - val_accuracy: 0.8867\nEpoch 9/30\n48000/48000 - 4s - loss: 0.2790 - accuracy: 0.8968 - val_loss: 0.3167 - val_accuracy: 0.8875\nEpoch 10/30\n48000/48000 - 4s - loss: 0.2700 - accuracy: 0.8975 - val_loss: 0.3043 - val_accuracy: 0.8928\nEpoch 11/30\n48000/48000 - 4s - loss: 0.2626 - accuracy: 0.9001 - val_loss: 0.3059 - val_accuracy: 0.8917\nEpoch 12/30\n48000/48000 - 5s - loss: 0.2548 - accuracy: 0.9036 - val_loss: 0.2942 - val_accuracy: 0.8969\nEpoch 13/30\n48000/48000 - 5s - loss: 0.2509 - accuracy: 0.9057 - val_loss: 0.3020 - val_accuracy: 0.8932\nEpoch 14/30\n48000/48000 - 4s - loss: 0.2444 - accuracy: 0.9078 - val_loss: 0.3040 - val_accuracy: 0.8957\nEpoch 15/30\n48000/48000 - 4s - loss: 0.2358 - accuracy: 0.9114 - val_loss: 0.3030 - val_accuracy: 0.8953\nEpoch 16/30\n48000/48000 - 4s - loss: 0.2338 - accuracy: 0.9115 - val_loss: 0.3063 - val_accuracy: 0.8957\nEpoch 17/30\n48000/48000 - 4s - loss: 0.2268 - accuracy: 0.9135 - val_loss: 0.3066 - val_accuracy: 0.8946\nEpoch 18/30\n48000/48000 - 4s - loss: 0.2224 - accuracy: 0.9151 - val_loss: 0.3101 - val_accuracy: 0.8927\nEpoch 19/30\n48000/48000 - 4s - loss: 0.2164 - accuracy: 0.9186 - val_loss: 0.2991 - val_accuracy: 0.8988\nEpoch 20/30\n48000/48000 - 4s - loss: 0.2149 - accuracy: 0.9187 - val_loss: 0.3061 - val_accuracy: 0.9003\nEpoch 21/30\n48000/48000 - 4s - loss: 0.2095 - accuracy: 0.9197 - val_loss: 0.3234 - val_accuracy: 0.8941\nEpoch 22/30\n48000/48000 - 4s - loss: 0.2054 - accuracy: 0.9222 - val_loss: 0.3055 - val_accuracy: 0.8969\nEpoch 23/30\n48000/48000 - 4s - loss: 0.2016 - accuracy: 0.9235 - val_loss: 0.3396 - val_accuracy: 0.8863\nEpoch 24/30\n48000/48000 - 4s - loss: 0.1994 - accuracy: 0.9246 - val_loss: 0.3282 - val_accuracy: 0.8960\nEpoch 25/30\n48000/48000 - 4s - loss: 0.1957 - accuracy: 0.9250 - val_loss: 0.3202 - val_accuracy: 0.8984\nEpoch 26/30\n48000/48000 - 4s - loss: 0.1912 - accuracy: 0.9263 - val_loss: 0.3117 - val_accuracy: 0.8988\nEpoch 27/30\n48000/48000 - 4s - loss: 0.1880 - accuracy: 0.9282 - val_loss: 0.3494 - val_accuracy: 0.8933\nEpoch 28/30\n48000/48000 - 4s - loss: 0.1853 - accuracy: 0.9294 - val_loss: 0.3247 - val_accuracy: 0.9003\nEpoch 29/30\n48000/48000 - 4s - loss: 0.1813 - accuracy: 0.9294 - val_loss: 0.3203 - val_accuracy: 0.9007\nEpoch 30/30\n48000/48000 - 4s - loss: 0.1800 - accuracy: 0.9309 - val_loss: 0.3413 - val_accuracy: 0.8940\n"
]
],
[
[
"**Plots after using dropout**",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(17,17))\n\nplt.subplot(2, 2, 1)\nplt.plot(history.history['loss'], label='Loss')\nplt.plot(history.history['val_loss'], label='Validation Loss')\nplt.legend()\nplt.title('Training - Loss Function')\n\nplt.subplot(2, 2, 2)\nplt.plot(history.history['accuracy'], label='Accuracy')\nplt.plot(history.history['val_accuracy'], label='Validation Accuracy')\nplt.legend()\nplt.title('Train - Accuracy')",
"_____no_output_____"
]
],
[
[
"# CNN Model",
"_____no_output_____"
]
],
[
[
"cnn_model = tf.keras.Sequential([\n tf.keras.layers.Conv2D(32, (3,3), padding='same', activation=tf.nn.relu,\n input_shape=(28, 28, 1)),\n tf.keras.layers.MaxPooling2D((2, 2), strides=2),\n tf.keras.layers.Conv2D(64, (3,3), padding='same', activation=tf.nn.relu),\n tf.keras.layers.MaxPooling2D((2, 2), strides=2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(128, activation=tf.nn.relu),\n tf.keras.layers.Dense(10, activation=tf.nn.softmax)\n])",
"_____no_output_____"
],
[
"cnn_model.summary()\ncnn_model.compile(optimizer = 'adam',\n loss = tf.keras.losses.SparseCategoricalCrossentropy(),\n metrics = ['accuracy'])",
"Model: \"sequential_2\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nconv2d (Conv2D) (None, 28, 28, 32) 320 \n_________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 14, 14, 32) 0 \n_________________________________________________________________\nconv2d_1 (Conv2D) (None, 14, 14, 64) 18496 \n_________________________________________________________________\nmax_pooling2d_1 (MaxPooling2 (None, 7, 7, 64) 0 \n_________________________________________________________________\nflatten_2 (Flatten) (None, 3136) 0 \n_________________________________________________________________\ndense_4 (Dense) (None, 128) 401536 \n_________________________________________________________________\ndense_5 (Dense) (None, 10) 1290 \n=================================================================\nTotal params: 421,642\nTrainable params: 421,642\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"history = cnn_model.fit(X_train, y_train, epochs = 30, verbose=2, validation_data=(X_validate, y_validate))",
"Train on 48000 samples, validate on 12000 samples\nEpoch 1/30\n48000/48000 - 8s - loss: 0.4294 - accuracy: 0.8461 - val_loss: 0.3328 - val_accuracy: 0.8806\nEpoch 2/30\n48000/48000 - 5s - loss: 0.2818 - accuracy: 0.8980 - val_loss: 0.2763 - val_accuracy: 0.8993\nEpoch 3/30\n48000/48000 - 5s - loss: 0.2347 - accuracy: 0.9141 - val_loss: 0.2479 - val_accuracy: 0.9129\nEpoch 4/30\n48000/48000 - 5s - loss: 0.2025 - accuracy: 0.9254 - val_loss: 0.2401 - val_accuracy: 0.9138\nEpoch 5/30\n48000/48000 - 5s - loss: 0.1735 - accuracy: 0.9351 - val_loss: 0.2252 - val_accuracy: 0.9201\nEpoch 6/30\n48000/48000 - 5s - loss: 0.1468 - accuracy: 0.9452 - val_loss: 0.2470 - val_accuracy: 0.9149\nEpoch 7/30\n48000/48000 - 5s - loss: 0.1234 - accuracy: 0.9544 - val_loss: 0.2401 - val_accuracy: 0.9235\nEpoch 8/30\n48000/48000 - 6s - loss: 0.1064 - accuracy: 0.9610 - val_loss: 0.2504 - val_accuracy: 0.9208\nEpoch 9/30\n48000/48000 - 5s - loss: 0.0882 - accuracy: 0.9676 - val_loss: 0.2493 - val_accuracy: 0.9253\nEpoch 10/30\n48000/48000 - 5s - loss: 0.0725 - accuracy: 0.9735 - val_loss: 0.2619 - val_accuracy: 0.9258\nEpoch 11/30\n48000/48000 - 5s - loss: 0.0623 - accuracy: 0.9761 - val_loss: 0.3119 - val_accuracy: 0.9218\nEpoch 12/30\n48000/48000 - 5s - loss: 0.0546 - accuracy: 0.9801 - val_loss: 0.3201 - val_accuracy: 0.9218\nEpoch 13/30\n48000/48000 - 5s - loss: 0.0441 - accuracy: 0.9834 - val_loss: 0.3549 - val_accuracy: 0.9184\nEpoch 14/30\n48000/48000 - 5s - loss: 0.0391 - accuracy: 0.9860 - val_loss: 0.3799 - val_accuracy: 0.9182\nEpoch 15/30\n48000/48000 - 5s - loss: 0.0335 - accuracy: 0.9877 - val_loss: 0.3877 - val_accuracy: 0.9206\nEpoch 16/30\n48000/48000 - 5s - loss: 0.0311 - accuracy: 0.9890 - val_loss: 0.4236 - val_accuracy: 0.9231\nEpoch 17/30\n48000/48000 - 5s - loss: 0.0291 - accuracy: 0.9891 - val_loss: 0.4567 - val_accuracy: 0.9170\nEpoch 18/30\n48000/48000 - 5s - loss: 0.0271 - accuracy: 0.9905 - val_loss: 0.4735 - val_accuracy: 0.9191\nEpoch 19/30\n48000/48000 - 5s - loss: 0.0227 - accuracy: 0.9921 - val_loss: 0.5177 - val_accuracy: 0.9193\nEpoch 20/30\n48000/48000 - 5s - loss: 0.0240 - accuracy: 0.9915 - val_loss: 0.4930 - val_accuracy: 0.9174\nEpoch 21/30\n48000/48000 - 5s - loss: 0.0218 - accuracy: 0.9923 - val_loss: 0.4744 - val_accuracy: 0.9219\nEpoch 22/30\n48000/48000 - 5s - loss: 0.0201 - accuracy: 0.9931 - val_loss: 0.5733 - val_accuracy: 0.9152\nEpoch 23/30\n48000/48000 - 5s - loss: 0.0186 - accuracy: 0.9939 - val_loss: 0.5311 - val_accuracy: 0.9197\nEpoch 24/30\n48000/48000 - 5s - loss: 0.0204 - accuracy: 0.9931 - val_loss: 0.5253 - val_accuracy: 0.9202\nEpoch 25/30\n48000/48000 - 5s - loss: 0.0157 - accuracy: 0.9946 - val_loss: 0.5985 - val_accuracy: 0.9225\nEpoch 26/30\n48000/48000 - 5s - loss: 0.0185 - accuracy: 0.9935 - val_loss: 0.5995 - val_accuracy: 0.9193\nEpoch 27/30\n48000/48000 - 5s - loss: 0.0174 - accuracy: 0.9939 - val_loss: 0.5684 - val_accuracy: 0.9222\nEpoch 28/30\n48000/48000 - 5s - loss: 0.0165 - accuracy: 0.9942 - val_loss: 0.6832 - val_accuracy: 0.9197\nEpoch 29/30\n48000/48000 - 5s - loss: 0.0179 - accuracy: 0.9935 - val_loss: 0.6576 - val_accuracy: 0.9160\nEpoch 30/30\n48000/48000 - 5s - loss: 0.0167 - accuracy: 0.9946 - val_loss: 0.7094 - val_accuracy: 0.9165\n"
],
[
"plt.figure(figsize=(17,17))\n\nplt.subplot(2, 2, 1)\nplt.plot(history.history['loss'], label='Loss')\nplt.plot(history.history['val_loss'], label='Validation Loss')\nplt.legend()\nplt.title('Training - Loss Function')\n\nplt.subplot(2, 2, 2)\nplt.plot(history.history['accuracy'], label='Accuracy')\nplt.plot(history.history['val_accuracy'], label='Validation Accuracy')\nplt.legend()\nplt.title('Train - Accuracy')",
"_____no_output_____"
]
],
[
[
"# Discussion\nThough the accuracy obtained on validation set using CNN was good this model still needs improvement and will be updating it in next versions!!\n\nDo upvote my Kernel if you liked it. It is my first implementation code on CNN",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a168e87f7f24a70e68ccb5ba6dd8f05a4c1b0c0
| 95,478 |
ipynb
|
Jupyter Notebook
|
3. Landmark Detection and Tracking.ipynb
|
wronaq/LandmarkDetection
|
db965d84f9ba3a2f497226828387169aa03c0a3c
|
[
"MIT"
] | null | null | null |
3. Landmark Detection and Tracking.ipynb
|
wronaq/LandmarkDetection
|
db965d84f9ba3a2f497226828387169aa03c0a3c
|
[
"MIT"
] | null | null | null |
3. Landmark Detection and Tracking.ipynb
|
wronaq/LandmarkDetection
|
db965d84f9ba3a2f497226828387169aa03c0a3c
|
[
"MIT"
] | null | null | null | 111.150175 | 31,080 | 0.812114 |
[
[
[
"# Project 3: Implement SLAM \n\n---\n\n## Project Overview\n\nIn this project, you'll implement SLAM for robot that moves and senses in a 2 dimensional, grid world!\n\nSLAM gives us a way to both localize a robot and build up a map of its environment as a robot moves and senses in real-time. This is an active area of research in the fields of robotics and autonomous systems. Since this localization and map-building relies on the visual sensing of landmarks, this is a computer vision problem. \n\nUsing what you've learned about robot motion, representations of uncertainty in motion and sensing, and localization techniques, you will be tasked with defining a function, `slam`, which takes in six parameters as input and returns the vector `mu`. \n> `mu` contains the (x,y) coordinate locations of the robot as it moves, and the positions of landmarks that it senses in the world\n\nYou can implement helper functions as you see fit, but your function must return `mu`. The vector, `mu`, should have (x, y) coordinates interlaced, for example, if there were 2 poses and 2 landmarks, `mu` will look like the following, where `P` is the robot position and `L` the landmark position:\n```\nmu = matrix([[Px0],\n [Py0],\n [Px1],\n [Py1],\n [Lx0],\n [Ly0],\n [Lx1],\n [Ly1]])\n```\n\nYou can see that `mu` holds the poses first `(x0, y0), (x1, y1), ...,` then the landmark locations at the end of the matrix; we consider a `nx1` matrix to be a vector.\n\n## Generating an environment\n\nIn a real SLAM problem, you may be given a map that contains information about landmark locations, and in this example, we will make our own data using the `make_data` function, which generates a world grid with landmarks in it and then generates data by placing a robot in that world and moving and sensing over some numer of time steps. The `make_data` function relies on a correct implementation of robot move/sense functions, which, at this point, should be complete and in the `robot_class.py` file. The data is collected as an instantiated robot moves and senses in a world. Your SLAM function will take in this data as input. So, let's first create this data and explore how it represents the movement and sensor measurements that our robot takes.\n\n---",
"_____no_output_____"
],
[
"## Create the world\n\nUse the code below to generate a world of a specified size with randomly generated landmark locations. You can change these parameters and see how your implementation of SLAM responds! \n\n`data` holds the sensors measurements and motion of your robot over time. It stores the measurements as `data[i][0]` and the motion as `data[i][1]`.\n\n#### Helper functions\n\nYou will be working with the `robot` class that may look familiar from the first notebook, \n\nIn fact, in the `helpers.py` file, you can read the details of how data is made with the `make_data` function. It should look very similar to the robot move/sense cycle you've seen in the first notebook.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom helpers import make_data\n\n# your implementation of slam should work with the following inputs\n# feel free to change these input values and see how it responds!\n\n# world parameters\nnum_landmarks = 5 # number of landmarks\nN = 20 # time steps\nworld_size = 100.0 # size of world (square)\n\n# robot parameters\nmeasurement_range = 50.0 # range at which we can sense landmarks\nmotion_noise = 2.0 # noise in robot motion\nmeasurement_noise = 2.0 # noise in the measurements\ndistance = 20.0 # distance by which robot (intends to) move each iteratation \n\n\n# make_data instantiates a robot, AND generates random landmarks for a given world size and number of landmarks\ndata = make_data(N, num_landmarks, world_size, measurement_range, motion_noise, measurement_noise, distance)",
" \nLandmarks: [[27, 49], [24, 80], [77, 55], [82, 94], [5, 84]]\nRobot: [x=52.54557 y=12.32299]\n"
]
],
[
[
"### A note on `make_data`\n\nThe function above, `make_data`, takes in so many world and robot motion/sensor parameters because it is responsible for:\n1. Instantiating a robot (using the robot class)\n2. Creating a grid world with landmarks in it\n\n**This function also prints out the true location of landmarks and the *final* robot location, which you should refer back to when you test your implementation of SLAM.**\n\nThe `data` this returns is an array that holds information about **robot sensor measurements** and **robot motion** `(dx, dy)` that is collected over a number of time steps, `N`. You will have to use *only* these readings about motion and measurements to track a robot over time and find the determine the location of the landmarks using SLAM. We only print out the true landmark locations for comparison, later.\n\n\nIn `data` the measurement and motion data can be accessed from the first and second index in the columns of the data array. See the following code for an example, where `i` is the time step:\n```\nmeasurement = data[i][0]\nmotion = data[i][1]\n```\n",
"_____no_output_____"
]
],
[
[
"# print out some stats about the data\ntime_step = 0\n\nprint('Example measurements: \\n', data[time_step][0])\nprint('\\n')\nprint('Example motion: \\n', data[time_step][1])",
"Example measurements: \n [[0, 17.723394310557538, -28.25038995820606], [1, 10.975839083495508, 3.805498536419904], [2, -32.69981051478427, 7.083564822359085], [3, -37.3785349968737, 19.761086436435537], [4, 32.92792899955666, -43.31952688274464]]\n\n\nExample motion: \n [12.74006472314289, 15.417222540072842]\n"
]
],
[
[
"Try changing the value of `time_step`, you should see that the list of measurements varies based on what in the world the robot sees after it moves. As you know from the first notebook, the robot can only sense so far and with a certain amount of accuracy in the measure of distance between its location and the location of landmarks. The motion of the robot always is a vector with two values: one for x and one for y displacement. This structure will be useful to keep in mind as you traverse this data in your implementation of slam.",
"_____no_output_____"
],
[
"## Initialize Constraints\n\nOne of the most challenging tasks here will be to create and modify the constraint matrix and vector: omega and xi. In the second notebook, you saw an example of how omega and xi could hold all the values the define the relationships between robot poses `xi` and landmark positions `Li` in a 1D world, as seen below, where omega is the blue matrix and xi is the pink vector.\n\n<img src='images/motion_constraint.png' width=50% height=50% />\n\n\nIn *this* project, you are tasked with implementing constraints for a 2D world. We are referring to robot poses as `Px, Py` and landmark positions as `Lx, Ly`, and one way to approach this challenge is to add *both* x and y locations in the constraint matrices.\n\n<img src='images/constraints2D.png' width=50% height=50% />\n\nYou may also choose to create two of each omega and xi (one for x and one for y positions).",
"_____no_output_____"
],
[
"### TODO: Write a function that initializes omega and xi\n\nComplete the function `initialize_constraints` so that it returns `omega` and `xi` constraints for the starting position of the robot. Any values that we do not yet know should be initialized with the value `0`. You may assume that our robot starts out in exactly the middle of the world with 100% confidence (no motion or measurement noise at this point). The inputs `N` time steps, `num_landmarks`, and `world_size` should give you all the information you need to construct intial constraints of the correct size and starting values.\n\n*Depending on your approach you may choose to return one omega and one xi that hold all (x,y) positions *or* two of each (one for x values and one for y); choose whichever makes most sense to you!*",
"_____no_output_____"
]
],
[
[
"def initialize_constraints(N, num_landmarks, world_size):\n ''' This function takes in a number of time steps N, number of landmarks, and a world_size,\n and returns initialized constraint matrices, omega and xi.'''\n \n ## Recommended: Define and store the size (rows/cols) of the constraint matrix in a variable\n omega_size = (N+num_landmarks)*2\n ## TODO: Define the constraint matrix, Omega, with two initial \"strength\" values\n ## for the initial x, y location of our robot\n omega = np.zeros((omega_size, omega_size))\n omega[0,0] = 1.\n omega[1,1] = 1.\n \n ## TODO: Define the constraint *vector*, xi\n ## you can assume that the robot starts out in the middle of the world with 100% confidence\n xi = np.zeros((omega_size, 1))\n xi[0,0] = world_size/2\n xi[1,0] = world_size/2\n \n return omega, xi\n ",
"_____no_output_____"
]
],
[
[
"### Test as you go\n\nIt's good practice to test out your code, as you go. Since `slam` relies on creating and updating constraint matrices, `omega` and `xi` to account for robot sensor measurements and motion, let's check that they initialize as expected for any given parameters.\n\nBelow, you'll find some test code that allows you to visualize the results of your function `initialize_constraints`. We are using the [seaborn](https://seaborn.pydata.org/) library for visualization.\n\n**Please change the test values of N, landmarks, and world_size and see the results**. Be careful not to use these values as input into your final smal function.\n\nThis code assumes that you have created one of each constraint: `omega` and `xi`, but you can change and add to this code, accordingly. The constraints should vary in size with the number of time steps and landmarks as these values affect the number of poses a robot will take `(Px0,Py0,...Pxn,Pyn)` and landmark locations `(Lx0,Ly0,...Lxn,Lyn)` whose relationships should be tracked in the constraint matrices. Recall that `omega` holds the weights of each variable and `xi` holds the value of the sum of these variables, as seen in Notebook 2. You'll need the `world_size` to determine the starting pose of the robot in the world and fill in the initial values for `xi`.",
"_____no_output_____"
]
],
[
[
"# import data viz resources\nimport matplotlib.pyplot as plt\nfrom pandas import DataFrame\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
],
[
"# define a small N and world_size (small for ease of visualization)\nN_test = 5\nnum_landmarks_test = 2\nsmall_world = 10\n\n# initialize the constraints\ninitial_omega, initial_xi = initialize_constraints(N_test, num_landmarks_test, small_world)",
"_____no_output_____"
],
[
"# define figure size\nplt.rcParams[\"figure.figsize\"] = (10,7)\n\n# display omega\nsns.heatmap(DataFrame(initial_omega), cmap='Blues', annot=True, linewidths=.5)",
"_____no_output_____"
],
[
"# define figure size\nplt.rcParams[\"figure.figsize\"] = (1,7)\n\n# display xi\nsns.heatmap(DataFrame(initial_xi), cmap='Oranges', annot=True, linewidths=.5)",
"_____no_output_____"
]
],
[
[
"---\n## SLAM inputs \n\nIn addition to `data`, your slam function will also take in:\n* N - The number of time steps that a robot will be moving and sensing\n* num_landmarks - The number of landmarks in the world\n* world_size - The size (w/h) of your world\n* motion_noise - The noise associated with motion; the update confidence for motion should be `1.0/motion_noise`\n* measurement_noise - The noise associated with measurement/sensing; the update weight for measurement should be `1.0/measurement_noise`\n\n#### A note on noise\n\nRecall that `omega` holds the relative \"strengths\" or weights for each position variable, and you can update these weights by accessing the correct index in omega `omega[row][col]` and *adding/subtracting* `1.0/noise` where `noise` is measurement or motion noise. `Xi` holds actual position values, and so to update `xi` you'll do a similar addition process only using the actual value of a motion or measurement. So for a vector index `xi[row][0]` you will end up adding/subtracting one measurement or motion divided by their respective `noise`.\n\n### TODO: Implement Graph SLAM\n\nFollow the TODO's below to help you complete this slam implementation (these TODO's are in the recommended order), then test out your implementation! \n\n#### Updating with motion and measurements\n\nWith a 2D omega and xi structure as shown above (in earlier cells), you'll have to be mindful about how you update the values in these constraint matrices to account for motion and measurement constraints in the x and y directions. Recall that the solution to these matrices (which holds all values for robot poses `P` and landmark locations `L`) is the vector, `mu`, which can be computed at the end of the construction of omega and xi as the inverse of omega times xi: $\\mu = \\Omega^{-1}\\xi$\n\n**You may also choose to return the values of `omega` and `xi` if you want to visualize their final state!**",
"_____no_output_____"
]
],
[
[
"## TODO: Complete the code to implement SLAM\n\n## slam takes in 6 arguments and returns mu, \n## mu is the entire path traversed by a robot (all x,y poses) *and* all landmarks locations\ndef slam(data, N, num_landmarks, world_size, motion_noise, measurement_noise):\n \n ## TODO: Use your initilization to create constraint matrices, omega and xi\n omega, xi = initialize_constraints(N, num_landmarks, world_size)\n \n ## TODO: Iterate through each time step in the data\n ## get all the motion and measurement data as you iterate\n \n ## TODO: update the constraint matrix/vector to account for all *measurements*\n ## this should be a series of additions that take into account the measurement noise\n \n for r, x in enumerate(data):\n # robot row/column index \n robot_x = r * 2 # 0,2,4,6\n robot_y = r * 2 + 1 # 1,3,5,7\n \n # extract data\n measurments = x[0]\n motions = x[1]\n\n # measurments row/column index\n for m in measurments: # for every landmark robot saw\n landmark_x = 2*(N + m[0]) # skip x and y for motions and add index of measurment\n landmark_y = 2*(N + m[0]) + 1 # i.e. if N=3 in omega indexes 0-5 are for motions\n # so 2*(3 + 0) (first landmark) = 6th and 7th index for \n # measurments to landmark no. 0\n # update omega \n # x diagonal positive\n omega[robot_x][robot_x] += 1./measurement_noise\n omega[landmark_x][landmark_x] += 1./measurement_noise\n # x off-diagonal negative\n omega[robot_x][landmark_x] -= 1./measurement_noise\n omega[landmark_x][robot_x] -= 1./measurement_noise\n # y diagonal positive\n omega[robot_y][robot_y] += 1./measurement_noise\n omega[landmark_y][landmark_y] += 1./measurement_noise\n # y off-diagonal negative\n omega[robot_y][landmark_y] -= 1./measurement_noise\n omega[landmark_y][robot_y] -= 1./measurement_noise\n \n # update xi\n # x\n xi[robot_x][0] -= m[1]/measurement_noise\n xi[landmark_x][0] += m[1]/measurement_noise\n # y\n xi[robot_y][0] -= m[2]/measurement_noise\n xi[landmark_y][0] += m[2]/measurement_noise\n \n \n ## TODO: update the constraint matrix/vector to account for all *motion* and motion noise\n \n # update omega\n # x diagonal positive\n omega[robot_x][robot_x] += 1./motion_noise\n omega[robot_x+2][robot_x+2] += 1./motion_noise\n # x off-diagonal negative\n omega[robot_x][robot_x+2] -= 1./motion_noise\n omega[robot_x+2][robot_x] -= 1./motion_noise\n # y diagonal positive\n omega[robot_y][robot_y] += 1./motion_noise\n omega[robot_y+2][robot_y+2] += 1./motion_noise\n # y off-diagonal negative\n omega[robot_y][robot_y+2] -= 1./motion_noise\n omega[robot_y+2][robot_y] -= 1./motion_noise\n\n # update xi\n # x\n xi[robot_x][0] -= motions[0]/motion_noise\n xi[robot_x+2][0] += motions[0]/motion_noise\n # y\n xi[robot_y][0] -= motions[1]/motion_noise\n xi[robot_y+2][0] += motions[1]/motion_noise\n \n \n ## TODO: After iterating through all the data\n ## Compute the best estimate of poses and landmark positions\n ## using the formula, omega_inverse * Xi\n mu = np.dot(np.linalg.inv(omega), xi)\n \n return mu # return `mu`\n",
"_____no_output_____"
]
],
[
[
"## Helper functions\n\nTo check that your implementation of SLAM works for various inputs, we have provided two helper functions that will help display the estimated pose and landmark locations that your function has produced. First, given a result `mu` and number of time steps, `N`, we define a function that extracts the poses and landmarks locations and returns those as their own, separate lists. \n\nThen, we define a function that nicely print out these lists; both of these we will call, in the next step.\n",
"_____no_output_____"
]
],
[
[
"# a helper function that creates a list of poses and of landmarks for ease of printing\n# this only works for the suggested constraint architecture of interlaced x,y poses\ndef get_poses_landmarks(mu, N):\n # create a list of poses\n poses = []\n for i in range(N):\n poses.append((mu[2*i].item(), mu[2*i+1].item()))\n\n # create a list of landmarks\n landmarks = []\n for i in range(num_landmarks):\n landmarks.append((mu[2*(N+i)].item(), mu[2*(N+i)+1].item()))\n\n # return completed lists\n return poses, landmarks\n",
"_____no_output_____"
],
[
"def print_all(poses, landmarks):\n print('\\n')\n print('Estimated Poses:')\n for i in range(len(poses)):\n print('['+', '.join('%.3f'%p for p in poses[i])+']')\n print('\\n')\n print('Estimated Landmarks:')\n for i in range(len(landmarks)):\n print('['+', '.join('%.3f'%l for l in landmarks[i])+']')\n",
"_____no_output_____"
]
],
[
[
"## Run SLAM\n\nOnce you've completed your implementation of `slam`, see what `mu` it returns for different world sizes and different landmarks!\n\n### What to Expect\n\nThe `data` that is generated is random, but you did specify the number, `N`, or time steps that the robot was expected to move and the `num_landmarks` in the world (which your implementation of `slam` should see and estimate a position for. Your robot should also start with an estimated pose in the very center of your square world, whose size is defined by `world_size`.\n\nWith these values in mind, you should expect to see a result that displays two lists:\n1. **Estimated poses**, a list of (x, y) pairs that is exactly `N` in length since this is how many motions your robot has taken. The very first pose should be the center of your world, i.e. `[50.000, 50.000]` for a world that is 100.0 in square size.\n2. **Estimated landmarks**, a list of landmark positions (x, y) that is exactly `num_landmarks` in length. \n\n#### Landmark Locations\n\nIf you refer back to the printout of *exact* landmark locations when this data was created, you should see values that are very similar to those coordinates, but not quite (since `slam` must account for noise in motion and measurement).",
"_____no_output_____"
]
],
[
[
"# call your implementation of slam, passing in the necessary parameters\nmu = slam(data, N, num_landmarks, world_size, motion_noise, measurement_noise)\n\n# print out the resulting landmarks and poses\nif(mu is not None):\n # get the lists of poses and landmarks\n # and print them out\n poses, landmarks = get_poses_landmarks(mu, N)\n print_all(poses, landmarks)",
"\n\nEstimated Poses:\n[50.000, 50.000]\n[66.657, 41.281]\n[84.150, 33.051]\n[81.917, 53.188]\n[80.070, 71.769]\n[77.585, 90.159]\n[95.390, 82.391]\n[76.620, 93.080]\n[95.932, 89.961]\n[77.360, 83.761]\n[59.537, 78.143]\n[40.114, 70.998]\n[20.583, 64.374]\n[38.378, 59.404]\n[55.523, 52.431]\n[74.689, 45.329]\n[93.677, 38.672]\n[93.011, 17.997]\n[72.604, 15.558]\n[52.828, 12.577]\n\n\nEstimated Landmarks:\n[27.523, 48.763]\n[25.068, 80.098]\n[77.024, 53.981]\n[82.903, 93.065]\n[5.899, 83.787]\n"
]
],
[
[
"## Visualize the constructed world\n\nFinally, using the `display_world` code from the `helpers.py` file (which was also used in the first notebook), we can actually visualize what you have coded with `slam`: the final position of the robot and the positon of landmarks, created from only motion and measurement data!\n\n**Note that these should be very similar to the printed *true* landmark locations and final pose from our call to `make_data` early in this notebook.**",
"_____no_output_____"
]
],
[
[
"# import the helper function\nfrom helpers import display_world\n\n# Display the final world!\n\n# define figure size\nplt.rcParams[\"figure.figsize\"] = (20,20)\n\n# check if poses has been created\nif 'poses' in locals():\n # print out the last pose\n print('Last pose: ', poses[-1])\n # display the last position of the robot *and* the landmark positions\n display_world(int(world_size), poses[-1], landmarks)",
"Last pose: (52.82752373068452, 12.577356769153198)\n"
]
],
[
[
"### Question: How far away is your final pose (as estimated by `slam`) compared to the *true* final pose? Why do you think these poses are different?\n\nYou can find the true value of the final pose in one of the first cells where `make_data` was called. You may also want to look at the true landmark locations and compare them to those that were estimated by `slam`. Ask yourself: what do you think would happen if we moved and sensed more (increased N)? Or if we had lower/higher noise parameters.",
"_____no_output_____"
],
[
"**Answer**: It is very close dx = 0.3 and dy = 0.25. This poses are different due to uncertainty in motion and sensing (noise). If we had lower noise parameters the differences would be even smaller and vice versa - higher noise - larger differences. Sensing and moving for more time should also give us more exact results. ",
"_____no_output_____"
],
[
"## Testing\n\nTo confirm that your slam code works before submitting your project, it is suggested that you run it on some test data and cases. A few such cases have been provided for you, in the cells below. When you are ready, uncomment the test cases in the next cells (there are two test cases, total); your output should be **close-to or exactly** identical to the given results. If there are minor discrepancies it could be a matter of floating point accuracy or in the calculation of the inverse matrix.\n\n### Submit your project\n\nIf you pass these tests, it is a good indication that your project will pass all the specifications in the project rubric. Follow the submission instructions to officially submit!",
"_____no_output_____"
]
],
[
[
"# Here is the data and estimated outputs for test case 1\n\ntest_data1 = [[[[1, 19.457599255548065, 23.8387362100849], [2, -13.195807561967236, 11.708840328458608], [3, -30.0954905279171, 15.387879242505843]], [-12.2607279422326, -15.801093326936487]], [[[2, -0.4659930049620491, 28.088559771215664], [4, -17.866382374890936, -16.384904503932]], [-12.2607279422326, -15.801093326936487]], [[[4, -6.202512900833806, -1.823403210274639]], [-12.2607279422326, -15.801093326936487]], [[[4, 7.412136480918645, 15.388585962142429]], [14.008259661173426, 14.274756084260822]], [[[4, -7.526138813444998, -0.4563942429717849]], [14.008259661173426, 14.274756084260822]], [[[2, -6.299793150150058, 29.047830407717623], [4, -21.93551130411791, -13.21956810989039]], [14.008259661173426, 14.274756084260822]], [[[1, 15.796300959032276, 30.65769689694247], [2, -18.64370821983482, 17.380022987031367]], [14.008259661173426, 14.274756084260822]], [[[1, 0.40311325410337906, 14.169429532679855], [2, -35.069349468466235, 2.4945558982439957]], [14.008259661173426, 14.274756084260822]], [[[1, -16.71340983241936, -2.777000269543834]], [-11.006096015782283, 16.699276945166858]], [[[1, -3.611096830835776, -17.954019226763958]], [-19.693482634035977, 3.488085684573048]], [[[1, 18.398273354362416, -22.705102332550947]], [-19.693482634035977, 3.488085684573048]], [[[2, 2.789312482883833, -39.73720193121324]], [12.849049222879723, -15.326510824972983]], [[[1, 21.26897046581808, -10.121029799040915], [2, -11.917698965880655, -23.17711662602097], [3, -31.81167947898398, -16.7985673023331]], [12.849049222879723, -15.326510824972983]], [[[1, 10.48157743234859, 5.692957082575485], [2, -22.31488473554935, -5.389184118551409], [3, -40.81803984305378, -2.4703329790238118]], [12.849049222879723, -15.326510824972983]], [[[0, 10.591050242096598, -39.2051798967113], [1, -3.5675572049297553, 22.849456408289125], [2, -38.39251065320351, 7.288990306029511]], [12.849049222879723, -15.326510824972983]], [[[0, -3.6225556479370766, -25.58006865235512]], [-7.8874682868419965, -18.379005523261092]], [[[0, 1.9784503557879374, -6.5025974151499]], [-7.8874682868419965, -18.379005523261092]], [[[0, 10.050665232782423, 11.026385307998742]], [-17.82919359778298, 9.062000642947142]], [[[0, 26.526838150174818, -0.22563393232425621], [4, -33.70303936886652, 2.880339841013677]], [-17.82919359778298, 9.062000642947142]]]\n\n# # Test Case 1\n# #\n# Estimated Pose(s):\n# [50.000, 50.000]\n# [37.858, 33.921]\n# [25.905, 18.268]\n# [13.524, 2.224]\n# [27.912, 16.886]\n# [42.250, 30.994]\n# [55.992, 44.886]\n# [70.749, 59.867]\n# [85.371, 75.230]\n# [73.831, 92.354]\n# [53.406, 96.465]\n# [34.370, 100.134]\n# [48.346, 83.952]\n# [60.494, 68.338]\n# [73.648, 53.082]\n# [86.733, 38.197]\n# [79.983, 20.324]\n# [72.515, 2.837]\n# [54.993, 13.221]\n# [37.164, 22.283]\n\n\n# Estimated Landmarks:\n# [82.679, 13.435]\n# [70.417, 74.203]\n# [36.688, 61.431]\n# [18.705, 66.136]\n# [20.437, 16.983]\n\n\n## Uncomment the following three lines for test case 1 and compare the output to the values above ###\n\nmu_1 = slam(test_data1, 20, 5, 100.0, 2.0, 2.0)\nposes, landmarks = get_poses_landmarks(mu_1, 20)\nprint_all(poses, landmarks)",
"\n\nEstimated Poses:\n[50.000, 50.000]\n[37.973, 33.652]\n[26.185, 18.155]\n[13.745, 2.116]\n[28.097, 16.783]\n[42.384, 30.902]\n[55.831, 44.497]\n[70.857, 59.699]\n[85.697, 75.543]\n[74.011, 92.434]\n[53.544, 96.454]\n[34.525, 100.080]\n[48.623, 83.953]\n[60.197, 68.107]\n[73.778, 52.935]\n[87.132, 38.538]\n[80.303, 20.508]\n[72.798, 2.945]\n[55.245, 13.255]\n[37.416, 22.317]\n\n\nEstimated Landmarks:\n[82.956, 13.539]\n[70.495, 74.141]\n[36.740, 61.281]\n[18.698, 66.060]\n[20.635, 16.875]\n"
],
[
"# Here is the data and estimated outputs for test case 2\n\ntest_data2 = [[[[0, 26.543274387283322, -6.262538160312672], [3, 9.937396825799755, -9.128540360867689]], [18.92765331253674, -6.460955043986683]], [[[0, 7.706544739722961, -3.758467215445748], [1, 17.03954411948937, 31.705489938553438], [3, -11.61731288777497, -6.64964096716416]], [18.92765331253674, -6.460955043986683]], [[[0, -12.35130507136378, 2.585119104239249], [1, -2.563534536165313, 38.22159657838369], [3, -26.961236804740935, -0.4802312626141525]], [-11.167066095509824, 16.592065417497455]], [[[0, 1.4138633151721272, -13.912454837810632], [1, 8.087721200818589, 20.51845934354381], [3, -17.091723454402302, -16.521500551709707], [4, -7.414211721400232, 38.09191602674439]], [-11.167066095509824, 16.592065417497455]], [[[0, 12.886743222179561, -28.703968411636318], [1, 21.660953298391387, 3.4912891084614914], [3, -6.401401414569506, -32.321583037341625], [4, 5.034079343639034, 23.102207946092893]], [-11.167066095509824, 16.592065417497455]], [[[1, 31.126317672358578, -10.036784369535214], [2, -38.70878528420893, 7.4987265861424595], [4, 17.977218575473767, 6.150889254289742]], [-6.595520680493778, -18.88118393939265]], [[[1, 41.82460922922086, 7.847527392202475], [3, 15.711709540417502, -30.34633659912818]], [-6.595520680493778, -18.88118393939265]], [[[0, 40.18454208294434, -6.710999804403755], [3, 23.019508919299156, -10.12110867290604]], [-6.595520680493778, -18.88118393939265]], [[[3, 27.18579315312821, 8.067219022708391]], [-6.595520680493778, -18.88118393939265]], [[], [11.492663265706092, 16.36822198838621]], [[[3, 24.57154567653098, 13.461499960708197]], [11.492663265706092, 16.36822198838621]], [[[0, 31.61945290413707, 0.4272295085799329], [3, 16.97392299158991, -5.274596836133088]], [11.492663265706092, 16.36822198838621]], [[[0, 22.407381798735177, -18.03500068379259], [1, 29.642444125196995, 17.3794951934614], [3, 4.7969752441371645, -21.07505361639969], [4, 14.726069092569372, 32.75999422300078]], [11.492663265706092, 16.36822198838621]], [[[0, 10.705527984670137, -34.589764174299596], [1, 18.58772336795603, -0.20109708164787765], [3, -4.839806195049413, -39.92208742305105], [4, 4.18824810165454, 14.146847823548889]], [11.492663265706092, 16.36822198838621]], [[[1, 5.878492140223764, -19.955352450942357], [4, -7.059505455306587, -0.9740849280550585]], [19.628527845173146, 3.83678180657467]], [[[1, -11.150789592446378, -22.736641053247872], [4, -28.832815721158255, -3.9462962046291388]], [-19.841703647091965, 2.5113335861604362]], [[[1, 8.64427397916182, -20.286336970889053], [4, -5.036917727942285, -6.311739993868336]], [-5.946642674882207, -19.09548221169787]], [[[0, 7.151866679283043, -39.56103232616369], [1, 16.01535401373368, -3.780995345194027], [4, -3.04801331832137, 13.697362774960865]], [-5.946642674882207, -19.09548221169787]], [[[0, 12.872879480504395, -19.707592098123207], [1, 22.236710716903136, 16.331770792606406], [3, -4.841206109583004, -21.24604435851242], [4, 4.27111163223552, 32.25309748614184]], [-5.946642674882207, -19.09548221169787]]] \n\n\n## Test Case 2\n##\n# Estimated Pose(s):\n# [50.000, 50.000]\n# [69.035, 45.061]\n# [87.655, 38.971]\n# [76.084, 55.541]\n# [64.283, 71.684]\n# [52.396, 87.887]\n# [44.674, 68.948]\n# [37.532, 49.680]\n# [31.392, 30.893]\n# [24.796, 12.012]\n# [33.641, 26.440]\n# [43.858, 43.560]\n# [54.735, 60.659]\n# [65.884, 77.791]\n# [77.413, 94.554]\n# [96.740, 98.020]\n# [76.149, 99.586]\n# [70.211, 80.580]\n# [64.130, 61.270]\n# [58.183, 42.175]\n\n\n# Estimated Landmarks:\n# [76.777, 42.415]\n# [85.109, 76.850]\n# [13.687, 95.386]\n# [59.488, 39.149]\n# [69.283, 93.654]\n\n\n## Uncomment the following three lines for test case 2 and compare to the values above ###\n\nmu_2 = slam(test_data2, 20, 5, 100.0, 2.0, 2.0)\nposes, landmarks = get_poses_landmarks(mu_2, 20)\nprint_all(poses, landmarks)\n",
"\n\nEstimated Poses:\n[50.000, 50.000]\n[69.181, 45.665]\n[87.743, 39.703]\n[76.270, 56.311]\n[64.317, 72.176]\n[52.257, 88.154]\n[44.059, 69.401]\n[37.002, 49.918]\n[30.924, 30.955]\n[23.508, 11.419]\n[34.180, 27.133]\n[44.155, 43.846]\n[54.806, 60.920]\n[65.698, 78.546]\n[77.468, 95.626]\n[96.802, 98.821]\n[75.957, 99.971]\n[70.200, 81.181]\n[64.054, 61.723]\n[58.107, 42.628]\n\n\nEstimated Landmarks:\n[76.779, 42.887]\n[85.065, 77.438]\n[13.548, 95.652]\n[59.449, 39.595]\n[69.263, 94.240]\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
]
] |
4a16accbceecb27e46a221c51e61f73c74777c46
| 35,906 |
ipynb
|
Jupyter Notebook
|
how-to-use-azureml/automated-machine-learning/local-run-classification-credit-card-fraud/auto-ml-classification-credit-card-fraud-local.ipynb
|
AaronZhangAus/MachineLearningNotebooks
|
aed952c561f8dcfbe2fcacf92b9afea6810961cc
|
[
"MIT"
] | 2 |
2021-06-25T17:45:54.000Z
|
2021-06-26T02:38:06.000Z
|
how-to-use-azureml/automated-machine-learning/local-run-classification-credit-card-fraud/auto-ml-classification-credit-card-fraud-local.ipynb
|
AaronZhangAus/MachineLearningNotebooks
|
aed952c561f8dcfbe2fcacf92b9afea6810961cc
|
[
"MIT"
] | null | null | null |
how-to-use-azureml/automated-machine-learning/local-run-classification-credit-card-fraud/auto-ml-classification-credit-card-fraud-local.ipynb
|
AaronZhangAus/MachineLearningNotebooks
|
aed952c561f8dcfbe2fcacf92b9afea6810961cc
|
[
"MIT"
] | 1 |
2021-02-04T04:45:45.000Z
|
2021-02-04T04:45:45.000Z
| 41.129439 | 429 | 0.590876 |
[
[
[
"Copyright (c) Microsoft Corporation. All rights reserved.\n\nLicensed under the MIT License.",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# Automated Machine Learning\n_**Classification of credit card fraudulent transactions with local run **_\n\n## Contents\n1. [Introduction](#Introduction)\n1. [Setup](#Setup)\n1. [Train](#Train)\n1. [Results](#Results)\n1. [Test](#Tests)\n1. [Explanation](#Explanation)\n1. [Acknowledgements](#Acknowledgements)",
"_____no_output_____"
],
[
"## Introduction\n\nIn this example we use the associated credit card dataset to showcase how you can use AutoML for a simple classification problem. The goal is to predict if a credit card transaction is considered a fraudulent charge.\n\nThis notebook is using the local machine compute to train the model.\n\nIf you are using an Azure Machine Learning Compute Instance, you are all set. Otherwise, go through the [configuration](../../../configuration.ipynb) notebook first if you haven't already to establish your connection to the AzureML Workspace. \n\nIn this notebook you will learn how to:\n1. Create an experiment using an existing workspace.\n2. Configure AutoML using `AutoMLConfig`.\n3. Train the model.\n4. Explore the results.\n5. Test the fitted model.\n6. Explore any model's explanation and explore feature importance in azure portal.\n7. Create an AKS cluster, deploy the webservice of AutoML scoring model and the explainer model to the AKS and consume the web service.",
"_____no_output_____"
],
[
"## Setup\n\nAs part of the setup you have already created an Azure ML `Workspace` object. For Automated ML you will need to create an `Experiment` object, which is a named object in a `Workspace` used to run experiments.",
"_____no_output_____"
]
],
[
[
"import logging\n\nfrom matplotlib import pyplot as plt\nimport pandas as pd\n\nimport azureml.core\nfrom azureml.core.experiment import Experiment\nfrom azureml.core.workspace import Workspace\nfrom azureml.core.dataset import Dataset\nfrom azureml.train.automl import AutoMLConfig\nfrom azureml.interpret import ExplanationClient",
"_____no_output_____"
]
],
[
[
"This sample notebook may use features that are not available in previous versions of the Azure ML SDK.",
"_____no_output_____"
]
],
[
[
"print(\"This notebook was created using version 1.20.0 of the Azure ML SDK\")\nprint(\"You are currently using version\", azureml.core.VERSION, \"of the Azure ML SDK\")",
"_____no_output_____"
],
[
"ws = Workspace.from_config()\n\n# choose a name for experiment\nexperiment_name = 'automl-classification-ccard-local'\n\nexperiment=Experiment(ws, experiment_name)\n\noutput = {}\noutput['Subscription ID'] = ws.subscription_id\noutput['Workspace'] = ws.name\noutput['Resource Group'] = ws.resource_group\noutput['Location'] = ws.location\noutput['Experiment Name'] = experiment.name\npd.set_option('display.max_colwidth', -1)\noutputDf = pd.DataFrame(data = output, index = [''])\noutputDf.T",
"_____no_output_____"
]
],
[
[
"### Load Data\n\nLoad the credit card dataset from a csv file containing both training features and labels. The features are inputs to the model, while the training labels represent the expected output of the model. Next, we'll split the data using random_split and extract the training data for the model.",
"_____no_output_____"
]
],
[
[
"data = \"https://automlsamplenotebookdata.blob.core.windows.net/automl-sample-notebook-data/creditcard.csv\"\ndataset = Dataset.Tabular.from_delimited_files(data)\ntraining_data, validation_data = dataset.random_split(percentage=0.8, seed=223)\nlabel_column_name = 'Class'",
"_____no_output_____"
]
],
[
[
"## Train\n\nInstantiate a AutoMLConfig object. This defines the settings and data used to run the experiment.\n\n|Property|Description|\n|-|-|\n|**task**|classification or regression|\n|**primary_metric**|This is the metric that you want to optimize. Classification supports the following primary metrics: <br><i>accuracy</i><br><i>AUC_weighted</i><br><i>average_precision_score_weighted</i><br><i>norm_macro_recall</i><br><i>precision_score_weighted</i>|\n|**enable_early_stopping**|Stop the run if the metric score is not showing improvement.|\n|**n_cross_validations**|Number of cross validation splits.|\n|**training_data**|Input dataset, containing both features and label column.|\n|**label_column_name**|The name of the label column.|\n\n**_You can find more information about primary metrics_** [here](https://docs.microsoft.com/en-us/azure/machine-learning/service/how-to-configure-auto-train#primary-metric)",
"_____no_output_____"
]
],
[
[
"automl_settings = {\n \"n_cross_validations\": 3,\n \"primary_metric\": 'average_precision_score_weighted',\n \"experiment_timeout_hours\": 0.25, # This is a time limit for testing purposes, remove it for real use cases, this will drastically limit ability to find the best model possible\n \"verbosity\": logging.INFO,\n \"enable_stack_ensemble\": False\n}\n\nautoml_config = AutoMLConfig(task = 'classification',\n debug_log = 'automl_errors.log',\n training_data = training_data,\n label_column_name = label_column_name,\n **automl_settings\n )",
"_____no_output_____"
]
],
[
[
"Call the `submit` method on the experiment object and pass the run configuration. Depending on the data and the number of iterations this can run for a while.\nIn this example, we specify `show_output = True` to print currently running iterations to the console.",
"_____no_output_____"
]
],
[
[
"local_run = experiment.submit(automl_config, show_output = True)",
"_____no_output_____"
],
[
"# If you need to retrieve a run that already started, use the following code\n#from azureml.train.automl.run import AutoMLRun\n#local_run = AutoMLRun(experiment = experiment, run_id = '<replace with your run id>')",
"_____no_output_____"
],
[
"local_run",
"_____no_output_____"
]
],
[
[
"## Results",
"_____no_output_____"
],
[
"#### Widget for Monitoring Runs\n\nThe widget will first report a \"loading\" status while running the first iteration. After completing the first iteration, an auto-updating graph and table will be shown. The widget will refresh once per minute, so you should see the graph update as child runs complete.\n\n**Note:** The widget displays a link at the bottom. Use this link to open a web interface to explore the individual run details",
"_____no_output_____"
]
],
[
[
"from azureml.widgets import RunDetails\nRunDetails(local_run).show()",
"_____no_output_____"
]
],
[
[
"### Analyze results\n\n#### Retrieve the Best Model\n\nBelow we select the best pipeline from our iterations. The `get_output` method on `automl_classifier` returns the best run and the fitted model for the last invocation. Overloads on `get_output` allow you to retrieve the best run and fitted model for *any* logged metric or for a particular *iteration*.",
"_____no_output_____"
]
],
[
[
"best_run, fitted_model = local_run.get_output()\nfitted_model",
"_____no_output_____"
]
],
[
[
"#### Print the properties of the model\nThe fitted_model is a python object and you can read the different properties of the object.\n",
"_____no_output_____"
],
[
"## Tests\n\nNow that the model is trained, split the data in the same way the data was split for training (The difference here is the data is being split locally) and then run the test data through the trained model to get the predicted values.",
"_____no_output_____"
]
],
[
[
"# convert the test data to dataframe\nX_test_df = validation_data.drop_columns(columns=[label_column_name]).to_pandas_dataframe()\ny_test_df = validation_data.keep_columns(columns=[label_column_name], validate=True).to_pandas_dataframe()",
"_____no_output_____"
],
[
"# call the predict functions on the model\ny_pred = fitted_model.predict(X_test_df)\ny_pred",
"_____no_output_____"
]
],
[
[
"### Calculate metrics for the prediction\n\nNow visualize the data on a scatter plot to show what our truth (actual) values are compared to the predicted values \nfrom the trained model that was returned.",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import confusion_matrix\nimport numpy as np\nimport itertools\n\ncf =confusion_matrix(y_test_df.values,y_pred)\nplt.imshow(cf,cmap=plt.cm.Blues,interpolation='nearest')\nplt.colorbar()\nplt.title('Confusion Matrix')\nplt.xlabel('Predicted')\nplt.ylabel('Actual')\nclass_labels = ['False','True']\ntick_marks = np.arange(len(class_labels))\nplt.xticks(tick_marks,class_labels)\nplt.yticks([-0.5,0,1,1.5],['','False','True',''])\n# plotting text value inside cells\nthresh = cf.max() / 2.\nfor i,j in itertools.product(range(cf.shape[0]),range(cf.shape[1])):\n plt.text(j,i,format(cf[i,j],'d'),horizontalalignment='center',color='white' if cf[i,j] >thresh else 'black')\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Explanation\nIn this section, we will show how to compute model explanations and visualize the explanations using azureml-interpret package. We will also show how to run the automl model and the explainer model through deploying an AKS web service.\n\nBesides retrieving an existing model explanation for an AutoML model, you can also explain your AutoML model with different test data. The following steps will allow you to compute and visualize engineered feature importance based on your test data.\n\n### Run the explanation\n#### Download the engineered feature importance from artifact store\nYou can use ExplanationClient to download the engineered feature explanations from the artifact store of the best_run. You can also use azure portal url to view the dash board visualization of the feature importance values of the engineered features.",
"_____no_output_____"
]
],
[
[
"client = ExplanationClient.from_run(best_run)\nengineered_explanations = client.download_model_explanation(raw=False)\nprint(engineered_explanations.get_feature_importance_dict())\nprint(\"You can visualize the engineered explanations under the 'Explanations (preview)' tab in the AutoML run at:-\\n\" + best_run.get_portal_url())",
"_____no_output_____"
]
],
[
[
"#### Download the raw feature importance from artifact store\nYou can use ExplanationClient to download the raw feature explanations from the artifact store of the best_run. You can also use azure portal url to view the dash board visualization of the feature importance values of the raw features.",
"_____no_output_____"
]
],
[
[
"raw_explanations = client.download_model_explanation(raw=True)\nprint(raw_explanations.get_feature_importance_dict())\nprint(\"You can visualize the raw explanations under the 'Explanations (preview)' tab in the AutoML run at:-\\n\" + best_run.get_portal_url())",
"_____no_output_____"
]
],
[
[
"#### Retrieve any other AutoML model from training",
"_____no_output_____"
]
],
[
[
"automl_run, fitted_model = local_run.get_output(metric='accuracy')",
"_____no_output_____"
]
],
[
[
"#### Setup the model explanations for AutoML models\nThe fitted_model can generate the following which will be used for getting the engineered explanations using automl_setup_model_explanations:-\n\n1. Featurized data from train samples/test samples\n2. Gather engineered name lists\n3. Find the classes in your labeled column in classification scenarios\n\nThe automl_explainer_setup_obj contains all the structures from above list.",
"_____no_output_____"
]
],
[
[
"X_train = training_data.drop_columns(columns=[label_column_name])\ny_train = training_data.keep_columns(columns=[label_column_name], validate=True)\nX_test = validation_data.drop_columns(columns=[label_column_name])",
"_____no_output_____"
],
[
"from azureml.train.automl.runtime.automl_explain_utilities import automl_setup_model_explanations\n\nautoml_explainer_setup_obj = automl_setup_model_explanations(fitted_model, X=X_train, \n X_test=X_test, y=y_train, \n task='classification')",
"_____no_output_____"
]
],
[
[
"#### Initialize the Mimic Explainer for feature importance\nFor explaining the AutoML models, use the MimicWrapper from azureml-interpret package. The MimicWrapper can be initialized with fields in automl_explainer_setup_obj, your workspace and a surrogate model to explain the AutoML model (fitted_model here). The MimicWrapper also takes the automl_run object where engineered explanations will be uploaded.",
"_____no_output_____"
]
],
[
[
"from interpret.ext.glassbox import LGBMExplainableModel\nfrom azureml.interpret.mimic_wrapper import MimicWrapper\nexplainer = MimicWrapper(ws, automl_explainer_setup_obj.automl_estimator,\n explainable_model=automl_explainer_setup_obj.surrogate_model, \n init_dataset=automl_explainer_setup_obj.X_transform, run=automl_run,\n features=automl_explainer_setup_obj.engineered_feature_names, \n feature_maps=[automl_explainer_setup_obj.feature_map],\n classes=automl_explainer_setup_obj.classes,\n explainer_kwargs=automl_explainer_setup_obj.surrogate_model_params)",
"_____no_output_____"
]
],
[
[
"#### Use Mimic Explainer for computing and visualizing engineered feature importance\nThe explain() method in MimicWrapper can be called with the transformed test samples to get the feature importance for the generated engineered features. You can also use azure portal url to view the dash board visualization of the feature importance values of the engineered features.",
"_____no_output_____"
]
],
[
[
"# Compute the engineered explanations\nengineered_explanations = explainer.explain(['local', 'global'], eval_dataset=automl_explainer_setup_obj.X_test_transform)\nprint(engineered_explanations.get_feature_importance_dict())\nprint(\"You can visualize the engineered explanations under the 'Explanations (preview)' tab in the AutoML run at:-\\n\" + automl_run.get_portal_url())",
"_____no_output_____"
]
],
[
[
"#### Use Mimic Explainer for computing and visualizing raw feature importance\nThe explain() method in MimicWrapper can be called with the transformed test samples to get the feature importance for the original features in your data. You can also use azure portal url to view the dash board visualization of the feature importance values of the original/raw features.",
"_____no_output_____"
]
],
[
[
"# Compute the raw explanations\nraw_explanations = explainer.explain(['local', 'global'], get_raw=True,\n raw_feature_names=automl_explainer_setup_obj.raw_feature_names,\n eval_dataset=automl_explainer_setup_obj.X_test_transform,\n raw_eval_dataset=automl_explainer_setup_obj.X_test_raw)\nprint(raw_explanations.get_feature_importance_dict())\nprint(\"You can visualize the raw explanations under the 'Explanations (preview)' tab in the AutoML run at:-\\n\" + automl_run.get_portal_url())",
"_____no_output_____"
]
],
[
[
"#### Initialize the scoring Explainer, save and upload it for later use in scoring explanation",
"_____no_output_____"
]
],
[
[
"from azureml.interpret.scoring.scoring_explainer import TreeScoringExplainer\nimport joblib\n\n# Initialize the ScoringExplainer\nscoring_explainer = TreeScoringExplainer(explainer.explainer, feature_maps=[automl_explainer_setup_obj.feature_map])\n\n# Pickle scoring explainer locally to './scoring_explainer.pkl'\nscoring_explainer_file_name = 'scoring_explainer.pkl'\nwith open(scoring_explainer_file_name, 'wb') as stream:\n joblib.dump(scoring_explainer, stream)\n\n# Upload the scoring explainer to the automl run\nautoml_run.upload_file('outputs/scoring_explainer.pkl', scoring_explainer_file_name)",
"_____no_output_____"
]
],
[
[
"### Deploying the scoring and explainer models to a web service to Azure Kubernetes Service (AKS)\n\nWe use the TreeScoringExplainer from azureml.interpret package to create the scoring explainer which will be used to compute the raw and engineered feature importances at the inference time. In the cell below, we register the AutoML model and the scoring explainer with the Model Management Service.",
"_____no_output_____"
]
],
[
[
"# Register trained automl model present in the 'outputs' folder in the artifacts\noriginal_model = automl_run.register_model(model_name='automl_model', \n model_path='outputs/model.pkl')\nscoring_explainer_model = automl_run.register_model(model_name='scoring_explainer',\n model_path='outputs/scoring_explainer.pkl')",
"_____no_output_____"
]
],
[
[
"#### Create the conda dependencies for setting up the service\n\nWe need to download the conda dependencies using the automl_run object.",
"_____no_output_____"
]
],
[
[
"from azureml.automl.core.shared import constants\nfrom azureml.core.environment import Environment\n\nautoml_run.download_file(constants.CONDA_ENV_FILE_PATH, 'myenv.yml')\nmyenv = Environment.from_conda_specification(name=\"myenv\", file_path=\"myenv.yml\")\nmyenv",
"_____no_output_____"
]
],
[
[
"#### Write the Entry Script\nWrite the script that will be used to predict on your model",
"_____no_output_____"
]
],
[
[
"%%writefile score.py\nimport joblib\nimport pandas as pd\nfrom azureml.core.model import Model\nfrom azureml.train.automl.runtime.automl_explain_utilities import automl_setup_model_explanations\n\n\ndef init():\n global automl_model\n global scoring_explainer\n\n # Retrieve the path to the model file using the model name\n # Assume original model is named original_prediction_model\n automl_model_path = Model.get_model_path('automl_model')\n scoring_explainer_path = Model.get_model_path('scoring_explainer')\n\n automl_model = joblib.load(automl_model_path)\n scoring_explainer = joblib.load(scoring_explainer_path)\n\n\ndef run(raw_data):\n data = pd.read_json(raw_data, orient='records') \n # Make prediction\n predictions = automl_model.predict(data)\n # Setup for inferencing explanations\n automl_explainer_setup_obj = automl_setup_model_explanations(automl_model,\n X_test=data, task='classification')\n # Retrieve model explanations for engineered explanations\n engineered_local_importance_values = scoring_explainer.explain(automl_explainer_setup_obj.X_test_transform)\n # Retrieve model explanations for raw explanations\n raw_local_importance_values = scoring_explainer.explain(automl_explainer_setup_obj.X_test_transform, get_raw=True)\n # You can return any data type as long as it is JSON-serializable\n return {'predictions': predictions.tolist(),\n 'engineered_local_importance_values': engineered_local_importance_values,\n 'raw_local_importance_values': raw_local_importance_values}\n",
"_____no_output_____"
]
],
[
[
"#### Create the InferenceConfig \nCreate the inference config that will be used when deploying the model",
"_____no_output_____"
]
],
[
[
"from azureml.core.model import InferenceConfig\n\ninf_config = InferenceConfig(entry_script='score.py', environment=myenv)",
"_____no_output_____"
]
],
[
[
"#### Provision the AKS Cluster\nThis is a one time setup. You can reuse this cluster for multiple deployments after it has been created. If you delete the cluster or the resource group that contains it, then you would have to recreate it.",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import ComputeTarget, AksCompute\nfrom azureml.core.compute_target import ComputeTargetException\n\n# Choose a name for your cluster.\naks_name = 'scoring-explain'\n\n# Verify that cluster does not exist already\ntry:\n aks_target = ComputeTarget(workspace=ws, name=aks_name)\n print('Found existing cluster, use it.')\nexcept ComputeTargetException:\n prov_config = AksCompute.provisioning_configuration(vm_size='STANDARD_D3_V2')\n aks_target = ComputeTarget.create(workspace=ws, \n name=aks_name,\n provisioning_configuration=prov_config)\naks_target.wait_for_completion(show_output=True)",
"_____no_output_____"
]
],
[
[
"#### Deploy web service to AKS",
"_____no_output_____"
]
],
[
[
"# Set the web service configuration (using default here)\nfrom azureml.core.webservice import AksWebservice\nfrom azureml.core.model import Model\n\naks_config = AksWebservice.deploy_configuration()",
"_____no_output_____"
],
[
"aks_service_name ='model-scoring-local-aks'\n\naks_service = Model.deploy(workspace=ws,\n name=aks_service_name,\n models=[scoring_explainer_model, original_model],\n inference_config=inf_config,\n deployment_config=aks_config,\n deployment_target=aks_target)\n\naks_service.wait_for_deployment(show_output = True)\nprint(aks_service.state)",
"_____no_output_____"
]
],
[
[
"#### View the service logs",
"_____no_output_____"
]
],
[
[
"aks_service.get_logs()",
"_____no_output_____"
]
],
[
[
"#### Consume the web service using run method to do the scoring and explanation of scoring.\nWe test the web sevice by passing data. Run() method retrieves API keys behind the scenes to make sure that call is authenticated.",
"_____no_output_____"
]
],
[
[
"# Serialize the first row of the test data into json\nX_test_json = X_test_df[:1].to_json(orient='records')\nprint(X_test_json)\n\n# Call the service to get the predictions and the engineered and raw explanations\noutput = aks_service.run(X_test_json)\n\n# Print the predicted value\nprint('predictions:\\n{}\\n'.format(output['predictions']))\n# Print the engineered feature importances for the predicted value\nprint('engineered_local_importance_values:\\n{}\\n'.format(output['engineered_local_importance_values']))\n# Print the raw feature importances for the predicted value\nprint('raw_local_importance_values:\\n{}\\n'.format(output['raw_local_importance_values']))\n",
"_____no_output_____"
]
],
[
[
"#### Clean up\nDelete the service.",
"_____no_output_____"
]
],
[
[
"aks_service.delete()",
"_____no_output_____"
]
],
[
[
"## Acknowledgements",
"_____no_output_____"
],
[
"This Credit Card fraud Detection dataset is made available under the Open Database License: http://opendatacommons.org/licenses/odbl/1.0/. Any rights in individual contents of the database are licensed under the Database Contents License: http://opendatacommons.org/licenses/dbcl/1.0/ and is available at: https://www.kaggle.com/mlg-ulb/creditcardfraud\n\n\nThe dataset has been collected and analysed during a research collaboration of Worldline and the Machine Learning Group (http://mlg.ulb.ac.be) of ULB (Université Libre de Bruxelles) on big data mining and fraud detection. More details on current and past projects on related topics are available on https://www.researchgate.net/project/Fraud-detection-5 and the page of the DefeatFraud project\nPlease cite the following works: \n•\tAndrea Dal Pozzolo, Olivier Caelen, Reid A. Johnson and Gianluca Bontempi. Calibrating Probability with Undersampling for Unbalanced Classification. In Symposium on Computational Intelligence and Data Mining (CIDM), IEEE, 2015\n•\tDal Pozzolo, Andrea; Caelen, Olivier; Le Borgne, Yann-Ael; Waterschoot, Serge; Bontempi, Gianluca. Learned lessons in credit card fraud detection from a practitioner perspective, Expert systems with applications,41,10,4915-4928,2014, Pergamon\n•\tDal Pozzolo, Andrea; Boracchi, Giacomo; Caelen, Olivier; Alippi, Cesare; Bontempi, Gianluca. Credit card fraud detection: a realistic modeling and a novel learning strategy, IEEE transactions on neural networks and learning systems,29,8,3784-3797,2018,IEEE\no\tDal Pozzolo, Andrea Adaptive Machine learning for credit card fraud detection ULB MLG PhD thesis (supervised by G. Bontempi)\n•\tCarcillo, Fabrizio; Dal Pozzolo, Andrea; Le Borgne, Yann-Aël; Caelen, Olivier; Mazzer, Yannis; Bontempi, Gianluca. Scarff: a scalable framework for streaming credit card fraud detection with Spark, Information fusion,41, 182-194,2018,Elsevier\n•\tCarcillo, Fabrizio; Le Borgne, Yann-Aël; Caelen, Olivier; Bontempi, Gianluca. Streaming active learning strategies for real-life credit card fraud detection: assessment and visualization, International Journal of Data Science and Analytics, 5,4,285-300,2018,Springer International Publishing",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a16adce6bc28d16620f4070d14cebb4875f06db
| 33,442 |
ipynb
|
Jupyter Notebook
|
datashader-work/datashader-examples/user_guide/1_Plotting_Pitfalls.ipynb
|
ventureBorbot/Data-Analysis
|
a44122aeb489fd97488c105b951c06d01d2db894
|
[
"MIT"
] | 4,358 |
2017-12-29T17:56:07.000Z
|
2022-03-30T15:14:57.000Z
|
datashader-work/datashader-examples/user_guide/1_Plotting_Pitfalls.ipynb
|
ventureBorbot/Data-Analysis
|
a44122aeb489fd97488c105b951c06d01d2db894
|
[
"MIT"
] | 61 |
2018-01-18T17:50:46.000Z
|
2022-03-09T20:16:01.000Z
|
datashader-work/datashader-examples/user_guide/1_Plotting_Pitfalls.ipynb
|
ventureBorbot/Data-Analysis
|
a44122aeb489fd97488c105b951c06d01d2db894
|
[
"MIT"
] | 3,689 |
2017-12-29T17:57:36.000Z
|
2022-03-29T12:26:03.000Z
| 66.353175 | 1,496 | 0.69021 |
[
[
[
"## Common plotting pitfalls that get worse with large data\n\nWhen working with large datasets, visualizations are often the only way available to understand the properties of that dataset -- there are simply too many data points to examine each one! Thus it is very important to be aware of some common plotting problems that are minor inconveniences with small datasets but very serious problems with larger ones.\n\nWe'll cover:\n\n1. [Overplotting](#1.-Overplotting)\n2. [Oversaturation](#2.-Oversaturation)\n3. [Undersampling](#3.-Undersampling)\n4. [Undersaturation](#4.-Undersaturation)\n5. [Underutilized range](#5.-Underutilized-range)\n6. [Nonuniform colormapping](#6.-Nonuniform-colormapping)\n\nYou can [skip to the end](#Summary) if you just want to see an illustration of these problems.\n\nThis notebook requires [HoloViews](http://holoviews.org), [colorcet](https://github.com/bokeh/colorcet), and matplotlib, and optionally scikit-image, which can be installed with:\n\n```\nconda install -c bokeh -c ioam holoviews colorcet matplotlib scikit-image\n```\n\nWe'll first load the plotting libraries and set up some defaults:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nnp.random.seed(42)\n\nimport holoviews as hv\nfrom holoviews.operation.datashader import datashade\nfrom holoviews import opts, dim\nhv.extension('matplotlib')\n\nfrom colorcet import fire\ndatashade.cmap=fire[50:]",
"_____no_output_____"
],
[
"opts.defaults(\n opts.Image(cmap=\"gray_r\", axiswise=True),\n opts.Points(cmap=\"bwr\", edgecolors='k', s=50, alpha=1.0), # Remove color_index=2\n opts.RGB(bgcolor=\"black\", show_grid=False),\n opts.Scatter3D(color=dim('c'), fig_size=250, cmap='bwr', edgecolor='k', s=50, alpha=1.0)) #color_index=3",
"_____no_output_____"
]
],
[
[
"### 1. Overplotting\n\nLet's consider plotting some 2D data points that come from two separate categories, here plotted as blue and red in **A** and **B** below. When the two categories are overlaid, the appearance of the result can be very different depending on which one is plotted first:",
"_____no_output_____"
]
],
[
[
"def blue_points(offset=0.5,pts=300):\n blues = (np.random.normal( offset,size=pts), np.random.normal( offset,size=pts), -1 * np.ones((pts)))\n return hv.Points(blues, vdims=['c']).opts(color=dim('c'))\n \ndef red_points(offset=0.5,pts=300):\n reds = (np.random.normal(-offset,size=pts), np.random.normal(-offset,size=pts), 1*np.ones((pts)))\n return hv.Points(reds, vdims=['c']).opts(color=dim('c'))\n\nblues, reds = blue_points(), red_points()\nblues + reds + (reds * blues) + (blues * reds)",
"_____no_output_____"
]
],
[
[
"Plots **C** and **D** shown the same distribution of points, yet they give a very different impression of which category is more common, which can lead to incorrect decisions based on this data. Of course, both are equally common in this case, so neither **C** nor **D** accurately reflects the data. The cause for this problem is simply occlusion:",
"_____no_output_____"
]
],
[
[
"hmap = hv.HoloMap({0:blues,0.000001:reds,1:blues,2:reds}, kdims=['level'])\nhv.Scatter3D(hmap.table(), kdims=['x','y','level'], vdims=['c'])",
"_____no_output_____"
]
],
[
[
"Occlusion of data by other data is called **overplotting** or **overdrawing**, and it occurs whenever a datapoint or curve is plotted on top of another datapoint or curve, obscuring it. It's thus a problem not just for scatterplots, as here, but for curve plots, 3D surface plots, 3D bar graphs, and any other plot type where data can be obscured.\n\n\n### 2. Oversaturation\n\nYou can reduce problems with overplotting by using transparency/opacity, via the alpha parameter provided to control opacity in most plotting programs. E.g. if alpha is 0.1, full color saturation will be achieved only when 10 points overlap, reducing the effects of plot ordering but making it harder to see individual points:",
"_____no_output_____"
]
],
[
[
"layout = blues + reds + (reds * blues) + (blues * reds)\nlayout.opts(opts.Points(s=50, alpha=0.1))",
"_____no_output_____"
]
],
[
[
"Here **C **and **D **look very similar (as they should, since the distributions are identical), but there are still a few locations with **oversaturation**, a problem that will occur when more than 10 points overlap. In this example the oversaturated points are located near the middle of the plot, but the only way to know whether they are there would be to plot both versions and compare, or to examine the pixel values to see if any have reached full saturation (a necessary but not sufficient condition for oversaturation). Locations where saturation has been reached have problems similar to overplotting, because only the last 10 points plotted will affect the final color (for alpha of 0.1).\n\nWorse, even if one has set the alpha value to approximately or usually avoid oversaturation, as in the plot above, the correct value depends on the dataset. If there are more points overlapping in that particular region, a manually adjusted alpha setting that worked well for a previous dataset will systematically misrepresent the new dataset:",
"_____no_output_____"
]
],
[
[
"blues, reds = blue_points(pts=600), red_points(pts=600)\nlayout = blues + reds + (reds * blues) + (blues * reds)\nlayout.opts(opts.Points(alpha=0.1))",
"_____no_output_____"
]
],
[
[
"Here **C **and **D **again look qualitatively different, yet still represent the same distributions. Since we're assuming that the point of the visualization is to reveal the underlying dataset, having to tune visualization parameters manually based on the properties of the dataset itself is a serious problem.\n\nTo make it even more complicated, the correct alpha also depends on the dot size, because smaller dots have less overlap for the same dataset. With smaller dots, **C **and **D **look more similar, but the color of the dots is now difficult to see in all cases because the dots are too transparent for this size:",
"_____no_output_____"
]
],
[
[
"layout = blues + reds + (reds * blues) + (blues * reds)\nlayout.opts(opts.Points(s=10, alpha=0.1, edgecolor=None))",
"_____no_output_____"
]
],
[
[
"As you can see, it is very difficult to find settings for the dotsize and alpha parameters that correctly reveal the data, even for relatively small and obvious datasets like these. With larger datasets with unknown contents, it is difficult to detect that such problems are occuring, leading to false conclusions based on inappropriately visualized data.\n\n### 3. Undersampling\n\nWith a single category instead of the multiple categories shown above, oversaturation simply obscures spatial differences in density. For instance, 10, 20, and 2000 single-category points overlapping will all look the same visually, for alpha=0.1. Let's again consider an example that has a sum of two normal distributions slightly offset from one another, but no longer using color to separate them into categories:",
"_____no_output_____"
]
],
[
[
"def gaussians(specs=[(1.5,0,1.0),(-1.5,0,1.0)],num=100):\n \"\"\"\n A concatenated list of points taken from 2D Gaussian distributions.\n Each distribution is specified as a tuple (x,y,s), where x,y is the mean\n and s is the standard deviation. Defaults to two horizontally\n offset unit-mean Gaussians.\n \"\"\"\n np.random.seed(1)\n dists = [(np.random.normal(x,s,num), np.random.normal(y,s,num)) for x,y,s in specs]\n return np.hstack([d[0] for d in dists]), np.hstack([d[1] for d in dists])\n \npoints = (hv.Points(gaussians(num=600), label=\"600 points\", group=\"Small dots\") +\n hv.Points(gaussians(num=60000), label=\"60000 points\", group=\"Small dots\") +\n hv.Points(gaussians(num=600), label=\"600 points\", group=\"Tiny dots\") +\n hv.Points(gaussians(num=60000), label=\"60000 points\", group=\"Tiny dots\"))\n\npoints.opts(\n opts.Points('Small_dots', s=1, alpha=1),\n opts.Points('Tiny_dots', s=0.1, alpha=0.1))",
"_____no_output_____"
]
],
[
[
"Just as shown for the multiple-category case above, finding settings to avoid overplotting and oversaturation is difficult. The \"Small dots\" setting (size 0.1, full alpha) works fairly well for a sample of 600 points **A,** but it has serious overplotting issues for larger datasets, obscuring the shape and density of the distribution **B.** Using the \"Tiny dots\" setting (10 times smaller dots, alpha 0.1) works well for the larger dataset **D,** but not at all for the 600-point dataset **C.** Clearly, not all of these settings are accurately conveying the underlying distribution, as they all appear quite different from one another. Similar problems occur for the same size of dataset, but with greater or lesser levels of overlap between points, which of course varies with every new dataset. \n\nIn any case, as dataset size increases, at some point plotting a full scatterplot like any of these will become impractical with current plotting software. At this point, people often simply subsample their dataset, plotting 10,000 or perhaps 100,000 randomly selected datapoints. But as panel **A **shows, the shape of an **undersampled** distribution can be very difficult or impossible to make out, leading to incorrect conclusions about the distribution. Such problems can occur even when taking very large numbers of samples, if examining sparsely populated regions of the space, which will approximate panel **A **for some plot settings and panel **C **for others. The actual shape of the distribution is only visible if sufficient datapoints are available in that region *and* appropriate plot settings are used, as in **D,** but ensuring that both conditions are true is a quite difficult process of trial and error, making it very likely that important features of the dataset will be missed.\n\nTo avoid undersampling large datasets, researchers often use 2D histograms visualized as heatmaps, rather than scatterplots showing individual points. A heatmap has a fixed-size grid regardless of the dataset size, so that they can make use of all the data. Heatmaps effectively approximate a probability density function over the specified space, with coarser heatmaps averaging out noise or irrelevant variations to reveal an underlying distribution, and finer heatmaps able to represent more details in the distribution.\n\nLet's look at some heatmaps with different numbers of bins for the same two-Gaussians distribution:",
"_____no_output_____"
]
],
[
[
"def heatmap(coords,bins=10,offset=0.0,transform=lambda d,m:d, label=None):\n \"\"\"\n Given a set of coordinates, bins them into a 2d histogram grid\n of the specified size, and optionally transforms the counts\n and/or compresses them into a visible range starting at a \n specified offset between 0 and 1.0.\n \"\"\"\n hist,xs,ys = np.histogram2d(coords[0], coords[1], bins=bins)\n counts = hist[:,::-1].T\n transformed = transform(counts,counts!=0)\n span = transformed.max()-transformed.min()\n compressed = np.where(counts!=0,offset+(1.0-offset)*transformed/span,0)\n args = dict(label=label) if label else {}\n return hv.Image(compressed,bounds=(xs[-1],ys[-1],xs[1],ys[1]),**args)\n\nhv.Layout([heatmap(gaussians(num=60000),bins) for bins in [8,20,200]])",
"_____no_output_____"
]
],
[
[
"As you can see, a too-coarse binning grid **A **cannot represent this distribution faithfully, but with enough bins **C,** the heatmap will approximate a tiny-dot scatterplot like plot **D **in the previous figure. For intermediate grid sizes **B **the heatmap can average out the effects of undersampling; **B **is actually a more faithful representation of the *distribution* than **C **is (which we know is two offset 2D Gaussians), while **C **more faithfully represents the *sampling* (i.e., the individual points drawn from this distribution). Thus choosing a good binning grid size for a heatmap does take some expertise and knowledge of the goals of the visualization, and it's always useful to look at multiple binning-grid spacings for comparison. Still, at least the binning parameter is something meaningful at the data level (how coarse a view of the data is desired?) rather than just a plotting detail (what size and transparency should I use for the points?) that must be determined arbitrarily.\n\nIn any case, at least in principle, the heatmap approach can entirely avoid the first three problems above: **overplotting** (since multiple data points sum arithmetically into the grid cell, without obscuring one another), **oversaturation** (because the minimum and maximum counts observed can automatically be mapped to the two ends of a visible color range), and **undersampling** (since the resulting plot size is independent of the number of data points, allowing it to use an unbounded amount of incoming data).\n\n\n\n### 4. Undersaturation\n\nOf course, heatmaps come with their own plotting pitfalls. One rarely appreciated issue common to both heatmaps and alpha-based scatterplots is **undersaturation**, where large numbers of data points can be missed entirely because they are spread over many different heatmap bins or many nearly transparent scatter points. To look at this problem, let's again consider a set of multiple 2D Gaussians, but this time with different amounts of spread (standard deviation):",
"_____no_output_____"
]
],
[
[
"dist = gaussians(specs=[(2,2,0.02), (2,-2,0.1), (-2,-2,0.5), (-2,2,1.0), (0,0,3)],num=10000)\nhv.Points(dist) + hv.Points(dist).opts(s=0.1) + hv.Points(dist).opts(s=0.01, alpha=0.05)",
"_____no_output_____"
]
],
[
[
"Plots **A,** **B,** and **C **are all scatterplots for the same data, which is a sum of 5 Gaussian distributions at different locations and with different standard deviations:\n\n1. Location (2,2): very narrow spread\n2. Location (2,-2): narrow spread\n3. Location (-2,-2): medium spread\n4. Location (-2,2): large spread\n5. Location (0,0): very large spread\n\nIn plot **A,** of course, the very large spread covers up everything else, completely obscuring the structure of this dataset by overplotting. Plots **B **and **C **reveal the structure better, but they required hand tuning and neither one is particularly satisfactory. In **B **there are four clearly visible Gaussians, but all but the largest appear to have the same density of points per pixel, which we know is not the case from how the dataset was constructed, and the smallest is nearly invisible. Each of the five Gaussians has the same number of data points (10000), but the second-largest looks like it has more than the others, and the narrowest one is likely to be overlooked altogether, which is thus a clear example of oversaturation obscuring important features. Yet if we try to combat the oversaturation by using transparency in **C,** we now get a clear problem with **undersaturation** -- the \"very large spread\" Gaussian is now essentially invisible. Again, there are just as many datapoints in that category, but we'd never even know they were there if only looking at **C.**\n\nSimilar problems occur for a heatmap view of the same data:",
"_____no_output_____"
]
],
[
[
"hv.Layout([heatmap(dist,bins) for bins in [8,20,200]])",
"_____no_output_____"
]
],
[
[
"Here the narrow-spread distributions lead to pixels with a very high count, and if the other pixels are linearly ramped into the available color range, from zero to that high count value, then the wider-spread values are obscured (as in **B **) or entirely invisible (as in **C **). \n\nTo avoid undersaturation, you can add an offset to ensure that low-count (but nonzero) bins are mapped into a visible color, with the remaining intensity scale used to indicate differences in counts:",
"_____no_output_____"
]
],
[
[
"hv.Layout([heatmap(dist,bins,offset=0.2) for bins in [8,20,200]]).cols(4)",
"_____no_output_____"
]
],
[
[
"Such mapping entirely avoids undersaturation, since all pixels are either clearly zero (in the background color, i.e. white in this case), or a non-background color taken from the colormap. The widest-spread Gaussian is now clearly visible in all cases. \n\nHowever, the actual structure (5 Gaussians of different spreads) is still not visible. In **A **the problem is clearly too-coarse binning, but in **B **the binning is also somewhat too coarse for this data, since the \"very narrow spread\" and \"narrow spread\" Gaussians show up identically, each mapping entirely into a single bin (the two black pixels). **C **shouldn't suffer from too-coarse binning, yet it still looks more like a plot of the \"very large spread\" distribution alone, than a plot of these five distributions of different spreads, and it is thus still highly misleading despite the correction for undersaturation.\n\n\n### 5. Underutilized range\n\nSo, what is the problem in plot **C **above? By construction, we've avoided the first four pitfalls: **overplotting**, **oversaturation**, **undersampling**, and **undersaturation**. But the problem is now more subtle: differences in datapoint density are not visible between the five Gaussians, because all or nearly all pixels end up being mapped into either the bottom end of the visible range (light gray), or the top end (black, used only for the single pixel holding the \"very narrow spread\" distribution). The entire rest of the visible colors in this gray colormap are unused, conveying no information to the viewer about the rich structure that we know this distribution contains. If the data were uniformly distributed over the range from minimum to maximum counts per pixel (0 to 10,000, in this case), then the above plot would work well, but that's not the case for this dataset or for most real-world datasets.\n\nSo, let's try transforming the data from its default linear representation (integer count values) into something that preserves relative differences in count values but maps them into visually distinct colors. A logarithmic transformation is one common choice:",
"_____no_output_____"
]
],
[
[
"hv.Layout([heatmap(dist,bins,offset=0.2,transform=lambda d,m: np.where(m,np.log1p(d),0)) for bins in [8,20,200]])",
"_____no_output_____"
]
],
[
[
"Aha! We can now see the full structure of the dataset, with all five Gaussians clearly visible in **B **and **C,** and the relative spreads also clearly visible in **C.** \n\nWe still have a problem, though. The choice of a logarithmic transform was fairly arbitrary, and it mainly works well because we happened to have used an approximately geometric progression of spread sizes when constructing the example. For large datasets with truly unknown structure, can we have a more principled approach to mapping the dataset values into a visible range? \n\nYes, if we think of the visualization problem in a different way. The underlying difficulty in plotting this dataset (as for very many real-world datasets) is that the values in each bin are numerically very different (ranging from 10,000, in the bin for the \"very narrow spread\" Gaussian, to 1 (for single datapoints from the \"very large spread\" Gaussian)). Given the 256 gray levels available in a normal monitor (and the similarly limited human ability to detect differences in gray values), numerically mapping the data values into the visible range is not going to work well. But given that we are already backing off from a direct numerical mapping in the above approaches for correcting undersaturation and for doing log transformations, what if we entirely abandon the numerical mapping approach, using the numbers only to form a partial ordering of the data values? Such an approach would be a rank-order plot, preserving order and not magnitudes. For 100 gray values, you can think of it as a percentile-based plot, with the lowest 1% of the data values mapping to the first visible gray value, the next 1% mapping to the next visible gray value, and so on to the top 1% of the data values mapping to the gray value 255 (black in this case). The actual data values would be ignored in such plots, but their relative magnitudes would still determine how they map onto colors on the screen, preserving the structure of the distribution rather than the numerical values.\n\nWe can approximate such a rank-order or percentile encoding using the histogram equalization function from an image-processing package, which makes sure that each gray level is used for about the same number of pixels in the plot:",
"_____no_output_____"
]
],
[
[
"try:\n from skimage.exposure import equalize_hist\n eq_hist = lambda d,m: equalize_hist(1000*d,nbins=100000,mask=m)\nexcept ImportError:\n eq_hist = lambda d,m: d\n print(\"scikit-image not installed; skipping histogram equalization\")\n \nhv.Layout([heatmap(dist,bins,transform=eq_hist) for bins in [8,20,200]])",
"_____no_output_____"
]
],
[
[
"Plot **C** now reveals the full structure that we know was in this dataset, i.e. five Gaussians with different spreads, with no arbitrary parameter choices. (Well, there is a \"number of bins\" parameter for building the histogram for equalizing, but for integer data like this even that parameter can be eliminated entirely.) The differences in counts between pixels are now very clearly visible, across the full (and very wide) range of counts in the original data.\n\nOf course, we've lost the actual counts themselves, and so we can no longer tell just how many datapoints are in the \"very narrow spread\" pixel in this case. So plot **C** is accurately conveying the structure, but additional information would need to be provided to show the actual counts, by adding a color key mapping from the visible gray values into the actual counts and/or by providing hovering value information.\n\nAt this point, one could also consider explicitly highlighting hotspots so that they cannot be overlooked. In plots B and C above, the two highest-density pixels are mapped to the two darkest pixel colors, which can reveal problems with your monitor settings if they were adjusted to make dark text appear blacker. Thus on those monitors, the highest values may not be clearly distinguishable from each other or from nearby grey values, which is a possible downside to fully utilizing the dynamic range available. But once the data is reliably and automatically mapped into a repeatable, reliable, fully utilized range for display, making explicit adjustments (e.g. based on wanting to make hotspots particularly clear) can be done in a principled way that doesn't depend on the actual data distribution (e.g. by just making the top few pixel values into a different color, or by stretching out those portions of the color map to show the extremes more safely across different monitors). Before getting into such specialized manipulations, there's a big pitfall to avoid first:\n\n### 6. Nonuniform colormapping\n\nLet's say you've managed avoid pitfalls 1-5 somehow. However, there is one more problem waiting to catch you at the last stage, ruining all of your work eliminating the other issues: using a perceptually non-uniform colormap. A heatmap requires a colormap before it can be visualized, i.e., a lookup table from a data value (typically a normalized magnitude in the range 0 to 1) to a pixel color. The goal of a scientific visualization is to reveal the underlying properties of the data to your visual system, and to do so it is necessary to choose colors for each pixel that lead the viewer to perceive that data faithfully. Unfortunately, most of the colormaps in common use in plotting programs are highly *non*uniform. \n\nFor instance, in \"jet\" (the default colormap for matlab and matplotlib until 2015), a large range of data values will all appear in shades of green that are perceptually indistinguishable, and similarly for the yellow regions of their \"hot\" colormaps:\n\n\n\nIn this image, a good colormap would have \"teeth\" equally visible at all data values, as for the perceptually uniform equivalents from the [colorcet](https://github.com/bokeh/colorcet) package:\n\n\n\nWe can easily see these effects if we look at our example dataset after histogram equalization, where all the different data levels are known to be distributed evenly in the array of normalized magnitudes:",
"_____no_output_____"
]
],
[
[
"hv.Layout([heatmap(dist,200,transform=eq_hist,label=cmap).opts(cmap=cmap) for cmap in [\"hot\",\"fire\"]]).cols(2)",
"_____no_output_____"
]
],
[
[
"Comparing **A ** to **B **it should be clear that the \"fire\" colormap is revealing much more of the data, accurately rendering the density differences between each of the different blobs. The unsuitable \"hot\" colormap is mapping all of the high density regions to perceptually indistinguishable shades of bright yellow/white, giving an \"oversaturated\" appearance even though we know the underlying heatmap array is *not* oversaturated (by construction). Luckily it is easy to avoid this problem; just use one of the 50 perceptually uniform colormaps available in the [colorcet](https://github.com/bokeh/colorcet) package, one of the four shipped with matplotlib [(viridis, plasma, inferno, or magma)](https://bids.github.io/colormap), or the Parula colormap shipped with Matlab.\n\n\n## Summary\n\nStarting with plots of specific datapoints, we showed how typical visualization techniques will systematically misrepresent the distribution of those points. Here's an example of each of those six problems, all for the same distribution:",
"_____no_output_____"
]
],
[
[
"layout = (hv.Points(dist,label=\"1. Overplotting\") + \n hv.Points(dist,label=\"2. Oversaturation\").opts(s=0.1,alpha=0.5) + \n hv.Points((dist[0][::200],dist[1][::200]),label=\"3. Undersampling\").opts(s=2,alpha=0.5) + \n hv.Points(dist,label=\"4. Undersaturation\").opts(s=0.01,alpha=0.05) + \n heatmap(dist,200,offset=0.2,label=\"5. Underutilized dynamic range\") +\n heatmap(dist,200,transform=eq_hist,label=\"6. Nonuniform colormapping\").opts(cmap=\"hot\"))\n\nlayout.opts(\n opts.Points(axiswise=False),\n opts.Layout(sublabel_format=\"\", tight=True)).cols(3)",
"_____no_output_____"
]
],
[
[
"Here we could avoid each of these problems by hand, using trial and error based on our knowledge about the underlying dataset, since we created it. But for big data in general, these issues are major problems, because you don't know what the data *should* look like. Thus:\n\n#### For big data, you don't know when the viz is lying\n\nI.e., visualization is supposed to help you explore and understand your data, but if your visualizations are systematically misrepresenting your data because of **overplotting**, **oversaturation**, **undersampling**, **undersaturation**, **underutilized range**, and **nonuniform colormapping**, then you won't be able to discover the real qualities of your data and will be unable to make the right decisions.\n\nLuckily, using the systematic approach outlined in this discussion, you can avoid *all* of these pitfalls, allowing you to render your data faithfully without requiring *any* \"magic parameters\" that depend on your dataset:",
"_____no_output_____"
]
],
[
[
"heatmap(dist,200,transform=eq_hist).opts(cmap=\"fire\")",
"_____no_output_____"
]
],
[
[
"### [Datashader](https://github.com/bokeh/datashader)\n\nThe steps above show how to avoid the six main plotting pitfalls by hand, but it can be awkward and relatively slow to do so. Luckily there is a new Python library available to automate and optimize these steps, named [Datashader](https://github.com/bokeh/datashader). Datashader avoids users having to make dataset-dependent decisions and parameter settings when visualizing a new dataset. Datashader makes it practical to create accurate visualizations of datasets too large to understand directly, up to a billion points on a normal laptop and larger datasets on a compute cluster. As a simple teaser, the above steps can be expressed very concisely using the Datashader interface provided by [HoloViews](http://holoviews.org):",
"_____no_output_____"
]
],
[
[
"hv.output(size=200)\ndatashade(hv.Points(dist))",
"_____no_output_____"
]
],
[
[
"Without any change to the settings, the same command will work with dataset sizes too large for most plotting programs, like this 50-million-point version of the distribution:",
"_____no_output_____"
]
],
[
[
"dist = gaussians(specs=[(2,2,0.02), (2,-2,0.1), (-2,-2,0.5), (-2,2,1.0), (0,0,3)], num=10000000)\ndatashade(hv.Points(dist))",
"_____no_output_____"
]
],
[
[
"See the [Datashader web site](https://raw.githubusercontent.com/bokeh/datashader/master/examples/README.md) for details and examples to help you get started.",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
] |
4a16ae0730cfd8c99437369270a3d2176e7aed02
| 466,802 |
ipynb
|
Jupyter Notebook
|
11_deep_learning.ipynb
|
otamilocintra/ml2gh
|
917bd7c7576dff094ea493f85e178e2ed5ef6e24
|
[
"Apache-2.0"
] | null | null | null |
11_deep_learning.ipynb
|
otamilocintra/ml2gh
|
917bd7c7576dff094ea493f85e178e2ed5ef6e24
|
[
"Apache-2.0"
] | null | null | null |
11_deep_learning.ipynb
|
otamilocintra/ml2gh
|
917bd7c7576dff094ea493f85e178e2ed5ef6e24
|
[
"Apache-2.0"
] | null | null | null | 69.320166 | 127,478 | 0.637782 |
[
[
[
"**Chapter 11 – Deep Learning**",
"_____no_output_____"
],
[
"_This notebook contains all the sample code and solutions to the exercises in chapter 11._",
"_____no_output_____"
],
[
"# Setup",
"_____no_output_____"
],
[
"First, let's make sure this notebook works well in both python 2 and 3, import a few common modules, ensure MatplotLib plots figures inline and prepare a function to save the figures:",
"_____no_output_____"
]
],
[
[
"# To support both python 2 and python 3\nfrom __future__ import division, print_function, unicode_literals\n\n# Common imports\nimport numpy as np\nimport os\n\n# to make this notebook's output stable across runs\ndef reset_graph(seed=42):\n tf.reset_default_graph()\n tf.set_random_seed(seed)\n np.random.seed(seed)\n\n# To plot pretty figures\n%matplotlib inline\nimport matplotlib\nimport matplotlib.pyplot as plt\nplt.rcParams['axes.labelsize'] = 14\nplt.rcParams['xtick.labelsize'] = 12\nplt.rcParams['ytick.labelsize'] = 12\n\n# Where to save the figures\nPROJECT_ROOT_DIR = \".\"\nCHAPTER_ID = \"deep\"\n\ndef save_fig(fig_id, tight_layout=True):\n path = os.path.join(PROJECT_ROOT_DIR, \"images\", CHAPTER_ID, fig_id + \".png\")\n print(\"Saving figure\", fig_id)\n if tight_layout:\n plt.tight_layout()\n plt.savefig(path, format='png', dpi=300)",
"_____no_output_____"
]
],
[
[
"# Vanishing/Exploding Gradients Problem",
"_____no_output_____"
]
],
[
[
"def logit(z):\n return 1 / (1 + np.exp(-z))",
"_____no_output_____"
],
[
"z = np.linspace(-5, 5, 200)\n\nplt.plot([-5, 5], [0, 0], 'k-')\nplt.plot([-5, 5], [1, 1], 'k--')\nplt.plot([0, 0], [-0.2, 1.2], 'k-')\nplt.plot([-5, 5], [-3/4, 7/4], 'g--')\nplt.plot(z, logit(z), \"b-\", linewidth=2)\nprops = dict(facecolor='black', shrink=0.1)\nplt.annotate('Saturating', xytext=(3.5, 0.7), xy=(5, 1), arrowprops=props, fontsize=14, ha=\"center\")\nplt.annotate('Saturating', xytext=(-3.5, 0.3), xy=(-5, 0), arrowprops=props, fontsize=14, ha=\"center\")\nplt.annotate('Linear', xytext=(2, 0.2), xy=(0, 0.5), arrowprops=props, fontsize=14, ha=\"center\")\nplt.grid(True)\nplt.title(\"Sigmoid activation function\", fontsize=14)\nplt.axis([-5, 5, -0.2, 1.2])\n\nsave_fig(\"sigmoid_saturation_plot\")\nplt.show()",
"Saving figure sigmoid_saturation_plot\n"
]
],
[
[
"## Xavier and He Initialization",
"_____no_output_____"
],
[
"Note: the book uses `tensorflow.contrib.layers.fully_connected()` rather than `tf.layers.dense()` (which did not exist when this chapter was written). It is now preferable to use `tf.layers.dense()`, because anything in the contrib module may change or be deleted without notice. The `dense()` function is almost identical to the `fully_connected()` function. The main differences relevant to this chapter are:\n* several parameters are renamed: `scope` becomes `name`, `activation_fn` becomes `activation` (and similarly the `_fn` suffix is removed from other parameters such as `normalizer_fn`), `weights_initializer` becomes `kernel_initializer`, etc.\n* the default `activation` is now `None` rather than `tf.nn.relu`.\n* it does not support `tensorflow.contrib.framework.arg_scope()` (introduced later in chapter 11).\n* it does not support regularizer params (introduced later in chapter 11).",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf",
"_____no_output_____"
],
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")",
"_____no_output_____"
],
[
"he_init = tf.variance_scaling_initializer()\nhidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu,\n kernel_initializer=he_init, name=\"hidden1\")",
"_____no_output_____"
]
],
[
[
"## Nonsaturating Activation Functions",
"_____no_output_____"
],
[
"### Leaky ReLU",
"_____no_output_____"
]
],
[
[
"def leaky_relu(z, alpha=0.01):\n return np.maximum(alpha*z, z)",
"_____no_output_____"
],
[
"plt.plot(z, leaky_relu(z, 0.05), \"b-\", linewidth=2)\nplt.plot([-5, 5], [0, 0], 'k-')\nplt.plot([0, 0], [-0.5, 4.2], 'k-')\nplt.grid(True)\nprops = dict(facecolor='black', shrink=0.1)\nplt.annotate('Leak', xytext=(-3.5, 0.5), xy=(-5, -0.2), arrowprops=props, fontsize=14, ha=\"center\")\nplt.title(\"Leaky ReLU activation function\", fontsize=14)\nplt.axis([-5, 5, -0.5, 4.2])\n\nsave_fig(\"leaky_relu_plot\")\nplt.show()",
"Saving figure leaky_relu_plot\n"
]
],
[
[
"Implementing Leaky ReLU in TensorFlow:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")",
"_____no_output_____"
],
[
"def leaky_relu(z, name=None):\n return tf.maximum(0.01 * z, z, name=name)\n\nhidden1 = tf.layers.dense(X, n_hidden1, activation=leaky_relu, name=\"hidden1\")",
"_____no_output_____"
]
],
[
[
"Let's train a neural network on MNIST using the Leaky ReLU. First let's create the graph:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\nn_hidden2 = 100\nn_outputs = 10",
"_____no_output_____"
],
[
"X = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")",
"_____no_output_____"
],
[
"with tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=leaky_relu, name=\"hidden1\")\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=leaky_relu, name=\"hidden2\")\n logits = tf.layers.dense(hidden2, n_outputs, name=\"outputs\")",
"_____no_output_____"
],
[
"with tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")",
"_____no_output_____"
],
[
"learning_rate = 0.01\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)",
"_____no_output_____"
],
[
"with tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))",
"_____no_output_____"
],
[
"init = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"Let's load the data:",
"_____no_output_____"
],
[
"**Warning**: `tf.examples.tutorials.mnist` is deprecated. We will use `tf.keras.datasets.mnist` instead.",
"_____no_output_____"
]
],
[
[
"(X_train, y_train), (X_test, y_test) = tf.keras.datasets.mnist.load_data()\nX_train = X_train.astype(np.float32).reshape(-1, 28*28) / 255.0\nX_test = X_test.astype(np.float32).reshape(-1, 28*28) / 255.0\ny_train = y_train.astype(np.int32)\ny_test = y_test.astype(np.int32)\nX_valid, X_train = X_train[:5000], X_train[5000:]\ny_valid, y_train = y_train[:5000], y_train[5000:]",
"_____no_output_____"
],
[
"def shuffle_batch(X, y, batch_size):\n rnd_idx = np.random.permutation(len(X))\n n_batches = len(X) // batch_size\n for batch_idx in np.array_split(rnd_idx, n_batches):\n X_batch, y_batch = X[batch_idx], y[batch_idx]\n yield X_batch, y_batch",
"_____no_output_____"
],
[
"n_epochs = 40\nbatch_size = 50\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n if epoch % 5 == 0:\n acc_batch = accuracy.eval(feed_dict={X: X_batch, y: y_batch})\n acc_valid = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Batch accuracy:\", acc_batch, \"Validation accuracy:\", acc_valid)\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\")",
"0 Batch accuracy: 0.86 Validation accuracy: 0.9044\n5 Batch accuracy: 0.94 Validation accuracy: 0.9496\n10 Batch accuracy: 0.92 Validation accuracy: 0.9654\n15 Batch accuracy: 0.94 Validation accuracy: 0.971\n20 Batch accuracy: 1.0 Validation accuracy: 0.9764\n25 Batch accuracy: 1.0 Validation accuracy: 0.9778\n30 Batch accuracy: 0.98 Validation accuracy: 0.978\n35 Batch accuracy: 1.0 Validation accuracy: 0.9788\n"
]
],
[
[
"### ELU",
"_____no_output_____"
]
],
[
[
"def elu(z, alpha=1):\n return np.where(z < 0, alpha * (np.exp(z) - 1), z)",
"_____no_output_____"
],
[
"plt.plot(z, elu(z), \"b-\", linewidth=2)\nplt.plot([-5, 5], [0, 0], 'k-')\nplt.plot([-5, 5], [-1, -1], 'k--')\nplt.plot([0, 0], [-2.2, 3.2], 'k-')\nplt.grid(True)\nplt.title(r\"ELU activation function ($\\alpha=1$)\", fontsize=14)\nplt.axis([-5, 5, -2.2, 3.2])\n\nsave_fig(\"elu_plot\")\nplt.show()",
"Saving figure elu_plot\n"
]
],
[
[
"Implementing ELU in TensorFlow is trivial, just specify the activation function when building each layer:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")",
"_____no_output_____"
],
[
"hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.elu, name=\"hidden1\")",
"_____no_output_____"
]
],
[
[
"### SELU",
"_____no_output_____"
],
[
"This activation function was proposed in this [great paper](https://arxiv.org/pdf/1706.02515.pdf) by Günter Klambauer, Thomas Unterthiner and Andreas Mayr, published in June 2017. During training, a neural network composed exclusively of a stack of dense layers using the SELU activation function and LeCun initialization will self-normalize: the output of each layer will tend to preserve the same mean and variance during training, which solves the vanishing/exploding gradients problem. As a result, this activation function outperforms the other activation functions very significantly for such neural nets, so you should really try it out. Unfortunately, the self-normalizing property of the SELU activation function is easily broken: you cannot use ℓ<sub>1</sub> or ℓ<sub>2</sub> regularization, regular dropout, max-norm, skip connections or other non-sequential topologies (so recurrent neural networks won't self-normalize). However, in practice it works quite well with sequential CNNs. If you break self-normalization, SELU will not necessarily outperform other activation functions.",
"_____no_output_____"
]
],
[
[
"from scipy.special import erfc\n\n# alpha and scale to self normalize with mean 0 and standard deviation 1\n# (see equation 14 in the paper):\nalpha_0_1 = -np.sqrt(2 / np.pi) / (erfc(1/np.sqrt(2)) * np.exp(1/2) - 1)\nscale_0_1 = (1 - erfc(1 / np.sqrt(2)) * np.sqrt(np.e)) * np.sqrt(2 * np.pi) * (2 * erfc(np.sqrt(2))*np.e**2 + np.pi*erfc(1/np.sqrt(2))**2*np.e - 2*(2+np.pi)*erfc(1/np.sqrt(2))*np.sqrt(np.e)+np.pi+2)**(-1/2)",
"_____no_output_____"
],
[
"def selu(z, scale=scale_0_1, alpha=alpha_0_1):\n return scale * elu(z, alpha)",
"_____no_output_____"
],
[
"plt.plot(z, selu(z), \"b-\", linewidth=2)\nplt.plot([-5, 5], [0, 0], 'k-')\nplt.plot([-5, 5], [-1.758, -1.758], 'k--')\nplt.plot([0, 0], [-2.2, 3.2], 'k-')\nplt.grid(True)\nplt.title(r\"SELU activation function\", fontsize=14)\nplt.axis([-5, 5, -2.2, 3.2])\n\nsave_fig(\"selu_plot\")\nplt.show()",
"Saving figure selu_plot\n"
]
],
[
[
"By default, the SELU hyperparameters (`scale` and `alpha`) are tuned in such a way that the mean output of each neuron remains close to 0, and the standard deviation remains close to 1 (assuming the inputs are standardized with mean 0 and standard deviation 1 too). Using this activation function, even a 1,000 layer deep neural network preserves roughly mean 0 and standard deviation 1 across all layers, avoiding the exploding/vanishing gradients problem:",
"_____no_output_____"
]
],
[
[
"np.random.seed(42)\nZ = np.random.normal(size=(500, 100)) # standardized inputs\nfor layer in range(1000):\n W = np.random.normal(size=(100, 100), scale=np.sqrt(1 / 100)) # LeCun initialization\n Z = selu(np.dot(Z, W))\n means = np.mean(Z, axis=0).mean()\n stds = np.std(Z, axis=0).mean()\n if layer % 100 == 0:\n print(\"Layer {}: mean {:.2f}, std deviation {:.2f}\".format(layer, means, stds))",
"Layer 0: mean -0.00, std deviation 1.00\nLayer 100: mean 0.02, std deviation 0.96\nLayer 200: mean 0.01, std deviation 0.90\nLayer 300: mean -0.02, std deviation 0.92\nLayer 400: mean 0.05, std deviation 0.89\nLayer 500: mean 0.01, std deviation 0.93\nLayer 600: mean 0.02, std deviation 0.92\nLayer 700: mean -0.02, std deviation 0.90\nLayer 800: mean 0.05, std deviation 0.83\nLayer 900: mean 0.02, std deviation 1.00\n"
]
],
[
[
"The `tf.nn.selu()` function was added in TensorFlow 1.4. For earlier versions, you can use the following implementation:",
"_____no_output_____"
]
],
[
[
"def selu(z, scale=alpha_0_1, alpha=scale_0_1):\n return scale * tf.where(z >= 0.0, z, alpha * tf.nn.elu(z))",
"_____no_output_____"
]
],
[
[
"However, the SELU activation function cannot be used along with regular Dropout (this would cancel the SELU activation function's self-normalizing property). Fortunately, there is a Dropout variant called Alpha Dropout proposed in the same paper. It is available in `tf.contrib.nn.alpha_dropout()` since TF 1.4 (or check out [this implementation](https://github.com/bioinf-jku/SNNs/blob/master/selu.py) by the Institute of Bioinformatics, Johannes Kepler University Linz).",
"_____no_output_____"
],
[
"Let's create a neural net for MNIST using the SELU activation function:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\nn_hidden2 = 100\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=selu, name=\"hidden1\")\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=selu, name=\"hidden2\")\n logits = tf.layers.dense(hidden2, n_outputs, name=\"outputs\")\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nlearning_rate = 0.01\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()\nn_epochs = 40\nbatch_size = 50",
"_____no_output_____"
]
],
[
[
"Now let's train it. Do not forget to scale the inputs to mean 0 and standard deviation 1:",
"_____no_output_____"
]
],
[
[
"means = X_train.mean(axis=0, keepdims=True)\nstds = X_train.std(axis=0, keepdims=True) + 1e-10\nX_val_scaled = (X_valid - means) / stds\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n X_batch_scaled = (X_batch - means) / stds\n sess.run(training_op, feed_dict={X: X_batch_scaled, y: y_batch})\n if epoch % 5 == 0:\n acc_batch = accuracy.eval(feed_dict={X: X_batch_scaled, y: y_batch})\n acc_valid = accuracy.eval(feed_dict={X: X_val_scaled, y: y_valid})\n print(epoch, \"Batch accuracy:\", acc_batch, \"Validation accuracy:\", acc_valid)\n\n save_path = saver.save(sess, \"./my_model_final_selu.ckpt\")",
"0 Batch accuracy: 0.88 Validation accuracy: 0.923\n5 Batch accuracy: 0.98 Validation accuracy: 0.9578\n10 Batch accuracy: 1.0 Validation accuracy: 0.9664\n15 Batch accuracy: 0.96 Validation accuracy: 0.9682\n20 Batch accuracy: 1.0 Validation accuracy: 0.9694\n25 Batch accuracy: 1.0 Validation accuracy: 0.9688\n30 Batch accuracy: 1.0 Validation accuracy: 0.9694\n35 Batch accuracy: 1.0 Validation accuracy: 0.97\n"
]
],
[
[
"# Batch Normalization",
"_____no_output_____"
],
[
"Note: the book uses `tensorflow.contrib.layers.batch_norm()` rather than `tf.layers.batch_normalization()` (which did not exist when this chapter was written). It is now preferable to use `tf.layers.batch_normalization()`, because anything in the contrib module may change or be deleted without notice. Instead of using the `batch_norm()` function as a regularizer parameter to the `fully_connected()` function, we now use `batch_normalization()` and we explicitly create a distinct layer. The parameters are a bit different, in particular:\n* `decay` is renamed to `momentum`,\n* `is_training` is renamed to `training`,\n* `updates_collections` is removed: the update operations needed by batch normalization are added to the `UPDATE_OPS` collection and you need to explicity run these operations during training (see the execution phase below),\n* we don't need to specify `scale=True`, as that is the default.\n\nAlso note that in order to run batch norm just _before_ each hidden layer's activation function, we apply the ELU activation function manually, right after the batch norm layer.\n\nNote: since the `tf.layers.dense()` function is incompatible with `tf.contrib.layers.arg_scope()` (which is used in the book), we now use python's `functools.partial()` function instead. It makes it easy to create a `my_dense_layer()` function that just calls `tf.layers.dense()` with the desired parameters automatically set (unless they are overridden when calling `my_dense_layer()`). As you can see, the code remains very similar.",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nimport tensorflow as tf\n\nn_inputs = 28 * 28\nn_hidden1 = 300\nn_hidden2 = 100\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\n\ntraining = tf.placeholder_with_default(False, shape=(), name='training')\n\nhidden1 = tf.layers.dense(X, n_hidden1, name=\"hidden1\")\nbn1 = tf.layers.batch_normalization(hidden1, training=training, momentum=0.9)\nbn1_act = tf.nn.elu(bn1)\n\nhidden2 = tf.layers.dense(bn1_act, n_hidden2, name=\"hidden2\")\nbn2 = tf.layers.batch_normalization(hidden2, training=training, momentum=0.9)\nbn2_act = tf.nn.elu(bn2)\n\nlogits_before_bn = tf.layers.dense(bn2_act, n_outputs, name=\"outputs\")\nlogits = tf.layers.batch_normalization(logits_before_bn, training=training,\n momentum=0.9)",
"_____no_output_____"
],
[
"reset_graph()\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ntraining = tf.placeholder_with_default(False, shape=(), name='training')",
"_____no_output_____"
]
],
[
[
"To avoid repeating the same parameters over and over again, we can use Python's `partial()` function:",
"_____no_output_____"
]
],
[
[
"from functools import partial\n\nmy_batch_norm_layer = partial(tf.layers.batch_normalization,\n training=training, momentum=0.9)\n\nhidden1 = tf.layers.dense(X, n_hidden1, name=\"hidden1\")\nbn1 = my_batch_norm_layer(hidden1)\nbn1_act = tf.nn.elu(bn1)\nhidden2 = tf.layers.dense(bn1_act, n_hidden2, name=\"hidden2\")\nbn2 = my_batch_norm_layer(hidden2)\nbn2_act = tf.nn.elu(bn2)\nlogits_before_bn = tf.layers.dense(bn2_act, n_outputs, name=\"outputs\")\nlogits = my_batch_norm_layer(logits_before_bn)",
"_____no_output_____"
]
],
[
[
"Let's build a neural net for MNIST, using the ELU activation function and Batch Normalization at each layer:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nbatch_norm_momentum = 0.9\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\ntraining = tf.placeholder_with_default(False, shape=(), name='training')\n\nwith tf.name_scope(\"dnn\"):\n he_init = tf.variance_scaling_initializer()\n\n my_batch_norm_layer = partial(\n tf.layers.batch_normalization,\n training=training,\n momentum=batch_norm_momentum)\n\n my_dense_layer = partial(\n tf.layers.dense,\n kernel_initializer=he_init)\n\n hidden1 = my_dense_layer(X, n_hidden1, name=\"hidden1\")\n bn1 = tf.nn.elu(my_batch_norm_layer(hidden1))\n hidden2 = my_dense_layer(bn1, n_hidden2, name=\"hidden2\")\n bn2 = tf.nn.elu(my_batch_norm_layer(hidden2))\n logits_before_bn = my_dense_layer(bn2, n_outputs, name=\"outputs\")\n logits = my_batch_norm_layer(logits_before_bn)\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n \ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"Note: since we are using `tf.layers.batch_normalization()` rather than `tf.contrib.layers.batch_norm()` (as in the book), we need to explicitly run the extra update operations needed by batch normalization (`sess.run([training_op, extra_update_ops],...`).",
"_____no_output_____"
]
],
[
[
"n_epochs = 20\nbatch_size = 200",
"_____no_output_____"
],
[
"extra_update_ops = tf.get_collection(tf.GraphKeys.UPDATE_OPS)\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run([training_op, extra_update_ops],\n feed_dict={training: True, X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\")",
"0 Validation accuracy: 0.8952\n1 Validation accuracy: 0.9202\n2 Validation accuracy: 0.9318\n3 Validation accuracy: 0.9422\n4 Validation accuracy: 0.9468\n5 Validation accuracy: 0.954\n6 Validation accuracy: 0.9568\n7 Validation accuracy: 0.96\n8 Validation accuracy: 0.962\n9 Validation accuracy: 0.9638\n10 Validation accuracy: 0.9662\n11 Validation accuracy: 0.9682\n12 Validation accuracy: 0.9672\n13 Validation accuracy: 0.9696\n14 Validation accuracy: 0.9706\n15 Validation accuracy: 0.9704\n16 Validation accuracy: 0.9718\n17 Validation accuracy: 0.9726\n18 Validation accuracy: 0.9738\n19 Validation accuracy: 0.9742\n"
]
],
[
[
"What!? That's not a great accuracy for MNIST. Of course, if you train for longer it will get much better accuracy, but with such a shallow network, Batch Norm and ELU are unlikely to have very positive impact: they shine mostly for much deeper nets.",
"_____no_output_____"
],
[
"Note that you could also make the training operation depend on the update operations:\n\n```python\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n extra_update_ops = tf.get_collection(tf.GraphKeys.UPDATE_OPS)\n with tf.control_dependencies(extra_update_ops):\n training_op = optimizer.minimize(loss)\n```\n\nThis way, you would just have to evaluate the `training_op` during training, TensorFlow would automatically run the update operations as well:\n\n```python\nsess.run(training_op, feed_dict={training: True, X: X_batch, y: y_batch})\n```",
"_____no_output_____"
],
[
"One more thing: notice that the list of trainable variables is shorter than the list of all global variables. This is because the moving averages are non-trainable variables. If you want to reuse a pretrained neural network (see below), you must not forget these non-trainable variables.",
"_____no_output_____"
]
],
[
[
"[v.name for v in tf.trainable_variables()]",
"_____no_output_____"
],
[
"[v.name for v in tf.global_variables()]",
"_____no_output_____"
]
],
[
[
"## Gradient Clipping",
"_____no_output_____"
],
[
"Let's create a simple neural net for MNIST and add gradient clipping. The first part is the same as earlier (except we added a few more layers to demonstrate reusing pretrained models, see below):",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\nn_hidden2 = 50\nn_hidden3 = 50\nn_hidden4 = 50\nn_hidden5 = 50\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\")\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu, name=\"hidden2\")\n hidden3 = tf.layers.dense(hidden2, n_hidden3, activation=tf.nn.relu, name=\"hidden3\")\n hidden4 = tf.layers.dense(hidden3, n_hidden4, activation=tf.nn.relu, name=\"hidden4\")\n hidden5 = tf.layers.dense(hidden4, n_hidden5, activation=tf.nn.relu, name=\"hidden5\")\n logits = tf.layers.dense(hidden5, n_outputs, name=\"outputs\")\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")",
"_____no_output_____"
],
[
"learning_rate = 0.01",
"_____no_output_____"
]
],
[
[
"Now we apply gradient clipping. For this, we need to get the gradients, use the `clip_by_value()` function to clip them, then apply them:",
"_____no_output_____"
]
],
[
[
"threshold = 1.0\n\noptimizer = tf.train.GradientDescentOptimizer(learning_rate)\ngrads_and_vars = optimizer.compute_gradients(loss)\ncapped_gvs = [(tf.clip_by_value(grad, -threshold, threshold), var)\n for grad, var in grads_and_vars]\ntraining_op = optimizer.apply_gradients(capped_gvs)",
"_____no_output_____"
]
],
[
[
"The rest is the same as usual:",
"_____no_output_____"
]
],
[
[
"with tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")",
"_____no_output_____"
],
[
"init = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 20\nbatch_size = 200",
"_____no_output_____"
],
[
"with tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\")",
"0 Validation accuracy: 0.288\n1 Validation accuracy: 0.7936\n2 Validation accuracy: 0.8798\n3 Validation accuracy: 0.906\n4 Validation accuracy: 0.9164\n5 Validation accuracy: 0.9218\n6 Validation accuracy: 0.9296\n7 Validation accuracy: 0.9358\n8 Validation accuracy: 0.9382\n9 Validation accuracy: 0.9414\n10 Validation accuracy: 0.9456\n11 Validation accuracy: 0.9474\n12 Validation accuracy: 0.9478\n13 Validation accuracy: 0.9534\n14 Validation accuracy: 0.9568\n15 Validation accuracy: 0.9566\n16 Validation accuracy: 0.9574\n17 Validation accuracy: 0.959\n18 Validation accuracy: 0.9622\n19 Validation accuracy: 0.9612\n"
]
],
[
[
"## Reusing Pretrained Layers",
"_____no_output_____"
],
[
"## Reusing a TensorFlow Model",
"_____no_output_____"
],
[
"First you need to load the graph's structure. The `import_meta_graph()` function does just that, loading the graph's operations into the default graph, and returning a `Saver` that you can then use to restore the model's state. Note that by default, a `Saver` saves the structure of the graph into a `.meta` file, so that's the file you should load:",
"_____no_output_____"
]
],
[
[
"reset_graph()",
"_____no_output_____"
],
[
"saver = tf.train.import_meta_graph(\"./my_model_final.ckpt.meta\")",
"_____no_output_____"
]
],
[
[
"Next you need to get a handle on all the operations you will need for training. If you don't know the graph's structure, you can list all the operations:",
"_____no_output_____"
]
],
[
[
"for op in tf.get_default_graph().get_operations():\n print(op.name)",
"X\ny\nhidden1/kernel/Initializer/random_uniform/shape\nhidden1/kernel/Initializer/random_uniform/min\nhidden1/kernel/Initializer/random_uniform/max\nhidden1/kernel/Initializer/random_uniform/RandomUniform\nhidden1/kernel/Initializer/random_uniform/sub\nhidden1/kernel/Initializer/random_uniform/mul\nhidden1/kernel/Initializer/random_uniform\nhidden1/kernel\nhidden1/kernel/Assign\nhidden1/kernel/read\nhidden1/bias/Initializer/zeros\nhidden1/bias\nhidden1/bias/Assign\nhidden1/bias/read\ndnn/hidden1/MatMul\ndnn/hidden1/BiasAdd\ndnn/hidden1/Relu\nhidden2/kernel/Initializer/random_uniform/shape\nhidden2/kernel/Initializer/random_uniform/min\nhidden2/kernel/Initializer/random_uniform/max\nhidden2/kernel/Initializer/random_uniform/RandomUniform\nhidden2/kernel/Initializer/random_uniform/sub\nhidden2/kernel/Initializer/random_uniform/mul\nhidden2/kernel/Initializer/random_uniform\nhidden2/kernel\nhidden2/kernel/Assign\nhidden2/kernel/read\nhidden2/bias/Initializer/zeros\nhidden2/bias\nhidden2/bias/Assign\nhidden2/bias/read\ndnn/hidden2/MatMul\ndnn/hidden2/BiasAdd\n<<208 more lines>>\nGradientDescent/update_hidden3/bias/ApplyGradientDescent\nGradientDescent/update_hidden4/kernel/ApplyGradientDescent\nGradientDescent/update_hidden4/bias/ApplyGradientDescent\nGradientDescent/update_hidden5/kernel/ApplyGradientDescent\nGradientDescent/update_hidden5/bias/ApplyGradientDescent\nGradientDescent/update_outputs/kernel/ApplyGradientDescent\nGradientDescent/update_outputs/bias/ApplyGradientDescent\nGradientDescent\neval/in_top_k/InTopKV2/k\neval/in_top_k/InTopKV2\neval/Cast\neval/Const\neval/accuracy\ninit\nsave/Const\nsave/SaveV2/tensor_names\nsave/SaveV2/shape_and_slices\nsave/SaveV2\nsave/control_dependency\nsave/RestoreV2/tensor_names\nsave/RestoreV2/shape_and_slices\nsave/RestoreV2\nsave/Assign\nsave/Assign_1\nsave/Assign_2\nsave/Assign_3\nsave/Assign_4\nsave/Assign_5\nsave/Assign_6\nsave/Assign_7\nsave/Assign_8\nsave/Assign_9\nsave/Assign_10\nsave/Assign_11\nsave/restore_all\n"
]
],
[
[
"Oops, that's a lot of operations! It's much easier to use TensorBoard to visualize the graph. The following hack will allow you to visualize the graph within Jupyter (if it does not work with your browser, you will need to use a `FileWriter` to save the graph and then visualize it in TensorBoard):",
"_____no_output_____"
]
],
[
[
"from tensorflow_graph_in_jupyter import show_graph",
"_____no_output_____"
],
[
"show_graph(tf.get_default_graph())",
"_____no_output_____"
]
],
[
[
"Once you know which operations you need, you can get a handle on them using the graph's `get_operation_by_name()` or `get_tensor_by_name()` methods:",
"_____no_output_____"
]
],
[
[
"X = tf.get_default_graph().get_tensor_by_name(\"X:0\")\ny = tf.get_default_graph().get_tensor_by_name(\"y:0\")\n\naccuracy = tf.get_default_graph().get_tensor_by_name(\"eval/accuracy:0\")\n\ntraining_op = tf.get_default_graph().get_operation_by_name(\"GradientDescent\")",
"_____no_output_____"
]
],
[
[
"If you are the author of the original model, you could make things easier for people who will reuse your model by giving operations very clear names and documenting them. Another approach is to create a collection containing all the important operations that people will want to get a handle on:",
"_____no_output_____"
]
],
[
[
"for op in (X, y, accuracy, training_op):\n tf.add_to_collection(\"my_important_ops\", op)",
"_____no_output_____"
]
],
[
[
"This way people who reuse your model will be able to simply write:",
"_____no_output_____"
]
],
[
[
"X, y, accuracy, training_op = tf.get_collection(\"my_important_ops\")",
"_____no_output_____"
]
],
[
[
"Now you can start a session, restore the model's state and continue training on your data:",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n saver.restore(sess, \"./my_model_final.ckpt\")\n # continue training the model...",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n"
]
],
[
[
"Actually, let's test this for real!",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n saver.restore(sess, \"./my_model_final.ckpt\")\n\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_new_model_final.ckpt\") ",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n0 Validation accuracy: 0.9636\n1 Validation accuracy: 0.9632\n2 Validation accuracy: 0.9658\n3 Validation accuracy: 0.9652\n4 Validation accuracy: 0.9646\n5 Validation accuracy: 0.965\n6 Validation accuracy: 0.969\n7 Validation accuracy: 0.9682\n8 Validation accuracy: 0.9682\n9 Validation accuracy: 0.9684\n10 Validation accuracy: 0.9704\n11 Validation accuracy: 0.971\n12 Validation accuracy: 0.9668\n13 Validation accuracy: 0.97\n14 Validation accuracy: 0.9712\n15 Validation accuracy: 0.9726\n16 Validation accuracy: 0.9718\n17 Validation accuracy: 0.971\n18 Validation accuracy: 0.9712\n19 Validation accuracy: 0.9712\n"
]
],
[
[
"Alternatively, if you have access to the Python code that built the original graph, you can use it instead of `import_meta_graph()`:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\nn_hidden2 = 50\nn_hidden3 = 50\nn_hidden4 = 50\nn_hidden5 = 50\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\")\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu, name=\"hidden2\")\n hidden3 = tf.layers.dense(hidden2, n_hidden3, activation=tf.nn.relu, name=\"hidden3\")\n hidden4 = tf.layers.dense(hidden3, n_hidden4, activation=tf.nn.relu, name=\"hidden4\")\n hidden5 = tf.layers.dense(hidden4, n_hidden5, activation=tf.nn.relu, name=\"hidden5\")\n logits = tf.layers.dense(hidden5, n_outputs, name=\"outputs\")\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\nlearning_rate = 0.01\nthreshold = 1.0\n\noptimizer = tf.train.GradientDescentOptimizer(learning_rate)\ngrads_and_vars = optimizer.compute_gradients(loss)\ncapped_gvs = [(tf.clip_by_value(grad, -threshold, threshold), var)\n for grad, var in grads_and_vars]\ntraining_op = optimizer.apply_gradients(capped_gvs)\n\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"And continue training:",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n saver.restore(sess, \"./my_model_final.ckpt\")\n\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_new_model_final.ckpt\") ",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n0 Validation accuracy: 0.9642\n1 Validation accuracy: 0.9632\n2 Validation accuracy: 0.9656\n3 Validation accuracy: 0.9652\n4 Validation accuracy: 0.9646\n5 Validation accuracy: 0.9652\n6 Validation accuracy: 0.9688\n7 Validation accuracy: 0.9686\n8 Validation accuracy: 0.9682\n9 Validation accuracy: 0.9686\n10 Validation accuracy: 0.9704\n11 Validation accuracy: 0.9712\n12 Validation accuracy: 0.967\n13 Validation accuracy: 0.9698\n14 Validation accuracy: 0.9708\n15 Validation accuracy: 0.9724\n16 Validation accuracy: 0.9718\n17 Validation accuracy: 0.9712\n18 Validation accuracy: 0.9708\n19 Validation accuracy: 0.9712\n"
]
],
[
[
"In general you will want to reuse only the lower layers. If you are using `import_meta_graph()` it will load the whole graph, but you can simply ignore the parts you do not need. In this example, we add a new 4th hidden layer on top of the pretrained 3rd layer (ignoring the old 4th hidden layer). We also build a new output layer, the loss for this new output, and a new optimizer to minimize it. We also need another saver to save the whole graph (containing both the entire old graph plus the new operations), and an initialization operation to initialize all the new variables:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_hidden4 = 20 # new layer\nn_outputs = 10 # new layer\n\nsaver = tf.train.import_meta_graph(\"./my_model_final.ckpt.meta\")\n\nX = tf.get_default_graph().get_tensor_by_name(\"X:0\")\ny = tf.get_default_graph().get_tensor_by_name(\"y:0\")\n\nhidden3 = tf.get_default_graph().get_tensor_by_name(\"dnn/hidden3/Relu:0\")\n\nnew_hidden4 = tf.layers.dense(hidden3, n_hidden4, activation=tf.nn.relu, name=\"new_hidden4\")\nnew_logits = tf.layers.dense(new_hidden4, n_outputs, name=\"new_outputs\")\n\nwith tf.name_scope(\"new_loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=new_logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"new_eval\"):\n correct = tf.nn.in_top_k(new_logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\nwith tf.name_scope(\"new_train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)\n\ninit = tf.global_variables_initializer()\nnew_saver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"And we can train this new model:",
"_____no_output_____"
]
],
[
[
"with tf.Session() as sess:\n init.run()\n saver.restore(sess, \"./my_model_final.ckpt\")\n\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = new_saver.save(sess, \"./my_new_model_final.ckpt\")",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n0 Validation accuracy: 0.9126\n1 Validation accuracy: 0.9374\n2 Validation accuracy: 0.946\n3 Validation accuracy: 0.9498\n4 Validation accuracy: 0.953\n5 Validation accuracy: 0.9528\n6 Validation accuracy: 0.9564\n7 Validation accuracy: 0.96\n8 Validation accuracy: 0.9616\n9 Validation accuracy: 0.9612\n10 Validation accuracy: 0.9634\n11 Validation accuracy: 0.9626\n12 Validation accuracy: 0.9648\n13 Validation accuracy: 0.9656\n14 Validation accuracy: 0.9664\n15 Validation accuracy: 0.967\n16 Validation accuracy: 0.968\n17 Validation accuracy: 0.9678\n18 Validation accuracy: 0.9684\n19 Validation accuracy: 0.9678\n"
]
],
[
[
"If you have access to the Python code that built the original graph, you can just reuse the parts you need and drop the rest:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300 # reused\nn_hidden2 = 50 # reused\nn_hidden3 = 50 # reused\nn_hidden4 = 20 # new!\nn_outputs = 10 # new!\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\") # reused\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu, name=\"hidden2\") # reused\n hidden3 = tf.layers.dense(hidden2, n_hidden3, activation=tf.nn.relu, name=\"hidden3\") # reused\n hidden4 = tf.layers.dense(hidden3, n_hidden4, activation=tf.nn.relu, name=\"hidden4\") # new!\n logits = tf.layers.dense(hidden4, n_outputs, name=\"outputs\") # new!\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)",
"_____no_output_____"
]
],
[
[
"However, you must create one `Saver` to restore the pretrained model (giving it the list of variables to restore, or else it will complain that the graphs don't match), and another `Saver` to save the new model, once it is trained:",
"_____no_output_____"
]
],
[
[
"reuse_vars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES,\n scope=\"hidden[123]\") # regular expression\nrestore_saver = tf.train.Saver(reuse_vars) # to restore layers 1-3\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_model_final.ckpt\")\n\n for epoch in range(n_epochs): # not shown in the book\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size): # not shown\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch}) # not shown\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid}) # not shown\n print(epoch, \"Validation accuracy:\", accuracy_val) # not shown\n\n save_path = saver.save(sess, \"./my_new_model_final.ckpt\")",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n0 Validation accuracy: 0.9024\n1 Validation accuracy: 0.9332\n2 Validation accuracy: 0.943\n3 Validation accuracy: 0.947\n4 Validation accuracy: 0.9516\n5 Validation accuracy: 0.9532\n6 Validation accuracy: 0.9558\n7 Validation accuracy: 0.9592\n8 Validation accuracy: 0.9586\n9 Validation accuracy: 0.9608\n10 Validation accuracy: 0.9626\n11 Validation accuracy: 0.962\n12 Validation accuracy: 0.964\n13 Validation accuracy: 0.9662\n14 Validation accuracy: 0.966\n15 Validation accuracy: 0.9662\n16 Validation accuracy: 0.9672\n17 Validation accuracy: 0.9674\n18 Validation accuracy: 0.9682\n19 Validation accuracy: 0.9678\n"
]
],
[
[
"## Reusing Models from Other Frameworks",
"_____no_output_____"
],
[
"In this example, for each variable we want to reuse, we find its initializer's assignment operation, and we get its second input, which corresponds to the initialization value. When we run the initializer, we replace the initialization values with the ones we want, using a `feed_dict`:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 2\nn_hidden1 = 3",
"_____no_output_____"
],
[
"original_w = [[1., 2., 3.], [4., 5., 6.]] # Load the weights from the other framework\noriginal_b = [7., 8., 9.] # Load the biases from the other framework\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\nhidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\")\n# [...] Build the rest of the model\n\n# Get a handle on the assignment nodes for the hidden1 variables\ngraph = tf.get_default_graph()\nassign_kernel = graph.get_operation_by_name(\"hidden1/kernel/Assign\")\nassign_bias = graph.get_operation_by_name(\"hidden1/bias/Assign\")\ninit_kernel = assign_kernel.inputs[1]\ninit_bias = assign_bias.inputs[1]\n\ninit = tf.global_variables_initializer()\n\nwith tf.Session() as sess:\n sess.run(init, feed_dict={init_kernel: original_w, init_bias: original_b})\n # [...] Train the model on your new task\n print(hidden1.eval(feed_dict={X: [[10.0, 11.0]]})) # not shown in the book",
"[[ 61. 83. 105.]]\n"
]
],
[
[
"Note: the weights variable created by the `tf.layers.dense()` function is called `\"kernel\"` (instead of `\"weights\"` when using the `tf.contrib.layers.fully_connected()`, as in the book), and the biases variable is called `bias` instead of `biases`.",
"_____no_output_____"
],
[
"Another approach (initially used in the book) would be to create dedicated assignment nodes and dedicated placeholders. This is more verbose and less efficient, but you may find this more explicit:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 2\nn_hidden1 = 3\n\noriginal_w = [[1., 2., 3.], [4., 5., 6.]] # Load the weights from the other framework\noriginal_b = [7., 8., 9.] # Load the biases from the other framework\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\nhidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\")\n# [...] Build the rest of the model\n\n# Get a handle on the variables of layer hidden1\nwith tf.variable_scope(\"\", default_name=\"\", reuse=True): # root scope\n hidden1_weights = tf.get_variable(\"hidden1/kernel\")\n hidden1_biases = tf.get_variable(\"hidden1/bias\")\n\n# Create dedicated placeholders and assignment nodes\noriginal_weights = tf.placeholder(tf.float32, shape=(n_inputs, n_hidden1))\noriginal_biases = tf.placeholder(tf.float32, shape=n_hidden1)\nassign_hidden1_weights = tf.assign(hidden1_weights, original_weights)\nassign_hidden1_biases = tf.assign(hidden1_biases, original_biases)\n\ninit = tf.global_variables_initializer()\n\nwith tf.Session() as sess:\n sess.run(init)\n sess.run(assign_hidden1_weights, feed_dict={original_weights: original_w})\n sess.run(assign_hidden1_biases, feed_dict={original_biases: original_b})\n # [...] Train the model on your new task\n print(hidden1.eval(feed_dict={X: [[10.0, 11.0]]}))",
"[[ 61. 83. 105.]]\n"
]
],
[
[
"Note that we could also get a handle on the variables using `get_collection()` and specifying the `scope`:",
"_____no_output_____"
]
],
[
[
"tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES, scope=\"hidden1\")",
"_____no_output_____"
]
],
[
[
"Or we could use the graph's `get_tensor_by_name()` method:",
"_____no_output_____"
]
],
[
[
"tf.get_default_graph().get_tensor_by_name(\"hidden1/kernel:0\")",
"_____no_output_____"
],
[
"tf.get_default_graph().get_tensor_by_name(\"hidden1/bias:0\")",
"_____no_output_____"
]
],
[
[
"### Freezing the Lower Layers",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300 # reused\nn_hidden2 = 50 # reused\nn_hidden3 = 50 # reused\nn_hidden4 = 20 # new!\nn_outputs = 10 # new!\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\") # reused\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu, name=\"hidden2\") # reused\n hidden3 = tf.layers.dense(hidden2, n_hidden3, activation=tf.nn.relu, name=\"hidden3\") # reused\n hidden4 = tf.layers.dense(hidden3, n_hidden4, activation=tf.nn.relu, name=\"hidden4\") # new!\n logits = tf.layers.dense(hidden4, n_outputs, name=\"outputs\") # new!\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")",
"_____no_output_____"
],
[
"with tf.name_scope(\"train\"): # not shown in the book\n optimizer = tf.train.GradientDescentOptimizer(learning_rate) # not shown\n train_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES,\n scope=\"hidden[34]|outputs\")\n training_op = optimizer.minimize(loss, var_list=train_vars)",
"_____no_output_____"
],
[
"reuse_vars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES,\n scope=\"hidden[123]\") # regular expression\nrestore_saver = tf.train.Saver(reuse_vars) # to restore layers 1-3\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_model_final.ckpt\")\n\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_new_model_final.ckpt\")",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n0 Validation accuracy: 0.8964\n1 Validation accuracy: 0.9298\n2 Validation accuracy: 0.94\n3 Validation accuracy: 0.9442\n4 Validation accuracy: 0.948\n5 Validation accuracy: 0.951\n6 Validation accuracy: 0.9508\n7 Validation accuracy: 0.9538\n8 Validation accuracy: 0.9554\n9 Validation accuracy: 0.957\n10 Validation accuracy: 0.9562\n11 Validation accuracy: 0.9566\n12 Validation accuracy: 0.9572\n13 Validation accuracy: 0.9578\n14 Validation accuracy: 0.959\n15 Validation accuracy: 0.9576\n16 Validation accuracy: 0.9574\n17 Validation accuracy: 0.9602\n18 Validation accuracy: 0.9592\n19 Validation accuracy: 0.9602\n"
],
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300 # reused\nn_hidden2 = 50 # reused\nn_hidden3 = 50 # reused\nn_hidden4 = 20 # new!\nn_outputs = 10 # new!\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")",
"_____no_output_____"
],
[
"with tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu,\n name=\"hidden1\") # reused frozen\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu,\n name=\"hidden2\") # reused frozen\n hidden2_stop = tf.stop_gradient(hidden2)\n hidden3 = tf.layers.dense(hidden2_stop, n_hidden3, activation=tf.nn.relu,\n name=\"hidden3\") # reused, not frozen\n hidden4 = tf.layers.dense(hidden3, n_hidden4, activation=tf.nn.relu,\n name=\"hidden4\") # new!\n logits = tf.layers.dense(hidden4, n_outputs, name=\"outputs\") # new!",
"_____no_output_____"
],
[
"with tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)",
"_____no_output_____"
]
],
[
[
"The training code is exactly the same as earlier:",
"_____no_output_____"
]
],
[
[
"reuse_vars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES,\n scope=\"hidden[123]\") # regular expression\nrestore_saver = tf.train.Saver(reuse_vars) # to restore layers 1-3\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_model_final.ckpt\")\n\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_new_model_final.ckpt\")",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n0 Validation accuracy: 0.902\n1 Validation accuracy: 0.9302\n2 Validation accuracy: 0.9438\n3 Validation accuracy: 0.9478\n4 Validation accuracy: 0.9514\n5 Validation accuracy: 0.9522\n6 Validation accuracy: 0.9524\n7 Validation accuracy: 0.9556\n8 Validation accuracy: 0.9556\n9 Validation accuracy: 0.9558\n10 Validation accuracy: 0.957\n11 Validation accuracy: 0.9552\n12 Validation accuracy: 0.9572\n13 Validation accuracy: 0.9582\n14 Validation accuracy: 0.9582\n15 Validation accuracy: 0.957\n16 Validation accuracy: 0.9566\n17 Validation accuracy: 0.9578\n18 Validation accuracy: 0.9594\n19 Validation accuracy: 0.958\n"
]
],
[
[
"### Caching the Frozen Layers",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300 # reused\nn_hidden2 = 50 # reused\nn_hidden3 = 50 # reused\nn_hidden4 = 20 # new!\nn_outputs = 10 # new!\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu,\n name=\"hidden1\") # reused frozen\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu,\n name=\"hidden2\") # reused frozen & cached\n hidden2_stop = tf.stop_gradient(hidden2)\n hidden3 = tf.layers.dense(hidden2_stop, n_hidden3, activation=tf.nn.relu,\n name=\"hidden3\") # reused, not frozen\n hidden4 = tf.layers.dense(hidden3, n_hidden4, activation=tf.nn.relu,\n name=\"hidden4\") # new!\n logits = tf.layers.dense(hidden4, n_outputs, name=\"outputs\") # new!\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)",
"_____no_output_____"
],
[
"reuse_vars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES,\n scope=\"hidden[123]\") # regular expression\nrestore_saver = tf.train.Saver(reuse_vars) # to restore layers 1-3\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"import numpy as np\n\nn_batches = len(X_train) // batch_size\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_model_final.ckpt\")\n \n h2_cache = sess.run(hidden2, feed_dict={X: X_train})\n h2_cache_valid = sess.run(hidden2, feed_dict={X: X_valid}) # not shown in the book\n\n for epoch in range(n_epochs):\n shuffled_idx = np.random.permutation(len(X_train))\n hidden2_batches = np.array_split(h2_cache[shuffled_idx], n_batches)\n y_batches = np.array_split(y_train[shuffled_idx], n_batches)\n for hidden2_batch, y_batch in zip(hidden2_batches, y_batches):\n sess.run(training_op, feed_dict={hidden2:hidden2_batch, y:y_batch})\n\n accuracy_val = accuracy.eval(feed_dict={hidden2: h2_cache_valid, # not shown\n y: y_valid}) # not shown\n print(epoch, \"Validation accuracy:\", accuracy_val) # not shown\n\n save_path = saver.save(sess, \"./my_new_model_final.ckpt\")",
"INFO:tensorflow:Restoring parameters from ./my_model_final.ckpt\n0 Validation accuracy: 0.902\n1 Validation accuracy: 0.9302\n2 Validation accuracy: 0.9438\n3 Validation accuracy: 0.9478\n4 Validation accuracy: 0.9514\n5 Validation accuracy: 0.9522\n6 Validation accuracy: 0.9524\n7 Validation accuracy: 0.9556\n8 Validation accuracy: 0.9556\n9 Validation accuracy: 0.9558\n10 Validation accuracy: 0.957\n11 Validation accuracy: 0.9552\n12 Validation accuracy: 0.9572\n13 Validation accuracy: 0.9582\n14 Validation accuracy: 0.9582\n15 Validation accuracy: 0.957\n16 Validation accuracy: 0.9566\n17 Validation accuracy: 0.9578\n18 Validation accuracy: 0.9594\n19 Validation accuracy: 0.958\n"
]
],
[
[
"# Faster Optimizers",
"_____no_output_____"
],
[
"## Momentum optimization",
"_____no_output_____"
]
],
[
[
"optimizer = tf.train.MomentumOptimizer(learning_rate=learning_rate,\n momentum=0.9)",
"_____no_output_____"
]
],
[
[
"## Nesterov Accelerated Gradient",
"_____no_output_____"
]
],
[
[
"optimizer = tf.train.MomentumOptimizer(learning_rate=learning_rate,\n momentum=0.9, use_nesterov=True)",
"_____no_output_____"
]
],
[
[
"## AdaGrad",
"_____no_output_____"
]
],
[
[
"optimizer = tf.train.AdagradOptimizer(learning_rate=learning_rate)",
"_____no_output_____"
]
],
[
[
"## RMSProp",
"_____no_output_____"
]
],
[
[
"optimizer = tf.train.RMSPropOptimizer(learning_rate=learning_rate,\n momentum=0.9, decay=0.9, epsilon=1e-10)",
"_____no_output_____"
]
],
[
[
"## Adam Optimization",
"_____no_output_____"
]
],
[
[
"optimizer = tf.train.AdamOptimizer(learning_rate=learning_rate)",
"_____no_output_____"
]
],
[
[
"## Learning Rate Scheduling",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\nn_hidden2 = 50\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\")\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu, name=\"hidden2\")\n logits = tf.layers.dense(hidden2, n_outputs, name=\"outputs\")\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")",
"_____no_output_____"
],
[
"with tf.name_scope(\"train\"): # not shown in the book\n initial_learning_rate = 0.1\n decay_steps = 10000\n decay_rate = 1/10\n global_step = tf.Variable(0, trainable=False, name=\"global_step\")\n learning_rate = tf.train.exponential_decay(initial_learning_rate, global_step,\n decay_steps, decay_rate)\n optimizer = tf.train.MomentumOptimizer(learning_rate, momentum=0.9)\n training_op = optimizer.minimize(loss, global_step=global_step)",
"_____no_output_____"
],
[
"init = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 5\nbatch_size = 50\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\")",
"0 Validation accuracy: 0.959\n1 Validation accuracy: 0.9688\n2 Validation accuracy: 0.9726\n3 Validation accuracy: 0.9804\n4 Validation accuracy: 0.982\n"
]
],
[
[
"# Avoiding Overfitting Through Regularization",
"_____no_output_____"
],
[
"## $\\ell_1$ and $\\ell_2$ regularization",
"_____no_output_____"
],
[
"Let's implement $\\ell_1$ regularization manually. First, we create the model, as usual (with just one hidden layer this time, for simplicity):",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\")\n logits = tf.layers.dense(hidden1, n_outputs, name=\"outputs\")",
"_____no_output_____"
]
],
[
[
"Next, we get a handle on the layer weights, and we compute the total loss, which is equal to the sum of the usual cross entropy loss and the $\\ell_1$ loss (i.e., the absolute values of the weights):",
"_____no_output_____"
]
],
[
[
"W1 = tf.get_default_graph().get_tensor_by_name(\"hidden1/kernel:0\")\nW2 = tf.get_default_graph().get_tensor_by_name(\"outputs/kernel:0\")\n\nscale = 0.001 # l1 regularization hyperparameter\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y,\n logits=logits)\n base_loss = tf.reduce_mean(xentropy, name=\"avg_xentropy\")\n reg_losses = tf.reduce_sum(tf.abs(W1)) + tf.reduce_sum(tf.abs(W2))\n loss = tf.add(base_loss, scale * reg_losses, name=\"loss\")",
"_____no_output_____"
]
],
[
[
"The rest is just as usual:",
"_____no_output_____"
]
],
[
[
"with tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\nlearning_rate = 0.01\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 20\nbatch_size = 200\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\")",
"0 Validation accuracy: 0.831\n1 Validation accuracy: 0.871\n2 Validation accuracy: 0.8838\n3 Validation accuracy: 0.8934\n4 Validation accuracy: 0.8966\n5 Validation accuracy: 0.8988\n6 Validation accuracy: 0.9016\n7 Validation accuracy: 0.9044\n8 Validation accuracy: 0.9058\n9 Validation accuracy: 0.906\n10 Validation accuracy: 0.9068\n11 Validation accuracy: 0.9054\n12 Validation accuracy: 0.907\n13 Validation accuracy: 0.9084\n14 Validation accuracy: 0.9088\n15 Validation accuracy: 0.9064\n16 Validation accuracy: 0.9066\n17 Validation accuracy: 0.9066\n18 Validation accuracy: 0.9066\n19 Validation accuracy: 0.9052\n"
]
],
[
[
"Alternatively, we can pass a regularization function to the `tf.layers.dense()` function, which will use it to create operations that will compute the regularization loss, and it adds these operations to the collection of regularization losses. The beginning is the same as above:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_hidden1 = 300\nn_hidden2 = 50\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")",
"_____no_output_____"
]
],
[
[
"Next, we will use Python's `partial()` function to avoid repeating the same arguments over and over again. Note that we set the `kernel_regularizer` argument:",
"_____no_output_____"
]
],
[
[
"scale = 0.001",
"_____no_output_____"
],
[
"my_dense_layer = partial(\n tf.layers.dense, activation=tf.nn.relu,\n kernel_regularizer=tf.contrib.layers.l1_regularizer(scale))\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = my_dense_layer(X, n_hidden1, name=\"hidden1\")\n hidden2 = my_dense_layer(hidden1, n_hidden2, name=\"hidden2\")\n logits = my_dense_layer(hidden2, n_outputs, activation=None,\n name=\"outputs\")",
"_____no_output_____"
]
],
[
[
"Next we must add the regularization losses to the base loss:",
"_____no_output_____"
]
],
[
[
"with tf.name_scope(\"loss\"): # not shown in the book\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits( # not shown\n labels=y, logits=logits) # not shown\n base_loss = tf.reduce_mean(xentropy, name=\"avg_xentropy\") # not shown\n reg_losses = tf.get_collection(tf.GraphKeys.REGULARIZATION_LOSSES)\n loss = tf.add_n([base_loss] + reg_losses, name=\"loss\")",
"_____no_output_____"
]
],
[
[
"And the rest is the same as usual:",
"_____no_output_____"
]
],
[
[
"with tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\nlearning_rate = 0.01\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.GradientDescentOptimizer(learning_rate)\n training_op = optimizer.minimize(loss)\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 20\nbatch_size = 200\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\")",
"0 Validation accuracy: 0.8274\n1 Validation accuracy: 0.8766\n2 Validation accuracy: 0.8952\n3 Validation accuracy: 0.9016\n4 Validation accuracy: 0.908\n5 Validation accuracy: 0.9096\n6 Validation accuracy: 0.9126\n7 Validation accuracy: 0.9154\n8 Validation accuracy: 0.9178\n9 Validation accuracy: 0.919\n10 Validation accuracy: 0.92\n11 Validation accuracy: 0.9224\n12 Validation accuracy: 0.9212\n13 Validation accuracy: 0.9228\n14 Validation accuracy: 0.9224\n15 Validation accuracy: 0.9216\n16 Validation accuracy: 0.9218\n17 Validation accuracy: 0.9228\n18 Validation accuracy: 0.9216\n19 Validation accuracy: 0.9214\n"
]
],
[
[
"## Dropout",
"_____no_output_____"
],
[
"Note: the book uses `tf.contrib.layers.dropout()` rather than `tf.layers.dropout()` (which did not exist when this chapter was written). It is now preferable to use `tf.layers.dropout()`, because anything in the contrib module may change or be deleted without notice. The `tf.layers.dropout()` function is almost identical to the `tf.contrib.layers.dropout()` function, except for a few minor differences. Most importantly:\n* you must specify the dropout rate (`rate`) rather than the keep probability (`keep_prob`), where `rate` is simply equal to `1 - keep_prob`,\n* the `is_training` parameter is renamed to `training`.",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")",
"_____no_output_____"
],
[
"training = tf.placeholder_with_default(False, shape=(), name='training')\n\ndropout_rate = 0.5 # == 1 - keep_prob\nX_drop = tf.layers.dropout(X, dropout_rate, training=training)\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X_drop, n_hidden1, activation=tf.nn.relu,\n name=\"hidden1\")\n hidden1_drop = tf.layers.dropout(hidden1, dropout_rate, training=training)\n hidden2 = tf.layers.dense(hidden1_drop, n_hidden2, activation=tf.nn.relu,\n name=\"hidden2\")\n hidden2_drop = tf.layers.dropout(hidden2, dropout_rate, training=training)\n logits = tf.layers.dense(hidden2_drop, n_outputs, name=\"outputs\")",
"_____no_output_____"
],
[
"with tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.MomentumOptimizer(learning_rate, momentum=0.9)\n training_op = optimizer.minimize(loss) \n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n \ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 20\nbatch_size = 50\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch, training: True})\n accuracy_val = accuracy.eval(feed_dict={X: X_valid, y: y_valid})\n print(epoch, \"Validation accuracy:\", accuracy_val)\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\")",
"0 Validation accuracy: 0.9264\n1 Validation accuracy: 0.9446\n2 Validation accuracy: 0.9488\n3 Validation accuracy: 0.9556\n4 Validation accuracy: 0.9612\n5 Validation accuracy: 0.9598\n6 Validation accuracy: 0.9616\n7 Validation accuracy: 0.9674\n8 Validation accuracy: 0.967\n9 Validation accuracy: 0.9706\n10 Validation accuracy: 0.9674\n11 Validation accuracy: 0.9678\n12 Validation accuracy: 0.9698\n13 Validation accuracy: 0.97\n14 Validation accuracy: 0.971\n15 Validation accuracy: 0.9702\n16 Validation accuracy: 0.9718\n17 Validation accuracy: 0.9716\n18 Validation accuracy: 0.9734\n19 Validation accuracy: 0.972\n"
]
],
[
[
"## Max norm",
"_____no_output_____"
],
[
"Let's go back to a plain and simple neural net for MNIST with just 2 hidden layers:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28\nn_hidden1 = 300\nn_hidden2 = 50\nn_outputs = 10\n\nlearning_rate = 0.01\nmomentum = 0.9\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu, name=\"hidden1\")\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu, name=\"hidden2\")\n logits = tf.layers.dense(hidden2, n_outputs, name=\"outputs\")\n\nwith tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)\n training_op = optimizer.minimize(loss) \n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))",
"_____no_output_____"
]
],
[
[
"Next, let's get a handle on the first hidden layer's weight and create an operation that will compute the clipped weights using the `clip_by_norm()` function. Then we create an assignment operation to assign the clipped weights to the weights variable:",
"_____no_output_____"
]
],
[
[
"threshold = 1.0\nweights = tf.get_default_graph().get_tensor_by_name(\"hidden1/kernel:0\")\nclipped_weights = tf.clip_by_norm(weights, clip_norm=threshold, axes=1)\nclip_weights = tf.assign(weights, clipped_weights)",
"_____no_output_____"
]
],
[
[
"We can do this as well for the second hidden layer:",
"_____no_output_____"
]
],
[
[
"weights2 = tf.get_default_graph().get_tensor_by_name(\"hidden2/kernel:0\")\nclipped_weights2 = tf.clip_by_norm(weights2, clip_norm=threshold, axes=1)\nclip_weights2 = tf.assign(weights2, clipped_weights2)",
"_____no_output_____"
]
],
[
[
"Let's add an initializer and a saver:",
"_____no_output_____"
]
],
[
[
"init = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"And now we can train the model. It's pretty much as usual, except that right after running the `training_op`, we run the `clip_weights` and `clip_weights2` operations:",
"_____no_output_____"
]
],
[
[
"n_epochs = 20\nbatch_size = 50",
"_____no_output_____"
],
[
"with tf.Session() as sess: # not shown in the book\n init.run() # not shown\n for epoch in range(n_epochs): # not shown\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size): # not shown\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n clip_weights.eval()\n clip_weights2.eval() # not shown\n acc_valid = accuracy.eval(feed_dict={X: X_valid, y: y_valid}) # not shown\n print(epoch, \"Validation accuracy:\", acc_valid) # not shown\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\") # not shown",
"0 Validation accuracy: 0.9568\n1 Validation accuracy: 0.9696\n2 Validation accuracy: 0.972\n3 Validation accuracy: 0.9768\n4 Validation accuracy: 0.9784\n5 Validation accuracy: 0.9786\n6 Validation accuracy: 0.9816\n7 Validation accuracy: 0.9808\n8 Validation accuracy: 0.981\n9 Validation accuracy: 0.983\n10 Validation accuracy: 0.9822\n11 Validation accuracy: 0.9854\n12 Validation accuracy: 0.9822\n13 Validation accuracy: 0.9842\n14 Validation accuracy: 0.984\n15 Validation accuracy: 0.9852\n16 Validation accuracy: 0.984\n17 Validation accuracy: 0.9844\n18 Validation accuracy: 0.9844\n19 Validation accuracy: 0.9844\n"
]
],
[
[
"The implementation above is straightforward and it works fine, but it is a bit messy. A better approach is to define a `max_norm_regularizer()` function:",
"_____no_output_____"
]
],
[
[
"def max_norm_regularizer(threshold, axes=1, name=\"max_norm\",\n collection=\"max_norm\"):\n def max_norm(weights):\n clipped = tf.clip_by_norm(weights, clip_norm=threshold, axes=axes)\n clip_weights = tf.assign(weights, clipped, name=name)\n tf.add_to_collection(collection, clip_weights)\n return None # there is no regularization loss term\n return max_norm",
"_____no_output_____"
]
],
[
[
"Then you can call this function to get a max norm regularizer (with the threshold you want). When you create a hidden layer, you can pass this regularizer to the `kernel_regularizer` argument:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28\nn_hidden1 = 300\nn_hidden2 = 50\nn_outputs = 10\n\nlearning_rate = 0.01\nmomentum = 0.9\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")",
"_____no_output_____"
],
[
"max_norm_reg = max_norm_regularizer(threshold=1.0)\n\nwith tf.name_scope(\"dnn\"):\n hidden1 = tf.layers.dense(X, n_hidden1, activation=tf.nn.relu,\n kernel_regularizer=max_norm_reg, name=\"hidden1\")\n hidden2 = tf.layers.dense(hidden1, n_hidden2, activation=tf.nn.relu,\n kernel_regularizer=max_norm_reg, name=\"hidden2\")\n logits = tf.layers.dense(hidden2, n_outputs, name=\"outputs\")",
"_____no_output_____"
],
[
"with tf.name_scope(\"loss\"):\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\nwith tf.name_scope(\"train\"):\n optimizer = tf.train.MomentumOptimizer(learning_rate, momentum)\n training_op = optimizer.minimize(loss) \n\nwith tf.name_scope(\"eval\"):\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"Training is as usual, except you must run the weights clipping operations after each training operation:",
"_____no_output_____"
]
],
[
[
"n_epochs = 20\nbatch_size = 50",
"_____no_output_____"
],
[
"clip_all_weights = tf.get_collection(\"max_norm\")\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for X_batch, y_batch in shuffle_batch(X_train, y_train, batch_size):\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n sess.run(clip_all_weights)\n acc_valid = accuracy.eval(feed_dict={X: X_valid, y: y_valid}) # not shown\n print(epoch, \"Validation accuracy:\", acc_valid) # not shown\n\n save_path = saver.save(sess, \"./my_model_final.ckpt\") # not shown",
"0 Validation accuracy: 0.9556\n1 Validation accuracy: 0.9698\n2 Validation accuracy: 0.9726\n3 Validation accuracy: 0.9744\n4 Validation accuracy: 0.9762\n5 Validation accuracy: 0.9772\n6 Validation accuracy: 0.979\n7 Validation accuracy: 0.9816\n8 Validation accuracy: 0.9814\n9 Validation accuracy: 0.9812\n10 Validation accuracy: 0.9818\n11 Validation accuracy: 0.9816\n12 Validation accuracy: 0.9802\n13 Validation accuracy: 0.9822\n14 Validation accuracy: 0.982\n15 Validation accuracy: 0.9812\n16 Validation accuracy: 0.9824\n17 Validation accuracy: 0.9836\n18 Validation accuracy: 0.9824\n19 Validation accuracy: 0.9826\n"
]
],
[
[
"# Exercise solutions",
"_____no_output_____"
],
[
"## 1. to 7.",
"_____no_output_____"
],
[
"See appendix A.",
"_____no_output_____"
],
[
"## 8. Deep Learning",
"_____no_output_____"
],
[
"### 8.1.",
"_____no_output_____"
],
[
"_Exercise: Build a DNN with five hidden layers of 100 neurons each, He initialization, and the ELU activation function._",
"_____no_output_____"
],
[
"We will need similar DNNs in the next exercises, so let's create a function to build this DNN:",
"_____no_output_____"
]
],
[
[
"he_init = tf.variance_scaling_initializer()\n\ndef dnn(inputs, n_hidden_layers=5, n_neurons=100, name=None,\n activation=tf.nn.elu, initializer=he_init):\n with tf.variable_scope(name, \"dnn\"):\n for layer in range(n_hidden_layers):\n inputs = tf.layers.dense(inputs, n_neurons, activation=activation,\n kernel_initializer=initializer,\n name=\"hidden%d\" % (layer + 1))\n return inputs",
"_____no_output_____"
],
[
"n_inputs = 28 * 28 # MNIST\nn_outputs = 5\n\nreset_graph()\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\ndnn_outputs = dnn(X)\n\nlogits = tf.layers.dense(dnn_outputs, n_outputs, kernel_initializer=he_init, name=\"logits\")\nY_proba = tf.nn.softmax(logits, name=\"Y_proba\")",
"_____no_output_____"
]
],
[
[
"### 8.2.",
"_____no_output_____"
],
[
"_Exercise: Using Adam optimization and early stopping, try training it on MNIST but only on digits 0 to 4, as we will use transfer learning for digits 5 to 9 in the next exercise. You will need a softmax output layer with five neurons, and as always make sure to save checkpoints at regular intervals and save the final model so you can reuse it later._",
"_____no_output_____"
],
[
"Let's complete the graph with the cost function, the training op, and all the other usual components:",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\n\nxentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\nloss = tf.reduce_mean(xentropy, name=\"loss\")\n\noptimizer = tf.train.AdamOptimizer(learning_rate)\ntraining_op = optimizer.minimize(loss, name=\"training_op\")\n\ncorrect = tf.nn.in_top_k(logits, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\ninit = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"Now let's create the training set, validation and test set (we need the validation set to implement early stopping):",
"_____no_output_____"
]
],
[
[
"X_train1 = X_train[y_train < 5]\ny_train1 = y_train[y_train < 5]\nX_valid1 = X_valid[y_valid < 5]\ny_valid1 = y_valid[y_valid < 5]\nX_test1 = X_test[y_test < 5]\ny_test1 = y_test[y_test < 5]",
"_____no_output_____"
],
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n\n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train1))\n for rnd_indices in np.array_split(rnd_idx, len(X_train1) // batch_size):\n X_batch, y_batch = X_train1[rnd_indices], y_train1[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid1, y: y_valid1})\n if loss_val < best_loss:\n save_path = saver.save(sess, \"./my_mnist_model_0_to_4.ckpt\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n saver.restore(sess, \"./my_mnist_model_0_to_4.ckpt\")\n acc_test = accuracy.eval(feed_dict={X: X_test1, y: y_test1})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"0\tValidation loss: 0.116407\tBest loss: 0.116407\tAccuracy: 97.58%\n1\tValidation loss: 0.180534\tBest loss: 0.116407\tAccuracy: 97.11%\n2\tValidation loss: 0.227535\tBest loss: 0.116407\tAccuracy: 93.86%\n3\tValidation loss: 0.107346\tBest loss: 0.107346\tAccuracy: 97.54%\n4\tValidation loss: 0.302668\tBest loss: 0.107346\tAccuracy: 95.35%\n5\tValidation loss: 1.631054\tBest loss: 0.107346\tAccuracy: 22.01%\n6\tValidation loss: 1.635262\tBest loss: 0.107346\tAccuracy: 18.73%\n7\tValidation loss: 1.671200\tBest loss: 0.107346\tAccuracy: 22.01%\n8\tValidation loss: 1.695277\tBest loss: 0.107346\tAccuracy: 19.27%\n9\tValidation loss: 1.744607\tBest loss: 0.107346\tAccuracy: 20.91%\n10\tValidation loss: 1.629857\tBest loss: 0.107346\tAccuracy: 22.01%\n11\tValidation loss: 1.810803\tBest loss: 0.107346\tAccuracy: 22.01%\n12\tValidation loss: 1.675703\tBest loss: 0.107346\tAccuracy: 18.73%\n13\tValidation loss: 1.633233\tBest loss: 0.107346\tAccuracy: 20.91%\n14\tValidation loss: 1.652905\tBest loss: 0.107346\tAccuracy: 20.91%\n15\tValidation loss: 1.635937\tBest loss: 0.107346\tAccuracy: 20.91%\n16\tValidation loss: 1.718919\tBest loss: 0.107346\tAccuracy: 19.08%\n17\tValidation loss: 1.682458\tBest loss: 0.107346\tAccuracy: 19.27%\n18\tValidation loss: 1.675366\tBest loss: 0.107346\tAccuracy: 18.73%\n19\tValidation loss: 1.645800\tBest loss: 0.107346\tAccuracy: 19.08%\n20\tValidation loss: 1.722334\tBest loss: 0.107346\tAccuracy: 22.01%\n21\tValidation loss: 1.656418\tBest loss: 0.107346\tAccuracy: 22.01%\n22\tValidation loss: 1.643529\tBest loss: 0.107346\tAccuracy: 18.73%\n23\tValidation loss: 1.644233\tBest loss: 0.107346\tAccuracy: 19.27%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_0_to_4.ckpt\nFinal test accuracy: 97.26%\n"
]
],
[
[
"This test accuracy is not too bad, but let's see if we can do better by tuning the hyperparameters.",
"_____no_output_____"
],
[
"### 8.3.",
"_____no_output_____"
],
[
"_Exercise: Tune the hyperparameters using cross-validation and see what precision you can achieve._",
"_____no_output_____"
],
[
"Let's create a `DNNClassifier` class, compatible with Scikit-Learn's `RandomizedSearchCV` class, to perform hyperparameter tuning. Here are the key points of this implementation:\n* the `__init__()` method (constructor) does nothing more than create instance variables for each of the hyperparameters.\n* the `fit()` method creates the graph, starts a session and trains the model:\n * it calls the `_build_graph()` method to build the graph (much lile the graph we defined earlier). Once this method is done creating the graph, it saves all the important operations as instance variables for easy access by other methods.\n * the `_dnn()` method builds the hidden layers, just like the `dnn()` function above, but also with support for batch normalization and dropout (for the next exercises).\n * if the `fit()` method is given a validation set (`X_valid` and `y_valid`), then it implements early stopping. This implementation does not save the best model to disk, but rather to memory: it uses the `_get_model_params()` method to get all the graph's variables and their values, and the `_restore_model_params()` method to restore the variable values (of the best model found). This trick helps speed up training.\n * After the `fit()` method has finished training the model, it keeps the session open so that predictions can be made quickly, without having to save a model to disk and restore it for every prediction. You can close the session by calling the `close_session()` method.\n* the `predict_proba()` method uses the trained model to predict the class probabilities.\n* the `predict()` method calls `predict_proba()` and returns the class with the highest probability, for each instance.",
"_____no_output_____"
]
],
[
[
"from sklearn.base import BaseEstimator, ClassifierMixin\nfrom sklearn.exceptions import NotFittedError\n\nclass DNNClassifier(BaseEstimator, ClassifierMixin):\n def __init__(self, n_hidden_layers=5, n_neurons=100, optimizer_class=tf.train.AdamOptimizer,\n learning_rate=0.01, batch_size=20, activation=tf.nn.elu, initializer=he_init,\n batch_norm_momentum=None, dropout_rate=None, random_state=None):\n \"\"\"Initialize the DNNClassifier by simply storing all the hyperparameters.\"\"\"\n self.n_hidden_layers = n_hidden_layers\n self.n_neurons = n_neurons\n self.optimizer_class = optimizer_class\n self.learning_rate = learning_rate\n self.batch_size = batch_size\n self.activation = activation\n self.initializer = initializer\n self.batch_norm_momentum = batch_norm_momentum\n self.dropout_rate = dropout_rate\n self.random_state = random_state\n self._session = None\n\n def _dnn(self, inputs):\n \"\"\"Build the hidden layers, with support for batch normalization and dropout.\"\"\"\n for layer in range(self.n_hidden_layers):\n if self.dropout_rate:\n inputs = tf.layers.dropout(inputs, self.dropout_rate, training=self._training)\n inputs = tf.layers.dense(inputs, self.n_neurons,\n kernel_initializer=self.initializer,\n name=\"hidden%d\" % (layer + 1))\n if self.batch_norm_momentum:\n inputs = tf.layers.batch_normalization(inputs, momentum=self.batch_norm_momentum,\n training=self._training)\n inputs = self.activation(inputs, name=\"hidden%d_out\" % (layer + 1))\n return inputs\n\n def _build_graph(self, n_inputs, n_outputs):\n \"\"\"Build the same model as earlier\"\"\"\n if self.random_state is not None:\n tf.set_random_seed(self.random_state)\n np.random.seed(self.random_state)\n\n X = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\n y = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\n if self.batch_norm_momentum or self.dropout_rate:\n self._training = tf.placeholder_with_default(False, shape=(), name='training')\n else:\n self._training = None\n\n dnn_outputs = self._dnn(X)\n\n logits = tf.layers.dense(dnn_outputs, n_outputs, kernel_initializer=he_init, name=\"logits\")\n Y_proba = tf.nn.softmax(logits, name=\"Y_proba\")\n\n xentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y,\n logits=logits)\n loss = tf.reduce_mean(xentropy, name=\"loss\")\n\n optimizer = self.optimizer_class(learning_rate=self.learning_rate)\n training_op = optimizer.minimize(loss)\n\n correct = tf.nn.in_top_k(logits, y, 1)\n accuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\n init = tf.global_variables_initializer()\n saver = tf.train.Saver()\n\n # Make the important operations available easily through instance variables\n self._X, self._y = X, y\n self._Y_proba, self._loss = Y_proba, loss\n self._training_op, self._accuracy = training_op, accuracy\n self._init, self._saver = init, saver\n\n def close_session(self):\n if self._session:\n self._session.close()\n\n def _get_model_params(self):\n \"\"\"Get all variable values (used for early stopping, faster than saving to disk)\"\"\"\n with self._graph.as_default():\n gvars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES)\n return {gvar.op.name: value for gvar, value in zip(gvars, self._session.run(gvars))}\n\n def _restore_model_params(self, model_params):\n \"\"\"Set all variables to the given values (for early stopping, faster than loading from disk)\"\"\"\n gvar_names = list(model_params.keys())\n assign_ops = {gvar_name: self._graph.get_operation_by_name(gvar_name + \"/Assign\")\n for gvar_name in gvar_names}\n init_values = {gvar_name: assign_op.inputs[1] for gvar_name, assign_op in assign_ops.items()}\n feed_dict = {init_values[gvar_name]: model_params[gvar_name] for gvar_name in gvar_names}\n self._session.run(assign_ops, feed_dict=feed_dict)\n\n def fit(self, X, y, n_epochs=100, X_valid=None, y_valid=None):\n \"\"\"Fit the model to the training set. If X_valid and y_valid are provided, use early stopping.\"\"\"\n self.close_session()\n\n # infer n_inputs and n_outputs from the training set.\n n_inputs = X.shape[1]\n self.classes_ = np.unique(y)\n n_outputs = len(self.classes_)\n \n # Translate the labels vector to a vector of sorted class indices, containing\n # integers from 0 to n_outputs - 1.\n # For example, if y is equal to [8, 8, 9, 5, 7, 6, 6, 6], then the sorted class\n # labels (self.classes_) will be equal to [5, 6, 7, 8, 9], and the labels vector\n # will be translated to [3, 3, 4, 0, 2, 1, 1, 1]\n self.class_to_index_ = {label: index\n for index, label in enumerate(self.classes_)}\n y = np.array([self.class_to_index_[label]\n for label in y], dtype=np.int32)\n \n self._graph = tf.Graph()\n with self._graph.as_default():\n self._build_graph(n_inputs, n_outputs)\n # extra ops for batch normalization\n extra_update_ops = tf.get_collection(tf.GraphKeys.UPDATE_OPS)\n\n # needed in case of early stopping\n max_checks_without_progress = 20\n checks_without_progress = 0\n best_loss = np.infty\n best_params = None\n \n # Now train the model!\n self._session = tf.Session(graph=self._graph)\n with self._session.as_default() as sess:\n self._init.run()\n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X))\n for rnd_indices in np.array_split(rnd_idx, len(X) // self.batch_size):\n X_batch, y_batch = X[rnd_indices], y[rnd_indices]\n feed_dict = {self._X: X_batch, self._y: y_batch}\n if self._training is not None:\n feed_dict[self._training] = True\n sess.run(self._training_op, feed_dict=feed_dict)\n if extra_update_ops:\n sess.run(extra_update_ops, feed_dict=feed_dict)\n if X_valid is not None and y_valid is not None:\n loss_val, acc_val = sess.run([self._loss, self._accuracy],\n feed_dict={self._X: X_valid,\n self._y: y_valid})\n if loss_val < best_loss:\n best_params = self._get_model_params()\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n else:\n loss_train, acc_train = sess.run([self._loss, self._accuracy],\n feed_dict={self._X: X_batch,\n self._y: y_batch})\n print(\"{}\\tLast training batch loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_train, acc_train * 100))\n # If we used early stopping then rollback to the best model found\n if best_params:\n self._restore_model_params(best_params)\n return self\n\n def predict_proba(self, X):\n if not self._session:\n raise NotFittedError(\"This %s instance is not fitted yet\" % self.__class__.__name__)\n with self._session.as_default() as sess:\n return self._Y_proba.eval(feed_dict={self._X: X})\n\n def predict(self, X):\n class_indices = np.argmax(self.predict_proba(X), axis=1)\n return np.array([[self.classes_[class_index]]\n for class_index in class_indices], np.int32)\n\n def save(self, path):\n self._saver.save(self._session, path)",
"_____no_output_____"
]
],
[
[
"Let's see if we get the exact same accuracy as earlier using this class (without dropout or batch norm):",
"_____no_output_____"
]
],
[
[
"dnn_clf = DNNClassifier(random_state=42)\ndnn_clf.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.116407\tBest loss: 0.116407\tAccuracy: 97.58%\n1\tValidation loss: 0.180534\tBest loss: 0.116407\tAccuracy: 97.11%\n2\tValidation loss: 0.227535\tBest loss: 0.116407\tAccuracy: 93.86%\n3\tValidation loss: 0.107346\tBest loss: 0.107346\tAccuracy: 97.54%\n4\tValidation loss: 0.302668\tBest loss: 0.107346\tAccuracy: 95.35%\n5\tValidation loss: 1.631054\tBest loss: 0.107346\tAccuracy: 22.01%\n6\tValidation loss: 1.635262\tBest loss: 0.107346\tAccuracy: 18.73%\n7\tValidation loss: 1.671200\tBest loss: 0.107346\tAccuracy: 22.01%\n8\tValidation loss: 1.695277\tBest loss: 0.107346\tAccuracy: 19.27%\n9\tValidation loss: 1.744607\tBest loss: 0.107346\tAccuracy: 20.91%\n10\tValidation loss: 1.629857\tBest loss: 0.107346\tAccuracy: 22.01%\n11\tValidation loss: 1.810803\tBest loss: 0.107346\tAccuracy: 22.01%\n12\tValidation loss: 1.675703\tBest loss: 0.107346\tAccuracy: 18.73%\n13\tValidation loss: 1.633233\tBest loss: 0.107346\tAccuracy: 20.91%\n14\tValidation loss: 1.652905\tBest loss: 0.107346\tAccuracy: 20.91%\n15\tValidation loss: 1.635937\tBest loss: 0.107346\tAccuracy: 20.91%\n16\tValidation loss: 1.718919\tBest loss: 0.107346\tAccuracy: 19.08%\n17\tValidation loss: 1.682458\tBest loss: 0.107346\tAccuracy: 19.27%\n18\tValidation loss: 1.675366\tBest loss: 0.107346\tAccuracy: 18.73%\n19\tValidation loss: 1.645800\tBest loss: 0.107346\tAccuracy: 19.08%\n20\tValidation loss: 1.722334\tBest loss: 0.107346\tAccuracy: 22.01%\n21\tValidation loss: 1.656418\tBest loss: 0.107346\tAccuracy: 22.01%\n22\tValidation loss: 1.643529\tBest loss: 0.107346\tAccuracy: 18.73%\n23\tValidation loss: 1.644233\tBest loss: 0.107346\tAccuracy: 19.27%\n24\tValidation loss: 1.690035\tBest loss: 0.107346\tAccuracy: 18.73%\nEarly stopping!\n"
]
],
[
[
"The model is trained, let's see if it gets the same accuracy as earlier:",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import accuracy_score\n\ny_pred = dnn_clf.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
]
],
[
[
"Yep! Working fine. Now we can use Scikit-Learn's `RandomizedSearchCV` class to search for better hyperparameters (this may take over an hour, depending on your system):",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV\n\ndef leaky_relu(alpha=0.01):\n def parametrized_leaky_relu(z, name=None):\n return tf.maximum(alpha * z, z, name=name)\n return parametrized_leaky_relu\n\nparam_distribs = {\n \"n_neurons\": [10, 30, 50, 70, 90, 100, 120, 140, 160],\n \"batch_size\": [10, 50, 100, 500],\n \"learning_rate\": [0.01, 0.02, 0.05, 0.1],\n \"activation\": [tf.nn.relu, tf.nn.elu, leaky_relu(alpha=0.01), leaky_relu(alpha=0.1)],\n # you could also try exploring different numbers of hidden layers, different optimizers, etc.\n #\"n_hidden_layers\": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10],\n #\"optimizer_class\": [tf.train.AdamOptimizer, partial(tf.train.MomentumOptimizer, momentum=0.95)],\n}\n\nrnd_search = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50,\n cv=3, random_state=42, verbose=2)\nrnd_search.fit(X_train1, y_train1, X_valid=X_valid1, y_valid=y_valid1, n_epochs=1000)\n\n# If you have Scikit-Learn 0.18 or earlier, you should upgrade, or use the fit_params argument:\n# fit_params = dict(X_valid=X_valid1, y_valid=y_valid1, n_epochs=1000)\n# rnd_search = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50,\n# fit_params=fit_params, random_state=42, verbose=2)\n# rnd_search.fit(X_train1, y_train1)\n",
"Fitting 3 folds for each of 50 candidates, totalling 150 fits\n[CV] n_neurons=10, learning_rate=0.05, batch_size=100, activation=<function elu at 0x1243639d8> \n"
],
[
"rnd_search.best_params_",
"_____no_output_____"
],
[
"y_pred = rnd_search.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
]
],
[
[
"Wonderful! Tuning the hyperparameters got us up to 98.91% accuracy! It may not sound like a great improvement to go from 97.26% to 98.91% accuracy, but consider the error rate: it went from roughly 2.6% to 1.1%. That's almost 60% reduction of the number of errors this model will produce!",
"_____no_output_____"
],
[
"It's a good idea to save this model:",
"_____no_output_____"
]
],
[
[
"rnd_search.best_estimator_.save(\"./my_best_mnist_model_0_to_4\")",
"_____no_output_____"
]
],
[
[
"### 8.4.",
"_____no_output_____"
],
[
"_Exercise: Now try adding Batch Normalization and compare the learning curves: is it converging faster than before? Does it produce a better model?_",
"_____no_output_____"
],
[
"Let's train the best model found, once again, to see how fast it converges (alternatively, you could tweak the code above to make it write summaries for TensorBoard, so you can visualize the learning curve):",
"_____no_output_____"
]
],
[
[
"dnn_clf = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,\n n_neurons=140, random_state=42)\ndnn_clf.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.083541\tBest loss: 0.083541\tAccuracy: 97.54%\n1\tValidation loss: 0.052198\tBest loss: 0.052198\tAccuracy: 98.40%\n2\tValidation loss: 0.044553\tBest loss: 0.044553\tAccuracy: 98.71%\n3\tValidation loss: 0.051113\tBest loss: 0.044553\tAccuracy: 98.48%\n4\tValidation loss: 0.046304\tBest loss: 0.044553\tAccuracy: 98.75%\n5\tValidation loss: 0.037796\tBest loss: 0.037796\tAccuracy: 98.91%\n6\tValidation loss: 0.048525\tBest loss: 0.037796\tAccuracy: 98.67%\n7\tValidation loss: 0.039877\tBest loss: 0.037796\tAccuracy: 98.75%\n8\tValidation loss: 0.038729\tBest loss: 0.037796\tAccuracy: 98.98%\n9\tValidation loss: 0.064167\tBest loss: 0.037796\tAccuracy: 98.24%\n10\tValidation loss: 0.057274\tBest loss: 0.037796\tAccuracy: 98.79%\n11\tValidation loss: 0.064388\tBest loss: 0.037796\tAccuracy: 98.55%\n12\tValidation loss: 0.056382\tBest loss: 0.037796\tAccuracy: 98.63%\n13\tValidation loss: 0.049408\tBest loss: 0.037796\tAccuracy: 98.91%\n14\tValidation loss: 0.038494\tBest loss: 0.037796\tAccuracy: 99.10%\n15\tValidation loss: 0.064619\tBest loss: 0.037796\tAccuracy: 98.67%\n16\tValidation loss: 0.055027\tBest loss: 0.037796\tAccuracy: 98.91%\n17\tValidation loss: 0.054773\tBest loss: 0.037796\tAccuracy: 98.91%\n18\tValidation loss: 0.076131\tBest loss: 0.037796\tAccuracy: 98.71%\n19\tValidation loss: 0.063031\tBest loss: 0.037796\tAccuracy: 98.59%\n20\tValidation loss: 0.120501\tBest loss: 0.037796\tAccuracy: 98.55%\n21\tValidation loss: 3.922006\tBest loss: 0.037796\tAccuracy: 94.14%\n22\tValidation loss: 0.395737\tBest loss: 0.037796\tAccuracy: 96.83%\n23\tValidation loss: 0.237014\tBest loss: 0.037796\tAccuracy: 96.56%\n24\tValidation loss: 0.159249\tBest loss: 0.037796\tAccuracy: 97.07%\n25\tValidation loss: 0.228444\tBest loss: 0.037796\tAccuracy: 95.74%\n26\tValidation loss: 0.134490\tBest loss: 0.037796\tAccuracy: 96.99%\nEarly stopping!\n"
]
],
[
[
"The best loss is reached at epoch 5.",
"_____no_output_____"
],
[
"Let's check that we do indeed get 98.9% accuracy on the test set:",
"_____no_output_____"
]
],
[
[
"y_pred = dnn_clf.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
]
],
[
[
"Good, now let's use the exact same model, but this time with batch normalization:",
"_____no_output_____"
]
],
[
[
"dnn_clf_bn = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,\n n_neurons=90, random_state=42,\n batch_norm_momentum=0.95)\ndnn_clf_bn.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.046685\tBest loss: 0.046685\tAccuracy: 98.63%\n1\tValidation loss: 0.040820\tBest loss: 0.040820\tAccuracy: 98.79%\n2\tValidation loss: 0.046557\tBest loss: 0.040820\tAccuracy: 98.67%\n3\tValidation loss: 0.032236\tBest loss: 0.032236\tAccuracy: 98.94%\n4\tValidation loss: 0.056148\tBest loss: 0.032236\tAccuracy: 98.44%\n5\tValidation loss: 0.035988\tBest loss: 0.032236\tAccuracy: 98.98%\n6\tValidation loss: 0.037958\tBest loss: 0.032236\tAccuracy: 98.94%\n7\tValidation loss: 0.034588\tBest loss: 0.032236\tAccuracy: 99.02%\n8\tValidation loss: 0.031261\tBest loss: 0.031261\tAccuracy: 99.34%\n9\tValidation loss: 0.050791\tBest loss: 0.031261\tAccuracy: 98.79%\n10\tValidation loss: 0.035324\tBest loss: 0.031261\tAccuracy: 99.02%\n11\tValidation loss: 0.039875\tBest loss: 0.031261\tAccuracy: 98.98%\n12\tValidation loss: 0.048575\tBest loss: 0.031261\tAccuracy: 98.94%\n13\tValidation loss: 0.028059\tBest loss: 0.028059\tAccuracy: 99.18%\n14\tValidation loss: 0.044112\tBest loss: 0.028059\tAccuracy: 99.14%\n15\tValidation loss: 0.039050\tBest loss: 0.028059\tAccuracy: 99.22%\n16\tValidation loss: 0.033278\tBest loss: 0.028059\tAccuracy: 99.14%\n17\tValidation loss: 0.031734\tBest loss: 0.028059\tAccuracy: 99.18%\n18\tValidation loss: 0.034500\tBest loss: 0.028059\tAccuracy: 99.14%\n19\tValidation loss: 0.032757\tBest loss: 0.028059\tAccuracy: 99.26%\n20\tValidation loss: 0.023842\tBest loss: 0.023842\tAccuracy: 99.53%\n21\tValidation loss: 0.026727\tBest loss: 0.023842\tAccuracy: 99.41%\n22\tValidation loss: 0.027016\tBest loss: 0.023842\tAccuracy: 99.41%\n23\tValidation loss: 0.033038\tBest loss: 0.023842\tAccuracy: 99.34%\n24\tValidation loss: 0.035490\tBest loss: 0.023842\tAccuracy: 99.18%\n25\tValidation loss: 0.060346\tBest loss: 0.023842\tAccuracy: 98.75%\n26\tValidation loss: 0.051341\tBest loss: 0.023842\tAccuracy: 99.26%\n27\tValidation loss: 0.033108\tBest loss: 0.023842\tAccuracy: 99.26%\n28\tValidation loss: 0.042162\tBest loss: 0.023842\tAccuracy: 99.18%\n29\tValidation loss: 0.036313\tBest loss: 0.023842\tAccuracy: 99.26%\n30\tValidation loss: 0.033812\tBest loss: 0.023842\tAccuracy: 99.26%\n31\tValidation loss: 0.038173\tBest loss: 0.023842\tAccuracy: 99.26%\n32\tValidation loss: 0.029853\tBest loss: 0.023842\tAccuracy: 99.37%\n33\tValidation loss: 0.026557\tBest loss: 0.023842\tAccuracy: 99.37%\n34\tValidation loss: 0.035003\tBest loss: 0.023842\tAccuracy: 99.37%\n35\tValidation loss: 0.027140\tBest loss: 0.023842\tAccuracy: 99.34%\n36\tValidation loss: 0.038988\tBest loss: 0.023842\tAccuracy: 99.34%\n37\tValidation loss: 0.048149\tBest loss: 0.023842\tAccuracy: 98.98%\n38\tValidation loss: 0.049070\tBest loss: 0.023842\tAccuracy: 99.02%\n39\tValidation loss: 0.041233\tBest loss: 0.023842\tAccuracy: 99.26%\n40\tValidation loss: 0.038571\tBest loss: 0.023842\tAccuracy: 99.26%\n41\tValidation loss: 0.036886\tBest loss: 0.023842\tAccuracy: 99.34%\nEarly stopping!\n"
]
],
[
[
"The best params are reached during epoch 20, that's actually a slower convergence than earlier. Let's check the accuracy:",
"_____no_output_____"
]
],
[
[
"y_pred = dnn_clf_bn.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
]
],
[
[
"Great, batch normalization improved accuracy! Let's see if we can find a good set of hyperparameters that will work even better with batch normalization:",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV\n\nparam_distribs = {\n \"n_neurons\": [10, 30, 50, 70, 90, 100, 120, 140, 160],\n \"batch_size\": [10, 50, 100, 500],\n \"learning_rate\": [0.01, 0.02, 0.05, 0.1],\n \"activation\": [tf.nn.relu, tf.nn.elu, leaky_relu(alpha=0.01), leaky_relu(alpha=0.1)],\n # you could also try exploring different numbers of hidden layers, different optimizers, etc.\n #\"n_hidden_layers\": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10],\n #\"optimizer_class\": [tf.train.AdamOptimizer, partial(tf.train.MomentumOptimizer, momentum=0.95)],\n \"batch_norm_momentum\": [0.9, 0.95, 0.98, 0.99, 0.999],\n}\n\nrnd_search_bn = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50, cv=3,\n random_state=42, verbose=2)\nrnd_search_bn.fit(X_train1, y_train1, X_valid=X_valid1, y_valid=y_valid1, n_epochs=1000)\n\n# If you have Scikit-Learn 0.18 or earlier, you should upgrade, or use the fit_params argument:\n# fit_params = dict(X_valid=X_valid1, y_valid=y_valid1, n_epochs=1000)\n# rnd_search_bn = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50,\n# fit_params=fit_params, random_state=42, verbose=2)\n# rnd_search_bn.fit(X_train1, y_train1)\n",
"Fitting 3 folds for each of 50 candidates, totalling 150 fits\n[CV] n_neurons=70, learning_rate=0.01, batch_size=50, batch_norm_momentum=0.99, activation=<function relu at 0x124366d08> \n"
],
[
"rnd_search_bn.best_params_",
"_____no_output_____"
],
[
"y_pred = rnd_search_bn.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
]
],
[
[
"Slightly better than earlier: 99.49% vs 99.42%. Let's see if dropout can do better.",
"_____no_output_____"
],
[
"### 8.5.",
"_____no_output_____"
],
[
"_Exercise: is the model overfitting the training set? Try adding dropout to every layer and try again. Does it help?_",
"_____no_output_____"
],
[
"Let's go back to the model we trained earlier and see how it performs on the training set:",
"_____no_output_____"
]
],
[
[
"y_pred = dnn_clf.predict(X_train1)\naccuracy_score(y_train1, y_pred)",
"_____no_output_____"
]
],
[
[
"The model performs significantly better on the training set than on the test set (99.51% vs 99.00%), which means it is overfitting the training set. A bit of regularization may help. Let's try adding dropout with a 50% dropout rate:",
"_____no_output_____"
]
],
[
[
"dnn_clf_dropout = DNNClassifier(activation=leaky_relu(alpha=0.1), batch_size=500, learning_rate=0.01,\n n_neurons=90, random_state=42,\n dropout_rate=0.5)\ndnn_clf_dropout.fit(X_train1, y_train1, n_epochs=1000, X_valid=X_valid1, y_valid=y_valid1)",
"0\tValidation loss: 0.131152\tBest loss: 0.131152\tAccuracy: 96.91%\n1\tValidation loss: 0.105306\tBest loss: 0.105306\tAccuracy: 97.46%\n2\tValidation loss: 0.091219\tBest loss: 0.091219\tAccuracy: 97.73%\n3\tValidation loss: 0.089638\tBest loss: 0.089638\tAccuracy: 97.85%\n4\tValidation loss: 0.091288\tBest loss: 0.089638\tAccuracy: 97.69%\n5\tValidation loss: 0.081112\tBest loss: 0.081112\tAccuracy: 98.05%\n6\tValidation loss: 0.075575\tBest loss: 0.075575\tAccuracy: 98.24%\n7\tValidation loss: 0.084841\tBest loss: 0.075575\tAccuracy: 97.77%\n8\tValidation loss: 0.075269\tBest loss: 0.075269\tAccuracy: 97.65%\n9\tValidation loss: 0.076625\tBest loss: 0.075269\tAccuracy: 98.12%\n10\tValidation loss: 0.072509\tBest loss: 0.072509\tAccuracy: 97.97%\n11\tValidation loss: 0.071006\tBest loss: 0.071006\tAccuracy: 98.44%\n12\tValidation loss: 0.073272\tBest loss: 0.071006\tAccuracy: 98.08%\n13\tValidation loss: 0.076293\tBest loss: 0.071006\tAccuracy: 98.16%\n14\tValidation loss: 0.074955\tBest loss: 0.071006\tAccuracy: 98.05%\n15\tValidation loss: 0.066207\tBest loss: 0.066207\tAccuracy: 98.20%\n16\tValidation loss: 0.067388\tBest loss: 0.066207\tAccuracy: 98.08%\n17\tValidation loss: 0.061916\tBest loss: 0.061916\tAccuracy: 98.40%\n18\tValidation loss: 0.064908\tBest loss: 0.061916\tAccuracy: 98.40%\n19\tValidation loss: 0.064921\tBest loss: 0.061916\tAccuracy: 98.40%\n20\tValidation loss: 0.069939\tBest loss: 0.061916\tAccuracy: 98.40%\n21\tValidation loss: 0.069870\tBest loss: 0.061916\tAccuracy: 98.32%\n22\tValidation loss: 0.062807\tBest loss: 0.061916\tAccuracy: 98.24%\n23\tValidation loss: 0.065312\tBest loss: 0.061916\tAccuracy: 98.44%\n24\tValidation loss: 0.067044\tBest loss: 0.061916\tAccuracy: 98.44%\n25\tValidation loss: 0.072251\tBest loss: 0.061916\tAccuracy: 98.16%\n26\tValidation loss: 0.064444\tBest loss: 0.061916\tAccuracy: 98.20%\n27\tValidation loss: 0.069022\tBest loss: 0.061916\tAccuracy: 98.44%\n28\tValidation loss: 0.069079\tBest loss: 0.061916\tAccuracy: 98.28%\n29\tValidation loss: 0.148266\tBest loss: 0.061916\tAccuracy: 96.52%\n30\tValidation loss: 0.119943\tBest loss: 0.061916\tAccuracy: 96.72%\n31\tValidation loss: 0.167303\tBest loss: 0.061916\tAccuracy: 96.68%\n32\tValidation loss: 0.131897\tBest loss: 0.061916\tAccuracy: 96.52%\n33\tValidation loss: 0.146681\tBest loss: 0.061916\tAccuracy: 95.43%\n34\tValidation loss: 0.125731\tBest loss: 0.061916\tAccuracy: 96.64%\n35\tValidation loss: 0.099879\tBest loss: 0.061916\tAccuracy: 97.89%\n36\tValidation loss: 0.096915\tBest loss: 0.061916\tAccuracy: 97.73%\n37\tValidation loss: 0.096422\tBest loss: 0.061916\tAccuracy: 97.85%\n38\tValidation loss: 0.108040\tBest loss: 0.061916\tAccuracy: 97.54%\nEarly stopping!\n"
]
],
[
[
"The best params are reached during epoch 17. Dropout somewhat slowed down convergence.",
"_____no_output_____"
],
[
"Let's check the accuracy:",
"_____no_output_____"
]
],
[
[
"y_pred = dnn_clf_dropout.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
]
],
[
[
"We are out of luck, dropout does not seem to help. Let's try tuning the hyperparameters, perhaps we can squeeze a bit more performance out of this model:",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import RandomizedSearchCV\n\nparam_distribs = {\n \"n_neurons\": [10, 30, 50, 70, 90, 100, 120, 140, 160],\n \"batch_size\": [10, 50, 100, 500],\n \"learning_rate\": [0.01, 0.02, 0.05, 0.1],\n \"activation\": [tf.nn.relu, tf.nn.elu, leaky_relu(alpha=0.01), leaky_relu(alpha=0.1)],\n # you could also try exploring different numbers of hidden layers, different optimizers, etc.\n #\"n_hidden_layers\": [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10],\n #\"optimizer_class\": [tf.train.AdamOptimizer, partial(tf.train.MomentumOptimizer, momentum=0.95)],\n \"dropout_rate\": [0.2, 0.3, 0.4, 0.5, 0.6],\n}\n\nrnd_search_dropout = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50,\n cv=3, random_state=42, verbose=2)\nrnd_search_dropout.fit(X_train1, y_train1, X_valid=X_valid1, y_valid=y_valid1, n_epochs=1000)\n\n# If you have Scikit-Learn 0.18 or earlier, you should upgrade, or use the fit_params argument:\n# fit_params = dict(X_valid=X_valid1, y_valid=y_valid1, n_epochs=1000)\n# rnd_search_dropout = RandomizedSearchCV(DNNClassifier(random_state=42), param_distribs, n_iter=50,\n# fit_params=fit_params, random_state=42, verbose=2)\n# rnd_search_dropout.fit(X_train1, y_train1)",
"[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.\n"
],
[
"rnd_search_dropout.best_params_",
"_____no_output_____"
],
[
"y_pred = rnd_search_dropout.predict(X_test1)\naccuracy_score(y_test1, y_pred)",
"_____no_output_____"
]
],
[
[
"Oh well, dropout did not improve the model. Better luck next time! :)",
"_____no_output_____"
],
[
"But that's okay, we have ourselves a nice DNN that achieves 99.49% accuracy on the test set using Batch Normalization, or 98.91% without BN. Let's see if some of this expertise on digits 0 to 4 can be transferred to the task of classifying digits 5 to 9. For the sake of simplicity we will reuse the DNN without BN.",
"_____no_output_____"
],
[
"## 9. Transfer learning",
"_____no_output_____"
],
[
"### 9.1.",
"_____no_output_____"
],
[
"_Exercise: create a new DNN that reuses all the pretrained hidden layers of the previous model, freezes them, and replaces the softmax output layer with a new one._",
"_____no_output_____"
],
[
"Let's load the best model's graph and get a handle on all the important operations we will need. Note that instead of creating a new softmax output layer, we will just reuse the existing one (since it has the same number of outputs as the existing one). We will reinitialize its parameters before training. ",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nrestore_saver = tf.train.import_meta_graph(\"./my_best_mnist_model_0_to_4.meta\")\n\nX = tf.get_default_graph().get_tensor_by_name(\"X:0\")\ny = tf.get_default_graph().get_tensor_by_name(\"y:0\")\nloss = tf.get_default_graph().get_tensor_by_name(\"loss:0\")\nY_proba = tf.get_default_graph().get_tensor_by_name(\"Y_proba:0\")\nlogits = Y_proba.op.inputs[0]\naccuracy = tf.get_default_graph().get_tensor_by_name(\"accuracy:0\")",
"_____no_output_____"
]
],
[
[
"To freeze the lower layers, we will exclude their variables from the optimizer's list of trainable variables, keeping only the output layer's trainable variables:",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\n\noutput_layer_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=\"logits\")\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam2\")\ntraining_op = optimizer.minimize(loss, var_list=output_layer_vars)",
"_____no_output_____"
],
[
"correct = tf.nn.in_top_k(logits, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")\n\ninit = tf.global_variables_initializer()\nfive_frozen_saver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"### 9.2.",
"_____no_output_____"
],
[
"_Exercise: train this new DNN on digits 5 to 9, using only 100 images per digit, and time how long it takes. Despite this small number of examples, can you achieve high precision?_",
"_____no_output_____"
],
[
"Let's create the training, validation and test sets. We need to subtract 5 from the labels because TensorFlow expects integers from 0 to `n_classes-1`.",
"_____no_output_____"
]
],
[
[
"X_train2_full = X_train[y_train >= 5]\ny_train2_full = y_train[y_train >= 5] - 5\nX_valid2_full = X_valid[y_valid >= 5]\ny_valid2_full = y_valid[y_valid >= 5] - 5\nX_test2 = X_test[y_test >= 5]\ny_test2 = y_test[y_test >= 5] - 5",
"_____no_output_____"
]
],
[
[
"Also, for the purpose of this exercise, we want to keep only 100 instances per class in the training set (and let's keep only 30 instances per class in the validation set). Let's create a small function to do that:",
"_____no_output_____"
]
],
[
[
"def sample_n_instances_per_class(X, y, n=100):\n Xs, ys = [], []\n for label in np.unique(y):\n idx = (y == label)\n Xc = X[idx][:n]\n yc = y[idx][:n]\n Xs.append(Xc)\n ys.append(yc)\n return np.concatenate(Xs), np.concatenate(ys)",
"_____no_output_____"
],
[
"X_train2, y_train2 = sample_n_instances_per_class(X_train2_full, y_train2_full, n=100)\nX_valid2, y_valid2 = sample_n_instances_per_class(X_valid2_full, y_valid2_full, n=30)",
"_____no_output_____"
]
],
[
[
"Now let's train the model. This is the same training code as earlier, using early stopping, except for the initialization: we first initialize all the variables, then we restore the best model trained earlier (on digits 0 to 4), and finally we reinitialize the output layer variables.",
"_____no_output_____"
]
],
[
[
"import time\n\nn_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_best_mnist_model_0_to_4\")\n t0 = time.time()\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = five_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_five_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\n t1 = time.time()\n print(\"Total training time: {:.1f}s\".format(t1 - t0))\n\nwith tf.Session() as sess:\n five_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_five_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_best_mnist_model_0_to_4\n0\tValidation loss: 1.361167\tBest loss: 1.361167\tAccuracy: 43.33%\n1\tValidation loss: 1.154602\tBest loss: 1.154602\tAccuracy: 57.33%\n2\tValidation loss: 1.054218\tBest loss: 1.054218\tAccuracy: 53.33%\n3\tValidation loss: 0.981128\tBest loss: 0.981128\tAccuracy: 62.67%\n4\tValidation loss: 0.995353\tBest loss: 0.981128\tAccuracy: 59.33%\n5\tValidation loss: 0.967000\tBest loss: 0.967000\tAccuracy: 65.33%\n6\tValidation loss: 0.955700\tBest loss: 0.955700\tAccuracy: 61.33%\n7\tValidation loss: 1.015331\tBest loss: 0.955700\tAccuracy: 58.67%\n8\tValidation loss: 0.978280\tBest loss: 0.955700\tAccuracy: 62.00%\n9\tValidation loss: 0.923389\tBest loss: 0.923389\tAccuracy: 69.33%\n10\tValidation loss: 0.996236\tBest loss: 0.923389\tAccuracy: 63.33%\n11\tValidation loss: 0.976757\tBest loss: 0.923389\tAccuracy: 62.67%\n12\tValidation loss: 0.969096\tBest loss: 0.923389\tAccuracy: 63.33%\n13\tValidation loss: 1.023069\tBest loss: 0.923389\tAccuracy: 63.33%\n14\tValidation loss: 1.104664\tBest loss: 0.923389\tAccuracy: 55.33%\n15\tValidation loss: 0.950175\tBest loss: 0.923389\tAccuracy: 65.33%\n16\tValidation loss: 1.002944\tBest loss: 0.923389\tAccuracy: 63.33%\n17\tValidation loss: 0.895543\tBest loss: 0.895543\tAccuracy: 70.67%\n18\tValidation loss: 0.961151\tBest loss: 0.895543\tAccuracy: 66.67%\n19\tValidation loss: 0.896372\tBest loss: 0.895543\tAccuracy: 67.33%\n20\tValidation loss: 0.911938\tBest loss: 0.895543\tAccuracy: 69.33%\n21\tValidation loss: 0.929007\tBest loss: 0.895543\tAccuracy: 68.00%\n22\tValidation loss: 0.939231\tBest loss: 0.895543\tAccuracy: 65.33%\n23\tValidation loss: 0.919057\tBest loss: 0.895543\tAccuracy: 68.67%\n24\tValidation loss: 0.994529\tBest loss: 0.895543\tAccuracy: 65.33%\n25\tValidation loss: 0.901279\tBest loss: 0.895543\tAccuracy: 68.67%\n26\tValidation loss: 0.916238\tBest loss: 0.895543\tAccuracy: 68.67%\n27\tValidation loss: 1.007434\tBest loss: 0.895543\tAccuracy: 65.33%\n28\tValidation loss: 0.924729\tBest loss: 0.895543\tAccuracy: 70.00%\n29\tValidation loss: 0.974399\tBest loss: 0.895543\tAccuracy: 66.00%\n30\tValidation loss: 0.899418\tBest loss: 0.895543\tAccuracy: 68.00%\n31\tValidation loss: 0.940563\tBest loss: 0.895543\tAccuracy: 66.00%\n32\tValidation loss: 0.920235\tBest loss: 0.895543\tAccuracy: 68.00%\n33\tValidation loss: 0.929848\tBest loss: 0.895543\tAccuracy: 68.67%\n34\tValidation loss: 0.930288\tBest loss: 0.895543\tAccuracy: 66.67%\n35\tValidation loss: 0.943884\tBest loss: 0.895543\tAccuracy: 64.67%\n36\tValidation loss: 0.939372\tBest loss: 0.895543\tAccuracy: 68.00%\n37\tValidation loss: 0.894239\tBest loss: 0.894239\tAccuracy: 67.33%\n38\tValidation loss: 0.888806\tBest loss: 0.888806\tAccuracy: 69.33%\n39\tValidation loss: 0.933829\tBest loss: 0.888806\tAccuracy: 66.00%\n40\tValidation loss: 0.911836\tBest loss: 0.888806\tAccuracy: 72.67%\n41\tValidation loss: 0.896729\tBest loss: 0.888806\tAccuracy: 70.00%\n42\tValidation loss: 0.929394\tBest loss: 0.888806\tAccuracy: 68.00%\n43\tValidation loss: 0.919418\tBest loss: 0.888806\tAccuracy: 69.33%\n44\tValidation loss: 0.907830\tBest loss: 0.888806\tAccuracy: 65.33%\n45\tValidation loss: 1.004304\tBest loss: 0.888806\tAccuracy: 71.33%\n46\tValidation loss: 0.871899\tBest loss: 0.871899\tAccuracy: 74.00%\n47\tValidation loss: 0.904889\tBest loss: 0.871899\tAccuracy: 67.33%\n48\tValidation loss: 0.914138\tBest loss: 0.871899\tAccuracy: 66.00%\n49\tValidation loss: 0.930001\tBest loss: 0.871899\tAccuracy: 69.33%\n50\tValidation loss: 0.962153\tBest loss: 0.871899\tAccuracy: 68.67%\n51\tValidation loss: 0.925021\tBest loss: 0.871899\tAccuracy: 65.33%\n52\tValidation loss: 0.974412\tBest loss: 0.871899\tAccuracy: 67.33%\n53\tValidation loss: 0.897499\tBest loss: 0.871899\tAccuracy: 68.67%\n54\tValidation loss: 0.933581\tBest loss: 0.871899\tAccuracy: 60.67%\n55\tValidation loss: 0.988574\tBest loss: 0.871899\tAccuracy: 68.67%\n56\tValidation loss: 0.927290\tBest loss: 0.871899\tAccuracy: 66.67%\n57\tValidation loss: 1.018713\tBest loss: 0.871899\tAccuracy: 64.00%\n58\tValidation loss: 0.964709\tBest loss: 0.871899\tAccuracy: 66.00%\n59\tValidation loss: 1.004696\tBest loss: 0.871899\tAccuracy: 59.33%\n60\tValidation loss: 1.008746\tBest loss: 0.871899\tAccuracy: 58.67%\n61\tValidation loss: 0.948558\tBest loss: 0.871899\tAccuracy: 68.00%\n62\tValidation loss: 0.966037\tBest loss: 0.871899\tAccuracy: 64.00%\n63\tValidation loss: 0.922541\tBest loss: 0.871899\tAccuracy: 68.00%\n64\tValidation loss: 0.892541\tBest loss: 0.871899\tAccuracy: 72.00%\n65\tValidation loss: 0.890340\tBest loss: 0.871899\tAccuracy: 70.67%\n66\tValidation loss: 0.957904\tBest loss: 0.871899\tAccuracy: 66.00%\nEarly stopping!\nTotal training time: 1.9s\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_five_frozen\nFinal test accuracy: 64.02%\n"
]
],
[
[
"Well that's not a great accuracy, is it? Of course with such a tiny training set, and with only one layer to tweak, we should not expect miracles.",
"_____no_output_____"
],
[
"### 9.3.",
"_____no_output_____"
],
[
"_Exercise: try caching the frozen layers, and train the model again: how much faster is it now?_",
"_____no_output_____"
],
[
"Let's start by getting a handle on the output of the last frozen layer:",
"_____no_output_____"
]
],
[
[
"hidden5_out = tf.get_default_graph().get_tensor_by_name(\"hidden5_out:0\")",
"_____no_output_____"
]
],
[
[
"Now let's train the model using roughly the same code as earlier. The difference is that we compute the output of the top frozen layer at the beginning (both for the training set and the validation set), and we cache it. This makes training roughly 1.5 to 3 times faster in this example (this may vary greatly, depending on your system): ",
"_____no_output_____"
]
],
[
[
"import time\n\nn_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_best_mnist_model_0_to_4\")\n t0 = time.time()\n \n hidden5_train = hidden5_out.eval(feed_dict={X: X_train2, y: y_train2})\n hidden5_valid = hidden5_out.eval(feed_dict={X: X_valid2, y: y_valid2})\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n h5_batch, y_batch = hidden5_train[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={hidden5_out: h5_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={hidden5_out: hidden5_valid, y: y_valid2})\n if loss_val < best_loss:\n save_path = five_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_five_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\n t1 = time.time()\n print(\"Total training time: {:.1f}s\".format(t1 - t0))\n\nwith tf.Session() as sess:\n five_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_five_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_best_mnist_model_0_to_4\n0\tValidation loss: 1.416103\tBest loss: 1.416103\tAccuracy: 44.00%\n1\tValidation loss: 1.099216\tBest loss: 1.099216\tAccuracy: 53.33%\n2\tValidation loss: 1.024954\tBest loss: 1.024954\tAccuracy: 59.33%\n3\tValidation loss: 0.969193\tBest loss: 0.969193\tAccuracy: 60.00%\n4\tValidation loss: 0.973461\tBest loss: 0.969193\tAccuracy: 64.67%\n5\tValidation loss: 0.949333\tBest loss: 0.949333\tAccuracy: 64.67%\n6\tValidation loss: 0.922953\tBest loss: 0.922953\tAccuracy: 66.67%\n7\tValidation loss: 0.957186\tBest loss: 0.922953\tAccuracy: 62.67%\n8\tValidation loss: 0.950264\tBest loss: 0.922953\tAccuracy: 68.00%\n9\tValidation loss: 1.053465\tBest loss: 0.922953\tAccuracy: 59.33%\n10\tValidation loss: 1.069949\tBest loss: 0.922953\tAccuracy: 54.00%\n11\tValidation loss: 0.965197\tBest loss: 0.922953\tAccuracy: 62.67%\n12\tValidation loss: 0.949233\tBest loss: 0.922953\tAccuracy: 63.33%\n13\tValidation loss: 0.926229\tBest loss: 0.922953\tAccuracy: 63.33%\n14\tValidation loss: 0.922854\tBest loss: 0.922854\tAccuracy: 67.33%\n15\tValidation loss: 0.965205\tBest loss: 0.922854\tAccuracy: 66.67%\n16\tValidation loss: 1.050026\tBest loss: 0.922854\tAccuracy: 59.33%\n17\tValidation loss: 0.946699\tBest loss: 0.922854\tAccuracy: 64.67%\n18\tValidation loss: 0.973966\tBest loss: 0.922854\tAccuracy: 64.00%\n19\tValidation loss: 0.902573\tBest loss: 0.902573\tAccuracy: 66.67%\n20\tValidation loss: 0.933625\tBest loss: 0.902573\tAccuracy: 65.33%\n21\tValidation loss: 0.938296\tBest loss: 0.902573\tAccuracy: 64.00%\n22\tValidation loss: 0.938790\tBest loss: 0.902573\tAccuracy: 66.67%\n23\tValidation loss: 0.936572\tBest loss: 0.902573\tAccuracy: 68.00%\n24\tValidation loss: 1.039109\tBest loss: 0.902573\tAccuracy: 65.33%\n25\tValidation loss: 1.146837\tBest loss: 0.902573\tAccuracy: 59.33%\n26\tValidation loss: 0.958702\tBest loss: 0.902573\tAccuracy: 68.67%\n27\tValidation loss: 0.915434\tBest loss: 0.902573\tAccuracy: 70.67%\n28\tValidation loss: 0.915402\tBest loss: 0.902573\tAccuracy: 66.00%\n29\tValidation loss: 0.920591\tBest loss: 0.902573\tAccuracy: 70.67%\n30\tValidation loss: 1.029216\tBest loss: 0.902573\tAccuracy: 64.67%\n31\tValidation loss: 1.039922\tBest loss: 0.902573\tAccuracy: 55.33%\n32\tValidation loss: 0.925041\tBest loss: 0.902573\tAccuracy: 64.00%\n33\tValidation loss: 0.944033\tBest loss: 0.902573\tAccuracy: 67.33%\n34\tValidation loss: 0.941914\tBest loss: 0.902573\tAccuracy: 66.67%\n35\tValidation loss: 0.866297\tBest loss: 0.866297\tAccuracy: 69.33%\n36\tValidation loss: 0.900787\tBest loss: 0.866297\tAccuracy: 70.67%\n37\tValidation loss: 0.889670\tBest loss: 0.866297\tAccuracy: 66.67%\n38\tValidation loss: 0.968139\tBest loss: 0.866297\tAccuracy: 62.00%\n39\tValidation loss: 0.929764\tBest loss: 0.866297\tAccuracy: 66.00%\n40\tValidation loss: 0.889130\tBest loss: 0.866297\tAccuracy: 68.00%\n41\tValidation loss: 0.940024\tBest loss: 0.866297\tAccuracy: 70.00%\n42\tValidation loss: 0.896472\tBest loss: 0.866297\tAccuracy: 69.33%\n43\tValidation loss: 0.893887\tBest loss: 0.866297\tAccuracy: 67.33%\n44\tValidation loss: 0.925727\tBest loss: 0.866297\tAccuracy: 68.67%\n45\tValidation loss: 0.945748\tBest loss: 0.866297\tAccuracy: 66.00%\n46\tValidation loss: 0.897087\tBest loss: 0.866297\tAccuracy: 70.00%\n47\tValidation loss: 0.923855\tBest loss: 0.866297\tAccuracy: 68.67%\n48\tValidation loss: 0.944244\tBest loss: 0.866297\tAccuracy: 66.67%\n49\tValidation loss: 0.975582\tBest loss: 0.866297\tAccuracy: 66.67%\n50\tValidation loss: 0.889869\tBest loss: 0.866297\tAccuracy: 68.67%\n51\tValidation loss: 0.895552\tBest loss: 0.866297\tAccuracy: 69.33%\n52\tValidation loss: 0.943707\tBest loss: 0.866297\tAccuracy: 66.00%\n53\tValidation loss: 0.902883\tBest loss: 0.866297\tAccuracy: 70.67%\n54\tValidation loss: 0.958292\tBest loss: 0.866297\tAccuracy: 68.67%\n55\tValidation loss: 0.917368\tBest loss: 0.866297\tAccuracy: 67.33%\nEarly stopping!\nTotal training time: 1.1s\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_five_frozen\nFinal test accuracy: 61.16%\n"
]
],
[
[
"### 9.4.",
"_____no_output_____"
],
[
"_Exercise: try again reusing just four hidden layers instead of five. Can you achieve a higher precision?_",
"_____no_output_____"
],
[
"Let's load the best model again, but this time we will create a new softmax output layer on top of the 4th hidden layer:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_outputs = 5\n\nrestore_saver = tf.train.import_meta_graph(\"./my_best_mnist_model_0_to_4.meta\")\n\nX = tf.get_default_graph().get_tensor_by_name(\"X:0\")\ny = tf.get_default_graph().get_tensor_by_name(\"y:0\")\n\nhidden4_out = tf.get_default_graph().get_tensor_by_name(\"hidden4_out:0\")\nlogits = tf.layers.dense(hidden4_out, n_outputs, kernel_initializer=he_init, name=\"new_logits\")\nY_proba = tf.nn.softmax(logits)\nxentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\nloss = tf.reduce_mean(xentropy)\ncorrect = tf.nn.in_top_k(logits, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32), name=\"accuracy\")",
"_____no_output_____"
]
],
[
[
"And now let's create the training operation. We want to freeze all the layers except for the new output layer:",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\n\noutput_layer_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=\"new_logits\")\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam2\")\ntraining_op = optimizer.minimize(loss, var_list=output_layer_vars)\n\ninit = tf.global_variables_initializer()\nfour_frozen_saver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"And once again we train the model with the same code as earlier. Note: we could of course write a function once and use it multiple times, rather than copying almost the same training code over and over again, but as we keep tweaking the code slightly, the function would need multiple arguments and `if` statements, and it would have to be at the beginning of the notebook, where it would not make much sense to readers. In short it would be very confusing, so we're better off with copy & paste.",
"_____no_output_____"
]
],
[
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_best_mnist_model_0_to_4\")\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = four_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_four_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n four_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_four_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_best_mnist_model_0_to_4\n0\tValidation loss: 1.073254\tBest loss: 1.073254\tAccuracy: 51.33%\n1\tValidation loss: 1.039487\tBest loss: 1.039487\tAccuracy: 64.00%\n2\tValidation loss: 0.991418\tBest loss: 0.991418\tAccuracy: 59.33%\n3\tValidation loss: 0.902691\tBest loss: 0.902691\tAccuracy: 64.67%\n4\tValidation loss: 0.919874\tBest loss: 0.902691\tAccuracy: 63.33%\n5\tValidation loss: 0.879734\tBest loss: 0.879734\tAccuracy: 72.00%\n6\tValidation loss: 0.877940\tBest loss: 0.877940\tAccuracy: 70.67%\n7\tValidation loss: 0.899513\tBest loss: 0.877940\tAccuracy: 71.33%\n8\tValidation loss: 0.879717\tBest loss: 0.877940\tAccuracy: 67.33%\n9\tValidation loss: 0.826527\tBest loss: 0.826527\tAccuracy: 75.33%\n10\tValidation loss: 0.890165\tBest loss: 0.826527\tAccuracy: 67.33%\n11\tValidation loss: 0.876235\tBest loss: 0.826527\tAccuracy: 68.67%\n12\tValidation loss: 0.877598\tBest loss: 0.826527\tAccuracy: 71.33%\n13\tValidation loss: 0.898070\tBest loss: 0.826527\tAccuracy: 74.67%\n14\tValidation loss: 0.923526\tBest loss: 0.826527\tAccuracy: 68.00%\n15\tValidation loss: 0.859624\tBest loss: 0.826527\tAccuracy: 70.00%\n16\tValidation loss: 0.896264\tBest loss: 0.826527\tAccuracy: 67.33%\n17\tValidation loss: 0.800813\tBest loss: 0.800813\tAccuracy: 73.33%\n18\tValidation loss: 0.811318\tBest loss: 0.800813\tAccuracy: 74.00%\n19\tValidation loss: 0.809687\tBest loss: 0.800813\tAccuracy: 75.33%\n20\tValidation loss: 0.807125\tBest loss: 0.800813\tAccuracy: 72.67%\n21\tValidation loss: 0.819150\tBest loss: 0.800813\tAccuracy: 71.33%\n22\tValidation loss: 0.849812\tBest loss: 0.800813\tAccuracy: 76.67%\n23\tValidation loss: 0.801709\tBest loss: 0.800813\tAccuracy: 74.67%\n24\tValidation loss: 0.832877\tBest loss: 0.800813\tAccuracy: 74.00%\n25\tValidation loss: 0.792853\tBest loss: 0.792853\tAccuracy: 72.67%\n26\tValidation loss: 0.842031\tBest loss: 0.792853\tAccuracy: 76.00%\n27\tValidation loss: 0.872236\tBest loss: 0.792853\tAccuracy: 71.33%\n28\tValidation loss: 0.782557\tBest loss: 0.782557\tAccuracy: 78.00%\n29\tValidation loss: 0.802515\tBest loss: 0.782557\tAccuracy: 73.33%\n30\tValidation loss: 0.812652\tBest loss: 0.782557\tAccuracy: 72.67%\n31\tValidation loss: 0.825467\tBest loss: 0.782557\tAccuracy: 76.00%\n32\tValidation loss: 0.791320\tBest loss: 0.782557\tAccuracy: 76.67%\n33\tValidation loss: 0.785207\tBest loss: 0.782557\tAccuracy: 77.33%\n34\tValidation loss: 0.815450\tBest loss: 0.782557\tAccuracy: 76.67%\n35\tValidation loss: 0.865081\tBest loss: 0.782557\tAccuracy: 71.33%\n36\tValidation loss: 0.852323\tBest loss: 0.782557\tAccuracy: 74.67%\n37\tValidation loss: 0.836967\tBest loss: 0.782557\tAccuracy: 72.00%\n38\tValidation loss: 0.807404\tBest loss: 0.782557\tAccuracy: 77.33%\n39\tValidation loss: 0.821566\tBest loss: 0.782557\tAccuracy: 75.33%\n40\tValidation loss: 0.817326\tBest loss: 0.782557\tAccuracy: 76.00%\n41\tValidation loss: 0.807987\tBest loss: 0.782557\tAccuracy: 70.67%\n42\tValidation loss: 0.838029\tBest loss: 0.782557\tAccuracy: 74.00%\n43\tValidation loss: 0.820425\tBest loss: 0.782557\tAccuracy: 76.00%\n44\tValidation loss: 0.785871\tBest loss: 0.782557\tAccuracy: 76.00%\n45\tValidation loss: 0.844337\tBest loss: 0.782557\tAccuracy: 78.67%\n46\tValidation loss: 0.764127\tBest loss: 0.764127\tAccuracy: 78.67%\n47\tValidation loss: 0.789726\tBest loss: 0.764127\tAccuracy: 77.33%\n48\tValidation loss: 0.839190\tBest loss: 0.764127\tAccuracy: 72.67%\n49\tValidation loss: 0.849353\tBest loss: 0.764127\tAccuracy: 75.33%\n50\tValidation loss: 0.869818\tBest loss: 0.764127\tAccuracy: 74.00%\n51\tValidation loss: 0.805526\tBest loss: 0.764127\tAccuracy: 76.67%\n52\tValidation loss: 0.850749\tBest loss: 0.764127\tAccuracy: 72.67%\n53\tValidation loss: 0.838693\tBest loss: 0.764127\tAccuracy: 71.33%\n54\tValidation loss: 0.791396\tBest loss: 0.764127\tAccuracy: 75.33%\n55\tValidation loss: 0.846888\tBest loss: 0.764127\tAccuracy: 76.00%\n56\tValidation loss: 0.826717\tBest loss: 0.764127\tAccuracy: 74.67%\n57\tValidation loss: 0.878286\tBest loss: 0.764127\tAccuracy: 70.67%\n58\tValidation loss: 0.878869\tBest loss: 0.764127\tAccuracy: 72.67%\n59\tValidation loss: 0.822241\tBest loss: 0.764127\tAccuracy: 72.67%\n60\tValidation loss: 0.864925\tBest loss: 0.764127\tAccuracy: 73.33%\n61\tValidation loss: 0.804545\tBest loss: 0.764127\tAccuracy: 73.33%\n62\tValidation loss: 0.891784\tBest loss: 0.764127\tAccuracy: 72.67%\n63\tValidation loss: 0.810186\tBest loss: 0.764127\tAccuracy: 74.00%\n64\tValidation loss: 0.810786\tBest loss: 0.764127\tAccuracy: 74.67%\n65\tValidation loss: 0.818044\tBest loss: 0.764127\tAccuracy: 74.00%\n66\tValidation loss: 0.853420\tBest loss: 0.764127\tAccuracy: 74.67%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_four_frozen\nFinal test accuracy: 69.10%\n"
]
],
[
[
"Still not fantastic, but much better.",
"_____no_output_____"
],
[
"### 9.5.",
"_____no_output_____"
],
[
"_Exercise: now unfreeze the top two hidden layers and continue training: can you get the model to perform even better?_",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\n\nunfrozen_vars = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope=\"hidden[34]|new_logits\")\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam3\")\ntraining_op = optimizer.minimize(loss, var_list=unfrozen_vars)\n\ninit = tf.global_variables_initializer()\ntwo_frozen_saver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n four_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_four_frozen\")\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = two_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_two_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n two_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_two_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_four_frozen\n0\tValidation loss: 1.054859\tBest loss: 1.054859\tAccuracy: 74.00%\n1\tValidation loss: 0.812410\tBest loss: 0.812410\tAccuracy: 78.00%\n2\tValidation loss: 0.750377\tBest loss: 0.750377\tAccuracy: 80.67%\n3\tValidation loss: 0.570973\tBest loss: 0.570973\tAccuracy: 84.67%\n4\tValidation loss: 0.805442\tBest loss: 0.570973\tAccuracy: 79.33%\n5\tValidation loss: 0.920925\tBest loss: 0.570973\tAccuracy: 80.00%\n6\tValidation loss: 0.817471\tBest loss: 0.570973\tAccuracy: 81.33%\n7\tValidation loss: 0.777876\tBest loss: 0.570973\tAccuracy: 84.00%\n8\tValidation loss: 1.030498\tBest loss: 0.570973\tAccuracy: 74.67%\n9\tValidation loss: 1.074356\tBest loss: 0.570973\tAccuracy: 81.33%\n10\tValidation loss: 0.912521\tBest loss: 0.570973\tAccuracy: 83.33%\n11\tValidation loss: 1.356695\tBest loss: 0.570973\tAccuracy: 79.33%\n12\tValidation loss: 0.918798\tBest loss: 0.570973\tAccuracy: 82.00%\n13\tValidation loss: 0.971029\tBest loss: 0.570973\tAccuracy: 82.67%\n14\tValidation loss: 0.860108\tBest loss: 0.570973\tAccuracy: 83.33%\n15\tValidation loss: 1.074813\tBest loss: 0.570973\tAccuracy: 82.00%\n16\tValidation loss: 0.867760\tBest loss: 0.570973\tAccuracy: 84.00%\n17\tValidation loss: 0.858290\tBest loss: 0.570973\tAccuracy: 85.33%\n18\tValidation loss: 0.996560\tBest loss: 0.570973\tAccuracy: 85.33%\n19\tValidation loss: 1.304507\tBest loss: 0.570973\tAccuracy: 83.33%\n20\tValidation loss: 1.134808\tBest loss: 0.570973\tAccuracy: 80.67%\n21\tValidation loss: 1.189581\tBest loss: 0.570973\tAccuracy: 82.00%\n22\tValidation loss: 1.131344\tBest loss: 0.570973\tAccuracy: 81.33%\n23\tValidation loss: 1.240507\tBest loss: 0.570973\tAccuracy: 82.67%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_two_frozen\nFinal test accuracy: 78.09%\n"
]
],
[
[
"Let's check what accuracy we can get by unfreezing all layers:",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\n\noptimizer = tf.train.AdamOptimizer(learning_rate, name=\"Adam4\")\ntraining_op = optimizer.minimize(loss)\n\ninit = tf.global_variables_initializer()\nno_frozen_saver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 1000\nbatch_size = 20\n\nmax_checks_without_progress = 20\nchecks_without_progress = 0\nbest_loss = np.infty\n\nwith tf.Session() as sess:\n init.run()\n two_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_two_frozen\")\n \n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n loss_val, acc_val = sess.run([loss, accuracy], feed_dict={X: X_valid2, y: y_valid2})\n if loss_val < best_loss:\n save_path = no_frozen_saver.save(sess, \"./my_mnist_model_5_to_9_no_frozen\")\n best_loss = loss_val\n checks_without_progress = 0\n else:\n checks_without_progress += 1\n if checks_without_progress > max_checks_without_progress:\n print(\"Early stopping!\")\n break\n print(\"{}\\tValidation loss: {:.6f}\\tBest loss: {:.6f}\\tAccuracy: {:.2f}%\".format(\n epoch, loss_val, best_loss, acc_val * 100))\n\nwith tf.Session() as sess:\n no_frozen_saver.restore(sess, \"./my_mnist_model_5_to_9_no_frozen\")\n acc_test = accuracy.eval(feed_dict={X: X_test2, y: y_test2})\n print(\"Final test accuracy: {:.2f}%\".format(acc_test * 100))",
"INFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_two_frozen\n0\tValidation loss: 0.863416\tBest loss: 0.863416\tAccuracy: 86.00%\n1\tValidation loss: 0.695079\tBest loss: 0.695079\tAccuracy: 90.00%\n2\tValidation loss: 0.402921\tBest loss: 0.402921\tAccuracy: 92.00%\n3\tValidation loss: 0.606936\tBest loss: 0.402921\tAccuracy: 92.00%\n4\tValidation loss: 0.354645\tBest loss: 0.354645\tAccuracy: 90.67%\n5\tValidation loss: 0.376935\tBest loss: 0.354645\tAccuracy: 90.67%\n6\tValidation loss: 0.593208\tBest loss: 0.354645\tAccuracy: 90.00%\n7\tValidation loss: 0.388302\tBest loss: 0.354645\tAccuracy: 92.67%\n8\tValidation loss: 0.503276\tBest loss: 0.354645\tAccuracy: 91.33%\n9\tValidation loss: 1.440716\tBest loss: 0.354645\tAccuracy: 80.00%\n10\tValidation loss: 0.464323\tBest loss: 0.354645\tAccuracy: 92.00%\n11\tValidation loss: 0.410302\tBest loss: 0.354645\tAccuracy: 93.33%\n12\tValidation loss: 1.131754\tBest loss: 0.354645\tAccuracy: 88.00%\n13\tValidation loss: 0.511544\tBest loss: 0.354645\tAccuracy: 92.00%\n14\tValidation loss: 0.402083\tBest loss: 0.354645\tAccuracy: 94.00%\n15\tValidation loss: 1.149943\tBest loss: 0.354645\tAccuracy: 92.00%\n16\tValidation loss: 0.405171\tBest loss: 0.354645\tAccuracy: 94.00%\n17\tValidation loss: 0.304346\tBest loss: 0.304346\tAccuracy: 94.67%\n18\tValidation loss: 0.386952\tBest loss: 0.304346\tAccuracy: 94.67%\n19\tValidation loss: 0.387063\tBest loss: 0.304346\tAccuracy: 94.67%\n20\tValidation loss: 0.384417\tBest loss: 0.304346\tAccuracy: 94.67%\n21\tValidation loss: 0.381116\tBest loss: 0.304346\tAccuracy: 94.67%\n22\tValidation loss: 0.379346\tBest loss: 0.304346\tAccuracy: 94.67%\n23\tValidation loss: 0.378128\tBest loss: 0.304346\tAccuracy: 94.67%\n24\tValidation loss: 0.376642\tBest loss: 0.304346\tAccuracy: 94.67%\n25\tValidation loss: 0.375432\tBest loss: 0.304346\tAccuracy: 94.67%\n26\tValidation loss: 0.374804\tBest loss: 0.304346\tAccuracy: 94.67%\n27\tValidation loss: 0.373952\tBest loss: 0.304346\tAccuracy: 94.67%\n28\tValidation loss: 0.373471\tBest loss: 0.304346\tAccuracy: 94.67%\n29\tValidation loss: 0.373027\tBest loss: 0.304346\tAccuracy: 94.67%\n30\tValidation loss: 0.373124\tBest loss: 0.304346\tAccuracy: 94.67%\n31\tValidation loss: 0.373098\tBest loss: 0.304346\tAccuracy: 94.67%\n32\tValidation loss: 0.373206\tBest loss: 0.304346\tAccuracy: 94.67%\n33\tValidation loss: 0.372812\tBest loss: 0.304346\tAccuracy: 94.67%\n34\tValidation loss: 0.373109\tBest loss: 0.304346\tAccuracy: 94.67%\n35\tValidation loss: 0.372616\tBest loss: 0.304346\tAccuracy: 94.67%\n36\tValidation loss: 0.372491\tBest loss: 0.304346\tAccuracy: 94.67%\n37\tValidation loss: 0.372270\tBest loss: 0.304346\tAccuracy: 94.67%\nEarly stopping!\nINFO:tensorflow:Restoring parameters from ./my_mnist_model_5_to_9_no_frozen\nFinal test accuracy: 91.34%\n"
]
],
[
[
"Let's compare that to a DNN trained from scratch:",
"_____no_output_____"
]
],
[
[
"dnn_clf_5_to_9 = DNNClassifier(n_hidden_layers=4, random_state=42)\ndnn_clf_5_to_9.fit(X_train2, y_train2, n_epochs=1000, X_valid=X_valid2, y_valid=y_valid2)",
"0\tValidation loss: 0.674618\tBest loss: 0.674618\tAccuracy: 80.67%\n1\tValidation loss: 0.584845\tBest loss: 0.584845\tAccuracy: 88.67%\n2\tValidation loss: 0.647296\tBest loss: 0.584845\tAccuracy: 84.00%\n3\tValidation loss: 0.530389\tBest loss: 0.530389\tAccuracy: 87.33%\n4\tValidation loss: 0.683215\tBest loss: 0.530389\tAccuracy: 90.67%\n5\tValidation loss: 0.538040\tBest loss: 0.530389\tAccuracy: 89.33%\n6\tValidation loss: 0.670196\tBest loss: 0.530389\tAccuracy: 90.67%\n7\tValidation loss: 0.836470\tBest loss: 0.530389\tAccuracy: 85.33%\n8\tValidation loss: 0.837684\tBest loss: 0.530389\tAccuracy: 92.67%\n9\tValidation loss: 0.588950\tBest loss: 0.530389\tAccuracy: 88.00%\n10\tValidation loss: 0.643213\tBest loss: 0.530389\tAccuracy: 90.67%\n11\tValidation loss: 1.010521\tBest loss: 0.530389\tAccuracy: 88.00%\n12\tValidation loss: 0.931423\tBest loss: 0.530389\tAccuracy: 90.00%\n13\tValidation loss: 1.563524\tBest loss: 0.530389\tAccuracy: 88.67%\n14\tValidation loss: 2.340119\tBest loss: 0.530389\tAccuracy: 89.33%\n15\tValidation loss: 1.402095\tBest loss: 0.530389\tAccuracy: 88.00%\n16\tValidation loss: 1.269974\tBest loss: 0.530389\tAccuracy: 86.00%\n17\tValidation loss: 1.036325\tBest loss: 0.530389\tAccuracy: 89.33%\n18\tValidation loss: 1.578565\tBest loss: 0.530389\tAccuracy: 88.67%\n19\tValidation loss: 0.993890\tBest loss: 0.530389\tAccuracy: 93.33%\n20\tValidation loss: 0.958130\tBest loss: 0.530389\tAccuracy: 87.33%\n21\tValidation loss: 1.505322\tBest loss: 0.530389\tAccuracy: 88.67%\n22\tValidation loss: 1.378772\tBest loss: 0.530389\tAccuracy: 89.33%\n23\tValidation loss: 0.999445\tBest loss: 0.530389\tAccuracy: 88.00%\n24\tValidation loss: 2.366345\tBest loss: 0.530389\tAccuracy: 90.00%\nEarly stopping!\n"
],
[
"y_pred = dnn_clf_5_to_9.predict(X_test2)\naccuracy_score(y_test2, y_pred)",
"_____no_output_____"
]
],
[
[
"Transfer learning allowed us to go from 84.8% accuracy to 91.3%. Not too bad!",
"_____no_output_____"
],
[
"## 10. Pretraining on an auxiliary task",
"_____no_output_____"
],
[
"In this exercise you will build a DNN that compares two MNIST digit images and predicts whether they represent the same digit or not. Then you will reuse the lower layers of this network to train an MNIST classifier using very little training data.",
"_____no_output_____"
],
[
"### 10.1.\nExercise: _Start by building two DNNs (let's call them DNN A and B), both similar to the one you built earlier but without the output layer: each DNN should have five hidden layers of 100 neurons each, He initialization, and ELU activation. Next, add one more hidden layer with 10 units on top of both DNNs. You should use TensorFlow's `concat()` function with `axis=1` to concatenate the outputs of both DNNs along the horizontal axis, then feed the result to the hidden layer. Finally, add an output layer with a single neuron using the logistic activation function._",
"_____no_output_____"
],
[
"**Warning**! There was an error in the book for this exercise: there was no instruction to add a top hidden layer. Without it, the neural network generally fails to start learning. If you have the latest version of the book, this error has been fixed.",
"_____no_output_____"
],
[
"You could have two input placeholders, `X1` and `X2`, one for the images that should be fed to the first DNN, and the other for the images that should be fed to the second DNN. It would work fine. However, another option is to have a single input placeholder to hold both sets of images (each row will hold a pair of images), and use `tf.unstack()` to split this tensor into two separate tensors, like this:",
"_____no_output_____"
]
],
[
[
"n_inputs = 28 * 28 # MNIST\n\nreset_graph()\n\nX = tf.placeholder(tf.float32, shape=(None, 2, n_inputs), name=\"X\")\nX1, X2 = tf.unstack(X, axis=1)",
"_____no_output_____"
]
],
[
[
"We also need the labels placeholder. Each label will be 0 if the images represent different digits, or 1 if they represent the same digit:",
"_____no_output_____"
]
],
[
[
"y = tf.placeholder(tf.int32, shape=[None, 1])",
"_____no_output_____"
]
],
[
[
"Now let's feed these inputs through two separate DNNs:",
"_____no_output_____"
]
],
[
[
"dnn1 = dnn(X1, name=\"DNN_A\")\ndnn2 = dnn(X2, name=\"DNN_B\")",
"_____no_output_____"
]
],
[
[
"And let's concatenate their outputs:",
"_____no_output_____"
]
],
[
[
"dnn_outputs = tf.concat([dnn1, dnn2], axis=1)",
"_____no_output_____"
]
],
[
[
"Each DNN outputs 100 activations (per instance), so the shape is `[None, 100]`:",
"_____no_output_____"
]
],
[
[
"dnn1.shape",
"_____no_output_____"
],
[
"dnn2.shape",
"_____no_output_____"
]
],
[
[
"And of course the concatenated outputs have a shape of `[None, 200]`:",
"_____no_output_____"
]
],
[
[
"dnn_outputs.shape",
"_____no_output_____"
]
],
[
[
"Now lets add an extra hidden layer with just 10 neurons, and the output layer, with a single neuron:",
"_____no_output_____"
]
],
[
[
"hidden = tf.layers.dense(dnn_outputs, units=10, activation=tf.nn.elu, kernel_initializer=he_init)\nlogits = tf.layers.dense(hidden, units=1, kernel_initializer=he_init)\ny_proba = tf.nn.sigmoid(logits)",
"_____no_output_____"
]
],
[
[
"The whole network predicts `1` if `y_proba >= 0.5` (i.e. the network predicts that the images represent the same digit), or `0` otherwise. We compute instead `logits >= 0`, which is equivalent but faster to compute: ",
"_____no_output_____"
]
],
[
[
"y_pred = tf.cast(tf.greater_equal(logits, 0), tf.int32)",
"_____no_output_____"
]
],
[
[
"Now let's add the cost function:",
"_____no_output_____"
]
],
[
[
"y_as_float = tf.cast(y, tf.float32)\nxentropy = tf.nn.sigmoid_cross_entropy_with_logits(labels=y_as_float, logits=logits)\nloss = tf.reduce_mean(xentropy)",
"_____no_output_____"
]
],
[
[
"And we can now create the training operation using an optimizer:",
"_____no_output_____"
]
],
[
[
"learning_rate = 0.01\nmomentum = 0.95\n\noptimizer = tf.train.MomentumOptimizer(learning_rate, momentum, use_nesterov=True)\ntraining_op = optimizer.minimize(loss)",
"_____no_output_____"
]
],
[
[
"We will want to measure our classifier's accuracy.",
"_____no_output_____"
]
],
[
[
"y_pred_correct = tf.equal(y_pred, y)\naccuracy = tf.reduce_mean(tf.cast(y_pred_correct, tf.float32))",
"_____no_output_____"
]
],
[
[
"And the usual `init` and `saver`:",
"_____no_output_____"
]
],
[
[
"init = tf.global_variables_initializer()\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"### 10.2.\n_Exercise: split the MNIST training set in two sets: split #1 should containing 55,000 images, and split #2 should contain contain 5,000 images. Create a function that generates a training batch where each instance is a pair of MNIST images picked from split #1. Half of the training instances should be pairs of images that belong to the same class, while the other half should be images from different classes. For each pair, the training label should be 0 if the images are from the same class, or 1 if they are from different classes._",
"_____no_output_____"
],
[
"The MNIST dataset returned by TensorFlow's `input_data()` function is already split into 3 parts: a training set (55,000 instances), a validation set (5,000 instances) and a test set (10,000 instances). Let's use the first set to generate the training set composed image pairs, and we will use the second set for the second phase of the exercise (to train a regular MNIST classifier). We will use the third set as the test set for both phases.",
"_____no_output_____"
]
],
[
[
"X_train1 = X_train\ny_train1 = y_train\n\nX_train2 = X_valid\ny_train2 = y_valid\n\nX_test = X_test\ny_test = y_test",
"_____no_output_____"
]
],
[
[
"Let's write a function that generates pairs of images: 50% representing the same digit, and 50% representing different digits. There are many ways to implement this. In this implementation, we first decide how many \"same\" pairs (i.e. pairs of images representing the same digit) we will generate, and how many \"different\" pairs (i.e. pairs of images representing different digits). We could just use `batch_size // 2` but we want to handle the case where it is odd (granted, that might be overkill!). Then we generate random pairs and we pick the right number of \"same\" pairs, then we generate the right number of \"different\" pairs. Finally we shuffle the batch and return it:",
"_____no_output_____"
]
],
[
[
"def generate_batch(images, labels, batch_size):\n size1 = batch_size // 2\n size2 = batch_size - size1\n if size1 != size2 and np.random.rand() > 0.5:\n size1, size2 = size2, size1\n X = []\n y = []\n while len(X) < size1:\n rnd_idx1, rnd_idx2 = np.random.randint(0, len(images), 2)\n if rnd_idx1 != rnd_idx2 and labels[rnd_idx1] == labels[rnd_idx2]:\n X.append(np.array([images[rnd_idx1], images[rnd_idx2]]))\n y.append([1])\n while len(X) < batch_size:\n rnd_idx1, rnd_idx2 = np.random.randint(0, len(images), 2)\n if labels[rnd_idx1] != labels[rnd_idx2]:\n X.append(np.array([images[rnd_idx1], images[rnd_idx2]]))\n y.append([0])\n rnd_indices = np.random.permutation(batch_size)\n return np.array(X)[rnd_indices], np.array(y)[rnd_indices]",
"_____no_output_____"
]
],
[
[
"Let's test it to generate a small batch of 5 image pairs:",
"_____no_output_____"
]
],
[
[
"batch_size = 5\nX_batch, y_batch = generate_batch(X_train1, y_train1, batch_size)",
"_____no_output_____"
]
],
[
[
"Each row in `X_batch` contains a pair of images:",
"_____no_output_____"
]
],
[
[
"X_batch.shape, X_batch.dtype",
"_____no_output_____"
]
],
[
[
"Let's look at these pairs:",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(3, 3 * batch_size))\nplt.subplot(121)\nplt.imshow(X_batch[:,0].reshape(28 * batch_size, 28), cmap=\"binary\", interpolation=\"nearest\")\nplt.axis('off')\nplt.subplot(122)\nplt.imshow(X_batch[:,1].reshape(28 * batch_size, 28), cmap=\"binary\", interpolation=\"nearest\")\nplt.axis('off')\nplt.show()",
"_____no_output_____"
]
],
[
[
"And let's look at the labels (0 means \"different\", 1 means \"same\"):",
"_____no_output_____"
]
],
[
[
"y_batch",
"_____no_output_____"
]
],
[
[
"Perfect!",
"_____no_output_____"
],
[
"### 10.3.\n_Exercise: train the DNN on this training set. For each image pair, you can simultaneously feed the first image to DNN A and the second image to DNN B. The whole network will gradually learn to tell whether two images belong to the same class or not._",
"_____no_output_____"
],
[
"Let's generate a test set composed of many pairs of images pulled from the MNIST test set:",
"_____no_output_____"
]
],
[
[
"X_test1, y_test1 = generate_batch(X_test, y_test, batch_size=len(X_test))",
"_____no_output_____"
]
],
[
[
"And now, let's train the model. There's really nothing special about this step, except for the fact that we need a fairly large `batch_size`, otherwise the model fails to learn anything and ends up with an accuracy of 50%:",
"_____no_output_____"
]
],
[
[
"n_epochs = 100\nbatch_size = 500\n\nwith tf.Session() as sess:\n init.run()\n for epoch in range(n_epochs):\n for iteration in range(len(X_train1) // batch_size):\n X_batch, y_batch = generate_batch(X_train1, y_train1, batch_size)\n loss_val, _ = sess.run([loss, training_op], feed_dict={X: X_batch, y: y_batch})\n print(epoch, \"Train loss:\", loss_val)\n if epoch % 5 == 0:\n acc_test = accuracy.eval(feed_dict={X: X_test1, y: y_test1})\n print(epoch, \"Test accuracy:\", acc_test)\n\n save_path = saver.save(sess, \"./my_digit_comparison_model.ckpt\")",
"0 Train loss: 0.69103277\n0 Test accuracy: 0.542\n1 Train loss: 0.6035354\n2 Train loss: 0.54946035\n3 Train loss: 0.47047246\n4 Train loss: 0.4060757\n5 Train loss: 0.38308156\n5 Test accuracy: 0.824\n6 Train loss: 0.39047274\n7 Train loss: 0.3390794\n8 Train loss: 0.3210671\n9 Train loss: 0.31792685\n10 Train loss: 0.24494292\n10 Test accuracy: 0.8881\n11 Train loss: 0.2929235\n12 Train loss: 0.23225449\n13 Train loss: 0.23180929\n14 Train loss: 0.19877923\n15 Train loss: 0.20065464\n15 Test accuracy: 0.9203\n16 Train loss: 0.19700499\n17 Train loss: 0.18893136\n18 Train loss: 0.19965452\n19 Train loss: 0.24071647\n20 Train loss: 0.18882024\n20 Test accuracy: 0.9367\n21 Train loss: 0.12419197\n22 Train loss: 0.14013417\n23 Train loss: 0.120789476\n24 Train loss: 0.15721135\n25 Train loss: 0.11507861\n25 Test accuracy: 0.948\n26 Train loss: 0.13891116\n27 Train loss: 0.1526081\n28 Train loss: 0.123436704\n<<50 more lines>>\n70 Test accuracy: 0.9743\n71 Train loss: 0.019732744\n72 Train loss: 0.039464083\n73 Train loss: 0.04187814\n74 Train loss: 0.05303406\n75 Train loss: 0.052625064\n75 Test accuracy: 0.9756\n76 Train loss: 0.038283084\n77 Train loss: 0.026332883\n78 Train loss: 0.07060841\n79 Train loss: 0.03239444\n80 Train loss: 0.03136283\n80 Test accuracy: 0.9731\n81 Train loss: 0.04390848\n82 Train loss: 0.015268046\n83 Train loss: 0.04875638\n84 Train loss: 0.029360933\n85 Train loss: 0.0418443\n85 Test accuracy: 0.9759\n86 Train loss: 0.018274888\n87 Train loss: 0.038872603\n88 Train loss: 0.02969683\n89 Train loss: 0.020990817\n90 Train loss: 0.045234833\n90 Test accuracy: 0.9769\n91 Train loss: 0.039237432\n92 Train loss: 0.031329047\n93 Train loss: 0.033414133\n94 Train loss: 0.025883088\n95 Train loss: 0.019567214\n95 Test accuracy: 0.9765\n96 Train loss: 0.020650322\n97 Train loss: 0.0339851\n98 Train loss: 0.047079965\n99 Train loss: 0.03125228\n"
]
],
[
[
"All right, we reach 97.6% accuracy on this digit comparison task. That's not too bad, this model knows a thing or two about comparing handwritten digits!\n\nLet's see if some of that knowledge can be useful for the regular MNIST classification task.",
"_____no_output_____"
],
[
"### 10.4.\n_Exercise: now create a new DNN by reusing and freezing the hidden layers of DNN A and adding a softmax output layer on top with 10 neurons. Train this network on split #2 and see if you can achieve high performance despite having only 500 images per class._",
"_____no_output_____"
],
[
"Let's create the model, it is pretty straightforward. There are many ways to freeze the lower layers, as explained in the book. In this example, we chose to use the `tf.stop_gradient()` function. Note that we need one `Saver` to restore the pretrained DNN A, and another `Saver` to save the final model: ",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\ndnn_outputs = dnn(X, name=\"DNN_A\")\nfrozen_outputs = tf.stop_gradient(dnn_outputs)\n\nlogits = tf.layers.dense(frozen_outputs, n_outputs, kernel_initializer=he_init)\nY_proba = tf.nn.softmax(logits)\n\nxentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\nloss = tf.reduce_mean(xentropy, name=\"loss\")\n\noptimizer = tf.train.MomentumOptimizer(learning_rate, momentum, use_nesterov=True)\ntraining_op = optimizer.minimize(loss)\n\ncorrect = tf.nn.in_top_k(logits, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n\ninit = tf.global_variables_initializer()\n\ndnn_A_vars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES, scope=\"DNN_A\")\nrestore_saver = tf.train.Saver(var_list={var.op.name: var for var in dnn_A_vars})\nsaver = tf.train.Saver()",
"_____no_output_____"
]
],
[
[
"Now on to training! We first initialize all variables (including the variables in the new output layer), then we restore the pretrained DNN A. Next, we just train the model on the small MNIST dataset (containing just 5,000 images):",
"_____no_output_____"
]
],
[
[
"n_epochs = 100\nbatch_size = 50\n\nwith tf.Session() as sess:\n init.run()\n restore_saver.restore(sess, \"./my_digit_comparison_model.ckpt\")\n\n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n if epoch % 10 == 0:\n acc_test = accuracy.eval(feed_dict={X: X_test, y: y_test})\n print(epoch, \"Test accuracy:\", acc_test)\n\n save_path = saver.save(sess, \"./my_mnist_model_final.ckpt\")",
"INFO:tensorflow:Restoring parameters from ./my_digit_comparison_model.ckpt\n0 Test accuracy: 0.9455\n10 Test accuracy: 0.9634\n20 Test accuracy: 0.9659\n30 Test accuracy: 0.9656\n40 Test accuracy: 0.9655\n50 Test accuracy: 0.9656\n60 Test accuracy: 0.9655\n70 Test accuracy: 0.9656\n80 Test accuracy: 0.9654\n90 Test accuracy: 0.9654\n"
]
],
[
[
"Well, 96.5% accuracy, that's not the best MNIST model we have trained so far, but recall that we are only using a small training set (just 500 images per digit). Let's compare this result with the same DNN trained from scratch, without using transfer learning:",
"_____no_output_____"
]
],
[
[
"reset_graph()\n\nn_inputs = 28 * 28 # MNIST\nn_outputs = 10\n\nX = tf.placeholder(tf.float32, shape=(None, n_inputs), name=\"X\")\ny = tf.placeholder(tf.int32, shape=(None), name=\"y\")\n\ndnn_outputs = dnn(X, name=\"DNN_A\")\n\nlogits = tf.layers.dense(dnn_outputs, n_outputs, kernel_initializer=he_init)\nY_proba = tf.nn.softmax(logits)\n\nxentropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=y, logits=logits)\nloss = tf.reduce_mean(xentropy, name=\"loss\")\n\noptimizer = tf.train.MomentumOptimizer(learning_rate, momentum, use_nesterov=True)\ntraining_op = optimizer.minimize(loss)\n\ncorrect = tf.nn.in_top_k(logits, y, 1)\naccuracy = tf.reduce_mean(tf.cast(correct, tf.float32))\n\ninit = tf.global_variables_initializer()\n\ndnn_A_vars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES, scope=\"DNN_A\")\nrestore_saver = tf.train.Saver(var_list={var.op.name: var for var in dnn_A_vars})\nsaver = tf.train.Saver()",
"_____no_output_____"
],
[
"n_epochs = 150\nbatch_size = 50\n\nwith tf.Session() as sess:\n init.run()\n\n for epoch in range(n_epochs):\n rnd_idx = np.random.permutation(len(X_train2))\n for rnd_indices in np.array_split(rnd_idx, len(X_train2) // batch_size):\n X_batch, y_batch = X_train2[rnd_indices], y_train2[rnd_indices]\n sess.run(training_op, feed_dict={X: X_batch, y: y_batch})\n if epoch % 10 == 0:\n acc_test = accuracy.eval(feed_dict={X: X_test, y: y_test})\n print(epoch, \"Test accuracy:\", acc_test)\n\n save_path = saver.save(sess, \"./my_mnist_model_final.ckpt\")",
"0 Test accuracy: 0.8694\n10 Test accuracy: 0.9276\n20 Test accuracy: 0.9299\n30 Test accuracy: 0.935\n40 Test accuracy: 0.942\n50 Test accuracy: 0.9435\n60 Test accuracy: 0.9442\n70 Test accuracy: 0.9447\n80 Test accuracy: 0.9448\n90 Test accuracy: 0.945\n100 Test accuracy: 0.945\n110 Test accuracy: 0.9458\n120 Test accuracy: 0.9456\n130 Test accuracy: 0.9458\n140 Test accuracy: 0.9458\n"
]
],
[
[
"Only 94.6% accuracy... So transfer learning helped us reduce the error rate from 5.4% to 3.5% (that's over 35% error reduction). Moreover, the model using transfer learning reached over 96% accuracy in less than 10 epochs.\n\nBottom line: transfer learning does not always work, but when it does it can make a big difference. So try it out!",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
] |
4a16b06989d27efa51760d85c833f0c0c80f71a4
| 225,630 |
ipynb
|
Jupyter Notebook
|
z-Analysis/01-Matplotlib/demo.ipynb
|
jiaxiaochu/Crawler
|
bb54d515dc217c27574b36124e16fd5b993775bd
|
[
"MIT"
] | null | null | null |
z-Analysis/01-Matplotlib/demo.ipynb
|
jiaxiaochu/Crawler
|
bb54d515dc217c27574b36124e16fd5b993775bd
|
[
"MIT"
] | 1 |
2020-08-27T10:25:38.000Z
|
2020-08-27T10:25:38.000Z
|
z-Analysis/01-Matplotlib/demo.ipynb
|
jiaxiaochu/Crawler
|
bb54d515dc217c27574b36124e16fd5b993775bd
|
[
"MIT"
] | null | null | null | 657.813411 | 87,932 | 0.953574 |
[
[
[
"## 引入matplotlib包",
"_____no_output_____"
]
],
[
[
"# 引入matplotlib包\n# from matplotlib import pyplot as plt\nimport matplotlib.pyplot as plt\nimport random\n# plt.plot([x轴坐标],[y轴坐标])\nplt.plot([1],[1])\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 绘制直线图",
"_____no_output_____"
]
],
[
[
"plt.plot([0,1,2,3],[1,2,3,4])\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(20,8),dpi=100)\nplt.plot([1,2,3,4],[1,2,3,4])\nplt.show()",
"_____no_output_____"
],
[
"import numpy as np\na = np.arange(10)\nprint(a)\nplt.plot(a,a*1.5,a,a*2.5,a,a*3.5,a,a*4.5)\nplt.show()",
"[0 1 2 3 4 5 6 7 8 9]\n"
]
],
[
[
"## 绘制散点图",
"_____no_output_____"
]
],
[
[
"plt.scatter([1],[1])\nplt.show()",
"_____no_output_____"
],
[
"# plt.scatter([1,2,3,4],[1,2,3,4])\nplt.scatter([1,6,2,3,4],[5,3,2,1,6])\nplt.show()",
"_____no_output_____"
]
],
[
[
"## 绘制折线图",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(20,10))\n#调用plot方法只需要传入所有的点的x和y的值的列表即可,可迭代对象也行\nplt.plot([1,2,3,4,5,6,7],[17,17,18,15,11,11,13])\nplt.show()\n# plt.figure(figsize=(10, 10))\n# #调用plot方法只需要传入所有的点的x和y的值的列表即可,可迭代对象也行\n# plt.plot([1, 2, 3, 4, 5, 6 ,7], [17,17,18,15,11,11,13])\n# plt.show()",
"_____no_output_____"
],
[
"# 准备数据\nx = range(1,31)\n# for i in x:\n# print(i)\n# print(\"#\" * 100)",
"_____no_output_____"
],
[
"y = [random.randint(10,20)for i in x]\nprint(y)",
"[16, 17, 18, 18, 12, 11, 18, 14, 11, 20, 13, 14, 20, 16, 16, 10, 12, 20, 16, 16, 15, 18, 15, 14, 14, 18, 13, 11, 13, 16]\n"
],
[
"# 开始绘制图形\nplt.figure(figsize=(20,8),dpi=100)\nplt.plot(x,y)\nplt.show()",
"_____no_output_____"
],
[
"# 设置图片大小\nplt.figure(figsize=(), dpi=)\n figsize:指定图的长宽\n dpi:图像的清晰度\n 返回fig对象\n\n# 保存为图片\nplt.savefig(path)",
"_____no_output_____"
],
[
"plt.figure(figsize=(20, 8), dpi=80)\nplt.scatter([1,6,2,3,4],[5,3,2,1,6])\nplt.savefig(\"test.png\")",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a16b11ee1676be17d6f5a7c037dc8049c8af3e5
| 8,137 |
ipynb
|
Jupyter Notebook
|
DeepLearningFrameworks/inference/ResNet50-MXNet.ipynb
|
gopala-kr/ds-notebooks
|
bc35430ecdd851f2ceab8f2437eec4d77cb59423
|
[
"MIT"
] | 1 |
2019-05-10T09:16:23.000Z
|
2019-05-10T09:16:23.000Z
|
DeepLearningFrameworks/inference/ResNet50-MXNet.ipynb
|
gopala-kr/ds-notebooks
|
bc35430ecdd851f2ceab8f2437eec4d77cb59423
|
[
"MIT"
] | null | null | null |
DeepLearningFrameworks/inference/ResNet50-MXNet.ipynb
|
gopala-kr/ds-notebooks
|
bc35430ecdd851f2ceab8f2437eec4d77cb59423
|
[
"MIT"
] | 1 |
2019-05-10T09:17:28.000Z
|
2019-05-10T09:17:28.000Z
| 22.921127 | 168 | 0.523289 |
[
[
[
"# GPU: 32*40 in 9.87s = 130/s\n# CPU: 32*8 in 31.9s = 8/s",
"_____no_output_____"
],
[
"import os\nimport sys\nimport numpy as np\nimport mxnet as mx\nfrom collections import namedtuple\nprint(\"OS: \", sys.platform)\nprint(\"Python: \", sys.version)\nprint(\"Numpy: \", np.__version__)\nprint(\"MXNet: \", mx.__version__)",
"OS: linux\nPython: 3.5.2 |Anaconda custom (64-bit)| (default, Jul 2 2016, 17:53:06) \n[GCC 4.4.7 20120313 (Red Hat 4.4.7-1)]\nNumpy: 1.13.3\nMXNet: 0.12.1\n"
],
[
"!cat /proc/cpuinfo | grep processor | wc -l",
"6\r\n"
],
[
"!nvidia-smi --query-gpu=gpu_name --format=csv",
"name\r\nTesla K80\r\n"
],
[
"Batch = namedtuple('Batch', ['data'])\nBATCH_SIZE = 32\nRESNET_FEATURES = 2048\nBATCHES_GPU = 40\nBATCHES_CPU = 8",
"_____no_output_____"
],
[
"def give_fake_data(batches):\n \"\"\" Create an array of fake data to run inference on\"\"\"\n np.random.seed(0)\n dta = np.random.rand(BATCH_SIZE*batches, 224, 224, 3).astype(np.float32)\n return dta, np.swapaxes(dta, 1, 3)",
"_____no_output_____"
],
[
"def yield_mb(X, batchsize):\n \"\"\" Function yield (complete) mini_batches of data\"\"\"\n for i in range(len(X)//batchsize):\n yield i, X[i*batchsize:(i+1)*batchsize]",
"_____no_output_____"
],
[
"# Create batches of fake data\nfake_input_data_cl, fake_input_data_cf = give_fake_data(BATCHES_GPU)\nprint(fake_input_data_cl.shape, fake_input_data_cf.shape)",
"(1280, 224, 224, 3) (1280, 3, 224, 224)\n"
],
[
"# Download Resnet weights\npath='http://data.mxnet.io/models/imagenet/'\n[mx.test_utils.download(path+'resnet/50-layers/resnet-50-symbol.json'),\n mx.test_utils.download(path+'resnet/50-layers/resnet-50-0000.params')]",
"_____no_output_____"
],
[
"# Load model\nsym, arg_params, aux_params = mx.model.load_checkpoint('resnet-50', 0)\n# List the last 10 layers\nall_layers = sym.get_internals()",
"_____no_output_____"
],
[
"print(all_layers.list_outputs()[-10:])",
"['bn1_moving_var', 'bn1_output', 'relu1_output', 'pool1_output', 'flatten0_output', 'fc1_weight', 'fc1_bias', 'fc1_output', 'softmax_label', 'softmax_output']\n"
],
[
"def predict_fn(classifier, data, batchsize):\n \"\"\" Return features from classifier \"\"\"\n out = np.zeros((len(data), RESNET_FEATURES), np.float32)\n for idx, dta in yield_mb(data, batchsize):\n classifier.forward(Batch(data=[mx.nd.array(dta)]))\n out[idx*batchsize:(idx+1)*batchsize] = classifier.get_outputs()[0].asnumpy().squeeze()\n return out",
"_____no_output_____"
]
],
[
[
"## 1. GPU",
"_____no_output_____"
]
],
[
[
"# Get last layer\nfe_sym = all_layers['flatten0_output']\n# Initialise GPU\nfe_mod = mx.mod.Module(symbol=fe_sym, context=[mx.gpu(0)], label_names=None)\nfe_mod.bind(for_training=False, inputs_need_grad=False,\n data_shapes=[('data', (BATCH_SIZE,3,224,224))])\nfe_mod.set_params(arg_params, aux_params)",
"_____no_output_____"
],
[
"cold_start = predict_fn(fe_mod, fake_input_data_cf, BATCH_SIZE)",
"_____no_output_____"
],
[
"%%time\n# GPU: 9.87s\nfeatures = predict_fn(fe_mod, fake_input_data_cf, BATCH_SIZE)",
"CPU times: user 8.08 s, sys: 1.7 s, total: 9.78 s\nWall time: 9.87 s\n"
]
],
[
[
"## 2. CPU",
"_____no_output_____"
]
],
[
[
"# Kill all GPUs ...\nos.environ['CUDA_VISIBLE_DEVICES'] = '-1'",
"_____no_output_____"
],
[
"# Get last layer\nfe_sym = all_layers['flatten0_output']\n# Initialise CPU\nfe_mod = mx.mod.Module(symbol=fe_sym, context=mx.cpu(), label_names=None)\nfe_mod.bind(for_training=False, inputs_need_grad=False,\n data_shapes=[('data', (BATCH_SIZE,3,224,224))])\nfe_mod.set_params(arg_params, aux_params)",
"_____no_output_____"
],
[
"# Create batches of fake data\nfake_input_data_cl, fake_input_data_cf = give_fake_data(BATCHES_CPU)\nprint(fake_input_data_cl.shape, fake_input_data_cf.shape)",
"(256, 224, 224, 3) (256, 3, 224, 224)\n"
],
[
"cold_start = predict_fn(fe_mod, fake_input_data_cf, BATCH_SIZE)",
"_____no_output_____"
],
[
"%%time\n# CPU: 31.9s\nfeatures = predict_fn(fe_mod, fake_input_data_cf, BATCH_SIZE)",
"CPU times: user 58.8 s, sys: 1.42 s, total: 1min\nWall time: 31.9 s\n"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a16be3afd805ec341e2e7c9839dcbf3e81b3c01
| 3,030 |
ipynb
|
Jupyter Notebook
|
ConvertTimeIntoMilitaryFormat.ipynb
|
KiChoudhary/python
|
bc2238481728841c59d92f9e96bb0794817820fe
|
[
"MIT"
] | null | null | null |
ConvertTimeIntoMilitaryFormat.ipynb
|
KiChoudhary/python
|
bc2238481728841c59d92f9e96bb0794817820fe
|
[
"MIT"
] | null | null | null |
ConvertTimeIntoMilitaryFormat.ipynb
|
KiChoudhary/python
|
bc2238481728841c59d92f9e96bb0794817820fe
|
[
"MIT"
] | null | null | null | 27.297297 | 246 | 0.457426 |
[
[
[
"<a href=\"https://colab.research.google.com/github/KiChoudhary/python/blob/master/ConvertTimeIntoMilitaryFormat.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"time = input(\"Enter the time: (HH:MM:SS AM/PM)\")",
"Enter the time: (HH:MM:SS AM/PM)10:40:44 PM\n"
],
[
"split_time = time.split(\" \")\nsplit_time2=split_time[0].split(\":\")",
"_____no_output_____"
],
[
"for i in range(0,len(split_time2)):\n split_time2[i]=int(split_time2[i])\n\n\nif split_time2[0] not in range(1,13):\n print(\"values entered are incorrect\")\nelif split_time2[1] not in range(0,60):\n print(\"values entered are incorrect\")\nelif split_time2[2] not in range(0,60):\n print(\"values entered are incorrect\")\nelse:\n d = {'am':0,'AM':0,'pm':1,'PM':1}\n if d[split_time[1]] == 0:\n print(split_time[0])\n else:\n split_time2[0] = int(split_time2[0])\n split_time2[0]+=12\n if split_time2[0]==24: splite_time2[0]=24\n print(split_time2[0],\":\",split_time2[1],\":\",split_time2[2])\n ",
"22 : 40 : 44\n"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code"
]
] |
4a16d483f98cdb403f1d1d2168e33e777a920fb2
| 23,377 |
ipynb
|
Jupyter Notebook
|
notebooks/SummarizeRELAX.ipynb
|
aglucaci/GeneInvestigator
|
2dc2dda0d7befab208b340c7800685739aced64a
|
[
"MIT"
] | null | null | null |
notebooks/SummarizeRELAX.ipynb
|
aglucaci/GeneInvestigator
|
2dc2dda0d7befab208b340c7800685739aced64a
|
[
"MIT"
] | null | null | null |
notebooks/SummarizeRELAX.ipynb
|
aglucaci/GeneInvestigator
|
2dc2dda0d7befab208b340c7800685739aced64a
|
[
"MIT"
] | null | null | null | 38.639669 | 1,740 | 0.467468 |
[
[
[
"import pandas as pd\nimport numpy as np\nimport os\nimport json\nimport altair as alt\nimport numpy as np\nimport glob",
"_____no_output_____"
],
[
"DATA_DIR = \"/Users/user/Documents/GeneInvestigator/results/BDNF/Recombinants\"\n\nRELAX_FILES = glob.glob(os.path.join(DATA_DIR, \"*.RELAX.json\"))\npvalue_threshold = 0.05",
"_____no_output_____"
],
[
"RELAX_FILES",
"_____no_output_____"
],
[
"def getRELAX_TR(JSON):\n with open(JSON, \"r\") as in_d:\n json_data = json.load(in_d)\n return json_data[\"test results\"]\n\ndef getRELAX_fits(JSON):\n with open(JSON, \"r\") as in_d:\n json_data = json.load(in_d)\n return json_data[\"fits\"]\n",
"_____no_output_____"
],
[
"# Main\npassedThreshold = []\n\nfor file in RELAX_FILES:\n pval = getRELAX_TR(file)[\"p-value\"]\n if pval <= pvalue_threshold:\n passedThreshold.append(file)\n print(\"# Passed p-value threshold:\", file.split(\"/\")[-1])\n",
"# Passed p-value threshold: BDNF_codons_RDP_recombinationFree.fas.Carnivora.RELAX.json\n# Passed p-value threshold: BDNF_codons_RDP_recombinationFree.fas.Primates.RELAX.json\n# Passed p-value threshold: BDNF_codons_RDP_recombinationFree.fas.Eulipotyphla.RELAX.json\n"
],
[
"# Process them separately\n\n#TEST_FILE = \"/Users/user/Documents/GeneInvestigator/results/BDNF/Recombinants/BDNF_codons_RDP_recombinationFree.fas.Carnivora.RELAX.json\"\n#TEST_FILE = \"/Users/user/Documents/GeneInvestigator/results/BDNF/Recombinants/BDNF_codons_RDP_recombinationFree.fas.Primates.RELAX.json\"\nTEST_FILE = '/Users/user/Documents/GeneInvestigator/results/BDNF/Recombinants/BDNF_codons_RDP_recombinationFree.fas.Eulipotyphla.RELAX.json'\n\nlabel = TEST_FILE.split(\"/\")[-1].split(\".\")[2]\nprint(\"Label:\", label)\nfits = getRELAX_fits(TEST_FILE)\nfits.keys()\n\n# print(fits[\"RELAX partitioned descriptive\"])",
"Label: Eulipotyphla\n"
],
[
"df = pd.DataFrame.from_dict(fits, orient='index')\ndf.T",
"_____no_output_____"
],
[
"df2_Ref = pd.DataFrame.from_dict(fits[\"RELAX partitioned descriptive\"][\"Rate Distributions\"][\"Reference\"], orient='index')\ndf2_Ref",
"_____no_output_____"
],
[
"df3_Test = pd.DataFrame.from_dict(fits[\"RELAX partitioned descriptive\"][\"Rate Distributions\"][\"Test\"], orient='index')\ndf3_Test",
"_____no_output_____"
],
[
"# Unconstrained model\n\nsource = df2_Ref\ndf2_Ref[\"log(omega)\"] = np.log10(df2_Ref[\"omega\"])\ndf2_Ref[\"Color\"] = \"Reference\"\n\nline = alt.Chart(source).mark_bar().encode(\n x='log(omega)',\n y='proportion',\n color=\"Color\"\n).properties(\n width=500,\n height=400,\n title=label\n)\n\nline.configure_title(\n fontSize=20,\n font='Courier',\n anchor='start',\n color='gray'\n)\n\ndf3_Test[\"log(omega)\"] = np.log10(df3_Test[\"omega\"])\ndf3_Test[\"Color\"] = \"Test\"\n\nsource = df3_Test\n\nline2 = alt.Chart(source).mark_bar().encode(\n x='log(omega)',\n y='proportion',\n color=\"Color\"\n).properties(\n width=500,\n height=400)\n\n\n\nline + line2",
"_____no_output_____"
],
[
"# Process them together as subplots",
"_____no_output_____"
],
[
"# RELAX Interpretation\n# http://hyphy.org/methods/selection-methods/#relax\n# method paper: https://academic.oup.com/mbe/article/32/3/820/981440\n\n",
"_____no_output_____"
]
],
[
[
"# Figure legend\nPatterns of natural selection across taxonomic groups under the Partitioned Descriptive model of the RELAX method. Selection profiles for BDNF are shown along Reference and Test branches for each taxonomic group. Three omega parameters and the relative proportion of sites they represent are plotted for Test (orange) and Referenc (blue) branches. Only omega categories representing nonzero proportions of sites are shown. Neutral selection corresponds to the omega=1.0 in this log10 scaled X-axis. These taxonomic groups represent datasets where significant (p<= 0.05) for relaxed selection was detected between test and reference branches.",
"_____no_output_____"
]
]
] |
[
"code",
"markdown"
] |
[
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
] |
4a16d568fd710fbc1fcf0adfe1588181cb683b55
| 57,871 |
ipynb
|
Jupyter Notebook
|
d2l-en/mxnet/chapter_convolutional-modern/resnet.ipynb
|
gr8khan/d2lai
|
7c10432f38c80e86978cd075d0024902b47842a0
|
[
"MIT"
] | null | null | null |
d2l-en/mxnet/chapter_convolutional-modern/resnet.ipynb
|
gr8khan/d2lai
|
7c10432f38c80e86978cd075d0024902b47842a0
|
[
"MIT"
] | null | null | null |
d2l-en/mxnet/chapter_convolutional-modern/resnet.ipynb
|
gr8khan/d2lai
|
7c10432f38c80e86978cd075d0024902b47842a0
|
[
"MIT"
] | null | null | null | 41.874819 | 420 | 0.525289 |
[
[
[
"# Residual Networks (ResNet)\n:label:`sec_resnet`\n\nAs we design increasingly deeper networks it becomes imperative to understand how adding layers can increase the complexity and expressiveness of the network.\nEven more important is the ability to design networks where adding layers makes networks strictly more expressive rather than just different.\nTo make some progress we need a bit of mathematics.\n\n\n## Function Classes\n\nConsider $\\mathcal{F}$, the class of functions that a specific network architecture (together with learning rates and other hyperparameter settings) can reach.\nThat is, for all $f \\in \\mathcal{F}$ there exists some set of parameters (e.g., weights and biases) that can be obtained through training on a suitable dataset.\nLet us assume that $f^*$ is the \"truth\" function that we really would like to find.\nIf it is in $\\mathcal{F}$, we are in good shape but typically we will not be quite so lucky.\nInstead, we will try to find some $f^*_\\mathcal{F}$ which is our best bet within $\\mathcal{F}$.\nFor instance, \ngiven a dataset with features $\\mathbf{X}$\nand labels $\\mathbf{y}$,\nwe might try finding it by solving the following optimization problem:\n\n$$f^*_\\mathcal{F} \\stackrel{\\mathrm{def}}{=} \\mathop{\\mathrm{argmin}}_f L(\\mathbf{X}, \\mathbf{y}, f) \\text{ subject to } f \\in \\mathcal{F}.$$\n\nIt is only reasonable to assume that if we design a different and more powerful architecture $\\mathcal{F}'$ we should arrive at a better outcome. In other words, we would expect that $f^*_{\\mathcal{F}'}$ is \"better\" than $f^*_{\\mathcal{F}}$. However, if $\\mathcal{F} \\not\\subseteq \\mathcal{F}'$ there is no guarantee that this should even happen. In fact, $f^*_{\\mathcal{F}'}$ might well be worse. \nAs illustrated by :numref:`fig_functionclasses`,\nfor non-nested function classes, a larger function class does not always move closer to the \"truth\" function $f^*$. For instance,\non the left of :numref:`fig_functionclasses`,\nthough $\\mathcal{F}_3$ is closer to $f^*$ than $\\mathcal{F}_1$, $\\mathcal{F}_6$ moves away and there is no guarantee that further increasing the complexity can reduce the distance from $f^*$.\nWith nested function classes\nwhere $\\mathcal{F}_1 \\subseteq \\ldots \\subseteq \\mathcal{F}_6$\non the right of :numref:`fig_functionclasses`,\nwe can avoid the aforementioned issue from the non-nested function classes.\n\n\n\n:label:`fig_functionclasses`\n\nThus,\nonly if larger function classes contain the smaller ones are we guaranteed that increasing them strictly increases the expressive power of the network.\nFor deep neural networks,\nif we can \ntrain the newly-added layer into an identity function $f(\\mathbf{x}) = \\mathbf{x}$, the new model will be as effective as the original model. As the new model may get a better solution to fit the training dataset, the added layer might make it easier to reduce training errors.\n\nThis is the question that He et al. considered when working on very deep computer vision models :cite:`He.Zhang.Ren.ea.2016`. \nAt the heart of their proposed *residual network* (*ResNet*) is the idea that every additional layer should \nmore easily\ncontain the identity function as one of its elements. \nThese considerations are rather profound but they led to a surprisingly simple\nsolution, a *residual block*.\nWith it, ResNet won the ImageNet Large Scale Visual Recognition Challenge in 2015. The design had a profound influence on how to\nbuild deep neural networks.\n\n\n\n## Residual Blocks\n\nLet us focus on a local part of a neural network, as depicted in :numref:`fig_residual_block`. Denote the input by $\\mathbf{x}$.\nWe assume that the desired underlying mapping we want to obtain by learning is $f(\\mathbf{x})$, to be used as the input to the activation function on the top.\nOn the left of :numref:`fig_residual_block`,\nthe portion within the dotted-line box \nmust directly learn the mapping $f(\\mathbf{x})$.\nOn the right,\nthe portion within the dotted-line box\nneeds to\nlearn the *residual mapping* $f(\\mathbf{x}) - \\mathbf{x}$,\nwhich is how the residual block derives its name.\nIf the identity mapping $f(\\mathbf{x}) = \\mathbf{x}$ is the desired underlying mapping,\nthe residual mapping is easier to learn:\nwe only need to push the weights and biases\nof the\nupper weight layer (e.g., fully-connected layer and convolutional layer)\nwithin the dotted-line box\nto zero.\nThe right figure in :numref:`fig_residual_block` illustrates the *residual block* of ResNet,\nwhere the solid line carrying the layer input \n$\\mathbf{x}$ to the addition operator\nis called a *residual connection* (or *shortcut connection*).\nWith residual blocks, inputs can \nforward propagate faster through the residual connections across layers.\n\n\n:label:`fig_residual_block`\n\n\nResNet follows VGG's full $3\\times 3$ convolutional layer design. The residual block has two $3\\times 3$ convolutional layers with the same number of output channels. Each convolutional layer is followed by a batch normalization layer and a ReLU activation function. Then, we skip these two convolution operations and add the input directly before the final ReLU activation function.\nThis kind of design requires that the output of the two convolutional layers has to be of the same shape as the input, so that they can be added together. If we want to change the number of channels, we need to introduce an additional $1\\times 1$ convolutional layer to transform the input into the desired shape for the addition operation. Let us have a look at the code below.\n",
"_____no_output_____"
]
],
[
[
"from d2l import mxnet as d2l\nfrom mxnet import np, npx\nfrom mxnet.gluon import nn\nnpx.set_np()\n\nclass Residual(nn.Block): #@save\n \"\"\"The Residual block of ResNet.\"\"\"\n def __init__(self, num_channels, use_1x1conv=False, strides=1, **kwargs):\n super().__init__(**kwargs)\n self.conv1 = nn.Conv2D(num_channels, kernel_size=3, padding=1,\n strides=strides)\n self.conv2 = nn.Conv2D(num_channels, kernel_size=3, padding=1)\n if use_1x1conv:\n self.conv3 = nn.Conv2D(num_channels, kernel_size=1,\n strides=strides)\n else:\n self.conv3 = None\n self.bn1 = nn.BatchNorm()\n self.bn2 = nn.BatchNorm()\n\n def forward(self, X):\n Y = npx.relu(self.bn1(self.conv1(X)))\n Y = self.bn2(self.conv2(Y))\n if self.conv3:\n X = self.conv3(X)\n return npx.relu(Y + X)",
"_____no_output_____"
]
],
[
[
"This code generates two types of networks: one where we add the input to the output before applying the ReLU nonlinearity whenever `use_1x1conv=False`, and one where we adjust channels and resolution by means of a $1 \\times 1$ convolution before adding. :numref:`fig_resnet_block` illustrates this:\n\n\n:label:`fig_resnet_block`\n\nNow let us look at a situation where the input and output are of the same shape.\n",
"_____no_output_____"
]
],
[
[
"blk = Residual(3)\nblk.initialize()\nX = np.random.uniform(size=(4, 3, 6, 6))\nblk(X).shape",
"_____no_output_____"
]
],
[
[
"We also have the option to halve the output height and width while increasing the number of output channels.\n",
"_____no_output_____"
]
],
[
[
"blk = Residual(6, use_1x1conv=True, strides=2)\nblk.initialize()\nblk(X).shape",
"_____no_output_____"
]
],
[
[
"## ResNet Model\n\nThe first two layers of ResNet are the same as those of the GoogLeNet we described before: the $7\\times 7$ convolutional layer with 64 output channels and a stride of 2 is followed by the $3\\times 3$ maximum pooling layer with a stride of 2. The difference is the batch normalization layer added after each convolutional layer in ResNet.\n",
"_____no_output_____"
]
],
[
[
"net = nn.Sequential()\nnet.add(nn.Conv2D(64, kernel_size=7, strides=2, padding=3),\n nn.BatchNorm(), nn.Activation('relu'),\n nn.MaxPool2D(pool_size=3, strides=2, padding=1))",
"_____no_output_____"
]
],
[
[
"GoogLeNet uses four modules made up of Inception blocks.\nHowever, ResNet uses four modules made up of residual blocks, each of which uses several residual blocks with the same number of output channels. \nThe number of channels in the first module is the same as the number of input channels. Since a maximum pooling layer with a stride of 2 has already been used, it is not necessary to reduce the height and width. In the first residual block for each of the subsequent modules, the number of channels is doubled compared with that of the previous module, and the height and width are halved.\n\nNow, we implement this module. Note that special processing has been performed on the first module.\n",
"_____no_output_____"
]
],
[
[
"def resnet_block(num_channels, num_residuals, first_block=False):\n blk = nn.Sequential()\n for i in range(num_residuals):\n if i == 0 and not first_block:\n blk.add(Residual(num_channels, use_1x1conv=True, strides=2))\n else:\n blk.add(Residual(num_channels))\n return blk",
"_____no_output_____"
]
],
[
[
"Then, we add all the modules to ResNet. Here, two residual blocks are used for each module.\n",
"_____no_output_____"
]
],
[
[
"net.add(resnet_block(64, 2, first_block=True),\n resnet_block(128, 2),\n resnet_block(256, 2),\n resnet_block(512, 2))",
"_____no_output_____"
]
],
[
[
"Finally, just like GoogLeNet, we add a global average pooling layer, followed by the fully-connected layer output.\n",
"_____no_output_____"
]
],
[
[
"net.add(nn.GlobalAvgPool2D(), nn.Dense(10))",
"_____no_output_____"
]
],
[
[
"There are 4 convolutional layers in each module (excluding the $1\\times 1$ convolutional layer). Together with the first $7\\times 7$ convolutional layer and the final fully-connected layer, there are 18 layers in total. Therefore, this model is commonly known as ResNet-18.\nBy configuring different numbers of channels and residual blocks in the module, we can create different ResNet models, such as the deeper 152-layer ResNet-152. Although the main architecture of ResNet is similar to that of GoogLeNet, ResNet's structure is simpler and easier to modify. All these factors have resulted in the rapid and widespread use of ResNet. :numref:`fig_resnet18` depicts the full ResNet-18.\n\n\n:label:`fig_resnet18`\n\nBefore training ResNet, let us observe how the input shape changes across different modules in ResNet. As in all the previous architectures, the resolution decreases while the number of channels increases up until the point where a global average pooling layer aggregates all features.\n",
"_____no_output_____"
]
],
[
[
"X = np.random.uniform(size=(1, 1, 224, 224))\nnet.initialize()\nfor layer in net:\n X = layer(X)\n print(layer.name, 'output shape:\\t', X.shape)",
"conv5 output shape:\t (1, 64, 112, 112)\nbatchnorm4 output shape:\t (1, 64, 112, 112)\nrelu0 output shape:\t (1, 64, 112, 112)\npool0 output shape:\t (1, 64, 56, 56)\nsequential1 output shape:\t (1, 64, 56, 56)\nsequential2 output shape:\t (1, 128, 28, 28)\nsequential3 output shape:\t (1, 256, 14, 14)\nsequential4 output shape:\t (1, 512, 7, 7)\npool1 output shape:\t (1, 512, 1, 1)\ndense0 output shape:\t (1, 10)\n"
]
],
[
[
"## Training\n\nWe train ResNet on the Fashion-MNIST dataset, just like before.\n",
"_____no_output_____"
]
],
[
[
"lr, num_epochs, batch_size = 0.05, 10, 256\ntrain_iter, test_iter = d2l.load_data_fashion_mnist(batch_size, resize=96)\nd2l.train_ch6(net, train_iter, test_iter, num_epochs, lr)",
"loss 0.012, train acc 0.997, test acc 0.881\n4852.1 examples/sec on gpu(0)\n"
]
],
[
[
"## Summary\n\n* Nested function classes are desirable. Learning an additional layer in deep neural networks as an identity function (though this is an extreme case) should be made easy.\n* The residual mapping can learn the identity function more easily, such as pushing parameters in the weight layer to zero.\n* We can train an effective deep neural network by having residual blocks. Inputs can forward propagate faster through the residual connections across layers.\n* ResNet had a major influence on the design of subsequent deep neural networks, both for convolutional and sequential nature.\n\n\n## Exercises\n\n1. What are the major differences between the Inception block in :numref:`fig_inception` and the residual block? After removing some paths in the Inception block, how are they related to each other?\n1. Refer to Table 1 in the ResNet paper :cite:`He.Zhang.Ren.ea.2016` to\n implement different variants.\n1. For deeper networks, ResNet introduces a \"bottleneck\" architecture to reduce\n model complexity. Try to implement it.\n1. In subsequent versions of ResNet, the authors changed the \"convolution, batch\n normalization, and activation\" structure to the \"batch normalization,\n activation, and convolution\" structure. Make this improvement\n yourself. See Figure 1 in :cite:`He.Zhang.Ren.ea.2016*1`\n for details.\n1. Why can't we just increase the complexity of functions without bound, even if the function classes are nested?\n",
"_____no_output_____"
],
[
"[Discussions](https://discuss.d2l.ai/t/85)\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
] |
4a16dc1636a9c9223fe286aaec0314c388bce269
| 26,509 |
ipynb
|
Jupyter Notebook
|
day3_simple_model.ipynb
|
AnettSz/dw_matrix_car
|
287f583356c438c1ffc43fe5c8698bd89976f593
|
[
"MIT"
] | null | null | null |
day3_simple_model.ipynb
|
AnettSz/dw_matrix_car
|
287f583356c438c1ffc43fe5c8698bd89976f593
|
[
"MIT"
] | null | null | null |
day3_simple_model.ipynb
|
AnettSz/dw_matrix_car
|
287f583356c438c1ffc43fe5c8698bd89976f593
|
[
"MIT"
] | null | null | null | 26,509 | 26,509 | 0.574258 |
[
[
[
"!pip install --upgrade tables\n!pip install eli5",
"Collecting tables\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/ed/c3/8fd9e3bb21872f9d69eb93b3014c86479864cca94e625fd03713ccacec80/tables-3.6.1-cp36-cp36m-manylinux1_x86_64.whl (4.3MB)\n\u001b[K |████████████████████████████████| 4.3MB 3.5MB/s \n\u001b[?25hRequirement already satisfied, skipping upgrade: numexpr>=2.6.2 in /usr/local/lib/python3.6/dist-packages (from tables) (2.7.1)\nRequirement already satisfied, skipping upgrade: numpy>=1.9.3 in /usr/local/lib/python3.6/dist-packages (from tables) (1.17.5)\nInstalling collected packages: tables\n Found existing installation: tables 3.4.4\n Uninstalling tables-3.4.4:\n Successfully uninstalled tables-3.4.4\nSuccessfully installed tables-3.6.1\nCollecting eli5\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/97/2f/c85c7d8f8548e460829971785347e14e45fa5c6617da374711dec8cb38cc/eli5-0.10.1-py2.py3-none-any.whl (105kB)\n\u001b[K |████████████████████████████████| 112kB 3.5MB/s \n\u001b[?25hRequirement already satisfied: scipy in /usr/local/lib/python3.6/dist-packages (from eli5) (1.4.1)\nRequirement already satisfied: scikit-learn>=0.18 in /usr/local/lib/python3.6/dist-packages (from eli5) (0.22.1)\nRequirement already satisfied: attrs>16.0.0 in /usr/local/lib/python3.6/dist-packages (from eli5) (19.3.0)\nRequirement already satisfied: tabulate>=0.7.7 in /usr/local/lib/python3.6/dist-packages (from eli5) (0.8.6)\nRequirement already satisfied: graphviz in /usr/local/lib/python3.6/dist-packages (from eli5) (0.10.1)\nRequirement already satisfied: jinja2 in /usr/local/lib/python3.6/dist-packages (from eli5) (2.11.1)\nRequirement already satisfied: numpy>=1.9.0 in /usr/local/lib/python3.6/dist-packages (from eli5) (1.17.5)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from eli5) (1.12.0)\nRequirement already satisfied: joblib>=0.11 in /usr/local/lib/python3.6/dist-packages (from scikit-learn>=0.18->eli5) (0.14.1)\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.6/dist-packages (from jinja2->eli5) (1.1.1)\nInstalling collected packages: eli5\nSuccessfully installed eli5-0.10.1\n"
],
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nfrom sklearn.dummy import DummyRegressor\nfrom sklearn.tree import DecisionTreeRegressor\n\nfrom sklearn.metrics import mean_absolute_error as mae\nfrom sklearn.model_selection import cross_val_score\n\nimport eli5\nfrom eli5.sklearn import PermutationImportance",
"_____no_output_____"
],
[
"cd \"/content/drive/My Drive/Colab Notebooks/dw_matrix/matrix_two/dw_matrix_car\"\n",
"/content/drive/My Drive/Colab Notebooks/dw_matrix/matrix_two/dw_matrix_car\n"
],
[
"\n!ls\n",
"cars_visualisation.ipynb data\tday3_simple_model.ipynb LICENSE README.md\n"
]
],
[
[
"## Reading data",
"_____no_output_____"
]
],
[
[
"df = pd.read_hdf('data/car.h5')\ndf.shape",
"_____no_output_____"
],
[
"df.columns.values",
"_____no_output_____"
]
],
[
[
"## Dummy Model",
"_____no_output_____"
]
],
[
[
"df. select_dtypes(np.number).columns",
"_____no_output_____"
],
[
" feats = ['car_id']\n X = df[ feats ].values\n y = df['price_value'].values\n\n model = DummyRegressor()\n model.fit(X, y)\n y_pred = model.predict(X)\n\n mae(y, y_pred)",
"_____no_output_____"
],
[
"# Remove prices in currencies different than PLN\ndf = df[ df['price_currency'] == 'PLN' ]\ndf.shape\n",
"_____no_output_____"
],
[
"SUFIX_CAT = '_cat'\n \nfor feat in df.columns:\n\n if isinstance(df[feat][0], list): continue\n\n factorized_values = df[ feat ].factorize()[0]\n\n if SUFIX_CAT in feat: \n df[feat] = factorized_value\n else: \n df[feat + SUFIX_CAT] = factorized_values",
"_____no_output_____"
],
[
"cat_feats = [x for x in df.columns if SUFIX_CAT in x]\ncat_feats = [x for x in cat_feats if 'price' not in x]\nlen(cat_feats)\n",
"_____no_output_____"
],
[
"\nX = df[ cat_feats ].values\ny = df['price_value'].values\n\nmodel = DecisionTreeRegressor(max_depth=5)\nscores = cross_val_score(model, X, y, cv=3, scoring='neg_mean_absolute_error')\n\nnp.mean(scores)\n\n#model.fit(X, y)\n#y_pred = model.predict(X)\n#mae(y, y_pred)",
"_____no_output_____"
],
[
"\nm = DecisionTreeRegressor(max_depth=5)\nm.fit(X, y)\n\nimp = PermutationImportance(m, random_state=0).fit(X, y)\neli5.show_weights(imp, feature_names=cat_feats)",
"_____no_output_____"
]
]
] |
[
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a16df0ca95af599b7dc35b6f3021eafd114228a
| 395,250 |
ipynb
|
Jupyter Notebook
|
technical_analysis/services/tki2hm002_3008_le_gall_bcy569_luokittelu_tuntitehtava.ipynb
|
mikalegall/flask
|
743faaf5ef6ef165612bbbc1b451613e522a5326
|
[
"MIT"
] | null | null | null |
technical_analysis/services/tki2hm002_3008_le_gall_bcy569_luokittelu_tuntitehtava.ipynb
|
mikalegall/flask
|
743faaf5ef6ef165612bbbc1b451613e522a5326
|
[
"MIT"
] | null | null | null |
technical_analysis/services/tki2hm002_3008_le_gall_bcy569_luokittelu_tuntitehtava.ipynb
|
mikalegall/flask
|
743faaf5ef6ef165612bbbc1b451613e522a5326
|
[
"MIT"
] | null | null | null | 199.520444 | 233,616 | 0.899823 |
[
[
[
"Perusprosessissa Data Wrangling \n<br>\nhttps://en.wikipedia.org/wiki/Data_wrangling\n<br>\neli datan valmistelu (ETL putsaus jne) vie yleensä 80% työajasta. Datan valmistelu koneoppimisen malleja varten:\n<br>\nhttps://nbviewer.jupyter.org/github/taanila/tilastoapu/blob/master/datan_valmistelu.ipynb\n<br>\nLuokittele sen jälkeen pystyrivit (sarakkeet) joko kategorisiin tai määrällisiin. Ja esitä analytiikan osa-alueen (perinteisen neliportaisen tason) mukaisia, siis ihmismielelle mielekkäitä, kysymyksiä aineistolle:\n",
"_____no_output_____"
],
[
"#### Ennakoiva analytiikka: Koneoppiminen\nhttps://tilastoapu.wordpress.com/2019/08/03/koneoppiminen-ja-scikit-learn-kirjasto/\n#### Lähestyminen valitaan sen mukaan<br>1. onko jo etukäteen saatavilla ennustettavan muuttujan todellisia arvoja (label) vahvistettuina tietoina (training set eli opetusdata, supervised learning)<br>2. vai ei (unsupervised learning) ja kolmantena mahdollisuutena on<br>3. algoritmin palkitseminen tai rankaiseminen (reinforcement learning) sen suorittaessa analytiikkaa\n\nKoneoppiminen opetusdatasta (supervised learning):\n<br>\nLähestyminen valitaan sen mukaan onko ennustettavan muuttujan (target) arvot kategorisia (discrete label) vai määrällisiä (continuous label):\n<br>\nKategoriselle muuttujalle luokittelumalli\n<br>\nMäärälliselle muuttujalle regressio-malli\n<br>\nFeature matrix tarkoittaa muuttujia, jotka selittävät ennustettavaa muuttujaa (target)\n<br>\n<font color=\"grey\">Koneoppiminen ilman opetusdataa (unsupervised learning):\n<br>\nKlusterointi-malli (pyritään löytämään yhteen kuuluvat havainnot, esim. K-means)\n<br>\nYksinkertaistamisen malli (core: pienin mahdollinen määrä muuttujia joka selittää riittävästi asiaa, esim. pääkomponenttianalyysi)\n<br>\n<i>Syvä oppiminen (deep learning) pohjautuu neuroverkkomenetelmiin, joilla pyritään myös jäljittelemään ihmisaivojenkin toimintaa</i>\n</font>",
"_____no_output_____"
],
[
"SUPERVISED LEARNING (<b><font color=\"red\">eri malleja pitää vaan työläästi kokeilla</font></b>)\n<br>\n<b>Luokittelumalleja kategoriselle target muuttujalle</b>:\n<br>\nK lähintä naapuria (K nearest neighbor), Päätöspuut (decision trees), Gaussian Naiivi Bayes, Logistinen regressio.\n<br>\nkäyttötarkoituksena esim:\n<br>\nOCR (Optical Character Recognition) eli kuvapikseleistä esim. auton rekisterinumeron muuttaminen tekstiksi, kuvapikseleistä esineen tunnistaminen, röntgenkuvasta sairauden tunnistaminen, maksuhäiriöön ajautumisen todennäköisyys, vakuutuspetos, roskapostin suodatus jne\n<br>\nHUOM! Mikäli kategorista targettia selittävään feature matrixiin otetaan mukaan kategorisia muuttujia, tulee ne kategoriset feature matrixin selittävät muuttujat muuttaa dikotomisiksi dummy muuttujiksi eli nolliksi ja ykkösiksi pandas kirjaston 'get_dummies()' komennolla\n<br>\ndf_dummies = pd.get_dummies(df)\n<br>\nHUOM! Yllä oleva komento muuttaa data framen kaikki tekstimuotoiset kategoriset muuttujat dummyiksi, mutta jos kategorinen muuttuja on muuta muotoa, niin silloin tulee antaa lisäparametreilla pandakselle tieto mitkä sarakkeet muutetaan \n<br>\n<br>\n<b>Useimmiten regressiomalli on target määrälliselle muuttujalle tarkoituksenmukaisin</b> (selittävä feature matrix on määrällinen muuttuja, koska korrelaatio):\n<br>\nlineaarinen regressio (etsitään suora viiva joka parhaiten kulkee havaintojen kautta)\n<br>\nkäyttötarkoituksena esim:\n<br>\nkysynnän ennustaminen, asunnon hinta (onko saunaa, parveketta tms), käytetyn auton hinta jne\n",
"_____no_output_____"
],
[
"LUOKITTELU",
"_____no_output_____"
]
],
[
[
"#Tuodaan käytettäväksi data-analytiikan kirjasto pandas ja\n#lyhennetään sitä kutsuttavaksi aliaksella pd\n## https://pandas.pydata.org/docs/user_guide/dsintro.html#dataframe\nimport pandas as pd\n\n#Tuodaan graafiseen esittämiseen matplotlib ja sen käyttöliittymäksi pyplot\n## https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.html?highlight=pyplot#module-matplotlib.pyplot\nimport matplotlib.pyplot as plt\n\n#Vaaditaan vanhoissa Juptter Notebook versioissa, jotta kaaviot tulostuvat\n%matplotlib inline\n\n#Katsotaan millaisia erilaisia muotoilutyylejä on saatavilla\nplt.style.available",
"_____no_output_____"
],
[
"#Valitaan graafinen esittäminen muotoiltavaksi tietyllä tyylillä\nplt.style.use('seaborn-whitegrid')\n\n#Asetetaan näytettävien rivien rajoite (ei rajoitetta)\npd.options.display.max_rows = None\n#Asetetaan näytettävien sarakkeiden rajoite (ei rajoitetta)\npd.options.display.max_columns = None\n\n#Tuodaan graafisen esittämisen tyylimäärittelyt erilaisilla kaaviolajeilla\n#ja käytetään tätä tarvittaessa matplotlib esitysten \"ylikirjoittamiseen\"\n#(korvaamiseen silloin kun on silmälle mukavampia vaihtoehtoja tarjolla)\nimport seaborn as sns\n\n# Harjoitteluun tarkoitettu datasetti Kurjenmiekat-kasveista (englanniksi iris) löytyy seaborn-kirjastosta\n## https://fi.wikipedia.org/wiki/Kurjenmiekat\niris = sns.load_dataset('iris')\n\n#Tarkistetaan lähdetiedoston metatiedoista monessako sarakkeessa on\n#minkäkin verran arvoja ja mitä tyyppiä ne sisältää\niris.info()\n",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 150 entries, 0 to 149\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 sepal_length 150 non-null float64\n 1 sepal_width 150 non-null float64\n 2 petal_length 150 non-null float64\n 3 petal_width 150 non-null float64\n 4 species 150 non-null object \ndtypes: float64(4), object(1)\nmemory usage: 6.0+ KB\n"
],
[
"#Kurkataan 5 vikaa riviä niinkuin Linuxissa ja nähdään sitenkin rivien määrä\niris.tail()\n##tai 5 ekaa riviä\n##iris.head()",
"_____no_output_____"
],
[
"#Näytetään kolme pienintä arvoa sarakkeesta 'petal_length' \niris.nsmallest(n=3,columns='petal_length')",
"_____no_output_____"
],
[
"#Näytetään kolme suurinta arvoa sarakkeesta 'petal_length' \niris.nlargest(n=3,columns='petal_length')",
"_____no_output_____"
],
[
"#Numeromuotoisen sarakkeen suodatus\n##Noudetaan data framesta Pandas kirjaston toiminnolla ne rivt, joissa\n#sarakkeen 'petal_length arvo on suurempi kuin 6.3 (suodatetaan rajoittimella > 6.3) \niris[ iris['petal_length'] > 6.3 ]\n##Uloimmat hakasulut viittaavat muuttujan iris \"alkioon\" / sarakkeeseen ja\n#niiden hakasulkujen sisällä suoritetaan suodatus",
"_____no_output_____"
]
],
[
[
"Yllä olevassa taulukossa\n<br>\nsepal = verholehti\n<br>\nhttps://fi.wikipedia.org/wiki/Verhi%C3%B6\n<br>\npetal = terälehti\n<br>\nhttps://fi.wikipedia.org/wiki/Teri%C3%B6\n<br>\nLajikkeet ovat Setosa, Versicolor ja Virginica",
"_____no_output_____"
]
],
[
[
"#Mahdollisten tyhjien arvojen tarkistus\niris.isnull().sum()",
"_____no_output_____"
],
[
"#Hajontakaaviossa voi kategorisen muuttujan eri arvoja esittää eri väreillä\nsns.pairplot(iris, hue='species')",
"_____no_output_____"
]
],
[
[
"Tiedon esittäminen visualisoituna (datasetin aineistosta graafisessa muodossa) paljastaa hyvin havainnollistavasti alarivillä Y-akselin 'petal_width' ja X-akselin 'petal_length' leikkauspisteen ruudukossa kuinka Setosa-lajike erottautuu selkeästi muista Kurjenmiekoista (iris) pelkästään petal (terälehti) pituuden ja leveyden osalta.\n<br>\n<br>\nYhtä selkeää eroa ei ole nähtävissä Versicolor ja Virginica lajikkeiden toisistaan erotteluun\n",
"_____no_output_____"
],
[
"Ottamalla kaikki verho- ja terälehti muuttujat ('sepal_length', 'sepal_width', 'petal_length' ja 'petal_width') mukaan feature matrixiin voitaneen koulutusdatasta kouluttaa hyvin osuva koneoppimisen malli, joka oppii lajittelemaan Kurjenmiekan sen verho- ja terälehtien mittojen mukaan tarkoituksenmukaiseen targettiin (lajikkeeseen). <b><font color=\"red\">Eri malleja pitää vaan työläästi kokeilla</font></b>",
"_____no_output_____"
],
[
"Datan valmistelua (Data Wrangling) sen jakamiseksi koulutusdataan ja testidataan",
"_____no_output_____"
]
],
[
[
"#Ei asenneta Anacondassa vaan kerrotaan mistä \"from foo.bar\" tuodaan käytettäväksi \"import lorem_ipsus\"\n##toiminnallisuus, joka jakaa datan\nfrom sklearn.model_selection import train_test_split\n\n#Vakiintuneen tavan mukaan feature matrix (selittävä muuttuja) on iso X\n##Poistetaan lajitieto\nX = iris.drop('species', axis=1)\n\n#Vakiintuneen tavan mukaan target (ennustettava muuttuja) on y\ny = iris['species']\n\n#random_state arvo määrittelee miten data erotellaan opetus- ja testidataan\n##eri kokeilukerroilla tulee käyttää samaa arvoa (arvolla itsellään ei ole mitään merkitystä)\n##sillä jos aineisto on jaettu eri tavalla osiin voi päätyä hyvinkin erilaiseen malliin\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=4)\n###Käytännössä datasetissä otoksen jakamisen 'random_state' arvolla on\n###jonkilainen vaikutus ennustusprosentin osumatarkkuuteen\n### X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=5)\n#HUOM! Ennen tuotantoon deployausta toistetaan splittausta moneen eri kertaan eri jaotusarvoilla\n#ja tuotantoon pääty niistä kokeiluista jonkilainen keskiarvon tyyppinen ratkaisu\n\n\n#Katsotaan millaisiin paloihin jako tapahtui\nprint(X_train.shape)\nprint(X_test.shape)\nprint(y_train.shape)\nprint(y_test.shape)",
"(112, 4)\n(38, 4)\n(112,)\n(38,)\n"
]
],
[
[
"### Ennakoivan analytiikan mallin sovitus datasettiin (1/4)\nKoneoppimisessa kokeillaan erilaisia algoritmeja samaan historiadataan ja katsotaan mikä niistä arvaa parhaiten, kun ennustetta verrataan jo tiedossa oleviin toteumiin\n\n#### 1. algoritmi kokeilu (ensimmäinen sovitusyritys: Miten malli sopii toteumaan) ",
"_____no_output_____"
],
[
"#### KNN eli K-nearest neighbors (K-lähimmät naapurit) -menetelmässä\n<b><font color=\"red\">eri \"K\"-arvoja pitää vaan työläästi kokeilla</font></b>, jotta luotavan koneoppimisen malli opetusdatasta (supervised learning) luokittelee riittävän tarkasti uudet havainnot tarkoituksenmukaisiin kategorioihin\n<br>\n<br>\nSuora copy+paste\n<br>\n\"Etsitään luokiteltavalle havainnolle K lähintä naapuria opetusdatasta.\n\nLuokiteltava havainto sijoitetaan siihen luokkaan, joka on enemmistönä K:n lähimmän naapurin joukossa.\"\n<br>\nhttps://nbviewer.jupyter.org/github/taanila/tilastoapu/blob/master/iris_knn.ipynb",
"_____no_output_____"
]
],
[
[
"#Ei asenneta Anacondassa vaan kerrotaan mistä \"from foo.bar\" tuodaan käytettäväksi \"import lorem_ipsus\"\n##toiminnallisuus, joka suorittaa KNN-vertailun\nfrom sklearn.neighbors import KNeighborsClassifier\n\n#Luodaan malli-olio, jossa parametrin 'n_neighbors' arvo on 'K' eli\n#syötteenä saatavan Kurjenmiekan verho- ja terälehtien pituuksien ja paksuuksien\n#perusteella verrataan sitä, tässä esimerkkitapauksessa viiteen, 5 lähimpään\n#samoilla mitoilla oleviin naapureihin \n##(\"Etsitään luokiteltavalle havainnolle K lähintä naapuria opetusdatasta\")\n#JA\n#katsotaan siitä (tässä esimerkkitapauksessa viiden) 5 naapurostosta mihin\n#kategorisen muuttujan luokkaan 'species' (Kurjenmiekka-kasvin laji) enemmistö\n# naapureista on luokiteltu, ja päätellään syötteenä saatavan Kurjenmiekan\n# kuuluvan myös siihen samaan luokkaan\n##\"Luokiteltava havainto sijoitetaan siihen luokkaan, joka on enemmistönä K:n lähimmän naapurin joukossa.\"\nmalli = KNeighborsClassifier(n_neighbors=5)\n\n#Ennustemallille annetaan parametreina:\n#feature matrix (muutosta selittävät muuttujat) ja\n#target (ennustettava muuttuja, joka muuttuu feature matrixin muutosten vuoksi)\nmalli.fit(X_train, y_train)\n###komennolla fit() sovitetaan malli dataan. Se on se komento, jolla konetta opetetaan\n\n#Olion sisältämät toiminnallisuudet saa IDEssä esiin kirjoittamalla 'malli.'\n#ja naputtelemalla tabulaattoria (tai CTRL+SPACE)\nmalli.get_params()",
"_____no_output_____"
]
],
[
[
"#### 1. algoritmin soveltuvuuden arviointi: Miten ennuste osuu toteumaan",
"_____no_output_____"
]
],
[
[
"#Malli-olion predict-toiminnolla voidaan laskea ennuste opetusdatalle\ny_train_malli = malli.predict(X_train)\n#Malli-olion predict-toiminnolla voidaan laskea ennuste testidatalle\ny_test_malli = malli.predict(X_test)\n\n#Ei asenneta Anacondassa vaan kerrotaan mistä \"from foo.bar\" tuodaan käytettäväksi \"import lorem_ipsus\"\n##toiminnallisuus, joka vertaa ennustuksen ja toteuman osumatarkkuutta\nfrom sklearn.metrics import accuracy_score\n\n#Opetusdatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_train, y_train_malli)",
"_____no_output_____"
],
[
"#Testidatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_test, y_test_malli)\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"_____no_output_____"
],
[
"#Ei asenneta Anacondassa vaan kerrotaan mistä \"from foo.bar\" tuodaan käytettäväksi \"import lorem_ipsus\"\n##toiminnallisuus, joka paljastaa epäonnistuneet ennustukset\nfrom sklearn.metrics import confusion_matrix\n\n#Opetusdatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_train, y_train_malli))",
"[[32 0 0]\n [ 0 40 2]\n [ 0 1 37]]\n"
]
],
[
[
"Yllä olevassa Kurjenmiekat-kasvin taulukossa ovat lajikkeet Setosa, Versicolor ja Virginica. Talukkoa luetaan siten, että luotu ennustemalli on onnistunut ennustamaan\n* ensimmäisellä rivillä (ja ensimmäisessä sarakkeessa) kaikki Setosat oikein\n* keskimmäisellä rivillä (ja keskimmäisessä sarakkeessa) Versicolorista lähes kaikki oikein, mutta osa oli mennyt vahingossa viimeiseen sarakkeeseen Virginicalle\n* viimeisellä rivillä (ja viimeisessä sarakkeessa) Virginicasta lähes kaikki oikein, mutta osa oli mennyt vahingossa keskimmäiseen sarakkeeseen Versicolorille",
"_____no_output_____"
]
],
[
[
"#Testidatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_test, y_test_malli))\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"[[18 0 0]\n [ 0 7 1]\n [ 0 0 12]]\n"
]
],
[
[
"Vaikuttaa siltä, että ei olla ylimallinnettu, koska algoritmiltä piilossa olleelle datalle saadaan ainoastaan yksi pieleen mennyt ennuste",
"_____no_output_____"
]
],
[
[
"#Täysin uuden Kurjenmiekka datasetin käyttöönotto, josta puuttuu\n#etukäteen tiedossa oleva 'species' label\n##Vakiintuneen tavan mukaan feature matrix (selittävä muuttuja) on iso X\nXnew = pd.read_excel('http://taanila.fi/irisnew.xlsx')\n# Jos CSV https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html\n#TARKISTA TARVITAANKO\n##sepstr, default ‘,’\n##usecolslist-like or callable, optional\n\n#Pandas kirjastolla noudetaan txt-tiedostot ikään kuin ne olisivat csv-tiedostoja\n##erotimerkin muoto tulee ilmaista\n### sep='\\s+'\n\n#Vakiintuneen tavan mukaan feature matrix (selittävä muuttuja) on iso X\n##Malli-olion predict-toiminnolla suoritetaan täysin uuden\n##syötteen luokittelu kategoriaan\nmalli.predict(Xnew)",
"_____no_output_____"
]
],
[
[
"Uudessa datasetissä ei yksinkertaisuuden vuoksi ollut kuin kolme havaintoa. Yllä olevasta tulosteesta näkee, että juuri koulutettu koneoppimisen algoritmi jakoi taulukon\n* ensimmäisen rivin osalta Setosaksi\n* toisen rivin osalta Virginicaksi\n* kolmannen rivin osalta Versicoloriksi",
"_____no_output_____"
],
[
"### Ennakoivan analytiikan mallin sovitus datasettiin (2/4)\nKoneoppimisessa kokeillaan erilaisia algoritmeja samaan historiadataan ja katsotaan mikä niistä arvaa parhaiten, kun ennustetta verrataan jo tiedossa oleviin toteumiin\n\n#### 2. algoritmi kokeilu (ensimmäinen sovitusyritys: Miten malli sopii toteumaan) ",
"_____no_output_____"
],
[
"#### Päätöspuu-menetelmässä\n<b><font color=\"red\">puun haarautumisen syvyyden eri arvoja pitää vaan työläästi kokeilla</font></b>, jotta luotavan koneoppimisen malli opetusdatasta (supervised learning) luokittelee riittävän tarkasti uudet havainnot tarkoituksenmukaisiin kategorioihin\n<br>\n<br>\nSuora copy+paste\n<br>\n\"Jokaisessa haarautumisessa algoritmi valitsee parhaiten erottelevan selittävän muuttujan ja siihen liittyvän rajakohdan.\n\nGini = millä todennäköisyydellä tehdään väärä luokittelu? Toimivassa päätöspuussa on haarautumisten jälkeen vain pieniä gini-arvoja.\"\n<br>\nhttps://nbviewer.jupyter.org/github/taanila/tilastoapu/blob/master/iris_dectree.ipynb",
"_____no_output_____"
]
],
[
[
"#random_state arvo määrittelee miten data erotellaan opetus- ja testidataan\n##eri kokeilukerroilla tulee käyttää samaa arvoa (arvolla itsellään ei ole mitään merkitystä)\n##sillä jos aineisto on jaettu eri tavalla osiin voi päätyä hyvinkin erilaiseen malliin\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=4)\n###Käytännössä datasetissä otoksen jakamisen 'random_state' arvolla on\n###jonkilainen vaikutus ennustusprosentin osumatarkkuuteen\n### X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=5)\n#HUOM! Ennen tuotantoon deployausta toistetaan splittausta moneen eri kertaan eri jaotusarvoilla\n#ja tuotantoon pääty niistä kokeiluista jonkilainen keskiarvon tyyppinen ratkaisu\n\n#Katsotaan millaisiin paloihin jako tapahtui\nprint(X_train.shape)\nprint(X_test.shape)\nprint(y_train.shape)\nprint(y_test.shape)",
"(112, 4)\n(38, 4)\n(112,)\n(38,)\n"
],
[
"#Ei asenneta Anacondassa vaan kerrotaan mistä \"from foo.bar\" tuodaan käytettäväksi \"import lorem_ipsus\"\n##toiminnallisuus, joka muodostaa päätöspuun\nfrom sklearn import tree\n\n#Kirjastosta saatavat puun erilaiset toiminnallisuudet saa IDEssä esiin kirjoittamalla 'tree.'\n#ja naputtelemalla tabulaattoria (tai CTRL+SPACE)\ntree.plot_tree",
"_____no_output_____"
],
[
"#Otetaan käyttöön perusmalli 'DecisionTreeClassifier'\n##ja annetaan päätöspuun haarautumisen enimmäissyvyys\n##parametrilla 'max_depth' \nmalli_2 = tree.DecisionTreeClassifier(max_depth = 4)\n#Syvyttä säätelemällä mallin osumatarkkuus vaihtelee, mutta ei kannata\n#ylimallintaa suurilla luvuilla, sillä päätöspuulla kyllä päästään\n#tarvittaessa 100%:n osumatarkkuuteen, joka ei tuntemattoman datan kanssa\n#sitten enää toimikaan\n\n#Ennustemallille annetaan parametreina:\n#feature matrix (muutosta selittävät muuttujat) ja\n#target (ennustettava muuttuja, joka muuttuu feature matrixin muutosten vuoksi)\nmalli_2.fit(X_train, y_train)\n###komennolla fit() sovitetaan malli dataan. Se on se komento, jolla konetta opetetaan\n\n\n#Malli-olion predict-toiminnolla voidaan laskea ennuste opetusdatalle\ny_train_malli_2 = malli_2.predict(X_train)\n#Malli-olion predict-toiminnolla voidaan laskea ennuste testidatalle\ny_test_malli_2 = malli_2.predict(X_test)",
"_____no_output_____"
],
[
"#Opetusdatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_train, y_train_malli_2)",
"_____no_output_____"
],
[
"#Testidatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_test, y_test_malli_2)\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"_____no_output_____"
],
[
"#Opetusdatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_train, y_train_malli_2))",
"[[32 0 0]\n [ 0 42 0]\n [ 0 0 38]]\n"
]
],
[
[
"Yllä olevassa Kurjenmiekat-kasvin taulukossa ovat lajikkeet Setosa, Versicolor ja Virginica. Talukkoa luetaan siten, että luotu ennustemalli on onnistunut ennustamaan\n* ensimmäisellä rivillä (ja ensimmäisessä sarakkeessa) kaikki Setosat oikein\n* keskimmäisellä rivillä (ja keskimmäisessä sarakkeessa) kaikki Versicolorista oikein\n* viimeisellä rivillä (ja viimeisessä sarakkeessa) kaikki Virginicasta oikein",
"_____no_output_____"
]
],
[
[
"#Testidatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_test, y_test_malli_2))\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"[[18 0 0]\n [ 0 7 1]\n [ 0 0 12]]\n"
]
],
[
[
"Yllä olevassa Kurjenmiekat-kasvin taulukossa ovat lajikkeet Setosa, Versicolor ja Virginica. Talukkoa luetaan siten, että luotu ennustemalli on onnistunut ennustamaan\n* ensimmäisellä rivillä (ja ensimmäisessä sarakkeessa) kaikki Setosat oikein\n* keskimmäisellä rivillä (ja keskimmäisessä sarakkeessa) Versicolorista lähes kaikki oikein, mutta osa oli mennyt vahingossa viimeiseen sarakkeeseen Virginicalle\n* viimeisellä rivillä (ja viimeisessä sarakkeessa) kaikki Virginicat oikein",
"_____no_output_____"
]
],
[
[
"#Suurennetaan esitysaluetta oletuskoostaan suuremmaksi\nplt.figure(figsize=(18, 10))\n\n#Taustalle piiloon luotu päätöspuumalli esitetään plotaten graafisena visualisointina\ntree.plot_tree(malli_2)\n#Kirjastosta saatavat puun erilaiset toiminnallisuudet saa IDEssä esiin kirjoittamalla 'tree.'\n#ja naputtelemalla tabulaattoria (tai CTRL+SPACE)",
"_____no_output_____"
]
],
[
[
"\"Jokaisessa haarautumisessa algoritmi valitsee parhaiten erottelevan selittävän muuttujan ja siihen liittyvän rajakohdan.\n<br>\nGini = millä todennäköisyydellä tehdään väärä luokittelu? Toimivassa päätöspuussa on haarautumisten jälkeen vain pieniä gini-arvoja.\"\n<br>\n<br>\nYllä olevassa päätöspuussa ylimmällä rivillä lähtötilanteessa X[2] viittaa terälehden pituuteen ('petal_length') If Then haaroituksessa booleania\n* vasemmalle mennään jos 'petal_length' on yhtäsuuri tai pienempi kuin 2,45 ja muutoin mennään oikealle (aiemmin rivillä 8 huomattiin tiedon visualisoinnissa kuinka kaikki Setosat olivat eroteltävissa terälehden avulla)",
"_____no_output_____"
]
],
[
[
"iris.head()",
"_____no_output_____"
],
[
"#Vakiintuneen tavan mukaan feature matrix (selittävä muuttuja) on iso X\n##Malli-olion predict-toiminnolla suoritetaan täysin uuden\n##syötteen luokittelu kategoriaan\nmalli_2.predict(Xnew)",
"_____no_output_____"
]
],
[
[
"Uudessa datasetissä ei yksinkertaisuuden vuoksi ollut kuin kolme havaintoa. Yllä olevasta tulosteesta näkee, että juuri koulutettu koneoppimisen algoritmi jakoi taulukon\n* ensimmäisen rivin osalta Setosaksi\n* toisen rivin osalta Virginicaksi\n* kolmannen rivin osalta Versicoloriksi",
"_____no_output_____"
],
[
"### Ennakoivan analytiikan mallin sovitus datasettiin (3/4)\nKoneoppimisessa kokeillaan erilaisia algoritmeja samaan historiadataan ja katsotaan mikä niistä arvaa parhaiten, kun ennustetta verrataan jo tiedossa oleviin toteumiin\n\n#### 3. algoritmi kokeilu (ensimmäinen sovitusyritys: Miten malli sopii toteumaan) ",
"_____no_output_____"
],
[
"#### Gaussian Naive Bayes -menetelmässä\n<br>\neri target kategorioihin luokittelussa feature matrixin selittävät muuttujat arvioidaan normaalijakauma-olettamalla, josta lasketaan todennäköisyys kategoriaan kuulumisesta\n<br>\n<br>\nSuora copy+paste\n<br>\n\"todennäköisyyksien lukuarvot eivät sellaisenaan ole luotettavia. Olennaista on mallin toteuttama luokittelu.\"\n<br>\nja\n<br>\n\"Mallin oletuksena on, että selittävien muuttujien arvot ovat kussakin luokassa toisistaan riippumattomia. Käytännössä Gaussian Naive Bayes toimii hyvin monenlaisten datojen kohdalla vaikka riippumattomuusoletus ei toteutuisikaan.\"\n<br>\nhttps://nbviewer.jupyter.org/github/taanila/tilastoapu/blob/master/iris_naive_bayes.ipynb",
"_____no_output_____"
]
],
[
[
"#random_state arvo määrittelee miten data erotellaan opetus- ja testidataan\n##eri kokeilukerroilla tulee käyttää samaa arvoa (arvolla itsellään ei ole mitään merkitystä)\n##sillä jos aineisto on jaettu eri tavalla osiin voi päätyä hyvinkin erilaiseen malliin\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=4)\n###Käytännössä datasetissä otoksen jakamisen 'random_state' arvolla on\n###jonkilainen vaikutus ennustusprosentin osumatarkkuuteen\n### X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=5)\n#HUOM! Ennen tuotantoon deployausta toistetaan splittausta moneen eri kertaan eri jaotusarvoilla\n#ja tuotantoon pääty niistä kokeiluista jonkilainen keskiarvon tyyppinen ratkaisu\n\n#Katsotaan millaisiin paloihin jako tapahtui\nprint(X_train.shape)\nprint(X_test.shape)\nprint(y_train.shape)\nprint(y_test.shape)",
"(112, 4)\n(38, 4)\n(112,)\n(38,)\n"
],
[
"#Ei asenneta Anacondassa vaan kerrotaan mistä \"from foo.bar\" tuodaan käytettäväksi \"import lorem_ipsus\"\n##toiminnallisuus, joka perustuu Bayesin teoreemaan (Gaussian Naive Bayes -menetelmä)\nfrom sklearn.naive_bayes import GaussianNB\n\n#Luodaan malli-olio\nmalli_3 = GaussianNB()\n\n#Ennustemallille annetaan parametreina:\n#feature matrix (muutosta selittävät muuttujat) ja\n#target (ennustettava muuttuja, joka muuttuu feature matrixin muutosten vuoksi)\nmalli_3.fit(X_train, y_train)\n###komennolla fit() sovitetaan malli dataan. Se on se komento, jolla konetta opetetaan\n\n#Olion sisältämät toiminnallisuudet saa IDEssä esiin kirjoittamalla 'malli.'\n#ja naputtelemalla tabulaattoria (tai CTRL+SPACE)\nmalli_3.get_params()\n",
"_____no_output_____"
],
[
"#Malli-olion predict-toiminnolla voidaan laskea ennuste opetusdatalle\ny_train_malli_3 = malli_3.predict(X_train)\n#Malli-olion predict-toiminnolla voidaan laskea ennuste testidatalle\ny_test_malli_3 = malli_3.predict(X_test)",
"_____no_output_____"
],
[
"#Opetusdatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_train, y_train_malli_3)",
"_____no_output_____"
],
[
"#Testidatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_test, y_test_malli_3)\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"_____no_output_____"
],
[
"#Opetusdatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_train, y_train_malli_3))",
"[[32 0 0]\n [ 0 40 2]\n [ 0 2 36]]\n"
]
],
[
[
"Yllä olevassa Kurjenmiekat-kasvin taulukossa ovat lajikkeet Setosa, Versicolor ja Virginica. Talukkoa luetaan siten, että luotu ennustemalli on onnistunut ennustamaan\n* ensimmäisellä rivillä (ja ensimmäisessä sarakkeessa) kaikki Setosat oikein\n* keskimmäisellä rivillä (ja keskimmäisessä sarakkeessa) Versicolorista lähes kaikki oikein, mutta osa oli mennyt vahingossa viimeiseen sarakkeeseen Virginicalle\n* viimeisellä rivillä (ja viimeisessä sarakkeessa) Virginicatista lähes kaikki oikein, mutta osa oli mennyt vahingossa keskimmäiseen sarakkeeseen Versicolorille",
"_____no_output_____"
]
],
[
[
"#Testidatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_test, y_test_malli_3))\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"[[18 0 0]\n [ 0 8 0]\n [ 0 1 11]]\n"
]
],
[
[
"Yllä olevassa Kurjenmiekat-kasvin taulukossa ovat lajikkeet Setosa, Versicolor ja Virginica. Talukkoa luetaan siten, että luotu ennustemalli on onnistunut ennustamaan\n* ensimmäisellä rivillä (ja ensimmäisessä sarakkeessa) kaikki Setosat oikein\n* keskimmäisellä rivillä (ja keskimmäisessä sarakkeessa) kaikki Versicolorista oikein\n* viimeisellä rivillä (ja viimeisessä sarakkeessa) Virginicatista lähes kaikki oikein, mutta osa oli mennyt vahingossa keskimmäiseen sarakkeeseen Versicolorille ",
"_____no_output_____"
]
],
[
[
"#Vakiintuneen tavan mukaan feature matrix (selittävä muuttuja) on iso X\n##Malli-olion predict-toiminnolla suoritetaan täysin uuden\n##syötteen luokittelu kategoriaan\nmalli_3.predict(Xnew)",
"_____no_output_____"
]
],
[
[
"Uudessa datasetissä ei yksinkertaisuuden vuoksi ollut kuin kolme havaintoa. Yllä olevasta tulosteesta näkee, että juuri koulutettu koneoppimisen algoritmi jakoi taulukon\n* ensimmäisen rivin osalta Setosaksi\n* toisen rivin osalta Virginicaksi\n* kolmannen rivin osalta Versicoloriksi",
"_____no_output_____"
],
[
"### Ennakoivan analytiikan mallin sovitus datasettiin (4/4)\nKoneoppimisessa kokeillaan erilaisia algoritmeja samaan historiadataan ja katsotaan mikä niistä arvaa parhaiten, kun ennustetta verrataan jo tiedossa oleviin toteumiin\n\n#### 4. algoritmi kokeilu (ensimmäinen sovitusyritys: Miten malli sopii toteumaan) ",
"_____no_output_____"
],
[
"#### Logistinen regressio -menetelmästä\nvoi lukea lisää sijainnissa https://tilastoapu.wordpress.com/2014/04/25/logistinen-regressio/\n<br>\nLogistisella regressiolla saadaan ennustavan luokittelun lisäksi myös hyviä todennäköisyyksiä luokittelun paikkaansapitävyydelle\n<br>\n<br>\nSuora copy+paste\n<br>\n\"Logistinen regressio on luokittelumenetelmä eikä sitä pidä sekoittaa tavalliseen regressioon.\"\n<br>\nhttps://nbviewer.jupyter.org/github/taanila/tilastoapu/blob/master/iris_logr.ipynb",
"_____no_output_____"
]
],
[
[
"#random_state arvo määrittelee miten data erotellaan opetus- ja testidataan\n##eri kokeilukerroilla tulee käyttää samaa arvoa (arvolla itsellään ei ole mitään merkitystä)\n##sillä jos aineisto on jaettu eri tavalla osiin voi päätyä hyvinkin erilaiseen malliin\nX_train, X_test, y_train, y_test = train_test_split(X, y, random_state=4)\n###Käytännössä datasetissä otoksen jakamisen 'random_state' arvolla on\n###jonkilainen vaikutus ennustusprosentin osumatarkkuuteen\n### X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=5)\n#HUOM! Ennen tuotantoon deployausta toistetaan splittausta moneen eri kertaan eri jaotusarvoilla\n#ja tuotantoon pääty niistä kokeiluista jonkilainen keskiarvon tyyppinen ratkaisu\n\n#Katsotaan millaisiin paloihin jako tapahtui\nprint(X_train.shape)\nprint(X_test.shape)\nprint(y_train.shape)\nprint(y_test.shape)",
"(112, 4)\n(38, 4)\n(112,)\n(38,)\n"
],
[
"#Ei asenneta Anacondassa vaan kerrotaan mistä \"from foo.bar\" tuodaan käytettäväksi \"import lorem_ipsus\"\n##toiminnallisuus, joka toteuttaa Logistisen regressiomallin\nfrom sklearn.linear_model import LogisticRegression\n\n#Luodaan malli-olio ja herjan vuoksi lisätään iteraatioita\nmalli_4 = LogisticRegression(max_iter = 10000)\n\n#Ennustemallille annetaan parametreina:\n#feature matrix (muutosta selittävät muuttujat) ja\n#target (ennustettava muuttuja, joka muuttuu feature matrixin muutosten vuoksi)\nmalli_4.fit(X_train, y_train)\n###komennolla fit() sovitetaan malli dataan. Se on se komento, jolla konetta opetetaan\n\n#Olion sisältämät toiminnallisuudet saa IDEssä esiin kirjoittamalla 'malli.'\n#ja naputtelemalla tabulaattoria (tai CTRL+SPACE)\nmalli_4.get_params()",
"_____no_output_____"
],
[
"#Malli-olion predict-toiminnolla voidaan laskea ennuste opetusdatalle\ny_train_malli_4 = malli_4.predict(X_train)\n#Malli-olion predict-toiminnolla voidaan laskea ennuste testidatalle\ny_test_malli_4 = malli_4.predict(X_test)",
"_____no_output_____"
],
[
"#Opetusdatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_train, y_train_malli_4)",
"_____no_output_____"
],
[
"#Testidatalle lasketun ennusteen vertaaminen tiedossa oleville\n#todellisille arvoille (label) ennustuksen osumatarkkuus prosentteina\naccuracy_score(y_test, y_test_malli_4)\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"_____no_output_____"
],
[
"#Opetusdatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_train, y_train_malli_4))",
"[[32 0 0]\n [ 0 41 1]\n [ 0 0 38]]\n"
],
[
"#Testidatan osalta väärin ennustettujen esiin nostaminen \nprint(confusion_matrix(y_test, y_test_malli_4))\n##Algoritmi ei ole nähnyt testidataa koulutusvaiheessa ja testidatalla\n##varmistetaan ettei ole ylimallinnettu koulutusvaiheessa",
"[[18 0 0]\n [ 0 7 1]\n [ 0 0 12]]\n"
]
],
[
[
"Yllä olevassa Kurjenmiekat-kasvin taulukossa ovat lajikkeet Setosa, Versicolor ja Virginica. Talukkoa luetaan siten, että luotu ennustemalli on onnistunut ennustamaan\n* ensimmäisellä rivillä (ja ensimmäisessä sarakkeessa) kaikki Setosat oikein\n* keskimmäisellä rivillä (ja keskimmäisessä sarakkeessa) Versicolorista lähes kaikki oikein, mutta osa oli mennyt vahingossa viimeiseen sarakkeeseen Virginicalle\n* viimeisellä rivillä (ja viimeisessä sarakkeessa) kaikki Virginicatit oikein ",
"_____no_output_____"
]
],
[
[
"#Vakiintuneen tavan mukaan feature matrix (selittävä muuttuja) on iso X\n##Malli-olion predict-toiminnolla suoritetaan täysin uuden\n##syötteen luokittelu kategoriaan\nmalli_4.predict(Xnew)",
"_____no_output_____"
]
],
[
[
"Uudessa datasetissä ei yksinkertaisuuden vuoksi ollut kuin kolme havaintoa. Yllä olevasta tulosteesta näkee, että juuri koulutettu koneoppimisen algoritmi jakoi taulukon\n* ensimmäisen rivin osalta Setosaksi\n* toisen rivin osalta Virginicaksi\n* kolmannen rivin osalta Versicoloriksi",
"_____no_output_____"
]
],
[
[
"#Logistisella regressiolla saadaan ennustavan luokittelun lisäksi\n#myös hyviä todennäköisyyksiä luokittelun paikkaansapitävyydelle \nmalli_4.predict_proba(Xnew)",
"_____no_output_____"
]
],
[
[
"Yllä olevaa taulukkoa luetaan siten, että taulukon ensimmäinen havainto on 96,63% todennäköisyydellä Setosa ja 3.37% todennäköisyydellä Versicolor sekä 0,00001% todennäköisyydellä Virginica",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\nprint(f'Lopeteltu {datetime.now()}')",
"Lopeteltu 2021-06-29 21:24:01.603072\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a16e00fc5cb765e40a3d563c995a85c9979d7f3
| 87,328 |
ipynb
|
Jupyter Notebook
|
fig6.ipynb
|
dvav/clonosGP
|
e7f9a08869df0e1857fe24e4e311999f7ba6560f
|
[
"MIT"
] | 9 |
2020-01-28T03:22:36.000Z
|
2022-01-11T09:38:22.000Z
|
fig6.ipynb
|
dvav/clonosGP
|
e7f9a08869df0e1857fe24e4e311999f7ba6560f
|
[
"MIT"
] | 2 |
2021-10-05T12:24:02.000Z
|
2022-01-09T12:33:23.000Z
|
fig6.ipynb
|
dvav/clonosGP
|
e7f9a08869df0e1857fe24e4e311999f7ba6560f
|
[
"MIT"
] | null | null | null | 516.733728 | 81,607 | 0.934282 |
[
[
[
"import pandas as pnd",
"_____no_output_____"
],
[
"%load_ext rpy2.ipython\n%R library(tidyverse)\n%R library(patchwork)",
"R[write to console]: ── \u001b[1mAttaching packages\u001b[22m ─────────────────────────────────────── tidyverse 1.3.0 ──\n\nR[write to console]: \u001b[32m✔\u001b[39m \u001b[34mggplot2\u001b[39m 3.3.0 \u001b[32m✔\u001b[39m \u001b[34mpurrr \u001b[39m 0.3.3\n\u001b[32m✔\u001b[39m \u001b[34mtibble \u001b[39m 3.0.0 \u001b[32m✔\u001b[39m \u001b[34mdplyr \u001b[39m 0.8.5\n\u001b[32m✔\u001b[39m \u001b[34mtidyr \u001b[39m 1.0.2 \u001b[32m✔\u001b[39m \u001b[34mstringr\u001b[39m 1.4.0\n\u001b[32m✔\u001b[39m \u001b[34mreadr \u001b[39m 1.3.1 \u001b[32m✔\u001b[39m \u001b[34mforcats\u001b[39m 0.5.0\n\nR[write to console]: ── \u001b[1mConflicts\u001b[22m ────────────────────────────────────────── tidyverse_conflicts() ──\n\u001b[31m✖\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mfilter()\u001b[39m masks \u001b[34mstats\u001b[39m::filter()\n\u001b[31m✖\u001b[39m \u001b[34mdplyr\u001b[39m::\u001b[32mlag()\u001b[39m masks \u001b[34mstats\u001b[39m::lag()\n\n"
],
[
"metrics = pnd.read_csv('results/simdata_metrics_Mat32_empirical.csv')",
"_____no_output_____"
],
[
"%%R -i metrics -w 7 -h 7 --units in\n\nauxfcn = function(metrics) {\n metrics %>%\n filter((LIK == 'Bin')) %>%\n mutate(PRIOR = if_else(PRIOR == 'Flat', PRIOR, str_c(PRIOR, COV, sep='-'))) %>%\n pivot_longer(c('ARI', 'AMI', 'FMI', 'RMSE'), names_to = 'METRIC', values_to = 'VALUE') %>%\n filter(METRIC %in% c('ARI')) %>% \n group_by(NCLUSTERS, NSAMPLES, NMUTS, PRIOR, METRIC) %>%\n summarise(LOC = mean(VALUE), SD = sd(VALUE), SE = SD / sqrt(n()), LO = max(0, LOC - 1.96*SE), HI = min(1, LOC + 1.96*SE)) %>%\n ungroup() %>%\n ggplot() +\n geom_line(aes(x = NCLUSTERS, y = LOC, color=PRIOR), position = position_dodge(width=1), show.legend = F, linetype = 'dashed') + \n geom_linerange(aes(x = NCLUSTERS, ymin = LO, ymax = HI, color = PRIOR), position = position_dodge(width=1), show.legend = F) + \n geom_point(aes(x = NCLUSTERS, y = LOC, fill = PRIOR, shape = PRIOR), size=2, position = position_dodge(width=1)) + \n facet_grid(NMUTS ~ NSAMPLES, labeller = labeller(NSAMPLES = function(val) str_c('# of samples = ', val),\n NMUTS = function(val) str_c('# of mutations = ', val))) +\n theme_bw() +\n theme(legend.position = 'bottom') +\n scale_x_continuous(breaks = c(2, 4, 8)) +\n scale_fill_brewer(palette = 'Set2') +\n scale_color_brewer(palette = 'Set2') + \n scale_shape_manual(values = c(23, 21, 22, 24, 25)) +\n labs(x = '# of clusters', y = 'Adjusted Rand Index', fill = 'model', shape = 'model') \n}\n\nauxfcn(metrics)\n\n# ggsave('tmp.pdf')",
"_____no_output_____"
],
[
"!git status",
"On branch master\r\nYour branch is up-to-date with 'origin/master'.\r\n\r\nChanges not staged for commit:\r\n (use \"git add <file>...\" to update what will be committed)\r\n (use \"git checkout -- <file>...\" to discard changes in working directory)\r\n\r\n\t\u001b[31mmodified: fig6.ipynb\u001b[m\r\n\r\nUntracked files:\r\n (use \"git add <file>...\" to include in what will be committed)\r\n\r\n\t\u001b[31mresults/simdata_metrics_Mat32_empirical.csv\u001b[m\r\n\r\nno changes added to commit (use \"git add\" and/or \"git commit -a\")\r\n"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
4a16e0800ea3d96cd49e3369cf6cfa6d067a11a1
| 4,478 |
ipynb
|
Jupyter Notebook
|
notebooks/experimental/torch_integration/Overriding PyTorch Autograd Example.ipynb
|
yanndupis/Grid
|
ce202add2a066eb6a1421d0391646b50e2d7f306
|
[
"Apache-2.0"
] | 1 |
2018-03-09T20:49:48.000Z
|
2018-03-09T20:49:48.000Z
|
notebooks/experimental/torch_integration/Overriding PyTorch Autograd Example.ipynb
|
yanndupis/Grid
|
ce202add2a066eb6a1421d0391646b50e2d7f306
|
[
"Apache-2.0"
] | 1 |
2018-03-28T09:08:28.000Z
|
2018-03-28T09:08:28.000Z
|
notebooks/experimental/torch_integration/Overriding PyTorch Autograd Example.ipynb
|
yanndupis/Grid
|
ce202add2a066eb6a1421d0391646b50e2d7f306
|
[
"Apache-2.0"
] | null | null | null | 22.39 | 85 | 0.489728 |
[
[
[
"import torch\nfrom torch.autograd import Variable, Function\nimport inspect\nimport random\nimport copy\n\n# import functools\n# TODO: Use functools.wrap to get original function/method dir attributes\n\nclass EncryptedAdd(Function):\n \n @staticmethod\n def forward(ctx, a, b):\n return a+b\n # compute a + b on encrypted data - they are regular PyTorch tensors\n \n @staticmethod\n def backward(ctx, grad_out):\n grad_out = VariableProxy(grad_out.data)\n return grad_out.var,grad_out.var\n # not grad_out operators are overloaded\n \nclass EncryptedMult(Function):\n \n @staticmethod\n def forward(ctx, a, b):\n ctx.save_for_backward(a,b) \n return a*b\n # compute a * b on encrypted data - they are regular PyTorch tensors\n \n @staticmethod\n def backward(ctx, grad_out):\n a,b = ctx.saved_tensors\n grad_out = grad_out\n return Variable(grad_out.data*b),Variable(grad_out.data*a)\n # not grad_out operators are overloaded\n\nclass VariableProxy(object):\n \n def __init__(self, var, requires_grad=True):\n self.var = Variable(var,requires_grad=requires_grad)\n\n def __add__(self, other):\n return (EncryptedAdd.apply(self.var, other.var))\n \n def __mul__(self,other):\n return (EncryptedMult.apply(self.var, other.var))\n \n def grad(self):\n return self.var.grad\n \nx = VariableProxy(torch.FloatTensor([1,1,1]),requires_grad=True)\ny = VariableProxy(torch.FloatTensor([2,3,4]),requires_grad=True)\n\nz = x * y\n\nz.backward(torch.FloatTensor([1]))\n\nx.grad()",
"_____no_output_____"
],
[
"z",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code"
]
] |
4a16e3ab02109395aceb448add5cbdcd247707b4
| 496,833 |
ipynb
|
Jupyter Notebook
|
Assignment_5.ipynb
|
Deepthi-01997264/cs480student
|
8cadc14bd184af905e4cd8e71774991cb8940e4c
|
[
"MIT"
] | null | null | null |
Assignment_5.ipynb
|
Deepthi-01997264/cs480student
|
8cadc14bd184af905e4cd8e71774991cb8940e4c
|
[
"MIT"
] | null | null | null |
Assignment_5.ipynb
|
Deepthi-01997264/cs480student
|
8cadc14bd184af905e4cd8e71774991cb8940e4c
|
[
"MIT"
] | null | null | null | 486.614104 | 244,753 | 0.928229 |
[
[
[
"<a href=\"https://colab.research.google.com/github/Deepthi-01997264/cs480student/blob/main/Assignment_5.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"\n#Assignment 5",
"_____no_output_____"
]
],
[
[
"# In this assignment, we will visualize and explore a CT scan!",
"_____no_output_____"
],
[
"# load numpy and matplotlib\n%pylab inline",
"Populating the interactive namespace from numpy and matplotlib\n"
],
[
"# we are using pydicom, so lets install it!\n!pip install pydicom",
"Collecting pydicom\n Downloading pydicom-2.3.0-py3-none-any.whl (2.0 MB)\n\u001b[K |████████████████████████████████| 2.0 MB 4.3 MB/s \n\u001b[?25hInstalling collected packages: pydicom\nSuccessfully installed pydicom-2.3.0\n"
]
],
[
[
"**Task 1**: Download and visualize data with SliceDrop! [20 Points]",
"_____no_output_____"
]
],
[
[
"# Please download https://cs480.org/data/ct.zip and extract it on your computer!\n# This is a CT scan of an arm in DICOM format.\n!wget https://cs480.org/data/ct.zip",
"--2022-04-12 02:57:31-- https://cs480.org/data/ct.zip\nResolving cs480.org (cs480.org)... 185.199.108.153, 185.199.109.153, 185.199.110.153, ...\nConnecting to cs480.org (cs480.org)|185.199.108.153|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 5847706 (5.6M) [application/zip]\nSaving to: ‘ct.zip’\n\nct.zip 100%[===================>] 5.58M --.-KB/s in 0.1s \n\n2022-04-12 02:57:32 (57.2 MB/s) - ‘ct.zip’ saved [5847706/5847706]\n\n"
],
[
"# 1) Let's explore the data without loading it.\n# TODO: Without loading the data, how many slices are there?",
"_____no_output_____"
],
[
"\"\"\"\nAnswer:\nDicom stores one slice as a single file, hence there are 220 slices as\nthere are 220 files present in the ct folder.\n\"\"\"",
"_____no_output_____"
],
[
"# 2) Let's visualize the data with SliceDrop! \n# Go to https://slicedrop.com and drag'n'drop all .dcm files into the browser.\n# Please use the 2D sliders to show axial, sagittal, and coronal slices in 3D.",
"_____no_output_____"
],
[
"# TODO Please post a screenshot of SliceDrop's 3D View in the text box below by \n# using the Upload image button after double-click.",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"**Task 2**: Load the data using pydicom as a 3D volume and then reslice it! [35 Points]",
"_____no_output_____"
]
],
[
[
"# TODO: Please upload ct.zip using the file panel on the left.\n# Then use the following snippet to extract the data.",
"_____no_output_____"
],
[
"import zipfile\nwith zipfile.ZipFile('ct.zip', 'r') as zip_ref:\n zip_ref.extractall('.')",
"_____no_output_____"
],
[
"# 1) Now loop through all the DICOM files and store them in a 3D numpy array.\n# Hint: You can either store them in a list first or read the dimensions of a\n# single image slice to properly create the 3D numpy array.\n# Hint 2: os.listdir(DIR) gives a list of filenames in a directory.\n# Hint 2b: This list is not sorted - make sure you sort it.\n# Hint 3: The dcmread function loads a single DICOM file.\n# Hint 4: You can then use .pixel_array to access the image data.",
"_____no_output_____"
],
[
"from pydicom import dcmread\nimport os\nimport numpy as np",
"_____no_output_____"
],
[
"# TODO: YOUR CODE FOR LOADING THE VOLUME AS A 3D NUMPY ARRAY\nfile_names = sorted(os.listdir(\"ct\"))\nfiles=[]\nfor fname in file_names:\n files.append(dcmread(\"ct/\"+fname))\n\n# skip files with no SliceLocation (eg scout views)\nslices = []\nskipcount = 0\nfor f in files:\n if hasattr(f, 'SliceLocation'):\n slices.append(f)\n else:\n skipcount = skipcount + 1\n# ensure they are in the correct order\nslices = sorted(slices, key=lambda s: s.SliceLocation)\n\n# pixel aspects, assuming all slices are the same\nps = slices[0].PixelSpacing\nss = slices[0].SliceThickness\nax_aspect = ps[1]/ps[0]\nsag_aspect = ps[1]/ss\ncor_aspect = ss/ps[0]\n\n# create 3D array\nimg_shape = list(slices[0].pixel_array.shape)\nimg_shape.append(len(slices))\nimg3d = np.zeros(img_shape)\n\n# creating a 3d volume from the slices\nfor i, s in enumerate(slices):\n img2d = s.pixel_array\n img3d[:, :, i] = img2d\n\n#img3d is the 3D volume made out of reading all slices\nprint(img3d.shape)",
"(454, 512, 220)\n"
],
[
"# 2) Now create and show axial, sagittal, and coronal slices from the 3D volume.\n# Hint: Please use imshow(XX, cmap='gray') to show the image.",
"_____no_output_____"
],
[
"# TODO: YOUR CODE FOR AXIAL\nplt.imshow(img3d[:, :, img_shape[2]//2], cmap='gray')",
"_____no_output_____"
],
[
"# TODO: YOUR CODE FOR SAGITTAL\nplt.imshow(img3d[:,img_shape[1]//2 , :], cmap='gray')",
"_____no_output_____"
],
[
"# TODO: YOUR CODE FOR CORONAL\n# plt.imshow(img3d[img_shape[0]//2, :, :].T, cmap='gray')\nplt.imshow(img3d[118, :, :].T, cmap='gray')",
"_____no_output_____"
]
],
[
[
"**Task 3**: Use the Window/Level-technique to visualize the data! [45 Points]",
"_____no_output_____"
]
],
[
[
"# We will now enhance the visualization from above by performing \n# Window/Level adjustment.\n# Here is one way of doing that:\n# vmin = level - window/2\n# vmax = level + window/2\n# plt.imshow(hu_pixels + rescale, cmap='gray', vmin=vmin, vmax=vmax)\n# plt.show()",
"_____no_output_____"
],
[
"# 1) Please load the Window/Level values from the DICOM file,\n# print these values, and then visualize one slice with window/level adjustment.\n# Hint: The DICOM header has the following tags.\n# (0028, 1050) Window Center \n# (0028, 1051) Window Width\n# Hint 2: You can use slice[key].value to access DICOM tag values.\n# Hint 3: (0028, 1052) Rescale Intercept might be important.",
"_____no_output_____"
],
[
"# TODO: YOUR CODE\nimg = img3d[:, :, 0]\nslice = slices[0]\nprint(slice[0x0028,0x1050].keyword, \"\\t\\t: \", slice[0x0028,0x1050].value)\nprint(slice[0x0028,0x1051].keyword, \"\\t\\t: \", slice[0x0028,0x1051].value)\nprint(slice[0x0028,0x1052].keyword, \"\\t: \", slice[0x0028,0x1052].value)",
"WindowCenter \t\t: 30\nWindowWidth \t\t: 410\nRescaleIntercept \t: -1024\n"
],
[
"# 2) Play around with different Window/Level values that enhance\n# the visualization.",
"_____no_output_____"
],
[
"# TODO: YOUR CODE\ndef convert_to_hu(slice, px):\n # convert slice into hounsfield units\n intercept = slice.RescaleIntercept\n slope = slice.RescaleSlope\n hu_image = px * slope + intercept\n return hu_image\n\ndef apply_window(img, window_center, window_width):\n # based on the center and width this function\n # will apply a window\n img_min = window_center - window_width // 2\n img_max = window_center + window_width // 2\n windowed = img.copy()\n windowed[windowed < img_min] = img_min\n windowed[windowed > img_max] = img_max\n return windowed\n\n# process the whole 3d volume\nct_windowed = np.zeros(img3d.shape)\nfor i in range(220):\n hu = convert_to_hu(slices[i], img3d[:,:,i])\n windowed = apply_window(hu, 400, 1000)\n ct_windowed[:, :, i] = windowed\n\n# displaying a random slice\nplt.subplot(\"131\")\nplt.title(\"original\")\nplt.imshow(img3d[118, :, :], cmap='gray')\nplt.subplot(\"132\")\nplt.title(\"After applying a window\")\nplt.imshow(ct_windowed[118,:,:], cmap='gray')\n",
"_____no_output_____"
],
[
"# Which values make sense and why?",
"_____no_output_____"
],
[
"# TODO: YOUR ANSWER\n\"\"\"\nTissue density is quantified as Hounsfield Units(HU).\nScale varies from -1000 for air to +1000 bone with 0 corresponding to water.\nThe general variation of order would be Air<Fat<Fluid<Softtissue<Bone<Metal.\nHence for bone, window center can be at HU=400 with a width of 1000, whereas\nfor soft tissue it would be 50 and 250 respectively.\n\"\"\"",
"_____no_output_____"
]
],
[
[
"**Bonus**: Create segmentations (label maps) for the volume using thresholding HU! [33 Points]",
"_____no_output_____"
]
],
[
[
"# Similar to Window/Level adjustment for visualization, we can threshold\n# the volume to highlight the following components using the Hounsfield Units:\n# 1) Fat\n# 2) Soft Tissue\n# 3) Bones\n#\n# Please create 3 segmentation masks for these structures.\n# Then, please visualize each 3 slices per structure to showcase the segmentation.\n# Hint: As a reminder, the following code allows thresholding of a numpy array.\n# new_mask = imagevolume.copy()\n# new_mask[new_mask < XXX] = 0\n# Hint2: You might need to cast new_mask to int16 not uint16.",
"_____no_output_____"
],
[
"# TODO: YOUR CODE TO SEGMENT FAT\ndef segment(img, window_center, window_width):\n # calculate threshold values\n img_min = window_center - window_width // 2\n img_max = window_center + window_width // 2\n mask = img.copy()\n # creating mask\n mask[mask < img_min] = 0\n mask[mask > img_max] = 0\n mask[mask != 0] = 1\n mask = mask.astype(int)\n segmented_image = mask * img\n return segmented_image, mask\n\nct_seg = np.zeros(img3d.shape)\nct_mask = np.zeros(img3d.shape)\n#fat\nfor i in range(220):\n hu = convert_to_hu(slices[i], img3d[:,:,i])\n seg, mask = segment(hu, -50, 40)\n ct_seg[:, :, i] = seg\n ct_mask[:, :, i] = mask\n\nplt.imshow(img3d[118,:,:], cmap='gray')\nplt.title('Original')\nplt.show()\n\nplt.imshow(ct_seg[118,:,:], cmap='gray')\nplt.title('Fat')\nplt.show()",
"_____no_output_____"
],
[
"# TODO: YOUR CODE TO SEGMENT SOFT TISSUE\nfor i in range(220):\n hu = convert_to_hu(slices[i], img3d[:,:,i])\n seg, mask = segment(hu, 50, 100)\n ct_seg[:, :, i] = seg\n ct_mask[:, :, i] = mask\n\nplt.imshow(img3d[118,:,:], cmap='gray')\nplt.title('Original/Windowed image')\nplt.show()\n\nplt.imshow(ct_seg[118,:,:], cmap='gray')\nplt.title('Soft Tissue')\nplt.show()",
"_____no_output_____"
],
[
"# TODO: YOUR CODE TO SEGMENT BONES\nfor i in range(220):\n hu = convert_to_hu(slices[i], img3d[:,:,i])\n seg, mask = segment(hu, 400, 700)\n ct_seg[:, :, i] = seg\n ct_mask[:, :, i] = mask\n\nplt.imshow(img3d[118,:,:], cmap='gray')\nplt.title('Original/Windowed image')\nplt.show()\n\nplt.imshow(ct_seg[118,:,:], cmap='gray')\nplt.title('Bone')\nplt.show()",
"_____no_output_____"
],
[
"# Are the segmentations good?",
"_____no_output_____"
],
[
"# TODO: YOUR ANSWER\n\"\"\"\nCurrently segmentation is fairly good but not yet perfect enough. Since we are\nhardcoding threshold HU values, segmentation quality depends on the values we \nchoose by trial and error.\n\"\"\"",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"#\n# Thank you and Great job!!\n#\n# _.---._\n# .' `.\n# :) (:\n# \\ (@) (@) /\n# \\ A /\n# ) (\n# \\\"\"\"\"\"/\n# `._.'\n# .=.\n# .---._.-.=.-._.---.\n# / ':-(_.-: :-._)-:` \\\n# / /' (__.-: :-.__) `\\ \\\n# / / (___.-` '-.___) \\ \\\n# / / (___.-'^`-.___) \\ \\\n# / / (___.-'=`-.___) \\ \\\n# / / (____.'=`.____) \\ \\\n# / / (___.'=`.___) \\ \\\n# (_.; `---'.=.`---' ;._)\n# ;|| __ _.=._ __ ||;\n# ;|| ( `.-.=.-.' ) ||;\n# ;|| \\ `.=.' / ||;\n# ;|| \\ .=. / ||;\n# ;|| .-`.`-._.-'.'-. ||;\n# .:::\\ ( ,): O O :(, ) /:::.\n# |||| ` / /'`--'--'`\\ \\ ' ||||\n# '''' / / \\ \\ ''''\n# / / \\ \\\n# / / \\ \\\n# / / \\ \\\n# / / \\ \\\n# / / \\ \\\n# /.' `.\\\n# (_)' `(_)\n# \\\\. .//\n# \\\\. .//\n# \\\\. .//\n# \\\\. .//\n# \\\\. .//\n# \\\\. .//\n# jgs \\\\. .//\n# ///) (\\\\\\\n# ,///' `\\\\\\,\n# ///' `\\\\\\\n# \"\"' '\"\"\n\n\n#Worked together with Anudeep Reddy Veerla\n",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a16ec6d0b84a747a0606190aa82ee5920e14e72
| 357,808 |
ipynb
|
Jupyter Notebook
|
docs/examples/Breast_Cancer.ipynb
|
artemmavrin/logitboost
|
a008fa6068e32bd0aa5980f47bfd2c2a646e9f3e
|
[
"MIT"
] | 10 |
2019-03-15T01:11:22.000Z
|
2021-12-23T07:37:22.000Z
|
docs/examples/Breast_Cancer.ipynb
|
artemmavrin/logitboost
|
a008fa6068e32bd0aa5980f47bfd2c2a646e9f3e
|
[
"MIT"
] | 2 |
2020-02-13T03:49:59.000Z
|
2021-10-31T20:00:33.000Z
|
docs/examples/Breast_Cancer.ipynb
|
artemmavrin/logitboost
|
a008fa6068e32bd0aa5980f47bfd2c2a646e9f3e
|
[
"MIT"
] | 8 |
2018-08-22T19:25:54.000Z
|
2021-02-17T16:13:27.000Z
| 892.289277 | 122,864 | 0.953715 |
[
[
[
"# Breast Cancer Diagnosis\n\nIn this notebook we will apply the LogitBoost algorithm to a toy dataset to classify cases of breast cancer as benign or malignant.",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nsns.set(style='darkgrid', palette='colorblind', color_codes=True)\n\nfrom sklearn.datasets import load_breast_cancer\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.metrics import accuracy_score, classification_report\nfrom sklearn.manifold import TSNE\n\nfrom logitboost import LogitBoost",
"_____no_output_____"
]
],
[
[
"## Loading the Data\n\nThe breast cancer dataset imported from [scikit-learn](http://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_breast_cancer.html) contains 569 samples with 30 real, positive features (including cancer mass attributes like mean radius, mean texture, mean perimeter, et cetera).\nOf the samples, 212 are labeled \"malignant\" and 357 are labeled \"benign\".\nWe load this data into a 569-by-30 feature matrix and a 569-dimensional target vector.\nThen we randomly shuffle the data and designate two thirds for training and one third for testing.",
"_____no_output_____"
]
],
[
[
"data = load_breast_cancer()\nX = data.data\ny = data.target_names[data.target]\nn_classes = data.target.size\n\n# Shuffle data and split it into training/testing samples\ntest_size = 1 / 3\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=test_size,\n shuffle=True, stratify=y,\n random_state=0)",
"_____no_output_____"
]
],
[
[
"## Visualizing the Training Set\n\nAlthough the features are 30-dimensional, we can visualize the training set by using [t-distributed stochastic neighbor embedding](https://en.wikipedia.org/wiki/T-distributed_stochastic_neighbor_embedding) (t-SNE) to project the features onto a 2-dimensional space.",
"_____no_output_____"
]
],
[
[
"tsne = TSNE(n_components=2, random_state=0)\nX_train_tsne = tsne.fit_transform(X_train)\n\nplt.figure(figsize=(10, 8))\nmask_benign = (y_train == 'benign')\nmask_malignant = (y_train == 'malignant')\n\nplt.scatter(X_train_tsne[mask_benign, 0], X_train_tsne[mask_benign, 1],\n marker='s', c='g', label='benign', edgecolor='k', alpha=0.7)\nplt.scatter(X_train_tsne[mask_malignant, 0], X_train_tsne[mask_malignant, 1],\n marker='o', c='r', label='malignant', edgecolor='k', alpha=0.7)\n\n\nplt.title('t-SNE plot of the training data')\nplt.xlabel('1st embedding axis')\nplt.ylabel('2nd embedding axis')\nplt.legend(loc='best', frameon=True, shadow=True)\n\nplt.tight_layout()\nplt.show()\nplt.close()",
"_____no_output_____"
]
],
[
[
"## Fitting the LogitBoost Model\n\nNext, we initialize a LogitBoost classifier and fit it to the training data.\nBy default, LogitBoost uses decision stumps (decision trees with depth 1, i.e., a single split) as its base estimator.",
"_____no_output_____"
]
],
[
[
"lboost = LogitBoost(n_estimators=200, random_state=0)\nlboost.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"## Prediction Accuracy\n\nAs a first indicator of how well the model predicts the correct labels, we can check its accuracy score (number of correct predictions over the number of total predictions) on the training and test data.\nIf the classifier is good, then the accuracy score should be close to 1.",
"_____no_output_____"
]
],
[
[
"y_pred_train = lboost.predict(X_train)\ny_pred_test = lboost.predict(X_test)\n\naccuracy_train = accuracy_score(y_train, y_pred_train)\naccuracy_test = accuracy_score(y_test, y_pred_test)\n\nprint('Training accuracy: %.4f' % accuracy_train)\nprint('Test accuracy: %.4f' % accuracy_test)",
"Training accuracy: 0.9947\nTest accuracy: 0.9737\n"
]
],
[
[
"## Precision and Recall\n\nWe can also report our LogitBoost model's [precision and recall](https://en.wikipedia.org/wiki/Precision_and_recall).",
"_____no_output_____"
]
],
[
[
"report_train = classification_report(y_train, y_pred_train)\nreport_test = classification_report(y_test, y_pred_test)\nprint('Training\\n%s' % report_train)\nprint('Testing\\n%s' % report_test)",
"Training\n precision recall f1-score support\n\n benign 0.99 1.00 1.00 238\n malignant 1.00 0.99 0.99 141\n\n accuracy 0.99 379\n macro avg 1.00 0.99 0.99 379\nweighted avg 0.99 0.99 0.99 379\n\nTesting\n precision recall f1-score support\n\n benign 0.97 0.98 0.98 119\n malignant 0.97 0.96 0.96 71\n\n accuracy 0.97 190\n macro avg 0.97 0.97 0.97 190\nweighted avg 0.97 0.97 0.97 190\n\n"
]
],
[
[
"## Visualizing Accuracy During Boosting",
"_____no_output_____"
]
],
[
[
"iterations = np.arange(1, lboost.n_estimators + 1)\nstaged_accuracy_train = list(lboost.staged_score(X_train, y_train))\nstaged_accuracy_test = list(lboost.staged_score(X_test, y_test))\n\nplt.figure(figsize=(10, 8))\nplt.plot(iterations, staged_accuracy_train, label='Training', marker='.')\nplt.plot(iterations, staged_accuracy_test, label='Test', marker='.')\n\nplt.xlabel('Iteration')\nplt.ylabel('Accuracy')\nplt.title('Ensemble accuracy during each boosting iteration')\nplt.legend(loc='best', shadow=True, frameon=True)\n\nplt.tight_layout()\nplt.show()\nplt.close()",
"_____no_output_____"
]
],
[
[
"## Contribution of Each Estimator in the Ensemble\n\nLike other ensemble models, the LogitBoost model can suffer from *over-specialization*: estimators added to the ensemble in later boosting iterations make relatively small or even negligible contributions toward improving the overall predictions on the training set.\nThis can be quantified by computing the mean of the absolute prediction of each estimator in the ensemble taken over the training set.",
"_____no_output_____"
]
],
[
[
"contrib_train = lboost.contributions(X_train)\n\nplt.figure(figsize=(10, 8))\nplt.plot(iterations, contrib_train, lw=2)\nplt.xlabel('Estimator Number')\nplt.ylabel('Average Absolute Contribution')\nplt.title('Average absolute contribution of the estimators in the ensemble')\nplt.show()\nplt.close()",
"_____no_output_____"
]
],
[
[
"## Appendix: System Information\n\nThis is included for replicability.",
"_____no_output_____"
]
],
[
[
"# sys_info.py is a file in the same directory as these example notebooks:\n# doc/source/examples\nimport sys_info",
"\nMachine\n=======\n Platform: Darwin-18.7.0-x86_64-i386-64bit\n Machine Type: x86_64\n Processor: i386\n\nPython\n======\n Version: 3.7.2 (v3.7.2:9a3ffc0492, Dec 24 2018, 02:44:43) \n [Clang 6.0 (clang-600.0.57)]\nImplementation: CPython\n\nPackages\n========\n numpy: 1.17.2\n scipy: 1.3.1\n matplotlib: 3.1.1\n seaborn: 0.9.0\n sklearn: 0.21.3\n"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
] |
[
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
] |
4a16ef823575bda570d18c84f39b198665cf2a87
| 238,645 |
ipynb
|
Jupyter Notebook
|
ab.ipynb
|
garrrychan/ab__kd
|
7e93251c96c5733b5dd605561fcc8f827dc5778a
|
[
"MIT"
] | 1 |
2019-06-12T21:50:43.000Z
|
2019-06-12T21:50:43.000Z
|
ab.ipynb
|
garrrychan/ab__kd
|
7e93251c96c5733b5dd605561fcc8f827dc5778a
|
[
"MIT"
] | null | null | null |
ab.ipynb
|
garrrychan/ab__kd
|
7e93251c96c5733b5dd605561fcc8f827dc5778a
|
[
"MIT"
] | null | null | null | 269.655367 | 101,238 | 0.894953 |
[
[
[
"Title: Are the Warriors better without Kevin Durant? \nDate: 2019-06-10 12:00\nTags: python\nSlug: ab_kd",
"_____no_output_____"
],
[
"In the media, there have been debates about whether or not the Golden State Warriors (GSW) are better without Kevin Durant (KD). From the eye-test, it's laughable to even suggest this, as he's one of the top 3 players in the league (Lebron, KD, Kawhi). Nonetheless, people argue that ball movement is better without him, and therefore make the GSW more lethal.\n\nBut, just because the Warriors won a title without KD, does not mean they don't need him more than ever. At the time of writing, the Toronto Raptors lead 3-1 in the Finals! #WeTheNorth 🦖🍁\n\nUsing Bayesian estimation, we can test this hypothesis, by comparing two treatment groups, games played with KD and without KD.\n\nBayesian statistics are an excellent tool to reach for when sample sizes are small, as we can introduce explicit assumptions into the model, when there aren't thousands of observations. ",
"_____no_output_____"
],
[
"---\n\n# Primer on Bayesian Statistics\n\n<img src=\"images/dist.png\" class=\"img-responsive\">\n\n$$P\\left(model\\;|\\;data\\right) = \\frac{P\\left(data\\;|\\;model\\right)}{P(data)}\\; P\\left(model\\right)$$\n\n---\n\n\n$$ \\text{prior} = P\\left(model\\right) $$\n> The **prior** is our belief in the model given no additional information. In our example, this is the mean win % with KD playing.\n\n\n$$ \\text{likelihood} = P\\left(data\\;|\\;model\\right) $$\n> The **likelihood** is the probability of the data we observed occurring given the model.\n\n$$ \\text{marginal probability of data} = P(data) $$\n> The **marginal probability** of the data is the probability that our data are observed regardless of what model we choose or believe in. \n\n$$ \\text{posterior} = P\\left(model\\;|\\;data\\right) $$\n> The **posterior** is our _updated_ belief in the model given the new data we have observed. Bayesian statistics are all about updating a prior belief we have about the world with new data, so we're transforming our _prior_ belief into this new _posterior_ belief about the world. <br><br> In this example, this is the GSW mean winning % with KD playing, given the game logs from the past three seasons.\n\n\nNote, a Bayesian approach is different from a Frequentist's. Rather than only testing whether two groups are different, we instead pursue an estimate of _how_ different they are, from the posterior distribution.",
"_____no_output_____"
],
[
"## Objective\nTo calculate the distribution of the posterior probability of GSW mean winning % with KD and without KD.\n\nMoreover, we can calculate the _delta_ between both probabilities to determine if the mean is statistically different from zero (i.e. no difference with or without him).",
"_____no_output_____"
],
[
"---\n# Observed Data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport scipy.stats as stats\nimport pymc3 as pm\nfrom IPython.display import HTML\nimport matplotlib.pyplot as plt\n%matplotlib inline\nplt.style.use('fivethirtyeight')\nfrom IPython.core.pylabtools import figsize\nimport matplotlib.pylab as pylab\nparams = {'legend.fontsize': 'x-large',\n 'figure.figsize': (15, 10),\n 'axes.labelsize': 'x-large',\n 'axes.titlesize':'x-large',\n 'xtick.labelsize':'x-large',\n 'ytick.labelsize':'x-large'}\npylab.rcParams.update(params)\n\nimport warnings\nwarnings.simplefilter(action='ignore', category=FutureWarning)",
"_____no_output_____"
]
],
[
[
"As the competition is much higher in the playoffs, let's analyze Playoff vs. Regular Season data separately. We can run one test on the regular season, and one test for the playoffs. \n\nData is from [Basketball Reference](https://www.basketball-reference.com/).",
"_____no_output_____"
],
[
"---\n# Regular Season",
"_____no_output_____"
],
[
"<table class=\"table\">\n <thead class=\"table-responsive table-bordered\">\n <tr>\n <th scope=\"col\">Regular Season</th>\n <th scope=\"col\">With Kevin Durant</th>\n <th scope=\"col\">No Kevin Durant</th>\n <th scope=\"col\">Notes</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <td>2019</td>\n <td>0.69 <br> {'W': 54, 'L': 24} </td>\n <td>0.75 <br> {'W': 3, 'L': 1} </td>\n <td>Record is better when KD is out, but small sample size.</td>\n </tr>\n <tr>\n <td>2018</td>\n <td>0.72 <br> {'W': 49, 'L': 19} </td>\n <td>0.64 <br> {'W': 9, 'L': 5} </td>\n <td>Record is better when KD plays</td>\n </tr>\n <tr>\n <td>2017</td>\n <td>0.82 <br> {'W': 51, 'L': 11} </td>\n <td>0.80 <br> {'W': 16, 'L': 4} </td>\n <td>Record is better when KD plays</td>\n </tr>\n <tr>\n <td>Total (3 seasons)</td>\n <td>0.740 <br> {'W': 154, 'L': 54} </td>\n <td>0.737 <br> {'W': 28, 'L': 10} </td>\n <td>Record is better when KD plays</td>\n </tr>\n </tbody>\n</table>",
"_____no_output_____"
],
[
"Over the last three seasons with the Warriors, KD has missed 38 games regular season games, and played in 208.",
"_____no_output_____"
]
],
[
[
"def occurrences(year, kd=True):\n '''occurences(2019, kd=True)\n By default, kd=True means with KD healthy'''\n # clean data\n # regular season\n data = pd.read_csv(f'./data/{year}.txt', sep=',')\n new_columns = ['Rk', 'G', 'Date', 'Age', 'Tm', 'Away', 'Opp', 'Result', 'GS',\n 'MP', 'FG', 'FGA', 'FG%', '3P', '3PA', '3P%', 'FT', 'FTA', 'FT%', 'ORB',\n 'DRB', 'TRB', 'AST', 'STL', 'BLK', 'TOV', 'PF', 'PTS', 'GmSc', '+/-']\n data.columns=new_columns\n # replace did not dress with inactive\n data.GS = np.where(data.GS == 'Did Not Dress','Inactive',data.GS)\n if kd == False:\n game_logs = list(data[data.GS=='Inactive'].Result)\n else:\n game_logs = list(data[data.GS!='Inactive'].Result)\n results = [game.split(' ')[0] for game in game_logs]\n occurrences = [1 if result == 'W' else 0 for result in results]\n return occurrences",
"_____no_output_____"
],
[
"regular_season_with_kd = occurrences(2019, kd=True)+occurrences(2018, kd=True)+occurrences(2017, kd=True)\nregular_season_no_kd = occurrences(2019, kd=False)+occurrences(2018, kd=False)+occurrences(2017, kd=False)\nprint(f'Observed win % when Kevin Durant plays: {round(np.mean(regular_season_with_kd),4)}')\nprint(f'Observed win % when Kevin Durant does not play: {round(np.mean(regular_season_no_kd),4)}')",
"Observed win % when Kevin Durant plays: 0.7404\nObserved win % when Kevin Durant does not play: 0.7368\n"
]
],
[
[
"* Note, we do not know the true win %, only the observed win %. We infer the true quantity from the observed data.\n\n* Notice the unequal sample sizes (208 vs. 38), but this is not problem in Bayesian analysis. We will see the uncertainty of the smaller sample size captured in the posterior distribution. ",
"_____no_output_____"
],
[
"---\n## Bayesian Tests with MCMC\n\n* Markov Chain Monte Carlo (MCMC) is a method to find the posterior distribution of our parameter of interest.\n> This type of algorithm generates Monte Carlo simulations in a way that relies on the Markov property, then accepts these simulations at a certain rate to get the posterior distribution.\n\n* We will use [PyMC3](https://docs.pymc.io/), a probabilistic library for Python to generate MC simulations.\n\n* Before seeing any of the data, my prior is that GSW will win between 50% - 90% of their games, because they are an above average basketball team, and no team has ever won more than 72 games.",
"_____no_output_____"
]
],
[
[
"# Instantiate\nobservations_A = regular_season_with_kd\nobservations_B = regular_season_no_kd\n\nwith pm.Model() as model:\n # Assume Uniform priors for p_A and p_B \n p_A = pm.Uniform(\"p_A\", 0.5, .9)\n p_B = pm.Uniform(\"p_B\", 0.5, .9)\n \n # Define the deterministic delta function. This is our unknown of interest.\n # Delta is deterministic, no uncertainty beyond p_A and p_B\n delta = pm.Deterministic(\"delta\", p_A - p_B)\n \n # We have two observation datasets: A, B\n # Posterior distribution is Bernoulli\n obs_A = pm.Bernoulli(\"obs_A\", p_A, observed=observations_A)\n obs_B = pm.Bernoulli(\"obs_B\", p_B, observed=observations_B)\n \n # Draw samples from the posterior distribution\n trace = pm.sample(20000)\n burned_trace=trace[1000:]",
"Auto-assigning NUTS sampler...\nInitializing NUTS using jitter+adapt_diag...\nMultiprocess sampling (2 chains in 2 jobs)\nNUTS: [p_B, p_A]\nSampling 2 chains: 100%|██████████| 41000/41000 [00:19<00:00, 2145.65draws/s]\n"
]
],
[
[
"* Using PyMC3, we generated a trace, or chain of values from the posterior distribution \n* Generated 20,000 samples from the posterior distribution (20,000 samples / chain / core)\n\nBecause this algorithm needs to converge, we set a number of tuning steps (1,000) to occur first and where the algorithm should \"start exploring.\" It's good to see the Markov Chains overlap, which suggests convergence.",
"_____no_output_____"
]
],
[
[
"pm.traceplot(trace);\n# plt.savefig('trace.svg');",
"_____no_output_____"
],
[
"df = pm.summary(burned_trace).round(2)[['mean', 'sd', 'hpd_2.5', 'hpd_97.5']]\nHTML(df.to_html(classes=\"table table-responsive table-striped table-bordered\"))",
"_____no_output_____"
]
],
[
[
"* Unlike with confidence intervals (frequentist), there is a measure of probability with the credible interval.\n* There is a 95% probability that the true win rate with KD is in the interval (0.68, 0.79).\n* There is a 95% probability that the true win rate with no KD is in the interval (0.59, 0.85).",
"_____no_output_____"
]
],
[
[
"p_A_samples = burned_trace[\"p_A\"]\np_B_samples = burned_trace[\"p_B\"]\ndelta_samples = burned_trace[\"delta\"]\n\nfigsize(15, 10)\nax = plt.subplot(311)\n\nplt.xlim(0, 1)\nplt.hist(p_A_samples, histtype='stepfilled', bins=25, alpha=0.85,\n label=\"posterior of $p_A$\", color=\"#006BB6\", density=True)\nplt.vlines(df.iloc[0][\"mean\"], 0, 12.5, color=\"white\", alpha=0.5,linestyle=\"--\",\n label=f'mean')\nplt.vlines(df.iloc[0][\"hpd_2.5\"], 0, 1.3, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='2.5%')\nplt.vlines(df.iloc[0][\"hpd_97.5\"], 0, 1.3, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='97.5%')\nplt.legend(loc=\"upper right\")\nplt.title(\"Regular Season \\n Posterior distributions of $p_A$, $p_B$, and delta unknowns \\n\\n $p_A$: Mean Win % with KD\")\n\nax = plt.subplot(312)\n\nplt.xlim(0, 1)\nplt.hist(p_B_samples, histtype='stepfilled', bins=25, alpha=0.85,\n label=\"posterior of $p_B$\", color=\"#FDB927\", density=True)\nplt.vlines(df.iloc[1][\"mean\"], 0, 5.5, color=\"white\", alpha=0.5,linestyle=\"--\",\n label=f'mean')\nplt.vlines(df.iloc[1][\"hpd_2.5\"], 0, .8, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='2.5%')\nplt.vlines(df.iloc[1][\"hpd_97.5\"], 0, .8, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='97.5%')\nplt.legend(loc=\"upper right\")\nplt.title(\"$p_B$: Mean Win % No KD\")\n\n\nax = plt.subplot(313)\nplt.xlim(-0.5, 0.5)\nplt.hist(delta_samples, histtype='stepfilled', bins=30, alpha=0.85,\n label=\"posterior of delta\", color=\"#BE3A34\", density=True)\n\nplt.vlines(df.iloc[2][\"mean\"], 0, 5, color=\"white\", alpha=0.5,linestyle=\"--\",\n label=f'mean delta')\nplt.vlines(df.iloc[2][\"hpd_2.5\"], 0, 1, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='2.5%')\nplt.vlines(df.iloc[2][\"hpd_97.5\"], 0, 1, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='97.5%')\n\nplt.legend(loc=\"upper right\");\nplt.title(\"$delta$ = $p_A - p_B$\")\nplt.savefig('reg_season.svg');",
"_____no_output_____"
]
],
[
[
"Note, the 2.5% and 97.5% markers indicate the quantiles for the credible interval, similar to the confidence interval in frequentist statistics.",
"_____no_output_____"
],
[
"---\n## Results\n\n* In the third graph, the posterior win rate is 1.2% higher when KD plays in the regular season.\n\n* Observe that because have less data for when KD is out, our posterior distribution of 𝑝𝐵 is wider, implying we are less certain about the true value of 𝑝𝐵 than we are of 𝑝𝐴. The 95% credible interval is much wider for $p_B$, as there is a smaller sample size, for when KD did not play. We are less certain that the GSW wins 73% of the time without KD.\n\n* The difference in sample sizes ($N_B$ < $N_A$) naturally fits into Bayesian analysis, whereas you need the same populations for frequentist approach!",
"_____no_output_____"
]
],
[
[
"# Count the number of samples less than 0, i.e. the area under the curve\nprint(\"Probability that GSW is worse with Kevin Durant in the regular season: %.2f\" % \\\n np.mean(delta_samples < 0))\n\nprint(\"Probability that GSW is better with Kevin Durant in the regular season: %.2f\" % \\\n np.mean(delta_samples > 0))",
"Probability that GSW is worse with Kevin Durant in the regular season: 0.45\nProbability that GSW is better with Kevin Durant in the regular season: 0.55\n"
]
],
[
[
"The probabilities are pretty close, so we can chalk this up to the Warriors having a experienced supporting cast. \n\nThere is significant overlap between the distribution pf posterior pA and posterior of pB, so one is not better than the other with high probability. The majority of the distribution of delta is around 0, so there is no statistically difference between the groups in the regular season.\n\nIdeally, we should perform more trials when KD is injured (as each data point for scenario B contributes more inferential power than each additional point for scenario A). One could do a similar analysis for when he played on the Oklahoma City Thunder.",
"_____no_output_____"
],
[
"---\n# Playoffs\n## Do superstars shine when the stakes are highest?",
"_____no_output_____"
],
[
"<table class=\"table\">\n <thead class=\"table-responsive table-bordered\">\n <tr>\n <th scope=\"col\">Playoffs</th>\n <th scope=\"col\">With Kevin Durant</th>\n <th scope=\"col\">No Kevin Durant</th>\n <th scope=\"col\">Notes</th>\n </tr>\n </thead>\n <tbody>\n <tr>\n <td>2019</td>\n <td>0.64 <br> {'W': 7, 'L': 4} </td>\n <td>0.66 <br> {'W': 6, 'L': 3} </td>\n <td>Record is marginally better when KD is out, but small sample size. Skewed by Portland series, which GSW won 4-0 with KD injured.</td>\n </tr>\n <tr>\n <td>2018</td>\n <td>0.76 <br> {'W': 16, 'L': 5} </td>\n <td>n/a <br> {'W': 0, 'L': 0} </td>\n <td>KD did not miss any games. Won Championship.</td>\n </tr>\n <tr>\n <td>2017</td>\n <td>0.82 <br> {'W': 14, 'L': 1} </td>\n <td>1 <br> {'W': 2, 'L': 0}. Small sample size. </td>\n <td>Won championship.</td>\n </tr>\n <td>Total (3 seasons)</td>\n <td>0.79 <br> {'W': 37, 'L': 10} </td>\n <td>0.73 <br> {'W': 8, 'L': 3} </td>\n <td>Record is better when KD plays</td>\n </tbody>\n</table>",
"_____no_output_____"
]
],
[
[
"playoffs_with_kd = occurrences('2019_playoffs', kd=True)+occurrences('2018_playoffs', kd=True)+occurrences('2017_playoffs', kd=True)\nplayoffs_no_kd = occurrences('2019_playoffs', kd=False)+occurrences('2018_playoffs', kd=False)+occurrences('2017_playoffs', kd=False)\nprint(f'Observed win % when Kevin Durant plays: {round(np.mean(playoffs_with_kd),2)}')\nprint(f'Observed win % when Kevin Durant does not play: {round(np.mean(playoffs_no_kd),2)}')",
"Observed win % when Kevin Durant plays: 0.79\nObserved win % when Kevin Durant does not play: 0.73\n"
]
],
[
[
"Over the last three playoff runs with the Warriors, KD has missed 11, and played in 47.\n\nSee how the difference is much more pronounced with more data across three seasons. Let's similar if the GSW has a higher win % with KD in the playoffs. ",
"_____no_output_____"
]
],
[
[
"playoff_obs_A = playoffs_with_kd\nplayoff_obs_B = playoffs_no_kd\n\nwith pm.Model() as playoff_model:\n playoff_p_A = pm.Uniform(\"playoff_p_A\", 0, 1)\n playoff_p_B = pm.Uniform(\"playoff_p_B\", 0, 1)\n \n playoff_delta = pm.Deterministic(\"playoff_delta\", playoff_p_A - playoff_p_B)\n \n playoff_obs_A = pm.Bernoulli(\"playoff_obs_A\", playoff_p_A, observed=playoff_obs_A)\n playoff_obs_B = pm.Bernoulli(\"playoff_obs_B\", playoff_p_B, observed=playoff_obs_B)\n\n playoff_trace = pm.sample(20000)\n playoff_burned_trace=playoff_trace[1000:]",
"Auto-assigning NUTS sampler...\nInitializing NUTS using jitter+adapt_diag...\nMultiprocess sampling (2 chains in 2 jobs)\nNUTS: [playoff_p_B, playoff_p_A]\nSampling 2 chains: 100%|██████████| 41000/41000 [00:20<00:00, 2004.28draws/s]\n"
],
[
"df2 = pm.summary(playoff_burned_trace).round(2)[['mean', 'sd', 'hpd_2.5', 'hpd_97.5']]\nHTML(df2.to_html(classes=\"table table-responsive table-striped table-bordered\"))",
"_____no_output_____"
],
[
"playoff_p_A_samples = playoff_burned_trace['playoff_p_A']\nplayoff_p_B_samples = playoff_burned_trace[\"playoff_p_B\"]\nplayoff_delta_samples = playoff_burned_trace[\"playoff_delta\"]\n\nfigsize(15, 10)\n\n#histogram of posteriors\n\nax = plt.subplot(311)\n\nplt.xlim(0, 1)\nplt.hist(playoff_p_A_samples, histtype='stepfilled', bins=25, alpha=0.85,\n label=\"posterior of $p_A$\", color=\"#006BB6\", density=True)\nplt.vlines(df2.iloc[0][\"mean\"], 0, 7.5, color=\"white\", alpha=0.5,linestyle=\"--\",\n label=f'mean delta')\nplt.vlines(df2.iloc[0][\"hpd_2.5\"], 0, 1, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='2.5%') \nplt.vlines(df2.iloc[0][\"hpd_97.5\"], 0, 1, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='97.5%')\nplt.legend(loc=\"upper right\")\nplt.title(\"Playoffs \\n Posterior distributions of $p_A$, $p_B$, and delta unknowns \\n\\n $p_A$: Mean Win % with KD\")\n\nax = plt.subplot(312)\n\nplt.xlim(0, 1)\nplt.hist(playoff_p_B_samples, histtype='stepfilled', bins=25, alpha=0.85,\n label=\"posterior of $p_B$\", color=\"#FDB927\", density=True)\nplt.vlines(df2.iloc[1][\"mean\"], 0, 3, color=\"white\", alpha=0.5,linestyle=\"--\",\n label=f'mean delta')\nplt.vlines(df2.iloc[1][\"hpd_2.5\"], 0, .8, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='2.5%')\nplt.vlines(df2.iloc[1][\"hpd_97.5\"], 0, .8, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='97.5%')\nplt.legend(loc=\"upper right\")\nplt.title(\"$p_B$: Mean Win % No KD\")\n\n\nax = plt.subplot(313)\nplt.xlim(-0.5, 0.5)\nplt.hist(playoff_delta_samples, histtype='stepfilled', bins=30, alpha=0.85,\n label=\"posterior of delta\", color=\"#BE3A34\", density=True)\n\nplt.vlines(df2.iloc[2][\"mean\"], 0, 3, color=\"white\", alpha=0.5,linestyle=\"--\",\n label=f'mean delta')\nplt.vlines(df2.iloc[2][\"hpd_2.5\"], 0, 0.25, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='2.5%')\nplt.vlines(df2.iloc[2][\"hpd_97.5\"], 0, 0.25, color=\"black\", alpha=0.5,linestyle=\"--\",\n label='97.5%')\n\nplt.legend(loc=\"upper right\");\nplt.title(\"$delta$: $p_A - p_B$\")\nplt.savefig('playoffs.svg');",
"_____no_output_____"
],
[
"# Count the number of samples less than 0, i.e. the area under the curve\nprint(\"Probability that GSW is worse with Kevin Durant in the playoffs: %.2f\" % \\\n np.mean(playoff_delta_samples < 0))\n\nprint(\"Probability that GSW is better with Kevin Durant in the playoffs: %.2f\" % \\\n np.mean(playoff_delta_samples > 0))",
"Probability that GSW is worse with Kevin Durant in the playoffs: 0.29\nProbability that GSW is better with Kevin Durant in the playoffs: 0.71\n"
]
],
[
[
"---\n## Are the Warriors better without Kevin Durant? No.\n\nBy combining results from the past three seasons, we obtain a larger test group, which allows us to observe a real change vs. looking at the pure stats for a single year.\n\nWe can see that while delta=0 (i.e. no effect when KD plays) is in the credible interval at 95%, the majority of the distribution is above delta=0, implying the treatment group with KD is likely better than the group without KD. In fact, the probability that GSW is better with Kevin Durant in the playoffs is 71%, a significant improvement than 55% in the regular season! \n\nSuperstars make a significant difference. The regular season is where you make your name, but the postseason is where you make your fame. The delta is 8% higher with KD. That's the advantage you gain with a player of his caliber, as he can hit clutch shots when it matters most.\n\nAs a basketball fan, I hope to see Kevin Durant healthy and back in action soon.",
"_____no_output_____"
],
[
"# References\n* https://multithreaded.stitchfix.com/blog/2015/05/26/significant-sample/\n* https://multithreaded.stitchfix.com/blog/2015/02/12/may-bayes-theorem-be-with-you/",
"_____no_output_____"
]
]
] |
[
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
] |
[
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
] |
4a16f8b6657b86f15332a9b03442b8ed7dd04109
| 21,411 |
ipynb
|
Jupyter Notebook
|
car_and_track_data_playground.ipynb
|
DamonDeng/deepracer_sample
|
8ed6a79b8185e802245eacec0e7553d0d9760f83
|
[
"MIT"
] | null | null | null |
car_and_track_data_playground.ipynb
|
DamonDeng/deepracer_sample
|
8ed6a79b8185e802245eacec0e7553d0d9760f83
|
[
"MIT"
] | null | null | null |
car_and_track_data_playground.ipynb
|
DamonDeng/deepracer_sample
|
8ed6a79b8185e802245eacec0e7553d0d9760f83
|
[
"MIT"
] | null | null | null | 23.976484 | 102 | 0.522769 |
[
[
[
"it is a playground notebook for related data, such as track length, waypoint distance, car size",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
],
[
"7*15",
"_____no_output_____"
],
[
"100/105",
"_____no_output_____"
],
[
"1/105 *10",
"_____no_output_____"
],
[
"100/300",
"_____no_output_____"
],
[
"1/2",
"_____no_output_____"
],
[
"from race_utils import SampleGenerator\ngenerator = SampleGenerator()",
"_____no_output_____"
],
[
"testing_param = generator.random_sample()",
"_____no_output_____"
],
[
"waypoints = testing_param['waypoints']",
"_____no_output_____"
],
[
"len(waypoints)",
"_____no_output_____"
],
[
"testing_param['track_length']/len(waypoints)",
"_____no_output_____"
],
[
"step_time = 1/15",
"_____no_output_____"
],
[
"1.4 * step_time",
"_____no_output_____"
],
[
"4.0 * step_time",
"_____no_output_____"
],
[
"testing_param['closest_waypoints']",
"_____no_output_____"
],
[
"params = testing_param",
"_____no_output_____"
],
[
"x = params['x']\ny = params['y']\nheading = params['heading']\nwaypoints = params['waypoints']\n\nwaypoints_length = len(waypoints)\n\nfront_waypoint = params['closest_waypoints'][1]\n\nrabbit_waypoint = front_waypoint + 1\n\nif (rabbit_waypoint >= waypoints_length):\n rabbit_waypoint = rabbit_waypoint % waypoints_length\n\nrabbit = [waypoints[rabbit_waypoint][0],waypoints[rabbit_waypoint][1]]\n\nradius = math.hypot(x - rabbit[0], y - rabbit[1])\n\npointing = [0,0]\npointing[0] = x + (radius * math.cos(heading))\npointing[1] = y + (radius * math.sin(heading))\n\nvector_delta = math.hypot(pointing[0] - rabbit[0], pointing[1] - rabbit[1])\n\nreward = 0.001\n\nreward += ( 1 - ( vector_delta / (radius * 2)))",
"_____no_output_____"
],
[
"reward",
"_____no_output_____"
],
[
"front_waypoint = params['closest_waypoints'][1]\n\nrabbit_waypoint = front_waypoint + 1\n\nrabbit = [waypoints[rabbit_waypoint][0],waypoints[rabbit_waypoint][1]]\n\n",
"_____no_output_____"
],
[
"rabbit",
"_____no_output_____"
],
[
"front_waypoint",
"_____no_output_____"
],
[
"rabbit_waypoint",
"_____no_output_____"
],
[
"\n\n\n\nrabbit_waypoint_data = waypoints[ribbit_waypoint]\n\nrabbit = [waypoints[rabbit_waypoint][0],waypoints[closest_waypoints[1]][1]]\n\n \nradius = math.hypot(x - rabbit[0], y - rabbit[1])\n\npointing[0] = x + (radius * math.cos(heading))\npointing[1] = y + (radius * math.sin(heading))\n\nvector_delta = math.hypot(pointing[0] - rabbit[0], pointing[1] - rabbit[1])\n\n# Max distance for pointing away will be the radius * 2\n# Min distance means we are pointing directly at the next waypoint\n# We can setup a reward that is a ratio to this max.\n\nif vector_delta == 0:\n reward += 1\nelse:\n reward += ( 1 - ( vector_delta / (radius * 2)))\n",
"_____no_output_____"
],
[
"len(waypoints)",
"_____no_output_____"
],
[
"waypoints[-1]",
"_____no_output_____"
],
[
"waypoints[0]",
"_____no_output_____"
],
[
"waypoints[118]",
"_____no_output_____"
],
[
"front_waypoint",
"_____no_output_____"
],
[
"rabbit = [waypoints[closest_waypoints+1][0],waypoints[closest_waypoints+1][1]]\n\nradius = math.hypot(x - rabbit[0], y - rabbit[1])\n\npointing[0] = x + (radius * math.cos(car_orientation))\npointing[1] = y + (radius * math.sin(car_orientation))\n\nvector_delta = math.hypot(pointing[0] - rabbit[0], pointing[1] - rabbit[1])",
"_____no_output_____"
],
[
"zhouju= 0.17\nlunju = 0.155",
"_____no_output_____"
],
[
"testing_param",
"_____no_output_____"
],
[
"def distance_between(x1, y1 , x2, y2):\n dx = x1 - x2\n dy = y1 - y2\n return math.sqrt(dx*dx + dy*dy)",
"_____no_output_____"
],
[
"waypoints = testing_param['waypoints']\nprev_waypoint = waypoints[-1]\n\ntotal_distance = 0\n\nfor waypoint in testing_param['waypoints']:\n \n distance = distance_between(prev_waypoint[0], prev_waypoint[1], waypoint[0], waypoint[1])\n total_distance = total_distance + distance\n prev_waypoint = waypoint",
"_____no_output_____"
],
[
"total_distance",
"_____no_output_____"
],
[
"pow(2,3)",
"_____no_output_____"
],
[
"pow(0.98, 105)",
"_____no_output_____"
],
[
"17.709159380834848",
"_____no_output_____"
]
]
] |
[
"markdown",
"code"
] |
[
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
] |
4a16fad32a013459e45bd0802819847ee5b97e54
| 7,413 |
ipynb
|
Jupyter Notebook
|
example-luis-entities.ipynb
|
joe-plumb/entities-from-luis-python
|
73e04fb74bd94b6d3761edfed2693df10068f3a7
|
[
"MIT"
] | null | null | null |
example-luis-entities.ipynb
|
joe-plumb/entities-from-luis-python
|
73e04fb74bd94b6d3761edfed2693df10068f3a7
|
[
"MIT"
] | null | null | null |
example-luis-entities.ipynb
|
joe-plumb/entities-from-luis-python
|
73e04fb74bd94b6d3761edfed2693df10068f3a7
|
[
"MIT"
] | null | null | null | 55.320896 | 1,525 | 0.535411 |
[
[
[
"import datetime, json, os, time\nimport pandas as pd\n\nfrom azure.cognitiveservices.language.luis.runtime import LUISRuntimeClient\nfrom msrest.authentication import CognitiveServicesCredentials\n\nfrom ratelimit import limits, sleep_and_retry\n\n# Load and set credentials\nwith open('credentials.json') as json_file:\n credentials = json.load(json_file)\n\nLUIS_RUNTIME_KEY=credentials['LUIS_RUNTIME_KEY']\nLUIS_RUNTIME_ENDPOINT=credentials['LUIS_RUNTIME_ENDPOINT']\nLUIS_RUNTIME_APPID=credentials['LUIS_RUNTIME_APPID']\n\n# Load data into dataframe\ndf = pd.read_csv(\"example.csv\", index_col='id')\n\n# view df from file\ndf.head()",
"_____no_output_____"
],
[
"# Instantiate a LUIS runtime client\nclientRuntime = LUISRuntimeClient(LUIS_RUNTIME_ENDPOINT, CognitiveServicesCredentials(LUIS_RUNTIME_KEY))",
"_____no_output_____"
],
[
"# Define functions for prediction and entity parsing from response\n\n@sleep_and_retry\n@limits(calls=5, period=1)\ndef predict(app_id, slot_name, query):\n request = { \"query\" : str(query) }\n # Note be sure to specify, using the slot_name parameter, whether your application is in staging or production.\n resp = clientRuntime.prediction.get_slot_prediction(app_id=app_id, slot_name=slot_name, prediction_request=request)\n return (resp)\n\ndef get_all_values(d):\n if isinstance(d, dict):\n for v in d.values():\n yield from get_all_values(v)\n elif isinstance(d, list):\n for v in d:\n yield from get_all_values(v)\n else:\n yield d \n\ndef get_results(resp):\n return ','.join(get_all_values(list(resp.prediction.entities.keys()))) , resp.prediction.sentiment.label, resp.prediction.sentiment.score\n",
"_____no_output_____"
],
[
"# Create new column with entities from LUIS\ndf[['entities', 'sentiment_label', 'sentiment_score']] = df.apply(lambda row: pd.Series(get_results(predict(LUIS_RUNTIME_APPID, 'production', row['text']))), axis=1)",
"_____no_output_____"
],
[
"# df with entities column\ndf.head()",
"_____no_output_____"
]
]
] |
[
"code"
] |
[
[
"code",
"code",
"code",
"code",
"code"
]
] |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.